GO analysis
ERM
2023-09-27
Last updated: 2023-09-27
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8169f19. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/41588_2018_171_MOESM3_ESMeQTL_ST2_for paper.csv
Ignored: data/Arr_GWAS.txt
Ignored: data/Arr_geneset.RDS
Ignored: data/BC_cell_lines.csv
Ignored: data/BurridgeDOXTOX.RDS
Ignored: data/CADGWASgene_table.csv
Ignored: data/CAD_geneset.RDS
Ignored: data/CALIMA_Data/
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/Counts_RNA_ERMatthews.txt
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDEMresp_list.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG-GO/
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/DRC48hoursdata.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/EPIstorelist24.RDS
Ignored: data/Ep24counts.txt
Ignored: data/Full_LD_rep.csv
Ignored: data/GOIsig.csv
Ignored: data/GOplots.R
Ignored: data/GTEX_setsimple.csv
Ignored: data/GTEX_sig24.RDS
Ignored: data/GTEx_gene_list.csv
Ignored: data/HFGWASgene_table.csv
Ignored: data/HF_geneset.RDS
Ignored: data/Heart_Left_Ventricle.v8.egenes.txt
Ignored: data/Heatmap_mat.RDS
Ignored: data/Heatmap_sig.RDS
Ignored: data/Hf_GWAS.txt
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/LD50_05via.csv
Ignored: data/LDH48hoursdata.csv
Ignored: data/Mt24counts.txt
Ignored: data/NoRespDEG_final.csv
Ignored: data/RINsamplelist.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/TMMnormed_x.RDS
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/TableS10.csv
Ignored: data/TableS11.csv
Ignored: data/TableS9.csv
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Var_test_list.RDS
Ignored: data/Var_test_list24.RDS
Ignored: data/Var_test_list24alt.RDS
Ignored: data/Var_test_list3.RDS
Ignored: data/Vargenes.RDS
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allfinal3hour.RDS
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/allmymatrix.RDS
Ignored: data/annotation_data_frame.RDS
Ignored: data/averageviabilitytable.RDS
Ignored: data/avgLD50.RDS
Ignored: data/avg_LD50.RDS
Ignored: data/backGL.txt
Ignored: data/burr_genes.RDS
Ignored: data/calcium_data.RDS
Ignored: data/clamp_summary.RDS
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/cormotifprobs.csv
Ignored: data/counts24hours.RDS
Ignored: data/cpmcount.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/crispr_genes.csv
Ignored: data/ctnnt_results.txt
Ignored: data/cvd_GWAS.txt
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/drug_noveh1.csv
Ignored: data/efit2.RDS
Ignored: data/efit2_final.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filcpm_counts.RDS
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/geneDoxonlyQTL.csv
Ignored: data/gene_corr_df.RDS
Ignored: data/gene_corr_frame.RDS
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/geneset_24.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/gtex/
Ignored: data/heartgenes.csv
Ignored: data/hsa_kegg_anno.RDS
Ignored: data/individualDRCfile.RDS
Ignored: data/individual_DRC48.RDS
Ignored: data/individual_LDH48.RDS
Ignored: data/indv_noveh1.csv
Ignored: data/kegglistDEG.RDS
Ignored: data/kegglistDEG24.RDS
Ignored: data/kegglistDEG3.RDS
Ignored: data/knowfig4.csv
Ignored: data/knowfig5.csv
Ignored: data/label_list.RDS
Ignored: data/ld50_table.csv
Ignored: data/mean_vardrug1.csv
Ignored: data/mean_varframe.csv
Ignored: data/mymatrix.RDS
Ignored: data/new_ld50avg.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/organized_drugframe.csv
Ignored: data/plan2plot.png
Ignored: data/plot_intv_list.RDS
Ignored: data/plot_list_DRC.RDS
Ignored: data/qval24hr.RDS
Ignored: data/qval3hr.RDS
Ignored: data/qvalueEPItemp.RDS
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/siglist_final.RDS
Ignored: data/siglist_old.RDS
Ignored: data/slope_table.csv
Ignored: data/supp10_24hlist.RDS
Ignored: data/supp10_3hlist.RDS
Ignored: data/supp_normLDH48.RDS
Ignored: data/supp_pca_all_anno.RDS
Ignored: data/table3a.omar
Ignored: data/testlist.txt
Ignored: data/toplistall.RDS
Ignored: data/trtonly_24h_genes.RDS
Ignored: data/trtonly_3h_genes.RDS
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Ignored: data/viability.RDS
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: .RDataTmp3
Untracked: 3hr all.pdf
Untracked: Code_files_list.csv
Untracked: Data_files_list.csv
Untracked: Doxorubicin_vehicle_3_24.csv
Untracked: Doxtoplist.csv
Untracked: EPIqvalue_analysis.Rmd
Untracked: GWAS_list_of_interest.xlsx
Untracked: KEGGpathwaylist.R
Untracked: OmicNavigator_learn.R
Untracked: SigDoxtoplist.csv
Untracked: analysis/ciFIT.R
Untracked: analysis/export_to_excel.R
Untracked: cleanupfiles_script.R
Untracked: code/biomart_gene_names.R
Untracked: code/constantcode.R
Untracked: code/corMotifcustom.R
Untracked: code/cpm_boxplot.R
Untracked: code/extracting_ggplot_data.R
Untracked: code/movingfilesto_ppl.R
Untracked: code/pearson_extract_func.R
Untracked: code/pearson_tox_extract.R
Untracked: code/plot1C.fun.R
Untracked: code/spearman_extract_func.R
Untracked: code/venndiagramcolor_control.R
Untracked: cormotif_p.post.list_4.csv
Untracked: figS1024h.pdf
Untracked: individual-legenddark2.png
Untracked: installed_old.rda
Untracked: motif_ER.txt
Untracked: motif_LR.txt
Untracked: motif_NR.txt
Untracked: motif_TI.txt
Untracked: output/DNR_DEGlist.csv
Untracked: output/DNRvenn.RDS
Untracked: output/DOX_DEGlist.csv
Untracked: output/DOXvenn.RDS
Untracked: output/EPI_DEGlist.csv
Untracked: output/EPIvenn.RDS
Untracked: output/Figures/
Untracked: output/MTX_DEGlist.csv
Untracked: output/MTXvenn.RDS
Untracked: output/TRZ_DEGlist.csv
Untracked: output/TableS8.csv
Untracked: output/Volcanoplot_10
Untracked: output/Volcanoplot_10.RDS
Untracked: output/allfinal_sup10.RDS
Untracked: output/cormotif_probability_genelist.csv
Untracked: output/endocytosisgenes.csv
Untracked: output/gene_corr_fig9.RDS
Untracked: output/legend_b.RDS
Untracked: output/motif_ERrep.RDS
Untracked: output/motif_LRrep.RDS
Untracked: output/motif_NRrep.RDS
Untracked: output/motif_TI_rep.RDS
Untracked: output/output-old/
Untracked: output/rank24genes.csv
Untracked: output/rank3genes.csv
Untracked: output/reneem@ls6.tacc.utexas.edu
Untracked: output/sequencinginformationforsupp.csv
Untracked: output/sequencinginformationforsupp.prn
Untracked: output/sigVDA24.txt
Untracked: output/sigVDA3.txt
Untracked: output/sigVDX24.txt
Untracked: output/sigVDX3.txt
Untracked: output/sigVEP24.txt
Untracked: output/sigVEP3.txt
Untracked: output/sigVMT24.txt
Untracked: output/sigVMT3.txt
Untracked: output/sigVTR24.txt
Untracked: output/sigVTR3.txt
Untracked: output/supplementary_motif_list_GO.RDS
Untracked: output/toptablebydrug.RDS
Untracked: output/tvl24hour.txt
Untracked: output/x_counts.RDS
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/Cormotifcluster_analysis.Rmd
Modified: analysis/Figure4.Rmd
Modified: analysis/Figure5.Rmd
Modified: analysis/GOI_plots.Rmd
Modified: analysis/Knowles2019.Rmd
Modified: analysis/Supplementary_figures.Rmd
Modified: analysis/run_all_analysis.Rmd
Modified: analysis/variance_scrip.Rmd
Modified: output/TNNI_LDH_RNAnormlist.txt
Modified: output/daplot.RDS
Modified: output/dxplot.RDS
Modified: output/epplot.RDS
Modified: output/mtplot.RDS
Modified: output/plan2plot.png
Modified: output/sequencing_info.txt
Modified: output/toplistall.csv
Modified: output/trplot.RDS
Modified: output/veplot.RDS
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/DEG-GO_analysis.Rmd) and
HTML (docs/DEG-GO_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8169f19 | reneeisnowhere | 2023-09-27 | updates and adding code |
| html | bcc7711 | reneeisnowhere | 2023-09-27 | Build site. |
| Rmd | bed3d02 | reneeisnowhere | 2023-09-27 | updates to links and code |
| Rmd | 06800c9 | reneeisnowhere | 2023-07-26 | Commits to small changes and edits |
| html | ae53b54 | reneeisnowhere | 2023-07-04 | Build site. |
| Rmd | 092462b | reneeisnowhere | 2023-07-04 | updates in name abbrv |
| html | b2bec16 | reneeisnowhere | 2023-06-30 | Build site. |
| Rmd | b106ddc | reneeisnowhere | 2023-06-30 | Final DEG-need to update KEGG heatmap |
| html | b205acd | reneeisnowhere | 2023-06-29 | Build site. |
| Rmd | ee83d9a | reneeisnowhere | 2023-06-29 | updates and fixes in code using final list |
| Rmd | f4bd5e1 | reneeisnowhere | 2023-06-27 | checking code changes overtime |
| Rmd | e94e8f2 | reneeisnowhere | 2023-06-19 | new code from Monday |
| html | 363f82e | reneeisnowhere | 2023-06-16 | Build site. |
| Rmd | 3d4ca64 | reneeisnowhere | 2023-06-16 | updates on Friday |
| html | 625d1d6 | reneeisnowhere | 2023-06-15 | Build site. |
| Rmd | 709354f | reneeisnowhere | 2023-06-15 | update in kdm4b! |
| html | a2aa7e4 | reneeisnowhere | 2023-06-15 | Build site. |
| Rmd | b5fb9e9 | reneeisnowhere | 2023-06-15 | adding kat6b and kdm5b |
| html | fc32961 | reneeisnowhere | 2023-06-02 | Build site. |
| Rmd | 8140265 | reneeisnowhere | 2023-06-02 | adding crispr genes |
| Rmd | 399ab8a | reneeisnowhere | 2023-06-02 | adding new genes in |
| html | d9ebe68 | reneeisnowhere | 2023-06-01 | Build site. |
| Rmd | 5415215 | reneeisnowhere | 2023-06-01 | logging GOIsig changes |
| Rmd | 786c2e2 | reneeisnowhere | 2023-06-01 | logging GOIsig changes |
| Rmd | 6250bc1 | reneeisnowhere | 2023-05-31 | FINALLY fixed the kegg heatmap |
| html | e0702ff | reneeisnowhere | 2023-05-26 | Build site. |
| Rmd | fd12426 | reneeisnowhere | 2023-05-26 | fixcorrelation break |
| html | 00442f9 | reneeisnowhere | 2023-05-26 | Build site. |
| Rmd | d215f0f | reneeisnowhere | 2023-05-26 | updating log2 and adding kegg 24 hour heatmap |
| Rmd | 38e7b3c | reneeisnowhere | 2023-05-26 | adding heatmap |
| Rmd | f3871a5 | reneeisnowhere | 2023-05-26 | updates |
| html | 10393fb | reneeisnowhere | 2023-05-25 | Build site. |
| Rmd | 0c372d0 | reneeisnowhere | 2023-05-25 | update expression correlation |
| html | 5907ad2 | reneeisnowhere | 2023-05-19 | Build site. |
| Rmd | 941b694 | reneeisnowhere | 2023-05-19 | adding corr analysis again |
| html | 860985f | reneeisnowhere | 2023-05-19 | Build site. |
| Rmd | 9f4b03a | reneeisnowhere | 2023-05-19 | adding corr analysis |
| Rmd | d55a810 | reneeisnowhere | 2023-05-18 | working on GWAS and correlation |
| Rmd | 8d87a52 | reneeisnowhere | 2023-05-17 | end of day |
| html | 021e130 | reneeisnowhere | 2023-04-20 | Build site. |
| Rmd | 9d976da | reneeisnowhere | 2023-04-20 | update broken image |
| html | 60af208 | reneeisnowhere | 2023-04-20 | Build site. |
| Rmd | 2b12884 | reneeisnowhere | 2023-04-20 | pheatmap color |
| html | a882f82 | reneeisnowhere | 2023-04-20 | Build site. |
| Rmd | 40ca8e6 | reneeisnowhere | 2023-04-20 | pheatmap color |
| html | 8ad428c | reneeisnowhere | 2023-04-20 | Build site. |
| Rmd | be18119 | reneeisnowhere | 2023-04-20 | updated adj pvalue |
| Rmd | f8eedbf | reneeisnowhere | 2023-04-19 | update after p value to 0.05 |
| html | 56e7c4f | reneeisnowhere | 2023-04-17 | Build site. |
| Rmd | b41cb80 | reneeisnowhere | 2023-04-17 | adding total counts percent graph |
| html | 353fc4a | reneeisnowhere | 2023-04-17 | Build site. |
| Rmd | 457bc7e | reneeisnowhere | 2023-04-17 | updated GO analysis |
| Rmd | 1e2e55e | reneeisnowhere | 2023-04-10 | before final data |
| Rmd | 4e52216 | reneeisnowhere | 2023-03-31 | End of week updates |
Analysis of Pairwise DEGs
I have created several files from the RNA analysis that contain the significant genes(determined by adj.P.val < 0.05) from each Time and Condition. The names of the files are in the following format: ‘sigV’+Drug(2 letters)+time.
example: ‘sigVDA3.txt’ means this file contains the significant DE genes from the DNR 3 hour compared to Vehicle Control 3 hour analysis
library(tidyverse)
library(gprofiler2)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(kableExtra)
library(scales)
library(ggVennDiagram)
library(cowplot)
library(ggpubr)
library(rstatix)
library(zoo)
library(ggsignif)
library(limma)
library(pheatmap)toplistall <- readRDS("data/toplistall.RDS")
siglist <- readRDS("data/siglist_final.RDS")
list2env(siglist, envir = .GlobalEnv)<environment: R_GlobalEnv>The analysis is based on all genes that passed the rowMeans>0 from the previous page which are about 14084 genes expressed in my RNA-seq data.
All analysis is completed with an adjusted p value of 0.05
drug_palNoVeh <-
c("#8B006D" , "#DF707E", "#F1B72B" , "#3386DD", "#707031")
toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
group_by(time, id) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
count(sigcount) %>%
pivot_wider(
id_cols = c(time, id),
names_from = sigcount,
values_from = n
) %>%
mutate(prop = sig / (sig + notsig) * 100) %>%
mutate(prop = if_else(is.na(prop), 0, prop)) %>%
ggplot(., aes(x = id, y = prop)) +
geom_col(aes(fill = id)) +
geom_text(aes(label = sprintf("%.2f", prop)),
position = position_dodge(0.9),
vjust = -.2) +
scale_fill_manual(values = drug_palNoVeh) +
guides(fill = guide_legend(title = "Treatment")) +
facet_wrap(id ~ time) + #labeller = (time = facettimelabel) )+
theme_bw() +
xlab("") +
ylab("Percentage of expressed genes") +
theme_bw() +
facet_wrap( ~ time) +
ggtitle("Percent DEGs (adj. P value <0.05)") +
scale_y_continuous(expand = expansion(c(0.02, .1))) +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
# axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "transparent"),
axis.text.x = element_text(
size = 8,
color = "white",
angle = 0
),
axis.text.y = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 12,
color = "black",
face = "bold"
)
)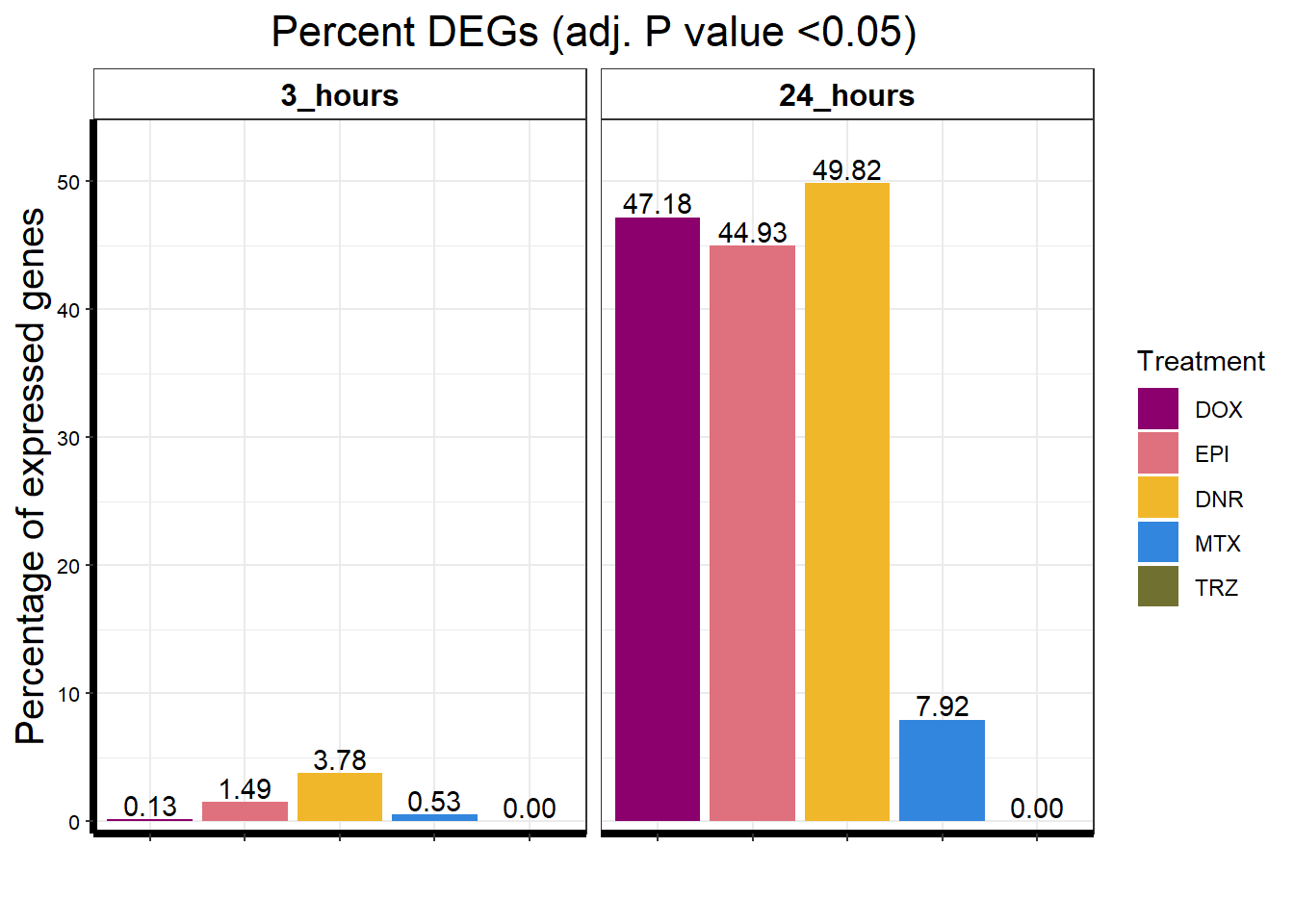
Knowels, et al. mentioned a relationship between transcriptional change level indicating cardiotoxicity. This may be an observation that supports that claim as well. ## DNR
This is analysis on the significantly differentially expressed genes for DNR by 3 and 24 hours.
# DAgostres3 <- gost(query = sigVDA3$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# numeric_ns = "ENTREZGENE",
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GP:BP","KEGG"))
# saveRDS(DAgostres3,"data/DEG-GO/DAgostres3.RDS")
DAgostres3 <- readRDS("data/DEG-GO/DAgostres3.RDS")
# DAp3 <- gostplot(DAgostres3, capped = FALSE, interactive = TRUE)
# publish_gostplot(DAp3, highlight_terms = c("KEGG:04115", "KEGG:05168", "KEGG:04110", "KEGG:03030", "KEGG:03410"))
# overlapKEGG <- hsa_kegg_anno %>%
# filter(eg %in%backGL$ENTREZID) #%>%
# filter(pathway %in% Path_of_interest)
# unique(overlapKEGG$pathway)
DAtable3 <- DAgostres3$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
# testframe <- DAgostres3star$meta
# testframeresult <- DAgostres3star$result
# testframeresult%>%
# filter(term_id %in% kegg)
# mutate_at(.vars = 6, .funs= scientific_format()) %>%
# kable(., captions=" fdr at 1") %>%
# kable_paper("striped", full_width = FALSE) %>%
# kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped","hover")) %>%
# scroll_box(width = "100%", height = "400px")
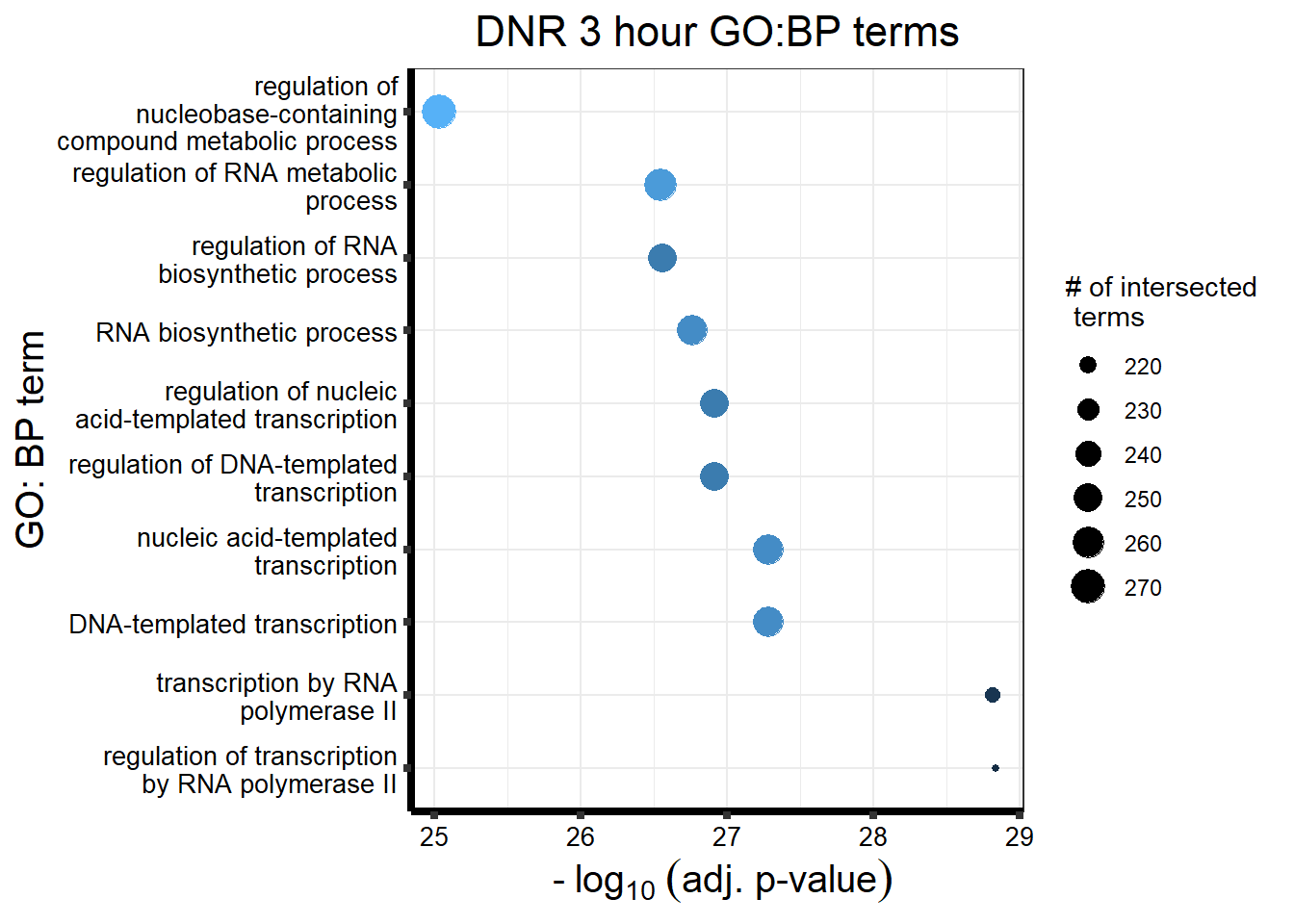
DAtable3 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('DNR 3 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DAtable3 %>% mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 212 | 1914 | 1.45e-29 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 217 | 1995 | 1.53e-29 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 256 | 2684 | 5.18e-28 |
| GO:BP | GO:0006351 | DNA-templated transcription | 256 | 2683 | 5.18e-28 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 249 | 2574 | 1.21e-27 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 249 | 2576 | 1.21e-27 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 256 | 2714 | 1.73e-27 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 249 | 2593 | 2.76e-27 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 262 | 2845 | 2.83e-27 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 271 | 3080 | 9.29e-26 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 263 | 3012 | 2.97e-24 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 262 | 3080 | 5.74e-22 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 260 | 3070 | 2.39e-21 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 265 | 3169 | 2.65e-21 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 262 | 3132 | 6.00e-21 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 261 | 3133 | 2.13e-20 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 264 | 3237 | 3.45e-19 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 283 | 3730 | 4.65e-18 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 306 | 4174 | 7.95e-17 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 276 | 3750 | 6.84e-15 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 312 | 4417 | 2.07e-14 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 305 | 4292 | 2.54e-14 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 87 | 740 | 2.36e-12 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 290 | 4245 | 2.13e-11 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 104 | 1015 | 2.29e-11 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 104 | 1017 | 2.43e-11 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 104 | 1026 | 4.60e-11 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 109 | 1119 | 8.53e-11 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 299 | 4539 | 5.91e-10 |
| GO:BP | GO:0009058 | biosynthetic process | 300 | 4590 | 1.83e-09 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 111 | 1209 | 1.98e-09 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 110 | 1214 | 2.76e-09 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 110 | 1239 | 1.24e-08 |
| GO:BP | GO:0010468 | regulation of gene expression | 275 | 3580 | 7.73e-08 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 110 | 1280 | 7.73e-08 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 136 | 1735 | 9.36e-08 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 110 | 1315 | 1.14e-06 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 110 | 1315 | 1.14e-06 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 110 | 1322 | 1.40e-06 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 116 | 1441 | 1.56e-06 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 117 | 1507 | 9.90e-06 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 52 | 4617 | 1.07e-05 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 120 | 1564 | 1.44e-05 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 119 | 1602 | 5.22e-05 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 133 | 1849 | 6.29e-05 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 120 | 1604 | 6.29e-05 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 81 | 937 | 9.99e-05 |
| GO:BP | GO:0019222 | regulation of metabolic process | 52 | 5017 | 1.97e-04 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 159 | 2412 | 1.36e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 8 | 67 | 1.54e-03 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 2 | 34 | 1.74e-03 |
| GO:BP | GO:0140467 | integrated stress response signaling | 7 | 35 | 2.02e-03 |
| GO:BP | GO:0003281 | ventricular septum development | 11 | 69 | 2.24e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 23 | 230 | 3.46e-03 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 5 | 84 | 3.64e-03 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 5 | 25 | 3.68e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 22 | 216 | 4.18e-03 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 2 | 67 | 4.59e-03 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 15 | 128 | 4.59e-03 |
| GO:BP | GO:0003162 | atrioventricular node development | 3 | 8 | 4.99e-03 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 15 | 129 | 5.68e-03 |
| GO:BP | GO:0021915 | neural tube development | 16 | 146 | 5.73e-03 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 3 | 4 | 5.73e-03 |
| GO:BP | GO:0003171 | atrioventricular valve development | 5 | 27 | 5.73e-03 |
| GO:BP | GO:0043009 | chordate embryonic development | 46 | 534 | 5.74e-03 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 2 | 77 | 5.92e-03 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 148 | 2270 | 6.88e-03 |
| GO:BP | GO:0019827 | stem cell population maintenance | 14 | 149 | 8.08e-03 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 44 | 4048 | 8.77e-03 |
| GO:BP | GO:0016070 | RNA metabolic process | 42 | 3600 | 8.83e-03 |
| GO:BP | GO:0045643 | regulation of eosinophil differentiation | 1 | 1 | 9.71e-03 |
| GO:BP | GO:0045645 | positive regulation of eosinophil differentiation | 1 | 1 | 9.71e-03 |
| GO:BP | GO:0045659 | negative regulation of neutrophil differentiation | 1 | 2 | 9.71e-03 |
| GO:BP | GO:0098727 | maintenance of cell number | 14 | 151 | 9.71e-03 |
| GO:BP | GO:0060840 | artery development | 11 | 83 | 9.71e-03 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 12 | 100 | 9.71e-03 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 46 | 551 | 1.03e-02 |
| GO:BP | GO:0030853 | negative regulation of granulocyte differentiation | 2 | 6 | 1.07e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 2 | 118 | 1.12e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 2 | 118 | 1.12e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 12 | 101 | 1.22e-02 |
| GO:BP | GO:0045658 | regulation of neutrophil differentiation | 1 | 2 | 1.34e-02 |
| GO:BP | GO:0090425 | acinar cell differentiation | 3 | 5 | 1.39e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 2 | 152 | 1.42e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 3 | 620 | 1.42e-02 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 10 | 68 | 1.44e-02 |
| GO:BP | GO:0001841 | neural tube formation | 11 | 101 | 1.54e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 2 | 151 | 1.54e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 5 | 123 | 1.62e-02 |
| GO:BP | GO:0055025 | positive regulation of cardiac muscle tissue development | 1 | 2 | 1.62e-02 |
| GO:BP | GO:0071772 | response to BMP | 11 | 131 | 1.80e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 11 | 131 | 1.80e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 7 | 40 | 1.80e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 8 | 80 | 1.84e-02 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 2 | 8 | 1.84e-02 |
| GO:BP | GO:0030222 | eosinophil differentiation | 1 | 2 | 1.90e-02 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 6 | 55 | 1.98e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 8 | 56 | 1.98e-02 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 17 | 179 | 2.13e-02 |
| GO:BP | GO:0045844 | positive regulation of striated muscle tissue development | 1 | 3 | 2.13e-02 |
| GO:BP | GO:0060430 | lung saccule development | 2 | 7 | 2.13e-02 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 3 | 11 | 2.13e-02 |
| GO:BP | GO:0048636 | positive regulation of muscle organ development | 1 | 3 | 2.13e-02 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 5 | 17 | 2.13e-02 |
| GO:BP | GO:0006473 | protein acetylation | 15 | 190 | 2.19e-02 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 4 | 17 | 2.21e-02 |
| GO:BP | GO:0001657 | ureteric bud development | 4 | 75 | 2.28e-02 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 4 | 76 | 2.32e-02 |
| GO:BP | GO:0072164 | mesonephric tubule development | 4 | 76 | 2.32e-02 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 3 | 45 | 2.34e-02 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 2 | 199 | 2.43e-02 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 5 | 56 | 2.44e-02 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 2 | 2 | 2.44e-02 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 2 | 4 | 2.44e-02 |
| GO:BP | GO:0003174 | mitral valve development | 3 | 12 | 2.44e-02 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 2 | 13 | 2.44e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 166 | 2646 | 2.44e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 10 | 94 | 2.44e-02 |
| GO:BP | GO:0030854 | positive regulation of granulocyte differentiation | 1 | 5 | 2.44e-02 |
| GO:BP | GO:0001823 | mesonephros development | 4 | 77 | 2.44e-02 |
| GO:BP | GO:0061614 | miRNA transcription | 5 | 57 | 2.49e-02 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 2 | 14 | 2.60e-02 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 8 | 60 | 2.60e-02 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 3 | 23 | 2.64e-02 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 6 | 35 | 2.64e-02 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 2 | 4 | 2.64e-02 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 2 | 11 | 2.77e-02 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 5 | 34 | 2.77e-02 |
| GO:BP | GO:0030223 | neutrophil differentiation | 1 | 8 | 2.81e-02 |
| GO:BP | GO:0055024 | regulation of cardiac muscle tissue development | 1 | 3 | 2.81e-02 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 3 | 10 | 2.86e-02 |
| GO:BP | GO:1904290 | negative regulation of mitotic DNA damage checkpoint | 1 | 1 | 3.05e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 8 | 109 | 3.23e-02 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 3 | 957 | 3.30e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 4 | 16 | 3.30e-02 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 44 | 4455 | 3.30e-02 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 2 | 279 | 3.33e-02 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 6 | 44 | 3.33e-02 |
| GO:BP | GO:0072175 | epithelial tube formation | 12 | 127 | 3.33e-02 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 2 | 310 | 3.37e-02 |
| GO:BP | GO:0014706 | striated muscle tissue development | 19 | 210 | 3.45e-02 |
| GO:BP | GO:0048844 | artery morphogenesis | 8 | 58 | 3.51e-02 |
| GO:BP | GO:0010720 | positive regulation of cell development | 2 | 317 | 3.52e-02 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 1 | 7 | 3.55e-02 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 6 | 39 | 3.61e-02 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 6 | 39 | 3.61e-02 |
| GO:BP | GO:1903674 | regulation of cap-dependent translational initiation | 1 | 1 | 3.65e-02 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 3 | 1014 | 3.65e-02 |
| GO:BP | GO:0003170 | heart valve development | 6 | 65 | 3.65e-02 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 18 | 198 | 3.65e-02 |
| GO:BP | GO:1903676 | positive regulation of cap-dependent translational initiation | 1 | 1 | 3.65e-02 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 3 | 20 | 3.80e-02 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 1 | 8 | 3.80e-02 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 1 | 12 | 3.80e-02 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 5 | 67 | 3.82e-02 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 2 | 4 | 3.84e-02 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 19 | 227 | 3.91e-02 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 11 | 116 | 4.08e-02 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 2 | 6 | 4.24e-02 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 22 | 303 | 4.26e-02 |
| GO:BP | GO:0007507 | heart development | 36 | 506 | 4.34e-02 |
| GO:BP | GO:0060325 | face morphogenesis | 5 | 28 | 4.43e-02 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 13 | 4570 | 4.43e-02 |
| GO:BP | GO:0001843 | neural tube closure | 9 | 90 | 4.52e-02 |
| GO:BP | GO:0035904 | aorta development | 7 | 50 | 4.58e-02 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 3 | 1137 | 4.59e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 132 | 2880 | 4.62e-02 |
| GO:BP | GO:0060976 | coronary vasculature development | 6 | 44 | 4.63e-02 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 13 | 4596 | 4.63e-02 |
| GO:BP | GO:0051170 | import into nucleus | 4 | 152 | 4.64e-02 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 3 | 15 | 4.65e-02 |
| GO:BP | GO:0060606 | tube closure | 9 | 91 | 4.65e-02 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 2 | 7 | 4.65e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 3 | 65 | 4.65e-02 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 2 | 9 | 4.66e-02 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 2 | 9 | 4.66e-02 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 7 | 62 | 4.69e-02 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 4.70e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 5 | 277 | 4.70e-02 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 21 | 265 | 4.77e-02 |
| GO:BP | GO:0060212 | negative regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 3 | 4.79e-02 |
| GO:BP | GO:0001701 | in utero embryonic development | 28 | 344 | 4.79e-02 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 2 | 55 | 4.79e-02 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 7 | 44 | 5.10e-02 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 1 | 15 | 5.12e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 2 | 398 | 5.12e-02 |
| GO:BP | GO:0003284 | septum primum development | 2 | 5 | 5.12e-02 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 2 | 5 | 5.12e-02 |
| GO:BP | GO:0002521 | leukocyte differentiation | 2 | 382 | 5.12e-02 |
| GO:BP | GO:0035905 | ascending aorta development | 2 | 5 | 5.12e-02 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 5 | 30 | 5.12e-02 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 5 | 28 | 5.12e-02 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 1 | 25 | 5.12e-02 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 15 | 160 | 5.12e-02 |
| GO:BP | GO:1904322 | cellular response to forskolin | 1 | 11 | 5.12e-02 |
| GO:BP | GO:0035264 | multicellular organism growth | 13 | 123 | 5.12e-02 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 5.12e-02 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 5 | 30 | 5.12e-02 |
| GO:BP | GO:0035148 | tube formation | 12 | 138 | 5.12e-02 |
| GO:BP | GO:1904321 | response to forskolin | 1 | 11 | 5.12e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 5 | 50 | 5.17e-02 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 4 | 65 | 5.17e-02 |
| GO:BP | GO:0006308 | DNA catabolic process | 4 | 19 | 5.17e-02 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 6 | 32 | 5.17e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 4 | 33 | 5.19e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 5 | 199 | 5.19e-02 |
| GO:BP | GO:0072073 | kidney epithelium development | 4 | 112 | 5.19e-02 |
| GO:BP | GO:0043627 | response to estrogen | 3 | 52 | 5.19e-02 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 1 | 3 | 5.24e-02 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 3 | 15 | 5.34e-02 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 13 | 4747 | 5.39e-02 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 18 | 222 | 5.44e-02 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 11 | 109 | 5.47e-02 |
| GO:BP | GO:0035909 | aorta morphogenesis | 5 | 26 | 5.51e-02 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 1 | 8 | 5.51e-02 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 3 | 17 | 5.51e-02 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 4 | 69 | 5.51e-02 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 3 | 10 | 5.51e-02 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 5.51e-02 |
| GO:BP | GO:0060591 | chondroblast differentiation | 2 | 3 | 5.53e-02 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 2 | 26 | 5.53e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 5 | 300 | 5.53e-02 |
| GO:BP | GO:0060010 | Sertoli cell fate commitment | 1 | 1 | 5.53e-02 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 2 | 9 | 5.53e-02 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 5.53e-02 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 4 | 124 | 5.54e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 5 | 81 | 5.58e-02 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 3 | 27 | 5.58e-02 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 5.58e-02 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 5.58e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 41 | 616 | 5.63e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 4 | 21 | 5.63e-02 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 1 | 13 | 5.65e-02 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 4 | 36 | 5.66e-02 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 3 | 16 | 5.66e-02 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 2 | 19 | 5.68e-02 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 5 | 45 | 5.72e-02 |
| GO:BP | GO:0060324 | face development | 6 | 45 | 5.72e-02 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 3 | 81 | 5.82e-02 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 5.87e-02 |
| GO:BP | GO:0090721 | primary adaptive immune response involving T cells and B cells | 1 | 1 | 5.87e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 11 | 114 | 5.87e-02 |
| GO:BP | GO:0090720 | primary adaptive immune response | 1 | 1 | 5.87e-02 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 13 | 4946 | 5.87e-02 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 5.87e-02 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 1 | 15 | 5.87e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 4 | 36 | 5.87e-02 |
| GO:BP | GO:0043543 | protein acylation | 15 | 228 | 5.87e-02 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 2 | 20 | 5.87e-02 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 4 | 51 | 5.87e-02 |
| GO:BP | GO:0060419 | heart growth | 9 | 72 | 6.01e-02 |
| GO:BP | GO:0033762 | response to glucagon | 1 | 13 | 6.09e-02 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 2 | 3 | 6.17e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 5 | 33 | 6.22e-02 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 4 | 36 | 6.28e-02 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 6.28e-02 |
| GO:BP | GO:0021983 | pituitary gland development | 3 | 22 | 6.28e-02 |
| GO:BP | GO:1903679 | positive regulation of cap-independent translational initiation | 1 | 3 | 6.35e-02 |
| GO:BP | GO:1903677 | regulation of cap-independent translational initiation | 1 | 3 | 6.35e-02 |
| GO:BP | GO:0003205 | cardiac chamber development | 4 | 149 | 6.38e-02 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 1 | 15 | 6.41e-02 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 3 | 17 | 6.41e-02 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 3 | 7 | 6.48e-02 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 6.64e-02 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 6.64e-02 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 1 | 33 | 6.64e-02 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 2 | 11 | 6.64e-02 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 6.64e-02 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 6.64e-02 |
| GO:BP | GO:0072739 | response to anisomycin | 2 | 4 | 6.64e-02 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 6.64e-02 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 6.64e-02 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 1 | 18 | 6.64e-02 |
| GO:BP | GO:0007613 | memory | 2 | 92 | 6.64e-02 |
| GO:BP | GO:1904003 | negative regulation of sebum secreting cell proliferation | 1 | 1 | 6.64e-02 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 6.64e-02 |
| GO:BP | GO:0060284 | regulation of cell development | 2 | 605 | 6.72e-02 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 3 | 108 | 6.72e-02 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 13 | 112 | 6.72e-02 |
| GO:BP | GO:0034405 | response to fluid shear stress | 2 | 31 | 6.77e-02 |
| GO:BP | GO:0045672 | positive regulation of osteoclast differentiation | 1 | 11 | 6.77e-02 |
| GO:BP | GO:0097676 | histone H3-K36 dimethylation | 2 | 4 | 6.80e-02 |
| GO:BP | GO:0035196 | miRNA processing | 6 | 41 | 6.95e-02 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 6.95e-02 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 3 | 11 | 6.96e-02 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 2 | 4 | 7.00e-02 |
| GO:BP | GO:0030225 | macrophage differentiation | 1 | 35 | 7.01e-02 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 1 | 18 | 7.08e-02 |
| GO:BP | GO:0060479 | lung cell differentiation | 1 | 18 | 7.08e-02 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 4 | 49 | 7.15e-02 |
| GO:BP | GO:0003161 | cardiac conduction system development | 4 | 33 | 7.17e-02 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 28 | 464 | 7.17e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 4 | 24 | 7.19e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 4 | 39 | 7.19e-02 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 2 | 5 | 7.19e-02 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 7 | 99 | 7.33e-02 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 2 | 4 | 7.33e-02 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 1 | 4 | 7.35e-02 |
| GO:BP | GO:2001144 | regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 7.51e-02 |
| GO:BP | GO:0033993 | response to lipid | 2 | 592 | 7.51e-02 |
| GO:BP | GO:2001145 | negative regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 7.51e-02 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 5 | 34 | 7.51e-02 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 1 | 36 | 7.56e-02 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 4 | 41 | 7.56e-02 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 7.56e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 5 | 344 | 7.56e-02 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 5 | 34 | 7.59e-02 |
| GO:BP | GO:0016573 | histone acetylation | 13 | 143 | 7.59e-02 |
| GO:BP | GO:0070848 | response to growth factor | 31 | 572 | 7.59e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 5 | 345 | 7.59e-02 |
| GO:BP | GO:0032526 | response to retinoic acid | 5 | 85 | 7.59e-02 |
| GO:BP | GO:0030279 | negative regulation of ossification | 2 | 25 | 7.59e-02 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 7.65e-02 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 1 | 22 | 7.65e-02 |
| GO:BP | GO:0030097 | hemopoiesis | 2 | 656 | 7.65e-02 |
| GO:BP | GO:0048568 | embryonic organ development | 29 | 316 | 7.73e-02 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 7.73e-02 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 2 | 10 | 7.83e-02 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 2 | 27 | 7.86e-02 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 7.86e-02 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 3 | 29 | 7.86e-02 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 7.94e-02 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 3 | 40 | 7.95e-02 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 8 | 65 | 8.01e-02 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 2 | 72 | 8.09e-02 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 7 | 50 | 8.22e-02 |
| GO:BP | GO:0007352 | zygotic specification of dorsal/ventral axis | 1 | 2 | 8.25e-02 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 8.25e-02 |
| GO:BP | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress | 1 | 5 | 8.27e-02 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 2 | 14 | 8.27e-02 |
| GO:BP | GO:1904690 | positive regulation of cytoplasmic translational initiation | 1 | 5 | 8.27e-02 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 3 | 126 | 8.32e-02 |
| GO:BP | GO:0002383 | immune response in brain or nervous system | 1 | 1 | 8.32e-02 |
| GO:BP | GO:0097281 | immune complex formation | 1 | 1 | 8.32e-02 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 3 | 40 | 8.32e-02 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 11 | 132 | 8.32e-02 |
| GO:BP | GO:0003283 | atrial septum development | 3 | 23 | 8.39e-02 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 3 | 38 | 8.49e-02 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 6 | 45 | 8.49e-02 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 3 | 40 | 8.49e-02 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 3 | 40 | 8.49e-02 |
| GO:BP | GO:0048538 | thymus development | 6 | 35 | 8.49e-02 |
| GO:BP | GO:0046832 | negative regulation of RNA export from nucleus | 1 | 2 | 8.49e-02 |
| GO:BP | GO:0019083 | viral transcription | 6 | 48 | 8.49e-02 |
| GO:BP | GO:0060420 | regulation of heart growth | 7 | 50 | 8.50e-02 |
| GO:BP | GO:0009894 | regulation of catabolic process | 48 | 875 | 8.56e-02 |
| GO:BP | GO:0061035 | regulation of cartilage development | 8 | 54 | 8.56e-02 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 19 | 336 | 8.56e-02 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 6 | 69 | 8.56e-02 |
| GO:BP | GO:0031018 | endocrine pancreas development | 4 | 32 | 8.62e-02 |
| GO:BP | GO:0050793 | regulation of developmental process | 3 | 1835 | 8.62e-02 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 8.66e-02 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 2 | 581 | 8.66e-02 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 6 | 53 | 8.66e-02 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 4 | 25 | 8.68e-02 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 4 | 25 | 8.68e-02 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 1 | 5 | 8.68e-02 |
| GO:BP | GO:0051169 | nuclear transport | 13 | 295 | 8.71e-02 |
| GO:BP | GO:0060428 | lung epithelium development | 1 | 30 | 8.71e-02 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 2 | 5 | 8.71e-02 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 2 | 6 | 8.71e-02 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 13 | 295 | 8.71e-02 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 8.76e-02 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 3 | 11 | 8.76e-02 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 8 | 61 | 8.79e-02 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 3 | 19 | 8.85e-02 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 2 | 8 | 8.85e-02 |
| GO:BP | GO:0071451 | cellular response to superoxide | 3 | 19 | 8.85e-02 |
| GO:BP | GO:0001568 | blood vessel development | 36 | 534 | 8.86e-02 |
| GO:BP | GO:0021998 | neural plate mediolateral regionalization | 1 | 1 | 8.86e-02 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 2 | 18 | 8.86e-02 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 13 | 148 | 8.86e-02 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 8.86e-02 |
| GO:BP | GO:0048372 | lateral mesodermal cell fate commitment | 1 | 1 | 8.86e-02 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 5 | 56 | 8.86e-02 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 1 | 9 | 8.86e-02 |
| GO:BP | GO:0048378 | regulation of lateral mesodermal cell fate specification | 1 | 1 | 8.86e-02 |
| GO:BP | GO:0048377 | lateral mesodermal cell fate specification | 1 | 1 | 8.86e-02 |
| GO:BP | GO:1904412 | regulation of cardiac ventricle development | 1 | 1 | 8.86e-02 |
| GO:BP | GO:1904414 | positive regulation of cardiac ventricle development | 1 | 1 | 8.86e-02 |
| GO:BP | GO:0048352 | paraxial mesoderm structural organization | 1 | 1 | 8.86e-02 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 89 | 8.86e-02 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 2 | 7 | 8.86e-02 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 35 | 8.86e-02 |
| GO:BP | GO:1905285 | fibrous ring of heart morphogenesis | 1 | 1 | 8.86e-02 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 1 | 69 | 8.86e-02 |
| GO:BP | GO:1904688 | regulation of cytoplasmic translational initiation | 1 | 6 | 8.86e-02 |
| GO:BP | GO:0048338 | mesoderm structural organization | 1 | 1 | 8.86e-02 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 12 | 8.89e-02 |
| GO:BP | GO:0032912 | negative regulation of transforming growth factor beta2 production | 1 | 2 | 8.89e-02 |
| GO:BP | GO:0007493 | endodermal cell fate determination | 1 | 1 | 8.89e-02 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 3 | 39 | 8.89e-02 |
| GO:BP | GO:0001949 | sebaceous gland cell differentiation | 1 | 1 | 8.89e-02 |
| GO:BP | GO:1904002 | regulation of sebum secreting cell proliferation | 1 | 1 | 8.89e-02 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 5 | 25 | 8.99e-02 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 9.01e-02 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 9.01e-02 |
| GO:BP | GO:0032535 | regulation of cellular component size | 3 | 305 | 9.01e-02 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 3 | 124 | 9.01e-02 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 9.01e-02 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 9.01e-02 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 1 | 2 | 9.01e-02 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 9.01e-02 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 9.01e-02 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 9.01e-02 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 9 | 192 | 9.18e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 2 | 36 | 9.18e-02 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 9.19e-02 |
| GO:BP | GO:0010043 | response to zinc ion | 1 | 31 | 9.19e-02 |
| GO:BP | GO:0001944 | vasculature development | 37 | 559 | 9.19e-02 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 9 | 84 | 9.19e-02 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 13 | 150 | 9.19e-02 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 9.19e-02 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 9.20e-02 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 1 | 7 | 9.20e-02 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 1 | 7 | 9.20e-02 |
| GO:BP | GO:0090084 | negative regulation of inclusion body assembly | 1 | 10 | 9.25e-02 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 2 | 4 | 9.25e-02 |
| GO:BP | GO:0009299 | mRNA transcription | 1 | 50 | 9.25e-02 |
| GO:BP | GO:0045175 | basal protein localization | 1 | 1 | 9.25e-02 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 3 | 9 | 9.25e-02 |
| GO:BP | GO:0060541 | respiratory system development | 4 | 163 | 9.25e-02 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 9.25e-02 |
| GO:BP | GO:0045445 | myoblast differentiation | 10 | 90 | 9.25e-02 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 3 | 42 | 9.25e-02 |
| GO:BP | GO:0036302 | atrioventricular canal development | 2 | 12 | 9.25e-02 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 9.26e-02 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 8 | 74 | 9.26e-02 |
| GO:BP | GO:0001822 | kidney development | 6 | 244 | 9.26e-02 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 3 | 13 | 9.26e-02 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 9 | 85 | 9.26e-02 |
| GO:BP | GO:0007614 | short-term memory | 1 | 9 | 9.26e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 2 | 159 | 9.26e-02 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 2 | 17 | 9.26e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 9.26e-02 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 9.26e-02 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 9.26e-02 |
| GO:BP | GO:0060914 | heart formation | 3 | 29 | 9.26e-02 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 1 | 35 | 9.26e-02 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 9.26e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 8 | 513 | 9.26e-02 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 1 | 103 | 9.26e-02 |
| GO:BP | GO:0021546 | rhombomere development | 2 | 4 | 9.29e-02 |
| GO:BP | GO:0007398 | ectoderm development | 2 | 15 | 9.29e-02 |
| GO:BP | GO:0048468 | cell development | 3 | 1963 | 9.37e-02 |
| GO:BP | GO:0001889 | liver development | 12 | 110 | 9.37e-02 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 9.40e-02 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 7 | 86 | 9.40e-02 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 29 | 551 | 9.41e-02 |
| GO:BP | GO:0016570 | histone modification | 29 | 433 | 9.42e-02 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 9.54e-02 |
| GO:BP | GO:0009790 | embryo development | 59 | 875 | 9.54e-02 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 1 | 88 | 9.54e-02 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 1 | 111 | 9.54e-02 |
| GO:BP | GO:0032240 | negative regulation of nucleobase-containing compound transport | 1 | 3 | 9.54e-02 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 5 | 274 | 9.54e-02 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 9.54e-02 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 1 | 8 | 9.54e-02 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 9.54e-02 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 9.54e-02 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 2 | 893 | 9.64e-02 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 3 | 135 | 9.64e-02 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 3 | 50 | 9.65e-02 |
| GO:BP | GO:0015074 | DNA integration | 2 | 9 | 9.65e-02 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 2 | 24 | 9.65e-02 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 2 | 16 | 9.73e-02 |
| GO:BP | GO:0033363 | secretory granule organization | 1 | 50 | 9.84e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 22 | 278 | 9.84e-02 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 4 | 26 | 9.87e-02 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 5 | 418 | 9.93e-02 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 6 | 482 | 9.93e-02 |
| GO:BP | GO:0072001 | renal system development | 6 | 251 | 9.96e-02 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 3 | 16 | 9.96e-02 |
| GO:BP | GO:0000303 | response to superoxide | 3 | 21 | 1.00e-01 |
| GO:BP | GO:0001842 | neural fold formation | 2 | 5 | 1.00e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 3 | 15 | 1.01e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 1.02e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 6 | 46 | 1.02e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 1 | 50 | 1.02e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 1 | 8 | 1.02e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 2 | 128 | 1.02e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 3 | 113 | 1.03e-01 |
| GO:BP | GO:0048645 | animal organ formation | 4 | 51 | 1.03e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 1.03e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 1 | 38 | 1.03e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 2 | 14 | 1.03e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 1 | 2 | 1.03e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 1 | 127 | 1.04e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 1.04e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 2 | 10 | 1.05e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 12 | 144 | 1.05e-01 |
| GO:BP | GO:0110024 | positive regulation of cardiac muscle myoblast proliferation | 1 | 3 | 1.05e-01 |
| GO:BP | GO:1901390 | positive regulation of transforming growth factor beta activation | 1 | 3 | 1.05e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 2 | 126 | 1.05e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 1.05e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 133 | 1.05e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 3 | 17 | 1.05e-01 |
| GO:BP | GO:0110021 | cardiac muscle myoblast proliferation | 1 | 3 | 1.05e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 5 | 45 | 1.05e-01 |
| GO:BP | GO:0110022 | regulation of cardiac muscle myoblast proliferation | 1 | 3 | 1.05e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 1.05e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 2 | 16 | 1.06e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 3 | 22 | 1.06e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 2 | 55 | 1.06e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 3 | 50 | 1.06e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 1.06e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 1 | 2 | 1.06e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 2 | 21 | 1.06e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 1 | 2 | 1.06e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 1.06e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 1.06e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 3 | 39 | 1.06e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 1.06e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 3 | 2035 | 1.06e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 1.06e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 1 | 2 | 1.06e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 3 | 33 | 1.06e-01 |
| GO:BP | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 1 | 1.06e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 1 | 2 | 1.07e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 3 | 55 | 1.07e-01 |
| GO:BP | GO:0021536 | diencephalon development | 1 | 41 | 1.08e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 3 | 18 | 1.08e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 6 | 63 | 1.08e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 2 | 113 | 1.08e-01 |
| GO:BP | GO:0006968 | cellular defense response | 1 | 15 | 1.08e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 4 | 22 | 1.08e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 7 | 57 | 1.08e-01 |
| GO:BP | GO:0021572 | rhombomere 6 development | 1 | 1 | 1.09e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 2 | 5 | 1.09e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 2 | 47 | 1.09e-01 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 13 | 1.09e-01 |
| GO:BP | GO:0021599 | abducens nerve formation | 1 | 1 | 1.09e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 1 | 4 | 1.09e-01 |
| GO:BP | GO:0021598 | abducens nerve morphogenesis | 1 | 1 | 1.09e-01 |
| GO:BP | GO:0021560 | abducens nerve development | 1 | 1 | 1.09e-01 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 1 | 16 | 1.10e-01 |
| GO:BP | GO:0001709 | cell fate determination | 2 | 25 | 1.10e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 1.10e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 1.10e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 1 | 11 | 1.10e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 7 | 70 | 1.10e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 10 | 1.10e-01 |
| GO:BP | GO:0097719 | neural tissue regeneration | 1 | 1 | 1.10e-01 |
| GO:BP | GO:0072359 | circulatory system development | 57 | 884 | 1.10e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 2 | 16 | 1.11e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 23 | 343 | 1.11e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.11e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.11e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 6 | 45 | 1.11e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 1.11e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 2 | 877 | 1.11e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 6 | 191 | 1.12e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 2 | 147 | 1.12e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 1.12e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 3 | 15 | 1.12e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 5 | 1074 | 1.12e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.12e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.12e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 1.12e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 1.12e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 5 | 1076 | 1.12e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 3 | 14 | 1.12e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 3 | 14 | 1.12e-01 |
| GO:BP | GO:0007254 | JNK cascade | 1 | 138 | 1.12e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 1 | 160 | 1.12e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 2 | 1167 | 1.13e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 2 | 267 | 1.13e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 1 | 138 | 1.13e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 2 | 8 | 1.13e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 1 | 60 | 1.13e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 2 | 8 | 1.13e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 2 | 8 | 1.13e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 2 | 150 | 1.13e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 1 | 37 | 1.15e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 4 | 33 | 1.16e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 1 | 5 | 1.16e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 1 | 141 | 1.16e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 4 | 31 | 1.17e-01 |
| GO:BP | GO:0060486 | club cell differentiation | 1 | 3 | 1.20e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 3 | 47 | 1.20e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 3 | 42 | 1.20e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 8 | 71 | 1.20e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 6 | 64 | 1.20e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 1 | 70 | 1.21e-01 |
| GO:BP | GO:2000761 | positive regulation of N-terminal peptidyl-lysine acetylation | 1 | 1 | 1.21e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 2 | 21 | 1.21e-01 |
| GO:BP | GO:2000759 | regulation of N-terminal peptidyl-lysine acetylation | 1 | 1 | 1.21e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 1.21e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 1 | 3 | 1.21e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 2 | 149 | 1.21e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 1.21e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 1.21e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 1 | 3 | 1.21e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 1 | 3 | 1.21e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 3 | 15 | 1.21e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 1 | 3 | 1.21e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 1 | 183 | 1.21e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 1 | 53 | 1.21e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 1.22e-01 |
| GO:BP | GO:0035265 | organ growth | 12 | 131 | 1.22e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 12 | 331 | 1.23e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 1 | 179 | 1.23e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 8 | 74 | 1.23e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 3 | 1.23e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 6 | 64 | 1.23e-01 |
| GO:BP | GO:0001756 | somitogenesis | 7 | 48 | 1.23e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 1.23e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 1 | 66 | 1.23e-01 |
| GO:BP | GO:0043041 | amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 1.23e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 1 | 2 | 1.23e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 1 | 4 | 1.23e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 1 | 6 | 1.23e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 1 | 6 | 1.23e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 2 | 21 | 1.24e-01 |
| GO:BP | GO:0003176 | aortic valve development | 3 | 37 | 1.24e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 35 | 504 | 1.24e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 1.24e-01 |
| GO:BP | GO:0035270 | endocrine system development | 3 | 86 | 1.24e-01 |
| GO:BP | GO:0051102 | DNA ligation involved in DNA recombination | 1 | 1 | 1.24e-01 |
| GO:BP | GO:0075713 | establishment of integrated proviral latency | 1 | 1 | 1.24e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 2 | 7 | 1.24e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 1.24e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 1.25e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 3 | 129 | 1.25e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 2 | 24 | 1.25e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 15 | 187 | 1.25e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 4 | 41 | 1.25e-01 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 1 | 7 | 1.25e-01 |
| GO:BP | GO:0002041 | intussusceptive angiogenesis | 1 | 1 | 1.25e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 2 | 38 | 1.25e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 1.27e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 1 | 86 | 1.27e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 1 | 6 | 1.27e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 1 | 14 | 1.27e-01 |
| GO:BP | GO:0070841 | inclusion body assembly | 1 | 22 | 1.27e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 2 | 57 | 1.27e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 1.27e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 2 | 12 | 1.27e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 1 | 2 | 1.28e-01 |
| GO:BP | GO:0010467 | gene expression | 10 | 4587 | 1.28e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 2 | 8 | 1.28e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 3 | 14 | 1.28e-01 |
| GO:BP | GO:0035781 | CD86 biosynthetic process | 1 | 2 | 1.28e-01 |
| GO:BP | GO:0007611 | learning or memory | 2 | 202 | 1.28e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 1 | 197 | 1.29e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 1.29e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 2 | 23 | 1.30e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 3 | 395 | 1.30e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 7 | 816 | 1.30e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 1 | 214 | 1.30e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 2 | 9 | 1.30e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 3 | 16 | 1.30e-01 |
| GO:BP | GO:0016310 | phosphorylation | 2 | 1372 | 1.30e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 1 | 4 | 1.30e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 1 | 190 | 1.30e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 3 | 16 | 1.30e-01 |
| GO:BP | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 1 | 4 | 1.30e-01 |
| GO:BP | GO:1902217 | erythrocyte apoptotic process | 1 | 1 | 1.30e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 1.30e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 2 | 1223 | 1.30e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 2 | 8 | 1.30e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 1 | 16 | 1.30e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 2 | 8 | 1.30e-01 |
| GO:BP | GO:0140742 | lncRNA transcription | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 6 | 216 | 1.31e-01 |
| GO:BP | GO:0140744 | negative regulation of lncRNA transcription | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0110064 | lncRNA catabolic process | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0140743 | regulation of lncRNA transcription | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0035261 | external genitalia morphogenesis | 1 | 1 | 1.31e-01 |
| GO:BP | GO:0061780 | mitotic cohesin loading | 1 | 1 | 1.31e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 1.31e-01 |
| GO:BP | GO:0035295 | tube development | 2 | 821 | 1.32e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 1 | 196 | 1.32e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 4 | 62 | 1.32e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 1 | 4 | 1.32e-01 |
| GO:BP | GO:0061053 | somite development | 8 | 63 | 1.32e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 1.32e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 7 | 320 | 1.32e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 6 | 2135 | 1.32e-01 |
| GO:BP | GO:0061009 | common bile duct development | 1 | 4 | 1.32e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 1.32e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 2 | 7 | 1.32e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 1.32e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 2 | 6 | 1.32e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 1.33e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 1 | 14 | 1.34e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 1 | 27 | 1.34e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 7 | 67 | 1.37e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 2 | 20 | 1.37e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 2 | 9 | 1.38e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 1.40e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 1 | 102 | 1.40e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 3 | 41 | 1.40e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 7 | 89 | 1.40e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 3 | 58 | 1.40e-01 |
| GO:BP | GO:0035026 | leading edge cell differentiation | 1 | 2 | 1.40e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 4 | 27 | 1.41e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 3 | 48 | 1.41e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 1 | 234 | 1.41e-01 |
| GO:BP | GO:0006325 | chromatin organization | 35 | 551 | 1.41e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 2 | 11 | 1.41e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 1.41e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 2 | 2 | 1.41e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 3 | 26 | 1.42e-01 |
| GO:BP | GO:0030879 | mammary gland development | 1 | 106 | 1.42e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 1 | 30 | 1.42e-01 |
| GO:BP | GO:0021503 | neural fold bending | 1 | 1 | 1.42e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 2 | 10 | 1.42e-01 |
| GO:BP | GO:0032915 | positive regulation of transforming growth factor beta2 production | 1 | 3 | 1.42e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 3 | 41 | 1.42e-01 |
| GO:BP | GO:0048371 | lateral mesodermal cell differentiation | 1 | 2 | 1.42e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 1 | 90 | 1.42e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 3 | 34 | 1.42e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 1.42e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 13 | 165 | 1.42e-01 |
| GO:BP | GO:0048511 | rhythmic process | 2 | 237 | 1.42e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 1 | 2 | 1.43e-01 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 1 | 3 | 1.43e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 2 | 24 | 1.43e-01 |
| GO:BP | GO:0060510 | type II pneumocyte differentiation | 1 | 5 | 1.43e-01 |
| GO:BP | GO:0070315 | G1 to G0 transition involved in cell differentiation | 1 | 4 | 1.43e-01 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 1 | 5 | 1.43e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 1 | 227 | 1.44e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 2 | 54 | 1.44e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 3 | 34 | 1.44e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 1 | 16 | 1.44e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 2 | 191 | 1.44e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 6 | 114 | 1.44e-01 |
| GO:BP | GO:0007492 | endoderm development | 7 | 65 | 1.44e-01 |
| GO:BP | GO:0045819 | positive regulation of glycogen catabolic process | 1 | 1 | 1.45e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 1 | 108 | 1.45e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 3 | 64 | 1.45e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 1 | 25 | 1.46e-01 |
| GO:BP | GO:0070898 | RNA polymerase III preinitiation complex assembly | 1 | 1 | 1.46e-01 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 2 | 18 | 1.46e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 1 | 4 | 1.46e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 1.46e-01 |
| GO:BP | GO:0050890 | cognition | 2 | 227 | 1.46e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 8 | 91 | 1.46e-01 |
| GO:BP | GO:0070982 | L-asparagine metabolic process | 1 | 2 | 1.46e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 1 | 2 | 1.47e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 2 | 1399 | 1.47e-01 |
| GO:BP | GO:0030900 | forebrain development | 7 | 281 | 1.47e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 3 | 55 | 1.47e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 3 | 21 | 1.47e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 2 | 258 | 1.47e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 1 | 108 | 1.47e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 3 | 17 | 1.47e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 1 | 236 | 1.47e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 5 | 189 | 1.47e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 5 | 45 | 1.47e-01 |
| GO:BP | GO:0046599 | regulation of centriole replication | 2 | 21 | 1.48e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 16 | 197 | 1.48e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 2 | 25 | 1.49e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 2 | 13 | 1.49e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 3 | 12 | 1.49e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 5 | 41 | 1.49e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 3 | 41 | 1.50e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 3 | 67 | 1.50e-01 |
| GO:BP | GO:0060429 | epithelium development | 2 | 850 | 1.50e-01 |
| GO:BP | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 2 | 1.52e-01 |
| GO:BP | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway | 1 | 2 | 1.52e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 5 | 37 | 1.52e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 1 | 35 | 1.52e-01 |
| GO:BP | GO:0035120 | post-embryonic appendage morphogenesis | 1 | 2 | 1.54e-01 |
| GO:BP | GO:0035127 | post-embryonic limb morphogenesis | 1 | 2 | 1.54e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 2 | 7 | 1.54e-01 |
| GO:BP | GO:0035128 | post-embryonic forelimb morphogenesis | 1 | 2 | 1.54e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 2 | 5 | 1.54e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 1.55e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 1.57e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 1 | 90 | 1.57e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 1.57e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 3 | 18 | 1.57e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 6 | 2293 | 1.57e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 1.57e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 3 | 26 | 1.58e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 1 | 265 | 1.59e-01 |
| GO:BP | GO:0048732 | gland development | 12 | 318 | 1.59e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 1 | 2 | 1.60e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 1.60e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 1 | 3 | 1.60e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 1 | 6 | 1.60e-01 |
| GO:BP | GO:0009798 | axis specification | 7 | 63 | 1.60e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 1 | 22 | 1.60e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 2 | 27 | 1.61e-01 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 1.61e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 1 | 23 | 1.61e-01 |
| GO:BP | GO:2000691 | negative regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 1.61e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 1.61e-01 |
| GO:BP | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 1.61e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 2 | 4 | 1.61e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 1.61e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 1.61e-01 |
| GO:BP | GO:0008406 | gonad development | 4 | 158 | 1.61e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 1 | 39 | 1.61e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 1 | 295 | 1.61e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 2 | 16 | 1.61e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0097198 | histone H3-K36 trimethylation | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 1 | 9 | 1.61e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 1.61e-01 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 2 | 21 | 1.61e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 3 | 20 | 1.61e-01 |
| GO:BP | GO:1905070 | anterior visceral endoderm cell migration | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0061865 | polarized secretion of basement membrane proteins in epithelium | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0061864 | basement membrane constituent secretion | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0036268 | swimming | 1 | 1 | 1.62e-01 |
| GO:BP | GO:0061921 | peptidyl-lysine propionylation | 1 | 1 | 1.62e-01 |
| GO:BP | GO:0140067 | peptidyl-lysine butyrylation | 1 | 1 | 1.62e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 1 | 282 | 1.63e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 5 | 53 | 1.63e-01 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 1 | 7 | 1.64e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 1.64e-01 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 1 | 4 | 1.64e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 3 | 100 | 1.64e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 5 | 40 | 1.64e-01 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 1 | 4 | 1.64e-01 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 1 | 4 | 1.64e-01 |
| GO:BP | GO:0001880 | Mullerian duct regression | 1 | 4 | 1.64e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 3 | 232 | 1.64e-01 |
| GO:BP | GO:0048369 | lateral mesoderm morphogenesis | 1 | 3 | 1.64e-01 |
| GO:BP | GO:1903215 | negative regulation of protein targeting to mitochondrion | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 1.64e-01 |
| GO:BP | GO:0048370 | lateral mesoderm formation | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 2 | 27 | 1.64e-01 |
| GO:BP | GO:0032197 | retrotransposition | 1 | 4 | 1.64e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 146 | 1.64e-01 |
| GO:BP | GO:2000174 | regulation of pro-T cell differentiation | 1 | 1 | 1.64e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 1.64e-01 |
| GO:BP | GO:2000176 | positive regulation of pro-T cell differentiation | 1 | 1 | 1.64e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 3 | 164 | 1.64e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 16 | 185 | 1.64e-01 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 1 | 8 | 1.65e-01 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 1.65e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 4 | 161 | 1.65e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.65e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 150 | 1.66e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 14 | 198 | 1.66e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 5 | 1123 | 1.67e-01 |
| GO:BP | GO:0003158 | endothelium development | 1 | 104 | 1.68e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 1 | 5 | 1.68e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 4 | 103 | 1.68e-01 |
| GO:BP | GO:0061114 | branching involved in pancreas morphogenesis | 1 | 1 | 1.68e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 6 | 73 | 1.68e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 1 | 40 | 1.68e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 1 | 5 | 1.68e-01 |
| GO:BP | GO:0002194 | hepatocyte cell migration | 1 | 1 | 1.68e-01 |
| GO:BP | GO:0043049 | otic placode formation | 1 | 1 | 1.68e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 5 | 39 | 1.68e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 8 | 86 | 1.68e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 4 | 809 | 1.68e-01 |
| GO:BP | GO:0051168 | nuclear export | 7 | 150 | 1.68e-01 |
| GO:BP | GO:2000979 | positive regulation of forebrain neuron differentiation | 1 | 1 | 1.68e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 1 | 7 | 1.68e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 1.69e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 1 | 24 | 1.69e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 1 | 139 | 1.69e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 1.69e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 1.69e-01 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 1 | 6 | 1.70e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 2 | 31 | 1.70e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 54 | 7552 | 1.71e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 3 | 13 | 1.71e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 14 | 132 | 1.71e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 1 | 6 | 1.71e-01 |
| GO:BP | GO:1904791 | negative regulation of shelterin complex assembly | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 5 | 53 | 1.72e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 2 | 49 | 1.73e-01 |
| GO:BP | GO:0072752 | cellular response to rapamycin | 1 | 2 | 1.73e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 2 | 20 | 1.74e-01 |
| GO:BP | GO:0035054 | embryonic heart tube anterior/posterior pattern specification | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 4 | 97 | 1.74e-01 |
| GO:BP | GO:0050821 | protein stabilization | 4 | 191 | 1.74e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 1.74e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 3 | 22 | 1.74e-01 |
| GO:BP | GO:0003285 | septum secundum development | 1 | 3 | 1.74e-01 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0048565 | digestive tract development | 10 | 92 | 1.74e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 1.74e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 1 | 8 | 1.74e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 1.74e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 2 | 15 | 1.74e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 1.75e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 6 | 47 | 1.75e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 1 | 13 | 1.75e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 1 | 27 | 1.75e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 1.75e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 2 | 7 | 1.75e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 1 | 8 | 1.76e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0016480 | negative regulation of transcription by RNA polymerase III | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 1.76e-01 |
| GO:BP | GO:0035878 | nail development | 2 | 8 | 1.76e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 3 | 2984 | 1.76e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 1 | 12 | 1.76e-01 |
| GO:BP | GO:0030154 | cell differentiation | 3 | 2958 | 1.76e-01 |
| GO:BP | GO:0044524 | protein sulfhydration | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 8 | 164 | 1.76e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 26 | 1.76e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0042110 | T cell activation | 7 | 313 | 1.76e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0018272 | protein-pyridoxal-5-phosphate linkage via peptidyl-N6-pyridoxal phosphate-L-lysine | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 1.76e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 1 | 8 | 1.76e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 2 | 9 | 1.76e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 1 | 13 | 1.76e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 2 | 50 | 1.76e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 3 | 17 | 1.76e-01 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 9 | 240 | 1.76e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 5 | 54 | 1.76e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0002457 | T cell antigen processing and presentation | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 6 | 71 | 1.76e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 2 | 6 | 1.76e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 2 | 5 | 1.76e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 29 | 555 | 1.76e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 2 | 35 | 1.76e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0031016 | pancreas development | 5 | 53 | 1.76e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 2 | 4 | 1.76e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 1 | 6 | 1.76e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 2 | 13 | 1.76e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 4 | 34 | 1.76e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 1.76e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 1 | 101 | 1.76e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 1 | 14 | 1.76e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 1 | 29 | 1.76e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 1 | 427 | 1.76e-01 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 1 | 23 | 1.76e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.76e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 3 | 24 | 1.76e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 8 | 101 | 1.76e-01 |
| GO:BP | GO:1902682 | protein localization to pericentric heterochromatin | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0042661 | regulation of mesodermal cell fate specification | 1 | 4 | 1.77e-01 |
| GO:BP | GO:0008584 | male gonad development | 3 | 99 | 1.77e-01 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 1 | 5 | 1.77e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 2 | 32 | 1.77e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 4 | 20 | 1.77e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 1.78e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 29 | 462 | 1.79e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 1.79e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 1.79e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 3 | 100 | 1.79e-01 |
| GO:BP | GO:0035284 | brain segmentation | 1 | 2 | 1.79e-01 |
| GO:BP | GO:0021571 | rhombomere 5 development | 1 | 2 | 1.79e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 1 | 12 | 1.79e-01 |
| GO:BP | GO:0048733 | sebaceous gland development | 1 | 6 | 1.79e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 4 | 30 | 1.79e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 3 | 17 | 1.79e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 2 | 11 | 1.79e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 1.79e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 1 | 7 | 1.79e-01 |
| GO:BP | GO:0001113 | transcription open complex formation at RNA polymerase II promoter | 1 | 1 | 1.80e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 6 | 120 | 1.80e-01 |
| GO:BP | GO:0061010 | gallbladder development | 1 | 1 | 1.80e-01 |
| GO:BP | GO:0070184 | mitochondrial tyrosyl-tRNA aminoacylation | 1 | 1 | 1.81e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 1.81e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 12 | 1173 | 1.81e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 2 | 7 | 1.81e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 2 | 33 | 1.81e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 22 | 378 | 1.81e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 18 | 1.81e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 4 | 276 | 1.81e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 29 | 1.81e-01 |
| GO:BP | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion | 1 | 5 | 1.82e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 3 | 138 | 1.82e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 1 | 9 | 1.82e-01 |
| GO:BP | GO:0097310 | cap2 mRNA methylation | 1 | 1 | 1.82e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 1.82e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 2 | 33 | 1.82e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 1 | 9 | 1.82e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 4 | 144 | 1.82e-01 |
| GO:BP | GO:1900119 | positive regulation of execution phase of apoptosis | 1 | 7 | 1.83e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 2 | 16 | 1.83e-01 |
| GO:BP | GO:0035601 | protein deacylation | 8 | 116 | 1.84e-01 |
| GO:BP | GO:0032736 | positive regulation of interleukin-13 production | 1 | 7 | 1.84e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 1 | 9 | 1.84e-01 |
| GO:BP | GO:0097305 | response to alcohol | 1 | 159 | 1.84e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 2 | 129 | 1.84e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 7 | 84 | 1.84e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 1 | 168 | 1.85e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 4 | 33 | 1.85e-01 |
| GO:BP | GO:0033301 | cell cycle comprising mitosis without cytokinesis | 1 | 1 | 1.85e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 1 | 5 | 1.85e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 3 | 21 | 1.85e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 5 | 211 | 1.85e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 6 | 665 | 1.86e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 1.86e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 1.86e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 1.86e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 3 | 140 | 1.86e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 2 | 18 | 1.86e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 1 | 29 | 1.86e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 43 | 663 | 1.86e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 3 | 14 | 1.86e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 11 | 1.86e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 3 | 40 | 1.86e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 2 | 222 | 1.86e-01 |
| GO:BP | GO:0061140 | lung secretory cell differentiation | 1 | 5 | 1.86e-01 |
| GO:BP | GO:2000257 | regulation of protein activation cascade | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 7 | 86 | 1.86e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 2 | 27 | 1.87e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 2 | 2065 | 1.87e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 9 | 101 | 1.87e-01 |
| GO:BP | GO:0071630 | nuclear protein quality control by the ubiquitin-proteasome system | 1 | 3 | 1.87e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 1.88e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 1 | 2 | 1.88e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 1 | 3 | 1.88e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 1.88e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 2 | 14 | 1.88e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 1 | 3 | 1.88e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 8 | 111 | 1.88e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 5 | 31 | 1.88e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 1.88e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 1 | 486 | 1.88e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 2 | 13 | 1.88e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 64 | 1.89e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 30 | 440 | 1.89e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 1.90e-01 |
| GO:BP | GO:0002376 | immune system process | 2 | 1447 | 1.90e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 3 | 36 | 1.90e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 1 | 6 | 1.90e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 1 | 5 | 1.90e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 2 | 8 | 1.90e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 5 | 1.90e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 12 | 110 | 1.90e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 3 | 3386 | 1.90e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 1 | 31 | 1.90e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 1.90e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 2 | 43 | 1.90e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 2 | 1869 | 1.90e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 17 | 251 | 1.90e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 2 | 9 | 1.90e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 1.91e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 1 | 14 | 1.91e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 2 | 7 | 1.91e-01 |
| GO:BP | GO:0021570 | rhombomere 4 development | 1 | 1 | 1.92e-01 |
| GO:BP | GO:0003175 | tricuspid valve development | 1 | 6 | 1.92e-01 |
| GO:BP | GO:0060897 | neural plate regionalization | 1 | 6 | 1.92e-01 |
| GO:BP | GO:0006556 | S-adenosylmethionine biosynthetic process | 1 | 3 | 1.92e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 1.92e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 4 | 67 | 1.92e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 2 | 103 | 1.92e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 7 | 91 | 1.92e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 3 | 26 | 1.92e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 3 | 20 | 1.92e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 2 | 9 | 1.92e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 8 | 89 | 1.92e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 2 | 2139 | 1.92e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 1 | 9 | 1.92e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 1 | 10 | 1.92e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 1 | 10 | 1.92e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 1 | 10 | 1.92e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 1 | 57 | 1.92e-01 |
| GO:BP | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition | 1 | 2 | 1.92e-01 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 1 | 3 | 1.92e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 28 | 418 | 1.92e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 2 | 10 | 1.94e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 1.94e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 2 | 11 | 1.94e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 2 | 2161 | 1.94e-01 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 1 | 5 | 1.95e-01 |
| GO:BP | GO:0030903 | notochord development | 3 | 17 | 1.95e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 1 | 199 | 1.96e-01 |
| GO:BP | GO:0010033 | response to organic substance | 2 | 1957 | 1.96e-01 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 1 | 7 | 1.96e-01 |
| GO:BP | GO:0007409 | axonogenesis | 2 | 359 | 1.96e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 1 | 34 | 1.96e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 4 | 1006 | 1.96e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 8 | 119 | 1.96e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 1 | 15 | 1.97e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 1.97e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 1 | 38 | 1.97e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 1.97e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 1 | 11 | 1.97e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 1 | 12 | 1.97e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 3 | 81 | 1.98e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 1.98e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 9 | 120 | 1.99e-01 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 1 | 7 | 1.99e-01 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 1 | 10 | 1.99e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 4 | 41 | 2.00e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 3 | 28 | 2.02e-01 |
| GO:BP | GO:1901301 | regulation of cargo loading into COPII-coated vesicle | 1 | 2 | 2.02e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 2 | 14 | 2.02e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 1 | 12 | 2.02e-01 |
| GO:BP | GO:0019482 | beta-alanine metabolic process | 1 | 2 | 2.02e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 2 | 20 | 2.02e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 2 | 147 | 2.02e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 1 | 12 | 2.02e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 1 | 479 | 2.02e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 2.02e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 2.02e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 2.02e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 1 | 30 | 2.02e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 1 | 30 | 2.02e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 2.02e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 7 | 90 | 2.04e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 1 | 3 | 2.04e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 3 | 76 | 2.04e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 6 | 80 | 2.04e-01 |
| GO:BP | GO:0035283 | central nervous system segmentation | 1 | 2 | 2.04e-01 |
| GO:BP | GO:0019043 | establishment of viral latency | 1 | 3 | 2.04e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 2 | 56 | 2.04e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 1 | 63 | 2.04e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 1 | 15 | 2.04e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 2 | 13 | 2.04e-01 |
| GO:BP | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1 | 3 | 2.04e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 1 | 19 | 2.04e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 1 | 19 | 2.04e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 1 | 290 | 2.04e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 1 | 3 | 2.04e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 2 | 53 | 2.04e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 2 | 13 | 2.04e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 1 | 215 | 2.04e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 2.04e-01 |
| GO:BP | GO:0060438 | trachea development | 1 | 13 | 2.04e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 1 | 11 | 2.04e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 1 | 6 | 2.04e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 215 | 2.04e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 2.04e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 1 | 19 | 2.04e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 2 | 22 | 2.04e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 2 | 12 | 2.04e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 5 | 155 | 2.04e-01 |
| GO:BP | GO:0051693 | actin filament capping | 1 | 34 | 2.05e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 1 | 478 | 2.05e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 2 | 15 | 2.05e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 20 | 260 | 2.05e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 8 | 114 | 2.05e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 3 | 19 | 2.05e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 3 | 22 | 2.05e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 2.05e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 2.05e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 8 | 114 | 2.05e-01 |
| GO:BP | GO:1905770 | regulation of mesodermal cell differentiation | 1 | 6 | 2.05e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 3 | 19 | 2.05e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 4 | 157 | 2.05e-01 |
| GO:BP | GO:0055123 | digestive system development | 6 | 100 | 2.05e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 2.05e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 2 | 7 | 2.05e-01 |
| GO:BP | GO:0070171 | negative regulation of tooth mineralization | 1 | 2 | 2.05e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 2.05e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 1 | 4 | 2.05e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 2.05e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 2.05e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 2 | 30 | 2.05e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 2 | 20 | 2.06e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 1 | 69 | 2.06e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 2 | 9 | 2.06e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 4 | 35 | 2.06e-01 |
| GO:BP | GO:0006974 | DNA damage response | 43 | 785 | 2.07e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 3 | 85 | 2.07e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 15 | 2.07e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 6 | 121 | 2.07e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 2.07e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 2 | 36 | 2.08e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 4 | 38 | 2.08e-01 |
| GO:BP | GO:0007548 | sex differentiation | 5 | 191 | 2.08e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 1 | 2 | 2.08e-01 |
| GO:BP | GO:0051028 | mRNA transport | 8 | 117 | 2.09e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 250 | 2.09e-01 |
| GO:BP | GO:0097368 | establishment of Sertoli cell barrier | 1 | 3 | 2.09e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 2.09e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 1 | 8 | 2.09e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 1 | 6 | 2.09e-01 |
| GO:BP | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 1 | 19 | 2.09e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 1 | 6 | 2.09e-01 |
| GO:BP | GO:0060173 | limb development | 12 | 142 | 2.09e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 2.09e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 2.09e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 2.09e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 2 | 14 | 2.09e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 6 | 898 | 2.09e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 1 | 12 | 2.09e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 7 | 71 | 2.09e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 2 | 14 | 2.09e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 7 | 3625 | 2.09e-01 |
| GO:BP | GO:0048736 | appendage development | 12 | 142 | 2.09e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 6 | 69 | 2.09e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 1 | 5 | 2.09e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 2.10e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 1 | 3 | 2.10e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 1 | 4 | 2.10e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 1 | 4 | 2.10e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 1 | 4 | 2.10e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 2 | 37 | 2.10e-01 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 1 | 11 | 2.10e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 5 | 37 | 2.10e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 4 | 39 | 2.10e-01 |
| GO:BP | GO:0007517 | muscle organ development | 1 | 277 | 2.10e-01 |
| GO:BP | GO:0001711 | endodermal cell fate commitment | 1 | 9 | 2.10e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 2.10e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 4 | 34 | 2.10e-01 |
| GO:BP | GO:0061564 | axon development | 2 | 398 | 2.10e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 1 | 11 | 2.10e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 1 | 615 | 2.10e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 2.10e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 3 | 23 | 2.10e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 2 | 36 | 2.10e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 1 | 20 | 2.10e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 1 | 8 | 2.10e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 1 | 8 | 2.10e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 2 | 12 | 2.10e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 2.10e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 1 | 7 | 2.10e-01 |
| GO:BP | GO:0051892 | negative regulation of cardioblast differentiation | 1 | 1 | 2.10e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 1 | 625 | 2.10e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 2 | 29 | 2.10e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 1 | 535 | 2.10e-01 |
| GO:BP | GO:0061159 | establishment of bipolar cell polarity involved in cell morphogenesis | 1 | 1 | 2.10e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 2.10e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 1 | 7 | 2.11e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 3 | 61 | 2.11e-01 |
| GO:BP | GO:0140066 | peptidyl-lysine crotonylation | 1 | 2 | 2.11e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 4 | 42 | 2.11e-01 |
| GO:BP | GO:0071921 | cohesin loading | 1 | 3 | 2.11e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 2 | 8 | 2.11e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 41 | 1199 | 2.12e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 1 | 637 | 2.12e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 4 | 37 | 2.12e-01 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 1 | 7 | 2.12e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 1 | 560 | 2.12e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 6 | 71 | 2.12e-01 |
| GO:BP | GO:0006528 | asparagine metabolic process | 1 | 5 | 2.12e-01 |
| GO:BP | GO:0010038 | response to metal ion | 1 | 264 | 2.12e-01 |
| GO:BP | GO:0016050 | vesicle organization | 1 | 320 | 2.12e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 236 | 2.12e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 1 | 6 | 2.12e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 2.12e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 2.12e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 6 | 2.12e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 3 | 16 | 2.12e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 2.12e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 2.12e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 5 | 234 | 2.12e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 1 | 8 | 2.12e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 1 | 15 | 2.12e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 2.12e-01 |
| GO:BP | GO:1904544 | positive regulation of free ubiquitin chain polymerization | 1 | 2 | 2.12e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 1 | 15 | 2.12e-01 |
| GO:BP | GO:1904542 | regulation of free ubiquitin chain polymerization | 1 | 2 | 2.12e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 2.12e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 1 | 40 | 2.12e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 1 | 11 | 2.13e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 5 | 50 | 2.13e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 1 | 36 | 2.13e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 2.13e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 1 | 4 | 2.13e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 1 | 89 | 2.14e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 2 | 13 | 2.14e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 1 | 44 | 2.14e-01 |
| GO:BP | GO:0097498 | endothelial tube lumen extension | 1 | 2 | 2.14e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 1 | 11 | 2.15e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 1 | 81 | 2.15e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 2.15e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 1 | 64 | 2.15e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 2 | 22 | 2.15e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 2 | 52 | 2.16e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 1 | 22 | 2.16e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 3 | 96 | 2.16e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 10 | 165 | 2.16e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 1 | 96 | 2.16e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 2.16e-01 |
| GO:BP | GO:0048589 | developmental growth | 30 | 522 | 2.16e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 2 | 47 | 2.17e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 53 | 956 | 2.17e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 4 | 76 | 2.17e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 2.18e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 2 | 303 | 2.18e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 1 | 556 | 2.18e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 1 | 93 | 2.18e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 1 | 92 | 2.18e-01 |
| GO:BP | GO:0046619 | lens placode formation involved in camera-type eye formation | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0001743 | lens placode formation | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 1 | 19 | 2.19e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 1 | 13 | 2.19e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 1 | 9 | 2.19e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 1 | 72 | 2.19e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 11 | 161 | 2.20e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 2 | 52 | 2.20e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 1 | 21 | 2.20e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 2.20e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 2 | 35 | 2.20e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 2 | 19 | 2.20e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 2 | 42 | 2.21e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 2 | 37 | 2.21e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 5 | 59 | 2.21e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 4 | 44 | 2.21e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 2.21e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 2.21e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 2.21e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 2.21e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 2 | 16 | 2.21e-01 |
| GO:BP | GO:0006428 | isoleucyl-tRNA aminoacylation | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 1 | 7 | 2.21e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 3 | 604 | 2.21e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 3 | 19 | 2.21e-01 |
| GO:BP | GO:0009888 | tissue development | 2 | 1425 | 2.21e-01 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 1 | 7 | 2.21e-01 |
| GO:BP | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 1 | 6 | 2.21e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 4 | 31 | 2.21e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 1 | 706 | 2.21e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 2 | 13 | 2.21e-01 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 1 | 6 | 2.21e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 5 | 79 | 2.22e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 2.22e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 2.22e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 2.22e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 2.22e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 2 | 23 | 2.22e-01 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 1 | 10 | 2.23e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 6 | 78 | 2.23e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 1 | 4 | 2.23e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 3 | 89 | 2.23e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 2.24e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 1 | 8 | 2.24e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 2 | 11 | 2.25e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 1 | 9 | 2.25e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 2 | 21 | 2.25e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 1 | 22 | 2.25e-01 |
| GO:BP | GO:0097403 | cellular response to raffinose | 1 | 1 | 2.25e-01 |
| GO:BP | GO:0003342 | proepicardium development | 1 | 2 | 2.25e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 1 | 6 | 2.25e-01 |
| GO:BP | GO:1904155 | DN2 thymocyte differentiation | 1 | 1 | 2.25e-01 |
| GO:BP | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 6 | 2.25e-01 |
| GO:BP | GO:1901545 | response to raffinose | 1 | 1 | 2.25e-01 |
| GO:BP | GO:0019042 | viral latency | 1 | 4 | 2.25e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 2.25e-01 |
| GO:BP | GO:0003343 | septum transversum development | 1 | 2 | 2.25e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 2 | 14 | 2.26e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 1 | 321 | 2.26e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 2 | 414 | 2.26e-01 |
| GO:BP | GO:0010909 | positive regulation of heparan sulfate proteoglycan biosynthetic process | 1 | 3 | 2.26e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 2 | 24 | 2.26e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 2 | 9 | 2.26e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 5 | 67 | 2.26e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0034773 | histone H4-K20 trimethylation | 1 | 5 | 2.26e-01 |
| GO:BP | GO:0035282 | segmentation | 8 | 75 | 2.26e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 4 | 212 | 2.26e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 1 | 5 | 2.26e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 2.27e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 2 | 23 | 2.28e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 2 | 54 | 2.28e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 1 | 279 | 2.28e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 2 | 14 | 2.28e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 4 | 57 | 2.28e-01 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 1 | 8 | 2.28e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 2 | 2565 | 2.28e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 2 | 2573 | 2.28e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 1 | 679 | 2.28e-01 |
| GO:BP | GO:1904831 | positive regulation of aortic smooth muscle cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0035887 | aortic smooth muscle cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0018352 | protein-pyridoxal-5-phosphate linkage | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0019343 | cysteine biosynthetic process via cystathionine | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0042723 | thiamine-containing compound metabolic process | 1 | 4 | 2.28e-01 |
| GO:BP | GO:1904829 | regulation of aortic smooth muscle cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0071934 | thiamine transmembrane transport | 1 | 3 | 2.28e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 2.29e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 1 | 52 | 2.29e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 2 | 130 | 2.29e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 1 | 26 | 2.29e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 1 | 2 | 2.29e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 1 | 4 | 2.29e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 1 | 4 | 2.29e-01 |
| GO:BP | GO:0061458 | reproductive system development | 4 | 215 | 2.29e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 1 | 2 | 2.29e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 1 | 17 | 2.29e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 1 | 5 | 2.29e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 2.29e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 4 | 127 | 2.29e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 2 | 280 | 2.30e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 1 | 8 | 2.30e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 2 | 81 | 2.30e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 1 | 8 | 2.30e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 1 | 18 | 2.30e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 1 | 3 | 2.30e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 4 | 39 | 2.30e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 2.30e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 2.30e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 3 | 95 | 2.30e-01 |
| GO:BP | GO:0038018 | Wnt receptor catabolic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 2.31e-01 |
| GO:BP | GO:0046794 | transport of virus | 1 | 19 | 2.31e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 2.31e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 1 | 4 | 2.31e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 3 | 35 | 2.32e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 2.32e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 3 | 16 | 2.32e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 2 | 8 | 2.32e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 38 | 2.32e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 2.32e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 13 | 207 | 2.33e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 2.33e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 12 | 161 | 2.33e-01 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 2.33e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 3 | 23 | 2.33e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 2 | 13 | 2.34e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 1 | 8 | 2.34e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 1 | 9 | 2.34e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 3 | 26 | 2.34e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 2 | 463 | 2.34e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 4 | 34 | 2.34e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 1 | 7 | 2.34e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 2 | 10 | 2.34e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 10 | 147 | 2.34e-01 |
| GO:BP | GO:0007099 | centriole replication | 2 | 38 | 2.34e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 1 | 18 | 2.34e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 2 | 7 | 2.34e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 1 | 14 | 2.34e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 2.34e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 1 | 13 | 2.34e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 1 | 14 | 2.34e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 2.35e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 3 | 28 | 2.35e-01 |
| GO:BP | GO:0061642 | chemoattraction of axon | 1 | 2 | 2.35e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.35e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 2.35e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 1 | 15 | 2.35e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 1 | 13 | 2.35e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 1 | 46 | 2.36e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 1 | 14 | 2.36e-01 |
| GO:BP | GO:0001112 | DNA-templated transcription open complex formation | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0001120 | protein-DNA complex remodeling | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 2.37e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 84 | 2.37e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 16 | 293 | 2.37e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 4 | 35 | 2.37e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 2.37e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 4 | 62 | 2.38e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 2.39e-01 |
| GO:BP | GO:0040008 | regulation of growth | 5 | 501 | 2.39e-01 |
| GO:BP | GO:0070914 | UV-damage excision repair | 1 | 13 | 2.40e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 3 | 4099 | 2.40e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 1 | 15 | 2.40e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0036211 | protein modification process | 2 | 2901 | 2.41e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 1 | 127 | 2.41e-01 |
| GO:BP | GO:0036451 | cap mRNA methylation | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 2.41e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0097309 | cap1 mRNA methylation | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 2.41e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 4 | 47 | 2.41e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 2.41e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 6 | 75 | 2.42e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 2 | 40 | 2.42e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 17 | 313 | 2.42e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 2.42e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 2 | 11 | 2.42e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 2.42e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 3 | 30 | 2.42e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 2 | 40 | 2.42e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 1 | 4 | 2.43e-01 |
| GO:BP | GO:0015888 | thiamine transport | 1 | 3 | 2.43e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 1 | 57 | 2.43e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 4 | 40 | 2.43e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 1 | 55 | 2.43e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 2.43e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 8 | 539 | 2.43e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 3 | 28 | 2.44e-01 |
| GO:BP | GO:0032877 | positive regulation of DNA endoreduplication | 1 | 2 | 2.44e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 1 | 3 | 2.44e-01 |
| GO:BP | GO:0001824 | blastocyst development | 7 | 94 | 2.44e-01 |
| GO:BP | GO:1901652 | response to peptide | 1 | 377 | 2.44e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 2 | 15 | 2.44e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 2 | 15 | 2.44e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 1 | 14 | 2.44e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 755 | 2.44e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 2 | 15 | 2.44e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 2 | 107 | 2.44e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 1 | 6 | 2.44e-01 |
| GO:BP | GO:0060032 | notochord regression | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 15 | 277 | 2.45e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 2 | 125 | 2.45e-01 |
| GO:BP | GO:0040007 | growth | 40 | 742 | 2.46e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 17 | 249 | 2.46e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 1 | 14 | 2.46e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 3 | 28 | 2.46e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 3 | 72 | 2.46e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 2.46e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 2 | 16 | 2.46e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 7 | 179 | 2.46e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 2.46e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 1 | 7 | 2.46e-01 |
| GO:BP | GO:0006417 | regulation of translation | 2 | 370 | 2.46e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 7 | 2.46e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 19 | 294 | 2.46e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 1 | 121 | 2.46e-01 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 1 | 5 | 2.47e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 1 | 4 | 2.47e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 6 | 67 | 2.47e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 2.47e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 14 | 256 | 2.47e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 1 | 4 | 2.47e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 3 | 28 | 2.47e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 2.47e-01 |
| GO:BP | GO:0051103 | DNA ligation involved in DNA repair | 1 | 5 | 2.47e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 1 | 4 | 2.47e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 2.47e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 2 | 318 | 2.47e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 3 | 26 | 2.47e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 2 | 12 | 2.47e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 2.47e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 1 | 2 | 2.47e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 2.47e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 1 | 3 | 2.47e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 2.47e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 4 | 108 | 2.47e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 901 | 2.47e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 1 | 82 | 2.47e-01 |
| GO:BP | GO:0043201 | response to leucine | 2 | 13 | 2.47e-01 |
| GO:BP | GO:0007498 | mesoderm development | 8 | 89 | 2.47e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 2.47e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 2.47e-01 |
| GO:BP | GO:0044029 | positive regulation of gene expression via CpG island demethylation | 1 | 3 | 2.48e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 2.48e-01 |
| GO:BP | GO:0071464 | cellular response to hydrostatic pressure | 1 | 3 | 2.48e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 1 | 26 | 2.48e-01 |
| GO:BP | GO:0070075 | tear secretion | 1 | 2 | 2.48e-01 |
| GO:BP | GO:0061171 | establishment of bipolar cell polarity | 1 | 1 | 2.48e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 2.48e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 42 | 2.48e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 4 | 51 | 2.48e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 2.48e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 3 | 36 | 2.48e-01 |
| GO:BP | GO:0001966 | thigmotaxis | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly | 1 | 7 | 2.49e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 1 | 23 | 2.49e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 1 | 23 | 2.49e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 1 | 8 | 2.49e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 1 | 11 | 2.49e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 3 | 165 | 2.49e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 1 | 23 | 2.49e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 10 | 160 | 2.49e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 336 | 2.49e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 8 | 742 | 2.50e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 1 | 61 | 2.50e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 12 | 216 | 2.50e-01 |
| GO:BP | GO:0007610 | behavior | 2 | 436 | 2.50e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 2 | 16 | 2.50e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 11 | 153 | 2.50e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 5 | 59 | 2.50e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 1 | 14 | 2.50e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 97 | 2.50e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 6 | 77 | 2.50e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 8 | 1147 | 2.50e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 1 | 796 | 2.50e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 3 | 27 | 2.50e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 2.50e-01 |
| GO:BP | GO:0098534 | centriole assembly | 2 | 41 | 2.50e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 2.50e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 1 | 14 | 2.50e-01 |
| GO:BP | GO:0044409 | entry into host | 2 | 113 | 2.50e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 2 | 512 | 2.50e-01 |
| GO:BP | GO:0061061 | muscle structure development | 30 | 546 | 2.50e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 2 | 3090 | 2.50e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 2 | 30 | 2.51e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 2 | 17 | 2.51e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 2 | 7 | 2.51e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 2 | 43 | 2.51e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 2 | 11 | 2.52e-01 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 1 | 8 | 2.52e-01 |
| GO:BP | GO:1905458 | positive regulation of lymphoid progenitor cell differentiation | 1 | 3 | 2.52e-01 |
| GO:BP | GO:0002572 | pro-T cell differentiation | 1 | 1 | 2.52e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 18 | 598 | 2.52e-01 |
| GO:BP | GO:0015884 | folic acid transport | 1 | 7 | 2.53e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 3 | 52 | 2.53e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 3 | 73 | 2.53e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 1 | 5 | 2.53e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 2 | 91 | 2.53e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 2.53e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 2.53e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 2.53e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 4 | 49 | 2.53e-01 |
| GO:BP | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process | 1 | 4 | 2.53e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 1 | 5 | 2.53e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 3 | 4128 | 2.53e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 1 | 382 | 2.53e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 2.53e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 1 | 96 | 2.53e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 2 | 516 | 2.54e-01 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 2 | 44 | 2.54e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 1 | 64 | 2.55e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 1 | 16 | 2.56e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 4 | 137 | 2.56e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 11 | 178 | 2.56e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 2 | 17 | 2.56e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 2 | 527 | 2.56e-01 |
| GO:BP | GO:0016076 | snRNA catabolic process | 1 | 5 | 2.56e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 2.56e-01 |
| GO:BP | GO:0003015 | heart process | 7 | 216 | 2.56e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 1 | 2 | 2.56e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 2.56e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 4 | 80 | 2.57e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 1 | 5 | 2.57e-01 |
| GO:BP | GO:0030910 | olfactory placode formation | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 2 | 531 | 2.57e-01 |
| GO:BP | GO:0071698 | olfactory placode development | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0071699 | olfactory placode morphogenesis | 1 | 2 | 2.57e-01 |
| GO:BP | GO:1904252 | negative regulation of bile acid metabolic process | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0055005 | ventricular cardiac myofibril assembly | 1 | 3 | 2.57e-01 |
| GO:BP | GO:0021675 | nerve development | 3 | 57 | 2.57e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 3 | 20 | 2.57e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 2 | 94 | 2.57e-01 |
| GO:BP | GO:0060838 | lymphatic endothelial cell fate commitment | 1 | 3 | 2.57e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 2 | 60 | 2.57e-01 |
| GO:BP | GO:0070858 | negative regulation of bile acid biosynthetic process | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 2.57e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 2 | 94 | 2.57e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 1 | 15 | 2.57e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 2.57e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 1 | 15 | 2.57e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 4 | 51 | 2.57e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 2 | 9 | 2.57e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 5 | 76 | 2.57e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 2.57e-01 |
| GO:BP | GO:0031103 | axon regeneration | 2 | 42 | 2.57e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 2 | 22 | 2.57e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 1 | 34 | 2.57e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 4 | 154 | 2.58e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 1 | 8 | 2.58e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 1 | 8 | 2.58e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 1 | 6 | 2.58e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 3 | 74 | 2.58e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 1 | 6 | 2.58e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 13 | 127 | 2.58e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 3 | 111 | 2.58e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 2 | 372 | 2.58e-01 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 1 | 3 | 2.58e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 2.59e-01 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 1 | 5 | 2.59e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 1 | 29 | 2.59e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 2.60e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 4 | 49 | 2.60e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 1 | 496 | 2.60e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 8 | 104 | 2.60e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 1 | 881 | 2.60e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 1 | 12 | 2.60e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 2.60e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 2.61e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 1 | 71 | 2.61e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 2 | 14 | 2.61e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 2.61e-01 |
| GO:BP | GO:0031529 | ruffle organization | 2 | 45 | 2.61e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 15 | 1974 | 2.61e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 1 | 3 | 2.61e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 3 | 111 | 2.61e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 7 | 99 | 2.61e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 1 | 19 | 2.61e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 2 | 17 | 2.61e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 4 | 57 | 2.61e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 3 | 27 | 2.61e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 4 | 157 | 2.61e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 2 | 551 | 2.61e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 2 | 47 | 2.61e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 1 | 28 | 2.61e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 1 | 28 | 2.61e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 1 | 3 | 2.61e-01 |
| GO:BP | GO:0051030 | snRNA transport | 1 | 6 | 2.61e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 1 | 6 | 2.61e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 1 | 3 | 2.61e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 3 | 27 | 2.61e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 4 | 40 | 2.61e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 27 | 2.61e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 8 | 117 | 2.62e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 1 | 4 | 2.62e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 4 | 135 | 2.62e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 1 | 9 | 2.62e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 418 | 2.62e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 2 | 34 | 2.62e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 2.62e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 1 | 6 | 2.62e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 7 | 150 | 2.62e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 110 | 2.62e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 3 | 46 | 2.62e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 10 | 161 | 2.63e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 6 | 134 | 2.63e-01 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 1 | 6 | 2.63e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 2 | 17 | 2.64e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 1 | 21 | 2.64e-01 |
| GO:BP | GO:1904582 | positive regulation of intracellular mRNA localization | 1 | 3 | 2.64e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 2 | 53 | 2.64e-01 |
| GO:BP | GO:0031175 | neuron projection development | 3 | 780 | 2.64e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 2 | 186 | 2.65e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 4 | 165 | 2.65e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 1 | 9 | 2.65e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 1 | 15 | 2.65e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 2.67e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 1 | 5 | 2.67e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 2.67e-01 |
| GO:BP | GO:0042221 | response to chemical | 2 | 2559 | 2.67e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 4 | 166 | 2.67e-01 |
| GO:BP | GO:0014004 | microglia differentiation | 1 | 6 | 2.67e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 15 | 227 | 2.67e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 1 | 13 | 2.67e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 1 | 6 | 2.67e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 2 | 78 | 2.67e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 1 | 5 | 2.67e-01 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 1 | 12 | 2.67e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 2.68e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 13 | 2.68e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 4 | 31 | 2.68e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 4 | 1248 | 2.68e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 2.68e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 2.68e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 2.68e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 4 | 134 | 2.68e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 2 | 14 | 2.68e-01 |
| GO:BP | GO:0019344 | cysteine biosynthetic process | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 2 | 24 | 2.68e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 3 | 120 | 2.68e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 2 | 22 | 2.68e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 5 | 295 | 2.68e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 2 | 424 | 2.68e-01 |
| GO:BP | GO:0007420 | brain development | 2 | 591 | 2.68e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 3 | 756 | 2.68e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 1 | 4 | 2.68e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 1 | 2 | 2.68e-01 |
| GO:BP | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | 3 | 2.68e-01 |
| GO:BP | GO:1900114 | positive regulation of histone H3-K9 trimethylation | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 3 | 29 | 2.69e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 2 | 55 | 2.69e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 1 | 14 | 2.69e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 14 | 262 | 2.69e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 2.69e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 2.69e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 3 | 2.69e-01 |
| GO:BP | GO:0019080 | viral gene expression | 6 | 96 | 2.69e-01 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 2.69e-01 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 1 | 3 | 2.69e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 7 | 1088 | 2.69e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 3 | 54 | 2.70e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 1 | 19 | 2.70e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0006096 | glycolytic process | 5 | 70 | 2.71e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 2 | 22 | 2.71e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 1 | 5 | 2.71e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 1 | 15 | 2.71e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 1 | 78 | 2.71e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 2 | 52 | 2.72e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 1 | 8 | 2.72e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 1 | 994 | 2.72e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 1 | 993 | 2.72e-01 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 2 | 17 | 2.72e-01 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 2 | 17 | 2.72e-01 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 3 | 50 | 2.72e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 1 | 16 | 2.72e-01 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 2 | 17 | 2.72e-01 |
| GO:BP | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway | 1 | 9 | 2.72e-01 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 1 | 3 | 2.72e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 2.72e-01 |
| GO:BP | GO:0044154 | histone H3-K14 acetylation | 1 | 12 | 2.72e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 1 | 22 | 2.72e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 1 | 10 | 2.73e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 1 | 9 | 2.73e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 2.73e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 2.74e-01 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 2 | 30 | 2.74e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 2 | 17 | 2.74e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 10 | 159 | 2.74e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 3 | 785 | 2.74e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 3 | 357 | 2.74e-01 |
| GO:BP | GO:1900245 | positive regulation of MDA-5 signaling pathway | 1 | 2 | 2.74e-01 |
| GO:BP | GO:0035441 | cell migration involved in vasculogenesis | 1 | 4 | 2.74e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 2.74e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 2 | 63 | 2.74e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 6 | 296 | 2.74e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 1 | 78 | 2.74e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 2 | 72 | 2.74e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 13 | 381 | 2.75e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 8 | 465 | 2.75e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 8 | 122 | 2.75e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 2.75e-01 |
| GO:BP | GO:0009725 | response to hormone | 6 | 642 | 2.75e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 2.75e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 9 | 234 | 2.76e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 2 | 19 | 2.76e-01 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 1 | 5 | 2.76e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 2.76e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 1 | 19 | 2.76e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 1 | 15 | 2.76e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 1 | 14 | 2.76e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 2.76e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 2 | 45 | 2.76e-01 |
| GO:BP | GO:0051216 | cartilage development | 2 | 151 | 2.77e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 2.77e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 2 | 15 | 2.78e-01 |
| GO:BP | GO:0061038 | uterus morphogenesis | 1 | 5 | 2.78e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 1 | 16 | 2.78e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 3 | 4579 | 2.78e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 3 | 56 | 2.78e-01 |
| GO:BP | GO:0001503 | ossification | 3 | 313 | 2.79e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 3 | 70 | 2.79e-01 |
| GO:BP | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | 1 | 5 | 2.79e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 2 | 75 | 2.79e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 12 | 358 | 2.80e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 2 | 1974 | 2.80e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0044000 | movement in host | 2 | 138 | 2.81e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 1 | 560 | 2.81e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 7 | 1111 | 2.82e-01 |
| GO:BP | GO:0030282 | bone mineralization | 6 | 82 | 2.82e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 1 | 24 | 2.82e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 1 | 24 | 2.82e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 1 | 21 | 2.82e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 2.82e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 1 | 23 | 2.82e-01 |
| GO:BP | GO:0060322 | head development | 2 | 633 | 2.82e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 1 | 37 | 2.82e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 2.82e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 2.82e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 2.82e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 1 | 35 | 2.82e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 3 | 27 | 2.82e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 2 | 124 | 2.82e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 2 | 16 | 2.82e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 2.83e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 2.83e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 2.83e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 2.83e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 13 | 213 | 2.84e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 2 | 420 | 2.84e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 1 | 6 | 2.84e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 2 | 21 | 2.84e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 2 | 11 | 2.84e-01 |
| GO:BP | GO:1990840 | response to lectin | 2 | 11 | 2.84e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 1 | 6 | 2.84e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 1 | 6 | 2.84e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 2 | 11 | 2.84e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 1 | 6 | 2.84e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 2 | 47 | 2.84e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 1 | 12 | 2.85e-01 |
| GO:BP | GO:0016255 | attachment of GPI anchor to protein | 1 | 6 | 2.85e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 1 | 120 | 2.86e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2 | 39 | 2.86e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 2.86e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 3 | 146 | 2.87e-01 |
| GO:BP | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification | 1 | 3 | 2.87e-01 |
| GO:BP | GO:0021508 | floor plate formation | 1 | 3 | 2.87e-01 |
| GO:BP | GO:0021776 | smoothened signaling pathway involved in spinal cord motor neuron cell fate specification | 1 | 3 | 2.87e-01 |
| GO:BP | GO:0021965 | spinal cord ventral commissure morphogenesis | 1 | 2 | 2.87e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 2 | 37 | 2.87e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 3 | 33 | 2.87e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 6 | 56 | 2.87e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 2 | 439 | 2.87e-01 |
| GO:BP | GO:0031099 | regeneration | 6 | 142 | 2.87e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 3 | 468 | 2.87e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 7 | 123 | 2.87e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 1 | 4 | 2.87e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 1 | 23 | 2.87e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 1 | 8 | 2.87e-01 |
| GO:BP | GO:0032502 | developmental process | 3 | 4494 | 2.87e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 2 | 15 | 2.88e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 2 | 14 | 2.88e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 3 | 20 | 2.88e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 2 | 28 | 2.88e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 2.88e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 6 | 67 | 2.88e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 4 | 2.88e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 1 | 22 | 2.88e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 2.89e-01 |
| GO:BP | GO:0016477 | cell migration | 1 | 1081 | 2.89e-01 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 1 | 4 | 2.89e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 9 | 554 | 2.89e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 1 | 16 | 2.89e-01 |
| GO:BP | GO:0061113 | pancreas morphogenesis | 1 | 3 | 2.89e-01 |
| GO:BP | GO:0010165 | response to X-ray | 2 | 29 | 2.89e-01 |
| GO:BP | GO:0060214 | endocardium formation | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0060298 | positive regulation of sarcomere organization | 1 | 4 | 2.89e-01 |
| GO:BP | GO:0021603 | cranial nerve formation | 1 | 4 | 2.89e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 1 | 16 | 2.89e-01 |
| GO:BP | GO:0034644 | cellular response to UV | 5 | 86 | 2.89e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 1 | 22 | 2.89e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 1 | 11 | 2.89e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 13 | 6996 | 2.89e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 2 | 25 | 2.89e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 2.89e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 3 | 25 | 2.89e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 3 | 20 | 2.89e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 3 | 32 | 2.90e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 4 | 68 | 2.90e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 5 | 561 | 2.90e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 1 | 3 | 2.90e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 135 | 2.90e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 2 | 71 | 2.90e-01 |
| GO:BP | GO:0001525 | angiogenesis | 1 | 390 | 2.90e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 2 | 639 | 2.91e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 2.92e-01 |
| GO:BP | GO:0090310 | negative regulation of DNA methylation-dependent heterochromatin formation | 1 | 2 | 2.92e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 1 | 36 | 2.92e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 2 | 25 | 2.93e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 6 | 70 | 2.93e-01 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 1 | 11 | 2.94e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 7 | 101 | 2.94e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 2 | 54 | 2.94e-01 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 3 | 36 | 2.94e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 2.95e-01 |
| GO:BP | GO:0065001 | specification of axis polarity | 1 | 2 | 2.95e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 2.95e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 1 | 17 | 2.95e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 2 | 12 | 2.95e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 2.95e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 1 | 9 | 2.96e-01 |
| GO:BP | GO:0048864 | stem cell development | 6 | 76 | 2.96e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 2 | 15 | 2.96e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 2 | 15 | 2.96e-01 |
| GO:BP | GO:0030850 | prostate gland development | 1 | 35 | 2.96e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 2 | 8 | 2.96e-01 |
| GO:BP | GO:0006301 | postreplication repair | 3 | 34 | 2.96e-01 |
| GO:BP | GO:0051707 | response to other organism | 1 | 820 | 2.96e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 1 | 820 | 2.97e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 2.97e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 5 | 53 | 2.97e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 1 | 9 | 2.97e-01 |
| GO:BP | GO:0032940 | secretion by cell | 1 | 569 | 2.98e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 2 | 192 | 2.98e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 2 | 23 | 2.98e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 2 | 17 | 2.98e-01 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 1 | 9 | 2.98e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 1 | 540 | 2.98e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 2 | 12 | 2.98e-01 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 1 | 4 | 2.98e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 1 | 14 | 2.98e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 1 | 21 | 2.98e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 1 | 22 | 2.98e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 1 | 5 | 2.98e-01 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 1 | 6 | 2.98e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 1 | 7 | 2.98e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 2 | 39 | 2.98e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 3 | 31 | 2.99e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 2 | 46 | 2.99e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 1 | 16 | 2.99e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 1 | 8 | 2.99e-01 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 11 | 2.99e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 7 | 763 | 2.99e-01 |
| GO:BP | GO:1904580 | regulation of intracellular mRNA localization | 1 | 4 | 2.99e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 1 | 5 | 2.99e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 2 | 17 | 2.99e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 2 | 21 | 2.99e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 1 | 850 | 2.99e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 3 | 665 | 3.00e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 1 | 6 | 3.00e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 1 | 20 | 3.00e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 3.00e-01 |
| GO:BP | GO:0048625 | myoblast fate commitment | 1 | 4 | 3.00e-01 |
| GO:BP | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process | 1 | 4 | 3.00e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 1 | 26 | 3.00e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 1 | 24 | 3.00e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 1 | 38 | 3.00e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 4 | 54 | 3.00e-01 |
| GO:BP | GO:0032815 | negative regulation of natural killer cell activation | 1 | 3 | 3.00e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 1 | 10 | 3.00e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 3.00e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 3 | 906 | 3.00e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 2 | 64 | 3.00e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 2 | 18 | 3.00e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 3.01e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 3.01e-01 |
| GO:BP | GO:0090244 | Wnt signaling pathway involved in somitogenesis | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 3 | 126 | 3.02e-01 |
| GO:BP | GO:0070509 | calcium ion import | 2 | 37 | 3.02e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 2 | 105 | 3.02e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 9 | 151 | 3.02e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 2 | 53 | 3.02e-01 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 1 | 6 | 3.02e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 4 | 52 | 3.02e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 1 | 142 | 3.02e-01 |
| GO:BP | GO:0045905 | positive regulation of translational termination | 1 | 4 | 3.02e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 1 | 6 | 3.02e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 7 | 3.02e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 1 | 5 | 3.03e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 3.03e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 2 | 22 | 3.04e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 1 | 4 | 3.04e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 2 | 83 | 3.04e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 1 | 4 | 3.04e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 3.04e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 1 | 17 | 3.04e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 1 | 15 | 3.04e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 8 | 3.04e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 1 | 41 | 3.04e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 6 | 101 | 3.04e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 2 | 23 | 3.04e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 3.04e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 2 | 59 | 3.04e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 2 | 23 | 3.04e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 3 | 117 | 3.04e-01 |
| GO:BP | GO:1905643 | positive regulation of DNA methylation | 1 | 2 | 3.04e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 1 | 7 | 3.04e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 5 | 152 | 3.05e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 1 | 8 | 3.05e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 11 | 3.05e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 1 | 10 | 3.05e-01 |
| GO:BP | GO:0048630 | skeletal muscle tissue growth | 1 | 8 | 3.05e-01 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 1 | 2 | 3.05e-01 |
| GO:BP | GO:2000653 | regulation of genetic imprinting | 1 | 4 | 3.05e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 6 | 94 | 3.06e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 2 | 16 | 3.06e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 13 | 203 | 3.06e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 3.06e-01 |
| GO:BP | GO:0048870 | cell motility | 1 | 1182 | 3.06e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 1 | 9 | 3.06e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 3.06e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 3.06e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 3 | 129 | 3.06e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 3.06e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 3.06e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 3 | 110 | 3.06e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 3.06e-01 |
| GO:BP | GO:0140352 | export from cell | 1 | 611 | 3.06e-01 |
| GO:BP | GO:0003129 | heart induction | 1 | 9 | 3.06e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 2 | 28 | 3.06e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 4 | 348 | 3.06e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 1 | 8 | 3.06e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 5 | 3.06e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 3.06e-01 |
| GO:BP | GO:0048666 | neuron development | 3 | 877 | 3.06e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 2 | 154 | 3.06e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 2 | 427 | 3.06e-01 |
| GO:BP | GO:0060039 | pericardium development | 2 | 17 | 3.06e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 1 | 14 | 3.09e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 1 | 12 | 3.09e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 1 | 105 | 3.09e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 1 | 963 | 3.10e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 3.10e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 5 | 206 | 3.10e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 1 | 103 | 3.10e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 1 | 5 | 3.10e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 4 | 112 | 3.10e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 1 | 13 | 3.11e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 13 | 3.11e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 2 | 53 | 3.11e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 6 | 121 | 3.12e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 7 | 106 | 3.12e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 1 | 24 | 3.12e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 3.12e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 3 | 43 | 3.12e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 2 | 62 | 3.12e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0045117 | azole transmembrane transport | 1 | 9 | 3.12e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 4 | 172 | 3.12e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 3.12e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 3.12e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 4 | 113 | 3.12e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 1 | 9 | 3.12e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 5 | 257 | 3.12e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 3.12e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 4 | 52 | 3.12e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 1 | 667 | 3.12e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 3.12e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 6 | 149 | 3.12e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 3.12e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 1 | 105 | 3.12e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 3 | 783 | 3.12e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 1 | 11 | 3.13e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 3 | 228 | 3.13e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 9 | 149 | 3.14e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 1 | 23 | 3.14e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 1 | 15 | 3.14e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 3.14e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 3.14e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 1 | 16 | 3.15e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 1 | 108 | 3.17e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 2 | 55 | 3.17e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 3.17e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 1 | 10 | 3.17e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 1 | 6 | 3.17e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 3.17e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 2 | 55 | 3.17e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 2 | 25 | 3.17e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 2 | 22 | 3.17e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 4 | 422 | 3.17e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 13 | 229 | 3.17e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 2 | 55 | 3.17e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 3.17e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 1 | 1380 | 3.17e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 1 | 5 | 3.17e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 1 | 10 | 3.17e-01 |
| GO:BP | GO:0046903 | secretion | 1 | 645 | 3.17e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 8 | 144 | 3.17e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 2 | 67 | 3.17e-01 |
| GO:BP | GO:0016180 | snRNA processing | 1 | 26 | 3.17e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 3.17e-01 |
| GO:BP | GO:0050658 | RNA transport | 8 | 144 | 3.17e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 2 | 55 | 3.18e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 1 | 5 | 3.18e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 1 | 7 | 3.18e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 6 | 631 | 3.18e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 1 | 6 | 3.18e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 2 | 14 | 3.18e-01 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 3.18e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 1 | 110 | 3.18e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 1 | 5 | 3.18e-01 |
| GO:BP | GO:0016571 | histone methylation | 8 | 117 | 3.18e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 1 | 10 | 3.18e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 2 | 23 | 3.19e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 2 | 23 | 3.19e-01 |
| GO:BP | GO:0035188 | hatching | 2 | 23 | 3.19e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 5 | 78 | 3.20e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 1 | 26 | 3.20e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 1 | 6 | 3.21e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 3.21e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 3 | 34 | 3.21e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 152 | 3.21e-01 |
| GO:BP | GO:0016049 | cell growth | 3 | 418 | 3.21e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 3.21e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 3 | 69 | 3.21e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 1 | 4 | 3.22e-01 |
| GO:BP | GO:0043583 | ear development | 9 | 153 | 3.22e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 2 | 18 | 3.22e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 1 | 31 | 3.23e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 1 | 29 | 3.23e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 3 | 36 | 3.23e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 1 | 4 | 3.24e-01 |
| GO:BP | GO:0006304 | DNA modification | 2 | 89 | 3.25e-01 |
| GO:BP | GO:0032528 | microvillus organization | 1 | 19 | 3.25e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 14 | 187 | 3.26e-01 |
| GO:BP | GO:0033505 | floor plate morphogenesis | 1 | 3 | 3.26e-01 |
| GO:BP | GO:0030902 | hindbrain development | 2 | 114 | 3.26e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 2 | 17 | 3.26e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 3.26e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 4 | 161 | 3.26e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 2 | 46 | 3.26e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 1 | 2 | 3.26e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 2 | 18 | 3.26e-01 |
| GO:BP | GO:0090398 | cellular senescence | 6 | 83 | 3.26e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 2 | 80 | 3.26e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 4 | 185 | 3.26e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 1 | 26 | 3.28e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 9 | 140 | 3.28e-01 |
| GO:BP | GO:0090118 | receptor-mediated endocytosis involved in cholesterol transport | 1 | 5 | 3.28e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 1 | 1681 | 3.28e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 2 | 16 | 3.28e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 3.28e-01 |
| GO:BP | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation | 1 | 5 | 3.28e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 20 | 3.28e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 3 | 21 | 3.28e-01 |
| GO:BP | GO:0032200 | telomere organization | 1 | 166 | 3.28e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 1 | 19 | 3.28e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 4 | 199 | 3.28e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 3.28e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 3.28e-01 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 2 | 18 | 3.28e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 1 | 10 | 3.28e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 3.28e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 1 | 13 | 3.28e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 1 | 19 | 3.28e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 3 | 42 | 3.28e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 1 | 17 | 3.29e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 3.29e-01 |
| GO:BP | GO:0002076 | osteoblast development | 2 | 12 | 3.29e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 3.29e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 1 | 9 | 3.30e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 3.30e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 3.30e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 2 | 80 | 3.30e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 8 | 147 | 3.30e-01 |
| GO:BP | GO:0071605 | monocyte chemotactic protein-1 production | 1 | 7 | 3.30e-01 |
| GO:BP | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 1 | 17 | 3.30e-01 |
| GO:BP | GO:0071637 | regulation of monocyte chemotactic protein-1 production | 1 | 7 | 3.30e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 2 | 22 | 3.30e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 1 | 168 | 3.30e-01 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 3.30e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 1 | 1378 | 3.30e-01 |
| GO:BP | GO:0060174 | limb bud formation | 1 | 10 | 3.30e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 3.30e-01 |
| GO:BP | GO:0007276 | gamete generation | 14 | 461 | 3.30e-01 |
| GO:BP | GO:0007369 | gastrulation | 2 | 146 | 3.30e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 3.30e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 1 | 10 | 3.31e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 3.31e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 3.31e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 3.31e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 11 | 330 | 3.31e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 3.31e-01 |
| GO:BP | GO:0051890 | regulation of cardioblast differentiation | 1 | 5 | 3.31e-01 |
| GO:BP | GO:0060462 | lung lobe development | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0060463 | lung lobe morphogenesis | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 1 | 6 | 3.31e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 1 | 167 | 3.32e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 3.32e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 1 | 6 | 3.32e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 2 | 26 | 3.32e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 3.32e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 3 | 38 | 3.32e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 2 | 22 | 3.32e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 1 | 782 | 3.33e-01 |
| GO:BP | GO:0060047 | heart contraction | 6 | 205 | 3.33e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 2 | 97 | 3.34e-01 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 1 | 29 | 3.34e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 4 | 47 | 3.34e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 3.34e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 1 | 40 | 3.34e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 2 | 7 | 3.34e-01 |
| GO:BP | GO:0019346 | transsulfuration | 1 | 5 | 3.34e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 2 | 18 | 3.34e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 3 | 41 | 3.34e-01 |
| GO:BP | GO:0009092 | homoserine metabolic process | 1 | 5 | 3.34e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 7 | 421 | 3.34e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 1 | 7 | 3.34e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 3 | 27 | 3.34e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 9 | 140 | 3.34e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 1 | 5 | 3.34e-01 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 1 | 18 | 3.34e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 7 | 719 | 3.34e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 18 | 326 | 3.34e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 7 | 4045 | 3.34e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 3.35e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 3.35e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 3.35e-01 |
| GO:BP | GO:0110154 | RNA decapping | 1 | 17 | 3.35e-01 |
| GO:BP | GO:0009749 | response to glucose | 9 | 137 | 3.36e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 1 | 68 | 3.36e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 1 | 5 | 3.36e-01 |
| GO:BP | GO:0019230 | proprioception | 1 | 3 | 3.36e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 1 | 23 | 3.36e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 1 | 7 | 3.37e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 1 | 13 | 3.37e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 1 | 6 | 3.37e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 7 | 1219 | 3.37e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 3 | 141 | 3.37e-01 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 1 | 8 | 3.37e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 1 | 20 | 3.37e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 1 | 9 | 3.37e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 1 | 9 | 3.37e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 1 | 9 | 3.37e-01 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 1 | 8 | 3.37e-01 |
| GO:BP | GO:0007417 | central nervous system development | 1 | 792 | 3.37e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 2 | 27 | 3.38e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 3.38e-01 |
| GO:BP | GO:0051098 | regulation of binding | 3 | 317 | 3.38e-01 |
| GO:BP | GO:0003188 | heart valve formation | 1 | 16 | 3.38e-01 |
| GO:BP | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation | 1 | 6 | 3.38e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 2 | 74 | 3.38e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 3 | 60 | 3.39e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 4 | 393 | 3.39e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 1 | 10 | 3.39e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 4 | 190 | 3.39e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 7 | 963 | 3.39e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 2 | 529 | 3.39e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 1 | 38 | 3.40e-01 |
| GO:BP | GO:0006621 | protein retention in ER lumen | 1 | 8 | 3.40e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 1 | 28 | 3.40e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 7 | 110 | 3.41e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 6 | 98 | 3.41e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 3 | 50 | 3.41e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 3.41e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 2 | 660 | 3.41e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 3.41e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 7 | 149 | 3.42e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 2 | 44 | 3.42e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 1 | 11 | 3.42e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 2 | 23 | 3.42e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 6 | 156 | 3.43e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 2 | 64 | 3.43e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 3.43e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 3.43e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 3.43e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 1 | 21 | 3.43e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 189 | 3.43e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 1 | 18 | 3.43e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 1 | 30 | 3.44e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 7 | 107 | 3.44e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 3.44e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 1 | 830 | 3.44e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 2 | 209 | 3.44e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 3.44e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 2 | 18 | 3.44e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 3.44e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 3.44e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 3 | 43 | 3.44e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 8 | 156 | 3.44e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 7 | 120 | 3.44e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 2 | 4241 | 3.44e-01 |
| GO:BP | GO:0010332 | response to gamma radiation | 4 | 50 | 3.44e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 1 | 17 | 3.44e-01 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 1 | 8 | 3.44e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 7 | 84 | 3.44e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 2 | 16 | 3.44e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 1 | 5 | 3.45e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 1 | 6 | 3.45e-01 |
| GO:BP | GO:0055009 | atrial cardiac muscle tissue morphogenesis | 1 | 5 | 3.45e-01 |
| GO:BP | GO:0003160 | endocardium morphogenesis | 1 | 3 | 3.45e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 19 | 418 | 3.45e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 1 | 5 | 3.45e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 2 | 541 | 3.45e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 25 | 1107 | 3.45e-01 |
| GO:BP | GO:0019985 | translesion synthesis | 2 | 24 | 3.45e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 4 | 58 | 3.45e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 1 | 7 | 3.45e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 1 | 7 | 3.45e-01 |
| GO:BP | GO:0061448 | connective tissue development | 2 | 208 | 3.46e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 3.46e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 4 | 717 | 3.46e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 12 | 378 | 3.46e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 1 | 5 | 3.46e-01 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 4 | 186 | 3.46e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 2 | 678 | 3.46e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 1 | 184 | 3.46e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 1 | 37 | 3.46e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 1 | 126 | 3.46e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 1 | 5 | 3.46e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 3.46e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 1 | 15 | 3.46e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 4 | 121 | 3.46e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 3 | 80 | 3.46e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 11 | 155 | 3.46e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 6 | 348 | 3.46e-01 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 1 | 30 | 3.46e-01 |
| GO:BP | GO:0048703 | embryonic viscerocranium morphogenesis | 1 | 8 | 3.46e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 1 | 8 | 3.46e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 12 | 3.46e-01 |
| GO:BP | GO:0001501 | skeletal system development | 15 | 377 | 3.47e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 3.47e-01 |
| GO:BP | GO:1905268 | negative regulation of chromatin organization | 1 | 4 | 3.47e-01 |
| GO:BP | GO:0031452 | negative regulation of heterochromatin formation | 1 | 4 | 3.47e-01 |
| GO:BP | GO:0120262 | negative regulation of heterochromatin organization | 1 | 4 | 3.47e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 1 | 31 | 3.48e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 1 | 12 | 3.48e-01 |
| GO:BP | GO:0060384 | innervation | 1 | 22 | 3.48e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 3 | 62 | 3.48e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 3 | 85 | 3.49e-01 |
| GO:BP | GO:0032682 | negative regulation of chemokine production | 1 | 12 | 3.49e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 3 | 36 | 3.49e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 13 | 204 | 3.49e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 1 | 127 | 3.49e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 3.49e-01 |
| GO:BP | GO:0039533 | regulation of MDA-5 signaling pathway | 1 | 5 | 3.49e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 1 | 22 | 3.49e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 1 | 187 | 3.49e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 5 | 57 | 3.49e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 3 | 324 | 3.49e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 3.49e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 4 | 399 | 3.50e-01 |
| GO:BP | GO:0010387 | COP9 signalosome assembly | 1 | 3 | 3.50e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 6 | 89 | 3.50e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 2 | 22 | 3.50e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 2 | 20 | 3.51e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 8 | 147 | 3.51e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 1 | 22 | 3.51e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 2 | 137 | 3.51e-01 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 1 | 30 | 3.51e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 17 | 3.51e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 3.51e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 1 | 7 | 3.51e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 7 | 133 | 3.51e-01 |
| GO:BP | GO:0009746 | response to hexose | 9 | 138 | 3.51e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 3.51e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 2 | 68 | 3.52e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 1 | 7 | 3.52e-01 |
| GO:BP | GO:0051867 | general adaptation syndrome, behavioral process | 1 | 3 | 3.52e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 3.52e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 13 | 406 | 3.52e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 3.52e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 1 | 6 | 3.53e-01 |
| GO:BP | GO:0048232 | male gamete generation | 11 | 343 | 3.53e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 10 | 185 | 3.53e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 3 | 22 | 3.53e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 2 | 31 | 3.53e-01 |
| GO:BP | GO:0021537 | telencephalon development | 6 | 201 | 3.53e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 3.53e-01 |
| GO:BP | GO:0000725 | recombinational repair | 7 | 153 | 3.53e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 12 | 3.53e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 3.53e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 3.53e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 1 | 7 | 3.53e-01 |
| GO:BP | GO:0090102 | cochlea development | 1 | 35 | 3.53e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 3.54e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 2 | 100 | 3.55e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 1 | 5 | 3.55e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 1 | 16 | 3.56e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 5 | 249 | 3.56e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 1 | 200 | 3.56e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 2 | 18 | 3.56e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 2 | 56 | 3.56e-01 |
| GO:BP | GO:0006298 | mismatch repair | 1 | 29 | 3.56e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 3.56e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 1 | 134 | 3.56e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 3.56e-01 |
| GO:BP | GO:2000674 | regulation of type B pancreatic cell apoptotic process | 1 | 6 | 3.57e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 3.57e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 4 | 53 | 3.57e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 5 | 89 | 3.57e-01 |
| GO:BP | GO:1990787 | negative regulation of hh target transcription factor activity | 1 | 4 | 3.57e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 3 | 75 | 3.57e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 3.57e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 2 | 35 | 3.57e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 1 | 19 | 3.57e-01 |
| GO:BP | GO:0036260 | RNA capping | 2 | 20 | 3.58e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 5 | 86 | 3.58e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 4 | 1075 | 3.58e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 2 | 14 | 3.59e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 3.59e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 1 | 6 | 3.59e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 1 | 19 | 3.59e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 1 | 11 | 3.59e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 1 | 6 | 3.59e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 1 | 11 | 3.59e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 3.59e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 3.59e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 2 | 20 | 3.59e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 3.59e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 8 | 130 | 3.60e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 3.61e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 4 | 71 | 3.61e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 1 | 39 | 3.61e-01 |
| GO:BP | GO:0006482 | protein demethylation | 2 | 27 | 3.61e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 2 | 27 | 3.61e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 1 | 151 | 3.61e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 3 | 192 | 3.61e-01 |
| GO:BP | GO:1905065 | positive regulation of vascular associated smooth muscle cell differentiation | 1 | 6 | 3.62e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 9 | 158 | 3.62e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 1 | 18 | 3.62e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 1 | 11 | 3.62e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 8 | 143 | 3.62e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 9 | 155 | 3.62e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 3.62e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 3 | 35 | 3.63e-01 |
| GO:BP | GO:1900112 | regulation of histone H3-K9 trimethylation | 1 | 5 | 3.63e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 3.63e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 3.64e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 1 | 29 | 3.64e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 7 | 89 | 3.64e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 7 | 89 | 3.64e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 5 | 2011 | 3.64e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 2 | 27 | 3.64e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 2 | 41 | 3.64e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 1 | 11 | 3.66e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 1 | 11 | 3.66e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 1 | 151 | 3.66e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 3.66e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 3 | 1028 | 3.66e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 6 | 112 | 3.66e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 5 | 68 | 3.66e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 3 | 4794 | 3.66e-01 |
| GO:BP | GO:0010917 | negative regulation of mitochondrial membrane potential | 1 | 9 | 3.66e-01 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 1 | 5 | 3.66e-01 |
| GO:BP | GO:0045837 | negative regulation of membrane potential | 1 | 9 | 3.66e-01 |
| GO:BP | GO:0001706 | endoderm formation | 1 | 46 | 3.66e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 1 | 14 | 3.66e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 3 | 45 | 3.67e-01 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 2 | 19 | 3.67e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 3.67e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 1 | 20 | 3.67e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 1 | 20 | 3.67e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 3.67e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 3.67e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 3.67e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 3 | 132 | 3.68e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 1 | 31 | 3.69e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 2 | 22 | 3.69e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 4 | 104 | 3.69e-01 |
| GO:BP | GO:0000012 | single strand break repair | 1 | 10 | 3.69e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 3.69e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 1 | 19 | 3.69e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 1 | 7 | 3.69e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 3.69e-01 |
| GO:BP | GO:0035461 | vitamin transmembrane transport | 1 | 14 | 3.69e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 39 | 787 | 3.69e-01 |
| GO:BP | GO:0009886 | post-embryonic animal morphogenesis | 1 | 10 | 3.69e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 4 | 538 | 3.69e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 1 | 7 | 3.69e-01 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 3.69e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 1 | 8 | 3.69e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 5 | 294 | 3.70e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 2 | 28 | 3.70e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 20 | 375 | 3.70e-01 |
| GO:BP | GO:0021561 | facial nerve development | 1 | 8 | 3.71e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 1 | 8 | 3.71e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 4 | 77 | 3.71e-01 |
| GO:BP | GO:0001955 | blood vessel maturation | 1 | 6 | 3.72e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 13 | 252 | 3.72e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 1 | 14 | 3.72e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 1 | 12 | 3.72e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 2 | 26 | 3.72e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 1 | 12 | 3.73e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 1 | 10 | 3.73e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 1 | 12 | 3.73e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 3.73e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 4 | 195 | 3.73e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 2 | 39 | 3.74e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 14 | 340 | 3.74e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 9 | 153 | 3.74e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 3.74e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 1 | 1939 | 3.74e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 4 | 222 | 3.74e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 3.74e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 3.74e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 1 | 60 | 3.74e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 1 | 7 | 3.75e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2088 | 3.75e-01 |
| GO:BP | GO:0001708 | cell fate specification | 2 | 56 | 3.75e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 3.75e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 1 | 7 | 3.75e-01 |
| GO:BP | GO:0033962 | P-body assembly | 1 | 20 | 3.75e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 3 | 259 | 3.75e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 15 | 273 | 3.75e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 20 | 376 | 3.75e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 3 | 43 | 3.76e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 1 | 10 | 3.76e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 1 | 76 | 3.77e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 1 | 47 | 3.77e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 1 | 12 | 3.78e-01 |
| GO:BP | GO:0035112 | genitalia morphogenesis | 1 | 5 | 3.78e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 1 | 12 | 3.78e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 3 | 50 | 3.78e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 1 | 50 | 3.78e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 2 | 452 | 3.79e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 3.80e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 1 | 42 | 3.80e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 1 | 13 | 3.80e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 1 | 5 | 3.81e-01 |
| GO:BP | GO:0001775 | cell activation | 9 | 655 | 3.81e-01 |
| GO:BP | GO:0021766 | hippocampus development | 1 | 62 | 3.81e-01 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 3.81e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 8 | 142 | 3.81e-01 |
| GO:BP | GO:0007165 | signal transduction | 2 | 3818 | 3.81e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 2 | 23 | 3.81e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 1 | 1061 | 3.81e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 3.81e-01 |
| GO:BP | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration | 1 | 6 | 3.81e-01 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 2 | 73 | 3.81e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 3.81e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 12 | 387 | 3.81e-01 |
| GO:BP | GO:0006412 | translation | 2 | 646 | 3.81e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 1 | 39 | 3.81e-01 |
| GO:BP | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 7 | 3.81e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.81e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 3.81e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 994 | 3.81e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 3.81e-01 |
| GO:BP | GO:0090132 | epithelium migration | 13 | 252 | 3.81e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 3.82e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 1 | 6 | 3.83e-01 |
| GO:BP | GO:0097283 | keratinocyte apoptotic process | 1 | 5 | 3.83e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 2 | 90 | 3.83e-01 |
| GO:BP | GO:1902172 | regulation of keratinocyte apoptotic process | 1 | 5 | 3.83e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 3.83e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 2 | 338 | 3.83e-01 |
| GO:BP | GO:0048678 | response to axon injury | 2 | 64 | 3.83e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 3.83e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 1 | 25 | 3.83e-01 |
| GO:BP | GO:0051866 | general adaptation syndrome | 1 | 4 | 3.83e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 12 | 204 | 3.83e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 5 | 2890 | 3.83e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 1 | 11 | 3.84e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 32 | 3.84e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 2 | 90 | 3.84e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 5 | 55 | 3.84e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 5 | 3.84e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 1 | 7 | 3.84e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 1 | 44 | 3.84e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 1 | 160 | 3.84e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 7 | 114 | 3.85e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 6 | 85 | 3.85e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 2 | 45 | 3.85e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 2 | 35 | 3.85e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 1 | 15 | 3.85e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 3 | 38 | 3.86e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 3.86e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 1 | 168 | 3.87e-01 |
| GO:BP | GO:0021591 | ventricular system development | 1 | 29 | 3.87e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 1 | 10 | 3.87e-01 |
| GO:BP | GO:0060718 | chorionic trophoblast cell differentiation | 1 | 5 | 3.87e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 6 | 143 | 3.87e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 24 | 504 | 3.87e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 1 | 7 | 3.87e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 3 | 162 | 3.88e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 2 | 41 | 3.88e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 1 | 8 | 3.88e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 1 | 29 | 3.88e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 3.88e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 1 | 4 | 3.88e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 1 | 22 | 3.88e-01 |
| GO:BP | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning | 1 | 5 | 3.89e-01 |
| GO:BP | GO:0007418 | ventral midline development | 1 | 4 | 3.89e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 2 | 74 | 3.89e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 2 | 30 | 3.89e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 1 | 35 | 3.89e-01 |
| GO:BP | GO:0070475 | rRNA base methylation | 1 | 8 | 3.89e-01 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 1 | 11 | 3.89e-01 |
| GO:BP | GO:0061744 | motor behavior | 1 | 17 | 3.89e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 2 | 41 | 3.89e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 3.89e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 3.89e-01 |
| GO:BP | GO:0042148 | strand invasion | 1 | 5 | 3.89e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 4 | 65 | 3.89e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 2 | 63 | 3.90e-01 |
| GO:BP | GO:0048699 | generation of neurons | 1 | 1119 | 3.90e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 2 | 25 | 3.90e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 3 | 284 | 3.90e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 3.90e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 4 | 229 | 3.90e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 1 | 7 | 3.90e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 1 | 7 | 3.90e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 1 | 43 | 3.90e-01 |
| GO:BP | GO:0048731 | system development | 2 | 2901 | 3.90e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 3.90e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 1 | 16 | 3.90e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 1 | 19 | 3.90e-01 |
| GO:BP | GO:0003128 | heart field specification | 1 | 16 | 3.90e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 1 | 10 | 3.90e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 1 | 4 | 3.90e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 1 | 174 | 3.90e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 5 | 82 | 3.91e-01 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 3.91e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 3.91e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 2 | 666 | 3.91e-01 |
| GO:BP | GO:0060836 | lymphatic endothelial cell differentiation | 1 | 5 | 3.91e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 1 | 17 | 3.91e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 3.91e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 37 | 7441 | 3.91e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 3.91e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 3.91e-01 |
| GO:BP | GO:0030916 | otic vesicle formation | 1 | 6 | 3.91e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 4 | 556 | 3.91e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 1 | 8 | 3.91e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 1 | 20 | 3.92e-01 |
| GO:BP | GO:0006950 | response to stress | 2 | 2780 | 3.92e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 400 | 3.92e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 1 | 28 | 3.92e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 1 | 26 | 3.92e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 4 | 2285 | 3.92e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 4 | 132 | 3.92e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 6 | 384 | 3.92e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 2 | 33 | 3.93e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 2 | 77 | 3.93e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 4 | 87 | 3.93e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 1 | 78 | 3.93e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 3.93e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 2 | 41 | 3.93e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 1 | 11 | 3.93e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1 | 1898 | 3.93e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 7 | 116 | 3.93e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 3.93e-01 |
| GO:BP | GO:0016081 | synaptic vesicle docking | 1 | 9 | 3.93e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 1 | 10 | 3.93e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 1 | 8 | 3.93e-01 |
| GO:BP | GO:0003097 | renal water transport | 1 | 6 | 3.93e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1201 | 3.94e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1719 | 3.94e-01 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 1 | 23 | 3.94e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 3 | 168 | 3.95e-01 |
| GO:BP | GO:0030901 | midbrain development | 4 | 69 | 3.95e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 2 | 441 | 3.95e-01 |
| GO:BP | GO:0001759 | organ induction | 2 | 17 | 3.95e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 1 | 19 | 3.95e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 4 | 126 | 3.95e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 1 | 19 | 3.95e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 1 | 19 | 3.95e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 1 | 246 | 3.95e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 1 | 13 | 3.95e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 2 | 30 | 3.96e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 3.96e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 1 | 47 | 3.96e-01 |
| GO:BP | GO:1902523 | positive regulation of protein K63-linked ubiquitination | 1 | 5 | 3.97e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 2 | 45 | 3.97e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 2 | 26 | 3.97e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 2 | 36 | 3.97e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 4 | 51 | 3.97e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 1 | 53 | 3.97e-01 |
| GO:BP | GO:0090130 | tissue migration | 13 | 256 | 3.97e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 3.97e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 3.97e-01 |
| GO:BP | GO:0035329 | hippo signaling | 3 | 41 | 3.97e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 1 | 270 | 3.99e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1233 | 3.99e-01 |
| GO:BP | GO:0035437 | maintenance of protein localization in endoplasmic reticulum | 1 | 11 | 3.99e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 6 | 88 | 3.99e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 4.00e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 4.00e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 1 | 5 | 4.00e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 1 | 15 | 4.00e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 1 | 8 | 4.00e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 1 | 52 | 4.00e-01 |
| GO:BP | GO:0007379 | segment specification | 1 | 14 | 4.00e-01 |
| GO:BP | GO:0006541 | glutamine metabolic process | 2 | 24 | 4.00e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 4.00e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 1 | 44 | 4.01e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 1 | 86 | 4.01e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 16 | 268 | 4.01e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 5 | 93 | 4.01e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 16 | 268 | 4.01e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 1 | 16 | 4.01e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 10 | 4.01e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 5 | 85 | 4.01e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 4 | 54 | 4.01e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 4.01e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 4.01e-01 |
| GO:BP | GO:0006353 | DNA-templated transcription termination | 1 | 22 | 4.02e-01 |
| GO:BP | GO:0032835 | glomerulus development | 1 | 54 | 4.03e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 1 | 7 | 4.03e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 4.04e-01 |
| GO:BP | GO:0021548 | pons development | 1 | 7 | 4.04e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 1 | 7 | 4.05e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 2 | 29 | 4.05e-01 |
| GO:BP | GO:0006266 | DNA ligation | 1 | 16 | 4.05e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 6 | 106 | 4.05e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 1 | 86 | 4.05e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 2 | 26 | 4.05e-01 |
| GO:BP | GO:0036075 | replacement ossification | 1 | 25 | 4.05e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 1 | 30 | 4.05e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 2 | 36 | 4.05e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 3 | 60 | 4.05e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 1 | 25 | 4.05e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 2 | 21 | 4.05e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2 | 4955 | 4.05e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 97 | 4.05e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 1 | 18 | 4.06e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 1 | 49 | 4.06e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 4.06e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 4.06e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 2 | 37 | 4.06e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 2 | 29 | 4.06e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 1 | 19 | 4.06e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 1 | 5 | 4.06e-01 |
| GO:BP | GO:0006983 | ER overload response | 1 | 12 | 4.06e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 2 | 116 | 4.06e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 2 | 69 | 4.07e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 2 | 51 | 4.09e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 4.09e-01 |
| GO:BP | GO:0023052 | signaling | 2 | 4141 | 4.09e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 1 | 6 | 4.09e-01 |
| GO:BP | GO:0048265 | response to pain | 1 | 17 | 4.09e-01 |
| GO:BP | GO:0001562 | response to protozoan | 1 | 11 | 4.09e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 1 | 50 | 4.09e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 2 | 28 | 4.09e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 7 | 119 | 4.09e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 3 | 156 | 4.09e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 4 | 83 | 4.09e-01 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 4.09e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 1 | 7 | 4.09e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 4 | 63 | 4.10e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 4.11e-01 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 1 | 7 | 4.12e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 1 | 8 | 4.13e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 2 | 91 | 4.13e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1 | 9 | 4.13e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 2 | 91 | 4.13e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 25 | 4.13e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 9 | 352 | 4.13e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 4.13e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 4.14e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 1 | 9 | 4.14e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 1 | 9 | 4.14e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 10 | 168 | 4.14e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 1 | 59 | 4.14e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 2 | 22 | 4.14e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 1 | 59 | 4.14e-01 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 1 | 8 | 4.14e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 2 | 23 | 4.14e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 1 | 5 | 4.14e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 2 | 186 | 4.14e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 1 | 7 | 4.14e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 3 | 299 | 4.14e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1138 | 4.15e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 3 | 59 | 4.15e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 1 | 16 | 4.15e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 4.15e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 4.15e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 1 | 17 | 4.15e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1294 | 4.15e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 4.15e-01 |
| GO:BP | GO:0021521 | ventral spinal cord interneuron specification | 1 | 3 | 4.15e-01 |
| GO:BP | GO:0060573 | cell fate specification involved in pattern specification | 1 | 3 | 4.15e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 2 | 29 | 4.15e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 4.15e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 1 | 9 | 4.16e-01 |
| GO:BP | GO:0007154 | cell communication | 2 | 4226 | 4.16e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 10 | 169 | 4.17e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 1 | 16 | 4.17e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 1 | 29 | 4.17e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 1 | 1278 | 4.17e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 4.18e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 1 | 13 | 4.18e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 870 | 4.18e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 1 | 13 | 4.18e-01 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 1 | 8 | 4.18e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 4 | 70 | 4.18e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 4 | 70 | 4.18e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 1 | 260 | 4.18e-01 |
| GO:BP | GO:0019058 | viral life cycle | 2 | 244 | 4.18e-01 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 1 | 13 | 4.19e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 1 | 10 | 4.19e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 2 | 223 | 4.19e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 8 | 160 | 4.19e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 4.19e-01 |
| GO:BP | GO:0006281 | DNA repair | 26 | 534 | 4.19e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 4.20e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 3 | 357 | 4.20e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 4.20e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 2 | 23 | 4.20e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 1 | 42 | 4.21e-01 |
| GO:BP | GO:0043038 | amino acid activation | 3 | 44 | 4.21e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 3 | 57 | 4.21e-01 |
| GO:BP | GO:0070997 | neuron death | 1 | 286 | 4.21e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 11 | 194 | 4.21e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 1 | 12 | 4.21e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 2 | 94 | 4.21e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 2 | 18 | 4.21e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 2 | 94 | 4.21e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 2 | 17 | 4.21e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 1 | 30 | 4.21e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 1 | 47 | 4.22e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 4.22e-01 |
| GO:BP | GO:0007033 | vacuole organization | 1 | 197 | 4.22e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 4.22e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 1 | 13 | 4.22e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 2 | 46 | 4.22e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 2 | 41 | 4.22e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 4.23e-01 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1267 | 4.23e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 2 | 23 | 4.23e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 1 | 21 | 4.23e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 1 | 41 | 4.24e-01 |
| GO:BP | GO:0042476 | odontogenesis | 2 | 86 | 4.25e-01 |
| GO:BP | GO:0048839 | inner ear development | 8 | 132 | 4.25e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 1 | 25 | 4.25e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 4.25e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 2 | 45 | 4.25e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 1 | 8 | 4.25e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 2 | 22 | 4.25e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 4.25e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 4.25e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 4.26e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 1 | 214 | 4.26e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 8 | 122 | 4.27e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 4.27e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 4.27e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 5 | 103 | 4.27e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 4.27e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 9 | 169 | 4.27e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 1 | 16 | 4.27e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 4.27e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 2 | 22 | 4.28e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 5 | 170 | 4.29e-01 |
| GO:BP | GO:0008213 | protein alkylation | 9 | 160 | 4.29e-01 |
| GO:BP | GO:0006479 | protein methylation | 9 | 160 | 4.29e-01 |
| GO:BP | GO:0039530 | MDA-5 signaling pathway | 1 | 8 | 4.29e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 44 | 884 | 4.29e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 3 | 292 | 4.29e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 1 | 97 | 4.29e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 4.29e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 4.29e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 4.29e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 8 | 4.29e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 8 | 4.29e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 1 | 57 | 4.30e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 1 | 54 | 4.30e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 1 | 16 | 4.30e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 4.30e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 3 | 43 | 4.30e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 1 | 24 | 4.30e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 4 | 211 | 4.31e-01 |
| GO:BP | GO:0021761 | limbic system development | 1 | 81 | 4.33e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 2 | 116 | 4.33e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 1 | 15 | 4.33e-01 |
| GO:BP | GO:0070170 | regulation of tooth mineralization | 1 | 4 | 4.34e-01 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 1 | 14 | 4.35e-01 |
| GO:BP | GO:0048664 | neuron fate determination | 1 | 4 | 4.35e-01 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 1 | 8 | 4.35e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 1 | 10 | 4.35e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 2 | 106 | 4.35e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 1 | 8 | 4.35e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 1 | 5 | 4.35e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 2 | 25 | 4.35e-01 |
| GO:BP | GO:0006915 | apoptotic process | 7 | 1443 | 4.35e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 4.36e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 1 | 59 | 4.36e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 4 | 108 | 4.36e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 4.36e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 4.37e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 4.37e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 1 | 25 | 4.37e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 3 | 51 | 4.37e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 1 | 13 | 4.38e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 4.38e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 1 | 8 | 4.38e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 1 | 20 | 4.38e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 4.39e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 4.39e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 2 | 31 | 4.39e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 1 | 9 | 4.39e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 1 | 10 | 4.39e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 3 | 48 | 4.39e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 4.39e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 1 | 10 | 4.39e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 3 | 188 | 4.39e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 13 | 249 | 4.40e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 2 | 90 | 4.40e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 2 | 105 | 4.40e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 4.40e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 2 | 771 | 4.40e-01 |
| GO:BP | GO:0032868 | response to insulin | 1 | 215 | 4.40e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 2 | 233 | 4.40e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 3 | 155 | 4.40e-01 |
| GO:BP | GO:0003407 | neural retina development | 1 | 48 | 4.40e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 1 | 61 | 4.40e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 4.40e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 4.40e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 4.40e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 1 | 12 | 4.42e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 2 | 28 | 4.42e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 4.42e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 1 | 8 | 4.42e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 1 | 25 | 4.42e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 4.42e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 4.42e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 1 | 25 | 4.42e-01 |
| GO:BP | GO:0061525 | hindgut development | 1 | 5 | 4.43e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 3 | 32 | 4.43e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 1 | 62 | 4.43e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 4 | 115 | 4.44e-01 |
| GO:BP | GO:0000423 | mitophagy | 2 | 37 | 4.44e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 4 | 115 | 4.44e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 2 | 37 | 4.44e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 2 | 100 | 4.45e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 4.46e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 8 | 4.47e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 1 | 17 | 4.47e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 2 | 775 | 4.47e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 4.47e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 1 | 16 | 4.47e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 1 | 27 | 4.47e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 1 | 14 | 4.47e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 1 | 15 | 4.48e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 4.48e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 6 | 220 | 4.48e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 1 | 4 | 4.48e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 4.48e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 2 | 25 | 4.48e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 8 | 146 | 4.48e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 2 | 72 | 4.48e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 1 | 21 | 4.48e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 7 | 131 | 4.48e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 1 | 104 | 4.48e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 2 | 272 | 4.48e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 2 | 37 | 4.48e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 4.48e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 3 | 43 | 4.51e-01 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 1 | 7 | 4.51e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 4.51e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 4.51e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 1 | 7 | 4.51e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 1 | 7 | 4.51e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 4.52e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 1 | 9 | 4.52e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 1 | 44 | 4.52e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 4.52e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 2 | 113 | 4.52e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 4.53e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 4.53e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 6 | 123 | 4.53e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 5 | 130 | 4.53e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 4.53e-01 |
| GO:BP | GO:1900363 | regulation of mRNA polyadenylation | 1 | 11 | 4.53e-01 |
| GO:BP | GO:0051258 | protein polymerization | 1 | 241 | 4.53e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 3 | 441 | 4.53e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 4.53e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 10 | 209 | 4.53e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 3 | 48 | 4.53e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 1 | 11 | 4.53e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 1 | 11 | 4.53e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 1 | 14 | 4.53e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 3 | 48 | 4.53e-01 |
| GO:BP | GO:0061303 | cornea development in camera-type eye | 1 | 4 | 4.53e-01 |
| GO:BP | GO:0007049 | cell cycle | 74 | 1529 | 4.53e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 1 | 8 | 4.53e-01 |
| GO:BP | GO:0045471 | response to ethanol | 1 | 73 | 4.53e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 5 | 99 | 4.53e-01 |
| GO:BP | GO:0070857 | regulation of bile acid biosynthetic process | 1 | 7 | 4.54e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 1 | 11 | 4.54e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 3 | 344 | 4.54e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 4.54e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 1 | 8 | 4.54e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 1 | 9 | 4.54e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 1 | 10 | 4.54e-01 |
| GO:BP | GO:0051591 | response to cAMP | 1 | 66 | 4.54e-01 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 1 | 8 | 4.55e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 2 | 33 | 4.55e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 9 | 182 | 4.55e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 4.56e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 1 | 10 | 4.56e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 2 | 28 | 4.58e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 2 | 28 | 4.58e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 1 | 14 | 4.58e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 4.58e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 2 | 32 | 4.58e-01 |
| GO:BP | GO:0012501 | programmed cell death | 7 | 1479 | 4.58e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 1 | 317 | 4.60e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 2 | 108 | 4.60e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 2 | 29 | 4.60e-01 |
| GO:BP | GO:0009411 | response to UV | 6 | 141 | 4.61e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 1 | 17 | 4.61e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 1 | 12 | 4.62e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 1 | 12 | 4.62e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 1 | 12 | 4.62e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 4.62e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 1 | 10 | 4.62e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 1 | 10 | 4.62e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 1 | 10 | 4.62e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 2 | 40 | 4.62e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 4.62e-01 |
| GO:BP | GO:0042116 | macrophage activation | 2 | 57 | 4.62e-01 |
| GO:BP | GO:0007399 | nervous system development | 3 | 1904 | 4.63e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 9 | 142 | 4.64e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 13 | 7069 | 4.64e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 4.64e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 2 | 28 | 4.64e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 4.64e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 2 | 90 | 4.66e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 4.66e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 4 | 42 | 4.66e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 65 | 4.67e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 1 | 19 | 4.68e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 2 | 118 | 4.68e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 1 | 25 | 4.68e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 1 | 37 | 4.68e-01 |
| GO:BP | GO:0060717 | chorion development | 1 | 8 | 4.68e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 6 | 118 | 4.68e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 4.69e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 4.70e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 2 | 28 | 4.70e-01 |
| GO:BP | GO:0001656 | metanephros development | 1 | 64 | 4.70e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 2 | 36 | 4.70e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 4.70e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 1 | 15 | 4.70e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 2 | 30 | 4.70e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 4.70e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 1 | 10 | 4.70e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 1 | 241 | 4.71e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 4.71e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 7 | 4.71e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 2 | 29 | 4.71e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 1 | 7 | 4.71e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 2 | 26 | 4.71e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 18 | 391 | 4.71e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 1 | 10 | 4.71e-01 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 1 | 14 | 4.71e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 4.72e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 5 | 300 | 4.72e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 4.72e-01 |
| GO:BP | GO:0006403 | RNA localization | 8 | 182 | 4.72e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 2 | 27 | 4.73e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 1 | 30 | 4.73e-01 |
| GO:BP | GO:0042552 | myelination | 3 | 111 | 4.74e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 13 | 308 | 4.75e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 1 | 18 | 4.75e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 3 | 1345 | 4.75e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 2 | 39 | 4.75e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 4 | 76 | 4.76e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 4.76e-01 |
| GO:BP | GO:0003157 | endocardium development | 1 | 9 | 4.76e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 1 | 31 | 4.76e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 2 | 23 | 4.77e-01 |
| GO:BP | GO:0050896 | response to stimulus | 3 | 5770 | 4.77e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 4.77e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 4.77e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 1 | 12 | 4.78e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 4.78e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 1 | 21 | 4.78e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 1 | 81 | 4.79e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 1 | 47 | 4.79e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 3 | 52 | 4.79e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 1 | 13 | 4.79e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 4.79e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 2 | 40 | 4.79e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 1 | 66 | 4.79e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 1 | 28 | 4.80e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 1 | 35 | 4.80e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 4.80e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 3 | 58 | 4.80e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 1 | 9 | 4.80e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 3 | 113 | 4.80e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 1 | 24 | 4.80e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 4.80e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 2 | 124 | 4.80e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 3 | 113 | 4.80e-01 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 1 | 11 | 4.81e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 3 | 49 | 4.81e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 1 | 10 | 4.81e-01 |
| GO:BP | GO:0051152 | positive regulation of smooth muscle cell differentiation | 1 | 10 | 4.81e-01 |
| GO:BP | GO:0043588 | skin development | 2 | 175 | 4.81e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 5 | 93 | 4.82e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 1 | 68 | 4.82e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 2 | 25 | 4.83e-01 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 1 | 11 | 4.84e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 1 | 28 | 4.84e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 1 | 71 | 4.84e-01 |
| GO:BP | GO:0070266 | necroptotic process | 2 | 32 | 4.84e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 4.84e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 1 | 31 | 4.84e-01 |
| GO:BP | GO:0002252 | immune effector process | 3 | 318 | 4.84e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2 | 4953 | 4.84e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 1 | 73 | 4.85e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 1 | 28 | 4.85e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 4.85e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 1 | 38 | 4.85e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 1 | 15 | 4.85e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 4.85e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 4.85e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 4.85e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 4.85e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 4.85e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 4.85e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 4.85e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 3 | 61 | 4.85e-01 |
| GO:BP | GO:0007431 | salivary gland development | 1 | 26 | 4.85e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 2 | 30 | 4.85e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 4 | 103 | 4.85e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 11 | 206 | 4.85e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 41 | 8061 | 4.85e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 4.86e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 11 | 217 | 4.86e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 22 | 459 | 4.86e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 2 | 36 | 4.86e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 2 | 55 | 4.86e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 4.87e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 4.87e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 1 | 15 | 4.87e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 2 | 1149 | 4.87e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 1 | 23 | 4.87e-01 |
| GO:BP | GO:0055001 | muscle cell development | 3 | 165 | 4.87e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 4.87e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 1 | 90 | 4.87e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 4.87e-01 |
| GO:BP | GO:0042311 | vasodilation | 1 | 39 | 4.88e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 4.88e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 12 | 237 | 4.88e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 6 | 111 | 4.88e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 15 | 4.88e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 1 | 33 | 4.89e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 4 | 59 | 4.89e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 4.89e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 68 | 4.89e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 5 | 94 | 4.89e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 2 | 74 | 4.89e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 1 | 12 | 4.89e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 4.89e-01 |
| GO:BP | GO:0043276 | anoikis | 2 | 32 | 4.89e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 7 | 157 | 4.89e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 1 | 17 | 4.89e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 2 | 62 | 4.89e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 1 | 20 | 4.89e-01 |
| GO:BP | GO:0042753 | positive regulation of circadian rhythm | 1 | 10 | 4.89e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 1 | 14 | 4.89e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 4 | 68 | 4.90e-01 |
| GO:BP | GO:0071472 | cellular response to salt stress | 1 | 11 | 4.92e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 4.92e-01 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 1 | 14 | 4.92e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 4.92e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 1 | 34 | 4.92e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 4 | 67 | 4.92e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 1 | 75 | 4.92e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 1 | 10 | 4.93e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 1 | 10 | 4.93e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 1 | 20 | 4.93e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 4.93e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 4.93e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 4.93e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 5 | 96 | 4.93e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 1 | 25 | 4.94e-01 |
| GO:BP | GO:0035640 | exploration behavior | 1 | 21 | 4.94e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 4.94e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 5 | 73 | 4.94e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 1 | 13 | 4.94e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 3 | 318 | 4.95e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 4.95e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 4.96e-01 |
| GO:BP | GO:0014029 | neural crest formation | 1 | 13 | 4.96e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 1 | 11 | 4.96e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 4 | 64 | 4.96e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 2 | 39 | 4.96e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 1 | 26 | 4.96e-01 |
| GO:BP | GO:0048873 | homeostasis of number of cells within a tissue | 1 | 24 | 4.96e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 1 | 105 | 4.96e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 4.97e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 4.97e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 4.97e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 4.97e-01 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 1 | 19 | 4.97e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 4.98e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 1 | 64 | 4.99e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 4.99e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 5 | 95 | 4.99e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 4.99e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 4 | 93 | 4.99e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 1 | 11 | 5.00e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 1 | 13 | 5.00e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 2 | 18 | 5.00e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 15 | 5.00e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 2 | 38 | 5.01e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 2 | 19 | 5.01e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 2 | 64 | 5.01e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1 | 26 | 5.01e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 1 | 29 | 5.01e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 1 | 16 | 5.02e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 1 | 91 | 5.02e-01 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 18 | 5.03e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 7 | 146 | 5.03e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 1 | 10 | 5.03e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 5.03e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 5.04e-01 |
| GO:BP | GO:0007584 | response to nutrient | 1 | 109 | 5.04e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 2 | 26 | 5.04e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 1 | 400 | 5.04e-01 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 1 | 36 | 5.04e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 1 | 12 | 5.04e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 2 | 197 | 5.04e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 2 | 114 | 5.05e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 1 | 21 | 5.05e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 5.05e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 1 | 12 | 5.06e-01 |
| GO:BP | GO:0000338 | protein deneddylation | 1 | 11 | 5.06e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 5 | 100 | 5.06e-01 |
| GO:BP | GO:0055057 | neuroblast division | 1 | 12 | 5.06e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 1 | 16 | 5.06e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 1 | 16 | 5.06e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 15 | 5.06e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 2 | 36 | 5.06e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 2 | 118 | 5.06e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 2 | 32 | 5.06e-01 |
| GO:BP | GO:0001867 | complement activation, lectin pathway | 1 | 5 | 5.06e-01 |
| GO:BP | GO:0032259 | methylation | 2 | 313 | 5.06e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 12 | 227 | 5.07e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 2 | 26 | 5.07e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 5.07e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 5.07e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 1 | 12 | 5.07e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 5.08e-01 |
| GO:BP | GO:0042594 | response to starvation | 8 | 178 | 5.08e-01 |
| GO:BP | GO:0016236 | macroautophagy | 1 | 308 | 5.10e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 5.10e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 1 | 11 | 5.11e-01 |
| GO:BP | GO:1904251 | regulation of bile acid metabolic process | 1 | 9 | 5.11e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 1 | 11 | 5.11e-01 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 1 | 15 | 5.11e-01 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 1 | 15 | 5.11e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 5.11e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 1 | 307 | 5.11e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 1 | 287 | 5.11e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 3 | 55 | 5.11e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 5 | 103 | 5.11e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 3 | 79 | 5.11e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 2 | 37 | 5.11e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 2 | 48 | 5.12e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 5.12e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 29 | 5.12e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 5.12e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 1 | 53 | 5.12e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 37 | 5.13e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 1 | 14 | 5.13e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 1 | 12 | 5.13e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 2 | 859 | 5.13e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 5.13e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 1 | 15 | 5.13e-01 |
| GO:BP | GO:0007015 | actin filament organization | 3 | 373 | 5.13e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 2 | 69 | 5.14e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 5.14e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 1 | 22 | 5.14e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 6 | 105 | 5.14e-01 |
| GO:BP | GO:0060581 | cell fate commitment involved in pattern specification | 1 | 3 | 5.14e-01 |
| GO:BP | GO:0033089 | positive regulation of T cell differentiation in thymus | 1 | 5 | 5.14e-01 |
| GO:BP | GO:0021520 | spinal cord motor neuron cell fate specification | 1 | 5 | 5.14e-01 |
| GO:BP | GO:0001890 | placenta development | 7 | 117 | 5.14e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 1 | 43 | 5.14e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 5.14e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 1 | 47 | 5.14e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 2 | 54 | 5.14e-01 |
| GO:BP | GO:0060579 | ventral spinal cord interneuron fate commitment | 1 | 3 | 5.14e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 2 | 30 | 5.14e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 2 | 52 | 5.14e-01 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 1 | 19 | 5.14e-01 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 1 | 19 | 5.14e-01 |
| GO:BP | GO:0035176 | social behavior | 1 | 34 | 5.14e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 1 | 13 | 5.14e-01 |
| GO:BP | GO:0033504 | floor plate development | 1 | 8 | 5.14e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 5.14e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 5.14e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 1 | 12 | 5.14e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 3 | 281 | 5.14e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 5.14e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 5.14e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 1 | 39 | 5.14e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 5.14e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 3 | 100 | 5.14e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 12 | 5.15e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 5.15e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 5.16e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 2 | 172 | 5.16e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 1 | 13 | 5.16e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 2 | 135 | 5.16e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 438 | 5.16e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 1 | 11 | 5.16e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 2 | 42 | 5.16e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 5.16e-01 |
| GO:BP | GO:0016197 | endosomal transport | 4 | 231 | 5.17e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 1 | 16 | 5.19e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 1 | 23 | 5.19e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 1 | 11 | 5.19e-01 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 1 | 8 | 5.19e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 2 | 113 | 5.19e-01 |
| GO:BP | GO:0060065 | uterus development | 1 | 13 | 5.19e-01 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 5.20e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 5 | 236 | 5.20e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 5.21e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 1 | 18 | 5.21e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 1 | 30 | 5.21e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 1 | 30 | 5.21e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 1 | 448 | 5.22e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 1 | 34 | 5.22e-01 |
| GO:BP | GO:0008219 | cell death | 5 | 1596 | 5.22e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 5.22e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 3 | 61 | 5.22e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 23 | 694 | 5.22e-01 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 1 | 41 | 5.22e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 5.22e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 1 | 21 | 5.22e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 2 | 123 | 5.22e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 3 | 47 | 5.23e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 1 | 10 | 5.23e-01 |
| GO:BP | GO:0006997 | nucleus organization | 1 | 126 | 5.23e-01 |
| GO:BP | GO:0016032 | viral process | 2 | 344 | 5.23e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 5.23e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 5 | 166 | 5.23e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 5.23e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 2 | 47 | 5.23e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 2 | 26 | 5.23e-01 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 1 | 21 | 5.23e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 11 | 5.23e-01 |
| GO:BP | GO:0099172 | presynapse organization | 2 | 42 | 5.23e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 4 | 60 | 5.23e-01 |
| GO:BP | GO:0031128 | developmental induction | 2 | 26 | 5.23e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 2 | 25 | 5.23e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 5.23e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 1 | 86 | 5.23e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 1 | 12 | 5.23e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 25 | 5.23e-01 |
| GO:BP | GO:0043584 | nose development | 1 | 10 | 5.23e-01 |
| GO:BP | GO:0048536 | spleen development | 2 | 28 | 5.23e-01 |
| GO:BP | GO:0071599 | otic vesicle development | 1 | 10 | 5.23e-01 |
| GO:BP | GO:0008585 | female gonad development | 1 | 77 | 5.24e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 5.24e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 17 | 5.25e-01 |
| GO:BP | GO:0060712 | spongiotrophoblast layer development | 1 | 11 | 5.25e-01 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 1 | 74 | 5.25e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 6 | 78 | 5.25e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 2 | 58 | 5.25e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 3 | 73 | 5.26e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 7 | 144 | 5.26e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 5.26e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 1 | 17 | 5.26e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 5.26e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 5.26e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 1 | 23 | 5.26e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 3 | 59 | 5.26e-01 |
| GO:BP | GO:0042303 | molting cycle | 3 | 78 | 5.26e-01 |
| GO:BP | GO:0042633 | hair cycle | 3 | 78 | 5.26e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 1 | 46 | 5.27e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 2 | 80 | 5.27e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 5.27e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 3 | 92 | 5.27e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 1 | 43 | 5.27e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 1 | 30 | 5.27e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 5.27e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 3 | 457 | 5.27e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 2 | 43 | 5.27e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 4 | 94 | 5.27e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 28 | 626 | 5.27e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 1 | 28 | 5.27e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 1 | 15 | 5.27e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 5.27e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 4 | 173 | 5.28e-01 |
| GO:BP | GO:0007389 | pattern specification process | 20 | 306 | 5.28e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 7 | 447 | 5.28e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 3 | 50 | 5.28e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 5 | 93 | 5.28e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 5.29e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 2 | 194 | 5.29e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 2 | 30 | 5.29e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 1 | 46 | 5.29e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 2 | 30 | 5.29e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 5.29e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 1 | 11 | 5.29e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 1 | 13 | 5.29e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 2 | 29 | 5.29e-01 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 1 | 11 | 5.30e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 1 | 15 | 5.30e-01 |
| GO:BP | GO:0090659 | walking behavior | 2 | 25 | 5.30e-01 |
| GO:BP | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 12 | 5.31e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 1 | 14 | 5.31e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 5.31e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 1 | 79 | 5.31e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 1 | 12 | 5.31e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 5.31e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 1 | 50 | 5.31e-01 |
| GO:BP | GO:0001947 | heart looping | 3 | 50 | 5.31e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 5.31e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 1 | 32 | 5.31e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 5.31e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 1 | 10 | 5.31e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 1 | 50 | 5.31e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 28 | 5.31e-01 |
| GO:BP | GO:0002520 | immune system development | 8 | 144 | 5.31e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 1 | 35 | 5.31e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 2 | 41 | 5.32e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 5.32e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 2 | 38 | 5.33e-01 |
| GO:BP | GO:0097484 | dendrite extension | 1 | 27 | 5.33e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 13 | 272 | 5.33e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 5.33e-01 |
| GO:BP | GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | 1 | 9 | 5.33e-01 |
| GO:BP | GO:0001816 | cytokine production | 7 | 451 | 5.33e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 1 | 18 | 5.33e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 5.34e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 2 | 70 | 5.34e-01 |
| GO:BP | GO:0007565 | female pregnancy | 1 | 122 | 5.34e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 5.35e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 1 | 26 | 5.35e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 1 | 252 | 5.35e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 6 | 123 | 5.36e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 3 | 59 | 5.36e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 1 | 96 | 5.36e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 1 | 11 | 5.36e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 1 | 11 | 5.36e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 3 | 68 | 5.36e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 5.36e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 1 | 23 | 5.36e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 9 | 271 | 5.36e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 5.36e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 5.36e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 1 | 13 | 5.36e-01 |
| GO:BP | GO:0021543 | pallium development | 1 | 139 | 5.36e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 5.37e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 1 | 18 | 5.37e-01 |
| GO:BP | GO:0051653 | spindle localization | 2 | 52 | 5.38e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 3 | 2102 | 5.38e-01 |
| GO:BP | GO:0032570 | response to progesterone | 1 | 27 | 5.38e-01 |
| GO:BP | GO:0090504 | epiboly | 2 | 31 | 5.38e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 1 | 13 | 5.38e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 2 | 39 | 5.39e-01 |
| GO:BP | GO:0032392 | DNA geometric change | 1 | 80 | 5.41e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 1 | 14 | 5.41e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 5.41e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 2 | 39 | 5.41e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 2 | 35 | 5.42e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 1 | 28 | 5.42e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 1 | 47 | 5.42e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 1 | 48 | 5.42e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 90 | 5.42e-01 |
| GO:BP | GO:0090208 | positive regulation of triglyceride metabolic process | 1 | 18 | 5.43e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 1 | 140 | 5.45e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 1 | 16 | 5.45e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 2 | 57 | 5.46e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 2 | 993 | 5.46e-01 |
| GO:BP | GO:0003002 | regionalization | 18 | 273 | 5.46e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1 | 72 | 5.46e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 1 | 27 | 5.46e-01 |
| GO:BP | GO:0031648 | protein destabilization | 2 | 45 | 5.47e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 2 | 30 | 5.48e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 5.48e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 1 | 51 | 5.48e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 5.48e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 3 | 159 | 5.48e-01 |
| GO:BP | GO:0055070 | copper ion homeostasis | 1 | 18 | 5.48e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 5.48e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 2 | 30 | 5.48e-01 |
| GO:BP | GO:0003016 | respiratory system process | 2 | 30 | 5.48e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 1 | 83 | 5.49e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 2 | 51 | 5.49e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 1 | 64 | 5.49e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 1 | 10 | 5.49e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 5.49e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 2 | 123 | 5.49e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 3 | 56 | 5.50e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 4 | 121 | 5.50e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 5.50e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 1 | 24 | 5.50e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2 | 26 | 5.50e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 1 | 20 | 5.51e-01 |
| GO:BP | GO:0030261 | chromosome condensation | 1 | 33 | 5.51e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 4 | 91 | 5.51e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 2 | 179 | 5.51e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 1 | 11 | 5.51e-01 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 1 | 5 | 5.52e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 5.52e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 1 | 10 | 5.52e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 1 | 11 | 5.52e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 14 | 5.52e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 3 | 62 | 5.52e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 1 | 10 | 5.52e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 1 | 20 | 5.52e-01 |
| GO:BP | GO:0001946 | lymphangiogenesis | 1 | 12 | 5.52e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 5.52e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 1 | 168 | 5.52e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 2 | 40 | 5.53e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 7 | 140 | 5.53e-01 |
| GO:BP | GO:0035272 | exocrine system development | 1 | 34 | 5.54e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 3 | 66 | 5.55e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 15 | 5.55e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 1 | 16 | 5.56e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 5.56e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 5.56e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 1 | 45 | 5.56e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 1 | 15 | 5.57e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 4 | 79 | 5.59e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 1 | 66 | 5.60e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 2 | 41 | 5.60e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 2 | 50 | 5.60e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 2 | 83 | 5.60e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 5.61e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 3 | 100 | 5.61e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 4 | 88 | 5.62e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 1 | 8 | 5.62e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 1 | 31 | 5.62e-01 |
| GO:BP | GO:0051642 | centrosome localization | 1 | 31 | 5.62e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 1 | 39 | 5.63e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 5.63e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 2 | 276 | 5.63e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 96 | 5.63e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 8 | 160 | 5.63e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 1 | 131 | 5.64e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 21 | 494 | 5.64e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 5.65e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 5.65e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 1 | 40 | 5.65e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 1 | 16 | 5.65e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 1 | 39 | 5.65e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 1 | 28 | 5.65e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 5.65e-01 |
| GO:BP | GO:0071103 | DNA conformation change | 1 | 87 | 5.65e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 1 | 91 | 5.65e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 3 | 59 | 5.65e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 2 | 77 | 5.65e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 2 | 30 | 5.65e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 1 | 21 | 5.65e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 3 | 70 | 5.66e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 543 | 5.66e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 5.66e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 1 | 22 | 5.66e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 1 | 19 | 5.66e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 1 | 141 | 5.67e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 1 | 30 | 5.67e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 5.67e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 1 | 7 | 5.68e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 1 | 11 | 5.68e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 1 | 11 | 5.68e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 1 | 13 | 5.68e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 11 | 246 | 5.68e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 8 | 194 | 5.68e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 5.68e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 1 | 14 | 5.68e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 5.68e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 1 | 16 | 5.68e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 10 | 288 | 5.69e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 1 | 17 | 5.69e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 5.69e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 2 | 27 | 5.69e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 5 | 98 | 5.69e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 5.69e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 1 | 42 | 5.70e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 5.70e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 1 | 13 | 5.70e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 2 | 38 | 5.70e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 2 | 50 | 5.70e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 4 | 95 | 5.70e-01 |
| GO:BP | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 14 | 5.71e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 1 | 33 | 5.71e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 4 | 71 | 5.71e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 496 | 5.71e-01 |
| GO:BP | GO:0008544 | epidermis development | 2 | 210 | 5.71e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 1 | 12 | 5.71e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 2 | 41 | 5.71e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 27 | 5.71e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 1 | 15 | 5.71e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 1 | 13 | 5.71e-01 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 1 | 15 | 5.72e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 1 | 14 | 5.72e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 1 | 15 | 5.74e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 5.74e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 1 | 48 | 5.75e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 1 | 20 | 5.75e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 1 | 135 | 5.75e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 1 | 49 | 5.75e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 3 | 52 | 5.75e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 2 | 61 | 5.78e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 1 | 52 | 5.78e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 4 | 58 | 5.78e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 5 | 358 | 5.78e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 1 | 55 | 5.78e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 2 | 98 | 5.80e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 3 | 45 | 5.80e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 5.80e-01 |
| GO:BP | GO:0006415 | translational termination | 1 | 17 | 5.80e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 5.81e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 3 | 107 | 5.82e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 3 | 57 | 5.82e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 4 | 276 | 5.82e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 4 | 107 | 5.82e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 5.82e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 5.82e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 2 | 34 | 5.82e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 2 | 55 | 5.82e-01 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 2 | 42 | 5.82e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 1 | 19 | 5.82e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 13 | 7806 | 5.82e-01 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 1 | 19 | 5.82e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 5.82e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 1 | 19 | 5.83e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 1 | 14 | 5.85e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 1 | 36 | 5.86e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 5.87e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 5.87e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 1 | 10 | 5.87e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 22 | 5.87e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 5.88e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 5.88e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 5.88e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 2 | 56 | 5.88e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 1 | 51 | 5.89e-01 |
| GO:BP | GO:0072006 | nephron development | 1 | 117 | 5.89e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 2 | 55 | 5.89e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 1 | 16 | 5.89e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 1 | 13 | 5.90e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 5.90e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 5.90e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 5.90e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 5.90e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 5.90e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 1 | 21 | 5.91e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 1 | 21 | 5.91e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 1 | 114 | 5.91e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 1 | 40 | 5.92e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 1 | 31 | 5.92e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 1 | 51 | 5.92e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 1 | 14 | 5.92e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 2 | 40 | 5.92e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 1 | 88 | 5.92e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 2 | 65 | 5.92e-01 |
| GO:BP | GO:0007040 | lysosome organization | 1 | 88 | 5.92e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 8 | 185 | 5.92e-01 |
| GO:BP | GO:0001654 | eye development | 2 | 272 | 5.93e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 5.94e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 5.94e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 4 | 78 | 5.95e-01 |
| GO:BP | GO:0003012 | muscle system process | 7 | 334 | 5.96e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 1 | 316 | 5.97e-01 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 1 | 27 | 5.97e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 1 | 17 | 5.98e-01 |
| GO:BP | GO:0150063 | visual system development | 2 | 273 | 5.98e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 1 | 41 | 5.98e-01 |
| GO:BP | GO:0080009 | mRNA methylation | 1 | 16 | 5.98e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 1 | 13 | 5.99e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 1 | 70 | 5.99e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 3 | 609 | 5.99e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 5.99e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 547 | 5.99e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 6.00e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 2 | 29 | 6.00e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 1 | 56 | 6.00e-01 |
| GO:BP | GO:0030220 | platelet formation | 1 | 17 | 6.00e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 3 | 59 | 6.00e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 6 | 116 | 6.00e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 6.01e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 1 | 20 | 6.01e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 1 | 14 | 6.01e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 1 | 23 | 6.01e-01 |
| GO:BP | GO:0060348 | bone development | 1 | 170 | 6.02e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 6.03e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 1 | 64 | 6.03e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 1 | 41 | 6.03e-01 |
| GO:BP | GO:0031167 | rRNA methylation | 1 | 24 | 6.03e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 2 | 86 | 6.03e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 1 | 34 | 6.03e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 2 | 40 | 6.03e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 1 | 69 | 6.03e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 3 | 49 | 6.03e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 6.03e-01 |
| GO:BP | GO:0048880 | sensory system development | 2 | 275 | 6.03e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 1 | 23 | 6.03e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 1 | 57 | 6.03e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 3 | 69 | 6.04e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 2 | 42 | 6.05e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 1 | 30 | 6.05e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 6 | 152 | 6.05e-01 |
| GO:BP | GO:0021762 | substantia nigra development | 1 | 41 | 6.05e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 2 | 110 | 6.05e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 1 | 16 | 6.06e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 1 | 184 | 6.06e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 6.06e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 2 | 35 | 6.06e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 6.07e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 6.07e-01 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 28 | 6.08e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 6.08e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 1 | 11 | 6.08e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 1 | 18 | 6.08e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 1 | 14 | 6.08e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 2 | 42 | 6.08e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 2 | 48 | 6.08e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 6.09e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 1 | 21 | 6.09e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 6.09e-01 |
| GO:BP | GO:0006996 | organelle organization | 1 | 3000 | 6.09e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 4 | 97 | 6.10e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 2 | 58 | 6.10e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 6 | 115 | 6.10e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 1 | 86 | 6.10e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 2 | 73 | 6.10e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 6.11e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 6.11e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 6.11e-01 |
| GO:BP | GO:0016577 | histone demethylation | 1 | 21 | 6.12e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 2 | 36 | 6.14e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 1 | 31 | 6.14e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 1 | 16 | 6.14e-01 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 2 | 42 | 6.14e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 6.16e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 31 | 6.18e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 6.18e-01 |
| GO:BP | GO:0031929 | TOR signaling | 5 | 115 | 6.18e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 1 | 14 | 6.19e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 6.19e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 1 | 18 | 6.19e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 3 | 95 | 6.20e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 1 | 36 | 6.20e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 1 | 16 | 6.20e-01 |
| GO:BP | GO:0006310 | DNA recombination | 9 | 257 | 6.20e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 1 | 18 | 6.20e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 1 | 63 | 6.20e-01 |
| GO:BP | GO:0046688 | response to copper ion | 1 | 29 | 6.20e-01 |
| GO:BP | GO:0090224 | regulation of spindle organization | 2 | 45 | 6.20e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 13 | 6.20e-01 |
| GO:BP | GO:0006833 | water transport | 1 | 10 | 6.20e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 6.20e-01 |
| GO:BP | GO:0097502 | mannosylation | 1 | 32 | 6.20e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 6.20e-01 |
| GO:BP | GO:0007530 | sex determination | 1 | 12 | 6.21e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 1 | 88 | 6.21e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 1 | 130 | 6.21e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 1 | 16 | 6.21e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 1 | 28 | 6.21e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 1 | 59 | 6.22e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 1 | 29 | 6.22e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 1 | 17 | 6.22e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 1 | 17 | 6.22e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 6.22e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 1 | 24 | 6.22e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 3 | 72 | 6.22e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 1 | 15 | 6.22e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 2 | 37 | 6.23e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 3 | 64 | 6.23e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 35 | 6.23e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 1 | 19 | 6.23e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 1 | 63 | 6.24e-01 |
| GO:BP | GO:0030073 | insulin secretion | 6 | 141 | 6.24e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 1 | 461 | 6.24e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 2 | 49 | 6.25e-01 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 1 | 7 | 6.25e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 3 | 158 | 6.25e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 2 | 47 | 6.25e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 597 | 6.25e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 597 | 6.25e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 2 | 41 | 6.25e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 1 | 17 | 6.26e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 6.26e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 6.26e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 1 | 65 | 6.26e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 6.26e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 6.26e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 3 | 67 | 6.27e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 1 | 21 | 6.27e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 198 | 6.27e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 11 | 228 | 6.28e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 2 | 50 | 6.29e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 1 | 22 | 6.29e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 1 | 16 | 6.29e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 5 | 119 | 6.29e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 1 | 30 | 6.29e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 1 | 469 | 6.29e-01 |
| GO:BP | GO:0007030 | Golgi organization | 2 | 137 | 6.30e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 6.30e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 7 | 500 | 6.30e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 6.30e-01 |
| GO:BP | GO:0042063 | gliogenesis | 5 | 235 | 6.31e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 6.32e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 6.32e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 1 | 15 | 6.32e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 1 | 21 | 6.32e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 1 | 22 | 6.32e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 691 | 6.32e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 6.32e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 2 | 136 | 6.32e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 1 | 37 | 6.32e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 1 | 40 | 6.32e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 6.32e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 4 | 84 | 6.33e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 1 | 94 | 6.34e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 2 | 75 | 6.34e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 5 | 121 | 6.34e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 1 | 27 | 6.34e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 1 | 27 | 6.34e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 1 | 17 | 6.34e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 7 | 258 | 6.35e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 5 | 116 | 6.37e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 3 | 43 | 6.37e-01 |
| GO:BP | GO:0006284 | base-excision repair | 2 | 44 | 6.37e-01 |
| GO:BP | GO:0006364 | rRNA processing | 6 | 221 | 6.38e-01 |
| GO:BP | GO:0071709 | membrane assembly | 1 | 53 | 6.38e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 2 | 323 | 6.38e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 1 | 19 | 6.38e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 2 | 36 | 6.39e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 6.39e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 2 | 92 | 6.39e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 1 | 37 | 6.40e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 6.40e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 6.40e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 6.40e-01 |
| GO:BP | GO:0007141 | male meiosis I | 1 | 15 | 6.41e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 1 | 18 | 6.41e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 1 | 23 | 6.41e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 2 | 43 | 6.41e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 1 | 30 | 6.41e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 3 | 631 | 6.41e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 1 | 126 | 6.41e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 1 | 20 | 6.42e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 3 | 1484 | 6.42e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 3 | 69 | 6.43e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 1 | 17 | 6.44e-01 |
| GO:BP | GO:0006260 | DNA replication | 11 | 255 | 6.45e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 1 | 22 | 6.46e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 1 | 57 | 6.46e-01 |
| GO:BP | GO:0065007 | biological regulation | 13 | 8320 | 6.46e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 3 | 70 | 6.46e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 1 | 18 | 6.46e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 1 | 18 | 6.46e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 1 | 21 | 6.46e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 1 | 36 | 6.46e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 1 | 18 | 6.46e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 6.46e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 2 | 40 | 6.46e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 1 | 18 | 6.46e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 6.46e-01 |
| GO:BP | GO:0007155 | cell adhesion | 9 | 1012 | 6.46e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 1 | 99 | 6.48e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 1 | 23 | 6.49e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 2 | 150 | 6.51e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 6.51e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 6.51e-01 |
| GO:BP | GO:0021756 | striatum development | 1 | 17 | 6.52e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 6.53e-01 |
| GO:BP | GO:0048469 | cell maturation | 1 | 108 | 6.54e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 1 | 42 | 6.55e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 8 | 428 | 6.55e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 1 | 21 | 6.55e-01 |
| GO:BP | GO:0022414 | reproductive process | 11 | 927 | 6.55e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 1 | 27 | 6.55e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 1 | 19 | 6.55e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 2 | 41 | 6.55e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 15 | 6.55e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 6.56e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 1 | 513 | 6.56e-01 |
| GO:BP | GO:0006914 | autophagy | 1 | 513 | 6.56e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 1 | 29 | 6.56e-01 |
| GO:BP | GO:0046697 | decidualization | 1 | 19 | 6.56e-01 |
| GO:BP | GO:0021511 | spinal cord patterning | 1 | 9 | 6.56e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 6.58e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 1 | 31 | 6.58e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 2 | 1133 | 6.58e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 2 | 48 | 6.59e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 1 | 73 | 6.59e-01 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 1 | 19 | 6.59e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 1 | 135 | 6.60e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 1 | 19 | 6.60e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 2 | 76 | 6.60e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 2 | 53 | 6.60e-01 |
| GO:BP | GO:0021510 | spinal cord development | 4 | 59 | 6.60e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 1 | 70 | 6.60e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 5 | 112 | 6.60e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 6.60e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 8 | 179 | 6.60e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 1 | 28 | 6.61e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 4 | 226 | 6.61e-01 |
| GO:BP | GO:0000003 | reproduction | 11 | 934 | 6.61e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 1 | 20 | 6.61e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 1 | 58 | 6.62e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 2 | 46 | 6.62e-01 |
| GO:BP | GO:0032602 | chemokine production | 2 | 46 | 6.62e-01 |
| GO:BP | GO:0008033 | tRNA processing | 1 | 128 | 6.63e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 2 | 106 | 6.64e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 1 | 485 | 6.65e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 3 | 71 | 6.65e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 6.65e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 6.65e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 6.65e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 9 | 185 | 6.65e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 1 | 25 | 6.65e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 1 | 22 | 6.65e-01 |
| GO:BP | GO:0006909 | phagocytosis | 2 | 148 | 6.65e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 1 | 22 | 6.65e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 2 | 90 | 6.65e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 6.66e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 1 | 55 | 6.66e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 1 | 26 | 6.66e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 1 | 68 | 6.66e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 1 | 57 | 6.66e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 6.66e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 1 | 68 | 6.66e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 1 | 24 | 6.66e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 1 | 27 | 6.66e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 5 | 114 | 6.67e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 2 | 73 | 6.67e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 2 | 115 | 6.67e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 6.67e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 1 | 21 | 6.67e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 2 | 73 | 6.67e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 1 | 36 | 6.67e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 2 | 43 | 6.68e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 2 | 58 | 6.68e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 32 | 798 | 6.69e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 29 | 6.69e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 3 | 69 | 6.70e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 676 | 6.70e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 1 | 44 | 6.70e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 1 | 44 | 6.72e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 2 | 222 | 6.72e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 827 | 6.73e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 2 | 52 | 6.74e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 6 | 172 | 6.74e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 6.74e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 4 | 98 | 6.74e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 1 | 25 | 6.75e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 1 | 67 | 6.76e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 6.76e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 1 | 26 | 6.76e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 2 | 67 | 6.76e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 6.76e-01 |
| GO:BP | GO:0061912 | selective autophagy | 3 | 90 | 6.76e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 1 | 22 | 6.76e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 6.77e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 3 | 88 | 6.77e-01 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 1 | 21 | 6.77e-01 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 1 | 21 | 6.77e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 2 | 76 | 6.77e-01 |
| GO:BP | GO:0008015 | blood circulation | 2 | 374 | 6.79e-01 |
| GO:BP | GO:0032506 | cytokinetic process | 1 | 39 | 6.79e-01 |
| GO:BP | GO:0017145 | stem cell division | 1 | 26 | 6.79e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 2 | 113 | 6.79e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 868 | 6.80e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 1 | 22 | 6.81e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 1 | 19 | 6.81e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 1 | 20 | 6.81e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 6.81e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 6.81e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 8 | 189 | 6.81e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 1 | 39 | 6.81e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 1 | 50 | 6.81e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 1 | 517 | 6.81e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 1 | 31 | 6.81e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 1 | 25 | 6.83e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 3 | 101 | 6.84e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 24 | 6.84e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 2 | 73 | 6.85e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 3 | 74 | 6.86e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 1 | 39 | 6.86e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 1 | 19 | 6.87e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 6.87e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 1 | 22 | 6.87e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 1 | 19 | 6.87e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 1 | 19 | 6.87e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 18 | 6.88e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 1 | 20 | 6.88e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 1 | 24 | 6.88e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 3 | 71 | 6.88e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 4 | 95 | 6.88e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 2 | 61 | 6.89e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 3 | 75 | 6.89e-01 |
| GO:BP | GO:0007423 | sensory organ development | 7 | 396 | 6.90e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 3 | 69 | 6.91e-01 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 3 | 104 | 6.92e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 2 | 38 | 6.92e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 1 | 27 | 6.92e-01 |
| GO:BP | GO:0014823 | response to activity | 2 | 46 | 6.92e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 5 | 188 | 6.93e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 3 | 54 | 6.93e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 1 | 15 | 6.94e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 1 | 18 | 6.94e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 1 | 34 | 6.94e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 1 | 612 | 6.94e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1 | 47 | 6.94e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 1 | 24 | 6.94e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 2 | 82 | 6.94e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 6 | 140 | 6.95e-01 |
| GO:BP | GO:0007281 | germ cell development | 1 | 187 | 6.96e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 1 | 44 | 6.96e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 1 | 39 | 6.97e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 1 | 39 | 6.97e-01 |
| GO:BP | GO:0045190 | isotype switching | 1 | 39 | 6.97e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 24 | 6.97e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 4 | 110 | 6.97e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 6.97e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 1 | 82 | 6.97e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 1 | 29 | 6.97e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 1 | 19 | 6.97e-01 |
| GO:BP | GO:0070988 | demethylation | 2 | 52 | 6.97e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 1 | 21 | 6.97e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 1 | 28 | 6.98e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 6.98e-01 |
| GO:BP | GO:0006936 | muscle contraction | 5 | 267 | 6.99e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 6.99e-01 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 2 | 47 | 6.99e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 3 | 79 | 7.00e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 2 | 42 | 7.00e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 3 | 70 | 7.00e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 1 | 29 | 7.00e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 1 | 44 | 7.00e-01 |
| GO:BP | GO:0015031 | protein transport | 2 | 1361 | 7.00e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 1 | 30 | 7.00e-01 |
| GO:BP | GO:0030168 | platelet activation | 5 | 91 | 7.01e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 2 | 55 | 7.01e-01 |
| GO:BP | GO:0000154 | rRNA modification | 1 | 35 | 7.01e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 28 | 710 | 7.01e-01 |
| GO:BP | GO:0022402 | cell cycle process | 41 | 1086 | 7.01e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 1 | 22 | 7.02e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 1 | 834 | 7.02e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 1 | 117 | 7.02e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 5 | 122 | 7.02e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 9 | 182 | 7.02e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 1 | 49 | 7.03e-01 |
| GO:BP | GO:0006414 | translational elongation | 1 | 65 | 7.03e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 1 | 61 | 7.04e-01 |
| GO:BP | GO:0048806 | genitalia development | 1 | 28 | 7.04e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 1 | 49 | 7.04e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 2 | 1229 | 7.04e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 1 | 24 | 7.04e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 7.04e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 1 | 20 | 7.04e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 1 | 31 | 7.06e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 7.07e-01 |
| GO:BP | GO:0046907 | intracellular transport | 2 | 1453 | 7.07e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 1 | 35 | 7.08e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 4 | 59 | 7.08e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 1 | 80 | 7.08e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 7.08e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 16 | 7.08e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 1 | 46 | 7.08e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 5 | 484 | 7.08e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 17 | 7.08e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 4 | 218 | 7.08e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 1 | 23 | 7.08e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 7.08e-01 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 2 | 52 | 7.09e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 1 | 16 | 7.10e-01 |
| GO:BP | GO:0030534 | adult behavior | 5 | 92 | 7.10e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 43 | 7.12e-01 |
| GO:BP | GO:0009651 | response to salt stress | 1 | 18 | 7.12e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 2 | 40 | 7.13e-01 |
| GO:BP | GO:0007612 | learning | 2 | 115 | 7.13e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 1 | 27 | 7.13e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 11 | 232 | 7.14e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 2 | 142 | 7.14e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 1 | 20 | 7.17e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 1 | 24 | 7.17e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 1 | 67 | 7.17e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 1 | 41 | 7.17e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 14 | 404 | 7.17e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 1 | 21 | 7.18e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 15 | 386 | 7.18e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 9 | 218 | 7.19e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1 | 21 | 7.19e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 1 | 68 | 7.20e-01 |
| GO:BP | GO:0016233 | telomere capping | 1 | 36 | 7.20e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 7.20e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 1 | 24 | 7.21e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 1 | 22 | 7.21e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 1 | 26 | 7.21e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 2 | 95 | 7.21e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 1 | 32 | 7.22e-01 |
| GO:BP | GO:0003013 | circulatory system process | 6 | 447 | 7.22e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 7.23e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 1 | 126 | 7.23e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 2 | 89 | 7.23e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 1 | 38 | 7.23e-01 |
| GO:BP | GO:0021544 | subpallium development | 1 | 18 | 7.23e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 1 | 25 | 7.23e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 1 | 25 | 7.23e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 5 | 129 | 7.23e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 2 | 97 | 7.23e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 1 | 24 | 7.24e-01 |
| GO:BP | GO:0009306 | protein secretion | 5 | 262 | 7.25e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 1 | 62 | 7.25e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 2 | 37 | 7.26e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 1 | 27 | 7.26e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 1 | 687 | 7.26e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 1 | 27 | 7.26e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 2 | 104 | 7.26e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 3 | 77 | 7.26e-01 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 1 | 37 | 7.26e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 5 | 262 | 7.26e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 4 | 98 | 7.26e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 2 | 76 | 7.26e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 1 | 665 | 7.26e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 1 | 26 | 7.27e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 2 | 60 | 7.27e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 2 | 31 | 7.27e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 2 | 44 | 7.27e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 7.27e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 2 | 104 | 7.28e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 1 | 27 | 7.29e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 3 | 120 | 7.29e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 2 | 31 | 7.30e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 1 | 47 | 7.30e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 7.31e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 1 | 25 | 7.31e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 7.31e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 7.31e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 2 | 106 | 7.32e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 7.32e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 1 | 30 | 7.32e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 7.34e-01 |
| GO:BP | GO:0032438 | melanosome organization | 1 | 36 | 7.34e-01 |
| GO:BP | GO:1903350 | response to dopamine | 1 | 48 | 7.34e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 2 | 52 | 7.34e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 1 | 21 | 7.34e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 1 | 15 | 7.34e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 2 | 53 | 7.34e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 2 | 104 | 7.34e-01 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 1 | 14 | 7.34e-01 |
| GO:BP | GO:0001764 | neuron migration | 3 | 133 | 7.34e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 7.34e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 1 | 96 | 7.35e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 1 | 40 | 7.35e-01 |
| GO:BP | GO:0006270 | DNA replication initiation | 1 | 35 | 7.35e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 1 | 36 | 7.35e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 2 | 58 | 7.35e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 1 | 36 | 7.35e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 1 | 47 | 7.35e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 39 | 7.35e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 1 | 31 | 7.36e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 3 | 116 | 7.36e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 1 | 23 | 7.36e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 7.36e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 7.36e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 1 | 87 | 7.36e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 7.36e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 5 | 123 | 7.36e-01 |
| GO:BP | GO:0021782 | glial cell development | 3 | 84 | 7.36e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 7.37e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 7.37e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 5 | 266 | 7.38e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 1 | 37 | 7.39e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 3 | 120 | 7.41e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 4 | 102 | 7.41e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 2 | 34 | 7.42e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 1 | 23 | 7.42e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 1 | 172 | 7.42e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 2 | 43 | 7.43e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 2 | 43 | 7.43e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 2 | 51 | 7.43e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 7.43e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 1 | 24 | 7.43e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 1 | 16 | 7.43e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 1 | 34 | 7.43e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 1 | 17 | 7.45e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 1 | 544 | 7.45e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 1 | 31 | 7.45e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 2 | 211 | 7.47e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 7.48e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 7.50e-01 |
| GO:BP | GO:0098754 | detoxification | 2 | 91 | 7.50e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 1 | 34 | 7.50e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 1 | 32 | 7.51e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 1 | 33 | 7.52e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 1 | 28 | 7.52e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 1 | 31 | 7.53e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 1 | 36 | 7.53e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 1 | 20 | 7.54e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 1 | 203 | 7.54e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 73 | 7.54e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 1 | 19 | 7.54e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 7.55e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 9 | 234 | 7.55e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 7.55e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 5 | 132 | 7.55e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 7.56e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 1 | 65 | 7.56e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 1 | 20 | 7.56e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 7 | 154 | 7.57e-01 |
| GO:BP | GO:0022037 | metencephalon development | 4 | 84 | 7.57e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 1 | 206 | 7.58e-01 |
| GO:BP | GO:0046323 | glucose import | 2 | 53 | 7.59e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 1 | 25 | 7.59e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 1 | 27 | 7.62e-01 |
| GO:BP | GO:0007052 | mitotic spindle organization | 4 | 129 | 7.63e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 1 | 22 | 7.63e-01 |
| GO:BP | GO:0014002 | astrocyte development | 1 | 27 | 7.63e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 7.64e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 1 | 92 | 7.64e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 1 | 92 | 7.64e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 7.64e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 1 | 53 | 7.64e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 7.64e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 7.65e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 2 | 121 | 7.66e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 1 | 25 | 7.66e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 5 | 134 | 7.66e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 1 | 37 | 7.67e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 1 | 38 | 7.67e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 1 | 31 | 7.67e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 7.67e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 1 | 33 | 7.67e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 1 | 33 | 7.67e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 2 | 121 | 7.68e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 7.68e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 1 | 20 | 7.69e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 1 | 59 | 7.69e-01 |
| GO:BP | GO:0098751 | bone cell development | 1 | 26 | 7.69e-01 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 1 | 60 | 7.70e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 65 | 7.70e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 1 | 34 | 7.71e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 3 | 258 | 7.71e-01 |
| GO:BP | GO:1901998 | toxin transport | 1 | 37 | 7.71e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 1 | 32 | 7.71e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 3 | 81 | 7.72e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 1 | 33 | 7.72e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 1 | 33 | 7.72e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 99 | 7.72e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 99 | 7.72e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 5 | 136 | 7.72e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 6 | 166 | 7.73e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 7.74e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 5 | 139 | 7.74e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 1 | 186 | 7.74e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 99 | 7.74e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 7.74e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 9 | 192 | 7.76e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 5 | 113 | 7.76e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 7.76e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 7.77e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 1 | 60 | 7.77e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 1 | 21 | 7.77e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 1 | 20 | 7.77e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 1 | 60 | 7.77e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 2 | 116 | 7.77e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 1 | 32 | 7.77e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 2 | 40 | 7.77e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 1 | 36 | 7.78e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 7.78e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 7.78e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 1 | 54 | 7.79e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 1 | 33 | 7.79e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 1 | 55 | 7.80e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 1 | 35 | 7.80e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 1 | 55 | 7.80e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 1 | 293 | 7.81e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 1 | 175 | 7.81e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 1 | 34 | 7.81e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 1 | 88 | 7.82e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 7.82e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 2 | 99 | 7.83e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 1 | 38 | 7.84e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 1 | 252 | 7.84e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 1 | 726 | 7.85e-01 |
| GO:BP | GO:0016556 | mRNA modification | 1 | 29 | 7.85e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 1 | 16 | 7.85e-01 |
| GO:BP | GO:0071867 | response to monoamine | 1 | 62 | 7.85e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 7.85e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 1 | 62 | 7.85e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 1 | 32 | 7.85e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 1 | 35 | 7.86e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 16 | 433 | 7.87e-01 |
| GO:BP | GO:0002790 | peptide secretion | 6 | 170 | 7.87e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 1 | 51 | 7.87e-01 |
| GO:BP | GO:0006959 | humoral immune response | 1 | 80 | 7.88e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 7.88e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 1 | 58 | 7.88e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 7.88e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 1 | 23 | 7.89e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 3 | 84 | 7.89e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 7.89e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 1 | 11 | 7.89e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 2 | 112 | 7.89e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 3 | 84 | 7.91e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 2 | 302 | 7.92e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 1 | 30 | 7.92e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 1 | 34 | 7.92e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 1 | 36 | 7.92e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 23 | 7.93e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 3 | 147 | 7.96e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 1 | 60 | 7.96e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 2 | 52 | 7.96e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 2 | 52 | 7.96e-01 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 1 | 18 | 7.96e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 2 | 51 | 7.96e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 2 | 158 | 7.96e-01 |
| GO:BP | GO:0051607 | defense response to virus | 2 | 202 | 7.96e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 7.96e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 7.96e-01 |
| GO:BP | GO:0007616 | long-term memory | 1 | 31 | 7.96e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 7.97e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 1 | 39 | 7.97e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 1 | 28 | 7.97e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 2 | 67 | 7.97e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 2 | 202 | 7.97e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 1 | 54 | 7.97e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 7.97e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 1 | 22 | 7.97e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 1 | 39 | 7.98e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 7.98e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 8 | 199 | 7.98e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 7.98e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 1 | 61 | 8.00e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 1 | 57 | 8.00e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 3 | 422 | 8.01e-01 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 1 | 55 | 8.01e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 3 | 85 | 8.01e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 1 | 37 | 8.03e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 1 | 44 | 8.03e-01 |
| GO:BP | GO:0021700 | developmental maturation | 1 | 211 | 8.04e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 8.06e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 1 | 353 | 8.06e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 1 | 30 | 8.06e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 1 | 31 | 8.06e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 1 | 62 | 8.06e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 2 | 1684 | 8.07e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 8.07e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 2 | 64 | 8.07e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 3 | 166 | 8.07e-01 |
| GO:BP | GO:0019079 | viral genome replication | 3 | 101 | 8.08e-01 |
| GO:BP | GO:0070925 | organelle assembly | 1 | 817 | 8.09e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 8.10e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 6 | 143 | 8.11e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 1 | 30 | 8.11e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 8.11e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 1 | 46 | 8.12e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 8.12e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 8.12e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 4 | 357 | 8.12e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 1 | 28 | 8.13e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 1 | 26 | 8.13e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 1 | 37 | 8.13e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 1 | 39 | 8.14e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 2 | 369 | 8.15e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 1 | 64 | 8.16e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 2 | 65 | 8.17e-01 |
| GO:BP | GO:0006605 | protein targeting | 2 | 307 | 8.17e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 1 | 42 | 8.17e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 4 | 137 | 8.17e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 4 | 116 | 8.17e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 5 | 275 | 8.17e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 1 | 50 | 8.18e-01 |
| GO:BP | GO:0000910 | cytokinesis | 6 | 164 | 8.18e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 6 | 194 | 8.18e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 10 | 252 | 8.18e-01 |
| GO:BP | GO:0007568 | aging | 1 | 115 | 8.18e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 1 | 39 | 8.18e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 1 | 43 | 8.19e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 2 | 66 | 8.19e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 7 | 178 | 8.20e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 1 | 66 | 8.20e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 8.20e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 1 | 38 | 8.20e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 1 | 28 | 8.20e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 3 | 97 | 8.20e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 1 | 29 | 8.20e-01 |
| GO:BP | GO:1902074 | response to salt | 5 | 271 | 8.20e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 1 | 27 | 8.21e-01 |
| GO:BP | GO:0042592 | homeostatic process | 1 | 1159 | 8.22e-01 |
| GO:BP | GO:1990868 | response to chemokine | 1 | 37 | 8.23e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 2 | 38 | 8.23e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 1 | 37 | 8.23e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 8.23e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 2 | 1836 | 8.24e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 1 | 31 | 8.24e-01 |
| GO:BP | GO:0006821 | chloride transport | 1 | 58 | 8.24e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 1 | 31 | 8.24e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 1 | 28 | 8.24e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 5 | 217 | 8.24e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 1 | 41 | 8.24e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 8.24e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 1 | 80 | 8.24e-01 |
| GO:BP | GO:0048284 | organelle fusion | 2 | 135 | 8.25e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 8 | 407 | 8.25e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 1 | 32 | 8.25e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 2 | 155 | 8.25e-01 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 1 | 15 | 8.27e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 1 | 14 | 8.27e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 8.27e-01 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 1 | 20 | 8.27e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 1 | 32 | 8.27e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 1 | 64 | 8.27e-01 |
| GO:BP | GO:0043200 | response to amino acid | 3 | 95 | 8.28e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 3 | 91 | 8.29e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 2 | 131 | 8.30e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 8.30e-01 |
| GO:BP | GO:0016485 | protein processing | 1 | 183 | 8.30e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 1 | 64 | 8.30e-01 |
| GO:BP | GO:0015833 | peptide transport | 6 | 179 | 8.30e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 8.32e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 2 | 1555 | 8.32e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 2 | 29 | 8.32e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 4 | 111 | 8.33e-01 |
| GO:BP | GO:0009615 | response to virus | 5 | 275 | 8.33e-01 |
| GO:BP | GO:0048477 | oogenesis | 1 | 64 | 8.35e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 8.35e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 48 | 8.36e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 1 | 61 | 8.36e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 1 | 87 | 8.38e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 1 | 57 | 8.38e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 1 | 46 | 8.40e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 2 | 116 | 8.41e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 1 | 51 | 8.41e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 1 | 32 | 8.41e-01 |
| GO:BP | GO:0042886 | amide transport | 4 | 235 | 8.41e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 1 | 43 | 8.41e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 1 | 316 | 8.41e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 1 | 66 | 8.42e-01 |
| GO:BP | GO:0048278 | vesicle docking | 1 | 57 | 8.42e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 1 | 60 | 8.43e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 2 | 67 | 8.44e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 1 | 52 | 8.45e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 8.45e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 3 | 173 | 8.45e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 1 | 97 | 8.46e-01 |
| GO:BP | GO:0043473 | pigmentation | 2 | 92 | 8.47e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 8.48e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 1 | 44 | 8.48e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 3 | 92 | 8.48e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 1 | 48 | 8.49e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 40 | 8.53e-01 |
| GO:BP | GO:0045010 | actin nucleation | 1 | 50 | 8.53e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 2 | 60 | 8.53e-01 |
| GO:BP | GO:0009314 | response to radiation | 11 | 352 | 8.53e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 2 | 151 | 8.53e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 1 | 56 | 8.53e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 1 | 52 | 8.53e-01 |
| GO:BP | GO:0021549 | cerebellum development | 3 | 78 | 8.55e-01 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 1 | 42 | 8.55e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 2 | 56 | 8.55e-01 |
| GO:BP | GO:0016358 | dendrite development | 3 | 202 | 8.55e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 1 | 101 | 8.56e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 1 | 49 | 8.56e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 8.57e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 2 | 124 | 8.58e-01 |
| GO:BP | GO:0015918 | sterol transport | 1 | 92 | 8.59e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 2 | 161 | 8.60e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 4 | 212 | 8.60e-01 |
| GO:BP | GO:0044057 | regulation of system process | 1 | 405 | 8.61e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 3 | 101 | 8.62e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 34 | 8.62e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 7 | 373 | 8.62e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 8.62e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 8 | 421 | 8.63e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 1 | 45 | 8.63e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 8.63e-01 |
| GO:BP | GO:0042551 | neuron maturation | 1 | 33 | 8.63e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 8.63e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 6 | 301 | 8.63e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 1 | 102 | 8.63e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 1 | 48 | 8.63e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 8.64e-01 |
| GO:BP | GO:0051310 | metaphase plate congression | 1 | 69 | 8.64e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 1 | 103 | 8.65e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 1 | 49 | 8.65e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 1 | 32 | 8.65e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 1 | 42 | 8.65e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 2 | 84 | 8.65e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 2 | 55 | 8.65e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 6 | 138 | 8.65e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 1 | 45 | 8.66e-01 |
| GO:BP | GO:0007051 | spindle organization | 5 | 188 | 8.67e-01 |
| GO:BP | GO:0051235 | maintenance of location | 5 | 260 | 8.70e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 1 | 29 | 8.70e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 1 | 115 | 8.70e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 1 | 40 | 8.70e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 4 | 75 | 8.70e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 2 | 133 | 8.72e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 3 | 94 | 8.72e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 58 | 8.72e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 28 | 8.72e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 8.73e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 8.73e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 8.73e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 70 | 8.74e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 8.74e-01 |
| GO:BP | GO:0051225 | spindle assembly | 3 | 120 | 8.74e-01 |
| GO:BP | GO:0006400 | tRNA modification | 1 | 90 | 8.74e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 1 | 122 | 8.74e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 2 | 92 | 8.74e-01 |
| GO:BP | GO:0015844 | monoamine transport | 2 | 45 | 8.74e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 8.75e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 3 | 200 | 8.76e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 1 | 46 | 8.77e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 2 | 93 | 8.77e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 843 | 8.77e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 1 | 43 | 8.77e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 8.77e-01 |
| GO:BP | GO:0051604 | protein maturation | 7 | 259 | 8.77e-01 |
| GO:BP | GO:0001508 | action potential | 3 | 105 | 8.77e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 8.77e-01 |
| GO:BP | GO:0022900 | electron transport chain | 2 | 142 | 8.78e-01 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 1 | 40 | 8.78e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 8.78e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 8.78e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 2 | 57 | 8.78e-01 |
| GO:BP | GO:0051302 | regulation of cell division | 1 | 145 | 8.79e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 8 | 429 | 8.80e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 1 | 269 | 8.80e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 1 | 82 | 8.81e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 2 | 105 | 8.81e-01 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 1 | 19 | 8.81e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 3 | 170 | 8.82e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 3 | 105 | 8.82e-01 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 1 | 44 | 8.82e-01 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 1 | 44 | 8.82e-01 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 1 | 44 | 8.82e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 8.83e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 8.85e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 1 | 49 | 8.85e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 8.85e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1 | 54 | 8.85e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 75 | 8.85e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 1 | 53 | 8.86e-01 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 1 | 45 | 8.88e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 8.88e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 1 | 80 | 8.88e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 6 | 285 | 8.88e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 1 | 46 | 8.88e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 8.92e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 8.93e-01 |
| GO:BP | GO:0061025 | membrane fusion | 2 | 137 | 8.93e-01 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 1 | 46 | 8.93e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 1 | 49 | 8.93e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 1 | 165 | 8.93e-01 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 1 | 40 | 8.95e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 1 | 41 | 8.95e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 2 | 191 | 8.95e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 2 | 119 | 8.95e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 1 | 167 | 8.96e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 1 | 54 | 8.96e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 8.96e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 2 | 142 | 8.97e-01 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 2 | 81 | 8.97e-01 |
| GO:BP | GO:0006955 | immune response | 4 | 882 | 8.97e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1 | 1226 | 8.97e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 3 | 242 | 8.97e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 1 | 48 | 8.97e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 2 | 129 | 8.97e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 1 | 79 | 8.97e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 8.97e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 8.97e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 3 | 86 | 8.97e-01 |
| GO:BP | GO:0007596 | blood coagulation | 3 | 150 | 8.97e-01 |
| GO:BP | GO:0006865 | amino acid transport | 1 | 102 | 8.97e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 1 | 163 | 8.97e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 1 | 46 | 8.98e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 1 | 40 | 8.98e-01 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 1 | 75 | 8.98e-01 |
| GO:BP | GO:0060271 | cilium assembly | 1 | 292 | 8.98e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 8.99e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 1 | 88 | 8.99e-01 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 1 | 47 | 8.99e-01 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 1 | 47 | 8.99e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 8 | 222 | 8.99e-01 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 1 | 47 | 8.99e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 1 | 74 | 8.99e-01 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 1 | 47 | 8.99e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 3 | 100 | 8.99e-01 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 9.00e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 7 | 246 | 9.00e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 1 | 63 | 9.00e-01 |
| GO:BP | GO:0006897 | endocytosis | 3 | 527 | 9.01e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 2 | 88 | 9.01e-01 |
| GO:BP | GO:0016043 | cellular component organization | 1 | 5076 | 9.01e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 9.03e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 1 | 36 | 9.03e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 3 | 214 | 9.03e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 1 | 50 | 9.03e-01 |
| GO:BP | GO:0006497 | protein lipidation | 1 | 89 | 9.03e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 1 | 39 | 9.03e-01 |
| GO:BP | GO:0001510 | RNA methylation | 2 | 87 | 9.03e-01 |
| GO:BP | GO:0050817 | coagulation | 3 | 153 | 9.03e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 1 | 68 | 9.05e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 1 | 112 | 9.05e-01 |
| GO:BP | GO:0007599 | hemostasis | 3 | 153 | 9.05e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 9.05e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 46 | 9.06e-01 |
| GO:BP | GO:0140029 | exocytic process | 1 | 69 | 9.07e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 4 | 121 | 9.07e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 2 | 81 | 9.07e-01 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 1 | 48 | 9.07e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 1 | 369 | 9.07e-01 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 1 | 48 | 9.07e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 82 | 9.08e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 3 | 98 | 9.08e-01 |
| GO:BP | GO:0071702 | organic substance transport | 2 | 2116 | 9.10e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 2 | 89 | 9.10e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 2 | 91 | 9.10e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1 | 5274 | 9.10e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 2 | 248 | 9.10e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 2 | 96 | 9.11e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 1 | 40 | 9.11e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 1 | 85 | 9.11e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 2 | 72 | 9.11e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 3 | 111 | 9.12e-01 |
| GO:BP | GO:0044782 | cilium organization | 1 | 313 | 9.12e-01 |
| GO:BP | GO:0050000 | chromosome localization | 1 | 82 | 9.13e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 1 | 92 | 9.13e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 9.14e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 1 | 90 | 9.14e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 2 | 211 | 9.14e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 2 | 73 | 9.14e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 1 | 70 | 9.14e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 2 | 73 | 9.17e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 9.18e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 1 | 180 | 9.19e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 9.19e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 2 | 92 | 9.19e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 8 | 532 | 9.19e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 2 | 93 | 9.19e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 1 | 88 | 9.19e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 9.19e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 1 | 39 | 9.19e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 1 | 66 | 9.20e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 2 | 154 | 9.20e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 1 | 149 | 9.20e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 1 | 60 | 9.20e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 1 | 88 | 9.20e-01 |
| GO:BP | GO:0009611 | response to wounding | 7 | 415 | 9.20e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 9.20e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 1 | 52 | 9.20e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 1 | 100 | 9.21e-01 |
| GO:BP | GO:0022406 | membrane docking | 1 | 84 | 9.21e-01 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 1 | 54 | 9.21e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 9.21e-01 |
| GO:BP | GO:0007286 | spermatid development | 3 | 105 | 9.21e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 1 | 51 | 9.21e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 1 | 67 | 9.21e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 1 | 51 | 9.21e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 9.21e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 2 | 64 | 9.21e-01 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 1 | 70 | 9.21e-01 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 2 | 79 | 9.21e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 4 | 701 | 9.21e-01 |
| GO:BP | GO:0042060 | wound healing | 6 | 320 | 9.22e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 1 | 64 | 9.23e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 1 | 55 | 9.23e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 1 | 58 | 9.23e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 1 | 47 | 9.23e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 2 | 72 | 9.24e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 1 | 35 | 9.25e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 1 | 49 | 9.25e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 9.26e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 1 | 77 | 9.26e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 2 | 84 | 9.26e-01 |
| GO:BP | GO:0051301 | cell division | 13 | 570 | 9.27e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 1 | 75 | 9.28e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 1 | 74 | 9.28e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 10 | 369 | 9.28e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 11 | 311 | 9.29e-01 |
| GO:BP | GO:0007292 | female gamete generation | 1 | 103 | 9.30e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 3 | 179 | 9.30e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 2 | 94 | 9.31e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 1 | 68 | 9.31e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 1 | 51 | 9.32e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 9 | 230 | 9.32e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 4 | 2466 | 9.33e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 2 | 49 | 9.34e-01 |
| GO:BP | GO:0098657 | import into cell | 2 | 166 | 9.34e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 9.35e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 1 | 69 | 9.36e-01 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 1 | 58 | 9.36e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 2 | 158 | 9.37e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 9.39e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 2 | 53 | 9.39e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 2 | 68 | 9.40e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 9.40e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 1 | 51 | 9.40e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 9.42e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 115 | 9.42e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 2 | 104 | 9.42e-01 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 1 | 57 | 9.42e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 9.44e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 1 | 47 | 9.44e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 1 | 99 | 9.45e-01 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 1 | 61 | 9.45e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 1 | 83 | 9.48e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 5 | 448 | 9.49e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 9.50e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 9.51e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 9.51e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 9.51e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 1 | 35 | 9.53e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 1 | 43 | 9.54e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 5 | 552 | 9.57e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 4 | 2724 | 9.58e-01 |
| GO:BP | GO:0051668 | localization within membrane | 6 | 605 | 9.58e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 1 | 42 | 9.58e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 3 | 123 | 9.58e-01 |
| GO:BP | GO:0009451 | RNA modification | 2 | 162 | 9.59e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 1 | 77 | 9.59e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 3 | 78 | 9.61e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 1 | 93 | 9.61e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 9 | 251 | 9.61e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 9 | 242 | 9.61e-01 |
| GO:BP | GO:0002253 | activation of immune response | 10 | 272 | 9.61e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 9.61e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 2 | 245 | 9.61e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 1 | 43 | 9.61e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 9.61e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 1 | 43 | 9.61e-01 |
| GO:BP | GO:0051641 | cellular localization | 2 | 2810 | 9.62e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 1 | 211 | 9.62e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 2 | 265 | 9.62e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 3 | 126 | 9.62e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 9.63e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 2 | 225 | 9.63e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 1 | 53 | 9.64e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 1 | 47 | 9.65e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 1 | 114 | 9.68e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 5 | 523 | 9.69e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 9.69e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 1 | 102 | 9.69e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 4 | 273 | 9.69e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 1 | 90 | 9.69e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 2 | 216 | 9.69e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 9.69e-01 |
| GO:BP | GO:0015849 | organic acid transport | 2 | 217 | 9.69e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 3 | 251 | 9.70e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 1 | 51 | 9.70e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 9.70e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 9.71e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 2 | 86 | 9.71e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 2 | 86 | 9.71e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 9.72e-01 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 1 | 71 | 9.72e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 122 | 9.73e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 3 | 114 | 9.73e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 1 | 96 | 9.74e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 9.74e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 1 | 170 | 9.74e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1 | 135 | 9.74e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 9.74e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 2 | 93 | 9.74e-01 |
| GO:BP | GO:0120193 | tight junction organization | 1 | 52 | 9.74e-01 |
| GO:BP | GO:0006954 | inflammatory response | 7 | 429 | 9.75e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 1 | 98 | 9.75e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 1 | 78 | 9.75e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 1 | 75 | 9.75e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 3 | 1496 | 9.78e-01 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 3 | 148 | 9.78e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 9.78e-01 |
| GO:BP | GO:0051304 | chromosome separation | 1 | 76 | 9.81e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 4 | 357 | 9.82e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 2 | 120 | 9.82e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 9.82e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 142 | 9.82e-01 |
| GO:BP | GO:0030258 | lipid modification | 4 | 162 | 9.82e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 4 | 425 | 9.82e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 2 | 130 | 9.83e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 9.83e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 1 | 104 | 9.83e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 2 | 145 | 9.83e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 1 | 104 | 9.83e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 1 | 72 | 9.84e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 4 | 362 | 9.84e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 2 | 98 | 9.84e-01 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 1 | 94 | 9.84e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 9.84e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 1 | 88 | 9.84e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 1 | 104 | 9.85e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 9.86e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 1 | 48 | 9.86e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 1 | 48 | 9.86e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 4 | 351 | 9.86e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 1 | 111 | 9.87e-01 |
| GO:BP | GO:0051648 | vesicle localization | 2 | 192 | 9.87e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 112 | 9.87e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 1 | 115 | 9.87e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 9.87e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 1 | 58 | 9.88e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 6 | 188 | 9.88e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 9.89e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 9.89e-01 |
| GO:BP | GO:0007127 | meiosis I | 1 | 77 | 9.89e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 2 | 317 | 9.90e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 70 | 9.90e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 9.90e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 9.91e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 1 | 113 | 9.91e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 2 | 109 | 9.91e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 1 | 64 | 9.91e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 2 | 209 | 9.91e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 2 | 135 | 9.91e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 174 | 9.91e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 1 | 81 | 9.91e-01 |
| GO:BP | GO:0030001 | metal ion transport | 1 | 562 | 9.91e-01 |
| GO:BP | GO:0003014 | renal system process | 1 | 78 | 9.91e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 61 | 9.91e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 9.91e-01 |
| GO:BP | GO:0015711 | organic anion transport | 2 | 264 | 9.91e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 9.91e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 9.91e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 61 | 9.91e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 3 | 128 | 9.92e-01 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 1 | 90 | 9.92e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 3 | 295 | 9.93e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 9.93e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 1 | 175 | 9.94e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 9.94e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 1 | 784 | 9.95e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 1 | 106 | 9.95e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 740 | 9.96e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 9.96e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 9.97e-01 |
| GO:BP | GO:0000280 | nuclear division | 6 | 344 | 9.97e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 1 | 56 | 9.97e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 998 | 9.97e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 1 | 56 | 9.97e-01 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 1 | 16 | 9.97e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 4 | 401 | 9.97e-01 |
| GO:BP | GO:0007338 | single fertilization | 1 | 70 | 9.97e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 1 | 75 | 9.98e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 9.98e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 4 | 408 | 9.99e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 11 | 337 | 9.99e-01 |
| GO:BP | GO:0061024 | membrane organization | 6 | 671 | 9.99e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 5 | 223 | 9.99e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 9.99e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 2 | 1335 | 9.99e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 5 | 224 | 9.99e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 74 | 9.99e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 3 | 475 | 9.99e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 2 | 162 | 9.99e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 74 | 9.99e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 1 | 669 | 9.99e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 1 | 106 | 1.00e+00 |
| GO:BP | GO:0006811 | monoatomic ion transport | 1 | 775 | 1.00e+00 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 1.00e+00 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 230 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0016054 | organic acid catabolic process | 1 | 187 | 1.00e+00 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 9 | 506 | 1.00e+00 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 1.00e+00 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 1.00e+00 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 138 | 1.00e+00 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 2 | 194 | 1.00e+00 |
| GO:BP | GO:0042330 | taxis | 3 | 389 | 1.00e+00 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 213 | 1.00e+00 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 1 | 133 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 1.00e+00 |
| GO:BP | GO:0006887 | exocytosis | 1 | 256 | 1.00e+00 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 250 | 1.00e+00 |
| GO:BP | GO:0007018 | microtubule-based movement | 1 | 300 | 1.00e+00 |
| GO:BP | GO:0007017 | microtubule-based process | 5 | 760 | 1.00e+00 |
| GO:BP | GO:0010970 | transport along microtubule | 1 | 162 | 1.00e+00 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 2 | 170 | 1.00e+00 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 1 | 154 | 1.00e+00 |
| GO:BP | GO:0010876 | lipid localization | 1 | 310 | 1.00e+00 |
| GO:BP | GO:0006869 | lipid transport | 1 | 272 | 1.00e+00 |
| GO:BP | GO:0006956 | complement activation | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 1.00e+00 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 3 | 389 | 1.00e+00 |
| GO:BP | GO:0006935 | chemotaxis | 3 | 389 | 1.00e+00 |
| GO:BP | GO:0042742 | defense response to bacterium | 1 | 95 | 1.00e+00 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 1.00e+00 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 3 | 632 | 1.00e+00 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 1.00e+00 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 1 | 145 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 1.00e+00 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 1.00e+00 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 4 | 151 | 1.00e+00 |
| GO:BP | GO:0060326 | cell chemotaxis | 1 | 162 | 1.00e+00 |
| GO:BP | GO:0099111 | microtubule-based transport | 1 | 191 | 1.00e+00 |
| GO:BP | GO:0048285 | organelle fission | 6 | 388 | 1.00e+00 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 1 | 376 | 1.00e+00 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 149 | 1.00e+00 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 1.00e+00 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 1.00e+00 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 1 | 187 | 1.00e+00 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 1.00e+00 |
| GO:BP | GO:0009566 | fertilization | 1 | 93 | 1.00e+00 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 681 | 1.00e+00 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 1 | 106 | 1.00e+00 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 378 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0051640 | organelle localization | 6 | 503 | 1.00e+00 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 1.00e+00 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 1.00e+00 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 1 | 111 | 1.00e+00 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 1.00e+00 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 1.00e+00 |
| GO:BP | GO:0007601 | visual perception | 2 | 96 | 1.00e+00 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 1 | 1238 | 1.00e+00 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 1.00e+00 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 1.00e+00 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 2 | 540 | 1.00e+00 |
| GO:BP | GO:0003341 | cilium movement | 1 | 97 | 1.00e+00 |
| GO:BP | GO:0044281 | small molecule metabolic process | 7 | 1389 | 1.00e+00 |
| GO:BP | GO:0044282 | small molecule catabolic process | 1 | 271 | 1.00e+00 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 1 | 403 | 1.00e+00 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 3 | 563 | 1.00e+00 |
| GO:BP | GO:0045055 | regulated exocytosis | 1 | 155 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 4 | 162 | 1.00e+00 |
| GO:BP | GO:0045087 | innate immune response | 3 | 488 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 1.00e+00 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 2 | 517 | 1.00e+00 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 1.00e+00 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 2 | 97 | 1.00e+00 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 1 | 157 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 66 | 415 | 1.78e-17 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 15 | 138 | 1.18e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 1 | 64 | 1.47e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 1 | 103 | 1.47e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 1 | 35 | 1.47e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 1 | 83 | 1.47e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 1 | 49 | 1.47e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 1.47e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 1.47e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 1.47e-01 |
| KEGG | KEGG:05224 | Breast cancer | 9 | 117 | 1.47e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 1.47e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 1 | 41 | 1.47e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 6 | 51 | 1.47e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 8 | 84 | 1.47e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 1 | 87 | 1.47e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 1.47e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 1.47e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 1 | 77 | 1.47e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 1 | 98 | 1.47e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 1.47e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 1.47e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 1 | 48 | 1.47e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 1 | 77 | 1.47e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 1.47e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 1 | 38 | 1.47e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 1.47e-01 |
| KEGG | KEGG:04924 | Renin secretion | 1 | 50 | 1.47e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 1.47e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 1 | 97 | 1.47e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 1 | 128 | 1.50e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 11 | 117 | 1.50e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 1 | 121 | 1.50e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 1.52e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 1 | 133 | 1.53e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 1 | 106 | 1.56e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 1 | 172 | 1.59e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 10 | 104 | 1.59e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 2 | 4 | 1.59e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 1.59e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 1 | 180 | 1.59e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 1.59e-01 |
| KEGG | KEGG:05034 | Alcoholism | 1 | 122 | 1.59e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 1 | 176 | 1.59e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 1 | 142 | 1.59e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 1.62e-01 |
| KEGG | KEGG:05020 | Prion disease | 1 | 220 | 1.86e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 11 | 145 | 2.00e-01 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 2.00e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 2.01e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 1 | 272 | 2.08e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 2.17e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 8 | 124 | 2.36e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 2.61e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 5 | 56 | 3.18e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 3.43e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 5 | 49 | 3.43e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 2 | 14 | 3.56e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 6 | 84 | 3.67e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 4 | 56 | 3.67e-01 |
| KEGG | KEGG:05164 | Influenza A | 1 | 103 | 3.71e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 1 | 70 | 3.71e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 1 | 161 | 3.71e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 4 | 65 | 3.71e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 4.30e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 9 | 134 | 4.53e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 4 | 66 | 4.53e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 4 | 70 | 4.92e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 5.17e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 4 | 57 | 5.22e-01 |
| KEGG | KEGG:04146 | Peroxisome | 1 | 68 | 5.25e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 5.41e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 1 | 48 | 5.41e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 5.42e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 1 | 12 | 5.45e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 3 | 35 | 5.53e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 1 | 150 | 5.53e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 3 | 40 | 5.53e-01 |
| KEGG | KEGG:04520 | Adherens junction | 4 | 67 | 5.59e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 5.59e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 2 | 53 | 6.01e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 6.16e-01 |
| KEGG | KEGG:04210 | Apoptosis | 7 | 118 | 6.16e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 3 | 86 | 6.43e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 1 | 44 | 6.67e-01 |
| KEGG | KEGG:04530 | Tight junction | 3 | 137 | 6.67e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 3 | 55 | 6.69e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 2 | 76 | 6.70e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 2 | 25 | 6.70e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 1 | 99 | 6.70e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 1 | 106 | 6.74e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 5 | 112 | 6.91e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 1 | 32 | 6.98e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 4 | 75 | 7.23e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 1 | 19 | 7.79e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 16 | 409 | 7.79e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 7.79e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 5 | 114 | 8.02e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 1 | 28 | 8.31e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 1 | 25 | 8.31e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 8.31e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 5 | 153 | 8.31e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 2 | 110 | 8.31e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 1 | 38 | 8.38e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 8.38e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 4 | 112 | 8.38e-01 |
| KEGG | KEGG:05133 | Pertussis | 2 | 45 | 8.49e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 6 | 134 | 8.49e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 3 | 69 | 8.49e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 8.65e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 2 | 115 | 8.65e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 1 | 24 | 8.65e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 3 | 67 | 8.65e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 8.65e-01 |
| KEGG | KEGG:05218 | Melanoma | 3 | 58 | 8.65e-01 |
| KEGG | KEGG:03040 | Spliceosome | 3 | 132 | 8.65e-01 |
| KEGG | KEGG:04110 | Cell cycle | 6 | 151 | 8.65e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 8.65e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 1 | 60 | 8.65e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 3 | 75 | 8.65e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 3 | 65 | 8.65e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 5 | 109 | 8.65e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 4 | 91 | 8.65e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 8.65e-01 |
| KEGG | KEGG:05142 | Chagas disease | 3 | 75 | 8.65e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 8.65e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 8.65e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 3 | 72 | 8.65e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 1 | 121 | 8.65e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 3 | 56 | 8.65e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 6 | 88 | 8.65e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 1 | 58 | 8.65e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 2 | 117 | 8.65e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 2 | 44 | 8.65e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 3 | 60 | 8.65e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 8.65e-01 |
| KEGG | KEGG:00000 | KEGG root term | 6 | 5558 | 8.65e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 2 | 131 | 8.65e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 2 | 20 | 8.66e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 4 | 76 | 8.66e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 8.66e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 3 | 89 | 8.66e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 8.75e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 8.95e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 9.08e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 1 | 85 | 9.25e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 1 | 84 | 9.27e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 9.27e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 1 | 91 | 9.27e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 6 | 214 | 9.27e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 1 | 26 | 9.27e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 1 | 27 | 9.27e-01 |
| KEGG | KEGG:04144 | Endocytosis | 3 | 232 | 9.27e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 2 | 115 | 9.42e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 3 | 87 | 9.42e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 9.42e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 1 | 52 | 9.51e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 5 | 145 | 9.52e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 1 | 35 | 9.52e-01 |
| KEGG | KEGG:05131 | Shigellosis | 2 | 210 | 9.56e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 4 | 135 | 9.56e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 1 | 26 | 9.56e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 1 | 31 | 9.56e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 1 | 19 | 9.58e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 1 | 110 | 9.60e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 1 | 31 | 9.60e-01 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 9.63e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 4 | 98 | 9.63e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 2 | 165 | 9.63e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 1 | 164 | 9.63e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 1 | 49 | 9.63e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 5 | 171 | 9.63e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 3 | 71 | 9.63e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 4 | 115 | 9.63e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 9.63e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 1 | 46 | 9.63e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 1 | 79 | 9.63e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 1 | 39 | 9.63e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 1 | 41 | 9.63e-01 |
| KEGG | KEGG:04217 | Necroptosis | 2 | 106 | 9.63e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 1 | 63 | 9.63e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 4 | 315 | 9.63e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 1 | 55 | 9.63e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 2 | 97 | 9.63e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 4 | 385 | 9.63e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 1 | 49 | 9.67e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 3 | 157 | 9.67e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 1 | 49 | 9.67e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 1 | 65 | 9.67e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 9.67e-01 |
| KEGG | KEGG:04730 | Long-term depression | 1 | 42 | 9.67e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 2 | 63 | 9.67e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 1 | 46 | 9.67e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 9.67e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 1 | 63 | 9.67e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 1 | 74 | 9.67e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 9.67e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 2 | 133 | 9.67e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 2 | 76 | 9.67e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 6 | 179 | 9.67e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 1 | 53 | 9.67e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 9.67e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 9.67e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 1 | 74 | 9.67e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 8 | 149 | 9.67e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 2 | 186 | 9.67e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 9.72e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 2 | 107 | 9.72e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 9.72e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 9.72e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 1 | 170 | 9.72e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 9.72e-01 |
| KEGG | KEGG:04611 | Platelet activation | 2 | 89 | 9.72e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 1 | 86 | 9.72e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 2 | 226 | 9.72e-01 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 2 | 169 | 9.94e-01 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 1 | 61 | 9.94e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 1 | 94 | 9.94e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 2 | 110 | 9.94e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 9.94e-01 |
DNR specific genes at 3 hours
How they may relate to 24 hour genes using GO analysis3 hours specifically (see venn below too):
total24 <-list(sigVDA24$ENTREZID,sigVDX24$ENTREZID,sigVEP24$ENTREZID,sigVMT24$ENTREZID)
total3 <- list(sigVDA3$ENTREZID,sigVDX3$ENTREZID,sigVEP3$ENTREZID,sigVMT3$ENTREZID)
list3totvenn3 <- VennDiagram::get.venn.partitions(total3)
list24totvenn24 <- VennDiagram::get.venn.partitions(total24)
Daun3sp <- list3totvenn3$..values..[[15]]
Daun24sp <- list24totvenn24$..values..[[15]]
# DAspgostres3 <- gost(query = Daun3sp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(DAspgostres3,"data/DEG-GO/DAspgostres3.RDS")
DAspgostres3 <- readRDS("data/DEG-GO/DAspgostres3.RDS")
# DAspp3 <- gostplot(DAspgostres3, capped = FALSE, interactive = TRUE)
# DAspp3
DAsptable3 <- DAspgostres3$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>% DNR specific genes at 24 hours
###no enrichment
# Daun24spgost <- gost(query = Daun24sp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(Daun24spgost,"data/DEG-GO/Daun24spgost.RDS")
Daun24spgost <- readRDS("data/DEG-GO/Daun24spgost.RDS")
#
# DAspp24h<- gostplot(Daun24spgost, capped = FALSE, interactive = TRUE)
# DAspp24h
DAspp24h <- Daun24spgost$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))# %>%
DAspp24h %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0061709 | reticulophagy | 5 | 17 | 2.17e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 10 | 52 | 2.52e-01 |
| GO:BP | GO:0007290 | spermatid nucleus elongation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 5 | 59 | 3.38e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 15 | 137 | 3.38e-01 |
| GO:BP | GO:1903168 | positive regulation of pyrroline-5-carboxylate reductase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0061960 | regulation of heme oxygenase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 3 | 31 | 3.38e-01 |
| GO:BP | GO:1903195 | regulation of L-dopa biosynthetic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 2 | 9 | 3.38e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 2 | 9 | 3.38e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 1 | 36 | 3.38e-01 |
| GO:BP | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport | 1 | 4 | 3.38e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 2 | 11 | 3.38e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 6 | 222 | 3.38e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 26 | 340 | 3.38e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 3 | 12 | 3.38e-01 |
| GO:BP | GO:1903197 | positive regulation of L-dopa biosynthetic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0007614 | short-term memory | 2 | 9 | 3.38e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 4 | 19 | 3.38e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 2 | 5 | 3.38e-01 |
| GO:BP | GO:1903190 | glyoxal catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903189 | glyoxal metabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903185 | L-dopa biosynthetic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 11 | 92 | 3.38e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 6 | 70 | 3.38e-01 |
| GO:BP | GO:1903141 | negative regulation of establishment of endothelial barrier | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 1 | 11 | 3.38e-01 |
| GO:BP | GO:1903073 | negative regulation of death-inducing signaling complex assembly | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 2 | 10 | 3.38e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 3 | 122 | 3.38e-01 |
| GO:BP | GO:1903176 | regulation of tyrosine 3-monooxygenase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903178 | positive regulation of tyrosine 3-monooxygenase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903072 | regulation of death-inducing signaling complex assembly | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 2 | 50 | 3.38e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0061753 | substrate localization to autophagosome | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 1 | 8 | 3.38e-01 |
| GO:BP | GO:1903184 | L-dopa metabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 3 | 58 | 3.38e-01 |
| GO:BP | GO:0061912 | selective autophagy | 7 | 90 | 3.38e-01 |
| GO:BP | GO:1903167 | regulation of pyrroline-5-carboxylate reductase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903122 | negative regulation of TRAIL-activated apoptotic signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 2 | 18 | 3.38e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 3 | 79 | 3.38e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 23 | 299 | 3.38e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 3 | 21 | 3.38e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 8 | 123 | 3.38e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 4 | 213 | 3.38e-01 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 1 | 74 | 3.38e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 2 | 22 | 3.38e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 1 | 9 | 3.38e-01 |
| GO:BP | GO:1902081 | negative regulation of calcium ion import into sarcoplasmic reticulum | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 9 | 80 | 3.38e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 8 | 58 | 3.38e-01 |
| GO:BP | GO:0032402 | melanosome transport | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 1 | 20 | 3.38e-01 |
| GO:BP | GO:0032400 | melanosome localization | 1 | 22 | 3.38e-01 |
| GO:BP | GO:0032392 | DNA geometric change | 1 | 80 | 3.38e-01 |
| GO:BP | GO:0009249 | protein lipoylation | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 2 | 39 | 3.38e-01 |
| GO:BP | GO:0009256 | 10-formyltetrahydrofolate metabolic process | 2 | 6 | 3.38e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 3.38e-01 |
| GO:BP | GO:1901895 | negative regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 3 | 3.38e-01 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0009397 | folic acid-containing compound catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1901877 | negative regulation of calcium ion binding | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1901876 | regulation of calcium ion binding | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 4 | 20 | 3.38e-01 |
| GO:BP | GO:0070925 | organelle assembly | 16 | 817 | 3.38e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0009444 | pyruvate oxidation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1901842 | negative regulation of high voltage-gated calcium channel activity | 2 | 4 | 3.38e-01 |
| GO:BP | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway | 1 | 3 | 3.38e-01 |
| GO:BP | GO:1902205 | regulation of interleukin-2-mediated signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1902206 | negative regulation of interleukin-2-mediated signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1902214 | regulation of interleukin-4-mediated signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0070103 | regulation of interleukin-6-mediated signaling pathway | 1 | 4 | 3.38e-01 |
| GO:BP | GO:1903198 | regulation of L-dopa decarboxylase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 1 | 30 | 3.38e-01 |
| GO:BP | GO:0032914 | positive regulation of transforming growth factor beta1 production | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 3 | 10 | 3.38e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 3 | 11 | 3.38e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 3 | 26 | 3.38e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 2 | 89 | 3.38e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 1 | 115 | 3.38e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 2 | 55 | 3.38e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 3.38e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0008015 | blood circulation | 1 | 374 | 3.38e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0070487 | monocyte aggregation | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 2 | 9 | 3.38e-01 |
| GO:BP | GO:0070494 | regulation of thrombin-activated receptor signaling pathway | 2 | 3 | 3.38e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 29 | 3.38e-01 |
| GO:BP | GO:1902233 | negative regulation of positive thymic T cell selection | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1902232 | regulation of positive thymic T cell selection | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0070495 | negative regulation of thrombin-activated receptor signaling pathway | 2 | 3 | 3.38e-01 |
| GO:BP | GO:1902227 | negative regulation of macrophage colony-stimulating factor signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1902224 | ketone body metabolic process | 2 | 7 | 3.38e-01 |
| GO:BP | GO:1902215 | negative regulation of interleukin-4-mediated signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0070327 | thyroid hormone transport | 1 | 7 | 3.38e-01 |
| GO:BP | GO:1903200 | positive regulation of L-dopa decarboxylase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903284 | positive regulation of glutathione peroxidase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 1 | 66 | 3.38e-01 |
| GO:BP | GO:0060167 | regulation of adenosine receptor signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 1 | 32 | 3.38e-01 |
| GO:BP | GO:0060168 | positive regulation of adenosine receptor signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1904273 | L-alanine import across plasma membrane | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 1 | 12 | 3.38e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 4 | 26 | 3.38e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 6 | 100 | 3.38e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 3.38e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 1 | 104 | 3.38e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 3.38e-01 |
| GO:BP | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 2 | 9 | 3.38e-01 |
| GO:BP | GO:0060192 | negative regulation of lipase activity | 3 | 13 | 3.38e-01 |
| GO:BP | GO:0035946 | mitochondrial mRNA surveillance | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0035945 | mitochondrial ncRNA surveillance | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 4 | 30 | 3.38e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 1 | 60 | 3.38e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 3.38e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 3.38e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 3 | 27 | 3.38e-01 |
| GO:BP | GO:1904021 | negative regulation of G protein-coupled receptor internalization | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1904020 | regulation of G protein-coupled receptor internalization | 1 | 3 | 3.38e-01 |
| GO:BP | GO:1904015 | cellular response to serotonin | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006713 | glucocorticoid catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036292 | DNA rewinding | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 4 | 73 | 3.38e-01 |
| GO:BP | GO:0036471 | cellular response to glyoxal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 4 | 25 | 3.38e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 2 | 7 | 3.38e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 2 | 10 | 3.38e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 4 | 214 | 3.38e-01 |
| GO:BP | GO:0006414 | translational elongation | 3 | 65 | 3.38e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0038145 | macrophage colony-stimulating factor signaling pathway | 2 | 3 | 3.38e-01 |
| GO:BP | GO:0038110 | interleukin-2-mediated signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1904736 | negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0038097 | positive regulation of mast cell activation by Fc-epsilon receptor signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1904735 | regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1904733 | negative regulation of electron transfer activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1904014 | response to serotonin | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 3 | 106 | 3.38e-01 |
| GO:BP | GO:0006478 | peptidyl-tyrosine sulfation | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 3 | 17 | 3.38e-01 |
| GO:BP | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway | 2 | 5 | 3.38e-01 |
| GO:BP | GO:0038029 | epidermal growth factor receptor signaling pathway via MAPK cascade | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0038020 | insulin receptor recycling | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0036531 | glutathione deglycation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036530 | protein deglycation, methylglyoxal removal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1904557 | L-alanine transmembrane transport | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0036529 | protein deglycation, glyoxal removal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036528 | peptidyl-lysine deglycation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036527 | peptidyl-arginine deglycation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036526 | peptidyl-cysteine deglycation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0036476 | neuron death in response to hydrogen peroxide | 2 | 5 | 3.38e-01 |
| GO:BP | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 4 | 23 | 3.38e-01 |
| GO:BP | GO:0060298 | positive regulation of sarcomere organization | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0035726 | common myeloid progenitor cell proliferation | 2 | 5 | 3.38e-01 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 2 | 8 | 3.38e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 1 | 16 | 3.38e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 1 | 63 | 3.38e-01 |
| GO:BP | GO:0097238 | cellular response to methylglyoxal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 1 | 110 | 3.38e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 2 | 28 | 3.38e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 3 | 83 | 3.38e-01 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 2 | 11 | 3.38e-01 |
| GO:BP | GO:0007032 | endosome organization | 7 | 92 | 3.38e-01 |
| GO:BP | GO:1903435 | positive regulation of constitutive secretory pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903433 | regulation of constitutive secretory pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 1 | 30 | 3.38e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 1 | 352 | 3.38e-01 |
| GO:BP | GO:0061004 | pattern specification involved in kidney development | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 1 | 20 | 3.38e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 1 | 26 | 3.38e-01 |
| GO:BP | GO:0061061 | muscle structure development | 12 | 546 | 3.38e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 12 | 149 | 3.38e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 16 | 179 | 3.38e-01 |
| GO:BP | GO:0061145 | lung smooth muscle development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1903285 | positive regulation of hydrogen peroxide catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0032328 | alanine transport | 2 | 14 | 3.38e-01 |
| GO:BP | GO:1903282 | regulation of glutathione peroxidase activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0034213 | quinolinate catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0061227 | pattern specification involved in mesonephros development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1903208 | negative regulation of hydrogen peroxide-induced neuron death | 2 | 5 | 3.38e-01 |
| GO:BP | GO:1903207 | regulation of hydrogen peroxide-induced neuron death | 2 | 5 | 3.38e-01 |
| GO:BP | GO:1903403 | negative regulation of renal phosphate excretion | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 1 | 29 | 3.38e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 2 | 7 | 3.38e-01 |
| GO:BP | GO:1903570 | regulation of protein kinase D signaling | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 2 | 23 | 3.38e-01 |
| GO:BP | GO:1903972 | regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 3 | 3.38e-01 |
| GO:BP | GO:1903969 | regulation of response to macrophage colony-stimulating factor | 1 | 3 | 3.38e-01 |
| GO:BP | GO:1903899 | positive regulation of PERK-mediated unfolded protein response | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0035601 | protein deacylation | 3 | 116 | 3.38e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 3 | 30 | 3.38e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 2 | 16 | 3.38e-01 |
| GO:BP | GO:1903801 | L-leucine import across plasma membrane | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 3.38e-01 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0035543 | positive regulation of SNARE complex assembly | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0035524 | proline transmembrane transport | 1 | 8 | 3.38e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 1 | 27 | 3.38e-01 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 3 | 9 | 3.38e-01 |
| GO:BP | GO:0035442 | dipeptide transmembrane transport | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 4 | 200 | 3.38e-01 |
| GO:BP | GO:0060532 | bronchus cartilage development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 8 | 103 | 3.38e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 5 | 60 | 3.38e-01 |
| GO:BP | GO:1903598 | positive regulation of gap junction assembly | 2 | 7 | 3.38e-01 |
| GO:BP | GO:1903596 | regulation of gap junction assembly | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 3 | 403 | 3.38e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 2 | 9 | 3.38e-01 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 1 | 3 | 3.38e-01 |
| GO:BP | GO:1903572 | positive regulation of protein kinase D signaling | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 5 | 362 | 3.38e-01 |
| GO:BP | GO:0070932 | histone H3 deacetylation | 1 | 12 | 3.38e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 1 | 7 | 3.38e-01 |
| GO:BP | GO:1901727 | positive regulation of histone deacetylase activity | 2 | 5 | 3.38e-01 |
| GO:BP | GO:0106044 | guanine deglycation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0021756 | striatum development | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0014904 | myotube cell development | 3 | 34 | 3.38e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 1 | 21 | 3.38e-01 |
| GO:BP | GO:0090091 | positive regulation of extracellular matrix disassembly | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0090094 | metanephric cap mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 7 | 59 | 3.38e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 2 | 15 | 3.38e-01 |
| GO:BP | GO:0021570 | rhombomere 4 development | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 4 | 163 | 3.38e-01 |
| GO:BP | GO:0090045 | positive regulation of deacetylase activity | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0015724 | formate transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 1 | 40 | 3.38e-01 |
| GO:BP | GO:0015734 | taurine transport | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0015735 | uronic acid transmembrane transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0015736 | hexuronate transmembrane transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0015738 | glucuronate transmembrane transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0021544 | subpallium development | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0090130 | tissue migration | 21 | 256 | 3.38e-01 |
| GO:BP | GO:0090132 | epithelium migration | 21 | 252 | 3.38e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 4 | 38 | 3.38e-01 |
| GO:BP | GO:0015797 | mannitol transmembrane transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0015798 | myo-inositol transport | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 1 | 29 | 3.38e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0015801 | aromatic amino acid transport | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0106045 | guanine deglycation, methylglyoxal removal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 2 | 39 | 3.38e-01 |
| GO:BP | GO:0120317 | sperm mitochondrial sheath assembly | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 1 | 84 | 3.38e-01 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 5 | 357 | 3.38e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 2 | 27 | 3.38e-01 |
| GO:BP | GO:0023058 | adaptation of signaling pathway | 1 | 16 | 3.38e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0010982 | regulation of high-density lipoprotein particle clearance | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0010983 | positive regulation of high-density lipoprotein particle clearance | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0010984 | regulation of lipoprotein particle clearance | 2 | 12 | 3.38e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 7 | 118 | 3.38e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 14 | 220 | 3.38e-01 |
| GO:BP | GO:0106046 | guanine deglycation, glyoxal removal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 4 | 52 | 3.38e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 31 | 427 | 3.38e-01 |
| GO:BP | GO:0072706 | response to sodium dodecyl sulfate | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072707 | cellular response to sodium dodecyl sulfate | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0022401 | negative adaptation of signaling pathway | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0021997 | neural plate axis specification | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0086092 | regulation of the force of heart contraction by cardiac conduction | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0021935 | cerebellar granule cell precursor tangential migration | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0089700 | protein kinase D signaling | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 2 | 28 | 3.38e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 4 | 94 | 3.38e-01 |
| GO:BP | GO:0140041 | cellular detoxification of methylglyoxal | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 1 | 14 | 3.38e-01 |
| GO:BP | GO:0015808 | L-alanine transport | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0016191 | synaptic vesicle uncoating | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0016197 | endosomal transport | 14 | 231 | 3.38e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 3 | 178 | 3.38e-01 |
| GO:BP | GO:0016237 | lysosomal microautophagy | 3 | 11 | 3.38e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 5 | 70 | 3.38e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 3 | 34 | 3.38e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 3 | 14 | 3.38e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0019083 | viral transcription | 4 | 48 | 3.38e-01 |
| GO:BP | GO:0097692 | histone H3-K4 monomethylation | 1 | 8 | 3.38e-01 |
| GO:BP | GO:0019068 | virion assembly | 1 | 35 | 3.38e-01 |
| GO:BP | GO:0016180 | snRNA processing | 1 | 26 | 3.38e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0097533 | cellular stress response to acid chemical | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0097532 | stress response to acid chemical | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0016570 | histone modification | 10 | 433 | 3.38e-01 |
| GO:BP | GO:0097494 | regulation of vesicle size | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 3 | 87 | 3.38e-01 |
| GO:BP | GO:0018277 | protein deamination | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0097195 | pilomotor reflex | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0097210 | response to gonadotropin-releasing hormone | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0018171 | peptidyl-cysteine oxidation | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0097213 | regulation of lysosomal membrane permeability | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 4 | 115 | 3.38e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 2 | 65 | 3.38e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 3 | 70 | 3.38e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 1 | 26 | 3.38e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 4 | 191 | 3.38e-01 |
| GO:BP | GO:0015816 | glycine transport | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0015820 | leucine transport | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0015827 | tryptophan transport | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0015829 | valine transport | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0090241 | negative regulation of histone H4 acetylation | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 1 | 16 | 3.38e-01 |
| GO:BP | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity | 3 | 12 | 3.38e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 2 | 53 | 3.38e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 2 | 28 | 3.38e-01 |
| GO:BP | GO:0046359 | butyrate catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 12 | 143 | 3.38e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 1 | 65 | 3.38e-01 |
| GO:BP | GO:0019518 | L-threonine catabolic process to glycine | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0098754 | detoxification | 1 | 91 | 3.38e-01 |
| GO:BP | GO:0016050 | vesicle organization | 4 | 320 | 3.38e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 3 | 133 | 3.38e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 3.38e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 3 | 119 | 3.38e-01 |
| GO:BP | GO:0098713 | leucine import across plasma membrane | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0019371 | cyclooxygenase pathway | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 3.38e-01 |
| GO:BP | GO:0090324 | negative regulation of oxidative phosphorylation | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 5 | 106 | 3.38e-01 |
| GO:BP | GO:0030043 | actin filament fragmentation | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0030049 | muscle filament sliding | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0071449 | cellular response to lipid hydroperoxide | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 2 | 11 | 3.38e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 2 | 11 | 3.38e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 2 | 14 | 3.38e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 2 | 14 | 3.38e-01 |
| GO:BP | GO:0010459 | negative regulation of heart rate | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 2 | 31 | 3.38e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 6 | 36 | 3.38e-01 |
| GO:BP | GO:0071549 | cellular response to dexamethasone stimulus | 4 | 24 | 3.38e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 3 | 13 | 3.38e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 2 | 13 | 3.38e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 13 | 496 | 3.38e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 13 | 313 | 3.38e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 6 | 133 | 3.38e-01 |
| GO:BP | GO:0010519 | negative regulation of phospholipase activity | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0010524 | positive regulation of calcium ion transport into cytosol | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0010526 | retrotransposon silencing | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 1 | 32 | 3.38e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 2 | 23 | 3.38e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 3 | 56 | 3.38e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 2 | 22 | 3.38e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 2 | 36 | 3.38e-01 |
| GO:BP | GO:0071701 | regulation of MAPK export from nucleus | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0031104 | dendrite regeneration | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0031048 | small ncRNA-mediated heterochromatin formation | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 16 | 277 | 3.38e-01 |
| GO:BP | GO:1900449 | regulation of glutamate receptor signaling pathway | 1 | 5 | 3.38e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 5 | 26 | 3.38e-01 |
| GO:BP | GO:0032200 | telomere organization | 1 | 166 | 3.38e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 1 | 158 | 3.38e-01 |
| GO:BP | GO:0071026 | cytoplasmic RNA surveillance | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1901551 | negative regulation of endothelial cell development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0071103 | DNA conformation change | 1 | 87 | 3.38e-01 |
| GO:BP | GO:1901494 | regulation of cysteine metabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1901373 | lipid hydroperoxide transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0071228 | cellular response to tumor cell | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 3 | 26 | 3.38e-01 |
| GO:BP | GO:1900276 | regulation of proteinase activated receptor activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1904978 | regulation of endosome organization | 2 | 5 | 3.38e-01 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 1 | 5 | 3.38e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 2 | 16 | 3.38e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 1 | 8 | 3.38e-01 |
| GO:BP | GO:0071352 | cellular response to interleukin-2 | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 2 | 21 | 3.38e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 2 | 5 | 3.38e-01 |
| GO:BP | GO:1900737 | negative regulation of phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 6 | 47 | 3.38e-01 |
| GO:BP | GO:1900625 | positive regulation of monocyte aggregation | 2 | 3 | 3.38e-01 |
| GO:BP | GO:1900623 | regulation of monocyte aggregation | 2 | 3 | 3.38e-01 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 6 | 40 | 3.38e-01 |
| GO:BP | GO:1900451 | positive regulation of glutamate receptor signaling pathway | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1901098 | positive regulation of autophagosome maturation | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0071765 | nuclear inner membrane organization | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 12 | 143 | 3.38e-01 |
| GO:BP | GO:0031038 | myosin II filament organization | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0010796 | regulation of multivesicular body size | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0072168 | specification of anterior mesonephric tubule identity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072169 | specification of posterior mesonephric tubule identity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 3 | 66 | 3.38e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 4 | 65 | 3.38e-01 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0072184 | renal vesicle progenitor cell differentiation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072185 | metanephric cap development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0072186 | metanephric cap morphogenesis | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 1 | 73 | 3.38e-01 |
| GO:BP | GO:0010849 | regulation of proton-transporting ATPase activity, rotational mechanism | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 1 | 20 | 3.38e-01 |
| GO:BP | GO:0072167 | specification of mesonephric tubule identity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072208 | metanephric smooth muscle tissue development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 1 | 26 | 3.38e-01 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 2 | 27 | 3.38e-01 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 2 | 20 | 3.38e-01 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 2 | 25 | 3.38e-01 |
| GO:BP | GO:0072258 | metanephric interstitial fibroblast differentiation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 19 | 211 | 3.38e-01 |
| GO:BP | GO:0072259 | metanephric interstitial fibroblast development | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072318 | clathrin coat disassembly | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0010900 | negative regulation of phosphatidylcholine catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072319 | vesicle uncoating | 2 | 9 | 3.38e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 2 | 36 | 3.38e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 3 | 145 | 3.38e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 3 | 19 | 3.38e-01 |
| GO:BP | GO:0072166 | posterior mesonephric tubule development | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072165 | anterior mesonephric tubule development | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 8 | 64 | 3.38e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 21 | 252 | 3.38e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 16 | 195 | 3.38e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 1 | 175 | 3.38e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 5 | 132 | 3.38e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 9 | 98 | 3.38e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 3 | 10 | 3.38e-01 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 2 | 10 | 3.38e-01 |
| GO:BP | GO:0150093 | amyloid-beta clearance by transcytosis | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 2 | 10 | 3.38e-01 |
| GO:BP | GO:0150063 | visual system development | 3 | 273 | 3.38e-01 |
| GO:BP | GO:0140924 | L-kynurenine transmembrane transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0010669 | epithelial structure maintenance | 2 | 15 | 3.38e-01 |
| GO:BP | GO:0072047 | proximal/distal pattern formation involved in nephron development | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0072048 | renal system pattern specification | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0140673 | transcription elongation-coupled chromatin remodeling | 2 | 2 | 3.38e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0072081 | specification of nephron tubule identity | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 3 | 30 | 3.38e-01 |
| GO:BP | GO:0072098 | anterior/posterior pattern specification involved in kidney development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0010755 | regulation of plasminogen activation | 3 | 16 | 3.38e-01 |
| GO:BP | GO:0010756 | positive regulation of plasminogen activation | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0010757 | negative regulation of plasminogen activation | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0032259 | methylation | 9 | 313 | 3.38e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 2 | 19 | 3.38e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 1 | 82 | 3.38e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 3 | 67 | 3.38e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 8 | 81 | 3.38e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 11 | 126 | 3.38e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 38 | 3.38e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 8 | 324 | 3.38e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 5 | 23 | 3.38e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 3.38e-01 |
| GO:BP | GO:1990074 | polyuridylation-dependent mRNA catabolic process | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 2 | 45 | 3.38e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 3 | 10 | 3.38e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 2 | 65 | 3.38e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 3.38e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 2 | 16 | 3.38e-01 |
| GO:BP | GO:0051454 | intracellular pH elevation | 2 | 4 | 3.38e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 1 | 76 | 3.38e-01 |
| GO:BP | GO:0048389 | intermediate mesoderm development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 2 | 26 | 3.38e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 3.38e-01 |
| GO:BP | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 1 | 14 | 3.38e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0042983 | amyloid precursor protein biosynthetic process | 1 | 14 | 3.38e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 1 | 69 | 3.38e-01 |
| GO:BP | GO:0042938 | dipeptide transport | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 12 | 93 | 3.38e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 1 | 87 | 3.38e-01 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0042874 | D-glucuronate transmembrane transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 2 | 8 | 3.38e-01 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 1 | 8 | 3.38e-01 |
| GO:BP | GO:1990679 | histone H4-K12 deacetylation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0046951 | ketone body biosynthetic process | 2 | 5 | 3.38e-01 |
| GO:BP | GO:0001654 | eye development | 3 | 272 | 3.38e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0000960 | regulation of mitochondrial RNA catabolic process | 1 | 5 | 3.38e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 2 | 6 | 3.38e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 2 | 13 | 3.38e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 2 | 14 | 3.38e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 2 | 71 | 3.38e-01 |
| GO:BP | GO:0000958 | mitochondrial mRNA catabolic process | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0000957 | mitochondrial RNA catabolic process | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0045903 | positive regulation of translational fidelity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0045915 | positive regulation of catecholamine metabolic process | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0046657 | folic acid catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 2 | 14 | 3.38e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 2 | 14 | 3.38e-01 |
| GO:BP | GO:0000725 | recombinational repair | 1 | 153 | 3.38e-01 |
| GO:BP | GO:0000962 | positive regulation of mitochondrial RNA catabolic process | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 11 | 113 | 3.38e-01 |
| GO:BP | GO:0042478 | regulation of eye photoreceptor cell development | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0000965 | mitochondrial RNA 3’-end processing | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 3 | 56 | 3.38e-01 |
| GO:BP | GO:0042560 | pteridine-containing compound catabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 4 | 35 | 3.38e-01 |
| GO:BP | GO:0046950 | cellular ketone body metabolic process | 2 | 7 | 3.38e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 5 | 32 | 3.38e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 12 | 112 | 3.38e-01 |
| GO:BP | GO:0046726 | positive regulation by virus of viral protein levels in host cell | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 4 | 63 | 3.38e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 3 | 21 | 3.38e-01 |
| GO:BP | GO:0003169 | coronary vein morphogenesis | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1905913 | negative regulation of calcium ion export across plasma membrane | 1 | 4 | 3.38e-01 |
| GO:BP | GO:1905912 | regulation of calcium ion export across plasma membrane | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0001193 | maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0001192 | maintenance of transcriptional fidelity during transcription elongation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 1 | 8 | 3.38e-01 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 2 | 23 | 3.38e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 3 | 83 | 3.38e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 4 | 19 | 3.38e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 2 | 44 | 3.38e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 4 | 55 | 3.38e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 2 | 156 | 3.38e-01 |
| GO:BP | GO:0044648 | histone H3-K4 dimethylation | 1 | 6 | 3.38e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 2 | 13 | 3.38e-01 |
| GO:BP | GO:0043519 | regulation of myosin II filament organization | 1 | 1 | 3.38e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0002031 | G protein-coupled receptor internalization | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | 1 | 14 | 3.38e-01 |
| GO:BP | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0051036 | regulation of endosome size | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0044788 | modulation by host of viral process | 1 | 40 | 3.38e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 1 | 49 | 3.38e-01 |
| GO:BP | GO:0002091 | negative regulation of receptor internalization | 1 | 14 | 3.38e-01 |
| GO:BP | GO:0050993 | dimethylallyl diphosphate metabolic process | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 15 | 182 | 3.38e-01 |
| GO:BP | GO:0043649 | dicarboxylic acid catabolic process | 3 | 14 | 3.38e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 8 | 74 | 3.38e-01 |
| GO:BP | GO:0050916 | sensory perception of sweet taste | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 5 | 35 | 3.38e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 5 | 50 | 3.38e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 2 | 15 | 3.38e-01 |
| GO:BP | GO:0050860 | negative regulation of T cell receptor signaling pathway | 3 | 17 | 3.38e-01 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 1 | 20 | 3.38e-01 |
| GO:BP | GO:0044241 | lipid digestion | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 3 | 40 | 3.38e-01 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 3 | 13 | 3.38e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 1 | 425 | 3.38e-01 |
| GO:BP | GO:2001026 | regulation of endothelial cell chemotaxis | 4 | 16 | 3.38e-01 |
| GO:BP | GO:0050992 | dimethylallyl diphosphate biosynthetic process | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 3 | 52 | 3.38e-01 |
| GO:BP | GO:2000827 | mitochondrial RNA surveillance | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 4 | 30 | 3.38e-01 |
| GO:BP | GO:0003400 | regulation of COPII vesicle coating | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0044804 | autophagy of nucleus | 4 | 18 | 3.38e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 3.38e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 3 | 89 | 3.38e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 6 | 113 | 3.38e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 3 | 30 | 3.38e-01 |
| GO:BP | GO:0045204 | MAPK export from nucleus | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 3 | 43 | 3.38e-01 |
| GO:BP | GO:2000284 | positive regulation of amino acid biosynthetic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 3 | 82 | 3.38e-01 |
| GO:BP | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity | 2 | 3 | 3.38e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 3 | 113 | 3.38e-01 |
| GO:BP | GO:2000257 | regulation of protein activation cascade | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 7 | 105 | 3.38e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 2 | 60 | 3.38e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 3 | 85 | 3.38e-01 |
| GO:BP | GO:2000192 | negative regulation of fatty acid transport | 2 | 7 | 3.38e-01 |
| GO:BP | GO:2000425 | regulation of apoptotic cell clearance | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0048769 | sarcomerogenesis | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0045061 | thymic T cell selection | 2 | 10 | 3.38e-01 |
| GO:BP | GO:2000470 | positive regulation of peroxidase activity | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0002574 | thrombocyte differentiation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0002575 | basophil chemotaxis | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0044849 | estrous cycle | 3 | 13 | 3.38e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 2 | 7 | 3.38e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0043363 | nucleate erythrocyte differentiation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 1 | 21 | 3.38e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 4 | 54 | 3.38e-01 |
| GO:BP | GO:0048880 | sensory system development | 3 | 275 | 3.38e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 7 | 233 | 3.38e-01 |
| GO:BP | GO:2000543 | positive regulation of gastrulation | 2 | 5 | 3.38e-01 |
| GO:BP | GO:0045056 | transcytosis | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 2 | 11 | 3.38e-01 |
| GO:BP | GO:0045059 | positive thymic T cell selection | 2 | 6 | 3.38e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 10 | 97 | 3.38e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 3.38e-01 |
| GO:BP | GO:0002931 | response to ischemia | 2 | 46 | 3.38e-01 |
| GO:BP | GO:0050917 | sensory perception of umami taste | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 7 | 59 | 3.38e-01 |
| GO:BP | GO:1905447 | negative regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 3 | 14 | 3.38e-01 |
| GO:BP | GO:1905123 | regulation of glucosylceramidase activity | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 1 | 30 | 3.38e-01 |
| GO:BP | GO:0042365 | water-soluble vitamin catabolic process | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 3.38e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 1 | 10 | 3.38e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 2 | 19 | 3.38e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 3 | 53 | 3.38e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 4 | 78 | 3.38e-01 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 1 | 22 | 3.38e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 11 | 92 | 3.38e-01 |
| GO:BP | GO:0006284 | base-excision repair | 1 | 44 | 3.38e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 9 | 103 | 3.38e-01 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 5 | 32 | 3.38e-01 |
| GO:BP | GO:0046293 | formaldehyde biosynthetic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 1 | 64 | 3.38e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0051622 | negative regulation of norepinephrine uptake | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0051935 | glutamate reuptake | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 1 | 29 | 3.38e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 20 | 3.38e-01 |
| GO:BP | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 5 | 47 | 3.38e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 3.38e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 4 | 68 | 3.38e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 1 | 22 | 3.38e-01 |
| GO:BP | GO:0045964 | positive regulation of dopamine metabolic process | 2 | 6 | 3.38e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0051621 | regulation of norepinephrine uptake | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 2 | 39 | 3.38e-01 |
| GO:BP | GO:0046016 | positive regulation of transcription by glucose | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 2 | 12 | 3.38e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 1 | 149 | 3.38e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0051945 | negative regulation of catecholamine uptake involved in synaptic transmission | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 3 | 45 | 3.38e-01 |
| GO:BP | GO:0003421 | growth plate cartilage axis specification | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0046292 | formaldehyde metabolic process | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 1 | 142 | 3.38e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 8 | 88 | 3.38e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 1 | 16 | 3.38e-01 |
| GO:BP | GO:0000053 | argininosuccinate metabolic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006107 | oxaloacetate metabolic process | 2 | 7 | 3.38e-01 |
| GO:BP | GO:0046532 | regulation of photoreceptor cell differentiation | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0055001 | muscle cell development | 6 | 165 | 3.38e-01 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 1 | 148 | 3.38e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 16 | 3.38e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 2 | 20 | 3.38e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 1 | 20 | 3.38e-01 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0046295 | glycolate biosynthetic process | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 2 | 79 | 3.38e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 3 | 67 | 3.38e-01 |
| GO:BP | GO:0000105 | histidine biosynthetic process | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 3 | 58 | 3.38e-01 |
| GO:BP | GO:0006298 | mismatch repair | 1 | 29 | 3.38e-01 |
| GO:BP | GO:0051946 | regulation of glutamate uptake involved in transmission of nerve impulse | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 4 | 26 | 3.38e-01 |
| GO:BP | GO:1903225 | negative regulation of endodermal cell differentiation | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 1 | 85 | 3.38e-01 |
| GO:BP | GO:0003013 | circulatory system process | 1 | 447 | 3.38e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 2 | 27 | 3.38e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 1 | 5 | 3.38e-01 |
| GO:BP | GO:0032438 | melanosome organization | 2 | 36 | 3.40e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 1 | 24 | 3.40e-01 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 2 | 12 | 3.40e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 3.40e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 2 | 22 | 3.40e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 1 | 6 | 3.40e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 1 | 10 | 3.40e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 3 | 170 | 3.40e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1 | 72 | 3.40e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 1 | 33 | 3.41e-01 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 2 | 11 | 3.42e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 1 | 134 | 3.42e-01 |
| GO:BP | GO:0031623 | receptor internalization | 1 | 95 | 3.42e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 2 | 112 | 3.43e-01 |
| GO:BP | GO:0006865 | amino acid transport | 2 | 102 | 3.44e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 4 | 180 | 3.44e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 3.44e-01 |
| GO:BP | GO:0072194 | kidney smooth muscle tissue development | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0036023 | embryonic skeletal limb joint morphogenesis | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0098629 | trans-Golgi network membrane organization | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0032468 | Golgi calcium ion homeostasis | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 5 | 186 | 3.44e-01 |
| GO:BP | GO:0032472 | Golgi calcium ion transport | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0072190 | ureter urothelium development | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 3.44e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 8 | 343 | 3.44e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 3.44e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 3.44e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 2 | 33 | 3.44e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 8 | 91 | 3.44e-01 |
| GO:BP | GO:0071377 | cellular response to glucagon stimulus | 2 | 4 | 3.44e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 2 | 37 | 3.44e-01 |
| GO:BP | GO:0045991 | carbon catabolite activation of transcription | 1 | 4 | 3.46e-01 |
| GO:BP | GO:0045650 | negative regulation of macrophage differentiation | 1 | 5 | 3.46e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 3.46e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 2 | 15 | 3.46e-01 |
| GO:BP | GO:0045990 | carbon catabolite regulation of transcription | 1 | 4 | 3.46e-01 |
| GO:BP | GO:0035771 | interleukin-4-mediated signaling pathway | 1 | 4 | 3.46e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 2 | 21 | 3.46e-01 |
| GO:BP | GO:0006409 | tRNA export from nucleus | 1 | 3 | 3.46e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 2 | 11 | 3.46e-01 |
| GO:BP | GO:0044790 | suppression of viral release by host | 2 | 16 | 3.46e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 8 | 285 | 3.46e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 1 | 10 | 3.46e-01 |
| GO:BP | GO:0016559 | peroxisome fission | 2 | 9 | 3.46e-01 |
| GO:BP | GO:0035600 | tRNA methylthiolation | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 1 | 16 | 3.46e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 10 | 92 | 3.46e-01 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 2 | 31 | 3.46e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 3 | 17 | 3.46e-01 |
| GO:BP | GO:0007030 | Golgi organization | 4 | 137 | 3.47e-01 |
| GO:BP | GO:0010522 | regulation of calcium ion transport into cytosol | 1 | 10 | 3.47e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 3 | 110 | 3.47e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 3 | 52 | 3.47e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 12 | 140 | 3.47e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 4 | 105 | 3.47e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 3 | 16 | 3.47e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 7 | 272 | 3.48e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 3 | 122 | 3.50e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 1 | 94 | 3.51e-01 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 1 | 11 | 3.51e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 4 | 151 | 3.51e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 3.51e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 3.51e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 3.51e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 3.51e-01 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 2 | 15 | 3.51e-01 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 2 | 15 | 3.51e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 3.51e-01 |
| GO:BP | GO:0097242 | amyloid-beta clearance | 2 | 27 | 3.51e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 2 | 14 | 3.52e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 8 | 94 | 3.52e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 2 | 14 | 3.52e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 3 | 23 | 3.54e-01 |
| GO:BP | GO:1901998 | toxin transport | 1 | 37 | 3.55e-01 |
| GO:BP | GO:0006857 | oligopeptide transport | 2 | 5 | 3.55e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 2 | 10 | 3.55e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 3.55e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 18 | 249 | 3.55e-01 |
| GO:BP | GO:0060746 | parental behavior | 1 | 7 | 3.55e-01 |
| GO:BP | GO:0007286 | spermatid development | 5 | 105 | 3.55e-01 |
| GO:BP | GO:0034635 | glutathione transport | 2 | 9 | 3.55e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 1 | 99 | 3.55e-01 |
| GO:BP | GO:0042711 | maternal behavior | 1 | 7 | 3.55e-01 |
| GO:BP | GO:0007638 | mechanosensory behavior | 2 | 8 | 3.55e-01 |
| GO:BP | GO:0035672 | oligopeptide transmembrane transport | 2 | 5 | 3.55e-01 |
| GO:BP | GO:0002441 | histamine secretion involved in inflammatory response | 1 | 8 | 3.56e-01 |
| GO:BP | GO:0002553 | histamine secretion by mast cell | 1 | 8 | 3.56e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 5 | 45 | 3.56e-01 |
| GO:BP | GO:0002349 | histamine production involved in inflammatory response | 1 | 8 | 3.56e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 2 | 16 | 3.56e-01 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 3.56e-01 |
| GO:BP | GO:0033169 | histone H3-K9 demethylation | 1 | 5 | 3.56e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 3 | 34 | 3.57e-01 |
| GO:BP | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity | 1 | 10 | 3.58e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 1 | 28 | 3.58e-01 |
| GO:BP | GO:2000402 | negative regulation of lymphocyte migration | 1 | 10 | 3.58e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 1 | 560 | 3.59e-01 |
| GO:BP | GO:0001881 | receptor recycling | 2 | 38 | 3.60e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 8 | 3.60e-01 |
| GO:BP | GO:0070669 | response to interleukin-2 | 1 | 3 | 3.60e-01 |
| GO:BP | GO:0033240 | positive regulation of amine metabolic process | 2 | 7 | 3.60e-01 |
| GO:BP | GO:0060336 | negative regulation of type II interferon-mediated signaling pathway | 1 | 6 | 3.60e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 2 | 121 | 3.60e-01 |
| GO:BP | GO:0060331 | negative regulation of response to type II interferon | 1 | 6 | 3.60e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 1 | 7 | 3.60e-01 |
| GO:BP | GO:0016571 | histone methylation | 4 | 117 | 3.60e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 11 | 3.60e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 2 | 19 | 3.60e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 3 | 19 | 3.62e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 12 | 109 | 3.62e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 2 | 118 | 3.62e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 3 | 19 | 3.63e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 1 | 30 | 3.63e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 3 | 441 | 3.63e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 2 | 26 | 3.63e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 2 | 119 | 3.63e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 2 | 29 | 3.63e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 1 | 106 | 3.63e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 3 | 19 | 3.64e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 1 | 16 | 3.65e-01 |
| GO:BP | GO:0007423 | sensory organ development | 3 | 396 | 3.65e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 2 | 26 | 3.65e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 1 | 4 | 3.65e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 1 | 16 | 3.65e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 1 | 30 | 3.65e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 2 | 53 | 3.65e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 2 | 88 | 3.65e-01 |
| GO:BP | GO:0071109 | superior temporal gyrus development | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 1 | 18 | 3.66e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 28 | 3.66e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 1 | 8 | 3.66e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 3.68e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 2 | 7 | 3.68e-01 |
| GO:BP | GO:0044029 | positive regulation of gene expression via CpG island demethylation | 1 | 3 | 3.68e-01 |
| GO:BP | GO:0071464 | cellular response to hydrostatic pressure | 1 | 3 | 3.68e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 2 | 39 | 3.68e-01 |
| GO:BP | GO:0046697 | decidualization | 3 | 19 | 3.68e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 3.70e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 25 | 358 | 3.70e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 3.70e-01 |
| GO:BP | GO:0001886 | endothelial cell morphogenesis | 2 | 10 | 3.70e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 12 | 740 | 3.70e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 5 | 111 | 3.71e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 3.71e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 2 | 12 | 3.73e-01 |
| GO:BP | GO:1901159 | xylulose 5-phosphate biosynthetic process | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0050765 | negative regulation of phagocytosis | 3 | 15 | 3.73e-01 |
| GO:BP | GO:0070486 | leukocyte aggregation | 2 | 9 | 3.73e-01 |
| GO:BP | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0097498 | endothelial tube lumen extension | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0038091 | positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway | 1 | 3 | 3.73e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 2 | 54 | 3.73e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 4 | 45 | 3.73e-01 |
| GO:BP | GO:0072133 | metanephric mesenchyme morphogenesis | 1 | 4 | 3.73e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 3.73e-01 |
| GO:BP | GO:1900275 | negative regulation of phospholipase C activity | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0044722 | renal phosphate excretion | 1 | 2 | 3.73e-01 |
| GO:BP | GO:1904715 | negative regulation of chaperone-mediated autophagy | 1 | 3 | 3.73e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 3.73e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 5 | 37 | 3.73e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 3 | 36 | 3.73e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 5 | 401 | 3.73e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 1 | 122 | 3.73e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 3 | 39 | 3.73e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 2 | 31 | 3.73e-01 |
| GO:BP | GO:0032197 | retrotransposition | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 4 | 45 | 3.73e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 1 | 5 | 3.73e-01 |
| GO:BP | GO:2000642 | negative regulation of early endosome to late endosome transport | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0070564 | positive regulation of vitamin D receptor signaling pathway | 1 | 5 | 3.73e-01 |
| GO:BP | GO:0071076 | RNA 3’ uridylation | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0072131 | kidney mesenchyme morphogenesis | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0032196 | transposition | 2 | 17 | 3.73e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 1 | 271 | 3.73e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 2 | 18 | 3.73e-01 |
| GO:BP | GO:1904105 | positive regulation of convergent extension involved in gastrulation | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 4 | 155 | 3.73e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 1 | 55 | 3.73e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 3 | 88 | 3.73e-01 |
| GO:BP | GO:0072268 | pattern specification involved in metanephros development | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0014889 | muscle atrophy | 2 | 9 | 3.73e-01 |
| GO:BP | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 1 | 188 | 3.73e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 4 | 26 | 3.73e-01 |
| GO:BP | GO:0021762 | substantia nigra development | 2 | 41 | 3.73e-01 |
| GO:BP | GO:0001821 | histamine secretion | 1 | 9 | 3.73e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 1 | 12 | 3.73e-01 |
| GO:BP | GO:0006108 | malate metabolic process | 1 | 7 | 3.73e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 3.73e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 3 | 172 | 3.73e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 2 | 14 | 3.73e-01 |
| GO:BP | GO:1903402 | regulation of renal phosphate excretion | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0061519 | macrophage homeostasis | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0010986 | positive regulation of lipoprotein particle clearance | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 4 | 161 | 3.73e-01 |
| GO:BP | GO:0098657 | import into cell | 3 | 166 | 3.73e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 2 | 17 | 3.73e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 3.73e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 2 | 15 | 3.73e-01 |
| GO:BP | GO:0051581 | negative regulation of neurotransmitter uptake | 1 | 2 | 3.73e-01 |
| GO:BP | GO:1904103 | regulation of convergent extension involved in gastrulation | 1 | 1 | 3.73e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 1 | 150 | 3.73e-01 |
| GO:BP | GO:1905279 | regulation of retrograde transport, endosome to Golgi | 1 | 6 | 3.73e-01 |
| GO:BP | GO:0006064 | glucuronate catabolic process | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 18 | 3.73e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 3 | 40 | 3.73e-01 |
| GO:BP | GO:1903392 | negative regulation of adherens junction organization | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 10 | 114 | 3.73e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.73e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 1 | 17 | 3.73e-01 |
| GO:BP | GO:0006954 | inflammatory response | 1 | 429 | 3.73e-01 |
| GO:BP | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | 1 | 4 | 3.73e-01 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 1 | 14 | 3.73e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 3.73e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 2 | 15 | 3.73e-01 |
| GO:BP | GO:0003422 | growth plate cartilage morphogenesis | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 4 | 269 | 3.73e-01 |
| GO:BP | GO:1905604 | negative regulation of blood-brain barrier permeability | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0072180 | mesonephric duct morphogenesis | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0051612 | negative regulation of serotonin uptake | 1 | 2 | 3.73e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 1 | 20 | 3.73e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 4 | 32 | 3.73e-01 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 2 | 17 | 3.73e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 6 | 49 | 3.73e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0090160 | Golgi to lysosome transport | 1 | 10 | 3.73e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 1 | 5 | 3.73e-01 |
| GO:BP | GO:2000392 | regulation of lamellipodium morphogenesis | 1 | 10 | 3.73e-01 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 1 | 38 | 3.73e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 1 | 7 | 3.73e-01 |
| GO:BP | GO:2000295 | regulation of hydrogen peroxide catabolic process | 1 | 2 | 3.73e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 1 | 18 | 3.73e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 2 | 22 | 3.73e-01 |
| GO:BP | GO:0060948 | cardiac vascular smooth muscle cell development | 1 | 3 | 3.73e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0007015 | actin filament organization | 9 | 373 | 3.73e-01 |
| GO:BP | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 1 | 39 | 3.73e-01 |
| GO:BP | GO:0034498 | early endosome to Golgi transport | 1 | 9 | 3.73e-01 |
| GO:BP | GO:0006260 | DNA replication | 1 | 255 | 3.73e-01 |
| GO:BP | GO:0051167 | xylulose 5-phosphate metabolic process | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 3.74e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 1 | 4 | 3.74e-01 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 1 | 18 | 3.74e-01 |
| GO:BP | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb | 1 | 4 | 3.74e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 2 | 55 | 3.74e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 1 | 8 | 3.74e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 27 | 3.74e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 1 | 23 | 3.74e-01 |
| GO:BP | GO:0006601 | creatine biosynthetic process | 1 | 2 | 3.74e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 1 | 7 | 3.74e-01 |
| GO:BP | GO:0038128 | ERBB2 signaling pathway | 2 | 14 | 3.74e-01 |
| GO:BP | GO:0007033 | vacuole organization | 14 | 197 | 3.74e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 3 | 50 | 3.74e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 3 | 35 | 3.74e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 4 | 39 | 3.75e-01 |
| GO:BP | GO:0010232 | vascular transport | 3 | 72 | 3.75e-01 |
| GO:BP | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly | 1 | 16 | 3.75e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 3 | 72 | 3.75e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 1 | 12 | 3.75e-01 |
| GO:BP | GO:0007270 | neuron-neuron synaptic transmission | 1 | 5 | 3.75e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 2 | 16 | 3.75e-01 |
| GO:BP | GO:0032570 | response to progesterone | 4 | 27 | 3.76e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 3.76e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 3.76e-01 |
| GO:BP | GO:0019464 | glycine decarboxylation via glycine cleavage system | 1 | 3 | 3.76e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 3.76e-01 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 1 | 14 | 3.76e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 1 | 19 | 3.76e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 8 | 126 | 3.76e-01 |
| GO:BP | GO:0071418 | cellular response to amine stimulus | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0032382 | positive regulation of intracellular sterol transport | 1 | 3 | 3.76e-01 |
| GO:BP | GO:0048172 | regulation of short-term neuronal synaptic plasticity | 2 | 9 | 3.76e-01 |
| GO:BP | GO:0032385 | positive regulation of intracellular cholesterol transport | 1 | 3 | 3.76e-01 |
| GO:BP | GO:0031639 | plasminogen activation | 1 | 19 | 3.76e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 14 | 172 | 3.77e-01 |
| GO:BP | GO:2001226 | negative regulation of chloride transport | 1 | 1 | 3.77e-01 |
| GO:BP | GO:0010985 | negative regulation of lipoprotein particle clearance | 2 | 6 | 3.78e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 1 | 19 | 3.79e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 5 | 408 | 3.79e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 1 | 20 | 3.79e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 1 | 206 | 3.79e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 3 | 20 | 3.79e-01 |
| GO:BP | GO:0006417 | regulation of translation | 12 | 370 | 3.79e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 1 | 28 | 3.79e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 681 | 3.80e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 2 | 17 | 3.81e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2 | 56 | 3.81e-01 |
| GO:BP | GO:0006310 | DNA recombination | 1 | 257 | 3.81e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 5 | 50 | 3.81e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 1 | 20 | 3.81e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 1 | 40 | 3.82e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 1 | 24 | 3.83e-01 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 1 | 19 | 3.83e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 1 | 43 | 3.84e-01 |
| GO:BP | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide | 1 | 2 | 3.84e-01 |
| GO:BP | GO:1905219 | regulation of platelet formation | 1 | 1 | 3.84e-01 |
| GO:BP | GO:1905221 | positive regulation of platelet formation | 1 | 1 | 3.84e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 3.84e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 3.85e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 3.85e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 3 | 43 | 3.85e-01 |
| GO:BP | GO:0062029 | positive regulation of stress granule assembly | 1 | 2 | 3.85e-01 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 2 | 35 | 3.85e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 1 | 38 | 3.85e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 1 | 129 | 3.85e-01 |
| GO:BP | GO:0048935 | peripheral nervous system neuron development | 1 | 9 | 3.85e-01 |
| GO:BP | GO:2000813 | negative regulation of barbed-end actin filament capping | 1 | 3 | 3.85e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 3.85e-01 |
| GO:BP | GO:0048934 | peripheral nervous system neuron differentiation | 1 | 9 | 3.85e-01 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 1 | 37 | 3.86e-01 |
| GO:BP | GO:0140588 | chromatin looping | 1 | 2 | 3.87e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 6 | 569 | 3.87e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 19 | 500 | 3.87e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 3.87e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 10 | 3.87e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 3 | 25 | 3.87e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 2 | 31 | 3.87e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 10 | 421 | 3.87e-01 |
| GO:BP | GO:0033085 | negative regulation of T cell differentiation in thymus | 1 | 5 | 3.87e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 1 | 76 | 3.87e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 1 | 16 | 3.87e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 3.87e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 1 | 17 | 3.89e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 740 | 3.89e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 7 | 72 | 3.89e-01 |
| GO:BP | GO:2000815 | regulation of mRNA stability involved in response to oxidative stress | 1 | 2 | 3.89e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 1 | 6 | 3.90e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 8 | 94 | 3.90e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 3 | 39 | 3.90e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 3.90e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 1 | 20 | 3.90e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 8 | 94 | 3.90e-01 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 28 | 3.90e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 1 | 18 | 3.90e-01 |
| GO:BP | GO:0043473 | pigmentation | 3 | 92 | 3.90e-01 |
| GO:BP | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 1 | 10 | 3.90e-01 |
| GO:BP | GO:0043316 | cytotoxic T cell degranulation | 1 | 2 | 3.90e-01 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 3.90e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 2 | 10 | 3.91e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 3 | 37 | 3.91e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 3.91e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 2 | 19 | 3.92e-01 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 1 | 9 | 3.92e-01 |
| GO:BP | GO:0090362 | positive regulation of platelet-derived growth factor production | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling | 2 | 12 | 3.92e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 3.92e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 3.92e-01 |
| GO:BP | GO:1990034 | calcium ion export across plasma membrane | 1 | 6 | 3.92e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 31 | 3.92e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 3 | 41 | 3.92e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 2 | 64 | 3.92e-01 |
| GO:BP | GO:0036022 | limb joint morphogenesis | 1 | 4 | 3.92e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 1 | 3 | 3.92e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 1 | 37 | 3.92e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 3.92e-01 |
| GO:BP | GO:0070269 | pyroptosis | 3 | 11 | 3.92e-01 |
| GO:BP | GO:0051608 | histamine transport | 1 | 11 | 3.93e-01 |
| GO:BP | GO:1903094 | negative regulation of protein K48-linked deubiquitination | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 3.93e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 1 | 24 | 3.93e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 1 | 139 | 3.93e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 13 | 3.93e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 1 | 136 | 3.93e-01 |
| GO:BP | GO:0150174 | negative regulation of phosphatidylcholine metabolic process | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0051014 | actin filament severing | 2 | 13 | 3.93e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 3 | 18 | 3.93e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 3.93e-01 |
| GO:BP | GO:0033198 | response to ATP | 1 | 24 | 3.93e-01 |
| GO:BP | GO:0051595 | response to methylglyoxal | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1903181 | positive regulation of dopamine biosynthetic process | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 1 | 11 | 3.93e-01 |
| GO:BP | GO:0050787 | detoxification of mercury ion | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 6 | 82 | 3.93e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 3.93e-01 |
| GO:BP | GO:0019605 | butyrate metabolic process | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 1 | 8 | 3.93e-01 |
| GO:BP | GO:0120197 | mucociliary clearance | 1 | 5 | 3.93e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 5 | 39 | 3.93e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 1 | 12 | 3.93e-01 |
| GO:BP | GO:2000157 | negative regulation of ubiquitin-specific protease activity | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 2 | 19 | 3.93e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 4 | 39 | 3.93e-01 |
| GO:BP | GO:0033523 | histone H2B ubiquitination | 1 | 10 | 3.93e-01 |
| GO:BP | GO:0046724 | oxalic acid secretion | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 5 | 162 | 3.93e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 3.93e-01 |
| GO:BP | GO:2000880 | positive regulation of dipeptide transport | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0071387 | cellular response to cortisol stimulus | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 7 | 75 | 3.93e-01 |
| GO:BP | GO:0002353 | plasma kallikrein-kinin cascade | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1903383 | regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0090089 | regulation of dipeptide transport | 1 | 2 | 3.93e-01 |
| GO:BP | GO:2001150 | positive regulation of dipeptide transmembrane transport | 1 | 2 | 3.93e-01 |
| GO:BP | GO:2001148 | regulation of dipeptide transmembrane transport | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0090088 | regulation of oligopeptide transport | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 2 | 13 | 3.93e-01 |
| GO:BP | GO:0006546 | glycine catabolic process | 1 | 3 | 3.93e-01 |
| GO:BP | GO:0070994 | detection of oxidative stress | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 1 | 16 | 3.93e-01 |
| GO:BP | GO:0036482 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 1 | 7 | 3.93e-01 |
| GO:BP | GO:0031335 | regulation of sulfur amino acid metabolic process | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0002930 | trabecular meshwork development | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 1 | 55 | 3.93e-01 |
| GO:BP | GO:0036525 | protein deglycation | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1905259 | negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0002254 | kinin cascade | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0045764 | positive regulation of amino acid metabolic process | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 3.93e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 1 | 16 | 3.93e-01 |
| GO:BP | GO:1903384 | negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0110061 | regulation of angiotensin-activated signaling pathway | 1 | 2 | 3.93e-01 |
| GO:BP | GO:1905258 | regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0021934 | hindbrain tangential cell migration | 1 | 3 | 3.93e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 2 | 64 | 3.93e-01 |
| GO:BP | GO:2000878 | positive regulation of oligopeptide transport | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 1 | 57 | 3.93e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 19 | 424 | 3.93e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 7 | 110 | 3.93e-01 |
| GO:BP | GO:0071676 | negative regulation of mononuclear cell migration | 1 | 16 | 3.94e-01 |
| GO:BP | GO:0032963 | collagen metabolic process | 2 | 74 | 3.94e-01 |
| GO:BP | GO:2000468 | regulation of peroxidase activity | 1 | 2 | 3.95e-01 |
| GO:BP | GO:0032308 | positive regulation of prostaglandin secretion | 2 | 6 | 3.95e-01 |
| GO:BP | GO:0051611 | regulation of serotonin uptake | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 3 | 100 | 3.96e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 1 | 60 | 3.96e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 3.97e-01 |
| GO:BP | GO:1900222 | negative regulation of amyloid-beta clearance | 1 | 7 | 3.97e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 2 | 16 | 3.97e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 1 | 3 | 3.97e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 1 | 17 | 3.97e-01 |
| GO:BP | GO:1990926 | short-term synaptic potentiation | 1 | 4 | 3.97e-01 |
| GO:BP | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation | 1 | 4 | 3.97e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 3.97e-01 |
| GO:BP | GO:1904751 | positive regulation of protein localization to nucleolus | 1 | 3 | 3.97e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 1 | 4 | 3.97e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 1 | 4 | 3.97e-01 |
| GO:BP | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process | 1 | 4 | 3.97e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 1 | 16 | 3.97e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 19 | 937 | 3.99e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 10 | 3.99e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 22 | 326 | 3.99e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 5 | 36 | 3.99e-01 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 1 | 5 | 3.99e-01 |
| GO:BP | GO:0043171 | peptide catabolic process | 1 | 16 | 3.99e-01 |
| GO:BP | GO:0007507 | heart development | 1 | 506 | 3.99e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 2 | 43 | 3.99e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 4 | 24 | 4.00e-01 |
| GO:BP | GO:0006897 | endocytosis | 9 | 527 | 4.00e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 4.01e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 2 | 20 | 4.01e-01 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 1 | 11 | 4.01e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 1 | 34 | 4.01e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 8 | 90 | 4.01e-01 |
| GO:BP | GO:0061377 | mammary gland lobule development | 2 | 13 | 4.01e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 8 | 97 | 4.01e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 15 | 4.01e-01 |
| GO:BP | GO:0060749 | mammary gland alveolus development | 2 | 13 | 4.01e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 2 | 28 | 4.01e-01 |
| GO:BP | GO:1901163 | regulation of trophoblast cell migration | 2 | 12 | 4.01e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 6 | 123 | 4.01e-01 |
| GO:BP | GO:0007613 | memory | 2 | 92 | 4.01e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 7 | 114 | 4.01e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 2 | 8 | 4.02e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 2 | 7 | 4.02e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 2 | 8 | 4.02e-01 |
| GO:BP | GO:0045058 | T cell selection | 2 | 22 | 4.02e-01 |
| GO:BP | GO:0006982 | response to lipid hydroperoxide | 1 | 2 | 4.02e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 2 | 8 | 4.02e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 11 | 140 | 4.02e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 4 | 65 | 4.03e-01 |
| GO:BP | GO:0051918 | negative regulation of fibrinolysis | 2 | 8 | 4.04e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 6 | 75 | 4.04e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 4.04e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 2 | 43 | 4.04e-01 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 2 | 13 | 4.04e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 1 | 29 | 4.04e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 1 | 12 | 4.04e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 17 | 4.04e-01 |
| GO:BP | GO:0022406 | membrane docking | 7 | 84 | 4.04e-01 |
| GO:BP | GO:0043461 | proton-transporting ATP synthase complex assembly | 1 | 8 | 4.05e-01 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 2 | 17 | 4.05e-01 |
| GO:BP | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly | 1 | 8 | 4.05e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 7 | 130 | 4.05e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 3 | 23 | 4.05e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 2 | 90 | 4.05e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 3 | 33 | 4.05e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 3 | 29 | 4.05e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 5 | 47 | 4.06e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 11 | 170 | 4.06e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 2 | 12 | 4.06e-01 |
| GO:BP | GO:0000910 | cytokinesis | 4 | 164 | 4.06e-01 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 1 | 10 | 4.07e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 18 | 464 | 4.07e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 4.07e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 1 | 15 | 4.07e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 2 | 10 | 4.07e-01 |
| GO:BP | GO:0072143 | mesangial cell development | 1 | 5 | 4.07e-01 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 4.07e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 4 | 185 | 4.07e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 2 | 21 | 4.07e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 3 | 34 | 4.07e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 3 | 39 | 4.07e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 20 | 300 | 4.08e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 4 | 37 | 4.08e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 3 | 49 | 4.08e-01 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 4.09e-01 |
| GO:BP | GO:0019080 | viral gene expression | 5 | 96 | 4.09e-01 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 1 | 10 | 4.09e-01 |
| GO:BP | GO:0006281 | DNA repair | 3 | 534 | 4.09e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 1 | 55 | 4.09e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 1 | 6 | 4.09e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 1 | 6 | 4.09e-01 |
| GO:BP | GO:1903650 | negative regulation of cytoplasmic transport | 1 | 3 | 4.09e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 18 | 303 | 4.09e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 1 | 8 | 4.09e-01 |
| GO:BP | GO:0006734 | NADH metabolic process | 1 | 12 | 4.09e-01 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 2 | 5 | 4.09e-01 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 2 | 5 | 4.09e-01 |
| GO:BP | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death | 1 | 3 | 4.09e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 3 | 25 | 4.09e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 3 | 103 | 4.09e-01 |
| GO:BP | GO:0032306 | regulation of prostaglandin secretion | 2 | 7 | 4.09e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 1 | 61 | 4.09e-01 |
| GO:BP | GO:0051031 | tRNA transport | 1 | 5 | 4.09e-01 |
| GO:BP | GO:0021546 | rhombomere development | 1 | 4 | 4.09e-01 |
| GO:BP | GO:2000653 | regulation of genetic imprinting | 1 | 4 | 4.10e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 8 | 97 | 4.10e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 1 | 13 | 4.12e-01 |
| GO:BP | GO:0030574 | collagen catabolic process | 1 | 26 | 4.13e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 2 | 124 | 4.13e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 4 | 62 | 4.13e-01 |
| GO:BP | GO:0009725 | response to hormone | 13 | 642 | 4.13e-01 |
| GO:BP | GO:1904353 | regulation of telomere capping | 1 | 23 | 4.13e-01 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 1 | 9 | 4.13e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 7 | 151 | 4.13e-01 |
| GO:BP | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 4 | 4.13e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 4.14e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 2 | 28 | 4.14e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 3 | 89 | 4.14e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 9 | 133 | 4.17e-01 |
| GO:BP | GO:0061450 | trophoblast cell migration | 2 | 13 | 4.17e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 5 | 45 | 4.17e-01 |
| GO:BP | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | 1 | 4 | 4.17e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 4 | 38 | 4.17e-01 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 1 | 4 | 4.17e-01 |
| GO:BP | GO:0051310 | metaphase plate congression | 1 | 69 | 4.17e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 2 | 60 | 4.17e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 17 | 4.17e-01 |
| GO:BP | GO:0015942 | formate metabolic process | 1 | 2 | 4.17e-01 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 1 | 17 | 4.17e-01 |
| GO:BP | GO:0001840 | neural plate development | 1 | 7 | 4.17e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 3 | 41 | 4.17e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 19 | 555 | 4.17e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 7 | 78 | 4.17e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 4 | 29 | 4.17e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 2 | 19 | 4.18e-01 |
| GO:BP | GO:0006600 | creatine metabolic process | 1 | 3 | 4.18e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1 | 26 | 4.18e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 2 | 191 | 4.18e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 4.18e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 4.18e-01 |
| GO:BP | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 1 | 4.18e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 2 | 15 | 4.18e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 1 | 3 | 4.18e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 4.18e-01 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 1 | 28 | 4.18e-01 |
| GO:BP | GO:0007517 | muscle organ development | 4 | 277 | 4.18e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 1 | 15 | 4.18e-01 |
| GO:BP | GO:0070562 | regulation of vitamin D receptor signaling pathway | 1 | 9 | 4.18e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 7 | 69 | 4.18e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 2 | 70 | 4.18e-01 |
| GO:BP | GO:0003165 | Purkinje myocyte development | 1 | 3 | 4.18e-01 |
| GO:BP | GO:0003167 | atrioventricular bundle cell differentiation | 1 | 3 | 4.18e-01 |
| GO:BP | GO:1903224 | regulation of endodermal cell differentiation | 1 | 6 | 4.19e-01 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 1 | 17 | 4.20e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 1 | 14 | 4.20e-01 |
| GO:BP | GO:0060912 | cardiac cell fate specification | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0006528 | asparagine metabolic process | 1 | 5 | 4.20e-01 |
| GO:BP | GO:0038156 | interleukin-3-mediated signaling pathway | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0071400 | cellular response to oleic acid | 1 | 3 | 4.21e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 2 | 12 | 4.21e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 1 | 3 | 4.21e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 4.21e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 1 | 39 | 4.21e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 2 | 73 | 4.21e-01 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 1 | 70 | 4.22e-01 |
| GO:BP | GO:0060433 | bronchus development | 1 | 6 | 4.24e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 1 | 5 | 4.24e-01 |
| GO:BP | GO:0043932 | ossification involved in bone remodeling | 1 | 2 | 4.24e-01 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 1 | 23 | 4.24e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 1 | 54 | 4.24e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 4.25e-01 |
| GO:BP | GO:0030001 | metal ion transport | 6 | 562 | 4.25e-01 |
| GO:BP | GO:0050812 | regulation of acyl-CoA biosynthetic process | 1 | 5 | 4.25e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 2 | 30 | 4.25e-01 |
| GO:BP | GO:0045039 | protein insertion into mitochondrial inner membrane | 1 | 13 | 4.26e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 4.26e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 3 | 523 | 4.27e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 4.27e-01 |
| GO:BP | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 2 | 48 | 4.27e-01 |
| GO:BP | GO:1903391 | regulation of adherens junction organization | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 4.27e-01 |
| GO:BP | GO:0002486 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 994 | 4.27e-01 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 2 | 21 | 4.27e-01 |
| GO:BP | GO:0060319 | primitive erythrocyte differentiation | 1 | 4 | 4.27e-01 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 2 | 21 | 4.27e-01 |
| GO:BP | GO:0051977 | lysophospholipid transport | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0044375 | regulation of peroxisome size | 1 | 3 | 4.28e-01 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 4.28e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 4 | 50 | 4.28e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 2 | 74 | 4.28e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 9 | 131 | 4.28e-01 |
| GO:BP | GO:0060330 | regulation of response to type II interferon | 1 | 12 | 4.28e-01 |
| GO:BP | GO:0072007 | mesangial cell differentiation | 1 | 5 | 4.28e-01 |
| GO:BP | GO:0060334 | regulation of type II interferon-mediated signaling pathway | 1 | 12 | 4.28e-01 |
| GO:BP | GO:0034462 | small-subunit processome assembly | 1 | 3 | 4.28e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 4 | 47 | 4.28e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 2 | 9 | 4.28e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 2 | 9 | 4.28e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 4.28e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 1 | 11 | 4.29e-01 |
| GO:BP | GO:0019079 | viral genome replication | 3 | 101 | 4.29e-01 |
| GO:BP | GO:2000812 | regulation of barbed-end actin filament capping | 1 | 3 | 4.29e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 1 | 50 | 4.29e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 1 | 28 | 4.29e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 1 | 7 | 4.29e-01 |
| GO:BP | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | 1 | 5 | 4.29e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 4.29e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 4.29e-01 |
| GO:BP | GO:1903413 | cellular response to bile acid | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 1 | 22 | 4.29e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 4.29e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 4.29e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 6 | 64 | 4.29e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 4.29e-01 |
| GO:BP | GO:2000391 | positive regulation of neutrophil extravasation | 1 | 6 | 4.29e-01 |
| GO:BP | GO:0045010 | actin nucleation | 3 | 50 | 4.29e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 5 | 51 | 4.29e-01 |
| GO:BP | GO:0090660 | cerebrospinal fluid circulation | 2 | 9 | 4.29e-01 |
| GO:BP | GO:0032808 | lacrimal gland development | 1 | 5 | 4.29e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 1 | 5 | 4.29e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 1 | 68 | 4.29e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 5 | 51 | 4.29e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 3 | 230 | 4.29e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1 | 21 | 4.30e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 1 | 25 | 4.30e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 2 | 172 | 4.31e-01 |
| GO:BP | GO:0006567 | threonine catabolic process | 1 | 3 | 4.31e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 2 | 131 | 4.32e-01 |
| GO:BP | GO:0034384 | high-density lipoprotein particle clearance | 1 | 7 | 4.32e-01 |
| GO:BP | GO:0031580 | membrane raft distribution | 1 | 3 | 4.32e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 2 | 89 | 4.32e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 38 | 612 | 4.32e-01 |
| GO:BP | GO:0048294 | negative regulation of isotype switching to IgE isotypes | 1 | 2 | 4.32e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 1 | 5 | 4.32e-01 |
| GO:BP | GO:0043046 | DNA methylation involved in gamete generation | 1 | 4 | 4.32e-01 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 1 | 11 | 4.32e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 6 | 107 | 4.32e-01 |
| GO:BP | GO:0045163 | clustering of voltage-gated potassium channels | 1 | 2 | 4.32e-01 |
| GO:BP | GO:0010610 | regulation of mRNA stability involved in response to stress | 1 | 3 | 4.32e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 3 | 292 | 4.32e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 4 | 45 | 4.32e-01 |
| GO:BP | GO:0019081 | viral translation | 2 | 18 | 4.32e-01 |
| GO:BP | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation | 1 | 3 | 4.32e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 1 | 40 | 4.32e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 1 | 252 | 4.32e-01 |
| GO:BP | GO:0071205 | protein localization to juxtaparanode region of axon | 1 | 3 | 4.32e-01 |
| GO:BP | GO:0140361 | cyclic-GMP-AMP transmembrane import across plasma membrane | 1 | 4 | 4.32e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 1 | 11 | 4.32e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 4.32e-01 |
| GO:BP | GO:0015677 | copper ion import | 1 | 5 | 4.32e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 3 | 49 | 4.33e-01 |
| GO:BP | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0140469 | GCN2-mediated signaling | 1 | 3 | 4.33e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 2 | 14 | 4.33e-01 |
| GO:BP | GO:0060279 | positive regulation of ovulation | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 1 | 16 | 4.33e-01 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 4.33e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 4.33e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 2 | 211 | 4.33e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 4.33e-01 |
| GO:BP | GO:0044546 | NLRP3 inflammasome complex assembly | 1 | 23 | 4.33e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 2 | 14 | 4.33e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 9 | 4.33e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 2 | 25 | 4.33e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 2 | 28 | 4.33e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 4.33e-01 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 1 | 75 | 4.34e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 2 | 76 | 4.34e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 4.34e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 3 | 89 | 4.34e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 2 | 23 | 4.34e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 13 | 168 | 4.34e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 4 | 34 | 4.34e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 4.34e-01 |
| GO:BP | GO:0045329 | carnitine biosynthetic process | 1 | 3 | 4.34e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 2 | 17 | 4.34e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 3 | 50 | 4.34e-01 |
| GO:BP | GO:0090361 | regulation of platelet-derived growth factor production | 1 | 2 | 4.34e-01 |
| GO:BP | GO:0090360 | platelet-derived growth factor production | 1 | 2 | 4.34e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 3 | 147 | 4.34e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 3 | 31 | 4.35e-01 |
| GO:BP | GO:0034030 | ribonucleoside bisphosphate biosynthetic process | 2 | 49 | 4.35e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 5 | 486 | 4.35e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 3 | 51 | 4.35e-01 |
| GO:BP | GO:0034033 | purine nucleoside bisphosphate biosynthetic process | 2 | 49 | 4.35e-01 |
| GO:BP | GO:0007431 | salivary gland development | 1 | 26 | 4.35e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 5 | 798 | 4.35e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 3 | 19 | 4.35e-01 |
| GO:BP | GO:0032967 | positive regulation of collagen biosynthetic process | 1 | 23 | 4.35e-01 |
| GO:BP | GO:0033866 | nucleoside bisphosphate biosynthetic process | 2 | 49 | 4.35e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 1 | 18 | 4.35e-01 |
| GO:BP | GO:0035752 | lysosomal lumen pH elevation | 1 | 3 | 4.35e-01 |
| GO:BP | GO:0010899 | regulation of phosphatidylcholine catabolic process | 1 | 3 | 4.36e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 1 | 6 | 4.37e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 4.37e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 2 | 11 | 4.37e-01 |
| GO:BP | GO:0051235 | maintenance of location | 3 | 260 | 4.37e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 14 | 771 | 4.37e-01 |
| GO:BP | GO:0006701 | progesterone biosynthetic process | 1 | 5 | 4.37e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 4 | 74 | 4.39e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 4.39e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 1 | 80 | 4.39e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 2 | 308 | 4.39e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 2 | 198 | 4.40e-01 |
| GO:BP | GO:0070102 | interleukin-6-mediated signaling pathway | 1 | 14 | 4.40e-01 |
| GO:BP | GO:0070561 | vitamin D receptor signaling pathway | 1 | 10 | 4.40e-01 |
| GO:BP | GO:0140632 | canonical inflammasome complex assembly | 1 | 23 | 4.40e-01 |
| GO:BP | GO:0071332 | cellular response to fructose stimulus | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0098902 | regulation of membrane depolarization during action potential | 1 | 5 | 4.40e-01 |
| GO:BP | GO:0060613 | fat pad development | 1 | 6 | 4.40e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 1 | 17 | 4.40e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 1 | 86 | 4.41e-01 |
| GO:BP | GO:0110076 | negative regulation of ferroptosis | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0048793 | pronephros development | 1 | 5 | 4.41e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 16 | 228 | 4.41e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 1 | 38 | 4.41e-01 |
| GO:BP | GO:1905205 | positive regulation of connective tissue replacement | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 3 | 31 | 4.41e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 1 | 91 | 4.41e-01 |
| GO:BP | GO:0010714 | positive regulation of collagen metabolic process | 1 | 24 | 4.41e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 3 | 28 | 4.41e-01 |
| GO:BP | GO:0071550 | death-inducing signaling complex assembly | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0010989 | negative regulation of low-density lipoprotein particle clearance | 1 | 4 | 4.41e-01 |
| GO:BP | GO:1900138 | negative regulation of phospholipase A2 activity | 1 | 4 | 4.41e-01 |
| GO:BP | GO:2000556 | positive regulation of T-helper 1 cell cytokine production | 1 | 2 | 4.41e-01 |
| GO:BP | GO:2000554 | regulation of T-helper 1 cell cytokine production | 1 | 2 | 4.41e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 1 | 15 | 4.41e-01 |
| GO:BP | GO:2000389 | regulation of neutrophil extravasation | 1 | 6 | 4.41e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 4.41e-01 |
| GO:BP | GO:2000659 | regulation of interleukin-1-mediated signaling pathway | 1 | 4 | 4.41e-01 |
| GO:BP | GO:0009258 | 10-formyltetrahydrofolate catabolic process | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 2 | 81 | 4.41e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 2 | 62 | 4.41e-01 |
| GO:BP | GO:1990442 | intrinsic apoptotic signaling pathway in response to nitrosative stress | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0070676 | intralumenal vesicle formation | 1 | 4 | 4.41e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 1 | 31 | 4.41e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 4.41e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 4.41e-01 |
| GO:BP | GO:0050000 | chromosome localization | 1 | 82 | 4.41e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 2 | 80 | 4.41e-01 |
| GO:BP | GO:2000211 | regulation of glutamate metabolic process | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0009441 | glycolate metabolic process | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0035744 | T-helper 1 cell cytokine production | 1 | 2 | 4.41e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 4.41e-01 |
| GO:BP | GO:1903093 | regulation of protein K48-linked deubiquitination | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 5 | 59 | 4.41e-01 |
| GO:BP | GO:0032765 | positive regulation of mast cell cytokine production | 1 | 3 | 4.41e-01 |
| GO:BP | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway | 1 | 3 | 4.41e-01 |
| GO:BP | GO:1903179 | regulation of dopamine biosynthetic process | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 1 | 30 | 4.41e-01 |
| GO:BP | GO:0040019 | positive regulation of embryonic development | 2 | 17 | 4.41e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0019835 | cytolysis | 1 | 6 | 4.41e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0061723 | glycophagy | 1 | 4 | 4.41e-01 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 6 | 4.41e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 1 | 119 | 4.41e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 4.41e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 4.41e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 4.41e-01 |
| GO:BP | GO:0070509 | calcium ion import | 1 | 37 | 4.41e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 2 | 152 | 4.41e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 22 | 4.41e-01 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 1 | 70 | 4.41e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 1 | 31 | 4.41e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 1 | 31 | 4.41e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 2 | 33 | 4.43e-01 |
| GO:BP | GO:0051665 | membrane raft localization | 1 | 4 | 4.43e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 1 | 7 | 4.43e-01 |
| GO:BP | GO:0060754 | positive regulation of mast cell chemotaxis | 1 | 5 | 4.43e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 1 | 7 | 4.43e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 1 | 64 | 4.43e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 7 | 123 | 4.43e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 1 | 13 | 4.43e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 9 | 126 | 4.43e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 1 | 25 | 4.43e-01 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 1 | 22 | 4.43e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 3 | 156 | 4.43e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 4.43e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 1 | 211 | 4.43e-01 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 1 | 29 | 4.43e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 1 | 15 | 4.43e-01 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 1 | 45 | 4.43e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 1 | 8 | 4.43e-01 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 1 | 79 | 4.43e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1 | 1147 | 4.43e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 7 | 90 | 4.43e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 1 | 165 | 4.43e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 4.43e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 3 | 39 | 4.44e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 20 | 1564 | 4.44e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 4 | 128 | 4.45e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 4 | 209 | 4.45e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 2 | 15 | 4.45e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 4 | 46 | 4.45e-01 |
| GO:BP | GO:1901289 | succinyl-CoA catabolic process | 1 | 6 | 4.45e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 2 | 19 | 4.45e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 3 | 26 | 4.45e-01 |
| GO:BP | GO:0006479 | protein methylation | 4 | 160 | 4.45e-01 |
| GO:BP | GO:0008213 | protein alkylation | 4 | 160 | 4.45e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 11 | 160 | 4.45e-01 |
| GO:BP | GO:1903412 | response to bile acid | 1 | 3 | 4.45e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 3 | 28 | 4.46e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 5 | 84 | 4.47e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 4.47e-01 |
| GO:BP | GO:0022010 | central nervous system myelination | 2 | 12 | 4.47e-01 |
| GO:BP | GO:0018149 | peptide cross-linking | 1 | 14 | 4.47e-01 |
| GO:BP | GO:0032291 | axon ensheathment in central nervous system | 2 | 12 | 4.47e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 1 | 12 | 4.47e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 1 | 13 | 4.47e-01 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 1 | 11 | 4.47e-01 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 1 | 11 | 4.47e-01 |
| GO:BP | GO:0045054 | constitutive secretory pathway | 1 | 7 | 4.48e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 1 | 13 | 4.48e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 2 | 198 | 4.48e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 2 | 22 | 4.48e-01 |
| GO:BP | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation | 1 | 2 | 4.48e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 1 | 6 | 4.48e-01 |
| GO:BP | GO:2001027 | negative regulation of endothelial cell chemotaxis | 1 | 2 | 4.48e-01 |
| GO:BP | GO:0140213 | negative regulation of long-chain fatty acid import into cell | 1 | 4 | 4.48e-01 |
| GO:BP | GO:0010748 | negative regulation of long-chain fatty acid import across plasma membrane | 1 | 4 | 4.48e-01 |
| GO:BP | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 1 | 10 | 4.49e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 3 | 28 | 4.49e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 1 | 92 | 4.50e-01 |
| GO:BP | GO:0060932 | His-Purkinje system cell differentiation | 1 | 4 | 4.50e-01 |
| GO:BP | GO:0072086 | specification of loop of Henle identity | 1 | 3 | 4.50e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 1 | 6 | 4.50e-01 |
| GO:BP | GO:0000055 | ribosomal large subunit export from nucleus | 1 | 8 | 4.50e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 4.50e-01 |
| GO:BP | GO:0031033 | myosin filament organization | 1 | 5 | 4.51e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 9 | 127 | 4.51e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 2 | 29 | 4.51e-01 |
| GO:BP | GO:0008584 | male gonad development | 1 | 99 | 4.51e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 2 | 35 | 4.51e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 4 | 40 | 4.51e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 2 | 11 | 4.51e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 3 | 29 | 4.51e-01 |
| GO:BP | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1 | 17 | 4.51e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 4.51e-01 |
| GO:BP | GO:0006306 | DNA methylation | 2 | 46 | 4.51e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 2 | 46 | 4.51e-01 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 1 | 6 | 4.51e-01 |
| GO:BP | GO:0034381 | plasma lipoprotein particle clearance | 2 | 25 | 4.51e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 2 | 84 | 4.51e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 4 | 129 | 4.51e-01 |
| GO:BP | GO:0050975 | sensory perception of touch | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 4 | 28 | 4.51e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 4.51e-01 |
| GO:BP | GO:0034375 | high-density lipoprotein particle remodeling | 2 | 11 | 4.51e-01 |
| GO:BP | GO:0072177 | mesonephric duct development | 1 | 7 | 4.51e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 1 | 13 | 4.51e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 7 | 126 | 4.51e-01 |
| GO:BP | GO:0021561 | facial nerve development | 1 | 8 | 4.51e-01 |
| GO:BP | GO:1905867 | epididymis development | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 1 | 100 | 4.51e-01 |
| GO:BP | GO:0032383 | regulation of intracellular cholesterol transport | 1 | 6 | 4.51e-01 |
| GO:BP | GO:0045905 | positive regulation of translational termination | 1 | 4 | 4.51e-01 |
| GO:BP | GO:0032026 | response to magnesium ion | 2 | 10 | 4.51e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 2 | 18 | 4.51e-01 |
| GO:BP | GO:0032380 | regulation of intracellular sterol transport | 1 | 6 | 4.51e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 3 | 24 | 4.51e-01 |
| GO:BP | GO:0051648 | vesicle localization | 3 | 192 | 4.51e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 1 | 8 | 4.51e-01 |
| GO:BP | GO:0051668 | localization within membrane | 3 | 605 | 4.51e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 5 | 89 | 4.51e-01 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 4.51e-01 |
| GO:BP | GO:0072239 | metanephric glomerulus vasculature development | 1 | 6 | 4.51e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 2 | 210 | 4.51e-01 |
| GO:BP | GO:0045901 | positive regulation of translational elongation | 1 | 4 | 4.51e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 4 | 151 | 4.51e-01 |
| GO:BP | GO:0002092 | positive regulation of receptor internalization | 2 | 21 | 4.51e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 4 | 53 | 4.51e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 1 | 13 | 4.51e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 4 | 351 | 4.51e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 1 | 33 | 4.51e-01 |
| GO:BP | GO:0051276 | chromosome organization | 5 | 543 | 4.51e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 3 | 43 | 4.51e-01 |
| GO:BP | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane | 1 | 19 | 4.51e-01 |
| GO:BP | GO:0006566 | threonine metabolic process | 1 | 4 | 4.51e-01 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 2 | 73 | 4.51e-01 |
| GO:BP | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport | 1 | 5 | 4.51e-01 |
| GO:BP | GO:0010501 | RNA secondary structure unwinding | 1 | 8 | 4.51e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 20 | 281 | 4.51e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 2 | 32 | 4.51e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 5 | 92 | 4.51e-01 |
| GO:BP | GO:0014732 | skeletal muscle atrophy | 1 | 5 | 4.51e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 5 | 92 | 4.51e-01 |
| GO:BP | GO:0098596 | imitative learning | 1 | 4 | 4.51e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 5 | 92 | 4.51e-01 |
| GO:BP | GO:0042297 | vocal learning | 1 | 4 | 4.51e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 4.51e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 3 | 104 | 4.52e-01 |
| GO:BP | GO:0060347 | heart trabecula formation | 1 | 13 | 4.52e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 4.52e-01 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 1 | 4 | 4.52e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 4.52e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 1 | 316 | 4.52e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 1 | 9 | 4.52e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 2 | 13 | 4.53e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 1 | 70 | 4.53e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 3 | 31 | 4.53e-01 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 1 | 18 | 4.53e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 1 | 1389 | 4.53e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 1 | 75 | 4.53e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 9 | 112 | 4.55e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 1 | 4 | 4.55e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 1 | 76 | 4.56e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 1 | 76 | 4.56e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 3 | 28 | 4.56e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 4.56e-01 |
| GO:BP | GO:0099500 | vesicle fusion to plasma membrane | 1 | 19 | 4.56e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 3 | 25 | 4.57e-01 |
| GO:BP | GO:0032310 | prostaglandin secretion | 2 | 8 | 4.57e-01 |
| GO:BP | GO:0048266 | behavioral response to pain | 2 | 11 | 4.57e-01 |
| GO:BP | GO:0044805 | late nucleophagy | 1 | 7 | 4.57e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 1 | 186 | 4.58e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 6 | 52 | 4.58e-01 |
| GO:BP | GO:0051412 | response to corticosterone | 1 | 10 | 4.58e-01 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 2 | 23 | 4.61e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 1 | 18 | 4.61e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 4.61e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 4.61e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 46 | 4.61e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 1 | 20 | 4.61e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 2 | 30 | 4.61e-01 |
| GO:BP | GO:0006826 | iron ion transport | 1 | 40 | 4.61e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 2 | 30 | 4.61e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 9 | 113 | 4.61e-01 |
| GO:BP | GO:0006936 | muscle contraction | 1 | 267 | 4.61e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 1 | 17 | 4.62e-01 |
| GO:BP | GO:0016233 | telomere capping | 1 | 36 | 4.62e-01 |
| GO:BP | GO:0120229 | protein localization to motile cilium | 1 | 4 | 4.62e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 1 | 6 | 4.62e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 1 | 8 | 4.62e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 2 | 17 | 4.62e-01 |
| GO:BP | GO:1901165 | positive regulation of trophoblast cell migration | 1 | 6 | 4.62e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 3 | 194 | 4.62e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 2 | 45 | 4.62e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 1 | 93 | 4.62e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 19 | 1507 | 4.63e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 3 | 29 | 4.63e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 9 | 117 | 4.63e-01 |
| GO:BP | GO:0001823 | mesonephros development | 1 | 77 | 4.63e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 3 | 24 | 4.64e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 1 | 30 | 4.64e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 41 | 687 | 4.64e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 103 | 4.64e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 1 | 46 | 4.64e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 2 | 87 | 4.64e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 4.64e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 2 | 198 | 4.64e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 3 | 38 | 4.64e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 2 | 70 | 4.64e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 28 | 459 | 4.64e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 2 | 77 | 4.64e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 1 | 122 | 4.64e-01 |
| GO:BP | GO:1904749 | regulation of protein localization to nucleolus | 1 | 7 | 4.64e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 1 | 1378 | 4.65e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 1 | 8 | 4.65e-01 |
| GO:BP | GO:0071376 | cellular response to corticotropin-releasing hormone stimulus | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 3 | 68 | 4.65e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 1 | 46 | 4.65e-01 |
| GO:BP | GO:0034587 | piRNA processing | 1 | 4 | 4.65e-01 |
| GO:BP | GO:0001947 | heart looping | 2 | 50 | 4.65e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 4.65e-01 |
| GO:BP | GO:0043435 | response to corticotropin-releasing hormone | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 19 | 294 | 4.65e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 4.65e-01 |
| GO:BP | GO:0060278 | regulation of ovulation | 1 | 2 | 4.65e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 1 | 50 | 4.65e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 4.65e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 1 | 6 | 4.65e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 4.66e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 1 | 49 | 4.66e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 7 | 484 | 4.66e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 1 | 32 | 4.66e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 4.66e-01 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 4.66e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 3 | 203 | 4.66e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 4 | 42 | 4.67e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 2 | 12 | 4.67e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 5 | 46 | 4.67e-01 |
| GO:BP | GO:0075525 | viral translational termination-reinitiation | 1 | 5 | 4.67e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 6 | 65 | 4.67e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 2 | 14 | 4.67e-01 |
| GO:BP | GO:0022615 | protein to membrane docking | 1 | 5 | 4.67e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 2 | 57 | 4.67e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 1 | 103 | 4.67e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 3 | 33 | 4.67e-01 |
| GO:BP | GO:0071603 | endothelial cell-cell adhesion | 1 | 4 | 4.67e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 2 | 91 | 4.67e-01 |
| GO:BP | GO:0006477 | protein sulfation | 1 | 6 | 4.67e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 6 | 76 | 4.67e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 5 | 92 | 4.67e-01 |
| GO:BP | GO:0034499 | late endosome to Golgi transport | 1 | 5 | 4.67e-01 |
| GO:BP | GO:0021915 | neural tube development | 6 | 146 | 4.67e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 5 | 93 | 4.67e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 2 | 250 | 4.67e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 3 | 52 | 4.67e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 17 | 1315 | 4.67e-01 |
| GO:BP | GO:0010916 | negative regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 4.67e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 17 | 1315 | 4.67e-01 |
| GO:BP | GO:0010915 | regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 4.67e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 318 | 4.67e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 7 | 4.67e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 2 | 61 | 4.67e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 1 | 114 | 4.67e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 2 | 20 | 4.67e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 4.67e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 1 | 30 | 4.67e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 1 | 8 | 4.67e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 1 | 44 | 4.67e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 1 | 32 | 4.67e-01 |
| GO:BP | GO:0007586 | digestion | 1 | 60 | 4.69e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 1 | 40 | 4.69e-01 |
| GO:BP | GO:0006547 | histidine metabolic process | 1 | 5 | 4.69e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 2 | 114 | 4.69e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 3 | 206 | 4.70e-01 |
| GO:BP | GO:1903140 | regulation of establishment of endothelial barrier | 2 | 12 | 4.71e-01 |
| GO:BP | GO:0022617 | extracellular matrix disassembly | 1 | 36 | 4.71e-01 |
| GO:BP | GO:0035272 | exocrine system development | 1 | 34 | 4.71e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 5 | 93 | 4.71e-01 |
| GO:BP | GO:0051917 | regulation of fibrinolysis | 2 | 10 | 4.71e-01 |
| GO:BP | GO:0072385 | minus-end-directed organelle transport along microtubule | 1 | 5 | 4.71e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 4.71e-01 |
| GO:BP | GO:1900736 | regulation of phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 2 | 4.71e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 6 | 71 | 4.71e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 20 | 1604 | 4.71e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 2 | 19 | 4.71e-01 |
| GO:BP | GO:0031648 | protein destabilization | 1 | 45 | 4.71e-01 |
| GO:BP | GO:1901550 | regulation of endothelial cell development | 2 | 12 | 4.71e-01 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 1 | 6 | 4.71e-01 |
| GO:BP | GO:0044070 | regulation of monoatomic anion transport | 2 | 12 | 4.71e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 2 | 26 | 4.71e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 4.71e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 2 | 26 | 4.71e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 17 | 1322 | 4.71e-01 |
| GO:BP | GO:0140459 | response to Gram-positive bacterium | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 24 | 1238 | 4.71e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 4.71e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 2 | 11 | 4.71e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 3 | 54 | 4.71e-01 |
| GO:BP | GO:0110075 | regulation of ferroptosis | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0002082 | regulation of oxidative phosphorylation | 2 | 19 | 4.71e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 1 | 48 | 4.71e-01 |
| GO:BP | GO:0070729 | cyclic nucleotide transport | 1 | 6 | 4.71e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 2 | 14 | 4.71e-01 |
| GO:BP | GO:0097707 | ferroptosis | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0032965 | regulation of collagen biosynthetic process | 1 | 30 | 4.71e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 1 | 18 | 4.71e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 2 | 13 | 4.71e-01 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 1 | 5 | 4.71e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 1 | 113 | 4.71e-01 |
| GO:BP | GO:0002580 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 1 | 4.71e-01 |
| GO:BP | GO:2000152 | regulation of ubiquitin-specific protease activity | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 2 | 39 | 4.71e-01 |
| GO:BP | GO:0002155 | regulation of thyroid hormone mediated signaling pathway | 1 | 5 | 4.71e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 4.71e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 2 | 69 | 4.71e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 2 | 16 | 4.71e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 3 | 20 | 4.71e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 1 | 21 | 4.71e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 2 | 13 | 4.71e-01 |
| GO:BP | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells | 1 | 4 | 4.71e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 1 | 16 | 4.71e-01 |
| GO:BP | GO:0009073 | aromatic amino acid family biosynthetic process | 1 | 3 | 4.71e-01 |
| GO:BP | GO:2000353 | positive regulation of endothelial cell apoptotic process | 2 | 13 | 4.71e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 1 | 8 | 4.71e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 1 | 51 | 4.71e-01 |
| GO:BP | GO:1904684 | negative regulation of metalloendopeptidase activity | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 2 | 125 | 4.71e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 2 | 21 | 4.71e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 14 | 859 | 4.71e-01 |
| GO:BP | GO:0043276 | anoikis | 2 | 32 | 4.71e-01 |
| GO:BP | GO:1903347 | negative regulation of bicellular tight junction assembly | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 1 | 13 | 4.71e-01 |
| GO:BP | GO:0090086 | negative regulation of protein deubiquitination | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 1 | 25 | 4.71e-01 |
| GO:BP | GO:1903790 | guanine nucleotide transmembrane transport | 1 | 5 | 4.71e-01 |
| GO:BP | GO:0099576 | regulation of protein catabolic process at postsynapse, modulating synaptic transmission | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0050909 | sensory perception of taste | 1 | 13 | 4.71e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 4.71e-01 |
| GO:BP | GO:0003383 | apical constriction | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 2 | 13 | 4.71e-01 |
| GO:BP | GO:1901671 | positive regulation of superoxide dismutase activity | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0072359 | circulatory system development | 1 | 884 | 4.71e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0042220 | response to cocaine | 1 | 28 | 4.71e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 6 | 165 | 4.71e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 2 | 12 | 4.71e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 1 | 25 | 4.71e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 2 | 21 | 4.71e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 2 | 107 | 4.71e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 2 | 86 | 4.71e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 1 | 7 | 4.71e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 9 | 330 | 4.72e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 20 | 513 | 4.72e-01 |
| GO:BP | GO:0060272 | embryonic skeletal joint morphogenesis | 1 | 7 | 4.72e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 1 | 17 | 4.72e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 29 | 4.72e-01 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 1 | 8 | 4.72e-01 |
| GO:BP | GO:1905603 | regulation of blood-brain barrier permeability | 1 | 6 | 4.72e-01 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 1 | 7 | 4.72e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 6 | 676 | 4.72e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 1 | 17 | 4.72e-01 |
| GO:BP | GO:0006914 | autophagy | 20 | 513 | 4.72e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 1 | 84 | 4.72e-01 |
| GO:BP | GO:1900221 | regulation of amyloid-beta clearance | 1 | 13 | 4.72e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 1 | 27 | 4.72e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 90 | 4.72e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 1 | 9 | 4.72e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 1 | 7 | 4.72e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 18 | 1441 | 4.72e-01 |
| GO:BP | GO:0098597 | observational learning | 1 | 5 | 4.72e-01 |
| GO:BP | GO:0060753 | regulation of mast cell chemotaxis | 1 | 7 | 4.72e-01 |
| GO:BP | GO:0007379 | segment specification | 2 | 14 | 4.72e-01 |
| GO:BP | GO:0014063 | negative regulation of serotonin secretion | 1 | 2 | 4.72e-01 |
| GO:BP | GO:0034201 | response to oleic acid | 1 | 5 | 4.73e-01 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 3 | 30 | 4.73e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 1 | 4 | 4.73e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 4 | 68 | 4.74e-01 |
| GO:BP | GO:0031638 | zymogen activation | 1 | 45 | 4.74e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 1 | 386 | 4.74e-01 |
| GO:BP | GO:0015917 | aminophospholipid transport | 1 | 8 | 4.75e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 3 | 27 | 4.75e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 4.75e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 7 | 639 | 4.75e-01 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 2 | 8 | 4.75e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 1 | 80 | 4.75e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 4.75e-01 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 3 | 24 | 4.75e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 1 | 10 | 4.75e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 1 | 37 | 4.75e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 4 | 42 | 4.75e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 1 | 317 | 4.75e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 3 | 58 | 4.75e-01 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 4.75e-01 |
| GO:BP | GO:0006578 | amino-acid betaine biosynthetic process | 1 | 5 | 4.75e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 1 | 5 | 4.75e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 3 | 129 | 4.75e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 1 | 13 | 4.75e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 2 | 104 | 4.75e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 1 | 51 | 4.75e-01 |
| GO:BP | GO:0035148 | tube formation | 6 | 138 | 4.75e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 1 | 47 | 4.75e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 1 | 17 | 4.75e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 2 | 173 | 4.75e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 5 | 64 | 4.75e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 3 | 275 | 4.76e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 2 | 93 | 4.76e-01 |
| GO:BP | GO:1902074 | response to salt | 3 | 271 | 4.76e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 4.76e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 4.76e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 2 | 37 | 4.78e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 2 | 56 | 4.80e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 2 | 52 | 4.80e-01 |
| GO:BP | GO:0042594 | response to starvation | 2 | 178 | 4.80e-01 |
| GO:BP | GO:0036016 | cellular response to interleukin-3 | 1 | 6 | 4.80e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 1 | 4 | 4.80e-01 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 4.80e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 1 | 4 | 4.80e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 2 | 22 | 4.80e-01 |
| GO:BP | GO:0051225 | spindle assembly | 1 | 120 | 4.80e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 2 | 106 | 4.80e-01 |
| GO:BP | GO:0036015 | response to interleukin-3 | 1 | 6 | 4.80e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 2 | 29 | 4.80e-01 |
| GO:BP | GO:0007052 | mitotic spindle organization | 2 | 129 | 4.80e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 4.80e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 4.80e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 2 | 71 | 4.80e-01 |
| GO:BP | GO:0032289 | central nervous system myelin formation | 1 | 4 | 4.80e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 1 | 4 | 4.80e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 1 | 15 | 4.80e-01 |
| GO:BP | GO:0060438 | trachea development | 1 | 13 | 4.80e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 3 | 28 | 4.80e-01 |
| GO:BP | GO:0060174 | limb bud formation | 1 | 10 | 4.81e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 5 | 65 | 4.81e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 4.81e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 4 | 52 | 4.81e-01 |
| GO:BP | GO:0033762 | response to glucagon | 2 | 13 | 4.81e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 2 | 97 | 4.81e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 4 | 138 | 4.81e-01 |
| GO:BP | GO:0014891 | striated muscle atrophy | 1 | 6 | 4.82e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 1 | 16 | 4.82e-01 |
| GO:BP | GO:0008272 | sulfate transport | 2 | 12 | 4.82e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 1 | 19 | 4.82e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 1 | 18 | 4.82e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 6 | 223 | 4.82e-01 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 1 | 5 | 4.82e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 1 | 18 | 4.82e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 6 | 121 | 4.83e-01 |
| GO:BP | GO:0050746 | regulation of lipoprotein metabolic process | 1 | 15 | 4.83e-01 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 1 | 11 | 4.83e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 2 | 12 | 4.83e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 2 | 10 | 4.83e-01 |
| GO:BP | GO:0019373 | epoxygenase P450 pathway | 2 | 4 | 4.83e-01 |
| GO:BP | GO:0006412 | translation | 16 | 646 | 4.85e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 1 | 67 | 4.85e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 1 | 23 | 4.85e-01 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 1 | 8 | 4.85e-01 |
| GO:BP | GO:0038134 | ERBB2-EGFR signaling pathway | 1 | 6 | 4.85e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 4 | 71 | 4.85e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 2 | 30 | 4.85e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 1 | 51 | 4.85e-01 |
| GO:BP | GO:0003012 | muscle system process | 1 | 334 | 4.85e-01 |
| GO:BP | GO:0032868 | response to insulin | 5 | 215 | 4.85e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 1 | 38 | 4.85e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 1 | 63 | 4.85e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 1 | 30 | 4.85e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 1 | 45 | 4.85e-01 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 2 | 25 | 4.85e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 2 | 30 | 4.85e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 7 | 93 | 4.85e-01 |
| GO:BP | GO:1905907 | negative regulation of amyloid fibril formation | 1 | 10 | 4.85e-01 |
| GO:BP | GO:0045829 | negative regulation of isotype switching | 1 | 4 | 4.86e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 2 | 43 | 4.86e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 4.86e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 1 | 32 | 4.86e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 1 | 50 | 4.86e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 4 | 165 | 4.86e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 2 | 19 | 4.86e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 32 | 4.86e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 5 | 517 | 4.86e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 1 | 21 | 4.86e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 4.86e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 3 | 19 | 4.86e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 4.86e-01 |
| GO:BP | GO:0009914 | hormone transport | 2 | 215 | 4.86e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 2 | 31 | 4.86e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 4.86e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 9 | 132 | 4.86e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 4.86e-01 |
| GO:BP | GO:0051620 | norepinephrine uptake | 1 | 4 | 4.86e-01 |
| GO:BP | GO:0051589 | negative regulation of neurotransmitter transport | 1 | 8 | 4.86e-01 |
| GO:BP | GO:0010712 | regulation of collagen metabolic process | 1 | 34 | 4.86e-01 |
| GO:BP | GO:0033572 | transferrin transport | 1 | 8 | 4.86e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 4.86e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 1 | 49 | 4.86e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 3 | 37 | 4.87e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 6 | 70 | 4.87e-01 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 1 | 23 | 4.87e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 1 | 18 | 4.87e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 1 | 76 | 4.87e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 2 | 391 | 4.87e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 2 | 246 | 4.87e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 4.87e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 3 | 32 | 4.87e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 1 | 19 | 4.87e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 1 | 9 | 4.88e-01 |
| GO:BP | GO:0060215 | primitive hemopoiesis | 1 | 7 | 4.88e-01 |
| GO:BP | GO:0046874 | quinolinate metabolic process | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0009086 | methionine biosynthetic process | 1 | 10 | 4.88e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 1 | 7 | 4.88e-01 |
| GO:BP | GO:0002154 | thyroid hormone mediated signaling pathway | 1 | 6 | 4.88e-01 |
| GO:BP | GO:0007220 | Notch receptor processing | 1 | 9 | 4.88e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 4.88e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 6 | 81 | 4.88e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 1 | 8 | 4.88e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 1 | 53 | 4.88e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 4 | 167 | 4.88e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 1 | 15 | 4.88e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 1 | 26 | 4.89e-01 |
| GO:BP | GO:0000423 | mitophagy | 3 | 37 | 4.89e-01 |
| GO:BP | GO:0070934 | CRD-mediated mRNA stabilization | 1 | 11 | 4.89e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 1 | 33 | 4.89e-01 |
| GO:BP | GO:0048565 | digestive tract development | 1 | 92 | 4.89e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 1 | 67 | 4.89e-01 |
| GO:BP | GO:0048232 | male gamete generation | 9 | 343 | 4.89e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 1 | 15 | 4.89e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 4.89e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 785 | 4.89e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 41 | 4.89e-01 |
| GO:BP | GO:1905150 | regulation of voltage-gated sodium channel activity | 1 | 6 | 4.89e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 2 | 24 | 4.89e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 4 | 48 | 4.89e-01 |
| GO:BP | GO:0031620 | regulation of fever generation | 1 | 4 | 4.89e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 134 | 4.89e-01 |
| GO:BP | GO:0018158 | protein oxidation | 1 | 12 | 4.90e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 6 | 669 | 4.90e-01 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 2 | 17 | 4.90e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 3 | 50 | 4.90e-01 |
| GO:BP | GO:0001408 | guanine nucleotide transport | 1 | 6 | 4.90e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 6 | 96 | 4.90e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 4.90e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 1 | 5 | 4.90e-01 |
| GO:BP | GO:1901164 | negative regulation of trophoblast cell migration | 1 | 4 | 4.90e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 2 | 192 | 4.90e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 1 | 8 | 4.90e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 3 | 27 | 4.90e-01 |
| GO:BP | GO:0031054 | pre-miRNA processing | 1 | 14 | 4.90e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 1 | 18 | 4.90e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 1 | 18 | 4.90e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 3 | 31 | 4.90e-01 |
| GO:BP | GO:0071051 | polyadenylation-dependent snoRNA 3’-end processing | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0060271 | cilium assembly | 6 | 292 | 4.91e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 1 | 43 | 4.91e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 1 | 41 | 4.91e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 3 | 37 | 4.91e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 4.91e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 1 | 12 | 4.91e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 4 | 41 | 4.91e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 7 | 321 | 4.91e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 4.91e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 2 | 77 | 4.91e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 3 | 179 | 4.93e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 1 | 9 | 4.94e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 4 | 72 | 4.94e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1 | 10 | 4.94e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 1 | 8 | 4.94e-01 |
| GO:BP | GO:0010746 | regulation of long-chain fatty acid import across plasma membrane | 1 | 5 | 4.94e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 1 | 4 | 4.94e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 3 | 37 | 4.94e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 2 | 13 | 4.94e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 1 | 49 | 4.94e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 1 | 20 | 4.94e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 2 | 11 | 4.95e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 9 | 461 | 4.95e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 1 | 137 | 4.96e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 4 | 153 | 4.96e-01 |
| GO:BP | GO:0003166 | bundle of His development | 1 | 6 | 4.96e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 2 | 188 | 4.96e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0051596 | methylglyoxal catabolic process | 1 | 5 | 4.96e-01 |
| GO:BP | GO:1903792 | negative regulation of monoatomic anion transport | 1 | 3 | 4.96e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 4.96e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 5 | 4.96e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0006526 | arginine biosynthetic process | 1 | 4 | 4.96e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 2 | 77 | 4.96e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 2 | 47 | 4.96e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 2 | 30 | 4.96e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 4.96e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 4.96e-01 |
| GO:BP | GO:2000825 | positive regulation of androgen receptor activity | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 2 | 418 | 4.96e-01 |
| GO:BP | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 1 | 3 | 4.96e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 1 | 4 | 4.96e-01 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 4 | 55 | 4.96e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0030220 | platelet formation | 2 | 17 | 4.96e-01 |
| GO:BP | GO:0016236 | macroautophagy | 4 | 308 | 4.96e-01 |
| GO:BP | GO:0140206 | dipeptide import across plasma membrane | 1 | 2 | 4.96e-01 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 4 | 55 | 4.96e-01 |
| GO:BP | GO:0140205 | oligopeptide import across plasma membrane | 1 | 2 | 4.96e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 7 | 439 | 4.96e-01 |
| GO:BP | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 2 | 194 | 4.96e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 4 | 53 | 4.96e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 11 | 665 | 4.96e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 1 | 28 | 4.96e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 3 | 81 | 4.96e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 2 | 9 | 4.96e-01 |
| GO:BP | GO:0061727 | methylglyoxal catabolic process to lactate | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 1 | 12 | 4.96e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 7 | 165 | 4.96e-01 |
| GO:BP | GO:1901668 | regulation of superoxide dismutase activity | 1 | 4 | 4.96e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 4.96e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 4.96e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0022614 | membrane to membrane docking | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 4.96e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 2 | 27 | 4.96e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 7 | 107 | 4.96e-01 |
| GO:BP | GO:0015739 | sialic acid transport | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway | 1 | 6 | 4.97e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 1 | 19 | 4.97e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 5 | 245 | 4.98e-01 |
| GO:BP | GO:0097278 | complement-dependent cytotoxicity | 1 | 4 | 4.98e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 2 | 179 | 4.99e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 2 | 79 | 4.99e-01 |
| GO:BP | GO:0016577 | histone demethylation | 1 | 21 | 4.99e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 1 | 31 | 4.99e-01 |
| GO:BP | GO:1990349 | gap junction-mediated intercellular transport | 1 | 2 | 4.99e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 15 | 4.99e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 1 | 13 | 4.99e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 2 | 75 | 4.99e-01 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 12 | 5.00e-01 |
| GO:BP | GO:0055123 | digestive system development | 1 | 100 | 5.00e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 3 | 50 | 5.00e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 5 | 540 | 5.00e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 3 | 31 | 5.00e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 4 | 174 | 5.00e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 1 | 5 | 5.00e-01 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 4 | 44 | 5.01e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 4 | 47 | 5.01e-01 |
| GO:BP | GO:0070900 | mitochondrial tRNA modification | 1 | 9 | 5.01e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 1 | 20 | 5.01e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1 | 1869 | 5.01e-01 |
| GO:BP | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 1 | 5 | 5.01e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 2 | 30 | 5.01e-01 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 1 | 7 | 5.01e-01 |
| GO:BP | GO:1902572 | negative regulation of serine-type peptidase activity | 1 | 4 | 5.01e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 1 | 10 | 5.01e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 1 | 78 | 5.01e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 70 | 5.01e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 1 | 25 | 5.01e-01 |
| GO:BP | GO:1900864 | mitochondrial RNA modification | 1 | 9 | 5.01e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 1 | 20 | 5.01e-01 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 5.01e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 2 | 35 | 5.01e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 1 | 62 | 5.01e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 1 | 30 | 5.01e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 9 | 121 | 5.01e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 2 | 17 | 5.01e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 3 | 42 | 5.01e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 114 | 5.01e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 1 | 149 | 5.01e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 1 | 53 | 5.01e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 1 | 10 | 5.01e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 1 | 10 | 5.01e-01 |
| GO:BP | GO:0008406 | gonad development | 1 | 158 | 5.01e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 3 | 70 | 5.01e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 2 | 17 | 5.01e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 37 | 5.01e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 4 | 133 | 5.01e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 2 | 111 | 5.01e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 1 | 30 | 5.01e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 1 | 112 | 5.01e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 1 | 13 | 5.01e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 2 | 16 | 5.01e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 114 | 5.01e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 1 | 51 | 5.01e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 5.01e-01 |
| GO:BP | GO:0002326 | B cell lineage commitment | 1 | 4 | 5.01e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 1 | 19 | 5.01e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 5.01e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 3 | 138 | 5.01e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 1 | 11 | 5.01e-01 |
| GO:BP | GO:0030901 | midbrain development | 2 | 69 | 5.01e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 1 | 6 | 5.01e-01 |
| GO:BP | GO:1900004 | negative regulation of serine-type endopeptidase activity | 1 | 4 | 5.01e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 1 | 6 | 5.01e-01 |
| GO:BP | GO:0006104 | succinyl-CoA metabolic process | 1 | 9 | 5.01e-01 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 1 | 7 | 5.01e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 2 | 60 | 5.01e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 1 | 75 | 5.01e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 13 | 775 | 5.01e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 5.01e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 2 | 21 | 5.02e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 9 | 469 | 5.02e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 2 | 19 | 5.02e-01 |
| GO:BP | GO:1905517 | macrophage migration | 4 | 34 | 5.02e-01 |
| GO:BP | GO:0072672 | neutrophil extravasation | 1 | 7 | 5.02e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 5.02e-01 |
| GO:BP | GO:0044062 | regulation of excretion | 2 | 12 | 5.03e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 1 | 86 | 5.03e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 2 | 42 | 5.03e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 1 | 35 | 5.03e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 35 | 5.03e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 1 | 19 | 5.03e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 19 | 1602 | 5.03e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 2 | 216 | 5.04e-01 |
| GO:BP | GO:0045656 | negative regulation of monocyte differentiation | 1 | 4 | 5.04e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 1 | 161 | 5.04e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 2 | 85 | 5.05e-01 |
| GO:BP | GO:0015849 | organic acid transport | 2 | 217 | 5.05e-01 |
| GO:BP | GO:0071305 | cellular response to vitamin D | 1 | 14 | 5.05e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 1 | 26 | 5.07e-01 |
| GO:BP | GO:0048280 | vesicle fusion with Golgi apparatus | 1 | 8 | 5.07e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 4 | 105 | 5.07e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 1 | 10 | 5.08e-01 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 1 | 8 | 5.08e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 5.08e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 3 | 50 | 5.08e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 2 | 56 | 5.08e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 1 | 14 | 5.08e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 1 | 39 | 5.08e-01 |
| GO:BP | GO:0098598 | learned vocalization behavior or vocal learning | 1 | 4 | 5.08e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 4 | 49 | 5.08e-01 |
| GO:BP | GO:0051693 | actin filament capping | 3 | 34 | 5.08e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 3 | 742 | 5.08e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4 | 100 | 5.08e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 2 | 88 | 5.08e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 4 | 51 | 5.09e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 6 | 357 | 5.09e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 5.09e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 3 | 140 | 5.09e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 7 | 94 | 5.09e-01 |
| GO:BP | GO:0033631 | cell-cell adhesion mediated by integrin | 1 | 12 | 5.09e-01 |
| GO:BP | GO:0061024 | membrane organization | 11 | 671 | 5.10e-01 |
| GO:BP | GO:0006837 | serotonin transport | 2 | 11 | 5.10e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 5.10e-01 |
| GO:BP | GO:1903649 | regulation of cytoplasmic transport | 1 | 25 | 5.10e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 5.10e-01 |
| GO:BP | GO:0042053 | regulation of dopamine metabolic process | 2 | 11 | 5.10e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 3 | 116 | 5.10e-01 |
| GO:BP | GO:0042069 | regulation of catecholamine metabolic process | 2 | 11 | 5.10e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 2 | 33 | 5.10e-01 |
| GO:BP | GO:0043320 | natural killer cell degranulation | 1 | 6 | 5.10e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 1 | 54 | 5.10e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 5.10e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 400 | 5.10e-01 |
| GO:BP | GO:0006950 | response to stress | 2 | 2780 | 5.10e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 1 | 6 | 5.10e-01 |
| GO:BP | GO:0071354 | cellular response to interleukin-6 | 1 | 23 | 5.10e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 1 | 10 | 5.10e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 2 | 71 | 5.10e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1 | 9 | 5.10e-01 |
| GO:BP | GO:0031340 | positive regulation of vesicle fusion | 1 | 9 | 5.10e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 2 | 74 | 5.10e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 1 | 12 | 5.10e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 1 | 13 | 5.10e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 1 | 10 | 5.10e-01 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 2 | 36 | 5.10e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 1 | 14 | 5.10e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 1 | 7 | 5.10e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 1 | 157 | 5.10e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 5.11e-01 |
| GO:BP | GO:0010988 | regulation of low-density lipoprotein particle clearance | 1 | 8 | 5.11e-01 |
| GO:BP | GO:0007399 | nervous system development | 17 | 1904 | 5.11e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 3 | 90 | 5.11e-01 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 1 | 5 | 5.11e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 2 | 47 | 5.11e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 16 | 666 | 5.11e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 1 | 884 | 5.12e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 1 | 10 | 5.13e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 2 | 11 | 5.13e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 2 | 11 | 5.13e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 1 | 11 | 5.13e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 3 | 49 | 5.13e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 5 | 78 | 5.13e-01 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 1 | 8 | 5.13e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 5.13e-01 |
| GO:BP | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 5.13e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 11 | 277 | 5.13e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 5.13e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 1 | 56 | 5.13e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 2 | 40 | 5.13e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 1 | 631 | 5.14e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 1 | 46 | 5.14e-01 |
| GO:BP | GO:0035265 | organ growth | 1 | 131 | 5.14e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 1 | 41 | 5.14e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 3 | 62 | 5.14e-01 |
| GO:BP | GO:0060348 | bone development | 2 | 170 | 5.14e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 1 | 60 | 5.14e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 2 | 59 | 5.14e-01 |
| GO:BP | GO:0034447 | very-low-density lipoprotein particle clearance | 1 | 4 | 5.14e-01 |
| GO:BP | GO:0003164 | His-Purkinje system development | 1 | 7 | 5.14e-01 |
| GO:BP | GO:0034382 | chylomicron remnant clearance | 1 | 2 | 5.14e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 77 | 5.14e-01 |
| GO:BP | GO:0051005 | negative regulation of lipoprotein lipase activity | 1 | 4 | 5.14e-01 |
| GO:BP | GO:0071830 | triglyceride-rich lipoprotein particle clearance | 1 | 2 | 5.14e-01 |
| GO:BP | GO:0007281 | germ cell development | 2 | 187 | 5.14e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 2 | 31 | 5.14e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 5.14e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 5.15e-01 |
| GO:BP | GO:0001660 | fever generation | 1 | 5 | 5.15e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 4 | 170 | 5.15e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 1 | 2102 | 5.15e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 2 | 11 | 5.15e-01 |
| GO:BP | GO:1904141 | positive regulation of microglial cell migration | 1 | 3 | 5.15e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 3 | 295 | 5.15e-01 |
| GO:BP | GO:0033869 | nucleoside bisphosphate catabolic process | 1 | 11 | 5.15e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 5.15e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 2 | 11 | 5.15e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 1 | 11 | 5.15e-01 |
| GO:BP | GO:0034034 | purine nucleoside bisphosphate catabolic process | 1 | 11 | 5.15e-01 |
| GO:BP | GO:0034031 | ribonucleoside bisphosphate catabolic process | 1 | 11 | 5.15e-01 |
| GO:BP | GO:0098779 | positive regulation of mitophagy in response to mitochondrial depolarization | 1 | 10 | 5.15e-01 |
| GO:BP | GO:0032483 | regulation of Rab protein signal transduction | 1 | 7 | 5.15e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 2 | 22 | 5.15e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 2 | 69 | 5.16e-01 |
| GO:BP | GO:0006531 | aspartate metabolic process | 1 | 4 | 5.16e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 15 | 227 | 5.16e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 2 | 21 | 5.16e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 3 | 827 | 5.16e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 5 | 59 | 5.16e-01 |
| GO:BP | GO:0009411 | response to UV | 1 | 141 | 5.16e-01 |
| GO:BP | GO:0070933 | histone H4 deacetylation | 1 | 7 | 5.16e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 3 | 44 | 5.16e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 4 | 75 | 5.16e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 1 | 23 | 5.17e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 1 | 544 | 5.17e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 7 | 5.17e-01 |
| GO:BP | GO:0009584 | detection of visible light | 1 | 20 | 5.17e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 1 | 8 | 5.17e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 3 | 37 | 5.18e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 1 | 8 | 5.19e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 1 | 12 | 5.20e-01 |
| GO:BP | GO:1905203 | regulation of connective tissue replacement | 1 | 5 | 5.20e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 2 | 252 | 5.20e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 1 | 16 | 5.20e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 1 | 64 | 5.21e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 5.21e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 5.21e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 12 | 5.21e-01 |
| GO:BP | GO:0072176 | nephric duct development | 1 | 12 | 5.21e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 1 | 11 | 5.21e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 1 | 475 | 5.21e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 2 | 23 | 5.21e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 1 | 34 | 5.21e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 3 | 39 | 5.21e-01 |
| GO:BP | GO:0098885 | modification of postsynaptic actin cytoskeleton | 1 | 10 | 5.21e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 5.21e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 7 | 106 | 5.21e-01 |
| GO:BP | GO:0009111 | vitamin catabolic process | 1 | 3 | 5.21e-01 |
| GO:BP | GO:0070741 | response to interleukin-6 | 1 | 23 | 5.22e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 5.22e-01 |
| GO:BP | GO:1902416 | positive regulation of mRNA binding | 1 | 8 | 5.22e-01 |
| GO:BP | GO:0071500 | cellular response to nitrosative stress | 1 | 6 | 5.22e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 2 | 26 | 5.22e-01 |
| GO:BP | GO:1905906 | regulation of amyloid fibril formation | 1 | 14 | 5.22e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 1 | 47 | 5.22e-01 |
| GO:BP | GO:0015811 | L-cystine transport | 1 | 3 | 5.22e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 5.22e-01 |
| GO:BP | GO:0033182 | regulation of histone ubiquitination | 1 | 6 | 5.22e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 1 | 13 | 5.22e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 1 | 5 | 5.22e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 3 | 32 | 5.22e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 5.22e-01 |
| GO:BP | GO:0090382 | phagosome maturation | 2 | 24 | 5.22e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 2 | 73 | 5.23e-01 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 3 | 186 | 5.23e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 4 | 191 | 5.23e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 1 | 63 | 5.23e-01 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 2 | 29 | 5.24e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 2 | 53 | 5.24e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 8 | 100 | 5.24e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 3 | 197 | 5.24e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 1 | 9 | 5.24e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 3 | 42 | 5.24e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 1 | 10 | 5.24e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 5.24e-01 |
| GO:BP | GO:0032790 | ribosome disassembly | 1 | 11 | 5.24e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 2 | 219 | 5.24e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 5 | 58 | 5.24e-01 |
| GO:BP | GO:0043697 | cell dedifferentiation | 1 | 6 | 5.25e-01 |
| GO:BP | GO:0043696 | dedifferentiation | 1 | 6 | 5.25e-01 |
| GO:BP | GO:0070358 | actin polymerization-dependent cell motility | 1 | 6 | 5.26e-01 |
| GO:BP | GO:0014062 | regulation of serotonin secretion | 1 | 3 | 5.26e-01 |
| GO:BP | GO:0099612 | protein localization to axon | 1 | 6 | 5.26e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 2 | 86 | 5.26e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 5.26e-01 |
| GO:BP | GO:0090102 | cochlea development | 1 | 35 | 5.26e-01 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 5.26e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 5.26e-01 |
| GO:BP | GO:0044782 | cilium organization | 6 | 313 | 5.26e-01 |
| GO:BP | GO:0007584 | response to nutrient | 3 | 109 | 5.26e-01 |
| GO:BP | GO:0002551 | mast cell chemotaxis | 1 | 9 | 5.26e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 22 | 344 | 5.26e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 5.26e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 1 | 11 | 5.26e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 1 | 33 | 5.26e-01 |
| GO:BP | GO:0006555 | methionine metabolic process | 1 | 13 | 5.26e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 3 | 513 | 5.26e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 2 | 13 | 5.26e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 1 | 71 | 5.28e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 1 | 5 | 5.28e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 1 | 51 | 5.28e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 1 | 41 | 5.28e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 1 | 9 | 5.28e-01 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 1 | 30 | 5.28e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 1 | 7 | 5.28e-01 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 1 | 22 | 5.28e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 5 | 74 | 5.28e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 2 | 18 | 5.29e-01 |
| GO:BP | GO:0051302 | regulation of cell division | 1 | 145 | 5.30e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 2 | 27 | 5.30e-01 |
| GO:BP | GO:0031650 | regulation of heat generation | 1 | 7 | 5.30e-01 |
| GO:BP | GO:0048293 | regulation of isotype switching to IgE isotypes | 1 | 5 | 5.31e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 1 | 64 | 5.31e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 5.31e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 1 | 21 | 5.31e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 26 | 2270 | 5.31e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 2 | 35 | 5.31e-01 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 2 | 43 | 5.31e-01 |
| GO:BP | GO:0048284 | organelle fusion | 2 | 135 | 5.31e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 2 | 30 | 5.31e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 1 | 168 | 5.31e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 1 | 27 | 5.31e-01 |
| GO:BP | GO:0048289 | isotype switching to IgE isotypes | 1 | 5 | 5.31e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 1 | 13 | 5.31e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 2 | 30 | 5.31e-01 |
| GO:BP | GO:0046949 | fatty-acyl-CoA biosynthetic process | 1 | 20 | 5.31e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 16 | 993 | 5.31e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 152 | 5.31e-01 |
| GO:BP | GO:0006482 | protein demethylation | 1 | 27 | 5.31e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 1 | 16 | 5.31e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 4 | 67 | 5.31e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 5 | 563 | 5.31e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 1 | 10 | 5.31e-01 |
| GO:BP | GO:0032713 | negative regulation of interleukin-4 production | 1 | 5 | 5.31e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 7 | 114 | 5.31e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 1 | 20 | 5.31e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 1 | 12 | 5.31e-01 |
| GO:BP | GO:0045453 | bone resorption | 1 | 40 | 5.31e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 5.31e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 3 | 59 | 5.31e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 3 | 35 | 5.31e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 5.31e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 1 | 14 | 5.31e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 2 | 48 | 5.32e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 1 | 175 | 5.32e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 5.32e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 2 | 147 | 5.32e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 1 | 604 | 5.32e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 2 | 39 | 5.32e-01 |
| GO:BP | GO:1905523 | positive regulation of macrophage migration | 2 | 13 | 5.33e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 1 | 20 | 5.33e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 1 | 6 | 5.34e-01 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 2 | 104 | 5.34e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 2 | 407 | 5.34e-01 |
| GO:BP | GO:0060468 | prevention of polyspermy | 1 | 2 | 5.34e-01 |
| GO:BP | GO:0060467 | negative regulation of fertilization | 1 | 2 | 5.34e-01 |
| GO:BP | GO:0048736 | appendage development | 1 | 142 | 5.34e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 3 | 57 | 5.34e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 1 | 24 | 5.34e-01 |
| GO:BP | GO:0006885 | regulation of pH | 6 | 74 | 5.34e-01 |
| GO:BP | GO:0140212 | regulation of long-chain fatty acid import into cell | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1 | 24 | 5.34e-01 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0060173 | limb development | 1 | 142 | 5.34e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 2 | 108 | 5.34e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 5.34e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 4 | 52 | 5.34e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 2 | 47 | 5.35e-01 |
| GO:BP | GO:0007548 | sex differentiation | 1 | 191 | 5.35e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 3 | 76 | 5.35e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 1 | 11 | 5.35e-01 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 1 | 15 | 5.35e-01 |
| GO:BP | GO:0033750 | ribosome localization | 1 | 15 | 5.35e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 2 | 84 | 5.36e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 5.36e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 5.36e-01 |
| GO:BP | GO:0072126 | positive regulation of glomerular mesangial cell proliferation | 1 | 7 | 5.36e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 3 | 57 | 5.36e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 1 | 10 | 5.36e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 6 | 150 | 5.37e-01 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 5.37e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 1 | 13 | 5.37e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 2 | 20 | 5.37e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 2 | 36 | 5.38e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 3 | 30 | 5.38e-01 |
| GO:BP | GO:0007051 | spindle organization | 1 | 188 | 5.38e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 1 | 12 | 5.38e-01 |
| GO:BP | GO:0001731 | formation of translation preinitiation complex | 1 | 11 | 5.38e-01 |
| GO:BP | GO:1904139 | regulation of microglial cell migration | 1 | 3 | 5.38e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 1 | 7 | 5.38e-01 |
| GO:BP | GO:0060539 | diaphragm development | 1 | 5 | 5.38e-01 |
| GO:BP | GO:1904124 | microglial cell migration | 1 | 3 | 5.38e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 7 | 110 | 5.38e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 5.38e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 1 | 5 | 5.38e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 5 | 241 | 5.38e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 1 | 7 | 5.38e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 1 | 22 | 5.38e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 2 | 128 | 5.38e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 97 | 5.38e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 3 | 33 | 5.38e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 2 | 85 | 5.38e-01 |
| GO:BP | GO:2000726 | negative regulation of cardiac muscle cell differentiation | 1 | 8 | 5.38e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 1 | 21 | 5.38e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 3 | 100 | 5.38e-01 |
| GO:BP | GO:0060696 | regulation of phospholipid catabolic process | 1 | 5 | 5.38e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 1 | 119 | 5.38e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 1 | 7 | 5.38e-01 |
| GO:BP | GO:2001225 | regulation of chloride transport | 1 | 4 | 5.38e-01 |
| GO:BP | GO:0072189 | ureter development | 1 | 10 | 5.38e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 1 | 35 | 5.38e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 4 | 53 | 5.38e-01 |
| GO:BP | GO:1903726 | negative regulation of phospholipid metabolic process | 1 | 7 | 5.38e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 5.38e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 104 | 5.38e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 1 | 4 | 5.38e-01 |
| GO:BP | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity | 1 | 4 | 5.39e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 5 | 127 | 5.39e-01 |
| GO:BP | GO:1900003 | regulation of serine-type endopeptidase activity | 1 | 4 | 5.39e-01 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 1 | 67 | 5.39e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 2 | 25 | 5.39e-01 |
| GO:BP | GO:0051170 | import into nucleus | 2 | 152 | 5.39e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 2 | 23 | 5.39e-01 |
| GO:BP | GO:0021535 | cell migration in hindbrain | 1 | 12 | 5.39e-01 |
| GO:BP | GO:1902571 | regulation of serine-type peptidase activity | 1 | 4 | 5.39e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 1 | 48 | 5.39e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 6 | 98 | 5.39e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 1 | 90 | 5.40e-01 |
| GO:BP | GO:1905216 | positive regulation of RNA binding | 1 | 9 | 5.40e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 1 | 91 | 5.40e-01 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 1 | 10 | 5.40e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 1 | 91 | 5.40e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 6 | 85 | 5.41e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 4 | 307 | 5.41e-01 |
| GO:BP | GO:0009750 | response to fructose | 1 | 3 | 5.41e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 5.41e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 3 | 59 | 5.41e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 2 | 26 | 5.41e-01 |
| GO:BP | GO:0002249 | lymphocyte anergy | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0002911 | regulation of lymphocyte anergy | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0002827 | positive regulation of T-helper 1 type immune response | 1 | 5 | 5.41e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 1 | 556 | 5.41e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0002870 | T cell anergy | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0002693 | positive regulation of cellular extravasation | 1 | 13 | 5.41e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 12 | 197 | 5.41e-01 |
| GO:BP | GO:0002667 | regulation of T cell anergy | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 1 | 12 | 5.41e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 6 | 418 | 5.41e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 1 | 17 | 5.41e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 1 | 149 | 5.42e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 1 | 25 | 5.42e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 9 | 302 | 5.42e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 3 | 23 | 5.43e-01 |
| GO:BP | GO:0035507 | regulation of myosin-light-chain-phosphatase activity | 1 | 5 | 5.43e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 5.43e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 2 | 12 | 5.43e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 4 | 80 | 5.43e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 1 | 57 | 5.43e-01 |
| GO:BP | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 5 | 5.43e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 1 | 28 | 5.43e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 1 | 8 | 5.43e-01 |
| GO:BP | GO:0030091 | protein repair | 1 | 7 | 5.43e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 1 | 22 | 5.43e-01 |
| GO:BP | GO:0043201 | response to leucine | 1 | 13 | 5.43e-01 |
| GO:BP | GO:1904717 | regulation of AMPA glutamate receptor clustering | 1 | 4 | 5.43e-01 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 1 | 7 | 5.43e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 3 | 40 | 5.43e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 1 | 212 | 5.43e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 2 | 217 | 5.43e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 1 | 5 | 5.43e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 2 | 14 | 5.43e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 7 | 187 | 5.43e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 7 | 187 | 5.43e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 2 | 97 | 5.43e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 5 | 5.43e-01 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 1 | 12 | 5.43e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 1 | 94 | 5.43e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 2 | 25 | 5.43e-01 |
| GO:BP | GO:0106104 | regulation of glutamate receptor clustering | 1 | 4 | 5.43e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 1 | 39 | 5.43e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 1 | 94 | 5.43e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 1 | 28 | 5.43e-01 |
| GO:BP | GO:0009438 | methylglyoxal metabolic process | 1 | 7 | 5.43e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 6 | 89 | 5.43e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 2 | 421 | 5.43e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 5.43e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 2 | 16 | 5.43e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 5.43e-01 |
| GO:BP | GO:0051098 | regulation of binding | 2 | 317 | 5.43e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 5.43e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 3 | 40 | 5.43e-01 |
| GO:BP | GO:0042448 | progesterone metabolic process | 1 | 13 | 5.43e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 1 | 8 | 5.43e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 2 | 104 | 5.43e-01 |
| GO:BP | GO:1905049 | negative regulation of metallopeptidase activity | 1 | 7 | 5.44e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 1 | 15 | 5.44e-01 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 1 | 14 | 5.44e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 2 | 504 | 5.44e-01 |
| GO:BP | GO:1904683 | regulation of metalloendopeptidase activity | 1 | 6 | 5.44e-01 |
| GO:BP | GO:0061458 | reproductive system development | 1 | 215 | 5.44e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 1 | 15 | 5.44e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 1 | 13 | 5.44e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 1 | 26 | 5.44e-01 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 3 | 42 | 5.44e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 1 | 23 | 5.44e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 4 | 55 | 5.44e-01 |
| GO:BP | GO:0009798 | axis specification | 2 | 63 | 5.44e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 1 | 19 | 5.44e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 1 | 19 | 5.44e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 1 | 25 | 5.44e-01 |
| GO:BP | GO:0060732 | positive regulation of inositol phosphate biosynthetic process | 1 | 5 | 5.44e-01 |
| GO:BP | GO:0060004 | reflex | 1 | 9 | 5.44e-01 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 1 | 8 | 5.45e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 5.47e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 5.47e-01 |
| GO:BP | GO:0006776 | vitamin A metabolic process | 1 | 3 | 5.48e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 1 | 57 | 5.49e-01 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 1 | 5 | 5.49e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 1 | 44 | 5.49e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 3 | 61 | 5.49e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 5.49e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 1 | 10 | 5.49e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 3 | 78 | 5.49e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 4 | 105 | 5.50e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 5.50e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 3 | 78 | 5.50e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 1 | 53 | 5.50e-01 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 2 | 38 | 5.50e-01 |
| GO:BP | GO:0002578 | negative regulation of antigen processing and presentation | 1 | 4 | 5.50e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 3 | 40 | 5.50e-01 |
| GO:BP | GO:0010155 | regulation of proton transport | 2 | 18 | 5.50e-01 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 1 | 10 | 5.50e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 1 | 40 | 5.50e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 5.50e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 5.50e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 2 | 20 | 5.50e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 6 | 91 | 5.50e-01 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 2 | 38 | 5.50e-01 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 1 | 11 | 5.50e-01 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 2 | 33 | 5.50e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 151 | 5.50e-01 |
| GO:BP | GO:0045654 | positive regulation of megakaryocyte differentiation | 1 | 9 | 5.50e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 6 | 96 | 5.50e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 5.50e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 70 | 5.50e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 1 | 21 | 5.51e-01 |
| GO:BP | GO:0006825 | copper ion transport | 1 | 13 | 5.51e-01 |
| GO:BP | GO:0015711 | organic anion transport | 2 | 264 | 5.51e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 2 | 429 | 5.52e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 1 | 14 | 5.52e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 1 | 47 | 5.52e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 2 | 35 | 5.52e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 7 | 93 | 5.52e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 2 | 73 | 5.52e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 3 | 85 | 5.52e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 1 | 17 | 5.52e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 1 | 14 | 5.52e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 1 | 14 | 5.52e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 3 | 62 | 5.52e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 6 | 161 | 5.52e-01 |
| GO:BP | GO:0071361 | cellular response to ethanol | 1 | 9 | 5.52e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 5.55e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 3 | 44 | 5.55e-01 |
| GO:BP | GO:1903350 | response to dopamine | 1 | 48 | 5.55e-01 |
| GO:BP | GO:0048268 | clathrin coat assembly | 1 | 13 | 5.56e-01 |
| GO:BP | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3’-5’ | 1 | 11 | 5.56e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 1 | 30 | 5.56e-01 |
| GO:BP | GO:0001824 | blastocyst development | 4 | 94 | 5.56e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 1 | 19 | 5.56e-01 |
| GO:BP | GO:1904732 | regulation of electron transfer activity | 1 | 8 | 5.56e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 1 | 45 | 5.56e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 4 | 956 | 5.56e-01 |
| GO:BP | GO:0031223 | auditory behavior | 1 | 6 | 5.56e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 5.56e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 8 | 134 | 5.56e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 5.56e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 1 | 23 | 5.57e-01 |
| GO:BP | GO:0007566 | embryo implantation | 3 | 39 | 5.57e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 1 | 29 | 5.57e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 8 | 438 | 5.57e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 5.57e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 2 | 25 | 5.57e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 1 | 10 | 5.57e-01 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 1 | 20 | 5.57e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 4 | 58 | 5.57e-01 |
| GO:BP | GO:0006400 | tRNA modification | 2 | 90 | 5.57e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 5.57e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 2 | 37 | 5.57e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 1 | 12 | 5.57e-01 |
| GO:BP | GO:0048680 | positive regulation of axon regeneration | 1 | 8 | 5.57e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 2 | 108 | 5.57e-01 |
| GO:BP | GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1 | 10 | 5.57e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 2 | 236 | 5.57e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 1 | 166 | 5.57e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 6 | 151 | 5.57e-01 |
| GO:BP | GO:0050427 | 3’-phosphoadenosine 5’-phosphosulfate metabolic process | 1 | 10 | 5.57e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 7 | 84 | 5.57e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 1 | 106 | 5.57e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 1 | 12 | 5.58e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 1 | 30 | 5.58e-01 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 1 | 12 | 5.58e-01 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 2 | 21 | 5.59e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 2 | 37 | 5.59e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 2 | 37 | 5.59e-01 |
| GO:BP | GO:0015732 | prostaglandin transport | 2 | 10 | 5.59e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 6 | 82 | 5.59e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 2 | 27 | 5.59e-01 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 2 | 14 | 5.59e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 2 | 26 | 5.59e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 1 | 40 | 5.61e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 1 | 25 | 5.61e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 1 | 33 | 5.61e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 75 | 5.62e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 5.62e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 2 | 35 | 5.62e-01 |
| GO:BP | GO:0036261 | 7-methylguanosine cap hypermethylation | 1 | 8 | 5.62e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 2 | 44 | 5.62e-01 |
| GO:BP | GO:0050930 | induction of positive chemotaxis | 1 | 12 | 5.62e-01 |
| GO:BP | GO:0097531 | mast cell migration | 1 | 12 | 5.62e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 2 | 39 | 5.62e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 1 | 96 | 5.62e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 1 | 12 | 5.62e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 1 | 15 | 5.62e-01 |
| GO:BP | GO:0042730 | fibrinolysis | 2 | 15 | 5.62e-01 |
| GO:BP | GO:0009583 | detection of light stimulus | 1 | 27 | 5.62e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 710 | 5.62e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 3 | 116 | 5.62e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 4 | 55 | 5.62e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 1 | 19 | 5.62e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 4 | 55 | 5.62e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 12 | 1015 | 5.63e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 1 | 77 | 5.63e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 1 | 14 | 5.63e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 1 | 561 | 5.64e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 5.64e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 1 | 40 | 5.64e-01 |
| GO:BP | GO:0071071 | regulation of phospholipid biosynthetic process | 1 | 15 | 5.64e-01 |
| GO:BP | GO:0022008 | neurogenesis | 12 | 1294 | 5.64e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 15 | 236 | 5.64e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 6 | 143 | 5.64e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 13 | 1119 | 5.64e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 1 | 30 | 5.64e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 12 | 1017 | 5.64e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 1 | 31 | 5.64e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 2 | 179 | 5.64e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 1 | 16 | 5.65e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 5.65e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 2 | 14 | 5.65e-01 |
| GO:BP | GO:1900223 | positive regulation of amyloid-beta clearance | 1 | 5 | 5.65e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 23 | 1995 | 5.65e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 3 | 60 | 5.65e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 1 | 74 | 5.65e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 1 | 25 | 5.65e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 3 | 89 | 5.65e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 3 | 149 | 5.66e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 99 | 5.66e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 1 | 14 | 5.66e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 99 | 5.66e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 5.66e-01 |
| GO:BP | GO:0015744 | succinate transport | 1 | 6 | 5.66e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 1 | 15 | 5.66e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 1 | 6 | 5.66e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 5.66e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 1 | 27 | 5.66e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 1 | 13 | 5.66e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 7 | 107 | 5.66e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 2 | 36 | 5.66e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 5.66e-01 |
| GO:BP | GO:1901654 | response to ketone | 3 | 147 | 5.66e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 3 | 54 | 5.66e-01 |
| GO:BP | GO:1903689 | regulation of wound healing, spreading of epidermal cells | 1 | 8 | 5.66e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 2 | 31 | 5.66e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 5.66e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 1 | 37 | 5.66e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 99 | 5.66e-01 |
| GO:BP | GO:0010918 | positive regulation of mitochondrial membrane potential | 1 | 9 | 5.66e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 2 | 104 | 5.66e-01 |
| GO:BP | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus | 1 | 6 | 5.66e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 3 | 490 | 5.66e-01 |
| GO:BP | GO:0033029 | regulation of neutrophil apoptotic process | 1 | 5 | 5.66e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 3 | 490 | 5.66e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 1 | 7 | 5.66e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 1 | 169 | 5.66e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 8 | 140 | 5.66e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 2 | 22 | 5.66e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 2 | 22 | 5.66e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 2 | 21 | 5.66e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 2 | 24 | 5.66e-01 |
| GO:BP | GO:0009314 | response to radiation | 2 | 352 | 5.66e-01 |
| GO:BP | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 1 | 9 | 5.67e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 1 | 32 | 5.67e-01 |
| GO:BP | GO:0015911 | long-chain fatty acid import across plasma membrane | 1 | 9 | 5.68e-01 |
| GO:BP | GO:1904925 | positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 1 | 14 | 5.68e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 1 | 140 | 5.68e-01 |
| GO:BP | GO:0009067 | aspartate family amino acid biosynthetic process | 1 | 18 | 5.68e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 5.68e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 5.68e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 1 | 31 | 5.69e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 1 | 9 | 5.69e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 1 | 4 | 5.69e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 5.69e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 1 | 105 | 5.69e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 5.69e-01 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 1 | 12 | 5.69e-01 |
| GO:BP | GO:0021794 | thalamus development | 1 | 6 | 5.70e-01 |
| GO:BP | GO:0006072 | glycerol-3-phosphate metabolic process | 1 | 7 | 5.70e-01 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 1 | 6 | 5.70e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 3 | 495 | 5.70e-01 |
| GO:BP | GO:0042420 | dopamine catabolic process | 1 | 5 | 5.70e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 1 | 8 | 5.70e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 22 | 1914 | 5.70e-01 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 1 | 6 | 5.70e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 1 | 12 | 5.70e-01 |
| GO:BP | GO:0032429 | regulation of phospholipase A2 activity | 1 | 8 | 5.71e-01 |
| GO:BP | GO:0070997 | neuron death | 2 | 286 | 5.71e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 1 | 21 | 5.71e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 2 | 18 | 5.71e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 2 | 88 | 5.71e-01 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 1 | 5 | 5.71e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 1 | 13 | 5.71e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 1 | 31 | 5.71e-01 |
| GO:BP | GO:0042118 | endothelial cell activation | 1 | 7 | 5.71e-01 |
| GO:BP | GO:0007616 | long-term memory | 1 | 31 | 5.71e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 12 | 1026 | 5.71e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 1 | 12 | 5.71e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 3 | 82 | 5.71e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 2 | 21 | 5.71e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 5 | 1214 | 5.71e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 3 | 51 | 5.71e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 2 | 21 | 5.71e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 6 | 252 | 5.71e-01 |
| GO:BP | GO:0097254 | renal tubular secretion | 2 | 15 | 5.71e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 4 | 53 | 5.71e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 1 | 33 | 5.71e-01 |
| GO:BP | GO:0035295 | tube development | 3 | 821 | 5.71e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 2 | 28 | 5.72e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 1 | 9 | 5.73e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 1 | 9 | 5.73e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 5.73e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 5.74e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 1 | 10 | 5.74e-01 |
| GO:BP | GO:0070572 | positive regulation of neuron projection regeneration | 1 | 8 | 5.75e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 1 | 9 | 5.75e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 2 | 22 | 5.75e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 5.75e-01 |
| GO:BP | GO:0007611 | learning or memory | 2 | 202 | 5.75e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 11 | 300 | 5.76e-01 |
| GO:BP | GO:1901724 | positive regulation of cell proliferation involved in kidney development | 1 | 8 | 5.77e-01 |
| GO:BP | GO:0006907 | pinocytosis | 1 | 18 | 5.77e-01 |
| GO:BP | GO:0006949 | syncytium formation | 2 | 39 | 5.77e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 1 | 123 | 5.78e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 5.78e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 1 | 68 | 5.78e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 5 | 726 | 5.78e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 1 | 209 | 5.78e-01 |
| GO:BP | GO:0010496 | intercellular transport | 1 | 4 | 5.78e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 2 | 150 | 5.78e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 1 | 16 | 5.78e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 4 | 76 | 5.78e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 5 | 122 | 5.81e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 1 | 30 | 5.81e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 2 | 36 | 5.81e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 2 | 22 | 5.81e-01 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 1 | 14 | 5.82e-01 |
| GO:BP | GO:0042953 | lipoprotein transport | 1 | 14 | 5.82e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 1 | 13 | 5.83e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 1 | 6 | 5.83e-01 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 1 | 11 | 5.83e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 1 | 60 | 5.83e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 1 | 17 | 5.83e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 6 | 775 | 5.83e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 1 | 41 | 5.83e-01 |
| GO:BP | GO:0043248 | proteasome assembly | 1 | 13 | 5.83e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 1 | 60 | 5.83e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 94 | 5.83e-01 |
| GO:BP | GO:1904923 | regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 1 | 15 | 5.83e-01 |
| GO:BP | GO:0046849 | bone remodeling | 1 | 59 | 5.83e-01 |
| GO:BP | GO:0061025 | membrane fusion | 2 | 137 | 5.83e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 1 | 108 | 5.83e-01 |
| GO:BP | GO:0061448 | connective tissue development | 7 | 208 | 5.83e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 82 | 5.83e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 9 | 168 | 5.83e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 1 | 46 | 5.83e-01 |
| GO:BP | GO:0007040 | lysosome organization | 5 | 88 | 5.83e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 5 | 88 | 5.83e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 2 | 45 | 5.83e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 3 | 511 | 5.83e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 2 | 28 | 5.83e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 2 | 43 | 5.83e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 3 | 717 | 5.83e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 2 | 43 | 5.83e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 2 | 19 | 5.83e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 2 | 19 | 5.83e-01 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 1 | 57 | 5.84e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 4 | 179 | 5.84e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 2 | 116 | 5.84e-01 |
| GO:BP | GO:0030321 | transepithelial chloride transport | 1 | 7 | 5.84e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 1 | 17 | 5.84e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 2 | 27 | 5.84e-01 |
| GO:BP | GO:0051591 | response to cAMP | 5 | 66 | 5.84e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 2 | 25 | 5.84e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 1 | 16 | 5.84e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 1 | 14 | 5.84e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 3 | 64 | 5.84e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 1 | 12 | 5.84e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 2 | 316 | 5.84e-01 |
| GO:BP | GO:0006304 | DNA modification | 2 | 89 | 5.84e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 3 | 234 | 5.84e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 2 | 16 | 5.84e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 5 | 632 | 5.84e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 5.84e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 5 | 121 | 5.84e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 2 | 25 | 5.84e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 3 | 162 | 5.84e-01 |
| GO:BP | GO:0045006 | DNA deamination | 1 | 9 | 5.84e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 2 | 25 | 5.84e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 2 | 30 | 5.85e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 3 | 46 | 5.85e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 5.85e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 1 | 12 | 5.85e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 1 | 62 | 5.85e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 1 | 15 | 5.85e-01 |
| GO:BP | GO:0010038 | response to metal ion | 18 | 264 | 5.85e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 2 | 311 | 5.85e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 1 | 100 | 5.85e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 2 | 293 | 5.85e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 5.85e-01 |
| GO:BP | GO:1901652 | response to peptide | 7 | 377 | 5.85e-01 |
| GO:BP | GO:0071867 | response to monoamine | 1 | 62 | 5.85e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 5.85e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 3 | 39 | 5.85e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 2 | 27 | 5.85e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 2 | 270 | 5.85e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 5 | 1239 | 5.85e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 1 | 15 | 5.85e-01 |
| GO:BP | GO:0031649 | heat generation | 1 | 11 | 5.85e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 4 | 43 | 5.85e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 5.85e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 1 | 35 | 5.85e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 1 | 14 | 5.85e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 4 | 71 | 5.85e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 5.85e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 3 | 40 | 5.85e-01 |
| GO:BP | GO:0002604 | regulation of dendritic cell antigen processing and presentation | 1 | 5 | 5.85e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 1 | 35 | 5.86e-01 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 8 | 102 | 5.86e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 4 | 83 | 5.86e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 5.86e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 10 | 418 | 5.87e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 3 | 38 | 5.87e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 8 | 5.88e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 8 | 5.88e-01 |
| GO:BP | GO:0110095 | cellular detoxification of aldehyde | 1 | 5 | 5.88e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 4 | 51 | 5.88e-01 |
| GO:BP | GO:0071360 | cellular response to exogenous dsRNA | 1 | 14 | 5.88e-01 |
| GO:BP | GO:0001781 | neutrophil apoptotic process | 1 | 5 | 5.88e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 1 | 124 | 5.88e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 2 | 96 | 5.88e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 5.88e-01 |
| GO:BP | GO:0033280 | response to vitamin D | 1 | 24 | 5.88e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 6 | 86 | 5.88e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 2 | 35 | 5.88e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 1 | 99 | 5.89e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 2 | 25 | 5.89e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 21 | 1278 | 5.89e-01 |
| GO:BP | GO:0044872 | lipoprotein localization | 1 | 15 | 5.89e-01 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 5 | 94 | 5.89e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 3 | 69 | 5.90e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 2 | 4794 | 5.90e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 3 | 29 | 5.90e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 2 | 26 | 5.90e-01 |
| GO:BP | GO:0007494 | midgut development | 1 | 7 | 5.90e-01 |
| GO:BP | GO:0000050 | urea cycle | 1 | 7 | 5.90e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 1 | 9 | 5.90e-01 |
| GO:BP | GO:1900272 | negative regulation of long-term synaptic potentiation | 1 | 8 | 5.90e-01 |
| GO:BP | GO:0021670 | lateral ventricle development | 1 | 11 | 5.90e-01 |
| GO:BP | GO:0019614 | catechol-containing compound catabolic process | 1 | 5 | 5.90e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 1 | 17 | 5.90e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 1 | 23 | 5.90e-01 |
| GO:BP | GO:0098751 | bone cell development | 1 | 26 | 5.90e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 1 | 29 | 5.90e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 1 | 11 | 5.90e-01 |
| GO:BP | GO:0032879 | regulation of localization | 7 | 1555 | 5.90e-01 |
| GO:BP | GO:0045190 | isotype switching | 3 | 39 | 5.90e-01 |
| GO:BP | GO:0051049 | regulation of transport | 8 | 1267 | 5.90e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 2 | 23 | 5.90e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 4 | 80 | 5.90e-01 |
| GO:BP | GO:0001820 | serotonin secretion | 1 | 5 | 5.90e-01 |
| GO:BP | GO:2000347 | positive regulation of hepatocyte proliferation | 1 | 9 | 5.90e-01 |
| GO:BP | GO:0042136 | neurotransmitter biosynthetic process | 1 | 5 | 5.90e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 3 | 39 | 5.90e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 3 | 39 | 5.90e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 2 | 44 | 5.90e-01 |
| GO:BP | GO:0042060 | wound healing | 21 | 320 | 5.90e-01 |
| GO:BP | GO:0042424 | catecholamine catabolic process | 1 | 5 | 5.90e-01 |
| GO:BP | GO:0006364 | rRNA processing | 10 | 221 | 5.90e-01 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 2 | 38 | 5.90e-01 |
| GO:BP | GO:0008542 | visual learning | 3 | 37 | 5.91e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 5.91e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 2 | 187 | 5.91e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 1 | 251 | 5.91e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 1 | 13 | 5.91e-01 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 1 | 16 | 5.92e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 1 | 17 | 5.92e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 1 | 16 | 5.92e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 1 | 19 | 5.92e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 3 | 57 | 5.92e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 6 | 79 | 5.93e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 7 | 998 | 5.93e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 2 | 23 | 5.93e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 1 | 12 | 5.94e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 1 | 12 | 5.94e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 1 | 23 | 5.94e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 2 | 24 | 5.94e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 1 | 116 | 5.94e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 5.94e-01 |
| GO:BP | GO:0030488 | tRNA methylation | 1 | 42 | 5.94e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | 40 | 5.94e-01 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 1 | 9 | 5.94e-01 |
| GO:BP | GO:0043588 | skin development | 6 | 175 | 5.94e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 2 | 24 | 5.94e-01 |
| GO:BP | GO:0048265 | response to pain | 2 | 17 | 5.94e-01 |
| GO:BP | GO:2001198 | regulation of dendritic cell differentiation | 1 | 8 | 5.95e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 4 | 91 | 5.95e-01 |
| GO:BP | GO:0015918 | sterol transport | 4 | 92 | 5.95e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 1 | 31 | 5.95e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 3 | 141 | 5.95e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 1 | 96 | 5.96e-01 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 1 | 13 | 5.97e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 1 | 14 | 5.97e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 843 | 5.97e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 1 | 17 | 5.98e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 5.98e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 4 | 71 | 5.98e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 8 | 189 | 5.98e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 1 | 34 | 5.98e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 6 | 100 | 5.98e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 2 | 29 | 5.98e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 2 | 29 | 5.98e-01 |
| GO:BP | GO:0021537 | telencephalon development | 1 | 201 | 5.98e-01 |
| GO:BP | GO:0150172 | regulation of phosphatidylcholine metabolic process | 1 | 10 | 5.98e-01 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 5.98e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 5.98e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 1 | 30 | 5.98e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 2 | 30 | 5.98e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 2 | 126 | 5.98e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 2 | 44 | 5.98e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 1 | 26 | 5.98e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 5.98e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 2 | 28 | 5.98e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 1 | 39 | 5.98e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 3 | 66 | 5.98e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 1 | 207 | 5.98e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 1 | 33 | 5.98e-01 |
| GO:BP | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | 1 | 12 | 5.98e-01 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 40 | 5.98e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 5.98e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 4 | 52 | 5.98e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 5.98e-01 |
| GO:BP | GO:0014048 | regulation of glutamate secretion | 1 | 8 | 5.98e-01 |
| GO:BP | GO:0016032 | viral process | 1 | 344 | 5.98e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 5.99e-01 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 2 | 15 | 5.99e-01 |
| GO:BP | GO:0042921 | glucocorticoid receptor signaling pathway | 1 | 13 | 5.99e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 1 | 67 | 5.99e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 1 | 225 | 5.99e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 3 | 373 | 5.99e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 1 | 106 | 5.99e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 6.00e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 1 | 32 | 6.01e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 2 | 395 | 6.01e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 8 | 457 | 6.01e-01 |
| GO:BP | GO:0019532 | oxalate transport | 1 | 5 | 6.01e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 4 | 154 | 6.01e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 3 | 48 | 6.01e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 4 | 72 | 6.01e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 6.01e-01 |
| GO:BP | GO:0006901 | vesicle coating | 1 | 36 | 6.01e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 5 | 1280 | 6.01e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 6 | 147 | 6.01e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 1 | 14 | 6.02e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 6.02e-01 |
| GO:BP | GO:0072070 | loop of Henle development | 1 | 8 | 6.02e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 5 | 96 | 6.02e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 1 | 11 | 6.02e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 1 | 13 | 6.02e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 1 | 16 | 6.02e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 6.02e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 4 | 62 | 6.02e-01 |
| GO:BP | GO:0001556 | oocyte maturation | 1 | 22 | 6.02e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 4 | 101 | 6.02e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 1 | 25 | 6.02e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 1 | 16 | 6.02e-01 |
| GO:BP | GO:0002726 | positive regulation of T cell cytokine production | 1 | 14 | 6.02e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 1 | 25 | 6.02e-01 |
| GO:BP | GO:0030030 | cell projection organization | 18 | 1233 | 6.02e-01 |
| GO:BP | GO:0060911 | cardiac cell fate commitment | 1 | 11 | 6.03e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 2 | 59 | 6.04e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 2 | 36 | 6.05e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 3 | 262 | 6.05e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 2 | 64 | 6.05e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 1 | 39 | 6.05e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 19 | 344 | 6.05e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 6.05e-01 |
| GO:BP | GO:0045576 | mast cell activation | 1 | 35 | 6.05e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 3 | 158 | 6.05e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 2 | 158 | 6.05e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 1 | 10 | 6.05e-01 |
| GO:BP | GO:0048666 | neuron development | 8 | 877 | 6.05e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 6.05e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 1 | 36 | 6.05e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 29 | 6.05e-01 |
| GO:BP | GO:0014075 | response to amine | 1 | 34 | 6.05e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 16 | 265 | 6.05e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 2 | 117 | 6.05e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 1 | 11 | 6.06e-01 |
| GO:BP | GO:0090385 | phagosome-lysosome fusion | 1 | 12 | 6.06e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 3 | 40 | 6.06e-01 |
| GO:BP | GO:0051409 | response to nitrosative stress | 1 | 10 | 6.06e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 1 | 9 | 6.06e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 3 | 422 | 6.06e-01 |
| GO:BP | GO:0097113 | AMPA glutamate receptor clustering | 1 | 7 | 6.06e-01 |
| GO:BP | GO:0097688 | glutamate receptor clustering | 1 | 7 | 6.06e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 1 | 57 | 6.06e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 13 | 301 | 6.07e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 6.07e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 6.07e-01 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 2 | 47 | 6.07e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 6.07e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 19 | 345 | 6.07e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 6.07e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 1 | 44 | 6.07e-01 |
| GO:BP | GO:0034383 | low-density lipoprotein particle clearance | 1 | 17 | 6.07e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 3 | 65 | 6.07e-01 |
| GO:BP | GO:0007049 | cell cycle | 1 | 1529 | 6.08e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 2 | 104 | 6.08e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 2 | 104 | 6.08e-01 |
| GO:BP | GO:0007099 | centriole replication | 1 | 38 | 6.08e-01 |
| GO:BP | GO:0097502 | mannosylation | 1 | 32 | 6.08e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 1 | 30 | 6.08e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 6.08e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 2 | 21 | 6.08e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 1 | 73 | 6.09e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 6.09e-01 |
| GO:BP | GO:0007343 | egg activation | 1 | 4 | 6.09e-01 |
| GO:BP | GO:0031175 | neuron projection development | 7 | 780 | 6.09e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 2 | 87 | 6.09e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 3 | 71 | 6.09e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 3 | 71 | 6.09e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 5 | 336 | 6.09e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 2 | 26 | 6.09e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 3 | 1484 | 6.09e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 1 | 11 | 6.09e-01 |
| GO:BP | GO:2001171 | positive regulation of ATP biosynthetic process | 1 | 10 | 6.09e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 1 | 73 | 6.09e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 2 | 18 | 6.09e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 7 | 123 | 6.09e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 2 | 46 | 6.09e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 2 | 46 | 6.09e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 5 | 551 | 6.09e-01 |
| GO:BP | GO:0070498 | interleukin-1-mediated signaling pathway | 1 | 20 | 6.09e-01 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 3 | 40 | 6.09e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 1 | 36 | 6.10e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 1 | 69 | 6.10e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 6 | 257 | 6.11e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 4 | 203 | 6.11e-01 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 1 | 10 | 6.11e-01 |
| GO:BP | GO:0048569 | post-embryonic animal organ development | 1 | 15 | 6.11e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 6.11e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 1 | 8 | 6.11e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 2 | 144 | 6.11e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 2 | 24 | 6.11e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 6.11e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 4 | 4539 | 6.12e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1 | 101 | 6.12e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 2 | 27 | 6.12e-01 |
| GO:BP | GO:0031958 | corticosteroid receptor signaling pathway | 1 | 14 | 6.13e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 1 | 48 | 6.13e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 1 | 10 | 6.13e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 3 | 47 | 6.13e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 4 | 214 | 6.13e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 3 | 45 | 6.13e-01 |
| GO:BP | GO:0070266 | necroptotic process | 2 | 32 | 6.13e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 1 | 16 | 6.13e-01 |
| GO:BP | GO:0097267 | omega-hydroxylase P450 pathway | 1 | 3 | 6.13e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 1 | 114 | 6.13e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 3 | 787 | 6.13e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 3 | 41 | 6.13e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 1 | 265 | 6.13e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 2 | 37 | 6.14e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 2 | 4245 | 6.14e-01 |
| GO:BP | GO:0034379 | very-low-density lipoprotein particle assembly | 1 | 8 | 6.15e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 1 | 8 | 6.15e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 6.15e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 2 | 67 | 6.15e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 227 | 6.15e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 2 | 38 | 6.16e-01 |
| GO:BP | GO:0050890 | cognition | 2 | 227 | 6.16e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 1 | 187 | 6.16e-01 |
| GO:BP | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | 1 | 10 | 6.17e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 1 | 18 | 6.18e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 1 | 34 | 6.18e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 3 | 232 | 6.18e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 6.18e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 2 | 32 | 6.18e-01 |
| GO:BP | GO:0042592 | homeostatic process | 8 | 1159 | 6.18e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 1 | 55 | 6.18e-01 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 1 | 16 | 6.18e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 3 | 58 | 6.18e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 1 | 12 | 6.18e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 1 | 10 | 6.18e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 6 | 185 | 6.18e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 1 | 14 | 6.18e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 1 | 13 | 6.19e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 32 | 6.19e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 14 | 803 | 6.19e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 1 | 20 | 6.19e-01 |
| GO:BP | GO:0007596 | blood coagulation | 11 | 150 | 6.19e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 7 | 199 | 6.19e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 3 | 155 | 6.19e-01 |
| GO:BP | GO:0042045 | epithelial fluid transport | 1 | 7 | 6.19e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 5 | 70 | 6.19e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 3 | 73 | 6.19e-01 |
| GO:BP | GO:0015791 | polyol transmembrane transport | 1 | 6 | 6.19e-01 |
| GO:BP | GO:0048731 | system development | 25 | 2901 | 6.19e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 3 | 41 | 6.19e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 2 | 336 | 6.19e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 1 | 273 | 6.19e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 1 | 17 | 6.19e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 4 | 4590 | 6.20e-01 |
| GO:BP | GO:0071941 | nitrogen cycle metabolic process | 1 | 7 | 6.20e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 1 | 189 | 6.20e-01 |
| GO:BP | GO:0019627 | urea metabolic process | 1 | 7 | 6.20e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 1 | 18 | 6.20e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 4 | 118 | 6.20e-01 |
| GO:BP | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | 1 | 10 | 6.20e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 1 | 16 | 6.20e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 1 | 10 | 6.20e-01 |
| GO:BP | GO:0034105 | positive regulation of tissue remodeling | 1 | 4 | 6.20e-01 |
| GO:BP | GO:0035640 | exploration behavior | 1 | 21 | 6.20e-01 |
| GO:BP | GO:0097709 | connective tissue replacement | 1 | 8 | 6.20e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 1 | 14 | 6.20e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 2 | 626 | 6.20e-01 |
| GO:BP | GO:0032060 | bleb assembly | 1 | 9 | 6.20e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 6.20e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 10 | 131 | 6.21e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 1 | 229 | 6.21e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 1 | 1681 | 6.21e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 3 | 61 | 6.21e-01 |
| GO:BP | GO:0140029 | exocytic process | 3 | 69 | 6.21e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 3 | 73 | 6.22e-01 |
| GO:BP | GO:1905522 | negative regulation of macrophage migration | 1 | 6 | 6.22e-01 |
| GO:BP | GO:1903580 | positive regulation of ATP metabolic process | 1 | 11 | 6.22e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 1 | 14 | 6.23e-01 |
| GO:BP | GO:0001822 | kidney development | 1 | 244 | 6.23e-01 |
| GO:BP | GO:0006996 | organelle organization | 42 | 3000 | 6.23e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 4 | 55 | 6.24e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 6.24e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 2 | 40 | 6.24e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 1 | 11 | 6.24e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 6.24e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 1 | 98 | 6.24e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 6.24e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 1 | 7 | 6.24e-01 |
| GO:BP | GO:0033234 | negative regulation of protein sumoylation | 1 | 8 | 6.24e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 6.24e-01 |
| GO:BP | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process | 1 | 16 | 6.25e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 1 | 25 | 6.25e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 6.25e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 1 | 11 | 6.25e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 2 | 49 | 6.25e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 5 | 287 | 6.26e-01 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 1 | 27 | 6.26e-01 |
| GO:BP | GO:0001832 | blastocyst growth | 1 | 19 | 6.26e-01 |
| GO:BP | GO:0098534 | centriole assembly | 1 | 41 | 6.26e-01 |
| GO:BP | GO:0043312 | neutrophil degranulation | 1 | 5 | 6.26e-01 |
| GO:BP | GO:0002664 | regulation of T cell tolerance induction | 1 | 6 | 6.26e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 1 | 19 | 6.26e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 1 | 9 | 6.26e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 1 | 132 | 6.26e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 2 | 32 | 6.26e-01 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 1 | 12 | 6.26e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 1 | 152 | 6.26e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 1 | 59 | 6.26e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 6.27e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 1 | 71 | 6.29e-01 |
| GO:BP | GO:0048538 | thymus development | 1 | 35 | 6.29e-01 |
| GO:BP | GO:0030073 | insulin secretion | 1 | 141 | 6.29e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 72 | 6.30e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 1 | 84 | 6.30e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 1 | 30 | 6.30e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 1 | 111 | 6.30e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 1 | 15 | 6.30e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 2 | 34 | 6.30e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 1 | 23 | 6.30e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 1 | 134 | 6.30e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 1 | 11 | 6.30e-01 |
| GO:BP | GO:0035188 | hatching | 1 | 23 | 6.30e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 1 | 23 | 6.30e-01 |
| GO:BP | GO:0072001 | renal system development | 1 | 251 | 6.30e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 1 | 1006 | 6.30e-01 |
| GO:BP | GO:0002468 | dendritic cell antigen processing and presentation | 1 | 6 | 6.30e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 4 | 56 | 6.30e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 389 | 6.30e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 6.31e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 1 | 10 | 6.31e-01 |
| GO:BP | GO:0007632 | visual behavior | 3 | 41 | 6.31e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 5 | 178 | 6.31e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 73 | 6.31e-01 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 2 | 66 | 6.31e-01 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 1 | 10 | 6.31e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 6.31e-01 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 2 | 66 | 6.31e-01 |
| GO:BP | GO:0071359 | cellular response to dsRNA | 1 | 18 | 6.31e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 6.31e-01 |
| GO:BP | GO:0042407 | cristae formation | 1 | 18 | 6.31e-01 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 1 | 81 | 6.31e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 1 | 136 | 6.31e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 1 | 102 | 6.31e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 5 | 68 | 6.32e-01 |
| GO:BP | GO:0009068 | aspartate family amino acid catabolic process | 1 | 12 | 6.32e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 13 | 1209 | 6.32e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 6.32e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 73 | 6.32e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 1 | 13 | 6.32e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 1 | 23 | 6.33e-01 |
| GO:BP | GO:0072015 | podocyte development | 1 | 9 | 6.33e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 2 | 376 | 6.33e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 6.33e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 1 | 49 | 6.33e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 4 | 113 | 6.33e-01 |
| GO:BP | GO:0090398 | cellular senescence | 2 | 83 | 6.34e-01 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 1 | 16 | 6.34e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 1 | 274 | 6.34e-01 |
| GO:BP | GO:0061326 | renal tubule development | 3 | 77 | 6.35e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 2 | 28 | 6.35e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 5 | 90 | 6.35e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 6.36e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 2 | 27 | 6.36e-01 |
| GO:BP | GO:0007216 | G protein-coupled glutamate receptor signaling pathway | 1 | 5 | 6.36e-01 |
| GO:BP | GO:0072350 | tricarboxylic acid metabolic process | 1 | 13 | 6.36e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 2 | 44 | 6.36e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 6.36e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 7 | 108 | 6.36e-01 |
| GO:BP | GO:0015874 | norepinephrine transport | 1 | 9 | 6.36e-01 |
| GO:BP | GO:0045838 | positive regulation of membrane potential | 1 | 12 | 6.36e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 8 | 216 | 6.36e-01 |
| GO:BP | GO:0007276 | gamete generation | 9 | 461 | 6.36e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 1 | 22 | 6.36e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 2 | 43 | 6.36e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 6.36e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 12 | 763 | 6.36e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 6.36e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 1 | 14 | 6.36e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 1 | 10 | 6.36e-01 |
| GO:BP | GO:0050817 | coagulation | 11 | 153 | 6.36e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 2 | 61 | 6.36e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 3 | 32 | 6.36e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 6.36e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 1 | 12 | 6.36e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 1 | 85 | 6.37e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 5 | 305 | 6.37e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 1 | 21 | 6.37e-01 |
| GO:BP | GO:0002638 | negative regulation of immunoglobulin production | 1 | 5 | 6.38e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 2 | 25 | 6.38e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 2 | 109 | 6.38e-01 |
| GO:BP | GO:0043299 | leukocyte degranulation | 1 | 41 | 6.38e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 2 | 694 | 6.38e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 6 | 120 | 6.38e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 4 | 107 | 6.38e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 6.38e-01 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 1 | 29 | 6.38e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 3 | 29 | 6.38e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 6.38e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 1 | 165 | 6.38e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 1 | 18 | 6.39e-01 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 1 | 16 | 6.39e-01 |
| GO:BP | GO:0051923 | sulfation | 1 | 13 | 6.39e-01 |
| GO:BP | GO:0006887 | exocytosis | 1 | 256 | 6.39e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 1 | 17 | 6.39e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 6.39e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 1 | 44 | 6.39e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 3 | 184 | 6.39e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 6.39e-01 |
| GO:BP | GO:0007599 | hemostasis | 11 | 153 | 6.39e-01 |
| GO:BP | GO:0045348 | positive regulation of MHC class II biosynthetic process | 1 | 8 | 6.39e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 6.39e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 6.39e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 6.39e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 6.39e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 3 | 81 | 6.41e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 1 | 53 | 6.41e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 1 | 13 | 6.41e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 1 | 26 | 6.41e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 1 | 23 | 6.41e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 10 | 166 | 6.42e-01 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 1 | 12 | 6.42e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 1 | 11 | 6.42e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 1 | 10 | 6.42e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 1 | 53 | 6.42e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 6.42e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 1 | 11 | 6.42e-01 |
| GO:BP | GO:0001732 | formation of cytoplasmic translation initiation complex | 1 | 16 | 6.42e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 17 | 1201 | 6.42e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 1 | 32 | 6.42e-01 |
| GO:BP | GO:0051641 | cellular localization | 2 | 2810 | 6.44e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 1 | 14 | 6.45e-01 |
| GO:BP | GO:0060324 | face development | 1 | 45 | 6.45e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 6.46e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 6.46e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 1 | 12 | 6.46e-01 |
| GO:BP | GO:0021591 | ventricular system development | 2 | 29 | 6.46e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 6 | 124 | 6.46e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 1 | 16 | 6.46e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 2 | 97 | 6.46e-01 |
| GO:BP | GO:0097722 | sperm motility | 2 | 61 | 6.46e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 2 | 61 | 6.46e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 6 | 159 | 6.47e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 1 | 57 | 6.47e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 6.47e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 1 | 10 | 6.47e-01 |
| GO:BP | GO:0019336 | phenol-containing compound catabolic process | 1 | 6 | 6.47e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 1 | 8 | 6.47e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 2 | 37 | 6.47e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 1 | 48 | 6.47e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 2 | 301 | 6.47e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 1 | 20 | 6.47e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 7 | 106 | 6.47e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 9 | 278 | 6.47e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 1 | 21 | 6.47e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 1 | 27 | 6.47e-01 |
| GO:BP | GO:0007369 | gastrulation | 9 | 146 | 6.47e-01 |
| GO:BP | GO:0007610 | behavior | 4 | 436 | 6.48e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 1 | 1074 | 6.48e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 1 | 1076 | 6.48e-01 |
| GO:BP | GO:0032418 | lysosome localization | 1 | 62 | 6.48e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 6.48e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 2 | 127 | 6.48e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 1 | 62 | 6.48e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 1 | 19 | 6.48e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 6.48e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 6.48e-01 |
| GO:BP | GO:2001256 | regulation of store-operated calcium entry | 1 | 11 | 6.48e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 4 | 69 | 6.48e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 2 | 28 | 6.49e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 8 | 149 | 6.49e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 2 | 52 | 6.49e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 6.50e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 1 | 31 | 6.50e-01 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 1 | 9 | 6.50e-01 |
| GO:BP | GO:0060026 | convergent extension | 1 | 14 | 6.50e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 1 | 58 | 6.50e-01 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 1 | 14 | 6.50e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 6.50e-01 |
| GO:BP | GO:0006379 | mRNA cleavage | 1 | 19 | 6.51e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 2 | 34 | 6.51e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 8 | 529 | 6.51e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 6.52e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 1 | 14 | 6.52e-01 |
| GO:BP | GO:0046321 | positive regulation of fatty acid oxidation | 1 | 15 | 6.52e-01 |
| GO:BP | GO:0006325 | chromatin organization | 7 | 551 | 6.52e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 4 | 265 | 6.52e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 1 | 16 | 6.52e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 3 | 52 | 6.53e-01 |
| GO:BP | GO:0007588 | excretion | 2 | 18 | 6.53e-01 |
| GO:BP | GO:0030902 | hindbrain development | 5 | 114 | 6.53e-01 |
| GO:BP | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 7 | 6.53e-01 |
| GO:BP | GO:0030050 | vesicle transport along actin filament | 1 | 13 | 6.53e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 1 | 7 | 6.53e-01 |
| GO:BP | GO:0071800 | podosome assembly | 1 | 13 | 6.53e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 6.53e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 1 | 14 | 6.53e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 1 | 218 | 6.53e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 2 | 46 | 6.53e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 1 | 15 | 6.53e-01 |
| GO:BP | GO:0002829 | negative regulation of type 2 immune response | 1 | 7 | 6.53e-01 |
| GO:BP | GO:0014047 | glutamate secretion | 1 | 14 | 6.53e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 6.54e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 6 | 92 | 6.54e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 6.54e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 2 | 21 | 6.54e-01 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 2 | 72 | 6.54e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 3 | 60 | 6.54e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 4 | 65 | 6.55e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2 | 26 | 6.55e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 2 | 25 | 6.55e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 4 | 72 | 6.55e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 25 | 1028 | 6.55e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 1 | 28 | 6.55e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 25 | 2412 | 6.55e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 5 | 1138 | 6.55e-01 |
| GO:BP | GO:0051451 | myoblast migration | 1 | 11 | 6.55e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 1 | 10 | 6.55e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 6.55e-01 |
| GO:BP | GO:0048278 | vesicle docking | 3 | 57 | 6.55e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 6.56e-01 |
| GO:BP | GO:0002517 | T cell tolerance induction | 1 | 6 | 6.56e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 1 | 17 | 6.56e-01 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 1 | 17 | 6.56e-01 |
| GO:BP | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic | 1 | 19 | 6.56e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 1 | 173 | 6.56e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 6.56e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 7 | 105 | 6.56e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 20 | 6.56e-01 |
| GO:BP | GO:0043628 | regulatory ncRNA 3’-end processing | 1 | 19 | 6.56e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 1 | 14 | 6.56e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 2 | 101 | 6.57e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 1 | 15 | 6.57e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 2 | 25 | 6.57e-01 |
| GO:BP | GO:0071625 | vocalization behavior | 1 | 13 | 6.58e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 1 | 116 | 6.58e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 4 | 161 | 6.58e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 1 | 13 | 6.59e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 1 | 11 | 6.59e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 1 | 12 | 6.59e-01 |
| GO:BP | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus | 1 | 8 | 6.59e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 6.59e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 1 | 10 | 6.59e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 2 | 33 | 6.59e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 6.59e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 6.60e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 2 | 59 | 6.60e-01 |
| GO:BP | GO:0051642 | centrosome localization | 1 | 31 | 6.60e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 1 | 31 | 6.60e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 2 | 31 | 6.60e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 3 | 259 | 6.60e-01 |
| GO:BP | GO:0044409 | entry into host | 4 | 113 | 6.60e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 6.60e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 3 | 39 | 6.60e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 3 | 39 | 6.60e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 3 | 197 | 6.60e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 1 | 11 | 6.60e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 6.60e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 1 | 16 | 6.61e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 1 | 234 | 6.61e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 6.61e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 2 | 34 | 6.61e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 3 | 168 | 6.63e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 2 | 80 | 6.63e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 6.63e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 3 | 194 | 6.63e-01 |
| GO:BP | GO:1902624 | positive regulation of neutrophil migration | 1 | 21 | 6.63e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 1 | 48 | 6.64e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 1 | 15 | 6.64e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 6 | 96 | 6.64e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 10 | 344 | 6.65e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 3 | 169 | 6.65e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 6.65e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 12 | 6.65e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 2 | 30 | 6.65e-01 |
| GO:BP | GO:0006541 | glutamine metabolic process | 1 | 24 | 6.65e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 2 | 127 | 6.65e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 1 | 26 | 6.66e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 5 | 870 | 6.67e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 1 | 20 | 6.67e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 5 | 90 | 6.67e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 7 | 110 | 6.67e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 1 | 115 | 6.67e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 1 | 15 | 6.67e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 1 | 115 | 6.67e-01 |
| GO:BP | GO:0003158 | endothelium development | 5 | 104 | 6.68e-01 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 2 | 29 | 6.68e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 8 | 541 | 6.68e-01 |
| GO:BP | GO:0035904 | aorta development | 1 | 50 | 6.68e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 2 | 102 | 6.68e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 6.68e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 2 | 30 | 6.68e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 1 | 37 | 6.68e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 3 | 62 | 6.68e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 1 | 246 | 6.68e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 2 | 39 | 6.68e-01 |
| GO:BP | GO:0001845 | phagolysosome assembly | 1 | 14 | 6.68e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 1 | 8 | 6.68e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 1 | 16 | 6.68e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 2 | 29 | 6.68e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 2 | 35 | 6.68e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 9 | 1061 | 6.69e-01 |
| GO:BP | GO:0008033 | tRNA processing | 2 | 128 | 6.69e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 3 | 57 | 6.69e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 1 | 46 | 6.69e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 1 | 45 | 6.69e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 1 | 166 | 6.70e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 2 | 103 | 6.70e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 3 | 30 | 6.70e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 1 | 15 | 6.70e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 3 | 30 | 6.70e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 3 | 90 | 6.70e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 2 | 33 | 6.70e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 6.70e-01 |
| GO:BP | GO:0051953 | negative regulation of amine transport | 1 | 15 | 6.70e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 8 | 267 | 6.70e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 2 | 55 | 6.70e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 1 | 111 | 6.70e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 2 | 27 | 6.70e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 6.70e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 4 | 227 | 6.70e-01 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 1 | 14 | 6.70e-01 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 4 | 60 | 6.70e-01 |
| GO:BP | GO:0032695 | negative regulation of interleukin-12 production | 1 | 6 | 6.70e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 3 | 43 | 6.70e-01 |
| GO:BP | GO:0051604 | protein maturation | 14 | 259 | 6.70e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 2 | 27 | 6.70e-01 |
| GO:BP | GO:0048589 | developmental growth | 2 | 522 | 6.70e-01 |
| GO:BP | GO:0006457 | protein folding | 5 | 192 | 6.70e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 2 | 233 | 6.71e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 2 | 49 | 6.71e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 16 | 534 | 6.71e-01 |
| GO:BP | GO:0071243 | cellular response to arsenic-containing substance | 1 | 18 | 6.71e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 2 | 75 | 6.71e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 1 | 14 | 6.72e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 1 | 14 | 6.72e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 2 | 234 | 6.72e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 6.72e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 1 | 49 | 6.73e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 1 | 47 | 6.73e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 1 | 20 | 6.73e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 2 | 69 | 6.73e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 4 | 73 | 6.73e-01 |
| GO:BP | GO:0001525 | angiogenesis | 3 | 390 | 6.73e-01 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 6.73e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 6.73e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 2 | 20 | 6.73e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 1 | 154 | 6.73e-01 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 170 | 6.73e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 2 | 38 | 6.73e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 6.73e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 2 | 28 | 6.73e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 2 | 34 | 6.73e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 2 | 234 | 6.73e-01 |
| GO:BP | GO:0010273 | detoxification of copper ion | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 6.74e-01 |
| GO:BP | GO:1990169 | stress response to copper ion | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 1 | 12 | 6.74e-01 |
| GO:BP | GO:0099515 | actin filament-based transport | 1 | 15 | 6.74e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 1 | 11 | 6.74e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 1 | 30 | 6.74e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 4 | 60 | 6.74e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 31 | 3012 | 6.74e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 6.74e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 6.74e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 1 | 369 | 6.74e-01 |
| GO:BP | GO:0044057 | regulation of system process | 2 | 405 | 6.75e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 6 | 539 | 6.75e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 2 | 64 | 6.75e-01 |
| GO:BP | GO:0030900 | forebrain development | 1 | 281 | 6.77e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 1 | 13 | 6.77e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 6.77e-01 |
| GO:BP | GO:0032288 | myelin assembly | 1 | 19 | 6.77e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 1 | 168 | 6.77e-01 |
| GO:BP | GO:0006415 | translational termination | 1 | 17 | 6.78e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 2 | 137 | 6.78e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 3 | 898 | 6.78e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 1 | 13 | 6.78e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 1 | 19 | 6.78e-01 |
| GO:BP | GO:1900006 | positive regulation of dendrite development | 1 | 14 | 6.78e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 6.79e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 1 | 9 | 6.79e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 1 | 61 | 6.79e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 3 | 288 | 6.79e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 1 | 41 | 6.79e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 3 | 62 | 6.80e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 1 | 37 | 6.80e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 2 | 48 | 6.81e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 1 | 14 | 6.81e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 1 | 101 | 6.81e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 6.81e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 1 | 44 | 6.81e-01 |
| GO:BP | GO:0046907 | intracellular transport | 22 | 1453 | 6.81e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 3 | 57 | 6.81e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 2 | 58 | 6.81e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 6.81e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 1 | 22 | 6.81e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 1 | 22 | 6.81e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 9 | 142 | 6.81e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 2 | 142 | 6.81e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 2 | 25 | 6.82e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 2 | 16 | 6.82e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 1 | 60 | 6.82e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 4 | 756 | 6.83e-01 |
| GO:BP | GO:0030810 | positive regulation of nucleotide biosynthetic process | 1 | 12 | 6.83e-01 |
| GO:BP | GO:1900373 | positive regulation of purine nucleotide biosynthetic process | 1 | 12 | 6.83e-01 |
| GO:BP | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 6.83e-01 |
| GO:BP | GO:0016045 | detection of bacterium | 1 | 8 | 6.83e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 6.83e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 3 | 211 | 6.83e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 6.83e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 30 | 2880 | 6.84e-01 |
| GO:BP | GO:0016049 | cell growth | 2 | 418 | 6.84e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 4 | 151 | 6.84e-01 |
| GO:BP | GO:0000280 | nuclear division | 1 | 344 | 6.84e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 6 | 98 | 6.84e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 6.85e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 4 | 72 | 6.85e-01 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 1 | 16 | 6.86e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 4 | 74 | 6.86e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 6.86e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 6.86e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 4 | 62 | 6.86e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 4 | 393 | 6.86e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 1 | 11 | 6.86e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 185 | 6.87e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 6.87e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 2 | 30 | 6.87e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 1 | 1248 | 6.88e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 1 | 54 | 6.88e-01 |
| GO:BP | GO:1903975 | regulation of glial cell migration | 1 | 12 | 6.88e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 378 | 6.88e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 6.88e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 1 | 18 | 6.88e-01 |
| GO:BP | GO:0071318 | cellular response to ATP | 1 | 12 | 6.88e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 1 | 21 | 6.88e-01 |
| GO:BP | GO:0015833 | peptide transport | 1 | 179 | 6.88e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 6 | 108 | 6.88e-01 |
| GO:BP | GO:0010876 | lipid localization | 12 | 310 | 6.89e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 6.90e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 14 | 6.90e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 8 | 157 | 6.90e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 1 | 19 | 6.90e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 1 | 13 | 6.90e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 6 | 701 | 6.90e-01 |
| GO:BP | GO:0072109 | glomerular mesangium development | 1 | 13 | 6.90e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 1 | 39 | 6.90e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 2 | 28 | 6.90e-01 |
| GO:BP | GO:0046185 | aldehyde catabolic process | 1 | 12 | 6.91e-01 |
| GO:BP | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus | 1 | 7 | 6.91e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 6.91e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 2 | 65 | 6.92e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 2 | 29 | 6.93e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 2 | 40 | 6.93e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 1 | 46 | 6.93e-01 |
| GO:BP | GO:1905048 | regulation of metallopeptidase activity | 1 | 14 | 6.93e-01 |
| GO:BP | GO:0060047 | heart contraction | 1 | 205 | 6.93e-01 |
| GO:BP | GO:0002691 | regulation of cellular extravasation | 1 | 24 | 6.93e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 2 | 24 | 6.93e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 3 | 51 | 6.93e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 1 | 69 | 6.93e-01 |
| GO:BP | GO:0031529 | ruffle organization | 2 | 45 | 6.94e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 6.94e-01 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 1 | 24 | 6.94e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 6.94e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 4 | 222 | 6.94e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 3 | 52 | 6.95e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 1 | 29 | 6.95e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 1 | 17 | 6.95e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 3 | 3730 | 6.95e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 1 | 14 | 6.95e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 1 | 21 | 6.95e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 6.95e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 1 | 631 | 6.97e-01 |
| GO:BP | GO:0140354 | lipid import into cell | 1 | 15 | 6.97e-01 |
| GO:BP | GO:0010759 | positive regulation of macrophage chemotaxis | 1 | 9 | 6.97e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 1 | 43 | 6.97e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 2 | 153 | 6.97e-01 |
| GO:BP | GO:0002890 | negative regulation of immunoglobulin mediated immune response | 1 | 9 | 6.97e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 35 | 2646 | 6.97e-01 |
| GO:BP | GO:0002713 | negative regulation of B cell mediated immunity | 1 | 9 | 6.97e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 3 | 86 | 6.97e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 1 | 68 | 6.97e-01 |
| GO:BP | GO:0002716 | negative regulation of natural killer cell mediated immunity | 1 | 10 | 6.97e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 6.97e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 3 | 75 | 6.97e-01 |
| GO:BP | GO:1904037 | positive regulation of epithelial cell apoptotic process | 2 | 26 | 6.98e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 6.98e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 6.98e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 9 | 153 | 6.98e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 1 | 13 | 6.98e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 3 | 170 | 6.98e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 2 | 34 | 6.98e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 17 | 1735 | 6.98e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 2 | 22 | 6.98e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 3 | 170 | 6.98e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 1 | 24 | 6.98e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 6.99e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 1 | 56 | 6.99e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 1 | 14 | 7.00e-01 |
| GO:BP | GO:0051301 | cell division | 7 | 570 | 7.00e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 7.00e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 1 | 16 | 7.00e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 10 | 556 | 7.00e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 1 | 18 | 7.00e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 1 | 11 | 7.00e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 7.00e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 9 | 719 | 7.00e-01 |
| GO:BP | GO:0070988 | demethylation | 1 | 52 | 7.00e-01 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 7.01e-01 |
| GO:BP | GO:0045956 | positive regulation of calcium ion-dependent exocytosis | 1 | 11 | 7.01e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 1 | 23 | 7.01e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 7.01e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 1 | 23 | 7.01e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 2 | 25 | 7.02e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 1 | 28 | 7.02e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 5 | 135 | 7.02e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 4 | 67 | 7.02e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 4 | 399 | 7.02e-01 |
| GO:BP | GO:0080154 | regulation of fertilization | 1 | 7 | 7.02e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 2 | 74 | 7.02e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 4 | 78 | 7.02e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 1 | 57 | 7.02e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 2 | 25 | 7.02e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 2 | 74 | 7.02e-01 |
| GO:BP | GO:0016485 | protein processing | 3 | 183 | 7.02e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 7.04e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1 | 1345 | 7.04e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 11 | 1229 | 7.04e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 4 | 706 | 7.04e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 2 | 156 | 7.04e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 2 | 63 | 7.04e-01 |
| GO:BP | GO:0003015 | heart process | 1 | 216 | 7.04e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 6 | 420 | 7.04e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 1 | 12 | 7.04e-01 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 7.04e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 3 | 66 | 7.05e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 4 | 227 | 7.05e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 1 | 11 | 7.05e-01 |
| GO:BP | GO:0042182 | ketone catabolic process | 1 | 9 | 7.05e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 4 | 66 | 7.05e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 81 | 7.05e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 6 | 105 | 7.05e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 1 | 22 | 7.05e-01 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 1 | 18 | 7.05e-01 |
| GO:BP | GO:0061548 | ganglion development | 1 | 12 | 7.05e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 5 | 478 | 7.05e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 7 | 83 | 7.05e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 4 | 73 | 7.05e-01 |
| GO:BP | GO:0008306 | associative learning | 4 | 65 | 7.05e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 1 | 52 | 7.05e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 7.05e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 1 | 18 | 7.05e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 7.05e-01 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 1 | 6 | 7.05e-01 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 3 | 66 | 7.05e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 7.05e-01 |
| GO:BP | GO:1902001 | fatty acid transmembrane transport | 1 | 15 | 7.05e-01 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 1 | 8 | 7.05e-01 |
| GO:BP | GO:0031929 | TOR signaling | 5 | 115 | 7.05e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 16 | 551 | 7.05e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 5 | 120 | 7.05e-01 |
| GO:BP | GO:0045346 | regulation of MHC class II biosynthetic process | 1 | 11 | 7.05e-01 |
| GO:BP | GO:0002544 | chronic inflammatory response | 1 | 6 | 7.05e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 4 | 63 | 7.05e-01 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 1 | 12 | 7.05e-01 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 4 | 104 | 7.05e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 7.05e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 3 | 63 | 7.05e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 5 | 348 | 7.05e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 338 | 7.05e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 7.05e-01 |
| GO:BP | GO:0048285 | organelle fission | 1 | 388 | 7.05e-01 |
| GO:BP | GO:0010039 | response to iron ion | 1 | 24 | 7.05e-01 |
| GO:BP | GO:0140353 | lipid export from cell | 2 | 28 | 7.05e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 1 | 316 | 7.05e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 22 | 1462 | 7.05e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 1 | 103 | 7.05e-01 |
| GO:BP | GO:0044321 | response to leptin | 1 | 19 | 7.06e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 7.06e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 2 | 31 | 7.06e-01 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 1 | 16 | 7.06e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 7.06e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 8 | 540 | 7.07e-01 |
| GO:BP | GO:0048699 | generation of neurons | 9 | 1119 | 7.08e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 1 | 52 | 7.08e-01 |
| GO:BP | GO:0046688 | response to copper ion | 2 | 29 | 7.08e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 2 | 43 | 7.08e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 2 | 353 | 7.08e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 1 | 17 | 7.09e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 1 | 14 | 7.09e-01 |
| GO:BP | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 1 | 10 | 7.09e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 2 | 69 | 7.09e-01 |
| GO:BP | GO:0032506 | cytokinetic process | 2 | 39 | 7.09e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 1 | 18 | 7.09e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 1 | 47 | 7.10e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 1 | 42 | 7.10e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 1 | 190 | 7.10e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 7.10e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 1 | 14 | 7.10e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 4 | 78 | 7.10e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 1 | 119 | 7.10e-01 |
| GO:BP | GO:0045684 | positive regulation of epidermis development | 1 | 20 | 7.10e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 2 | 36 | 7.10e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 1 | 13 | 7.11e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 2 | 609 | 7.11e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 1 | 16 | 7.11e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 84 | 7.11e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 7.11e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 7.11e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 2 | 57 | 7.12e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 5 | 284 | 7.12e-01 |
| GO:BP | GO:2000311 | regulation of AMPA receptor activity | 1 | 15 | 7.12e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 2 | 27 | 7.12e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 3 | 121 | 7.12e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 1 | 71 | 7.12e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 7.12e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 8 | 248 | 7.12e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 3 | 64 | 7.12e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 7.12e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 2 | 64 | 7.12e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 2 | 357 | 7.12e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 12 | 290 | 7.12e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 1 | 35 | 7.12e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 2 | 149 | 7.12e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 1 | 64 | 7.12e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 1 | 38 | 7.15e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 2 | 42 | 7.15e-01 |
| GO:BP | GO:0030258 | lipid modification | 9 | 162 | 7.15e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 7.15e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 3 | 868 | 7.15e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 3 | 84 | 7.15e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 1 | 494 | 7.15e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 4 | 90 | 7.15e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 3 | 65 | 7.15e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 10 | 258 | 7.15e-01 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 1 | 21 | 7.15e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 1 | 76 | 7.16e-01 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 1 | 23 | 7.16e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 4 | 101 | 7.16e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 7.16e-01 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 1 | 12 | 7.16e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 1 | 11 | 7.16e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 4 | 64 | 7.16e-01 |
| GO:BP | GO:0110096 | cellular response to aldehyde | 1 | 10 | 7.16e-01 |
| GO:BP | GO:0045342 | MHC class II biosynthetic process | 1 | 12 | 7.16e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 85 | 7.16e-01 |
| GO:BP | GO:0051169 | nuclear transport | 6 | 295 | 7.16e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 6 | 295 | 7.16e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 7.16e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 1 | 12 | 7.16e-01 |
| GO:BP | GO:0002115 | store-operated calcium entry | 1 | 15 | 7.16e-01 |
| GO:BP | GO:0032835 | glomerulus development | 3 | 54 | 7.16e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 3 | 234 | 7.16e-01 |
| GO:BP | GO:0002643 | regulation of tolerance induction | 1 | 10 | 7.17e-01 |
| GO:BP | GO:0098543 | detection of other organism | 1 | 9 | 7.17e-01 |
| GO:BP | GO:0051168 | nuclear export | 4 | 150 | 7.17e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 11 | 1173 | 7.17e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 7.18e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 1 | 51 | 7.18e-01 |
| GO:BP | GO:0051004 | regulation of lipoprotein lipase activity | 1 | 11 | 7.18e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 9 | 221 | 7.18e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 2 | 166 | 7.18e-01 |
| GO:BP | GO:0032309 | icosanoid secretion | 2 | 18 | 7.19e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 1 | 199 | 7.19e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4 | 230 | 7.19e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 7 | 146 | 7.20e-01 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 237 | 7.20e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 1 | 20 | 7.20e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 7.20e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 1 | 28 | 7.20e-01 |
| GO:BP | GO:0006953 | acute-phase response | 2 | 18 | 7.20e-01 |
| GO:BP | GO:0035813 | regulation of renal sodium excretion | 1 | 10 | 7.20e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 3 | 55 | 7.20e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 2 | 151 | 7.20e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 4 | 127 | 7.21e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 27 | 7.21e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 7.21e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 1 | 27 | 7.21e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 4 | 527 | 7.21e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 1 | 8 | 7.21e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 1 | 65 | 7.21e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 7.21e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 3 | 116 | 7.21e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 1 | 56 | 7.21e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 2 | 276 | 7.21e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 4 | 162 | 7.21e-01 |
| GO:BP | GO:1900544 | positive regulation of purine nucleotide metabolic process | 1 | 16 | 7.21e-01 |
| GO:BP | GO:0045981 | positive regulation of nucleotide metabolic process | 1 | 16 | 7.21e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 1 | 36 | 7.21e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 1 | 35 | 7.21e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 2 | 42 | 7.21e-01 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 1 | 19 | 7.21e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 1 | 31 | 7.22e-01 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 1 | 17 | 7.22e-01 |
| GO:BP | GO:0002283 | neutrophil activation involved in immune response | 1 | 7 | 7.22e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 7.23e-01 |
| GO:BP | GO:2000738 | positive regulation of stem cell differentiation | 1 | 17 | 7.23e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 1 | 68 | 7.23e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 1 | 9 | 7.23e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 3 | 53 | 7.23e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 1 | 10 | 7.23e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 2 | 742 | 7.23e-01 |
| GO:BP | GO:0006085 | acetyl-CoA biosynthetic process | 1 | 16 | 7.24e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 1 | 115 | 7.24e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 4 | 531 | 7.24e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 1 | 14 | 7.24e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 1 | 17 | 7.24e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 1 | 25 | 7.24e-01 |
| GO:BP | GO:0045333 | cellular respiration | 8 | 211 | 7.24e-01 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 1 | 17 | 7.24e-01 |
| GO:BP | GO:0098930 | axonal transport | 1 | 60 | 7.24e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 1 | 25 | 7.24e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 1 | 26 | 7.26e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 1 | 28 | 7.26e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 1 | 200 | 7.26e-01 |
| GO:BP | GO:0120193 | tight junction organization | 1 | 52 | 7.26e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 7.26e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 4 | 75 | 7.26e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 1 | 129 | 7.26e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 5 | 61 | 7.26e-01 |
| GO:BP | GO:0009749 | response to glucose | 4 | 137 | 7.26e-01 |
| GO:BP | GO:0048535 | lymph node development | 1 | 11 | 7.26e-01 |
| GO:BP | GO:0048864 | stem cell development | 1 | 76 | 7.26e-01 |
| GO:BP | GO:0016573 | histone acetylation | 1 | 143 | 7.26e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 7.26e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 1 | 816 | 7.27e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 11 | 242 | 7.28e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 2 | 377 | 7.28e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 2 | 404 | 7.28e-01 |
| GO:BP | GO:0061687 | detoxification of inorganic compound | 1 | 12 | 7.28e-01 |
| GO:BP | GO:0097501 | stress response to metal ion | 1 | 11 | 7.28e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 1 | 19 | 7.28e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 1 | 24 | 7.28e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 1 | 26 | 7.28e-01 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 1 | 24 | 7.29e-01 |
| GO:BP | GO:0002520 | immune system development | 8 | 144 | 7.29e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 10 | 620 | 7.29e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 1 | 69 | 7.29e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 1 | 49 | 7.29e-01 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 3 | 74 | 7.29e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 1 | 10 | 7.29e-01 |
| GO:BP | GO:0034097 | response to cytokine | 9 | 607 | 7.30e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 7.31e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 18 | 1974 | 7.32e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 1 | 19 | 7.32e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 211 | 7.33e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 1 | 19 | 7.33e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 1 | 76 | 7.33e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 1 | 129 | 7.33e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 2 | 67 | 7.33e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 2 | 192 | 7.33e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 1 | 34 | 7.33e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 2 | 166 | 7.33e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 1 | 13 | 7.34e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 2 | 31 | 7.34e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 1 | 20 | 7.34e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 1 | 55 | 7.34e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 8 | 280 | 7.34e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 4 | 83 | 7.35e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 3 | 71 | 7.35e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 1 | 45 | 7.35e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 1 | 102 | 7.35e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 2 | 289 | 7.36e-01 |
| GO:BP | GO:0006706 | steroid catabolic process | 1 | 13 | 7.36e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 5 | 90 | 7.36e-01 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 2 | 56 | 7.36e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 1 | 18 | 7.36e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 3 | 83 | 7.36e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 15 | 691 | 7.36e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 10 | 504 | 7.36e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 2 | 34 | 7.36e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 22 | 1836 | 7.37e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 7.37e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 5 | 139 | 7.38e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 39 | 7.38e-01 |
| GO:BP | GO:0044000 | movement in host | 4 | 138 | 7.38e-01 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 1 | 23 | 7.38e-01 |
| GO:BP | GO:0031998 | regulation of fatty acid beta-oxidation | 1 | 16 | 7.38e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 1 | 148 | 7.38e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 2 | 40 | 7.38e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 3 | 129 | 7.38e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 2 | 73 | 7.38e-01 |
| GO:BP | GO:0048599 | oocyte development | 1 | 38 | 7.38e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 7.38e-01 |
| GO:BP | GO:0060429 | epithelium development | 2 | 850 | 7.38e-01 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 1 | 15 | 7.38e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 92 | 7.38e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 3 | 462 | 7.38e-01 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 1 | 28 | 7.38e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 7.39e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 5 | 49 | 7.39e-01 |
| GO:BP | GO:0009746 | response to hexose | 4 | 138 | 7.39e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 2 | 161 | 7.39e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 7.39e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 2 | 763 | 7.39e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 2 | 55 | 7.40e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 1 | 20 | 7.40e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 1 | 150 | 7.40e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 1 | 22 | 7.40e-01 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 1 | 31 | 7.40e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 1 | 15 | 7.41e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 1 | 29 | 7.41e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 7.41e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 9 | 158 | 7.41e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 7.41e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 2 | 96 | 7.41e-01 |
| GO:BP | GO:0051258 | protein polymerization | 8 | 241 | 7.41e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 1 | 50 | 7.41e-01 |
| GO:BP | GO:0035812 | renal sodium excretion | 1 | 11 | 7.41e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 1 | 113 | 7.42e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 7 | 875 | 7.42e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 1 | 19 | 7.42e-01 |
| GO:BP | GO:0009611 | response to wounding | 24 | 415 | 7.42e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 1 | 19 | 7.42e-01 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 3 | 63 | 7.42e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 468 | 7.43e-01 |
| GO:BP | GO:0043200 | response to amino acid | 5 | 95 | 7.44e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 7.44e-01 |
| GO:BP | GO:0040007 | growth | 3 | 742 | 7.44e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 1 | 26 | 7.44e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 2 | 39 | 7.44e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 1 | 31 | 7.44e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 2 | 428 | 7.44e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 7.44e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 1 | 80 | 7.44e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 1 | 27 | 7.44e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 1 | 27 | 7.44e-01 |
| GO:BP | GO:0010224 | response to UV-B | 1 | 17 | 7.44e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 9 | 538 | 7.44e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 7.45e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 7 | 551 | 7.45e-01 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 1 | 10 | 7.45e-01 |
| GO:BP | GO:0035282 | segmentation | 2 | 75 | 7.45e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 3 | 64 | 7.45e-01 |
| GO:BP | GO:0071702 | organic substance transport | 8 | 2116 | 7.45e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 1 | 14 | 7.45e-01 |
| GO:BP | GO:0030728 | ovulation | 1 | 14 | 7.46e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 1 | 35 | 7.47e-01 |
| GO:BP | GO:0051640 | organelle localization | 1 | 503 | 7.47e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 2 | 164 | 7.47e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 1 | 39 | 7.47e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 1 | 130 | 7.47e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 16 | 372 | 7.47e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 2 | 34 | 7.47e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 2 | 55 | 7.47e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 2 | 41 | 7.47e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 1 | 35 | 7.47e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 3 | 226 | 7.47e-01 |
| GO:BP | GO:0007498 | mesoderm development | 3 | 89 | 7.47e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 1 | 35 | 7.47e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 1 | 51 | 7.47e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3386 | 7.48e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 2 | 99 | 7.48e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 1 | 15 | 7.48e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 1 | 31 | 7.48e-01 |
| GO:BP | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | 1 | 11 | 7.48e-01 |
| GO:BP | GO:2000810 | regulation of bicellular tight junction assembly | 1 | 15 | 7.48e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 1 | 77 | 7.49e-01 |
| GO:BP | GO:1903307 | positive regulation of regulated secretory pathway | 2 | 32 | 7.49e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 2 | 47 | 7.49e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 3 | 1219 | 7.49e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 15 | 318 | 7.49e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 1 | 42 | 7.49e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 1 | 138 | 7.49e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 1 | 50 | 7.50e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 1 | 50 | 7.50e-01 |
| GO:BP | GO:0042886 | amide transport | 1 | 235 | 7.51e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 3 | 748 | 7.51e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 1 | 147 | 7.51e-01 |
| GO:BP | GO:0021766 | hippocampus development | 1 | 62 | 7.51e-01 |
| GO:BP | GO:0048468 | cell development | 25 | 1963 | 7.51e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 1 | 9 | 7.51e-01 |
| GO:BP | GO:0001706 | endoderm formation | 1 | 46 | 7.53e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 7.53e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 1 | 39 | 7.53e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 295 | 7.53e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 1 | 27 | 7.53e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 3 | 93 | 7.53e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 7.53e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 3 | 59 | 7.53e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 1 | 18 | 7.53e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 1 | 21 | 7.53e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 1 | 18 | 7.53e-01 |
| GO:BP | GO:0001510 | RNA methylation | 1 | 87 | 7.53e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 1 | 160 | 7.53e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 4 | 663 | 7.53e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 3 | 56 | 7.53e-01 |
| GO:BP | GO:0001503 | ossification | 2 | 313 | 7.54e-01 |
| GO:BP | GO:0006909 | phagocytosis | 12 | 148 | 7.54e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 7.54e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 1 | 23 | 7.55e-01 |
| GO:BP | GO:0036260 | RNA capping | 1 | 20 | 7.55e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 4 | 496 | 7.55e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 2 | 33 | 7.55e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 5 | 68 | 7.55e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 1 | 146 | 7.55e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 8 | 168 | 7.55e-01 |
| GO:BP | GO:0006869 | lipid transport | 10 | 272 | 7.56e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 1 | 41 | 7.56e-01 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 7.56e-01 |
| GO:BP | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 1 | 21 | 7.56e-01 |
| GO:BP | GO:0009306 | protein secretion | 1 | 262 | 7.57e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 3 | 73 | 7.57e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 1 | 23 | 7.57e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 1 | 262 | 7.57e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 2 | 34 | 7.57e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 1 | 18 | 7.58e-01 |
| GO:BP | GO:0009451 | RNA modification | 2 | 162 | 7.58e-01 |
| GO:BP | GO:0032940 | secretion by cell | 2 | 569 | 7.58e-01 |
| GO:BP | GO:0001764 | neuron migration | 2 | 133 | 7.58e-01 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 7.58e-01 |
| GO:BP | GO:0009888 | tissue development | 3 | 1425 | 7.58e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 1 | 147 | 7.58e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 4 | 142 | 7.58e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 4 | 441 | 7.60e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 3 | 295 | 7.60e-01 |
| GO:BP | GO:0072006 | nephron development | 6 | 117 | 7.60e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 1 | 20 | 7.60e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 1 | 18 | 7.60e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 1 | 23 | 7.60e-01 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 1 | 26 | 7.60e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 1 | 13 | 7.60e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 4 | 58 | 7.61e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 2 | 42 | 7.61e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 605 | 7.61e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 7.61e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 3 | 400 | 7.61e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 1 | 22 | 7.62e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 1 | 266 | 7.62e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 1 | 17 | 7.62e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 4 | 77 | 7.62e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 7 | 755 | 7.62e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 2 | 31 | 7.62e-01 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 1 | 30 | 7.62e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 1 | 10 | 7.62e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 2 | 35 | 7.62e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 1 | 80 | 7.62e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 2 | 21 | 7.62e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 1 | 19 | 7.63e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 7.63e-01 |
| GO:BP | GO:0060419 | heart growth | 3 | 72 | 7.63e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 3 | 117 | 7.63e-01 |
| GO:BP | GO:0030534 | adult behavior | 6 | 92 | 7.63e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 3 | 51 | 7.63e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 7.63e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 2 | 256 | 7.63e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 7.63e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 2 | 34 | 7.63e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 3 | 54 | 7.63e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 1 | 21 | 7.63e-01 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 1 | 41 | 7.63e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 31 | 3169 | 7.63e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 2 | 44 | 7.63e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 1 | 54 | 7.63e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 4 | 67 | 7.64e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 1 | 32 | 7.64e-01 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 3 | 82 | 7.64e-01 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 3 | 82 | 7.64e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 1 | 29 | 7.65e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 1 | 19 | 7.65e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 10 | 222 | 7.65e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 2 | 78 | 7.66e-01 |
| GO:BP | GO:0001843 | neural tube closure | 2 | 90 | 7.66e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 7.66e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 1 | 160 | 7.66e-01 |
| GO:BP | GO:0033260 | nuclear DNA replication | 1 | 35 | 7.66e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 1 | 667 | 7.66e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 7.67e-01 |
| GO:BP | GO:0003341 | cilium movement | 2 | 97 | 7.67e-01 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 1 | 11 | 7.67e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 1 | 58 | 7.68e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 7.68e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 5 | 1014 | 7.69e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 7.69e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 1 | 56 | 7.69e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 8 | 139 | 7.70e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 1 | 50 | 7.70e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 5 | 560 | 7.70e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 1 | 86 | 7.70e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 30 | 3080 | 7.70e-01 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 1 | 23 | 7.70e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 7.70e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 7.70e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 7.70e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 2 | 38 | 7.70e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 3 | 79 | 7.70e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 1 | 59 | 7.70e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 2 | 57 | 7.70e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 1 | 23 | 7.70e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 2 | 48 | 7.70e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 2 | 95 | 7.70e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 7.70e-01 |
| GO:BP | GO:0006270 | DNA replication initiation | 1 | 35 | 7.70e-01 |
| GO:BP | GO:0060606 | tube closure | 2 | 91 | 7.70e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 3 | 116 | 7.70e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 7.70e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 2 | 145 | 7.70e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 11 | 258 | 7.70e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 7.70e-01 |
| GO:BP | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 12 | 7.70e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 1 | 440 | 7.70e-01 |
| GO:BP | GO:0035739 | CD4-positive, alpha-beta T cell proliferation | 1 | 12 | 7.70e-01 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 9 | 131 | 7.70e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 1 | 12 | 7.70e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 1 | 22 | 7.70e-01 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 1 | 28 | 7.70e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 5 | 66 | 7.70e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 2 | 36 | 7.71e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 1 | 552 | 7.71e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 1 | 26 | 7.71e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 1 | 42 | 7.71e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 4 | 51 | 7.71e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 10 | 1123 | 7.72e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 5 | 341 | 7.72e-01 |
| GO:BP | GO:0070848 | response to growth factor | 7 | 572 | 7.72e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 2 | 78 | 7.72e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 7.72e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 7.72e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 1 | 18 | 7.72e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 5 | 342 | 7.72e-01 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 1 | 16 | 7.72e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 43 | 7.72e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 1 | 16 | 7.72e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 11 | 782 | 7.72e-01 |
| GO:BP | GO:0012501 | programmed cell death | 13 | 1479 | 7.72e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 1 | 24 | 7.72e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 2 | 24 | 7.72e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 3 | 97 | 7.72e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 3 | 79 | 7.72e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 1 | 18 | 7.72e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 1 | 26 | 7.72e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 1 | 20 | 7.73e-01 |
| GO:BP | GO:0006891 | intra-Golgi vesicle-mediated transport | 1 | 33 | 7.73e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 3 | 68 | 7.73e-01 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 1 | 19 | 7.73e-01 |
| GO:BP | GO:0031342 | negative regulation of cell killing | 1 | 11 | 7.73e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 1 | 86 | 7.74e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 111 | 7.74e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 16 | 761 | 7.75e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 20 | 7.75e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 4 | 433 | 7.75e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 4 | 57 | 7.76e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 1 | 40 | 7.76e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 7.76e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 1 | 56 | 7.76e-01 |
| GO:BP | GO:0006935 | chemotaxis | 3 | 389 | 7.76e-01 |
| GO:BP | GO:0042110 | T cell activation | 5 | 313 | 7.77e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 1 | 40 | 7.77e-01 |
| GO:BP | GO:0001655 | urogenital system development | 2 | 49 | 7.78e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 1 | 13 | 7.78e-01 |
| GO:BP | GO:0042330 | taxis | 3 | 389 | 7.78e-01 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 2 | 57 | 7.79e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 1 | 598 | 7.81e-01 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 1 | 240 | 7.81e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 1 | 21 | 7.81e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 4 | 105 | 7.81e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 2 | 53 | 7.82e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 7.82e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 3 | 320 | 7.82e-01 |
| GO:BP | GO:0060840 | artery development | 1 | 83 | 7.82e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 7.82e-01 |
| GO:BP | GO:0016358 | dendrite development | 8 | 202 | 7.83e-01 |
| GO:BP | GO:0008219 | cell death | 14 | 1596 | 7.83e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 1 | 21 | 7.83e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 1 | 22 | 7.83e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 2 | 38 | 7.83e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 1 | 137 | 7.83e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 7.83e-01 |
| GO:BP | GO:0140352 | export from cell | 2 | 611 | 7.83e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 1 | 23 | 7.83e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 7.83e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 17 | 1849 | 7.83e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 7.83e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 4 | 834 | 7.84e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 2 | 46 | 7.84e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 1 | 40 | 7.84e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 2 | 250 | 7.84e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 1 | 60 | 7.85e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 2 | 40 | 7.85e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 7.85e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 4 | 906 | 7.86e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 2 | 41 | 7.86e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 1 | 21 | 7.86e-01 |
| GO:BP | GO:0090314 | positive regulation of protein targeting to membrane | 1 | 22 | 7.86e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 1 | 28 | 7.86e-01 |
| GO:BP | GO:0006884 | cell volume homeostasis | 1 | 24 | 7.86e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1 | 2011 | 7.86e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 7.86e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 1 | 43 | 7.86e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 2 | 94 | 7.86e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 5 | 98 | 7.86e-01 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 1 | 36 | 7.87e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 7.87e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 7.88e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 5 | 71 | 7.89e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 7.89e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 7.91e-01 |
| GO:BP | GO:0007612 | learning | 6 | 115 | 7.91e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 1 | 111 | 7.92e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 1 | 27 | 7.92e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 2 | 51 | 7.92e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 2 | 31 | 7.93e-01 |
| GO:BP | GO:0008544 | epidermis development | 2 | 210 | 7.93e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 3 | 126 | 7.93e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 13 | 220 | 7.93e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 3 | 64 | 7.94e-01 |
| GO:BP | GO:0001916 | positive regulation of T cell mediated cytotoxicity | 1 | 13 | 7.95e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 7.95e-01 |
| GO:BP | GO:0010332 | response to gamma radiation | 2 | 50 | 7.95e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 3 | 81 | 7.95e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 1 | 15 | 7.96e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 1 | 296 | 7.96e-01 |
| GO:BP | GO:0001568 | blood vessel development | 3 | 534 | 7.96e-01 |
| GO:BP | GO:0040011 | locomotion | 5 | 954 | 7.96e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 2 | 40 | 7.96e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 1 | 17 | 7.96e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 1 | 21 | 7.96e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 25 | 2574 | 7.96e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 3 | 1684 | 7.96e-01 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 1 | 40 | 7.96e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 1 | 85 | 7.96e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 15 | 1107 | 7.96e-01 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 1 | 28 | 7.96e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 25 | 2576 | 7.96e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 7.96e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 4 | 68 | 7.97e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 4 | 154 | 7.97e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 18 | 369 | 7.97e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 7.97e-01 |
| GO:BP | GO:0015872 | dopamine transport | 2 | 29 | 7.97e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 1 | 33 | 7.97e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 2 | 186 | 7.98e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 2 | 179 | 7.98e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 2 | 24 | 7.98e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 3 | 64 | 7.99e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 2 | 1088 | 7.99e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 1 | 87 | 7.99e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 2 | 159 | 7.99e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 1 | 33 | 7.99e-01 |
| GO:BP | GO:0009615 | response to virus | 2 | 275 | 7.99e-01 |
| GO:BP | GO:0000413 | protein peptidyl-prolyl isomerization | 1 | 26 | 7.99e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 7 | 271 | 7.99e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 1 | 17 | 8.00e-01 |
| GO:BP | GO:0007601 | visual perception | 1 | 96 | 8.00e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 3 | 101 | 8.01e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 6 | 174 | 8.02e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 3 | 132 | 8.02e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 8.02e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 3 | 274 | 8.02e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 1 | 64 | 8.02e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 1 | 95 | 8.02e-01 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 1 | 25 | 8.02e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 4 | 547 | 8.02e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 3 | 57 | 8.02e-01 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 1 | 19 | 8.02e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 1 | 22 | 8.02e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 8.02e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1 | 3070 | 8.02e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 8.02e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 1 | 18 | 8.02e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 8.02e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 1 | 97 | 8.02e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 8.02e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 132 | 8.02e-01 |
| GO:BP | GO:0006497 | protein lipidation | 2 | 89 | 8.03e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 1 | 25 | 8.03e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1 | 18 | 8.03e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 2 | 41 | 8.04e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 2 | 29 | 8.04e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 8.04e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 2 | 29 | 8.04e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 2 | 62 | 8.04e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 1 | 24 | 8.04e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 8.04e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 1 | 23 | 8.04e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 1 | 33 | 8.04e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 25 | 2593 | 8.04e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 660 | 8.04e-01 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 1 | 29 | 8.04e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 1 | 37 | 8.04e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 8.05e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 1 | 22 | 8.05e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 2 | 40 | 8.05e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 2 | 1111 | 8.05e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 1 | 39 | 8.06e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 1 | 42 | 8.06e-01 |
| GO:BP | GO:0021761 | limbic system development | 4 | 81 | 8.06e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 1 | 49 | 8.06e-01 |
| GO:BP | GO:0015031 | protein transport | 19 | 1361 | 8.06e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 1 | 3132 | 8.06e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 7 | 796 | 8.06e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 2 | 53 | 8.06e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 1 | 3133 | 8.07e-01 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 1 | 50 | 8.07e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 8.07e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 16 | 1226 | 8.07e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1 | 2065 | 8.07e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 1 | 34 | 8.07e-01 |
| GO:BP | GO:0099068 | postsynapse assembly | 1 | 32 | 8.07e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 1 | 160 | 8.08e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 1 | 94 | 8.08e-01 |
| GO:BP | GO:0072678 | T cell migration | 1 | 38 | 8.09e-01 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5770 | 8.10e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 3 | 51 | 8.10e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 678 | 8.10e-01 |
| GO:BP | GO:0046903 | secretion | 2 | 645 | 8.10e-01 |
| GO:BP | GO:0007565 | female pregnancy | 6 | 122 | 8.11e-01 |
| GO:BP | GO:0042311 | vasodilation | 1 | 39 | 8.11e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 1 | 19 | 8.12e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 1 | 19 | 8.12e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 1 | 162 | 8.13e-01 |
| GO:BP | GO:0001944 | vasculature development | 3 | 559 | 8.13e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 1 | 15 | 8.13e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 2 | 161 | 8.13e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 1 | 22 | 8.13e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 8.13e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 2 | 120 | 8.13e-01 |
| GO:BP | GO:0001841 | neural tube formation | 2 | 101 | 8.13e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 8.13e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 3 | 85 | 8.13e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 1 | 22 | 8.14e-01 |
| GO:BP | GO:0030168 | platelet activation | 5 | 91 | 8.14e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 1 | 50 | 8.14e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 1 | 20 | 8.14e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 27 | 2845 | 8.14e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 3 | 67 | 8.14e-01 |
| GO:BP | GO:0022402 | cell cycle process | 2 | 1086 | 8.15e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 8.17e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 1 | 93 | 8.17e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 4 | 485 | 8.17e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 1 | 22 | 8.17e-01 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 1 | 14 | 8.17e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 1 | 18 | 8.17e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 1 | 23 | 8.18e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 1 | 3237 | 8.18e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 1 | 4128 | 8.18e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 2 | 55 | 8.18e-01 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 1 | 29 | 8.19e-01 |
| GO:BP | GO:0071715 | icosanoid transport | 2 | 24 | 8.19e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 3 | 105 | 8.19e-01 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 2 | 191 | 8.19e-01 |
| GO:BP | GO:0099171 | presynaptic modulation of chemical synaptic transmission | 1 | 26 | 8.19e-01 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 1 | 30 | 8.19e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 2 | 45 | 8.19e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 1 | 39 | 8.19e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 2 | 45 | 8.19e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 9 | 184 | 8.19e-01 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 3 | 89 | 8.19e-01 |
| GO:BP | GO:0006915 | apoptotic process | 12 | 1443 | 8.19e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 2 | 45 | 8.19e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 1 | 36 | 8.19e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 8.19e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 1 | 45 | 8.19e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 2 | 204 | 8.19e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 8.19e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 2 | 92 | 8.19e-01 |
| GO:BP | GO:0051179 | localization | 2 | 3969 | 8.20e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 1 | 36 | 8.20e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 4 | 82 | 8.20e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 8.20e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 2 | 53 | 8.20e-01 |
| GO:BP | GO:0002438 | acute inflammatory response to antigenic stimulus | 1 | 10 | 8.20e-01 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 1 | 11 | 8.20e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 1 | 28 | 8.20e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 1 | 33 | 8.21e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 8.22e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 8.23e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 1 | 40 | 8.23e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 1 | 86 | 8.23e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 8.24e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 2 | 1137 | 8.24e-01 |
| GO:BP | GO:0001756 | somitogenesis | 1 | 48 | 8.24e-01 |
| GO:BP | GO:0002507 | tolerance induction | 1 | 15 | 8.24e-01 |
| GO:BP | GO:0099172 | presynapse organization | 1 | 42 | 8.24e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 8.24e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 9 | 1974 | 8.24e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 1 | 106 | 8.26e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 1 | 19 | 8.26e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 4 | 230 | 8.27e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 8.27e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 1 | 26 | 8.27e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 7 | 155 | 8.27e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 1 | 70 | 8.27e-01 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 1 | 30 | 8.27e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 3 | 68 | 8.27e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 3 | 512 | 8.27e-01 |
| GO:BP | GO:0048675 | axon extension | 3 | 110 | 8.27e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 1 | 293 | 8.27e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 5 | 108 | 8.27e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 1 | 127 | 8.27e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 1 | 26 | 8.27e-01 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 1 | 21 | 8.27e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 1 | 70 | 8.27e-01 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 2 | 197 | 8.28e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 1 | 17 | 8.28e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 1 | 48 | 8.28e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1 | 1898 | 8.29e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 22 | 414 | 8.29e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 8.29e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 1 | 348 | 8.29e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 1 | 24 | 8.29e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 1 | 33 | 8.29e-01 |
| GO:BP | GO:0045923 | positive regulation of fatty acid metabolic process | 1 | 26 | 8.29e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 1 | 44 | 8.29e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 1 | 33 | 8.29e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 3 | 77 | 8.30e-01 |
| GO:BP | GO:0051607 | defense response to virus | 3 | 202 | 8.30e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 1 | 19 | 8.30e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 1 | 27 | 8.30e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 1 | 88 | 8.31e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 1 | 22 | 8.31e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 11 | 830 | 8.31e-01 |
| GO:BP | GO:0007420 | brain development | 1 | 591 | 8.31e-01 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 4 | 122 | 8.31e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 3 | 202 | 8.32e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 153 | 8.32e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 1 | 109 | 8.32e-01 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 3 | 87 | 8.32e-01 |
| GO:BP | GO:0043113 | receptor clustering | 2 | 45 | 8.35e-01 |
| GO:BP | GO:0031297 | replication fork processing | 1 | 42 | 8.35e-01 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 1 | 30 | 8.35e-01 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 1 | 36 | 8.35e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 8.35e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 1 | 50 | 8.35e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 1 | 23 | 8.35e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 1 | 22 | 8.35e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 1 | 63 | 8.35e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 3 | 144 | 8.36e-01 |
| GO:BP | GO:0050658 | RNA transport | 3 | 144 | 8.36e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 2 | 49 | 8.36e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 1 | 36 | 8.36e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 1 | 55 | 8.36e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 8.36e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 9 | 187 | 8.36e-01 |
| GO:BP | GO:0021675 | nerve development | 1 | 57 | 8.36e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 1 | 58 | 8.36e-01 |
| GO:BP | GO:0046034 | ATP metabolic process | 4 | 116 | 8.37e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 24 | 8.37e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 8.37e-01 |
| GO:BP | GO:0015844 | monoamine transport | 3 | 45 | 8.37e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 25 | 8.37e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 1 | 23 | 8.37e-01 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 1 | 31 | 8.38e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 3 | 194 | 8.38e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 228 | 8.38e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 3 | 116 | 8.38e-01 |
| GO:BP | GO:0006821 | chloride transport | 4 | 58 | 8.38e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1 | 2285 | 8.38e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 1 | 112 | 8.38e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 8.39e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 8.39e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 8.40e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 34 | 8.40e-01 |
| GO:BP | GO:0031424 | keratinization | 1 | 13 | 8.40e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 8.41e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 35 | 3750 | 8.42e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 8.42e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 2 | 317 | 8.42e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 4 | 121 | 8.43e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 2 | 28 | 8.43e-01 |
| GO:BP | GO:0007409 | axonogenesis | 2 | 359 | 8.43e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 3 | 147 | 8.44e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 1 | 14 | 8.44e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 1 | 14 | 8.44e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 8.44e-01 |
| GO:BP | GO:0007417 | central nervous system development | 4 | 792 | 8.44e-01 |
| GO:BP | GO:0007492 | endoderm development | 1 | 65 | 8.44e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 1 | 24 | 8.44e-01 |
| GO:BP | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 19 | 8.44e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 1 | 31 | 8.44e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 1 | 41 | 8.44e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 25 | 2683 | 8.44e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 1 | 42 | 8.44e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 25 | 2684 | 8.44e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 8.44e-01 |
| GO:BP | GO:0001508 | action potential | 1 | 105 | 8.44e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 7 | 212 | 8.44e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 2 | 38 | 8.44e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 4 | 517 | 8.44e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 3 | 64 | 8.45e-01 |
| GO:BP | GO:0060322 | head development | 1 | 633 | 8.45e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 4 | 300 | 8.45e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 8.46e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 1 | 20 | 8.46e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 4 | 597 | 8.46e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 4 | 597 | 8.46e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 3 | 104 | 8.46e-01 |
| GO:BP | GO:0032502 | developmental process | 1 | 4494 | 8.47e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 1 | 384 | 8.47e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 1 | 111 | 8.47e-01 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 5 | 209 | 8.47e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 1 | 24 | 8.48e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 2 | 61 | 8.48e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 1 | 41 | 8.48e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 7 | 135 | 8.48e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 1 | 53 | 8.48e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 1 | 358 | 8.51e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 2 | 256 | 8.52e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 8.52e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 1 | 31 | 8.54e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 2 | 258 | 8.54e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 4 | 242 | 8.54e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 1 | 506 | 8.54e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 8.54e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 39 | 4174 | 8.54e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 2 | 34 | 8.54e-01 |
| GO:BP | GO:0021782 | glial cell development | 3 | 84 | 8.54e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 8.55e-01 |
| GO:BP | GO:0050877 | nervous system process | 6 | 634 | 8.56e-01 |
| GO:BP | GO:0006997 | nucleus organization | 1 | 126 | 8.56e-01 |
| GO:BP | GO:0042551 | neuron maturation | 1 | 33 | 8.56e-01 |
| GO:BP | GO:0014002 | astrocyte development | 1 | 27 | 8.56e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 1 | 447 | 8.56e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 1 | 93 | 8.56e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 1 | 21 | 8.57e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 3 | 76 | 8.58e-01 |
| GO:BP | GO:0001816 | cytokine production | 1 | 451 | 8.58e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 1 | 29 | 8.58e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 1 | 19 | 8.58e-01 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 1 | 20 | 8.58e-01 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 1 | 36 | 8.58e-01 |
| GO:BP | GO:0007254 | JNK cascade | 1 | 138 | 8.59e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 7 | 640 | 8.59e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 1 | 31 | 8.59e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 1 | 382 | 8.59e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 357 | 8.59e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 25 | 2714 | 8.59e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 2 | 51 | 8.59e-01 |
| GO:BP | GO:0050821 | protein stabilization | 4 | 191 | 8.59e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 3 | 69 | 8.59e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 1 | 39 | 8.59e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 1 | 94 | 8.59e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 8.59e-01 |
| GO:BP | GO:0010458 | exit from mitosis | 1 | 30 | 8.59e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 1 | 760 | 8.59e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 2 | 47 | 8.59e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 9 | 273 | 8.59e-01 |
| GO:BP | GO:0016486 | peptide hormone processing | 1 | 27 | 8.60e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 1 | 20 | 8.60e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 2 | 47 | 8.60e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 17 | 8.60e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 7 | 172 | 8.60e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 1 | 36 | 8.60e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 1 | 43 | 8.60e-01 |
| GO:BP | GO:0000266 | mitochondrial fission | 1 | 38 | 8.61e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 8.61e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 2 | 216 | 8.61e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 1 | 40 | 8.61e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 1 | 95 | 8.62e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 1 | 28 | 8.62e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 8.62e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3 | 62 | 8.62e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 17 | 3625 | 8.62e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 1 | 44 | 8.62e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 1 | 21 | 8.62e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 5 | 679 | 8.62e-01 |
| GO:BP | GO:0001914 | regulation of T cell mediated cytotoxicity | 1 | 17 | 8.62e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 4 | 91 | 8.62e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 8.62e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 1 | 112 | 8.63e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 1 | 29 | 8.63e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 9 | 809 | 8.64e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 5 | 218 | 8.64e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 8.64e-01 |
| GO:BP | GO:0035270 | endocrine system development | 1 | 86 | 8.64e-01 |
| GO:BP | GO:0043038 | amino acid activation | 1 | 44 | 8.64e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 8.64e-01 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 1 | 48 | 8.65e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 1 | 29 | 8.65e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 8 | 164 | 8.65e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 1 | 665 | 8.65e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 8.65e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 2 | 34 | 8.65e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 8 | 994 | 8.65e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 4 | 135 | 8.65e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 6 | 1380 | 8.65e-01 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 1 | 38 | 8.65e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 8.65e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 2 | 40 | 8.65e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 8 | 993 | 8.65e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 28 | 3080 | 8.65e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 8.65e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 1 | 42 | 8.65e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 8.65e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 3 | 303 | 8.65e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 1 | 24 | 8.65e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 7 | 881 | 8.65e-01 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 1 | 25 | 8.65e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 1 | 282 | 8.65e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 6 | 532 | 8.65e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 1 | 217 | 8.65e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 8.65e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 2 | 42 | 8.65e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 4 | 113 | 8.65e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 4 | 113 | 8.65e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 8 | 156 | 8.65e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 2 | 50 | 8.66e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 8.66e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1081 | 8.67e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 2 | 45 | 8.67e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 2 | 183 | 8.68e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 1 | 2565 | 8.68e-01 |
| GO:BP | GO:0001990 | regulation of systemic arterial blood pressure by hormone | 1 | 22 | 8.68e-01 |
| GO:BP | GO:0048477 | oogenesis | 1 | 64 | 8.68e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 1 | 30 | 8.68e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 1 | 40 | 8.68e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 6 | 90 | 8.68e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 11 | 1223 | 8.68e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 6 | 131 | 8.68e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 8.68e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 1 | 378 | 8.68e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 3 | 140 | 8.68e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 8.68e-01 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 3 | 96 | 8.68e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 1 | 381 | 8.68e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 1 | 2573 | 8.68e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 3 | 77 | 8.68e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 1 | 23 | 8.70e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 8.70e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 1 | 38 | 8.70e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 2 | 96 | 8.70e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 3 | 71 | 8.70e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 3 | 121 | 8.70e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 4 | 378 | 8.70e-01 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 3 | 97 | 8.71e-01 |
| GO:BP | GO:0007155 | cell adhesion | 2 | 1012 | 8.71e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 4 | 335 | 8.71e-01 |
| GO:BP | GO:0061564 | axon development | 2 | 398 | 8.72e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 1 | 31 | 8.72e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 8.72e-01 |
| GO:BP | GO:0002253 | activation of immune response | 9 | 272 | 8.72e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 4 | 147 | 8.73e-01 |
| GO:BP | GO:0140448 | signaling receptor ligand precursor processing | 1 | 29 | 8.73e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 1 | 448 | 8.73e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 8.73e-01 |
| GO:BP | GO:0001656 | metanephros development | 1 | 64 | 8.73e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 1 | 32 | 8.73e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 1 | 31 | 8.73e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 4 | 95 | 8.73e-01 |
| GO:BP | GO:0033574 | response to testosterone | 1 | 29 | 8.73e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 2 | 50 | 8.73e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 1 | 57 | 8.74e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 3 | 130 | 8.74e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 2 | 98 | 8.74e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 6 | 185 | 8.74e-01 |
| GO:BP | GO:0061053 | somite development | 1 | 63 | 8.74e-01 |
| GO:BP | GO:0021549 | cerebellum development | 2 | 78 | 8.74e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 1 | 105 | 8.75e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 1 | 387 | 8.75e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 418 | 8.75e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 3 | 70 | 8.75e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 3 | 66 | 8.80e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 9 | 963 | 8.80e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 1 | 59 | 8.80e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 2 | 101 | 8.80e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 6 | 104 | 8.81e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 8.82e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 1 | 88 | 8.82e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 1 | 43 | 8.83e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 8.83e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 1 | 110 | 8.83e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 1 | 39 | 8.83e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 1 | 113 | 8.83e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 1 | 1149 | 8.83e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 8.83e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 3 | 58 | 8.83e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 8.83e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 1 | 38 | 8.83e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 2 | 784 | 8.83e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 2 | 42 | 8.83e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 8.85e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 5 | 83 | 8.85e-01 |
| GO:BP | GO:0051028 | mRNA transport | 2 | 117 | 8.85e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 1 | 63 | 8.85e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 2 | 251 | 8.85e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 1 | 37 | 8.86e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 8.86e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 1 | 33 | 8.86e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 1 | 28 | 8.86e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 1 | 209 | 8.86e-01 |
| GO:BP | GO:0035176 | social behavior | 1 | 34 | 8.86e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 2 | 63 | 8.86e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 1 | 36 | 8.86e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 1 | 69 | 8.86e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 1 | 151 | 8.86e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 1 | 155 | 8.86e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 11 | 211 | 8.87e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 3 | 102 | 8.87e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 2 | 79 | 8.87e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 8.87e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 2 | 55 | 8.87e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 8.87e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 8.87e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 2 | 185 | 8.87e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 1 | 29 | 8.87e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 5 | 228 | 8.88e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 8.88e-01 |
| GO:BP | GO:0010033 | response to organic substance | 27 | 1957 | 8.88e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 2 | 60 | 8.89e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 8.89e-01 |
| GO:BP | GO:0048469 | cell maturation | 3 | 108 | 8.89e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 8.89e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 1 | 406 | 8.89e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 9 | 1167 | 8.89e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 3 | 84 | 8.89e-01 |
| GO:BP | GO:0060541 | respiratory system development | 7 | 163 | 8.89e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 1 | 38 | 8.89e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 1 | 72 | 8.90e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 1 | 36 | 8.90e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 4 | 479 | 8.90e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 1 | 36 | 8.90e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 5 | 97 | 8.91e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 1 | 33 | 8.91e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 8.91e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 6 | 135 | 8.91e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 35 | 8.92e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 2 | 249 | 8.93e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 3 | 92 | 8.93e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 8.93e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 1 | 34 | 8.93e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 8.94e-01 |
| GO:BP | GO:0030282 | bone mineralization | 3 | 82 | 8.95e-01 |
| GO:BP | GO:0001708 | cell fate specification | 2 | 56 | 8.95e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 3 | 535 | 8.95e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 4 | 114 | 8.95e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 2 | 240 | 8.95e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 1 | 149 | 8.95e-01 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 1 | 18 | 8.95e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 1 | 29 | 8.96e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 1 | 16 | 8.97e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 8.97e-01 |
| GO:BP | GO:0030324 | lung development | 6 | 146 | 8.97e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 1 | 54 | 8.97e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 1 | 39 | 8.97e-01 |
| GO:BP | GO:0006810 | transport | 11 | 3298 | 8.98e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 1 | 32 | 8.98e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 2 | 241 | 8.98e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 1 | 43 | 8.98e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 1 | 45 | 8.98e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 39 | 4292 | 8.98e-01 |
| GO:BP | GO:0048732 | gland development | 1 | 318 | 8.98e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 8.99e-01 |
| GO:BP | GO:0097028 | dendritic cell differentiation | 1 | 23 | 9.00e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 1 | 183 | 9.00e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 4 | 159 | 9.01e-01 |
| GO:BP | GO:0022037 | metencephalon development | 2 | 84 | 9.02e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 1 | 7552 | 9.02e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 1 | 11 | 9.02e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 9.05e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 2 | 1399 | 9.05e-01 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 7 | 120 | 9.05e-01 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 4 | 137 | 9.05e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 1 | 30 | 9.05e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 2 | 331 | 9.05e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 3 | 100 | 9.05e-01 |
| GO:BP | GO:0007389 | pattern specification process | 6 | 306 | 9.06e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1182 | 9.06e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 5 | 213 | 9.06e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 172 | 9.06e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 2 | 52 | 9.06e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 2 | 123 | 9.06e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 1 | 69 | 9.06e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 9.07e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 9.07e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 6 | 150 | 9.07e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 1 | 46 | 9.07e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 1 | 46 | 9.07e-01 |
| GO:BP | GO:0050892 | intestinal absorption | 1 | 22 | 9.07e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 38 | 4417 | 9.08e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1 | 2890 | 9.08e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 9.08e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 9.09e-01 |
| GO:BP | GO:0009790 | embryo development | 1 | 875 | 9.09e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 1 | 41 | 9.09e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 2 | 101 | 9.11e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 9.11e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 1 | 43 | 9.11e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 65 | 9.11e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 1 | 25 | 9.11e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 1 | 42 | 9.11e-01 |
| GO:BP | GO:0006403 | RNA localization | 3 | 182 | 9.13e-01 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 1 | 18 | 9.13e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 1 | 36 | 9.13e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 1 | 52 | 9.13e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 2 | 57 | 9.13e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 1 | 963 | 9.13e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 1 | 154 | 9.13e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 1 | 27 | 9.13e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 6 | 2135 | 9.13e-01 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 1 | 36 | 9.13e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 1 | 39 | 9.13e-01 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 1 | 20 | 9.13e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 39 | 2984 | 9.13e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 3 | 61 | 9.14e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 3 | 82 | 9.14e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 9.14e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 1 | 185 | 9.14e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 2 | 463 | 9.15e-01 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 4 | 142 | 9.15e-01 |
| GO:BP | GO:0033993 | response to lipid | 6 | 592 | 9.15e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 3 | 111 | 9.15e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 1 | 157 | 9.15e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 189 | 9.15e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 8 | 371 | 9.15e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 1 | 22 | 9.15e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 8 | 375 | 9.15e-01 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 1 | 46 | 9.15e-01 |
| GO:BP | GO:0021536 | diencephalon development | 2 | 41 | 9.15e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 1 | 36 | 9.16e-01 |
| GO:BP | GO:0021543 | pallium development | 1 | 139 | 9.16e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 3 | 1835 | 9.16e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 1 | 24 | 9.17e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 8 | 376 | 9.17e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 40 | 4099 | 9.18e-01 |
| GO:BP | GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | 1 | 30 | 9.18e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 1 | 190 | 9.18e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 9.18e-01 |
| GO:BP | GO:0030879 | mammary gland development | 2 | 106 | 9.18e-01 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 4 | 144 | 9.18e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 1 | 6996 | 9.19e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 2 | 120 | 9.19e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 9.19e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 2 | 131 | 9.20e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 1 | 13 | 9.20e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 6 | 295 | 9.20e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 9.21e-01 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 1 | 47 | 9.21e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 9.21e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 1 | 54 | 9.21e-01 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 1 | 47 | 9.21e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 12 | 236 | 9.21e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 9.21e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 1 | 54 | 9.21e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 1 | 21 | 9.21e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 9.21e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 9 | 423 | 9.21e-01 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 1 | 47 | 9.21e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 9.21e-01 |
| GO:BP | GO:0007602 | phototransduction | 1 | 17 | 9.21e-01 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 1 | 47 | 9.21e-01 |
| GO:BP | GO:0051234 | establishment of localization | 11 | 3433 | 9.21e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 4 | 121 | 9.21e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 9.21e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 9.23e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 1 | 52 | 9.23e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 1 | 196 | 9.23e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 1 | 44 | 9.24e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 9.25e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 4 | 414 | 9.25e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 7 | 133 | 9.25e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 1 | 191 | 9.26e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 5 | 106 | 9.27e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 3 | 111 | 9.27e-01 |
| GO:BP | GO:0046717 | acid secretion | 1 | 25 | 9.27e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 2 | 108 | 9.27e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 2 | 88 | 9.27e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 1 | 74 | 9.28e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 9.28e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 1 | 204 | 9.28e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 1 | 60 | 9.28e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 1 | 39 | 9.28e-01 |
| GO:BP | GO:0003002 | regionalization | 5 | 273 | 9.28e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 3 | 120 | 9.28e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 1 | 83 | 9.28e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 1 | 27 | 9.28e-01 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 1 | 48 | 9.28e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 9.28e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 9.28e-01 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 1 | 48 | 9.28e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 1 | 50 | 9.29e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 6 | 160 | 9.29e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 1 | 36 | 9.29e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 1 | 41 | 9.29e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 501 | 9.29e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 6 | 260 | 9.29e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 9.29e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 1 | 67 | 9.29e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 9.30e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 1 | 337 | 9.30e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 3 | 893 | 9.30e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 1 | 214 | 9.30e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 9.30e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 9 | 429 | 9.31e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 2 | 67 | 9.31e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 2 | 55 | 9.31e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 1 | 22 | 9.32e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 4 | 111 | 9.32e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 3 | 616 | 9.33e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 1 | 64 | 9.33e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 9.33e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 9.33e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 2 | 89 | 9.33e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 2 | 91 | 9.33e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 3 | 68 | 9.34e-01 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 1 | 20 | 9.34e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 9.34e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 9.35e-01 |
| GO:BP | GO:0007568 | aging | 4 | 115 | 9.35e-01 |
| GO:BP | GO:0071709 | membrane assembly | 1 | 53 | 9.35e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 1 | 26 | 9.36e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 1 | 74 | 9.36e-01 |
| GO:BP | GO:0034644 | cellular response to UV | 2 | 86 | 9.36e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 1 | 31 | 9.37e-01 |
| GO:BP | GO:0008347 | glial cell migration | 1 | 47 | 9.37e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 1 | 31 | 9.37e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 1 | 36 | 9.37e-01 |
| GO:BP | GO:0022900 | electron transport chain | 5 | 142 | 9.37e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 1 | 41 | 9.37e-01 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 1 | 52 | 9.37e-01 |
| GO:BP | GO:0035094 | response to nicotine | 1 | 28 | 9.37e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 1 | 60 | 9.37e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 1 | 35 | 9.37e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 9.37e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 2 | 90 | 9.38e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 2 | 68 | 9.38e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 6 | 311 | 9.38e-01 |
| GO:BP | GO:0016310 | phosphorylation | 11 | 1372 | 9.38e-01 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 1 | 84 | 9.38e-01 |
| GO:BP | GO:0042116 | macrophage activation | 2 | 57 | 9.38e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 2 | 48 | 9.38e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 6 | 2293 | 9.38e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 1 | 198 | 9.38e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 1 | 52 | 9.38e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 2 | 57 | 9.38e-01 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 1 | 54 | 9.38e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 2 | 92 | 9.38e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 2 | 269 | 9.38e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 9.38e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 2 | 126 | 9.38e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 1 | 45 | 9.38e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 3 | 390 | 9.38e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 2 | 43 | 9.39e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 2 | 43 | 9.39e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 1 | 55 | 9.39e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 2 | 62 | 9.39e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 2 | 67 | 9.39e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 2 | 144 | 9.39e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 26 | 9.41e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 1 | 52 | 9.41e-01 |
| GO:BP | GO:0042552 | myelination | 3 | 111 | 9.41e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 1 | 47 | 9.42e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 9.42e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 39 | 783 | 9.44e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 9.44e-01 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 1 | 36 | 9.44e-01 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 1 | 36 | 9.44e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 4 | 159 | 9.45e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 1 | 199 | 9.46e-01 |
| GO:BP | GO:0001889 | liver development | 3 | 110 | 9.46e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 6 | 319 | 9.46e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 1 | 47 | 9.46e-01 |
| GO:BP | GO:0007292 | female gamete generation | 1 | 103 | 9.46e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 1 | 23 | 9.47e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 9.47e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 1 | 58 | 9.47e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 1 | 46 | 9.47e-01 |
| GO:BP | GO:0097305 | response to alcohol | 1 | 159 | 9.50e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 63 | 4579 | 9.50e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 9.50e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 9.51e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 3 | 112 | 9.52e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 2 | 135 | 9.52e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 1 | 56 | 9.52e-01 |
| GO:BP | GO:0048002 | antigen processing and presentation of peptide antigen | 1 | 41 | 9.52e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 1 | 101 | 9.52e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 5 | 279 | 9.53e-01 |
| GO:BP | GO:0008585 | female gonad development | 1 | 77 | 9.53e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 1 | 55 | 9.53e-01 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 1 | 58 | 9.53e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 1 | 52 | 9.53e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 1 | 40 | 9.54e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 9.54e-01 |
| GO:BP | GO:0030154 | cell differentiation | 37 | 2958 | 9.54e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 1 | 38 | 9.55e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 1 | 27 | 9.55e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 1 | 39 | 9.56e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 1 | 73 | 9.56e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 2 | 99 | 9.56e-01 |
| GO:BP | GO:0009617 | response to bacterium | 3 | 321 | 9.56e-01 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 1 | 23 | 9.56e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 1 | 59 | 9.57e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 4 | 121 | 9.57e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 3 | 293 | 9.57e-01 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 1 | 57 | 9.57e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 1 | 79 | 9.58e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 7 | 1939 | 9.59e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 9.59e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 1 | 38 | 9.59e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 9.59e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4048 | 9.59e-01 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 1 | 33 | 9.59e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 2 | 111 | 9.59e-01 |
| GO:BP | GO:0016043 | cellular component organization | 2 | 5076 | 9.59e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 3 | 95 | 9.59e-01 |
| GO:BP | GO:0001890 | placenta development | 3 | 117 | 9.59e-01 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 1 | 61 | 9.59e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 3 | 116 | 9.60e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 9.61e-01 |
| GO:BP | GO:0035825 | homologous recombination | 1 | 43 | 9.61e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 1 | 48 | 9.61e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 1 | 52 | 9.61e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 11 | 279 | 9.61e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 9.61e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 9.61e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 6 | 398 | 9.61e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 9.61e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 9.61e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 2 | 310 | 9.62e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 3 | 130 | 9.62e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 1 | 8061 | 9.62e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 3 | 486 | 9.63e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 1 | 54 | 9.63e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 1 | 55 | 9.63e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 9.63e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 2 | 78 | 9.64e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 2 | 70 | 9.64e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 6 | 1199 | 9.64e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 9.64e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 3 | 139 | 9.65e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 2 | 71 | 9.65e-01 |
| GO:BP | GO:0006413 | translational initiation | 2 | 116 | 9.65e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4241 | 9.65e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 2 | 154 | 9.65e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2 | 5274 | 9.65e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 2 | 260 | 9.65e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 1 | 232 | 9.65e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 2 | 72 | 9.65e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 9.65e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 1 | 46 | 9.67e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 10 | 369 | 9.67e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 1 | 55 | 9.67e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 5 | 2035 | 9.67e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 2 | 157 | 9.67e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 96 | 9.68e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 9.68e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 9.69e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 9.69e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 4 | 111 | 9.71e-01 |
| GO:BP | GO:0051653 | spindle localization | 1 | 52 | 9.71e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4455 | 9.71e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 1 | 103 | 9.71e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 9.72e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 9.72e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 1 | 114 | 9.72e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 2 | 141 | 9.72e-01 |
| GO:BP | GO:0009056 | catabolic process | 13 | 2088 | 9.72e-01 |
| GO:BP | GO:0031016 | pancreas development | 1 | 53 | 9.72e-01 |
| GO:BP | GO:1901616 | organic hydroxy compound catabolic process | 1 | 31 | 9.72e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 9.72e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 1 | 65 | 9.72e-01 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 1 | 31 | 9.72e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 1 | 142 | 9.72e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 9.72e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 9 | 487 | 9.72e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 1 | 91 | 9.72e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 3 | 77 | 9.72e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 1 | 194 | 9.72e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 9.72e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 85 | 9.72e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 1 | 7806 | 9.72e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 3 | 111 | 9.73e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 9.73e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4570 | 9.73e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 2 | 4617 | 9.73e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4596 | 9.74e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 2 | 53 | 9.74e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 1 | 877 | 9.74e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 1 | 237 | 9.74e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 1 | 88 | 9.75e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 1 | 58 | 9.75e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 7 | 850 | 9.75e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1 | 4045 | 9.76e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 1 | 61 | 9.76e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 1 | 58 | 9.76e-01 |
| GO:BP | GO:0045471 | response to ethanol | 1 | 73 | 9.78e-01 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 1 | 71 | 9.78e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 2 | 59 | 9.78e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 1 | 59 | 9.78e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 5 | 118 | 9.79e-01 |
| GO:BP | GO:0007165 | signal transduction | 1 | 3818 | 9.79e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 1 | 68 | 9.79e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 1 | 249 | 9.79e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 3 | 113 | 9.79e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4747 | 9.79e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 1 | 84 | 9.79e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 1 | 69 | 9.79e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 2 | 294 | 9.79e-01 |
| GO:BP | GO:0009408 | response to heat | 2 | 84 | 9.79e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 9 | 957 | 9.79e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 9.79e-01 |
| GO:BP | GO:0065007 | biological regulation | 1 | 8320 | 9.79e-01 |
| GO:BP | GO:0003014 | renal system process | 2 | 78 | 9.79e-01 |
| GO:BP | GO:0006508 | proteolysis | 7 | 1318 | 9.79e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 2 | 167 | 9.79e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 1 | 39 | 9.79e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 1 | 55 | 9.80e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 11 | 1719 | 9.80e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 2 | 93 | 9.80e-01 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 1 | 30 | 9.81e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 1 | 70 | 9.81e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4946 | 9.81e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 3 | 615 | 9.81e-01 |
| GO:BP | GO:0009566 | fertilization | 1 | 93 | 9.81e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 9.81e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 2 | 144 | 9.81e-01 |
| GO:BP | GO:0003170 | heart valve development | 1 | 65 | 9.81e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 2 | 86 | 9.81e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 5 | 130 | 9.81e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 9.81e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 9.81e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 9.81e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 9.82e-01 |
| GO:BP | GO:0051304 | chromosome separation | 1 | 76 | 9.82e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 1 | 61 | 9.82e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 2 | 3580 | 9.82e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 3 | 118 | 9.82e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 9.82e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 2 | 121 | 9.83e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 3 | 625 | 9.83e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 9.83e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 4 | 199 | 9.83e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 2 | 99 | 9.83e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 258 | 9.83e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 1 | 75 | 9.83e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 3 | 1101 | 9.83e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 9.83e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 1 | 123 | 9.83e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 9.83e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 3 | 123 | 9.83e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 1 | 4955 | 9.83e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 1 | 272 | 9.83e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2 | 5017 | 9.83e-01 |
| GO:BP | GO:0023052 | signaling | 1 | 4141 | 9.84e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 3 | 637 | 9.84e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1 | 114 | 9.84e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 9.85e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 3 | 608 | 9.85e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 9.85e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 5 | 901 | 9.85e-01 |
| GO:BP | GO:0007154 | cell communication | 1 | 4226 | 9.85e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 52 | 9.88e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 9.88e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 4 | 170 | 9.89e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 1 | 131 | 9.89e-01 |
| GO:BP | GO:0071772 | response to BMP | 1 | 131 | 9.89e-01 |
| GO:BP | GO:0050886 | endocrine process | 1 | 57 | 9.89e-01 |
| GO:BP | GO:0051707 | response to other organism | 6 | 820 | 9.89e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 6 | 820 | 9.90e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 9.90e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 9.90e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 9.90e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 1 | 182 | 9.90e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 2 | 98 | 9.91e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 4 | 115 | 9.91e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 3 | 665 | 9.91e-01 |
| GO:BP | GO:0045087 | innate immune response | 1 | 488 | 9.91e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 10 | 2139 | 9.91e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 4 | 706 | 9.91e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 1 | 66 | 9.91e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 3 | 215 | 9.92e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 10 | 2161 | 9.92e-01 |
| GO:BP | GO:0036211 | protein modification process | 24 | 2901 | 9.92e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 4 | 448 | 9.92e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 7 | 204 | 9.92e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 1 | 484 | 9.92e-01 |
| GO:BP | GO:0031341 | regulation of cell killing | 1 | 36 | 9.92e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 1 | 66 | 9.92e-01 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 1 | 90 | 9.92e-01 |
| GO:BP | GO:0021700 | developmental maturation | 3 | 211 | 9.92e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 9.92e-01 |
| GO:BP | GO:0001906 | cell killing | 1 | 69 | 9.92e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 1 | 101 | 9.92e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 9.92e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 9.92e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 1 | 192 | 9.93e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 9.93e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 2 | 155 | 9.93e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 1 | 90 | 9.93e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 9.93e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 3 | 160 | 9.93e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 27 | 3090 | 9.94e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 1 | 86 | 9.94e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 3 | 2466 | 9.94e-01 |
| GO:BP | GO:0031099 | regeneration | 3 | 142 | 9.94e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 9.94e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 2 | 76 | 9.94e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 2 | 103 | 9.94e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 9.94e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 1 | 57 | 9.94e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 2 | 77 | 9.95e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 133 | 9.96e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 2 | 382 | 9.96e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 9.96e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 1 | 64 | 9.96e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 1 | 427 | 9.97e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 3 | 199 | 9.97e-01 |
| GO:BP | GO:0002252 | immune effector process | 1 | 318 | 9.97e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 3 | 142 | 9.97e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 3 | 160 | 9.97e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 3 | 2724 | 9.97e-01 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 1 | 27 | 9.97e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 4 | 179 | 9.97e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 9.97e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 1 | 581 | 9.97e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3 | 161 | 9.98e-01 |
| GO:BP | GO:0022600 | digestive system process | 1 | 54 | 9.98e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 3 | 128 | 9.98e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 7 | 190 | 9.98e-01 |
| GO:BP | GO:0042063 | gliogenesis | 5 | 235 | 9.98e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 1 | 64 | 9.98e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 95 | 9.99e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 2 | 656 | 9.99e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 2 | 118 | 9.99e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 2 | 118 | 9.99e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0010467 | gene expression | 1 | 4587 | 1.00e+00 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 5 | 268 | 1.00e+00 |
| GO:BP | GO:0140013 | meiotic nuclear division | 1 | 120 | 1.00e+00 |
| GO:BP | GO:0016070 | RNA metabolic process | 1 | 3600 | 1.00e+00 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 10 | 276 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0043933 | protein-containing complex organization | 4 | 1496 | 1.00e+00 |
| GO:BP | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition | 1 | 17 | 1.00e+00 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 206 | 1.00e+00 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 1.00e+00 |
| GO:BP | GO:0032635 | interleukin-6 production | 1 | 86 | 1.00e+00 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 1 | 86 | 1.00e+00 |
| GO:BP | GO:0031349 | positive regulation of defense response | 1 | 251 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.00e+00 |
| GO:BP | GO:0002250 | adaptive immune response | 3 | 218 | 1.00e+00 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1335 | 1.00e+00 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 1 | 81 | 1.00e+00 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 2 | 138 | 1.00e+00 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 8 | 222 | 1.00e+00 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.00e+00 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 1 | 7441 | 1.00e+00 |
| GO:BP | GO:0071478 | cellular response to radiation | 2 | 156 | 1.00e+00 |
| GO:BP | GO:0006955 | immune response | 2 | 882 | 1.00e+00 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 1.00e+00 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 1 | 108 | 1.00e+00 |
| GO:BP | GO:0080134 | regulation of response to stress | 3 | 1075 | 1.00e+00 |
| GO:BP | GO:0009266 | response to temperature stimulus | 2 | 133 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 1.00e+00 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 1 | 135 | 1.00e+00 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 3 | 229 | 1.00e+00 |
| GO:BP | GO:0007338 | single fertilization | 2 | 70 | 1.00e+00 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 1 | 81 | 1.00e+00 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 1 | 102 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0007127 | meiosis I | 1 | 77 | 1.00e+00 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 1 | 7069 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 5 | 268 | 1.00e+00 |
| KEGG | KEGG:05219 | Bladder cancer | 6 | 37 | 2.36e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 2 | 48 | 2.36e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 1 | 39 | 2.36e-01 |
| KEGG | KEGG:03430 | Mismatch repair | 1 | 22 | 2.36e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 1 | 43 | 2.36e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 1 | 63 | 2.36e-01 |
| KEGG | KEGG:04611 | Platelet activation | 1 | 89 | 2.36e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 6 | 133 | 2.36e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 1 | 26 | 2.36e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 5 | 28 | 2.36e-01 |
| KEGG | KEGG:03030 | DNA replication | 1 | 35 | 2.36e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 1 | 41 | 2.36e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 3 | 77 | 2.36e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 5 | 115 | 2.42e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 2 | 20 | 2.42e-01 |
| KEGG | KEGG:03060 | Protein export | 1 | 23 | 3.00e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 2 | 38 | 3.00e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 8 | 91 | 3.00e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 2 | 58 | 3.18e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 3 | 157 | 3.18e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 15 | 176 | 3.18e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 3 | 59 | 3.18e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 2 | 69 | 3.18e-01 |
| KEGG | KEGG:05218 | Melanoma | 7 | 58 | 3.32e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 4 | 94 | 3.32e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 2 | 87 | 3.32e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 3 | 52 | 3.32e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 2 | 48 | 3.68e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 7 | 74 | 4.00e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 2 | 124 | 4.06e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 2 | 55 | 4.06e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 6 | 67 | 4.06e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 15 | 179 | 4.06e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 4.06e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 5 | 76 | 4.06e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 2 | 61 | 4.06e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 2 | 18 | 4.06e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 3 | 145 | 4.06e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 1 | 86 | 4.14e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 1 | 41 | 4.20e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 14 | 186 | 4.20e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 3 | 84 | 4.20e-01 |
| KEGG | KEGG:04530 | Tight junction | 1 | 137 | 4.20e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 1 | 21 | 4.20e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 2 | 25 | 4.20e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 2 | 64 | 4.20e-01 |
| KEGG | KEGG:05214 | Glioma | 6 | 67 | 4.20e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 5 | 49 | 4.20e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 3 | 128 | 4.20e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 1 | 44 | 4.20e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 13 | 177 | 4.26e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 4.30e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 9 | 114 | 4.32e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 1 | 107 | 4.32e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 13 | 164 | 4.32e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 7 | 165 | 4.32e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 2 | 82 | 4.32e-01 |
| KEGG | KEGG:04144 | Endocytosis | 5 | 232 | 4.32e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 5 | 112 | 4.32e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 6 | 78 | 4.32e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 9 | 145 | 4.32e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 2 | 51 | 4.32e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 3 | 106 | 4.32e-01 |
| KEGG | KEGG:03010 | Ribosome | 3 | 127 | 4.32e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 3 | 117 | 4.32e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 2 | 83 | 4.32e-01 |
| KEGG | KEGG:05131 | Shigellosis | 5 | 210 | 4.32e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 2 | 49 | 4.32e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 3 | 103 | 4.32e-01 |
| KEGG | KEGG:04730 | Long-term depression | 1 | 42 | 4.32e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 4 | 72 | 4.32e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 3 | 97 | 4.32e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 2 | 112 | 4.32e-01 |
| KEGG | KEGG:04924 | Renin secretion | 2 | 50 | 4.32e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 2 | 55 | 4.32e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 4 | 162 | 4.32e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 2 | 98 | 4.32e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 6 | 74 | 4.32e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 5 | 75 | 4.32e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 11 | 172 | 4.32e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 3 | 28 | 4.32e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 5 | 57 | 4.32e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 4 | 23 | 4.32e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 4 | 43 | 4.32e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 7 | 86 | 4.32e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 2 | 38 | 4.32e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 1 | 9 | 4.32e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 1 | 28 | 4.32e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 4.32e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 7 | 84 | 4.32e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 6 | 69 | 4.59e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 4.59e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 3 | 128 | 4.59e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 1 | 53 | 4.59e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 5 | 75 | 4.59e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 2 | 121 | 4.59e-01 |
| KEGG | KEGG:05133 | Pertussis | 2 | 45 | 4.59e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 1 | 15 | 4.59e-01 |
| KEGG | KEGG:05164 | Influenza A | 2 | 103 | 4.59e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 12 | 170 | 4.59e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 4 | 49 | 4.71e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 3 | 32 | 4.73e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 2 | 55 | 5.21e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 3 | 121 | 5.21e-01 |
| KEGG | KEGG:04742 | Taste transduction | 1 | 22 | 5.21e-01 |
| KEGG | KEGG:04540 | Gap junction | 1 | 73 | 5.25e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 5.25e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 2 | 76 | 5.25e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 5.25e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 8 | 133 | 5.25e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 1 | 35 | 5.30e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 2 | 138 | 5.30e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 5 | 78 | 5.33e-01 |
| KEGG | KEGG:04360 | Axon guidance | 10 | 162 | 5.45e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 2 | 75 | 5.51e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 1 | 78 | 5.55e-01 |
| KEGG | KEGG:04744 | Phototransduction | 1 | 12 | 5.55e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 2 | 77 | 5.55e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 4 | 87 | 5.55e-01 |
| KEGG | KEGG:04142 | Lysosome | 5 | 118 | 5.55e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 5 | 69 | 5.55e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 2 | 303 | 5.55e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 2 | 12 | 5.55e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 3 | 31 | 5.55e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 8 | 115 | 5.55e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 8 | 110 | 5.55e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 4 | 65 | 5.55e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 1 | 74 | 5.55e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 5.58e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 4 | 65 | 5.58e-01 |
| KEGG | KEGG:03040 | Spliceosome | 1 | 132 | 5.58e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 3 | 32 | 5.64e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 1 | 11 | 5.64e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 1 | 108 | 5.84e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 2 | 31 | 5.84e-01 |
| KEGG | KEGG:04976 | Bile secretion | 1 | 42 | 5.86e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 2 | 151 | 5.86e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 2 | 110 | 5.86e-01 |
| KEGG | KEGG:04210 | Apoptosis | 1 | 118 | 5.86e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 4 | 70 | 5.90e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 1 | 33 | 6.05e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 7 | 104 | 6.05e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 1 | 99 | 6.11e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 2 | 17 | 6.11e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 6.19e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 2 | 88 | 6.34e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 4 | 62 | 6.35e-01 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 2 | 36 | 6.35e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 6 | 79 | 6.42e-01 |
| KEGG | KEGG:04520 | Adherens junction | 4 | 67 | 6.52e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 3 | 38 | 6.52e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 4 | 149 | 6.55e-01 |
| KEGG | KEGG:05016 | Huntington disease | 12 | 256 | 6.55e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 2 | 385 | 6.55e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 1 | 50 | 6.67e-01 |
| KEGG | KEGG:05020 | Prion disease | 2 | 220 | 6.67e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 4 | 56 | 6.67e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 2 | 22 | 6.67e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 6 | 109 | 6.67e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 1 | 17 | 6.67e-01 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 1 | 12 | 6.67e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 2 | 29 | 6.67e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 2 | 26 | 6.67e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 2 | 103 | 6.69e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 1 | 24 | 6.69e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 1 | 12 | 6.69e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 6.87e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 6.90e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 3 | 226 | 6.90e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 2 | 66 | 6.90e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 1 | 33 | 6.90e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 1 | 31 | 6.90e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 3 | 56 | 6.90e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 3 | 55 | 6.90e-01 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 1 | 169 | 6.90e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 4 | 84 | 6.90e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 5 | 180 | 6.96e-01 |
| KEGG | KEGG:05134 | Legionellosis | 1 | 39 | 7.00e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 1 | 12 | 7.00e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 4 | 240 | 7.00e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 6 | 171 | 7.00e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 3 | 195 | 7.00e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 2 | 409 | 7.00e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 3 | 65 | 7.00e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 7.00e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 2 | 48 | 7.00e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 2 | 35 | 7.00e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 3 | 115 | 7.00e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 7.00e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 2 | 31 | 7.00e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 1 | 20 | 7.00e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 1 | 22 | 7.00e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 5 | 89 | 7.00e-01 |
| KEGG | KEGG:05224 | Breast cancer | 6 | 117 | 7.00e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 7.00e-01 |
| KEGG | KEGG:01200 | Carbon metabolism | 3 | 98 | 7.00e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 2 | 272 | 7.00e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 3 | 46 | 7.00e-01 |
| KEGG | KEGG:04217 | Necroptosis | 1 | 106 | 7.06e-01 |
| KEGG | KEGG:04146 | Peroxisome | 2 | 68 | 7.06e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 3 | 56 | 7.06e-01 |
| KEGG | KEGG:05034 | Alcoholism | 2 | 122 | 7.06e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 5 | 63 | 7.14e-01 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 5 | 7.15e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 2 | 64 | 7.26e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 2 | 33 | 7.26e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 1 | 37 | 7.26e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 4 | 91 | 7.26e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 7.34e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 1 | 16 | 7.36e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 1 | 16 | 7.44e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 4 | 70 | 7.44e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 15 | 315 | 7.59e-01 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 1 | 26 | 7.64e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 1 | 35 | 7.78e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 6 | 145 | 7.78e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 2 | 64 | 7.78e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 3 | 55 | 7.78e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 2 | 99 | 7.78e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 1 | 34 | 7.78e-01 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 1 | 9 | 7.78e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 7.78e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 2 | 28 | 7.78e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 7.78e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 1 | 40 | 7.78e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 8 | 142 | 7.78e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 7.78e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 5 | 131 | 7.80e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 2 | 49 | 7.80e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 1 | 50 | 7.99e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 13 | 254 | 8.37e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 4 | 106 | 8.41e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 2 | 134 | 8.57e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 5 | 135 | 8.57e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 2 | 75 | 8.57e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 3 | 85 | 8.57e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 2 | 41 | 8.60e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 7 | 150 | 8.60e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 8.60e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 2 | 19 | 8.60e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 1 | 46 | 8.60e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 1 | 44 | 8.67e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 8.82e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 8.82e-01 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 2 | 23 | 8.82e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 2 | 44 | 8.82e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 3 | 84 | 8.82e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 1 | 15 | 8.82e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 1 | 49 | 8.82e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 1 | 28 | 8.82e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 12 | 8.82e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 10 | 415 | 8.82e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 5 | 83 | 8.82e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 8.83e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 8.85e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 3 | 153 | 8.86e-01 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 1 | 13 | 8.86e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 1 | 106 | 8.86e-01 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 2 | 21 | 8.86e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 1 | 51 | 8.86e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 1 | 71 | 8.99e-01 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 9.04e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 1 | 44 | 9.05e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 1 | 161 | 9.14e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 9.14e-01 |
| KEGG | KEGG:04110 | Cell cycle | 4 | 151 | 9.15e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 1 | 49 | 9.15e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 9.18e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 1 | 86 | 9.18e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 2 | 76 | 9.18e-01 |
| KEGG | KEGG:05142 | Chagas disease | 1 | 75 | 9.18e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 9.18e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 9.18e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 9.18e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 1 | 214 | 9.39e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 4 | 110 | 9.42e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 9.47e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 1 | 44 | 9.51e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 1 | 60 | 9.64e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 1 | 82 | 9.85e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 1 | 73 | 9.85e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 1 | 25 | 9.85e-01 |
| KEGG | KEGG:04145 | Phagosome | 2 | 99 | 9.85e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 9.85e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 3 | 134 | 9.85e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 1 | 56 | 9.85e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 9.85e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 2 | 97 | 9.85e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 1 | 67 | 9.85e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 1 | 98 | 9.92e-01 |
All DNR specifc genes adj. p value < 0.05
DAsptable3 <- DAspgostres3$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DAsptable3 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('DNR specific 3 hour GO:BP \nterms n = 335') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)
DAsptable3 %>% mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006351 | DNA-templated transcription | 140 | 2683 | 3.52e-10 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 140 | 2684 | 3.52e-10 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 112 | 1914 | 3.52e-10 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 116 | 1995 | 3.52e-10 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 140 | 2714 | 6.55e-10 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 135 | 2574 | 7.79e-10 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 135 | 2576 | 7.79e-10 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 143 | 2845 | 9.22e-10 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 135 | 2593 | 1.02e-09 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 148 | 3080 | 5.23e-09 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 145 | 3012 | 6.38e-09 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 145 | 3080 | 4.79e-08 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 146 | 3169 | 1.76e-07 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 144 | 3132 | 2.91e-07 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 142 | 3070 | 2.99e-07 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 143 | 3133 | 6.34e-07 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 146 | 3237 | 9.61e-07 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 159 | 3730 | 1.04e-06 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 173 | 4174 | 3.58e-06 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 153 | 3750 | 9.75e-05 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 45 | 740 | 5.05e-04 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 165 | 4245 | 5.57e-04 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 59 | 1119 | 7.94e-04 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 55 | 1015 | 9.20e-04 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 55 | 1017 | 9.24e-04 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 171 | 4417 | 9.59e-04 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 172 | 4539 | 1.10e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 55 | 1026 | 1.13e-03 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 166 | 4292 | 1.42e-03 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 61 | 1209 | 1.60e-03 |
| GO:BP | GO:0009058 | biosynthetic process | 172 | 4590 | 2.33e-03 |
| GO:BP | GO:0140467 | integrated stress response signaling | 5 | 35 | 4.78e-03 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 11 | 128 | 5.84e-03 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 11 | 129 | 6.88e-03 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 58 | 1214 | 8.14e-03 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 71 | 1564 | 8.78e-03 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 62 | 1315 | 1.18e-02 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 3 | 16 | 1.18e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 62 | 1315 | 1.18e-02 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 68 | 1507 | 1.25e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 62 | 1322 | 1.27e-02 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 58 | 1239 | 1.34e-02 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 74 | 1735 | 1.75e-02 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 71 | 1604 | 1.75e-02 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 5 | 39 | 2.46e-02 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 5 | 39 | 2.46e-02 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 64 | 1441 | 2.71e-02 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 58 | 1280 | 2.71e-02 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 14 | 227 | 2.94e-02 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 12 | 179 | 3.05e-02 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 3.22e-02 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 4 | 108 | 3.59e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 14 | 230 | 3.79e-02 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 4 | 135 | 4.24e-02 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 3 | 17 | 4.26e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 7 | 240 | 4.99e-02 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 2 | 4 | 5.03e-02 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 4 | 195 | 5.71e-02 |
| GO:BP | GO:0072739 | response to anisomycin | 2 | 4 | 5.86e-02 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 13 | 222 | 6.23e-02 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 2 | 3 | 6.24e-02 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 3 | 36 | 6.38e-02 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 4 | 28 | 6.53e-02 |
| GO:BP | GO:0001701 | in utero embryonic development | 20 | 344 | 6.84e-02 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 7.06e-02 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 4 | 30 | 7.30e-02 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 4 | 30 | 7.30e-02 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 5 | 53 | 7.96e-02 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 7.96e-02 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 4 | 22 | 7.96e-02 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 6 | 156 | 8.04e-02 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 3 | 41 | 8.53e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 28 | 534 | 8.68e-02 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 3 | 9 | 9.02e-02 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 7 | 100 | 9.97e-02 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 3 | 14 | 9.97e-02 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 3 | 14 | 9.97e-02 |
| GO:BP | GO:0010468 | regulation of gene expression | 150 | 3580 | 1.03e-01 |
| GO:BP | GO:0021915 | neural tube development | 9 | 146 | 1.03e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 4 | 45 | 1.04e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 4 | 34 | 1.04e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 7 | 293 | 1.05e-01 |
| GO:BP | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 1 | 1.08e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 2 | 29 | 1.08e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 3 | 26 | 1.08e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 1 | 2 | 1.08e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 4 | 252 | 1.08e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 44 | 937 | 1.08e-01 |
| GO:BP | GO:0070848 | response to growth factor | 19 | 572 | 1.10e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 28 | 551 | 1.10e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.10e-01 |
| GO:BP | GO:0090132 | epithelium migration | 4 | 252 | 1.10e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 65 | 1602 | 1.10e-01 |
| GO:BP | GO:0090130 | tissue migration | 4 | 256 | 1.11e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 2 | 35 | 1.11e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 1 | 2 | 1.12e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 3 | 132 | 1.12e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 2 | 30 | 1.13e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 1.13e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 1.13e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 1.13e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.13e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 2 | 143 | 1.13e-01 |
| GO:BP | GO:1902217 | erythrocyte apoptotic process | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.13e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 38 | 1.13e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 2 | 15 | 1.13e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 5 | 64 | 1.13e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 5 | 64 | 1.13e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.13e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 1.13e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 2 | 40 | 1.13e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 1.18e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 27 | 616 | 1.18e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 1.19e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 42 | 1.19e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 10 | 265 | 1.21e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 15 | 303 | 1.21e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 2 | 43 | 1.21e-01 |
| GO:BP | GO:0035026 | leading edge cell differentiation | 1 | 2 | 1.21e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 3 | 55 | 1.30e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 1.30e-01 |
| GO:BP | GO:2000761 | positive regulation of N-terminal peptidyl-lysine acetylation | 1 | 1 | 1.32e-01 |
| GO:BP | GO:2000759 | regulation of N-terminal peptidyl-lysine acetylation | 1 | 1 | 1.32e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 18 | 1.34e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 1.34e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 2 | 10 | 1.34e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 1.34e-01 |
| GO:BP | GO:0070982 | L-asparagine metabolic process | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 1.38e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 87 | 2270 | 1.38e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 2 | 182 | 1.41e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 4 | 99 | 1.41e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 12 | 464 | 1.43e-01 |
| GO:BP | GO:0016480 | negative regulation of transcription by RNA polymerase III | 1 | 4 | 1.44e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 5 | 59 | 1.44e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 2 | 19 | 1.45e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 1.45e-01 |
| GO:BP | GO:0002041 | intussusceptive angiogenesis | 1 | 1 | 1.45e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 4 | 34 | 1.45e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 3 | 46 | 1.45e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 2 | 19 | 1.45e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 2 | 6 | 1.45e-01 |
| GO:BP | GO:0001841 | neural tube formation | 6 | 101 | 1.45e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 1.45e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 1 | 13 | 1.45e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 1.45e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 1 | 5 | 1.47e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 2 | 54 | 1.47e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 4 | 25 | 1.47e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 6 | 61 | 1.47e-01 |
| GO:BP | GO:0051028 | mRNA transport | 7 | 117 | 1.47e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 3 | 25 | 1.47e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 3 | 25 | 1.47e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 1.48e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 1.48e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 5 | 73 | 1.48e-01 |
| GO:BP | GO:0072752 | cellular response to rapamycin | 1 | 2 | 1.51e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 1.51e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 1.51e-01 |
| GO:BP | GO:0051169 | nuclear transport | 7 | 295 | 1.51e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 7 | 295 | 1.51e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 4 | 62 | 1.54e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 1 | 2 | 1.54e-01 |
| GO:BP | GO:0003283 | atrial septum development | 2 | 23 | 1.56e-01 |
| GO:BP | GO:0000303 | response to superoxide | 2 | 21 | 1.57e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 1.57e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 1.57e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 1.57e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 1 | 16 | 1.57e-01 |
| GO:BP | GO:0003170 | heart valve development | 3 | 65 | 1.57e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 4 | 121 | 1.57e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 1.57e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 71 | 1849 | 1.57e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 1.57e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 3 | 17 | 1.58e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 1.59e-01 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 4 | 1.61e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 2 | 13 | 1.61e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 7 | 352 | 1.61e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 4 | 35 | 1.62e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 1.62e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 4 | 164 | 1.62e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 1.62e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 1 | 7 | 1.62e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 2 | 22 | 1.62e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 2 | 25 | 1.62e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 3 | 14 | 1.62e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 2 | 46 | 1.62e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 1.62e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 1.62e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 4 | 598 | 1.63e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 4 | 358 | 1.64e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 17 | 551 | 1.64e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 3 | 17 | 1.66e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 1.66e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 1.66e-01 |
| GO:BP | GO:0006556 | S-adenosylmethionine biosynthetic process | 1 | 3 | 1.66e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 2 | 64 | 1.66e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 1.66e-01 |
| GO:BP | GO:0021503 | neural fold bending | 1 | 1 | 1.66e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 7 | 149 | 1.66e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 2 | 14 | 1.67e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 1 | 4 | 1.67e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 2 | 13 | 1.67e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 3 | 45 | 1.68e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 10 | 1.68e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 1 | 7 | 1.70e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 5 | 80 | 1.74e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 3 | 67 | 1.74e-01 |
| GO:BP | GO:0045819 | positive regulation of glycogen catabolic process | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 2 | 21 | 1.74e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 1.75e-01 |
| GO:BP | GO:0071630 | nuclear protein quality control by the ubiquitin-proteasome system | 1 | 3 | 1.75e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 2 | 15 | 1.75e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 1.75e-01 |
| GO:BP | GO:0035196 | miRNA processing | 4 | 41 | 1.75e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 1 | 5 | 1.75e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 1.75e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 1.75e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 1.75e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 5 | 134 | 1.75e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 2 | 26 | 1.75e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 1 | 22 | 1.75e-01 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 1 | 6 | 1.75e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 27 | 620 | 1.75e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 2 | 41 | 1.75e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 2 | 27 | 1.75e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 7 | 90 | 1.75e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 1 | 3 | 1.75e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 1 | 3 | 1.75e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 5 | 133 | 1.76e-01 |
| GO:BP | GO:0030903 | notochord development | 3 | 17 | 1.76e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 11 | 1.76e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 1 | 8 | 1.76e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 3 | 31 | 1.76e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 11 | 706 | 1.76e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 1.76e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 1.76e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 3 | 19 | 1.76e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 3 | 19 | 1.76e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 5 | 89 | 1.76e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 1.76e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 2 | 41 | 1.76e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 5 | 70 | 1.76e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 2 | 7 | 1.76e-01 |
| GO:BP | GO:0006528 | asparagine metabolic process | 1 | 5 | 1.76e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 2 | 14 | 1.76e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 1.76e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 1 | 7 | 1.76e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 2 | 7 | 1.76e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 4 | 56 | 1.77e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 8 | 277 | 1.77e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 1.77e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 2 | 28 | 1.78e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 5 | 67 | 1.78e-01 |
| GO:BP | GO:0097198 | histone H3-K36 trimethylation | 1 | 1 | 1.78e-01 |
| GO:BP | GO:0019083 | viral transcription | 2 | 48 | 1.78e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 2 | 13 | 1.80e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 6 | 84 | 1.80e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 1.80e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 1.80e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 3 | 51 | 1.80e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 3 | 76 | 1.80e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 6 | 116 | 1.82e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 17 | 555 | 1.84e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 6 | 85 | 1.85e-01 |
| GO:BP | GO:0048538 | thymus development | 4 | 35 | 1.86e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 1.86e-01 |
| GO:BP | GO:2000174 | regulation of pro-T cell differentiation | 1 | 1 | 1.87e-01 |
| GO:BP | GO:2000176 | positive regulation of pro-T cell differentiation | 1 | 1 | 1.87e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 1 | 12 | 1.87e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 1.87e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 1 | 3 | 1.87e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 1.87e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 1 | 4 | 1.87e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 1.87e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 1 | 4 | 1.87e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 1 | 4 | 1.87e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 4 | 45 | 1.87e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 3 | 330 | 1.87e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 2 | 90 | 1.87e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 1.87e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 2 | 8 | 1.87e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 6 | 71 | 1.91e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 2 | 15 | 1.92e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 1.92e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 1.92e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 1.92e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 1.92e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 1 | 40 | 1.92e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 5 | 149 | 1.93e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 1 | 3 | 1.93e-01 |
| GO:BP | GO:0048232 | male gamete generation | 3 | 343 | 1.93e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 4 | 39 | 1.93e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 3 | 26 | 1.93e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 2 | 11 | 1.93e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 1.93e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 4 | 60 | 1.93e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 4 | 37 | 1.93e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 1 | 3 | 1.94e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 1 | 9 | 1.94e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 1 | 2 | 1.94e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 1.94e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 3 | 80 | 1.94e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 1.94e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 1 | 14 | 1.94e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 1.96e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 7 | 1.96e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 5 | 78 | 1.96e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 1 | 7 | 1.96e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 1 | 5 | 1.96e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 3 | 35 | 1.96e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 5 | 94 | 1.96e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 12 | 227 | 1.96e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 1.96e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 2 | 34 | 1.96e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 17 | 875 | 1.96e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 8 | 158 | 1.96e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 7 | 144 | 1.97e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 1.97e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 11 | 742 | 1.97e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 3 | 42 | 1.97e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 1.97e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 1.97e-01 |
| GO:BP | GO:0021570 | rhombomere 4 development | 1 | 1 | 1.98e-01 |
| GO:BP | GO:0044524 | protein sulfhydration | 1 | 1 | 1.98e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 1.98e-01 |
| GO:BP | GO:0018272 | protein-pyridoxal-5-phosphate linkage via peptidyl-N6-pyridoxal phosphate-L-lysine | 1 | 1 | 1.98e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 8 | 462 | 1.99e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 1 | 5 | 1.99e-01 |
| GO:BP | GO:1902682 | protein localization to pericentric heterochromatin | 1 | 1 | 1.99e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 3 | 27 | 1.99e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 3 | 23 | 1.99e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 2 | 13 | 1.99e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 1 | 4 | 1.99e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 6 | 109 | 1.99e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 7 | 160 | 1.99e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 1.99e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 1.99e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 5 | 69 | 1.99e-01 |
| GO:BP | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 2 | 17 | 1.99e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 3 | 34 | 1.99e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 1 | 4 | 1.99e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 1.99e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 2 | 90 | 1.99e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 1 | 12 | 1.99e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 1 | 4 | 2.00e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 2.00e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 7 | 144 | 2.01e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 1 | 9 | 2.01e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 6 | 111 | 2.01e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 2 | 14 | 2.01e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 2.01e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 1 | 53 | 2.01e-01 |
| GO:BP | GO:0050658 | RNA transport | 7 | 144 | 2.01e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 2.01e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 1 | 8 | 2.01e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 1 | 8 | 2.01e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 2.04e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 6 | 101 | 2.04e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 6 | 127 | 2.04e-01 |
| GO:BP | GO:0001113 | transcription open complex formation at RNA polymerase II promoter | 1 | 1 | 2.04e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 10 | 198 | 2.04e-01 |
| GO:BP | GO:0070184 | mitochondrial tyrosyl-tRNA aminoacylation | 1 | 1 | 2.05e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 5 | 86 | 2.06e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 1 | 4 | 2.06e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 2.08e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 1 | 10 | 2.08e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 1 | 9 | 2.08e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 3 | 20 | 2.08e-01 |
| GO:BP | GO:0097310 | cap2 mRNA methylation | 1 | 1 | 2.08e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 7 | 147 | 2.09e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 4 | 50 | 2.09e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 2.11e-01 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 2.11e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 2.11e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 2.11e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 2.11e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 2 | 16 | 2.12e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 2 | 113 | 2.12e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 5 | 74 | 2.12e-01 |
| GO:BP | GO:0033301 | cell cycle comprising mitosis without cytokinesis | 1 | 1 | 2.12e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 1 | 5 | 2.14e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 3 | 249 | 2.14e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 1 | 9 | 2.15e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 2.15e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 2 | 114 | 2.15e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 2 | 17 | 2.16e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 1 | 3 | 2.16e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 11 | 763 | 2.18e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 1 | 6 | 2.18e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 1 | 6 | 2.18e-01 |
| GO:BP | GO:0016570 | histone modification | 18 | 433 | 2.18e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 2.18e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 4 | 69 | 2.19e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 2.19e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 2.19e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 1 | 4 | 2.19e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 2.19e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 2.19e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 2.19e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 2.19e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 2.19e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 2 | 9 | 2.19e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 2.19e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 2.19e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 5 | 74 | 2.19e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 2.19e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 5 | 54 | 2.19e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 3 | 234 | 2.20e-01 |
| GO:BP | GO:0051168 | nuclear export | 4 | 150 | 2.21e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 13 | 2.22e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 1 | 13 | 2.22e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 86 | 2412 | 2.23e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 1 | 9 | 2.23e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 1 | 6 | 2.24e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 2.24e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 1 | 20 | 2.24e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 8 | 132 | 2.26e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 1 | 17 | 2.26e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 2 | 40 | 2.26e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 4 | 67 | 2.26e-01 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 2.26e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 3 | 100 | 2.26e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 2 | 97 | 2.26e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 2.27e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 1 | 12 | 2.27e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 2.27e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 2.27e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 2 | 7 | 2.27e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 1 | 8 | 2.27e-01 |
| GO:BP | GO:1904542 | regulation of free ubiquitin chain polymerization | 1 | 2 | 2.27e-01 |
| GO:BP | GO:1904544 | positive regulation of free ubiquitin chain polymerization | 1 | 2 | 2.27e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 4 | 58 | 2.27e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 1 | 3 | 2.27e-01 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 4 | 84 | 2.28e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 1 | 9 | 2.28e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 2.29e-01 |
| GO:BP | GO:0010909 | positive regulation of heparan sulfate proteoglycan biosynthetic process | 1 | 3 | 2.31e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 5 | 124 | 2.32e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 1 | 22 | 2.32e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 2 | 11 | 2.33e-01 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 2.33e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 172 | 4617 | 2.33e-01 |
| GO:BP | GO:0007276 | gamete generation | 3 | 461 | 2.33e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 12 | 313 | 2.33e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 1 | 8 | 2.33e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 5 | 103 | 2.33e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 1 | 44 | 2.33e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 11 | 2.33e-01 |
| GO:BP | GO:0097498 | endothelial tube lumen extension | 1 | 2 | 2.33e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 5 | 67 | 2.33e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 3 | 27 | 2.33e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 2 | 11 | 2.34e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 2 | 12 | 2.34e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 2.34e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 3 | 32 | 2.35e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 2 | 9 | 2.36e-01 |
| GO:BP | GO:0006301 | postreplication repair | 3 | 34 | 2.36e-01 |
| GO:BP | GO:0035148 | tube formation | 6 | 138 | 2.36e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 1 | 9 | 2.36e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 2.36e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 1 | 5 | 2.36e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 1 | 8 | 2.38e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 2.38e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 3 | 38 | 2.38e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 1 | 4 | 2.40e-01 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 2 | 17 | 2.41e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 1 | 2 | 2.41e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 1 | 13 | 2.41e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 2.41e-01 |
| GO:BP | GO:0016477 | cell migration | 14 | 1081 | 2.41e-01 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 2 | 17 | 2.41e-01 |
| GO:BP | GO:0001889 | liver development | 2 | 110 | 2.41e-01 |
| GO:BP | GO:0051030 | snRNA transport | 1 | 6 | 2.41e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 1 | 6 | 2.41e-01 |
| GO:BP | GO:0006428 | isoleucyl-tRNA aminoacylation | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 2.41e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 3 | 30 | 2.41e-01 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 2 | 17 | 2.41e-01 |
| GO:BP | GO:0035295 | tube development | 11 | 821 | 2.42e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 1 | 17 | 2.42e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 1 | 21 | 2.43e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 5 | 91 | 2.43e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 1 | 5 | 2.43e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 1 | 6 | 2.43e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 1 | 5 | 2.43e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 1 | 10 | 2.43e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 1 | 2 | 2.43e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 85 | 2.43e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 2 | 112 | 2.44e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 2 | 34 | 2.44e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 1 | 13 | 2.44e-01 |
| GO:BP | GO:1901545 | response to raffinose | 1 | 1 | 2.45e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 1 | 8 | 2.45e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 10 | 262 | 2.45e-01 |
| GO:BP | GO:0097403 | cellular response to raffinose | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 2.45e-01 |
| GO:BP | GO:0003129 | heart induction | 1 | 9 | 2.45e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 1 | 11 | 2.45e-01 |
| GO:BP | GO:0003342 | proepicardium development | 1 | 2 | 2.45e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 1 | 10 | 2.45e-01 |
| GO:BP | GO:0003343 | septum transversum development | 1 | 2 | 2.45e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 1 | 19 | 2.46e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 1 | 3 | 2.46e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 1 | 18 | 2.46e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 2.46e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 2 | 21 | 2.46e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 1 | 28 | 2.46e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 1 | 3 | 2.46e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 2 | 21 | 2.46e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 2.46e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 7 | 160 | 2.46e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 1 | 6 | 2.46e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 1 | 20 | 2.46e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 1 | 3 | 2.46e-01 |
| GO:BP | GO:0035887 | aortic smooth muscle cell differentiation | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 13 | 2.46e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 1 | 6 | 2.46e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 1 | 26 | 2.46e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 1 | 6 | 2.46e-01 |
| GO:BP | GO:1904829 | regulation of aortic smooth muscle cell differentiation | 1 | 2 | 2.46e-01 |
| GO:BP | GO:1904831 | positive regulation of aortic smooth muscle cell differentiation | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 2.46e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 1 | 6 | 2.46e-01 |
| GO:BP | GO:0019343 | cysteine biosynthetic process via cystathionine | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 16 | 787 | 2.46e-01 |
| GO:BP | GO:0018352 | protein-pyridoxal-5-phosphate linkage | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 1 | 10 | 2.46e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 1 | 4 | 2.46e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 3 | 88 | 2.46e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 1 | 6 | 2.46e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 2 | 14 | 2.46e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 16 | 717 | 2.46e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 2 | 36 | 2.46e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 1 | 5 | 2.46e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 24 | 2880 | 2.46e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 4 | 179 | 2.46e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 1 | 12 | 2.46e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 17 | 316 | 2.46e-01 |
| GO:BP | GO:0071772 | response to BMP | 4 | 131 | 2.47e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 4 | 131 | 2.47e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 2.47e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 1 | 10 | 2.47e-01 |
| GO:BP | GO:0022414 | reproductive process | 4 | 927 | 2.47e-01 |
| GO:BP | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process | 1 | 4 | 2.47e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 1 | 3 | 2.47e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 2.47e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 2.47e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 2.47e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 2.47e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 2.47e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 2 | 15 | 2.47e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 2 | 18 | 2.47e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 5 | 94 | 2.47e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 2.48e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 2 | 16 | 2.48e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 3 | 300 | 2.49e-01 |
| GO:BP | GO:0000003 | reproduction | 4 | 934 | 2.49e-01 |
| GO:BP | GO:0001568 | blood vessel development | 8 | 534 | 2.49e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 8 | 160 | 2.49e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 13 | 957 | 2.51e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 2.51e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 5 | 86 | 2.51e-01 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 2.51e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 7 | 130 | 2.51e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 1 | 3 | 2.52e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 3 | 45 | 2.52e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 1 | 6 | 2.52e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 2 | 45 | 2.52e-01 |
| GO:BP | GO:0001112 | DNA-templated transcription open complex formation | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0008584 | male gonad development | 1 | 99 | 2.52e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 2.52e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 5 | 91 | 2.52e-01 |
| GO:BP | GO:0001120 | protein-DNA complex remodeling | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 1 | 15 | 2.52e-01 |
| GO:BP | GO:0019080 | viral gene expression | 2 | 96 | 2.52e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 6 | 517 | 2.52e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 1 | 100 | 2.52e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 3 | 37 | 2.52e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 2.52e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 2 | 115 | 2.52e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 10 | 203 | 2.52e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 2 | 18 | 2.52e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 2.52e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 2.52e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 1 | 4 | 2.52e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 3 | 39 | 2.52e-01 |
| GO:BP | GO:0002572 | pro-T cell differentiation | 1 | 1 | 2.52e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 2.52e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 3 | 538 | 2.52e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 2.52e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 2 | 17 | 2.52e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 5 | 414 | 2.52e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 1 | 3 | 2.52e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 1 | 6 | 2.52e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 1 | 13 | 2.52e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 1 | 7 | 2.52e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 2 | 8 | 2.52e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1 | 72 | 2.52e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 2 | 143 | 2.52e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 2 | 30 | 2.52e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 1 | 6 | 2.52e-01 |
| GO:BP | GO:0006473 | protein acetylation | 9 | 190 | 2.52e-01 |
| GO:BP | GO:1905458 | positive regulation of lymphoid progenitor cell differentiation | 1 | 3 | 2.52e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 2 | 150 | 2.52e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 2 | 16 | 2.52e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 1 | 30 | 2.52e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 5 | 251 | 2.52e-01 |
| GO:BP | GO:0036075 | replacement ossification | 1 | 25 | 2.52e-01 |
| GO:BP | GO:0036451 | cap mRNA methylation | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 2 | 14 | 2.52e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 1 | 25 | 2.52e-01 |
| GO:BP | GO:0097309 | cap1 mRNA methylation | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0061642 | chemoattraction of axon | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 7 | 258 | 2.52e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 2 | 16 | 2.52e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 2.52e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 2.52e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 2.52e-01 |
| GO:BP | GO:0071464 | cellular response to hydrostatic pressure | 1 | 3 | 2.52e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 1 | 25 | 2.52e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 1 | 4 | 2.52e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 8 | 2.52e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 5 | 216 | 2.52e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 6 | 126 | 2.52e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 3 | 34 | 2.52e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 1 | 6 | 2.52e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 6 | 117 | 2.52e-01 |
| GO:BP | GO:0044029 | positive regulation of gene expression via CpG island demethylation | 1 | 3 | 2.52e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 1 | 16 | 2.52e-01 |
| GO:BP | GO:0048265 | response to pain | 1 | 17 | 2.52e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 1 | 10 | 2.52e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 1 | 10 | 2.52e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 1 | 25 | 2.52e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 2.52e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 3 | 46 | 2.52e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 1 | 6 | 2.52e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 2.52e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 1 | 24 | 2.53e-01 |
| GO:BP | GO:0032877 | positive regulation of DNA endoreduplication | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 11 | 249 | 2.53e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 2 | 28 | 2.53e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 134 | 2.53e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 2.53e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 3 | 32 | 2.53e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 4 | 65 | 2.53e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 3 | 21 | 2.53e-01 |
| GO:BP | GO:0060032 | notochord regression | 1 | 1 | 2.53e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 13 | 278 | 2.53e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 8 | 153 | 2.53e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 8 | 165 | 2.53e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 2 | 17 | 2.53e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 1 | 29 | 2.53e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 1 | 25 | 2.54e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 2.54e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 9 | 663 | 2.54e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 2.54e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 2 | 16 | 2.54e-01 |
| GO:BP | GO:0001824 | blastocyst development | 5 | 94 | 2.54e-01 |
| GO:BP | GO:0001562 | response to protozoan | 1 | 11 | 2.54e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 6 | 120 | 2.54e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 4 | 47 | 2.54e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 2.54e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 2 | 24 | 2.54e-01 |
| GO:BP | GO:0001944 | vasculature development | 8 | 559 | 2.54e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 1 | 9 | 2.54e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 2.54e-01 |
| GO:BP | GO:0060039 | pericardium development | 2 | 17 | 2.54e-01 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 5 | 2.54e-01 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 1 | 5 | 2.54e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 2 | 110 | 2.54e-01 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 1 | 3 | 2.54e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 2.54e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 2 | 26 | 2.54e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 2.54e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 2.54e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 3 | 556 | 2.54e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 12 | 2.54e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 2.54e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 1 | 6 | 2.55e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 20 | 5017 | 2.56e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 10 | 560 | 2.56e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 2.56e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 12 | 2.56e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 2.56e-01 |
| GO:BP | GO:0060174 | limb bud formation | 1 | 10 | 2.56e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 2.56e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 1 | 11 | 2.56e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 1 | 4 | 2.56e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 1 | 3 | 2.58e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 3 | 31 | 2.58e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 2 | 18 | 2.58e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 4 | 54 | 2.61e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 2 | 12 | 2.61e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 6 | 294 | 2.61e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 2.61e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 1 | 114 | 2.61e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 20 | 2.61e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 1 | 5 | 2.61e-01 |
| GO:BP | GO:1904582 | positive regulation of intracellular mRNA localization | 1 | 3 | 2.61e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 1 | 19 | 2.62e-01 |
| GO:BP | GO:0035441 | cell migration involved in vasculogenesis | 1 | 4 | 2.62e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 4 | 500 | 2.62e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 7 | 2.62e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 2.62e-01 |
| GO:BP | GO:0097676 | histone H3-K36 dimethylation | 1 | 4 | 2.62e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 2 | 96 | 2.62e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 1 | 25 | 2.63e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 1 | 25 | 2.63e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 1 | 51 | 2.64e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 1 | 5 | 2.65e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 1 | 5 | 2.65e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 3 | 78 | 2.65e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 59 | 2646 | 2.66e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 1 | 14 | 2.66e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 1 | 17 | 2.66e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 2.66e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 2 | 22 | 2.66e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 2 | 16 | 2.66e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 1 | 10 | 2.66e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 5 | 112 | 2.66e-01 |
| GO:BP | GO:0003174 | mitral valve development | 1 | 12 | 2.66e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 2.67e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 2.67e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 2.67e-01 |
| GO:BP | GO:0001843 | neural tube closure | 4 | 90 | 2.67e-01 |
| GO:BP | GO:0006364 | rRNA processing | 6 | 221 | 2.68e-01 |
| GO:BP | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process | 1 | 4 | 2.68e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 2.68e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 2 | 30 | 2.68e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 2 | 29 | 2.68e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 1 | 94 | 2.68e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 2.68e-01 |
| GO:BP | GO:0051098 | regulation of binding | 6 | 317 | 2.68e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 3 | 22 | 2.68e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 2.68e-01 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 1 | 6 | 2.68e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 2 | 18 | 2.68e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 3 | 35 | 2.68e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 2.68e-01 |
| GO:BP | GO:0060606 | tube closure | 4 | 91 | 2.68e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 2 | 43 | 2.68e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 6 | 151 | 2.68e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 9 | 132 | 2.68e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 2 | 67 | 2.68e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 2.68e-01 |
| GO:BP | GO:0006482 | protein demethylation | 2 | 27 | 2.68e-01 |
| GO:BP | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 1 | 25 | 2.68e-01 |
| GO:BP | GO:0006403 | RNA localization | 7 | 182 | 2.68e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 3 | 34 | 2.68e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 2 | 13 | 2.68e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0016573 | histone acetylation | 7 | 143 | 2.68e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 1 | 6 | 2.68e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 3 | 49 | 2.68e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 2 | 55 | 2.68e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 2 | 22 | 2.68e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 10 | 2.68e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 1 | 7 | 2.68e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 2 | 56 | 2.68e-01 |
| GO:BP | GO:0090398 | cellular senescence | 5 | 83 | 2.68e-01 |
| GO:BP | GO:0019985 | translesion synthesis | 2 | 24 | 2.68e-01 |
| GO:BP | GO:1900114 | positive regulation of histone H3-K9 trimethylation | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0019344 | cysteine biosynthetic process | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 2 | 26 | 2.68e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 2 | 27 | 2.68e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 2 | 46 | 2.68e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 2 | 37 | 2.68e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 2 | 27 | 2.68e-01 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 1 | 6 | 2.68e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 1 | 28 | 2.68e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 21 | 1199 | 2.68e-01 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 1 | 3 | 2.68e-01 |
| GO:BP | GO:0048625 | myoblast fate commitment | 1 | 4 | 2.68e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 2 | 54 | 2.68e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 2 | 80 | 2.68e-01 |
| GO:BP | GO:0042311 | vasodilation | 1 | 39 | 2.68e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 1 | 9 | 2.68e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 1 | 19 | 2.68e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 3 | 236 | 2.68e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 1 | 9 | 2.68e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 1 | 9 | 2.68e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 6 | 155 | 2.68e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 1 | 11 | 2.68e-01 |
| GO:BP | GO:0003128 | heart field specification | 1 | 16 | 2.69e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 1 | 13 | 2.69e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 1 | 16 | 2.69e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 1 | 34 | 2.69e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 1 | 30 | 2.69e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 1 | 4 | 2.69e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 3 | 35 | 2.69e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 1 | 35 | 2.70e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 2.70e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 2.71e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 1 | 10 | 2.71e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 4 | 2.72e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 2 | 104 | 2.72e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 2.72e-01 |
| GO:BP | GO:0008033 | tRNA processing | 1 | 128 | 2.72e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 1 | 35 | 2.72e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 1 | 20 | 2.73e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 32 | 2.74e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 1 | 5 | 2.77e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 1 | 14 | 2.77e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 2.77e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 2 | 185 | 2.77e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 1 | 15 | 2.79e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 1 | 21 | 2.79e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 1 | 97 | 2.79e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 2.79e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 3 | 631 | 2.79e-01 |
| GO:BP | GO:0021546 | rhombomere development | 1 | 4 | 2.80e-01 |
| GO:BP | GO:0090310 | negative regulation of DNA methylation-dependent heterochromatin formation | 1 | 2 | 2.80e-01 |
| GO:BP | GO:0002076 | osteoblast development | 2 | 12 | 2.80e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 2.80e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 6 | 161 | 2.80e-01 |
| GO:BP | GO:0007423 | sensory organ development | 4 | 396 | 2.80e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 1 | 5 | 2.80e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 1 | 7 | 2.80e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 1 | 6 | 2.80e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 1 | 27 | 2.80e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 5 | 123 | 2.80e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 1 | 5 | 2.80e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 1 | 4 | 2.81e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 4 | 86 | 2.82e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 1 | 100 | 2.84e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 1 | 11 | 2.84e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 2 | 41 | 2.84e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 31 | 898 | 2.84e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 2.85e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 4 | 61 | 2.85e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 2.85e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 8 | 185 | 2.85e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 8 | 227 | 2.85e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 1 | 24 | 2.85e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 2 | 20 | 2.85e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 1 | 17 | 2.85e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 1 | 8 | 2.85e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 2.85e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 2 | 50 | 2.85e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 4 | 220 | 2.85e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 1 | 7 | 2.85e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 2 | 151 | 2.85e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 2.85e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 1 | 42 | 2.85e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 7 | 148 | 2.85e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 1 | 37 | 2.85e-01 |
| GO:BP | GO:0008406 | gonad development | 1 | 158 | 2.85e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 4 | 494 | 2.85e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 1 | 16 | 2.85e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 6 | 172 | 2.85e-01 |
| GO:BP | GO:0021508 | floor plate formation | 1 | 3 | 2.85e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 1 | 5 | 2.85e-01 |
| GO:BP | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification | 1 | 3 | 2.85e-01 |
| GO:BP | GO:0021776 | smoothened signaling pathway involved in spinal cord motor neuron cell fate specification | 1 | 3 | 2.85e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 1 | 8 | 2.85e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 6 | 373 | 2.85e-01 |
| GO:BP | GO:0021965 | spinal cord ventral commissure morphogenesis | 1 | 2 | 2.85e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 4 | 427 | 2.85e-01 |
| GO:BP | GO:0009790 | embryo development | 34 | 875 | 2.85e-01 |
| GO:BP | GO:1904580 | regulation of intracellular mRNA localization | 1 | 4 | 2.85e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 1 | 19 | 2.85e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 1 | 30 | 2.85e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 2 | 26 | 2.85e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 1 | 30 | 2.85e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 1 | 8 | 2.85e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 3 | 63 | 2.85e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 2.85e-01 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 1 | 7 | 2.85e-01 |
| GO:BP | GO:0035329 | hippo signaling | 3 | 41 | 2.85e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 1 | 18 | 2.85e-01 |
| GO:BP | GO:2000653 | regulation of genetic imprinting | 1 | 4 | 2.85e-01 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 1 | 36 | 2.85e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 1 | 16 | 2.85e-01 |
| GO:BP | GO:0040011 | locomotion | 12 | 954 | 2.85e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 1 | 4 | 2.85e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 1 | 43 | 2.86e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 2.86e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 4 | 3600 | 2.86e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 2.86e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 1 | 136 | 2.86e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 2 | 23 | 2.86e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 6 | 424 | 2.86e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 1 | 40 | 2.86e-01 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 1 | 8 | 2.86e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 3 | 40 | 2.86e-01 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 1 | 8 | 2.86e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 1 | 7 | 2.86e-01 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 2 | 19 | 2.86e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 2.86e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 1 | 47 | 2.86e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 10 | 237 | 2.86e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 3 | 101 | 2.86e-01 |
| GO:BP | GO:0045905 | positive regulation of translational termination | 1 | 4 | 2.86e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 1 | 11 | 2.86e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 1 | 11 | 2.86e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 2.86e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 1 | 16 | 2.86e-01 |
| GO:BP | GO:0008219 | cell death | 17 | 1596 | 2.86e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 2.86e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 1 | 12 | 2.86e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 4 | 76 | 2.86e-01 |
| GO:BP | GO:0001525 | angiogenesis | 6 | 390 | 2.86e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 1 | 12 | 2.86e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 1 | 11 | 2.86e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 7 | 168 | 2.86e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 1 | 4 | 2.86e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 1 | 99 | 2.86e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 4 | 89 | 2.86e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 5 | 798 | 2.86e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 3 | 45 | 2.86e-01 |
| GO:BP | GO:0060429 | epithelium development | 36 | 850 | 2.86e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 2 | 14 | 2.86e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0048645 | animal organ formation | 4 | 51 | 2.86e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 2.86e-01 |
| GO:BP | GO:0031929 | TOR signaling | 2 | 115 | 2.86e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 2.86e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 2 | 28 | 2.86e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 7 | 169 | 2.86e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 1 | 30 | 2.86e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 2 | 29 | 2.86e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 9 | 194 | 2.86e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 1 | 7 | 2.86e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 2.86e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 1 | 11 | 2.86e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 1 | 6 | 2.86e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 8 | 393 | 2.86e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 7 | 150 | 2.86e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 6 | 147 | 2.86e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 3 | 188 | 2.86e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 5 | 89 | 2.86e-01 |
| GO:BP | GO:1905643 | positive regulation of DNA methylation | 1 | 2 | 2.86e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 1 | 49 | 2.86e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 1 | 25 | 2.86e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 6 | 407 | 2.86e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 1 | 54 | 2.86e-01 |
| GO:BP | GO:0036260 | RNA capping | 2 | 20 | 2.86e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 2.86e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 2.86e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 3 | 36 | 2.86e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 2 | 32 | 2.86e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 11 | 280 | 2.86e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 13 | 418 | 2.86e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 1 | 161 | 2.86e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 1 | 39 | 2.86e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 1 | 7 | 2.86e-01 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 2.86e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 1 | 13 | 2.87e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 3 | 41 | 2.87e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 2 | 16 | 2.87e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 11 | 272 | 2.87e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 1 | 5 | 2.87e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 9 | 267 | 2.87e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 2.88e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 2.88e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 2.88e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 2 | 45 | 2.89e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 1 | 20 | 2.89e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 2 | 29 | 2.89e-01 |
| GO:BP | GO:0048870 | cell motility | 14 | 1182 | 2.89e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 9 | 656 | 2.89e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 3 | 48 | 2.89e-01 |
| GO:BP | GO:0001759 | organ induction | 1 | 17 | 2.89e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 1 | 21 | 2.89e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 3 | 120 | 2.89e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 1 | 19 | 2.89e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 1 | 17 | 2.89e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 1 | 39 | 2.89e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 10 | 1107 | 2.90e-01 |
| GO:BP | GO:0001503 | ossification | 5 | 313 | 2.90e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 2.90e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 2.90e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 2.92e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 9 | 486 | 2.92e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 7 | 161 | 2.92e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 7 | 185 | 2.92e-01 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 2.92e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 2 | 83 | 2.92e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 2 | 104 | 2.92e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 2 | 29 | 2.92e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 2.92e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 2.92e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 2.92e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 2 | 34 | 2.92e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 2.92e-01 |
| GO:BP | GO:0099172 | presynapse organization | 1 | 42 | 2.92e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 2 | 28 | 2.92e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 1 | 3 | 2.92e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 1 | 15 | 2.92e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 2.92e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 2.92e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 1 | 8 | 2.92e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1 | 18 | 2.92e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 2.92e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 1 | 7 | 2.92e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 1 | 48 | 2.92e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 1 | 40 | 2.92e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 8 | 399 | 2.92e-01 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 1 | 14 | 2.93e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 4 | 67 | 2.94e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 2.94e-01 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 2 | 50 | 2.94e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 1 | 10 | 2.94e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 5 | 93 | 2.94e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 1 | 28 | 2.94e-01 |
| GO:BP | GO:0007431 | salivary gland development | 1 | 26 | 2.94e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 1 | 29 | 2.94e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 2.94e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 1 | 37 | 2.94e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 4 | 57 | 2.94e-01 |
| GO:BP | GO:0031529 | ruffle organization | 1 | 45 | 2.94e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 1 | 5 | 2.94e-01 |
| GO:BP | GO:0001654 | eye development | 3 | 272 | 2.94e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 1 | 6 | 2.94e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 2.94e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 1 | 17 | 2.94e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 3 | 46 | 2.94e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 2.94e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 4 | 75 | 2.94e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 4 | 236 | 2.94e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 2 | 73 | 2.94e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 2.94e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 2.94e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 2.94e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 8 | 179 | 2.94e-01 |
| GO:BP | GO:0150063 | visual system development | 3 | 273 | 2.94e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 5 | 110 | 2.94e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 1 | 39 | 2.94e-01 |
| GO:BP | GO:2000674 | regulation of type B pancreatic cell apoptotic process | 1 | 6 | 2.94e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 2.95e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 1 | 51 | 2.95e-01 |
| GO:BP | GO:0045837 | negative regulation of membrane potential | 1 | 9 | 2.96e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 3 | 52 | 2.96e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 2 | 33 | 2.96e-01 |
| GO:BP | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation | 1 | 6 | 2.96e-01 |
| GO:BP | GO:0010917 | negative regulation of mitochondrial membrane potential | 1 | 9 | 2.96e-01 |
| GO:BP | GO:0007548 | sex differentiation | 1 | 191 | 2.96e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 10 | 260 | 2.96e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 8 | 427 | 2.96e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2 | 56 | 2.96e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 1 | 16 | 2.96e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 1 | 7 | 2.96e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 1 | 7 | 2.96e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 1 | 17 | 2.96e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 3 | 48 | 2.96e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 6 | 421 | 2.97e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 6 | 131 | 2.98e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 1 | 23 | 2.99e-01 |
| GO:BP | GO:0048880 | sensory system development | 3 | 275 | 2.99e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 3.01e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 1 | 8 | 3.01e-01 |
| GO:BP | GO:0009092 | homoserine metabolic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 3 | 170 | 3.01e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 2 | 80 | 3.01e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 3.01e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 3 | 44 | 3.01e-01 |
| GO:BP | GO:0006541 | glutamine metabolic process | 2 | 24 | 3.01e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 2 | 139 | 3.01e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 1 | 186 | 3.01e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 4 | 150 | 3.01e-01 |
| GO:BP | GO:0021561 | facial nerve development | 1 | 8 | 3.01e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 2 | 75 | 3.01e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 7 | 185 | 3.01e-01 |
| GO:BP | GO:0019346 | transsulfuration | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 4 | 84 | 3.01e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 3.01e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 3.01e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 3 | 79 | 3.01e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 2 | 22 | 3.01e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 1 | 20 | 3.01e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 25 | 3.01e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 3.01e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 6 | 118 | 3.01e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 6 | 118 | 3.01e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 7 | 194 | 3.01e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 1 | 13 | 3.01e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 3.01e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 1 | 13 | 3.01e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 3.01e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 1 | 212 | 3.01e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 3.01e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 5 | 116 | 3.01e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 2 | 86 | 3.01e-01 |
| GO:BP | GO:0019230 | proprioception | 1 | 3 | 3.01e-01 |
| GO:BP | GO:0033505 | floor plate morphogenesis | 1 | 3 | 3.01e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 1 | 2 | 3.01e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 1 | 7 | 3.01e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 3 | 58 | 3.01e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 3.01e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 3.01e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 1 | 55 | 3.01e-01 |
| GO:BP | GO:0043543 | protein acylation | 6 | 228 | 3.01e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 3.01e-01 |
| GO:BP | GO:0061458 | reproductive system development | 1 | 215 | 3.01e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 3.01e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 1 | 17 | 3.01e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 1 | 6 | 3.02e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 1 | 6 | 3.02e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 5 | 513 | 3.02e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 1 | 55 | 3.02e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 2 | 28 | 3.02e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 1 | 56 | 3.02e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 1 | 47 | 3.02e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 2 | 31 | 3.02e-01 |
| GO:BP | GO:0006914 | autophagy | 5 | 513 | 3.02e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 17 | 418 | 3.02e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 3.02e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 12 | 268 | 3.02e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 12 | 268 | 3.02e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 1 | 7 | 3.02e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 3.02e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 3 | 95 | 3.02e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 4 | 59 | 3.02e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 2 | 37 | 3.02e-01 |
| GO:BP | GO:0031452 | negative regulation of heterochromatin formation | 1 | 4 | 3.02e-01 |
| GO:BP | GO:0120262 | negative regulation of heterochromatin organization | 1 | 4 | 3.02e-01 |
| GO:BP | GO:0000423 | mitophagy | 2 | 37 | 3.02e-01 |
| GO:BP | GO:1905268 | negative regulation of chromatin organization | 1 | 4 | 3.02e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 2 | 73 | 3.02e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 2 | 65 | 3.03e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 3.03e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 3.03e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 7 | 479 | 3.03e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 2 | 31 | 3.03e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 2 | 22 | 3.03e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 2 | 23 | 3.03e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 3.03e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 1 | 22 | 3.03e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 6 | 513 | 3.04e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 6 | 429 | 3.05e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 2 | 19 | 3.06e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 1 | 9 | 3.06e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 1 | 9 | 3.06e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 5 | 756 | 3.07e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 3.07e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 3.07e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 1 | 6 | 3.07e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 1 | 6 | 3.07e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 1 | 11 | 3.08e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 1 | 23 | 3.08e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 3 | 49 | 3.08e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 2 | 36 | 3.09e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 3.09e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 2 | 206 | 3.09e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 4 | 55 | 3.09e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 4 | 4048 | 3.09e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 1 | 20 | 3.09e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 2 | 21 | 3.09e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 1 | 8 | 3.09e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 5 | 485 | 3.09e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 1 | 5 | 3.09e-01 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 1 | 11 | 3.10e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 6 | 161 | 3.10e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 4 | 123 | 3.10e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 1 | 56 | 3.10e-01 |
| GO:BP | GO:0006915 | apoptotic process | 15 | 1443 | 3.10e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 1 | 27 | 3.10e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 3.10e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 1 | 21 | 3.10e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 1 | 28 | 3.11e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 1 | 64 | 3.11e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 3.11e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 2 | 57 | 3.11e-01 |
| GO:BP | GO:0035640 | exploration behavior | 1 | 21 | 3.11e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 1 | 45 | 3.11e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 1 | 265 | 3.11e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 1 | 57 | 3.11e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 10 | 276 | 3.11e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 2 | 167 | 3.11e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 2 | 167 | 3.11e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 3.11e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 1 | 16 | 3.11e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 6 | 442 | 3.11e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 3.11e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 2 | 52 | 3.11e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 3.11e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 3.11e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 1 | 61 | 3.11e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 3.11e-01 |
| GO:BP | GO:0042552 | myelination | 2 | 111 | 3.11e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 1 | 43 | 3.11e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 1 | 16 | 3.11e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 3.11e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 1 | 11 | 3.11e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 1 | 15 | 3.11e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.11e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 1 | 8 | 3.11e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 3.11e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 4 | 178 | 3.11e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 2 | 36 | 3.11e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 3.12e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 1 | 7 | 3.12e-01 |
| GO:BP | GO:0051049 | regulation of transport | 9 | 1267 | 3.12e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 1 | 7 | 3.12e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0048732 | gland development | 3 | 318 | 3.12e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 2 | 36 | 3.12e-01 |
| GO:BP | GO:0060914 | heart formation | 1 | 29 | 3.12e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 3.12e-01 |
| GO:BP | GO:0007498 | mesoderm development | 5 | 89 | 3.12e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 3.12e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 3.12e-01 |
| GO:BP | GO:0048565 | digestive tract development | 2 | 92 | 3.12e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 5 | 128 | 3.12e-01 |
| GO:BP | GO:0070475 | rRNA base methylation | 1 | 8 | 3.12e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 2 | 113 | 3.12e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 2 | 113 | 3.12e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 1 | 60 | 3.14e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 1 | 25 | 3.14e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 3.14e-01 |
| GO:BP | GO:0010387 | COP9 signalosome assembly | 1 | 3 | 3.14e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 3 | 56 | 3.14e-01 |
| GO:BP | GO:1905065 | positive regulation of vascular associated smooth muscle cell differentiation | 1 | 6 | 3.14e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 4 | 357 | 3.14e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 1 | 63 | 3.14e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 3.14e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 2 | 28 | 3.14e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 3.14e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 2 | 112 | 3.14e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 3 | 45 | 3.14e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 3 | 114 | 3.14e-01 |
| GO:BP | GO:0051867 | general adaptation syndrome, behavioral process | 1 | 3 | 3.15e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 1 | 57 | 3.15e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 2 | 33 | 3.15e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 2 | 113 | 3.15e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 3 | 77 | 3.15e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 3.15e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 3 | 31 | 3.15e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 2 | 67 | 3.15e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 2 | 44 | 3.15e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 3.15e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 1 | 80 | 3.15e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 29 | 3.15e-01 |
| GO:BP | GO:1900112 | regulation of histone H3-K9 trimethylation | 1 | 5 | 3.15e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 3.15e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 3.16e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 1 | 62 | 3.17e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 1 | 65 | 3.17e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 2 | 192 | 3.17e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 2 | 26 | 3.18e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 2 | 43 | 3.18e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 2 | 28 | 3.18e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 1 | 26 | 3.18e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 1 | 7 | 3.18e-01 |
| GO:BP | GO:0031128 | developmental induction | 1 | 26 | 3.18e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 1 | 48 | 3.18e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 3.19e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 1 | 172 | 3.19e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 1 | 5 | 3.19e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 1 | 10 | 3.19e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 1 | 14 | 3.20e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 3 | 47 | 3.20e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 1 | 211 | 3.20e-01 |
| GO:BP | GO:0035272 | exocrine system development | 1 | 34 | 3.20e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 1 | 41 | 3.20e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 1 | 52 | 3.20e-01 |
| GO:BP | GO:0006983 | ER overload response | 1 | 12 | 3.20e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 3.20e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 2 | 23 | 3.20e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 1 | 7 | 3.20e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 1 | 13 | 3.21e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 1 | 39 | 3.21e-01 |
| GO:BP | GO:1990787 | negative regulation of hh target transcription factor activity | 1 | 4 | 3.21e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 1 | 40 | 3.21e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 3.21e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 3 | 147 | 3.21e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 9 | 204 | 3.21e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 3.21e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 1 | 7 | 3.21e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 3.21e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 2 | 16 | 3.21e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 2 | 36 | 3.21e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 1 | 8 | 3.21e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 3 | 68 | 3.21e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 3.21e-01 |
| GO:BP | GO:0060047 | heart contraction | 3 | 205 | 3.22e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 4 | 76 | 3.23e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 1 | 85 | 3.23e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 5 | 448 | 3.23e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 4 | 93 | 3.23e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 2 | 36 | 3.23e-01 |
| GO:BP | GO:0097283 | keratinocyte apoptotic process | 1 | 5 | 3.23e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 2 | 50 | 3.23e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 2 | 73 | 3.23e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 2 | 73 | 3.23e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 1 | 57 | 3.23e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 2 | 29 | 3.23e-01 |
| GO:BP | GO:0032879 | regulation of localization | 9 | 1555 | 3.23e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 5 | 110 | 3.23e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 3.23e-01 |
| GO:BP | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 7 | 3.23e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 8 | 216 | 3.23e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 1 | 26 | 3.23e-01 |
| GO:BP | GO:1902172 | regulation of keratinocyte apoptotic process | 1 | 5 | 3.23e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 1 | 68 | 3.23e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 1 | 10 | 3.23e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 1 | 48 | 3.23e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 1 | 18 | 3.23e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 2 | 158 | 3.23e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 1 | 16 | 3.23e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 2 | 39 | 3.23e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 1 | 10 | 3.23e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 1 | 8 | 3.23e-01 |
| GO:BP | GO:0003097 | renal water transport | 1 | 6 | 3.23e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | 7 | 3.23e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 1 | 22 | 3.23e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 3 | 404 | 3.23e-01 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 1 | 13 | 3.23e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 1 | 30 | 3.23e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 3.23e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 2 | 40 | 3.23e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 1 | 4 | 3.23e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 1 | 23 | 3.23e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 10 | 3.23e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 1 | 33 | 3.23e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 3.23e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 5 | 3.23e-01 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 1 | 19 | 3.23e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 5 | 107 | 3.23e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 5 | 105 | 3.23e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 3.23e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 3.23e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 1 | 53 | 3.24e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 3 | 71 | 3.24e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 1 | 19 | 3.24e-01 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 3.24e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 2 | 30 | 3.25e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 4 | 71 | 3.25e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 1 | 12 | 3.25e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 4 | 94 | 3.25e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 7 | 207 | 3.26e-01 |
| GO:BP | GO:0012501 | programmed cell death | 15 | 1479 | 3.26e-01 |
| GO:BP | GO:0055123 | digestive system development | 2 | 100 | 3.26e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 5 | 123 | 3.27e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 4 | 70 | 3.27e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 1 | 8 | 3.27e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 1 | 179 | 3.27e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 10 | 679 | 3.27e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 13 | 1219 | 3.27e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 1 | 73 | 3.27e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 3.28e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 3.28e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 2 | 50 | 3.28e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 1 | 55 | 3.28e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 1 | 15 | 3.30e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 2 | 76 | 3.30e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 41 | 1111 | 3.30e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 2 | 121 | 3.30e-01 |
| GO:BP | GO:0006325 | chromatin organization | 20 | 551 | 3.30e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 6 | 147 | 3.30e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 2 | 20 | 3.30e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 3.31e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 1 | 32 | 3.31e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 1 | 51 | 3.31e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 4 | 95 | 3.31e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 1 | 98 | 3.31e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 3.31e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 27 | 1123 | 3.31e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 2 | 47 | 3.31e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 4 | 78 | 3.31e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 1 | 7 | 3.32e-01 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 1 | 14 | 3.32e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 6 | 382 | 3.32e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 2 | 27 | 3.32e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 8 | 667 | 3.32e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 1 | 2 | 3.32e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 6 | 144 | 3.32e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 1 | 7 | 3.32e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 1 | 17 | 3.32e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0097484 | dendrite extension | 1 | 27 | 3.32e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 2 | 83 | 3.32e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 1 | 20 | 3.32e-01 |
| GO:BP | GO:0009749 | response to glucose | 5 | 137 | 3.32e-01 |
| GO:BP | GO:0003015 | heart process | 3 | 216 | 3.33e-01 |
| GO:BP | GO:0001501 | skeletal system development | 8 | 377 | 3.33e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 1 | 10 | 3.33e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 2 | 25 | 3.33e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 2 | 25 | 3.33e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 3 | 59 | 3.33e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 1 | 56 | 3.33e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 1 | 82 | 3.33e-01 |
| GO:BP | GO:0051866 | general adaptation syndrome | 1 | 4 | 3.33e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 3 | 37 | 3.33e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 4 | 4455 | 3.35e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 1 | 41 | 3.35e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 3.35e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 1 | 127 | 3.35e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 3 | 185 | 3.35e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 2 | 165 | 3.35e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 3 | 428 | 3.35e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 1 | 57 | 3.35e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 4 | 81 | 3.35e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 3.35e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 1 | 59 | 3.36e-01 |
| GO:BP | GO:0043276 | anoikis | 2 | 32 | 3.36e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 3.36e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 2 | 30 | 3.36e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 2 | 40 | 3.36e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 1 | 7 | 3.36e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 3.36e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 1 | 61 | 3.36e-01 |
| GO:BP | GO:0060718 | chorionic trophoblast cell differentiation | 1 | 5 | 3.36e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 2 | 25 | 3.36e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 3.36e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 1 | 22 | 3.36e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 2 | 55 | 3.36e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 2 | 135 | 3.36e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 1 | 5 | 3.36e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 18 | 710 | 3.36e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 40 | 1088 | 3.36e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 1 | 7 | 3.36e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 3 | 50 | 3.36e-01 |
| GO:BP | GO:0001890 | placenta development | 6 | 117 | 3.36e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 2 | 90 | 3.36e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 1 | 5 | 3.37e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 1 | 4 | 3.37e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 1 | 12 | 3.37e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 1 | 18 | 3.37e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 1 | 5 | 3.37e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 2 | 200 | 3.37e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 3.37e-01 |
| GO:BP | GO:0007418 | ventral midline development | 1 | 4 | 3.37e-01 |
| GO:BP | GO:1902523 | positive regulation of protein K63-linked ubiquitination | 1 | 5 | 3.37e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 3.37e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 3.37e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 3.37e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 15 | 901 | 3.37e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 3.37e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 1 | 4 | 3.37e-01 |
| GO:BP | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning | 1 | 5 | 3.37e-01 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 2 | 36 | 3.37e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 1 | 55 | 3.37e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 4 | 114 | 3.38e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 4 | 114 | 3.38e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 2 | 199 | 3.38e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 7 | 893 | 3.38e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 1 | 70 | 3.39e-01 |
| GO:BP | GO:0042148 | strand invasion | 1 | 5 | 3.39e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 3.39e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 3.39e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 3 | 226 | 3.39e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 2 | 43 | 3.40e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 12 | 336 | 3.40e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 4 | 275 | 3.40e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 19 | 504 | 3.40e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 3.40e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 1 | 68 | 3.40e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 1 | 82 | 3.40e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 4 | 78 | 3.40e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 1 | 301 | 3.40e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 1 | 10 | 3.40e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 3.40e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 3.40e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 3.40e-01 |
| GO:BP | GO:0009746 | response to hexose | 5 | 138 | 3.40e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 8 | 441 | 3.40e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 3 | 98 | 3.42e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 1 | 14 | 3.42e-01 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 1 | 8 | 3.42e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 1 | 37 | 3.43e-01 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 1 | 7 | 3.44e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 1 | 58 | 3.44e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 4 | 4570 | 3.44e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 4 | 56 | 3.44e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 3 | 50 | 3.44e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 1 | 11 | 3.45e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 1 | 137 | 3.45e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 1 | 81 | 3.45e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 7 | 209 | 3.45e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 7 | 155 | 3.45e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 3.45e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 3.45e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 3.45e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 3.45e-01 |
| GO:BP | GO:0035601 | protein deacylation | 5 | 116 | 3.45e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 3.45e-01 |
| GO:BP | GO:0006096 | glycolytic process | 1 | 70 | 3.45e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 37 | 3.45e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 6 | 144 | 3.45e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 2 | 29 | 3.45e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 1 | 23 | 3.45e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 8 | 189 | 3.45e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 4 | 331 | 3.45e-01 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 1 | 8 | 3.45e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 1 | 5 | 3.45e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 3 | 59 | 3.46e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 1 | 8 | 3.46e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 1 | 8 | 3.47e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 4 | 4596 | 3.47e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 1 | 21 | 3.47e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 4 | 375 | 3.47e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 1 | 12 | 3.47e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 3.47e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 3.47e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 1 | 11 | 3.48e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 5 | 114 | 3.48e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 2 | 26 | 3.48e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 3.48e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 4 | 376 | 3.48e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 2 | 41 | 3.48e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 2 | 36 | 3.48e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 1 | 68 | 3.48e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 1 | 96 | 3.48e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 39 | 3.48e-01 |
| GO:BP | GO:0048864 | stem cell development | 4 | 76 | 3.48e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 2 | 25 | 3.48e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 1 | 23 | 3.49e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 3.49e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 2 | 132 | 3.49e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 2 | 30 | 3.49e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 3 | 77 | 3.49e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 6 | 381 | 3.49e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 3.49e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 3.49e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 1 | 85 | 3.49e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 1 | 22 | 3.49e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 2 | 30 | 3.49e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 6 | 120 | 3.49e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 1 | 7 | 3.49e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 1 | 16 | 3.49e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 1 | 23 | 3.49e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 3.49e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 1 | 98 | 3.49e-01 |
| GO:BP | GO:0034644 | cellular response to UV | 3 | 86 | 3.49e-01 |
| GO:BP | GO:0061053 | somite development | 4 | 63 | 3.50e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 5 | 400 | 3.50e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 1 | 40 | 3.50e-01 |
| GO:BP | GO:0072359 | circulatory system development | 31 | 884 | 3.50e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 1 | 67 | 3.50e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 1 | 12 | 3.50e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 11 | 320 | 3.50e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 2 | 37 | 3.50e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 1 | 13 | 3.50e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 16 | 452 | 3.50e-01 |
| GO:BP | GO:0030282 | bone mineralization | 2 | 82 | 3.50e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 3.51e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 8 | 3.51e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 8 | 3.51e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 1 | 21 | 3.51e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 2 | 242 | 3.51e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 3.51e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 3.51e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 3.51e-01 |
| GO:BP | GO:0070509 | calcium ion import | 1 | 37 | 3.52e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 2 | 378 | 3.52e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 1 | 17 | 3.52e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 1 | 14 | 3.52e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 4 | 796 | 3.52e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 3 | 121 | 3.52e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 2 | 41 | 3.53e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 1 | 7 | 3.54e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 3.54e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 2 | 39 | 3.54e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 5 | 152 | 3.54e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 1 | 31 | 3.55e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 3.55e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 1 | 28 | 3.55e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 1 | 44 | 3.55e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 1 | 8 | 3.55e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 5 | 142 | 3.55e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 5 | 132 | 3.55e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 4 | 100 | 3.55e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0060573 | cell fate specification involved in pattern specification | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 3.55e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 2 | 109 | 3.55e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 1 | 68 | 3.55e-01 |
| GO:BP | GO:0021521 | ventral spinal cord interneuron specification | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 1 | 7 | 3.55e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 3 | 51 | 3.55e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 50 | 1681 | 3.55e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 3.56e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 3.56e-01 |
| GO:BP | GO:0006974 | DNA damage response | 25 | 785 | 3.56e-01 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 1 | 9 | 3.56e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 2 | 51 | 3.56e-01 |
| GO:BP | GO:0098754 | detoxification | 1 | 91 | 3.56e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 2 | 31 | 3.56e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 5 | 119 | 3.56e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 2 | 133 | 3.56e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 10 | 615 | 3.56e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 2 | 39 | 3.56e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 3.56e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 2 | 215 | 3.57e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 1 | 25 | 3.57e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 5 | 122 | 3.57e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 2 | 186 | 3.57e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 3.58e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 1 | 34 | 3.58e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 1 | 9 | 3.58e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 1 | 65 | 3.58e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 6 | 153 | 3.58e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 1 | 9 | 3.58e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 1 | 6 | 3.58e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 1 | 9 | 3.58e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 3.58e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 6 | 159 | 3.58e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 6 | 213 | 3.58e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 2 | 34 | 3.58e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 3.58e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 1 | 7 | 3.58e-01 |
| GO:BP | GO:0030073 | insulin secretion | 5 | 141 | 3.58e-01 |
| GO:BP | GO:0042753 | positive regulation of circadian rhythm | 1 | 10 | 3.58e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 1 | 87 | 3.58e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 1 | 8 | 3.58e-01 |
| GO:BP | GO:1900363 | regulation of mRNA polyadenylation | 1 | 11 | 3.58e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 2 | 69 | 3.59e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 2 | 27 | 3.59e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 1 | 14 | 3.59e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 1 | 4 | 3.59e-01 |
| GO:BP | GO:0016049 | cell growth | 4 | 418 | 3.59e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 5 | 134 | 3.59e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 1 | 24 | 3.60e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 31 | 956 | 3.60e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 1 | 22 | 3.60e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 4 | 4747 | 3.60e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 1 | 12 | 3.60e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 2 | 258 | 3.60e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 3.60e-01 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 1 | 7 | 3.60e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 1 | 12 | 3.60e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 1 | 28 | 3.60e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 1 | 11 | 3.60e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 1 | 92 | 3.60e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 2 | 75 | 3.60e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 1 | 9 | 3.60e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 2 | 19 | 3.61e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 99 | 3.61e-01 |
| GO:BP | GO:0048536 | spleen development | 2 | 28 | 3.61e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 3 | 55 | 3.61e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 3 | 59 | 3.61e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 99 | 3.61e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 1 | 62 | 3.61e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 25 | 660 | 3.61e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 1 | 71 | 3.61e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 99 | 3.62e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 2 | 33 | 3.62e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 17 | 561 | 3.62e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 1 | 10 | 3.62e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 1 | 77 | 3.62e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 1 | 28 | 3.62e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 1 | 76 | 3.62e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 3.62e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 5 | 136 | 3.62e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 1 | 22 | 3.62e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 3.62e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 1 | 12 | 3.62e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 3.62e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 3 | 72 | 3.62e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 3.62e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 3 | 433 | 3.62e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 3.62e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 3.62e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 2 | 71 | 3.62e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 1 | 67 | 3.62e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 3.62e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 10 | 625 | 3.62e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 3.62e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 1 | 12 | 3.62e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 3.62e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 2 | 25 | 3.63e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 5 | 123 | 3.63e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 2 | 36 | 3.63e-01 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 18 | 3.63e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 2 | 41 | 3.63e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 25 | 3.63e-01 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 1 | 18 | 3.63e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 2 | 28 | 3.63e-01 |
| GO:BP | GO:0051642 | centrosome localization | 1 | 31 | 3.63e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 3.63e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 1 | 16 | 3.63e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 1 | 51 | 3.63e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 1 | 164 | 3.63e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 1 | 31 | 3.63e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 3 | 42 | 3.63e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 2 | 41 | 3.63e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 1 | 46 | 3.64e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 1 | 14 | 3.64e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 3.65e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 1 | 10 | 3.66e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 3.66e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 1 | 13 | 3.67e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 1 | 12 | 3.67e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 10 | 1278 | 3.69e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 3 | 160 | 3.69e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 1 | 34 | 3.69e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 15 | 3.69e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 11 | 440 | 3.69e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 1 | 7 | 3.69e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 1 | 7 | 3.69e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 7 | 288 | 3.69e-01 |
| GO:BP | GO:0042633 | hair cycle | 2 | 78 | 3.70e-01 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 1 | 19 | 3.70e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 1 | 10 | 3.70e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 2 | 30 | 3.70e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 2 | 58 | 3.70e-01 |
| GO:BP | GO:0016358 | dendrite development | 3 | 202 | 3.70e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 4 | 4946 | 3.70e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 3 | 59 | 3.70e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 1 | 10 | 3.70e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 1 | 81 | 3.70e-01 |
| GO:BP | GO:0061525 | hindgut development | 1 | 5 | 3.70e-01 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 1 | 19 | 3.70e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 5 | 116 | 3.70e-01 |
| GO:BP | GO:0042303 | molting cycle | 2 | 78 | 3.70e-01 |
| GO:BP | GO:0042063 | gliogenesis | 3 | 235 | 3.70e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 1 | 10 | 3.70e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 5 | 78 | 3.70e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 1 | 16 | 3.70e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 3.70e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 5 | 1167 | 3.70e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 2 | 30 | 3.70e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 2 | 29 | 3.70e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 1 | 9 | 3.70e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 1 | 7 | 3.70e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 2 | 57 | 3.70e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 36 | 3.70e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 3 | 69 | 3.70e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 2 | 30 | 3.71e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 3.71e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 1 | 52 | 3.71e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 1 | 44 | 3.71e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 2 | 35 | 3.71e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 3.71e-01 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 1 | 14 | 3.71e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 1 | 14 | 3.71e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 4 | 425 | 3.71e-01 |
| GO:BP | GO:0003016 | respiratory system process | 2 | 30 | 3.71e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 2 | 421 | 3.71e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 11 | 706 | 3.71e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 1 | 74 | 3.72e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 15 | 3.72e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 2 | 20 | 3.72e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2 | 26 | 3.72e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 1 | 13 | 3.72e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 3 | 64 | 3.72e-01 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 1 | 8 | 3.72e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 3 | 45 | 3.73e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 10 | 637 | 3.73e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 3 | 108 | 3.73e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 1 | 18 | 3.73e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 27 | 3.73e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 1 | 27 | 3.73e-01 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 1 | 11 | 3.73e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 1 | 36 | 3.73e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 103 | 3.73e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 3 | 57 | 3.73e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 2 | 395 | 3.73e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 1 | 16 | 3.75e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 3.75e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 1 | 50 | 3.75e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 3 | 57 | 3.75e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 1 | 107 | 3.75e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 3 | 53 | 3.75e-01 |
| GO:BP | GO:0090659 | walking behavior | 2 | 25 | 3.75e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 3.75e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 15 | 438 | 3.76e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 1 | 281 | 3.76e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 1 | 39 | 3.76e-01 |
| GO:BP | GO:0090504 | epiboly | 2 | 31 | 3.76e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 1 | 15 | 3.76e-01 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 1 | 21 | 3.76e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 3.76e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 1 | 13 | 3.76e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 8 | 206 | 3.76e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 2 | 155 | 3.77e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 4 | 123 | 3.77e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 1 | 93 | 3.77e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 5 | 119 | 3.77e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 3.77e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 2 | 41 | 3.80e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 1 | 8 | 3.80e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 3 | 69 | 3.80e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 4 | 97 | 3.80e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 2 | 292 | 3.80e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 7 | 157 | 3.81e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 8 | 204 | 3.81e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 1 | 16 | 3.82e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 1 | 16 | 3.82e-01 |
| GO:BP | GO:0051152 | positive regulation of smooth muscle cell differentiation | 1 | 10 | 3.82e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 3 | 834 | 3.82e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 1 | 18 | 3.82e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 7 | 229 | 3.82e-01 |
| GO:BP | GO:0043201 | response to leucine | 1 | 13 | 3.83e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 3.83e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 3.83e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 6 | 157 | 3.85e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 3.85e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 2 | 42 | 3.85e-01 |
| GO:BP | GO:0060717 | chorion development | 1 | 8 | 3.85e-01 |
| GO:BP | GO:0071472 | cellular response to salt stress | 1 | 11 | 3.85e-01 |
| GO:BP | GO:0009306 | protein secretion | 2 | 262 | 3.85e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 3.85e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 2 | 262 | 3.85e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 1 | 22 | 3.85e-01 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 1 | 11 | 3.85e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 3.85e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 1 | 12 | 3.85e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 3.85e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 3 | 120 | 3.85e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 2 | 63 | 3.85e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 3.85e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 3.85e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 1 | 9 | 3.85e-01 |
| GO:BP | GO:0042116 | macrophage activation | 2 | 57 | 3.85e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 2 | 149 | 3.85e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 5 | 270 | 3.85e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 2 | 27 | 3.85e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 3.85e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 1 | 92 | 3.85e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 3.85e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 3 | 70 | 3.85e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 3.85e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 3.85e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 1 | 20 | 3.85e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 4 | 881 | 3.85e-01 |
| GO:BP | GO:0031016 | pancreas development | 2 | 53 | 3.85e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 3.85e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 2 | 44 | 3.85e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 2 | 246 | 3.85e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 25 | 678 | 3.85e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 3.85e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 3.85e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 2 | 151 | 3.86e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 2 | 30 | 3.86e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 1 | 9 | 3.86e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 1 | 19 | 3.86e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 1 | 47 | 3.86e-01 |
| GO:BP | GO:0030168 | platelet activation | 5 | 91 | 3.86e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 1 | 7 | 3.86e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 2 | 108 | 3.86e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 3 | 302 | 3.86e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 9 | 870 | 3.86e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 4 | 256 | 3.86e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 8 | 197 | 3.86e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 7 | 3.86e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 3.86e-01 |
| GO:BP | GO:0055057 | neuroblast division | 1 | 12 | 3.86e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 2 | 36 | 3.86e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 3.87e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 5 | 1028 | 3.87e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 1 | 15 | 3.87e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 15 | 504 | 3.87e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 27 | 3.87e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 1 | 110 | 3.87e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 2 | 43 | 3.87e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 3.87e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 1 | 70 | 3.87e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 2 | 156 | 3.87e-01 |
| GO:BP | GO:0060840 | artery development | 3 | 83 | 3.87e-01 |
| GO:BP | GO:1903350 | response to dopamine | 1 | 48 | 3.87e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 2 | 266 | 3.87e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 3 | 88 | 3.87e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 12 | 3.87e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 3 | 149 | 3.87e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 2 | 40 | 3.87e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 3 | 69 | 3.87e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 1 | 14 | 3.87e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 17 | 3.87e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 2 | 151 | 3.87e-01 |
| GO:BP | GO:0090208 | positive regulation of triglyceride metabolic process | 1 | 18 | 3.87e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 3.87e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 3.87e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 1 | 13 | 3.87e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 3 | 138 | 3.88e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 3.88e-01 |
| GO:BP | GO:0007416 | synapse assembly | 2 | 152 | 3.89e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 2 | 185 | 3.89e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 1 | 17 | 3.90e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 5 | 123 | 3.90e-01 |
| GO:BP | GO:0006417 | regulation of translation | 10 | 370 | 3.90e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 1 | 44 | 3.90e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 3.90e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 1 | 12 | 3.90e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 4 | 99 | 3.90e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 1 | 20 | 3.90e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 6 | 406 | 3.91e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 4 | 103 | 3.91e-01 |
| GO:BP | GO:0046688 | response to copper ion | 1 | 29 | 3.91e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 1 | 11 | 3.91e-01 |
| GO:BP | GO:0010467 | gene expression | 4 | 4587 | 3.91e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 3.91e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 3 | 72 | 3.91e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 2 | 126 | 3.91e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 2 | 43 | 3.91e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 3.91e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 4 | 282 | 3.91e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 3.93e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 12 | 1137 | 3.93e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 2 | 48 | 3.93e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 3.94e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 11 | 307 | 3.95e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 2 | 42 | 3.95e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 1 | 10 | 3.95e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 1 | 12 | 3.95e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 1 | 18 | 3.96e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 1 | 11 | 3.96e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 1 | 13 | 3.97e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 1 | 36 | 3.97e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 3.97e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 4 | 386 | 3.97e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 3.98e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 2 | 62 | 3.98e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 2 | 67 | 3.99e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 1 | 11 | 3.99e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 3.99e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 1 | 30 | 3.99e-01 |
| GO:BP | GO:0007399 | nervous system development | 7 | 1904 | 3.99e-01 |
| GO:BP | GO:0007492 | endoderm development | 3 | 65 | 3.99e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 3.99e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 2 | 50 | 3.99e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 4 | 122 | 3.99e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 3.99e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 1 | 15 | 3.99e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 1 | 15 | 3.99e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 1 | 28 | 3.99e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 4 | 89 | 3.99e-01 |
| GO:BP | GO:0050890 | cognition | 3 | 227 | 3.99e-01 |
| GO:BP | GO:0050821 | protein stabilization | 3 | 191 | 3.99e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 1 | 10 | 3.99e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 1 | 12 | 3.99e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 1 | 32 | 3.99e-01 |
| GO:BP | GO:0001867 | complement activation, lectin pathway | 1 | 5 | 3.99e-01 |
| GO:BP | GO:0016236 | macroautophagy | 2 | 308 | 3.99e-01 |
| GO:BP | GO:0009411 | response to UV | 4 | 141 | 3.99e-01 |
| GO:BP | GO:0042594 | response to starvation | 5 | 178 | 3.99e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 1 | 38 | 3.99e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 3.99e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 1 | 23 | 4.00e-01 |
| GO:BP | GO:0016032 | viral process | 3 | 344 | 4.00e-01 |
| GO:BP | GO:0006414 | translational elongation | 1 | 65 | 4.00e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 4.01e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 4.01e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 1 | 78 | 4.01e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 4.02e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 4.02e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 3 | 65 | 4.02e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 1 | 17 | 4.03e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 1 | 16 | 4.04e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 1 | 50 | 4.04e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 1 | 103 | 4.04e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 4.04e-01 |
| GO:BP | GO:0008544 | epidermis development | 5 | 210 | 4.04e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 4.04e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 2 | 168 | 4.05e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 4.05e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 3 | 91 | 4.05e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 3 | 134 | 4.05e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 1 | 28 | 4.05e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 4.05e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 3 | 73 | 4.05e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 1 | 37 | 4.05e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 4.05e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 3 | 69 | 4.05e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 1 | 14 | 4.05e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 1 | 53 | 4.05e-01 |
| GO:BP | GO:0000338 | protein deneddylation | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 1 | 44 | 4.05e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 3 | 97 | 4.05e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 79 | 4.05e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 1 | 15 | 4.05e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 1 | 40 | 4.05e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 1 | 31 | 4.05e-01 |
| GO:BP | GO:1990840 | response to lectin | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0001756 | somitogenesis | 3 | 48 | 4.05e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 1 | 17 | 4.05e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0060712 | spongiotrophoblast layer development | 1 | 11 | 4.05e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 4.05e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 1 | 15 | 4.05e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 1 | 15 | 4.05e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 1 | 23 | 4.05e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 1 | 16 | 4.06e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 4.06e-01 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 1 | 8 | 4.06e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 1 | 86 | 4.06e-01 |
| GO:BP | GO:0061912 | selective autophagy | 1 | 90 | 4.06e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 1 | 126 | 4.07e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 4.07e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 1 | 37 | 4.07e-01 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 28 | 4.07e-01 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 1 | 11 | 4.07e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 4.08e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 1 | 49 | 4.08e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 4.08e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 1 | 22 | 4.08e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 2 | 38 | 4.08e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 4.08e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 4.08e-01 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 1 | 8 | 4.08e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 89 | 4.08e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 4.08e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 4.08e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 3 | 152 | 4.08e-01 |
| GO:BP | GO:0008015 | blood circulation | 2 | 374 | 4.08e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 1 | 10 | 4.09e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 1 | 92 | 4.09e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 1 | 127 | 4.09e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 4 | 127 | 4.09e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 1 | 92 | 4.09e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 1 | 60 | 4.10e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 1 | 60 | 4.10e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 1 | 45 | 4.10e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 4.10e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 1 | 46 | 4.10e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 1 | 173 | 4.10e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 1 | 36 | 4.11e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 3 | 75 | 4.11e-01 |
| GO:BP | GO:0060173 | limb development | 4 | 142 | 4.11e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 5 | 358 | 4.11e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 3 | 160 | 4.11e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 4 | 104 | 4.11e-01 |
| GO:BP | GO:0048736 | appendage development | 4 | 142 | 4.11e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 31 | 4.11e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 5 | 1380 | 4.11e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 3 | 82 | 4.11e-01 |
| GO:BP | GO:0097502 | mannosylation | 1 | 32 | 4.11e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 2 | 49 | 4.11e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 4.11e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 1 | 11 | 4.11e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 3 | 74 | 4.11e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 4 | 317 | 4.11e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 1 | 10 | 4.11e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 4 | 105 | 4.11e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 1 | 217 | 4.13e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 15 | 4.13e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 2 | 40 | 4.13e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 4.14e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 1 | 42 | 4.14e-01 |
| GO:BP | GO:0033089 | positive regulation of T cell differentiation in thymus | 1 | 5 | 4.14e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 2 | 115 | 4.14e-01 |
| GO:BP | GO:0033504 | floor plate development | 1 | 8 | 4.14e-01 |
| GO:BP | GO:0021520 | spinal cord motor neuron cell fate specification | 1 | 5 | 4.14e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 1 | 62 | 4.14e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 2 | 115 | 4.14e-01 |
| GO:BP | GO:0060581 | cell fate commitment involved in pattern specification | 1 | 3 | 4.14e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 1 | 126 | 4.14e-01 |
| GO:BP | GO:0071867 | response to monoamine | 1 | 62 | 4.14e-01 |
| GO:BP | GO:0060579 | ventral spinal cord interneuron fate commitment | 1 | 3 | 4.14e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 1 | 17 | 4.14e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 1 | 10 | 4.14e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 1 | 21 | 4.14e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 1 | 14 | 4.14e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 3 | 95 | 4.14e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 2 | 73 | 4.14e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 4.14e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 4.14e-01 |
| GO:BP | GO:0021537 | telencephalon development | 3 | 201 | 4.15e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 1 | 21 | 4.16e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 1 | 134 | 4.17e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 4.17e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 1 | 87 | 4.17e-01 |
| GO:BP | GO:0010038 | response to metal ion | 2 | 264 | 4.17e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 4 | 106 | 4.17e-01 |
| GO:BP | GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | 1 | 9 | 4.17e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 2 | 31 | 4.17e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 4.17e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 3 | 101 | 4.17e-01 |
| GO:BP | GO:0070997 | neuron death | 2 | 286 | 4.17e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 1 | 16 | 4.17e-01 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 1 | 27 | 4.17e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 1 | 49 | 4.17e-01 |
| GO:BP | GO:0035188 | hatching | 1 | 23 | 4.17e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 1 | 23 | 4.17e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 1 | 23 | 4.17e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 4.17e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 3 | 86 | 4.18e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 1 | 51 | 4.19e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 8 | 279 | 4.19e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 3 | 221 | 4.19e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 2 | 29 | 4.21e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 5 | 318 | 4.21e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 4.21e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 1 | 33 | 4.21e-01 |
| GO:BP | GO:0035904 | aorta development | 1 | 50 | 4.21e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 1 | 151 | 4.21e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 1 | 16 | 4.21e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 4.21e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 5 | 146 | 4.21e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 1 | 18 | 4.21e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 1 | 21 | 4.21e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 10 | 295 | 4.21e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 4.22e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 4.22e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 7 | 210 | 4.22e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 4.22e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 35 | 4.22e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 4.24e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 1 | 27 | 4.24e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 3 | 101 | 4.24e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 1 | 10 | 4.24e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 1 | 17 | 4.24e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 2 | 86 | 4.24e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 3 | 69 | 4.24e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 1 | 27 | 4.24e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 2 | 41 | 4.24e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 3 | 67 | 4.24e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 4.25e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 1 | 102 | 4.26e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 1 | 35 | 4.26e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 2 | 53 | 4.26e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 1 | 72 | 4.26e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 4.26e-01 |
| GO:BP | GO:0036211 | protein modification process | 54 | 2901 | 4.26e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 1 | 11 | 4.26e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 22 | 4.26e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 4.26e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 2 | 36 | 4.27e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 4.27e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 6 | 830 | 4.27e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 4.27e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 1 | 61 | 4.28e-01 |
| GO:BP | GO:0070988 | demethylation | 2 | 52 | 4.29e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 1 | 65 | 4.29e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 1 | 44 | 4.29e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 9 | 273 | 4.29e-01 |
| GO:BP | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 12 | 4.29e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 1 | 13 | 4.29e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 4.29e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 14 | 4.29e-01 |
| GO:BP | GO:0031167 | rRNA methylation | 1 | 24 | 4.29e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 3 | 121 | 4.29e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 1 | 57 | 4.29e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 2 | 265 | 4.29e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 1 | 10 | 4.29e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 2 | 58 | 4.29e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 2 | 76 | 4.29e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 4.29e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 1 | 11 | 4.29e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 4.29e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 2 | 136 | 4.30e-01 |
| GO:BP | GO:0043583 | ear development | 3 | 153 | 4.30e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 1 | 47 | 4.31e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 1 | 17 | 4.31e-01 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 4.32e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 1 | 8 | 4.32e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 1 | 33 | 4.32e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 1 | 19 | 4.32e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 4.32e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 4 | 317 | 4.33e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 4.33e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 2 | 318 | 4.33e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 22 | 2035 | 4.33e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 14 | 719 | 4.33e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1 | 47 | 4.33e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 2 | 111 | 4.33e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 2 | 133 | 4.33e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 4.33e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 4.33e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 156 | 4.34e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 5 | 165 | 4.34e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 1 | 19 | 4.34e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 4.34e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 2 | 55 | 4.34e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 1 | 14 | 4.35e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 1 | 42 | 4.35e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 2 | 184 | 4.35e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 2 | 101 | 4.35e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 3 | 81 | 4.36e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 4 | 993 | 4.36e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 4 | 994 | 4.37e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 4.37e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 4.37e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 1 | 23 | 4.37e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 1 | 15 | 4.37e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 1 | 28 | 4.37e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 3 | 60 | 4.37e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 1 | 21 | 4.38e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 4.38e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 1 | 23 | 4.38e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 7 | 691 | 4.38e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 4.39e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 1 | 18 | 4.39e-01 |
| GO:BP | GO:0016310 | phosphorylation | 5 | 1372 | 4.39e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 2 | 43 | 4.39e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 4.39e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 1 | 113 | 4.40e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 4.40e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 4.41e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 3 | 523 | 4.41e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 1 | 20 | 4.41e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 1 | 29 | 4.41e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 1 | 14 | 4.41e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 1 | 166 | 4.41e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 1 | 22 | 4.41e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 1 | 11 | 4.41e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 1 | 11 | 4.41e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 1 | 10 | 4.41e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 1 | 13 | 4.41e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 1 | 21 | 4.41e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 4.41e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 5 | 140 | 4.41e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 3 | 199 | 4.41e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 3 | 77 | 4.41e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 4.41e-01 |
| GO:BP | GO:0060419 | heart growth | 3 | 72 | 4.41e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 4.41e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 172 | 4.41e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 1 | 165 | 4.41e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 1 | 14 | 4.41e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 3 | 71 | 4.41e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 2 | 187 | 4.41e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 4.41e-01 |
| GO:BP | GO:0006821 | chloride transport | 1 | 58 | 4.41e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 172 | 4.41e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 1 | 15 | 4.41e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 1 | 40 | 4.41e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 4.41e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 4.41e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 5 | 166 | 4.41e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 8 | 228 | 4.41e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 4.41e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 1 | 50 | 4.41e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 4.41e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 4.41e-01 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 1 | 5 | 4.41e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 1 | 167 | 4.41e-01 |
| GO:BP | GO:0007507 | heart development | 15 | 506 | 4.41e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 3 | 81 | 4.41e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 6 | 168 | 4.41e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 1 | 12 | 4.42e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 6 | 539 | 4.42e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 1 | 71 | 4.42e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 1 | 160 | 4.42e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 2 | 39 | 4.42e-01 |
| GO:BP | GO:0043038 | amino acid activation | 2 | 44 | 4.42e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 9 | 249 | 4.43e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 1 | 16 | 4.43e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 1 | 7 | 4.43e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 1 | 18 | 4.43e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 1 | 11 | 4.43e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 198 | 4.43e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 1 | 27 | 4.43e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 1 | 163 | 4.43e-01 |
| GO:BP | GO:0006415 | translational termination | 1 | 17 | 4.43e-01 |
| GO:BP | GO:0040008 | regulation of growth | 4 | 501 | 4.44e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 1 | 30 | 4.44e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 4.44e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 4 | 142 | 4.45e-01 |
| GO:BP | GO:0016577 | histone demethylation | 1 | 21 | 4.45e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 2 | 52 | 4.46e-01 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 1 | 60 | 4.46e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 4 | 140 | 4.46e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 1 | 116 | 4.47e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 4 | 372 | 4.48e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 1 | 24 | 4.49e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 9 | 326 | 4.49e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 1 | 166 | 4.49e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 4.50e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 1 | 99 | 4.50e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 1 | 13 | 4.50e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 17 | 1835 | 4.50e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 1 | 62 | 4.51e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 1 | 21 | 4.52e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 1 | 26 | 4.52e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 4.52e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 4.52e-01 |
| GO:BP | GO:0002790 | peptide secretion | 5 | 170 | 4.53e-01 |
| GO:BP | GO:0030879 | mammary gland development | 2 | 106 | 4.53e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 1 | 14 | 4.53e-01 |
| GO:BP | GO:0003013 | circulatory system process | 2 | 447 | 4.53e-01 |
| GO:BP | GO:0006833 | water transport | 1 | 10 | 4.53e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 4 | 140 | 4.53e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 1 | 50 | 4.53e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 13 | 4.53e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 1 | 58 | 4.53e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 3 | 79 | 4.53e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 4.54e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 2 | 60 | 4.54e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 1 | 66 | 4.55e-01 |
| GO:BP | GO:0060348 | bone development | 1 | 170 | 4.55e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 1 | 13 | 4.55e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 90 | 4.55e-01 |
| GO:BP | GO:0032506 | cytokinetic process | 1 | 39 | 4.56e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 2 | 48 | 4.56e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 5 | 378 | 4.56e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 1 | 14 | 4.56e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 1 | 15 | 4.56e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 2 | 105 | 4.56e-01 |
| GO:BP | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 14 | 4.56e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 14 | 1133 | 4.56e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 2 | 439 | 4.56e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 1 | 27 | 4.56e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 4.56e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 1 | 17 | 4.56e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 1 | 22 | 4.56e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 4.56e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 2 | 129 | 4.57e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 1 | 91 | 4.58e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 1 | 21 | 4.58e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 2 | 103 | 4.58e-01 |
| GO:BP | GO:0006281 | DNA repair | 16 | 534 | 4.58e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 1 | 22 | 4.60e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 2 | 38 | 4.61e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 1 | 90 | 4.62e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 4.63e-01 |
| GO:BP | GO:0007141 | male meiosis I | 1 | 15 | 4.63e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 1 | 14 | 4.64e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 4.64e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 1 | 29 | 4.65e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 1 | 30 | 4.65e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 1 | 35 | 4.65e-01 |
| GO:BP | GO:0051235 | maintenance of location | 4 | 260 | 4.65e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 2 | 87 | 4.65e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 1 | 31 | 4.65e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 2 | 52 | 4.65e-01 |
| GO:BP | GO:0007530 | sex determination | 1 | 12 | 4.65e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 5 | 168 | 4.65e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 1 | 40 | 4.65e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 4 | 84 | 4.65e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 4.65e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 1 | 13 | 4.66e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 1 | 28 | 4.66e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 12 | 1101 | 4.67e-01 |
| GO:BP | GO:0009611 | response to wounding | 5 | 415 | 4.67e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 3 | 71 | 4.67e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 8 | 1061 | 4.67e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 4.67e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 4 | 156 | 4.67e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 1 | 16 | 4.67e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 1 | 180 | 4.68e-01 |
| GO:BP | GO:0003158 | endothelium development | 3 | 104 | 4.68e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 1 | 20 | 4.69e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 1 | 31 | 4.69e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 3 | 90 | 4.69e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 3 | 604 | 4.69e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 1 | 16 | 4.70e-01 |
| GO:BP | GO:0007596 | blood coagulation | 3 | 150 | 4.70e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 3 | 100 | 4.70e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 3 | 232 | 4.70e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 36 | 1075 | 4.70e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 20 | 868 | 4.70e-01 |
| GO:BP | GO:0007030 | Golgi organization | 1 | 137 | 4.70e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 4 | 104 | 4.70e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 1 | 122 | 4.70e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 4.70e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 4.71e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 2 | 72 | 4.71e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 4.71e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 422 | 4.71e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 2 | 147 | 4.71e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 5 | 140 | 4.72e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 4.72e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 4.72e-01 |
| GO:BP | GO:0080009 | mRNA methylation | 1 | 16 | 4.73e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 1 | 41 | 4.74e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 7 | 340 | 4.74e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 3 | 172 | 4.74e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 3 | 84 | 4.74e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 1 | 27 | 4.75e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 1 | 21 | 4.75e-01 |
| GO:BP | GO:0042060 | wound healing | 4 | 320 | 4.75e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 4.75e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 2 | 310 | 4.75e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 1 | 23 | 4.75e-01 |
| GO:BP | GO:0033993 | response to lipid | 2 | 592 | 4.75e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 1 | 19 | 4.75e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 1 | 15 | 4.75e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 1 | 31 | 4.75e-01 |
| GO:BP | GO:0044057 | regulation of system process | 4 | 405 | 4.75e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 4.77e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 1 | 20 | 4.77e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 2 | 52 | 4.77e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 11 | 2293 | 4.78e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 2 | 75 | 4.79e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 5 | 214 | 4.79e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 4.79e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 2 | 40 | 4.79e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 4 | 260 | 4.79e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 1 | 22 | 4.79e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 5 | 742 | 4.80e-01 |
| GO:BP | GO:0050817 | coagulation | 3 | 153 | 4.80e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 2 | 111 | 4.80e-01 |
| GO:BP | GO:0061024 | membrane organization | 3 | 671 | 4.80e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 4.80e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 6 | 187 | 4.81e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 29 | 1006 | 4.81e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0007599 | hemostasis | 3 | 153 | 4.82e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 3 | 137 | 4.82e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 5 | 387 | 4.82e-01 |
| GO:BP | GO:0051170 | import into nucleus | 2 | 152 | 4.82e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 5 | 228 | 4.82e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 3 | 85 | 4.82e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 1 | 26 | 4.82e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 1 | 12 | 4.82e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 4.83e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 1 | 37 | 4.83e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 1 | 14 | 4.83e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 4.83e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 4.83e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 1 | 16 | 4.83e-01 |
| GO:BP | GO:0061448 | connective tissue development | 8 | 208 | 4.83e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 2 | 54 | 4.83e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 4.83e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 1 | 38 | 4.83e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 1 | 68 | 4.83e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 1 | 69 | 4.83e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 29 | 4.83e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 1 | 18 | 4.84e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 1 | 23 | 4.84e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 1 | 37 | 4.85e-01 |
| GO:BP | GO:1990868 | response to chemokine | 1 | 37 | 4.85e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 1 | 11 | 4.85e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 1 | 16 | 4.85e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 1 | 70 | 4.85e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 1 | 45 | 4.85e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 1 | 8 | 4.85e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 1 | 29 | 4.85e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 4.85e-01 |
| GO:BP | GO:0016197 | endosomal transport | 1 | 231 | 4.85e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 1 | 34 | 4.85e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 1 | 23 | 4.86e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 4.86e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 1 | 76 | 4.86e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 2 | 184 | 4.87e-01 |
| GO:BP | GO:0051668 | localization within membrane | 3 | 605 | 4.87e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 1 | 27 | 4.87e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 2 | 222 | 4.88e-01 |
| GO:BP | GO:0030900 | forebrain development | 7 | 281 | 4.88e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 1 | 23 | 4.88e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 2 | 154 | 4.88e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 17 | 763 | 4.88e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 4.89e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 4.90e-01 |
| GO:BP | GO:0017145 | stem cell division | 1 | 26 | 4.91e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 4.91e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 16 | 694 | 4.92e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 3 | 91 | 4.92e-01 |
| GO:BP | GO:0046697 | decidualization | 1 | 19 | 4.92e-01 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 2 | 47 | 4.92e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 68 | 4.93e-01 |
| GO:BP | GO:0015833 | peptide transport | 5 | 179 | 4.93e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 2 | 64 | 4.93e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 1 | 26 | 4.95e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 1 | 17 | 4.95e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 4.95e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 1 | 15 | 4.95e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 4.95e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 4.95e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 4.95e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 1 | 17 | 4.95e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 1 | 15 | 4.96e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 4.97e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 4 | 1173 | 4.97e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 2 | 64 | 4.97e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 48 | 4.97e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 1 | 36 | 4.98e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 1 | 16 | 4.98e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 3 | 58 | 4.98e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 3 | 73 | 4.98e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 6 | 198 | 4.99e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 4 | 146 | 4.99e-01 |
| GO:BP | GO:0060324 | face development | 1 | 45 | 5.00e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 1 | 11 | 5.00e-01 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 31 | 1076 | 5.01e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 1 | 142 | 5.01e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 31 | 1074 | 5.01e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 14 | 3625 | 5.01e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 2 | 65 | 5.01e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 1 | 34 | 5.02e-01 |
| GO:BP | GO:0000154 | rRNA modification | 1 | 35 | 5.02e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 1 | 18 | 5.02e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 39 | 5.02e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 43 | 5.02e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 1 | 25 | 5.02e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 1 | 25 | 5.03e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 5.03e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 2 | 61 | 5.03e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 1 | 22 | 5.03e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 2 | 68 | 5.03e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 1 | 22 | 5.03e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 1 | 60 | 5.03e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 1 | 36 | 5.03e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 2 | 223 | 5.03e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 5.03e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 5.03e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 7 | 218 | 5.03e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 1 | 19 | 5.03e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 5.03e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 2 | 66 | 5.03e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 2 | 42 | 5.03e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 1 | 19 | 5.03e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 2 | 31 | 5.03e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 3 | 1149 | 5.03e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 1 | 25 | 5.03e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 5.03e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 5.05e-01 |
| GO:BP | GO:0006897 | endocytosis | 2 | 527 | 5.05e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 5.06e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 5.06e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 5.06e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 1 | 17 | 5.06e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 1 | 18 | 5.06e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 18 | 5.06e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 5.06e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 1 | 58 | 5.07e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 5.07e-01 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 1 | 42 | 5.07e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 1 | 32 | 5.07e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 2 | 121 | 5.07e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 4 | 400 | 5.07e-01 |
| GO:BP | GO:0048699 | generation of neurons | 8 | 1119 | 5.08e-01 |
| GO:BP | GO:0003012 | muscle system process | 3 | 334 | 5.08e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 1 | 26 | 5.08e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 1 | 27 | 5.08e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 7 | 199 | 5.09e-01 |
| GO:BP | GO:0002520 | immune system development | 5 | 144 | 5.09e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 1 | 26 | 5.10e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 5.10e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 3 | 114 | 5.10e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 2 | 64 | 5.10e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 1 | 113 | 5.10e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 1 | 20 | 5.10e-01 |
| GO:BP | GO:0022008 | neurogenesis | 9 | 1294 | 5.10e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 1 | 34 | 5.10e-01 |
| GO:BP | GO:0098657 | import into cell | 2 | 166 | 5.10e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 5.10e-01 |
| GO:BP | GO:0046599 | regulation of centriole replication | 1 | 21 | 5.10e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 5.10e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 5.10e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 1 | 22 | 5.10e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 1 | 121 | 5.10e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 5.10e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 6 | 3386 | 5.10e-01 |
| GO:BP | GO:0001947 | heart looping | 2 | 50 | 5.10e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 4 | 252 | 5.10e-01 |
| GO:BP | GO:0051216 | cartilage development | 6 | 151 | 5.10e-01 |
| GO:BP | GO:0043200 | response to amino acid | 1 | 95 | 5.10e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 8 | 1223 | 5.10e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 1 | 43 | 5.10e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 1 | 90 | 5.10e-01 |
| GO:BP | GO:1902074 | response to salt | 3 | 271 | 5.10e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 1 | 43 | 5.10e-01 |
| GO:BP | GO:0030534 | adult behavior | 4 | 92 | 5.10e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 1 | 26 | 5.10e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 3 | 293 | 5.10e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 5.10e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 2 | 382 | 5.10e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 5.10e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 1 | 159 | 5.10e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 1 | 19 | 5.10e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 5.11e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 7 | 234 | 5.12e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 1 | 22 | 5.12e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 1 | 34 | 5.13e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 3 | 108 | 5.13e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 1 | 29 | 5.13e-01 |
| GO:BP | GO:0048731 | system development | 9 | 2901 | 5.13e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 1 | 23 | 5.13e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 70 | 5.13e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 1 | 36 | 5.13e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 6 | 189 | 5.13e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 1 | 23 | 5.13e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 33 | 4579 | 5.13e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 1 | 25 | 5.13e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 7 | 209 | 5.13e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 1 | 36 | 5.13e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 2 | 83 | 5.13e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 1 | 33 | 5.13e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 1 | 17 | 5.13e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 5.13e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 1 | 25 | 5.13e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 1 | 23 | 5.13e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 2 | 170 | 5.13e-01 |
| GO:BP | GO:0009615 | response to virus | 4 | 275 | 5.13e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 2 | 59 | 5.13e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 2 | 31 | 5.13e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 3 | 97 | 5.13e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 1 | 86 | 5.15e-01 |
| GO:BP | GO:0051258 | protein polymerization | 1 | 241 | 5.15e-01 |
| GO:BP | GO:0051225 | spindle assembly | 1 | 120 | 5.15e-01 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 1 | 19 | 5.15e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 5.16e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 1 | 33 | 5.17e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 1 | 33 | 5.17e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 24 | 5.18e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 2 | 626 | 5.18e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 5 | 782 | 5.18e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 1 | 211 | 5.18e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 2 | 461 | 5.18e-01 |
| GO:BP | GO:0007015 | actin filament organization | 8 | 373 | 5.18e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 17 | 5.19e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 16 | 5.19e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 1 | 64 | 5.19e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 5.19e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 1 | 151 | 5.19e-01 |
| GO:BP | GO:0021511 | spinal cord patterning | 1 | 9 | 5.19e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 6 | 299 | 5.19e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 2 | 57 | 5.19e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 3 | 115 | 5.20e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 1 | 13 | 5.20e-01 |
| GO:BP | GO:0006298 | mismatch repair | 1 | 29 | 5.20e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 5.20e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 5.20e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 55 | 3090 | 5.21e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 1 | 64 | 5.21e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 5.21e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 2 | 194 | 5.21e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 5.21e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 5 | 149 | 5.22e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 3 | 122 | 5.23e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 5.23e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 2 | 75 | 5.23e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 5.23e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 2 | 53 | 5.23e-01 |
| GO:BP | GO:0016571 | histone methylation | 4 | 117 | 5.23e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 5.23e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 1 | 34 | 5.23e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 2 | 56 | 5.23e-01 |
| GO:BP | GO:0021510 | spinal cord development | 1 | 59 | 5.23e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 97 | 5.23e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 1 | 38 | 5.23e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 1 | 28 | 5.23e-01 |
| GO:BP | GO:0048511 | rhythmic process | 6 | 237 | 5.24e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 2 | 469 | 5.24e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 1 | 27 | 5.24e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 2 | 47 | 5.24e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 1 | 21 | 5.24e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 75 | 5.24e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 1 | 38 | 5.25e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 17 | 5.26e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 4 | 116 | 5.26e-01 |
| GO:BP | GO:0032438 | melanosome organization | 1 | 36 | 5.28e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 2 | 158 | 5.28e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 1 | 22 | 5.28e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 1 | 36 | 5.28e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1 | 21 | 5.28e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 7 | 234 | 5.28e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2 | 114 | 5.28e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 1 | 31 | 5.28e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 9 | 300 | 5.28e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 3 | 80 | 5.28e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 2 | 295 | 5.29e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 2 | 60 | 5.29e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 3 | 214 | 5.29e-01 |
| GO:BP | GO:0010332 | response to gamma radiation | 2 | 50 | 5.30e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 4 | 157 | 5.30e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 2 | 127 | 5.30e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 1 | 18 | 5.30e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 5.30e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 1 | 85 | 5.30e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 1 | 15 | 5.30e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 6 | 284 | 5.31e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 1 | 27 | 5.31e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 1 | 61 | 5.31e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 5.32e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 1 | 257 | 5.32e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 1 | 26 | 5.32e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 1 | 37 | 5.33e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 5.34e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 2 | 50 | 5.35e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 1 | 19 | 5.35e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 1 | 22 | 5.35e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 1 | 24 | 5.35e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 2 | 71 | 5.35e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 1 | 105 | 5.35e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 1 | 21 | 5.35e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 13 | 1229 | 5.35e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 3 | 84 | 5.35e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 2 | 72 | 5.35e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 6 | 1939 | 5.35e-01 |
| GO:BP | GO:0043473 | pigmentation | 2 | 92 | 5.35e-01 |
| GO:BP | GO:0032606 | type I interferon production | 2 | 76 | 5.35e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 2 | 76 | 5.35e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 1 | 20 | 5.35e-01 |
| GO:BP | GO:0000725 | recombinational repair | 5 | 153 | 5.35e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 5.35e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 5.35e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 1 | 83 | 5.36e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 5.36e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 3 | 113 | 5.36e-01 |
| GO:BP | GO:0006270 | DNA replication initiation | 1 | 35 | 5.36e-01 |
| GO:BP | GO:0009651 | response to salt stress | 1 | 18 | 5.37e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 133 | 5.37e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 4 | 348 | 5.38e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 1 | 44 | 5.38e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 2 | 107 | 5.38e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 2 | 56 | 5.38e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 29 | 4099 | 5.38e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 2 | 52 | 5.38e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 5.38e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 2 | 40 | 5.39e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 146 | 5.39e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 1 | 20 | 5.39e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 1 | 24 | 5.39e-01 |
| GO:BP | GO:0021675 | nerve development | 2 | 57 | 5.39e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 5.39e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 3 | 108 | 5.40e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 2 | 101 | 5.41e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 1 | 36 | 5.41e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 2 | 37 | 5.41e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 1 | 24 | 5.41e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 1 | 52 | 5.42e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 5.42e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 2 | 53 | 5.44e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 2 | 51 | 5.44e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 2 | 54 | 5.45e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 132 | 5.45e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 5 | 147 | 5.45e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 150 | 5.45e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 1 | 46 | 5.46e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 2 | 418 | 5.46e-01 |
| GO:BP | GO:0006479 | protein methylation | 5 | 160 | 5.47e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 8 | 252 | 5.47e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 6 | 197 | 5.47e-01 |
| GO:BP | GO:0008213 | protein alkylation | 5 | 160 | 5.47e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 1 | 82 | 5.50e-01 |
| GO:BP | GO:0001508 | action potential | 3 | 105 | 5.51e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 1 | 24 | 5.51e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 1 | 20 | 5.51e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 5.51e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 1 | 26 | 5.51e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 82 | 5.54e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 5.54e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 1 | 19 | 5.55e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 1 | 16 | 5.55e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 2 | 92 | 5.55e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 1 | 80 | 5.55e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 7 | 369 | 5.56e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 2 | 189 | 5.56e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 1 | 32 | 5.56e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 1 | 31 | 5.56e-01 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 1 | 52 | 5.56e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 3 | 96 | 5.57e-01 |
| GO:BP | GO:0022402 | cell cycle process | 13 | 1086 | 5.57e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 14 | 687 | 5.57e-01 |
| GO:BP | GO:0044409 | entry into host | 2 | 113 | 5.58e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 2 | 93 | 5.58e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 5.58e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 3 | 133 | 5.59e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 2 | 52 | 5.59e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 2 | 52 | 5.59e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 1 | 141 | 5.59e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 1 | 112 | 5.59e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 11 | 459 | 5.59e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 1 | 27 | 5.59e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 5 | 232 | 5.59e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 2 | 89 | 5.59e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 2 | 49 | 5.59e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 2 | 89 | 5.59e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 7 | 308 | 5.59e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 1 | 90 | 5.60e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 3 | 95 | 5.60e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 2 | 78 | 5.62e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 78 | 2135 | 5.62e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 5.62e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 16 | 1974 | 5.63e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 5.63e-01 |
| GO:BP | GO:0035270 | endocrine system development | 4 | 86 | 5.63e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 1 | 294 | 5.64e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 3 | 113 | 5.64e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 1 | 25 | 5.64e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 1 | 27 | 5.64e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 6 | 127 | 5.65e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 5.66e-01 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 2 | 104 | 5.67e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 1 | 28 | 5.67e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 5.67e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 6 | 211 | 5.67e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 1 | 197 | 5.67e-01 |
| GO:BP | GO:0007612 | learning | 1 | 115 | 5.68e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 5.69e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 5.69e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 1 | 23 | 5.69e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 2 | 414 | 5.69e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 3 | 105 | 5.69e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 4 | 130 | 5.69e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 2 | 219 | 5.69e-01 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 1 | 55 | 5.69e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 1 | 48 | 5.69e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 7 | 761 | 5.70e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 1 | 85 | 5.70e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 3 | 279 | 5.70e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 1 | 31 | 5.70e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 3 | 119 | 5.70e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 1 | 30 | 5.71e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 5.72e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 1 | 35 | 5.73e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 1 | 32 | 5.73e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 163 | 5.73e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 3 | 290 | 5.73e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 2 | 376 | 5.73e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 5.73e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 5.74e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 11 | 1869 | 5.74e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 1 | 31 | 5.75e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 1 | 54 | 5.75e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 5.76e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 2 | 81 | 5.76e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 5.77e-01 |
| GO:BP | GO:0097305 | response to alcohol | 1 | 159 | 5.77e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 5.77e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 2 | 70 | 5.77e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 2 | 50 | 5.77e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 5.78e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 4 | 101 | 5.78e-01 |
| GO:BP | GO:0030001 | metal ion transport | 4 | 562 | 5.78e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 1 | 17 | 5.78e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 1 | 36 | 5.78e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 5.79e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 2 | 56 | 5.79e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 1 | 32 | 5.79e-01 |
| GO:BP | GO:0009798 | axis specification | 2 | 63 | 5.81e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 305 | 5.82e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 1 | 33 | 5.82e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 1 | 28 | 5.82e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 5.82e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 2 | 67 | 5.82e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 6 | 609 | 5.82e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 1 | 33 | 5.82e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 1 | 25 | 5.83e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 6 | 216 | 5.84e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 2 | 57 | 5.84e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 3 | 91 | 5.84e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 1 | 187 | 5.84e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 1 | 117 | 5.84e-01 |
| GO:BP | GO:0031648 | protein destabilization | 1 | 45 | 5.84e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 1 | 35 | 5.84e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 1 | 52 | 5.84e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 2 | 116 | 5.84e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 2 | 209 | 5.84e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 3 | 91 | 5.84e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 5.86e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 6 | 2011 | 5.86e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 4 | 182 | 5.86e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 9 | 1014 | 5.87e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 2 | 38 | 5.87e-01 |
| GO:BP | GO:0006400 | tRNA modification | 1 | 90 | 5.87e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 1 | 18 | 5.87e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 1 | 15 | 5.87e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 1 | 21 | 5.87e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 1 | 35 | 5.87e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 2 | 79 | 5.87e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 5.88e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 5.88e-01 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 1 | 14 | 5.88e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 1 | 20 | 5.88e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 5.89e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 2 | 55 | 5.89e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 1 | 31 | 5.89e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 5.89e-01 |
| GO:BP | GO:0051653 | spindle localization | 1 | 52 | 5.89e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 1 | 222 | 5.89e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 1 | 31 | 5.89e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 2 | 84 | 5.89e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 1 | 80 | 5.89e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 1 | 27 | 5.89e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 1 | 37 | 5.89e-01 |
| GO:BP | GO:0046879 | hormone secretion | 5 | 210 | 5.89e-01 |
| GO:BP | GO:0070266 | necroptotic process | 1 | 32 | 5.89e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 6 | 2139 | 5.89e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 14 | 1345 | 5.89e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 4 | 124 | 5.90e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 1 | 191 | 5.91e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 1 | 23 | 5.91e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 115 | 5.92e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 5.92e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 1 | 36 | 5.92e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 5.92e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 1 | 49 | 5.92e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 7 | 222 | 5.92e-01 |
| GO:BP | GO:0000910 | cytokinesis | 3 | 164 | 5.92e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 1 | 23 | 5.92e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 5.93e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 1 | 24 | 5.95e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 1 | 16 | 5.95e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 1 | 39 | 5.95e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 5.95e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 4 | 7552 | 5.95e-01 |
| GO:BP | GO:0035265 | organ growth | 4 | 131 | 5.95e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 5.96e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 1 | 50 | 5.97e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 6 | 2161 | 5.97e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 1 | 68 | 5.97e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 1 | 39 | 5.98e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 5.98e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 5.98e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 3 | 94 | 5.98e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 3 | 94 | 5.98e-01 |
| GO:BP | GO:0051591 | response to cAMP | 2 | 66 | 5.98e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 14 | 1256 | 5.98e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 1 | 106 | 5.99e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 5.99e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 1 | 60 | 5.99e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 2 | 85 | 5.99e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 5.99e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 5.99e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 3 | 118 | 5.99e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 3 | 106 | 5.99e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 14 | 1255 | 5.99e-01 |
| GO:BP | GO:0021591 | ventricular system development | 1 | 29 | 5.99e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 1 | 126 | 5.99e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 1 | 11 | 5.99e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 4 | 640 | 5.99e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 2 | 66 | 5.99e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 1 | 50 | 6.00e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 6.00e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 1 | 55 | 6.00e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 1 | 88 | 6.00e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 6.00e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 1 | 20 | 6.00e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 1 | 49 | 6.00e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 1 | 52 | 6.00e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 6.00e-01 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 1 | 29 | 6.00e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 1 | 37 | 6.00e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 1 | 19 | 6.00e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 8 | 256 | 6.00e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 24 | 783 | 6.02e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 2 | 203 | 6.02e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 4 | 138 | 6.02e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 14 | 2285 | 6.02e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 2 | 65 | 6.03e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 2 | 57 | 6.03e-01 |
| GO:BP | GO:0006954 | inflammatory response | 2 | 429 | 6.03e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 1 | 38 | 6.03e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 1 | 44 | 6.03e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 4 | 94 | 6.03e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 1 | 34 | 6.03e-01 |
| GO:BP | GO:0009888 | tissue development | 20 | 1425 | 6.03e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 1 | 30 | 6.03e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 1 | 20 | 6.03e-01 |
| GO:BP | GO:0006260 | DNA replication | 7 | 255 | 6.04e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 8 | 258 | 6.04e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 1 | 27 | 6.04e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 1 | 22 | 6.04e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 1 | 51 | 6.04e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 6.04e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 1 | 32 | 6.04e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 2 | 68 | 6.04e-01 |
| GO:BP | GO:0010033 | response to organic substance | 11 | 1957 | 6.05e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 1 | 35 | 6.05e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 6.05e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 1 | 277 | 6.05e-01 |
| GO:BP | GO:0040007 | growth | 13 | 742 | 6.06e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 2 | 69 | 6.06e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 1 | 32 | 6.06e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 1 | 28 | 6.06e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 58 | 6.06e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 2 | 206 | 6.06e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 1 | 28 | 6.06e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 1 | 211 | 6.07e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 1 | 39 | 6.07e-01 |
| GO:BP | GO:0009914 | hormone transport | 5 | 215 | 6.08e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 6.09e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 7 | 1974 | 6.09e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 1 | 48 | 6.09e-01 |
| GO:BP | GO:0007049 | cell cycle | 4 | 1529 | 6.09e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 1 | 25 | 6.09e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 1 | 29 | 6.09e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 6.09e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 8 | 1399 | 6.09e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 2 | 140 | 6.09e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 6.10e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 2 | 89 | 6.10e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 1 | 552 | 6.10e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 1 | 30 | 6.10e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 6.10e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 6.10e-01 |
| GO:BP | GO:0035282 | segmentation | 3 | 75 | 6.10e-01 |
| GO:BP | GO:0007155 | cell adhesion | 9 | 1012 | 6.10e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 6.10e-01 |
| GO:BP | GO:0043588 | skin development | 3 | 175 | 6.10e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 6.10e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 6 | 771 | 6.10e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 6.10e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 1 | 23 | 6.10e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 1 | 42 | 6.10e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 1 | 212 | 6.10e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 1 | 15 | 6.10e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 6.10e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 1 | 22 | 6.10e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 6.11e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 1 | 116 | 6.11e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 6.11e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 1 | 225 | 6.11e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 2 | 84 | 6.11e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 1 | 38 | 6.12e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 1 | 26 | 6.12e-01 |
| GO:BP | GO:0044000 | movement in host | 2 | 138 | 6.13e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 1 | 42 | 6.14e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 6.14e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 1 | 57 | 6.14e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 5 | 138 | 6.14e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 3 | 102 | 6.14e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 7 | 229 | 6.15e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 15 | 1145 | 6.15e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 7 | 384 | 6.16e-01 |
| GO:BP | GO:0007051 | spindle organization | 1 | 188 | 6.16e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 1 | 16 | 6.16e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 6.17e-01 |
| GO:BP | GO:0007616 | long-term memory | 1 | 31 | 6.17e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 2 | 64 | 6.17e-01 |
| GO:BP | GO:0001764 | neuron migration | 1 | 133 | 6.17e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 6.17e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 2 | 269 | 6.19e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 1 | 21 | 6.19e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 344 | 6.19e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 338 | 6.19e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 5 | 154 | 6.20e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 1 | 285 | 6.20e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 1 | 13 | 6.20e-01 |
| GO:BP | GO:0016556 | mRNA modification | 1 | 29 | 6.20e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 8 | 726 | 6.20e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 6 | 775 | 6.21e-01 |
| GO:BP | GO:0006936 | muscle contraction | 2 | 267 | 6.21e-01 |
| GO:BP | GO:0051310 | metaphase plate congression | 1 | 69 | 6.22e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 3 | 141 | 6.24e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 4 | 192 | 6.26e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 1 | 39 | 6.27e-01 |
| GO:BP | GO:0030901 | midbrain development | 1 | 69 | 6.27e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 2 | 81 | 6.27e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 1 | 22 | 6.27e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 2 | 92 | 6.27e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 6.27e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 1 | 48 | 6.27e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 2 | 93 | 6.27e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 19 | 2102 | 6.27e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 96 | 6.28e-01 |
| GO:BP | GO:0045010 | actin nucleation | 1 | 50 | 6.28e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 3 | 369 | 6.28e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 3 | 155 | 6.30e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 6.30e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 2 | 341 | 6.30e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 4 | 129 | 6.30e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 2 | 96 | 6.31e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 2 | 342 | 6.31e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 1 | 39 | 6.31e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 2 | 116 | 6.31e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 6.33e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 1 | 41 | 6.33e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 4 | 475 | 6.33e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 6.33e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 6.33e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 1 | 38 | 6.33e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 6.33e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 92 | 6.33e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 6.33e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 1 | 43 | 6.33e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 4 | 316 | 6.33e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 6.35e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 6.35e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 3 | 357 | 6.35e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 1 | 56 | 6.36e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 1 | 42 | 6.36e-01 |
| GO:BP | GO:0015844 | monoamine transport | 2 | 45 | 6.36e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 3 | 90 | 6.36e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 6.37e-01 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 1 | 18 | 6.37e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 5 | 1389 | 6.38e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 6.38e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 6.38e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 4 | 6996 | 6.38e-01 |
| GO:BP | GO:0048513 | animal organ development | 13 | 2189 | 6.38e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 5 | 775 | 6.38e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 1 | 39 | 6.39e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 1 | 104 | 6.39e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 1 | 31 | 6.40e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 8 | 274 | 6.40e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 1 | 44 | 6.40e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 9 | 335 | 6.40e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 6.41e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 1 | 248 | 6.41e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 1 | 37 | 6.42e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 5 | 666 | 6.43e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 9 | 316 | 6.43e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 40 | 6.44e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 6.44e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 4 | 529 | 6.44e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 1 | 43 | 6.44e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 6.45e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 6.45e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 6.46e-01 |
| GO:BP | GO:0001706 | endoderm formation | 1 | 46 | 6.46e-01 |
| GO:BP | GO:0031175 | neuron projection development | 6 | 780 | 6.46e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 1 | 506 | 6.47e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 24 | 884 | 6.47e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 1 | 32 | 6.47e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 6.47e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 4 | 155 | 6.47e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 1 | 95 | 6.48e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 2 | 71 | 6.48e-01 |
| GO:BP | GO:0010165 | response to X-ray | 1 | 29 | 6.48e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 1 | 38 | 6.48e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 29 | 6.49e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 12 | 2065 | 6.49e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 6.49e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 1 | 29 | 6.49e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 1 | 200 | 6.49e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 1 | 28 | 6.49e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 1 | 99 | 6.49e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 1 | 30 | 6.50e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 1 | 90 | 6.50e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 1 | 102 | 6.50e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 6 | 344 | 6.51e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 391 | 6.51e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 6.51e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 34 | 6.51e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 9 | 311 | 6.51e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 6.51e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 6.51e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 2 | 126 | 6.51e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 6 | 345 | 6.52e-01 |
| GO:BP | GO:0007099 | centriole replication | 1 | 38 | 6.52e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 1 | 28 | 6.53e-01 |
| GO:BP | GO:0032940 | secretion by cell | 2 | 569 | 6.53e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 6.54e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 2 | 81 | 6.54e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 6.54e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 6.55e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 5 | 236 | 6.56e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 6.56e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 6.56e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 4 | 541 | 6.56e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 1 | 420 | 6.56e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 3 | 138 | 6.58e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 2 | 129 | 6.59e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 7 | 265 | 6.59e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 1 | 45 | 6.59e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 122 | 6.59e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 1 | 32 | 6.59e-01 |
| GO:BP | GO:0009314 | response to radiation | 7 | 352 | 6.59e-01 |
| GO:BP | GO:0001709 | cell fate determination | 1 | 25 | 6.59e-01 |
| GO:BP | GO:0032502 | developmental process | 7 | 4494 | 6.59e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 2 | 84 | 6.59e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 1 | 29 | 6.59e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 1 | 45 | 6.59e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 5 | 605 | 6.60e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 11 | 1484 | 6.60e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 6.61e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 1 | 154 | 6.62e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 6 | 246 | 6.62e-01 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 1 | 75 | 6.63e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 2 | 183 | 6.64e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 6.64e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 5 | 816 | 6.64e-01 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 2 | 73 | 6.64e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 1 | 49 | 6.65e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 2 | 90 | 6.65e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 5 | 547 | 6.65e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 4 | 174 | 6.65e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 6.65e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 5 | 247 | 6.65e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 1 | 79 | 6.65e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 4 | 234 | 6.65e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 2 | 105 | 6.66e-01 |
| GO:BP | GO:0016050 | vesicle organization | 1 | 320 | 6.67e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 1 | 29 | 6.69e-01 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 1 | 15 | 6.69e-01 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 1 | 20 | 6.69e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 2 | 101 | 6.69e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 1 | 14 | 6.69e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 1 | 403 | 6.69e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 1 | 214 | 6.70e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 1 | 70 | 6.72e-01 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 1 | 42 | 6.72e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 1 | 79 | 6.72e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 3 | 75 | 6.72e-01 |
| GO:BP | GO:0001656 | metanephros development | 2 | 64 | 6.72e-01 |
| GO:BP | GO:0042886 | amide transport | 5 | 235 | 6.72e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 3 | 138 | 6.72e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 1 | 43 | 6.72e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 6.72e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 6.72e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 6.74e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 4 | 669 | 6.74e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 15 | 2565 | 6.75e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 2 | 96 | 6.75e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 8 | 296 | 6.75e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 2 | 250 | 6.76e-01 |
| GO:BP | GO:0098534 | centriole assembly | 1 | 41 | 6.77e-01 |
| GO:BP | GO:0009451 | RNA modification | 2 | 162 | 6.77e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 1 | 124 | 6.77e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 1 | 29 | 6.77e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 6.77e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 1 | 82 | 6.77e-01 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 1 | 42 | 6.77e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 15 | 2573 | 6.78e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 111 | 6.78e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 3 | 149 | 6.78e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 7 | 272 | 6.78e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 142 | 6.80e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 3 | 563 | 6.81e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 1 | 40 | 6.81e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 1 | 32 | 6.81e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 3 | 179 | 6.81e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 2 | 91 | 6.81e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 2 | 227 | 6.81e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1 | 54 | 6.81e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 6.81e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 3 | 140 | 6.81e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1 | 135 | 6.82e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 1 | 55 | 6.82e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 6.82e-01 |
| GO:BP | GO:0042551 | neuron maturation | 1 | 33 | 6.82e-01 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 1 | 44 | 6.82e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 2 | 104 | 6.83e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 3 | 137 | 6.83e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 1 | 40 | 6.83e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 6.83e-01 |
| GO:BP | GO:0140352 | export from cell | 2 | 611 | 6.83e-01 |
| GO:BP | GO:0050000 | chromosome localization | 1 | 82 | 6.83e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 1 | 68 | 6.83e-01 |
| GO:BP | GO:0021543 | pallium development | 2 | 139 | 6.85e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 6.85e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 441 | 6.85e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 1 | 114 | 6.85e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 6.85e-01 |
| GO:BP | GO:0001510 | RNA methylation | 2 | 87 | 6.85e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 1 | 37 | 6.86e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 1 | 75 | 6.86e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 2 | 68 | 6.86e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 5 | 336 | 6.87e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 1 | 49 | 6.87e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 2 | 94 | 6.87e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 2 | 94 | 6.87e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 35 | 6.88e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 6.89e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 1 | 63 | 6.90e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 6.90e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 6.90e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 6.90e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 6.90e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 1 | 112 | 6.90e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 6.92e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 2 | 295 | 6.94e-01 |
| GO:BP | GO:0090224 | regulation of spindle organization | 1 | 45 | 6.94e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 6.94e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 3 | 418 | 6.94e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 22 | 1712 | 6.94e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 3 | 175 | 6.96e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 1 | 53 | 6.96e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 1 | 37 | 6.96e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 2 | 141 | 6.96e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 6.98e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 6.98e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 1 | 130 | 6.98e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 15 | 4128 | 6.98e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 6.98e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 6.98e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 1 | 115 | 6.98e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 6 | 230 | 6.98e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 9 | 1147 | 7.00e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 1 | 41 | 7.01e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 1 | 131 | 7.01e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 2 | 75 | 7.01e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 5 | 293 | 7.02e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 7.02e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 7.02e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 7.02e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 1 | 41 | 7.02e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 5 | 199 | 7.02e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 1 | 53 | 7.02e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 5 | 259 | 7.02e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 1 | 1238 | 7.02e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 7.03e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 1 | 32 | 7.03e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 28 | 7.03e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 2 | 556 | 7.04e-01 |
| GO:BP | GO:0048284 | organelle fusion | 1 | 135 | 7.04e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 4 | 516 | 7.04e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 2 | 76 | 7.04e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 2 | 76 | 7.04e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 1 | 126 | 7.05e-01 |
| GO:BP | GO:0006304 | DNA modification | 1 | 89 | 7.05e-01 |
| GO:BP | GO:0042476 | odontogenesis | 3 | 86 | 7.06e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 7.06e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 7.06e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 1 | 77 | 7.06e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 2 | 258 | 7.07e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 7 | 993 | 7.07e-01 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 1 | 40 | 7.08e-01 |
| GO:BP | GO:0061564 | axon development | 2 | 398 | 7.09e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 1 | 64 | 7.09e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 16 | 827 | 7.10e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 1 | 760 | 7.10e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 1 | 68 | 7.11e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 1 | 31 | 7.11e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 7.12e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 1 | 317 | 7.13e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 3 | 125 | 7.13e-01 |
| GO:BP | GO:0007568 | aging | 1 | 115 | 7.13e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 7.13e-01 |
| GO:BP | GO:0034097 | response to cytokine | 2 | 607 | 7.14e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 2 | 303 | 7.14e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 7.14e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 7.14e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 2 | 87 | 7.14e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 1 | 60 | 7.14e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 5 | 631 | 7.15e-01 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 7.15e-01 |
| GO:BP | GO:0046903 | secretion | 2 | 645 | 7.15e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 4 | 143 | 7.18e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 12 | 597 | 7.19e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 12 | 597 | 7.19e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 1 | 86 | 7.19e-01 |
| GO:BP | GO:0046907 | intracellular transport | 4 | 1453 | 7.19e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 2 | 63 | 7.19e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 1 | 99 | 7.19e-01 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 1 | 40 | 7.20e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 496 | 7.20e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 7.20e-01 |
| GO:BP | GO:0001823 | mesonephros development | 2 | 77 | 7.20e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 1 | 135 | 7.21e-01 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 2 | 79 | 7.21e-01 |
| GO:BP | GO:0007420 | brain development | 3 | 591 | 7.21e-01 |
| GO:BP | GO:0006284 | base-excision repair | 1 | 44 | 7.21e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 7.21e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 1 | 47 | 7.21e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 8 | 859 | 7.22e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 1 | 77 | 7.22e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 8 | 343 | 7.22e-01 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 1 | 34 | 7.22e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 1 | 22 | 7.24e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 1 | 46 | 7.24e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 2 | 217 | 7.24e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 7.24e-01 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 1 | 19 | 7.25e-01 |
| GO:BP | GO:0035176 | social behavior | 1 | 34 | 7.25e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 1 | 46 | 7.25e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 17 | 2890 | 7.25e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 1 | 39 | 7.25e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 7.26e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 1 | 161 | 7.28e-01 |
| GO:BP | GO:0051301 | cell division | 1 | 570 | 7.28e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 7.28e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 7 | 755 | 7.28e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 1 | 48 | 7.28e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 35 | 1248 | 7.29e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 7.29e-01 |
| GO:BP | GO:0009056 | catabolic process | 26 | 2088 | 7.29e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 4 | 569 | 7.29e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 7.30e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 7.32e-01 |
| GO:BP | GO:0048589 | developmental growth | 14 | 522 | 7.33e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 1 | 66 | 7.33e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 1 | 121 | 7.34e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 10 | 612 | 7.35e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 1 | 66 | 7.36e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 1 | 34 | 7.36e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 4 | 7441 | 7.37e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 3 | 632 | 7.37e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 46 | 7.37e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 7.37e-01 |
| GO:BP | GO:0006412 | translation | 3 | 646 | 7.41e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 6 | 242 | 7.41e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 7.42e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 1 | 36 | 7.42e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 2 | 96 | 7.44e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 7.44e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 2 | 301 | 7.45e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 7.45e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 3 | 1201 | 7.45e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 7.45e-01 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 1 | 81 | 7.46e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 1 | 50 | 7.46e-01 |
| GO:BP | GO:0048666 | neuron development | 2 | 877 | 7.49e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 1 | 83 | 7.49e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 2 | 120 | 7.50e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 250 | 7.50e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 1 | 116 | 7.50e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 3 | 486 | 7.51e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 7.51e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 7.51e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 2 | 496 | 7.51e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 2 | 110 | 7.51e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 1 | 58 | 7.51e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 7.51e-01 |
| GO:BP | GO:0051607 | defense response to virus | 2 | 202 | 7.51e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 2 | 86 | 7.52e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 2 | 86 | 7.52e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 2 | 202 | 7.52e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 1 | 51 | 7.52e-01 |
| GO:BP | GO:0022900 | electron transport chain | 1 | 142 | 7.52e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 1 | 52 | 7.52e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 2 | 162 | 7.53e-01 |
| GO:BP | GO:0060322 | head development | 3 | 633 | 7.53e-01 |
| GO:BP | GO:0061061 | muscle structure development | 12 | 546 | 7.53e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 3 | 532 | 7.53e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 7.53e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 2 | 906 | 7.53e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 1 | 47 | 7.55e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 1 | 39 | 7.55e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 1 | 271 | 7.55e-01 |
| GO:BP | GO:0003002 | regionalization | 9 | 273 | 7.56e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 7.56e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 9 | 560 | 7.56e-01 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 7.56e-01 |
| GO:BP | GO:0030030 | cell projection organization | 3 | 1233 | 7.56e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 213 | 7.56e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 3 | 169 | 7.56e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 3 | 963 | 7.56e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 2 | 1226 | 7.56e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 7.56e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 7.57e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 7.59e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 1 | 64 | 7.59e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 1 | 51 | 7.60e-01 |
| GO:BP | GO:0030850 | prostate gland development | 1 | 35 | 7.60e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 7.60e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 1 | 51 | 7.60e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 7.60e-01 |
| GO:BP | GO:0006935 | chemotaxis | 3 | 389 | 7.60e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 1 | 27 | 7.60e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 7.60e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 16 | 1555 | 7.60e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 2 | 168 | 7.61e-01 |
| GO:BP | GO:0042330 | taxis | 3 | 389 | 7.61e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 7.61e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 7.62e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 7.62e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 1 | 52 | 7.63e-01 |
| GO:BP | GO:0007286 | spermatid development | 2 | 105 | 7.63e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 5 | 241 | 7.63e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 1 | 51 | 7.63e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 4 | 7069 | 7.63e-01 |
| GO:BP | GO:0007369 | gastrulation | 3 | 146 | 7.65e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 7.65e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 2 | 681 | 7.65e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 6 | 287 | 7.66e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 1 | 93 | 7.66e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 1 | 53 | 7.66e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 1 | 69 | 7.68e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 1 | 54 | 7.68e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 1 | 173 | 7.69e-01 |
| GO:BP | GO:0090102 | cochlea development | 1 | 35 | 7.71e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 7.72e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 1 | 59 | 7.73e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 1 | 59 | 7.73e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 2 | 357 | 7.73e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 7.74e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 3 | 121 | 7.74e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 7.74e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 2 | 93 | 7.74e-01 |
| GO:BP | GO:0007517 | muscle organ development | 6 | 277 | 7.75e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 7.75e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 1 | 162 | 7.75e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 2 | 696 | 7.75e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 1 | 49 | 7.75e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 1 | 276 | 7.76e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 2 | 110 | 7.76e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 2 | 111 | 7.76e-01 |
| GO:BP | GO:0006952 | defense response | 3 | 944 | 7.76e-01 |
| GO:BP | GO:0006310 | DNA recombination | 1 | 257 | 7.76e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 2 | 700 | 7.77e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 1 | 353 | 7.78e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 1 | 58 | 7.78e-01 |
| GO:BP | GO:0051302 | regulation of cell division | 2 | 145 | 7.78e-01 |
| GO:BP | GO:0021549 | cerebellum development | 2 | 78 | 7.78e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 2 | 100 | 7.79e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 1 | 120 | 7.79e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 2 | 145 | 7.79e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 1 | 784 | 7.79e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 7.80e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 7.80e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 7.81e-01 |
| GO:BP | GO:0045087 | innate immune response | 1 | 488 | 7.81e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 7.82e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 7.82e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 1 | 52 | 7.82e-01 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 3 | 148 | 7.82e-01 |
| GO:BP | GO:0030154 | cell differentiation | 2 | 2958 | 7.82e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 2 | 135 | 7.82e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 1 | 35 | 7.82e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 7.82e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 1 | 51 | 7.82e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 7.82e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 7.82e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 2 | 108 | 7.82e-01 |
| GO:BP | GO:0030902 | hindbrain development | 3 | 114 | 7.83e-01 |
| GO:BP | GO:0007052 | mitotic spindle organization | 2 | 129 | 7.83e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 7.84e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 7.84e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 7.84e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 2 | 2984 | 7.84e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 4 | 8061 | 7.84e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 2 | 357 | 7.84e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 3 | 186 | 7.84e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 1 | 665 | 7.84e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 2 | 97 | 7.84e-01 |
| GO:BP | GO:0051276 | chromosome organization | 5 | 543 | 7.85e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 7.85e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 3 | 348 | 7.86e-01 |
| GO:BP | GO:0001822 | kidney development | 6 | 244 | 7.89e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 1 | 448 | 7.89e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 4 | 251 | 7.89e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 2 | 114 | 7.89e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 2 | 94 | 7.90e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 7.91e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 2 | 362 | 7.92e-01 |
| GO:BP | GO:0007417 | central nervous system development | 4 | 792 | 7.92e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 7.92e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 1 | 34 | 7.93e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 7.93e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 1 | 104 | 7.97e-01 |
| GO:BP | GO:0032259 | methylation | 7 | 313 | 7.97e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 14 | 4045 | 7.97e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 112 | 7.98e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 6 | 809 | 7.98e-01 |
| GO:BP | GO:0002253 | activation of immune response | 6 | 272 | 7.99e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 2 | 351 | 7.99e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 2 | 101 | 7.99e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 1 | 47 | 7.99e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 2 | 188 | 7.99e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 1 | 68 | 7.99e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 2 | 490 | 8.00e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 2 | 490 | 8.00e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 8.00e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 1 | 43 | 8.01e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 1 | 43 | 8.01e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 1 | 35 | 8.02e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 3 | 512 | 8.03e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 2 | 495 | 8.03e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 1 | 72 | 8.05e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 1 | 166 | 8.07e-01 |
| GO:BP | GO:0021700 | developmental maturation | 2 | 211 | 8.08e-01 |
| GO:BP | GO:0032868 | response to insulin | 4 | 215 | 8.08e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 1 | 33 | 8.09e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 4 | 665 | 8.10e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 2 | 102 | 8.11e-01 |
| GO:BP | GO:0006997 | nucleus organization | 1 | 126 | 8.11e-01 |
| GO:BP | GO:0072001 | renal system development | 6 | 251 | 8.11e-01 |
| GO:BP | GO:0022037 | metencephalon development | 2 | 84 | 8.12e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 1 | 88 | 8.13e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 2 | 517 | 8.13e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 2 | 103 | 8.13e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 2 | 527 | 8.15e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 2 | 511 | 8.16e-01 |
| GO:BP | GO:0030258 | lipid modification | 2 | 162 | 8.16e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 1 | 49 | 8.16e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 1 | 108 | 8.16e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 2 | 531 | 8.18e-01 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 8.20e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 1 | 50 | 8.20e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 1 | 75 | 8.21e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 8.22e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 11 | 1898 | 8.22e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 8.23e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 8.23e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 1 | 53 | 8.24e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 2 | 130 | 8.25e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 14 | 963 | 8.25e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 8.26e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 1 | 57 | 8.27e-01 |
| GO:BP | GO:0050877 | nervous system process | 6 | 634 | 8.27e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 2 | 540 | 8.28e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 1 | 544 | 8.28e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 1 | 93 | 8.28e-01 |
| GO:BP | GO:0051707 | response to other organism | 2 | 820 | 8.28e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 2 | 820 | 8.29e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 2 | 551 | 8.30e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 1 | 401 | 8.31e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 2 | 98 | 8.31e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 4 | 7806 | 8.32e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 2 | 109 | 8.33e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 2 | 129 | 8.34e-01 |
| GO:BP | GO:0007389 | pattern specification process | 9 | 306 | 8.34e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 5 | 337 | 8.35e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 6 | 1138 | 8.36e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 8.36e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 1 | 408 | 8.36e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 1 | 57 | 8.36e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 1 | 119 | 8.36e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 8.38e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 65 | 8.38e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 2 | 850 | 8.38e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 6 | 4794 | 8.38e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 1 | 103 | 8.39e-01 |
| GO:BP | GO:0051604 | protein maturation | 1 | 259 | 8.40e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 5 | 188 | 8.41e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 8.42e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 128 | 8.42e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 8.42e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 1 | 115 | 8.42e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 8.43e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 1 | 113 | 8.43e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 8.43e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 1 | 111 | 8.45e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 4 | 196 | 8.46e-01 |
| GO:BP | GO:0048469 | cell maturation | 1 | 108 | 8.46e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 9 | 1378 | 8.47e-01 |
| GO:BP | GO:0007127 | meiosis I | 1 | 77 | 8.47e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 1 | 57 | 8.48e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 1 | 69 | 8.48e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 8.49e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 8.50e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 2 | 123 | 8.52e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 1 | 29 | 8.54e-01 |
| GO:BP | GO:0023061 | signal release | 5 | 321 | 8.54e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 2 | 112 | 8.54e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 2 | 273 | 8.54e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 1 | 59 | 8.54e-01 |
| GO:BP | GO:0015031 | protein transport | 3 | 1361 | 8.54e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 1 | 110 | 8.54e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 230 | 8.55e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 61 | 8.56e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 61 | 8.56e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 8.57e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 1 | 135 | 8.57e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 1 | 81 | 8.58e-01 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 1 | 94 | 8.59e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 2 | 126 | 8.59e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 1 | 58 | 8.59e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 2 | 139 | 8.59e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 1 | 61 | 8.60e-01 |
| GO:BP | GO:0019079 | viral genome replication | 1 | 101 | 8.62e-01 |
| GO:BP | GO:0065007 | biological regulation | 4 | 8320 | 8.62e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 1 | 62 | 8.63e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 8.64e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 1 | 107 | 8.64e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 3 | 165 | 8.65e-01 |
| GO:BP | GO:0003014 | renal system process | 1 | 78 | 8.65e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 2 | 540 | 8.67e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 4 | 1836 | 8.69e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 8.70e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 1 | 74 | 8.72e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 1 | 48 | 8.76e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 1 | 48 | 8.76e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 8.76e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 3 | 535 | 8.76e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 1 | 179 | 8.76e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 8.78e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 8.80e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 1 | 145 | 8.80e-01 |
| GO:BP | GO:1901652 | response to peptide | 7 | 377 | 8.81e-01 |
| GO:BP | GO:0021536 | diencephalon development | 1 | 41 | 8.81e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 2 | 639 | 8.81e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 8.81e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 3 | 1462 | 8.81e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 8.81e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 1 | 124 | 8.81e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 8.81e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 8.83e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 8.83e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 8.86e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 1 | 447 | 8.89e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 8.89e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 3 | 478 | 8.91e-01 |
| GO:BP | GO:0001816 | cytokine production | 1 | 451 | 8.91e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 359 | 8.91e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 3 | 151 | 8.92e-01 |
| GO:BP | GO:0042113 | B cell activation | 4 | 170 | 8.92e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 72 | 8.94e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 70 | 8.94e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 1 | 130 | 8.94e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 73 | 8.96e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 1 | 64 | 8.96e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 8.96e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 8.96e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 8.96e-01 |
| GO:BP | GO:0007040 | lysosome organization | 1 | 88 | 8.96e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 74 | 8.96e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 1 | 88 | 8.96e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 74 | 8.96e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 73 | 8.97e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 8.98e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 2 | 162 | 8.98e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 1 | 149 | 9.02e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 9.03e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 3 | 468 | 9.04e-01 |
| GO:BP | GO:0007601 | visual perception | 2 | 96 | 9.04e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 2 | 463 | 9.05e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 1 | 73 | 9.05e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 3 | 676 | 9.08e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 9.08e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 2 | 97 | 9.08e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 1 | 106 | 9.10e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 1 | 77 | 9.11e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 1 | 56 | 9.13e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 1 | 56 | 9.13e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 2 | 114 | 9.13e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 1 | 233 | 9.14e-01 |
| GO:BP | GO:0051641 | cellular localization | 6 | 2810 | 9.14e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 9.14e-01 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 1 | 16 | 9.15e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 3 | 482 | 9.17e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 2 | 170 | 9.19e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 8 | 4241 | 9.19e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 3 | 162 | 9.19e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 1 | 64 | 9.19e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 1 | 72 | 9.19e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 2 | 994 | 9.21e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 9.21e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 3 | 998 | 9.22e-01 |
| GO:BP | GO:0003008 | system process | 8 | 1189 | 9.22e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 1 | 245 | 9.23e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 1 | 129 | 9.23e-01 |
| GO:BP | GO:0000280 | nuclear division | 3 | 344 | 9.26e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 1 | 75 | 9.27e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 1 | 154 | 9.27e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 211 | 9.27e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 2 | 153 | 9.27e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 3 | 1335 | 9.28e-01 |
| GO:BP | GO:0008104 | protein localization | 4 | 2129 | 9.29e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 9.29e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 9.29e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 1 | 157 | 9.29e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 3 | 190 | 9.30e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 4 | 2138 | 9.30e-01 |
| GO:BP | GO:0006950 | response to stress | 7 | 2780 | 9.30e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 7 | 369 | 9.30e-01 |
| GO:BP | GO:0007613 | memory | 1 | 92 | 9.30e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 11 | 581 | 9.30e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 2 | 457 | 9.30e-01 |
| GO:BP | GO:0007254 | JNK cascade | 2 | 138 | 9.30e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 5 | 321 | 9.32e-01 |
| GO:BP | GO:0007281 | germ cell development | 2 | 187 | 9.32e-01 |
| GO:BP | GO:0051648 | vesicle localization | 1 | 192 | 9.33e-01 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 9.34e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 9.35e-01 |
| GO:BP | GO:0003341 | cilium movement | 1 | 97 | 9.35e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 9.35e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 843 | 9.36e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 1 | 105 | 9.37e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 1 | 192 | 9.37e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 1 | 86 | 9.39e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 4 | 484 | 9.40e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 1 | 170 | 9.41e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 3 | 785 | 9.43e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 3 | 1684 | 9.43e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 1 | 65 | 9.43e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 9.43e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 6 | 877 | 9.43e-01 |
| GO:BP | GO:0007033 | vacuole organization | 2 | 197 | 9.43e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 3 | 1496 | 9.43e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 9.43e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 9.43e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 9.43e-01 |
| GO:BP | GO:0006996 | organelle organization | 5 | 3000 | 9.44e-01 |
| GO:BP | GO:0006956 | complement activation | 1 | 23 | 9.46e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 9.46e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 1 | 69 | 9.49e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 2 | 276 | 9.50e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 9.50e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 13 | 1719 | 9.50e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 1 | 251 | 9.51e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 9.51e-01 |
| GO:BP | GO:0048285 | organelle fission | 3 | 388 | 9.51e-01 |
| GO:BP | GO:0042221 | response to chemical | 11 | 2559 | 9.54e-01 |
| GO:BP | GO:0001708 | cell fate specification | 1 | 56 | 9.55e-01 |
| GO:BP | GO:0006810 | transport | 12 | 3298 | 9.57e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 9.57e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 9.57e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 2 | 106 | 9.58e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 9.58e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 9 | 4955 | 9.58e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 9.60e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 1 | 190 | 9.61e-01 |
| GO:BP | GO:0006508 | proteolysis | 13 | 1318 | 9.61e-01 |
| GO:BP | GO:0007565 | female pregnancy | 1 | 122 | 9.62e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 9.63e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 1 | 96 | 9.63e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 1 | 324 | 9.64e-01 |
| GO:BP | GO:0007292 | female gamete generation | 1 | 103 | 9.64e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 1 | 98 | 9.65e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 1 | 133 | 9.65e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 1 | 98 | 9.66e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 9.67e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 2 | 166 | 9.69e-01 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 2 | 186 | 9.70e-01 |
| GO:BP | GO:0051234 | establishment of localization | 12 | 3433 | 9.71e-01 |
| GO:BP | GO:0051640 | organelle localization | 3 | 503 | 9.73e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 4 | 2450 | 9.74e-01 |
| GO:BP | GO:0072006 | nephron development | 1 | 117 | 9.74e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 1 | 106 | 9.74e-01 |
| GO:BP | GO:0042592 | homeostatic process | 5 | 1159 | 9.74e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 1 | 131 | 9.74e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 1 | 86 | 9.74e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 9.75e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 9.76e-01 |
| GO:BP | GO:0070925 | organelle assembly | 1 | 817 | 9.78e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 1 | 88 | 9.78e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 1 | 135 | 9.79e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 1 | 194 | 9.84e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 2 | 252 | 9.85e-01 |
| GO:BP | GO:0071702 | organic substance transport | 8 | 2116 | 9.87e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 8 | 740 | 9.87e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 1 | 162 | 9.88e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 9.88e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 2 | 389 | 9.90e-01 |
| GO:BP | GO:0051179 | localization | 10 | 3969 | 9.90e-01 |
| GO:BP | GO:0009725 | response to hormone | 2 | 642 | 9.91e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 2 | 174 | 9.91e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 9.91e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 1 | 90 | 9.91e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 14 | 4953 | 9.91e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 9.91e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 9.91e-01 |
| GO:BP | GO:0010876 | lipid localization | 1 | 310 | 9.91e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 9.91e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 9.92e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 9.92e-01 |
| GO:BP | GO:0060271 | cilium assembly | 2 | 292 | 9.93e-01 |
| GO:BP | GO:0044782 | cilium organization | 4 | 313 | 9.93e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 138 | 9.94e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 9.94e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 1 | 168 | 9.94e-01 |
| GO:BP | GO:0007165 | signal transduction | 1 | 3818 | 9.94e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 378 | 9.95e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 3 | 2466 | 9.95e-01 |
| GO:BP | GO:0006909 | phagocytosis | 1 | 148 | 9.95e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 9.95e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 1 | 154 | 9.95e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 149 | 9.95e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 1 | 157 | 9.96e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 1 | 194 | 9.96e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 1 | 191 | 9.97e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 1 | 105 | 9.97e-01 |
| GO:BP | GO:0023052 | signaling | 1 | 4141 | 9.97e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 9.98e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 9.98e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 2 | 142 | 9.98e-01 |
| GO:BP | GO:0007154 | cell communication | 1 | 4226 | 9.98e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 161 | 9.98e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 7 | 665 | 9.98e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 13 | 5274 | 9.98e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 1 | 300 | 9.99e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 1 | 271 | 1.00e+00 |
| GO:BP | GO:0006397 | mRNA processing | 5 | 450 | 1.00e+00 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 1.00e+00 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 1.00e+00 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 1.00e+00 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 1.00e+00 |
| GO:BP | GO:0010721 | negative regulation of cell development | 1 | 191 | 1.00e+00 |
| GO:BP | GO:0016043 | cellular component organization | 12 | 5076 | 1.00e+00 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5770 | 1.00e+00 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0006959 | humoral immune response | 1 | 80 | 1.00e+00 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 1.00e+00 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 1.00e+00 |
| GO:BP | GO:0002252 | immune effector process | 1 | 318 | 1.00e+00 |
| GO:BP | GO:0006869 | lipid transport | 1 | 272 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 37 | 415 | 1.34e-08 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 8 | 104 | 3.21e-01 |
| KEGG | KEGG:05224 | Breast cancer | 6 | 117 | 3.21e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 3 | 56 | 3.21e-01 |
| KEGG | KEGG:04210 | Apoptosis | 7 | 118 | 3.21e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 4 | 66 | 3.21e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 4 | 86 | 3.21e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 3 | 121 | 3.21e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 2 | 131 | 3.21e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 4 | 138 | 3.21e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 2 | 112 | 3.21e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 2 | 35 | 3.21e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 2 | 14 | 3.21e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 3 | 65 | 3.21e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 4 | 49 | 3.21e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 7 | 117 | 3.21e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 5 | 84 | 3.21e-01 |
| KEGG | KEGG:03040 | Spliceosome | 3 | 132 | 3.21e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 2 | 104 | 3.21e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 5 | 153 | 3.21e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 3.21e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 2 | 97 | 3.21e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 2 | 76 | 3.21e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 3 | 40 | 3.21e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 2 | 49 | 3.21e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 2 | 110 | 3.21e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 2 | 48 | 3.21e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 2 | 150 | 3.21e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 2 | 87 | 3.21e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 5 | 124 | 3.21e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 3 | 86 | 3.21e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 3 | 145 | 3.21e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 3.21e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 2 | 115 | 3.21e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 3.21e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 3.21e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 3 | 172 | 3.21e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 2 | 70 | 3.21e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 2 | 136 | 3.21e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 6 | 409 | 3.26e-01 |
| KEGG | KEGG:04144 | Endocytosis | 3 | 232 | 3.26e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 6 | 180 | 3.44e-01 |
| KEGG | KEGG:05164 | Influenza A | 5 | 103 | 3.44e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 2 | 214 | 3.75e-01 |
| KEGG | KEGG:05131 | Shigellosis | 2 | 210 | 3.75e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 1 | 28 | 3.97e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 3.98e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 3.98e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 2 | 70 | 3.98e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 2 | 83 | 3.98e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 3 | 72 | 3.98e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 3 | 69 | 3.98e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 3.98e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 3.98e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 3.98e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 3.98e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 3 | 60 | 3.98e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 3.98e-01 |
| KEGG | KEGG:04520 | Adherens junction | 3 | 67 | 3.98e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 3 | 84 | 3.98e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 3.98e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 3.98e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 5 | 87 | 3.98e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 2 | 25 | 3.98e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 1 | 60 | 3.98e-01 |
| KEGG | KEGG:05142 | Chagas disease | 3 | 75 | 3.98e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 2 | 99 | 3.98e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 1 | 19 | 3.98e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 3 | 77 | 3.98e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 1 | 38 | 3.98e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 5 | 145 | 3.98e-01 |
| KEGG | KEGG:05133 | Pertussis | 2 | 45 | 3.98e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 3.98e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 3.98e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 3 | 91 | 3.98e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 2 | 142 | 3.98e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 4 | 84 | 3.98e-01 |
| KEGG | KEGG:04360 | Axon guidance | 2 | 162 | 3.98e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 1 | 58 | 3.98e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 3 | 57 | 3.98e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 6 | 134 | 3.98e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 3.98e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 2 | 56 | 3.98e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 4 | 114 | 3.98e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 3.99e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 2 | 20 | 3.99e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 2 | 115 | 3.99e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 1 | 85 | 3.99e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 1 | 84 | 3.99e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 1 | 91 | 4.02e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 4.03e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 4.03e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 2 | 165 | 4.03e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 4 | 98 | 4.03e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 1 | 64 | 4.12e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 2 | 35 | 4.13e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 1 | 110 | 4.13e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 5 | 134 | 4.13e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 4.13e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 2 | 176 | 4.13e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 4.17e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 1 | 77 | 4.19e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 2 | 35 | 4.19e-01 |
| KEGG | KEGG:05218 | Melanoma | 3 | 58 | 4.19e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 3 | 56 | 4.19e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 1 | 24 | 4.22e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 4.22e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 4.26e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 3 | 315 | 4.26e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 2 | 44 | 4.26e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 2 | 161 | 4.26e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 3 | 75 | 4.26e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 4.26e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 3 | 75 | 4.26e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 4 | 121 | 4.46e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 1 | 103 | 4.92e-01 |
| KEGG | KEGG:04530 | Tight junction | 1 | 137 | 4.92e-01 |
| KEGG | KEGG:04924 | Renin secretion | 1 | 50 | 4.95e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 1 | 171 | 5.11e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 5.11e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 1 | 52 | 5.24e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 5.28e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 2 | 385 | 5.28e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 3 | 95 | 5.28e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 4 | 109 | 5.28e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 5.28e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 1 | 128 | 5.28e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 2 | 186 | 5.38e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 1 | 133 | 5.38e-01 |
| KEGG | KEGG:04217 | Necroptosis | 2 | 106 | 5.41e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 1 | 79 | 5.44e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 5.48e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 5.48e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 5.53e-01 |
| KEGG | KEGG:00000 | KEGG root term | 2 | 5558 | 5.53e-01 |
| KEGG | KEGG:05034 | Alcoholism | 1 | 122 | 5.54e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 4 | 88 | 5.56e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 2 | 97 | 5.70e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 1 | 26 | 5.70e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 3 | 71 | 5.70e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 3 | 98 | 5.70e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 1 | 26 | 5.75e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 1 | 19 | 5.94e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 2 | 89 | 5.97e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 1 | 65 | 5.97e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 3 | 157 | 5.97e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 2 | 226 | 5.97e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 1 | 27 | 5.97e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 5 | 272 | 6.15e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 4 | 145 | 6.18e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 1 | 31 | 6.26e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 1 | 35 | 6.26e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 2 | 67 | 6.37e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 2 | 112 | 6.39e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 1 | 135 | 6.40e-01 |
| KEGG | KEGG:05020 | Prion disease | 1 | 220 | 6.40e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 1 | 51 | 6.40e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 2 | 63 | 6.40e-01 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 6.41e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 6.51e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 1 | 31 | 6.51e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 1 | 63 | 6.59e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 1 | 41 | 6.82e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 1 | 49 | 6.86e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 1 | 48 | 6.87e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 3 | 115 | 6.87e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 6.87e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 1 | 46 | 6.87e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 1 | 55 | 6.88e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 6.90e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 2 | 133 | 6.90e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 1 | 46 | 6.90e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 1 | 53 | 6.90e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 1 | 179 | 6.90e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 4 | 149 | 6.90e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 1 | 170 | 6.90e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 6.90e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 6.97e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 1 | 63 | 6.97e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 2 | 87 | 6.97e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 1 | 39 | 7.00e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 1 | 65 | 7.01e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 1 | 74 | 7.25e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 2 | 76 | 7.29e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 1 | 74 | 7.29e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 3 | 1146 | 7.45e-01 |
| KEGG | KEGG:04110 | Cell cycle | 3 | 151 | 7.45e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 1 | 49 | 7.94e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 7.94e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 1 | 86 | 7.97e-01 |
| KEGG | KEGG:04730 | Long-term depression | 1 | 42 | 7.97e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 7.97e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 1 | 49 | 8.08e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 2 | 107 | 8.10e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 2 | 164 | 8.14e-01 |
| KEGG | KEGG:04611 | Platelet activation | 2 | 89 | 8.14e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 8.33e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 8.33e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 8.33e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 3 | 128 | 8.33e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 2 | 106 | 8.44e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 1 | 106 | 8.84e-01 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 2 | 169 | 9.05e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 2 | 110 | 9.17e-01 |
| KEGG | KEGG:05016 | Huntington disease | 2 | 256 | 9.33e-01 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 1 | 61 | 9.40e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 1 | 94 | 9.53e-01 |
# length(union(Daun3sp, sigVDX3$ENTREZID))
ggVennDiagram::ggVennDiagram(
list(Daun3sp, sigVDA24$ENTREZID),
category.names = c("3 hour\n DNR- specific ", "24 hours\n DEG p < 0.05"),
show_intersect = FALSE,
set_color = "black",
label = "count",
label_percent_digit = 1,
label_size = 5,
label_alpha = 0,
label_color = "black",
edge_lty = "solid"
) +
scale_x_continuous(expand = expansion(mult = .2)) +
scale_y_continuous(expand = expansion(mult = .1)) +
scale_fill_gradient(low = "red4", high = "tan1") +
labs(title = "3 hr DNR-sp with 24 hour DNR", caption = paste("n =", length(union(
Daun3sp, sigVDX3$ENTREZID
)), "genes")) +
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5))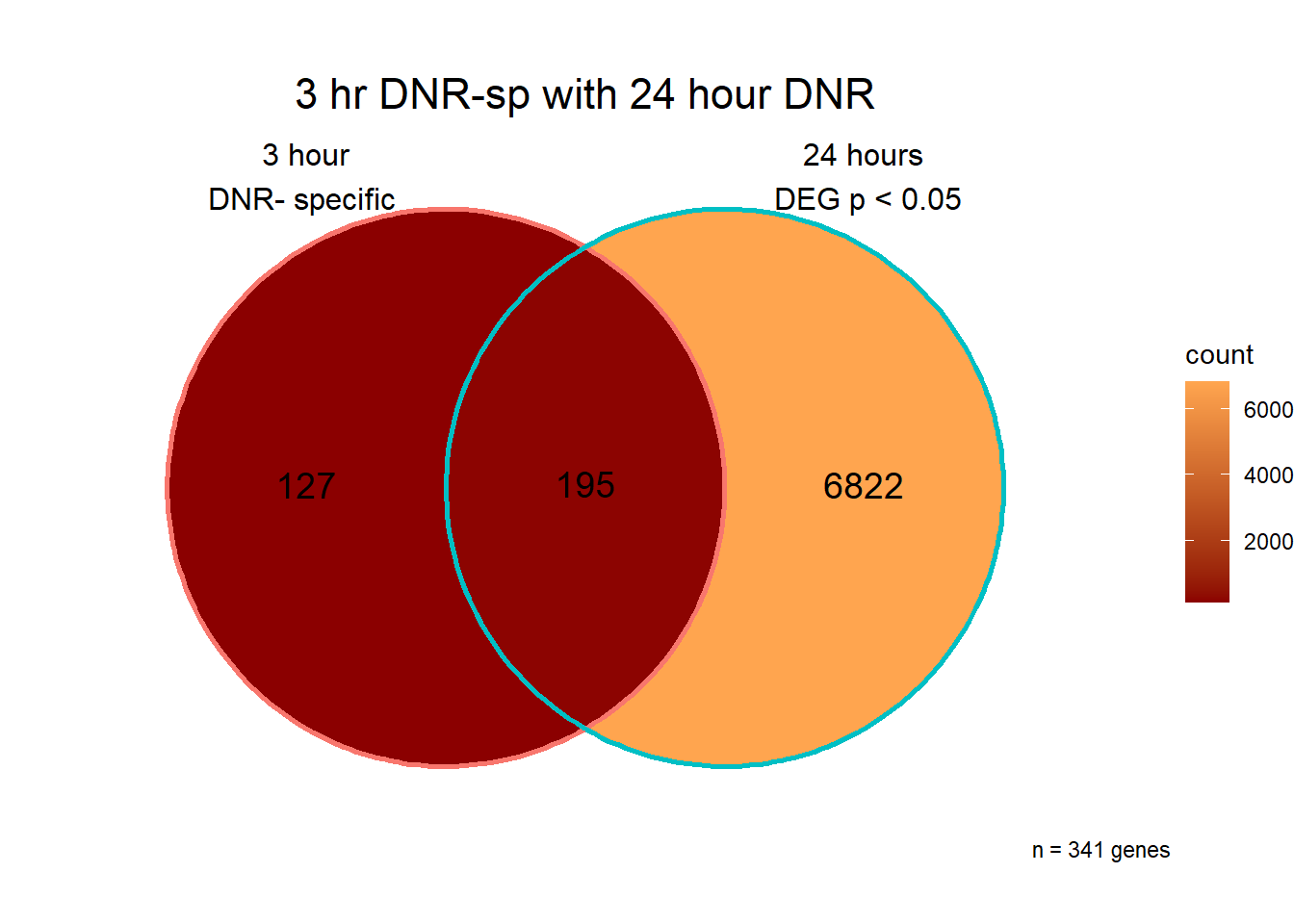
Interestingly, Over half of the DNR specific genes are in the DNR 24 hour group.
When I look at those genes, I do not see a significant enrichment in GO:BP terms, only the KEGG pathway connection to Herpes simplex virus
list324venn <-
VennDiagram::get.venn.partitions(list(Daun3sp, sigVDA24$ENTREZID))
over3_24daun <- list324venn$..values..[[1]]
#
# over3_24gost <- gost(query = over3_24daun,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(over3_24gost, "data/DEG-GO/over3_24gost.RDS")
over3_24gost <- readRDS("data/DEG-GO/over3_24gost.RDS")
# DA3_24table <- gostplot(over3_24gost, capped = FALSE, interactive = TRUE)
# DA3_24table
plDA3_24table <- over3_24gost$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
plDA3_24table %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "Overlap of 3hr and 24 hour DNR sigDEGs and all GO:BP and KEGG pathways") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 55 | 1995 | 1.12e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 55 | 1914 | 1.12e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 27 | 740 | 1.14e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 70 | 2845 | 1.25e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 67 | 2684 | 1.25e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 67 | 2683 | 1.25e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 65 | 2593 | 1.25e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 67 | 2714 | 1.25e-01 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 2 | 3 | 1.25e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 65 | 2576 | 1.25e-01 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 1.25e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 65 | 2574 | 1.25e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 35 | 1119 | 1.44e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 32 | 1015 | 1.74e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 32 | 1017 | 1.74e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.74e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 3 | 108 | 1.74e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 71 | 3080 | 1.74e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 3 | 46 | 1.74e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 73 | 3080 | 1.74e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 1.74e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 3 | 17 | 1.74e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 2 | 143 | 1.74e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 2 | 10 | 1.74e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 35 | 1209 | 1.74e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 1.74e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.74e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 1.74e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 70 | 3012 | 1.74e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 3 | 14 | 1.74e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.74e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 1.74e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 32 | 1026 | 1.74e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 1.74e-01 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 1.74e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 1.74e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 2 | 19 | 1.77e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 2 | 19 | 1.77e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.77e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 1.77e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 1.77e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 1 | 2 | 1.91e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 3 | 30 | 1.95e-01 |
| GO:BP | GO:0000303 | response to superoxide | 2 | 21 | 1.95e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 2 | 182 | 1.95e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 34 | 1214 | 1.95e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 2 | 22 | 1.95e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 1.95e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 1.95e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 2 | 13 | 1.95e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 71 | 3169 | 1.95e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 1.95e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 2 | 13 | 1.95e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 3 | 30 | 1.95e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 2 | 14 | 1.95e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 10 | 3132 | 1.95e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 2 | 195 | 1.95e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 10 | 3133 | 1.95e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 1.95e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 1 | 5 | 1.95e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 1.95e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 11 | 1.95e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 4 | 67 | 1.95e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 3 | 23 | 1.95e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 2 | 28 | 1.95e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 7 | 158 | 1.95e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 1.95e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 10 | 3237 | 1.95e-01 |
| GO:BP | GO:0045819 | positive regulation of glycogen catabolic process | 1 | 1 | 1.95e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 1.95e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 2 | 17 | 1.95e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 1.95e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 39 | 1564 | 1.95e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 1 | 4 | 1.95e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 34 | 1239 | 1.95e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 1.95e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 3 | 20 | 1.95e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 3 | 39 | 1.95e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 44 | 1735 | 1.95e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 3 | 42 | 1.95e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 1 | 7 | 1.95e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 1 | 13 | 1.95e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 3 | 156 | 1.95e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 3 | 39 | 1.95e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 1 | 16 | 1.95e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 1.95e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 2 | 14 | 1.95e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 1.99e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 1.99e-01 |
| GO:BP | GO:0090130 | tissue migration | 2 | 256 | 1.99e-01 |
| GO:BP | GO:0090132 | epithelium migration | 2 | 252 | 1.99e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 1 | 3 | 1.99e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 1 | 7 | 1.99e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 1.99e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 1.99e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 1.99e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 2 | 252 | 1.99e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 1.99e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 70 | 3070 | 1.99e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 1 | 2 | 2.02e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 1 | 8 | 2.02e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 2.02e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 90 | 4174 | 2.02e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 2.02e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 2.03e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 2.03e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 1 | 22 | 2.03e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 2 | 14 | 2.04e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 2 | 17 | 2.06e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 39 | 1604 | 2.08e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 34 | 1280 | 2.08e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 2.09e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 2.10e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 2 | 14 | 2.15e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 2 | 7 | 2.15e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 2.15e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 2.15e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 1 | 2 | 2.15e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 1 | 8 | 2.15e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 1 | 4 | 2.15e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 2.15e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 1 | 4 | 2.15e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 2.15e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 2.15e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 1 | 4 | 2.15e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 1 | 3 | 2.15e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 6 | 172 | 2.15e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 1 | 8 | 2.15e-01 |
| GO:BP | GO:0021570 | rhombomere 4 development | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 2 | 14 | 2.15e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 2.15e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 2.15e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 1 | 12 | 2.15e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 1 | 40 | 2.15e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 1 | 5 | 2.15e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 34 | 1507 | 2.15e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 2 | 11 | 2.15e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 2.15e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 3 | 25 | 2.15e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 1 | 3 | 2.15e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 2 | 17 | 2.15e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 2 | 16 | 2.15e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 5 | 89 | 2.15e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 2.15e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 1 | 3 | 2.15e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 2.15e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 1 | 5 | 2.15e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 1 | 4 | 2.15e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 1 | 4 | 2.15e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 2 | 26 | 2.15e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 1 | 14 | 2.15e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 5 | 123 | 2.16e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 7 | 165 | 2.17e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 2 | 9 | 2.20e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 1 | 4 | 2.23e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 3 | 46 | 2.30e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 2 | 358 | 2.30e-01 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 2 | 17 | 2.32e-01 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 2 | 17 | 2.32e-01 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 2 | 17 | 2.32e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 3 | 45 | 2.32e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 2.34e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 2 | 21 | 2.34e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 2.34e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 1 | 53 | 2.36e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 2.38e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 10 | 3750 | 2.40e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 2 | 14 | 2.40e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 1 | 6 | 2.40e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 1 | 2 | 2.40e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 1 | 3 | 2.40e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 1 | 5 | 2.40e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 5 | 203 | 2.40e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 2.40e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 3 | 121 | 2.40e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 1 | 6 | 2.40e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 3 | 53 | 2.40e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 2.41e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 79 | 3730 | 2.41e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 2 | 18 | 2.41e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 1 | 9 | 2.41e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 2 | 36 | 2.41e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 2 | 16 | 2.41e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 2.41e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 6 | 227 | 2.41e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 6 | 258 | 2.41e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 4 | 153 | 2.41e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 1 | 4 | 2.41e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 34 | 1315 | 2.41e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 1 | 6 | 2.41e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 1 | 9 | 2.41e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 2.41e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 34 | 1315 | 2.41e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 2 | 8 | 2.41e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 2 | 17 | 2.41e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 2.41e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 2 | 110 | 2.41e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 5 | 149 | 2.41e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 2.41e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 2.41e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 2.41e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 2.41e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 1 | 8 | 2.41e-01 |
| GO:BP | GO:0097498 | endothelial tube lumen extension | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 4 | 89 | 2.43e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 8 | 706 | 2.43e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 34 | 1322 | 2.43e-01 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 4 | 240 | 2.43e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 1 | 10 | 2.43e-01 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 2.44e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 2.44e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 1 | 20 | 2.44e-01 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 2.45e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 4 | 74 | 2.48e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 1 | 13 | 2.49e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 1 | 10 | 2.50e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 2 | 45 | 2.50e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 2 | 35 | 2.50e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 2.50e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 1 | 8 | 2.50e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 1 | 5 | 2.53e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 2.53e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 2 | 12 | 2.53e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 2.53e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 20 | 2.53e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 1 | 6 | 2.53e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 1 | 12 | 2.53e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 4 | 84 | 2.53e-01 |
| GO:BP | GO:0071464 | cellular response to hydrostatic pressure | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 2.53e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 1 | 10 | 2.53e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 2 | 22 | 2.53e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 2 | 135 | 2.53e-01 |
| GO:BP | GO:0006482 | protein demethylation | 2 | 27 | 2.53e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 2.53e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 1 | 6 | 2.53e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 1 | 16 | 2.53e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 2.53e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 39 | 2270 | 2.53e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 1 | 6 | 2.53e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 1 | 6 | 2.53e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 1 | 22 | 2.53e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 2 | 13 | 2.53e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 2 | 26 | 2.53e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 3 | 64 | 2.53e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 2 | 35 | 2.53e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 2.53e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 1 | 10 | 2.53e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 1 | 6 | 2.53e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 2 | 27 | 2.53e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 3 | 41 | 2.53e-01 |
| GO:BP | GO:0044029 | positive regulation of gene expression via CpG island demethylation | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 1 | 5 | 2.53e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 2.53e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 4 | 76 | 2.53e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0061642 | chemoattraction of axon | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 3 | 64 | 2.53e-01 |
| GO:BP | GO:0003129 | heart induction | 1 | 9 | 2.53e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 4 | 236 | 2.53e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 5 | 128 | 2.53e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 1 | 15 | 2.53e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 2 | 80 | 2.53e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 1 | 6 | 2.53e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 2.53e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 3 | 598 | 2.53e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 2.53e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 1 | 8 | 2.53e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 2.53e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 1 | 44 | 2.53e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 4 | 227 | 2.53e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.53e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 32 | 2.54e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 9 | 280 | 2.54e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 2 | 43 | 2.54e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 1 | 20 | 2.54e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 1 | 10 | 2.54e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 2.54e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4 | 100 | 2.54e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 2.54e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 8 | 742 | 2.54e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 1 | 3 | 2.54e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 5 | 129 | 2.54e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 2 | 37 | 2.54e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 1 | 26 | 2.54e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 4 | 103 | 2.54e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 2 | 29 | 2.54e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 1 | 4 | 2.54e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 1 | 28 | 2.54e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 1 | 4 | 2.54e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 6 | 147 | 2.55e-01 |
| GO:BP | GO:0030903 | notochord development | 2 | 17 | 2.55e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 11 | 344 | 2.55e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 2 | 26 | 2.55e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 2 | 55 | 2.57e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 2.57e-01 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 2.57e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 8 | 2.57e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 2.57e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 1 | 6 | 2.57e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 3 | 120 | 2.57e-01 |
| GO:BP | GO:0006364 | rRNA processing | 5 | 221 | 2.57e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 1 | 6 | 2.57e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 31 | 1441 | 2.57e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 1 | 6 | 2.57e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 2 | 34 | 2.57e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 1 | 13 | 2.57e-01 |
| GO:BP | GO:0048265 | response to pain | 1 | 17 | 2.59e-01 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 5 | 2.59e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 4 | 160 | 2.59e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 2.59e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 2.59e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 2.59e-01 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 1 | 3 | 2.59e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 3 | 50 | 2.60e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 1 | 4 | 2.60e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 1 | 17 | 2.60e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 2 | 45 | 2.65e-01 |
| GO:BP | GO:0072739 | response to anisomycin | 1 | 4 | 2.65e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 12 | 2.65e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 2.65e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 2.65e-01 |
| GO:BP | GO:0008584 | male gonad development | 1 | 99 | 2.67e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 2.67e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 1 | 5 | 2.67e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 1 | 3 | 2.67e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1 | 72 | 2.67e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 12 | 2.67e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 1 | 6 | 2.67e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 2 | 23 | 2.67e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 1 | 100 | 2.67e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 1 | 5 | 2.67e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 1 | 25 | 2.67e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 1 | 24 | 2.69e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 1 | 14 | 2.70e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 2.70e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 1 | 25 | 2.70e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 2.70e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 1 | 2 | 2.70e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 2 | 16 | 2.70e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 2 | 29 | 2.70e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 2 | 29 | 2.70e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 7 | 2.70e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 2.70e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 3 | 2.70e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 2 | 28 | 2.70e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 2 | 32 | 2.70e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 2.70e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 2.70e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 5 | 119 | 2.70e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 2.70e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 8 | 763 | 2.70e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 2.70e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 3 | 95 | 2.71e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 5 | 152 | 2.71e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 2 | 34 | 2.71e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 2.71e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 10 | 2.71e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 4 | 717 | 2.71e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 2.71e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 4 | 293 | 2.71e-01 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 2 | 28 | 2.71e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 3 | 59 | 2.71e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 2 | 29 | 2.71e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 1 | 21 | 2.71e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 25 | 2.71e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 4 | 265 | 2.71e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 2 | 67 | 2.71e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 1 | 67 | 2.71e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 2.71e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 2.71e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 2.71e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 3 | 70 | 2.71e-01 |
| GO:BP | GO:0060047 | heart contraction | 6 | 205 | 2.71e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 1 | 114 | 2.71e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 2.72e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 3 | 73 | 2.73e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 2.74e-01 |
| GO:BP | GO:0001759 | organ induction | 2 | 17 | 2.74e-01 |
| GO:BP | GO:0030168 | platelet activation | 5 | 91 | 2.74e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 1 | 13 | 2.74e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 1 | 5 | 2.74e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 10 | 4245 | 2.74e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 1 | 4 | 2.74e-01 |
| GO:BP | GO:0003128 | heart field specification | 1 | 16 | 2.74e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 45 | 4539 | 2.74e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 4 | 2.74e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 1 | 9 | 2.74e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 1 | 51 | 2.74e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 1 | 11 | 2.74e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 1 | 16 | 2.74e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 4 | 126 | 2.74e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 1 | 9 | 2.74e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 1 | 9 | 2.74e-01 |
| GO:BP | GO:0051168 | nuclear export | 3 | 150 | 2.74e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 25 | 2.75e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 3 | 68 | 2.75e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 1 | 10 | 2.79e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 2.79e-01 |
| GO:BP | GO:0090398 | cellular senescence | 4 | 83 | 2.79e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 2 | 22 | 2.79e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 1 | 28 | 2.80e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 2 | 73 | 2.80e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 1 | 4 | 2.80e-01 |
| GO:BP | GO:0021546 | rhombomere development | 1 | 4 | 2.80e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 2 | 19 | 2.80e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 3 | 71 | 2.80e-01 |
| GO:BP | GO:0035295 | tube development | 7 | 821 | 2.80e-01 |
| GO:BP | GO:0070848 | response to growth factor | 12 | 572 | 2.81e-01 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 1 | 7 | 2.81e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 1 | 15 | 2.82e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 2 | 26 | 2.82e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 2 | 22 | 2.82e-01 |
| GO:BP | GO:0042311 | vasodilation | 1 | 39 | 2.82e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 2 | 31 | 2.82e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 4 | 112 | 2.82e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 2.82e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 1 | 5 | 2.82e-01 |
| GO:BP | GO:2000653 | regulation of genetic imprinting | 1 | 4 | 2.82e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 2 | 31 | 2.82e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 2.85e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 3 | 50 | 2.85e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 1 | 34 | 2.85e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 1 | 4 | 2.87e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 4 | 164 | 2.87e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 2.87e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 1 | 30 | 2.87e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 7 | 464 | 2.87e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 2 | 76 | 2.87e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 12 | 4590 | 2.87e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 1 | 16 | 2.87e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 1 | 19 | 2.89e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 5 | 373 | 2.90e-01 |
| GO:BP | GO:0003015 | heart process | 6 | 216 | 2.90e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 1 | 30 | 2.90e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 2 | 23 | 2.90e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 1 | 6 | 2.90e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 1 | 94 | 2.90e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 2 | 86 | 2.90e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 2 | 65 | 2.91e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 5 | 277 | 2.91e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 1 | 11 | 2.91e-01 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 2.91e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 1 | 11 | 2.91e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 2.91e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 42 | 1849 | 2.91e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 2.91e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 4 | 787 | 2.91e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 2 | 34 | 2.91e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 1 | 7 | 2.91e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 2.91e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 2 | 44 | 2.91e-01 |
| GO:BP | GO:0007431 | salivary gland development | 1 | 26 | 2.91e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 2 | 45 | 2.91e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 1 | 7 | 2.91e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 1 | 11 | 2.91e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 2 | 51 | 2.91e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 2 | 36 | 2.91e-01 |
| GO:BP | GO:0008033 | tRNA processing | 1 | 128 | 2.91e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 2 | 33 | 2.91e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 2.91e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 1 | 3 | 2.91e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 15 | 534 | 2.91e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 1 | 30 | 2.91e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 3 | 80 | 2.91e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 36 | 2.91e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 2 | 27 | 2.91e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 2.91e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 1 | 30 | 2.91e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 1 | 11 | 2.91e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 1 | 13 | 2.91e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 1 | 19 | 2.91e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 2.91e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 2.91e-01 |
| GO:BP | GO:0008406 | gonad development | 1 | 158 | 2.91e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 2.91e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 2.91e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 1 | 16 | 2.91e-01 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 1 | 14 | 2.91e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 1 | 17 | 2.91e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 1 | 35 | 2.91e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 1 | 43 | 2.93e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 1 | 161 | 2.95e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 2 | 56 | 2.95e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 2 | 29 | 2.96e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 2.96e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 2 | 25 | 2.97e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 2 | 25 | 2.97e-01 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 2.97e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 1 | 97 | 2.97e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 1 | 5 | 2.98e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 1 | 8 | 2.99e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 1 | 7 | 2.99e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 87 | 4292 | 3.00e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 1 | 7 | 3.00e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 1 | 6 | 3.00e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 1 | 15 | 3.00e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 3.00e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1 | 18 | 3.00e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 1 | 100 | 3.00e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 1 | 47 | 3.00e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 1 | 2 | 3.00e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 3.00e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 1 | 7 | 3.00e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 1 | 38 | 3.00e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 2 | 36 | 3.00e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 3.00e-01 |
| GO:BP | GO:0048565 | digestive tract development | 2 | 92 | 3.00e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 3.00e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 1 | 10 | 3.02e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 3.03e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 1 | 5 | 3.03e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 1 | 7 | 3.03e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 1 | 6 | 3.03e-01 |
| GO:BP | GO:0043276 | anoikis | 2 | 32 | 3.03e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 1 | 17 | 3.03e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 1 | 16 | 3.03e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 3.03e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 2 | 1602 | 3.03e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 2 | 60 | 3.03e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 2 | 56 | 3.03e-01 |
| GO:BP | GO:0007548 | sex differentiation | 1 | 191 | 3.03e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 4 | 295 | 3.03e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 1 | 30 | 3.03e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 15 | 551 | 3.03e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 1 | 9 | 3.03e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 3 | 121 | 3.03e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 3.03e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 2 | 50 | 3.03e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 1 | 20 | 3.03e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 2 | 73 | 3.03e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 13 | 555 | 3.03e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 3.03e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 1 | 6 | 3.03e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 1 | 15 | 3.03e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 11 | 3580 | 3.03e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 1 | 8 | 3.03e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 3.03e-01 |
| GO:BP | GO:0021561 | facial nerve development | 1 | 8 | 3.03e-01 |
| GO:BP | GO:0048232 | male gamete generation | 2 | 343 | 3.03e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 3.03e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 1 | 6 | 3.03e-01 |
| GO:BP | GO:0040011 | locomotion | 9 | 954 | 3.03e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 3 | 108 | 3.03e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 1 | 42 | 3.03e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 2 | 226 | 3.03e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 1 | 22 | 3.03e-01 |
| GO:BP | GO:0051169 | nuclear transport | 4 | 295 | 3.03e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 3.03e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 2 | 76 | 3.03e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 1 | 13 | 3.03e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 3 | 89 | 3.03e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 3.03e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 2 | 40 | 3.03e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 3.03e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 2 | 59 | 3.03e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 3.03e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 2 | 330 | 3.03e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 2 | 73 | 3.03e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 3.03e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 2 | 50 | 3.03e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 1 | 37 | 3.03e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 2 | 55 | 3.03e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 3.03e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 3 | 59 | 3.03e-01 |
| GO:BP | GO:0000423 | mitophagy | 1 | 37 | 3.03e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 1 | 5 | 3.03e-01 |
| GO:BP | GO:0001843 | neural tube closure | 1 | 90 | 3.03e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 1 | 43 | 3.03e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 1 | 6 | 3.03e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 1 | 13 | 3.03e-01 |
| GO:BP | GO:0060606 | tube closure | 1 | 91 | 3.03e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 1 | 136 | 3.03e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 1 | 17 | 3.03e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 3.03e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 1 | 40 | 3.03e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 2 | 78 | 3.03e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 3.03e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 3.03e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 89 | 4417 | 3.03e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 3 | 57 | 3.03e-01 |
| GO:BP | GO:0048645 | animal organ formation | 3 | 51 | 3.03e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 1 | 20 | 3.04e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 2 | 41 | 3.04e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 3 | 91 | 3.04e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 3.04e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 3.04e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 1 | 94 | 3.04e-01 |
| GO:BP | GO:0035640 | exploration behavior | 1 | 21 | 3.04e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 2 | 85 | 3.04e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 1 | 11 | 3.04e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 10 | 316 | 3.04e-01 |
| GO:BP | GO:0060914 | heart formation | 1 | 29 | 3.04e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 1 | 132 | 3.04e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 1 | 11 | 3.04e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 1 | 21 | 3.04e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 5 | 140 | 3.04e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 2 | 55 | 3.04e-01 |
| GO:BP | GO:0061458 | reproductive system development | 1 | 215 | 3.04e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 3.04e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 3.04e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 2 | 26 | 3.04e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 3.04e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 5 | 216 | 3.04e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 3.04e-01 |
| GO:BP | GO:0035601 | protein deacylation | 3 | 116 | 3.04e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 1 | 23 | 3.04e-01 |
| GO:BP | GO:0006541 | glutamine metabolic process | 1 | 24 | 3.04e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 3.04e-01 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 1 | 16 | 3.04e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 1 | 33 | 3.04e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 1 | 15 | 3.04e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 2 | 32 | 3.04e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 3.04e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 38 | 2412 | 3.04e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 3.04e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 3.04e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 18 | 3.04e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 1 | 7 | 3.04e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 1 | 212 | 3.04e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 1 | 185 | 3.04e-01 |
| GO:BP | GO:0055123 | digestive system development | 2 | 100 | 3.04e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 6 | 179 | 3.04e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 3 | 120 | 3.04e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 3.04e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 2 | 37 | 3.04e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 1 | 45 | 3.04e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 7 | 234 | 3.04e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 3.04e-01 |
| GO:BP | GO:0035272 | exocrine system development | 1 | 34 | 3.04e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 3.04e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 2 | 41 | 3.04e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 1 | 7 | 3.04e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 2 | 19 | 3.04e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 1 | 8 | 3.04e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 3.04e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.04e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 3.04e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 1 | 8 | 3.04e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 3.04e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 2 | 29 | 3.04e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 1 | 16 | 3.06e-01 |
| GO:BP | GO:0001841 | neural tube formation | 1 | 101 | 3.07e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 1 | 7 | 3.07e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 3.08e-01 |
| GO:BP | GO:0022414 | reproductive process | 3 | 927 | 3.08e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 3 | 61 | 3.08e-01 |
| GO:BP | GO:0006983 | ER overload response | 1 | 12 | 3.08e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 2 | 34 | 3.08e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 1 | 25 | 3.08e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 2 | 88 | 3.08e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 3.08e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 4 | 352 | 3.08e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 6 | 189 | 3.09e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 3.09e-01 |
| GO:BP | GO:0000003 | reproduction | 3 | 934 | 3.09e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 2 | 82 | 3.09e-01 |
| GO:BP | GO:0001756 | somitogenesis | 3 | 48 | 3.09e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 1 | 186 | 3.09e-01 |
| GO:BP | GO:0031128 | developmental induction | 1 | 26 | 3.09e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 2 | 31 | 3.09e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 1 | 26 | 3.09e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 3 | 43 | 3.10e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 4 | 234 | 3.10e-01 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 3.12e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 3.13e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 2 | 36 | 3.13e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 10 | 3.13e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 1 | 8 | 3.14e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 3 | 119 | 3.14e-01 |
| GO:BP | GO:0003097 | renal water transport | 1 | 6 | 3.14e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 3.14e-01 |
| GO:BP | GO:0019083 | viral transcription | 2 | 48 | 3.14e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 3 | 88 | 3.14e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 1 | 54 | 3.14e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 5 | 3.14e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 4 | 898 | 3.14e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 1 | 4 | 3.14e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 3.14e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 1 | 13 | 3.14e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 3.14e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 2 | 25 | 3.14e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 25 | 3.14e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 3.14e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 1 | 26 | 3.15e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 3.15e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 5 | 462 | 3.16e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 1 | 10 | 3.16e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 1 | 10 | 3.16e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 4 | 260 | 3.16e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 7 | 237 | 3.19e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 1 | 16 | 3.19e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 3 | 54 | 3.20e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 3.20e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 1 | 23 | 3.20e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 3.20e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 2 | 58 | 3.20e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 1 | 22 | 3.20e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 1 | 18 | 3.20e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 1 | 57 | 3.21e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 1 | 80 | 3.21e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 2 | 133 | 3.21e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 1 | 7 | 3.21e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 1 | 2 | 3.21e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 1 | 7 | 3.21e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 1 | 46 | 3.21e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 3.22e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 3.22e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 1 | 4 | 3.22e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 3 | 61 | 3.23e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1 | 9 | 3.24e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 3.24e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 1 | 19 | 3.24e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 1 | 55 | 3.24e-01 |
| GO:BP | GO:0042148 | strand invasion | 1 | 5 | 3.24e-01 |
| GO:BP | GO:0048538 | thymus development | 2 | 35 | 3.25e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 2 | 49 | 3.26e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 3 | 427 | 3.26e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 1 | 15 | 3.26e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 4 | 116 | 3.26e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 4 | 117 | 3.27e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 36 | 3.27e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 2 | 62 | 3.28e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 3.28e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 9 | 400 | 3.28e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2 | 26 | 3.28e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 1 | 7 | 3.28e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 1 | 4 | 3.29e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 7 | 3.29e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 4 | 101 | 3.29e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 3.29e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 1 | 57 | 3.29e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 1 | 68 | 3.29e-01 |
| GO:BP | GO:0097484 | dendrite extension | 1 | 27 | 3.29e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 1 | 20 | 3.29e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 3 | 67 | 3.29e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 3.29e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 3.29e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 3.29e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 5 | 161 | 3.29e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 3.29e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 3 | 220 | 3.29e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 5 | 301 | 3.31e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 2 | 378 | 3.32e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 2 | 41 | 3.32e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 3 | 75 | 3.32e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 1 | 14 | 3.34e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 1 | 9 | 3.34e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 3 | 126 | 3.35e-01 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 1 | 7 | 3.35e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 3.35e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 6 | 494 | 3.35e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 1 | 22 | 3.35e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 3.35e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 3.35e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 3.36e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 3.36e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 3.36e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 3.36e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 1 | 8 | 3.36e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 35 | 3.37e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 3 | 69 | 3.37e-01 |
| GO:BP | GO:0098754 | detoxification | 2 | 91 | 3.37e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 2 | 42 | 3.37e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 1 | 211 | 3.37e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 2 | 27 | 3.37e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 2 | 34 | 3.37e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 1 | 64 | 3.37e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 14 | 616 | 3.38e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 11 | 438 | 3.38e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 1 | 98 | 3.38e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 1 | 127 | 3.38e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0007276 | gamete generation | 2 | 461 | 3.38e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 1 | 37 | 3.38e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 1 | 11 | 3.38e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 6 | 227 | 3.38e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 1 | 172 | 3.38e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 5 | 381 | 3.38e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 6 | 194 | 3.38e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 1 | 16 | 3.38e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 3.38e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 7 | 249 | 3.38e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 3.38e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 3.38e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 3.39e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 1 | 82 | 3.39e-01 |
| GO:BP | GO:0050890 | cognition | 3 | 227 | 3.39e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 1 | 12 | 3.40e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 8 | 3.40e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 5 | 179 | 3.40e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 1 | 8 | 3.40e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 1 | 7 | 3.40e-01 |
| GO:BP | GO:0007596 | blood coagulation | 6 | 150 | 3.40e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 1 | 68 | 3.40e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 8 | 278 | 3.40e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 3.40e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 1 | 14 | 3.40e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 1 | 13 | 3.40e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 1 | 7 | 3.40e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 3.40e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 3.40e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 1 | 23 | 3.40e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 8 | 3.40e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 2 | 62 | 3.40e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 1 | 127 | 3.40e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 13 | 504 | 3.40e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 1 | 21 | 3.40e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 2 | 31 | 3.40e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 3.40e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 3.40e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 39 | 3.40e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 2 | 67 | 3.40e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 6 | 663 | 3.40e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 3.40e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 1 | 8 | 3.40e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 3.40e-01 |
| GO:BP | GO:0016477 | cell migration | 9 | 1081 | 3.40e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 3.40e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 1 | 13 | 3.40e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 3.40e-01 |
| GO:BP | GO:0035196 | miRNA processing | 2 | 41 | 3.40e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 1 | 61 | 3.40e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 1 | 51 | 3.41e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 3.41e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 1 | 23 | 3.41e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 3.41e-01 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 1 | 9 | 3.41e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 3.41e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 1 | 40 | 3.41e-01 |
| GO:BP | GO:0035148 | tube formation | 1 | 138 | 3.41e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 3.41e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 1 | 21 | 3.41e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 1 | 132 | 3.42e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 1 | 17 | 3.42e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 2 | 107 | 3.42e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 3.42e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 1 | 6 | 3.42e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 3 | 55 | 3.42e-01 |
| GO:BP | GO:0070509 | calcium ion import | 1 | 37 | 3.42e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 1 | 179 | 3.42e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 1 | 14 | 3.42e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 1 | 9 | 3.42e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 1 | 9 | 3.42e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 3 | 81 | 3.42e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 1 | 9 | 3.42e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 1 | 41 | 3.42e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 1 | 68 | 3.42e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 41 | 2646 | 3.43e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 3.43e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 1 | 4 | 3.45e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 4 | 78 | 3.45e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 2 | 265 | 3.45e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 1 | 12 | 3.45e-01 |
| GO:BP | GO:0050817 | coagulation | 6 | 153 | 3.45e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 1 | 294 | 3.45e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 1 | 12 | 3.45e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 3.45e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 1 | 14 | 3.45e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 3 | 71 | 3.45e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 1 | 9 | 3.45e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 3.45e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 6 | 230 | 3.45e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 3.45e-01 |
| GO:BP | GO:0021537 | telencephalon development | 4 | 201 | 3.45e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 3 | 111 | 3.45e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 3 | 221 | 3.45e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 1 | 28 | 3.45e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 3.45e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 2 | 50 | 3.45e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 3.45e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 2 | 185 | 3.45e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 1 | 41 | 3.45e-01 |
| GO:BP | GO:0007599 | hemostasis | 6 | 153 | 3.45e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 2 | 29 | 3.45e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 2 | 34 | 3.45e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 1 | 25 | 3.45e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 3.45e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 1 | 137 | 3.46e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 1 | 96 | 3.47e-01 |
| GO:BP | GO:0001824 | blastocyst development | 3 | 94 | 3.47e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 1 | 12 | 3.47e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 1 | 48 | 3.47e-01 |
| GO:BP | GO:0021915 | neural tube development | 1 | 146 | 3.47e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 1 | 50 | 3.47e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 3.47e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 2 | 38 | 3.48e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 1 | 34 | 3.48e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 4 | 123 | 3.48e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 44 | 2880 | 3.50e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 1 | 7 | 3.50e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 1 | 7 | 3.50e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 15 | 3.50e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 1 | 22 | 3.50e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 1 | 24 | 3.50e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 11 | 418 | 3.51e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 2 | 73 | 3.52e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 1 | 16 | 3.52e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 1 | 28 | 3.53e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 1 | 12 | 3.53e-01 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 1 | 9 | 3.53e-01 |
| GO:BP | GO:0051098 | regulation of binding | 4 | 317 | 3.53e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 1 | 7 | 3.54e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 1 | 49 | 3.54e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 1 | 10 | 3.54e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 1 | 14 | 3.54e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 3.54e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 1 | 10 | 3.54e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 1 | 10 | 3.54e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 6 | 262 | 3.54e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 1 | 13 | 3.55e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 15 | 3.55e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 3.56e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 1 | 31 | 3.57e-01 |
| GO:BP | GO:0051642 | centrosome localization | 1 | 31 | 3.57e-01 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 1 | 14 | 3.57e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 35 | 3.60e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 2 | 31 | 3.60e-01 |
| GO:BP | GO:0051028 | mRNA transport | 2 | 117 | 3.60e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 6 | 204 | 3.62e-01 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 1 | 11 | 3.63e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 3 | 78 | 3.64e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 2 | 31 | 3.64e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 2 | 538 | 3.64e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 3.64e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 3.64e-01 |
| GO:BP | GO:0001568 | blood vessel development | 5 | 534 | 3.65e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 1 | 55 | 3.65e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 2 | 104 | 3.65e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 1 | 7 | 3.65e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 2 | 76 | 3.65e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 3.65e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 7 | 3.65e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 1 | 18 | 3.65e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 1 | 13 | 3.65e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 99 | 3.66e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 99 | 3.66e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 1 | 16 | 3.66e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 1 | 16 | 3.66e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 1 | 34 | 3.66e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 3.66e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 1 | 8 | 3.66e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 1 | 44 | 3.66e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 3.66e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 1 | 16 | 3.66e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 1 | 11 | 3.66e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 2 | 37 | 3.66e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 3.66e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 2 | 36 | 3.66e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 2 | 204 | 3.66e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 1 | 52 | 3.66e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 99 | 3.66e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 3.67e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 3.67e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 1 | 33 | 3.67e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 1 | 16 | 3.67e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 1 | 18 | 3.67e-01 |
| GO:BP | GO:0043201 | response to leucine | 1 | 13 | 3.67e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 3.67e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 1 | 36 | 3.67e-01 |
| GO:BP | GO:0044057 | regulation of system process | 4 | 405 | 3.67e-01 |
| GO:BP | GO:0048864 | stem cell development | 3 | 76 | 3.67e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 3 | 81 | 3.67e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 10 | 551 | 3.67e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 1 | 9 | 3.67e-01 |
| GO:BP | GO:0030900 | forebrain development | 5 | 281 | 3.68e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 2 | 28 | 3.68e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 3 | 84 | 3.68e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 6 | 414 | 3.68e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 17 | 3.68e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 3.68e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 2 | 40 | 3.68e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 2 | 115 | 3.68e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 3.68e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 1 | 30 | 3.68e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 2 | 115 | 3.68e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 4 | 131 | 3.68e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 3.68e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 5 | 256 | 3.68e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 11 | 561 | 3.68e-01 |
| GO:BP | GO:0070988 | demethylation | 2 | 52 | 3.68e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 5 | 207 | 3.68e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 1 | 90 | 3.68e-01 |
| GO:BP | GO:0014823 | response to activity | 2 | 46 | 3.68e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 2 | 58 | 3.68e-01 |
| GO:BP | GO:0061053 | somite development | 3 | 63 | 3.68e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 3 | 85 | 3.68e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 1 | 39 | 3.68e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 1 | 15 | 3.68e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 3.68e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 2 | 556 | 3.69e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 3 | 4617 | 3.69e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 3.69e-01 |
| GO:BP | GO:0055057 | neuroblast division | 1 | 12 | 3.69e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 6 | 218 | 3.69e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 1 | 164 | 3.69e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 3 | 78 | 3.69e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 5 | 406 | 3.69e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 11 | 3.70e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 3 | 91 | 3.70e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 2 | 55 | 3.70e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 77 | 3.70e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 3.70e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 3.70e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 3.70e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 2 | 39 | 3.70e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 12 | 3.70e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 1 | 14 | 3.71e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 17 | 875 | 3.71e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 1 | 13 | 3.71e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 7 | 252 | 3.71e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 1 | 382 | 3.71e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 1 | 18 | 3.71e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 3.71e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 1 | 19 | 3.71e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 1 | 20 | 3.71e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 3.72e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 1 | 11 | 3.72e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 7 | 272 | 3.72e-01 |
| GO:BP | GO:0001525 | angiogenesis | 4 | 390 | 3.72e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 1 | 12 | 3.73e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 1 | 36 | 3.74e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 1 | 10 | 3.75e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 2 | 37 | 3.75e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 1 | 90 | 3.75e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 1 | 12 | 3.75e-01 |
| GO:BP | GO:0001944 | vasculature development | 5 | 559 | 3.78e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 3.78e-01 |
| GO:BP | GO:0061912 | selective autophagy | 1 | 90 | 3.78e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 1 | 20 | 3.79e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 7 | 691 | 3.79e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 1 | 36 | 3.79e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 1 | 12 | 3.79e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 3.79e-01 |
| GO:BP | GO:0006325 | chromatin organization | 9 | 551 | 3.79e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 3 | 3600 | 3.79e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 3.79e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 3.79e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 1 | 23 | 3.79e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 1 | 15 | 3.79e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 1 | 92 | 3.79e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 1 | 16 | 3.79e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 1 | 15 | 3.79e-01 |
| GO:BP | GO:0001867 | complement activation, lectin pathway | 1 | 5 | 3.79e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 1 | 92 | 3.79e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 1 | 44 | 3.79e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 3.79e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 3.79e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 1 | 10 | 3.79e-01 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 1 | 8 | 3.80e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 1 | 11 | 3.80e-01 |
| GO:BP | GO:0046688 | response to copper ion | 1 | 29 | 3.81e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 2 | 136 | 3.81e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 1 | 34 | 3.81e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 2 | 52 | 3.85e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 3.85e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 2 | 133 | 3.86e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 4 | 160 | 3.87e-01 |
| GO:BP | GO:0003170 | heart valve development | 1 | 65 | 3.87e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 5 | 155 | 3.87e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 3.87e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 1 | 22 | 3.87e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 1 | 36 | 3.87e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 3.87e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 3.87e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 3.87e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 3.87e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 1 | 51 | 3.87e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 3.88e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 1 | 14 | 3.88e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 5 | 560 | 3.88e-01 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 3.88e-01 |
| GO:BP | GO:0006414 | translational elongation | 1 | 65 | 3.88e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 3.88e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 1 | 16 | 3.88e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 172 | 3.88e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 3 | 80 | 3.88e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 1 | 38 | 3.89e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 1 | 28 | 3.89e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 1 | 54 | 3.89e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 1 | 10 | 3.89e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 3.89e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 1 | 97 | 3.89e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 3.89e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 2 | 38 | 3.89e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 2 | 79 | 3.89e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 3.89e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 1 | 39 | 3.89e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 3 | 128 | 3.89e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 8 | 957 | 3.89e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 6 | 187 | 3.90e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 2 | 52 | 3.90e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 1 | 114 | 3.90e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 1 | 53 | 3.90e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 19 | 783 | 3.91e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 1 | 44 | 3.91e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 1 | 23 | 3.91e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 3.92e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 1 | 61 | 3.92e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 1 | 21 | 3.92e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 2 | 188 | 3.92e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 3 | 98 | 3.92e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 1 | 40 | 3.92e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 3 | 123 | 3.92e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 1 | 37 | 3.92e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 2 | 58 | 3.92e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 2 | 173 | 3.93e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 3.93e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 3.93e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 5 | 210 | 3.94e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 15 | 3.94e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 1 | 45 | 3.96e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 1 | 69 | 3.96e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 3.96e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 1 | 281 | 3.96e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 1 | 10 | 3.96e-01 |
| GO:BP | GO:0097502 | mannosylation | 1 | 32 | 3.96e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 1 | 31 | 3.96e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 18 | 1123 | 3.96e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 3 | 331 | 3.96e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 1 | 37 | 3.96e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 1 | 23 | 3.96e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 3.96e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 1 | 72 | 3.96e-01 |
| GO:BP | GO:0035188 | hatching | 1 | 23 | 3.96e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 1 | 17 | 3.96e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 1 | 50 | 3.96e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 3 | 275 | 3.96e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 1 | 46 | 3.96e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 1 | 113 | 3.96e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 1 | 23 | 3.96e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 1 | 49 | 3.96e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 2 | 631 | 3.96e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 3 | 101 | 3.97e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 1 | 21 | 3.97e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 3.98e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 86 | 3.99e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 3.99e-01 |
| GO:BP | GO:0042060 | wound healing | 9 | 320 | 3.99e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 1 | 12 | 4.00e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 1 | 36 | 4.01e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 2 | 40 | 4.01e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 2 | 105 | 4.01e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 22 | 4.01e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 1 | 16 | 4.01e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 4.02e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 4.02e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 1 | 73 | 4.02e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 1 | 13 | 4.02e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 1 | 42 | 4.02e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 1 | 21 | 4.02e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 1 | 115 | 4.02e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 1 | 10 | 4.02e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 3 | 5017 | 4.02e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 2 | 129 | 4.05e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 1 | 40 | 4.05e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 1 | 17 | 4.05e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 2 | 84 | 4.06e-01 |
| GO:BP | GO:0006821 | chloride transport | 1 | 58 | 4.06e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 3 | 99 | 4.06e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 3 | 122 | 4.06e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 2 | 72 | 4.06e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 4.07e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 4.07e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 4 | 144 | 4.07e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 1 | 48 | 4.07e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 1 | 74 | 4.07e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 6 | 276 | 4.07e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 14 | 4.07e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 1 | 11 | 4.07e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 4 | 147 | 4.07e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 1 | 18 | 4.08e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 1 | 27 | 4.09e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 1 | 27 | 4.09e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 4.09e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 3 | 282 | 4.09e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 1 | 8 | 4.09e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 1 | 33 | 4.09e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 2 | 133 | 4.09e-01 |
| GO:BP | GO:0050658 | RNA transport | 2 | 144 | 4.10e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 2 | 144 | 4.10e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 4.10e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 8 | 316 | 4.10e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 4.10e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 4.10e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 4.10e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 1 | 23 | 4.10e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 1 | 18 | 4.10e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 2 | 75 | 4.10e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 1 | 19 | 4.11e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 1 | 49 | 4.11e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 6 | 265 | 4.11e-01 |
| GO:BP | GO:0003012 | muscle system process | 3 | 334 | 4.11e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 4.12e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 1 | 19 | 4.12e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 1 | 23 | 4.12e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 2 | 65 | 4.12e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1182 | 4.12e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 1 | 59 | 4.12e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 1 | 14 | 4.12e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 2 | 47 | 4.12e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 4.12e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 1 | 14 | 4.13e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 1 | 35 | 4.13e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 1 | 22 | 4.13e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 2 | 42 | 4.13e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 7 | 284 | 4.13e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 1 | 66 | 4.13e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 2 | 147 | 4.13e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 2 | 66 | 4.13e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 4.13e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 1 | 29 | 4.13e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 16 | 660 | 4.13e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 1 | 33 | 4.13e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 1 | 67 | 4.13e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 3 | 4048 | 4.13e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 3 | 293 | 4.13e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 1 | 11 | 4.15e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 1 | 18 | 4.15e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 2 | 61 | 4.15e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 4.15e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 4.15e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 4.15e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 1 | 47 | 4.15e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 1 | 14 | 4.15e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 4.15e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 1 | 44 | 4.15e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 1 | 7 | 4.15e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 13 | 4.15e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 2 | 64 | 4.15e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 1 | 34 | 4.15e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 3 | 123 | 4.15e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 1 | 20 | 4.15e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 4.17e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 1 | 217 | 4.17e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 1 | 15 | 4.17e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 1 | 12 | 4.17e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 4.17e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 4.17e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 3 | 103 | 4.17e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 6 | 358 | 4.17e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1 | 47 | 4.17e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 4.17e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 4 | 140 | 4.17e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 2 | 52 | 4.17e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 5 | 229 | 4.18e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 1 | 15 | 4.18e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 1 | 13 | 4.18e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 1 | 21 | 4.19e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 4.20e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 1 | 42 | 4.20e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 4.20e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 4.20e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 4.20e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 4.20e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 2 | 479 | 4.21e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 4.22e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 2 | 56 | 4.22e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 4.22e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 2 | 43 | 4.22e-01 |
| GO:BP | GO:0001508 | action potential | 3 | 105 | 4.22e-01 |
| GO:BP | GO:0016577 | histone demethylation | 1 | 21 | 4.22e-01 |
| GO:BP | GO:0036211 | protein modification process | 36 | 2901 | 4.22e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 1 | 51 | 4.22e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 4.22e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 2 | 43 | 4.22e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 1 | 18 | 4.22e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 4 | 140 | 4.22e-01 |
| GO:BP | GO:0001947 | heart looping | 2 | 50 | 4.22e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 1 | 15 | 4.22e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 2 | 184 | 4.22e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 2 | 42 | 4.24e-01 |
| GO:BP | GO:0010332 | response to gamma radiation | 2 | 50 | 4.24e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 1 | 14 | 4.24e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 1 | 21 | 4.24e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 2 | 154 | 4.24e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 2 | 64 | 4.24e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 1 | 13 | 4.24e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 4.24e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 4.24e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 13 | 4.24e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 1 | 39 | 4.24e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 4.24e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 1 | 22 | 4.24e-01 |
| GO:BP | GO:0006833 | water transport | 1 | 10 | 4.24e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 3 | 425 | 4.24e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 1 | 40 | 4.24e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 1 | 64 | 4.24e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 3 | 90 | 4.26e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 2 | 192 | 4.26e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 2 | 59 | 4.26e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 2 | 295 | 4.26e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 1 | 16 | 4.28e-01 |
| GO:BP | GO:0007141 | male meiosis I | 1 | 15 | 4.28e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 1 | 16 | 4.28e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 4.28e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 1 | 20 | 4.28e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 1 | 158 | 4.29e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 1 | 21 | 4.29e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 2 | 72 | 4.29e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 6 | 267 | 4.29e-01 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 1 | 60 | 4.29e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 2 | 132 | 4.29e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 5 | 216 | 4.29e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 5 | 222 | 4.29e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 2 | 147 | 4.30e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 1 | 26 | 4.30e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 3 | 105 | 4.31e-01 |
| GO:BP | GO:0002076 | osteoblast development | 1 | 12 | 4.32e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 1 | 41 | 4.33e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 4 | 198 | 4.33e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 1 | 18 | 4.33e-01 |
| GO:BP | GO:0007530 | sex determination | 1 | 12 | 4.33e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 16 | 678 | 4.34e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 1 | 320 | 4.34e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 2 | 37 | 4.35e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 2 | 60 | 4.35e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 4.35e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 1 | 656 | 4.35e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 4 | 159 | 4.35e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 2 | 53 | 4.35e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 2 | 56 | 4.35e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 4.36e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 2 | 486 | 4.36e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 2 | 83 | 4.37e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 1 | 90 | 4.37e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 1 | 16 | 4.37e-01 |
| GO:BP | GO:0043473 | pigmentation | 2 | 92 | 4.37e-01 |
| GO:BP | GO:0032506 | cytokinetic process | 1 | 39 | 4.37e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 1 | 43 | 4.37e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 4.37e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 1 | 137 | 4.37e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 4.37e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 1 | 21 | 4.37e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 1 | 35 | 4.37e-01 |
| GO:BP | GO:0006915 | apoptotic process | 31 | 1443 | 4.37e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 172 | 4.38e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 2 | 47 | 4.38e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 3 | 150 | 4.38e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 1 | 19 | 4.38e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 1 | 8 | 4.39e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 4.39e-01 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 1 | 19 | 4.40e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 1 | 19 | 4.40e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 1 | 19 | 4.40e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 1 | 16 | 4.40e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 2 | 75 | 4.40e-01 |
| GO:BP | GO:0098657 | import into cell | 2 | 166 | 4.40e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 1 | 15 | 4.40e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 1 | 27 | 4.40e-01 |
| GO:BP | GO:0060039 | pericardium development | 1 | 17 | 4.40e-01 |
| GO:BP | GO:0019080 | viral gene expression | 1 | 96 | 4.40e-01 |
| GO:BP | GO:0071772 | response to BMP | 2 | 131 | 4.41e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 2 | 131 | 4.41e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 1 | 26 | 4.41e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 3 | 105 | 4.41e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 2 | 40 | 4.41e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 198 | 4.41e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 1 | 23 | 4.41e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 2 | 376 | 4.41e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 1 | 28 | 4.41e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 4.42e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 1 | 14 | 4.42e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 29 | 4.42e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 1 | 421 | 4.42e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 1 | 20 | 4.42e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 4.42e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 4.42e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 5 | 127 | 4.42e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 1 | 20 | 4.42e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 2 | 500 | 4.42e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 3 | 91 | 4.42e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 2 | 101 | 4.42e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 15 | 620 | 4.42e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 3 | 91 | 4.42e-01 |
| GO:BP | GO:0035282 | segmentation | 3 | 75 | 4.42e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 1 | 17 | 4.42e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 2 | 71 | 4.42e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 2 | 72 | 4.42e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 2 | 56 | 4.42e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 2 | 52 | 4.42e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 1 | 17 | 4.42e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 4.44e-01 |
| GO:BP | GO:0090659 | walking behavior | 1 | 25 | 4.45e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 3 | 4455 | 4.45e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 2 | 185 | 4.45e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2 | 56 | 4.45e-01 |
| GO:BP | GO:0021675 | nerve development | 2 | 57 | 4.45e-01 |
| GO:BP | GO:0016358 | dendrite development | 3 | 202 | 4.45e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 4.46e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 4.46e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 4.46e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 5 | 236 | 4.47e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 2 | 53 | 4.47e-01 |
| GO:BP | GO:0007498 | mesoderm development | 3 | 89 | 4.47e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 4.47e-01 |
| GO:BP | GO:0017145 | stem cell division | 1 | 26 | 4.47e-01 |
| GO:BP | GO:0032879 | regulation of localization | 11 | 1555 | 4.47e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 1 | 28 | 4.47e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 2 | 51 | 4.47e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 4.48e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 1 | 25 | 4.48e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 12 | 1199 | 4.48e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 1 | 25 | 4.48e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 2 | 54 | 4.48e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 1 | 23 | 4.48e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 1 | 15 | 4.49e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 4 | 214 | 4.49e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 4.49e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 407 | 4.49e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 4.49e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 1 | 23 | 4.49e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 4 | 141 | 4.49e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 4.49e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 2 | 45 | 4.50e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 2 | 158 | 4.50e-01 |
| GO:BP | GO:0046697 | decidualization | 1 | 19 | 4.50e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 1 | 17 | 4.50e-01 |
| GO:BP | GO:0016570 | histone modification | 6 | 433 | 4.50e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 4.50e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 1 | 38 | 4.50e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 2 | 52 | 4.50e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 2 | 52 | 4.50e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 1 | 45 | 4.51e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 2 | 107 | 4.52e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 4.52e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 3 | 94 | 4.52e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 3 | 94 | 4.52e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 4 | 378 | 4.52e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 2 | 55 | 4.52e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 4.52e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 2 | 513 | 4.52e-01 |
| GO:BP | GO:0006914 | autophagy | 2 | 513 | 4.52e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 1 | 26 | 4.52e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 1 | 27 | 4.52e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 1 | 69 | 4.52e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 1 | 16 | 4.52e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 1 | 68 | 4.52e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 3 | 107 | 4.52e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 1 | 27 | 4.52e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 2 | 150 | 4.52e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 1 | 36 | 4.53e-01 |
| GO:BP | GO:1990868 | response to chemokine | 1 | 37 | 4.53e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 1 | 37 | 4.53e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 1 | 151 | 4.53e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 4 | 155 | 4.53e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 4.53e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 2 | 63 | 4.54e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 2 | 57 | 4.54e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 3 | 4570 | 4.54e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 1 | 93 | 4.55e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 13 | 710 | 4.55e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 4.55e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 18 | 1167 | 4.55e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 4.55e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 1 | 18 | 4.55e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 1 | 70 | 4.56e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 4.56e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 4.56e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 4.57e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 1 | 155 | 4.59e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 4.59e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 4.59e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 68 | 4.59e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 1 | 17 | 4.59e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 4.59e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 4.59e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 4.59e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 4.59e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 1 | 25 | 4.59e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 2 | 78 | 4.59e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 3 | 4596 | 4.59e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 4.59e-01 |
| GO:BP | GO:0060324 | face development | 1 | 45 | 4.60e-01 |
| GO:BP | GO:0006403 | RNA localization | 2 | 182 | 4.60e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 4.60e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 4.60e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 1 | 25 | 4.60e-01 |
| GO:BP | GO:0072359 | circulatory system development | 6 | 884 | 4.61e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 48 | 4.62e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 1 | 80 | 4.62e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 2 | 251 | 4.62e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 7 | 307 | 4.62e-01 |
| GO:BP | GO:0044409 | entry into host | 2 | 113 | 4.62e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 1 | 13 | 4.62e-01 |
| GO:BP | GO:0046599 | regulation of centriole replication | 1 | 21 | 4.62e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 1 | 26 | 4.62e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 4.62e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 1 | 27 | 4.62e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 1 | 20 | 4.62e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 3 | 120 | 4.62e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 18 | 4.62e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 1 | 17 | 4.62e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 5 | 679 | 4.62e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 4.62e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 6 | 313 | 4.64e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 1 | 25 | 4.64e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 1 | 26 | 4.64e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 1 | 25 | 4.64e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 4.65e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 1 | 185 | 4.67e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 1 | 19 | 4.67e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 1 | 26 | 4.67e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 2 | 50 | 4.67e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 2 | 53 | 4.67e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 5 | 206 | 4.67e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 2 | 189 | 4.67e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 2 | 81 | 4.67e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 1 | 60 | 4.67e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 4.67e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 2 | 116 | 4.67e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 17 | 4.67e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 39 | 4.67e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 3 | 146 | 4.67e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 4.67e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 1 | 104 | 4.67e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 1 | 29 | 4.67e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 4.67e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 1 | 23 | 4.67e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 1 | 23 | 4.67e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 4 | 387 | 4.67e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 1 | 23 | 4.67e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 2 | 160 | 4.67e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 1 | 22 | 4.67e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 4.67e-01 |
| GO:BP | GO:0009798 | axis specification | 2 | 63 | 4.69e-01 |
| GO:BP | GO:0008015 | blood circulation | 3 | 374 | 4.69e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 1 | 58 | 4.69e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 4.69e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 4.69e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 2 | 56 | 4.69e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 4.69e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 2 | 165 | 4.69e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 4.69e-01 |
| GO:BP | GO:0009790 | embryo development | 19 | 875 | 4.69e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 1 | 34 | 4.69e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 3 | 122 | 4.69e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 3 | 270 | 4.69e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 4.71e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 1 | 14 | 4.71e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 24 | 4.71e-01 |
| GO:BP | GO:0001889 | liver development | 3 | 110 | 4.72e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 16 | 4.72e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 146 | 4.72e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 5 | 197 | 4.72e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 17 | 4.72e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 4.72e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 1 | 90 | 4.73e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 4.73e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 1 | 33 | 4.73e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 3 | 4747 | 4.73e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 3 | 317 | 4.73e-01 |
| GO:BP | GO:0032438 | melanosome organization | 1 | 36 | 4.73e-01 |
| GO:BP | GO:0012501 | programmed cell death | 31 | 1479 | 4.73e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 1 | 31 | 4.73e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 1 | 34 | 4.73e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 1 | 20 | 4.73e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 4.73e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 70 | 4.73e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 6 | 249 | 4.74e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 2 | 73 | 4.75e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 3 | 114 | 4.75e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 150 | 4.75e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 1 | 33 | 4.76e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 1 | 33 | 4.76e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 2 | 69 | 4.76e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 1 | 71 | 4.76e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 37 | 4.76e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 1 | 27 | 4.76e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 1 | 21 | 4.77e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 1 | 37 | 4.77e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 6 | 424 | 4.77e-01 |
| GO:BP | GO:0003158 | endothelium development | 3 | 104 | 4.77e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 2 | 55 | 4.77e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 2 | 84 | 4.77e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 1 | 22 | 4.77e-01 |
| GO:BP | GO:0051225 | spindle assembly | 1 | 120 | 4.77e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 1 | 36 | 4.77e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 3 | 112 | 4.78e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 24 | 1111 | 4.78e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 3 | 448 | 4.78e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 1 | 52 | 4.78e-01 |
| GO:BP | GO:0048731 | system development | 4 | 2901 | 4.81e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 3 | 138 | 4.82e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 1 | 27 | 4.83e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 1 | 31 | 4.83e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 1 | 34 | 4.83e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 4.83e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 4.83e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 1 | 38 | 4.83e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 1 | 19 | 4.84e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 1 | 64 | 4.85e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 1 | 38 | 4.85e-01 |
| GO:BP | GO:0006936 | muscle contraction | 6 | 267 | 4.85e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 1 | 26 | 4.85e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 1 | 21 | 4.85e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 75 | 4.85e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 4.85e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 1 | 58 | 4.86e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 2 | 85 | 4.86e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 3 | 4946 | 4.86e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 2 | 75 | 4.86e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 6 | 870 | 4.86e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 1 | 36 | 4.86e-01 |
| GO:BP | GO:0003002 | regionalization | 8 | 273 | 4.86e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 2 | 74 | 4.86e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 1 | 64 | 4.86e-01 |
| GO:BP | GO:0006270 | DNA replication initiation | 1 | 35 | 4.86e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 4.86e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 2 | 65 | 4.86e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 1 | 24 | 4.89e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 1 | 20 | 4.89e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 3 | 372 | 4.89e-01 |
| GO:BP | GO:0030534 | adult behavior | 2 | 92 | 4.90e-01 |
| GO:BP | GO:0009611 | response to wounding | 10 | 415 | 4.90e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 2 | 57 | 4.91e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 1 | 24 | 4.92e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 2 | 69 | 4.92e-01 |
| GO:BP | GO:0051049 | regulation of transport | 9 | 1267 | 4.92e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 1 | 27 | 4.92e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 1 | 35 | 4.93e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 97 | 4.93e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 4.93e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 3 | 442 | 4.95e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 2 | 194 | 4.95e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 1 | 29 | 4.96e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 4.96e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 1 | 85 | 4.97e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 4.97e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 1 | 16 | 4.97e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 1 | 19 | 4.97e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 1 | 25 | 4.97e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 4.97e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 163 | 4.97e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 1 | 20 | 4.97e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 1 | 24 | 4.97e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 2 | 91 | 4.98e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 5.00e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 1 | 232 | 5.00e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 1 | 83 | 5.00e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 5 | 318 | 5.00e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 5.00e-01 |
| GO:BP | GO:0050821 | protein stabilization | 2 | 191 | 5.00e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 1 | 197 | 5.02e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 5.02e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 1 | 22 | 5.02e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 1 | 21 | 5.02e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 3 | 357 | 5.02e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 2 | 86 | 5.04e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 1 | 37 | 5.04e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 1 | 31 | 5.05e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 2 | 57 | 5.05e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 2 | 64 | 5.05e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 1 | 32 | 5.06e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 5.06e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 5.07e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 5.07e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 6 | 303 | 5.08e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 5.09e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 2 | 203 | 5.13e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 3 | 138 | 5.13e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 1 | 80 | 5.13e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 2 | 95 | 5.13e-01 |
| GO:BP | GO:0016032 | viral process | 2 | 344 | 5.14e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 5.14e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 82 | 5.14e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 2 | 186 | 5.14e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 1 | 39 | 5.15e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 2 | 81 | 5.15e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 5.15e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 1 | 28 | 5.15e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 5.15e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 1 | 25 | 5.15e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 1 | 25 | 5.15e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 2 | 97 | 5.16e-01 |
| GO:BP | GO:0044000 | movement in host | 2 | 138 | 5.16e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 1 | 47 | 5.16e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 5.16e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 1 | 130 | 5.16e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 1 | 35 | 5.17e-01 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 1 | 14 | 5.17e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 5.17e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 1 | 54 | 5.17e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 2 | 206 | 5.17e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 5.17e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 43 | 5.17e-01 |
| GO:BP | GO:0010467 | gene expression | 3 | 4587 | 5.17e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 5.17e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 2 | 71 | 5.17e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 124 | 5.17e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 1 | 80 | 5.17e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 1 | 31 | 5.17e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 1 | 30 | 5.17e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 1 | 90 | 5.17e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 5.17e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 23 | 1088 | 5.17e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 9 | 719 | 5.17e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 2 | 101 | 5.17e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 7 | 311 | 5.17e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 89 | 5.17e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 1 | 82 | 5.18e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 3 | 140 | 5.18e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 2 | 96 | 5.18e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 1 | 57 | 5.20e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 5.20e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 3 | 114 | 5.20e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 1 | 222 | 5.21e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 1 | 17 | 5.22e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 1 | 539 | 5.22e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 2 | 92 | 5.22e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 15 | 2035 | 5.22e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 1 | 31 | 5.22e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 1 | 23 | 5.22e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 2 | 60 | 5.22e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 1 | 52 | 5.22e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 7 | 506 | 5.22e-01 |
| GO:BP | GO:0007612 | learning | 1 | 115 | 5.22e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 1 | 23 | 5.22e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 1 | 28 | 5.23e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 1 | 26 | 5.23e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 1 | 28 | 5.23e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 1 | 39 | 5.23e-01 |
| GO:BP | GO:0048536 | spleen development | 1 | 28 | 5.23e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 1 | 16 | 5.23e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 5.23e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 1 | 36 | 5.23e-01 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 2 | 73 | 5.23e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 1 | 24 | 5.23e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 1 | 33 | 5.23e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 64 | 5.23e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 5 | 228 | 5.23e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 2 | 269 | 5.23e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 1 | 249 | 5.24e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 5.24e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 1 | 85 | 5.24e-01 |
| GO:BP | GO:0042552 | myelination | 1 | 111 | 5.24e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 36 | 3090 | 5.24e-01 |
| GO:BP | GO:0006897 | endocytosis | 1 | 527 | 5.25e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 1 | 33 | 5.25e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 5.26e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 1 | 35 | 5.26e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 1 | 29 | 5.26e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 1 | 37 | 5.26e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 1 | 41 | 5.26e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 3 | 123 | 5.26e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 1 | 113 | 5.27e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 5.27e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 1 | 113 | 5.27e-01 |
| GO:BP | GO:0099172 | presynapse organization | 1 | 42 | 5.27e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 1 | 19 | 5.27e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 1 | 32 | 5.27e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 1 | 112 | 5.29e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 1 | 28 | 5.31e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 1 | 35 | 5.31e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 1 | 113 | 5.31e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 1 | 50 | 5.33e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 5.33e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 1 | 22 | 5.33e-01 |
| GO:BP | GO:0006417 | regulation of translation | 5 | 370 | 5.33e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 5.33e-01 |
| GO:BP | GO:0070266 | necroptotic process | 1 | 32 | 5.33e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 3 | 563 | 5.34e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 5.35e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 1 | 30 | 5.35e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 1 | 28 | 5.36e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 1 | 28 | 5.36e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 1 | 47 | 5.36e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 5.36e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 1 | 39 | 5.36e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 2 | 139 | 5.36e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 1 | 117 | 5.36e-01 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 1 | 29 | 5.36e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 1 | 36 | 5.36e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 5.36e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 1 | 37 | 5.36e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 10 | 687 | 5.36e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 1 | 55 | 5.36e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 5.36e-01 |
| GO:BP | GO:0003013 | circulatory system process | 3 | 447 | 5.37e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 1 | 25 | 5.37e-01 |
| GO:BP | GO:0009615 | response to virus | 3 | 275 | 5.38e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 5.39e-01 |
| GO:BP | GO:0051216 | cartilage development | 4 | 151 | 5.39e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 5.40e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 7 | 386 | 5.40e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 1 | 15 | 5.40e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 1 | 71 | 5.40e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 1 | 27 | 5.40e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 1 | 11 | 5.40e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 6 | 256 | 5.40e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 1 | 39 | 5.40e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 5.40e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 16 | 756 | 5.41e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 1 | 30 | 5.41e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 1 | 26 | 5.41e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 1 | 43 | 5.41e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 1 | 49 | 5.41e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 1 | 27 | 5.41e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 5.41e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 5.41e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 2 | 85 | 5.41e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 115 | 5.41e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 2 | 90 | 5.42e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 2 | 84 | 5.42e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 2 | 67 | 5.42e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 1 | 29 | 5.42e-01 |
| GO:BP | GO:0001656 | metanephros development | 2 | 64 | 5.42e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 5.42e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 5.42e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 6 | 300 | 5.42e-01 |
| GO:BP | GO:0006400 | tRNA modification | 1 | 90 | 5.42e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 6 | 258 | 5.42e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 1 | 40 | 5.43e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 4 | 288 | 5.43e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 12 | 796 | 5.43e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 94 | 5.44e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 1 | 35 | 5.44e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 1 | 44 | 5.44e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 2 | 126 | 5.45e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 1 | 50 | 5.45e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 1 | 32 | 5.45e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 6 | 268 | 5.46e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 6 | 268 | 5.46e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 22 | 1075 | 5.46e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 5.46e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 1 | 121 | 5.47e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 2 | 105 | 5.47e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 5.47e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 1 | 22 | 5.47e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 1 | 49 | 5.47e-01 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 1 | 47 | 5.48e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 5.48e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 2 | 116 | 5.48e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 2 | 199 | 5.48e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 5.48e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 2 | 101 | 5.48e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 1 | 39 | 5.48e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 1 | 106 | 5.49e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 1 | 126 | 5.49e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 5.49e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 1 | 52 | 5.49e-01 |
| GO:BP | GO:0061448 | connective tissue development | 5 | 208 | 5.49e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 2 | 129 | 5.49e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 2 | 93 | 5.50e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 1 | 39 | 5.51e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 2 | 154 | 5.51e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 1 | 42 | 5.51e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 1 | 43 | 5.51e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 5.51e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 5.51e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 1 | 103 | 5.51e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 13 | 881 | 5.52e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 3 | 161 | 5.53e-01 |
| GO:BP | GO:0021543 | pallium development | 2 | 139 | 5.54e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 1 | 440 | 5.54e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 5.56e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 3 | 170 | 5.56e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 2 | 94 | 5.56e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 2 | 91 | 5.56e-01 |
| GO:BP | GO:0008219 | cell death | 32 | 1596 | 5.57e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 1 | 90 | 5.57e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 58 | 5.57e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 1 | 42 | 5.57e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 2 | 100 | 5.58e-01 |
| GO:BP | GO:0030001 | metal ion transport | 4 | 562 | 5.58e-01 |
| GO:BP | GO:0045010 | actin nucleation | 1 | 50 | 5.58e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 3 | 513 | 5.58e-01 |
| GO:BP | GO:0016310 | phosphorylation | 19 | 1372 | 5.58e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 1 | 48 | 5.59e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 7 | 357 | 5.59e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 1 | 48 | 5.59e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 2 | 91 | 5.59e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 1 | 13 | 5.60e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 5.61e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 1 | 22 | 5.61e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 1 | 31 | 5.61e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 2 | 95 | 5.61e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 5.62e-01 |
| GO:BP | GO:0060429 | epithelium development | 19 | 850 | 5.62e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 2 | 94 | 5.63e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 2 | 94 | 5.63e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 25 | 1219 | 5.63e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 1 | 53 | 5.64e-01 |
| GO:BP | GO:0007389 | pattern specification process | 8 | 306 | 5.64e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 5.65e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 5.65e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 2 | 706 | 5.65e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 5.67e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 4 | 167 | 5.67e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 1 | 78 | 5.67e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 1 | 111 | 5.67e-01 |
| GO:BP | GO:0048732 | gland development | 2 | 318 | 5.67e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 1 | 41 | 5.67e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 5.67e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 2 | 114 | 5.67e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 2 | 113 | 5.67e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 2 | 114 | 5.67e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 1 | 38 | 5.67e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 1 | 300 | 5.67e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 1 | 39 | 5.67e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 5.67e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 1 | 39 | 5.67e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 5.67e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 2 | 75 | 5.67e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 1 | 56 | 5.67e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 5.67e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 1 | 42 | 5.67e-01 |
| GO:BP | GO:0007051 | spindle organization | 1 | 188 | 5.67e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 2 | 143 | 5.67e-01 |
| GO:BP | GO:0001764 | neuron migration | 1 | 133 | 5.67e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 5.67e-01 |
| GO:BP | GO:0010165 | response to X-ray | 1 | 29 | 5.67e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 29 | 5.67e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 5.67e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 5 | 384 | 5.67e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 13 | 798 | 5.67e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 5.67e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 3 | 120 | 5.67e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 1 | 30 | 5.68e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 4 | 197 | 5.68e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 5 | 188 | 5.68e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 6 | 404 | 5.68e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 8 | 1101 | 5.68e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 3 | 140 | 5.68e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 5.68e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 3 | 155 | 5.68e-01 |
| GO:BP | GO:0030901 | midbrain development | 1 | 69 | 5.68e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 1 | 132 | 5.68e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 1 | 38 | 5.68e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 2 | 76 | 5.68e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 2 | 76 | 5.68e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 5.68e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 5.71e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 1 | 43 | 5.71e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 3 | 168 | 5.72e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 1 | 31 | 5.72e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 5.72e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 40 | 5.74e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 3 | 169 | 5.74e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 5.74e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2 | 4099 | 5.75e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 5.76e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 5.76e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 2 | 111 | 5.76e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 5.76e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 1 | 41 | 5.76e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 5.76e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 5.76e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 5.76e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 1 | 37 | 5.76e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 4 | 3386 | 5.76e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 1 | 44 | 5.76e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 1 | 28 | 5.76e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 1 | 29 | 5.76e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 1 | 29 | 5.76e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 3 | 138 | 5.76e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 2 | 149 | 5.76e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 1 | 43 | 5.77e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 1 | 42 | 5.77e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 1 | 32 | 5.77e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 60 | 5.77e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 5.79e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 2 | 161 | 5.79e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 3 | 632 | 5.80e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 1 | 292 | 5.80e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 40 | 5.80e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 2 | 115 | 5.80e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 5.81e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 2 | 141 | 5.81e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 1 | 57 | 5.82e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 5.82e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 1 | 30 | 5.82e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 5.82e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 5.82e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 2 | 84 | 5.82e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 1 | 246 | 5.82e-01 |
| GO:BP | GO:0001823 | mesonephros development | 2 | 77 | 5.82e-01 |
| GO:BP | GO:0007099 | centriole replication | 1 | 38 | 5.82e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 5.82e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 1 | 21 | 5.83e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 34 | 5.83e-01 |
| GO:BP | GO:0031529 | ruffle organization | 1 | 45 | 5.83e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 1 | 44 | 5.83e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 2 | 151 | 5.85e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 5.85e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 1 | 104 | 5.85e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 2 | 317 | 5.85e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 17 | 956 | 5.86e-01 |
| GO:BP | GO:0007015 | actin filament organization | 7 | 373 | 5.87e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 2 | 105 | 5.87e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 1 | 552 | 5.88e-01 |
| GO:BP | GO:0060348 | bone development | 2 | 170 | 5.88e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 5.88e-01 |
| GO:BP | GO:0001709 | cell fate determination | 1 | 25 | 5.88e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 1 | 86 | 5.89e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 1 | 32 | 5.89e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 5.89e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 5.90e-01 |
| GO:BP | GO:0051170 | import into nucleus | 1 | 152 | 5.90e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 2 | 109 | 5.90e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 2 | 96 | 5.90e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 1 | 89 | 5.91e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 9 | 1229 | 5.91e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 1 | 98 | 5.91e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 5.91e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 6 | 326 | 5.92e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 1 | 45 | 5.93e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 5.93e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 2 | 375 | 5.93e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 1 | 108 | 5.93e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 4 | 775 | 5.93e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 1 | 32 | 5.93e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 5.93e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 5.93e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 1 | 90 | 5.93e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 5.93e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 2 | 376 | 5.93e-01 |
| GO:BP | GO:0016236 | macroautophagy | 1 | 308 | 5.93e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 1 | 102 | 5.93e-01 |
| GO:BP | GO:0007416 | synapse assembly | 2 | 152 | 5.94e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 4 | 209 | 5.94e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 250 | 5.95e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 1 | 49 | 5.95e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 2 | 110 | 5.96e-01 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 1 | 84 | 5.97e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 1 | 99 | 5.97e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 1 | 79 | 5.97e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 1 | 127 | 5.97e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 2 | 113 | 5.97e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 5.98e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 3 | 165 | 5.98e-01 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 1 | 42 | 5.98e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 1 | 59 | 5.99e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 1 | 92 | 6.00e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 1 | 93 | 6.00e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 1 | 29 | 6.00e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 2 | 97 | 6.00e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 6.00e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 2 | 90 | 6.01e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 6 | 428 | 6.01e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 1 | 236 | 6.01e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 1 | 45 | 6.01e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 122 | 6.02e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 133 | 6.02e-01 |
| GO:BP | GO:0042116 | macrophage activation | 1 | 57 | 6.02e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 6 | 726 | 6.02e-01 |
| GO:BP | GO:0007420 | brain development | 8 | 591 | 6.02e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 6.02e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 1 | 22 | 6.02e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 3 | 1380 | 6.02e-01 |
| GO:BP | GO:0098534 | centriole assembly | 1 | 41 | 6.03e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 2 | 84 | 6.03e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 2 | 303 | 6.03e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 2 | 89 | 6.04e-01 |
| GO:BP | GO:0050877 | nervous system process | 6 | 634 | 6.06e-01 |
| GO:BP | GO:1902074 | response to salt | 2 | 271 | 6.06e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 1 | 68 | 6.06e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 2 | 120 | 6.06e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 2 | 108 | 6.07e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 6.07e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 2 | 68 | 6.07e-01 |
| GO:BP | GO:0001501 | skeletal system development | 4 | 377 | 6.08e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 1 | 37 | 6.08e-01 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 1 | 44 | 6.09e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 6 | 418 | 6.10e-01 |
| GO:BP | GO:0002520 | immune system development | 3 | 144 | 6.10e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 6.10e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 3 | 186 | 6.10e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 1 | 40 | 6.10e-01 |
| GO:BP | GO:0030282 | bone mineralization | 2 | 82 | 6.10e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 1 | 70 | 6.11e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 6.11e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1 | 54 | 6.11e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 1 | 151 | 6.11e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 6.11e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 18 | 1006 | 6.12e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 626 | 6.13e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 1 | 32 | 6.14e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 2 | 122 | 6.14e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 35 | 6.16e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 4 | 605 | 6.17e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 1 | 126 | 6.17e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 1 | 55 | 6.18e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 228 | 6.18e-01 |
| GO:BP | GO:0016049 | cell growth | 2 | 418 | 6.18e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 1 | 75 | 6.18e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 5 | 277 | 6.19e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 1 | 82 | 6.19e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 6.19e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 6.19e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 6.20e-01 |
| GO:BP | GO:0090224 | regulation of spindle organization | 1 | 45 | 6.20e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 2 | 86 | 6.20e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 2 | 86 | 6.20e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 1 | 49 | 6.20e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 2 | 96 | 6.20e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 1 | 35 | 6.20e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 142 | 6.21e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 1 | 36 | 6.21e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 7 | 1137 | 6.21e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 6.23e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 6.23e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 28 | 6.23e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 2 | 106 | 6.23e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 1 | 49 | 6.23e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 6.23e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 6.23e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 6.25e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 2 | 252 | 6.25e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 1 | 41 | 6.27e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 6.27e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 6.28e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 6.28e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 6.28e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 1 | 427 | 6.29e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 5 | 274 | 6.29e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 1 | 41 | 6.29e-01 |
| GO:BP | GO:0035329 | hippo signaling | 1 | 41 | 6.29e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 4 | 667 | 6.29e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 2 | 110 | 6.30e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 3 | 185 | 6.30e-01 |
| GO:BP | GO:0007517 | muscle organ development | 5 | 277 | 6.30e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 1 | 53 | 6.31e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 2 | 116 | 6.31e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 6.31e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 1 | 75 | 6.32e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 1 | 155 | 6.32e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 1 | 64 | 6.32e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 8 | 504 | 6.32e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 1 | 50 | 6.32e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 2 | 301 | 6.32e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 1 | 41 | 6.32e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 1 | 114 | 6.32e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 6.32e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 1 | 46 | 6.32e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1 | 135 | 6.32e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 2 | 104 | 6.33e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 1 | 60 | 6.33e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 2 | 162 | 6.33e-01 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 1 | 19 | 6.33e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 5 | 287 | 6.35e-01 |
| GO:BP | GO:0006479 | protein methylation | 3 | 160 | 6.35e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 6 | 340 | 6.35e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 6.35e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2 | 4579 | 6.35e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 2 | 94 | 6.35e-01 |
| GO:BP | GO:0008213 | protein alkylation | 3 | 160 | 6.35e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 6.35e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 6.35e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 3 | 2139 | 6.36e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 1 | 168 | 6.36e-01 |
| GO:BP | GO:0001654 | eye development | 1 | 272 | 6.36e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 1 | 115 | 6.37e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 2 | 144 | 6.37e-01 |
| GO:BP | GO:0006284 | base-excision repair | 1 | 44 | 6.37e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 1 | 130 | 6.38e-01 |
| GO:BP | GO:0150063 | visual system development | 1 | 273 | 6.39e-01 |
| GO:BP | GO:0040008 | regulation of growth | 9 | 501 | 6.39e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 3 | 178 | 6.40e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 2 | 116 | 6.40e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 1 | 46 | 6.40e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 6.40e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 3 | 174 | 6.40e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 6 | 344 | 6.40e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 6.41e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 3 | 2161 | 6.41e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 2 | 279 | 6.41e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 2 | 168 | 6.41e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 2 | 135 | 6.42e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 6 | 345 | 6.42e-01 |
| GO:BP | GO:0048880 | sensory system development | 1 | 275 | 6.42e-01 |
| GO:BP | GO:0048736 | appendage development | 2 | 142 | 6.42e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 15 | 1173 | 6.42e-01 |
| GO:BP | GO:0060173 | limb development | 2 | 142 | 6.42e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 6.42e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 6.42e-01 |
| GO:BP | GO:0048284 | organelle fusion | 1 | 135 | 6.43e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 2 | 114 | 6.43e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 1 | 47 | 6.43e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 2 | 290 | 6.45e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 1 | 48 | 6.46e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 1 | 49 | 6.46e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 19 | 1076 | 6.46e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 19 | 1074 | 6.46e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 6.46e-01 |
| GO:BP | GO:0006304 | DNA modification | 1 | 89 | 6.46e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 2 | 184 | 6.46e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 1 | 66 | 6.47e-01 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 1 | 104 | 6.47e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 6.47e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 6.47e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 2 | 99 | 6.47e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 7 | 560 | 6.47e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 1 | 46 | 6.47e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 6.47e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 1 | 48 | 6.47e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 6.48e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 6.48e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 6.48e-01 |
| GO:BP | GO:0060322 | head development | 6 | 633 | 6.48e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 3 | 529 | 6.48e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 4 | 669 | 6.49e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 2 | 145 | 6.49e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 2 | 97 | 6.50e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 6 | 335 | 6.50e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 1 | 39 | 6.50e-01 |
| GO:BP | GO:0007568 | aging | 1 | 115 | 6.50e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 3 | 143 | 6.52e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 1 | 160 | 6.52e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 2 | 151 | 6.52e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 1 | 152 | 6.52e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 2 | 279 | 6.52e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 4 | 200 | 6.53e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 2 | 116 | 6.53e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 1 | 86 | 6.54e-01 |
| GO:BP | GO:0007049 | cell cycle | 26 | 1529 | 6.54e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 2 | 187 | 6.54e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 3 | 232 | 6.54e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 6.54e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 3 | 512 | 6.54e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 1 | 77 | 6.54e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 6.55e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 1 | 310 | 6.55e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 3 | 517 | 6.55e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 1 | 36 | 6.55e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 6.55e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 46 | 6.55e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 2 | 142 | 6.55e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 2 | 400 | 6.55e-01 |
| GO:BP | GO:0031929 | TOR signaling | 2 | 115 | 6.57e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 1 | 66 | 6.57e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 2 | 901 | 6.57e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 6.58e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 3 | 541 | 6.58e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 5 | 295 | 6.58e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 1 | 57 | 6.59e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 6.59e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 1 | 118 | 6.59e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 1 | 83 | 6.60e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 1 | 68 | 6.62e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 1 | 81 | 6.62e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 4 | 234 | 6.63e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 2 | 104 | 6.63e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 6 | 1014 | 6.63e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 1 | 27 | 6.63e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 6.63e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 1 | 67 | 6.63e-01 |
| GO:BP | GO:0030879 | mammary gland development | 1 | 106 | 6.63e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 2 | 101 | 6.63e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 1 | 161 | 6.63e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 6.63e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 6.63e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 1 | 48 | 6.64e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 3 | 527 | 6.67e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 6.67e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 3 | 138 | 6.67e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 1 | 58 | 6.67e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 3 | 241 | 6.68e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 1 | 118 | 6.68e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 1 | 118 | 6.68e-01 |
| GO:BP | GO:0035270 | endocrine system development | 2 | 86 | 6.69e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 1 | 121 | 6.69e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 1 | 58 | 6.69e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 1 | 439 | 6.70e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 3 | 531 | 6.70e-01 |
| GO:BP | GO:0019058 | viral life cycle | 3 | 244 | 6.70e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 1 | 52 | 6.70e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 3 | 223 | 6.72e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 6.72e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 1 | 51 | 6.72e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 6.73e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 1 | 68 | 6.73e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 2 | 102 | 6.73e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 2 | 994 | 6.73e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 1 | 59 | 6.73e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 6.73e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 1 | 69 | 6.73e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 2 | 993 | 6.73e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 6.73e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 4 | 318 | 6.73e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 6.74e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 1 | 43 | 6.74e-01 |
| GO:BP | GO:1901652 | response to peptide | 7 | 377 | 6.75e-01 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 6.75e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 1 | 116 | 6.75e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 6 | 344 | 6.75e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 1 | 87 | 6.76e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 45 | 2135 | 6.76e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 2 | 103 | 6.76e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 1 | 760 | 6.76e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 1 | 51 | 6.76e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 6.76e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 1 | 51 | 6.76e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 1012 | 6.76e-01 |
| GO:BP | GO:0009749 | response to glucose | 2 | 137 | 6.77e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 2 | 188 | 6.78e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 6.78e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 6.78e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 1 | 64 | 6.78e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 7 | 393 | 6.78e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 1 | 93 | 6.78e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 1 | 433 | 6.79e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 1 | 65 | 6.79e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 1 | 183 | 6.80e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 3 | 179 | 6.81e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 5 | 761 | 6.81e-01 |
| GO:BP | GO:0030258 | lipid modification | 2 | 162 | 6.81e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 6.83e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 6.83e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 3 | 209 | 6.83e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 3 | 551 | 6.83e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 3 | 517 | 6.83e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 1 | 56 | 6.84e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 213 | 6.85e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 1 | 51 | 6.85e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 6.85e-01 |
| GO:BP | GO:0007399 | nervous system development | 2 | 1904 | 6.85e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 1 | 54 | 6.85e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 1 | 52 | 6.85e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 6.85e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 5 | 343 | 6.86e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 1 | 49 | 6.86e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 1 | 31 | 6.86e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 3 | 154 | 6.86e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 2 | 92 | 6.86e-01 |
| GO:BP | GO:0009746 | response to hexose | 2 | 138 | 6.86e-01 |
| GO:BP | GO:0009451 | RNA modification | 1 | 162 | 6.87e-01 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 6.87e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 4 | 228 | 6.87e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 1 | 59 | 6.87e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 1 | 59 | 6.87e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 5 | 299 | 6.88e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 1 | 58 | 6.88e-01 |
| GO:BP | GO:0006935 | chemotaxis | 2 | 389 | 6.89e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 1 | 27 | 6.89e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 2 | 93 | 6.89e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 16 | 868 | 6.89e-01 |
| GO:BP | GO:0042330 | taxis | 2 | 389 | 6.91e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 3 | 157 | 6.91e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 2 | 223 | 6.92e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 6.92e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 10 | 1835 | 6.92e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 2 | 87 | 6.92e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 1 | 47 | 6.93e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 3 | 296 | 6.93e-01 |
| GO:BP | GO:0040007 | growth | 10 | 742 | 6.94e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 6.94e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 1 | 51 | 6.94e-01 |
| GO:BP | GO:0042063 | gliogenesis | 3 | 235 | 6.95e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 27 | 1681 | 6.95e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 1 | 1238 | 6.95e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 6.95e-01 |
| GO:BP | GO:0051591 | response to cAMP | 1 | 66 | 6.95e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 2 | 224 | 6.95e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 1 | 65 | 6.95e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 1 | 222 | 6.95e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 2 | 146 | 6.95e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 1 | 71 | 6.95e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 11 | 2102 | 6.96e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 1 | 123 | 6.96e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 6.97e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 6.97e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 8 | 1133 | 6.97e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 6.98e-01 |
| GO:BP | GO:0001890 | placenta development | 2 | 117 | 6.98e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 2 | 132 | 6.98e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 6.98e-01 |
| GO:BP | GO:0042594 | response to starvation | 1 | 178 | 6.98e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 7 | 399 | 6.98e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 6.98e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 3 | 7552 | 6.98e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 3 | 540 | 6.99e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 6.99e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 6.99e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 2 | 142 | 6.99e-01 |
| GO:BP | GO:0016571 | histone methylation | 2 | 117 | 7.01e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 1 | 67 | 7.01e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 1 | 56 | 7.02e-01 |
| GO:BP | GO:0030073 | insulin secretion | 2 | 141 | 7.03e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 1 | 162 | 7.03e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 1 | 57 | 7.03e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 7.03e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 7 | 612 | 7.04e-01 |
| GO:BP | GO:0051235 | maintenance of location | 2 | 260 | 7.04e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 2 | 134 | 7.05e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 2 | 316 | 7.05e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 1 | 57 | 7.06e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 1 | 64 | 7.06e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 336 | 7.06e-01 |
| GO:BP | GO:0060419 | heart growth | 1 | 72 | 7.06e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 258 | 7.07e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 1 | 33 | 7.07e-01 |
| GO:BP | GO:0031175 | neuron projection development | 4 | 780 | 7.08e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 2 | 136 | 7.08e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 7.08e-01 |
| GO:BP | GO:0001822 | kidney development | 4 | 244 | 7.08e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 1 | 69 | 7.08e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 2 | 1028 | 7.08e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 1 | 47 | 7.09e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 2 | 112 | 7.09e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 2 | 1974 | 7.09e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 7.09e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 2 | 260 | 7.09e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 2 | 149 | 7.10e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 2 | 166 | 7.11e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 1 | 170 | 7.13e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 7.13e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 1 | 67 | 7.14e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 1 | 68 | 7.14e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 7.14e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 2 | 125 | 7.15e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 134 | 7.18e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 1 | 35 | 7.18e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 1 | 43 | 7.18e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 1 | 43 | 7.18e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 47 | 2293 | 7.19e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 1 | 88 | 7.19e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 2 | 137 | 7.21e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 1 | 82 | 7.21e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 7.22e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 7 | 459 | 7.22e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 7.22e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 1 | 194 | 7.22e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 1 | 71 | 7.23e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 1 | 106 | 7.24e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 1 | 214 | 7.25e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 1 | 53 | 7.25e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 4 | 230 | 7.25e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 2 | 106 | 7.25e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 1 | 357 | 7.25e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 1 | 104 | 7.25e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 1 | 49 | 7.26e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 8 | 597 | 7.27e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 8 | 597 | 7.27e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 3 | 167 | 7.27e-01 |
| GO:BP | GO:0072001 | renal system development | 4 | 251 | 7.27e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 1 | 104 | 7.29e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 7.30e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 1 | 50 | 7.31e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 2 | 199 | 7.32e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 1 | 615 | 7.33e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 112 | 7.33e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 1 | 55 | 7.34e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 3 | 189 | 7.35e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 2 | 130 | 7.36e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 2 | 149 | 7.36e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 1 | 57 | 7.36e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 1 | 103 | 7.36e-01 |
| GO:BP | GO:0007423 | sensory organ development | 1 | 396 | 7.36e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 1 | 784 | 7.36e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 1 | 57 | 7.36e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 1 | 625 | 7.36e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 2 | 110 | 7.36e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 7.36e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 1 | 37 | 7.38e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 2 | 128 | 7.38e-01 |
| GO:BP | GO:0045087 | innate immune response | 1 | 488 | 7.38e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 1 | 76 | 7.38e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 16 | 1278 | 7.40e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 4 | 4128 | 7.40e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 3 | 6996 | 7.41e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 1 | 637 | 7.41e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 1 | 34 | 7.42e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 7.42e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 1 | 75 | 7.42e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 2 | 101 | 7.42e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 1 | 108 | 7.43e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 1 | 119 | 7.44e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 1 | 144 | 7.44e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 2 | 111 | 7.44e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 7.46e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 2 | 475 | 7.46e-01 |
| GO:BP | GO:0048589 | developmental growth | 6 | 522 | 7.46e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 1 | 38 | 7.46e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 3 | 199 | 7.47e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 1 | 67 | 7.47e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 1 | 93 | 7.48e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 7.48e-01 |
| GO:BP | GO:0007507 | heart development | 7 | 506 | 7.48e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 1 | 369 | 7.49e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 23 | 1974 | 7.51e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 2 | 151 | 7.52e-01 |
| GO:BP | GO:0030154 | cell differentiation | 2 | 2958 | 7.53e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 1 | 69 | 7.53e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 7.53e-01 |
| GO:BP | GO:0034330 | cell junction organization | 9 | 575 | 7.53e-01 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 7.54e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 7.54e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 65 | 7.54e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 2 | 348 | 7.54e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 3 | 639 | 7.54e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 4 | 1107 | 7.55e-01 |
| GO:BP | GO:0060840 | artery development | 1 | 83 | 7.55e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 9 | 763 | 7.55e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 2 | 2984 | 7.55e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 7.55e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 2 | 182 | 7.55e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 7.55e-01 |
| GO:BP | GO:0010038 | response to metal ion | 2 | 264 | 7.55e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 1 | 113 | 7.55e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 1 | 70 | 7.57e-01 |
| GO:BP | GO:0050808 | synapse organization | 3 | 365 | 7.58e-01 |
| GO:BP | GO:0007127 | meiosis I | 1 | 77 | 7.58e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 7 | 547 | 7.58e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 7.58e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 1 | 68 | 7.58e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 4 | 242 | 7.58e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 1 | 57 | 7.59e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1086 | 7.59e-01 |
| GO:BP | GO:0016197 | endosomal transport | 1 | 231 | 7.59e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 2 | 126 | 7.59e-01 |
| GO:BP | GO:0032868 | response to insulin | 3 | 215 | 7.60e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 2 | 129 | 7.60e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 3 | 293 | 7.61e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 7.61e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 7.61e-01 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 2 | 148 | 7.62e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 1 | 111 | 7.62e-01 |
| GO:BP | GO:0007492 | endoderm development | 1 | 65 | 7.62e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 7.63e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 1 | 209 | 7.63e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 2 | 121 | 7.65e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 3 | 196 | 7.65e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 5 | 771 | 7.65e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 1 | 78 | 7.65e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 1 | 59 | 7.65e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 1 | 65 | 7.65e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 2 | 127 | 7.66e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 3 | 348 | 7.66e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 7.66e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 61 | 7.67e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 61 | 7.67e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 7.67e-01 |
| GO:BP | GO:0000910 | cytokinesis | 1 | 164 | 7.68e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 5 | 906 | 7.68e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 1 | 834 | 7.68e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 1 | 81 | 7.69e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 97 | 7.69e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 2 | 106 | 7.69e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 2 | 1145 | 7.70e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 1 | 101 | 7.71e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 1 | 61 | 7.71e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 7.73e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 2 | 169 | 7.74e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 7.75e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 2 | 222 | 7.75e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 1 | 62 | 7.75e-01 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 1 | 94 | 7.76e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 1 | 523 | 7.76e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 3 | 1939 | 7.78e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 7.78e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 302 | 7.78e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 95 | 7.80e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 2 | 166 | 7.81e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 16 | 1345 | 7.81e-01 |
| GO:BP | GO:0019079 | viral genome replication | 1 | 101 | 7.81e-01 |
| GO:BP | GO:0043200 | response to amino acid | 1 | 95 | 7.82e-01 |
| GO:BP | GO:0007601 | visual perception | 2 | 96 | 7.82e-01 |
| GO:BP | GO:0015844 | monoamine transport | 1 | 45 | 7.82e-01 |
| GO:BP | GO:0007369 | gastrulation | 2 | 146 | 7.82e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 3 | 998 | 7.83e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 4 | 246 | 7.85e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 1 | 135 | 7.85e-01 |
| GO:BP | GO:0003014 | renal system process | 1 | 78 | 7.86e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 2 | 192 | 7.86e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 1 | 107 | 7.86e-01 |
| GO:BP | GO:0061024 | membrane organization | 2 | 671 | 7.87e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 15 | 1147 | 7.87e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 2 | 97 | 7.87e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 1 | 71 | 7.88e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 8 | 1898 | 7.88e-01 |
| GO:BP | GO:0002790 | peptide secretion | 2 | 170 | 7.88e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 7.89e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 2 | 229 | 7.89e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 90 | 7.89e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 7.90e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 5 | 321 | 7.90e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 1 | 48 | 7.91e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 230 | 7.91e-01 |
| GO:BP | GO:0003008 | system process | 7 | 1189 | 7.91e-01 |
| GO:BP | GO:0007417 | central nervous system development | 13 | 792 | 7.91e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 1 | 81 | 7.91e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 1 | 48 | 7.91e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 5 | 1061 | 7.92e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 7 | 461 | 7.92e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 1 | 96 | 7.92e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 7 | 441 | 7.92e-01 |
| GO:BP | GO:0007254 | JNK cascade | 2 | 138 | 7.93e-01 |
| GO:BP | GO:0048666 | neuron development | 4 | 877 | 7.93e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 2 | 168 | 7.95e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 2 | 170 | 7.95e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 7.95e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 7.96e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 153 | 7.97e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 6 | 496 | 7.97e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 1 | 604 | 7.99e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 7.99e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 2 | 556 | 8.00e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 1 | 86 | 8.00e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 2 | 463 | 8.01e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 8.01e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 4 | 1149 | 8.03e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 2 | 147 | 8.03e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 2 | 640 | 8.03e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 1 | 98 | 8.03e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 8 | 1256 | 8.03e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 8.03e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 8 | 1255 | 8.03e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 20 | 1248 | 8.04e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1 | 1226 | 8.05e-01 |
| GO:BP | GO:0051668 | localization within membrane | 1 | 605 | 8.06e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 4 | 666 | 8.06e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 2 | 149 | 8.06e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 7 | 469 | 8.06e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 8.07e-01 |
| GO:BP | GO:0002253 | activation of immune response | 4 | 272 | 8.07e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 6 | 420 | 8.07e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 1 | 893 | 8.07e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 70 | 8.07e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 2 | 153 | 8.08e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 8.08e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 1 | 145 | 8.08e-01 |
| GO:BP | GO:0032502 | developmental process | 4 | 4494 | 8.09e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 1 | 96 | 8.09e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 2 | 185 | 8.09e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 1 | 109 | 8.09e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 74 | 8.10e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 3 | 609 | 8.10e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 74 | 8.10e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 1 | 124 | 8.10e-01 |
| GO:BP | GO:0015833 | peptide transport | 2 | 179 | 8.10e-01 |
| GO:BP | GO:0051302 | regulation of cell division | 1 | 145 | 8.10e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 1 | 52 | 8.10e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 1 | 105 | 8.10e-01 |
| GO:BP | GO:0000725 | recombinational repair | 2 | 153 | 8.13e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 1 | 114 | 8.13e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 8.13e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 8.13e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 2 | 485 | 8.13e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 4 | 269 | 8.13e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 1 | 88 | 8.13e-01 |
| GO:BP | GO:0007040 | lysosome organization | 1 | 88 | 8.13e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 63 | 3625 | 8.14e-01 |
| GO:BP | GO:0016573 | histone acetylation | 1 | 143 | 8.14e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 1 | 90 | 8.14e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 1 | 93 | 8.16e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 1 | 119 | 8.16e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 8.16e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 3 | 242 | 8.17e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 1 | 108 | 8.19e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 1 | 73 | 8.19e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 2 | 742 | 8.19e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 1 | 100 | 8.19e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 3 | 7441 | 8.20e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 1 | 130 | 8.21e-01 |
| GO:BP | GO:0048699 | generation of neurons | 5 | 1119 | 8.21e-01 |
| GO:BP | GO:0021510 | spinal cord development | 1 | 59 | 8.21e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 8.21e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 8.24e-01 |
| GO:BP | GO:0006260 | DNA replication | 3 | 255 | 8.24e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 1 | 91 | 8.25e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 1 | 148 | 8.26e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 1 | 72 | 8.26e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 8.29e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 1 | 150 | 8.31e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 8.31e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 359 | 8.31e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1 | 2065 | 8.31e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 2 | 285 | 8.32e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 8.32e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 1 | 56 | 8.33e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 8 | 1484 | 8.33e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 1 | 56 | 8.33e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 6 | 2011 | 8.33e-01 |
| GO:BP | GO:0021700 | developmental maturation | 1 | 211 | 8.33e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 2 | 276 | 8.34e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 1 | 106 | 8.35e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 1 | 295 | 8.36e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 3 | 273 | 8.37e-01 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 1 | 16 | 8.37e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 2 | 175 | 8.38e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 8.41e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 3 | 258 | 8.42e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 2 | 516 | 8.44e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 2 | 276 | 8.44e-01 |
| GO:BP | GO:0016050 | vesicle organization | 2 | 320 | 8.44e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 3 | 7069 | 8.44e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 4 | 1712 | 8.44e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 2 | 165 | 8.44e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 38 | 2285 | 8.44e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 1 | 233 | 8.44e-01 |
| GO:BP | GO:0046907 | intracellular transport | 2 | 1453 | 8.44e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 2 | 160 | 8.44e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 6 | 662 | 8.45e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 211 | 8.45e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 1 | 163 | 8.45e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 1 | 160 | 8.45e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 1 | 75 | 8.47e-01 |
| GO:BP | GO:0006396 | RNA processing | 2 | 929 | 8.47e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 103 | 8.47e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 1 | 129 | 8.47e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 8.48e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 8.48e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 2 | 165 | 8.48e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 8.50e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 8.50e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 2 | 167 | 8.52e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 10 | 1555 | 8.52e-01 |
| GO:BP | GO:0070997 | neuron death | 4 | 286 | 8.52e-01 |
| GO:BP | GO:0009888 | tissue development | 4 | 1425 | 8.52e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 4 | 448 | 8.52e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 543 | 8.53e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 1 | 337 | 8.56e-01 |
| GO:BP | GO:0006954 | inflammatory response | 3 | 429 | 8.56e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 398 | 8.56e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 1 | 109 | 8.56e-01 |
| GO:BP | GO:0051648 | vesicle localization | 1 | 192 | 8.57e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 1 | 134 | 8.57e-01 |
| GO:BP | GO:0043588 | skin development | 2 | 175 | 8.57e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 8.60e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 2 | 198 | 8.60e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 96 | 8.61e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 1 | 65 | 8.61e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 1 | 820 | 8.61e-01 |
| GO:BP | GO:0003341 | cilium movement | 1 | 97 | 8.61e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 5 | 418 | 8.61e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 1 | 317 | 8.61e-01 |
| GO:BP | GO:0051707 | response to other organism | 1 | 820 | 8.61e-01 |
| GO:BP | GO:0007286 | spermatid development | 1 | 105 | 8.61e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 441 | 8.61e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 1 | 154 | 8.61e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 3 | 8061 | 8.61e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 2 | 165 | 8.62e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 211 | 8.62e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 7 | 631 | 8.62e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 8 | 850 | 8.62e-01 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 8.62e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 1 | 157 | 8.64e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 1 | 86 | 8.64e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 2 | 179 | 8.64e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 8 | 532 | 8.64e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 2 | 259 | 8.64e-01 |
| GO:BP | GO:0046879 | hormone secretion | 2 | 210 | 8.64e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 8.64e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 2 | 1389 | 8.64e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 1 | 114 | 8.64e-01 |
| GO:BP | GO:0051607 | defense response to virus | 1 | 202 | 8.64e-01 |
| GO:BP | GO:0006508 | proteolysis | 2 | 1318 | 8.64e-01 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 2 | 186 | 8.64e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 8.64e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 1 | 202 | 8.65e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 8.65e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 1 | 170 | 8.65e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 8.65e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 1 | 111 | 8.65e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 1 | 69 | 8.65e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 1 | 75 | 8.65e-01 |
| GO:BP | GO:0006974 | DNA damage response | 10 | 785 | 8.65e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 1 | 127 | 8.65e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 1 | 105 | 8.65e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 1 | 180 | 8.66e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 1 | 496 | 8.66e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 2 | 830 | 8.67e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 2 | 159 | 8.67e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 8.69e-01 |
| GO:BP | GO:0007052 | mitotic spindle organization | 1 | 129 | 8.69e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 2 | 213 | 8.69e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 8.69e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 1 | 102 | 8.72e-01 |
| GO:BP | GO:0009914 | hormone transport | 2 | 215 | 8.74e-01 |
| GO:BP | GO:0035265 | organ growth | 1 | 131 | 8.74e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 4 | 484 | 8.75e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 3 | 257 | 8.75e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 1 | 192 | 8.75e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 2 | 694 | 8.75e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 8.77e-01 |
| GO:BP | GO:0044782 | cilium organization | 4 | 313 | 8.78e-01 |
| GO:BP | GO:0006956 | complement activation | 1 | 23 | 8.78e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 2 | 569 | 8.79e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 2 | 166 | 8.79e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 1 | 149 | 8.79e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 1 | 113 | 8.79e-01 |
| GO:BP | GO:0006412 | translation | 5 | 646 | 8.79e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 3 | 272 | 8.80e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 8.80e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 1 | 129 | 8.80e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 2 | 251 | 8.82e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 1 | 963 | 8.82e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 4 | 775 | 8.82e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 8.84e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 8.84e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 8.86e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 8.87e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 2 | 540 | 8.87e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1223 | 8.89e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 1 | 124 | 8.89e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 8.90e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 1 | 96 | 8.90e-01 |
| GO:BP | GO:0001503 | ossification | 4 | 313 | 8.91e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1 | 4045 | 8.91e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 2 | 336 | 8.91e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 1 | 120 | 8.91e-01 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 8.91e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 3 | 7806 | 8.91e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 4 | 391 | 8.91e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 42 | 2565 | 8.91e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 2 | 1335 | 8.91e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 1 | 98 | 8.92e-01 |
| GO:BP | GO:0022008 | neurogenesis | 5 | 1294 | 8.92e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 1 | 357 | 8.92e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 2 | 544 | 8.92e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 8.93e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 2 | 190 | 8.93e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 3 | 212 | 8.93e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 1 | 142 | 8.93e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 42 | 2573 | 8.93e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 8.93e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 8.93e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 3 | 993 | 8.93e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 8.93e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 1 | 362 | 8.94e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 8.94e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 1 | 139 | 8.94e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 8.95e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 8.96e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 3 | 4241 | 8.97e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 1 | 351 | 8.97e-01 |
| GO:BP | GO:0007565 | female pregnancy | 1 | 122 | 8.97e-01 |
| GO:BP | GO:0006281 | DNA repair | 6 | 534 | 8.98e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 3 | 484 | 8.98e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 3 | 665 | 8.98e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 1 | 353 | 8.99e-01 |
| GO:BP | GO:0000280 | nuclear division | 2 | 344 | 9.00e-01 |
| GO:BP | GO:0042886 | amide transport | 2 | 235 | 9.01e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 9.01e-01 |
| GO:BP | GO:0009056 | catabolic process | 14 | 2088 | 9.02e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 9.02e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 1 | 133 | 9.02e-01 |
| GO:BP | GO:0007292 | female gamete generation | 1 | 103 | 9.02e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 9.03e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 1 | 190 | 9.05e-01 |
| GO:BP | GO:0006997 | nucleus organization | 1 | 126 | 9.05e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 2 | 482 | 9.07e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 2 | 1496 | 9.07e-01 |
| GO:BP | GO:0009306 | protein secretion | 2 | 262 | 9.07e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 1 | 110 | 9.07e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 1 | 324 | 9.07e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 9.07e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 2 | 262 | 9.08e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 2 | 294 | 9.08e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 9.10e-01 |
| GO:BP | GO:0065007 | biological regulation | 3 | 8320 | 9.10e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 9.11e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 1 | 157 | 9.13e-01 |
| GO:BP | GO:0008544 | epidermis development | 2 | 210 | 9.13e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 2 | 266 | 9.14e-01 |
| GO:BP | GO:0072006 | nephron development | 1 | 117 | 9.14e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 10 | 809 | 9.14e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 4 | 4794 | 9.15e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 1 | 126 | 9.16e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 1 | 422 | 9.17e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 2 | 816 | 9.17e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 5 | 447 | 9.17e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 10 | 1201 | 9.17e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 1 | 401 | 9.18e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 1 | 131 | 9.18e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 1 | 86 | 9.18e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 10 | 755 | 9.18e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 3 | 2724 | 9.20e-01 |
| GO:BP | GO:0001816 | cytokine production | 5 | 451 | 9.20e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 1 | 408 | 9.21e-01 |
| GO:BP | GO:0051301 | cell division | 3 | 570 | 9.22e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 2 | 1836 | 9.22e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 4 | 884 | 9.24e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 5 | 4955 | 9.24e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 1 | 88 | 9.26e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 1 | 135 | 9.26e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 5 | 877 | 9.26e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 2 | 827 | 9.26e-01 |
| GO:BP | GO:0048285 | organelle fission | 2 | 388 | 9.27e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 1 | 135 | 9.27e-01 |
| GO:BP | GO:0061061 | muscle structure development | 6 | 546 | 9.27e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 4 | 369 | 9.28e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 9.29e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 15 | 1378 | 9.29e-01 |
| GO:BP | GO:0032259 | methylation | 3 | 313 | 9.29e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 1 | 94 | 9.29e-01 |
| GO:BP | GO:0030030 | cell projection organization | 10 | 1233 | 9.30e-01 |
| GO:BP | GO:0051640 | organelle localization | 1 | 503 | 9.30e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 1 | 403 | 9.31e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 9.32e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 2 | 199 | 9.32e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 1 | 116 | 9.32e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 2 | 785 | 9.32e-01 |
| GO:BP | GO:0060271 | cilium assembly | 3 | 292 | 9.32e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 1 | 151 | 9.33e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 994 | 9.33e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 7 | 782 | 9.33e-01 |
| GO:BP | GO:0070925 | organelle assembly | 1 | 817 | 9.33e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 3 | 338 | 9.34e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 1 | 162 | 9.35e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 4 | 1138 | 9.38e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 6 | 740 | 9.38e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 1 | 206 | 9.38e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 1 | 90 | 9.39e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 2 | 308 | 9.39e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 1 | 457 | 9.39e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 1 | 194 | 9.39e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 5 | 665 | 9.39e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 1 | 134 | 9.39e-01 |
| GO:BP | GO:0048511 | rhythmic process | 2 | 237 | 9.41e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 1 | 225 | 9.42e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 2 | 389 | 9.42e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 8 | 963 | 9.44e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 3 | 581 | 9.45e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 9.47e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 9.48e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 9.52e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 9.53e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 2 | 859 | 9.53e-01 |
| GO:BP | GO:0007281 | germ cell development | 1 | 187 | 9.53e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 2 | 215 | 9.53e-01 |
| GO:BP | GO:0051641 | cellular localization | 3 | 2810 | 9.54e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 9.54e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 9.55e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 9.55e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 1 | 156 | 9.55e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 1 | 168 | 9.56e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 9.56e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 681 | 9.56e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 468 | 9.56e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 9.56e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 138 | 9.57e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 1 | 154 | 9.58e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 2 | 433 | 9.58e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 2 | 271 | 9.58e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 2 | 250 | 9.58e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 9.58e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 9.58e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 9.58e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 9.58e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 1 | 157 | 9.58e-01 |
| GO:BP | GO:0010876 | lipid localization | 2 | 310 | 9.59e-01 |
| GO:BP | GO:0007033 | vacuole organization | 1 | 197 | 9.59e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 2 | 398 | 9.61e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 4 | 1869 | 9.61e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 9.61e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 1 | 535 | 9.61e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 149 | 9.62e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1361 | 9.62e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 9.63e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 1 | 194 | 9.63e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 1 | 105 | 9.63e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 1 | 168 | 9.63e-01 |
| GO:BP | GO:0023061 | signal release | 2 | 321 | 9.63e-01 |
| GO:BP | GO:0006909 | phagocytosis | 1 | 148 | 9.63e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 9.64e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 1 | 191 | 9.64e-01 |
| GO:BP | GO:0010033 | response to organic substance | 1 | 1957 | 9.64e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 1 | 187 | 9.66e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 9.66e-01 |
| GO:BP | GO:0006310 | DNA recombination | 2 | 257 | 9.69e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1719 | 9.69e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1462 | 9.69e-01 |
| GO:BP | GO:0006996 | organelle organization | 3 | 3000 | 9.69e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 9.71e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 8 | 2890 | 9.72e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 227 | 9.72e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 1 | 245 | 9.73e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 9.75e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 2 | 2466 | 9.76e-01 |
| GO:BP | GO:0071702 | organic substance transport | 5 | 2116 | 9.76e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 1 | 211 | 9.76e-01 |
| GO:BP | GO:0042592 | homeostatic process | 3 | 1159 | 9.76e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 4 | 422 | 9.76e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 9.76e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 5 | 676 | 9.77e-01 |
| GO:BP | GO:0051179 | localization | 6 | 3969 | 9.78e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 1 | 300 | 9.78e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 9.78e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 9.78e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 2 | 305 | 9.79e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 2 | 252 | 9.81e-01 |
| GO:BP | GO:0009725 | response to hormone | 1 | 642 | 9.83e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 2 | 843 | 9.83e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 9.83e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 9.85e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 2 | 382 | 9.85e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 9.85e-01 |
| GO:BP | GO:0007165 | signal transduction | 1 | 3818 | 9.86e-01 |
| GO:BP | GO:0002252 | immune effector process | 1 | 318 | 9.86e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1684 | 9.86e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 9.86e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 9.86e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 174 | 9.87e-01 |
| GO:BP | GO:0006950 | response to stress | 17 | 2780 | 9.88e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 4 | 478 | 9.89e-01 |
| GO:BP | GO:0023052 | signaling | 1 | 4141 | 9.92e-01 |
| GO:BP | GO:0051258 | protein polymerization | 1 | 241 | 9.92e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 9.92e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 9.92e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 9.92e-01 |
| GO:BP | GO:0006869 | lipid transport | 1 | 272 | 9.92e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 2 | 418 | 9.92e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 9.92e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 1 | 191 | 9.92e-01 |
| GO:BP | GO:0006959 | humoral immune response | 1 | 80 | 9.92e-01 |
| GO:BP | GO:0007154 | cell communication | 1 | 4226 | 9.93e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 9.93e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 9.95e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 9.95e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 9.95e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 1 | 142 | 9.96e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 1 | 200 | 9.96e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 5 | 5274 | 9.96e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 9.96e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 9.96e-01 |
| GO:BP | GO:0009314 | response to radiation | 2 | 352 | 9.96e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 9.96e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 11 | 1399 | 9.97e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 1 | 214 | 9.98e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 9.98e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 9.98e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 9.98e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 9.98e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 9.99e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 3 | 103 | 9.99e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 665 | 1.00e+00 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 1.00e+00 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0032940 | secretion by cell | 2 | 569 | 1.00e+00 |
| GO:BP | GO:0140352 | export from cell | 2 | 611 | 1.00e+00 |
| GO:BP | GO:0046903 | secretion | 2 | 645 | 1.00e+00 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5770 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0016043 | cellular component organization | 8 | 5076 | 1.00e+00 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 19 | 415 | 2.59e-03 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 4 | 66 | 3.67e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 3 | 86 | 4.26e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 6 | 104 | 4.26e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 6 | 117 | 4.32e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 4.32e-01 |
| KEGG | KEGG:03040 | Spliceosome | 3 | 132 | 4.80e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 4.80e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 3 | 145 | 4.80e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 2 | 99 | 5.71e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 1 | 48 | 5.71e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 2 | 35 | 5.71e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 2 | 121 | 5.71e-01 |
| KEGG | KEGG:05164 | Influenza A | 5 | 103 | 5.71e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 3 | 56 | 5.71e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 5 | 409 | 5.71e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 3 | 84 | 5.71e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 3 | 75 | 5.71e-01 |
| KEGG | KEGG:05218 | Melanoma | 3 | 58 | 5.71e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 3 | 153 | 5.71e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 3 | 75 | 5.71e-01 |
| KEGG | KEGG:05224 | Breast cancer | 4 | 117 | 5.71e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 5.71e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 5.71e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 3 | 72 | 5.71e-01 |
| KEGG | KEGG:04360 | Axon guidance | 2 | 162 | 5.71e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 4 | 91 | 5.71e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 2 | 138 | 5.71e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 1 | 19 | 5.71e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 1 | 40 | 5.71e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 5.71e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 2 | 20 | 5.71e-01 |
| KEGG | KEGG:04210 | Apoptosis | 4 | 118 | 5.71e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 5.71e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 2 | 56 | 5.87e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 4 | 134 | 6.06e-01 |
| KEGG | KEGG:04144 | Endocytosis | 2 | 232 | 6.15e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 1 | 115 | 6.15e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 6.30e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 6.43e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 1 | 24 | 6.43e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 1 | 56 | 6.43e-01 |
| KEGG | KEGG:04217 | Necroptosis | 4 | 106 | 6.43e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 3 | 114 | 6.43e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 2 | 57 | 6.81e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 2 | 69 | 6.81e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 1 | 14 | 6.81e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 6.81e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 3 | 157 | 6.85e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 3 | 87 | 6.85e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 3 | 98 | 6.85e-01 |
| KEGG | KEGG:04924 | Renin secretion | 1 | 50 | 6.87e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 3 | 87 | 6.87e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 5 | 179 | 6.87e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 3 | 121 | 6.88e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 2 | 60 | 6.88e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 2 | 70 | 6.88e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 1 | 52 | 6.88e-01 |
| KEGG | KEGG:05142 | Chagas disease | 2 | 75 | 6.88e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 3 | 115 | 7.01e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 2 | 67 | 7.01e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 1 | 26 | 7.01e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 1 | 35 | 7.01e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 1 | 124 | 7.01e-01 |
| KEGG | KEGG:05133 | Pertussis | 2 | 45 | 7.01e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 2 | 91 | 7.01e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 2 | 65 | 7.01e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 1 | 31 | 7.01e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 3 | 109 | 7.01e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 3 | 110 | 7.01e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 2 | 112 | 7.01e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 1 | 26 | 7.01e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 2 | 315 | 7.01e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 3 | 134 | 7.01e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 2 | 63 | 7.01e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 1 | 79 | 7.01e-01 |
| KEGG | KEGG:05214 | Glioma | 2 | 67 | 7.01e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 3 | 88 | 7.01e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 2 | 83 | 7.01e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 7.01e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 3 | 136 | 7.01e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 1 | 77 | 7.01e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 7.01e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 1 | 19 | 7.01e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 1 | 27 | 7.01e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 7.08e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 1 | 39 | 7.08e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 2 | 76 | 7.08e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 1 | 65 | 7.08e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 2 | 107 | 7.08e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 1 | 74 | 7.08e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 1 | 46 | 7.08e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 7.08e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 2 | 95 | 7.08e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 1 | 49 | 7.08e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 1 | 170 | 7.08e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 1 | 172 | 7.08e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 3 | 150 | 7.08e-01 |
| KEGG | KEGG:04530 | Tight junction | 2 | 137 | 7.08e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 3 | 128 | 7.08e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 1 | 165 | 7.08e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 1 | 63 | 7.08e-01 |
| KEGG | KEGG:04611 | Platelet activation | 2 | 89 | 7.08e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 2 | 145 | 7.08e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 7.08e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 1 | 41 | 7.08e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 2 | 97 | 7.08e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 2 | 76 | 7.08e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 1 | 53 | 7.08e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 3 | 180 | 7.08e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 2 | 98 | 7.08e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 2 | 272 | 7.08e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 7.08e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 1 | 176 | 7.08e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 1 | 97 | 7.08e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 2 | 385 | 7.08e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 1 | 46 | 7.08e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 1 | 65 | 7.08e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 2 | 84 | 7.08e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 1 | 74 | 7.08e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 1 | 115 | 7.08e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 1 | 161 | 7.08e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 2 | 87 | 7.08e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 1 | 135 | 7.08e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 3 | 145 | 7.08e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 1 | 55 | 7.08e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 2 | 84 | 7.08e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 7.08e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 1 | 70 | 7.08e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 7.08e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 7.11e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 7.11e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 7.13e-01 |
| KEGG | KEGG:04520 | Adherens junction | 1 | 67 | 7.13e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 7.13e-01 |
| KEGG | KEGG:04730 | Long-term depression | 1 | 42 | 7.13e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 1 | 85 | 7.13e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 1 | 186 | 7.13e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 2 | 164 | 7.13e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 1 | 49 | 7.13e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 2 | 106 | 7.13e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 2 | 142 | 7.13e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 7.13e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 2 | 131 | 7.13e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 7.13e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 1 | 86 | 7.19e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 7.19e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 7.26e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 2 | 171 | 7.26e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 7.26e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 1 | 89 | 7.33e-01 |
| KEGG | KEGG:04110 | Cell cycle | 2 | 151 | 7.40e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 2 | 214 | 7.41e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 7.41e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 1 | 110 | 7.41e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 7.41e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 1 | 226 | 7.41e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 1 | 112 | 7.41e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 7.41e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 3 | 149 | 7.47e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 7.58e-01 |
| KEGG | KEGG:05131 | Shigellosis | 2 | 210 | 7.66e-01 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 2 | 169 | 7.67e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 1 | 133 | 7.70e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 7.70e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 2 | 110 | 7.70e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 7.70e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 7.97e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 3 | 254 | 7.97e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 3 | 240 | 8.03e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 8.04e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 8.04e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 2 | 162 | 8.04e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 8.41e-01 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 1 | 61 | 8.48e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 8.52e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 1 | 94 | 8.63e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 8.71e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 1 | 133 | 9.10e-01 |
| KEGG | KEGG:05020 | Prion disease | 1 | 220 | 9.12e-01 |
| KEGG | KEGG:05034 | Alcoholism | 1 | 122 | 9.13e-01 |
DNR 24 sigDEG
Looking at the DNR 24 hour geneset using adj. P value of 0.05, I have enrichment in the terms cell cycle and chromosome segregation.
# DAgostres24 <- gost(query = sigVDA24$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(DAgostres24, "data/DAgostres24.RDS")
DAgostres24 <- readRDS("data/DAgostres24.RDS")
# DAp24 <- gostplot(DAgostres24, capped = FALSE, interactive = TRUE)
# DAp24
DAtable124 <- DAgostres24$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DAtable124 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('DNR 24 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)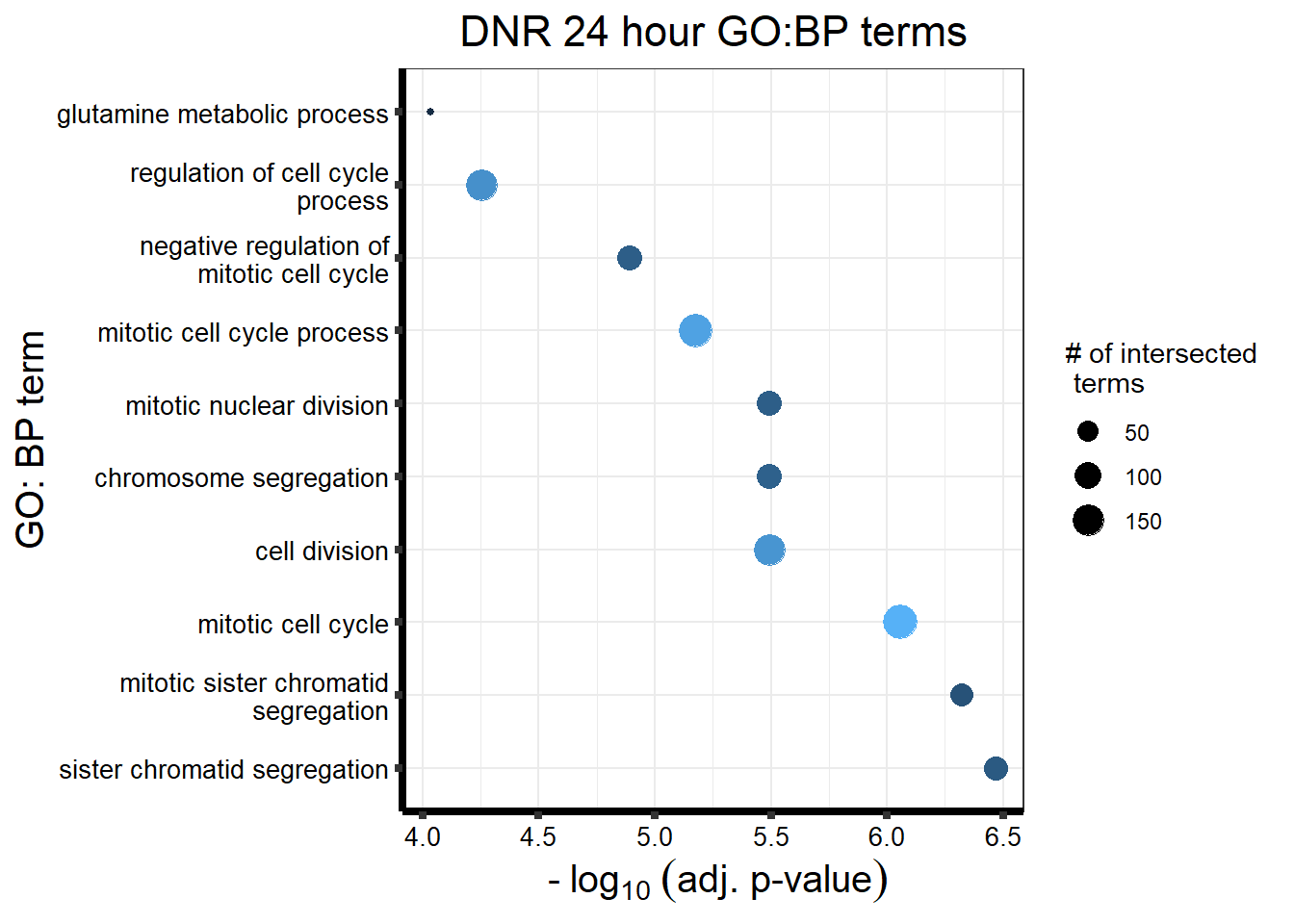
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DAtable124 %>% mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "DNR 24 hour sig DEGs enrichment, all KEGG and GO:BP") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0000819 | sister chromatid segregation | 72 | 217 | 3.40e-07 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 62 | 179 | 4.74e-07 |
| GO:BP | GO:0000278 | mitotic cell cycle | 185 | 827 | 8.76e-07 |
| GO:BP | GO:0007059 | chromosome segregation | 82 | 369 | 3.20e-06 |
| GO:BP | GO:0140014 | mitotic nuclear division | 78 | 251 | 3.20e-06 |
| GO:BP | GO:0051301 | cell division | 150 | 570 | 3.20e-06 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 167 | 694 | 6.67e-06 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 78 | 211 | 1.28e-05 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 144 | 626 | 5.55e-05 |
| GO:BP | GO:0006541 | glutamine metabolic process | 4 | 24 | 9.25e-05 |
| GO:BP | GO:0022402 | cell cycle process | 227 | 1086 | 2.07e-04 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 42 | 139 | 2.07e-04 |
| GO:BP | GO:0000280 | nuclear division | 102 | 344 | 2.07e-04 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 82 | 273 | 2.35e-04 |
| GO:BP | GO:0051726 | regulation of cell cycle | 202 | 956 | 2.75e-04 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 20 | 42 | 3.04e-04 |
| GO:BP | GO:0007052 | mitotic spindle organization | 37 | 129 | 3.50e-04 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 36 | 121 | 3.99e-04 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 110 | 338 | 4.11e-04 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 50 | 183 | 4.24e-04 |
| GO:BP | GO:0051276 | chromosome organization | 135 | 543 | 4.26e-04 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 18 | 52 | 4.26e-04 |
| GO:BP | GO:0007051 | spindle organization | 55 | 188 | 4.62e-04 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 25 | 71 | 4.62e-04 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 32 | 79 | 4.62e-04 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 18 | 54 | 4.62e-04 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 12 | 18 | 4.62e-04 |
| GO:BP | GO:0051225 | spindle assembly | 35 | 120 | 4.62e-04 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 7 | 7 | 4.64e-04 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 42 | 157 | 4.92e-04 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 89 | 273 | 6.73e-04 |
| GO:BP | GO:0048285 | organelle fission | 107 | 388 | 6.73e-04 |
| GO:BP | GO:0006974 | DNA damage response | 193 | 785 | 6.81e-04 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 27 | 88 | 7.25e-04 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 18 | 55 | 7.86e-04 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 16 | 46 | 8.14e-04 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 46 | 90 | 1.06e-03 |
| GO:BP | GO:0051255 | spindle midzone assembly | 9 | 12 | 1.12e-03 |
| GO:BP | GO:0051231 | spindle elongation | 9 | 12 | 1.12e-03 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 18 | 58 | 1.12e-03 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 29 | 99 | 1.12e-03 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 16 | 47 | 1.12e-03 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 16 | 47 | 1.12e-03 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 16 | 47 | 1.12e-03 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 9 | 1.12e-03 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 27 | 89 | 1.12e-03 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 16 | 47 | 1.12e-03 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 27 | 91 | 1.12e-03 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 27 | 100 | 1.13e-03 |
| GO:BP | GO:0051304 | chromosome separation | 25 | 76 | 1.17e-03 |
| GO:BP | GO:0051310 | metaphase plate congression | 20 | 69 | 1.27e-03 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 8 | 8 | 1.27e-03 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 17 | 44 | 1.37e-03 |
| GO:BP | GO:0006177 | GMP biosynthetic process | 3 | 13 | 1.37e-03 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 17 | 44 | 1.37e-03 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 3 | 14 | 1.37e-03 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 17 | 44 | 1.37e-03 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 18 | 57 | 1.41e-03 |
| GO:BP | GO:0007049 | cell cycle | 297 | 1529 | 1.41e-03 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 4 | 60 | 1.41e-03 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 98 | 441 | 1.41e-03 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 16 | 48 | 1.49e-03 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 27 | 92 | 1.49e-03 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 16 | 48 | 1.49e-03 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 17 | 45 | 1.62e-03 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 18 | 61 | 1.62e-03 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 10 | 1.76e-03 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 76 | 235 | 1.80e-03 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 10 | 69 | 1.84e-03 |
| GO:BP | GO:0006281 | DNA repair | 127 | 534 | 1.92e-03 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 56 | 209 | 2.69e-03 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 17 | 27 | 2.69e-03 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 11 | 21 | 2.93e-03 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 21 | 75 | 3.02e-03 |
| GO:BP | GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 3 | 19 | 3.22e-03 |
| GO:BP | GO:0050000 | chromosome localization | 22 | 82 | 3.25e-03 |
| GO:BP | GO:0046060 | dATP metabolic process | 2 | 3 | 3.40e-03 |
| GO:BP | GO:0009127 | purine nucleoside monophosphate biosynthetic process | 3 | 21 | 4.17e-03 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 22 | 70 | 4.19e-03 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 52 | 166 | 4.44e-03 |
| GO:BP | GO:0051783 | regulation of nuclear division | 29 | 116 | 5.53e-03 |
| GO:BP | GO:0046033 | AMP metabolic process | 3 | 22 | 6.65e-03 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 8 | 11 | 6.77e-03 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 28 | 81 | 6.80e-03 |
| GO:BP | GO:0046037 | GMP metabolic process | 3 | 23 | 7.20e-03 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 7.30e-03 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 7.30e-03 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 79 | 148 | 7.30e-03 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 7 | 11 | 7.83e-03 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 120 | 418 | 8.19e-03 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 56 | 236 | 8.71e-03 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 14 | 161 | 8.71e-03 |
| GO:BP | GO:0044209 | AMP salvage | 2 | 5 | 9.24e-03 |
| GO:BP | GO:0032263 | GMP salvage | 2 | 4 | 9.24e-03 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 68 | 308 | 9.41e-03 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 84 | 391 | 1.01e-02 |
| GO:BP | GO:0051302 | regulation of cell division | 47 | 145 | 1.03e-02 |
| GO:BP | GO:0051383 | kinetochore organization | 11 | 23 | 1.07e-02 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 3 | 30 | 1.24e-02 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 3 | 27 | 1.24e-02 |
| GO:BP | GO:0036166 | phenotypic switching | 5 | 7 | 1.26e-02 |
| GO:BP | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process | 2 | 6 | 1.28e-02 |
| GO:BP | GO:0010212 | response to ionizing radiation | 39 | 130 | 1.28e-02 |
| GO:BP | GO:0006270 | DNA replication initiation | 15 | 35 | 1.41e-02 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 15 | 178 | 1.65e-02 |
| GO:BP | GO:0046053 | dAMP metabolic process | 2 | 5 | 1.72e-02 |
| GO:BP | GO:0044770 | cell cycle phase transition | 102 | 504 | 1.77e-02 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 6 | 43 | 1.89e-02 |
| GO:BP | GO:0000910 | cytokinesis | 43 | 164 | 1.89e-02 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 9 | 42 | 1.93e-02 |
| GO:BP | GO:0106380 | purine ribonucleotide salvage | 2 | 7 | 2.15e-02 |
| GO:BP | GO:0035878 | nail development | 2 | 8 | 2.15e-02 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 2 | 8 | 2.15e-02 |
| GO:BP | GO:0006260 | DNA replication | 96 | 255 | 2.15e-02 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 3 | 37 | 2.16e-02 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 3 | 37 | 2.16e-02 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 43 | 129 | 2.23e-02 |
| GO:BP | GO:0010332 | response to gamma radiation | 17 | 50 | 2.33e-02 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 28 | 85 | 2.33e-02 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 6 | 8 | 2.33e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 14 | 268 | 2.33e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 14 | 268 | 2.33e-02 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 3 | 39 | 2.34e-02 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 16 | 28 | 2.36e-02 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 12 | 40 | 2.39e-02 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 14 | 36 | 2.63e-02 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 6 | 9 | 2.66e-02 |
| GO:BP | GO:1902969 | mitotic DNA replication | 9 | 14 | 2.68e-02 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 4 | 151 | 2.68e-02 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 104 | 552 | 2.73e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 5 | 7 | 2.89e-02 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 24 | 47 | 2.90e-02 |
| GO:BP | GO:0009170 | purine deoxyribonucleoside monophosphate metabolic process | 2 | 7 | 2.92e-02 |
| GO:BP | GO:0090224 | regulation of spindle organization | 9 | 45 | 3.12e-02 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 9 | 81 | 3.14e-02 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 3.25e-02 |
| GO:BP | GO:0006284 | base-excision repair | 11 | 44 | 3.31e-02 |
| GO:BP | GO:0009411 | response to UV | 12 | 141 | 3.31e-02 |
| GO:BP | GO:0032261 | purine nucleotide salvage | 2 | 10 | 3.31e-02 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 2 | 11 | 3.31e-02 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 2 | 11 | 3.31e-02 |
| GO:BP | GO:0007098 | centrosome cycle | 33 | 127 | 3.31e-02 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 2 | 11 | 3.31e-02 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 8 | 11 | 3.35e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 3 | 35 | 3.35e-02 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 2 | 16 | 3.37e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 3 | 36 | 3.52e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 3 | 36 | 3.52e-02 |
| GO:BP | GO:1990227 | paranodal junction maintenance | 1 | 2 | 3.79e-02 |
| GO:BP | GO:0019985 | translesion synthesis | 10 | 24 | 3.97e-02 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 65 | 274 | 4.07e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 14 | 39 | 4.10e-02 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 2 | 18 | 4.10e-02 |
| GO:BP | GO:0061642 | chemoattraction of axon | 2 | 2 | 4.45e-02 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 3 | 50 | 4.57e-02 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 10 | 15 | 4.76e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 1 | 8 | 4.77e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 10 | 36 | 4.77e-02 |
| GO:BP | GO:0033260 | nuclear DNA replication | 9 | 35 | 4.89e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 3 | 13 | 4.98e-02 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 17 | 20 | 5.01e-02 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 30 | 121 | 5.44e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 2 | 15 | 5.44e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 2 | 15 | 5.44e-02 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 2 | 14 | 5.44e-02 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 6 | 28 | 5.47e-02 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 77 | 5.47e-02 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 11 | 103 | 5.47e-02 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 7 | 13 | 5.82e-02 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 2 | 4 | 5.84e-02 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 4 | 15 | 5.84e-02 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 8 | 12 | 5.84e-02 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 8 | 12 | 5.84e-02 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 31 | 104 | 5.87e-02 |
| GO:BP | GO:0009151 | purine deoxyribonucleotide metabolic process | 2 | 12 | 5.87e-02 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 7 | 13 | 6.18e-02 |
| GO:BP | GO:0071478 | cellular response to radiation | 31 | 156 | 6.18e-02 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 38 | 134 | 6.18e-02 |
| GO:BP | GO:0042148 | strand invasion | 4 | 5 | 6.27e-02 |
| GO:BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 4 | 18 | 6.61e-02 |
| GO:BP | GO:0015691 | cadmium ion transport | 4 | 5 | 6.66e-02 |
| GO:BP | GO:0070574 | cadmium ion transmembrane transport | 4 | 5 | 6.66e-02 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 34 | 137 | 6.71e-02 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 15 | 33 | 6.71e-02 |
| GO:BP | GO:0043173 | nucleotide salvage | 2 | 15 | 6.97e-02 |
| GO:BP | GO:0006520 | amino acid metabolic process | 4 | 222 | 6.97e-02 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 7 | 40 | 6.97e-02 |
| GO:BP | GO:0099068 | postsynapse assembly | 9 | 32 | 6.97e-02 |
| GO:BP | GO:0044210 | ‘de novo’ CTP biosynthetic process | 1 | 2 | 6.97e-02 |
| GO:BP | GO:0051299 | centrosome separation | 10 | 14 | 7.03e-02 |
| GO:BP | GO:1990170 | stress response to cadmium ion | 2 | 2 | 7.28e-02 |
| GO:BP | GO:0034502 | protein localization to chromosome | 8 | 110 | 7.28e-02 |
| GO:BP | GO:0071585 | detoxification of cadmium ion | 2 | 2 | 7.28e-02 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 7.28e-02 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 7.30e-02 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 2 | 19 | 7.41e-02 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 31 | 48 | 7.58e-02 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 3 | 64 | 7.58e-02 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 21 | 270 | 7.58e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 12 | 34 | 7.61e-02 |
| GO:BP | GO:0048368 | lateral mesoderm development | 2 | 10 | 7.61e-02 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 10 | 42 | 7.61e-02 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 23 | 318 | 7.82e-02 |
| GO:BP | GO:0035611 | protein branching point deglutamylation | 1 | 1 | 7.82e-02 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 41 | 149 | 7.82e-02 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 2 | 15 | 7.82e-02 |
| GO:BP | GO:0098792 | xenophagy | 9 | 12 | 7.93e-02 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 40 | 74 | 8.09e-02 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 8.09e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 13 | 38 | 8.09e-02 |
| GO:BP | GO:0051795 | positive regulation of timing of catagen | 1 | 3 | 8.09e-02 |
| GO:BP | GO:2000233 | negative regulation of rRNA processing | 3 | 3 | 8.09e-02 |
| GO:BP | GO:0061912 | selective autophagy | 60 | 90 | 8.09e-02 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 9 | 19 | 8.09e-02 |
| GO:BP | GO:0006312 | mitotic recombination | 8 | 22 | 8.24e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 26 | 73 | 8.24e-02 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 8.24e-02 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 19 | 50 | 8.35e-02 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 10 | 29 | 8.35e-02 |
| GO:BP | GO:1900262 | regulation of DNA-directed DNA polymerase activity | 11 | 13 | 8.35e-02 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 28 | 92 | 8.35e-02 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 11 | 13 | 8.35e-02 |
| GO:BP | GO:0006259 | DNA metabolic process | 192 | 884 | 8.35e-02 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 27 | 68 | 8.39e-02 |
| GO:BP | GO:0072201 | negative regulation of mesenchymal cell proliferation | 5 | 7 | 8.43e-02 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 14 | 186 | 8.43e-02 |
| GO:BP | GO:0034508 | centromere complex assembly | 18 | 28 | 8.44e-02 |
| GO:BP | GO:0034644 | cellular response to UV | 6 | 86 | 8.44e-02 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 2 | 16 | 8.52e-02 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 6 | 23 | 8.62e-02 |
| GO:BP | GO:0006301 | postreplication repair | 13 | 34 | 8.62e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 2 | 84 | 8.86e-02 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 7 | 681 | 8.94e-02 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 7 | 14 | 9.17e-02 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 6 | 21 | 9.18e-02 |
| GO:BP | GO:0097114 | NMDA glutamate receptor clustering | 2 | 5 | 9.18e-02 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 9.18e-02 |
| GO:BP | GO:0051794 | regulation of timing of catagen | 1 | 4 | 9.22e-02 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 12 | 17 | 9.22e-02 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 12 | 17 | 9.22e-02 |
| GO:BP | GO:0044314 | protein K27-linked ubiquitination | 5 | 8 | 9.22e-02 |
| GO:BP | GO:0042637 | catagen | 1 | 4 | 9.22e-02 |
| GO:BP | GO:0097106 | postsynaptic density organization | 7 | 30 | 9.22e-02 |
| GO:BP | GO:0060364 | frontal suture morphogenesis | 1 | 4 | 9.22e-02 |
| GO:BP | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | 2 | 5 | 9.22e-02 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 9.22e-02 |
| GO:BP | GO:0006189 | ‘de novo’ IMP biosynthetic process | 5 | 6 | 9.22e-02 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 12 | 17 | 9.22e-02 |
| GO:BP | GO:0033554 | cellular response to stress | 58 | 1681 | 9.27e-02 |
| GO:BP | GO:0060169 | negative regulation of adenosine receptor signaling pathway | 1 | 1 | 9.30e-02 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 1 | 5 | 9.30e-02 |
| GO:BP | GO:0046111 | xanthine biosynthetic process | 1 | 1 | 9.30e-02 |
| GO:BP | GO:0060407 | negative regulation of penile erection | 1 | 1 | 9.30e-02 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 7 | 696 | 9.36e-02 |
| GO:BP | GO:0032392 | DNA geometric change | 42 | 80 | 9.53e-02 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 4 | 6 | 9.53e-02 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 4 | 6 | 9.53e-02 |
| GO:BP | GO:0006082 | organic acid metabolic process | 7 | 700 | 9.55e-02 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 11 | 41 | 9.55e-02 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 15 | 160 | 9.58e-02 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 8 | 17 | 9.59e-02 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 5 | 7 | 9.59e-02 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 2 | 18 | 9.59e-02 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 4 | 14 | 9.69e-02 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 26 | 464 | 9.69e-02 |
| GO:BP | GO:0106014 | regulation of inflammatory response to wounding | 2 | 4 | 9.69e-02 |
| GO:BP | GO:0071168 | protein localization to chromatin | 6 | 48 | 9.82e-02 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 9.84e-02 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 12 | 22 | 9.86e-02 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 14 | 103 | 9.86e-02 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 8 | 19 | 9.94e-02 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 14 | 41 | 9.94e-02 |
| GO:BP | GO:0097065 | anterior head development | 6 | 7 | 9.94e-02 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 8 | 19 | 9.94e-02 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 3 | 5 | 1.00e-01 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 3 | 5 | 1.00e-01 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 3 | 5 | 1.00e-01 |
| GO:BP | GO:0048818 | positive regulation of hair follicle maturation | 1 | 5 | 1.02e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 2 | 80 | 1.02e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 2 | 2 | 1.03e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 6 | 25 | 1.03e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 1.03e-01 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 2 | 20 | 1.04e-01 |
| GO:BP | GO:0051095 | regulation of helicase activity | 8 | 12 | 1.04e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 46 | 168 | 1.06e-01 |
| GO:BP | GO:0002076 | osteoblast development | 2 | 12 | 1.06e-01 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 5 | 11 | 1.06e-01 |
| GO:BP | GO:0061724 | lipophagy | 2 | 7 | 1.06e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 8 | 21 | 1.07e-01 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 3 | 4 | 1.12e-01 |
| GO:BP | GO:1903925 | response to bisphenol A | 3 | 4 | 1.13e-01 |
| GO:BP | GO:0006175 | dATP biosynthetic process | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 3 | 17 | 1.13e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 1.13e-01 |
| GO:BP | GO:0046599 | regulation of centriole replication | 10 | 21 | 1.13e-01 |
| GO:BP | GO:0032792 | negative regulation of CREB transcription factor activity | 1 | 4 | 1.13e-01 |
| GO:BP | GO:1903926 | cellular response to bisphenol A | 3 | 4 | 1.13e-01 |
| GO:BP | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 1.13e-01 |
| GO:BP | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 1.13e-01 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 1 | 7 | 1.13e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 5 | 7 | 1.14e-01 |
| GO:BP | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process | 6 | 6 | 1.15e-01 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 3 | 3 | 1.16e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 7 | 9 | 1.17e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 14 | 35 | 1.17e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 2 | 14 | 1.18e-01 |
| GO:BP | GO:0043094 | cellular metabolic compound salvage | 2 | 25 | 1.18e-01 |
| GO:BP | GO:0006298 | mismatch repair | 4 | 29 | 1.18e-01 |
| GO:BP | GO:0051640 | organelle localization | 92 | 503 | 1.18e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 18 | 68 | 1.19e-01 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 2 | 9 | 1.19e-01 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 1.20e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 1.20e-01 |
| GO:BP | GO:0032506 | cytokinetic process | 13 | 39 | 1.21e-01 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 6 | 26 | 1.23e-01 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 15 | 18 | 1.23e-01 |
| GO:BP | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | 3 | 7 | 1.23e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 5 | 28 | 1.24e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 1.24e-01 |
| GO:BP | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2 | 2 | 1.25e-01 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 4 | 191 | 1.29e-01 |
| GO:BP | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process | 5 | 15 | 1.30e-01 |
| GO:BP | GO:0035973 | aggrephagy | 2 | 8 | 1.30e-01 |
| GO:BP | GO:0021694 | cerebellar Purkinje cell layer formation | 1 | 12 | 1.31e-01 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 16 | 22 | 1.31e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 22 | 352 | 1.31e-01 |
| GO:BP | GO:0009439 | cyanate metabolic process | 2 | 2 | 1.31e-01 |
| GO:BP | GO:0009440 | cyanate catabolic process | 2 | 2 | 1.31e-01 |
| GO:BP | GO:0006655 | phosphatidylglycerol biosynthetic process | 9 | 9 | 1.31e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 54 | 116 | 1.31e-01 |
| GO:BP | GO:0038101 | sequestering of nodal from receptor via nodal binding | 1 | 1 | 1.33e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 1.33e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 19 | 331 | 1.33e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 11 | 45 | 1.34e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 6 | 28 | 1.34e-01 |
| GO:BP | GO:0090215 | regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 4 | 4 | 1.35e-01 |
| GO:BP | GO:0043103 | hypoxanthine salvage | 1 | 2 | 1.35e-01 |
| GO:BP | GO:0002636 | positive regulation of germinal center formation | 1 | 1 | 1.35e-01 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 4 | 4 | 1.35e-01 |
| GO:BP | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | 5 | 16 | 1.35e-01 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 7 | 19 | 1.35e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 19 | 60 | 1.35e-01 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 4 | 197 | 1.35e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 4 | 6 | 1.35e-01 |
| GO:BP | GO:0070256 | negative regulation of mucus secretion | 1 | 2 | 1.35e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 2 | 123 | 1.35e-01 |
| GO:BP | GO:0046110 | xanthine metabolic process | 1 | 1 | 1.35e-01 |
| GO:BP | GO:0046061 | dATP catabolic process | 1 | 2 | 1.35e-01 |
| GO:BP | GO:0009314 | response to radiation | 20 | 352 | 1.35e-01 |
| GO:BP | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1 | 8 | 1.35e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 1.35e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 6 | 425 | 1.35e-01 |
| GO:BP | GO:0045217 | cell-cell junction maintenance | 1 | 13 | 1.35e-01 |
| GO:BP | GO:0072033 | renal vesicle formation | 6 | 8 | 1.35e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 6 | 14 | 1.39e-01 |
| GO:BP | GO:0044851 | hair cycle phase | 1 | 9 | 1.39e-01 |
| GO:BP | GO:0044848 | biological phase | 1 | 9 | 1.39e-01 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 10 | 18 | 1.39e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 12 | 13 | 1.39e-01 |
| GO:BP | GO:0046386 | deoxyribose phosphate catabolic process | 2 | 22 | 1.39e-01 |
| GO:BP | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process | 13 | 17 | 1.39e-01 |
| GO:BP | GO:0032020 | ISG15-protein conjugation | 1 | 6 | 1.39e-01 |
| GO:BP | GO:0032223 | negative regulation of synaptic transmission, cholinergic | 1 | 1 | 1.39e-01 |
| GO:BP | GO:0007032 | endosome organization | 15 | 92 | 1.39e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 5 | 69 | 1.39e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 10 | 82 | 1.39e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 11 | 127 | 1.39e-01 |
| GO:BP | GO:0045617 | negative regulation of keratinocyte differentiation | 1 | 3 | 1.39e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 1.39e-01 |
| GO:BP | GO:0009264 | deoxyribonucleotide catabolic process | 2 | 22 | 1.39e-01 |
| GO:BP | GO:0048819 | regulation of hair follicle maturation | 1 | 9 | 1.39e-01 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 1 | 8 | 1.39e-01 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 4 | 23 | 1.39e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 6 | 10 | 1.39e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 11 | 44 | 1.39e-01 |
| GO:BP | GO:0048341 | paraxial mesoderm formation | 5 | 6 | 1.39e-01 |
| GO:BP | GO:0019856 | pyrimidine nucleobase biosynthetic process | 1 | 8 | 1.39e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 12 | 19 | 1.39e-01 |
| GO:BP | GO:0071772 | response to BMP | 2 | 131 | 1.40e-01 |
| GO:BP | GO:0021571 | rhombomere 5 development | 2 | 2 | 1.40e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 11 | 132 | 1.40e-01 |
| GO:BP | GO:0086016 | AV node cell action potential | 7 | 9 | 1.40e-01 |
| GO:BP | GO:0006543 | glutamine catabolic process | 1 | 3 | 1.40e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 2 | 131 | 1.40e-01 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 1 | 16 | 1.40e-01 |
| GO:BP | GO:0035284 | brain segmentation | 2 | 2 | 1.40e-01 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 7 | 9 | 1.40e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 7 | 9 | 1.40e-01 |
| GO:BP | GO:0009092 | homoserine metabolic process | 4 | 5 | 1.41e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 41 | 875 | 1.41e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 2 | 6 | 1.41e-01 |
| GO:BP | GO:0072308 | negative regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0060346 | bone trabecula formation | 1 | 9 | 1.41e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 3 | 440 | 1.41e-01 |
| GO:BP | GO:0043928 | exonucleolytic catabolism of deadenylated mRNA | 2 | 13 | 1.41e-01 |
| GO:BP | GO:0019346 | transsulfuration | 4 | 5 | 1.41e-01 |
| GO:BP | GO:0021591 | ventricular system development | 20 | 29 | 1.41e-01 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 4 | 4 | 1.41e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 6 | 52 | 1.41e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 1.41e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 3 | 69 | 1.44e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 3 | 4 | 1.44e-01 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 1.44e-01 |
| GO:BP | GO:0097113 | AMPA glutamate receptor clustering | 3 | 7 | 1.44e-01 |
| GO:BP | GO:0097688 | glutamate receptor clustering | 3 | 7 | 1.44e-01 |
| GO:BP | GO:0007099 | centriole replication | 3 | 38 | 1.44e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 10 | 14 | 1.44e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 8 | 11 | 1.44e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 2 | 116 | 1.44e-01 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 2 | 3 | 1.45e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 42 | 868 | 1.45e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 4 | 194 | 1.45e-01 |
| GO:BP | GO:0060157 | urinary bladder development | 2 | 2 | 1.46e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 2 | 59 | 1.49e-01 |
| GO:BP | GO:0061709 | reticulophagy | 14 | 17 | 1.49e-01 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 8 | 9 | 1.50e-01 |
| GO:BP | GO:0097293 | XMP biosynthetic process | 5 | 7 | 1.52e-01 |
| GO:BP | GO:0097294 | ‘de novo’ XMP biosynthetic process | 5 | 7 | 1.52e-01 |
| GO:BP | GO:1990936 | vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 1.52e-01 |
| GO:BP | GO:1905174 | regulation of vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 1.52e-01 |
| GO:BP | GO:0097292 | XMP metabolic process | 5 | 7 | 1.52e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 4 | 76 | 1.52e-01 |
| GO:BP | GO:0090071 | negative regulation of ribosome biogenesis | 3 | 4 | 1.52e-01 |
| GO:BP | GO:0090678 | cell dedifferentiation involved in phenotypic switching | 2 | 2 | 1.52e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 8 | 20 | 1.53e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 8 | 16 | 1.56e-01 |
| GO:BP | GO:0045828 | positive regulation of isoprenoid metabolic process | 1 | 1 | 1.56e-01 |
| GO:BP | GO:1900054 | positive regulation of retinoic acid biosynthetic process | 1 | 1 | 1.56e-01 |
| GO:BP | GO:0061820 | telomeric D-loop disassembly | 5 | 8 | 1.57e-01 |
| GO:BP | GO:0051798 | positive regulation of hair follicle development | 1 | 9 | 1.57e-01 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 3 | 41 | 1.57e-01 |
| GO:BP | GO:0070212 | protein poly-ADP-ribosylation | 5 | 6 | 1.57e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 52 | 1028 | 1.57e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 19 | 62 | 1.57e-01 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 4 | 5 | 1.58e-01 |
| GO:BP | GO:0046101 | hypoxanthine biosynthetic process | 1 | 3 | 1.58e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 6 | 65 | 1.58e-01 |
| GO:BP | GO:0042454 | ribonucleoside catabolic process | 3 | 12 | 1.58e-01 |
| GO:BP | GO:0046090 | deoxyadenosine metabolic process | 1 | 2 | 1.58e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 29 | 85 | 1.58e-01 |
| GO:BP | GO:0031297 | replication fork processing | 25 | 42 | 1.58e-01 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 2 | 8 | 1.58e-01 |
| GO:BP | GO:0043096 | purine nucleobase salvage | 1 | 3 | 1.58e-01 |
| GO:BP | GO:0006157 | deoxyadenosine catabolic process | 1 | 2 | 1.58e-01 |
| GO:BP | GO:0046103 | inosine biosynthetic process | 1 | 3 | 1.58e-01 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 10 | 18 | 1.59e-01 |
| GO:BP | GO:0032901 | positive regulation of neurotrophin production | 1 | 1 | 1.59e-01 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 4 | 30 | 1.59e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 12 | 39 | 1.59e-01 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 10 | 18 | 1.59e-01 |
| GO:BP | GO:0072719 | cellular response to cisplatin | 4 | 4 | 1.60e-01 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 4 | 12 | 1.61e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 1.61e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 1.61e-01 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 1 | 12 | 1.61e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 12 | 93 | 1.62e-01 |
| GO:BP | GO:0007617 | mating behavior | 2 | 21 | 1.63e-01 |
| GO:BP | GO:0098534 | centriole assembly | 3 | 41 | 1.65e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 1 | 22 | 1.65e-01 |
| GO:BP | GO:0019276 | UDP-N-acetylgalactosamine metabolic process | 2 | 2 | 1.65e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 2 | 64 | 1.65e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 8 | 186 | 1.66e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 1.66e-01 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 2 | 7 | 1.66e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 16 | 219 | 1.66e-01 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 5 | 1.66e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 1 | 6 | 1.66e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 39 | 120 | 1.66e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 8 | 14 | 1.66e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 1.66e-01 |
| GO:BP | GO:1901491 | negative regulation of lymphangiogenesis | 3 | 3 | 1.66e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 1 | 6 | 1.66e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 9 | 80 | 1.66e-01 |
| GO:BP | GO:0090677 | reversible differentiation | 3 | 5 | 1.66e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 5 | 9 | 1.66e-01 |
| GO:BP | GO:0035609 | C-terminal protein deglutamylation | 1 | 2 | 1.67e-01 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 4 | 30 | 1.67e-01 |
| GO:BP | GO:0014029 | neural crest formation | 4 | 13 | 1.67e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 4 | 22 | 1.68e-01 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 4 | 6 | 1.68e-01 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 1 | 24 | 1.69e-01 |
| GO:BP | GO:0000921 | septin ring assembly | 1 | 1 | 1.69e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 2 | 2 | 1.69e-01 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 9 | 240 | 1.69e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 3 | 74 | 1.69e-01 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 5 | 19 | 1.69e-01 |
| GO:BP | GO:0030879 | mammary gland development | 2 | 106 | 1.69e-01 |
| GO:BP | GO:1900121 | negative regulation of receptor binding | 1 | 8 | 1.69e-01 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 2 | 13 | 1.69e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 41 | 135 | 1.71e-01 |
| GO:BP | GO:0048820 | hair follicle maturation | 1 | 15 | 1.71e-01 |
| GO:BP | GO:0046521 | sphingoid catabolic process | 1 | 1 | 1.71e-01 |
| GO:BP | GO:1905364 | regulation of endosomal vesicle fusion | 1 | 1 | 1.71e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 1 | 7 | 1.71e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 2 | 14 | 1.71e-01 |
| GO:BP | GO:0022616 | DNA strand elongation | 11 | 35 | 1.71e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 2 | 7 | 1.71e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 9 | 26 | 1.71e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 3 | 40 | 1.71e-01 |
| GO:BP | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential | 3 | 3 | 1.71e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 6 | 6 | 1.71e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 5 | 35 | 1.71e-01 |
| GO:BP | GO:0086017 | Purkinje myocyte action potential | 3 | 4 | 1.72e-01 |
| GO:BP | GO:1901490 | regulation of lymphangiogenesis | 4 | 5 | 1.72e-01 |
| GO:BP | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation | 2 | 5 | 1.72e-01 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 7 | 19 | 1.72e-01 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 7 | 19 | 1.72e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 66 | 175 | 1.72e-01 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 7 | 19 | 1.72e-01 |
| GO:BP | GO:1900176 | negative regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 1 | 1.73e-01 |
| GO:BP | GO:0140131 | positive regulation of lymphocyte chemotaxis | 4 | 10 | 1.73e-01 |
| GO:BP | GO:1900146 | negative regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 1 | 1.73e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 6 | 6 | 1.73e-01 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 1 | 5 | 1.74e-01 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 4 | 1.74e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 8 | 39 | 1.76e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 3 | 5 | 1.76e-01 |
| GO:BP | GO:0002905 | regulation of mature B cell apoptotic process | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 8 | 9 | 1.76e-01 |
| GO:BP | GO:0006154 | adenosine catabolic process | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0051382 | kinetochore assembly | 7 | 18 | 1.76e-01 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 1 | 12 | 1.76e-01 |
| GO:BP | GO:0060167 | regulation of adenosine receptor signaling pathway | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0002901 | mature B cell apoptotic process | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0002906 | negative regulation of mature B cell apoptotic process | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0051418 | microtubule nucleation by microtubule organizing center | 5 | 5 | 1.76e-01 |
| GO:BP | GO:0009217 | purine deoxyribonucleoside triphosphate catabolic process | 1 | 4 | 1.76e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 7 | 40 | 1.76e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 4 | 36 | 1.76e-01 |
| GO:BP | GO:0046124 | purine deoxyribonucleoside catabolic process | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0046100 | hypoxanthine metabolic process | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 10 | 29 | 1.76e-01 |
| GO:BP | GO:1903546 | protein localization to photoreceptor outer segment | 4 | 5 | 1.76e-01 |
| GO:BP | GO:0046059 | dAMP catabolic process | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0016236 | macroautophagy | 109 | 308 | 1.76e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 6 | 99 | 1.76e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 19 | 32 | 1.76e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 11 | 12 | 1.76e-01 |
| GO:BP | GO:0035610 | protein side chain deglutamylation | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 10 | 15 | 1.76e-01 |
| GO:BP | GO:0090222 | centrosome-templated microtubule nucleation | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 15 | 55 | 1.76e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 1.76e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 1.76e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 11 | 160 | 1.76e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 1.78e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 10 | 293 | 1.78e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 24 | 45 | 1.78e-01 |
| GO:BP | GO:0006206 | pyrimidine nucleobase metabolic process | 1 | 13 | 1.79e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 17 | 86 | 1.79e-01 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 1 | 13 | 1.79e-01 |
| GO:BP | GO:0010459 | negative regulation of heart rate | 2 | 7 | 1.80e-01 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 9 | 15 | 1.80e-01 |
| GO:BP | GO:0071103 | DNA conformation change | 43 | 87 | 1.81e-01 |
| GO:BP | GO:0090129 | positive regulation of synapse maturation | 1 | 8 | 1.81e-01 |
| GO:BP | GO:0009171 | purine deoxyribonucleoside monophosphate biosynthetic process | 1 | 3 | 1.81e-01 |
| GO:BP | GO:0106381 | purine deoxyribonucleotide salvage | 1 | 3 | 1.81e-01 |
| GO:BP | GO:0006170 | dAMP biosynthetic process | 1 | 3 | 1.81e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 5 | 20 | 1.81e-01 |
| GO:BP | GO:0106383 | dAMP salvage | 1 | 3 | 1.81e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 4 | 240 | 1.81e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 6 | 75 | 1.81e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 1.82e-01 |
| GO:BP | GO:0070925 | organelle assembly | 69 | 817 | 1.82e-01 |
| GO:BP | GO:1902065 | response to L-glutamate | 7 | 11 | 1.82e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 1.82e-01 |
| GO:BP | GO:0046036 | CTP metabolic process | 1 | 14 | 1.82e-01 |
| GO:BP | GO:0001507 | acetylcholine catabolic process in synaptic cleft | 1 | 2 | 1.82e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 14 | 25 | 1.82e-01 |
| GO:BP | GO:0035445 | borate transmembrane transport | 1 | 1 | 1.82e-01 |
| GO:BP | GO:0035283 | central nervous system segmentation | 2 | 2 | 1.82e-01 |
| GO:BP | GO:0003094 | glomerular filtration | 7 | 21 | 1.82e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 32 | 50 | 1.82e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 2 | 26 | 1.82e-01 |
| GO:BP | GO:0046713 | borate transport | 1 | 1 | 1.82e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 1.82e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 29 | 88 | 1.82e-01 |
| GO:BP | GO:0006244 | pyrimidine nucleotide catabolic process | 5 | 19 | 1.82e-01 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 7 | 46 | 1.82e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 4 | 241 | 1.82e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 1 | 23 | 1.82e-01 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 1 | 15 | 1.82e-01 |
| GO:BP | GO:0032202 | telomere assembly | 3 | 4 | 1.82e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 1.82e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 28 | 103 | 1.82e-01 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 5 | 6 | 1.82e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 3 | 13 | 1.82e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 2 | 65 | 1.82e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 1.83e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 1.83e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 4 | 717 | 1.83e-01 |
| GO:BP | GO:0008592 | regulation of Toll signaling pathway | 3 | 3 | 1.83e-01 |
| GO:BP | GO:0097012 | response to granulocyte macrophage colony-stimulating factor | 2 | 6 | 1.83e-01 |
| GO:BP | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus | 2 | 6 | 1.83e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 12 | 16 | 1.83e-01 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 11 | 28 | 1.87e-01 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 1.87e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 2 | 4 | 1.87e-01 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 1 | 16 | 1.87e-01 |
| GO:BP | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 1.87e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 5 | 39 | 1.87e-01 |
| GO:BP | GO:0035608 | protein deglutamylation | 1 | 2 | 1.87e-01 |
| GO:BP | GO:0046725 | negative regulation by virus of viral protein levels in host cell | 1 | 2 | 1.87e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 8 | 89 | 1.87e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 12 | 19 | 1.87e-01 |
| GO:BP | GO:0009146 | purine nucleoside triphosphate catabolic process | 1 | 5 | 1.91e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 23 | 30 | 1.91e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 7 | 16 | 1.91e-01 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 8 | 18 | 1.91e-01 |
| GO:BP | GO:0009204 | deoxyribonucleoside triphosphate catabolic process | 1 | 5 | 1.91e-01 |
| GO:BP | GO:0006196 | AMP catabolic process | 1 | 3 | 1.91e-01 |
| GO:BP | GO:0046102 | inosine metabolic process | 1 | 4 | 1.91e-01 |
| GO:BP | GO:0046122 | purine deoxyribonucleoside metabolic process | 1 | 3 | 1.91e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 11 | 23 | 1.91e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 4 | 36 | 1.91e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 1.92e-01 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 5 | 14 | 1.92e-01 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 1 | 16 | 1.92e-01 |
| GO:BP | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic | 2 | 19 | 1.92e-01 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 5 | 1.92e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 6 | 31 | 1.92e-01 |
| GO:BP | GO:0045008 | depyrimidination | 3 | 8 | 1.92e-01 |
| GO:BP | GO:0010651 | negative regulation of cell communication by electrical coupling | 1 | 1 | 1.93e-01 |
| GO:BP | GO:1901845 | negative regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 1.93e-01 |
| GO:BP | GO:1903780 | negative regulation of cardiac conduction | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0110154 | RNA decapping | 2 | 17 | 1.93e-01 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 4 | 4 | 1.93e-01 |
| GO:BP | GO:0046075 | dTTP metabolic process | 4 | 4 | 1.93e-01 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 4 | 4 | 1.93e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 3 | 17 | 1.94e-01 |
| GO:BP | GO:0015791 | polyol transmembrane transport | 4 | 6 | 1.95e-01 |
| GO:BP | GO:0021756 | striatum development | 3 | 17 | 1.95e-01 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 6 | 10 | 1.95e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 13 | 23 | 1.95e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 1.95e-01 |
| GO:BP | GO:0070166 | enamel mineralization | 1 | 11 | 1.95e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 21 | 58 | 1.95e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 7 | 68 | 1.96e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 9 | 15 | 1.96e-01 |
| GO:BP | GO:0044208 | ‘de novo’ AMP biosynthetic process | 4 | 8 | 1.96e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 7 | 22 | 1.97e-01 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 8 | 12 | 1.98e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 50 | 1145 | 1.98e-01 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 6 | 13 | 1.98e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 3 | 7 | 1.98e-01 |
| GO:BP | GO:0009116 | nucleoside metabolic process | 2 | 42 | 1.98e-01 |
| GO:BP | GO:2000809 | positive regulation of synaptic vesicle clustering | 2 | 3 | 1.98e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 2 | 71 | 1.98e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 3 | 45 | 1.98e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 4 | 32 | 1.98e-01 |
| GO:BP | GO:0036245 | cellular response to menadione | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0098735 | positive regulation of the force of heart contraction | 2 | 4 | 1.99e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 12 | 192 | 2.00e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 5 | 9 | 2.00e-01 |
| GO:BP | GO:0009164 | nucleoside catabolic process | 3 | 15 | 2.00e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 13 | 214 | 2.00e-01 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 3 | 4 | 2.00e-01 |
| GO:BP | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 2 | 2.00e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 2.00e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 6 | 107 | 2.01e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 5 | 76 | 2.01e-01 |
| GO:BP | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | 2 | 4 | 2.01e-01 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 2 | 11 | 2.01e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 2 | 11 | 2.01e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 3 | 21 | 2.02e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 3 | 4 | 2.02e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 10 | 16 | 2.02e-01 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 8 | 29 | 2.02e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 230 | 2.02e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 1 | 29 | 2.02e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 4 | 25 | 2.03e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 7 | 15 | 2.05e-01 |
| GO:BP | GO:0010458 | exit from mitosis | 20 | 30 | 2.05e-01 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 3 | 20 | 2.07e-01 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 6 | 11 | 2.07e-01 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 1 | 23 | 2.07e-01 |
| GO:BP | GO:0009172 | purine deoxyribonucleoside monophosphate catabolic process | 1 | 3 | 2.07e-01 |
| GO:BP | GO:0046121 | deoxyribonucleoside catabolic process | 1 | 4 | 2.07e-01 |
| GO:BP | GO:0046130 | purine ribonucleoside catabolic process | 1 | 5 | 2.07e-01 |
| GO:BP | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway | 3 | 3 | 2.08e-01 |
| GO:BP | GO:0072703 | cellular response to methyl methanesulfonate | 1 | 1 | 2.08e-01 |
| GO:BP | GO:0072702 | response to methyl methanesulfonate | 1 | 1 | 2.08e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 27 | 48 | 2.08e-01 |
| GO:BP | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3’-5’ | 5 | 11 | 2.08e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 7 | 105 | 2.08e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 6 | 17 | 2.08e-01 |
| GO:BP | GO:0008544 | epidermis development | 2 | 210 | 2.08e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 1 | 33 | 2.08e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 3 | 48 | 2.08e-01 |
| GO:BP | GO:1903551 | regulation of extracellular exosome assembly | 4 | 6 | 2.09e-01 |
| GO:BP | GO:0097719 | neural tissue regeneration | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 4 | 787 | 2.09e-01 |
| GO:BP | GO:2001189 | negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 2.09e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 2.09e-01 |
| GO:BP | GO:2000521 | negative regulation of immunological synapse formation | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 7 | 29 | 2.09e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 1 | 16 | 2.09e-01 |
| GO:BP | GO:0090069 | regulation of ribosome biogenesis | 3 | 5 | 2.09e-01 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 2 | 8 | 2.09e-01 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 7 | 21 | 2.09e-01 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 5 | 22 | 2.09e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 2 | 60 | 2.09e-01 |
| GO:BP | GO:0043556 | regulation of translation in response to oxidative stress | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0032938 | negative regulation of translation in response to oxidative stress | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 3 | 48 | 2.10e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 19 | 33 | 2.10e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 26 | 135 | 2.10e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 60 | 271 | 2.10e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 7 | 11 | 2.10e-01 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 5 | 8 | 2.11e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 6 | 94 | 2.11e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 9 | 111 | 2.12e-01 |
| GO:BP | GO:0097205 | renal filtration | 7 | 23 | 2.12e-01 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 3 | 33 | 2.12e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 2.12e-01 |
| GO:BP | GO:0003166 | bundle of His development | 6 | 6 | 2.12e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 3 | 17 | 2.13e-01 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 4 | 26 | 2.13e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 2.13e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 6 | 56 | 2.13e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 2.13e-01 |
| GO:BP | GO:0060067 | cervix development | 2 | 2 | 2.13e-01 |
| GO:BP | GO:0044806 | G-quadruplex DNA unwinding | 4 | 7 | 2.13e-01 |
| GO:BP | GO:0006581 | acetylcholine catabolic process | 1 | 3 | 2.13e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 6 | 52 | 2.13e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 2 | 17 | 2.13e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 3 | 23 | 2.13e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 13 | 33 | 2.13e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 22 | 72 | 2.13e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 21 | 393 | 2.13e-01 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 2.13e-01 |
| GO:BP | GO:0097186 | amelogenesis | 1 | 14 | 2.13e-01 |
| GO:BP | GO:0098928 | presynaptic signal transduction | 2 | 2 | 2.13e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 2.13e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 2.13e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 1 | 17 | 2.13e-01 |
| GO:BP | GO:0099526 | presynapse to nucleus signaling pathway | 2 | 2 | 2.13e-01 |
| GO:BP | GO:0034627 | ‘de novo’ NAD biosynthetic process | 2 | 2 | 2.13e-01 |
| GO:BP | GO:0046338 | phosphatidylethanolamine catabolic process | 2 | 4 | 2.13e-01 |
| GO:BP | GO:1901594 | response to capsazepine | 1 | 1 | 2.13e-01 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 12 | 21 | 2.13e-01 |
| GO:BP | GO:0036438 | maintenance of lens transparency | 3 | 6 | 2.14e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 9 | 26 | 2.15e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 1 | 34 | 2.15e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 58 | 378 | 2.15e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 2 | 13 | 2.15e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 7 | 23 | 2.15e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 2.17e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 11 | 152 | 2.17e-01 |
| GO:BP | GO:1904172 | positive regulation of bleb assembly | 1 | 1 | 2.17e-01 |
| GO:BP | GO:0031106 | septin ring organization | 1 | 2 | 2.17e-01 |
| GO:BP | GO:1904170 | regulation of bleb assembly | 1 | 1 | 2.17e-01 |
| GO:BP | GO:0002949 | tRNA threonylcarbamoyladenosine modification | 3 | 5 | 2.17e-01 |
| GO:BP | GO:0060405 | regulation of penile erection | 1 | 4 | 2.17e-01 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 6 | 7 | 2.17e-01 |
| GO:BP | GO:0072092 | ureteric bud invasion | 3 | 3 | 2.17e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 4 | 11 | 2.18e-01 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 2 | 6 | 2.18e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 9 | 25 | 2.19e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 64 | 2.19e-01 |
| GO:BP | GO:0009153 | purine deoxyribonucleotide biosynthetic process | 1 | 4 | 2.21e-01 |
| GO:BP | GO:0072136 | metanephric mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0072135 | kidney mesenchymal cell proliferation | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0048789 | cytoskeletal matrix organization at active zone | 1 | 1 | 2.21e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 11 | 36 | 2.21e-01 |
| GO:BP | GO:0097115 | neurexin clustering involved in presynaptic membrane assembly | 1 | 1 | 2.21e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 4 | 226 | 2.21e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 4 | 22 | 2.22e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 80 | 357 | 2.22e-01 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 17 | 33 | 2.23e-01 |
| GO:BP | GO:0035721 | intraciliary retrograde transport | 9 | 14 | 2.23e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 2.23e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 2.23e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 2.23e-01 |
| GO:BP | GO:0038109 | Kit signaling pathway | 4 | 4 | 2.23e-01 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 12 | 44 | 2.23e-01 |
| GO:BP | GO:0036216 | cellular response to stem cell factor stimulus | 4 | 4 | 2.23e-01 |
| GO:BP | GO:0036215 | response to stem cell factor | 4 | 4 | 2.23e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 6 | 82 | 2.23e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 8 | 92 | 2.23e-01 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 3 | 5 | 2.23e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 5 | 51 | 2.23e-01 |
| GO:BP | GO:0090038 | negative regulation of protein kinase C signaling | 4 | 4 | 2.23e-01 |
| GO:BP | GO:0015750 | pentose transmembrane transport | 2 | 2 | 2.23e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 143 | 234 | 2.23e-01 |
| GO:BP | GO:1900108 | negative regulation of nodal signaling pathway | 1 | 2 | 2.23e-01 |
| GO:BP | GO:0000017 | alpha-glucoside transport | 2 | 2 | 2.23e-01 |
| GO:BP | GO:0015756 | fucose transmembrane transport | 2 | 2 | 2.23e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 2 | 265 | 2.23e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 3 | 159 | 2.23e-01 |
| GO:BP | GO:0035582 | sequestering of BMP in extracellular matrix | 1 | 2 | 2.23e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 17 | 20 | 2.23e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 21 | 77 | 2.23e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 14 | 158 | 2.24e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 5 | 82 | 2.24e-01 |
| GO:BP | GO:0032049 | cardiolipin biosynthetic process | 7 | 7 | 2.24e-01 |
| GO:BP | GO:1901753 | leukotriene A4 biosynthetic process | 1 | 1 | 2.24e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 3 | 53 | 2.24e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 4 | 101 | 2.24e-01 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 21 | 30 | 2.24e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 2.24e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 9 | 88 | 2.24e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 20 | 50 | 2.24e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 2.24e-01 |
| GO:BP | GO:0071029 | nuclear ncRNA surveillance | 3 | 7 | 2.24e-01 |
| GO:BP | GO:0106354 | tRNA surveillance | 3 | 7 | 2.24e-01 |
| GO:BP | GO:0051097 | negative regulation of helicase activity | 4 | 5 | 2.24e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 6 | 103 | 2.24e-01 |
| GO:BP | GO:0051126 | negative regulation of actin nucleation | 2 | 6 | 2.24e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 7 | 106 | 2.24e-01 |
| GO:BP | GO:0090217 | negative regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 3 | 3 | 2.24e-01 |
| GO:BP | GO:0051152 | positive regulation of smooth muscle cell differentiation | 6 | 10 | 2.24e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 2.24e-01 |
| GO:BP | GO:0048642 | negative regulation of skeletal muscle tissue development | 1 | 3 | 2.24e-01 |
| GO:BP | GO:0036376 | sodium ion export across plasma membrane | 3 | 12 | 2.24e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 36 | 2.24e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 20 | 27 | 2.24e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 2.24e-01 |
| GO:BP | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process | 3 | 7 | 2.24e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 3 | 5 | 2.24e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 2.24e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 2.24e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 2 | 5 | 2.24e-01 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 4 | 28 | 2.24e-01 |
| GO:BP | GO:0071046 | nuclear polyadenylation-dependent ncRNA catabolic process | 3 | 7 | 2.24e-01 |
| GO:BP | GO:0071505 | response to mycophenolic acid | 2 | 2 | 2.24e-01 |
| GO:BP | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process | 3 | 3 | 2.24e-01 |
| GO:BP | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process | 3 | 7 | 2.24e-01 |
| GO:BP | GO:0071506 | cellular response to mycophenolic acid | 2 | 2 | 2.24e-01 |
| GO:BP | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process | 3 | 3 | 2.24e-01 |
| GO:BP | GO:0002634 | regulation of germinal center formation | 1 | 6 | 2.24e-01 |
| GO:BP | GO:0006152 | purine nucleoside catabolic process | 1 | 6 | 2.24e-01 |
| GO:BP | GO:0003162 | atrioventricular node development | 6 | 8 | 2.24e-01 |
| GO:BP | GO:0070255 | regulation of mucus secretion | 1 | 6 | 2.24e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 30 | 46 | 2.25e-01 |
| GO:BP | GO:0090657 | telomeric loop disassembly | 5 | 9 | 2.25e-01 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 2.25e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 2 | 2 | 2.25e-01 |
| GO:BP | GO:0032918 | spermidine acetylation | 2 | 2 | 2.25e-01 |
| GO:BP | GO:0032917 | polyamine acetylation | 2 | 2 | 2.25e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 3 | 15 | 2.25e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 8 | 11 | 2.25e-01 |
| GO:BP | GO:0070459 | prolactin secretion | 1 | 4 | 2.25e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 5 | 7 | 2.25e-01 |
| GO:BP | GO:0035845 | photoreceptor cell outer segment organization | 3 | 10 | 2.25e-01 |
| GO:BP | GO:0098754 | detoxification | 10 | 91 | 2.25e-01 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 2 | 8 | 2.25e-01 |
| GO:BP | GO:0035904 | aorta development | 3 | 50 | 2.25e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 7 | 8 | 2.25e-01 |
| GO:BP | GO:0033082 | regulation of extrathymic T cell differentiation | 1 | 1 | 2.25e-01 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 8 | 10 | 2.25e-01 |
| GO:BP | GO:1901623 | regulation of lymphocyte chemotaxis | 4 | 13 | 2.25e-01 |
| GO:BP | GO:0061146 | Peyer’s patch morphogenesis | 3 | 3 | 2.26e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 2.27e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 17 | 67 | 2.27e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 3 | 14 | 2.27e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 2.27e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 5 | 39 | 2.27e-01 |
| GO:BP | GO:0019606 | 2-oxobutyrate catabolic process | 1 | 1 | 2.28e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 2.28e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 3 | 16 | 2.28e-01 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 13 | 25 | 2.28e-01 |
| GO:BP | GO:0061152 | trachea submucosa development | 2 | 2 | 2.28e-01 |
| GO:BP | GO:0061153 | trachea gland development | 2 | 2 | 2.28e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 7 | 8 | 2.28e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 5 | 39 | 2.29e-01 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 9 | 30 | 2.29e-01 |
| GO:BP | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation | 2 | 2 | 2.29e-01 |
| GO:BP | GO:1900052 | regulation of retinoic acid biosynthetic process | 1 | 3 | 2.29e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 2.31e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 2.31e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 2.31e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 2 | 278 | 2.31e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 7 | 12 | 2.31e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 3 | 25 | 2.31e-01 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 4 | 16 | 2.31e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 3 | 277 | 2.31e-01 |
| GO:BP | GO:1904997 | regulation of leukocyte adhesion to arterial endothelial cell | 3 | 3 | 2.32e-01 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 4 | 8 | 2.32e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 2.35e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 4 | 28 | 2.35e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 2.35e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 2.35e-01 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 4 | 23 | 2.35e-01 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 1 | 25 | 2.35e-01 |
| GO:BP | GO:0000255 | allantoin metabolic process | 1 | 4 | 2.35e-01 |
| GO:BP | GO:0048537 | mucosa-associated lymphoid tissue development | 1 | 6 | 2.35e-01 |
| GO:BP | GO:0048541 | Peyer’s patch development | 1 | 6 | 2.35e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 3 | 26 | 2.35e-01 |
| GO:BP | GO:0002314 | germinal center B cell differentiation | 1 | 3 | 2.35e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 9 | 27 | 2.35e-01 |
| GO:BP | GO:0060058 | positive regulation of apoptotic process involved in mammary gland involution | 2 | 3 | 2.35e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 1 | 41 | 2.35e-01 |
| GO:BP | GO:0060057 | apoptotic process involved in mammary gland involution | 2 | 3 | 2.35e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 9 | 27 | 2.35e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 21 | 399 | 2.36e-01 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 5 | 8 | 2.36e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 5 | 72 | 2.36e-01 |
| GO:BP | GO:0032899 | regulation of neurotrophin production | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 10 | 16 | 2.36e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 2.36e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 2 | 17 | 2.37e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 4 | 8 | 2.37e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 4 | 8 | 2.37e-01 |
| GO:BP | GO:0008063 | Toll signaling pathway | 4 | 6 | 2.37e-01 |
| GO:BP | GO:0006188 | IMP biosynthetic process | 1 | 10 | 2.37e-01 |
| GO:BP | GO:0042245 | RNA repair | 2 | 2 | 2.37e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 8 | 31 | 2.37e-01 |
| GO:BP | GO:0010820 | positive regulation of T cell chemotaxis | 3 | 7 | 2.37e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 5 | 155 | 2.37e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 6 | 57 | 2.37e-01 |
| GO:BP | GO:0010225 | response to UV-C | 9 | 13 | 2.37e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 2.37e-01 |
| GO:BP | GO:0034656 | nucleobase-containing small molecule catabolic process | 3 | 20 | 2.37e-01 |
| GO:BP | GO:0060453 | regulation of gastric acid secretion | 2 | 8 | 2.37e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 1 | 34 | 2.38e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 6 | 18 | 2.38e-01 |
| GO:BP | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 1 | 3 | 2.38e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 1 | 15 | 2.38e-01 |
| GO:BP | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 2 | 2.38e-01 |
| GO:BP | GO:0090272 | negative regulation of fibroblast growth factor production | 2 | 2 | 2.38e-01 |
| GO:BP | GO:1900175 | regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 2 | 2.38e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 2.38e-01 |
| GO:BP | GO:1904613 | cellular response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 2.38e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 4 | 12 | 2.38e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 4 | 12 | 2.38e-01 |
| GO:BP | GO:0060775 | planar cell polarity pathway involved in gastrula mediolateral intercalation | 2 | 2 | 2.38e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 7 | 47 | 2.38e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 3 | 11 | 2.38e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 5 | 5 | 2.38e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 2 | 214 | 2.38e-01 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 5 | 7 | 2.38e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 2.38e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 6 | 50 | 2.38e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 13 | 51 | 2.38e-01 |
| GO:BP | GO:0060031 | mediolateral intercalation | 2 | 2 | 2.38e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 5 | 73 | 2.38e-01 |
| GO:BP | GO:0006915 | apoptotic process | 8 | 1443 | 2.38e-01 |
| GO:BP | GO:2000821 | regulation of grooming behavior | 2 | 3 | 2.38e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 2.38e-01 |
| GO:BP | GO:0018410 | C-terminal protein amino acid modification | 1 | 10 | 2.38e-01 |
| GO:BP | GO:0070266 | necroptotic process | 5 | 32 | 2.38e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 6 | 28 | 2.38e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 20 | 439 | 2.38e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 5 | 65 | 2.38e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 120 | 760 | 2.39e-01 |
| GO:BP | GO:0061525 | hindgut development | 2 | 5 | 2.39e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 3 | 3 | 2.39e-01 |
| GO:BP | GO:0034476 | U5 snRNA 3’-end processing | 2 | 3 | 2.40e-01 |
| GO:BP | GO:0034473 | U1 snRNA 3’-end processing | 2 | 3 | 2.40e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 5 | 40 | 2.40e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 5 | 73 | 2.41e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 1 | 36 | 2.41e-01 |
| GO:BP | GO:0070244 | negative regulation of thymocyte apoptotic process | 1 | 6 | 2.42e-01 |
| GO:BP | GO:0009155 | purine deoxyribonucleotide catabolic process | 1 | 7 | 2.42e-01 |
| GO:BP | GO:0035641 | locomotory exploration behavior | 3 | 11 | 2.42e-01 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 1 | 10 | 2.42e-01 |
| GO:BP | GO:0009169 | purine ribonucleoside monophosphate catabolic process | 1 | 7 | 2.42e-01 |
| GO:BP | GO:0002138 | retinoic acid biosynthetic process | 2 | 8 | 2.42e-01 |
| GO:BP | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 4 | 4 | 2.42e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 41 | 113 | 2.42e-01 |
| GO:BP | GO:0051305 | chromosome movement towards spindle pole | 3 | 7 | 2.42e-01 |
| GO:BP | GO:0009128 | purine nucleoside monophosphate catabolic process | 1 | 7 | 2.42e-01 |
| GO:BP | GO:0006362 | transcription elongation by RNA polymerase I | 3 | 4 | 2.42e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 5 | 7 | 2.42e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 1 | 10 | 2.42e-01 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 2.42e-01 |
| GO:BP | GO:0051105 | regulation of DNA ligation | 5 | 6 | 2.42e-01 |
| GO:BP | GO:0043200 | response to amino acid | 2 | 95 | 2.42e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 2.42e-01 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 2.42e-01 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 1 | 7 | 2.42e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 8 | 16 | 2.42e-01 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 2 | 13 | 2.42e-01 |
| GO:BP | GO:0032230 | positive regulation of synaptic transmission, GABAergic | 2 | 10 | 2.42e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 7 | 9 | 2.42e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 19 | 31 | 2.42e-01 |
| GO:BP | GO:0051098 | regulation of binding | 2 | 317 | 2.42e-01 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 6 | 72 | 2.43e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 7 | 76 | 2.43e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 14 | 22 | 2.43e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 11 | 262 | 2.43e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 4 | 20 | 2.44e-01 |
| GO:BP | GO:0051835 | positive regulation of synapse structural plasticity | 2 | 2 | 2.44e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 24 | 459 | 2.44e-01 |
| GO:BP | GO:0032402 | melanosome transport | 6 | 19 | 2.44e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 4 | 311 | 2.45e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 6 | 110 | 2.45e-01 |
| GO:BP | GO:0070293 | renal absorption | 5 | 24 | 2.45e-01 |
| GO:BP | GO:1904550 | response to arachidonic acid | 1 | 1 | 2.45e-01 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 2 | 2 | 2.45e-01 |
| GO:BP | GO:0043084 | penile erection | 1 | 6 | 2.45e-01 |
| GO:BP | GO:0036337 | Fas signaling pathway | 2 | 5 | 2.45e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 3 | 179 | 2.45e-01 |
| GO:BP | GO:1903995 | regulation of non-membrane spanning protein tyrosine kinase activity | 3 | 3 | 2.45e-01 |
| GO:BP | GO:0043650 | dicarboxylic acid biosynthetic process | 1 | 9 | 2.45e-01 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 2 | 2 | 2.45e-01 |
| GO:BP | GO:0036486 | ventral trunk neural crest cell migration | 3 | 3 | 2.45e-01 |
| GO:BP | GO:1903553 | positive regulation of extracellular exosome assembly | 3 | 4 | 2.45e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 1 | 20 | 2.45e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 7 | 22 | 2.45e-01 |
| GO:BP | GO:1902340 | negative regulation of chromosome condensation | 2 | 2 | 2.45e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 8 | 10 | 2.45e-01 |
| GO:BP | GO:0090179 | planar cell polarity pathway involved in neural tube closure | 4 | 12 | 2.45e-01 |
| GO:BP | GO:0009445 | putrescine metabolic process | 2 | 6 | 2.45e-01 |
| GO:BP | GO:0090663 | galanin-activated signaling pathway | 1 | 1 | 2.45e-01 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 5 | 24 | 2.45e-01 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 1 | 33 | 2.45e-01 |
| GO:BP | GO:0097490 | sympathetic neuron projection extension | 4 | 4 | 2.45e-01 |
| GO:BP | GO:1900029 | positive regulation of ruffle assembly | 5 | 12 | 2.45e-01 |
| GO:BP | GO:1902151 | regulation of response to DNA integrity checkpoint signaling | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0010819 | regulation of T cell chemotaxis | 3 | 7 | 2.45e-01 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 6 | 14 | 2.45e-01 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 10 | 16 | 2.45e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 5 | 30 | 2.45e-01 |
| GO:BP | GO:0046340 | diacylglycerol catabolic process | 2 | 3 | 2.45e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 3 | 17 | 2.45e-01 |
| GO:BP | GO:1902145 | regulation of response to cell cycle checkpoint signaling | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 1 | 9 | 2.45e-01 |
| GO:BP | GO:0033089 | positive regulation of T cell differentiation in thymus | 1 | 5 | 2.45e-01 |
| GO:BP | GO:0021544 | subpallium development | 3 | 18 | 2.45e-01 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 2 | 62 | 2.45e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 28 | 53 | 2.45e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 9 | 132 | 2.45e-01 |
| GO:BP | GO:1905213 | negative regulation of mitotic chromosome condensation | 2 | 2 | 2.45e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 12 | 80 | 2.45e-01 |
| GO:BP | GO:0033632 | regulation of cell-cell adhesion mediated by integrin | 1 | 6 | 2.45e-01 |
| GO:BP | GO:1902153 | regulation of response to DNA damage checkpoint signaling | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0086054 | bundle of His cell to Purkinje myocyte communication by electrical coupling | 2 | 2 | 2.45e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 303 | 2.45e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 4 | 6 | 2.45e-01 |
| GO:BP | GO:0071288 | cellular response to mercury ion | 2 | 2 | 2.45e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 32 | 86 | 2.45e-01 |
| GO:BP | GO:0046085 | adenosine metabolic process | 1 | 5 | 2.45e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 8 | 1389 | 2.45e-01 |
| GO:BP | GO:1903379 | regulation of mitotic chromosome condensation | 2 | 2 | 2.45e-01 |
| GO:BP | GO:1902608 | positive regulation of large conductance calcium-activated potassium channel activity | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 4 | 6 | 2.45e-01 |
| GO:BP | GO:0071307 | cellular response to vitamin K | 1 | 4 | 2.45e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 6 | 30 | 2.45e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 6 | 30 | 2.45e-01 |
| GO:BP | GO:0097491 | sympathetic neuron projection guidance | 4 | 4 | 2.45e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 2 | 31 | 2.45e-01 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 2 | 7 | 2.45e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 4 | 898 | 2.45e-01 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 6 | 14 | 2.45e-01 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 4 | 9 | 2.45e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 9 | 35 | 2.45e-01 |
| GO:BP | GO:0070342 | brown fat cell proliferation | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0000423 | mitophagy | 9 | 37 | 2.45e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 5 | 45 | 2.45e-01 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 6 | 14 | 2.45e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 2 | 8 | 2.46e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 2 | 23 | 2.46e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 4 | 319 | 2.47e-01 |
| GO:BP | GO:0012501 | programmed cell death | 8 | 1479 | 2.47e-01 |
| GO:BP | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER | 2 | 2 | 2.48e-01 |
| GO:BP | GO:0032764 | negative regulation of mast cell cytokine production | 3 | 3 | 2.49e-01 |
| GO:BP | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process | 2 | 5 | 2.49e-01 |
| GO:BP | GO:0046136 | positive regulation of vitamin metabolic process | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0060431 | primary lung bud formation | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0046391 | 5-phosphoribose 1-diphosphate metabolic process | 2 | 5 | 2.49e-01 |
| GO:BP | GO:0019747 | regulation of isoprenoid metabolic process | 1 | 4 | 2.49e-01 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 2 | 12 | 2.50e-01 |
| GO:BP | GO:0090594 | inflammatory response to wounding | 2 | 12 | 2.50e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 3 | 513 | 2.50e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 18 | 187 | 2.50e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 13 | 38 | 2.50e-01 |
| GO:BP | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation | 3 | 5 | 2.50e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 3 | 223 | 2.50e-01 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 20 | 36 | 2.50e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 2.50e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 5 | 17 | 2.50e-01 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 5 | 10 | 2.50e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 5 | 9 | 2.50e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 2.50e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 2.50e-01 |
| GO:BP | GO:0097117 | guanylate kinase-associated protein clustering | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 1 | 46 | 2.50e-01 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 3 | 23 | 2.50e-01 |
| GO:BP | GO:0045004 | DNA replication proofreading | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0021768 | nucleus accumbens development | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0008050 | female courtship behavior | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 3 | 3 | 2.50e-01 |
| GO:BP | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 11 | 31 | 2.50e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 9 | 175 | 2.50e-01 |
| GO:BP | GO:0045196 | establishment or maintenance of neuroblast polarity | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 7 | 13 | 2.50e-01 |
| GO:BP | GO:0045200 | establishment of neuroblast polarity | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 2 | 18 | 2.50e-01 |
| GO:BP | GO:0090656 | t-circle formation | 7 | 13 | 2.50e-01 |
| GO:BP | GO:0044790 | suppression of viral release by host | 3 | 16 | 2.50e-01 |
| GO:BP | GO:0030592 | DNA ADP-ribosylation | 2 | 3 | 2.50e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 5 | 506 | 2.50e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 1 | 13 | 2.50e-01 |
| GO:BP | GO:0032898 | neurotrophin production | 3 | 4 | 2.50e-01 |
| GO:BP | GO:1900451 | positive regulation of glutamate receptor signaling pathway | 1 | 2 | 2.50e-01 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 2 | 10 | 2.50e-01 |
| GO:BP | GO:1904716 | positive regulation of chaperone-mediated autophagy | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 3 | 6 | 2.50e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 8 | 21 | 2.50e-01 |
| GO:BP | GO:0045760 | positive regulation of action potential | 2 | 4 | 2.50e-01 |
| GO:BP | GO:1904717 | regulation of AMPA glutamate receptor clustering | 2 | 4 | 2.50e-01 |
| GO:BP | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell communication | 2 | 6 | 2.50e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 2 | 19 | 2.50e-01 |
| GO:BP | GO:0007174 | epidermal growth factor catabolic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling | 2 | 6 | 2.50e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 3 | 19 | 2.50e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 3 | 19 | 2.50e-01 |
| GO:BP | GO:0070997 | neuron death | 17 | 286 | 2.50e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 2.50e-01 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 5 | 6 | 2.50e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 9 | 28 | 2.50e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 4 | 64 | 2.50e-01 |
| GO:BP | GO:0070254 | mucus secretion | 1 | 8 | 2.50e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 16 | 19 | 2.50e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 4 | 18 | 2.50e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 6 | 20 | 2.50e-01 |
| GO:BP | GO:1904979 | negative regulation of endosome organization | 1 | 2 | 2.50e-01 |
| GO:BP | GO:1901658 | glycosyl compound catabolic process | 3 | 23 | 2.50e-01 |
| GO:BP | GO:2000520 | regulation of immunological synapse formation | 1 | 2 | 2.50e-01 |
| GO:BP | GO:1904976 | cellular response to bleomycin | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0021549 | cerebellum development | 1 | 78 | 2.50e-01 |
| GO:BP | GO:1904975 | response to bleomycin | 2 | 2 | 2.50e-01 |
| GO:BP | GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 2.50e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 6 | 10 | 2.50e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 2.50e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 33 | 185 | 2.50e-01 |
| GO:BP | GO:0035128 | post-embryonic forelimb morphogenesis | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0072529 | pyrimidine-containing compound catabolic process | 5 | 29 | 2.50e-01 |
| GO:BP | GO:0035127 | post-embryonic limb morphogenesis | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0035120 | post-embryonic appendage morphogenesis | 1 | 2 | 2.50e-01 |
| GO:BP | GO:1900239 | regulation of phenotypic switching | 2 | 3 | 2.50e-01 |
| GO:BP | GO:0070194 | synaptonemal complex disassembly | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 2 | 11 | 2.50e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 5 | 10 | 2.50e-01 |
| GO:BP | GO:1904934 | negative regulation of cell proliferation in midbrain | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0015793 | glycerol transmembrane transport | 2 | 4 | 2.50e-01 |
| GO:BP | GO:0061350 | planar cell polarity pathway involved in cardiac muscle tissue morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0061354 | planar cell polarity pathway involved in pericardium morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:1902228 | positive regulation of macrophage colony-stimulating factor signaling pathway | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 3 | 3 | 2.50e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 15 | 155 | 2.50e-01 |
| GO:BP | GO:0060611 | mammary gland fat development | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0016557 | peroxisome membrane biogenesis | 3 | 3 | 2.50e-01 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 1 | 10 | 2.50e-01 |
| GO:BP | GO:0033693 | neurofilament bundle assembly | 3 | 3 | 2.50e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 36 | 149 | 2.50e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 9 | 47 | 2.50e-01 |
| GO:BP | GO:1904024 | negative regulation of glucose catabolic process to lactate via pyruvate | 2 | 3 | 2.50e-01 |
| GO:BP | GO:0036309 | protein localization to M-band | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0009143 | nucleoside triphosphate catabolic process | 1 | 10 | 2.50e-01 |
| GO:BP | GO:0043111 | replication fork arrest | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0061349 | planar cell polarity pathway involved in cardiac right atrium morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 2 | 105 | 2.50e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 11 | 94 | 2.50e-01 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 7 | 13 | 2.50e-01 |
| GO:BP | GO:0006272 | leading strand elongation | 2 | 4 | 2.50e-01 |
| GO:BP | GO:0098925 | retrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0036164 | cell-abiotic substrate adhesion | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 7 | 8 | 2.50e-01 |
| GO:BP | GO:0043174 | nucleoside salvage | 1 | 9 | 2.50e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 13 | 134 | 2.50e-01 |
| GO:BP | GO:0036517 | chemoattraction of serotonergic neuron axon | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 1 | 246 | 2.50e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 3 | 224 | 2.50e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 5 | 31 | 2.50e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 4 | 5 | 2.50e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 5 | 70 | 2.50e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 2 | 6 | 2.50e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 6 | 22 | 2.50e-01 |
| GO:BP | GO:1905366 | negative regulation of intralumenal vesicle formation | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 5 | 249 | 2.50e-01 |
| GO:BP | GO:1905365 | regulation of intralumenal vesicle formation | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0071694 | maintenance of protein location in extracellular region | 1 | 4 | 2.50e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 37 | 204 | 2.50e-01 |
| GO:BP | GO:0106104 | regulation of glutamate receptor clustering | 2 | 4 | 2.50e-01 |
| GO:BP | GO:0046755 | viral budding | 16 | 26 | 2.50e-01 |
| GO:BP | GO:0060542 | regulation of strand invasion | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 2.50e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0016102 | diterpenoid biosynthetic process | 2 | 8 | 2.50e-01 |
| GO:BP | GO:0033566 | gamma-tubulin complex localization | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 6 | 11 | 2.50e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 34 | 101 | 2.50e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0071971 | extracellular exosome assembly | 4 | 6 | 2.50e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 3 | 160 | 2.50e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 16 | 279 | 2.50e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 7 | 28 | 2.50e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 11 | 94 | 2.50e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 6 | 94 | 2.50e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 11 | 29 | 2.50e-01 |
| GO:BP | GO:0043634 | polyadenylation-dependent ncRNA catabolic process | 3 | 8 | 2.50e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 1 | 3 | 2.50e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 8 | 13 | 2.50e-01 |
| GO:BP | GO:2001170 | negative regulation of ATP biosynthetic process | 2 | 3 | 2.50e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 3 | 19 | 2.50e-01 |
| GO:BP | GO:0098530 | positive regulation of strand invasion | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0033078 | extrathymic T cell differentiation | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 6 | 57 | 2.50e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 33 | 1214 | 2.50e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 3 | 19 | 2.50e-01 |
| GO:BP | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 6 | 19 | 2.50e-01 |
| GO:BP | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 4 | 48 | 2.50e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 2.50e-01 |
| GO:BP | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0061346 | planar cell polarity pathway involved in heart morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0061348 | planar cell polarity pathway involved in ventricular septum morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 2 | 15 | 2.50e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 5 | 172 | 2.50e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 3 | 30 | 2.51e-01 |
| GO:BP | GO:0009790 | embryo development | 3 | 875 | 2.51e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 8 | 52 | 2.51e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 3 | 79 | 2.51e-01 |
| GO:BP | GO:0045794 | negative regulation of cell volume | 2 | 4 | 2.51e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 2.51e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 2.51e-01 |
| GO:BP | GO:0090244 | Wnt signaling pathway involved in somitogenesis | 2 | 5 | 2.51e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 5 | 40 | 2.52e-01 |
| GO:BP | GO:0051648 | vesicle localization | 11 | 192 | 2.52e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 2.52e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 5 | 70 | 2.52e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 19 | 420 | 2.52e-01 |
| GO:BP | GO:0042594 | response to starvation | 3 | 178 | 2.52e-01 |
| GO:BP | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 2.52e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 3 | 22 | 2.52e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 24 | 44 | 2.52e-01 |
| GO:BP | GO:0060454 | positive regulation of gastric acid secretion | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0051974 | negative regulation of telomerase activity | 4 | 11 | 2.52e-01 |
| GO:BP | GO:0090212 | negative regulation of establishment of blood-brain barrier | 1 | 1 | 2.52e-01 |
| GO:BP | GO:2000791 | negative regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 2.52e-01 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 3 | 5 | 2.52e-01 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 3 | 5 | 2.52e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 22 | 68 | 2.52e-01 |
| GO:BP | GO:0070889 | platelet alpha granule organization | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0003241 | growth involved in heart morphogenesis | 3 | 4 | 2.53e-01 |
| GO:BP | GO:0003273 | cell migration involved in endocardial cushion formation | 3 | 4 | 2.53e-01 |
| GO:BP | GO:0001560 | regulation of cell growth by extracellular stimulus | 3 | 3 | 2.53e-01 |
| GO:BP | GO:0031291 | Ran protein signal transduction | 2 | 3 | 2.53e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 12 | 217 | 2.54e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 3 | 5 | 2.54e-01 |
| GO:BP | GO:0010643 | cell communication by chemical coupling | 2 | 2 | 2.54e-01 |
| GO:BP | GO:0014839 | myoblast migration involved in skeletal muscle regeneration | 2 | 3 | 2.54e-01 |
| GO:BP | GO:0043201 | response to leucine | 1 | 13 | 2.54e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 38 | 61 | 2.55e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 5 | 7 | 2.55e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 5 | 7 | 2.55e-01 |
| GO:BP | GO:0055123 | digestive system development | 9 | 100 | 2.55e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 2 | 24 | 2.55e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 9 | 18 | 2.55e-01 |
| GO:BP | GO:2000232 | regulation of rRNA processing | 5 | 16 | 2.55e-01 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 1 | 12 | 2.55e-01 |
| GO:BP | GO:0032185 | septin cytoskeleton organization | 1 | 4 | 2.55e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 6 | 47 | 2.55e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 4 | 260 | 2.55e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 3 | 42 | 2.55e-01 |
| GO:BP | GO:0009159 | deoxyribonucleoside monophosphate catabolic process | 1 | 8 | 2.55e-01 |
| GO:BP | GO:0002467 | germinal center formation | 1 | 10 | 2.55e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 3 | 113 | 2.56e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 12 | 424 | 2.56e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 2.56e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 117 | 189 | 2.56e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 2.56e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 2 | 8 | 2.56e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 13 | 98 | 2.56e-01 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 2 | 13 | 2.56e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 2.56e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 2 | 8 | 2.56e-01 |
| GO:BP | GO:1901339 | regulation of store-operated calcium channel activity | 3 | 8 | 2.56e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 10 | 2139 | 2.56e-01 |
| GO:BP | GO:0071494 | cellular response to UV-C | 2 | 4 | 2.56e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 1 | 52 | 2.56e-01 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 2 | 13 | 2.56e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 4 | 22 | 2.56e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 3 | 870 | 2.56e-01 |
| GO:BP | GO:0019048 | modulation by virus of host process | 3 | 5 | 2.56e-01 |
| GO:BP | GO:0071582 | negative regulation of zinc ion transport | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0034402 | recruitment of 3’-end processing factors to RNA polymerase II holoenzyme complex | 1 | 2 | 2.56e-01 |
| GO:BP | GO:0071581 | regulation of zinc ion transmembrane import | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 4 | 8 | 2.56e-01 |
| GO:BP | GO:0019054 | modulation by virus of host cellular process | 3 | 5 | 2.56e-01 |
| GO:BP | GO:0046131 | pyrimidine ribonucleoside metabolic process | 2 | 7 | 2.56e-01 |
| GO:BP | GO:0071580 | regulation of zinc ion transmembrane transport | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0046133 | pyrimidine ribonucleoside catabolic process | 2 | 7 | 2.56e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 14 | 28 | 2.56e-01 |
| GO:BP | GO:0016540 | protein autoprocessing | 6 | 16 | 2.56e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 8 | 67 | 2.56e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 7 | 67 | 2.56e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 6 | 59 | 2.56e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 10 | 2161 | 2.56e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 17 | 381 | 2.56e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 4 | 9 | 2.56e-01 |
| GO:BP | GO:0071584 | negative regulation of zinc ion transmembrane import | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0021546 | rhombomere development | 2 | 4 | 2.56e-01 |
| GO:BP | GO:1903954 | positive regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 2 | 19 | 2.56e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 31 | 60 | 2.56e-01 |
| GO:BP | GO:2001015 | negative regulation of skeletal muscle cell differentiation | 2 | 3 | 2.56e-01 |
| GO:BP | GO:0009158 | ribonucleoside monophosphate catabolic process | 1 | 10 | 2.56e-01 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 1 | 9 | 2.56e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 2.56e-01 |
| GO:BP | GO:1900107 | regulation of nodal signaling pathway | 1 | 3 | 2.56e-01 |
| GO:BP | GO:0071583 | negative regulation of zinc ion transmembrane transport | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0043157 | response to cation stress | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0060013 | righting reflex | 3 | 6 | 2.56e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 6 | 423 | 2.56e-01 |
| GO:BP | GO:0003170 | heart valve development | 3 | 65 | 2.56e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 1 | 25 | 2.56e-01 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 3 | 23 | 2.56e-01 |
| GO:BP | GO:0044878 | mitotic cytokinesis checkpoint signaling | 2 | 4 | 2.56e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 3 | 69 | 2.56e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 1 | 38 | 2.56e-01 |
| GO:BP | GO:0032077 | positive regulation of deoxyribonuclease activity | 3 | 5 | 2.56e-01 |
| GO:BP | GO:0032060 | bleb assembly | 2 | 9 | 2.56e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 7 | 50 | 2.56e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 4 | 9 | 2.56e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 10 | 34 | 2.56e-01 |
| GO:BP | GO:1904415 | regulation of xenophagy | 6 | 7 | 2.56e-01 |
| GO:BP | GO:1903952 | regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.56e-01 |
| GO:BP | GO:1904417 | positive regulation of xenophagy | 6 | 7 | 2.56e-01 |
| GO:BP | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 2.56e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 7 | 7 | 2.56e-01 |
| GO:BP | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway | 1 | 3 | 2.56e-01 |
| GO:BP | GO:1903806 | L-isoleucine import across plasma membrane | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 2 | 8 | 2.56e-01 |
| GO:BP | GO:1903714 | isoleucine transmembrane transport | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 1 | 24 | 2.56e-01 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 2 | 15 | 2.56e-01 |
| GO:BP | GO:0031990 | mRNA export from nucleus in response to heat stress | 1 | 2 | 2.56e-01 |
| GO:BP | GO:0000301 | retrograde transport, vesicle recycling within Golgi | 6 | 9 | 2.56e-01 |
| GO:BP | GO:1901165 | positive regulation of trophoblast cell migration | 2 | 6 | 2.56e-01 |
| GO:BP | GO:0030903 | notochord development | 9 | 17 | 2.56e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 2 | 18 | 2.56e-01 |
| GO:BP | GO:0090125 | cell-cell adhesion involved in synapse maturation | 1 | 2 | 2.56e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 8 | 28 | 2.56e-01 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 4 | 8 | 2.56e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 2.56e-01 |
| GO:BP | GO:0044003 | modulation by symbiont of host process | 3 | 5 | 2.56e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 6 | 69 | 2.56e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 4 | 13 | 2.56e-01 |
| GO:BP | GO:0044068 | modulation by symbiont of host cellular process | 3 | 5 | 2.56e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 1 | 11 | 2.56e-01 |
| GO:BP | GO:0019731 | antibacterial humoral response | 2 | 15 | 2.56e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 2.56e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 7 | 50 | 2.56e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 6 | 90 | 2.56e-01 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 3 | 20 | 2.56e-01 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 2 | 30 | 2.57e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 5 | 29 | 2.57e-01 |
| GO:BP | GO:0046040 | IMP metabolic process | 1 | 15 | 2.57e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 24 | 32 | 2.57e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 11 | 21 | 2.57e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 8 | 116 | 2.57e-01 |
| GO:BP | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 4 | 2.57e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 2.57e-01 |
| GO:BP | GO:0071476 | cellular hypotonic response | 6 | 8 | 2.57e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 17 | 26 | 2.58e-01 |
| GO:BP | GO:0033326 | cerebrospinal fluid secretion | 2 | 3 | 2.58e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 3 | 26 | 2.58e-01 |
| GO:BP | GO:0048850 | hypophysis morphogenesis | 2 | 2 | 2.58e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 5 | 64 | 2.59e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 2.59e-01 |
| GO:BP | GO:0008291 | acetylcholine metabolic process | 1 | 3 | 2.59e-01 |
| GO:BP | GO:1900619 | acetate ester metabolic process | 1 | 3 | 2.59e-01 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 10 | 37 | 2.60e-01 |
| GO:BP | GO:1903416 | response to glycoside | 3 | 6 | 2.60e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 2.60e-01 |
| GO:BP | GO:0099606 | microtubule plus-end directed mitotic chromosome migration | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 30 | 58 | 2.60e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 11 | 41 | 2.60e-01 |
| GO:BP | GO:0021599 | abducens nerve formation | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0021598 | abducens nerve morphogenesis | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0021560 | abducens nerve development | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0021572 | rhombomere 6 development | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0072709 | cellular response to sorbitol | 2 | 5 | 2.60e-01 |
| GO:BP | GO:0003336 | corneocyte desquamation | 1 | 1 | 2.60e-01 |
| GO:BP | GO:2001188 | regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 2 | 2.60e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 10 | 17 | 2.61e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 2.61e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 11 | 293 | 2.61e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 6 | 20 | 2.61e-01 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 2 | 12 | 2.61e-01 |
| GO:BP | GO:0042822 | pyridoxal phosphate metabolic process | 4 | 4 | 2.61e-01 |
| GO:BP | GO:1904999 | positive regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 2.61e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 4 | 64 | 2.61e-01 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 9 | 11 | 2.62e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 1 | 45 | 2.62e-01 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 2 | 19 | 2.62e-01 |
| GO:BP | GO:0042631 | cellular response to water deprivation | 1 | 1 | 2.62e-01 |
| GO:BP | GO:0070455 | positive regulation of heme biosynthetic process | 3 | 3 | 2.62e-01 |
| GO:BP | GO:1901465 | positive regulation of tetrapyrrole biosynthetic process | 3 | 3 | 2.62e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 6 | 429 | 2.62e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 2.63e-01 |
| GO:BP | GO:0099554 | trans-synaptic signaling by soluble gas, modulating synaptic transmission | 2 | 2 | 2.63e-01 |
| GO:BP | GO:0099555 | trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 2 | 2 | 2.63e-01 |
| GO:BP | GO:0035176 | social behavior | 4 | 34 | 2.63e-01 |
| GO:BP | GO:0140450 | protein targeting to Golgi apparatus | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0042126 | nitrate metabolic process | 4 | 4 | 2.64e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 2 | 2 | 2.64e-01 |
| GO:BP | GO:1990523 | bone regeneration | 2 | 2 | 2.64e-01 |
| GO:BP | GO:0001561 | fatty acid alpha-oxidation | 2 | 5 | 2.64e-01 |
| GO:BP | GO:0046465 | dolichyl diphosphate metabolic process | 3 | 5 | 2.64e-01 |
| GO:BP | GO:0060429 | epithelium development | 3 | 850 | 2.64e-01 |
| GO:BP | GO:0006489 | dolichyl diphosphate biosynthetic process | 3 | 5 | 2.64e-01 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 1 | 2 | 2.64e-01 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 3 | 9 | 2.64e-01 |
| GO:BP | GO:0006043 | glucosamine catabolic process | 2 | 2 | 2.64e-01 |
| GO:BP | GO:0050893 | sensory processing | 4 | 4 | 2.64e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 7 | 83 | 2.64e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 2.64e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 4 | 4 | 2.64e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 2.64e-01 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 3 | 5 | 2.64e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 1 | 48 | 2.64e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 2.64e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 1 | 54 | 2.64e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 3 | 22 | 2.65e-01 |
| GO:BP | GO:0070106 | interleukin-27-mediated signaling pathway | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 4 | 12 | 2.65e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 2 | 4 | 2.65e-01 |
| GO:BP | GO:0006228 | UTP biosynthetic process | 7 | 8 | 2.65e-01 |
| GO:BP | GO:0043633 | polyadenylation-dependent RNA catabolic process | 3 | 9 | 2.65e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 30 | 142 | 2.65e-01 |
| GO:BP | GO:0000725 | recombinational repair | 74 | 153 | 2.65e-01 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 3 | 8 | 2.65e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic process | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0001947 | heart looping | 5 | 50 | 2.65e-01 |
| GO:BP | GO:0022614 | membrane to membrane docking | 3 | 5 | 2.65e-01 |
| GO:BP | GO:0071047 | polyadenylation-dependent mRNA catabolic process | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 2.65e-01 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 5 | 12 | 2.65e-01 |
| GO:BP | GO:2000208 | positive regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 33 | 52 | 2.65e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 6 | 39 | 2.65e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 36 | 70 | 2.65e-01 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 4 | 9 | 2.65e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 5 | 47 | 2.65e-01 |
| GO:BP | GO:0072257 | metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 2.65e-01 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 34 | 1280 | 2.65e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 6 | 91 | 2.65e-01 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 9 | 2.65e-01 |
| GO:BP | GO:2000202 | positive regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:2000199 | positive regulation of ribonucleoprotein complex localization | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 2 | 284 | 2.65e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 1 | 67 | 2.65e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 5 | 60 | 2.65e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 7 | 33 | 2.65e-01 |
| GO:BP | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity | 1 | 3 | 2.65e-01 |
| GO:BP | GO:2000200 | regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 3 | 19 | 2.65e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 6 | 7 | 2.65e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 78 | 133 | 2.65e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 6 | 7 | 2.65e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 22 | 441 | 2.65e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 9 | 11 | 2.65e-01 |
| GO:BP | GO:0046361 | 2-oxobutyrate metabolic process | 1 | 2 | 2.65e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 12 | 17 | 2.65e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 5 | 6 | 2.65e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 5 | 6 | 2.65e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 20 | 36 | 2.65e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 4 | 112 | 2.65e-01 |
| GO:BP | GO:0046051 | UTP metabolic process | 8 | 10 | 2.65e-01 |
| GO:BP | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0000459 | exonucleolytic trimming involved in rRNA processing | 3 | 9 | 2.65e-01 |
| GO:BP | GO:2000206 | regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 3 | 18 | 2.65e-01 |
| GO:BP | GO:0080170 | hydrogen peroxide transmembrane transport | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0033631 | cell-cell adhesion mediated by integrin | 1 | 12 | 2.65e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 4 | 15 | 2.65e-01 |
| GO:BP | GO:0003192 | mitral valve formation | 3 | 3 | 2.65e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 4 | 70 | 2.65e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 13 | 142 | 2.65e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 2 | 5 | 2.65e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 9 | 55 | 2.65e-01 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 12 | 17 | 2.65e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 8 | 358 | 2.65e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 2 | 23 | 2.65e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 26 | 71 | 2.65e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 11 | 16 | 2.65e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 11 | 28 | 2.65e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 4 | 113 | 2.66e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 2 | 8 | 2.66e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 16 | 295 | 2.66e-01 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 4 | 20 | 2.66e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 3 | 271 | 2.67e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 5 | 6 | 2.68e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 18 | 30 | 2.68e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 2.70e-01 |
| GO:BP | GO:1905938 | positive regulation of germ cell proliferation | 2 | 2 | 2.70e-01 |
| GO:BP | GO:0032222 | regulation of synaptic transmission, cholinergic | 1 | 3 | 2.70e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 2.70e-01 |
| GO:BP | GO:2000256 | positive regulation of male germ cell proliferation | 2 | 2 | 2.70e-01 |
| GO:BP | GO:1990646 | cellular response to prolactin | 2 | 2 | 2.70e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 6 | 41 | 2.70e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 8 | 51 | 2.70e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 2.70e-01 |
| GO:BP | GO:0046073 | dTMP metabolic process | 3 | 7 | 2.70e-01 |
| GO:BP | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential | 3 | 10 | 2.70e-01 |
| GO:BP | GO:0060039 | pericardium development | 8 | 17 | 2.70e-01 |
| GO:BP | GO:0006885 | regulation of pH | 5 | 74 | 2.70e-01 |
| GO:BP | GO:0008219 | cell death | 8 | 1596 | 2.70e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 33 | 1239 | 2.71e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 13 | 102 | 2.72e-01 |
| GO:BP | GO:2000431 | regulation of cytokinesis, actomyosin contractile ring assembly | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 2.72e-01 |
| GO:BP | GO:0003097 | renal water transport | 3 | 6 | 2.72e-01 |
| GO:BP | GO:0071569 | protein ufmylation | 4 | 6 | 2.72e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 15 | 2.72e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 6 | 21 | 2.72e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 22 | 33 | 2.72e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 2 | 17 | 2.72e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 2.72e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 55 | 119 | 2.72e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 5 | 544 | 2.72e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 5 | 89 | 2.72e-01 |
| GO:BP | GO:1900060 | negative regulation of ceramide biosynthetic process | 1 | 5 | 2.72e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 2 | 6 | 2.72e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 10 | 183 | 2.72e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 14 | 44 | 2.72e-01 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 3 | 209 | 2.72e-01 |
| GO:BP | GO:0046444 | FMN metabolic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0003283 | atrial septum development | 7 | 23 | 2.72e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 1 | 31 | 2.72e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 2 | 4 | 2.72e-01 |
| GO:BP | GO:0000303 | response to superoxide | 3 | 21 | 2.72e-01 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 15 | 27 | 2.72e-01 |
| GO:BP | GO:0009231 | riboflavin biosynthetic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0021700 | developmental maturation | 2 | 211 | 2.72e-01 |
| GO:BP | GO:0140961 | cellular detoxification of metal ion | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0033320 | UDP-D-xylose biosynthetic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 5 | 66 | 2.72e-01 |
| GO:BP | GO:0052648 | ribitol phosphate metabolic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0033319 | UDP-D-xylose metabolic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 4 | 34 | 2.72e-01 |
| GO:BP | GO:0009398 | FMN biosynthetic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 5 | 39 | 2.72e-01 |
| GO:BP | GO:0052647 | pentitol phosphate metabolic process | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 5 | 66 | 2.72e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 3 | 14 | 2.72e-01 |
| GO:BP | GO:0048873 | homeostasis of number of cells within a tissue | 3 | 24 | 2.72e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 2 | 3 | 2.72e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 7 | 7 | 2.72e-01 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 4 | 7 | 2.72e-01 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 3 | 13 | 2.72e-01 |
| GO:BP | GO:0098957 | anterograde axonal transport of mitochondrion | 1 | 5 | 2.72e-01 |
| GO:BP | GO:1902990 | mitotic telomere maintenance via semi-conservative replication | 1 | 2 | 2.72e-01 |
| GO:BP | GO:0098849 | cellular detoxification of cadmium ion | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 1 | 26 | 2.72e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 29 | 73 | 2.72e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 35 | 55 | 2.72e-01 |
| GO:BP | GO:0033387 | putrescine biosynthetic process from ornithine | 1 | 3 | 2.72e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 8 | 12 | 2.72e-01 |
| GO:BP | GO:0048854 | brain morphogenesis | 9 | 28 | 2.72e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 5 | 16 | 2.72e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 8 | 97 | 2.72e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 2.72e-01 |
| GO:BP | GO:0001880 | Mullerian duct regression | 3 | 4 | 2.72e-01 |
| GO:BP | GO:0046135 | pyrimidine nucleoside catabolic process | 2 | 8 | 2.72e-01 |
| GO:BP | GO:0070932 | histone H3 deacetylation | 8 | 12 | 2.72e-01 |
| GO:BP | GO:0071468 | cellular response to acidic pH | 2 | 10 | 2.72e-01 |
| GO:BP | GO:0140042 | lipid droplet formation | 9 | 12 | 2.72e-01 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 1 | 15 | 2.72e-01 |
| GO:BP | GO:0060028 | convergent extension involved in axis elongation | 3 | 6 | 2.72e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 7 | 42 | 2.72e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 14 | 37 | 2.73e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 10 | 19 | 2.74e-01 |
| GO:BP | GO:1903613 | regulation of protein tyrosine phosphatase activity | 1 | 4 | 2.74e-01 |
| GO:BP | GO:0086092 | regulation of the force of heart contraction by cardiac conduction | 1 | 2 | 2.74e-01 |
| GO:BP | GO:1903281 | positive regulation of calcium:sodium antiporter activity | 1 | 2 | 2.74e-01 |
| GO:BP | GO:0035581 | sequestering of extracellular ligand from receptor | 1 | 7 | 2.74e-01 |
| GO:BP | GO:0106015 | negative regulation of inflammatory response to wounding | 1 | 2 | 2.74e-01 |
| GO:BP | GO:0010288 | response to lead ion | 7 | 18 | 2.74e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 5 | 22 | 2.75e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 6 | 165 | 2.75e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 6 | 10 | 2.75e-01 |
| GO:BP | GO:0099179 | regulation of synaptic membrane adhesion | 4 | 4 | 2.75e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 4 | 14 | 2.75e-01 |
| GO:BP | GO:0009125 | nucleoside monophosphate catabolic process | 1 | 13 | 2.75e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 2.75e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 2.75e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 8 | 114 | 2.75e-01 |
| GO:BP | GO:0046069 | cGMP catabolic process | 1 | 4 | 2.75e-01 |
| GO:BP | GO:0006833 | water transport | 3 | 10 | 2.75e-01 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 26 | 67 | 2.75e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 2 | 2 | 2.75e-01 |
| GO:BP | GO:0032400 | melanosome localization | 6 | 22 | 2.75e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 12 | 14 | 2.76e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 2.76e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 3 | 22 | 2.76e-01 |
| GO:BP | GO:0000722 | telomere maintenance via recombination | 4 | 14 | 2.76e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 1 | 8 | 2.76e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 2.76e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 2.76e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 13 | 2.76e-01 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 7 | 11 | 2.76e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 2 | 18 | 2.76e-01 |
| GO:BP | GO:0039536 | negative regulation of RIG-I signaling pathway | 2 | 7 | 2.76e-01 |
| GO:BP | GO:0032571 | response to vitamin K | 1 | 5 | 2.76e-01 |
| GO:BP | GO:0035166 | post-embryonic hemopoiesis | 3 | 5 | 2.76e-01 |
| GO:BP | GO:0061757 | leukocyte adhesion to arterial endothelial cell | 3 | 4 | 2.76e-01 |
| GO:BP | GO:0060219 | camera-type eye photoreceptor cell differentiation | 11 | 18 | 2.77e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 2.77e-01 |
| GO:BP | GO:0060049 | regulation of protein glycosylation | 2 | 10 | 2.77e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 2 | 18 | 2.77e-01 |
| GO:BP | GO:0014034 | neural crest cell fate commitment | 2 | 4 | 2.77e-01 |
| GO:BP | GO:0042946 | glucoside transport | 3 | 3 | 2.77e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 26 | 44 | 2.77e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 6 | 45 | 2.78e-01 |
| GO:BP | GO:0045007 | depurination | 4 | 4 | 2.78e-01 |
| GO:BP | GO:0003164 | His-Purkinje system development | 5 | 7 | 2.78e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 8 | 48 | 2.78e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 10 | 14 | 2.78e-01 |
| GO:BP | GO:0001843 | neural tube closure | 2 | 90 | 2.78e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 2 | 11 | 2.78e-01 |
| GO:BP | GO:0036372 | opsin transport | 1 | 2 | 2.78e-01 |
| GO:BP | GO:0001967 | suckling behavior | 1 | 10 | 2.78e-01 |
| GO:BP | GO:2000210 | positive regulation of anoikis | 4 | 6 | 2.79e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 2 | 16 | 2.79e-01 |
| GO:BP | GO:0046337 | phosphatidylethanolamine metabolic process | 6 | 19 | 2.79e-01 |
| GO:BP | GO:2000310 | regulation of NMDA receptor activity | 4 | 19 | 2.79e-01 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 4 | 20 | 2.79e-01 |
| GO:BP | GO:0016570 | histone modification | 1 | 433 | 2.80e-01 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 6 | 12 | 2.80e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 2.80e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 2 | 418 | 2.80e-01 |
| GO:BP | GO:0060449 | bud elongation involved in lung branching | 1 | 7 | 2.80e-01 |
| GO:BP | GO:0032458 | slow endocytic recycling | 2 | 2 | 2.80e-01 |
| GO:BP | GO:0061485 | memory T cell proliferation | 1 | 1 | 2.80e-01 |
| GO:BP | GO:0043605 | amide catabolic process | 1 | 10 | 2.80e-01 |
| GO:BP | GO:0006218 | uridine catabolic process | 1 | 1 | 2.80e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 3 | 218 | 2.80e-01 |
| GO:BP | GO:0046108 | uridine metabolic process | 1 | 1 | 2.80e-01 |
| GO:BP | GO:0014886 | transition between slow and fast fiber | 1 | 1 | 2.80e-01 |
| GO:BP | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 2 | 2.80e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 6 | 353 | 2.80e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 6 | 11 | 2.80e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 10 | 15 | 2.80e-01 |
| GO:BP | GO:0060606 | tube closure | 2 | 91 | 2.80e-01 |
| GO:BP | GO:0046398 | UDP-glucuronate metabolic process | 1 | 2 | 2.80e-01 |
| GO:BP | GO:0007144 | female meiosis I | 6 | 9 | 2.81e-01 |
| GO:BP | GO:0071895 | odontoblast differentiation | 2 | 5 | 2.81e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 2.81e-01 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 1 | 2 | 2.81e-01 |
| GO:BP | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain | 1 | 2 | 2.81e-01 |
| GO:BP | GO:0060083 | smooth muscle contraction involved in micturition | 1 | 2 | 2.81e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 15 | 27 | 2.81e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 2.81e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 4 | 20 | 2.81e-01 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 1 | 17 | 2.82e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 2.82e-01 |
| GO:BP | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation | 1 | 1 | 2.82e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 6 | 18 | 2.82e-01 |
| GO:BP | GO:0036497 | eIF2alpha dephosphorylation in response to endoplasmic reticulum stress | 1 | 1 | 2.82e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 7 | 13 | 2.82e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 2 | 12 | 2.82e-01 |
| GO:BP | GO:1905443 | regulation of clathrin coat assembly | 1 | 1 | 2.82e-01 |
| GO:BP | GO:1903917 | positive regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 2.82e-01 |
| GO:BP | GO:1902074 | response to salt | 21 | 271 | 2.82e-01 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 3 | 5 | 2.82e-01 |
| GO:BP | GO:1903916 | regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0071955 | recycling endosome to Golgi transport | 1 | 2 | 2.82e-01 |
| GO:BP | GO:1905445 | positive regulation of clathrin coat assembly | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0010609 | mRNA localization resulting in post-transcriptional regulation of gene expression | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 3 | 22 | 2.82e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 2 | 144 | 2.82e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 9 | 14 | 2.83e-01 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 2 | 4 | 2.83e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 7 | 26 | 2.83e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 2.83e-01 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 1 | 9 | 2.83e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 11 | 41 | 2.83e-01 |
| GO:BP | GO:1904747 | positive regulation of apoptotic process involved in development | 4 | 5 | 2.83e-01 |
| GO:BP | GO:1902339 | positive regulation of apoptotic process involved in morphogenesis | 4 | 5 | 2.83e-01 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 4 | 15 | 2.83e-01 |
| GO:BP | GO:0021594 | rhombomere formation | 1 | 1 | 2.83e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 4 | 25 | 2.83e-01 |
| GO:BP | GO:0021595 | rhombomere structural organization | 1 | 1 | 2.83e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 7 | 1484 | 2.83e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 10 | 20 | 2.83e-01 |
| GO:BP | GO:0021659 | rhombomere 3 structural organization | 1 | 1 | 2.83e-01 |
| GO:BP | GO:0021660 | rhombomere 3 formation | 1 | 1 | 2.83e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 3 | 27 | 2.83e-01 |
| GO:BP | GO:0021664 | rhombomere 5 morphogenesis | 1 | 1 | 2.83e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 57 | 206 | 2.83e-01 |
| GO:BP | GO:0021666 | rhombomere 5 formation | 1 | 1 | 2.83e-01 |
| GO:BP | GO:0048742 | regulation of skeletal muscle fiber development | 1 | 8 | 2.83e-01 |
| GO:BP | GO:0021665 | rhombomere 5 structural organization | 1 | 1 | 2.83e-01 |
| GO:BP | GO:1901207 | regulation of heart looping | 2 | 2 | 2.83e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 14 | 2.83e-01 |
| GO:BP | GO:0060682 | primary ureteric bud growth | 1 | 1 | 2.83e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 50 | 80 | 2.83e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 29 | 80 | 2.83e-01 |
| GO:BP | GO:1905340 | regulation of protein localization to kinetochore | 3 | 4 | 2.83e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 2.83e-01 |
| GO:BP | GO:1905342 | positive regulation of protein localization to kinetochore | 3 | 4 | 2.83e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 14 | 138 | 2.83e-01 |
| GO:BP | GO:0000296 | spermine transport | 1 | 3 | 2.83e-01 |
| GO:BP | GO:1904046 | negative regulation of vascular endothelial growth factor production | 1 | 1 | 2.83e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 20 | 30 | 2.83e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 6 | 101 | 2.83e-01 |
| GO:BP | GO:0060080 | inhibitory postsynaptic potential | 1 | 8 | 2.83e-01 |
| GO:BP | GO:0035623 | renal glucose absorption | 2 | 2 | 2.83e-01 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 14 | 23 | 2.84e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 5 | 26 | 2.84e-01 |
| GO:BP | GO:0090148 | membrane fission | 24 | 44 | 2.84e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 7 | 137 | 2.85e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 2 | 32 | 2.85e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 6 | 8 | 2.85e-01 |
| GO:BP | GO:0090398 | cellular senescence | 28 | 83 | 2.86e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 4 | 89 | 2.86e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 2 | 32 | 2.86e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 2.86e-01 |
| GO:BP | GO:0046065 | dCTP metabolic process | 1 | 1 | 2.86e-01 |
| GO:BP | GO:0006253 | dCTP catabolic process | 1 | 1 | 2.86e-01 |
| GO:BP | GO:0097338 | response to clozapine | 2 | 2 | 2.86e-01 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 3 | 6 | 2.87e-01 |
| GO:BP | GO:0043622 | cortical microtubule organization | 3 | 4 | 2.87e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 1 | 33 | 2.87e-01 |
| GO:BP | GO:0090155 | negative regulation of sphingolipid biosynthetic process | 1 | 6 | 2.87e-01 |
| GO:BP | GO:0002091 | negative regulation of receptor internalization | 5 | 14 | 2.87e-01 |
| GO:BP | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 2.88e-01 |
| GO:BP | GO:1900075 | positive regulation of neuromuscular synaptic transmission | 1 | 2 | 2.88e-01 |
| GO:BP | GO:2000302 | positive regulation of synaptic vesicle exocytosis | 1 | 3 | 2.88e-01 |
| GO:BP | GO:1900073 | regulation of neuromuscular synaptic transmission | 1 | 2 | 2.88e-01 |
| GO:BP | GO:1901503 | ether biosynthetic process | 4 | 11 | 2.88e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 19 | 75 | 2.88e-01 |
| GO:BP | GO:0006421 | asparaginyl-tRNA aminoacylation | 3 | 3 | 2.88e-01 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 2 | 4 | 2.88e-01 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 2 | 4 | 2.88e-01 |
| GO:BP | GO:0019660 | glycolytic fermentation | 2 | 4 | 2.88e-01 |
| GO:BP | GO:1901003 | negative regulation of fermentation | 2 | 4 | 2.88e-01 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 2 | 4 | 2.88e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 12 | 21 | 2.88e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 14 | 158 | 2.89e-01 |
| GO:BP | GO:0043584 | nose development | 2 | 10 | 2.89e-01 |
| GO:BP | GO:0010652 | positive regulation of cell communication by chemical coupling | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0098906 | regulation of Purkinje myocyte action potential | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0010645 | regulation of cell communication by chemical coupling | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 25 | 377 | 2.89e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 2 | 94 | 2.89e-01 |
| GO:BP | GO:0007620 | copulation | 1 | 10 | 2.89e-01 |
| GO:BP | GO:1990029 | vasomotion | 1 | 1 | 2.89e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 1 | 33 | 2.89e-01 |
| GO:BP | GO:0086044 | atrial cardiac muscle cell to AV node cell communication by electrical coupling | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0086055 | Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0048565 | digestive tract development | 8 | 92 | 2.89e-01 |
| GO:BP | GO:0035601 | protein deacylation | 53 | 116 | 2.89e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 13 | 40 | 2.89e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 3 | 13 | 2.89e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 3 | 18 | 2.89e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 21 | 41 | 2.90e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 8 | 146 | 2.91e-01 |
| GO:BP | GO:0021889 | olfactory bulb interneuron differentiation | 6 | 7 | 2.91e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 2.91e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 5 | 479 | 2.91e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 10 | 75 | 2.92e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 4 | 121 | 2.92e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 2 | 7 | 2.92e-01 |
| GO:BP | GO:0071932 | replication fork reversal | 4 | 5 | 2.92e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 6 | 14 | 2.92e-01 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 2 | 11 | 2.92e-01 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 1 | 9 | 2.92e-01 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 1 | 9 | 2.92e-01 |
| GO:BP | GO:0035499 | carnosine biosynthetic process | 1 | 1 | 2.93e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 2.93e-01 |
| GO:BP | GO:0044557 | relaxation of smooth muscle | 2 | 9 | 2.93e-01 |
| GO:BP | GO:0097698 | telomere maintenance via base-excision repair | 1 | 1 | 2.93e-01 |
| GO:BP | GO:0106230 | protein depropionylation | 2 | 2 | 2.94e-01 |
| GO:BP | GO:0060995 | cell-cell signaling involved in kidney development | 1 | 1 | 2.94e-01 |
| GO:BP | GO:0061290 | canonical Wnt signaling pathway involved in metanephric kidney development | 1 | 1 | 2.94e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 3 | 23 | 2.94e-01 |
| GO:BP | GO:0099172 | presynapse organization | 6 | 42 | 2.94e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 6 | 22 | 2.94e-01 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 3 | 10 | 2.94e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 6 | 23 | 2.94e-01 |
| GO:BP | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis | 2 | 2 | 2.94e-01 |
| GO:BP | GO:1900235 | negative regulation of Kit signaling pathway | 2 | 2 | 2.94e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 2.94e-01 |
| GO:BP | GO:0061289 | Wnt signaling pathway involved in kidney development | 1 | 1 | 2.94e-01 |
| GO:BP | GO:0071051 | polyadenylation-dependent snoRNA 3’-end processing | 6 | 7 | 2.94e-01 |
| GO:BP | GO:1900234 | regulation of Kit signaling pathway | 2 | 2 | 2.94e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 32 | 103 | 2.94e-01 |
| GO:BP | GO:0031393 | negative regulation of prostaglandin biosynthetic process | 2 | 2 | 2.94e-01 |
| GO:BP | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process | 4 | 6 | 2.94e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 6 | 22 | 2.94e-01 |
| GO:BP | GO:0072204 | cell-cell signaling involved in metanephros development | 1 | 1 | 2.94e-01 |
| GO:BP | GO:0030011 | maintenance of cell polarity | 2 | 16 | 2.94e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 3 | 31 | 2.94e-01 |
| GO:BP | GO:1990743 | protein sialylation | 1 | 5 | 2.94e-01 |
| GO:BP | GO:1903929 | primary palate development | 1 | 2 | 2.94e-01 |
| GO:BP | GO:0051877 | pigment granule aggregation in cell center | 1 | 2 | 2.94e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 3 | 206 | 2.95e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 41 | 200 | 2.95e-01 |
| GO:BP | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential | 2 | 3 | 2.95e-01 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 4 | 13 | 2.95e-01 |
| GO:BP | GO:0086048 | membrane depolarization during bundle of His cell action potential | 2 | 3 | 2.95e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.95e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 2.95e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 2.95e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 3 | 7 | 2.95e-01 |
| GO:BP | GO:0006498 | N-terminal protein lipidation | 4 | 7 | 2.96e-01 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 4 | 9 | 2.96e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 6 | 35 | 2.96e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 4 | 50 | 2.96e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 2 | 3 | 2.96e-01 |
| GO:BP | GO:0009611 | response to wounding | 2 | 415 | 2.97e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 2.97e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 2.97e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 4 | 29 | 2.97e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 2.97e-01 |
| GO:BP | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response | 1 | 2 | 2.97e-01 |
| GO:BP | GO:0002540 | leukotriene production involved in inflammatory response | 1 | 2 | 2.97e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 14 | 140 | 2.97e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 25 | 64 | 2.97e-01 |
| GO:BP | GO:0021891 | olfactory bulb interneuron development | 4 | 5 | 2.97e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 2 | 6 | 2.97e-01 |
| GO:BP | GO:1901656 | glycoside transport | 4 | 5 | 2.97e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 4 | 38 | 2.97e-01 |
| GO:BP | GO:0018352 | protein-pyridoxal-5-phosphate linkage | 1 | 2 | 2.97e-01 |
| GO:BP | GO:0006540 | glutamate decarboxylation to succinate | 1 | 1 | 2.97e-01 |
| GO:BP | GO:1903542 | negative regulation of exosomal secretion | 1 | 4 | 2.98e-01 |
| GO:BP | GO:0070676 | intralumenal vesicle formation | 1 | 4 | 2.98e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 5 | 66 | 2.98e-01 |
| GO:BP | GO:0035927 | RNA import into mitochondrion | 3 | 4 | 2.98e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 2 | 84 | 2.98e-01 |
| GO:BP | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis | 3 | 4 | 2.98e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 1 | 24 | 2.99e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 2.99e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 1 | 8 | 2.99e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 3.00e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 2 | 15 | 3.00e-01 |
| GO:BP | GO:0032200 | telomere organization | 34 | 166 | 3.00e-01 |
| GO:BP | GO:0001696 | gastric acid secretion | 2 | 10 | 3.00e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 48 | 188 | 3.00e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 13 | 23 | 3.00e-01 |
| GO:BP | GO:0021773 | striatal medium spiny neuron differentiation | 1 | 3 | 3.01e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 4 | 6 | 3.01e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0051136 | regulation of NK T cell differentiation | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth | 1 | 2 | 3.01e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 1 | 7 | 3.01e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 4 | 93 | 3.01e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 4 | 92 | 3.01e-01 |
| GO:BP | GO:0032691 | negative regulation of interleukin-1 beta production | 2 | 13 | 3.01e-01 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 2 | 18 | 3.01e-01 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0045951 | positive regulation of mitotic recombination | 1 | 1 | 3.01e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 3 | 13 | 3.01e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 3 | 13 | 3.01e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 7 | 11 | 3.01e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 3 | 69 | 3.02e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 12 | 27 | 3.02e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 3 | 14 | 3.03e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 14 | 21 | 3.03e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 3.03e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 132 | 3.04e-01 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 6 | 7 | 3.04e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 3.04e-01 |
| GO:BP | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | 1 | 2 | 3.04e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 8 | 127 | 3.04e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 3.04e-01 |
| GO:BP | GO:0018009 | N-terminal peptidyl-L-cysteine N-palmitoylation | 2 | 2 | 3.04e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 8 | 36 | 3.05e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 2 | 4 | 3.05e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 3.05e-01 |
| GO:BP | GO:0070295 | renal water absorption | 3 | 5 | 3.05e-01 |
| GO:BP | GO:0090399 | replicative senescence | 2 | 13 | 3.05e-01 |
| GO:BP | GO:0070233 | negative regulation of T cell apoptotic process | 1 | 14 | 3.05e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 5 | 10 | 3.05e-01 |
| GO:BP | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction | 2 | 2 | 3.05e-01 |
| GO:BP | GO:1901862 | negative regulation of muscle tissue development | 2 | 9 | 3.05e-01 |
| GO:BP | GO:0090031 | positive regulation of steroid hormone biosynthetic process | 3 | 5 | 3.05e-01 |
| GO:BP | GO:0000495 | box H/ACA RNA 3’-end processing | 2 | 2 | 3.05e-01 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 5 | 21 | 3.05e-01 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 2 | 3 | 3.05e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 3 | 129 | 3.05e-01 |
| GO:BP | GO:0034964 | box H/ACA RNA processing | 2 | 2 | 3.05e-01 |
| GO:BP | GO:2000565 | negative regulation of CD8-positive, alpha-beta T cell proliferation | 2 | 2 | 3.05e-01 |
| GO:BP | GO:0006424 | glutamyl-tRNA aminoacylation | 1 | 2 | 3.05e-01 |
| GO:BP | GO:0006433 | prolyl-tRNA aminoacylation | 1 | 2 | 3.05e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 2 | 22 | 3.05e-01 |
| GO:BP | GO:0007033 | vacuole organization | 8 | 197 | 3.05e-01 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 1 | 3 | 3.05e-01 |
| GO:BP | GO:0030034 | microvillar actin bundle assembly | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0000730 | DNA recombinase assembly | 2 | 3 | 3.05e-01 |
| GO:BP | GO:0090735 | DNA repair complex assembly | 2 | 3 | 3.05e-01 |
| GO:BP | GO:0003408 | optic cup formation involved in camera-type eye development | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 9 | 21 | 3.05e-01 |
| GO:BP | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 2 | 8 | 3.05e-01 |
| GO:BP | GO:1904933 | regulation of cell proliferation in midbrain | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0051885 | positive regulation of timing of anagen | 1 | 2 | 3.05e-01 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 3 | 10 | 3.05e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 20 | 3.05e-01 |
| GO:BP | GO:0036518 | chemorepulsion of dopaminergic neuron axon | 1 | 2 | 3.05e-01 |
| GO:BP | GO:1905438 | non-canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0001841 | neural tube formation | 2 | 101 | 3.05e-01 |
| GO:BP | GO:0036515 | serotonergic neuron axon guidance | 1 | 2 | 3.05e-01 |
| GO:BP | GO:0035754 | B cell chemotaxis | 3 | 5 | 3.06e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 5 | 38 | 3.06e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 4 | 40 | 3.06e-01 |
| GO:BP | GO:0030261 | chromosome condensation | 12 | 33 | 3.06e-01 |
| GO:BP | GO:2000311 | regulation of AMPA receptor activity | 2 | 15 | 3.06e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 3.06e-01 |
| GO:BP | GO:0021670 | lateral ventricle development | 5 | 11 | 3.07e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 10 | 113 | 3.07e-01 |
| GO:BP | GO:0035552 | oxidative single-stranded DNA demethylation | 2 | 3 | 3.07e-01 |
| GO:BP | GO:1902045 | negative regulation of Fas signaling pathway | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0051106 | positive regulation of DNA ligation | 4 | 5 | 3.08e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 1 | 25 | 3.08e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 4 | 317 | 3.08e-01 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 4 | 7 | 3.08e-01 |
| GO:BP | GO:0060945 | cardiac neuron differentiation | 1 | 1 | 3.08e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 2 | 15 | 3.08e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 16 | 378 | 3.08e-01 |
| GO:BP | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis | 1 | 1 | 3.08e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 3.09e-01 |
| GO:BP | GO:0006666 | 3-keto-sphinganine metabolic process | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 4 | 12 | 3.09e-01 |
| GO:BP | GO:0043602 | nitrate catabolic process | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0046210 | nitric oxide catabolic process | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0098924 | retrograde trans-synaptic signaling by nitric oxide | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0098923 | retrograde trans-synaptic signaling by soluble gas | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 6 | 487 | 3.09e-01 |
| GO:BP | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 3 | 14 | 3.09e-01 |
| GO:BP | GO:0072718 | response to cisplatin | 3 | 6 | 3.09e-01 |
| GO:BP | GO:0097695 | establishment of protein-containing complex localization to telomere | 3 | 3 | 3.09e-01 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 2 | 2 | 3.09e-01 |
| GO:BP | GO:0097694 | establishment of RNA localization to telomere | 3 | 3 | 3.09e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 3 | 15 | 3.09e-01 |
| GO:BP | GO:1905463 | negative regulation of DNA duplex unwinding | 3 | 4 | 3.09e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 1 | 10 | 3.10e-01 |
| GO:BP | GO:0052040 | modulation by symbiont of host programmed cell death | 3 | 3 | 3.10e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 24 | 47 | 3.10e-01 |
| GO:BP | GO:0008614 | pyridoxine metabolic process | 1 | 2 | 3.10e-01 |
| GO:BP | GO:1902723 | negative regulation of skeletal muscle satellite cell proliferation | 2 | 2 | 3.10e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 2 | 42 | 3.10e-01 |
| GO:BP | GO:0042823 | pyridoxal phosphate biosynthetic process | 1 | 2 | 3.10e-01 |
| GO:BP | GO:0039526 | modulation by virus of host apoptotic process | 3 | 3 | 3.10e-01 |
| GO:BP | GO:0008615 | pyridoxine biosynthetic process | 1 | 2 | 3.10e-01 |
| GO:BP | GO:0052150 | modulation by symbiont of host apoptotic process | 3 | 3 | 3.10e-01 |
| GO:BP | GO:0048732 | gland development | 3 | 318 | 3.10e-01 |
| GO:BP | GO:0003138 | primary heart field specification | 2 | 3 | 3.10e-01 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 5 | 41 | 3.10e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 3.11e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 3.11e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 16 | 70 | 3.11e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 31 | 604 | 3.11e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 2 | 41 | 3.12e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 2 | 18 | 3.12e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 6 | 7 | 3.12e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 8 | 147 | 3.12e-01 |
| GO:BP | GO:1903566 | positive regulation of protein localization to cilium | 5 | 7 | 3.12e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 8 | 51 | 3.12e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 10 | 89 | 3.13e-01 |
| GO:BP | GO:0072126 | positive regulation of glomerular mesangial cell proliferation | 5 | 7 | 3.13e-01 |
| GO:BP | GO:0006598 | polyamine catabolic process | 3 | 4 | 3.13e-01 |
| GO:BP | GO:0043276 | anoikis | 12 | 32 | 3.13e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 1 | 21 | 3.13e-01 |
| GO:BP | GO:0090210 | regulation of establishment of blood-brain barrier | 1 | 3 | 3.13e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 4 | 19 | 3.13e-01 |
| GO:BP | GO:1905198 | manchette assembly | 4 | 4 | 3.13e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 1 | 97 | 3.13e-01 |
| GO:BP | GO:1903774 | positive regulation of viral budding via host ESCRT complex | 2 | 3 | 3.13e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 8 | 17 | 3.13e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 6 | 22 | 3.14e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 6 | 13 | 3.14e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 2 | 111 | 3.14e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 10 | 14 | 3.14e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 4 | 4 | 3.14e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 4 | 4 | 3.14e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 4 | 4 | 3.14e-01 |
| GO:BP | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration | 2 | 6 | 3.14e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 2 | 343 | 3.14e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 2 | 75 | 3.14e-01 |
| GO:BP | GO:0060173 | limb development | 2 | 142 | 3.14e-01 |
| GO:BP | GO:0030952 | establishment or maintenance of cytoskeleton polarity | 3 | 7 | 3.14e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 21 | 740 | 3.14e-01 |
| GO:BP | GO:0048736 | appendage development | 2 | 142 | 3.14e-01 |
| GO:BP | GO:0002505 | antigen processing and presentation of polysaccharide antigen via MHC class II | 1 | 1 | 3.14e-01 |
| GO:BP | GO:0046754 | viral exocytosis | 1 | 1 | 3.14e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 2 | 14 | 3.14e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 3.14e-01 |
| GO:BP | GO:0002747 | antigen processing and presentation following phagocytosis | 1 | 1 | 3.14e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 2 | 12 | 3.15e-01 |
| GO:BP | GO:0009174 | pyrimidine ribonucleoside monophosphate biosynthetic process | 4 | 9 | 3.15e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 3 | 40 | 3.15e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 8 | 10 | 3.15e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 4 | 15 | 3.15e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 3 | 18 | 3.15e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 3 | 18 | 3.15e-01 |
| GO:BP | GO:0044205 | ‘de novo’ UMP biosynthetic process | 2 | 3 | 3.15e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 1 | 29 | 3.15e-01 |
| GO:BP | GO:1905222 | atrioventricular canal morphogenesis | 3 | 4 | 3.15e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 8 | 10 | 3.15e-01 |
| GO:BP | GO:0035935 | androgen secretion | 1 | 1 | 3.15e-01 |
| GO:BP | GO:2000836 | positive regulation of androgen secretion | 1 | 1 | 3.15e-01 |
| GO:BP | GO:2000834 | regulation of androgen secretion | 1 | 1 | 3.15e-01 |
| GO:BP | GO:0010165 | response to X-ray | 11 | 29 | 3.16e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 6 | 30 | 3.16e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 5 | 31 | 3.16e-01 |
| GO:BP | GO:0051694 | pointed-end actin filament capping | 3 | 6 | 3.16e-01 |
| GO:BP | GO:0016114 | terpenoid biosynthetic process | 2 | 12 | 3.16e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 9 | 68 | 3.16e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 3.16e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 6 | 53 | 3.16e-01 |
| GO:BP | GO:0007522 | visceral muscle development | 1 | 1 | 3.16e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 30 | 72 | 3.16e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 3 | 26 | 3.16e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 3.16e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 153 | 3.16e-01 |
| GO:BP | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 3 | 3.16e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 3.16e-01 |
| GO:BP | GO:0006265 | DNA topological change | 1 | 10 | 3.16e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 3.16e-01 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 4 | 7 | 3.16e-01 |
| GO:BP | GO:0021856 | hypothalamic tangential migration using cell-axon interactions | 5 | 6 | 3.16e-01 |
| GO:BP | GO:0021828 | gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 5 | 6 | 3.16e-01 |
| GO:BP | GO:1905301 | regulation of macropinocytosis | 3 | 3 | 3.16e-01 |
| GO:BP | GO:1903279 | regulation of calcium:sodium antiporter activity | 1 | 3 | 3.16e-01 |
| GO:BP | GO:0045955 | negative regulation of calcium ion-dependent exocytosis | 4 | 6 | 3.16e-01 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 5 | 5 | 3.16e-01 |
| GO:BP | GO:1990261 | pre-mRNA catabolic process | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 2 | 37 | 3.17e-01 |
| GO:BP | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 2 | 3 | 3.17e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 2 | 504 | 3.17e-01 |
| GO:BP | GO:0097291 | renal phosphate ion absorption | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 2 | 63 | 3.17e-01 |
| GO:BP | GO:0034478 | phosphatidylglycerol catabolic process | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0014832 | urinary bladder smooth muscle contraction | 2 | 4 | 3.17e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 3 | 55 | 3.17e-01 |
| GO:BP | GO:1905462 | regulation of DNA duplex unwinding | 3 | 7 | 3.17e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 100 | 3.17e-01 |
| GO:BP | GO:0001100 | negative regulation of exit from mitosis | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0048703 | embryonic viscerocranium morphogenesis | 1 | 8 | 3.17e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 1 | 64 | 3.18e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 3 | 45 | 3.18e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 1 | 4 | 3.18e-01 |
| GO:BP | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 1 | 2 | 3.18e-01 |
| GO:BP | GO:1905026 | positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.18e-01 |
| GO:BP | GO:1905033 | positive regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 2 | 3.18e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 4 | 22 | 3.18e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 28 | 414 | 3.18e-01 |
| GO:BP | GO:0045769 | negative regulation of asymmetric cell division | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0035513 | oxidative RNA demethylation | 3 | 5 | 3.19e-01 |
| GO:BP | GO:0010224 | response to UV-B | 2 | 17 | 3.19e-01 |
| GO:BP | GO:0031175 | neuron projection development | 3 | 780 | 3.19e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 10 | 133 | 3.19e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 3 | 763 | 3.19e-01 |
| GO:BP | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 3.19e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 3.19e-01 |
| GO:BP | GO:2000080 | negative regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 2 | 31 | 3.19e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 2 | 11 | 3.19e-01 |
| GO:BP | GO:0044345 | stromal-epithelial cell signaling involved in prostate gland development | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0042746 | circadian sleep/wake cycle, wakefulness | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0016080 | synaptic vesicle targeting | 1 | 3 | 3.19e-01 |
| GO:BP | GO:0090246 | convergent extension involved in somitogenesis | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0060322 | head development | 12 | 633 | 3.19e-01 |
| GO:BP | GO:0048852 | diencephalon morphogenesis | 2 | 2 | 3.19e-01 |
| GO:BP | GO:0006266 | DNA ligation | 9 | 16 | 3.19e-01 |
| GO:BP | GO:0150018 | basal dendrite development | 3 | 3 | 3.20e-01 |
| GO:BP | GO:0150019 | basal dendrite morphogenesis | 3 | 3 | 3.20e-01 |
| GO:BP | GO:0150020 | basal dendrite arborization | 3 | 3 | 3.20e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 9 | 13 | 3.20e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 2 | 7 | 3.20e-01 |
| GO:BP | GO:0007616 | long-term memory | 8 | 31 | 3.20e-01 |
| GO:BP | GO:0106034 | protein maturation by [2Fe-2S] cluster transfer | 1 | 3 | 3.20e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 2 | 21 | 3.20e-01 |
| GO:BP | GO:0006222 | UMP biosynthetic process | 2 | 8 | 3.20e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 104 | 3.20e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 9 | 46 | 3.20e-01 |
| GO:BP | GO:0060932 | His-Purkinje system cell differentiation | 4 | 4 | 3.20e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 6 | 1229 | 3.20e-01 |
| GO:BP | GO:0036211 | protein modification process | 7 | 2901 | 3.20e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 11 | 113 | 3.20e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 8 | 234 | 3.20e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 19 | 204 | 3.20e-01 |
| GO:BP | GO:2000538 | positive regulation of B cell chemotaxis | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0006400 | tRNA modification | 54 | 90 | 3.20e-01 |
| GO:BP | GO:2000537 | regulation of B cell chemotaxis | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0033599 | regulation of mammary gland epithelial cell proliferation | 10 | 14 | 3.20e-01 |
| GO:BP | GO:0035694 | mitochondrial protein catabolic process | 2 | 6 | 3.20e-01 |
| GO:BP | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | 1 | 4 | 3.20e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 3.21e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 3.21e-01 |
| GO:BP | GO:0002313 | mature B cell differentiation involved in immune response | 3 | 17 | 3.21e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 3.21e-01 |
| GO:BP | GO:0035695 | mitophagy by induced vacuole formation | 1 | 1 | 3.21e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 3.21e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 3.21e-01 |
| GO:BP | GO:1903966 | monounsaturated fatty acid biosynthetic process | 1 | 2 | 3.21e-01 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 4 | 27 | 3.21e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 4 | 18 | 3.21e-01 |
| GO:BP | GO:1904978 | regulation of endosome organization | 1 | 5 | 3.22e-01 |
| GO:BP | GO:0022615 | protein to membrane docking | 1 | 5 | 3.22e-01 |
| GO:BP | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 3 | 6 | 3.22e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 2 | 8 | 3.22e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 1 | 109 | 3.23e-01 |
| GO:BP | GO:0060856 | establishment of blood-brain barrier | 2 | 9 | 3.23e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.23e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 20 | 64 | 3.23e-01 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 4 | 14 | 3.23e-01 |
| GO:BP | GO:0010312 | detoxification of zinc ion | 1 | 1 | 3.23e-01 |
| GO:BP | GO:1990359 | stress response to zinc ion | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0046074 | dTMP catabolic process | 1 | 3 | 3.23e-01 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 14 | 21 | 3.23e-01 |
| GO:BP | GO:0021894 | cerebral cortex GABAergic interneuron development | 1 | 3 | 3.23e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 7 | 197 | 3.23e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 5 | 56 | 3.24e-01 |
| GO:BP | GO:0030091 | protein repair | 2 | 7 | 3.24e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 8 | 73 | 3.24e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 10 | 24 | 3.24e-01 |
| GO:BP | GO:0046068 | cGMP metabolic process | 2 | 11 | 3.25e-01 |
| GO:BP | GO:0033499 | galactose catabolic process via UDP-galactose | 2 | 4 | 3.25e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 3 | 139 | 3.25e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 2 | 2 | 3.25e-01 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 1 | 9 | 3.26e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 3.26e-01 |
| GO:BP | GO:2000639 | negative regulation of SREBP signaling pathway | 3 | 3 | 3.26e-01 |
| GO:BP | GO:0051607 | defense response to virus | 2 | 202 | 3.26e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 3.26e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 1 | 98 | 3.26e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 2 | 202 | 3.27e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 2 | 2 | 3.27e-01 |
| GO:BP | GO:0009446 | putrescine biosynthetic process | 1 | 4 | 3.27e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 11 | 357 | 3.27e-01 |
| GO:BP | GO:0051653 | spindle localization | 8 | 52 | 3.28e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 3.28e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 3 | 3 | 3.28e-01 |
| GO:BP | GO:0009447 | putrescine catabolic process | 1 | 2 | 3.28e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 3 | 12 | 3.28e-01 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 2 | 3 | 3.28e-01 |
| GO:BP | GO:0048627 | myoblast development | 1 | 3 | 3.28e-01 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 1 | 4 | 3.28e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 2 | 23 | 3.28e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 2 | 11 | 3.28e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 7 | 39 | 3.28e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 4 | 20 | 3.28e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 2 | 128 | 3.29e-01 |
| GO:BP | GO:0099576 | regulation of protein catabolic process at postsynapse, modulating synaptic transmission | 1 | 4 | 3.29e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 5 | 10 | 3.29e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 3 | 60 | 3.29e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 5 | 10 | 3.29e-01 |
| GO:BP | GO:0038129 | ERBB3 signaling pathway | 4 | 4 | 3.29e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 3 | 27 | 3.29e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 2 | 156 | 3.29e-01 |
| GO:BP | GO:0030576 | Cajal body organization | 2 | 4 | 3.29e-01 |
| GO:BP | GO:0003335 | corneocyte development | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1 | 10 | 3.29e-01 |
| GO:BP | GO:0032902 | nerve growth factor production | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0015680 | protein maturation by copper ion transfer | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0044849 | estrous cycle | 2 | 13 | 3.30e-01 |
| GO:BP | GO:1902527 | positive regulation of protein monoubiquitination | 4 | 5 | 3.30e-01 |
| GO:BP | GO:1904875 | regulation of DNA ligase activity | 2 | 2 | 3.30e-01 |
| GO:BP | GO:0030913 | paranodal junction assembly | 4 | 6 | 3.30e-01 |
| GO:BP | GO:1903518 | positive regulation of single strand break repair | 2 | 2 | 3.30e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 4 | 39 | 3.30e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 3.31e-01 |
| GO:BP | GO:1902729 | negative regulation of proteoglycan biosynthetic process | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0032683 | negative regulation of connective tissue growth factor production | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 2 | 13 | 3.32e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 12 | 61 | 3.32e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 3.32e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 1 | 69 | 3.32e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 1 | 45 | 3.32e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 6 | 560 | 3.32e-01 |
| GO:BP | GO:1901859 | negative regulation of mitochondrial DNA metabolic process | 1 | 2 | 3.32e-01 |
| GO:BP | GO:0061687 | detoxification of inorganic compound | 3 | 12 | 3.32e-01 |
| GO:BP | GO:0097501 | stress response to metal ion | 3 | 11 | 3.32e-01 |
| GO:BP | GO:0051257 | meiotic spindle midzone assembly | 1 | 2 | 3.32e-01 |
| GO:BP | GO:0032237 | activation of store-operated calcium channel activity | 4 | 5 | 3.32e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 17 | 406 | 3.32e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 2 | 2 | 3.33e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 14 | 326 | 3.33e-01 |
| GO:BP | GO:0097531 | mast cell migration | 9 | 12 | 3.33e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 2 | 551 | 3.33e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 10 | 92 | 3.33e-01 |
| GO:BP | GO:0000425 | pexophagy | 1 | 6 | 3.33e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 5 | 108 | 3.33e-01 |
| GO:BP | GO:0006895 | Golgi to endosome transport | 10 | 17 | 3.33e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 10 | 92 | 3.33e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 8 | 23 | 3.33e-01 |
| GO:BP | GO:1903361 | protein localization to basolateral plasma membrane | 1 | 7 | 3.33e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 3 | 5 | 3.33e-01 |
| GO:BP | GO:0060840 | artery development | 3 | 83 | 3.33e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 9 | 24 | 3.33e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 8 | 13 | 3.33e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 2 | 2 | 3.33e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 10 | 12 | 3.33e-01 |
| GO:BP | GO:0006183 | GTP biosynthetic process | 7 | 9 | 3.33e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 2 | 7 | 3.33e-01 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 3 | 5 | 3.33e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 2 | 8 | 3.33e-01 |
| GO:BP | GO:1990535 | neuron projection maintenance | 4 | 9 | 3.33e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 3 | 33 | 3.33e-01 |
| GO:BP | GO:0000160 | phosphorelay signal transduction system | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 3.34e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 3.34e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 7 | 17 | 3.34e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 3.34e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 11 | 21 | 3.34e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 3.34e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 3.34e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 8 | 387 | 3.34e-01 |
| GO:BP | GO:0070159 | mitochondrial threonyl-tRNA aminoacylation | 1 | 1 | 3.34e-01 |
| GO:BP | GO:0051660 | establishment of centrosome localization | 4 | 8 | 3.34e-01 |
| GO:BP | GO:0043490 | malate-aspartate shuttle | 2 | 4 | 3.35e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 8 | 15 | 3.35e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 3.35e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 3.35e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 2 | 35 | 3.35e-01 |
| GO:BP | GO:0075071 | modulation by symbiont of host autophagy | 1 | 1 | 3.35e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 2 | 5 | 3.35e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 3.35e-01 |
| GO:BP | GO:0071275 | cellular response to aluminum ion | 1 | 1 | 3.35e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 6 | 10 | 3.35e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 10 | 14 | 3.35e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 2 | 40 | 3.35e-01 |
| GO:BP | GO:0039521 | suppression by virus of host autophagy | 1 | 1 | 3.35e-01 |
| GO:BP | GO:0006971 | hypotonic response | 2 | 10 | 3.35e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 2 | 5 | 3.35e-01 |
| GO:BP | GO:0002157 | positive regulation of thyroid hormone mediated signaling pathway | 2 | 3 | 3.35e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 3.35e-01 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 2 | 5 | 3.35e-01 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 2 | 3 | 3.35e-01 |
| GO:BP | GO:0001782 | B cell homeostasis | 1 | 24 | 3.35e-01 |
| GO:BP | GO:0140321 | suppression of host autophagy | 1 | 1 | 3.35e-01 |
| GO:BP | GO:0007619 | courtship behavior | 2 | 3 | 3.35e-01 |
| GO:BP | GO:1905672 | negative regulation of lysosome organization | 1 | 1 | 3.35e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 11 | 3.35e-01 |
| GO:BP | GO:1905775 | negative regulation of DNA helicase activity | 2 | 2 | 3.36e-01 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 7 | 9 | 3.36e-01 |
| GO:BP | GO:0097510 | base-excision repair, AP site formation via deaminated base removal | 1 | 1 | 3.36e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 16 | 186 | 3.36e-01 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 3 | 3.36e-01 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 3.36e-01 |
| GO:BP | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | 1 | 2 | 3.36e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 3.36e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 16 | 358 | 3.36e-01 |
| GO:BP | GO:0090400 | stress-induced premature senescence | 4 | 7 | 3.36e-01 |
| GO:BP | GO:0160024 | Leydig cell proliferation | 2 | 2 | 3.36e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 3.36e-01 |
| GO:BP | GO:0099178 | regulation of retrograde trans-synaptic signaling by endocanabinoid | 2 | 3 | 3.37e-01 |
| GO:BP | GO:0000379 | tRNA-type intron splice site recognition and cleavage | 1 | 3 | 3.37e-01 |
| GO:BP | GO:0071321 | cellular response to cGMP | 1 | 7 | 3.37e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 11 | 362 | 3.38e-01 |
| GO:BP | GO:0046208 | spermine catabolic process | 2 | 2 | 3.38e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 12 | 19 | 3.38e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 6 | 7 | 3.38e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 2 | 13 | 3.38e-01 |
| GO:BP | GO:0050960 | detection of temperature stimulus involved in thermoception | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 24 | 49 | 3.38e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 4 | 34 | 3.38e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 3 | 7 | 3.38e-01 |
| GO:BP | GO:0006386 | termination of RNA polymerase III transcription | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 4 | 149 | 3.38e-01 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 2 | 4 | 3.38e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 1 | 8 | 3.38e-01 |
| GO:BP | GO:1901506 | regulation of acylglycerol transport | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0030656 | regulation of vitamin metabolic process | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0097089 | methyl-branched fatty acid metabolic process | 1 | 3 | 3.38e-01 |
| GO:BP | GO:0036146 | cellular response to mycotoxin | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 2 | 41 | 3.38e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 12 | 19 | 3.38e-01 |
| GO:BP | GO:0072721 | cellular response to dithiothreitol | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 3.38e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 1 | 16 | 3.38e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 3.38e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 12 | 19 | 3.38e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 3.38e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 2 | 42 | 3.38e-01 |
| GO:BP | GO:0140018 | regulation of cytoplasmic translational fidelity | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0021685 | cerebellar granular layer structural organization | 1 | 1 | 3.38e-01 |
| GO:BP | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway | 4 | 4 | 3.38e-01 |
| GO:BP | GO:0071502 | cellular response to temperature stimulus | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1905883 | regulation of triglyceride transport | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1905885 | positive regulation of triglyceride transport | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0006304 | DNA modification | 2 | 89 | 3.38e-01 |
| GO:BP | GO:1901508 | positive regulation of acylglycerol transport | 1 | 2 | 3.38e-01 |
| GO:BP | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 4 | 3.38e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 43 | 3.38e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 26 | 124 | 3.39e-01 |
| GO:BP | GO:0090166 | Golgi disassembly | 2 | 6 | 3.39e-01 |
| GO:BP | GO:0014041 | regulation of neuron maturation | 1 | 6 | 3.39e-01 |
| GO:BP | GO:1904439 | negative regulation of iron ion import across plasma membrane | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0006936 | muscle contraction | 8 | 267 | 3.39e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 13 | 21 | 3.39e-01 |
| GO:BP | GO:0014849 | ureter smooth muscle contraction | 2 | 2 | 3.39e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 2 | 22 | 3.39e-01 |
| GO:BP | GO:0035364 | thymine transport | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0032692 | negative regulation of interleukin-1 production | 2 | 15 | 3.39e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 12 | 139 | 3.39e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 3 | 4 | 3.39e-01 |
| GO:BP | GO:1903723 | negative regulation of centriole elongation | 1 | 2 | 3.39e-01 |
| GO:BP | GO:0003360 | brainstem development | 3 | 7 | 3.39e-01 |
| GO:BP | GO:0072105 | ureteric peristalsis | 2 | 2 | 3.39e-01 |
| GO:BP | GO:0003431 | growth plate cartilage chondrocyte development | 3 | 5 | 3.39e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 10 | 84 | 3.39e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 4 | 44 | 3.39e-01 |
| GO:BP | GO:0070848 | response to growth factor | 2 | 572 | 3.39e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 3.39e-01 |
| GO:BP | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process | 3 | 8 | 3.39e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 15 | 38 | 3.39e-01 |
| GO:BP | GO:0006113 | fermentation | 2 | 5 | 3.39e-01 |
| GO:BP | GO:0031860 | telomeric 3’ overhang formation | 1 | 4 | 3.39e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 2 | 42 | 3.39e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 1 | 102 | 3.39e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 24 | 63 | 3.39e-01 |
| GO:BP | GO:0043465 | regulation of fermentation | 2 | 5 | 3.39e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 4 | 5 | 3.39e-01 |
| GO:BP | GO:0009838 | abscission | 2 | 6 | 3.39e-01 |
| GO:BP | GO:0043063 | intercellular bridge organization | 1 | 1 | 3.39e-01 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 4 | 5 | 3.39e-01 |
| GO:BP | GO:0071504 | cellular response to heparin | 3 | 4 | 3.39e-01 |
| GO:BP | GO:0061009 | common bile duct development | 3 | 4 | 3.39e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 4 | 35 | 3.39e-01 |
| GO:BP | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 3 | 4 | 3.39e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 3.39e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 2 | 20 | 3.39e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 2 | 20 | 3.39e-01 |
| GO:BP | GO:0015761 | mannose transmembrane transport | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0061205 | paramesonephric duct development | 2 | 4 | 3.39e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 13 | 252 | 3.39e-01 |
| GO:BP | GO:0015848 | spermidine transport | 1 | 3 | 3.39e-01 |
| GO:BP | GO:0001822 | kidney development | 12 | 244 | 3.39e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 38 | 756 | 3.39e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 17 | 1712 | 3.39e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 17 | 490 | 3.39e-01 |
| GO:BP | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation | 6 | 7 | 3.39e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 4 | 159 | 3.39e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 17 | 490 | 3.39e-01 |
| GO:BP | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron development | 6 | 7 | 3.39e-01 |
| GO:BP | GO:0060166 | olfactory pit development | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway | 4 | 5 | 3.39e-01 |
| GO:BP | GO:0000320 | re-entry into mitotic cell cycle | 2 | 2 | 3.40e-01 |
| GO:BP | GO:0046600 | negative regulation of centriole replication | 4 | 7 | 3.40e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 19 | 48 | 3.40e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 1 | 76 | 3.40e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 3.40e-01 |
| GO:BP | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 2 | 4 | 3.40e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 5 | 18 | 3.40e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 67 | 251 | 3.41e-01 |
| GO:BP | GO:1904911 | negative regulation of establishment of RNA localization to telomere | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0039689 | negative stranded viral RNA replication | 2 | 3 | 3.41e-01 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 2 | 5 | 3.41e-01 |
| GO:BP | GO:1904913 | regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 8 | 12 | 3.41e-01 |
| GO:BP | GO:1904850 | negative regulation of establishment of protein localization to telomere | 1 | 1 | 3.41e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 6 | 7 | 3.41e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 4 | 59 | 3.41e-01 |
| GO:BP | GO:1900372 | negative regulation of purine nucleotide biosynthetic process | 2 | 3 | 3.41e-01 |
| GO:BP | GO:0030809 | negative regulation of nucleotide biosynthetic process | 2 | 3 | 3.41e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 4 | 63 | 3.41e-01 |
| GO:BP | GO:1904914 | negative regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 3.41e-01 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 1 | 8 | 3.41e-01 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 2 | 5 | 3.41e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 19 | 64 | 3.41e-01 |
| GO:BP | GO:1904910 | regulation of establishment of RNA localization to telomere | 1 | 1 | 3.41e-01 |
| GO:BP | GO:1904792 | positive regulation of shelterin complex assembly | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 12 | 17 | 3.41e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 2 | 4 | 3.41e-01 |
| GO:BP | GO:0055009 | atrial cardiac muscle tissue morphogenesis | 2 | 5 | 3.41e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 3.41e-01 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 2 | 9 | 3.41e-01 |
| GO:BP | GO:0099171 | presynaptic modulation of chemical synaptic transmission | 2 | 26 | 3.41e-01 |
| GO:BP | GO:0032528 | microvillus organization | 5 | 19 | 3.41e-01 |
| GO:BP | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process | 1 | 5 | 3.41e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 2 | 35 | 3.41e-01 |
| GO:BP | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | 2 | 4 | 3.41e-01 |
| GO:BP | GO:0048022 | negative regulation of melanin biosynthetic process | 2 | 4 | 3.41e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 5 | 10 | 3.41e-01 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 4 | 4 | 3.41e-01 |
| GO:BP | GO:0046710 | GDP metabolic process | 5 | 10 | 3.42e-01 |
| GO:BP | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation | 3 | 3 | 3.42e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 1 | 10 | 3.42e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 1 | 12 | 3.42e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 5 | 11 | 3.42e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 11 | 35 | 3.42e-01 |
| GO:BP | GO:1990953 | intramanchette transport | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 114 | 3.43e-01 |
| GO:BP | GO:0008090 | retrograde axonal transport | 2 | 19 | 3.43e-01 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 3 | 5 | 3.43e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 9 | 88 | 3.43e-01 |
| GO:BP | GO:1904799 | regulation of neuron remodeling | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0072034 | renal vesicle induction | 3 | 3 | 3.43e-01 |
| GO:BP | GO:1904800 | negative regulation of neuron remodeling | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 114 | 3.43e-01 |
| GO:BP | GO:2000173 | negative regulation of branching morphogenesis of a nerve | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 17 | 60 | 3.43e-01 |
| GO:BP | GO:0050925 | negative regulation of negative chemotaxis | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 6 | 147 | 3.43e-01 |
| GO:BP | GO:1900449 | regulation of glutamate receptor signaling pathway | 1 | 5 | 3.43e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 5 | 51 | 3.43e-01 |
| GO:BP | GO:0061519 | macrophage homeostasis | 1 | 3 | 3.43e-01 |
| GO:BP | GO:0035702 | monocyte homeostasis | 1 | 2 | 3.43e-01 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 5 | 7 | 3.43e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 3 | 25 | 3.43e-01 |
| GO:BP | GO:0035351 | heme transmembrane transport | 1 | 2 | 3.43e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 3 | 9 | 3.43e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 1 | 69 | 3.43e-01 |
| GO:BP | GO:0009197 | pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 16 | 40 | 3.43e-01 |
| GO:BP | GO:0042727 | flavin-containing compound biosynthetic process | 1 | 2 | 3.43e-01 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 6 | 18 | 3.43e-01 |
| GO:BP | GO:0001705 | ectoderm formation | 3 | 3 | 3.43e-01 |
| GO:BP | GO:0046072 | dTDP metabolic process | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway | 2 | 6 | 3.43e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 5 | 26 | 3.43e-01 |
| GO:BP | GO:0046077 | dUDP metabolic process | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0006233 | dTDP biosynthetic process | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0055070 | copper ion homeostasis | 4 | 18 | 3.43e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 1 | 101 | 3.43e-01 |
| GO:BP | GO:1904631 | response to glucoside | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 10 | 23 | 3.43e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 5 | 11 | 3.43e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 3.43e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 3.43e-01 |
| GO:BP | GO:0002931 | response to ischemia | 4 | 46 | 3.43e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 1 | 187 | 3.43e-01 |
| GO:BP | GO:0006227 | dUDP biosynthetic process | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0009196 | pyrimidine deoxyribonucleoside diphosphate metabolic process | 2 | 2 | 3.43e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 12 | 184 | 3.44e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 5 | 11 | 3.44e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 40 | 87 | 3.44e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 3 | 22 | 3.44e-01 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 7 | 10 | 3.44e-01 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 7 | 10 | 3.44e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 2 | 12 | 3.45e-01 |
| GO:BP | GO:0090132 | epithelium migration | 13 | 252 | 3.45e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 13 | 25 | 3.45e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 7 | 13 | 3.45e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 10 | 167 | 3.45e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 10 | 182 | 3.45e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 3 | 4 | 3.45e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 27 | 1026 | 3.45e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 4 | 57 | 3.45e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 4 | 65 | 3.46e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 4 | 161 | 3.46e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 4 | 40 | 3.46e-01 |
| GO:BP | GO:0038135 | ERBB2-ERBB4 signaling pathway | 3 | 4 | 3.46e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 2 | 15 | 3.46e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 3 | 19 | 3.46e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 17 | 495 | 3.47e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 10 | 121 | 3.47e-01 |
| GO:BP | GO:1905116 | positive regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 3.47e-01 |
| GO:BP | GO:1901647 | positive regulation of synoviocyte proliferation | 1 | 3 | 3.47e-01 |
| GO:BP | GO:1905115 | regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 2 | 20 | 3.47e-01 |
| GO:BP | GO:0002941 | synoviocyte proliferation | 1 | 3 | 3.47e-01 |
| GO:BP | GO:1901645 | regulation of synoviocyte proliferation | 1 | 3 | 3.47e-01 |
| GO:BP | GO:0014901 | satellite cell activation involved in skeletal muscle regeneration | 2 | 3 | 3.47e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 3 | 71 | 3.47e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 79 | 287 | 3.47e-01 |
| GO:BP | GO:0000917 | division septum assembly | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation | 1 | 2 | 3.47e-01 |
| GO:BP | GO:1903837 | regulation of mRNA 3’-UTR binding | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0007182 | common-partner SMAD protein phosphorylation | 1 | 6 | 3.47e-01 |
| GO:BP | GO:1904100 | positive regulation of protein O-linked glycosylation | 1 | 2 | 3.47e-01 |
| GO:BP | GO:1904098 | regulation of protein O-linked glycosylation | 1 | 2 | 3.47e-01 |
| GO:BP | GO:0036514 | dopaminergic neuron axon guidance | 1 | 3 | 3.47e-01 |
| GO:BP | GO:0003213 | cardiac right atrium morphogenesis | 1 | 2 | 3.47e-01 |
| GO:BP | GO:0030578 | PML body organization | 1 | 6 | 3.47e-01 |
| GO:BP | GO:1903839 | positive regulation of mRNA 3’-UTR binding | 1 | 1 | 3.47e-01 |
| GO:BP | GO:1905359 | protein localization to meiotic spindle | 1 | 1 | 3.47e-01 |
| GO:BP | GO:1903096 | protein localization to meiotic spindle midzone | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0090529 | cell septum assembly | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 5 | 7 | 3.47e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 6 | 12 | 3.47e-01 |
| GO:BP | GO:0021540 | corpus callosum morphogenesis | 4 | 5 | 3.47e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 3 | 4 | 3.47e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 1 | 23 | 3.47e-01 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 11 | 19 | 3.47e-01 |
| GO:BP | GO:1905774 | regulation of DNA helicase activity | 2 | 5 | 3.47e-01 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 1 | 1 | 3.47e-01 |
| GO:BP | GO:1904938 | planar cell polarity pathway involved in axon guidance | 1 | 3 | 3.47e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 6 | 67 | 3.47e-01 |
| GO:BP | GO:0035148 | tube formation | 8 | 138 | 3.47e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 1 | 121 | 3.47e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 12 | 341 | 3.47e-01 |
| GO:BP | GO:1904274 | tricellular tight junction assembly | 1 | 1 | 3.47e-01 |
| GO:BP | GO:1903444 | negative regulation of brown fat cell differentiation | 3 | 3 | 3.48e-01 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 1 | 3 | 3.48e-01 |
| GO:BP | GO:0048864 | stem cell development | 26 | 76 | 3.48e-01 |
| GO:BP | GO:2000753 | positive regulation of glucosylceramide catabolic process | 1 | 1 | 3.48e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 2 | 187 | 3.48e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 1 | 13 | 3.48e-01 |
| GO:BP | GO:1900163 | positive regulation of phospholipid scramblase activity | 1 | 1 | 3.48e-01 |
| GO:BP | GO:2000752 | regulation of glucosylceramide catabolic process | 1 | 1 | 3.48e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 4 | 57 | 3.48e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 3.48e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 3 | 12 | 3.48e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 5 | 20 | 3.48e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 2 | 187 | 3.48e-01 |
| GO:BP | GO:1900161 | regulation of phospholipid scramblase activity | 1 | 1 | 3.48e-01 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 4 | 15 | 3.48e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 13 | 25 | 3.48e-01 |
| GO:BP | GO:0072402 | response to DNA integrity checkpoint signaling | 3 | 8 | 3.48e-01 |
| GO:BP | GO:0072423 | response to DNA damage checkpoint signaling | 3 | 8 | 3.48e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 12 | 342 | 3.48e-01 |
| GO:BP | GO:0072396 | response to cell cycle checkpoint signaling | 3 | 8 | 3.48e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 11 | 26 | 3.48e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 13 | 47 | 3.48e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 3.49e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 5 | 16 | 3.49e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 5 | 16 | 3.49e-01 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 4 | 10 | 3.49e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 1 | 65 | 3.49e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 3.50e-01 |
| GO:BP | GO:0002335 | mature B cell differentiation | 1 | 21 | 3.50e-01 |
| GO:BP | GO:0060914 | heart formation | 13 | 29 | 3.50e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 2 | 9 | 3.50e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 7 | 9 | 3.50e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 4 | 57 | 3.50e-01 |
| GO:BP | GO:0099548 | trans-synaptic signaling by nitric oxide | 1 | 2 | 3.50e-01 |
| GO:BP | GO:0099543 | trans-synaptic signaling by soluble gas | 1 | 2 | 3.50e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 19 | 73 | 3.51e-01 |
| GO:BP | GO:0060056 | mammary gland involution | 3 | 8 | 3.51e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 3 | 906 | 3.51e-01 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 3 | 17 | 3.51e-01 |
| GO:BP | GO:0062026 | negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process | 1 | 1 | 3.51e-01 |
| GO:BP | GO:0062025 | regulation of SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 1 | 1 | 3.51e-01 |
| GO:BP | GO:0010868 | negative regulation of triglyceride biosynthetic process | 2 | 3 | 3.51e-01 |
| GO:BP | GO:0061044 | negative regulation of vascular wound healing | 1 | 4 | 3.51e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 3.51e-01 |
| GO:BP | GO:0035265 | organ growth | 1 | 131 | 3.51e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 7 | 17 | 3.51e-01 |
| GO:BP | GO:0034058 | endosomal vesicle fusion | 1 | 10 | 3.51e-01 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 2 | 9 | 3.51e-01 |
| GO:BP | GO:0019262 | N-acetylneuraminate catabolic process | 4 | 6 | 3.51e-01 |
| GO:BP | GO:1901628 | positive regulation of postsynaptic membrane organization | 1 | 2 | 3.51e-01 |
| GO:BP | GO:0015798 | myo-inositol transport | 2 | 3 | 3.51e-01 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 1 | 5 | 3.51e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 2 | 14 | 3.51e-01 |
| GO:BP | GO:1990550 | mitochondrial alpha-ketoglutarate transmembrane transport | 1 | 1 | 3.51e-01 |
| GO:BP | GO:0009751 | response to salicylic acid | 1 | 1 | 3.51e-01 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 2 | 16 | 3.51e-01 |
| GO:BP | GO:0000916 | actomyosin contractile ring contraction | 1 | 1 | 3.51e-01 |
| GO:BP | GO:0036213 | contractile ring contraction | 1 | 1 | 3.51e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 4 | 18 | 3.51e-01 |
| GO:BP | GO:0021979 | hypothalamus cell differentiation | 1 | 8 | 3.51e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 5 | 37 | 3.51e-01 |
| GO:BP | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 1 | 5 | 3.51e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 7 | 3090 | 3.51e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 20 | 26 | 3.51e-01 |
| GO:BP | GO:0003373 | dynamin family protein polymerization involved in membrane fission | 1 | 2 | 3.51e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 2 | 14 | 3.51e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 6 | 8 | 3.51e-01 |
| GO:BP | GO:0090149 | mitochondrial membrane fission | 1 | 2 | 3.51e-01 |
| GO:BP | GO:0035377 | transepithelial water transport | 2 | 2 | 3.51e-01 |
| GO:BP | GO:1904719 | positive regulation of AMPA glutamate receptor clustering | 1 | 2 | 3.51e-01 |
| GO:BP | GO:0003374 | dynamin family protein polymerization involved in mitochondrial fission | 1 | 2 | 3.51e-01 |
| GO:BP | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion | 1 | 5 | 3.51e-01 |
| GO:BP | GO:0042819 | vitamin B6 biosynthetic process | 1 | 3 | 3.51e-01 |
| GO:BP | GO:0072255 | metanephric glomerular mesangial cell development | 1 | 1 | 3.52e-01 |
| GO:BP | GO:1905176 | positive regulation of vascular associated smooth muscle cell dedifferentiation | 1 | 1 | 3.52e-01 |
| GO:BP | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway | 2 | 3 | 3.52e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 3.52e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 3.52e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 83 | 143 | 3.52e-01 |
| GO:BP | GO:1904835 | dorsal root ganglion morphogenesis | 2 | 2 | 3.52e-01 |
| GO:BP | GO:0021649 | vestibulocochlear nerve structural organization | 2 | 2 | 3.52e-01 |
| GO:BP | GO:0007420 | brain development | 11 | 591 | 3.52e-01 |
| GO:BP | GO:0061552 | ganglion morphogenesis | 2 | 2 | 3.52e-01 |
| GO:BP | GO:1903375 | facioacoustic ganglion development | 2 | 2 | 3.52e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 13 | 22 | 3.54e-01 |
| GO:BP | GO:1904748 | regulation of apoptotic process involved in development | 3 | 9 | 3.54e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 6 | 93 | 3.54e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 3.54e-01 |
| GO:BP | GO:1902337 | regulation of apoptotic process involved in morphogenesis | 3 | 9 | 3.54e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 2 | 23 | 3.54e-01 |
| GO:BP | GO:0016233 | telomere capping | 14 | 36 | 3.54e-01 |
| GO:BP | GO:0035825 | homologous recombination | 16 | 43 | 3.54e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 1 | 77 | 3.54e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 2 | 8 | 3.55e-01 |
| GO:BP | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 5 | 3.55e-01 |
| GO:BP | GO:0035329 | hippo signaling | 17 | 41 | 3.55e-01 |
| GO:BP | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 4 | 6 | 3.55e-01 |
| GO:BP | GO:1903126 | negative regulation of centriole-centriole cohesion | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 3 | 4 | 3.56e-01 |
| GO:BP | GO:0018171 | peptidyl-cysteine oxidation | 3 | 3 | 3.56e-01 |
| GO:BP | GO:1904679 | myo-inositol import across plasma membrane | 1 | 1 | 3.56e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 3.57e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 3 | 33 | 3.57e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 3.57e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 15 | 569 | 3.57e-01 |
| GO:BP | GO:2000393 | negative regulation of lamellipodium morphogenesis | 2 | 2 | 3.58e-01 |
| GO:BP | GO:2000744 | positive regulation of anterior head development | 2 | 2 | 3.58e-01 |
| GO:BP | GO:2000742 | regulation of anterior head development | 2 | 2 | 3.58e-01 |
| GO:BP | GO:0016237 | lysosomal microautophagy | 8 | 11 | 3.58e-01 |
| GO:BP | GO:2000386 | positive regulation of ovarian follicle development | 2 | 2 | 3.58e-01 |
| GO:BP | GO:0006950 | response to stress | 6 | 2780 | 3.58e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 3 | 295 | 3.58e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 12 | 32 | 3.58e-01 |
| GO:BP | GO:0019068 | virion assembly | 7 | 35 | 3.58e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 1 | 17 | 3.59e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 1 | 17 | 3.59e-01 |
| GO:BP | GO:0032075 | positive regulation of nuclease activity | 3 | 7 | 3.59e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 2 | 39 | 3.59e-01 |
| GO:BP | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation | 1 | 1 | 3.59e-01 |
| GO:BP | GO:0048666 | neuron development | 3 | 877 | 3.59e-01 |
| GO:BP | GO:0090383 | phagosome acidification | 1 | 5 | 3.59e-01 |
| GO:BP | GO:0036115 | fatty-acyl-CoA catabolic process | 5 | 6 | 3.59e-01 |
| GO:BP | GO:0015939 | pantothenate metabolic process | 2 | 2 | 3.59e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 3.60e-01 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 19 | 64 | 3.60e-01 |
| GO:BP | GO:0062196 | regulation of lysosome size | 5 | 8 | 3.60e-01 |
| GO:BP | GO:0010886 | positive regulation of cholesterol storage | 4 | 5 | 3.60e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 3.60e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 4 | 15 | 3.60e-01 |
| GO:BP | GO:0045204 | MAPK export from nucleus | 3 | 3 | 3.60e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 1 | 111 | 3.60e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 4 | 12 | 3.60e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 30 | 809 | 3.60e-01 |
| GO:BP | GO:0021593 | rhombomere morphogenesis | 1 | 1 | 3.60e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 4 | 94 | 3.60e-01 |
| GO:BP | GO:2001311 | lysobisphosphatidic acid metabolic process | 2 | 2 | 3.60e-01 |
| GO:BP | GO:0014040 | positive regulation of Schwann cell differentiation | 1 | 2 | 3.60e-01 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 1 | 10 | 3.60e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 3.60e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 3 | 12 | 3.60e-01 |
| GO:BP | GO:0014859 | negative regulation of skeletal muscle cell proliferation | 2 | 3 | 3.60e-01 |
| GO:BP | GO:0021658 | rhombomere 3 morphogenesis | 1 | 1 | 3.60e-01 |
| GO:BP | GO:0090130 | tissue migration | 13 | 256 | 3.60e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 2 | 2 | 3.61e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 2 | 13 | 3.61e-01 |
| GO:BP | GO:0003285 | septum secundum development | 3 | 3 | 3.61e-01 |
| GO:BP | GO:0010038 | response to metal ion | 19 | 264 | 3.61e-01 |
| GO:BP | GO:0072001 | renal system development | 12 | 251 | 3.61e-01 |
| GO:BP | GO:0009414 | response to water deprivation | 1 | 4 | 3.61e-01 |
| GO:BP | GO:0014806 | smooth muscle hyperplasia | 2 | 2 | 3.61e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 8 | 133 | 3.61e-01 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 2 | 142 | 3.61e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 1 | 138 | 3.61e-01 |
| GO:BP | GO:0051823 | regulation of synapse structural plasticity | 1 | 5 | 3.61e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 2 | 14 | 3.61e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 4 | 9 | 3.61e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 5 | 58 | 3.61e-01 |
| GO:BP | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway | 2 | 4 | 3.61e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 4 | 24 | 3.61e-01 |
| GO:BP | GO:0042551 | neuron maturation | 6 | 33 | 3.61e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 3.62e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 9 | 90 | 3.62e-01 |
| GO:BP | GO:0032352 | positive regulation of hormone metabolic process | 1 | 11 | 3.62e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 2 | 34 | 3.63e-01 |
| GO:BP | GO:0090102 | cochlea development | 9 | 35 | 3.63e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 4 | 63 | 3.63e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 34 | 167 | 3.63e-01 |
| GO:BP | GO:0070305 | response to cGMP | 1 | 7 | 3.63e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 2 | 742 | 3.63e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 2 | 8 | 3.63e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 12 | 187 | 3.63e-01 |
| GO:BP | GO:1905024 | regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.63e-01 |
| GO:BP | GO:0009213 | pyrimidine deoxyribonucleoside triphosphate catabolic process | 1 | 2 | 3.63e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 3 | 32 | 3.63e-01 |
| GO:BP | GO:0072347 | response to anesthetic | 1 | 1 | 3.63e-01 |
| GO:BP | GO:0046548 | retinal rod cell development | 3 | 6 | 3.63e-01 |
| GO:BP | GO:0042044 | fluid transport | 3 | 17 | 3.64e-01 |
| GO:BP | GO:1902038 | positive regulation of hematopoietic stem cell differentiation | 1 | 1 | 3.64e-01 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 1 | 7 | 3.64e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 7 | 97 | 3.64e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 12 | 14 | 3.64e-01 |
| GO:BP | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 1 | 10 | 3.64e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 3 | 32 | 3.64e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 3 | 46 | 3.64e-01 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 2 | 7 | 3.64e-01 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 407 | 3.64e-01 |
| GO:BP | GO:0072553 | terminal button organization | 1 | 5 | 3.64e-01 |
| GO:BP | GO:0014848 | urinary tract smooth muscle contraction | 2 | 6 | 3.64e-01 |
| GO:BP | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway | 3 | 4 | 3.64e-01 |
| GO:BP | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | 1 | 3 | 3.64e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 5 | 115 | 3.64e-01 |
| GO:BP | GO:1900244 | positive regulation of synaptic vesicle endocytosis | 1 | 4 | 3.64e-01 |
| GO:BP | GO:0070509 | calcium ion import | 4 | 37 | 3.65e-01 |
| GO:BP | GO:0019388 | galactose catabolic process | 2 | 5 | 3.65e-01 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 2 | 9 | 3.65e-01 |
| GO:BP | GO:1990049 | retrograde neuronal dense core vesicle transport | 1 | 4 | 3.65e-01 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 15 | 23 | 3.65e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 1 | 5 | 3.65e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 9 | 36 | 3.65e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 3 | 35 | 3.65e-01 |
| GO:BP | GO:0035865 | cellular response to potassium ion | 2 | 9 | 3.65e-01 |
| GO:BP | GO:0046168 | glycerol-3-phosphate catabolic process | 2 | 2 | 3.65e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 4 | 8 | 3.65e-01 |
| GO:BP | GO:0010424 | DNA methylation on cytosine within a CG sequence | 1 | 1 | 3.65e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 3 | 33 | 3.65e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 12 | 33 | 3.65e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 2 | 3 | 3.65e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 13 | 162 | 3.65e-01 |
| GO:BP | GO:0036072 | direct ossification | 1 | 7 | 3.65e-01 |
| GO:BP | GO:0001957 | intramembranous ossification | 1 | 7 | 3.65e-01 |
| GO:BP | GO:0071467 | cellular response to pH | 2 | 16 | 3.65e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 6 | 13 | 3.65e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 18 | 30 | 3.66e-01 |
| GO:BP | GO:0010818 | T cell chemotaxis | 3 | 12 | 3.66e-01 |
| GO:BP | GO:0061956 | penetration of cumulus oophorus | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0071279 | cellular response to cobalt ion | 1 | 4 | 3.66e-01 |
| GO:BP | GO:0099551 | trans-synaptic signaling by neuropeptide, modulating synaptic transmission | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0006041 | glucosamine metabolic process | 2 | 3 | 3.66e-01 |
| GO:BP | GO:0003193 | pulmonary valve formation | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0098905 | regulation of bundle of His cell action potential | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 2 | 14 | 3.66e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 11 | 66 | 3.66e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 16 | 31 | 3.66e-01 |
| GO:BP | GO:0042311 | vasodilation | 5 | 39 | 3.66e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 3 | 4 | 3.66e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 26 | 1015 | 3.66e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 13 | 81 | 3.66e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 9 | 16 | 3.67e-01 |
| GO:BP | GO:0072739 | response to anisomycin | 1 | 4 | 3.67e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 2 | 27 | 3.67e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 6 | 14 | 3.67e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 6 | 14 | 3.67e-01 |
| GO:BP | GO:1903964 | monounsaturated fatty acid metabolic process | 1 | 2 | 3.67e-01 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 12 | 26 | 3.67e-01 |
| GO:BP | GO:0006646 | phosphatidylethanolamine biosynthetic process | 7 | 8 | 3.67e-01 |
| GO:BP | GO:0061952 | midbody abscission | 5 | 17 | 3.67e-01 |
| GO:BP | GO:0003330 | regulation of extracellular matrix constituent secretion | 5 | 7 | 3.67e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 24 | 227 | 3.67e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 18 | 69 | 3.67e-01 |
| GO:BP | GO:0032070 | regulation of deoxyribonuclease activity | 6 | 9 | 3.67e-01 |
| GO:BP | GO:1900208 | regulation of cardiolipin metabolic process | 3 | 3 | 3.67e-01 |
| GO:BP | GO:0034199 | activation of protein kinase A activity | 2 | 3 | 3.68e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 2 | 6 | 3.68e-01 |
| GO:BP | GO:0007498 | mesoderm development | 2 | 89 | 3.68e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 26 | 1017 | 3.69e-01 |
| GO:BP | GO:0097601 | retina blood vessel maintenance | 1 | 1 | 3.69e-01 |
| GO:BP | GO:0015755 | fructose transmembrane transport | 3 | 5 | 3.69e-01 |
| GO:BP | GO:0140882 | zinc export across plasma membrane | 1 | 3 | 3.69e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 2 | 42 | 3.69e-01 |
| GO:BP | GO:0071579 | regulation of zinc ion transport | 1 | 2 | 3.69e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 2 | 52 | 3.69e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 9 | 23 | 3.69e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 3 | 3 | 3.69e-01 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 5 | 16 | 3.69e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 11 | 172 | 3.70e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 3.70e-01 |
| GO:BP | GO:0035498 | carnosine metabolic process | 1 | 2 | 3.70e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 6 | 10 | 3.70e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 10 | 14 | 3.70e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 3.70e-01 |
| GO:BP | GO:0000706 | meiotic DNA double-strand break processing | 2 | 2 | 3.70e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 2 | 23 | 3.70e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 6 | 11 | 3.70e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 4 | 19 | 3.70e-01 |
| GO:BP | GO:0080163 | regulation of protein serine/threonine phosphatase activity | 2 | 2 | 3.70e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 3.70e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 3.70e-01 |
| GO:BP | GO:0032876 | negative regulation of DNA endoreduplication | 1 | 1 | 3.70e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 3.70e-01 |
| GO:BP | GO:0035928 | rRNA import into mitochondrion | 2 | 3 | 3.70e-01 |
| GO:BP | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 3 | 3.71e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 3.71e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 11 | 194 | 3.71e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 1 | 13 | 3.71e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 3.71e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 3.71e-01 |
| GO:BP | GO:0048645 | animal organ formation | 4 | 51 | 3.71e-01 |
| GO:BP | GO:0034498 | early endosome to Golgi transport | 3 | 9 | 3.71e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0060677 | ureteric bud elongation | 7 | 7 | 3.71e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 3.72e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 3.72e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 16 | 52 | 3.72e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 6 | 72 | 3.72e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 3.72e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 23 | 56 | 3.72e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 4 | 5 | 3.72e-01 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 6 | 9 | 3.72e-01 |
| GO:BP | GO:1900452 | regulation of long-term synaptic depression | 2 | 10 | 3.72e-01 |
| GO:BP | GO:0014824 | artery smooth muscle contraction | 2 | 7 | 3.72e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 1 | 11 | 3.72e-01 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 3.72e-01 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 3.72e-01 |
| GO:BP | GO:0014816 | skeletal muscle satellite cell differentiation | 3 | 6 | 3.72e-01 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 3.72e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 3.72e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 2 | 34 | 3.72e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 2 | 23 | 3.72e-01 |
| GO:BP | GO:0071709 | membrane assembly | 4 | 53 | 3.72e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 4 | 400 | 3.72e-01 |
| GO:BP | GO:0051697 | protein delipidation | 1 | 5 | 3.72e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 17 | 511 | 3.72e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 1 | 19 | 3.72e-01 |
| GO:BP | GO:0009650 | UV protection | 2 | 11 | 3.72e-01 |
| GO:BP | GO:1903621 | protein localization to photoreceptor connecting cilium | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 13 | 228 | 3.72e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 10 | 29 | 3.72e-01 |
| GO:BP | GO:1905373 | ceramide phosphoethanolamine biosynthetic process | 2 | 2 | 3.72e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 3.72e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 3.72e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 3.72e-01 |
| GO:BP | GO:1905371 | ceramide phosphoethanolamine metabolic process | 2 | 2 | 3.72e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 3 | 21 | 3.72e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 47 | 106 | 3.72e-01 |
| GO:BP | GO:0097428 | protein maturation by iron-sulfur cluster transfer | 3 | 17 | 3.72e-01 |
| GO:BP | GO:0009609 | response to symbiotic bacterium | 1 | 2 | 3.72e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 3.72e-01 |
| GO:BP | GO:0061865 | polarized secretion of basement membrane proteins in epithelium | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 3 | 54 | 3.72e-01 |
| GO:BP | GO:0072220 | metanephric descending thin limb development | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 3.72e-01 |
| GO:BP | GO:0072230 | metanephric proximal straight tubule development | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0046688 | response to copper ion | 2 | 29 | 3.72e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 11 | 19 | 3.72e-01 |
| GO:BP | GO:1905070 | anterior visceral endoderm cell migration | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0000493 | box H/ACA snoRNP assembly | 1 | 2 | 3.72e-01 |
| GO:BP | GO:0061864 | basement membrane constituent secretion | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 2 | 18 | 3.72e-01 |
| GO:BP | GO:0051664 | nuclear pore localization | 2 | 4 | 3.72e-01 |
| GO:BP | GO:0072022 | descending thin limb development | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0072032 | proximal convoluted tubule segment 2 development | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0009608 | response to symbiont | 1 | 2 | 3.72e-01 |
| GO:BP | GO:2000691 | negative regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0006021 | inositol biosynthetic process | 1 | 3 | 3.72e-01 |
| GO:BP | GO:0070384 | Harderian gland development | 1 | 4 | 3.72e-01 |
| GO:BP | GO:0072232 | metanephric proximal convoluted tubule segment 2 development | 1 | 1 | 3.72e-01 |
| GO:BP | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0001771 | immunological synapse formation | 1 | 7 | 3.72e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 3.72e-01 |
| GO:BP | GO:0072020 | proximal straight tubule development | 1 | 1 | 3.72e-01 |
| GO:BP | GO:1901660 | calcium ion export | 3 | 3 | 3.72e-01 |
| GO:BP | GO:0034499 | late endosome to Golgi transport | 2 | 5 | 3.73e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 2 | 2 | 3.73e-01 |
| GO:BP | GO:1903753 | negative regulation of p38MAPK cascade | 3 | 5 | 3.73e-01 |
| GO:BP | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 2 | 6 | 3.73e-01 |
| GO:BP | GO:0021892 | cerebral cortex GABAergic interneuron differentiation | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 2 | 33 | 3.73e-01 |
| GO:BP | GO:0006046 | N-acetylglucosamine catabolic process | 3 | 3 | 3.73e-01 |
| GO:BP | GO:0071403 | cellular response to high density lipoprotein particle stimulus | 3 | 3 | 3.73e-01 |
| GO:BP | GO:0038018 | Wnt receptor catabolic process | 2 | 2 | 3.73e-01 |
| GO:BP | GO:0098532 | histone H3-K27 trimethylation | 1 | 4 | 3.73e-01 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 4 | 8 | 3.73e-01 |
| GO:BP | GO:0006434 | seryl-tRNA aminoacylation | 2 | 2 | 3.74e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 5 | 141 | 3.74e-01 |
| GO:BP | GO:0006269 | DNA replication, synthesis of RNA primer | 2 | 7 | 3.74e-01 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 7 | 8 | 3.74e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 3 | 18 | 3.74e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 1 | 59 | 3.74e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 1 | 64 | 3.75e-01 |
| GO:BP | GO:0072239 | metanephric glomerulus vasculature development | 3 | 6 | 3.75e-01 |
| GO:BP | GO:0071578 | zinc ion import across plasma membrane | 2 | 6 | 3.75e-01 |
| GO:BP | GO:2000631 | regulation of pre-miRNA processing | 3 | 3 | 3.75e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 2 | 119 | 3.75e-01 |
| GO:BP | GO:0043041 | amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1 | 3.75e-01 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 3 | 30 | 3.75e-01 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 1 | 4 | 3.75e-01 |
| GO:BP | GO:1990637 | response to prolactin | 2 | 3 | 3.75e-01 |
| GO:BP | GO:1903997 | positive regulation of non-membrane spanning protein tyrosine kinase activity | 2 | 2 | 3.75e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 3.75e-01 |
| GO:BP | GO:0086098 | angiotensin-activated signaling pathway involved in heart process | 2 | 3 | 3.75e-01 |
| GO:BP | GO:0051036 | regulation of endosome size | 7 | 13 | 3.76e-01 |
| GO:BP | GO:1905719 | protein localization to perinuclear region of cytoplasm | 1 | 8 | 3.76e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 19 | 74 | 3.76e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 3 | 11 | 3.76e-01 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 2 | 18 | 3.76e-01 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 2 | 10 | 3.76e-01 |
| GO:BP | GO:0001656 | metanephros development | 2 | 64 | 3.76e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 14 | 92 | 3.76e-01 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 3 | 3 | 3.76e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 3 | 37 | 3.76e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 13 | 144 | 3.77e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 8 | 784 | 3.77e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 1 | 19 | 3.77e-01 |
| GO:BP | GO:0110155 | NAD-cap decapping | 2 | 3 | 3.77e-01 |
| GO:BP | GO:0070495 | negative regulation of thrombin-activated receptor signaling pathway | 3 | 3 | 3.77e-01 |
| GO:BP | GO:0075506 | entry of viral genome into host nucleus through nuclear pore complex via importin | 2 | 2 | 3.77e-01 |
| GO:BP | GO:0055057 | neuroblast division | 4 | 12 | 3.77e-01 |
| GO:BP | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 5 | 5 | 3.77e-01 |
| GO:BP | GO:0070494 | regulation of thrombin-activated receptor signaling pathway | 3 | 3 | 3.77e-01 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 3 | 9 | 3.77e-01 |
| GO:BP | GO:2000969 | positive regulation of AMPA receptor activity | 1 | 4 | 3.77e-01 |
| GO:BP | GO:0060763 | mammary duct terminal end bud growth | 1 | 3 | 3.77e-01 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 1 | 144 | 3.77e-01 |
| GO:BP | GO:1905667 | negative regulation of protein localization to endosome | 1 | 1 | 3.77e-01 |
| GO:BP | GO:0015823 | phenylalanine transport | 2 | 4 | 3.77e-01 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 12 | 24 | 3.78e-01 |
| GO:BP | GO:0048102 | autophagic cell death | 1 | 6 | 3.78e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 7 | 140 | 3.78e-01 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 1 | 11 | 3.79e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 5 | 42 | 3.79e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 154 | 3.79e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 1 | 30 | 3.79e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 3.79e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 18 | 494 | 3.79e-01 |
| GO:BP | GO:0060948 | cardiac vascular smooth muscle cell development | 3 | 3 | 3.79e-01 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 11 | 16 | 3.79e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 11 | 206 | 3.79e-01 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 1 | 3 | 3.79e-01 |
| GO:BP | GO:0106217 | tRNA C3-cytosine methylation | 1 | 2 | 3.80e-01 |
| GO:BP | GO:0050859 | negative regulation of B cell receptor signaling pathway | 1 | 3 | 3.80e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 3.80e-01 |
| GO:BP | GO:1901321 | positive regulation of heart induction | 2 | 4 | 3.81e-01 |
| GO:BP | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | 2 | 4 | 3.81e-01 |
| GO:BP | GO:0010621 | negative regulation of transcription by transcription factor localization | 1 | 3 | 3.81e-01 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 3 | 6 | 3.81e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 2 | 25 | 3.81e-01 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 3 | 46 | 3.81e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 20 | 49 | 3.81e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 17 | 48 | 3.81e-01 |
| GO:BP | GO:1904762 | positive regulation of myofibroblast differentiation | 1 | 1 | 3.81e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 14 | 53 | 3.81e-01 |
| GO:BP | GO:0010738 | regulation of protein kinase A signaling | 1 | 13 | 3.81e-01 |
| GO:BP | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 5 | 5 | 3.81e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 15 | 70 | 3.82e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 9 | 23 | 3.82e-01 |
| GO:BP | GO:0014004 | microglia differentiation | 1 | 6 | 3.82e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 4 | 6 | 3.82e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 1 | 93 | 3.82e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 3.82e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 3.82e-01 |
| GO:BP | GO:0006248 | CMP catabolic process | 1 | 3 | 3.82e-01 |
| GO:BP | GO:0046050 | UMP catabolic process | 1 | 3 | 3.82e-01 |
| GO:BP | GO:0009888 | tissue development | 4 | 1425 | 3.82e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 2 | 663 | 3.82e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 11 | 207 | 3.83e-01 |
| GO:BP | GO:0061743 | motor learning | 1 | 5 | 3.83e-01 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 1 | 8 | 3.83e-01 |
| GO:BP | GO:0060068 | vagina development | 3 | 8 | 3.83e-01 |
| GO:BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 4 | 5 | 3.83e-01 |
| GO:BP | GO:0097494 | regulation of vesicle size | 11 | 19 | 3.83e-01 |
| GO:BP | GO:1904782 | negative regulation of NMDA glutamate receptor activity | 2 | 2 | 3.84e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 228 | 3.84e-01 |
| GO:BP | GO:0000712 | resolution of meiotic recombination intermediates | 1 | 13 | 3.84e-01 |
| GO:BP | GO:1903932 | regulation of DNA primase activity | 4 | 5 | 3.84e-01 |
| GO:BP | GO:0051767 | nitric-oxide synthase biosynthetic process | 1 | 13 | 3.84e-01 |
| GO:BP | GO:1903934 | positive regulation of DNA primase activity | 4 | 5 | 3.84e-01 |
| GO:BP | GO:1903772 | regulation of viral budding via host ESCRT complex | 2 | 4 | 3.84e-01 |
| GO:BP | GO:0051769 | regulation of nitric-oxide synthase biosynthetic process | 1 | 13 | 3.84e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 1 | 17 | 3.84e-01 |
| GO:BP | GO:0006595 | polyamine metabolic process | 2 | 16 | 3.84e-01 |
| GO:BP | GO:0009615 | response to virus | 2 | 275 | 3.84e-01 |
| GO:BP | GO:1901403 | positive regulation of tetrapyrrole metabolic process | 3 | 4 | 3.84e-01 |
| GO:BP | GO:0006722 | triterpenoid metabolic process | 2 | 2 | 3.84e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 3 | 120 | 3.84e-01 |
| GO:BP | GO:0061002 | negative regulation of dendritic spine morphogenesis | 1 | 6 | 3.84e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 4 | 47 | 3.84e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 4 | 54 | 3.84e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 151 | 3.84e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 2 | 10 | 3.84e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 2 | 15 | 3.85e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 7 | 73 | 3.85e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 2 | 15 | 3.85e-01 |
| GO:BP | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | 1 | 3 | 3.85e-01 |
| GO:BP | GO:0060686 | negative regulation of prostatic bud formation | 1 | 4 | 3.85e-01 |
| GO:BP | GO:1904322 | cellular response to forskolin | 4 | 11 | 3.85e-01 |
| GO:BP | GO:1904321 | response to forskolin | 4 | 11 | 3.85e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 9 | 12 | 3.85e-01 |
| GO:BP | GO:1904293 | negative regulation of ERAD pathway | 1 | 6 | 3.85e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 17 | 22 | 3.85e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 1 | 27 | 3.85e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 11 | 31 | 3.85e-01 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 8 | 29 | 3.85e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 5 | 21 | 3.85e-01 |
| GO:BP | GO:0061586 | positive regulation of transcription by transcription factor localization | 1 | 1 | 3.85e-01 |
| GO:BP | GO:0099156 | cell-cell signaling via exosome | 1 | 1 | 3.85e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 5 | 21 | 3.85e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 3 | 6 | 3.85e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 4 | 45 | 3.85e-01 |
| GO:BP | GO:0009194 | pyrimidine ribonucleoside diphosphate biosynthetic process | 2 | 4 | 3.85e-01 |
| GO:BP | GO:0006225 | UDP biosynthetic process | 2 | 4 | 3.85e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 8 | 18 | 3.86e-01 |
| GO:BP | GO:0060383 | positive regulation of DNA strand elongation | 1 | 1 | 3.86e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 7 | 140 | 3.86e-01 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 4 | 14 | 3.86e-01 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 6 | 21 | 3.86e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 2 | 28 | 3.86e-01 |
| GO:BP | GO:1902178 | fibroblast growth factor receptor apoptotic signaling pathway | 1 | 1 | 3.86e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 19 | 133 | 3.86e-01 |
| GO:BP | GO:0150037 | regulation of calcium-dependent activation of synaptic vesicle fusion | 1 | 2 | 3.87e-01 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 6 | 7 | 3.87e-01 |
| GO:BP | GO:0140039 | cell-cell adhesion in response to extracellular stimulus | 1 | 2 | 3.87e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 1 | 121 | 3.87e-01 |
| GO:BP | GO:0048311 | mitochondrion distribution | 1 | 14 | 3.87e-01 |
| GO:BP | GO:0061668 | mitochondrial ribosome assembly | 4 | 8 | 3.88e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 2 | 53 | 3.88e-01 |
| GO:BP | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing | 5 | 7 | 3.88e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 3 | 47 | 3.88e-01 |
| GO:BP | GO:0060047 | heart contraction | 19 | 205 | 3.88e-01 |
| GO:BP | GO:0000212 | meiotic spindle organization | 7 | 14 | 3.88e-01 |
| GO:BP | GO:0021637 | trigeminal nerve structural organization | 3 | 4 | 3.88e-01 |
| GO:BP | GO:0050923 | regulation of negative chemotaxis | 3 | 4 | 3.88e-01 |
| GO:BP | GO:0035640 | exploration behavior | 3 | 21 | 3.88e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 33 | 901 | 3.88e-01 |
| GO:BP | GO:0046886 | positive regulation of hormone biosynthetic process | 4 | 9 | 3.88e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 2 | 53 | 3.88e-01 |
| GO:BP | GO:0090346 | cellular organofluorine metabolic process | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 14 | 21 | 3.88e-01 |
| GO:BP | GO:0090345 | cellular organohalogen metabolic process | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 2 | 30 | 3.88e-01 |
| GO:BP | GO:0016926 | protein desumoylation | 6 | 9 | 3.88e-01 |
| GO:BP | GO:0045110 | intermediate filament bundle assembly | 5 | 6 | 3.88e-01 |
| GO:BP | GO:0021636 | trigeminal nerve morphogenesis | 3 | 4 | 3.88e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 8 | 123 | 3.88e-01 |
| GO:BP | GO:2000504 | positive regulation of blood vessel remodeling | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway | 1 | 2 | 3.88e-01 |
| GO:BP | GO:0099163 | synaptic signaling by nitric oxide | 1 | 3 | 3.88e-01 |
| GO:BP | GO:0048865 | stem cell fate commitment | 2 | 5 | 3.88e-01 |
| GO:BP | GO:0060243 | negative regulation of cell growth involved in contact inhibition | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 3.88e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 131 | 218 | 3.88e-01 |
| GO:BP | GO:2001016 | positive regulation of skeletal muscle cell differentiation | 2 | 5 | 3.88e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 47 | 234 | 3.88e-01 |
| GO:BP | GO:0000098 | sulfur amino acid catabolic process | 2 | 7 | 3.88e-01 |
| GO:BP | GO:0046967 | cytosol to endoplasmic reticulum transport | 2 | 4 | 3.89e-01 |
| GO:BP | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1 | 17 | 3.89e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 3.89e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 2 | 34 | 3.89e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 7 | 9 | 3.89e-01 |
| GO:BP | GO:0006884 | cell volume homeostasis | 2 | 24 | 3.89e-01 |
| GO:BP | GO:0060916 | mesenchymal cell proliferation involved in lung development | 3 | 5 | 3.89e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 3 | 555 | 3.89e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 8 | 165 | 3.90e-01 |
| GO:BP | GO:0000707 | meiotic DNA recombinase assembly | 1 | 1 | 3.90e-01 |
| GO:BP | GO:0007066 | female meiosis sister chromatid cohesion | 1 | 1 | 3.90e-01 |
| GO:BP | GO:0000050 | urea cycle | 3 | 7 | 3.90e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 13 | 147 | 3.90e-01 |
| GO:BP | GO:0010447 | response to acidic pH | 2 | 16 | 3.91e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 2 | 4 | 3.91e-01 |
| GO:BP | GO:0000451 | rRNA 2’-O-methylation | 3 | 3 | 3.91e-01 |
| GO:BP | GO:1990564 | protein polyufmylation | 3 | 5 | 3.91e-01 |
| GO:BP | GO:1990592 | protein K69-linked ufmylation | 3 | 5 | 3.91e-01 |
| GO:BP | GO:1901341 | positive regulation of store-operated calcium channel activity | 2 | 6 | 3.91e-01 |
| GO:BP | GO:2000346 | negative regulation of hepatocyte proliferation | 1 | 3 | 3.92e-01 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 1 | 10 | 3.92e-01 |
| GO:BP | GO:2001205 | negative regulation of osteoclast development | 1 | 2 | 3.92e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 3 | 36 | 3.92e-01 |
| GO:BP | GO:0090520 | sphingolipid mediated signaling pathway | 3 | 11 | 3.92e-01 |
| GO:BP | GO:0033198 | response to ATP | 3 | 24 | 3.92e-01 |
| GO:BP | GO:0021915 | neural tube development | 1 | 146 | 3.92e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 52 | 173 | 3.92e-01 |
| GO:BP | GO:0038133 | ERBB2-ERBB3 signaling pathway | 3 | 3 | 3.92e-01 |
| GO:BP | GO:0048672 | positive regulation of collateral sprouting | 4 | 10 | 3.92e-01 |
| GO:BP | GO:0061369 | negative regulation of testicular blood vessel morphogenesis | 1 | 1 | 3.92e-01 |
| GO:BP | GO:2001014 | regulation of skeletal muscle cell differentiation | 4 | 16 | 3.92e-01 |
| GO:BP | GO:0048792 | spontaneous exocytosis of neurotransmitter | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 1 | 198 | 3.92e-01 |
| GO:BP | GO:0042940 | D-amino acid transport | 2 | 5 | 3.92e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 6 | 10 | 3.92e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 12 | 229 | 3.92e-01 |
| GO:BP | GO:0031948 | positive regulation of glucocorticoid biosynthetic process | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0031945 | positive regulation of glucocorticoid metabolic process | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 11 | 16 | 3.92e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 15 | 45 | 3.92e-01 |
| GO:BP | GO:2000019 | negative regulation of male gonad development | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0044751 | cellular response to human chorionic gonadotropin stimulus | 1 | 2 | 3.92e-01 |
| GO:BP | GO:2000066 | positive regulation of cortisol biosynthetic process | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0034514 | mitochondrial unfolded protein response | 2 | 2 | 3.92e-01 |
| GO:BP | GO:2000180 | negative regulation of androgen biosynthetic process | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 14 | 36 | 3.92e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 4 | 17 | 3.92e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 4 | 106 | 3.92e-01 |
| GO:BP | GO:0044154 | histone H3-K14 acetylation | 3 | 12 | 3.92e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 1 | 18 | 3.92e-01 |
| GO:BP | GO:0006656 | phosphatidylcholine biosynthetic process | 12 | 25 | 3.92e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 3 | 5 | 3.92e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 7 | 67 | 3.92e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 28 | 1119 | 3.92e-01 |
| GO:BP | GO:0007382 | specification of segmental identity, maxillary segment | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 8 | 11 | 3.92e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 38 | 3.92e-01 |
| GO:BP | GO:1905871 | regulation of protein localization to cell leading edge | 1 | 2 | 3.92e-01 |
| GO:BP | GO:0050955 | thermoception | 1 | 3 | 3.92e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 4 | 18 | 3.92e-01 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 1 | 10 | 3.92e-01 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 3 | 4 | 3.93e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 3 | 28 | 3.93e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 5 | 14 | 3.93e-01 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 3 | 4 | 3.93e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 19 | 28 | 3.93e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 10 | 16 | 3.93e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 4 | 10 | 3.93e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 13 | 246 | 3.93e-01 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 3 | 4 | 3.93e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 1 | 127 | 3.93e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 8 | 71 | 3.93e-01 |
| GO:BP | GO:0016189 | synaptic vesicle to endosome fusion | 1 | 3 | 3.93e-01 |
| GO:BP | GO:0033212 | iron import into cell | 3 | 7 | 3.93e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 3 | 121 | 3.93e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 3 | 21 | 3.93e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 1 | 17 | 3.93e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 3 | 26 | 3.93e-01 |
| GO:BP | GO:0006741 | NADP biosynthetic process | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0031915 | positive regulation of synaptic plasticity | 5 | 6 | 3.93e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 11 | 47 | 3.93e-01 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 2 | 4 | 3.93e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 3 | 6 | 3.93e-01 |
| GO:BP | GO:0046049 | UMP metabolic process | 2 | 10 | 3.93e-01 |
| GO:BP | GO:1903996 | negative regulation of non-membrane spanning protein tyrosine kinase activity | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 1 | 17 | 3.93e-01 |
| GO:BP | GO:0071839 | apoptotic process in bone marrow cell | 4 | 4 | 3.93e-01 |
| GO:BP | GO:0070102 | interleukin-6-mediated signaling pathway | 10 | 14 | 3.93e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 18 | 27 | 3.93e-01 |
| GO:BP | GO:1990048 | anterograde neuronal dense core vesicle transport | 1 | 4 | 3.93e-01 |
| GO:BP | GO:0090117 | endosome to lysosome transport of low-density lipoprotein particle | 1 | 2 | 3.93e-01 |
| GO:BP | GO:0003129 | heart induction | 4 | 9 | 3.93e-01 |
| GO:BP | GO:0042816 | vitamin B6 metabolic process | 4 | 5 | 3.93e-01 |
| GO:BP | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process | 1 | 7 | 3.93e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 1 | 7 | 3.93e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 1 | 179 | 3.93e-01 |
| GO:BP | GO:0014820 | tonic smooth muscle contraction | 2 | 8 | 3.93e-01 |
| GO:BP | GO:0019290 | siderophore biosynthetic process | 1 | 1 | 3.93e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 1 | 33 | 3.93e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 3 | 25 | 3.93e-01 |
| GO:BP | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process | 3 | 3 | 3.93e-01 |
| GO:BP | GO:0009237 | siderophore metabolic process | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0006003 | fructose 2,6-bisphosphate metabolic process | 1 | 4 | 3.93e-01 |
| GO:BP | GO:1903644 | regulation of chaperone-mediated protein folding | 3 | 3 | 3.93e-01 |
| GO:BP | GO:0034755 | iron ion transmembrane transport | 7 | 16 | 3.93e-01 |
| GO:BP | GO:1904394 | negative regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0070593 | dendrite self-avoidance | 1 | 11 | 3.93e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 8 | 167 | 3.94e-01 |
| GO:BP | GO:1903434 | negative regulation of constitutive secretory pathway | 1 | 1 | 3.94e-01 |
| GO:BP | GO:1903936 | cellular response to sodium arsenite | 1 | 5 | 3.95e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 4 | 16 | 3.95e-01 |
| GO:BP | GO:0035519 | protein K29-linked ubiquitination | 5 | 6 | 3.95e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 2 | 6 | 3.95e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 1 | 17 | 3.95e-01 |
| GO:BP | GO:0140212 | regulation of long-chain fatty acid import into cell | 3 | 7 | 3.95e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 4 | 23 | 3.95e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 3 | 48 | 3.95e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 3 | 48 | 3.95e-01 |
| GO:BP | GO:0009597 | detection of virus | 4 | 5 | 3.95e-01 |
| GO:BP | GO:0090168 | Golgi reassembly | 4 | 5 | 3.95e-01 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 1 | 11 | 3.96e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 2 | 58 | 3.96e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 2 | 2 | 3.96e-01 |
| GO:BP | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis | 2 | 3 | 3.96e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 10 | 15 | 3.96e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 3 | 18 | 3.96e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 7 | 22 | 3.96e-01 |
| GO:BP | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development | 1 | 3 | 3.97e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 3 | 14 | 3.97e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 46 | 174 | 3.97e-01 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 3 | 13 | 3.97e-01 |
| GO:BP | GO:0016078 | tRNA catabolic process | 3 | 14 | 3.97e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 9 | 43 | 3.97e-01 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 9 | 19 | 3.98e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 1 | 316 | 3.98e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 6 | 8 | 3.98e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 3 | 114 | 3.98e-01 |
| GO:BP | GO:0008215 | spermine metabolic process | 3 | 5 | 3.98e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 5 | 24 | 3.98e-01 |
| GO:BP | GO:1903402 | regulation of renal phosphate excretion | 1 | 2 | 3.98e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 6 | 14 | 3.98e-01 |
| GO:BP | GO:0044722 | renal phosphate excretion | 1 | 2 | 3.98e-01 |
| GO:BP | GO:0006914 | autophagy | 19 | 513 | 3.98e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 3.98e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 19 | 513 | 3.98e-01 |
| GO:BP | GO:0032416 | negative regulation of sodium:proton antiporter activity | 1 | 1 | 3.98e-01 |
| GO:BP | GO:0060929 | atrioventricular node cell fate commitment | 2 | 2 | 3.98e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 2 | 665 | 3.98e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 3 | 36 | 3.98e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 3 | 30 | 3.98e-01 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 6 | 14 | 3.98e-01 |
| GO:BP | GO:0043038 | amino acid activation | 23 | 44 | 3.98e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 2 | 28 | 3.98e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 3 | 36 | 3.99e-01 |
| GO:BP | GO:0016358 | dendrite development | 13 | 202 | 3.99e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 18 | 103 | 3.99e-01 |
| GO:BP | GO:0046753 | non-lytic viral release | 3 | 14 | 3.99e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 2 | 57 | 3.99e-01 |
| GO:BP | GO:0048855 | adenohypophysis morphogenesis | 1 | 1 | 3.99e-01 |
| GO:BP | GO:1904381 | Golgi apparatus mannose trimming | 3 | 3 | 3.99e-01 |
| GO:BP | GO:0034729 | histone H3-K79 methylation | 2 | 2 | 3.99e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 10 | 13 | 4.00e-01 |
| GO:BP | GO:0003064 | regulation of heart rate by hormone | 1 | 2 | 4.00e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 2 | 38 | 4.00e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 8 | 81 | 4.00e-01 |
| GO:BP | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 3 | 4.00e-01 |
| GO:BP | GO:0060348 | bone development | 1 | 170 | 4.00e-01 |
| GO:BP | GO:0098930 | axonal transport | 6 | 60 | 4.00e-01 |
| GO:BP | GO:0061833 | protein localization to tricellular tight junction | 1 | 2 | 4.01e-01 |
| GO:BP | GO:0033024 | mast cell apoptotic process | 3 | 3 | 4.01e-01 |
| GO:BP | GO:0033025 | regulation of mast cell apoptotic process | 3 | 3 | 4.01e-01 |
| GO:BP | GO:0048290 | isotype switching to IgA isotypes | 1 | 4 | 4.01e-01 |
| GO:BP | GO:0036335 | intestinal stem cell homeostasis | 1 | 3 | 4.01e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 4.01e-01 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 4 | 4.01e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 4.01e-01 |
| GO:BP | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 4.01e-01 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 5 | 8 | 4.01e-01 |
| GO:BP | GO:1903423 | positive regulation of synaptic vesicle recycling | 1 | 6 | 4.01e-01 |
| GO:BP | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 1 | 2 | 4.01e-01 |
| GO:BP | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 1 | 2 | 4.01e-01 |
| GO:BP | GO:2000041 | negative regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 4.01e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 1 | 16 | 4.01e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 19 | 49 | 4.01e-01 |
| GO:BP | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification | 1 | 2 | 4.01e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 1 | 210 | 4.01e-01 |
| GO:BP | GO:0044343 | canonical Wnt signaling pathway involved in regulation of type B pancreatic cell proliferation | 1 | 1 | 4.01e-01 |
| GO:BP | GO:2000079 | regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 4.01e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 3 | 47 | 4.01e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 3 | 21 | 4.01e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 3 | 34 | 4.01e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 5 | 21 | 4.01e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 5 | 56 | 4.02e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 4.02e-01 |
| GO:BP | GO:0060065 | uterus development | 3 | 13 | 4.02e-01 |
| GO:BP | GO:0055011 | atrial cardiac muscle cell differentiation | 3 | 3 | 4.02e-01 |
| GO:BP | GO:0110051 | metabolite repair | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0055014 | atrial cardiac muscle cell development | 3 | 3 | 4.02e-01 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 4 | 6 | 4.02e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 7 | 82 | 4.02e-01 |
| GO:BP | GO:0070318 | positive regulation of G0 to G1 transition | 1 | 5 | 4.02e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 7 | 11 | 4.03e-01 |
| GO:BP | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 2 | 2 | 4.03e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 5 | 112 | 4.03e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 10 | 17 | 4.03e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 13 | 36 | 4.03e-01 |
| GO:BP | GO:0003032 | detection of oxygen | 2 | 5 | 4.03e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 31 | 130 | 4.03e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 3 | 20 | 4.04e-01 |
| GO:BP | GO:1900222 | negative regulation of amyloid-beta clearance | 6 | 7 | 4.04e-01 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 3 | 282 | 4.04e-01 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0046684 | response to pyrethroid | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 4 | 58 | 4.04e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 2 | 6 | 4.04e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 4 | 69 | 4.04e-01 |
| GO:BP | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development | 5 | 13 | 4.04e-01 |
| GO:BP | GO:0045618 | positive regulation of keratinocyte differentiation | 10 | 12 | 4.04e-01 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 44 | 1199 | 4.04e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 3 | 24 | 4.04e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 9 | 371 | 4.04e-01 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 2 | 7 | 4.04e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 9 | 39 | 4.04e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 4.04e-01 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 4.04e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 7 | 100 | 4.04e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 4 | 28 | 4.04e-01 |
| GO:BP | GO:2000397 | positive regulation of ubiquitin-dependent endocytosis | 2 | 2 | 4.05e-01 |
| GO:BP | GO:0072014 | proximal tubule development | 1 | 7 | 4.05e-01 |
| GO:BP | GO:0032364 | intracellular oxygen homeostasis | 1 | 6 | 4.05e-01 |
| GO:BP | GO:1903975 | regulation of glial cell migration | 2 | 12 | 4.05e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 6 | 10 | 4.05e-01 |
| GO:BP | GO:2000395 | regulation of ubiquitin-dependent endocytosis | 2 | 2 | 4.05e-01 |
| GO:BP | GO:0009992 | intracellular water homeostasis | 1 | 3 | 4.05e-01 |
| GO:BP | GO:0006417 | regulation of translation | 9 | 370 | 4.05e-01 |
| GO:BP | GO:2000620 | positive regulation of histone H4-K16 acetylation | 2 | 2 | 4.05e-01 |
| GO:BP | GO:0032203 | telomere formation via telomerase | 1 | 1 | 4.05e-01 |
| GO:BP | GO:0034337 | RNA folding | 1 | 1 | 4.05e-01 |
| GO:BP | GO:0048245 | eosinophil chemotaxis | 1 | 5 | 4.05e-01 |
| GO:BP | GO:0060754 | positive regulation of mast cell chemotaxis | 1 | 5 | 4.05e-01 |
| GO:BP | GO:1901626 | regulation of postsynaptic membrane organization | 1 | 2 | 4.05e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 20 | 35 | 4.05e-01 |
| GO:BP | GO:0009175 | pyrimidine ribonucleoside monophosphate catabolic process | 1 | 4 | 4.05e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 37 | 65 | 4.05e-01 |
| GO:BP | GO:0046079 | dUMP catabolic process | 1 | 4 | 4.05e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 6 | 76 | 4.05e-01 |
| GO:BP | GO:0019878 | lysine biosynthetic process via aminoadipic acid | 2 | 2 | 4.05e-01 |
| GO:BP | GO:0051601 | exocyst localization | 4 | 4 | 4.05e-01 |
| GO:BP | GO:0032262 | pyrimidine nucleotide salvage | 1 | 4 | 4.05e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 4 | 145 | 4.05e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 20 | 35 | 4.05e-01 |
| GO:BP | GO:0010138 | pyrimidine ribonucleotide salvage | 1 | 4 | 4.05e-01 |
| GO:BP | GO:0140495 | migracytosis | 1 | 3 | 4.05e-01 |
| GO:BP | GO:0046063 | dCMP metabolic process | 1 | 3 | 4.05e-01 |
| GO:BP | GO:1900247 | regulation of cytoplasmic translational elongation | 7 | 10 | 4.05e-01 |
| GO:BP | GO:0160040 | mitocytosis | 1 | 3 | 4.05e-01 |
| GO:BP | GO:0006249 | dCMP catabolic process | 1 | 3 | 4.05e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 27 | 58 | 4.05e-01 |
| GO:BP | GO:0044206 | UMP salvage | 1 | 4 | 4.05e-01 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 4 | 32 | 4.05e-01 |
| GO:BP | GO:0009085 | lysine biosynthetic process | 2 | 2 | 4.05e-01 |
| GO:BP | GO:0003307 | regulation of Wnt signaling pathway involved in heart development | 3 | 7 | 4.05e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 1 | 17 | 4.05e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 4.05e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 10 | 30 | 4.05e-01 |
| GO:BP | GO:2000493 | negative regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 4.07e-01 |
| GO:BP | GO:0051642 | centrosome localization | 17 | 31 | 4.07e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 17 | 31 | 4.07e-01 |
| GO:BP | GO:0019441 | tryptophan catabolic process to kynurenine | 1 | 1 | 4.07e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 22 | 34 | 4.07e-01 |
| GO:BP | GO:0019240 | citrulline biosynthetic process | 3 | 4 | 4.07e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 6 | 67 | 4.07e-01 |
| GO:BP | GO:0048769 | sarcomerogenesis | 3 | 3 | 4.07e-01 |
| GO:BP | GO:0009052 | pentose-phosphate shunt, non-oxidative branch | 4 | 6 | 4.07e-01 |
| GO:BP | GO:1902389 | ceramide 1-phosphate transport | 2 | 7 | 4.08e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 1 | 107 | 4.08e-01 |
| GO:BP | GO:2000822 | regulation of behavioral fear response | 1 | 7 | 4.08e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 9 | 24 | 4.08e-01 |
| GO:BP | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway | 2 | 4 | 4.08e-01 |
| GO:BP | GO:0043628 | regulatory ncRNA 3’-end processing | 12 | 19 | 4.08e-01 |
| GO:BP | GO:0060073 | micturition | 1 | 2 | 4.08e-01 |
| GO:BP | GO:0043697 | cell dedifferentiation | 2 | 6 | 4.08e-01 |
| GO:BP | GO:0043696 | dedifferentiation | 2 | 6 | 4.08e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 16 | 4955 | 4.08e-01 |
| GO:BP | GO:0062232 | prostanoid catabolic process | 1 | 2 | 4.08e-01 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 4 | 31 | 4.08e-01 |
| GO:BP | GO:1905344 | prostaglandin catabolic process | 1 | 2 | 4.08e-01 |
| GO:BP | GO:0006203 | dGTP catabolic process | 2 | 2 | 4.08e-01 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 3 | 3 | 4.08e-01 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 1 | 3 | 4.08e-01 |
| GO:BP | GO:1904425 | negative regulation of GTP binding | 3 | 3 | 4.08e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 156 | 4.09e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 10 | 58 | 4.09e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 11 | 64 | 4.09e-01 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 4.09e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 3 | 27 | 4.09e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 6 | 16 | 4.09e-01 |
| GO:BP | GO:0045578 | negative regulation of B cell differentiation | 3 | 6 | 4.10e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 11 | 27 | 4.10e-01 |
| GO:BP | GO:0072414 | response to mitotic cell cycle checkpoint signaling | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 4.10e-01 |
| GO:BP | GO:0046511 | sphinganine biosynthetic process | 2 | 2 | 4.10e-01 |
| GO:BP | GO:0072432 | response to G1 DNA damage checkpoint signaling | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 17 | 26 | 4.10e-01 |
| GO:BP | GO:0003057 | regulation of the force of heart contraction by chemical signal | 3 | 5 | 4.10e-01 |
| GO:BP | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis | 2 | 3 | 4.10e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 2 | 529 | 4.11e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 24 | 120 | 4.11e-01 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 1 | 10 | 4.11e-01 |
| GO:BP | GO:0031441 | negative regulation of mRNA 3’-end processing | 1 | 2 | 4.11e-01 |
| GO:BP | GO:0048308 | organelle inheritance | 10 | 15 | 4.11e-01 |
| GO:BP | GO:0048313 | Golgi inheritance | 10 | 15 | 4.11e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 30 | 164 | 4.11e-01 |
| GO:BP | GO:0008216 | spermidine metabolic process | 4 | 7 | 4.11e-01 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 3 | 11 | 4.11e-01 |
| GO:BP | GO:1904219 | positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 4.12e-01 |
| GO:BP | GO:1904222 | positive regulation of serine C-palmitoyltransferase activity | 1 | 2 | 4.12e-01 |
| GO:BP | GO:0006657 | CDP-choline pathway | 4 | 6 | 4.12e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 1 | 10 | 4.12e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 13 | 26 | 4.12e-01 |
| GO:BP | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 4.12e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 4.12e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 4 | 49 | 4.12e-01 |
| GO:BP | GO:0032915 | positive regulation of transforming growth factor beta2 production | 2 | 3 | 4.12e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 4.12e-01 |
| GO:BP | GO:1905065 | positive regulation of vascular associated smooth muscle cell differentiation | 3 | 6 | 4.12e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 4 | 16 | 4.12e-01 |
| GO:BP | GO:0035385 | Roundabout signaling pathway | 3 | 5 | 4.12e-01 |
| GO:BP | GO:1904482 | cellular response to tetrahydrofolate | 1 | 1 | 4.12e-01 |
| GO:BP | GO:1904481 | response to tetrahydrofolate | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0036018 | cellular response to erythropoietin | 3 | 4 | 4.12e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 3 | 25 | 4.12e-01 |
| GO:BP | GO:0030842 | regulation of intermediate filament depolymerization | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0030844 | positive regulation of intermediate filament depolymerization | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0045106 | intermediate filament depolymerization | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 6 | 679 | 4.12e-01 |
| GO:BP | GO:0045108 | regulation of intermediate filament polymerization or depolymerization | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0099627 | neurotransmitter receptor cycle | 1 | 1 | 4.12e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 5 | 267 | 4.12e-01 |
| GO:BP | GO:0099630 | postsynaptic neurotransmitter receptor cycle | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0046898 | response to cycloheximide | 4 | 4 | 4.12e-01 |
| GO:BP | GO:0042640 | anagen | 1 | 5 | 4.13e-01 |
| GO:BP | GO:2000322 | regulation of glucocorticoid receptor signaling pathway | 2 | 7 | 4.13e-01 |
| GO:BP | GO:0003167 | atrioventricular bundle cell differentiation | 3 | 3 | 4.13e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 1 | 30 | 4.13e-01 |
| GO:BP | GO:0014038 | regulation of Schwann cell differentiation | 1 | 3 | 4.13e-01 |
| GO:BP | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1 | 4 | 4.13e-01 |
| GO:BP | GO:0060744 | mammary gland branching involved in thelarche | 1 | 5 | 4.13e-01 |
| GO:BP | GO:0042695 | thelarche | 1 | 5 | 4.13e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 1 | 216 | 4.13e-01 |
| GO:BP | GO:0051107 | negative regulation of DNA ligation | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 50 | 1219 | 4.13e-01 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 1 | 8 | 4.13e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 5 | 41 | 4.13e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 3 | 20 | 4.13e-01 |
| GO:BP | GO:1901163 | regulation of trophoblast cell migration | 2 | 12 | 4.13e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 3 | 7 | 4.13e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 4.13e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 7 | 90 | 4.13e-01 |
| GO:BP | GO:0060751 | branch elongation involved in mammary gland duct branching | 1 | 4 | 4.13e-01 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 1 | 5 | 4.13e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 7 | 111 | 4.13e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 2 | 19 | 4.13e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 4.13e-01 |
| GO:BP | GO:0034182 | regulation of maintenance of mitotic sister chromatid cohesion | 1 | 8 | 4.13e-01 |
| GO:BP | GO:1904876 | negative regulation of DNA ligase activity | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 3 | 7 | 4.13e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 4.13e-01 |
| GO:BP | GO:1903645 | negative regulation of chaperone-mediated protein folding | 2 | 2 | 4.13e-01 |
| GO:BP | GO:0034091 | regulation of maintenance of sister chromatid cohesion | 1 | 8 | 4.13e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 4.13e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 3 | 3 | 4.13e-01 |
| GO:BP | GO:0009826 | unidimensional cell growth | 2 | 3 | 4.13e-01 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 4 | 15 | 4.13e-01 |
| GO:BP | GO:0006391 | transcription initiation at mitochondrial promoter | 3 | 4 | 4.13e-01 |
| GO:BP | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 5 | 7 | 4.13e-01 |
| GO:BP | GO:0042942 | D-serine transport | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 4.13e-01 |
| GO:BP | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 2 | 4 | 4.13e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 4 | 9 | 4.13e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 4.13e-01 |
| GO:BP | GO:0051884 | regulation of timing of anagen | 1 | 5 | 4.13e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 4.13e-01 |
| GO:BP | GO:0097325 | melanocyte proliferation | 1 | 2 | 4.13e-01 |
| GO:BP | GO:0035863 | dITP catabolic process | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 4 | 9 | 4.13e-01 |
| GO:BP | GO:0035847 | uterine epithelium development | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0097379 | dorsal spinal cord interneuron posterior axon guidance | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0035862 | dITP metabolic process | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0035849 | nephric duct elongation | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0009186 | deoxyribonucleoside diphosphate metabolic process | 3 | 4 | 4.13e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 1 | 4 | 4.13e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 2 | 15 | 4.13e-01 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 2 | 12 | 4.13e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 2 | 21 | 4.13e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 1 | 18 | 4.13e-01 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 9 | 18 | 4.14e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 1 | 20 | 4.14e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 8 | 13 | 4.14e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 4.14e-01 |
| GO:BP | GO:0060134 | prepulse inhibition | 1 | 7 | 4.14e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 10 | 31 | 4.14e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 4.14e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 2 | 541 | 4.14e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 3 | 51 | 4.14e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 3 | 5 | 4.15e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 2 | 9 | 4.16e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 4 | 44 | 4.16e-01 |
| GO:BP | GO:0044375 | regulation of peroxisome size | 3 | 3 | 4.16e-01 |
| GO:BP | GO:0038138 | ERBB4-ERBB4 signaling pathway | 3 | 3 | 4.17e-01 |
| GO:BP | GO:0015675 | nickel cation transport | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0035444 | nickel cation transmembrane transport | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0015676 | vanadium ion transport | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 2 | 29 | 4.17e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 2 | 25 | 4.17e-01 |
| GO:BP | GO:0017055 | negative regulation of RNA polymerase II transcription preinitiation complex assembly | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0060035 | notochord cell development | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0030954 | astral microtubule nucleation | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 7 | 28 | 4.17e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 2 | 755 | 4.17e-01 |
| GO:BP | GO:1905936 | regulation of germ cell proliferation | 3 | 5 | 4.17e-01 |
| GO:BP | GO:1904636 | response to ionomycin | 1 | 3 | 4.17e-01 |
| GO:BP | GO:1904637 | cellular response to ionomycin | 1 | 3 | 4.17e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 2 | 16 | 4.17e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 3 | 53 | 4.17e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 36 | 179 | 4.17e-01 |
| GO:BP | GO:0006825 | copper ion transport | 1 | 13 | 4.17e-01 |
| GO:BP | GO:1903978 | regulation of microglial cell activation | 1 | 9 | 4.17e-01 |
| GO:BP | GO:0010967 | regulation of polyamine biosynthetic process | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 4.17e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 4.17e-01 |
| GO:BP | GO:1905474 | canonical Wnt signaling pathway involved in stem cell proliferation | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0051415 | microtubule nucleation by interphase microtubule organizing center | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0060064 | Spemann organizer formation at the anterior end of the primitive streak | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0072223 | metanephric glomerular mesangium development | 2 | 3 | 4.17e-01 |
| GO:BP | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 1 | 4 | 4.17e-01 |
| GO:BP | GO:0051548 | negative regulation of keratinocyte migration | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0032423 | regulation of mismatch repair | 3 | 3 | 4.17e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 33 | 678 | 4.17e-01 |
| GO:BP | GO:0023041 | neuronal signal transduction | 1 | 6 | 4.17e-01 |
| GO:BP | GO:0019075 | virus maturation | 1 | 2 | 4.17e-01 |
| GO:BP | GO:0001759 | organ induction | 2 | 17 | 4.17e-01 |
| GO:BP | GO:0045105 | intermediate filament polymerization or depolymerization | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0015692 | lead ion transport | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0060034 | notochord cell differentiation | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0071318 | cellular response to ATP | 2 | 12 | 4.18e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 40 | 300 | 4.18e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 1 | 30 | 4.18e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 4.18e-01 |
| GO:BP | GO:0097104 | postsynaptic membrane assembly | 2 | 8 | 4.18e-01 |
| GO:BP | GO:0030208 | dermatan sulfate biosynthetic process | 2 | 5 | 4.18e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 5 | 87 | 4.18e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 9 | 14 | 4.18e-01 |
| GO:BP | GO:1902897 | regulation of postsynaptic density protein 95 clustering | 1 | 2 | 4.19e-01 |
| GO:BP | GO:0009648 | photoperiodism | 15 | 28 | 4.19e-01 |
| GO:BP | GO:0010808 | positive regulation of synaptic vesicle priming | 1 | 1 | 4.19e-01 |
| GO:BP | GO:1903301 | positive regulation of hexokinase activity | 1 | 5 | 4.19e-01 |
| GO:BP | GO:0043456 | regulation of pentose-phosphate shunt | 1 | 6 | 4.19e-01 |
| GO:BP | GO:0009099 | valine biosynthetic process | 2 | 3 | 4.19e-01 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 3 | 17 | 4.19e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 4 | 12 | 4.20e-01 |
| GO:BP | GO:0098904 | regulation of AV node cell action potential | 1 | 3 | 4.20e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 28 | 265 | 4.20e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 11 | 401 | 4.20e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 2 | 35 | 4.20e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 61 | 4.20e-01 |
| GO:BP | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | 3 | 4 | 4.20e-01 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 1 | 3 | 4.20e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 1 | 13 | 4.20e-01 |
| GO:BP | GO:0014856 | skeletal muscle cell proliferation | 4 | 14 | 4.20e-01 |
| GO:BP | GO:0140374 | antiviral innate immune response | 13 | 17 | 4.20e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 1 | 258 | 4.20e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 4.20e-01 |
| GO:BP | GO:1903935 | response to sodium arsenite | 1 | 6 | 4.20e-01 |
| GO:BP | GO:0006500 | N-terminal protein palmitoylation | 2 | 3 | 4.20e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 61 | 4.20e-01 |
| GO:BP | GO:0010610 | regulation of mRNA stability involved in response to stress | 3 | 3 | 4.20e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 2 | 26 | 4.20e-01 |
| GO:BP | GO:0009097 | isoleucine biosynthetic process | 1 | 2 | 4.20e-01 |
| GO:BP | GO:1901585 | regulation of acid-sensing ion channel activity | 1 | 1 | 4.20e-01 |
| GO:BP | GO:1904181 | positive regulation of membrane depolarization | 3 | 8 | 4.20e-01 |
| GO:BP | GO:1903365 | regulation of fear response | 1 | 8 | 4.20e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 10 | 42 | 4.20e-01 |
| GO:BP | GO:0021855 | hypothalamus cell migration | 5 | 6 | 4.20e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 3 | 62 | 4.20e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 1 | 16 | 4.21e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 8 | 131 | 4.21e-01 |
| GO:BP | GO:0003014 | renal system process | 10 | 78 | 4.21e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 5 | 10 | 4.21e-01 |
| GO:BP | GO:0008052 | sensory organ boundary specification | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0021698 | cerebellar cortex structural organization | 1 | 2 | 4.21e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 4.21e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 3 | 14 | 4.21e-01 |
| GO:BP | GO:0021693 | cerebellar Purkinje cell layer structural organization | 1 | 2 | 4.21e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 3 | 433 | 4.21e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 23 | 4.21e-01 |
| GO:BP | GO:0061193 | taste bud development | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 4.21e-01 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 18 | 35 | 4.21e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 60 | 4.21e-01 |
| GO:BP | GO:0010650 | positive regulation of cell communication by electrical coupling | 2 | 3 | 4.21e-01 |
| GO:BP | GO:0033979 | box H/ACA RNA metabolic process | 2 | 3 | 4.21e-01 |
| GO:BP | GO:0010160 | formation of animal organ boundary | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 1 | 31 | 4.21e-01 |
| GO:BP | GO:0031120 | snRNA pseudouridine synthesis | 2 | 3 | 4.21e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 5 | 10 | 4.21e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 12 | 59 | 4.21e-01 |
| GO:BP | GO:0140469 | GCN2-mediated signaling | 3 | 3 | 4.21e-01 |
| GO:BP | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells | 1 | 4 | 4.21e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 2 | 16 | 4.21e-01 |
| GO:BP | GO:1904044 | response to aldosterone | 3 | 9 | 4.21e-01 |
| GO:BP | GO:0010917 | negative regulation of mitochondrial membrane potential | 1 | 9 | 4.21e-01 |
| GO:BP | GO:0045837 | negative regulation of membrane potential | 1 | 9 | 4.21e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 2 | 16 | 4.21e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 4 | 96 | 4.21e-01 |
| GO:BP | GO:0006591 | ornithine metabolic process | 1 | 6 | 4.21e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 13 | 35 | 4.21e-01 |
| GO:BP | GO:0007494 | midgut development | 5 | 7 | 4.21e-01 |
| GO:BP | GO:0097421 | liver regeneration | 5 | 23 | 4.22e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 4.22e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 7 | 110 | 4.22e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 3 | 39 | 4.22e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 3 | 8 | 4.22e-01 |
| GO:BP | GO:0061448 | connective tissue development | 1 | 208 | 4.22e-01 |
| GO:BP | GO:0030397 | membrane disassembly | 1 | 11 | 4.22e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 27 | 45 | 4.22e-01 |
| GO:BP | GO:0006829 | zinc ion transport | 6 | 23 | 4.22e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 2 | 8 | 4.22e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 1 | 130 | 4.22e-01 |
| GO:BP | GO:0001906 | cell killing | 3 | 69 | 4.22e-01 |
| GO:BP | GO:0033026 | negative regulation of mast cell apoptotic process | 1 | 1 | 4.22e-01 |
| GO:BP | GO:1903800 | positive regulation of miRNA processing | 3 | 3 | 4.22e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 5 | 8 | 4.22e-01 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 3 | 6 | 4.22e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 8 | 19 | 4.22e-01 |
| GO:BP | GO:0095500 | acetylcholine receptor signaling pathway | 1 | 18 | 4.22e-01 |
| GO:BP | GO:0072190 | ureter urothelium development | 3 | 3 | 4.22e-01 |
| GO:BP | GO:0002545 | chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.22e-01 |
| GO:BP | GO:0002880 | regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.22e-01 |
| GO:BP | GO:0002882 | positive regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.22e-01 |
| GO:BP | GO:0101030 | tRNA-guanine transglycosylation | 2 | 2 | 4.22e-01 |
| GO:BP | GO:1902068 | regulation of sphingolipid mediated signaling pathway | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0048630 | skeletal muscle tissue growth | 1 | 8 | 4.23e-01 |
| GO:BP | GO:0016050 | vesicle organization | 9 | 320 | 4.23e-01 |
| GO:BP | GO:0062009 | secondary palate development | 6 | 20 | 4.23e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 2 | 8 | 4.23e-01 |
| GO:BP | GO:1904438 | regulation of iron ion import across plasma membrane | 1 | 1 | 4.24e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 2 | 23 | 4.24e-01 |
| GO:BP | GO:0015854 | guanine transport | 1 | 2 | 4.24e-01 |
| GO:BP | GO:0044313 | protein K6-linked deubiquitination | 2 | 3 | 4.24e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 12 | 448 | 4.24e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 3 | 36 | 4.24e-01 |
| GO:BP | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway | 1 | 6 | 4.24e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 2 | 4 | 4.24e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 2 | 4 | 4.24e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 2 | 4 | 4.24e-01 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 1 | 11 | 4.24e-01 |
| GO:BP | GO:0021873 | forebrain neuroblast division | 2 | 3 | 4.24e-01 |
| GO:BP | GO:0035344 | hypoxanthine transport | 1 | 2 | 4.24e-01 |
| GO:BP | GO:0001951 | intestinal D-glucose absorption | 2 | 3 | 4.24e-01 |
| GO:BP | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway | 2 | 6 | 4.24e-01 |
| GO:BP | GO:0033327 | Leydig cell differentiation | 6 | 9 | 4.24e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 15 | 191 | 4.24e-01 |
| GO:BP | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0048641 | regulation of skeletal muscle tissue development | 1 | 20 | 4.25e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 2 | 41 | 4.25e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 2 | 50 | 4.25e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 9 | 29 | 4.25e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 14 | 272 | 4.25e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 10 | 4.25e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 3 | 70 | 4.25e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 5 | 6 | 4.25e-01 |
| GO:BP | GO:0015707 | nitrite transport | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 3 | 8 | 4.26e-01 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 1 | 8 | 4.26e-01 |
| GO:BP | GO:1902685 | positive regulation of receptor localization to synapse | 2 | 3 | 4.26e-01 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 4.26e-01 |
| GO:BP | GO:0061734 | parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 4 | 6 | 4.26e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 4.26e-01 |
| GO:BP | GO:0055071 | manganese ion homeostasis | 1 | 2 | 4.26e-01 |
| GO:BP | GO:0072303 | positive regulation of glomerular metanephric mesangial cell proliferation | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0009173 | pyrimidine ribonucleoside monophosphate metabolic process | 2 | 12 | 4.26e-01 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0018215 | protein phosphopantetheinylation | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 53 | 122 | 4.26e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 5 | 154 | 4.26e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 3 | 1061 | 4.26e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 4.26e-01 |
| GO:BP | GO:0008611 | ether lipid biosynthetic process | 3 | 10 | 4.26e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 4 | 25 | 4.26e-01 |
| GO:BP | GO:0009730 | detection of carbohydrate stimulus | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0008655 | pyrimidine-containing compound salvage | 1 | 5 | 4.26e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 1 | 36 | 4.26e-01 |
| GO:BP | GO:0009732 | detection of hexose stimulus | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0009749 | response to glucose | 12 | 137 | 4.26e-01 |
| GO:BP | GO:0046504 | glycerol ether biosynthetic process | 3 | 10 | 4.26e-01 |
| GO:BP | GO:0034287 | detection of monosaccharide stimulus | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0046204 | nor-spermidine metabolic process | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 5 | 35 | 4.26e-01 |
| GO:BP | GO:1905337 | positive regulation of aggrephagy | 2 | 3 | 4.26e-01 |
| GO:BP | GO:0032919 | spermine acetylation | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0032920 | putrescine acetylation | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 3 | 10 | 4.26e-01 |
| GO:BP | GO:0097384 | cellular lipid biosynthetic process | 3 | 10 | 4.26e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 4 | 71 | 4.26e-01 |
| GO:BP | GO:0051594 | detection of glucose | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1905335 | regulation of aggrephagy | 2 | 3 | 4.26e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 3 | 3 | 4.26e-01 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 3 | 13 | 4.26e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 32 | 96 | 4.26e-01 |
| GO:BP | GO:0007271 | synaptic transmission, cholinergic | 1 | 15 | 4.26e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 4.26e-01 |
| GO:BP | GO:0040009 | regulation of growth rate | 1 | 3 | 4.26e-01 |
| GO:BP | GO:0006171 | cAMP biosynthetic process | 3 | 8 | 4.26e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 39 | 151 | 4.26e-01 |
| GO:BP | GO:0044528 | regulation of mitochondrial mRNA stability | 5 | 7 | 4.26e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 2 | 11 | 4.26e-01 |
| GO:BP | GO:0002317 | plasma cell differentiation | 1 | 5 | 4.26e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 7 | 203 | 4.26e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 4 | 28 | 4.26e-01 |
| GO:BP | GO:0061364 | apoptotic process involved in luteolysis | 2 | 3 | 4.27e-01 |
| GO:BP | GO:0046697 | decidualization | 6 | 19 | 4.27e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 9 | 72 | 4.27e-01 |
| GO:BP | GO:0035290 | trunk segmentation | 3 | 4 | 4.27e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 2 | 7 | 4.27e-01 |
| GO:BP | GO:0036484 | trunk neural crest cell migration | 3 | 4 | 4.27e-01 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 1 | 8 | 4.27e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 22 | 48 | 4.27e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 3 | 14 | 4.27e-01 |
| GO:BP | GO:1905839 | negative regulation of telomeric D-loop disassembly | 1 | 2 | 4.27e-01 |
| GO:BP | GO:1905838 | regulation of telomeric D-loop disassembly | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 6 | 10 | 4.27e-01 |
| GO:BP | GO:1903949 | positive regulation of atrial cardiac muscle cell action potential | 1 | 1 | 4.27e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 1 | 63 | 4.27e-01 |
| GO:BP | GO:0061739 | protein lipidation involved in autophagosome assembly | 2 | 3 | 4.27e-01 |
| GO:BP | GO:1905778 | negative regulation of exonuclease activity | 1 | 2 | 4.27e-01 |
| GO:BP | GO:1904199 | positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.27e-01 |
| GO:BP | GO:1905777 | regulation of exonuclease activity | 1 | 2 | 4.27e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 23 | 386 | 4.27e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0009449 | gamma-aminobutyric acid biosynthetic process | 1 | 4 | 4.27e-01 |
| GO:BP | GO:0032214 | negative regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0032213 | regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 3 | 631 | 4.27e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 24 | 236 | 4.27e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 2 | 21 | 4.27e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 4.27e-01 |
| GO:BP | GO:1904534 | negative regulation of telomeric loop disassembly | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 4.27e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 1 | 74 | 4.27e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0061450 | trophoblast cell migration | 2 | 13 | 4.27e-01 |
| GO:BP | GO:0048671 | negative regulation of collateral sprouting | 5 | 9 | 4.27e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 49 | 84 | 4.27e-01 |
| GO:BP | GO:1902261 | positive regulation of delayed rectifier potassium channel activity | 1 | 5 | 4.27e-01 |
| GO:BP | GO:1990736 | regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.27e-01 |
| GO:BP | GO:1903764 | regulation of potassium ion export across plasma membrane | 3 | 5 | 4.27e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 6 | 40 | 4.27e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 6 | 83 | 4.28e-01 |
| GO:BP | GO:0072677 | eosinophil migration | 1 | 7 | 4.28e-01 |
| GO:BP | GO:0097151 | positive regulation of inhibitory postsynaptic potential | 3 | 4 | 4.29e-01 |
| GO:BP | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity | 1 | 6 | 4.29e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 7 | 9 | 4.29e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 4.29e-01 |
| GO:BP | GO:0060324 | face development | 3 | 45 | 4.29e-01 |
| GO:BP | GO:0003271 | smoothened signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 5 | 18 | 4.29e-01 |
| GO:BP | GO:0120077 | angiogenic sprout fusion | 2 | 2 | 4.29e-01 |
| GO:BP | GO:0120078 | cell adhesion involved in sprouting angiogenesis | 2 | 2 | 4.29e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 1 | 26 | 4.29e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 4.29e-01 |
| GO:BP | GO:1903348 | positive regulation of bicellular tight junction assembly | 4 | 5 | 4.29e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 4.29e-01 |
| GO:BP | GO:0097105 | presynaptic membrane assembly | 1 | 8 | 4.29e-01 |
| GO:BP | GO:0018277 | protein deamination | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 16 | 58 | 4.31e-01 |
| GO:BP | GO:1905145 | cellular response to acetylcholine | 1 | 20 | 4.31e-01 |
| GO:BP | GO:0071243 | cellular response to arsenic-containing substance | 9 | 18 | 4.31e-01 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 6 | 11 | 4.31e-01 |
| GO:BP | GO:0120041 | positive regulation of macrophage proliferation | 1 | 4 | 4.31e-01 |
| GO:BP | GO:0014740 | negative regulation of muscle hyperplasia | 1 | 1 | 4.31e-01 |
| GO:BP | GO:0032463 | negative regulation of protein homooligomerization | 1 | 2 | 4.31e-01 |
| GO:BP | GO:0042482 | positive regulation of odontogenesis | 1 | 5 | 4.31e-01 |
| GO:BP | GO:1903969 | regulation of response to macrophage colony-stimulating factor | 1 | 3 | 4.31e-01 |
| GO:BP | GO:0032460 | negative regulation of protein oligomerization | 1 | 2 | 4.31e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 6 | 86 | 4.31e-01 |
| GO:BP | GO:0009056 | catabolic process | 19 | 2088 | 4.31e-01 |
| GO:BP | GO:1903972 | regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 3 | 4.31e-01 |
| GO:BP | GO:0097154 | GABAergic neuron differentiation | 1 | 5 | 4.31e-01 |
| GO:BP | GO:0021678 | third ventricle development | 3 | 4 | 4.31e-01 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 1 | 6 | 4.31e-01 |
| GO:BP | GO:0060442 | branching involved in prostate gland morphogenesis | 2 | 7 | 4.31e-01 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 1 | 23 | 4.31e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 4.31e-01 |
| GO:BP | GO:0015818 | isoleucine transport | 1 | 5 | 4.31e-01 |
| GO:BP | GO:1904555 | L-proline transmembrane transport | 1 | 5 | 4.31e-01 |
| GO:BP | GO:1903804 | glycine import across plasma membrane | 1 | 4 | 4.31e-01 |
| GO:BP | GO:1905647 | proline import across plasma membrane | 1 | 5 | 4.31e-01 |
| GO:BP | GO:1904271 | L-proline import across plasma membrane | 1 | 5 | 4.31e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 6 | 85 | 4.32e-01 |
| GO:BP | GO:0042732 | D-xylose metabolic process | 1 | 2 | 4.32e-01 |
| GO:BP | GO:0090618 | DNA clamp unloading | 1 | 2 | 4.32e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 11 | 408 | 4.32e-01 |
| GO:BP | GO:0090216 | positive regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 1 | 4.32e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 4.32e-01 |
| GO:BP | GO:1902983 | DNA strand elongation involved in mitotic DNA replication | 1 | 1 | 4.32e-01 |
| GO:BP | GO:1902296 | DNA strand elongation involved in cell cycle DNA replication | 1 | 1 | 4.32e-01 |
| GO:BP | GO:0060419 | heart growth | 42 | 72 | 4.32e-01 |
| GO:BP | GO:1902319 | DNA strand elongation involved in nuclear cell cycle DNA replication | 1 | 1 | 4.32e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 6 | 14 | 4.32e-01 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 2 | 12 | 4.32e-01 |
| GO:BP | GO:0070778 | L-aspartate transmembrane transport | 3 | 11 | 4.32e-01 |
| GO:BP | GO:0007423 | sensory organ development | 1 | 396 | 4.32e-01 |
| GO:BP | GO:0014904 | myotube cell development | 1 | 34 | 4.32e-01 |
| GO:BP | GO:0090188 | negative regulation of pancreatic juice secretion | 1 | 4 | 4.32e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 7 | 16 | 4.33e-01 |
| GO:BP | GO:0070383 | DNA cytosine deamination | 1 | 5 | 4.33e-01 |
| GO:BP | GO:0001830 | trophectodermal cell fate commitment | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 2 | 32 | 4.33e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 4.33e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 7 | 10 | 4.33e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 5 | 60 | 4.33e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 7 | 10 | 4.34e-01 |
| GO:BP | GO:0001660 | fever generation | 1 | 5 | 4.34e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 2 | 14 | 4.34e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 3 | 55 | 4.35e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 3 | 55 | 4.35e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 1 | 19 | 4.35e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 1 | 19 | 4.35e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 1 | 27 | 4.35e-01 |
| GO:BP | GO:0060789 | hair follicle placode formation | 3 | 5 | 4.35e-01 |
| GO:BP | GO:0042415 | norepinephrine metabolic process | 2 | 7 | 4.35e-01 |
| GO:BP | GO:0003015 | heart process | 19 | 216 | 4.35e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 12 | 38 | 4.35e-01 |
| GO:BP | GO:0006883 | intracellular sodium ion homeostasis | 3 | 13 | 4.35e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 3 | 1123 | 4.35e-01 |
| GO:BP | GO:0035295 | tube development | 2 | 821 | 4.35e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 4.36e-01 |
| GO:BP | GO:1901329 | regulation of odontoblast differentiation | 1 | 2 | 4.36e-01 |
| GO:BP | GO:1905303 | positive regulation of macropinocytosis | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 4 | 10 | 4.36e-01 |
| GO:BP | GO:0035864 | response to potassium ion | 3 | 12 | 4.36e-01 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 8 | 12 | 4.36e-01 |
| GO:BP | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion | 4 | 7 | 4.36e-01 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 4.36e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 15 | 26 | 4.36e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 4.36e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 2 | 21 | 4.36e-01 |
| GO:BP | GO:0014829 | vascular associated smooth muscle contraction | 3 | 20 | 4.36e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 21 | 108 | 4.36e-01 |
| GO:BP | GO:0042592 | homeostatic process | 26 | 1159 | 4.36e-01 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 16 | 26 | 4.36e-01 |
| GO:BP | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0051455 | monopolar spindle attachment to meiosis I kinetochore | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0120094 | negative regulation of peptidyl-lysine crotonylation | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0120093 | regulation of peptidyl-lysine crotonylation | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 3 | 42 | 4.36e-01 |
| GO:BP | GO:0071787 | endoplasmic reticulum tubular network formation | 3 | 4 | 4.36e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 27 | 119 | 4.36e-01 |
| GO:BP | GO:0150093 | amyloid-beta clearance by transcytosis | 4 | 7 | 4.36e-01 |
| GO:BP | GO:0045054 | constitutive secretory pathway | 3 | 7 | 4.36e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 15 | 26 | 4.36e-01 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 6 | 14 | 4.36e-01 |
| GO:BP | GO:1901463 | regulation of tetrapyrrole biosynthetic process | 3 | 7 | 4.36e-01 |
| GO:BP | GO:0044873 | lipoprotein localization to membrane | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0006096 | glycolytic process | 1 | 70 | 4.36e-01 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 8 | 17 | 4.36e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 1 | 23 | 4.36e-01 |
| GO:BP | GO:1904868 | telomerase catalytic core complex assembly | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0014028 | notochord formation | 2 | 3 | 4.36e-01 |
| GO:BP | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0060685 | regulation of prostatic bud formation | 1 | 5 | 4.36e-01 |
| GO:BP | GO:1903062 | regulation of reverse cholesterol transport | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0042413 | carnitine catabolic process | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 11 | 20 | 4.36e-01 |
| GO:BP | GO:0001907 | killing by symbiont of host cells | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0071317 | cellular response to isoquinoline alkaloid | 1 | 2 | 4.36e-01 |
| GO:BP | GO:1904884 | positive regulation of telomerase catalytic core complex assembly | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0071315 | cellular response to morphine | 1 | 2 | 4.36e-01 |
| GO:BP | GO:1904882 | regulation of telomerase catalytic core complex assembly | 2 | 2 | 4.36e-01 |
| GO:BP | GO:1903064 | positive regulation of reverse cholesterol transport | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 1 | 19 | 4.36e-01 |
| GO:BP | GO:0090381 | regulation of heart induction | 1 | 4 | 4.36e-01 |
| GO:BP | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation | 4 | 6 | 4.36e-01 |
| GO:BP | GO:0090022 | regulation of neutrophil chemotaxis | 3 | 19 | 4.36e-01 |
| GO:BP | GO:0016197 | endosomal transport | 6 | 231 | 4.36e-01 |
| GO:BP | GO:0043588 | skin development | 1 | 175 | 4.36e-01 |
| GO:BP | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade | 1 | 4 | 4.36e-01 |
| GO:BP | GO:0051666 | actin cortical patch localization | 1 | 2 | 4.36e-01 |
| GO:BP | GO:0033668 | suppression by symbiont of host apoptotic process | 2 | 2 | 4.36e-01 |
| GO:BP | GO:1990983 | tRNA demethylation | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0060613 | fat pad development | 1 | 6 | 4.36e-01 |
| GO:BP | GO:0044565 | dendritic cell proliferation | 2 | 3 | 4.36e-01 |
| GO:BP | GO:0052041 | suppression by symbiont of host programmed cell death | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0052042 | induction by symbiont of host programmed cell death | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0052151 | positive regulation by symbiont of host apoptotic process | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 1 | 2 | 4.36e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 19 | 36 | 4.36e-01 |
| GO:BP | GO:0120222 | regulation of blastocyst development | 2 | 3 | 4.36e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 50 | 1226 | 4.36e-01 |
| GO:BP | GO:1902474 | positive regulation of protein localization to synapse | 1 | 6 | 4.36e-01 |
| GO:BP | GO:0070453 | regulation of heme biosynthetic process | 3 | 7 | 4.36e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 29 | 1209 | 4.36e-01 |
| GO:BP | GO:0019051 | induction by virus of host apoptotic process | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0019050 | suppression by virus of host apoptotic process | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0100012 | regulation of heart induction by canonical Wnt signaling pathway | 1 | 4 | 4.36e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 8 | 15 | 4.36e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 2 | 52 | 4.36e-01 |
| GO:BP | GO:0031128 | developmental induction | 15 | 26 | 4.36e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 2 | 3 | 4.36e-01 |
| GO:BP | GO:1905395 | response to flavonoid | 2 | 2 | 4.37e-01 |
| GO:BP | GO:0046606 | negative regulation of centrosome cycle | 4 | 12 | 4.37e-01 |
| GO:BP | GO:0010826 | negative regulation of centrosome duplication | 4 | 12 | 4.37e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 4.37e-01 |
| GO:BP | GO:0010232 | vascular transport | 1 | 72 | 4.37e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 1 | 72 | 4.37e-01 |
| GO:BP | GO:0062043 | positive regulation of cardiac epithelial to mesenchymal transition | 2 | 9 | 4.37e-01 |
| GO:BP | GO:1900248 | negative regulation of cytoplasmic translational elongation | 2 | 2 | 4.37e-01 |
| GO:BP | GO:1902954 | regulation of early endosome to recycling endosome transport | 1 | 2 | 4.37e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 43 | 1111 | 4.37e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 4 | 8 | 4.37e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 22 | 43 | 4.37e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 7 | 14 | 4.37e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 3 | 1107 | 4.37e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 3 | 5 | 4.37e-01 |
| GO:BP | GO:2000754 | regulation of sphingomyelin catabolic process | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 1 | 9 | 4.38e-01 |
| GO:BP | GO:2000755 | positive regulation of sphingomyelin catabolic process | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 11 | 222 | 4.38e-01 |
| GO:BP | GO:0110113 | positive regulation of lipid transporter activity | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0051591 | response to cAMP | 13 | 66 | 4.38e-01 |
| GO:BP | GO:0033504 | floor plate development | 4 | 8 | 4.38e-01 |
| GO:BP | GO:0098902 | regulation of membrane depolarization during action potential | 4 | 5 | 4.38e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 4.38e-01 |
| GO:BP | GO:0033306 | phytol metabolic process | 2 | 2 | 4.38e-01 |
| GO:BP | GO:1903173 | fatty alcohol metabolic process | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 7 | 96 | 4.38e-01 |
| GO:BP | GO:0061764 | late endosome to lysosome transport via multivesicular body sorting pathway | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 2 | 15 | 4.38e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 5 | 4.38e-01 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 1 | 4 | 4.38e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 7 | 24 | 4.39e-01 |
| GO:BP | GO:0098760 | response to interleukin-7 | 3 | 12 | 4.39e-01 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 3 | 12 | 4.39e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 4.39e-01 |
| GO:BP | GO:2001176 | regulation of mediator complex assembly | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0001928 | regulation of exocyst assembly | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0001927 | exocyst assembly | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0060178 | regulation of exocyst localization | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0006451 | translational readthrough | 2 | 10 | 4.39e-01 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 6 | 25 | 4.39e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 2 | 10 | 4.39e-01 |
| GO:BP | GO:0036034 | mediator complex assembly | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 3 | 9 | 4.39e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 2 | 12 | 4.39e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 7 | 10 | 4.39e-01 |
| GO:BP | GO:2001178 | positive regulation of mediator complex assembly | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 1 | 2 | 4.39e-01 |
| GO:BP | GO:0044854 | plasma membrane raft assembly | 1 | 6 | 4.39e-01 |
| GO:BP | GO:0070221 | sulfide oxidation, using sulfide:quinone oxidoreductase | 1 | 3 | 4.39e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 7 | 108 | 4.39e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 2 | 4 | 4.39e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 32 | 105 | 4.39e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0009190 | cyclic nucleotide biosynthetic process | 6 | 14 | 4.39e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 2 | 8 | 4.39e-01 |
| GO:BP | GO:0019418 | sulfide oxidation | 1 | 3 | 4.39e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 4.40e-01 |
| GO:BP | GO:0008033 | tRNA processing | 73 | 128 | 4.40e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 74 | 4.40e-01 |
| GO:BP | GO:0033197 | response to vitamin E | 2 | 9 | 4.40e-01 |
| GO:BP | GO:0140454 | protein aggregate center assembly | 1 | 1 | 4.40e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 5 | 7 | 4.40e-01 |
| GO:BP | GO:0097070 | ductus arteriosus closure | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 9 | 54 | 4.40e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 1 | 107 | 4.40e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 74 | 4.40e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 9 | 390 | 4.40e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 7 | 44 | 4.40e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 2 | 8 | 4.40e-01 |
| GO:BP | GO:0002023 | reduction of food intake in response to dietary excess | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 1 | 9 | 4.41e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6 | 7 | 4.41e-01 |
| GO:BP | GO:0043048 | dolichyl monophosphate biosynthetic process | 1 | 1 | 4.41e-01 |
| GO:BP | GO:1900141 | regulation of oligodendrocyte apoptotic process | 2 | 2 | 4.41e-01 |
| GO:BP | GO:0071455 | cellular response to hyperoxia | 1 | 5 | 4.41e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 4.41e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 2 | 8 | 4.41e-01 |
| GO:BP | GO:0021854 | hypothalamus development | 1 | 15 | 4.41e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 3 | 16 | 4.41e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 12 | 46 | 4.41e-01 |
| GO:BP | GO:0007231 | osmosensory signaling pathway | 3 | 3 | 4.41e-01 |
| GO:BP | GO:0034196 | acylglycerol transport | 1 | 4 | 4.41e-01 |
| GO:BP | GO:1905144 | response to acetylcholine | 1 | 22 | 4.41e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 12 | 46 | 4.41e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 11 | 39 | 4.41e-01 |
| GO:BP | GO:0034197 | triglyceride transport | 1 | 4 | 4.41e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 2 | 19 | 4.41e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1081 | 4.41e-01 |
| GO:BP | GO:0060753 | regulation of mast cell chemotaxis | 1 | 7 | 4.41e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 8 | 13 | 4.41e-01 |
| GO:BP | GO:1901723 | negative regulation of cell proliferation involved in kidney development | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0035627 | ceramide transport | 3 | 15 | 4.41e-01 |
| GO:BP | GO:0062111 | zinc ion import into organelle | 2 | 7 | 4.41e-01 |
| GO:BP | GO:0048699 | generation of neurons | 3 | 1119 | 4.41e-01 |
| GO:BP | GO:0034330 | cell junction organization | 2 | 575 | 4.41e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 4.41e-01 |
| GO:BP | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade | 2 | 5 | 4.41e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 4 | 27 | 4.41e-01 |
| GO:BP | GO:0060745 | mammary gland branching involved in pregnancy | 2 | 5 | 4.41e-01 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 19 | 4.41e-01 |
| GO:BP | GO:0097090 | presynaptic membrane organization | 1 | 8 | 4.41e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 46 | 106 | 4.41e-01 |
| GO:BP | GO:1900063 | regulation of peroxisome organization | 1 | 4 | 4.41e-01 |
| GO:BP | GO:1904048 | regulation of spontaneous neurotransmitter secretion | 2 | 2 | 4.42e-01 |
| GO:BP | GO:0072209 | metanephric mesangial cell differentiation | 1 | 1 | 4.42e-01 |
| GO:BP | GO:0046078 | dUMP metabolic process | 1 | 6 | 4.42e-01 |
| GO:BP | GO:0072254 | metanephric glomerular mesangial cell differentiation | 1 | 1 | 4.42e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 4.42e-01 |
| GO:BP | GO:0046035 | CMP metabolic process | 1 | 6 | 4.42e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 4.42e-01 |
| GO:BP | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 4.42e-01 |
| GO:BP | GO:0009178 | pyrimidine deoxyribonucleoside monophosphate catabolic process | 1 | 5 | 4.42e-01 |
| GO:BP | GO:0002894 | positive regulation of type II hypersensitivity | 2 | 3 | 4.42e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 3 | 1173 | 4.42e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 4.42e-01 |
| GO:BP | GO:0060708 | spongiotrophoblast differentiation | 2 | 2 | 4.42e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 4 | 53 | 4.42e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 4.42e-01 |
| GO:BP | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 6 | 4.42e-01 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 2 | 5 | 4.42e-01 |
| GO:BP | GO:0001798 | positive regulation of type IIa hypersensitivity | 2 | 3 | 4.42e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 44 | 1088 | 4.42e-01 |
| GO:BP | GO:0022406 | membrane docking | 21 | 84 | 4.42e-01 |
| GO:BP | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 6 | 4.42e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 2 | 41 | 4.42e-01 |
| GO:BP | GO:0019377 | glycolipid catabolic process | 5 | 12 | 4.42e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 3 | 12 | 4.42e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 3 | 12 | 4.42e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 3 | 12 | 4.42e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 6 | 7 | 4.43e-01 |
| GO:BP | GO:0071941 | nitrogen cycle metabolic process | 3 | 7 | 4.43e-01 |
| GO:BP | GO:0009746 | response to hexose | 12 | 138 | 4.43e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 4 | 85 | 4.43e-01 |
| GO:BP | GO:0097503 | sialylation | 1 | 18 | 4.43e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 5 | 121 | 4.43e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 3 | 18 | 4.43e-01 |
| GO:BP | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis | 1 | 3 | 4.43e-01 |
| GO:BP | GO:0070483 | detection of hypoxia | 1 | 2 | 4.43e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 3 | 76 | 4.43e-01 |
| GO:BP | GO:0042305 | specification of segmental identity, mandibular segment | 1 | 3 | 4.43e-01 |
| GO:BP | GO:0007613 | memory | 14 | 92 | 4.43e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 3 | 12 | 4.43e-01 |
| GO:BP | GO:0086101 | endothelin receptor signaling pathway involved in heart process | 1 | 3 | 4.43e-01 |
| GO:BP | GO:0019627 | urea metabolic process | 3 | 7 | 4.43e-01 |
| GO:BP | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | 6 | 10 | 4.43e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 37 | 85 | 4.43e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 1 | 9 | 4.43e-01 |
| GO:BP | GO:0015819 | lysine transport | 2 | 2 | 4.43e-01 |
| GO:BP | GO:0061945 | regulation of protein K48-linked ubiquitination | 2 | 4 | 4.43e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 1 | 14 | 4.44e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 42 | 1849 | 4.44e-01 |
| GO:BP | GO:0003096 | renal sodium ion transport | 2 | 10 | 4.44e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 4.44e-01 |
| GO:BP | GO:0032025 | response to cobalt ion | 3 | 10 | 4.44e-01 |
| GO:BP | GO:0050860 | negative regulation of T cell receptor signaling pathway | 1 | 17 | 4.44e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 3 | 15 | 4.44e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 4.44e-01 |
| GO:BP | GO:1904232 | regulation of aconitate hydratase activity | 2 | 2 | 4.44e-01 |
| GO:BP | GO:1904234 | positive regulation of aconitate hydratase activity | 2 | 2 | 4.44e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 4.44e-01 |
| GO:BP | GO:0000012 | single strand break repair | 6 | 10 | 4.44e-01 |
| GO:BP | GO:0061900 | glial cell activation | 2 | 23 | 4.44e-01 |
| GO:BP | GO:1904862 | inhibitory synapse assembly | 7 | 13 | 4.44e-01 |
| GO:BP | GO:1900365 | positive regulation of mRNA polyadenylation | 3 | 4 | 4.44e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 6 | 91 | 4.44e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 5 | 51 | 4.44e-01 |
| GO:BP | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 2 | 5 | 4.44e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 3 | 56 | 4.44e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 3 | 12 | 4.44e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 3 | 56 | 4.44e-01 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 4 | 5 | 4.44e-01 |
| GO:BP | GO:0060980 | cell migration involved in coronary vasculogenesis | 1 | 1 | 4.44e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 11 | 97 | 4.44e-01 |
| GO:BP | GO:1903781 | positive regulation of cardiac conduction | 1 | 1 | 4.44e-01 |
| GO:BP | GO:1903810 | L-histidine import across plasma membrane | 2 | 2 | 4.44e-01 |
| GO:BP | GO:1901726 | negative regulation of histone deacetylase activity | 1 | 2 | 4.44e-01 |
| GO:BP | GO:0006600 | creatine metabolic process | 1 | 3 | 4.44e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 4.44e-01 |
| GO:BP | GO:0150066 | negative regulation of deacetylase activity | 1 | 2 | 4.44e-01 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 1 | 13 | 4.44e-01 |
| GO:BP | GO:0032058 | positive regulation of translational initiation in response to stress | 1 | 4 | 4.44e-01 |
| GO:BP | GO:1902308 | regulation of peptidyl-serine dephosphorylation | 1 | 3 | 4.44e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 1 | 75 | 4.44e-01 |
| GO:BP | GO:0035733 | hepatic stellate cell activation | 6 | 8 | 4.44e-01 |
| GO:BP | GO:0050808 | synapse organization | 19 | 365 | 4.44e-01 |
| GO:BP | GO:0070842 | aggresome assembly | 3 | 5 | 4.45e-01 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 1 | 26 | 4.45e-01 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 1 | 26 | 4.45e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 4.45e-01 |
| GO:BP | GO:0046929 | negative regulation of neurotransmitter secretion | 3 | 6 | 4.45e-01 |
| GO:BP | GO:1990927 | calcium ion regulated lysosome exocytosis | 1 | 3 | 4.45e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 10 | 18 | 4.45e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 10 | 55 | 4.45e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 7 | 9 | 4.45e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 20 | 4.45e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 7 | 98 | 4.45e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 4 | 216 | 4.45e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 3 | 54 | 4.45e-01 |
| GO:BP | GO:0038130 | ERBB4 signaling pathway | 4 | 6 | 4.45e-01 |
| GO:BP | GO:1904428 | negative regulation of tubulin deacetylation | 3 | 6 | 4.45e-01 |
| GO:BP | GO:0007566 | embryo implantation | 3 | 39 | 4.46e-01 |
| GO:BP | GO:0036113 | very long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0061817 | endoplasmic reticulum-plasma membrane tethering | 1 | 5 | 4.46e-01 |
| GO:BP | GO:0014841 | skeletal muscle satellite cell proliferation | 3 | 10 | 4.46e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 7 | 11 | 4.46e-01 |
| GO:BP | GO:0044057 | regulation of system process | 26 | 405 | 4.46e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 2 | 41 | 4.46e-01 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 5 | 9 | 4.47e-01 |
| GO:BP | GO:0032700 | negative regulation of interleukin-17 production | 1 | 9 | 4.47e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 4.47e-01 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 2 | 6 | 4.47e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 49 | 85 | 4.47e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 4.47e-01 |
| GO:BP | GO:0090262 | regulation of transcription-coupled nucleotide-excision repair | 1 | 1 | 4.47e-01 |
| GO:BP | GO:1903516 | regulation of single strand break repair | 2 | 3 | 4.47e-01 |
| GO:BP | GO:0090385 | phagosome-lysosome fusion | 1 | 12 | 4.47e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 16 | 299 | 4.47e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 9 | 179 | 4.47e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 2 | 17 | 4.47e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 64 | 241 | 4.47e-01 |
| GO:BP | GO:0097711 | ciliary basal body-plasma membrane docking | 2 | 2 | 4.47e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 6 | 57 | 4.47e-01 |
| GO:BP | GO:1901006 | ubiquinone-6 biosynthetic process | 1 | 1 | 4.47e-01 |
| GO:BP | GO:1901004 | ubiquinone-6 metabolic process | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 22 | 36 | 4.47e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 2 | 13 | 4.47e-01 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 3 | 15 | 4.47e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 4.47e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 7 | 97 | 4.47e-01 |
| GO:BP | GO:0035732 | nitric oxide storage | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 31 | 50 | 4.47e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 2 | 16 | 4.47e-01 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 2 | 14 | 4.47e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0002481 | antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent | 2 | 2 | 4.47e-01 |
| GO:BP | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 3 | 3 | 4.47e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.47e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.47e-01 |
| GO:BP | GO:1902047 | polyamine transmembrane transport | 1 | 10 | 4.47e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.47e-01 |
| GO:BP | GO:0006431 | methionyl-tRNA aminoacylation | 2 | 2 | 4.47e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 12 | 184 | 4.47e-01 |
| GO:BP | GO:0006566 | threonine metabolic process | 4 | 4 | 4.48e-01 |
| GO:BP | GO:1901392 | regulation of transforming growth factor beta1 activation | 1 | 1 | 4.48e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 10 | 20 | 4.48e-01 |
| GO:BP | GO:0032329 | serine transport | 2 | 10 | 4.48e-01 |
| GO:BP | GO:1901394 | positive regulation of transforming growth factor beta1 activation | 1 | 1 | 4.48e-01 |
| GO:BP | GO:0090472 | dibasic protein processing | 1 | 1 | 4.48e-01 |
| GO:BP | GO:0090241 | negative regulation of histone H4 acetylation | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 13 | 88 | 4.48e-01 |
| GO:BP | GO:0036316 | SREBP-SCAP complex retention in endoplasmic reticulum | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 15 | 30 | 4.48e-01 |
| GO:BP | GO:0039534 | negative regulation of MDA-5 signaling pathway | 1 | 3 | 4.49e-01 |
| GO:BP | GO:0034354 | ‘de novo’ NAD biosynthetic process from tryptophan | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0016559 | peroxisome fission | 7 | 9 | 4.49e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 2 | 8 | 4.49e-01 |
| GO:BP | GO:0097252 | oligodendrocyte apoptotic process | 3 | 4 | 4.49e-01 |
| GO:BP | GO:1900045 | negative regulation of protein K63-linked ubiquitination | 1 | 7 | 4.50e-01 |
| GO:BP | GO:0010561 | negative regulation of glycoprotein biosynthetic process | 2 | 10 | 4.50e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 4 | 30 | 4.51e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 3 | 3 | 4.51e-01 |
| GO:BP | GO:0038145 | macrophage colony-stimulating factor signaling pathway | 1 | 3 | 4.51e-01 |
| GO:BP | GO:0120040 | regulation of macrophage proliferation | 1 | 4 | 4.51e-01 |
| GO:BP | GO:1904141 | positive regulation of microglial cell migration | 1 | 3 | 4.51e-01 |
| GO:BP | GO:1903569 | positive regulation of protein localization to ciliary membrane | 1 | 1 | 4.51e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 6 | 843 | 4.51e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 2 | 236 | 4.51e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 3 | 23 | 4.51e-01 |
| GO:BP | GO:0016104 | triterpenoid biosynthetic process | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 24 | 4.51e-01 |
| GO:BP | GO:0071361 | cellular response to ethanol | 1 | 9 | 4.52e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 3 | 81 | 4.52e-01 |
| GO:BP | GO:1901166 | neural crest cell migration involved in autonomic nervous system development | 4 | 5 | 4.52e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 3 | 1201 | 4.52e-01 |
| GO:BP | GO:1903765 | negative regulation of potassium ion export across plasma membrane | 1 | 1 | 4.52e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 4 | 21 | 4.52e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 12 | 137 | 4.52e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 11 | 123 | 4.52e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 4.52e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 10 | 40 | 4.52e-01 |
| GO:BP | GO:0035589 | G protein-coupled purinergic nucleotide receptor signaling pathway | 2 | 4 | 4.52e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 8 | 12 | 4.52e-01 |
| GO:BP | GO:0098926 | postsynaptic signal transduction | 1 | 25 | 4.52e-01 |
| GO:BP | GO:1901254 | positive regulation of intracellular transport of viral material | 1 | 1 | 4.52e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 2 | 49 | 4.52e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 2 | 662 | 4.52e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 2 | 59 | 4.52e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 7 | 40 | 4.52e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 2 | 57 | 4.52e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 62 | 114 | 4.53e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 3 | 29 | 4.53e-01 |
| GO:BP | GO:0032053 | ciliary basal body organization | 1 | 4 | 4.53e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 2 | 83 | 4.53e-01 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 6 | 4.54e-01 |
| GO:BP | GO:1902257 | negative regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 2 | 4.54e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 4.54e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 4.54e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 4.54e-01 |
| GO:BP | GO:0071625 | vocalization behavior | 2 | 13 | 4.54e-01 |
| GO:BP | GO:0090435 | protein localization to nuclear envelope | 1 | 14 | 4.54e-01 |
| GO:BP | GO:1903545 | cellular response to butyrate | 1 | 1 | 4.54e-01 |
| GO:BP | GO:0061772 | xenobiotic transport across blood-nerve barrier | 1 | 1 | 4.54e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 4.54e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 11 | 227 | 4.55e-01 |
| GO:BP | GO:0051409 | response to nitrosative stress | 6 | 10 | 4.55e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 5 | 9 | 4.55e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 22 | 37 | 4.55e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 2 | 12 | 4.55e-01 |
| GO:BP | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process | 5 | 9 | 4.55e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 3 | 357 | 4.55e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 5 | 11 | 4.55e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 12 | 19 | 4.55e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 4.55e-01 |
| GO:BP | GO:0018057 | peptidyl-lysine oxidation | 4 | 5 | 4.55e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 2 | 41 | 4.56e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 4.56e-01 |
| GO:BP | GO:1902775 | mitochondrial large ribosomal subunit assembly | 3 | 4 | 4.56e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 5 | 7 | 4.56e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 6 | 15 | 4.56e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 3 | 48 | 4.56e-01 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 3 | 13 | 4.56e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 11 | 70 | 4.56e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 4.56e-01 |
| GO:BP | GO:0031119 | tRNA pseudouridine synthesis | 1 | 6 | 4.56e-01 |
| GO:BP | GO:0019082 | viral protein processing | 2 | 31 | 4.56e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 4.56e-01 |
| GO:BP | GO:0071257 | cellular response to electrical stimulus | 4 | 8 | 4.56e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 11 | 70 | 4.56e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 25 | 637 | 4.56e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 1 | 78 | 4.56e-01 |
| GO:BP | GO:1903347 | negative regulation of bicellular tight junction assembly | 1 | 3 | 4.56e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 6 | 37 | 4.56e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 3 | 136 | 4.56e-01 |
| GO:BP | GO:0019322 | pentose biosynthetic process | 1 | 2 | 4.56e-01 |
| GO:BP | GO:0019520 | aldonic acid metabolic process | 1 | 2 | 4.56e-01 |
| GO:BP | GO:0019521 | D-gluconate metabolic process | 1 | 2 | 4.56e-01 |
| GO:BP | GO:0046177 | D-gluconate catabolic process | 1 | 2 | 4.56e-01 |
| GO:BP | GO:0034486 | vacuolar transmembrane transport | 2 | 3 | 4.56e-01 |
| GO:BP | GO:0046176 | aldonic acid catabolic process | 1 | 2 | 4.56e-01 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 2 | 6 | 4.56e-01 |
| GO:BP | GO:2000348 | regulation of CD40 signaling pathway | 2 | 4 | 4.57e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 2 | 6 | 4.57e-01 |
| GO:BP | GO:0032960 | regulation of inositol trisphosphate biosynthetic process | 1 | 4 | 4.57e-01 |
| GO:BP | GO:0031048 | small ncRNA-mediated heterochromatin formation | 2 | 2 | 4.57e-01 |
| GO:BP | GO:0070512 | positive regulation of histone H4-K20 methylation | 1 | 1 | 4.57e-01 |
| GO:BP | GO:2000392 | regulation of lamellipodium morphogenesis | 5 | 10 | 4.57e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 3 | 28 | 4.57e-01 |
| GO:BP | GO:0003105 | negative regulation of glomerular filtration | 1 | 3 | 4.57e-01 |
| GO:BP | GO:0098693 | regulation of synaptic vesicle cycle | 8 | 13 | 4.57e-01 |
| GO:BP | GO:0036445 | neuronal stem cell division | 1 | 1 | 4.57e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 4.57e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 1 | 19 | 4.57e-01 |
| GO:BP | GO:0140251 | regulation protein catabolic process at presynapse | 2 | 2 | 4.57e-01 |
| GO:BP | GO:0099540 | trans-synaptic signaling by neuropeptide | 1 | 1 | 4.57e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 4.57e-01 |
| GO:BP | GO:0010891 | negative regulation of sequestering of triglyceride | 1 | 5 | 4.57e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 79 | 4.57e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 2 | 15 | 4.57e-01 |
| GO:BP | GO:0006425 | glutaminyl-tRNA aminoacylation | 1 | 2 | 4.57e-01 |
| GO:BP | GO:0006429 | leucyl-tRNA aminoacylation | 1 | 2 | 4.57e-01 |
| GO:BP | GO:1905608 | positive regulation of presynapse assembly | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0097069 | cellular response to thyroxine stimulus | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 6 | 34 | 4.58e-01 |
| GO:BP | GO:2000490 | negative regulation of hepatic stellate cell activation | 2 | 2 | 4.58e-01 |
| GO:BP | GO:1904387 | cellular response to L-phenylalanine derivative | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 4.58e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 11 | 16 | 4.58e-01 |
| GO:BP | GO:0034263 | positive regulation of autophagy in response to ER overload | 1 | 1 | 4.58e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 1 | 6 | 4.58e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 4.58e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 1 | 6 | 4.58e-01 |
| GO:BP | GO:0003188 | heart valve formation | 9 | 16 | 4.58e-01 |
| GO:BP | GO:0009408 | response to heat | 4 | 84 | 4.58e-01 |
| GO:BP | GO:0060638 | mesenchymal-epithelial cell signaling | 1 | 4 | 4.58e-01 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 1 | 6 | 4.58e-01 |
| GO:BP | GO:0045063 | T-helper 1 cell differentiation | 6 | 14 | 4.58e-01 |
| GO:BP | GO:0070257 | positive regulation of mucus secretion | 1 | 4 | 4.58e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 13 | 237 | 4.59e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 8 | 4241 | 4.59e-01 |
| GO:BP | GO:0030238 | male sex determination | 5 | 8 | 4.59e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 4.59e-01 |
| GO:BP | GO:1905232 | cellular response to L-glutamate | 2 | 4 | 4.59e-01 |
| GO:BP | GO:1903429 | regulation of cell maturation | 3 | 13 | 4.59e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 1 | 28 | 4.60e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 2 | 20 | 4.60e-01 |
| GO:BP | GO:0051958 | methotrexate transport | 1 | 2 | 4.60e-01 |
| GO:BP | GO:0019227 | neuronal action potential propagation | 2 | 5 | 4.60e-01 |
| GO:BP | GO:0009222 | pyrimidine ribonucleotide catabolic process | 1 | 7 | 4.60e-01 |
| GO:BP | GO:0010737 | protein kinase A signaling | 1 | 23 | 4.60e-01 |
| GO:BP | GO:0009131 | pyrimidine nucleoside monophosphate catabolic process | 1 | 6 | 4.60e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 12 | 19 | 4.60e-01 |
| GO:BP | GO:0098870 | action potential propagation | 2 | 5 | 4.60e-01 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 3 | 9 | 4.60e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 7 | 22 | 4.60e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 8 | 54 | 4.60e-01 |
| GO:BP | GO:1901724 | positive regulation of cell proliferation involved in kidney development | 5 | 8 | 4.60e-01 |
| GO:BP | GO:0018160 | peptidyl-pyrromethane cofactor linkage | 1 | 1 | 4.61e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 2 | 5 | 4.61e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 4.62e-01 |
| GO:BP | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration | 1 | 1 | 4.62e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 4.62e-01 |
| GO:BP | GO:2000632 | negative regulation of pre-miRNA processing | 2 | 2 | 4.62e-01 |
| GO:BP | GO:0060596 | mammary placode formation | 2 | 2 | 4.62e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 7 | 27 | 4.62e-01 |
| GO:BP | GO:0009110 | vitamin biosynthetic process | 2 | 14 | 4.62e-01 |
| GO:BP | GO:1905072 | cardiac jelly development | 2 | 3 | 4.62e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 1 | 16 | 4.62e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 6 | 8 | 4.62e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 4.62e-01 |
| GO:BP | GO:0030030 | cell projection organization | 3 | 1233 | 4.62e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 57 | 103 | 4.62e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 2 | 9 | 4.62e-01 |
| GO:BP | GO:0044321 | response to leptin | 2 | 19 | 4.62e-01 |
| GO:BP | GO:0006432 | phenylalanyl-tRNA aminoacylation | 3 | 4 | 4.62e-01 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 4.62e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 4.63e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 20 | 40 | 4.63e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 4 | 13 | 4.63e-01 |
| GO:BP | GO:0060087 | relaxation of vascular associated smooth muscle | 1 | 7 | 4.63e-01 |
| GO:BP | GO:0031033 | myosin filament organization | 4 | 5 | 4.63e-01 |
| GO:BP | GO:0060532 | bronchus cartilage development | 2 | 2 | 4.63e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 6 | 11 | 4.63e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 4.63e-01 |
| GO:BP | GO:0061145 | lung smooth muscle development | 2 | 2 | 4.63e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 2 | 81 | 4.63e-01 |
| GO:BP | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | 4 | 5 | 4.63e-01 |
| GO:BP | GO:0097211 | cellular response to gonadotropin-releasing hormone | 2 | 3 | 4.64e-01 |
| GO:BP | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib | 3 | 4 | 4.64e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 4.64e-01 |
| GO:BP | GO:1902722 | positive regulation of prolactin secretion | 1 | 1 | 4.64e-01 |
| GO:BP | GO:1900186 | negative regulation of clathrin-dependent endocytosis | 2 | 4 | 4.65e-01 |
| GO:BP | GO:0060398 | regulation of growth hormone receptor signaling pathway | 1 | 5 | 4.65e-01 |
| GO:BP | GO:0006771 | riboflavin metabolic process | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 1 | 5 | 4.65e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 1 | 67 | 4.65e-01 |
| GO:BP | GO:1902866 | regulation of retina development in camera-type eye | 1 | 1 | 4.65e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 4 | 211 | 4.65e-01 |
| GO:BP | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis | 1 | 1 | 4.66e-01 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 5 | 10 | 4.66e-01 |
| GO:BP | GO:0098939 | dendritic transport of mitochondrion | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 3 | 62 | 4.66e-01 |
| GO:BP | GO:0003174 | mitral valve development | 6 | 12 | 4.66e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 1 | 14 | 4.66e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 7 | 11 | 4.66e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 9 | 16 | 4.66e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 2 | 6 | 4.67e-01 |
| GO:BP | GO:0038180 | nerve growth factor signaling pathway | 3 | 10 | 4.67e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 3 | 84 | 4.67e-01 |
| GO:BP | GO:0009256 | 10-formyltetrahydrofolate metabolic process | 2 | 6 | 4.67e-01 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 7 | 9 | 4.67e-01 |
| GO:BP | GO:0062042 | regulation of cardiac epithelial to mesenchymal transition | 2 | 10 | 4.67e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 24 | 71 | 4.67e-01 |
| GO:BP | GO:0048853 | forebrain morphogenesis | 3 | 7 | 4.67e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 2 | 8 | 4.67e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 4.67e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 4.67e-01 |
| GO:BP | GO:0014719 | skeletal muscle satellite cell activation | 2 | 5 | 4.67e-01 |
| GO:BP | GO:0071503 | response to heparin | 3 | 5 | 4.67e-01 |
| GO:BP | GO:0021780 | glial cell fate specification | 3 | 3 | 4.67e-01 |
| GO:BP | GO:0072197 | ureter morphogenesis | 3 | 5 | 4.67e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 3 | 9 | 4.67e-01 |
| GO:BP | GO:0042758 | long-chain fatty acid catabolic process | 3 | 3 | 4.67e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 5 | 14 | 4.67e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 2 | 55 | 4.67e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 4 | 2011 | 4.67e-01 |
| GO:BP | GO:0046380 | N-acetylneuraminate biosynthetic process | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0070544 | histone H3-K36 demethylation | 3 | 5 | 4.67e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 5 | 4.68e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 4.68e-01 |
| GO:BP | GO:0003012 | muscle system process | 8 | 334 | 4.68e-01 |
| GO:BP | GO:0006610 | ribosomal protein import into nucleus | 1 | 4 | 4.68e-01 |
| GO:BP | GO:0006404 | RNA import into nucleus | 1 | 4 | 4.68e-01 |
| GO:BP | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade | 7 | 16 | 4.68e-01 |
| GO:BP | GO:0106058 | positive regulation of calcineurin-mediated signaling | 7 | 16 | 4.68e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 4 | 73 | 4.68e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 4.68e-01 |
| GO:BP | GO:0007495 | visceral mesoderm-endoderm interaction involved in midgut development | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0051309 | female meiosis chromosome separation | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 2 | 330 | 4.68e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 3 | 22 | 4.68e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 1 | 30 | 4.68e-01 |
| GO:BP | GO:0072229 | metanephric proximal convoluted tubule development | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0072019 | proximal convoluted tubule development | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0060232 | delamination | 1 | 5 | 4.68e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 6 | 25 | 4.68e-01 |
| GO:BP | GO:0061159 | establishment of bipolar cell polarity involved in cell morphogenesis | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0051892 | negative regulation of cardioblast differentiation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 2 | 24 | 4.68e-01 |
| GO:BP | GO:0019045 | latent virus replication | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 1 | 8 | 4.68e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 1 | 532 | 4.68e-01 |
| GO:BP | GO:0061000 | negative regulation of dendritic spine development | 1 | 10 | 4.68e-01 |
| GO:BP | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0019046 | release from viral latency | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 4.68e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 23 | 64 | 4.68e-01 |
| GO:BP | GO:1905454 | negative regulation of myeloid progenitor cell differentiation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0033630 | positive regulation of cell adhesion mediated by integrin | 1 | 14 | 4.68e-01 |
| GO:BP | GO:0032601 | connective tissue growth factor production | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 5 | 23 | 4.68e-01 |
| GO:BP | GO:0106047 | polyamine deacetylation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0033962 | P-body assembly | 1 | 20 | 4.68e-01 |
| GO:BP | GO:0071313 | cellular response to caffeine | 5 | 10 | 4.68e-01 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 1 | 15 | 4.68e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 4.68e-01 |
| GO:BP | GO:0032643 | regulation of connective tissue growth factor production | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0071228 | cellular response to tumor cell | 3 | 4 | 4.68e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 2 | 28 | 4.68e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 7 | 143 | 4.68e-01 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 1 | 30 | 4.68e-01 |
| GO:BP | GO:0061072 | iris morphogenesis | 1 | 5 | 4.68e-01 |
| GO:BP | GO:2001045 | negative regulation of integrin-mediated signaling pathway | 1 | 3 | 4.68e-01 |
| GO:BP | GO:0060354 | negative regulation of cell adhesion molecule production | 1 | 5 | 4.68e-01 |
| GO:BP | GO:0048219 | inter-Golgi cisterna vesicle-mediated transport | 1 | 1 | 4.68e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 11 | 19 | 4.68e-01 |
| GO:BP | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | 7 | 10 | 4.68e-01 |
| GO:BP | GO:0106048 | spermidine deacetylation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 4.68e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 6 | 50 | 4.68e-01 |
| GO:BP | GO:0045925 | positive regulation of female receptivity | 1 | 1 | 4.69e-01 |
| GO:BP | GO:0006538 | glutamate catabolic process | 1 | 7 | 4.69e-01 |
| GO:BP | GO:0009448 | gamma-aminobutyric acid metabolic process | 1 | 6 | 4.69e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 13 | 22 | 4.69e-01 |
| GO:BP | GO:0051593 | response to folic acid | 4 | 9 | 4.69e-01 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 5 | 4.70e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 1 | 21 | 4.70e-01 |
| GO:BP | GO:0010585 | glutamine secretion | 2 | 2 | 4.70e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 3 | 18 | 4.70e-01 |
| GO:BP | GO:0036017 | response to erythropoietin | 3 | 4 | 4.71e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 1 | 12 | 4.71e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 1 | 12 | 4.71e-01 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 4.71e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 6 | 16 | 4.71e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 2 | 29 | 4.71e-01 |
| GO:BP | GO:0006482 | protein demethylation | 12 | 27 | 4.71e-01 |
| GO:BP | GO:0001712 | ectodermal cell fate commitment | 2 | 2 | 4.71e-01 |
| GO:BP | GO:0098920 | retrograde trans-synaptic signaling by lipid | 2 | 4 | 4.71e-01 |
| GO:BP | GO:0034201 | response to oleic acid | 4 | 5 | 4.71e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 1 | 19 | 4.71e-01 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 2 | 5 | 4.71e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 1 | 19 | 4.71e-01 |
| GO:BP | GO:0036510 | trimming of terminal mannose on C branch | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0003195 | tricuspid valve formation | 2 | 2 | 4.71e-01 |
| GO:BP | GO:0098921 | retrograde trans-synaptic signaling by endocannabinoid | 2 | 4 | 4.71e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 10 | 73 | 4.71e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 12 | 27 | 4.71e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 2 | 7 | 4.71e-01 |
| GO:BP | GO:0048304 | positive regulation of isotype switching to IgG isotypes | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 10 | 46 | 4.71e-01 |
| GO:BP | GO:0071415 | cellular response to purine-containing compound | 6 | 11 | 4.71e-01 |
| GO:BP | GO:1903464 | negative regulation of mitotic cell cycle DNA replication | 1 | 1 | 4.71e-01 |
| GO:BP | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation | 1 | 1 | 4.71e-01 |
| GO:BP | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol | 1 | 3 | 4.71e-01 |
| GO:BP | GO:1902576 | negative regulation of nuclear cell cycle DNA replication | 1 | 1 | 4.71e-01 |
| GO:BP | GO:0003341 | cilium movement | 1 | 97 | 4.71e-01 |
| GO:BP | GO:0016180 | snRNA processing | 5 | 26 | 4.71e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 7 | 101 | 4.71e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 15 | 20 | 4.72e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 2 | 16 | 4.72e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 4 | 12 | 4.72e-01 |
| GO:BP | GO:0007286 | spermatid development | 1 | 105 | 4.72e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 3 | 20 | 4.72e-01 |
| GO:BP | GO:2000808 | negative regulation of synaptic vesicle clustering | 1 | 1 | 4.72e-01 |
| GO:BP | GO:0140029 | exocytic process | 3 | 69 | 4.72e-01 |
| GO:BP | GO:1904627 | response to phorbol 13-acetate 12-myristate | 2 | 4 | 4.72e-01 |
| GO:BP | GO:1904628 | cellular response to phorbol 13-acetate 12-myristate | 2 | 4 | 4.72e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 7 | 54 | 4.72e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 1 | 12 | 4.72e-01 |
| GO:BP | GO:1990547 | mitochondrial phosphate ion transmembrane transport | 1 | 1 | 4.72e-01 |
| GO:BP | GO:0090135 | actin filament branching | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0036153 | triglyceride acyl-chain remodeling | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 12 | 131 | 4.73e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 4 | 1974 | 4.73e-01 |
| GO:BP | GO:1904139 | regulation of microglial cell migration | 1 | 3 | 4.73e-01 |
| GO:BP | GO:1904124 | microglial cell migration | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0061518 | microglial cell proliferation | 1 | 4 | 4.73e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 29 | 486 | 4.73e-01 |
| GO:BP | GO:0035891 | exit from host cell | 2 | 28 | 4.73e-01 |
| GO:BP | GO:0019076 | viral release from host cell | 2 | 28 | 4.73e-01 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 2 | 4.74e-01 |
| GO:BP | GO:2000394 | positive regulation of lamellipodium morphogenesis | 3 | 6 | 4.74e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 1 | 12 | 4.74e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 17 | 23 | 4.74e-01 |
| GO:BP | GO:0006435 | threonyl-tRNA aminoacylation | 1 | 3 | 4.74e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 4 | 5 | 4.74e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 65 | 213 | 4.74e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 4.74e-01 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 6 | 12 | 4.74e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 4.74e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 1 | 32 | 4.74e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 4.74e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 8 | 14 | 4.74e-01 |
| GO:BP | GO:0019550 | glutamate catabolic process to aspartate | 2 | 2 | 4.74e-01 |
| GO:BP | GO:0019551 | glutamate catabolic process to 2-oxoglutarate | 2 | 2 | 4.74e-01 |
| GO:BP | GO:0015846 | polyamine transport | 1 | 12 | 4.74e-01 |
| GO:BP | GO:0055111 | ingression involved in gastrulation with mouth forming second | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0070291 | N-acylethanolamine metabolic process | 1 | 4 | 4.74e-01 |
| GO:BP | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 1 | 4 | 4.74e-01 |
| GO:BP | GO:1901290 | succinyl-CoA biosynthetic process | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0051793 | medium-chain fatty acid catabolic process | 2 | 3 | 4.74e-01 |
| GO:BP | GO:0019678 | propionate metabolic process, methylmalonyl pathway | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 4.74e-01 |
| GO:BP | GO:0009972 | cytidine deamination | 1 | 6 | 4.74e-01 |
| GO:BP | GO:1904998 | negative regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0046087 | cytidine metabolic process | 1 | 6 | 4.74e-01 |
| GO:BP | GO:0006307 | DNA dealkylation involved in DNA repair | 3 | 9 | 4.74e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 18 | 67 | 4.74e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 2 | 22 | 4.74e-01 |
| GO:BP | GO:0032253 | dense core granule localization | 1 | 7 | 4.74e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 7 | 35 | 4.74e-01 |
| GO:BP | GO:1900130 | regulation of lipid binding | 1 | 1 | 4.74e-01 |
| GO:BP | GO:1901950 | dense core granule transport | 1 | 7 | 4.74e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 3 | 40 | 4.74e-01 |
| GO:BP | GO:0006216 | cytidine catabolic process | 1 | 6 | 4.74e-01 |
| GO:BP | GO:1900131 | negative regulation of lipid binding | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0016554 | cytidine to uridine editing | 1 | 6 | 4.74e-01 |
| GO:BP | GO:0099519 | dense core granule cytoskeletal transport | 1 | 7 | 4.74e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 3 | 85 | 4.74e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 6 | 20 | 4.74e-01 |
| GO:BP | GO:0007612 | learning | 6 | 115 | 4.74e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 4.74e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 4 | 51 | 4.74e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 24 | 615 | 4.75e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 4.75e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 2 | 14 | 4.75e-01 |
| GO:BP | GO:0048232 | male gamete generation | 2 | 343 | 4.75e-01 |
| GO:BP | GO:0061880 | regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0061881 | positive regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly | 1 | 2 | 4.75e-01 |
| GO:BP | GO:0071864 | positive regulation of cell proliferation in bone marrow | 2 | 5 | 4.75e-01 |
| GO:BP | GO:1902497 | iron-sulfur cluster transmembrane transport | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0033624 | negative regulation of integrin activation | 2 | 3 | 4.75e-01 |
| GO:BP | GO:1903329 | regulation of iron-sulfur cluster assembly | 1 | 1 | 4.75e-01 |
| GO:BP | GO:1903331 | positive regulation of iron-sulfur cluster assembly | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0140466 | iron-sulfur cluster export from the mitochondrion | 1 | 1 | 4.75e-01 |
| GO:BP | GO:1902031 | regulation of NADP metabolic process | 1 | 8 | 4.75e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 4.75e-01 |
| GO:BP | GO:0036371 | protein localization to T-tubule | 1 | 2 | 4.75e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 1 | 8 | 4.75e-01 |
| GO:BP | GO:0106089 | negative regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0106088 | regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0030497 | fatty acid elongation | 4 | 12 | 4.75e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 8 | 14 | 4.76e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 19 | 41 | 4.76e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 2 | 269 | 4.76e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 9 | 13 | 4.76e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 5 | 97 | 4.76e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 12 | 142 | 4.76e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1182 | 4.76e-01 |
| GO:BP | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis | 3 | 6 | 4.76e-01 |
| GO:BP | GO:1901081 | negative regulation of relaxation of smooth muscle | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 3 | 93 | 4.76e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 1 | 96 | 4.76e-01 |
| GO:BP | GO:0007416 | synapse assembly | 9 | 152 | 4.76e-01 |
| GO:BP | GO:0003108 | negative regulation of the force of heart contraction by chemical signal | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0046292 | formaldehyde metabolic process | 3 | 3 | 4.76e-01 |
| GO:BP | GO:0006273 | lagging strand elongation | 2 | 4 | 4.76e-01 |
| GO:BP | GO:0032286 | central nervous system myelin maintenance | 3 | 3 | 4.76e-01 |
| GO:BP | GO:0045010 | actin nucleation | 3 | 50 | 4.76e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 10 | 84 | 4.76e-01 |
| GO:BP | GO:1903860 | negative regulation of dendrite extension | 2 | 2 | 4.76e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 4.76e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 4.76e-01 |
| GO:BP | GO:0031284 | positive regulation of guanylate cyclase activity | 2 | 2 | 4.76e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 1 | 111 | 4.76e-01 |
| GO:BP | GO:0070086 | ubiquitin-dependent endocytosis | 2 | 4 | 4.76e-01 |
| GO:BP | GO:0046057 | dADP catabolic process | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0021561 | facial nerve development | 5 | 8 | 4.76e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 5 | 8 | 4.76e-01 |
| GO:BP | GO:0009192 | deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0009184 | purine deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0051901 | positive regulation of mitochondrial depolarization | 2 | 6 | 4.76e-01 |
| GO:BP | GO:0046067 | dGDP catabolic process | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway | 3 | 17 | 4.76e-01 |
| GO:BP | GO:0003139 | secondary heart field specification | 3 | 10 | 4.76e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 12 | 46 | 4.76e-01 |
| GO:BP | GO:0098903 | regulation of membrane repolarization during action potential | 1 | 6 | 4.76e-01 |
| GO:BP | GO:0006573 | valine metabolic process | 4 | 7 | 4.76e-01 |
| GO:BP | GO:2001035 | regulation of tongue muscle cell differentiation | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0042264 | peptidyl-aspartic acid hydroxylation | 2 | 2 | 4.76e-01 |
| GO:BP | GO:2001037 | positive regulation of tongue muscle cell differentiation | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0035981 | tongue muscle cell differentiation | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0006853 | carnitine shuttle | 1 | 3 | 4.76e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 4.77e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 1 | 93 | 4.77e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 2 | 21 | 4.77e-01 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 4 | 25 | 4.77e-01 |
| GO:BP | GO:0032487 | regulation of Rap protein signal transduction | 1 | 3 | 4.77e-01 |
| GO:BP | GO:1905223 | epicardium morphogenesis | 1 | 2 | 4.77e-01 |
| GO:BP | GO:0016182 | synaptic vesicle budding from endosome | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0001508 | action potential | 11 | 105 | 4.77e-01 |
| GO:BP | GO:1900240 | negative regulation of phenotypic switching | 1 | 2 | 4.77e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 3 | 4 | 4.77e-01 |
| GO:BP | GO:0006565 | L-serine catabolic process | 4 | 4 | 4.77e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 2 | 104 | 4.77e-01 |
| GO:BP | GO:1905931 | negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 2 | 4.77e-01 |
| GO:BP | GO:0001503 | ossification | 1 | 313 | 4.77e-01 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 2 | 8 | 4.77e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 10 | 31 | 4.77e-01 |
| GO:BP | GO:1905916 | negative regulation of cell differentiation involved in phenotypic switching | 1 | 2 | 4.77e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 2 | 620 | 4.77e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 9 | 41 | 4.78e-01 |
| GO:BP | GO:0042407 | cristae formation | 2 | 18 | 4.78e-01 |
| GO:BP | GO:0003219 | cardiac right ventricle formation | 2 | 4 | 4.78e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 6 | 149 | 4.78e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:0000105 | histidine biosynthetic process | 1 | 2 | 4.78e-01 |
| GO:BP | GO:1905331 | negative regulation of morphogenesis of an epithelium | 1 | 6 | 4.78e-01 |
| GO:BP | GO:0010700 | negative regulation of norepinephrine secretion | 2 | 2 | 4.78e-01 |
| GO:BP | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis | 2 | 4 | 4.78e-01 |
| GO:BP | GO:0006310 | DNA recombination | 123 | 257 | 4.78e-01 |
| GO:BP | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process | 1 | 2 | 4.78e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 3 | 4 | 4.78e-01 |
| GO:BP | GO:0033278 | cell proliferation in midbrain | 1 | 4 | 4.78e-01 |
| GO:BP | GO:0060050 | positive regulation of protein glycosylation | 1 | 6 | 4.78e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 32 | 547 | 4.78e-01 |
| GO:BP | GO:0009957 | epidermal cell fate specification | 2 | 4 | 4.78e-01 |
| GO:BP | GO:0051964 | negative regulation of synapse assembly | 1 | 6 | 4.78e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 6 | 25 | 4.78e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 13 | 56 | 4.78e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 2 | 140 | 4.78e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 19 | 32 | 4.78e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 13 | 19 | 4.78e-01 |
| GO:BP | GO:0060857 | establishment of glial blood-brain barrier | 2 | 2 | 4.78e-01 |
| GO:BP | GO:0061744 | motor behavior | 3 | 17 | 4.78e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 6 | 200 | 4.78e-01 |
| GO:BP | GO:0055129 | L-proline biosynthetic process | 2 | 5 | 4.79e-01 |
| GO:BP | GO:0006561 | proline biosynthetic process | 2 | 5 | 4.79e-01 |
| GO:BP | GO:1905663 | positive regulation of telomerase RNA reverse transcriptase activity | 2 | 2 | 4.79e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 31 | 61 | 4.79e-01 |
| GO:BP | GO:0042779 | tRNA 3’-trailer cleavage | 1 | 3 | 4.79e-01 |
| GO:BP | GO:0006207 | ‘de novo’ pyrimidine nucleobase biosynthetic process | 3 | 6 | 4.79e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 28 | 496 | 4.79e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 4.79e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 9 | 16 | 4.79e-01 |
| GO:BP | GO:0030046 | parallel actin filament bundle assembly | 2 | 5 | 4.79e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 3 | 5 | 4.79e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 5 | 12 | 4.79e-01 |
| GO:BP | GO:0034760 | negative regulation of iron ion transmembrane transport | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0001778 | plasma membrane repair | 2 | 28 | 4.79e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 5 | 12 | 4.79e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 5 | 25 | 4.79e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 3 | 5 | 4.79e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 3 | 4 | 4.79e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 6 | 39 | 4.79e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0061564 | axon development | 6 | 398 | 4.79e-01 |
| GO:BP | GO:0036089 | cleavage furrow formation | 2 | 7 | 4.79e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 4 | 993 | 4.79e-01 |
| GO:BP | GO:0034757 | negative regulation of iron ion transport | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 33 | 57 | 4.79e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 8 | 1719 | 4.79e-01 |
| GO:BP | GO:1905450 | negative regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 2 | 2 | 4.79e-01 |
| GO:BP | GO:1903910 | negative regulation of receptor clustering | 2 | 2 | 4.79e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 1 | 13 | 4.79e-01 |
| GO:BP | GO:0007625 | grooming behavior | 2 | 10 | 4.79e-01 |
| GO:BP | GO:0032238 | adenosine transport | 1 | 2 | 4.80e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 17 | 26 | 4.80e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 2 | 82 | 4.80e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 4.80e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 10 | 112 | 4.80e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 5 | 54 | 4.80e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 4.80e-01 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 4 | 10 | 4.80e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 6 | 40 | 4.80e-01 |
| GO:BP | GO:0055089 | fatty acid homeostasis | 1 | 12 | 4.80e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 4 | 28 | 4.80e-01 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 2 | 20 | 4.81e-01 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 3 | 16 | 4.81e-01 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 3 | 16 | 4.81e-01 |
| GO:BP | GO:0006294 | nucleotide-excision repair, preincision complex assembly | 1 | 1 | 4.81e-01 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 10 | 29 | 4.82e-01 |
| GO:BP | GO:0106101 | ER-dependent peroxisome localization | 1 | 1 | 4.82e-01 |
| GO:BP | GO:1903943 | regulation of hepatocyte apoptotic process | 4 | 7 | 4.82e-01 |
| GO:BP | GO:0032581 | ER-dependent peroxisome organization | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0032815 | negative regulation of natural killer cell activation | 1 | 3 | 4.82e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 4.82e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 6 | 43 | 4.82e-01 |
| GO:BP | GO:0033564 | anterior/posterior axon guidance | 2 | 4 | 4.82e-01 |
| GO:BP | GO:0003419 | growth plate cartilage chondrocyte proliferation | 1 | 2 | 4.82e-01 |
| GO:BP | GO:0061326 | renal tubule development | 4 | 77 | 4.82e-01 |
| GO:BP | GO:0106035 | protein maturation by [4Fe-4S] cluster transfer | 4 | 7 | 4.82e-01 |
| GO:BP | GO:0016254 | preassembly of GPI anchor in ER membrane | 5 | 8 | 4.82e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 9 | 24 | 4.82e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 10 | 280 | 4.82e-01 |
| GO:BP | GO:1902044 | regulation of Fas signaling pathway | 1 | 4 | 4.82e-01 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 2 | 4 | 4.82e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 1 | 7 | 4.82e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 1 | 7 | 4.82e-01 |
| GO:BP | GO:0090675 | intermicrovillar adhesion | 1 | 1 | 4.83e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 10 | 172 | 4.83e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 9 | 28 | 4.83e-01 |
| GO:BP | GO:0042539 | hypotonic salinity response | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 8 | 12 | 4.83e-01 |
| GO:BP | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 2 | 17 | 4.83e-01 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 25 | 4.83e-01 |
| GO:BP | GO:0042167 | heme catabolic process | 1 | 6 | 4.83e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 7 | 149 | 4.83e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 4.83e-01 |
| GO:BP | GO:0021592 | fourth ventricle development | 1 | 1 | 4.83e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 3 | 29 | 4.83e-01 |
| GO:BP | GO:0021993 | initiation of neural tube closure | 1 | 1 | 4.83e-01 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 2 | 8 | 4.83e-01 |
| GO:BP | GO:0046149 | pigment catabolic process | 1 | 6 | 4.83e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0071477 | cellular hypotonic salinity response | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0097527 | necroptotic signaling pathway | 1 | 3 | 4.83e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 2 | 22 | 4.83e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 2 | 3 | 4.83e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 5 | 9 | 4.83e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 2 | 14 | 4.83e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 2 | 3 | 4.83e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 4.83e-01 |
| GO:BP | GO:1905609 | positive regulation of smooth muscle cell-matrix adhesion | 1 | 1 | 4.83e-01 |
| GO:BP | GO:0097374 | sensory neuron axon guidance | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0032912 | negative regulation of transforming growth factor beta2 production | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 2 | 31 | 4.83e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 5 | 719 | 4.83e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 8 | 179 | 4.83e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 11 | 73 | 4.83e-01 |
| GO:BP | GO:0048549 | positive regulation of pinocytosis | 5 | 7 | 4.83e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 2 | 276 | 4.83e-01 |
| GO:BP | GO:0003400 | regulation of COPII vesicle coating | 3 | 3 | 4.83e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 11 | 73 | 4.83e-01 |
| GO:BP | GO:0009886 | post-embryonic animal morphogenesis | 1 | 10 | 4.83e-01 |
| GO:BP | GO:0098917 | retrograde trans-synaptic signaling | 3 | 7 | 4.83e-01 |
| GO:BP | GO:0021785 | branchiomotor neuron axon guidance | 3 | 5 | 4.83e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 4 | 14 | 4.83e-01 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 5 | 19 | 4.83e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 9 | 16 | 4.83e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 6 | 10 | 4.84e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 6 | 10 | 4.84e-01 |
| GO:BP | GO:0003002 | regionalization | 1 | 273 | 4.84e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 2 | 2 | 4.84e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 17 | 340 | 4.84e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 23 | 42 | 4.84e-01 |
| GO:BP | GO:0051350 | negative regulation of lyase activity | 3 | 15 | 4.84e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 4.84e-01 |
| GO:BP | GO:1902977 | mitotic DNA replication preinitiation complex assembly | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 5 | 8 | 4.84e-01 |
| GO:BP | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway | 2 | 5 | 4.84e-01 |
| GO:BP | GO:0006535 | cysteine biosynthetic process from serine | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 3 | 1147 | 4.84e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 2 | 39 | 4.84e-01 |
| GO:BP | GO:0045006 | DNA deamination | 1 | 9 | 4.84e-01 |
| GO:BP | GO:1904618 | positive regulation of actin binding | 1 | 2 | 4.84e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 1 | 5 | 4.84e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 6 | 85 | 4.84e-01 |
| GO:BP | GO:2001108 | positive regulation of Rho guanyl-nucleotide exchange factor activity | 1 | 2 | 4.84e-01 |
| GO:BP | GO:1905099 | positive regulation of guanyl-nucleotide exchange factor activity | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0006419 | alanyl-tRNA aminoacylation | 2 | 2 | 4.84e-01 |
| GO:BP | GO:1904531 | positive regulation of actin filament binding | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0046726 | positive regulation by virus of viral protein levels in host cell | 3 | 4 | 4.84e-01 |
| GO:BP | GO:0106001 | intestinal hexose absorption | 2 | 3 | 4.85e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 23 | 170 | 4.85e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 38 | 197 | 4.85e-01 |
| GO:BP | GO:0043113 | receptor clustering | 2 | 45 | 4.85e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 7 | 102 | 4.86e-01 |
| GO:BP | GO:1904533 | regulation of telomeric loop disassembly | 1 | 2 | 4.86e-01 |
| GO:BP | GO:0015770 | sucrose transport | 3 | 4 | 4.86e-01 |
| GO:BP | GO:0015766 | disaccharide transport | 3 | 4 | 4.86e-01 |
| GO:BP | GO:0015772 | oligosaccharide transport | 3 | 4 | 4.86e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 17 | 29 | 4.86e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 20 | 4.86e-01 |
| GO:BP | GO:0001815 | positive regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 4.86e-01 |
| GO:BP | GO:0035803 | egg coat formation | 2 | 2 | 4.86e-01 |
| GO:BP | GO:2000225 | negative regulation of testosterone biosynthetic process | 1 | 1 | 4.86e-01 |
| GO:BP | GO:0072011 | glomerular endothelium development | 1 | 2 | 4.86e-01 |
| GO:BP | GO:0035287 | head segmentation | 1 | 3 | 4.86e-01 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 3 | 8 | 4.86e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 4.86e-01 |
| GO:BP | GO:1903210 | podocyte apoptotic process | 1 | 3 | 4.86e-01 |
| GO:BP | GO:0097018 | renal albumin absorption | 1 | 3 | 4.86e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 4 | 1939 | 4.86e-01 |
| GO:BP | GO:0035289 | posterior head segmentation | 1 | 3 | 4.86e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 10 | 15 | 4.86e-01 |
| GO:BP | GO:0007380 | specification of segmental identity, head | 1 | 3 | 4.86e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 2 | 40 | 4.86e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 7 | 35 | 4.86e-01 |
| GO:BP | GO:0030237 | female sex determination | 1 | 1 | 4.86e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 4.86e-01 |
| GO:BP | GO:0097492 | sympathetic neuron axon guidance | 1 | 4 | 4.86e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 4.86e-01 |
| GO:BP | GO:0035524 | proline transmembrane transport | 3 | 8 | 4.86e-01 |
| GO:BP | GO:0034350 | regulation of glial cell apoptotic process | 6 | 10 | 4.86e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 2 | 14 | 4.86e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 3 | 25 | 4.86e-01 |
| GO:BP | GO:0036298 | recombinational interstrand cross-link repair | 1 | 2 | 4.86e-01 |
| GO:BP | GO:0060456 | positive regulation of digestive system process | 1 | 8 | 4.86e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 3 | 25 | 4.86e-01 |
| GO:BP | GO:0001554 | luteolysis | 3 | 7 | 4.86e-01 |
| GO:BP | GO:0009438 | methylglyoxal metabolic process | 4 | 7 | 4.86e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 26 | 706 | 4.86e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 1 | 39 | 4.86e-01 |
| GO:BP | GO:0021935 | cerebellar granule cell precursor tangential migration | 2 | 2 | 4.86e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 6 | 35 | 4.86e-01 |
| GO:BP | GO:0070716 | mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication | 1 | 2 | 4.86e-01 |
| GO:BP | GO:0042365 | water-soluble vitamin catabolic process | 2 | 2 | 4.86e-01 |
| GO:BP | GO:0098885 | modification of postsynaptic actin cytoskeleton | 4 | 10 | 4.86e-01 |
| GO:BP | GO:0044058 | regulation of digestive system process | 3 | 22 | 4.86e-01 |
| GO:BP | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | 1 | 6 | 4.87e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 2 | 14 | 4.87e-01 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 1 | 12 | 4.87e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 4.87e-01 |
| GO:BP | GO:0061682 | seminal vesicle morphogenesis | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 29 | 185 | 4.87e-01 |
| GO:BP | GO:1904192 | regulation of cholangiocyte apoptotic process | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0007072 | positive regulation of transcription involved in exit from mitosis | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 5 | 7 | 4.87e-01 |
| GO:BP | GO:0061727 | methylglyoxal catabolic process to lactate | 4 | 5 | 4.87e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 4.87e-01 |
| GO:BP | GO:0039654 | fusion of virus membrane with host endosome membrane | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0051596 | methylglyoxal catabolic process | 4 | 5 | 4.87e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 2 | 17 | 4.87e-01 |
| GO:BP | GO:1904193 | negative regulation of cholangiocyte apoptotic process | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0060318 | definitive erythrocyte differentiation | 3 | 4 | 4.87e-01 |
| GO:BP | GO:0015881 | creatine transmembrane transport | 1 | 4 | 4.87e-01 |
| GO:BP | GO:0090270 | regulation of fibroblast growth factor production | 2 | 4 | 4.87e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 2 | 6 | 4.87e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 4.87e-01 |
| GO:BP | GO:0090269 | fibroblast growth factor production | 2 | 4 | 4.87e-01 |
| GO:BP | GO:0071409 | cellular response to cycloheximide | 2 | 2 | 4.87e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 4 | 7 | 4.87e-01 |
| GO:BP | GO:0014043 | negative regulation of neuron maturation | 1 | 2 | 4.87e-01 |
| GO:BP | GO:1902488 | cholangiocyte apoptotic process | 1 | 1 | 4.87e-01 |
| GO:BP | GO:2000584 | negative regulation of platelet-derived growth factor receptor-alpha signaling pathway | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 2 | 7 | 4.87e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 4.87e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 3 | 20 | 4.87e-01 |
| GO:BP | GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 1 | 14 | 4.87e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 1 | 37 | 4.87e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 4 | 11 | 4.87e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 5 | 96 | 4.87e-01 |
| GO:BP | GO:0048297 | negative regulation of isotype switching to IgA isotypes | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0098828 | modulation of inhibitory postsynaptic potential | 3 | 4 | 4.87e-01 |
| GO:BP | GO:0003175 | tricuspid valve development | 4 | 6 | 4.87e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 7 | 103 | 4.87e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 4.87e-01 |
| GO:BP | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 1 | 3 | 4.87e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 3 | 27 | 4.87e-01 |
| GO:BP | GO:0035781 | CD86 biosynthetic process | 2 | 2 | 4.87e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 3 | 27 | 4.87e-01 |
| GO:BP | GO:0060633 | negative regulation of transcription initiation by RNA polymerase II | 4 | 6 | 4.87e-01 |
| GO:BP | GO:2000727 | positive regulation of cardiac muscle cell differentiation | 1 | 6 | 4.87e-01 |
| GO:BP | GO:0042983 | amyloid precursor protein biosynthetic process | 1 | 14 | 4.87e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 1 | 33 | 4.87e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 3 | 11 | 4.87e-01 |
| GO:BP | GO:0014031 | mesenchymal cell development | 4 | 6 | 4.87e-01 |
| GO:BP | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 15 | 281 | 4.87e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 4.87e-01 |
| GO:BP | GO:0034351 | negative regulation of glial cell apoptotic process | 5 | 8 | 4.87e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 16 | 54 | 4.87e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 2 | 84 | 4.87e-01 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 17 | 31 | 4.87e-01 |
| GO:BP | GO:0022008 | neurogenesis | 3 | 1294 | 4.87e-01 |
| GO:BP | GO:0098749 | cerebellar neuron development | 2 | 3 | 4.87e-01 |
| GO:BP | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 1 | 6 | 4.87e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 24 | 625 | 4.87e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 1 | 33 | 4.88e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 5 | 191 | 4.88e-01 |
| GO:BP | GO:0060397 | growth hormone receptor signaling pathway via JAK-STAT | 5 | 9 | 4.88e-01 |
| GO:BP | GO:0042726 | flavin-containing compound metabolic process | 1 | 4 | 4.88e-01 |
| GO:BP | GO:0014900 | muscle hyperplasia | 2 | 2 | 4.88e-01 |
| GO:BP | GO:1990683 | DNA double-strand break attachment to nuclear envelope | 2 | 2 | 4.88e-01 |
| GO:BP | GO:0006824 | cobalt ion transport | 2 | 5 | 4.88e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 2 | 28 | 4.88e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 1 | 10 | 4.88e-01 |
| GO:BP | GO:1903433 | regulation of constitutive secretory pathway | 1 | 2 | 4.88e-01 |
| GO:BP | GO:1904021 | negative regulation of G protein-coupled receptor internalization | 2 | 2 | 4.88e-01 |
| GO:BP | GO:0009082 | branched-chain amino acid biosynthetic process | 1 | 4 | 4.89e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 9 | 16 | 4.89e-01 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 11 | 20 | 4.89e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 4 | 13 | 4.90e-01 |
| GO:BP | GO:0008037 | cell recognition | 3 | 87 | 4.90e-01 |
| GO:BP | GO:0140214 | positive regulation of long-chain fatty acid import into cell | 2 | 2 | 4.90e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 1 | 7 | 4.90e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 1 | 15 | 4.90e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0072356 | chromosome passenger complex localization to kinetochore | 1 | 1 | 4.90e-01 |
| GO:BP | GO:0006671 | phytosphingosine metabolic process | 1 | 1 | 4.90e-01 |
| GO:BP | GO:0071602 | phytosphingosine biosynthetic process | 1 | 1 | 4.90e-01 |
| GO:BP | GO:0034031 | ribonucleoside bisphosphate catabolic process | 7 | 11 | 4.90e-01 |
| GO:BP | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 2 | 6 | 4.90e-01 |
| GO:BP | GO:0033869 | nucleoside bisphosphate catabolic process | 7 | 11 | 4.90e-01 |
| GO:BP | GO:0034034 | purine nucleoside bisphosphate catabolic process | 7 | 11 | 4.90e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 8 | 85 | 4.90e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 3 | 113 | 4.90e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 6 | 16 | 4.90e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 17 | 30 | 4.91e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 2 | 39 | 4.91e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 4 | 34 | 4.91e-01 |
| GO:BP | GO:0009268 | response to pH | 2 | 26 | 4.91e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 1 | 8 | 4.91e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 1 | 21 | 4.91e-01 |
| GO:BP | GO:1904442 | negative regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 4.92e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 3 | 1278 | 4.92e-01 |
| GO:BP | GO:1990789 | thyroid gland epithelial cell proliferation | 1 | 1 | 4.92e-01 |
| GO:BP | GO:1903697 | negative regulation of microvillus assembly | 1 | 1 | 4.92e-01 |
| GO:BP | GO:1904441 | regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 4.92e-01 |
| GO:BP | GO:1905843 | regulation of cellular response to gamma radiation | 1 | 1 | 4.92e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 4 | 22 | 4.92e-01 |
| GO:BP | GO:2001228 | regulation of response to gamma radiation | 1 | 1 | 4.92e-01 |
| GO:BP | GO:0031640 | killing of cells of another organism | 1 | 10 | 4.92e-01 |
| GO:BP | GO:1901098 | positive regulation of autophagosome maturation | 5 | 6 | 4.92e-01 |
| GO:BP | GO:1902888 | protein localization to astral microtubule | 1 | 1 | 4.92e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 1 | 10 | 4.92e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 5 | 7 | 4.92e-01 |
| GO:BP | GO:0016077 | sno(s)RNA catabolic process | 2 | 3 | 4.93e-01 |
| GO:BP | GO:0006012 | galactose metabolic process | 2 | 9 | 4.93e-01 |
| GO:BP | GO:0001834 | trophectodermal cell proliferation | 1 | 2 | 4.93e-01 |
| GO:BP | GO:0001845 | phagolysosome assembly | 1 | 14 | 4.93e-01 |
| GO:BP | GO:1903544 | response to butyrate | 2 | 2 | 4.93e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 189 | 4.93e-01 |
| GO:BP | GO:0022013 | pallium cell proliferation in forebrain | 1 | 1 | 4.94e-01 |
| GO:BP | GO:0016139 | glycoside catabolic process | 3 | 8 | 4.94e-01 |
| GO:BP | GO:0019321 | pentose metabolic process | 6 | 9 | 4.94e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 4.94e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 2 | 13 | 4.94e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 2 | 317 | 4.94e-01 |
| GO:BP | GO:0010255 | glucose mediated signaling pathway | 1 | 5 | 4.94e-01 |
| GO:BP | GO:0010182 | sugar mediated signaling pathway | 1 | 5 | 4.94e-01 |
| GO:BP | GO:0120187 | positive regulation of protein localization to chromatin | 1 | 3 | 4.94e-01 |
| GO:BP | GO:1900454 | positive regulation of long-term synaptic depression | 1 | 3 | 4.94e-01 |
| GO:BP | GO:0015827 | tryptophan transport | 4 | 6 | 4.94e-01 |
| GO:BP | GO:0009757 | hexose mediated signaling | 1 | 5 | 4.94e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 2 | 3 | 4.94e-01 |
| GO:BP | GO:0032069 | regulation of nuclease activity | 3 | 17 | 4.94e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 26 | 150 | 4.95e-01 |
| GO:BP | GO:0038026 | reelin-mediated signaling pathway | 6 | 9 | 4.95e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 5 | 36 | 4.95e-01 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 1 | 3 | 4.95e-01 |
| GO:BP | GO:0060723 | regulation of cell proliferation involved in embryonic placenta development | 1 | 1 | 4.95e-01 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0060721 | regulation of spongiotrophoblast cell proliferation | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 1 | 30 | 4.95e-01 |
| GO:BP | GO:1904370 | regulation of protein localization to actin cortical patch | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 2 | 26 | 4.95e-01 |
| GO:BP | GO:0044379 | protein localization to actin cortical patch | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 38 | 1735 | 4.95e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 2 | 14 | 4.95e-01 |
| GO:BP | GO:1903119 | protein localization to actin cytoskeleton | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0000729 | DNA double-strand break processing | 5 | 18 | 4.95e-01 |
| GO:BP | GO:0031443 | fast-twitch skeletal muscle fiber contraction | 1 | 3 | 4.95e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 3 | 30 | 4.95e-01 |
| GO:BP | GO:0043473 | pigmentation | 15 | 92 | 4.95e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 3 | 21 | 4.95e-01 |
| GO:BP | GO:1904373 | response to kainic acid | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 13 | 249 | 4.95e-01 |
| GO:BP | GO:1904372 | positive regulation of protein localization to actin cortical patch | 1 | 1 | 4.95e-01 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 1 | 2 | 4.95e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 7 | 169 | 4.95e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 4 | 10 | 4.95e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 4 | 10 | 4.95e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 4.95e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 1 | 7 | 4.95e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 2 | 17 | 4.95e-01 |
| GO:BP | GO:0046294 | formaldehyde catabolic process | 2 | 2 | 4.95e-01 |
| GO:BP | GO:1902463 | protein localization to cell leading edge | 2 | 5 | 4.95e-01 |
| GO:BP | GO:0000710 | meiotic mismatch repair | 2 | 2 | 4.95e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 4.95e-01 |
| GO:BP | GO:0061107 | seminal vesicle development | 2 | 2 | 4.95e-01 |
| GO:BP | GO:0070203 | regulation of establishment of protein localization to telomere | 4 | 11 | 4.95e-01 |
| GO:BP | GO:0070084 | protein initiator methionine removal | 2 | 4 | 4.95e-01 |
| GO:BP | GO:0031649 | heat generation | 1 | 11 | 4.95e-01 |
| GO:BP | GO:0030900 | forebrain development | 4 | 281 | 4.95e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 13 | 52 | 4.95e-01 |
| GO:BP | GO:0070078 | histone H3-R2 demethylation | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 4.96e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 2 | 17 | 4.96e-01 |
| GO:BP | GO:0018395 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0070079 | histone H4-R3 demethylation | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 13 | 161 | 4.96e-01 |
| GO:BP | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 3 | 266 | 4.96e-01 |
| GO:BP | GO:0060697 | positive regulation of phospholipid catabolic process | 1 | 1 | 4.97e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 36 | 111 | 4.97e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 4.97e-01 |
| GO:BP | GO:0007158 | neuron cell-cell adhesion | 1 | 13 | 4.97e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 4.97e-01 |
| GO:BP | GO:0006680 | glucosylceramide catabolic process | 1 | 3 | 4.97e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 29 | 616 | 4.97e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 6 | 14 | 4.97e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 29 | 51 | 4.97e-01 |
| GO:BP | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway | 3 | 3 | 4.97e-01 |
| GO:BP | GO:0023021 | termination of signal transduction | 1 | 3 | 4.97e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 1 | 11 | 4.97e-01 |
| GO:BP | GO:0071865 | regulation of apoptotic process in bone marrow cell | 3 | 3 | 4.97e-01 |
| GO:BP | GO:0071866 | negative regulation of apoptotic process in bone marrow cell | 3 | 3 | 4.97e-01 |
| GO:BP | GO:1904777 | negative regulation of protein localization to cell cortex | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0098725 | symmetric cell division | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0003242 | cardiac chamber ballooning | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0070668 | positive regulation of mast cell proliferation | 2 | 4 | 4.97e-01 |
| GO:BP | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development | 2 | 2 | 4.97e-01 |
| GO:BP | GO:1902034 | negative regulation of hematopoietic stem cell proliferation | 1 | 2 | 4.97e-01 |
| GO:BP | GO:1902767 | isoprenoid biosynthetic process via mevalonate | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 4.97e-01 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 3 | 4 | 4.97e-01 |
| GO:BP | GO:0007584 | response to nutrient | 2 | 109 | 4.97e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 3 | 10 | 4.97e-01 |
| GO:BP | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 25 | 427 | 4.97e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 4.97e-01 |
| GO:BP | GO:0003182 | coronary sinus valve morphogenesis | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0003178 | coronary sinus valve development | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 5 | 40 | 4.97e-01 |
| GO:BP | GO:0018282 | metal incorporation into metallo-sulfur cluster | 2 | 2 | 4.97e-01 |
| GO:BP | GO:0061551 | trigeminal ganglion development | 3 | 3 | 4.97e-01 |
| GO:BP | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration | 2 | 2 | 4.97e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 4.97e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 6 | 31 | 4.97e-01 |
| GO:BP | GO:0003270 | Notch signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 4.97e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 3 | 7 | 4.97e-01 |
| GO:BP | GO:1990575 | mitochondrial L-ornithine transmembrane transport | 1 | 2 | 4.97e-01 |
| GO:BP | GO:0018283 | iron incorporation into metallo-sulfur cluster | 2 | 2 | 4.97e-01 |
| GO:BP | GO:0060843 | venous endothelial cell differentiation | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 17 | 31 | 4.97e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 2 | 94 | 4.97e-01 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 4 | 5 | 4.97e-01 |
| GO:BP | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling | 2 | 8 | 4.97e-01 |
| GO:BP | GO:2000657 | negative regulation of apolipoprotein binding | 1 | 1 | 4.97e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 2 | 39 | 4.97e-01 |
| GO:BP | GO:2000656 | regulation of apolipoprotein binding | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 3 | 6 | 4.97e-01 |
| GO:BP | GO:0061713 | anterior neural tube closure | 1 | 2 | 4.97e-01 |
| GO:BP | GO:0021769 | orbitofrontal cortex development | 2 | 2 | 4.97e-01 |
| GO:BP | GO:0001765 | membrane raft assembly | 1 | 9 | 4.97e-01 |
| GO:BP | GO:0050890 | cognition | 11 | 227 | 4.97e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 4.98e-01 |
| GO:BP | GO:0007389 | pattern specification process | 1 | 306 | 4.98e-01 |
| GO:BP | GO:1905216 | positive regulation of RNA binding | 5 | 9 | 4.98e-01 |
| GO:BP | GO:0007000 | nucleolus organization | 10 | 17 | 4.98e-01 |
| GO:BP | GO:1903019 | negative regulation of glycoprotein metabolic process | 1 | 12 | 4.98e-01 |
| GO:BP | GO:1901210 | regulation of cardiac chamber formation | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 2 | 19 | 4.98e-01 |
| GO:BP | GO:1903141 | negative regulation of establishment of endothelial barrier | 2 | 2 | 4.98e-01 |
| GO:BP | GO:0097533 | cellular stress response to acid chemical | 2 | 2 | 4.98e-01 |
| GO:BP | GO:0003065 | positive regulation of heart rate by epinephrine | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 5 | 8 | 4.98e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 4.98e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 20 | 133 | 4.98e-01 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 4.98e-01 |
| GO:BP | GO:1901211 | negative regulation of cardiac chamber formation | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0003169 | coronary vein morphogenesis | 2 | 2 | 4.98e-01 |
| GO:BP | GO:1903033 | positive regulation of microtubule plus-end binding | 2 | 2 | 4.98e-01 |
| GO:BP | GO:1901551 | negative regulation of endothelial cell development | 2 | 2 | 4.98e-01 |
| GO:BP | GO:1901208 | negative regulation of heart looping | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 18 | 57 | 4.98e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 4.98e-01 |
| GO:BP | GO:0062176 | R-loop processing | 4 | 6 | 4.98e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 2 | 2 | 4.98e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 4.98e-01 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 1 | 16 | 4.98e-01 |
| GO:BP | GO:0097532 | stress response to acid chemical | 2 | 2 | 4.98e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 3 | 38 | 4.98e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 4.98e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 4.98e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 3 | 26 | 4.98e-01 |
| GO:BP | GO:1903031 | regulation of microtubule plus-end binding | 2 | 2 | 4.98e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 24 | 41 | 4.98e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 3 | 53 | 4.98e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 7 | 151 | 4.98e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 12 | 19 | 4.98e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 8 | 43 | 4.98e-01 |
| GO:BP | GO:0001824 | blastocyst development | 1 | 94 | 4.98e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 7 | 52 | 4.98e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 5 | 51 | 4.98e-01 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 13 | 29 | 4.99e-01 |
| GO:BP | GO:0042552 | myelination | 2 | 111 | 4.99e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 3 | 46 | 4.99e-01 |
| GO:BP | GO:0035963 | cellular response to interleukin-13 | 1 | 1 | 4.99e-01 |
| GO:BP | GO:0071472 | cellular response to salt stress | 5 | 11 | 4.99e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 17 | 26 | 4.99e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 1 | 31 | 4.99e-01 |
| GO:BP | GO:0002551 | mast cell chemotaxis | 1 | 9 | 4.99e-01 |
| GO:BP | GO:0110022 | regulation of cardiac muscle myoblast proliferation | 2 | 3 | 4.99e-01 |
| GO:BP | GO:0110021 | cardiac muscle myoblast proliferation | 2 | 3 | 4.99e-01 |
| GO:BP | GO:1901390 | positive regulation of transforming growth factor beta activation | 2 | 3 | 4.99e-01 |
| GO:BP | GO:0110024 | positive regulation of cardiac muscle myoblast proliferation | 2 | 3 | 4.99e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 5 | 60 | 5.00e-01 |
| GO:BP | GO:0032105 | negative regulation of response to extracellular stimulus | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0032096 | negative regulation of response to food | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0014721 | twitch skeletal muscle contraction | 4 | 5 | 5.00e-01 |
| GO:BP | GO:0032108 | negative regulation of response to nutrient levels | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0003010 | voluntary skeletal muscle contraction | 4 | 5 | 5.00e-01 |
| GO:BP | GO:0033210 | leptin-mediated signaling pathway | 1 | 10 | 5.00e-01 |
| GO:BP | GO:0032099 | negative regulation of appetite | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0110008 | ncRNA deadenylation | 1 | 1 | 5.00e-01 |
| GO:BP | GO:0036015 | response to interleukin-3 | 3 | 6 | 5.00e-01 |
| GO:BP | GO:0036016 | cellular response to interleukin-3 | 3 | 6 | 5.00e-01 |
| GO:BP | GO:0061738 | late endosomal microautophagy | 1 | 3 | 5.00e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 3 | 12 | 5.00e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 3 | 7 | 5.00e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 35 | 81 | 5.00e-01 |
| GO:BP | GO:0030202 | heparin metabolic process | 2 | 14 | 5.00e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 18 | 29 | 5.00e-01 |
| GO:BP | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity | 1 | 10 | 5.00e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 1 | 26 | 5.00e-01 |
| GO:BP | GO:2000591 | positive regulation of metanephric mesenchymal cell migration | 1 | 3 | 5.00e-01 |
| GO:BP | GO:0003104 | positive regulation of glomerular filtration | 1 | 3 | 5.00e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 1 | 26 | 5.00e-01 |
| GO:BP | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 1 | 3 | 5.00e-01 |
| GO:BP | GO:1900238 | regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 1 | 3 | 5.00e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 3 | 775 | 5.00e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 5.00e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 13 | 34 | 5.00e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 2 | 29 | 5.00e-01 |
| GO:BP | GO:1905271 | regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 5.00e-01 |
| GO:BP | GO:1905273 | positive regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 5.00e-01 |
| GO:BP | GO:0070143 | mitochondrial alanyl-tRNA aminoacylation | 1 | 1 | 5.00e-01 |
| GO:BP | GO:0031929 | TOR signaling | 1 | 115 | 5.00e-01 |
| GO:BP | GO:1905706 | regulation of mitochondrial ATP synthesis coupled proton transport | 2 | 3 | 5.00e-01 |
| GO:BP | GO:0048369 | lateral mesoderm morphogenesis | 3 | 3 | 5.00e-01 |
| GO:BP | GO:0048370 | lateral mesoderm formation | 3 | 3 | 5.00e-01 |
| GO:BP | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 5.01e-01 |
| GO:BP | GO:0030210 | heparin biosynthetic process | 1 | 9 | 5.01e-01 |
| GO:BP | GO:0002159 | desmosome assembly | 2 | 3 | 5.02e-01 |
| GO:BP | GO:0071603 | endothelial cell-cell adhesion | 2 | 4 | 5.02e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 10 | 16 | 5.02e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 1 | 9 | 5.02e-01 |
| GO:BP | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing | 1 | 1 | 5.02e-01 |
| GO:BP | GO:1905090 | negative regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 5.02e-01 |
| GO:BP | GO:0036149 | phosphatidylinositol acyl-chain remodeling | 2 | 3 | 5.02e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 2 | 2 | 5.02e-01 |
| GO:BP | GO:0051890 | regulation of cardioblast differentiation | 2 | 5 | 5.02e-01 |
| GO:BP | GO:1904058 | positive regulation of sensory perception of pain | 2 | 3 | 5.02e-01 |
| GO:BP | GO:1902915 | negative regulation of protein polyubiquitination | 1 | 9 | 5.02e-01 |
| GO:BP | GO:0003128 | heart field specification | 5 | 16 | 5.02e-01 |
| GO:BP | GO:1900006 | positive regulation of dendrite development | 5 | 14 | 5.02e-01 |
| GO:BP | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | 1 | 8 | 5.02e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 5.02e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 5.02e-01 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 5 | 7 | 5.02e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 2 | 113 | 5.02e-01 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 5 | 7 | 5.02e-01 |
| GO:BP | GO:0060026 | convergent extension | 2 | 14 | 5.02e-01 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 1 | 7 | 5.02e-01 |
| GO:BP | GO:0031129 | inductive cell-cell signaling | 1 | 1 | 5.02e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 2 | 113 | 5.02e-01 |
| GO:BP | GO:1903232 | melanosome assembly | 10 | 19 | 5.02e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 302 | 5.02e-01 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 1 | 5 | 5.02e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 5.02e-01 |
| GO:BP | GO:0006788 | heme oxidation | 2 | 2 | 5.02e-01 |
| GO:BP | GO:0071707 | immunoglobulin heavy chain V-D-J recombination | 2 | 2 | 5.02e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 22 | 265 | 5.02e-01 |
| GO:BP | GO:1902725 | negative regulation of satellite cell differentiation | 1 | 2 | 5.02e-01 |
| GO:BP | GO:0090669 | telomerase RNA stabilization | 2 | 4 | 5.02e-01 |
| GO:BP | GO:1904273 | L-alanine import across plasma membrane | 3 | 4 | 5.03e-01 |
| GO:BP | GO:0099140 | presynaptic actin cytoskeleton organization | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0099187 | presynaptic cytoskeleton organization | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 5.03e-01 |
| GO:BP | GO:0071236 | cellular response to antibiotic | 1 | 11 | 5.03e-01 |
| GO:BP | GO:0070417 | cellular response to cold | 1 | 8 | 5.03e-01 |
| GO:BP | GO:0090672 | telomerase RNA localization | 6 | 19 | 5.03e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 1 | 29 | 5.03e-01 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 6 | 19 | 5.03e-01 |
| GO:BP | GO:0050914 | sensory perception of salty taste | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 6 | 19 | 5.03e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 4 | 28 | 5.03e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 2 | 39 | 5.03e-01 |
| GO:BP | GO:0060766 | negative regulation of androgen receptor signaling pathway | 6 | 12 | 5.03e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 5.03e-01 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 6 | 19 | 5.03e-01 |
| GO:BP | GO:0045339 | farnesyl diphosphate catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:1902247 | geranylgeranyl diphosphate catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 4 | 55 | 5.03e-01 |
| GO:BP | GO:1902001 | fatty acid transmembrane transport | 5 | 15 | 5.03e-01 |
| GO:BP | GO:0050655 | dermatan sulfate proteoglycan metabolic process | 3 | 12 | 5.03e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 5 | 5 | 5.03e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 5 | 7 | 5.03e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 25 | 77 | 5.03e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 3 | 1974 | 5.03e-01 |
| GO:BP | GO:0150077 | regulation of neuroinflammatory response | 1 | 13 | 5.04e-01 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 4 | 20 | 5.04e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 10 | 13 | 5.04e-01 |
| GO:BP | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 1 | 2 | 5.04e-01 |
| GO:BP | GO:0006499 | N-terminal protein myristoylation | 2 | 4 | 5.04e-01 |
| GO:BP | GO:0018377 | protein myristoylation | 2 | 4 | 5.04e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 3 | 3 | 5.04e-01 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 1 | 17 | 5.04e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 3 | 91 | 5.04e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 1 | 43 | 5.04e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 4 | 42 | 5.04e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 2 | 86 | 5.04e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 2 | 33 | 5.04e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 2 | 11 | 5.04e-01 |
| GO:BP | GO:0048338 | mesoderm structural organization | 1 | 1 | 5.04e-01 |
| GO:BP | GO:1905285 | fibrous ring of heart morphogenesis | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0021998 | neural plate mediolateral regionalization | 1 | 1 | 5.04e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 5 | 198 | 5.04e-01 |
| GO:BP | GO:2000655 | negative regulation of cellular response to testosterone stimulus | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0060897 | neural plate regionalization | 3 | 6 | 5.04e-01 |
| GO:BP | GO:0048352 | paraxial mesoderm structural organization | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development | 2 | 3 | 5.04e-01 |
| GO:BP | GO:0048372 | lateral mesodermal cell fate commitment | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0060520 | activation of prostate induction by androgen receptor signaling pathway | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0060515 | prostate field specification | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0048377 | lateral mesodermal cell fate specification | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0060514 | prostate induction | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0048378 | regulation of lateral mesodermal cell fate specification | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0033015 | tetrapyrrole catabolic process | 1 | 7 | 5.04e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 12 | 34 | 5.04e-01 |
| GO:BP | GO:0007417 | central nervous system development | 12 | 792 | 5.04e-01 |
| GO:BP | GO:0002885 | positive regulation of hypersensitivity | 3 | 5 | 5.04e-01 |
| GO:BP | GO:1904414 | positive regulation of cardiac ventricle development | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0003306 | Wnt signaling pathway involved in heart development | 3 | 8 | 5.04e-01 |
| GO:BP | GO:0035600 | tRNA methylthiolation | 2 | 2 | 5.04e-01 |
| GO:BP | GO:1904412 | regulation of cardiac ventricle development | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 7 | 38 | 5.04e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 2 | 33 | 5.04e-01 |
| GO:BP | GO:0051643 | endoplasmic reticulum localization | 1 | 8 | 5.04e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 5.04e-01 |
| GO:BP | GO:0019102 | male somatic sex determination | 1 | 1 | 5.04e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 7 | 20 | 5.04e-01 |
| GO:BP | GO:0015911 | long-chain fatty acid import across plasma membrane | 3 | 9 | 5.04e-01 |
| GO:BP | GO:0016049 | cell growth | 3 | 418 | 5.04e-01 |
| GO:BP | GO:0006787 | porphyrin-containing compound catabolic process | 1 | 7 | 5.04e-01 |
| GO:BP | GO:1904220 | regulation of serine C-palmitoyltransferase activity | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0098942 | retrograde trans-synaptic signaling by trans-synaptic protein complex | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 4 | 40 | 5.05e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 18 | 5.05e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 2 | 122 | 5.05e-01 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 1 | 56 | 5.05e-01 |
| GO:BP | GO:0150099 | neuron-glial cell signaling | 2 | 4 | 5.05e-01 |
| GO:BP | GO:0090118 | receptor-mediated endocytosis involved in cholesterol transport | 1 | 5 | 5.05e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 5 | 80 | 5.05e-01 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 2 | 4 | 5.05e-01 |
| GO:BP | GO:0034383 | low-density lipoprotein particle clearance | 1 | 17 | 5.05e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0060517 | epithelial cell proliferation involved in prostatic bud elongation | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0006560 | proline metabolic process | 4 | 7 | 5.05e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 1 | 7 | 5.05e-01 |
| GO:BP | GO:0019264 | glycine biosynthetic process from serine | 1 | 2 | 5.05e-01 |
| GO:BP | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 23 | 55 | 5.06e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 3 | 18 | 5.06e-01 |
| GO:BP | GO:0035750 | protein localization to myelin sheath abaxonal region | 1 | 1 | 5.06e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 13 | 152 | 5.07e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 6 | 103 | 5.07e-01 |
| GO:BP | GO:0003058 | hormonal regulation of the force of heart contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 2 | 5 | 5.08e-01 |
| GO:BP | GO:0009932 | cell tip growth | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 2 | 10 | 5.08e-01 |
| GO:BP | GO:0051352 | negative regulation of ligase activity | 1 | 2 | 5.08e-01 |
| GO:BP | GO:2000627 | positive regulation of miRNA catabolic process | 1 | 4 | 5.08e-01 |
| GO:BP | GO:2000553 | positive regulation of T-helper 2 cell cytokine production | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 5.08e-01 |
| GO:BP | GO:1990765 | colon smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 5.08e-01 |
| GO:BP | GO:1904343 | positive regulation of colon smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0014828 | distal stomach smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 3 | 74 | 5.08e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 2 | 46 | 5.08e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 2 | 46 | 5.08e-01 |
| GO:BP | GO:0097326 | melanocyte adhesion | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0061303 | cornea development in camera-type eye | 1 | 4 | 5.08e-01 |
| GO:BP | GO:0010522 | regulation of calcium ion transport into cytosol | 1 | 10 | 5.08e-01 |
| GO:BP | GO:0120072 | positive regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0120071 | regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 1 | 13 | 5.08e-01 |
| GO:BP | GO:1904341 | regulation of colon smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0120064 | stomach pylorus smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0120063 | stomach smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0120065 | pyloric antrum smooth muscle contraction | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0097378 | dorsal spinal cord interneuron axon guidance | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0032775 | DNA methylation on adenine | 1 | 2 | 5.08e-01 |
| GO:BP | GO:0018872 | arsonoacetate metabolic process | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0032485 | regulation of Ral protein signal transduction | 2 | 3 | 5.08e-01 |
| GO:BP | GO:0097502 | mannosylation | 6 | 32 | 5.08e-01 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 4 | 24 | 5.08e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 17 | 53 | 5.08e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 4 | 56 | 5.08e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 12 | 115 | 5.08e-01 |
| GO:BP | GO:0042256 | cytosolic ribosome assembly | 1 | 5 | 5.08e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 11 | 104 | 5.09e-01 |
| GO:BP | GO:0035654 | clathrin-coated vesicle cargo loading, AP-3-mediated | 5 | 6 | 5.09e-01 |
| GO:BP | GO:0035652 | clathrin-coated vesicle cargo loading | 5 | 6 | 5.09e-01 |
| GO:BP | GO:2000301 | negative regulation of synaptic vesicle exocytosis | 2 | 4 | 5.09e-01 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 1 | 8 | 5.09e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 6 | 110 | 5.09e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 5.09e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 14 | 27 | 5.09e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 5.09e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 15 | 166 | 5.10e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 5 | 112 | 5.10e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 10 | 24 | 5.10e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 24 | 197 | 5.11e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 31 | 321 | 5.11e-01 |
| GO:BP | GO:0014860 | neurotransmitter secretion involved in regulation of skeletal muscle contraction | 1 | 1 | 5.11e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 1 | 39 | 5.11e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 2 | 22 | 5.11e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 1 | 31 | 5.11e-01 |
| GO:BP | GO:0048468 | cell development | 3 | 1963 | 5.12e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 5.12e-01 |
| GO:BP | GO:0048266 | behavioral response to pain | 1 | 11 | 5.12e-01 |
| GO:BP | GO:0099532 | synaptic vesicle endosomal processing | 3 | 7 | 5.12e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 6 | 21 | 5.12e-01 |
| GO:BP | GO:0071348 | cellular response to interleukin-11 | 1 | 1 | 5.12e-01 |
| GO:BP | GO:1905605 | positive regulation of blood-brain barrier permeability | 1 | 4 | 5.12e-01 |
| GO:BP | GO:0051029 | rRNA transport | 2 | 5 | 5.12e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 3 | 5 | 5.12e-01 |
| GO:BP | GO:0032071 | regulation of endodeoxyribonuclease activity | 3 | 7 | 5.12e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 3 | 83 | 5.12e-01 |
| GO:BP | GO:1903712 | cysteine transmembrane transport | 1 | 3 | 5.12e-01 |
| GO:BP | GO:0055113 | epiboly involved in gastrulation with mouth forming second | 1 | 1 | 5.12e-01 |
| GO:BP | GO:0042883 | cysteine transport | 1 | 3 | 5.12e-01 |
| GO:BP | GO:0042262 | DNA protection | 1 | 6 | 5.12e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 5.12e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 5.12e-01 |
| GO:BP | GO:0051031 | tRNA transport | 4 | 5 | 5.12e-01 |
| GO:BP | GO:0097210 | response to gonadotropin-releasing hormone | 2 | 4 | 5.12e-01 |
| GO:BP | GO:1901740 | negative regulation of myoblast fusion | 2 | 2 | 5.12e-01 |
| GO:BP | GO:0090186 | regulation of pancreatic juice secretion | 1 | 6 | 5.12e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 12 | 84 | 5.12e-01 |
| GO:BP | GO:0010979 | regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 5.12e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 5.12e-01 |
| GO:BP | GO:0051340 | regulation of ligase activity | 7 | 11 | 5.12e-01 |
| GO:BP | GO:0010980 | positive regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 5.12e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 5.12e-01 |
| GO:BP | GO:0048278 | vesicle docking | 12 | 57 | 5.13e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 1 | 117 | 5.13e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 5.13e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 5 | 43 | 5.13e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 2 | 6 | 5.13e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 3 | 19 | 5.13e-01 |
| GO:BP | GO:0010746 | regulation of long-chain fatty acid import across plasma membrane | 2 | 5 | 5.13e-01 |
| GO:BP | GO:0032696 | negative regulation of interleukin-13 production | 2 | 3 | 5.13e-01 |
| GO:BP | GO:0120158 | positive regulation of collagen catabolic process | 1 | 1 | 5.13e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 6 | 53 | 5.13e-01 |
| GO:BP | GO:0003275 | apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 5.13e-01 |
| GO:BP | GO:1902256 | regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 5.13e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 5.14e-01 |
| GO:BP | GO:0061386 | closure of optic fissure | 2 | 2 | 5.14e-01 |
| GO:BP | GO:0048697 | positive regulation of collateral sprouting in absence of injury | 1 | 1 | 5.14e-01 |
| GO:BP | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission | 2 | 3 | 5.14e-01 |
| GO:BP | GO:0044857 | plasma membrane raft organization | 1 | 9 | 5.14e-01 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 1 | 12 | 5.14e-01 |
| GO:BP | GO:0042753 | positive regulation of circadian rhythm | 1 | 10 | 5.15e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 6 | 85 | 5.15e-01 |
| GO:BP | GO:0070934 | CRD-mediated mRNA stabilization | 7 | 11 | 5.15e-01 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 4 | 10 | 5.15e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 18 | 34 | 5.15e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 5.16e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 6 | 5.16e-01 |
| GO:BP | GO:0015910 | long-chain fatty acid import into peroxisome | 2 | 3 | 5.16e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 5.16e-01 |
| GO:BP | GO:0018158 | protein oxidation | 1 | 12 | 5.16e-01 |
| GO:BP | GO:0090557 | establishment of endothelial intestinal barrier | 3 | 10 | 5.16e-01 |
| GO:BP | GO:0070673 | response to interleukin-18 | 3 | 5 | 5.16e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 24 | 44 | 5.16e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 4 | 111 | 5.16e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 11 | 23 | 5.16e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 1 | 27 | 5.16e-01 |
| GO:BP | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol | 2 | 2 | 5.16e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 2 | 4 | 5.16e-01 |
| GO:BP | GO:0032252 | secretory granule localization | 1 | 9 | 5.16e-01 |
| GO:BP | GO:2001279 | regulation of unsaturated fatty acid biosynthetic process | 4 | 7 | 5.16e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 8 | 168 | 5.16e-01 |
| GO:BP | GO:1904020 | regulation of G protein-coupled receptor internalization | 3 | 3 | 5.16e-01 |
| GO:BP | GO:0071863 | regulation of cell proliferation in bone marrow | 2 | 6 | 5.16e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 7 | 55 | 5.16e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 12 | 5.16e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 3 | 1380 | 5.16e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 5.16e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 1 | 86 | 5.16e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 15 | 300 | 5.17e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 3 | 21 | 5.17e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 3 | 40 | 5.17e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 3 | 49 | 5.17e-01 |
| GO:BP | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine | 1 | 2 | 5.17e-01 |
| GO:BP | GO:2000056 | regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 2 | 13 | 5.17e-01 |
| GO:BP | GO:0031444 | slow-twitch skeletal muscle fiber contraction | 2 | 2 | 5.17e-01 |
| GO:BP | GO:2000057 | negative regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 15 | 23 | 5.17e-01 |
| GO:BP | GO:0002315 | marginal zone B cell differentiation | 1 | 6 | 5.17e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 1 | 6 | 5.17e-01 |
| GO:BP | GO:0006924 | activation-induced cell death of T cells | 1 | 8 | 5.17e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 5.17e-01 |
| GO:BP | GO:0051351 | positive regulation of ligase activity | 6 | 9 | 5.17e-01 |
| GO:BP | GO:0042930 | enterobactin transport | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 3 | 8 | 5.17e-01 |
| GO:BP | GO:1904184 | positive regulation of pyruvate dehydrogenase activity | 1 | 3 | 5.17e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 6 | 161 | 5.17e-01 |
| GO:BP | GO:0120322 | lipid modification by small protein conjugation | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0120323 | lipid ubiquitination | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 5 | 8 | 5.17e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 6 | 214 | 5.17e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 5.17e-01 |
| GO:BP | GO:0051006 | positive regulation of lipoprotein lipase activity | 1 | 4 | 5.17e-01 |
| GO:BP | GO:0010898 | positive regulation of triglyceride catabolic process | 1 | 4 | 5.17e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 5.17e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 2 | 5 | 5.17e-01 |
| GO:BP | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | 1 | 2 | 5.17e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 9 | 34 | 5.18e-01 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 5.18e-01 |
| GO:BP | GO:0045176 | apical protein localization | 1 | 13 | 5.18e-01 |
| GO:BP | GO:0002155 | regulation of thyroid hormone mediated signaling pathway | 2 | 5 | 5.18e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 3 | 85 | 5.18e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 18 | 34 | 5.18e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 5.18e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 12 | 43 | 5.18e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 5.18e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 5.18e-01 |
| GO:BP | GO:0046717 | acid secretion | 2 | 25 | 5.18e-01 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 1 | 17 | 5.18e-01 |
| GO:BP | GO:0000056 | ribosomal small subunit export from nucleus | 1 | 8 | 5.18e-01 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 1 | 17 | 5.18e-01 |
| GO:BP | GO:2001204 | regulation of osteoclast development | 1 | 4 | 5.18e-01 |
| GO:BP | GO:0090209 | negative regulation of triglyceride metabolic process | 1 | 7 | 5.18e-01 |
| GO:BP | GO:0061517 | macrophage proliferation | 1 | 7 | 5.19e-01 |
| GO:BP | GO:0002158 | osteoclast proliferation | 1 | 7 | 5.19e-01 |
| GO:BP | GO:0051451 | myoblast migration | 2 | 11 | 5.19e-01 |
| GO:BP | GO:0071374 | cellular response to parathyroid hormone stimulus | 1 | 7 | 5.19e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 4 | 30 | 5.19e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 27 | 186 | 5.19e-01 |
| GO:BP | GO:0042487 | regulation of odontogenesis of dentin-containing tooth | 1 | 6 | 5.19e-01 |
| GO:BP | GO:0061300 | cerebellum vasculature development | 2 | 2 | 5.19e-01 |
| GO:BP | GO:0060022 | hard palate development | 2 | 5 | 5.19e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 10 | 56 | 5.19e-01 |
| GO:BP | GO:0036359 | renal potassium excretion | 2 | 2 | 5.19e-01 |
| GO:BP | GO:0006592 | ornithine biosynthetic process | 1 | 1 | 5.19e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 1 | 7 | 5.20e-01 |
| GO:BP | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation | 2 | 2 | 5.20e-01 |
| GO:BP | GO:0060061 | Spemann organizer formation | 2 | 3 | 5.20e-01 |
| GO:BP | GO:1903689 | regulation of wound healing, spreading of epidermal cells | 1 | 8 | 5.20e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 8 | 68 | 5.20e-01 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 5 | 27 | 5.20e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 5.20e-01 |
| GO:BP | GO:0060301 | positive regulation of cytokine activity | 1 | 1 | 5.20e-01 |
| GO:BP | GO:1905040 | otic placode development | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0060510 | type II pneumocyte differentiation | 4 | 5 | 5.20e-01 |
| GO:BP | GO:1904645 | response to amyloid-beta | 8 | 32 | 5.20e-01 |
| GO:BP | GO:1902378 | VEGF-activated neuropilin signaling pathway involved in axon guidance | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 3 | 35 | 5.20e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 7 | 194 | 5.20e-01 |
| GO:BP | GO:0007402 | ganglion mother cell fate determination | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 2 | 26 | 5.20e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 2 | 2 | 5.20e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 2 | 1399 | 5.21e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 2 | 17 | 5.21e-01 |
| GO:BP | GO:1903387 | positive regulation of homophilic cell adhesion | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0046479 | glycosphingolipid catabolic process | 4 | 10 | 5.21e-01 |
| GO:BP | GO:1902426 | deactivation of mitotic spindle assembly checkpoint | 2 | 3 | 5.21e-01 |
| GO:BP | GO:0051589 | negative regulation of neurotransmitter transport | 1 | 8 | 5.21e-01 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 3 | 5.21e-01 |
| GO:BP | GO:1905179 | negative regulation of cardiac muscle tissue regeneration | 1 | 2 | 5.21e-01 |
| GO:BP | GO:1905178 | regulation of cardiac muscle tissue regeneration | 1 | 2 | 5.21e-01 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 3 | 5.21e-01 |
| GO:BP | GO:0007635 | chemosensory behavior | 1 | 10 | 5.21e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 6 | 98 | 5.21e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 8 | 115 | 5.21e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 8 | 115 | 5.21e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 1 | 8 | 5.21e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 5.22e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 5.22e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 2 | 6 | 5.22e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 5.22e-01 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 6 | 18 | 5.22e-01 |
| GO:BP | GO:0061458 | reproductive system development | 3 | 215 | 5.22e-01 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 3 | 35 | 5.22e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 1 | 540 | 5.22e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 5.23e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 10 | 22 | 5.23e-01 |
| GO:BP | GO:0035426 | extracellular matrix-cell signaling | 3 | 4 | 5.23e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 2 | 20 | 5.23e-01 |
| GO:BP | GO:0090251 | protein localization involved in establishment of planar polarity | 2 | 2 | 5.23e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 4 | 15 | 5.23e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 5 | 50 | 5.23e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 4 | 256 | 5.23e-01 |
| GO:BP | GO:0006547 | histidine metabolic process | 2 | 5 | 5.23e-01 |
| GO:BP | GO:2000642 | negative regulation of early endosome to late endosome transport | 2 | 2 | 5.23e-01 |
| GO:BP | GO:1905517 | macrophage migration | 3 | 34 | 5.23e-01 |
| GO:BP | GO:0006887 | exocytosis | 8 | 256 | 5.23e-01 |
| GO:BP | GO:0048560 | establishment of anatomical structure orientation | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0017085 | response to insecticide | 1 | 4 | 5.25e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 2 | 100 | 5.25e-01 |
| GO:BP | GO:0034759 | regulation of iron ion transmembrane transport | 1 | 2 | 5.25e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 4 | 17 | 5.25e-01 |
| GO:BP | GO:0060271 | cilium assembly | 4 | 292 | 5.25e-01 |
| GO:BP | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity | 2 | 2 | 5.25e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 5.25e-01 |
| GO:BP | GO:0015860 | purine nucleoside transmembrane transport | 1 | 3 | 5.25e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 1 | 40 | 5.25e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 30 | 660 | 5.26e-01 |
| GO:BP | GO:1901908 | diadenosine hexaphosphate metabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 5 | 7 | 5.26e-01 |
| GO:BP | GO:1901910 | adenosine 5’-(hexahydrogen pentaphosphate) metabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 5 | 120 | 5.26e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 3 | 21 | 5.26e-01 |
| GO:BP | GO:0071543 | diphosphoinositol polyphosphate metabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:1901911 | adenosine 5’-(hexahydrogen pentaphosphate) catabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0051792 | medium-chain fatty acid biosynthetic process | 2 | 4 | 5.26e-01 |
| GO:BP | GO:0019085 | early viral transcription | 2 | 2 | 5.26e-01 |
| GO:BP | GO:1901907 | diadenosine pentaphosphate catabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 44 | 140 | 5.26e-01 |
| GO:BP | GO:0042439 | ethanolamine-containing compound metabolic process | 1 | 5 | 5.26e-01 |
| GO:BP | GO:0007207 | phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | 2 | 6 | 5.26e-01 |
| GO:BP | GO:1901909 | diadenosine hexaphosphate catabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:1901906 | diadenosine pentaphosphate metabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 3 | 16 | 5.26e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 3 | 12 | 5.26e-01 |
| GO:BP | GO:0048874 | host-mediated regulation of intestinal microbiota composition | 1 | 2 | 5.26e-01 |
| GO:BP | GO:1905341 | negative regulation of protein localization to kinetochore | 1 | 1 | 5.26e-01 |
| GO:BP | GO:0061622 | glycolytic process through glucose-1-phosphate | 1 | 2 | 5.26e-01 |
| GO:BP | GO:0071262 | regulation of translational initiation in response to starvation | 2 | 2 | 5.26e-01 |
| GO:BP | GO:0071264 | positive regulation of translational initiation in response to starvation | 2 | 2 | 5.26e-01 |
| GO:BP | GO:0046849 | bone remodeling | 4 | 59 | 5.26e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 7 | 37 | 5.26e-01 |
| GO:BP | GO:0019254 | carnitine metabolic process, CoA-linked | 2 | 3 | 5.26e-01 |
| GO:BP | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis | 1 | 3 | 5.26e-01 |
| GO:BP | GO:0006527 | arginine catabolic process | 1 | 7 | 5.26e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 5 | 13 | 5.26e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 3 | 13 | 5.26e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 2 | 109 | 5.27e-01 |
| GO:BP | GO:1903911 | positive regulation of receptor clustering | 1 | 6 | 5.27e-01 |
| GO:BP | GO:1990262 | anti-Mullerian hormone signaling pathway | 1 | 2 | 5.27e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 6 | 27 | 5.27e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 7 | 10 | 5.27e-01 |
| GO:BP | GO:0048103 | somatic stem cell division | 4 | 10 | 5.27e-01 |
| GO:BP | GO:0030103 | vasopressin secretion | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0003358 | noradrenergic neuron development | 1 | 2 | 5.27e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 13 | 40 | 5.27e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 5.27e-01 |
| GO:BP | GO:1904226 | regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 5.27e-01 |
| GO:BP | GO:1904227 | negative regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 5.27e-01 |
| GO:BP | GO:1902951 | negative regulation of dendritic spine maintenance | 1 | 2 | 5.27e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 8 | 22 | 5.27e-01 |
| GO:BP | GO:0071879 | positive regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 5.27e-01 |
| GO:BP | GO:0040007 | growth | 3 | 742 | 5.27e-01 |
| GO:BP | GO:0010905 | negative regulation of UDP-glucose catabolic process | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0032480 | negative regulation of type I interferon production | 10 | 23 | 5.27e-01 |
| GO:BP | GO:0050894 | determination of affect | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 5.27e-01 |
| GO:BP | GO:0006837 | serotonin transport | 1 | 11 | 5.27e-01 |
| GO:BP | GO:0048312 | intracellular distribution of mitochondria | 1 | 5 | 5.27e-01 |
| GO:BP | GO:0010904 | regulation of UDP-glucose catabolic process | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 5.27e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 5.27e-01 |
| GO:BP | GO:0097497 | blood vessel endothelial cell delamination | 1 | 1 | 5.27e-01 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 6 | 9 | 5.27e-01 |
| GO:BP | GO:0044027 | negative regulation of gene expression via CpG island methylation | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0046084 | adenine biosynthetic process | 1 | 2 | 5.27e-01 |
| GO:BP | GO:1904257 | zinc ion import into Golgi lumen | 1 | 3 | 5.27e-01 |
| GO:BP | GO:1902938 | regulation of intracellular calcium activated chloride channel activity | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 2 | 4 | 5.27e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 113 | 376 | 5.28e-01 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 10 | 23 | 5.28e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 13 | 52 | 5.28e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 2 | 7 | 5.28e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 3 | 25 | 5.28e-01 |
| GO:BP | GO:0030393 | fructoselysine metabolic process | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0001731 | formation of translation preinitiation complex | 3 | 11 | 5.28e-01 |
| GO:BP | GO:2000975 | positive regulation of pro-B cell differentiation | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 10 | 28 | 5.28e-01 |
| GO:BP | GO:0019367 | fatty acid elongation, saturated fatty acid | 2 | 6 | 5.28e-01 |
| GO:BP | GO:0019368 | fatty acid elongation, unsaturated fatty acid | 2 | 6 | 5.28e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 3 | 25 | 5.28e-01 |
| GO:BP | GO:0061171 | establishment of bipolar cell polarity | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0016322 | neuron remodeling | 2 | 11 | 5.28e-01 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 2 | 6 | 5.28e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 3 | 122 | 5.28e-01 |
| GO:BP | GO:0008584 | male gonad development | 10 | 99 | 5.28e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 8 | 47 | 5.28e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 2 | 41 | 5.28e-01 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 2 | 20 | 5.28e-01 |
| GO:BP | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity | 1 | 3 | 5.28e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 3 | 538 | 5.28e-01 |
| GO:BP | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery | 4 | 6 | 5.28e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 5.28e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 5.28e-01 |
| GO:BP | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid | 2 | 6 | 5.28e-01 |
| GO:BP | GO:0070075 | tear secretion | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0031439 | positive regulation of mRNA cleavage | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0019043 | establishment of viral latency | 1 | 3 | 5.28e-01 |
| GO:BP | GO:0031437 | regulation of mRNA cleavage | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0034625 | fatty acid elongation, monounsaturated fatty acid | 2 | 6 | 5.28e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 1 | 12 | 5.28e-01 |
| GO:BP | GO:0045829 | negative regulation of isotype switching | 2 | 4 | 5.28e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 9 | 276 | 5.28e-01 |
| GO:BP | GO:0035378 | carbon dioxide transmembrane transport | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 4 | 605 | 5.28e-01 |
| GO:BP | GO:1905704 | positive regulation of inhibitory synapse assembly | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0072237 | metanephric proximal tubule development | 1 | 3 | 5.28e-01 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 1 | 11 | 5.28e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 3 | 18 | 5.28e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 2 | 31 | 5.28e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 2 | 5 | 5.28e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 1 | 25 | 5.28e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 3 | 52 | 5.28e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 29 | 5.28e-01 |
| GO:BP | GO:0007517 | muscle organ development | 3 | 277 | 5.28e-01 |
| GO:BP | GO:0046878 | positive regulation of saliva secretion | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0060353 | regulation of cell adhesion molecule production | 1 | 6 | 5.28e-01 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 1 | 11 | 5.28e-01 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 5.28e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 3 | 89 | 5.29e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 3 | 18 | 5.29e-01 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 4 | 9 | 5.29e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 3 | 18 | 5.29e-01 |
| GO:BP | GO:1904386 | response to L-phenylalanine derivative | 4 | 5 | 5.29e-01 |
| GO:BP | GO:0003331 | positive regulation of extracellular matrix constituent secretion | 1 | 4 | 5.29e-01 |
| GO:BP | GO:1905786 | positive regulation of anaphase-promoting complex-dependent catabolic process | 1 | 2 | 5.29e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 1 | 272 | 5.29e-01 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 2 | 13 | 5.29e-01 |
| GO:BP | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion | 1 | 1 | 5.30e-01 |
| GO:BP | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation | 3 | 4 | 5.30e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 4 | 76 | 5.30e-01 |
| GO:BP | GO:0060004 | reflex | 2 | 9 | 5.31e-01 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 1 | 16 | 5.31e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 3 | 1898 | 5.31e-01 |
| GO:BP | GO:0097473 | retinal rod cell apoptotic process | 2 | 2 | 5.31e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 8 | 18 | 5.31e-01 |
| GO:BP | GO:0015861 | cytidine transport | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0019408 | dolichol biosynthetic process | 1 | 2 | 5.31e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 1 | 12 | 5.31e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 5.31e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 1 | 12 | 5.31e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 1 | 7 | 5.31e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 6 | 34 | 5.31e-01 |
| GO:BP | GO:0034352 | positive regulation of glial cell apoptotic process | 2 | 2 | 5.31e-01 |
| GO:BP | GO:0009786 | regulation of asymmetric cell division | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0019482 | beta-alanine metabolic process | 1 | 2 | 5.31e-01 |
| GO:BP | GO:0006182 | cGMP biosynthetic process | 1 | 6 | 5.31e-01 |
| GO:BP | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 7 | 30 | 5.31e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 4 | 17 | 5.32e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 2 | 3 | 5.32e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 2 | 3 | 5.32e-01 |
| GO:BP | GO:0097043 | histone H3-K56 acetylation | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 1 | 8 | 5.32e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 1 | 9 | 5.32e-01 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 5.32e-01 |
| GO:BP | GO:0001796 | regulation of type IIa hypersensitivity | 2 | 3 | 5.32e-01 |
| GO:BP | GO:0002892 | regulation of type II hypersensitivity | 2 | 3 | 5.32e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 4 | 11 | 5.32e-01 |
| GO:BP | GO:0019860 | uracil metabolic process | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 9 | 62 | 5.32e-01 |
| GO:BP | GO:0007127 | meiosis I | 33 | 77 | 5.32e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 7 | 40 | 5.32e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 7 | 38 | 5.32e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 3 | 11 | 5.33e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 2 | 9 | 5.33e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 35 | 5.33e-01 |
| GO:BP | GO:0010724 | regulation of definitive erythrocyte differentiation | 2 | 2 | 5.33e-01 |
| GO:BP | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation | 1 | 5 | 5.33e-01 |
| GO:BP | GO:0042749 | regulation of circadian sleep/wake cycle | 1 | 9 | 5.33e-01 |
| GO:BP | GO:0060687 | regulation of branching involved in prostate gland morphogenesis | 1 | 3 | 5.33e-01 |
| GO:BP | GO:0062029 | positive regulation of stress granule assembly | 2 | 2 | 5.33e-01 |
| GO:BP | GO:0090245 | axis elongation involved in somitogenesis | 1 | 4 | 5.33e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 10 | 100 | 5.33e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 3 | 4 | 5.33e-01 |
| GO:BP | GO:0010722 | regulation of ferrochelatase activity | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0060311 | negative regulation of elastin catabolic process | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0060310 | regulation of elastin catabolic process | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0007276 | gamete generation | 2 | 461 | 5.33e-01 |
| GO:BP | GO:0070189 | kynurenine metabolic process | 1 | 6 | 5.33e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 3 | 4 | 5.33e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 5 | 796 | 5.33e-01 |
| GO:BP | GO:0140673 | transcription elongation-coupled chromatin remodeling | 2 | 2 | 5.33e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 3 | 4 | 5.33e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 7 | 54 | 5.33e-01 |
| GO:BP | GO:0010711 | negative regulation of collagen catabolic process | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0060927 | cardiac pacemaker cell fate commitment | 2 | 3 | 5.33e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 5 | 68 | 5.33e-01 |
| GO:BP | GO:0140588 | chromatin looping | 2 | 2 | 5.33e-01 |
| GO:BP | GO:0051728 | cell cycle switching, mitotic to meiotic cell cycle | 1 | 2 | 5.33e-01 |
| GO:BP | GO:0060184 | cell cycle switching | 1 | 2 | 5.33e-01 |
| GO:BP | GO:0051729 | germline cell cycle switching, mitotic to meiotic cell cycle | 1 | 2 | 5.33e-01 |
| GO:BP | GO:0071788 | endoplasmic reticulum tubular network maintenance | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0007507 | heart development | 1 | 506 | 5.33e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 5.33e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 8 | 48 | 5.33e-01 |
| GO:BP | GO:0014738 | regulation of muscle hyperplasia | 1 | 1 | 5.34e-01 |
| GO:BP | GO:0010710 | regulation of collagen catabolic process | 2 | 3 | 5.34e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 4 | 9 | 5.34e-01 |
| GO:BP | GO:0019694 | alkanesulfonate metabolic process | 5 | 6 | 5.34e-01 |
| GO:BP | GO:0060309 | elastin catabolic process | 2 | 2 | 5.34e-01 |
| GO:BP | GO:0007220 | Notch receptor processing | 6 | 9 | 5.34e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 5.34e-01 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0046048 | UDP metabolic process | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0019530 | taurine metabolic process | 5 | 6 | 5.34e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 4 | 9 | 5.34e-01 |
| GO:BP | GO:1904491 | protein localization to ciliary transition zone | 2 | 7 | 5.34e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 2 | 104 | 5.34e-01 |
| GO:BP | GO:0014906 | myotube cell development involved in skeletal muscle regeneration | 1 | 1 | 5.34e-01 |
| GO:BP | GO:0072678 | T cell migration | 5 | 38 | 5.34e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 5.34e-01 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 8 | 27 | 5.34e-01 |
| GO:BP | GO:0019236 | response to pheromone | 1 | 1 | 5.34e-01 |
| GO:BP | GO:0042074 | cell migration involved in gastrulation | 5 | 10 | 5.34e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 7 | 75 | 5.34e-01 |
| GO:BP | GO:0001889 | liver development | 1 | 110 | 5.34e-01 |
| GO:BP | GO:0046452 | dihydrofolate metabolic process | 1 | 3 | 5.35e-01 |
| GO:BP | GO:2000815 | regulation of mRNA stability involved in response to oxidative stress | 2 | 2 | 5.35e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 8 | 23 | 5.35e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 10 | 5.35e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 10 | 129 | 5.35e-01 |
| GO:BP | GO:1904877 | positive regulation of DNA ligase activity | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 5.35e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 2 | 39 | 5.35e-01 |
| GO:BP | GO:0045190 | isotype switching | 2 | 39 | 5.35e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 2 | 39 | 5.35e-01 |
| GO:BP | GO:1903093 | regulation of protein K48-linked deubiquitination | 1 | 3 | 5.35e-01 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 2 | 13 | 5.35e-01 |
| GO:BP | GO:0051490 | negative regulation of filopodium assembly | 4 | 6 | 5.35e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 9 | 40 | 5.35e-01 |
| GO:BP | GO:0061365 | positive regulation of triglyceride lipase activity | 1 | 4 | 5.35e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 2 | 16 | 5.35e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 5.36e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 2 | 26 | 5.36e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 4 | 6 | 5.36e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 5.36e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 4 | 6 | 5.36e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 4 | 6 | 5.36e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 4 | 6 | 5.36e-01 |
| GO:BP | GO:1905267 | endonucleolytic cleavage involved in tRNA processing | 2 | 3 | 5.36e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 4 | 6 | 5.36e-01 |
| GO:BP | GO:1905552 | positive regulation of protein localization to endoplasmic reticulum | 2 | 2 | 5.36e-01 |
| GO:BP | GO:0039533 | regulation of MDA-5 signaling pathway | 1 | 5 | 5.36e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 3 | 52 | 5.36e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 1 | 17 | 5.36e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 26 | 71 | 5.36e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 5.36e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 10 | 21 | 5.36e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0089700 | protein kinase D signaling | 3 | 4 | 5.36e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1902073 | positive regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 11 | 22 | 5.36e-01 |
| GO:BP | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 5.37e-01 |
| GO:BP | GO:2001223 | negative regulation of neuron migration | 3 | 8 | 5.37e-01 |
| GO:BP | GO:0008355 | olfactory learning | 2 | 2 | 5.37e-01 |
| GO:BP | GO:0007270 | neuron-neuron synaptic transmission | 4 | 5 | 5.37e-01 |
| GO:BP | GO:0050930 | induction of positive chemotaxis | 1 | 12 | 5.37e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 5 | 25 | 5.37e-01 |
| GO:BP | GO:2000312 | regulation of kainate selective glutamate receptor activity | 1 | 3 | 5.37e-01 |
| GO:BP | GO:0140569 | extraction of mislocalized protein from ER membrane | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0010836 | negative regulation of protein ADP-ribosylation | 2 | 4 | 5.38e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 3 | 59 | 5.38e-01 |
| GO:BP | GO:0036369 | transcription factor catabolic process | 3 | 5 | 5.38e-01 |
| GO:BP | GO:0072098 | anterior/posterior pattern specification involved in kidney development | 2 | 2 | 5.38e-01 |
| GO:BP | GO:0070202 | regulation of establishment of protein localization to chromosome | 4 | 12 | 5.38e-01 |
| GO:BP | GO:0030220 | platelet formation | 1 | 17 | 5.38e-01 |
| GO:BP | GO:0061227 | pattern specification involved in mesonephros development | 2 | 2 | 5.38e-01 |
| GO:BP | GO:0048389 | intermediate mesoderm development | 2 | 2 | 5.38e-01 |
| GO:BP | GO:0071918 | urea transmembrane transport | 1 | 2 | 5.38e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 3 | 104 | 5.38e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 1 | 112 | 5.38e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 2 | 6 | 5.38e-01 |
| GO:BP | GO:0140157 | ammonium import across plasma membrane | 1 | 3 | 5.38e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 4 | 16 | 5.38e-01 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 1 | 7 | 5.38e-01 |
| GO:BP | GO:0106227 | peptidyl-lysine glutarylation | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 3 | 5 | 5.38e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 1 | 82 | 5.38e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 2 | 22 | 5.38e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 4 | 6 | 5.39e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 30 | 90 | 5.39e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 1 | 39 | 5.39e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 5.39e-01 |
| GO:BP | GO:0033169 | histone H3-K9 demethylation | 1 | 5 | 5.39e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 7 | 15 | 5.39e-01 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0140066 | peptidyl-lysine crotonylation | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 3 | 36 | 5.39e-01 |
| GO:BP | GO:1902811 | positive regulation of skeletal muscle fiber differentiation | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 37 | 96 | 5.39e-01 |
| GO:BP | GO:0090679 | cell differentiation involved in phenotypic switching | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0090088 | regulation of oligopeptide transport | 2 | 2 | 5.39e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 6 | 17 | 5.39e-01 |
| GO:BP | GO:2000625 | regulation of miRNA catabolic process | 1 | 5 | 5.39e-01 |
| GO:BP | GO:1905930 | regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0046041 | ITP metabolic process | 2 | 2 | 5.39e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 13 | 53 | 5.39e-01 |
| GO:BP | GO:0090089 | regulation of dipeptide transport | 2 | 2 | 5.39e-01 |
| GO:BP | GO:0009756 | carbohydrate mediated signaling | 1 | 6 | 5.39e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 16 | 53 | 5.39e-01 |
| GO:BP | GO:1905915 | regulation of cell differentiation involved in phenotypic switching | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0051900 | regulation of mitochondrial depolarization | 5 | 15 | 5.39e-01 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 1 | 8 | 5.39e-01 |
| GO:BP | GO:0061090 | positive regulation of sequestering of zinc ion | 1 | 1 | 5.39e-01 |
| GO:BP | GO:2000878 | positive regulation of oligopeptide transport | 2 | 2 | 5.39e-01 |
| GO:BP | GO:0009203 | ribonucleoside triphosphate catabolic process | 2 | 2 | 5.39e-01 |
| GO:BP | GO:2000880 | positive regulation of dipeptide transport | 2 | 2 | 5.39e-01 |
| GO:BP | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential | 3 | 4 | 5.39e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 2 | 20 | 5.39e-01 |
| GO:BP | GO:0035655 | interleukin-18-mediated signaling pathway | 2 | 3 | 5.39e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 5.39e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 5 | 22 | 5.39e-01 |
| GO:BP | GO:2001150 | positive regulation of dipeptide transmembrane transport | 2 | 2 | 5.39e-01 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 2 | 20 | 5.39e-01 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 1 | 8 | 5.39e-01 |
| GO:BP | GO:1905460 | negative regulation of vascular associated smooth muscle cell apoptotic process | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 1 | 45 | 5.39e-01 |
| GO:BP | GO:0002101 | tRNA wobble cytosine modification | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0002182 | cytoplasmic translational elongation | 6 | 12 | 5.39e-01 |
| GO:BP | GO:2001148 | regulation of dipeptide transmembrane transport | 2 | 2 | 5.39e-01 |
| GO:BP | GO:1905420 | vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0140914 | zinc ion import into secretory vesicle | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0140917 | zinc ion import into mitochondrion | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0140915 | zinc ion import into zymogen granule | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0071351 | cellular response to interleukin-18 | 2 | 3 | 5.39e-01 |
| GO:BP | GO:0140916 | zinc ion import into lysosome | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0001946 | lymphangiogenesis | 5 | 12 | 5.40e-01 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 1 | 10 | 5.40e-01 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 1 | 16 | 5.40e-01 |
| GO:BP | GO:0007371 | ventral midline determination | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0072285 | mesenchymal to epithelial transition involved in metanephric renal vesicle formation | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0072036 | mesenchymal to epithelial transition involved in renal vesicle formation | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 5.40e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 1 | 14 | 5.40e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 2 | 2 | 5.40e-01 |
| GO:BP | GO:1901289 | succinyl-CoA catabolic process | 4 | 6 | 5.40e-01 |
| GO:BP | GO:1905533 | negative regulation of leucine import across plasma membrane | 1 | 2 | 5.40e-01 |
| GO:BP | GO:0060358 | negative regulation of leucine import | 1 | 2 | 5.40e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 6 | 21 | 5.40e-01 |
| GO:BP | GO:0060357 | regulation of leucine import | 1 | 2 | 5.40e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 5.40e-01 |
| GO:BP | GO:0015918 | sterol transport | 3 | 92 | 5.40e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 1 | 10 | 5.40e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 112 | 375 | 5.40e-01 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 5.40e-01 |
| GO:BP | GO:1990379 | lipid transport across blood-brain barrier | 2 | 6 | 5.40e-01 |
| GO:BP | GO:1905809 | negative regulation of synapse organization | 1 | 8 | 5.40e-01 |
| GO:BP | GO:0034635 | glutathione transport | 6 | 9 | 5.40e-01 |
| GO:BP | GO:0071072 | negative regulation of phospholipid biosynthetic process | 2 | 6 | 5.40e-01 |
| GO:BP | GO:0036525 | protein deglycation | 2 | 2 | 5.40e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 5.40e-01 |
| GO:BP | GO:1905259 | negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 2 | 2 | 5.40e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 3 | 9 | 5.40e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | 7 | 5.40e-01 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 1 | 2 | 5.40e-01 |
| GO:BP | GO:1905258 | regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 2 | 2 | 5.40e-01 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 1 | 2 | 5.40e-01 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 5.40e-01 |
| GO:BP | GO:0006882 | intracellular zinc ion homeostasis | 5 | 28 | 5.40e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 2 | 1075 | 5.40e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 4 | 86 | 5.40e-01 |
| GO:BP | GO:1901307 | positive regulation of spermidine biosynthetic process | 1 | 1 | 5.40e-01 |
| GO:BP | GO:1901304 | regulation of spermidine biosynthetic process | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway | 5 | 8 | 5.40e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 5 | 68 | 5.40e-01 |
| GO:BP | GO:0035507 | regulation of myosin-light-chain-phosphatase activity | 1 | 5 | 5.41e-01 |
| GO:BP | GO:0045041 | protein import into mitochondrial intermembrane space | 2 | 3 | 5.41e-01 |
| GO:BP | GO:0001890 | placenta development | 1 | 117 | 5.41e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 2 | 29 | 5.41e-01 |
| GO:BP | GO:0061502 | early endosome to recycling endosome transport | 1 | 3 | 5.41e-01 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 2 | 3 | 5.41e-01 |
| GO:BP | GO:0006559 | L-phenylalanine catabolic process | 2 | 3 | 5.41e-01 |
| GO:BP | GO:0061824 | cytosolic ciliogenesis | 1 | 1 | 5.41e-01 |
| GO:BP | GO:0106300 | protein-DNA covalent cross-linking repair | 3 | 7 | 5.41e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 9 | 14 | 5.41e-01 |
| GO:BP | GO:1902222 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process | 2 | 3 | 5.41e-01 |
| GO:BP | GO:0006558 | L-phenylalanine metabolic process | 2 | 3 | 5.41e-01 |
| GO:BP | GO:1905464 | positive regulation of DNA duplex unwinding | 1 | 3 | 5.41e-01 |
| GO:BP | GO:0009386 | translational attenuation | 1 | 2 | 5.41e-01 |
| GO:BP | GO:1905776 | positive regulation of DNA helicase activity | 1 | 3 | 5.41e-01 |
| GO:BP | GO:0090023 | positive regulation of neutrophil chemotaxis | 2 | 14 | 5.42e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 3 | 195 | 5.42e-01 |
| GO:BP | GO:0110112 | regulation of lipid transporter activity | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 1 | 7 | 5.42e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 15 | 24 | 5.42e-01 |
| GO:BP | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway | 2 | 3 | 5.42e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 3 | 25 | 5.42e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 5.42e-01 |
| GO:BP | GO:0042136 | neurotransmitter biosynthetic process | 1 | 5 | 5.42e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 3 | 32 | 5.42e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 2 | 52 | 5.42e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 82 | 273 | 5.42e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 55 | 257 | 5.42e-01 |
| GO:BP | GO:0097576 | vacuole fusion | 1 | 12 | 5.42e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 5 | 90 | 5.42e-01 |
| GO:BP | GO:0010936 | negative regulation of macrophage cytokine production | 1 | 7 | 5.43e-01 |
| GO:BP | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 4 | 5.43e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 3 | 106 | 5.43e-01 |
| GO:BP | GO:0099502 | calcium-dependent activation of synaptic vesicle fusion | 1 | 4 | 5.43e-01 |
| GO:BP | GO:1901634 | positive regulation of synaptic vesicle membrane organization | 1 | 4 | 5.43e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 2 | 28 | 5.43e-01 |
| GO:BP | GO:0006549 | isoleucine metabolic process | 1 | 6 | 5.43e-01 |
| GO:BP | GO:0000348 | mRNA branch site recognition | 1 | 1 | 5.43e-01 |
| GO:BP | GO:0099041 | vesicle tethering to Golgi | 3 | 5 | 5.43e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 5.43e-01 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 2 | 44 | 5.43e-01 |
| GO:BP | GO:0051646 | mitochondrion localization | 1 | 47 | 5.43e-01 |
| GO:BP | GO:0043278 | response to morphine | 2 | 16 | 5.43e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 3 | 19 | 5.43e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 2 | 16 | 5.43e-01 |
| GO:BP | GO:0046070 | dGTP metabolic process | 2 | 3 | 5.43e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 1 | 19 | 5.44e-01 |
| GO:BP | GO:0106020 | regulation of vesicle docking | 1 | 2 | 5.44e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 2 | 4 | 5.44e-01 |
| GO:BP | GO:0072733 | response to staurosporine | 1 | 3 | 5.44e-01 |
| GO:BP | GO:0072734 | cellular response to staurosporine | 1 | 3 | 5.44e-01 |
| GO:BP | GO:0007386 | compartment pattern specification | 2 | 5 | 5.44e-01 |
| GO:BP | GO:0072144 | glomerular mesangial cell development | 2 | 4 | 5.44e-01 |
| GO:BP | GO:0021603 | cranial nerve formation | 1 | 4 | 5.44e-01 |
| GO:BP | GO:0016553 | base conversion or substitution editing | 1 | 10 | 5.44e-01 |
| GO:BP | GO:0006553 | lysine metabolic process | 2 | 7 | 5.44e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 5.44e-01 |
| GO:BP | GO:0035965 | cardiolipin acyl-chain remodeling | 1 | 4 | 5.44e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 2 | 433 | 5.44e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 8 | 24 | 5.44e-01 |
| GO:BP | GO:1905794 | response to puromycin | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 11 | 486 | 5.44e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 10 | 68 | 5.44e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 1 | 6 | 5.44e-01 |
| GO:BP | GO:0015744 | succinate transport | 1 | 6 | 5.44e-01 |
| GO:BP | GO:1905795 | cellular response to puromycin | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process | 3 | 4 | 5.44e-01 |
| GO:BP | GO:0061470 | T follicular helper cell differentiation | 2 | 4 | 5.44e-01 |
| GO:BP | GO:0060312 | regulation of blood vessel remodeling | 1 | 4 | 5.45e-01 |
| GO:BP | GO:1904210 | VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.45e-01 |
| GO:BP | GO:1904240 | negative regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.45e-01 |
| GO:BP | GO:1904239 | regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0006880 | intracellular sequestering of iron ion | 3 | 3 | 5.45e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 3 | 556 | 5.45e-01 |
| GO:BP | GO:0001661 | conditioned taste aversion | 1 | 2 | 5.45e-01 |
| GO:BP | GO:0006590 | thyroid hormone generation | 4 | 10 | 5.45e-01 |
| GO:BP | GO:0006667 | sphinganine metabolic process | 2 | 2 | 5.45e-01 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 5.45e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 11 | 23 | 5.45e-01 |
| GO:BP | GO:0070408 | carbamoyl phosphate metabolic process | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0070409 | carbamoyl phosphate biosynthetic process | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 5.45e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 5.45e-01 |
| GO:BP | GO:1903945 | positive regulation of hepatocyte apoptotic process | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0015779 | glucuronoside transport | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0051866 | general adaptation syndrome | 2 | 4 | 5.45e-01 |
| GO:BP | GO:0046866 | tetraterpenoid transport | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0046867 | carotenoid transport | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 3 | 9 | 5.45e-01 |
| GO:BP | GO:0071929 | alpha-tubulin acetylation | 2 | 3 | 5.45e-01 |
| GO:BP | GO:0070649 | formin-nucleated actin cable assembly | 2 | 3 | 5.46e-01 |
| GO:BP | GO:0110009 | formin-nucleated actin cable organization | 2 | 3 | 5.46e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 5.46e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 3 | 51 | 5.46e-01 |
| GO:BP | GO:0061944 | negative regulation of protein K48-linked ubiquitination | 1 | 2 | 5.46e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 1 | 24 | 5.46e-01 |
| GO:BP | GO:0035962 | response to interleukin-13 | 1 | 1 | 5.46e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 3 | 1378 | 5.47e-01 |
| GO:BP | GO:0032534 | regulation of microvillus assembly | 3 | 6 | 5.47e-01 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 3 | 4 | 5.47e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 6 | 213 | 5.47e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 3 | 29 | 5.47e-01 |
| GO:BP | GO:0016198 | axon choice point recognition | 3 | 5 | 5.47e-01 |
| GO:BP | GO:0098751 | bone cell development | 2 | 26 | 5.47e-01 |
| GO:BP | GO:0070100 | negative regulation of chemokine-mediated signaling pathway | 3 | 6 | 5.47e-01 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 2 | 17 | 5.47e-01 |
| GO:BP | GO:0036302 | atrioventricular canal development | 6 | 12 | 5.47e-01 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 11 | 20 | 5.47e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 1 | 8 | 5.47e-01 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 7 | 10 | 5.47e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 27 | 99 | 5.47e-01 |
| GO:BP | GO:1904556 | L-tryptophan transmembrane transport | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0015824 | proline transport | 2 | 11 | 5.47e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 5.48e-01 |
| GO:BP | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 1 | 2 | 5.48e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 2 | 17 | 5.48e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 2 | 17 | 5.48e-01 |
| GO:BP | GO:0098779 | positive regulation of mitophagy in response to mitochondrial depolarization | 5 | 10 | 5.48e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 4 | 72 | 5.48e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 7 | 10 | 5.48e-01 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 3 | 22 | 5.48e-01 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 1 | 14 | 5.48e-01 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 5.48e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 5.48e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 4 | 70 | 5.48e-01 |
| GO:BP | GO:2000589 | regulation of metanephric mesenchymal cell migration | 1 | 3 | 5.48e-01 |
| GO:BP | GO:0035789 | metanephric mesenchymal cell migration | 1 | 3 | 5.48e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 6 | 19 | 5.48e-01 |
| GO:BP | GO:1905673 | positive regulation of lysosome organization | 1 | 1 | 5.48e-01 |
| GO:BP | GO:0097466 | ubiquitin-dependent glycoprotein ERAD pathway | 1 | 5 | 5.49e-01 |
| GO:BP | GO:1904529 | regulation of actin filament binding | 1 | 3 | 5.49e-01 |
| GO:BP | GO:1904382 | mannose trimming involved in glycoprotein ERAD pathway | 1 | 5 | 5.49e-01 |
| GO:BP | GO:0007281 | germ cell development | 1 | 187 | 5.49e-01 |
| GO:BP | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process | 1 | 5 | 5.49e-01 |
| GO:BP | GO:1904426 | positive regulation of GTP binding | 1 | 3 | 5.49e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 9 | 40 | 5.49e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 5.49e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 1 | 28 | 5.49e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 3 | 16 | 5.49e-01 |
| GO:BP | GO:1905603 | regulation of blood-brain barrier permeability | 5 | 6 | 5.49e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 4 | 18 | 5.49e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 1 | 26 | 5.49e-01 |
| GO:BP | GO:0140647 | P450-containing electron transport chain | 1 | 2 | 5.49e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 10 | 30 | 5.49e-01 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 1 | 5 | 5.49e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 7 | 14 | 5.49e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 8 | 15 | 5.49e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 5.50e-01 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 5.50e-01 |
| GO:BP | GO:0040038 | polar body extrusion after meiotic divisions | 3 | 5 | 5.50e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 5.50e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 9 | 99 | 5.50e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 9 | 99 | 5.50e-01 |
| GO:BP | GO:0045046 | protein import into peroxisome membrane | 3 | 6 | 5.50e-01 |
| GO:BP | GO:1902268 | negative regulation of polyamine transmembrane transport | 1 | 3 | 5.50e-01 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 9 | 33 | 5.50e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 6 | 11 | 5.50e-01 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 9 | 33 | 5.50e-01 |
| GO:BP | GO:0062036 | sensory perception of hot stimulus | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0120168 | detection of hot stimulus involved in thermoception | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 3 | 62 | 5.50e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 1 | 5 | 5.50e-01 |
| GO:BP | GO:1903537 | meiotic cell cycle process involved in oocyte maturation | 1 | 5 | 5.50e-01 |
| GO:BP | GO:0006567 | threonine catabolic process | 3 | 3 | 5.51e-01 |
| GO:BP | GO:0032349 | positive regulation of aldosterone biosynthetic process | 1 | 3 | 5.51e-01 |
| GO:BP | GO:0032346 | positive regulation of aldosterone metabolic process | 1 | 3 | 5.51e-01 |
| GO:BP | GO:2000224 | regulation of testosterone biosynthetic process | 1 | 2 | 5.51e-01 |
| GO:BP | GO:0061184 | positive regulation of dermatome development | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0006996 | organelle organization | 62 | 3000 | 5.51e-01 |
| GO:BP | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis | 2 | 4 | 5.51e-01 |
| GO:BP | GO:0009566 | fertilization | 2 | 93 | 5.51e-01 |
| GO:BP | GO:0021796 | cerebral cortex regionalization | 1 | 2 | 5.51e-01 |
| GO:BP | GO:0010832 | negative regulation of myotube differentiation | 6 | 10 | 5.51e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 2 | 834 | 5.51e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 20 | 893 | 5.51e-01 |
| GO:BP | GO:0000389 | mRNA 3’-splice site recognition | 1 | 6 | 5.51e-01 |
| GO:BP | GO:0030324 | lung development | 2 | 146 | 5.52e-01 |
| GO:BP | GO:0072682 | eosinophil extravasation | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 1 | 104 | 5.52e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 5.52e-01 |
| GO:BP | GO:0016255 | attachment of GPI anchor to protein | 3 | 6 | 5.52e-01 |
| GO:BP | GO:0033342 | negative regulation of collagen binding | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0002334 | transitional two stage B cell differentiation | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0098907 | regulation of SA node cell action potential | 3 | 7 | 5.52e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 5 | 23 | 5.52e-01 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 1 | 36 | 5.52e-01 |
| GO:BP | GO:0003218 | cardiac left ventricle formation | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 1 | 36 | 5.52e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 1 | 19 | 5.52e-01 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 1 | 36 | 5.52e-01 |
| GO:BP | GO:0036093 | germ cell proliferation | 3 | 7 | 5.52e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 4 | 30 | 5.52e-01 |
| GO:BP | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation | 3 | 3 | 5.52e-01 |
| GO:BP | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 5 | 23 | 5.52e-01 |
| GO:BP | GO:0045730 | respiratory burst | 1 | 21 | 5.52e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 1 | 4 | 5.52e-01 |
| GO:BP | GO:0048268 | clathrin coat assembly | 1 | 13 | 5.52e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 5 | 23 | 5.52e-01 |
| GO:BP | GO:2000419 | regulation of eosinophil extravasation | 1 | 1 | 5.52e-01 |
| GO:BP | GO:2000420 | negative regulation of eosinophil extravasation | 1 | 1 | 5.52e-01 |
| GO:BP | GO:2000609 | regulation of thyroid hormone generation | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0038173 | interleukin-17A-mediated signaling pathway | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 10 | 31 | 5.52e-01 |
| GO:BP | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration | 4 | 6 | 5.52e-01 |
| GO:BP | GO:0071873 | response to norepinephrine | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0071874 | cellular response to norepinephrine stimulus | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0043972 | histone H3-K23 acetylation | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0110012 | protein localization to P-body | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 5.52e-01 |
| GO:BP | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure | 3 | 14 | 5.52e-01 |
| GO:BP | GO:0010359 | regulation of anion channel activity | 4 | 5 | 5.52e-01 |
| GO:BP | GO:0099022 | vesicle tethering | 15 | 31 | 5.52e-01 |
| GO:BP | GO:0035247 | peptidyl-arginine omega-N-methylation | 1 | 3 | 5.52e-01 |
| GO:BP | GO:0031064 | negative regulation of histone deacetylation | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0045653 | negative regulation of megakaryocyte differentiation | 1 | 17 | 5.52e-01 |
| GO:BP | GO:0003158 | endothelium development | 10 | 104 | 5.52e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 17 | 56 | 5.52e-01 |
| GO:BP | GO:0035212 | cell competition in a multicellular organism | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0032714 | negative regulation of interleukin-5 production | 2 | 2 | 5.52e-01 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 5.52e-01 |
| GO:BP | GO:0046321 | positive regulation of fatty acid oxidation | 10 | 15 | 5.53e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 5 | 107 | 5.53e-01 |
| GO:BP | GO:0002414 | immunoglobulin transcytosis in epithelial cells | 1 | 2 | 5.53e-01 |
| GO:BP | GO:0090667 | cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 5.53e-01 |
| GO:BP | GO:0090668 | endothelial cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 5.53e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 2 | 39 | 5.53e-01 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 4 | 8 | 5.53e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 4 | 5 | 5.53e-01 |
| GO:BP | GO:2000454 | positive regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 5.54e-01 |
| GO:BP | GO:0033023 | mast cell homeostasis | 3 | 4 | 5.54e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 6 | 37 | 5.54e-01 |
| GO:BP | GO:0032425 | positive regulation of mismatch repair | 2 | 2 | 5.54e-01 |
| GO:BP | GO:0070899 | mitochondrial tRNA wobble uridine modification | 1 | 1 | 5.54e-01 |
| GO:BP | GO:0007611 | learning or memory | 5 | 202 | 5.54e-01 |
| GO:BP | GO:0003332 | negative regulation of extracellular matrix constituent secretion | 2 | 2 | 5.55e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 5.55e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 15 | 30 | 5.55e-01 |
| GO:BP | GO:0000390 | spliceosomal complex disassembly | 2 | 3 | 5.55e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 5 | 133 | 5.55e-01 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 5.55e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 9 | 99 | 5.55e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 41 | 99 | 5.55e-01 |
| GO:BP | GO:0022414 | reproductive process | 3 | 927 | 5.55e-01 |
| GO:BP | GO:0110135 | Norrin signaling pathway | 2 | 2 | 5.55e-01 |
| GO:BP | GO:0001544 | initiation of primordial ovarian follicle growth | 1 | 1 | 5.55e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 5.55e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 2 | 41 | 5.55e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 4 | 6 | 5.55e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 3 | 5 | 5.56e-01 |
| GO:BP | GO:0015840 | urea transport | 1 | 2 | 5.56e-01 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 2 | 9 | 5.56e-01 |
| GO:BP | GO:0071838 | cell proliferation in bone marrow | 2 | 7 | 5.56e-01 |
| GO:BP | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 1 | 6 | 5.56e-01 |
| GO:BP | GO:0051013 | microtubule severing | 1 | 7 | 5.56e-01 |
| GO:BP | GO:0033762 | response to glucagon | 2 | 13 | 5.56e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 3 | 48 | 5.56e-01 |
| GO:BP | GO:0042745 | circadian sleep/wake cycle | 1 | 11 | 5.56e-01 |
| GO:BP | GO:0090107 | regulation of high-density lipoprotein particle assembly | 1 | 3 | 5.56e-01 |
| GO:BP | GO:2000234 | positive regulation of rRNA processing | 1 | 10 | 5.56e-01 |
| GO:BP | GO:0044782 | cilium organization | 4 | 313 | 5.56e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 3 | 37 | 5.56e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 7 | 165 | 5.56e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 5.56e-01 |
| GO:BP | GO:0032259 | methylation | 1 | 313 | 5.57e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 5.57e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 5.57e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 17 | 31 | 5.57e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 3 | 57 | 5.57e-01 |
| GO:BP | GO:0045002 | double-strand break repair via single-strand annealing | 4 | 7 | 5.57e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 4 | 160 | 5.57e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 16 | 36 | 5.57e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 3 | 7 | 5.57e-01 |
| GO:BP | GO:0044860 | protein localization to plasma membrane raft | 1 | 4 | 5.58e-01 |
| GO:BP | GO:0035752 | lysosomal lumen pH elevation | 3 | 3 | 5.58e-01 |
| GO:BP | GO:0000003 | reproduction | 3 | 934 | 5.58e-01 |
| GO:BP | GO:0110017 | cap-independent translational initiation of linear mRNA | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 7 | 25 | 5.58e-01 |
| GO:BP | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway | 3 | 6 | 5.58e-01 |
| GO:BP | GO:0030889 | negative regulation of B cell proliferation | 3 | 9 | 5.58e-01 |
| GO:BP | GO:1902747 | negative regulation of lens fiber cell differentiation | 3 | 5 | 5.58e-01 |
| GO:BP | GO:0002192 | IRES-dependent translational initiation of linear mRNA | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0098967 | exocytic insertion of neurotransmitter receptor to postsynaptic membrane | 3 | 5 | 5.58e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 5.58e-01 |
| GO:BP | GO:0038154 | interleukin-11-mediated signaling pathway | 2 | 3 | 5.58e-01 |
| GO:BP | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process | 4 | 5 | 5.58e-01 |
| GO:BP | GO:0072137 | condensed mesenchymal cell proliferation | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 7 | 310 | 5.58e-01 |
| GO:BP | GO:0006659 | phosphatidylserine biosynthetic process | 2 | 4 | 5.58e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 5.58e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 2 | 117 | 5.58e-01 |
| GO:BP | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process | 1 | 2 | 5.58e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 3 | 28 | 5.58e-01 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 1 | 6 | 5.58e-01 |
| GO:BP | GO:0060086 | circadian temperature homeostasis | 2 | 3 | 5.58e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 8 | 15 | 5.58e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 3 | 11 | 5.58e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 6 | 18 | 5.58e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0060479 | lung cell differentiation | 6 | 18 | 5.58e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 5.58e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 2 | 150 | 5.58e-01 |
| GO:BP | GO:0014047 | glutamate secretion | 1 | 14 | 5.58e-01 |
| GO:BP | GO:0046951 | ketone body biosynthetic process | 4 | 5 | 5.59e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 5 | 33 | 5.59e-01 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 1 | 13 | 5.59e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 2 | 24 | 5.59e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 71 | 344 | 5.59e-01 |
| GO:BP | GO:0048265 | response to pain | 2 | 17 | 5.59e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 52 | 452 | 5.59e-01 |
| GO:BP | GO:0072680 | extracellular matrix-dependent thymocyte migration | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 6 | 10 | 5.59e-01 |
| GO:BP | GO:0038163 | thrombopoietin-mediated signaling pathway | 2 | 3 | 5.59e-01 |
| GO:BP | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay | 1 | 3 | 5.59e-01 |
| GO:BP | GO:0072681 | fibronectin-dependent thymocyte migration | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 7 | 131 | 5.59e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 5.59e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 1 | 15 | 5.59e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 12 | 90 | 5.59e-01 |
| GO:BP | GO:0032765 | positive regulation of mast cell cytokine production | 3 | 3 | 5.59e-01 |
| GO:BP | GO:2000415 | positive regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 8 | 83 | 5.59e-01 |
| GO:BP | GO:0045186 | zonula adherens assembly | 1 | 1 | 5.59e-01 |
| GO:BP | GO:2000413 | regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0032967 | positive regulation of collagen biosynthetic process | 4 | 23 | 5.59e-01 |
| GO:BP | GO:0010747 | positive regulation of long-chain fatty acid import across plasma membrane | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0042117 | monocyte activation | 1 | 8 | 5.60e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 5.60e-01 |
| GO:BP | GO:1903567 | regulation of protein localization to ciliary membrane | 1 | 2 | 5.60e-01 |
| GO:BP | GO:2000344 | positive regulation of acrosome reaction | 1 | 6 | 5.60e-01 |
| GO:BP | GO:0009249 | protein lipoylation | 4 | 6 | 5.60e-01 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 1 | 97 | 5.60e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 11 | 114 | 5.60e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 5.60e-01 |
| GO:BP | GO:0030328 | prenylcysteine catabolic process | 1 | 2 | 5.60e-01 |
| GO:BP | GO:0042335 | cuticle development | 1 | 2 | 5.60e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 4 | 69 | 5.60e-01 |
| GO:BP | GO:0097500 | receptor localization to non-motile cilium | 3 | 4 | 5.60e-01 |
| GO:BP | GO:0032289 | central nervous system myelin formation | 1 | 4 | 5.60e-01 |
| GO:BP | GO:0016577 | histone demethylation | 9 | 21 | 5.60e-01 |
| GO:BP | GO:0000019 | regulation of mitotic recombination | 1 | 5 | 5.60e-01 |
| GO:BP | GO:0030329 | prenylcysteine metabolic process | 1 | 2 | 5.60e-01 |
| GO:BP | GO:0070586 | cell-cell adhesion involved in gastrulation | 3 | 7 | 5.60e-01 |
| GO:BP | GO:0007379 | segment specification | 2 | 14 | 5.60e-01 |
| GO:BP | GO:2000388 | positive regulation of antral ovarian follicle growth | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0001809 | positive regulation of type IV hypersensitivity | 1 | 1 | 5.60e-01 |
| GO:BP | GO:2000387 | regulation of antral ovarian follicle growth | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0021547 | midbrain-hindbrain boundary initiation | 1 | 1 | 5.60e-01 |
| GO:BP | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 3 | 5.60e-01 |
| GO:BP | GO:0021501 | prechordal plate formation | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0003062 | regulation of heart rate by chemical signal | 1 | 6 | 5.61e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 1 | 303 | 5.61e-01 |
| GO:BP | GO:0019470 | 4-hydroxyproline catabolic process | 2 | 4 | 5.61e-01 |
| GO:BP | GO:0019471 | 4-hydroxyproline metabolic process | 2 | 4 | 5.61e-01 |
| GO:BP | GO:0098711 | iron ion import across plasma membrane | 1 | 4 | 5.61e-01 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 5.61e-01 |
| GO:BP | GO:0015886 | heme transport | 2 | 9 | 5.61e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 5 | 5.61e-01 |
| GO:BP | GO:0009629 | response to gravity | 2 | 5 | 5.61e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 7 | 40 | 5.61e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 7 | 23 | 5.61e-01 |
| GO:BP | GO:0035553 | oxidative single-stranded RNA demethylation | 1 | 3 | 5.61e-01 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 2 | 21 | 5.61e-01 |
| GO:BP | GO:0015855 | pyrimidine nucleobase transport | 1 | 3 | 5.61e-01 |
| GO:BP | GO:0031034 | myosin filament assembly | 3 | 4 | 5.61e-01 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 10 | 17 | 5.61e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 4 | 10 | 5.61e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 2 | 18 | 5.61e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 12 | 37 | 5.61e-01 |
| GO:BP | GO:0070350 | regulation of white fat cell proliferation | 1 | 2 | 5.61e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 5 | 190 | 5.61e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 151 | 5.61e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 8 | 156 | 5.61e-01 |
| GO:BP | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process | 2 | 3 | 5.61e-01 |
| GO:BP | GO:0015853 | adenine transport | 1 | 5 | 5.61e-01 |
| GO:BP | GO:0033183 | negative regulation of histone ubiquitination | 3 | 4 | 5.61e-01 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 2 | 21 | 5.61e-01 |
| GO:BP | GO:1902766 | skeletal muscle satellite cell migration | 1 | 2 | 5.61e-01 |
| GO:BP | GO:1901187 | regulation of ephrin receptor signaling pathway | 1 | 2 | 5.61e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 10 | 19 | 5.61e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 5.61e-01 |
| GO:BP | GO:0015862 | uridine transport | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 2 | 57 | 5.61e-01 |
| GO:BP | GO:0015864 | pyrimidine nucleoside transport | 1 | 4 | 5.61e-01 |
| GO:BP | GO:1901315 | negative regulation of histone H2A K63-linked ubiquitination | 3 | 4 | 5.61e-01 |
| GO:BP | GO:1901314 | regulation of histone H2A K63-linked ubiquitination | 3 | 4 | 5.61e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 5 | 10 | 5.61e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 10 | 19 | 5.61e-01 |
| GO:BP | GO:0070343 | white fat cell proliferation | 1 | 2 | 5.61e-01 |
| GO:BP | GO:0097474 | retinal cone cell apoptotic process | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0072717 | cellular response to actinomycin D | 1 | 4 | 5.62e-01 |
| GO:BP | GO:0097281 | immune complex formation | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0002383 | immune response in brain or nervous system | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0032930 | positive regulation of superoxide anion generation | 4 | 10 | 5.62e-01 |
| GO:BP | GO:0061847 | response to cholecystokinin | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0060527 | prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis | 1 | 3 | 5.62e-01 |
| GO:BP | GO:0032095 | regulation of response to food | 1 | 8 | 5.62e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 8 | 23 | 5.62e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 4 | 597 | 5.62e-01 |
| GO:BP | GO:1990859 | cellular response to endothelin | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0060526 | prostate glandular acinus morphogenesis | 1 | 3 | 5.62e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 161 | 313 | 5.62e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 4 | 597 | 5.62e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 5 | 73 | 5.63e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 4 | 13 | 5.63e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 5 | 60 | 5.63e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 2 | 84 | 5.63e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 2 | 90 | 5.63e-01 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 7 | 34 | 5.63e-01 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 4 | 15 | 5.63e-01 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 4 | 15 | 5.63e-01 |
| GO:BP | GO:0060992 | response to fungicide | 1 | 4 | 5.64e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 2 | 6 | 5.64e-01 |
| GO:BP | GO:0007565 | female pregnancy | 6 | 122 | 5.64e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 6 | 17 | 5.64e-01 |
| GO:BP | GO:1905661 | regulation of telomerase RNA reverse transcriptase activity | 1 | 3 | 5.64e-01 |
| GO:BP | GO:2000064 | regulation of cortisol biosynthetic process | 2 | 7 | 5.64e-01 |
| GO:BP | GO:1904826 | regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 5.64e-01 |
| GO:BP | GO:1990428 | miRNA transport | 1 | 3 | 5.64e-01 |
| GO:BP | GO:1990678 | histone H4-K16 deacetylation | 1 | 1 | 5.64e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 5.64e-01 |
| GO:BP | GO:1904828 | positive regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 5.64e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 3 | 17 | 5.64e-01 |
| GO:BP | GO:0010831 | positive regulation of myotube differentiation | 4 | 10 | 5.64e-01 |
| GO:BP | GO:0060180 | female mating behavior | 2 | 2 | 5.64e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 5 | 534 | 5.64e-01 |
| GO:BP | GO:0045924 | regulation of female receptivity | 2 | 2 | 5.64e-01 |
| GO:BP | GO:0002154 | thyroid hormone mediated signaling pathway | 2 | 6 | 5.64e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 3 | 44 | 5.64e-01 |
| GO:BP | GO:0010795 | regulation of ubiquinone biosynthetic process | 1 | 2 | 5.64e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 3 | 30 | 5.64e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 7 | 469 | 5.65e-01 |
| GO:BP | GO:0009051 | pentose-phosphate shunt, oxidative branch | 1 | 4 | 5.65e-01 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 4 | 6 | 5.65e-01 |
| GO:BP | GO:0071394 | cellular response to testosterone stimulus | 4 | 11 | 5.65e-01 |
| GO:BP | GO:1904057 | negative regulation of sensory perception of pain | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0015838 | amino-acid betaine transport | 1 | 7 | 5.65e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 10 | 24 | 5.65e-01 |
| GO:BP | GO:0042868 | antisense RNA metabolic process | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 2 | 11 | 5.65e-01 |
| GO:BP | GO:0071040 | nuclear polyadenylation-dependent antisense transcript catabolic process | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0071041 | antisense RNA transcript catabolic process | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0061053 | somite development | 22 | 63 | 5.65e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 3 | 76 | 5.66e-01 |
| GO:BP | GO:0060632 | regulation of microtubule-based movement | 1 | 31 | 5.66e-01 |
| GO:BP | GO:0010759 | positive regulation of macrophage chemotaxis | 2 | 9 | 5.66e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 5.66e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 5.66e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 5.66e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 5.66e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 5.66e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 5.66e-01 |
| GO:BP | GO:1904182 | regulation of pyruvate dehydrogenase activity | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 5.66e-01 |
| GO:BP | GO:1905584 | outer hair cell apoptotic process | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0032287 | peripheral nervous system myelin maintenance | 3 | 8 | 5.66e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 18 | 96 | 5.66e-01 |
| GO:BP | GO:0040016 | embryonic cleavage | 2 | 7 | 5.66e-01 |
| GO:BP | GO:0009443 | pyridoxal 5’-phosphate salvage | 1 | 1 | 5.66e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 9 | 15 | 5.66e-01 |
| GO:BP | GO:0007015 | actin filament organization | 13 | 373 | 5.67e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 9 | 19 | 5.67e-01 |
| GO:BP | GO:0031442 | positive regulation of mRNA 3’-end processing | 5 | 10 | 5.67e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 4 | 71 | 5.67e-01 |
| GO:BP | GO:0031449 | regulation of slow-twitch skeletal muscle fiber contraction | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 3 | 70 | 5.67e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 2 | 61 | 5.67e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 52 | 5.67e-01 |
| GO:BP | GO:2000635 | negative regulation of primary miRNA processing | 2 | 2 | 5.67e-01 |
| GO:BP | GO:0106004 | tRNA (guanine-N7)-methylation | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0060023 | soft palate development | 1 | 3 | 5.67e-01 |
| GO:BP | GO:2000634 | regulation of primary miRNA processing | 2 | 2 | 5.67e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 12 | 21 | 5.67e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 7 | 71 | 5.67e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 14 | 62 | 5.67e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 86 | 5.67e-01 |
| GO:BP | GO:0034334 | adherens junction maintenance | 4 | 6 | 5.67e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 4 | 8 | 5.68e-01 |
| GO:BP | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 7 | 5.68e-01 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 4 | 14 | 5.68e-01 |
| GO:BP | GO:1904797 | negative regulation of core promoter binding | 1 | 1 | 5.68e-01 |
| GO:BP | GO:0090382 | phagosome maturation | 1 | 24 | 5.68e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 2 | 881 | 5.68e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 2 | 19 | 5.68e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 46 | 81 | 5.68e-01 |
| GO:BP | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly | 1 | 3 | 5.69e-01 |
| GO:BP | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly | 1 | 8 | 5.69e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 35 | 114 | 5.69e-01 |
| GO:BP | GO:0038203 | TORC2 signaling | 5 | 12 | 5.69e-01 |
| GO:BP | GO:0043461 | proton-transporting ATP synthase complex assembly | 1 | 8 | 5.69e-01 |
| GO:BP | GO:0098529 | neuromuscular junction development, skeletal muscle fiber | 1 | 1 | 5.69e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 17 | 32 | 5.69e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 4 | 29 | 5.69e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 3 | 4 | 5.69e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 5.69e-01 |
| GO:BP | GO:0002100 | tRNA wobble adenosine to inosine editing | 1 | 1 | 5.69e-01 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 10 | 19 | 5.69e-01 |
| GO:BP | GO:0007604 | phototransduction, UV | 1 | 1 | 5.69e-01 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 2 | 16 | 5.69e-01 |
| GO:BP | GO:2000810 | regulation of bicellular tight junction assembly | 6 | 15 | 5.69e-01 |
| GO:BP | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 1 | 5.69e-01 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 3 | 14 | 5.70e-01 |
| GO:BP | GO:0140361 | cyclic-GMP-AMP transmembrane import across plasma membrane | 1 | 4 | 5.70e-01 |
| GO:BP | GO:0060374 | mast cell differentiation | 3 | 4 | 5.70e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 27 | 49 | 5.70e-01 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 10 | 35 | 5.70e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 710 | 5.70e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 44 | 74 | 5.70e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 7 | 21 | 5.70e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 16 | 73 | 5.70e-01 |
| GO:BP | GO:0044805 | late nucleophagy | 1 | 7 | 5.70e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 6 | 50 | 5.70e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 8 | 27 | 5.70e-01 |
| GO:BP | GO:1902174 | positive regulation of keratinocyte apoptotic process | 1 | 3 | 5.71e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 1 | 15 | 5.71e-01 |
| GO:BP | GO:0006548 | histidine catabolic process | 1 | 3 | 5.71e-01 |
| GO:BP | GO:1990386 | mitotic cleavage furrow ingression | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 5.71e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 5.71e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 5.71e-01 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 4 | 8 | 5.71e-01 |
| GO:BP | GO:1990108 | protein linear deubiquitination | 1 | 3 | 5.71e-01 |
| GO:BP | GO:0043435 | response to corticotropin-releasing hormone | 2 | 3 | 5.71e-01 |
| GO:BP | GO:0071376 | cellular response to corticotropin-releasing hormone stimulus | 2 | 3 | 5.71e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 4 | 8 | 5.71e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 5.71e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 46 | 127 | 5.71e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 1 | 146 | 5.72e-01 |
| GO:BP | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | 1 | 13 | 5.72e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 5.72e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 20 | 42 | 5.72e-01 |
| GO:BP | GO:0009624 | response to nematode | 1 | 4 | 5.72e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 5.72e-01 |
| GO:BP | GO:0016062 | adaptation of rhodopsin mediated signaling | 1 | 1 | 5.72e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 5.72e-01 |
| GO:BP | GO:1904003 | negative regulation of sebum secreting cell proliferation | 1 | 1 | 5.72e-01 |
| GO:BP | GO:2000172 | regulation of branching morphogenesis of a nerve | 1 | 3 | 5.72e-01 |
| GO:BP | GO:0010748 | negative regulation of long-chain fatty acid import across plasma membrane | 3 | 4 | 5.72e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 3 | 67 | 5.72e-01 |
| GO:BP | GO:0140213 | negative regulation of long-chain fatty acid import into cell | 3 | 4 | 5.72e-01 |
| GO:BP | GO:0045606 | positive regulation of epidermal cell differentiation | 5 | 16 | 5.72e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 2 | 42 | 5.72e-01 |
| GO:BP | GO:1902524 | positive regulation of protein K48-linked ubiquitination | 1 | 2 | 5.72e-01 |
| GO:BP | GO:0030321 | transepithelial chloride transport | 1 | 7 | 5.72e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 6 | 27 | 5.72e-01 |
| GO:BP | GO:0090238 | positive regulation of arachidonic acid secretion | 1 | 2 | 5.72e-01 |
| GO:BP | GO:0071107 | response to parathyroid hormone | 1 | 9 | 5.72e-01 |
| GO:BP | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport | 4 | 5 | 5.72e-01 |
| GO:BP | GO:2000551 | regulation of T-helper 2 cell cytokine production | 1 | 3 | 5.72e-01 |
| GO:BP | GO:0035745 | T-helper 2 cell cytokine production | 1 | 3 | 5.72e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 4 | 4579 | 5.72e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 14 | 656 | 5.73e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 5 | 50 | 5.73e-01 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 2 | 7 | 5.73e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 5.73e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 2 | 23 | 5.73e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 3 | 111 | 5.73e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 15 | 25 | 5.73e-01 |
| GO:BP | GO:0009450 | gamma-aminobutyric acid catabolic process | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 7 | 161 | 5.73e-01 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 5 | 16 | 5.73e-01 |
| GO:BP | GO:0021849 | neuroblast division in subventricular zone | 1 | 3 | 5.73e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 1 | 118 | 5.73e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 2 | 5 | 5.73e-01 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 2 | 29 | 5.73e-01 |
| GO:BP | GO:1903793 | positive regulation of monoatomic anion transport | 2 | 2 | 5.73e-01 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 4 | 134 | 5.73e-01 |
| GO:BP | GO:1904448 | regulation of aspartate secretion | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 2 | 5.73e-01 |
| GO:BP | GO:1904450 | positive regulation of aspartate secretion | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0061528 | aspartate secretion | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0098935 | dendritic transport | 2 | 13 | 5.73e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 5.73e-01 |
| GO:BP | GO:0097202 | activation of cysteine-type endopeptidase activity | 7 | 16 | 5.73e-01 |
| GO:BP | GO:1904685 | positive regulation of metalloendopeptidase activity | 1 | 1 | 5.73e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 1 | 118 | 5.73e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 3 | 30 | 5.73e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 3 | 4 | 5.73e-01 |
| GO:BP | GO:1901529 | positive regulation of anion channel activity | 2 | 2 | 5.73e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 5.73e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 3 | 3 | 5.73e-01 |
| GO:BP | GO:1903961 | positive regulation of anion transmembrane transport | 2 | 2 | 5.73e-01 |
| GO:BP | GO:0060384 | innervation | 3 | 22 | 5.74e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 5.74e-01 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 2 | 13 | 5.74e-01 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 7 | 11 | 5.74e-01 |
| GO:BP | GO:0003356 | regulation of cilium beat frequency | 1 | 10 | 5.74e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 5.74e-01 |
| GO:BP | GO:0021740 | principal sensory nucleus of trigeminal nerve development | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0034381 | plasma lipoprotein particle clearance | 1 | 25 | 5.74e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 10 | 62 | 5.74e-01 |
| GO:BP | GO:0021730 | trigeminal sensory nucleus development | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 2 | 14 | 5.75e-01 |
| GO:BP | GO:0061302 | smooth muscle cell-matrix adhesion | 2 | 5 | 5.75e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 6 | 113 | 5.75e-01 |
| GO:BP | GO:0090666 | scaRNA localization to Cajal body | 2 | 5 | 5.76e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 422 | 5.76e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 8 | 30 | 5.76e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 7 | 70 | 5.76e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 1 | 4 | 5.76e-01 |
| GO:BP | GO:0090309 | positive regulation of DNA methylation-dependent heterochromatin formation | 2 | 12 | 5.76e-01 |
| GO:BP | GO:0071624 | positive regulation of granulocyte chemotaxis | 2 | 14 | 5.76e-01 |
| GO:BP | GO:2000035 | regulation of stem cell division | 5 | 9 | 5.76e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 2 | 10 | 5.76e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 2 | 22 | 5.76e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 5.76e-01 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 8 | 13 | 5.76e-01 |
| GO:BP | GO:0032877 | positive regulation of DNA endoreduplication | 1 | 2 | 5.76e-01 |
| GO:BP | GO:0019086 | late viral transcription | 3 | 4 | 5.76e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 3 | 4 | 5.76e-01 |
| GO:BP | GO:1904353 | regulation of telomere capping | 3 | 23 | 5.76e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 6 | 101 | 5.76e-01 |
| GO:BP | GO:0140820 | cytosol to Golgi apparatus transport | 1 | 4 | 5.77e-01 |
| GO:BP | GO:0006564 | L-serine biosynthetic process | 1 | 6 | 5.77e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 2 | 74 | 5.77e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 17 | 323 | 5.77e-01 |
| GO:BP | GO:1901073 | glucosamine-containing compound biosynthetic process | 1 | 2 | 5.77e-01 |
| GO:BP | GO:0006045 | N-acetylglucosamine biosynthetic process | 1 | 2 | 5.77e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 2 | 26 | 5.78e-01 |
| GO:BP | GO:0018146 | keratan sulfate biosynthetic process | 5 | 13 | 5.78e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 2 | 3 | 5.78e-01 |
| GO:BP | GO:0055118 | negative regulation of cardiac muscle contraction | 1 | 4 | 5.78e-01 |
| GO:BP | GO:0070376 | regulation of ERK5 cascade | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0031642 | negative regulation of myelination | 2 | 8 | 5.78e-01 |
| GO:BP | GO:0007610 | behavior | 18 | 436 | 5.78e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 4 | 68 | 5.78e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 5 | 24 | 5.78e-01 |
| GO:BP | GO:0040008 | regulation of growth | 22 | 501 | 5.78e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 5 | 24 | 5.78e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 6 | 290 | 5.78e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 15 | 4245 | 5.78e-01 |
| GO:BP | GO:0001579 | medium-chain fatty acid transport | 1 | 2 | 5.78e-01 |
| GO:BP | GO:0003334 | keratinocyte development | 1 | 8 | 5.78e-01 |
| GO:BP | GO:0010714 | positive regulation of collagen metabolic process | 4 | 24 | 5.78e-01 |
| GO:BP | GO:0075136 | response to host | 1 | 1 | 5.78e-01 |
| GO:BP | GO:1905135 | biotin import across plasma membrane | 1 | 2 | 5.78e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 5.78e-01 |
| GO:BP | GO:0052200 | response to host defenses | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0030682 | mitigation of host defenses by symbiont | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0052173 | response to defenses of other organism | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0051258 | protein polymerization | 2 | 241 | 5.78e-01 |
| GO:BP | GO:0051902 | negative regulation of mitochondrial depolarization | 2 | 5 | 5.78e-01 |
| GO:BP | GO:2000254 | regulation of male germ cell proliferation | 2 | 4 | 5.78e-01 |
| GO:BP | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration | 1 | 3 | 5.78e-01 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 4 | 6 | 5.79e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 2 | 126 | 5.79e-01 |
| GO:BP | GO:0032745 | positive regulation of interleukin-21 production | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0032625 | interleukin-21 production | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | 5 | 9 | 5.79e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 1 | 9 | 5.79e-01 |
| GO:BP | GO:0042428 | serotonin metabolic process | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0032665 | regulation of interleukin-21 production | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 8 | 25 | 5.79e-01 |
| GO:BP | GO:0019042 | viral latency | 1 | 4 | 5.79e-01 |
| GO:BP | GO:0030157 | pancreatic juice secretion | 2 | 11 | 5.79e-01 |
| GO:BP | GO:0072488 | ammonium transmembrane transport | 2 | 6 | 5.79e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 8 | 25 | 5.79e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 5 | 232 | 5.79e-01 |
| GO:BP | GO:0097017 | renal protein absorption | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 4 | 32 | 5.79e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 1 | 155 | 5.79e-01 |
| GO:BP | GO:0090403 | oxidative stress-induced premature senescence | 1 | 3 | 5.79e-01 |
| GO:BP | GO:1903959 | regulation of monoatomic anion transmembrane transport | 5 | 6 | 5.79e-01 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 5.80e-01 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 5.80e-01 |
| GO:BP | GO:0071377 | cellular response to glucagon stimulus | 1 | 4 | 5.80e-01 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 6 | 10 | 5.80e-01 |
| GO:BP | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 2 | 5 | 5.80e-01 |
| GO:BP | GO:0097377 | spinal cord interneuron axon guidance | 1 | 1 | 5.80e-01 |
| GO:BP | GO:0035846 | oviduct epithelium development | 1 | 2 | 5.80e-01 |
| GO:BP | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation | 1 | 3 | 5.80e-01 |
| GO:BP | GO:0097477 | lateral motor column neuron migration | 1 | 3 | 5.80e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 1 | 41 | 5.80e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 10 | 771 | 5.80e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 8 | 19 | 5.80e-01 |
| GO:BP | GO:1905550 | regulation of protein localization to endoplasmic reticulum | 3 | 4 | 5.80e-01 |
| GO:BP | GO:0030258 | lipid modification | 1 | 162 | 5.80e-01 |
| GO:BP | GO:2000143 | negative regulation of DNA-templated transcription initiation | 4 | 6 | 5.80e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 8 | 16 | 5.80e-01 |
| GO:BP | GO:0007258 | JUN phosphorylation | 1 | 2 | 5.80e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 2 | 22 | 5.80e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 2 | 26 | 5.80e-01 |
| GO:BP | GO:0072319 | vesicle uncoating | 2 | 9 | 5.80e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 1 | 30 | 5.81e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 4 | 191 | 5.81e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 4 | 11 | 5.82e-01 |
| GO:BP | GO:0042921 | glucocorticoid receptor signaling pathway | 2 | 13 | 5.82e-01 |
| GO:BP | GO:1902462 | positive regulation of mesenchymal stem cell proliferation | 3 | 4 | 5.82e-01 |
| GO:BP | GO:1902460 | regulation of mesenchymal stem cell proliferation | 3 | 4 | 5.82e-01 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 2 | 7 | 5.82e-01 |
| GO:BP | GO:2000458 | regulation of astrocyte chemotaxis | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 1 | 10 | 5.82e-01 |
| GO:BP | GO:0036145 | dendritic cell homeostasis | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0035700 | astrocyte chemotaxis | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0035196 | miRNA processing | 22 | 41 | 5.82e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 18 | 816 | 5.82e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 2 | 30 | 5.82e-01 |
| GO:BP | GO:0006422 | aspartyl-tRNA aminoacylation | 1 | 2 | 5.82e-01 |
| GO:BP | GO:2000360 | negative regulation of binding of sperm to zona pellucida | 2 | 2 | 5.82e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 5 | 111 | 5.82e-01 |
| GO:BP | GO:1904693 | midbrain morphogenesis | 1 | 3 | 5.82e-01 |
| GO:BP | GO:0070933 | histone H4 deacetylation | 3 | 7 | 5.82e-01 |
| GO:BP | GO:0106028 | neuron projection retraction | 1 | 1 | 5.82e-01 |
| GO:BP | GO:1905458 | positive regulation of lymphoid progenitor cell differentiation | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 5 | 8 | 5.83e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 6 | 126 | 5.83e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 1 | 16 | 5.83e-01 |
| GO:BP | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 5.83e-01 |
| GO:BP | GO:1903463 | regulation of mitotic cell cycle DNA replication | 1 | 2 | 5.83e-01 |
| GO:BP | GO:1900099 | negative regulation of plasma cell differentiation | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0019742 | pentacyclic triterpenoid metabolic process | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 14 | 37 | 5.83e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 5 | 551 | 5.83e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 4 | 6 | 5.83e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 6 | 10 | 5.83e-01 |
| GO:BP | GO:2000078 | positive regulation of type B pancreatic cell development | 2 | 2 | 5.83e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 3 | 378 | 5.84e-01 |
| GO:BP | GO:0015961 | diadenosine polyphosphate catabolic process | 2 | 6 | 5.84e-01 |
| GO:BP | GO:0006204 | IMP catabolic process | 3 | 3 | 5.84e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 3 | 66 | 5.84e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 7 | 90 | 5.84e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 5.84e-01 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 1 | 11 | 5.84e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 5.84e-01 |
| GO:BP | GO:0021761 | limbic system development | 4 | 81 | 5.84e-01 |
| GO:BP | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | 2 | 8 | 5.85e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 1 | 13 | 5.85e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 8 | 116 | 5.85e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 2 | 26 | 5.85e-01 |
| GO:BP | GO:2000369 | regulation of clathrin-dependent endocytosis | 1 | 18 | 5.85e-01 |
| GO:BP | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development | 2 | 2 | 5.85e-01 |
| GO:BP | GO:0060701 | negative regulation of ribonuclease activity | 1 | 2 | 5.85e-01 |
| GO:BP | GO:0060702 | negative regulation of endoribonuclease activity | 1 | 2 | 5.85e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 5.85e-01 |
| GO:BP | GO:1900259 | regulation of RNA-dependent RNA polymerase activity | 1 | 1 | 5.85e-01 |
| GO:BP | GO:1900260 | negative regulation of RNA-dependent RNA polymerase activity | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 2 | 51 | 5.85e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 5 | 199 | 5.85e-01 |
| GO:BP | GO:1905243 | cellular response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 10 | 49 | 5.85e-01 |
| GO:BP | GO:0006747 | FAD biosynthetic process | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0072388 | flavin adenine dinucleotide biosynthetic process | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 10 | 13 | 5.85e-01 |
| GO:BP | GO:0046443 | FAD metabolic process | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0048210 | Golgi vesicle fusion to target membrane | 3 | 4 | 5.86e-01 |
| GO:BP | GO:0090219 | negative regulation of lipid kinase activity | 5 | 8 | 5.86e-01 |
| GO:BP | GO:0001955 | blood vessel maturation | 2 | 6 | 5.86e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 1 | 44 | 5.86e-01 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 1 | 12 | 5.86e-01 |
| GO:BP | GO:0010713 | negative regulation of collagen metabolic process | 6 | 8 | 5.86e-01 |
| GO:BP | GO:1905313 | transforming growth factor beta receptor signaling pathway involved in heart development | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0014006 | regulation of microglia differentiation | 1 | 1 | 5.86e-01 |
| GO:BP | GO:1903564 | regulation of protein localization to cilium | 5 | 11 | 5.86e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 2 | 76 | 5.86e-01 |
| GO:BP | GO:0014008 | positive regulation of microglia differentiation | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0060242 | contact inhibition | 5 | 8 | 5.86e-01 |
| GO:BP | GO:1901232 | regulation of convergent extension involved in axis elongation | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0044338 | canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation | 1 | 3 | 5.86e-01 |
| GO:BP | GO:1901229 | regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 5.86e-01 |
| GO:BP | GO:1901231 | positive regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0048696 | regulation of collateral sprouting in absence of injury | 1 | 2 | 5.86e-01 |
| GO:BP | GO:0001787 | natural killer cell proliferation | 1 | 6 | 5.86e-01 |
| GO:BP | GO:1901233 | negative regulation of convergent extension involved in axis elongation | 1 | 1 | 5.86e-01 |
| GO:BP | GO:1902416 | positive regulation of mRNA binding | 5 | 8 | 5.86e-01 |
| GO:BP | GO:0003013 | circulatory system process | 2 | 447 | 5.86e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 7 | 269 | 5.86e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 2 | 15 | 5.86e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1 | 24 | 5.86e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 1 | 24 | 5.86e-01 |
| GO:BP | GO:0014805 | smooth muscle adaptation | 2 | 3 | 5.87e-01 |
| GO:BP | GO:0072695 | regulation of DNA recombination at telomere | 1 | 2 | 5.87e-01 |
| GO:BP | GO:0048239 | negative regulation of DNA recombination at telomere | 1 | 2 | 5.87e-01 |
| GO:BP | GO:0045986 | negative regulation of smooth muscle contraction | 1 | 12 | 5.87e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 5.87e-01 |
| GO:BP | GO:0048320 | axial mesoderm formation | 1 | 2 | 5.87e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 5.87e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 20 | 35 | 5.88e-01 |
| GO:BP | GO:0019449 | L-cysteine catabolic process to hypotaurine | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 5.88e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 5.88e-01 |
| GO:BP | GO:0019452 | L-cysteine catabolic process to taurine | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 5.88e-01 |
| GO:BP | GO:0001764 | neuron migration | 7 | 133 | 5.88e-01 |
| GO:BP | GO:0032532 | regulation of microvillus length | 2 | 4 | 5.88e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 5.89e-01 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 1 | 17 | 5.89e-01 |
| GO:BP | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 5 | 5.89e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 5.89e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 5.89e-01 |
| GO:BP | GO:0006526 | arginine biosynthetic process | 4 | 4 | 5.89e-01 |
| GO:BP | GO:0010166 | wax metabolic process | 1 | 2 | 5.89e-01 |
| GO:BP | GO:0010025 | wax biosynthetic process | 1 | 2 | 5.89e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 26 | 55 | 5.89e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 1 | 35 | 5.89e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 3 | 740 | 5.89e-01 |
| GO:BP | GO:0140448 | signaling receptor ligand precursor processing | 1 | 29 | 5.89e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 4 | 39 | 5.89e-01 |
| GO:BP | GO:0071139 | resolution of recombination intermediates | 4 | 6 | 5.89e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 5.89e-01 |
| GO:BP | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity | 3 | 3 | 5.89e-01 |
| GO:BP | GO:0099628 | neurotransmitter receptor diffusion trapping | 1 | 4 | 5.89e-01 |
| GO:BP | GO:0032868 | response to insulin | 53 | 215 | 5.89e-01 |
| GO:BP | GO:0061789 | dense core granule priming | 1 | 1 | 5.89e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 5.89e-01 |
| GO:BP | GO:0090086 | negative regulation of protein deubiquitination | 3 | 4 | 5.89e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 5.89e-01 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 3 | 3 | 5.89e-01 |
| GO:BP | GO:0003430 | growth plate cartilage chondrocyte growth | 2 | 3 | 5.89e-01 |
| GO:BP | GO:0032415 | regulation of sodium:proton antiporter activity | 1 | 4 | 5.89e-01 |
| GO:BP | GO:0048806 | genitalia development | 2 | 28 | 5.89e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 2 | 73 | 5.89e-01 |
| GO:BP | GO:0098953 | receptor diffusion trapping | 1 | 4 | 5.89e-01 |
| GO:BP | GO:0032288 | myelin assembly | 8 | 19 | 5.89e-01 |
| GO:BP | GO:0098970 | postsynaptic neurotransmitter receptor diffusion trapping | 1 | 4 | 5.89e-01 |
| GO:BP | GO:1990966 | ATP generation from poly-ADP-D-ribose | 1 | 4 | 5.89e-01 |
| GO:BP | GO:2000670 | positive regulation of dendritic cell apoptotic process | 1 | 2 | 5.89e-01 |
| GO:BP | GO:0006102 | isocitrate metabolic process | 4 | 6 | 5.90e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 2 | 10 | 5.90e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 6 | 138 | 5.90e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 1 | 12 | 5.90e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 2 | 80 | 5.90e-01 |
| GO:BP | GO:0042045 | epithelial fluid transport | 3 | 7 | 5.90e-01 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 1 | 4 | 5.90e-01 |
| GO:BP | GO:1901080 | regulation of relaxation of smooth muscle | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0060541 | respiratory system development | 2 | 163 | 5.90e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 5.90e-01 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 5 | 28 | 5.90e-01 |
| GO:BP | GO:0010511 | regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 8 | 154 | 5.90e-01 |
| GO:BP | GO:0036075 | replacement ossification | 4 | 25 | 5.90e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 8 | 1076 | 5.90e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 8 | 1074 | 5.90e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 4 | 25 | 5.90e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 2 | 52 | 5.90e-01 |
| GO:BP | GO:0045654 | positive regulation of megakaryocyte differentiation | 2 | 9 | 5.90e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 5 | 31 | 5.90e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 1 | 55 | 5.90e-01 |
| GO:BP | GO:0040010 | positive regulation of growth rate | 2 | 2 | 5.90e-01 |
| GO:BP | GO:0007530 | sex determination | 6 | 12 | 5.90e-01 |
| GO:BP | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 1 | 9 | 5.90e-01 |
| GO:BP | GO:0035788 | cell migration involved in metanephros development | 1 | 4 | 5.90e-01 |
| GO:BP | GO:0006855 | xenobiotic transmembrane transport | 2 | 11 | 5.90e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 2 | 5 | 5.90e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 14 | 34 | 5.90e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 9 | 19 | 5.91e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 9 | 19 | 5.91e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 9 | 19 | 5.91e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 15 | 5.91e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 8 | 42 | 5.91e-01 |
| GO:BP | GO:1903726 | negative regulation of phospholipid metabolic process | 2 | 7 | 5.91e-01 |
| GO:BP | GO:0046712 | GDP catabolic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0046056 | dADP metabolic process | 1 | 1 | 5.91e-01 |
| GO:BP | GO:0046066 | dGDP metabolic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 3 | 782 | 5.91e-01 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 5 | 8 | 5.92e-01 |
| GO:BP | GO:0042635 | positive regulation of hair cycle | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 2 | 485 | 5.92e-01 |
| GO:BP | GO:1990009 | retinal cell apoptotic process | 4 | 5 | 5.92e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 2 | 54 | 5.92e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 5.92e-01 |
| GO:BP | GO:0072537 | fibroblast activation | 7 | 11 | 5.92e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 2 | 6 | 5.92e-01 |
| GO:BP | GO:0098789 | pre-mRNA cleavage required for polyadenylation | 1 | 12 | 5.92e-01 |
| GO:BP | GO:1905007 | positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 5 | 5.92e-01 |
| GO:BP | GO:0022009 | central nervous system vasculogenesis | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0006901 | vesicle coating | 9 | 36 | 5.92e-01 |
| GO:BP | GO:0021793 | chemorepulsion of branchiomotor axon | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0002184 | cytoplasmic translational termination | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 3 | 41 | 5.93e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 23 | 43 | 5.93e-01 |
| GO:BP | GO:0002520 | immune system development | 1 | 144 | 5.94e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 5.94e-01 |
| GO:BP | GO:0015816 | glycine transport | 2 | 9 | 5.94e-01 |
| GO:BP | GO:1904515 | positive regulation of TORC2 signaling | 2 | 4 | 5.94e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 5.94e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 7 | 42 | 5.94e-01 |
| GO:BP | GO:0046851 | negative regulation of bone remodeling | 2 | 9 | 5.94e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 2 | 158 | 5.94e-01 |
| GO:BP | GO:0016310 | phosphorylation | 7 | 1372 | 5.94e-01 |
| GO:BP | GO:0030644 | intracellular chloride ion homeostasis | 1 | 7 | 5.94e-01 |
| GO:BP | GO:0030002 | intracellular monoatomic anion homeostasis | 1 | 7 | 5.94e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 1 | 13 | 5.94e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 798 | 5.94e-01 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 5 | 8 | 5.94e-01 |
| GO:BP | GO:1901523 | icosanoid catabolic process | 1 | 2 | 5.94e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 5.94e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 6 | 127 | 5.95e-01 |
| GO:BP | GO:0090314 | positive regulation of protein targeting to membrane | 5 | 22 | 5.95e-01 |
| GO:BP | GO:0033625 | positive regulation of integrin activation | 3 | 6 | 5.95e-01 |
| GO:BP | GO:0097067 | cellular response to thyroid hormone stimulus | 5 | 14 | 5.95e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 1 | 8 | 5.95e-01 |
| GO:BP | GO:0006501 | C-terminal protein lipidation | 2 | 6 | 5.95e-01 |
| GO:BP | GO:0060574 | intestinal epithelial cell maturation | 2 | 3 | 5.95e-01 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 1 | 7 | 5.95e-01 |
| GO:BP | GO:0046031 | ADP metabolic process | 5 | 7 | 5.95e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 5.95e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 1 | 88 | 5.95e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 2 | 5 | 5.95e-01 |
| GO:BP | GO:1900142 | negative regulation of oligodendrocyte apoptotic process | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0002265 | astrocyte activation involved in immune response | 2 | 4 | 5.95e-01 |
| GO:BP | GO:2000533 | negative regulation of renal albumin absorption | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0034447 | very-low-density lipoprotein particle clearance | 2 | 4 | 5.95e-01 |
| GO:BP | GO:0098657 | import into cell | 23 | 166 | 5.95e-01 |
| GO:BP | GO:0031054 | pre-miRNA processing | 8 | 14 | 5.96e-01 |
| GO:BP | GO:1905209 | positive regulation of cardiocyte differentiation | 1 | 11 | 5.96e-01 |
| GO:BP | GO:1904528 | positive regulation of microtubule binding | 2 | 6 | 5.96e-01 |
| GO:BP | GO:0060377 | negative regulation of mast cell differentiation | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 5.96e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 3 | 114 | 5.96e-01 |
| GO:BP | GO:0080058 | protein deglutathionylation | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0035246 | peptidyl-arginine N-methylation | 1 | 4 | 5.96e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 4 | 158 | 5.96e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 18 | 39 | 5.96e-01 |
| GO:BP | GO:0051597 | response to methylmercury | 2 | 5 | 5.97e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 5.97e-01 |
| GO:BP | GO:0003168 | Purkinje myocyte differentiation | 1 | 1 | 5.97e-01 |
| GO:BP | GO:1901652 | response to peptide | 35 | 377 | 5.97e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 5.97e-01 |
| GO:BP | GO:0070267 | oncosis | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 2 | 57 | 5.97e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 3 | 8 | 5.97e-01 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 3 | 4 | 5.98e-01 |
| GO:BP | GO:0097742 | de novo centriole assembly | 3 | 4 | 5.98e-01 |
| GO:BP | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation | 2 | 2 | 5.98e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 5.98e-01 |
| GO:BP | GO:0010896 | regulation of triglyceride catabolic process | 1 | 7 | 5.99e-01 |
| GO:BP | GO:0006426 | glycyl-tRNA aminoacylation | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0021629 | olfactory nerve structural organization | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 3 | 114 | 5.99e-01 |
| GO:BP | GO:0051707 | response to other organism | 3 | 820 | 5.99e-01 |
| GO:BP | GO:0070150 | mitochondrial glycyl-tRNA aminoacylation | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 6 | 84 | 5.99e-01 |
| GO:BP | GO:1903182 | regulation of SUMO transferase activity | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 8 | 18 | 5.99e-01 |
| GO:BP | GO:1903755 | positive regulation of SUMO transferase activity | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 2 | 16 | 5.99e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 4 | 170 | 5.99e-01 |
| GO:BP | GO:0021686 | cerebellar granular layer maturation | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0021933 | radial glia guided migration of cerebellar granule cell | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0072684 | mitochondrial tRNA 3’-trailer cleavage, endonucleolytic | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 4 | 12 | 5.99e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 3 | 820 | 5.99e-01 |
| GO:BP | GO:0010815 | bradykinin catabolic process | 4 | 6 | 5.99e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 5.99e-01 |
| GO:BP | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process | 1 | 2 | 5.99e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 7 | 21 | 5.99e-01 |
| GO:BP | GO:0080021 | response to benzoic acid | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0014842 | regulation of skeletal muscle satellite cell proliferation | 2 | 8 | 6.00e-01 |
| GO:BP | GO:1904116 | response to vasopressin | 2 | 5 | 6.00e-01 |
| GO:BP | GO:0061031 | endodermal digestive tract morphogenesis | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 5 | 461 | 6.00e-01 |
| GO:BP | GO:0046477 | glycosylceramide catabolic process | 2 | 5 | 6.00e-01 |
| GO:BP | GO:1904117 | cellular response to vasopressin | 2 | 5 | 6.00e-01 |
| GO:BP | GO:1902214 | regulation of interleukin-4-mediated signaling pathway | 2 | 2 | 6.00e-01 |
| GO:BP | GO:0007624 | ultradian rhythm | 1 | 2 | 6.00e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 2 | 30 | 6.00e-01 |
| GO:BP | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway | 4 | 7 | 6.00e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 1 | 62 | 6.00e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 3 | 37 | 6.00e-01 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 5 | 17 | 6.00e-01 |
| GO:BP | GO:0038003 | G protein-coupled opioid receptor signaling pathway | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0090237 | regulation of arachidonic acid secretion | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0046469 | platelet activating factor metabolic process | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 6.00e-01 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 2 | 23 | 6.00e-01 |
| GO:BP | GO:0030961 | peptidyl-arginine hydroxylation | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0097350 | neutrophil clearance | 3 | 5 | 6.00e-01 |
| GO:BP | GO:2000182 | regulation of progesterone biosynthetic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 3 | 13 | 6.00e-01 |
| GO:BP | GO:0006420 | arginyl-tRNA aminoacylation | 2 | 3 | 6.00e-01 |
| GO:BP | GO:0042761 | very long-chain fatty acid biosynthetic process | 5 | 12 | 6.00e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 3 | 61 | 6.00e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 8 | 560 | 6.00e-01 |
| GO:BP | GO:0035711 | T-helper 1 cell activation | 2 | 2 | 6.00e-01 |
| GO:BP | GO:0009188 | ribonucleoside diphosphate biosynthetic process | 1 | 8 | 6.00e-01 |
| GO:BP | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 6 | 6.00e-01 |
| GO:BP | GO:0061966 | establishment of left/right asymmetry | 1 | 3 | 6.00e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 1 | 4 | 6.00e-01 |
| GO:BP | GO:1990134 | epithelial cell apoptotic process involved in palatal shelf morphogenesis | 1 | 1 | 6.00e-01 |
| GO:BP | GO:1903577 | cellular response to L-arginine | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0098758 | response to interleukin-8 | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0060160 | negative regulation of dopamine receptor signaling pathway | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0043931 | ossification involved in bone maturation | 1 | 17 | 6.00e-01 |
| GO:BP | GO:0072262 | metanephric glomerular mesangial cell proliferation involved in metanephros development | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 6.00e-01 |
| GO:BP | GO:0098759 | cellular response to interleukin-8 | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0009258 | 10-formyltetrahydrofolate catabolic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 16 | 90 | 6.00e-01 |
| GO:BP | GO:0046083 | adenine metabolic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:1990504 | dense core granule exocytosis | 2 | 7 | 6.00e-01 |
| GO:BP | GO:0032978 | protein insertion into membrane from inner side | 2 | 4 | 6.00e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 1 | 31 | 6.00e-01 |
| GO:BP | GO:0032979 | protein insertion into mitochondrial inner membrane from matrix | 2 | 4 | 6.00e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 6 | 16 | 6.01e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 5 | 30 | 6.01e-01 |
| GO:BP | GO:0032074 | negative regulation of nuclease activity | 2 | 6 | 6.01e-01 |
| GO:BP | GO:0003150 | muscular septum morphogenesis | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0060486 | club cell differentiation | 2 | 3 | 6.01e-01 |
| GO:BP | GO:0051410 | detoxification of nitrogen compound | 2 | 5 | 6.01e-01 |
| GO:BP | GO:0032736 | positive regulation of interleukin-13 production | 1 | 7 | 6.01e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 1 | 9 | 6.01e-01 |
| GO:BP | GO:0070458 | cellular detoxification of nitrogen compound | 2 | 5 | 6.01e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 13 | 265 | 6.01e-01 |
| GO:BP | GO:0070236 | negative regulation of activation-induced cell death of T cells | 2 | 3 | 6.01e-01 |
| GO:BP | GO:1903944 | negative regulation of hepatocyte apoptotic process | 3 | 6 | 6.01e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 2 | 34 | 6.01e-01 |
| GO:BP | GO:0007601 | visual perception | 2 | 96 | 6.01e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 5 | 9 | 6.01e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 3 | 23 | 6.02e-01 |
| GO:BP | GO:0009744 | response to sucrose | 2 | 4 | 6.02e-01 |
| GO:BP | GO:0034285 | response to disaccharide | 2 | 4 | 6.02e-01 |
| GO:BP | GO:0002946 | tRNA C5-cytosine methylation | 2 | 3 | 6.02e-01 |
| GO:BP | GO:0055001 | muscle cell development | 33 | 165 | 6.02e-01 |
| GO:BP | GO:2000510 | positive regulation of dendritic cell chemotaxis | 1 | 3 | 6.02e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 2 | 598 | 6.02e-01 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 3 | 5 | 6.02e-01 |
| GO:BP | GO:0032802 | low-density lipoprotein particle receptor catabolic process | 2 | 5 | 6.02e-01 |
| GO:BP | GO:0018106 | peptidyl-histidine phosphorylation | 1 | 1 | 6.02e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 3 | 18 | 6.02e-01 |
| GO:BP | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process | 2 | 5 | 6.02e-01 |
| GO:BP | GO:0007097 | nuclear migration | 3 | 23 | 6.02e-01 |
| GO:BP | GO:0032799 | low-density lipoprotein receptor particle metabolic process | 2 | 5 | 6.02e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 8 | 157 | 6.02e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 2 | 33 | 6.02e-01 |
| GO:BP | GO:1904829 | regulation of aortic smooth muscle cell differentiation | 1 | 2 | 6.02e-01 |
| GO:BP | GO:1904831 | positive regulation of aortic smooth muscle cell differentiation | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0035887 | aortic smooth muscle cell differentiation | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 4 | 12 | 6.02e-01 |
| GO:BP | GO:0035022 | positive regulation of Rac protein signal transduction | 4 | 5 | 6.02e-01 |
| GO:BP | GO:0010739 | positive regulation of protein kinase A signaling | 1 | 6 | 6.02e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 2 | 31 | 6.02e-01 |
| GO:BP | GO:0007518 | myoblast fate determination | 1 | 1 | 6.02e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 13 | 27 | 6.02e-01 |
| GO:BP | GO:0097484 | dendrite extension | 11 | 27 | 6.03e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 6.03e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 1 | 170 | 6.03e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 185 | 6.03e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 7 | 6.03e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 6.03e-01 |
| GO:BP | GO:0070158 | mitochondrial seryl-tRNA aminoacylation | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 17 | 43 | 6.03e-01 |
| GO:BP | GO:0070460 | thyroid-stimulating hormone secretion | 1 | 2 | 6.03e-01 |
| GO:BP | GO:1904891 | positive regulation of excitatory synapse assembly | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0015828 | tyrosine transport | 1 | 3 | 6.03e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 1 | 56 | 6.03e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 6 | 18 | 6.03e-01 |
| GO:BP | GO:0008272 | sulfate transport | 3 | 12 | 6.03e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 2 | 957 | 6.03e-01 |
| GO:BP | GO:0002339 | B cell selection | 2 | 4 | 6.03e-01 |
| GO:BP | GO:0046709 | IDP catabolic process | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0019343 | cysteine biosynthetic process via cystathionine | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0018206 | peptidyl-methionine modification | 4 | 13 | 6.03e-01 |
| GO:BP | GO:0046707 | IDP metabolic process | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 4 | 8 | 6.03e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 6.04e-01 |
| GO:BP | GO:1904616 | regulation of actin binding | 1 | 3 | 6.04e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 45 | 249 | 6.04e-01 |
| GO:BP | GO:0043649 | dicarboxylic acid catabolic process | 1 | 14 | 6.04e-01 |
| GO:BP | GO:0001545 | primary ovarian follicle growth | 1 | 1 | 6.04e-01 |
| GO:BP | GO:0016031 | tRNA import into mitochondrion | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 4 | 40 | 6.05e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 1 | 4 | 6.05e-01 |
| GO:BP | GO:0070777 | D-aspartate transport | 1 | 4 | 6.05e-01 |
| GO:BP | GO:0070779 | D-aspartate import across plasma membrane | 1 | 4 | 6.05e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 5 | 8 | 6.05e-01 |
| GO:BP | GO:0032639 | TRAIL production | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0001813 | regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0035945 | mitochondrial ncRNA surveillance | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1903457 | lactate catabolic process | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0035946 | mitochondrial mRNA surveillance | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 1 | 7 | 6.05e-01 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 2 | 8 | 6.05e-01 |
| GO:BP | GO:2000827 | mitochondrial RNA surveillance | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 20 | 31 | 6.05e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 5 | 71 | 6.05e-01 |
| GO:BP | GO:0099044 | vesicle tethering to endoplasmic reticulum | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0042352 | GDP-L-fucose salvage | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0032730 | positive regulation of interleukin-1 alpha production | 1 | 3 | 6.05e-01 |
| GO:BP | GO:0032679 | regulation of TRAIL production | 1 | 1 | 6.05e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 19 | 67 | 6.05e-01 |
| GO:BP | GO:0002537 | nitric oxide production involved in inflammatory response | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0016094 | polyprenol biosynthetic process | 1 | 3 | 6.05e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 3 | 38 | 6.05e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 8 | 19 | 6.05e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 9 | 14 | 6.05e-01 |
| GO:BP | GO:0043095 | regulation of GTP cyclohydrolase I activity | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 5 | 155 | 6.05e-01 |
| GO:BP | GO:0043105 | negative regulation of GTP cyclohydrolase I activity | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0071026 | cytoplasmic RNA surveillance | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0032759 | positive regulation of TRAIL production | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 4 | 348 | 6.05e-01 |
| GO:BP | GO:0034696 | response to prostaglandin F | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 13 | 31 | 6.05e-01 |
| GO:BP | GO:0031958 | corticosteroid receptor signaling pathway | 2 | 14 | 6.05e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 3 | 27 | 6.05e-01 |
| GO:BP | GO:0003348 | cardiac endothelial cell differentiation | 2 | 4 | 6.05e-01 |
| GO:BP | GO:0061183 | regulation of dermatome development | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 2 | 3 | 6.05e-01 |
| GO:BP | GO:0006848 | pyruvate transport | 1 | 5 | 6.05e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 1 | 5 | 6.05e-01 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 2 | 11 | 6.05e-01 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 1 | 22 | 6.05e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 2 | 11 | 6.05e-01 |
| GO:BP | GO:0021943 | formation of radial glial scaffolds | 1 | 3 | 6.05e-01 |
| GO:BP | GO:0046604 | positive regulation of mitotic centrosome separation | 1 | 3 | 6.05e-01 |
| GO:BP | GO:0060956 | endocardial cell differentiation | 2 | 4 | 6.05e-01 |
| GO:BP | GO:1901475 | pyruvate transmembrane transport | 1 | 5 | 6.05e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 10 | 21 | 6.05e-01 |
| GO:BP | GO:0061054 | dermatome development | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 6 | 462 | 6.05e-01 |
| GO:BP | GO:0072143 | mesangial cell development | 2 | 5 | 6.05e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 1 | 36 | 6.05e-01 |
| GO:BP | GO:0072008 | glomerular mesangial cell differentiation | 2 | 4 | 6.05e-01 |
| GO:BP | GO:0014807 | regulation of somitogenesis | 2 | 3 | 6.05e-01 |
| GO:BP | GO:1903801 | L-leucine import across plasma membrane | 3 | 5 | 6.06e-01 |
| GO:BP | GO:0098713 | leucine import across plasma membrane | 3 | 5 | 6.06e-01 |
| GO:BP | GO:1990961 | xenobiotic detoxification by transmembrane export across the plasma membrane | 4 | 7 | 6.06e-01 |
| GO:BP | GO:0098787 | mRNA cleavage involved in mRNA processing | 1 | 13 | 6.06e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 2 | 24 | 6.06e-01 |
| GO:BP | GO:0033604 | negative regulation of catecholamine secretion | 3 | 6 | 6.06e-01 |
| GO:BP | GO:1904055 | negative regulation of cholangiocyte proliferation | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0018335 | protein succinylation | 2 | 4 | 6.07e-01 |
| GO:BP | GO:0106077 | histone succinylation | 2 | 4 | 6.07e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 6.07e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 10 | 50 | 6.07e-01 |
| GO:BP | GO:1903636 | regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 6.07e-01 |
| GO:BP | GO:1903638 | positive regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0002296 | T-helper 1 cell lineage commitment | 1 | 3 | 6.07e-01 |
| GO:BP | GO:0032682 | negative regulation of chemokine production | 5 | 12 | 6.07e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 5 | 28 | 6.07e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 2 | 25 | 6.07e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 4 | 15 | 6.07e-01 |
| GO:BP | GO:0006463 | steroid hormone receptor complex assembly | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0034230 | enkephalin processing | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 14 | 47 | 6.07e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 1 | 252 | 6.07e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 3 | 850 | 6.07e-01 |
| GO:BP | GO:0001788 | antibody-dependent cellular cytotoxicity | 2 | 2 | 6.07e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 9 | 29 | 6.07e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 7 | 13 | 6.07e-01 |
| GO:BP | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | 2 | 5 | 6.08e-01 |
| GO:BP | GO:0061698 | protein deglutarylation | 1 | 2 | 6.08e-01 |
| GO:BP | GO:0036048 | protein desuccinylation | 1 | 2 | 6.08e-01 |
| GO:BP | GO:0036049 | peptidyl-lysine desuccinylation | 1 | 2 | 6.08e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 6.08e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 6.08e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 13 | 500 | 6.08e-01 |
| GO:BP | GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1 | 8 | 6.08e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 6.08e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 14 | 35 | 6.08e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 2 | 49 | 6.08e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 3 | 78 | 6.08e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 11 | 37 | 6.08e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 1 | 9 | 6.08e-01 |
| GO:BP | GO:0048680 | positive regulation of axon regeneration | 1 | 8 | 6.08e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 2 | 993 | 6.08e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 2 | 13 | 6.08e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 6.08e-01 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 5 | 8 | 6.08e-01 |
| GO:BP | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 3 | 9 | 6.08e-01 |
| GO:BP | GO:0099093 | calcium export from the mitochondrion | 1 | 2 | 6.08e-01 |
| GO:BP | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway | 7 | 9 | 6.08e-01 |
| GO:BP | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis | 1 | 2 | 6.08e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 2 | 994 | 6.08e-01 |
| GO:BP | GO:0031392 | regulation of prostaglandin biosynthetic process | 3 | 6 | 6.08e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 2 | 8 | 6.08e-01 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 1 | 14 | 6.08e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 7 | 16 | 6.08e-01 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 5 | 18 | 6.09e-01 |
| GO:BP | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination | 2 | 3 | 6.09e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 1 | 118 | 6.09e-01 |
| GO:BP | GO:0031645 | negative regulation of nervous system process | 3 | 9 | 6.09e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 2 | 8 | 6.09e-01 |
| GO:BP | GO:0042976 | activation of Janus kinase activity | 1 | 7 | 6.09e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 2 | 8 | 6.09e-01 |
| GO:BP | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 4 | 6.09e-01 |
| GO:BP | GO:0006508 | proteolysis | 2 | 1318 | 6.09e-01 |
| GO:BP | GO:0032808 | lacrimal gland development | 4 | 5 | 6.09e-01 |
| GO:BP | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity | 1 | 3 | 6.10e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 6.10e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 2 | 20 | 6.10e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 6.10e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 4 | 6 | 6.10e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 6.10e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 6.10e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 4 | 7 | 6.10e-01 |
| GO:BP | GO:0043973 | histone H3-K4 acetylation | 1 | 1 | 6.10e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 6.10e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 3 | 97 | 6.10e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 4 | 5 | 6.10e-01 |
| GO:BP | GO:0070926 | regulation of ATP:ADP antiporter activity | 1 | 1 | 6.10e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 53 | 100 | 6.10e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 2 | 517 | 6.10e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 36 | 162 | 6.10e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 1 | 64 | 6.10e-01 |
| GO:BP | GO:0033182 | regulation of histone ubiquitination | 4 | 6 | 6.10e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 1 | 14 | 6.10e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 1 | 14 | 6.10e-01 |
| GO:BP | GO:0032072 | regulation of restriction endodeoxyribonuclease activity | 1 | 1 | 6.10e-01 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 2 | 6 | 6.10e-01 |
| GO:BP | GO:1903442 | response to lipoic acid | 1 | 1 | 6.10e-01 |
| GO:BP | GO:2000074 | regulation of type B pancreatic cell development | 3 | 8 | 6.10e-01 |
| GO:BP | GO:0036255 | response to methylamine | 1 | 1 | 6.10e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 4 | 4 | 6.10e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 17 | 40 | 6.10e-01 |
| GO:BP | GO:0000350 | generation of catalytic spliceosome for second transesterification step | 1 | 2 | 6.10e-01 |
| GO:BP | GO:0030643 | intracellular phosphate ion homeostasis | 3 | 5 | 6.10e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 19 | 90 | 6.10e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 6.10e-01 |
| GO:BP | GO:0061301 | cerebellum vasculature morphogenesis | 1 | 1 | 6.10e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 4 | 6 | 6.11e-01 |
| GO:BP | GO:0006769 | nicotinamide metabolic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1901509 | regulation of endothelial tube morphogenesis | 3 | 3 | 6.11e-01 |
| GO:BP | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 6.11e-01 |
| GO:BP | GO:0018197 | peptidyl-aspartic acid modification | 3 | 5 | 6.11e-01 |
| GO:BP | GO:1900114 | positive regulation of histone H3-K9 trimethylation | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 14 | 276 | 6.12e-01 |
| GO:BP | GO:0072177 | mesonephric duct development | 2 | 7 | 6.12e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 6.12e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 2 | 31 | 6.12e-01 |
| GO:BP | GO:0035922 | foramen ovale closure | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0061550 | cranial ganglion development | 3 | 4 | 6.12e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 25 | 50 | 6.12e-01 |
| GO:BP | GO:0099542 | trans-synaptic signaling by endocannabinoid | 2 | 6 | 6.12e-01 |
| GO:BP | GO:0099541 | trans-synaptic signaling by lipid | 2 | 6 | 6.12e-01 |
| GO:BP | GO:0036306 | embryonic heart tube elongation | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0140706 | protein-containing complex localization to centriolar satellite | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0060577 | pulmonary vein morphogenesis | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 31 | 95 | 6.12e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 3 | 38 | 6.12e-01 |
| GO:BP | GO:0002051 | osteoblast fate commitment | 2 | 2 | 6.13e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 2 | 11 | 6.13e-01 |
| GO:BP | GO:0097577 | sequestering of iron ion | 1 | 3 | 6.13e-01 |
| GO:BP | GO:1990792 | cellular response to glial cell derived neurotrophic factor | 1 | 1 | 6.13e-01 |
| GO:BP | GO:0009224 | CMP biosynthetic process | 1 | 3 | 6.13e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 24 | 37 | 6.13e-01 |
| GO:BP | GO:1990790 | response to glial cell derived neurotrophic factor | 1 | 1 | 6.13e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 11 | 136 | 6.13e-01 |
| GO:BP | GO:0032197 | retrotransposition | 3 | 4 | 6.13e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 6.13e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 6.13e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 7 | 41 | 6.13e-01 |
| GO:BP | GO:1904447 | folate import across plasma membrane | 1 | 4 | 6.13e-01 |
| GO:BP | GO:0060755 | negative regulation of mast cell chemotaxis | 1 | 1 | 6.13e-01 |
| GO:BP | GO:1905755 | protein localization to cytoplasmic microtubule | 1 | 2 | 6.14e-01 |
| GO:BP | GO:1902889 | protein localization to spindle microtubule | 1 | 2 | 6.14e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 1 | 252 | 6.14e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 6.14e-01 |
| GO:BP | GO:1904015 | cellular response to serotonin | 1 | 1 | 6.14e-01 |
| GO:BP | GO:1904014 | response to serotonin | 1 | 1 | 6.14e-01 |
| GO:BP | GO:0010757 | negative regulation of plasminogen activation | 5 | 7 | 6.14e-01 |
| GO:BP | GO:0048511 | rhythmic process | 9 | 237 | 6.14e-01 |
| GO:BP | GO:0042538 | hyperosmotic salinity response | 4 | 8 | 6.14e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 4 | 66 | 6.14e-01 |
| GO:BP | GO:0097370 | protein O-GlcNAcylation via threonine | 1 | 1 | 6.14e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 1 | 26 | 6.14e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 5 | 230 | 6.15e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 32 | 60 | 6.15e-01 |
| GO:BP | GO:0050821 | protein stabilization | 4 | 191 | 6.15e-01 |
| GO:BP | GO:0060696 | regulation of phospholipid catabolic process | 3 | 5 | 6.15e-01 |
| GO:BP | GO:0035780 | CD80 biosynthetic process | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0016203 | muscle attachment | 1 | 1 | 6.15e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 6.15e-01 |
| GO:BP | GO:0036035 | osteoclast development | 1 | 10 | 6.15e-01 |
| GO:BP | GO:0017183 | peptidyl-diphthamide biosynthetic process from peptidyl-histidine | 3 | 7 | 6.15e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 3 | 23 | 6.15e-01 |
| GO:BP | GO:0017182 | peptidyl-diphthamide metabolic process | 3 | 7 | 6.15e-01 |
| GO:BP | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 2 | 6.15e-01 |
| GO:BP | GO:0060024 | rhythmic synaptic transmission | 2 | 3 | 6.15e-01 |
| GO:BP | GO:0120179 | adherens junction disassembly | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0007292 | female gamete generation | 25 | 103 | 6.15e-01 |
| GO:BP | GO:0090386 | phagosome maturation involved in apoptotic cell clearance | 1 | 2 | 6.15e-01 |
| GO:BP | GO:0090387 | phagolysosome assembly involved in apoptotic cell clearance | 1 | 2 | 6.15e-01 |
| GO:BP | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion | 2 | 3 | 6.15e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 6.15e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 5 | 61 | 6.15e-01 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 1 | 8 | 6.15e-01 |
| GO:BP | GO:0015712 | hexose phosphate transport | 1 | 3 | 6.16e-01 |
| GO:BP | GO:0015760 | glucose-6-phosphate transport | 1 | 3 | 6.16e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 12 | 31 | 6.16e-01 |
| GO:BP | GO:0008406 | gonad development | 2 | 158 | 6.16e-01 |
| GO:BP | GO:0060092 | regulation of synaptic transmission, glycinergic | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0140719 | constitutive heterochromatin formation | 5 | 10 | 6.16e-01 |
| GO:BP | GO:0061087 | positive regulation of histone H3-K27 methylation | 1 | 4 | 6.16e-01 |
| GO:BP | GO:0070537 | histone H2A K63-linked deubiquitination | 2 | 8 | 6.16e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 4 | 1167 | 6.16e-01 |
| GO:BP | GO:0032098 | regulation of appetite | 1 | 8 | 6.16e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 7 | 23 | 6.16e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 5 | 63 | 6.16e-01 |
| GO:BP | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation | 2 | 3 | 6.16e-01 |
| GO:BP | GO:0060720 | spongiotrophoblast cell proliferation | 1 | 2 | 6.16e-01 |
| GO:BP | GO:0060722 | cell proliferation involved in embryonic placenta development | 1 | 2 | 6.16e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 2 | 20 | 6.16e-01 |
| GO:BP | GO:0031648 | protein destabilization | 2 | 45 | 6.16e-01 |
| GO:BP | GO:0051394 | regulation of nerve growth factor receptor activity | 1 | 1 | 6.16e-01 |
| GO:BP | GO:1905532 | regulation of leucine import across plasma membrane | 1 | 3 | 6.16e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 6 | 9 | 6.16e-01 |
| GO:BP | GO:0090214 | spongiotrophoblast layer developmental growth | 1 | 2 | 6.16e-01 |
| GO:BP | GO:0006579 | amino-acid betaine catabolic process | 1 | 1 | 6.16e-01 |
| GO:BP | GO:1900826 | negative regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0060299 | negative regulation of sarcomere organization | 1 | 1 | 6.16e-01 |
| GO:BP | GO:1903547 | regulation of growth hormone activity | 1 | 2 | 6.16e-01 |
| GO:BP | GO:0008306 | associative learning | 3 | 65 | 6.16e-01 |
| GO:BP | GO:0038009 | regulation of signal transduction by receptor internalization | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 6 | 12 | 6.16e-01 |
| GO:BP | GO:0071043 | CUT metabolic process | 1 | 5 | 6.16e-01 |
| GO:BP | GO:0071034 | CUT catabolic process | 1 | 5 | 6.16e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 1 | 168 | 6.17e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 3 | 78 | 6.17e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 3 | 26 | 6.17e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 4 | 21 | 6.17e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 5 | 31 | 6.17e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 9 | 54 | 6.17e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 5 | 211 | 6.17e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 3 | 1914 | 6.17e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 5 | 25 | 6.17e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 2 | 35 | 6.17e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 1 | 35 | 6.17e-01 |
| GO:BP | GO:1904692 | positive regulation of type B pancreatic cell proliferation | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 2 | 6 | 6.17e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 2 | 1869 | 6.17e-01 |
| GO:BP | GO:0015734 | taurine transport | 2 | 6 | 6.17e-01 |
| GO:BP | GO:0002318 | myeloid progenitor cell differentiation | 3 | 7 | 6.17e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 1 | 169 | 6.17e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 8 | 17 | 6.17e-01 |
| GO:BP | GO:0019464 | glycine decarboxylation via glycine cleavage system | 3 | 3 | 6.17e-01 |
| GO:BP | GO:0030205 | dermatan sulfate metabolic process | 2 | 9 | 6.17e-01 |
| GO:BP | GO:0052779 | amino disaccharide metabolic process | 2 | 9 | 6.17e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 3 | 60 | 6.18e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 4 | 1248 | 6.18e-01 |
| GO:BP | GO:0060197 | cloacal septation | 1 | 2 | 6.18e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 6.18e-01 |
| GO:BP | GO:0070445 | regulation of oligodendrocyte progenitor proliferation | 1 | 5 | 6.18e-01 |
| GO:BP | GO:0070444 | oligodendrocyte progenitor proliferation | 1 | 5 | 6.18e-01 |
| GO:BP | GO:0035995 | detection of muscle stretch | 1 | 7 | 6.18e-01 |
| GO:BP | GO:0033567 | DNA replication, Okazaki fragment processing | 1 | 2 | 6.18e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 1 | 20 | 6.18e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 7 | 11 | 6.18e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 7 | 11 | 6.18e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 6.18e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 6.18e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 6.18e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 3 | 2646 | 6.18e-01 |
| GO:BP | GO:0051458 | corticotropin secretion | 2 | 4 | 6.18e-01 |
| GO:BP | GO:1903367 | positive regulation of fear response | 2 | 4 | 6.18e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 6 | 55 | 6.18e-01 |
| GO:BP | GO:0072660 | maintenance of protein location in plasma membrane | 1 | 2 | 6.18e-01 |
| GO:BP | GO:0090156 | intracellular sphingolipid homeostasis | 1 | 5 | 6.18e-01 |
| GO:BP | GO:0072658 | maintenance of protein location in membrane | 1 | 2 | 6.18e-01 |
| GO:BP | GO:0061091 | regulation of phospholipid translocation | 1 | 5 | 6.18e-01 |
| GO:BP | GO:0006678 | glucosylceramide metabolic process | 1 | 5 | 6.18e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 5 | 130 | 6.18e-01 |
| GO:BP | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 2 | 6.18e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 2 | 109 | 6.18e-01 |
| GO:BP | GO:0000709 | meiotic joint molecule formation | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 23 | 188 | 6.19e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 1 | 535 | 6.19e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 5 | 14 | 6.19e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 6.19e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 3 | 830 | 6.19e-01 |
| GO:BP | GO:0036261 | 7-methylguanosine cap hypermethylation | 5 | 8 | 6.19e-01 |
| GO:BP | GO:0006563 | L-serine metabolic process | 7 | 10 | 6.19e-01 |
| GO:BP | GO:0000708 | meiotic strand invasion | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0010774 | meiotic strand invasion involved in reciprocal meiotic recombination | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 2 | 101 | 6.19e-01 |
| GO:BP | GO:0039530 | MDA-5 signaling pathway | 1 | 8 | 6.19e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 6.19e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 6 | 41 | 6.19e-01 |
| GO:BP | GO:0030540 | female genitalia development | 3 | 14 | 6.19e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 6.19e-01 |
| GO:BP | GO:1903352 | L-ornithine transmembrane transport | 1 | 4 | 6.19e-01 |
| GO:BP | GO:1904923 | regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 8 | 15 | 6.19e-01 |
| GO:BP | GO:0045338 | farnesyl diphosphate metabolic process | 2 | 5 | 6.19e-01 |
| GO:BP | GO:1902859 | propionyl-CoA catabolic process | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0071140 | resolution of mitotic recombination intermediates | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0019389 | glucuronoside metabolic process | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0019391 | glucuronoside catabolic process | 1 | 2 | 6.19e-01 |
| GO:BP | GO:1904557 | L-alanine transmembrane transport | 3 | 4 | 6.19e-01 |
| GO:BP | GO:0070977 | bone maturation | 1 | 19 | 6.19e-01 |
| GO:BP | GO:1903849 | positive regulation of aorta morphogenesis | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0003252 | negative regulation of cell proliferation involved in heart valve morphogenesis | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0021944 | neuronal-glial interaction involved in hindbrain glial-mediated radial cell migration | 1 | 1 | 6.19e-01 |
| GO:BP | GO:1903847 | regulation of aorta morphogenesis | 1 | 2 | 6.19e-01 |
| GO:BP | GO:1901072 | glucosamine-containing compound catabolic process | 4 | 5 | 6.19e-01 |
| GO:BP | GO:0045337 | farnesyl diphosphate biosynthetic process | 1 | 3 | 6.19e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 51 | 1137 | 6.19e-01 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 2 | 39 | 6.19e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 6.20e-01 |
| GO:BP | GO:0071528 | tRNA re-export from nucleus | 1 | 1 | 6.20e-01 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 2 | 11 | 6.21e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 6.21e-01 |
| GO:BP | GO:0002069 | columnar/cuboidal epithelial cell maturation | 5 | 9 | 6.21e-01 |
| GO:BP | GO:0030185 | nitric oxide transport | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 12 | 39 | 6.22e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 3 | 12 | 6.22e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 5 | 13 | 6.22e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 6.22e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 6.22e-01 |
| GO:BP | GO:0072675 | osteoclast fusion | 1 | 5 | 6.22e-01 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 2 | 14 | 6.22e-01 |
| GO:BP | GO:0007521 | muscle cell fate determination | 1 | 2 | 6.22e-01 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 2 | 5 | 6.22e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 2 | 8 | 6.22e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 6.22e-01 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 6 | 14 | 6.22e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 4 | 7 | 6.22e-01 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 3 | 45 | 6.22e-01 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 1 | 14 | 6.22e-01 |
| GO:BP | GO:0016320 | endoplasmic reticulum membrane fusion | 1 | 2 | 6.22e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 6.22e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 1 | 12 | 6.22e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 6 | 134 | 6.22e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 6.22e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 6.22e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 6.22e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 2 | 161 | 6.22e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 6.22e-01 |
| GO:BP | GO:2000644 | regulation of receptor catabolic process | 3 | 9 | 6.22e-01 |
| GO:BP | GO:0006107 | oxaloacetate metabolic process | 5 | 7 | 6.22e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 9 | 115 | 6.22e-01 |
| GO:BP | GO:1905578 | regulation of ERBB3 signaling pathway | 1 | 1 | 6.22e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 6 | 25 | 6.22e-01 |
| GO:BP | GO:1905580 | positive regulation of ERBB3 signaling pathway | 1 | 1 | 6.22e-01 |
| GO:BP | GO:1900112 | regulation of histone H3-K9 trimethylation | 2 | 5 | 6.22e-01 |
| GO:BP | GO:0051573 | negative regulation of histone H3-K9 methylation | 2 | 5 | 6.22e-01 |
| GO:BP | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 1 | 16 | 6.23e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 6.23e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 5 | 17 | 6.23e-01 |
| GO:BP | GO:0002432 | granuloma formation | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 3 | 16 | 6.24e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 32 | 6.24e-01 |
| GO:BP | GO:0009093 | cysteine catabolic process | 2 | 3 | 6.24e-01 |
| GO:BP | GO:0046439 | L-cysteine metabolic process | 2 | 3 | 6.24e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 6 | 8 | 6.24e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 7 | 199 | 6.24e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 6 | 8 | 6.24e-01 |
| GO:BP | GO:0061088 | regulation of sequestering of zinc ion | 2 | 4 | 6.24e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 6 | 8 | 6.24e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 6.24e-01 |
| GO:BP | GO:1990786 | cellular response to dsDNA | 1 | 2 | 6.24e-01 |
| GO:BP | GO:0021536 | diencephalon development | 1 | 41 | 6.24e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 2 | 12 | 6.24e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 8 | 52 | 6.24e-01 |
| GO:BP | GO:1990784 | response to dsDNA | 1 | 2 | 6.24e-01 |
| GO:BP | GO:0019448 | L-cysteine catabolic process | 2 | 3 | 6.24e-01 |
| GO:BP | GO:0046794 | transport of virus | 9 | 19 | 6.24e-01 |
| GO:BP | GO:0009750 | response to fructose | 1 | 3 | 6.24e-01 |
| GO:BP | GO:0097332 | response to antipsychotic drug | 1 | 2 | 6.24e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 3 | 9 | 6.24e-01 |
| GO:BP | GO:2000489 | regulation of hepatic stellate cell activation | 4 | 6 | 6.24e-01 |
| GO:BP | GO:0006702 | androgen biosynthetic process | 2 | 4 | 6.24e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 1 | 40 | 6.24e-01 |
| GO:BP | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis | 3 | 9 | 6.24e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 7 | 59 | 6.24e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 3 | 86 | 6.24e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 3 | 75 | 6.24e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 3 | 73 | 6.24e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 1 | 64 | 6.24e-01 |
| GO:BP | GO:0015742 | alpha-ketoglutarate transport | 1 | 3 | 6.25e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 209 | 6.25e-01 |
| GO:BP | GO:0001770 | establishment of natural killer cell polarity | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0140354 | lipid import into cell | 1 | 15 | 6.26e-01 |
| GO:BP | GO:1901160 | primary amino compound metabolic process | 1 | 8 | 6.26e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 2 | 1014 | 6.26e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 6 | 19 | 6.26e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 7 | 29 | 6.26e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 6 | 25 | 6.26e-01 |
| GO:BP | GO:0034756 | regulation of iron ion transport | 1 | 4 | 6.26e-01 |
| GO:BP | GO:0019060 | intracellular transport of viral protein in host cell | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 12 | 38 | 6.26e-01 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 2 | 9 | 6.26e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 6 | 19 | 6.26e-01 |
| GO:BP | GO:1905857 | positive regulation of pentose-phosphate shunt | 2 | 3 | 6.26e-01 |
| GO:BP | GO:0035787 | cell migration involved in kidney development | 1 | 5 | 6.26e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 6 | 25 | 6.26e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 2 | 6 | 6.26e-01 |
| GO:BP | GO:0030581 | symbiont intracellular protein transport in host | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0030488 | tRNA methylation | 19 | 42 | 6.26e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 3 | 16 | 6.26e-01 |
| GO:BP | GO:0046533 | negative regulation of photoreceptor cell differentiation | 2 | 3 | 6.26e-01 |
| GO:BP | GO:0045819 | positive regulation of glycogen catabolic process | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0072174 | metanephric tubule formation | 2 | 3 | 6.26e-01 |
| GO:BP | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway | 2 | 3 | 6.26e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 6.26e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 6.26e-01 |
| GO:BP | GO:2000492 | regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation | 2 | 3 | 6.26e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 1 | 67 | 6.26e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 8 | 233 | 6.26e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0060020 | Bergmann glial cell differentiation | 4 | 9 | 6.26e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0072382 | minus-end-directed vesicle transport along microtubule | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1901678 | iron coordination entity transport | 4 | 11 | 6.27e-01 |
| GO:BP | GO:1905089 | regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 2 | 6.27e-01 |
| GO:BP | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 6 | 6.27e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 1 | 10 | 6.27e-01 |
| GO:BP | GO:0015938 | coenzyme A catabolic process | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0044580 | butyryl-CoA catabolic process | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 6.28e-01 |
| GO:BP | GO:0060385 | axonogenesis involved in innervation | 1 | 7 | 6.28e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 2 | 63 | 6.28e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 6.28e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 5 | 32 | 6.28e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 5 | 225 | 6.28e-01 |
| GO:BP | GO:1901492 | positive regulation of lymphangiogenesis | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 9 | 102 | 6.28e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 7 | 19 | 6.28e-01 |
| GO:BP | GO:0061034 | olfactory bulb mitral cell layer development | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0031118 | rRNA pseudouridine synthesis | 3 | 10 | 6.28e-01 |
| GO:BP | GO:1904851 | positive regulation of establishment of protein localization to telomere | 3 | 10 | 6.28e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 6.28e-01 |
| GO:BP | GO:0006106 | fumarate metabolic process | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0021934 | hindbrain tangential cell migration | 3 | 3 | 6.28e-01 |
| GO:BP | GO:0003100 | regulation of systemic arterial blood pressure by endothelin | 2 | 5 | 6.28e-01 |
| GO:BP | GO:0010033 | response to organic substance | 2 | 1957 | 6.28e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 2 | 20 | 6.28e-01 |
| GO:BP | GO:0006532 | aspartate biosynthetic process | 2 | 2 | 6.28e-01 |
| GO:BP | GO:0032570 | response to progesterone | 1 | 27 | 6.28e-01 |
| GO:BP | GO:0140060 | axon arborization | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0070572 | positive regulation of neuron projection regeneration | 1 | 8 | 6.28e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 7 | 10 | 6.28e-01 |
| GO:BP | GO:0006412 | translation | 2 | 646 | 6.29e-01 |
| GO:BP | GO:0033623 | regulation of integrin activation | 4 | 11 | 6.29e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 3 | 76 | 6.29e-01 |
| GO:BP | GO:1901423 | response to benzene | 1 | 1 | 6.29e-01 |
| GO:BP | GO:1904710 | positive regulation of granulosa cell apoptotic process | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0046687 | response to chromate | 1 | 1 | 6.29e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 2 | 86 | 6.29e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 3 | 76 | 6.29e-01 |
| GO:BP | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0031117 | positive regulation of microtubule depolymerization | 2 | 4 | 6.29e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 2 | 86 | 6.29e-01 |
| GO:BP | GO:0071247 | cellular response to chromate | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 6.29e-01 |
| GO:BP | GO:0060931 | sinoatrial node cell development | 2 | 4 | 6.29e-01 |
| GO:BP | GO:1905396 | cellular response to flavonoid | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0033385 | geranylgeranyl diphosphate metabolic process | 1 | 2 | 6.29e-01 |
| GO:BP | GO:0048668 | collateral sprouting | 3 | 25 | 6.29e-01 |
| GO:BP | GO:1904045 | cellular response to aldosterone | 1 | 5 | 6.29e-01 |
| GO:BP | GO:0061025 | membrane fusion | 4 | 137 | 6.29e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 6 | 131 | 6.29e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 11 | 212 | 6.29e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 1 | 159 | 6.29e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 12 | 203 | 6.29e-01 |
| GO:BP | GO:0002939 | tRNA N1-guanine methylation | 2 | 3 | 6.29e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 6.29e-01 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 6 | 12 | 6.29e-01 |
| GO:BP | GO:1901837 | negative regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 4 | 6.29e-01 |
| GO:BP | GO:1990264 | peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | 2 | 4 | 6.29e-01 |
| GO:BP | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation | 1 | 2 | 6.29e-01 |
| GO:BP | GO:0035229 | positive regulation of glutamate-cysteine ligase activity | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0035227 | regulation of glutamate-cysteine ligase activity | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0035282 | segmentation | 7 | 75 | 6.29e-01 |
| GO:BP | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway | 1 | 2 | 6.29e-01 |
| GO:BP | GO:0098937 | anterograde dendritic transport | 1 | 9 | 6.29e-01 |
| GO:BP | GO:0016081 | synaptic vesicle docking | 2 | 9 | 6.30e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 6.30e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 6.30e-01 |
| GO:BP | GO:0031282 | regulation of guanylate cyclase activity | 2 | 3 | 6.30e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 6.30e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 6.30e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 4 | 29 | 6.30e-01 |
| GO:BP | GO:0000055 | ribosomal large subunit export from nucleus | 3 | 8 | 6.30e-01 |
| GO:BP | GO:1904180 | negative regulation of membrane depolarization | 2 | 6 | 6.30e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 9 | 31 | 6.30e-01 |
| GO:BP | GO:0046458 | hexadecanal metabolic process | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0050832 | defense response to fungus | 1 | 14 | 6.30e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 4 | 11 | 6.30e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 2 | 13 | 6.30e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 11 | 37 | 6.30e-01 |
| GO:BP | GO:0018993 | somatic sex determination | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0045720 | negative regulation of integrin biosynthetic process | 1 | 1 | 6.31e-01 |
| GO:BP | GO:2000654 | regulation of cellular response to testosterone stimulus | 1 | 2 | 6.31e-01 |
| GO:BP | GO:0042595 | behavioral response to starvation | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 14 | 45 | 6.31e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 6 | 39 | 6.31e-01 |
| GO:BP | GO:0000958 | mitochondrial mRNA catabolic process | 2 | 3 | 6.32e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 18 | 39 | 6.32e-01 |
| GO:BP | GO:0015916 | fatty-acyl-CoA transport | 1 | 2 | 6.32e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 18 | 39 | 6.32e-01 |
| GO:BP | GO:1901337 | thioester transport | 1 | 2 | 6.32e-01 |
| GO:BP | GO:1903059 | regulation of protein lipidation | 1 | 11 | 6.32e-01 |
| GO:BP | GO:0021615 | glossopharyngeal nerve morphogenesis | 2 | 2 | 6.32e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 9 | 31 | 6.32e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 1 | 144 | 6.32e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 1 | 31 | 6.33e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 12 | 115 | 6.33e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 9 | 22 | 6.33e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 6.33e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 3 | 128 | 6.33e-01 |
| GO:BP | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity | 3 | 5 | 6.33e-01 |
| GO:BP | GO:1900143 | positive regulation of oligodendrocyte apoptotic process | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 4 | 26 | 6.33e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 9 | 40 | 6.33e-01 |
| GO:BP | GO:0018992 | germ-line sex determination | 1 | 2 | 6.33e-01 |
| GO:BP | GO:0014036 | neural crest cell fate specification | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0007542 | primary sex determination, germ-line | 1 | 2 | 6.33e-01 |
| GO:BP | GO:0006959 | humoral immune response | 2 | 80 | 6.33e-01 |
| GO:BP | GO:0060516 | primary prostatic bud elongation | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0019100 | male germ-line sex determination | 1 | 2 | 6.33e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 2 | 33 | 6.33e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0080120 | CAAX-box protein maturation | 1 | 3 | 6.33e-01 |
| GO:BP | GO:1904864 | negative regulation of beta-catenin-TCF complex assembly | 1 | 2 | 6.33e-01 |
| GO:BP | GO:1904863 | regulation of beta-catenin-TCF complex assembly | 1 | 2 | 6.33e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 6.33e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 6.33e-01 |
| GO:BP | GO:0035622 | intrahepatic bile duct development | 1 | 2 | 6.33e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 1 | 6 | 6.33e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 9 | 17 | 6.33e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 1 | 6 | 6.33e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 1 | 6 | 6.33e-01 |
| GO:BP | GO:0009133 | nucleoside diphosphate biosynthetic process | 7 | 11 | 6.33e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 6 | 27 | 6.33e-01 |
| GO:BP | GO:0035279 | miRNA-mediated gene silencing by mRNA destabilization | 1 | 3 | 6.34e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 17 | 46 | 6.34e-01 |
| GO:BP | GO:1905302 | negative regulation of macropinocytosis | 1 | 1 | 6.34e-01 |
| GO:BP | GO:1904050 | positive regulation of spontaneous neurotransmitter secretion | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0010526 | retrotransposon silencing | 1 | 3 | 6.34e-01 |
| GO:BP | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium | 1 | 1 | 6.34e-01 |
| GO:BP | GO:1902216 | positive regulation of interleukin-4-mediated signaling pathway | 1 | 1 | 6.34e-01 |
| GO:BP | GO:1901380 | negative regulation of potassium ion transmembrane transport | 2 | 18 | 6.34e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 3 | 8 | 6.34e-01 |
| GO:BP | GO:0048284 | organelle fusion | 3 | 135 | 6.34e-01 |
| GO:BP | GO:1990074 | polyuridylation-dependent mRNA catabolic process | 2 | 3 | 6.34e-01 |
| GO:BP | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process | 1 | 3 | 6.34e-01 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 3 | 8 | 6.34e-01 |
| GO:BP | GO:1902809 | regulation of skeletal muscle fiber differentiation | 1 | 4 | 6.34e-01 |
| GO:BP | GO:0034653 | retinoic acid catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0016103 | diterpenoid catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 25 | 91 | 6.34e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 9 | 33 | 6.34e-01 |
| GO:BP | GO:0035350 | FAD transmembrane transport | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0015880 | coenzyme A transport | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0015883 | FAD transport | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0080121 | AMP transport | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0071106 | adenosine 3’,5’-bisphosphate transmembrane transport | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0035349 | coenzyme A transmembrane transport | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 7 | 44 | 6.35e-01 |
| GO:BP | GO:0035873 | lactate transmembrane transport | 1 | 5 | 6.35e-01 |
| GO:BP | GO:0015727 | lactate transport | 1 | 5 | 6.35e-01 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 4 | 28 | 6.35e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 7 | 12 | 6.35e-01 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 2 | 9 | 6.36e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 9 | 16 | 6.36e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 3 | 12 | 6.36e-01 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 6 | 11 | 6.36e-01 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 1 | 10 | 6.36e-01 |
| GO:BP | GO:0032488 | Cdc42 protein signal transduction | 1 | 10 | 6.36e-01 |
| GO:BP | GO:0018364 | peptidyl-glutamine methylation | 1 | 4 | 6.36e-01 |
| GO:BP | GO:0060155 | platelet dense granule organization | 11 | 23 | 6.36e-01 |
| GO:BP | GO:0008015 | blood circulation | 31 | 374 | 6.36e-01 |
| GO:BP | GO:0097376 | interneuron axon guidance | 1 | 1 | 6.36e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 6.36e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 3 | 1995 | 6.36e-01 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 1 | 7 | 6.36e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 9 | 16 | 6.36e-01 |
| GO:BP | GO:0072278 | metanephric comma-shaped body morphogenesis | 1 | 3 | 6.36e-01 |
| GO:BP | GO:0036364 | transforming growth factor beta1 activation | 1 | 1 | 6.36e-01 |
| GO:BP | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 2 | 5 | 6.36e-01 |
| GO:BP | GO:2000645 | negative regulation of receptor catabolic process | 1 | 3 | 6.36e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 10 | 448 | 6.36e-01 |
| GO:BP | GO:0097476 | spinal cord motor neuron migration | 1 | 3 | 6.36e-01 |
| GO:BP | GO:0010814 | substance P catabolic process | 2 | 3 | 6.36e-01 |
| GO:BP | GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 2 | 5 | 6.36e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 4 | 21 | 6.36e-01 |
| GO:BP | GO:0046081 | dUTP catabolic process | 1 | 1 | 6.36e-01 |
| GO:BP | GO:0046080 | dUTP metabolic process | 1 | 1 | 6.36e-01 |
| GO:BP | GO:2000508 | regulation of dendritic cell chemotaxis | 1 | 3 | 6.36e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 3 | 8 | 6.36e-01 |
| GO:BP | GO:0032194 | ubiquinone biosynthetic process via 3,4-dihydroxy-5-polyprenylbenzoate | 1 | 1 | 6.36e-01 |
| GO:BP | GO:0042371 | vitamin K biosynthetic process | 1 | 1 | 6.36e-01 |
| GO:BP | GO:0099152 | regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane | 2 | 3 | 6.36e-01 |
| GO:BP | GO:0009234 | menaquinone biosynthetic process | 1 | 1 | 6.36e-01 |
| GO:BP | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus | 3 | 6 | 6.36e-01 |
| GO:BP | GO:1903811 | L-asparagine import across plasma membrane | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 14 | 61 | 6.37e-01 |
| GO:BP | GO:0055024 | regulation of cardiac muscle tissue development | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 6.37e-01 |
| GO:BP | GO:0051891 | positive regulation of cardioblast differentiation | 2 | 4 | 6.37e-01 |
| GO:BP | GO:1903937 | response to acrylamide | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 6.37e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 5 | 54 | 6.37e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 5 | 126 | 6.37e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 6 | 11 | 6.37e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 6.37e-01 |
| GO:BP | GO:0034436 | glycoprotein transport | 2 | 3 | 6.38e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 2 | 15 | 6.38e-01 |
| GO:BP | GO:0070859 | positive regulation of bile acid biosynthetic process | 2 | 3 | 6.38e-01 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 3 | 7 | 6.38e-01 |
| GO:BP | GO:1904253 | positive regulation of bile acid metabolic process | 2 | 3 | 6.38e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 6.38e-01 |
| GO:BP | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 4 | 21 | 6.38e-01 |
| GO:BP | GO:1902215 | negative regulation of interleukin-4-mediated signaling pathway | 1 | 1 | 6.38e-01 |
| GO:BP | GO:1902233 | negative regulation of positive thymic T cell selection | 1 | 1 | 6.38e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 4 | 12 | 6.38e-01 |
| GO:BP | GO:1902206 | negative regulation of interleukin-2-mediated signaling pathway | 1 | 1 | 6.38e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 7 | 1006 | 6.38e-01 |
| GO:BP | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling | 9 | 12 | 6.38e-01 |
| GO:BP | GO:0048371 | lateral mesodermal cell differentiation | 2 | 2 | 6.38e-01 |
| GO:BP | GO:0030327 | prenylated protein catabolic process | 1 | 3 | 6.38e-01 |
| GO:BP | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis | 2 | 6 | 6.38e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 1 | 60 | 6.38e-01 |
| GO:BP | GO:0014852 | regulation of skeletal muscle contraction by neural stimulation via neuromuscular junction | 1 | 2 | 6.38e-01 |
| GO:BP | GO:1902205 | regulation of interleukin-2-mediated signaling pathway | 1 | 1 | 6.38e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 8 | 20 | 6.38e-01 |
| GO:BP | GO:0006689 | ganglioside catabolic process | 2 | 5 | 6.39e-01 |
| GO:BP | GO:0010272 | response to silver ion | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0097188 | dentin mineralization | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0071105 | response to interleukin-11 | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis | 1 | 3 | 6.39e-01 |
| GO:BP | GO:0016199 | axon midline choice point recognition | 2 | 3 | 6.39e-01 |
| GO:BP | GO:0009098 | leucine biosynthetic process | 1 | 2 | 6.39e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 2 | 88 | 6.39e-01 |
| GO:BP | GO:1901252 | regulation of intracellular transport of viral material | 1 | 2 | 6.39e-01 |
| GO:BP | GO:0072046 | establishment of planar polarity involved in nephron morphogenesis | 1 | 1 | 6.39e-01 |
| GO:BP | GO:1901175 | lycopene metabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:1901176 | lycopene catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0031000 | response to caffeine | 2 | 13 | 6.39e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 1 | 5 | 6.39e-01 |
| GO:BP | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 2 | 3 | 6.39e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 2 | 8 | 6.39e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 3 | 963 | 6.39e-01 |
| GO:BP | GO:0062172 | lutein catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0097068 | response to thyroxine | 3 | 4 | 6.39e-01 |
| GO:BP | GO:0031167 | rRNA methylation | 12 | 24 | 6.39e-01 |
| GO:BP | GO:0035585 | calcium-mediated signaling using extracellular calcium source | 1 | 2 | 6.39e-01 |
| GO:BP | GO:0070564 | positive regulation of vitamin D receptor signaling pathway | 1 | 5 | 6.39e-01 |
| GO:BP | GO:1900453 | negative regulation of long-term synaptic depression | 1 | 3 | 6.39e-01 |
| GO:BP | GO:0071283 | cellular response to iron(III) ion | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0062170 | lutein metabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0016124 | xanthophyll catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0140009 | L-aspartate import across plasma membrane | 1 | 5 | 6.39e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 21 | 198 | 6.39e-01 |
| GO:BP | GO:0016118 | carotenoid catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0008065 | establishment of blood-nerve barrier | 2 | 3 | 6.39e-01 |
| GO:BP | GO:0019682 | glyceraldehyde-3-phosphate metabolic process | 2 | 5 | 6.39e-01 |
| GO:BP | GO:1901826 | zeaxanthin catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process | 3 | 16 | 6.39e-01 |
| GO:BP | GO:0016110 | tetraterpenoid catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0042116 | macrophage activation | 2 | 57 | 6.39e-01 |
| GO:BP | GO:1903703 | enterocyte differentiation | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 2 | 41 | 6.39e-01 |
| GO:BP | GO:0045344 | negative regulation of MHC class I biosynthetic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0006768 | biotin metabolic process | 2 | 3 | 6.39e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 10 | 32 | 6.39e-01 |
| GO:BP | GO:0007398 | ectoderm development | 3 | 15 | 6.40e-01 |
| GO:BP | GO:0019080 | viral gene expression | 2 | 96 | 6.40e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 2 | 666 | 6.40e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 1 | 6 | 6.40e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 6 | 4292 | 6.40e-01 |
| GO:BP | GO:0021627 | olfactory nerve morphogenesis | 2 | 3 | 6.40e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 28 | 52 | 6.40e-01 |
| GO:BP | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis | 1 | 3 | 6.40e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 2 | 43 | 6.40e-01 |
| GO:BP | GO:1905381 | negative regulation of snRNA transcription by RNA polymerase II | 1 | 1 | 6.40e-01 |
| GO:BP | GO:1904746 | negative regulation of apoptotic process involved in development | 1 | 3 | 6.40e-01 |
| GO:BP | GO:0071712 | ER-associated misfolded protein catabolic process | 1 | 12 | 6.40e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 1 | 19 | 6.40e-01 |
| GO:BP | GO:1901303 | negative regulation of cargo loading into COPII-coated vesicle | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 3 | 1835 | 6.40e-01 |
| GO:BP | GO:0021502 | neural fold elevation formation | 1 | 3 | 6.40e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 2 | 108 | 6.40e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 5 | 35 | 6.40e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 3 | 70 | 6.40e-01 |
| GO:BP | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0035624 | receptor transactivation | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0032811 | negative regulation of epinephrine secretion | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0032928 | regulation of superoxide anion generation | 4 | 11 | 6.41e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 6.41e-01 |
| GO:BP | GO:0009617 | response to bacterium | 4 | 321 | 6.41e-01 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 1 | 7 | 6.41e-01 |
| GO:BP | GO:1903576 | response to L-arginine | 1 | 3 | 6.41e-01 |
| GO:BP | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process | 2 | 3 | 6.41e-01 |
| GO:BP | GO:0043248 | proteasome assembly | 1 | 13 | 6.41e-01 |
| GO:BP | GO:1900864 | mitochondrial RNA modification | 5 | 9 | 6.42e-01 |
| GO:BP | GO:0070900 | mitochondrial tRNA modification | 5 | 9 | 6.42e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 2 | 32 | 6.42e-01 |
| GO:BP | GO:0140192 | regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 6.42e-01 |
| GO:BP | GO:0048319 | axial mesoderm morphogenesis | 1 | 3 | 6.42e-01 |
| GO:BP | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 6.42e-01 |
| GO:BP | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 1 | 16 | 6.42e-01 |
| GO:BP | GO:2000124 | regulation of endocannabinoid signaling pathway | 1 | 3 | 6.42e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 5 | 9 | 6.43e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 16 | 121 | 6.43e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 6.43e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 2 | 58 | 6.44e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 6.44e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 5 | 41 | 6.44e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 8 | 45 | 6.44e-01 |
| GO:BP | GO:0140754 | reorganization of cellular membranes to establish viral sites of replication | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0015808 | L-alanine transport | 6 | 9 | 6.44e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 11 | 149 | 6.44e-01 |
| GO:BP | GO:1904205 | negative regulation of skeletal muscle hypertrophy | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0006409 | tRNA export from nucleus | 2 | 3 | 6.44e-01 |
| GO:BP | GO:0007588 | excretion | 1 | 18 | 6.44e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 1 | 12 | 6.44e-01 |
| GO:BP | GO:0001823 | mesonephros development | 3 | 77 | 6.44e-01 |
| GO:BP | GO:0034983 | peptidyl-lysine deacetylation | 3 | 8 | 6.45e-01 |
| GO:BP | GO:0006627 | protein processing involved in protein targeting to mitochondrion | 2 | 6 | 6.45e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 9 | 38 | 6.45e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 2 | 170 | 6.45e-01 |
| GO:BP | GO:0036155 | acylglycerol acyl-chain remodeling | 2 | 2 | 6.45e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 1 | 128 | 6.45e-01 |
| GO:BP | GO:0045342 | MHC class II biosynthetic process | 5 | 12 | 6.46e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 9 | 70 | 6.46e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 21 | 665 | 6.46e-01 |
| GO:BP | GO:0010916 | negative regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 6.46e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 1 | 36 | 6.46e-01 |
| GO:BP | GO:0010915 | regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 6.46e-01 |
| GO:BP | GO:2000230 | negative regulation of pancreatic stellate cell proliferation | 1 | 1 | 6.46e-01 |
| GO:BP | GO:0033353 | S-adenosylmethionine cycle | 2 | 7 | 6.46e-01 |
| GO:BP | GO:0099538 | synaptic signaling via neuropeptide | 1 | 2 | 6.46e-01 |
| GO:BP | GO:0051004 | regulation of lipoprotein lipase activity | 1 | 11 | 6.46e-01 |
| GO:BP | GO:0048799 | animal organ maturation | 1 | 20 | 6.46e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 2 | 15 | 6.46e-01 |
| GO:BP | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration | 3 | 5 | 6.46e-01 |
| GO:BP | GO:0001756 | somitogenesis | 16 | 48 | 6.46e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 10 | 15 | 6.46e-01 |
| GO:BP | GO:2000535 | regulation of entry of bacterium into host cell | 1 | 6 | 6.46e-01 |
| GO:BP | GO:0016191 | synaptic vesicle uncoating | 4 | 7 | 6.46e-01 |
| GO:BP | GO:0070602 | regulation of centromeric sister chromatid cohesion | 2 | 4 | 6.47e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 1 | 12 | 6.47e-01 |
| GO:BP | GO:0007387 | anterior compartment pattern formation | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0007388 | posterior compartment specification | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0033750 | ribosome localization | 1 | 15 | 6.47e-01 |
| GO:BP | GO:0009067 | aspartate family amino acid biosynthetic process | 10 | 18 | 6.47e-01 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 1 | 19 | 6.47e-01 |
| GO:BP | GO:0014728 | regulation of the force of skeletal muscle contraction | 1 | 3 | 6.47e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 3 | 20 | 6.47e-01 |
| GO:BP | GO:1903527 | positive regulation of membrane tubulation | 2 | 3 | 6.47e-01 |
| GO:BP | GO:0033079 | immature T cell proliferation | 2 | 7 | 6.47e-01 |
| GO:BP | GO:0033080 | immature T cell proliferation in thymus | 2 | 7 | 6.47e-01 |
| GO:BP | GO:1902624 | positive regulation of neutrophil migration | 2 | 21 | 6.47e-01 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 1 | 15 | 6.47e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 6.47e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 4 | 31 | 6.47e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 5 | 11 | 6.47e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 2 | 10 | 6.47e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 6.47e-01 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 1 | 5 | 6.47e-01 |
| GO:BP | GO:0097341 | zymogen inhibition | 2 | 5 | 6.47e-01 |
| GO:BP | GO:0097340 | inhibition of cysteine-type endopeptidase activity | 2 | 5 | 6.47e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 2 | 30 | 6.47e-01 |
| GO:BP | GO:0014862 | regulation of skeletal muscle contraction by chemo-mechanical energy conversion | 1 | 3 | 6.47e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 15 | 23 | 6.47e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 2 | 39 | 6.47e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 6 | 10 | 6.47e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 4 | 7 | 6.47e-01 |
| GO:BP | GO:0070637 | pyridine nucleoside metabolic process | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0046495 | nicotinamide riboside metabolic process | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0006738 | nicotinamide riboside catabolic process | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0070638 | pyridine nucleoside catabolic process | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0033523 | histone H2B ubiquitination | 5 | 10 | 6.47e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 2 | 228 | 6.48e-01 |
| GO:BP | GO:1902847 | regulation of neuronal signal transduction | 1 | 1 | 6.48e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 1 | 5 | 6.48e-01 |
| GO:BP | GO:1902998 | positive regulation of neurofibrillary tangle assembly | 1 | 1 | 6.48e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 5 | 11 | 6.48e-01 |
| GO:BP | GO:0050812 | regulation of acyl-CoA biosynthetic process | 3 | 5 | 6.48e-01 |
| GO:BP | GO:1905590 | fibronectin fibril organization | 1 | 1 | 6.48e-01 |
| GO:BP | GO:0046293 | formaldehyde biosynthetic process | 1 | 1 | 6.48e-01 |
| GO:BP | GO:0021932 | hindbrain radial glia guided cell migration | 4 | 8 | 6.48e-01 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 8 | 18 | 6.48e-01 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 1 | 5 | 6.48e-01 |
| GO:BP | GO:0050916 | sensory perception of sweet taste | 3 | 3 | 6.48e-01 |
| GO:BP | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate | 2 | 4 | 6.48e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 2 | 44 | 6.48e-01 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 2 | 9 | 6.48e-01 |
| GO:BP | GO:0031453 | positive regulation of heterochromatin formation | 2 | 15 | 6.48e-01 |
| GO:BP | GO:0120263 | positive regulation of heterochromatin organization | 2 | 15 | 6.48e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 17 | 91 | 6.48e-01 |
| GO:BP | GO:0070294 | renal sodium ion absorption | 1 | 9 | 6.48e-01 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 10 | 6.48e-01 |
| GO:BP | GO:0097102 | endothelial tip cell fate specification | 1 | 2 | 6.48e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 6.48e-01 |
| GO:BP | GO:0071921 | cohesin loading | 2 | 3 | 6.48e-01 |
| GO:BP | GO:0061441 | renal artery morphogenesis | 1 | 2 | 6.48e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 7 | 4617 | 6.49e-01 |
| GO:BP | GO:0060126 | somatotropin secreting cell differentiation | 1 | 2 | 6.49e-01 |
| GO:BP | GO:0070729 | cyclic nucleotide transport | 1 | 6 | 6.49e-01 |
| GO:BP | GO:1903790 | guanine nucleotide transmembrane transport | 1 | 5 | 6.49e-01 |
| GO:BP | GO:1903393 | positive regulation of adherens junction organization | 1 | 2 | 6.49e-01 |
| GO:BP | GO:0051168 | nuclear export | 11 | 150 | 6.49e-01 |
| GO:BP | GO:0044795 | trans-Golgi network to recycling endosome transport | 1 | 1 | 6.49e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 1 | 129 | 6.49e-01 |
| GO:BP | GO:0042339 | keratan sulfate metabolic process | 5 | 15 | 6.49e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 3 | 4 | 6.49e-01 |
| GO:BP | GO:1905602 | positive regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 2 | 6.49e-01 |
| GO:BP | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation | 3 | 5 | 6.50e-01 |
| GO:BP | GO:0060319 | primitive erythrocyte differentiation | 3 | 4 | 6.50e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 6.50e-01 |
| GO:BP | GO:0099118 | microtubule-based protein transport | 1 | 14 | 6.50e-01 |
| GO:BP | GO:0098840 | protein transport along microtubule | 1 | 14 | 6.50e-01 |
| GO:BP | GO:1901340 | negative regulation of store-operated calcium channel activity | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 19 | 64 | 6.50e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 6.50e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 6.50e-01 |
| GO:BP | GO:0015822 | ornithine transport | 1 | 5 | 6.50e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 12 | 42 | 6.50e-01 |
| GO:BP | GO:1903512 | phytanic acid metabolic process | 1 | 2 | 6.50e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 74 | 3625 | 6.50e-01 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 5 | 12 | 6.50e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 12 | 307 | 6.50e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 9 | 14 | 6.51e-01 |
| GO:BP | GO:0033572 | transferrin transport | 4 | 8 | 6.51e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 2 | 27 | 6.51e-01 |
| GO:BP | GO:1900364 | negative regulation of mRNA polyadenylation | 1 | 1 | 6.51e-01 |
| GO:BP | GO:0002280 | monocyte activation involved in immune response | 1 | 1 | 6.51e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 2 | 28 | 6.51e-01 |
| GO:BP | GO:1903818 | positive regulation of voltage-gated potassium channel activity | 1 | 11 | 6.51e-01 |
| GO:BP | GO:0007344 | pronuclear fusion | 1 | 1 | 6.51e-01 |
| GO:BP | GO:2000250 | negative regulation of actin cytoskeleton reorganization | 1 | 1 | 6.51e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 32 | 70 | 6.51e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 20 | 4539 | 6.51e-01 |
| GO:BP | GO:0000741 | karyogamy | 1 | 1 | 6.51e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 7 | 19 | 6.51e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 3 | 149 | 6.51e-01 |
| GO:BP | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation | 1 | 6 | 6.51e-01 |
| GO:BP | GO:0042780 | tRNA 3’-end processing | 1 | 7 | 6.51e-01 |
| GO:BP | GO:0060525 | prostate glandular acinus development | 5 | 5 | 6.51e-01 |
| GO:BP | GO:0051645 | Golgi localization | 7 | 14 | 6.51e-01 |
| GO:BP | GO:0002678 | positive regulation of chronic inflammatory response | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 1 | 23 | 6.52e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 9 | 190 | 6.52e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 2 | 1149 | 6.52e-01 |
| GO:BP | GO:0002943 | tRNA dihydrouridine synthesis | 2 | 4 | 6.52e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 3 | 104 | 6.52e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 4 | 24 | 6.52e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 3 | 104 | 6.52e-01 |
| GO:BP | GO:0150045 | regulation of synaptic signaling by nitric oxide | 1 | 1 | 6.52e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 6 | 47 | 6.52e-01 |
| GO:BP | GO:0050652 | dermatan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0048104 | establishment of body hair or bristle planar orientation | 1 | 3 | 6.52e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 3 | 21 | 6.52e-01 |
| GO:BP | GO:0048105 | establishment of body hair planar orientation | 1 | 3 | 6.52e-01 |
| GO:BP | GO:0021541 | ammon gyrus development | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0021510 | spinal cord development | 6 | 59 | 6.52e-01 |
| GO:BP | GO:0032497 | detection of lipopolysaccharide | 2 | 2 | 6.52e-01 |
| GO:BP | GO:0007399 | nervous system development | 3 | 1904 | 6.52e-01 |
| GO:BP | GO:2001198 | regulation of dendritic cell differentiation | 1 | 8 | 6.53e-01 |
| GO:BP | GO:0009912 | auditory receptor cell fate commitment | 3 | 4 | 6.53e-01 |
| GO:BP | GO:0060120 | inner ear receptor cell fate commitment | 3 | 4 | 6.53e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 3 | 56 | 6.53e-01 |
| GO:BP | GO:0050915 | sensory perception of sour taste | 1 | 4 | 6.53e-01 |
| GO:BP | GO:0072383 | plus-end-directed vesicle transport along microtubule | 2 | 6 | 6.53e-01 |
| GO:BP | GO:0035583 | sequestering of TGFbeta in extracellular matrix | 1 | 3 | 6.53e-01 |
| GO:BP | GO:1902480 | protein localization to mitotic spindle | 1 | 1 | 6.53e-01 |
| GO:BP | GO:0061960 | regulation of heme oxygenase activity | 1 | 1 | 6.53e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 8 | 38 | 6.53e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 1 | 20 | 6.53e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 6 | 43 | 6.53e-01 |
| GO:BP | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion | 1 | 3 | 6.53e-01 |
| GO:BP | GO:0017196 | N-terminal peptidyl-methionine acetylation | 2 | 8 | 6.53e-01 |
| GO:BP | GO:0031529 | ruffle organization | 16 | 45 | 6.53e-01 |
| GO:BP | GO:0003412 | establishment of epithelial cell apical/basal polarity involved in camera-type eye morphogenesis | 1 | 1 | 6.53e-01 |
| GO:BP | GO:0034030 | ribonucleoside bisphosphate biosynthetic process | 11 | 49 | 6.54e-01 |
| GO:BP | GO:0033866 | nucleoside bisphosphate biosynthetic process | 11 | 49 | 6.54e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 1 | 12 | 6.54e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 6.54e-01 |
| GO:BP | GO:0046865 | terpenoid transport | 2 | 3 | 6.54e-01 |
| GO:BP | GO:0045988 | negative regulation of striated muscle contraction | 2 | 7 | 6.54e-01 |
| GO:BP | GO:0046864 | isoprenoid transport | 2 | 3 | 6.54e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 4 | 9 | 6.54e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 4 | 58 | 6.54e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 6.54e-01 |
| GO:BP | GO:0034033 | purine nucleoside bisphosphate biosynthetic process | 11 | 49 | 6.54e-01 |
| GO:BP | GO:0030539 | male genitalia development | 1 | 13 | 6.54e-01 |
| GO:BP | GO:0010133 | proline catabolic process to glutamate | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 10 | 18 | 6.54e-01 |
| GO:BP | GO:1990758 | mitotic sister chromatid biorientation | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 2 | 111 | 6.54e-01 |
| GO:BP | GO:1902595 | regulation of DNA replication origin binding | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0032650 | regulation of interleukin-1 alpha production | 1 | 4 | 6.54e-01 |
| GO:BP | GO:0032610 | interleukin-1 alpha production | 1 | 4 | 6.54e-01 |
| GO:BP | GO:0044357 | regulation of rRNA stability | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0018344 | protein geranylgeranylation | 1 | 6 | 6.54e-01 |
| GO:BP | GO:0010265 | SCF complex assembly | 1 | 2 | 6.54e-01 |
| GO:BP | GO:1902374 | regulation of rRNA catabolic process | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 1 | 13 | 6.54e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 6.55e-01 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 4 | 15 | 6.55e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 10 | 18 | 6.55e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 1 | 13 | 6.55e-01 |
| GO:BP | GO:0150032 | positive regulation of protein localization to lysosome | 1 | 2 | 6.55e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 6.55e-01 |
| GO:BP | GO:0021508 | floor plate formation | 1 | 3 | 6.55e-01 |
| GO:BP | GO:2000615 | regulation of histone H3-K9 acetylation | 2 | 7 | 6.55e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 2 | 1133 | 6.55e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 9 | 38 | 6.55e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 26 | 1138 | 6.55e-01 |
| GO:BP | GO:0002309 | T cell proliferation involved in immune response | 1 | 3 | 6.55e-01 |
| GO:BP | GO:0031623 | receptor internalization | 1 | 95 | 6.55e-01 |
| GO:BP | GO:0035821 | modulation of process of another organism | 3 | 5 | 6.55e-01 |
| GO:BP | GO:0071500 | cellular response to nitrosative stress | 2 | 6 | 6.56e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 5 | 18 | 6.56e-01 |
| GO:BP | GO:0043977 | histone H2A-K5 acetylation | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0042665 | regulation of ectodermal cell fate specification | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0043980 | histone H2B-K12 acetylation | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0038001 | paracrine signaling | 1 | 3 | 6.56e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 6.56e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 6.56e-01 |
| GO:BP | GO:0006258 | UDP-glucose catabolic process | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 3 | 61 | 6.56e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 6.56e-01 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 6.56e-01 |
| GO:BP | GO:2000723 | negative regulation of cardiac vascular smooth muscle cell differentiation | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0072007 | mesangial cell differentiation | 1 | 5 | 6.56e-01 |
| GO:BP | GO:0006554 | lysine catabolic process | 1 | 6 | 6.56e-01 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 1 | 6 | 6.56e-01 |
| GO:BP | GO:1903939 | regulation of TORC2 signaling | 2 | 5 | 6.56e-01 |
| GO:BP | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle | 1 | 3 | 6.56e-01 |
| GO:BP | GO:0001715 | ectodermal cell fate specification | 1 | 1 | 6.56e-01 |
| GO:BP | GO:1902896 | terminal web assembly | 1 | 2 | 6.56e-01 |
| GO:BP | GO:2000077 | negative regulation of type B pancreatic cell development | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 6.56e-01 |
| GO:BP | GO:0006574 | valine catabolic process | 2 | 4 | 6.56e-01 |
| GO:BP | GO:0015825 | L-serine transport | 1 | 7 | 6.56e-01 |
| GO:BP | GO:0042666 | negative regulation of ectodermal cell fate specification | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0006669 | sphinganine-1-phosphate biosynthetic process | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0045887 | positive regulation of synaptic assembly at neuromuscular junction | 1 | 1 | 6.56e-01 |
| GO:BP | GO:1902656 | calcium ion import into cytosol | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 1 | 10 | 6.56e-01 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 1 | 10 | 6.56e-01 |
| GO:BP | GO:0071642 | positive regulation of macrophage inflammatory protein 1 alpha production | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0015677 | copper ion import | 1 | 5 | 6.56e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 3 | 101 | 6.56e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 5 | 8 | 6.56e-01 |
| GO:BP | GO:0000023 | maltose metabolic process | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 2 | 108 | 6.56e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 1 | 15 | 6.56e-01 |
| GO:BP | GO:1903333 | negative regulation of protein folding | 2 | 4 | 6.56e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 1 | 15 | 6.56e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 1 | 13 | 6.56e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 1 | 15 | 6.56e-01 |
| GO:BP | GO:0051641 | cellular localization | 2 | 2810 | 6.56e-01 |
| GO:BP | GO:1990046 | stress-induced mitochondrial fusion | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | 1 | 10 | 6.56e-01 |
| GO:BP | GO:0043586 | tongue development | 5 | 15 | 6.56e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 2 | 12 | 6.56e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 2 | 24 | 6.56e-01 |
| GO:BP | GO:0090143 | nucleoid organization | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 6 | 135 | 6.56e-01 |
| GO:BP | GO:0090144 | mitochondrial nucleoid organization | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0060718 | chorionic trophoblast cell differentiation | 2 | 5 | 6.56e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 2 | 4 | 6.56e-01 |
| GO:BP | GO:2000543 | positive regulation of gastrulation | 4 | 5 | 6.56e-01 |
| GO:BP | GO:0072359 | circulatory system development | 1 | 884 | 6.56e-01 |
| GO:BP | GO:0140051 | positive regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 6.56e-01 |
| GO:BP | GO:2000800 | regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 6.56e-01 |
| GO:BP | GO:2000802 | positive regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0140049 | regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0034772 | histone H4-K20 dimethylation | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0072236 | metanephric loop of Henle development | 1 | 3 | 6.56e-01 |
| GO:BP | GO:1905764 | regulation of protection from non-homologous end joining at telomere | 2 | 3 | 6.56e-01 |
| GO:BP | GO:0015878 | biotin transport | 1 | 3 | 6.56e-01 |
| GO:BP | GO:0031627 | telomeric loop formation | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 6 | 135 | 6.56e-01 |
| GO:BP | GO:0045763 | negative regulation of cellular amino acid metabolic process | 2 | 2 | 6.56e-01 |
| GO:BP | GO:1905765 | negative regulation of protection from non-homologous end joining at telomere | 2 | 3 | 6.56e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 6.56e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 41 | 6.56e-01 |
| GO:BP | GO:0061819 | telomeric DNA-containing double minutes formation | 2 | 3 | 6.56e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 3 | 2880 | 6.56e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 14 | 33 | 6.56e-01 |
| GO:BP | GO:0030389 | fructosamine metabolic process | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0090152 | establishment of protein localization to mitochondrial membrane involved in mitochondrial fission | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0042797 | tRNA transcription by RNA polymerase III | 1 | 7 | 6.57e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 7 | 39 | 6.57e-01 |
| GO:BP | GO:1900221 | regulation of amyloid-beta clearance | 1 | 13 | 6.57e-01 |
| GO:BP | GO:0010764 | negative regulation of fibroblast migration | 1 | 9 | 6.57e-01 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 18 | 30 | 6.57e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 2 | 34 | 6.57e-01 |
| GO:BP | GO:1905702 | regulation of inhibitory synapse assembly | 1 | 3 | 6.57e-01 |
| GO:BP | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis | 1 | 1 | 6.58e-01 |
| GO:BP | GO:1905453 | regulation of myeloid progenitor cell differentiation | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 5 | 6.58e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 10 | 17 | 6.58e-01 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 5 | 9 | 6.58e-01 |
| GO:BP | GO:0071691 | cardiac muscle thin filament assembly | 1 | 3 | 6.58e-01 |
| GO:BP | GO:0031340 | positive regulation of vesicle fusion | 1 | 9 | 6.58e-01 |
| GO:BP | GO:1903382 | negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 6.59e-01 |
| GO:BP | GO:1903394 | protein localization to kinetochore involved in kinetochore assembly | 1 | 1 | 6.59e-01 |
| GO:BP | GO:1905281 | positive regulation of retrograde transport, endosome to Golgi | 1 | 2 | 6.59e-01 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 6.59e-01 |
| GO:BP | GO:1903381 | regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine | 2 | 5 | 6.59e-01 |
| GO:BP | GO:0009229 | thiamine diphosphate biosynthetic process | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0042724 | thiamine-containing compound biosynthetic process | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 4 | 7 | 6.59e-01 |
| GO:BP | GO:0021999 | neural plate anterior/posterior regionalization | 1 | 5 | 6.59e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 5 | 82 | 6.59e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 4 | 102 | 6.59e-01 |
| GO:BP | GO:1902528 | regulation of protein linear polyubiquitination | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0021537 | telencephalon development | 54 | 201 | 6.59e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 4 | 34 | 6.59e-01 |
| GO:BP | GO:1902530 | positive regulation of protein linear polyubiquitination | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0060245 | detection of cell density | 5 | 9 | 6.59e-01 |
| GO:BP | GO:0055107 | Golgi to secretory granule transport | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0060939 | epicardium-derived cardiac fibroblast cell development | 2 | 3 | 6.59e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 2 | 35 | 6.59e-01 |
| GO:BP | GO:0060938 | epicardium-derived cardiac fibroblast cell differentiation | 2 | 3 | 6.59e-01 |
| GO:BP | GO:0061140 | lung secretory cell differentiation | 1 | 5 | 6.59e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 5 | 6.59e-01 |
| GO:BP | GO:0048861 | leukemia inhibitory factor signaling pathway | 2 | 6 | 6.59e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 2 | 28 | 6.59e-01 |
| GO:BP | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | 4 | 14 | 6.59e-01 |
| GO:BP | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib | 3 | 5 | 6.59e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 4 | 89 | 6.59e-01 |
| GO:BP | GO:2001113 | negative regulation of cellular response to hepatocyte growth factor stimulus | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 5 | 165 | 6.59e-01 |
| GO:BP | GO:0001802 | type III hypersensitivity | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0001803 | regulation of type III hypersensitivity | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0010587 | miRNA catabolic process | 5 | 10 | 6.59e-01 |
| GO:BP | GO:0051944 | positive regulation of catecholamine uptake involved in synaptic transmission | 2 | 2 | 6.59e-01 |
| GO:BP | GO:0042820 | vitamin B6 catabolic process | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 5 | 11 | 6.59e-01 |
| GO:BP | GO:1902203 | negative regulation of hepatocyte growth factor receptor signaling pathway | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0032361 | pyridoxal phosphate catabolic process | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 6 | 37 | 6.59e-01 |
| GO:BP | GO:0035437 | maintenance of protein localization in endoplasmic reticulum | 6 | 11 | 6.59e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 11 | 42 | 6.59e-01 |
| GO:BP | GO:0001805 | positive regulation of type III hypersensitivity | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0021986 | habenula development | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0036138 | peptidyl-histidine hydroxylation | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0035607 | fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in hemopoiesis | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow cell | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 10 | 418 | 6.59e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 4 | 138 | 6.59e-01 |
| GO:BP | GO:0016095 | polyprenol catabolic process | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 1 | 316 | 6.59e-01 |
| GO:BP | GO:0034477 | U6 snRNA 3’-end processing | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0071701 | regulation of MAPK export from nucleus | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0009106 | lipoate metabolic process | 1 | 1 | 6.59e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 1 | 60 | 6.59e-01 |
| GO:BP | GO:0009107 | lipoate biosynthetic process | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 1 | 7 | 6.59e-01 |
| GO:BP | GO:0021538 | epithalamus development | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0034105 | positive regulation of tissue remodeling | 1 | 4 | 6.59e-01 |
| GO:BP | GO:1902259 | regulation of delayed rectifier potassium channel activity | 1 | 12 | 6.59e-01 |
| GO:BP | GO:0051586 | positive regulation of dopamine uptake involved in synaptic transmission | 2 | 2 | 6.59e-01 |
| GO:BP | GO:0001806 | type IV hypersensitivity | 2 | 2 | 6.59e-01 |
| GO:BP | GO:2000359 | regulation of binding of sperm to zona pellucida | 2 | 2 | 6.59e-01 |
| GO:BP | GO:0061890 | positive regulation of astrocyte activation | 1 | 1 | 6.59e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 6.60e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 6.60e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 1 | 6 | 6.60e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 10 | 32 | 6.60e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 1 | 75 | 6.60e-01 |
| GO:BP | GO:1904587 | response to glycoprotein | 1 | 5 | 6.60e-01 |
| GO:BP | GO:0048294 | negative regulation of isotype switching to IgE isotypes | 1 | 2 | 6.60e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 7 | 18 | 6.60e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 4 | 123 | 6.60e-01 |
| GO:BP | GO:0072006 | nephron development | 4 | 117 | 6.60e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 4 | 6 | 6.60e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 2 | 30 | 6.60e-01 |
| GO:BP | GO:0034959 | endothelin maturation | 1 | 1 | 6.60e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 16 | 54 | 6.60e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 50 | 101 | 6.60e-01 |
| GO:BP | GO:0010816 | calcitonin catabolic process | 1 | 1 | 6.60e-01 |
| GO:BP | GO:0106134 | positive regulation of cardiac muscle cell contraction | 1 | 1 | 6.60e-01 |
| GO:BP | GO:0036466 | synaptic vesicle recycling via endosome | 3 | 4 | 6.60e-01 |
| GO:BP | GO:0035964 | COPI-coated vesicle budding | 1 | 6 | 6.60e-01 |
| GO:BP | GO:0048200 | Golgi transport vesicle coating | 1 | 6 | 6.60e-01 |
| GO:BP | GO:0048205 | COPI coating of Golgi vesicle | 1 | 6 | 6.60e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 2 | 10 | 6.60e-01 |
| GO:BP | GO:0070498 | interleukin-1-mediated signaling pathway | 4 | 20 | 6.60e-01 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 3 | 7 | 6.60e-01 |
| GO:BP | GO:0090160 | Golgi to lysosome transport | 6 | 10 | 6.60e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 2 | 2 | 6.60e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 1 | 311 | 6.60e-01 |
| GO:BP | GO:0033088 | negative regulation of immature T cell proliferation in thymus | 1 | 3 | 6.61e-01 |
| GO:BP | GO:0033087 | negative regulation of immature T cell proliferation | 1 | 3 | 6.61e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 8 | 14 | 6.61e-01 |
| GO:BP | GO:1904585 | response to putrescine | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 25 | 344 | 6.61e-01 |
| GO:BP | GO:0060313 | negative regulation of blood vessel remodeling | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0046745 | viral capsid secondary envelopment | 1 | 1 | 6.61e-01 |
| GO:BP | GO:1990828 | hepatocyte dedifferentiation | 1 | 1 | 6.61e-01 |
| GO:BP | GO:1904586 | cellular response to putrescine | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 8 | 16 | 6.61e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 4 | 1101 | 6.61e-01 |
| GO:BP | GO:0003407 | neural retina development | 10 | 48 | 6.61e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 6.61e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 5 | 35 | 6.61e-01 |
| GO:BP | GO:0099158 | regulation of recycling endosome localization within postsynapse | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0071672 | negative regulation of smooth muscle cell chemotaxis | 1 | 2 | 6.61e-01 |
| GO:BP | GO:1905242 | response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0099527 | postsynapse to nucleus signaling pathway | 1 | 6 | 6.61e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 6 | 4417 | 6.61e-01 |
| GO:BP | GO:1903430 | negative regulation of cell maturation | 1 | 4 | 6.61e-01 |
| GO:BP | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway | 1 | 7 | 6.62e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 2 | 39 | 6.62e-01 |
| GO:BP | GO:1903608 | protein localization to cytoplasmic stress granule | 1 | 7 | 6.62e-01 |
| GO:BP | GO:1901143 | insulin catabolic process | 1 | 3 | 6.62e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 6 | 63 | 6.62e-01 |
| GO:BP | GO:0034773 | histone H4-K20 trimethylation | 3 | 5 | 6.62e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 8 | 80 | 6.62e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 9 | 25 | 6.62e-01 |
| GO:BP | GO:0030854 | positive regulation of granulocyte differentiation | 3 | 5 | 6.62e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 4 | 155 | 6.62e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 2 | 17 | 6.62e-01 |
| GO:BP | GO:0009620 | response to fungus | 1 | 22 | 6.62e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 9 | 16 | 6.62e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 1 | 14 | 6.62e-01 |
| GO:BP | GO:0072429 | response to intra-S DNA damage checkpoint signaling | 1 | 4 | 6.62e-01 |
| GO:BP | GO:0036285 | SAGA complex assembly | 1 | 1 | 6.62e-01 |
| GO:BP | GO:1904431 | positive regulation of t-circle formation | 1 | 3 | 6.62e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 64 | 123 | 6.62e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 2 | 10 | 6.62e-01 |
| GO:BP | GO:0060003 | copper ion export | 1 | 2 | 6.63e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 5 | 120 | 6.63e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 4 | 27 | 6.63e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 4 | 163 | 6.63e-01 |
| GO:BP | GO:0106091 | glial cell projection elongation | 1 | 2 | 6.63e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 4 | 47 | 6.63e-01 |
| GO:BP | GO:0052472 | modulation by host of symbiont transcription | 1 | 2 | 6.63e-01 |
| GO:BP | GO:1902440 | protein localization to mitotic spindle pole body | 1 | 1 | 6.63e-01 |
| GO:BP | GO:0002713 | negative regulation of B cell mediated immunity | 4 | 9 | 6.64e-01 |
| GO:BP | GO:0002890 | negative regulation of immunoglobulin mediated immune response | 4 | 9 | 6.64e-01 |
| GO:BP | GO:0048513 | animal organ development | 2 | 2189 | 6.64e-01 |
| GO:BP | GO:2000435 | negative regulation of protein neddylation | 1 | 4 | 6.64e-01 |
| GO:BP | GO:2000436 | positive regulation of protein neddylation | 1 | 5 | 6.64e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 19 | 64 | 6.64e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 1 | 8 | 6.64e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 6.64e-01 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 1 | 11 | 6.64e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 6.64e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 1 | 8 | 6.64e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 6.64e-01 |
| GO:BP | GO:0007500 | mesodermal cell fate determination | 1 | 2 | 6.64e-01 |
| GO:BP | GO:1905512 | regulation of short-term synaptic potentiation | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0070317 | negative regulation of G0 to G1 transition | 1 | 5 | 6.64e-01 |
| GO:BP | GO:0098886 | modification of dendritic spine | 1 | 2 | 6.64e-01 |
| GO:BP | GO:1905513 | negative regulation of short-term synaptic potentiation | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 4 | 135 | 6.64e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 2 | 11 | 6.64e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 2 | 27 | 6.64e-01 |
| GO:BP | GO:1902910 | positive regulation of melanosome transport | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0060901 | regulation of hair cycle by canonical Wnt signaling pathway | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0051796 | negative regulation of timing of catagen | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0072168 | specification of anterior mesonephric tubule identity | 1 | 1 | 6.65e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 2 | 8 | 6.65e-01 |
| GO:BP | GO:0072167 | specification of mesonephric tubule identity | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0071907 | determination of digestive tract left/right asymmetry | 3 | 5 | 6.65e-01 |
| GO:BP | GO:0072165 | anterior mesonephric tubule development | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0072169 | specification of posterior mesonephric tubule identity | 1 | 1 | 6.65e-01 |
| GO:BP | GO:1902605 | heterotrimeric G-protein complex assembly | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0097039 | protein linear polyubiquitination | 2 | 5 | 6.65e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 71 | 6.65e-01 |
| GO:BP | GO:0072259 | metanephric interstitial fibroblast development | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0072258 | metanephric interstitial fibroblast differentiation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0072184 | renal vesicle progenitor cell differentiation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0034104 | negative regulation of tissue remodeling | 2 | 13 | 6.65e-01 |
| GO:BP | GO:0099514 | synaptic vesicle cytoskeletal transport | 2 | 16 | 6.65e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 6.65e-01 |
| GO:BP | GO:0051235 | maintenance of location | 5 | 260 | 6.65e-01 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 2 | 5 | 6.65e-01 |
| GO:BP | GO:1903676 | positive regulation of cap-dependent translational initiation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 5 | 10 | 6.65e-01 |
| GO:BP | GO:0006364 | rRNA processing | 6 | 221 | 6.65e-01 |
| GO:BP | GO:0048490 | anterograde synaptic vesicle transport | 2 | 16 | 6.65e-01 |
| GO:BP | GO:1903674 | regulation of cap-dependent translational initiation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0099517 | synaptic vesicle transport along microtubule | 2 | 16 | 6.65e-01 |
| GO:BP | GO:0060545 | positive regulation of necroptotic process | 1 | 4 | 6.65e-01 |
| GO:BP | GO:0045659 | negative regulation of neutrophil differentiation | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0001974 | blood vessel remodeling | 4 | 32 | 6.65e-01 |
| GO:BP | GO:1905604 | negative regulation of blood-brain barrier permeability | 2 | 2 | 6.65e-01 |
| GO:BP | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0036148 | phosphatidylglycerol acyl-chain remodeling | 1 | 5 | 6.65e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 6.65e-01 |
| GO:BP | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway | 1 | 1 | 6.65e-01 |
| GO:BP | GO:2001145 | negative regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 6.66e-01 |
| GO:BP | GO:2001144 | regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0071475 | cellular hyperosmotic salinity response | 2 | 6 | 6.66e-01 |
| GO:BP | GO:1905269 | positive regulation of chromatin organization | 2 | 15 | 6.66e-01 |
| GO:BP | GO:0042168 | heme metabolic process | 9 | 40 | 6.66e-01 |
| GO:BP | GO:0140568 | extraction of mislocalized protein from membrane | 1 | 2 | 6.66e-01 |
| GO:BP | GO:0007512 | adult heart development | 7 | 9 | 6.66e-01 |
| GO:BP | GO:1904429 | regulation of t-circle formation | 2 | 5 | 6.66e-01 |
| GO:BP | GO:0060010 | Sertoli cell fate commitment | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0021675 | nerve development | 5 | 57 | 6.66e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 20 | 4590 | 6.66e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 7 | 76 | 6.66e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 5 | 52 | 6.66e-01 |
| GO:BP | GO:0007141 | male meiosis I | 4 | 15 | 6.66e-01 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 7 | 14 | 6.66e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 2 | 25 | 6.66e-01 |
| GO:BP | GO:0014857 | regulation of skeletal muscle cell proliferation | 2 | 10 | 6.66e-01 |
| GO:BP | GO:0045684 | positive regulation of epidermis development | 5 | 20 | 6.66e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 3 | 62 | 6.66e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 61 | 118 | 6.66e-01 |
| GO:BP | GO:0001568 | blood vessel development | 7 | 534 | 6.66e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 6.66e-01 |
| GO:BP | GO:0030070 | insulin processing | 2 | 6 | 6.66e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 2 | 100 | 6.66e-01 |
| GO:BP | GO:0016076 | snRNA catabolic process | 2 | 5 | 6.66e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 11 | 6.67e-01 |
| GO:BP | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | 1 | 8 | 6.67e-01 |
| GO:BP | GO:0045626 | negative regulation of T-helper 1 cell differentiation | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0090226 | regulation of microtubule nucleation by Ran protein signal transduction | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0009397 | folic acid-containing compound catabolic process | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 2 | 14 | 6.67e-01 |
| GO:BP | GO:0042560 | pteridine-containing compound catabolic process | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0046657 | folic acid catabolic process | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 4 | 3070 | 6.67e-01 |
| GO:BP | GO:0060558 | regulation of calcidiol 1-monooxygenase activity | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 4 | 49 | 6.67e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 1 | 29 | 6.67e-01 |
| GO:BP | GO:2000467 | positive regulation of glycogen (starch) synthase activity | 2 | 4 | 6.67e-01 |
| GO:BP | GO:0010668 | ectodermal cell differentiation | 1 | 5 | 6.67e-01 |
| GO:BP | GO:0048280 | vesicle fusion with Golgi apparatus | 1 | 8 | 6.67e-01 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 2 | 10 | 6.67e-01 |
| GO:BP | GO:0006116 | NADH oxidation | 2 | 4 | 6.67e-01 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 6 | 11 | 6.67e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 1 | 154 | 6.67e-01 |
| GO:BP | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 4 | 11 | 6.67e-01 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 2 | 10 | 6.67e-01 |
| GO:BP | GO:1901142 | insulin metabolic process | 3 | 11 | 6.67e-01 |
| GO:BP | GO:0002445 | type II hypersensitivity | 2 | 3 | 6.67e-01 |
| GO:BP | GO:0001794 | type IIa hypersensitivity | 2 | 3 | 6.67e-01 |
| GO:BP | GO:0035336 | long-chain fatty-acyl-CoA metabolic process | 5 | 19 | 6.67e-01 |
| GO:BP | GO:0021978 | telencephalon regionalization | 1 | 4 | 6.67e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 4 | 46 | 6.67e-01 |
| GO:BP | GO:0070836 | caveola assembly | 2 | 4 | 6.67e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 7 | 13 | 6.67e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 2 | 13 | 6.67e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 2 | 19 | 6.67e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 3 | 54 | 6.67e-01 |
| GO:BP | GO:1903044 | protein localization to membrane raft | 1 | 8 | 6.68e-01 |
| GO:BP | GO:0035261 | external genitalia morphogenesis | 1 | 1 | 6.68e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 5 | 21 | 6.68e-01 |
| GO:BP | GO:0061780 | mitotic cohesin loading | 1 | 1 | 6.68e-01 |
| GO:BP | GO:1901373 | lipid hydroperoxide transport | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 23 | 859 | 6.68e-01 |
| GO:BP | GO:0000154 | rRNA modification | 18 | 35 | 6.68e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 2 | 23 | 6.68e-01 |
| GO:BP | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 10 | 71 | 6.68e-01 |
| GO:BP | GO:0032965 | regulation of collagen biosynthetic process | 8 | 30 | 6.69e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 5 | 16 | 6.69e-01 |
| GO:BP | GO:0160025 | sensory perception of itch | 1 | 1 | 6.69e-01 |
| GO:BP | GO:2000843 | regulation of testosterone secretion | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0160023 | sneeze reflex | 1 | 1 | 6.69e-01 |
| GO:BP | GO:2000845 | positive regulation of testosterone secretion | 1 | 2 | 6.69e-01 |
| GO:BP | GO:2000626 | negative regulation of miRNA catabolic process | 1 | 1 | 6.69e-01 |
| GO:BP | GO:0140235 | RNA polyadenylation at postsynapse | 1 | 1 | 6.69e-01 |
| GO:BP | GO:0035936 | testosterone secretion | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0070175 | positive regulation of enamel mineralization | 1 | 2 | 6.69e-01 |
| GO:BP | GO:1900210 | positive regulation of cardiolipin metabolic process | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 2 | 57 | 6.70e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 10 | 23 | 6.70e-01 |
| GO:BP | GO:0048569 | post-embryonic animal organ development | 4 | 15 | 6.70e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 2 | 12 | 6.70e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 9 | 6.70e-01 |
| GO:BP | GO:0015821 | methionine transport | 1 | 4 | 6.70e-01 |
| GO:BP | GO:0048132 | female germ-line stem cell asymmetric division | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 1 | 185 | 6.70e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 1 | 157 | 6.70e-01 |
| GO:BP | GO:0018095 | protein polyglutamylation | 1 | 9 | 6.70e-01 |
| GO:BP | GO:2000833 | positive regulation of steroid hormone secretion | 1 | 6 | 6.70e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 2 | 19 | 6.70e-01 |
| GO:BP | GO:0051103 | DNA ligation involved in DNA repair | 3 | 5 | 6.71e-01 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 1 | 6 | 6.71e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 6.71e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 1 | 8 | 6.71e-01 |
| GO:BP | GO:0046203 | spermidine catabolic process | 1 | 2 | 6.71e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 4 | 18 | 6.71e-01 |
| GO:BP | GO:0097048 | dendritic cell apoptotic process | 2 | 7 | 6.71e-01 |
| GO:BP | GO:2000668 | regulation of dendritic cell apoptotic process | 2 | 7 | 6.71e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 12 | 51 | 6.72e-01 |
| GO:BP | GO:0048731 | system development | 3 | 2901 | 6.72e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 2 | 49 | 6.72e-01 |
| GO:BP | GO:0031652 | positive regulation of heat generation | 2 | 4 | 6.72e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 6.72e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 11 | 28 | 6.72e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 8 | 316 | 6.72e-01 |
| GO:BP | GO:0034463 | 90S preribosome assembly | 1 | 1 | 6.72e-01 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 2 | 21 | 6.72e-01 |
| GO:BP | GO:0032966 | negative regulation of collagen biosynthetic process | 5 | 7 | 6.72e-01 |
| GO:BP | GO:2000764 | positive regulation of semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 6.72e-01 |
| GO:BP | GO:2000763 | positive regulation of transcription from RNA polymerase II promoter involved in norepinephrine biosynthetic process | 1 | 1 | 6.72e-01 |
| GO:BP | GO:2001262 | positive regulation of semaphorin-plexin signaling pathway | 1 | 1 | 6.72e-01 |
| GO:BP | GO:0035844 | cloaca development | 1 | 3 | 6.72e-01 |
| GO:BP | GO:1904897 | regulation of hepatic stellate cell proliferation | 1 | 1 | 6.72e-01 |
| GO:BP | GO:0071527 | semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 6.72e-01 |
| GO:BP | GO:1990922 | hepatic stellate cell proliferation | 1 | 1 | 6.72e-01 |
| GO:BP | GO:1904899 | positive regulation of hepatic stellate cell proliferation | 1 | 1 | 6.72e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 551 | 6.73e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 4 | 10 | 6.73e-01 |
| GO:BP | GO:1903435 | positive regulation of constitutive secretory pathway | 1 | 1 | 6.73e-01 |
| GO:BP | GO:0071922 | regulation of cohesin loading | 1 | 2 | 6.73e-01 |
| GO:BP | GO:1901835 | positive regulation of deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 1 | 6.73e-01 |
| GO:BP | GO:1904246 | negative regulation of polynucleotide adenylyltransferase activity | 1 | 1 | 6.73e-01 |
| GO:BP | GO:1901834 | regulation of deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 1 | 6.73e-01 |
| GO:BP | GO:0030001 | metal ion transport | 12 | 562 | 6.73e-01 |
| GO:BP | GO:0007497 | posterior midgut development | 1 | 2 | 6.73e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 2 | 111 | 6.73e-01 |
| GO:BP | GO:0033031 | positive regulation of neutrophil apoptotic process | 1 | 3 | 6.73e-01 |
| GO:BP | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing | 4 | 6 | 6.73e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 4 | 236 | 6.73e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 9 | 196 | 6.73e-01 |
| GO:BP | GO:0030505 | inorganic diphosphate transport | 1 | 3 | 6.73e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 6.73e-01 |
| GO:BP | GO:0006685 | sphingomyelin catabolic process | 1 | 6 | 6.73e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 6.73e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 2 | 48 | 6.73e-01 |
| GO:BP | GO:0061017 | hepatoblast differentiation | 1 | 1 | 6.73e-01 |
| GO:BP | GO:1905711 | response to phosphatidylethanolamine | 1 | 1 | 6.73e-01 |
| GO:BP | GO:0038128 | ERBB2 signaling pathway | 9 | 14 | 6.73e-01 |
| GO:BP | GO:1903598 | positive regulation of gap junction assembly | 2 | 7 | 6.73e-01 |
| GO:BP | GO:0006379 | mRNA cleavage | 1 | 19 | 6.73e-01 |
| GO:BP | GO:0006863 | purine nucleobase transport | 1 | 7 | 6.73e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 6.73e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 6.73e-01 |
| GO:BP | GO:0071926 | endocannabinoid signaling pathway | 2 | 5 | 6.73e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 2 | 69 | 6.74e-01 |
| GO:BP | GO:1902022 | L-lysine transport | 1 | 6 | 6.74e-01 |
| GO:BP | GO:1903401 | L-lysine transmembrane transport | 1 | 6 | 6.74e-01 |
| GO:BP | GO:1901843 | positive regulation of high voltage-gated calcium channel activity | 2 | 4 | 6.74e-01 |
| GO:BP | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0001971 | negative regulation of activation of membrane attack complex | 1 | 1 | 6.74e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 9 | 17 | 6.74e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 10 | 438 | 6.74e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 6.74e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 6.74e-01 |
| GO:BP | GO:1901632 | regulation of synaptic vesicle membrane organization | 1 | 8 | 6.74e-01 |
| GO:BP | GO:1900081 | regulation of arginine catabolic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 73 | 6.74e-01 |
| GO:BP | GO:1900082 | negative regulation of arginine catabolic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0033241 | regulation of amine catabolic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:2000283 | negative regulation of amino acid biosynthetic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0098736 | negative regulation of the force of heart contraction | 1 | 1 | 6.74e-01 |
| GO:BP | GO:1903248 | regulation of citrulline biosynthetic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:1903249 | negative regulation of citrulline biosynthetic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0033242 | negative regulation of amine catabolic process | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0061108 | seminal vesicle epithelium development | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 6.74e-01 |
| GO:BP | GO:0007040 | lysosome organization | 3 | 88 | 6.74e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 3 | 88 | 6.74e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 1 | 107 | 6.74e-01 |
| GO:BP | GO:0032782 | bile acid secretion | 1 | 5 | 6.74e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 41 | 2102 | 6.74e-01 |
| GO:BP | GO:1901654 | response to ketone | 3 | 147 | 6.74e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 4 | 4099 | 6.74e-01 |
| GO:BP | GO:0042628 | mating plug formation | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0009306 | protein secretion | 2 | 262 | 6.74e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 3 | 8 | 6.74e-01 |
| GO:BP | GO:0070375 | ERK5 cascade | 1 | 3 | 6.74e-01 |
| GO:BP | GO:1905523 | positive regulation of macrophage migration | 2 | 13 | 6.74e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 7 | 13 | 6.74e-01 |
| GO:BP | GO:0032835 | glomerulus development | 9 | 54 | 6.74e-01 |
| GO:BP | GO:0043363 | nucleate erythrocyte differentiation | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0002574 | thrombocyte differentiation | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 10 | 22 | 6.74e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 5 | 38 | 6.74e-01 |
| GO:BP | GO:0008585 | female gonad development | 24 | 77 | 6.74e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 6.75e-01 |
| GO:BP | GO:0006783 | heme biosynthetic process | 7 | 30 | 6.75e-01 |
| GO:BP | GO:0018125 | peptidyl-cysteine methylation | 1 | 2 | 6.75e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 5 | 58 | 6.75e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 5 | 169 | 6.75e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 6.75e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 17 | 61 | 6.75e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 2 | 262 | 6.75e-01 |
| GO:BP | GO:2000633 | positive regulation of pre-miRNA processing | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0051295 | establishment of meiotic spindle localization | 1 | 4 | 6.75e-01 |
| GO:BP | GO:0007411 | axon guidance | 2 | 185 | 6.75e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 2 | 50 | 6.75e-01 |
| GO:BP | GO:0045932 | negative regulation of muscle contraction | 1 | 20 | 6.75e-01 |
| GO:BP | GO:0060457 | negative regulation of digestive system process | 1 | 7 | 6.75e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 7 | 34 | 6.76e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 5 | 10 | 6.76e-01 |
| GO:BP | GO:1904970 | brush border assembly | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0035543 | positive regulation of SNARE complex assembly | 1 | 1 | 6.76e-01 |
| GO:BP | GO:1903284 | positive regulation of glutathione peroxidase activity | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 2 | 14 | 6.76e-01 |
| GO:BP | GO:0007409 | axonogenesis | 4 | 359 | 6.76e-01 |
| GO:BP | GO:0035771 | interleukin-4-mediated signaling pathway | 2 | 4 | 6.76e-01 |
| GO:BP | GO:0043508 | negative regulation of JUN kinase activity | 4 | 13 | 6.76e-01 |
| GO:BP | GO:0016256 | N-glycan processing to lysosome | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 6.76e-01 |
| GO:BP | GO:0015920 | lipopolysaccharide transport | 1 | 1 | 6.76e-01 |
| GO:BP | GO:1903285 | positive regulation of hydrogen peroxide catabolic process | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 6.76e-01 |
| GO:BP | GO:0051945 | negative regulation of catecholamine uptake involved in synaptic transmission | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0001408 | guanine nucleotide transport | 1 | 6 | 6.76e-01 |
| GO:BP | GO:0051622 | negative regulation of norepinephrine uptake | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0035669 | TRAM-dependent toll-like receptor 4 signaling pathway | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 5 | 174 | 6.76e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 6.76e-01 |
| GO:BP | GO:0098838 | folate transmembrane transport | 1 | 6 | 6.76e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 2 | 14 | 6.76e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 1 | 19 | 6.76e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 5 | 8 | 6.76e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 1 | 19 | 6.76e-01 |
| GO:BP | GO:0000413 | protein peptidyl-prolyl isomerization | 6 | 26 | 6.76e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 8 | 179 | 6.76e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 2 | 186 | 6.76e-01 |
| GO:BP | GO:0060066 | oviduct development | 1 | 4 | 6.76e-01 |
| GO:BP | GO:1904992 | positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0021535 | cell migration in hindbrain | 8 | 12 | 6.76e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 1 | 40 | 6.76e-01 |
| GO:BP | GO:0045672 | positive regulation of osteoclast differentiation | 1 | 11 | 6.76e-01 |
| GO:BP | GO:0097283 | keratinocyte apoptotic process | 1 | 5 | 6.76e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 6.76e-01 |
| GO:BP | GO:1904990 | regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 6.76e-01 |
| GO:BP | GO:1902172 | regulation of keratinocyte apoptotic process | 1 | 5 | 6.76e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 4 | 3132 | 6.76e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 1 | 17 | 6.76e-01 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 14 | 34 | 6.76e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 16 | 45 | 6.76e-01 |
| GO:BP | GO:1903570 | regulation of protein kinase D signaling | 1 | 1 | 6.76e-01 |
| GO:BP | GO:1903572 | positive regulation of protein kinase D signaling | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0002575 | basophil chemotaxis | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 2 | 22 | 6.76e-01 |
| GO:BP | GO:0035441 | cell migration involved in vasculogenesis | 2 | 4 | 6.77e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 6.77e-01 |
| GO:BP | GO:0032713 | negative regulation of interleukin-4 production | 3 | 5 | 6.77e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 5 | 3750 | 6.77e-01 |
| GO:BP | GO:0110096 | cellular response to aldehyde | 4 | 10 | 6.77e-01 |
| GO:BP | GO:0002070 | epithelial cell maturation | 8 | 16 | 6.77e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 3 | 29 | 6.78e-01 |
| GO:BP | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity | 1 | 3 | 6.78e-01 |
| GO:BP | GO:1902645 | tertiary alcohol biosynthetic process | 2 | 8 | 6.78e-01 |
| GO:BP | GO:0006050 | mannosamine metabolic process | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 28 | 6.78e-01 |
| GO:BP | GO:0045087 | innate immune response | 7 | 488 | 6.78e-01 |
| GO:BP | GO:0021825 | substrate-dependent cerebral cortex tangential migration | 1 | 2 | 6.78e-01 |
| GO:BP | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | 1 | 4 | 6.78e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 1 | 33 | 6.78e-01 |
| GO:BP | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0006051 | N-acetylmannosamine metabolic process | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 2 | 47 | 6.78e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 4 | 3133 | 6.78e-01 |
| GO:BP | GO:0021827 | postnatal olfactory bulb interneuron migration | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0106016 | positive regulation of inflammatory response to wounding | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0030282 | bone mineralization | 13 | 82 | 6.78e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 4 | 94 | 6.78e-01 |
| GO:BP | GO:0019344 | cysteine biosynthetic process | 1 | 3 | 6.78e-01 |
| GO:BP | GO:0034651 | cortisol biosynthetic process | 2 | 8 | 6.78e-01 |
| GO:BP | GO:1903334 | positive regulation of protein folding | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0061085 | regulation of histone H3-K27 methylation | 1 | 4 | 6.78e-01 |
| GO:BP | GO:0043418 | homocysteine catabolic process | 1 | 3 | 6.78e-01 |
| GO:BP | GO:0071896 | protein localization to adherens junction | 3 | 6 | 6.78e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 13 | 56 | 6.78e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 9 | 15 | 6.78e-01 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 4 | 7 | 6.78e-01 |
| GO:BP | GO:0016561 | protein import into peroxisome matrix, translocation | 2 | 3 | 6.78e-01 |
| GO:BP | GO:0042669 | regulation of inner ear auditory receptor cell fate specification | 1 | 1 | 6.78e-01 |
| GO:BP | GO:0032353 | negative regulation of hormone biosynthetic process | 2 | 8 | 6.78e-01 |
| GO:BP | GO:0033227 | dsRNA transport | 2 | 2 | 6.78e-01 |
| GO:BP | GO:0015801 | aromatic amino acid transport | 6 | 11 | 6.79e-01 |
| GO:BP | GO:0007548 | sex differentiation | 2 | 191 | 6.79e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 189 | 6.79e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 1 | 199 | 6.79e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 6.79e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 6.79e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 8 | 24 | 6.80e-01 |
| GO:BP | GO:0007350 | blastoderm segmentation | 2 | 17 | 6.80e-01 |
| GO:BP | GO:0001556 | oocyte maturation | 2 | 22 | 6.80e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 2 | 49 | 6.80e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 6.80e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 2 | 52 | 6.80e-01 |
| GO:BP | GO:0021511 | spinal cord patterning | 1 | 9 | 6.80e-01 |
| GO:BP | GO:0008300 | isoprenoid catabolic process | 2 | 5 | 6.80e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 43 | 180 | 6.80e-01 |
| GO:BP | GO:0048073 | regulation of eye pigmentation | 1 | 2 | 6.80e-01 |
| GO:BP | GO:0051764 | actin crosslink formation | 5 | 10 | 6.80e-01 |
| GO:BP | GO:0036506 | maintenance of unfolded protein | 1 | 3 | 6.80e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 11 | 517 | 6.80e-01 |
| GO:BP | GO:0061549 | sympathetic ganglion development | 3 | 6 | 6.80e-01 |
| GO:BP | GO:0038029 | epidermal growth factor receptor signaling pathway via MAPK cascade | 1 | 1 | 6.80e-01 |
| GO:BP | GO:1904378 | maintenance of unfolded protein involved in ERAD pathway | 1 | 3 | 6.80e-01 |
| GO:BP | GO:0051001 | negative regulation of nitric-oxide synthase activity | 1 | 6 | 6.80e-01 |
| GO:BP | GO:0002519 | natural killer cell tolerance induction | 1 | 1 | 6.80e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 3 | 15 | 6.80e-01 |
| GO:BP | GO:0006742 | NADP catabolic process | 1 | 3 | 6.80e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 3 | 10 | 6.80e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 5 | 185 | 6.80e-01 |
| GO:BP | GO:0019677 | NAD catabolic process | 1 | 3 | 6.80e-01 |
| GO:BP | GO:0071765 | nuclear inner membrane organization | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0009304 | tRNA transcription | 1 | 8 | 6.81e-01 |
| GO:BP | GO:0030916 | otic vesicle formation | 4 | 6 | 6.81e-01 |
| GO:BP | GO:0044349 | DNA excision | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0000718 | nucleotide-excision repair, DNA damage removal | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation | 1 | 4 | 6.81e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 2 | 80 | 6.81e-01 |
| GO:BP | GO:0097712 | vesicle targeting, trans-Golgi to periciliary membrane compartment | 1 | 1 | 6.82e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 6 | 37 | 6.82e-01 |
| GO:BP | GO:1903741 | negative regulation of phosphatidate phosphatase activity | 1 | 1 | 6.83e-01 |
| GO:BP | GO:1903730 | regulation of phosphatidate phosphatase activity | 1 | 1 | 6.83e-01 |
| GO:BP | GO:0048669 | collateral sprouting in absence of injury | 1 | 4 | 6.83e-01 |
| GO:BP | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation | 1 | 4 | 6.83e-01 |
| GO:BP | GO:0006982 | response to lipid hydroperoxide | 2 | 2 | 6.83e-01 |
| GO:BP | GO:0043060 | meiotic metaphase I plate congression | 1 | 1 | 6.83e-01 |
| GO:BP | GO:0006983 | ER overload response | 4 | 12 | 6.84e-01 |
| GO:BP | GO:2000417 | negative regulation of eosinophil migration | 1 | 1 | 6.84e-01 |
| GO:BP | GO:0045903 | positive regulation of translational fidelity | 1 | 1 | 6.84e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 6.84e-01 |
| GO:BP | GO:0060741 | prostate gland stromal morphogenesis | 1 | 2 | 6.84e-01 |
| GO:BP | GO:2001288 | positive regulation of caveolin-mediated endocytosis | 2 | 3 | 6.84e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 2 | 52 | 6.84e-01 |
| GO:BP | GO:0061061 | muscle structure development | 1 | 546 | 6.84e-01 |
| GO:BP | GO:0090410 | malonate catabolic process | 1 | 1 | 6.84e-01 |
| GO:BP | GO:0000821 | regulation of arginine metabolic process | 2 | 3 | 6.84e-01 |
| GO:BP | GO:1990390 | protein K33-linked ubiquitination | 1 | 2 | 6.84e-01 |
| GO:BP | GO:0002344 | B cell affinity maturation | 1 | 2 | 6.84e-01 |
| GO:BP | GO:0002332 | transitional stage B cell differentiation | 1 | 2 | 6.84e-01 |
| GO:BP | GO:0002343 | peripheral B cell selection | 1 | 2 | 6.84e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 9 | 37 | 6.84e-01 |
| GO:BP | GO:0033341 | regulation of collagen binding | 1 | 2 | 6.84e-01 |
| GO:BP | GO:0007418 | ventral midline development | 2 | 4 | 6.84e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 3 | 118 | 6.85e-01 |
| GO:BP | GO:0021702 | cerebellar Purkinje cell differentiation | 5 | 10 | 6.85e-01 |
| GO:BP | GO:0072385 | minus-end-directed organelle transport along microtubule | 3 | 5 | 6.85e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 172 | 6.85e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 1 | 29 | 6.85e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 1 | 35 | 6.85e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 14 | 125 | 6.85e-01 |
| GO:BP | GO:0006891 | intra-Golgi vesicle-mediated transport | 1 | 33 | 6.85e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 1 | 45 | 6.85e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 6.85e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 1 | 47 | 6.86e-01 |
| GO:BP | GO:2000774 | positive regulation of cellular senescence | 3 | 9 | 6.86e-01 |
| GO:BP | GO:0010637 | negative regulation of mitochondrial fusion | 4 | 8 | 6.86e-01 |
| GO:BP | GO:1901622 | positive regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 1 | 6.86e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 2 | 12 | 6.86e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 6 | 295 | 6.86e-01 |
| GO:BP | GO:0015964 | diadenosine triphosphate catabolic process | 1 | 1 | 6.86e-01 |
| GO:BP | GO:0015962 | diadenosine triphosphate metabolic process | 1 | 1 | 6.86e-01 |
| GO:BP | GO:1900623 | regulation of monocyte aggregation | 2 | 3 | 6.86e-01 |
| GO:BP | GO:1900625 | positive regulation of monocyte aggregation | 2 | 3 | 6.86e-01 |
| GO:BP | GO:0051562 | negative regulation of mitochondrial calcium ion concentration | 1 | 2 | 6.86e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1 | 135 | 6.86e-01 |
| GO:BP | GO:0032613 | interleukin-10 production | 2 | 24 | 6.86e-01 |
| GO:BP | GO:0051647 | nucleus localization | 2 | 28 | 6.86e-01 |
| GO:BP | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.86e-01 |
| GO:BP | GO:0036292 | DNA rewinding | 1 | 1 | 6.86e-01 |
| GO:BP | GO:2000452 | regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.86e-01 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 2 | 24 | 6.86e-01 |
| GO:BP | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 6.86e-01 |
| GO:BP | GO:1903225 | negative regulation of endodermal cell differentiation | 2 | 3 | 6.86e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 2 | 11 | 6.87e-01 |
| GO:BP | GO:0070911 | global genome nucleotide-excision repair | 1 | 2 | 6.87e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 9 | 403 | 6.87e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 2 | 4 | 6.87e-01 |
| GO:BP | GO:0036116 | long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 3 | 7 | 6.87e-01 |
| GO:BP | GO:0001192 | maintenance of transcriptional fidelity during transcription elongation | 1 | 1 | 6.87e-01 |
| GO:BP | GO:1900535 | palmitic acid biosynthetic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0008626 | granzyme-mediated apoptotic signaling pathway | 1 | 1 | 6.87e-01 |
| GO:BP | GO:1990791 | dorsal root ganglion development | 3 | 6 | 6.87e-01 |
| GO:BP | GO:1904925 | positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 7 | 14 | 6.87e-01 |
| GO:BP | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process | 1 | 2 | 6.87e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 6.87e-01 |
| GO:BP | GO:0001193 | maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0043970 | histone H3-K9 acetylation | 2 | 8 | 6.87e-01 |
| GO:BP | GO:0050428 | 3’-phosphoadenosine 5’-phosphosulfate biosynthetic process | 1 | 2 | 6.87e-01 |
| GO:BP | GO:1900533 | palmitic acid metabolic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0071919 | G-quadruplex DNA formation | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0000103 | sulfate assimilation | 1 | 2 | 6.87e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 3 | 25 | 6.87e-01 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 6 | 15 | 6.87e-01 |
| GO:BP | GO:0003056 | regulation of vascular associated smooth muscle contraction | 3 | 8 | 6.87e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 1 | 28 | 6.87e-01 |
| GO:BP | GO:0002883 | regulation of hypersensitivity | 3 | 5 | 6.87e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 3 | 21 | 6.87e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 2 | 963 | 6.87e-01 |
| GO:BP | GO:0032596 | protein transport into membrane raft | 2 | 4 | 6.87e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 1 | 114 | 6.87e-01 |
| GO:BP | GO:2000659 | regulation of interleukin-1-mediated signaling pathway | 4 | 4 | 6.87e-01 |
| GO:BP | GO:0051391 | tRNA acetylation | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0071688 | striated muscle myosin thick filament assembly | 2 | 3 | 6.87e-01 |
| GO:BP | GO:0009210 | pyrimidine ribonucleoside triphosphate catabolic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:1903766 | positive regulation of potassium ion export across plasma membrane | 1 | 3 | 6.87e-01 |
| GO:BP | GO:1904812 | rRNA acetylation involved in maturation of SSU-rRNA | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0035281 | pre-miRNA export from nucleus | 1 | 2 | 6.87e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 11 | 36 | 6.87e-01 |
| GO:BP | GO:0032484 | Ral protein signal transduction | 2 | 5 | 6.87e-01 |
| GO:BP | GO:0110010 | basolateral protein secretion | 2 | 3 | 6.87e-01 |
| GO:BP | GO:1990882 | rRNA acetylation | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0046052 | UTP catabolic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0040031 | snRNA modification | 2 | 7 | 6.87e-01 |
| GO:BP | GO:1990884 | RNA acetylation | 1 | 1 | 6.87e-01 |
| GO:BP | GO:0061015 | snRNA import into nucleus | 1 | 2 | 6.87e-01 |
| GO:BP | GO:0030241 | skeletal muscle myosin thick filament assembly | 2 | 3 | 6.87e-01 |
| GO:BP | GO:0006254 | CTP catabolic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:1903650 | negative regulation of cytoplasmic transport | 2 | 3 | 6.87e-01 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 9 | 17 | 6.87e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 2 | 14 | 6.88e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 2 | 95 | 6.88e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 2 | 27 | 6.88e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 3 | 7 | 6.88e-01 |
| GO:BP | GO:0048148 | behavioral response to cocaine | 3 | 9 | 6.88e-01 |
| GO:BP | GO:0061198 | fungiform papilla formation | 1 | 3 | 6.88e-01 |
| GO:BP | GO:0006997 | nucleus organization | 4 | 126 | 6.88e-01 |
| GO:BP | GO:0009451 | RNA modification | 43 | 162 | 6.88e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 6.88e-01 |
| GO:BP | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death | 2 | 3 | 6.88e-01 |
| GO:BP | GO:1901687 | glutathione derivative biosynthetic process | 1 | 3 | 6.88e-01 |
| GO:BP | GO:1901685 | glutathione derivative metabolic process | 1 | 3 | 6.88e-01 |
| GO:BP | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | 1 | 5 | 6.88e-01 |
| GO:BP | GO:0099040 | ceramide translocation | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 6.88e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 4 | 10 | 6.88e-01 |
| GO:BP | GO:0031038 | myosin II filament organization | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0070988 | demethylation | 4 | 52 | 6.89e-01 |
| GO:BP | GO:0043519 | regulation of myosin II filament organization | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0051709 | regulation of killing of cells of another organism | 2 | 3 | 6.89e-01 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 6.89e-01 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 1 | 24 | 6.89e-01 |
| GO:BP | GO:0072706 | response to sodium dodecyl sulfate | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 38 | 6.89e-01 |
| GO:BP | GO:0072707 | cellular response to sodium dodecyl sulfate | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0060921 | sinoatrial node cell differentiation | 2 | 5 | 6.89e-01 |
| GO:BP | GO:0021750 | vestibular nucleus development | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0034115 | negative regulation of heterotypic cell-cell adhesion | 1 | 6 | 6.89e-01 |
| GO:BP | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation | 1 | 6 | 6.89e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 4 | 35 | 6.89e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 4 | 92 | 6.89e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 20 | 39 | 6.89e-01 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 5 | 11 | 6.89e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 2 | 99 | 6.89e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 4 | 10 | 6.89e-01 |
| GO:BP | GO:0010839 | negative regulation of keratinocyte proliferation | 1 | 19 | 6.89e-01 |
| GO:BP | GO:0035037 | sperm entry | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 9 | 114 | 6.89e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 8 | 29 | 6.89e-01 |
| GO:BP | GO:0006101 | citrate metabolic process | 3 | 5 | 6.89e-01 |
| GO:BP | GO:0019518 | L-threonine catabolic process to glycine | 1 | 1 | 6.89e-01 |
| GO:BP | GO:1905528 | positive regulation of Golgi lumen acidification | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0019284 | L-methionine salvage from S-adenosylmethionine | 1 | 3 | 6.89e-01 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 1 | 12 | 6.89e-01 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 1 | 12 | 6.89e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 2 | 3 | 6.89e-01 |
| GO:BP | GO:0090298 | negative regulation of mitochondrial DNA replication | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0006288 | base-excision repair, DNA ligation | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 13 | 147 | 6.90e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 14 | 92 | 6.90e-01 |
| GO:BP | GO:0002331 | pre-B cell allelic exclusion | 1 | 2 | 6.90e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 6.90e-01 |
| GO:BP | GO:0036250 | peroxisome transport along microtubule | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 4 | 16 | 6.90e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 6.90e-01 |
| GO:BP | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway | 5 | 9 | 6.90e-01 |
| GO:BP | GO:0006953 | acute-phase response | 1 | 18 | 6.90e-01 |
| GO:BP | GO:0006651 | diacylglycerol biosynthetic process | 4 | 7 | 6.90e-01 |
| GO:BP | GO:0071344 | diphosphate metabolic process | 1 | 2 | 6.90e-01 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 1 | 14 | 6.90e-01 |
| GO:BP | GO:0090208 | positive regulation of triglyceride metabolic process | 1 | 18 | 6.91e-01 |
| GO:BP | GO:1904251 | regulation of bile acid metabolic process | 1 | 9 | 6.91e-01 |
| GO:BP | GO:0009812 | flavonoid metabolic process | 1 | 4 | 6.91e-01 |
| GO:BP | GO:0033027 | positive regulation of mast cell apoptotic process | 1 | 1 | 6.91e-01 |
| GO:BP | GO:0046039 | GTP metabolic process | 10 | 21 | 6.91e-01 |
| GO:BP | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission | 1 | 1 | 6.91e-01 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 7 | 19 | 6.92e-01 |
| GO:BP | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 1 | 6 | 6.92e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 3 | 15 | 6.92e-01 |
| GO:BP | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 6.92e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 1 | 187 | 6.92e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 2 | 40 | 6.92e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 1 | 187 | 6.92e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 10 | 79 | 6.92e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 6.92e-01 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 1 | 9 | 6.92e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 2 | 3 | 6.92e-01 |
| GO:BP | GO:1901334 | lactone metabolic process | 1 | 9 | 6.92e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 1 | 8 | 6.92e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 2 | 15 | 6.93e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 2 | 15 | 6.93e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 15 | 62 | 6.93e-01 |
| GO:BP | GO:0003165 | Purkinje myocyte development | 2 | 3 | 6.93e-01 |
| GO:BP | GO:0071449 | cellular response to lipid hydroperoxide | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 2 | 1256 | 6.93e-01 |
| GO:BP | GO:0035635 | entry of bacterium into host cell | 1 | 8 | 6.93e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 4 | 93 | 6.93e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 3 | 49 | 6.93e-01 |
| GO:BP | GO:0160007 | glutathione import into mitochondrion | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 6.93e-01 |
| GO:BP | GO:2000643 | positive regulation of early endosome to late endosome transport | 4 | 7 | 6.93e-01 |
| GO:BP | GO:0017126 | nucleologenesis | 1 | 2 | 6.93e-01 |
| GO:BP | GO:1900079 | regulation of arginine biosynthetic process | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0090384 | phagosome-lysosome docking | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 2 | 1255 | 6.93e-01 |
| GO:BP | GO:0071386 | cellular response to corticosterone stimulus | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 5 | 16 | 6.93e-01 |
| GO:BP | GO:0006782 | protoporphyrinogen IX biosynthetic process | 2 | 9 | 6.93e-01 |
| GO:BP | GO:0071400 | cellular response to oleic acid | 2 | 3 | 6.93e-01 |
| GO:BP | GO:2000368 | positive regulation of acrosomal vesicle exocytosis | 1 | 2 | 6.93e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 6.93e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 12 | 168 | 6.94e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 49 | 294 | 6.94e-01 |
| GO:BP | GO:0007568 | aging | 9 | 115 | 6.94e-01 |
| GO:BP | GO:0001944 | vasculature development | 7 | 559 | 6.94e-01 |
| GO:BP | GO:2000474 | regulation of opioid receptor signaling pathway | 1 | 2 | 6.94e-01 |
| GO:BP | GO:1900373 | positive regulation of purine nucleotide biosynthetic process | 3 | 12 | 6.94e-01 |
| GO:BP | GO:0017145 | stem cell division | 8 | 26 | 6.94e-01 |
| GO:BP | GO:0030810 | positive regulation of nucleotide biosynthetic process | 3 | 12 | 6.94e-01 |
| GO:BP | GO:0044828 | negative regulation by host of viral genome replication | 1 | 7 | 6.94e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 2 | 608 | 6.94e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 2 | 67 | 6.94e-01 |
| GO:BP | GO:0003235 | sinus venosus development | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 1 | 13 | 6.94e-01 |
| GO:BP | GO:0038097 | positive regulation of mast cell activation by Fc-epsilon receptor signaling pathway | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 6.94e-01 |
| GO:BP | GO:0006713 | glucocorticoid catabolic process | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0003236 | sinus venosus morphogenesis | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 1 | 12 | 6.94e-01 |
| GO:BP | GO:0003260 | cardioblast migration | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0042664 | negative regulation of endodermal cell fate specification | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0070368 | positive regulation of hepatocyte differentiation | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0060975 | cardioblast migration to the midline involved in heart field formation | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0032474 | otolith morphogenesis | 1 | 3 | 6.94e-01 |
| GO:BP | GO:0003259 | cardioblast anterior-lateral migration | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 6 | 31 | 6.94e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 7 | 160 | 6.94e-01 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 7 | 120 | 6.94e-01 |
| GO:BP | GO:0015851 | nucleobase transport | 1 | 7 | 6.94e-01 |
| GO:BP | GO:0003318 | cell migration to the midline involved in heart development | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 6.94e-01 |
| GO:BP | GO:0009444 | pyruvate oxidation | 1 | 1 | 6.94e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 6.94e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 2 | 31 | 6.94e-01 |
| GO:BP | GO:0120127 | response to zinc ion starvation | 3 | 6 | 6.94e-01 |
| GO:BP | GO:2000065 | negative regulation of cortisol biosynthetic process | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0032348 | negative regulation of aldosterone biosynthetic process | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0072189 | ureter development | 5 | 10 | 6.94e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 6.94e-01 |
| GO:BP | GO:0032345 | negative regulation of aldosterone metabolic process | 1 | 4 | 6.94e-01 |
| GO:BP | GO:1905069 | allantois development | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0034224 | cellular response to zinc ion starvation | 3 | 6 | 6.94e-01 |
| GO:BP | GO:0072109 | glomerular mesangium development | 6 | 13 | 6.94e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 6.94e-01 |
| GO:BP | GO:0045056 | transcytosis | 8 | 17 | 6.94e-01 |
| GO:BP | GO:0060490 | lateral sprouting involved in lung morphogenesis | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0060489 | planar dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0007621 | negative regulation of female receptivity | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 2 | 78 | 6.94e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 13 | 676 | 6.95e-01 |
| GO:BP | GO:0030850 | prostate gland development | 3 | 35 | 6.95e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 6.95e-01 |
| GO:BP | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway | 1 | 1 | 6.95e-01 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 8 | 16 | 6.95e-01 |
| GO:BP | GO:0062100 | positive regulation of programmed necrotic cell death | 1 | 5 | 6.95e-01 |
| GO:BP | GO:0042748 | circadian sleep/wake cycle, non-REM sleep | 1 | 3 | 6.95e-01 |
| GO:BP | GO:0035685 | helper T cell diapedesis | 1 | 2 | 6.95e-01 |
| GO:BP | GO:1990839 | response to endothelin | 1 | 2 | 6.95e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 2 | 66 | 6.95e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 19 | 40 | 6.95e-01 |
| GO:BP | GO:0010849 | regulation of proton-transporting ATPase activity, rotational mechanism | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 1 | 132 | 6.95e-01 |
| GO:BP | GO:0002579 | positive regulation of antigen processing and presentation | 2 | 3 | 6.95e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 1 | 57 | 6.95e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 5 | 59 | 6.95e-01 |
| GO:BP | GO:1904338 | regulation of dopaminergic neuron differentiation | 1 | 7 | 6.95e-01 |
| GO:BP | GO:0051930 | regulation of sensory perception of pain | 1 | 13 | 6.95e-01 |
| GO:BP | GO:0051931 | regulation of sensory perception | 1 | 13 | 6.95e-01 |
| GO:BP | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron | 1 | 2 | 6.95e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 1 | 10 | 6.95e-01 |
| GO:BP | GO:0003226 | right ventricular compact myocardium morphogenesis | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 3 | 23 | 6.96e-01 |
| GO:BP | GO:1903883 | positive regulation of interleukin-17-mediated signaling pathway | 1 | 1 | 6.96e-01 |
| GO:BP | GO:1903881 | regulation of interleukin-17-mediated signaling pathway | 1 | 1 | 6.96e-01 |
| GO:BP | GO:1903886 | positive regulation of chemokine (C-C motif) ligand 20 production | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0036392 | chemokine (C-C motif) ligand 20 production | 1 | 1 | 6.96e-01 |
| GO:BP | GO:1903884 | regulation of chemokine (C-C motif) ligand 20 production | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0007014 | actin ubiquitination | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0034462 | small-subunit processome assembly | 2 | 3 | 6.96e-01 |
| GO:BP | GO:0032000 | positive regulation of fatty acid beta-oxidation | 2 | 9 | 6.96e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 4 | 65 | 6.96e-01 |
| GO:BP | GO:0034975 | protein folding in endoplasmic reticulum | 1 | 10 | 6.96e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 6 | 64 | 6.96e-01 |
| GO:BP | GO:1905799 | regulation of intraciliary retrograde transport | 1 | 2 | 6.96e-01 |
| GO:BP | GO:0042670 | retinal cone cell differentiation | 3 | 6 | 6.96e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 3 | 151 | 6.96e-01 |
| GO:BP | GO:0046356 | acetyl-CoA catabolic process | 1 | 3 | 6.96e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 6.96e-01 |
| GO:BP | GO:1904568 | cellular response to wortmannin | 1 | 2 | 6.96e-01 |
| GO:BP | GO:1904567 | response to wortmannin | 1 | 2 | 6.96e-01 |
| GO:BP | GO:1900276 | regulation of proteinase activated receptor activity | 1 | 1 | 6.96e-01 |
| GO:BP | GO:1900737 | negative regulation of phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 1 | 6.96e-01 |
| GO:BP | GO:1903999 | negative regulation of eating behavior | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0003421 | growth plate cartilage axis specification | 1 | 1 | 6.96e-01 |
| GO:BP | GO:1903403 | negative regulation of renal phosphate excretion | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0003249 | cell proliferation involved in heart valve morphogenesis | 1 | 3 | 6.96e-01 |
| GO:BP | GO:0003250 | regulation of cell proliferation involved in heart valve morphogenesis | 1 | 3 | 6.96e-01 |
| GO:BP | GO:1903721 | positive regulation of I-kappaB phosphorylation | 1 | 5 | 6.97e-01 |
| GO:BP | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0060672 | epithelial cell morphogenesis involved in placental branching | 1 | 2 | 6.97e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 4 | 3237 | 6.97e-01 |
| GO:BP | GO:0000957 | mitochondrial RNA catabolic process | 3 | 6 | 6.97e-01 |
| GO:BP | GO:0022027 | interkinetic nuclear migration | 2 | 4 | 6.97e-01 |
| GO:BP | GO:1903893 | positive regulation of ATF6-mediated unfolded protein response | 1 | 1 | 6.97e-01 |
| GO:BP | GO:1903054 | negative regulation of extracellular matrix organization | 3 | 6 | 6.97e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 6.97e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 2 | 126 | 6.97e-01 |
| GO:BP | GO:0006423 | cysteinyl-tRNA aminoacylation | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0090027 | negative regulation of monocyte chemotaxis | 1 | 4 | 6.97e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 6.97e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 4 | 11 | 6.97e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 2 | 6 | 6.97e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 3 | 13 | 6.97e-01 |
| GO:BP | GO:0071037 | nuclear polyadenylation-dependent snRNA catabolic process | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0045792 | negative regulation of cell size | 2 | 9 | 6.97e-01 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 6 | 12 | 6.97e-01 |
| GO:BP | GO:0071036 | nuclear polyadenylation-dependent snoRNA catabolic process | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0031509 | subtelomeric heterochromatin formation | 3 | 7 | 6.97e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 6.97e-01 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 4 | 7 | 6.97e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 6.97e-01 |
| GO:BP | GO:0003406 | retinal pigment epithelium development | 1 | 4 | 6.98e-01 |
| GO:BP | GO:0072674 | multinuclear osteoclast differentiation | 1 | 6 | 6.98e-01 |
| GO:BP | GO:0044804 | autophagy of nucleus | 10 | 18 | 6.98e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 6.98e-01 |
| GO:BP | GO:0060268 | negative regulation of respiratory burst | 2 | 4 | 6.98e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 6 | 17 | 6.98e-01 |
| GO:BP | GO:0002071 | glandular epithelial cell maturation | 3 | 7 | 6.98e-01 |
| GO:BP | GO:0006127 | glycerophosphate shuttle | 1 | 2 | 6.98e-01 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 3 | 11 | 6.98e-01 |
| GO:BP | GO:0006104 | succinyl-CoA metabolic process | 5 | 9 | 6.98e-01 |
| GO:BP | GO:0000053 | argininosuccinate metabolic process | 1 | 1 | 6.98e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 6.98e-01 |
| GO:BP | GO:0120193 | tight junction organization | 4 | 52 | 6.98e-01 |
| GO:BP | GO:0010725 | regulation of primitive erythrocyte differentiation | 1 | 1 | 6.98e-01 |
| GO:BP | GO:0016183 | synaptic vesicle coating | 3 | 4 | 6.98e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 2 | 212 | 6.98e-01 |
| GO:BP | GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | 4 | 14 | 6.98e-01 |
| GO:BP | GO:0035854 | eosinophil fate commitment | 1 | 1 | 6.98e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 5 | 215 | 6.98e-01 |
| GO:BP | GO:1990679 | histone H4-K12 deacetylation | 1 | 1 | 6.98e-01 |
| GO:BP | GO:1905770 | regulation of mesodermal cell differentiation | 4 | 6 | 6.99e-01 |
| GO:BP | GO:0060592 | mammary gland formation | 4 | 6 | 6.99e-01 |
| GO:BP | GO:0019079 | viral genome replication | 1 | 101 | 6.99e-01 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 1 | 9 | 6.99e-01 |
| GO:BP | GO:0031104 | dendrite regeneration | 1 | 1 | 6.99e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 1 | 27 | 6.99e-01 |
| GO:BP | GO:0006497 | protein lipidation | 27 | 89 | 6.99e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 1 | 18 | 6.99e-01 |
| GO:BP | GO:1903307 | positive regulation of regulated secretory pathway | 1 | 32 | 6.99e-01 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 3 | 17 | 6.99e-01 |
| GO:BP | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay | 1 | 1 | 6.99e-01 |
| GO:BP | GO:1903744 | positive regulation of anterograde synaptic vesicle transport | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0008582 | regulation of synaptic assembly at neuromuscular junction | 3 | 4 | 6.99e-01 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 3 | 17 | 6.99e-01 |
| GO:BP | GO:1902805 | positive regulation of synaptic vesicle transport | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 1 | 18 | 6.99e-01 |
| GO:BP | GO:0014724 | regulation of twitch skeletal muscle contraction | 1 | 3 | 6.99e-01 |
| GO:BP | GO:0070651 | nonfunctional rRNA decay | 1 | 1 | 6.99e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 4 | 3012 | 6.99e-01 |
| GO:BP | GO:0042760 | very long-chain fatty acid catabolic process | 2 | 5 | 6.99e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 6 | 26 | 6.99e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 7 | 54 | 6.99e-01 |
| GO:BP | GO:0110095 | cellular detoxification of aldehyde | 3 | 5 | 6.99e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 6.99e-01 |
| GO:BP | GO:0042118 | endothelial cell activation | 3 | 7 | 6.99e-01 |
| GO:BP | GO:0140249 | protein catabolic process at postsynapse | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0140246 | protein catabolic process at synapse | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0099533 | positive regulation of presynaptic cytosolic calcium concentration | 1 | 1 | 7.00e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 1 | 19 | 7.00e-01 |
| GO:BP | GO:0072386 | plus-end-directed organelle transport along microtubule | 2 | 7 | 7.00e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 3 | 4 | 7.00e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 6 | 11 | 7.00e-01 |
| GO:BP | GO:0060681 | branch elongation involved in ureteric bud branching | 3 | 3 | 7.00e-01 |
| GO:BP | GO:0019083 | viral transcription | 6 | 48 | 7.00e-01 |
| GO:BP | GO:0045141 | meiotic telomere clustering | 1 | 5 | 7.00e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 18 | 71 | 7.00e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 3 | 9 | 7.00e-01 |
| GO:BP | GO:0021897 | forebrain astrocyte development | 2 | 2 | 7.01e-01 |
| GO:BP | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity | 4 | 4 | 7.01e-01 |
| GO:BP | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis | 3 | 5 | 7.01e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 7.01e-01 |
| GO:BP | GO:0046348 | amino sugar catabolic process | 5 | 9 | 7.01e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 7.01e-01 |
| GO:BP | GO:0021896 | forebrain astrocyte differentiation | 2 | 2 | 7.01e-01 |
| GO:BP | GO:0002337 | B-1a B cell differentiation | 1 | 2 | 7.01e-01 |
| GO:BP | GO:2000097 | regulation of smooth muscle cell-matrix adhesion | 2 | 3 | 7.01e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 9 | 20 | 7.01e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 2 | 1223 | 7.01e-01 |
| GO:BP | GO:0007084 | mitotic nuclear membrane reassembly | 1 | 9 | 7.01e-01 |
| GO:BP | GO:0101024 | mitotic nuclear membrane organization | 1 | 9 | 7.01e-01 |
| GO:BP | GO:0009589 | detection of UV | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0038020 | insulin receptor recycling | 2 | 4 | 7.01e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 3 | 5 | 7.01e-01 |
| GO:BP | GO:0032695 | negative regulation of interleukin-12 production | 1 | 6 | 7.01e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 5 | 37 | 7.01e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 4 | 124 | 7.01e-01 |
| GO:BP | GO:0001575 | globoside metabolic process | 1 | 2 | 7.01e-01 |
| GO:BP | GO:1903048 | regulation of acetylcholine-gated cation channel activity | 1 | 1 | 7.01e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 2 | 11 | 7.01e-01 |
| GO:BP | GO:0071389 | cellular response to mineralocorticoid stimulus | 4 | 9 | 7.01e-01 |
| GO:BP | GO:0031179 | peptide modification | 1 | 4 | 7.01e-01 |
| GO:BP | GO:1903049 | negative regulation of acetylcholine-gated cation channel activity | 1 | 1 | 7.01e-01 |
| GO:BP | GO:0001576 | globoside biosynthetic process | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 2 | 14 | 7.02e-01 |
| GO:BP | GO:1905772 | positive regulation of mesodermal cell differentiation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0038116 | chemokine (C-C motif) ligand 21 signaling pathway | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 1 | 40 | 7.02e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 7.02e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 3 | 53 | 7.02e-01 |
| GO:BP | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1905449 | regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 3 | 5 | 7.02e-01 |
| GO:BP | GO:1903167 | regulation of pyrroline-5-carboxylate reductase activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0072192 | ureter epithelial cell differentiation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 10 | 60 | 7.02e-01 |
| GO:BP | GO:2000825 | positive regulation of androgen receptor activity | 3 | 5 | 7.02e-01 |
| GO:BP | GO:0030035 | microspike assembly | 3 | 5 | 7.02e-01 |
| GO:BP | GO:0060567 | negative regulation of termination of DNA-templated transcription | 1 | 2 | 7.02e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 31 | 65 | 7.02e-01 |
| GO:BP | GO:2000111 | positive regulation of macrophage apoptotic process | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0010900 | negative regulation of phosphatidylcholine catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0010899 | regulation of phosphatidylcholine catabolic process | 2 | 3 | 7.02e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 22 | 41 | 7.02e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 1 | 126 | 7.02e-01 |
| GO:BP | GO:0072200 | negative regulation of mesenchymal cell proliferation involved in ureter development | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903200 | positive regulation of L-dopa decarboxylase activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903073 | negative regulation of death-inducing signaling complex assembly | 1 | 1 | 7.02e-01 |
| GO:BP | GO:2000805 | negative regulation of termination of RNA polymerase II transcription, poly(A)-coupled | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0035272 | exocrine system development | 4 | 34 | 7.02e-01 |
| GO:BP | GO:0097264 | self proteolysis | 2 | 7 | 7.02e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 2 | 7 | 7.02e-01 |
| GO:BP | GO:0006546 | glycine catabolic process | 3 | 3 | 7.02e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 3 | 81 | 7.02e-01 |
| GO:BP | GO:0090194 | negative regulation of glomerulus development | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0097238 | cellular response to methylglyoxal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0030154 | cell differentiation | 3 | 2958 | 7.02e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 15 | 43 | 7.02e-01 |
| GO:BP | GO:0072377 | blood coagulation, common pathway | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0033240 | positive regulation of amine metabolic process | 5 | 7 | 7.02e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 5 | 10 | 7.02e-01 |
| GO:BP | GO:0048242 | epinephrine secretion | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 19 | 43 | 7.02e-01 |
| GO:BP | GO:0046359 | butyrate catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0045915 | positive regulation of catecholamine metabolic process | 4 | 6 | 7.02e-01 |
| GO:BP | GO:1903198 | regulation of L-dopa decarboxylase activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0046836 | glycolipid transport | 1 | 7 | 7.02e-01 |
| GO:BP | GO:0010159 | specification of animal organ position | 2 | 2 | 7.02e-01 |
| GO:BP | GO:1903178 | positive regulation of tyrosine 3-monooxygenase activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0033366 | protein localization to secretory granule | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0001949 | sebaceous gland cell differentiation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0060152 | microtubule-based peroxisome localization | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0060151 | peroxisome localization | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1903176 | regulation of tyrosine 3-monooxygenase activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 1 | 4 | 7.02e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 2 | 100 | 7.02e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 7 | 351 | 7.02e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 7.02e-01 |
| GO:BP | GO:0002934 | desmosome organization | 3 | 9 | 7.02e-01 |
| GO:BP | GO:0033383 | geranyl diphosphate metabolic process | 1 | 3 | 7.02e-01 |
| GO:BP | GO:2000284 | positive regulation of amino acid biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0033505 | floor plate morphogenesis | 1 | 3 | 7.02e-01 |
| GO:BP | GO:0034213 | quinolinate catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903122 | negative regulation of TRAIL-activated apoptotic signaling pathway | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 3 | 12 | 7.02e-01 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 12 | 563 | 7.02e-01 |
| GO:BP | GO:0014843 | growth factor dependent regulation of skeletal muscle satellite cell proliferation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1903184 | L-dopa metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903168 | positive regulation of pyrroline-5-carboxylate reductase activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0055020 | positive regulation of cardiac muscle fiber development | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0033602 | negative regulation of dopamine secretion | 3 | 4 | 7.02e-01 |
| GO:BP | GO:1904736 | negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1904735 | regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0046295 | glycolate biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1904733 | negative regulation of electron transfer activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0055018 | regulation of cardiac muscle fiber development | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0060235 | lens induction in camera-type eye | 2 | 3 | 7.02e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 7 | 12 | 7.02e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 55 | 277 | 7.02e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 211 | 7.02e-01 |
| GO:BP | GO:1901964 | positive regulation of cell proliferation involved in outflow tract morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1905426 | positive regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1901494 | regulation of cysteine metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0140924 | L-kynurenine transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0014060 | regulation of epinephrine secretion | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1905424 | regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0015782 | CMP-N-acetylneuraminate transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0015789 | UDP-N-acetylgalactosamine transmembrane transport | 2 | 4 | 7.02e-01 |
| GO:BP | GO:0002726 | positive regulation of T cell cytokine production | 1 | 14 | 7.02e-01 |
| GO:BP | GO:0015797 | mannitol transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 4 | 7 | 7.02e-01 |
| GO:BP | GO:0060503 | bud dilation involved in lung branching | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0015787 | UDP-glucuronic acid transmembrane transport | 2 | 4 | 7.02e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 3 | 78 | 7.02e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 5 | 21 | 7.02e-01 |
| GO:BP | GO:1904022 | positive regulation of G protein-coupled receptor internalization | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0015724 | formate transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903185 | L-dopa biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0045964 | positive regulation of dopamine metabolic process | 4 | 6 | 7.02e-01 |
| GO:BP | GO:0042063 | gliogenesis | 1 | 235 | 7.02e-01 |
| GO:BP | GO:0015735 | uronic acid transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0015736 | hexuronate transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0015738 | glucuronate transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 12 | 33 | 7.02e-01 |
| GO:BP | GO:1903189 | glyoxal metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903190 | glyoxal catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 7 | 39 | 7.02e-01 |
| GO:BP | GO:1903195 | regulation of L-dopa biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1904430 | negative regulation of t-circle formation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 8 | 63 | 7.02e-01 |
| GO:BP | GO:1903197 | positive regulation of L-dopa biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0045726 | positive regulation of integrin biosynthetic process | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1904002 | regulation of sebum secreting cell proliferation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1990442 | intrinsic apoptotic signaling pathway in response to nitrosative stress | 2 | 3 | 7.02e-01 |
| GO:BP | GO:0120191 | negative regulation of termination of RNA polymerase II transcription | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1902748 | positive regulation of lens fiber cell differentiation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0021563 | glossopharyngeal nerve development | 2 | 3 | 7.02e-01 |
| GO:BP | GO:2000004 | regulation of metanephric S-shaped body morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:2000005 | negative regulation of metanephric S-shaped body morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036150 | phosphatidylserine acyl-chain remodeling | 1 | 7 | 7.02e-01 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 1 | 57 | 7.02e-01 |
| GO:BP | GO:2000006 | regulation of metanephric comma-shaped body morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:2000007 | negative regulation of metanephric comma-shaped body morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 1 | 27 | 7.02e-01 |
| GO:BP | GO:0071893 | BMP signaling pathway involved in nephric duct formation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036367 | light adaption | 1 | 3 | 7.02e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 2 | 26 | 7.02e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 4 | 2890 | 7.02e-01 |
| GO:BP | GO:1990034 | calcium ion export across plasma membrane | 4 | 6 | 7.02e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 6 | 135 | 7.02e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 1 | 55 | 7.02e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 7.02e-01 |
| GO:BP | GO:1903072 | regulation of death-inducing signaling complex assembly | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0106046 | guanine deglycation, glyoxal removal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0106045 | guanine deglycation, methylglyoxal removal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0106044 | guanine deglycation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 9 | 142 | 7.02e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 2 | 57 | 7.02e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 7.02e-01 |
| GO:BP | GO:0048599 | oocyte development | 6 | 38 | 7.02e-01 |
| GO:BP | GO:0001525 | angiogenesis | 28 | 390 | 7.02e-01 |
| GO:BP | GO:0007493 | endodermal cell fate determination | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 3 | 42 | 7.02e-01 |
| GO:BP | GO:0006562 | proline catabolic process | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 3 | 12 | 7.02e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 7 | 64 | 7.02e-01 |
| GO:BP | GO:0071550 | death-inducing signaling complex assembly | 2 | 3 | 7.02e-01 |
| GO:BP | GO:0038002 | endocrine signaling | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036531 | glutathione deglycation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036530 | protein deglycation, methylglyoxal removal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036529 | protein deglycation, glyoxal removal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 1 | 78 | 7.02e-01 |
| GO:BP | GO:0002352 | B cell negative selection | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0036528 | peptidyl-lysine deglycation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036527 | peptidyl-arginine deglycation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0070085 | glycosylation | 1 | 202 | 7.02e-01 |
| GO:BP | GO:0070235 | regulation of activation-induced cell death of T cells | 2 | 3 | 7.02e-01 |
| GO:BP | GO:0006580 | ethanolamine metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0140041 | cellular detoxification of methylglyoxal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 13 | 67 | 7.02e-01 |
| GO:BP | GO:0042874 | D-glucuronate transmembrane transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0006193 | ITP catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1900113 | negative regulation of histone H3-K9 trimethylation | 1 | 3 | 7.02e-01 |
| GO:BP | GO:0016132 | brassinosteroid biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0016131 | brassinosteroid metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0016129 | phytosteroid biosynthetic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0016128 | phytosteroid metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036471 | cellular response to glyoxal | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0070099 | regulation of chemokine-mediated signaling pathway | 3 | 8 | 7.02e-01 |
| GO:BP | GO:0036526 | peptidyl-cysteine deglycation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0005997 | xylulose metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 8 | 15 | 7.02e-01 |
| GO:BP | GO:0048391 | intermediate mesoderm formation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0072101 | specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0061151 | BMP signaling pathway involved in renal system segmentation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0072096 | negative regulation of branch elongation involved in ureteric bud branching | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0006954 | inflammatory response | 12 | 429 | 7.02e-01 |
| GO:BP | GO:0061155 | pulmonary artery endothelial tube morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0061150 | renal system segmentation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0048390 | intermediate mesoderm morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 15 | 43 | 7.02e-01 |
| GO:BP | GO:0072097 | negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0009207 | purine ribonucleoside triphosphate catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0061149 | BMP signaling pathway involved in ureter morphogenesis | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0048392 | intermediate mesodermal cell differentiation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0072099 | anterior/posterior pattern specification involved in ureteric bud development | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 2 | 7 | 7.02e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 5 | 22 | 7.02e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 2 | 7 | 7.02e-01 |
| GO:BP | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 2 | 12 | 7.02e-01 |
| GO:BP | GO:0071332 | cellular response to fructose stimulus | 2 | 2 | 7.02e-01 |
| GO:BP | GO:0072100 | specification of ureteric bud anterior/posterior symmetry | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0061141 | lung ciliated cell differentiation | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 3 | 7 | 7.02e-01 |
| GO:BP | GO:1905949 | negative regulation of calcium ion import across plasma membrane | 1 | 4 | 7.02e-01 |
| GO:BP | GO:0030302 | deoxynucleotide transport | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0046949 | fatty-acyl-CoA biosynthetic process | 5 | 20 | 7.02e-01 |
| GO:BP | GO:2000525 | positive regulation of T cell costimulation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 3 | 15 | 7.02e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 27 | 52 | 7.02e-01 |
| GO:BP | GO:0016036 | cellular response to phosphate starvation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 18 | 376 | 7.02e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 2 | 9 | 7.02e-01 |
| GO:BP | GO:2001136 | negative regulation of endocytic recycling | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1903140 | regulation of establishment of endothelial barrier | 1 | 12 | 7.02e-01 |
| GO:BP | GO:1901550 | regulation of endothelial cell development | 1 | 12 | 7.02e-01 |
| GO:BP | GO:0071492 | cellular response to UV-A | 1 | 8 | 7.02e-01 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 2 | 11 | 7.02e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 11 | 540 | 7.02e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 2 | 52 | 7.03e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 2 | 52 | 7.03e-01 |
| GO:BP | GO:0002176 | male germ cell proliferation | 2 | 6 | 7.03e-01 |
| GO:BP | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration | 4 | 7 | 7.03e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 3 | 2984 | 7.03e-01 |
| GO:BP | GO:1990773 | matrix metallopeptidase secretion | 1 | 5 | 7.03e-01 |
| GO:BP | GO:1904464 | regulation of matrix metallopeptidase secretion | 1 | 5 | 7.03e-01 |
| GO:BP | GO:0030049 | muscle filament sliding | 4 | 10 | 7.03e-01 |
| GO:BP | GO:1904837 | beta-catenin-TCF complex assembly | 1 | 3 | 7.04e-01 |
| GO:BP | GO:0007538 | primary sex determination | 1 | 2 | 7.04e-01 |
| GO:BP | GO:0003176 | aortic valve development | 15 | 37 | 7.04e-01 |
| GO:BP | GO:0007431 | salivary gland development | 3 | 26 | 7.04e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 12 | 40 | 7.04e-01 |
| GO:BP | GO:0019064 | fusion of virus membrane with host plasma membrane | 2 | 4 | 7.04e-01 |
| GO:BP | GO:0046877 | regulation of saliva secretion | 1 | 3 | 7.04e-01 |
| GO:BP | GO:0039663 | membrane fusion involved in viral entry into host cell | 2 | 4 | 7.04e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 7.05e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 7.05e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 1 | 14 | 7.05e-01 |
| GO:BP | GO:0034720 | histone H3-K4 demethylation | 1 | 5 | 7.05e-01 |
| GO:BP | GO:0009180 | purine ribonucleoside diphosphate biosynthetic process | 3 | 4 | 7.05e-01 |
| GO:BP | GO:0006172 | ADP biosynthetic process | 3 | 4 | 7.05e-01 |
| GO:BP | GO:1905430 | cellular response to glycine | 1 | 2 | 7.05e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 1 | 72 | 7.05e-01 |
| GO:BP | GO:0032438 | melanosome organization | 4 | 36 | 7.05e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 7.05e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 1 | 59 | 7.05e-01 |
| GO:BP | GO:0035551 | protein initiator methionine removal involved in protein maturation | 1 | 3 | 7.05e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 7 | 5017 | 7.05e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 18 | 120 | 7.05e-01 |
| GO:BP | GO:0030263 | apoptotic chromosome condensation | 1 | 4 | 7.05e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 7.05e-01 |
| GO:BP | GO:0003099 | positive regulation of the force of heart contraction by chemical signal | 1 | 2 | 7.05e-01 |
| GO:BP | GO:0000294 | nuclear-transcribed mRNA catabolic process, RNase MRP-dependent | 1 | 2 | 7.05e-01 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 2 | 17 | 7.05e-01 |
| GO:BP | GO:0048496 | maintenance of animal organ identity | 1 | 4 | 7.05e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 7.06e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 3 | 27 | 7.06e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 5 | 19 | 7.06e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 5 | 17 | 7.06e-01 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 6 | 30 | 7.07e-01 |
| GO:BP | GO:0006097 | glyoxylate cycle | 1 | 2 | 7.07e-01 |
| GO:BP | GO:1903845 | negative regulation of cellular response to transforming growth factor beta stimulus | 2 | 2 | 7.07e-01 |
| GO:BP | GO:1904347 | regulation of small intestine smooth muscle contraction | 1 | 1 | 7.07e-01 |
| GO:BP | GO:1904349 | positive regulation of small intestine smooth muscle contraction | 1 | 1 | 7.07e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 5 | 17 | 7.07e-01 |
| GO:BP | GO:0035502 | metanephric part of ureteric bud development | 1 | 3 | 7.07e-01 |
| GO:BP | GO:0072284 | metanephric S-shaped body morphogenesis | 1 | 4 | 7.07e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 19 | 320 | 7.07e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 4 | 12 | 7.07e-01 |
| GO:BP | GO:0045898 | regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 7 | 7.07e-01 |
| GO:BP | GO:0099039 | sphingolipid translocation | 2 | 4 | 7.07e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 1 | 80 | 7.07e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 1 | 37 | 7.07e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 5 | 16 | 7.07e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 4 | 10 | 7.07e-01 |
| GO:BP | GO:0072584 | caveolin-mediated endocytosis | 3 | 9 | 7.07e-01 |
| GO:BP | GO:0006621 | protein retention in ER lumen | 4 | 8 | 7.07e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 1 | 15 | 7.08e-01 |
| GO:BP | GO:0097400 | interleukin-17-mediated signaling pathway | 3 | 8 | 7.08e-01 |
| GO:BP | GO:0019371 | cyclooxygenase pathway | 2 | 7 | 7.08e-01 |
| GO:BP | GO:2001293 | malonyl-CoA metabolic process | 3 | 6 | 7.08e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 4 | 3080 | 7.08e-01 |
| GO:BP | GO:0051412 | response to corticosterone | 5 | 10 | 7.08e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 14 | 199 | 7.08e-01 |
| GO:BP | GO:0048632 | negative regulation of skeletal muscle tissue growth | 1 | 1 | 7.08e-01 |
| GO:BP | GO:0002115 | store-operated calcium entry | 2 | 15 | 7.08e-01 |
| GO:BP | GO:0034516 | response to vitamin B6 | 1 | 1 | 7.08e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 1 | 31 | 7.08e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 4 | 116 | 7.09e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 6 | 23 | 7.09e-01 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 7 | 36 | 7.09e-01 |
| GO:BP | GO:1900028 | negative regulation of ruffle assembly | 2 | 4 | 7.09e-01 |
| GO:BP | GO:0051170 | import into nucleus | 13 | 152 | 7.10e-01 |
| GO:BP | GO:0099525 | presynaptic dense core vesicle exocytosis | 2 | 4 | 7.10e-01 |
| GO:BP | GO:1905912 | regulation of calcium ion export across plasma membrane | 2 | 4 | 7.10e-01 |
| GO:BP | GO:1905913 | negative regulation of calcium ion export across plasma membrane | 2 | 4 | 7.10e-01 |
| GO:BP | GO:1904179 | positive regulation of adipose tissue development | 2 | 6 | 7.10e-01 |
| GO:BP | GO:0015711 | organic anion transport | 1 | 264 | 7.11e-01 |
| GO:BP | GO:0015847 | putrescine transport | 2 | 2 | 7.11e-01 |
| GO:BP | GO:1904772 | response to tetrachloromethane | 2 | 4 | 7.11e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 32 | 691 | 7.11e-01 |
| GO:BP | GO:0071305 | cellular response to vitamin D | 2 | 14 | 7.11e-01 |
| GO:BP | GO:0045629 | negative regulation of T-helper 2 cell differentiation | 1 | 3 | 7.11e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 7.11e-01 |
| GO:BP | GO:1990167 | protein K27-linked deubiquitination | 2 | 3 | 7.11e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 5 | 275 | 7.12e-01 |
| GO:BP | GO:1901569 | fatty acid derivative catabolic process | 1 | 10 | 7.12e-01 |
| GO:BP | GO:0021551 | central nervous system morphogenesis | 1 | 2 | 7.12e-01 |
| GO:BP | GO:0097155 | fasciculation of sensory neuron axon | 1 | 3 | 7.12e-01 |
| GO:BP | GO:0030043 | actin filament fragmentation | 2 | 4 | 7.13e-01 |
| GO:BP | GO:0010273 | detoxification of copper ion | 1 | 8 | 7.13e-01 |
| GO:BP | GO:1990169 | stress response to copper ion | 1 | 8 | 7.13e-01 |
| GO:BP | GO:0038134 | ERBB2-EGFR signaling pathway | 1 | 6 | 7.13e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 1 | 43 | 7.13e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 7.13e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 7.13e-01 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 6 | 14 | 7.13e-01 |
| GO:BP | GO:0060699 | regulation of endoribonuclease activity | 1 | 4 | 7.13e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 14 | 79 | 7.13e-01 |
| GO:BP | GO:1990145 | maintenance of translational fidelity | 1 | 2 | 7.13e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 4 | 19 | 7.13e-01 |
| GO:BP | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 1 | 14 | 7.13e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 3 | 7.13e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 2 | 12 | 7.13e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 2 | 17 | 7.13e-01 |
| GO:BP | GO:1905664 | regulation of calcium ion import across plasma membrane | 2 | 8 | 7.13e-01 |
| GO:BP | GO:0072560 | type B pancreatic cell maturation | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0071888 | macrophage apoptotic process | 5 | 9 | 7.13e-01 |
| GO:BP | GO:0060717 | chorion development | 3 | 8 | 7.13e-01 |
| GO:BP | GO:1900126 | negative regulation of hyaluronan biosynthetic process | 1 | 2 | 7.13e-01 |
| GO:BP | GO:2000416 | regulation of eosinophil migration | 2 | 2 | 7.13e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 13 | 632 | 7.13e-01 |
| GO:BP | GO:0072387 | flavin adenine dinucleotide metabolic process | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 2 | 9 | 7.14e-01 |
| GO:BP | GO:0008354 | germ cell migration | 1 | 6 | 7.14e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 2 | 19 | 7.15e-01 |
| GO:BP | GO:0048203 | vesicle targeting, trans-Golgi to endosome | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 10 | 31 | 7.15e-01 |
| GO:BP | GO:0038192 | gastric inhibitory peptide signaling pathway | 1 | 1 | 7.15e-01 |
| GO:BP | GO:0090248 | cell migration involved in somitogenic axis elongation | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0090249 | regulation of cell migration involved in somitogenic axis elongation | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 10 | 46 | 7.16e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 2 | 16 | 7.16e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 29 | 7.16e-01 |
| GO:BP | GO:0010796 | regulation of multivesicular body size | 1 | 2 | 7.16e-01 |
| GO:BP | GO:1902744 | negative regulation of lamellipodium organization | 2 | 4 | 7.16e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 10 | 28 | 7.16e-01 |
| GO:BP | GO:0060712 | spongiotrophoblast layer development | 3 | 11 | 7.16e-01 |
| GO:BP | GO:2000354 | regulation of ovarian follicle development | 2 | 5 | 7.16e-01 |
| GO:BP | GO:1902836 | positive regulation of proline import across plasma membrane | 1 | 2 | 7.16e-01 |
| GO:BP | GO:1902834 | regulation of proline import across plasma membrane | 1 | 2 | 7.16e-01 |
| GO:BP | GO:1905735 | regulation of L-proline import across plasma membrane | 1 | 2 | 7.16e-01 |
| GO:BP | GO:1905737 | positive regulation of L-proline import across plasma membrane | 1 | 2 | 7.16e-01 |
| GO:BP | GO:0070881 | regulation of proline transport | 1 | 2 | 7.16e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 3 | 2285 | 7.16e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 7.16e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 7.17e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 3 | 23 | 7.17e-01 |
| GO:BP | GO:0019541 | propionate metabolic process | 1 | 2 | 7.17e-01 |
| GO:BP | GO:0034397 | telomere localization | 1 | 5 | 7.17e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 1 | 26 | 7.17e-01 |
| GO:BP | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation | 1 | 5 | 7.17e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 1 | 45 | 7.17e-01 |
| GO:BP | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 2 | 7.17e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 1 | 26 | 7.17e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 1 | 22 | 7.17e-01 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 2 | 9 | 7.17e-01 |
| GO:BP | GO:0097164 | ammonium ion metabolic process | 1 | 14 | 7.17e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 8 | 252 | 7.18e-01 |
| GO:BP | GO:0080009 | mRNA methylation | 1 | 16 | 7.18e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 4 | 57 | 7.18e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 2 | 11 | 7.18e-01 |
| GO:BP | GO:0048213 | Golgi vesicle prefusion complex stabilization | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 2 | 34 | 7.18e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 7.18e-01 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 1 | 10 | 7.18e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 9 | 23 | 7.18e-01 |
| GO:BP | GO:0072318 | clathrin coat disassembly | 4 | 8 | 7.19e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 13 | 27 | 7.19e-01 |
| GO:BP | GO:0001743 | lens placode formation | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0046619 | lens placode formation involved in camera-type eye formation | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 5 | 47 | 7.19e-01 |
| GO:BP | GO:0019532 | oxalate transport | 1 | 5 | 7.19e-01 |
| GO:BP | GO:0060426 | lung vasculature development | 1 | 3 | 7.20e-01 |
| GO:BP | GO:0021781 | glial cell fate commitment | 3 | 5 | 7.20e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 1 | 101 | 7.20e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 3 | 6 | 7.20e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 7 | 13 | 7.20e-01 |
| GO:BP | GO:0070662 | mast cell proliferation | 2 | 6 | 7.20e-01 |
| GO:BP | GO:0015891 | siderophore transport | 1 | 2 | 7.20e-01 |
| GO:BP | GO:0070666 | regulation of mast cell proliferation | 2 | 6 | 7.20e-01 |
| GO:BP | GO:0045346 | regulation of MHC class II biosynthetic process | 8 | 11 | 7.20e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 3 | 994 | 7.20e-01 |
| GO:BP | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway | 1 | 10 | 7.20e-01 |
| GO:BP | GO:1904527 | negative regulation of microtubule binding | 1 | 2 | 7.21e-01 |
| GO:BP | GO:1902511 | negative regulation of apoptotic DNA fragmentation | 1 | 2 | 7.21e-01 |
| GO:BP | GO:0045348 | positive regulation of MHC class II biosynthetic process | 6 | 8 | 7.21e-01 |
| GO:BP | GO:0032076 | negative regulation of deoxyribonuclease activity | 1 | 1 | 7.21e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 7.21e-01 |
| GO:BP | GO:0051867 | general adaptation syndrome, behavioral process | 1 | 3 | 7.21e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 6 | 285 | 7.21e-01 |
| GO:BP | GO:0045453 | bone resorption | 1 | 40 | 7.21e-01 |
| GO:BP | GO:0051230 | spindle disassembly | 1 | 2 | 7.21e-01 |
| GO:BP | GO:0043984 | histone H4-K16 acetylation | 6 | 14 | 7.21e-01 |
| GO:BP | GO:0051228 | mitotic spindle disassembly | 1 | 2 | 7.21e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 15 | 162 | 7.21e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 4 | 37 | 7.21e-01 |
| GO:BP | GO:0032836 | glomerular basement membrane development | 2 | 9 | 7.22e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 2 | 61 | 7.22e-01 |
| GO:BP | GO:0021527 | spinal cord association neuron differentiation | 2 | 3 | 7.22e-01 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 2 | 4 | 7.22e-01 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 2 | 4 | 7.22e-01 |
| GO:BP | GO:0048755 | branching morphogenesis of a nerve | 1 | 8 | 7.22e-01 |
| GO:BP | GO:1903719 | regulation of I-kappaB phosphorylation | 1 | 6 | 7.22e-01 |
| GO:BP | GO:0046449 | creatinine metabolic process | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 2 | 10 | 7.23e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 3 | 16 | 7.23e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 4 | 114 | 7.23e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 1 | 209 | 7.23e-01 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 1 | 88 | 7.23e-01 |
| GO:BP | GO:0070268 | cornification | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 3 | 11 | 7.23e-01 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0098528 | skeletal muscle fiber differentiation | 1 | 5 | 7.23e-01 |
| GO:BP | GO:0061073 | ciliary body morphogenesis | 1 | 3 | 7.23e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 1 | 24 | 7.23e-01 |
| GO:BP | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment | 1 | 1 | 7.23e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 4 | 155 | 7.23e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 7 | 26 | 7.23e-01 |
| GO:BP | GO:0048548 | regulation of pinocytosis | 6 | 11 | 7.23e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 3 | 9 | 7.23e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 6 | 116 | 7.23e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 80 | 324 | 7.24e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 2 | 16 | 7.24e-01 |
| GO:BP | GO:0021644 | vagus nerve morphogenesis | 1 | 1 | 7.24e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 3 | 131 | 7.24e-01 |
| GO:BP | GO:0097101 | blood vessel endothelial cell fate specification | 1 | 3 | 7.24e-01 |
| GO:BP | GO:0060846 | blood vessel endothelial cell fate commitment | 1 | 3 | 7.24e-01 |
| GO:BP | GO:2000252 | negative regulation of feeding behavior | 2 | 2 | 7.24e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 9 | 26 | 7.25e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 1 | 199 | 7.25e-01 |
| GO:BP | GO:0051138 | positive regulation of NK T cell differentiation | 1 | 3 | 7.25e-01 |
| GO:BP | GO:0048625 | myoblast fate commitment | 2 | 4 | 7.25e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 3 | 30 | 7.25e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 3 | 30 | 7.25e-01 |
| GO:BP | GO:0015884 | folic acid transport | 1 | 7 | 7.25e-01 |
| GO:BP | GO:1903385 | regulation of homophilic cell adhesion | 1 | 3 | 7.25e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 7 | 29 | 7.25e-01 |
| GO:BP | GO:0006413 | translational initiation | 2 | 116 | 7.25e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 1 | 166 | 7.25e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 3 | 79 | 7.25e-01 |
| GO:BP | GO:0006427 | histidyl-tRNA aminoacylation | 1 | 2 | 7.25e-01 |
| GO:BP | GO:1905581 | positive regulation of low-density lipoprotein particle clearance | 1 | 2 | 7.25e-01 |
| GO:BP | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 2 | 7.25e-01 |
| GO:BP | GO:0050746 | regulation of lipoprotein metabolic process | 1 | 15 | 7.25e-01 |
| GO:BP | GO:0048702 | embryonic neurocranium morphogenesis | 1 | 7 | 7.25e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 2 | 45 | 7.25e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 7.26e-01 |
| GO:BP | GO:0060018 | astrocyte fate commitment | 2 | 2 | 7.26e-01 |
| GO:BP | GO:1903332 | regulation of protein folding | 3 | 8 | 7.26e-01 |
| GO:BP | GO:0072289 | metanephric nephron tubule formation | 2 | 2 | 7.26e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 3 | 4 | 7.26e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 7.26e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 10 | 46 | 7.26e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 1 | 25 | 7.26e-01 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 2 | 89 | 7.26e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 8 | 391 | 7.26e-01 |
| GO:BP | GO:0045039 | protein insertion into mitochondrial inner membrane | 2 | 13 | 7.26e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 7.26e-01 |
| GO:BP | GO:0055025 | positive regulation of cardiac muscle tissue development | 1 | 2 | 7.27e-01 |
| GO:BP | GO:0044070 | regulation of monoatomic anion transport | 7 | 12 | 7.27e-01 |
| GO:BP | GO:2000532 | regulation of renal albumin absorption | 1 | 1 | 7.27e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 12 | 227 | 7.27e-01 |
| GO:BP | GO:0097368 | establishment of Sertoli cell barrier | 2 | 3 | 7.27e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 7.27e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 8 | 16 | 7.28e-01 |
| GO:BP | GO:1905349 | ciliary transition zone assembly | 1 | 2 | 7.28e-01 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 1 | 9 | 7.28e-01 |
| GO:BP | GO:0044691 | tooth eruption | 3 | 5 | 7.28e-01 |
| GO:BP | GO:0010753 | positive regulation of cGMP-mediated signaling | 1 | 3 | 7.28e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 4 | 3169 | 7.28e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 7 | 105 | 7.28e-01 |
| GO:BP | GO:0003343 | septum transversum development | 1 | 2 | 7.28e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 2 | 15 | 7.28e-01 |
| GO:BP | GO:0003350 | pulmonary myocardium development | 1 | 2 | 7.28e-01 |
| GO:BP | GO:0003342 | proepicardium development | 1 | 2 | 7.28e-01 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 5 | 13 | 7.29e-01 |
| GO:BP | GO:0032536 | regulation of cell projection size | 3 | 8 | 7.29e-01 |
| GO:BP | GO:1902498 | regulation of protein autoubiquitination | 3 | 5 | 7.29e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 2 | 18 | 7.29e-01 |
| GO:BP | GO:0046544 | development of secondary male sexual characteristics | 1 | 1 | 7.29e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 2 | 9 | 7.29e-01 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 1 | 10 | 7.29e-01 |
| GO:BP | GO:0051461 | positive regulation of corticotropin secretion | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0009651 | response to salt stress | 1 | 18 | 7.29e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 7.29e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 10 | 98 | 7.29e-01 |
| GO:BP | GO:0007528 | neuromuscular junction development | 6 | 39 | 7.29e-01 |
| GO:BP | GO:0031144 | proteasome localization | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0071360 | cellular response to exogenous dsRNA | 5 | 14 | 7.29e-01 |
| GO:BP | GO:0046618 | xenobiotic export from cell | 4 | 8 | 7.29e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 3 | 19 | 7.29e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 1 | 19 | 7.29e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 7.29e-01 |
| GO:BP | GO:2001137 | positive regulation of endocytic recycling | 1 | 7 | 7.29e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 4 | 7 | 7.29e-01 |
| GO:BP | GO:0015960 | diadenosine polyphosphate biosynthetic process | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0015965 | diadenosine tetraphosphate metabolic process | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0015966 | diadenosine tetraphosphate biosynthetic process | 1 | 2 | 7.29e-01 |
| GO:BP | GO:2000793 | cell proliferation involved in heart valve development | 2 | 4 | 7.29e-01 |
| GO:BP | GO:0090500 | endocardial cushion to mesenchymal transition | 2 | 4 | 7.29e-01 |
| GO:BP | GO:0050996 | positive regulation of lipid catabolic process | 1 | 16 | 7.29e-01 |
| GO:BP | GO:1903850 | regulation of cristae formation | 2 | 4 | 7.29e-01 |
| GO:BP | GO:0046903 | secretion | 2 | 645 | 7.29e-01 |
| GO:BP | GO:0045213 | neurotransmitter receptor metabolic process | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 7 | 373 | 7.29e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 6 | 9 | 7.29e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 7.29e-01 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 170 | 7.29e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 6 | 295 | 7.30e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 1 | 105 | 7.30e-01 |
| GO:BP | GO:0034414 | tRNA 3’-trailer cleavage, endonucleolytic | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1904694 | negative regulation of vascular associated smooth muscle contraction | 1 | 3 | 7.30e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 10 | 29 | 7.30e-01 |
| GO:BP | GO:0051169 | nuclear transport | 6 | 295 | 7.30e-01 |
| GO:BP | GO:1902803 | regulation of synaptic vesicle transport | 2 | 5 | 7.30e-01 |
| GO:BP | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1902858 | propionyl-CoA metabolic process | 2 | 4 | 7.30e-01 |
| GO:BP | GO:1902269 | positive regulation of polyamine transmembrane transport | 1 | 2 | 7.30e-01 |
| GO:BP | GO:2001294 | malonyl-CoA catabolic process | 2 | 4 | 7.30e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 2 | 64 | 7.30e-01 |
| GO:BP | GO:0043378 | positive regulation of CD8-positive, alpha-beta T cell differentiation | 2 | 2 | 7.30e-01 |
| GO:BP | GO:0006572 | tyrosine catabolic process | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0006479 | protein methylation | 2 | 160 | 7.30e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 9 | 18 | 7.30e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 7.30e-01 |
| GO:BP | GO:0008213 | protein alkylation | 2 | 160 | 7.30e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 29 | 7.30e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 2 | 60 | 7.30e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 1 | 140 | 7.30e-01 |
| GO:BP | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone | 3 | 5 | 7.30e-01 |
| GO:BP | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 1 | 5 | 7.30e-01 |
| GO:BP | GO:0006533 | aspartate catabolic process | 2 | 2 | 7.30e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 8 | 64 | 7.30e-01 |
| GO:BP | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 1 | 10 | 7.31e-01 |
| GO:BP | GO:0150031 | regulation of protein localization to lysosome | 1 | 3 | 7.31e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 6 | 51 | 7.31e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 5 | 25 | 7.31e-01 |
| GO:BP | GO:0018175 | protein nucleotidylation | 1 | 2 | 7.31e-01 |
| GO:BP | GO:1902024 | L-histidine transport | 2 | 4 | 7.31e-01 |
| GO:BP | GO:1902688 | regulation of NAD metabolic process | 1 | 1 | 7.31e-01 |
| GO:BP | GO:0018117 | protein adenylylation | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0007252 | I-kappaB phosphorylation | 2 | 14 | 7.31e-01 |
| GO:BP | GO:0015829 | valine transport | 2 | 4 | 7.31e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 6 | 612 | 7.31e-01 |
| GO:BP | GO:0015817 | histidine transport | 2 | 4 | 7.31e-01 |
| GO:BP | GO:0010712 | regulation of collagen metabolic process | 21 | 34 | 7.31e-01 |
| GO:BP | GO:0090184 | positive regulation of kidney development | 3 | 9 | 7.31e-01 |
| GO:BP | GO:0060192 | negative regulation of lipase activity | 1 | 13 | 7.31e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 1 | 279 | 7.31e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 7.32e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 16 | 59 | 7.32e-01 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 1 | 21 | 7.32e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 3 | 9 | 7.32e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 3 | 9 | 7.32e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 25 | 7.32e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 1 | 25 | 7.32e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 3 | 9 | 7.32e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 19 | 65 | 7.32e-01 |
| GO:BP | GO:0009227 | nucleotide-sugar catabolic process | 1 | 3 | 7.32e-01 |
| GO:BP | GO:1902081 | negative regulation of calcium ion import into sarcoplasmic reticulum | 1 | 1 | 7.33e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 2 | 77 | 7.33e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 2 | 24 | 7.33e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 10 | 43 | 7.33e-01 |
| GO:BP | GO:0051582 | positive regulation of neurotransmitter uptake | 3 | 4 | 7.33e-01 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 9 | 15 | 7.33e-01 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 1 | 8 | 7.33e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 7 | 48 | 7.33e-01 |
| GO:BP | GO:0045959 | negative regulation of complement activation, classical pathway | 2 | 4 | 7.34e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 7.34e-01 |
| GO:BP | GO:0060764 | cell-cell signaling involved in mammary gland development | 1 | 1 | 7.34e-01 |
| GO:BP | GO:1990120 | messenger ribonucleoprotein complex assembly | 1 | 3 | 7.34e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 5 | 294 | 7.34e-01 |
| GO:BP | GO:0015844 | monoamine transport | 2 | 45 | 7.34e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 7.34e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 32 | 55 | 7.34e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 3 | 7 | 7.34e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 3 | 22 | 7.34e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 7 | 70 | 7.34e-01 |
| GO:BP | GO:0006865 | amino acid transport | 2 | 102 | 7.35e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 3 | 7 | 7.35e-01 |
| GO:BP | GO:0048477 | oogenesis | 3 | 64 | 7.35e-01 |
| GO:BP | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway | 3 | 4 | 7.35e-01 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 1 | 4 | 7.35e-01 |
| GO:BP | GO:1904028 | positive regulation of collagen fibril organization | 1 | 4 | 7.35e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 12 | 40 | 7.35e-01 |
| GO:BP | GO:0032094 | response to food | 1 | 22 | 7.35e-01 |
| GO:BP | GO:2000738 | positive regulation of stem cell differentiation | 5 | 17 | 7.35e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 16 | 41 | 7.35e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 11 | 21 | 7.36e-01 |
| GO:BP | GO:0033280 | response to vitamin D | 3 | 24 | 7.36e-01 |
| GO:BP | GO:1901727 | positive regulation of histone deacetylase activity | 3 | 5 | 7.36e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 3 | 24 | 7.36e-01 |
| GO:BP | GO:0001654 | eye development | 4 | 272 | 7.36e-01 |
| GO:BP | GO:0008347 | glial cell migration | 2 | 47 | 7.36e-01 |
| GO:BP | GO:0090660 | cerebrospinal fluid circulation | 1 | 9 | 7.36e-01 |
| GO:BP | GO:0070173 | regulation of enamel mineralization | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0015697 | quaternary ammonium group transport | 1 | 17 | 7.36e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 4 | 20 | 7.36e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 31 | 53 | 7.36e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 4 | 7.36e-01 |
| GO:BP | GO:0046032 | ADP catabolic process | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0060737 | prostate gland morphogenetic growth | 2 | 2 | 7.36e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 6 | 23 | 7.36e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 2 | 8 | 7.36e-01 |
| GO:BP | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0072350 | tricarboxylic acid metabolic process | 7 | 13 | 7.37e-01 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 2 | 6 | 7.37e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 2 | 21 | 7.37e-01 |
| GO:BP | GO:0099074 | mitochondrion to lysosome transport | 1 | 3 | 7.37e-01 |
| GO:BP | GO:0032351 | negative regulation of hormone metabolic process | 2 | 10 | 7.37e-01 |
| GO:BP | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | 3 | 7.37e-01 |
| GO:BP | GO:0099075 | mitochondrion-derived vesicle mediated transport | 1 | 3 | 7.37e-01 |
| GO:BP | GO:0010636 | positive regulation of mitochondrial fusion | 1 | 2 | 7.37e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 2 | 54 | 7.37e-01 |
| GO:BP | GO:1902283 | negative regulation of primary amine oxidase activity | 1 | 1 | 7.37e-01 |
| GO:BP | GO:0061055 | myotome development | 2 | 2 | 7.37e-01 |
| GO:BP | GO:0034650 | cortisol metabolic process | 2 | 9 | 7.37e-01 |
| GO:BP | GO:0046813 | receptor-mediated virion attachment to host cell | 1 | 6 | 7.37e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 2 | 9 | 7.37e-01 |
| GO:BP | GO:0006002 | fructose 6-phosphate metabolic process | 2 | 8 | 7.37e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 1 | 50 | 7.38e-01 |
| GO:BP | GO:0046373 | L-arabinose metabolic process | 1 | 1 | 7.38e-01 |
| GO:BP | GO:0031591 | wybutosine biosynthetic process | 2 | 4 | 7.38e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 3 | 2412 | 7.38e-01 |
| GO:BP | GO:0019566 | arabinose metabolic process | 1 | 1 | 7.38e-01 |
| GO:BP | GO:0031590 | wybutosine metabolic process | 2 | 4 | 7.38e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 3 | 13 | 7.38e-01 |
| GO:BP | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum | 2 | 2 | 7.38e-01 |
| GO:BP | GO:1904606 | fat cell apoptotic process | 1 | 1 | 7.38e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 15 | 7.38e-01 |
| GO:BP | GO:1904649 | regulation of fat cell apoptotic process | 1 | 1 | 7.38e-01 |
| GO:BP | GO:1903971 | positive regulation of response to macrophage colony-stimulating factor | 1 | 1 | 7.38e-01 |
| GO:BP | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c | 1 | 7 | 7.38e-01 |
| GO:BP | GO:0070562 | regulation of vitamin D receptor signaling pathway | 1 | 9 | 7.38e-01 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 1 | 4 | 7.38e-01 |
| GO:BP | GO:1903974 | positive regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 1 | 7.38e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 3 | 204 | 7.38e-01 |
| GO:BP | GO:0032707 | negative regulation of interleukin-23 production | 1 | 2 | 7.38e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 1 | 69 | 7.38e-01 |
| GO:BP | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 2 | 7.38e-01 |
| GO:BP | GO:0098908 | regulation of neuronal action potential | 1 | 3 | 7.38e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 7.38e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 7 | 382 | 7.39e-01 |
| GO:BP | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | 4 | 6 | 7.39e-01 |
| GO:BP | GO:1903651 | positive regulation of cytoplasmic transport | 3 | 11 | 7.39e-01 |
| GO:BP | GO:0010635 | regulation of mitochondrial fusion | 3 | 11 | 7.39e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 1 | 73 | 7.39e-01 |
| GO:BP | GO:0031947 | negative regulation of glucocorticoid biosynthetic process | 1 | 5 | 7.39e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 3 | 36 | 7.39e-01 |
| GO:BP | GO:0031944 | negative regulation of glucocorticoid metabolic process | 1 | 5 | 7.39e-01 |
| GO:BP | GO:0010813 | neuropeptide catabolic process | 2 | 4 | 7.39e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 2 | 14 | 7.39e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 15 | 59 | 7.40e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 15 | 59 | 7.40e-01 |
| GO:BP | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | 1 | 4 | 7.40e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 7.40e-01 |
| GO:BP | GO:1900104 | regulation of hyaluranon cable assembly | 1 | 3 | 7.40e-01 |
| GO:BP | GO:1900106 | positive regulation of hyaluranon cable assembly | 1 | 3 | 7.40e-01 |
| GO:BP | GO:0046379 | extracellular polysaccharide metabolic process | 1 | 2 | 7.40e-01 |
| GO:BP | GO:0045226 | extracellular polysaccharide biosynthetic process | 1 | 2 | 7.40e-01 |
| GO:BP | GO:0036118 | hyaluranon cable assembly | 1 | 3 | 7.40e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 9 | 25 | 7.40e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 7.40e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 10 | 21 | 7.40e-01 |
| GO:BP | GO:0071284 | cellular response to lead ion | 1 | 4 | 7.40e-01 |
| GO:BP | GO:0002524 | hypersensitivity | 4 | 7 | 7.40e-01 |
| GO:BP | GO:1905551 | negative regulation of protein localization to endoplasmic reticulum | 1 | 2 | 7.40e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 2 | 468 | 7.40e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 1 | 108 | 7.40e-01 |
| GO:BP | GO:0150063 | visual system development | 4 | 273 | 7.40e-01 |
| GO:BP | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 2 | 19 | 7.40e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 1 | 260 | 7.40e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 7.40e-01 |
| GO:BP | GO:2000439 | positive regulation of monocyte extravasation | 2 | 3 | 7.40e-01 |
| GO:BP | GO:1904231 | positive regulation of succinate dehydrogenase activity | 1 | 2 | 7.41e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 7.41e-01 |
| GO:BP | GO:1904229 | regulation of succinate dehydrogenase activity | 1 | 2 | 7.41e-01 |
| GO:BP | GO:0010716 | negative regulation of extracellular matrix disassembly | 1 | 2 | 7.41e-01 |
| GO:BP | GO:0045793 | positive regulation of cell size | 1 | 11 | 7.41e-01 |
| GO:BP | GO:0006545 | glycine biosynthetic process | 1 | 4 | 7.42e-01 |
| GO:BP | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition | 2 | 3 | 7.42e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 12 | 427 | 7.42e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 1 | 2 | 7.42e-01 |
| GO:BP | GO:0071716 | leukotriene transport | 3 | 5 | 7.42e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 7 | 41 | 7.43e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 7.43e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 3 | 27 | 7.43e-01 |
| GO:BP | GO:0097168 | mesenchymal stem cell proliferation | 3 | 4 | 7.43e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 4 | 86 | 7.43e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 4 | 86 | 7.43e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 6 | 152 | 7.43e-01 |
| GO:BP | GO:1903826 | L-arginine transmembrane transport | 2 | 12 | 7.43e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4048 | 7.43e-01 |
| GO:BP | GO:0090240 | positive regulation of histone H4 acetylation | 3 | 6 | 7.43e-01 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 1 | 3 | 7.43e-01 |
| GO:BP | GO:1901000 | regulation of response to salt stress | 1 | 4 | 7.43e-01 |
| GO:BP | GO:0015802 | basic amino acid transport | 10 | 17 | 7.43e-01 |
| GO:BP | GO:0036228 | protein localization to nuclear inner membrane | 1 | 3 | 7.43e-01 |
| GO:BP | GO:1900069 | regulation of cellular hyperosmotic salinity response | 1 | 4 | 7.43e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 3 | 3386 | 7.43e-01 |
| GO:BP | GO:0099105 | ion channel modulating, G protein-coupled receptor signaling pathway | 1 | 7 | 7.43e-01 |
| GO:BP | GO:0021578 | hindbrain maturation | 2 | 4 | 7.43e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 45 | 164 | 7.43e-01 |
| GO:BP | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | 1 | 10 | 7.43e-01 |
| GO:BP | GO:0009753 | response to jasmonic acid | 2 | 3 | 7.44e-01 |
| GO:BP | GO:0071395 | cellular response to jasmonic acid stimulus | 2 | 3 | 7.44e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 1 | 13 | 7.44e-01 |
| GO:BP | GO:0006072 | glycerol-3-phosphate metabolic process | 3 | 7 | 7.44e-01 |
| GO:BP | GO:0007341 | penetration of zona pellucida | 1 | 4 | 7.44e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 4 | 51 | 7.45e-01 |
| GO:BP | GO:1900118 | negative regulation of execution phase of apoptosis | 3 | 6 | 7.45e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 6 | 11 | 7.45e-01 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 4 | 7 | 7.45e-01 |
| GO:BP | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity | 1 | 6 | 7.45e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 1 | 59 | 7.45e-01 |
| GO:BP | GO:0015833 | peptide transport | 1 | 179 | 7.45e-01 |
| GO:BP | GO:0036071 | N-glycan fucosylation | 1 | 3 | 7.45e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 7.45e-01 |
| GO:BP | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching | 2 | 4 | 7.45e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 7.45e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 318 | 7.46e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 3 | 12 | 7.46e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 3 | 2574 | 7.46e-01 |
| GO:BP | GO:0097343 | ripoptosome assembly | 1 | 2 | 7.46e-01 |
| GO:BP | GO:1901026 | ripoptosome assembly involved in necroptotic process | 1 | 2 | 7.46e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 3 | 2576 | 7.46e-01 |
| GO:BP | GO:2000426 | negative regulation of apoptotic cell clearance | 1 | 1 | 7.46e-01 |
| GO:BP | GO:0030449 | regulation of complement activation | 1 | 11 | 7.46e-01 |
| GO:BP | GO:0045163 | clustering of voltage-gated potassium channels | 1 | 2 | 7.46e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 6 | 15 | 7.46e-01 |
| GO:BP | GO:0016137 | glycoside metabolic process | 4 | 16 | 7.47e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 6 | 64 | 7.47e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 32 | 7.47e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 7.47e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 7.47e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 13 | 669 | 7.47e-01 |
| GO:BP | GO:0045658 | regulation of neutrophil differentiation | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0042541 | hemoglobin biosynthetic process | 5 | 13 | 7.47e-01 |
| GO:BP | GO:1990399 | epithelium regeneration | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 8 | 38 | 7.47e-01 |
| GO:BP | GO:0045016 | mitochondrial magnesium ion transmembrane transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:1905041 | regulation of epithelium regeneration | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 8 | 38 | 7.47e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 1 | 33 | 7.47e-01 |
| GO:BP | GO:0043654 | recognition of apoptotic cell | 2 | 4 | 7.47e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 7.48e-01 |
| GO:BP | GO:0090234 | regulation of kinetochore assembly | 1 | 3 | 7.48e-01 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 2 | 4 | 7.48e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 7.48e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 2 | 38 | 7.48e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 7.48e-01 |
| GO:BP | GO:0042701 | progesterone secretion | 2 | 3 | 7.48e-01 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 2 | 19 | 7.49e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 20 | 7.49e-01 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 2 | 19 | 7.49e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 2 | 95 | 7.49e-01 |
| GO:BP | GO:0002384 | hepatic immune response | 1 | 1 | 7.49e-01 |
| GO:BP | GO:0048880 | sensory system development | 4 | 275 | 7.49e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 9 | 17 | 7.49e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 12 | 40 | 7.49e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 12 | 40 | 7.49e-01 |
| GO:BP | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion | 2 | 5 | 7.49e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 7 | 418 | 7.49e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 6 | 55 | 7.50e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 1 | 14 | 7.50e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 3 | 2593 | 7.50e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 3 | 91 | 7.50e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 10 | 185 | 7.50e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 2 | 63 | 7.50e-01 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1 | 9 | 7.50e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 30 | 292 | 7.50e-01 |
| GO:BP | GO:0002457 | T cell antigen processing and presentation | 2 | 4 | 7.51e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 8 | 123 | 7.51e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 7.51e-01 |
| GO:BP | GO:0060406 | positive regulation of penile erection | 1 | 3 | 7.51e-01 |
| GO:BP | GO:0016115 | terpenoid catabolic process | 1 | 4 | 7.51e-01 |
| GO:BP | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process | 1 | 3 | 7.51e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 4 | 9 | 7.51e-01 |
| GO:BP | GO:0045728 | respiratory burst after phagocytosis | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 1 | 37 | 7.51e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 3 | 14 | 7.51e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 3 | 14 | 7.51e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 1 | 222 | 7.52e-01 |
| GO:BP | GO:0021586 | pons maturation | 1 | 1 | 7.52e-01 |
| GO:BP | GO:0021722 | superior olivary nucleus maturation | 1 | 1 | 7.52e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 1 | 18 | 7.52e-01 |
| GO:BP | GO:0021718 | superior olivary nucleus development | 1 | 1 | 7.52e-01 |
| GO:BP | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly | 1 | 7 | 7.52e-01 |
| GO:BP | GO:0016340 | calcium-dependent cell-matrix adhesion | 1 | 2 | 7.52e-01 |
| GO:BP | GO:0061086 | negative regulation of histone H3-K27 methylation | 1 | 2 | 7.52e-01 |
| GO:BP | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | 1 | 2 | 7.53e-01 |
| GO:BP | GO:0008039 | synaptic target recognition | 1 | 2 | 7.53e-01 |
| GO:BP | GO:0060168 | positive regulation of adenosine receptor signaling pathway | 1 | 1 | 7.53e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 7.53e-01 |
| GO:BP | GO:0060264 | regulation of respiratory burst involved in inflammatory response | 2 | 3 | 7.53e-01 |
| GO:BP | GO:1903596 | regulation of gap junction assembly | 5 | 8 | 7.53e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 25 | 153 | 7.54e-01 |
| GO:BP | GO:0007228 | positive regulation of hh target transcription factor activity | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0007227 | signal transduction downstream of smoothened | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0003352 | regulation of cilium movement | 1 | 17 | 7.54e-01 |
| GO:BP | GO:0060438 | trachea development | 4 | 13 | 7.54e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 7.54e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 12 | 68 | 7.54e-01 |
| GO:BP | GO:0043007 | maintenance of rDNA | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0035726 | common myeloid progenitor cell proliferation | 4 | 5 | 7.54e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 5 | 17 | 7.55e-01 |
| GO:BP | GO:0046501 | protoporphyrinogen IX metabolic process | 2 | 11 | 7.55e-01 |
| GO:BP | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway | 1 | 1 | 7.55e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 2 | 11 | 7.55e-01 |
| GO:BP | GO:0046104 | thymidine metabolic process | 1 | 5 | 7.55e-01 |
| GO:BP | GO:0009409 | response to cold | 9 | 41 | 7.55e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 8 | 382 | 7.55e-01 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 1 | 5 | 7.55e-01 |
| GO:BP | GO:0051695 | actin filament uncapping | 1 | 2 | 7.55e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 9 | 23 | 7.55e-01 |
| GO:BP | GO:0035094 | response to nicotine | 1 | 28 | 7.55e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 27 | 242 | 7.55e-01 |
| GO:BP | GO:0032382 | positive regulation of intracellular sterol transport | 2 | 3 | 7.55e-01 |
| GO:BP | GO:0032385 | positive regulation of intracellular cholesterol transport | 2 | 3 | 7.55e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 7.55e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 9 | 90 | 7.55e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 1 | 50 | 7.56e-01 |
| GO:BP | GO:1903286 | regulation of potassium ion import | 3 | 8 | 7.56e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 31 | 667 | 7.56e-01 |
| GO:BP | GO:0048014 | Tie signaling pathway | 2 | 3 | 7.56e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 10 | 105 | 7.56e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 7.56e-01 |
| GO:BP | GO:0006085 | acetyl-CoA biosynthetic process | 1 | 16 | 7.56e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 1 | 11 | 7.56e-01 |
| GO:BP | GO:1990840 | response to lectin | 1 | 11 | 7.56e-01 |
| GO:BP | GO:0010501 | RNA secondary structure unwinding | 4 | 8 | 7.56e-01 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 8 | 17 | 7.56e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 1 | 11 | 7.56e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 6 | 10 | 7.57e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 6 | 258 | 7.57e-01 |
| GO:BP | GO:0099011 | neuronal dense core vesicle exocytosis | 1 | 4 | 7.57e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 1 | 71 | 7.57e-01 |
| GO:BP | GO:0072300 | positive regulation of metanephric glomerulus development | 1 | 1 | 7.57e-01 |
| GO:BP | GO:0072298 | regulation of metanephric glomerulus development | 1 | 1 | 7.57e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 5 | 3580 | 7.57e-01 |
| GO:BP | GO:0014042 | positive regulation of neuron maturation | 1 | 3 | 7.57e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 1 | 71 | 7.57e-01 |
| GO:BP | GO:0072216 | positive regulation of metanephros development | 1 | 1 | 7.57e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 2 | 58 | 7.57e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 5 | 4174 | 7.57e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 4 | 28 | 7.57e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 2 | 7 | 7.57e-01 |
| GO:BP | GO:0050765 | negative regulation of phagocytosis | 11 | 15 | 7.57e-01 |
| GO:BP | GO:0060215 | primitive hemopoiesis | 3 | 7 | 7.58e-01 |
| GO:BP | GO:0001542 | ovulation from ovarian follicle | 2 | 8 | 7.58e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 16 | 7.58e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 7.58e-01 |
| GO:BP | GO:0002074 | extraocular skeletal muscle development | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 7.58e-01 |
| GO:BP | GO:0038172 | interleukin-33-mediated signaling pathway | 1 | 1 | 7.58e-01 |
| GO:BP | GO:0061188 | negative regulation of rDNA heterochromatin formation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0061187 | regulation of rDNA heterochromatin formation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0030311 | poly-N-acetyllactosamine biosynthetic process | 2 | 5 | 7.58e-01 |
| GO:BP | GO:0045347 | negative regulation of MHC class II biosynthetic process | 1 | 3 | 7.58e-01 |
| GO:BP | GO:0071359 | cellular response to dsRNA | 6 | 18 | 7.58e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 7 | 10 | 7.58e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 2 | 2 | 7.58e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 7.58e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 2 | 211 | 7.58e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 2 | 2 | 7.58e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 2 | 2 | 7.58e-01 |
| GO:BP | GO:0023035 | CD40 signaling pathway | 2 | 11 | 7.58e-01 |
| GO:BP | GO:0001905 | activation of membrane attack complex | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0001969 | regulation of activation of membrane attack complex | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 1 | 57 | 7.58e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 1 | 55 | 7.59e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 4 | 34 | 7.59e-01 |
| GO:BP | GO:1905382 | positive regulation of snRNA transcription by RNA polymerase II | 1 | 3 | 7.59e-01 |
| GO:BP | GO:0014075 | response to amine | 1 | 34 | 7.59e-01 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 3 | 20 | 7.59e-01 |
| GO:BP | GO:2001027 | negative regulation of endothelial cell chemotaxis | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0060820 | inactivation of X chromosome by heterochromatin formation | 1 | 1 | 7.59e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 7 | 11 | 7.59e-01 |
| GO:BP | GO:0021553 | olfactory nerve development | 4 | 5 | 7.59e-01 |
| GO:BP | GO:0071420 | cellular response to histamine | 3 | 4 | 7.59e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 7.59e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 7.59e-01 |
| GO:BP | GO:1905653 | positive regulation of artery morphogenesis | 2 | 5 | 7.59e-01 |
| GO:BP | GO:1905651 | regulation of artery morphogenesis | 2 | 5 | 7.59e-01 |
| GO:BP | GO:0090170 | regulation of Golgi inheritance | 1 | 4 | 7.60e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 8 | 19 | 7.60e-01 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 1 | 5 | 7.60e-01 |
| GO:BP | GO:0016032 | viral process | 3 | 344 | 7.60e-01 |
| GO:BP | GO:0043587 | tongue morphogenesis | 2 | 7 | 7.60e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 1 | 229 | 7.61e-01 |
| GO:BP | GO:0035356 | intracellular triglyceride homeostasis | 3 | 5 | 7.61e-01 |
| GO:BP | GO:0044381 | glucose import in response to insulin stimulus | 1 | 4 | 7.61e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 31 | 631 | 7.61e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 3 | 70 | 7.61e-01 |
| GO:BP | GO:0043474 | pigment metabolic process involved in pigmentation | 1 | 3 | 7.61e-01 |
| GO:BP | GO:0060465 | pharynx development | 1 | 3 | 7.61e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 3 | 70 | 7.61e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 3 | 42 | 7.61e-01 |
| GO:BP | GO:1902732 | positive regulation of chondrocyte proliferation | 2 | 5 | 7.62e-01 |
| GO:BP | GO:0048859 | formation of anatomical boundary | 1 | 3 | 7.62e-01 |
| GO:BP | GO:1904237 | positive regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.62e-01 |
| GO:BP | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.62e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 4 | 21 | 7.62e-01 |
| GO:BP | GO:0046833 | positive regulation of RNA export from nucleus | 2 | 5 | 7.62e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 27 | 57 | 7.62e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 3 | 20 | 7.62e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 1 | 50 | 7.62e-01 |
| GO:BP | GO:2001213 | negative regulation of vasculogenesis | 1 | 1 | 7.62e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 7.62e-01 |
| GO:BP | GO:1904580 | regulation of intracellular mRNA localization | 2 | 4 | 7.62e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 2 | 64 | 7.63e-01 |
| GO:BP | GO:1905524 | negative regulation of protein autoubiquitination | 1 | 2 | 7.63e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 12 | 116 | 7.63e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 2 | 4 | 7.63e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 9 | 17 | 7.63e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 7.63e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 7.63e-01 |
| GO:BP | GO:0070141 | response to UV-A | 1 | 11 | 7.63e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 6 | 20 | 7.63e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 7.63e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1 | 2065 | 7.63e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 2 | 9 | 7.63e-01 |
| GO:BP | GO:0034163 | regulation of toll-like receptor 9 signaling pathway | 4 | 6 | 7.64e-01 |
| GO:BP | GO:0006000 | fructose metabolic process | 1 | 10 | 7.64e-01 |
| GO:BP | GO:0035455 | response to interferon-alpha | 9 | 16 | 7.64e-01 |
| GO:BP | GO:0061669 | spontaneous neurotransmitter secretion | 2 | 5 | 7.64e-01 |
| GO:BP | GO:0015074 | DNA integration | 4 | 9 | 7.64e-01 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 3 | 10 | 7.65e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 3 | 21 | 7.65e-01 |
| GO:BP | GO:0055095 | lipoprotein particle mediated signaling | 1 | 3 | 7.65e-01 |
| GO:BP | GO:0055096 | low-density lipoprotein particle mediated signaling | 1 | 3 | 7.65e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 6 | 97 | 7.65e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 45 | 147 | 7.65e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 56 | 204 | 7.65e-01 |
| GO:BP | GO:0034371 | chylomicron remodeling | 1 | 1 | 7.65e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 7.65e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 7.65e-01 |
| GO:BP | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration | 1 | 2 | 7.65e-01 |
| GO:BP | GO:1904526 | regulation of microtubule binding | 2 | 10 | 7.65e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 6 | 22 | 7.65e-01 |
| GO:BP | GO:0060304 | regulation of phosphatidylinositol dephosphorylation | 2 | 5 | 7.65e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 3 | 10 | 7.65e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 7.65e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 8 | 15 | 7.65e-01 |
| GO:BP | GO:0002604 | regulation of dendritic cell antigen processing and presentation | 2 | 5 | 7.65e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 1 | 230 | 7.66e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 1 | 55 | 7.66e-01 |
| GO:BP | GO:0045723 | positive regulation of fatty acid biosynthetic process | 3 | 12 | 7.66e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 3 | 17 | 7.66e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 3 | 2683 | 7.66e-01 |
| GO:BP | GO:1904506 | negative regulation of telomere maintenance in response to DNA damage | 2 | 3 | 7.66e-01 |
| GO:BP | GO:0043011 | myeloid dendritic cell differentiation | 1 | 10 | 7.66e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 3 | 2684 | 7.66e-01 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 1 | 43 | 7.66e-01 |
| GO:BP | GO:0042221 | response to chemical | 2 | 2559 | 7.66e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 14 | 56 | 7.66e-01 |
| GO:BP | GO:0002940 | tRNA N2-guanine methylation | 1 | 4 | 7.66e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 7.66e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 1 | 11 | 7.66e-01 |
| GO:BP | GO:1900098 | regulation of plasma cell differentiation | 1 | 2 | 7.66e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 2 | 6 | 7.66e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 2 | 53 | 7.67e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 4 | 12 | 7.67e-01 |
| GO:BP | GO:0006403 | RNA localization | 55 | 182 | 7.67e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 7.67e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 8 | 14 | 7.67e-01 |
| GO:BP | GO:0048469 | cell maturation | 2 | 108 | 7.68e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 3 | 29 | 7.68e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 7.68e-01 |
| GO:BP | GO:0070922 | RISC complex assembly | 3 | 9 | 7.68e-01 |
| GO:BP | GO:1903327 | negative regulation of tRNA metabolic process | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0090206 | negative regulation of cholesterol metabolic process | 2 | 5 | 7.68e-01 |
| GO:BP | GO:0042473 | outer ear morphogenesis | 3 | 5 | 7.68e-01 |
| GO:BP | GO:0036416 | tRNA stabilization | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 2 | 50 | 7.68e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 295 | 7.68e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 3 | 45 | 7.68e-01 |
| GO:BP | GO:0106119 | negative regulation of sterol biosynthetic process | 2 | 5 | 7.68e-01 |
| GO:BP | GO:0140447 | cytokine precursor processing | 1 | 1 | 7.68e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 2 | 10 | 7.68e-01 |
| GO:BP | GO:0036415 | regulation of tRNA stability | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0031650 | regulation of heat generation | 2 | 7 | 7.68e-01 |
| GO:BP | GO:1902370 | regulation of tRNA catabolic process | 1 | 2 | 7.68e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 2 | 13 | 7.68e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 11 | 20 | 7.68e-01 |
| GO:BP | GO:0060936 | cardiac fibroblast cell development | 2 | 4 | 7.68e-01 |
| GO:BP | GO:0060935 | cardiac fibroblast cell differentiation | 2 | 4 | 7.68e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 3 | 29 | 7.68e-01 |
| GO:BP | GO:1903326 | regulation of tRNA metabolic process | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0045541 | negative regulation of cholesterol biosynthetic process | 2 | 5 | 7.68e-01 |
| GO:BP | GO:0035457 | cellular response to interferon-alpha | 5 | 7 | 7.68e-01 |
| GO:BP | GO:1902371 | negative regulation of tRNA catabolic process | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 18 | 73 | 7.68e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 5 | 31 | 7.69e-01 |
| GO:BP | GO:0070172 | positive regulation of tooth mineralization | 1 | 2 | 7.69e-01 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 1 | 11 | 7.69e-01 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 1 | 12 | 7.69e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4455 | 7.69e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 2 | 53 | 7.69e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 4 | 34 | 7.69e-01 |
| GO:BP | GO:0075525 | viral translational termination-reinitiation | 1 | 5 | 7.69e-01 |
| GO:BP | GO:0016571 | histone methylation | 11 | 117 | 7.70e-01 |
| GO:BP | GO:0035747 | natural killer cell chemotaxis | 2 | 3 | 7.70e-01 |
| GO:BP | GO:0019509 | L-methionine salvage from methylthioadenosine | 2 | 5 | 7.70e-01 |
| GO:BP | GO:0060267 | positive regulation of respiratory burst | 2 | 3 | 7.70e-01 |
| GO:BP | GO:0006701 | progesterone biosynthetic process | 1 | 5 | 7.70e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 7.70e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 2 | 28 | 7.71e-01 |
| GO:BP | GO:0051541 | elastin metabolic process | 2 | 5 | 7.71e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 2 | 108 | 7.71e-01 |
| GO:BP | GO:1902559 | 3’-phospho-5’-adenylyl sulfate transmembrane transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:1902558 | 5’-adenylyl sulfate transmembrane transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0048172 | regulation of short-term neuronal synaptic plasticity | 1 | 9 | 7.71e-01 |
| GO:BP | GO:0046963 | 3’-phosphoadenosine 5’-phosphosulfate transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 2 | 25 | 7.71e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 2 | 28 | 7.71e-01 |
| GO:BP | GO:2001286 | regulation of caveolin-mediated endocytosis | 3 | 5 | 7.71e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 2 | 28 | 7.71e-01 |
| GO:BP | GO:0043585 | nose morphogenesis | 1 | 3 | 7.71e-01 |
| GO:BP | GO:1990959 | eosinophil homeostasis | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0033133 | positive regulation of glucokinase activity | 2 | 4 | 7.71e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 20 | 998 | 7.71e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 3 | 2714 | 7.71e-01 |
| GO:BP | GO:1901537 | positive regulation of DNA demethylation | 1 | 7 | 7.71e-01 |
| GO:BP | GO:2000192 | negative regulation of fatty acid transport | 4 | 7 | 7.71e-01 |
| GO:BP | GO:0022600 | digestive system process | 2 | 54 | 7.71e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 1 | 59 | 7.71e-01 |
| GO:BP | GO:2000546 | positive regulation of endothelial cell chemotaxis to fibroblast growth factor | 1 | 3 | 7.71e-01 |
| GO:BP | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor | 1 | 3 | 7.71e-01 |
| GO:BP | GO:0050896 | response to stimulus | 3 | 5770 | 7.71e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 2 | 4 | 7.72e-01 |
| GO:BP | GO:0045981 | positive regulation of nucleotide metabolic process | 3 | 16 | 7.72e-01 |
| GO:BP | GO:0071380 | cellular response to prostaglandin E stimulus | 1 | 14 | 7.72e-01 |
| GO:BP | GO:1900544 | positive regulation of purine nucleotide metabolic process | 3 | 16 | 7.72e-01 |
| GO:BP | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell | 1 | 2 | 7.72e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 2 | 3 | 7.72e-01 |
| GO:BP | GO:0051028 | mRNA transport | 35 | 117 | 7.72e-01 |
| GO:BP | GO:1990926 | short-term synaptic potentiation | 2 | 4 | 7.72e-01 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 12 | 23 | 7.72e-01 |
| GO:BP | GO:2000674 | regulation of type B pancreatic cell apoptotic process | 2 | 6 | 7.72e-01 |
| GO:BP | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 3 | 82 | 7.72e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 3 | 26 | 7.72e-01 |
| GO:BP | GO:0072156 | distal tubule morphogenesis | 1 | 3 | 7.72e-01 |
| GO:BP | GO:0021648 | vestibulocochlear nerve morphogenesis | 2 | 3 | 7.72e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 4 | 199 | 7.72e-01 |
| GO:BP | GO:0033029 | regulation of neutrophil apoptotic process | 1 | 5 | 7.72e-01 |
| GO:BP | GO:0090598 | male anatomical structure morphogenesis | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0048808 | male genitalia morphogenesis | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0006642 | triglyceride mobilization | 1 | 3 | 7.72e-01 |
| GO:BP | GO:0042048 | olfactory behavior | 5 | 7 | 7.72e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 3 | 14 | 7.72e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 3 | 7 | 7.72e-01 |
| GO:BP | GO:0090158 | endoplasmic reticulum membrane organization | 3 | 14 | 7.72e-01 |
| GO:BP | GO:0035697 | CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.72e-01 |
| GO:BP | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.72e-01 |
| GO:BP | GO:2000409 | positive regulation of T cell extravasation | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0000266 | mitochondrial fission | 4 | 38 | 7.73e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 25 | 50 | 7.73e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 7.73e-01 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 5 | 21 | 7.73e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 7.73e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 2 | 5 | 7.73e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 3 | 14 | 7.73e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 1 | 34 | 7.73e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 5 | 14 | 7.73e-01 |
| GO:BP | GO:0033633 | negative regulation of cell-cell adhesion mediated by integrin | 1 | 2 | 7.73e-01 |
| GO:BP | GO:0006363 | termination of RNA polymerase I transcription | 1 | 4 | 7.73e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 7.73e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 15 | 36 | 7.73e-01 |
| GO:BP | GO:0043171 | peptide catabolic process | 9 | 16 | 7.73e-01 |
| GO:BP | GO:0043029 | T cell homeostasis | 8 | 27 | 7.73e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 3 | 344 | 7.73e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 15 | 36 | 7.73e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 2 | 72 | 7.73e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 2 | 89 | 7.73e-01 |
| GO:BP | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 2 | 4 | 7.73e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 15 | 159 | 7.73e-01 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 2 | 4 | 7.74e-01 |
| GO:BP | GO:0061868 | hepatic stellate cell migration | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0061869 | regulation of hepatic stellate cell migration | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0051234 | establishment of localization | 2 | 3433 | 7.74e-01 |
| GO:BP | GO:0061870 | positive regulation of hepatic stellate cell migration | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 7.74e-01 |
| GO:BP | GO:1904880 | response to hydrogen sulfide | 1 | 3 | 7.74e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 24 | 62 | 7.74e-01 |
| GO:BP | GO:0072193 | ureter smooth muscle cell differentiation | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0072191 | ureter smooth muscle development | 1 | 2 | 7.74e-01 |
| GO:BP | GO:2000741 | positive regulation of mesenchymal stem cell differentiation | 1 | 4 | 7.74e-01 |
| GO:BP | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 5 | 7.74e-01 |
| GO:BP | GO:1990834 | response to odorant | 1 | 3 | 7.74e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 3 | 345 | 7.74e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 1 | 30 | 7.74e-01 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 1 | 6 | 7.74e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 5 | 7 | 7.75e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 7.75e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 2 | 18 | 7.75e-01 |
| GO:BP | GO:0046950 | cellular ketone body metabolic process | 2 | 7 | 7.75e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 2 | 49 | 7.75e-01 |
| GO:BP | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1 | 3 | 7.75e-01 |
| GO:BP | GO:1903422 | negative regulation of synaptic vesicle recycling | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0033206 | meiotic cytokinesis | 1 | 7 | 7.75e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 2 | 93 | 7.75e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 3 | 6 | 7.75e-01 |
| GO:BP | GO:0070535 | histone H2A K63-linked ubiquitination | 3 | 6 | 7.75e-01 |
| GO:BP | GO:1905429 | response to glycine | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0016056 | rhodopsin mediated signaling pathway | 3 | 6 | 7.76e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 4 | 29 | 7.76e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 2 | 3 | 7.76e-01 |
| GO:BP | GO:0060585 | positive regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 2 | 3 | 7.76e-01 |
| GO:BP | GO:0051026 | chiasma assembly | 1 | 3 | 7.76e-01 |
| GO:BP | GO:1904683 | regulation of metalloendopeptidase activity | 2 | 6 | 7.76e-01 |
| GO:BP | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0014826 | vein smooth muscle contraction | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 3 | 51 | 7.76e-01 |
| GO:BP | GO:0035352 | NAD transmembrane transport | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 1 | 4 | 7.76e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4570 | 7.76e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 3 | 37 | 7.77e-01 |
| GO:BP | GO:0038162 | erythropoietin-mediated signaling pathway | 1 | 2 | 7.77e-01 |
| GO:BP | GO:1990770 | small intestine smooth muscle contraction | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0046959 | habituation | 2 | 5 | 7.77e-01 |
| GO:BP | GO:0048793 | pronephros development | 1 | 5 | 7.77e-01 |
| GO:BP | GO:0043045 | DNA methylation involved in embryo development | 1 | 7 | 7.77e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 7.77e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 7.77e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 2 | 5 | 7.78e-01 |
| GO:BP | GO:0001547 | antral ovarian follicle growth | 3 | 5 | 7.78e-01 |
| GO:BP | GO:0006226 | dUMP biosynthetic process | 1 | 2 | 7.78e-01 |
| GO:BP | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine | 2 | 7 | 7.79e-01 |
| GO:BP | GO:0071352 | cellular response to interleukin-2 | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0032483 | regulation of Rab protein signal transduction | 1 | 7 | 7.79e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 1 | 242 | 7.79e-01 |
| GO:BP | GO:0038110 | interleukin-2-mediated signaling pathway | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 7 | 110 | 7.79e-01 |
| GO:BP | GO:2000487 | positive regulation of glutamine transport | 1 | 2 | 7.79e-01 |
| GO:BP | GO:1903713 | asparagine transmembrane transport | 1 | 1 | 7.79e-01 |
| GO:BP | GO:0051365 | cellular response to potassium ion starvation | 1 | 1 | 7.79e-01 |
| GO:BP | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH | 1 | 1 | 7.79e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 2 | 13 | 7.79e-01 |
| GO:BP | GO:0036378 | calcitriol biosynthetic process from calciol | 2 | 2 | 7.79e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4596 | 7.79e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 5 | 150 | 7.80e-01 |
| GO:BP | GO:0072535 | tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 7.80e-01 |
| GO:BP | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 7.80e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 7.80e-01 |
| GO:BP | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 7.80e-01 |
| GO:BP | GO:2000412 | positive regulation of thymocyte migration | 1 | 1 | 7.80e-01 |
| GO:BP | GO:2000307 | regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 7.80e-01 |
| GO:BP | GO:0018002 | N-terminal peptidyl-glutamic acid acetylation | 1 | 2 | 7.80e-01 |
| GO:BP | GO:0017190 | N-terminal peptidyl-aspartic acid acetylation | 1 | 2 | 7.80e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 2 | 2 | 7.80e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 2 | 119 | 7.80e-01 |
| GO:BP | GO:1900272 | negative regulation of long-term synaptic potentiation | 1 | 8 | 7.80e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 1 | 7 | 7.80e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 4 | 39 | 7.80e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 6 | 24 | 7.80e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 1 | 90 | 7.81e-01 |
| GO:BP | GO:2000353 | positive regulation of endothelial cell apoptotic process | 1 | 13 | 7.81e-01 |
| GO:BP | GO:0046185 | aldehyde catabolic process | 5 | 12 | 7.81e-01 |
| GO:BP | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway | 1 | 2 | 7.81e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 1 | 25 | 7.81e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 39 | 145 | 7.81e-01 |
| GO:BP | GO:2000367 | regulation of acrosomal vesicle exocytosis | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0001807 | regulation of type IV hypersensitivity | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0006550 | isoleucine catabolic process | 1 | 4 | 7.82e-01 |
| GO:BP | GO:0051459 | regulation of corticotropin secretion | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0035483 | gastric emptying | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0097354 | prenylation | 1 | 10 | 7.82e-01 |
| GO:BP | GO:0018342 | protein prenylation | 1 | 10 | 7.82e-01 |
| GO:BP | GO:0038183 | bile acid signaling pathway | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 12 | 58 | 7.82e-01 |
| GO:BP | GO:0045875 | negative regulation of sister chromatid cohesion | 1 | 5 | 7.82e-01 |
| GO:BP | GO:1904437 | positive regulation of transferrin receptor binding | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0070561 | vitamin D receptor signaling pathway | 1 | 10 | 7.82e-01 |
| GO:BP | GO:1904432 | regulation of ferrous iron binding | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0043305 | negative regulation of mast cell degranulation | 2 | 2 | 7.82e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2565 | 7.82e-01 |
| GO:BP | GO:0051935 | glutamate reuptake | 2 | 4 | 7.82e-01 |
| GO:BP | GO:0044830 | modulation by host of viral RNA genome replication | 1 | 6 | 7.82e-01 |
| GO:BP | GO:0016119 | carotene metabolic process | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0016121 | carotene catabolic process | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0016122 | xanthophyll metabolic process | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 7.82e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 7.82e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 3 | 74 | 7.82e-01 |
| GO:BP | GO:1901825 | zeaxanthin metabolic process | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 2 | 5 | 7.82e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 1 | 56 | 7.82e-01 |
| GO:BP | GO:0046549 | retinal cone cell development | 2 | 5 | 7.82e-01 |
| GO:BP | GO:0006952 | defense response | 4 | 944 | 7.82e-01 |
| GO:BP | GO:1904434 | positive regulation of ferrous iron binding | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0046247 | terpene catabolic process | 1 | 1 | 7.82e-01 |
| GO:BP | GO:1904435 | regulation of transferrin receptor binding | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 7.83e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 7 | 57 | 7.83e-01 |
| GO:BP | GO:0003157 | endocardium development | 3 | 9 | 7.83e-01 |
| GO:BP | GO:1901301 | regulation of cargo loading into COPII-coated vesicle | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 7.83e-01 |
| GO:BP | GO:0007197 | adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway | 1 | 3 | 7.83e-01 |
| GO:BP | GO:0001553 | luteinization | 4 | 10 | 7.83e-01 |
| GO:BP | GO:0090032 | negative regulation of steroid hormone biosynthetic process | 1 | 6 | 7.83e-01 |
| GO:BP | GO:0035054 | embryonic heart tube anterior/posterior pattern specification | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2573 | 7.83e-01 |
| GO:BP | GO:0002544 | chronic inflammatory response | 1 | 6 | 7.83e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 3 | 17 | 7.84e-01 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 1 | 10 | 7.84e-01 |
| GO:BP | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | 1 | 12 | 7.84e-01 |
| GO:BP | GO:0045843 | negative regulation of striated muscle tissue development | 4 | 5 | 7.84e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 4 | 22 | 7.84e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 2 | 40 | 7.84e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 2 | 7 | 7.84e-01 |
| GO:BP | GO:0097240 | chromosome attachment to the nuclear envelope | 1 | 3 | 7.84e-01 |
| GO:BP | GO:0035684 | helper T cell extravasation | 1 | 3 | 7.84e-01 |
| GO:BP | GO:0048561 | establishment of animal organ orientation | 1 | 1 | 7.84e-01 |
| GO:BP | GO:0048241 | epinephrine transport | 1 | 4 | 7.84e-01 |
| GO:BP | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation | 3 | 4 | 7.84e-01 |
| GO:BP | GO:0032119 | sequestering of zinc ion | 2 | 5 | 7.85e-01 |
| GO:BP | GO:0030422 | siRNA processing | 1 | 7 | 7.85e-01 |
| GO:BP | GO:0021766 | hippocampus development | 2 | 62 | 7.85e-01 |
| GO:BP | GO:0035504 | regulation of myosin light chain kinase activity | 1 | 1 | 7.85e-01 |
| GO:BP | GO:0035505 | positive regulation of myosin light chain kinase activity | 1 | 1 | 7.85e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 7.85e-01 |
| GO:BP | GO:1903649 | regulation of cytoplasmic transport | 6 | 25 | 7.85e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 2 | 15 | 7.85e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 15 | 775 | 7.85e-01 |
| GO:BP | GO:0018201 | peptidyl-glycine modification | 1 | 4 | 7.85e-01 |
| GO:BP | GO:0097028 | dendritic cell differentiation | 2 | 23 | 7.86e-01 |
| GO:BP | GO:0014858 | positive regulation of skeletal muscle cell proliferation | 1 | 2 | 7.87e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 3 | 46 | 7.87e-01 |
| GO:BP | GO:0038016 | insulin receptor internalization | 1 | 3 | 7.87e-01 |
| GO:BP | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 3 | 7.87e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 2 | 12 | 7.87e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 4 | 43 | 7.87e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 7.87e-01 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 1 | 17 | 7.88e-01 |
| GO:BP | GO:1904204 | regulation of skeletal muscle hypertrophy | 1 | 1 | 7.88e-01 |
| GO:BP | GO:0070103 | regulation of interleukin-6-mediated signaling pathway | 2 | 4 | 7.88e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 3 | 122 | 7.88e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 7 | 14 | 7.88e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 7 | 14 | 7.88e-01 |
| GO:BP | GO:0110091 | negative regulation of hippocampal neuron apoptotic process | 1 | 2 | 7.88e-01 |
| GO:BP | GO:1990258 | histone glutamine methylation | 1 | 2 | 7.88e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 3 | 22 | 7.89e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 2 | 28 | 7.89e-01 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 7.89e-01 |
| GO:BP | GO:0051454 | intracellular pH elevation | 4 | 4 | 7.89e-01 |
| GO:BP | GO:0070476 | rRNA (guanine-N7)-methylation | 1 | 2 | 7.89e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 1 | 63 | 7.89e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 4 | 32 | 7.89e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 2 | 1555 | 7.90e-01 |
| GO:BP | GO:0097402 | neuroblast migration | 1 | 2 | 7.90e-01 |
| GO:BP | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway | 1 | 3 | 7.90e-01 |
| GO:BP | GO:0072343 | pancreatic stellate cell proliferation | 1 | 1 | 7.90e-01 |
| GO:BP | GO:0051953 | negative regulation of amine transport | 4 | 15 | 7.90e-01 |
| GO:BP | GO:0060694 | regulation of cholesterol transporter activity | 1 | 1 | 7.90e-01 |
| GO:BP | GO:1904597 | negative regulation of connective tissue replacement involved in inflammatory response wound healing | 1 | 1 | 7.90e-01 |
| GO:BP | GO:1904596 | regulation of connective tissue replacement involved in inflammatory response wound healing | 1 | 1 | 7.90e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 3 | 38 | 7.90e-01 |
| GO:BP | GO:1905204 | negative regulation of connective tissue replacement | 1 | 1 | 7.90e-01 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 2 | 5 | 7.90e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 19 | 97 | 7.90e-01 |
| GO:BP | GO:0033085 | negative regulation of T cell differentiation in thymus | 1 | 5 | 7.90e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 7.90e-01 |
| GO:BP | GO:1900223 | positive regulation of amyloid-beta clearance | 1 | 5 | 7.90e-01 |
| GO:BP | GO:1901558 | response to metformin | 1 | 1 | 7.90e-01 |
| GO:BP | GO:0071306 | cellular response to vitamin E | 1 | 2 | 7.90e-01 |
| GO:BP | GO:2000229 | regulation of pancreatic stellate cell proliferation | 1 | 1 | 7.90e-01 |
| GO:BP | GO:0009086 | methionine biosynthetic process | 5 | 10 | 7.91e-01 |
| GO:BP | GO:1903742 | regulation of anterograde synaptic vesicle transport | 1 | 3 | 7.91e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 6 | 93 | 7.91e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 7.91e-01 |
| GO:BP | GO:0070948 | regulation of neutrophil mediated cytotoxicity | 1 | 2 | 7.91e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 7 | 36 | 7.91e-01 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 2 | 31 | 7.91e-01 |
| GO:BP | GO:1904780 | negative regulation of protein localization to centrosome | 1 | 2 | 7.91e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 3 | 37 | 7.91e-01 |
| GO:BP | GO:0009136 | purine nucleoside diphosphate biosynthetic process | 3 | 5 | 7.91e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 7.91e-01 |
| GO:BP | GO:1903525 | regulation of membrane tubulation | 2 | 5 | 7.91e-01 |
| GO:BP | GO:0009313 | oligosaccharide catabolic process | 3 | 11 | 7.91e-01 |
| GO:BP | GO:0006149 | deoxyinosine catabolic process | 1 | 1 | 7.91e-01 |
| GO:BP | GO:0006148 | inosine catabolic process | 1 | 1 | 7.91e-01 |
| GO:BP | GO:0046094 | deoxyinosine metabolic process | 1 | 1 | 7.91e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 63 | 421 | 7.91e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4747 | 7.91e-01 |
| GO:BP | GO:0034418 | urate biosynthetic process | 1 | 2 | 7.91e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 3 | 28 | 7.91e-01 |
| GO:BP | GO:1905380 | regulation of snRNA transcription by RNA polymerase II | 2 | 4 | 7.91e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 35 | 7.91e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 3 | 2845 | 7.91e-01 |
| GO:BP | GO:0019065 | receptor-mediated endocytosis of virus by host cell | 2 | 9 | 7.91e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 215 | 7.91e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 10 | 51 | 7.91e-01 |
| GO:BP | GO:0015867 | ATP transport | 3 | 9 | 7.92e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 7 | 19 | 7.92e-01 |
| GO:BP | GO:1902996 | regulation of neurofibrillary tangle assembly | 1 | 2 | 7.92e-01 |
| GO:BP | GO:1901998 | toxin transport | 2 | 37 | 7.92e-01 |
| GO:BP | GO:0000961 | negative regulation of mitochondrial RNA catabolic process | 1 | 2 | 7.92e-01 |
| GO:BP | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production | 3 | 5 | 7.92e-01 |
| GO:BP | GO:0035523 | protein K29-linked deubiquitination | 1 | 4 | 7.92e-01 |
| GO:BP | GO:1905035 | negative regulation of antifungal innate immune response | 1 | 2 | 7.92e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 7.92e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 6 | 13 | 7.92e-01 |
| GO:BP | GO:0055062 | phosphate ion homeostasis | 1 | 8 | 7.93e-01 |
| GO:BP | GO:0019915 | lipid storage | 8 | 66 | 7.93e-01 |
| GO:BP | GO:0060847 | endothelial cell fate specification | 1 | 3 | 7.93e-01 |
| GO:BP | GO:0060300 | regulation of cytokine activity | 1 | 3 | 7.93e-01 |
| GO:BP | GO:0071205 | protein localization to juxtaparanode region of axon | 2 | 3 | 7.93e-01 |
| GO:BP | GO:0033030 | negative regulation of neutrophil apoptotic process | 1 | 1 | 7.93e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 7.93e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 4 | 19 | 7.93e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 7 | 16 | 7.93e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 7 | 16 | 7.93e-01 |
| GO:BP | GO:0061357 | positive regulation of Wnt protein secretion | 1 | 4 | 7.93e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 7.93e-01 |
| GO:BP | GO:0016560 | protein import into peroxisome matrix, docking | 2 | 3 | 7.93e-01 |
| GO:BP | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly | 1 | 1 | 7.93e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 3 | 19 | 7.94e-01 |
| GO:BP | GO:1905600 | regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 3 | 7.94e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 7 | 496 | 7.94e-01 |
| GO:BP | GO:0060183 | apelin receptor signaling pathway | 1 | 1 | 7.94e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 11 | 61 | 7.94e-01 |
| GO:BP | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway | 1 | 6 | 7.95e-01 |
| GO:BP | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway | 1 | 2 | 7.95e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 2 | 10 | 7.95e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 2 | 12 | 7.95e-01 |
| GO:BP | GO:0060330 | regulation of response to type II interferon | 6 | 12 | 7.95e-01 |
| GO:BP | GO:0060334 | regulation of type II interferon-mediated signaling pathway | 6 | 12 | 7.95e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 1 | 124 | 7.95e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 7.95e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 1 | 65 | 7.95e-01 |
| GO:BP | GO:0060174 | limb bud formation | 5 | 10 | 7.95e-01 |
| GO:BP | GO:0035021 | negative regulation of Rac protein signal transduction | 2 | 4 | 7.96e-01 |
| GO:BP | GO:0035811 | negative regulation of urine volume | 1 | 2 | 7.96e-01 |
| GO:BP | GO:0070669 | response to interleukin-2 | 3 | 3 | 7.96e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 5 | 22 | 7.96e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 1 | 21 | 7.96e-01 |
| GO:BP | GO:1905050 | positive regulation of metallopeptidase activity | 3 | 5 | 7.96e-01 |
| GO:BP | GO:0045329 | carnitine biosynthetic process | 2 | 3 | 7.96e-01 |
| GO:BP | GO:1990544 | mitochondrial ATP transmembrane transport | 2 | 4 | 7.96e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 7.96e-01 |
| GO:BP | GO:0140021 | mitochondrial ADP transmembrane transport | 2 | 4 | 7.96e-01 |
| GO:BP | GO:0060749 | mammary gland alveolus development | 5 | 13 | 7.96e-01 |
| GO:BP | GO:0061377 | mammary gland lobule development | 5 | 13 | 7.96e-01 |
| GO:BP | GO:0033034 | positive regulation of myeloid cell apoptotic process | 2 | 6 | 7.96e-01 |
| GO:BP | GO:0021997 | neural plate axis specification | 1 | 2 | 7.97e-01 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 1 | 8 | 7.97e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 3 | 35 | 7.97e-01 |
| GO:BP | GO:0046054 | dGMP metabolic process | 2 | 2 | 7.97e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 2 | 78 | 7.97e-01 |
| GO:BP | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline | 3 | 9 | 7.97e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 1 | 13 | 7.97e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 5 | 13 | 7.98e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 8 | 51 | 7.98e-01 |
| GO:BP | GO:0034143 | regulation of toll-like receptor 4 signaling pathway | 3 | 15 | 7.98e-01 |
| GO:BP | GO:0046167 | glycerol-3-phosphate biosynthetic process | 1 | 2 | 7.98e-01 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 2 | 5 | 7.98e-01 |
| GO:BP | GO:0002322 | B cell proliferation involved in immune response | 2 | 3 | 7.98e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 29 | 50 | 7.98e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 1 | 49 | 7.99e-01 |
| GO:BP | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering | 1 | 9 | 7.99e-01 |
| GO:BP | GO:0051693 | actin filament capping | 1 | 34 | 7.99e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 5 | 75 | 7.99e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 3 | 165 | 8.00e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4946 | 8.00e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 7 | 18 | 8.00e-01 |
| GO:BP | GO:0007614 | short-term memory | 5 | 9 | 8.00e-01 |
| GO:BP | GO:0098964 | anterograde dendritic transport of messenger ribonucleoprotein complex | 1 | 2 | 8.00e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 2 | 30 | 8.00e-01 |
| GO:BP | GO:0010912 | positive regulation of isomerase activity | 2 | 5 | 8.00e-01 |
| GO:BP | GO:0010911 | regulation of isomerase activity | 2 | 5 | 8.00e-01 |
| GO:BP | GO:1903489 | positive regulation of lactation | 1 | 1 | 8.00e-01 |
| GO:BP | GO:0034375 | high-density lipoprotein particle remodeling | 2 | 11 | 8.00e-01 |
| GO:BP | GO:0045923 | positive regulation of fatty acid metabolic process | 5 | 26 | 8.00e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 6 | 94 | 8.00e-01 |
| GO:BP | GO:1904958 | positive regulation of midbrain dopaminergic neuron differentiation | 1 | 3 | 8.00e-01 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 1 | 5 | 8.01e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 8.01e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 2 | 9 | 8.01e-01 |
| GO:BP | GO:0060462 | lung lobe development | 1 | 4 | 8.01e-01 |
| GO:BP | GO:0002253 | activation of immune response | 1 | 272 | 8.01e-01 |
| GO:BP | GO:0001781 | neutrophil apoptotic process | 1 | 5 | 8.01e-01 |
| GO:BP | GO:0001510 | RNA methylation | 36 | 87 | 8.01e-01 |
| GO:BP | GO:0060463 | lung lobe morphogenesis | 1 | 4 | 8.01e-01 |
| GO:BP | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity | 3 | 4 | 8.01e-01 |
| GO:BP | GO:0050917 | sensory perception of umami taste | 3 | 3 | 8.01e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 65 | 8.01e-01 |
| GO:BP | GO:0046832 | negative regulation of RNA export from nucleus | 1 | 2 | 8.01e-01 |
| GO:BP | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification | 1 | 9 | 8.01e-01 |
| GO:BP | GO:0061197 | fungiform papilla morphogenesis | 1 | 4 | 8.01e-01 |
| GO:BP | GO:0021871 | forebrain regionalization | 1 | 8 | 8.01e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 8 | 10 | 8.01e-01 |
| GO:BP | GO:0051005 | negative regulation of lipoprotein lipase activity | 3 | 4 | 8.01e-01 |
| GO:BP | GO:0140250 | regulation protein catabolic process at synapse | 2 | 5 | 8.01e-01 |
| GO:BP | GO:1904594 | regulation of termination of RNA polymerase II transcription | 1 | 3 | 8.01e-01 |
| GO:BP | GO:2000804 | regulation of termination of RNA polymerase II transcription, poly(A)-coupled | 1 | 3 | 8.01e-01 |
| GO:BP | GO:0030846 | termination of RNA polymerase II transcription, poly(A)-coupled | 1 | 3 | 8.01e-01 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 5 | 12 | 8.01e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 8.01e-01 |
| GO:BP | GO:0051030 | snRNA transport | 3 | 6 | 8.02e-01 |
| GO:BP | GO:0006668 | sphinganine-1-phosphate metabolic process | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0044793 | negative regulation by host of viral process | 1 | 10 | 8.02e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 3 | 18 | 8.02e-01 |
| GO:BP | GO:0030309 | poly-N-acetyllactosamine metabolic process | 2 | 6 | 8.02e-01 |
| GO:BP | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0061027 | umbilical cord development | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0036304 | umbilical cord morphogenesis | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0106071 | positive regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 3 | 8.02e-01 |
| GO:BP | GO:0072176 | nephric duct development | 2 | 12 | 8.03e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 8.03e-01 |
| GO:BP | GO:0031016 | pancreas development | 2 | 53 | 8.03e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 2 | 78 | 8.03e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 2 | 19 | 8.03e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 1 | 13 | 8.03e-01 |
| GO:BP | GO:0021543 | pallium development | 34 | 139 | 8.03e-01 |
| GO:BP | GO:0009631 | cold acclimation | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0040019 | positive regulation of embryonic development | 3 | 17 | 8.03e-01 |
| GO:BP | GO:0071671 | regulation of smooth muscle cell chemotaxis | 2 | 4 | 8.03e-01 |
| GO:BP | GO:1903292 | protein localization to Golgi membrane | 1 | 4 | 8.03e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 1 | 39 | 8.03e-01 |
| GO:BP | GO:0002252 | immune effector process | 4 | 318 | 8.03e-01 |
| GO:BP | GO:0031930 | mitochondria-nucleus signaling pathway | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0002329 | pre-B cell differentiation | 1 | 4 | 8.04e-01 |
| GO:BP | GO:1903109 | positive regulation of mitochondrial transcription | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 8 | 17 | 8.04e-01 |
| GO:BP | GO:0002536 | respiratory burst involved in inflammatory response | 2 | 4 | 8.04e-01 |
| GO:BP | GO:1905908 | positive regulation of amyloid fibril formation | 2 | 5 | 8.04e-01 |
| GO:BP | GO:0006850 | mitochondrial pyruvate transmembrane transport | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0038065 | collagen-activated signaling pathway | 1 | 13 | 8.04e-01 |
| GO:BP | GO:0072070 | loop of Henle development | 1 | 8 | 8.04e-01 |
| GO:BP | GO:0045779 | negative regulation of bone resorption | 1 | 7 | 8.04e-01 |
| GO:BP | GO:0051639 | actin filament network formation | 1 | 7 | 8.04e-01 |
| GO:BP | GO:1905203 | regulation of connective tissue replacement | 3 | 5 | 8.04e-01 |
| GO:BP | GO:0000960 | regulation of mitochondrial RNA catabolic process | 2 | 5 | 8.04e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 6 | 687 | 8.04e-01 |
| GO:BP | GO:0003084 | positive regulation of systemic arterial blood pressure | 1 | 7 | 8.04e-01 |
| GO:BP | GO:0042426 | choline catabolic process | 1 | 3 | 8.04e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 7 | 14 | 8.04e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 6 | 11 | 8.05e-01 |
| GO:BP | GO:0036476 | neuron death in response to hydrogen peroxide | 3 | 5 | 8.05e-01 |
| GO:BP | GO:0032972 | regulation of muscle filament sliding speed | 1 | 2 | 8.05e-01 |
| GO:BP | GO:1903207 | regulation of hydrogen peroxide-induced neuron death | 3 | 5 | 8.05e-01 |
| GO:BP | GO:0036301 | macrophage colony-stimulating factor production | 1 | 2 | 8.05e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 6 | 11 | 8.05e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 6 | 11 | 8.05e-01 |
| GO:BP | GO:1903208 | negative regulation of hydrogen peroxide-induced neuron death | 3 | 5 | 8.05e-01 |
| GO:BP | GO:1901033 | positive regulation of response to reactive oxygen species | 1 | 6 | 8.05e-01 |
| GO:BP | GO:1901256 | regulation of macrophage colony-stimulating factor production | 1 | 2 | 8.05e-01 |
| GO:BP | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death | 1 | 6 | 8.05e-01 |
| GO:BP | GO:0007603 | phototransduction, visible light | 4 | 9 | 8.05e-01 |
| GO:BP | GO:0051598 | meiotic recombination checkpoint signaling | 1 | 2 | 8.05e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 1 | 40 | 8.06e-01 |
| GO:BP | GO:0007340 | acrosome reaction | 1 | 20 | 8.06e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 8.06e-01 |
| GO:BP | GO:0002478 | antigen processing and presentation of exogenous peptide antigen | 2 | 24 | 8.06e-01 |
| GO:BP | GO:0021884 | forebrain neuron development | 10 | 16 | 8.07e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 7 | 31 | 8.07e-01 |
| GO:BP | GO:0042357 | thiamine diphosphate metabolic process | 1 | 2 | 8.07e-01 |
| GO:BP | GO:0010985 | negative regulation of lipoprotein particle clearance | 2 | 6 | 8.07e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 4 | 12 | 8.07e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 8.07e-01 |
| GO:BP | GO:0098795 | global gene silencing by mRNA cleavage | 1 | 4 | 8.08e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 48 | 300 | 8.08e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 8.08e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 4 | 23 | 8.08e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 8.08e-01 |
| GO:BP | GO:0050992 | dimethylallyl diphosphate biosynthetic process | 1 | 2 | 8.09e-01 |
| GO:BP | GO:2001112 | regulation of cellular response to hepatocyte growth factor stimulus | 1 | 1 | 8.09e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 6 | 75 | 8.09e-01 |
| GO:BP | GO:0060667 | branch elongation involved in salivary gland morphogenesis | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 6 | 95 | 8.09e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 5 | 20 | 8.09e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 8.09e-01 |
| GO:BP | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development | 1 | 1 | 8.09e-01 |
| GO:BP | GO:0060595 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0060915 | mesenchymal cell differentiation involved in lung development | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0050993 | dimethylallyl diphosphate metabolic process | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0060615 | mammary gland bud formation | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0021847 | ventricular zone neuroblast division | 1 | 1 | 8.09e-01 |
| GO:BP | GO:0110077 | vesicle-mediated intercellular transport | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 1 | 4 | 8.09e-01 |
| GO:BP | GO:0031247 | actin rod assembly | 1 | 2 | 8.09e-01 |
| GO:BP | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 5 | 58 | 8.10e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 1 | 20 | 8.10e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 8.10e-01 |
| GO:BP | GO:0036066 | protein O-linked fucosylation | 2 | 5 | 8.10e-01 |
| GO:BP | GO:0051659 | maintenance of mitochondrion location | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 37 | 74 | 8.10e-01 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 6 | 63 | 8.10e-01 |
| GO:BP | GO:0031448 | positive regulation of fast-twitch skeletal muscle fiber contraction | 1 | 2 | 8.10e-01 |
| GO:BP | GO:2000864 | regulation of estradiol secretion | 1 | 2 | 8.10e-01 |
| GO:BP | GO:1902082 | positive regulation of calcium ion import into sarcoplasmic reticulum | 1 | 1 | 8.10e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 13 | 40 | 8.10e-01 |
| GO:BP | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0035938 | estradiol secretion | 1 | 2 | 8.10e-01 |
| GO:BP | GO:1903116 | positive regulation of actin filament-based movement | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0090084 | negative regulation of inclusion body assembly | 1 | 10 | 8.10e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 23 | 539 | 8.10e-01 |
| GO:BP | GO:1990822 | basic amino acid transmembrane transport | 2 | 14 | 8.10e-01 |
| GO:BP | GO:0060923 | cardiac muscle cell fate commitment | 3 | 7 | 8.10e-01 |
| GO:BP | GO:2001138 | regulation of phospholipid transport | 1 | 12 | 8.10e-01 |
| GO:BP | GO:1903727 | positive regulation of phospholipid metabolic process | 1 | 9 | 8.10e-01 |
| GO:BP | GO:0032594 | protein transport within lipid bilayer | 2 | 6 | 8.10e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 2 | 10 | 8.11e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 4 | 28 | 8.11e-01 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 4 | 45 | 8.11e-01 |
| GO:BP | GO:0048243 | norepinephrine secretion | 2 | 5 | 8.11e-01 |
| GO:BP | GO:0014061 | regulation of norepinephrine secretion | 2 | 5 | 8.11e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 3 | 6 | 8.11e-01 |
| GO:BP | GO:1903597 | negative regulation of gap junction assembly | 1 | 1 | 8.11e-01 |
| GO:BP | GO:0060177 | regulation of angiotensin metabolic process | 1 | 1 | 8.11e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 21 | 84 | 8.11e-01 |
| GO:BP | GO:0060336 | negative regulation of type II interferon-mediated signaling pathway | 3 | 6 | 8.11e-01 |
| GO:BP | GO:0002246 | wound healing involved in inflammatory response | 3 | 6 | 8.11e-01 |
| GO:BP | GO:0060331 | negative regulation of response to type II interferon | 3 | 6 | 8.11e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 1 | 296 | 8.11e-01 |
| GO:BP | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization | 1 | 4 | 8.12e-01 |
| GO:BP | GO:0097242 | amyloid-beta clearance | 1 | 27 | 8.12e-01 |
| GO:BP | GO:0002387 | immune response in gut-associated lymphoid tissue | 1 | 1 | 8.12e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 2 | 142 | 8.12e-01 |
| GO:BP | GO:1905867 | epididymis development | 2 | 2 | 8.12e-01 |
| GO:BP | GO:0150094 | amyloid-beta clearance by cellular catabolic process | 1 | 7 | 8.12e-01 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 3 | 19 | 8.12e-01 |
| GO:BP | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway | 2 | 4 | 8.12e-01 |
| GO:BP | GO:0001709 | cell fate determination | 5 | 25 | 8.12e-01 |
| GO:BP | GO:0002395 | immune response in nasopharyngeal-associated lymphoid tissue | 1 | 1 | 8.12e-01 |
| GO:BP | GO:0010887 | negative regulation of cholesterol storage | 4 | 9 | 8.12e-01 |
| GO:BP | GO:0019835 | cytolysis | 1 | 6 | 8.12e-01 |
| GO:BP | GO:0021570 | rhombomere 4 development | 1 | 1 | 8.12e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 2 | 7 | 8.12e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 3 | 77 | 8.12e-01 |
| GO:BP | GO:1990402 | embryonic liver development | 1 | 3 | 8.12e-01 |
| GO:BP | GO:0006414 | translational elongation | 8 | 65 | 8.12e-01 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 2 | 4 | 8.12e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 1 | 111 | 8.12e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 7 | 26 | 8.12e-01 |
| GO:BP | GO:0014732 | skeletal muscle atrophy | 3 | 5 | 8.13e-01 |
| GO:BP | GO:0015871 | choline transport | 3 | 6 | 8.13e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 3 | 27 | 8.13e-01 |
| GO:BP | GO:0140352 | export from cell | 14 | 611 | 8.13e-01 |
| GO:BP | GO:0032291 | axon ensheathment in central nervous system | 1 | 12 | 8.13e-01 |
| GO:BP | GO:0022010 | central nervous system myelination | 1 | 12 | 8.13e-01 |
| GO:BP | GO:0050882 | voluntary musculoskeletal movement | 2 | 7 | 8.13e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 1 | 22 | 8.13e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 1 | 1315 | 8.13e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1315 | 8.13e-01 |
| GO:BP | GO:0098814 | spontaneous synaptic transmission | 3 | 10 | 8.13e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 2 | 20 | 8.13e-01 |
| GO:BP | GO:0071988 | protein localization to spindle pole body | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0032761 | positive regulation of lymphotoxin A production | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 1 | 12 | 8.14e-01 |
| GO:BP | GO:0070781 | response to biotin | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0032681 | regulation of lymphotoxin A production | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 7 | 13 | 8.14e-01 |
| GO:BP | GO:0032641 | lymphotoxin A production | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0021628 | olfactory nerve formation | 1 | 2 | 8.14e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 3 | 7 | 8.14e-01 |
| GO:BP | GO:0032914 | positive regulation of transforming growth factor beta1 production | 1 | 6 | 8.14e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1 | 1322 | 8.14e-01 |
| GO:BP | GO:0048318 | axial mesoderm development | 1 | 6 | 8.14e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 2 | 60 | 8.14e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 1 | 100 | 8.14e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 2 | 4 | 8.14e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 2 | 60 | 8.14e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 4 | 11 | 8.14e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 2 | 20 | 8.14e-01 |
| GO:BP | GO:0042886 | amide transport | 1 | 235 | 8.14e-01 |
| GO:BP | GO:1902224 | ketone body metabolic process | 2 | 7 | 8.14e-01 |
| GO:BP | GO:0046016 | positive regulation of transcription by glucose | 2 | 3 | 8.14e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 5 | 23 | 8.14e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 1 | 18 | 8.14e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 1 | 20 | 8.14e-01 |
| GO:BP | GO:0006415 | translational termination | 5 | 17 | 8.14e-01 |
| GO:BP | GO:0090094 | metanephric cap mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0072208 | metanephric smooth muscle tissue development | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0048686 | regulation of sprouting of injured axon | 1 | 1 | 8.14e-01 |
| GO:BP | GO:1902908 | regulation of melanosome transport | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 4 | 8 | 8.14e-01 |
| GO:BP | GO:0071358 | cellular response to type III interferon | 1 | 4 | 8.14e-01 |
| GO:BP | GO:0002475 | antigen processing and presentation via MHC class Ib | 5 | 8 | 8.14e-01 |
| GO:BP | GO:0072185 | metanephric cap development | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0006605 | protein targeting | 8 | 307 | 8.14e-01 |
| GO:BP | GO:0072186 | metanephric cap morphogenesis | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0048688 | negative regulation of sprouting of injured axon | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0048690 | regulation of axon extension involved in regeneration | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0048692 | negative regulation of axon extension involved in regeneration | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 2 | 11 | 8.14e-01 |
| GO:BP | GO:0032771 | regulation of tyrosinase activity | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0038196 | type III interferon-mediated signaling pathway | 1 | 4 | 8.14e-01 |
| GO:BP | GO:0032773 | positive regulation of tyrosinase activity | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0048817 | negative regulation of hair follicle maturation | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0072166 | posterior mesonephric tubule development | 1 | 1 | 8.14e-01 |
| GO:BP | GO:0050658 | RNA transport | 43 | 144 | 8.14e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 43 | 144 | 8.14e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 8.14e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 8.14e-01 |
| GO:BP | GO:0070315 | G1 to G0 transition involved in cell differentiation | 2 | 4 | 8.14e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 8 | 65 | 8.14e-01 |
| GO:BP | GO:2000870 | regulation of progesterone secretion | 1 | 1 | 8.14e-01 |
| GO:BP | GO:1903803 | L-glutamine import across plasma membrane | 2 | 4 | 8.14e-01 |
| GO:BP | GO:0010669 | epithelial structure maintenance | 1 | 15 | 8.14e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 2 | 22 | 8.14e-01 |
| GO:BP | GO:0060478 | acrosomal vesicle exocytosis | 1 | 5 | 8.14e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 2 | 22 | 8.14e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 8.14e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 11 | 26 | 8.14e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 2 | 250 | 8.14e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 1 | 92 | 8.14e-01 |
| GO:BP | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly | 1 | 5 | 8.14e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 1 | 92 | 8.14e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 8.14e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 1 | 92 | 8.14e-01 |
| GO:BP | GO:0022400 | regulation of rhodopsin mediated signaling pathway | 2 | 5 | 8.14e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 4 | 68 | 8.14e-01 |
| GO:BP | GO:0006529 | asparagine biosynthetic process | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II | 1 | 17 | 8.14e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 17 | 25 | 8.14e-01 |
| GO:BP | GO:0090230 | regulation of centromere complex assembly | 1 | 4 | 8.14e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 8.14e-01 |
| GO:BP | GO:0034159 | regulation of toll-like receptor 8 signaling pathway | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0034161 | positive regulation of toll-like receptor 8 signaling pathway | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 4 | 3730 | 8.14e-01 |
| GO:BP | GO:0001711 | endodermal cell fate commitment | 4 | 9 | 8.15e-01 |
| GO:BP | GO:1903625 | negative regulation of DNA catabolic process | 1 | 3 | 8.15e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 7 | 19 | 8.15e-01 |
| GO:BP | GO:0034139 | regulation of toll-like receptor 3 signaling pathway | 4 | 8 | 8.15e-01 |
| GO:BP | GO:2001295 | malonyl-CoA biosynthetic process | 1 | 2 | 8.15e-01 |
| GO:BP | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration | 2 | 4 | 8.15e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 4 | 113 | 8.15e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 5 | 10 | 8.15e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 5 | 10 | 8.15e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 1 | 67 | 8.16e-01 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 1 | 9 | 8.16e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 11 | 31 | 8.16e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 14 | 50 | 8.16e-01 |
| GO:BP | GO:1900425 | negative regulation of defense response to bacterium | 1 | 4 | 8.16e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 8.16e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 7 | 11 | 8.17e-01 |
| GO:BP | GO:0072338 | lactam metabolic process | 1 | 2 | 8.17e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 10 | 28 | 8.17e-01 |
| GO:BP | GO:0061792 | secretory granule maturation | 1 | 3 | 8.17e-01 |
| GO:BP | GO:1905413 | regulation of dense core granule exocytosis | 1 | 2 | 8.17e-01 |
| GO:BP | GO:1990502 | dense core granule maturation | 1 | 3 | 8.17e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 8.17e-01 |
| GO:BP | GO:2000251 | positive regulation of actin cytoskeleton reorganization | 4 | 14 | 8.17e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 8.17e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 3 | 77 | 8.17e-01 |
| GO:BP | GO:0006909 | phagocytosis | 3 | 148 | 8.17e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 2 | 31 | 8.18e-01 |
| GO:BP | GO:0061010 | gallbladder development | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 1 | 5 | 8.18e-01 |
| GO:BP | GO:0070126 | mitochondrial translational termination | 1 | 4 | 8.18e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 1 | 28 | 8.18e-01 |
| GO:BP | GO:0021564 | vagus nerve development | 1 | 2 | 8.18e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 8.18e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 1 | 39 | 8.18e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 8.18e-01 |
| GO:BP | GO:0070358 | actin polymerization-dependent cell motility | 1 | 6 | 8.18e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 1 | 39 | 8.18e-01 |
| GO:BP | GO:1903630 | regulation of aminoacyl-tRNA ligase activity | 1 | 2 | 8.18e-01 |
| GO:BP | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | 5 | 11 | 8.18e-01 |
| GO:BP | GO:0032929 | negative regulation of superoxide anion generation | 1 | 1 | 8.18e-01 |
| GO:BP | GO:1902362 | melanocyte apoptotic process | 1 | 1 | 8.19e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 3 | 11 | 8.19e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 3 | 11 | 8.19e-01 |
| GO:BP | GO:0072095 | regulation of branch elongation involved in ureteric bud branching | 2 | 2 | 8.19e-01 |
| GO:BP | GO:0061325 | cell proliferation involved in outflow tract morphogenesis | 2 | 4 | 8.19e-01 |
| GO:BP | GO:0090290 | positive regulation of osteoclast proliferation | 1 | 1 | 8.19e-01 |
| GO:BP | GO:1903942 | positive regulation of respiratory gaseous exchange | 1 | 1 | 8.19e-01 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 8.19e-01 |
| GO:BP | GO:0120229 | protein localization to motile cilium | 2 | 4 | 8.19e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 3 | 44 | 8.19e-01 |
| GO:BP | GO:0061113 | pancreas morphogenesis | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0006114 | glycerol biosynthetic process | 2 | 3 | 8.19e-01 |
| GO:BP | GO:0019401 | alditol biosynthetic process | 2 | 3 | 8.19e-01 |
| GO:BP | GO:0048137 | spermatocyte division | 1 | 1 | 8.19e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 1 | 48 | 8.19e-01 |
| GO:BP | GO:0043415 | positive regulation of skeletal muscle tissue regeneration | 2 | 4 | 8.20e-01 |
| GO:BP | GO:0046532 | regulation of photoreceptor cell differentiation | 3 | 4 | 8.20e-01 |
| GO:BP | GO:2000491 | positive regulation of hepatic stellate cell activation | 1 | 4 | 8.20e-01 |
| GO:BP | GO:2000831 | regulation of steroid hormone secretion | 1 | 13 | 8.20e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 5 | 8 | 8.20e-01 |
| GO:BP | GO:0046541 | saliva secretion | 1 | 4 | 8.20e-01 |
| GO:BP | GO:0061975 | articular cartilage development | 1 | 3 | 8.20e-01 |
| GO:BP | GO:2000257 | regulation of protein activation cascade | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0001698 | gastrin-induced gastric acid secretion | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0045844 | positive regulation of striated muscle tissue development | 1 | 3 | 8.20e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 4 | 8 | 8.20e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 2 | 27 | 8.20e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 8.20e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 2 | 4 | 8.20e-01 |
| GO:BP | GO:0048636 | positive regulation of muscle organ development | 1 | 3 | 8.20e-01 |
| GO:BP | GO:0008295 | spermidine biosynthetic process | 1 | 3 | 8.20e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 1 | 14 | 8.20e-01 |
| GO:BP | GO:2001256 | regulation of store-operated calcium entry | 1 | 11 | 8.20e-01 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 40 | 8.21e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 1 | 105 | 8.21e-01 |
| GO:BP | GO:0043335 | protein unfolding | 1 | 5 | 8.21e-01 |
| GO:BP | GO:0060375 | regulation of mast cell differentiation | 1 | 2 | 8.21e-01 |
| GO:BP | GO:0070995 | NADPH oxidation | 2 | 3 | 8.21e-01 |
| GO:BP | GO:2001074 | regulation of metanephric ureteric bud development | 1 | 1 | 8.21e-01 |
| GO:BP | GO:2001076 | positive regulation of metanephric ureteric bud development | 1 | 1 | 8.21e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 2 | 62 | 8.21e-01 |
| GO:BP | GO:0071867 | response to monoamine | 2 | 62 | 8.21e-01 |
| GO:BP | GO:0060484 | lung-associated mesenchyme development | 1 | 5 | 8.21e-01 |
| GO:BP | GO:0061032 | visceral serous pericardium development | 1 | 1 | 8.21e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 8.21e-01 |
| GO:BP | GO:0060347 | heart trabecula formation | 6 | 13 | 8.22e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 4 | 12 | 8.22e-01 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 1 | 5 | 8.22e-01 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 1 | 5 | 8.22e-01 |
| GO:BP | GO:0055005 | ventricular cardiac myofibril assembly | 1 | 3 | 8.22e-01 |
| GO:BP | GO:0018026 | peptidyl-lysine monomethylation | 4 | 11 | 8.22e-01 |
| GO:BP | GO:1904245 | regulation of polynucleotide adenylyltransferase activity | 1 | 2 | 8.22e-01 |
| GO:BP | GO:2000347 | positive regulation of hepatocyte proliferation | 5 | 9 | 8.22e-01 |
| GO:BP | GO:0035799 | ureter maturation | 1 | 1 | 8.22e-01 |
| GO:BP | GO:2000987 | positive regulation of behavioral fear response | 1 | 3 | 8.22e-01 |
| GO:BP | GO:0045113 | regulation of integrin biosynthetic process | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0060455 | negative regulation of gastric acid secretion | 1 | 3 | 8.22e-01 |
| GO:BP | GO:1900194 | negative regulation of oocyte maturation | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0003383 | apical constriction | 2 | 4 | 8.22e-01 |
| GO:BP | GO:0006677 | glycosylceramide metabolic process | 1 | 13 | 8.22e-01 |
| GO:BP | GO:1905123 | regulation of glucosylceramidase activity | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 3 | 84 | 8.22e-01 |
| GO:BP | GO:0032264 | IMP salvage | 2 | 4 | 8.23e-01 |
| GO:BP | GO:0009073 | aromatic amino acid family biosynthetic process | 2 | 3 | 8.23e-01 |
| GO:BP | GO:0007165 | signal transduction | 5 | 3818 | 8.23e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 8.23e-01 |
| GO:BP | GO:0006826 | iron ion transport | 6 | 40 | 8.23e-01 |
| GO:BP | GO:0006478 | peptidyl-tyrosine sulfation | 1 | 2 | 8.23e-01 |
| GO:BP | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane | 1 | 19 | 8.23e-01 |
| GO:BP | GO:0042661 | regulation of mesodermal cell fate specification | 1 | 4 | 8.23e-01 |
| GO:BP | GO:0045292 | mRNA cis splicing, via spliceosome | 1 | 26 | 8.23e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 5 | 12 | 8.23e-01 |
| GO:BP | GO:0021590 | cerebellum maturation | 1 | 3 | 8.24e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 4 | 23 | 8.24e-01 |
| GO:BP | GO:0021699 | cerebellar cortex maturation | 1 | 3 | 8.24e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 17 | 404 | 8.24e-01 |
| GO:BP | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis | 2 | 4 | 8.24e-01 |
| GO:BP | GO:0075509 | endocytosis involved in viral entry into host cell | 4 | 11 | 8.24e-01 |
| GO:BP | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway | 2 | 5 | 8.24e-01 |
| GO:BP | GO:0061888 | regulation of astrocyte activation | 2 | 3 | 8.25e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 10 | 56 | 8.25e-01 |
| GO:BP | GO:0051791 | medium-chain fatty acid metabolic process | 2 | 9 | 8.25e-01 |
| GO:BP | GO:0070171 | negative regulation of tooth mineralization | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0033007 | negative regulation of mast cell activation involved in immune response | 4 | 4 | 8.25e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 27 | 66 | 8.25e-01 |
| GO:BP | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | 2 | 4 | 8.25e-01 |
| GO:BP | GO:0001655 | urogenital system development | 3 | 49 | 8.25e-01 |
| GO:BP | GO:0051167 | xylulose 5-phosphate metabolic process | 2 | 4 | 8.25e-01 |
| GO:BP | GO:1901159 | xylulose 5-phosphate biosynthetic process | 2 | 4 | 8.25e-01 |
| GO:BP | GO:0006064 | glucuronate catabolic process | 2 | 4 | 8.25e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 4 | 53 | 8.25e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 6 | 92 | 8.25e-01 |
| GO:BP | GO:0051179 | localization | 2 | 3969 | 8.25e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 5 | 51 | 8.25e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 3 | 3080 | 8.25e-01 |
| GO:BP | GO:0060428 | lung epithelium development | 6 | 30 | 8.25e-01 |
| GO:BP | GO:1903226 | positive regulation of endodermal cell differentiation | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0032940 | secretion by cell | 12 | 569 | 8.25e-01 |
| GO:BP | GO:0071071 | regulation of phospholipid biosynthetic process | 8 | 15 | 8.25e-01 |
| GO:BP | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production | 2 | 3 | 8.25e-01 |
| GO:BP | GO:0033083 | regulation of immature T cell proliferation | 1 | 6 | 8.26e-01 |
| GO:BP | GO:0050747 | positive regulation of lipoprotein metabolic process | 1 | 4 | 8.26e-01 |
| GO:BP | GO:0020027 | hemoglobin metabolic process | 6 | 16 | 8.26e-01 |
| GO:BP | GO:1903061 | positive regulation of protein lipidation | 1 | 4 | 8.26e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 6 | 10 | 8.26e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 2 | 104 | 8.26e-01 |
| GO:BP | GO:0033084 | regulation of immature T cell proliferation in thymus | 1 | 6 | 8.26e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 8.26e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 1 | 11 | 8.26e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 2 | 11 | 8.26e-01 |
| GO:BP | GO:0070973 | protein localization to endoplasmic reticulum exit site | 1 | 6 | 8.26e-01 |
| GO:BP | GO:0038165 | oncostatin-M-mediated signaling pathway | 1 | 4 | 8.26e-01 |
| GO:BP | GO:0043132 | NAD transport | 1 | 3 | 8.26e-01 |
| GO:BP | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate | 1 | 4 | 8.26e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 2 | 109 | 8.26e-01 |
| GO:BP | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway | 2 | 3 | 8.26e-01 |
| GO:BP | GO:1903282 | regulation of glutathione peroxidase activity | 1 | 1 | 8.26e-01 |
| GO:BP | GO:2000470 | positive regulation of peroxidase activity | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 6 | 132 | 8.26e-01 |
| GO:BP | GO:0070391 | response to lipoteichoic acid | 3 | 6 | 8.26e-01 |
| GO:BP | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 2 | 8.26e-01 |
| GO:BP | GO:1905447 | negative regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0071223 | cellular response to lipoteichoic acid | 3 | 6 | 8.26e-01 |
| GO:BP | GO:0051621 | regulation of norepinephrine uptake | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0007254 | JNK cascade | 4 | 138 | 8.26e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 8 | 96 | 8.26e-01 |
| GO:BP | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 8.26e-01 |
| GO:BP | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 8.26e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 2 | 24 | 8.27e-01 |
| GO:BP | GO:0060742 | epithelial cell differentiation involved in prostate gland development | 2 | 8 | 8.27e-01 |
| GO:BP | GO:0030534 | adult behavior | 4 | 92 | 8.27e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 8.27e-01 |
| GO:BP | GO:1905749 | regulation of endosome to plasma membrane protein transport | 2 | 5 | 8.27e-01 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 4 | 14 | 8.27e-01 |
| GO:BP | GO:0010706 | ganglioside biosynthetic process via lactosylceramide | 1 | 5 | 8.27e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 4 | 9 | 8.27e-01 |
| GO:BP | GO:2000437 | regulation of monocyte extravasation | 2 | 4 | 8.27e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 8.28e-01 |
| GO:BP | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine | 1 | 3 | 8.28e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 6 | 155 | 8.28e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 3 | 79 | 8.28e-01 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 6 | 27 | 8.28e-01 |
| GO:BP | GO:0032241 | positive regulation of nucleobase-containing compound transport | 2 | 6 | 8.28e-01 |
| GO:BP | GO:1900363 | regulation of mRNA polyadenylation | 4 | 11 | 8.28e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 59 | 149 | 8.29e-01 |
| GO:BP | GO:0031133 | regulation of axon diameter | 1 | 4 | 8.29e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 8.29e-01 |
| GO:BP | GO:0031581 | hemidesmosome assembly | 2 | 6 | 8.29e-01 |
| GO:BP | GO:0071379 | cellular response to prostaglandin stimulus | 1 | 16 | 8.29e-01 |
| GO:BP | GO:0070902 | mitochondrial tRNA pseudouridine synthesis | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1 | 1441 | 8.29e-01 |
| GO:BP | GO:0019676 | ammonia assimilation cycle | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0042450 | arginine biosynthetic process via ornithine | 1 | 1 | 8.29e-01 |
| GO:BP | GO:1901535 | regulation of DNA demethylation | 1 | 8 | 8.29e-01 |
| GO:BP | GO:0042667 | auditory receptor cell fate specification | 1 | 1 | 8.29e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 19 | 395 | 8.29e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 3 | 20 | 8.29e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 7 | 247 | 8.29e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 3 | 18 | 8.29e-01 |
| GO:BP | GO:0048002 | antigen processing and presentation of peptide antigen | 3 | 41 | 8.29e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 4 | 25 | 8.30e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 2 | 87 | 8.30e-01 |
| GO:BP | GO:0097021 | lymphocyte migration into lymphoid organs | 2 | 6 | 8.30e-01 |
| GO:BP | GO:0048675 | axon extension | 26 | 110 | 8.31e-01 |
| GO:BP | GO:0070475 | rRNA base methylation | 1 | 8 | 8.31e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 4 | 112 | 8.31e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 8.31e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 3 | 173 | 8.31e-01 |
| GO:BP | GO:0060736 | prostate gland growth | 2 | 7 | 8.31e-01 |
| GO:BP | GO:0032472 | Golgi calcium ion transport | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0032468 | Golgi calcium ion homeostasis | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0098629 | trans-Golgi network membrane organization | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0048069 | eye pigmentation | 1 | 4 | 8.31e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 23 | 78 | 8.31e-01 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 1 | 9 | 8.31e-01 |
| GO:BP | GO:0061753 | substrate localization to autophagosome | 1 | 3 | 8.31e-01 |
| GO:BP | GO:0042350 | GDP-L-fucose biosynthetic process | 1 | 3 | 8.32e-01 |
| GO:BP | GO:1904466 | positive regulation of matrix metallopeptidase secretion | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0016046 | detection of fungus | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0070430 | positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 6 | 83 | 8.32e-01 |
| GO:BP | GO:0002841 | negative regulation of T cell mediated immune response to tumor cell | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0030073 | insulin secretion | 6 | 141 | 8.32e-01 |
| GO:BP | GO:0099500 | vesicle fusion to plasma membrane | 1 | 19 | 8.32e-01 |
| GO:BP | GO:0000962 | positive regulation of mitochondrial RNA catabolic process | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0048635 | negative regulation of muscle organ development | 1 | 6 | 8.33e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 1 | 32 | 8.33e-01 |
| GO:BP | GO:0010360 | negative regulation of anion channel activity | 1 | 2 | 8.34e-01 |
| GO:BP | GO:1903960 | negative regulation of anion transmembrane transport | 1 | 2 | 8.34e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 13 | 8.34e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 2 | 30 | 8.34e-01 |
| GO:BP | GO:0090131 | mesenchyme migration | 1 | 4 | 8.34e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 4 | 118 | 8.34e-01 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 2 | 5 | 8.35e-01 |
| GO:BP | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress | 2 | 5 | 8.35e-01 |
| GO:BP | GO:0051311 | meiotic metaphase plate congression | 1 | 2 | 8.35e-01 |
| GO:BP | GO:1990868 | response to chemokine | 3 | 37 | 8.35e-01 |
| GO:BP | GO:0044062 | regulation of excretion | 1 | 12 | 8.35e-01 |
| GO:BP | GO:0060872 | semicircular canal development | 2 | 7 | 8.35e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 2 | 15 | 8.35e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 2 | 5 | 8.35e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 3 | 37 | 8.35e-01 |
| GO:BP | GO:0003284 | septum primum development | 2 | 5 | 8.35e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 1 | 4 | 8.35e-01 |
| GO:BP | GO:0042363 | fat-soluble vitamin catabolic process | 1 | 1 | 8.35e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 6 | 13 | 8.35e-01 |
| GO:BP | GO:1902871 | positive regulation of amacrine cell differentiation | 1 | 1 | 8.35e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 27 | 139 | 8.35e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 6 | 134 | 8.36e-01 |
| GO:BP | GO:0051238 | sequestering of metal ion | 7 | 17 | 8.36e-01 |
| GO:BP | GO:0014905 | myoblast fusion involved in skeletal muscle regeneration | 2 | 3 | 8.36e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 9 | 22 | 8.37e-01 |
| GO:BP | GO:0043380 | regulation of memory T cell differentiation | 2 | 4 | 8.37e-01 |
| GO:BP | GO:0035766 | cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.37e-01 |
| GO:BP | GO:0035768 | endothelial cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.37e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 50 | 192 | 8.37e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 5 | 222 | 8.37e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 8.37e-01 |
| GO:BP | GO:2000544 | regulation of endothelial cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.37e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 24 | 44 | 8.37e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 8.37e-01 |
| GO:BP | GO:1904847 | regulation of cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.37e-01 |
| GO:BP | GO:0031508 | pericentric heterochromatin formation | 1 | 3 | 8.37e-01 |
| GO:BP | GO:0051956 | negative regulation of amino acid transport | 2 | 10 | 8.37e-01 |
| GO:BP | GO:0045124 | regulation of bone resorption | 5 | 24 | 8.37e-01 |
| GO:BP | GO:0015876 | acetyl-CoA transport | 1 | 1 | 8.37e-01 |
| GO:BP | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 5 | 8.37e-01 |
| GO:BP | GO:1905323 | telomerase holoenzyme complex assembly | 2 | 6 | 8.37e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 1 | 30 | 8.37e-01 |
| GO:BP | GO:1990569 | UDP-N-acetylglucosamine transmembrane transport | 1 | 6 | 8.37e-01 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 2 | 20 | 8.37e-01 |
| GO:BP | GO:0043097 | pyrimidine nucleoside salvage | 1 | 2 | 8.37e-01 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 2 | 11 | 8.37e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 1 | 126 | 8.37e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 11 | 53 | 8.38e-01 |
| GO:BP | GO:0071801 | regulation of podosome assembly | 2 | 8 | 8.38e-01 |
| GO:BP | GO:0060399 | positive regulation of growth hormone receptor signaling pathway | 1 | 2 | 8.38e-01 |
| GO:BP | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase | 1 | 2 | 8.39e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 8.39e-01 |
| GO:BP | GO:0048934 | peripheral nervous system neuron differentiation | 1 | 9 | 8.39e-01 |
| GO:BP | GO:0006704 | glucocorticoid biosynthetic process | 2 | 11 | 8.39e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 3 | 10 | 8.39e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 3 | 10 | 8.39e-01 |
| GO:BP | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | 1 | 5 | 8.39e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 8.39e-01 |
| GO:BP | GO:0048935 | peripheral nervous system neuron development | 1 | 9 | 8.39e-01 |
| GO:BP | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 1 | 7 | 8.39e-01 |
| GO:BP | GO:1904844 | response to L-glutamine | 1 | 3 | 8.39e-01 |
| GO:BP | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway | 1 | 2 | 8.39e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 2 | 8 | 8.40e-01 |
| GO:BP | GO:0043373 | CD4-positive, alpha-beta T cell lineage commitment | 4 | 9 | 8.40e-01 |
| GO:BP | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | 2 | 5 | 8.40e-01 |
| GO:BP | GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 1 | 4 | 8.40e-01 |
| GO:BP | GO:0034372 | very-low-density lipoprotein particle remodeling | 1 | 4 | 8.40e-01 |
| GO:BP | GO:1904751 | positive regulation of protein localization to nucleolus | 2 | 3 | 8.40e-01 |
| GO:BP | GO:0006734 | NADH metabolic process | 2 | 12 | 8.40e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 3 | 18 | 8.40e-01 |
| GO:BP | GO:2000405 | negative regulation of T cell migration | 1 | 4 | 8.40e-01 |
| GO:BP | GO:0061196 | fungiform papilla development | 1 | 5 | 8.40e-01 |
| GO:BP | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway | 2 | 7 | 8.40e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 16 | 72 | 8.40e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 8.40e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1 | 1507 | 8.41e-01 |
| GO:BP | GO:0033622 | integrin activation | 4 | 18 | 8.41e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 4 | 25 | 8.41e-01 |
| GO:BP | GO:1904396 | regulation of neuromuscular junction development | 3 | 5 | 8.41e-01 |
| GO:BP | GO:0001915 | negative regulation of T cell mediated cytotoxicity | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0051124 | synaptic assembly at neuromuscular junction | 3 | 6 | 8.41e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 4 | 108 | 8.41e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 6 | 136 | 8.41e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 6 | 42 | 8.41e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 8.41e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 4 | 123 | 8.41e-01 |
| GO:BP | GO:0021550 | medulla oblongata development | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0030322 | stabilization of membrane potential | 1 | 6 | 8.41e-01 |
| GO:BP | GO:0015732 | prostaglandin transport | 1 | 10 | 8.41e-01 |
| GO:BP | GO:0016598 | protein arginylation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 2 | 31 | 8.41e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 60 | 151 | 8.41e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 8.41e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 10 | 305 | 8.41e-01 |
| GO:BP | GO:2001111 | positive regulation of lens epithelial cell proliferation | 1 | 1 | 8.41e-01 |
| GO:BP | GO:0097166 | lens epithelial cell proliferation | 1 | 1 | 8.41e-01 |
| GO:BP | GO:2001109 | regulation of lens epithelial cell proliferation | 1 | 1 | 8.41e-01 |
| GO:BP | GO:0019062 | virion attachment to host cell | 1 | 9 | 8.41e-01 |
| GO:BP | GO:0021524 | visceral motor neuron differentiation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0048293 | regulation of isotype switching to IgE isotypes | 1 | 5 | 8.41e-01 |
| GO:BP | GO:0048289 | isotype switching to IgE isotypes | 1 | 5 | 8.41e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 4 | 512 | 8.41e-01 |
| GO:BP | GO:0006014 | D-ribose metabolic process | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 8.41e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 2 | 22 | 8.41e-01 |
| GO:BP | GO:0061889 | negative regulation of astrocyte activation | 1 | 2 | 8.42e-01 |
| GO:BP | GO:0071109 | superior temporal gyrus development | 1 | 2 | 8.42e-01 |
| GO:BP | GO:0071461 | cellular response to redox state | 1 | 1 | 8.42e-01 |
| GO:BP | GO:0061356 | regulation of Wnt protein secretion | 1 | 4 | 8.42e-01 |
| GO:BP | GO:1903391 | regulation of adherens junction organization | 1 | 5 | 8.42e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 5 | 233 | 8.42e-01 |
| GO:BP | GO:0044721 | protein import into peroxisome matrix, substrate release | 1 | 2 | 8.42e-01 |
| GO:BP | GO:0042069 | regulation of catecholamine metabolic process | 4 | 11 | 8.43e-01 |
| GO:BP | GO:0042053 | regulation of dopamine metabolic process | 4 | 11 | 8.43e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 1 | 348 | 8.43e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 3 | 6 | 8.43e-01 |
| GO:BP | GO:0034342 | response to type III interferon | 1 | 5 | 8.43e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 3 | 6 | 8.43e-01 |
| GO:BP | GO:0009068 | aspartate family amino acid catabolic process | 1 | 12 | 8.43e-01 |
| GO:BP | GO:0021559 | trigeminal nerve development | 3 | 5 | 8.43e-01 |
| GO:BP | GO:0010533 | regulation of activation of Janus kinase activity | 1 | 3 | 8.43e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 2 | 6996 | 8.43e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 12 | 42 | 8.43e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 8.43e-01 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 2 | 11 | 8.44e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 5 | 234 | 8.44e-01 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 8 | 39 | 8.44e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 3 | 12 | 8.44e-01 |
| GO:BP | GO:1904383 | response to sodium phosphate | 3 | 5 | 8.44e-01 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 2 | 11 | 8.44e-01 |
| GO:BP | GO:2000410 | regulation of thymocyte migration | 1 | 1 | 8.44e-01 |
| GO:BP | GO:2000418 | positive regulation of eosinophil migration | 1 | 1 | 8.44e-01 |
| GO:BP | GO:0010155 | regulation of proton transport | 1 | 18 | 8.44e-01 |
| GO:BP | GO:0042220 | response to cocaine | 3 | 28 | 8.44e-01 |
| GO:BP | GO:0007597 | blood coagulation, intrinsic pathway | 1 | 3 | 8.44e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 8 | 35 | 8.44e-01 |
| GO:BP | GO:0036031 | recruitment of mRNA capping enzyme to RNA polymerase II holoenzyme complex | 1 | 1 | 8.44e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 2 | 43 | 8.44e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 2 | 43 | 8.44e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 4 | 46 | 8.44e-01 |
| GO:BP | GO:0002676 | regulation of chronic inflammatory response | 1 | 2 | 8.44e-01 |
| GO:BP | GO:0045916 | negative regulation of complement activation | 3 | 8 | 8.44e-01 |
| GO:BP | GO:0017014 | protein nitrosylation | 1 | 11 | 8.44e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 1 | 124 | 8.44e-01 |
| GO:BP | GO:0015746 | citrate transport | 1 | 3 | 8.44e-01 |
| GO:BP | GO:0036265 | RNA (guanine-N7)-methylation | 3 | 6 | 8.44e-01 |
| GO:BP | GO:0006842 | tricarboxylic acid transport | 1 | 3 | 8.44e-01 |
| GO:BP | GO:1990044 | protein localization to lipid droplet | 1 | 4 | 8.44e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 2 | 49 | 8.44e-01 |
| GO:BP | GO:0018119 | peptidyl-cysteine S-nitrosylation | 1 | 11 | 8.44e-01 |
| GO:BP | GO:2001171 | positive regulation of ATP biosynthetic process | 1 | 10 | 8.44e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 5 | 301 | 8.44e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 5 | 21 | 8.44e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 4 | 192 | 8.44e-01 |
| GO:BP | GO:0150003 | regulation of spontaneous synaptic transmission | 2 | 6 | 8.44e-01 |
| GO:BP | GO:0016243 | regulation of autophagosome size | 1 | 2 | 8.44e-01 |
| GO:BP | GO:2000503 | positive regulation of natural killer cell chemotaxis | 1 | 1 | 8.44e-01 |
| GO:BP | GO:0045907 | positive regulation of vasoconstriction | 1 | 15 | 8.44e-01 |
| GO:BP | GO:0001773 | myeloid dendritic cell activation | 1 | 12 | 8.44e-01 |
| GO:BP | GO:0021942 | radial glia guided migration of Purkinje cell | 1 | 3 | 8.44e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 13 | 30 | 8.44e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 5 | 52 | 8.44e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 13 | 30 | 8.44e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 3 | 276 | 8.45e-01 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 3 | 17 | 8.45e-01 |
| GO:BP | GO:0006935 | chemotaxis | 4 | 389 | 8.45e-01 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 2 | 12 | 8.45e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 1 | 31 | 8.45e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 2 | 8 | 8.46e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 4 | 55 | 8.46e-01 |
| GO:BP | GO:2000653 | regulation of genetic imprinting | 2 | 4 | 8.46e-01 |
| GO:BP | GO:0002638 | negative regulation of immunoglobulin production | 2 | 5 | 8.46e-01 |
| GO:BP | GO:0097360 | chorionic trophoblast cell proliferation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0003210 | cardiac atrium formation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0048840 | otolith development | 1 | 6 | 8.46e-01 |
| GO:BP | GO:0070366 | regulation of hepatocyte differentiation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0042663 | regulation of endodermal cell fate specification | 1 | 2 | 8.46e-01 |
| GO:BP | GO:1901382 | regulation of chorionic trophoblast cell proliferation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 4 | 12 | 8.46e-01 |
| GO:BP | GO:0060974 | cell migration involved in heart formation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 5 | 234 | 8.46e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 1 | 71 | 8.46e-01 |
| GO:BP | GO:0002468 | dendritic cell antigen processing and presentation | 2 | 6 | 8.46e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 12 | 29 | 8.46e-01 |
| GO:BP | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | 2 | 3 | 8.46e-01 |
| GO:BP | GO:0042330 | taxis | 4 | 389 | 8.46e-01 |
| GO:BP | GO:1905384 | regulation of protein localization to presynapse | 1 | 2 | 8.46e-01 |
| GO:BP | GO:1905386 | positive regulation of protein localization to presynapse | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0014022 | neural plate elongation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 1 | 21 | 8.46e-01 |
| GO:BP | GO:0022007 | convergent extension involved in neural plate elongation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0000338 | protein deneddylation | 2 | 11 | 8.46e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 7 | 26 | 8.46e-01 |
| GO:BP | GO:0043615 | astrocyte cell migration | 2 | 6 | 8.47e-01 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 1 | 10 | 8.47e-01 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 4 | 19 | 8.47e-01 |
| GO:BP | GO:1903224 | regulation of endodermal cell differentiation | 3 | 6 | 8.47e-01 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 3 | 13 | 8.47e-01 |
| GO:BP | GO:0097498 | endothelial tube lumen extension | 1 | 2 | 8.48e-01 |
| GO:BP | GO:0033239 | negative regulation of amine metabolic process | 2 | 2 | 8.48e-01 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 8.48e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 3 | 24 | 8.49e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 2 | 23 | 8.49e-01 |
| GO:BP | GO:1901078 | negative regulation of relaxation of muscle | 1 | 4 | 8.49e-01 |
| GO:BP | GO:0002032 | desensitization of G protein-coupled receptor signaling pathway by arrestin | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0061884 | regulation of mini excitatory postsynaptic potential | 1 | 2 | 8.49e-01 |
| GO:BP | GO:1904105 | positive regulation of convergent extension involved in gastrulation | 1 | 1 | 8.49e-01 |
| GO:BP | GO:0098816 | mini excitatory postsynaptic potential | 1 | 2 | 8.49e-01 |
| GO:BP | GO:1904103 | regulation of convergent extension involved in gastrulation | 1 | 1 | 8.49e-01 |
| GO:BP | GO:0061885 | positive regulation of mini excitatory postsynaptic potential | 1 | 2 | 8.49e-01 |
| GO:BP | GO:1900275 | negative regulation of phospholipase C activity | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0089709 | L-histidine transmembrane transport | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1 | 1564 | 8.49e-01 |
| GO:BP | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 1 | 21 | 8.49e-01 |
| GO:BP | GO:0003422 | growth plate cartilage morphogenesis | 1 | 1 | 8.49e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 8.49e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 6 | 15 | 8.49e-01 |
| GO:BP | GO:0002826 | negative regulation of T-helper 1 type immune response | 1 | 3 | 8.49e-01 |
| GO:BP | GO:2000340 | positive regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0003160 | endocardium morphogenesis | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0060842 | arterial endothelial cell differentiation | 1 | 5 | 8.49e-01 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 3 | 14 | 8.49e-01 |
| GO:BP | GO:0038171 | cannabinoid signaling pathway | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0043950 | positive regulation of cAMP-mediated signaling | 3 | 5 | 8.49e-01 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 2 | 11 | 8.49e-01 |
| GO:BP | GO:0060729 | intestinal epithelial structure maintenance | 2 | 4 | 8.49e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 8.49e-01 |
| GO:BP | GO:1903108 | regulation of mitochondrial transcription | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 4 | 10 | 8.49e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 8.49e-01 |
| GO:BP | GO:0006105 | succinate metabolic process | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0006601 | creatine biosynthetic process | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0110104 | mRNA alternative polyadenylation | 1 | 5 | 8.49e-01 |
| GO:BP | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus | 3 | 8 | 8.50e-01 |
| GO:BP | GO:1901483 | regulation of transcription factor catabolic process | 1 | 3 | 8.50e-01 |
| GO:BP | GO:1901485 | positive regulation of transcription factor catabolic process | 1 | 3 | 8.50e-01 |
| GO:BP | GO:0032715 | negative regulation of interleukin-6 production | 1 | 24 | 8.50e-01 |
| GO:BP | GO:0006528 | asparagine metabolic process | 2 | 5 | 8.50e-01 |
| GO:BP | GO:0032418 | lysosome localization | 9 | 62 | 8.50e-01 |
| GO:BP | GO:2000617 | positive regulation of histone H3-K9 acetylation | 1 | 6 | 8.50e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 9 | 62 | 8.50e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 5 | 232 | 8.50e-01 |
| GO:BP | GO:0045218 | zonula adherens maintenance | 1 | 3 | 8.50e-01 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 1 | 34 | 8.50e-01 |
| GO:BP | GO:0015949 | nucleobase-containing small molecule interconversion | 3 | 8 | 8.51e-01 |
| GO:BP | GO:0071418 | cellular response to amine stimulus | 1 | 2 | 8.51e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 13 | 25 | 8.51e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 6 | 27 | 8.51e-01 |
| GO:BP | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome | 1 | 2 | 8.51e-01 |
| GO:BP | GO:0048007 | antigen processing and presentation, exogenous lipid antigen via MHC class Ib | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 4 | 527 | 8.51e-01 |
| GO:BP | GO:2001226 | negative regulation of chloride transport | 1 | 1 | 8.51e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1602 | 8.52e-01 |
| GO:BP | GO:1901610 | positive regulation of vesicle transport along microtubule | 1 | 3 | 8.52e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 1 | 2 | 8.52e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 12 | 66 | 8.52e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 3 | 49 | 8.52e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 4 | 11 | 8.52e-01 |
| GO:BP | GO:0051712 | positive regulation of killing of cells of another organism | 1 | 2 | 8.52e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 2 | 18 | 8.52e-01 |
| GO:BP | GO:0030853 | negative regulation of granulocyte differentiation | 3 | 6 | 8.53e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 8.53e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 4 | 9 | 8.53e-01 |
| GO:BP | GO:0002038 | positive regulation of L-glutamate import across plasma membrane | 1 | 3 | 8.53e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 8.53e-01 |
| GO:BP | GO:1905219 | regulation of platelet formation | 1 | 1 | 8.53e-01 |
| GO:BP | GO:1904823 | purine nucleobase transmembrane transport | 1 | 3 | 8.53e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 8.53e-01 |
| GO:BP | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide | 1 | 2 | 8.53e-01 |
| GO:BP | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway | 2 | 5 | 8.53e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 2 | 58 | 8.53e-01 |
| GO:BP | GO:1905221 | positive regulation of platelet formation | 1 | 1 | 8.53e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 8 | 478 | 8.53e-01 |
| GO:BP | GO:0002175 | protein localization to paranode region of axon | 1 | 3 | 8.53e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 5 | 19 | 8.54e-01 |
| GO:BP | GO:1905906 | regulation of amyloid fibril formation | 6 | 14 | 8.54e-01 |
| GO:BP | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration | 1 | 3 | 8.54e-01 |
| GO:BP | GO:0009798 | axis specification | 2 | 63 | 8.54e-01 |
| GO:BP | GO:0043578 | nuclear matrix organization | 1 | 3 | 8.54e-01 |
| GO:BP | GO:1903003 | positive regulation of protein deubiquitination | 1 | 6 | 8.54e-01 |
| GO:BP | GO:0090292 | nuclear matrix anchoring at nuclear membrane | 1 | 3 | 8.54e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 3 | 17 | 8.54e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 4 | 531 | 8.55e-01 |
| GO:BP | GO:0048936 | peripheral nervous system neuron axonogenesis | 1 | 4 | 8.55e-01 |
| GO:BP | GO:0007586 | digestion | 2 | 60 | 8.55e-01 |
| GO:BP | GO:0035482 | gastric motility | 1 | 3 | 8.55e-01 |
| GO:BP | GO:0035026 | leading edge cell differentiation | 1 | 2 | 8.55e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 4 | 8 | 8.55e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 1 | 43 | 8.55e-01 |
| GO:BP | GO:1904177 | regulation of adipose tissue development | 2 | 8 | 8.55e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 2 | 26 | 8.55e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1 | 1604 | 8.55e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 59 | 8.55e-01 |
| GO:BP | GO:0016485 | protein processing | 2 | 183 | 8.56e-01 |
| GO:BP | GO:1901860 | positive regulation of mitochondrial DNA metabolic process | 1 | 3 | 8.57e-01 |
| GO:BP | GO:1900119 | positive regulation of execution phase of apoptosis | 1 | 7 | 8.57e-01 |
| GO:BP | GO:0006531 | aspartate metabolic process | 3 | 4 | 8.57e-01 |
| GO:BP | GO:0019230 | proprioception | 1 | 3 | 8.57e-01 |
| GO:BP | GO:0006857 | oligopeptide transport | 4 | 5 | 8.57e-01 |
| GO:BP | GO:0035672 | oligopeptide transmembrane transport | 4 | 5 | 8.57e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 11 | 55 | 8.58e-01 |
| GO:BP | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0070170 | regulation of tooth mineralization | 1 | 4 | 8.59e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 8.59e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 38 | 148 | 8.59e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 9 | 55 | 8.59e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 2 | 66 | 8.59e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 2 | 7 | 8.59e-01 |
| GO:BP | GO:0042660 | positive regulation of cell fate specification | 1 | 1 | 8.59e-01 |
| GO:BP | GO:1903580 | positive regulation of ATP metabolic process | 1 | 11 | 8.59e-01 |
| GO:BP | GO:0045345 | positive regulation of MHC class I biosynthetic process | 1 | 2 | 8.59e-01 |
| GO:BP | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 2 | 17 | 8.59e-01 |
| GO:BP | GO:0061047 | positive regulation of branching involved in lung morphogenesis | 1 | 2 | 8.59e-01 |
| GO:BP | GO:1902913 | positive regulation of neuroepithelial cell differentiation | 1 | 2 | 8.59e-01 |
| GO:BP | GO:0090362 | positive regulation of platelet-derived growth factor production | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0045609 | positive regulation of inner ear auditory receptor cell differentiation | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0045633 | positive regulation of mechanoreceptor differentiation | 1 | 1 | 8.59e-01 |
| GO:BP | GO:2000982 | positive regulation of inner ear receptor cell differentiation | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0048597 | post-embryonic camera-type eye morphogenesis | 1 | 4 | 8.59e-01 |
| GO:BP | GO:1904708 | regulation of granulosa cell apoptotic process | 1 | 2 | 8.59e-01 |
| GO:BP | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis | 1 | 2 | 8.59e-01 |
| GO:BP | GO:1904700 | granulosa cell apoptotic process | 1 | 2 | 8.59e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 8.59e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 5 | 31 | 8.59e-01 |
| GO:BP | GO:1903729 | regulation of plasma membrane organization | 1 | 15 | 8.59e-01 |
| GO:BP | GO:0071387 | cellular response to cortisol stimulus | 1 | 2 | 8.59e-01 |
| GO:BP | GO:1902565 | positive regulation of neutrophil activation | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0002930 | trabecular meshwork development | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0014052 | regulation of gamma-aminobutyric acid secretion | 1 | 5 | 8.59e-01 |
| GO:BP | GO:0070474 | positive regulation of uterine smooth muscle contraction | 1 | 4 | 8.59e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 1 | 384 | 8.60e-01 |
| GO:BP | GO:0150174 | negative regulation of phosphatidylcholine metabolic process | 1 | 2 | 8.60e-01 |
| GO:BP | GO:0140285 | endosome fission | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0034343 | type III interferon production | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0034344 | regulation of type III interferon production | 1 | 3 | 8.60e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 1 | 22 | 8.60e-01 |
| GO:BP | GO:0035332 | positive regulation of hippo signaling | 2 | 6 | 8.60e-01 |
| GO:BP | GO:0006390 | mitochondrial transcription | 7 | 18 | 8.60e-01 |
| GO:BP | GO:0048733 | sebaceous gland development | 1 | 6 | 8.60e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 3 | 50 | 8.60e-01 |
| GO:BP | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport | 1 | 4 | 8.60e-01 |
| GO:BP | GO:0006714 | sesquiterpenoid metabolic process | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0046724 | oxalic acid secretion | 1 | 1 | 8.61e-01 |
| GO:BP | GO:0046968 | peptide antigen transport | 1 | 1 | 8.61e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 1 | 26 | 8.61e-01 |
| GO:BP | GO:0019605 | butyrate metabolic process | 1 | 1 | 8.61e-01 |
| GO:BP | GO:0070994 | detection of oxidative stress | 1 | 1 | 8.61e-01 |
| GO:BP | GO:0045764 | positive regulation of amino acid metabolic process | 1 | 1 | 8.61e-01 |
| GO:BP | GO:0043379 | memory T cell differentiation | 2 | 5 | 8.61e-01 |
| GO:BP | GO:1903384 | negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0050787 | detoxification of mercury ion | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0045112 | integrin biosynthetic process | 1 | 3 | 8.61e-01 |
| GO:BP | GO:1903094 | negative regulation of protein K48-linked deubiquitination | 1 | 2 | 8.61e-01 |
| GO:BP | GO:1903383 | regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 7 | 259 | 8.61e-01 |
| GO:BP | GO:2000157 | negative regulation of ubiquitin-specific protease activity | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0051595 | response to methylglyoxal | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0043306 | positive regulation of mast cell degranulation | 2 | 10 | 8.61e-01 |
| GO:BP | GO:0036482 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 2 | 8.61e-01 |
| GO:BP | GO:1903181 | positive regulation of dopamine biosynthetic process | 1 | 2 | 8.61e-01 |
| GO:BP | GO:0110061 | regulation of angiotensin-activated signaling pathway | 1 | 2 | 8.61e-01 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 1 | 6 | 8.61e-01 |
| GO:BP | GO:0033008 | positive regulation of mast cell activation involved in immune response | 2 | 10 | 8.61e-01 |
| GO:BP | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway | 1 | 4 | 8.61e-01 |
| GO:BP | GO:0031335 | regulation of sulfur amino acid metabolic process | 1 | 1 | 8.61e-01 |
| GO:BP | GO:0031554 | regulation of termination of DNA-templated transcription | 1 | 4 | 8.61e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 2 | 22 | 8.61e-01 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 8.62e-01 |
| GO:BP | GO:2001280 | positive regulation of unsaturated fatty acid biosynthetic process | 1 | 4 | 8.62e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 2 | 15 | 8.62e-01 |
| GO:BP | GO:0030974 | thiamine pyrophosphate transmembrane transport | 1 | 1 | 8.62e-01 |
| GO:BP | GO:2000523 | regulation of T cell costimulation | 1 | 2 | 8.62e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 3 | 20 | 8.62e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 8.62e-01 |
| GO:BP | GO:1990962 | xenobiotic transport across blood-brain barrier | 2 | 3 | 8.62e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 8.62e-01 |
| GO:BP | GO:0032328 | alanine transport | 7 | 14 | 8.62e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 13 | 33 | 8.62e-01 |
| GO:BP | GO:2000646 | positive regulation of receptor catabolic process | 1 | 5 | 8.62e-01 |
| GO:BP | GO:0048866 | stem cell fate specification | 1 | 2 | 8.63e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 3 | 7069 | 8.63e-01 |
| GO:BP | GO:1905451 | positive regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 3 | 8.63e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 7 | 191 | 8.63e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 7 | 31 | 8.63e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 1 | 369 | 8.63e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 2 | 137 | 8.63e-01 |
| GO:BP | GO:0035929 | steroid hormone secretion | 1 | 18 | 8.63e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 35 | 116 | 8.63e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 6 | 12 | 8.63e-01 |
| GO:BP | GO:0061046 | regulation of branching involved in lung morphogenesis | 1 | 3 | 8.63e-01 |
| GO:BP | GO:0015670 | carbon dioxide transport | 1 | 2 | 8.64e-01 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 2 | 29 | 8.64e-01 |
| GO:BP | GO:0060025 | regulation of synaptic activity | 1 | 3 | 8.64e-01 |
| GO:BP | GO:0060629 | regulation of homologous chromosome segregation | 1 | 1 | 8.64e-01 |
| GO:BP | GO:0060011 | Sertoli cell proliferation | 1 | 4 | 8.65e-01 |
| GO:BP | GO:0034695 | response to prostaglandin E | 1 | 17 | 8.65e-01 |
| GO:BP | GO:0009111 | vitamin catabolic process | 1 | 3 | 8.65e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 2 | 6 | 8.65e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 3 | 4128 | 8.65e-01 |
| GO:BP | GO:1903842 | response to arsenite ion | 1 | 4 | 8.65e-01 |
| GO:BP | GO:0021626 | central nervous system maturation | 2 | 5 | 8.65e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 7 | 8.65e-01 |
| GO:BP | GO:2001225 | regulation of chloride transport | 2 | 4 | 8.65e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 8.65e-01 |
| GO:BP | GO:1903843 | cellular response to arsenite ion | 1 | 4 | 8.65e-01 |
| GO:BP | GO:0050904 | diapedesis | 2 | 6 | 8.65e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 18 | 78 | 8.65e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 5 | 45 | 8.65e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 8.66e-01 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 1 | 8 | 8.66e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 27 | 8.67e-01 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 3 | 6 | 8.67e-01 |
| GO:BP | GO:1901842 | negative regulation of high voltage-gated calcium channel activity | 2 | 4 | 8.67e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 6 | 38 | 8.67e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 8.67e-01 |
| GO:BP | GO:1902644 | tertiary alcohol metabolic process | 3 | 17 | 8.67e-01 |
| GO:BP | GO:0015820 | leucine transport | 1 | 10 | 8.67e-01 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 2 | 36 | 8.67e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 8 | 2270 | 8.68e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 8.68e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 8.68e-01 |
| GO:BP | GO:2000485 | regulation of glutamine transport | 1 | 2 | 8.68e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 39 | 8.68e-01 |
| GO:BP | GO:0001886 | endothelial cell morphogenesis | 5 | 10 | 8.68e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 4 | 551 | 8.68e-01 |
| GO:BP | GO:0071529 | cementum mineralization | 1 | 3 | 8.68e-01 |
| GO:BP | GO:0016556 | mRNA modification | 1 | 29 | 8.68e-01 |
| GO:BP | GO:0070070 | proton-transporting V-type ATPase complex assembly | 1 | 7 | 8.68e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 8.68e-01 |
| GO:BP | GO:0060159 | regulation of dopamine receptor signaling pathway | 1 | 7 | 8.68e-01 |
| GO:BP | GO:0048538 | thymus development | 1 | 35 | 8.68e-01 |
| GO:BP | GO:0006968 | cellular defense response | 1 | 15 | 8.68e-01 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 4 | 22 | 8.68e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 14 | 28 | 8.68e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 1 | 6 | 8.68e-01 |
| GO:BP | GO:0050427 | 3’-phosphoadenosine 5’-phosphosulfate metabolic process | 1 | 10 | 8.68e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 8.68e-01 |
| GO:BP | GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1 | 10 | 8.68e-01 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 2 | 13 | 8.68e-01 |
| GO:BP | GO:1902232 | regulation of positive thymic T cell selection | 1 | 2 | 8.69e-01 |
| GO:BP | GO:0038195 | urokinase plasminogen activator signaling pathway | 1 | 2 | 8.69e-01 |
| GO:BP | GO:1902227 | negative regulation of macrophage colony-stimulating factor signaling pathway | 1 | 1 | 8.69e-01 |
| GO:BP | GO:0019884 | antigen processing and presentation of exogenous antigen | 2 | 28 | 8.69e-01 |
| GO:BP | GO:0060437 | lung growth | 1 | 5 | 8.69e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 3 | 7 | 8.69e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 1 | 9 | 8.69e-01 |
| GO:BP | GO:0046784 | viral mRNA export from host cell nucleus | 1 | 6 | 8.70e-01 |
| GO:BP | GO:0090193 | positive regulation of glomerulus development | 1 | 3 | 8.70e-01 |
| GO:BP | GO:0045117 | azole transmembrane transport | 5 | 9 | 8.70e-01 |
| GO:BP | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process | 1 | 4 | 8.70e-01 |
| GO:BP | GO:0036343 | psychomotor behavior | 1 | 2 | 8.71e-01 |
| GO:BP | GO:0014002 | astrocyte development | 1 | 27 | 8.71e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 8.71e-01 |
| GO:BP | GO:0017185 | peptidyl-lysine hydroxylation | 3 | 9 | 8.71e-01 |
| GO:BP | GO:0042214 | terpene metabolic process | 1 | 1 | 8.71e-01 |
| GO:BP | GO:0030431 | sleep | 2 | 13 | 8.71e-01 |
| GO:BP | GO:0016116 | carotenoid metabolic process | 1 | 1 | 8.71e-01 |
| GO:BP | GO:0016108 | tetraterpenoid metabolic process | 1 | 1 | 8.71e-01 |
| GO:BP | GO:0120253 | hydrocarbon catabolic process | 1 | 1 | 8.71e-01 |
| GO:BP | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway | 1 | 3 | 8.71e-01 |
| GO:BP | GO:0023052 | signaling | 5 | 4141 | 8.71e-01 |
| GO:BP | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly | 1 | 6 | 8.71e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 5 | 55 | 8.71e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 1 | 49 | 8.71e-01 |
| GO:BP | GO:0071670 | smooth muscle cell chemotaxis | 1 | 6 | 8.71e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 8.71e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 8.71e-01 |
| GO:BP | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration | 1 | 3 | 8.71e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 34 | 2035 | 8.71e-01 |
| GO:BP | GO:0034380 | high-density lipoprotein particle assembly | 1 | 10 | 8.71e-01 |
| GO:BP | GO:0032849 | positive regulation of cellular pH reduction | 2 | 2 | 8.71e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 7 | 32 | 8.71e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 8.71e-01 |
| GO:BP | GO:0003011 | involuntary skeletal muscle contraction | 1 | 3 | 8.72e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 9 | 87 | 8.72e-01 |
| GO:BP | GO:0018242 | protein O-linked glycosylation via serine | 1 | 7 | 8.72e-01 |
| GO:BP | GO:0061520 | Langerhans cell differentiation | 1 | 3 | 8.72e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 12 | 28 | 8.72e-01 |
| GO:BP | GO:0001732 | formation of cytoplasmic translation initiation complex | 7 | 16 | 8.72e-01 |
| GO:BP | GO:0031580 | membrane raft distribution | 1 | 3 | 8.72e-01 |
| GO:BP | GO:0007525 | somatic muscle development | 1 | 4 | 8.72e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 1 | 13 | 8.72e-01 |
| GO:BP | GO:0010986 | positive regulation of lipoprotein particle clearance | 2 | 4 | 8.73e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 1 | 7 | 8.74e-01 |
| GO:BP | GO:0045471 | response to ethanol | 1 | 73 | 8.74e-01 |
| GO:BP | GO:0120045 | stereocilium maintenance | 1 | 2 | 8.74e-01 |
| GO:BP | GO:0140291 | peptidyl-glutamate ADP-deribosylation | 1 | 3 | 8.74e-01 |
| GO:BP | GO:0071267 | L-methionine salvage | 1 | 5 | 8.74e-01 |
| GO:BP | GO:0071265 | L-methionine biosynthetic process | 1 | 5 | 8.74e-01 |
| GO:BP | GO:0046305 | alkanesulfonate biosynthetic process | 1 | 2 | 8.74e-01 |
| GO:BP | GO:0042412 | taurine biosynthetic process | 1 | 2 | 8.74e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 1 | 50 | 8.74e-01 |
| GO:BP | GO:0043102 | amino acid salvage | 1 | 5 | 8.74e-01 |
| GO:BP | GO:1903288 | positive regulation of potassium ion import across plasma membrane | 2 | 7 | 8.74e-01 |
| GO:BP | GO:0001706 | endoderm formation | 5 | 46 | 8.74e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 1 | 29 | 8.74e-01 |
| GO:BP | GO:0035442 | dipeptide transmembrane transport | 3 | 4 | 8.74e-01 |
| GO:BP | GO:0042938 | dipeptide transport | 3 | 4 | 8.74e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 3 | 5 | 8.74e-01 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 13 | 28 | 8.74e-01 |
| GO:BP | GO:0010989 | negative regulation of low-density lipoprotein particle clearance | 3 | 4 | 8.74e-01 |
| GO:BP | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient | 1 | 7 | 8.74e-01 |
| GO:BP | GO:0090045 | positive regulation of deacetylase activity | 3 | 7 | 8.75e-01 |
| GO:BP | GO:0035621 | ER to Golgi ceramide transport | 1 | 3 | 8.75e-01 |
| GO:BP | GO:0014734 | skeletal muscle hypertrophy | 1 | 1 | 8.75e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 2 | 53 | 8.75e-01 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 5 | 12 | 8.75e-01 |
| GO:BP | GO:0036123 | histone H3-K9 dimethylation | 1 | 3 | 8.75e-01 |
| GO:BP | GO:0033967 | box C/D RNA metabolic process | 1 | 3 | 8.75e-01 |
| GO:BP | GO:0000494 | box C/D RNA 3’-end processing | 1 | 3 | 8.75e-01 |
| GO:BP | GO:0071934 | thiamine transmembrane transport | 3 | 3 | 8.75e-01 |
| GO:BP | GO:0034963 | box C/D RNA processing | 1 | 3 | 8.75e-01 |
| GO:BP | GO:0042723 | thiamine-containing compound metabolic process | 3 | 4 | 8.75e-01 |
| GO:BP | GO:0002092 | positive regulation of receptor internalization | 1 | 21 | 8.75e-01 |
| GO:BP | GO:0061548 | ganglion development | 6 | 12 | 8.75e-01 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 1 | 13 | 8.75e-01 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 1 | 7 | 8.75e-01 |
| GO:BP | GO:0044650 | adhesion of symbiont to host cell | 1 | 9 | 8.76e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 7 | 61 | 8.76e-01 |
| GO:BP | GO:0019317 | fucose catabolic process | 1 | 3 | 8.76e-01 |
| GO:BP | GO:0042355 | L-fucose catabolic process | 1 | 3 | 8.76e-01 |
| GO:BP | GO:0060012 | synaptic transmission, glycinergic | 1 | 2 | 8.77e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 8.77e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 4 | 15 | 8.77e-01 |
| GO:BP | GO:0007217 | tachykinin receptor signaling pathway | 1 | 3 | 8.77e-01 |
| GO:BP | GO:0070165 | positive regulation of adiponectin secretion | 1 | 1 | 8.77e-01 |
| GO:BP | GO:1902564 | negative regulation of neutrophil activation | 1 | 2 | 8.77e-01 |
| GO:BP | GO:0002282 | microglial cell activation involved in immune response | 1 | 1 | 8.77e-01 |
| GO:BP | GO:1903215 | negative regulation of protein targeting to mitochondrion | 1 | 3 | 8.78e-01 |
| GO:BP | GO:0060179 | male mating behavior | 1 | 5 | 8.78e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 11 | 523 | 8.78e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 5 | 17 | 8.78e-01 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 1 | 7 | 8.78e-01 |
| GO:BP | GO:0007549 | dosage compensation | 4 | 29 | 8.78e-01 |
| GO:BP | GO:0035066 | positive regulation of histone acetylation | 1 | 25 | 8.78e-01 |
| GO:BP | GO:0070487 | monocyte aggregation | 2 | 4 | 8.79e-01 |
| GO:BP | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway | 1 | 12 | 8.79e-01 |
| GO:BP | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy | 1 | 3 | 8.79e-01 |
| GO:BP | GO:1902988 | neurofibrillary tangle assembly | 1 | 3 | 8.79e-01 |
| GO:BP | GO:0009996 | negative regulation of cell fate specification | 2 | 4 | 8.79e-01 |
| GO:BP | GO:1990127 | intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 8.79e-01 |
| GO:BP | GO:1902238 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 8.79e-01 |
| GO:BP | GO:0016486 | peptide hormone processing | 8 | 27 | 8.80e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 1 | 43 | 8.80e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 5 | 22 | 8.80e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 8.80e-01 |
| GO:BP | GO:0070672 | response to interleukin-15 | 2 | 3 | 8.80e-01 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 4 | 11 | 8.80e-01 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 1 | 10 | 8.80e-01 |
| GO:BP | GO:0045625 | regulation of T-helper 1 cell differentiation | 2 | 5 | 8.80e-01 |
| GO:BP | GO:0061355 | Wnt protein secretion | 1 | 5 | 8.81e-01 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 2 | 6 | 8.81e-01 |
| GO:BP | GO:1905686 | positive regulation of plasma membrane repair | 1 | 4 | 8.81e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 8.81e-01 |
| GO:BP | GO:0006555 | methionine metabolic process | 1 | 13 | 8.81e-01 |
| GO:BP | GO:0010960 | magnesium ion homeostasis | 2 | 9 | 8.81e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 46 | 2135 | 8.82e-01 |
| GO:BP | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway | 1 | 3 | 8.82e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 8.82e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 8 | 258 | 8.82e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 2 | 9 | 8.82e-01 |
| GO:BP | GO:0002840 | regulation of T cell mediated immune response to tumor cell | 3 | 5 | 8.82e-01 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 1 | 31 | 8.83e-01 |
| GO:BP | GO:0007154 | cell communication | 5 | 4226 | 8.83e-01 |
| GO:BP | GO:0071000 | response to magnetism | 1 | 2 | 8.83e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 2 | 105 | 8.83e-01 |
| GO:BP | GO:1905665 | positive regulation of calcium ion import across plasma membrane | 1 | 6 | 8.83e-01 |
| GO:BP | GO:0030047 | actin modification | 2 | 5 | 8.83e-01 |
| GO:BP | GO:0038007 | netrin-activated signaling pathway | 4 | 8 | 8.84e-01 |
| GO:BP | GO:0060440 | trachea formation | 1 | 7 | 8.84e-01 |
| GO:BP | GO:0044351 | macropinocytosis | 3 | 8 | 8.84e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 8.84e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 10 | 44 | 8.84e-01 |
| GO:BP | GO:0009725 | response to hormone | 18 | 642 | 8.84e-01 |
| GO:BP | GO:2000407 | regulation of T cell extravasation | 1 | 3 | 8.85e-01 |
| GO:BP | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity | 3 | 12 | 8.85e-01 |
| GO:BP | GO:0051946 | regulation of glutamate uptake involved in transmission of nerve impulse | 1 | 3 | 8.85e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 7 | 15 | 8.85e-01 |
| GO:BP | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission | 1 | 3 | 8.85e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 3 | 19 | 8.85e-01 |
| GO:BP | GO:0010983 | positive regulation of high-density lipoprotein particle clearance | 1 | 2 | 8.86e-01 |
| GO:BP | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 1 | 4 | 8.86e-01 |
| GO:BP | GO:0090715 | immunological memory formation process | 2 | 6 | 8.86e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 2 | 336 | 8.86e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 1 | 37 | 8.87e-01 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 5 | 25 | 8.87e-01 |
| GO:BP | GO:0021548 | pons development | 3 | 7 | 8.87e-01 |
| GO:BP | GO:1905150 | regulation of voltage-gated sodium channel activity | 3 | 6 | 8.87e-01 |
| GO:BP | GO:0031134 | sister chromatid biorientation | 1 | 3 | 8.87e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 7 | 95 | 8.87e-01 |
| GO:BP | GO:0002295 | T-helper cell lineage commitment | 5 | 7 | 8.87e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 1 | 37 | 8.87e-01 |
| GO:BP | GO:1903487 | regulation of lactation | 1 | 1 | 8.87e-01 |
| GO:BP | GO:1900100 | positive regulation of plasma cell differentiation | 1 | 1 | 8.87e-01 |
| GO:BP | GO:1990418 | response to insulin-like growth factor stimulus | 1 | 2 | 8.87e-01 |
| GO:BP | GO:0072081 | specification of nephron tubule identity | 2 | 4 | 8.87e-01 |
| GO:BP | GO:0072047 | proximal/distal pattern formation involved in nephron development | 2 | 4 | 8.87e-01 |
| GO:BP | GO:0006071 | glycerol metabolic process | 1 | 12 | 8.87e-01 |
| GO:BP | GO:2000370 | positive regulation of clathrin-dependent endocytosis | 1 | 5 | 8.88e-01 |
| GO:BP | GO:0032240 | negative regulation of nucleobase-containing compound transport | 1 | 3 | 8.88e-01 |
| GO:BP | GO:2001026 | regulation of endothelial cell chemotaxis | 9 | 16 | 8.88e-01 |
| GO:BP | GO:0070370 | cellular heat acclimation | 2 | 5 | 8.88e-01 |
| GO:BP | GO:0010286 | heat acclimation | 2 | 5 | 8.88e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 3 | 223 | 8.88e-01 |
| GO:BP | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 4 | 8.88e-01 |
| GO:BP | GO:1901896 | positive regulation of ATPase-coupled calcium transmembrane transporter activity | 2 | 6 | 8.89e-01 |
| GO:BP | GO:0043969 | histone H2B acetylation | 1 | 3 | 8.89e-01 |
| GO:BP | GO:0010430 | fatty acid omega-oxidation | 1 | 2 | 8.89e-01 |
| GO:BP | GO:0072566 | chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 8.89e-01 |
| GO:BP | GO:1901877 | negative regulation of calcium ion binding | 1 | 2 | 8.89e-01 |
| GO:BP | GO:1901876 | regulation of calcium ion binding | 1 | 2 | 8.89e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 2 | 33 | 8.89e-01 |
| GO:BP | GO:0006011 | UDP-glucose metabolic process | 1 | 6 | 8.89e-01 |
| GO:BP | GO:0071608 | macrophage inflammatory protein-1 alpha production | 1 | 3 | 8.89e-01 |
| GO:BP | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | 3 | 8.89e-01 |
| GO:BP | GO:2000338 | regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 8.89e-01 |
| GO:BP | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production | 1 | 3 | 8.89e-01 |
| GO:BP | GO:1904398 | positive regulation of neuromuscular junction development | 1 | 2 | 8.89e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 2 | 194 | 8.90e-01 |
| GO:BP | GO:0048631 | regulation of skeletal muscle tissue growth | 2 | 7 | 8.90e-01 |
| GO:BP | GO:0033299 | secretion of lysosomal enzymes | 2 | 7 | 8.90e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 1 | 64 | 8.90e-01 |
| GO:BP | GO:1903852 | positive regulation of cristae formation | 1 | 3 | 8.90e-01 |
| GO:BP | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 3 | 8.91e-01 |
| GO:BP | GO:0019477 | L-lysine catabolic process | 1 | 4 | 8.91e-01 |
| GO:BP | GO:0019474 | L-lysine catabolic process to acetyl-CoA | 1 | 4 | 8.91e-01 |
| GO:BP | GO:0046440 | L-lysine metabolic process | 1 | 4 | 8.91e-01 |
| GO:BP | GO:0019695 | choline metabolic process | 1 | 4 | 8.92e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 5 | 15 | 8.93e-01 |
| GO:BP | GO:0036260 | RNA capping | 9 | 20 | 8.93e-01 |
| GO:BP | GO:0097254 | renal tubular secretion | 1 | 15 | 8.93e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 1 | 129 | 8.93e-01 |
| GO:BP | GO:0070327 | thyroid hormone transport | 1 | 7 | 8.93e-01 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 2 | 5 | 8.93e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 2 | 11 | 8.93e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 28 | 72 | 8.93e-01 |
| GO:BP | GO:0048295 | positive regulation of isotype switching to IgE isotypes | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0045991 | carbon catabolite activation of transcription | 2 | 4 | 8.94e-01 |
| GO:BP | GO:0045990 | carbon catabolite regulation of transcription | 2 | 4 | 8.94e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 8.94e-01 |
| GO:BP | GO:0007599 | hemostasis | 2 | 153 | 8.94e-01 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 16 | 37 | 8.94e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 7 | 44 | 8.95e-01 |
| GO:BP | GO:0046368 | GDP-L-fucose metabolic process | 1 | 4 | 8.96e-01 |
| GO:BP | GO:0002729 | positive regulation of natural killer cell cytokine production | 1 | 4 | 8.96e-01 |
| GO:BP | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 10 | 8.96e-01 |
| GO:BP | GO:1903963 | arachidonate transport | 1 | 11 | 8.96e-01 |
| GO:BP | GO:0050482 | arachidonic acid secretion | 1 | 11 | 8.96e-01 |
| GO:BP | GO:0071354 | cellular response to interleukin-6 | 11 | 23 | 8.96e-01 |
| GO:BP | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 3 | 8.96e-01 |
| GO:BP | GO:0046958 | nonassociative learning | 2 | 6 | 8.96e-01 |
| GO:BP | GO:0060594 | mammary gland specification | 1 | 3 | 8.96e-01 |
| GO:BP | GO:0060523 | prostate epithelial cord elongation | 1 | 1 | 8.96e-01 |
| GO:BP | GO:0060648 | mammary gland bud morphogenesis | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 1 | 38 | 8.96e-01 |
| GO:BP | GO:0071549 | cellular response to dexamethasone stimulus | 9 | 24 | 8.96e-01 |
| GO:BP | GO:0021523 | somatic motor neuron differentiation | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0090025 | regulation of monocyte chemotaxis | 2 | 12 | 8.96e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 2 | 58 | 8.96e-01 |
| GO:BP | GO:0071698 | olfactory placode development | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0071699 | olfactory placode morphogenesis | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0010842 | retina layer formation | 2 | 16 | 8.96e-01 |
| GO:BP | GO:0031998 | regulation of fatty acid beta-oxidation | 2 | 16 | 8.96e-01 |
| GO:BP | GO:0051799 | negative regulation of hair follicle development | 2 | 5 | 8.96e-01 |
| GO:BP | GO:0030910 | olfactory placode formation | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0072672 | neutrophil extravasation | 1 | 7 | 8.96e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 6 | 422 | 8.96e-01 |
| GO:BP | GO:0018094 | protein polyglycylation | 1 | 1 | 8.97e-01 |
| GO:BP | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 12 | 8.97e-01 |
| GO:BP | GO:0014891 | striated muscle atrophy | 2 | 6 | 8.97e-01 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 1 | 16 | 8.97e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 5 | 7 | 8.97e-01 |
| GO:BP | GO:1904054 | regulation of cholangiocyte proliferation | 1 | 2 | 8.97e-01 |
| GO:BP | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing | 1 | 5 | 8.97e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 8.97e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 4 | 10 | 8.97e-01 |
| GO:BP | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 14 | 29 | 8.97e-01 |
| GO:BP | GO:0002019 | regulation of renal output by angiotensin | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0097195 | pilomotor reflex | 1 | 2 | 8.98e-01 |
| GO:BP | GO:0060700 | regulation of ribonuclease activity | 1 | 5 | 8.98e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 8.98e-01 |
| GO:BP | GO:2001245 | regulation of phosphatidylcholine biosynthetic process | 1 | 5 | 8.98e-01 |
| GO:BP | GO:1904669 | ATP export | 1 | 1 | 8.98e-01 |
| GO:BP | GO:0071673 | positive regulation of smooth muscle cell chemotaxis | 1 | 2 | 8.98e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 8.98e-01 |
| GO:BP | GO:1903371 | regulation of endoplasmic reticulum tubular network organization | 1 | 6 | 8.98e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 8.98e-01 |
| GO:BP | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis | 1 | 7 | 8.98e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 2 | 13 | 8.98e-01 |
| GO:BP | GO:0042447 | hormone catabolic process | 1 | 7 | 8.98e-01 |
| GO:BP | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 2 | 6 | 8.98e-01 |
| GO:BP | GO:0044597 | daunorubicin metabolic process | 3 | 7 | 8.98e-01 |
| GO:BP | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 2 | 6 | 8.98e-01 |
| GO:BP | GO:0097035 | regulation of membrane lipid distribution | 2 | 43 | 8.98e-01 |
| GO:BP | GO:0120317 | sperm mitochondrial sheath assembly | 2 | 2 | 8.98e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 2 | 3 | 8.98e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 2 | 7441 | 8.99e-01 |
| GO:BP | GO:0051668 | localization within membrane | 12 | 605 | 8.99e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 3 | 49 | 9.00e-01 |
| GO:BP | GO:0071676 | negative regulation of mononuclear cell migration | 3 | 16 | 9.00e-01 |
| GO:BP | GO:0044417 | translocation of molecules into host | 1 | 7 | 9.00e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 9.00e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 3 | 39 | 9.00e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 10 | 88 | 9.00e-01 |
| GO:BP | GO:0006949 | syncytium formation | 1 | 39 | 9.00e-01 |
| GO:BP | GO:0140331 | aminophospholipid translocation | 1 | 6 | 9.00e-01 |
| GO:BP | GO:0097213 | regulation of lysosomal membrane permeability | 1 | 2 | 9.00e-01 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 4 | 11 | 9.01e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 2 | 113 | 9.01e-01 |
| GO:BP | GO:0030222 | eosinophil differentiation | 1 | 2 | 9.01e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 3 | 16 | 9.01e-01 |
| GO:BP | GO:0010626 | negative regulation of Schwann cell proliferation | 3 | 6 | 9.01e-01 |
| GO:BP | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan | 1 | 4 | 9.01e-01 |
| GO:BP | GO:0048677 | axon extension involved in regeneration | 1 | 2 | 9.01e-01 |
| GO:BP | GO:2001200 | positive regulation of dendritic cell differentiation | 1 | 3 | 9.01e-01 |
| GO:BP | GO:0048682 | sprouting of injured axon | 1 | 2 | 9.01e-01 |
| GO:BP | GO:1901608 | regulation of vesicle transport along microtubule | 1 | 4 | 9.01e-01 |
| GO:BP | GO:0001574 | ganglioside biosynthetic process | 1 | 14 | 9.01e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 1 | 98 | 9.01e-01 |
| GO:BP | GO:0050689 | negative regulation of defense response to virus by host | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0072194 | kidney smooth muscle tissue development | 1 | 2 | 9.02e-01 |
| GO:BP | GO:0036023 | embryonic skeletal limb joint morphogenesis | 1 | 2 | 9.02e-01 |
| GO:BP | GO:1903677 | regulation of cap-independent translational initiation | 1 | 3 | 9.02e-01 |
| GO:BP | GO:1903679 | positive regulation of cap-independent translational initiation | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0001555 | oocyte growth | 1 | 2 | 9.02e-01 |
| GO:BP | GO:0061038 | uterus morphogenesis | 2 | 5 | 9.02e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 7 | 90 | 9.02e-01 |
| GO:BP | GO:0051665 | membrane raft localization | 1 | 4 | 9.02e-01 |
| GO:BP | GO:1901668 | regulation of superoxide dismutase activity | 2 | 4 | 9.02e-01 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 1 | 9 | 9.02e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 9.02e-01 |
| GO:BP | GO:0031017 | exocrine pancreas development | 1 | 4 | 9.03e-01 |
| GO:BP | GO:0150172 | regulation of phosphatidylcholine metabolic process | 2 | 10 | 9.03e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 2 | 28 | 9.03e-01 |
| GO:BP | GO:0001923 | B-1 B cell differentiation | 1 | 3 | 9.03e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 9.03e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 2 | 21 | 9.03e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 9.03e-01 |
| GO:BP | GO:0045650 | negative regulation of macrophage differentiation | 2 | 5 | 9.03e-01 |
| GO:BP | GO:1904037 | positive regulation of epithelial cell apoptotic process | 4 | 26 | 9.03e-01 |
| GO:BP | GO:1900038 | negative regulation of cellular response to hypoxia | 3 | 8 | 9.04e-01 |
| GO:BP | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | 3 | 11 | 9.04e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 6 | 29 | 9.04e-01 |
| GO:BP | GO:0044788 | modulation by host of viral process | 5 | 40 | 9.04e-01 |
| GO:BP | GO:0032687 | negative regulation of interferon-alpha production | 1 | 2 | 9.04e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 16 | 29 | 9.05e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 2 | 65 | 9.05e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 2 | 11 | 9.05e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 9.06e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 9.06e-01 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 2 | 36 | 9.06e-01 |
| GO:BP | GO:0021682 | nerve maturation | 1 | 2 | 9.06e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 8 | 15 | 9.06e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 10 | 88 | 9.07e-01 |
| GO:BP | GO:0023058 | adaptation of signaling pathway | 6 | 16 | 9.07e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 7 | 16 | 9.07e-01 |
| GO:BP | GO:0030050 | vesicle transport along actin filament | 7 | 13 | 9.07e-01 |
| GO:BP | GO:0019400 | alditol metabolic process | 1 | 14 | 9.07e-01 |
| GO:BP | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | 1 | 8 | 9.07e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 1 | 16 | 9.07e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 3 | 18 | 9.07e-01 |
| GO:BP | GO:0090289 | regulation of osteoclast proliferation | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0071485 | cellular response to absence of light | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0009646 | response to absence of light | 1 | 3 | 9.07e-01 |
| GO:BP | GO:1990603 | dark adaptation | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0070142 | synaptic vesicle budding | 1 | 8 | 9.07e-01 |
| GO:BP | GO:0072215 | regulation of metanephros development | 2 | 2 | 9.07e-01 |
| GO:BP | GO:0060684 | epithelial-mesenchymal cell signaling | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 2 | 14 | 9.07e-01 |
| GO:BP | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning | 1 | 5 | 9.07e-01 |
| GO:BP | GO:1903979 | negative regulation of microglial cell activation | 2 | 5 | 9.07e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 1 | 25 | 9.07e-01 |
| GO:BP | GO:2000501 | regulation of natural killer cell chemotaxis | 1 | 2 | 9.07e-01 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 3 | 8 | 9.08e-01 |
| GO:BP | GO:0042908 | xenobiotic transport | 10 | 30 | 9.08e-01 |
| GO:BP | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus | 3 | 7 | 9.08e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 9.08e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 1 | 35 | 9.08e-01 |
| GO:BP | GO:0070472 | regulation of uterine smooth muscle contraction | 1 | 5 | 9.08e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 7 | 32 | 9.08e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 13 | 27 | 9.09e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 4 | 11 | 9.09e-01 |
| GO:BP | GO:0070346 | positive regulation of fat cell proliferation | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0034204 | lipid translocation | 1 | 38 | 9.10e-01 |
| GO:BP | GO:0110089 | regulation of hippocampal neuron apoptotic process | 1 | 2 | 9.10e-01 |
| GO:BP | GO:0110088 | hippocampal neuron apoptotic process | 1 | 2 | 9.10e-01 |
| GO:BP | GO:0120197 | mucociliary clearance | 3 | 5 | 9.10e-01 |
| GO:BP | GO:1904340 | positive regulation of dopaminergic neuron differentiation | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0090324 | negative regulation of oxidative phosphorylation | 3 | 6 | 9.10e-01 |
| GO:BP | GO:0046940 | nucleoside monophosphate phosphorylation | 4 | 9 | 9.10e-01 |
| GO:BP | GO:0030252 | growth hormone secretion | 1 | 12 | 9.10e-01 |
| GO:BP | GO:1903593 | regulation of histamine secretion by mast cell | 1 | 4 | 9.10e-01 |
| GO:BP | GO:1903595 | positive regulation of histamine secretion by mast cell | 1 | 4 | 9.10e-01 |
| GO:BP | GO:1904582 | positive regulation of intracellular mRNA localization | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0071464 | cellular response to hydrostatic pressure | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0060452 | positive regulation of cardiac muscle contraction | 1 | 7 | 9.10e-01 |
| GO:BP | GO:1905907 | negative regulation of amyloid fibril formation | 4 | 10 | 9.10e-01 |
| GO:BP | GO:0044029 | positive regulation of gene expression via CpG island demethylation | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0090192 | regulation of glomerulus development | 1 | 4 | 9.10e-01 |
| GO:BP | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 12 | 194 | 9.10e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 36 | 726 | 9.10e-01 |
| GO:BP | GO:0006369 | termination of RNA polymerase II transcription | 3 | 8 | 9.10e-01 |
| GO:BP | GO:0048752 | semicircular canal morphogenesis | 1 | 5 | 9.10e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 2 | 5 | 9.10e-01 |
| GO:BP | GO:0016043 | cellular component organization | 3 | 5076 | 9.10e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 9.11e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 6 | 147 | 9.11e-01 |
| GO:BP | GO:1902499 | positive regulation of protein autoubiquitination | 1 | 2 | 9.12e-01 |
| GO:BP | GO:1903276 | regulation of sodium ion export across plasma membrane | 1 | 4 | 9.12e-01 |
| GO:BP | GO:1903278 | positive regulation of sodium ion export across plasma membrane | 1 | 4 | 9.12e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 9.12e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 9.12e-01 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 1 | 22 | 9.12e-01 |
| GO:BP | GO:1905155 | positive regulation of membrane invagination | 3 | 11 | 9.13e-01 |
| GO:BP | GO:0060100 | positive regulation of phagocytosis, engulfment | 3 | 11 | 9.13e-01 |
| GO:BP | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation | 1 | 2 | 9.13e-01 |
| GO:BP | GO:2000328 | regulation of T-helper 17 cell lineage commitment | 1 | 1 | 9.13e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 15 | 30 | 9.13e-01 |
| GO:BP | GO:0045797 | positive regulation of intestinal cholesterol absorption | 1 | 1 | 9.13e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 18 | 41 | 9.14e-01 |
| GO:BP | GO:0061737 | leukotriene signaling pathway | 1 | 1 | 9.14e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 9.14e-01 |
| GO:BP | GO:1904715 | negative regulation of chaperone-mediated autophagy | 1 | 3 | 9.14e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 11 | 47 | 9.14e-01 |
| GO:BP | GO:0032502 | developmental process | 3 | 4494 | 9.14e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 4 | 121 | 9.14e-01 |
| GO:BP | GO:0044467 | glial cell-derived neurotrophic factor production | 1 | 2 | 9.14e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 1 | 484 | 9.14e-01 |
| GO:BP | GO:1990809 | endoplasmic reticulum tubular network membrane organization | 1 | 4 | 9.14e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 1 | 13 | 9.14e-01 |
| GO:BP | GO:2000295 | regulation of hydrogen peroxide catabolic process | 1 | 2 | 9.14e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 2 | 4 | 9.14e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 3 | 45 | 9.14e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 2 | 6 | 9.14e-01 |
| GO:BP | GO:0051581 | negative regulation of neurotransmitter uptake | 1 | 2 | 9.14e-01 |
| GO:BP | GO:1900166 | regulation of glial cell-derived neurotrophic factor production | 1 | 2 | 9.14e-01 |
| GO:BP | GO:0051612 | negative regulation of serotonin uptake | 1 | 2 | 9.14e-01 |
| GO:BP | GO:1900168 | positive regulation of glial cell-derived neurotrophic factor production | 1 | 2 | 9.14e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 1 | 147 | 9.15e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 9 | 160 | 9.15e-01 |
| GO:BP | GO:0042478 | regulation of eye photoreceptor cell development | 1 | 1 | 9.15e-01 |
| GO:BP | GO:0006776 | vitamin A metabolic process | 2 | 3 | 9.15e-01 |
| GO:BP | GO:1903392 | negative regulation of adherens junction organization | 1 | 3 | 9.15e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 2 | 2293 | 9.15e-01 |
| GO:BP | GO:1905279 | regulation of retrograde transport, endosome to Golgi | 2 | 6 | 9.15e-01 |
| GO:BP | GO:0038091 | positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway | 1 | 3 | 9.15e-01 |
| GO:BP | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway | 1 | 3 | 9.15e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 2 | 18 | 9.15e-01 |
| GO:BP | GO:2000726 | negative regulation of cardiac muscle cell differentiation | 3 | 8 | 9.15e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 2 | 69 | 9.15e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 1 | 17 | 9.15e-01 |
| GO:BP | GO:0032743 | positive regulation of interleukin-2 production | 2 | 20 | 9.15e-01 |
| GO:BP | GO:0035701 | hematopoietic stem cell migration | 2 | 6 | 9.15e-01 |
| GO:BP | GO:0015917 | aminophospholipid transport | 3 | 8 | 9.15e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 2 | 89 | 9.15e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 11 | 118 | 9.15e-01 |
| GO:BP | GO:0097638 | L-arginine import across plasma membrane | 2 | 4 | 9.16e-01 |
| GO:BP | GO:0097749 | membrane tubulation | 1 | 4 | 9.16e-01 |
| GO:BP | GO:0006353 | DNA-templated transcription termination | 1 | 22 | 9.16e-01 |
| GO:BP | GO:0071715 | icosanoid transport | 2 | 24 | 9.16e-01 |
| GO:BP | GO:1903412 | response to bile acid | 2 | 3 | 9.16e-01 |
| GO:BP | GO:0061589 | calcium activated phosphatidylserine scrambling | 1 | 3 | 9.16e-01 |
| GO:BP | GO:0061591 | calcium activated galactosylceramide scrambling | 1 | 2 | 9.16e-01 |
| GO:BP | GO:0021800 | cerebral cortex tangential migration | 1 | 4 | 9.16e-01 |
| GO:BP | GO:0022417 | protein maturation by protein folding | 2 | 8 | 9.17e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 12 | 148 | 9.17e-01 |
| GO:BP | GO:2000297 | negative regulation of synapse maturation | 1 | 1 | 9.17e-01 |
| GO:BP | GO:0009635 | response to herbicide | 1 | 3 | 9.17e-01 |
| GO:BP | GO:0019563 | glycerol catabolic process | 1 | 5 | 9.17e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 4 | 49 | 9.18e-01 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 6 | 19 | 9.18e-01 |
| GO:BP | GO:0030728 | ovulation | 3 | 14 | 9.18e-01 |
| GO:BP | GO:0002370 | natural killer cell cytokine production | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0002727 | regulation of natural killer cell cytokine production | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 9.19e-01 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 6 | 13 | 9.19e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 2 | 93 | 9.19e-01 |
| GO:BP | GO:0002001 | renin secretion into blood stream | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 1 | 36 | 9.19e-01 |
| GO:BP | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb | 2 | 4 | 9.19e-01 |
| GO:BP | GO:0045627 | positive regulation of T-helper 1 cell differentiation | 1 | 3 | 9.20e-01 |
| GO:BP | GO:0140650 | radial glia-guided pyramidal neuron migration | 1 | 4 | 9.20e-01 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 1 | 18 | 9.20e-01 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 1 | 24 | 9.20e-01 |
| GO:BP | GO:0033091 | positive regulation of immature T cell proliferation | 1 | 2 | 9.20e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 11 | 119 | 9.20e-01 |
| GO:BP | GO:0033092 | positive regulation of immature T cell proliferation in thymus | 1 | 2 | 9.20e-01 |
| GO:BP | GO:1903792 | negative regulation of monoatomic anion transport | 1 | 3 | 9.20e-01 |
| GO:BP | GO:0045343 | regulation of MHC class I biosynthetic process | 1 | 3 | 9.20e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 1 | 74 | 9.20e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 9.20e-01 |
| GO:BP | GO:0045341 | MHC class I biosynthetic process | 1 | 3 | 9.20e-01 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 2 | 8 | 9.20e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 50 | 1345 | 9.20e-01 |
| GO:BP | GO:0044406 | adhesion of symbiont to host | 1 | 10 | 9.20e-01 |
| GO:BP | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome | 4 | 13 | 9.20e-01 |
| GO:BP | GO:1905167 | positive regulation of lysosomal protein catabolic process | 1 | 4 | 9.21e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 1 | 17 | 9.21e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 2 | 36 | 9.21e-01 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 2 | 4 | 9.21e-01 |
| GO:BP | GO:0070640 | vitamin D3 metabolic process | 2 | 2 | 9.21e-01 |
| GO:BP | GO:0150007 | clathrin-dependent synaptic vesicle endocytosis | 1 | 2 | 9.21e-01 |
| GO:BP | GO:0001502 | cartilage condensation | 3 | 12 | 9.22e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 609 | 9.22e-01 |
| GO:BP | GO:0042832 | defense response to protozoan | 3 | 10 | 9.22e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 1 | 20 | 9.22e-01 |
| GO:BP | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity | 2 | 10 | 9.22e-01 |
| GO:BP | GO:0034694 | response to prostaglandin | 1 | 21 | 9.22e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 9.22e-01 |
| GO:BP | GO:0046055 | dGMP catabolic process | 1 | 1 | 9.23e-01 |
| GO:BP | GO:0044211 | CTP salvage | 1 | 3 | 9.23e-01 |
| GO:BP | GO:0002082 | regulation of oxidative phosphorylation | 2 | 19 | 9.23e-01 |
| GO:BP | GO:0090026 | positive regulation of monocyte chemotaxis | 1 | 7 | 9.23e-01 |
| GO:BP | GO:0035461 | vitamin transmembrane transport | 1 | 14 | 9.23e-01 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 1 | 10 | 9.23e-01 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 1 | 7 | 9.24e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 9.24e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3 | 5274 | 9.24e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 4 | 10 | 9.24e-01 |
| GO:BP | GO:0034158 | toll-like receptor 8 signaling pathway | 1 | 3 | 9.24e-01 |
| GO:BP | GO:0050892 | intestinal absorption | 2 | 22 | 9.24e-01 |
| GO:BP | GO:0009195 | pyrimidine ribonucleoside diphosphate catabolic process | 1 | 3 | 9.25e-01 |
| GO:BP | GO:0042360 | vitamin E metabolic process | 1 | 2 | 9.25e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 8 | 24 | 9.25e-01 |
| GO:BP | GO:0009140 | pyrimidine nucleoside diphosphate catabolic process | 1 | 3 | 9.25e-01 |
| GO:BP | GO:0006256 | UDP catabolic process | 1 | 3 | 9.25e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 12 | 150 | 9.25e-01 |
| GO:BP | GO:1905048 | regulation of metallopeptidase activity | 6 | 14 | 9.25e-01 |
| GO:BP | GO:0002693 | positive regulation of cellular extravasation | 6 | 13 | 9.25e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 2 | 18 | 9.25e-01 |
| GO:BP | GO:0035723 | interleukin-15-mediated signaling pathway | 1 | 2 | 9.25e-01 |
| GO:BP | GO:0071350 | cellular response to interleukin-15 | 1 | 2 | 9.25e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 9.25e-01 |
| GO:BP | GO:1904732 | regulation of electron transfer activity | 3 | 8 | 9.25e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 9.25e-01 |
| GO:BP | GO:0002674 | negative regulation of acute inflammatory response | 1 | 5 | 9.25e-01 |
| GO:BP | GO:0021801 | cerebral cortex radial glia-guided migration | 9 | 22 | 9.26e-01 |
| GO:BP | GO:0035188 | hatching | 10 | 23 | 9.26e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 10 | 23 | 9.26e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 2 | 19 | 9.26e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 14 | 43 | 9.26e-01 |
| GO:BP | GO:0002353 | plasma kallikrein-kinin cascade | 1 | 2 | 9.26e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 10 | 23 | 9.26e-01 |
| GO:BP | GO:0002254 | kinin cascade | 1 | 2 | 9.26e-01 |
| GO:BP | GO:0022030 | telencephalon glial cell migration | 9 | 22 | 9.26e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 4 | 4045 | 9.26e-01 |
| GO:BP | GO:1904959 | regulation of cytochrome-c oxidase activity | 2 | 7 | 9.26e-01 |
| GO:BP | GO:1905526 | regulation of Golgi lumen acidification | 1 | 3 | 9.26e-01 |
| GO:BP | GO:0070471 | uterine smooth muscle contraction | 1 | 6 | 9.26e-01 |
| GO:BP | GO:0014827 | intestine smooth muscle contraction | 1 | 3 | 9.27e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 4 | 639 | 9.27e-01 |
| GO:BP | GO:0032226 | positive regulation of synaptic transmission, dopaminergic | 1 | 3 | 9.27e-01 |
| GO:BP | GO:0044872 | lipoprotein localization | 2 | 15 | 9.27e-01 |
| GO:BP | GO:0072540 | T-helper 17 cell lineage commitment | 2 | 3 | 9.27e-01 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 1 | 8 | 9.27e-01 |
| GO:BP | GO:2000556 | positive regulation of T-helper 1 cell cytokine production | 1 | 2 | 9.27e-01 |
| GO:BP | GO:2000554 | regulation of T-helper 1 cell cytokine production | 1 | 2 | 9.27e-01 |
| GO:BP | GO:0035744 | T-helper 1 cell cytokine production | 1 | 2 | 9.27e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 9.27e-01 |
| GO:BP | GO:0006030 | chitin metabolic process | 1 | 2 | 9.27e-01 |
| GO:BP | GO:0006032 | chitin catabolic process | 1 | 2 | 9.27e-01 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 3 | 9 | 9.28e-01 |
| GO:BP | GO:0006867 | asparagine transport | 1 | 3 | 9.28e-01 |
| GO:BP | GO:0015866 | ADP transport | 2 | 6 | 9.28e-01 |
| GO:BP | GO:1900148 | negative regulation of Schwann cell migration | 1 | 3 | 9.28e-01 |
| GO:BP | GO:0014012 | peripheral nervous system axon regeneration | 1 | 6 | 9.28e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 9.28e-01 |
| GO:BP | GO:0030647 | aminoglycoside antibiotic metabolic process | 1 | 7 | 9.28e-01 |
| GO:BP | GO:0030638 | polyketide metabolic process | 1 | 8 | 9.28e-01 |
| GO:BP | GO:0044598 | doxorubicin metabolic process | 1 | 8 | 9.28e-01 |
| GO:BP | GO:0097241 | hematopoietic stem cell migration to bone marrow | 1 | 5 | 9.28e-01 |
| GO:BP | GO:0007608 | sensory perception of smell | 1 | 16 | 9.28e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 4 | 8 | 9.29e-01 |
| GO:BP | GO:0106005 | RNA 5’-cap (guanine-N7)-methylation | 1 | 2 | 9.29e-01 |
| GO:BP | GO:1903980 | positive regulation of microglial cell activation | 1 | 2 | 9.29e-01 |
| GO:BP | GO:0002424 | T cell mediated immune response to tumor cell | 3 | 6 | 9.29e-01 |
| GO:BP | GO:1903899 | positive regulation of PERK-mediated unfolded protein response | 1 | 3 | 9.29e-01 |
| GO:BP | GO:0030259 | lipid glycosylation | 1 | 6 | 9.29e-01 |
| GO:BP | GO:0090091 | positive regulation of extracellular matrix disassembly | 3 | 6 | 9.29e-01 |
| GO:BP | GO:1905162 | regulation of phagosome maturation | 1 | 2 | 9.29e-01 |
| GO:BP | GO:0051918 | negative regulation of fibrinolysis | 5 | 8 | 9.29e-01 |
| GO:BP | GO:0022900 | electron transport chain | 6 | 142 | 9.29e-01 |
| GO:BP | GO:0035709 | memory T cell activation | 1 | 4 | 9.29e-01 |
| GO:BP | GO:0071073 | positive regulation of phospholipid biosynthetic process | 2 | 7 | 9.29e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 10 | 112 | 9.30e-01 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 1 | 5 | 9.30e-01 |
| GO:BP | GO:0034776 | response to histamine | 4 | 5 | 9.30e-01 |
| GO:BP | GO:0015888 | thiamine transport | 3 | 3 | 9.30e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 1 | 13 | 9.30e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 4 | 11 | 9.30e-01 |
| GO:BP | GO:0010041 | response to iron(III) ion | 1 | 4 | 9.30e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 9.30e-01 |
| GO:BP | GO:0044241 | lipid digestion | 1 | 10 | 9.30e-01 |
| GO:BP | GO:1905771 | negative regulation of mesodermal cell differentiation | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0042662 | negative regulation of mesodermal cell fate specification | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 5 | 15 | 9.30e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 2 | 37 | 9.30e-01 |
| GO:BP | GO:0035562 | negative regulation of chromatin binding | 3 | 8 | 9.30e-01 |
| GO:BP | GO:0071800 | podosome assembly | 3 | 13 | 9.31e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 9 | 22 | 9.31e-01 |
| GO:BP | GO:0045656 | negative regulation of monocyte differentiation | 1 | 4 | 9.31e-01 |
| GO:BP | GO:0043353 | enucleate erythrocyte differentiation | 1 | 9 | 9.31e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 5 | 33 | 9.31e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 10 | 52 | 9.31e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 3 | 68 | 9.32e-01 |
| GO:BP | GO:0009311 | oligosaccharide metabolic process | 10 | 43 | 9.32e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 9.32e-01 |
| GO:BP | GO:0043382 | positive regulation of memory T cell differentiation | 1 | 3 | 9.32e-01 |
| GO:BP | GO:0097752 | regulation of DNA stability | 1 | 3 | 9.32e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 9.32e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 1 | 53 | 9.32e-01 |
| GO:BP | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells | 1 | 4 | 9.32e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 12 | 457 | 9.32e-01 |
| GO:BP | GO:0098597 | observational learning | 2 | 5 | 9.32e-01 |
| GO:BP | GO:1903976 | negative regulation of glial cell migration | 2 | 5 | 9.32e-01 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 7 | 21 | 9.33e-01 |
| GO:BP | GO:0071798 | response to prostaglandin D | 1 | 2 | 9.33e-01 |
| GO:BP | GO:0071799 | cellular response to prostaglandin D stimulus | 1 | 2 | 9.33e-01 |
| GO:BP | GO:0032225 | regulation of synaptic transmission, dopaminergic | 1 | 5 | 9.33e-01 |
| GO:BP | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 4 | 9.33e-01 |
| GO:BP | GO:0006578 | amino-acid betaine biosynthetic process | 2 | 5 | 9.33e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 9.33e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 1 | 47 | 9.34e-01 |
| GO:BP | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response | 1 | 2 | 9.34e-01 |
| GO:BP | GO:0060298 | positive regulation of sarcomere organization | 1 | 4 | 9.34e-01 |
| GO:BP | GO:0042308 | negative regulation of protein import into nucleus | 1 | 13 | 9.34e-01 |
| GO:BP | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 1 | 9.34e-01 |
| GO:BP | GO:0051604 | protein maturation | 4 | 259 | 9.35e-01 |
| GO:BP | GO:0010039 | response to iron ion | 1 | 24 | 9.35e-01 |
| GO:BP | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain | 1 | 1 | 9.35e-01 |
| GO:BP | GO:0090258 | negative regulation of mitochondrial fission | 1 | 3 | 9.36e-01 |
| GO:BP | GO:0019740 | nitrogen utilization | 1 | 4 | 9.36e-01 |
| GO:BP | GO:0043932 | ossification involved in bone remodeling | 1 | 2 | 9.36e-01 |
| GO:BP | GO:1903350 | response to dopamine | 1 | 48 | 9.36e-01 |
| GO:BP | GO:0071803 | positive regulation of podosome assembly | 1 | 6 | 9.37e-01 |
| GO:BP | GO:0070671 | response to interleukin-12 | 2 | 6 | 9.37e-01 |
| GO:BP | GO:0002486 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent | 1 | 3 | 9.37e-01 |
| GO:BP | GO:0051977 | lysophospholipid transport | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0032790 | ribosome disassembly | 4 | 11 | 9.37e-01 |
| GO:BP | GO:0071486 | cellular response to high light intensity | 1 | 1 | 9.37e-01 |
| GO:BP | GO:0048563 | post-embryonic animal organ morphogenesis | 1 | 8 | 9.37e-01 |
| GO:BP | GO:0002716 | negative regulation of natural killer cell mediated immunity | 2 | 10 | 9.37e-01 |
| GO:BP | GO:0031914 | negative regulation of synaptic plasticity | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 6 | 91 | 9.37e-01 |
| GO:BP | GO:0016573 | histone acetylation | 11 | 143 | 9.37e-01 |
| GO:BP | GO:0008078 | mesodermal cell migration | 2 | 3 | 9.37e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 2 | 20 | 9.37e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 22 | 95 | 9.37e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 2 | 7 | 9.37e-01 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 1 | 11 | 9.37e-01 |
| GO:BP | GO:0061439 | kidney vasculature morphogenesis | 1 | 7 | 9.37e-01 |
| GO:BP | GO:0061438 | renal system vasculature morphogenesis | 1 | 7 | 9.37e-01 |
| GO:BP | GO:0070841 | inclusion body assembly | 4 | 22 | 9.37e-01 |
| GO:BP | GO:0019293 | tyrosine biosynthetic process, by oxidation of phenylalanine | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0032819 | positive regulation of natural killer cell proliferation | 1 | 4 | 9.37e-01 |
| GO:BP | GO:0006571 | tyrosine biosynthetic process | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 9.37e-01 |
| GO:BP | GO:0009095 | aromatic amino acid family biosynthetic process, prephenate pathway | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0000965 | mitochondrial RNA 3’-end processing | 1 | 5 | 9.37e-01 |
| GO:BP | GO:0045989 | positive regulation of striated muscle contraction | 2 | 12 | 9.37e-01 |
| GO:BP | GO:0071211 | protein targeting to vacuole involved in autophagy | 1 | 4 | 9.37e-01 |
| GO:BP | GO:0010755 | regulation of plasminogen activation | 7 | 16 | 9.37e-01 |
| GO:BP | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination | 1 | 4 | 9.37e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 6 | 29 | 9.37e-01 |
| GO:BP | GO:0006069 | ethanol oxidation | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0032489 | regulation of Cdc42 protein signal transduction | 1 | 6 | 9.37e-01 |
| GO:BP | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity | 1 | 3 | 9.37e-01 |
| GO:BP | GO:0007290 | spermatid nucleus elongation | 1 | 1 | 9.37e-01 |
| GO:BP | GO:0072017 | distal tubule development | 2 | 6 | 9.37e-01 |
| GO:BP | GO:0002921 | negative regulation of humoral immune response | 3 | 10 | 9.37e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 6 | 447 | 9.37e-01 |
| GO:BP | GO:0000432 | positive regulation of transcription from RNA polymerase II promoter by glucose | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly | 1 | 3 | 9.37e-01 |
| GO:BP | GO:1903431 | positive regulation of cell maturation | 1 | 6 | 9.37e-01 |
| GO:BP | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose | 1 | 2 | 9.37e-01 |
| GO:BP | GO:2000857 | positive regulation of mineralocorticoid secretion | 1 | 3 | 9.37e-01 |
| GO:BP | GO:1900193 | regulation of oocyte maturation | 1 | 5 | 9.37e-01 |
| GO:BP | GO:2000860 | positive regulation of aldosterone secretion | 1 | 3 | 9.37e-01 |
| GO:BP | GO:1901164 | negative regulation of trophoblast cell migration | 1 | 4 | 9.37e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 2 | 8 | 9.37e-01 |
| GO:BP | GO:0032380 | regulation of intracellular sterol transport | 1 | 6 | 9.37e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 17 | 96 | 9.37e-01 |
| GO:BP | GO:0032383 | regulation of intracellular cholesterol transport | 1 | 6 | 9.37e-01 |
| GO:BP | GO:0044060 | regulation of endocrine process | 1 | 24 | 9.37e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 1 | 39 | 9.37e-01 |
| GO:BP | GO:0035469 | determination of pancreatic left/right asymmetry | 1 | 3 | 9.37e-01 |
| GO:BP | GO:1990705 | cholangiocyte proliferation | 1 | 3 | 9.37e-01 |
| GO:BP | GO:0050434 | positive regulation of viral transcription | 1 | 3 | 9.37e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 9.37e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 2 | 47 | 9.37e-01 |
| GO:BP | GO:0072015 | podocyte development | 1 | 9 | 9.37e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 11 | 51 | 9.38e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 3 | 245 | 9.38e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 1 | 14 | 9.38e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 2 | 68 | 9.38e-01 |
| GO:BP | GO:1902396 | protein localization to bicellular tight junction | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 2 | 10 | 9.38e-01 |
| GO:BP | GO:0000478 | endonucleolytic cleavage involved in rRNA processing | 2 | 17 | 9.38e-01 |
| GO:BP | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 17 | 9.38e-01 |
| GO:BP | GO:0090713 | immunological memory process | 2 | 7 | 9.38e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 22 | 106 | 9.38e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 10 | 27 | 9.38e-01 |
| GO:BP | GO:0051135 | positive regulation of NK T cell activation | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0030901 | midbrain development | 3 | 69 | 9.38e-01 |
| GO:BP | GO:0090361 | regulation of platelet-derived growth factor production | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0003130 | BMP signaling pathway involved in heart induction | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0090360 | platelet-derived growth factor production | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0072125 | negative regulation of glomerular mesangial cell proliferation | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0003133 | endodermal-mesodermal cell signaling | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0035990 | tendon cell differentiation | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0003134 | endodermal-mesodermal cell signaling involved in heart induction | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0035993 | deltoid tuberosity development | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 9.38e-01 |
| GO:BP | GO:1905310 | regulation of cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0048143 | astrocyte activation | 7 | 12 | 9.38e-01 |
| GO:BP | GO:0060128 | corticotropin hormone secreting cell differentiation | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0019405 | alditol catabolic process | 1 | 6 | 9.38e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 8 | 33 | 9.38e-01 |
| GO:BP | GO:0060279 | positive regulation of ovulation | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0072198 | mesenchymal cell proliferation involved in ureter development | 1 | 1 | 9.38e-01 |
| GO:BP | GO:0035992 | tendon formation | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0072199 | regulation of mesenchymal cell proliferation involved in ureter development | 1 | 1 | 9.38e-01 |
| GO:BP | GO:0001816 | cytokine production | 6 | 451 | 9.38e-01 |
| GO:BP | GO:1905312 | positive regulation of cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0001562 | response to protozoan | 2 | 11 | 9.38e-01 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 4 | 15 | 9.38e-01 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 4 | 15 | 9.38e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 3 | 47 | 9.38e-01 |
| GO:BP | GO:2000391 | positive regulation of neutrophil extravasation | 2 | 6 | 9.38e-01 |
| GO:BP | GO:0010982 | regulation of high-density lipoprotein particle clearance | 1 | 2 | 9.38e-01 |
| GO:BP | GO:0014051 | gamma-aminobutyric acid secretion | 1 | 8 | 9.38e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 3 | 9 | 9.38e-01 |
| GO:BP | GO:0022401 | negative adaptation of signaling pathway | 5 | 15 | 9.38e-01 |
| GO:BP | GO:0045906 | negative regulation of vasoconstriction | 1 | 6 | 9.38e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 3 | 42 | 9.38e-01 |
| GO:BP | GO:0019511 | peptidyl-proline hydroxylation | 3 | 14 | 9.38e-01 |
| GO:BP | GO:0071316 | cellular response to nicotine | 2 | 4 | 9.38e-01 |
| GO:BP | GO:0046034 | ATP metabolic process | 5 | 116 | 9.38e-01 |
| GO:BP | GO:0007030 | Golgi organization | 3 | 137 | 9.39e-01 |
| GO:BP | GO:0098963 | dendritic transport of messenger ribonucleoprotein complex | 1 | 4 | 9.39e-01 |
| GO:BP | GO:0098961 | dendritic transport of ribonucleoprotein complex | 1 | 4 | 9.39e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 6 | 78 | 9.39e-01 |
| GO:BP | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway | 2 | 4 | 9.39e-01 |
| GO:BP | GO:0031342 | negative regulation of cell killing | 3 | 11 | 9.39e-01 |
| GO:BP | GO:0010524 | positive regulation of calcium ion transport into cytosol | 3 | 7 | 9.39e-01 |
| GO:BP | GO:0006583 | melanin biosynthetic process from tyrosine | 1 | 1 | 9.39e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 9.39e-01 |
| GO:BP | GO:1903179 | regulation of dopamine biosynthetic process | 1 | 3 | 9.39e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 1 | 581 | 9.39e-01 |
| GO:BP | GO:0048535 | lymph node development | 4 | 11 | 9.39e-01 |
| GO:BP | GO:0010624 | regulation of Schwann cell proliferation | 3 | 7 | 9.39e-01 |
| GO:BP | GO:1905205 | positive regulation of connective tissue replacement | 1 | 3 | 9.39e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 10 | 62 | 9.39e-01 |
| GO:BP | GO:0014010 | Schwann cell proliferation | 3 | 7 | 9.39e-01 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 1 | 11 | 9.39e-01 |
| GO:BP | GO:0009441 | glycolate metabolic process | 1 | 1 | 9.39e-01 |
| GO:BP | GO:1901387 | positive regulation of voltage-gated calcium channel activity | 5 | 13 | 9.39e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 9.39e-01 |
| GO:BP | GO:0044000 | movement in host | 1 | 138 | 9.39e-01 |
| GO:BP | GO:2000109 | regulation of macrophage apoptotic process | 1 | 6 | 9.39e-01 |
| GO:BP | GO:0110076 | negative regulation of ferroptosis | 1 | 3 | 9.39e-01 |
| GO:BP | GO:2000211 | regulation of glutamate metabolic process | 1 | 1 | 9.39e-01 |
| GO:BP | GO:0043416 | regulation of skeletal muscle tissue regeneration | 1 | 5 | 9.39e-01 |
| GO:BP | GO:0071484 | cellular response to light intensity | 2 | 4 | 9.39e-01 |
| GO:BP | GO:1903238 | positive regulation of leukocyte tethering or rolling | 1 | 5 | 9.40e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 9.40e-01 |
| GO:BP | GO:0097052 | L-kynurenine metabolic process | 1 | 4 | 9.40e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 3 | 141 | 9.40e-01 |
| GO:BP | GO:0005984 | disaccharide metabolic process | 1 | 5 | 9.40e-01 |
| GO:BP | GO:0010984 | regulation of lipoprotein particle clearance | 3 | 12 | 9.40e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 2 | 9 | 9.41e-01 |
| GO:BP | GO:1902931 | negative regulation of alcohol biosynthetic process | 1 | 10 | 9.41e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 2 | 76 | 9.41e-01 |
| GO:BP | GO:0070741 | response to interleukin-6 | 6 | 23 | 9.41e-01 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 2 | 5 | 9.41e-01 |
| GO:BP | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 2 | 10 | 9.41e-01 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 6 | 137 | 9.41e-01 |
| GO:BP | GO:0009584 | detection of visible light | 1 | 20 | 9.41e-01 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 2 | 11 | 9.41e-01 |
| GO:BP | GO:0015669 | gas transport | 1 | 6 | 9.41e-01 |
| GO:BP | GO:0060263 | regulation of respiratory burst | 3 | 10 | 9.41e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 9 | 106 | 9.41e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 2 | 46 | 9.41e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 6 | 168 | 9.41e-01 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 1 | 104 | 9.41e-01 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 11 | 9.41e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 1 | 220 | 9.41e-01 |
| GO:BP | GO:0010988 | regulation of low-density lipoprotein particle clearance | 2 | 8 | 9.41e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 1 | 7 | 9.41e-01 |
| GO:BP | GO:0032971 | regulation of muscle filament sliding | 1 | 3 | 9.41e-01 |
| GO:BP | GO:0150147 | cell-cell junction disassembly | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0031204 | post-translational protein targeting to membrane, translocation | 1 | 7 | 9.41e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 6 | 16 | 9.41e-01 |
| GO:BP | GO:0035815 | positive regulation of renal sodium excretion | 1 | 7 | 9.41e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 2 | 17 | 9.42e-01 |
| GO:BP | GO:0071224 | cellular response to peptidoglycan | 1 | 2 | 9.42e-01 |
| GO:BP | GO:0070901 | mitochondrial tRNA methylation | 1 | 4 | 9.42e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 9.42e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 9.42e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 2 | 7806 | 9.42e-01 |
| GO:BP | GO:0006772 | thiamine metabolic process | 1 | 2 | 9.43e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 9.43e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 5 | 271 | 9.43e-01 |
| GO:BP | GO:0002447 | eosinophil mediated immunity | 1 | 5 | 9.43e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 3 | 27 | 9.43e-01 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 7 | 18 | 9.43e-01 |
| GO:BP | GO:0032693 | negative regulation of interleukin-10 production | 1 | 7 | 9.43e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 2 | 77 | 9.43e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 9.44e-01 |
| GO:BP | GO:1904783 | positive regulation of NMDA glutamate receptor activity | 1 | 4 | 9.44e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 4 | 24 | 9.44e-01 |
| GO:BP | GO:0021631 | optic nerve morphogenesis | 1 | 2 | 9.44e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 10 | 400 | 9.44e-01 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 2 | 8 | 9.44e-01 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 2 | 5 | 9.45e-01 |
| GO:BP | GO:0072683 | T cell extravasation | 2 | 8 | 9.45e-01 |
| GO:BP | GO:0002827 | positive regulation of T-helper 1 type immune response | 3 | 5 | 9.45e-01 |
| GO:BP | GO:0034162 | toll-like receptor 9 signaling pathway | 1 | 12 | 9.45e-01 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 4 | 8 | 9.45e-01 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 4 | 10 | 9.45e-01 |
| GO:BP | GO:0007168 | receptor guanylyl cyclase signaling pathway | 1 | 5 | 9.46e-01 |
| GO:BP | GO:0035814 | negative regulation of renal sodium excretion | 1 | 2 | 9.46e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 2 | 250 | 9.46e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 9.46e-01 |
| GO:BP | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus | 2 | 7 | 9.46e-01 |
| GO:BP | GO:0048521 | negative regulation of behavior | 2 | 5 | 9.47e-01 |
| GO:BP | GO:0061024 | membrane organization | 9 | 671 | 9.47e-01 |
| GO:BP | GO:0060435 | bronchiole development | 1 | 2 | 9.47e-01 |
| GO:BP | GO:0002689 | negative regulation of leukocyte chemotaxis | 1 | 10 | 9.48e-01 |
| GO:BP | GO:0042693 | muscle cell fate commitment | 3 | 11 | 9.48e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 12 | 58 | 9.48e-01 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 1 | 9 | 9.48e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 3 | 61 | 9.48e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 2 | 317 | 9.48e-01 |
| GO:BP | GO:0006570 | tyrosine metabolic process | 1 | 6 | 9.48e-01 |
| GO:BP | GO:0001920 | negative regulation of receptor recycling | 1 | 3 | 9.49e-01 |
| GO:BP | GO:0001922 | B-1 B cell homeostasis | 1 | 4 | 9.49e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 2 | 129 | 9.49e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 1 | 190 | 9.49e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 2 | 4 | 9.49e-01 |
| GO:BP | GO:2000110 | negative regulation of macrophage apoptotic process | 1 | 3 | 9.49e-01 |
| GO:BP | GO:0072268 | pattern specification involved in metanephros development | 1 | 4 | 9.49e-01 |
| GO:BP | GO:0072131 | kidney mesenchyme morphogenesis | 1 | 4 | 9.49e-01 |
| GO:BP | GO:0072133 | metanephric mesenchyme morphogenesis | 1 | 4 | 9.49e-01 |
| GO:BP | GO:0072180 | mesonephric duct morphogenesis | 1 | 2 | 9.49e-01 |
| GO:BP | GO:0042448 | progesterone metabolic process | 1 | 13 | 9.49e-01 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 4 | 9.49e-01 |
| GO:BP | GO:1905153 | regulation of membrane invagination | 3 | 11 | 9.49e-01 |
| GO:BP | GO:0060099 | regulation of phagocytosis, engulfment | 3 | 11 | 9.49e-01 |
| GO:BP | GO:0098743 | cell aggregation | 1 | 12 | 9.49e-01 |
| GO:BP | GO:0015874 | norepinephrine transport | 2 | 9 | 9.49e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 10 | 81 | 9.49e-01 |
| GO:BP | GO:1990384 | hyaloid vascular plexus regression | 2 | 3 | 9.50e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 6 | 70 | 9.50e-01 |
| GO:BP | GO:0072679 | thymocyte migration | 1 | 1 | 9.50e-01 |
| GO:BP | GO:0035270 | endocrine system development | 2 | 86 | 9.50e-01 |
| GO:BP | GO:0032667 | regulation of interleukin-23 production | 2 | 6 | 9.50e-01 |
| GO:BP | GO:0032627 | interleukin-23 production | 2 | 6 | 9.50e-01 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 1 | 27 | 9.50e-01 |
| GO:BP | GO:0034157 | positive regulation of toll-like receptor 7 signaling pathway | 1 | 2 | 9.50e-01 |
| GO:BP | GO:0070197 | meiotic attachment of telomere to nuclear envelope | 1 | 1 | 9.50e-01 |
| GO:BP | GO:0033574 | response to testosterone | 1 | 29 | 9.50e-01 |
| GO:BP | GO:0044821 | meiotic telomere tethering at nuclear periphery | 1 | 1 | 9.50e-01 |
| GO:BP | GO:0030473 | nuclear migration along microtubule | 1 | 5 | 9.50e-01 |
| GO:BP | GO:0034398 | telomere tethering at nuclear periphery | 1 | 1 | 9.50e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 2 | 68 | 9.50e-01 |
| GO:BP | GO:0061590 | calcium activated phosphatidylcholine scrambling | 1 | 3 | 9.50e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 9.50e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 9.50e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 2 | 13 | 9.50e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 2 | 5 | 9.50e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 9.50e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 3 | 69 | 9.51e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 9.52e-01 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 1 | 31 | 9.52e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 22 | 122 | 9.52e-01 |
| GO:BP | GO:0033033 | negative regulation of myeloid cell apoptotic process | 5 | 10 | 9.53e-01 |
| GO:BP | GO:0001878 | response to yeast | 1 | 3 | 9.53e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 9 | 40 | 9.53e-01 |
| GO:BP | GO:0042756 | drinking behavior | 2 | 6 | 9.54e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 9.54e-01 |
| GO:BP | GO:0097305 | response to alcohol | 10 | 159 | 9.54e-01 |
| GO:BP | GO:2000813 | negative regulation of barbed-end actin filament capping | 1 | 3 | 9.54e-01 |
| GO:BP | GO:0021798 | forebrain dorsal/ventral pattern formation | 1 | 2 | 9.54e-01 |
| GO:BP | GO:0140367 | antibacterial innate immune response | 1 | 2 | 9.54e-01 |
| GO:BP | GO:0046907 | intracellular transport | 32 | 1453 | 9.54e-01 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 5 | 29 | 9.56e-01 |
| GO:BP | GO:0043316 | cytotoxic T cell degranulation | 1 | 2 | 9.56e-01 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 4 | 11 | 9.56e-01 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 4 | 11 | 9.56e-01 |
| GO:BP | GO:0046884 | follicle-stimulating hormone secretion | 1 | 4 | 9.57e-01 |
| GO:BP | GO:0099515 | actin filament-based transport | 7 | 15 | 9.57e-01 |
| GO:BP | GO:1900424 | regulation of defense response to bacterium | 3 | 12 | 9.57e-01 |
| GO:BP | GO:0008594 | photoreceptor cell morphogenesis | 1 | 4 | 9.57e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 3 | 70 | 9.57e-01 |
| GO:BP | GO:0021877 | forebrain neuron fate commitment | 1 | 1 | 9.57e-01 |
| GO:BP | GO:1990051 | activation of protein kinase C activity | 1 | 3 | 9.57e-01 |
| GO:BP | GO:1903659 | regulation of complement-dependent cytotoxicity | 1 | 2 | 9.57e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 2 | 178 | 9.57e-01 |
| GO:BP | GO:2001054 | negative regulation of mesenchymal cell apoptotic process | 1 | 3 | 9.58e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 9.58e-01 |
| GO:BP | GO:0034378 | chylomicron assembly | 1 | 3 | 9.58e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 5 | 51 | 9.58e-01 |
| GO:BP | GO:0061004 | pattern specification involved in kidney development | 3 | 6 | 9.58e-01 |
| GO:BP | GO:1904480 | positive regulation of intestinal absorption | 1 | 1 | 9.58e-01 |
| GO:BP | GO:0072048 | renal system pattern specification | 3 | 6 | 9.58e-01 |
| GO:BP | GO:1904731 | positive regulation of intestinal lipid absorption | 1 | 1 | 9.58e-01 |
| GO:BP | GO:0033004 | negative regulation of mast cell activation | 1 | 4 | 9.58e-01 |
| GO:BP | GO:0048550 | negative regulation of pinocytosis | 1 | 4 | 9.58e-01 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 1 | 5 | 9.58e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 2 | 18 | 9.58e-01 |
| GO:BP | GO:0006382 | adenosine to inosine editing | 1 | 4 | 9.58e-01 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 2 | 122 | 9.59e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 9 | 160 | 9.59e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 1 | 154 | 9.59e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 4 | 15 | 9.59e-01 |
| GO:BP | GO:0014889 | muscle atrophy | 4 | 9 | 9.59e-01 |
| GO:BP | GO:2000468 | regulation of peroxidase activity | 1 | 2 | 9.59e-01 |
| GO:BP | GO:0042321 | negative regulation of circadian sleep/wake cycle, sleep | 1 | 2 | 9.59e-01 |
| GO:BP | GO:1901895 | negative regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 3 | 9.59e-01 |
| GO:BP | GO:0032277 | negative regulation of gonadotropin secretion | 1 | 4 | 9.59e-01 |
| GO:BP | GO:0051611 | regulation of serotonin uptake | 1 | 3 | 9.59e-01 |
| GO:BP | GO:0070857 | regulation of bile acid biosynthetic process | 3 | 7 | 9.59e-01 |
| GO:BP | GO:0048712 | negative regulation of astrocyte differentiation | 3 | 9 | 9.59e-01 |
| GO:BP | GO:0014850 | response to muscle activity | 5 | 17 | 9.59e-01 |
| GO:BP | GO:0060732 | positive regulation of inositol phosphate biosynthetic process | 1 | 5 | 9.59e-01 |
| GO:BP | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process | 1 | 4 | 9.59e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 9.60e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 2 | 15 | 9.60e-01 |
| GO:BP | GO:0002553 | histamine secretion by mast cell | 2 | 8 | 9.60e-01 |
| GO:BP | GO:2000425 | regulation of apoptotic cell clearance | 2 | 7 | 9.60e-01 |
| GO:BP | GO:0048251 | elastic fiber assembly | 1 | 12 | 9.60e-01 |
| GO:BP | GO:0140507 | granzyme-mediated programmed cell death signaling pathway | 2 | 3 | 9.60e-01 |
| GO:BP | GO:0002349 | histamine production involved in inflammatory response | 2 | 8 | 9.60e-01 |
| GO:BP | GO:0009233 | menaquinone metabolic process | 1 | 1 | 9.60e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 10 | 71 | 9.60e-01 |
| GO:BP | GO:0002441 | histamine secretion involved in inflammatory response | 2 | 8 | 9.60e-01 |
| GO:BP | GO:0048633 | positive regulation of skeletal muscle tissue growth | 1 | 6 | 9.60e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 6 | 27 | 9.60e-01 |
| GO:BP | GO:0018243 | protein O-linked glycosylation via threonine | 2 | 7 | 9.60e-01 |
| GO:BP | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 6 | 9.60e-01 |
| GO:BP | GO:0006897 | endocytosis | 3 | 527 | 9.60e-01 |
| GO:BP | GO:0021819 | layer formation in cerebral cortex | 4 | 12 | 9.60e-01 |
| GO:BP | GO:1905247 | positive regulation of aspartic-type peptidase activity | 1 | 8 | 9.60e-01 |
| GO:BP | GO:0007492 | endoderm development | 25 | 65 | 9.61e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 9.61e-01 |
| GO:BP | GO:1902563 | regulation of neutrophil activation | 2 | 3 | 9.61e-01 |
| GO:BP | GO:0045851 | pH reduction | 1 | 1 | 9.61e-01 |
| GO:BP | GO:0061314 | Notch signaling involved in heart development | 1 | 9 | 9.61e-01 |
| GO:BP | GO:0072378 | blood coagulation, fibrin clot formation | 1 | 7 | 9.61e-01 |
| GO:BP | GO:0060439 | trachea morphogenesis | 1 | 7 | 9.61e-01 |
| GO:BP | GO:0010961 | intracellular magnesium ion homeostasis | 1 | 4 | 9.62e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 1 | 55 | 9.62e-01 |
| GO:BP | GO:0032817 | regulation of natural killer cell proliferation | 1 | 4 | 9.62e-01 |
| GO:BP | GO:0002835 | negative regulation of response to tumor cell | 1 | 2 | 9.62e-01 |
| GO:BP | GO:0002838 | negative regulation of immune response to tumor cell | 1 | 2 | 9.62e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 21 | 1238 | 9.62e-01 |
| GO:BP | GO:0070092 | regulation of glucagon secretion | 1 | 3 | 9.62e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 1 | 11 | 9.62e-01 |
| GO:BP | GO:0070091 | glucagon secretion | 1 | 3 | 9.62e-01 |
| GO:BP | GO:0009312 | oligosaccharide biosynthetic process | 3 | 15 | 9.62e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 9 | 28 | 9.62e-01 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 2 | 9 | 9.62e-01 |
| GO:BP | GO:0061092 | positive regulation of phospholipid translocation | 1 | 4 | 9.62e-01 |
| GO:BP | GO:0035696 | monocyte extravasation | 2 | 5 | 9.62e-01 |
| GO:BP | GO:0007060 | male meiosis chromosome segregation | 1 | 1 | 9.63e-01 |
| GO:BP | GO:0042026 | protein refolding | 1 | 26 | 9.63e-01 |
| GO:BP | GO:0071910 | determination of liver left/right asymmetry | 1 | 4 | 9.63e-01 |
| GO:BP | GO:0045950 | negative regulation of mitotic recombination | 1 | 2 | 9.63e-01 |
| GO:BP | GO:0031424 | keratinization | 1 | 13 | 9.63e-01 |
| GO:BP | GO:0002692 | negative regulation of cellular extravasation | 1 | 5 | 9.63e-01 |
| GO:BP | GO:0032309 | icosanoid secretion | 1 | 18 | 9.63e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 9 | 28 | 9.63e-01 |
| GO:BP | GO:0051180 | vitamin transport | 2 | 28 | 9.63e-01 |
| GO:BP | GO:0060911 | cardiac cell fate commitment | 1 | 11 | 9.63e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 2 | 20 | 9.63e-01 |
| GO:BP | GO:0071281 | cellular response to iron ion | 2 | 6 | 9.63e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 389 | 9.64e-01 |
| GO:BP | GO:0072102 | glomerulus morphogenesis | 1 | 7 | 9.64e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 9.64e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 9.64e-01 |
| GO:BP | GO:0032494 | response to peptidoglycan | 2 | 5 | 9.64e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 9.64e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 9.64e-01 |
| GO:BP | GO:0048254 | snoRNA localization | 1 | 5 | 9.64e-01 |
| GO:BP | GO:0060912 | cardiac cell fate specification | 1 | 2 | 9.65e-01 |
| GO:BP | GO:0038156 | interleukin-3-mediated signaling pathway | 1 | 1 | 9.65e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 3 | 211 | 9.65e-01 |
| GO:BP | GO:0018199 | peptidyl-glutamine modification | 1 | 3 | 9.65e-01 |
| GO:BP | GO:1905563 | negative regulation of vascular endothelial cell proliferation | 1 | 5 | 9.65e-01 |
| GO:BP | GO:1905599 | positive regulation of low-density lipoprotein receptor activity | 1 | 4 | 9.65e-01 |
| GO:BP | GO:0034135 | regulation of toll-like receptor 2 signaling pathway | 1 | 7 | 9.65e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 1 | 19 | 9.65e-01 |
| GO:BP | GO:0140353 | lipid export from cell | 1 | 28 | 9.66e-01 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 2 | 14 | 9.66e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 1 | 166 | 9.66e-01 |
| GO:BP | GO:2000389 | regulation of neutrophil extravasation | 2 | 6 | 9.66e-01 |
| GO:BP | GO:0031638 | zymogen activation | 1 | 45 | 9.66e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 9.66e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 2 | 187 | 9.67e-01 |
| GO:BP | GO:0002434 | immune complex clearance | 1 | 1 | 9.67e-01 |
| GO:BP | GO:0090261 | positive regulation of inclusion body assembly | 1 | 5 | 9.67e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 1 | 57 | 9.67e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 4 | 24 | 9.67e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 3 | 336 | 9.67e-01 |
| GO:BP | GO:1905034 | regulation of antifungal innate immune response | 1 | 3 | 9.67e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 22 | 216 | 9.67e-01 |
| GO:BP | GO:0007567 | parturition | 1 | 8 | 9.67e-01 |
| GO:BP | GO:0002829 | negative regulation of type 2 immune response | 1 | 7 | 9.67e-01 |
| GO:BP | GO:0090296 | regulation of mitochondrial DNA replication | 1 | 4 | 9.67e-01 |
| GO:BP | GO:0007155 | cell adhesion | 6 | 1012 | 9.68e-01 |
| GO:BP | GO:2000318 | positive regulation of T-helper 17 type immune response | 2 | 7 | 9.68e-01 |
| GO:BP | GO:0070667 | negative regulation of mast cell proliferation | 1 | 2 | 9.68e-01 |
| GO:BP | GO:0060335 | positive regulation of type II interferon-mediated signaling pathway | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0060332 | positive regulation of response to type II interferon | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0043503 | skeletal muscle fiber adaptation | 1 | 3 | 9.68e-01 |
| GO:BP | GO:1900147 | regulation of Schwann cell migration | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0036135 | Schwann cell migration | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 2 | 14 | 9.68e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 2 | 14 | 9.68e-01 |
| GO:BP | GO:0015849 | organic acid transport | 22 | 217 | 9.68e-01 |
| GO:BP | GO:0043323 | positive regulation of natural killer cell degranulation | 1 | 3 | 9.69e-01 |
| GO:BP | GO:0002752 | cell surface pattern recognition receptor signaling pathway | 1 | 27 | 9.69e-01 |
| GO:BP | GO:0043321 | regulation of natural killer cell degranulation | 1 | 3 | 9.69e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 2 | 7 | 9.69e-01 |
| GO:BP | GO:0002513 | tolerance induction to self antigen | 1 | 4 | 9.69e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 1 | 4794 | 9.69e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 8 | 24 | 9.69e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 5 | 8061 | 9.69e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 4 | 15 | 9.69e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 9.69e-01 |
| GO:BP | GO:0035630 | bone mineralization involved in bone maturation | 2 | 6 | 9.69e-01 |
| GO:BP | GO:0060433 | bronchus development | 1 | 6 | 9.69e-01 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 2 | 96 | 9.69e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 1 | 20 | 9.69e-01 |
| GO:BP | GO:0060913 | cardiac cell fate determination | 1 | 2 | 9.70e-01 |
| GO:BP | GO:1900138 | negative regulation of phospholipase A2 activity | 1 | 4 | 9.70e-01 |
| GO:BP | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 4 | 9.70e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 4 | 7552 | 9.70e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 4 | 16 | 9.70e-01 |
| GO:BP | GO:0006869 | lipid transport | 16 | 272 | 9.70e-01 |
| GO:BP | GO:0061723 | glycophagy | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0071609 | chemokine (C-C motif) ligand 5 production | 2 | 6 | 9.71e-01 |
| GO:BP | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production | 2 | 6 | 9.71e-01 |
| GO:BP | GO:1905268 | negative regulation of chromatin organization | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0120262 | negative regulation of heterochromatin organization | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 4 | 90 | 9.71e-01 |
| GO:BP | GO:1903711 | spermidine transmembrane transport | 1 | 2 | 9.71e-01 |
| GO:BP | GO:0034155 | regulation of toll-like receptor 7 signaling pathway | 1 | 2 | 9.71e-01 |
| GO:BP | GO:0031452 | negative regulation of heterochromatin formation | 1 | 4 | 9.71e-01 |
| GO:BP | GO:1902523 | positive regulation of protein K63-linked ubiquitination | 1 | 5 | 9.71e-01 |
| GO:BP | GO:0030168 | platelet activation | 7 | 91 | 9.71e-01 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 3 | 20 | 9.71e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 9.71e-01 |
| GO:BP | GO:0034138 | toll-like receptor 3 signaling pathway | 1 | 12 | 9.71e-01 |
| GO:BP | GO:1990911 | response to psychosocial stress | 1 | 2 | 9.71e-01 |
| GO:BP | GO:0010876 | lipid localization | 18 | 310 | 9.71e-01 |
| GO:BP | GO:0002025 | norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure | 1 | 3 | 9.71e-01 |
| GO:BP | GO:1903998 | regulation of eating behavior | 1 | 1 | 9.71e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 3 | 31 | 9.72e-01 |
| GO:BP | GO:1903891 | regulation of ATF6-mediated unfolded protein response | 1 | 4 | 9.72e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 3 | 31 | 9.72e-01 |
| GO:BP | GO:0045333 | cellular respiration | 4 | 211 | 9.72e-01 |
| GO:BP | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 3 | 9.72e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.72e-01 |
| GO:BP | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation | 1 | 2 | 9.72e-01 |
| GO:BP | GO:0072086 | specification of loop of Henle identity | 1 | 3 | 9.72e-01 |
| GO:BP | GO:0044648 | histone H3-K4 dimethylation | 1 | 6 | 9.72e-01 |
| GO:BP | GO:0050975 | sensory perception of touch | 1 | 2 | 9.72e-01 |
| GO:BP | GO:0042420 | dopamine catabolic process | 1 | 5 | 9.72e-01 |
| GO:BP | GO:0045901 | positive regulation of translational elongation | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0045905 | positive regulation of translational termination | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0000967 | rRNA 5’-end processing | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0034471 | ncRNA 5’-end processing | 1 | 5 | 9.73e-01 |
| GO:BP | GO:1905684 | regulation of plasma membrane repair | 1 | 7 | 9.73e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 10 | 84 | 9.73e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 9.73e-01 |
| GO:BP | GO:0000472 | endonucleolytic cleavage to generate mature 5’-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0035900 | response to isolation stress | 1 | 2 | 9.73e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 1 | 27 | 9.73e-01 |
| GO:BP | GO:0006491 | N-glycan processing | 3 | 18 | 9.73e-01 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 3 | 48 | 9.73e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 8 | 19 | 9.73e-01 |
| GO:BP | GO:0046325 | negative regulation of glucose import | 2 | 10 | 9.73e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 10 | 37 | 9.73e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 4 | 63 | 9.74e-01 |
| GO:BP | GO:0061588 | calcium activated phospholipid scrambling | 1 | 4 | 9.74e-01 |
| GO:BP | GO:0035812 | renal sodium excretion | 2 | 11 | 9.74e-01 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 1 | 102 | 9.74e-01 |
| GO:BP | GO:0021869 | forebrain ventricular zone progenitor cell division | 1 | 2 | 9.74e-01 |
| GO:BP | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter | 1 | 3 | 9.74e-01 |
| GO:BP | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter | 1 | 3 | 9.74e-01 |
| GO:BP | GO:0034112 | positive regulation of homotypic cell-cell adhesion | 1 | 10 | 9.74e-01 |
| GO:BP | GO:1905806 | regulation of synapse pruning | 2 | 5 | 9.74e-01 |
| GO:BP | GO:0071076 | RNA 3’ uridylation | 1 | 4 | 9.74e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 3 | 138 | 9.74e-01 |
| GO:BP | GO:0021859 | pyramidal neuron differentiation | 1 | 11 | 9.74e-01 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 1 | 82 | 9.74e-01 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 1 | 19 | 9.74e-01 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 1 | 82 | 9.74e-01 |
| GO:BP | GO:0048643 | positive regulation of skeletal muscle tissue development | 1 | 14 | 9.74e-01 |
| GO:BP | GO:0071599 | otic vesicle development | 4 | 10 | 9.75e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0072334 | UDP-galactose transmembrane transport | 1 | 4 | 9.75e-01 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 15 | 9.75e-01 |
| GO:BP | GO:0060278 | regulation of ovulation | 1 | 2 | 9.76e-01 |
| GO:BP | GO:0018916 | nitrobenzene metabolic process | 1 | 4 | 9.76e-01 |
| GO:BP | GO:0033234 | negative regulation of protein sumoylation | 1 | 8 | 9.76e-01 |
| GO:BP | GO:0001708 | cell fate specification | 18 | 56 | 9.76e-01 |
| GO:BP | GO:0051969 | regulation of transmission of nerve impulse | 1 | 6 | 9.76e-01 |
| GO:BP | GO:0048050 | post-embryonic eye morphogenesis | 1 | 7 | 9.76e-01 |
| GO:BP | GO:0061209 | cell proliferation involved in mesonephros development | 1 | 3 | 9.76e-01 |
| GO:BP | GO:1904977 | lymphatic endothelial cell migration | 1 | 3 | 9.76e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 7 | 26 | 9.76e-01 |
| GO:BP | GO:0010593 | negative regulation of lamellipodium assembly | 1 | 2 | 9.76e-01 |
| GO:BP | GO:1904352 | positive regulation of protein catabolic process in the vacuole | 1 | 6 | 9.76e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 9.76e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 7 | 39 | 9.76e-01 |
| GO:BP | GO:0140459 | response to Gram-positive bacterium | 1 | 3 | 9.77e-01 |
| GO:BP | GO:0097707 | ferroptosis | 1 | 4 | 9.77e-01 |
| GO:BP | GO:2000152 | regulation of ubiquitin-specific protease activity | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 1 | 5 | 9.77e-01 |
| GO:BP | GO:0035112 | genitalia morphogenesis | 1 | 5 | 9.77e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 9.77e-01 |
| GO:BP | GO:1901671 | positive regulation of superoxide dismutase activity | 1 | 3 | 9.77e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 2 | 16 | 9.77e-01 |
| GO:BP | GO:0110075 | regulation of ferroptosis | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0036022 | limb joint morphogenesis | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0071650 | negative regulation of chemokine (C-C motif) ligand 5 production | 1 | 3 | 9.77e-01 |
| GO:BP | GO:0033489 | cholesterol biosynthetic process via desmosterol | 1 | 4 | 9.77e-01 |
| GO:BP | GO:1904690 | positive regulation of cytoplasmic translational initiation | 1 | 5 | 9.77e-01 |
| GO:BP | GO:0033490 | cholesterol biosynthetic process via lathosterol | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | 4 | 14 | 9.77e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 6 | 16 | 9.77e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 30 | 9.77e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 8 | 100 | 9.77e-01 |
| GO:BP | GO:1904684 | negative regulation of metalloendopeptidase activity | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 1 | 12 | 9.77e-01 |
| GO:BP | GO:0009583 | detection of light stimulus | 1 | 27 | 9.77e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 2 | 19 | 9.77e-01 |
| GO:BP | GO:0014063 | negative regulation of serotonin secretion | 1 | 2 | 9.77e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 5 | 75 | 9.77e-01 |
| GO:BP | GO:0030574 | collagen catabolic process | 7 | 26 | 9.77e-01 |
| GO:BP | GO:0034587 | piRNA processing | 1 | 4 | 9.78e-01 |
| GO:BP | GO:0036065 | fucosylation | 1 | 10 | 9.78e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 5 | 23 | 9.78e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 9.79e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 2 | 83 | 9.79e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 5 | 191 | 9.79e-01 |
| GO:BP | GO:0007596 | blood coagulation | 1 | 150 | 9.79e-01 |
| GO:BP | GO:0046174 | polyol catabolic process | 1 | 7 | 9.79e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 9.79e-01 |
| GO:BP | GO:0046487 | glyoxylate metabolic process | 1 | 5 | 9.79e-01 |
| GO:BP | GO:0015671 | oxygen transport | 1 | 4 | 9.79e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 9.79e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 3 | 10 | 9.80e-01 |
| GO:BP | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | 6 | 13 | 9.80e-01 |
| GO:BP | GO:0035934 | corticosterone secretion | 1 | 3 | 9.80e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 1 | 14 | 9.80e-01 |
| GO:BP | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly | 4 | 16 | 9.80e-01 |
| GO:BP | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation | 2 | 8 | 9.80e-01 |
| GO:BP | GO:0061762 | CAMKK-AMPK signaling cascade | 1 | 5 | 9.80e-01 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 1 | 38 | 9.80e-01 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 4 | 18 | 9.80e-01 |
| GO:BP | GO:0007356 | thorax and anterior abdomen determination | 1 | 1 | 9.80e-01 |
| GO:BP | GO:0032490 | detection of molecule of bacterial origin | 2 | 4 | 9.81e-01 |
| GO:BP | GO:0051014 | actin filament severing | 1 | 13 | 9.81e-01 |
| GO:BP | GO:0033006 | regulation of mast cell activation involved in immune response | 3 | 21 | 9.81e-01 |
| GO:BP | GO:0050817 | coagulation | 1 | 153 | 9.81e-01 |
| GO:BP | GO:0032275 | luteinizing hormone secretion | 1 | 6 | 9.81e-01 |
| GO:BP | GO:0042989 | sequestering of actin monomers | 1 | 9 | 9.81e-01 |
| GO:BP | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 2 | 10 | 9.81e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 14 | 64 | 9.81e-01 |
| GO:BP | GO:0060986 | endocrine hormone secretion | 1 | 35 | 9.82e-01 |
| GO:BP | GO:0090659 | walking behavior | 1 | 25 | 9.82e-01 |
| GO:BP | GO:2000402 | negative regulation of lymphocyte migration | 1 | 10 | 9.82e-01 |
| GO:BP | GO:0065001 | specification of axis polarity | 1 | 2 | 9.82e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 3 | 248 | 9.82e-01 |
| GO:BP | GO:0010536 | positive regulation of activation of Janus kinase activity | 1 | 2 | 9.83e-01 |
| GO:BP | GO:0032847 | regulation of cellular pH reduction | 1 | 5 | 9.83e-01 |
| GO:BP | GO:0070508 | cholesterol import | 1 | 6 | 9.83e-01 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 9.83e-01 |
| GO:BP | GO:0042182 | ketone catabolic process | 3 | 9 | 9.83e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 5 | 16 | 9.83e-01 |
| GO:BP | GO:0090325 | regulation of locomotion involved in locomotory behavior | 1 | 3 | 9.83e-01 |
| GO:BP | GO:0006707 | cholesterol catabolic process | 1 | 8 | 9.83e-01 |
| GO:BP | GO:0016127 | sterol catabolic process | 1 | 8 | 9.83e-01 |
| GO:BP | GO:0007216 | G protein-coupled glutamate receptor signaling pathway | 1 | 5 | 9.83e-01 |
| GO:BP | GO:0060161 | positive regulation of dopamine receptor signaling pathway | 1 | 3 | 9.83e-01 |
| GO:BP | GO:0048539 | bone marrow development | 2 | 7 | 9.83e-01 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 1 | 5 | 9.83e-01 |
| GO:BP | GO:1903236 | regulation of leukocyte tethering or rolling | 1 | 6 | 9.83e-01 |
| GO:BP | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia | 2 | 9 | 9.83e-01 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 1 | 42 | 9.83e-01 |
| GO:BP | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin | 1 | 1 | 9.83e-01 |
| GO:BP | GO:1903413 | cellular response to bile acid | 1 | 2 | 9.84e-01 |
| GO:BP | GO:0043320 | natural killer cell degranulation | 3 | 6 | 9.84e-01 |
| GO:BP | GO:0019614 | catechol-containing compound catabolic process | 1 | 5 | 9.84e-01 |
| GO:BP | GO:0042424 | catecholamine catabolic process | 1 | 5 | 9.84e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 1 | 10 | 9.84e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 1 | 877 | 9.84e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 2 | 88 | 9.84e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 1 | 10 | 9.84e-01 |
| GO:BP | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing | 1 | 4 | 9.84e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 5 | 122 | 9.84e-01 |
| GO:BP | GO:0043302 | positive regulation of leukocyte degranulation | 2 | 15 | 9.84e-01 |
| GO:BP | GO:0097278 | complement-dependent cytotoxicity | 3 | 4 | 9.84e-01 |
| GO:BP | GO:0099612 | protein localization to axon | 1 | 6 | 9.85e-01 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 1 | 21 | 9.85e-01 |
| GO:BP | GO:1904749 | regulation of protein localization to nucleolus | 2 | 7 | 9.85e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 2 | 26 | 9.85e-01 |
| GO:BP | GO:1900150 | regulation of defense response to fungus | 1 | 3 | 9.85e-01 |
| GO:BP | GO:0030277 | maintenance of gastrointestinal epithelium | 2 | 8 | 9.85e-01 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 1 | 5 | 9.85e-01 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 9.85e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 6 | 21 | 9.85e-01 |
| GO:BP | GO:0006907 | pinocytosis | 7 | 18 | 9.85e-01 |
| GO:BP | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 5 | 9.85e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 13 | 561 | 9.85e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 7 | 41 | 9.85e-01 |
| GO:BP | GO:0060430 | lung saccule development | 1 | 7 | 9.86e-01 |
| GO:BP | GO:0006821 | chloride transport | 5 | 58 | 9.86e-01 |
| GO:BP | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell | 1 | 3 | 9.86e-01 |
| GO:BP | GO:0051049 | regulation of transport | 56 | 1267 | 9.86e-01 |
| GO:BP | GO:1905383 | protein localization to presynapse | 3 | 11 | 9.87e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 19 | 337 | 9.87e-01 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 3 | 9 | 9.87e-01 |
| GO:BP | GO:0010519 | negative regulation of phospholipase activity | 2 | 7 | 9.87e-01 |
| GO:BP | GO:0003072 | renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure | 1 | 3 | 9.87e-01 |
| GO:BP | GO:1902603 | carnitine transmembrane transport | 1 | 4 | 9.87e-01 |
| GO:BP | GO:0002034 | maintenance of blood vessel diameter homeostasis by renin-angiotensin | 1 | 3 | 9.87e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 9 | 1335 | 9.88e-01 |
| GO:BP | GO:0007369 | gastrulation | 7 | 146 | 9.88e-01 |
| GO:BP | GO:0045579 | positive regulation of B cell differentiation | 4 | 10 | 9.88e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 8 | 25 | 9.88e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 2 | 115 | 9.88e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 4 | 13 | 9.88e-01 |
| GO:BP | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 9.88e-01 |
| GO:BP | GO:0071637 | regulation of monocyte chemotactic protein-1 production | 1 | 7 | 9.88e-01 |
| GO:BP | GO:0072044 | collecting duct development | 2 | 6 | 9.88e-01 |
| GO:BP | GO:0071605 | monocyte chemotactic protein-1 production | 1 | 7 | 9.88e-01 |
| GO:BP | GO:1904672 | regulation of somatic stem cell population maintenance | 1 | 6 | 9.88e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 19 | 44 | 9.88e-01 |
| GO:BP | GO:0010756 | positive regulation of plasminogen activation | 1 | 7 | 9.88e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.88e-01 |
| GO:BP | GO:0060539 | diaphragm development | 2 | 5 | 9.88e-01 |
| GO:BP | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway | 1 | 4 | 9.88e-01 |
| GO:BP | GO:0010387 | COP9 signalosome assembly | 1 | 3 | 9.88e-01 |
| GO:BP | GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | 1 | 9 | 9.88e-01 |
| GO:BP | GO:0098596 | imitative learning | 1 | 4 | 9.88e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.88e-01 |
| GO:BP | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 6 | 9.88e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 1 | 35 | 9.88e-01 |
| GO:BP | GO:0042297 | vocal learning | 1 | 4 | 9.88e-01 |
| GO:BP | GO:0001821 | histamine secretion | 2 | 9 | 9.88e-01 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 1 | 87 | 9.88e-01 |
| GO:BP | GO:0045576 | mast cell activation | 2 | 35 | 9.88e-01 |
| GO:BP | GO:0006868 | glutamine transport | 2 | 8 | 9.88e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 14 | 556 | 9.88e-01 |
| GO:BP | GO:0042373 | vitamin K metabolic process | 2 | 6 | 9.88e-01 |
| GO:BP | GO:0021764 | amygdala development | 1 | 3 | 9.88e-01 |
| GO:BP | GO:0045059 | positive thymic T cell selection | 3 | 6 | 9.88e-01 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 19 | 66 | 9.89e-01 |
| GO:BP | GO:0048149 | behavioral response to ethanol | 2 | 5 | 9.89e-01 |
| GO:BP | GO:0034134 | toll-like receptor 2 signaling pathway | 2 | 11 | 9.89e-01 |
| GO:BP | GO:0010918 | positive regulation of mitochondrial membrane potential | 3 | 9 | 9.89e-01 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 2 | 9 | 9.90e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 17 | 41 | 9.90e-01 |
| GO:BP | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | 1 | 13 | 9.90e-01 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 1 | 18 | 9.90e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 21 | 47 | 9.90e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 1 | 30 | 9.90e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 1 | 30 | 9.90e-01 |
| GO:BP | GO:0009642 | response to light intensity | 1 | 11 | 9.90e-01 |
| GO:BP | GO:0002438 | acute inflammatory response to antigenic stimulus | 4 | 10 | 9.90e-01 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 4 | 11 | 9.90e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 13 | 803 | 9.90e-01 |
| GO:BP | GO:0016045 | detection of bacterium | 1 | 8 | 9.90e-01 |
| GO:BP | GO:0034384 | high-density lipoprotein particle clearance | 1 | 7 | 9.90e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 17 | 65 | 9.90e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 9 | 19 | 9.90e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 6 | 288 | 9.90e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 3 | 130 | 9.90e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 2 | 14 | 9.90e-01 |
| GO:BP | GO:0035813 | regulation of renal sodium excretion | 1 | 10 | 9.90e-01 |
| GO:BP | GO:1900736 | regulation of phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 2 | 9.90e-01 |
| GO:BP | GO:0071349 | cellular response to interleukin-12 | 1 | 5 | 9.90e-01 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 5 | 14 | 9.90e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 5 | 211 | 9.90e-01 |
| GO:BP | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 1 | 15 | 9.90e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 1 | 42 | 9.90e-01 |
| GO:BP | GO:0019081 | viral translation | 1 | 18 | 9.90e-01 |
| GO:BP | GO:0042953 | lipoprotein transport | 1 | 14 | 9.91e-01 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 1 | 15 | 9.91e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 1 | 16 | 9.91e-01 |
| GO:BP | GO:0002580 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 1 | 9.91e-01 |
| GO:BP | GO:0035095 | behavioral response to nicotine | 1 | 3 | 9.91e-01 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 4 | 19 | 9.91e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 1 | 8 | 9.91e-01 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 16 | 57 | 9.91e-01 |
| GO:BP | GO:0006688 | glycosphingolipid biosynthetic process | 1 | 25 | 9.91e-01 |
| GO:BP | GO:0022617 | extracellular matrix disassembly | 5 | 36 | 9.91e-01 |
| GO:BP | GO:1904688 | regulation of cytoplasmic translational initiation | 1 | 6 | 9.91e-01 |
| GO:BP | GO:0021957 | corticospinal tract morphogenesis | 1 | 7 | 9.91e-01 |
| GO:BP | GO:0009644 | response to high light intensity | 1 | 2 | 9.91e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 20 | 46 | 9.91e-01 |
| GO:BP | GO:0009820 | alkaloid metabolic process | 1 | 2 | 9.91e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 6 | 33 | 9.91e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 23 | 139 | 9.91e-01 |
| GO:BP | GO:0010467 | gene expression | 1 | 4587 | 9.91e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 7 | 27 | 9.91e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 1 | 31 | 9.91e-01 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 2 | 19 | 9.92e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 7 | 47 | 9.92e-01 |
| GO:BP | GO:0097187 | dentinogenesis | 1 | 4 | 9.92e-01 |
| GO:BP | GO:0097709 | connective tissue replacement | 2 | 8 | 9.92e-01 |
| GO:BP | GO:0007196 | adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway | 1 | 1 | 9.92e-01 |
| GO:BP | GO:0015942 | formate metabolic process | 1 | 2 | 9.92e-01 |
| GO:BP | GO:0071286 | cellular response to magnesium ion | 1 | 3 | 9.92e-01 |
| GO:BP | GO:0002031 | G protein-coupled receptor internalization | 2 | 10 | 9.92e-01 |
| GO:BP | GO:0120252 | hydrocarbon metabolic process | 1 | 2 | 9.92e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 4 | 44 | 9.92e-01 |
| GO:BP | GO:0044409 | entry into host | 17 | 113 | 9.92e-01 |
| GO:BP | GO:0043031 | negative regulation of macrophage activation | 2 | 9 | 9.92e-01 |
| GO:BP | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 7 | 9.92e-01 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 3 | 10 | 9.92e-01 |
| GO:BP | GO:0021794 | thalamus development | 1 | 6 | 9.93e-01 |
| GO:BP | GO:0046874 | quinolinate metabolic process | 1 | 1 | 9.93e-01 |
| GO:BP | GO:0072179 | nephric duct formation | 1 | 1 | 9.93e-01 |
| GO:BP | GO:0046668 | regulation of retinal cell programmed cell death | 1 | 2 | 9.93e-01 |
| GO:BP | GO:0015705 | iodide transport | 1 | 2 | 9.93e-01 |
| GO:BP | GO:0045956 | positive regulation of calcium ion-dependent exocytosis | 2 | 11 | 9.93e-01 |
| GO:BP | GO:2000812 | regulation of barbed-end actin filament capping | 1 | 3 | 9.93e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 11 | 36 | 9.93e-01 |
| GO:BP | GO:0065007 | biological regulation | 5 | 8320 | 9.93e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 2 | 475 | 9.93e-01 |
| GO:BP | GO:1901317 | regulation of flagellated sperm motility | 1 | 6 | 9.93e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 2 | 168 | 9.93e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 16 | 107 | 9.93e-01 |
| GO:BP | GO:1902260 | negative regulation of delayed rectifier potassium channel activity | 1 | 5 | 9.93e-01 |
| GO:BP | GO:0140206 | dipeptide import across plasma membrane | 1 | 2 | 9.93e-01 |
| GO:BP | GO:0090527 | actin filament reorganization | 2 | 10 | 9.93e-01 |
| GO:BP | GO:0015739 | sialic acid transport | 1 | 1 | 9.93e-01 |
| GO:BP | GO:0140205 | oligopeptide import across plasma membrane | 1 | 2 | 9.93e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 9.93e-01 |
| GO:BP | GO:0070162 | adiponectin secretion | 1 | 4 | 9.93e-01 |
| GO:BP | GO:0070163 | regulation of adiponectin secretion | 1 | 4 | 9.93e-01 |
| GO:BP | GO:1905516 | positive regulation of fertilization | 1 | 3 | 9.93e-01 |
| GO:BP | GO:0072205 | metanephric collecting duct development | 1 | 5 | 9.94e-01 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 1 | 35 | 9.94e-01 |
| GO:BP | GO:0035025 | positive regulation of Rho protein signal transduction | 1 | 22 | 9.94e-01 |
| GO:BP | GO:0002858 | regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0097692 | histone H3-K4 monomethylation | 1 | 8 | 9.94e-01 |
| GO:BP | GO:0032026 | response to magnesium ion | 2 | 10 | 9.94e-01 |
| GO:BP | GO:0015879 | carnitine transport | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0021853 | cerebral cortex GABAergic interneuron migration | 1 | 2 | 9.94e-01 |
| GO:BP | GO:0002002 | regulation of angiotensin levels in blood | 2 | 6 | 9.95e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 5 | 30 | 9.95e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 2 | 38 | 9.95e-01 |
| GO:BP | GO:0032963 | collagen metabolic process | 6 | 74 | 9.95e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 6 | 34 | 9.95e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 2 | 3600 | 9.95e-01 |
| GO:BP | GO:1905245 | regulation of aspartic-type peptidase activity | 1 | 11 | 9.95e-01 |
| GO:BP | GO:0010496 | intercellular transport | 1 | 4 | 9.95e-01 |
| GO:BP | GO:0099004 | calmodulin dependent kinase signaling pathway | 1 | 6 | 9.95e-01 |
| GO:BP | GO:0150078 | positive regulation of neuroinflammatory response | 1 | 4 | 9.95e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 227 | 9.95e-01 |
| GO:BP | GO:0099641 | anterograde axonal protein transport | 2 | 9 | 9.96e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 9 | 1496 | 9.96e-01 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 1 | 20 | 9.96e-01 |
| GO:BP | GO:0098664 | G protein-coupled serotonin receptor signaling pathway | 1 | 8 | 9.96e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 9 | 35 | 9.96e-01 |
| GO:BP | GO:0032274 | gonadotropin secretion | 2 | 10 | 9.97e-01 |
| GO:BP | GO:0042362 | fat-soluble vitamin biosynthetic process | 3 | 6 | 9.97e-01 |
| GO:BP | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production | 1 | 4 | 9.97e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 1 | 29 | 9.97e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 7 | 23 | 9.97e-01 |
| GO:BP | GO:0032276 | regulation of gonadotropin secretion | 1 | 6 | 9.98e-01 |
| GO:BP | GO:0048536 | spleen development | 1 | 28 | 9.98e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 5 | 142 | 9.98e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 1 | 47 | 9.98e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 9.98e-01 |
| GO:BP | GO:0007602 | phototransduction | 1 | 17 | 9.98e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 4 | 58 | 9.98e-01 |
| GO:BP | GO:0150079 | negative regulation of neuroinflammatory response | 2 | 6 | 9.98e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 9.98e-01 |
| GO:BP | GO:0140894 | endolysosomal toll-like receptor signaling pathway | 2 | 24 | 9.99e-01 |
| GO:BP | GO:0061535 | glutamate secretion, neurotransmission | 1 | 5 | 9.99e-01 |
| GO:BP | GO:0071702 | organic substance transport | 2 | 2116 | 9.99e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 4 | 59 | 9.99e-01 |
| GO:BP | GO:0098543 | detection of other organism | 1 | 9 | 9.99e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 10 | 34 | 9.99e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 3 | 22 | 9.99e-01 |
| GO:BP | GO:0002283 | neutrophil activation involved in immune response | 1 | 7 | 9.99e-01 |
| GO:BP | GO:0042368 | vitamin D biosynthetic process | 2 | 4 | 9.99e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 7 | 182 | 9.99e-01 |
| GO:BP | GO:0045332 | phospholipid translocation | 1 | 33 | 9.99e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 10 | 39 | 9.99e-01 |
| GO:BP | GO:0006457 | protein folding | 2 | 192 | 9.99e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 1 | 16 | 9.99e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 2 | 10 | 9.99e-01 |
| GO:BP | GO:0007638 | mechanosensory behavior | 3 | 8 | 9.99e-01 |
| GO:BP | GO:1900426 | positive regulation of defense response to bacterium | 1 | 6 | 9.99e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 9.99e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 1 | 91 | 9.99e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 1 | 91 | 9.99e-01 |
| GO:BP | GO:2000321 | positive regulation of T-helper 17 cell differentiation | 1 | 3 | 9.99e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 4 | 15 | 9.99e-01 |
| GO:BP | GO:0001839 | neural plate morphogenesis | 1 | 4 | 9.99e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 1 | 251 | 9.99e-01 |
| GO:BP | GO:1903028 | positive regulation of opsonization | 1 | 1 | 9.99e-01 |
| GO:BP | GO:0070207 | protein homotrimerization | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0042711 | maternal behavior | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0070206 | protein trimerization | 2 | 12 | 1.00e+00 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0001714 | endodermal cell fate specification | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0032645 | regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 7 | 23 | 1.00e+00 |
| GO:BP | GO:0000365 | mRNA trans splicing, via spliceosome | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0000353 | formation of quadruple SL/U4/U5/U6 snRNP | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0007343 | egg activation | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0007342 | fusion of sperm to egg plasma membrane involved in single fertilization | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0099640 | axo-dendritic protein transport | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0007308 | oocyte construction | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0006376 | mRNA splice site selection | 2 | 31 | 1.00e+00 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0007339 | binding of sperm to zona pellucida | 7 | 20 | 1.00e+00 |
| GO:BP | GO:0070269 | pyroptosis | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 5 | 30 | 1.00e+00 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 1 | 74 | 1.00e+00 |
| GO:BP | GO:0007338 | single fertilization | 1 | 70 | 1.00e+00 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 1.00e+00 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0007320 | insemination | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0007309 | oocyte axis specification | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 9 | 24 | 1.00e+00 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 10 | 25 | 1.00e+00 |
| GO:BP | GO:0015721 | bile acid and bile salt transport | 3 | 11 | 1.00e+00 |
| GO:BP | GO:0042730 | fibrinolysis | 5 | 15 | 1.00e+00 |
| GO:BP | GO:0060746 | parental behavior | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0045291 | mRNA trans splicing, SL addition | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:1903317 | regulation of protein maturation | 7 | 50 | 1.00e+00 |
| GO:BP | GO:0030299 | intestinal cholesterol absorption | 4 | 8 | 1.00e+00 |
| GO:BP | GO:0034382 | chylomicron remnant clearance | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0019336 | phenol-containing compound catabolic process | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0043046 | DNA methylation involved in gamete generation | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0032604 | granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:2001140 | positive regulation of phospholipid transport | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0034379 | very-low-density lipoprotein particle assembly | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0034374 | low-density lipoprotein particle remodeling | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0006477 | protein sulfation | 1 | 6 | 1.00e+00 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0030300 | regulation of intestinal cholesterol absorption | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0002507 | tolerance induction | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0099024 | plasma membrane invagination | 3 | 42 | 1.00e+00 |
| GO:BP | GO:0019373 | epoxygenase P450 pathway | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 1.00e+00 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 1.00e+00 |
| GO:BP | GO:0001914 | regulation of T cell mediated cytotoxicity | 1 | 17 | 1.00e+00 |
| GO:BP | GO:0002517 | T cell tolerance induction | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0006397 | mRNA processing | 3 | 450 | 1.00e+00 |
| GO:BP | GO:0042791 | 5S class rRNA transcription by RNA polymerase III | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 3 | 14 | 1.00e+00 |
| GO:BP | GO:0001916 | positive regulation of T cell mediated cytotoxicity | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0070486 | leukocyte aggregation | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 1.00e+00 |
| GO:BP | GO:2000858 | regulation of aldosterone secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 2 | 3 | 1.00e+00 |
| GO:BP | GO:0002503 | peptide antigen assembly with MHC class II protein complex | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0042359 | vitamin D metabolic process | 3 | 9 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0021554 | optic nerve development | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 7 | 19 | 1.00e+00 |
| GO:BP | GO:0021843 | substrate-independent telencephalic tangential interneuron migration | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0021830 | interneuron migration from the subpallium to the cortex | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0002423 | natural killer cell mediated immune response to tumor cell | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0006108 | malate metabolic process | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 2 | 18 | 1.00e+00 |
| GO:BP | GO:0140632 | canonical inflammasome complex assembly | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0021826 | substrate-independent telencephalic tangential migration | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0035036 | sperm-egg recognition | 8 | 22 | 1.00e+00 |
| GO:BP | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 2 | 20 | 1.00e+00 |
| GO:BP | GO:0042178 | xenobiotic catabolic process | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0002418 | immune response to tumor cell | 3 | 12 | 1.00e+00 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 3 | 38 | 1.00e+00 |
| GO:BP | GO:0061760 | antifungal innate immune response | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0002399 | MHC class II protein complex assembly | 2 | 4 | 1.00e+00 |
| GO:BP | GO:1990349 | gap junction-mediated intercellular transport | 1 | 2 | 1.00e+00 |
| GO:BP | GO:1900004 | negative regulation of serine-type endopeptidase activity | 1 | 4 | 1.00e+00 |
| GO:BP | GO:1900003 | regulation of serine-type endopeptidase activity | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0021762 | substantia nigra development | 1 | 41 | 1.00e+00 |
| GO:BP | GO:0001820 | serotonin secretion | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002396 | MHC protein complex assembly | 2 | 8 | 1.00e+00 |
| GO:BP | GO:1903817 | negative regulation of voltage-gated potassium channel activity | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.00e+00 |
| GO:BP | GO:0045838 | positive regulation of membrane potential | 4 | 12 | 1.00e+00 |
| GO:BP | GO:1905920 | positive regulation of CoA-transferase activity | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002501 | peptide antigen assembly with MHC protein complex | 2 | 8 | 1.00e+00 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 1 | 23 | 1.00e+00 |
| GO:BP | GO:2000848 | positive regulation of corticosteroid hormone secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 22 | 55 | 1.00e+00 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0060468 | prevention of polyspermy | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0050909 | sensory perception of taste | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0050907 | detection of chemical stimulus involved in sensory perception | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0008380 | RNA splicing | 3 | 419 | 1.00e+00 |
| GO:BP | GO:0050886 | endocrine process | 1 | 57 | 1.00e+00 |
| GO:BP | GO:2000846 | regulation of corticosteroid hormone secretion | 1 | 9 | 1.00e+00 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 1 | 94 | 1.00e+00 |
| GO:BP | GO:0008343 | adult feeding behavior | 1 | 1 | 1.00e+00 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 3 | 33 | 1.00e+00 |
| GO:BP | GO:2000855 | regulation of mineralocorticoid secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0042421 | norepinephrine biosynthetic process | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 3 | 6 | 1.00e+00 |
| GO:BP | GO:0001867 | complement activation, lectin pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002326 | B cell lineage commitment | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0001840 | neural plate development | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0008228 | opsonization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0021860 | pyramidal neuron development | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 3 | 32 | 1.00e+00 |
| GO:BP | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 1.00e+00 |
| GO:BP | GO:1905918 | regulation of CoA-transferase activity | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 1.00e+00 |
| GO:BP | GO:1905522 | negative regulation of macrophage migration | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 22 | 55 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0046666 | retinal cell programmed cell death | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0035933 | glucocorticoid secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0002911 | regulation of lymphocyte anergy | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0051917 | regulation of fibrinolysis | 5 | 10 | 1.00e+00 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 2 | 23 | 1.00e+00 |
| GO:BP | GO:0006910 | phagocytosis, recognition | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0072104 | glomerular capillary formation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 2 | 33 | 1.00e+00 |
| GO:BP | GO:1904936 | interneuron migration | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0031341 | regulation of cell killing | 7 | 36 | 1.00e+00 |
| GO:BP | GO:0072103 | glomerulus vasculature morphogenesis | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0046415 | urate metabolic process | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0035989 | tendon development | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0015889 | cobalamin transport | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0002922 | positive regulation of humoral immune response | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0031223 | auditory behavior | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0001991 | regulation of systemic arterial blood pressure by circulatory renin-angiotensin | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 1 | 16 | 1.00e+00 |
| GO:BP | GO:1905049 | negative regulation of metallopeptidase activity | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0034154 | toll-like receptor 7 signaling pathway | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0002717 | positive regulation of natural killer cell mediated immunity | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 2 | 18 | 1.00e+00 |
| GO:BP | GO:1902571 | regulation of serine-type peptidase activity | 1 | 4 | 1.00e+00 |
| GO:BP | GO:1902572 | negative regulation of serine-type peptidase activity | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0000480 | endonucleolytic cleavage in 5’-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 8 | 1.00e+00 |
| GO:BP | GO:1901616 | organic hydroxy compound catabolic process | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0006956 | complement activation | 2 | 23 | 1.00e+00 |
| GO:BP | GO:0016071 | mRNA metabolic process | 4 | 665 | 1.00e+00 |
| GO:BP | GO:0035932 | aldosterone secretion | 1 | 5 | 1.00e+00 |
| GO:BP | GO:1904729 | regulation of intestinal lipid absorption | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0002834 | regulation of response to tumor cell | 3 | 11 | 1.00e+00 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 7 | 192 | 1.00e+00 |
| GO:BP | GO:0080154 | regulation of fertilization | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0015031 | protein transport | 35 | 1361 | 1.00e+00 |
| GO:BP | GO:0002836 | positive regulation of response to tumor cell | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0002837 | regulation of immune response to tumor cell | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0031639 | plasminogen activation | 7 | 19 | 1.00e+00 |
| GO:BP | GO:0051957 | positive regulation of amino acid transport | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0002839 | positive regulation of immune response to tumor cell | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 7 | 24 | 1.00e+00 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 2 | 28 | 1.00e+00 |
| GO:BP | GO:2000380 | regulation of mesoderm development | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0015811 | L-cystine transport | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0051923 | sulfation | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0052695 | cellular glucuronidation | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0006810 | transport | 4 | 3298 | 1.00e+00 |
| GO:BP | GO:0006706 | steroid catabolic process | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0002016 | regulation of blood volume by renin-angiotensin | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0046641 | positive regulation of alpha-beta T cell proliferation | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 10 | 36 | 1.00e+00 |
| GO:BP | GO:0006068 | ethanol catabolic process | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0002003 | angiotensin maturation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0014062 | regulation of serotonin secretion | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0014049 | positive regulation of glutamate secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0014048 | regulation of glutamate secretion | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0002870 | T cell anergy | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0035930 | corticosteroid hormone secretion | 2 | 12 | 1.00e+00 |
| GO:BP | GO:0035931 | mineralocorticoid secretion | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0035739 | CD4-positive, alpha-beta T cell proliferation | 2 | 12 | 1.00e+00 |
| GO:BP | GO:0002855 | regulation of natural killer cell mediated immune response to tumor cell | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0036305 | ameloblast differentiation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0044546 | NLRP3 inflammasome complex assembly | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0001990 | regulation of systemic arterial blood pressure by hormone | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0046164 | alcohol catabolic process | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0045058 | T cell selection | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 35 | 94 | 1.00e+00 |
| GO:BP | GO:0045061 | thymic T cell selection | 4 | 10 | 1.00e+00 |
| GO:BP | GO:0017038 | protein import | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0017121 | plasma membrane phospholipid scrambling | 4 | 13 | 1.00e+00 |
| GO:BP | GO:0043300 | regulation of leukocyte degranulation | 3 | 27 | 1.00e+00 |
| GO:BP | GO:0043299 | leukocyte degranulation | 1 | 41 | 1.00e+00 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0002578 | negative regulation of antigen processing and presentation | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0032429 | regulation of phospholipase A2 activity | 2 | 8 | 1.00e+00 |
| GO:BP | GO:1903027 | regulation of opsonization | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0098598 | learned vocalization behavior or vocal learning | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0045088 | regulation of innate immune response | 4 | 232 | 1.00e+00 |
| GO:BP | GO:0043312 | neutrophil degranulation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0010455 | positive regulation of cell fate commitment | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 19 | 40 | 1.00e+00 |
| GO:BP | GO:0018149 | peptide cross-linking | 1 | 14 | 1.00e+00 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0045109 | intermediate filament organization | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0010324 | membrane invagination | 3 | 49 | 1.00e+00 |
| GO:BP | GO:0000395 | mRNA 5’-splice site recognition | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0002249 | lymphocyte anergy | 1 | 4 | 1.00e+00 |
| GO:BP | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation | 2 | 12 | 1.00e+00 |
| GO:BP | GO:0031620 | regulation of fever generation | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0007210 | serotonin receptor signaling pathway | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 4 | 23 | 1.00e+00 |
| GO:BP | GO:0060467 | negative regulation of fertilization | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0098856 | intestinal lipid absorption | 4 | 10 | 1.00e+00 |
| GO:BP | GO:0098883 | synapse pruning | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0034310 | primary alcohol catabolic process | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 2 | 118 | 1.00e+00 |
| GO:BP | GO:0003081 | regulation of systemic arterial blood pressure by renin-angiotensin | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0002643 | regulation of tolerance induction | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0051620 | norepinephrine uptake | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0097267 | omega-hydroxylase P450 pathway | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0051608 | histamine transport | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0071830 | triglyceride-rich lipoprotein particle clearance | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition | 1 | 17 | 1.00e+00 |
| GO:BP | GO:0060123 | regulation of growth hormone secretion | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0032306 | regulation of prostaglandin secretion | 3 | 7 | 1.00e+00 |
| GO:BP | GO:0010572 | positive regulation of platelet activation | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0032308 | positive regulation of prostaglandin secretion | 3 | 6 | 1.00e+00 |
| GO:BP | GO:0002691 | regulation of cellular extravasation | 1 | 24 | 1.00e+00 |
| GO:BP | GO:0032310 | prostaglandin secretion | 3 | 8 | 1.00e+00 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 2 | 27 | 1.00e+00 |
| GO:BP | GO:0060124 | positive regulation of growth hormone secretion | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 2 | 29 | 1.00e+00 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 2 | 18 | 1.00e+00 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 1 | 94 | 1.00e+00 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 2 | 18 | 1.00e+00 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 1.00e+00 |
| GO:BP | GO:0071705 | nitrogen compound transport | 31 | 1684 | 1.00e+00 |
| GO:BP | GO:0002667 | regulation of T cell anergy | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0002664 | regulation of T cell tolerance induction | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0060272 | embryonic skeletal joint morphogenesis | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | 4 | 30 | 1.00e+00 |
| GO:BP | GO:1904478 | regulation of intestinal absorption | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 1 | 131 | 1.00e+00 |
| GO:BP | GO:0045026 | plasma membrane fusion | 3 | 5 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0006067 | ethanol metabolic process | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:04115 | p53 signaling pathway | 15 | 65 | 6.44e-06 |
| KEGG | KEGG:03030 | DNA replication | 22 | 35 | 5.18e-04 |
| KEGG | KEGG:04110 | Cell cycle | 63 | 151 | 2.49e-03 |
| KEGG | KEGG:03430 | Mismatch repair | 16 | 22 | 7.67e-03 |
| KEGG | KEGG:03410 | Base excision repair | 26 | 33 | 9.65e-03 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 6 | 61 | 1.21e-02 |
| KEGG | KEGG:05034 | Alcoholism | 7 | 122 | 2.65e-02 |
| KEGG | KEGG:04217 | Necroptosis | 6 | 106 | 2.65e-02 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 2 | 20 | 2.65e-02 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 6 | 110 | 4.57e-02 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 3 | 69 | 4.57e-02 |
| KEGG | KEGG:01524 | Platinum drug resistance | 21 | 65 | 4.57e-02 |
| KEGG | KEGG:00230 | Purine metabolism | 4 | 97 | 5.85e-02 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 4 | 43 | 6.72e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 6.72e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 4 | 75 | 6.72e-02 |
| KEGG | KEGG:04210 | Apoptosis | 29 | 118 | 6.72e-02 |
| KEGG | KEGG:03440 | Homologous recombination | 10 | 39 | 6.94e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 1 | 57 | 7.89e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 8.14e-02 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 8.18e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 8.18e-02 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 8.18e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 8.18e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 1 | 84 | 8.89e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 9.14e-02 |
| KEGG | KEGG:00920 | Sulfur metabolism | 4 | 10 | 9.17e-02 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 1.05e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 27 | 48 | 1.14e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 1.19e-01 |
| KEGG | KEGG:05224 | Breast cancer | 1 | 117 | 1.19e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 1.19e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 2 | 78 | 1.25e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 1.25e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 1 | 145 | 1.25e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 1.38e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 1.38e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 4 | 44 | 1.59e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 8 | 24 | 2.02e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 6 | 46 | 2.14e-01 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 2 | 6 | 2.33e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 1 | 16 | 2.54e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 6 | 19 | 2.95e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 1 | 409 | 3.01e-01 |
| KEGG | KEGG:04530 | Tight junction | 5 | 137 | 3.01e-01 |
| KEGG | KEGG:04540 | Gap junction | 3 | 73 | 3.01e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 20 | 77 | 3.01e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 2 | 66 | 3.30e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 1 | 29 | 3.62e-01 |
| KEGG | KEGG:04145 | Phagosome | 5 | 99 | 3.83e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 1 | 84 | 3.84e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 22 | 87 | 4.07e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 3 | 106 | 4.12e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 10 | 115 | 4.12e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 4.12e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 4.13e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 2 | 91 | 4.39e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 35 | 109 | 4.39e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 2 | 42 | 4.39e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 2 | 8 | 4.39e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 2 | 34 | 4.39e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 2 | 49 | 4.39e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 2 | 91 | 4.39e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 27 | 56 | 4.44e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 2 | 73 | 4.44e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 5 | 18 | 4.44e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 1 | 180 | 4.44e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 4.44e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 2 | 6 | 4.44e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 2 | 55 | 4.44e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 8 | 86 | 4.44e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 4.44e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 7 | 30 | 4.44e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 8 | 145 | 4.44e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 28 | 56 | 4.66e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 4.70e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 2 | 112 | 4.88e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 12 | 55 | 4.88e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 9 | 107 | 5.06e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 6 | 58 | 5.06e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 4 | 157 | 5.06e-01 |
| KEGG | KEGG:05016 | Huntington disease | 5 | 256 | 5.06e-01 |
| KEGG | KEGG:03018 | RNA degradation | 2 | 74 | 5.06e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 2 | 22 | 5.06e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 7 | 1146 | 5.06e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 4 | 35 | 5.06e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 5.06e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 5.09e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 8 | 214 | 5.09e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 3 | 19 | 5.09e-01 |
| KEGG | KEGG:04976 | Bile secretion | 1 | 42 | 5.09e-01 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 1 | 5 | 5.09e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 3 | 74 | 5.09e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 4 | 114 | 5.09e-01 |
| KEGG | KEGG:05164 | Influenza A | 8 | 103 | 5.18e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 6 | 303 | 5.26e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 12 | 76 | 5.26e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 5.26e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 12 | 70 | 5.26e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 44 | 74 | 5.26e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 1 | 58 | 5.27e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 3 | 46 | 5.30e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 5.45e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 20 | 133 | 5.45e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 7 | 24 | 5.45e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 8 | 164 | 5.63e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 16 | 86 | 5.64e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 5.76e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 22 | 135 | 5.76e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 4 | 35 | 5.76e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 5.76e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 2 | 172 | 5.76e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 2 | 56 | 5.76e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 5.76e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 20 | 44 | 5.76e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 3 | 35 | 5.80e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 4 | 38 | 5.80e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 18 | 31 | 5.84e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 8 | 128 | 5.84e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 4 | 56 | 5.93e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 5.98e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 3 | 26 | 5.98e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 2 | 53 | 5.98e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 4 | 43 | 5.98e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 9 | 32 | 5.98e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 8 | 134 | 5.98e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 1 | 22 | 6.04e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 5 | 44 | 6.04e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 7 | 33 | 6.04e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 31 | 51 | 6.04e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 7 | 170 | 6.04e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 3 | 16 | 6.06e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 20 | 82 | 6.06e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 1 | 41 | 6.06e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 1 | 121 | 6.06e-01 |
| KEGG | KEGG:04146 | Peroxisome | 39 | 68 | 6.06e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 3 | 10 | 6.06e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 3 | 61 | 6.06e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 1 | 97 | 6.06e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 3 | 150 | 6.06e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 32 | 108 | 6.06e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 1 | 131 | 6.06e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 10 | 37 | 6.06e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 1 | 20 | 6.06e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 1 | 115 | 6.06e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 2 | 22 | 6.06e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 11 | 41 | 6.06e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 22 | 98 | 6.06e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 3 | 151 | 6.06e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 6 | 63 | 6.06e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 14 | 385 | 6.06e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 2 | 15 | 6.06e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 12 | 50 | 6.07e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 29 | 133 | 6.15e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 6.16e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 12 | 6.16e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 14 | 76 | 6.30e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 23 | 121 | 6.31e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 6.31e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 3 | 40 | 6.33e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 4 | 87 | 6.33e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 1 | 16 | 6.33e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 1 | 8 | 6.33e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 6.33e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 2 | 4 | 6.33e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 3 | 106 | 6.33e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 4 | 44 | 6.33e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 21 | 85 | 6.33e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1 | 69 | 6.66e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 12 | 56 | 6.66e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 1 | 21 | 6.66e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 3 | 46 | 6.66e-01 |
| KEGG | KEGG:04144 | Endocytosis | 4 | 232 | 6.66e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 1 | 63 | 6.85e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 14 | 72 | 6.85e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 1 | 83 | 6.85e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 6.85e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 2 | 51 | 6.85e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 1 | 40 | 6.85e-01 |
| KEGG | KEGG:04924 | Renin secretion | 12 | 50 | 6.85e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 2 | 22 | 6.85e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 6.85e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 1 | 28 | 6.85e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 2 | 18 | 6.85e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 4 | 28 | 6.85e-01 |
| KEGG | KEGG:04142 | Lysosome | 1 | 118 | 6.85e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 6.85e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 10 | 6.85e-01 |
| KEGG | KEGG:00232 | Caffeine metabolism | 1 | 1 | 6.85e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 6.85e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 6.85e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 2 | 22 | 6.85e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 3 | 82 | 6.91e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 36 | 99 | 7.04e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 1 | 67 | 7.04e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 11 | 17 | 7.04e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 16 | 28 | 7.06e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 2 | 23 | 7.06e-01 |
| KEGG | KEGG:05020 | Prion disease | 3 | 220 | 7.06e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 12 | 84 | 7.06e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 2 | 11 | 7.06e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 2 | 7 | 7.06e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 12 | 64 | 7.06e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 8 | 110 | 7.06e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 7.06e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 4 | 104 | 7.06e-01 |
| KEGG | KEGG:02010 | ABC transporters | 13 | 29 | 7.06e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 1 | 49 | 7.06e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 3 | 240 | 7.06e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 1 | 78 | 7.06e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 2 | 37 | 7.06e-01 |
| KEGG | KEGG:04520 | Adherens junction | 1 | 67 | 7.08e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 10 | 17 | 7.08e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 2 | 272 | 7.08e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 9 | 48 | 7.12e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 2 | 113 | 7.17e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 24 | 138 | 7.17e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 1 | 17 | 7.19e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 7.27e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 4 | 315 | 7.28e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 7.32e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 7.35e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 7 | 14 | 7.35e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 7.37e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 4 | 25 | 7.37e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 7.37e-01 |
| KEGG | KEGG:05142 | Chagas disease | 5 | 75 | 7.39e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 1 | 27 | 7.39e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 3 | 161 | 7.39e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 1 | 3 | 7.39e-01 |
| KEGG | KEGG:03040 | Spliceosome | 38 | 132 | 7.39e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 1 | 14 | 7.39e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 6 | 165 | 7.39e-01 |
| KEGG | KEGG:04730 | Long-term depression | 12 | 42 | 7.39e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 1 | 115 | 7.45e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 12 | 70 | 7.45e-01 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 7.45e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 2 | 38 | 7.51e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 7.66e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 1 | 88 | 7.66e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 12 | 49 | 7.69e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 10 | 78 | 7.69e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 3 | 49 | 7.77e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 7.77e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 13 | 77 | 7.79e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 2 | 171 | 7.79e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 7.79e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 8 | 65 | 7.79e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 7.79e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 8 | 25 | 7.79e-01 |
| KEGG | KEGG:04611 | Platelet activation | 6 | 89 | 7.79e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 7.81e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 1 | 98 | 7.81e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 1 | 12 | 7.81e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 7.91e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 7.93e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 1 | 23 | 7.93e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 7.93e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 3 | 55 | 7.93e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 3 | 83 | 7.93e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 2 | 20 | 7.93e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 8 | 15 | 7.93e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 3 | 79 | 7.93e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 7.93e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 1 | 12 | 7.93e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 7.93e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 9 | 110 | 8.02e-01 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 1 | 3 | 8.02e-01 |
| KEGG | KEGG:05131 | Shigellosis | 21 | 210 | 8.03e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 30 | 49 | 8.03e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 3 | 226 | 8.06e-01 |
| KEGG | KEGG:03050 | Proteasome | 7 | 42 | 8.10e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 5 | 24 | 8.12e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 2 | 33 | 8.12e-01 |
| KEGG | KEGG:03060 | Protein export | 5 | 23 | 8.12e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 6 | 24 | 8.12e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 8.12e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 9 | 52 | 8.12e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 4 | 104 | 8.15e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 3 | 13 | 8.15e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 1 | 112 | 8.25e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 11 | 50 | 8.26e-01 |
| KEGG | KEGG:00511 | Other glycan degradation | 3 | 16 | 8.28e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 8.29e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 1 | 124 | 8.37e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 3 | 415 | 8.48e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 8.53e-01 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 8.55e-01 |
| KEGG | KEGG:03265 | Virion - Lyssavirus | 1 | 3 | 8.55e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 8.55e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 1 | 106 | 8.55e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 1 | 69 | 8.60e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 6 | 10 | 8.61e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 2 | 74 | 8.72e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 21 | 43 | 8.74e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 2 | 179 | 8.75e-01 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 1 | 12 | 8.75e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 9 | 176 | 8.77e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 18 | 38 | 8.89e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 3 | 12 | 8.96e-01 |
| KEGG | KEGG:00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 2 | 2 | 9.06e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 5 | 94 | 9.11e-01 |
| KEGG | KEGG:05133 | Pertussis | 1 | 45 | 9.12e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 9.12e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 14 | 28 | 9.14e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 9.17e-01 |
| KEGG | KEGG:00780 | Biotin metabolism | 1 | 3 | 9.17e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 1 | 63 | 9.17e-01 |
| KEGG | KEGG:04744 | Phototransduction | 1 | 12 | 9.27e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 2 | 44 | 9.29e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 9.31e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 2 | 9 | 9.34e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 3 | 108 | 9.36e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 4 | 31 | 9.36e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 2 | 12 | 9.37e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 14 | 97 | 9.45e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 5 | 53 | 9.50e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 1 | 32 | 9.53e-01 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 3 | 23 | 9.54e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 26 | 86 | 9.54e-01 |
| KEGG | KEGG:05134 | Legionellosis | 4 | 39 | 9.55e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 1 | 31 | 9.61e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 6 | 60 | 9.61e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 2 | 25 | 9.61e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 16 | 103 | 9.61e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 19 | 142 | 9.61e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 3 | 99 | 9.69e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 1 | 186 | 9.96e-01 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 1 | 19 | 9.96e-01 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 2 | 26 | 9.99e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 4 | 41 | 9.99e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 3 | 33 | 9.99e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 2 | 26 | 9.99e-01 |
| KEGG | KEGG:04742 | Taste transduction | 1 | 22 | 9.99e-01 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 2 | 9 | 9.99e-01 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 5 | 13 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 14 | 31 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 17 | 127 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 4 | 75 | 1.00e+00 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 4 | 19 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 4 | 36 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 2 | 98 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 6 | 89 | 1.00e+00 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 5 | 1.00e+00 |
| KEGG | KEGG:05310 | Asthma | 1 | 3 | 1.00e+00 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 3 | 9 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 2 | 169 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 2 | 12 | 1.00e+00 |
###DNR 3 v 24 intersection
All 322 3 hour DNR-specific genes are in the intersection of the DNR sigDEGs for 3 hours and 24 hours. This could support the idea that DNR’s more toxic effect is related to earlier impacts on cell function. Note the enriched go terms for the 304 genes that they share. I need to see if there is a trend in EP and DX. I do see in Mito 3 hour (looked a little later).
daun3v24int <- intersect(sigVDA3$ENTREZID, sigVDA24$ENTREZID)
length(union(sigVDA3$ENTREZID, sigVDA24$ENTREZID))[1] 7237ggVennDiagram::ggVennDiagram(
list(sigVDA3$ENTREZID, sigVDA24$ENTREZID),
category.names = c("3 hour DNR\nn = 532", "24 hour DNR\n n = 7017"),
show_intersect = FALSE,
set_color = "black",
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid"
) +
scale_x_continuous(expand = expansion(mult = .2)) +
scale_y_continuous(expand = expansion(mult = .1)) +
scale_fill_gradient(low = "red4", high = "tan1") +
labs(title = "DEG p < 0.05, All DNR 3hour and 24 hours sets",
caption = paste("n =", length(
union(sigVDA3$ENTREZID, sigVDA24$ENTREZID)
), "genes")) +
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5))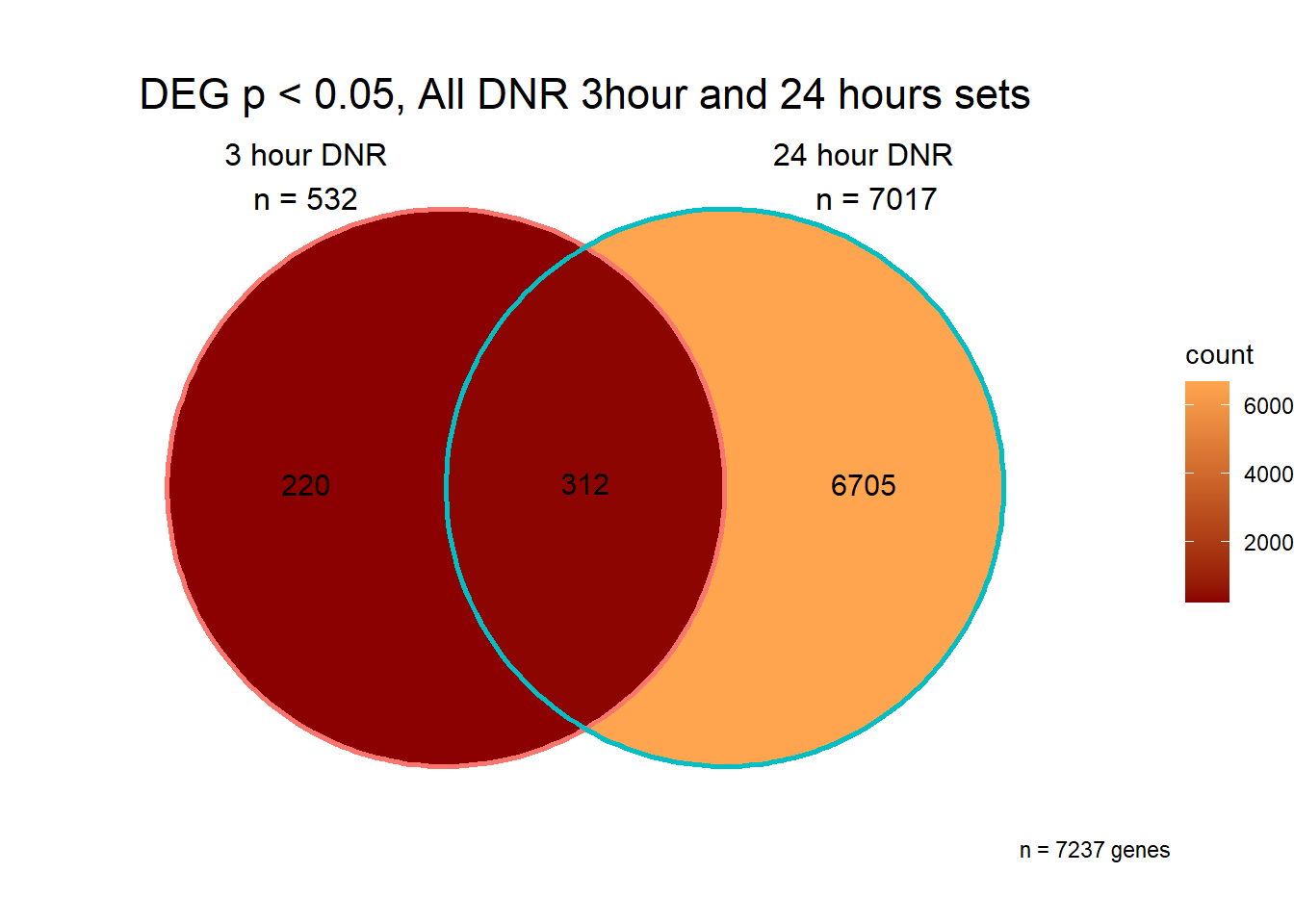
length(intersect(over3_24daun, intersect(sigVDA3$ENTREZID, sigVDA24$ENTREZID)))[1] 195# DAgostresint <- gost(query = daun3v24int,
# organism = "hsapiens", significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(DAgostresint,"data/DEG-GO/DAgostresint.RDS")
DAgostresint <- readRDS("data/DEG-GO/DAgostresint.RDS")
# DApint <- gostplot(DAgostresint, capped = FALSE, interactive = TRUE)
# DApint
DAtable_int <- DAgostresint$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DAtable_int %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('DNR intersect 3 v 24 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)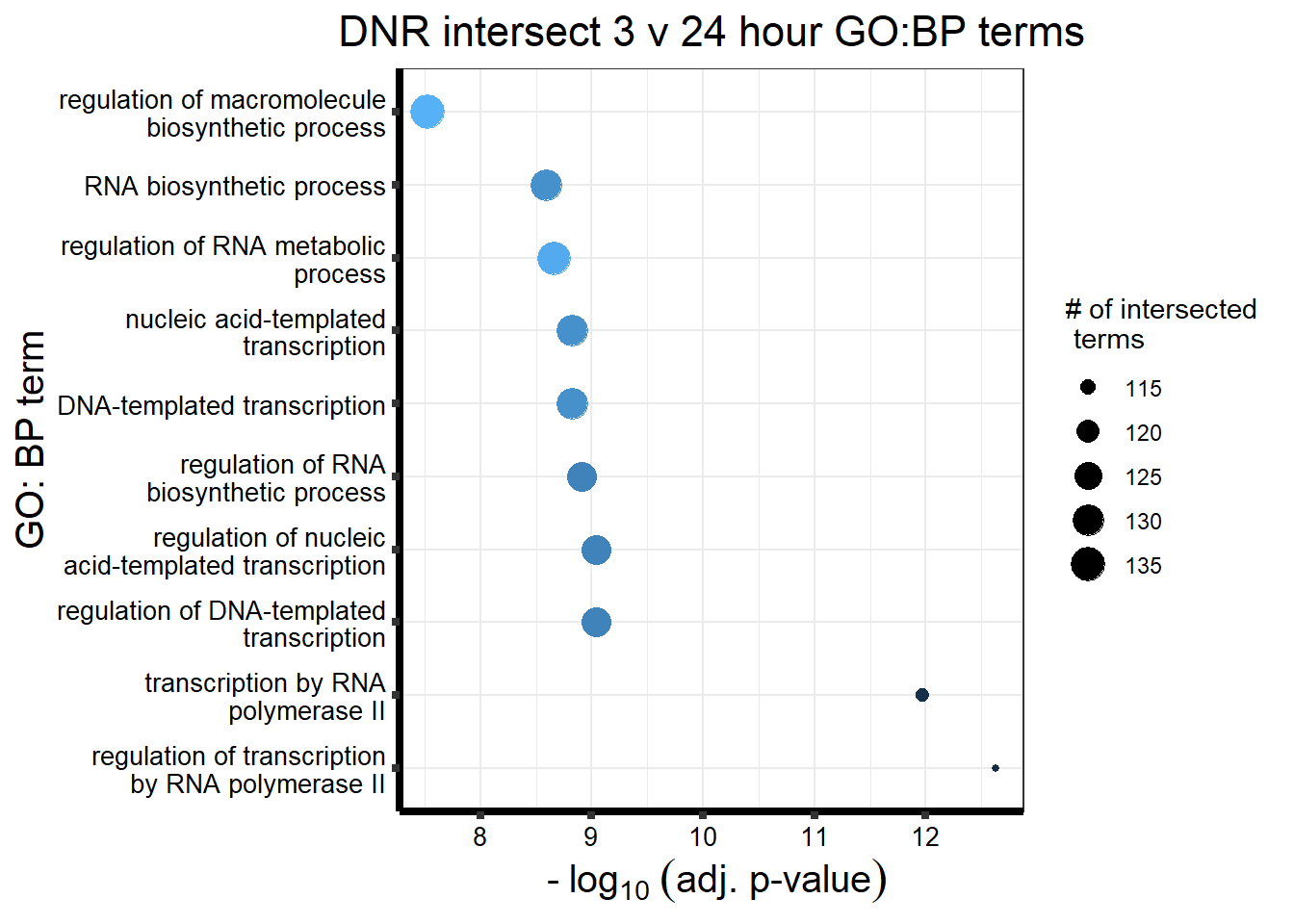
DAtable_int %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 113 | 1914 | 2.36e-13 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 114 | 1995 | 1.07e-12 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 128 | 2576 | 9.23e-10 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 128 | 2574 | 9.23e-10 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 128 | 2593 | 1.23e-09 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 130 | 2684 | 1.51e-09 |
| GO:BP | GO:0006351 | DNA-templated transcription | 130 | 2683 | 1.51e-09 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 134 | 2845 | 2.19e-09 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 130 | 2714 | 2.56e-09 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 135 | 3012 | 2.97e-08 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 53 | 740 | 3.65e-08 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 139 | 3080 | 1.07e-07 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 135 | 3080 | 1.63e-07 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 137 | 3169 | 2.49e-07 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 67 | 3132 | 5.86e-07 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 67 | 3133 | 6.21e-07 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 66 | 3070 | 6.98e-07 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 68 | 3237 | 9.00e-07 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 33 | 4292 | 2.22e-06 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 36 | 4417 | 2.45e-06 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 35 | 4174 | 2.62e-06 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 60 | 1015 | 7.87e-06 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 60 | 1017 | 7.95e-06 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 60 | 1026 | 1.11e-05 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 61 | 1119 | 1.28e-05 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 32 | 3730 | 1.34e-05 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 69 | 3750 | 6.81e-05 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 60 | 1214 | 1.20e-04 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 61 | 1209 | 1.60e-04 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 34 | 4617 | 1.77e-04 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 60 | 1239 | 2.47e-04 |
| GO:BP | GO:0010468 | regulation of gene expression | 29 | 3580 | 3.58e-04 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 78 | 1735 | 5.22e-04 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 60 | 1280 | 6.14e-04 |
| GO:BP | GO:0019222 | regulation of metabolic process | 34 | 5017 | 1.32e-03 |
| GO:BP | GO:0003162 | atrioventricular node development | 3 | 8 | 1.36e-03 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 71 | 4245 | 1.49e-03 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 29 | 4539 | 1.61e-03 |
| GO:BP | GO:0009058 | biosynthetic process | 29 | 4590 | 2.11e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 17 | 216 | 4.10e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 5 | 67 | 6.64e-03 |
| GO:BP | GO:0014706 | striated muscle tissue development | 15 | 210 | 1.05e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 58 | 1315 | 1.32e-02 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 58 | 1315 | 1.32e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 58 | 1322 | 1.44e-02 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 14 | 198 | 1.61e-02 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 2 | 4 | 2.03e-02 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 2 | 2 | 2.03e-02 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 75 | 2412 | 2.09e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 6 | 80 | 2.12e-02 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 65 | 1564 | 2.12e-02 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 3 | 20 | 2.12e-02 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 60 | 1441 | 2.69e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 22 | 937 | 2.69e-02 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 75 | 1849 | 2.75e-02 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 62 | 1507 | 2.77e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 5 | 101 | 3.52e-02 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 2 | 6 | 3.54e-02 |
| GO:BP | GO:1903674 | regulation of cap-dependent translational initiation | 1 | 1 | 3.77e-02 |
| GO:BP | GO:1903676 | positive regulation of cap-dependent translational initiation | 1 | 1 | 3.77e-02 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 65 | 1604 | 3.77e-02 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 3 | 9 | 3.77e-02 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 3 | 15 | 3.77e-02 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 3 | 9 | 3.77e-02 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 2 | 7 | 3.77e-02 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 3 | 27 | 3.77e-02 |
| GO:BP | GO:0016070 | RNA metabolic process | 29 | 3600 | 4.08e-02 |
| GO:BP | GO:0003161 | cardiac conduction system development | 4 | 33 | 4.12e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 3 | 40 | 4.14e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 3 | 50 | 4.32e-02 |
| GO:BP | GO:0006308 | DNA catabolic process | 3 | 19 | 4.36e-02 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 3 | 17 | 4.36e-02 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 4.36e-02 |
| GO:BP | GO:0003197 | endocardial cushion development | 6 | 46 | 4.36e-02 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 1 | 8 | 4.40e-02 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 3 | 11 | 4.83e-02 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 4 | 34 | 4.83e-02 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 28 | 4048 | 4.83e-02 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 7 | 68 | 4.83e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 5 | 109 | 4.90e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 33 | 415 | 5.40e-06 |
DOX 3 hour and 24 hour
# DXgostres <- gost(query = sigVDX3$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(DXgostres,"data/DEG-GO/DXgostres.RDS")
DXgostres <- readRDS("data/DEG-GO/DXgostres.RDS")
# DXp <- gostplot(DXgostres, capped = FALSE, interactive = TRUE)
# DXp
DXtable1 <- DXgostres$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DXtable1 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 8 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('DOX 3 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 9,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)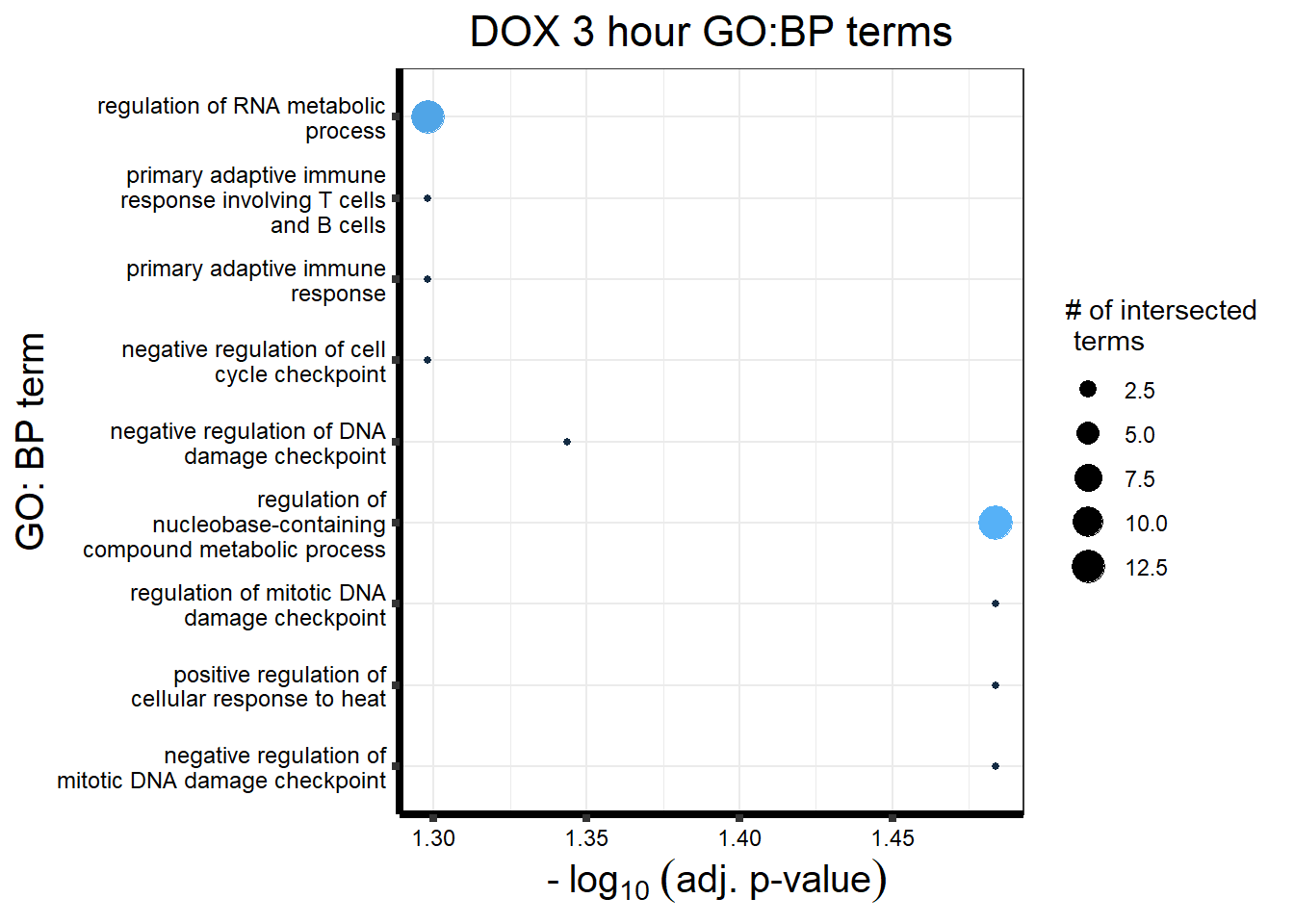
DXtable1 %>% mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "3 hour list of DOX KEGG and GO:BP terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 3.28e-02 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 13 | 3080 | 3.28e-02 |
| GO:BP | GO:1904290 | negative regulation of mitotic DNA damage checkpoint | 1 | 1 | 3.28e-02 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 3.28e-02 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 4.53e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 5.03e-02 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 12 | 2845 | 5.03e-02 |
| GO:BP | GO:0090720 | primary adaptive immune response | 1 | 1 | 5.03e-02 |
| GO:BP | GO:0090721 | primary adaptive immune response involving T cells and B cells | 1 | 1 | 5.03e-02 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 9 | 1995 | 5.39e-02 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 11 | 2576 | 5.39e-02 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 11 | 2714 | 5.39e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 5.39e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 1 | 33 | 5.39e-02 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 1 | 3 | 5.39e-02 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 11 | 2574 | 5.39e-02 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 11 | 2684 | 5.39e-02 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 16 | 4048 | 5.39e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 5.39e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 1 | 36 | 5.39e-02 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 14 | 4417 | 5.39e-02 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 1 | 36 | 5.39e-02 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 1 | 7 | 5.39e-02 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 14 | 4292 | 5.39e-02 |
| GO:BP | GO:0006351 | DNA-templated transcription | 11 | 2683 | 5.39e-02 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 5.39e-02 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 11 | 2593 | 5.39e-02 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 1 | 25 | 5.39e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 1 | 39 | 5.47e-02 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 3 | 1602 | 5.87e-02 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 5.97e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 6.01e-02 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 1 | 60 | 6.87e-02 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 1 | 13 | 6.87e-02 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 1 | 64 | 6.87e-02 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 7 | 2412 | 6.87e-02 |
| GO:BP | GO:0015074 | DNA integration | 1 | 9 | 6.87e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 1 | 56 | 6.87e-02 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 11 | 3012 | 6.87e-02 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 1 | 14 | 7.07e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 2 | 616 | 7.08e-02 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 11 | 3070 | 7.08e-02 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 11 | 3080 | 7.08e-02 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 1 | 85 | 7.08e-02 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 1 | 19 | 7.08e-02 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 1 | 9 | 7.08e-02 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 16 | 4455 | 7.08e-02 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 13 | 4174 | 7.08e-02 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 11 | 3133 | 7.08e-02 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 1 | 81 | 7.08e-02 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 1 | 19 | 7.08e-02 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 1 | 19 | 7.08e-02 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 11 | 3169 | 7.08e-02 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 11 | 3132 | 7.08e-02 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 8 | 1914 | 7.08e-02 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 1 | 96 | 7.08e-02 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 11 | 3237 | 7.19e-02 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 7.19e-02 |
| GO:BP | GO:0051170 | import into nucleus | 2 | 152 | 7.19e-02 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 1 | 113 | 7.19e-02 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 7.19e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 7 | 2646 | 7.19e-02 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 1 | 20 | 7.19e-02 |
| GO:BP | GO:0007614 | short-term memory | 1 | 9 | 7.19e-02 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 13 | 4596 | 7.19e-02 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 7.19e-02 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 13 | 4570 | 7.19e-02 |
| GO:BP | GO:0043574 | peroxisomal transport | 1 | 22 | 7.19e-02 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 1 | 103 | 7.19e-02 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 7.34e-02 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 1 | 26 | 7.50e-02 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 3 | 2270 | 7.50e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 7.50e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 1 | 142 | 8.16e-02 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 8.36e-02 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 8.36e-02 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 1 | 36 | 8.36e-02 |
| GO:BP | GO:0034504 | protein localization to nucleus | 2 | 276 | 8.36e-02 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 8.36e-02 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 13 | 8.36e-02 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 13 | 4747 | 8.36e-02 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 178 | 8.36e-02 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 8.36e-02 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 8.36e-02 |
| GO:BP | GO:0007031 | peroxisome organization | 1 | 34 | 8.36e-02 |
| GO:BP | GO:0032200 | telomere organization | 1 | 166 | 8.36e-02 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 8.36e-02 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 2 | 274 | 8.36e-02 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 8.66e-02 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 8.66e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 7 | 2880 | 8.66e-02 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 8.66e-02 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 14 | 4617 | 8.66e-02 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 8.66e-02 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 8.66e-02 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 8.66e-02 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 8.66e-02 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 13 | 4946 | 8.79e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 8.81e-02 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 9.55e-02 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 1 | 209 | 9.57e-02 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 9.59e-02 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 1 | 52 | 1.00e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 14 | 3600 | 1.01e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 1 | 55 | 1.06e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 1.06e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 1.06e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 12 | 3580 | 1.06e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 1.06e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 2 | 937 | 1.09e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 1.09e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 11 | 3730 | 1.09e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 1 | 295 | 1.09e-01 |
| GO:BP | GO:0051169 | nuclear transport | 1 | 295 | 1.09e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 1.09e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 1.11e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 3 | 859 | 1.12e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 11 | 3750 | 1.12e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 4 | 1315 | 1.20e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 338 | 1.20e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 4 | 1315 | 1.20e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 1 | 22 | 1.20e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 4 | 1322 | 1.20e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 1.21e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 1 | 21 | 1.21e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 1.24e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 2 | 1075 | 1.26e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 391 | 1.28e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 1.29e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 14 | 5017 | 1.33e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 1 | 44 | 1.33e-01 |
| GO:BP | GO:0016180 | snRNA processing | 1 | 26 | 1.33e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 1.33e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 1.36e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 1.38e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 441 | 1.38e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 4 | 1441 | 1.38e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 438 | 1.40e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 1 | 439 | 1.40e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 1.42e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 1.47e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 504 | 1.49e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 4 | 1507 | 1.53e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 1 | 108 | 1.57e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 1.57e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 1.60e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 543 | 1.63e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 1.66e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 4 | 1564 | 1.68e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 1 | 126 | 1.72e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 1.72e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 1 | 62 | 1.72e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 626 | 1.78e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 4 | 1604 | 1.78e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 1.83e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 694 | 1.83e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 2 | 1681 | 1.83e-01 |
| GO:BP | GO:0048511 | rhythmic process | 2 | 237 | 1.83e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 1.91e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 1.93e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 2 | 258 | 1.93e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 1 | 72 | 1.99e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 785 | 2.00e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 2.00e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 11 | 4245 | 2.00e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 2.00e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 660 | 2.00e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 678 | 2.03e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 2 | 400 | 2.04e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 1 | 55 | 2.05e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 827 | 2.05e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 2.08e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 3 | 4099 | 2.09e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 2.18e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 1 | 756 | 2.18e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 1 | 884 | 2.20e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 84 | 2.20e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 2 | 400 | 2.20e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 8 | 4579 | 2.20e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 1 | 191 | 2.20e-01 |
| GO:BP | GO:0007049 | cell cycle | 3 | 1529 | 2.26e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 2.26e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 1 | 85 | 2.26e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1 | 91 | 2.27e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 1 | 906 | 2.27e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 2.27e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 1 | 956 | 2.27e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 2.27e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 1 | 82 | 2.28e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 2.30e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 97 | 2.33e-01 |
| GO:BP | GO:0006325 | chromatin organization | 2 | 551 | 2.33e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 1 | 1028 | 2.33e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 2.36e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 1 | 232 | 2.37e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 1 | 1006 | 2.41e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 2.41e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1086 | 2.43e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 1 | 99 | 2.43e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 2.43e-01 |
| GO:BP | GO:0007613 | memory | 1 | 92 | 2.45e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 110 | 2.49e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 11 | 4539 | 2.51e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 1 | 1076 | 2.51e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 1 | 106 | 2.51e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 1 | 1074 | 2.51e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 2.58e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 1 | 1088 | 2.65e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 1 | 96 | 2.66e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 1 | 120 | 2.66e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 1 | 1111 | 2.66e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 11 | 4590 | 2.66e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 2.67e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 1 | 122 | 2.76e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 1 | 131 | 2.77e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 3 | 1147 | 2.77e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 2.77e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 1 | 120 | 2.77e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 1 | 1248 | 2.79e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 2.82e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 1 | 1219 | 2.82e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 1 | 149 | 2.83e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 135 | 2.83e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 2.83e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 1 | 151 | 2.86e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1 | 1345 | 2.88e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 2.88e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 95 | 2.91e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1361 | 2.98e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1453 | 3.00e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 3.00e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 3.01e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 1 | 127 | 3.01e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 2 | 561 | 3.01e-01 |
| GO:BP | GO:0006915 | apoptotic process | 1 | 1443 | 3.06e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1462 | 3.07e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 159 | 3.08e-01 |
| GO:BP | GO:0012501 | programmed cell death | 1 | 1479 | 3.11e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 2 | 679 | 3.23e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 3.23e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 152 | 3.23e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 2 | 631 | 3.24e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 1 | 96 | 3.24e-01 |
| GO:BP | GO:0008219 | cell death | 1 | 1596 | 3.29e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 1 | 168 | 3.36e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 1 | 167 | 3.37e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 3.37e-01 |
| GO:BP | GO:0007498 | mesoderm development | 1 | 89 | 3.41e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1684 | 3.47e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 3.50e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 3 | 1378 | 3.50e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 1 | 164 | 3.50e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 189 | 3.53e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 3.53e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 1 | 161 | 3.53e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 1 | 123 | 3.53e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 1 | 184 | 3.55e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 3.55e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 1 | 187 | 3.56e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 3.56e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 1 | 280 | 3.56e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1 | 2011 | 3.56e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 2 | 719 | 3.56e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 3.58e-01 |
| GO:BP | GO:0040008 | regulation of growth | 2 | 501 | 3.58e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 213 | 3.58e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 3.59e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 1 | 200 | 3.62e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 1 | 124 | 3.63e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 3.64e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 3.64e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 2 | 816 | 3.64e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1 | 2065 | 3.67e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 1 | 331 | 3.67e-01 |
| GO:BP | GO:0048589 | developmental growth | 2 | 522 | 3.67e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2129 | 3.73e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2138 | 3.73e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 3.74e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 3.81e-01 |
| GO:BP | GO:0006950 | response to stress | 2 | 2780 | 3.83e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2 | 2890 | 3.83e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 3.83e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 14 | 7441 | 3.85e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 251 | 3.93e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 4 | 1849 | 3.93e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1 | 2285 | 3.96e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2116 | 3.97e-01 |
| GO:BP | GO:0010467 | gene expression | 14 | 4587 | 3.98e-01 |
| GO:BP | GO:0007369 | gastrulation | 1 | 146 | 4.00e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 4.00e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 2 | 834 | 4.00e-01 |
| GO:BP | GO:0050890 | cognition | 1 | 227 | 4.04e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 1 | 277 | 4.05e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 868 | 4.05e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 1 | 246 | 4.06e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 273 | 4.06e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 4.07e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 1 | 270 | 4.07e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2450 | 4.07e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 4.13e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 1 | 300 | 4.25e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 1 | 303 | 4.25e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 1 | 2565 | 4.25e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 1 | 2573 | 4.25e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 14 | 7069 | 4.34e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 1 | 260 | 4.34e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 4.36e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 4.36e-01 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 317 | 4.36e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 305 | 4.36e-01 |
| GO:BP | GO:0070997 | neuron death | 1 | 286 | 4.36e-01 |
| GO:BP | GO:0006996 | organelle organization | 1 | 3000 | 4.37e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 4.37e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2810 | 4.37e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 1 | 717 | 4.40e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 206 | 4.40e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 1 | 267 | 4.42e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 4.45e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 1 | 344 | 4.57e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 1 | 345 | 4.57e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 2 | 957 | 4.59e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 1 | 787 | 4.65e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 357 | 4.75e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 4.75e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 4.78e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 4.78e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 1 | 317 | 4.78e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 344 | 4.79e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 1 | 375 | 4.81e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 4.81e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 1 | 376 | 4.82e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 4.87e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 359 | 4.87e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 4.97e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 2 | 665 | 4.97e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 1 | 418 | 5.00e-01 |
| GO:BP | GO:0040007 | growth | 2 | 742 | 5.00e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 2 | 1123 | 5.00e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 1 | 898 | 5.00e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 5.00e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 5.00e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 3 | 1974 | 5.00e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 13 | 6996 | 5.03e-01 |
| GO:BP | GO:0016049 | cell growth | 1 | 418 | 5.07e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 5.07e-01 |
| GO:BP | GO:0043588 | skin development | 1 | 175 | 5.07e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 398 | 5.07e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 1 | 420 | 5.07e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 2 | 1173 | 5.07e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 5.08e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1 | 3625 | 5.11e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 5.14e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 2 | 1149 | 5.14e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 5.29e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 2 | 1137 | 5.32e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 1 | 414 | 5.36e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 1 | 448 | 5.47e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 5.54e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 5.69e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 1 | 479 | 5.69e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 3 | 1735 | 5.69e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 14 | 7806 | 5.69e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 5.79e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 5.80e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 5.80e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 5.83e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 1 | 512 | 5.83e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 5.85e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 5.87e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 5.87e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 1 | 527 | 5.87e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 1 | 531 | 5.89e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 2 | 901 | 5.96e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1 | 4045 | 5.98e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 1 | 551 | 5.98e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 6.03e-01 |
| GO:BP | GO:0007165 | signal transduction | 1 | 3818 | 6.04e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 496 | 6.05e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 2 | 1119 | 6.05e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 2 | 1380 | 6.08e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 1 | 560 | 6.09e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 998 | 6.13e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 1 | 875 | 6.14e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 6.15e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 6.16e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 4 | 2135 | 6.16e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 6.16e-01 |
| GO:BP | GO:0006396 | RNA processing | 2 | 929 | 6.16e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 6.18e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 6.21e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 6.22e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 6.22e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 6.22e-01 |
| GO:BP | GO:0023052 | signaling | 1 | 4141 | 6.22e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 547 | 6.22e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 6.25e-01 |
| GO:BP | GO:0007154 | cell communication | 1 | 4226 | 6.27e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 1 | 639 | 6.27e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 6.27e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 2 | 1209 | 6.27e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 2 | 785 | 6.31e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 605 | 6.41e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 6.43e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 1 | 620 | 6.46e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 6.47e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 597 | 6.47e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 597 | 6.47e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 691 | 6.54e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 4 | 2293 | 6.54e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 1 | 538 | 6.64e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 3 | 7552 | 6.82e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 556 | 6.83e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 6.88e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 1 | 809 | 6.91e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 6.93e-01 |
| GO:BP | GO:0031175 | neuron projection development | 1 | 780 | 6.97e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 755 | 6.98e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 1 | 440 | 6.98e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 676 | 6.98e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 7.04e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 1 | 504 | 7.05e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1199 | 7.05e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 740 | 7.13e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 1 | 796 | 7.20e-01 |
| GO:BP | GO:0016043 | cellular component organization | 1 | 5076 | 7.23e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1 | 5274 | 7.40e-01 |
| GO:BP | GO:0048666 | neuron development | 1 | 877 | 7.45e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 2 | 1835 | 7.48e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 1 | 881 | 7.54e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 1 | 615 | 7.54e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 1 | 625 | 7.59e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 1 | 637 | 7.63e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 963 | 7.63e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 2 | 1939 | 7.67e-01 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5770 | 7.88e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 1 | 994 | 7.97e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 1 | 993 | 7.97e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 468 | 8.00e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 1 | 706 | 8.02e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 1014 | 8.16e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 1 | 1061 | 8.16e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 1 | 1017 | 8.18e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 1 | 1015 | 8.18e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 1 | 1026 | 8.21e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 8.24e-01 |
| GO:BP | GO:0048699 | generation of neurons | 1 | 1119 | 8.29e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 8.34e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 8.34e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 8.34e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 8.34e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1201 | 8.34e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 1 | 1167 | 8.35e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 2 | 2035 | 8.37e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1233 | 8.38e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 8.45e-01 |
| GO:BP | GO:0042592 | homeostatic process | 1 | 1159 | 8.58e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 8.58e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1 | 1214 | 8.58e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 1 | 742 | 8.59e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1294 | 8.59e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 3 | 8061 | 8.59e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 1 | 1239 | 8.59e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1138 | 8.59e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 8.60e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 8.60e-01 |
| GO:BP | GO:0036211 | protein modification process | 1 | 2901 | 8.64e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1 | 1280 | 8.64e-01 |
| GO:BP | GO:0016310 | phosphorylation | 1 | 1372 | 8.64e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1223 | 8.64e-01 |
| GO:BP | GO:0009790 | embryo development | 1 | 875 | 8.68e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 870 | 8.75e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 1 | 3090 | 8.80e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2088 | 8.87e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 9.13e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 9.15e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 9.15e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 9.15e-01 |
| GO:BP | GO:0065007 | biological regulation | 3 | 8320 | 9.17e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 9.30e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 9.33e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 9.39e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 3 | 4241 | 9.39e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1904 | 9.54e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 2 | 1974 | 9.54e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 665 | 9.60e-01 |
| GO:BP | GO:0030154 | cell differentiation | 2 | 2958 | 9.60e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 2 | 2984 | 9.60e-01 |
| GO:BP | GO:0010033 | response to organic substance | 1 | 1957 | 9.63e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1 | 2139 | 9.63e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1 | 2161 | 9.63e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 9.63e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 9.65e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1 | 1898 | 9.65e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1719 | 9.65e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 9.65e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 1 | 2102 | 9.65e-01 |
| GO:BP | GO:0009888 | tissue development | 1 | 1425 | 9.74e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2 | 4955 | 9.81e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0007275 | multicellular organism development | 3 | 3386 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 1.00e+00 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 1.00e+00 |
| GO:BP | GO:0032501 | multicellular organismal process | 2 | 4794 | 1.00e+00 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1335 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 1.00e+00 |
| GO:BP | GO:0048731 | system development | 2 | 2901 | 1.00e+00 |
| GO:BP | GO:0048856 | anatomical structure development | 3 | 4128 | 1.00e+00 |
| GO:BP | GO:0032502 | developmental process | 2 | 4494 | 1.00e+00 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1496 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 1 | 106 | 7.37e-02 |
| KEGG | KEGG:04146 | Peroxisome | 1 | 68 | 7.37e-02 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 7.42e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 2 | 415 | 1.47e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 8.41e-01 |
# DXgostres24 <- gost(query = sigVDX24$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(DXgostres24,"data/DEG-GO/gostres24.RDS")
DXgostres24 <- readRDS("data/DEG-GO/gostres24.RDS")
# gostres24 <- readRDS("data/DEG-GO/gostres24.RDS")
# p24 <- gostplot(DXgostres24, capped = FALSE, interactive = TRUE)
# p24
DXtable124 <- DXgostres24$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DXtable124 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 8 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected\n terms")) +
ggtitle('DOX 24 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)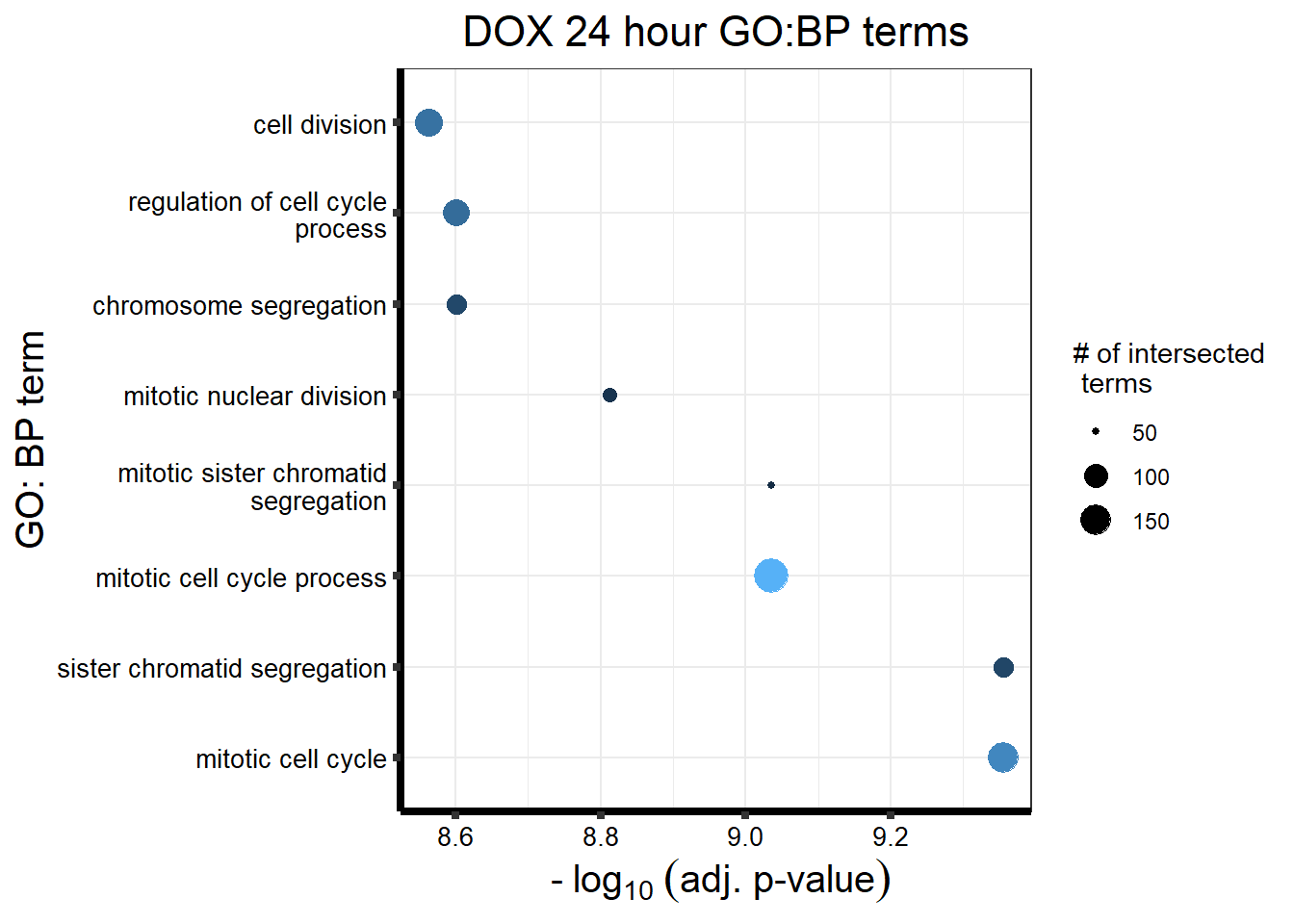
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DXtable124 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "24 hour list of DOX KEGG and GO:BP terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0000278 | mitotic cell cycle | 146 | 827 | 4.42e-10 |
| GO:BP | GO:0000819 | sister chromatid segregation | 80 | 217 | 4.42e-10 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 50 | 179 | 9.23e-10 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 185 | 694 | 9.23e-10 |
| GO:BP | GO:0140014 | mitotic nuclear division | 59 | 251 | 1.54e-09 |
| GO:BP | GO:0007059 | chromosome segregation | 82 | 369 | 2.50e-09 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 119 | 626 | 2.50e-09 |
| GO:BP | GO:0051301 | cell division | 125 | 570 | 2.73e-09 |
| GO:BP | GO:0051276 | chromosome organization | 104 | 543 | 7.34e-08 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 47 | 139 | 1.24e-07 |
| GO:BP | GO:0051726 | regulation of cell cycle | 158 | 956 | 1.50e-07 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 59 | 211 | 1.99e-07 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 50 | 183 | 2.53e-07 |
| GO:BP | GO:0022402 | cell cycle process | 168 | 1086 | 3.19e-07 |
| GO:BP | GO:0000280 | nuclear division | 78 | 344 | 3.38e-07 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 62 | 273 | 9.80e-07 |
| GO:BP | GO:0007051 | spindle organization | 48 | 188 | 1.73e-06 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 12 | 18 | 2.76e-06 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 39 | 157 | 2.77e-06 |
| GO:BP | GO:0007052 | mitotic spindle organization | 34 | 129 | 2.95e-06 |
| GO:BP | GO:0007049 | cell cycle | 217 | 1529 | 3.06e-06 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 28 | 71 | 4.57e-06 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 31 | 121 | 4.86e-06 |
| GO:BP | GO:0048285 | organelle fission | 80 | 388 | 4.86e-06 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 39 | 90 | 5.95e-06 |
| GO:BP | GO:0006974 | DNA damage response | 121 | 785 | 6.00e-06 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 19 | 42 | 6.00e-06 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 59 | 209 | 6.01e-06 |
| GO:BP | GO:0051225 | spindle assembly | 35 | 120 | 7.17e-06 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 20 | 55 | 8.98e-06 |
| GO:BP | GO:0006281 | DNA repair | 79 | 534 | 9.43e-06 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 82 | 441 | 9.52e-06 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 96 | 338 | 1.10e-05 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 92 | 273 | 1.68e-05 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 72 | 148 | 1.68e-05 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 65 | 235 | 1.68e-05 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 68 | 236 | 1.84e-05 |
| GO:BP | GO:0051304 | chromosome separation | 28 | 76 | 1.84e-05 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 71 | 391 | 2.20e-05 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 28 | 79 | 2.27e-05 |
| GO:BP | GO:0006260 | DNA replication | 125 | 255 | 2.52e-05 |
| GO:BP | GO:0051310 | metaphase plate congression | 22 | 69 | 2.64e-05 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 7 | 7 | 2.64e-05 |
| GO:BP | GO:0051231 | spindle elongation | 9 | 12 | 3.60e-05 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 20 | 52 | 4.21e-05 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 9 | 4.30e-05 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 104 | 552 | 4.55e-05 |
| GO:BP | GO:0006541 | glutamine metabolic process | 4 | 24 | 4.66e-05 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 36 | 91 | 4.72e-05 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 20 | 54 | 5.34e-05 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 34 | 100 | 6.03e-05 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 14 | 27 | 6.51e-05 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 46 | 129 | 7.20e-05 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 8 | 8 | 7.24e-05 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 14 | 44 | 7.24e-05 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 14 | 44 | 7.24e-05 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 14 | 44 | 7.24e-05 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 18 | 46 | 7.98e-05 |
| GO:BP | GO:0044770 | cell cycle phase transition | 81 | 504 | 8.41e-05 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 14 | 45 | 8.84e-05 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 34 | 47 | 8.84e-05 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 10 | 8.84e-05 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 34 | 47 | 8.84e-05 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 36 | 92 | 8.84e-05 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 34 | 47 | 8.84e-05 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 35 | 99 | 8.84e-05 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 22 | 58 | 9.13e-05 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 28 | 88 | 1.18e-04 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 56 | 308 | 1.27e-04 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 18 | 47 | 1.34e-04 |
| GO:BP | GO:0051255 | spindle midzone assembly | 9 | 12 | 1.38e-04 |
| GO:BP | GO:0010212 | response to ionizing radiation | 41 | 130 | 1.47e-04 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 25 | 81 | 1.61e-04 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 29 | 85 | 1.73e-04 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 10 | 21 | 1.82e-04 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 46 | 166 | 1.82e-04 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 22 | 61 | 2.05e-04 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 28 | 89 | 2.14e-04 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 18 | 48 | 2.28e-04 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 18 | 48 | 2.28e-04 |
| GO:BP | GO:0051302 | regulation of cell division | 37 | 145 | 2.41e-04 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 22 | 75 | 2.52e-04 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 69 | 418 | 2.59e-04 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 18 | 57 | 2.72e-04 |
| GO:BP | GO:0050000 | chromosome localization | 23 | 82 | 2.73e-04 |
| GO:BP | GO:0051783 | regulation of nuclear division | 25 | 116 | 3.34e-04 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 15 | 70 | 3.53e-04 |
| GO:BP | GO:0046037 | GMP metabolic process | 4 | 23 | 4.11e-04 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 31 | 121 | 4.31e-04 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 7 | 11 | 5.27e-04 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 3 | 14 | 5.27e-04 |
| GO:BP | GO:0006177 | GMP biosynthetic process | 3 | 13 | 5.27e-04 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 66 | 274 | 5.40e-04 |
| GO:BP | GO:0007017 | microtubule-based process | 50 | 760 | 5.41e-04 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 6.32e-04 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 22 | 73 | 6.32e-04 |
| GO:BP | GO:0006270 | DNA replication initiation | 12 | 35 | 6.32e-04 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 6.32e-04 |
| GO:BP | GO:0000910 | cytokinesis | 40 | 164 | 6.55e-04 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 24 | 81 | 7.53e-04 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 31 | 178 | 7.63e-04 |
| GO:BP | GO:0007098 | centrosome cycle | 29 | 127 | 7.96e-04 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 11 | 28 | 8.85e-04 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 28 | 104 | 1.17e-03 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 10 | 69 | 1.24e-03 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 27 | 47 | 1.24e-03 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 16 | 48 | 1.25e-03 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 12 | 36 | 1.35e-03 |
| GO:BP | GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 3 | 19 | 1.37e-03 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 30 | 137 | 1.69e-03 |
| GO:BP | GO:0046060 | dATP metabolic process | 2 | 3 | 1.70e-03 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 4 | 60 | 1.77e-03 |
| GO:BP | GO:0009127 | purine nucleoside monophosphate biosynthetic process | 3 | 21 | 1.77e-03 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 8 | 11 | 1.80e-03 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 8 | 42 | 1.82e-03 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 4 | 37 | 2.10e-03 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 24 | 68 | 2.26e-03 |
| GO:BP | GO:0051383 | kinetochore organization | 13 | 23 | 2.26e-03 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 4 | 39 | 2.43e-03 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 6 | 9 | 2.43e-03 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 28 | 134 | 2.64e-03 |
| GO:BP | GO:0046033 | AMP metabolic process | 3 | 22 | 2.76e-03 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 30 | 149 | 2.85e-03 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 2.94e-03 |
| GO:BP | GO:0071478 | cellular response to radiation | 46 | 156 | 3.18e-03 |
| GO:BP | GO:0090224 | regulation of spindle organization | 8 | 45 | 3.18e-03 |
| GO:BP | GO:0006259 | DNA metabolic process | 108 | 884 | 3.18e-03 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 8 | 30 | 3.24e-03 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 7 | 13 | 3.36e-03 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 23 | 36 | 3.96e-03 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 23 | 36 | 3.96e-03 |
| GO:BP | GO:0019985 | translesion synthesis | 10 | 24 | 4.29e-03 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 15 | 33 | 4.34e-03 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 25 | 318 | 4.47e-03 |
| GO:BP | GO:0006301 | postreplication repair | 12 | 34 | 4.47e-03 |
| GO:BP | GO:0044209 | AMP salvage | 2 | 5 | 4.57e-03 |
| GO:BP | GO:0032263 | GMP salvage | 2 | 4 | 4.57e-03 |
| GO:BP | GO:0033554 | cellular response to stress | 181 | 1681 | 5.02e-03 |
| GO:BP | GO:0046599 | regulation of centriole replication | 9 | 21 | 5.28e-03 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 3 | 27 | 5.46e-03 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 3 | 8 | 5.67e-03 |
| GO:BP | GO:0006275 | regulation of DNA replication | 46 | 116 | 5.73e-03 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 14 | 161 | 5.85e-03 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 12 | 15 | 6.13e-03 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 23 | 160 | 6.13e-03 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 21 | 35 | 6.13e-03 |
| GO:BP | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process | 2 | 6 | 6.37e-03 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 10 | 15 | 6.53e-03 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 4 | 50 | 6.57e-03 |
| GO:BP | GO:1900262 | regulation of DNA-directed DNA polymerase activity | 11 | 13 | 7.81e-03 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 17 | 103 | 7.81e-03 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 11 | 13 | 7.81e-03 |
| GO:BP | GO:0046053 | dAMP metabolic process | 2 | 5 | 8.56e-03 |
| GO:BP | GO:0009411 | response to UV | 18 | 141 | 8.70e-03 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 31 | 92 | 9.09e-03 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 3 | 37 | 9.97e-03 |
| GO:BP | GO:0007099 | centriole replication | 12 | 38 | 1.02e-02 |
| GO:BP | GO:0051298 | centrosome duplication | 9 | 68 | 1.04e-02 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 16 | 36 | 1.08e-02 |
| GO:BP | GO:0106380 | purine ribonucleotide salvage | 2 | 7 | 1.09e-02 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 4 | 15 | 1.12e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 3 | 84 | 1.18e-02 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 13 | 45 | 1.19e-02 |
| GO:BP | GO:0006284 | base-excision repair | 10 | 44 | 1.22e-02 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 19 | 74 | 1.27e-02 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 6 | 12 | 1.34e-02 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 37 | 186 | 1.34e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 71 | 1028 | 1.34e-02 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 6 | 12 | 1.34e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 69 | 268 | 1.43e-02 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 19 | 50 | 1.43e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 69 | 268 | 1.43e-02 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 6 | 8 | 1.43e-02 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 5 | 7 | 1.44e-02 |
| GO:BP | GO:0010332 | response to gamma radiation | 18 | 50 | 1.49e-02 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 4 | 64 | 1.52e-02 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 7 | 11 | 1.53e-02 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 5 | 14 | 1.53e-02 |
| GO:BP | GO:0009170 | purine deoxyribonucleoside monophosphate metabolic process | 2 | 7 | 1.56e-02 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 7 | 28 | 1.56e-02 |
| GO:BP | GO:0034644 | cellular response to UV | 18 | 86 | 1.58e-02 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 2 | 14 | 1.69e-02 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 2 | 11 | 1.83e-02 |
| GO:BP | GO:0032261 | purine nucleotide salvage | 2 | 10 | 1.83e-02 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 2 | 11 | 1.83e-02 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 2 | 11 | 1.83e-02 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 43 | 357 | 1.84e-02 |
| GO:BP | GO:0006175 | dATP biosynthetic process | 1 | 1 | 1.92e-02 |
| GO:BP | GO:0034508 | centromere complex assembly | 12 | 28 | 1.92e-02 |
| GO:BP | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 1.92e-02 |
| GO:BP | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 1.92e-02 |
| GO:BP | GO:0031297 | replication fork processing | 19 | 42 | 1.95e-02 |
| GO:BP | GO:1902969 | mitotic DNA replication | 10 | 14 | 2.00e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 12 | 82 | 2.04e-02 |
| GO:BP | GO:0098534 | centriole assembly | 12 | 41 | 2.10e-02 |
| GO:BP | GO:0042148 | strand invasion | 4 | 5 | 2.11e-02 |
| GO:BP | GO:0070925 | organelle assembly | 145 | 817 | 2.19e-02 |
| GO:BP | GO:0051299 | centrosome separation | 8 | 14 | 2.28e-02 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 9 | 22 | 2.31e-02 |
| GO:BP | GO:0006189 | ‘de novo’ IMP biosynthetic process | 5 | 6 | 2.31e-02 |
| GO:BP | GO:0036166 | phenotypic switching | 4 | 7 | 2.38e-02 |
| GO:BP | GO:0035878 | nail development | 2 | 8 | 2.39e-02 |
| GO:BP | GO:0033260 | nuclear DNA replication | 6 | 35 | 2.39e-02 |
| GO:BP | GO:0140013 | meiotic nuclear division | 43 | 120 | 2.39e-02 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 47 | 135 | 2.41e-02 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 14 | 40 | 2.44e-02 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 12 | 44 | 2.45e-02 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 34 | 48 | 2.47e-02 |
| GO:BP | GO:0009314 | response to radiation | 65 | 352 | 2.47e-02 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 14 | 41 | 2.49e-02 |
| GO:BP | GO:0032506 | cytokinetic process | 12 | 39 | 2.49e-02 |
| GO:BP | GO:0051656 | establishment of organelle localization | 51 | 378 | 2.54e-02 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 2.67e-02 |
| GO:BP | GO:0032392 | DNA geometric change | 19 | 80 | 2.67e-02 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 8 | 40 | 2.70e-02 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 6 | 43 | 2.73e-02 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 6 | 9 | 2.73e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 4 | 7 | 2.73e-02 |
| GO:BP | GO:2000233 | negative regulation of rRNA processing | 3 | 3 | 2.73e-02 |
| GO:BP | GO:0038202 | TORC1 signaling | 41 | 58 | 2.77e-02 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 27 | 175 | 2.83e-02 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 9 | 19 | 2.84e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 2 | 15 | 2.86e-02 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 2 | 14 | 2.86e-02 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 7 | 19 | 2.86e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 2 | 15 | 2.86e-02 |
| GO:BP | GO:0006312 | mitotic recombination | 8 | 22 | 2.87e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 3 | 123 | 2.90e-02 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 21 | 60 | 2.98e-02 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 12 | 18 | 2.99e-02 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 12 | 18 | 2.99e-02 |
| GO:BP | GO:0006979 | response to oxidative stress | 43 | 352 | 2.99e-02 |
| GO:BP | GO:0071103 | DNA conformation change | 20 | 87 | 3.00e-02 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 6 | 13 | 3.07e-02 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 14 | 35 | 3.10e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 1 | 8 | 3.11e-02 |
| GO:BP | GO:0009151 | purine deoxyribonucleotide metabolic process | 2 | 12 | 3.12e-02 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 4 | 151 | 3.33e-02 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 12 | 37 | 3.33e-02 |
| GO:BP | GO:0071772 | response to BMP | 3 | 131 | 3.33e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 3 | 131 | 3.33e-02 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 2 | 16 | 3.37e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 32 | 240 | 3.37e-02 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 11 | 14 | 3.50e-02 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 4 | 5 | 3.52e-02 |
| GO:BP | GO:0006302 | double-strand break repair | 37 | 271 | 3.53e-02 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 3 | 4 | 3.56e-02 |
| GO:BP | GO:0051640 | organelle localization | 62 | 503 | 3.61e-02 |
| GO:BP | GO:0043173 | nucleotide salvage | 2 | 15 | 3.73e-02 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 11 | 29 | 3.73e-02 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 9 | 42 | 3.73e-02 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 2 | 18 | 3.73e-02 |
| GO:BP | GO:0034502 | protein localization to chromosome | 17 | 110 | 3.76e-02 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 10 | 12 | 3.88e-02 |
| GO:BP | GO:1903926 | cellular response to bisphenol A | 3 | 4 | 3.91e-02 |
| GO:BP | GO:1903925 | response to bisphenol A | 3 | 4 | 3.91e-02 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 7 | 65 | 3.91e-02 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 2 | 19 | 4.06e-02 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 22 | 103 | 4.07e-02 |
| GO:BP | GO:0106383 | dAMP salvage | 1 | 3 | 4.12e-02 |
| GO:BP | GO:0009092 | homoserine metabolic process | 3 | 5 | 4.12e-02 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 21 | 62 | 4.12e-02 |
| GO:BP | GO:0019346 | transsulfuration | 3 | 5 | 4.12e-02 |
| GO:BP | GO:0006170 | dAMP biosynthetic process | 1 | 3 | 4.12e-02 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 15 | 20 | 4.12e-02 |
| GO:BP | GO:0106381 | purine deoxyribonucleotide salvage | 1 | 3 | 4.12e-02 |
| GO:BP | GO:0009171 | purine deoxyribonucleoside monophosphate biosynthetic process | 1 | 3 | 4.12e-02 |
| GO:BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 9 | 18 | 4.15e-02 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 2 | 15 | 4.32e-02 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 3 | 3 | 4.40e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 1 | 13 | 4.40e-02 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 59 | 262 | 4.43e-02 |
| GO:BP | GO:0051293 | establishment of spindle localization | 18 | 48 | 4.54e-02 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 6 | 21 | 4.59e-02 |
| GO:BP | GO:0031053 | primary miRNA processing | 13 | 16 | 4.59e-02 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 7 | 19 | 4.59e-02 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 7 | 19 | 4.59e-02 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 7 | 19 | 4.59e-02 |
| GO:BP | GO:0038101 | sequestering of nodal from receptor via nodal binding | 1 | 1 | 4.60e-02 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 13 | 31 | 4.63e-02 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 5 | 4.74e-02 |
| GO:BP | GO:0051653 | spindle localization | 19 | 52 | 4.75e-02 |
| GO:BP | GO:0061642 | chemoattraction of axon | 2 | 2 | 4.78e-02 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 4.84e-02 |
| GO:BP | GO:0070212 | protein poly-ADP-ribosylation | 5 | 6 | 4.90e-02 |
| GO:BP | GO:0097294 | ‘de novo’ XMP biosynthetic process | 5 | 7 | 4.90e-02 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 2 | 16 | 4.90e-02 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 7 | 9 | 4.90e-02 |
| GO:BP | GO:0097293 | XMP biosynthetic process | 5 | 7 | 4.90e-02 |
| GO:BP | GO:0097292 | XMP metabolic process | 5 | 7 | 4.90e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 35 | 293 | 5.06e-02 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 23 | 113 | 5.06e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 45 | 142 | 5.11e-02 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 6 | 23 | 5.35e-02 |
| GO:BP | GO:0043928 | exonucleolytic catabolism of deadenylated mRNA | 5 | 13 | 5.36e-02 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 16 | 270 | 5.60e-02 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 5 | 7 | 5.63e-02 |
| GO:BP | GO:0099068 | postsynapse assembly | 7 | 32 | 5.63e-02 |
| GO:BP | GO:0035611 | protein branching point deglutamylation | 1 | 1 | 5.81e-02 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 5.81e-02 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 9 | 18 | 5.99e-02 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 2 | 18 | 5.99e-02 |
| GO:BP | GO:1990227 | paranodal junction maintenance | 1 | 2 | 5.99e-02 |
| GO:BP | GO:0009439 | cyanate metabolic process | 2 | 2 | 5.99e-02 |
| GO:BP | GO:0009440 | cyanate catabolic process | 2 | 2 | 5.99e-02 |
| GO:BP | GO:0009153 | purine deoxyribonucleotide biosynthetic process | 1 | 4 | 5.99e-02 |
| GO:BP | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | 3 | 7 | 6.06e-02 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 6 | 9 | 6.17e-02 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 16 | 217 | 6.17e-02 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 6 | 7 | 6.17e-02 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 12 | 51 | 6.17e-02 |
| GO:BP | GO:0072708 | response to sorbitol | 7 | 7 | 6.29e-02 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 10 | 25 | 6.38e-02 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 18 | 30 | 6.40e-02 |
| GO:BP | GO:0022616 | DNA strand elongation | 18 | 35 | 6.59e-02 |
| GO:BP | GO:0009217 | purine deoxyribonucleoside triphosphate catabolic process | 4 | 4 | 6.69e-02 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 2 | 20 | 6.71e-02 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 13 | 21 | 6.72e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 4 | 6.72e-02 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 6.75e-02 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 7 | 10 | 6.77e-02 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 9 | 14 | 6.80e-02 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 9 | 14 | 6.80e-02 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 9 | 14 | 6.80e-02 |
| GO:BP | GO:0015691 | cadmium ion transport | 3 | 5 | 6.86e-02 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 4 | 191 | 6.86e-02 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 5 | 8 | 6.86e-02 |
| GO:BP | GO:0070574 | cadmium ion transmembrane transport | 3 | 5 | 6.86e-02 |
| GO:BP | GO:0097065 | anterior head development | 6 | 7 | 6.86e-02 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 5 | 8 | 6.86e-02 |
| GO:BP | GO:0072719 | cellular response to cisplatin | 3 | 4 | 6.86e-02 |
| GO:BP | GO:1990748 | cellular detoxification | 5 | 76 | 6.86e-02 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 10 | 11 | 6.86e-02 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 1 | 5 | 6.86e-02 |
| GO:BP | GO:0090071 | negative regulation of ribosome biogenesis | 3 | 4 | 6.86e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 12 | 21 | 7.16e-02 |
| GO:BP | GO:0031503 | protein-containing complex localization | 31 | 168 | 7.16e-02 |
| GO:BP | GO:0090677 | reversible differentiation | 3 | 5 | 7.16e-02 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 14 | 33 | 7.23e-02 |
| GO:BP | GO:0051795 | positive regulation of timing of catagen | 1 | 3 | 7.24e-02 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 3 | 3 | 7.24e-02 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 7.24e-02 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 7.30e-02 |
| GO:BP | GO:1903546 | protein localization to photoreceptor outer segment | 4 | 5 | 7.30e-02 |
| GO:BP | GO:1900146 | negative regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 1 | 7.30e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 8 | 92 | 7.30e-02 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 4 | 197 | 7.30e-02 |
| GO:BP | GO:0006298 | mismatch repair | 24 | 29 | 7.30e-02 |
| GO:BP | GO:0072308 | negative regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 1 | 7.30e-02 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 1 | 7 | 7.30e-02 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 2 | 2 | 7.30e-02 |
| GO:BP | GO:1900176 | negative regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 1 | 7.30e-02 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 28 | 85 | 7.32e-02 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 11 | 13 | 7.32e-02 |
| GO:BP | GO:0043094 | cellular metabolic compound salvage | 2 | 25 | 7.40e-02 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 9 | 29 | 7.44e-02 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 14 | 36 | 7.44e-02 |
| GO:BP | GO:0046075 | dTTP metabolic process | 4 | 4 | 7.48e-02 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 4 | 4 | 7.48e-02 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 4 | 4 | 7.48e-02 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 77 | 7.51e-02 |
| GO:BP | GO:0071585 | detoxification of cadmium ion | 2 | 2 | 7.71e-02 |
| GO:BP | GO:0046111 | xanthine biosynthetic process | 1 | 1 | 7.71e-02 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 7.71e-02 |
| GO:BP | GO:1990170 | stress response to cadmium ion | 2 | 2 | 7.71e-02 |
| GO:BP | GO:0060407 | negative regulation of penile erection | 1 | 1 | 7.71e-02 |
| GO:BP | GO:0044314 | protein K27-linked ubiquitination | 5 | 8 | 7.71e-02 |
| GO:BP | GO:0060169 | negative regulation of adenosine receptor signaling pathway | 1 | 1 | 7.71e-02 |
| GO:BP | GO:0035721 | intraciliary retrograde transport | 12 | 14 | 7.71e-02 |
| GO:BP | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process | 5 | 6 | 7.72e-02 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 5 | 21 | 7.74e-02 |
| GO:BP | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | 5 | 16 | 7.74e-02 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 7 | 425 | 7.76e-02 |
| GO:BP | GO:0098792 | xenophagy | 9 | 12 | 7.81e-02 |
| GO:BP | GO:0006520 | amino acid metabolic process | 4 | 222 | 7.81e-02 |
| GO:BP | GO:0051382 | kinetochore assembly | 8 | 18 | 7.95e-02 |
| GO:BP | GO:0048368 | lateral mesoderm development | 2 | 10 | 7.98e-02 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 5 | 8 | 8.06e-02 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 4 | 4 | 8.10e-02 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 18 | 86 | 8.10e-02 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 6 | 26 | 8.13e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 3 | 230 | 8.26e-02 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 1 | 7 | 8.35e-02 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 18 | 30 | 8.35e-02 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 5 | 82 | 8.35e-02 |
| GO:BP | GO:0060364 | frontal suture morphogenesis | 1 | 4 | 8.40e-02 |
| GO:BP | GO:0051794 | regulation of timing of catagen | 1 | 4 | 8.40e-02 |
| GO:BP | GO:0044210 | ‘de novo’ CTP biosynthetic process | 1 | 2 | 8.40e-02 |
| GO:BP | GO:0043174 | nucleoside salvage | 1 | 9 | 8.40e-02 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 9 | 15 | 8.40e-02 |
| GO:BP | GO:0042637 | catagen | 1 | 4 | 8.40e-02 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 8.40e-02 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 12 | 219 | 8.40e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 7 | 36 | 8.44e-02 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 5 | 51 | 8.44e-02 |
| GO:BP | GO:0044208 | ‘de novo’ AMP biosynthetic process | 5 | 8 | 8.44e-02 |
| GO:BP | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2 | 2 | 8.61e-02 |
| GO:BP | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process | 13 | 17 | 8.61e-02 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 19 | 46 | 8.68e-02 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 6 | 14 | 8.68e-02 |
| GO:BP | GO:0046755 | viral budding | 7 | 26 | 8.76e-02 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 3 | 5 | 8.77e-02 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 3 | 5 | 8.77e-02 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 3 | 5 | 8.77e-02 |
| GO:BP | GO:0006543 | glutamine catabolic process | 3 | 3 | 8.89e-02 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 9.01e-02 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 1 | 9 | 9.01e-02 |
| GO:BP | GO:0048818 | positive regulation of hair follicle maturation | 2 | 5 | 9.14e-02 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 11 | 194 | 9.14e-02 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 2 | 23 | 9.17e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 7 | 77 | 9.17e-02 |
| GO:BP | GO:0046386 | deoxyribose phosphate catabolic process | 14 | 22 | 9.21e-02 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 9.21e-02 |
| GO:BP | GO:0006310 | DNA recombination | 37 | 257 | 9.21e-02 |
| GO:BP | GO:0009264 | deoxyribonucleotide catabolic process | 14 | 22 | 9.21e-02 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 3 | 9 | 9.21e-02 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 5 | 25 | 9.21e-02 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 5 | 6 | 9.21e-02 |
| GO:BP | GO:0086016 | AV node cell action potential | 3 | 9 | 9.21e-02 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 10 | 14 | 9.23e-02 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 28 | 88 | 9.30e-02 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 3 | 8 | 9.32e-02 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 3 | 11 | 9.32e-02 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 17 | 25 | 9.73e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 10 | 152 | 9.76e-02 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 9.80e-02 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 6 | 47 | 9.85e-02 |
| GO:BP | GO:0019856 | pyrimidine nucleobase biosynthetic process | 2 | 8 | 9.85e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 9 | 39 | 9.89e-02 |
| GO:BP | GO:0097176 | epoxide metabolic process | 3 | 4 | 9.97e-02 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 1.00e-01 |
| GO:BP | GO:0061820 | telomeric D-loop disassembly | 5 | 8 | 1.00e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 56 | 188 | 1.00e-01 |
| GO:BP | GO:1905174 | regulation of vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 1.02e-01 |
| GO:BP | GO:1990936 | vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 1.02e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 51 | 206 | 1.02e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 73 | 1226 | 1.02e-01 |
| GO:BP | GO:0090678 | cell dedifferentiation involved in phenotypic switching | 2 | 2 | 1.02e-01 |
| GO:BP | GO:0031291 | Ran protein signal transduction | 2 | 3 | 1.04e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 1.04e-01 |
| GO:BP | GO:0002076 | osteoblast development | 2 | 12 | 1.04e-01 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 2 | 9 | 1.05e-01 |
| GO:BP | GO:0061912 | selective autophagy | 40 | 90 | 1.05e-01 |
| GO:BP | GO:1903416 | response to glycoside | 3 | 6 | 1.05e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 3 | 265 | 1.05e-01 |
| GO:BP | GO:0045008 | depyrimidination | 6 | 8 | 1.06e-01 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 8 | 36 | 1.06e-01 |
| GO:BP | GO:0014029 | neural crest formation | 6 | 13 | 1.07e-01 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 6 | 10 | 1.08e-01 |
| GO:BP | GO:0035845 | photoreceptor cell outer segment organization | 3 | 10 | 1.08e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 11 | 55 | 1.08e-01 |
| GO:BP | GO:0032792 | negative regulation of CREB transcription factor activity | 1 | 4 | 1.09e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 1.09e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 8 | 27 | 1.10e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 6 | 28 | 1.10e-01 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 5 | 16 | 1.10e-01 |
| GO:BP | GO:0035582 | sequestering of BMP in extracellular matrix | 1 | 2 | 1.11e-01 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 1.11e-01 |
| GO:BP | GO:1900108 | negative regulation of nodal signaling pathway | 1 | 2 | 1.11e-01 |
| GO:BP | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 1.11e-01 |
| GO:BP | GO:0046725 | negative regulation by virus of viral protein levels in host cell | 1 | 2 | 1.11e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 54 | 439 | 1.11e-01 |
| GO:BP | GO:0090069 | regulation of ribosome biogenesis | 3 | 5 | 1.13e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 4 | 79 | 1.14e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 20 | 56 | 1.14e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 3 | 17 | 1.16e-01 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 3 | 6 | 1.16e-01 |
| GO:BP | GO:0051095 | regulation of helicase activity | 6 | 12 | 1.16e-01 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 3 | 6 | 1.16e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 10 | 12 | 1.17e-01 |
| GO:BP | GO:0072201 | negative regulation of mesenchymal cell proliferation | 4 | 7 | 1.17e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 2 | 31 | 1.18e-01 |
| GO:BP | GO:0098754 | detoxification | 8 | 91 | 1.18e-01 |
| GO:BP | GO:0002636 | positive regulation of germinal center formation | 1 | 1 | 1.18e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 9 | 25 | 1.18e-01 |
| GO:BP | GO:0070256 | negative regulation of mucus secretion | 1 | 2 | 1.18e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 4 | 32 | 1.18e-01 |
| GO:BP | GO:0090148 | membrane fission | 9 | 44 | 1.18e-01 |
| GO:BP | GO:0046061 | dATP catabolic process | 1 | 2 | 1.18e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 18 | 64 | 1.18e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 8 | 35 | 1.18e-01 |
| GO:BP | GO:0046110 | xanthine metabolic process | 1 | 1 | 1.18e-01 |
| GO:BP | GO:0043103 | hypoxanthine salvage | 1 | 2 | 1.18e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 1.19e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 4 | 240 | 1.19e-01 |
| GO:BP | GO:0019068 | virion assembly | 8 | 35 | 1.19e-01 |
| GO:BP | GO:1901490 | regulation of lymphangiogenesis | 4 | 5 | 1.19e-01 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 3 | 4 | 1.19e-01 |
| GO:BP | GO:0090222 | centrosome-templated microtubule nucleation | 1 | 2 | 1.19e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 5 | 28 | 1.19e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 6 | 311 | 1.19e-01 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 3 | 7 | 1.20e-01 |
| GO:BP | GO:0048854 | brain morphogenesis | 12 | 28 | 1.21e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 4 | 241 | 1.21e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 12 | 76 | 1.22e-01 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 15 | 22 | 1.22e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 12 | 14 | 1.22e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 5 | 31 | 1.22e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 22 | 149 | 1.23e-01 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 1.23e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 31 | 103 | 1.23e-01 |
| GO:BP | GO:0000921 | septin ring assembly | 1 | 1 | 1.24e-01 |
| GO:BP | GO:0032200 | telomere organization | 39 | 166 | 1.24e-01 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 3 | 27 | 1.24e-01 |
| GO:BP | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway | 3 | 3 | 1.24e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 15 | 93 | 1.24e-01 |
| GO:BP | GO:0032223 | negative regulation of synaptic transmission, cholinergic | 1 | 1 | 1.25e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 11 | 16 | 1.26e-01 |
| GO:BP | GO:1900175 | regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 2 | 1.26e-01 |
| GO:BP | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 2 | 1.26e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 2 | 2 | 1.26e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 6 | 319 | 1.26e-01 |
| GO:BP | GO:0035825 | homologous recombination | 13 | 43 | 1.26e-01 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 10 | 12 | 1.26e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 2 | 101 | 1.26e-01 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 5 | 8 | 1.26e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 86 | 494 | 1.26e-01 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 3 | 4 | 1.26e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 5 | 20 | 1.26e-01 |
| GO:BP | GO:0086017 | Purkinje myocyte action potential | 3 | 4 | 1.26e-01 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 5 | 23 | 1.26e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 7 | 11 | 1.26e-01 |
| GO:BP | GO:0051305 | chromosome movement towards spindle pole | 3 | 7 | 1.29e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 5 | 45 | 1.30e-01 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 6 | 22 | 1.33e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 9 | 19 | 1.35e-01 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 11 | 26 | 1.35e-01 |
| GO:BP | GO:0030903 | notochord development | 9 | 17 | 1.35e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 6 | 30 | 1.35e-01 |
| GO:BP | GO:0090215 | regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 4 | 4 | 1.35e-01 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 17 | 30 | 1.35e-01 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 5 | 13 | 1.35e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 3 | 17 | 1.35e-01 |
| GO:BP | GO:0048341 | paraxial mesoderm formation | 5 | 6 | 1.35e-01 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 4 | 4 | 1.35e-01 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 4 | 11 | 1.35e-01 |
| GO:BP | GO:0044848 | biological phase | 1 | 9 | 1.35e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 6 | 12 | 1.35e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 12 | 68 | 1.35e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 6 | 17 | 1.35e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 6 | 9 | 1.35e-01 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 8 | 17 | 1.35e-01 |
| GO:BP | GO:0044851 | hair cycle phase | 1 | 9 | 1.35e-01 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 1 | 8 | 1.35e-01 |
| GO:BP | GO:0090217 | negative regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 3 | 3 | 1.35e-01 |
| GO:BP | GO:0048819 | regulation of hair follicle maturation | 1 | 9 | 1.35e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 32 | 48 | 1.35e-01 |
| GO:BP | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process | 3 | 3 | 1.35e-01 |
| GO:BP | GO:0045617 | negative regulation of keratinocyte differentiation | 1 | 3 | 1.35e-01 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 3 | 10 | 1.35e-01 |
| GO:BP | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process | 3 | 3 | 1.35e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 4 | 6 | 1.35e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 1.35e-01 |
| GO:BP | GO:1901491 | negative regulation of lymphangiogenesis | 3 | 3 | 1.35e-01 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 14 | 18 | 1.35e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 1.35e-01 |
| GO:BP | GO:0072135 | kidney mesenchymal cell proliferation | 1 | 2 | 1.36e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 16 | 26 | 1.36e-01 |
| GO:BP | GO:1990535 | neuron projection maintenance | 5 | 9 | 1.36e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 3 | 5 | 1.36e-01 |
| GO:BP | GO:1904976 | cellular response to bleomycin | 2 | 2 | 1.36e-01 |
| GO:BP | GO:1904975 | response to bleomycin | 2 | 2 | 1.36e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 6 | 56 | 1.36e-01 |
| GO:BP | GO:0072136 | metanephric mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 1.36e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 7 | 20 | 1.36e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 6 | 52 | 1.36e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 3 | 303 | 1.36e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 7 | 9 | 1.36e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 1.36e-01 |
| GO:BP | GO:0009445 | putrescine metabolic process | 2 | 6 | 1.36e-01 |
| GO:BP | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process | 5 | 15 | 1.36e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 4 | 21 | 1.36e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 7 | 32 | 1.36e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 7 | 23 | 1.36e-01 |
| GO:BP | GO:0044806 | G-quadruplex DNA unwinding | 4 | 7 | 1.37e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 25 | 73 | 1.37e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 13 | 22 | 1.37e-01 |
| GO:BP | GO:0016236 | macroautophagy | 123 | 308 | 1.37e-01 |
| GO:BP | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process | 4 | 7 | 1.38e-01 |
| GO:BP | GO:0071029 | nuclear ncRNA surveillance | 4 | 7 | 1.38e-01 |
| GO:BP | GO:0071046 | nuclear polyadenylation-dependent ncRNA catabolic process | 4 | 7 | 1.38e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 64 | 1.38e-01 |
| GO:BP | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process | 4 | 7 | 1.38e-01 |
| GO:BP | GO:0106354 | tRNA surveillance | 4 | 7 | 1.38e-01 |
| GO:BP | GO:0000725 | recombinational repair | 22 | 153 | 1.38e-01 |
| GO:BP | GO:0008063 | Toll signaling pathway | 4 | 6 | 1.40e-01 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 5 | 7 | 1.40e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 6 | 34 | 1.40e-01 |
| GO:BP | GO:0060346 | bone trabecula formation | 1 | 9 | 1.40e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 25 | 52 | 1.40e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 17 | 72 | 1.41e-01 |
| GO:BP | GO:1903551 | regulation of extracellular exosome assembly | 4 | 6 | 1.41e-01 |
| GO:BP | GO:0043096 | purine nucleobase salvage | 1 | 3 | 1.42e-01 |
| GO:BP | GO:0006157 | deoxyadenosine catabolic process | 1 | 2 | 1.42e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 7 | 186 | 1.42e-01 |
| GO:BP | GO:0044790 | suppression of viral release by host | 3 | 16 | 1.42e-01 |
| GO:BP | GO:0045828 | positive regulation of isoprenoid metabolic process | 1 | 1 | 1.42e-01 |
| GO:BP | GO:0046090 | deoxyadenosine metabolic process | 1 | 2 | 1.42e-01 |
| GO:BP | GO:0046101 | hypoxanthine biosynthetic process | 1 | 3 | 1.42e-01 |
| GO:BP | GO:0046103 | inosine biosynthetic process | 1 | 3 | 1.42e-01 |
| GO:BP | GO:1900054 | positive regulation of retinoic acid biosynthetic process | 1 | 1 | 1.42e-01 |
| GO:BP | GO:0097113 | AMPA glutamate receptor clustering | 3 | 7 | 1.42e-01 |
| GO:BP | GO:0097688 | glutamate receptor clustering | 3 | 7 | 1.42e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 5 | 7 | 1.42e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 8 | 9 | 1.42e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 3 | 7 | 1.43e-01 |
| GO:BP | GO:0035609 | C-terminal protein deglutamylation | 1 | 2 | 1.43e-01 |
| GO:BP | GO:0051418 | microtubule nucleation by microtubule organizing center | 4 | 5 | 1.44e-01 |
| GO:BP | GO:0007144 | female meiosis I | 5 | 9 | 1.45e-01 |
| GO:BP | GO:0009204 | deoxyribonucleoside triphosphate catabolic process | 4 | 5 | 1.45e-01 |
| GO:BP | GO:0009146 | purine nucleoside triphosphate catabolic process | 4 | 5 | 1.45e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 1.45e-01 |
| GO:BP | GO:0071694 | maintenance of protein location in extracellular region | 1 | 4 | 1.45e-01 |
| GO:BP | GO:0048820 | hair follicle maturation | 3 | 15 | 1.45e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 1.45e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 1.45e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 22 | 64 | 1.45e-01 |
| GO:BP | GO:0072033 | renal vesicle formation | 6 | 8 | 1.46e-01 |
| GO:BP | GO:0006188 | IMP biosynthetic process | 3 | 10 | 1.48e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 4 | 48 | 1.48e-01 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 8 | 12 | 1.48e-01 |
| GO:BP | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 2 | 1.48e-01 |
| GO:BP | GO:0008592 | regulation of Toll signaling pathway | 3 | 3 | 1.48e-01 |
| GO:BP | GO:0009116 | nucleoside metabolic process | 2 | 42 | 1.48e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 4 | 506 | 1.48e-01 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 23 | 64 | 1.48e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 7 | 33 | 1.48e-01 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 3 | 41 | 1.49e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 6 | 57 | 1.49e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 2 | 2 | 1.50e-01 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 3 | 18 | 1.50e-01 |
| GO:BP | GO:0051105 | regulation of DNA ligation | 4 | 6 | 1.50e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 12 | 75 | 1.50e-01 |
| GO:BP | GO:0090179 | planar cell polarity pathway involved in neural tube closure | 4 | 12 | 1.50e-01 |
| GO:BP | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic | 2 | 19 | 1.50e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 5 | 25 | 1.50e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 8 | 30 | 1.53e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 10 | 146 | 1.53e-01 |
| GO:BP | GO:0060633 | negative regulation of transcription initiation by RNA polymerase II | 6 | 6 | 1.53e-01 |
| GO:BP | GO:0051798 | positive regulation of hair follicle development | 1 | 9 | 1.54e-01 |
| GO:BP | GO:0072702 | response to methyl methanesulfonate | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0072703 | cellular response to methyl methanesulfonate | 1 | 1 | 1.54e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 2 | 59 | 1.56e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 9 | 277 | 1.56e-01 |
| GO:BP | GO:0032901 | positive regulation of neurotrophin production | 1 | 1 | 1.57e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 1.57e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 5 | 39 | 1.57e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 1.57e-01 |
| GO:BP | GO:1900107 | regulation of nodal signaling pathway | 1 | 3 | 1.57e-01 |
| GO:BP | GO:0072718 | response to cisplatin | 3 | 6 | 1.57e-01 |
| GO:BP | GO:0021571 | rhombomere 5 development | 2 | 2 | 1.57e-01 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 6 | 19 | 1.57e-01 |
| GO:BP | GO:0035284 | brain segmentation | 2 | 2 | 1.57e-01 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 6 | 19 | 1.57e-01 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 8 | 10 | 1.58e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 12 | 41 | 1.59e-01 |
| GO:BP | GO:0035610 | protein side chain deglutamylation | 1 | 2 | 1.59e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 11 | 158 | 1.59e-01 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 3 | 19 | 1.59e-01 |
| GO:BP | GO:0036438 | maintenance of lens transparency | 4 | 6 | 1.59e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 2 | 14 | 1.59e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 4 | 22 | 1.59e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 1.61e-01 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 1 | 12 | 1.61e-01 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 4 | 7 | 1.61e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 33 | 604 | 1.61e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 21 | 30 | 1.62e-01 |
| GO:BP | GO:0010288 | response to lead ion | 5 | 18 | 1.62e-01 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 2 | 8 | 1.62e-01 |
| GO:BP | GO:1905364 | regulation of endosomal vesicle fusion | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0046521 | sphingoid catabolic process | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0032202 | telomere assembly | 3 | 4 | 1.63e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 2 | 12 | 1.63e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 17 | 49 | 1.63e-01 |
| GO:BP | GO:0032077 | positive regulation of deoxyribonuclease activity | 3 | 5 | 1.63e-01 |
| GO:BP | GO:1900121 | negative regulation of receptor binding | 1 | 8 | 1.64e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 3 | 6 | 1.64e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 10 | 464 | 1.64e-01 |
| GO:BP | GO:0046059 | dAMP catabolic process | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 35 | 134 | 1.64e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 2 | 214 | 1.64e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 27 | 133 | 1.64e-01 |
| GO:BP | GO:0046100 | hypoxanthine metabolic process | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0106014 | regulation of inflammatory response to wounding | 2 | 4 | 1.64e-01 |
| GO:BP | GO:0002906 | negative regulation of mature B cell apoptotic process | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0002905 | regulation of mature B cell apoptotic process | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0006154 | adenosine catabolic process | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 3 | 159 | 1.64e-01 |
| GO:BP | GO:0002901 | mature B cell apoptotic process | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0060167 | regulation of adenosine receptor signaling pathway | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0046124 | purine deoxyribonucleoside catabolic process | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 3 | 76 | 1.64e-01 |
| GO:BP | GO:0030592 | DNA ADP-ribosylation | 2 | 3 | 1.64e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 3 | 69 | 1.64e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 17 | 34 | 1.65e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 1 | 6 | 1.65e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 3 | 15 | 1.65e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 1 | 6 | 1.65e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 1 | 99 | 1.67e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 26 | 459 | 1.67e-01 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 5 | 12 | 1.68e-01 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 3 | 32 | 1.69e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 3 | 5 | 1.69e-01 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 1 | 5 | 1.69e-01 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 4 | 10 | 1.69e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 5 | 29 | 1.70e-01 |
| GO:BP | GO:1904550 | response to arachidonic acid | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 11 | 14 | 1.71e-01 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 2 | 2 | 1.71e-01 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 2 | 2 | 1.71e-01 |
| GO:BP | GO:0035608 | protein deglutamylation | 1 | 2 | 1.71e-01 |
| GO:BP | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing | 7 | 7 | 1.71e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 4 | 9 | 1.71e-01 |
| GO:BP | GO:0000212 | meiotic spindle organization | 6 | 14 | 1.71e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 33 | 51 | 1.71e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 1 | 7 | 1.72e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 2 | 64 | 1.72e-01 |
| GO:BP | GO:1904172 | positive regulation of bleb assembly | 1 | 1 | 1.73e-01 |
| GO:BP | GO:1904170 | regulation of bleb assembly | 1 | 1 | 1.73e-01 |
| GO:BP | GO:0072709 | cellular response to sorbitol | 5 | 5 | 1.73e-01 |
| GO:BP | GO:0031106 | septin ring organization | 1 | 2 | 1.73e-01 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 7 | 15 | 1.73e-01 |
| GO:BP | GO:0071932 | replication fork reversal | 5 | 5 | 1.73e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 7 | 14 | 1.73e-01 |
| GO:BP | GO:0090657 | telomeric loop disassembly | 5 | 9 | 1.73e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 8 | 88 | 1.73e-01 |
| GO:BP | GO:0001507 | acetylcholine catabolic process in synaptic cleft | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0061525 | hindgut development | 2 | 5 | 1.74e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 5 | 15 | 1.74e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 15 | 23 | 1.74e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 1 | 3 | 1.74e-01 |
| GO:BP | GO:0071288 | cellular response to mercury ion | 2 | 2 | 1.74e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 6 | 147 | 1.74e-01 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 8 | 38 | 1.74e-01 |
| GO:BP | GO:0035581 | sequestering of extracellular ligand from receptor | 1 | 7 | 1.74e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 1.74e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 6 | 7 | 1.75e-01 |
| GO:BP | GO:0035445 | borate transmembrane transport | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 4 | 13 | 1.76e-01 |
| GO:BP | GO:0070194 | synaptonemal complex disassembly | 1 | 2 | 1.76e-01 |
| GO:BP | GO:0046713 | borate transport | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 33 | 868 | 1.76e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 1.76e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 11 | 331 | 1.77e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 11 | 114 | 1.77e-01 |
| GO:BP | GO:0042454 | ribonucleoside catabolic process | 3 | 12 | 1.77e-01 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 4 | 16 | 1.78e-01 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 5 | 13 | 1.78e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 6 | 28 | 1.78e-01 |
| GO:BP | GO:0071569 | protein ufmylation | 5 | 6 | 1.78e-01 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 2 | 11 | 1.78e-01 |
| GO:BP | GO:0036337 | Fas signaling pathway | 3 | 5 | 1.78e-01 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 5 | 8 | 1.78e-01 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 1.79e-01 |
| GO:BP | GO:0036245 | cellular response to menadione | 1 | 2 | 1.79e-01 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 1.81e-01 |
| GO:BP | GO:0021694 | cerebellar Purkinje cell layer formation | 1 | 12 | 1.81e-01 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 12 | 28 | 1.82e-01 |
| GO:BP | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | 2 | 5 | 1.83e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 1.84e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 1.84e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 35 | 875 | 1.84e-01 |
| GO:BP | GO:1903774 | positive regulation of viral budding via host ESCRT complex | 2 | 3 | 1.84e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 1.84e-01 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 16 | 24 | 1.84e-01 |
| GO:BP | GO:0006196 | AMP catabolic process | 1 | 3 | 1.84e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 2 | 5 | 1.84e-01 |
| GO:BP | GO:0046102 | inosine metabolic process | 1 | 4 | 1.84e-01 |
| GO:BP | GO:0046122 | purine deoxyribonucleoside metabolic process | 1 | 3 | 1.84e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 8 | 42 | 1.84e-01 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 1 | 9 | 1.84e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 10 | 14 | 1.84e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 24 | 185 | 1.85e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 19 | 260 | 1.86e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 2 | 11 | 1.87e-01 |
| GO:BP | GO:0043634 | polyadenylation-dependent ncRNA catabolic process | 4 | 8 | 1.87e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 21 | 44 | 1.87e-01 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 1 | 10 | 1.87e-01 |
| GO:BP | GO:2000821 | regulation of grooming behavior | 2 | 3 | 1.87e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 6 | 22 | 1.87e-01 |
| GO:BP | GO:1902045 | negative regulation of Fas signaling pathway | 2 | 2 | 1.87e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 3 | 440 | 1.87e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 4 | 22 | 1.88e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 7 | 28 | 1.88e-01 |
| GO:BP | GO:0032020 | ISG15-protein conjugation | 1 | 6 | 1.88e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 23 | 300 | 1.90e-01 |
| GO:BP | GO:0110154 | RNA decapping | 2 | 17 | 1.90e-01 |
| GO:BP | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation | 2 | 2 | 1.90e-01 |
| GO:BP | GO:0006206 | pyrimidine nucleobase metabolic process | 2 | 13 | 1.90e-01 |
| GO:BP | GO:1902065 | response to L-glutamate | 7 | 11 | 1.90e-01 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 3 | 23 | 1.90e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 29 | 81 | 1.90e-01 |
| GO:BP | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation | 2 | 5 | 1.91e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 14 | 132 | 1.92e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 2 | 63 | 1.92e-01 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 1 | 9 | 1.92e-01 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 1 | 9 | 1.92e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 28 | 96 | 1.93e-01 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 1 | 15 | 1.93e-01 |
| GO:BP | GO:1903780 | negative regulation of cardiac conduction | 1 | 1 | 1.93e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 1.93e-01 |
| GO:BP | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1 | 8 | 1.93e-01 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 6 | 8 | 1.93e-01 |
| GO:BP | GO:0045217 | cell-cell junction maintenance | 1 | 13 | 1.93e-01 |
| GO:BP | GO:0008090 | retrograde axonal transport | 2 | 19 | 1.93e-01 |
| GO:BP | GO:1901845 | negative regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 21 | 63 | 1.93e-01 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 4 | 7 | 1.93e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 2 | 149 | 1.93e-01 |
| GO:BP | GO:0035329 | hippo signaling | 12 | 41 | 1.93e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 9 | 21 | 1.93e-01 |
| GO:BP | GO:0010651 | negative regulation of cell communication by electrical coupling | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0042594 | response to starvation | 10 | 178 | 1.95e-01 |
| GO:BP | GO:0048642 | negative regulation of skeletal muscle tissue development | 1 | 3 | 1.95e-01 |
| GO:BP | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3’-5’ | 6 | 11 | 1.95e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 3 | 74 | 1.95e-01 |
| GO:BP | GO:0090272 | negative regulation of fibroblast growth factor production | 2 | 2 | 1.96e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 1.96e-01 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 6 | 13 | 1.96e-01 |
| GO:BP | GO:0090656 | t-circle formation | 6 | 13 | 1.96e-01 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 3 | 23 | 1.96e-01 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 6 | 13 | 1.96e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 2 | 10 | 1.96e-01 |
| GO:BP | GO:0060031 | mediolateral intercalation | 2 | 2 | 1.96e-01 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 3 | 5 | 1.96e-01 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 1.96e-01 |
| GO:BP | GO:0006222 | UMP biosynthetic process | 2 | 8 | 1.96e-01 |
| GO:BP | GO:0060775 | planar cell polarity pathway involved in gastrula mediolateral intercalation | 2 | 2 | 1.96e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 4 | 5 | 1.96e-01 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 2 | 10 | 1.96e-01 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 2 | 3 | 1.96e-01 |
| GO:BP | GO:0036376 | sodium ion export across plasma membrane | 4 | 12 | 1.96e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 2 | 116 | 1.97e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 1.97e-01 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 1 | 16 | 1.97e-01 |
| GO:BP | GO:0034473 | U1 snRNA 3’-end processing | 2 | 3 | 1.97e-01 |
| GO:BP | GO:0034476 | U5 snRNA 3’-end processing | 2 | 3 | 1.97e-01 |
| GO:BP | GO:1903553 | positive regulation of extracellular exosome assembly | 3 | 4 | 1.97e-01 |
| GO:BP | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell communication | 2 | 6 | 1.97e-01 |
| GO:BP | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling | 2 | 6 | 1.97e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 2 | 6 | 1.97e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 2 | 3 | 1.97e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 3 | 179 | 1.97e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 2 | 111 | 1.97e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 22 | 86 | 1.97e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 10 | 55 | 1.97e-01 |
| GO:BP | GO:0010458 | exit from mitosis | 21 | 30 | 1.97e-01 |
| GO:BP | GO:0021591 | ventricular system development | 14 | 29 | 1.97e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 4 | 11 | 1.97e-01 |
| GO:BP | GO:0044205 | ‘de novo’ UMP biosynthetic process | 2 | 3 | 1.97e-01 |
| GO:BP | GO:1903995 | regulation of non-membrane spanning protein tyrosine kinase activity | 3 | 3 | 1.97e-01 |
| GO:BP | GO:0060013 | righting reflex | 3 | 6 | 1.98e-01 |
| GO:BP | GO:1905366 | negative regulation of intralumenal vesicle formation | 2 | 2 | 1.98e-01 |
| GO:BP | GO:1904979 | negative regulation of endosome organization | 2 | 2 | 1.98e-01 |
| GO:BP | GO:1905365 | regulation of intralumenal vesicle formation | 2 | 2 | 1.98e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 2 | 40 | 1.98e-01 |
| GO:BP | GO:0046121 | deoxyribonucleoside catabolic process | 1 | 4 | 1.98e-01 |
| GO:BP | GO:0046130 | purine ribonucleoside catabolic process | 1 | 5 | 1.98e-01 |
| GO:BP | GO:0009172 | purine deoxyribonucleoside monophosphate catabolic process | 1 | 3 | 1.98e-01 |
| GO:BP | GO:0071971 | extracellular exosome assembly | 4 | 6 | 1.99e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 4 | 14 | 2.00e-01 |
| GO:BP | GO:1905198 | manchette assembly | 3 | 4 | 2.00e-01 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 2 | 13 | 2.00e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 31 | 89 | 2.00e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 2.00e-01 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 2 | 13 | 2.00e-01 |
| GO:BP | GO:0010165 | response to X-ray | 7 | 29 | 2.00e-01 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 1 | 16 | 2.00e-01 |
| GO:BP | GO:0099179 | regulation of synaptic membrane adhesion | 4 | 4 | 2.00e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 2 | 13 | 2.00e-01 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 15 | 30 | 2.01e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 12 | 19 | 2.01e-01 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 1 | 12 | 2.01e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 19 | 103 | 2.01e-01 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 5 | 9 | 2.01e-01 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 3 | 8 | 2.02e-01 |
| GO:BP | GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 2.02e-01 |
| GO:BP | GO:0072257 | metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 2.02e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 8 | 18 | 2.02e-01 |
| GO:BP | GO:0070106 | interleukin-27-mediated signaling pathway | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0048742 | regulation of skeletal muscle fiber development | 1 | 8 | 2.02e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 2.02e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 24 | 162 | 2.02e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 6 | 159 | 2.02e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 6 | 69 | 2.02e-01 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 13 | 17 | 2.03e-01 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 13 | 17 | 2.03e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 29 | 112 | 2.03e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 2.03e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 13 | 35 | 2.03e-01 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 2 | 62 | 2.03e-01 |
| GO:BP | GO:0006266 | DNA ligation | 12 | 16 | 2.03e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 2 | 11 | 2.03e-01 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 7 | 41 | 2.03e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 5 | 22 | 2.03e-01 |
| GO:BP | GO:0060049 | regulation of protein glycosylation | 2 | 10 | 2.03e-01 |
| GO:BP | GO:0070166 | enamel mineralization | 1 | 11 | 2.03e-01 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 3 | 5 | 2.03e-01 |
| GO:BP | GO:0045007 | depurination | 4 | 4 | 2.03e-01 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 13 | 17 | 2.03e-01 |
| GO:BP | GO:0032060 | bleb assembly | 2 | 9 | 2.03e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 7 | 39 | 2.03e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 34 | 170 | 2.03e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 7 | 11 | 2.03e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 3 | 12 | 2.05e-01 |
| GO:BP | GO:0051835 | positive regulation of synapse structural plasticity | 2 | 2 | 2.05e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 5 | 41 | 2.06e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 20 | 64 | 2.06e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 9 | 80 | 2.06e-01 |
| GO:BP | GO:0061724 | lipophagy | 2 | 7 | 2.06e-01 |
| GO:BP | GO:0006244 | pyrimidine nucleotide catabolic process | 5 | 19 | 2.06e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 3 | 11 | 2.06e-01 |
| GO:BP | GO:0006581 | acetylcholine catabolic process | 1 | 3 | 2.07e-01 |
| GO:BP | GO:0006421 | asparaginyl-tRNA aminoacylation | 3 | 3 | 2.07e-01 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 4 | 5 | 2.07e-01 |
| GO:BP | GO:0035973 | aggrephagy | 2 | 8 | 2.07e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 4 | 36 | 2.07e-01 |
| GO:BP | GO:0099606 | microtubule plus-end directed mitotic chromosome migration | 1 | 1 | 2.07e-01 |
| GO:BP | GO:0000722 | telomere maintenance via recombination | 10 | 14 | 2.07e-01 |
| GO:BP | GO:0098735 | positive regulation of the force of heart contraction | 2 | 4 | 2.07e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 2.08e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 2.08e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 4 | 38 | 2.08e-01 |
| GO:BP | GO:0046073 | dTMP metabolic process | 3 | 7 | 2.08e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 4 | 155 | 2.08e-01 |
| GO:BP | GO:0015756 | fucose transmembrane transport | 2 | 2 | 2.09e-01 |
| GO:BP | GO:1902153 | regulation of response to DNA damage checkpoint signaling | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0009174 | pyrimidine ribonucleoside monophosphate biosynthetic process | 2 | 9 | 2.09e-01 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 2 | 6 | 2.09e-01 |
| GO:BP | GO:0015750 | pentose transmembrane transport | 2 | 2 | 2.09e-01 |
| GO:BP | GO:0000017 | alpha-glucoside transport | 2 | 2 | 2.09e-01 |
| GO:BP | GO:1902145 | regulation of response to cell cycle checkpoint signaling | 1 | 1 | 2.09e-01 |
| GO:BP | GO:1902151 | regulation of response to DNA integrity checkpoint signaling | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0060449 | bud elongation involved in lung branching | 4 | 7 | 2.10e-01 |
| GO:BP | GO:0035283 | central nervous system segmentation | 2 | 2 | 2.10e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 4 | 45 | 2.10e-01 |
| GO:BP | GO:0060405 | regulation of penile erection | 1 | 4 | 2.10e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 2 | 2 | 2.10e-01 |
| GO:BP | GO:0019606 | 2-oxobutyrate catabolic process | 1 | 1 | 2.10e-01 |
| GO:BP | GO:0097114 | NMDA glutamate receptor clustering | 2 | 5 | 2.10e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 4 | 20 | 2.10e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 6 | 161 | 2.10e-01 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 2 | 8 | 2.10e-01 |
| GO:BP | GO:0046040 | IMP metabolic process | 1 | 15 | 2.10e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 18 | 24 | 2.10e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 25 | 204 | 2.11e-01 |
| GO:BP | GO:1904717 | regulation of AMPA glutamate receptor clustering | 2 | 4 | 2.11e-01 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 3 | 5 | 2.11e-01 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 7 | 35 | 2.11e-01 |
| GO:BP | GO:0060542 | regulation of strand invasion | 1 | 1 | 2.11e-01 |
| GO:BP | GO:0098530 | positive regulation of strand invasion | 1 | 1 | 2.11e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 10 | 25 | 2.11e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 3 | 14 | 2.11e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 6 | 40 | 2.11e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 2 | 284 | 2.11e-01 |
| GO:BP | GO:0106104 | regulation of glutamate receptor clustering | 2 | 4 | 2.11e-01 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 21 | 67 | 2.11e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 3 | 89 | 2.11e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 5 | 353 | 2.11e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 7 | 31 | 2.11e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 14 | 16 | 2.11e-01 |
| GO:BP | GO:0007127 | meiosis I | 27 | 77 | 2.11e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 2.11e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 10 | 67 | 2.11e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 1 | 8 | 2.11e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 2.11e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 3 | 3 | 2.12e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 1 | 41 | 2.12e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 5 | 295 | 2.12e-01 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 1 | 23 | 2.12e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 2 | 12 | 2.12e-01 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 10 | 19 | 2.12e-01 |
| GO:BP | GO:0021685 | cerebellar granular layer structural organization | 1 | 1 | 2.12e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 10 | 103 | 2.13e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 6 | 19 | 2.13e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 6 | 19 | 2.13e-01 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 3 | 13 | 2.13e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 11 | 27 | 2.15e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 1 | 17 | 2.15e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 12 | 52 | 2.15e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 2.15e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 10 | 16 | 2.15e-01 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 4 | 8 | 2.15e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 24 | 39 | 2.15e-01 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 4 | 8 | 2.15e-01 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 2 | 6 | 2.15e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 2 | 6 | 2.15e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 33 | 60 | 2.15e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 5 | 28 | 2.15e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 13 | 80 | 2.15e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 22 | 45 | 2.15e-01 |
| GO:BP | GO:2000256 | positive regulation of male germ cell proliferation | 2 | 2 | 2.16e-01 |
| GO:BP | GO:1990646 | cellular response to prolactin | 2 | 2 | 2.16e-01 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 3 | 2.16e-01 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 8 | 19 | 2.16e-01 |
| GO:BP | GO:1905938 | positive regulation of germ cell proliferation | 2 | 2 | 2.16e-01 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 2.16e-01 |
| GO:BP | GO:0046391 | 5-phosphoribose 1-diphosphate metabolic process | 2 | 5 | 2.16e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 5 | 132 | 2.16e-01 |
| GO:BP | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation | 2 | 3 | 2.16e-01 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 9 | 29 | 2.16e-01 |
| GO:BP | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process | 2 | 5 | 2.16e-01 |
| GO:BP | GO:0060067 | cervix development | 2 | 2 | 2.16e-01 |
| GO:BP | GO:0097012 | response to granulocyte macrophage colony-stimulating factor | 2 | 6 | 2.16e-01 |
| GO:BP | GO:1904934 | negative regulation of cell proliferation in midbrain | 1 | 1 | 2.16e-01 |
| GO:BP | GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus | 2 | 6 | 2.16e-01 |
| GO:BP | GO:0061354 | planar cell polarity pathway involved in pericardium morphogenesis | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0071505 | response to mycophenolic acid | 2 | 2 | 2.16e-01 |
| GO:BP | GO:0036517 | chemoattraction of serotonergic neuron axon | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0061350 | planar cell polarity pathway involved in cardiac muscle tissue morphogenesis | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0061346 | planar cell polarity pathway involved in heart morphogenesis | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0033212 | iron import into cell | 3 | 7 | 2.16e-01 |
| GO:BP | GO:0061348 | planar cell polarity pathway involved in ventricular septum morphogenesis | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0061349 | planar cell polarity pathway involved in cardiac right atrium morphogenesis | 1 | 1 | 2.16e-01 |
| GO:BP | GO:0071506 | cellular response to mycophenolic acid | 2 | 2 | 2.16e-01 |
| GO:BP | GO:0072092 | ureteric bud invasion | 3 | 3 | 2.16e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 5 | 18 | 2.17e-01 |
| GO:BP | GO:0006655 | phosphatidylglycerol biosynthetic process | 8 | 9 | 2.18e-01 |
| GO:BP | GO:0001947 | heart looping | 5 | 50 | 2.18e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 14 | 26 | 2.18e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 2 | 13 | 2.18e-01 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 7 | 22 | 2.19e-01 |
| GO:BP | GO:0036309 | protein localization to M-band | 2 | 2 | 2.19e-01 |
| GO:BP | GO:0006152 | purine nucleoside catabolic process | 1 | 6 | 2.19e-01 |
| GO:BP | GO:0070255 | regulation of mucus secretion | 1 | 6 | 2.19e-01 |
| GO:BP | GO:0002634 | regulation of germinal center formation | 1 | 6 | 2.19e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 8 | 16 | 2.20e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 2 | 6 | 2.20e-01 |
| GO:BP | GO:0032458 | slow endocytic recycling | 2 | 2 | 2.20e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 2.20e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 2 | 6 | 2.20e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 10 | 15 | 2.20e-01 |
| GO:BP | GO:0036164 | cell-abiotic substrate adhesion | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 4 | 9 | 2.20e-01 |
| GO:BP | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | 2 | 4 | 2.20e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 1 | 8 | 2.20e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 1 | 226 | 2.20e-01 |
| GO:BP | GO:0097186 | amelogenesis | 1 | 14 | 2.20e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 2.20e-01 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 1 | 12 | 2.20e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 24 | 124 | 2.20e-01 |
| GO:BP | GO:2001189 | negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 2 | 3 | 2.20e-01 |
| GO:BP | GO:0071584 | negative regulation of zinc ion transmembrane import | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0009231 | riboflavin biosynthetic process | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 10 | 27 | 2.20e-01 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 3 | 25 | 2.20e-01 |
| GO:BP | GO:1903806 | L-isoleucine import across plasma membrane | 1 | 1 | 2.20e-01 |
| GO:BP | GO:1904024 | negative regulation of glucose catabolic process to lactate via pyruvate | 2 | 3 | 2.20e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 2.20e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 2.20e-01 |
| GO:BP | GO:2000521 | negative regulation of immunological synapse formation | 1 | 1 | 2.20e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 10 | 27 | 2.20e-01 |
| GO:BP | GO:1903714 | isoleucine transmembrane transport | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0043633 | polyadenylation-dependent RNA catabolic process | 4 | 9 | 2.20e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 2.20e-01 |
| GO:BP | GO:0000459 | exonucleolytic trimming involved in rRNA processing | 4 | 9 | 2.20e-01 |
| GO:BP | GO:0052647 | pentitol phosphate metabolic process | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 4 | 9 | 2.20e-01 |
| GO:BP | GO:0052648 | ribitol phosphate metabolic process | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0071580 | regulation of zinc ion transmembrane transport | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0046444 | FMN metabolic process | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0071581 | regulation of zinc ion transmembrane import | 1 | 1 | 2.20e-01 |
| GO:BP | GO:2001170 | negative regulation of ATP biosynthetic process | 2 | 3 | 2.20e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 8 | 45 | 2.20e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 13 | 40 | 2.20e-01 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 3 | 9 | 2.20e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 3 | 11 | 2.20e-01 |
| GO:BP | GO:0071583 | negative regulation of zinc ion transmembrane transport | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 15 | 46 | 2.20e-01 |
| GO:BP | GO:0045760 | positive regulation of action potential | 2 | 4 | 2.20e-01 |
| GO:BP | GO:0030913 | paranodal junction assembly | 5 | 6 | 2.20e-01 |
| GO:BP | GO:0030261 | chromosome condensation | 12 | 33 | 2.20e-01 |
| GO:BP | GO:0009398 | FMN biosynthetic process | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0071582 | negative regulation of zinc ion transport | 1 | 1 | 2.20e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 15 | 46 | 2.20e-01 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 4 | 6 | 2.21e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 2.22e-01 |
| GO:BP | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER | 2 | 2 | 2.22e-01 |
| GO:BP | GO:0015791 | polyol transmembrane transport | 4 | 6 | 2.22e-01 |
| GO:BP | GO:0061146 | Peyer’s patch morphogenesis | 3 | 3 | 2.22e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 2 | 48 | 2.22e-01 |
| GO:BP | GO:0007032 | endosome organization | 17 | 92 | 2.22e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 2 | 35 | 2.22e-01 |
| GO:BP | GO:0018410 | C-terminal protein amino acid modification | 1 | 10 | 2.22e-01 |
| GO:BP | GO:0071051 | polyadenylation-dependent snoRNA 3’-end processing | 4 | 7 | 2.22e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 6 | 423 | 2.22e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 5 | 47 | 2.22e-01 |
| GO:BP | GO:0050893 | sensory processing | 3 | 4 | 2.23e-01 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 2 | 5 | 2.23e-01 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 1 | 13 | 2.23e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 2 | 17 | 2.23e-01 |
| GO:BP | GO:1903379 | regulation of mitotic chromosome condensation | 2 | 2 | 2.23e-01 |
| GO:BP | GO:1905213 | negative regulation of mitotic chromosome condensation | 2 | 2 | 2.23e-01 |
| GO:BP | GO:0070295 | renal water absorption | 2 | 5 | 2.23e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 1 | 22 | 2.23e-01 |
| GO:BP | GO:0009164 | nucleoside catabolic process | 3 | 15 | 2.23e-01 |
| GO:BP | GO:1902340 | negative regulation of chromosome condensation | 2 | 2 | 2.23e-01 |
| GO:BP | GO:0046398 | UDP-glucuronate metabolic process | 1 | 2 | 2.23e-01 |
| GO:BP | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 2 | 2.23e-01 |
| GO:BP | GO:0032185 | septin cytoskeleton organization | 1 | 4 | 2.24e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 9 | 46 | 2.25e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 3 | 26 | 2.25e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 4 | 89 | 2.26e-01 |
| GO:BP | GO:0003283 | atrial septum development | 4 | 23 | 2.26e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 12 | 33 | 2.26e-01 |
| GO:BP | GO:0002314 | germinal center B cell differentiation | 1 | 3 | 2.26e-01 |
| GO:BP | GO:1900052 | regulation of retinoic acid biosynthetic process | 1 | 3 | 2.26e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 59 | 257 | 2.26e-01 |
| GO:BP | GO:0016233 | telomere capping | 12 | 36 | 2.26e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 11 | 23 | 2.26e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 2.26e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 3 | 50 | 2.26e-01 |
| GO:BP | GO:0000255 | allantoin metabolic process | 1 | 4 | 2.26e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 9 | 13 | 2.26e-01 |
| GO:BP | GO:0048541 | Peyer’s patch development | 1 | 6 | 2.26e-01 |
| GO:BP | GO:0048537 | mucosa-associated lymphoid tissue development | 1 | 6 | 2.26e-01 |
| GO:BP | GO:0045004 | DNA replication proofreading | 1 | 2 | 2.26e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 19 | 33 | 2.26e-01 |
| GO:BP | GO:0048313 | Golgi inheritance | 7 | 15 | 2.26e-01 |
| GO:BP | GO:0048308 | organelle inheritance | 7 | 15 | 2.26e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 7 | 58 | 2.26e-01 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 11 | 19 | 2.26e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 1 | 8 | 2.26e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 9 | 17 | 2.26e-01 |
| GO:BP | GO:2000232 | regulation of rRNA processing | 5 | 16 | 2.27e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 6 | 59 | 2.27e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 5 | 377 | 2.27e-01 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 2 | 17 | 2.27e-01 |
| GO:BP | GO:0036372 | opsin transport | 1 | 2 | 2.27e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 6 | 8 | 2.27e-01 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 2 | 4 | 2.27e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 7 | 10 | 2.28e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 2.29e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 2 | 8 | 2.29e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 4 | 172 | 2.29e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 21 | 317 | 2.29e-01 |
| GO:BP | GO:0046036 | CTP metabolic process | 1 | 14 | 2.29e-01 |
| GO:BP | GO:0070266 | necroptotic process | 10 | 32 | 2.29e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 2.29e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 2.29e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 2 | 6 | 2.29e-01 |
| GO:BP | GO:0014886 | transition between slow and fast fiber | 1 | 1 | 2.30e-01 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 1 | 24 | 2.30e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 2.30e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 6 | 429 | 2.30e-01 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 8 | 10 | 2.30e-01 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 8 | 10 | 2.30e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 22 | 32 | 2.30e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 13 | 121 | 2.30e-01 |
| GO:BP | GO:0044878 | mitotic cytokinesis checkpoint signaling | 2 | 4 | 2.31e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 6 | 343 | 2.31e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 18 | 45 | 2.31e-01 |
| GO:BP | GO:0043556 | regulation of translation in response to oxidative stress | 1 | 1 | 2.31e-01 |
| GO:BP | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | 1 | 2 | 2.31e-01 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 2.31e-01 |
| GO:BP | GO:0032938 | negative regulation of translation in response to oxidative stress | 1 | 1 | 2.31e-01 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 4 | 26 | 2.31e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 5 | 33 | 2.31e-01 |
| GO:BP | GO:0071307 | cellular response to vitamin K | 1 | 4 | 2.31e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 2.31e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 2.31e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 5 | 28 | 2.31e-01 |
| GO:BP | GO:0001560 | regulation of cell growth by extracellular stimulus | 3 | 3 | 2.31e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 2.31e-01 |
| GO:BP | GO:1905340 | regulation of protein localization to kinetochore | 3 | 4 | 2.31e-01 |
| GO:BP | GO:1905342 | positive regulation of protein localization to kinetochore | 3 | 4 | 2.31e-01 |
| GO:BP | GO:1903772 | regulation of viral budding via host ESCRT complex | 2 | 4 | 2.32e-01 |
| GO:BP | GO:0036486 | ventral trunk neural crest cell migration | 3 | 3 | 2.35e-01 |
| GO:BP | GO:0043111 | replication fork arrest | 2 | 2 | 2.35e-01 |
| GO:BP | GO:0010225 | response to UV-C | 5 | 13 | 2.35e-01 |
| GO:BP | GO:0046340 | diacylglycerol catabolic process | 2 | 3 | 2.35e-01 |
| GO:BP | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process | 4 | 6 | 2.35e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 7 | 10 | 2.35e-01 |
| GO:BP | GO:0016557 | peroxisome membrane biogenesis | 3 | 3 | 2.36e-01 |
| GO:BP | GO:0051974 | negative regulation of telomerase activity | 5 | 11 | 2.36e-01 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 2 | 209 | 2.36e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 5 | 39 | 2.36e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 4 | 15 | 2.36e-01 |
| GO:BP | GO:0060028 | convergent extension involved in axis elongation | 3 | 6 | 2.37e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 4 | 6 | 2.37e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 5 | 9 | 2.37e-01 |
| GO:BP | GO:0070244 | negative regulation of thymocyte apoptotic process | 1 | 6 | 2.37e-01 |
| GO:BP | GO:0009128 | purine nucleoside monophosphate catabolic process | 1 | 7 | 2.37e-01 |
| GO:BP | GO:0009169 | purine ribonucleoside monophosphate catabolic process | 1 | 7 | 2.37e-01 |
| GO:BP | GO:0009155 | purine deoxyribonucleotide catabolic process | 1 | 7 | 2.37e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 2 | 5 | 2.37e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 15 | 279 | 2.37e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 2 | 192 | 2.37e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 2 | 7 | 2.37e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 2 | 22 | 2.37e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 2.39e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 2 | 53 | 2.39e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 4 | 92 | 2.39e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 4 | 93 | 2.39e-01 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 3 | 3 | 2.39e-01 |
| GO:BP | GO:1903954 | positive regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 7 | 11 | 2.39e-01 |
| GO:BP | GO:1903952 | regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 2 | 11 | 2.39e-01 |
| GO:BP | GO:1904716 | positive regulation of chaperone-mediated autophagy | 1 | 1 | 2.39e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 6 | 18 | 2.39e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 2.40e-01 |
| GO:BP | GO:0022614 | membrane to membrane docking | 3 | 5 | 2.40e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 7 | 10 | 2.41e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 9 | 15 | 2.41e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 16 | 32 | 2.41e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 16 | 65 | 2.41e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 5 | 26 | 2.41e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 2.41e-01 |
| GO:BP | GO:0090038 | negative regulation of protein kinase C signaling | 4 | 4 | 2.41e-01 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 1 | 16 | 2.41e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 3 | 6 | 2.41e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 2.41e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 2.41e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 12 | 19 | 2.41e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 2 | 218 | 2.42e-01 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 5 | 10 | 2.42e-01 |
| GO:BP | GO:0043201 | response to leucine | 2 | 13 | 2.42e-01 |
| GO:BP | GO:0099526 | presynapse to nucleus signaling pathway | 1 | 2 | 2.42e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 16 | 40 | 2.42e-01 |
| GO:BP | GO:0098928 | presynaptic signal transduction | 1 | 2 | 2.42e-01 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 5 | 10 | 2.42e-01 |
| GO:BP | GO:0015793 | glycerol transmembrane transport | 3 | 4 | 2.42e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 3 | 32 | 2.43e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 1 | 246 | 2.43e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 6 | 7 | 2.43e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 6 | 7 | 2.43e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 7 | 41 | 2.43e-01 |
| GO:BP | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway | 1 | 3 | 2.44e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 2.45e-01 |
| GO:BP | GO:0051607 | defense response to virus | 3 | 202 | 2.45e-01 |
| GO:BP | GO:0032899 | regulation of neurotrophin production | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0003336 | corneocyte desquamation | 1 | 1 | 2.46e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 2.46e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 3 | 202 | 2.46e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 12 | 158 | 2.46e-01 |
| GO:BP | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential | 3 | 3 | 2.46e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 6 | 67 | 2.46e-01 |
| GO:BP | GO:0019262 | N-acetylneuraminate catabolic process | 4 | 6 | 2.46e-01 |
| GO:BP | GO:0090244 | Wnt signaling pathway involved in somitogenesis | 2 | 5 | 2.47e-01 |
| GO:BP | GO:0033632 | regulation of cell-cell adhesion mediated by integrin | 1 | 6 | 2.47e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 2 | 22 | 2.47e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 3 | 14 | 2.47e-01 |
| GO:BP | GO:0033089 | positive regulation of T cell differentiation in thymus | 1 | 5 | 2.47e-01 |
| GO:BP | GO:0046085 | adenosine metabolic process | 1 | 5 | 2.47e-01 |
| GO:BP | GO:0043084 | penile erection | 1 | 6 | 2.47e-01 |
| GO:BP | GO:0051126 | negative regulation of actin nucleation | 2 | 6 | 2.47e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 14 | 98 | 2.47e-01 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 1 | 9 | 2.47e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 6 | 7 | 2.47e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 3 | 19 | 2.47e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 2.47e-01 |
| GO:BP | GO:0046049 | UMP metabolic process | 2 | 10 | 2.47e-01 |
| GO:BP | GO:0009159 | deoxyribonucleoside monophosphate catabolic process | 2 | 8 | 2.47e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 9 | 23 | 2.48e-01 |
| GO:BP | GO:0035641 | locomotory exploration behavior | 3 | 11 | 2.48e-01 |
| GO:BP | GO:0071047 | polyadenylation-dependent mRNA catabolic process | 2 | 4 | 2.49e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 7 | 681 | 2.49e-01 |
| GO:BP | GO:0032918 | spermidine acetylation | 2 | 2 | 2.49e-01 |
| GO:BP | GO:0032917 | polyamine acetylation | 2 | 2 | 2.49e-01 |
| GO:BP | GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic process | 2 | 4 | 2.49e-01 |
| GO:BP | GO:0006273 | lagging strand elongation | 4 | 4 | 2.49e-01 |
| GO:BP | GO:0009194 | pyrimidine ribonucleoside diphosphate biosynthetic process | 2 | 4 | 2.49e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 3 | 160 | 2.49e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 12 | 21 | 2.49e-01 |
| GO:BP | GO:2000431 | regulation of cytokinesis, actomyosin contractile ring assembly | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0003273 | cell migration involved in endocardial cushion formation | 3 | 4 | 2.49e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0000423 | mitophagy | 9 | 37 | 2.49e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 9 | 73 | 2.49e-01 |
| GO:BP | GO:0006272 | leading strand elongation | 2 | 4 | 2.49e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 2.49e-01 |
| GO:BP | GO:1901660 | calcium ion export | 3 | 3 | 2.49e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 12 | 194 | 2.49e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 1 | 34 | 2.49e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 3 | 249 | 2.49e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 38 | 414 | 2.49e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 18 | 50 | 2.49e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 22 | 108 | 2.49e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 5 | 94 | 2.49e-01 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 4 | 28 | 2.49e-01 |
| GO:BP | GO:0006225 | UDP biosynthetic process | 2 | 4 | 2.49e-01 |
| GO:BP | GO:0003241 | growth involved in heart morphogenesis | 3 | 4 | 2.49e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 2.49e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 5 | 20 | 2.49e-01 |
| GO:BP | GO:1903518 | positive regulation of single strand break repair | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 2 | 26 | 2.50e-01 |
| GO:BP | GO:1904875 | regulation of DNA ligase activity | 2 | 2 | 2.50e-01 |
| GO:BP | GO:0043622 | cortical microtubule organization | 3 | 4 | 2.50e-01 |
| GO:BP | GO:0042126 | nitrate metabolic process | 4 | 4 | 2.50e-01 |
| GO:BP | GO:0021756 | striatum development | 3 | 17 | 2.51e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 11 | 31 | 2.51e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 2.51e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 2.51e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 1 | 21 | 2.51e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 11 | 67 | 2.52e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 5 | 50 | 2.52e-01 |
| GO:BP | GO:0060057 | apoptotic process involved in mammary gland involution | 2 | 3 | 2.52e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 2 | 65 | 2.52e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 12 | 31 | 2.52e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1 | 10 | 2.52e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 2 | 18 | 2.52e-01 |
| GO:BP | GO:0032075 | positive regulation of nuclease activity | 3 | 7 | 2.52e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 11 | 39 | 2.52e-01 |
| GO:BP | GO:0060058 | positive regulation of apoptotic process involved in mammary gland involution | 2 | 3 | 2.52e-01 |
| GO:BP | GO:0043650 | dicarboxylic acid biosynthetic process | 1 | 9 | 2.52e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 4 | 11 | 2.53e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 6 | 8 | 2.53e-01 |
| GO:BP | GO:0090102 | cochlea development | 9 | 35 | 2.53e-01 |
| GO:BP | GO:0010643 | cell communication by chemical coupling | 2 | 2 | 2.53e-01 |
| GO:BP | GO:0001843 | neural tube closure | 9 | 90 | 2.53e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 5 | 7 | 2.53e-01 |
| GO:BP | GO:0060431 | primary lung bud formation | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0046136 | positive regulation of vitamin metabolic process | 1 | 2 | 2.53e-01 |
| GO:BP | GO:0019747 | regulation of isoprenoid metabolic process | 1 | 4 | 2.53e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 2 | 69 | 2.53e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 5 | 7 | 2.53e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 1 | 36 | 2.53e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 2.53e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 1 | 23 | 2.53e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 2.53e-01 |
| GO:BP | GO:0090399 | replicative senescence | 2 | 13 | 2.53e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 4 | 28 | 2.53e-01 |
| GO:BP | GO:0140450 | protein targeting to Golgi apparatus | 1 | 1 | 2.53e-01 |
| GO:BP | GO:0009143 | nucleoside triphosphate catabolic process | 1 | 10 | 2.53e-01 |
| GO:BP | GO:0003094 | glomerular filtration | 5 | 21 | 2.53e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 11 | 26 | 2.54e-01 |
| GO:BP | GO:0045769 | negative regulation of asymmetric cell division | 1 | 1 | 2.54e-01 |
| GO:BP | GO:0045951 | positive regulation of mitotic recombination | 1 | 1 | 2.55e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 7 | 358 | 2.55e-01 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 3 | 10 | 2.56e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 5 | 50 | 2.57e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 5 | 50 | 2.57e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 4 | 103 | 2.57e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 11 | 72 | 2.57e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 2.57e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 9 | 156 | 2.57e-01 |
| GO:BP | GO:0060219 | camera-type eye photoreceptor cell differentiation | 11 | 18 | 2.58e-01 |
| GO:BP | GO:0019276 | UDP-N-acetylgalactosamine metabolic process | 2 | 2 | 2.58e-01 |
| GO:BP | GO:0055009 | atrial cardiac muscle tissue morphogenesis | 2 | 5 | 2.58e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 2 | 2 | 2.59e-01 |
| GO:BP | GO:0033693 | neurofilament bundle assembly | 3 | 3 | 2.59e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 2 | 17 | 2.59e-01 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 3 | 3 | 2.59e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 7 | 696 | 2.59e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 14 | 155 | 2.59e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 3 | 20 | 2.59e-01 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 2.59e-01 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 2 | 5 | 2.59e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 72 | 189 | 2.59e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 10 | 23 | 2.59e-01 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 3 | 13 | 2.59e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 2 | 11 | 2.59e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 31 | 111 | 2.59e-01 |
| GO:BP | GO:1902228 | positive regulation of macrophage colony-stimulating factor signaling pathway | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0060611 | mammary gland fat development | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0001100 | negative regulation of exit from mitosis | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0060606 | tube closure | 9 | 91 | 2.59e-01 |
| GO:BP | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 1 | 3 | 2.59e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 4 | 69 | 2.59e-01 |
| GO:BP | GO:0009158 | ribonucleoside monophosphate catabolic process | 2 | 10 | 2.60e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 100 | 234 | 2.60e-01 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 13 | 21 | 2.60e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 5 | 64 | 2.60e-01 |
| GO:BP | GO:0001561 | fatty acid alpha-oxidation | 2 | 5 | 2.60e-01 |
| GO:BP | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 5 | 5 | 2.60e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 3 | 21 | 2.60e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 3 | 59 | 2.61e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 13 | 47 | 2.61e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 162 | 452 | 2.61e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 6 | 94 | 2.62e-01 |
| GO:BP | GO:0019240 | citrulline biosynthetic process | 4 | 4 | 2.62e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 7 | 700 | 2.62e-01 |
| GO:BP | GO:1901207 | regulation of heart looping | 2 | 2 | 2.62e-01 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 3 | 5 | 2.62e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 6 | 18 | 2.62e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 2.62e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 10 | 16 | 2.62e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 2 | 70 | 2.62e-01 |
| GO:BP | GO:0070254 | mucus secretion | 1 | 8 | 2.62e-01 |
| GO:BP | GO:0042822 | pyridoxal phosphate metabolic process | 4 | 4 | 2.62e-01 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 3 | 5 | 2.62e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 15 | 27 | 2.62e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 2.62e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 2 | 278 | 2.63e-01 |
| GO:BP | GO:0019731 | antibacterial humoral response | 2 | 15 | 2.64e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 6 | 8 | 2.64e-01 |
| GO:BP | GO:0051257 | meiotic spindle midzone assembly | 1 | 2 | 2.64e-01 |
| GO:BP | GO:0003360 | brainstem development | 2 | 7 | 2.64e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 8 | 23 | 2.64e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 15 | 381 | 2.65e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 3 | 57 | 2.65e-01 |
| GO:BP | GO:1902339 | positive regulation of apoptotic process involved in morphogenesis | 4 | 5 | 2.65e-01 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 3 | 33 | 2.65e-01 |
| GO:BP | GO:1904747 | positive regulation of apoptotic process involved in development | 4 | 5 | 2.65e-01 |
| GO:BP | GO:0140131 | positive regulation of lymphocyte chemotaxis | 4 | 10 | 2.65e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 3 | 4 | 2.65e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 12 | 49 | 2.65e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 3 | 17 | 2.65e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 3 | 111 | 2.66e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 7 | 26 | 2.66e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 3 | 551 | 2.66e-01 |
| GO:BP | GO:1990564 | protein polyufmylation | 4 | 5 | 2.66e-01 |
| GO:BP | GO:1990592 | protein K69-linked ufmylation | 4 | 5 | 2.66e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 11 | 16 | 2.66e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 10 | 13 | 2.66e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 10 | 16 | 2.66e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 16 | 38 | 2.66e-01 |
| GO:BP | GO:0003032 | detection of oxygen | 2 | 5 | 2.67e-01 |
| GO:BP | GO:0046600 | negative regulation of centriole replication | 4 | 7 | 2.67e-01 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 8 | 23 | 2.67e-01 |
| GO:BP | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 4 | 4 | 2.67e-01 |
| GO:BP | GO:0034964 | box H/ACA RNA processing | 2 | 2 | 2.67e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 6 | 15 | 2.67e-01 |
| GO:BP | GO:0000495 | box H/ACA RNA 3’-end processing | 2 | 2 | 2.67e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 1 | 12 | 2.67e-01 |
| GO:BP | GO:0071476 | cellular hypotonic response | 5 | 8 | 2.67e-01 |
| GO:BP | GO:0006595 | polyamine metabolic process | 2 | 16 | 2.67e-01 |
| GO:BP | GO:0009447 | putrescine catabolic process | 1 | 2 | 2.67e-01 |
| GO:BP | GO:0061152 | trachea submucosa development | 2 | 2 | 2.67e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 5 | 56 | 2.67e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 1 | 11 | 2.67e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 1 | 10 | 2.67e-01 |
| GO:BP | GO:0061153 | trachea gland development | 2 | 2 | 2.67e-01 |
| GO:BP | GO:1904417 | positive regulation of xenophagy | 5 | 7 | 2.67e-01 |
| GO:BP | GO:1904415 | regulation of xenophagy | 5 | 7 | 2.67e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 3 | 28 | 2.68e-01 |
| GO:BP | GO:0046361 | 2-oxobutyrate metabolic process | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0008544 | epidermis development | 2 | 210 | 2.68e-01 |
| GO:BP | GO:1901753 | leukotriene A4 biosynthetic process | 1 | 1 | 2.68e-01 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 5 | 18 | 2.68e-01 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 2.68e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 6 | 15 | 2.68e-01 |
| GO:BP | GO:1990029 | vasomotion | 1 | 1 | 2.68e-01 |
| GO:BP | GO:0033326 | cerebrospinal fluid secretion | 2 | 3 | 2.68e-01 |
| GO:BP | GO:0086044 | atrial cardiac muscle cell to AV node cell communication by electrical coupling | 1 | 1 | 2.68e-01 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 2.68e-01 |
| GO:BP | GO:0021768 | nucleus accumbens development | 1 | 1 | 2.68e-01 |
| GO:BP | GO:0010652 | positive regulation of cell communication by chemical coupling | 1 | 1 | 2.68e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 2.68e-01 |
| GO:BP | GO:0086055 | Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling | 1 | 1 | 2.68e-01 |
| GO:BP | GO:0010645 | regulation of cell communication by chemical coupling | 1 | 1 | 2.68e-01 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 2.68e-01 |
| GO:BP | GO:0034755 | iron ion transmembrane transport | 4 | 16 | 2.68e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 2.68e-01 |
| GO:BP | GO:0098906 | regulation of Purkinje myocyte action potential | 1 | 1 | 2.68e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 2 | 2 | 2.68e-01 |
| GO:BP | GO:0140042 | lipid droplet formation | 9 | 12 | 2.68e-01 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 3 | 12 | 2.68e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 2 | 19 | 2.68e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 2.68e-01 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 1 | 4 | 2.68e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 20 | 560 | 2.69e-01 |
| GO:BP | GO:0051152 | positive regulation of smooth muscle cell differentiation | 6 | 10 | 2.69e-01 |
| GO:BP | GO:0002467 | germinal center formation | 1 | 10 | 2.69e-01 |
| GO:BP | GO:0033082 | regulation of extrathymic T cell differentiation | 1 | 1 | 2.69e-01 |
| GO:BP | GO:0000303 | response to superoxide | 6 | 21 | 2.69e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 2 | 11 | 2.69e-01 |
| GO:BP | GO:0032898 | neurotrophin production | 3 | 4 | 2.69e-01 |
| GO:BP | GO:1990049 | retrograde neuronal dense core vesicle transport | 1 | 4 | 2.69e-01 |
| GO:BP | GO:0034656 | nucleobase-containing small molecule catabolic process | 3 | 20 | 2.70e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 3 | 36 | 2.70e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 10 | 14 | 2.70e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 2.71e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 2 | 5 | 2.71e-01 |
| GO:BP | GO:1901594 | response to capsazepine | 1 | 1 | 2.71e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 3 | 25 | 2.71e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 4 | 9 | 2.72e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 4 | 9 | 2.72e-01 |
| GO:BP | GO:0009173 | pyrimidine ribonucleoside monophosphate metabolic process | 2 | 12 | 2.72e-01 |
| GO:BP | GO:0051106 | positive regulation of DNA ligation | 3 | 5 | 2.72e-01 |
| GO:BP | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 3 | 2.72e-01 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 5 | 20 | 2.72e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 2 | 76 | 2.73e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 9 | 41 | 2.73e-01 |
| GO:BP | GO:0086054 | bundle of His cell to Purkinje myocyte communication by electrical coupling | 2 | 2 | 2.73e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 7 | 33 | 2.73e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 3 | 9 | 2.74e-01 |
| GO:BP | GO:0090129 | positive regulation of synapse maturation | 1 | 8 | 2.74e-01 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 5 | 6 | 2.74e-01 |
| GO:BP | GO:0021693 | cerebellar Purkinje cell layer structural organization | 1 | 2 | 2.75e-01 |
| GO:BP | GO:0021698 | cerebellar cortex structural organization | 1 | 2 | 2.75e-01 |
| GO:BP | GO:0061485 | memory T cell proliferation | 1 | 1 | 2.75e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 2 | 26 | 2.75e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 3 | 69 | 2.75e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 5 | 5 | 2.76e-01 |
| GO:BP | GO:0019264 | glycine biosynthetic process from serine | 2 | 2 | 2.77e-01 |
| GO:BP | GO:0036216 | cellular response to stem cell factor stimulus | 4 | 4 | 2.77e-01 |
| GO:BP | GO:0038109 | Kit signaling pathway | 4 | 4 | 2.77e-01 |
| GO:BP | GO:0036215 | response to stem cell factor | 4 | 4 | 2.77e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 2.77e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 4 | 110 | 2.79e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 2.79e-01 |
| GO:BP | GO:0032049 | cardiolipin biosynthetic process | 5 | 7 | 2.79e-01 |
| GO:BP | GO:0070848 | response to growth factor | 3 | 572 | 2.79e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 4 | 8 | 2.79e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 2.79e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 2.79e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 19 | 59 | 2.80e-01 |
| GO:BP | GO:0006722 | triterpenoid metabolic process | 2 | 2 | 2.80e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 2.80e-01 |
| GO:BP | GO:2000143 | negative regulation of DNA-templated transcription initiation | 6 | 6 | 2.80e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 1 | 46 | 2.80e-01 |
| GO:BP | GO:1903934 | positive regulation of DNA primase activity | 4 | 5 | 2.80e-01 |
| GO:BP | GO:1903932 | regulation of DNA primase activity | 4 | 5 | 2.80e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 2.81e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 8 | 67 | 2.81e-01 |
| GO:BP | GO:0042946 | glucoside transport | 3 | 3 | 2.81e-01 |
| GO:BP | GO:0043157 | response to cation stress | 1 | 1 | 2.81e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 2 | 84 | 2.81e-01 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 1 | 97 | 2.81e-01 |
| GO:BP | GO:0033387 | putrescine biosynthetic process from ornithine | 1 | 3 | 2.81e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 6 | 57 | 2.82e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 2.82e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 11 | 116 | 2.82e-01 |
| GO:BP | GO:0090735 | DNA repair complex assembly | 2 | 3 | 2.82e-01 |
| GO:BP | GO:1901465 | positive regulation of tetrapyrrole biosynthetic process | 3 | 3 | 2.82e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 2.82e-01 |
| GO:BP | GO:1902990 | mitotic telomere maintenance via semi-conservative replication | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 6 | 12 | 2.82e-01 |
| GO:BP | GO:0070455 | positive regulation of heme biosynthetic process | 3 | 3 | 2.82e-01 |
| GO:BP | GO:0000730 | DNA recombinase assembly | 2 | 3 | 2.82e-01 |
| GO:BP | GO:0051098 | regulation of binding | 2 | 317 | 2.82e-01 |
| GO:BP | GO:0046465 | dolichyl diphosphate metabolic process | 3 | 5 | 2.82e-01 |
| GO:BP | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0036518 | chemorepulsion of dopaminergic neuron axon | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0051885 | positive regulation of timing of anagen | 1 | 2 | 2.82e-01 |
| GO:BP | GO:1905438 | non-canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0070086 | ubiquitin-dependent endocytosis | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 1 | 29 | 2.82e-01 |
| GO:BP | GO:0008291 | acetylcholine metabolic process | 1 | 3 | 2.82e-01 |
| GO:BP | GO:0003408 | optic cup formation involved in camera-type eye development | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 6 | 487 | 2.82e-01 |
| GO:BP | GO:1904933 | regulation of cell proliferation in midbrain | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0009650 | UV protection | 2 | 11 | 2.82e-01 |
| GO:BP | GO:0042245 | RNA repair | 2 | 2 | 2.82e-01 |
| GO:BP | GO:0006489 | dolichyl diphosphate biosynthetic process | 3 | 5 | 2.82e-01 |
| GO:BP | GO:0060080 | inhibitory postsynaptic potential | 1 | 8 | 2.82e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 5 | 39 | 2.82e-01 |
| GO:BP | GO:1900619 | acetate ester metabolic process | 1 | 3 | 2.82e-01 |
| GO:BP | GO:0036515 | serotonergic neuron axon guidance | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 15 | 33 | 2.82e-01 |
| GO:BP | GO:0086048 | membrane depolarization during bundle of His cell action potential | 2 | 3 | 2.82e-01 |
| GO:BP | GO:0006218 | uridine catabolic process | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 4 | 12 | 2.82e-01 |
| GO:BP | GO:2000202 | positive regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.82e-01 |
| GO:BP | GO:2000200 | regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 4 | 112 | 2.82e-01 |
| GO:BP | GO:0097205 | renal filtration | 5 | 23 | 2.82e-01 |
| GO:BP | GO:0046108 | uridine metabolic process | 1 | 1 | 2.82e-01 |
| GO:BP | GO:2000199 | positive regulation of ribonucleoprotein complex localization | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential | 2 | 3 | 2.82e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 4 | 12 | 2.82e-01 |
| GO:BP | GO:2000208 | positive regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 2.82e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 12 | 22 | 2.82e-01 |
| GO:BP | GO:0006885 | regulation of pH | 2 | 74 | 2.82e-01 |
| GO:BP | GO:2000206 | regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.82e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 2.82e-01 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 3 | 5 | 2.82e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 2.82e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 3 | 129 | 2.82e-01 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 2 | 14 | 2.82e-01 |
| GO:BP | GO:0019660 | glycolytic fermentation | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 2 | 4 | 2.82e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 2 | 6 | 2.82e-01 |
| GO:BP | GO:0014839 | myoblast migration involved in skeletal muscle regeneration | 2 | 3 | 2.82e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 9 | 75 | 2.82e-01 |
| GO:BP | GO:0031990 | mRNA export from nucleus in response to heat stress | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 2 | 4 | 2.82e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 31 | 147 | 2.82e-01 |
| GO:BP | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway | 6 | 6 | 2.82e-01 |
| GO:BP | GO:1901003 | negative regulation of fermentation | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0016570 | histone modification | 1 | 433 | 2.82e-01 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0071578 | zinc ion import across plasma membrane | 2 | 6 | 2.82e-01 |
| GO:BP | GO:0043628 | regulatory ncRNA 3’-end processing | 7 | 19 | 2.82e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 4 | 113 | 2.82e-01 |
| GO:BP | GO:0003138 | primary heart field specification | 2 | 3 | 2.82e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 2.82e-01 |
| GO:BP | GO:0034402 | recruitment of 3’-end processing factors to RNA polymerase II holoenzyme complex | 1 | 2 | 2.82e-01 |
| GO:BP | GO:0030952 | establishment or maintenance of cytoskeleton polarity | 3 | 7 | 2.82e-01 |
| GO:BP | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation | 2 | 5 | 2.82e-01 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 1 | 7 | 2.82e-01 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 2 | 5 | 2.83e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 14 | 53 | 2.83e-01 |
| GO:BP | GO:0006046 | N-acetylglucosamine catabolic process | 3 | 3 | 2.83e-01 |
| GO:BP | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning | 1 | 1 | 2.83e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 15 | 44 | 2.84e-01 |
| GO:BP | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 1 | 10 | 2.84e-01 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 2.84e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 5 | 58 | 2.84e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 24 | 393 | 2.84e-01 |
| GO:BP | GO:0051660 | establishment of centrosome localization | 5 | 8 | 2.84e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 7 | 89 | 2.84e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 2 | 65 | 2.84e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 2.84e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 2 | 19 | 2.84e-01 |
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 2.84e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 2 | 4 | 2.84e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 2.84e-01 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 2 | 4 | 2.84e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 5 | 17 | 2.84e-01 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 2 | 4 | 2.84e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 2 | 7 | 2.84e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 5 | 40 | 2.84e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 32 | 72 | 2.84e-01 |
| GO:BP | GO:0002138 | retinoic acid biosynthetic process | 2 | 8 | 2.84e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 24 | 101 | 2.84e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 1 | 33 | 2.84e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 10 | 43 | 2.85e-01 |
| GO:BP | GO:0000729 | DNA double-strand break processing | 5 | 18 | 2.85e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 7 | 21 | 2.85e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 13 | 30 | 2.85e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 25 | 55 | 2.85e-01 |
| GO:BP | GO:0042727 | flavin-containing compound biosynthetic process | 1 | 2 | 2.85e-01 |
| GO:BP | GO:1904271 | L-proline import across plasma membrane | 3 | 5 | 2.86e-01 |
| GO:BP | GO:0097491 | sympathetic neuron projection guidance | 4 | 4 | 2.86e-01 |
| GO:BP | GO:1904555 | L-proline transmembrane transport | 3 | 5 | 2.86e-01 |
| GO:BP | GO:0003192 | mitral valve formation | 3 | 3 | 2.86e-01 |
| GO:BP | GO:1905647 | proline import across plasma membrane | 3 | 5 | 2.86e-01 |
| GO:BP | GO:0097490 | sympathetic neuron projection extension | 4 | 4 | 2.86e-01 |
| GO:BP | GO:0021670 | lateral ventricle development | 6 | 11 | 2.86e-01 |
| GO:BP | GO:0032571 | response to vitamin K | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 7 | 47 | 2.86e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 6 | 101 | 2.86e-01 |
| GO:BP | GO:2000809 | positive regulation of synaptic vesicle clustering | 2 | 3 | 2.86e-01 |
| GO:BP | GO:1904613 | cellular response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 2.86e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 4 | 784 | 2.86e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 24 | 1229 | 2.86e-01 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 4 | 4 | 2.86e-01 |
| GO:BP | GO:2000520 | regulation of immunological synapse formation | 1 | 2 | 2.87e-01 |
| GO:BP | GO:1990359 | stress response to zinc ion | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0010312 | detoxification of zinc ion | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 15 | 28 | 2.87e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 9 | 94 | 2.87e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 2 | 4 | 2.87e-01 |
| GO:BP | GO:0033631 | cell-cell adhesion mediated by integrin | 1 | 12 | 2.88e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 6 | 105 | 2.88e-01 |
| GO:BP | GO:0006362 | transcription elongation by RNA polymerase I | 3 | 4 | 2.89e-01 |
| GO:BP | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway | 3 | 4 | 2.89e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 6 | 22 | 2.89e-01 |
| GO:BP | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis | 3 | 4 | 2.89e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 3 | 72 | 2.89e-01 |
| GO:BP | GO:0009608 | response to symbiont | 1 | 2 | 2.89e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 1 | 52 | 2.89e-01 |
| GO:BP | GO:0009609 | response to symbiotic bacterium | 1 | 2 | 2.89e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 2.89e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 1 | 13 | 2.89e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 2 | 4 | 2.89e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 36 | 119 | 2.89e-01 |
| GO:BP | GO:1902608 | positive regulation of large conductance calcium-activated potassium channel activity | 1 | 2 | 2.90e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 5 | 13 | 2.90e-01 |
| GO:BP | GO:0045196 | establishment or maintenance of neuroblast polarity | 1 | 3 | 2.90e-01 |
| GO:BP | GO:0090663 | galanin-activated signaling pathway | 1 | 1 | 2.90e-01 |
| GO:BP | GO:0003097 | renal water transport | 4 | 6 | 2.90e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 2.90e-01 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 1 | 1 | 2.90e-01 |
| GO:BP | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane | 1 | 3 | 2.90e-01 |
| GO:BP | GO:1905359 | protein localization to meiotic spindle | 1 | 1 | 2.90e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 2.90e-01 |
| GO:BP | GO:1903096 | protein localization to meiotic spindle midzone | 1 | 1 | 2.90e-01 |
| GO:BP | GO:0045200 | establishment of neuroblast polarity | 1 | 3 | 2.90e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 4 | 14 | 2.90e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 2.90e-01 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 12 | 16 | 2.90e-01 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 5 | 27 | 2.90e-01 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 1 | 3 | 2.90e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 2.90e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 15 | 21 | 2.90e-01 |
| GO:BP | GO:1901859 | negative regulation of mitochondrial DNA metabolic process | 2 | 2 | 2.90e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 10 | 14 | 2.90e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 10 | 207 | 2.91e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 3 | 73 | 2.91e-01 |
| GO:BP | GO:1905463 | negative regulation of DNA duplex unwinding | 2 | 4 | 2.91e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 2 | 101 | 2.91e-01 |
| GO:BP | GO:1990048 | anterograde neuronal dense core vesicle transport | 1 | 4 | 2.92e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 3 | 5 | 2.92e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 7 | 60 | 2.92e-01 |
| GO:BP | GO:0006434 | seryl-tRNA aminoacylation | 2 | 2 | 2.92e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 5 | 17 | 2.92e-01 |
| GO:BP | GO:0006424 | glutamyl-tRNA aminoacylation | 1 | 2 | 2.92e-01 |
| GO:BP | GO:0006433 | prolyl-tRNA aminoacylation | 1 | 2 | 2.92e-01 |
| GO:BP | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter | 1 | 1 | 2.92e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 2.92e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 2.92e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 2 | 42 | 2.92e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 2.92e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 1 | 5 | 2.92e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 1 | 5 | 2.92e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 23 | 77 | 2.92e-01 |
| GO:BP | GO:0071504 | cellular response to heparin | 3 | 4 | 2.92e-01 |
| GO:BP | GO:0032222 | regulation of synaptic transmission, cholinergic | 1 | 3 | 2.92e-01 |
| GO:BP | GO:0061009 | common bile duct development | 3 | 4 | 2.92e-01 |
| GO:BP | GO:0035364 | thymine transport | 1 | 1 | 2.92e-01 |
| GO:BP | GO:0051648 | vesicle localization | 23 | 192 | 2.92e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 3 | 24 | 2.92e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 3 | 35 | 2.92e-01 |
| GO:BP | GO:0070889 | platelet alpha granule organization | 1 | 2 | 2.93e-01 |
| GO:BP | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential | 2 | 10 | 2.93e-01 |
| GO:BP | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 4 | 2.93e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 10 | 35 | 2.93e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 3 | 73 | 2.93e-01 |
| GO:BP | GO:0046710 | GDP metabolic process | 1 | 10 | 2.94e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 2 | 55 | 2.94e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 2.94e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 4 | 7 | 2.94e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 3 | 36 | 2.94e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 2 | 29 | 2.94e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 15 | 37 | 2.94e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 2 | 30 | 2.94e-01 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 6 | 72 | 2.94e-01 |
| GO:BP | GO:1901658 | glycosyl compound catabolic process | 3 | 23 | 2.94e-01 |
| GO:BP | GO:0046133 | pyrimidine ribonucleoside catabolic process | 2 | 7 | 2.94e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 5 | 371 | 2.94e-01 |
| GO:BP | GO:0046131 | pyrimidine ribonucleoside metabolic process | 2 | 7 | 2.94e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 8 | 165 | 2.94e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 6 | 29 | 2.94e-01 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 2 | 5 | 2.94e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 2 | 54 | 2.94e-01 |
| GO:BP | GO:0033566 | gamma-tubulin complex localization | 2 | 2 | 2.95e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 11 | 29 | 2.95e-01 |
| GO:BP | GO:2000210 | positive regulation of anoikis | 1 | 6 | 2.95e-01 |
| GO:BP | GO:0070997 | neuron death | 17 | 286 | 2.95e-01 |
| GO:BP | GO:0014816 | skeletal muscle satellite cell differentiation | 3 | 6 | 2.95e-01 |
| GO:BP | GO:0008050 | female courtship behavior | 2 | 2 | 2.95e-01 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 1 | 17 | 2.96e-01 |
| GO:BP | GO:0006203 | dGTP catabolic process | 2 | 2 | 2.96e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 1 | 34 | 2.96e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 2.96e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 7 | 378 | 2.96e-01 |
| GO:BP | GO:0048789 | cytoskeletal matrix organization at active zone | 1 | 1 | 2.97e-01 |
| GO:BP | GO:0097115 | neurexin clustering involved in presynaptic membrane assembly | 1 | 1 | 2.97e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 9 | 41 | 2.97e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 2 | 23 | 2.97e-01 |
| GO:BP | GO:1905115 | regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 2.97e-01 |
| GO:BP | GO:1905116 | positive regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 2.97e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 5 | 70 | 2.97e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 11 | 19 | 2.98e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 10 | 18 | 2.98e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 11 | 19 | 2.98e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 11 | 19 | 2.98e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 18 | 34 | 2.98e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 2.98e-01 |
| GO:BP | GO:0038129 | ERBB3 signaling pathway | 4 | 4 | 2.98e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 13 | 27 | 2.99e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 3 | 139 | 2.99e-01 |
| GO:BP | GO:1904098 | regulation of protein O-linked glycosylation | 1 | 2 | 2.99e-01 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 7 | 19 | 2.99e-01 |
| GO:BP | GO:0051767 | nitric-oxide synthase biosynthetic process | 1 | 13 | 2.99e-01 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 7 | 19 | 2.99e-01 |
| GO:BP | GO:0097698 | telomere maintenance via base-excision repair | 1 | 1 | 2.99e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 26 | 479 | 2.99e-01 |
| GO:BP | GO:0051769 | regulation of nitric-oxide synthase biosynthetic process | 1 | 13 | 2.99e-01 |
| GO:BP | GO:0009125 | nucleoside monophosphate catabolic process | 1 | 13 | 2.99e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 9 | 97 | 2.99e-01 |
| GO:BP | GO:0090672 | telomerase RNA localization | 7 | 19 | 2.99e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 2.99e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 1 | 17 | 2.99e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 2 | 97 | 2.99e-01 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 1 | 3 | 2.99e-01 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 3 | 6 | 2.99e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 6 | 21 | 2.99e-01 |
| GO:BP | GO:1904100 | positive regulation of protein O-linked glycosylation | 1 | 2 | 2.99e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 2.99e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 2 | 60 | 2.99e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 7 | 197 | 2.99e-01 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 7 | 19 | 2.99e-01 |
| GO:BP | GO:0062196 | regulation of lysosome size | 5 | 8 | 2.99e-01 |
| GO:BP | GO:0070459 | prolactin secretion | 1 | 4 | 2.99e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 1 | 45 | 3.00e-01 |
| GO:BP | GO:0072032 | proximal convoluted tubule segment 2 development | 1 | 1 | 3.00e-01 |
| GO:BP | GO:1901339 | regulation of store-operated calcium channel activity | 3 | 8 | 3.00e-01 |
| GO:BP | GO:0007617 | mating behavior | 2 | 21 | 3.00e-01 |
| GO:BP | GO:0072230 | metanephric proximal straight tubule development | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0072022 | descending thin limb development | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0072232 | metanephric proximal convoluted tubule segment 2 development | 1 | 1 | 3.00e-01 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 7 | 10 | 3.00e-01 |
| GO:BP | GO:0035623 | renal glucose absorption | 2 | 2 | 3.00e-01 |
| GO:BP | GO:0070342 | brown fat cell proliferation | 1 | 1 | 3.00e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 10 | 16 | 3.00e-01 |
| GO:BP | GO:0072020 | proximal straight tubule development | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0140018 | regulation of cytoplasmic translational fidelity | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 4 | 30 | 3.00e-01 |
| GO:BP | GO:0072220 | metanephric descending thin limb development | 1 | 1 | 3.00e-01 |
| GO:BP | GO:1900060 | negative regulation of ceramide biosynthetic process | 1 | 5 | 3.00e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 2 | 71 | 3.01e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 5 | 22 | 3.01e-01 |
| GO:BP | GO:1903126 | negative regulation of centriole-centriole cohesion | 1 | 1 | 3.01e-01 |
| GO:BP | GO:0016358 | dendrite development | 18 | 202 | 3.01e-01 |
| GO:BP | GO:1903929 | primary palate development | 1 | 2 | 3.01e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 5 | 22 | 3.01e-01 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 2 | 11 | 3.01e-01 |
| GO:BP | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | 2 | 2 | 3.01e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 2 | 14 | 3.01e-01 |
| GO:BP | GO:0030576 | Cajal body organization | 4 | 4 | 3.01e-01 |
| GO:BP | GO:0051877 | pigment granule aggregation in cell center | 1 | 2 | 3.01e-01 |
| GO:BP | GO:0072529 | pyrimidine-containing compound catabolic process | 6 | 29 | 3.01e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 2 | 8 | 3.02e-01 |
| GO:BP | GO:1903723 | negative regulation of centriole elongation | 1 | 2 | 3.02e-01 |
| GO:BP | GO:0006307 | DNA dealkylation involved in DNA repair | 7 | 9 | 3.02e-01 |
| GO:BP | GO:0000098 | sulfur amino acid catabolic process | 2 | 7 | 3.02e-01 |
| GO:BP | GO:0016077 | sno(s)RNA catabolic process | 2 | 3 | 3.02e-01 |
| GO:BP | GO:0016102 | diterpenoid biosynthetic process | 2 | 8 | 3.02e-01 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 4 | 15 | 3.02e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 2 | 108 | 3.02e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 3 | 69 | 3.02e-01 |
| GO:BP | GO:0000706 | meiotic DNA double-strand break processing | 2 | 2 | 3.02e-01 |
| GO:BP | GO:2000080 | negative regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 3.02e-01 |
| GO:BP | GO:0090435 | protein localization to nuclear envelope | 7 | 14 | 3.02e-01 |
| GO:BP | GO:0007522 | visceral muscle development | 1 | 1 | 3.02e-01 |
| GO:BP | GO:0090246 | convergent extension involved in somitogenesis | 1 | 1 | 3.02e-01 |
| GO:BP | GO:0044345 | stromal-epithelial cell signaling involved in prostate gland development | 1 | 1 | 3.02e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 1 | 48 | 3.02e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 5 | 7 | 3.02e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 9 | 21 | 3.02e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 2 | 13 | 3.02e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 4 | 6 | 3.02e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 3 | 127 | 3.02e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 8 | 15 | 3.02e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 3.02e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 3 | 4 | 3.02e-01 |
| GO:BP | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process | 2 | 2 | 3.02e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 6 | 20 | 3.02e-01 |
| GO:BP | GO:0007616 | long-term memory | 8 | 31 | 3.04e-01 |
| GO:BP | GO:0043605 | amide catabolic process | 1 | 10 | 3.04e-01 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 2 | 5 | 3.04e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 6 | 40 | 3.04e-01 |
| GO:BP | GO:0035891 | exit from host cell | 5 | 28 | 3.04e-01 |
| GO:BP | GO:0019076 | viral release from host cell | 5 | 28 | 3.04e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 23 | 400 | 3.04e-01 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 2 | 8 | 3.04e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 2 | 2 | 3.04e-01 |
| GO:BP | GO:0097695 | establishment of protein-containing complex localization to telomere | 2 | 3 | 3.05e-01 |
| GO:BP | GO:0097694 | establishment of RNA localization to telomere | 2 | 3 | 3.05e-01 |
| GO:BP | GO:0046338 | phosphatidylethanolamine catabolic process | 2 | 4 | 3.05e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 8 | 56 | 3.05e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 6 | 29 | 3.05e-01 |
| GO:BP | GO:1904997 | regulation of leukocyte adhesion to arterial endothelial cell | 3 | 3 | 3.05e-01 |
| GO:BP | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | 1 | 2 | 3.05e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 2 | 14 | 3.05e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 19 | 64 | 3.06e-01 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 1 | 31 | 3.06e-01 |
| GO:BP | GO:0021894 | cerebral cortex GABAergic interneuron development | 1 | 3 | 3.06e-01 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 6 | 7 | 3.06e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 1 | 67 | 3.06e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 7 | 43 | 3.06e-01 |
| GO:BP | GO:1905026 | positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.06e-01 |
| GO:BP | GO:1905033 | positive regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 2 | 3.06e-01 |
| GO:BP | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 1 | 2 | 3.06e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 3 | 18 | 3.06e-01 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 8 | 11 | 3.07e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 3.07e-01 |
| GO:BP | GO:1905775 | negative regulation of DNA helicase activity | 1 | 2 | 3.07e-01 |
| GO:BP | GO:0035927 | RNA import into mitochondrion | 3 | 4 | 3.07e-01 |
| GO:BP | GO:0071494 | cellular response to UV-C | 2 | 4 | 3.07e-01 |
| GO:BP | GO:0061668 | mitochondrial ribosome assembly | 5 | 8 | 3.07e-01 |
| GO:BP | GO:0090166 | Golgi disassembly | 3 | 6 | 3.07e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 3.07e-01 |
| GO:BP | GO:0036089 | cleavage furrow formation | 2 | 7 | 3.08e-01 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 7 | 30 | 3.08e-01 |
| GO:BP | GO:0009196 | pyrimidine deoxyribonucleoside diphosphate metabolic process | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0009197 | pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0016197 | endosomal transport | 25 | 231 | 3.08e-01 |
| GO:BP | GO:0010459 | negative regulation of heart rate | 2 | 7 | 3.08e-01 |
| GO:BP | GO:0060453 | regulation of gastric acid secretion | 2 | 8 | 3.08e-01 |
| GO:BP | GO:0046072 | dTDP metabolic process | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0046077 | dUDP metabolic process | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0010224 | response to UV-B | 2 | 17 | 3.08e-01 |
| GO:BP | GO:0006227 | dUDP biosynthetic process | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 3 | 19 | 3.08e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 3 | 27 | 3.08e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 3 | 19 | 3.08e-01 |
| GO:BP | GO:0006233 | dTDP biosynthetic process | 2 | 2 | 3.08e-01 |
| GO:BP | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process | 3 | 8 | 3.08e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 4 | 121 | 3.08e-01 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 6 | 8 | 3.08e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 3 | 133 | 3.08e-01 |
| GO:BP | GO:1903429 | regulation of cell maturation | 2 | 13 | 3.08e-01 |
| GO:BP | GO:0021636 | trigeminal nerve morphogenesis | 3 | 4 | 3.09e-01 |
| GO:BP | GO:2001188 | regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 2 | 3.09e-01 |
| GO:BP | GO:0033078 | extrathymic T cell differentiation | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 3.09e-01 |
| GO:BP | GO:0021637 | trigeminal nerve structural organization | 3 | 4 | 3.09e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 5 | 97 | 3.09e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 3.09e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 3.09e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 3 | 13 | 3.09e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 3 | 13 | 3.09e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 21 | 175 | 3.09e-01 |
| GO:BP | GO:1902723 | negative regulation of skeletal muscle satellite cell proliferation | 2 | 2 | 3.10e-01 |
| GO:BP | GO:0070932 | histone H3 deacetylation | 8 | 12 | 3.10e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 3.10e-01 |
| GO:BP | GO:0086098 | angiotensin-activated signaling pathway involved in heart process | 2 | 3 | 3.10e-01 |
| GO:BP | GO:1990637 | response to prolactin | 2 | 3 | 3.10e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 3.10e-01 |
| GO:BP | GO:0098849 | cellular detoxification of cadmium ion | 1 | 1 | 3.10e-01 |
| GO:BP | GO:0140961 | cellular detoxification of metal ion | 1 | 1 | 3.10e-01 |
| GO:BP | GO:0000379 | tRNA-type intron splice site recognition and cleavage | 1 | 3 | 3.10e-01 |
| GO:BP | GO:0003431 | growth plate cartilage chondrocyte development | 3 | 5 | 3.10e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 20 | 206 | 3.10e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 2 | 7 | 3.11e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 2 | 20 | 3.12e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 2 | 20 | 3.12e-01 |
| GO:BP | GO:0097211 | cellular response to gonadotropin-releasing hormone | 2 | 3 | 3.12e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 1 | 69 | 3.12e-01 |
| GO:BP | GO:1900029 | positive regulation of ruffle assembly | 4 | 12 | 3.12e-01 |
| GO:BP | GO:0035513 | oxidative RNA demethylation | 3 | 5 | 3.12e-01 |
| GO:BP | GO:0072402 | response to DNA integrity checkpoint signaling | 3 | 8 | 3.12e-01 |
| GO:BP | GO:0072396 | response to cell cycle checkpoint signaling | 3 | 8 | 3.12e-01 |
| GO:BP | GO:0098957 | anterograde axonal transport of mitochondrion | 1 | 5 | 3.12e-01 |
| GO:BP | GO:0072423 | response to DNA damage checkpoint signaling | 3 | 8 | 3.12e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 8 | 137 | 3.12e-01 |
| GO:BP | GO:0003335 | corneocyte development | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0060945 | cardiac neuron differentiation | 1 | 1 | 3.12e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 26 | 144 | 3.12e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 24 | 399 | 3.12e-01 |
| GO:BP | GO:1903810 | L-histidine import across plasma membrane | 2 | 2 | 3.12e-01 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 2 | 18 | 3.12e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 2 | 2 | 3.12e-01 |
| GO:BP | GO:0002313 | mature B cell differentiation involved in immune response | 2 | 17 | 3.12e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 1 | 15 | 3.12e-01 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 5 | 21 | 3.12e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 3 | 113 | 3.12e-01 |
| GO:BP | GO:0006666 | 3-keto-sphinganine metabolic process | 1 | 1 | 3.14e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 5 | 7 | 3.14e-01 |
| GO:BP | GO:0021773 | striatal medium spiny neuron differentiation | 2 | 3 | 3.14e-01 |
| GO:BP | GO:0033624 | negative regulation of integrin activation | 1 | 3 | 3.14e-01 |
| GO:BP | GO:0032487 | regulation of Rap protein signal transduction | 1 | 3 | 3.14e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 3 | 48 | 3.15e-01 |
| GO:BP | GO:0009615 | response to virus | 3 | 275 | 3.15e-01 |
| GO:BP | GO:0007620 | copulation | 1 | 10 | 3.15e-01 |
| GO:BP | GO:0032025 | response to cobalt ion | 3 | 10 | 3.15e-01 |
| GO:BP | GO:0009838 | abscission | 2 | 6 | 3.15e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 5 | 22 | 3.15e-01 |
| GO:BP | GO:0006043 | glucosamine catabolic process | 2 | 2 | 3.15e-01 |
| GO:BP | GO:0042823 | pyridoxal phosphate biosynthetic process | 1 | 2 | 3.15e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 5 | 28 | 3.15e-01 |
| GO:BP | GO:0008615 | pyridoxine biosynthetic process | 1 | 2 | 3.15e-01 |
| GO:BP | GO:0008614 | pyridoxine metabolic process | 1 | 2 | 3.15e-01 |
| GO:BP | GO:0006386 | termination of RNA polymerase III transcription | 1 | 1 | 3.15e-01 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 1 | 4 | 3.15e-01 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 1 | 142 | 3.16e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 4 | 26 | 3.16e-01 |
| GO:BP | GO:1905445 | positive regulation of clathrin coat assembly | 1 | 1 | 3.16e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 7 | 387 | 3.16e-01 |
| GO:BP | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 2 | 3 | 3.16e-01 |
| GO:BP | GO:1905443 | regulation of clathrin coat assembly | 1 | 1 | 3.16e-01 |
| GO:BP | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery | 5 | 6 | 3.16e-01 |
| GO:BP | GO:0014904 | myotube cell development | 1 | 34 | 3.16e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 3 | 206 | 3.16e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 3.16e-01 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 12 | 29 | 3.16e-01 |
| GO:BP | GO:0021546 | rhombomere development | 2 | 4 | 3.16e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 10 | 17 | 3.16e-01 |
| GO:BP | GO:0000320 | re-entry into mitotic cell cycle | 1 | 2 | 3.16e-01 |
| GO:BP | GO:1901862 | negative regulation of muscle tissue development | 1 | 9 | 3.16e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 4 | 69 | 3.16e-01 |
| GO:BP | GO:0071257 | cellular response to electrical stimulus | 4 | 8 | 3.16e-01 |
| GO:BP | GO:1904439 | negative regulation of iron ion import across plasma membrane | 1 | 1 | 3.16e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 7 | 267 | 3.16e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 20 | 31 | 3.16e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 2 | 32 | 3.16e-01 |
| GO:BP | GO:2000639 | negative regulation of SREBP signaling pathway | 3 | 3 | 3.16e-01 |
| GO:BP | GO:0051642 | centrosome localization | 20 | 31 | 3.16e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 7 | 113 | 3.16e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 6 | 216 | 3.16e-01 |
| GO:BP | GO:0021544 | subpallium development | 3 | 18 | 3.16e-01 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 4 | 66 | 3.17e-01 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 4 | 66 | 3.17e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 3.17e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 1 | 17 | 3.17e-01 |
| GO:BP | GO:0021664 | rhombomere 5 morphogenesis | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0021660 | rhombomere 3 formation | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0021659 | rhombomere 3 structural organization | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 1 | 18 | 3.17e-01 |
| GO:BP | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | 4 | 5 | 3.17e-01 |
| GO:BP | GO:0021665 | rhombomere 5 structural organization | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0021595 | rhombomere structural organization | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0021666 | rhombomere 5 formation | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 3.17e-01 |
| GO:BP | GO:0021594 | rhombomere formation | 1 | 1 | 3.17e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 2 | 6 | 3.17e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 1 | 16 | 3.17e-01 |
| GO:BP | GO:0090168 | Golgi reassembly | 3 | 5 | 3.17e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 5 | 390 | 3.17e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 26 | 191 | 3.17e-01 |
| GO:BP | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway | 1 | 4 | 3.17e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 23 | 52 | 3.17e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 5 | 43 | 3.17e-01 |
| GO:BP | GO:0048850 | hypophysis morphogenesis | 2 | 2 | 3.17e-01 |
| GO:BP | GO:0097501 | stress response to metal ion | 3 | 11 | 3.18e-01 |
| GO:BP | GO:0061687 | detoxification of inorganic compound | 3 | 12 | 3.18e-01 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 1 | 10 | 3.18e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 3.18e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 8 | 14 | 3.18e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 4 | 120 | 3.18e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 22 | 48 | 3.18e-01 |
| GO:BP | GO:0071955 | recycling endosome to Golgi transport | 1 | 2 | 3.18e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 7 | 48 | 3.19e-01 |
| GO:BP | GO:0090155 | negative regulation of sphingolipid biosynthetic process | 1 | 6 | 3.19e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 22 | 679 | 3.19e-01 |
| GO:BP | GO:0010820 | positive regulation of T cell chemotaxis | 3 | 7 | 3.20e-01 |
| GO:BP | GO:0042631 | cellular response to water deprivation | 1 | 1 | 3.20e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 4 | 8 | 3.20e-01 |
| GO:BP | GO:0014041 | regulation of neuron maturation | 1 | 6 | 3.20e-01 |
| GO:BP | GO:1904046 | negative regulation of vascular endothelial growth factor production | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process | 1 | 5 | 3.20e-01 |
| GO:BP | GO:0051697 | protein delipidation | 1 | 5 | 3.21e-01 |
| GO:BP | GO:1904938 | planar cell polarity pathway involved in axon guidance | 1 | 3 | 3.21e-01 |
| GO:BP | GO:0036514 | dopaminergic neuron axon guidance | 1 | 3 | 3.21e-01 |
| GO:BP | GO:0003213 | cardiac right atrium morphogenesis | 1 | 2 | 3.21e-01 |
| GO:BP | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation | 1 | 2 | 3.21e-01 |
| GO:BP | GO:0072721 | cellular response to dithiothreitol | 1 | 1 | 3.22e-01 |
| GO:BP | GO:0036146 | cellular response to mycotoxin | 1 | 1 | 3.22e-01 |
| GO:BP | GO:0046135 | pyrimidine nucleoside catabolic process | 2 | 8 | 3.22e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 6 | 16 | 3.22e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 7 | 16 | 3.22e-01 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 4 | 17 | 3.22e-01 |
| GO:BP | GO:0002949 | tRNA threonylcarbamoyladenosine modification | 3 | 5 | 3.22e-01 |
| GO:BP | GO:0046074 | dTMP catabolic process | 1 | 3 | 3.22e-01 |
| GO:BP | GO:1990953 | intramanchette transport | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0046051 | UTP metabolic process | 5 | 10 | 3.23e-01 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 5 | 15 | 3.23e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 20 | 427 | 3.23e-01 |
| GO:BP | GO:0000296 | spermine transport | 1 | 3 | 3.23e-01 |
| GO:BP | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 3 | 3.23e-01 |
| GO:BP | GO:0032700 | negative regulation of interleukin-17 production | 1 | 9 | 3.23e-01 |
| GO:BP | GO:0048627 | myoblast development | 1 | 3 | 3.23e-01 |
| GO:BP | GO:1905936 | regulation of germ cell proliferation | 3 | 5 | 3.23e-01 |
| GO:BP | GO:0090398 | cellular senescence | 20 | 83 | 3.23e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 1 | 77 | 3.23e-01 |
| GO:BP | GO:0021599 | abducens nerve formation | 1 | 1 | 3.24e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 16 | 50 | 3.24e-01 |
| GO:BP | GO:0021598 | abducens nerve morphogenesis | 1 | 1 | 3.24e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 3.24e-01 |
| GO:BP | GO:0034199 | activation of protein kinase A activity | 3 | 3 | 3.24e-01 |
| GO:BP | GO:0021572 | rhombomere 6 development | 1 | 1 | 3.24e-01 |
| GO:BP | GO:0021560 | abducens nerve development | 1 | 1 | 3.24e-01 |
| GO:BP | GO:2000302 | positive regulation of synaptic vesicle exocytosis | 2 | 3 | 3.24e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 3.24e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 3.24e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 6 | 30 | 3.24e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 8 | 18 | 3.24e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 3.24e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 8 | 18 | 3.24e-01 |
| GO:BP | GO:0015680 | protein maturation by copper ion transfer | 2 | 2 | 3.24e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 29 | 68 | 3.24e-01 |
| GO:BP | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway | 2 | 3 | 3.24e-01 |
| GO:BP | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 2 | 4 | 3.24e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 17 | 39 | 3.24e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 14 | 61 | 3.24e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 42 | 172 | 3.24e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 1 | 17 | 3.24e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 1 | 17 | 3.24e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 12 | 35 | 3.25e-01 |
| GO:BP | GO:0035695 | mitophagy by induced vacuole formation | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0070159 | mitochondrial threonyl-tRNA aminoacylation | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 3.26e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 2 | 40 | 3.26e-01 |
| GO:BP | GO:0055057 | neuroblast division | 2 | 12 | 3.27e-01 |
| GO:BP | GO:1903281 | positive regulation of calcium:sodium antiporter activity | 1 | 2 | 3.27e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 3 | 544 | 3.27e-01 |
| GO:BP | GO:0016080 | synaptic vesicle targeting | 3 | 3 | 3.27e-01 |
| GO:BP | GO:0086092 | regulation of the force of heart contraction by cardiac conduction | 1 | 2 | 3.27e-01 |
| GO:BP | GO:1900240 | negative regulation of phenotypic switching | 1 | 2 | 3.27e-01 |
| GO:BP | GO:1905916 | negative regulation of cell differentiation involved in phenotypic switching | 1 | 2 | 3.27e-01 |
| GO:BP | GO:1905931 | negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 2 | 3.27e-01 |
| GO:BP | GO:0010424 | DNA methylation on cytosine within a CG sequence | 1 | 1 | 3.27e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 3.27e-01 |
| GO:BP | GO:0006914 | autophagy | 80 | 513 | 3.28e-01 |
| GO:BP | GO:1902729 | negative regulation of proteoglycan biosynthetic process | 1 | 1 | 3.28e-01 |
| GO:BP | GO:0003166 | bundle of His development | 5 | 6 | 3.28e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 80 | 513 | 3.28e-01 |
| GO:BP | GO:0032683 | negative regulation of connective tissue growth factor production | 1 | 1 | 3.28e-01 |
| GO:BP | GO:0046684 | response to pyrethroid | 1 | 1 | 3.28e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 9 | 17 | 3.28e-01 |
| GO:BP | GO:1904631 | response to glucoside | 1 | 1 | 3.28e-01 |
| GO:BP | GO:1904425 | negative regulation of GTP binding | 3 | 3 | 3.28e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 4 | 11 | 3.28e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 5 | 39 | 3.29e-01 |
| GO:BP | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0070233 | negative regulation of T cell apoptotic process | 1 | 14 | 3.29e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 2 | 12 | 3.29e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 2 | 11 | 3.29e-01 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 2 | 15 | 3.29e-01 |
| GO:BP | GO:1900239 | regulation of phenotypic switching | 2 | 3 | 3.29e-01 |
| GO:BP | GO:0099554 | trans-synaptic signaling by soluble gas, modulating synaptic transmission | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0099555 | trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 2 | 2 | 3.29e-01 |
| GO:BP | GO:1905462 | regulation of DNA duplex unwinding | 3 | 7 | 3.29e-01 |
| GO:BP | GO:0035166 | post-embryonic hemopoiesis | 4 | 5 | 3.29e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 2 | 5 | 3.29e-01 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 2 | 18 | 3.29e-01 |
| GO:BP | GO:1900451 | positive regulation of glutamate receptor signaling pathway | 1 | 2 | 3.29e-01 |
| GO:BP | GO:1900372 | negative regulation of purine nucleotide biosynthetic process | 2 | 3 | 3.29e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 16 | 424 | 3.29e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 53 | 265 | 3.29e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 11 | 3.29e-01 |
| GO:BP | GO:0015755 | fructose transmembrane transport | 3 | 5 | 3.29e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 3.29e-01 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 4 | 20 | 3.29e-01 |
| GO:BP | GO:0140882 | zinc export across plasma membrane | 1 | 3 | 3.29e-01 |
| GO:BP | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0030809 | negative regulation of nucleotide biosynthetic process | 2 | 3 | 3.29e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 3 | 12 | 3.29e-01 |
| GO:BP | GO:0045618 | positive regulation of keratinocyte differentiation | 10 | 12 | 3.29e-01 |
| GO:BP | GO:1903613 | regulation of protein tyrosine phosphatase activity | 1 | 4 | 3.29e-01 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 2 | 5 | 3.29e-01 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 1 | 3 | 3.29e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 17 | 204 | 3.29e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 4 | 121 | 3.29e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 8 | 11 | 3.29e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 1 | 16 | 3.29e-01 |
| GO:BP | GO:1904762 | positive regulation of myofibroblast differentiation | 1 | 1 | 3.29e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 3.29e-01 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 2 | 20 | 3.29e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 3.29e-01 |
| GO:BP | GO:0071579 | regulation of zinc ion transport | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0046168 | glycerol-3-phosphate catabolic process | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0097117 | guanylate kinase-associated protein clustering | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0007174 | epidermal growth factor catabolic process | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0030397 | membrane disassembly | 1 | 11 | 3.29e-01 |
| GO:BP | GO:1903434 | negative regulation of constitutive secretory pathway | 1 | 1 | 3.29e-01 |
| GO:BP | GO:0006498 | N-terminal protein lipidation | 5 | 7 | 3.29e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 7 | 26 | 3.29e-01 |
| GO:BP | GO:1904048 | regulation of spontaneous neurotransmitter secretion | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 27 | 420 | 3.29e-01 |
| GO:BP | GO:2000744 | positive regulation of anterior head development | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 5 | 22 | 3.29e-01 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 3 | 9 | 3.29e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 6 | 16 | 3.29e-01 |
| GO:BP | GO:2000742 | regulation of anterior head development | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0009446 | putrescine biosynthetic process | 1 | 4 | 3.29e-01 |
| GO:BP | GO:0043465 | regulation of fermentation | 2 | 5 | 3.29e-01 |
| GO:BP | GO:0006113 | fermentation | 2 | 5 | 3.29e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 7 | 9 | 3.30e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 2 | 8 | 3.30e-01 |
| GO:BP | GO:0001841 | neural tube formation | 9 | 101 | 3.30e-01 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 2 | 18 | 3.30e-01 |
| GO:BP | GO:0034627 | ‘de novo’ NAD biosynthetic process | 2 | 2 | 3.31e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 60 | 251 | 3.31e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 3 | 223 | 3.31e-01 |
| GO:BP | GO:0006741 | NADP biosynthetic process | 1 | 2 | 3.31e-01 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 1 | 15 | 3.31e-01 |
| GO:BP | GO:0033319 | UDP-D-xylose metabolic process | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0033320 | UDP-D-xylose biosynthetic process | 1 | 1 | 3.31e-01 |
| GO:BP | GO:1904719 | positive regulation of AMPA glutamate receptor clustering | 1 | 2 | 3.31e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 2 | 26 | 3.31e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 3.31e-01 |
| GO:BP | GO:1901628 | positive regulation of postsynaptic membrane organization | 1 | 2 | 3.31e-01 |
| GO:BP | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity | 1 | 3 | 3.31e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 3.31e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 3.31e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 16 | 23 | 3.31e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 3.32e-01 |
| GO:BP | GO:0035499 | carnosine biosynthetic process | 1 | 1 | 3.32e-01 |
| GO:BP | GO:0046898 | response to cycloheximide | 4 | 4 | 3.32e-01 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 5 | 3.32e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 27 | 135 | 3.32e-01 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 5 | 13 | 3.32e-01 |
| GO:BP | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | 1 | 3 | 3.32e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 3.32e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 2 | 26 | 3.32e-01 |
| GO:BP | GO:0043063 | intercellular bridge organization | 1 | 1 | 3.32e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 3.32e-01 |
| GO:BP | GO:1905533 | negative regulation of leucine import across plasma membrane | 2 | 2 | 3.32e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 2 | 295 | 3.32e-01 |
| GO:BP | GO:0060358 | negative regulation of leucine import | 2 | 2 | 3.32e-01 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 2 | 3.32e-01 |
| GO:BP | GO:0060357 | regulation of leucine import | 2 | 2 | 3.32e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 4 | 40 | 3.32e-01 |
| GO:BP | GO:0015692 | lead ion transport | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 6 | 16 | 3.33e-01 |
| GO:BP | GO:0015676 | vanadium ion transport | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0071895 | odontoblast differentiation | 1 | 5 | 3.33e-01 |
| GO:BP | GO:0015675 | nickel cation transport | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0009888 | tissue development | 5 | 1425 | 3.33e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 3 | 224 | 3.33e-01 |
| GO:BP | GO:0035444 | nickel cation transmembrane transport | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0006207 | ‘de novo’ pyrimidine nucleobase biosynthetic process | 2 | 6 | 3.34e-01 |
| GO:BP | GO:0009790 | embryo development | 3 | 875 | 3.35e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 6 | 14 | 3.35e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 17 | 31 | 3.35e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 2 | 2 | 3.35e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 2 | 2 | 3.35e-01 |
| GO:BP | GO:1903966 | monounsaturated fatty acid biosynthetic process | 1 | 2 | 3.35e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 3 | 128 | 3.35e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 75 | 218 | 3.35e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 22 | 35 | 3.35e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 22 | 35 | 3.35e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 14 | 69 | 3.35e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 1 | 10 | 3.35e-01 |
| GO:BP | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells | 2 | 4 | 3.35e-01 |
| GO:BP | GO:0045054 | constitutive secretory pathway | 2 | 7 | 3.36e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 2 | 18 | 3.36e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 13 | 58 | 3.36e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 25 | 61 | 3.36e-01 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 5 | 6 | 3.36e-01 |
| GO:BP | GO:1901623 | regulation of lymphocyte chemotaxis | 4 | 13 | 3.36e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 12 | 22 | 3.36e-01 |
| GO:BP | GO:0072347 | response to anesthetic | 1 | 1 | 3.36e-01 |
| GO:BP | GO:0010609 | mRNA localization resulting in post-transcriptional regulation of gene expression | 1 | 2 | 3.36e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 5 | 21 | 3.36e-01 |
| GO:BP | GO:1905176 | positive regulation of vascular associated smooth muscle cell dedifferentiation | 1 | 1 | 3.36e-01 |
| GO:BP | GO:0072255 | metanephric glomerular mesangial cell development | 1 | 1 | 3.36e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 1 | 19 | 3.36e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 1 | 21 | 3.37e-01 |
| GO:BP | GO:0071275 | cellular response to aluminum ion | 1 | 1 | 3.37e-01 |
| GO:BP | GO:0140321 | suppression of host autophagy | 1 | 1 | 3.37e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 7 | 66 | 3.37e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 3.37e-01 |
| GO:BP | GO:0039521 | suppression by virus of host autophagy | 1 | 1 | 3.37e-01 |
| GO:BP | GO:1905672 | negative regulation of lysosome organization | 1 | 1 | 3.37e-01 |
| GO:BP | GO:0075071 | modulation by symbiont of host autophagy | 1 | 1 | 3.37e-01 |
| GO:BP | GO:0006598 | polyamine catabolic process | 1 | 4 | 3.37e-01 |
| GO:BP | GO:0106230 | protein depropionylation | 2 | 2 | 3.38e-01 |
| GO:BP | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development | 8 | 13 | 3.38e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 2 | 93 | 3.38e-01 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 6 | 9 | 3.38e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 32 | 73 | 3.38e-01 |
| GO:BP | GO:1902044 | regulation of Fas signaling pathway | 2 | 4 | 3.39e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 15 | 406 | 3.39e-01 |
| GO:BP | GO:0010819 | regulation of T cell chemotaxis | 3 | 7 | 3.39e-01 |
| GO:BP | GO:0034337 | RNA folding | 1 | 1 | 3.39e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 3 | 81 | 3.39e-01 |
| GO:BP | GO:0140251 | regulation protein catabolic process at presynapse | 2 | 2 | 3.39e-01 |
| GO:BP | GO:1904999 | positive regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0032203 | telomere formation via telomerase | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0071279 | cellular response to cobalt ion | 1 | 4 | 3.39e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 11 | 127 | 3.39e-01 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 2 | 19 | 3.40e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 1 | 717 | 3.40e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 6 | 23 | 3.40e-01 |
| GO:BP | GO:0060039 | pericardium development | 7 | 17 | 3.40e-01 |
| GO:BP | GO:0062043 | positive regulation of cardiac epithelial to mesenchymal transition | 2 | 9 | 3.40e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 3.40e-01 |
| GO:BP | GO:0003193 | pulmonary valve formation | 1 | 2 | 3.40e-01 |
| GO:BP | GO:0098905 | regulation of bundle of His cell action potential | 1 | 2 | 3.40e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 7 | 17 | 3.40e-01 |
| GO:BP | GO:0016540 | protein autoprocessing | 11 | 16 | 3.40e-01 |
| GO:BP | GO:1901165 | positive regulation of trophoblast cell migration | 2 | 6 | 3.40e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 1 | 27 | 3.40e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 1 | 19 | 3.40e-01 |
| GO:BP | GO:0048102 | autophagic cell death | 1 | 6 | 3.40e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 11 | 123 | 3.40e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 4 | 13 | 3.40e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 3.40e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 1 | 13 | 3.40e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 3.40e-01 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0030011 | maintenance of cell polarity | 7 | 16 | 3.40e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 1 | 19 | 3.40e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 8 | 83 | 3.40e-01 |
| GO:BP | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 4 | 6 | 3.40e-01 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 1 | 2 | 3.40e-01 |
| GO:BP | GO:1904482 | cellular response to tetrahydrofolate | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 7 | 11 | 3.40e-01 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 3.40e-01 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 11 | 39 | 3.40e-01 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 6 | 16 | 3.40e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 1 | 18 | 3.40e-01 |
| GO:BP | GO:0097089 | methyl-branched fatty acid metabolic process | 1 | 3 | 3.40e-01 |
| GO:BP | GO:1904481 | response to tetrahydrofolate | 1 | 1 | 3.40e-01 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0030954 | astral microtubule nucleation | 1 | 1 | 3.40e-01 |
| GO:BP | GO:1990743 | protein sialylation | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0036213 | contractile ring contraction | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0000916 | actomyosin contractile ring contraction | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0003175 | tricuspid valve development | 5 | 6 | 3.40e-01 |
| GO:BP | GO:0070318 | positive regulation of G0 to G1 transition | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 5 | 10 | 3.41e-01 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 2 | 4 | 3.41e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 22 | 53 | 3.41e-01 |
| GO:BP | GO:0090400 | stress-induced premature senescence | 3 | 7 | 3.42e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 33 | 61 | 3.42e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 8 | 70 | 3.42e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 3 | 870 | 3.42e-01 |
| GO:BP | GO:0018009 | N-terminal peptidyl-L-cysteine N-palmitoylation | 2 | 2 | 3.42e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 3 | 28 | 3.42e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 3.42e-01 |
| GO:BP | GO:0039526 | modulation by virus of host apoptotic process | 3 | 3 | 3.42e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 3.42e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 3.42e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 3.42e-01 |
| GO:BP | GO:0052040 | modulation by symbiont of host programmed cell death | 3 | 3 | 3.42e-01 |
| GO:BP | GO:0090212 | negative regulation of establishment of blood-brain barrier | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 3.42e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 1 | 25 | 3.42e-01 |
| GO:BP | GO:0060454 | positive regulation of gastric acid secretion | 1 | 2 | 3.42e-01 |
| GO:BP | GO:0052150 | modulation by symbiont of host apoptotic process | 3 | 3 | 3.42e-01 |
| GO:BP | GO:0032237 | activation of store-operated calcium channel activity | 2 | 5 | 3.42e-01 |
| GO:BP | GO:0002894 | positive regulation of type II hypersensitivity | 2 | 3 | 3.43e-01 |
| GO:BP | GO:0001798 | positive regulation of type IIa hypersensitivity | 2 | 3 | 3.43e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 5 | 104 | 3.43e-01 |
| GO:BP | GO:0006996 | organelle organization | 149 | 3000 | 3.43e-01 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 4 | 3.43e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 1 | 7 | 3.43e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 7 | 11 | 3.43e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 8 | 182 | 3.43e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 2 | 32 | 3.43e-01 |
| GO:BP | GO:0002505 | antigen processing and presentation of polysaccharide antigen via MHC class II | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 1 | 25 | 3.43e-01 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 4 | 8 | 3.43e-01 |
| GO:BP | GO:0046754 | viral exocytosis | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 7 | 12 | 3.43e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 44 | 1145 | 3.43e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 2 | 3 | 3.43e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 1 | 22 | 3.43e-01 |
| GO:BP | GO:0002747 | antigen processing and presentation following phagocytosis | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0003401 | axis elongation | 9 | 24 | 3.43e-01 |
| GO:BP | GO:0039689 | negative stranded viral RNA replication | 2 | 3 | 3.43e-01 |
| GO:BP | GO:0032764 | negative regulation of mast cell cytokine production | 3 | 3 | 3.43e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 9 | 107 | 3.43e-01 |
| GO:BP | GO:0097719 | neural tissue regeneration | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 5 | 10 | 3.43e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 4 | 24 | 3.44e-01 |
| GO:BP | GO:0035627 | ceramide transport | 2 | 15 | 3.44e-01 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 2 | 3 | 3.44e-01 |
| GO:BP | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | 1 | 4 | 3.44e-01 |
| GO:BP | GO:0097601 | retina blood vessel maintenance | 1 | 1 | 3.44e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 6 | 16 | 3.44e-01 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 4 | 14 | 3.44e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 6 | 23 | 3.44e-01 |
| GO:BP | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 3 | 6 | 3.45e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 5 | 6 | 3.45e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 2 | 21 | 3.45e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 3 | 12 | 3.45e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 3 | 665 | 3.45e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 5 | 6 | 3.45e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 3.45e-01 |
| GO:BP | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 4 | 5 | 3.45e-01 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 6 | 10 | 3.45e-01 |
| GO:BP | GO:0000301 | retrograde transport, vesicle recycling within Golgi | 6 | 9 | 3.45e-01 |
| GO:BP | GO:0106015 | negative regulation of inflammatory response to wounding | 1 | 2 | 3.45e-01 |
| GO:BP | GO:1903712 | cysteine transmembrane transport | 3 | 3 | 3.45e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 2 | 33 | 3.45e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 5 | 9 | 3.45e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 17 | 24 | 3.45e-01 |
| GO:BP | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion | 4 | 5 | 3.45e-01 |
| GO:BP | GO:0046604 | positive regulation of mitotic centrosome separation | 3 | 3 | 3.45e-01 |
| GO:BP | GO:0046606 | negative regulation of centrosome cycle | 5 | 12 | 3.45e-01 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 7 | 15 | 3.45e-01 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 5 | 7 | 3.45e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 2 | 2 | 3.45e-01 |
| GO:BP | GO:1903997 | positive regulation of non-membrane spanning protein tyrosine kinase activity | 2 | 2 | 3.45e-01 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 2 | 20 | 3.45e-01 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 11 | 34 | 3.45e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 3.45e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 3 | 31 | 3.45e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 5 | 6 | 3.45e-01 |
| GO:BP | GO:0042883 | cysteine transport | 3 | 3 | 3.45e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 21 | 94 | 3.45e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 6 | 10 | 3.45e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 9 | 40 | 3.45e-01 |
| GO:BP | GO:0036115 | fatty-acyl-CoA catabolic process | 5 | 6 | 3.45e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 9 | 82 | 3.45e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 2 | 418 | 3.45e-01 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 5 | 5 | 3.45e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 21 | 94 | 3.45e-01 |
| GO:BP | GO:0048703 | embryonic viscerocranium morphogenesis | 1 | 8 | 3.45e-01 |
| GO:BP | GO:0060056 | mammary gland involution | 3 | 8 | 3.45e-01 |
| GO:BP | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | 6 | 10 | 3.45e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 3 | 25 | 3.45e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 5 | 6 | 3.45e-01 |
| GO:BP | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 3.45e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 6 | 7 | 3.45e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 4 | 18 | 3.45e-01 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 2 | 6 | 3.45e-01 |
| GO:BP | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 9 | 14 | 3.45e-01 |
| GO:BP | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 3.45e-01 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 4 | 5 | 3.45e-01 |
| GO:BP | GO:0046967 | cytosol to endoplasmic reticulum transport | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0051664 | nuclear pore localization | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 3.45e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 4 | 18 | 3.45e-01 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 3 | 30 | 3.45e-01 |
| GO:BP | GO:1901321 | positive regulation of heart induction | 2 | 4 | 3.45e-01 |
| GO:BP | GO:1904219 | positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0022406 | membrane docking | 19 | 84 | 3.45e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 3 | 78 | 3.45e-01 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 4 | 7 | 3.45e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 1 | 23 | 3.45e-01 |
| GO:BP | GO:0002091 | negative regulation of receptor internalization | 5 | 14 | 3.45e-01 |
| GO:BP | GO:0071139 | resolution of recombination intermediates | 5 | 6 | 3.45e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 5 | 6 | 3.45e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 4 | 29 | 3.45e-01 |
| GO:BP | GO:0035519 | protein K29-linked ubiquitination | 3 | 6 | 3.45e-01 |
| GO:BP | GO:1904222 | positive regulation of serine C-palmitoyltransferase activity | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0010826 | negative regulation of centrosome duplication | 5 | 12 | 3.45e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 2 | 55 | 3.45e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 1 | 787 | 3.45e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 12 | 1389 | 3.45e-01 |
| GO:BP | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 2 | 2 | 3.45e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 3 | 4 | 3.45e-01 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 8 | 29 | 3.45e-01 |
| GO:BP | GO:0072034 | renal vesicle induction | 3 | 3 | 3.46e-01 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 6 | 26 | 3.46e-01 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 6 | 26 | 3.46e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 5 | 11 | 3.46e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 26 | 44 | 3.47e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 7 | 24 | 3.47e-01 |
| GO:BP | GO:2000537 | regulation of B cell chemotaxis | 1 | 1 | 3.47e-01 |
| GO:BP | GO:2000538 | positive regulation of B cell chemotaxis | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0015823 | phenylalanine transport | 2 | 4 | 3.47e-01 |
| GO:BP | GO:0006304 | DNA modification | 1 | 89 | 3.47e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 3.47e-01 |
| GO:BP | GO:0033499 | galactose catabolic process via UDP-galactose | 2 | 4 | 3.47e-01 |
| GO:BP | GO:2000386 | positive regulation of ovarian follicle development | 2 | 2 | 3.47e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 3.47e-01 |
| GO:BP | GO:1905024 | regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.47e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 11 | 173 | 3.47e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 7 | 9 | 3.47e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 3 | 12 | 3.47e-01 |
| GO:BP | GO:0046065 | dCTP metabolic process | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0006253 | dCTP catabolic process | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0021828 | gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 5 | 6 | 3.47e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 1 | 22 | 3.47e-01 |
| GO:BP | GO:0021856 | hypothalamic tangential migration using cell-axon interactions | 5 | 6 | 3.47e-01 |
| GO:BP | GO:0016078 | tRNA catabolic process | 3 | 14 | 3.47e-01 |
| GO:BP | GO:0051309 | female meiosis chromosome separation | 1 | 2 | 3.47e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 2 | 119 | 3.47e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 4 | 6 | 3.47e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 3 | 14 | 3.47e-01 |
| GO:BP | GO:0014901 | satellite cell activation involved in skeletal muscle regeneration | 2 | 3 | 3.48e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 11 | 19 | 3.48e-01 |
| GO:BP | GO:2000310 | regulation of NMDA receptor activity | 3 | 19 | 3.48e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 4 | 183 | 3.48e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 12 | 16 | 3.48e-01 |
| GO:BP | GO:0097210 | response to gonadotropin-releasing hormone | 2 | 4 | 3.48e-01 |
| GO:BP | GO:0006833 | water transport | 7 | 10 | 3.48e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 28 | 105 | 3.48e-01 |
| GO:BP | GO:0006971 | hypotonic response | 5 | 10 | 3.48e-01 |
| GO:BP | GO:0032902 | nerve growth factor production | 2 | 2 | 3.49e-01 |
| GO:BP | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | 1 | 3 | 3.49e-01 |
| GO:BP | GO:0048852 | diencephalon morphogenesis | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0060157 | urinary bladder development | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0038133 | ERBB2-ERBB3 signaling pathway | 3 | 3 | 3.49e-01 |
| GO:BP | GO:0002335 | mature B cell differentiation | 2 | 21 | 3.49e-01 |
| GO:BP | GO:0097510 | base-excision repair, AP site formation via deaminated base removal | 1 | 1 | 3.49e-01 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 2 | 30 | 3.49e-01 |
| GO:BP | GO:0060686 | negative regulation of prostatic bud formation | 1 | 4 | 3.49e-01 |
| GO:BP | GO:0007566 | embryo implantation | 3 | 39 | 3.49e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 50 | 213 | 3.50e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 4 | 293 | 3.51e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 20 | 386 | 3.51e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 1 | 7 | 3.51e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 3 | 38 | 3.51e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 4 | 25 | 3.51e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 3 | 4 | 3.51e-01 |
| GO:BP | GO:1903516 | regulation of single strand break repair | 2 | 3 | 3.51e-01 |
| GO:BP | GO:0014034 | neural crest cell fate commitment | 3 | 4 | 3.52e-01 |
| GO:BP | GO:1903542 | negative regulation of exosomal secretion | 2 | 4 | 3.52e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 20 | 48 | 3.52e-01 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 3.52e-01 |
| GO:BP | GO:0032253 | dense core granule localization | 1 | 7 | 3.52e-01 |
| GO:BP | GO:0099519 | dense core granule cytoskeletal transport | 1 | 7 | 3.52e-01 |
| GO:BP | GO:0003104 | positive regulation of glomerular filtration | 3 | 3 | 3.52e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 14 | 60 | 3.52e-01 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 1 | 4 | 3.52e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 9 | 555 | 3.52e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 1 | 64 | 3.52e-01 |
| GO:BP | GO:1901950 | dense core granule transport | 1 | 7 | 3.52e-01 |
| GO:BP | GO:0070676 | intralumenal vesicle formation | 2 | 4 | 3.52e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 6 | 10 | 3.52e-01 |
| GO:BP | GO:0030034 | microvillar actin bundle assembly | 1 | 1 | 3.52e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 3.52e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 5 | 165 | 3.52e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 9 | 140 | 3.52e-01 |
| GO:BP | GO:0014740 | negative regulation of muscle hyperplasia | 1 | 1 | 3.52e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 4 | 6 | 3.52e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 4 | 6 | 3.52e-01 |
| GO:BP | GO:1901503 | ether biosynthetic process | 3 | 11 | 3.52e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 33 | 130 | 3.52e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 19 | 203 | 3.52e-01 |
| GO:BP | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway | 1 | 6 | 3.52e-01 |
| GO:BP | GO:0021892 | cerebral cortex GABAergic interneuron differentiation | 1 | 3 | 3.53e-01 |
| GO:BP | GO:0060429 | epithelium development | 3 | 850 | 3.53e-01 |
| GO:BP | GO:0008219 | cell death | 48 | 1596 | 3.53e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 3 | 27 | 3.53e-01 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 4 | 15 | 3.54e-01 |
| GO:BP | GO:0006404 | RNA import into nucleus | 3 | 4 | 3.54e-01 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 3 | 9 | 3.54e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 2 | 8 | 3.54e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 1 | 15 | 3.54e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 3 | 8 | 3.54e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 15 | 3.54e-01 |
| GO:BP | GO:0010621 | negative regulation of transcription by transcription factor localization | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration | 2 | 6 | 3.56e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 2 | 16 | 3.56e-01 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 2 | 20 | 3.56e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 5 | 187 | 3.56e-01 |
| GO:BP | GO:0106034 | protein maturation by [2Fe-2S] cluster transfer | 1 | 3 | 3.56e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 1 | 23 | 3.56e-01 |
| GO:BP | GO:1902775 | mitochondrial large ribosomal subunit assembly | 3 | 4 | 3.56e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 3.56e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 3.56e-01 |
| GO:BP | GO:0098925 | retrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 1 | 1 | 3.56e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 7 | 133 | 3.56e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 25 | 80 | 3.56e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 3.56e-01 |
| GO:BP | GO:0019075 | virus maturation | 1 | 2 | 3.56e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 4 | 42 | 3.57e-01 |
| GO:BP | GO:1902954 | regulation of early endosome to recycling endosome transport | 1 | 2 | 3.57e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 25 | 151 | 3.57e-01 |
| GO:BP | GO:1903566 | positive regulation of protein localization to cilium | 5 | 7 | 3.57e-01 |
| GO:BP | GO:0032070 | regulation of deoxyribonuclease activity | 3 | 9 | 3.57e-01 |
| GO:BP | GO:0015761 | mannose transmembrane transport | 1 | 1 | 3.58e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 5 | 167 | 3.58e-01 |
| GO:BP | GO:0002939 | tRNA N1-guanine methylation | 3 | 3 | 3.58e-01 |
| GO:BP | GO:0003002 | regionalization | 2 | 273 | 3.58e-01 |
| GO:BP | GO:0099172 | presynapse organization | 5 | 42 | 3.58e-01 |
| GO:BP | GO:0044565 | dendritic cell proliferation | 2 | 3 | 3.58e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 3.58e-01 |
| GO:BP | GO:0001782 | B cell homeostasis | 1 | 24 | 3.58e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 2 | 17 | 3.58e-01 |
| GO:BP | GO:0032053 | ciliary basal body organization | 4 | 4 | 3.58e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 14 | 31 | 3.58e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 21 | 47 | 3.59e-01 |
| GO:BP | GO:1902527 | positive regulation of protein monoubiquitination | 4 | 5 | 3.59e-01 |
| GO:BP | GO:0042819 | vitamin B6 biosynthetic process | 1 | 3 | 3.59e-01 |
| GO:BP | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation | 4 | 6 | 3.59e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 44 | 71 | 3.59e-01 |
| GO:BP | GO:0090309 | positive regulation of DNA methylation-dependent heterochromatin formation | 2 | 12 | 3.59e-01 |
| GO:BP | GO:0032402 | melanosome transport | 4 | 19 | 3.59e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 10 | 14 | 3.59e-01 |
| GO:BP | GO:0062025 | regulation of SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 1 | 1 | 3.59e-01 |
| GO:BP | GO:0062026 | negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process | 1 | 1 | 3.59e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 1 | 2 | 3.60e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 4 | 14 | 3.60e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 10 | 18 | 3.60e-01 |
| GO:BP | GO:0098930 | axonal transport | 8 | 60 | 3.60e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 2 | 52 | 3.61e-01 |
| GO:BP | GO:0014859 | negative regulation of skeletal muscle cell proliferation | 2 | 3 | 3.61e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 10 | 252 | 3.61e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 25 | 42 | 3.61e-01 |
| GO:BP | GO:0046069 | cGMP catabolic process | 1 | 4 | 3.61e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 54 | 119 | 3.61e-01 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 6 | 13 | 3.61e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 4 | 21 | 3.61e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 2 | 30 | 3.61e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 1 | 11 | 3.62e-01 |
| GO:BP | GO:0062042 | regulation of cardiac epithelial to mesenchymal transition | 2 | 10 | 3.62e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 3.62e-01 |
| GO:BP | GO:0000012 | single strand break repair | 6 | 10 | 3.62e-01 |
| GO:BP | GO:0042940 | D-amino acid transport | 2 | 5 | 3.62e-01 |
| GO:BP | GO:0090031 | positive regulation of steroid hormone biosynthetic process | 3 | 5 | 3.63e-01 |
| GO:BP | GO:0097531 | mast cell migration | 9 | 12 | 3.63e-01 |
| GO:BP | GO:0036497 | eIF2alpha dephosphorylation in response to endoplasmic reticulum stress | 1 | 1 | 3.63e-01 |
| GO:BP | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development | 1 | 3 | 3.63e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 3.63e-01 |
| GO:BP | GO:1903916 | regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 3.63e-01 |
| GO:BP | GO:1903917 | positive regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 3.63e-01 |
| GO:BP | GO:2000348 | regulation of CD40 signaling pathway | 2 | 4 | 3.63e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 4 | 54 | 3.63e-01 |
| GO:BP | GO:0033026 | negative regulation of mast cell apoptotic process | 1 | 1 | 3.63e-01 |
| GO:BP | GO:0120093 | regulation of peptidyl-lysine crotonylation | 1 | 1 | 3.63e-01 |
| GO:BP | GO:0120094 | negative regulation of peptidyl-lysine crotonylation | 1 | 1 | 3.63e-01 |
| GO:BP | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation | 1 | 1 | 3.63e-01 |
| GO:BP | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process | 1 | 7 | 3.63e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 3 | 12 | 3.63e-01 |
| GO:BP | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis | 3 | 3 | 3.63e-01 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 12 | 36 | 3.63e-01 |
| GO:BP | GO:0060856 | establishment of blood-brain barrier | 2 | 9 | 3.63e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 1 | 16 | 3.63e-01 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 12 | 36 | 3.63e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 5 | 6 | 3.63e-01 |
| GO:BP | GO:0009052 | pentose-phosphate shunt, non-oxidative branch | 4 | 6 | 3.63e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 3.63e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 8 | 14 | 3.63e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 12 | 229 | 3.63e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 3 | 23 | 3.63e-01 |
| GO:BP | GO:1904636 | response to ionomycin | 1 | 3 | 3.63e-01 |
| GO:BP | GO:1904637 | cellular response to ionomycin | 1 | 3 | 3.63e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 100 | 3.64e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 16 | 36 | 3.64e-01 |
| GO:BP | GO:0006003 | fructose 2,6-bisphosphate metabolic process | 1 | 4 | 3.64e-01 |
| GO:BP | GO:0006269 | DNA replication, synthesis of RNA primer | 6 | 7 | 3.64e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 6 | 91 | 3.64e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0006573 | valine metabolic process | 4 | 7 | 3.64e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 8 | 39 | 3.64e-01 |
| GO:BP | GO:0055071 | manganese ion homeostasis | 1 | 2 | 3.64e-01 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 1 | 17 | 3.64e-01 |
| GO:BP | GO:0007517 | muscle organ development | 4 | 277 | 3.64e-01 |
| GO:BP | GO:0021549 | cerebellum development | 1 | 78 | 3.64e-01 |
| GO:BP | GO:0009186 | deoxyribonucleoside diphosphate metabolic process | 3 | 4 | 3.64e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 1 | 17 | 3.64e-01 |
| GO:BP | GO:0044313 | protein K6-linked deubiquitination | 2 | 3 | 3.64e-01 |
| GO:BP | GO:0043200 | response to amino acid | 2 | 95 | 3.64e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 90 | 3.64e-01 |
| GO:BP | GO:0046048 | UDP metabolic process | 1 | 7 | 3.64e-01 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 1 | 2 | 3.64e-01 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 1 | 7 | 3.64e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 4 | 5 | 3.65e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 3 | 763 | 3.65e-01 |
| GO:BP | GO:0009611 | response to wounding | 2 | 415 | 3.65e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 3.65e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 3.65e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 3.65e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 3.65e-01 |
| GO:BP | GO:0061002 | negative regulation of dendritic spine morphogenesis | 3 | 6 | 3.65e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 3.65e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 2 | 513 | 3.65e-01 |
| GO:BP | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 6 | 3.65e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 8 | 24 | 3.66e-01 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 4 | 7 | 3.66e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 3 | 160 | 3.66e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 3.66e-01 |
| GO:BP | GO:0099576 | regulation of protein catabolic process at postsynapse, modulating synaptic transmission | 1 | 4 | 3.66e-01 |
| GO:BP | GO:0055123 | digestive system development | 7 | 100 | 3.66e-01 |
| GO:BP | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 2 | 7 | 3.66e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 9 | 23 | 3.67e-01 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 2 | 12 | 3.67e-01 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 5 | 14 | 3.67e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 1 | 898 | 3.67e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 104 | 3.67e-01 |
| GO:BP | GO:0043584 | nose development | 2 | 10 | 3.67e-01 |
| GO:BP | GO:0097338 | response to clozapine | 2 | 2 | 3.67e-01 |
| GO:BP | GO:1903943 | regulation of hepatocyte apoptotic process | 4 | 7 | 3.67e-01 |
| GO:BP | GO:0090132 | epithelium migration | 10 | 252 | 3.67e-01 |
| GO:BP | GO:0043696 | dedifferentiation | 2 | 6 | 3.67e-01 |
| GO:BP | GO:1903375 | facioacoustic ganglion development | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0046753 | non-lytic viral release | 3 | 14 | 3.67e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 2 | 41 | 3.67e-01 |
| GO:BP | GO:1904835 | dorsal root ganglion morphogenesis | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0106217 | tRNA C3-cytosine methylation | 1 | 2 | 3.67e-01 |
| GO:BP | GO:0030656 | regulation of vitamin metabolic process | 1 | 7 | 3.67e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 3.67e-01 |
| GO:BP | GO:0001906 | cell killing | 4 | 69 | 3.67e-01 |
| GO:BP | GO:0061552 | ganglion morphogenesis | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 26 | 41 | 3.67e-01 |
| GO:BP | GO:0031441 | negative regulation of mRNA 3’-end processing | 1 | 2 | 3.67e-01 |
| GO:BP | GO:0043697 | cell dedifferentiation | 2 | 6 | 3.67e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 12 | 21 | 3.67e-01 |
| GO:BP | GO:0015854 | guanine transport | 1 | 2 | 3.67e-01 |
| GO:BP | GO:0140212 | regulation of long-chain fatty acid import into cell | 4 | 7 | 3.67e-01 |
| GO:BP | GO:1903837 | regulation of mRNA 3’-UTR binding | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0035344 | hypoxanthine transport | 1 | 2 | 3.67e-01 |
| GO:BP | GO:1903839 | positive regulation of mRNA 3’-UTR binding | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 5 | 80 | 3.67e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 12 | 115 | 3.67e-01 |
| GO:BP | GO:0097291 | renal phosphate ion absorption | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 2 | 16 | 3.67e-01 |
| GO:BP | GO:0035863 | dITP catabolic process | 1 | 1 | 3.67e-01 |
| GO:BP | GO:1902747 | negative regulation of lens fiber cell differentiation | 3 | 5 | 3.67e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 3.67e-01 |
| GO:BP | GO:0048565 | digestive tract development | 6 | 92 | 3.67e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 3.67e-01 |
| GO:BP | GO:0021649 | vestibulocochlear nerve structural organization | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0035862 | dITP metabolic process | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0035935 | androgen secretion | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 5 | 34 | 3.67e-01 |
| GO:BP | GO:0061364 | apoptotic process involved in luteolysis | 2 | 3 | 3.67e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 12 | 115 | 3.67e-01 |
| GO:BP | GO:2000836 | positive regulation of androgen secretion | 1 | 1 | 3.67e-01 |
| GO:BP | GO:1903348 | positive regulation of bicellular tight junction assembly | 4 | 5 | 3.67e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 16 | 2139 | 3.67e-01 |
| GO:BP | GO:1905663 | positive regulation of telomerase RNA reverse transcriptase activity | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 5 | 227 | 3.67e-01 |
| GO:BP | GO:2000834 | regulation of androgen secretion | 1 | 1 | 3.67e-01 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 1 | 9 | 3.67e-01 |
| GO:BP | GO:0071403 | cellular response to high density lipoprotein particle stimulus | 3 | 3 | 3.67e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 2 | 75 | 3.67e-01 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 5 | 10 | 3.67e-01 |
| GO:BP | GO:0015798 | myo-inositol transport | 2 | 3 | 3.67e-01 |
| GO:BP | GO:0035377 | transepithelial water transport | 2 | 2 | 3.67e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 1 | 109 | 3.68e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 3.68e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 13 | 3.68e-01 |
| GO:BP | GO:0003129 | heart induction | 4 | 9 | 3.68e-01 |
| GO:BP | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 7 | 18 | 3.68e-01 |
| GO:BP | GO:0071072 | negative regulation of phospholipid biosynthetic process | 5 | 6 | 3.68e-01 |
| GO:BP | GO:0006540 | glutamate decarboxylation to succinate | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0009751 | response to salicylic acid | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0018352 | protein-pyridoxal-5-phosphate linkage | 1 | 2 | 3.68e-01 |
| GO:BP | GO:0039536 | negative regulation of RIG-I signaling pathway | 2 | 7 | 3.68e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 9 | 65 | 3.68e-01 |
| GO:BP | GO:0046208 | spermine catabolic process | 2 | 2 | 3.68e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 4 | 10 | 3.68e-01 |
| GO:BP | GO:0070509 | calcium ion import | 6 | 37 | 3.68e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 12 | 53 | 3.68e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 2 | 19 | 3.69e-01 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 6 | 8 | 3.69e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 4 | 30 | 3.69e-01 |
| GO:BP | GO:0007066 | female meiosis sister chromatid cohesion | 1 | 1 | 3.69e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 4 | 8 | 3.69e-01 |
| GO:BP | GO:0000707 | meiotic DNA recombinase assembly | 1 | 1 | 3.69e-01 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 7 | 23 | 3.69e-01 |
| GO:BP | GO:1905930 | regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 3.70e-01 |
| GO:BP | GO:1905915 | regulation of cell differentiation involved in phenotypic switching | 1 | 3 | 3.70e-01 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 1 | 9 | 3.70e-01 |
| GO:BP | GO:1905420 | vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 3.70e-01 |
| GO:BP | GO:0090679 | cell differentiation involved in phenotypic switching | 1 | 3 | 3.70e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 5 | 14 | 3.70e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 3 | 71 | 3.70e-01 |
| GO:BP | GO:1903279 | regulation of calcium:sodium antiporter activity | 1 | 3 | 3.70e-01 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 1 | 8 | 3.70e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 30 | 70 | 3.70e-01 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 5 | 14 | 3.70e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 5 | 14 | 3.70e-01 |
| GO:BP | GO:1905460 | negative regulation of vascular associated smooth muscle cell apoptotic process | 1 | 2 | 3.70e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 46 | 114 | 3.70e-01 |
| GO:BP | GO:1902178 | fibroblast growth factor receptor apoptotic signaling pathway | 1 | 1 | 3.70e-01 |
| GO:BP | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 5 | 7 | 3.70e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 3.70e-01 |
| GO:BP | GO:1902047 | polyamine transmembrane transport | 2 | 10 | 3.70e-01 |
| GO:BP | GO:0016189 | synaptic vesicle to endosome fusion | 1 | 3 | 3.70e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 8 | 86 | 3.70e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 46 | 809 | 3.70e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 1 | 98 | 3.70e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 3.70e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 6 | 8 | 3.70e-01 |
| GO:BP | GO:0006915 | apoptotic process | 8 | 1443 | 3.71e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 2 | 36 | 3.71e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 32 | 74 | 3.71e-01 |
| GO:BP | GO:0033979 | box H/ACA RNA metabolic process | 2 | 3 | 3.71e-01 |
| GO:BP | GO:0090125 | cell-cell adhesion involved in synapse maturation | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0030046 | parallel actin filament bundle assembly | 2 | 5 | 3.71e-01 |
| GO:BP | GO:0090529 | cell septum assembly | 1 | 1 | 3.71e-01 |
| GO:BP | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 3.71e-01 |
| GO:BP | GO:0000917 | division septum assembly | 1 | 1 | 3.71e-01 |
| GO:BP | GO:0010738 | regulation of protein kinase A signaling | 1 | 13 | 3.71e-01 |
| GO:BP | GO:2000791 | negative regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 3.71e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 3.71e-01 |
| GO:BP | GO:1904274 | tricellular tight junction assembly | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 2 | 8 | 3.72e-01 |
| GO:BP | GO:0051601 | exocyst localization | 2 | 4 | 3.73e-01 |
| GO:BP | GO:1902038 | positive regulation of hematopoietic stem cell differentiation | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0150037 | regulation of calcium-dependent activation of synaptic vesicle fusion | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0035702 | monocyte homeostasis | 1 | 2 | 3.73e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 28 | 637 | 3.73e-01 |
| GO:BP | GO:0061519 | macrophage homeostasis | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 4 | 21 | 3.73e-01 |
| GO:BP | GO:1901585 | regulation of acid-sensing ion channel activity | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0035754 | B cell chemotaxis | 3 | 5 | 3.73e-01 |
| GO:BP | GO:1990789 | thyroid gland epithelial cell proliferation | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 8 | 19 | 3.73e-01 |
| GO:BP | GO:1903697 | negative regulation of microvillus assembly | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 3.73e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 86 | 421 | 3.73e-01 |
| GO:BP | GO:0033197 | response to vitamin E | 1 | 9 | 3.73e-01 |
| GO:BP | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 1 | 11 | 3.73e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 4 | 16 | 3.73e-01 |
| GO:BP | GO:0045794 | negative regulation of cell volume | 2 | 4 | 3.73e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 7 | 76 | 3.73e-01 |
| GO:BP | GO:1904442 | negative regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0051593 | response to folic acid | 1 | 9 | 3.73e-01 |
| GO:BP | GO:1904441 | regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 3.73e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 4 | 5 | 3.73e-01 |
| GO:BP | GO:0048290 | isotype switching to IgA isotypes | 1 | 4 | 3.74e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 16 | 2161 | 3.74e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 9 | 73 | 3.74e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 1 | 23 | 3.74e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 5 | 67 | 3.74e-01 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 1 | 3 | 3.74e-01 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 4 | 12 | 3.74e-01 |
| GO:BP | GO:0032528 | microvillus organization | 7 | 19 | 3.74e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 4 | 19 | 3.75e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 4 | 42 | 3.75e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 4 | 20 | 3.75e-01 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 1 | 6 | 3.76e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 3.76e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 3.76e-01 |
| GO:BP | GO:0001880 | Mullerian duct regression | 2 | 4 | 3.76e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 11 | 186 | 3.76e-01 |
| GO:BP | GO:0062232 | prostanoid catabolic process | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0031393 | negative regulation of prostaglandin biosynthetic process | 2 | 2 | 3.76e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 6 | 42 | 3.76e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 7 | 149 | 3.76e-01 |
| GO:BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0042695 | thelarche | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 3 | 17 | 3.76e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 2 | 5 | 3.76e-01 |
| GO:BP | GO:0150019 | basal dendrite morphogenesis | 3 | 3 | 3.76e-01 |
| GO:BP | GO:0042640 | anagen | 1 | 5 | 3.76e-01 |
| GO:BP | GO:1901341 | positive regulation of store-operated calcium channel activity | 2 | 6 | 3.76e-01 |
| GO:BP | GO:0150018 | basal dendrite development | 3 | 3 | 3.76e-01 |
| GO:BP | GO:0048022 | negative regulation of melanin biosynthetic process | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0097325 | melanocyte proliferation | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 7 | 17 | 3.76e-01 |
| GO:BP | GO:0010038 | response to metal ion | 29 | 264 | 3.76e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 14 | 3.76e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 11 | 131 | 3.76e-01 |
| GO:BP | GO:0018282 | metal incorporation into metallo-sulfur cluster | 2 | 2 | 3.76e-01 |
| GO:BP | GO:1901626 | regulation of postsynaptic membrane organization | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0070512 | positive regulation of histone H4-K20 methylation | 1 | 1 | 3.76e-01 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0072229 | metanephric proximal convoluted tubule development | 1 | 2 | 3.76e-01 |
| GO:BP | GO:1905344 | prostaglandin catabolic process | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0060744 | mammary gland branching involved in thelarche | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0018283 | iron incorporation into metallo-sulfur cluster | 2 | 2 | 3.76e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 1 | 32 | 3.76e-01 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 2 | 3 | 3.76e-01 |
| GO:BP | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0051884 | regulation of timing of anagen | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0090618 | DNA clamp unloading | 2 | 2 | 3.76e-01 |
| GO:BP | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0072019 | proximal convoluted tubule development | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0070293 | renal absorption | 7 | 24 | 3.76e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 2 | 53 | 3.76e-01 |
| GO:BP | GO:0150020 | basal dendrite arborization | 3 | 3 | 3.76e-01 |
| GO:BP | GO:0010868 | negative regulation of triglyceride biosynthetic process | 1 | 3 | 3.76e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 5 | 12 | 3.76e-01 |
| GO:BP | GO:0060751 | branch elongation involved in mammary gland duct branching | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 3.77e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 1 | 102 | 3.77e-01 |
| GO:BP | GO:0044003 | modulation by symbiont of host process | 4 | 5 | 3.77e-01 |
| GO:BP | GO:0035552 | oxidative single-stranded DNA demethylation | 3 | 3 | 3.77e-01 |
| GO:BP | GO:0044068 | modulation by symbiont of host cellular process | 4 | 5 | 3.77e-01 |
| GO:BP | GO:0090130 | tissue migration | 10 | 256 | 3.77e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 16 | 92 | 3.77e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 16 | 92 | 3.77e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 50 | 282 | 3.77e-01 |
| GO:BP | GO:0001771 | immunological synapse formation | 2 | 7 | 3.77e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 7 | 60 | 3.77e-01 |
| GO:BP | GO:0019048 | modulation by virus of host process | 4 | 5 | 3.77e-01 |
| GO:BP | GO:0019054 | modulation by virus of host cellular process | 4 | 5 | 3.77e-01 |
| GO:BP | GO:2000620 | positive regulation of histone H4-K16 acetylation | 2 | 2 | 3.77e-01 |
| GO:BP | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0007389 | pattern specification process | 2 | 306 | 3.77e-01 |
| GO:BP | GO:2000079 | regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 3.77e-01 |
| GO:BP | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 1 | 2 | 3.77e-01 |
| GO:BP | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0046050 | UMP catabolic process | 1 | 3 | 3.77e-01 |
| GO:BP | GO:0009597 | detection of virus | 4 | 5 | 3.77e-01 |
| GO:BP | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction | 2 | 2 | 3.77e-01 |
| GO:BP | GO:2000041 | negative regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0006248 | CMP catabolic process | 1 | 3 | 3.77e-01 |
| GO:BP | GO:0090594 | inflammatory response to wounding | 2 | 12 | 3.77e-01 |
| GO:BP | GO:0051823 | regulation of synapse structural plasticity | 2 | 5 | 3.77e-01 |
| GO:BP | GO:0044343 | canonical Wnt signaling pathway involved in regulation of type B pancreatic cell proliferation | 1 | 1 | 3.77e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 13 | 45 | 3.77e-01 |
| GO:BP | GO:0015848 | spermidine transport | 1 | 3 | 3.77e-01 |
| GO:BP | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation | 6 | 7 | 3.77e-01 |
| GO:BP | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron development | 6 | 7 | 3.77e-01 |
| GO:BP | GO:0038018 | Wnt receptor catabolic process | 2 | 2 | 3.78e-01 |
| GO:BP | GO:1990575 | mitochondrial L-ornithine transmembrane transport | 1 | 2 | 3.78e-01 |
| GO:BP | GO:0046511 | sphinganine biosynthetic process | 2 | 2 | 3.78e-01 |
| GO:BP | GO:0031120 | snRNA pseudouridine synthesis | 2 | 3 | 3.78e-01 |
| GO:BP | GO:0072739 | response to anisomycin | 1 | 4 | 3.78e-01 |
| GO:BP | GO:0061205 | paramesonephric duct development | 2 | 4 | 3.78e-01 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 1 | 30 | 3.78e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 4 | 40 | 3.79e-01 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 2 | 23 | 3.79e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 1 | 130 | 3.79e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 3.79e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 3.79e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 3.79e-01 |
| GO:BP | GO:1903964 | monounsaturated fatty acid metabolic process | 1 | 2 | 3.79e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 3.79e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 3.79e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 3.79e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 1 | 24 | 3.79e-01 |
| GO:BP | GO:0035601 | protein deacylation | 52 | 116 | 3.80e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 4 | 51 | 3.80e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 37 | 179 | 3.80e-01 |
| GO:BP | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway | 4 | 5 | 3.80e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 3.80e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 2 | 45 | 3.80e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 38 | 140 | 3.81e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 3 | 3 | 3.81e-01 |
| GO:BP | GO:2000565 | negative regulation of CD8-positive, alpha-beta T cell proliferation | 2 | 2 | 3.81e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 4 | 25 | 3.81e-01 |
| GO:BP | GO:0110051 | metabolite repair | 1 | 1 | 3.81e-01 |
| GO:BP | GO:0030879 | mammary gland development | 1 | 106 | 3.82e-01 |
| GO:BP | GO:0035928 | rRNA import into mitochondrion | 2 | 3 | 3.82e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 3 | 20 | 3.82e-01 |
| GO:BP | GO:1902389 | ceramide 1-phosphate transport | 2 | 7 | 3.82e-01 |
| GO:BP | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 228 | 3.82e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 2 | 2 | 3.82e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 1 | 39 | 3.82e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 67 | 287 | 3.82e-01 |
| GO:BP | GO:0006564 | L-serine biosynthetic process | 4 | 6 | 3.82e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 1 | 35 | 3.82e-01 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 7 | 12 | 3.82e-01 |
| GO:BP | GO:0000105 | histidine biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 6 | 11 | 3.82e-01 |
| GO:BP | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 1 | 4 | 3.82e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 4 | 4 | 3.82e-01 |
| GO:BP | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | 3 | 4 | 3.82e-01 |
| GO:BP | GO:0031860 | telomeric 3’ overhang formation | 1 | 4 | 3.82e-01 |
| GO:BP | GO:0060419 | heart growth | 29 | 72 | 3.82e-01 |
| GO:BP | GO:0044873 | lipoprotein localization to membrane | 1 | 1 | 3.82e-01 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 1 | 10 | 3.82e-01 |
| GO:BP | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 3 | 3.82e-01 |
| GO:BP | GO:0001778 | plasma membrane repair | 2 | 28 | 3.82e-01 |
| GO:BP | GO:0003064 | regulation of heart rate by hormone | 2 | 2 | 3.82e-01 |
| GO:BP | GO:0060047 | heart contraction | 12 | 205 | 3.82e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 2 | 4 | 3.82e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 3 | 18 | 3.82e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 22 | 49 | 3.82e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 4 | 42 | 3.82e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 24 | 54 | 3.82e-01 |
| GO:BP | GO:0051409 | response to nitrosative stress | 6 | 10 | 3.83e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 7 | 31 | 3.83e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 3.83e-01 |
| GO:BP | GO:0090251 | protein localization involved in establishment of planar polarity | 2 | 2 | 3.83e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 3 | 5 | 3.83e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 3 | 32 | 3.83e-01 |
| GO:BP | GO:0070483 | detection of hypoxia | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 1 | 8 | 3.83e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 3 | 5 | 3.83e-01 |
| GO:BP | GO:0043456 | regulation of pentose-phosphate shunt | 1 | 6 | 3.83e-01 |
| GO:BP | GO:0002540 | leukotriene production involved in inflammatory response | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0046688 | response to copper ion | 5 | 29 | 3.83e-01 |
| GO:BP | GO:1903804 | glycine import across plasma membrane | 1 | 4 | 3.83e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 3.83e-01 |
| GO:BP | GO:0015818 | isoleucine transport | 1 | 5 | 3.83e-01 |
| GO:BP | GO:1903301 | positive regulation of hexokinase activity | 1 | 5 | 3.83e-01 |
| GO:BP | GO:1904782 | negative regulation of NMDA glutamate receptor activity | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0099630 | postsynaptic neurotransmitter receptor cycle | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0099627 | neurotransmitter receptor cycle | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0006228 | UTP biosynthetic process | 6 | 8 | 3.83e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 2 | 18 | 3.83e-01 |
| GO:BP | GO:0098904 | regulation of AV node cell action potential | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0009085 | lysine biosynthetic process | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0099156 | cell-cell signaling via exosome | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0110155 | NAD-cap decapping | 3 | 3 | 3.83e-01 |
| GO:BP | GO:0060166 | olfactory pit development | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0061586 | positive regulation of transcription by transcription factor localization | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0019878 | lysine biosynthetic process via aminoadipic acid | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 23 | 441 | 3.83e-01 |
| GO:BP | GO:0006771 | riboflavin metabolic process | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0036371 | protein localization to T-tubule | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 1 | 30 | 3.83e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 25 | 43 | 3.83e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 2 | 504 | 3.84e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 3.84e-01 |
| GO:BP | GO:1990261 | pre-mRNA catabolic process | 1 | 1 | 3.84e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 27 | 615 | 3.85e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 6 | 43 | 3.85e-01 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 3 | 5 | 3.85e-01 |
| GO:BP | GO:0061764 | late endosome to lysosome transport via multivesicular body sorting pathway | 2 | 2 | 3.85e-01 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 2 | 33 | 3.86e-01 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 6 | 18 | 3.86e-01 |
| GO:BP | GO:0051136 | regulation of NK T cell differentiation | 1 | 4 | 3.86e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 7 | 28 | 3.86e-01 |
| GO:BP | GO:0060173 | limb development | 2 | 142 | 3.86e-01 |
| GO:BP | GO:0048736 | appendage development | 2 | 142 | 3.86e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 7 | 42 | 3.87e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 2 | 18 | 3.87e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 6 | 9 | 3.87e-01 |
| GO:BP | GO:1905065 | positive regulation of vascular associated smooth muscle cell differentiation | 3 | 6 | 3.87e-01 |
| GO:BP | GO:0034058 | endosomal vesicle fusion | 1 | 10 | 3.87e-01 |
| GO:BP | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway | 2 | 3 | 3.87e-01 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 5 | 24 | 3.87e-01 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 4 | 6 | 3.87e-01 |
| GO:BP | GO:0060914 | heart formation | 11 | 29 | 3.87e-01 |
| GO:BP | GO:0032252 | secretory granule localization | 1 | 9 | 3.87e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 3.87e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 5 | 114 | 3.87e-01 |
| GO:BP | GO:0034499 | late endosome to Golgi transport | 2 | 5 | 3.87e-01 |
| GO:BP | GO:0061952 | midbody abscission | 4 | 17 | 3.87e-01 |
| GO:BP | GO:1903621 | protein localization to photoreceptor connecting cilium | 1 | 1 | 3.87e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 6 | 91 | 3.88e-01 |
| GO:BP | GO:0002941 | synoviocyte proliferation | 1 | 3 | 3.88e-01 |
| GO:BP | GO:1901645 | regulation of synoviocyte proliferation | 1 | 3 | 3.88e-01 |
| GO:BP | GO:0035351 | heme transmembrane transport | 1 | 2 | 3.88e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 1 | 19 | 3.88e-01 |
| GO:BP | GO:0044557 | relaxation of smooth muscle | 6 | 9 | 3.88e-01 |
| GO:BP | GO:1901647 | positive regulation of synoviocyte proliferation | 1 | 3 | 3.88e-01 |
| GO:BP | GO:0035694 | mitochondrial protein catabolic process | 2 | 6 | 3.88e-01 |
| GO:BP | GO:0012501 | programmed cell death | 8 | 1479 | 3.88e-01 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 3 | 11 | 3.88e-01 |
| GO:BP | GO:0001928 | regulation of exocyst assembly | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0070221 | sulfide oxidation, using sulfide:quinone oxidoreductase | 1 | 3 | 3.88e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 11 | 20 | 3.88e-01 |
| GO:BP | GO:0060083 | smooth muscle contraction involved in micturition | 1 | 2 | 3.88e-01 |
| GO:BP | GO:0001927 | exocyst assembly | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0019418 | sulfide oxidation | 1 | 3 | 3.88e-01 |
| GO:BP | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process | 3 | 3 | 3.88e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 3.88e-01 |
| GO:BP | GO:0040009 | regulation of growth rate | 1 | 3 | 3.88e-01 |
| GO:BP | GO:0060178 | regulation of exocyst localization | 1 | 1 | 3.88e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 2 | 6 | 3.88e-01 |
| GO:BP | GO:0006768 | biotin metabolic process | 3 | 3 | 3.88e-01 |
| GO:BP | GO:0061757 | leukocyte adhesion to arterial endothelial cell | 3 | 4 | 3.88e-01 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 2 | 2 | 3.88e-01 |
| GO:BP | GO:0030208 | dermatan sulfate biosynthetic process | 2 | 5 | 3.88e-01 |
| GO:BP | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain | 1 | 2 | 3.88e-01 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 5 | 7 | 3.88e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 2 | 105 | 3.88e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 1 | 86 | 3.88e-01 |
| GO:BP | GO:0016254 | preassembly of GPI anchor in ER membrane | 6 | 8 | 3.88e-01 |
| GO:BP | GO:0071409 | cellular response to cycloheximide | 2 | 2 | 3.88e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 3 | 25 | 3.88e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 5 | 236 | 3.88e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 3 | 18 | 3.88e-01 |
| GO:BP | GO:0060354 | negative regulation of cell adhesion molecule production | 3 | 5 | 3.88e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 4 | 25 | 3.88e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 5 | 3.88e-01 |
| GO:BP | GO:0061945 | regulation of protein K48-linked ubiquitination | 2 | 4 | 3.88e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 17 | 29 | 3.88e-01 |
| GO:BP | GO:0140039 | cell-cell adhesion in response to extracellular stimulus | 1 | 2 | 3.88e-01 |
| GO:BP | GO:0035733 | hepatic stellate cell activation | 6 | 8 | 3.88e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 3 | 63 | 3.88e-01 |
| GO:BP | GO:2000631 | regulation of pre-miRNA processing | 3 | 3 | 3.88e-01 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 6 | 3.88e-01 |
| GO:BP | GO:0046337 | phosphatidylethanolamine metabolic process | 7 | 19 | 3.89e-01 |
| GO:BP | GO:0160024 | Leydig cell proliferation | 2 | 2 | 3.89e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 3.89e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 6 | 97 | 3.89e-01 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 19 | 3.89e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 3 | 16 | 3.89e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 1 | 36 | 3.89e-01 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 2 | 12 | 3.89e-01 |
| GO:BP | GO:0098760 | response to interleukin-7 | 2 | 12 | 3.89e-01 |
| GO:BP | GO:0006895 | Golgi to endosome transport | 10 | 17 | 3.89e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 3 | 40 | 3.89e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 40 | 84 | 3.89e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 4 | 20 | 3.89e-01 |
| GO:BP | GO:1905222 | atrioventricular canal morphogenesis | 3 | 4 | 3.89e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 4 | 83 | 3.90e-01 |
| GO:BP | GO:0060682 | primary ureteric bud growth | 1 | 1 | 3.90e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 8 | 58 | 3.90e-01 |
| GO:BP | GO:0006041 | glucosamine metabolic process | 3 | 3 | 3.90e-01 |
| GO:BP | GO:0002317 | plasma cell differentiation | 1 | 5 | 3.90e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 10 | 15 | 3.90e-01 |
| GO:BP | GO:0043276 | anoikis | 12 | 32 | 3.90e-01 |
| GO:BP | GO:0140647 | P450-containing electron transport chain | 1 | 2 | 3.90e-01 |
| GO:BP | GO:0019388 | galactose catabolic process | 2 | 5 | 3.90e-01 |
| GO:BP | GO:0009110 | vitamin biosynthetic process | 2 | 14 | 3.91e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 3.91e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 114 | 3.91e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 3 | 5 | 3.91e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 114 | 3.91e-01 |
| GO:BP | GO:0036359 | renal potassium excretion | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 3.91e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 2 | 75 | 3.91e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 3 | 12 | 3.91e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 3 | 14 | 3.92e-01 |
| GO:BP | GO:0032534 | regulation of microvillus assembly | 2 | 6 | 3.92e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 7 | 10 | 3.92e-01 |
| GO:BP | GO:0021785 | branchiomotor neuron axon guidance | 3 | 5 | 3.92e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 3.93e-01 |
| GO:BP | GO:0016114 | terpenoid biosynthetic process | 2 | 12 | 3.93e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 4 | 48 | 3.93e-01 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 5 | 9 | 3.93e-01 |
| GO:BP | GO:0016180 | snRNA processing | 2 | 26 | 3.93e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 3 | 54 | 3.93e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 6 | 8 | 3.93e-01 |
| GO:BP | GO:0032876 | negative regulation of DNA endoreduplication | 1 | 1 | 3.93e-01 |
| GO:BP | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 2 | 6 | 3.94e-01 |
| GO:BP | GO:0033169 | histone H3-K9 demethylation | 2 | 5 | 3.94e-01 |
| GO:BP | GO:0016926 | protein desumoylation | 5 | 9 | 3.94e-01 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 1 | 9 | 3.94e-01 |
| GO:BP | GO:0032352 | positive regulation of hormone metabolic process | 1 | 11 | 3.94e-01 |
| GO:BP | GO:0072239 | metanephric glomerulus vasculature development | 2 | 6 | 3.94e-01 |
| GO:BP | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway | 2 | 4 | 3.94e-01 |
| GO:BP | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 4 | 3.94e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 13 | 29 | 3.95e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 9 | 14 | 3.95e-01 |
| GO:BP | GO:1904438 | regulation of iron ion import across plasma membrane | 1 | 1 | 3.96e-01 |
| GO:BP | GO:0042816 | vitamin B6 metabolic process | 4 | 5 | 3.96e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 15 | 30 | 3.96e-01 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 3 | 3 | 3.96e-01 |
| GO:BP | GO:0010160 | formation of animal organ boundary | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0008052 | sensory organ boundary specification | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway | 2 | 6 | 3.97e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 6 | 121 | 3.97e-01 |
| GO:BP | GO:0061193 | taste bud development | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 1 | 18 | 3.97e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 3.97e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 19 | 188 | 3.97e-01 |
| GO:BP | GO:0051591 | response to cAMP | 7 | 66 | 3.97e-01 |
| GO:BP | GO:0035148 | tube formation | 11 | 138 | 3.97e-01 |
| GO:BP | GO:0031915 | positive regulation of synaptic plasticity | 5 | 6 | 3.97e-01 |
| GO:BP | GO:0021658 | rhombomere 3 morphogenesis | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0044154 | histone H3-K14 acetylation | 5 | 12 | 3.97e-01 |
| GO:BP | GO:0072105 | ureteric peristalsis | 2 | 2 | 3.97e-01 |
| GO:BP | GO:0046929 | negative regulation of neurotransmitter secretion | 3 | 6 | 3.97e-01 |
| GO:BP | GO:0021593 | rhombomere morphogenesis | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0014849 | ureter smooth muscle contraction | 2 | 2 | 3.97e-01 |
| GO:BP | GO:0019082 | viral protein processing | 2 | 31 | 3.97e-01 |
| GO:BP | GO:0014040 | positive regulation of Schwann cell differentiation | 1 | 2 | 3.97e-01 |
| GO:BP | GO:0051097 | negative regulation of helicase activity | 3 | 5 | 3.98e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 2 | 3 | 3.98e-01 |
| GO:BP | GO:0044854 | plasma membrane raft assembly | 1 | 6 | 3.98e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 3 | 6 | 3.98e-01 |
| GO:BP | GO:0060613 | fat pad development | 1 | 6 | 3.98e-01 |
| GO:BP | GO:0098967 | exocytic insertion of neurotransmitter receptor to postsynaptic membrane | 4 | 5 | 3.98e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 26 | 65 | 3.98e-01 |
| GO:BP | GO:0006419 | alanyl-tRNA aminoacylation | 2 | 2 | 3.98e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 1 | 121 | 3.98e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 2 | 28 | 3.98e-01 |
| GO:BP | GO:0001967 | suckling behavior | 1 | 10 | 3.99e-01 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 1 | 5 | 3.99e-01 |
| GO:BP | GO:1904428 | negative regulation of tubulin deacetylation | 3 | 6 | 3.99e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 2 | 42 | 3.99e-01 |
| GO:BP | GO:0043602 | nitrate catabolic process | 1 | 1 | 3.99e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 2 | 22 | 3.99e-01 |
| GO:BP | GO:0055070 | copper ion homeostasis | 5 | 18 | 3.99e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 3.99e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 19 | 27 | 3.99e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 2 | 2 | 3.99e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 8 | 15 | 3.99e-01 |
| GO:BP | GO:0046210 | nitric oxide catabolic process | 1 | 1 | 3.99e-01 |
| GO:BP | GO:1903361 | protein localization to basolateral plasma membrane | 1 | 7 | 3.99e-01 |
| GO:BP | GO:0051793 | medium-chain fatty acid catabolic process | 2 | 3 | 3.99e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 5 | 44 | 3.99e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 3 | 37 | 3.99e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 8 | 24 | 3.99e-01 |
| GO:BP | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation | 1 | 1 | 3.99e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 3.99e-01 |
| GO:BP | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0048278 | vesicle docking | 13 | 57 | 4.00e-01 |
| GO:BP | GO:0021979 | hypothalamus cell differentiation | 1 | 8 | 4.00e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 4.00e-01 |
| GO:BP | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay | 1 | 3 | 4.00e-01 |
| GO:BP | GO:0071468 | cellular response to acidic pH | 2 | 10 | 4.00e-01 |
| GO:BP | GO:1900161 | regulation of phospholipid scramblase activity | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0001951 | intestinal D-glucose absorption | 2 | 3 | 4.00e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 4.00e-01 |
| GO:BP | GO:0006829 | zinc ion transport | 4 | 23 | 4.00e-01 |
| GO:BP | GO:0090381 | regulation of heart induction | 1 | 4 | 4.00e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 4 | 28 | 4.00e-01 |
| GO:BP | GO:1900163 | positive regulation of phospholipid scramblase activity | 1 | 1 | 4.00e-01 |
| GO:BP | GO:2000493 | negative regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 39 | 74 | 4.00e-01 |
| GO:BP | GO:0051351 | positive regulation of ligase activity | 7 | 9 | 4.00e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 4.00e-01 |
| GO:BP | GO:2000752 | regulation of glucosylceramide catabolic process | 1 | 1 | 4.00e-01 |
| GO:BP | GO:2000753 | positive regulation of glucosylceramide catabolic process | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 6 | 256 | 4.00e-01 |
| GO:BP | GO:1904353 | regulation of telomere capping | 3 | 23 | 4.00e-01 |
| GO:BP | GO:1902474 | positive regulation of protein localization to synapse | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0035524 | proline transmembrane transport | 4 | 8 | 4.00e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 1 | 14 | 4.00e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 4 | 44 | 4.00e-01 |
| GO:BP | GO:0046452 | dihydrofolate metabolic process | 1 | 3 | 4.00e-01 |
| GO:BP | GO:1904679 | myo-inositol import across plasma membrane | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0100012 | regulation of heart induction by canonical Wnt signaling pathway | 1 | 4 | 4.00e-01 |
| GO:BP | GO:0060685 | regulation of prostatic bud formation | 1 | 5 | 4.00e-01 |
| GO:BP | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade | 1 | 4 | 4.00e-01 |
| GO:BP | GO:0021891 | olfactory bulb interneuron development | 1 | 5 | 4.00e-01 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 2 | 3 | 4.00e-01 |
| GO:BP | GO:0035904 | aorta development | 3 | 50 | 4.00e-01 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 2 | 4 | 4.01e-01 |
| GO:BP | GO:1905301 | regulation of macropinocytosis | 3 | 3 | 4.01e-01 |
| GO:BP | GO:0009175 | pyrimidine ribonucleoside monophosphate catabolic process | 1 | 4 | 4.01e-01 |
| GO:BP | GO:0006249 | dCMP catabolic process | 1 | 3 | 4.01e-01 |
| GO:BP | GO:1902463 | protein localization to cell leading edge | 4 | 5 | 4.01e-01 |
| GO:BP | GO:0001554 | luteolysis | 3 | 7 | 4.01e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 4.01e-01 |
| GO:BP | GO:0010138 | pyrimidine ribonucleotide salvage | 1 | 4 | 4.01e-01 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 6 | 11 | 4.01e-01 |
| GO:BP | GO:0046063 | dCMP metabolic process | 1 | 3 | 4.01e-01 |
| GO:BP | GO:0032262 | pyrimidine nucleotide salvage | 1 | 4 | 4.01e-01 |
| GO:BP | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation | 1 | 1 | 4.01e-01 |
| GO:BP | GO:0044206 | UMP salvage | 1 | 4 | 4.01e-01 |
| GO:BP | GO:0046079 | dUMP catabolic process | 1 | 4 | 4.01e-01 |
| GO:BP | GO:1905371 | ceramide phosphoethanolamine metabolic process | 2 | 2 | 4.01e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 4.01e-01 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 4 | 4 | 4.01e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 2 | 11 | 4.01e-01 |
| GO:BP | GO:1905373 | ceramide phosphoethanolamine biosynthetic process | 2 | 2 | 4.01e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 2 | 14 | 4.01e-01 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 5 | 14 | 4.01e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 7 | 19 | 4.01e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 9 | 43 | 4.01e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 4.01e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 14 | 84 | 4.01e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 21 | 486 | 4.01e-01 |
| GO:BP | GO:2001176 | regulation of mediator complex assembly | 1 | 1 | 4.01e-01 |
| GO:BP | GO:0036034 | mediator complex assembly | 1 | 1 | 4.01e-01 |
| GO:BP | GO:2001178 | positive regulation of mediator complex assembly | 1 | 1 | 4.01e-01 |
| GO:BP | GO:0046380 | N-acetylneuraminate biosynthetic process | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 37 | 90 | 4.02e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 4.02e-01 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 5 | 19 | 4.02e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 2 | 33 | 4.02e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 27 | 625 | 4.02e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 13 | 27 | 4.02e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 3 | 69 | 4.02e-01 |
| GO:BP | GO:0035265 | organ growth | 1 | 131 | 4.03e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 38 | 901 | 4.03e-01 |
| GO:BP | GO:0033183 | negative regulation of histone ubiquitination | 3 | 4 | 4.04e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 2 | 12 | 4.04e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 4.04e-01 |
| GO:BP | GO:0010808 | positive regulation of synaptic vesicle priming | 1 | 1 | 4.04e-01 |
| GO:BP | GO:2000019 | negative regulation of male gonad development | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 2 | 12 | 4.04e-01 |
| GO:BP | GO:0018158 | protein oxidation | 1 | 12 | 4.04e-01 |
| GO:BP | GO:0071625 | vocalization behavior | 2 | 13 | 4.04e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 106 | 2011 | 4.04e-01 |
| GO:BP | GO:1901315 | negative regulation of histone H2A K63-linked ubiquitination | 3 | 4 | 4.04e-01 |
| GO:BP | GO:0061369 | negative regulation of testicular blood vessel morphogenesis | 1 | 1 | 4.04e-01 |
| GO:BP | GO:1901314 | regulation of histone H2A K63-linked ubiquitination | 3 | 4 | 4.04e-01 |
| GO:BP | GO:0000389 | mRNA 3’-splice site recognition | 1 | 6 | 4.04e-01 |
| GO:BP | GO:0031948 | positive regulation of glucocorticoid biosynthetic process | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 2 | 6 | 4.04e-01 |
| GO:BP | GO:0031945 | positive regulation of glucocorticoid metabolic process | 1 | 1 | 4.04e-01 |
| GO:BP | GO:2000180 | negative regulation of androgen biosynthetic process | 1 | 1 | 4.04e-01 |
| GO:BP | GO:2000066 | positive regulation of cortisol biosynthetic process | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 4.04e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 4.04e-01 |
| GO:BP | GO:1901403 | positive regulation of tetrapyrrole metabolic process | 3 | 4 | 4.04e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 55 | 234 | 4.04e-01 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 2 | 12 | 4.04e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 1 | 21 | 4.04e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 4 | 10 | 4.04e-01 |
| GO:BP | GO:0032400 | melanosome localization | 2 | 22 | 4.04e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 12 | 19 | 4.04e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 1 | 24 | 4.05e-01 |
| GO:BP | GO:0071787 | endoplasmic reticulum tubular network formation | 3 | 4 | 4.05e-01 |
| GO:BP | GO:1905550 | regulation of protein localization to endoplasmic reticulum | 3 | 4 | 4.05e-01 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 4 | 4 | 4.05e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 5 | 23 | 4.05e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 7 | 16 | 4.05e-01 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 3 | 4 | 4.06e-01 |
| GO:BP | GO:0001822 | kidney development | 9 | 244 | 4.06e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 1 | 13 | 4.06e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 4.06e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 4.06e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 7 | 93 | 4.06e-01 |
| GO:BP | GO:0006825 | copper ion transport | 3 | 13 | 4.06e-01 |
| GO:BP | GO:0071361 | cellular response to ethanol | 1 | 9 | 4.07e-01 |
| GO:BP | GO:1905395 | response to flavonoid | 2 | 2 | 4.07e-01 |
| GO:BP | GO:0045110 | intermediate filament bundle assembly | 5 | 6 | 4.07e-01 |
| GO:BP | GO:0015846 | polyamine transport | 2 | 12 | 4.07e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 4.07e-01 |
| GO:BP | GO:1900247 | regulation of cytoplasmic translational elongation | 7 | 10 | 4.07e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 4.07e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 1 | 30 | 4.07e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 1 | 19 | 4.07e-01 |
| GO:BP | GO:0120078 | cell adhesion involved in sprouting angiogenesis | 2 | 2 | 4.07e-01 |
| GO:BP | GO:0120077 | angiogenic sprout fusion | 2 | 2 | 4.07e-01 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 2 | 16 | 4.07e-01 |
| GO:BP | GO:0002157 | positive regulation of thyroid hormone mediated signaling pathway | 2 | 3 | 4.07e-01 |
| GO:BP | GO:0007619 | courtship behavior | 2 | 3 | 4.07e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 7 | 16 | 4.07e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 4.07e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 5 | 81 | 4.07e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 2 | 11 | 4.07e-01 |
| GO:BP | GO:0072197 | ureter morphogenesis | 3 | 5 | 4.07e-01 |
| GO:BP | GO:1905552 | positive regulation of protein localization to endoplasmic reticulum | 2 | 2 | 4.07e-01 |
| GO:BP | GO:0071503 | response to heparin | 3 | 5 | 4.07e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 41 | 1214 | 4.07e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 4 | 16 | 4.07e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 4 | 16 | 4.07e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 46 | 834 | 4.07e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 2 | 14 | 4.07e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 2 | 138 | 4.07e-01 |
| GO:BP | GO:0046070 | dGTP metabolic process | 2 | 3 | 4.07e-01 |
| GO:BP | GO:0097154 | GABAergic neuron differentiation | 1 | 5 | 4.07e-01 |
| GO:BP | GO:2000808 | negative regulation of synaptic vesicle clustering | 1 | 1 | 4.08e-01 |
| GO:BP | GO:0010886 | positive regulation of cholesterol storage | 2 | 5 | 4.08e-01 |
| GO:BP | GO:1904978 | regulation of endosome organization | 2 | 5 | 4.08e-01 |
| GO:BP | GO:0071707 | immunoglobulin heavy chain V-D-J recombination | 2 | 2 | 4.08e-01 |
| GO:BP | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 2 | 5 | 4.08e-01 |
| GO:BP | GO:1903936 | cellular response to sodium arsenite | 1 | 5 | 4.08e-01 |
| GO:BP | GO:0061000 | negative regulation of dendritic spine development | 5 | 10 | 4.08e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 30 | 300 | 4.08e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 6 | 101 | 4.08e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 10 | 31 | 4.08e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 2 | 94 | 4.08e-01 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 4 | 15 | 4.08e-01 |
| GO:BP | GO:1903433 | regulation of constitutive secretory pathway | 1 | 2 | 4.08e-01 |
| GO:BP | GO:0042726 | flavin-containing compound metabolic process | 1 | 4 | 4.08e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 8 | 25 | 4.08e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 38 | 85 | 4.08e-01 |
| GO:BP | GO:0009188 | ribonucleoside diphosphate biosynthetic process | 1 | 8 | 4.08e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 2 | 144 | 4.08e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 37 | 58 | 4.08e-01 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 4 | 5 | 4.09e-01 |
| GO:BP | GO:0031453 | positive regulation of heterochromatin formation | 2 | 15 | 4.09e-01 |
| GO:BP | GO:0060763 | mammary duct terminal end bud growth | 1 | 3 | 4.09e-01 |
| GO:BP | GO:0120263 | positive regulation of heterochromatin organization | 2 | 15 | 4.09e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 7 | 112 | 4.09e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 9 | 169 | 4.09e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 7 | 20 | 4.09e-01 |
| GO:BP | GO:1903569 | positive regulation of protein localization to ciliary membrane | 1 | 1 | 4.09e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 1 | 25 | 4.09e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 4.09e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 34 | 58 | 4.09e-01 |
| GO:BP | GO:0003174 | mitral valve development | 8 | 12 | 4.09e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 4.09e-01 |
| GO:BP | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway | 1 | 2 | 4.09e-01 |
| GO:BP | GO:0070593 | dendrite self-avoidance | 1 | 11 | 4.09e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 5 | 30 | 4.09e-01 |
| GO:BP | GO:0046697 | decidualization | 5 | 19 | 4.09e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 4.09e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 4.09e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 1 | 4 | 4.09e-01 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 7 | 29 | 4.09e-01 |
| GO:BP | GO:0061713 | anterior neural tube closure | 1 | 2 | 4.10e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 1 | 40 | 4.10e-01 |
| GO:BP | GO:0060035 | notochord cell development | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0060034 | notochord cell differentiation | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 1 | 3 | 4.10e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 12 | 68 | 4.10e-01 |
| GO:BP | GO:0061881 | positive regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0048641 | regulation of skeletal muscle tissue development | 1 | 20 | 4.10e-01 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 1 | 11 | 4.10e-01 |
| GO:BP | GO:0061880 | regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 4.10e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 3 | 20 | 4.10e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 2 | 9 | 4.10e-01 |
| GO:BP | GO:0072432 | response to G1 DNA damage checkpoint signaling | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0072414 | response to mitotic cell cycle checkpoint signaling | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 3 | 179 | 4.11e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 1 | 111 | 4.11e-01 |
| GO:BP | GO:2001311 | lysobisphosphatidic acid metabolic process | 2 | 2 | 4.11e-01 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 3 | 8 | 4.11e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 4 | 16 | 4.11e-01 |
| GO:BP | GO:1903753 | negative regulation of p38MAPK cascade | 3 | 5 | 4.11e-01 |
| GO:BP | GO:2000173 | negative regulation of branching morphogenesis of a nerve | 1 | 1 | 4.11e-01 |
| GO:BP | GO:1904799 | regulation of neuron remodeling | 1 | 1 | 4.11e-01 |
| GO:BP | GO:1904800 | negative regulation of neuron remodeling | 1 | 1 | 4.11e-01 |
| GO:BP | GO:0006547 | histidine metabolic process | 2 | 5 | 4.11e-01 |
| GO:BP | GO:1902261 | positive regulation of delayed rectifier potassium channel activity | 1 | 5 | 4.11e-01 |
| GO:BP | GO:0048864 | stem cell development | 3 | 76 | 4.11e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 1 | 27 | 4.11e-01 |
| GO:BP | GO:1901506 | regulation of acylglycerol transport | 1 | 2 | 4.11e-01 |
| GO:BP | GO:1901508 | positive regulation of acylglycerol transport | 1 | 2 | 4.11e-01 |
| GO:BP | GO:1905885 | positive regulation of triglyceride transport | 1 | 2 | 4.11e-01 |
| GO:BP | GO:1905883 | regulation of triglyceride transport | 1 | 2 | 4.11e-01 |
| GO:BP | GO:0060050 | positive regulation of protein glycosylation | 1 | 6 | 4.11e-01 |
| GO:BP | GO:0051340 | regulation of ligase activity | 8 | 11 | 4.11e-01 |
| GO:BP | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway | 2 | 2 | 4.11e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 19 | 31 | 4.11e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 31 | 197 | 4.11e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 5 | 37 | 4.11e-01 |
| GO:BP | GO:0071839 | apoptotic process in bone marrow cell | 4 | 4 | 4.11e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 4 | 214 | 4.11e-01 |
| GO:BP | GO:2001205 | negative regulation of osteoclast development | 1 | 2 | 4.12e-01 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 5 | 8 | 4.12e-01 |
| GO:BP | GO:0072014 | proximal tubule development | 2 | 7 | 4.12e-01 |
| GO:BP | GO:0009992 | intracellular water homeostasis | 2 | 3 | 4.12e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 5 | 133 | 4.12e-01 |
| GO:BP | GO:1905774 | regulation of DNA helicase activity | 1 | 5 | 4.12e-01 |
| GO:BP | GO:0070203 | regulation of establishment of protein localization to telomere | 4 | 11 | 4.12e-01 |
| GO:BP | GO:2000346 | negative regulation of hepatocyte proliferation | 1 | 3 | 4.12e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 5 | 23 | 4.12e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 5 | 23 | 4.12e-01 |
| GO:BP | GO:0010836 | negative regulation of protein ADP-ribosylation | 3 | 4 | 4.12e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 5 | 23 | 4.12e-01 |
| GO:BP | GO:0007613 | memory | 19 | 92 | 4.13e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 3 | 271 | 4.13e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 12 | 38 | 4.13e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 8 | 55 | 4.13e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 2 | 60 | 4.14e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 2 | 22 | 4.14e-01 |
| GO:BP | GO:0001696 | gastric acid secretion | 2 | 10 | 4.14e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 4.14e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 4.14e-01 |
| GO:BP | GO:1901656 | glycoside transport | 4 | 5 | 4.14e-01 |
| GO:BP | GO:0035498 | carnosine metabolic process | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 7 | 13 | 4.14e-01 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 1 | 2 | 4.14e-01 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0008216 | spermidine metabolic process | 4 | 7 | 4.14e-01 |
| GO:BP | GO:0014856 | skeletal muscle cell proliferation | 5 | 14 | 4.14e-01 |
| GO:BP | GO:0009648 | photoperiodism | 13 | 28 | 4.14e-01 |
| GO:BP | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion | 4 | 7 | 4.14e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 4.14e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 9 | 44 | 4.14e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 2 | 7 | 4.14e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 2 | 7 | 4.14e-01 |
| GO:BP | GO:1902308 | regulation of peptidyl-serine dephosphorylation | 3 | 3 | 4.14e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 4.15e-01 |
| GO:BP | GO:1904199 | positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.15e-01 |
| GO:BP | GO:1903949 | positive regulation of atrial cardiac muscle cell action potential | 1 | 1 | 4.15e-01 |
| GO:BP | GO:1990736 | regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.15e-01 |
| GO:BP | GO:1902319 | DNA strand elongation involved in nuclear cell cycle DNA replication | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0030210 | heparin biosynthetic process | 1 | 9 | 4.15e-01 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 9 | 25 | 4.15e-01 |
| GO:BP | GO:1902983 | DNA strand elongation involved in mitotic DNA replication | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 407 | 4.15e-01 |
| GO:BP | GO:1902296 | DNA strand elongation involved in cell cycle DNA replication | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 38 | 90 | 4.15e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 16 | 26 | 4.15e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 3 | 37 | 4.15e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 4 | 14 | 4.16e-01 |
| GO:BP | GO:0043038 | amino acid activation | 9 | 44 | 4.16e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 4 | 9 | 4.16e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 4 | 9 | 4.16e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 1 | 9 | 4.16e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 2 | 43 | 4.16e-01 |
| GO:BP | GO:0009099 | valine biosynthetic process | 1 | 3 | 4.16e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 4.16e-01 |
| GO:BP | GO:0009097 | isoleucine biosynthetic process | 1 | 2 | 4.16e-01 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 2 | 18 | 4.16e-01 |
| GO:BP | GO:1900186 | negative regulation of clathrin-dependent endocytosis | 3 | 4 | 4.16e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 3 | 11 | 4.16e-01 |
| GO:BP | GO:0002892 | regulation of type II hypersensitivity | 2 | 3 | 4.16e-01 |
| GO:BP | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib | 2 | 4 | 4.16e-01 |
| GO:BP | GO:0001796 | regulation of type IIa hypersensitivity | 2 | 3 | 4.16e-01 |
| GO:BP | GO:1900045 | negative regulation of protein K63-linked ubiquitination | 1 | 7 | 4.16e-01 |
| GO:BP | GO:1901254 | positive regulation of intracellular transport of viral material | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0032815 | negative regulation of natural killer cell activation | 1 | 3 | 4.17e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 16 | 37 | 4.17e-01 |
| GO:BP | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0006646 | phosphatidylethanolamine biosynthetic process | 4 | 8 | 4.17e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 3 | 26 | 4.17e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 4.17e-01 |
| GO:BP | GO:0101030 | tRNA-guanine transglycosylation | 2 | 2 | 4.18e-01 |
| GO:BP | GO:2000504 | positive regulation of blood vessel remodeling | 1 | 1 | 4.18e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 1 | 42 | 4.18e-01 |
| GO:BP | GO:0098749 | cerebellar neuron development | 1 | 3 | 4.18e-01 |
| GO:BP | GO:0006610 | ribosomal protein import into nucleus | 1 | 4 | 4.18e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 5 | 7 | 4.18e-01 |
| GO:BP | GO:0033327 | Leydig cell differentiation | 6 | 9 | 4.18e-01 |
| GO:BP | GO:0048853 | forebrain morphogenesis | 4 | 7 | 4.18e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 4.18e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 5 | 35 | 4.18e-01 |
| GO:BP | GO:0032238 | adenosine transport | 1 | 2 | 4.18e-01 |
| GO:BP | GO:1902268 | negative regulation of polyamine transmembrane transport | 1 | 3 | 4.18e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 1 | 258 | 4.18e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 4.18e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 4.18e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 4.18e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 5 | 211 | 4.19e-01 |
| GO:BP | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 5 | 4.19e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 29 | 52 | 4.19e-01 |
| GO:BP | GO:1901726 | negative regulation of histone deacetylase activity | 2 | 2 | 4.19e-01 |
| GO:BP | GO:0150066 | negative regulation of deacetylase activity | 2 | 2 | 4.19e-01 |
| GO:BP | GO:0043490 | malate-aspartate shuttle | 2 | 4 | 4.19e-01 |
| GO:BP | GO:0060677 | ureteric bud elongation | 5 | 7 | 4.19e-01 |
| GO:BP | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 4.19e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 2 | 2 | 4.19e-01 |
| GO:BP | GO:0072209 | metanephric mesangial cell differentiation | 1 | 1 | 4.19e-01 |
| GO:BP | GO:0006853 | carnitine shuttle | 1 | 3 | 4.19e-01 |
| GO:BP | GO:0072254 | metanephric glomerular mesangial cell differentiation | 1 | 1 | 4.19e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 45 | 87 | 4.19e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 1 | 4 | 4.19e-01 |
| GO:BP | GO:0003015 | heart process | 12 | 216 | 4.19e-01 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 4 | 8 | 4.19e-01 |
| GO:BP | GO:0021540 | corpus callosum morphogenesis | 4 | 5 | 4.19e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 38 | 4.19e-01 |
| GO:BP | GO:0106300 | protein-DNA covalent cross-linking repair | 3 | 7 | 4.20e-01 |
| GO:BP | GO:0010917 | negative regulation of mitochondrial membrane potential | 2 | 9 | 4.20e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 2 | 5 | 4.20e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 2 | 5 | 4.20e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 7 | 13 | 4.20e-01 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 1 | 9 | 4.20e-01 |
| GO:BP | GO:0045837 | negative regulation of membrane potential | 2 | 9 | 4.20e-01 |
| GO:BP | GO:0038135 | ERBB2-ERBB4 signaling pathway | 3 | 4 | 4.20e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 2 | 2 | 4.20e-01 |
| GO:BP | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process | 5 | 9 | 4.20e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 8 | 39 | 4.20e-01 |
| GO:BP | GO:0016104 | triterpenoid biosynthetic process | 1 | 1 | 4.20e-01 |
| GO:BP | GO:0090022 | regulation of neutrophil chemotaxis | 1 | 19 | 4.20e-01 |
| GO:BP | GO:0071455 | cellular response to hyperoxia | 1 | 5 | 4.20e-01 |
| GO:BP | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion | 1 | 1 | 4.20e-01 |
| GO:BP | GO:0036298 | recombinational interstrand cross-link repair | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0070716 | mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 6 | 4.20e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 1 | 6 | 4.20e-01 |
| GO:BP | GO:0060638 | mesenchymal-epithelial cell signaling | 1 | 4 | 4.20e-01 |
| GO:BP | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade | 1 | 5 | 4.20e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 5 | 50 | 4.20e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 1 | 6 | 4.20e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 4.20e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 12 | 66 | 4.20e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 27 | 167 | 4.20e-01 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 3 | 25 | 4.20e-01 |
| GO:BP | GO:0099022 | vesicle tethering | 13 | 31 | 4.20e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 2 | 8 | 4.20e-01 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 2 | 13 | 4.21e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 10 | 51 | 4.21e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 25 | 50 | 4.21e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 5 | 23 | 4.21e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 4.21e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 4.21e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 7 | 156 | 4.21e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 2 | 4 | 4.21e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 4.21e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 2 | 4 | 4.21e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 2 | 4 | 4.21e-01 |
| GO:BP | GO:2000810 | regulation of bicellular tight junction assembly | 8 | 15 | 4.21e-01 |
| GO:BP | GO:0008655 | pyrimidine-containing compound salvage | 1 | 5 | 4.21e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 1 | 33 | 4.22e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 2 | 36 | 4.22e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 5 | 13 | 4.22e-01 |
| GO:BP | GO:0061956 | penetration of cumulus oophorus | 1 | 1 | 4.22e-01 |
| GO:BP | GO:0070384 | Harderian gland development | 1 | 4 | 4.22e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 3 | 3 | 4.22e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 5 | 4.22e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 3 | 29 | 4.22e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 6 | 102 | 4.22e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 2 | 16 | 4.22e-01 |
| GO:BP | GO:0003105 | negative regulation of glomerular filtration | 1 | 3 | 4.22e-01 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 4.23e-01 |
| GO:BP | GO:1900073 | regulation of neuromuscular synaptic transmission | 1 | 2 | 4.23e-01 |
| GO:BP | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0021561 | facial nerve development | 5 | 8 | 4.23e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 9 | 12 | 4.23e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 5 | 53 | 4.23e-01 |
| GO:BP | GO:0010967 | regulation of polyamine biosynthetic process | 2 | 2 | 4.23e-01 |
| GO:BP | GO:0001830 | trophectodermal cell fate commitment | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0097069 | cellular response to thyroxine stimulus | 2 | 2 | 4.23e-01 |
| GO:BP | GO:1900075 | positive regulation of neuromuscular synaptic transmission | 1 | 2 | 4.23e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 8 | 120 | 4.23e-01 |
| GO:BP | GO:2000490 | negative regulation of hepatic stellate cell activation | 2 | 2 | 4.23e-01 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 4 | 6 | 4.23e-01 |
| GO:BP | GO:0044849 | estrous cycle | 6 | 13 | 4.23e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 5 | 8 | 4.23e-01 |
| GO:BP | GO:1904387 | cellular response to L-phenylalanine derivative | 2 | 2 | 4.23e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 4.23e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 4.23e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 4.23e-01 |
| GO:BP | GO:1903996 | negative regulation of non-membrane spanning protein tyrosine kinase activity | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0000390 | spliceosomal complex disassembly | 2 | 3 | 4.23e-01 |
| GO:BP | GO:0036113 | very long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0006591 | ornithine metabolic process | 1 | 6 | 4.24e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 11 | 20 | 4.24e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 29 | 77 | 4.24e-01 |
| GO:BP | GO:0035640 | exploration behavior | 3 | 21 | 4.24e-01 |
| GO:BP | GO:0061833 | protein localization to tricellular tight junction | 1 | 2 | 4.24e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 22 | 198 | 4.24e-01 |
| GO:BP | GO:0021700 | developmental maturation | 3 | 211 | 4.24e-01 |
| GO:BP | GO:0048672 | positive regulation of collateral sprouting | 6 | 10 | 4.25e-01 |
| GO:BP | GO:1905269 | positive regulation of chromatin organization | 2 | 15 | 4.25e-01 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 2 | 31 | 4.25e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 6 | 39 | 4.25e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 4 | 33 | 4.25e-01 |
| GO:BP | GO:0072001 | renal system development | 9 | 251 | 4.25e-01 |
| GO:BP | GO:0061289 | Wnt signaling pathway involved in kidney development | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0061290 | canonical Wnt signaling pathway involved in metanephric kidney development | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0031119 | tRNA pseudouridine synthesis | 1 | 6 | 4.25e-01 |
| GO:BP | GO:0060995 | cell-cell signaling involved in kidney development | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0072204 | cell-cell signaling involved in metanephros development | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 2 | 4 | 4.25e-01 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 4 | 5 | 4.25e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 4 | 110 | 4.25e-01 |
| GO:BP | GO:0036211 | protein modification process | 3 | 2901 | 4.25e-01 |
| GO:BP | GO:0097332 | response to antipsychotic drug | 2 | 2 | 4.25e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 6 | 134 | 4.25e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 6 | 103 | 4.25e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 6 | 46 | 4.25e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 4.25e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 4 | 11 | 4.26e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 2 | 9 | 4.26e-01 |
| GO:BP | GO:0045063 | T-helper 1 cell differentiation | 7 | 14 | 4.26e-01 |
| GO:BP | GO:0050655 | dermatan sulfate proteoglycan metabolic process | 1 | 12 | 4.26e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 1 | 32 | 4.26e-01 |
| GO:BP | GO:2000397 | positive regulation of ubiquitin-dependent endocytosis | 1 | 2 | 4.26e-01 |
| GO:BP | GO:2000395 | regulation of ubiquitin-dependent endocytosis | 1 | 2 | 4.26e-01 |
| GO:BP | GO:1904044 | response to aldosterone | 3 | 9 | 4.26e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 3 | 87 | 4.26e-01 |
| GO:BP | GO:0090210 | regulation of establishment of blood-brain barrier | 1 | 3 | 4.26e-01 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 2 | 9 | 4.26e-01 |
| GO:BP | GO:0030578 | PML body organization | 1 | 6 | 4.26e-01 |
| GO:BP | GO:0007182 | common-partner SMAD protein phosphorylation | 1 | 6 | 4.26e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 1 | 45 | 4.26e-01 |
| GO:BP | GO:1903975 | regulation of glial cell migration | 2 | 12 | 4.26e-01 |
| GO:BP | GO:1904911 | negative regulation of establishment of RNA localization to telomere | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1904913 | regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1904914 | negative regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1904850 | negative regulation of establishment of protein localization to telomere | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 10 | 4.26e-01 |
| GO:BP | GO:1904792 | positive regulation of shelterin complex assembly | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1904910 | regulation of establishment of RNA localization to telomere | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0051548 | negative regulation of keratinocyte migration | 2 | 2 | 4.26e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 4.26e-01 |
| GO:BP | GO:0048645 | animal organ formation | 3 | 51 | 4.27e-01 |
| GO:BP | GO:0070079 | histone H4-R3 demethylation | 1 | 1 | 4.27e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 1 | 121 | 4.27e-01 |
| GO:BP | GO:0034478 | phosphatidylglycerol catabolic process | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0018395 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 8 | 35 | 4.27e-01 |
| GO:BP | GO:0071043 | CUT metabolic process | 2 | 5 | 4.27e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 26 | 200 | 4.27e-01 |
| GO:BP | GO:0007507 | heart development | 2 | 506 | 4.27e-01 |
| GO:BP | GO:0071034 | CUT catabolic process | 2 | 5 | 4.27e-01 |
| GO:BP | GO:0002182 | cytoplasmic translational elongation | 8 | 12 | 4.27e-01 |
| GO:BP | GO:0070078 | histone H3-R2 demethylation | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0007625 | grooming behavior | 2 | 10 | 4.27e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 39 | 174 | 4.27e-01 |
| GO:BP | GO:0071709 | membrane assembly | 3 | 53 | 4.27e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 7 | 38 | 4.27e-01 |
| GO:BP | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation | 2 | 2 | 4.27e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 2 | 6 | 4.27e-01 |
| GO:BP | GO:0017085 | response to insecticide | 1 | 4 | 4.27e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 3 | 15 | 4.27e-01 |
| GO:BP | GO:0032230 | positive regulation of synaptic transmission, GABAergic | 1 | 10 | 4.27e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 6 | 10 | 4.27e-01 |
| GO:BP | GO:0000425 | pexophagy | 1 | 6 | 4.27e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 2 | 36 | 4.27e-01 |
| GO:BP | GO:0090117 | endosome to lysosome transport of low-density lipoprotein particle | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 6 | 68 | 4.27e-01 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 1 | 10 | 4.27e-01 |
| GO:BP | GO:0060271 | cilium assembly | 7 | 292 | 4.27e-01 |
| GO:BP | GO:0034201 | response to oleic acid | 4 | 5 | 4.27e-01 |
| GO:BP | GO:0046878 | positive regulation of saliva secretion | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0035378 | carbon dioxide transmembrane transport | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0035127 | post-embryonic limb morphogenesis | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0035120 | post-embryonic appendage morphogenesis | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0035128 | post-embryonic forelimb morphogenesis | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0072237 | metanephric proximal tubule development | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 3 | 74 | 4.27e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 3 | 74 | 4.27e-01 |
| GO:BP | GO:1990550 | mitochondrial alpha-ketoglutarate transmembrane transport | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 13 | 25 | 4.27e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 4 | 40 | 4.27e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 10 | 21 | 4.27e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 4.27e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 4.28e-01 |
| GO:BP | GO:0035176 | social behavior | 5 | 34 | 4.28e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 23 | 4.28e-01 |
| GO:BP | GO:0003164 | His-Purkinje system development | 5 | 7 | 4.28e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 22 | 91 | 4.28e-01 |
| GO:BP | GO:1900248 | negative regulation of cytoplasmic translational elongation | 2 | 2 | 4.28e-01 |
| GO:BP | GO:0010561 | negative regulation of glycoprotein biosynthetic process | 1 | 10 | 4.28e-01 |
| GO:BP | GO:0003307 | regulation of Wnt signaling pathway involved in heart development | 3 | 7 | 4.28e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 27 | 71 | 4.28e-01 |
| GO:BP | GO:0033599 | regulation of mammary gland epithelial cell proliferation | 1 | 14 | 4.28e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 4.28e-01 |
| GO:BP | GO:1904394 | negative regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 1 | 4.28e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 2 | 8 | 4.28e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 2 | 40 | 4.28e-01 |
| GO:BP | GO:0034514 | mitochondrial unfolded protein response | 2 | 2 | 4.29e-01 |
| GO:BP | GO:1900449 | regulation of glutamate receptor signaling pathway | 2 | 5 | 4.29e-01 |
| GO:BP | GO:1904748 | regulation of apoptotic process involved in development | 3 | 9 | 4.29e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 4.29e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 4.29e-01 |
| GO:BP | GO:0140214 | positive regulation of long-chain fatty acid import into cell | 2 | 2 | 4.29e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 3 | 8 | 4.29e-01 |
| GO:BP | GO:0071321 | cellular response to cGMP | 1 | 7 | 4.29e-01 |
| GO:BP | GO:1902337 | regulation of apoptotic process involved in morphogenesis | 3 | 9 | 4.29e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0000710 | meiotic mismatch repair | 2 | 2 | 4.29e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 12 | 26 | 4.29e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 2 | 34 | 4.29e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 5 | 33 | 4.30e-01 |
| GO:BP | GO:0071041 | antisense RNA transcript catabolic process | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0071040 | nuclear polyadenylation-dependent antisense transcript catabolic process | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0042868 | antisense RNA metabolic process | 1 | 1 | 4.30e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 11 | 167 | 4.30e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 5 | 30 | 4.30e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 9 | 106 | 4.30e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 14 | 184 | 4.30e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 4 | 21 | 4.30e-01 |
| GO:BP | GO:0062176 | R-loop processing | 3 | 6 | 4.30e-01 |
| GO:BP | GO:0032069 | regulation of nuclease activity | 5 | 17 | 4.30e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 6 | 8 | 4.30e-01 |
| GO:BP | GO:0021761 | limbic system development | 4 | 81 | 4.30e-01 |
| GO:BP | GO:1905609 | positive regulation of smooth muscle cell-matrix adhesion | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 154 | 4.30e-01 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 2 | 13 | 4.31e-01 |
| GO:BP | GO:1900141 | regulation of oligodendrocyte apoptotic process | 2 | 2 | 4.31e-01 |
| GO:BP | GO:0018377 | protein myristoylation | 3 | 4 | 4.31e-01 |
| GO:BP | GO:0006499 | N-terminal protein myristoylation | 3 | 4 | 4.31e-01 |
| GO:BP | GO:0008584 | male gonad development | 5 | 99 | 4.32e-01 |
| GO:BP | GO:0075506 | entry of viral genome into host nucleus through nuclear pore complex via importin | 2 | 2 | 4.32e-01 |
| GO:BP | GO:2000322 | regulation of glucocorticoid receptor signaling pathway | 2 | 7 | 4.32e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 14 | 36 | 4.32e-01 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 4.32e-01 |
| GO:BP | GO:1903935 | response to sodium arsenite | 1 | 6 | 4.32e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 13 | 59 | 4.32e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 2 | 8 | 4.32e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 2 | 22 | 4.32e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 7 | 13 | 4.32e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 15 | 36 | 4.32e-01 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 10 | 17 | 4.32e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 2 | 22 | 4.32e-01 |
| GO:BP | GO:0007584 | response to nutrient | 3 | 109 | 4.32e-01 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 1 | 8 | 4.32e-01 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 1 | 8 | 4.32e-01 |
| GO:BP | GO:1902685 | positive regulation of receptor localization to synapse | 2 | 3 | 4.32e-01 |
| GO:BP | GO:0021915 | neural tube development | 1 | 146 | 4.32e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 2 | 17 | 4.32e-01 |
| GO:BP | GO:2001228 | regulation of response to gamma radiation | 1 | 1 | 4.33e-01 |
| GO:BP | GO:1905843 | regulation of cellular response to gamma radiation | 1 | 1 | 4.33e-01 |
| GO:BP | GO:1904862 | inhibitory synapse assembly | 7 | 13 | 4.33e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 6 | 21 | 4.33e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 3 | 47 | 4.33e-01 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 6 | 9 | 4.34e-01 |
| GO:BP | GO:0036369 | transcription factor catabolic process | 2 | 5 | 4.34e-01 |
| GO:BP | GO:1904220 | regulation of serine C-palmitoyltransferase activity | 1 | 3 | 4.34e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 18 | 1712 | 4.34e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 9 | 12 | 4.34e-01 |
| GO:BP | GO:0021873 | forebrain neuroblast division | 1 | 3 | 4.34e-01 |
| GO:BP | GO:0042942 | D-serine transport | 1 | 1 | 4.34e-01 |
| GO:BP | GO:0009786 | regulation of asymmetric cell division | 1 | 1 | 4.34e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 4 | 12 | 4.34e-01 |
| GO:BP | GO:0003330 | regulation of extracellular matrix constituent secretion | 2 | 7 | 4.34e-01 |
| GO:BP | GO:0097421 | liver regeneration | 5 | 23 | 4.34e-01 |
| GO:BP | GO:0050859 | negative regulation of B cell receptor signaling pathway | 1 | 3 | 4.34e-01 |
| GO:BP | GO:0150099 | neuron-glial cell signaling | 4 | 4 | 4.35e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 8 | 56 | 4.36e-01 |
| GO:BP | GO:1904876 | negative regulation of DNA ligase activity | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 1 | 13 | 4.36e-01 |
| GO:BP | GO:0051107 | negative regulation of DNA ligation | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 4 | 8 | 4.36e-01 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 4 | 5 | 4.36e-01 |
| GO:BP | GO:0038026 | reelin-mediated signaling pathway | 5 | 9 | 4.36e-01 |
| GO:BP | GO:0014028 | notochord formation | 2 | 3 | 4.36e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 4 | 20 | 4.36e-01 |
| GO:BP | GO:0048311 | mitochondrion distribution | 1 | 14 | 4.36e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 12 | 22 | 4.36e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 2 | 24 | 4.36e-01 |
| GO:BP | GO:0030035 | microspike assembly | 4 | 5 | 4.36e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 4.36e-01 |
| GO:BP | GO:0009826 | unidimensional cell growth | 2 | 3 | 4.36e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0051103 | DNA ligation involved in DNA repair | 4 | 5 | 4.36e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 5 | 100 | 4.36e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 35 | 166 | 4.36e-01 |
| GO:BP | GO:0002931 | response to ischemia | 4 | 46 | 4.36e-01 |
| GO:BP | GO:0007498 | mesoderm development | 1 | 89 | 4.36e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 2 | 11 | 4.36e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 29 | 47 | 4.36e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 2 | 45 | 4.36e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 3 | 53 | 4.36e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 4.36e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 151 | 4.36e-01 |
| GO:BP | GO:0031128 | developmental induction | 13 | 26 | 4.36e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 1 | 31 | 4.36e-01 |
| GO:BP | GO:0001656 | metanephros development | 2 | 64 | 4.36e-01 |
| GO:BP | GO:0009213 | pyrimidine deoxyribonucleoside triphosphate catabolic process | 1 | 2 | 4.36e-01 |
| GO:BP | GO:0051694 | pointed-end actin filament capping | 3 | 6 | 4.37e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 7 | 210 | 4.37e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 1 | 88 | 4.38e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 16 | 85 | 4.38e-01 |
| GO:BP | GO:0097379 | dorsal spinal cord interneuron posterior axon guidance | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0035849 | nephric duct elongation | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0035847 | uterine epithelium development | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 2 | 23 | 4.38e-01 |
| GO:BP | GO:0009414 | response to water deprivation | 1 | 4 | 4.38e-01 |
| GO:BP | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination | 1 | 1 | 4.38e-01 |
| GO:BP | GO:1902031 | regulation of NADP metabolic process | 1 | 8 | 4.38e-01 |
| GO:BP | GO:0042413 | carnitine catabolic process | 1 | 1 | 4.38e-01 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0022615 | protein to membrane docking | 1 | 5 | 4.39e-01 |
| GO:BP | GO:0097252 | oligodendrocyte apoptotic process | 3 | 4 | 4.39e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 2 | 5 | 4.39e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 2 | 5 | 4.39e-01 |
| GO:BP | GO:0048369 | lateral mesoderm morphogenesis | 3 | 3 | 4.39e-01 |
| GO:BP | GO:0072303 | positive regulation of glomerular metanephric mesangial cell proliferation | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0048370 | lateral mesoderm formation | 3 | 3 | 4.39e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0010891 | negative regulation of sequestering of triglyceride | 1 | 5 | 4.39e-01 |
| GO:BP | GO:0007033 | vacuole organization | 6 | 197 | 4.39e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0051964 | negative regulation of synapse assembly | 1 | 6 | 4.39e-01 |
| GO:BP | GO:0006045 | N-acetylglucosamine biosynthetic process | 2 | 2 | 4.39e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 3 | 57 | 4.39e-01 |
| GO:BP | GO:1905331 | negative regulation of morphogenesis of an epithelium | 1 | 6 | 4.39e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 4.39e-01 |
| GO:BP | GO:0033278 | cell proliferation in midbrain | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 5 | 9 | 4.39e-01 |
| GO:BP | GO:1901073 | glucosamine-containing compound biosynthetic process | 2 | 2 | 4.39e-01 |
| GO:BP | GO:0071283 | cellular response to iron(III) ion | 1 | 1 | 4.39e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 2 | 6 | 4.39e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 1 | 26 | 4.39e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 3 | 4 | 4.39e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 15 | 326 | 4.39e-01 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 3 | 4 | 4.40e-01 |
| GO:BP | GO:0072223 | metanephric glomerular mesangium development | 2 | 3 | 4.40e-01 |
| GO:BP | GO:0003057 | regulation of the force of heart contraction by chemical signal | 3 | 5 | 4.40e-01 |
| GO:BP | GO:0051455 | monopolar spindle attachment to meiosis I kinetochore | 1 | 1 | 4.40e-01 |
| GO:BP | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation | 1 | 1 | 4.40e-01 |
| GO:BP | GO:0060789 | hair follicle placode formation | 3 | 5 | 4.40e-01 |
| GO:BP | GO:0097742 | de novo centriole assembly | 3 | 4 | 4.40e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 4.40e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 10 | 16 | 4.40e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 20 | 1239 | 4.40e-01 |
| GO:BP | GO:0009178 | pyrimidine deoxyribonucleoside monophosphate catabolic process | 1 | 5 | 4.40e-01 |
| GO:BP | GO:0003373 | dynamin family protein polymerization involved in membrane fission | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0090149 | mitochondrial membrane fission | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0046035 | CMP metabolic process | 1 | 6 | 4.40e-01 |
| GO:BP | GO:0003374 | dynamin family protein polymerization involved in mitochondrial fission | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0046078 | dUMP metabolic process | 1 | 6 | 4.40e-01 |
| GO:BP | GO:1903944 | negative regulation of hepatocyte apoptotic process | 3 | 6 | 4.40e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 4 | 28 | 4.40e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 5 | 7 | 4.40e-01 |
| GO:BP | GO:0003010 | voluntary skeletal muscle contraction | 2 | 5 | 4.40e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 5 | 265 | 4.40e-01 |
| GO:BP | GO:0014721 | twitch skeletal muscle contraction | 2 | 5 | 4.40e-01 |
| GO:BP | GO:0003331 | positive regulation of extracellular matrix constituent secretion | 1 | 4 | 4.40e-01 |
| GO:BP | GO:0036093 | germ cell proliferation | 3 | 7 | 4.40e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 2 | 34 | 4.40e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 8 | 16 | 4.40e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 6 | 23 | 4.40e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 2 | 8 | 4.40e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 4.40e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 5 | 9 | 4.40e-01 |
| GO:BP | GO:0060383 | positive regulation of DNA strand elongation | 1 | 1 | 4.40e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 2 | 4 | 4.40e-01 |
| GO:BP | GO:0045925 | positive regulation of female receptivity | 1 | 1 | 4.40e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 2 | 17 | 4.40e-01 |
| GO:BP | GO:0048304 | positive regulation of isotype switching to IgG isotypes | 1 | 4 | 4.40e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 3 | 33 | 4.40e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 3 | 51 | 4.40e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 4.40e-01 |
| GO:BP | GO:0017055 | negative regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 13 | 358 | 4.41e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 5 | 21 | 4.41e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 1 | 128 | 4.41e-01 |
| GO:BP | GO:0034486 | vacuolar transmembrane transport | 2 | 3 | 4.41e-01 |
| GO:BP | GO:0080163 | regulation of protein serine/threonine phosphatase activity | 2 | 2 | 4.41e-01 |
| GO:BP | GO:0042758 | long-chain fatty acid catabolic process | 3 | 3 | 4.41e-01 |
| GO:BP | GO:0014806 | smooth muscle hyperplasia | 1 | 2 | 4.41e-01 |
| GO:BP | GO:0014738 | regulation of muscle hyperplasia | 1 | 1 | 4.41e-01 |
| GO:BP | GO:1902908 | regulation of melanosome transport | 2 | 2 | 4.42e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 2 | 20 | 4.43e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 1 | 37 | 4.43e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 4.43e-01 |
| GO:BP | GO:0036316 | SREBP-SCAP complex retention in endoplasmic reticulum | 2 | 2 | 4.43e-01 |
| GO:BP | GO:0033630 | positive regulation of cell adhesion mediated by integrin | 1 | 14 | 4.43e-01 |
| GO:BP | GO:0000493 | box H/ACA snoRNP assembly | 1 | 2 | 4.43e-01 |
| GO:BP | GO:0045955 | negative regulation of calcium ion-dependent exocytosis | 5 | 6 | 4.43e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 1 | 25 | 4.43e-01 |
| GO:BP | GO:0099163 | synaptic signaling by nitric oxide | 3 | 3 | 4.43e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 1 | 10 | 4.43e-01 |
| GO:BP | GO:0045105 | intermediate filament polymerization or depolymerization | 2 | 2 | 4.43e-01 |
| GO:BP | GO:0048855 | adenohypophysis morphogenesis | 1 | 1 | 4.43e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 24 | 1280 | 4.43e-01 |
| GO:BP | GO:0046886 | positive regulation of hormone biosynthetic process | 3 | 9 | 4.43e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 2 | 8 | 4.43e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 2 | 7 | 4.43e-01 |
| GO:BP | GO:1905786 | positive regulation of anaphase-promoting complex-dependent catabolic process | 1 | 2 | 4.43e-01 |
| GO:BP | GO:0042311 | vasodilation | 6 | 39 | 4.43e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 4 | 7 | 4.43e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 4.43e-01 |
| GO:BP | GO:0015939 | pantothenate metabolic process | 1 | 2 | 4.44e-01 |
| GO:BP | GO:0036149 | phosphatidylinositol acyl-chain remodeling | 2 | 3 | 4.44e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 52 | 156 | 4.44e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 1 | 19 | 4.44e-01 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 1 | 17 | 4.44e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 1 | 18 | 4.44e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 1 | 17 | 4.44e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 52 | 313 | 4.44e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 4 | 20 | 4.44e-01 |
| GO:BP | GO:0060232 | delamination | 1 | 5 | 4.44e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 16 | 37 | 4.44e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 3 | 14 | 4.44e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 4 | 51 | 4.45e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 17 | 81 | 4.45e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 4 | 7 | 4.45e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 1 | 129 | 4.45e-01 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 1 | 16 | 4.45e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 7 | 47 | 4.45e-01 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 11 | 16 | 4.45e-01 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 11 | 16 | 4.45e-01 |
| GO:BP | GO:0036484 | trunk neural crest cell migration | 3 | 4 | 4.45e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 4.45e-01 |
| GO:BP | GO:0035290 | trunk segmentation | 3 | 4 | 4.45e-01 |
| GO:BP | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | 1 | 6 | 4.45e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 3 | 9 | 4.45e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 10 | 46 | 4.45e-01 |
| GO:BP | GO:1903764 | regulation of potassium ion export across plasma membrane | 3 | 5 | 4.45e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 7 | 26 | 4.45e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 3 | 13 | 4.45e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 2 | 19 | 4.45e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 1 | 198 | 4.45e-01 |
| GO:BP | GO:0046548 | retinal rod cell development | 1 | 6 | 4.45e-01 |
| GO:BP | GO:0098924 | retrograde trans-synaptic signaling by nitric oxide | 1 | 1 | 4.45e-01 |
| GO:BP | GO:0098923 | retrograde trans-synaptic signaling by soluble gas | 1 | 1 | 4.45e-01 |
| GO:BP | GO:0006600 | creatine metabolic process | 1 | 3 | 4.45e-01 |
| GO:BP | GO:0061502 | early endosome to recycling endosome transport | 1 | 3 | 4.46e-01 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 4.46e-01 |
| GO:BP | GO:0006656 | phosphatidylcholine biosynthetic process | 11 | 25 | 4.46e-01 |
| GO:BP | GO:0051792 | medium-chain fatty acid biosynthetic process | 2 | 4 | 4.46e-01 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 2 | 29 | 4.46e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 5 | 34 | 4.46e-01 |
| GO:BP | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity | 1 | 6 | 4.46e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 7 | 30 | 4.46e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 28 | 138 | 4.46e-01 |
| GO:BP | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway | 2 | 4 | 4.46e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 2 | 39 | 4.46e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 1 | 127 | 4.47e-01 |
| GO:BP | GO:0032691 | negative regulation of interleukin-1 beta production | 2 | 13 | 4.47e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 2 | 14 | 4.47e-01 |
| GO:BP | GO:0040038 | polar body extrusion after meiotic divisions | 4 | 5 | 4.48e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 8 | 14 | 4.48e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 59 | 294 | 4.48e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 12 | 4.48e-01 |
| GO:BP | GO:0008215 | spermine metabolic process | 3 | 5 | 4.48e-01 |
| GO:BP | GO:0060004 | reflex | 3 | 9 | 4.48e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 6 | 70 | 4.48e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0014841 | skeletal muscle satellite cell proliferation | 3 | 10 | 4.48e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 4.48e-01 |
| GO:BP | GO:1905608 | positive regulation of presynapse assembly | 1 | 2 | 4.48e-01 |
| GO:BP | GO:0060857 | establishment of glial blood-brain barrier | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 4.48e-01 |
| GO:BP | GO:0042779 | tRNA 3’-trailer cleavage | 1 | 3 | 4.48e-01 |
| GO:BP | GO:0099543 | trans-synaptic signaling by soluble gas | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0099548 | trans-synaptic signaling by nitric oxide | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 132 | 4.48e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 16 | 1719 | 4.48e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 4.48e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 4.48e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 4.48e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 1 | 118 | 4.48e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 4.48e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 3 | 3090 | 4.48e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 3 | 30 | 4.48e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 4.48e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 9 | 13 | 4.48e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 4.48e-01 |
| GO:BP | GO:1901740 | negative regulation of myoblast fusion | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 3 | 49 | 4.48e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 7 | 24 | 4.49e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 55 | 241 | 4.49e-01 |
| GO:BP | GO:1904777 | negative regulation of protein localization to cell cortex | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0006429 | leucyl-tRNA aminoacylation | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0006425 | glutaminyl-tRNA aminoacylation | 1 | 2 | 4.49e-01 |
| GO:BP | GO:2000393 | negative regulation of lamellipodium morphogenesis | 2 | 2 | 4.49e-01 |
| GO:BP | GO:0098725 | symmetric cell division | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 4.49e-01 |
| GO:BP | GO:0072733 | response to staurosporine | 1 | 3 | 4.49e-01 |
| GO:BP | GO:1903062 | regulation of reverse cholesterol transport | 1 | 1 | 4.49e-01 |
| GO:BP | GO:1904234 | positive regulation of aconitate hydratase activity | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 5 | 10 | 4.49e-01 |
| GO:BP | GO:0090269 | fibroblast growth factor production | 2 | 4 | 4.49e-01 |
| GO:BP | GO:0019046 | release from viral latency | 1 | 2 | 4.49e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 5 | 10 | 4.49e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 4.49e-01 |
| GO:BP | GO:0072734 | cellular response to staurosporine | 1 | 3 | 4.49e-01 |
| GO:BP | GO:1903064 | positive regulation of reverse cholesterol transport | 1 | 1 | 4.49e-01 |
| GO:BP | GO:1904232 | regulation of aconitate hydratase activity | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0070143 | mitochondrial alanyl-tRNA aminoacylation | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0090270 | regulation of fibroblast growth factor production | 2 | 4 | 4.49e-01 |
| GO:BP | GO:1905667 | negative regulation of protein localization to endosome | 1 | 1 | 4.49e-01 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 4 | 8 | 4.49e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 2 | 7 | 4.49e-01 |
| GO:BP | GO:0070537 | histone H2A K63-linked deubiquitination | 2 | 8 | 4.49e-01 |
| GO:BP | GO:0000023 | maltose metabolic process | 1 | 2 | 4.49e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 9 | 23 | 4.49e-01 |
| GO:BP | GO:0019045 | latent virus replication | 1 | 2 | 4.49e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 4.49e-01 |
| GO:BP | GO:0006561 | proline biosynthetic process | 3 | 5 | 4.49e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 8 | 24 | 4.49e-01 |
| GO:BP | GO:0055129 | L-proline biosynthetic process | 3 | 5 | 4.49e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 24 | 50 | 4.49e-01 |
| GO:BP | GO:0006882 | intracellular zinc ion homeostasis | 9 | 28 | 4.49e-01 |
| GO:BP | GO:0006883 | intracellular sodium ion homeostasis | 4 | 13 | 4.49e-01 |
| GO:BP | GO:0006183 | GTP biosynthetic process | 6 | 9 | 4.49e-01 |
| GO:BP | GO:0062111 | zinc ion import into organelle | 1 | 7 | 4.49e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 2 | 33 | 4.49e-01 |
| GO:BP | GO:0034760 | negative regulation of iron ion transmembrane transport | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0034757 | negative regulation of iron ion transport | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0006021 | inositol biosynthetic process | 1 | 3 | 4.49e-01 |
| GO:BP | GO:0010737 | protein kinase A signaling | 1 | 23 | 4.49e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 17 | 34 | 4.49e-01 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 2 | 20 | 4.50e-01 |
| GO:BP | GO:1902725 | negative regulation of satellite cell differentiation | 1 | 2 | 4.50e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 2 | 20 | 4.51e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 4.51e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 1 | 45 | 4.51e-01 |
| GO:BP | GO:0070202 | regulation of establishment of protein localization to chromosome | 4 | 12 | 4.51e-01 |
| GO:BP | GO:0002051 | osteoblast fate commitment | 2 | 2 | 4.51e-01 |
| GO:BP | GO:0051029 | rRNA transport | 1 | 5 | 4.51e-01 |
| GO:BP | GO:0036445 | neuronal stem cell division | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0140066 | peptidyl-lysine crotonylation | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 1 | 14 | 4.51e-01 |
| GO:BP | GO:0042983 | amyloid precursor protein biosynthetic process | 1 | 14 | 4.51e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 11 | 357 | 4.51e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 4.51e-01 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 1 | 2 | 4.51e-01 |
| GO:BP | GO:1902977 | mitotic DNA replication preinitiation complex assembly | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0060754 | positive regulation of mast cell chemotaxis | 1 | 5 | 4.51e-01 |
| GO:BP | GO:0090669 | telomerase RNA stabilization | 2 | 4 | 4.51e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 53 | 96 | 4.51e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 4.51e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0014038 | regulation of Schwann cell differentiation | 1 | 3 | 4.51e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 6 | 14 | 4.51e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 1 | 4 | 4.51e-01 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 3 | 11 | 4.51e-01 |
| GO:BP | GO:0007495 | visceral mesoderm-endoderm interaction involved in midgut development | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 4.51e-01 |
| GO:BP | GO:1902888 | protein localization to astral microtubule | 1 | 1 | 4.51e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 22 | 108 | 4.51e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 17 | 120 | 4.51e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 14 | 187 | 4.51e-01 |
| GO:BP | GO:0001765 | membrane raft assembly | 1 | 9 | 4.51e-01 |
| GO:BP | GO:0033306 | phytol metabolic process | 2 | 2 | 4.52e-01 |
| GO:BP | GO:1903173 | fatty alcohol metabolic process | 2 | 2 | 4.52e-01 |
| GO:BP | GO:1904627 | response to phorbol 13-acetate 12-myristate | 1 | 4 | 4.52e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 4.52e-01 |
| GO:BP | GO:0061044 | negative regulation of vascular wound healing | 1 | 4 | 4.52e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 1 | 19 | 4.52e-01 |
| GO:BP | GO:1904628 | cellular response to phorbol 13-acetate 12-myristate | 1 | 4 | 4.52e-01 |
| GO:BP | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 1 | 6 | 4.52e-01 |
| GO:BP | GO:0110009 | formin-nucleated actin cable organization | 1 | 3 | 4.52e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 8 | 140 | 4.52e-01 |
| GO:BP | GO:0072126 | positive regulation of glomerular mesangial cell proliferation | 5 | 7 | 4.52e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 4.52e-01 |
| GO:BP | GO:0070649 | formin-nucleated actin cable assembly | 1 | 3 | 4.52e-01 |
| GO:BP | GO:0042167 | heme catabolic process | 1 | 6 | 4.52e-01 |
| GO:BP | GO:0046149 | pigment catabolic process | 1 | 6 | 4.52e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 3 | 10 | 4.53e-01 |
| GO:BP | GO:0006451 | translational readthrough | 2 | 10 | 4.53e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 2 | 10 | 4.53e-01 |
| GO:BP | GO:0040016 | embryonic cleavage | 2 | 7 | 4.53e-01 |
| GO:BP | GO:0095500 | acetylcholine receptor signaling pathway | 1 | 18 | 4.53e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 6 | 64 | 4.53e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 3 | 26 | 4.53e-01 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.53e-01 |
| GO:BP | GO:0009133 | nucleoside diphosphate biosynthetic process | 1 | 11 | 4.53e-01 |
| GO:BP | GO:1905532 | regulation of leucine import across plasma membrane | 2 | 3 | 4.53e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 8 | 14 | 4.53e-01 |
| GO:BP | GO:1903978 | regulation of microglial cell activation | 1 | 9 | 4.53e-01 |
| GO:BP | GO:0090520 | sphingolipid mediated signaling pathway | 1 | 11 | 4.53e-01 |
| GO:BP | GO:0007000 | nucleolus organization | 1 | 17 | 4.53e-01 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.53e-01 |
| GO:BP | GO:0060348 | bone development | 1 | 170 | 4.53e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 2 | 17 | 4.53e-01 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 1 | 2 | 4.53e-01 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 1 | 2 | 4.53e-01 |
| GO:BP | GO:0019551 | glutamate catabolic process to 2-oxoglutarate | 2 | 2 | 4.53e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 8 | 90 | 4.53e-01 |
| GO:BP | GO:0097151 | positive regulation of inhibitory postsynaptic potential | 3 | 4 | 4.53e-01 |
| GO:BP | GO:0019550 | glutamate catabolic process to aspartate | 2 | 2 | 4.53e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 3 | 33 | 4.55e-01 |
| GO:BP | GO:0021592 | fourth ventricle development | 1 | 1 | 4.56e-01 |
| GO:BP | GO:0021993 | initiation of neural tube closure | 1 | 1 | 4.56e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 4.56e-01 |
| GO:BP | GO:0099041 | vesicle tethering to Golgi | 3 | 5 | 4.56e-01 |
| GO:BP | GO:1902497 | iron-sulfur cluster transmembrane transport | 1 | 1 | 4.56e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 1 | 36 | 4.56e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 3 | 18 | 4.56e-01 |
| GO:BP | GO:0070673 | response to interleukin-18 | 4 | 5 | 4.56e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0140466 | iron-sulfur cluster export from the mitochondrion | 1 | 1 | 4.56e-01 |
| GO:BP | GO:1903331 | positive regulation of iron-sulfur cluster assembly | 1 | 1 | 4.56e-01 |
| GO:BP | GO:1903329 | regulation of iron-sulfur cluster assembly | 1 | 1 | 4.56e-01 |
| GO:BP | GO:0036072 | direct ossification | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0001957 | intramembranous ossification | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0001889 | liver development | 4 | 110 | 4.56e-01 |
| GO:BP | GO:2000301 | negative regulation of synaptic vesicle exocytosis | 2 | 4 | 4.57e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 1 | 3 | 4.57e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 18 | 237 | 4.57e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 4.58e-01 |
| GO:BP | GO:0051958 | methotrexate transport | 1 | 2 | 4.58e-01 |
| GO:BP | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 4.58e-01 |
| GO:BP | GO:2000691 | negative regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0032416 | negative regulation of sodium:proton antiporter activity | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0035196 | miRNA processing | 21 | 41 | 4.58e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 3 | 6 | 4.58e-01 |
| GO:BP | GO:0009222 | pyrimidine ribonucleotide catabolic process | 1 | 7 | 4.58e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 2 | 8 | 4.58e-01 |
| GO:BP | GO:0044722 | renal phosphate excretion | 1 | 2 | 4.58e-01 |
| GO:BP | GO:1903402 | regulation of renal phosphate excretion | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0061865 | polarized secretion of basement membrane proteins in epithelium | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0061864 | basement membrane constituent secretion | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0009131 | pyrimidine nucleoside monophosphate catabolic process | 1 | 6 | 4.58e-01 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 4.58e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 33 | 76 | 4.58e-01 |
| GO:BP | GO:1905070 | anterior visceral endoderm cell migration | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 4 | 10 | 4.58e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 6 | 19 | 4.58e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 2 | 5 | 4.58e-01 |
| GO:BP | GO:0060456 | positive regulation of digestive system process | 2 | 8 | 4.58e-01 |
| GO:BP | GO:0007271 | synaptic transmission, cholinergic | 1 | 15 | 4.58e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 4.58e-01 |
| GO:BP | GO:1901394 | positive regulation of transforming growth factor beta1 activation | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0015860 | purine nucleoside transmembrane transport | 1 | 3 | 4.58e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 2 | 15 | 4.58e-01 |
| GO:BP | GO:0046176 | aldonic acid catabolic process | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 5 | 10 | 4.58e-01 |
| GO:BP | GO:0098903 | regulation of membrane repolarization during action potential | 1 | 6 | 4.58e-01 |
| GO:BP | GO:0046177 | D-gluconate catabolic process | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 2 | 41 | 4.58e-01 |
| GO:BP | GO:1901392 | regulation of transforming growth factor beta1 activation | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0050960 | detection of temperature stimulus involved in thermoception | 1 | 3 | 4.58e-01 |
| GO:BP | GO:0019520 | aldonic acid metabolic process | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0019521 | D-gluconate metabolic process | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0021855 | hypothalamus cell migration | 5 | 6 | 4.58e-01 |
| GO:BP | GO:0071502 | cellular response to temperature stimulus | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0019322 | pentose biosynthetic process | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0090472 | dibasic protein processing | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0003358 | noradrenergic neuron development | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 2 | 28 | 4.58e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 5 | 10 | 4.58e-01 |
| GO:BP | GO:0055089 | fatty acid homeostasis | 1 | 12 | 4.58e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 26 | 496 | 4.58e-01 |
| GO:BP | GO:0032096 | negative regulation of response to food | 1 | 7 | 4.58e-01 |
| GO:BP | GO:0032099 | negative regulation of appetite | 1 | 7 | 4.58e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 6 | 22 | 4.58e-01 |
| GO:BP | GO:0032105 | negative regulation of response to extracellular stimulus | 1 | 7 | 4.58e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 3 | 13 | 4.58e-01 |
| GO:BP | GO:0033210 | leptin-mediated signaling pathway | 1 | 10 | 4.58e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 2 | 4 | 4.58e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 11 | 19 | 4.58e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 39 | 203 | 4.58e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 1 | 102 | 4.58e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 6 | 15 | 4.58e-01 |
| GO:BP | GO:0032108 | negative regulation of response to nutrient levels | 1 | 7 | 4.58e-01 |
| GO:BP | GO:0022013 | pallium cell proliferation in forebrain | 1 | 1 | 4.59e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 2 | 15 | 4.59e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 7 | 85 | 4.59e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 4.59e-01 |
| GO:BP | GO:0106001 | intestinal hexose absorption | 2 | 3 | 4.60e-01 |
| GO:BP | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions | 1 | 1 | 4.60e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 1 | 38 | 4.60e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 2 | 228 | 4.60e-01 |
| GO:BP | GO:0035426 | extracellular matrix-cell signaling | 3 | 4 | 4.60e-01 |
| GO:BP | GO:0000712 | resolution of meiotic recombination intermediates | 11 | 13 | 4.60e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 4 | 532 | 4.60e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 4.60e-01 |
| GO:BP | GO:0006527 | arginine catabolic process | 2 | 7 | 4.60e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 1 | 59 | 4.61e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 2 | 663 | 4.61e-01 |
| GO:BP | GO:0009408 | response to heat | 4 | 84 | 4.61e-01 |
| GO:BP | GO:1902805 | positive regulation of synaptic vesicle transport | 2 | 2 | 4.61e-01 |
| GO:BP | GO:0097527 | necroptotic signaling pathway | 1 | 3 | 4.61e-01 |
| GO:BP | GO:1903744 | positive regulation of anterograde synaptic vesicle transport | 2 | 2 | 4.61e-01 |
| GO:BP | GO:1904527 | negative regulation of microtubule binding | 2 | 2 | 4.61e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 3 | 29 | 4.62e-01 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 4 | 10 | 4.62e-01 |
| GO:BP | GO:0061772 | xenobiotic transport across blood-nerve barrier | 1 | 1 | 4.62e-01 |
| GO:BP | GO:0006435 | threonyl-tRNA aminoacylation | 1 | 3 | 4.62e-01 |
| GO:BP | GO:1903545 | cellular response to butyrate | 1 | 1 | 4.62e-01 |
| GO:BP | GO:0070102 | interleukin-6-mediated signaling pathway | 5 | 14 | 4.62e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 1 | 7 | 4.62e-01 |
| GO:BP | GO:1903019 | negative regulation of glycoprotein metabolic process | 1 | 12 | 4.62e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 2 | 41 | 4.62e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 1 | 7 | 4.62e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 3 | 4 | 4.62e-01 |
| GO:BP | GO:0001815 | positive regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 4.62e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 5 | 14 | 4.62e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 2 | 28 | 4.62e-01 |
| GO:BP | GO:2001015 | negative regulation of skeletal muscle cell differentiation | 2 | 3 | 4.62e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 3 | 23 | 4.62e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 2 | 17 | 4.62e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 7 | 98 | 4.62e-01 |
| GO:BP | GO:0007097 | nuclear migration | 5 | 23 | 4.62e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 4.62e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 7 | 29 | 4.62e-01 |
| GO:BP | GO:0006560 | proline metabolic process | 4 | 7 | 4.62e-01 |
| GO:BP | GO:0098942 | retrograde trans-synaptic signaling by trans-synaptic protein complex | 2 | 2 | 4.63e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 12 | 101 | 4.63e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 4.63e-01 |
| GO:BP | GO:2000656 | regulation of apolipoprotein binding | 1 | 1 | 4.63e-01 |
| GO:BP | GO:2000657 | negative regulation of apolipoprotein binding | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0048219 | inter-Golgi cisterna vesicle-mediated transport | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0039654 | fusion of virus membrane with host endosome membrane | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 4 | 11 | 4.63e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 4.63e-01 |
| GO:BP | GO:1905145 | cellular response to acetylcholine | 1 | 20 | 4.63e-01 |
| GO:BP | GO:0098693 | regulation of synaptic vesicle cycle | 1 | 13 | 4.63e-01 |
| GO:BP | GO:0030643 | intracellular phosphate ion homeostasis | 3 | 5 | 4.63e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0042357 | thiamine diphosphate metabolic process | 2 | 2 | 4.63e-01 |
| GO:BP | GO:0071140 | resolution of mitotic recombination intermediates | 2 | 2 | 4.63e-01 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 5 | 16 | 4.63e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 6 | 184 | 4.63e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 4 | 35 | 4.63e-01 |
| GO:BP | GO:0060026 | convergent extension | 2 | 14 | 4.63e-01 |
| GO:BP | GO:2001045 | negative regulation of integrin-mediated signaling pathway | 1 | 3 | 4.63e-01 |
| GO:BP | GO:0032601 | connective tissue growth factor production | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway | 5 | 6 | 4.63e-01 |
| GO:BP | GO:0032643 | regulation of connective tissue growth factor production | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 5 | 90 | 4.63e-01 |
| GO:BP | GO:0098902 | regulation of membrane depolarization during action potential | 3 | 5 | 4.64e-01 |
| GO:BP | GO:0042746 | circadian sleep/wake cycle, wakefulness | 1 | 1 | 4.64e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 4.64e-01 |
| GO:BP | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 4.64e-01 |
| GO:BP | GO:0033273 | response to vitamin | 2 | 61 | 4.64e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 4 | 57 | 4.64e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 4.64e-01 |
| GO:BP | GO:1900006 | positive regulation of dendrite development | 2 | 14 | 4.64e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 29 | 706 | 4.64e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 16 | 30 | 4.64e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 4 | 112 | 4.64e-01 |
| GO:BP | GO:0030202 | heparin metabolic process | 1 | 14 | 4.64e-01 |
| GO:BP | GO:0061739 | protein lipidation involved in autophagosome assembly | 2 | 3 | 4.65e-01 |
| GO:BP | GO:0006265 | DNA topological change | 1 | 10 | 4.65e-01 |
| GO:BP | GO:0035780 | CD80 biosynthetic process | 1 | 1 | 4.65e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 4.65e-01 |
| GO:BP | GO:1902247 | geranylgeranyl diphosphate catabolic process | 1 | 1 | 4.65e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 4 | 4955 | 4.65e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 13 | 46 | 4.65e-01 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 3 | 5 | 4.65e-01 |
| GO:BP | GO:0051666 | actin cortical patch localization | 1 | 2 | 4.65e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 2 | 42 | 4.65e-01 |
| GO:BP | GO:0045339 | farnesyl diphosphate catabolic process | 1 | 1 | 4.65e-01 |
| GO:BP | GO:0006500 | N-terminal protein palmitoylation | 2 | 3 | 4.66e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 5 | 52 | 4.66e-01 |
| GO:BP | GO:0048210 | Golgi vesicle fusion to target membrane | 3 | 4 | 4.66e-01 |
| GO:BP | GO:1902915 | negative regulation of protein polyubiquitination | 1 | 9 | 4.67e-01 |
| GO:BP | GO:1903972 | regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 3 | 4.67e-01 |
| GO:BP | GO:0033567 | DNA replication, Okazaki fragment processing | 2 | 2 | 4.67e-01 |
| GO:BP | GO:1903969 | regulation of response to macrophage colony-stimulating factor | 1 | 3 | 4.67e-01 |
| GO:BP | GO:1905719 | protein localization to perinuclear region of cytoplasm | 2 | 8 | 4.67e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 1 | 19 | 4.67e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 1 | 62 | 4.67e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 6 | 16 | 4.67e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 1 | 19 | 4.67e-01 |
| GO:BP | GO:0042482 | positive regulation of odontogenesis | 1 | 5 | 4.67e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 16 | 139 | 4.67e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 1 | 4 | 4.67e-01 |
| GO:BP | GO:0019051 | induction by virus of host apoptotic process | 2 | 2 | 4.67e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 1 | 20 | 4.67e-01 |
| GO:BP | GO:0019050 | suppression by virus of host apoptotic process | 2 | 2 | 4.67e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 153 | 4.67e-01 |
| GO:BP | GO:0033668 | suppression by symbiont of host apoptotic process | 2 | 2 | 4.67e-01 |
| GO:BP | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | 1 | 8 | 4.67e-01 |
| GO:BP | GO:0001731 | formation of translation preinitiation complex | 3 | 11 | 4.67e-01 |
| GO:BP | GO:0032920 | putrescine acetylation | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0032919 | spermine acetylation | 1 | 1 | 4.67e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 5 | 39 | 4.67e-01 |
| GO:BP | GO:0070305 | response to cGMP | 1 | 7 | 4.67e-01 |
| GO:BP | GO:0052042 | induction by symbiont of host programmed cell death | 2 | 2 | 4.67e-01 |
| GO:BP | GO:0046204 | nor-spermidine metabolic process | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0052151 | positive regulation by symbiont of host apoptotic process | 2 | 2 | 4.67e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 15 | 53 | 4.67e-01 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 13 | 20 | 4.67e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 6 | 27 | 4.67e-01 |
| GO:BP | GO:0120041 | positive regulation of macrophage proliferation | 1 | 4 | 4.67e-01 |
| GO:BP | GO:0052041 | suppression by symbiont of host programmed cell death | 2 | 2 | 4.67e-01 |
| GO:BP | GO:0003271 | smoothened signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 4.67e-01 |
| GO:BP | GO:1902257 | negative regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 2 | 4.67e-01 |
| GO:BP | GO:0001907 | killing by symbiont of host cells | 2 | 2 | 4.67e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 4.67e-01 |
| GO:BP | GO:0007420 | brain development | 19 | 591 | 4.67e-01 |
| GO:BP | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development | 2 | 2 | 4.67e-01 |
| GO:BP | GO:1990792 | cellular response to glial cell derived neurotrophic factor | 1 | 1 | 4.67e-01 |
| GO:BP | GO:1990790 | response to glial cell derived neurotrophic factor | 1 | 1 | 4.67e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 4 | 22 | 4.67e-01 |
| GO:BP | GO:0051589 | negative regulation of neurotransmitter transport | 1 | 8 | 4.67e-01 |
| GO:BP | GO:0014719 | skeletal muscle satellite cell activation | 2 | 5 | 4.67e-01 |
| GO:BP | GO:0034350 | regulation of glial cell apoptotic process | 6 | 10 | 4.67e-01 |
| GO:BP | GO:0061072 | iris morphogenesis | 1 | 5 | 4.68e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 4.68e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 1 | 216 | 4.68e-01 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 1 | 11 | 4.68e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 4.68e-01 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 1 | 11 | 4.68e-01 |
| GO:BP | GO:0044857 | plasma membrane raft organization | 1 | 9 | 4.68e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 2 | 16 | 4.68e-01 |
| GO:BP | GO:0071313 | cellular response to caffeine | 5 | 10 | 4.68e-01 |
| GO:BP | GO:0006788 | heme oxidation | 2 | 2 | 4.68e-01 |
| GO:BP | GO:0060322 | head development | 20 | 633 | 4.69e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 4.69e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 2 | 14 | 4.69e-01 |
| GO:BP | GO:0060745 | mammary gland branching involved in pregnancy | 2 | 5 | 4.69e-01 |
| GO:BP | GO:1990683 | DNA double-strand break attachment to nuclear envelope | 2 | 2 | 4.69e-01 |
| GO:BP | GO:0080170 | hydrogen peroxide transmembrane transport | 1 | 1 | 4.69e-01 |
| GO:BP | GO:0007494 | midgut development | 5 | 7 | 4.69e-01 |
| GO:BP | GO:2001014 | regulation of skeletal muscle cell differentiation | 5 | 16 | 4.69e-01 |
| GO:BP | GO:0051036 | regulation of endosome size | 6 | 13 | 4.69e-01 |
| GO:BP | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation | 1 | 6 | 4.69e-01 |
| GO:BP | GO:0098532 | histone H3-K27 trimethylation | 1 | 4 | 4.69e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 4.69e-01 |
| GO:BP | GO:1903444 | negative regulation of brown fat cell differentiation | 3 | 3 | 4.69e-01 |
| GO:BP | GO:1904877 | positive regulation of DNA ligase activity | 1 | 1 | 4.69e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 3 | 843 | 4.69e-01 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 25 | 4.69e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 4.70e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 4.70e-01 |
| GO:BP | GO:0010759 | positive regulation of macrophage chemotaxis | 2 | 9 | 4.70e-01 |
| GO:BP | GO:0061448 | connective tissue development | 9 | 208 | 4.70e-01 |
| GO:BP | GO:0003170 | heart valve development | 3 | 65 | 4.70e-01 |
| GO:BP | GO:0048792 | spontaneous exocytosis of neurotransmitter | 1 | 1 | 4.70e-01 |
| GO:BP | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 6 | 4.70e-01 |
| GO:BP | GO:1901463 | regulation of tetrapyrrole biosynthetic process | 4 | 7 | 4.70e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 7 | 54 | 4.70e-01 |
| GO:BP | GO:1903703 | enterocyte differentiation | 1 | 1 | 4.70e-01 |
| GO:BP | GO:0070453 | regulation of heme biosynthetic process | 4 | 7 | 4.70e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 2 | 27 | 4.70e-01 |
| GO:BP | GO:1905341 | negative regulation of protein localization to kinetochore | 1 | 1 | 4.70e-01 |
| GO:BP | GO:0097473 | retinal rod cell apoptotic process | 2 | 2 | 4.71e-01 |
| GO:BP | GO:0071415 | cellular response to purine-containing compound | 6 | 11 | 4.71e-01 |
| GO:BP | GO:0045578 | negative regulation of B cell differentiation | 3 | 6 | 4.71e-01 |
| GO:BP | GO:0044782 | cilium organization | 7 | 313 | 4.71e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 1 | 28 | 4.71e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 2 | 6 | 4.71e-01 |
| GO:BP | GO:1904780 | negative regulation of protein localization to centrosome | 2 | 2 | 4.71e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 6 | 22 | 4.71e-01 |
| GO:BP | GO:0001508 | action potential | 7 | 105 | 4.72e-01 |
| GO:BP | GO:0006417 | regulation of translation | 5 | 370 | 4.72e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 28 | 140 | 4.72e-01 |
| GO:BP | GO:0060243 | negative regulation of cell growth involved in contact inhibition | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 4.73e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 18 | 340 | 4.73e-01 |
| GO:BP | GO:0060068 | vagina development | 1 | 8 | 4.73e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 4.73e-01 |
| GO:BP | GO:1901006 | ubiquinone-6 biosynthetic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:1901004 | ubiquinone-6 metabolic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0034287 | detection of monosaccharide stimulus | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0051594 | detection of glucose | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0009732 | detection of hexose stimulus | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0009730 | detection of carbohydrate stimulus | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 1 | 12 | 4.74e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 28 | 547 | 4.74e-01 |
| GO:BP | GO:0046041 | ITP metabolic process | 2 | 2 | 4.74e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 3 | 26 | 4.75e-01 |
| GO:BP | GO:0014832 | urinary bladder smooth muscle contraction | 2 | 4 | 4.75e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 4.75e-01 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 7 | 18 | 4.75e-01 |
| GO:BP | GO:0033962 | P-body assembly | 2 | 20 | 4.76e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 11 | 29 | 4.76e-01 |
| GO:BP | GO:0003128 | heart field specification | 5 | 16 | 4.76e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 2 | 15 | 4.76e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 6 | 10 | 4.76e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 5 | 21 | 4.76e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 5 | 50 | 4.76e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 1 | 74 | 4.76e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 1 | 63 | 4.76e-01 |
| GO:BP | GO:1904231 | positive regulation of succinate dehydrogenase activity | 2 | 2 | 4.76e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 29 | 616 | 4.76e-01 |
| GO:BP | GO:1904229 | regulation of succinate dehydrogenase activity | 2 | 2 | 4.76e-01 |
| GO:BP | GO:0055111 | ingression involved in gastrulation with mouth forming second | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0098939 | dendritic transport of mitochondrion | 1 | 2 | 4.77e-01 |
| GO:BP | GO:1903352 | L-ornithine transmembrane transport | 1 | 4 | 4.77e-01 |
| GO:BP | GO:0043048 | dolichyl monophosphate biosynthetic process | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 2 | 906 | 4.77e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 2 | 117 | 4.78e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 12 | 121 | 4.78e-01 |
| GO:BP | GO:0099187 | presynaptic cytoskeleton organization | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0099140 | presynaptic actin cytoskeleton organization | 1 | 1 | 4.78e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 4 | 6 | 4.78e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 26 | 64 | 4.78e-01 |
| GO:BP | GO:0030393 | fructoselysine metabolic process | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0015824 | proline transport | 4 | 11 | 4.78e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 4 | 18 | 4.78e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 5 | 64 | 4.78e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 3 | 12 | 4.78e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 2 | 14 | 4.78e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 4.78e-01 |
| GO:BP | GO:1903093 | regulation of protein K48-linked deubiquitination | 1 | 3 | 4.78e-01 |
| GO:BP | GO:0061743 | motor learning | 1 | 5 | 4.78e-01 |
| GO:BP | GO:0035864 | response to potassium ion | 3 | 12 | 4.78e-01 |
| GO:BP | GO:0034498 | early endosome to Golgi transport | 2 | 9 | 4.78e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 1 | 52 | 4.78e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 2 | 24 | 4.78e-01 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 1 | 8 | 4.78e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 7 | 10 | 4.78e-01 |
| GO:BP | GO:0045108 | regulation of intermediate filament polymerization or depolymerization | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration | 2 | 2 | 4.78e-01 |
| GO:BP | GO:0045106 | intermediate filament depolymerization | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0008272 | sulfate transport | 3 | 12 | 4.78e-01 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 2 | 14 | 4.78e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 4.78e-01 |
| GO:BP | GO:1905144 | response to acetylcholine | 1 | 22 | 4.78e-01 |
| GO:BP | GO:0048245 | eosinophil chemotaxis | 1 | 5 | 4.78e-01 |
| GO:BP | GO:0030842 | regulation of intermediate filament depolymerization | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0015827 | tryptophan transport | 2 | 6 | 4.78e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 12 | 73 | 4.78e-01 |
| GO:BP | GO:0035803 | egg coat formation | 2 | 2 | 4.78e-01 |
| GO:BP | GO:0034351 | negative regulation of glial cell apoptotic process | 4 | 8 | 4.78e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 2 | 17 | 4.78e-01 |
| GO:BP | GO:0043041 | amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0030844 | positive regulation of intermediate filament depolymerization | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 1 | 2 | 4.78e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 3 | 16 | 4.78e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 12 | 100 | 4.79e-01 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 5 | 13 | 4.79e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 82 | 375 | 4.80e-01 |
| GO:BP | GO:2000591 | positive regulation of metanephric mesenchymal cell migration | 1 | 3 | 4.80e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 4 | 48 | 4.80e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 4 | 48 | 4.80e-01 |
| GO:BP | GO:0034139 | regulation of toll-like receptor 3 signaling pathway | 6 | 8 | 4.80e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 2 | 46 | 4.80e-01 |
| GO:BP | GO:1902068 | regulation of sphingolipid mediated signaling pathway | 1 | 1 | 4.80e-01 |
| GO:BP | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 1 | 3 | 4.80e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 10 | 28 | 4.80e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 34 | 1015 | 4.80e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 2 | 46 | 4.80e-01 |
| GO:BP | GO:0008585 | female gonad development | 23 | 77 | 4.80e-01 |
| GO:BP | GO:0106101 | ER-dependent peroxisome localization | 1 | 1 | 4.80e-01 |
| GO:BP | GO:0032581 | ER-dependent peroxisome organization | 1 | 1 | 4.80e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 7 | 11 | 4.80e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 4 | 41 | 4.80e-01 |
| GO:BP | GO:1900238 | regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 1 | 3 | 4.80e-01 |
| GO:BP | GO:0006563 | L-serine metabolic process | 2 | 10 | 4.80e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 4.80e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 18 | 164 | 4.80e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 2 | 34 | 4.81e-01 |
| GO:BP | GO:0045344 | negative regulation of MHC class I biosynthetic process | 1 | 1 | 4.81e-01 |
| GO:BP | GO:1902568 | positive regulation of eosinophil activation | 2 | 2 | 4.81e-01 |
| GO:BP | GO:0043311 | positive regulation of eosinophil degranulation | 2 | 2 | 4.81e-01 |
| GO:BP | GO:0033015 | tetrapyrrole catabolic process | 1 | 7 | 4.81e-01 |
| GO:BP | GO:0006787 | porphyrin-containing compound catabolic process | 1 | 7 | 4.81e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 1 | 11 | 4.81e-01 |
| GO:BP | GO:0021796 | cerebral cortex regionalization | 1 | 2 | 4.81e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 8 | 12 | 4.81e-01 |
| GO:BP | GO:0030157 | pancreatic juice secretion | 2 | 11 | 4.81e-01 |
| GO:BP | GO:0035865 | cellular response to potassium ion | 2 | 9 | 4.81e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 3 | 15 | 4.81e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 4 | 30 | 4.82e-01 |
| GO:BP | GO:0042761 | very long-chain fatty acid biosynthetic process | 8 | 12 | 4.82e-01 |
| GO:BP | GO:1901211 | negative regulation of cardiac chamber formation | 1 | 1 | 4.82e-01 |
| GO:BP | GO:1901208 | negative regulation of heart looping | 1 | 1 | 4.82e-01 |
| GO:BP | GO:1901210 | regulation of cardiac chamber formation | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0030497 | fatty acid elongation | 8 | 12 | 4.82e-01 |
| GO:BP | GO:1905099 | positive regulation of guanyl-nucleotide exchange factor activity | 1 | 2 | 4.82e-01 |
| GO:BP | GO:1904618 | positive regulation of actin binding | 1 | 2 | 4.82e-01 |
| GO:BP | GO:1904531 | positive regulation of actin filament binding | 1 | 2 | 4.82e-01 |
| GO:BP | GO:2001108 | positive regulation of Rho guanyl-nucleotide exchange factor activity | 1 | 2 | 4.82e-01 |
| GO:BP | GO:1901329 | regulation of odontoblast differentiation | 1 | 2 | 4.82e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 4.82e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 4 | 85 | 4.82e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 3 | 81 | 4.82e-01 |
| GO:BP | GO:0001788 | antibody-dependent cellular cytotoxicity | 2 | 2 | 4.82e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 34 | 1017 | 4.82e-01 |
| GO:BP | GO:0019083 | viral transcription | 1 | 48 | 4.82e-01 |
| GO:BP | GO:0110008 | ncRNA deadenylation | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 4.82e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 4.83e-01 |
| GO:BP | GO:0090557 | establishment of endothelial intestinal barrier | 4 | 10 | 4.83e-01 |
| GO:BP | GO:0003195 | tricuspid valve formation | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 9 | 75 | 4.83e-01 |
| GO:BP | GO:0062029 | positive regulation of stress granule assembly | 1 | 2 | 4.83e-01 |
| GO:BP | GO:0031648 | protein destabilization | 16 | 45 | 4.83e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 7 | 34 | 4.83e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 10 | 19 | 4.84e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 20 | 4.84e-01 |
| GO:BP | GO:1903726 | negative regulation of phospholipid metabolic process | 5 | 7 | 4.84e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 8 | 19 | 4.84e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 10 | 19 | 4.84e-01 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 3 | 6 | 4.84e-01 |
| GO:BP | GO:0071528 | tRNA re-export from nucleus | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0070668 | positive regulation of mast cell proliferation | 2 | 4 | 4.84e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 4.84e-01 |
| GO:BP | GO:0060087 | relaxation of vascular associated smooth muscle | 5 | 7 | 4.84e-01 |
| GO:BP | GO:0003341 | cilium movement | 5 | 97 | 4.84e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 4 | 40 | 4.84e-01 |
| GO:BP | GO:0097576 | vacuole fusion | 1 | 12 | 4.84e-01 |
| GO:BP | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis | 2 | 3 | 4.84e-01 |
| GO:BP | GO:1904322 | cellular response to forskolin | 4 | 11 | 4.84e-01 |
| GO:BP | GO:1904321 | response to forskolin | 4 | 11 | 4.84e-01 |
| GO:BP | GO:0051596 | methylglyoxal catabolic process | 3 | 5 | 4.84e-01 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 1 | 11 | 4.84e-01 |
| GO:BP | GO:0040008 | regulation of growth | 33 | 501 | 4.84e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 3 | 44 | 4.84e-01 |
| GO:BP | GO:0061727 | methylglyoxal catabolic process to lactate | 3 | 5 | 4.84e-01 |
| GO:BP | GO:2000615 | regulation of histone H3-K9 acetylation | 2 | 7 | 4.84e-01 |
| GO:BP | GO:0071243 | cellular response to arsenic-containing substance | 10 | 18 | 4.85e-01 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 4.85e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 1 | 64 | 4.85e-01 |
| GO:BP | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis | 1 | 1 | 4.85e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 4.85e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 2 | 3 | 4.85e-01 |
| GO:BP | GO:0060517 | epithelial cell proliferation involved in prostatic bud elongation | 1 | 1 | 4.85e-01 |
| GO:BP | GO:0071500 | cellular response to nitrosative stress | 2 | 6 | 4.85e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 1 | 50 | 4.85e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 4.85e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 29 | 71 | 4.85e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 4.85e-01 |
| GO:BP | GO:0060024 | rhythmic synaptic transmission | 2 | 3 | 4.85e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 2 | 3 | 4.85e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 1 | 272 | 4.85e-01 |
| GO:BP | GO:0009957 | epidermal cell fate specification | 2 | 4 | 4.85e-01 |
| GO:BP | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis | 2 | 4 | 4.85e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 2 | 3 | 4.85e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 4.85e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 2 | 5 | 4.85e-01 |
| GO:BP | GO:0061709 | reticulophagy | 6 | 17 | 4.85e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 5 | 143 | 4.85e-01 |
| GO:BP | GO:0030091 | protein repair | 3 | 7 | 4.85e-01 |
| GO:BP | GO:0003219 | cardiac right ventricle formation | 2 | 4 | 4.85e-01 |
| GO:BP | GO:0070150 | mitochondrial glycyl-tRNA aminoacylation | 1 | 1 | 4.85e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 1 | 26 | 4.85e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 2 | 3 | 4.85e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 5 | 39 | 4.85e-01 |
| GO:BP | GO:0006426 | glycyl-tRNA aminoacylation | 1 | 1 | 4.85e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 8 | 15 | 4.85e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 3 | 29 | 4.85e-01 |
| GO:BP | GO:0030329 | prenylcysteine metabolic process | 1 | 2 | 4.85e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 4.85e-01 |
| GO:BP | GO:0061682 | seminal vesicle morphogenesis | 1 | 1 | 4.85e-01 |
| GO:BP | GO:0021766 | hippocampus development | 3 | 62 | 4.85e-01 |
| GO:BP | GO:0030328 | prenylcysteine catabolic process | 1 | 2 | 4.85e-01 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 2 | 6 | 4.85e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 20 | 37 | 4.85e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 6 | 21 | 4.86e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 6 | 82 | 4.86e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 13 | 96 | 4.86e-01 |
| GO:BP | GO:0061738 | late endosomal microautophagy | 1 | 3 | 4.86e-01 |
| GO:BP | GO:0014848 | urinary tract smooth muscle contraction | 5 | 6 | 4.86e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 12 | 113 | 4.86e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 4 | 12 | 4.86e-01 |
| GO:BP | GO:0048549 | positive regulation of pinocytosis | 5 | 7 | 4.86e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 4.86e-01 |
| GO:BP | GO:1900365 | positive regulation of mRNA polyadenylation | 3 | 4 | 4.87e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 27 | 45 | 4.87e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 6 | 23 | 4.87e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 1 | 31 | 4.87e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 3 | 27 | 4.87e-01 |
| GO:BP | GO:0000019 | regulation of mitotic recombination | 1 | 5 | 4.87e-01 |
| GO:BP | GO:0060766 | negative regulation of androgen receptor signaling pathway | 7 | 12 | 4.87e-01 |
| GO:BP | GO:0032289 | central nervous system myelin formation | 1 | 4 | 4.87e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 2 | 3 | 4.87e-01 |
| GO:BP | GO:0000160 | phosphorelay signal transduction system | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 3 | 18 | 4.88e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 6 | 34 | 4.89e-01 |
| GO:BP | GO:0006657 | CDP-choline pathway | 3 | 6 | 4.89e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 1 | 5 | 4.89e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 8 | 40 | 4.89e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 4 | 8 | 4.90e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 11 | 61 | 4.90e-01 |
| GO:BP | GO:0015744 | succinate transport | 1 | 6 | 4.90e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0021889 | olfactory bulb interneuron differentiation | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 4.90e-01 |
| GO:BP | GO:0003139 | secondary heart field specification | 1 | 10 | 4.90e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 4.90e-01 |
| GO:BP | GO:0106020 | regulation of vesicle docking | 1 | 2 | 4.90e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 4.90e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 1 | 27 | 4.90e-01 |
| GO:BP | GO:0050894 | determination of affect | 1 | 1 | 4.90e-01 |
| GO:BP | GO:0002352 | B cell negative selection | 2 | 2 | 4.90e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 1 | 6 | 4.90e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 1 | 5 | 4.90e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 4.90e-01 |
| GO:BP | GO:0060397 | growth hormone receptor signaling pathway via JAK-STAT | 5 | 9 | 4.90e-01 |
| GO:BP | GO:0070586 | cell-cell adhesion involved in gastrulation | 2 | 7 | 4.90e-01 |
| GO:BP | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | 5 | 9 | 4.90e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 2 | 18 | 4.90e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 3 | 145 | 4.90e-01 |
| GO:BP | GO:0009749 | response to glucose | 12 | 137 | 4.90e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 1 | 28 | 4.91e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 20 | 4.91e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 4 | 7 | 4.91e-01 |
| GO:BP | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway | 3 | 6 | 4.92e-01 |
| GO:BP | GO:0016062 | adaptation of rhodopsin mediated signaling | 1 | 1 | 4.92e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 4.92e-01 |
| GO:BP | GO:1905232 | cellular response to L-glutamate | 2 | 4 | 4.92e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 4 | 106 | 4.92e-01 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 10 | 19 | 4.92e-01 |
| GO:BP | GO:1990927 | calcium ion regulated lysosome exocytosis | 1 | 3 | 4.92e-01 |
| GO:BP | GO:0015881 | creatine transmembrane transport | 1 | 4 | 4.93e-01 |
| GO:BP | GO:0060980 | cell migration involved in coronary vasculogenesis | 1 | 1 | 4.93e-01 |
| GO:BP | GO:1903781 | positive regulation of cardiac conduction | 1 | 1 | 4.93e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 9 | 123 | 4.93e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 7 | 107 | 4.93e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 2 | 8 | 4.93e-01 |
| GO:BP | GO:0042264 | peptidyl-aspartic acid hydroxylation | 2 | 2 | 4.93e-01 |
| GO:BP | GO:0098926 | postsynaptic signal transduction | 1 | 25 | 4.93e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 2 | 27 | 4.93e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 15 | 34 | 4.93e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 82 | 376 | 4.93e-01 |
| GO:BP | GO:0031642 | negative regulation of myelination | 4 | 8 | 4.93e-01 |
| GO:BP | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway | 3 | 3 | 4.93e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 9 | 88 | 4.93e-01 |
| GO:BP | GO:0090383 | phagosome acidification | 1 | 5 | 4.93e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 4.93e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 4.93e-01 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 4 | 10 | 4.93e-01 |
| GO:BP | GO:0044528 | regulation of mitochondrial mRNA stability | 5 | 7 | 4.93e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 4.93e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 5 | 194 | 4.93e-01 |
| GO:BP | GO:0051902 | negative regulation of mitochondrial depolarization | 2 | 5 | 4.93e-01 |
| GO:BP | GO:2000254 | regulation of male germ cell proliferation | 2 | 4 | 4.93e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 19 | 41 | 4.93e-01 |
| GO:BP | GO:1903811 | L-asparagine import across plasma membrane | 1 | 1 | 4.94e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 15 | 161 | 4.94e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 10 | 51 | 4.94e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 2 | 4 | 4.94e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 2 | 5 | 4.94e-01 |
| GO:BP | GO:0048297 | negative regulation of isotype switching to IgA isotypes | 1 | 1 | 4.94e-01 |
| GO:BP | GO:1904141 | positive regulation of microglial cell migration | 1 | 3 | 4.94e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 6 | 14 | 4.94e-01 |
| GO:BP | GO:0038145 | macrophage colony-stimulating factor signaling pathway | 1 | 3 | 4.94e-01 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 2 | 7 | 4.94e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 4.94e-01 |
| GO:BP | GO:0120040 | regulation of macrophage proliferation | 1 | 4 | 4.94e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 3 | 27 | 4.94e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 3 | 35 | 4.94e-01 |
| GO:BP | GO:0071788 | endoplasmic reticulum tubular network maintenance | 1 | 1 | 4.94e-01 |
| GO:BP | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure | 2 | 14 | 4.94e-01 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 3 | 13 | 4.94e-01 |
| GO:BP | GO:1900235 | negative regulation of Kit signaling pathway | 2 | 2 | 4.95e-01 |
| GO:BP | GO:0060929 | atrioventricular node cell fate commitment | 2 | 2 | 4.95e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 4.95e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 2 | 535 | 4.95e-01 |
| GO:BP | GO:0009082 | branched-chain amino acid biosynthetic process | 1 | 4 | 4.95e-01 |
| GO:BP | GO:1900234 | regulation of Kit signaling pathway | 2 | 2 | 4.95e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 6 | 20 | 4.95e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 52 | 106 | 4.95e-01 |
| GO:BP | GO:1990009 | retinal cell apoptotic process | 4 | 5 | 4.95e-01 |
| GO:BP | GO:0015766 | disaccharide transport | 3 | 4 | 4.96e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 24 | 46 | 4.96e-01 |
| GO:BP | GO:0099551 | trans-synaptic signaling by neuropeptide, modulating synaptic transmission | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0015770 | sucrose transport | 3 | 4 | 4.96e-01 |
| GO:BP | GO:0006431 | methionyl-tRNA aminoacylation | 2 | 2 | 4.96e-01 |
| GO:BP | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 3 | 84 | 4.96e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 4 | 13 | 4.96e-01 |
| GO:BP | GO:0001946 | lymphangiogenesis | 5 | 12 | 4.96e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 8 | 39 | 4.96e-01 |
| GO:BP | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | 4 | 8 | 4.96e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 4.96e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 4.96e-01 |
| GO:BP | GO:0090385 | phagosome-lysosome fusion | 3 | 12 | 4.96e-01 |
| GO:BP | GO:1905090 | negative regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 7 | 63 | 4.96e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 2 | 6 | 4.96e-01 |
| GO:BP | GO:0043474 | pigment metabolic process involved in pigmentation | 3 | 3 | 4.96e-01 |
| GO:BP | GO:0048873 | homeostasis of number of cells within a tissue | 4 | 24 | 4.96e-01 |
| GO:BP | GO:0015772 | oligosaccharide transport | 3 | 4 | 4.96e-01 |
| GO:BP | GO:0061817 | endoplasmic reticulum-plasma membrane tethering | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0030220 | platelet formation | 3 | 17 | 4.96e-01 |
| GO:BP | GO:0019742 | pentacyclic triterpenoid metabolic process | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 10 | 19 | 4.96e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 1 | 7 | 4.96e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 4.96e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 2 | 15 | 4.96e-01 |
| GO:BP | GO:0015855 | pyrimidine nucleobase transport | 1 | 3 | 4.96e-01 |
| GO:BP | GO:0015853 | adenine transport | 1 | 5 | 4.96e-01 |
| GO:BP | GO:0015862 | uridine transport | 1 | 4 | 4.96e-01 |
| GO:BP | GO:0015864 | pyrimidine nucleoside transport | 1 | 4 | 4.96e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 1 | 29 | 4.96e-01 |
| GO:BP | GO:0021510 | spinal cord development | 6 | 59 | 4.96e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 11 | 21 | 4.96e-01 |
| GO:BP | GO:1990547 | mitochondrial phosphate ion transmembrane transport | 1 | 1 | 4.97e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 2 | 742 | 4.97e-01 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 1 | 5 | 4.97e-01 |
| GO:BP | GO:0002885 | positive regulation of hypersensitivity | 3 | 5 | 4.97e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 1 | 12 | 4.97e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 7 | 21 | 4.97e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 1 | 12 | 4.97e-01 |
| GO:BP | GO:0019321 | pentose metabolic process | 6 | 9 | 4.97e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 3 | 23 | 4.97e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 1 | 73 | 4.97e-01 |
| GO:BP | GO:1902766 | skeletal muscle satellite cell migration | 1 | 2 | 4.97e-01 |
| GO:BP | GO:0060708 | spongiotrophoblast differentiation | 2 | 2 | 4.97e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 5 | 25 | 4.97e-01 |
| GO:BP | GO:0051001 | negative regulation of nitric-oxide synthase activity | 4 | 6 | 4.97e-01 |
| GO:BP | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing | 1 | 1 | 4.97e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 3 | 12 | 4.98e-01 |
| GO:BP | GO:0034759 | regulation of iron ion transmembrane transport | 1 | 2 | 4.98e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 10 | 4.98e-01 |
| GO:BP | GO:0051900 | regulation of mitochondrial depolarization | 5 | 15 | 4.98e-01 |
| GO:BP | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis | 2 | 3 | 4.98e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 2 | 187 | 4.98e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 2 | 187 | 4.98e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 21 | 92 | 4.98e-01 |
| GO:BP | GO:0090314 | positive regulation of protein targeting to membrane | 5 | 22 | 4.98e-01 |
| GO:BP | GO:1990983 | tRNA demethylation | 1 | 1 | 4.99e-01 |
| GO:BP | GO:0007402 | ganglion mother cell fate determination | 1 | 1 | 4.99e-01 |
| GO:BP | GO:1903911 | positive regulation of receptor clustering | 1 | 6 | 4.99e-01 |
| GO:BP | GO:0001759 | organ induction | 6 | 17 | 4.99e-01 |
| GO:BP | GO:0002481 | antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent | 2 | 2 | 4.99e-01 |
| GO:BP | GO:1904058 | positive regulation of sensory perception of pain | 3 | 3 | 4.99e-01 |
| GO:BP | GO:0080021 | response to benzoic acid | 1 | 1 | 4.99e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 42 | 242 | 4.99e-01 |
| GO:BP | GO:0060753 | regulation of mast cell chemotaxis | 1 | 7 | 4.99e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 3 | 1107 | 4.99e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 4.99e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 1 | 67 | 4.99e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 4 | 96 | 4.99e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 5.00e-01 |
| GO:BP | GO:0060134 | prepulse inhibition | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0097202 | activation of cysteine-type endopeptidase activity | 7 | 16 | 5.00e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 5.00e-01 |
| GO:BP | GO:0006924 | activation-induced cell death of T cells | 1 | 8 | 5.00e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 34 | 1026 | 5.00e-01 |
| GO:BP | GO:0018160 | peptidyl-pyrromethane cofactor linkage | 1 | 1 | 5.00e-01 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 4 | 16 | 5.00e-01 |
| GO:BP | GO:0032692 | negative regulation of interleukin-1 production | 2 | 15 | 5.00e-01 |
| GO:BP | GO:0061053 | somite development | 21 | 63 | 5.00e-01 |
| GO:BP | GO:0055001 | muscle cell development | 1 | 165 | 5.00e-01 |
| GO:BP | GO:0035965 | cardiolipin acyl-chain remodeling | 2 | 4 | 5.00e-01 |
| GO:BP | GO:0033504 | floor plate development | 2 | 8 | 5.00e-01 |
| GO:BP | GO:0042415 | norepinephrine metabolic process | 3 | 7 | 5.00e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 2 | 22 | 5.00e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 5.00e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 5.00e-01 |
| GO:BP | GO:1903031 | regulation of microtubule plus-end binding | 2 | 2 | 5.00e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 1 | 14 | 5.00e-01 |
| GO:BP | GO:1903033 | positive regulation of microtubule plus-end binding | 2 | 2 | 5.00e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 4 | 13 | 5.00e-01 |
| GO:BP | GO:0051646 | mitochondrion localization | 8 | 47 | 5.00e-01 |
| GO:BP | GO:0032329 | serine transport | 2 | 10 | 5.00e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 7 | 160 | 5.00e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 3 | 12 | 5.00e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 2 | 47 | 5.00e-01 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 2 | 4 | 5.00e-01 |
| GO:BP | GO:0032960 | regulation of inositol trisphosphate biosynthetic process | 1 | 4 | 5.00e-01 |
| GO:BP | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 1 | 4 | 5.00e-01 |
| GO:BP | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 3 | 5.00e-01 |
| GO:BP | GO:0031175 | neuron projection development | 2 | 780 | 5.00e-01 |
| GO:BP | GO:0038138 | ERBB4-ERBB4 signaling pathway | 3 | 3 | 5.01e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 25 | 756 | 5.01e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 3 | 57 | 5.01e-01 |
| GO:BP | GO:0003242 | cardiac chamber ballooning | 1 | 1 | 5.01e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 2 | 2 | 5.01e-01 |
| GO:BP | GO:2000056 | regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0043588 | skin development | 1 | 175 | 5.01e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 11 | 198 | 5.01e-01 |
| GO:BP | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.01e-01 |
| GO:BP | GO:2000057 | negative regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 4 | 56 | 5.01e-01 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 6 | 9 | 5.01e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 1 | 26 | 5.01e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 4 | 10 | 5.01e-01 |
| GO:BP | GO:1904180 | negative regulation of membrane depolarization | 3 | 6 | 5.01e-01 |
| GO:BP | GO:0061300 | cerebellum vasculature development | 2 | 2 | 5.01e-01 |
| GO:BP | GO:0061302 | smooth muscle cell-matrix adhesion | 2 | 5 | 5.01e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 4 | 10 | 5.01e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 16 | 299 | 5.01e-01 |
| GO:BP | GO:0018095 | protein polyglutamylation | 2 | 9 | 5.02e-01 |
| GO:BP | GO:0090346 | cellular organofluorine metabolic process | 1 | 1 | 5.02e-01 |
| GO:BP | GO:0090345 | cellular organohalogen metabolic process | 1 | 1 | 5.02e-01 |
| GO:BP | GO:0032071 | regulation of endodeoxyribonuclease activity | 2 | 7 | 5.02e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 5.02e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 5.02e-01 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 6 | 9 | 5.02e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 5.02e-01 |
| GO:BP | GO:0015822 | ornithine transport | 1 | 5 | 5.02e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 5.02e-01 |
| GO:BP | GO:0032438 | melanosome organization | 4 | 36 | 5.02e-01 |
| GO:BP | GO:0061622 | glycolytic process through glucose-1-phosphate | 1 | 2 | 5.02e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 22 | 249 | 5.02e-01 |
| GO:BP | GO:0070383 | DNA cytosine deamination | 1 | 5 | 5.03e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 22 | 36 | 5.03e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 5.03e-01 |
| GO:BP | GO:0002551 | mast cell chemotaxis | 4 | 9 | 5.03e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 3 | 22 | 5.03e-01 |
| GO:BP | GO:1902722 | positive regulation of prolactin secretion | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0110113 | positive regulation of lipid transporter activity | 1 | 1 | 5.03e-01 |
| GO:BP | GO:2000755 | positive regulation of sphingomyelin catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 1 | 6 | 5.03e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 5.03e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 60 | 5.03e-01 |
| GO:BP | GO:0006423 | cysteinyl-tRNA aminoacylation | 2 | 2 | 5.03e-01 |
| GO:BP | GO:1905661 | regulation of telomerase RNA reverse transcriptase activity | 2 | 3 | 5.03e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 5 | 605 | 5.03e-01 |
| GO:BP | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration | 1 | 1 | 5.03e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 1 | 57 | 5.03e-01 |
| GO:BP | GO:0097374 | sensory neuron axon guidance | 2 | 2 | 5.03e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 22 | 185 | 5.03e-01 |
| GO:BP | GO:0030889 | negative regulation of B cell proliferation | 4 | 9 | 5.03e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 2 | 755 | 5.03e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 5.03e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 5.03e-01 |
| GO:BP | GO:0010898 | positive regulation of triglyceride catabolic process | 1 | 4 | 5.03e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 2 | 6 | 5.03e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 1 | 12 | 5.03e-01 |
| GO:BP | GO:0006592 | ornithine biosynthetic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0021854 | hypothalamus development | 1 | 15 | 5.03e-01 |
| GO:BP | GO:0051006 | positive regulation of lipoprotein lipase activity | 1 | 4 | 5.03e-01 |
| GO:BP | GO:2000754 | regulation of sphingomyelin catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 7 | 11 | 5.03e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 5 | 25 | 5.03e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 5 | 25 | 5.03e-01 |
| GO:BP | GO:1905809 | negative regulation of synapse organization | 1 | 8 | 5.03e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 1 | 10 | 5.03e-01 |
| GO:BP | GO:0003306 | Wnt signaling pathway involved in heart development | 1 | 8 | 5.03e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 1 | 179 | 5.03e-01 |
| GO:BP | GO:0042407 | cristae formation | 8 | 18 | 5.04e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 41 | 136 | 5.04e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 2 | 9 | 5.04e-01 |
| GO:BP | GO:0003252 | negative regulation of cell proliferation involved in heart valve morphogenesis | 2 | 2 | 5.04e-01 |
| GO:BP | GO:0061386 | closure of optic fissure | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 2 | 9 | 5.05e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 3 | 136 | 5.05e-01 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 9 | 36 | 5.05e-01 |
| GO:BP | GO:0031640 | killing of cells of another organism | 1 | 10 | 5.05e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 3 | 85 | 5.05e-01 |
| GO:BP | GO:0034729 | histone H3-K79 methylation | 2 | 2 | 5.05e-01 |
| GO:BP | GO:1903347 | negative regulation of bicellular tight junction assembly | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0140374 | antiviral innate immune response | 1 | 17 | 5.05e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 35 | 5.05e-01 |
| GO:BP | GO:0160040 | mitocytosis | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0048103 | somatic stem cell division | 4 | 10 | 5.05e-01 |
| GO:BP | GO:0072429 | response to intra-S DNA damage checkpoint signaling | 3 | 4 | 5.05e-01 |
| GO:BP | GO:0120063 | stomach smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 8 | 13 | 5.05e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 4 | 9 | 5.05e-01 |
| GO:BP | GO:0120064 | stomach pylorus smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0030900 | forebrain development | 16 | 281 | 5.05e-01 |
| GO:BP | GO:1904341 | regulation of colon smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0072553 | terminal button organization | 2 | 5 | 5.05e-01 |
| GO:BP | GO:1990765 | colon smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0014828 | distal stomach smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0120065 | pyloric antrum smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0097326 | melanocyte adhesion | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0120072 | positive regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0140495 | migracytosis | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0120071 | regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:1904343 | positive regulation of colon smooth muscle contraction | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0106227 | peptidyl-lysine glutarylation | 1 | 1 | 5.05e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 5.06e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 1 | 8 | 5.06e-01 |
| GO:BP | GO:0019694 | alkanesulfonate metabolic process | 5 | 6 | 5.06e-01 |
| GO:BP | GO:0106088 | regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 5.06e-01 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 1 | 24 | 5.06e-01 |
| GO:BP | GO:0106089 | negative regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 5.06e-01 |
| GO:BP | GO:0019530 | taurine metabolic process | 5 | 6 | 5.06e-01 |
| GO:BP | GO:0032613 | interleukin-10 production | 1 | 24 | 5.06e-01 |
| GO:BP | GO:0003270 | Notch signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0060843 | venous endothelial cell differentiation | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 18 | 46 | 5.07e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 3 | 5 | 5.07e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 5.07e-01 |
| GO:BP | GO:0003182 | coronary sinus valve morphogenesis | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0003178 | coronary sinus valve development | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 3 | 63 | 5.07e-01 |
| GO:BP | GO:0099171 | presynaptic modulation of chemical synaptic transmission | 2 | 26 | 5.07e-01 |
| GO:BP | GO:0014900 | muscle hyperplasia | 1 | 2 | 5.07e-01 |
| GO:BP | GO:2000454 | positive regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 7 | 21 | 5.08e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 18 | 42 | 5.08e-01 |
| GO:BP | GO:0030103 | vasopressin secretion | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0097497 | blood vessel endothelial cell delamination | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 5.08e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 2 | 10 | 5.08e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 4 | 82 | 5.08e-01 |
| GO:BP | GO:0002280 | monocyte activation involved in immune response | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0042044 | fluid transport | 3 | 17 | 5.08e-01 |
| GO:BP | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 1 | 16 | 5.08e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 4 | 37 | 5.08e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 12 | 61 | 5.09e-01 |
| GO:BP | GO:0030237 | female sex determination | 1 | 1 | 5.09e-01 |
| GO:BP | GO:0006824 | cobalt ion transport | 2 | 5 | 5.09e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 5 | 114 | 5.09e-01 |
| GO:BP | GO:0006288 | base-excision repair, DNA ligation | 1 | 1 | 5.09e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 5.09e-01 |
| GO:BP | GO:0090298 | negative regulation of mitochondrial DNA replication | 1 | 1 | 5.09e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 6 | 12 | 5.09e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 29 | 153 | 5.09e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 1 | 59 | 5.09e-01 |
| GO:BP | GO:2000225 | negative regulation of testosterone biosynthetic process | 1 | 1 | 5.09e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 7 | 99 | 5.09e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 3 | 16 | 5.09e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 6 | 16 | 5.09e-01 |
| GO:BP | GO:0035732 | nitric oxide storage | 1 | 1 | 5.09e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 3 | 67 | 5.09e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 4 | 6 | 5.09e-01 |
| GO:BP | GO:0030185 | nitric oxide transport | 1 | 2 | 5.09e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 1 | 78 | 5.10e-01 |
| GO:BP | GO:1902595 | regulation of DNA replication origin binding | 2 | 2 | 5.10e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 3 | 40 | 5.10e-01 |
| GO:BP | GO:0007258 | JUN phosphorylation | 2 | 2 | 5.10e-01 |
| GO:BP | GO:0006400 | tRNA modification | 36 | 90 | 5.10e-01 |
| GO:BP | GO:0072677 | eosinophil migration | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 6 | 22 | 5.10e-01 |
| GO:BP | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development | 2 | 3 | 5.10e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 5.10e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 5 | 49 | 5.10e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 1 | 15 | 5.10e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 3 | 35 | 5.10e-01 |
| GO:BP | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade | 3 | 16 | 5.10e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0106058 | positive regulation of calcineurin-mediated signaling | 3 | 16 | 5.10e-01 |
| GO:BP | GO:0097503 | sialylation | 1 | 18 | 5.11e-01 |
| GO:BP | GO:0014031 | mesenchymal cell development | 4 | 6 | 5.11e-01 |
| GO:BP | GO:0061734 | parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 3 | 6 | 5.12e-01 |
| GO:BP | GO:0045342 | MHC class II biosynthetic process | 7 | 12 | 5.12e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 9 | 142 | 5.12e-01 |
| GO:BP | GO:0032912 | negative regulation of transforming growth factor beta2 production | 2 | 2 | 5.12e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 5.13e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 3 | 37 | 5.13e-01 |
| GO:BP | GO:0032730 | positive regulation of interleukin-1 alpha production | 1 | 3 | 5.13e-01 |
| GO:BP | GO:0009746 | response to hexose | 12 | 138 | 5.13e-01 |
| GO:BP | GO:0006950 | response to stress | 5 | 2780 | 5.13e-01 |
| GO:BP | GO:0032259 | methylation | 1 | 313 | 5.13e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 15 | 70 | 5.13e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 15 | 70 | 5.13e-01 |
| GO:BP | GO:0033625 | positive regulation of integrin activation | 3 | 6 | 5.13e-01 |
| GO:BP | GO:0060992 | response to fungicide | 1 | 4 | 5.13e-01 |
| GO:BP | GO:1905517 | macrophage migration | 4 | 34 | 5.13e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 3 | 23 | 5.13e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 5.13e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 15 | 54 | 5.14e-01 |
| GO:BP | GO:0010818 | T cell chemotaxis | 3 | 12 | 5.14e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 2 | 3 | 5.14e-01 |
| GO:BP | GO:0099532 | synaptic vesicle endosomal processing | 1 | 7 | 5.14e-01 |
| GO:BP | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation | 1 | 5 | 5.14e-01 |
| GO:BP | GO:0060687 | regulation of branching involved in prostate gland morphogenesis | 1 | 3 | 5.14e-01 |
| GO:BP | GO:0090245 | axis elongation involved in somitogenesis | 1 | 4 | 5.14e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 1 | 187 | 5.14e-01 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 1 | 13 | 5.15e-01 |
| GO:BP | GO:0051415 | microtubule nucleation by interphase microtubule organizing center | 1 | 1 | 5.15e-01 |
| GO:BP | GO:0019080 | viral gene expression | 4 | 96 | 5.15e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 14 | 26 | 5.15e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 1 | 4 | 5.15e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 2 | 10 | 5.15e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 5.15e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 16 | 816 | 5.15e-01 |
| GO:BP | GO:0015838 | amino-acid betaine transport | 1 | 7 | 5.15e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 4 | 37 | 5.15e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 5.16e-01 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 9 | 17 | 5.16e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 9 | 17 | 5.16e-01 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 5 | 8 | 5.16e-01 |
| GO:BP | GO:1901290 | succinyl-CoA biosynthetic process | 1 | 1 | 5.16e-01 |
| GO:BP | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission | 1 | 3 | 5.16e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 5.16e-01 |
| GO:BP | GO:0019678 | propionate metabolic process, methylmalonyl pathway | 1 | 1 | 5.16e-01 |
| GO:BP | GO:0007292 | female gamete generation | 26 | 103 | 5.16e-01 |
| GO:BP | GO:0046851 | negative regulation of bone remodeling | 2 | 9 | 5.16e-01 |
| GO:BP | GO:0002100 | tRNA wobble adenosine to inosine editing | 1 | 1 | 5.16e-01 |
| GO:BP | GO:0097711 | ciliary basal body-plasma membrane docking | 2 | 2 | 5.16e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 11 | 115 | 5.16e-01 |
| GO:BP | GO:1902034 | negative regulation of hematopoietic stem cell proliferation | 1 | 2 | 5.17e-01 |
| GO:BP | GO:0014022 | neural plate elongation | 2 | 2 | 5.17e-01 |
| GO:BP | GO:0022007 | convergent extension involved in neural plate elongation | 2 | 2 | 5.17e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 2 | 8 | 5.17e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 7 | 63 | 5.17e-01 |
| GO:BP | GO:0032868 | response to insulin | 2 | 215 | 5.17e-01 |
| GO:BP | GO:0045204 | MAPK export from nucleus | 1 | 3 | 5.17e-01 |
| GO:BP | GO:0090500 | endocardial cushion to mesenchymal transition | 3 | 4 | 5.17e-01 |
| GO:BP | GO:1903567 | regulation of protein localization to ciliary membrane | 1 | 2 | 5.17e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 17 | 30 | 5.17e-01 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 11 | 19 | 5.17e-01 |
| GO:BP | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration | 5 | 6 | 5.17e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 22 | 57 | 5.17e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 6 | 25 | 5.17e-01 |
| GO:BP | GO:0009386 | translational attenuation | 1 | 2 | 5.17e-01 |
| GO:BP | GO:0061518 | microglial cell proliferation | 1 | 4 | 5.17e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 6 | 25 | 5.17e-01 |
| GO:BP | GO:1904139 | regulation of microglial cell migration | 1 | 3 | 5.17e-01 |
| GO:BP | GO:0043970 | histone H3-K9 acetylation | 2 | 8 | 5.17e-01 |
| GO:BP | GO:0061944 | negative regulation of protein K48-linked ubiquitination | 1 | 2 | 5.17e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 15 | 111 | 5.17e-01 |
| GO:BP | GO:1904124 | microglial cell migration | 1 | 3 | 5.17e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 6 | 26 | 5.18e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 1 | 20 | 5.18e-01 |
| GO:BP | GO:1903645 | negative regulation of chaperone-mediated protein folding | 2 | 2 | 5.18e-01 |
| GO:BP | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process | 2 | 3 | 5.18e-01 |
| GO:BP | GO:0015961 | diadenosine polyphosphate catabolic process | 4 | 6 | 5.18e-01 |
| GO:BP | GO:0006171 | cAMP biosynthetic process | 3 | 8 | 5.18e-01 |
| GO:BP | GO:2000969 | positive regulation of AMPA receptor activity | 1 | 4 | 5.18e-01 |
| GO:BP | GO:0002339 | B cell selection | 3 | 4 | 5.18e-01 |
| GO:BP | GO:0110135 | Norrin signaling pathway | 2 | 2 | 5.18e-01 |
| GO:BP | GO:0009203 | ribonucleoside triphosphate catabolic process | 2 | 2 | 5.18e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 5 | 55 | 5.18e-01 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 1 | 144 | 5.19e-01 |
| GO:BP | GO:0050808 | synapse organization | 24 | 365 | 5.19e-01 |
| GO:BP | GO:0006391 | transcription initiation at mitochondrial promoter | 3 | 4 | 5.19e-01 |
| GO:BP | GO:0007231 | osmosensory signaling pathway | 3 | 3 | 5.19e-01 |
| GO:BP | GO:0019043 | establishment of viral latency | 1 | 3 | 5.19e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 1 | 12 | 5.19e-01 |
| GO:BP | GO:0072675 | osteoclast fusion | 1 | 5 | 5.20e-01 |
| GO:BP | GO:1990966 | ATP generation from poly-ADP-D-ribose | 1 | 4 | 5.20e-01 |
| GO:BP | GO:2000632 | negative regulation of pre-miRNA processing | 2 | 2 | 5.20e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 15 | 90 | 5.20e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 5.20e-01 |
| GO:BP | GO:0061303 | cornea development in camera-type eye | 1 | 4 | 5.20e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 7 | 28 | 5.20e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 6 | 113 | 5.20e-01 |
| GO:BP | GO:0021537 | telencephalon development | 15 | 201 | 5.20e-01 |
| GO:BP | GO:0090262 | regulation of transcription-coupled nucleotide-excision repair | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0007072 | positive regulation of transcription involved in exit from mitosis | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 20 | 32 | 5.21e-01 |
| GO:BP | GO:0010650 | positive regulation of cell communication by electrical coupling | 2 | 3 | 5.21e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 5 | 31 | 5.21e-01 |
| GO:BP | GO:1903689 | regulation of wound healing, spreading of epidermal cells | 2 | 8 | 5.21e-01 |
| GO:BP | GO:1905474 | canonical Wnt signaling pathway involved in stem cell proliferation | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 9 | 469 | 5.21e-01 |
| GO:BP | GO:1902576 | negative regulation of nuclear cell cycle DNA replication | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 4 | 5 | 5.21e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 4 | 67 | 5.21e-01 |
| GO:BP | GO:0032463 | negative regulation of protein homooligomerization | 1 | 2 | 5.21e-01 |
| GO:BP | GO:0031449 | regulation of slow-twitch skeletal muscle fiber contraction | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0002023 | reduction of food intake in response to dietary excess | 1 | 2 | 5.21e-01 |
| GO:BP | GO:1903464 | negative regulation of mitotic cell cycle DNA replication | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0032460 | negative regulation of protein oligomerization | 1 | 2 | 5.21e-01 |
| GO:BP | GO:0060064 | Spemann organizer formation at the anterior end of the primitive streak | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 4 | 72 | 5.21e-01 |
| GO:BP | GO:0033025 | regulation of mast cell apoptotic process | 1 | 3 | 5.21e-01 |
| GO:BP | GO:0033024 | mast cell apoptotic process | 1 | 3 | 5.21e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 1 | 11 | 5.21e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 4 | 662 | 5.21e-01 |
| GO:BP | GO:0036018 | cellular response to erythropoietin | 3 | 4 | 5.22e-01 |
| GO:BP | GO:0006667 | sphinganine metabolic process | 2 | 2 | 5.22e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 5.23e-01 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 1 | 16 | 5.23e-01 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 5.23e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 1 | 17 | 5.23e-01 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 5 | 16 | 5.23e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 2 | 16 | 5.23e-01 |
| GO:BP | GO:0044751 | cellular response to human chorionic gonadotropin stimulus | 1 | 2 | 5.23e-01 |
| GO:BP | GO:1905259 | negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 5.23e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 5.23e-01 |
| GO:BP | GO:0031444 | slow-twitch skeletal muscle fiber contraction | 2 | 2 | 5.23e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 7 | 34 | 5.23e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 5 | 19 | 5.23e-01 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 2 | 14 | 5.23e-01 |
| GO:BP | GO:0007382 | specification of segmental identity, maxillary segment | 1 | 1 | 5.23e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 2 | 26 | 5.23e-01 |
| GO:BP | GO:1905871 | regulation of protein localization to cell leading edge | 1 | 2 | 5.23e-01 |
| GO:BP | GO:1905007 | positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 5 | 5.23e-01 |
| GO:BP | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 2 | 3 | 5.23e-01 |
| GO:BP | GO:0022009 | central nervous system vasculogenesis | 1 | 4 | 5.23e-01 |
| GO:BP | GO:1905258 | regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 5.23e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 1 | 31 | 5.23e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 23 | 186 | 5.23e-01 |
| GO:BP | GO:0045763 | negative regulation of cellular amino acid metabolic process | 2 | 2 | 5.23e-01 |
| GO:BP | GO:1900452 | regulation of long-term synaptic depression | 3 | 10 | 5.23e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 7 | 12 | 5.23e-01 |
| GO:BP | GO:0140192 | regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 2 | 3 | 5.23e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 3 | 1123 | 5.23e-01 |
| GO:BP | GO:0042732 | D-xylose metabolic process | 1 | 2 | 5.23e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 3 | 30 | 5.23e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 4 | 15 | 5.23e-01 |
| GO:BP | GO:0032074 | negative regulation of nuclease activity | 2 | 6 | 5.23e-01 |
| GO:BP | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 1 | 1 | 5.23e-01 |
| GO:BP | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 1 | 5.23e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 5.23e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 5 | 30 | 5.23e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 1 | 12 | 5.23e-01 |
| GO:BP | GO:0060632 | regulation of microtubule-based movement | 2 | 31 | 5.23e-01 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 1 | 13 | 5.23e-01 |
| GO:BP | GO:0061365 | positive regulation of triglyceride lipase activity | 1 | 4 | 5.23e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 8 | 96 | 5.23e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 5.23e-01 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 3 | 4 | 5.23e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 2 | 131 | 5.23e-01 |
| GO:BP | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 6 | 5.23e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 5.23e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 1 | 13 | 5.23e-01 |
| GO:BP | GO:1904528 | positive regulation of microtubule binding | 2 | 6 | 5.23e-01 |
| GO:BP | GO:0060932 | His-Purkinje system cell differentiation | 3 | 4 | 5.23e-01 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 3 | 4 | 5.23e-01 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 3 | 4 | 5.23e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 5.23e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 9 | 62 | 5.23e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 5 | 30 | 5.23e-01 |
| GO:BP | GO:0090188 | negative regulation of pancreatic juice secretion | 1 | 4 | 5.23e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 2 | 13 | 5.23e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 5.24e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 5.24e-01 |
| GO:BP | GO:0048671 | negative regulation of collateral sprouting | 4 | 9 | 5.24e-01 |
| GO:BP | GO:1990167 | protein K27-linked deubiquitination | 2 | 3 | 5.24e-01 |
| GO:BP | GO:0032095 | regulation of response to food | 1 | 8 | 5.24e-01 |
| GO:BP | GO:1904117 | cellular response to vasopressin | 2 | 5 | 5.24e-01 |
| GO:BP | GO:1904116 | response to vasopressin | 2 | 5 | 5.24e-01 |
| GO:BP | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0090666 | scaRNA localization to Cajal body | 2 | 5 | 5.25e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 2 | 4 | 5.25e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 2 | 40 | 5.25e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 2 | 5 | 5.25e-01 |
| GO:BP | GO:0071865 | regulation of apoptotic process in bone marrow cell | 3 | 3 | 5.25e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 6 | 10 | 5.25e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 26 | 91 | 5.25e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 4 | 41 | 5.25e-01 |
| GO:BP | GO:0071866 | negative regulation of apoptotic process in bone marrow cell | 3 | 3 | 5.25e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 2 | 4 | 5.25e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 5.25e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 2 | 4 | 5.25e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 25 | 740 | 5.25e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 6 | 195 | 5.25e-01 |
| GO:BP | GO:0046084 | adenine biosynthetic process | 1 | 2 | 5.25e-01 |
| GO:BP | GO:0071467 | cellular response to pH | 2 | 16 | 5.25e-01 |
| GO:BP | GO:0002545 | chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0002882 | positive regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0098828 | modulation of inhibitory postsynaptic potential | 3 | 4 | 5.25e-01 |
| GO:BP | GO:0002880 | regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0035781 | CD86 biosynthetic process | 2 | 2 | 5.25e-01 |
| GO:BP | GO:0006997 | nucleus organization | 5 | 126 | 5.25e-01 |
| GO:BP | GO:0033564 | anterior/posterior axon guidance | 1 | 4 | 5.25e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 5.25e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 95 | 485 | 5.26e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 1 | 65 | 5.26e-01 |
| GO:BP | GO:1904181 | positive regulation of membrane depolarization | 3 | 8 | 5.26e-01 |
| GO:BP | GO:0007565 | female pregnancy | 5 | 122 | 5.26e-01 |
| GO:BP | GO:1903401 | L-lysine transmembrane transport | 1 | 6 | 5.26e-01 |
| GO:BP | GO:0006096 | glycolytic process | 1 | 70 | 5.26e-01 |
| GO:BP | GO:1902022 | L-lysine transport | 1 | 6 | 5.26e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 9 | 26 | 5.27e-01 |
| GO:BP | GO:2000584 | negative regulation of platelet-derived growth factor receptor-alpha signaling pathway | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0061107 | seminal vesicle development | 2 | 2 | 5.27e-01 |
| GO:BP | GO:0048104 | establishment of body hair or bristle planar orientation | 2 | 3 | 5.27e-01 |
| GO:BP | GO:0048105 | establishment of body hair planar orientation | 2 | 3 | 5.27e-01 |
| GO:BP | GO:0045002 | double-strand break repair via single-strand annealing | 5 | 7 | 5.27e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 8 | 246 | 5.27e-01 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 2 | 6 | 5.27e-01 |
| GO:BP | GO:0003108 | negative regulation of the force of heart contraction by chemical signal | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0015886 | heme transport | 3 | 9 | 5.28e-01 |
| GO:BP | GO:1901081 | negative regulation of relaxation of smooth muscle | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 4 | 22 | 5.28e-01 |
| GO:BP | GO:0006422 | aspartyl-tRNA aminoacylation | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 5.28e-01 |
| GO:BP | GO:1900130 | regulation of lipid binding | 1 | 1 | 5.28e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 6 | 24 | 5.28e-01 |
| GO:BP | GO:1900131 | negative regulation of lipid binding | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 5 | 17 | 5.28e-01 |
| GO:BP | GO:0030238 | male sex determination | 4 | 8 | 5.28e-01 |
| GO:BP | GO:0039534 | negative regulation of MDA-5 signaling pathway | 2 | 3 | 5.28e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 14 | 32 | 5.28e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 36 | 1119 | 5.28e-01 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 5 | 18 | 5.28e-01 |
| GO:BP | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 1 | 6 | 5.28e-01 |
| GO:BP | GO:0045046 | protein import into peroxisome membrane | 3 | 6 | 5.28e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 21 | 40 | 5.28e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 2 | 25 | 5.28e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 6 | 1484 | 5.28e-01 |
| GO:BP | GO:0035295 | tube development | 2 | 821 | 5.28e-01 |
| GO:BP | GO:0010232 | vascular transport | 1 | 72 | 5.28e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 1 | 72 | 5.28e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 5.28e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 2 | 5 | 5.28e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 3 | 25 | 5.28e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 1 | 16 | 5.28e-01 |
| GO:BP | GO:0031034 | myosin filament assembly | 3 | 4 | 5.29e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 28 | 150 | 5.29e-01 |
| GO:BP | GO:1905303 | positive regulation of macropinocytosis | 2 | 2 | 5.29e-01 |
| GO:BP | GO:0006101 | citrate metabolic process | 1 | 5 | 5.29e-01 |
| GO:BP | GO:0097500 | receptor localization to non-motile cilium | 3 | 4 | 5.29e-01 |
| GO:BP | GO:0030321 | transepithelial chloride transport | 3 | 7 | 5.29e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 2 | 66 | 5.29e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 5.29e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 2 | 4 | 5.29e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 13 | 29 | 5.29e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 6 | 11 | 5.29e-01 |
| GO:BP | GO:0072537 | fibroblast activation | 7 | 11 | 5.29e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 14 | 52 | 5.29e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 18 | 303 | 5.29e-01 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 5.29e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 2 | 31 | 5.29e-01 |
| GO:BP | GO:0120187 | positive regulation of protein localization to chromatin | 1 | 3 | 5.29e-01 |
| GO:BP | GO:0001845 | phagolysosome assembly | 4 | 14 | 5.29e-01 |
| GO:BP | GO:0033623 | regulation of integrin activation | 4 | 11 | 5.30e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 1 | 86 | 5.30e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 1 | 25 | 5.30e-01 |
| GO:BP | GO:0035789 | metanephric mesenchymal cell migration | 1 | 3 | 5.30e-01 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 5.30e-01 |
| GO:BP | GO:2000589 | regulation of metanephric mesenchymal cell migration | 1 | 3 | 5.30e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 3 | 3 | 5.30e-01 |
| GO:BP | GO:0010610 | regulation of mRNA stability involved in response to stress | 2 | 3 | 5.30e-01 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 4 | 7 | 5.30e-01 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 4 | 7 | 5.30e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 5 | 10 | 5.30e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 40 | 179 | 5.30e-01 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 4 | 6 | 5.30e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 3 | 19 | 5.30e-01 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 3 | 13 | 5.31e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 5 | 11 | 5.31e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 5.31e-01 |
| GO:BP | GO:0048496 | maintenance of animal organ identity | 3 | 4 | 5.31e-01 |
| GO:BP | GO:1902897 | regulation of postsynaptic density protein 95 clustering | 1 | 2 | 5.31e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 18 | 5.31e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 1 | 54 | 5.31e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 4 | 12 | 5.31e-01 |
| GO:BP | GO:1902528 | regulation of protein linear polyubiquitination | 1 | 2 | 5.31e-01 |
| GO:BP | GO:1903382 | negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 5.31e-01 |
| GO:BP | GO:1905458 | positive regulation of lymphoid progenitor cell differentiation | 2 | 3 | 5.31e-01 |
| GO:BP | GO:1904192 | regulation of cholangiocyte apoptotic process | 1 | 1 | 5.31e-01 |
| GO:BP | GO:1904193 | negative regulation of cholangiocyte apoptotic process | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 1 | 67 | 5.31e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 40 | 1199 | 5.31e-01 |
| GO:BP | GO:1902530 | positive regulation of protein linear polyubiquitination | 1 | 2 | 5.31e-01 |
| GO:BP | GO:1903381 | regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 5.31e-01 |
| GO:BP | GO:1905281 | positive regulation of retrograde transport, endosome to Golgi | 1 | 2 | 5.31e-01 |
| GO:BP | GO:0009449 | gamma-aminobutyric acid biosynthetic process | 1 | 4 | 5.31e-01 |
| GO:BP | GO:1902488 | cholangiocyte apoptotic process | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0044057 | regulation of system process | 22 | 405 | 5.31e-01 |
| GO:BP | GO:0034352 | positive regulation of glial cell apoptotic process | 2 | 2 | 5.31e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 41 | 1209 | 5.31e-01 |
| GO:BP | GO:0060916 | mesenchymal cell proliferation involved in lung development | 4 | 5 | 5.31e-01 |
| GO:BP | GO:0060061 | Spemann organizer formation | 2 | 3 | 5.31e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 7 | 122 | 5.31e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 3 | 14 | 5.31e-01 |
| GO:BP | GO:1901166 | neural crest cell migration involved in autonomic nervous system development | 3 | 5 | 5.31e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 23 | 79 | 5.31e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 13 | 30 | 5.31e-01 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 7 | 26 | 5.31e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 1 | 10 | 5.31e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 3 | 18 | 5.31e-01 |
| GO:BP | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity | 1 | 10 | 5.31e-01 |
| GO:BP | GO:0021678 | third ventricle development | 3 | 4 | 5.31e-01 |
| GO:BP | GO:0050860 | negative regulation of T cell receptor signaling pathway | 1 | 17 | 5.32e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 1 | 21 | 5.32e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 5 | 11 | 5.32e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 6 | 8 | 5.32e-01 |
| GO:BP | GO:0009192 | deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 5.32e-01 |
| GO:BP | GO:0009184 | purine deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 5.32e-01 |
| GO:BP | GO:0046067 | dGDP catabolic process | 1 | 1 | 5.32e-01 |
| GO:BP | GO:1902269 | positive regulation of polyamine transmembrane transport | 2 | 2 | 5.32e-01 |
| GO:BP | GO:0046057 | dADP catabolic process | 1 | 1 | 5.32e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 14 | 88 | 5.32e-01 |
| GO:BP | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol | 1 | 3 | 5.32e-01 |
| GO:BP | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway | 2 | 5 | 5.32e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 2 | 53 | 5.32e-01 |
| GO:BP | GO:0036510 | trimming of terminal mannose on C branch | 1 | 3 | 5.32e-01 |
| GO:BP | GO:0018364 | peptidyl-glutamine methylation | 2 | 4 | 5.32e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 3 | 95 | 5.33e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 7 | 35 | 5.33e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 21 | 137 | 5.33e-01 |
| GO:BP | GO:0051394 | regulation of nerve growth factor receptor activity | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 1 | 75 | 5.33e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 13 | 22 | 5.33e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 50 | 1149 | 5.33e-01 |
| GO:BP | GO:0110017 | cap-independent translational initiation of linear mRNA | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 1 | 11 | 5.33e-01 |
| GO:BP | GO:0002192 | IRES-dependent translational initiation of linear mRNA | 1 | 1 | 5.33e-01 |
| GO:BP | GO:1904184 | positive regulation of pyruvate dehydrogenase activity | 1 | 3 | 5.33e-01 |
| GO:BP | GO:0060299 | negative regulation of sarcomere organization | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0038009 | regulation of signal transduction by receptor internalization | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 7 | 17 | 5.33e-01 |
| GO:BP | GO:1900826 | negative regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 1 | 5.33e-01 |
| GO:BP | GO:1902767 | isoprenoid biosynthetic process via mevalonate | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0019471 | 4-hydroxyproline metabolic process | 2 | 4 | 5.33e-01 |
| GO:BP | GO:0045730 | respiratory burst | 1 | 21 | 5.33e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 5.33e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 5.33e-01 |
| GO:BP | GO:0019470 | 4-hydroxyproline catabolic process | 2 | 4 | 5.33e-01 |
| GO:BP | GO:0035441 | cell migration involved in vasculogenesis | 3 | 4 | 5.34e-01 |
| GO:BP | GO:0003356 | regulation of cilium beat frequency | 1 | 10 | 5.34e-01 |
| GO:BP | GO:1901163 | regulation of trophoblast cell migration | 1 | 12 | 5.35e-01 |
| GO:BP | GO:1900244 | positive regulation of synaptic vesicle endocytosis | 1 | 4 | 5.35e-01 |
| GO:BP | GO:0032799 | low-density lipoprotein receptor particle metabolic process | 2 | 5 | 5.35e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 4 | 60 | 5.35e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 5 | 7 | 5.35e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 4 | 34 | 5.35e-01 |
| GO:BP | GO:0070158 | mitochondrial seryl-tRNA aminoacylation | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 7 | 57 | 5.35e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 5.35e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 5.35e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 6 | 17 | 5.35e-01 |
| GO:BP | GO:0060712 | spongiotrophoblast layer development | 5 | 11 | 5.35e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 5.35e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 5.35e-01 |
| GO:BP | GO:1990428 | miRNA transport | 2 | 3 | 5.35e-01 |
| GO:BP | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process | 2 | 5 | 5.35e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 5.35e-01 |
| GO:BP | GO:0032802 | low-density lipoprotein particle receptor catabolic process | 2 | 5 | 5.35e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 5.35e-01 |
| GO:BP | GO:0003065 | positive regulation of heart rate by epinephrine | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0070291 | N-acylethanolamine metabolic process | 1 | 4 | 5.35e-01 |
| GO:BP | GO:0120222 | regulation of blastocyst development | 1 | 3 | 5.35e-01 |
| GO:BP | GO:0070934 | CRD-mediated mRNA stabilization | 5 | 11 | 5.36e-01 |
| GO:BP | GO:0019441 | tryptophan catabolic process to kynurenine | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0006012 | galactose metabolic process | 2 | 9 | 5.36e-01 |
| GO:BP | GO:0006294 | nucleotide-excision repair, preincision complex assembly | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0048490 | anterograde synaptic vesicle transport | 2 | 16 | 5.36e-01 |
| GO:BP | GO:0099517 | synaptic vesicle transport along microtubule | 2 | 16 | 5.36e-01 |
| GO:BP | GO:0099514 | synaptic vesicle cytoskeletal transport | 2 | 16 | 5.36e-01 |
| GO:BP | GO:0043473 | pigmentation | 11 | 92 | 5.36e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 2 | 15 | 5.36e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 2 | 16 | 5.36e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 15 | 31 | 5.36e-01 |
| GO:BP | GO:0003058 | hormonal regulation of the force of heart contraction | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0009932 | cell tip growth | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0072356 | chromosome passenger complex localization to kinetochore | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0080058 | protein deglutathionylation | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 5 | 5.36e-01 |
| GO:BP | GO:0098711 | iron ion import across plasma membrane | 1 | 4 | 5.36e-01 |
| GO:BP | GO:0006565 | L-serine catabolic process | 2 | 4 | 5.36e-01 |
| GO:BP | GO:0048732 | gland development | 3 | 318 | 5.36e-01 |
| GO:BP | GO:1902256 | regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 3 | 10 | 5.36e-01 |
| GO:BP | GO:2000435 | negative regulation of protein neddylation | 2 | 4 | 5.36e-01 |
| GO:BP | GO:0060324 | face development | 3 | 45 | 5.36e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 7 | 783 | 5.36e-01 |
| GO:BP | GO:1903232 | melanosome assembly | 2 | 19 | 5.36e-01 |
| GO:BP | GO:0120168 | detection of hot stimulus involved in thermoception | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 4 | 5.36e-01 |
| GO:BP | GO:0099502 | calcium-dependent activation of synaptic vesicle fusion | 1 | 4 | 5.36e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 4 | 16 | 5.36e-01 |
| GO:BP | GO:0062036 | sensory perception of hot stimulus | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1901634 | positive regulation of synaptic vesicle membrane organization | 1 | 4 | 5.36e-01 |
| GO:BP | GO:0003275 | apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0018057 | peptidyl-lysine oxidation | 3 | 5 | 5.36e-01 |
| GO:BP | GO:0070544 | histone H3-K36 demethylation | 2 | 5 | 5.36e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 3 | 10 | 5.36e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 2 | 104 | 5.37e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 8 | 34 | 5.37e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 3 | 3 | 5.37e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 17 | 551 | 5.37e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 3 | 8 | 5.37e-01 |
| GO:BP | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis | 3 | 4 | 5.37e-01 |
| GO:BP | GO:2000793 | cell proliferation involved in heart valve development | 3 | 4 | 5.37e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 19 | 53 | 5.37e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 44 | 197 | 5.37e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 5.37e-01 |
| GO:BP | GO:2001288 | positive regulation of caveolin-mediated endocytosis | 2 | 3 | 5.37e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 2 | 3 | 5.37e-01 |
| GO:BP | GO:0006432 | phenylalanyl-tRNA aminoacylation | 1 | 4 | 5.37e-01 |
| GO:BP | GO:0000821 | regulation of arginine metabolic process | 2 | 3 | 5.37e-01 |
| GO:BP | GO:0060721 | regulation of spongiotrophoblast cell proliferation | 1 | 1 | 5.37e-01 |
| GO:BP | GO:0098657 | import into cell | 19 | 166 | 5.37e-01 |
| GO:BP | GO:0060723 | regulation of cell proliferation involved in embryonic placenta development | 1 | 1 | 5.37e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 2 | 39 | 5.37e-01 |
| GO:BP | GO:1905178 | regulation of cardiac muscle tissue regeneration | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 3 | 5 | 5.37e-01 |
| GO:BP | GO:1905179 | negative regulation of cardiac muscle tissue regeneration | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development | 2 | 2 | 5.37e-01 |
| GO:BP | GO:0071351 | cellular response to interleukin-18 | 2 | 3 | 5.37e-01 |
| GO:BP | GO:0060020 | Bergmann glial cell differentiation | 4 | 9 | 5.37e-01 |
| GO:BP | GO:0035655 | interleukin-18-mediated signaling pathway | 2 | 3 | 5.37e-01 |
| GO:BP | GO:0019860 | uracil metabolic process | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | 5 | 10 | 5.37e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 3 | 448 | 5.37e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 5.37e-01 |
| GO:BP | GO:0009056 | catabolic process | 38 | 2088 | 5.37e-01 |
| GO:BP | GO:0097384 | cellular lipid biosynthetic process | 2 | 10 | 5.37e-01 |
| GO:BP | GO:0008611 | ether lipid biosynthetic process | 2 | 10 | 5.37e-01 |
| GO:BP | GO:0046504 | glycerol ether biosynthetic process | 2 | 10 | 5.37e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 1 | 36 | 5.37e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 8 | 15 | 5.37e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 10 | 16 | 5.37e-01 |
| GO:BP | GO:0060510 | type II pneumocyte differentiation | 3 | 5 | 5.37e-01 |
| GO:BP | GO:0032213 | regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 11 | 120 | 5.37e-01 |
| GO:BP | GO:0032214 | negative regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 5.37e-01 |
| GO:BP | GO:1902416 | positive regulation of mRNA binding | 2 | 8 | 5.37e-01 |
| GO:BP | GO:1905838 | regulation of telomeric D-loop disassembly | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 19 | 272 | 5.37e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 7 | 31 | 5.37e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 5.37e-01 |
| GO:BP | GO:1905839 | negative regulation of telomeric D-loop disassembly | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 2 | 8 | 5.37e-01 |
| GO:BP | GO:0050955 | thermoception | 1 | 3 | 5.37e-01 |
| GO:BP | GO:1904534 | negative regulation of telomeric loop disassembly | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0016049 | cell growth | 26 | 418 | 5.37e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 9 | 41 | 5.37e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 5 | 10 | 5.37e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 2 | 14 | 5.37e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 5.37e-01 |
| GO:BP | GO:1905778 | negative regulation of exonuclease activity | 1 | 2 | 5.37e-01 |
| GO:BP | GO:1905777 | regulation of exonuclease activity | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 5.37e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 6 | 9 | 5.37e-01 |
| GO:BP | GO:0001787 | natural killer cell proliferation | 1 | 6 | 5.37e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 1 | 36 | 5.37e-01 |
| GO:BP | GO:1903765 | negative regulation of potassium ion export across plasma membrane | 1 | 1 | 5.37e-01 |
| GO:BP | GO:0061090 | positive regulation of sequestering of zinc ion | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 59 | 273 | 5.38e-01 |
| GO:BP | GO:2000644 | regulation of receptor catabolic process | 3 | 9 | 5.38e-01 |
| GO:BP | GO:0140915 | zinc ion import into zymogen granule | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0016050 | vesicle organization | 55 | 320 | 5.38e-01 |
| GO:BP | GO:0051295 | establishment of meiotic spindle localization | 1 | 4 | 5.38e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 12 | 25 | 5.38e-01 |
| GO:BP | GO:0140916 | zinc ion import into lysosome | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 5.38e-01 |
| GO:BP | GO:0140914 | zinc ion import into secretory vesicle | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0140917 | zinc ion import into mitochondrion | 1 | 1 | 5.38e-01 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 2 | 6 | 5.38e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 38 | 344 | 5.38e-01 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 1 | 9 | 5.38e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 14 | 53 | 5.38e-01 |
| GO:BP | GO:0002445 | type II hypersensitivity | 2 | 3 | 5.38e-01 |
| GO:BP | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib | 2 | 5 | 5.38e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 27 | 157 | 5.38e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 5.38e-01 |
| GO:BP | GO:0034091 | regulation of maintenance of sister chromatid cohesion | 1 | 8 | 5.38e-01 |
| GO:BP | GO:0034182 | regulation of maintenance of mitotic sister chromatid cohesion | 1 | 8 | 5.38e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 9 | 172 | 5.38e-01 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 5 | 10 | 5.38e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 4 | 71 | 5.38e-01 |
| GO:BP | GO:0001794 | type IIa hypersensitivity | 2 | 3 | 5.38e-01 |
| GO:BP | GO:0072584 | caveolin-mediated endocytosis | 5 | 9 | 5.38e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 10 | 28 | 5.38e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 12 | 20 | 5.38e-01 |
| GO:BP | GO:0060701 | negative regulation of ribonuclease activity | 1 | 2 | 5.38e-01 |
| GO:BP | GO:0060702 | negative regulation of endoribonuclease activity | 1 | 2 | 5.38e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 5 | 72 | 5.38e-01 |
| GO:BP | GO:0090206 | negative regulation of cholesterol metabolic process | 3 | 5 | 5.39e-01 |
| GO:BP | GO:0045541 | negative regulation of cholesterol biosynthetic process | 3 | 5 | 5.39e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 2 | 269 | 5.39e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 4 | 51 | 5.39e-01 |
| GO:BP | GO:0106119 | negative regulation of sterol biosynthetic process | 3 | 5 | 5.39e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 11 | 228 | 5.39e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 5 | 85 | 5.39e-01 |
| GO:BP | GO:0009617 | response to bacterium | 5 | 321 | 5.39e-01 |
| GO:BP | GO:0072236 | metanephric loop of Henle development | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 5.39e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 22 | 80 | 5.40e-01 |
| GO:BP | GO:0038203 | TORC2 signaling | 3 | 12 | 5.40e-01 |
| GO:BP | GO:1905454 | negative regulation of myeloid progenitor cell differentiation | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 9 | 97 | 5.40e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 3 | 24 | 5.40e-01 |
| GO:BP | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate | 2 | 4 | 5.40e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 2 | 7 | 5.40e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 5.40e-01 |
| GO:BP | GO:0021849 | neuroblast division in subventricular zone | 1 | 3 | 5.40e-01 |
| GO:BP | GO:1903860 | negative regulation of dendrite extension | 2 | 2 | 5.41e-01 |
| GO:BP | GO:0019408 | dolichol biosynthetic process | 1 | 2 | 5.41e-01 |
| GO:BP | GO:0060596 | mammary placode formation | 2 | 2 | 5.41e-01 |
| GO:BP | GO:0003013 | circulatory system process | 3 | 447 | 5.41e-01 |
| GO:BP | GO:0003167 | atrioventricular bundle cell differentiation | 2 | 3 | 5.41e-01 |
| GO:BP | GO:1905072 | cardiac jelly development | 2 | 3 | 5.41e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 6 | 97 | 5.41e-01 |
| GO:BP | GO:1903910 | negative regulation of receptor clustering | 1 | 2 | 5.41e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 5.41e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 5.41e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 5.41e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 1 | 107 | 5.41e-01 |
| GO:BP | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome | 1 | 1 | 5.42e-01 |
| GO:BP | GO:0006884 | cell volume homeostasis | 5 | 24 | 5.42e-01 |
| GO:BP | GO:0070836 | caveola assembly | 2 | 4 | 5.42e-01 |
| GO:BP | GO:1904556 | L-tryptophan transmembrane transport | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 5.42e-01 |
| GO:BP | GO:0071348 | cellular response to interleukin-11 | 1 | 1 | 5.42e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 5.42e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 8 | 31 | 5.42e-01 |
| GO:BP | GO:0003332 | negative regulation of extracellular matrix constituent secretion | 2 | 2 | 5.42e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 2 | 7 | 5.42e-01 |
| GO:BP | GO:0048666 | neuron development | 2 | 877 | 5.42e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 1 | 28 | 5.42e-01 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 3 | 3 | 5.42e-01 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 3 | 3 | 5.42e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 16 | 32 | 5.42e-01 |
| GO:BP | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process | 3 | 4 | 5.42e-01 |
| GO:BP | GO:1905040 | otic placode development | 1 | 1 | 5.42e-01 |
| GO:BP | GO:2000553 | positive regulation of T-helper 2 cell cytokine production | 1 | 3 | 5.42e-01 |
| GO:BP | GO:0060301 | positive regulation of cytokine activity | 1 | 1 | 5.42e-01 |
| GO:BP | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 1 | 3 | 5.42e-01 |
| GO:BP | GO:1902378 | VEGF-activated neuropilin signaling pathway involved in axon guidance | 1 | 1 | 5.42e-01 |
| GO:BP | GO:1990961 | xenobiotic detoxification by transmembrane export across the plasma membrane | 4 | 7 | 5.42e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 10 | 52 | 5.42e-01 |
| GO:BP | GO:0060527 | prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis | 1 | 3 | 5.42e-01 |
| GO:BP | GO:1904829 | regulation of aortic smooth muscle cell differentiation | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0032765 | positive regulation of mast cell cytokine production | 2 | 3 | 5.42e-01 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 1 | 16 | 5.42e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 42 | 96 | 5.42e-01 |
| GO:BP | GO:0061450 | trophoblast cell migration | 1 | 13 | 5.42e-01 |
| GO:BP | GO:0071036 | nuclear polyadenylation-dependent snoRNA catabolic process | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0061847 | response to cholecystokinin | 1 | 1 | 5.42e-01 |
| GO:BP | GO:1903800 | positive regulation of miRNA processing | 2 | 3 | 5.42e-01 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 2 | 7 | 5.42e-01 |
| GO:BP | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0014906 | myotube cell development involved in skeletal muscle regeneration | 1 | 1 | 5.42e-01 |
| GO:BP | GO:0006936 | muscle contraction | 14 | 267 | 5.42e-01 |
| GO:BP | GO:1905302 | negative regulation of macropinocytosis | 1 | 1 | 5.42e-01 |
| GO:BP | GO:0034756 | regulation of iron ion transport | 2 | 4 | 5.42e-01 |
| GO:BP | GO:1990859 | cellular response to endothelin | 1 | 1 | 5.42e-01 |
| GO:BP | GO:1904050 | positive regulation of spontaneous neurotransmitter secretion | 1 | 1 | 5.42e-01 |
| GO:BP | GO:0099039 | sphingolipid translocation | 3 | 4 | 5.42e-01 |
| GO:BP | GO:0001824 | blastocyst development | 1 | 94 | 5.42e-01 |
| GO:BP | GO:0035887 | aortic smooth muscle cell differentiation | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 5.42e-01 |
| GO:BP | GO:0071037 | nuclear polyadenylation-dependent snRNA catabolic process | 1 | 2 | 5.42e-01 |
| GO:BP | GO:1902866 | regulation of retina development in camera-type eye | 1 | 1 | 5.42e-01 |
| GO:BP | GO:1904831 | positive regulation of aortic smooth muscle cell differentiation | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0016255 | attachment of GPI anchor to protein | 3 | 6 | 5.42e-01 |
| GO:BP | GO:0060526 | prostate glandular acinus morphogenesis | 1 | 3 | 5.42e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 1 | 222 | 5.42e-01 |
| GO:BP | GO:1904515 | positive regulation of TORC2 signaling | 3 | 4 | 5.42e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 3 | 11 | 5.43e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 5.43e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 5 | 14 | 5.43e-01 |
| GO:BP | GO:0021501 | prechordal plate formation | 1 | 1 | 5.43e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 3 | 4 | 5.43e-01 |
| GO:BP | GO:0034196 | acylglycerol transport | 1 | 4 | 5.43e-01 |
| GO:BP | GO:0034197 | triglyceride transport | 1 | 4 | 5.43e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 2 | 3 | 5.43e-01 |
| GO:BP | GO:0021547 | midbrain-hindbrain boundary initiation | 1 | 1 | 5.43e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 5.43e-01 |
| GO:BP | GO:0032423 | regulation of mismatch repair | 3 | 3 | 5.43e-01 |
| GO:BP | GO:0060398 | regulation of growth hormone receptor signaling pathway | 1 | 5 | 5.43e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 3 | 4 | 5.43e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 3 | 4 | 5.43e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 1 | 30 | 5.43e-01 |
| GO:BP | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway | 2 | 2 | 5.43e-01 |
| GO:BP | GO:0044828 | negative regulation by host of viral genome replication | 2 | 7 | 5.43e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 12 | 142 | 5.44e-01 |
| GO:BP | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion | 2 | 3 | 5.44e-01 |
| GO:BP | GO:1904645 | response to amyloid-beta | 7 | 32 | 5.44e-01 |
| GO:BP | GO:0001705 | ectoderm formation | 2 | 3 | 5.44e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 14 | 20 | 5.44e-01 |
| GO:BP | GO:0090386 | phagosome maturation involved in apoptotic cell clearance | 1 | 2 | 5.44e-01 |
| GO:BP | GO:0060442 | branching involved in prostate gland morphogenesis | 2 | 7 | 5.44e-01 |
| GO:BP | GO:0090387 | phagolysosome assembly involved in apoptotic cell clearance | 1 | 2 | 5.44e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 4 | 6 | 5.44e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 2 | 22 | 5.44e-01 |
| GO:BP | GO:1904998 | negative regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0045338 | farnesyl diphosphate metabolic process | 2 | 5 | 5.44e-01 |
| GO:BP | GO:0032775 | DNA methylation on adenine | 1 | 2 | 5.44e-01 |
| GO:BP | GO:0018872 | arsonoacetate metabolic process | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 11 | 20 | 5.44e-01 |
| GO:BP | GO:0032967 | positive regulation of collagen biosynthetic process | 3 | 23 | 5.44e-01 |
| GO:BP | GO:1904884 | positive regulation of telomerase catalytic core complex assembly | 1 | 2 | 5.45e-01 |
| GO:BP | GO:1904868 | telomerase catalytic core complex assembly | 1 | 2 | 5.45e-01 |
| GO:BP | GO:1904882 | regulation of telomerase catalytic core complex assembly | 1 | 2 | 5.45e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 4 | 11 | 5.45e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 3 | 5 | 5.45e-01 |
| GO:BP | GO:1901307 | positive regulation of spermidine biosynthetic process | 1 | 1 | 5.45e-01 |
| GO:BP | GO:1901304 | regulation of spermidine biosynthetic process | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 6 | 9 | 5.45e-01 |
| GO:BP | GO:1905605 | positive regulation of blood-brain barrier permeability | 1 | 4 | 5.45e-01 |
| GO:BP | GO:0043508 | negative regulation of JUN kinase activity | 5 | 13 | 5.45e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 8 | 47 | 5.45e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 5.45e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 2 | 61 | 5.45e-01 |
| GO:BP | GO:0034030 | ribonucleoside bisphosphate biosynthetic process | 29 | 49 | 5.46e-01 |
| GO:BP | GO:0034033 | purine nucleoside bisphosphate biosynthetic process | 29 | 49 | 5.46e-01 |
| GO:BP | GO:1903785 | L-valine transmembrane transport | 1 | 1 | 5.46e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 2 | 78 | 5.46e-01 |
| GO:BP | GO:0033866 | nucleoside bisphosphate biosynthetic process | 29 | 49 | 5.46e-01 |
| GO:BP | GO:0040007 | growth | 46 | 742 | 5.46e-01 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 5 | 12 | 5.46e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 6 | 32 | 5.46e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 1 | 227 | 5.46e-01 |
| GO:BP | GO:0097484 | dendrite extension | 19 | 27 | 5.46e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 1 | 10 | 5.46e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 11 | 19 | 5.46e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 5 | 13 | 5.46e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 11 | 19 | 5.46e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 11 | 19 | 5.46e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 2 | 279 | 5.46e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 18 | 106 | 5.46e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 2 | 293 | 5.47e-01 |
| GO:BP | GO:0061087 | positive regulation of histone H3-K27 methylation | 1 | 4 | 5.47e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 2 | 26 | 5.47e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 5.47e-01 |
| GO:BP | GO:0070408 | carbamoyl phosphate metabolic process | 1 | 1 | 5.47e-01 |
| GO:BP | GO:0070409 | carbamoyl phosphate biosynthetic process | 1 | 1 | 5.47e-01 |
| GO:BP | GO:0051352 | negative regulation of ligase activity | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 2 | 22 | 5.47e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 14 | 34 | 5.47e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 2 | 22 | 5.47e-01 |
| GO:BP | GO:0015707 | nitrite transport | 1 | 1 | 5.47e-01 |
| GO:BP | GO:0034263 | positive regulation of autophagy in response to ER overload | 1 | 1 | 5.47e-01 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 1 | 14 | 5.47e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 6 | 26 | 5.47e-01 |
| GO:BP | GO:0021793 | chemorepulsion of branchiomotor axon | 1 | 1 | 5.47e-01 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 5.47e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 5 | 9 | 5.47e-01 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 7 | 17 | 5.47e-01 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 4 | 8 | 5.47e-01 |
| GO:BP | GO:0001955 | blood vessel maturation | 2 | 6 | 5.47e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 8 | 68 | 5.47e-01 |
| GO:BP | GO:1990262 | anti-Mullerian hormone signaling pathway | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0009256 | 10-formyltetrahydrofolate metabolic process | 1 | 6 | 5.47e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 15 | 56 | 5.47e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 3 | 24 | 5.47e-01 |
| GO:BP | GO:0001503 | ossification | 1 | 313 | 5.47e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 1 | 13 | 5.47e-01 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 3 | 4 | 5.48e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 8 | 43 | 5.48e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 3 | 69 | 5.48e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 3 | 6 | 5.48e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 3 | 17 | 5.48e-01 |
| GO:BP | GO:0043045 | DNA methylation involved in embryo development | 1 | 7 | 5.49e-01 |
| GO:BP | GO:1904529 | regulation of actin filament binding | 1 | 3 | 5.49e-01 |
| GO:BP | GO:1904426 | positive regulation of GTP binding | 1 | 3 | 5.49e-01 |
| GO:BP | GO:0008306 | associative learning | 2 | 65 | 5.49e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 3 | 80 | 5.49e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 5.49e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 4 | 748 | 5.49e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 5.49e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 6 | 10 | 5.49e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 3 | 12 | 5.49e-01 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 6 | 12 | 5.49e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 5.50e-01 |
| GO:BP | GO:0010637 | negative regulation of mitochondrial fusion | 3 | 8 | 5.50e-01 |
| GO:BP | GO:2000655 | negative regulation of cellular response to testosterone stimulus | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0006549 | isoleucine metabolic process | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0048865 | stem cell fate commitment | 3 | 5 | 5.50e-01 |
| GO:BP | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0001545 | primary ovarian follicle growth | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0051258 | protein polymerization | 2 | 241 | 5.50e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 5.50e-01 |
| GO:BP | GO:0009972 | cytidine deamination | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0046087 | cytidine metabolic process | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 5.50e-01 |
| GO:BP | GO:0006216 | cytidine catabolic process | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0016554 | cytidine to uridine editing | 1 | 6 | 5.50e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 5.50e-01 |
| GO:BP | GO:0036153 | triglyceride acyl-chain remodeling | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0090240 | positive regulation of histone H4 acetylation | 3 | 6 | 5.50e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 2 | 60 | 5.50e-01 |
| GO:BP | GO:0097378 | dorsal spinal cord interneuron axon guidance | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0071874 | cellular response to norepinephrine stimulus | 1 | 2 | 5.50e-01 |
| GO:BP | GO:0015816 | glycine transport | 1 | 9 | 5.50e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 9 | 38 | 5.50e-01 |
| GO:BP | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 7 | 5.50e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 5 | 15 | 5.50e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 10 | 33 | 5.50e-01 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 1 | 14 | 5.50e-01 |
| GO:BP | GO:0044058 | regulation of digestive system process | 4 | 22 | 5.50e-01 |
| GO:BP | GO:0071873 | response to norepinephrine | 1 | 2 | 5.50e-01 |
| GO:BP | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly | 1 | 7 | 5.50e-01 |
| GO:BP | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process | 3 | 5 | 5.50e-01 |
| GO:BP | GO:1904382 | mannose trimming involved in glycoprotein ERAD pathway | 3 | 5 | 5.50e-01 |
| GO:BP | GO:0097466 | ubiquitin-dependent glycoprotein ERAD pathway | 3 | 5 | 5.50e-01 |
| GO:BP | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0008406 | gonad development | 6 | 158 | 5.51e-01 |
| GO:BP | GO:0042350 | GDP-L-fucose biosynthetic process | 2 | 3 | 5.51e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 5 | 26 | 5.51e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 2 | 14 | 5.51e-01 |
| GO:BP | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 1 | 56 | 5.51e-01 |
| GO:BP | GO:0140454 | protein aggregate center assembly | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 3 | 22 | 5.51e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 3 | 14 | 5.51e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 2 | 4 | 5.51e-01 |
| GO:BP | GO:0051597 | response to methylmercury | 1 | 5 | 5.51e-01 |
| GO:BP | GO:0042074 | cell migration involved in gastrulation | 1 | 10 | 5.51e-01 |
| GO:BP | GO:1904434 | positive regulation of ferrous iron binding | 1 | 2 | 5.51e-01 |
| GO:BP | GO:1904435 | regulation of transferrin receptor binding | 1 | 2 | 5.51e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 5.51e-01 |
| GO:BP | GO:1904437 | positive regulation of transferrin receptor binding | 1 | 2 | 5.51e-01 |
| GO:BP | GO:0036075 | replacement ossification | 4 | 25 | 5.51e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 10 | 5.51e-01 |
| GO:BP | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process | 4 | 5 | 5.51e-01 |
| GO:BP | GO:1904432 | regulation of ferrous iron binding | 1 | 2 | 5.51e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 5.51e-01 |
| GO:BP | GO:0021541 | ammon gyrus development | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 1 | 8 | 5.51e-01 |
| GO:BP | GO:0032286 | central nervous system myelin maintenance | 2 | 3 | 5.51e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 4 | 25 | 5.51e-01 |
| GO:BP | GO:0106048 | spermidine deacetylation | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0106047 | polyamine deacetylation | 1 | 1 | 5.51e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 1 | 114 | 5.51e-01 |
| GO:BP | GO:1904851 | positive regulation of establishment of protein localization to telomere | 3 | 10 | 5.51e-01 |
| GO:BP | GO:0044321 | response to leptin | 2 | 19 | 5.51e-01 |
| GO:BP | GO:0097492 | sympathetic neuron axon guidance | 3 | 4 | 5.51e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 4 | 56 | 5.51e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 4 | 56 | 5.51e-01 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 5 | 17 | 5.51e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 3 | 8 | 5.51e-01 |
| GO:BP | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 2 | 5.51e-01 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 1 | 5 | 5.51e-01 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 1 | 7 | 5.51e-01 |
| GO:BP | GO:0010447 | response to acidic pH | 2 | 16 | 5.52e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 5.52e-01 |
| GO:BP | GO:0032610 | interleukin-1 alpha production | 1 | 4 | 5.52e-01 |
| GO:BP | GO:1900063 | regulation of peroxisome organization | 1 | 4 | 5.52e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 6 | 40 | 5.52e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 2 | 32 | 5.52e-01 |
| GO:BP | GO:0030327 | prenylated protein catabolic process | 1 | 3 | 5.52e-01 |
| GO:BP | GO:0032650 | regulation of interleukin-1 alpha production | 1 | 4 | 5.52e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 31 | 1219 | 5.52e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 4 | 25 | 5.52e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 8 | 47 | 5.52e-01 |
| GO:BP | GO:2000822 | regulation of behavioral fear response | 1 | 7 | 5.52e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 37 | 140 | 5.52e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 10 | 16 | 5.52e-01 |
| GO:BP | GO:0050923 | regulation of negative chemotaxis | 2 | 4 | 5.52e-01 |
| GO:BP | GO:1902480 | protein localization to mitotic spindle | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 4 | 25 | 5.52e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 2 | 104 | 5.52e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 8 | 51 | 5.52e-01 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 5.52e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 5.52e-01 |
| GO:BP | GO:0036255 | response to methylamine | 1 | 1 | 5.52e-01 |
| GO:BP | GO:1903442 | response to lipoic acid | 1 | 1 | 5.52e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 3 | 9 | 5.52e-01 |
| GO:BP | GO:1903423 | positive regulation of synaptic vesicle recycling | 3 | 6 | 5.52e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 4 | 71 | 5.52e-01 |
| GO:BP | GO:0060353 | regulation of cell adhesion molecule production | 3 | 6 | 5.52e-01 |
| GO:BP | GO:0000056 | ribosomal small subunit export from nucleus | 1 | 8 | 5.52e-01 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 8 | 19 | 5.54e-01 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 4 | 21 | 5.54e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 26 | 81 | 5.54e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 3 | 18 | 5.54e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 1 | 102 | 5.54e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 18 | 67 | 5.54e-01 |
| GO:BP | GO:0014805 | smooth muscle adaptation | 1 | 3 | 5.55e-01 |
| GO:BP | GO:0003062 | regulation of heart rate by chemical signal | 3 | 6 | 5.55e-01 |
| GO:BP | GO:0038130 | ERBB4 signaling pathway | 4 | 6 | 5.56e-01 |
| GO:BP | GO:0003150 | muscular septum morphogenesis | 1 | 4 | 5.56e-01 |
| GO:BP | GO:0048560 | establishment of anatomical structure orientation | 2 | 2 | 5.56e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 41 | 73 | 5.56e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 3 | 16 | 5.56e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 16 | 38 | 5.56e-01 |
| GO:BP | GO:0071602 | phytosphingosine biosynthetic process | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 4 | 35 | 5.56e-01 |
| GO:BP | GO:0010021 | amylopectin biosynthetic process | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0006671 | phytosphingosine metabolic process | 1 | 1 | 5.56e-01 |
| GO:BP | GO:2000896 | amylopectin metabolic process | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 1 | 52 | 5.56e-01 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 1 | 4 | 5.56e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 2 | 57 | 5.56e-01 |
| GO:BP | GO:0098694 | regulation of synaptic vesicle budding from presynaptic endocytic zone membrane | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 3 | 13 | 5.56e-01 |
| GO:BP | GO:0060073 | micturition | 1 | 2 | 5.56e-01 |
| GO:BP | GO:0045010 | actin nucleation | 1 | 50 | 5.57e-01 |
| GO:BP | GO:0072144 | glomerular mesangial cell development | 2 | 4 | 5.57e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 3 | 35 | 5.57e-01 |
| GO:BP | GO:0042592 | homeostatic process | 7 | 1159 | 5.57e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 2 | 4 | 5.57e-01 |
| GO:BP | GO:2001204 | regulation of osteoclast development | 1 | 4 | 5.57e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 5.57e-01 |
| GO:BP | GO:0007386 | compartment pattern specification | 2 | 5 | 5.57e-01 |
| GO:BP | GO:0090209 | negative regulation of triglyceride metabolic process | 1 | 7 | 5.57e-01 |
| GO:BP | GO:0006102 | isocitrate metabolic process | 4 | 6 | 5.57e-01 |
| GO:BP | GO:2000234 | positive regulation of rRNA processing | 5 | 10 | 5.57e-01 |
| GO:BP | GO:0048680 | positive regulation of axon regeneration | 1 | 8 | 5.57e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 5.58e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 1 | 68 | 5.58e-01 |
| GO:BP | GO:2000635 | negative regulation of primary miRNA processing | 2 | 2 | 5.58e-01 |
| GO:BP | GO:2000634 | regulation of primary miRNA processing | 2 | 2 | 5.58e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 1 | 14 | 5.58e-01 |
| GO:BP | GO:0099628 | neurotransmitter receptor diffusion trapping | 1 | 4 | 5.58e-01 |
| GO:BP | GO:0098953 | receptor diffusion trapping | 1 | 4 | 5.58e-01 |
| GO:BP | GO:0098970 | postsynaptic neurotransmitter receptor diffusion trapping | 1 | 4 | 5.58e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 1 | 33 | 5.58e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 5.58e-01 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 5 | 8 | 5.58e-01 |
| GO:BP | GO:0070084 | protein initiator methionine removal | 2 | 4 | 5.58e-01 |
| GO:BP | GO:1903649 | regulation of cytoplasmic transport | 13 | 25 | 5.58e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 14 | 36 | 5.58e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 6 | 52 | 5.58e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 1 | 38 | 5.58e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 17 | 96 | 5.58e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 1 | 23 | 5.58e-01 |
| GO:BP | GO:0003162 | atrioventricular node development | 5 | 8 | 5.58e-01 |
| GO:BP | GO:0010722 | regulation of ferrochelatase activity | 1 | 1 | 5.58e-01 |
| GO:BP | GO:1901523 | icosanoid catabolic process | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0003334 | keratinocyte development | 1 | 8 | 5.58e-01 |
| GO:BP | GO:0099178 | regulation of retrograde trans-synaptic signaling by endocanabinoid | 2 | 3 | 5.58e-01 |
| GO:BP | GO:1904797 | negative regulation of core promoter binding | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 8 | 24 | 5.59e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 4 | 7 | 5.59e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 10 | 22 | 5.59e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 6 | 20 | 5.59e-01 |
| GO:BP | GO:0070778 | L-aspartate transmembrane transport | 2 | 11 | 5.59e-01 |
| GO:BP | GO:0010635 | regulation of mitochondrial fusion | 4 | 11 | 5.59e-01 |
| GO:BP | GO:0051490 | negative regulation of filopodium assembly | 4 | 6 | 5.59e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 3 | 1278 | 5.59e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 2 | 22 | 5.59e-01 |
| GO:BP | GO:0032480 | negative regulation of type I interferon production | 10 | 23 | 5.59e-01 |
| GO:BP | GO:0035212 | cell competition in a multicellular organism | 1 | 1 | 5.59e-01 |
| GO:BP | GO:1903527 | positive regulation of membrane tubulation | 3 | 3 | 5.59e-01 |
| GO:BP | GO:1903945 | positive regulation of hepatocyte apoptotic process | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0060065 | uterus development | 5 | 13 | 5.59e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 5 | 104 | 5.59e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 11 | 68 | 5.59e-01 |
| GO:BP | GO:0010714 | positive regulation of collagen metabolic process | 3 | 24 | 5.59e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 1 | 67 | 5.59e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 6 | 22 | 5.59e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 79 | 5.59e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 5.59e-01 |
| GO:BP | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway | 2 | 2 | 5.59e-01 |
| GO:BP | GO:0051350 | negative regulation of lyase activity | 3 | 15 | 5.59e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 5.59e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 1 | 15 | 5.59e-01 |
| GO:BP | GO:1902524 | positive regulation of protein K48-linked ubiquitination | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0051391 | tRNA acetylation | 1 | 1 | 5.60e-01 |
| GO:BP | GO:1990882 | rRNA acetylation | 1 | 1 | 5.60e-01 |
| GO:BP | GO:1904812 | rRNA acetylation involved in maturation of SSU-rRNA | 1 | 1 | 5.60e-01 |
| GO:BP | GO:1990884 | RNA acetylation | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 16 | 5.60e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 7 | 54 | 5.60e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 5 | 8 | 5.60e-01 |
| GO:BP | GO:0150077 | regulation of neuroinflammatory response | 1 | 13 | 5.60e-01 |
| GO:BP | GO:0070842 | aggresome assembly | 2 | 5 | 5.60e-01 |
| GO:BP | GO:0061031 | endodermal digestive tract morphogenesis | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 5.61e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 2 | 11 | 5.61e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 5.61e-01 |
| GO:BP | GO:2000388 | positive regulation of antral ovarian follicle growth | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0031000 | response to caffeine | 2 | 13 | 5.61e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 1 | 86 | 5.61e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 1 | 252 | 5.61e-01 |
| GO:BP | GO:1901142 | insulin metabolic process | 3 | 11 | 5.61e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 4 | 49 | 5.61e-01 |
| GO:BP | GO:2000387 | regulation of antral ovarian follicle growth | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0001809 | positive regulation of type IV hypersensitivity | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane | 12 | 19 | 5.61e-01 |
| GO:BP | GO:0002337 | B-1a B cell differentiation | 1 | 2 | 5.62e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 2 | 20 | 5.62e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 5.62e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 5.62e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 1 | 7 | 5.62e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 1 | 7 | 5.62e-01 |
| GO:BP | GO:0036506 | maintenance of unfolded protein | 2 | 3 | 5.62e-01 |
| GO:BP | GO:1901610 | positive regulation of vesicle transport along microtubule | 3 | 3 | 5.62e-01 |
| GO:BP | GO:0097105 | presynaptic membrane assembly | 4 | 8 | 5.62e-01 |
| GO:BP | GO:1904378 | maintenance of unfolded protein involved in ERAD pathway | 2 | 3 | 5.62e-01 |
| GO:BP | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation | 2 | 3 | 5.62e-01 |
| GO:BP | GO:0031129 | inductive cell-cell signaling | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 5.62e-01 |
| GO:BP | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation | 2 | 2 | 5.62e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 5.63e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 2 | 19 | 5.63e-01 |
| GO:BP | GO:1902426 | deactivation of mitotic spindle assembly checkpoint | 2 | 3 | 5.64e-01 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 3 | 5.64e-01 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 3 | 5.64e-01 |
| GO:BP | GO:0051643 | endoplasmic reticulum localization | 1 | 8 | 5.64e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 8 | 21 | 5.64e-01 |
| GO:BP | GO:1905795 | cellular response to puromycin | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 6 | 161 | 5.65e-01 |
| GO:BP | GO:1905794 | response to puromycin | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway | 1 | 2 | 5.65e-01 |
| GO:BP | GO:0035669 | TRAM-dependent toll-like receptor 4 signaling pathway | 1 | 2 | 5.65e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 8 | 84 | 5.65e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 3 | 29 | 5.65e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1081 | 5.65e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 30 | 114 | 5.65e-01 |
| GO:BP | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway | 1 | 2 | 5.65e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 23 | 4245 | 5.65e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 16 | 59 | 5.66e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 1 | 31 | 5.66e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 5.66e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 3 | 22 | 5.66e-01 |
| GO:BP | GO:0042551 | neuron maturation | 1 | 33 | 5.66e-01 |
| GO:BP | GO:0045006 | DNA deamination | 1 | 9 | 5.66e-01 |
| GO:BP | GO:0035750 | protein localization to myelin sheath abaxonal region | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 5.66e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 5.66e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0046068 | cGMP metabolic process | 1 | 11 | 5.66e-01 |
| GO:BP | GO:0060515 | prostate field specification | 1 | 1 | 5.66e-01 |
| GO:BP | GO:1905223 | epicardium morphogenesis | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0060520 | activation of prostate induction by androgen receptor signaling pathway | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0060514 | prostate induction | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0010980 | positive regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 6 | 23 | 5.66e-01 |
| GO:BP | GO:0010979 | regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0070779 | D-aspartate import across plasma membrane | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0070777 | D-aspartate transport | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 8 | 16 | 5.66e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 6 | 40 | 5.66e-01 |
| GO:BP | GO:1904055 | negative regulation of cholangiocyte proliferation | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0019102 | male somatic sex determination | 1 | 1 | 5.66e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 3 | 7 | 5.66e-01 |
| GO:BP | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway | 3 | 17 | 5.66e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1 | 24 | 5.66e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 1 | 24 | 5.66e-01 |
| GO:BP | GO:0060267 | positive regulation of respiratory burst | 2 | 3 | 5.66e-01 |
| GO:BP | GO:0010511 | regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 302 | 5.66e-01 |
| GO:BP | GO:0035788 | cell migration involved in metanephros development | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 6 | 14 | 5.66e-01 |
| GO:BP | GO:0019042 | viral latency | 1 | 4 | 5.67e-01 |
| GO:BP | GO:0045186 | zonula adherens assembly | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0035385 | Roundabout signaling pathway | 2 | 5 | 5.67e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 1 | 112 | 5.67e-01 |
| GO:BP | GO:0061301 | cerebellum vasculature morphogenesis | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0040010 | positive regulation of growth rate | 2 | 2 | 5.67e-01 |
| GO:BP | GO:0051458 | corticotropin secretion | 1 | 4 | 5.67e-01 |
| GO:BP | GO:0098935 | dendritic transport | 6 | 13 | 5.68e-01 |
| GO:BP | GO:0000050 | urea cycle | 4 | 7 | 5.68e-01 |
| GO:BP | GO:0015825 | L-serine transport | 1 | 7 | 5.68e-01 |
| GO:BP | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 2 | 5 | 5.68e-01 |
| GO:BP | GO:0031118 | rRNA pseudouridine synthesis | 3 | 10 | 5.69e-01 |
| GO:BP | GO:0090219 | negative regulation of lipid kinase activity | 3 | 8 | 5.70e-01 |
| GO:BP | GO:0032058 | positive regulation of translational initiation in response to stress | 1 | 4 | 5.70e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 19 | 1111 | 5.70e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 3 | 29 | 5.70e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 13 | 73 | 5.70e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 7 | 20 | 5.70e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 9 | 23 | 5.70e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 5.70e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 2 | 18 | 5.70e-01 |
| GO:BP | GO:0035589 | G protein-coupled purinergic nucleotide receptor signaling pathway | 2 | 4 | 5.70e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 5 | 17 | 5.70e-01 |
| GO:BP | GO:0001805 | positive regulation of type III hypersensitivity | 1 | 2 | 5.70e-01 |
| GO:BP | GO:0001803 | regulation of type III hypersensitivity | 1 | 2 | 5.70e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 13 | 73 | 5.70e-01 |
| GO:BP | GO:0001802 | type III hypersensitivity | 1 | 2 | 5.70e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 1 | 84 | 5.70e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 5 | 18 | 5.70e-01 |
| GO:BP | GO:0033023 | mast cell homeostasis | 1 | 4 | 5.70e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 5 | 106 | 5.70e-01 |
| GO:BP | GO:1902889 | protein localization to spindle microtubule | 1 | 2 | 5.70e-01 |
| GO:BP | GO:1905755 | protein localization to cytoplasmic microtubule | 1 | 2 | 5.70e-01 |
| GO:BP | GO:0051031 | tRNA transport | 3 | 5 | 5.70e-01 |
| GO:BP | GO:0010166 | wax metabolic process | 2 | 2 | 5.70e-01 |
| GO:BP | GO:0010025 | wax biosynthetic process | 2 | 2 | 5.70e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 2 | 9 | 5.70e-01 |
| GO:BP | GO:0032098 | regulation of appetite | 1 | 8 | 5.70e-01 |
| GO:BP | GO:0098779 | positive regulation of mitophagy in response to mitochondrial depolarization | 4 | 10 | 5.71e-01 |
| GO:BP | GO:0002315 | marginal zone B cell differentiation | 1 | 6 | 5.71e-01 |
| GO:BP | GO:0097067 | cellular response to thyroid hormone stimulus | 8 | 14 | 5.71e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 1 | 6 | 5.71e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 1 | 233 | 5.71e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 3 | 76 | 5.71e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 2 | 30 | 5.71e-01 |
| GO:BP | GO:0016031 | tRNA import into mitochondrion | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0071315 | cellular response to morphine | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 2 | 39 | 5.71e-01 |
| GO:BP | GO:0071317 | cellular response to isoquinoline alkaloid | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly | 1 | 3 | 5.71e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 7 | 130 | 5.72e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 9 | 113 | 5.72e-01 |
| GO:BP | GO:0045347 | negative regulation of MHC class II biosynthetic process | 2 | 3 | 5.72e-01 |
| GO:BP | GO:0071374 | cellular response to parathyroid hormone stimulus | 1 | 7 | 5.72e-01 |
| GO:BP | GO:0061517 | macrophage proliferation | 1 | 7 | 5.72e-01 |
| GO:BP | GO:0120179 | adherens junction disassembly | 1 | 1 | 5.72e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 30 | 51 | 5.72e-01 |
| GO:BP | GO:0002158 | osteoclast proliferation | 1 | 7 | 5.72e-01 |
| GO:BP | GO:0042487 | regulation of odontogenesis of dentin-containing tooth | 1 | 6 | 5.72e-01 |
| GO:BP | GO:0044860 | protein localization to plasma membrane raft | 1 | 4 | 5.72e-01 |
| GO:BP | GO:0007548 | sex differentiation | 7 | 191 | 5.72e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 2 | 26 | 5.72e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 3 | 9 | 5.72e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 10 | 276 | 5.72e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 4 | 21 | 5.72e-01 |
| GO:BP | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway | 7 | 9 | 5.72e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 1 | 41 | 5.72e-01 |
| GO:BP | GO:1903365 | regulation of fear response | 1 | 8 | 5.72e-01 |
| GO:BP | GO:0006574 | valine catabolic process | 2 | 4 | 5.72e-01 |
| GO:BP | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity | 1 | 1 | 5.73e-01 |
| GO:BP | GO:1901340 | negative regulation of store-operated calcium channel activity | 1 | 1 | 5.73e-01 |
| GO:BP | GO:0032287 | peripheral nervous system myelin maintenance | 5 | 8 | 5.73e-01 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 1 | 6 | 5.73e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 16 | 534 | 5.73e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 5.73e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 5 | 13 | 5.73e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 4 | 11 | 5.73e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 4 | 49 | 5.73e-01 |
| GO:BP | GO:0032485 | regulation of Ral protein signal transduction | 2 | 3 | 5.73e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 5.73e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 5 | 3070 | 5.73e-01 |
| GO:BP | GO:0007281 | germ cell development | 7 | 187 | 5.73e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 3 | 109 | 5.74e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 3 | 12 | 5.74e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 13 | 161 | 5.74e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 5 | 11 | 5.74e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 5 | 37 | 5.74e-01 |
| GO:BP | GO:1905337 | positive regulation of aggrephagy | 2 | 3 | 5.74e-01 |
| GO:BP | GO:1905335 | regulation of aggrephagy | 2 | 3 | 5.74e-01 |
| GO:BP | GO:1903394 | protein localization to kinetochore involved in kinetochore assembly | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 4 | 94 | 5.74e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 5.74e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 5.74e-01 |
| GO:BP | GO:1990108 | protein linear deubiquitination | 1 | 3 | 5.74e-01 |
| GO:BP | GO:1902216 | positive regulation of interleukin-4-mediated signaling pathway | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 2 | 31 | 5.74e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 3 | 6 | 5.74e-01 |
| GO:BP | GO:1990523 | bone regeneration | 2 | 2 | 5.74e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 1 | 55 | 5.74e-01 |
| GO:BP | GO:0072006 | nephron development | 1 | 117 | 5.74e-01 |
| GO:BP | GO:2001286 | regulation of caveolin-mediated endocytosis | 3 | 5 | 5.74e-01 |
| GO:BP | GO:1905523 | positive regulation of macrophage migration | 2 | 13 | 5.74e-01 |
| GO:BP | GO:0060697 | positive regulation of phospholipid catabolic process | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0006680 | glucosylceramide catabolic process | 1 | 3 | 5.74e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 26 | 154 | 5.74e-01 |
| GO:BP | GO:0023021 | termination of signal transduction | 1 | 3 | 5.74e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 3 | 5 | 5.75e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 1 | 93 | 5.75e-01 |
| GO:BP | GO:0030581 | symbiont intracellular protein transport in host | 1 | 2 | 5.76e-01 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 8 | 13 | 5.76e-01 |
| GO:BP | GO:1905216 | positive regulation of RNA binding | 2 | 9 | 5.76e-01 |
| GO:BP | GO:0019060 | intracellular transport of viral protein in host cell | 1 | 2 | 5.76e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 1 | 31 | 5.76e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 5.77e-01 |
| GO:BP | GO:0046031 | ADP metabolic process | 5 | 7 | 5.77e-01 |
| GO:BP | GO:0090216 | positive regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 1 | 5.77e-01 |
| GO:BP | GO:0000350 | generation of catalytic spliceosome for second transesterification step | 1 | 2 | 5.77e-01 |
| GO:BP | GO:0090118 | receptor-mediated endocytosis involved in cholesterol transport | 1 | 5 | 5.77e-01 |
| GO:BP | GO:1901837 | negative regulation of transcription of nucleolar large rRNA by RNA polymerase I | 2 | 4 | 5.77e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 8 | 14 | 5.77e-01 |
| GO:BP | GO:0036335 | intestinal stem cell homeostasis | 1 | 3 | 5.77e-01 |
| GO:BP | GO:0140361 | cyclic-GMP-AMP transmembrane import across plasma membrane | 1 | 4 | 5.77e-01 |
| GO:BP | GO:0007371 | ventral midline determination | 1 | 1 | 5.77e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 5.77e-01 |
| GO:BP | GO:0072036 | mesenchymal to epithelial transition involved in renal vesicle formation | 1 | 1 | 5.77e-01 |
| GO:BP | GO:0072285 | mesenchymal to epithelial transition involved in metanephric renal vesicle formation | 1 | 1 | 5.77e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 3 | 15 | 5.77e-01 |
| GO:BP | GO:0060840 | artery development | 8 | 83 | 5.77e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 6 | 18 | 5.78e-01 |
| GO:BP | GO:0048870 | cell motility | 16 | 1182 | 5.78e-01 |
| GO:BP | GO:0046443 | FAD metabolic process | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 7 | 12 | 5.78e-01 |
| GO:BP | GO:0106004 | tRNA (guanine-N7)-methylation | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0051892 | negative regulation of cardioblast differentiation | 1 | 1 | 5.78e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 3 | 17 | 5.78e-01 |
| GO:BP | GO:0006747 | FAD biosynthetic process | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0072388 | flavin adenine dinucleotide biosynthetic process | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0061159 | establishment of bipolar cell polarity involved in cell morphogenesis | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0060310 | regulation of elastin catabolic process | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0060311 | negative regulation of elastin catabolic process | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0010711 | negative regulation of collagen catabolic process | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 5 | 8 | 5.78e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 4 | 55 | 5.78e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 5 | 13 | 5.78e-01 |
| GO:BP | GO:0097722 | sperm motility | 2 | 61 | 5.78e-01 |
| GO:BP | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport | 3 | 5 | 5.78e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 2 | 61 | 5.78e-01 |
| GO:BP | GO:0009051 | pentose-phosphate shunt, oxidative branch | 1 | 4 | 5.79e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 5.79e-01 |
| GO:BP | GO:1904373 | response to kainic acid | 1 | 1 | 5.79e-01 |
| GO:BP | GO:1903119 | protein localization to actin cytoskeleton | 1 | 1 | 5.79e-01 |
| GO:BP | GO:1904372 | positive regulation of protein localization to actin cortical patch | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 6 | 10 | 5.79e-01 |
| GO:BP | GO:0044379 | protein localization to actin cortical patch | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 38 | 5.79e-01 |
| GO:BP | GO:1904370 | regulation of protein localization to actin cortical patch | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0097341 | zymogen inhibition | 2 | 5 | 5.79e-01 |
| GO:BP | GO:0097340 | inhibition of cysteine-type endopeptidase activity | 2 | 5 | 5.79e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process | 1 | 2 | 5.79e-01 |
| GO:BP | GO:0072684 | mitochondrial tRNA 3’-trailer cleavage, endonucleolytic | 1 | 1 | 5.79e-01 |
| GO:BP | GO:1904685 | positive regulation of metalloendopeptidase activity | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0070572 | positive regulation of neuron projection regeneration | 1 | 8 | 5.79e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 1 | 28 | 5.80e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 11 | 48 | 5.80e-01 |
| GO:BP | GO:0045829 | negative regulation of isotype switching | 2 | 4 | 5.80e-01 |
| GO:BP | GO:0120322 | lipid modification by small protein conjugation | 1 | 1 | 5.80e-01 |
| GO:BP | GO:0120323 | lipid ubiquitination | 1 | 1 | 5.80e-01 |
| GO:BP | GO:0007601 | visual perception | 1 | 96 | 5.81e-01 |
| GO:BP | GO:1905704 | positive regulation of inhibitory synapse assembly | 1 | 2 | 5.81e-01 |
| GO:BP | GO:1904586 | cellular response to putrescine | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1904585 | response to putrescine | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 5.81e-01 |
| GO:BP | GO:1990504 | dense core granule exocytosis | 2 | 7 | 5.81e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 5.81e-01 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 1 | 7 | 5.81e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 1 | 29 | 5.81e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 1 | 7 | 5.81e-01 |
| GO:BP | GO:1990828 | hepatocyte dedifferentiation | 1 | 1 | 5.81e-01 |
| GO:BP | GO:2000489 | regulation of hepatic stellate cell activation | 4 | 6 | 5.81e-01 |
| GO:BP | GO:1900098 | regulation of plasma cell differentiation | 2 | 2 | 5.81e-01 |
| GO:BP | GO:0061789 | dense core granule priming | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0061184 | positive regulation of dermatome development | 1 | 1 | 5.81e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 2 | 16 | 5.81e-01 |
| GO:BP | GO:0018117 | protein adenylylation | 1 | 2 | 5.81e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 3 | 11 | 5.81e-01 |
| GO:BP | GO:2000224 | regulation of testosterone biosynthetic process | 1 | 2 | 5.81e-01 |
| GO:BP | GO:0032346 | positive regulation of aldosterone metabolic process | 1 | 3 | 5.81e-01 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 4 | 15 | 5.81e-01 |
| GO:BP | GO:0006880 | intracellular sequestering of iron ion | 3 | 3 | 5.81e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 2 | 17 | 5.81e-01 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 4 | 15 | 5.81e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 1 | 28 | 5.81e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 11 | 37 | 5.81e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 8 | 15 | 5.81e-01 |
| GO:BP | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis | 4 | 6 | 5.81e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 1 | 116 | 5.81e-01 |
| GO:BP | GO:0018175 | protein nucleotidylation | 1 | 2 | 5.81e-01 |
| GO:BP | GO:0002184 | cytoplasmic translational termination | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0072174 | metanephric tubule formation | 2 | 3 | 5.81e-01 |
| GO:BP | GO:0032349 | positive regulation of aldosterone biosynthetic process | 1 | 3 | 5.81e-01 |
| GO:BP | GO:0034696 | response to prostaglandin F | 1 | 2 | 5.81e-01 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 10 | 18 | 5.81e-01 |
| GO:BP | GO:0035229 | positive regulation of glutamate-cysteine ligase activity | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 26 | 1849 | 5.81e-01 |
| GO:BP | GO:0006538 | glutamate catabolic process | 1 | 7 | 5.81e-01 |
| GO:BP | GO:0035227 | regulation of glutamate-cysteine ligase activity | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0009448 | gamma-aminobutyric acid metabolic process | 1 | 6 | 5.81e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 3 | 24 | 5.81e-01 |
| GO:BP | GO:0098529 | neuromuscular junction development, skeletal muscle fiber | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 3 | 24 | 5.81e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 5 | 131 | 5.81e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 4 | 7 | 5.81e-01 |
| GO:BP | GO:0010896 | regulation of triglyceride catabolic process | 1 | 7 | 5.82e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 3 | 9 | 5.82e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 710 | 5.82e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 3 | 165 | 5.82e-01 |
| GO:BP | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration | 1 | 3 | 5.82e-01 |
| GO:BP | GO:0006659 | phosphatidylserine biosynthetic process | 2 | 4 | 5.82e-01 |
| GO:BP | GO:0032679 | regulation of TRAIL production | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0098907 | regulation of SA node cell action potential | 1 | 7 | 5.82e-01 |
| GO:BP | GO:1905273 | positive regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 14 | 149 | 5.82e-01 |
| GO:BP | GO:0007604 | phototransduction, UV | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0032759 | positive regulation of TRAIL production | 1 | 1 | 5.82e-01 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 4 | 15 | 5.82e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 5.82e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 5.82e-01 |
| GO:BP | GO:0001813 | regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 5.82e-01 |
| GO:BP | GO:1905706 | regulation of mitochondrial ATP synthesis coupled proton transport | 2 | 3 | 5.82e-01 |
| GO:BP | GO:1905267 | endonucleolytic cleavage involved in tRNA processing | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0032639 | TRAIL production | 1 | 1 | 5.82e-01 |
| GO:BP | GO:1905271 | regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 5.82e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0018215 | protein phosphopantetheinylation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 11 | 31 | 5.82e-01 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 4 | 40 | 5.83e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 5.83e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 5.83e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 19 | 57 | 5.83e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 18 | 45 | 5.83e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 5.83e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 2 | 70 | 5.83e-01 |
| GO:BP | GO:0071864 | positive regulation of cell proliferation in bone marrow | 2 | 5 | 5.83e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 7 | 1006 | 5.83e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 9 | 54 | 5.83e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 2 | 316 | 5.83e-01 |
| GO:BP | GO:1902938 | regulation of intracellular calcium activated chloride channel activity | 1 | 1 | 5.83e-01 |
| GO:BP | GO:1902951 | negative regulation of dendritic spine maintenance | 1 | 2 | 5.83e-01 |
| GO:BP | GO:1904182 | regulation of pyruvate dehydrogenase activity | 1 | 4 | 5.83e-01 |
| GO:BP | GO:0000708 | meiotic strand invasion | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0061055 | myotome development | 1 | 2 | 5.83e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 5.83e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 2 | 5 | 5.83e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 4 | 56 | 5.83e-01 |
| GO:BP | GO:0090403 | oxidative stress-induced premature senescence | 1 | 3 | 5.83e-01 |
| GO:BP | GO:1900143 | positive regulation of oligodendrocyte apoptotic process | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0034230 | enkephalin processing | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0000709 | meiotic joint molecule formation | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0007612 | learning | 3 | 115 | 5.83e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 3 | 10 | 5.83e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 1 | 10 | 5.83e-01 |
| GO:BP | GO:0055113 | epiboly involved in gastrulation with mouth forming second | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0010774 | meiotic strand invasion involved in reciprocal meiotic recombination | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 10 | 234 | 5.83e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 6 | 14 | 5.83e-01 |
| GO:BP | GO:0035652 | clathrin-coated vesicle cargo loading | 1 | 6 | 5.83e-01 |
| GO:BP | GO:0035654 | clathrin-coated vesicle cargo loading, AP-3-mediated | 1 | 6 | 5.83e-01 |
| GO:BP | GO:0010587 | miRNA catabolic process | 4 | 10 | 5.83e-01 |
| GO:BP | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0034104 | negative regulation of tissue remodeling | 2 | 13 | 5.84e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 1 | 24 | 5.84e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 2 | 34 | 5.84e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 5 | 3132 | 5.85e-01 |
| GO:BP | GO:1900364 | negative regulation of mRNA polyadenylation | 1 | 1 | 5.85e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 1 | 4 | 5.85e-01 |
| GO:BP | GO:0070911 | global genome nucleotide-excision repair | 1 | 2 | 5.85e-01 |
| GO:BP | GO:2000975 | positive regulation of pro-B cell differentiation | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 11 | 54 | 5.85e-01 |
| GO:BP | GO:1905765 | negative regulation of protection from non-homologous end joining at telomere | 2 | 3 | 5.85e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 4 | 8 | 5.85e-01 |
| GO:BP | GO:0031443 | fast-twitch skeletal muscle fiber contraction | 1 | 3 | 5.85e-01 |
| GO:BP | GO:1905764 | regulation of protection from non-homologous end joining at telomere | 2 | 3 | 5.85e-01 |
| GO:BP | GO:0031439 | positive regulation of mRNA cleavage | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 1 | 10 | 5.85e-01 |
| GO:BP | GO:0061819 | telomeric DNA-containing double minutes formation | 2 | 3 | 5.85e-01 |
| GO:BP | GO:0031437 | regulation of mRNA cleavage | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 1 | 122 | 5.85e-01 |
| GO:BP | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 7 | 17 | 5.85e-01 |
| GO:BP | GO:0030540 | female genitalia development | 1 | 14 | 5.85e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 2 | 7 | 5.85e-01 |
| GO:BP | GO:0051451 | myoblast migration | 2 | 11 | 5.85e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 4 | 1835 | 5.85e-01 |
| GO:BP | GO:0070933 | histone H4 deacetylation | 4 | 7 | 5.86e-01 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 14 | 23 | 5.86e-01 |
| GO:BP | GO:0010904 | regulation of UDP-glucose catabolic process | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0010905 | negative regulation of UDP-glucose catabolic process | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 5.86e-01 |
| GO:BP | GO:0071879 | positive regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 2 | 21 | 5.86e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 1 | 27 | 5.86e-01 |
| GO:BP | GO:0044027 | negative regulation of gene expression via CpG island methylation | 1 | 1 | 5.86e-01 |
| GO:BP | GO:1904227 | negative regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 5.86e-01 |
| GO:BP | GO:1904226 | regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 5.86e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 23 | 123 | 5.86e-01 |
| GO:BP | GO:0046745 | viral capsid secondary envelopment | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0050925 | negative regulation of negative chemotaxis | 1 | 2 | 5.86e-01 |
| GO:BP | GO:0090135 | actin filament branching | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 5 | 3133 | 5.86e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 2 | 5 | 5.86e-01 |
| GO:BP | GO:2000035 | regulation of stem cell division | 5 | 9 | 5.86e-01 |
| GO:BP | GO:0032288 | myelin assembly | 2 | 19 | 5.86e-01 |
| GO:BP | GO:0033385 | geranylgeranyl diphosphate metabolic process | 1 | 2 | 5.86e-01 |
| GO:BP | GO:1901187 | regulation of ephrin receptor signaling pathway | 1 | 2 | 5.86e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 6 | 28 | 5.86e-01 |
| GO:BP | GO:0051647 | nucleus localization | 5 | 28 | 5.87e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 2 | 108 | 5.87e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 9 | 49 | 5.87e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 5.87e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 5 | 107 | 5.87e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 7 | 13 | 5.87e-01 |
| GO:BP | GO:0061669 | spontaneous neurotransmitter secretion | 3 | 5 | 5.87e-01 |
| GO:BP | GO:0030070 | insulin processing | 2 | 6 | 5.87e-01 |
| GO:BP | GO:0032915 | positive regulation of transforming growth factor beta2 production | 2 | 3 | 5.87e-01 |
| GO:BP | GO:0048371 | lateral mesodermal cell differentiation | 2 | 2 | 5.87e-01 |
| GO:BP | GO:1904990 | regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1904992 | positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 1 | 14 | 5.88e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 5.88e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 5.88e-01 |
| GO:BP | GO:0035336 | long-chain fatty-acyl-CoA metabolic process | 13 | 19 | 5.88e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 8 | 20 | 5.88e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 8 | 40 | 5.88e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 17 | 55 | 5.88e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 5 | 93 | 5.89e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 1 | 39 | 5.89e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 3 | 21 | 5.89e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 5.89e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 1 | 9 | 5.89e-01 |
| GO:BP | GO:1901724 | positive regulation of cell proliferation involved in kidney development | 3 | 8 | 5.89e-01 |
| GO:BP | GO:1990264 | peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | 2 | 4 | 5.89e-01 |
| GO:BP | GO:0042539 | hypotonic salinity response | 2 | 2 | 5.89e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 6 | 49 | 5.89e-01 |
| GO:BP | GO:0071477 | cellular hypotonic salinity response | 2 | 2 | 5.89e-01 |
| GO:BP | GO:0048320 | axial mesoderm formation | 1 | 2 | 5.89e-01 |
| GO:BP | GO:0021978 | telencephalon regionalization | 1 | 4 | 5.89e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 5 | 31 | 5.89e-01 |
| GO:BP | GO:1904923 | regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 4 | 15 | 5.89e-01 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 1 | 34 | 5.89e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 1 | 86 | 5.89e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 3 | 128 | 5.89e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 3 | 101 | 5.89e-01 |
| GO:BP | GO:0042473 | outer ear morphogenesis | 4 | 5 | 5.90e-01 |
| GO:BP | GO:0060545 | positive regulation of necroptotic process | 1 | 4 | 5.90e-01 |
| GO:BP | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 5.90e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 4 | 9 | 5.90e-01 |
| GO:BP | GO:0072674 | multinuclear osteoclast differentiation | 1 | 6 | 5.90e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 10 | 41 | 5.90e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 6 | 620 | 5.90e-01 |
| GO:BP | GO:0033242 | negative regulation of amine catabolic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0099093 | calcium export from the mitochondrion | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0033241 | regulation of amine catabolic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0046707 | IDP metabolic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:2000283 | negative regulation of amino acid biosynthetic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0036017 | response to erythropoietin | 3 | 4 | 5.90e-01 |
| GO:BP | GO:0015828 | tyrosine transport | 1 | 3 | 5.90e-01 |
| GO:BP | GO:0035995 | detection of muscle stretch | 3 | 7 | 5.90e-01 |
| GO:BP | GO:0046709 | IDP catabolic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 23 | 42 | 5.90e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 3 | 33 | 5.90e-01 |
| GO:BP | GO:0052779 | amino disaccharide metabolic process | 2 | 9 | 5.90e-01 |
| GO:BP | GO:2000311 | regulation of AMPA receptor activity | 2 | 15 | 5.90e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 29 | 59 | 5.90e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 5.90e-01 |
| GO:BP | GO:0030205 | dermatan sulfate metabolic process | 2 | 9 | 5.90e-01 |
| GO:BP | GO:0098736 | negative regulation of the force of heart contraction | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 8 | 66 | 5.90e-01 |
| GO:BP | GO:0090226 | regulation of microtubule nucleation by Ran protein signal transduction | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 32 | 150 | 5.90e-01 |
| GO:BP | GO:1900082 | negative regulation of arginine catabolic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:1903249 | negative regulation of citrulline biosynthetic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0070460 | thyroid-stimulating hormone secretion | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 5.90e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 6 | 101 | 5.90e-01 |
| GO:BP | GO:0014728 | regulation of the force of skeletal muscle contraction | 1 | 3 | 5.90e-01 |
| GO:BP | GO:0014862 | regulation of skeletal muscle contraction by chemo-mechanical energy conversion | 1 | 3 | 5.90e-01 |
| GO:BP | GO:1903248 | regulation of citrulline biosynthetic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:1900081 | regulation of arginine catabolic process | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0014043 | negative regulation of neuron maturation | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0038163 | thrombopoietin-mediated signaling pathway | 1 | 3 | 5.91e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 2 | 22 | 5.91e-01 |
| GO:BP | GO:0038180 | nerve growth factor signaling pathway | 3 | 10 | 5.91e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 4 | 6 | 5.91e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 2 | 8 | 5.91e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 5.91e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 3 | 17 | 5.91e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 48 | 307 | 5.91e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 20 | 31 | 5.91e-01 |
| GO:BP | GO:0019377 | glycolipid catabolic process | 5 | 12 | 5.91e-01 |
| GO:BP | GO:0006855 | xenobiotic transmembrane transport | 2 | 11 | 5.91e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 2 | 14 | 5.91e-01 |
| GO:BP | GO:0001890 | placenta development | 1 | 117 | 5.91e-01 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 6 | 37 | 5.91e-01 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 5.91e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 5.91e-01 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0051890 | regulation of cardioblast differentiation | 2 | 5 | 5.91e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 10 | 97 | 5.91e-01 |
| GO:BP | GO:0120158 | positive regulation of collagen catabolic process | 1 | 1 | 5.91e-01 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 4 | 8 | 5.92e-01 |
| GO:BP | GO:0048265 | response to pain | 2 | 17 | 5.92e-01 |
| GO:BP | GO:0044805 | late nucleophagy | 1 | 7 | 5.92e-01 |
| GO:BP | GO:0043029 | T cell homeostasis | 5 | 27 | 5.92e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 5.92e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 1 | 27 | 5.92e-01 |
| GO:BP | GO:0046877 | regulation of saliva secretion | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0086101 | endothelin receptor signaling pathway involved in heart process | 1 | 3 | 5.92e-01 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 1 | 17 | 5.92e-01 |
| GO:BP | GO:0042305 | specification of segmental identity, mandibular segment | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0017196 | N-terminal peptidyl-methionine acetylation | 2 | 8 | 5.92e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 8 | 29 | 5.92e-01 |
| GO:BP | GO:0002383 | immune response in brain or nervous system | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 9 | 15 | 5.92e-01 |
| GO:BP | GO:0097281 | immune complex formation | 1 | 1 | 5.92e-01 |
| GO:BP | GO:1902174 | positive regulation of keratinocyte apoptotic process | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 5.92e-01 |
| GO:BP | GO:1904491 | protein localization to ciliary transition zone | 2 | 7 | 5.92e-01 |
| GO:BP | GO:1990442 | intrinsic apoptotic signaling pathway in response to nitrosative stress | 1 | 3 | 5.92e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 2 | 10 | 5.93e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.93e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.93e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 9 | 29 | 5.93e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 4 | 28 | 5.93e-01 |
| GO:BP | GO:0009237 | siderophore metabolic process | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0019290 | siderophore biosynthetic process | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0036285 | SAGA complex assembly | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0042352 | GDP-L-fucose salvage | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0010916 | negative regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0010915 | regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0060490 | lateral sprouting involved in lung morphogenesis | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0060489 | planar dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0072137 | condensed mesenchymal cell proliferation | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 35 | 103 | 5.93e-01 |
| GO:BP | GO:0045346 | regulation of MHC class II biosynthetic process | 9 | 11 | 5.93e-01 |
| GO:BP | GO:0106028 | neuron projection retraction | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 2 | 21 | 5.93e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 1 | 13 | 5.94e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 3 | 54 | 5.94e-01 |
| GO:BP | GO:0090675 | intermicrovillar adhesion | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 1 | 140 | 5.94e-01 |
| GO:BP | GO:0048874 | host-mediated regulation of intestinal microbiota composition | 1 | 2 | 5.94e-01 |
| GO:BP | GO:0042439 | ethanolamine-containing compound metabolic process | 1 | 5 | 5.94e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 6 | 252 | 5.94e-01 |
| GO:BP | GO:0001660 | fever generation | 1 | 5 | 5.95e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 5.95e-01 |
| GO:BP | GO:1904210 | VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.95e-01 |
| GO:BP | GO:1904239 | regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.95e-01 |
| GO:BP | GO:1904240 | negative regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0034354 | ‘de novo’ NAD biosynthetic process from tryptophan | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0061824 | cytosolic ciliogenesis | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 15 | 40 | 5.95e-01 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 1 | 6 | 5.95e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 5.95e-01 |
| GO:BP | GO:0060160 | negative regulation of dopamine receptor signaling pathway | 1 | 3 | 5.95e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 1 | 88 | 5.95e-01 |
| GO:BP | GO:0051235 | maintenance of location | 1 | 260 | 5.95e-01 |
| GO:BP | GO:0006901 | vesicle coating | 15 | 36 | 5.95e-01 |
| GO:BP | GO:0016198 | axon choice point recognition | 2 | 5 | 5.95e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 8 | 35 | 5.95e-01 |
| GO:BP | GO:1990034 | calcium ion export across plasma membrane | 4 | 6 | 5.95e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 3 | 40 | 5.95e-01 |
| GO:BP | GO:1904693 | midbrain morphogenesis | 1 | 3 | 5.95e-01 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 1 | 35 | 5.96e-01 |
| GO:BP | GO:0046083 | adenine metabolic process | 1 | 3 | 5.96e-01 |
| GO:BP | GO:0072383 | plus-end-directed vesicle transport along microtubule | 1 | 6 | 5.96e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 11 | 41 | 5.96e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 2 | 86 | 5.96e-01 |
| GO:BP | GO:2000609 | regulation of thyroid hormone generation | 1 | 2 | 5.96e-01 |
| GO:BP | GO:1900114 | positive regulation of histone H3-K9 trimethylation | 1 | 2 | 5.96e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 2 | 31 | 5.96e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 5.96e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 11 | 39 | 5.96e-01 |
| GO:BP | GO:0006837 | serotonin transport | 1 | 11 | 5.97e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 2 | 35 | 5.97e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 3 | 14 | 5.97e-01 |
| GO:BP | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | 3 | 5 | 5.97e-01 |
| GO:BP | GO:0016203 | muscle attachment | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0060197 | cloacal septation | 1 | 2 | 5.97e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 5.97e-01 |
| GO:BP | GO:1905857 | positive regulation of pentose-phosphate shunt | 2 | 3 | 5.97e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 7 | 13 | 5.97e-01 |
| GO:BP | GO:0033182 | regulation of histone ubiquitination | 3 | 6 | 5.97e-01 |
| GO:BP | GO:0070535 | histone H2A K63-linked ubiquitination | 3 | 6 | 5.97e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 18 | 46 | 5.97e-01 |
| GO:BP | GO:0009234 | menaquinone biosynthetic process | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0042371 | vitamin K biosynthetic process | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0032194 | ubiquinone biosynthetic process via 3,4-dihydroxy-5-polyprenylbenzoate | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine | 1 | 2 | 5.97e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 12 | 31 | 5.97e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 40 | 292 | 5.97e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 2 | 39 | 5.97e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 9 | 42 | 5.97e-01 |
| GO:BP | GO:0031929 | TOR signaling | 60 | 115 | 5.97e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 5 | 37 | 5.97e-01 |
| GO:BP | GO:0036525 | protein deglycation | 1 | 2 | 5.97e-01 |
| GO:BP | GO:1901492 | positive regulation of lymphangiogenesis | 1 | 2 | 5.97e-01 |
| GO:BP | GO:0030389 | fructosamine metabolic process | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0062009 | secondary palate development | 6 | 20 | 5.97e-01 |
| GO:BP | GO:0070417 | cellular response to cold | 1 | 8 | 5.98e-01 |
| GO:BP | GO:0071236 | cellular response to antibiotic | 1 | 11 | 5.98e-01 |
| GO:BP | GO:1901252 | regulation of intracellular transport of viral material | 1 | 2 | 5.98e-01 |
| GO:BP | GO:1900453 | negative regulation of long-term synaptic depression | 1 | 3 | 5.98e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 9 | 30 | 5.98e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 1 | 16 | 5.98e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 84 | 517 | 5.98e-01 |
| GO:BP | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | 1 | 10 | 5.98e-01 |
| GO:BP | GO:0043095 | regulation of GTP cyclohydrolase I activity | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 5.99e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 5.99e-01 |
| GO:BP | GO:0043105 | negative regulation of GTP cyclohydrolase I activity | 1 | 1 | 5.99e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 1 | 20 | 5.99e-01 |
| GO:BP | GO:0003407 | neural retina development | 3 | 48 | 5.99e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 5.99e-01 |
| GO:BP | GO:0060312 | regulation of blood vessel remodeling | 1 | 4 | 5.99e-01 |
| GO:BP | GO:0071929 | alpha-tubulin acetylation | 2 | 3 | 5.99e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 1 | 30 | 5.99e-01 |
| GO:BP | GO:0010746 | regulation of long-chain fatty acid import across plasma membrane | 2 | 5 | 5.99e-01 |
| GO:BP | GO:0010936 | negative regulation of macrophage cytokine production | 1 | 7 | 5.99e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 11 | 23 | 5.99e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 6.00e-01 |
| GO:BP | GO:0050930 | induction of positive chemotaxis | 2 | 12 | 6.00e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 6.00e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 3 | 433 | 6.00e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 4 | 5 | 6.00e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 4 | 41 | 6.00e-01 |
| GO:BP | GO:0006863 | purine nucleobase transport | 1 | 7 | 6.00e-01 |
| GO:BP | GO:0014824 | artery smooth muscle contraction | 2 | 7 | 6.00e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 6.00e-01 |
| GO:BP | GO:0140157 | ammonium import across plasma membrane | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0001544 | initiation of primordial ovarian follicle growth | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 2 | 85 | 6.00e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 2 | 13 | 6.00e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 11 | 30 | 6.00e-01 |
| GO:BP | GO:0099074 | mitochondrion to lysosome transport | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 6.00e-01 |
| GO:BP | GO:0006566 | threonine metabolic process | 3 | 4 | 6.00e-01 |
| GO:BP | GO:0006559 | L-phenylalanine catabolic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0006558 | L-phenylalanine metabolic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 6.00e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 2 | 1147 | 6.00e-01 |
| GO:BP | GO:0072717 | cellular response to actinomycin D | 1 | 4 | 6.00e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 6 | 19 | 6.00e-01 |
| GO:BP | GO:0048702 | embryonic neurocranium morphogenesis | 1 | 7 | 6.00e-01 |
| GO:BP | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process | 1 | 2 | 6.00e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 6.00e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 6.00e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 9 | 963 | 6.00e-01 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 2 | 17 | 6.00e-01 |
| GO:BP | GO:0060577 | pulmonary vein morphogenesis | 1 | 1 | 6.00e-01 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 3 | 23 | 6.00e-01 |
| GO:BP | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 2 | 3 | 6.00e-01 |
| GO:BP | GO:0140009 | L-aspartate import across plasma membrane | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0060525 | prostate glandular acinus development | 3 | 5 | 6.00e-01 |
| GO:BP | GO:1905735 | regulation of L-proline import across plasma membrane | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0048132 | female germ-line stem cell asymmetric division | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0090384 | phagosome-lysosome docking | 1 | 1 | 6.00e-01 |
| GO:BP | GO:1900079 | regulation of arginine biosynthetic process | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0015787 | UDP-glucuronic acid transmembrane transport | 2 | 4 | 6.00e-01 |
| GO:BP | GO:0015789 | UDP-N-acetylgalactosamine transmembrane transport | 2 | 4 | 6.00e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 1 | 295 | 6.00e-01 |
| GO:BP | GO:1905737 | positive regulation of L-proline import across plasma membrane | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0090241 | negative regulation of histone H4 acetylation | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 5 | 19 | 6.00e-01 |
| GO:BP | GO:0042256 | cytosolic ribosome assembly | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0010636 | positive regulation of mitochondrial fusion | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0060309 | elastin catabolic process | 2 | 2 | 6.00e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 2 | 5 | 6.00e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 5 | 18 | 6.00e-01 |
| GO:BP | GO:0070881 | regulation of proline transport | 1 | 2 | 6.00e-01 |
| GO:BP | GO:1902283 | negative regulation of primary amine oxidase activity | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0042749 | regulation of circadian sleep/wake cycle | 2 | 9 | 6.00e-01 |
| GO:BP | GO:0035922 | foramen ovale closure | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0051169 | nuclear transport | 1 | 295 | 6.00e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 6 | 37 | 6.00e-01 |
| GO:BP | GO:1902222 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0099075 | mitochondrion-derived vesicle mediated transport | 1 | 3 | 6.00e-01 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 1 | 3 | 6.00e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0072143 | mesangial cell development | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0035787 | cell migration involved in kidney development | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 12 | 52 | 6.00e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 3 | 38 | 6.00e-01 |
| GO:BP | GO:0019254 | carnitine metabolic process, CoA-linked | 2 | 3 | 6.00e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 6 | 19 | 6.00e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 12 | 149 | 6.00e-01 |
| GO:BP | GO:0070236 | negative regulation of activation-induced cell death of T cells | 2 | 3 | 6.00e-01 |
| GO:BP | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 1 | 4 | 6.00e-01 |
| GO:BP | GO:0050914 | sensory perception of salty taste | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 6.00e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 6.00e-01 |
| GO:BP | GO:0039530 | MDA-5 signaling pathway | 4 | 8 | 6.00e-01 |
| GO:BP | GO:0072008 | glomerular mesangial cell differentiation | 1 | 4 | 6.00e-01 |
| GO:BP | GO:0036306 | embryonic heart tube elongation | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 1 | 719 | 6.00e-01 |
| GO:BP | GO:1902836 | positive regulation of proline import across plasma membrane | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0016139 | glycoside catabolic process | 3 | 8 | 6.00e-01 |
| GO:BP | GO:1902834 | regulation of proline import across plasma membrane | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 2 | 15 | 6.00e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 2 | 14 | 6.01e-01 |
| GO:BP | GO:1904616 | regulation of actin binding | 1 | 3 | 6.01e-01 |
| GO:BP | GO:0007521 | muscle cell fate determination | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 10 | 152 | 6.01e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 2 | 58 | 6.01e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 8 | 461 | 6.01e-01 |
| GO:BP | GO:0016182 | synaptic vesicle budding from endosome | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0034625 | fatty acid elongation, monounsaturated fatty acid | 4 | 6 | 6.01e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 6.01e-01 |
| GO:BP | GO:0019367 | fatty acid elongation, saturated fatty acid | 4 | 6 | 6.01e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 798 | 6.01e-01 |
| GO:BP | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid | 4 | 6 | 6.01e-01 |
| GO:BP | GO:0019368 | fatty acid elongation, unsaturated fatty acid | 4 | 6 | 6.01e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 9 | 13 | 6.01e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 6 | 115 | 6.01e-01 |
| GO:BP | GO:0070257 | positive regulation of mucus secretion | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0042723 | thiamine-containing compound metabolic process | 4 | 4 | 6.01e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 3 | 9 | 6.01e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 2 | 51 | 6.01e-01 |
| GO:BP | GO:1904828 | positive regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:1904826 | regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 4 | 56 | 6.02e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 9 | 71 | 6.02e-01 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 3 | 5 | 6.02e-01 |
| GO:BP | GO:0051707 | response to other organism | 5 | 820 | 6.02e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 1 | 6 | 6.02e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 12 | 6.03e-01 |
| GO:BP | GO:0035507 | regulation of myosin-light-chain-phosphatase activity | 1 | 5 | 6.03e-01 |
| GO:BP | GO:1904381 | Golgi apparatus mannose trimming | 2 | 3 | 6.03e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 3 | 159 | 6.03e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 2 | 5 | 6.03e-01 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 2 | 13 | 6.03e-01 |
| GO:BP | GO:0061551 | trigeminal ganglion development | 2 | 3 | 6.03e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 1 | 33 | 6.03e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 1 | 33 | 6.03e-01 |
| GO:BP | GO:0045606 | positive regulation of epidermal cell differentiation | 11 | 16 | 6.03e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 3 | 70 | 6.04e-01 |
| GO:BP | GO:0015801 | aromatic amino acid transport | 4 | 11 | 6.04e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 6 | 17 | 6.04e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 5 | 820 | 6.04e-01 |
| GO:BP | GO:0060574 | intestinal epithelial cell maturation | 2 | 3 | 6.04e-01 |
| GO:BP | GO:1900222 | negative regulation of amyloid-beta clearance | 3 | 7 | 6.04e-01 |
| GO:BP | GO:1904257 | zinc ion import into Golgi lumen | 1 | 3 | 6.04e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 6.04e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 5 | 135 | 6.04e-01 |
| GO:BP | GO:0090667 | cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0090668 | endothelial cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 1 | 31 | 6.05e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 67 | 143 | 6.05e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 8 | 15 | 6.05e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 8 | 70 | 6.05e-01 |
| GO:BP | GO:0014842 | regulation of skeletal muscle satellite cell proliferation | 2 | 8 | 6.05e-01 |
| GO:BP | GO:0015960 | diadenosine polyphosphate biosynthetic process | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 28 | 597 | 6.05e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 6.05e-01 |
| GO:BP | GO:0061145 | lung smooth muscle development | 1 | 2 | 6.05e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 1 | 6 | 6.05e-01 |
| GO:BP | GO:0015965 | diadenosine tetraphosphate metabolic process | 1 | 2 | 6.05e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 5 | 24 | 6.05e-01 |
| GO:BP | GO:2000633 | positive regulation of pre-miRNA processing | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 28 | 597 | 6.05e-01 |
| GO:BP | GO:0060516 | primary prostatic bud elongation | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0007542 | primary sex determination, germ-line | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | 7 | 6.05e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 6.05e-01 |
| GO:BP | GO:0060532 | bronchus cartilage development | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0014036 | neural crest cell fate specification | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1904863 | regulation of beta-catenin-TCF complex assembly | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 2 | 223 | 6.05e-01 |
| GO:BP | GO:1904864 | negative regulation of beta-catenin-TCF complex assembly | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0023041 | neuronal signal transduction | 1 | 6 | 6.05e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 1 | 316 | 6.05e-01 |
| GO:BP | GO:0018992 | germ-line sex determination | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0019100 | male germ-line sex determination | 1 | 2 | 6.05e-01 |
| GO:BP | GO:2000074 | regulation of type B pancreatic cell development | 2 | 8 | 6.05e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 9 | 94 | 6.05e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 21 | 678 | 6.05e-01 |
| GO:BP | GO:0060010 | Sertoli cell fate commitment | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0015966 | diadenosine tetraphosphate biosynthetic process | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 12 | 27 | 6.05e-01 |
| GO:BP | GO:0035622 | intrahepatic bile duct development | 1 | 2 | 6.05e-01 |
| GO:BP | GO:1904431 | positive regulation of t-circle formation | 1 | 3 | 6.05e-01 |
| GO:BP | GO:0001834 | trophectodermal cell proliferation | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0072488 | ammonium transmembrane transport | 2 | 6 | 6.05e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 10 | 236 | 6.05e-01 |
| GO:BP | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 5 | 3237 | 6.06e-01 |
| GO:BP | GO:1904205 | negative regulation of skeletal muscle hypertrophy | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1901337 | thioester transport | 1 | 2 | 6.06e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 3 | 111 | 6.06e-01 |
| GO:BP | GO:0015916 | fatty-acyl-CoA transport | 1 | 2 | 6.06e-01 |
| GO:BP | GO:1905949 | negative regulation of calcium ion import across plasma membrane | 1 | 4 | 6.06e-01 |
| GO:BP | GO:0035037 | sperm entry | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1905528 | positive regulation of Golgi lumen acidification | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0050832 | defense response to fungus | 1 | 14 | 6.06e-01 |
| GO:BP | GO:0006051 | N-acetylmannosamine metabolic process | 1 | 2 | 6.06e-01 |
| GO:BP | GO:0006050 | mannosamine metabolic process | 1 | 2 | 6.06e-01 |
| GO:BP | GO:0035094 | response to nicotine | 1 | 28 | 6.06e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 3 | 4241 | 6.06e-01 |
| GO:BP | GO:0140354 | lipid import into cell | 1 | 15 | 6.06e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 10 | 36 | 6.06e-01 |
| GO:BP | GO:0046294 | formaldehyde catabolic process | 2 | 2 | 6.06e-01 |
| GO:BP | GO:0098885 | modification of postsynaptic actin cytoskeleton | 3 | 10 | 6.07e-01 |
| GO:BP | GO:0071907 | determination of digestive tract left/right asymmetry | 3 | 5 | 6.07e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 1 | 311 | 6.07e-01 |
| GO:BP | GO:1905413 | regulation of dense core granule exocytosis | 2 | 2 | 6.07e-01 |
| GO:BP | GO:0007270 | neuron-neuron synaptic transmission | 2 | 5 | 6.07e-01 |
| GO:BP | GO:0097043 | histone H3-K56 acetylation | 1 | 2 | 6.07e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 2 | 1061 | 6.07e-01 |
| GO:BP | GO:0032532 | regulation of microvillus length | 2 | 4 | 6.08e-01 |
| GO:BP | GO:0021780 | glial cell fate specification | 3 | 3 | 6.08e-01 |
| GO:BP | GO:0006590 | thyroid hormone generation | 4 | 10 | 6.08e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 6.08e-01 |
| GO:BP | GO:0046185 | aldehyde catabolic process | 7 | 12 | 6.08e-01 |
| GO:BP | GO:0099500 | vesicle fusion to plasma membrane | 12 | 19 | 6.08e-01 |
| GO:BP | GO:0035745 | T-helper 2 cell cytokine production | 1 | 3 | 6.08e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 6.08e-01 |
| GO:BP | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly | 1 | 8 | 6.08e-01 |
| GO:BP | GO:0060374 | mast cell differentiation | 3 | 4 | 6.08e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 3 | 6 | 6.08e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 9 | 21 | 6.08e-01 |
| GO:BP | GO:0007220 | Notch receptor processing | 5 | 9 | 6.08e-01 |
| GO:BP | GO:0043461 | proton-transporting ATP synthase complex assembly | 1 | 8 | 6.08e-01 |
| GO:BP | GO:0090144 | mitochondrial nucleoid organization | 1 | 1 | 6.08e-01 |
| GO:BP | GO:0090143 | nucleoid organization | 1 | 1 | 6.08e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 8 | 45 | 6.08e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 4 | 47 | 6.08e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 2 | 21 | 6.08e-01 |
| GO:BP | GO:2000551 | regulation of T-helper 2 cell cytokine production | 1 | 3 | 6.08e-01 |
| GO:BP | GO:0032361 | pyridoxal phosphate catabolic process | 1 | 1 | 6.08e-01 |
| GO:BP | GO:0042820 | vitamin B6 catabolic process | 1 | 1 | 6.08e-01 |
| GO:BP | GO:0007530 | sex determination | 4 | 12 | 6.09e-01 |
| GO:BP | GO:0061085 | regulation of histone H3-K27 methylation | 1 | 4 | 6.09e-01 |
| GO:BP | GO:0006325 | chromatin organization | 8 | 551 | 6.09e-01 |
| GO:BP | GO:0098751 | bone cell development | 2 | 26 | 6.09e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 1 | 266 | 6.10e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 11 | 68 | 6.10e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 7 | 79 | 6.10e-01 |
| GO:BP | GO:1903650 | negative regulation of cytoplasmic transport | 2 | 3 | 6.10e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 1 | 19 | 6.10e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 6.11e-01 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 1 | 10 | 6.11e-01 |
| GO:BP | GO:0035700 | astrocyte chemotaxis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 6 | 12 | 6.11e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0036145 | dendritic cell homeostasis | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0071941 | nitrogen cycle metabolic process | 1 | 7 | 6.11e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 6.11e-01 |
| GO:BP | GO:0036150 | phosphatidylserine acyl-chain remodeling | 1 | 7 | 6.11e-01 |
| GO:BP | GO:0019627 | urea metabolic process | 1 | 7 | 6.11e-01 |
| GO:BP | GO:1904533 | regulation of telomeric loop disassembly | 1 | 2 | 6.11e-01 |
| GO:BP | GO:1904526 | regulation of microtubule binding | 3 | 10 | 6.11e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 6 | 12 | 6.11e-01 |
| GO:BP | GO:2000458 | regulation of astrocyte chemotaxis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 14 | 35 | 6.11e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 2 | 14 | 6.11e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 1 | 31 | 6.11e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 18 | 1088 | 6.11e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 1 | 36 | 6.11e-01 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 1 | 10 | 6.11e-01 |
| GO:BP | GO:0072359 | circulatory system development | 2 | 884 | 6.11e-01 |
| GO:BP | GO:0002296 | T-helper 1 cell lineage commitment | 1 | 3 | 6.11e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 3 | 58 | 6.11e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 5 | 8 | 6.11e-01 |
| GO:BP | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 2 | 5 | 6.11e-01 |
| GO:BP | GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 2 | 5 | 6.11e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 1 | 20 | 6.11e-01 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 2 | 20 | 6.11e-01 |
| GO:BP | GO:1905396 | cellular response to flavonoid | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0042118 | endothelial cell activation | 4 | 7 | 6.11e-01 |
| GO:BP | GO:0034773 | histone H4-K20 trimethylation | 3 | 5 | 6.11e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 6.11e-01 |
| GO:BP | GO:0003188 | heart valve formation | 8 | 16 | 6.11e-01 |
| GO:BP | GO:0046348 | amino sugar catabolic process | 8 | 9 | 6.11e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 4 | 17 | 6.11e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 28 | 61 | 6.11e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 7 | 40 | 6.12e-01 |
| GO:BP | GO:0007624 | ultradian rhythm | 1 | 2 | 6.12e-01 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0097494 | regulation of vesicle size | 7 | 19 | 6.12e-01 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0007611 | learning or memory | 40 | 202 | 6.12e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 13 | 993 | 6.12e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 4 | 16 | 6.12e-01 |
| GO:BP | GO:0150093 | amyloid-beta clearance by transcytosis | 4 | 7 | 6.12e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 1 | 27 | 6.12e-01 |
| GO:BP | GO:1905580 | positive regulation of ERBB3 signaling pathway | 1 | 1 | 6.12e-01 |
| GO:BP | GO:1905578 | regulation of ERBB3 signaling pathway | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0048312 | intracellular distribution of mitochondria | 3 | 5 | 6.12e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 3 | 23 | 6.12e-01 |
| GO:BP | GO:0036367 | light adaption | 1 | 3 | 6.12e-01 |
| GO:BP | GO:1990758 | mitotic sister chromatid biorientation | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 3 | 15 | 6.12e-01 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 1 | 9 | 6.13e-01 |
| GO:BP | GO:1901652 | response to peptide | 71 | 377 | 6.13e-01 |
| GO:BP | GO:2000394 | positive regulation of lamellipodium morphogenesis | 3 | 6 | 6.13e-01 |
| GO:BP | GO:0070899 | mitochondrial tRNA wobble uridine modification | 1 | 1 | 6.13e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 2 | 19 | 6.13e-01 |
| GO:BP | GO:2000312 | regulation of kainate selective glutamate receptor activity | 1 | 3 | 6.13e-01 |
| GO:BP | GO:0006106 | fumarate metabolic process | 2 | 3 | 6.13e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 11 | 39 | 6.13e-01 |
| GO:BP | GO:0006532 | aspartate biosynthetic process | 2 | 2 | 6.13e-01 |
| GO:BP | GO:0008015 | blood circulation | 2 | 374 | 6.13e-01 |
| GO:BP | GO:0003218 | cardiac left ventricle formation | 1 | 2 | 6.13e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 9 | 49 | 6.14e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 10 | 42 | 6.14e-01 |
| GO:BP | GO:0097104 | postsynaptic membrane assembly | 2 | 8 | 6.14e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 2 | 7 | 6.14e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 2 | 36 | 6.14e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 7 | 15 | 6.14e-01 |
| GO:BP | GO:0097090 | presynaptic membrane organization | 4 | 8 | 6.14e-01 |
| GO:BP | GO:0002159 | desmosome assembly | 2 | 3 | 6.14e-01 |
| GO:BP | GO:0043060 | meiotic metaphase I plate congression | 1 | 1 | 6.14e-01 |
| GO:BP | GO:0051365 | cellular response to potassium ion starvation | 1 | 1 | 6.14e-01 |
| GO:BP | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH | 1 | 1 | 6.14e-01 |
| GO:BP | GO:2000487 | positive regulation of glutamine transport | 1 | 2 | 6.14e-01 |
| GO:BP | GO:0010585 | glutamine secretion | 1 | 2 | 6.14e-01 |
| GO:BP | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway | 1 | 1 | 6.14e-01 |
| GO:BP | GO:1903713 | asparagine transmembrane transport | 1 | 1 | 6.14e-01 |
| GO:BP | GO:0016094 | polyprenol biosynthetic process | 1 | 3 | 6.14e-01 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 2 | 9 | 6.14e-01 |
| GO:BP | GO:0007416 | synapse assembly | 10 | 152 | 6.15e-01 |
| GO:BP | GO:0035963 | cellular response to interleukin-13 | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 1 | 14 | 6.16e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 3 | 108 | 6.16e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 4 | 13 | 6.16e-01 |
| GO:BP | GO:0042117 | monocyte activation | 1 | 8 | 6.16e-01 |
| GO:BP | GO:0010272 | response to silver ion | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 4 | 11 | 6.16e-01 |
| GO:BP | GO:0097188 | dentin mineralization | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 2 | 27 | 6.16e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 5 | 99 | 6.16e-01 |
| GO:BP | GO:1990134 | epithelial cell apoptotic process involved in palatal shelf morphogenesis | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 2 | 27 | 6.16e-01 |
| GO:BP | GO:0071318 | cellular response to ATP | 3 | 12 | 6.16e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 6.16e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 6.16e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 6.16e-01 |
| GO:BP | GO:0050812 | regulation of acyl-CoA biosynthetic process | 2 | 5 | 6.16e-01 |
| GO:BP | GO:0006621 | protein retention in ER lumen | 4 | 8 | 6.17e-01 |
| GO:BP | GO:0042780 | tRNA 3’-end processing | 1 | 7 | 6.17e-01 |
| GO:BP | GO:0099540 | trans-synaptic signaling by neuropeptide | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0000718 | nucleotide-excision repair, DNA damage removal | 1 | 1 | 6.17e-01 |
| GO:BP | GO:1905089 | regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0042748 | circadian sleep/wake cycle, non-REM sleep | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 6.17e-01 |
| GO:BP | GO:2000078 | positive regulation of type B pancreatic cell development | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0099118 | microtubule-based protein transport | 1 | 14 | 6.17e-01 |
| GO:BP | GO:1904447 | folate import across plasma membrane | 1 | 4 | 6.17e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 31 | 76 | 6.17e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 5 | 850 | 6.17e-01 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 2 | 10 | 6.17e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 3 | 41 | 6.17e-01 |
| GO:BP | GO:0044349 | DNA excision | 1 | 1 | 6.17e-01 |
| GO:BP | GO:1903651 | positive regulation of cytoplasmic transport | 6 | 11 | 6.17e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 6.17e-01 |
| GO:BP | GO:0003012 | muscle system process | 16 | 334 | 6.17e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 3 | 4 | 6.17e-01 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 3 | 7 | 6.17e-01 |
| GO:BP | GO:0098840 | protein transport along microtubule | 1 | 14 | 6.17e-01 |
| GO:BP | GO:0001764 | neuron migration | 8 | 133 | 6.17e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 10 | 1137 | 6.17e-01 |
| GO:BP | GO:0009224 | CMP biosynthetic process | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0062100 | positive regulation of programmed necrotic cell death | 1 | 5 | 6.17e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 6.17e-01 |
| GO:BP | GO:0002432 | granuloma formation | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 6.17e-01 |
| GO:BP | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0042921 | glucocorticoid receptor signaling pathway | 2 | 13 | 6.18e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 13 | 84 | 6.18e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 1 | 54 | 6.18e-01 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 7 | 18 | 6.18e-01 |
| GO:BP | GO:0006204 | IMP catabolic process | 3 | 3 | 6.18e-01 |
| GO:BP | GO:0060479 | lung cell differentiation | 7 | 18 | 6.18e-01 |
| GO:BP | GO:0060956 | endocardial cell differentiation | 2 | 4 | 6.18e-01 |
| GO:BP | GO:0003348 | cardiac endothelial cell differentiation | 2 | 4 | 6.18e-01 |
| GO:BP | GO:0060377 | negative regulation of mast cell differentiation | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0014807 | regulation of somitogenesis | 2 | 3 | 6.18e-01 |
| GO:BP | GO:0061198 | fungiform papilla formation | 2 | 3 | 6.18e-01 |
| GO:BP | GO:0048268 | clathrin coat assembly | 1 | 13 | 6.18e-01 |
| GO:BP | GO:0046533 | negative regulation of photoreceptor cell differentiation | 2 | 3 | 6.18e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 70 | 321 | 6.18e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 6.19e-01 |
| GO:BP | GO:0015851 | nucleobase transport | 1 | 7 | 6.19e-01 |
| GO:BP | GO:0010747 | positive regulation of long-chain fatty acid import across plasma membrane | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 4 | 31 | 6.19e-01 |
| GO:BP | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 6.19e-01 |
| GO:BP | GO:1902073 | positive regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 2 | 13 | 6.19e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 2 | 13 | 6.19e-01 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 13 | 24 | 6.19e-01 |
| GO:BP | GO:0035437 | maintenance of protein localization in endoplasmic reticulum | 6 | 11 | 6.19e-01 |
| GO:BP | GO:0009107 | lipoate biosynthetic process | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0002176 | male germ cell proliferation | 2 | 6 | 6.19e-01 |
| GO:BP | GO:0035635 | entry of bacterium into host cell | 2 | 8 | 6.19e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 7 | 21 | 6.19e-01 |
| GO:BP | GO:0009106 | lipoate metabolic process | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0070317 | negative regulation of G0 to G1 transition | 1 | 5 | 6.19e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 7 | 19 | 6.19e-01 |
| GO:BP | GO:0090382 | phagosome maturation | 5 | 24 | 6.19e-01 |
| GO:BP | GO:0021629 | olfactory nerve structural organization | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 8 | 83 | 6.19e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 6.19e-01 |
| GO:BP | GO:0071922 | regulation of cohesin loading | 1 | 2 | 6.19e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 5 | 12 | 6.19e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 122 | 438 | 6.19e-01 |
| GO:BP | GO:0140250 | regulation protein catabolic process at synapse | 3 | 5 | 6.19e-01 |
| GO:BP | GO:0042635 | positive regulation of hair cycle | 1 | 1 | 6.20e-01 |
| GO:BP | GO:0051030 | snRNA transport | 3 | 6 | 6.20e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 4 | 10 | 6.20e-01 |
| GO:BP | GO:1905590 | fibronectin fibril organization | 1 | 1 | 6.20e-01 |
| GO:BP | GO:0072177 | mesonephric duct development | 2 | 7 | 6.20e-01 |
| GO:BP | GO:0061458 | reproductive system development | 7 | 215 | 6.20e-01 |
| GO:BP | GO:0090156 | intracellular sphingolipid homeostasis | 1 | 5 | 6.20e-01 |
| GO:BP | GO:0002331 | pre-B cell allelic exclusion | 1 | 2 | 6.20e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 11 | 170 | 6.20e-01 |
| GO:BP | GO:2000774 | positive regulation of cellular senescence | 4 | 9 | 6.20e-01 |
| GO:BP | GO:0003014 | renal system process | 13 | 78 | 6.20e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 3 | 40 | 6.21e-01 |
| GO:BP | GO:0002101 | tRNA wobble cytosine modification | 1 | 2 | 6.21e-01 |
| GO:BP | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential | 2 | 4 | 6.21e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 6 | 13 | 6.21e-01 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 2 | 8 | 6.21e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 4 | 30 | 6.21e-01 |
| GO:BP | GO:0035624 | receptor transactivation | 3 | 3 | 6.21e-01 |
| GO:BP | GO:0060092 | regulation of synaptic transmission, glycinergic | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0045348 | positive regulation of MHC class II biosynthetic process | 2 | 8 | 6.21e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 6.22e-01 |
| GO:BP | GO:0050890 | cognition | 11 | 227 | 6.22e-01 |
| GO:BP | GO:0033366 | protein localization to secretory granule | 1 | 1 | 6.22e-01 |
| GO:BP | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 1 | 1 | 6.22e-01 |
| GO:BP | GO:1903742 | regulation of anterograde synaptic vesicle transport | 2 | 3 | 6.22e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 2 | 25 | 6.22e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 40 | 227 | 6.22e-01 |
| GO:BP | GO:0010795 | regulation of ubiquinone biosynthetic process | 1 | 2 | 6.23e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 5 | 17 | 6.23e-01 |
| GO:BP | GO:0006669 | sphinganine-1-phosphate biosynthetic process | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 6.23e-01 |
| GO:BP | GO:0090186 | regulation of pancreatic juice secretion | 1 | 6 | 6.23e-01 |
| GO:BP | GO:0043977 | histone H2A-K5 acetylation | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0043980 | histone H2B-K12 acetylation | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing | 4 | 6 | 6.23e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 6.23e-01 |
| GO:BP | GO:0030961 | peptidyl-arginine hydroxylation | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 2 | 15 | 6.23e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 2 | 15 | 6.23e-01 |
| GO:BP | GO:0010832 | negative regulation of myotube differentiation | 6 | 10 | 6.23e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 13 | 53 | 6.23e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 6.23e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 3 | 20 | 6.23e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 1 | 11 | 6.23e-01 |
| GO:BP | GO:1901483 | regulation of transcription factor catabolic process | 1 | 3 | 6.23e-01 |
| GO:BP | GO:1901485 | positive regulation of transcription factor catabolic process | 1 | 3 | 6.23e-01 |
| GO:BP | GO:0099527 | postsynapse to nucleus signaling pathway | 1 | 6 | 6.23e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 1 | 19 | 6.23e-01 |
| GO:BP | GO:0035971 | peptidyl-histidine dephosphorylation | 1 | 1 | 6.24e-01 |
| GO:BP | GO:0021999 | neural plate anterior/posterior regionalization | 2 | 5 | 6.24e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 2 | 9 | 6.24e-01 |
| GO:BP | GO:2000984 | negative regulation of ATP citrate synthase activity | 1 | 1 | 6.24e-01 |
| GO:BP | GO:1905512 | regulation of short-term synaptic potentiation | 1 | 1 | 6.24e-01 |
| GO:BP | GO:1905513 | negative regulation of short-term synaptic potentiation | 1 | 1 | 6.24e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 8 | 27 | 6.24e-01 |
| GO:BP | GO:0009229 | thiamine diphosphate biosynthetic process | 1 | 1 | 6.25e-01 |
| GO:BP | GO:0042724 | thiamine-containing compound biosynthetic process | 1 | 1 | 6.25e-01 |
| GO:BP | GO:0018146 | keratan sulfate biosynthetic process | 5 | 13 | 6.25e-01 |
| GO:BP | GO:0033206 | meiotic cytokinesis | 1 | 7 | 6.25e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 1 | 12 | 6.25e-01 |
| GO:BP | GO:0009651 | response to salt stress | 2 | 18 | 6.25e-01 |
| GO:BP | GO:0010710 | regulation of collagen catabolic process | 2 | 3 | 6.25e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 23 | 881 | 6.25e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 2 | 8 | 6.25e-01 |
| GO:BP | GO:0046951 | ketone body biosynthetic process | 2 | 5 | 6.25e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 1 | 12 | 6.25e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 398 | 6.25e-01 |
| GO:BP | GO:0048699 | generation of neurons | 2 | 1119 | 6.25e-01 |
| GO:BP | GO:0072386 | plus-end-directed organelle transport along microtubule | 1 | 7 | 6.25e-01 |
| GO:BP | GO:0001661 | conditioned taste aversion | 1 | 2 | 6.25e-01 |
| GO:BP | GO:0007286 | spermatid development | 4 | 105 | 6.25e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 6 | 83 | 6.25e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 1 | 118 | 6.25e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 1 | 118 | 6.25e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 2 | 26 | 6.25e-01 |
| GO:BP | GO:1904386 | response to L-phenylalanine derivative | 3 | 5 | 6.25e-01 |
| GO:BP | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 5 | 21 | 6.25e-01 |
| GO:BP | GO:0042262 | DNA protection | 1 | 6 | 6.25e-01 |
| GO:BP | GO:0050821 | protein stabilization | 3 | 191 | 6.25e-01 |
| GO:BP | GO:1990169 | stress response to copper ion | 3 | 8 | 6.25e-01 |
| GO:BP | GO:0010273 | detoxification of copper ion | 3 | 8 | 6.25e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 6.25e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 8 | 34 | 6.26e-01 |
| GO:BP | GO:0003419 | growth plate cartilage chondrocyte proliferation | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0008039 | synaptic target recognition | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 4 | 28 | 6.26e-01 |
| GO:BP | GO:0032877 | positive regulation of DNA endoreduplication | 1 | 2 | 6.26e-01 |
| GO:BP | GO:0016036 | cellular response to phosphate starvation | 1 | 2 | 6.26e-01 |
| GO:BP | GO:1901678 | iron coordination entity transport | 3 | 11 | 6.26e-01 |
| GO:BP | GO:0070376 | regulation of ERK5 cascade | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 4 | 19 | 6.26e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 35 | 1380 | 6.26e-01 |
| GO:BP | GO:0006403 | RNA localization | 40 | 182 | 6.27e-01 |
| GO:BP | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus | 3 | 6 | 6.27e-01 |
| GO:BP | GO:1905069 | allantois development | 2 | 4 | 6.27e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 3 | 1914 | 6.27e-01 |
| GO:BP | GO:0097377 | spinal cord interneuron axon guidance | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 4 | 1898 | 6.27e-01 |
| GO:BP | GO:0018106 | peptidyl-histidine phosphorylation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation | 1 | 3 | 6.27e-01 |
| GO:BP | GO:1901723 | negative regulation of cell proliferation involved in kidney development | 1 | 3 | 6.27e-01 |
| GO:BP | GO:1904450 | positive regulation of aspartate secretion | 1 | 2 | 6.27e-01 |
| GO:BP | GO:1904448 | regulation of aspartate secretion | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0009450 | gamma-aminobutyric acid catabolic process | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0097477 | lateral motor column neuron migration | 1 | 3 | 6.27e-01 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 5 | 10 | 6.27e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 4 | 111 | 6.27e-01 |
| GO:BP | GO:0006579 | amino-acid betaine catabolic process | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0006769 | nicotinamide metabolic process | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0035846 | oviduct epithelium development | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0061528 | aspartate secretion | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0061744 | motor behavior | 2 | 17 | 6.27e-01 |
| GO:BP | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis | 4 | 6 | 6.27e-01 |
| GO:BP | GO:0071921 | cohesin loading | 2 | 3 | 6.27e-01 |
| GO:BP | GO:2000492 | regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 3 | 86 | 6.27e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 5 | 38 | 6.27e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 6.27e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 7 | 196 | 6.27e-01 |
| GO:BP | GO:0072262 | metanephric glomerular mesangial cell proliferation involved in metanephros development | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 6.27e-01 |
| GO:BP | GO:0110112 | regulation of lipid transporter activity | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration | 3 | 5 | 6.27e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 3 | 26 | 6.27e-01 |
| GO:BP | GO:0051004 | regulation of lipoprotein lipase activity | 1 | 11 | 6.27e-01 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 2 | 25 | 6.27e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 2 | 39 | 6.27e-01 |
| GO:BP | GO:0045190 | isotype switching | 2 | 39 | 6.27e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 2 | 39 | 6.27e-01 |
| GO:BP | GO:0098758 | response to interleukin-8 | 1 | 1 | 6.27e-01 |
| GO:BP | GO:2000182 | regulation of progesterone biosynthetic process | 1 | 3 | 6.27e-01 |
| GO:BP | GO:0060086 | circadian temperature homeostasis | 1 | 3 | 6.27e-01 |
| GO:BP | GO:0098759 | cellular response to interleukin-8 | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 3 | 7 | 6.27e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 2 | 28 | 6.27e-01 |
| GO:BP | GO:1902771 | positive regulation of choline O-acetyltransferase activity | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1902769 | regulation of choline O-acetyltransferase activity | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1902997 | negative regulation of neurofibrillary tangle assembly | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1902955 | positive regulation of early endosome to recycling endosome transport | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1901303 | negative regulation of cargo loading into COPII-coated vesicle | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 1 | 17 | 6.27e-01 |
| GO:BP | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 6.27e-01 |
| GO:BP | GO:0070651 | nonfunctional rRNA decay | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 6.27e-01 |
| GO:BP | GO:1903801 | L-leucine import across plasma membrane | 2 | 5 | 6.27e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 2 | 12 | 6.27e-01 |
| GO:BP | GO:0098713 | leucine import across plasma membrane | 2 | 5 | 6.27e-01 |
| GO:BP | GO:0021944 | neuronal-glial interaction involved in hindbrain glial-mediated radial cell migration | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0098917 | retrograde trans-synaptic signaling | 5 | 7 | 6.28e-01 |
| GO:BP | GO:0071918 | urea transmembrane transport | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0001743 | lens placode formation | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0008626 | granzyme-mediated apoptotic signaling pathway | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0046619 | lens placode formation involved in camera-type eye formation | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 15 | 81 | 6.28e-01 |
| GO:BP | GO:1901911 | adenosine 5’-(hexahydrogen pentaphosphate) catabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 8 | 154 | 6.28e-01 |
| GO:BP | GO:1901910 | adenosine 5’-(hexahydrogen pentaphosphate) metabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:1901906 | diadenosine pentaphosphate metabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0060939 | epicardium-derived cardiac fibroblast cell development | 2 | 3 | 6.28e-01 |
| GO:BP | GO:1901909 | diadenosine hexaphosphate catabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:1901908 | diadenosine hexaphosphate metabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0060938 | epicardium-derived cardiac fibroblast cell differentiation | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0071543 | diphosphoinositol polyphosphate metabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:1901907 | diadenosine pentaphosphate catabolic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 5 | 8 | 6.28e-01 |
| GO:BP | GO:0071107 | response to parathyroid hormone | 1 | 9 | 6.28e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 1 | 324 | 6.28e-01 |
| GO:BP | GO:0106035 | protein maturation by [4Fe-4S] cluster transfer | 2 | 7 | 6.28e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 11 | 31 | 6.28e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 5 | 60 | 6.28e-01 |
| GO:BP | GO:0014820 | tonic smooth muscle contraction | 2 | 8 | 6.28e-01 |
| GO:BP | GO:0072007 | mesangial cell differentiation | 1 | 5 | 6.28e-01 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 1 | 6 | 6.28e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 2 | 126 | 6.28e-01 |
| GO:BP | GO:0072660 | maintenance of protein location in plasma membrane | 1 | 2 | 6.28e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 2 | 276 | 6.28e-01 |
| GO:BP | GO:0072658 | maintenance of protein location in membrane | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 3 | 18 | 6.28e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 3 | 18 | 6.28e-01 |
| GO:BP | GO:1901509 | regulation of endothelial tube morphogenesis | 3 | 3 | 6.28e-01 |
| GO:BP | GO:0038154 | interleukin-11-mediated signaling pathway | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 6.28e-01 |
| GO:BP | GO:0038001 | paracrine signaling | 1 | 3 | 6.28e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 5 | 109 | 6.28e-01 |
| GO:BP | GO:0002155 | regulation of thyroid hormone mediated signaling pathway | 2 | 5 | 6.28e-01 |
| GO:BP | GO:0032808 | lacrimal gland development | 2 | 5 | 6.28e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 1 | 19 | 6.29e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 1 | 19 | 6.29e-01 |
| GO:BP | GO:0043973 | histone H3-K4 acetylation | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 26 | 141 | 6.29e-01 |
| GO:BP | GO:1905584 | outer hair cell apoptotic process | 1 | 2 | 6.29e-01 |
| GO:BP | GO:0048200 | Golgi transport vesicle coating | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0035964 | COPI-coated vesicle budding | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0048205 | COPI coating of Golgi vesicle | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0061890 | positive regulation of astrocyte activation | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 6.29e-01 |
| GO:BP | GO:0018206 | peptidyl-methionine modification | 3 | 13 | 6.29e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 8 | 85 | 6.29e-01 |
| GO:BP | GO:0032240 | negative regulation of nucleobase-containing compound transport | 2 | 3 | 6.29e-01 |
| GO:BP | GO:1905464 | positive regulation of DNA duplex unwinding | 1 | 3 | 6.29e-01 |
| GO:BP | GO:1905776 | positive regulation of DNA helicase activity | 1 | 3 | 6.29e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 6.29e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 11 | 23 | 6.29e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 146 | 6.29e-01 |
| GO:BP | GO:0039533 | regulation of MDA-5 signaling pathway | 1 | 5 | 6.29e-01 |
| GO:BP | GO:0010522 | regulation of calcium ion transport into cytosol | 1 | 10 | 6.29e-01 |
| GO:BP | GO:0060384 | innervation | 4 | 22 | 6.29e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 2 | 84 | 6.29e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 3 | 111 | 6.30e-01 |
| GO:BP | GO:1903849 | positive regulation of aorta morphogenesis | 1 | 2 | 6.30e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 6.30e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 3 | 12 | 6.30e-01 |
| GO:BP | GO:0048203 | vesicle targeting, trans-Golgi to endosome | 1 | 2 | 6.30e-01 |
| GO:BP | GO:0003169 | coronary vein morphogenesis | 1 | 2 | 6.30e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 6.30e-01 |
| GO:BP | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling | 2 | 8 | 6.30e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 3 | 12 | 6.30e-01 |
| GO:BP | GO:1903847 | regulation of aorta morphogenesis | 1 | 2 | 6.30e-01 |
| GO:BP | GO:0003100 | regulation of systemic arterial blood pressure by endothelin | 1 | 5 | 6.30e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 7 | 26 | 6.30e-01 |
| GO:BP | GO:0050434 | positive regulation of viral transcription | 2 | 3 | 6.30e-01 |
| GO:BP | GO:2000452 | regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0031284 | positive regulation of guanylate cyclase activity | 1 | 2 | 6.30e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 2 | 26 | 6.30e-01 |
| GO:BP | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0042552 | myelination | 1 | 111 | 6.30e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 6.30e-01 |
| GO:BP | GO:0071863 | regulation of cell proliferation in bone marrow | 2 | 6 | 6.31e-01 |
| GO:BP | GO:0032696 | negative regulation of interleukin-13 production | 2 | 3 | 6.31e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 2 | 6 | 6.31e-01 |
| GO:BP | GO:0017014 | protein nitrosylation | 1 | 11 | 6.31e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 5 | 41 | 6.31e-01 |
| GO:BP | GO:0018119 | peptidyl-cysteine S-nitrosylation | 1 | 11 | 6.31e-01 |
| GO:BP | GO:0071642 | positive regulation of macrophage inflammatory protein 1 alpha production | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 6.31e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 15 | 44 | 6.31e-01 |
| GO:BP | GO:1903939 | regulation of TORC2 signaling | 3 | 5 | 6.31e-01 |
| GO:BP | GO:1902656 | calcium ion import into cytosol | 1 | 1 | 6.31e-01 |
| GO:BP | GO:1900142 | negative regulation of oligodendrocyte apoptotic process | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0048377 | lateral mesodermal cell fate specification | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0048372 | lateral mesodermal cell fate commitment | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0030682 | mitigation of host defenses by symbiont | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0048378 | regulation of lateral mesodermal cell fate specification | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0052173 | response to defenses of other organism | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0034031 | ribonucleoside bisphosphate catabolic process | 6 | 11 | 6.31e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 1 | 26 | 6.31e-01 |
| GO:BP | GO:0008033 | tRNA processing | 47 | 128 | 6.31e-01 |
| GO:BP | GO:1905285 | fibrous ring of heart morphogenesis | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0052200 | response to host defenses | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0061183 | regulation of dermatome development | 1 | 2 | 6.31e-01 |
| GO:BP | GO:2000533 | negative regulation of renal albumin absorption | 1 | 1 | 6.31e-01 |
| GO:BP | GO:2001223 | negative regulation of neuron migration | 3 | 8 | 6.31e-01 |
| GO:BP | GO:0048338 | mesoderm structural organization | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0048352 | paraxial mesoderm structural organization | 1 | 1 | 6.31e-01 |
| GO:BP | GO:1905603 | regulation of blood-brain barrier permeability | 1 | 6 | 6.31e-01 |
| GO:BP | GO:0075136 | response to host | 1 | 1 | 6.31e-01 |
| GO:BP | GO:1904412 | regulation of cardiac ventricle development | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0061054 | dermatome development | 1 | 2 | 6.31e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 1 | 540 | 6.31e-01 |
| GO:BP | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1 | 17 | 6.31e-01 |
| GO:BP | GO:0033869 | nucleoside bisphosphate catabolic process | 6 | 11 | 6.31e-01 |
| GO:BP | GO:0042335 | cuticle development | 1 | 2 | 6.31e-01 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 3 | 9 | 6.31e-01 |
| GO:BP | GO:0034034 | purine nucleoside bisphosphate catabolic process | 6 | 11 | 6.31e-01 |
| GO:BP | GO:1901143 | insulin catabolic process | 1 | 3 | 6.31e-01 |
| GO:BP | GO:1904414 | positive regulation of cardiac ventricle development | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0021998 | neural plate mediolateral regionalization | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 3 | 40 | 6.31e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 3 | 280 | 6.31e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 21 | 67 | 6.31e-01 |
| GO:BP | GO:0002175 | protein localization to paranode region of axon | 2 | 3 | 6.31e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 6.31e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 1 | 5 | 6.31e-01 |
| GO:BP | GO:0006364 | rRNA processing | 3 | 221 | 6.31e-01 |
| GO:BP | GO:1901233 | negative regulation of convergent extension involved in axis elongation | 1 | 1 | 6.32e-01 |
| GO:BP | GO:1901232 | regulation of convergent extension involved in axis elongation | 1 | 1 | 6.32e-01 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 6.32e-01 |
| GO:BP | GO:1901231 | positive regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 7 | 71 | 6.32e-01 |
| GO:BP | GO:1901229 | regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 2 | 82 | 6.32e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 2 | 52 | 6.32e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 2 | 1201 | 6.32e-01 |
| GO:BP | GO:1903818 | positive regulation of voltage-gated potassium channel activity | 1 | 11 | 6.32e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 1 | 113 | 6.32e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 1 | 30 | 6.32e-01 |
| GO:BP | GO:0099040 | ceramide translocation | 1 | 2 | 6.32e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 2 | 26 | 6.32e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 3 | 90 | 6.32e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 2 | 11 | 6.32e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 1 | 113 | 6.32e-01 |
| GO:BP | GO:0014857 | regulation of skeletal muscle cell proliferation | 6 | 10 | 6.32e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 18 | 56 | 6.32e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 150 | 6.32e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 6.32e-01 |
| GO:BP | GO:0001654 | eye development | 11 | 272 | 6.32e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 4 | 25 | 6.33e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 6.33e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 4 | 25 | 6.33e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 6.33e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 6.33e-01 |
| GO:BP | GO:0140719 | constitutive heterochromatin formation | 6 | 10 | 6.33e-01 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 3 | 11 | 6.33e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 54 | 95 | 6.33e-01 |
| GO:BP | GO:0097428 | protein maturation by iron-sulfur cluster transfer | 1 | 17 | 6.33e-01 |
| GO:BP | GO:0061061 | muscle structure development | 4 | 546 | 6.33e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 3 | 640 | 6.33e-01 |
| GO:BP | GO:0021686 | cerebellar granular layer maturation | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0090234 | regulation of kinetochore assembly | 1 | 3 | 6.33e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 1 | 26 | 6.33e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 2 | 6 | 6.33e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 1 | 18 | 6.33e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 7 | 1076 | 6.33e-01 |
| GO:BP | GO:0018197 | peptidyl-aspartic acid modification | 3 | 5 | 6.33e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 9 | 230 | 6.33e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 2 | 6 | 6.33e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 7 | 1074 | 6.33e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 2 | 6 | 6.33e-01 |
| GO:BP | GO:0016553 | base conversion or substitution editing | 1 | 10 | 6.33e-01 |
| GO:BP | GO:0009227 | nucleotide-sugar catabolic process | 2 | 3 | 6.33e-01 |
| GO:BP | GO:0150045 | regulation of synaptic signaling by nitric oxide | 1 | 1 | 6.33e-01 |
| GO:BP | GO:2001136 | negative regulation of endocytic recycling | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0021933 | radial glia guided migration of cerebellar granule cell | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0006702 | androgen biosynthetic process | 2 | 4 | 6.33e-01 |
| GO:BP | GO:0006651 | diacylglycerol biosynthetic process | 5 | 7 | 6.33e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 3 | 8 | 6.33e-01 |
| GO:BP | GO:0023035 | CD40 signaling pathway | 2 | 11 | 6.33e-01 |
| GO:BP | GO:0021935 | cerebellar granule cell precursor tangential migration | 1 | 2 | 6.34e-01 |
| GO:BP | GO:0106091 | glial cell projection elongation | 1 | 2 | 6.34e-01 |
| GO:BP | GO:0030263 | apoptotic chromosome condensation | 1 | 4 | 6.34e-01 |
| GO:BP | GO:0098937 | anterograde dendritic transport | 5 | 9 | 6.34e-01 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 2 | 11 | 6.34e-01 |
| GO:BP | GO:0009912 | auditory receptor cell fate commitment | 3 | 4 | 6.34e-01 |
| GO:BP | GO:1901176 | lycopene catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 2 | 18 | 6.34e-01 |
| GO:BP | GO:1901826 | zeaxanthin catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0021615 | glossopharyngeal nerve morphogenesis | 2 | 2 | 6.34e-01 |
| GO:BP | GO:0072189 | ureter development | 5 | 10 | 6.34e-01 |
| GO:BP | GO:1901175 | lycopene metabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0071360 | cellular response to exogenous dsRNA | 6 | 14 | 6.34e-01 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 3 | 21 | 6.34e-01 |
| GO:BP | GO:0016124 | xanthophyll catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0140569 | extraction of mislocalized protein from ER membrane | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0032736 | positive regulation of interleukin-13 production | 1 | 7 | 6.34e-01 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 3 | 21 | 6.34e-01 |
| GO:BP | GO:0060120 | inner ear receptor cell fate commitment | 3 | 4 | 6.34e-01 |
| GO:BP | GO:0062172 | lutein catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0006545 | glycine biosynthetic process | 1 | 4 | 6.34e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 5 | 23 | 6.34e-01 |
| GO:BP | GO:0062170 | lutein metabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0016110 | tetraterpenoid catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0016118 | carotenoid catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0097502 | mannosylation | 6 | 32 | 6.34e-01 |
| GO:BP | GO:0021508 | floor plate formation | 1 | 3 | 6.34e-01 |
| GO:BP | GO:1903564 | regulation of protein localization to cilium | 3 | 11 | 6.34e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 19 | 37 | 6.34e-01 |
| GO:BP | GO:0098789 | pre-mRNA cleavage required for polyadenylation | 1 | 12 | 6.34e-01 |
| GO:BP | GO:0019449 | L-cysteine catabolic process to hypotaurine | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity | 3 | 5 | 6.34e-01 |
| GO:BP | GO:0019452 | L-cysteine catabolic process to taurine | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 6 | 17 | 6.34e-01 |
| GO:BP | GO:0042840 | D-glucuronate catabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 11 | 60 | 6.34e-01 |
| GO:BP | GO:0042839 | D-glucuronate metabolic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 3 | 20 | 6.34e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 2 | 5 | 6.34e-01 |
| GO:BP | GO:0015677 | copper ion import | 1 | 5 | 6.34e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 16 | 43 | 6.34e-01 |
| GO:BP | GO:0110012 | protein localization to P-body | 1 | 1 | 6.34e-01 |
| GO:BP | GO:1903826 | L-arginine transmembrane transport | 1 | 12 | 6.34e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 16 | 43 | 6.34e-01 |
| GO:BP | GO:0038173 | interleukin-17A-mediated signaling pathway | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0070926 | regulation of ATP:ADP antiporter activity | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0030505 | inorganic diphosphate transport | 1 | 3 | 6.34e-01 |
| GO:BP | GO:1901380 | negative regulation of potassium ion transmembrane transport | 3 | 18 | 6.34e-01 |
| GO:BP | GO:0007423 | sensory organ development | 1 | 396 | 6.34e-01 |
| GO:BP | GO:0097474 | retinal cone cell apoptotic process | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0002334 | transitional two stage B cell differentiation | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 1 | 10 | 6.34e-01 |
| GO:BP | GO:0048468 | cell development | 4 | 1963 | 6.35e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 3 | 5 | 6.35e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 2 | 80 | 6.36e-01 |
| GO:BP | GO:1902074 | response to salt | 8 | 271 | 6.36e-01 |
| GO:BP | GO:0060897 | neural plate regionalization | 3 | 6 | 6.36e-01 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 6.37e-01 |
| GO:BP | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0009620 | response to fungus | 1 | 22 | 6.37e-01 |
| GO:BP | GO:0031144 | proteasome localization | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0036071 | N-glycan fucosylation | 2 | 3 | 6.37e-01 |
| GO:BP | GO:0002524 | hypersensitivity | 3 | 7 | 6.37e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 9 | 62 | 6.38e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 4 | 111 | 6.38e-01 |
| GO:BP | GO:1990791 | dorsal root ganglion development | 3 | 6 | 6.38e-01 |
| GO:BP | GO:0043113 | receptor clustering | 6 | 45 | 6.38e-01 |
| GO:BP | GO:0071472 | cellular response to salt stress | 4 | 11 | 6.38e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 3 | 12 | 6.38e-01 |
| GO:BP | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering | 2 | 4 | 6.39e-01 |
| GO:BP | GO:1903638 | positive regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 6.39e-01 |
| GO:BP | GO:1903636 | regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 6.39e-01 |
| GO:BP | GO:0031958 | corticosteroid receptor signaling pathway | 2 | 14 | 6.39e-01 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 1 | 12 | 6.39e-01 |
| GO:BP | GO:0060901 | regulation of hair cycle by canonical Wnt signaling pathway | 1 | 1 | 6.39e-01 |
| GO:BP | GO:1902910 | positive regulation of melanosome transport | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 3 | 33 | 6.39e-01 |
| GO:BP | GO:0051796 | negative regulation of timing of catagen | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 5 | 15 | 6.39e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 8 | 27 | 6.39e-01 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 3 | 33 | 6.39e-01 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 5 | 19 | 6.39e-01 |
| GO:BP | GO:1903608 | protein localization to cytoplasmic stress granule | 3 | 7 | 6.39e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 5 | 13 | 6.39e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 17 | 30 | 6.39e-01 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway | 2 | 3 | 6.39e-01 |
| GO:BP | GO:0009438 | methylglyoxal metabolic process | 3 | 7 | 6.39e-01 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0070126 | mitochondrial translational termination | 1 | 4 | 6.39e-01 |
| GO:BP | GO:0048469 | cell maturation | 2 | 108 | 6.39e-01 |
| GO:BP | GO:0042428 | serotonin metabolic process | 1 | 6 | 6.39e-01 |
| GO:BP | GO:0043309 | regulation of eosinophil degranulation | 2 | 2 | 6.39e-01 |
| GO:BP | GO:0019236 | response to pheromone | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0030030 | cell projection organization | 2 | 1233 | 6.40e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 6.40e-01 |
| GO:BP | GO:0048319 | axial mesoderm morphogenesis | 1 | 3 | 6.40e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 4 | 8 | 6.40e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 28 | 49 | 6.40e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 1 | 39 | 6.40e-01 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 1 | 12 | 6.40e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 6.40e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 19 | 52 | 6.40e-01 |
| GO:BP | GO:0002520 | immune system development | 1 | 144 | 6.40e-01 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 1 | 12 | 6.40e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 6.40e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 6.40e-01 |
| GO:BP | GO:0042753 | positive regulation of circadian rhythm | 3 | 10 | 6.40e-01 |
| GO:BP | GO:0051901 | positive regulation of mitochondrial depolarization | 1 | 6 | 6.40e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 3 | 11 | 6.40e-01 |
| GO:BP | GO:0150063 | visual system development | 11 | 273 | 6.40e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 6.40e-01 |
| GO:BP | GO:1902896 | terminal web assembly | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 4 | 25 | 6.40e-01 |
| GO:BP | GO:1902259 | regulation of delayed rectifier potassium channel activity | 1 | 12 | 6.40e-01 |
| GO:BP | GO:0033240 | positive regulation of amine metabolic process | 3 | 7 | 6.40e-01 |
| GO:BP | GO:1903463 | regulation of mitotic cell cycle DNA replication | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0061698 | protein deglutarylation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0051410 | detoxification of nitrogen compound | 2 | 5 | 6.40e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 5 | 9 | 6.40e-01 |
| GO:BP | GO:0140706 | protein-containing complex localization to centriolar satellite | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0014829 | vascular associated smooth muscle contraction | 7 | 20 | 6.40e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0030854 | positive regulation of granulocyte differentiation | 3 | 5 | 6.40e-01 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 3 | 22 | 6.40e-01 |
| GO:BP | GO:0070458 | cellular detoxification of nitrogen compound | 2 | 5 | 6.40e-01 |
| GO:BP | GO:0048697 | positive regulation of collateral sprouting in absence of injury | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 1 | 18 | 6.40e-01 |
| GO:BP | GO:1904692 | positive regulation of type B pancreatic cell proliferation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 1 | 23 | 6.40e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 16 | 78 | 6.40e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 5 | 9 | 6.40e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 2 | 12 | 6.40e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 3 | 1995 | 6.40e-01 |
| GO:BP | GO:0010265 | SCF complex assembly | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation | 2 | 3 | 6.40e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 25 | 120 | 6.40e-01 |
| GO:BP | GO:0010839 | negative regulation of keratinocyte proliferation | 1 | 19 | 6.40e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 22 | 252 | 6.40e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 16 | 341 | 6.40e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 6.40e-01 |
| GO:BP | GO:0014724 | regulation of twitch skeletal muscle contraction | 1 | 3 | 6.40e-01 |
| GO:BP | GO:0001756 | somitogenesis | 15 | 48 | 6.40e-01 |
| GO:BP | GO:1905913 | negative regulation of calcium ion export across plasma membrane | 2 | 4 | 6.40e-01 |
| GO:BP | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity | 1 | 3 | 6.40e-01 |
| GO:BP | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity | 2 | 3 | 6.40e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 3 | 52 | 6.40e-01 |
| GO:BP | GO:0036048 | protein desuccinylation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0040031 | snRNA modification | 2 | 7 | 6.40e-01 |
| GO:BP | GO:1905912 | regulation of calcium ion export across plasma membrane | 2 | 4 | 6.40e-01 |
| GO:BP | GO:1902440 | protein localization to mitotic spindle pole body | 1 | 1 | 6.40e-01 |
| GO:BP | GO:1900099 | negative regulation of plasma cell differentiation | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0036049 | peptidyl-lysine desuccinylation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 6.40e-01 |
| GO:BP | GO:0060948 | cardiac vascular smooth muscle cell development | 2 | 3 | 6.40e-01 |
| GO:BP | GO:0031033 | myosin filament organization | 3 | 5 | 6.40e-01 |
| GO:BP | GO:0060242 | contact inhibition | 4 | 8 | 6.40e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 6.40e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 5 | 35 | 6.40e-01 |
| GO:BP | GO:0046849 | bone remodeling | 5 | 59 | 6.41e-01 |
| GO:BP | GO:0045141 | meiotic telomere clustering | 2 | 5 | 6.41e-01 |
| GO:BP | GO:1903489 | positive regulation of lactation | 1 | 1 | 6.41e-01 |
| GO:BP | GO:1904980 | positive regulation of endosome organization | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0042745 | circadian sleep/wake cycle | 2 | 11 | 6.41e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 5 | 199 | 6.41e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 6.41e-01 |
| GO:BP | GO:0007141 | male meiosis I | 6 | 15 | 6.41e-01 |
| GO:BP | GO:0051645 | Golgi localization | 2 | 14 | 6.41e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 3 | 100 | 6.41e-01 |
| GO:BP | GO:0015808 | L-alanine transport | 3 | 9 | 6.41e-01 |
| GO:BP | GO:0097018 | renal albumin absorption | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0009757 | hexose mediated signaling | 1 | 5 | 6.41e-01 |
| GO:BP | GO:0016310 | phosphorylation | 8 | 1372 | 6.41e-01 |
| GO:BP | GO:0035287 | head segmentation | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0010255 | glucose mediated signaling pathway | 1 | 5 | 6.41e-01 |
| GO:BP | GO:0010182 | sugar mediated signaling pathway | 1 | 5 | 6.41e-01 |
| GO:BP | GO:1900454 | positive regulation of long-term synaptic depression | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0019389 | glucuronoside metabolic process | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0019391 | glucuronoside catabolic process | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 3 | 19 | 6.41e-01 |
| GO:BP | GO:0007380 | specification of segmental identity, head | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0035289 | posterior head segmentation | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 422 | 6.41e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 6.41e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 15 | 281 | 6.41e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 16 | 342 | 6.41e-01 |
| GO:BP | GO:1901632 | regulation of synaptic vesicle membrane organization | 6 | 8 | 6.41e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 6.41e-01 |
| GO:BP | GO:0035356 | intracellular triglyceride homeostasis | 4 | 5 | 6.41e-01 |
| GO:BP | GO:0035771 | interleukin-4-mediated signaling pathway | 2 | 4 | 6.41e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 5 | 105 | 6.41e-01 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 1 | 5 | 6.41e-01 |
| GO:BP | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 4 | 9 | 6.41e-01 |
| GO:BP | GO:0072011 | glomerular endothelium development | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0001971 | negative regulation of activation of membrane attack complex | 1 | 1 | 6.41e-01 |
| GO:BP | GO:1903210 | podocyte apoptotic process | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 6 | 11 | 6.41e-01 |
| GO:BP | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic active zone membrane | 6 | 8 | 6.41e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 6.41e-01 |
| GO:BP | GO:0009443 | pyridoxal 5’-phosphate salvage | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 4 | 37 | 6.41e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 12 | 37 | 6.41e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 6 | 69 | 6.41e-01 |
| GO:BP | GO:0051639 | actin filament network formation | 1 | 7 | 6.42e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 8 | 30 | 6.42e-01 |
| GO:BP | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway | 1 | 7 | 6.42e-01 |
| GO:BP | GO:0031340 | positive regulation of vesicle fusion | 1 | 9 | 6.42e-01 |
| GO:BP | GO:1905280 | negative regulation of retrograde transport, endosome to Golgi | 1 | 1 | 6.42e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 2 | 15 | 6.42e-01 |
| GO:BP | GO:0099161 | regulation of presynaptic dense core granule exocytosis | 1 | 1 | 6.43e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 6.43e-01 |
| GO:BP | GO:0061670 | evoked neurotransmitter secretion | 1 | 1 | 6.43e-01 |
| GO:BP | GO:0071389 | cellular response to mineralocorticoid stimulus | 4 | 9 | 6.43e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 12 | 64 | 6.43e-01 |
| GO:BP | GO:0006526 | arginine biosynthetic process | 3 | 4 | 6.43e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 5 | 105 | 6.43e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 3 | 56 | 6.43e-01 |
| GO:BP | GO:0006501 | C-terminal protein lipidation | 2 | 6 | 6.44e-01 |
| GO:BP | GO:0006548 | histidine catabolic process | 1 | 3 | 6.44e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 9 | 448 | 6.44e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 5 | 9 | 6.44e-01 |
| GO:BP | GO:0016128 | phytosteroid metabolic process | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0015840 | urea transport | 1 | 2 | 6.44e-01 |
| GO:BP | GO:0016129 | phytosteroid biosynthetic process | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 6 | 775 | 6.44e-01 |
| GO:BP | GO:0016131 | brassinosteroid metabolic process | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0016132 | brassinosteroid biosynthetic process | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 2 | 69 | 6.44e-01 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 1 | 14 | 6.44e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 2 | 11 | 6.44e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 6.44e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 3 | 8 | 6.44e-01 |
| GO:BP | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | 1 | 4 | 6.44e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 9 | 51 | 6.44e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 3 | 8 | 6.44e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 14 | 71 | 6.44e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 3 | 8 | 6.44e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 7 | 1167 | 6.45e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 27 | 59 | 6.45e-01 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 3 | 7 | 6.45e-01 |
| GO:BP | GO:1902214 | regulation of interleukin-4-mediated signaling pathway | 1 | 2 | 6.45e-01 |
| GO:BP | GO:0044795 | trans-Golgi network to recycling endosome transport | 1 | 1 | 6.45e-01 |
| GO:BP | GO:0060155 | platelet dense granule organization | 9 | 23 | 6.45e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 122 | 6.45e-01 |
| GO:BP | GO:0016322 | neuron remodeling | 3 | 11 | 6.45e-01 |
| GO:BP | GO:0045041 | protein import into mitochondrial intermembrane space | 2 | 3 | 6.45e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 11 | 138 | 6.45e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 2 | 41 | 6.45e-01 |
| GO:BP | GO:0046458 | hexadecanal metabolic process | 1 | 1 | 6.45e-01 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 7 | 11 | 6.46e-01 |
| GO:BP | GO:0045654 | positive regulation of megakaryocyte differentiation | 2 | 9 | 6.46e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 3 | 1974 | 6.46e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 23 | 782 | 6.46e-01 |
| GO:BP | GO:0006627 | protein processing involved in protein targeting to mitochondrion | 4 | 6 | 6.46e-01 |
| GO:BP | GO:0036138 | peptidyl-histidine hydroxylation | 1 | 1 | 6.46e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 17 | 67 | 6.46e-01 |
| GO:BP | GO:0031048 | small ncRNA-mediated heterochromatin formation | 1 | 2 | 6.46e-01 |
| GO:BP | GO:0097039 | protein linear polyubiquitination | 2 | 5 | 6.46e-01 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 2 | 11 | 6.46e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 6.46e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 13 | 23 | 6.46e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 1 | 12 | 6.46e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 1 | 21 | 6.46e-01 |
| GO:BP | GO:0046379 | extracellular polysaccharide metabolic process | 2 | 2 | 6.47e-01 |
| GO:BP | GO:0045226 | extracellular polysaccharide biosynthetic process | 2 | 2 | 6.47e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 2 | 28 | 6.47e-01 |
| GO:BP | GO:0006412 | translation | 5 | 646 | 6.47e-01 |
| GO:BP | GO:0035844 | cloaca development | 1 | 3 | 6.47e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 1 | 29 | 6.47e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 6.47e-01 |
| GO:BP | GO:2000369 | regulation of clathrin-dependent endocytosis | 1 | 18 | 6.47e-01 |
| GO:BP | GO:0098886 | modification of dendritic spine | 1 | 2 | 6.47e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 2 | 50 | 6.47e-01 |
| GO:BP | GO:0030311 | poly-N-acetyllactosamine biosynthetic process | 4 | 5 | 6.47e-01 |
| GO:BP | GO:1903790 | guanine nucleotide transmembrane transport | 1 | 5 | 6.47e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 95 | 796 | 6.47e-01 |
| GO:BP | GO:0070729 | cyclic nucleotide transport | 1 | 6 | 6.47e-01 |
| GO:BP | GO:2000802 | positive regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0042797 | tRNA transcription by RNA polymerase III | 1 | 7 | 6.47e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 8 | 89 | 6.47e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 3 | 104 | 6.47e-01 |
| GO:BP | GO:0016183 | synaptic vesicle coating | 3 | 4 | 6.47e-01 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 13 | 28 | 6.47e-01 |
| GO:BP | GO:0140049 | regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0140051 | positive regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 6.47e-01 |
| GO:BP | GO:0060755 | negative regulation of mast cell chemotaxis | 1 | 1 | 6.47e-01 |
| GO:BP | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death | 2 | 3 | 6.47e-01 |
| GO:BP | GO:2000800 | regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 6.47e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 2 | 232 | 6.47e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 2 | 3 | 6.47e-01 |
| GO:BP | GO:1903059 | regulation of protein lipidation | 1 | 11 | 6.47e-01 |
| GO:BP | GO:0016237 | lysosomal microautophagy | 1 | 11 | 6.47e-01 |
| GO:BP | GO:1900208 | regulation of cardiolipin metabolic process | 2 | 3 | 6.47e-01 |
| GO:BP | GO:1901080 | regulation of relaxation of smooth muscle | 1 | 1 | 6.48e-01 |
| GO:BP | GO:0042045 | epithelial fluid transport | 1 | 7 | 6.48e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 8 | 1248 | 6.48e-01 |
| GO:BP | GO:0070075 | tear secretion | 1 | 2 | 6.48e-01 |
| GO:BP | GO:0061171 | establishment of bipolar cell polarity | 1 | 1 | 6.48e-01 |
| GO:BP | GO:0098787 | mRNA cleavage involved in mRNA processing | 1 | 13 | 6.48e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 2 | 117 | 6.48e-01 |
| GO:BP | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 1 | 6 | 6.49e-01 |
| GO:BP | GO:0015821 | methionine transport | 1 | 4 | 6.49e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 9 | 27 | 6.49e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 3 | 86 | 6.49e-01 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 5 | 12 | 6.49e-01 |
| GO:BP | GO:0007512 | adult heart development | 7 | 9 | 6.49e-01 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 7 | 31 | 6.49e-01 |
| GO:BP | GO:2000659 | regulation of interleukin-1-mediated signaling pathway | 1 | 4 | 6.49e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 6.49e-01 |
| GO:BP | GO:0036302 | atrioventricular canal development | 7 | 12 | 6.49e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 6.50e-01 |
| GO:BP | GO:1901390 | positive regulation of transforming growth factor beta activation | 1 | 3 | 6.50e-01 |
| GO:BP | GO:2000645 | negative regulation of receptor catabolic process | 1 | 3 | 6.50e-01 |
| GO:BP | GO:0007497 | posterior midgut development | 1 | 2 | 6.50e-01 |
| GO:BP | GO:0000055 | ribosomal large subunit export from nucleus | 3 | 8 | 6.50e-01 |
| GO:BP | GO:0072682 | eosinophil extravasation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 6 | 38 | 6.50e-01 |
| GO:BP | GO:0009190 | cyclic nucleotide biosynthetic process | 3 | 14 | 6.50e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 2 | 15 | 6.50e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 4 | 29 | 6.50e-01 |
| GO:BP | GO:0001806 | type IV hypersensitivity | 2 | 2 | 6.50e-01 |
| GO:BP | GO:2000419 | regulation of eosinophil extravasation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0045792 | negative regulation of cell size | 3 | 9 | 6.50e-01 |
| GO:BP | GO:0036364 | transforming growth factor beta1 activation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0071691 | cardiac muscle thin filament assembly | 2 | 3 | 6.50e-01 |
| GO:BP | GO:2000420 | negative regulation of eosinophil extravasation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 2 | 13 | 6.50e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 4 | 290 | 6.50e-01 |
| GO:BP | GO:1903850 | regulation of cristae formation | 2 | 4 | 6.50e-01 |
| GO:BP | GO:0035585 | calcium-mediated signaling using extracellular calcium source | 1 | 2 | 6.50e-01 |
| GO:BP | GO:0010133 | proline catabolic process to glutamate | 1 | 2 | 6.50e-01 |
| GO:BP | GO:1901072 | glucosamine-containing compound catabolic process | 3 | 5 | 6.50e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 6.50e-01 |
| GO:BP | GO:0016561 | protein import into peroxisome matrix, translocation | 2 | 3 | 6.50e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 8 | 42 | 6.50e-01 |
| GO:BP | GO:0090410 | malonate catabolic process | 1 | 1 | 6.51e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 1 | 129 | 6.51e-01 |
| GO:BP | GO:0048880 | sensory system development | 11 | 275 | 6.51e-01 |
| GO:BP | GO:1904891 | positive regulation of excitatory synapse assembly | 1 | 2 | 6.51e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 2 | 40 | 6.51e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 163 | 6.51e-01 |
| GO:BP | GO:0045887 | positive regulation of synaptic assembly at neuromuscular junction | 1 | 1 | 6.51e-01 |
| GO:BP | GO:2000525 | positive regulation of T cell costimulation | 1 | 2 | 6.51e-01 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 1 | 7 | 6.51e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 2 | 1378 | 6.51e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 5 | 25 | 6.51e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 6.51e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 9 | 42 | 6.52e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 6 | 98 | 6.52e-01 |
| GO:BP | GO:0002537 | nitric oxide production involved in inflammatory response | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 2 | 8 | 6.52e-01 |
| GO:BP | GO:1901475 | pyruvate transmembrane transport | 2 | 5 | 6.52e-01 |
| GO:BP | GO:0006848 | pyruvate transport | 2 | 5 | 6.52e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 4 | 31 | 6.52e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 4 | 63 | 6.52e-01 |
| GO:BP | GO:1905673 | positive regulation of lysosome organization | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0060318 | definitive erythrocyte differentiation | 3 | 4 | 6.52e-01 |
| GO:BP | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway | 1 | 10 | 6.52e-01 |
| GO:BP | GO:0090107 | regulation of high-density lipoprotein particle assembly | 2 | 3 | 6.52e-01 |
| GO:BP | GO:0051026 | chiasma assembly | 1 | 3 | 6.52e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 6 | 29 | 6.52e-01 |
| GO:BP | GO:0003285 | septum secundum development | 2 | 3 | 6.52e-01 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 1 | 9 | 6.52e-01 |
| GO:BP | GO:0015819 | lysine transport | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0060927 | cardiac pacemaker cell fate commitment | 2 | 3 | 6.52e-01 |
| GO:BP | GO:0055014 | atrial cardiac muscle cell development | 2 | 3 | 6.52e-01 |
| GO:BP | GO:0055011 | atrial cardiac muscle cell differentiation | 2 | 3 | 6.52e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 2 | 88 | 6.52e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 9 | 24 | 6.52e-01 |
| GO:BP | GO:0009624 | response to nematode | 1 | 4 | 6.52e-01 |
| GO:BP | GO:1900260 | negative regulation of RNA-dependent RNA polymerase activity | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0043378 | positive regulation of CD8-positive, alpha-beta T cell differentiation | 2 | 2 | 6.52e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 8 | 24 | 6.52e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 2 | 12 | 6.52e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 2 | 9 | 6.52e-01 |
| GO:BP | GO:1900259 | regulation of RNA-dependent RNA polymerase activity | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0033198 | response to ATP | 2 | 24 | 6.52e-01 |
| GO:BP | GO:1905035 | negative regulation of antifungal innate immune response | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0046368 | GDP-L-fucose metabolic process | 2 | 4 | 6.52e-01 |
| GO:BP | GO:0006497 | protein lipidation | 46 | 89 | 6.53e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 6.53e-01 |
| GO:BP | GO:0036261 | 7-methylguanosine cap hypermethylation | 4 | 8 | 6.53e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 1 | 40 | 6.53e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 3 | 20 | 6.53e-01 |
| GO:BP | GO:0001525 | angiogenesis | 26 | 390 | 6.53e-01 |
| GO:BP | GO:0034975 | protein folding in endoplasmic reticulum | 1 | 10 | 6.53e-01 |
| GO:BP | GO:0089709 | L-histidine transmembrane transport | 2 | 5 | 6.53e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 10 | 18 | 6.53e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 2 | 236 | 6.53e-01 |
| GO:BP | GO:0036035 | osteoclast development | 1 | 10 | 6.53e-01 |
| GO:BP | GO:0070175 | positive regulation of enamel mineralization | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 6.54e-01 |
| GO:BP | GO:0002519 | natural killer cell tolerance induction | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0034436 | glycoprotein transport | 2 | 3 | 6.54e-01 |
| GO:BP | GO:1903048 | regulation of acetylcholine-gated cation channel activity | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0046712 | GDP catabolic process | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0015861 | cytidine transport | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 6.54e-01 |
| GO:BP | GO:0046066 | dGDP metabolic process | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 3 | 13 | 6.54e-01 |
| GO:BP | GO:1903049 | negative regulation of acetylcholine-gated cation channel activity | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 2 | 91 | 6.54e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 6.54e-01 |
| GO:BP | GO:0019227 | neuronal action potential propagation | 1 | 5 | 6.54e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 11 | 25 | 6.54e-01 |
| GO:BP | GO:0007518 | myoblast fate determination | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0046056 | dADP metabolic process | 1 | 1 | 6.54e-01 |
| GO:BP | GO:2000535 | regulation of entry of bacterium into host cell | 1 | 6 | 6.54e-01 |
| GO:BP | GO:0098870 | action potential propagation | 1 | 5 | 6.54e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 6 | 25 | 6.54e-01 |
| GO:BP | GO:0006959 | humoral immune response | 2 | 80 | 6.54e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 7 | 6.54e-01 |
| GO:BP | GO:0044381 | glucose import in response to insulin stimulus | 2 | 4 | 6.54e-01 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 5 | 17 | 6.54e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 1 | 10 | 6.54e-01 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 5 | 17 | 6.54e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 6.54e-01 |
| GO:BP | GO:1904899 | positive regulation of hepatic stellate cell proliferation | 1 | 1 | 6.54e-01 |
| GO:BP | GO:0009629 | response to gravity | 2 | 5 | 6.54e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 3 | 115 | 6.54e-01 |
| GO:BP | GO:1990922 | hepatic stellate cell proliferation | 1 | 1 | 6.54e-01 |
| GO:BP | GO:1904897 | regulation of hepatic stellate cell proliferation | 1 | 1 | 6.54e-01 |
| GO:BP | GO:1990784 | response to dsDNA | 1 | 2 | 6.55e-01 |
| GO:BP | GO:1990786 | cellular response to dsDNA | 1 | 2 | 6.55e-01 |
| GO:BP | GO:2001016 | positive regulation of skeletal muscle cell differentiation | 1 | 5 | 6.55e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 2 | 1399 | 6.55e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 6.55e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 6.55e-01 |
| GO:BP | GO:0035261 | external genitalia morphogenesis | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0061780 | mitotic cohesin loading | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0045337 | farnesyl diphosphate biosynthetic process | 1 | 3 | 6.55e-01 |
| GO:BP | GO:0008037 | cell recognition | 3 | 87 | 6.55e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 6.55e-01 |
| GO:BP | GO:0071394 | cellular response to testosterone stimulus | 4 | 11 | 6.55e-01 |
| GO:BP | GO:0140820 | cytosol to Golgi apparatus transport | 1 | 4 | 6.55e-01 |
| GO:BP | GO:0000741 | karyogamy | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0007344 | pronuclear fusion | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0002154 | thyroid hormone mediated signaling pathway | 1 | 6 | 6.56e-01 |
| GO:BP | GO:0030241 | skeletal muscle myosin thick filament assembly | 2 | 3 | 6.56e-01 |
| GO:BP | GO:0048769 | sarcomerogenesis | 2 | 3 | 6.56e-01 |
| GO:BP | GO:0045924 | regulation of female receptivity | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 2 | 44 | 6.56e-01 |
| GO:BP | GO:0071688 | striated muscle myosin thick filament assembly | 2 | 3 | 6.56e-01 |
| GO:BP | GO:2000392 | regulation of lamellipodium morphogenesis | 4 | 10 | 6.56e-01 |
| GO:BP | GO:0060180 | female mating behavior | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0071247 | cellular response to chromate | 1 | 1 | 6.57e-01 |
| GO:BP | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 1 | 6.57e-01 |
| GO:BP | GO:1901423 | response to benzene | 1 | 1 | 6.57e-01 |
| GO:BP | GO:1904710 | positive regulation of granulosa cell apoptotic process | 1 | 1 | 6.57e-01 |
| GO:BP | GO:0046479 | glycosphingolipid catabolic process | 4 | 10 | 6.57e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 4 | 21 | 6.57e-01 |
| GO:BP | GO:0046687 | response to chromate | 1 | 1 | 6.57e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 4 | 62 | 6.57e-01 |
| GO:BP | GO:0017182 | peptidyl-diphthamide metabolic process | 4 | 7 | 6.58e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0045915 | positive regulation of catecholamine metabolic process | 2 | 6 | 6.58e-01 |
| GO:BP | GO:0097350 | neutrophil clearance | 3 | 5 | 6.58e-01 |
| GO:BP | GO:1903387 | positive regulation of homophilic cell adhesion | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0045964 | positive regulation of dopamine metabolic process | 2 | 6 | 6.58e-01 |
| GO:BP | GO:0017183 | peptidyl-diphthamide biosynthetic process from peptidyl-histidine | 4 | 7 | 6.58e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 14 | 26 | 6.58e-01 |
| GO:BP | GO:1990678 | histone H4-K16 deacetylation | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0015962 | diadenosine triphosphate metabolic process | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0015964 | diadenosine triphosphate catabolic process | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 31 | 70 | 6.58e-01 |
| GO:BP | GO:0140291 | peptidyl-glutamate ADP-deribosylation | 2 | 3 | 6.58e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 6 | 39 | 6.58e-01 |
| GO:BP | GO:0032243 | negative regulation of nucleoside transport | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0015910 | long-chain fatty acid import into peroxisome | 1 | 3 | 6.58e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 6.59e-01 |
| GO:BP | GO:0071386 | cellular response to corticosterone stimulus | 1 | 1 | 6.59e-01 |
| GO:BP | GO:1903959 | regulation of monoatomic anion transmembrane transport | 5 | 6 | 6.59e-01 |
| GO:BP | GO:0033383 | geranyl diphosphate metabolic process | 1 | 3 | 6.59e-01 |
| GO:BP | GO:1990258 | histone glutamine methylation | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0010831 | positive regulation of myotube differentiation | 4 | 10 | 6.59e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 6 | 155 | 6.59e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 6.59e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 2 | 19 | 6.59e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 2 | 17 | 6.59e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 2 | 17 | 6.59e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 6.60e-01 |
| GO:BP | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway | 3 | 7 | 6.60e-01 |
| GO:BP | GO:0021536 | diencephalon development | 2 | 41 | 6.60e-01 |
| GO:BP | GO:1904746 | negative regulation of apoptotic process involved in development | 1 | 3 | 6.61e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 1 | 6 | 6.61e-01 |
| GO:BP | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis | 1 | 3 | 6.61e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 11 | 59 | 6.61e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 11 | 59 | 6.61e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 9 | 37 | 6.61e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 15 | 2646 | 6.61e-01 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 5 | 13 | 6.61e-01 |
| GO:BP | GO:1904251 | regulation of bile acid metabolic process | 1 | 9 | 6.61e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 1 | 36 | 6.61e-01 |
| GO:BP | GO:0032488 | Cdc42 protein signal transduction | 1 | 10 | 6.61e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 2 | 151 | 6.61e-01 |
| GO:BP | GO:0014075 | response to amine | 1 | 34 | 6.61e-01 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 2 | 31 | 6.61e-01 |
| GO:BP | GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1 | 8 | 6.61e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 6.61e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 1 | 18 | 6.61e-01 |
| GO:BP | GO:0010815 | bradykinin catabolic process | 4 | 6 | 6.61e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 29 | 6.61e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 3 | 36 | 6.61e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 2 | 41 | 6.61e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 2 | 14 | 6.61e-01 |
| GO:BP | GO:0021989 | olfactory cortex development | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 2 | 100 | 6.61e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 3 | 115 | 6.61e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 3 | 8 | 6.61e-01 |
| GO:BP | GO:1903280 | negative regulation of calcium:sodium antiporter activity | 1 | 1 | 6.61e-01 |
| GO:BP | GO:1905799 | regulation of intraciliary retrograde transport | 1 | 2 | 6.61e-01 |
| GO:BP | GO:1903999 | negative regulation of eating behavior | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0090214 | spongiotrophoblast layer developmental growth | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0060720 | spongiotrophoblast cell proliferation | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 30 | 190 | 6.61e-01 |
| GO:BP | GO:1903547 | regulation of growth hormone activity | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0060722 | cell proliferation involved in embryonic placenta development | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0010700 | negative regulation of norepinephrine secretion | 2 | 2 | 6.62e-01 |
| GO:BP | GO:2000353 | positive regulation of endothelial cell apoptotic process | 1 | 13 | 6.62e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 9 | 17 | 6.62e-01 |
| GO:BP | GO:0009207 | purine ribonucleoside triphosphate catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0006193 | ITP catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0097712 | vesicle targeting, trans-Golgi to periciliary membrane compartment | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 13 | 61 | 6.62e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 6 | 265 | 6.62e-01 |
| GO:BP | GO:0009098 | leucine biosynthetic process | 1 | 2 | 6.63e-01 |
| GO:BP | GO:0030309 | poly-N-acetyllactosamine metabolic process | 5 | 6 | 6.63e-01 |
| GO:BP | GO:0006049 | UDP-N-acetylglucosamine catabolic process | 1 | 1 | 6.63e-01 |
| GO:BP | GO:1901098 | positive regulation of autophagosome maturation | 1 | 6 | 6.63e-01 |
| GO:BP | GO:0006420 | arginyl-tRNA aminoacylation | 2 | 3 | 6.63e-01 |
| GO:BP | GO:1905664 | regulation of calcium ion import across plasma membrane | 2 | 8 | 6.63e-01 |
| GO:BP | GO:0070350 | regulation of white fat cell proliferation | 1 | 2 | 6.63e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 23 | 4539 | 6.63e-01 |
| GO:BP | GO:0035553 | oxidative single-stranded RNA demethylation | 1 | 3 | 6.63e-01 |
| GO:BP | GO:0048668 | collateral sprouting | 13 | 25 | 6.63e-01 |
| GO:BP | GO:0070343 | white fat cell proliferation | 1 | 2 | 6.63e-01 |
| GO:BP | GO:0060718 | chorionic trophoblast cell differentiation | 2 | 5 | 6.63e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 8 | 71 | 6.63e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 10 | 54 | 6.63e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 35 | 162 | 6.63e-01 |
| GO:BP | GO:0042976 | activation of Janus kinase activity | 1 | 7 | 6.63e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 8 | 64 | 6.63e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 4 | 23 | 6.64e-01 |
| GO:BP | GO:0051562 | negative regulation of mitochondrial calcium ion concentration | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0003400 | regulation of COPII vesicle coating | 2 | 3 | 6.65e-01 |
| GO:BP | GO:1904293 | negative regulation of ERAD pathway | 1 | 6 | 6.65e-01 |
| GO:BP | GO:1905062 | positive regulation of cardioblast proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0034477 | U6 snRNA 3’-end processing | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0021543 | pallium development | 8 | 139 | 6.65e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 3 | 28 | 6.65e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 2 | 301 | 6.65e-01 |
| GO:BP | GO:2000727 | positive regulation of cardiac muscle cell differentiation | 2 | 6 | 6.65e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 2 | 9 | 6.65e-01 |
| GO:BP | GO:0046495 | nicotinamide riboside metabolic process | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 3 | 7 | 6.65e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 3 | 13 | 6.65e-01 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 3 | 120 | 6.65e-01 |
| GO:BP | GO:0006738 | nicotinamide riboside catabolic process | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 5 | 126 | 6.65e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 8 | 560 | 6.65e-01 |
| GO:BP | GO:0070637 | pyridine nucleoside metabolic process | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0060003 | copper ion export | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0003250 | regulation of cell proliferation involved in heart valve morphogenesis | 2 | 3 | 6.65e-01 |
| GO:BP | GO:0070638 | pyridine nucleoside catabolic process | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 2 | 3 | 6.65e-01 |
| GO:BP | GO:0007207 | phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | 3 | 6 | 6.65e-01 |
| GO:BP | GO:0003249 | cell proliferation involved in heart valve morphogenesis | 2 | 3 | 6.65e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 2 | 348 | 6.65e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 8 | 38 | 6.65e-01 |
| GO:BP | GO:0060699 | regulation of endoribonuclease activity | 1 | 4 | 6.66e-01 |
| GO:BP | GO:0097264 | self proteolysis | 2 | 7 | 6.66e-01 |
| GO:BP | GO:0022008 | neurogenesis | 2 | 1294 | 6.66e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 3 | 10 | 6.66e-01 |
| GO:BP | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | 1 | 5 | 6.66e-01 |
| GO:BP | GO:0031627 | telomeric loop formation | 1 | 2 | 6.66e-01 |
| GO:BP | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine | 3 | 5 | 6.66e-01 |
| GO:BP | GO:1903937 | response to acrylamide | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0014860 | neurotransmitter secretion involved in regulation of skeletal muscle contraction | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0010724 | regulation of definitive erythrocyte differentiation | 2 | 2 | 6.66e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 1 | 6 | 6.66e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 8 | 17 | 6.66e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 3 | 15 | 6.66e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 2 | 14 | 6.66e-01 |
| GO:BP | GO:0097577 | sequestering of iron ion | 3 | 3 | 6.66e-01 |
| GO:BP | GO:0014004 | microglia differentiation | 1 | 6 | 6.66e-01 |
| GO:BP | GO:0051573 | negative regulation of histone H3-K9 methylation | 1 | 5 | 6.66e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 7 | 12 | 6.66e-01 |
| GO:BP | GO:0070963 | positive regulation of neutrophil mediated killing of gram-negative bacterium | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0061966 | establishment of left/right asymmetry | 1 | 3 | 6.66e-01 |
| GO:BP | GO:0072070 | loop of Henle development | 1 | 8 | 6.66e-01 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 2 | 7 | 6.66e-01 |
| GO:BP | GO:0070962 | positive regulation of neutrophil mediated killing of bacterium | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 5 | 17 | 6.66e-01 |
| GO:BP | GO:0030302 | deoxynucleotide transport | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0099109 | potassium channel activating, G protein-coupled receptor signaling pathway | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0046618 | xenobiotic export from cell | 4 | 8 | 6.66e-01 |
| GO:BP | GO:0002883 | regulation of hypersensitivity | 2 | 5 | 6.67e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 24 | 55 | 6.67e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 6.67e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 4 | 16 | 6.67e-01 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 8 | 12 | 6.67e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 6.67e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 6.67e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 1 | 47 | 6.67e-01 |
| GO:BP | GO:0008065 | establishment of blood-nerve barrier | 2 | 3 | 6.67e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 6.67e-01 |
| GO:BP | GO:2000415 | positive regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 6.67e-01 |
| GO:BP | GO:2000413 | regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0072681 | fibronectin-dependent thymocyte migration | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0072680 | extracellular matrix-dependent thymocyte migration | 1 | 1 | 6.67e-01 |
| GO:BP | GO:0034447 | very-low-density lipoprotein particle clearance | 1 | 4 | 6.67e-01 |
| GO:BP | GO:0016095 | polyprenol catabolic process | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 9 | 316 | 6.67e-01 |
| GO:BP | GO:0010668 | ectodermal cell differentiation | 1 | 5 | 6.67e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 2 | 47 | 6.67e-01 |
| GO:BP | GO:2001246 | negative regulation of phosphatidylcholine biosynthetic process | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0035962 | response to interleukin-13 | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0071400 | cellular response to oleic acid | 2 | 3 | 6.68e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 1 | 49 | 6.68e-01 |
| GO:BP | GO:0033572 | transferrin transport | 5 | 8 | 6.68e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 6 | 37 | 6.68e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 6 | 37 | 6.68e-01 |
| GO:BP | GO:2000723 | negative regulation of cardiac vascular smooth muscle cell differentiation | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0070267 | oncosis | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 5 | 666 | 6.68e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 1 | 27 | 6.68e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 1 | 11 | 6.68e-01 |
| GO:BP | GO:0016577 | histone demethylation | 6 | 21 | 6.68e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 2 | 13 | 6.68e-01 |
| GO:BP | GO:0071105 | response to interleukin-11 | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0099145 | regulation of exocytic insertion of neurotransmitter receptor to postsynaptic membrane | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 3 | 15 | 6.69e-01 |
| GO:BP | GO:0033227 | dsRNA transport | 2 | 2 | 6.69e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 4 | 212 | 6.69e-01 |
| GO:BP | GO:0048477 | oogenesis | 5 | 64 | 6.69e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 3 | 15 | 6.69e-01 |
| GO:BP | GO:0006826 | iron ion transport | 4 | 40 | 6.69e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 2 | 54 | 6.69e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 4 | 17 | 6.69e-01 |
| GO:BP | GO:0072695 | regulation of DNA recombination at telomere | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0048239 | negative regulation of DNA recombination at telomere | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0061441 | renal artery morphogenesis | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0043931 | ossification involved in bone maturation | 1 | 17 | 6.69e-01 |
| GO:BP | GO:0042136 | neurotransmitter biosynthetic process | 1 | 5 | 6.69e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 6 | 13 | 6.69e-01 |
| GO:BP | GO:0097102 | endothelial tip cell fate specification | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 3 | 9 | 6.69e-01 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 1 | 10 | 6.69e-01 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 1 | 7 | 6.69e-01 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 2 | 17 | 6.69e-01 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 4 | 46 | 6.69e-01 |
| GO:BP | GO:1900022 | regulation of D-erythro-sphingosine kinase activity | 1 | 1 | 6.69e-01 |
| GO:BP | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 2 | 4 | 6.70e-01 |
| GO:BP | GO:2000097 | regulation of smooth muscle cell-matrix adhesion | 1 | 3 | 6.70e-01 |
| GO:BP | GO:0032425 | positive regulation of mismatch repair | 2 | 2 | 6.70e-01 |
| GO:BP | GO:0044580 | butyryl-CoA catabolic process | 2 | 3 | 6.70e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 3 | 10 | 6.70e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 2 | 3 | 6.70e-01 |
| GO:BP | GO:2001113 | negative regulation of cellular response to hepatocyte growth factor stimulus | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0046866 | tetraterpenoid transport | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0046867 | carotenoid transport | 1 | 1 | 6.70e-01 |
| GO:BP | GO:1903422 | negative regulation of synaptic vesicle recycling | 1 | 2 | 6.70e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 4 | 1075 | 6.70e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 11 | 20 | 6.70e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 1 | 10 | 6.70e-01 |
| GO:BP | GO:0006127 | glycerophosphate shuttle | 1 | 2 | 6.70e-01 |
| GO:BP | GO:1904028 | positive regulation of collagen fibril organization | 1 | 4 | 6.70e-01 |
| GO:BP | GO:0048284 | organelle fusion | 2 | 135 | 6.70e-01 |
| GO:BP | GO:0015938 | coenzyme A catabolic process | 2 | 3 | 6.70e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 2 | 3 | 6.70e-01 |
| GO:BP | GO:1902203 | negative regulation of hepatocyte growth factor receptor signaling pathway | 1 | 1 | 6.70e-01 |
| GO:BP | GO:1990822 | basic amino acid transmembrane transport | 1 | 14 | 6.70e-01 |
| GO:BP | GO:0014006 | regulation of microglia differentiation | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0051168 | nuclear export | 17 | 150 | 6.70e-01 |
| GO:BP | GO:1904338 | regulation of dopaminergic neuron differentiation | 1 | 7 | 6.70e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 2 | 12 | 6.70e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 2 | 126 | 6.70e-01 |
| GO:BP | GO:1905313 | transforming growth factor beta receptor signaling pathway involved in heart development | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0072560 | type B pancreatic cell maturation | 1 | 2 | 6.70e-01 |
| GO:BP | GO:0035711 | T-helper 1 cell activation | 2 | 2 | 6.70e-01 |
| GO:BP | GO:1990839 | response to endothelin | 1 | 2 | 6.70e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 3 | 10 | 6.70e-01 |
| GO:BP | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 4 | 187 | 6.70e-01 |
| GO:BP | GO:0035685 | helper T cell diapedesis | 1 | 2 | 6.70e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 2 | 4 | 6.70e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 2 | 8 | 6.70e-01 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 1 | 4 | 6.70e-01 |
| GO:BP | GO:0010526 | retrotransposon silencing | 1 | 3 | 6.70e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 2 | 8 | 6.70e-01 |
| GO:BP | GO:0035279 | miRNA-mediated gene silencing by mRNA destabilization | 1 | 3 | 6.70e-01 |
| GO:BP | GO:0014008 | positive regulation of microglia differentiation | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 4 | 187 | 6.70e-01 |
| GO:BP | GO:0031064 | negative regulation of histone deacetylation | 2 | 2 | 6.70e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 6 | 10 | 6.70e-01 |
| GO:BP | GO:0038172 | interleukin-33-mediated signaling pathway | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 2 | 10 | 6.70e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 6.70e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 6.71e-01 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 3 | 10 | 6.71e-01 |
| GO:BP | GO:0035281 | pre-miRNA export from nucleus | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0090208 | positive regulation of triglyceride metabolic process | 1 | 18 | 6.71e-01 |
| GO:BP | GO:0061015 | snRNA import into nucleus | 1 | 2 | 6.71e-01 |
| GO:BP | GO:1902498 | regulation of protein autoubiquitination | 3 | 5 | 6.71e-01 |
| GO:BP | GO:0110021 | cardiac muscle myoblast proliferation | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0015883 | FAD transport | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0021603 | cranial nerve formation | 1 | 4 | 6.71e-01 |
| GO:BP | GO:0015880 | coenzyme A transport | 1 | 2 | 6.71e-01 |
| GO:BP | GO:1905381 | negative regulation of snRNA transcription by RNA polymerase II | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0018125 | peptidyl-cysteine methylation | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 9 | 22 | 6.71e-01 |
| GO:BP | GO:0032976 | release of matrix enzymes from mitochondria | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0018171 | peptidyl-cysteine oxidation | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0110022 | regulation of cardiac muscle myoblast proliferation | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0062035 | sensory perception of cold stimulus | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0110024 | positive regulation of cardiac muscle myoblast proliferation | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0034397 | telomere localization | 2 | 5 | 6.71e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 2 | 50 | 6.71e-01 |
| GO:BP | GO:0140060 | axon arborization | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 5 | 90 | 6.71e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 1 | 147 | 6.71e-01 |
| GO:BP | GO:0015697 | quaternary ammonium group transport | 1 | 17 | 6.71e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 7 | 25 | 6.71e-01 |
| GO:BP | GO:0120169 | detection of cold stimulus involved in thermoception | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0021627 | olfactory nerve morphogenesis | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 80 | 500 | 6.71e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 8 | 22 | 6.71e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 2 | 3 | 6.71e-01 |
| GO:BP | GO:1990046 | stress-induced mitochondrial fusion | 1 | 2 | 6.71e-01 |
| GO:BP | GO:1900210 | positive regulation of cardiolipin metabolic process | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0032714 | negative regulation of interleukin-5 production | 2 | 2 | 6.71e-01 |
| GO:BP | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0035350 | FAD transmembrane transport | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0035349 | coenzyme A transmembrane transport | 1 | 2 | 6.71e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 4 | 17 | 6.71e-01 |
| GO:BP | GO:0071838 | cell proliferation in bone marrow | 2 | 7 | 6.71e-01 |
| GO:BP | GO:0071106 | adenosine 3’,5’-bisphosphate transmembrane transport | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion | 2 | 3 | 6.71e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 4 | 14 | 6.71e-01 |
| GO:BP | GO:2000158 | positive regulation of ubiquitin-specific protease activity | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0034285 | response to disaccharide | 1 | 4 | 6.71e-01 |
| GO:BP | GO:0031508 | pericentric heterochromatin formation | 2 | 3 | 6.71e-01 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 3 | 5 | 6.71e-01 |
| GO:BP | GO:0009744 | response to sucrose | 1 | 4 | 6.71e-01 |
| GO:BP | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation | 2 | 5 | 6.71e-01 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 3 | 6 | 6.71e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 2 | 4 | 6.71e-01 |
| GO:BP | GO:0080121 | AMP transport | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 5 | 9 | 6.71e-01 |
| GO:BP | GO:0009750 | response to fructose | 1 | 3 | 6.71e-01 |
| GO:BP | GO:0019482 | beta-alanine metabolic process | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0046203 | spermidine catabolic process | 1 | 2 | 6.71e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 5 | 8 | 6.71e-01 |
| GO:BP | GO:0044691 | tooth eruption | 3 | 5 | 6.71e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 6.71e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 6 | 608 | 6.71e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 6.71e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 23 | 2880 | 6.71e-01 |
| GO:BP | GO:0060126 | somatotropin secreting cell differentiation | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 6 | 40 | 6.71e-01 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 1 | 8 | 6.71e-01 |
| GO:BP | GO:0002318 | myeloid progenitor cell differentiation | 3 | 7 | 6.71e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 2 | 58 | 6.71e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 56 | 163 | 6.71e-01 |
| GO:BP | GO:0043248 | proteasome assembly | 1 | 13 | 6.71e-01 |
| GO:BP | GO:0032570 | response to progesterone | 6 | 27 | 6.72e-01 |
| GO:BP | GO:0036250 | peroxisome transport along microtubule | 1 | 1 | 6.72e-01 |
| GO:BP | GO:0009304 | tRNA transcription | 1 | 8 | 6.72e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 1 | 18 | 6.72e-01 |
| GO:BP | GO:0007350 | blastoderm segmentation | 1 | 17 | 6.72e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 3 | 6 | 6.72e-01 |
| GO:BP | GO:0031649 | heat generation | 1 | 11 | 6.72e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 6.72e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 3 | 20 | 6.72e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 32 | 6.72e-01 |
| GO:BP | GO:0042669 | regulation of inner ear auditory receptor cell fate specification | 1 | 1 | 6.73e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 3 | 35 | 6.73e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 5 | 13 | 6.73e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 17 | 98 | 6.73e-01 |
| GO:BP | GO:0071603 | endothelial cell-cell adhesion | 2 | 4 | 6.73e-01 |
| GO:BP | GO:0060022 | hard palate development | 2 | 5 | 6.73e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 2 | 60 | 6.74e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 8 | 100 | 6.74e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 2 | 39 | 6.74e-01 |
| GO:BP | GO:0021644 | vagus nerve morphogenesis | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 2 | 4 | 6.74e-01 |
| GO:BP | GO:0003168 | Purkinje myocyte differentiation | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly | 1 | 5 | 6.74e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 3 | 126 | 6.74e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 5 | 19 | 6.74e-01 |
| GO:BP | GO:0071359 | cellular response to dsRNA | 7 | 18 | 6.74e-01 |
| GO:BP | GO:0009086 | methionine biosynthetic process | 1 | 10 | 6.74e-01 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 2 | 45 | 6.74e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 6.74e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 6.74e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 6 | 15 | 6.74e-01 |
| GO:BP | GO:0035607 | fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0098508 | endothelial to hematopoietic transition | 2 | 2 | 6.74e-01 |
| GO:BP | GO:1903679 | positive regulation of cap-independent translational initiation | 2 | 3 | 6.74e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 11 | 21 | 6.74e-01 |
| GO:BP | GO:0140543 | positive regulation of piRNA transcription | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 9 | 179 | 6.74e-01 |
| GO:BP | GO:1903677 | regulation of cap-independent translational initiation | 2 | 3 | 6.74e-01 |
| GO:BP | GO:0140542 | regulation of piRNA transcription | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 2 | 14 | 6.74e-01 |
| GO:BP | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in hemopoiesis | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow cell | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 14 | 63 | 6.74e-01 |
| GO:BP | GO:0016076 | snRNA catabolic process | 3 | 5 | 6.74e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 3 | 259 | 6.74e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 6.74e-01 |
| GO:BP | GO:0006529 | asparagine biosynthetic process | 1 | 2 | 6.74e-01 |
| GO:BP | GO:0055024 | regulation of cardiac muscle tissue development | 2 | 3 | 6.74e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 4 | 34 | 6.74e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 6.75e-01 |
| GO:BP | GO:0007600 | sensory perception | 2 | 269 | 6.75e-01 |
| GO:BP | GO:1905426 | positive regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 6.75e-01 |
| GO:BP | GO:1905424 | regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 6.75e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 6.75e-01 |
| GO:BP | GO:1900535 | palmitic acid biosynthetic process | 1 | 1 | 6.75e-01 |
| GO:BP | GO:1900533 | palmitic acid metabolic process | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0006014 | D-ribose metabolic process | 2 | 3 | 6.75e-01 |
| GO:BP | GO:0036116 | long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 7 | 16 | 6.75e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 24 | 830 | 6.75e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 9 | 51 | 6.75e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 41 | 6.75e-01 |
| GO:BP | GO:0015779 | glucuronoside transport | 1 | 1 | 6.75e-01 |
| GO:BP | GO:1903644 | regulation of chaperone-mediated protein folding | 2 | 3 | 6.75e-01 |
| GO:BP | GO:0006414 | translational elongation | 6 | 65 | 6.75e-01 |
| GO:BP | GO:0009195 | pyrimidine ribonucleoside diphosphate catabolic process | 2 | 3 | 6.75e-01 |
| GO:BP | GO:0006256 | UDP catabolic process | 2 | 3 | 6.75e-01 |
| GO:BP | GO:0045684 | positive regulation of epidermis development | 12 | 20 | 6.75e-01 |
| GO:BP | GO:0080120 | CAAX-box protein maturation | 1 | 3 | 6.75e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 3 | 64 | 6.75e-01 |
| GO:BP | GO:1901569 | fatty acid derivative catabolic process | 1 | 10 | 6.75e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 19 | 3750 | 6.75e-01 |
| GO:BP | GO:0003412 | establishment of epithelial cell apical/basal polarity involved in camera-type eye morphogenesis | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 6.75e-01 |
| GO:BP | GO:0042595 | behavioral response to starvation | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 21 | 51 | 6.75e-01 |
| GO:BP | GO:0009140 | pyrimidine nucleoside diphosphate catabolic process | 2 | 3 | 6.75e-01 |
| GO:BP | GO:2000654 | regulation of cellular response to testosterone stimulus | 1 | 2 | 6.75e-01 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 1 | 23 | 6.75e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 6.75e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 6.76e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 2 | 158 | 6.76e-01 |
| GO:BP | GO:1905243 | cellular response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0097476 | spinal cord motor neuron migration | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0061550 | cranial ganglion development | 2 | 4 | 6.76e-01 |
| GO:BP | GO:0006463 | steroid hormone receptor complex assembly | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0097376 | interneuron axon guidance | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0072278 | metanephric comma-shaped body morphogenesis | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0032713 | negative regulation of interleukin-4 production | 3 | 5 | 6.76e-01 |
| GO:BP | GO:1903182 | regulation of SUMO transferase activity | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0001408 | guanine nucleotide transport | 1 | 6 | 6.76e-01 |
| GO:BP | GO:1903755 | positive regulation of SUMO transferase activity | 1 | 1 | 6.76e-01 |
| GO:BP | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation | 3 | 4 | 6.76e-01 |
| GO:BP | GO:0098838 | folate transmembrane transport | 1 | 6 | 6.76e-01 |
| GO:BP | GO:1904606 | fat cell apoptotic process | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 3 | 57 | 6.76e-01 |
| GO:BP | GO:0060683 | regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0003096 | renal sodium ion transport | 2 | 10 | 6.76e-01 |
| GO:BP | GO:1904649 | regulation of fat cell apoptotic process | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0060438 | trachea development | 4 | 13 | 6.76e-01 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 1 | 5 | 6.76e-01 |
| GO:BP | GO:1901608 | regulation of vesicle transport along microtubule | 3 | 4 | 6.76e-01 |
| GO:BP | GO:1903044 | protein localization to membrane raft | 1 | 8 | 6.76e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 1 | 144 | 6.76e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 4 | 29 | 6.76e-01 |
| GO:BP | GO:0042538 | hyperosmotic salinity response | 4 | 8 | 6.76e-01 |
| GO:BP | GO:0051170 | import into nucleus | 50 | 152 | 6.76e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 11 | 46 | 6.76e-01 |
| GO:BP | GO:0032415 | regulation of sodium:proton antiporter activity | 1 | 4 | 6.76e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 2 | 4 | 6.76e-01 |
| GO:BP | GO:0038020 | insulin receptor recycling | 2 | 4 | 6.76e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 10 | 38 | 6.76e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 24 | 133 | 6.76e-01 |
| GO:BP | GO:1903727 | positive regulation of phospholipid metabolic process | 4 | 9 | 6.76e-01 |
| GO:BP | GO:0006409 | tRNA export from nucleus | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0045959 | negative regulation of complement activation, classical pathway | 2 | 4 | 6.76e-01 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0010814 | substance P catabolic process | 2 | 3 | 6.76e-01 |
| GO:BP | GO:0060975 | cardioblast migration to the midline involved in heart field formation | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0003318 | cell migration to the midline involved in heart development | 1 | 1 | 6.76e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 13 | 28 | 6.76e-01 |
| GO:BP | GO:1903457 | lactate catabolic process | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0042664 | negative regulation of endodermal cell fate specification | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0070368 | positive regulation of hepatocyte differentiation | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0003236 | sinus venosus morphogenesis | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0010764 | negative regulation of fibroblast migration | 2 | 9 | 6.76e-01 |
| GO:BP | GO:0003259 | cardioblast anterior-lateral migration | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0003235 | sinus venosus development | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0035752 | lysosomal lumen pH elevation | 2 | 3 | 6.76e-01 |
| GO:BP | GO:0003260 | cardioblast migration | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0098814 | spontaneous synaptic transmission | 4 | 10 | 6.76e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 5 | 57 | 6.76e-01 |
| GO:BP | GO:0060184 | cell cycle switching | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0051728 | cell cycle switching, mitotic to meiotic cell cycle | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0051729 | germline cell cycle switching, mitotic to meiotic cell cycle | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0010041 | response to iron(III) ion | 1 | 4 | 6.77e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 3 | 18 | 6.77e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 5 | 10 | 6.77e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 6 | 62 | 6.77e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 2 | 38 | 6.77e-01 |
| GO:BP | GO:0009268 | response to pH | 2 | 26 | 6.77e-01 |
| GO:BP | GO:0150003 | regulation of spontaneous synaptic transmission | 2 | 6 | 6.78e-01 |
| GO:BP | GO:0007538 | primary sex determination | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0003430 | growth plate cartilage chondrocyte growth | 1 | 3 | 6.78e-01 |
| GO:BP | GO:0097198 | histone H3-K36 trimethylation | 1 | 1 | 6.78e-01 |
| GO:BP | GO:1904837 | beta-catenin-TCF complex assembly | 1 | 3 | 6.78e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 2 | 13 | 6.78e-01 |
| GO:BP | GO:0072190 | ureter urothelium development | 1 | 3 | 6.78e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 6 | 47 | 6.78e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 15 | 74 | 6.78e-01 |
| GO:BP | GO:0002414 | immunoglobulin transcytosis in epithelial cells | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0046541 | saliva secretion | 1 | 4 | 6.78e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 6.78e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 6.78e-01 |
| GO:BP | GO:1902803 | regulation of synaptic vesicle transport | 3 | 5 | 6.79e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 23 | 4590 | 6.79e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 4 | 27 | 6.79e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 10 | 38 | 6.79e-01 |
| GO:BP | GO:0033080 | immature T cell proliferation in thymus | 2 | 7 | 6.79e-01 |
| GO:BP | GO:0033079 | immature T cell proliferation | 2 | 7 | 6.79e-01 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 3 | 21 | 6.79e-01 |
| GO:BP | GO:0060741 | prostate gland stromal morphogenesis | 1 | 2 | 6.79e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 2 | 11 | 6.79e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 1 | 8 | 6.79e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 2 | 357 | 6.79e-01 |
| GO:BP | GO:0018201 | peptidyl-glycine modification | 2 | 4 | 6.79e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 3 | 127 | 6.79e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 9 | 23 | 6.79e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 9 | 44 | 6.80e-01 |
| GO:BP | GO:0032965 | regulation of collagen biosynthetic process | 3 | 30 | 6.80e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 12 | 23 | 6.80e-01 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 1 | 15 | 6.80e-01 |
| GO:BP | GO:0033750 | ribosome localization | 1 | 15 | 6.80e-01 |
| GO:BP | GO:0021943 | formation of radial glial scaffolds | 1 | 3 | 6.80e-01 |
| GO:BP | GO:0101024 | mitotic nuclear membrane organization | 1 | 9 | 6.80e-01 |
| GO:BP | GO:0007084 | mitotic nuclear membrane reassembly | 1 | 9 | 6.80e-01 |
| GO:BP | GO:0072319 | vesicle uncoating | 1 | 9 | 6.80e-01 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 4 | 8 | 6.80e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 6.80e-01 |
| GO:BP | GO:0045898 | regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 7 | 6.80e-01 |
| GO:BP | GO:0090184 | positive regulation of kidney development | 3 | 9 | 6.80e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 151 | 6.80e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 7 | 771 | 6.80e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 3 | 27 | 6.80e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 2 | 209 | 6.81e-01 |
| GO:BP | GO:0051764 | actin crosslink formation | 5 | 10 | 6.81e-01 |
| GO:BP | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 1 | 2 | 6.81e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 6.81e-01 |
| GO:BP | GO:0055095 | lipoprotein particle mediated signaling | 1 | 3 | 6.81e-01 |
| GO:BP | GO:0071888 | macrophage apoptotic process | 4 | 9 | 6.81e-01 |
| GO:BP | GO:0032094 | response to food | 1 | 22 | 6.81e-01 |
| GO:BP | GO:2000467 | positive regulation of glycogen (starch) synthase activity | 3 | 4 | 6.81e-01 |
| GO:BP | GO:0055096 | low-density lipoprotein particle mediated signaling | 1 | 3 | 6.81e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 1 | 1173 | 6.81e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 6.81e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 6 | 146 | 6.81e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 3 | 11 | 6.81e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 4 | 10 | 6.81e-01 |
| GO:BP | GO:0043972 | histone H3-K23 acetylation | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 25 | 50 | 6.81e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 14 | 74 | 6.81e-01 |
| GO:BP | GO:2000436 | positive regulation of protein neddylation | 2 | 5 | 6.81e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 2 | 15 | 6.81e-01 |
| GO:BP | GO:0005997 | xylulose metabolic process | 1 | 1 | 6.81e-01 |
| GO:BP | GO:2000172 | regulation of branching morphogenesis of a nerve | 1 | 3 | 6.81e-01 |
| GO:BP | GO:0034371 | chylomicron remodeling | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0099525 | presynaptic dense core vesicle exocytosis | 3 | 4 | 6.81e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 1 | 40 | 6.81e-01 |
| GO:BP | GO:0046080 | dUTP metabolic process | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0046081 | dUTP catabolic process | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0002943 | tRNA dihydrouridine synthesis | 1 | 4 | 6.81e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 9 | 29 | 6.81e-01 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 1 | 8 | 6.81e-01 |
| GO:BP | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death | 3 | 6 | 6.81e-01 |
| GO:BP | GO:1901033 | positive regulation of response to reactive oxygen species | 3 | 6 | 6.81e-01 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 3 | 9 | 6.81e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 12 | 25 | 6.81e-01 |
| GO:BP | GO:1902605 | heterotrimeric G-protein complex assembly | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0070922 | RISC complex assembly | 3 | 9 | 6.81e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 85 | 164 | 6.81e-01 |
| GO:BP | GO:0001770 | establishment of natural killer cell polarity | 1 | 1 | 6.81e-01 |
| GO:BP | GO:0016032 | viral process | 2 | 344 | 6.82e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 4 | 20 | 6.82e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 1 | 132 | 6.82e-01 |
| GO:BP | GO:1903721 | positive regulation of I-kappaB phosphorylation | 1 | 5 | 6.82e-01 |
| GO:BP | GO:0015918 | sterol transport | 9 | 92 | 6.82e-01 |
| GO:BP | GO:1990386 | mitotic cleavage furrow ingression | 1 | 1 | 6.82e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 1 | 40 | 6.82e-01 |
| GO:BP | GO:1901160 | primary amino compound metabolic process | 1 | 8 | 6.82e-01 |
| GO:BP | GO:0060936 | cardiac fibroblast cell development | 1 | 4 | 6.82e-01 |
| GO:BP | GO:0060935 | cardiac fibroblast cell differentiation | 1 | 4 | 6.82e-01 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 1 | 19 | 6.82e-01 |
| GO:BP | GO:1904506 | negative regulation of telomere maintenance in response to DNA damage | 2 | 3 | 6.82e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 6.82e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 6.82e-01 |
| GO:BP | GO:2000764 | positive regulation of semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 6.83e-01 |
| GO:BP | GO:2000763 | positive regulation of transcription from RNA polymerase II promoter involved in norepinephrine biosynthetic process | 1 | 1 | 6.83e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 6.83e-01 |
| GO:BP | GO:2001262 | positive regulation of semaphorin-plexin signaling pathway | 1 | 1 | 6.83e-01 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 1 | 17 | 6.83e-01 |
| GO:BP | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration | 4 | 7 | 6.83e-01 |
| GO:BP | GO:0071527 | semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 6.83e-01 |
| GO:BP | GO:0042168 | heme metabolic process | 19 | 40 | 6.83e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 6.83e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 3 | 5 | 6.84e-01 |
| GO:BP | GO:0032072 | regulation of restriction endodeoxyribonuclease activity | 1 | 1 | 6.84e-01 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 4 | 8 | 6.84e-01 |
| GO:BP | GO:0071492 | cellular response to UV-A | 1 | 8 | 6.84e-01 |
| GO:BP | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 1 | 3 | 6.84e-01 |
| GO:BP | GO:0000348 | mRNA branch site recognition | 1 | 1 | 6.84e-01 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 2 | 6 | 6.84e-01 |
| GO:BP | GO:0015734 | taurine transport | 2 | 6 | 6.84e-01 |
| GO:BP | GO:0046717 | acid secretion | 2 | 25 | 6.85e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 2 | 362 | 6.85e-01 |
| GO:BP | GO:0140246 | protein catabolic process at synapse | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0140249 | protein catabolic process at postsynapse | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0007549 | dosage compensation | 5 | 29 | 6.85e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 1 | 26 | 6.85e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 1 | 26 | 6.85e-01 |
| GO:BP | GO:0007398 | ectoderm development | 1 | 15 | 6.85e-01 |
| GO:BP | GO:0072109 | glomerular mesangium development | 4 | 13 | 6.85e-01 |
| GO:BP | GO:0000451 | rRNA 2’-O-methylation | 2 | 3 | 6.85e-01 |
| GO:BP | GO:1904003 | negative regulation of sebum secreting cell proliferation | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 7 | 13 | 6.85e-01 |
| GO:BP | GO:0030539 | male genitalia development | 2 | 13 | 6.86e-01 |
| GO:BP | GO:0034772 | histone H4-K20 dimethylation | 1 | 2 | 6.86e-01 |
| GO:BP | GO:0090230 | regulation of centromere complex assembly | 1 | 4 | 6.86e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 3 | 7 | 6.86e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 2 | 10 | 6.86e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 8 | 80 | 6.87e-01 |
| GO:BP | GO:0051866 | general adaptation syndrome | 2 | 4 | 6.87e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 4 | 8 | 6.87e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 8 | 17 | 6.87e-01 |
| GO:BP | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation | 1 | 2 | 6.87e-01 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 3 | 7 | 6.87e-01 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 1 | 7 | 6.87e-01 |
| GO:BP | GO:0006085 | acetyl-CoA biosynthetic process | 1 | 16 | 6.87e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 15 | 62 | 6.87e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 3 | 24 | 6.87e-01 |
| GO:BP | GO:1902688 | regulation of NAD metabolic process | 1 | 1 | 6.87e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 4 | 70 | 6.87e-01 |
| GO:BP | GO:0006553 | lysine metabolic process | 3 | 7 | 6.88e-01 |
| GO:BP | GO:2000485 | regulation of glutamine transport | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 4 | 21 | 6.88e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 2 | 66 | 6.88e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 6.88e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 6.88e-01 |
| GO:BP | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0042339 | keratan sulfate metabolic process | 5 | 15 | 6.88e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 17 | 36 | 6.88e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 6.88e-01 |
| GO:BP | GO:0070977 | bone maturation | 1 | 19 | 6.88e-01 |
| GO:BP | GO:0032658 | regulation of interleukin-15 production | 1 | 1 | 6.88e-01 |
| GO:BP | GO:0032738 | positive regulation of interleukin-15 production | 1 | 1 | 6.88e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 7 | 34 | 6.88e-01 |
| GO:BP | GO:0032618 | interleukin-15 production | 1 | 1 | 6.88e-01 |
| GO:BP | GO:2000368 | positive regulation of acrosomal vesicle exocytosis | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0035665 | TIRAP-dependent toll-like receptor 4 signaling pathway | 1 | 1 | 6.88e-01 |
| GO:BP | GO:0035664 | TIRAP-dependent toll-like receptor signaling pathway | 1 | 1 | 6.88e-01 |
| GO:BP | GO:0033342 | negative regulation of collagen binding | 1 | 1 | 6.88e-01 |
| GO:BP | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis | 1 | 3 | 6.88e-01 |
| GO:BP | GO:0033505 | floor plate morphogenesis | 1 | 3 | 6.88e-01 |
| GO:BP | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly | 4 | 6 | 6.88e-01 |
| GO:BP | GO:0035246 | peptidyl-arginine N-methylation | 2 | 4 | 6.89e-01 |
| GO:BP | GO:0021503 | neural fold bending | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 12 | 89 | 6.89e-01 |
| GO:BP | GO:0071919 | G-quadruplex DNA formation | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 4 | 135 | 6.89e-01 |
| GO:BP | GO:0140029 | exocytic process | 35 | 69 | 6.89e-01 |
| GO:BP | GO:0071073 | positive regulation of phospholipid biosynthetic process | 3 | 7 | 6.90e-01 |
| GO:BP | GO:0006254 | CTP catabolic process | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 6.90e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 5 | 27 | 6.90e-01 |
| GO:BP | GO:0061034 | olfactory bulb mitral cell layer development | 1 | 2 | 6.90e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 7 | 131 | 6.90e-01 |
| GO:BP | GO:0009210 | pyrimidine ribonucleoside triphosphate catabolic process | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0046052 | UTP catabolic process | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 1 | 107 | 6.90e-01 |
| GO:BP | GO:0046477 | glycosylceramide catabolic process | 2 | 5 | 6.90e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 6.90e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 2 | 146 | 6.90e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 59 | 6.90e-01 |
| GO:BP | GO:0007014 | actin ubiquitination | 1 | 1 | 6.90e-01 |
| GO:BP | GO:1903884 | regulation of chemokine (C-C motif) ligand 20 production | 1 | 1 | 6.90e-01 |
| GO:BP | GO:1903881 | regulation of interleukin-17-mediated signaling pathway | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 6 | 55 | 6.90e-01 |
| GO:BP | GO:1903886 | positive regulation of chemokine (C-C motif) ligand 20 production | 1 | 1 | 6.90e-01 |
| GO:BP | GO:1903883 | positive regulation of interleukin-17-mediated signaling pathway | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0036392 | chemokine (C-C motif) ligand 20 production | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 3 | 258 | 6.90e-01 |
| GO:BP | GO:0045626 | negative regulation of T-helper 1 cell differentiation | 1 | 2 | 6.90e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 660 | 6.90e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 3 | 78 | 6.90e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 12 | 67 | 6.90e-01 |
| GO:BP | GO:0003158 | endothelium development | 3 | 104 | 6.90e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 6.90e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 1 | 43 | 6.90e-01 |
| GO:BP | GO:0098908 | regulation of neuronal action potential | 1 | 3 | 6.90e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 2 | 5 | 6.90e-01 |
| GO:BP | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | 1 | 2 | 6.90e-01 |
| GO:BP | GO:1902859 | propionyl-CoA catabolic process | 1 | 2 | 6.90e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 3 | 14 | 6.90e-01 |
| GO:BP | GO:0010816 | calcitonin catabolic process | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0072046 | establishment of planar polarity involved in nephron morphogenesis | 1 | 1 | 6.90e-01 |
| GO:BP | GO:1900112 | regulation of histone H3-K9 trimethylation | 2 | 5 | 6.90e-01 |
| GO:BP | GO:0034959 | endothelin maturation | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 3 | 114 | 6.90e-01 |
| GO:BP | GO:2000252 | negative regulation of feeding behavior | 2 | 2 | 6.90e-01 |
| GO:BP | GO:0045875 | negative regulation of sister chromatid cohesion | 1 | 5 | 6.91e-01 |
| GO:BP | GO:1905135 | biotin import across plasma membrane | 1 | 2 | 6.91e-01 |
| GO:BP | GO:0001579 | medium-chain fatty acid transport | 1 | 2 | 6.91e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 6.91e-01 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 4 | 8 | 6.91e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 8 | 135 | 6.91e-01 |
| GO:BP | GO:1901537 | positive regulation of DNA demethylation | 1 | 7 | 6.91e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 12 | 46 | 6.91e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 4 | 133 | 6.91e-01 |
| GO:BP | GO:0046836 | glycolipid transport | 1 | 7 | 6.91e-01 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 6.91e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 16 | 26 | 6.91e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 1 | 126 | 6.91e-01 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 2 | 14 | 6.91e-01 |
| GO:BP | GO:0001556 | oocyte maturation | 4 | 22 | 6.91e-01 |
| GO:BP | GO:0048861 | leukemia inhibitory factor signaling pathway | 2 | 6 | 6.91e-01 |
| GO:BP | GO:0016081 | synaptic vesicle docking | 2 | 9 | 6.91e-01 |
| GO:BP | GO:0046833 | positive regulation of RNA export from nucleus | 2 | 5 | 6.91e-01 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 1 | 23 | 6.91e-01 |
| GO:BP | GO:0009249 | protein lipoylation | 1 | 6 | 6.91e-01 |
| GO:BP | GO:0006580 | ethanolamine metabolic process | 1 | 1 | 6.91e-01 |
| GO:BP | GO:0060304 | regulation of phosphatidylinositol dephosphorylation | 3 | 5 | 6.91e-01 |
| GO:BP | GO:0032364 | intracellular oxygen homeostasis | 1 | 6 | 6.91e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 6.91e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 13 | 84 | 6.91e-01 |
| GO:BP | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium | 1 | 1 | 6.91e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 13 | 64 | 6.91e-01 |
| GO:BP | GO:0140708 | CAT tailing | 1 | 1 | 6.91e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 6 | 34 | 6.91e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 2 | 16 | 6.92e-01 |
| GO:BP | GO:0022027 | interkinetic nuclear migration | 2 | 4 | 6.92e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 26 | 665 | 6.92e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 5 | 135 | 6.92e-01 |
| GO:BP | GO:0070564 | positive regulation of vitamin D receptor signaling pathway | 1 | 5 | 6.92e-01 |
| GO:BP | GO:0018993 | somatic sex determination | 1 | 1 | 6.92e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 1 | 7 | 6.92e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 3 | 124 | 6.92e-01 |
| GO:BP | GO:0045720 | negative regulation of integrin biosynthetic process | 1 | 1 | 6.92e-01 |
| GO:BP | GO:0070189 | kynurenine metabolic process | 1 | 6 | 6.92e-01 |
| GO:BP | GO:0015712 | hexose phosphate transport | 1 | 3 | 6.92e-01 |
| GO:BP | GO:0015760 | glucose-6-phosphate transport | 1 | 3 | 6.92e-01 |
| GO:BP | GO:0046321 | positive regulation of fatty acid oxidation | 3 | 15 | 6.92e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 6.93e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 6 | 462 | 6.93e-01 |
| GO:BP | GO:0044793 | negative regulation by host of viral process | 2 | 10 | 6.93e-01 |
| GO:BP | GO:0005984 | disaccharide metabolic process | 1 | 5 | 6.93e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 2 | 13 | 6.93e-01 |
| GO:BP | GO:0110095 | cellular detoxification of aldehyde | 4 | 5 | 6.93e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 2 | 260 | 6.93e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 3 | 27 | 6.93e-01 |
| GO:BP | GO:1902001 | fatty acid transmembrane transport | 1 | 15 | 6.93e-01 |
| GO:BP | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0071420 | cellular response to histamine | 3 | 4 | 6.93e-01 |
| GO:BP | GO:0072382 | minus-end-directed vesicle transport along microtubule | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 2 | 4 | 6.93e-01 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 2 | 4 | 6.93e-01 |
| GO:BP | GO:0070294 | renal sodium ion absorption | 5 | 9 | 6.93e-01 |
| GO:BP | GO:0003226 | right ventricular compact myocardium morphogenesis | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 5 | 16 | 6.93e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 2 | 53 | 6.93e-01 |
| GO:BP | GO:0051412 | response to corticosterone | 5 | 10 | 6.93e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 8 | 172 | 6.93e-01 |
| GO:BP | GO:1901687 | glutathione derivative biosynthetic process | 2 | 3 | 6.93e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 9 | 30 | 6.93e-01 |
| GO:BP | GO:0000154 | rRNA modification | 18 | 35 | 6.93e-01 |
| GO:BP | GO:1901685 | glutathione derivative metabolic process | 2 | 3 | 6.93e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 6.93e-01 |
| GO:BP | GO:0048266 | behavioral response to pain | 1 | 11 | 6.93e-01 |
| GO:BP | GO:0097283 | keratinocyte apoptotic process | 1 | 5 | 6.93e-01 |
| GO:BP | GO:1902172 | regulation of keratinocyte apoptotic process | 1 | 5 | 6.93e-01 |
| GO:BP | GO:1903778 | protein localization to vacuolar membrane | 1 | 1 | 6.93e-01 |
| GO:BP | GO:0140213 | negative regulation of long-chain fatty acid import into cell | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0010748 | negative regulation of long-chain fatty acid import across plasma membrane | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0033239 | negative regulation of amine metabolic process | 2 | 2 | 6.94e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 3 | 7 | 6.94e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 6.94e-01 |
| GO:BP | GO:0055118 | negative regulation of cardiac muscle contraction | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0006508 | proteolysis | 3 | 1318 | 6.94e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 1 | 53 | 6.94e-01 |
| GO:BP | GO:0044357 | regulation of rRNA stability | 1 | 1 | 6.94e-01 |
| GO:BP | GO:1902374 | regulation of rRNA catabolic process | 1 | 1 | 6.94e-01 |
| GO:BP | GO:0033353 | S-adenosylmethionine cycle | 3 | 7 | 6.94e-01 |
| GO:BP | GO:0140448 | signaling receptor ligand precursor processing | 7 | 29 | 6.94e-01 |
| GO:BP | GO:2000510 | positive regulation of dendritic cell chemotaxis | 1 | 3 | 6.94e-01 |
| GO:BP | GO:1904683 | regulation of metalloendopeptidase activity | 3 | 6 | 6.94e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 3 | 7 | 6.94e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 6.94e-01 |
| GO:BP | GO:0086096 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway involved in heart process | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 14 | 25 | 6.95e-01 |
| GO:BP | GO:0061108 | seminal vesicle epithelium development | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 1 | 18 | 6.95e-01 |
| GO:BP | GO:1903741 | negative regulation of phosphatidate phosphatase activity | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 4 | 73 | 6.95e-01 |
| GO:BP | GO:1903730 | regulation of phosphatidate phosphatase activity | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 10 | 123 | 6.95e-01 |
| GO:BP | GO:0006172 | ADP biosynthetic process | 3 | 4 | 6.95e-01 |
| GO:BP | GO:0042628 | mating plug formation | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 5 | 32 | 6.95e-01 |
| GO:BP | GO:0097035 | regulation of membrane lipid distribution | 2 | 43 | 6.95e-01 |
| GO:BP | GO:0071877 | regulation of adenylate cyclase-inhibiting adrenergic receptor signaling pathway | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0140193 | regulation of adenylate cyclase-inhibiting adrenergic receptor signaling pathway involved in heart process | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 52 | 1735 | 6.95e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 2 | 2 | 6.95e-01 |
| GO:BP | GO:0009180 | purine ribonucleoside diphosphate biosynthetic process | 3 | 4 | 6.95e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 2 | 2 | 6.95e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 6.95e-01 |
| GO:BP | GO:0006116 | NADH oxidation | 2 | 4 | 6.95e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 2 | 2 | 6.95e-01 |
| GO:BP | GO:0140194 | negative regulation of adenylate cyclase-inhibiting adrenergic receptor signaling pathway involved in heart process | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0070666 | regulation of mast cell proliferation | 1 | 6 | 6.95e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 11 | 21 | 6.95e-01 |
| GO:BP | GO:0070662 | mast cell proliferation | 1 | 6 | 6.95e-01 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 6 | 17 | 6.95e-01 |
| GO:BP | GO:0030049 | muscle filament sliding | 2 | 10 | 6.95e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 6.95e-01 |
| GO:BP | GO:0007387 | anterior compartment pattern formation | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0007388 | posterior compartment specification | 1 | 1 | 6.95e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 3 | 9 | 6.95e-01 |
| GO:BP | GO:0032682 | negative regulation of chemokine production | 3 | 12 | 6.95e-01 |
| GO:BP | GO:0003352 | regulation of cilium movement | 1 | 17 | 6.95e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 103 | 249 | 6.96e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 1 | 8 | 6.96e-01 |
| GO:BP | GO:0009756 | carbohydrate mediated signaling | 1 | 6 | 6.96e-01 |
| GO:BP | GO:0060245 | detection of cell density | 4 | 9 | 6.96e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 1 | 8 | 6.96e-01 |
| GO:BP | GO:0048513 | animal organ development | 4 | 2189 | 6.96e-01 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 7 | 18 | 6.96e-01 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 1 | 11 | 6.96e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 8 | 43 | 6.96e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 6.96e-01 |
| GO:BP | GO:0034463 | 90S preribosome assembly | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 3 | 27 | 6.96e-01 |
| GO:BP | GO:0042930 | enterobactin transport | 1 | 1 | 6.96e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 2 | 37 | 6.96e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 12 | 75 | 6.96e-01 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 1 | 7 | 6.96e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 29 | 129 | 6.96e-01 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 2 | 9 | 6.96e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 16 | 27 | 6.96e-01 |
| GO:BP | GO:0032197 | retrotransposition | 2 | 4 | 6.96e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 18 | 43 | 6.96e-01 |
| GO:BP | GO:0006689 | ganglioside catabolic process | 2 | 5 | 6.96e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 4 | 18 | 6.96e-01 |
| GO:BP | GO:0038116 | chemokine (C-C motif) ligand 21 signaling pathway | 1 | 2 | 6.96e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 15 | 27 | 6.96e-01 |
| GO:BP | GO:0034115 | negative regulation of heterotypic cell-cell adhesion | 1 | 6 | 6.97e-01 |
| GO:BP | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation | 1 | 6 | 6.97e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 16 | 66 | 6.97e-01 |
| GO:BP | GO:0097370 | protein O-GlcNAcylation via threonine | 1 | 1 | 6.97e-01 |
| GO:BP | GO:1903630 | regulation of aminoacyl-tRNA ligase activity | 1 | 2 | 6.98e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 2 | 16 | 6.98e-01 |
| GO:BP | GO:0099533 | positive regulation of presynaptic cytosolic calcium concentration | 1 | 1 | 6.98e-01 |
| GO:BP | GO:0032000 | positive regulation of fatty acid beta-oxidation | 3 | 9 | 6.98e-01 |
| GO:BP | GO:0015802 | basic amino acid transport | 1 | 17 | 6.98e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 1 | 21 | 6.99e-01 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 1 | 12 | 6.99e-01 |
| GO:BP | GO:0003056 | regulation of vascular associated smooth muscle contraction | 3 | 8 | 6.99e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 1 | 155 | 6.99e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 1 | 14 | 6.99e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 7 | 28 | 6.99e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 1 | 14 | 6.99e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 1 | 40 | 7.00e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 7.00e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 7.00e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0021934 | hindbrain tangential cell migration | 2 | 3 | 7.00e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 20 | 39 | 7.00e-01 |
| GO:BP | GO:0099044 | vesicle tethering to endoplasmic reticulum | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0032625 | interleukin-21 production | 1 | 1 | 7.00e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 2 | 19 | 7.00e-01 |
| GO:BP | GO:0021871 | forebrain regionalization | 1 | 8 | 7.00e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 1 | 35 | 7.00e-01 |
| GO:BP | GO:0031016 | pancreas development | 2 | 53 | 7.00e-01 |
| GO:BP | GO:0030258 | lipid modification | 1 | 162 | 7.00e-01 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 4 | 28 | 7.00e-01 |
| GO:BP | GO:0032745 | positive regulation of interleukin-21 production | 1 | 1 | 7.00e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 2 | 19 | 7.00e-01 |
| GO:BP | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity | 2 | 6 | 7.00e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 4 | 14 | 7.00e-01 |
| GO:BP | GO:0014732 | skeletal muscle atrophy | 2 | 5 | 7.00e-01 |
| GO:BP | GO:0032665 | regulation of interleukin-21 production | 1 | 1 | 7.00e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 5 | 23 | 7.00e-01 |
| GO:BP | GO:0048014 | Tie signaling pathway | 2 | 3 | 7.00e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 7.00e-01 |
| GO:BP | GO:0061091 | regulation of phospholipid translocation | 1 | 5 | 7.00e-01 |
| GO:BP | GO:0006678 | glucosylceramide metabolic process | 1 | 5 | 7.00e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 7.00e-01 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 2 | 8 | 7.00e-01 |
| GO:BP | GO:0052472 | modulation by host of symbiont transcription | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 12 | 46 | 7.01e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 12 | 38 | 7.01e-01 |
| GO:BP | GO:0002115 | store-operated calcium entry | 2 | 15 | 7.01e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 7.01e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 12 | 85 | 7.01e-01 |
| GO:BP | GO:0006949 | syncytium formation | 6 | 39 | 7.01e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 2 | 52 | 7.01e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 13 | 28 | 7.01e-01 |
| GO:BP | GO:0010739 | positive regulation of protein kinase A signaling | 5 | 6 | 7.01e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 13 | 30 | 7.01e-01 |
| GO:BP | GO:0071712 | ER-associated misfolded protein catabolic process | 1 | 12 | 7.01e-01 |
| GO:BP | GO:0070235 | regulation of activation-induced cell death of T cells | 2 | 3 | 7.01e-01 |
| GO:BP | GO:0036265 | RNA (guanine-N7)-methylation | 3 | 6 | 7.01e-01 |
| GO:BP | GO:2000627 | positive regulation of miRNA catabolic process | 1 | 4 | 7.01e-01 |
| GO:BP | GO:1903577 | cellular response to L-arginine | 1 | 3 | 7.02e-01 |
| GO:BP | GO:0061792 | secretory granule maturation | 1 | 3 | 7.02e-01 |
| GO:BP | GO:1990502 | dense core granule maturation | 1 | 3 | 7.02e-01 |
| GO:BP | GO:0031623 | receptor internalization | 1 | 95 | 7.02e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 1 | 12 | 7.03e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 7.03e-01 |
| GO:BP | GO:0002071 | glandular epithelial cell maturation | 4 | 7 | 7.04e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 2 | 317 | 7.04e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 5 | 48 | 7.04e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 5 | 29 | 7.04e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 7.04e-01 |
| GO:BP | GO:1903766 | positive regulation of potassium ion export across plasma membrane | 1 | 3 | 7.04e-01 |
| GO:BP | GO:0033088 | negative regulation of immature T cell proliferation in thymus | 1 | 3 | 7.04e-01 |
| GO:BP | GO:0033087 | negative regulation of immature T cell proliferation | 1 | 3 | 7.04e-01 |
| GO:BP | GO:1904429 | regulation of t-circle formation | 3 | 5 | 7.04e-01 |
| GO:BP | GO:0072387 | flavin adenine dinucleotide metabolic process | 1 | 2 | 7.04e-01 |
| GO:BP | GO:1904349 | positive regulation of small intestine smooth muscle contraction | 1 | 1 | 7.04e-01 |
| GO:BP | GO:1904347 | regulation of small intestine smooth muscle contraction | 1 | 1 | 7.04e-01 |
| GO:BP | GO:0080009 | mRNA methylation | 1 | 16 | 7.04e-01 |
| GO:BP | GO:0060313 | negative regulation of blood vessel remodeling | 1 | 2 | 7.04e-01 |
| GO:BP | GO:0033762 | response to glucagon | 2 | 13 | 7.05e-01 |
| GO:BP | GO:2001279 | regulation of unsaturated fatty acid biosynthetic process | 4 | 7 | 7.05e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 7 | 123 | 7.05e-01 |
| GO:BP | GO:0007635 | chemosensory behavior | 1 | 10 | 7.06e-01 |
| GO:BP | GO:1905209 | positive regulation of cardiocyte differentiation | 3 | 11 | 7.06e-01 |
| GO:BP | GO:0070085 | glycosylation | 4 | 202 | 7.06e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 5 | 18 | 7.06e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 33 | 1974 | 7.06e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 9 | 17 | 7.06e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 2 | 12 | 7.06e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 22 | 55 | 7.06e-01 |
| GO:BP | GO:0030916 | otic vesicle formation | 4 | 6 | 7.06e-01 |
| GO:BP | GO:0002069 | columnar/cuboidal epithelial cell maturation | 6 | 9 | 7.06e-01 |
| GO:BP | GO:0070498 | interleukin-1-mediated signaling pathway | 2 | 20 | 7.06e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 2 | 3 | 7.06e-01 |
| GO:BP | GO:0034414 | tRNA 3’-trailer cleavage, endonucleolytic | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0032483 | regulation of Rab protein signal transduction | 2 | 7 | 7.06e-01 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 6 | 11 | 7.06e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 2 | 29 | 7.06e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 2 | 29 | 7.06e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 3 | 13 | 7.06e-01 |
| GO:BP | GO:0030002 | intracellular monoatomic anion homeostasis | 1 | 7 | 7.06e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 2 | 38 | 7.06e-01 |
| GO:BP | GO:0006479 | protein methylation | 4 | 160 | 7.06e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 2 | 16 | 7.06e-01 |
| GO:BP | GO:0035697 | CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.06e-01 |
| GO:BP | GO:0030644 | intracellular chloride ion homeostasis | 1 | 7 | 7.06e-01 |
| GO:BP | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.06e-01 |
| GO:BP | GO:0071550 | death-inducing signaling complex assembly | 1 | 3 | 7.06e-01 |
| GO:BP | GO:2000409 | positive regulation of T cell extravasation | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 1 | 31 | 7.06e-01 |
| GO:BP | GO:0001542 | ovulation from ovarian follicle | 1 | 8 | 7.06e-01 |
| GO:BP | GO:0055025 | positive regulation of cardiac muscle tissue development | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0008213 | protein alkylation | 4 | 160 | 7.06e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 7 | 82 | 7.06e-01 |
| GO:BP | GO:0110096 | cellular response to aldehyde | 8 | 10 | 7.06e-01 |
| GO:BP | GO:0006562 | proline catabolic process | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0032811 | negative regulation of epinephrine secretion | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 12 | 76 | 7.06e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 12 | 76 | 7.06e-01 |
| GO:BP | GO:1905702 | regulation of inhibitory synapse assembly | 1 | 3 | 7.06e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 56 | 277 | 7.07e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 7.07e-01 |
| GO:BP | GO:0007040 | lysosome organization | 2 | 88 | 7.07e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 2 | 4 | 7.07e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 3 | 11 | 7.07e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 7.07e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 2 | 88 | 7.07e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 7.07e-01 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 4 | 19 | 7.07e-01 |
| GO:BP | GO:1902558 | 5’-adenylyl sulfate transmembrane transport | 1 | 2 | 7.07e-01 |
| GO:BP | GO:1902559 | 3’-phospho-5’-adenylyl sulfate transmembrane transport | 1 | 2 | 7.07e-01 |
| GO:BP | GO:1901529 | positive regulation of anion channel activity | 2 | 2 | 7.07e-01 |
| GO:BP | GO:1903793 | positive regulation of monoatomic anion transport | 2 | 2 | 7.07e-01 |
| GO:BP | GO:1903961 | positive regulation of anion transmembrane transport | 2 | 2 | 7.07e-01 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 1 | 12 | 7.07e-01 |
| GO:BP | GO:0003084 | positive regulation of systemic arterial blood pressure | 1 | 7 | 7.07e-01 |
| GO:BP | GO:0046963 | 3’-phosphoadenosine 5’-phosphosulfate transport | 1 | 2 | 7.07e-01 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 4 | 19 | 7.07e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 3 | 330 | 7.07e-01 |
| GO:BP | GO:0045343 | regulation of MHC class I biosynthetic process | 2 | 3 | 7.07e-01 |
| GO:BP | GO:0045341 | MHC class I biosynthetic process | 2 | 3 | 7.07e-01 |
| GO:BP | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process | 1 | 3 | 7.07e-01 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 7.07e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 1 | 51 | 7.08e-01 |
| GO:BP | GO:0006783 | heme biosynthetic process | 13 | 30 | 7.08e-01 |
| GO:BP | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation | 1 | 4 | 7.08e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 7.08e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 8 | 24 | 7.09e-01 |
| GO:BP | GO:0046292 | formaldehyde metabolic process | 2 | 3 | 7.09e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 7 | 30 | 7.09e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 7 | 30 | 7.09e-01 |
| GO:BP | GO:0007252 | I-kappaB phosphorylation | 2 | 14 | 7.09e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 1 | 33 | 7.09e-01 |
| GO:BP | GO:0018344 | protein geranylgeranylation | 1 | 6 | 7.09e-01 |
| GO:BP | GO:0061549 | sympathetic ganglion development | 3 | 6 | 7.09e-01 |
| GO:BP | GO:0002713 | negative regulation of B cell mediated immunity | 4 | 9 | 7.09e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 3 | 14 | 7.09e-01 |
| GO:BP | GO:0046832 | negative regulation of RNA export from nucleus | 1 | 2 | 7.09e-01 |
| GO:BP | GO:0050996 | positive regulation of lipid catabolic process | 1 | 16 | 7.09e-01 |
| GO:BP | GO:0002890 | negative regulation of immunoglobulin mediated immune response | 4 | 9 | 7.09e-01 |
| GO:BP | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | 2 | 5 | 7.09e-01 |
| GO:BP | GO:0022010 | central nervous system myelination | 1 | 12 | 7.09e-01 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 1 | 22 | 7.09e-01 |
| GO:BP | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway | 1 | 3 | 7.09e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 41 | 691 | 7.09e-01 |
| GO:BP | GO:0032291 | axon ensheathment in central nervous system | 1 | 12 | 7.09e-01 |
| GO:BP | GO:0009589 | detection of UV | 1 | 2 | 7.09e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 4 | 12 | 7.09e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 2 | 6 | 7.09e-01 |
| GO:BP | GO:1902732 | positive regulation of chondrocyte proliferation | 2 | 5 | 7.09e-01 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 7.09e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 2 | 2065 | 7.09e-01 |
| GO:BP | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 2 | 7.10e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 3 | 40 | 7.10e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 2 | 111 | 7.10e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 5 | 32 | 7.10e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 2 | 15 | 7.10e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 2 | 15 | 7.10e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 4 | 93 | 7.10e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 6 | 212 | 7.10e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 2 | 15 | 7.10e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 21 | 114 | 7.10e-01 |
| GO:BP | GO:1901334 | lactone metabolic process | 1 | 9 | 7.10e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 51 | 127 | 7.10e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 7.10e-01 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 1 | 9 | 7.10e-01 |
| GO:BP | GO:0060023 | soft palate development | 1 | 3 | 7.10e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 6 | 37 | 7.10e-01 |
| GO:BP | GO:0032930 | positive regulation of superoxide anion generation | 4 | 10 | 7.10e-01 |
| GO:BP | GO:1904925 | positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 3 | 14 | 7.10e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 6 | 24 | 7.10e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 4 | 34 | 7.10e-01 |
| GO:BP | GO:1903719 | regulation of I-kappaB phosphorylation | 1 | 6 | 7.10e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 4 | 116 | 7.10e-01 |
| GO:BP | GO:0021563 | glossopharyngeal nerve development | 2 | 3 | 7.10e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 4 | 170 | 7.10e-01 |
| GO:BP | GO:0034105 | positive regulation of tissue remodeling | 1 | 4 | 7.10e-01 |
| GO:BP | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway | 3 | 8 | 7.10e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 3 | 32 | 7.10e-01 |
| GO:BP | GO:0090170 | regulation of Golgi inheritance | 1 | 4 | 7.11e-01 |
| GO:BP | GO:2000354 | regulation of ovarian follicle development | 2 | 5 | 7.11e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 8 | 39 | 7.11e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 7.11e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 7.11e-01 |
| GO:BP | GO:0061870 | positive regulation of hepatic stellate cell migration | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0061868 | hepatic stellate cell migration | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0032474 | otolith morphogenesis | 1 | 3 | 7.11e-01 |
| GO:BP | GO:0061869 | regulation of hepatic stellate cell migration | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 7.11e-01 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 2 | 8 | 7.11e-01 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 2 | 30 | 7.11e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 6 | 24 | 7.11e-01 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 2 | 4 | 7.11e-01 |
| GO:BP | GO:0021740 | principal sensory nucleus of trigeminal nerve development | 1 | 1 | 7.11e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0021730 | trigeminal sensory nucleus development | 1 | 1 | 7.11e-01 |
| GO:BP | GO:0097368 | establishment of Sertoli cell barrier | 2 | 3 | 7.12e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 2 | 55 | 7.12e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 2 | 55 | 7.12e-01 |
| GO:BP | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | 9 | 14 | 7.12e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 2 | 44 | 7.12e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 2 | 7 | 7.13e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 7 | 56 | 7.13e-01 |
| GO:BP | GO:1904045 | cellular response to aldosterone | 4 | 5 | 7.13e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 8 | 29 | 7.13e-01 |
| GO:BP | GO:0032978 | protein insertion into membrane from inner side | 2 | 4 | 7.13e-01 |
| GO:BP | GO:0032979 | protein insertion into mitochondrial inner membrane from matrix | 2 | 4 | 7.13e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 14 | 45 | 7.14e-01 |
| GO:BP | GO:1905349 | ciliary transition zone assembly | 1 | 2 | 7.14e-01 |
| GO:BP | GO:0090238 | positive regulation of arachidonic acid secretion | 1 | 2 | 7.14e-01 |
| GO:BP | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 5 | 10 | 7.14e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 24 | 993 | 7.14e-01 |
| GO:BP | GO:0007158 | neuron cell-cell adhesion | 1 | 13 | 7.14e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 4 | 7.14e-01 |
| GO:BP | GO:0010985 | negative regulation of lipoprotein particle clearance | 3 | 6 | 7.15e-01 |
| GO:BP | GO:2000077 | negative regulation of type B pancreatic cell development | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 5 | 23 | 7.15e-01 |
| GO:BP | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient | 1 | 7 | 7.15e-01 |
| GO:BP | GO:0006258 | UDP-glucose catabolic process | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway | 1 | 1 | 7.15e-01 |
| GO:BP | GO:0051695 | actin filament uncapping | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 24 | 994 | 7.15e-01 |
| GO:BP | GO:0031590 | wybutosine metabolic process | 2 | 4 | 7.15e-01 |
| GO:BP | GO:0031591 | wybutosine biosynthetic process | 2 | 4 | 7.15e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 1 | 50 | 7.15e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 1 | 168 | 7.15e-01 |
| GO:BP | GO:0072377 | blood coagulation, common pathway | 1 | 1 | 7.15e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 8 | 28 | 7.15e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 47 | 92 | 7.15e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 7 | 39 | 7.15e-01 |
| GO:BP | GO:0048799 | animal organ maturation | 1 | 20 | 7.15e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 2 | 8 | 7.15e-01 |
| GO:BP | GO:1990074 | polyuridylation-dependent mRNA catabolic process | 1 | 3 | 7.15e-01 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 4 | 24 | 7.16e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 2 | 6 | 7.16e-01 |
| GO:BP | GO:0006533 | aspartate catabolic process | 2 | 2 | 7.16e-01 |
| GO:BP | GO:0043007 | maintenance of rDNA | 1 | 2 | 7.16e-01 |
| GO:BP | GO:2001137 | positive regulation of endocytic recycling | 1 | 7 | 7.16e-01 |
| GO:BP | GO:0030810 | positive regulation of nucleotide biosynthetic process | 2 | 12 | 7.16e-01 |
| GO:BP | GO:1900373 | positive regulation of purine nucleotide biosynthetic process | 2 | 12 | 7.16e-01 |
| GO:BP | GO:0001502 | cartilage condensation | 2 | 12 | 7.16e-01 |
| GO:BP | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | 1 | 8 | 7.16e-01 |
| GO:BP | GO:1990379 | lipid transport across blood-brain barrier | 2 | 6 | 7.16e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 29 | 60 | 7.16e-01 |
| GO:BP | GO:0045056 | transcytosis | 7 | 17 | 7.16e-01 |
| GO:BP | GO:0061088 | regulation of sequestering of zinc ion | 2 | 4 | 7.16e-01 |
| GO:BP | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 2 | 7.16e-01 |
| GO:BP | GO:0002074 | extraocular skeletal muscle development | 1 | 2 | 7.16e-01 |
| GO:BP | GO:0002329 | pre-B cell differentiation | 1 | 4 | 7.16e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 5 | 11 | 7.16e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 12 | 276 | 7.17e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 3 | 32 | 7.17e-01 |
| GO:BP | GO:2001198 | regulation of dendritic cell differentiation | 1 | 8 | 7.17e-01 |
| GO:BP | GO:0072350 | tricarboxylic acid metabolic process | 1 | 13 | 7.17e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 61 | 180 | 7.17e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 1 | 154 | 7.17e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 1 | 19 | 7.17e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 7.17e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 2 | 12 | 7.17e-01 |
| GO:BP | GO:0071071 | regulation of phospholipid biosynthetic process | 6 | 15 | 7.17e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 2 | 21 | 7.17e-01 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 2 | 10 | 7.17e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 4 | 94 | 7.17e-01 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 2 | 5 | 7.17e-01 |
| GO:BP | GO:0060764 | cell-cell signaling involved in mammary gland development | 1 | 1 | 7.17e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 7.17e-01 |
| GO:BP | GO:1903367 | positive regulation of fear response | 1 | 4 | 7.17e-01 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 2 | 10 | 7.17e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 7 | 250 | 7.17e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 4 | 7 | 7.17e-01 |
| GO:BP | GO:0001915 | negative regulation of T cell mediated cytotoxicity | 1 | 3 | 7.17e-01 |
| GO:BP | GO:0017126 | nucleologenesis | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 4 | 19 | 7.18e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 2 | 5 | 7.18e-01 |
| GO:BP | GO:0060717 | chorion development | 3 | 8 | 7.18e-01 |
| GO:BP | GO:0016256 | N-glycan processing to lysosome | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0006097 | glyoxylate cycle | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 7.18e-01 |
| GO:BP | GO:0008355 | olfactory learning | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 19 | 47 | 7.18e-01 |
| GO:BP | GO:2000670 | positive regulation of dendritic cell apoptotic process | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0045986 | negative regulation of smooth muscle contraction | 6 | 12 | 7.19e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 2 | 490 | 7.19e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 44 | 103 | 7.19e-01 |
| GO:BP | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor | 1 | 3 | 7.19e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 2 | 490 | 7.19e-01 |
| GO:BP | GO:2000546 | positive regulation of endothelial cell chemotaxis to fibroblast growth factor | 1 | 3 | 7.19e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 4 | 116 | 7.19e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 8 | 41 | 7.19e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 3 | 28 | 7.19e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 3 | 10 | 7.19e-01 |
| GO:BP | GO:0060931 | sinoatrial node cell development | 2 | 4 | 7.19e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 26 | 147 | 7.19e-01 |
| GO:BP | GO:0006379 | mRNA cleavage | 1 | 19 | 7.19e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 11 | 37 | 7.19e-01 |
| GO:BP | GO:1905430 | cellular response to glycine | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 1 | 185 | 7.20e-01 |
| GO:BP | GO:0071896 | protein localization to adherens junction | 2 | 6 | 7.20e-01 |
| GO:BP | GO:0002841 | negative regulation of T cell mediated immune response to tumor cell | 1 | 1 | 7.20e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 1 | 157 | 7.20e-01 |
| GO:BP | GO:0046032 | ADP catabolic process | 1 | 2 | 7.20e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 7 | 104 | 7.20e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 7 | 104 | 7.20e-01 |
| GO:BP | GO:1900069 | regulation of cellular hyperosmotic salinity response | 1 | 4 | 7.20e-01 |
| GO:BP | GO:0048696 | regulation of collateral sprouting in absence of injury | 1 | 2 | 7.20e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 9 | 7.20e-01 |
| GO:BP | GO:1901000 | regulation of response to salt stress | 1 | 4 | 7.20e-01 |
| GO:BP | GO:0044338 | canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation | 1 | 3 | 7.20e-01 |
| GO:BP | GO:0051013 | microtubule severing | 4 | 7 | 7.20e-01 |
| GO:BP | GO:1901843 | positive regulation of high voltage-gated calcium channel activity | 2 | 4 | 7.20e-01 |
| GO:BP | GO:0070173 | regulation of enamel mineralization | 1 | 2 | 7.21e-01 |
| GO:BP | GO:0030422 | siRNA processing | 1 | 7 | 7.21e-01 |
| GO:BP | GO:0048294 | negative regulation of isotype switching to IgE isotypes | 1 | 2 | 7.21e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 8 | 190 | 7.21e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 18 | 1869 | 7.21e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 4 | 34 | 7.21e-01 |
| GO:BP | GO:0048232 | male gamete generation | 3 | 343 | 7.21e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 2 | 225 | 7.21e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 7.21e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 7.21e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 2 | 25 | 7.21e-01 |
| GO:BP | GO:0038065 | collagen-activated signaling pathway | 1 | 13 | 7.21e-01 |
| GO:BP | GO:0045779 | negative regulation of bone resorption | 1 | 7 | 7.21e-01 |
| GO:BP | GO:0071332 | cellular response to fructose stimulus | 1 | 2 | 7.21e-01 |
| GO:BP | GO:1990418 | response to insulin-like growth factor stimulus | 1 | 2 | 7.21e-01 |
| GO:BP | GO:1903487 | regulation of lactation | 1 | 1 | 7.21e-01 |
| GO:BP | GO:1900100 | positive regulation of plasma cell differentiation | 1 | 1 | 7.21e-01 |
| GO:BP | GO:0021511 | spinal cord patterning | 1 | 9 | 7.21e-01 |
| GO:BP | GO:1990544 | mitochondrial ATP transmembrane transport | 2 | 4 | 7.22e-01 |
| GO:BP | GO:0140021 | mitochondrial ADP transmembrane transport | 2 | 4 | 7.22e-01 |
| GO:BP | GO:0001905 | activation of membrane attack complex | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 15 | 40 | 7.22e-01 |
| GO:BP | GO:0070375 | ERK5 cascade | 1 | 3 | 7.22e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 2 | 495 | 7.22e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 8 | 893 | 7.22e-01 |
| GO:BP | GO:0001969 | regulation of activation of membrane attack complex | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 7 | 15 | 7.22e-01 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 5 | 15 | 7.22e-01 |
| GO:BP | GO:0060151 | peroxisome localization | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0060152 | microtubule-based peroxisome localization | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 17 | 34 | 7.22e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 4 | 68 | 7.22e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 7.23e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 3 | 21 | 7.23e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 1 | 64 | 7.23e-01 |
| GO:BP | GO:2000617 | positive regulation of histone H3-K9 acetylation | 1 | 6 | 7.23e-01 |
| GO:BP | GO:1903537 | meiotic cell cycle process involved in oocyte maturation | 1 | 5 | 7.23e-01 |
| GO:BP | GO:0015670 | carbon dioxide transport | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0060066 | oviduct development | 1 | 4 | 7.23e-01 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 5 | 31 | 7.23e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 3 | 147 | 7.23e-01 |
| GO:BP | GO:0015727 | lactate transport | 1 | 5 | 7.23e-01 |
| GO:BP | GO:0035873 | lactate transmembrane transport | 1 | 5 | 7.23e-01 |
| GO:BP | GO:2000347 | positive regulation of hepatocyte proliferation | 4 | 9 | 7.23e-01 |
| GO:BP | GO:0042670 | retinal cone cell differentiation | 3 | 6 | 7.24e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 7.24e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 5 | 92 | 7.24e-01 |
| GO:BP | GO:0060268 | negative regulation of respiratory burst | 1 | 4 | 7.24e-01 |
| GO:BP | GO:0002265 | astrocyte activation involved in immune response | 2 | 4 | 7.24e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 2 | 31 | 7.24e-01 |
| GO:BP | GO:1900223 | positive regulation of amyloid-beta clearance | 1 | 5 | 7.25e-01 |
| GO:BP | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching | 1 | 2 | 7.25e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 2 | 15 | 7.25e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 2 | 357 | 7.25e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 4 | 13 | 7.25e-01 |
| GO:BP | GO:2001295 | malonyl-CoA biosynthetic process | 1 | 2 | 7.25e-01 |
| GO:BP | GO:0055062 | phosphate ion homeostasis | 3 | 8 | 7.25e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 3 | 72 | 7.25e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 4 | 95 | 7.25e-01 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 1 | 16 | 7.25e-01 |
| GO:BP | GO:0018335 | protein succinylation | 2 | 4 | 7.25e-01 |
| GO:BP | GO:0106077 | histone succinylation | 2 | 4 | 7.25e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 7.25e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 1 | 26 | 7.26e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 2 | 401 | 7.26e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 185 | 7.26e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 5 | 17 | 7.26e-01 |
| GO:BP | GO:0070070 | proton-transporting V-type ATPase complex assembly | 1 | 7 | 7.26e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 40 | 145 | 7.26e-01 |
| GO:BP | GO:0060558 | regulation of calcidiol 1-monooxygenase activity | 1 | 2 | 7.27e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 1 | 170 | 7.27e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 1 | 4 | 7.27e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 4 | 9 | 7.27e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 3 | 113 | 7.27e-01 |
| GO:BP | GO:0015911 | long-chain fatty acid import across plasma membrane | 3 | 9 | 7.27e-01 |
| GO:BP | GO:0045453 | bone resorption | 3 | 40 | 7.27e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 7.28e-01 |
| GO:BP | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity | 3 | 4 | 7.28e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 5 | 538 | 7.28e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 1 | 11 | 7.28e-01 |
| GO:BP | GO:0016559 | peroxisome fission | 1 | 9 | 7.28e-01 |
| GO:BP | GO:2000815 | regulation of mRNA stability involved in response to oxidative stress | 1 | 2 | 7.28e-01 |
| GO:BP | GO:1903430 | negative regulation of cell maturation | 2 | 4 | 7.28e-01 |
| GO:BP | GO:1902462 | positive regulation of mesenchymal stem cell proliferation | 2 | 4 | 7.28e-01 |
| GO:BP | GO:1902460 | regulation of mesenchymal stem cell proliferation | 2 | 4 | 7.28e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 3 | 58 | 7.28e-01 |
| GO:BP | GO:0006772 | thiamine metabolic process | 2 | 2 | 7.28e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 1 | 60 | 7.28e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 5 | 19 | 7.29e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 3 | 73 | 7.29e-01 |
| GO:BP | GO:0061025 | membrane fusion | 2 | 137 | 7.29e-01 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 1 | 15 | 7.29e-01 |
| GO:BP | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | 1 | 13 | 7.29e-01 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 4 | 11 | 7.29e-01 |
| GO:BP | GO:0048859 | formation of anatomical boundary | 1 | 3 | 7.29e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 5 | 93 | 7.29e-01 |
| GO:BP | GO:0051791 | medium-chain fatty acid metabolic process | 2 | 9 | 7.29e-01 |
| GO:BP | GO:0016340 | calcium-dependent cell-matrix adhesion | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 1 | 11 | 7.30e-01 |
| GO:BP | GO:0006390 | mitochondrial transcription | 8 | 18 | 7.30e-01 |
| GO:BP | GO:0009067 | aspartate family amino acid biosynthetic process | 7 | 18 | 7.30e-01 |
| GO:BP | GO:0099152 | regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane | 1 | 3 | 7.30e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 47 | 185 | 7.30e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 7.30e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 7.30e-01 |
| GO:BP | GO:0015884 | folic acid transport | 1 | 7 | 7.31e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0090131 | mesenchyme migration | 1 | 4 | 7.31e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 3 | 11 | 7.31e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 1 | 149 | 7.31e-01 |
| GO:BP | GO:0044830 | modulation by host of viral RNA genome replication | 1 | 6 | 7.31e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 2 | 8 | 7.31e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 3 | 11 | 7.31e-01 |
| GO:BP | GO:0031282 | regulation of guanylate cyclase activity | 1 | 3 | 7.31e-01 |
| GO:BP | GO:0017145 | stem cell division | 11 | 26 | 7.31e-01 |
| GO:BP | GO:0048630 | skeletal muscle tissue growth | 1 | 8 | 7.31e-01 |
| GO:BP | GO:0034383 | low-density lipoprotein particle clearance | 1 | 17 | 7.31e-01 |
| GO:BP | GO:0006567 | threonine catabolic process | 2 | 3 | 7.32e-01 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 1 | 17 | 7.32e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 7.32e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 1 | 631 | 7.32e-01 |
| GO:BP | GO:0033031 | positive regulation of neutrophil apoptotic process | 1 | 3 | 7.32e-01 |
| GO:BP | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | 1 | 10 | 7.32e-01 |
| GO:BP | GO:0034635 | glutathione transport | 1 | 9 | 7.32e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 5 | 24 | 7.32e-01 |
| GO:BP | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0009451 | RNA modification | 83 | 162 | 7.32e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 2 | 8 | 7.32e-01 |
| GO:BP | GO:0097533 | cellular stress response to acid chemical | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0097532 | stress response to acid chemical | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0033280 | response to vitamin D | 2 | 24 | 7.32e-01 |
| GO:BP | GO:0060334 | regulation of type II interferon-mediated signaling pathway | 3 | 12 | 7.32e-01 |
| GO:BP | GO:0060330 | regulation of response to type II interferon | 3 | 12 | 7.32e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 7.32e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 32 | 7.32e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 1 | 199 | 7.32e-01 |
| GO:BP | GO:0045176 | apical protein localization | 1 | 13 | 7.32e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 189 | 7.32e-01 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 1 | 12 | 7.32e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 2 | 10 | 7.32e-01 |
| GO:BP | GO:0016320 | endoplasmic reticulum membrane fusion | 1 | 2 | 7.32e-01 |
| GO:BP | GO:2000845 | positive regulation of testosterone secretion | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0160025 | sensory perception of itch | 1 | 1 | 7.32e-01 |
| GO:BP | GO:0160023 | sneeze reflex | 1 | 1 | 7.32e-01 |
| GO:BP | GO:0035936 | testosterone secretion | 1 | 2 | 7.32e-01 |
| GO:BP | GO:2000843 | regulation of testosterone secretion | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 1 | 44 | 7.33e-01 |
| GO:BP | GO:0072300 | positive regulation of metanephric glomerulus development | 1 | 1 | 7.33e-01 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 3 | 7 | 7.33e-01 |
| GO:BP | GO:0072216 | positive regulation of metanephros development | 1 | 1 | 7.33e-01 |
| GO:BP | GO:0048548 | regulation of pinocytosis | 6 | 11 | 7.33e-01 |
| GO:BP | GO:0014042 | positive regulation of neuron maturation | 1 | 3 | 7.33e-01 |
| GO:BP | GO:0072298 | regulation of metanephric glomerulus development | 1 | 1 | 7.33e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 2 | 294 | 7.33e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 2 | 408 | 7.33e-01 |
| GO:BP | GO:0061470 | T follicular helper cell differentiation | 2 | 4 | 7.33e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 3 | 6 | 7.33e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 98 | 3625 | 7.33e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 2 | 511 | 7.33e-01 |
| GO:BP | GO:1905450 | negative regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 2 | 7.33e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 1 | 61 | 7.33e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 2 | 35 | 7.33e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 1 | 7 | 7.33e-01 |
| GO:BP | GO:1905602 | positive regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 2 | 7.33e-01 |
| GO:BP | GO:2001256 | regulation of store-operated calcium entry | 1 | 11 | 7.34e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 2 | 276 | 7.34e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 4 | 15 | 7.34e-01 |
| GO:BP | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development | 1 | 2 | 7.34e-01 |
| GO:BP | GO:0060672 | epithelial cell morphogenesis involved in placental branching | 1 | 2 | 7.34e-01 |
| GO:BP | GO:0006850 | mitochondrial pyruvate transmembrane transport | 1 | 2 | 7.34e-01 |
| GO:BP | GO:0021559 | trigeminal nerve development | 3 | 5 | 7.34e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 2 | 3 | 7.34e-01 |
| GO:BP | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation | 1 | 4 | 7.34e-01 |
| GO:BP | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling | 2 | 12 | 7.35e-01 |
| GO:BP | GO:0007015 | actin filament organization | 4 | 373 | 7.35e-01 |
| GO:BP | GO:0016243 | regulation of autophagosome size | 1 | 2 | 7.35e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 7.35e-01 |
| GO:BP | GO:0010712 | regulation of collagen metabolic process | 3 | 34 | 7.35e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 7.35e-01 |
| GO:BP | GO:0051598 | meiotic recombination checkpoint signaling | 1 | 2 | 7.35e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 2 | 57 | 7.35e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 7.36e-01 |
| GO:BP | GO:0006555 | methionine metabolic process | 1 | 13 | 7.36e-01 |
| GO:BP | GO:0097055 | agmatine biosynthetic process | 1 | 1 | 7.36e-01 |
| GO:BP | GO:1905653 | positive regulation of artery morphogenesis | 2 | 5 | 7.36e-01 |
| GO:BP | GO:1905651 | regulation of artery morphogenesis | 2 | 5 | 7.36e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 4 | 16 | 7.36e-01 |
| GO:BP | GO:0043649 | dicarboxylic acid catabolic process | 1 | 14 | 7.36e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 14 | 323 | 7.36e-01 |
| GO:BP | GO:0090023 | positive regulation of neutrophil chemotaxis | 2 | 14 | 7.36e-01 |
| GO:BP | GO:1902238 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0098629 | trans-Golgi network membrane organization | 1 | 2 | 7.36e-01 |
| GO:BP | GO:2000508 | regulation of dendritic cell chemotaxis | 1 | 3 | 7.36e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 8 | 43 | 7.36e-01 |
| GO:BP | GO:1990127 | intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0045672 | positive regulation of osteoclast differentiation | 1 | 11 | 7.36e-01 |
| GO:BP | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0071228 | cellular response to tumor cell | 2 | 4 | 7.36e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 2 | 22 | 7.36e-01 |
| GO:BP | GO:0051582 | positive regulation of neurotransmitter uptake | 1 | 4 | 7.36e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 3 | 11 | 7.36e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 4 | 30 | 7.36e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 15 | 91 | 7.36e-01 |
| GO:BP | GO:0036228 | protein localization to nuclear inner membrane | 1 | 3 | 7.36e-01 |
| GO:BP | GO:0060696 | regulation of phospholipid catabolic process | 1 | 5 | 7.36e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 7.36e-01 |
| GO:BP | GO:0060465 | pharynx development | 1 | 3 | 7.37e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 9 | 16 | 7.37e-01 |
| GO:BP | GO:0007418 | ventral midline development | 2 | 4 | 7.37e-01 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 2 | 20 | 7.37e-01 |
| GO:BP | GO:0050882 | voluntary musculoskeletal movement | 3 | 7 | 7.37e-01 |
| GO:BP | GO:0045932 | negative regulation of muscle contraction | 10 | 20 | 7.37e-01 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 1 | 3 | 7.37e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 7.37e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 38 | 78 | 7.37e-01 |
| GO:BP | GO:0060921 | sinoatrial node cell differentiation | 3 | 5 | 7.37e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 7.37e-01 |
| GO:BP | GO:1904567 | response to wortmannin | 1 | 2 | 7.38e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 3 | 7 | 7.38e-01 |
| GO:BP | GO:0038192 | gastric inhibitory peptide signaling pathway | 1 | 1 | 7.38e-01 |
| GO:BP | GO:1904568 | cellular response to wortmannin | 1 | 2 | 7.38e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 5 | 23 | 7.38e-01 |
| GO:BP | GO:0070141 | response to UV-A | 1 | 11 | 7.38e-01 |
| GO:BP | GO:0001974 | blood vessel remodeling | 5 | 32 | 7.38e-01 |
| GO:BP | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process | 1 | 2 | 7.38e-01 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 12 | 7.38e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 1 | 13 | 7.38e-01 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 1 | 9 | 7.38e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 1 | 30 | 7.38e-01 |
| GO:BP | GO:0001923 | B-1 B cell differentiation | 1 | 3 | 7.38e-01 |
| GO:BP | GO:1904587 | response to glycoprotein | 1 | 5 | 7.38e-01 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 7.38e-01 |
| GO:BP | GO:0061017 | hepatoblast differentiation | 1 | 1 | 7.39e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 2 | 433 | 7.39e-01 |
| GO:BP | GO:0051028 | mRNA transport | 1 | 117 | 7.40e-01 |
| GO:BP | GO:0098795 | global gene silencing by mRNA cleavage | 1 | 4 | 7.40e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 6 | 204 | 7.40e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 9 | 47 | 7.40e-01 |
| GO:BP | GO:0021702 | cerebellar Purkinje cell differentiation | 5 | 10 | 7.40e-01 |
| GO:BP | GO:1905551 | negative regulation of protein localization to endoplasmic reticulum | 1 | 2 | 7.40e-01 |
| GO:BP | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 5 | 7.40e-01 |
| GO:BP | GO:1990569 | UDP-N-acetylglucosamine transmembrane transport | 2 | 6 | 7.40e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 9 | 740 | 7.40e-01 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 4 | 7 | 7.40e-01 |
| GO:BP | GO:0046544 | development of secondary male sexual characteristics | 1 | 1 | 7.41e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 1 | 12 | 7.41e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 7.42e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 6 | 169 | 7.42e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 3 | 25 | 7.42e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 1 | 63 | 7.42e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 7.42e-01 |
| GO:BP | GO:0046039 | GTP metabolic process | 5 | 21 | 7.42e-01 |
| GO:BP | GO:0070973 | protein localization to endoplasmic reticulum exit site | 3 | 6 | 7.42e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 2 | 34 | 7.42e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 9 | 52 | 7.42e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 6 | 14 | 7.43e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 2 | 111 | 7.43e-01 |
| GO:BP | GO:0072289 | metanephric nephron tubule formation | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0072191 | ureter smooth muscle development | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0072193 | ureter smooth muscle cell differentiation | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0060737 | prostate gland morphogenetic growth | 1 | 2 | 7.43e-01 |
| GO:BP | GO:2000741 | positive regulation of mesenchymal stem cell differentiation | 1 | 4 | 7.43e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 7.43e-01 |
| GO:BP | GO:0060018 | astrocyte fate commitment | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0000961 | negative regulation of mitochondrial RNA catabolic process | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0098920 | retrograde trans-synaptic signaling by lipid | 2 | 4 | 7.43e-01 |
| GO:BP | GO:0098921 | retrograde trans-synaptic signaling by endocannabinoid | 2 | 4 | 7.43e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 7.43e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 4 | 32 | 7.43e-01 |
| GO:BP | GO:0001712 | ectodermal cell fate commitment | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0031054 | pre-miRNA processing | 6 | 14 | 7.43e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 4 | 19 | 7.44e-01 |
| GO:BP | GO:0071380 | cellular response to prostaglandin E stimulus | 1 | 14 | 7.44e-01 |
| GO:BP | GO:1904204 | regulation of skeletal muscle hypertrophy | 1 | 1 | 7.44e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 1 | 22 | 7.44e-01 |
| GO:BP | GO:0033967 | box C/D RNA metabolic process | 1 | 3 | 7.45e-01 |
| GO:BP | GO:0034963 | box C/D RNA processing | 1 | 3 | 7.45e-01 |
| GO:BP | GO:0000494 | box C/D RNA 3’-end processing | 1 | 3 | 7.45e-01 |
| GO:BP | GO:0003099 | positive regulation of the force of heart contraction by chemical signal | 1 | 2 | 7.45e-01 |
| GO:BP | GO:0001823 | mesonephros development | 12 | 77 | 7.45e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 7.45e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 1 | 44 | 7.45e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 1 | 168 | 7.45e-01 |
| GO:BP | GO:2000064 | regulation of cortisol biosynthetic process | 1 | 7 | 7.45e-01 |
| GO:BP | GO:0042220 | response to cocaine | 3 | 28 | 7.45e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 8 | 189 | 7.45e-01 |
| GO:BP | GO:0007568 | aging | 3 | 115 | 7.45e-01 |
| GO:BP | GO:0038003 | G protein-coupled opioid receptor signaling pathway | 4 | 5 | 7.45e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 3 | 6 | 7.45e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 1 | 43 | 7.45e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 4 | 43 | 7.45e-01 |
| GO:BP | GO:2000625 | regulation of miRNA catabolic process | 1 | 5 | 7.45e-01 |
| GO:BP | GO:0099011 | neuronal dense core vesicle exocytosis | 1 | 4 | 7.45e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 10 | 55 | 7.45e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 5 | 20 | 7.45e-01 |
| GO:BP | GO:0048148 | behavioral response to cocaine | 3 | 9 | 7.45e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 5 | 110 | 7.45e-01 |
| GO:BP | GO:0008354 | germ cell migration | 1 | 6 | 7.45e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 3 | 20 | 7.46e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 3 | 118 | 7.46e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 1 | 60 | 7.46e-01 |
| GO:BP | GO:0045659 | negative regulation of neutrophil differentiation | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0035247 | peptidyl-arginine omega-N-methylation | 1 | 3 | 7.46e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 1 | 71 | 7.46e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 1 | 71 | 7.46e-01 |
| GO:BP | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine | 4 | 7 | 7.46e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 1 | 169 | 7.46e-01 |
| GO:BP | GO:0042365 | water-soluble vitamin catabolic process | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0090237 | regulation of arachidonic acid secretion | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 14 | 200 | 7.46e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 7.46e-01 |
| GO:BP | GO:0046469 | platelet activating factor metabolic process | 1 | 5 | 7.46e-01 |
| GO:BP | GO:0031247 | actin rod assembly | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0010501 | RNA secondary structure unwinding | 3 | 8 | 7.46e-01 |
| GO:BP | GO:2000738 | positive regulation of stem cell differentiation | 4 | 17 | 7.47e-01 |
| GO:BP | GO:0097070 | ductus arteriosus closure | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 5 | 73 | 7.47e-01 |
| GO:BP | GO:0002726 | positive regulation of T cell cytokine production | 1 | 14 | 7.47e-01 |
| GO:BP | GO:1903109 | positive regulation of mitochondrial transcription | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0048280 | vesicle fusion with Golgi apparatus | 1 | 8 | 7.47e-01 |
| GO:BP | GO:0031930 | mitochondria-nucleus signaling pathway | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 5 | 30 | 7.47e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 47 | 147 | 7.47e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 7 | 23 | 7.47e-01 |
| GO:BP | GO:0045988 | negative regulation of striated muscle contraction | 2 | 7 | 7.48e-01 |
| GO:BP | GO:0061140 | lung secretory cell differentiation | 1 | 5 | 7.48e-01 |
| GO:BP | GO:0035455 | response to interferon-alpha | 10 | 16 | 7.48e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 9 | 118 | 7.48e-01 |
| GO:BP | GO:0036031 | recruitment of mRNA capping enzyme to RNA polymerase II holoenzyme complex | 1 | 1 | 7.49e-01 |
| GO:BP | GO:0006982 | response to lipid hydroperoxide | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 1 | 22 | 7.49e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 2 | 41 | 7.49e-01 |
| GO:BP | GO:0006072 | glycerol-3-phosphate metabolic process | 4 | 7 | 7.49e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 1 | 45 | 7.50e-01 |
| GO:BP | GO:0035583 | sequestering of TGFbeta in extracellular matrix | 1 | 3 | 7.50e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 3 | 79 | 7.50e-01 |
| GO:BP | GO:0042351 | ‘de novo’ GDP-L-fucose biosynthetic process | 1 | 2 | 7.50e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 7.50e-01 |
| GO:BP | GO:0071377 | cellular response to glucagon stimulus | 1 | 4 | 7.50e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 7.50e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 9 | 17 | 7.50e-01 |
| GO:BP | GO:0009566 | fertilization | 2 | 93 | 7.51e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 15 | 556 | 7.51e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 2 | 25 | 7.51e-01 |
| GO:BP | GO:2000344 | positive regulation of acrosome reaction | 1 | 6 | 7.51e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 33 | 631 | 7.51e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 2 | 29 | 7.51e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 2 | 6 | 7.51e-01 |
| GO:BP | GO:1902566 | regulation of eosinophil activation | 2 | 2 | 7.51e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 2 | 39 | 7.52e-01 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 7.52e-01 |
| GO:BP | GO:0000957 | mitochondrial RNA catabolic process | 3 | 6 | 7.52e-01 |
| GO:BP | GO:0045728 | respiratory burst after phagocytosis | 1 | 2 | 7.52e-01 |
| GO:BP | GO:2001213 | negative regulation of vasculogenesis | 1 | 1 | 7.52e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 2 | 124 | 7.52e-01 |
| GO:BP | GO:0032849 | positive regulation of cellular pH reduction | 2 | 2 | 7.52e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 9 | 14 | 7.53e-01 |
| GO:BP | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process | 4 | 16 | 7.53e-01 |
| GO:BP | GO:1901535 | regulation of DNA demethylation | 1 | 8 | 7.53e-01 |
| GO:BP | GO:0050746 | regulation of lipoprotein metabolic process | 1 | 15 | 7.54e-01 |
| GO:BP | GO:0021553 | olfactory nerve development | 4 | 5 | 7.54e-01 |
| GO:BP | GO:0070171 | negative regulation of tooth mineralization | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 4 | 10 | 7.54e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 8 | 14 | 7.54e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 5 | 18 | 7.54e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 5 | 18 | 7.54e-01 |
| GO:BP | GO:0015742 | alpha-ketoglutarate transport | 1 | 3 | 7.54e-01 |
| GO:BP | GO:0051891 | positive regulation of cardioblast differentiation | 1 | 4 | 7.54e-01 |
| GO:BP | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 2 | 14 | 7.54e-01 |
| GO:BP | GO:0032596 | protein transport into membrane raft | 2 | 4 | 7.54e-01 |
| GO:BP | GO:0035272 | exocrine system development | 5 | 34 | 7.54e-01 |
| GO:BP | GO:0060437 | lung growth | 1 | 5 | 7.54e-01 |
| GO:BP | GO:1905453 | regulation of myeloid progenitor cell differentiation | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0048561 | establishment of animal organ orientation | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 7 | 957 | 7.54e-01 |
| GO:BP | GO:0006867 | asparagine transport | 1 | 3 | 7.55e-01 |
| GO:BP | GO:2001293 | malonyl-CoA metabolic process | 3 | 6 | 7.55e-01 |
| GO:BP | GO:1901289 | succinyl-CoA catabolic process | 3 | 6 | 7.55e-01 |
| GO:BP | GO:0032835 | glomerulus development | 10 | 54 | 7.55e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 1 | 2 | 7.55e-01 |
| GO:BP | GO:0061377 | mammary gland lobule development | 6 | 13 | 7.55e-01 |
| GO:BP | GO:0046813 | receptor-mediated virion attachment to host cell | 1 | 6 | 7.55e-01 |
| GO:BP | GO:0051311 | meiotic metaphase plate congression | 1 | 2 | 7.55e-01 |
| GO:BP | GO:1902024 | L-histidine transport | 1 | 4 | 7.55e-01 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 1 | 8 | 7.55e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 7 | 61 | 7.55e-01 |
| GO:BP | GO:0015817 | histidine transport | 1 | 4 | 7.55e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 8 | 199 | 7.55e-01 |
| GO:BP | GO:0060749 | mammary gland alveolus development | 6 | 13 | 7.55e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 71 | 7.55e-01 |
| GO:BP | GO:0070172 | positive regulation of tooth mineralization | 1 | 2 | 7.55e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 4 | 10 | 7.55e-01 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 2 | 5 | 7.55e-01 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 1 | 10 | 7.55e-01 |
| GO:BP | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | 1 | 12 | 7.55e-01 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 3 | 17 | 7.56e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 4 | 70 | 7.56e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 2 | 17 | 7.56e-01 |
| GO:BP | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II | 1 | 17 | 7.56e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 19 | 50 | 7.56e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 7.56e-01 |
| GO:BP | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization | 1 | 4 | 7.56e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 21 | 50 | 7.56e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 2 | 49 | 7.56e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 8 | 36 | 7.56e-01 |
| GO:BP | GO:0036155 | acylglycerol acyl-chain remodeling | 2 | 2 | 7.56e-01 |
| GO:BP | GO:0032681 | regulation of lymphotoxin A production | 1 | 1 | 7.56e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 5 | 22 | 7.56e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 7.56e-01 |
| GO:BP | GO:0031117 | positive regulation of microtubule depolymerization | 2 | 4 | 7.56e-01 |
| GO:BP | GO:0032761 | positive regulation of lymphotoxin A production | 1 | 1 | 7.56e-01 |
| GO:BP | GO:0032641 | lymphotoxin A production | 1 | 1 | 7.56e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 7.56e-01 |
| GO:BP | GO:0031017 | exocrine pancreas development | 1 | 4 | 7.56e-01 |
| GO:BP | GO:1903576 | response to L-arginine | 1 | 3 | 7.56e-01 |
| GO:BP | GO:1903598 | positive regulation of gap junction assembly | 2 | 7 | 7.57e-01 |
| GO:BP | GO:0008104 | protein localization | 3 | 2129 | 7.57e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 7.57e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 209 | 7.57e-01 |
| GO:BP | GO:0060846 | blood vessel endothelial cell fate commitment | 1 | 3 | 7.57e-01 |
| GO:BP | GO:1903054 | negative regulation of extracellular matrix organization | 3 | 6 | 7.57e-01 |
| GO:BP | GO:0097101 | blood vessel endothelial cell fate specification | 1 | 3 | 7.57e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 1 | 2 | 7.57e-01 |
| GO:BP | GO:0010713 | negative regulation of collagen metabolic process | 5 | 8 | 7.57e-01 |
| GO:BP | GO:0016486 | peptide hormone processing | 6 | 27 | 7.57e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4048 | 7.57e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 3 | 78 | 7.57e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 15 | 40 | 7.57e-01 |
| GO:BP | GO:0001553 | luteinization | 4 | 10 | 7.58e-01 |
| GO:BP | GO:0032497 | detection of lipopolysaccharide | 2 | 2 | 7.58e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 7.58e-01 |
| GO:BP | GO:0010359 | regulation of anion channel activity | 4 | 5 | 7.58e-01 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 1 | 10 | 7.58e-01 |
| GO:BP | GO:0035684 | helper T cell extravasation | 1 | 3 | 7.58e-01 |
| GO:BP | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c | 1 | 7 | 7.58e-01 |
| GO:BP | GO:0042053 | regulation of dopamine metabolic process | 5 | 11 | 7.58e-01 |
| GO:BP | GO:0030449 | regulation of complement activation | 2 | 11 | 7.58e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 3 | 301 | 7.58e-01 |
| GO:BP | GO:0042069 | regulation of catecholamine metabolic process | 5 | 11 | 7.58e-01 |
| GO:BP | GO:0070391 | response to lipoteichoic acid | 1 | 6 | 7.58e-01 |
| GO:BP | GO:0032298 | positive regulation of DNA-templated DNA replication initiation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 3 | 2138 | 7.58e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 5 | 144 | 7.58e-01 |
| GO:BP | GO:0050658 | RNA transport | 5 | 144 | 7.58e-01 |
| GO:BP | GO:0071223 | cellular response to lipoteichoic acid | 1 | 6 | 7.58e-01 |
| GO:BP | GO:0060025 | regulation of synaptic activity | 1 | 3 | 7.58e-01 |
| GO:BP | GO:0014047 | glutamate secretion | 1 | 14 | 7.59e-01 |
| GO:BP | GO:1904379 | protein localization to cytosolic proteasome complex involved in ERAD pathway | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 7.59e-01 |
| GO:BP | GO:1904327 | protein localization to cytosolic proteasome complex | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 7.59e-01 |
| GO:BP | GO:0061857 | endoplasmic reticulum stress-induced pre-emptive quality control | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0051867 | general adaptation syndrome, behavioral process | 1 | 3 | 7.59e-01 |
| GO:BP | GO:0033633 | negative regulation of cell-cell adhesion mediated by integrin | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0006782 | protoporphyrinogen IX biosynthetic process | 4 | 9 | 7.59e-01 |
| GO:BP | GO:0007417 | central nervous system development | 20 | 792 | 7.59e-01 |
| GO:BP | GO:0032484 | Ral protein signal transduction | 2 | 5 | 7.59e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 7 | 20 | 7.59e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 1 | 21 | 7.59e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 2 | 12 | 7.59e-01 |
| GO:BP | GO:0001576 | globoside biosynthetic process | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 3 | 2574 | 7.59e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 5 | 12 | 7.59e-01 |
| GO:BP | GO:0001575 | globoside metabolic process | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 5 | 16 | 7.59e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 3 | 2576 | 7.59e-01 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 1 | 9 | 7.59e-01 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 1 | 8 | 7.59e-01 |
| GO:BP | GO:0031529 | ruffle organization | 4 | 45 | 7.59e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 1 | 10 | 7.59e-01 |
| GO:BP | GO:0019284 | L-methionine salvage from S-adenosylmethionine | 1 | 3 | 7.59e-01 |
| GO:BP | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway | 2 | 4 | 7.59e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 5 | 25 | 7.59e-01 |
| GO:BP | GO:0043382 | positive regulation of memory T cell differentiation | 1 | 3 | 7.59e-01 |
| GO:BP | GO:0006182 | cGMP biosynthetic process | 1 | 6 | 7.59e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 7.59e-01 |
| GO:BP | GO:0045213 | neurotransmitter receptor metabolic process | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 7.60e-01 |
| GO:BP | GO:1904021 | negative regulation of G protein-coupled receptor internalization | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 7 | 39 | 7.60e-01 |
| GO:BP | GO:0061073 | ciliary body morphogenesis | 1 | 3 | 7.60e-01 |
| GO:BP | GO:2001148 | regulation of dipeptide transmembrane transport | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0019566 | arabinose metabolic process | 1 | 1 | 7.60e-01 |
| GO:BP | GO:2001150 | positive regulation of dipeptide transmembrane transport | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0090088 | regulation of oligopeptide transport | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0090089 | regulation of dipeptide transport | 1 | 2 | 7.60e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 49 | 90 | 7.60e-01 |
| GO:BP | GO:0007500 | mesodermal cell fate determination | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 7 | 39 | 7.60e-01 |
| GO:BP | GO:0046373 | L-arabinose metabolic process | 1 | 1 | 7.60e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 2 | 320 | 7.60e-01 |
| GO:BP | GO:2000878 | positive regulation of oligopeptide transport | 1 | 2 | 7.60e-01 |
| GO:BP | GO:2000880 | positive regulation of dipeptide transport | 1 | 2 | 7.60e-01 |
| GO:BP | GO:1905323 | telomerase holoenzyme complex assembly | 1 | 6 | 7.60e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 7 | 19 | 7.61e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 17 | 64 | 7.61e-01 |
| GO:BP | GO:0042116 | macrophage activation | 2 | 57 | 7.61e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 3 | 21 | 7.61e-01 |
| GO:BP | GO:0097017 | renal protein absorption | 1 | 5 | 7.61e-01 |
| GO:BP | GO:0046950 | cellular ketone body metabolic process | 2 | 7 | 7.61e-01 |
| GO:BP | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | 3 | 5 | 7.62e-01 |
| GO:BP | GO:0048597 | post-embryonic camera-type eye morphogenesis | 2 | 4 | 7.62e-01 |
| GO:BP | GO:0097068 | response to thyroxine | 2 | 4 | 7.62e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 4 | 10 | 7.62e-01 |
| GO:BP | GO:0060264 | regulation of respiratory burst involved in inflammatory response | 1 | 3 | 7.62e-01 |
| GO:BP | GO:0019532 | oxalate transport | 1 | 5 | 7.62e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 1 | 6 | 7.63e-01 |
| GO:BP | GO:1905772 | positive regulation of mesodermal cell differentiation | 1 | 1 | 7.63e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 3 | 2593 | 7.63e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 7 | 155 | 7.63e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 7 | 18 | 7.63e-01 |
| GO:BP | GO:0006668 | sphinganine-1-phosphate metabolic process | 1 | 2 | 7.64e-01 |
| GO:BP | GO:1903108 | regulation of mitochondrial transcription | 2 | 5 | 7.64e-01 |
| GO:BP | GO:0043586 | tongue development | 4 | 15 | 7.64e-01 |
| GO:BP | GO:0032241 | positive regulation of nucleobase-containing compound transport | 2 | 6 | 7.64e-01 |
| GO:BP | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process | 2 | 3 | 7.64e-01 |
| GO:BP | GO:0071284 | cellular response to lead ion | 1 | 4 | 7.64e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 11 | 90 | 7.64e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 7.64e-01 |
| GO:BP | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 1 | 4 | 7.64e-01 |
| GO:BP | GO:0007525 | somatic muscle development | 2 | 4 | 7.64e-01 |
| GO:BP | GO:0021986 | habenula development | 1 | 1 | 7.64e-01 |
| GO:BP | GO:0061520 | Langerhans cell differentiation | 2 | 3 | 7.64e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 5 | 114 | 7.64e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 9 | 27 | 7.64e-01 |
| GO:BP | GO:0006482 | protein demethylation | 9 | 27 | 7.64e-01 |
| GO:BP | GO:0021538 | epithalamus development | 1 | 1 | 7.64e-01 |
| GO:BP | GO:0043373 | CD4-positive, alpha-beta T cell lineage commitment | 4 | 9 | 7.64e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 3 | 54 | 7.64e-01 |
| GO:BP | GO:0021564 | vagus nerve development | 1 | 2 | 7.64e-01 |
| GO:BP | GO:0036148 | phosphatidylglycerol acyl-chain remodeling | 1 | 5 | 7.64e-01 |
| GO:BP | GO:1903292 | protein localization to Golgi membrane | 2 | 4 | 7.64e-01 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 2 | 14 | 7.64e-01 |
| GO:BP | GO:2000983 | regulation of ATP citrate synthase activity | 1 | 2 | 7.64e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 2 | 28 | 7.64e-01 |
| GO:BP | GO:0097400 | interleukin-17-mediated signaling pathway | 1 | 8 | 7.65e-01 |
| GO:BP | GO:1904179 | positive regulation of adipose tissue development | 3 | 6 | 7.65e-01 |
| GO:BP | GO:0016560 | protein import into peroxisome matrix, docking | 2 | 3 | 7.65e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 2 | 5 | 7.65e-01 |
| GO:BP | GO:0044804 | autophagy of nucleus | 1 | 18 | 7.65e-01 |
| GO:BP | GO:0050915 | sensory perception of sour taste | 1 | 4 | 7.65e-01 |
| GO:BP | GO:0071475 | cellular hyperosmotic salinity response | 2 | 6 | 7.66e-01 |
| GO:BP | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone | 3 | 5 | 7.66e-01 |
| GO:BP | GO:1903525 | regulation of membrane tubulation | 2 | 5 | 7.66e-01 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 7.66e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 8 | 39 | 7.66e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 3 | 57 | 7.66e-01 |
| GO:BP | GO:1904958 | positive regulation of midbrain dopaminergic neuron differentiation | 1 | 3 | 7.66e-01 |
| GO:BP | GO:0070142 | synaptic vesicle budding | 3 | 8 | 7.66e-01 |
| GO:BP | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | 3 | 8 | 7.66e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 3 | 15 | 7.66e-01 |
| GO:BP | GO:0019064 | fusion of virus membrane with host plasma membrane | 1 | 4 | 7.66e-01 |
| GO:BP | GO:0039663 | membrane fusion involved in viral entry into host cell | 1 | 4 | 7.66e-01 |
| GO:BP | GO:1903286 | regulation of potassium ion import | 3 | 8 | 7.66e-01 |
| GO:BP | GO:0097240 | chromosome attachment to the nuclear envelope | 1 | 3 | 7.66e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 211 | 7.67e-01 |
| GO:BP | GO:0060820 | inactivation of X chromosome by heterochromatin formation | 1 | 1 | 7.67e-01 |
| GO:BP | GO:1905665 | positive regulation of calcium ion import across plasma membrane | 1 | 6 | 7.67e-01 |
| GO:BP | GO:0043097 | pyrimidine nucleoside salvage | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 2 | 36 | 7.67e-01 |
| GO:BP | GO:1900221 | regulation of amyloid-beta clearance | 3 | 13 | 7.67e-01 |
| GO:BP | GO:1902744 | negative regulation of lamellipodium organization | 1 | 4 | 7.67e-01 |
| GO:BP | GO:0006427 | histidyl-tRNA aminoacylation | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0140141 | mitochondrial potassium ion transmembrane transport | 1 | 2 | 7.67e-01 |
| GO:BP | GO:1900113 | negative regulation of histone H3-K9 trimethylation | 1 | 3 | 7.67e-01 |
| GO:BP | GO:2000111 | positive regulation of macrophage apoptotic process | 1 | 2 | 7.67e-01 |
| GO:BP | GO:1990773 | matrix metallopeptidase secretion | 1 | 5 | 7.67e-01 |
| GO:BP | GO:1904464 | regulation of matrix metallopeptidase secretion | 1 | 5 | 7.67e-01 |
| GO:BP | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0018277 | protein deamination | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 7.67e-01 |
| GO:BP | GO:0009409 | response to cold | 7 | 41 | 7.67e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 29 | 59 | 7.68e-01 |
| GO:BP | GO:0120127 | response to zinc ion starvation | 3 | 6 | 7.68e-01 |
| GO:BP | GO:0034224 | cellular response to zinc ion starvation | 3 | 6 | 7.68e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 3 | 50 | 7.68e-01 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 2 | 17 | 7.68e-01 |
| GO:BP | GO:1905242 | response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 7.68e-01 |
| GO:BP | GO:0035502 | metanephric part of ureteric bud development | 1 | 3 | 7.68e-01 |
| GO:BP | GO:0072284 | metanephric S-shaped body morphogenesis | 1 | 4 | 7.68e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 7.68e-01 |
| GO:BP | GO:0035811 | negative regulation of urine volume | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 5 | 50 | 7.68e-01 |
| GO:BP | GO:0045039 | protein insertion into mitochondrial inner membrane | 2 | 13 | 7.68e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 142 | 7.68e-01 |
| GO:BP | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation | 2 | 3 | 7.68e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 6 | 612 | 7.68e-01 |
| GO:BP | GO:0035551 | protein initiator methionine removal involved in protein maturation | 1 | 3 | 7.68e-01 |
| GO:BP | GO:0031167 | rRNA methylation | 11 | 24 | 7.68e-01 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 3 | 5 | 7.69e-01 |
| GO:BP | GO:0016199 | axon midline choice point recognition | 1 | 3 | 7.69e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 2 | 14 | 7.69e-01 |
| GO:BP | GO:0048213 | Golgi vesicle prefusion complex stabilization | 1 | 2 | 7.69e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 7 | 76 | 7.69e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 4 | 58 | 7.69e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 7.69e-01 |
| GO:BP | GO:0090204 | protein localization to nuclear pore | 1 | 3 | 7.69e-01 |
| GO:BP | GO:0007603 | phototransduction, visible light | 2 | 9 | 7.69e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 7.69e-01 |
| GO:BP | GO:0006685 | sphingomyelin catabolic process | 1 | 6 | 7.69e-01 |
| GO:BP | GO:0014858 | positive regulation of skeletal muscle cell proliferation | 1 | 2 | 7.69e-01 |
| GO:BP | GO:1901301 | regulation of cargo loading into COPII-coated vesicle | 1 | 2 | 7.69e-01 |
| GO:BP | GO:1902996 | regulation of neurofibrillary tangle assembly | 1 | 2 | 7.69e-01 |
| GO:BP | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process | 1 | 2 | 7.69e-01 |
| GO:BP | GO:1902963 | negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process | 1 | 2 | 7.69e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 6 | 418 | 7.70e-01 |
| GO:BP | GO:0002478 | antigen processing and presentation of exogenous peptide antigen | 6 | 24 | 7.70e-01 |
| GO:BP | GO:0048806 | genitalia development | 1 | 28 | 7.70e-01 |
| GO:BP | GO:0048686 | regulation of sprouting of injured axon | 1 | 1 | 7.70e-01 |
| GO:BP | GO:0048688 | negative regulation of sprouting of injured axon | 1 | 1 | 7.70e-01 |
| GO:BP | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol | 1 | 2 | 7.70e-01 |
| GO:BP | GO:0048690 | regulation of axon extension involved in regeneration | 1 | 1 | 7.70e-01 |
| GO:BP | GO:0048692 | negative regulation of axon extension involved in regeneration | 1 | 1 | 7.70e-01 |
| GO:BP | GO:0007431 | salivary gland development | 3 | 26 | 7.71e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 3 | 7 | 7.71e-01 |
| GO:BP | GO:2000360 | negative regulation of binding of sperm to zona pellucida | 2 | 2 | 7.71e-01 |
| GO:BP | GO:1905050 | positive regulation of metallopeptidase activity | 2 | 5 | 7.71e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 2 | 93 | 7.71e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 6 | 1315 | 7.72e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 6 | 1315 | 7.72e-01 |
| GO:BP | GO:1990399 | epithelium regeneration | 1 | 2 | 7.72e-01 |
| GO:BP | GO:1905041 | regulation of epithelium regeneration | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 1 | 9 | 7.72e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 7.72e-01 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 1 | 10 | 7.72e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 3 | 160 | 7.72e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 7.72e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 3 | 11 | 7.72e-01 |
| GO:BP | GO:0045163 | clustering of voltage-gated potassium channels | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 15 | 40 | 7.72e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 1 | 14 | 7.72e-01 |
| GO:BP | GO:0033007 | negative regulation of mast cell activation involved in immune response | 3 | 4 | 7.72e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 15 | 40 | 7.72e-01 |
| GO:BP | GO:2000643 | positive regulation of early endosome to late endosome transport | 3 | 7 | 7.72e-01 |
| GO:BP | GO:0038134 | ERBB2-EGFR signaling pathway | 1 | 6 | 7.72e-01 |
| GO:BP | GO:1990770 | small intestine smooth muscle contraction | 1 | 1 | 7.73e-01 |
| GO:BP | GO:0038162 | erythropoietin-mediated signaling pathway | 1 | 2 | 7.73e-01 |
| GO:BP | GO:0009798 | axis specification | 2 | 63 | 7.73e-01 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 3 | 16 | 7.73e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 2 | 45 | 7.73e-01 |
| GO:BP | GO:0070444 | oligodendrocyte progenitor proliferation | 1 | 5 | 7.74e-01 |
| GO:BP | GO:0070445 | regulation of oligodendrocyte progenitor proliferation | 1 | 5 | 7.74e-01 |
| GO:BP | GO:1900126 | negative regulation of hyaluronan biosynthetic process | 1 | 2 | 7.74e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 8 | 36 | 7.74e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 5 | 39 | 7.74e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 2 | 57 | 7.75e-01 |
| GO:BP | GO:0010911 | regulation of isomerase activity | 2 | 5 | 7.75e-01 |
| GO:BP | GO:1905279 | regulation of retrograde transport, endosome to Golgi | 1 | 6 | 7.75e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 5 | 21 | 7.75e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 6 | 1322 | 7.75e-01 |
| GO:BP | GO:0070476 | rRNA (guanine-N7)-methylation | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0010039 | response to iron ion | 2 | 24 | 7.75e-01 |
| GO:BP | GO:0010912 | positive regulation of isomerase activity | 2 | 5 | 7.75e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 7.75e-01 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 3 | 14 | 7.75e-01 |
| GO:BP | GO:2001027 | negative regulation of endothelial cell chemotaxis | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 5 | 11 | 7.75e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 1 | 65 | 7.75e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 6 | 28 | 7.75e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 7.76e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 6 | 14 | 7.76e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 6 | 14 | 7.76e-01 |
| GO:BP | GO:0008300 | isoprenoid catabolic process | 2 | 5 | 7.76e-01 |
| GO:BP | GO:0030850 | prostate gland development | 1 | 35 | 7.76e-01 |
| GO:BP | GO:0061197 | fungiform papilla morphogenesis | 1 | 4 | 7.76e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 2 | 108 | 7.76e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 15 | 45 | 7.77e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 12 | 51 | 7.77e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 7.77e-01 |
| GO:BP | GO:0043324 | pigment metabolic process involved in developmental pigmentation | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0042441 | eye pigment metabolic process | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0010033 | response to organic substance | 2 | 1957 | 7.77e-01 |
| GO:BP | GO:0006726 | eye pigment biosynthetic process | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 1 | 310 | 7.77e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 3 | 65 | 7.77e-01 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 5 | 20 | 7.77e-01 |
| GO:BP | GO:0150094 | amyloid-beta clearance by cellular catabolic process | 1 | 7 | 7.77e-01 |
| GO:BP | GO:2000532 | regulation of renal albumin absorption | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0090296 | regulation of mitochondrial DNA replication | 1 | 4 | 7.77e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 7.77e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 7.77e-01 |
| GO:BP | GO:0046104 | thymidine metabolic process | 1 | 5 | 7.77e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 7.77e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 5 | 23 | 7.77e-01 |
| GO:BP | GO:0045916 | negative regulation of complement activation | 3 | 8 | 7.77e-01 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 1 | 5 | 7.77e-01 |
| GO:BP | GO:0048632 | negative regulation of skeletal muscle tissue growth | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0048625 | myoblast fate commitment | 2 | 4 | 7.77e-01 |
| GO:BP | GO:0001568 | blood vessel development | 13 | 534 | 7.77e-01 |
| GO:BP | GO:0015829 | valine transport | 2 | 4 | 7.77e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 2 | 19 | 7.77e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 17 | 64 | 7.77e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 1 | 8 | 7.77e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 1 | 8 | 7.77e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 7.77e-01 |
| GO:BP | GO:1904273 | L-alanine import across plasma membrane | 2 | 4 | 7.77e-01 |
| GO:BP | GO:1904430 | negative regulation of t-circle formation | 1 | 2 | 7.78e-01 |
| GO:BP | GO:0034462 | small-subunit processome assembly | 1 | 3 | 7.78e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 3 | 2683 | 7.78e-01 |
| GO:BP | GO:0035821 | modulation of process of another organism | 2 | 5 | 7.78e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 3 | 2684 | 7.78e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 3 | 8 | 7.78e-01 |
| GO:BP | GO:0090249 | regulation of cell migration involved in somitogenic axis elongation | 1 | 2 | 7.78e-01 |
| GO:BP | GO:0090248 | cell migration involved in somitogenic axis elongation | 1 | 2 | 7.78e-01 |
| GO:BP | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway | 4 | 9 | 7.78e-01 |
| GO:BP | GO:0002940 | tRNA N2-guanine methylation | 1 | 4 | 7.78e-01 |
| GO:BP | GO:0031509 | subtelomeric heterochromatin formation | 3 | 7 | 7.78e-01 |
| GO:BP | GO:0002934 | desmosome organization | 3 | 9 | 7.79e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 7.79e-01 |
| GO:BP | GO:0071624 | positive regulation of granulocyte chemotaxis | 2 | 14 | 7.79e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 1 | 75 | 7.79e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 3 | 10 | 7.79e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 4 | 45 | 7.79e-01 |
| GO:BP | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | 1 | 4 | 7.79e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 2 | 9 | 7.79e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 7.80e-01 |
| GO:BP | GO:0009136 | purine nucleoside diphosphate biosynthetic process | 3 | 5 | 7.80e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 59 | 404 | 7.80e-01 |
| GO:BP | GO:0110010 | basolateral protein secretion | 2 | 3 | 7.80e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 5 | 344 | 7.80e-01 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 1 | 57 | 7.80e-01 |
| GO:BP | GO:0032782 | bile acid secretion | 1 | 5 | 7.80e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 3 | 63 | 7.80e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 3 | 162 | 7.80e-01 |
| GO:BP | GO:0070995 | NADPH oxidation | 2 | 3 | 7.80e-01 |
| GO:BP | GO:2000833 | positive regulation of steroid hormone secretion | 1 | 6 | 7.81e-01 |
| GO:BP | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.81e-01 |
| GO:BP | GO:1904237 | positive regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.81e-01 |
| GO:BP | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation | 1 | 2 | 7.81e-01 |
| GO:BP | GO:2000668 | regulation of dendritic cell apoptotic process | 2 | 7 | 7.81e-01 |
| GO:BP | GO:0042760 | very long-chain fatty acid catabolic process | 1 | 5 | 7.81e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 4 | 55 | 7.81e-01 |
| GO:BP | GO:0072678 | T cell migration | 4 | 38 | 7.81e-01 |
| GO:BP | GO:0098743 | cell aggregation | 2 | 12 | 7.81e-01 |
| GO:BP | GO:0010989 | negative regulation of low-density lipoprotein particle clearance | 2 | 4 | 7.81e-01 |
| GO:BP | GO:0021675 | nerve development | 10 | 57 | 7.81e-01 |
| GO:BP | GO:0007276 | gamete generation | 4 | 461 | 7.81e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 2 | 34 | 7.81e-01 |
| GO:BP | GO:0097048 | dendritic cell apoptotic process | 2 | 7 | 7.81e-01 |
| GO:BP | GO:0031392 | regulation of prostaglandin biosynthetic process | 3 | 6 | 7.81e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 2 | 64 | 7.81e-01 |
| GO:BP | GO:0034163 | regulation of toll-like receptor 9 signaling pathway | 5 | 6 | 7.81e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 5 | 14 | 7.81e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 5 | 345 | 7.81e-01 |
| GO:BP | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 5 | 7.81e-01 |
| GO:BP | GO:1903226 | positive regulation of endodermal cell differentiation | 1 | 2 | 7.81e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4455 | 7.81e-01 |
| GO:BP | GO:0019682 | glyceraldehyde-3-phosphate metabolic process | 2 | 5 | 7.81e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 1 | 46 | 7.81e-01 |
| GO:BP | GO:1901825 | zeaxanthin metabolic process | 1 | 1 | 7.81e-01 |
| GO:BP | GO:0016122 | xanthophyll metabolic process | 1 | 1 | 7.81e-01 |
| GO:BP | GO:0046247 | terpene catabolic process | 1 | 1 | 7.81e-01 |
| GO:BP | GO:0140447 | cytokine precursor processing | 1 | 1 | 7.81e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 23 | 84 | 7.81e-01 |
| GO:BP | GO:0016119 | carotene metabolic process | 1 | 1 | 7.81e-01 |
| GO:BP | GO:0016121 | carotene catabolic process | 1 | 1 | 7.81e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 18 | 68 | 7.81e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 3 | 25 | 7.81e-01 |
| GO:BP | GO:0140568 | extraction of mislocalized protein from membrane | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 19 | 7.82e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 3 | 83 | 7.82e-01 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 8 | 36 | 7.82e-01 |
| GO:BP | GO:0001807 | regulation of type IV hypersensitivity | 1 | 1 | 7.82e-01 |
| GO:BP | GO:2000367 | regulation of acrosomal vesicle exocytosis | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 1 | 5 | 7.82e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 5 | 97 | 7.82e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 2 | 19 | 7.82e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 4 | 110 | 7.82e-01 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 1 | 5 | 7.82e-01 |
| GO:BP | GO:0038128 | ERBB2 signaling pathway | 5 | 14 | 7.82e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 3 | 7 | 7.83e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 4 | 30 | 7.83e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 2 | 131 | 7.83e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 59 | 300 | 7.83e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 13 | 39 | 7.83e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 2 | 12 | 7.83e-01 |
| GO:BP | GO:0002322 | B cell proliferation involved in immune response | 1 | 3 | 7.83e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 3 | 2714 | 7.83e-01 |
| GO:BP | GO:0097343 | ripoptosome assembly | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0045981 | positive regulation of nucleotide metabolic process | 2 | 16 | 7.83e-01 |
| GO:BP | GO:1901026 | ripoptosome assembly involved in necroptotic process | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0002344 | B cell affinity maturation | 1 | 2 | 7.83e-01 |
| GO:BP | GO:1900544 | positive regulation of purine nucleotide metabolic process | 2 | 16 | 7.83e-01 |
| GO:BP | GO:0002332 | transitional stage B cell differentiation | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0007621 | negative regulation of female receptivity | 1 | 1 | 7.83e-01 |
| GO:BP | GO:0002343 | peripheral B cell selection | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 7.83e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 7 | 403 | 7.83e-01 |
| GO:BP | GO:0007254 | JNK cascade | 3 | 138 | 7.83e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 7.83e-01 |
| GO:BP | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering | 4 | 9 | 7.83e-01 |
| GO:BP | GO:0051167 | xylulose 5-phosphate metabolic process | 2 | 4 | 7.84e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 2 | 165 | 7.84e-01 |
| GO:BP | GO:0019541 | propionate metabolic process | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | 2 | 4 | 7.84e-01 |
| GO:BP | GO:0016191 | synaptic vesicle uncoating | 3 | 7 | 7.84e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 5 | 50 | 7.84e-01 |
| GO:BP | GO:0015867 | ATP transport | 4 | 9 | 7.84e-01 |
| GO:BP | GO:0008347 | glial cell migration | 2 | 47 | 7.84e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 9 | 275 | 7.84e-01 |
| GO:BP | GO:1901159 | xylulose 5-phosphate biosynthetic process | 2 | 4 | 7.84e-01 |
| GO:BP | GO:0006064 | glucuronate catabolic process | 2 | 4 | 7.84e-01 |
| GO:BP | GO:0045793 | positive regulation of cell size | 1 | 11 | 7.84e-01 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 1 | 10 | 7.84e-01 |
| GO:BP | GO:0046726 | positive regulation by virus of viral protein levels in host cell | 2 | 4 | 7.84e-01 |
| GO:BP | GO:0046356 | acetyl-CoA catabolic process | 1 | 3 | 7.84e-01 |
| GO:BP | GO:0060426 | lung vasculature development | 1 | 3 | 7.84e-01 |
| GO:BP | GO:0021551 | central nervous system morphogenesis | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0048569 | post-embryonic animal organ development | 7 | 15 | 7.84e-01 |
| GO:BP | GO:0048086 | negative regulation of developmental pigmentation | 1 | 1 | 7.84e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 1 | 39 | 7.84e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 1 | 39 | 7.84e-01 |
| GO:BP | GO:0000294 | nuclear-transcribed mRNA catabolic process, RNase MRP-dependent | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 7.84e-01 |
| GO:BP | GO:0015878 | biotin transport | 1 | 3 | 7.85e-01 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 2 | 11 | 7.85e-01 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 2 | 11 | 7.85e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 2 | 77 | 7.85e-01 |
| GO:BP | GO:1990390 | protein K33-linked ubiquitination | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0002070 | epithelial cell maturation | 3 | 16 | 7.85e-01 |
| GO:BP | GO:1901550 | regulation of endothelial cell development | 1 | 12 | 7.85e-01 |
| GO:BP | GO:0071352 | cellular response to interleukin-2 | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0038110 | interleukin-2-mediated signaling pathway | 1 | 2 | 7.85e-01 |
| GO:BP | GO:1903140 | regulation of establishment of endothelial barrier | 1 | 12 | 7.85e-01 |
| GO:BP | GO:0021502 | neural fold elevation formation | 1 | 3 | 7.85e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 45 | 105 | 7.85e-01 |
| GO:BP | GO:0006363 | termination of RNA polymerase I transcription | 1 | 4 | 7.85e-01 |
| GO:BP | GO:0051641 | cellular localization | 4 | 2810 | 7.85e-01 |
| GO:BP | GO:0014060 | regulation of epinephrine secretion | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0048242 | epinephrine secretion | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 3 | 7 | 7.85e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 3 | 25 | 7.85e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 1 | 72 | 7.85e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 1 | 12 | 7.85e-01 |
| GO:BP | GO:0032773 | positive regulation of tyrosinase activity | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0048817 | negative regulation of hair follicle maturation | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0032771 | regulation of tyrosinase activity | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 16 | 43 | 7.85e-01 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 3 | 39 | 7.85e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 5 | 1014 | 7.85e-01 |
| GO:BP | GO:0055107 | Golgi to secretory granule transport | 1 | 1 | 7.85e-01 |
| GO:BP | GO:0002032 | desensitization of G protein-coupled receptor signaling pathway by arrestin | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0061884 | regulation of mini excitatory postsynaptic potential | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 2 | 14 | 7.86e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 7 | 22 | 7.86e-01 |
| GO:BP | GO:1902858 | propionyl-CoA metabolic process | 2 | 4 | 7.86e-01 |
| GO:BP | GO:0035600 | tRNA methylthiolation | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 2 | 14 | 7.86e-01 |
| GO:BP | GO:0043654 | recognition of apoptotic cell | 1 | 4 | 7.86e-01 |
| GO:BP | GO:0061885 | positive regulation of mini excitatory postsynaptic potential | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 4 | 27 | 7.86e-01 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 1 | 22 | 7.86e-01 |
| GO:BP | GO:1900194 | negative regulation of oocyte maturation | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 1 | 35 | 7.86e-01 |
| GO:BP | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development | 1 | 2 | 7.86e-01 |
| GO:BP | GO:1990145 | maintenance of translational fidelity | 1 | 2 | 7.86e-01 |
| GO:BP | GO:2001294 | malonyl-CoA catabolic process | 2 | 4 | 7.86e-01 |
| GO:BP | GO:0045726 | positive regulation of integrin biosynthetic process | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0098816 | mini excitatory postsynaptic potential | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 16 | 114 | 7.86e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 7 | 1101 | 7.86e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 1 | 199 | 7.86e-01 |
| GO:BP | GO:1904772 | response to tetrachloromethane | 2 | 4 | 7.86e-01 |
| GO:BP | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation | 1 | 5 | 7.86e-01 |
| GO:BP | GO:0035523 | protein K29-linked deubiquitination | 1 | 4 | 7.86e-01 |
| GO:BP | GO:0034143 | regulation of toll-like receptor 4 signaling pathway | 3 | 15 | 7.86e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 3 | 22 | 7.86e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 27 | 687 | 7.86e-01 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 1 | 13 | 7.86e-01 |
| GO:BP | GO:0070602 | regulation of centromeric sister chromatid cohesion | 2 | 4 | 7.86e-01 |
| GO:BP | GO:0006554 | lysine catabolic process | 1 | 6 | 7.86e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 7.86e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 1 | 55 | 7.86e-01 |
| GO:BP | GO:0050904 | diapedesis | 2 | 6 | 7.86e-01 |
| GO:BP | GO:0038165 | oncostatin-M-mediated signaling pathway | 1 | 4 | 7.86e-01 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 1 | 7 | 7.86e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 3 | 22 | 7.86e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 10 | 29 | 7.86e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 48 | 125 | 7.86e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 2 | 89 | 7.86e-01 |
| GO:BP | GO:1905429 | response to glycine | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 7 | 77 | 7.86e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 1 | 11 | 7.86e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 2 | 486 | 7.86e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4570 | 7.86e-01 |
| GO:BP | GO:0060486 | club cell differentiation | 2 | 3 | 7.86e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 3 | 44 | 7.86e-01 |
| GO:BP | GO:0071934 | thiamine transmembrane transport | 3 | 3 | 7.87e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 2 | 43 | 7.87e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 4 | 9 | 7.87e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 2 | 43 | 7.87e-01 |
| GO:BP | GO:1902871 | positive regulation of amacrine cell differentiation | 1 | 1 | 7.87e-01 |
| GO:BP | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching | 2 | 4 | 7.87e-01 |
| GO:BP | GO:0071988 | protein localization to spindle pole body | 1 | 2 | 7.87e-01 |
| GO:BP | GO:0002447 | eosinophil mediated immunity | 3 | 5 | 7.87e-01 |
| GO:BP | GO:0045625 | regulation of T-helper 1 cell differentiation | 2 | 5 | 7.87e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 5 | 26 | 7.87e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 5 | 23 | 7.87e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 2 | 82 | 7.87e-01 |
| GO:BP | GO:0097402 | neuroblast migration | 1 | 2 | 7.87e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 7.87e-01 |
| GO:BP | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 2 | 7.87e-01 |
| GO:BP | GO:0006002 | fructose 6-phosphate metabolic process | 2 | 8 | 7.87e-01 |
| GO:BP | GO:0045629 | negative regulation of T-helper 2 cell differentiation | 1 | 3 | 7.87e-01 |
| GO:BP | GO:0014905 | myoblast fusion involved in skeletal muscle regeneration | 2 | 3 | 7.87e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 4 | 17 | 7.87e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 7.87e-01 |
| GO:BP | GO:2001111 | positive regulation of lens epithelial cell proliferation | 1 | 1 | 7.87e-01 |
| GO:BP | GO:0097166 | lens epithelial cell proliferation | 1 | 1 | 7.87e-01 |
| GO:BP | GO:2001109 | regulation of lens epithelial cell proliferation | 1 | 1 | 7.87e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 3 | 39 | 7.87e-01 |
| GO:BP | GO:0007528 | neuromuscular junction development | 6 | 39 | 7.87e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 2 | 13 | 7.88e-01 |
| GO:BP | GO:0006393 | termination of mitochondrial transcription | 1 | 2 | 7.88e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 2 | 25 | 7.88e-01 |
| GO:BP | GO:0043585 | nose morphogenesis | 1 | 3 | 7.88e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 2 | 17 | 7.88e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 7.88e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 7.88e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 3 | 676 | 7.88e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 11 | 115 | 7.88e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 3 | 65 | 7.88e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 7.88e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 3 | 8 | 7.88e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 7.88e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4596 | 7.88e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 6 | 20 | 7.88e-01 |
| GO:BP | GO:0021750 | vestibular nucleus development | 1 | 1 | 7.88e-01 |
| GO:BP | GO:0045087 | innate immune response | 4 | 488 | 7.89e-01 |
| GO:BP | GO:0050428 | 3’-phosphoadenosine 5’-phosphosulfate biosynthetic process | 1 | 2 | 7.89e-01 |
| GO:BP | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process | 1 | 2 | 7.89e-01 |
| GO:BP | GO:0000103 | sulfate assimilation | 1 | 2 | 7.89e-01 |
| GO:BP | GO:0002092 | positive regulation of receptor internalization | 3 | 21 | 7.89e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 2 | 13 | 7.89e-01 |
| GO:BP | GO:0031442 | positive regulation of mRNA 3’-end processing | 4 | 10 | 7.90e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 6 | 28 | 7.90e-01 |
| GO:BP | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 1 | 16 | 7.90e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 6 | 13 | 7.90e-01 |
| GO:BP | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion | 1 | 5 | 7.90e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 7.90e-01 |
| GO:BP | GO:0019835 | cytolysis | 1 | 6 | 7.90e-01 |
| GO:BP | GO:0032536 | regulation of cell projection size | 3 | 8 | 7.90e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 7.90e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 8 | 70 | 7.90e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 2 | 7 | 7.90e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 7.90e-01 |
| GO:BP | GO:0002536 | respiratory burst involved in inflammatory response | 1 | 4 | 7.90e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 52 | 7.90e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 3 | 12 | 7.90e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 7 | 21 | 7.90e-01 |
| GO:BP | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 2 | 5 | 7.90e-01 |
| GO:BP | GO:0060263 | regulation of respiratory burst | 2 | 10 | 7.90e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 3 | 7 | 7.91e-01 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 1 | 6 | 7.91e-01 |
| GO:BP | GO:0046054 | dGMP metabolic process | 2 | 2 | 7.91e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 7.91e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 7.91e-01 |
| GO:BP | GO:0006952 | defense response | 4 | 944 | 7.91e-01 |
| GO:BP | GO:0048599 | oocyte development | 10 | 38 | 7.91e-01 |
| GO:BP | GO:1903845 | negative regulation of cellular response to transforming growth factor beta stimulus | 1 | 2 | 7.91e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 4 | 7 | 7.91e-01 |
| GO:BP | GO:0006891 | intra-Golgi vesicle-mediated transport | 2 | 33 | 7.92e-01 |
| GO:BP | GO:0034553 | mitochondrial respiratory chain complex II assembly | 2 | 4 | 7.92e-01 |
| GO:BP | GO:0034552 | respiratory chain complex II assembly | 2 | 4 | 7.92e-01 |
| GO:BP | GO:1903333 | negative regulation of protein folding | 2 | 4 | 7.92e-01 |
| GO:BP | GO:0003008 | system process | 5 | 1189 | 7.92e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 1 | 55 | 7.93e-01 |
| GO:BP | GO:0002475 | antigen processing and presentation via MHC class Ib | 2 | 8 | 7.93e-01 |
| GO:BP | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus | 2 | 8 | 7.93e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 7 | 859 | 7.93e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 7.93e-01 |
| GO:BP | GO:2000642 | negative regulation of early endosome to late endosome transport | 1 | 2 | 7.93e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 2 | 38 | 7.93e-01 |
| GO:BP | GO:0150032 | positive regulation of protein localization to lysosome | 1 | 2 | 7.93e-01 |
| GO:BP | GO:0099105 | ion channel modulating, G protein-coupled receptor signaling pathway | 1 | 7 | 7.94e-01 |
| GO:BP | GO:0006572 | tyrosine catabolic process | 1 | 2 | 7.94e-01 |
| GO:BP | GO:0014891 | striated muscle atrophy | 2 | 6 | 7.94e-01 |
| GO:BP | GO:0018002 | N-terminal peptidyl-glutamic acid acetylation | 1 | 2 | 7.94e-01 |
| GO:BP | GO:0017190 | N-terminal peptidyl-aspartic acid acetylation | 1 | 2 | 7.94e-01 |
| GO:BP | GO:0002678 | positive regulation of chronic inflammatory response | 1 | 1 | 7.95e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 2 | 64 | 7.95e-01 |
| GO:BP | GO:0021932 | hindbrain radial glia guided cell migration | 4 | 8 | 7.95e-01 |
| GO:BP | GO:0044788 | modulation by host of viral process | 5 | 40 | 7.95e-01 |
| GO:BP | GO:0021527 | spinal cord association neuron differentiation | 2 | 3 | 7.95e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 8 | 211 | 7.95e-01 |
| GO:BP | GO:0010813 | neuropeptide catabolic process | 2 | 4 | 7.95e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 4 | 99 | 7.95e-01 |
| GO:BP | GO:0097164 | ammonium ion metabolic process | 1 | 14 | 7.95e-01 |
| GO:BP | GO:0032928 | regulation of superoxide anion generation | 4 | 11 | 7.95e-01 |
| GO:BP | GO:0015876 | acetyl-CoA transport | 1 | 1 | 7.95e-01 |
| GO:BP | GO:0060484 | lung-associated mesenchyme development | 1 | 5 | 7.96e-01 |
| GO:BP | GO:1905524 | negative regulation of protein autoubiquitination | 1 | 2 | 7.96e-01 |
| GO:BP | GO:0006983 | ER overload response | 4 | 12 | 7.96e-01 |
| GO:BP | GO:0035021 | negative regulation of Rac protein signal transduction | 1 | 4 | 7.96e-01 |
| GO:BP | GO:1901654 | response to ketone | 4 | 147 | 7.96e-01 |
| GO:BP | GO:1904694 | negative regulation of vascular associated smooth muscle contraction | 1 | 3 | 7.97e-01 |
| GO:BP | GO:0035854 | eosinophil fate commitment | 1 | 1 | 7.97e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 7.97e-01 |
| GO:BP | GO:0010725 | regulation of primitive erythrocyte differentiation | 1 | 1 | 7.97e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 3 | 21 | 7.97e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 3 | 23 | 7.97e-01 |
| GO:BP | GO:0007228 | positive regulation of hh target transcription factor activity | 1 | 2 | 7.97e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 2 | 10 | 7.97e-01 |
| GO:BP | GO:0007227 | signal transduction downstream of smoothened | 1 | 2 | 7.97e-01 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 1 | 7 | 7.97e-01 |
| GO:BP | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity | 1 | 2 | 7.97e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 7.98e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 1 | 74 | 7.98e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 10 | 57 | 7.98e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 9 | 26 | 7.98e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 1 | 80 | 7.98e-01 |
| GO:BP | GO:0044241 | lipid digestion | 1 | 10 | 7.98e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 5 | 9 | 7.98e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 5 | 22 | 7.98e-01 |
| GO:BP | GO:1902224 | ketone body metabolic process | 2 | 7 | 7.98e-01 |
| GO:BP | GO:0010716 | negative regulation of extracellular matrix disassembly | 1 | 2 | 7.98e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 56 | 204 | 7.98e-01 |
| GO:BP | GO:0048675 | axon extension | 36 | 110 | 7.98e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 2 | 22 | 7.99e-01 |
| GO:BP | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification | 1 | 9 | 7.99e-01 |
| GO:BP | GO:0030853 | negative regulation of granulocyte differentiation | 3 | 6 | 7.99e-01 |
| GO:BP | GO:0050896 | response to stimulus | 10 | 5770 | 8.00e-01 |
| GO:BP | GO:2000474 | regulation of opioid receptor signaling pathway | 1 | 2 | 8.00e-01 |
| GO:BP | GO:0042063 | gliogenesis | 1 | 235 | 8.00e-01 |
| GO:BP | GO:0046959 | habituation | 1 | 5 | 8.00e-01 |
| GO:BP | GO:0048731 | system development | 4 | 2901 | 8.00e-01 |
| GO:BP | GO:0035799 | ureter maturation | 1 | 1 | 8.00e-01 |
| GO:BP | GO:2000417 | negative regulation of eosinophil migration | 1 | 1 | 8.00e-01 |
| GO:BP | GO:1902437 | positive regulation of male mating behavior | 1 | 2 | 8.00e-01 |
| GO:BP | GO:1902435 | regulation of male mating behavior | 1 | 2 | 8.00e-01 |
| GO:BP | GO:0045907 | positive regulation of vasoconstriction | 1 | 15 | 8.01e-01 |
| GO:BP | GO:0048636 | positive regulation of muscle organ development | 1 | 3 | 8.01e-01 |
| GO:BP | GO:0045844 | positive regulation of striated muscle tissue development | 1 | 3 | 8.01e-01 |
| GO:BP | GO:0002268 | follicular dendritic cell differentiation | 1 | 2 | 8.01e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 8.01e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 67 | 149 | 8.01e-01 |
| GO:BP | GO:0035282 | segmentation | 21 | 75 | 8.01e-01 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 3 | 7 | 8.01e-01 |
| GO:BP | GO:1900028 | negative regulation of ruffle assembly | 1 | 4 | 8.01e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 10 | 70 | 8.01e-01 |
| GO:BP | GO:0110091 | negative regulation of hippocampal neuron apoptotic process | 1 | 2 | 8.01e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4747 | 8.02e-01 |
| GO:BP | GO:0002790 | peptide secretion | 28 | 170 | 8.02e-01 |
| GO:BP | GO:2000523 | regulation of T cell costimulation | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 15 | 8.02e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 8.02e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 3 | 2845 | 8.02e-01 |
| GO:BP | GO:0002544 | chronic inflammatory response | 1 | 6 | 8.02e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 3 | 7 | 8.02e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 3 | 2285 | 8.02e-01 |
| GO:BP | GO:1903974 | positive regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 1 | 8.02e-01 |
| GO:BP | GO:0032707 | negative regulation of interleukin-23 production | 1 | 2 | 8.02e-01 |
| GO:BP | GO:1903971 | positive regulation of response to macrophage colony-stimulating factor | 1 | 1 | 8.02e-01 |
| GO:BP | GO:0018342 | protein prenylation | 2 | 10 | 8.02e-01 |
| GO:BP | GO:0097354 | prenylation | 2 | 10 | 8.02e-01 |
| GO:BP | GO:0021648 | vestibulocochlear nerve morphogenesis | 2 | 3 | 8.03e-01 |
| GO:BP | GO:1903334 | positive regulation of protein folding | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0106016 | positive regulation of inflammatory response to wounding | 1 | 2 | 8.03e-01 |
| GO:BP | GO:2000543 | positive regulation of gastrulation | 1 | 5 | 8.03e-01 |
| GO:BP | GO:0060478 | acrosomal vesicle exocytosis | 1 | 5 | 8.03e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 17 | 37 | 8.03e-01 |
| GO:BP | GO:0071305 | cellular response to vitamin D | 1 | 14 | 8.03e-01 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 1 | 9 | 8.03e-01 |
| GO:BP | GO:0071379 | cellular response to prostaglandin stimulus | 1 | 16 | 8.03e-01 |
| GO:BP | GO:1904809 | regulation of dense core granule transport | 1 | 1 | 8.03e-01 |
| GO:BP | GO:1904811 | positive regulation of dense core granule transport | 1 | 1 | 8.03e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 8.03e-01 |
| GO:BP | GO:1901951 | regulation of anterograde dense core granule transport | 1 | 1 | 8.03e-01 |
| GO:BP | GO:1901953 | positive regulation of anterograde dense core granule transport | 1 | 1 | 8.03e-01 |
| GO:BP | GO:0044375 | regulation of peroxisome size | 1 | 3 | 8.03e-01 |
| GO:BP | GO:1905386 | positive regulation of protein localization to presynapse | 1 | 2 | 8.03e-01 |
| GO:BP | GO:1905384 | regulation of protein localization to presynapse | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination | 1 | 4 | 8.03e-01 |
| GO:BP | GO:0034983 | peptidyl-lysine deacetylation | 2 | 8 | 8.03e-01 |
| GO:BP | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline | 3 | 9 | 8.03e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 2 | 66 | 8.03e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 3 | 87 | 8.03e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 3 | 53 | 8.03e-01 |
| GO:BP | GO:0006701 | progesterone biosynthetic process | 1 | 5 | 8.03e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 1 | 92 | 8.03e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 1 | 92 | 8.03e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 1 | 92 | 8.03e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 8.04e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 5 | 36 | 8.04e-01 |
| GO:BP | GO:0001944 | vasculature development | 6 | 559 | 8.04e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 4 | 101 | 8.04e-01 |
| GO:BP | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 1 | 86 | 8.04e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 1 | 86 | 8.04e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 7 | 23 | 8.04e-01 |
| GO:BP | GO:0071395 | cellular response to jasmonic acid stimulus | 2 | 3 | 8.04e-01 |
| GO:BP | GO:0003157 | endocardium development | 3 | 9 | 8.04e-01 |
| GO:BP | GO:1903803 | L-glutamine import across plasma membrane | 1 | 4 | 8.04e-01 |
| GO:BP | GO:0009753 | response to jasmonic acid | 2 | 3 | 8.04e-01 |
| GO:BP | GO:0006642 | triglyceride mobilization | 2 | 3 | 8.04e-01 |
| GO:BP | GO:0009258 | 10-formyltetrahydrofolate catabolic process | 1 | 3 | 8.04e-01 |
| GO:BP | GO:0045117 | azole transmembrane transport | 6 | 9 | 8.04e-01 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 5 | 19 | 8.05e-01 |
| GO:BP | GO:2000825 | positive regulation of androgen receptor activity | 2 | 5 | 8.05e-01 |
| GO:BP | GO:0019401 | alditol biosynthetic process | 2 | 3 | 8.05e-01 |
| GO:BP | GO:0006114 | glycerol biosynthetic process | 2 | 3 | 8.05e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 5 | 155 | 8.06e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 4 | 19 | 8.06e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 2 | 7 | 8.06e-01 |
| GO:BP | GO:0034720 | histone H3-K4 demethylation | 1 | 5 | 8.06e-01 |
| GO:BP | GO:1903512 | phytanic acid metabolic process | 1 | 2 | 8.06e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 8.06e-01 |
| GO:BP | GO:0060319 | primitive erythrocyte differentiation | 2 | 4 | 8.06e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 4 | 12 | 8.06e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 8.06e-01 |
| GO:BP | GO:0097155 | fasciculation of sensory neuron axon | 1 | 3 | 8.06e-01 |
| GO:BP | GO:0032836 | glomerular basement membrane development | 1 | 9 | 8.06e-01 |
| GO:BP | GO:0001698 | gastrin-induced gastric acid secretion | 1 | 1 | 8.06e-01 |
| GO:BP | GO:0043415 | positive regulation of skeletal muscle tissue regeneration | 2 | 4 | 8.06e-01 |
| GO:BP | GO:0036015 | response to interleukin-3 | 1 | 6 | 8.06e-01 |
| GO:BP | GO:0036016 | cellular response to interleukin-3 | 1 | 6 | 8.06e-01 |
| GO:BP | GO:1901998 | toxin transport | 3 | 37 | 8.06e-01 |
| GO:BP | GO:0035022 | positive regulation of Rac protein signal transduction | 2 | 5 | 8.06e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 9 | 53 | 8.06e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 6 | 17 | 8.06e-01 |
| GO:BP | GO:0061010 | gallbladder development | 1 | 1 | 8.06e-01 |
| GO:BP | GO:1990834 | response to odorant | 1 | 3 | 8.06e-01 |
| GO:BP | GO:0060457 | negative regulation of digestive system process | 1 | 7 | 8.06e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 8.06e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 8.07e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 7 | 33 | 8.07e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 8 | 10 | 8.07e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 318 | 8.07e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 8 | 122 | 8.07e-01 |
| GO:BP | GO:0002384 | hepatic immune response | 1 | 1 | 8.07e-01 |
| GO:BP | GO:1905382 | positive regulation of snRNA transcription by RNA polymerase II | 1 | 3 | 8.08e-01 |
| GO:BP | GO:0061723 | glycophagy | 2 | 4 | 8.08e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 4 | 84 | 8.08e-01 |
| GO:BP | GO:0048295 | positive regulation of isotype switching to IgE isotypes | 2 | 3 | 8.09e-01 |
| GO:BP | GO:0009635 | response to herbicide | 1 | 3 | 8.09e-01 |
| GO:BP | GO:1990120 | messenger ribonucleoprotein complex assembly | 1 | 3 | 8.09e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 22 | 569 | 8.09e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 2 | 6 | 8.09e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 8.09e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 8.09e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4946 | 8.09e-01 |
| GO:BP | GO:0032264 | IMP salvage | 2 | 4 | 8.09e-01 |
| GO:BP | GO:1903393 | positive regulation of adherens junction organization | 1 | 2 | 8.09e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 11 | 21 | 8.09e-01 |
| GO:BP | GO:0006415 | translational termination | 2 | 17 | 8.09e-01 |
| GO:BP | GO:0007399 | nervous system development | 2 | 1904 | 8.09e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 1 | 13 | 8.09e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 13 | 60 | 8.09e-01 |
| GO:BP | GO:0032242 | regulation of nucleoside transport | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 7 | 10 | 8.09e-01 |
| GO:BP | GO:0060592 | mammary gland formation | 3 | 6 | 8.09e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 3 | 23 | 8.09e-01 |
| GO:BP | GO:0015746 | citrate transport | 1 | 3 | 8.09e-01 |
| GO:BP | GO:0070562 | regulation of vitamin D receptor signaling pathway | 1 | 9 | 8.09e-01 |
| GO:BP | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition | 2 | 3 | 8.09e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 1 | 126 | 8.09e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 7 | 10 | 8.09e-01 |
| GO:BP | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0006842 | tricarboxylic acid transport | 1 | 3 | 8.09e-01 |
| GO:BP | GO:0071672 | negative regulation of smooth muscle cell chemotaxis | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 8.09e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 3 | 101 | 8.09e-01 |
| GO:BP | GO:1902565 | positive regulation of neutrophil activation | 1 | 1 | 8.09e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 26 | 97 | 8.10e-01 |
| GO:BP | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 8.10e-01 |
| GO:BP | GO:1905048 | regulation of metallopeptidase activity | 5 | 14 | 8.10e-01 |
| GO:BP | GO:0060585 | positive regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0014826 | vein smooth muscle contraction | 1 | 3 | 8.10e-01 |
| GO:BP | GO:0045471 | response to ethanol | 1 | 73 | 8.10e-01 |
| GO:BP | GO:1903615 | positive regulation of protein tyrosine phosphatase activity | 1 | 1 | 8.10e-01 |
| GO:BP | GO:0048073 | regulation of eye pigmentation | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0043011 | myeloid dendritic cell differentiation | 1 | 10 | 8.10e-01 |
| GO:BP | GO:0019858 | cytosine metabolic process | 1 | 1 | 8.10e-01 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 4 | 11 | 8.10e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 3 | 25 | 8.10e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 20 | 8.10e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 8.11e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 2 | 30 | 8.11e-01 |
| GO:BP | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production | 4 | 5 | 8.11e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 12 | 41 | 8.11e-01 |
| GO:BP | GO:0072176 | nephric duct development | 2 | 12 | 8.11e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 3 | 16 | 8.11e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 8.11e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 8.11e-01 |
| GO:BP | GO:0031581 | hemidesmosome assembly | 2 | 6 | 8.11e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 17 | 31 | 8.11e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 3 | 8 | 8.11e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 3 | 9 | 8.11e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 3 | 100 | 8.11e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 1 | 100 | 8.11e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 1 | 165 | 8.11e-01 |
| GO:BP | GO:0071926 | endocannabinoid signaling pathway | 2 | 5 | 8.12e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 2 | 656 | 8.12e-01 |
| GO:BP | GO:0070988 | demethylation | 4 | 52 | 8.12e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 3 | 75 | 8.12e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 1 | 222 | 8.12e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 2 | 90 | 8.12e-01 |
| GO:BP | GO:0051931 | regulation of sensory perception | 1 | 13 | 8.12e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 7 | 102 | 8.12e-01 |
| GO:BP | GO:0051930 | regulation of sensory perception of pain | 1 | 13 | 8.12e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 2 | 45 | 8.12e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 8.12e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 6 | 16 | 8.12e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 1 | 108 | 8.13e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 2 | 7 | 8.13e-01 |
| GO:BP | GO:0007165 | signal transduction | 6 | 3818 | 8.13e-01 |
| GO:BP | GO:0048318 | axial mesoderm development | 1 | 6 | 8.13e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 8.13e-01 |
| GO:BP | GO:0006550 | isoleucine catabolic process | 1 | 4 | 8.13e-01 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 1 | 9 | 8.14e-01 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1 | 9 | 8.14e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 20 | 51 | 8.14e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 2 | 1939 | 8.15e-01 |
| GO:BP | GO:0007597 | blood coagulation, intrinsic pathway | 1 | 3 | 8.15e-01 |
| GO:BP | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | 1 | 2 | 8.15e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 8.15e-01 |
| GO:BP | GO:0048002 | antigen processing and presentation of peptide antigen | 3 | 41 | 8.15e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 16 | 42 | 8.15e-01 |
| GO:BP | GO:0044070 | regulation of monoatomic anion transport | 8 | 12 | 8.15e-01 |
| GO:BP | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination | 1 | 3 | 8.15e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 5 | 4579 | 8.15e-01 |
| GO:BP | GO:0046794 | transport of virus | 8 | 19 | 8.15e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 4 | 9 | 8.16e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 1 | 3 | 8.16e-01 |
| GO:BP | GO:1902511 | negative regulation of apoptotic DNA fragmentation | 1 | 2 | 8.16e-01 |
| GO:BP | GO:0003406 | retinal pigment epithelium development | 2 | 4 | 8.16e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 2 | 20 | 8.16e-01 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 1 | 8 | 8.16e-01 |
| GO:BP | GO:0032076 | negative regulation of deoxyribonuclease activity | 1 | 1 | 8.16e-01 |
| GO:BP | GO:0061196 | fungiform papilla development | 1 | 5 | 8.16e-01 |
| GO:BP | GO:0045843 | negative regulation of striated muscle tissue development | 3 | 5 | 8.16e-01 |
| GO:BP | GO:0006742 | NADP catabolic process | 1 | 3 | 8.17e-01 |
| GO:BP | GO:0006149 | deoxyinosine catabolic process | 1 | 1 | 8.17e-01 |
| GO:BP | GO:0019677 | NAD catabolic process | 1 | 3 | 8.17e-01 |
| GO:BP | GO:0046094 | deoxyinosine metabolic process | 1 | 1 | 8.17e-01 |
| GO:BP | GO:0006148 | inosine catabolic process | 1 | 1 | 8.17e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 1 | 48 | 8.17e-01 |
| GO:BP | GO:2001138 | regulation of phospholipid transport | 2 | 12 | 8.17e-01 |
| GO:BP | GO:0034418 | urate biosynthetic process | 1 | 2 | 8.17e-01 |
| GO:BP | GO:0060385 | axonogenesis involved in innervation | 4 | 7 | 8.17e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 2 | 8 | 8.17e-01 |
| GO:BP | GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 2 | 4 | 8.17e-01 |
| GO:BP | GO:0034372 | very-low-density lipoprotein particle remodeling | 2 | 4 | 8.17e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 3 | 48 | 8.17e-01 |
| GO:BP | GO:1904880 | response to hydrogen sulfide | 1 | 3 | 8.17e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 2 | 4 | 8.17e-01 |
| GO:BP | GO:0032695 | negative regulation of interleukin-12 production | 1 | 6 | 8.17e-01 |
| GO:BP | GO:2000251 | positive regulation of actin cytoskeleton reorganization | 9 | 14 | 8.17e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 8.17e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 7 | 31 | 8.17e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 2 | 382 | 8.17e-01 |
| GO:BP | GO:0071716 | leukotriene transport | 1 | 5 | 8.17e-01 |
| GO:BP | GO:0014852 | regulation of skeletal muscle contraction by neural stimulation via neuromuscular junction | 1 | 2 | 8.17e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 8.17e-01 |
| GO:BP | GO:1905770 | regulation of mesodermal cell differentiation | 3 | 6 | 8.18e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 28 | 199 | 8.18e-01 |
| GO:BP | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway | 1 | 3 | 8.18e-01 |
| GO:BP | GO:0070950 | regulation of neutrophil mediated killing of bacterium | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0070960 | positive regulation of neutrophil mediated cytotoxicity | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 4 | 19 | 8.18e-01 |
| GO:BP | GO:0070951 | regulation of neutrophil mediated killing of gram-negative bacterium | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0030974 | thiamine pyrophosphate transmembrane transport | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0070961 | positive regulation of neutrophil mediated killing of symbiont cell | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0044000 | movement in host | 19 | 138 | 8.18e-01 |
| GO:BP | GO:0015669 | gas transport | 1 | 6 | 8.18e-01 |
| GO:BP | GO:1900135 | positive regulation of renin secretion into blood stream | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0033602 | negative regulation of dopamine secretion | 1 | 4 | 8.18e-01 |
| GO:BP | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | 2 | 6 | 8.18e-01 |
| GO:BP | GO:2000491 | positive regulation of hepatic stellate cell activation | 2 | 4 | 8.18e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 2 | 31 | 8.18e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 14 | 214 | 8.18e-01 |
| GO:BP | GO:0035938 | estradiol secretion | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0120193 | tight junction organization | 19 | 52 | 8.19e-01 |
| GO:BP | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0038196 | type III interferon-mediated signaling pathway | 1 | 4 | 8.19e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 8.19e-01 |
| GO:BP | GO:0007154 | cell communication | 7 | 4226 | 8.19e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 1 | 22 | 8.19e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 8.19e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 8.19e-01 |
| GO:BP | GO:0034516 | response to vitamin B6 | 1 | 1 | 8.19e-01 |
| GO:BP | GO:0036304 | umbilical cord morphogenesis | 1 | 2 | 8.19e-01 |
| GO:BP | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 8.19e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 68 | 151 | 8.19e-01 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 4 | 9 | 8.19e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 9 | 400 | 8.19e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 6 | 1441 | 8.19e-01 |
| GO:BP | GO:1904580 | regulation of intracellular mRNA localization | 1 | 4 | 8.19e-01 |
| GO:BP | GO:0110077 | vesicle-mediated intercellular transport | 1 | 2 | 8.19e-01 |
| GO:BP | GO:2000864 | regulation of estradiol secretion | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0042541 | hemoglobin biosynthetic process | 5 | 13 | 8.19e-01 |
| GO:BP | GO:0061086 | negative regulation of histone H3-K27 methylation | 1 | 2 | 8.19e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 7 | 44 | 8.19e-01 |
| GO:BP | GO:0061027 | umbilical cord development | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 5 | 12 | 8.19e-01 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 9 | 20 | 8.19e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 2 | 15 | 8.19e-01 |
| GO:BP | GO:0150174 | negative regulation of phosphatidylcholine metabolic process | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 21 | 112 | 8.19e-01 |
| GO:BP | GO:0071358 | cellular response to type III interferon | 1 | 4 | 8.19e-01 |
| GO:BP | GO:1904970 | brush border assembly | 1 | 2 | 8.19e-01 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 2 | 13 | 8.19e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 8.19e-01 |
| GO:BP | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 1 | 9 | 8.19e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 2 | 9 | 8.19e-01 |
| GO:BP | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0034159 | regulation of toll-like receptor 8 signaling pathway | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 3 | 21 | 8.19e-01 |
| GO:BP | GO:0034161 | positive regulation of toll-like receptor 8 signaling pathway | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 1 | 229 | 8.19e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 7 | 62 | 8.19e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 3 | 14 | 8.19e-01 |
| GO:BP | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway | 2 | 4 | 8.19e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 2 | 111 | 8.19e-01 |
| GO:BP | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 8.19e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 9 | 56 | 8.19e-01 |
| GO:BP | GO:0043921 | modulation by host of viral transcription | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0002457 | T cell antigen processing and presentation | 2 | 4 | 8.19e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 1 | 4 | 8.19e-01 |
| GO:BP | GO:0060847 | endothelial cell fate specification | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 4 | 213 | 8.19e-01 |
| GO:BP | GO:0051228 | mitotic spindle disassembly | 1 | 2 | 8.19e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0051230 | spindle disassembly | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0060300 | regulation of cytokine activity | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0006491 | N-glycan processing | 1 | 18 | 8.19e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 27 | 166 | 8.20e-01 |
| GO:BP | GO:0034381 | plasma lipoprotein particle clearance | 1 | 25 | 8.20e-01 |
| GO:BP | GO:0017185 | peptidyl-lysine hydroxylation | 4 | 9 | 8.20e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 3 | 18 | 8.20e-01 |
| GO:BP | GO:0033133 | positive regulation of glucokinase activity | 2 | 4 | 8.20e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 2 | 6 | 8.20e-01 |
| GO:BP | GO:0000973 | post-transcriptional tethering of RNA polymerase II gene DNA at nuclear periphery | 1 | 2 | 8.20e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 3 | 26 | 8.20e-01 |
| GO:BP | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 4 | 27 | 8.20e-01 |
| GO:BP | GO:0015920 | lipopolysaccharide transport | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 2 | 4 | 8.20e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 2 | 4 | 8.20e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 8 | 53 | 8.20e-01 |
| GO:BP | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 2 | 109 | 8.20e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 8.20e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 1 | 73 | 8.20e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 8.20e-01 |
| GO:BP | GO:0043615 | astrocyte cell migration | 2 | 6 | 8.21e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 1 | 3 | 8.21e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 3 | 21 | 8.21e-01 |
| GO:BP | GO:2001112 | regulation of cellular response to hepatocyte growth factor stimulus | 1 | 1 | 8.21e-01 |
| GO:BP | GO:0035981 | tongue muscle cell differentiation | 1 | 1 | 8.21e-01 |
| GO:BP | GO:2001037 | positive regulation of tongue muscle cell differentiation | 1 | 1 | 8.21e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 7 | 64 | 8.21e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 1 | 43 | 8.21e-01 |
| GO:BP | GO:0045992 | negative regulation of embryonic development | 1 | 2 | 8.21e-01 |
| GO:BP | GO:2001035 | regulation of tongue muscle cell differentiation | 1 | 1 | 8.21e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 6 | 158 | 8.21e-01 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 20 | 38 | 8.21e-01 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 20 | 38 | 8.21e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 8 | 66 | 8.21e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 8.22e-01 |
| GO:BP | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway | 2 | 7 | 8.22e-01 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 4 | 8 | 8.22e-01 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 1 | 5 | 8.22e-01 |
| GO:BP | GO:0071354 | cellular response to interleukin-6 | 5 | 23 | 8.22e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 1 | 10 | 8.22e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 8.22e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 8 | 376 | 8.22e-01 |
| GO:BP | GO:0043380 | regulation of memory T cell differentiation | 1 | 4 | 8.22e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 3 | 33 | 8.22e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 8.22e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 3 | 3012 | 8.22e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 8 | 83 | 8.22e-01 |
| GO:BP | GO:2000407 | regulation of T cell extravasation | 1 | 3 | 8.22e-01 |
| GO:BP | GO:0046532 | regulation of photoreceptor cell differentiation | 2 | 4 | 8.22e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 2 | 10 | 8.22e-01 |
| GO:BP | GO:0032966 | negative regulation of collagen biosynthetic process | 2 | 7 | 8.22e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 2 | 71 | 8.22e-01 |
| GO:BP | GO:1990117 | B cell receptor apoptotic signaling pathway | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0071264 | positive regulation of translational initiation in response to starvation | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0048250 | iron import into the mitochondrion | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 1 | 230 | 8.22e-01 |
| GO:BP | GO:0071262 | regulation of translational initiation in response to starvation | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0002358 | B cell homeostatic proliferation | 1 | 1 | 8.22e-01 |
| GO:BP | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 2 | 8.22e-01 |
| GO:BP | GO:0006000 | fructose metabolic process | 1 | 10 | 8.22e-01 |
| GO:BP | GO:2000673 | positive regulation of motor neuron apoptotic process | 1 | 1 | 8.22e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 8 | 20 | 8.22e-01 |
| GO:BP | GO:2000340 | positive regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 3 | 8.22e-01 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 1 | 5 | 8.23e-01 |
| GO:BP | GO:0048137 | spermatocyte division | 1 | 1 | 8.23e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 3 | 16 | 8.23e-01 |
| GO:BP | GO:0008295 | spermidine biosynthetic process | 1 | 3 | 8.23e-01 |
| GO:BP | GO:0035352 | NAD transmembrane transport | 1 | 3 | 8.23e-01 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 6 | 19 | 8.23e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 8.23e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 2 | 35 | 8.23e-01 |
| GO:BP | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 8.24e-01 |
| GO:BP | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 8.24e-01 |
| GO:BP | GO:1902645 | tertiary alcohol biosynthetic process | 1 | 8 | 8.24e-01 |
| GO:BP | GO:0034651 | cortisol biosynthetic process | 1 | 8 | 8.24e-01 |
| GO:BP | GO:0032353 | negative regulation of hormone biosynthetic process | 1 | 8 | 8.24e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 5 | 11 | 8.24e-01 |
| GO:BP | GO:0140588 | chromatin looping | 1 | 2 | 8.24e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 2 | 27 | 8.24e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 3 | 30 | 8.24e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 3 | 30 | 8.24e-01 |
| GO:BP | GO:0031645 | negative regulation of nervous system process | 4 | 9 | 8.24e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 6 | 21 | 8.24e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 2 | 52 | 8.24e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 2 | 52 | 8.24e-01 |
| GO:BP | GO:1905581 | positive regulation of low-density lipoprotein particle clearance | 1 | 2 | 8.24e-01 |
| GO:BP | GO:0044721 | protein import into peroxisome matrix, substrate release | 1 | 2 | 8.24e-01 |
| GO:BP | GO:0090027 | negative regulation of monocyte chemotaxis | 2 | 4 | 8.24e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 295 | 8.24e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 1 | 44 | 8.24e-01 |
| GO:BP | GO:0050877 | nervous system process | 3 | 634 | 8.24e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 4 | 10 | 8.24e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 3 | 15 | 8.24e-01 |
| GO:BP | GO:0046865 | terpenoid transport | 1 | 3 | 8.25e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 8.25e-01 |
| GO:BP | GO:0042667 | auditory receptor cell fate specification | 1 | 1 | 8.25e-01 |
| GO:BP | GO:0046864 | isoprenoid transport | 1 | 3 | 8.25e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 3 | 9 | 8.25e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 3 | 9 | 8.25e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 3 | 9 | 8.25e-01 |
| GO:BP | GO:1905162 | regulation of phagosome maturation | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 4 | 24 | 8.25e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 4 | 7 | 8.25e-01 |
| GO:BP | GO:0044597 | daunorubicin metabolic process | 4 | 7 | 8.25e-01 |
| GO:BP | GO:0090108 | positive regulation of high-density lipoprotein particle assembly | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0003342 | proepicardium development | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0006784 | heme A biosynthetic process | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0046160 | heme a metabolic process | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0021524 | visceral motor neuron differentiation | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0003350 | pulmonary myocardium development | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0003343 | septum transversum development | 1 | 2 | 8.25e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 3 | 10 | 8.25e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 3 | 7 | 8.25e-01 |
| GO:BP | GO:0002309 | T cell proliferation involved in immune response | 1 | 3 | 8.25e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 2 | 55 | 8.25e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 3 | 10 | 8.25e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 3 | 22 | 8.25e-01 |
| GO:BP | GO:2000674 | regulation of type B pancreatic cell apoptotic process | 1 | 6 | 8.26e-01 |
| GO:BP | GO:0060595 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production | 2 | 3 | 8.26e-01 |
| GO:BP | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 4 | 8.26e-01 |
| GO:BP | GO:0140541 | piRNA transcription | 1 | 1 | 8.26e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 8.26e-01 |
| GO:BP | GO:0060615 | mammary gland bud formation | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0021769 | orbitofrontal cortex development | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0021847 | ventricular zone neuroblast division | 1 | 1 | 8.26e-01 |
| GO:BP | GO:0060915 | mesenchymal cell differentiation involved in lung development | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0060529 | squamous basal epithelial stem cell differentiation involved in prostate gland acinus development | 1 | 1 | 8.26e-01 |
| GO:BP | GO:0060667 | branch elongation involved in salivary gland morphogenesis | 1 | 2 | 8.26e-01 |
| GO:BP | GO:1900864 | mitochondrial RNA modification | 4 | 9 | 8.26e-01 |
| GO:BP | GO:0070900 | mitochondrial tRNA modification | 4 | 9 | 8.26e-01 |
| GO:BP | GO:0071461 | cellular response to redox state | 1 | 1 | 8.26e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 8.27e-01 |
| GO:BP | GO:1900195 | positive regulation of oocyte maturation | 1 | 3 | 8.27e-01 |
| GO:BP | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste | 1 | 1 | 8.27e-01 |
| GO:BP | GO:0061187 | regulation of rDNA heterochromatin formation | 1 | 2 | 8.27e-01 |
| GO:BP | GO:0061188 | negative regulation of rDNA heterochromatin formation | 1 | 2 | 8.27e-01 |
| GO:BP | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 2 | 14 | 8.28e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 99 | 8.28e-01 |
| GO:BP | GO:0035469 | determination of pancreatic left/right asymmetry | 1 | 3 | 8.28e-01 |
| GO:BP | GO:0009886 | post-embryonic animal morphogenesis | 1 | 10 | 8.28e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 3 | 2450 | 8.28e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 99 | 8.28e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 47 | 116 | 8.28e-01 |
| GO:BP | GO:0051461 | positive regulation of corticotropin secretion | 1 | 2 | 8.28e-01 |
| GO:BP | GO:0021718 | superior olivary nucleus development | 1 | 1 | 8.28e-01 |
| GO:BP | GO:0021586 | pons maturation | 1 | 1 | 8.28e-01 |
| GO:BP | GO:0021722 | superior olivary nucleus maturation | 1 | 1 | 8.28e-01 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 4 | 11 | 8.28e-01 |
| GO:BP | GO:0002295 | T-helper cell lineage commitment | 3 | 7 | 8.28e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 12 | 35 | 8.29e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 1 | 11 | 8.29e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 5 | 28 | 8.29e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 1 | 47 | 8.29e-01 |
| GO:BP | GO:1990840 | response to lectin | 1 | 11 | 8.29e-01 |
| GO:BP | GO:0010159 | specification of animal organ position | 1 | 2 | 8.29e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 1 | 11 | 8.29e-01 |
| GO:BP | GO:0048793 | pronephros development | 1 | 5 | 8.29e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 8.29e-01 |
| GO:BP | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent | 1 | 2 | 8.29e-01 |
| GO:BP | GO:1990401 | embryonic lung development | 1 | 1 | 8.29e-01 |
| GO:BP | GO:0060177 | regulation of angiotensin metabolic process | 1 | 1 | 8.29e-01 |
| GO:BP | GO:1903597 | negative regulation of gap junction assembly | 1 | 1 | 8.29e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 99 | 8.29e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 8 | 23 | 8.29e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 5 | 11 | 8.29e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 8.30e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 8.30e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 11 | 2412 | 8.30e-01 |
| GO:BP | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation | 1 | 2 | 8.30e-01 |
| GO:BP | GO:0035457 | cellular response to interferon-alpha | 5 | 7 | 8.31e-01 |
| GO:BP | GO:0070366 | regulation of hepatocyte differentiation | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0070358 | actin polymerization-dependent cell motility | 1 | 6 | 8.31e-01 |
| GO:BP | GO:0042663 | regulation of endodermal cell fate specification | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0060974 | cell migration involved in heart formation | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 1 | 55 | 8.31e-01 |
| GO:BP | GO:0003210 | cardiac atrium formation | 1 | 2 | 8.31e-01 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 10 | 29 | 8.31e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 2 | 49 | 8.31e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 2 | 77 | 8.31e-01 |
| GO:BP | GO:0022400 | regulation of rhodopsin mediated signaling pathway | 4 | 5 | 8.31e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 5 | 65 | 8.31e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 8.31e-01 |
| GO:BP | GO:0043435 | response to corticotropin-releasing hormone | 2 | 3 | 8.31e-01 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 1 | 31 | 8.31e-01 |
| GO:BP | GO:0032914 | positive regulation of transforming growth factor beta1 production | 1 | 6 | 8.31e-01 |
| GO:BP | GO:0071376 | cellular response to corticotropin-releasing hormone stimulus | 2 | 3 | 8.31e-01 |
| GO:BP | GO:0071316 | cellular response to nicotine | 1 | 4 | 8.31e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 7 | 49 | 8.31e-01 |
| GO:BP | GO:0048172 | regulation of short-term neuronal synaptic plasticity | 1 | 9 | 8.31e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 3 | 7 | 8.32e-01 |
| GO:BP | GO:0046449 | creatinine metabolic process | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis | 3 | 9 | 8.32e-01 |
| GO:BP | GO:0048866 | stem cell fate specification | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0061046 | regulation of branching involved in lung morphogenesis | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 8.32e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 8.32e-01 |
| GO:BP | GO:0007379 | segment specification | 1 | 14 | 8.32e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 1 | 104 | 8.32e-01 |
| GO:BP | GO:0042426 | choline catabolic process | 1 | 3 | 8.32e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 3 | 16 | 8.32e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 5 | 269 | 8.32e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 8.32e-01 |
| GO:BP | GO:0021535 | cell migration in hindbrain | 7 | 12 | 8.32e-01 |
| GO:BP | GO:0070948 | regulation of neutrophil mediated cytotoxicity | 2 | 2 | 8.32e-01 |
| GO:BP | GO:2001280 | positive regulation of unsaturated fatty acid biosynthetic process | 1 | 4 | 8.32e-01 |
| GO:BP | GO:0014734 | skeletal muscle hypertrophy | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0006909 | phagocytosis | 1 | 148 | 8.32e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 12 | 43 | 8.32e-01 |
| GO:BP | GO:1905526 | regulation of Golgi lumen acidification | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0036066 | protein O-linked fucosylation | 2 | 5 | 8.32e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 8.32e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 5 | 24 | 8.33e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 3 | 3080 | 8.33e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 3 | 3080 | 8.33e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 4 | 186 | 8.33e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2 | 4099 | 8.34e-01 |
| GO:BP | GO:0000960 | regulation of mitochondrial RNA catabolic process | 2 | 5 | 8.34e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 1 | 59 | 8.34e-01 |
| GO:BP | GO:2001245 | regulation of phosphatidylcholine biosynthetic process | 2 | 5 | 8.35e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 1 | 242 | 8.35e-01 |
| GO:BP | GO:0045658 | regulation of neutrophil differentiation | 1 | 2 | 8.35e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 1 | 25 | 8.35e-01 |
| GO:BP | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity | 1 | 2 | 8.35e-01 |
| GO:BP | GO:0010887 | negative regulation of cholesterol storage | 4 | 9 | 8.35e-01 |
| GO:BP | GO:0090086 | negative regulation of protein deubiquitination | 1 | 4 | 8.35e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 3 | 11 | 8.35e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 1 | 2 | 8.35e-01 |
| GO:BP | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation | 1 | 4 | 8.35e-01 |
| GO:BP | GO:2000646 | positive regulation of receptor catabolic process | 1 | 5 | 8.35e-01 |
| GO:BP | GO:0006083 | acetate metabolic process | 1 | 1 | 8.35e-01 |
| GO:BP | GO:0000413 | protein peptidyl-prolyl isomerization | 3 | 26 | 8.35e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 4 | 994 | 8.35e-01 |
| GO:BP | GO:1902659 | regulation of glucose mediated signaling pathway | 1 | 3 | 8.35e-01 |
| GO:BP | GO:0019427 | acetyl-CoA biosynthetic process from acetate | 1 | 1 | 8.35e-01 |
| GO:BP | GO:0048669 | collateral sprouting in absence of injury | 1 | 4 | 8.35e-01 |
| GO:BP | GO:0046501 | protoporphyrinogen IX metabolic process | 2 | 11 | 8.35e-01 |
| GO:BP | GO:1902661 | positive regulation of glucose mediated signaling pathway | 1 | 3 | 8.35e-01 |
| GO:BP | GO:1903852 | positive regulation of cristae formation | 1 | 3 | 8.35e-01 |
| GO:BP | GO:0006226 | dUMP biosynthetic process | 1 | 2 | 8.35e-01 |
| GO:BP | GO:0019085 | early viral transcription | 1 | 2 | 8.36e-01 |
| GO:BP | GO:0043335 | protein unfolding | 1 | 5 | 8.36e-01 |
| GO:BP | GO:0070092 | regulation of glucagon secretion | 1 | 3 | 8.36e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 2 | 7 | 8.36e-01 |
| GO:BP | GO:0070091 | glucagon secretion | 1 | 3 | 8.36e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 8.36e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 14 | 95 | 8.36e-01 |
| GO:BP | GO:0033029 | regulation of neutrophil apoptotic process | 1 | 5 | 8.36e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 3 | 12 | 8.36e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 13 | 151 | 8.36e-01 |
| GO:BP | GO:0098964 | anterograde dendritic transport of messenger ribonucleoprotein complex | 1 | 2 | 8.36e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 5 | 9 | 8.36e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 2 | 2135 | 8.36e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 8.36e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 8.36e-01 |
| GO:BP | GO:0048069 | eye pigmentation | 2 | 4 | 8.37e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 2 | 5 | 8.37e-01 |
| GO:BP | GO:0034695 | response to prostaglandin E | 1 | 17 | 8.37e-01 |
| GO:BP | GO:0106005 | RNA 5’-cap (guanine-N7)-methylation | 1 | 2 | 8.37e-01 |
| GO:BP | GO:0060440 | trachea formation | 1 | 7 | 8.37e-01 |
| GO:BP | GO:0030473 | nuclear migration along microtubule | 2 | 5 | 8.37e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 4 | 233 | 8.37e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 1 | 2 | 8.37e-01 |
| GO:BP | GO:0061032 | visceral serous pericardium development | 1 | 1 | 8.38e-01 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 10 | 8.38e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 1 | 11 | 8.38e-01 |
| GO:BP | GO:1905167 | positive regulation of lysosomal protein catabolic process | 1 | 4 | 8.38e-01 |
| GO:BP | GO:2000426 | negative regulation of apoptotic cell clearance | 1 | 1 | 8.38e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 2 | 48 | 8.38e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 8.38e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 3 | 35 | 8.38e-01 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 4 | 8 | 8.38e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 1 | 87 | 8.39e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 4 | 234 | 8.39e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 8.39e-01 |
| GO:BP | GO:0043379 | memory T cell differentiation | 1 | 5 | 8.39e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 22 | 52 | 8.39e-01 |
| GO:BP | GO:0015949 | nucleobase-containing small molecule interconversion | 3 | 8 | 8.39e-01 |
| GO:BP | GO:1904037 | positive regulation of epithelial cell apoptotic process | 1 | 26 | 8.39e-01 |
| GO:BP | GO:0015833 | peptide transport | 29 | 179 | 8.39e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 2 | 5 | 8.39e-01 |
| GO:BP | GO:0002038 | positive regulation of L-glutamate import across plasma membrane | 1 | 3 | 8.39e-01 |
| GO:BP | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus | 2 | 7 | 8.39e-01 |
| GO:BP | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 10 | 8.39e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 26 | 106 | 8.39e-01 |
| GO:BP | GO:0007493 | endodermal cell fate determination | 1 | 1 | 8.39e-01 |
| GO:BP | GO:0031134 | sister chromatid biorientation | 1 | 3 | 8.39e-01 |
| GO:BP | GO:1904002 | regulation of sebum secreting cell proliferation | 1 | 1 | 8.39e-01 |
| GO:BP | GO:0001949 | sebaceous gland cell differentiation | 1 | 1 | 8.39e-01 |
| GO:BP | GO:0060700 | regulation of ribonuclease activity | 1 | 5 | 8.39e-01 |
| GO:BP | GO:0021628 | olfactory nerve formation | 1 | 2 | 8.39e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 2 | 14 | 8.39e-01 |
| GO:BP | GO:1900193 | regulation of oocyte maturation | 2 | 5 | 8.39e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 122 | 285 | 8.39e-01 |
| GO:BP | GO:0033085 | negative regulation of T cell differentiation in thymus | 1 | 5 | 8.39e-01 |
| GO:BP | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate | 1 | 4 | 8.39e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 5 | 74 | 8.39e-01 |
| GO:BP | GO:0032328 | alanine transport | 3 | 14 | 8.39e-01 |
| GO:BP | GO:0036123 | histone H3-K9 dimethylation | 1 | 3 | 8.39e-01 |
| GO:BP | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development | 1 | 2 | 8.39e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 1 | 13 | 8.39e-01 |
| GO:BP | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly | 1 | 1 | 8.39e-01 |
| GO:BP | GO:0036466 | synaptic vesicle recycling via endosome | 2 | 4 | 8.39e-01 |
| GO:BP | GO:1903431 | positive regulation of cell maturation | 2 | 6 | 8.39e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 2 | 14 | 8.39e-01 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 2 | 122 | 8.39e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 5 | 23 | 8.39e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 7 | 50 | 8.39e-01 |
| GO:BP | GO:0071801 | regulation of podosome assembly | 2 | 8 | 8.39e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 8.39e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 6 | 4128 | 8.40e-01 |
| GO:BP | GO:0032972 | regulation of muscle filament sliding speed | 1 | 2 | 8.40e-01 |
| GO:BP | GO:0051541 | elastin metabolic process | 2 | 5 | 8.40e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 3 | 52 | 8.40e-01 |
| GO:BP | GO:0019915 | lipid storage | 2 | 66 | 8.40e-01 |
| GO:BP | GO:0003011 | involuntary skeletal muscle contraction | 1 | 3 | 8.40e-01 |
| GO:BP | GO:1900363 | regulation of mRNA polyadenylation | 4 | 11 | 8.40e-01 |
| GO:BP | GO:1901551 | negative regulation of endothelial cell development | 1 | 2 | 8.40e-01 |
| GO:BP | GO:1903141 | negative regulation of establishment of endothelial barrier | 1 | 2 | 8.40e-01 |
| GO:BP | GO:2001171 | positive regulation of ATP biosynthetic process | 1 | 10 | 8.40e-01 |
| GO:BP | GO:1904847 | regulation of cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.40e-01 |
| GO:BP | GO:0035766 | cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.40e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 8.40e-01 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 1 | 43 | 8.40e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 8.40e-01 |
| GO:BP | GO:2000544 | regulation of endothelial cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.40e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 3 | 46 | 8.40e-01 |
| GO:BP | GO:0035768 | endothelial cell chemotaxis to fibroblast growth factor | 2 | 4 | 8.40e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 4 | 234 | 8.40e-01 |
| GO:BP | GO:0002395 | immune response in nasopharyngeal-associated lymphoid tissue | 1 | 1 | 8.40e-01 |
| GO:BP | GO:0002387 | immune response in gut-associated lymphoid tissue | 1 | 1 | 8.40e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 1 | 2 | 8.40e-01 |
| GO:BP | GO:0070170 | regulation of tooth mineralization | 1 | 4 | 8.40e-01 |
| GO:BP | GO:0033341 | regulation of collagen binding | 1 | 2 | 8.41e-01 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 9 | 63 | 8.41e-01 |
| GO:BP | GO:0042989 | sequestering of actin monomers | 1 | 9 | 8.41e-01 |
| GO:BP | GO:1903538 | regulation of meiotic cell cycle process involved in oocyte maturation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 18 | 116 | 8.41e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 6 | 138 | 8.41e-01 |
| GO:BP | GO:0019740 | nitrogen utilization | 2 | 4 | 8.41e-01 |
| GO:BP | GO:1905380 | regulation of snRNA transcription by RNA polymerase II | 2 | 4 | 8.41e-01 |
| GO:BP | GO:0019303 | D-ribose catabolic process | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0070781 | response to biotin | 1 | 1 | 8.41e-01 |
| GO:BP | GO:1905749 | regulation of endosome to plasma membrane protein transport | 1 | 5 | 8.41e-01 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 1 | 102 | 8.41e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 14 | 598 | 8.41e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 6 | 1507 | 8.41e-01 |
| GO:BP | GO:1903371 | regulation of endoplasmic reticulum tubular network organization | 1 | 6 | 8.41e-01 |
| GO:BP | GO:0060174 | limb bud formation | 2 | 10 | 8.41e-01 |
| GO:BP | GO:0072318 | clathrin coat disassembly | 3 | 8 | 8.41e-01 |
| GO:BP | GO:1904383 | response to sodium phosphate | 1 | 5 | 8.41e-01 |
| GO:BP | GO:0018293 | protein-FAD linkage | 1 | 1 | 8.41e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 7 | 4617 | 8.41e-01 |
| GO:BP | GO:0048512 | circadian behavior | 6 | 30 | 8.41e-01 |
| GO:BP | GO:2000124 | regulation of endocannabinoid signaling pathway | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0070370 | cellular heat acclimation | 2 | 5 | 8.41e-01 |
| GO:BP | GO:0010286 | heat acclimation | 2 | 5 | 8.41e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 1 | 25 | 8.41e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 25 | 8.41e-01 |
| GO:BP | GO:0035526 | retrograde transport, plasma membrane to Golgi | 1 | 2 | 8.42e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 2 | 70 | 8.42e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 7 | 33 | 8.42e-01 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 1 | 11 | 8.42e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 2 | 4 | 8.42e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 5 | 15 | 8.42e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 1 | 9 | 8.42e-01 |
| GO:BP | GO:0048511 | rhythmic process | 3 | 237 | 8.43e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 2 | 20 | 8.43e-01 |
| GO:BP | GO:0099538 | synaptic signaling via neuropeptide | 1 | 2 | 8.43e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 5 | 468 | 8.43e-01 |
| GO:BP | GO:0090660 | cerebrospinal fluid circulation | 1 | 9 | 8.43e-01 |
| GO:BP | GO:0000266 | mitochondrial fission | 4 | 38 | 8.43e-01 |
| GO:BP | GO:0015820 | leucine transport | 4 | 10 | 8.43e-01 |
| GO:BP | GO:0030252 | growth hormone secretion | 1 | 12 | 8.44e-01 |
| GO:BP | GO:0032345 | negative regulation of aldosterone metabolic process | 1 | 4 | 8.44e-01 |
| GO:BP | GO:0032348 | negative regulation of aldosterone biosynthetic process | 1 | 4 | 8.44e-01 |
| GO:BP | GO:0034342 | response to type III interferon | 1 | 5 | 8.44e-01 |
| GO:BP | GO:0070669 | response to interleukin-2 | 1 | 3 | 8.44e-01 |
| GO:BP | GO:2000065 | negative regulation of cortisol biosynthetic process | 1 | 4 | 8.44e-01 |
| GO:BP | GO:0060375 | regulation of mast cell differentiation | 1 | 2 | 8.44e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 2 | 4 | 8.44e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 2 | 11 | 8.44e-01 |
| GO:BP | GO:0043984 | histone H4-K16 acetylation | 6 | 14 | 8.44e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 5 | 134 | 8.45e-01 |
| GO:BP | GO:0015711 | organic anion transport | 1 | 264 | 8.45e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 3 | 3169 | 8.45e-01 |
| GO:BP | GO:0001547 | antral ovarian follicle growth | 3 | 5 | 8.45e-01 |
| GO:BP | GO:0090193 | positive regulation of glomerulus development | 1 | 3 | 8.45e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 8.45e-01 |
| GO:BP | GO:1900118 | negative regulation of execution phase of apoptosis | 1 | 6 | 8.45e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 8.45e-01 |
| GO:BP | GO:0070561 | vitamin D receptor signaling pathway | 1 | 10 | 8.45e-01 |
| GO:BP | GO:0038183 | bile acid signaling pathway | 1 | 3 | 8.45e-01 |
| GO:BP | GO:0045292 | mRNA cis splicing, via spliceosome | 1 | 26 | 8.45e-01 |
| GO:BP | GO:0048808 | male genitalia morphogenesis | 1 | 2 | 8.45e-01 |
| GO:BP | GO:0090598 | male anatomical structure morphogenesis | 1 | 2 | 8.45e-01 |
| GO:BP | GO:1900272 | negative regulation of long-term synaptic potentiation | 1 | 8 | 8.46e-01 |
| GO:BP | GO:1904054 | regulation of cholangiocyte proliferation | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0019509 | L-methionine salvage from methylthioadenosine | 2 | 5 | 8.46e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 20 | 36 | 8.46e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 5 | 20 | 8.46e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 20 | 36 | 8.46e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 19 | 50 | 8.46e-01 |
| GO:BP | GO:0015888 | thiamine transport | 3 | 3 | 8.46e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 7 | 222 | 8.46e-01 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 1 | 8 | 8.46e-01 |
| GO:BP | GO:0036118 | hyaluranon cable assembly | 1 | 3 | 8.46e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 8.46e-01 |
| GO:BP | GO:0006457 | protein folding | 2 | 192 | 8.46e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 8.46e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 2 | 18 | 8.46e-01 |
| GO:BP | GO:1900106 | positive regulation of hyaluranon cable assembly | 1 | 3 | 8.46e-01 |
| GO:BP | GO:1900104 | regulation of hyaluranon cable assembly | 1 | 3 | 8.46e-01 |
| GO:BP | GO:1902624 | positive regulation of neutrophil migration | 2 | 21 | 8.46e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 4 | 11 | 8.46e-01 |
| GO:BP | GO:0042701 | progesterone secretion | 1 | 3 | 8.46e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 8.47e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 1 | 11 | 8.47e-01 |
| GO:BP | GO:0071549 | cellular response to dexamethasone stimulus | 1 | 24 | 8.47e-01 |
| GO:BP | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis | 2 | 4 | 8.47e-01 |
| GO:BP | GO:2000192 | negative regulation of fatty acid transport | 1 | 7 | 8.47e-01 |
| GO:BP | GO:0060742 | epithelial cell differentiation involved in prostate gland development | 3 | 8 | 8.48e-01 |
| GO:BP | GO:0006597 | spermine biosynthetic process | 1 | 2 | 8.48e-01 |
| GO:BP | GO:1905034 | regulation of antifungal innate immune response | 1 | 3 | 8.48e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 5 | 15 | 8.48e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 5 | 69 | 8.48e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 8 | 18 | 8.48e-01 |
| GO:BP | GO:1902616 | acyl carnitine transmembrane transport | 1 | 1 | 8.48e-01 |
| GO:BP | GO:0046958 | nonassociative learning | 3 | 6 | 8.48e-01 |
| GO:BP | GO:0097168 | mesenchymal stem cell proliferation | 2 | 4 | 8.49e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 10 | 35 | 8.49e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 7 | 478 | 8.49e-01 |
| GO:BP | GO:0043969 | histone H2B acetylation | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 27 | 2102 | 8.49e-01 |
| GO:BP | GO:0006954 | inflammatory response | 6 | 429 | 8.49e-01 |
| GO:BP | GO:1900924 | negative regulation of glycine import across plasma membrane | 1 | 2 | 8.49e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 3 | 16 | 8.49e-01 |
| GO:BP | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway | 1 | 1 | 8.49e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 8.49e-01 |
| GO:BP | GO:2001074 | regulation of metanephric ureteric bud development | 1 | 1 | 8.49e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 4 | 3580 | 8.49e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 3 | 16 | 8.49e-01 |
| GO:BP | GO:2001076 | positive regulation of metanephric ureteric bud development | 1 | 1 | 8.49e-01 |
| GO:BP | GO:0006857 | oligopeptide transport | 1 | 5 | 8.49e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 2 | 28 | 8.49e-01 |
| GO:BP | GO:0035672 | oligopeptide transmembrane transport | 1 | 5 | 8.49e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 2 | 28 | 8.49e-01 |
| GO:BP | GO:1900923 | regulation of glycine import across plasma membrane | 1 | 2 | 8.49e-01 |
| GO:BP | GO:2000359 | regulation of binding of sperm to zona pellucida | 2 | 2 | 8.49e-01 |
| GO:BP | GO:0070902 | mitochondrial tRNA pseudouridine synthesis | 1 | 2 | 8.49e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 5 | 35 | 8.49e-01 |
| GO:BP | GO:1900425 | negative regulation of defense response to bacterium | 1 | 4 | 8.49e-01 |
| GO:BP | GO:0051799 | negative regulation of hair follicle development | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0021578 | hindbrain maturation | 2 | 4 | 8.49e-01 |
| GO:BP | GO:0015871 | choline transport | 2 | 6 | 8.50e-01 |
| GO:BP | GO:0019062 | virion attachment to host cell | 1 | 9 | 8.50e-01 |
| GO:BP | GO:0035332 | positive regulation of hippo signaling | 3 | 6 | 8.50e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 3 | 71 | 8.50e-01 |
| GO:BP | GO:0016556 | mRNA modification | 1 | 29 | 8.50e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 1 | 20 | 8.50e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 2 | 13 | 8.50e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 2 | 18 | 8.50e-01 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 4 | 15 | 8.50e-01 |
| GO:BP | GO:0045016 | mitochondrial magnesium ion transmembrane transport | 1 | 2 | 8.50e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 5 | 42 | 8.50e-01 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 4 | 15 | 8.50e-01 |
| GO:BP | GO:0016137 | glycoside metabolic process | 2 | 16 | 8.50e-01 |
| GO:BP | GO:1902499 | positive regulation of protein autoubiquitination | 1 | 2 | 8.50e-01 |
| GO:BP | GO:0002579 | positive regulation of antigen processing and presentation | 2 | 3 | 8.50e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 2 | 19 | 8.51e-01 |
| GO:BP | GO:0042026 | protein refolding | 3 | 26 | 8.51e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 11 | 191 | 8.51e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 8.51e-01 |
| GO:BP | GO:1902931 | negative regulation of alcohol biosynthetic process | 4 | 10 | 8.51e-01 |
| GO:BP | GO:1904057 | negative regulation of sensory perception of pain | 1 | 1 | 8.51e-01 |
| GO:BP | GO:0060428 | lung epithelium development | 3 | 30 | 8.51e-01 |
| GO:BP | GO:0007341 | penetration of zona pellucida | 1 | 4 | 8.51e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 8.51e-01 |
| GO:BP | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis | 2 | 5 | 8.51e-01 |
| GO:BP | GO:1904823 | purine nucleobase transmembrane transport | 2 | 3 | 8.51e-01 |
| GO:BP | GO:0030282 | bone mineralization | 17 | 82 | 8.51e-01 |
| GO:BP | GO:0048293 | regulation of isotype switching to IgE isotypes | 3 | 5 | 8.51e-01 |
| GO:BP | GO:0045218 | zonula adherens maintenance | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0048289 | isotype switching to IgE isotypes | 3 | 5 | 8.51e-01 |
| GO:BP | GO:0044211 | CTP salvage | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 9 | 26 | 8.51e-01 |
| GO:BP | GO:0033604 | negative regulation of catecholamine secretion | 3 | 6 | 8.52e-01 |
| GO:BP | GO:0009584 | detection of visible light | 1 | 20 | 8.52e-01 |
| GO:BP | GO:0002253 | activation of immune response | 1 | 272 | 8.52e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 1 | 14 | 8.52e-01 |
| GO:BP | GO:0007588 | excretion | 1 | 18 | 8.52e-01 |
| GO:BP | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 4 | 12 | 8.52e-01 |
| GO:BP | GO:0006438 | valyl-tRNA aminoacylation | 1 | 2 | 8.52e-01 |
| GO:BP | GO:0034471 | ncRNA 5’-end processing | 1 | 5 | 8.52e-01 |
| GO:BP | GO:0000472 | endonucleolytic cleavage to generate mature 5’-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 5 | 8.52e-01 |
| GO:BP | GO:0090715 | immunological memory formation process | 1 | 6 | 8.52e-01 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 5 | 45 | 8.52e-01 |
| GO:BP | GO:0000967 | rRNA 5’-end processing | 1 | 5 | 8.52e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 4 | 65 | 8.52e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 8.52e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 8.52e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 1 | 105 | 8.52e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 7 | 44 | 8.52e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1223 | 8.52e-01 |
| GO:BP | GO:1901382 | regulation of chorionic trophoblast cell proliferation | 1 | 2 | 8.52e-01 |
| GO:BP | GO:0060336 | negative regulation of type II interferon-mediated signaling pathway | 1 | 6 | 8.52e-01 |
| GO:BP | GO:0060462 | lung lobe development | 1 | 4 | 8.52e-01 |
| GO:BP | GO:0002252 | immune effector process | 2 | 318 | 8.52e-01 |
| GO:BP | GO:0060463 | lung lobe morphogenesis | 1 | 4 | 8.52e-01 |
| GO:BP | GO:0042661 | regulation of mesodermal cell fate specification | 2 | 4 | 8.52e-01 |
| GO:BP | GO:0090158 | endoplasmic reticulum membrane organization | 4 | 14 | 8.52e-01 |
| GO:BP | GO:0097360 | chorionic trophoblast cell proliferation | 1 | 2 | 8.52e-01 |
| GO:BP | GO:0060331 | negative regulation of response to type II interferon | 1 | 6 | 8.52e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 1 | 457 | 8.53e-01 |
| GO:BP | GO:0044467 | glial cell-derived neurotrophic factor production | 1 | 2 | 8.53e-01 |
| GO:BP | GO:1902960 | negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process | 1 | 3 | 8.53e-01 |
| GO:BP | GO:0046949 | fatty-acyl-CoA biosynthetic process | 11 | 20 | 8.53e-01 |
| GO:BP | GO:0048254 | snoRNA localization | 1 | 5 | 8.53e-01 |
| GO:BP | GO:1902948 | negative regulation of tau-protein kinase activity | 1 | 2 | 8.53e-01 |
| GO:BP | GO:1900168 | positive regulation of glial cell-derived neurotrophic factor production | 1 | 2 | 8.53e-01 |
| GO:BP | GO:0019065 | receptor-mediated endocytosis of virus by host cell | 2 | 9 | 8.53e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 8.53e-01 |
| GO:BP | GO:1900166 | regulation of glial cell-derived neurotrophic factor production | 1 | 2 | 8.53e-01 |
| GO:BP | GO:1903580 | positive regulation of ATP metabolic process | 1 | 11 | 8.53e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2565 | 8.53e-01 |
| GO:BP | GO:1902988 | neurofibrillary tangle assembly | 1 | 3 | 8.53e-01 |
| GO:BP | GO:1904557 | L-alanine transmembrane transport | 2 | 4 | 8.53e-01 |
| GO:BP | GO:0016571 | histone methylation | 1 | 117 | 8.53e-01 |
| GO:BP | GO:0048755 | branching morphogenesis of a nerve | 1 | 8 | 8.53e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 2 | 5 | 8.53e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 2 | 5 | 8.53e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 3 | 24 | 8.53e-01 |
| GO:BP | GO:0048682 | sprouting of injured axon | 1 | 2 | 8.54e-01 |
| GO:BP | GO:0071281 | cellular response to iron ion | 1 | 6 | 8.54e-01 |
| GO:BP | GO:0010960 | magnesium ion homeostasis | 4 | 9 | 8.54e-01 |
| GO:BP | GO:0048677 | axon extension involved in regeneration | 1 | 2 | 8.54e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 4 | 118 | 8.54e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 1 | 57 | 8.54e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 1 | 11 | 8.54e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 8.54e-01 |
| GO:BP | GO:0044598 | doxorubicin metabolic process | 1 | 8 | 8.54e-01 |
| GO:BP | GO:0019884 | antigen processing and presentation of exogenous antigen | 2 | 28 | 8.54e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2573 | 8.54e-01 |
| GO:BP | GO:0009306 | protein secretion | 4 | 262 | 8.54e-01 |
| GO:BP | GO:0022417 | protein maturation by protein folding | 3 | 8 | 8.54e-01 |
| GO:BP | GO:0030638 | polyketide metabolic process | 1 | 8 | 8.54e-01 |
| GO:BP | GO:0009068 | aspartate family amino acid catabolic process | 6 | 12 | 8.54e-01 |
| GO:BP | GO:0030647 | aminoglycoside antibiotic metabolic process | 1 | 7 | 8.54e-01 |
| GO:BP | GO:0061113 | pancreas morphogenesis | 1 | 3 | 8.54e-01 |
| GO:BP | GO:0006104 | succinyl-CoA metabolic process | 4 | 9 | 8.54e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 3 | 31 | 8.54e-01 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 5 | 8.55e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 1 | 93 | 8.55e-01 |
| GO:BP | GO:0033091 | positive regulation of immature T cell proliferation | 1 | 2 | 8.55e-01 |
| GO:BP | GO:0045627 | positive regulation of T-helper 1 cell differentiation | 1 | 3 | 8.55e-01 |
| GO:BP | GO:0033092 | positive regulation of immature T cell proliferation in thymus | 1 | 2 | 8.55e-01 |
| GO:BP | GO:0035747 | natural killer cell chemotaxis | 2 | 3 | 8.55e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 172 | 8.55e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 4 | 262 | 8.55e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 1 | 37 | 8.55e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 8 | 26 | 8.55e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 8 | 41 | 8.55e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 24 | 2270 | 8.56e-01 |
| GO:BP | GO:0140331 | aminophospholipid translocation | 2 | 6 | 8.56e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 2 | 2293 | 8.56e-01 |
| GO:BP | GO:0045116 | protein neddylation | 8 | 26 | 8.56e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 4 | 119 | 8.57e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 8.57e-01 |
| GO:BP | GO:0045345 | positive regulation of MHC class I biosynthetic process | 1 | 2 | 8.57e-01 |
| GO:BP | GO:0031652 | positive regulation of heat generation | 2 | 4 | 8.57e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 1 | 123 | 8.57e-01 |
| GO:BP | GO:0003160 | endocardium morphogenesis | 1 | 3 | 8.57e-01 |
| GO:BP | GO:0060842 | arterial endothelial cell differentiation | 1 | 5 | 8.57e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 2 | 173 | 8.57e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 8.57e-01 |
| GO:BP | GO:0097242 | amyloid-beta clearance | 5 | 27 | 8.58e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 7 | 32 | 8.58e-01 |
| GO:BP | GO:0048241 | epinephrine transport | 1 | 4 | 8.58e-01 |
| GO:BP | GO:0035621 | ER to Golgi ceramide transport | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0001781 | neutrophil apoptotic process | 1 | 5 | 8.58e-01 |
| GO:BP | GO:2000338 | regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 8.58e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 2 | 18 | 8.58e-01 |
| GO:BP | GO:0070100 | negative regulation of chemokine-mediated signaling pathway | 1 | 6 | 8.58e-01 |
| GO:BP | GO:0030154 | cell differentiation | 3 | 2958 | 8.58e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 2 | 69 | 8.58e-01 |
| GO:BP | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | 2 | 3 | 8.58e-01 |
| GO:BP | GO:0072566 | chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 8.58e-01 |
| GO:BP | GO:0071608 | macrophage inflammatory protein-1 alpha production | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 3 | 54 | 8.58e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 8.58e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 2 | 74 | 8.58e-01 |
| GO:BP | GO:0046549 | retinal cone cell development | 2 | 5 | 8.59e-01 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 5 | 12 | 8.59e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 8 | 168 | 8.59e-01 |
| GO:BP | GO:0060159 | regulation of dopamine receptor signaling pathway | 1 | 7 | 8.59e-01 |
| GO:BP | GO:0042447 | hormone catabolic process | 1 | 7 | 8.59e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 10 | 62 | 8.59e-01 |
| GO:BP | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1 | 3 | 8.59e-01 |
| GO:BP | GO:0097752 | regulation of DNA stability | 1 | 3 | 8.59e-01 |
| GO:BP | GO:0060736 | prostate gland growth | 3 | 7 | 8.59e-01 |
| GO:BP | GO:0031133 | regulation of axon diameter | 1 | 4 | 8.59e-01 |
| GO:BP | GO:0006865 | amino acid transport | 4 | 102 | 8.59e-01 |
| GO:BP | GO:0034650 | cortisol metabolic process | 1 | 9 | 8.60e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 3 | 42 | 8.60e-01 |
| GO:BP | GO:0032351 | negative regulation of hormone metabolic process | 1 | 10 | 8.60e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 1 | 9 | 8.60e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 2 | 13 | 8.60e-01 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 1 | 5 | 8.60e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 8.60e-01 |
| GO:BP | GO:0071205 | protein localization to juxtaparanode region of axon | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0010669 | epithelial structure maintenance | 1 | 15 | 8.60e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 2 | 194 | 8.60e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 1 | 56 | 8.60e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 1 | 194 | 8.60e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 8.60e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 3 | 72 | 8.60e-01 |
| GO:BP | GO:1904253 | positive regulation of bile acid metabolic process | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0070859 | positive regulation of bile acid biosynthetic process | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0006107 | oxaloacetate metabolic process | 3 | 7 | 8.61e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 3 | 2984 | 8.61e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 1 | 296 | 8.61e-01 |
| GO:BP | GO:1990962 | xenobiotic transport across blood-brain barrier | 1 | 3 | 8.61e-01 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 1 | 12 | 8.61e-01 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 5 | 22 | 8.61e-01 |
| GO:BP | GO:0002729 | positive regulation of natural killer cell cytokine production | 1 | 4 | 8.61e-01 |
| GO:BP | GO:0060215 | primitive hemopoiesis | 3 | 7 | 8.61e-01 |
| GO:BP | GO:0045723 | positive regulation of fatty acid biosynthetic process | 2 | 12 | 8.61e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 8.61e-01 |
| GO:BP | GO:0045653 | negative regulation of megakaryocyte differentiation | 1 | 17 | 8.61e-01 |
| GO:BP | GO:0023052 | signaling | 6 | 4141 | 8.61e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 2 | 52 | 8.61e-01 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 7 | 27 | 8.61e-01 |
| GO:BP | GO:2000405 | negative regulation of T cell migration | 1 | 4 | 8.61e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 3 | 29 | 8.61e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 7 | 15 | 8.61e-01 |
| GO:BP | GO:0070741 | response to interleukin-6 | 5 | 23 | 8.61e-01 |
| GO:BP | GO:0048635 | negative regulation of muscle organ development | 3 | 6 | 8.61e-01 |
| GO:BP | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis | 1 | 7 | 8.62e-01 |
| GO:BP | GO:0060729 | intestinal epithelial structure maintenance | 1 | 4 | 8.62e-01 |
| GO:BP | GO:0015866 | ADP transport | 2 | 6 | 8.62e-01 |
| GO:BP | GO:0021699 | cerebellar cortex maturation | 1 | 3 | 8.62e-01 |
| GO:BP | GO:0021590 | cerebellum maturation | 1 | 3 | 8.62e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 8.62e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 8.62e-01 |
| GO:BP | GO:0016485 | protein processing | 4 | 183 | 8.62e-01 |
| GO:BP | GO:0051709 | regulation of killing of cells of another organism | 2 | 3 | 8.63e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 3 | 10 | 8.63e-01 |
| GO:BP | GO:0051956 | negative regulation of amino acid transport | 3 | 10 | 8.63e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 2 | 105 | 8.63e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 6 | 1564 | 8.63e-01 |
| GO:BP | GO:0048840 | otolith development | 1 | 6 | 8.63e-01 |
| GO:BP | GO:1905867 | epididymis development | 1 | 2 | 8.63e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 11 | 62 | 8.63e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 1 | 137 | 8.63e-01 |
| GO:BP | GO:0072156 | distal tubule morphogenesis | 1 | 3 | 8.63e-01 |
| GO:BP | GO:0061141 | lung ciliated cell differentiation | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0120253 | hydrocarbon catabolic process | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0016116 | carotenoid metabolic process | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0042214 | terpene metabolic process | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0019676 | ammonia assimilation cycle | 1 | 2 | 8.63e-01 |
| GO:BP | GO:0016108 | tetraterpenoid metabolic process | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0042450 | arginine biosynthetic process via ornithine | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 2 | 29 | 8.63e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 3 | 8 | 8.63e-01 |
| GO:BP | GO:0071910 | determination of liver left/right asymmetry | 1 | 4 | 8.63e-01 |
| GO:BP | GO:0021884 | forebrain neuron development | 2 | 16 | 8.63e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 6 | 15 | 8.63e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 3 | 15 | 8.64e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 4 | 4292 | 8.64e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 3 | 70 | 8.64e-01 |
| GO:BP | GO:0019853 | L-ascorbic acid biosynthetic process | 1 | 3 | 8.64e-01 |
| GO:BP | GO:0048550 | negative regulation of pinocytosis | 1 | 4 | 8.64e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 3 | 70 | 8.64e-01 |
| GO:BP | GO:1901336 | lactone biosynthetic process | 1 | 3 | 8.64e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 13 | 33 | 8.64e-01 |
| GO:BP | GO:0000338 | protein deneddylation | 3 | 11 | 8.64e-01 |
| GO:BP | GO:0043306 | positive regulation of mast cell degranulation | 5 | 10 | 8.64e-01 |
| GO:BP | GO:0033008 | positive regulation of mast cell activation involved in immune response | 5 | 10 | 8.64e-01 |
| GO:BP | GO:0097021 | lymphocyte migration into lymphoid organs | 2 | 6 | 8.64e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 1 | 36 | 8.64e-01 |
| GO:BP | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway | 1 | 2 | 8.64e-01 |
| GO:BP | GO:1990959 | eosinophil homeostasis | 1 | 2 | 8.64e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 63 | 192 | 8.65e-01 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 2 | 8 | 8.65e-01 |
| GO:BP | GO:0106071 | positive regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 3 | 8.65e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 185 | 8.65e-01 |
| GO:BP | GO:0042048 | olfactory behavior | 3 | 7 | 8.66e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 26 | 62 | 8.66e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 2 | 5 | 8.66e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 8.66e-01 |
| GO:BP | GO:0009313 | oligosaccharide catabolic process | 3 | 11 | 8.66e-01 |
| GO:BP | GO:0032594 | protein transport within lipid bilayer | 2 | 6 | 8.67e-01 |
| GO:BP | GO:0048633 | positive regulation of skeletal muscle tissue growth | 1 | 6 | 8.67e-01 |
| GO:BP | GO:0033034 | positive regulation of myeloid cell apoptotic process | 2 | 6 | 8.67e-01 |
| GO:BP | GO:0022600 | digestive system process | 5 | 54 | 8.67e-01 |
| GO:BP | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell | 1 | 2 | 8.67e-01 |
| GO:BP | GO:0006011 | UDP-glucose metabolic process | 2 | 6 | 8.67e-01 |
| GO:BP | GO:0009812 | flavonoid metabolic process | 1 | 4 | 8.67e-01 |
| GO:BP | GO:1904690 | positive regulation of cytoplasmic translational initiation | 2 | 5 | 8.67e-01 |
| GO:BP | GO:1900119 | positive regulation of execution phase of apoptosis | 1 | 7 | 8.67e-01 |
| GO:BP | GO:0051693 | actin filament capping | 1 | 34 | 8.67e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 18 | 73 | 8.67e-01 |
| GO:BP | GO:0043132 | NAD transport | 1 | 3 | 8.67e-01 |
| GO:BP | GO:0032687 | negative regulation of interferon-alpha production | 1 | 2 | 8.67e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 23 | 484 | 8.68e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 8.68e-01 |
| GO:BP | GO:1901387 | positive regulation of voltage-gated calcium channel activity | 1 | 13 | 8.69e-01 |
| GO:BP | GO:0034344 | regulation of type III interferon production | 1 | 3 | 8.69e-01 |
| GO:BP | GO:1904959 | regulation of cytochrome-c oxidase activity | 2 | 7 | 8.69e-01 |
| GO:BP | GO:0034343 | type III interferon production | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0045124 | regulation of bone resorption | 6 | 24 | 8.69e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 8 | 220 | 8.69e-01 |
| GO:BP | GO:0016056 | rhodopsin mediated signaling pathway | 1 | 6 | 8.69e-01 |
| GO:BP | GO:0008582 | regulation of synaptic assembly at neuromuscular junction | 2 | 4 | 8.69e-01 |
| GO:BP | GO:0033083 | regulation of immature T cell proliferation | 2 | 6 | 8.70e-01 |
| GO:BP | GO:0033084 | regulation of immature T cell proliferation in thymus | 2 | 6 | 8.70e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 3 | 6 | 8.70e-01 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 4 | 14 | 8.70e-01 |
| GO:BP | GO:0030047 | actin modification | 2 | 5 | 8.70e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 2 | 44 | 8.70e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 8.70e-01 |
| GO:BP | GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | 8 | 14 | 8.70e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 8.70e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 6 | 1602 | 8.70e-01 |
| GO:BP | GO:0030050 | vesicle transport along actin filament | 6 | 13 | 8.70e-01 |
| GO:BP | GO:0010706 | ganglioside biosynthetic process via lactosylceramide | 2 | 5 | 8.70e-01 |
| GO:BP | GO:0060011 | Sertoli cell proliferation | 1 | 4 | 8.70e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 2 | 15 | 8.70e-01 |
| GO:BP | GO:0070327 | thyroid hormone transport | 1 | 7 | 8.71e-01 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 1 | 6 | 8.71e-01 |
| GO:BP | GO:0002838 | negative regulation of immune response to tumor cell | 1 | 2 | 8.71e-01 |
| GO:BP | GO:0002835 | negative regulation of response to tumor cell | 1 | 2 | 8.71e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 258 | 8.71e-01 |
| GO:BP | GO:0042355 | L-fucose catabolic process | 1 | 3 | 8.71e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 13 | 8.71e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 8.71e-01 |
| GO:BP | GO:0019317 | fucose catabolic process | 1 | 3 | 8.71e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 20 | 66 | 8.71e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 7 | 19 | 8.71e-01 |
| GO:BP | GO:0043686 | co-translational protein modification | 1 | 3 | 8.72e-01 |
| GO:BP | GO:0006528 | asparagine metabolic process | 1 | 5 | 8.72e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 9 | 295 | 8.72e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 25 | 512 | 8.72e-01 |
| GO:BP | GO:1904340 | positive regulation of dopaminergic neuron differentiation | 1 | 3 | 8.72e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 4 | 98 | 8.72e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 2 | 447 | 8.72e-01 |
| GO:BP | GO:0033033 | negative regulation of myeloid cell apoptotic process | 1 | 10 | 8.72e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 1 | 22 | 8.72e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 3 | 26 | 8.72e-01 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 1 | 4 | 8.72e-01 |
| GO:BP | GO:0046784 | viral mRNA export from host cell nucleus | 1 | 6 | 8.72e-01 |
| GO:BP | GO:1903596 | regulation of gap junction assembly | 2 | 8 | 8.72e-01 |
| GO:BP | GO:0035270 | endocrine system development | 2 | 86 | 8.72e-01 |
| GO:BP | GO:1904844 | response to L-glutamine | 1 | 3 | 8.73e-01 |
| GO:BP | GO:0048538 | thymus development | 3 | 35 | 8.73e-01 |
| GO:BP | GO:0051665 | membrane raft localization | 2 | 4 | 8.73e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 18 | 344 | 8.73e-01 |
| GO:BP | GO:0042221 | response to chemical | 3 | 2559 | 8.73e-01 |
| GO:BP | GO:0051459 | regulation of corticotropin secretion | 1 | 3 | 8.73e-01 |
| GO:BP | GO:0035483 | gastric emptying | 1 | 2 | 8.73e-01 |
| GO:BP | GO:0034334 | adherens junction maintenance | 2 | 6 | 8.73e-01 |
| GO:BP | GO:0071803 | positive regulation of podosome assembly | 1 | 6 | 8.73e-01 |
| GO:BP | GO:0019464 | glycine decarboxylation via glycine cleavage system | 2 | 3 | 8.73e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 1 | 50 | 8.73e-01 |
| GO:BP | GO:0002826 | negative regulation of T-helper 1 type immune response | 1 | 3 | 8.73e-01 |
| GO:BP | GO:0060235 | lens induction in camera-type eye | 1 | 3 | 8.74e-01 |
| GO:BP | GO:0006353 | DNA-templated transcription termination | 1 | 22 | 8.74e-01 |
| GO:BP | GO:0031998 | regulation of fatty acid beta-oxidation | 7 | 16 | 8.74e-01 |
| GO:BP | GO:0001816 | cytokine production | 2 | 451 | 8.75e-01 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 4 | 27 | 8.75e-01 |
| GO:BP | GO:0002921 | negative regulation of humoral immune response | 3 | 10 | 8.76e-01 |
| GO:BP | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration | 1 | 4 | 8.76e-01 |
| GO:BP | GO:0072385 | minus-end-directed organelle transport along microtubule | 2 | 5 | 8.76e-01 |
| GO:BP | GO:0007340 | acrosome reaction | 5 | 20 | 8.76e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 21 | 72 | 8.76e-01 |
| GO:BP | GO:0061038 | uterus morphogenesis | 2 | 5 | 8.77e-01 |
| GO:BP | GO:0001510 | RNA methylation | 39 | 87 | 8.77e-01 |
| GO:BP | GO:0019477 | L-lysine catabolic process | 1 | 4 | 8.77e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 11 | 62 | 8.77e-01 |
| GO:BP | GO:0019058 | viral life cycle | 31 | 244 | 8.77e-01 |
| GO:BP | GO:0046440 | L-lysine metabolic process | 1 | 4 | 8.77e-01 |
| GO:BP | GO:0019474 | L-lysine catabolic process to acetyl-CoA | 1 | 4 | 8.77e-01 |
| GO:BP | GO:0150031 | regulation of protein localization to lysosome | 1 | 3 | 8.77e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 1 | 10 | 8.77e-01 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 1 | 13 | 8.77e-01 |
| GO:BP | GO:1903061 | positive regulation of protein lipidation | 1 | 4 | 8.77e-01 |
| GO:BP | GO:0050747 | positive regulation of lipoprotein metabolic process | 1 | 4 | 8.77e-01 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 1 | 14 | 8.77e-01 |
| GO:BP | GO:1900038 | negative regulation of cellular response to hypoxia | 2 | 8 | 8.77e-01 |
| GO:BP | GO:1904177 | regulation of adipose tissue development | 1 | 8 | 8.77e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 16 | 57 | 8.78e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 4 | 3386 | 8.78e-01 |
| GO:BP | GO:0006714 | sesquiterpenoid metabolic process | 1 | 2 | 8.78e-01 |
| GO:BP | GO:2000328 | regulation of T-helper 17 cell lineage commitment | 1 | 1 | 8.78e-01 |
| GO:BP | GO:0009725 | response to hormone | 2 | 642 | 8.78e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 3 | 10 | 8.78e-01 |
| GO:BP | GO:0010753 | positive regulation of cGMP-mediated signaling | 1 | 3 | 8.79e-01 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 1 | 11 | 8.79e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 8.79e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 8.79e-01 |
| GO:BP | GO:1903288 | positive regulation of potassium ion import across plasma membrane | 2 | 7 | 8.79e-01 |
| GO:BP | GO:0072540 | T-helper 17 cell lineage commitment | 2 | 3 | 8.79e-01 |
| GO:BP | GO:0060012 | synaptic transmission, glycinergic | 1 | 2 | 8.79e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 23 | 88 | 8.79e-01 |
| GO:BP | GO:1902644 | tertiary alcohol metabolic process | 2 | 17 | 8.79e-01 |
| GO:BP | GO:0003176 | aortic valve development | 7 | 37 | 8.79e-01 |
| GO:BP | GO:0018242 | protein O-linked glycosylation via serine | 1 | 7 | 8.79e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 2 | 75 | 8.79e-01 |
| GO:BP | GO:0021682 | nerve maturation | 1 | 2 | 8.79e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 6 | 1604 | 8.79e-01 |
| GO:BP | GO:0021781 | glial cell fate commitment | 3 | 5 | 8.79e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 2 | 7 | 8.80e-01 |
| GO:BP | GO:0031179 | peptide modification | 1 | 4 | 8.80e-01 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 4 | 24 | 8.80e-01 |
| GO:BP | GO:0048936 | peripheral nervous system neuron axonogenesis | 1 | 4 | 8.80e-01 |
| GO:BP | GO:0010842 | retina layer formation | 3 | 16 | 8.80e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 11 | 64 | 8.80e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 9 | 47 | 8.80e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 3 | 232 | 8.80e-01 |
| GO:BP | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 3 | 8.80e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 4 | 14 | 8.81e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 418 | 8.81e-01 |
| GO:BP | GO:0046940 | nucleoside monophosphate phosphorylation | 2 | 9 | 8.81e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 2 | 60 | 8.81e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 2 | 60 | 8.81e-01 |
| GO:BP | GO:0034204 | lipid translocation | 1 | 38 | 8.81e-01 |
| GO:BP | GO:0060399 | positive regulation of growth hormone receptor signaling pathway | 1 | 2 | 8.81e-01 |
| GO:BP | GO:0070495 | negative regulation of thrombin-activated receptor signaling pathway | 1 | 3 | 8.81e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 2 | 58 | 8.81e-01 |
| GO:BP | GO:0032119 | sequestering of zinc ion | 2 | 5 | 8.81e-01 |
| GO:BP | GO:0070494 | regulation of thrombin-activated receptor signaling pathway | 1 | 3 | 8.81e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 8.81e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 5 | 20 | 8.81e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 2 | 18 | 8.81e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 2 | 1836 | 8.81e-01 |
| GO:BP | GO:0038195 | urokinase plasminogen activator signaling pathway | 1 | 2 | 8.82e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 3 | 28 | 8.82e-01 |
| GO:BP | GO:0043353 | enucleate erythrocyte differentiation | 2 | 9 | 8.82e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 4 | 4417 | 8.82e-01 |
| GO:BP | GO:1903942 | positive regulation of respiratory gaseous exchange | 1 | 1 | 8.82e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 3 | 7 | 8.82e-01 |
| GO:BP | GO:0090290 | positive regulation of osteoclast proliferation | 1 | 1 | 8.82e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 2 | 211 | 8.82e-01 |
| GO:BP | GO:0001773 | myeloid dendritic cell activation | 1 | 12 | 8.82e-01 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 1 | 3 | 8.82e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 3 | 112 | 8.82e-01 |
| GO:BP | GO:0072215 | regulation of metanephros development | 1 | 2 | 8.82e-01 |
| GO:BP | GO:0051005 | negative regulation of lipoprotein lipase activity | 2 | 4 | 8.82e-01 |
| GO:BP | GO:0090192 | regulation of glomerulus development | 1 | 4 | 8.82e-01 |
| GO:BP | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 17 | 8.82e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 1 | 32 | 8.82e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 3 | 113 | 8.82e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 9 | 61 | 8.82e-01 |
| GO:BP | GO:0000478 | endonucleolytic cleavage involved in rRNA processing | 3 | 17 | 8.82e-01 |
| GO:BP | GO:0140367 | antibacterial innate immune response | 1 | 2 | 8.82e-01 |
| GO:BP | GO:0002266 | follicular dendritic cell activation | 1 | 2 | 8.82e-01 |
| GO:BP | GO:0032382 | positive regulation of intracellular sterol transport | 1 | 3 | 8.82e-01 |
| GO:BP | GO:0032385 | positive regulation of intracellular cholesterol transport | 1 | 3 | 8.82e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 4 | 29 | 8.82e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 4 | 8 | 8.82e-01 |
| GO:BP | GO:1905600 | regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 3 | 8.82e-01 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 8.82e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 4 | 28 | 8.82e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 20 | 154 | 8.83e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 52 | 139 | 8.83e-01 |
| GO:BP | GO:0019230 | proprioception | 1 | 3 | 8.83e-01 |
| GO:BP | GO:1904398 | positive regulation of neuromuscular junction development | 1 | 2 | 8.83e-01 |
| GO:BP | GO:1904466 | positive regulation of matrix metallopeptidase secretion | 1 | 2 | 8.83e-01 |
| GO:BP | GO:0070430 | positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 2 | 8.83e-01 |
| GO:BP | GO:0051935 | glutamate reuptake | 2 | 4 | 8.83e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 8.83e-01 |
| GO:BP | GO:0002840 | regulation of T cell mediated immune response to tumor cell | 3 | 5 | 8.83e-01 |
| GO:BP | GO:1905908 | positive regulation of amyloid fibril formation | 1 | 5 | 8.83e-01 |
| GO:BP | GO:0016046 | detection of fungus | 1 | 1 | 8.83e-01 |
| GO:BP | GO:0014889 | muscle atrophy | 2 | 9 | 8.83e-01 |
| GO:BP | GO:0044650 | adhesion of symbiont to host cell | 1 | 9 | 8.83e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 2 | 4 | 8.85e-01 |
| GO:BP | GO:0045329 | carnitine biosynthetic process | 1 | 3 | 8.85e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 1 | 27 | 8.85e-01 |
| GO:BP | GO:1900150 | regulation of defense response to fungus | 1 | 3 | 8.85e-01 |
| GO:BP | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway | 2 | 4 | 8.85e-01 |
| GO:BP | GO:0003383 | apical constriction | 1 | 4 | 8.85e-01 |
| GO:BP | GO:0015844 | monoamine transport | 2 | 45 | 8.85e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 2 | 119 | 8.86e-01 |
| GO:BP | GO:0051138 | positive regulation of NK T cell differentiation | 1 | 3 | 8.86e-01 |
| GO:BP | GO:0019079 | viral genome replication | 28 | 101 | 8.86e-01 |
| GO:BP | GO:0006413 | translational initiation | 2 | 116 | 8.86e-01 |
| GO:BP | GO:0002370 | natural killer cell cytokine production | 1 | 4 | 8.86e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 8.86e-01 |
| GO:BP | GO:0030043 | actin filament fragmentation | 1 | 4 | 8.86e-01 |
| GO:BP | GO:0097028 | dendritic cell differentiation | 2 | 23 | 8.86e-01 |
| GO:BP | GO:0002727 | regulation of natural killer cell cytokine production | 1 | 4 | 8.86e-01 |
| GO:BP | GO:0007614 | short-term memory | 4 | 9 | 8.86e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 8.86e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 5 | 33 | 8.86e-01 |
| GO:BP | GO:0036260 | RNA capping | 6 | 20 | 8.86e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 2 | 17 | 8.86e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 1 | 28 | 8.86e-01 |
| GO:BP | GO:0070672 | response to interleukin-15 | 2 | 3 | 8.88e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 1 | 199 | 8.88e-01 |
| GO:BP | GO:0071344 | diphosphate metabolic process | 1 | 2 | 8.88e-01 |
| GO:BP | GO:0071224 | cellular response to peptidoglycan | 1 | 2 | 8.88e-01 |
| GO:BP | GO:0070671 | response to interleukin-12 | 1 | 6 | 8.88e-01 |
| GO:BP | GO:0034138 | toll-like receptor 3 signaling pathway | 7 | 12 | 8.88e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 8.88e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 1 | 6 | 8.88e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 4 | 16 | 8.88e-01 |
| GO:BP | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity | 3 | 12 | 8.88e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 140 | 1345 | 8.88e-01 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 5 | 9 | 8.88e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 9 | 96 | 8.88e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 5 | 68 | 8.88e-01 |
| GO:BP | GO:0042308 | negative regulation of protein import into nucleus | 4 | 13 | 8.88e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 1 | 25 | 8.88e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 8.88e-01 |
| GO:BP | GO:0020027 | hemoglobin metabolic process | 6 | 16 | 8.89e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 23 | 88 | 8.89e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 8.89e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 2 | 62 | 8.89e-01 |
| GO:BP | GO:0071867 | response to monoamine | 2 | 62 | 8.89e-01 |
| GO:BP | GO:1904700 | granulosa cell apoptotic process | 1 | 2 | 8.90e-01 |
| GO:BP | GO:1904708 | regulation of granulosa cell apoptotic process | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0007030 | Golgi organization | 3 | 137 | 8.90e-01 |
| GO:BP | GO:0031944 | negative regulation of glucocorticoid metabolic process | 1 | 5 | 8.90e-01 |
| GO:BP | GO:0031947 | negative regulation of glucocorticoid biosynthetic process | 1 | 5 | 8.90e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 16 | 81 | 8.91e-01 |
| GO:BP | GO:0006369 | termination of RNA polymerase II transcription | 1 | 8 | 8.91e-01 |
| GO:BP | GO:1903385 | regulation of homophilic cell adhesion | 1 | 3 | 8.91e-01 |
| GO:BP | GO:0030728 | ovulation | 1 | 14 | 8.91e-01 |
| GO:BP | GO:0002001 | renin secretion into blood stream | 2 | 4 | 8.91e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 1 | 122 | 8.91e-01 |
| GO:BP | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure | 2 | 4 | 8.91e-01 |
| GO:BP | GO:0016115 | terpenoid catabolic process | 1 | 4 | 8.91e-01 |
| GO:BP | GO:0018026 | peptidyl-lysine monomethylation | 1 | 11 | 8.91e-01 |
| GO:BP | GO:0002278 | eosinophil activation involved in immune response | 2 | 4 | 8.91e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 1 | 20 | 8.91e-01 |
| GO:BP | GO:0034694 | response to prostaglandin | 1 | 21 | 8.91e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 6 | 16 | 8.91e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 1 | 32 | 8.91e-01 |
| GO:BP | GO:0043308 | eosinophil degranulation | 2 | 4 | 8.91e-01 |
| GO:BP | GO:0045113 | regulation of integrin biosynthetic process | 1 | 2 | 8.91e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 6 | 16 | 8.91e-01 |
| GO:BP | GO:1990705 | cholangiocyte proliferation | 1 | 3 | 8.91e-01 |
| GO:BP | GO:2000653 | regulation of genetic imprinting | 1 | 4 | 8.91e-01 |
| GO:BP | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway | 2 | 12 | 8.91e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 9 | 155 | 8.91e-01 |
| GO:BP | GO:0035066 | positive regulation of histone acetylation | 12 | 25 | 8.91e-01 |
| GO:BP | GO:1903332 | regulation of protein folding | 3 | 8 | 8.91e-01 |
| GO:BP | GO:0032693 | negative regulation of interleukin-10 production | 1 | 7 | 8.92e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 8.92e-01 |
| GO:BP | GO:0061975 | articular cartilage development | 1 | 3 | 8.92e-01 |
| GO:BP | GO:0006556 | S-adenosylmethionine biosynthetic process | 1 | 3 | 8.92e-01 |
| GO:BP | GO:0090084 | negative regulation of inclusion body assembly | 4 | 10 | 8.92e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 22 | 40 | 8.92e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 8.92e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 1 | 10 | 8.92e-01 |
| GO:BP | GO:2000416 | regulation of eosinophil migration | 2 | 2 | 8.92e-01 |
| GO:BP | GO:0071486 | cellular response to high light intensity | 1 | 1 | 8.92e-01 |
| GO:BP | GO:0090713 | immunological memory process | 1 | 7 | 8.92e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 1 | 348 | 8.93e-01 |
| GO:BP | GO:0002638 | negative regulation of immunoglobulin production | 2 | 5 | 8.93e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 8.93e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 2 | 17 | 8.93e-01 |
| GO:BP | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | 1 | 11 | 8.93e-01 |
| GO:BP | GO:0043950 | positive regulation of cAMP-mediated signaling | 4 | 5 | 8.93e-01 |
| GO:BP | GO:0040019 | positive regulation of embryonic development | 8 | 17 | 8.93e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 3 | 59 | 8.93e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 8.93e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 8.94e-01 |
| GO:BP | GO:0006734 | NADH metabolic process | 3 | 12 | 8.94e-01 |
| GO:BP | GO:0033523 | histone H2B ubiquitination | 3 | 10 | 8.94e-01 |
| GO:BP | GO:0060406 | positive regulation of penile erection | 1 | 3 | 8.94e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 2 | 12 | 8.94e-01 |
| GO:BP | GO:0030488 | tRNA methylation | 17 | 42 | 8.94e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 8.94e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 6 | 47 | 8.95e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 2 | 124 | 8.95e-01 |
| GO:BP | GO:0046476 | glycosylceramide biosynthetic process | 3 | 6 | 8.95e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 8.95e-01 |
| GO:BP | GO:0032929 | negative regulation of superoxide anion generation | 1 | 1 | 8.95e-01 |
| GO:BP | GO:0070475 | rRNA base methylation | 1 | 8 | 8.96e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 8 | 24 | 8.96e-01 |
| GO:BP | GO:0046167 | glycerol-3-phosphate biosynthetic process | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 3 | 8.96e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 8.96e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 4 | 37 | 8.96e-01 |
| GO:BP | GO:0097638 | L-arginine import across plasma membrane | 2 | 4 | 8.96e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 17 | 84 | 8.96e-01 |
| GO:BP | GO:1905604 | negative regulation of blood-brain barrier permeability | 1 | 2 | 8.96e-01 |
| GO:BP | GO:0006776 | vitamin A metabolic process | 2 | 3 | 8.96e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 16 | 38 | 8.97e-01 |
| GO:BP | GO:1903625 | negative regulation of DNA catabolic process | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0150147 | cell-cell junction disassembly | 1 | 4 | 8.97e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 25 | 527 | 8.97e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 26 | 551 | 8.97e-01 |
| GO:BP | GO:0070346 | positive regulation of fat cell proliferation | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 3 | 11 | 8.97e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 3 | 11 | 8.97e-01 |
| GO:BP | GO:1990809 | endoplasmic reticulum tubular network membrane organization | 1 | 4 | 8.97e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 8.97e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 8.97e-01 |
| GO:BP | GO:2000380 | regulation of mesoderm development | 2 | 3 | 8.99e-01 |
| GO:BP | GO:0140252 | regulation protein catabolic process at postsynapse | 1 | 3 | 8.99e-01 |
| GO:BP | GO:1905289 | regulation of CAMKK-AMPK signaling cascade | 1 | 2 | 8.99e-01 |
| GO:BP | GO:0006382 | adenosine to inosine editing | 2 | 4 | 8.99e-01 |
| GO:BP | GO:0060629 | regulation of homologous chromosome segregation | 1 | 1 | 8.99e-01 |
| GO:BP | GO:0061762 | CAMKK-AMPK signaling cascade | 2 | 5 | 8.99e-01 |
| GO:BP | GO:0048643 | positive regulation of skeletal muscle tissue development | 1 | 14 | 8.99e-01 |
| GO:BP | GO:0070949 | regulation of neutrophil mediated killing of symbiont cell | 1 | 1 | 8.99e-01 |
| GO:BP | GO:1900133 | regulation of renin secretion into blood stream | 1 | 2 | 8.99e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 39 | 148 | 8.99e-01 |
| GO:BP | GO:2000307 | regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 9.00e-01 |
| GO:BP | GO:0072535 | tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 9.00e-01 |
| GO:BP | GO:0060872 | semicircular canal development | 3 | 7 | 9.00e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 7 | 57 | 9.00e-01 |
| GO:BP | GO:0001574 | ganglioside biosynthetic process | 1 | 14 | 9.00e-01 |
| GO:BP | GO:2000412 | positive regulation of thymocyte migration | 1 | 1 | 9.00e-01 |
| GO:BP | GO:0045950 | negative regulation of mitotic recombination | 1 | 2 | 9.00e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 1 | 49 | 9.00e-01 |
| GO:BP | GO:0007060 | male meiosis chromosome segregation | 1 | 1 | 9.00e-01 |
| GO:BP | GO:1903350 | response to dopamine | 1 | 48 | 9.00e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 9.00e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 7 | 5017 | 9.00e-01 |
| GO:BP | GO:0000962 | positive regulation of mitochondrial RNA catabolic process | 1 | 3 | 9.00e-01 |
| GO:BP | GO:1903003 | positive regulation of protein deubiquitination | 2 | 6 | 9.00e-01 |
| GO:BP | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | 3 | 9.01e-01 |
| GO:BP | GO:0034375 | high-density lipoprotein particle remodeling | 2 | 11 | 9.01e-01 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 14 | 28 | 9.01e-01 |
| GO:BP | GO:0044417 | translocation of molecules into host | 1 | 7 | 9.01e-01 |
| GO:BP | GO:0035709 | memory T cell activation | 1 | 4 | 9.01e-01 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 1 | 12 | 9.02e-01 |
| GO:BP | GO:0099542 | trans-synaptic signaling by endocannabinoid | 2 | 6 | 9.02e-01 |
| GO:BP | GO:0099541 | trans-synaptic signaling by lipid | 2 | 6 | 9.02e-01 |
| GO:BP | GO:0006935 | chemotaxis | 3 | 389 | 9.02e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 2 | 18 | 9.02e-01 |
| GO:BP | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 3 | 10 | 9.02e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0090292 | nuclear matrix anchoring at nuclear membrane | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0043578 | nuclear matrix organization | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 3 | 19 | 9.03e-01 |
| GO:BP | GO:0090026 | positive regulation of monocyte chemotaxis | 1 | 7 | 9.03e-01 |
| GO:BP | GO:0014052 | regulation of gamma-aminobutyric acid secretion | 3 | 5 | 9.03e-01 |
| GO:BP | GO:0010899 | regulation of phosphatidylcholine catabolic process | 1 | 3 | 9.03e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 1 | 49 | 9.03e-01 |
| GO:BP | GO:0042330 | taxis | 3 | 389 | 9.03e-01 |
| GO:BP | GO:0042886 | amide transport | 37 | 235 | 9.03e-01 |
| GO:BP | GO:0048251 | elastic fiber assembly | 1 | 12 | 9.03e-01 |
| GO:BP | GO:2000503 | positive regulation of natural killer cell chemotaxis | 1 | 1 | 9.03e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 3 | 15 | 9.03e-01 |
| GO:BP | GO:1990402 | embryonic liver development | 1 | 3 | 9.03e-01 |
| GO:BP | GO:0032790 | ribosome disassembly | 3 | 11 | 9.03e-01 |
| GO:BP | GO:1990051 | activation of protein kinase C activity | 1 | 3 | 9.03e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 9.03e-01 |
| GO:BP | GO:0006953 | acute-phase response | 1 | 18 | 9.04e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 9.04e-01 |
| GO:BP | GO:0150007 | clathrin-dependent synaptic vesicle endocytosis | 1 | 2 | 9.04e-01 |
| GO:BP | GO:2000381 | negative regulation of mesoderm development | 1 | 1 | 9.04e-01 |
| GO:BP | GO:1902811 | positive regulation of skeletal muscle fiber differentiation | 1 | 2 | 9.04e-01 |
| GO:BP | GO:0032502 | developmental process | 6 | 4494 | 9.04e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 9.04e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 25 | 531 | 9.04e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 3 | 7 | 9.04e-01 |
| GO:BP | GO:1900625 | positive regulation of monocyte aggregation | 1 | 3 | 9.05e-01 |
| GO:BP | GO:1900623 | regulation of monocyte aggregation | 1 | 3 | 9.05e-01 |
| GO:BP | GO:0075509 | endocytosis involved in viral entry into host cell | 2 | 11 | 9.05e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 14 | 496 | 9.05e-01 |
| GO:BP | GO:1904020 | regulation of G protein-coupled receptor internalization | 1 | 3 | 9.05e-01 |
| GO:BP | GO:0031204 | post-translational protein targeting to membrane, translocation | 1 | 7 | 9.05e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 14 | 37 | 9.05e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 5 | 15 | 9.05e-01 |
| GO:BP | GO:0002438 | acute inflammatory response to antigenic stimulus | 3 | 10 | 9.05e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 9.05e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 3 | 51 | 9.05e-01 |
| GO:BP | GO:0140469 | GCN2-mediated signaling | 1 | 3 | 9.05e-01 |
| GO:BP | GO:0006808 | regulation of nitrogen utilization | 1 | 2 | 9.05e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 1 | 12 | 9.05e-01 |
| GO:BP | GO:0002904 | positive regulation of B cell apoptotic process | 1 | 2 | 9.05e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 2 | 142 | 9.06e-01 |
| GO:BP | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 1 | 82 | 9.07e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 17 | 95 | 9.07e-01 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 1 | 82 | 9.07e-01 |
| GO:BP | GO:0010533 | regulation of activation of Janus kinase activity | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 9.07e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 3 | 3730 | 9.07e-01 |
| GO:BP | GO:0042908 | xenobiotic transport | 8 | 30 | 9.07e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 1 | 215 | 9.07e-01 |
| GO:BP | GO:0033004 | negative regulation of mast cell activation | 3 | 4 | 9.07e-01 |
| GO:BP | GO:1905155 | positive regulation of membrane invagination | 1 | 11 | 9.08e-01 |
| GO:BP | GO:0060100 | positive regulation of phagocytosis, engulfment | 1 | 11 | 9.08e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 5 | 29 | 9.08e-01 |
| GO:BP | GO:0030901 | midbrain development | 1 | 69 | 9.08e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 2 | 18 | 9.08e-01 |
| GO:BP | GO:0032715 | negative regulation of interleukin-6 production | 1 | 24 | 9.08e-01 |
| GO:BP | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing | 1 | 5 | 9.08e-01 |
| GO:BP | GO:0032819 | positive regulation of natural killer cell proliferation | 1 | 4 | 9.08e-01 |
| GO:BP | GO:0032847 | regulation of cellular pH reduction | 2 | 5 | 9.08e-01 |
| GO:BP | GO:2001225 | regulation of chloride transport | 2 | 4 | 9.08e-01 |
| GO:BP | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus | 3 | 7 | 9.08e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 2 | 71 | 9.08e-01 |
| GO:BP | GO:0006032 | chitin catabolic process | 2 | 2 | 9.08e-01 |
| GO:BP | GO:0006030 | chitin metabolic process | 2 | 2 | 9.08e-01 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 1 | 34 | 9.08e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 5 | 61 | 9.08e-01 |
| GO:BP | GO:0003165 | Purkinje myocyte development | 1 | 3 | 9.08e-01 |
| GO:BP | GO:0055005 | ventricular cardiac myofibril assembly | 1 | 3 | 9.08e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 1 | 58 | 9.08e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 2 | 4 | 9.08e-01 |
| GO:BP | GO:1990926 | short-term synaptic potentiation | 1 | 4 | 9.08e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 10 | 33 | 9.08e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 9.08e-01 |
| GO:BP | GO:1903711 | spermidine transmembrane transport | 1 | 2 | 9.09e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 1 | 29 | 9.09e-01 |
| GO:BP | GO:0099612 | protein localization to axon | 2 | 6 | 9.09e-01 |
| GO:BP | GO:0061548 | ganglion development | 4 | 12 | 9.09e-01 |
| GO:BP | GO:0060523 | prostate epithelial cord elongation | 1 | 1 | 9.09e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 9.09e-01 |
| GO:BP | GO:0060594 | mammary gland specification | 1 | 3 | 9.09e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 18 | 95 | 9.09e-01 |
| GO:BP | GO:0060648 | mammary gland bud morphogenesis | 1 | 2 | 9.09e-01 |
| GO:BP | GO:0009583 | detection of light stimulus | 1 | 27 | 9.09e-01 |
| GO:BP | GO:0051586 | positive regulation of dopamine uptake involved in synaptic transmission | 1 | 2 | 9.09e-01 |
| GO:BP | GO:0015732 | prostaglandin transport | 1 | 10 | 9.09e-01 |
| GO:BP | GO:0051944 | positive regulation of catecholamine uptake involved in synaptic transmission | 1 | 2 | 9.09e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 1 | 384 | 9.09e-01 |
| GO:BP | GO:0044872 | lipoprotein localization | 1 | 15 | 9.09e-01 |
| GO:BP | GO:0030431 | sleep | 1 | 13 | 9.09e-01 |
| GO:BP | GO:0060684 | epithelial-mesenchymal cell signaling | 2 | 3 | 9.09e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 9.09e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 9.09e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 3 | 19 | 9.09e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 27 | 51 | 9.09e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 7 | 14 | 9.10e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 2 | 539 | 9.10e-01 |
| GO:BP | GO:0060192 | negative regulation of lipase activity | 1 | 13 | 9.10e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 9.10e-01 |
| GO:BP | GO:0018199 | peptidyl-glutamine modification | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0060455 | negative regulation of gastric acid secretion | 1 | 3 | 9.10e-01 |
| GO:BP | GO:2000987 | positive regulation of behavioral fear response | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 4 | 47 | 9.10e-01 |
| GO:BP | GO:1904732 | regulation of electron transfer activity | 2 | 8 | 9.10e-01 |
| GO:BP | GO:0072683 | T cell extravasation | 2 | 8 | 9.10e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 3 | 31 | 9.10e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 4 | 138 | 9.10e-01 |
| GO:BP | GO:0070474 | positive regulation of uterine smooth muscle contraction | 1 | 4 | 9.10e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 4 | 192 | 9.10e-01 |
| GO:BP | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0008078 | mesodermal cell migration | 2 | 3 | 9.10e-01 |
| GO:BP | GO:0006571 | tyrosine biosynthetic process | 1 | 2 | 9.10e-01 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 1 | 8 | 9.10e-01 |
| GO:BP | GO:0046968 | peptide antigen transport | 1 | 1 | 9.10e-01 |
| GO:BP | GO:0110104 | mRNA alternative polyadenylation | 1 | 5 | 9.10e-01 |
| GO:BP | GO:0015074 | DNA integration | 1 | 9 | 9.10e-01 |
| GO:BP | GO:1901078 | negative regulation of relaxation of muscle | 1 | 4 | 9.10e-01 |
| GO:BP | GO:0030073 | insulin secretion | 20 | 141 | 9.10e-01 |
| GO:BP | GO:0019293 | tyrosine biosynthetic process, by oxidation of phenylalanine | 1 | 2 | 9.10e-01 |
| GO:BP | GO:0002019 | regulation of renal output by angiotensin | 1 | 3 | 9.10e-01 |
| GO:BP | GO:0009095 | aromatic amino acid family biosynthetic process, prephenate pathway | 1 | 2 | 9.10e-01 |
| GO:BP | GO:0071800 | podosome assembly | 3 | 13 | 9.10e-01 |
| GO:BP | GO:0002604 | regulation of dendritic cell antigen processing and presentation | 2 | 5 | 9.10e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 5 | 8 | 9.11e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 9.11e-01 |
| GO:BP | GO:0002946 | tRNA C5-cytosine methylation | 1 | 3 | 9.11e-01 |
| GO:BP | GO:0006531 | aspartate metabolic process | 2 | 4 | 9.11e-01 |
| GO:BP | GO:0006677 | glycosylceramide metabolic process | 1 | 13 | 9.11e-01 |
| GO:BP | GO:1903593 | regulation of histamine secretion by mast cell | 1 | 4 | 9.11e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 2 | 465 | 9.11e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 2 | 7 | 9.11e-01 |
| GO:BP | GO:1904684 | negative regulation of metalloendopeptidase activity | 1 | 4 | 9.11e-01 |
| GO:BP | GO:1905246 | negative regulation of aspartic-type peptidase activity | 1 | 3 | 9.11e-01 |
| GO:BP | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport | 1 | 4 | 9.11e-01 |
| GO:BP | GO:1903595 | positive regulation of histamine secretion by mast cell | 1 | 4 | 9.11e-01 |
| GO:BP | GO:0042662 | negative regulation of mesodermal cell fate specification | 1 | 3 | 9.11e-01 |
| GO:BP | GO:1903225 | negative regulation of endodermal cell differentiation | 1 | 3 | 9.11e-01 |
| GO:BP | GO:1905771 | negative regulation of mesodermal cell differentiation | 1 | 3 | 9.11e-01 |
| GO:BP | GO:0034465 | response to carbon monoxide | 1 | 3 | 9.11e-01 |
| GO:BP | GO:0006583 | melanin biosynthetic process from tyrosine | 1 | 1 | 9.11e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 1 | 369 | 9.11e-01 |
| GO:BP | GO:1903278 | positive regulation of sodium ion export across plasma membrane | 1 | 4 | 9.11e-01 |
| GO:BP | GO:1903276 | regulation of sodium ion export across plasma membrane | 1 | 4 | 9.11e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 3 | 33 | 9.12e-01 |
| GO:BP | GO:0019371 | cyclooxygenase pathway | 1 | 7 | 9.12e-01 |
| GO:BP | GO:2000726 | negative regulation of cardiac muscle cell differentiation | 1 | 8 | 9.12e-01 |
| GO:BP | GO:0060161 | positive regulation of dopamine receptor signaling pathway | 1 | 3 | 9.12e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 9.12e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 4 | 10 | 9.12e-01 |
| GO:BP | GO:0050916 | sensory perception of sweet taste | 1 | 3 | 9.12e-01 |
| GO:BP | GO:0072338 | lactam metabolic process | 1 | 2 | 9.12e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 5 | 99 | 9.12e-01 |
| GO:BP | GO:0110089 | regulation of hippocampal neuron apoptotic process | 1 | 2 | 9.12e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 2 | 11 | 9.12e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 78 | 726 | 9.12e-01 |
| GO:BP | GO:0110088 | hippocampal neuron apoptotic process | 1 | 2 | 9.12e-01 |
| GO:BP | GO:0019695 | choline metabolic process | 1 | 4 | 9.12e-01 |
| GO:BP | GO:0043171 | peptide catabolic process | 4 | 16 | 9.13e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 7 | 21 | 9.13e-01 |
| GO:BP | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin | 1 | 6 | 9.13e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 5 | 90 | 9.13e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 4 | 13 | 9.13e-01 |
| GO:BP | GO:0014827 | intestine smooth muscle contraction | 1 | 3 | 9.13e-01 |
| GO:BP | GO:0006546 | glycine catabolic process | 1 | 3 | 9.13e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 10 | 61 | 9.14e-01 |
| GO:BP | GO:1905684 | regulation of plasma membrane repair | 2 | 7 | 9.14e-01 |
| GO:BP | GO:0045923 | positive regulation of fatty acid metabolic process | 4 | 26 | 9.14e-01 |
| GO:BP | GO:0030259 | lipid glycosylation | 1 | 6 | 9.14e-01 |
| GO:BP | GO:0060179 | male mating behavior | 2 | 5 | 9.14e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 9.14e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 3 | 12 | 9.14e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 9.15e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 4 | 91 | 9.16e-01 |
| GO:BP | GO:0043587 | tongue morphogenesis | 1 | 7 | 9.16e-01 |
| GO:BP | GO:0031424 | keratinization | 2 | 13 | 9.16e-01 |
| GO:BP | GO:0042832 | defense response to protozoan | 1 | 10 | 9.16e-01 |
| GO:BP | GO:0006887 | exocytosis | 19 | 256 | 9.16e-01 |
| GO:BP | GO:0007608 | sensory perception of smell | 1 | 16 | 9.17e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 4 | 62 | 9.17e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 13 | 517 | 9.17e-01 |
| GO:BP | GO:1904691 | negative regulation of type B pancreatic cell proliferation | 1 | 2 | 9.17e-01 |
| GO:BP | GO:0032418 | lysosome localization | 4 | 62 | 9.17e-01 |
| GO:BP | GO:0032226 | positive regulation of synaptic transmission, dopaminergic | 1 | 3 | 9.17e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 2 | 19 | 9.17e-01 |
| GO:BP | GO:0035726 | common myeloid progenitor cell proliferation | 1 | 5 | 9.17e-01 |
| GO:BP | GO:0000958 | mitochondrial mRNA catabolic process | 1 | 3 | 9.17e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 2 | 220 | 9.17e-01 |
| GO:BP | GO:0051238 | sequestering of metal ion | 8 | 17 | 9.17e-01 |
| GO:BP | GO:0051454 | intracellular pH elevation | 2 | 4 | 9.17e-01 |
| GO:BP | GO:0006968 | cellular defense response | 2 | 15 | 9.17e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 8 | 27 | 9.17e-01 |
| GO:BP | GO:0018343 | protein farnesylation | 1 | 2 | 9.17e-01 |
| GO:BP | GO:0006704 | glucocorticoid biosynthetic process | 1 | 11 | 9.17e-01 |
| GO:BP | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | 1 | 11 | 9.17e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 13 | 427 | 9.17e-01 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 2 | 11 | 9.17e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 1 | 7 | 9.17e-01 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 1 | 131 | 9.17e-01 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 2 | 10 | 9.17e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 1 | 29 | 9.17e-01 |
| GO:BP | GO:0071671 | regulation of smooth muscle cell chemotaxis | 2 | 4 | 9.17e-01 |
| GO:BP | GO:1904352 | positive regulation of protein catabolic process in the vacuole | 1 | 6 | 9.17e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 3 | 2890 | 9.18e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 3 | 30 | 9.18e-01 |
| GO:BP | GO:0035461 | vitamin transmembrane transport | 1 | 14 | 9.18e-01 |
| GO:BP | GO:0038016 | insulin receptor internalization | 1 | 3 | 9.19e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 3 | 33 | 9.19e-01 |
| GO:BP | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway | 2 | 3 | 9.19e-01 |
| GO:BP | GO:0001709 | cell fate determination | 3 | 25 | 9.19e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 9.19e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 2 | 53 | 9.19e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 963 | 9.19e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 21 | 120 | 9.19e-01 |
| GO:BP | GO:0032225 | regulation of synaptic transmission, dopaminergic | 1 | 5 | 9.19e-01 |
| GO:BP | GO:2000831 | regulation of steroid hormone secretion | 1 | 13 | 9.19e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 5 | 22 | 9.20e-01 |
| GO:BP | GO:0034158 | toll-like receptor 8 signaling pathway | 1 | 3 | 9.20e-01 |
| GO:BP | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response | 1 | 2 | 9.20e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 2 | 4 | 9.20e-01 |
| GO:BP | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress | 1 | 5 | 9.20e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 2 | 168 | 9.20e-01 |
| GO:BP | GO:0048521 | negative regulation of behavior | 2 | 5 | 9.20e-01 |
| GO:BP | GO:0070315 | G1 to G0 transition involved in cell differentiation | 2 | 4 | 9.20e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 2 | 248 | 9.20e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 9.20e-01 |
| GO:BP | GO:1903980 | positive regulation of microglial cell activation | 2 | 2 | 9.20e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 1 | 30 | 9.20e-01 |
| GO:BP | GO:2000297 | negative regulation of synapse maturation | 1 | 1 | 9.20e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 11 | 78 | 9.20e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 9.21e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 4 | 15 | 9.21e-01 |
| GO:BP | GO:0097052 | L-kynurenine metabolic process | 1 | 4 | 9.21e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 2 | 46 | 9.21e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 1 | 55 | 9.21e-01 |
| GO:BP | GO:0019448 | L-cysteine catabolic process | 1 | 3 | 9.21e-01 |
| GO:BP | GO:0009093 | cysteine catabolic process | 1 | 3 | 9.21e-01 |
| GO:BP | GO:0046305 | alkanesulfonate biosynthetic process | 1 | 2 | 9.21e-01 |
| GO:BP | GO:0046439 | L-cysteine metabolic process | 1 | 3 | 9.21e-01 |
| GO:BP | GO:0042412 | taurine biosynthetic process | 1 | 2 | 9.21e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 2 | 8 | 9.21e-01 |
| GO:BP | GO:0030168 | platelet activation | 8 | 91 | 9.21e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 70 | 148 | 9.21e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 3 | 27 | 9.21e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 9.22e-01 |
| GO:BP | GO:0021801 | cerebral cortex radial glia-guided migration | 10 | 22 | 9.22e-01 |
| GO:BP | GO:0022030 | telencephalon glial cell migration | 10 | 22 | 9.22e-01 |
| GO:BP | GO:0021798 | forebrain dorsal/ventral pattern formation | 1 | 2 | 9.22e-01 |
| GO:BP | GO:0009233 | menaquinone metabolic process | 1 | 1 | 9.22e-01 |
| GO:BP | GO:0006679 | glucosylceramide biosynthetic process | 1 | 2 | 9.22e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 1 | 5 | 9.22e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 17 | 159 | 9.22e-01 |
| GO:BP | GO:1905203 | regulation of connective tissue replacement | 1 | 5 | 9.23e-01 |
| GO:BP | GO:0043627 | response to estrogen | 2 | 52 | 9.23e-01 |
| GO:BP | GO:0001839 | neural plate morphogenesis | 2 | 4 | 9.23e-01 |
| GO:BP | GO:0015847 | putrescine transport | 1 | 2 | 9.23e-01 |
| GO:BP | GO:1902603 | carnitine transmembrane transport | 1 | 4 | 9.23e-01 |
| GO:BP | GO:1904688 | regulation of cytoplasmic translational initiation | 2 | 6 | 9.23e-01 |
| GO:BP | GO:0048631 | regulation of skeletal muscle tissue growth | 1 | 7 | 9.23e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 8 | 27 | 9.23e-01 |
| GO:BP | GO:0044406 | adhesion of symbiont to host | 1 | 10 | 9.23e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 2 | 141 | 9.24e-01 |
| GO:BP | GO:0060347 | heart trabecula formation | 5 | 13 | 9.24e-01 |
| GO:BP | GO:0090032 | negative regulation of steroid hormone biosynthetic process | 1 | 6 | 9.24e-01 |
| GO:BP | GO:0048243 | norepinephrine secretion | 1 | 5 | 9.24e-01 |
| GO:BP | GO:0014061 | regulation of norepinephrine secretion | 1 | 5 | 9.24e-01 |
| GO:BP | GO:0006955 | immune response | 3 | 882 | 9.25e-01 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 4 | 17 | 9.25e-01 |
| GO:BP | GO:0140285 | endosome fission | 1 | 3 | 9.25e-01 |
| GO:BP | GO:0043305 | negative regulation of mast cell degranulation | 2 | 2 | 9.25e-01 |
| GO:BP | GO:0045112 | integrin biosynthetic process | 1 | 3 | 9.25e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 8 | 16 | 9.25e-01 |
| GO:BP | GO:0002025 | norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure | 1 | 3 | 9.25e-01 |
| GO:BP | GO:0060439 | trachea morphogenesis | 1 | 7 | 9.25e-01 |
| GO:BP | GO:1990911 | response to psychosocial stress | 1 | 2 | 9.25e-01 |
| GO:BP | GO:0030001 | metal ion transport | 14 | 562 | 9.25e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 9.26e-01 |
| GO:BP | GO:0048733 | sebaceous gland development | 1 | 6 | 9.26e-01 |
| GO:BP | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 1 | 7 | 9.26e-01 |
| GO:BP | GO:0034776 | response to histamine | 3 | 5 | 9.26e-01 |
| GO:BP | GO:0010961 | intracellular magnesium ion homeostasis | 1 | 4 | 9.26e-01 |
| GO:BP | GO:0010984 | regulation of lipoprotein particle clearance | 2 | 12 | 9.26e-01 |
| GO:BP | GO:0099515 | actin filament-based transport | 6 | 15 | 9.26e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 1 | 31 | 9.26e-01 |
| GO:BP | GO:0002424 | T cell mediated immune response to tumor cell | 3 | 6 | 9.26e-01 |
| GO:BP | GO:0046055 | dGMP catabolic process | 1 | 1 | 9.26e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 4 | 95 | 9.26e-01 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 5 | 14 | 9.26e-01 |
| GO:BP | GO:0090160 | Golgi to lysosome transport | 4 | 10 | 9.26e-01 |
| GO:BP | GO:0010360 | negative regulation of anion channel activity | 1 | 2 | 9.26e-01 |
| GO:BP | GO:0006682 | galactosylceramide biosynthetic process | 2 | 4 | 9.26e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 9.26e-01 |
| GO:BP | GO:0019375 | galactolipid biosynthetic process | 2 | 4 | 9.26e-01 |
| GO:BP | GO:1903960 | negative regulation of anion transmembrane transport | 1 | 2 | 9.26e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 1 | 6996 | 9.26e-01 |
| GO:BP | GO:0048712 | negative regulation of astrocyte differentiation | 2 | 9 | 9.27e-01 |
| GO:BP | GO:1901860 | positive regulation of mitochondrial DNA metabolic process | 1 | 3 | 9.27e-01 |
| GO:BP | GO:0044062 | regulation of excretion | 1 | 12 | 9.27e-01 |
| GO:BP | GO:0006844 | acyl carnitine transport | 1 | 2 | 9.27e-01 |
| GO:BP | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity | 2 | 4 | 9.27e-01 |
| GO:BP | GO:0061523 | cilium disassembly | 1 | 4 | 9.27e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 1 | 19 | 9.27e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 2 | 108 | 9.27e-01 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 11 | 9.27e-01 |
| GO:BP | GO:0043416 | regulation of skeletal muscle tissue regeneration | 2 | 5 | 9.27e-01 |
| GO:BP | GO:0043503 | skeletal muscle fiber adaptation | 1 | 3 | 9.27e-01 |
| GO:BP | GO:0070099 | regulation of chemokine-mediated signaling pathway | 1 | 8 | 9.28e-01 |
| GO:BP | GO:1903659 | regulation of complement-dependent cytotoxicity | 1 | 2 | 9.28e-01 |
| GO:BP | GO:0021550 | medulla oblongata development | 1 | 2 | 9.28e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 1 | 57 | 9.28e-01 |
| GO:BP | GO:0002082 | regulation of oxidative phosphorylation | 3 | 19 | 9.28e-01 |
| GO:BP | GO:0009642 | response to light intensity | 1 | 11 | 9.28e-01 |
| GO:BP | GO:0060681 | branch elongation involved in ureteric bud branching | 1 | 3 | 9.28e-01 |
| GO:BP | GO:0015891 | siderophore transport | 1 | 2 | 9.28e-01 |
| GO:BP | GO:0009631 | cold acclimation | 1 | 2 | 9.29e-01 |
| GO:BP | GO:0001562 | response to protozoan | 1 | 11 | 9.29e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 2 | 10 | 9.29e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 5 | 135 | 9.29e-01 |
| GO:BP | GO:0001878 | response to yeast | 1 | 3 | 9.29e-01 |
| GO:BP | GO:0061753 | substrate localization to autophagosome | 1 | 3 | 9.29e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 1 | 33 | 9.29e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 2 | 15 | 9.29e-01 |
| GO:BP | GO:0048752 | semicircular canal morphogenesis | 2 | 5 | 9.29e-01 |
| GO:BP | GO:1903238 | positive regulation of leukocyte tethering or rolling | 1 | 5 | 9.29e-01 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 1 | 18 | 9.29e-01 |
| GO:BP | GO:0048563 | post-embryonic animal organ morphogenesis | 2 | 8 | 9.29e-01 |
| GO:BP | GO:0015917 | aminophospholipid transport | 2 | 8 | 9.29e-01 |
| GO:BP | GO:0050975 | sensory perception of touch | 1 | 2 | 9.29e-01 |
| GO:BP | GO:0060099 | regulation of phagocytosis, engulfment | 1 | 11 | 9.29e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 2 | 58 | 9.29e-01 |
| GO:BP | GO:1905153 | regulation of membrane invagination | 1 | 11 | 9.29e-01 |
| GO:BP | GO:0061589 | calcium activated phosphatidylserine scrambling | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 2 | 11 | 9.30e-01 |
| GO:BP | GO:0031914 | negative regulation of synaptic plasticity | 1 | 2 | 9.30e-01 |
| GO:BP | GO:0061591 | calcium activated galactosylceramide scrambling | 1 | 2 | 9.30e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 2 | 92 | 9.30e-01 |
| GO:BP | GO:0050689 | negative regulation of defense response to virus by host | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 37 | 667 | 9.30e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 3 | 22 | 9.30e-01 |
| GO:BP | GO:0033490 | cholesterol biosynthetic process via lathosterol | 1 | 4 | 9.30e-01 |
| GO:BP | GO:0033489 | cholesterol biosynthetic process via desmosterol | 1 | 4 | 9.30e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0035562 | negative regulation of chromatin binding | 3 | 8 | 9.30e-01 |
| GO:BP | GO:2000321 | positive regulation of T-helper 17 cell differentiation | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 2 | 2035 | 9.30e-01 |
| GO:BP | GO:0071265 | L-methionine biosynthetic process | 1 | 5 | 9.30e-01 |
| GO:BP | GO:0043102 | amino acid salvage | 1 | 5 | 9.30e-01 |
| GO:BP | GO:0071267 | L-methionine salvage | 1 | 5 | 9.30e-01 |
| GO:BP | GO:2001054 | negative regulation of mesenchymal cell apoptotic process | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 9.31e-01 |
| GO:BP | GO:0036301 | macrophage colony-stimulating factor production | 1 | 2 | 9.31e-01 |
| GO:BP | GO:1901256 | regulation of macrophage colony-stimulating factor production | 1 | 2 | 9.31e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 1 | 45 | 9.31e-01 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 1 | 27 | 9.33e-01 |
| GO:BP | GO:0050765 | negative regulation of phagocytosis | 5 | 15 | 9.33e-01 |
| GO:BP | GO:1901896 | positive regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 6 | 9.33e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 9.33e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 9.33e-01 |
| GO:BP | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning | 1 | 5 | 9.34e-01 |
| GO:BP | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity | 1 | 3 | 9.34e-01 |
| GO:BP | GO:1905449 | regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 2 | 5 | 9.34e-01 |
| GO:BP | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway | 2 | 5 | 9.34e-01 |
| GO:BP | GO:0043307 | eosinophil activation | 2 | 4 | 9.34e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 3 | 82 | 9.34e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 1 | 49 | 9.34e-01 |
| GO:BP | GO:0007217 | tachykinin receptor signaling pathway | 1 | 3 | 9.34e-01 |
| GO:BP | GO:0021523 | somatic motor neuron differentiation | 1 | 2 | 9.34e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 1 | 19 | 9.34e-01 |
| GO:BP | GO:1990868 | response to chemokine | 2 | 37 | 9.34e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 2 | 37 | 9.34e-01 |
| GO:BP | GO:0075525 | viral translational termination-reinitiation | 1 | 5 | 9.34e-01 |
| GO:BP | GO:0035701 | hematopoietic stem cell migration | 1 | 6 | 9.34e-01 |
| GO:BP | GO:0010155 | regulation of proton transport | 1 | 18 | 9.34e-01 |
| GO:BP | GO:0036378 | calcitriol biosynthetic process from calciol | 1 | 2 | 9.34e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 9.34e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 9.35e-01 |
| GO:BP | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 1 | 6 | 9.35e-01 |
| GO:BP | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 1 | 6 | 9.35e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 1 | 4 | 9.35e-01 |
| GO:BP | GO:0051135 | positive regulation of NK T cell activation | 1 | 3 | 9.35e-01 |
| GO:BP | GO:1905686 | positive regulation of plasma membrane repair | 1 | 4 | 9.35e-01 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 2 | 4 | 9.35e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 3 | 18 | 9.35e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 2 | 7 | 9.35e-01 |
| GO:BP | GO:0030277 | maintenance of gastrointestinal epithelium | 1 | 8 | 9.35e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 9.35e-01 |
| GO:BP | GO:0150172 | regulation of phosphatidylcholine metabolic process | 3 | 10 | 9.35e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.35e-01 |
| GO:BP | GO:0045989 | positive regulation of striated muscle contraction | 5 | 12 | 9.35e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 9.35e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 1 | 14 | 9.35e-01 |
| GO:BP | GO:0038007 | netrin-activated signaling pathway | 2 | 8 | 9.35e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 1 | 22 | 9.36e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 1 | 17 | 9.36e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 1 | 53 | 9.36e-01 |
| GO:BP | GO:0035482 | gastric motility | 1 | 3 | 9.36e-01 |
| GO:BP | GO:0072378 | blood coagulation, fibrin clot formation | 1 | 7 | 9.36e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 1 | 68 | 9.36e-01 |
| GO:BP | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 3 | 9.36e-01 |
| GO:BP | GO:0030534 | adult behavior | 4 | 92 | 9.36e-01 |
| GO:BP | GO:0051953 | negative regulation of amine transport | 2 | 15 | 9.36e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 6 | 49 | 9.37e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 1 | 31 | 9.37e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 19 | 132 | 9.37e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 2 | 17 | 9.37e-01 |
| GO:BP | GO:0044351 | macropinocytosis | 2 | 8 | 9.37e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 3 | 110 | 9.37e-01 |
| GO:BP | GO:0065001 | specification of axis polarity | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 13 | 540 | 9.37e-01 |
| GO:BP | GO:0060923 | cardiac muscle cell fate commitment | 1 | 7 | 9.37e-01 |
| GO:BP | GO:0061092 | positive regulation of phospholipid translocation | 1 | 4 | 9.37e-01 |
| GO:BP | GO:0001732 | formation of cytoplasmic translation initiation complex | 1 | 16 | 9.38e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 2 | 19 | 9.38e-01 |
| GO:BP | GO:0031650 | regulation of heat generation | 2 | 7 | 9.38e-01 |
| GO:BP | GO:0031342 | negative regulation of cell killing | 1 | 11 | 9.38e-01 |
| GO:BP | GO:0032817 | regulation of natural killer cell proliferation | 1 | 4 | 9.38e-01 |
| GO:BP | GO:0048539 | bone marrow development | 2 | 7 | 9.38e-01 |
| GO:BP | GO:0002503 | peptide antigen assembly with MHC class II protein complex | 1 | 4 | 9.38e-01 |
| GO:BP | GO:0002399 | MHC class II protein complex assembly | 1 | 4 | 9.38e-01 |
| GO:BP | GO:0006868 | glutamine transport | 1 | 8 | 9.38e-01 |
| GO:BP | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death | 1 | 4 | 9.38e-01 |
| GO:BP | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process | 1 | 4 | 9.38e-01 |
| GO:BP | GO:0045851 | pH reduction | 1 | 1 | 9.39e-01 |
| GO:BP | GO:0019511 | peptidyl-proline hydroxylation | 3 | 14 | 9.39e-01 |
| GO:BP | GO:0034587 | piRNA processing | 1 | 4 | 9.40e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 1 | 20 | 9.40e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 1 | 20 | 9.40e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 9.40e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 3 | 7 | 9.40e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 15 | 47 | 9.40e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 14 | 563 | 9.40e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 4 | 16 | 9.40e-01 |
| GO:BP | GO:0016573 | histone acetylation | 66 | 143 | 9.40e-01 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 4 | 11 | 9.41e-01 |
| GO:BP | GO:1903729 | regulation of plasma membrane organization | 1 | 15 | 9.41e-01 |
| GO:BP | GO:0097254 | renal tubular secretion | 3 | 15 | 9.41e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 9.41e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 31 | 639 | 9.41e-01 |
| GO:BP | GO:0048149 | behavioral response to ethanol | 1 | 5 | 9.41e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 5 | 63 | 9.41e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 70 | 150 | 9.41e-01 |
| GO:BP | GO:1990044 | protein localization to lipid droplet | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0050892 | intestinal absorption | 4 | 22 | 9.41e-01 |
| GO:BP | GO:1905383 | protein localization to presynapse | 3 | 11 | 9.41e-01 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 1 | 8 | 9.41e-01 |
| GO:BP | GO:0061357 | positive regulation of Wnt protein secretion | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0000480 | endonucleolytic cleavage in 5’-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 8 | 9.41e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 9.42e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 12 | 75 | 9.42e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 6 | 15 | 9.42e-01 |
| GO:BP | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway | 1 | 4 | 9.42e-01 |
| GO:BP | GO:0099641 | anterograde axonal protein transport | 2 | 9 | 9.42e-01 |
| GO:BP | GO:0060452 | positive regulation of cardiac muscle contraction | 3 | 7 | 9.42e-01 |
| GO:BP | GO:0072672 | neutrophil extravasation | 1 | 7 | 9.42e-01 |
| GO:BP | GO:0097305 | response to alcohol | 1 | 159 | 9.42e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 29 | 58 | 9.42e-01 |
| GO:BP | GO:0010626 | negative regulation of Schwann cell proliferation | 2 | 6 | 9.42e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 9.42e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 12 | 39 | 9.42e-01 |
| GO:BP | GO:0035095 | behavioral response to nicotine | 1 | 3 | 9.42e-01 |
| GO:BP | GO:0002693 | positive regulation of cellular extravasation | 1 | 13 | 9.42e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 9.43e-01 |
| GO:BP | GO:0046903 | secretion | 3 | 645 | 9.43e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 9.43e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 2 | 877 | 9.43e-01 |
| GO:BP | GO:0044409 | entry into host | 20 | 113 | 9.43e-01 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 5 | 9.43e-01 |
| GO:BP | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan | 1 | 4 | 9.43e-01 |
| GO:BP | GO:0051969 | regulation of transmission of nerve impulse | 1 | 6 | 9.43e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 15 | 52 | 9.44e-01 |
| GO:BP | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission | 1 | 3 | 9.44e-01 |
| GO:BP | GO:0051946 | regulation of glutamate uptake involved in transmission of nerve impulse | 1 | 3 | 9.44e-01 |
| GO:BP | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 12 | 9.44e-01 |
| GO:BP | GO:1903998 | regulation of eating behavior | 1 | 1 | 9.44e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 4 | 68 | 9.44e-01 |
| GO:BP | GO:0001708 | cell fate specification | 1 | 56 | 9.44e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 3 | 998 | 9.44e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 9.44e-01 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 1 | 31 | 9.44e-01 |
| GO:BP | GO:1903307 | positive regulation of regulated secretory pathway | 1 | 32 | 9.44e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 9.45e-01 |
| GO:BP | GO:1902260 | negative regulation of delayed rectifier potassium channel activity | 3 | 5 | 9.45e-01 |
| GO:BP | GO:0035025 | positive regulation of Rho protein signal transduction | 2 | 22 | 9.45e-01 |
| GO:BP | GO:0015879 | carnitine transport | 1 | 4 | 9.45e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 8 | 147 | 9.45e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 6 | 13 | 9.45e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 11 | 23 | 9.45e-01 |
| GO:BP | GO:1903224 | regulation of endodermal cell differentiation | 1 | 6 | 9.45e-01 |
| GO:BP | GO:0050917 | sensory perception of umami taste | 2 | 3 | 9.45e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 9 | 23 | 9.45e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 2 | 58 | 9.46e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 11 | 61 | 9.46e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 9.46e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 9.46e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 19 | 134 | 9.46e-01 |
| GO:BP | GO:0060433 | bronchus development | 1 | 6 | 9.46e-01 |
| GO:BP | GO:0042420 | dopamine catabolic process | 1 | 5 | 9.46e-01 |
| GO:BP | GO:0021626 | central nervous system maturation | 2 | 5 | 9.47e-01 |
| GO:BP | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly | 1 | 3 | 9.47e-01 |
| GO:BP | GO:0042360 | vitamin E metabolic process | 1 | 2 | 9.47e-01 |
| GO:BP | GO:0035812 | renal sodium excretion | 2 | 11 | 9.47e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 9.47e-01 |
| GO:BP | GO:0014012 | peripheral nervous system axon regeneration | 1 | 6 | 9.47e-01 |
| GO:BP | GO:0018916 | nitrobenzene metabolic process | 1 | 4 | 9.47e-01 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 6 | 21 | 9.47e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 1 | 50 | 9.47e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 3 | 271 | 9.47e-01 |
| GO:BP | GO:0022900 | electron transport chain | 6 | 142 | 9.48e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 2 | 5 | 9.48e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 10 | 391 | 9.48e-01 |
| GO:BP | GO:0070472 | regulation of uterine smooth muscle contraction | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0097029 | mature conventional dendritic cell differentiation | 1 | 1 | 9.48e-01 |
| GO:BP | GO:0048934 | peripheral nervous system neuron differentiation | 3 | 9 | 9.48e-01 |
| GO:BP | GO:0048935 | peripheral nervous system neuron development | 3 | 9 | 9.48e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 3 | 174 | 9.48e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 9.48e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 9.48e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 3 | 114 | 9.48e-01 |
| GO:BP | GO:2000418 | positive regulation of eosinophil migration | 1 | 1 | 9.48e-01 |
| GO:BP | GO:2000410 | regulation of thymocyte migration | 1 | 1 | 9.48e-01 |
| GO:BP | GO:0021631 | optic nerve morphogenesis | 1 | 2 | 9.48e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 5 | 382 | 9.48e-01 |
| GO:BP | GO:1904396 | regulation of neuromuscular junction development | 2 | 5 | 9.48e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 4 | 68 | 9.48e-01 |
| GO:BP | GO:1904783 | positive regulation of NMDA glutamate receptor activity | 1 | 4 | 9.48e-01 |
| GO:BP | GO:0042693 | muscle cell fate commitment | 2 | 11 | 9.48e-01 |
| GO:BP | GO:0051124 | synaptic assembly at neuromuscular junction | 2 | 6 | 9.48e-01 |
| GO:BP | GO:0010897 | negative regulation of triglyceride catabolic process | 1 | 3 | 9.48e-01 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0071529 | cementum mineralization | 1 | 3 | 9.48e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 3 | 8 | 9.48e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 11 | 43 | 9.48e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 10 | 373 | 9.48e-01 |
| GO:BP | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 4 | 9.48e-01 |
| GO:BP | GO:0051957 | positive regulation of amino acid transport | 3 | 16 | 9.48e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 22 | 351 | 9.48e-01 |
| GO:BP | GO:0035815 | positive regulation of renal sodium excretion | 1 | 7 | 9.48e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 3 | 47 | 9.48e-01 |
| GO:BP | GO:0034157 | positive regulation of toll-like receptor 7 signaling pathway | 1 | 2 | 9.48e-01 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 15 | 9.48e-01 |
| GO:BP | GO:2001140 | positive regulation of phospholipid transport | 1 | 11 | 9.48e-01 |
| GO:BP | GO:0061439 | kidney vasculature morphogenesis | 1 | 7 | 9.49e-01 |
| GO:BP | GO:0061438 | renal system vasculature morphogenesis | 1 | 7 | 9.49e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 9.49e-01 |
| GO:BP | GO:0032489 | regulation of Cdc42 protein signal transduction | 1 | 6 | 9.49e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 1 | 484 | 9.49e-01 |
| GO:BP | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 5 | 9.49e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 74 | 160 | 9.49e-01 |
| GO:BP | GO:0070901 | mitochondrial tRNA methylation | 1 | 4 | 9.49e-01 |
| GO:BP | GO:0090025 | regulation of monocyte chemotaxis | 1 | 12 | 9.49e-01 |
| GO:BP | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 3 | 9.49e-01 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 1 | 42 | 9.49e-01 |
| GO:BP | GO:2000501 | regulation of natural killer cell chemotaxis | 1 | 2 | 9.49e-01 |
| GO:BP | GO:1903842 | response to arsenite ion | 1 | 4 | 9.50e-01 |
| GO:BP | GO:1903843 | cellular response to arsenite ion | 1 | 4 | 9.50e-01 |
| GO:BP | GO:0061888 | regulation of astrocyte activation | 1 | 3 | 9.50e-01 |
| GO:BP | GO:1903207 | regulation of hydrogen peroxide-induced neuron death | 1 | 5 | 9.50e-01 |
| GO:BP | GO:1903208 | negative regulation of hydrogen peroxide-induced neuron death | 1 | 5 | 9.50e-01 |
| GO:BP | GO:0036476 | neuron death in response to hydrogen peroxide | 1 | 5 | 9.50e-01 |
| GO:BP | GO:0036065 | fucosylation | 3 | 10 | 9.50e-01 |
| GO:BP | GO:0008594 | photoreceptor cell morphogenesis | 1 | 4 | 9.50e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1453 | 9.50e-01 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 9.50e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 3 | 4174 | 9.50e-01 |
| GO:BP | GO:0010988 | regulation of low-density lipoprotein particle clearance | 3 | 8 | 9.50e-01 |
| GO:BP | GO:0051180 | vitamin transport | 3 | 28 | 9.50e-01 |
| GO:BP | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 1 | 3 | 9.50e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 19 | 136 | 9.50e-01 |
| GO:BP | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation | 2 | 8 | 9.50e-01 |
| GO:BP | GO:2000152 | regulation of ubiquitin-specific protease activity | 1 | 4 | 9.51e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 1 | 78 | 9.51e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 3 | 13 | 9.51e-01 |
| GO:BP | GO:0035188 | hatching | 10 | 23 | 9.51e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 10 | 23 | 9.51e-01 |
| GO:BP | GO:0001777 | T cell homeostatic proliferation | 1 | 1 | 9.51e-01 |
| GO:BP | GO:0035929 | steroid hormone secretion | 1 | 18 | 9.51e-01 |
| GO:BP | GO:0090091 | positive regulation of extracellular matrix disassembly | 2 | 6 | 9.51e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 10 | 23 | 9.51e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 7 | 40 | 9.51e-01 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 40 | 9.51e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 2 | 554 | 9.51e-01 |
| GO:BP | GO:1905150 | regulation of voltage-gated sodium channel activity | 2 | 6 | 9.51e-01 |
| GO:BP | GO:0090289 | regulation of osteoclast proliferation | 1 | 3 | 9.51e-01 |
| GO:BP | GO:0097241 | hematopoietic stem cell migration to bone marrow | 1 | 5 | 9.51e-01 |
| GO:BP | GO:0099004 | calmodulin dependent kinase signaling pathway | 2 | 6 | 9.51e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 25 | 88 | 9.52e-01 |
| GO:BP | GO:0002752 | cell surface pattern recognition receptor signaling pathway | 1 | 27 | 9.52e-01 |
| GO:BP | GO:0010986 | positive regulation of lipoprotein particle clearance | 1 | 4 | 9.52e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 10 | 561 | 9.52e-01 |
| GO:BP | GO:0071798 | response to prostaglandin D | 1 | 2 | 9.53e-01 |
| GO:BP | GO:0071799 | cellular response to prostaglandin D stimulus | 1 | 2 | 9.53e-01 |
| GO:BP | GO:0051712 | positive regulation of killing of cells of another organism | 1 | 2 | 9.53e-01 |
| GO:BP | GO:0042373 | vitamin K metabolic process | 2 | 6 | 9.54e-01 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 1 | 9 | 9.54e-01 |
| GO:BP | GO:0006578 | amino-acid betaine biosynthetic process | 1 | 5 | 9.54e-01 |
| GO:BP | GO:1901162 | primary amino compound biosynthetic process | 1 | 2 | 9.54e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 1 | 96 | 9.54e-01 |
| GO:BP | GO:0038171 | cannabinoid signaling pathway | 2 | 5 | 9.54e-01 |
| GO:BP | GO:0002246 | wound healing involved in inflammatory response | 2 | 6 | 9.54e-01 |
| GO:BP | GO:0046487 | glyoxylate metabolic process | 2 | 5 | 9.55e-01 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 2 | 7 | 9.55e-01 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 4 | 25 | 9.55e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 609 | 9.55e-01 |
| GO:BP | GO:1901668 | regulation of superoxide dismutase activity | 1 | 4 | 9.55e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 8 | 26 | 9.55e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 9.55e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 1 | 7441 | 9.56e-01 |
| GO:BP | GO:0008343 | adult feeding behavior | 1 | 1 | 9.56e-01 |
| GO:BP | GO:0070103 | regulation of interleukin-6-mediated signaling pathway | 1 | 4 | 9.56e-01 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 1 | 4 | 9.56e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 9 | 305 | 9.56e-01 |
| GO:BP | GO:0035814 | negative regulation of renal sodium excretion | 1 | 2 | 9.56e-01 |
| GO:BP | GO:0009073 | aromatic amino acid family biosynthetic process | 1 | 3 | 9.56e-01 |
| GO:BP | GO:0035630 | bone mineralization involved in bone maturation | 2 | 6 | 9.56e-01 |
| GO:BP | GO:0034162 | toll-like receptor 9 signaling pathway | 1 | 12 | 9.56e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 9.57e-01 |
| GO:BP | GO:0045576 | mast cell activation | 1 | 35 | 9.57e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 3 | 42 | 9.57e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 11 | 58 | 9.57e-01 |
| GO:BP | GO:0036343 | psychomotor behavior | 1 | 2 | 9.57e-01 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 2 | 6 | 9.57e-01 |
| GO:BP | GO:0060913 | cardiac cell fate determination | 1 | 2 | 9.57e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 30 | 9.57e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 9.57e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 3 | 12 | 9.58e-01 |
| GO:BP | GO:0071350 | cellular response to interleukin-15 | 1 | 2 | 9.58e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 15 | 166 | 9.58e-01 |
| GO:BP | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 6 | 9.58e-01 |
| GO:BP | GO:0033299 | secretion of lysosomal enzymes | 2 | 7 | 9.58e-01 |
| GO:BP | GO:0097676 | histone H3-K36 dimethylation | 1 | 4 | 9.58e-01 |
| GO:BP | GO:0035723 | interleukin-15-mediated signaling pathway | 1 | 2 | 9.58e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 2 | 10 | 9.58e-01 |
| GO:BP | GO:0016043 | cellular component organization | 5 | 5076 | 9.58e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 3 | 9 | 9.58e-01 |
| GO:BP | GO:0072102 | glomerulus morphogenesis | 1 | 7 | 9.58e-01 |
| GO:BP | GO:0034134 | toll-like receptor 2 signaling pathway | 2 | 11 | 9.58e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 1 | 39 | 9.59e-01 |
| GO:BP | GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | 1 | 9 | 9.59e-01 |
| GO:BP | GO:0045579 | positive regulation of B cell differentiation | 5 | 10 | 9.60e-01 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 19 | 37 | 9.60e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 9.60e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 9 | 69 | 9.60e-01 |
| GO:BP | GO:0021942 | radial glia guided migration of Purkinje cell | 1 | 3 | 9.60e-01 |
| GO:BP | GO:1990384 | hyaloid vascular plexus regression | 1 | 3 | 9.60e-01 |
| GO:BP | GO:0019086 | late viral transcription | 1 | 4 | 9.61e-01 |
| GO:BP | GO:0006681 | galactosylceramide metabolic process | 2 | 5 | 9.61e-01 |
| GO:BP | GO:0070471 | uterine smooth muscle contraction | 1 | 6 | 9.61e-01 |
| GO:BP | GO:0098961 | dendritic transport of ribonucleoprotein complex | 1 | 4 | 9.61e-01 |
| GO:BP | GO:0098963 | dendritic transport of messenger ribonucleoprotein complex | 1 | 4 | 9.61e-01 |
| GO:BP | GO:2000110 | negative regulation of macrophage apoptotic process | 1 | 3 | 9.61e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 2 | 1138 | 9.61e-01 |
| GO:BP | GO:0019374 | galactolipid metabolic process | 2 | 5 | 9.61e-01 |
| GO:BP | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration | 1 | 3 | 9.61e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 5 | 10 | 9.62e-01 |
| GO:BP | GO:2000439 | positive regulation of monocyte extravasation | 1 | 3 | 9.62e-01 |
| GO:BP | GO:0048050 | post-embryonic eye morphogenesis | 1 | 7 | 9.62e-01 |
| GO:BP | GO:0071673 | positive regulation of smooth muscle cell chemotaxis | 1 | 2 | 9.62e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.62e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.62e-01 |
| GO:BP | GO:0061325 | cell proliferation involved in outflow tract morphogenesis | 1 | 4 | 9.62e-01 |
| GO:BP | GO:2000109 | regulation of macrophage apoptotic process | 2 | 6 | 9.62e-01 |
| GO:BP | GO:2001200 | positive regulation of dendritic cell differentiation | 1 | 3 | 9.62e-01 |
| GO:BP | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process | 1 | 4 | 9.62e-01 |
| GO:BP | GO:0016480 | negative regulation of transcription by RNA polymerase III | 1 | 4 | 9.62e-01 |
| GO:BP | GO:0042424 | catecholamine catabolic process | 1 | 5 | 9.63e-01 |
| GO:BP | GO:0019614 | catechol-containing compound catabolic process | 1 | 5 | 9.63e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 4 | 14 | 9.63e-01 |
| GO:BP | GO:0061590 | calcium activated phosphatidylcholine scrambling | 1 | 3 | 9.63e-01 |
| GO:BP | GO:0099640 | axo-dendritic protein transport | 2 | 10 | 9.63e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 3 | 11 | 9.63e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 9.63e-01 |
| GO:BP | GO:0051604 | protein maturation | 3 | 259 | 9.63e-01 |
| GO:BP | GO:0021819 | layer formation in cerebral cortex | 5 | 12 | 9.63e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 1 | 7069 | 9.64e-01 |
| GO:BP | GO:0032971 | regulation of muscle filament sliding | 1 | 3 | 9.64e-01 |
| GO:BP | GO:1902563 | regulation of neutrophil activation | 1 | 3 | 9.64e-01 |
| GO:BP | GO:0035900 | response to isolation stress | 1 | 2 | 9.64e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 2 | 106 | 9.64e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 2 | 6 | 9.64e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 2 | 6 | 9.64e-01 |
| GO:BP | GO:0007586 | digestion | 5 | 60 | 9.64e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 4 | 16 | 9.64e-01 |
| GO:BP | GO:0002468 | dendritic cell antigen processing and presentation | 2 | 6 | 9.64e-01 |
| GO:BP | GO:0014850 | response to muscle activity | 1 | 17 | 9.64e-01 |
| GO:BP | GO:0061314 | Notch signaling involved in heart development | 1 | 9 | 9.64e-01 |
| GO:BP | GO:0002676 | regulation of chronic inflammatory response | 1 | 2 | 9.64e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 3 | 41 | 9.64e-01 |
| GO:BP | GO:0007196 | adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway | 1 | 1 | 9.64e-01 |
| GO:BP | GO:0019323 | pentose catabolic process | 1 | 2 | 9.65e-01 |
| GO:BP | GO:0042182 | ketone catabolic process | 3 | 9 | 9.65e-01 |
| GO:BP | GO:0042448 | progesterone metabolic process | 1 | 13 | 9.65e-01 |
| GO:BP | GO:0030222 | eosinophil differentiation | 1 | 2 | 9.65e-01 |
| GO:BP | GO:0045333 | cellular respiration | 2 | 211 | 9.65e-01 |
| GO:BP | GO:0007168 | receptor guanylyl cyclase signaling pathway | 1 | 5 | 9.66e-01 |
| GO:BP | GO:0010624 | regulation of Schwann cell proliferation | 2 | 7 | 9.66e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 5 | 11 | 9.66e-01 |
| GO:BP | GO:0014010 | Schwann cell proliferation | 2 | 7 | 9.66e-01 |
| GO:BP | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | 1 | 13 | 9.66e-01 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 1 | 40 | 9.66e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 12 | 107 | 9.67e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 6 | 178 | 9.67e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 9 | 70 | 9.67e-01 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 1 | 11 | 9.67e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 2 | 37 | 9.67e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 1 | 49 | 9.67e-01 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 1 | 11 | 9.67e-01 |
| GO:BP | GO:0070841 | inclusion body assembly | 6 | 22 | 9.67e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 2 | 30 | 9.68e-01 |
| GO:BP | GO:0001886 | endothelial cell morphogenesis | 3 | 10 | 9.68e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 3 | 288 | 9.69e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 1 | 53 | 9.69e-01 |
| GO:BP | GO:0023058 | adaptation of signaling pathway | 1 | 16 | 9.69e-01 |
| GO:BP | GO:0002282 | microglial cell activation involved in immune response | 1 | 1 | 9.69e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 4 | 13 | 9.69e-01 |
| GO:BP | GO:2000318 | positive regulation of T-helper 17 type immune response | 3 | 7 | 9.69e-01 |
| GO:BP | GO:0060986 | endocrine hormone secretion | 1 | 35 | 9.69e-01 |
| GO:BP | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production | 2 | 6 | 9.69e-01 |
| GO:BP | GO:0071609 | chemokine (C-C motif) ligand 5 production | 2 | 6 | 9.69e-01 |
| GO:BP | GO:1902564 | negative regulation of neutrophil activation | 1 | 2 | 9.69e-01 |
| GO:BP | GO:2000370 | positive regulation of clathrin-dependent endocytosis | 1 | 5 | 9.69e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 7 | 26 | 9.69e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 3 | 30 | 9.69e-01 |
| GO:BP | GO:0089700 | protein kinase D signaling | 1 | 4 | 9.69e-01 |
| GO:BP | GO:1901164 | negative regulation of trophoblast cell migration | 1 | 4 | 9.69e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 1 | 38 | 9.69e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 5 | 5274 | 9.69e-01 |
| GO:BP | GO:0021548 | pons development | 3 | 7 | 9.69e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 3 | 28 | 9.70e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 7 | 47 | 9.70e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 2 | 9 | 9.70e-01 |
| GO:BP | GO:1902523 | positive regulation of protein K63-linked ubiquitination | 1 | 5 | 9.70e-01 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 2 | 18 | 9.70e-01 |
| GO:BP | GO:1901727 | positive regulation of histone deacetylase activity | 1 | 5 | 9.70e-01 |
| GO:BP | GO:0034155 | regulation of toll-like receptor 7 signaling pathway | 1 | 2 | 9.70e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 9.70e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 3 | 14 | 9.70e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 3 | 14 | 9.70e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 26 | 160 | 9.70e-01 |
| GO:BP | GO:0097749 | membrane tubulation | 1 | 4 | 9.70e-01 |
| GO:BP | GO:1905451 | positive regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 3 | 9.70e-01 |
| GO:BP | GO:0006897 | endocytosis | 2 | 527 | 9.70e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 9.70e-01 |
| GO:BP | GO:0035934 | corticosterone secretion | 1 | 3 | 9.70e-01 |
| GO:BP | GO:0018243 | protein O-linked glycosylation via threonine | 3 | 7 | 9.70e-01 |
| GO:BP | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell | 1 | 3 | 9.70e-01 |
| GO:BP | GO:1903215 | negative regulation of protein targeting to mitochondrion | 1 | 3 | 9.70e-01 |
| GO:BP | GO:0071676 | negative regulation of mononuclear cell migration | 5 | 16 | 9.70e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 1 | 211 | 9.70e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 4 | 13 | 9.70e-01 |
| GO:BP | GO:0007567 | parturition | 1 | 8 | 9.71e-01 |
| GO:BP | GO:0050427 | 3’-phosphoadenosine 5’-phosphosulfate metabolic process | 1 | 10 | 9.71e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 1 | 581 | 9.71e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 12 | 1335 | 9.71e-01 |
| GO:BP | GO:1903450 | regulation of G1 to G0 transition | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0120229 | protein localization to motile cilium | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1 | 10 | 9.71e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 8 | 78 | 9.71e-01 |
| GO:BP | GO:0014051 | gamma-aminobutyric acid secretion | 1 | 8 | 9.71e-01 |
| GO:BP | GO:1903391 | regulation of adherens junction organization | 1 | 5 | 9.71e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 1 | 8 | 9.71e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 9.71e-01 |
| GO:BP | GO:0061356 | regulation of Wnt protein secretion | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0031580 | membrane raft distribution | 1 | 3 | 9.71e-01 |
| GO:BP | GO:0140352 | export from cell | 1 | 611 | 9.72e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 3 | 4045 | 9.72e-01 |
| GO:BP | GO:0002396 | MHC protein complex assembly | 1 | 8 | 9.73e-01 |
| GO:BP | GO:1903976 | negative regulation of glial cell migration | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0009644 | response to high light intensity | 1 | 2 | 9.73e-01 |
| GO:BP | GO:0001706 | endoderm formation | 6 | 46 | 9.73e-01 |
| GO:BP | GO:0002501 | peptide antigen assembly with MHC protein complex | 1 | 8 | 9.73e-01 |
| GO:BP | GO:0045906 | negative regulation of vasoconstriction | 1 | 6 | 9.73e-01 |
| GO:BP | GO:0051179 | localization | 4 | 3969 | 9.73e-01 |
| GO:BP | GO:0097278 | complement-dependent cytotoxicity | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0010430 | fatty acid omega-oxidation | 1 | 2 | 9.73e-01 |
| GO:BP | GO:1904751 | positive regulation of protein localization to nucleolus | 1 | 3 | 9.73e-01 |
| GO:BP | GO:0021764 | amygdala development | 1 | 3 | 9.73e-01 |
| GO:BP | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome | 4 | 13 | 9.73e-01 |
| GO:BP | GO:0034112 | positive regulation of homotypic cell-cell adhesion | 1 | 10 | 9.73e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 9.73e-01 |
| GO:BP | GO:0006071 | glycerol metabolic process | 2 | 12 | 9.73e-01 |
| GO:BP | GO:0071286 | cellular response to magnesium ion | 1 | 3 | 9.73e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 9.73e-01 |
| GO:BP | GO:0046325 | negative regulation of glucose import | 4 | 10 | 9.73e-01 |
| GO:BP | GO:0030322 | stabilization of membrane potential | 2 | 6 | 9.73e-01 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 1 | 13 | 9.74e-01 |
| GO:BP | GO:0033574 | response to testosterone | 9 | 29 | 9.74e-01 |
| GO:BP | GO:0072017 | distal tubule development | 1 | 6 | 9.74e-01 |
| GO:BP | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome | 1 | 2 | 9.74e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 9.74e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 4 | 9 | 9.74e-01 |
| GO:BP | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib | 1 | 3 | 9.74e-01 |
| GO:BP | GO:0033006 | regulation of mast cell activation involved in immune response | 10 | 21 | 9.74e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 9.74e-01 |
| GO:BP | GO:0048007 | antigen processing and presentation, exogenous lipid antigen via MHC class Ib | 1 | 3 | 9.74e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 9.74e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 9.74e-01 |
| GO:BP | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway | 1 | 4 | 9.74e-01 |
| GO:BP | GO:0071605 | monocyte chemotactic protein-1 production | 2 | 7 | 9.74e-01 |
| GO:BP | GO:0071637 | regulation of monocyte chemotactic protein-1 production | 2 | 7 | 9.74e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 1 | 42 | 9.74e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 6 | 27 | 9.74e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 19 | 64 | 9.74e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 2 | 10 | 9.74e-01 |
| GO:BP | GO:0002674 | negative regulation of acute inflammatory response | 1 | 5 | 9.74e-01 |
| GO:BP | GO:0032743 | positive regulation of interleukin-2 production | 2 | 20 | 9.74e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 2 | 317 | 9.74e-01 |
| GO:BP | GO:0007197 | adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway | 1 | 3 | 9.74e-01 |
| GO:BP | GO:0140894 | endolysosomal toll-like receptor signaling pathway | 14 | 24 | 9.74e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 9.75e-01 |
| GO:BP | GO:0015671 | oxygen transport | 1 | 4 | 9.75e-01 |
| GO:BP | GO:0001775 | cell activation | 2 | 655 | 9.75e-01 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 9.75e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 2 | 10 | 9.75e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 2 | 395 | 9.75e-01 |
| GO:BP | GO:0090659 | walking behavior | 1 | 25 | 9.75e-01 |
| GO:BP | GO:0060332 | positive regulation of response to type II interferon | 1 | 4 | 9.76e-01 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 9.76e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 9.76e-01 |
| GO:BP | GO:0060335 | positive regulation of type II interferon-mediated signaling pathway | 1 | 4 | 9.76e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 1 | 16 | 9.76e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 15 | 669 | 9.76e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 1 | 26 | 9.76e-01 |
| GO:BP | GO:1903236 | regulation of leukocyte tethering or rolling | 1 | 6 | 9.77e-01 |
| GO:BP | GO:0001920 | negative regulation of receptor recycling | 1 | 3 | 9.77e-01 |
| GO:BP | GO:0007409 | axonogenesis | 5 | 359 | 9.77e-01 |
| GO:BP | GO:0034398 | telomere tethering at nuclear periphery | 1 | 1 | 9.77e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 1 | 18 | 9.77e-01 |
| GO:BP | GO:0043299 | leukocyte degranulation | 1 | 41 | 9.77e-01 |
| GO:BP | GO:0044821 | meiotic telomere tethering at nuclear periphery | 1 | 1 | 9.77e-01 |
| GO:BP | GO:0070197 | meiotic attachment of telomere to nuclear envelope | 1 | 1 | 9.77e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 2 | 35 | 9.77e-01 |
| GO:BP | GO:0071670 | smooth muscle cell chemotaxis | 2 | 6 | 9.77e-01 |
| GO:BP | GO:0034380 | high-density lipoprotein particle assembly | 1 | 10 | 9.77e-01 |
| GO:BP | GO:0001916 | positive regulation of T cell mediated cytotoxicity | 3 | 13 | 9.77e-01 |
| GO:BP | GO:0045656 | negative regulation of monocyte differentiation | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0030574 | collagen catabolic process | 4 | 26 | 9.77e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 9.77e-01 |
| GO:BP | GO:0034653 | retinoic acid catabolic process | 1 | 1 | 9.77e-01 |
| GO:BP | GO:0090045 | positive regulation of deacetylase activity | 2 | 7 | 9.77e-01 |
| GO:BP | GO:0016103 | diterpenoid catabolic process | 1 | 1 | 9.77e-01 |
| GO:BP | GO:1902809 | regulation of skeletal muscle fiber differentiation | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0006570 | tyrosine metabolic process | 1 | 6 | 9.77e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 9.77e-01 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 2 | 11 | 9.78e-01 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 1 | 14 | 9.78e-01 |
| GO:BP | GO:0070487 | monocyte aggregation | 1 | 4 | 9.78e-01 |
| GO:BP | GO:0010387 | COP9 signalosome assembly | 1 | 3 | 9.78e-01 |
| GO:BP | GO:0019673 | GDP-mannose metabolic process | 1 | 7 | 9.79e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 6 | 187 | 9.79e-01 |
| GO:BP | GO:0019563 | glycerol catabolic process | 1 | 5 | 9.79e-01 |
| GO:BP | GO:0001711 | endodermal cell fate commitment | 2 | 9 | 9.79e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 9.79e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 22 | 139 | 9.79e-01 |
| GO:BP | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity | 2 | 10 | 9.79e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 1 | 18 | 9.79e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 4 | 121 | 9.79e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 1 | 7806 | 9.79e-01 |
| GO:BP | GO:2001026 | regulation of endothelial cell chemotaxis | 2 | 16 | 9.79e-01 |
| GO:BP | GO:0061024 | membrane organization | 55 | 671 | 9.79e-01 |
| GO:BP | GO:0120197 | mucociliary clearance | 2 | 5 | 9.79e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 6 | 21 | 9.80e-01 |
| GO:BP | GO:0038027 | apolipoprotein A-I-mediated signaling pathway | 1 | 5 | 9.80e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 6 | 25 | 9.80e-01 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 1 | 104 | 9.80e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 3 | 16 | 9.80e-01 |
| GO:BP | GO:0032494 | response to peptidoglycan | 2 | 5 | 9.80e-01 |
| GO:BP | GO:1903817 | negative regulation of voltage-gated potassium channel activity | 3 | 6 | 9.80e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 2 | 43 | 9.80e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 3 | 129 | 9.80e-01 |
| GO:BP | GO:0021869 | forebrain ventricular zone progenitor cell division | 1 | 2 | 9.80e-01 |
| GO:BP | GO:1903792 | negative regulation of monoatomic anion transport | 1 | 3 | 9.80e-01 |
| GO:BP | GO:0009312 | oligosaccharide biosynthetic process | 2 | 15 | 9.80e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 5 | 11 | 9.80e-01 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 2 | 8 | 9.81e-01 |
| GO:BP | GO:0097187 | dentinogenesis | 1 | 4 | 9.81e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 1 | 26 | 9.81e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 8 | 19 | 9.81e-01 |
| GO:BP | GO:0098664 | G protein-coupled serotonin receptor signaling pathway | 1 | 8 | 9.81e-01 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 9 | 48 | 9.81e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 4 | 16 | 9.81e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 1 | 20 | 9.81e-01 |
| GO:BP | GO:0034135 | regulation of toll-like receptor 2 signaling pathway | 1 | 7 | 9.81e-01 |
| GO:BP | GO:1903412 | response to bile acid | 1 | 3 | 9.81e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 2 | 130 | 9.82e-01 |
| GO:BP | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 4 | 15 | 9.82e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 3 | 36 | 9.82e-01 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 3 | 35 | 9.82e-01 |
| GO:BP | GO:0001990 | regulation of systemic arterial blood pressure by hormone | 1 | 22 | 9.82e-01 |
| GO:BP | GO:0061588 | calcium activated phospholipid scrambling | 1 | 4 | 9.82e-01 |
| GO:BP | GO:0009996 | negative regulation of cell fate specification | 1 | 4 | 9.82e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 215 | 9.82e-01 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 9 | 20 | 9.82e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 6 | 16 | 9.82e-01 |
| GO:BP | GO:0071599 | otic vesicle development | 4 | 10 | 9.82e-01 |
| GO:BP | GO:0006605 | protein targeting | 2 | 307 | 9.82e-01 |
| GO:BP | GO:0032026 | response to magnesium ion | 1 | 10 | 9.83e-01 |
| GO:BP | GO:0022617 | extracellular matrix disassembly | 1 | 36 | 9.83e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 1 | 47 | 9.83e-01 |
| GO:BP | GO:0042938 | dipeptide transport | 1 | 4 | 9.83e-01 |
| GO:BP | GO:0035442 | dipeptide transmembrane transport | 1 | 4 | 9.83e-01 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 2 | 7 | 9.83e-01 |
| GO:BP | GO:0002858 | regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 9.83e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 5 | 55 | 9.83e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 5 | 35 | 9.83e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 6 | 24 | 9.83e-01 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 8 | 29 | 9.83e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 14 | 632 | 9.83e-01 |
| GO:BP | GO:0001914 | regulation of T cell mediated cytotoxicity | 3 | 17 | 9.83e-01 |
| GO:BP | GO:0032277 | negative regulation of gonadotropin secretion | 1 | 4 | 9.83e-01 |
| GO:BP | GO:0031638 | zymogen activation | 12 | 45 | 9.83e-01 |
| GO:BP | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain | 1 | 1 | 9.83e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 10 | 24 | 9.83e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 4 | 28 | 9.83e-01 |
| GO:BP | GO:0070857 | regulation of bile acid biosynthetic process | 2 | 7 | 9.83e-01 |
| GO:BP | GO:0010757 | negative regulation of plasminogen activation | 2 | 7 | 9.84e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 5 | 22 | 9.84e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 1 | 91 | 9.84e-01 |
| GO:BP | GO:0070640 | vitamin D3 metabolic process | 1 | 2 | 9.84e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 1 | 91 | 9.84e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 9.84e-01 |
| GO:BP | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia | 1 | 9 | 9.84e-01 |
| GO:BP | GO:0019400 | alditol metabolic process | 2 | 14 | 9.84e-01 |
| GO:BP | GO:2000437 | regulation of monocyte extravasation | 1 | 4 | 9.84e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 2 | 245 | 9.84e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 1 | 15 | 9.84e-01 |
| GO:BP | GO:0032383 | regulation of intracellular cholesterol transport | 1 | 6 | 9.85e-01 |
| GO:BP | GO:0032380 | regulation of intracellular sterol transport | 1 | 6 | 9.85e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 9.85e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 7 | 50 | 9.85e-01 |
| GO:BP | GO:0035744 | T-helper 1 cell cytokine production | 1 | 2 | 9.85e-01 |
| GO:BP | GO:0002716 | negative regulation of natural killer cell mediated immunity | 2 | 10 | 9.85e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 2 | 13 | 9.85e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 2 | 10 | 9.85e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 2 | 10 | 9.85e-01 |
| GO:BP | GO:2000554 | regulation of T-helper 1 cell cytokine production | 1 | 2 | 9.85e-01 |
| GO:BP | GO:0032963 | collagen metabolic process | 1 | 74 | 9.85e-01 |
| GO:BP | GO:2000556 | positive regulation of T-helper 1 cell cytokine production | 1 | 2 | 9.85e-01 |
| GO:BP | GO:0048535 | lymph node development | 5 | 11 | 9.85e-01 |
| GO:BP | GO:1905049 | negative regulation of metallopeptidase activity | 1 | 7 | 9.86e-01 |
| GO:BP | GO:0023061 | signal release | 16 | 321 | 9.86e-01 |
| GO:BP | GO:0009311 | oligosaccharide metabolic process | 3 | 43 | 9.86e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 9.86e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 389 | 9.86e-01 |
| GO:BP | GO:0140507 | granzyme-mediated programmed cell death signaling pathway | 1 | 3 | 9.86e-01 |
| GO:BP | GO:0002827 | positive regulation of T-helper 1 type immune response | 3 | 5 | 9.86e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 1 | 94 | 9.86e-01 |
| GO:BP | GO:0007596 | blood coagulation | 1 | 150 | 9.86e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 1 | 94 | 9.86e-01 |
| GO:BP | GO:0015874 | norepinephrine transport | 1 | 9 | 9.86e-01 |
| GO:BP | GO:0046884 | follicle-stimulating hormone secretion | 1 | 4 | 9.86e-01 |
| GO:BP | GO:0072044 | collecting duct development | 2 | 6 | 9.86e-01 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 3 | 10 | 9.86e-01 |
| GO:BP | GO:0015705 | iodide transport | 1 | 2 | 9.86e-01 |
| GO:BP | GO:1905906 | regulation of amyloid fibril formation | 2 | 14 | 9.86e-01 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 2 | 9 | 9.86e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 7 | 27 | 9.86e-01 |
| GO:BP | GO:0006105 | succinate metabolic process | 1 | 5 | 9.86e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 12 | 1496 | 9.86e-01 |
| GO:BP | GO:0007155 | cell adhesion | 6 | 1012 | 9.86e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 2 | 336 | 9.87e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 8 | 122 | 9.87e-01 |
| GO:BP | GO:0061355 | Wnt protein secretion | 1 | 5 | 9.87e-01 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 1 | 89 | 9.87e-01 |
| GO:BP | GO:0007599 | hemostasis | 2 | 153 | 9.87e-01 |
| GO:BP | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production | 1 | 4 | 9.88e-01 |
| GO:BP | GO:0050817 | coagulation | 1 | 153 | 9.88e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 9.88e-01 |
| GO:BP | GO:0002376 | immune system process | 7 | 1447 | 9.88e-01 |
| GO:BP | GO:0071484 | cellular response to light intensity | 1 | 4 | 9.88e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 3 | 34 | 9.88e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 2 | 39 | 9.88e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 6 | 51 | 9.88e-01 |
| GO:BP | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin | 1 | 1 | 9.89e-01 |
| GO:BP | GO:2000402 | negative regulation of lymphocyte migration | 1 | 10 | 9.89e-01 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 2 | 5 | 9.89e-01 |
| GO:BP | GO:0007602 | phototransduction | 2 | 17 | 9.89e-01 |
| GO:BP | GO:0007356 | thorax and anterior abdomen determination | 1 | 1 | 9.89e-01 |
| GO:BP | GO:0043302 | positive regulation of leukocyte degranulation | 7 | 15 | 9.90e-01 |
| GO:BP | GO:0019405 | alditol catabolic process | 1 | 6 | 9.90e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 7 | 115 | 9.90e-01 |
| GO:BP | GO:0120252 | hydrocarbon metabolic process | 1 | 2 | 9.90e-01 |
| GO:BP | GO:0006069 | ethanol oxidation | 2 | 2 | 9.90e-01 |
| GO:BP | GO:0019336 | phenol-containing compound catabolic process | 1 | 6 | 9.90e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 5 | 31 | 9.90e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 5 | 31 | 9.90e-01 |
| GO:BP | GO:0044060 | regulation of endocrine process | 1 | 24 | 9.90e-01 |
| GO:BP | GO:0071650 | negative regulation of chemokine (C-C motif) ligand 5 production | 1 | 3 | 9.90e-01 |
| GO:BP | GO:0045956 | positive regulation of calcium ion-dependent exocytosis | 2 | 11 | 9.90e-01 |
| GO:BP | GO:0120317 | sperm mitochondrial sheath assembly | 1 | 2 | 9.90e-01 |
| GO:BP | GO:0060911 | cardiac cell fate commitment | 1 | 11 | 9.91e-01 |
| GO:BP | GO:1903963 | arachidonate transport | 1 | 11 | 9.91e-01 |
| GO:BP | GO:0006688 | glycosphingolipid biosynthetic process | 1 | 25 | 9.91e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 16 | 1238 | 9.91e-01 |
| GO:BP | GO:0050482 | arachidonic acid secretion | 1 | 11 | 9.91e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 5 | 23 | 9.91e-01 |
| GO:BP | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | 1 | 13 | 9.91e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 2 | 29 | 9.91e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 2 | 29 | 9.91e-01 |
| GO:BP | GO:0010876 | lipid localization | 9 | 310 | 9.91e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 7 | 216 | 9.91e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 10 | 100 | 9.91e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 9.91e-01 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 9.91e-01 |
| GO:BP | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 2 | 3 | 9.91e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 9 | 142 | 9.91e-01 |
| GO:BP | GO:1905907 | negative regulation of amyloid fibril formation | 2 | 10 | 9.91e-01 |
| GO:BP | GO:0015849 | organic acid transport | 7 | 217 | 9.91e-01 |
| GO:BP | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 9.91e-01 |
| GO:BP | GO:0002855 | regulation of natural killer cell mediated immune response to tumor cell | 1 | 4 | 9.91e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 9.92e-01 |
| GO:BP | GO:0060435 | bronchiole development | 1 | 2 | 9.92e-01 |
| GO:BP | GO:0002829 | negative regulation of type 2 immune response | 1 | 7 | 9.92e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 6 | 15 | 9.92e-01 |
| GO:BP | GO:0006907 | pinocytosis | 4 | 18 | 9.92e-01 |
| GO:BP | GO:0048536 | spleen development | 1 | 28 | 9.92e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 3 | 28 | 9.92e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 3 | 36 | 9.92e-01 |
| GO:BP | GO:0007210 | serotonin receptor signaling pathway | 1 | 8 | 9.92e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 3 | 190 | 9.93e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 9 | 39 | 9.93e-01 |
| GO:BP | GO:0035813 | regulation of renal sodium excretion | 1 | 10 | 9.93e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 9.93e-01 |
| GO:BP | GO:0022401 | negative adaptation of signaling pathway | 5 | 15 | 9.93e-01 |
| GO:BP | GO:1904936 | interneuron migration | 1 | 6 | 9.93e-01 |
| GO:BP | GO:0032627 | interleukin-23 production | 2 | 6 | 9.93e-01 |
| GO:BP | GO:0032667 | regulation of interleukin-23 production | 2 | 6 | 9.93e-01 |
| GO:BP | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 1 | 10 | 9.93e-01 |
| GO:BP | GO:0007309 | oocyte axis specification | 1 | 3 | 9.93e-01 |
| GO:BP | GO:0007308 | oocyte construction | 1 | 3 | 9.93e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 2 | 25 | 9.93e-01 |
| GO:BP | GO:0097709 | connective tissue replacement | 1 | 8 | 9.93e-01 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 6 | 23 | 9.94e-01 |
| GO:BP | GO:0048143 | astrocyte activation | 3 | 12 | 9.94e-01 |
| GO:BP | GO:0031452 | negative regulation of heterochromatin formation | 1 | 4 | 9.94e-01 |
| GO:BP | GO:1905268 | negative regulation of chromatin organization | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0120262 | negative regulation of heterochromatin organization | 1 | 4 | 9.94e-01 |
| GO:BP | GO:1904749 | regulation of protein localization to nucleolus | 2 | 7 | 9.94e-01 |
| GO:BP | GO:0021877 | forebrain neuron fate commitment | 1 | 1 | 9.94e-01 |
| GO:BP | GO:0045650 | negative regulation of macrophage differentiation | 1 | 5 | 9.94e-01 |
| GO:BP | GO:0042756 | drinking behavior | 1 | 6 | 9.94e-01 |
| GO:BP | GO:0009820 | alkaloid metabolic process | 1 | 2 | 9.94e-01 |
| GO:BP | GO:0072205 | metanephric collecting duct development | 1 | 5 | 9.94e-01 |
| GO:BP | GO:0042791 | 5S class rRNA transcription by RNA polymerase III | 1 | 6 | 9.94e-01 |
| GO:BP | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 2 | 10 | 9.95e-01 |
| GO:BP | GO:0140650 | radial glia-guided pyramidal neuron migration | 1 | 4 | 9.95e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 9.95e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 2 | 20 | 9.95e-01 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 3 | 38 | 9.95e-01 |
| GO:BP | GO:0032940 | secretion by cell | 29 | 569 | 9.95e-01 |
| GO:BP | GO:0007369 | gastrulation | 1 | 146 | 9.95e-01 |
| GO:BP | GO:0046641 | positive regulation of alpha-beta T cell proliferation | 2 | 11 | 9.95e-01 |
| GO:BP | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity | 1 | 3 | 9.96e-01 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 1 | 19 | 9.97e-01 |
| GO:BP | GO:0021859 | pyramidal neuron differentiation | 1 | 11 | 9.97e-01 |
| GO:BP | GO:0002002 | regulation of angiotensin levels in blood | 2 | 6 | 9.97e-01 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 4 | 16 | 9.97e-01 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 1 | 18 | 9.98e-01 |
| GO:BP | GO:0010918 | positive regulation of mitochondrial membrane potential | 2 | 9 | 9.98e-01 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 1 | 10 | 9.98e-01 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 4 | 19 | 9.98e-01 |
| GO:BP | GO:0010536 | positive regulation of activation of Janus kinase activity | 1 | 2 | 9.98e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 13 | 59 | 9.98e-01 |
| GO:BP | GO:1904672 | regulation of somatic stem cell population maintenance | 1 | 6 | 9.98e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 9.98e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 1 | 336 | 9.98e-01 |
| GO:BP | GO:0010496 | intercellular transport | 1 | 4 | 9.98e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 10 | 90 | 9.98e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 6 | 30 | 9.98e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 11 | 775 | 9.98e-01 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 2 | 23 | 9.98e-01 |
| GO:BP | GO:0071220 | cellular response to bacterial lipoprotein | 1 | 2 | 9.98e-01 |
| GO:BP | GO:0071221 | cellular response to bacterial lipopeptide | 1 | 2 | 9.98e-01 |
| GO:BP | GO:0002423 | natural killer cell mediated immune response to tumor cell | 1 | 4 | 9.98e-01 |
| GO:BP | GO:0070339 | response to bacterial lipopeptide | 1 | 2 | 9.98e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 9.98e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 2 | 27 | 9.98e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 3 | 3600 | 9.98e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 1 | 90 | 9.98e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 5 | 24 | 9.98e-01 |
| GO:BP | GO:0010524 | positive regulation of calcium ion transport into cytosol | 3 | 7 | 9.98e-01 |
| GO:BP | GO:0051014 | actin filament severing | 1 | 13 | 9.98e-01 |
| GO:BP | GO:0006869 | lipid transport | 6 | 272 | 9.98e-01 |
| GO:BP | GO:0007216 | G protein-coupled glutamate receptor signaling pathway | 2 | 5 | 9.98e-01 |
| GO:BP | GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | 1 | 30 | 9.99e-01 |
| GO:BP | GO:1903028 | positive regulation of opsonization | 1 | 1 | 9.99e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 1 | 44 | 9.99e-01 |
| GO:BP | GO:0001991 | regulation of systemic arterial blood pressure by circulatory renin-angiotensin | 4 | 10 | 9.99e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 250 | 9.99e-01 |
| GO:BP | GO:0042178 | xenobiotic catabolic process | 2 | 12 | 9.99e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 9 | 41 | 9.99e-01 |
| GO:BP | GO:0035112 | genitalia morphogenesis | 1 | 5 | 9.99e-01 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 6 | 23 | 9.99e-01 |
| GO:BP | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin | 1 | 2 | 9.99e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 14 | 40 | 1.00e+00 |
| GO:BP | GO:2000391 | positive regulation of neutrophil extravasation | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0072679 | thymocyte migration | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 1.00e+00 |
| GO:BP | GO:0032490 | detection of molecule of bacterial origin | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0032493 | response to bacterial lipoprotein | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0006397 | mRNA processing | 3 | 450 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 9 | 22 | 1.00e+00 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 3 | 38 | 1.00e+00 |
| GO:BP | GO:0046174 | polyol catabolic process | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0006376 | mRNA splice site selection | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0032501 | multicellular organismal process | 3 | 4794 | 1.00e+00 |
| GO:BP | GO:0070206 | protein trimerization | 1 | 12 | 1.00e+00 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0034374 | low-density lipoprotein particle remodeling | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0070207 | protein homotrimerization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0045061 | thymic T cell selection | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0006477 | protein sulfation | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 5 | 20 | 1.00e+00 |
| GO:BP | GO:0000353 | formation of quadruple SL/U4/U5/U6 snRNP | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 10 | 25 | 1.00e+00 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 1 | 94 | 1.00e+00 |
| GO:BP | GO:0000165 | MAPK cascade | 3 | 556 | 1.00e+00 |
| GO:BP | GO:0046034 | ATP metabolic process | 6 | 116 | 1.00e+00 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 5 | 14 | 1.00e+00 |
| GO:BP | GO:0050909 | sensory perception of taste | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 3 | 65 | 1.00e+00 |
| GO:BP | GO:0050907 | detection of chemical stimulus involved in sensory perception | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0045088 | regulation of innate immune response | 2 | 232 | 1.00e+00 |
| GO:BP | GO:1905247 | positive regulation of aspartic-type peptidase activity | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0061760 | antifungal innate immune response | 3 | 7 | 1.00e+00 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 8 | 24 | 1.00e+00 |
| GO:BP | GO:1905245 | regulation of aspartic-type peptidase activity | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0050886 | endocrine process | 3 | 57 | 1.00e+00 |
| GO:BP | GO:0007342 | fusion of sperm to egg plasma membrane involved in single fertilization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0007339 | binding of sperm to zona pellucida | 5 | 20 | 1.00e+00 |
| GO:BP | GO:0007338 | single fertilization | 2 | 70 | 1.00e+00 |
| GO:BP | GO:0006067 | ethanol metabolic process | 3 | 8 | 1.00e+00 |
| GO:BP | GO:0007320 | insemination | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0006068 | ethanol catabolic process | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0032429 | regulation of phospholipase A2 activity | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0033234 | negative regulation of protein sumoylation | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0044546 | NLRP3 inflammasome complex assembly | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 11 | 192 | 1.00e+00 |
| GO:BP | GO:0065007 | biological regulation | 1 | 8320 | 1.00e+00 |
| GO:BP | GO:0034384 | high-density lipoprotein particle clearance | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 2 | 16 | 1.00e+00 |
| GO:BP | GO:0032604 | granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 3 | 293 | 1.00e+00 |
| GO:BP | GO:0002834 | regulation of response to tumor cell | 3 | 11 | 1.00e+00 |
| GO:BP | GO:0046164 | alcohol catabolic process | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 2 | 475 | 1.00e+00 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0045058 | T cell selection | 3 | 22 | 1.00e+00 |
| GO:BP | GO:0003081 | regulation of systemic arterial blood pressure by renin-angiotensin | 4 | 15 | 1.00e+00 |
| GO:BP | GO:0006821 | chloride transport | 6 | 58 | 1.00e+00 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 3 | 289 | 1.00e+00 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 2 | 32 | 1.00e+00 |
| GO:BP | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0007492 | endoderm development | 8 | 65 | 1.00e+00 |
| GO:BP | GO:0006886 | intracellular protein transport | 74 | 803 | 1.00e+00 |
| GO:BP | GO:0070508 | cholesterol import | 1 | 6 | 1.00e+00 |
| GO:BP | GO:1905920 | positive regulation of CoA-transferase activity | 1 | 5 | 1.00e+00 |
| GO:BP | GO:1905918 | regulation of CoA-transferase activity | 1 | 5 | 1.00e+00 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 2 | 33 | 1.00e+00 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 16 | 34 | 1.00e+00 |
| GO:BP | GO:0034310 | primary alcohol catabolic process | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0006910 | phagocytosis, recognition | 2 | 12 | 1.00e+00 |
| GO:BP | GO:1905806 | regulation of synapse pruning | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0050794 | regulation of cellular process | 5 | 7552 | 1.00e+00 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0050789 | regulation of biological process | 7 | 8061 | 1.00e+00 |
| GO:BP | GO:0006956 | complement activation | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0002922 | positive regulation of humoral immune response | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0002839 | positive regulation of immune response to tumor cell | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0000365 | mRNA trans splicing, via spliceosome | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0034154 | toll-like receptor 7 signaling pathway | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0006810 | transport | 2 | 3298 | 1.00e+00 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 15 | 55 | 1.00e+00 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0070268 | cornification | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0070269 | pyroptosis | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 3 | 289 | 1.00e+00 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 1.00e+00 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition | 2 | 17 | 1.00e+00 |
| GO:BP | GO:0006640 | monoacylglycerol biosynthetic process | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0002836 | positive regulation of response to tumor cell | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0002837 | regulation of immune response to tumor cell | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0032645 | regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0045026 | plasma membrane fusion | 2 | 5 | 1.00e+00 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 4 | 211 | 1.00e+00 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 4 | 191 | 1.00e+00 |
| GO:BP | GO:0006706 | steroid catabolic process | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0045059 | positive thymic T cell selection | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0006707 | cholesterol catabolic process | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 3 | 87 | 1.00e+00 |
| GO:BP | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 1 | 20 | 1.00e+00 |
| GO:BP | GO:1903979 | negative regulation of microglial cell activation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0000395 | mRNA 5’-splice site recognition | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0070486 | leukocyte aggregation | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0002031 | G protein-coupled receptor internalization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 11 | 182 | 1.00e+00 |
| GO:BP | GO:0014048 | regulation of glutamate secretion | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0014049 | positive regulation of glutamate secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0060539 | diaphragm development | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0001840 | neural plate development | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0051917 | regulation of fibrinolysis | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0051918 | negative regulation of fibrinolysis | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0051923 | sulfation | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0001867 | complement activation, lectin pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:2000425 | regulation of apoptotic cell clearance | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0150079 | negative regulation of neuroinflammatory response | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0150078 | positive regulation of neuroinflammatory response | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0021885 | forebrain cell migration | 21 | 46 | 1.00e+00 |
| GO:BP | GO:0021860 | pyramidal neuron development | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 4 | 28 | 1.00e+00 |
| GO:BP | GO:0045332 | phospholipid translocation | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0002349 | histamine production involved in inflammatory response | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0021794 | thalamus development | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0140632 | canonical inflammasome complex assembly | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0021957 | corticospinal tract morphogenesis | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 2 | 20 | 1.00e+00 |
| GO:BP | GO:0045291 | mRNA trans splicing, SL addition | 1 | 6 | 1.00e+00 |
| GO:BP | GO:2000389 | regulation of neutrophil extravasation | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0042363 | fat-soluble vitamin catabolic process | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0002418 | immune response to tumor cell | 3 | 12 | 1.00e+00 |
| GO:BP | GO:0042362 | fat-soluble vitamin biosynthetic process | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0060693 | regulation of branching involved in salivary gland morphogenesis | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0080154 | regulation of fertilization | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0042359 | vitamin D metabolic process | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0010755 | regulation of plasminogen activation | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0051608 | histamine transport | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0051668 | localization within membrane | 6 | 605 | 1.00e+00 |
| GO:BP | GO:1900426 | positive regulation of defense response to bacterium | 2 | 6 | 1.00e+00 |
| GO:BP | GO:1900424 | regulation of defense response to bacterium | 2 | 12 | 1.00e+00 |
| GO:BP | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly | 2 | 16 | 1.00e+00 |
| GO:BP | GO:0042119 | neutrophil activation | 3 | 16 | 1.00e+00 |
| GO:BP | GO:0001821 | histamine secretion | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 6 | 168 | 1.00e+00 |
| GO:BP | GO:0022029 | telencephalon cell migration | 18 | 44 | 1.00e+00 |
| GO:BP | GO:0014002 | astrocyte development | 10 | 27 | 1.00e+00 |
| GO:BP | GO:0140353 | lipid export from cell | 1 | 28 | 1.00e+00 |
| GO:BP | GO:0015031 | protein transport | 129 | 1361 | 1.00e+00 |
| GO:BP | GO:0021762 | substantia nigra development | 5 | 41 | 1.00e+00 |
| GO:BP | GO:0099024 | plasma membrane invagination | 3 | 42 | 1.00e+00 |
| GO:BP | GO:0035696 | monocyte extravasation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 3 | 38 | 1.00e+00 |
| GO:BP | GO:0017121 | plasma membrane phospholipid scrambling | 3 | 13 | 1.00e+00 |
| GO:BP | GO:0098883 | synapse pruning | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0035933 | glucocorticoid secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0098856 | intestinal lipid absorption | 4 | 10 | 1.00e+00 |
| GO:BP | GO:0035930 | corticosteroid hormone secretion | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0018149 | peptide cross-linking | 1 | 14 | 1.00e+00 |
| GO:BP | GO:0019373 | epoxygenase P450 pathway | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0035739 | CD4-positive, alpha-beta T cell proliferation | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0002003 | angiotensin maturation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 1.00e+00 |
| GO:BP | GO:0002016 | regulation of blood volume by renin-angiotensin | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0098543 | detection of other organism | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0098528 | skeletal muscle fiber differentiation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | 4 | 14 | 1.00e+00 |
| GO:BP | GO:0046666 | retinal cell programmed cell death | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0060124 | positive regulation of growth hormone secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0060123 | regulation of growth hormone secretion | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0052695 | cellular glucuronidation | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0090324 | negative regulation of oxidative phosphorylation | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0015721 | bile acid and bile salt transport | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0021554 | optic nerve development | 3 | 7 | 1.00e+00 |
| GO:BP | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 12 | 1.00e+00 |
| GO:BP | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0021520 | spinal cord motor neuron cell fate specification | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0042368 | vitamin D biosynthetic process | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 2 | 5 | 1.00e+00 |
| GO:BP | GO:0002283 | neutrophil activation involved in immune response | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0060430 | lung saccule development | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 19 | 66 | 1.00e+00 |
| GO:BP | GO:0016045 | detection of bacterium | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0016071 | mRNA metabolic process | 7 | 665 | 1.00e+00 |
| GO:BP | GO:0016127 | sterol catabolic process | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 7 | 14 | 1.00e+00 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 1 | 74 | 1.00e+00 |
| GO:BP | GO:0036230 | granulocyte activation | 3 | 19 | 1.00e+00 |
| GO:BP | GO:0015889 | cobalamin transport | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0030299 | intestinal cholesterol absorption | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0043300 | regulation of leukocyte degranulation | 11 | 27 | 1.00e+00 |
| GO:BP | GO:0071705 | nitrogen compound transport | 154 | 1684 | 1.00e+00 |
| GO:BP | GO:0071715 | icosanoid transport | 1 | 24 | 1.00e+00 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 2 | 28 | 1.00e+00 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 3 | 29 | 1.00e+00 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 3 | 27 | 1.00e+00 |
| GO:BP | GO:0008228 | opsonization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1267 | 1.00e+00 |
| GO:BP | GO:0019081 | viral translation | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0008380 | RNA splicing | 3 | 419 | 1.00e+00 |
| GO:BP | GO:0002692 | negative regulation of cellular extravasation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002691 | regulation of cellular extravasation | 2 | 24 | 1.00e+00 |
| GO:BP | GO:0002689 | negative regulation of leukocyte chemotaxis | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0031639 | plasminogen activation | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0043031 | negative regulation of macrophage activation | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0009111 | vitamin catabolic process | 2 | 3 | 1.00e+00 |
| GO:BP | GO:0071702 | organic substance transport | 2 | 2116 | 1.00e+00 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0032310 | prostaglandin secretion | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0032309 | icosanoid secretion | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0032308 | positive regulation of prostaglandin secretion | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0032306 | regulation of prostaglandin secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 13 | 39 | 1.00e+00 |
| GO:BP | GO:0045109 | intermediate filament organization | 6 | 21 | 1.00e+00 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 2 | 5 | 1.00e+00 |
| GO:BP | GO:0032276 | regulation of gonadotropin secretion | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0045838 | positive regulation of membrane potential | 3 | 12 | 1.00e+00 |
| GO:BP | GO:0032275 | luteinizing hormone secretion | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0032274 | gonadotropin secretion | 2 | 10 | 1.00e+00 |
| GO:BP | GO:1903027 | regulation of opsonization | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0043320 | natural killer cell degranulation | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0043312 | neutrophil degranulation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002717 | positive regulation of natural killer cell mediated immunity | 1 | 11 | 1.00e+00 |
| GO:BP | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 24 | 337 | 1.00e+00 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 2 | 29 | 1.00e+00 |
| GO:BP | GO:0002643 | regulation of tolerance induction | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 3 | 137 | 1.00e+00 |
| GO:BP | GO:0035036 | sperm-egg recognition | 2 | 22 | 1.00e+00 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0010324 | membrane invagination | 3 | 49 | 1.00e+00 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 12 | 58 | 1.00e+00 |
| GO:BP | GO:0001714 | endodermal cell fate specification | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 4 | 18 | 1.00e+00 |
| GO:BP | GO:0010467 | gene expression | 4 | 4587 | 1.00e+00 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0010519 | negative regulation of phospholipase activity | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0002441 | histamine secretion involved in inflammatory response | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.00e+00 |
| GO:BP | GO:1901616 | organic hydroxy compound catabolic process | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 21 | 227 | 1.00e+00 |
| GO:BP | GO:0010572 | positive regulation of platelet activation | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0072657 | protein localization to membrane | 5 | 523 | 1.00e+00 |
| GO:BP | GO:0060746 | parental behavior | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0042421 | norepinephrine biosynthetic process | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0060732 | positive regulation of inositol phosphate biosynthetic process | 2 | 5 | 1.00e+00 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 15 | 55 | 1.00e+00 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 1.00e+00 |
| GO:BP | GO:0002507 | tolerance induction | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0060836 | lymphatic endothelial cell differentiation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0072015 | podocyte development | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 4 | 96 | 1.00e+00 |
| GO:BP | GO:0042953 | lipoprotein transport | 1 | 14 | 1.00e+00 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 4 | 27 | 1.00e+00 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 2 | 20 | 1.00e+00 |
| GO:BP | GO:0031349 | positive regulation of defense response | 16 | 251 | 1.00e+00 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0002553 | histamine secretion by mast cell | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 2 | 23 | 1.00e+00 |
| GO:BP | GO:1901317 | regulation of flagellated sperm motility | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0031341 | regulation of cell killing | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 15 | 57 | 1.00e+00 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0042730 | fibrinolysis | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0042711 | maternal behavior | 1 | 7 | 1.00e+00 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0002513 | tolerance induction to self antigen | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 4 | 18 | 1.00e+00 |
| GO:BP | GO:0046415 | urate metabolic process | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:04115 | p53 signaling pathway | 14 | 65 | 7.52e-08 |
| KEGG | KEGG:03410 | Base excision repair | 26 | 33 | 3.26e-05 |
| KEGG | KEGG:04110 | Cell cycle | 53 | 151 | 3.26e-05 |
| KEGG | KEGG:03030 | DNA replication | 25 | 35 | 6.57e-05 |
| KEGG | KEGG:03440 | Homologous recombination | 13 | 39 | 1.38e-03 |
| KEGG | KEGG:03430 | Mismatch repair | 17 | 22 | 3.99e-03 |
| KEGG | KEGG:01524 | Platinum drug resistance | 22 | 65 | 7.91e-03 |
| KEGG | KEGG:04217 | Necroptosis | 11 | 106 | 2.87e-02 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 14 | 48 | 2.87e-02 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 3 | 69 | 3.29e-02 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 2 | 61 | 4.24e-02 |
| KEGG | KEGG:00230 | Purine metabolism | 4 | 97 | 4.88e-02 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 2 | 20 | 4.93e-02 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 9 | 43 | 4.93e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 4 | 75 | 5.14e-02 |
| KEGG | KEGG:05034 | Alcoholism | 2 | 122 | 5.32e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 5.32e-02 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 2 | 110 | 5.32e-02 |
| KEGG | KEGG:04210 | Apoptosis | 31 | 118 | 6.37e-02 |
| KEGG | KEGG:00480 | Glutathione metabolism | 4 | 44 | 6.94e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 1 | 57 | 7.14e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 7.40e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 7.55e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 7.55e-02 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 7.55e-02 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 7.55e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 1 | 84 | 8.23e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 8.49e-02 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 2 | 6 | 1.10e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 1.18e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 3 | 10 | 1.18e-01 |
| KEGG | KEGG:05224 | Breast cancer | 1 | 117 | 1.18e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 1.18e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 1.18e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 1.19e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 1 | 145 | 1.19e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 1.34e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 1.35e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 8 | 24 | 1.42e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 4 | 46 | 1.73e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 16 | 70 | 1.96e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 6 | 86 | 2.09e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 21 | 87 | 2.23e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 21 | 66 | 2.27e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 8 | 145 | 2.27e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 2 | 78 | 2.35e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 16 | 56 | 2.37e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 6 | 19 | 2.37e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 9 | 115 | 2.37e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 30 | 56 | 2.37e-01 |
| KEGG | KEGG:04530 | Tight junction | 3 | 137 | 2.37e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 2 | 8 | 2.37e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 13 | 35 | 2.37e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 1 | 409 | 2.53e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 1 | 16 | 2.64e-01 |
| KEGG | KEGG:03018 | RNA degradation | 11 | 74 | 2.66e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 4 | 35 | 3.30e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 43 | 180 | 3.34e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 1 | 34 | 3.48e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 15 | 107 | 3.52e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 1 | 29 | 3.77e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 7 | 42 | 3.77e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 5 | 18 | 3.77e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 27 | 109 | 3.77e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 14 | 76 | 3.82e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 9 | 44 | 3.84e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 3 | 6 | 3.84e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 1 | 84 | 3.91e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 4.07e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 2 | 91 | 4.07e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 2 | 91 | 4.20e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 12 | 30 | 4.32e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 4.32e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 28 | 133 | 4.46e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 2 | 49 | 4.46e-01 |
| KEGG | KEGG:04540 | Gap junction | 4 | 73 | 4.67e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 3 | 40 | 4.99e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 49 | 134 | 4.99e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 14 | 55 | 4.99e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 2 | 112 | 5.07e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 2 | 55 | 5.07e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 2 | 44 | 5.09e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 18 | 69 | 5.09e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 3 | 172 | 5.09e-01 |
| KEGG | KEGG:05164 | Influenza A | 2 | 103 | 5.09e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 5.09e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 1 | 56 | 5.09e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 5.09e-01 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 1 | 5 | 5.09e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 5.09e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 5.09e-01 |
| KEGG | KEGG:05016 | Huntington disease | 5 | 256 | 5.09e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 6 | 1146 | 5.09e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 10 | 128 | 5.09e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 5.09e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 24 | 98 | 5.09e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 5.09e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 1 | 55 | 5.63e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 17 | 74 | 5.69e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 26 | 85 | 5.69e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 5.69e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 18 | 31 | 5.69e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 5.69e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 5.69e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 9 | 164 | 5.69e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 2 | 38 | 5.69e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 1 | 21 | 5.85e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 15 | 51 | 5.85e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 16 | 32 | 5.85e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 5 | 22 | 5.85e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 11 | 58 | 5.85e-01 |
| KEGG | KEGG:04144 | Endocytosis | 62 | 232 | 5.85e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 5.85e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 12 | 19 | 5.85e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 5.85e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 2 | 63 | 5.85e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 5.85e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 5.85e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 1 | 74 | 5.85e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 5 | 114 | 5.85e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 1 | 82 | 5.86e-01 |
| KEGG | KEGG:05131 | Shigellosis | 43 | 210 | 6.04e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 2 | 22 | 6.07e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 21 | 135 | 6.07e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 20 | 82 | 6.07e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 3 | 4 | 6.07e-01 |
| KEGG | KEGG:04976 | Bile secretion | 1 | 42 | 6.07e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 4 | 25 | 6.07e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 1 | 87 | 6.07e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 3 | 106 | 6.07e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 51 | 99 | 6.07e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 10 | 214 | 6.07e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 1 | 73 | 6.07e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 2 | 97 | 6.13e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 2 | 37 | 6.13e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 9 | 272 | 6.23e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 1 | 58 | 6.23e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 15 | 76 | 6.23e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 3 | 16 | 6.23e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 6.23e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 6.23e-01 |
| KEGG | KEGG:04145 | Phagosome | 4 | 99 | 6.23e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 3 | 23 | 6.23e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 6.24e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 9 | 37 | 6.24e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 3 | 56 | 6.38e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 26 | 121 | 6.46e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 6.46e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 9 | 17 | 6.46e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 21 | 77 | 6.46e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 14 | 86 | 6.46e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 1 | 115 | 6.46e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 3 | 38 | 6.46e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 2 | 28 | 6.46e-01 |
| KEGG | KEGG:04146 | Peroxisome | 35 | 68 | 6.46e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 18 | 108 | 6.46e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 6.46e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 1 | 88 | 6.48e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 1 | 26 | 6.55e-01 |
| KEGG | KEGG:04142 | Lysosome | 1 | 118 | 6.59e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 18 | 46 | 6.59e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 6.59e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 1 | 22 | 6.59e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 1 | 20 | 6.59e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 6 | 170 | 6.59e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 23 | 133 | 6.59e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 6.59e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 1 | 40 | 6.59e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 1 | 106 | 6.59e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 15 | 71 | 6.69e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 6.69e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 6.73e-01 |
| KEGG | KEGG:04520 | Adherens junction | 13 | 67 | 6.73e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 1 | 3 | 6.73e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 2 | 18 | 6.73e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 4 | 157 | 6.76e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 1 | 131 | 6.76e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 3 | 46 | 6.76e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 1 | 115 | 6.76e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 6 | 33 | 6.77e-01 |
| KEGG | KEGG:02010 | ABC transporters | 8 | 29 | 6.77e-01 |
| KEGG | KEGG:00000 | KEGG root term | 9 | 5558 | 6.77e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 3 | 16 | 6.77e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 9 | 17 | 6.77e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 3 | 35 | 6.79e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 4 | 10 | 6.85e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 6 | 165 | 6.85e-01 |
| KEGG | KEGG:04730 | Long-term depression | 6 | 42 | 6.87e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 4 | 121 | 6.87e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 4 | 303 | 6.90e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 12 | 385 | 6.92e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 6.98e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 12 | 72 | 7.08e-01 |
| KEGG | KEGG:05142 | Chagas disease | 3 | 75 | 7.10e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 12 | 7.10e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 2 | 41 | 7.10e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 4 | 104 | 7.14e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 1 | 27 | 7.15e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 3 | 161 | 7.15e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 2 | 11 | 7.19e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 2 | 44 | 7.20e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 7.25e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 5 | 150 | 7.30e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 10 | 49 | 7.30e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 7.30e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 16 | 56 | 7.30e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 2 | 28 | 7.30e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 2 | 15 | 7.30e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 7 | 10 | 7.30e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 1 | 83 | 7.30e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 13 | 24 | 7.30e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 17 | 49 | 7.30e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 15 | 78 | 7.30e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 2 | 151 | 7.30e-01 |
| KEGG | KEGG:00780 | Biotin metabolism | 2 | 3 | 7.30e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 7.30e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 7 | 17 | 7.30e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 7 | 25 | 7.31e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 1 | 41 | 7.52e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 3 | 79 | 7.52e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 7.52e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 1 | 8 | 7.56e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 1 | 84 | 7.56e-01 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 7.56e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 2 | 15 | 7.56e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 11 | 65 | 7.56e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 10 | 7.61e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 5 | 28 | 7.63e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 17 | 110 | 7.63e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 1 | 43 | 7.70e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 7.70e-01 |
| KEGG | KEGG:05133 | Pertussis | 2 | 45 | 7.74e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 2 | 50 | 7.79e-01 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 2 | 3 | 7.79e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 37 | 176 | 7.87e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 4 | 67 | 7.94e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 7.94e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 2 | 22 | 7.96e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 3 | 240 | 7.96e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 8.05e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 2 | 14 | 8.05e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 7 | 14 | 8.05e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 1 | 49 | 8.05e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 2 | 98 | 8.05e-01 |
| KEGG | KEGG:00232 | Caffeine metabolism | 1 | 1 | 8.05e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 8 | 70 | 8.05e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 26 | 49 | 8.05e-01 |
| KEGG | KEGG:03040 | Spliceosome | 2 | 132 | 8.05e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 8.05e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 6 | 12 | 8.05e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 8.05e-01 |
| KEGG | KEGG:04924 | Renin secretion | 16 | 50 | 8.05e-01 |
| KEGG | KEGG:03265 | Virion - Lyssavirus | 1 | 3 | 8.07e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 2 | 69 | 8.07e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 1 | 14 | 8.08e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 6 | 61 | 8.08e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 9 | 24 | 8.12e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 1 | 112 | 8.12e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 4 | 20 | 8.16e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 1 | 63 | 8.18e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 8 | 48 | 8.20e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 2 | 38 | 8.22e-01 |
| KEGG | KEGG:03050 | Proteasome | 4 | 42 | 8.26e-01 |
| KEGG | KEGG:04744 | Phototransduction | 1 | 12 | 8.26e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 2 | 52 | 8.26e-01 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 8.26e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 8 | 77 | 8.26e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 3 | 138 | 8.27e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 2 | 33 | 8.27e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 8.27e-01 |
| KEGG | KEGG:04611 | Platelet activation | 9 | 89 | 8.27e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 9 | 110 | 8.43e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 10 | 78 | 8.43e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 1 | 12 | 8.43e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 1 | 415 | 8.43e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 2 | 50 | 8.43e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 1 | 51 | 8.43e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 20 | 104 | 8.43e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 25 | 171 | 8.43e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 7 | 315 | 8.44e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 3 | 13 | 8.69e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 1 | 69 | 8.69e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 8.80e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 9 | 64 | 8.89e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 11 | 24 | 8.89e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 1 | 32 | 8.91e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 1 | 12 | 8.91e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 7 | 83 | 8.91e-01 |
| KEGG | KEGG:03060 | Protein export | 9 | 23 | 8.91e-01 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 1 | 12 | 8.91e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 8.91e-01 |
| KEGG | KEGG:00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 2 | 2 | 8.91e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 7 | 124 | 9.08e-01 |
| KEGG | KEGG:00511 | Other glycan degradation | 3 | 16 | 9.20e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 4 | 226 | 9.20e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 5 | 28 | 9.20e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 23 | 60 | 9.20e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 1 | 179 | 9.20e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 2 | 7 | 9.20e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 9.20e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 2 | 103 | 9.24e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 3 | 86 | 9.24e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 2 | 44 | 9.24e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 3 | 63 | 9.24e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 9 | 43 | 9.24e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 5 | 28 | 9.28e-01 |
| KEGG | KEGG:05134 | Legionellosis | 3 | 39 | 9.31e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 29 | 142 | 9.54e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 4 | 26 | 9.58e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 1 | 106 | 9.58e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 2 | 99 | 9.58e-01 |
| KEGG | KEGG:05020 | Prion disease | 2 | 220 | 9.60e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 2 | 12 | 9.77e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 1 | 74 | 9.77e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 1 | 23 | 9.78e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 2 | 9 | 9.79e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 7 | 53 | 9.79e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 4 | 31 | 9.80e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 1 | 33 | 9.81e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 2 | 64 | 9.83e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 17 | 97 | 9.83e-01 |
| KEGG | KEGG:01200 | Carbon metabolism | 6 | 98 | 9.93e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 4 | 55 | 9.95e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 1 | 31 | 9.95e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 9.95e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 1 | 31 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 4 | 36 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 2 | 40 | 1.00e+00 |
| KEGG | KEGG:04740 | Olfactory transduction | 1 | 25 | 1.00e+00 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 1.00e+00 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 2 | 9 | 1.00e+00 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 3 | 41 | 1.00e+00 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 4 | 254 | 1.00e+00 |
| KEGG | KEGG:04742 | Taste transduction | 2 | 22 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 16 | 127 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 2 | 75 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 11 | 89 | 1.00e+00 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 10 | 94 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 1 | 186 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 1 | 19 | 1.00e+00 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 3 | 9 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 2 | 23 | 1.00e+00 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 4 | 19 | 1.00e+00 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 3 | 13 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 2 | 12 | 1.00e+00 |
| KEGG | KEGG:05310 | Asthma | 1 | 3 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 3 | 169 | 1.00e+00 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 5 | 1.00e+00 |
EPI
All Epi DEGs adj. p value < 0.05
EPspecific genes set now does not show enrichment in any terms.
# EPsp3 <- list3totvenn3$..values..[[12]]
# EPspgostres <- gost(query = EPsp3,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EPspgostres,"data/DEG-GO/EPspgostres.RDS")
# EPgostres <- gost(query = sigVEP3$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
#
# saveRDS(EPgostres,"data/DEG-GO/EPgostres.RDS")
EPspc3h <- readRDS("data/DEG-GO/EPspgostres.RDS")
EPgostres <- readRDS("data/DEG-GO/EPgostres.RDS")
# EPp <- gostplot(EPgostres, capped = FALSE, interactive = TRUE)
# EPp
EPtable1 <- EPgostres$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
EPtable1 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('EPI 3 hour top ten GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)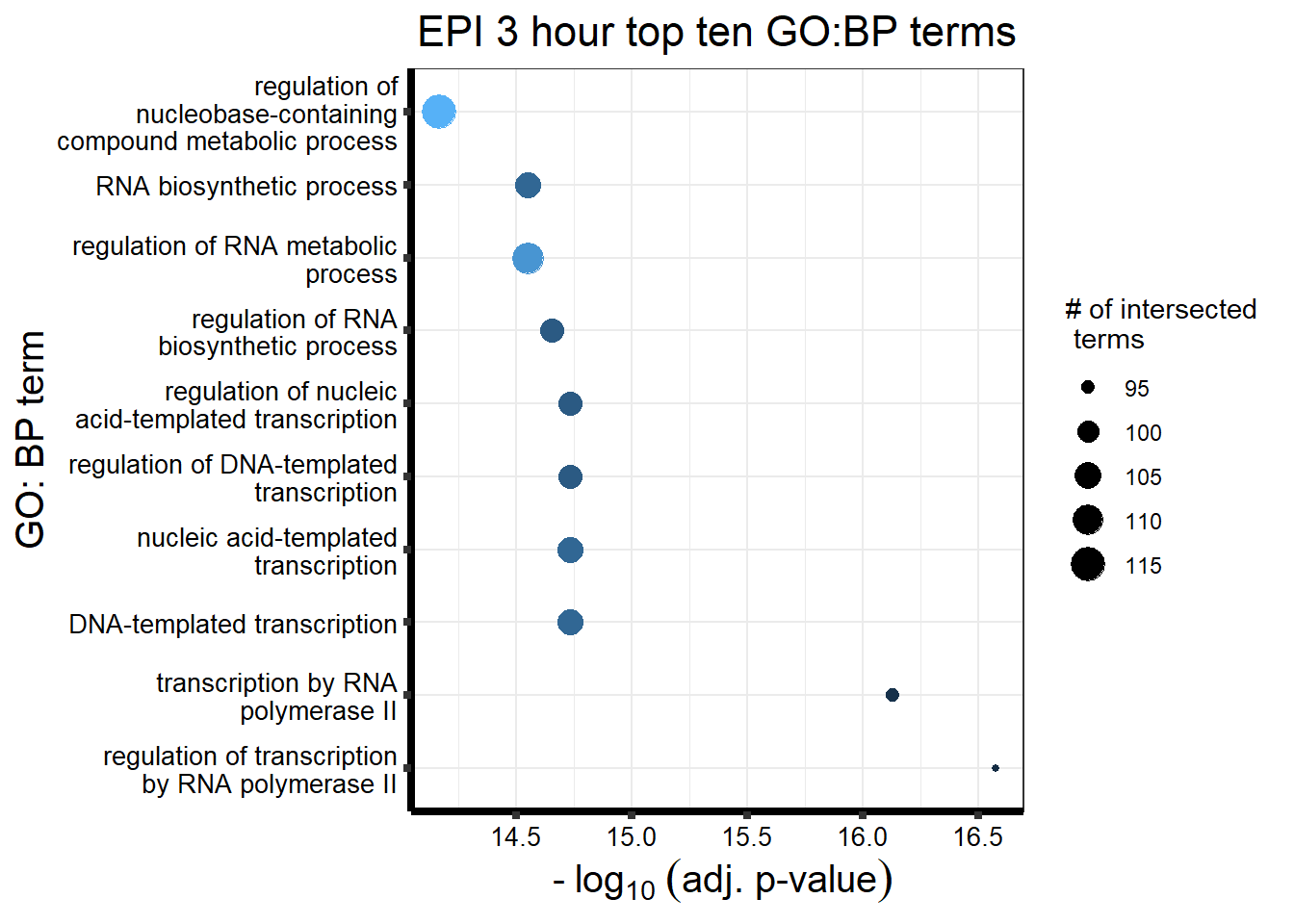
EPtable1 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "3hour EPI full enrichment list") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 94 | 1914 | 2.65e-17 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 95 | 1995 | 7.43e-17 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 102 | 2576 | 1.84e-15 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 102 | 2574 | 1.84e-15 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 104 | 2684 | 1.84e-15 |
| GO:BP | GO:0006351 | DNA-templated transcription | 104 | 2683 | 1.84e-15 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 102 | 2593 | 2.20e-15 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 104 | 2714 | 2.81e-15 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 111 | 2845 | 2.81e-15 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 115 | 3080 | 6.81e-15 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 106 | 3012 | 1.52e-13 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 106 | 3070 | 8.60e-13 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 105 | 3080 | 2.82e-12 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 106 | 3132 | 2.82e-12 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 106 | 3133 | 3.21e-12 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 106 | 3169 | 5.78e-12 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 132 | 4292 | 2.54e-11 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 106 | 3237 | 3.39e-11 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 113 | 3730 | 9.15e-11 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 133 | 4417 | 1.08e-10 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 112 | 3750 | 5.52e-10 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 125 | 4174 | 2.28e-09 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 109 | 4245 | 1.09e-06 |
| GO:BP | GO:0010468 | regulation of gene expression | 117 | 3580 | 2.83e-06 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 134 | 4617 | 1.15e-05 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 111 | 4539 | 1.15e-05 |
| GO:BP | GO:0009058 | biosynthetic process | 111 | 4590 | 2.46e-05 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 35 | 740 | 9.32e-05 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 40 | 1017 | 3.73e-04 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 29 | 4048 | 3.73e-04 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 40 | 1015 | 3.73e-04 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 48 | 1441 | 4.67e-04 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 40 | 1026 | 4.67e-04 |
| GO:BP | GO:0019222 | regulation of metabolic process | 135 | 5017 | 8.76e-04 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 41 | 1214 | 1.11e-03 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 68 | 2412 | 1.12e-03 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 41 | 1119 | 1.14e-03 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 50 | 1602 | 1.24e-03 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 44 | 1315 | 1.37e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 44 | 1315 | 1.37e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 44 | 1322 | 1.48e-03 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 29 | 4455 | 1.57e-03 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 41 | 1239 | 1.64e-03 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 54 | 1735 | 1.99e-03 |
| GO:BP | GO:0016070 | RNA metabolic process | 27 | 3600 | 2.17e-03 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 29 | 4570 | 2.50e-03 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 25 | 4596 | 2.88e-03 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 39 | 1280 | 3.00e-03 |
| GO:BP | GO:1904290 | negative regulation of mitotic DNA damage checkpoint | 1 | 1 | 4.64e-03 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 39 | 1209 | 4.79e-03 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 25 | 4747 | 5.22e-03 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 25 | 4946 | 8.06e-03 |
| GO:BP | GO:0019827 | stem cell population maintenance | 10 | 149 | 8.65e-03 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 33 | 937 | 9.00e-03 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 45 | 1507 | 9.00e-03 |
| GO:BP | GO:0098727 | maintenance of cell number | 10 | 151 | 9.82e-03 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 1.16e-02 |
| GO:BP | GO:1903676 | positive regulation of cap-dependent translational initiation | 1 | 1 | 1.16e-02 |
| GO:BP | GO:1903674 | regulation of cap-dependent translational initiation | 1 | 1 | 1.16e-02 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 54 | 1849 | 1.24e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 2 | 16 | 1.40e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 69 | 2646 | 1.87e-02 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 45 | 1564 | 1.99e-02 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 2.13e-02 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 2 | 34 | 2.35e-02 |
| GO:BP | GO:0055025 | positive regulation of cardiac muscle tissue development | 1 | 2 | 2.76e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 2 | 5 | 2.95e-02 |
| GO:BP | GO:1903677 | regulation of cap-independent translational initiation | 1 | 3 | 2.97e-02 |
| GO:BP | GO:1903679 | positive regulation of cap-independent translational initiation | 1 | 3 | 2.97e-02 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 6 | 68 | 3.06e-02 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 19 | 1604 | 3.06e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 3.16e-02 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 5 | 62 | 3.34e-02 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 2 | 8 | 3.48e-02 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 1 | 4 | 3.50e-02 |
| GO:BP | GO:0045844 | positive regulation of striated muscle tissue development | 1 | 3 | 3.50e-02 |
| GO:BP | GO:0048636 | positive regulation of muscle organ development | 1 | 3 | 3.50e-02 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 4 | 3.50e-02 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 14 | 336 | 4.09e-02 |
| GO:BP | GO:1904690 | positive regulation of cytoplasmic translational initiation | 1 | 5 | 4.16e-02 |
| GO:BP | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress | 1 | 5 | 4.16e-02 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 5 | 4.16e-02 |
| GO:BP | GO:0045659 | negative regulation of neutrophil differentiation | 1 | 2 | 4.23e-02 |
| GO:BP | GO:0045645 | positive regulation of eosinophil differentiation | 1 | 1 | 4.23e-02 |
| GO:BP | GO:0045643 | regulation of eosinophil differentiation | 1 | 1 | 4.23e-02 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 4.23e-02 |
| GO:BP | GO:1904688 | regulation of cytoplasmic translational initiation | 1 | 6 | 4.62e-02 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 1 | 6 | 4.62e-02 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 4 | 44 | 4.62e-02 |
| GO:BP | GO:0055024 | regulation of cardiac muscle tissue development | 1 | 3 | 5.00e-02 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 2 | 67 | 5.00e-02 |
| GO:BP | GO:0006473 | protein acetylation | 9 | 190 | 5.13e-02 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 1 | 7 | 5.13e-02 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 1 | 7 | 5.13e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 5.27e-02 |
| GO:BP | GO:0090721 | primary adaptive immune response involving T cells and B cells | 1 | 1 | 5.40e-02 |
| GO:BP | GO:0090720 | primary adaptive immune response | 1 | 1 | 5.40e-02 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 5.40e-02 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 1 | 8 | 5.40e-02 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 5.40e-02 |
| GO:BP | GO:0045658 | regulation of neutrophil differentiation | 1 | 2 | 5.40e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 5.57e-02 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 2 | 10 | 5.69e-02 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 2 | 11 | 5.69e-02 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 1 | 8 | 5.77e-02 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 1 | 25 | 5.77e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 11 | 216 | 5.77e-02 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 5 | 50 | 5.97e-02 |
| GO:BP | GO:0006968 | cellular defense response | 2 | 15 | 5.97e-02 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 2 | 77 | 5.97e-02 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 10 | 6.14e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 4 | 40 | 6.15e-02 |
| GO:BP | GO:0030853 | negative regulation of granulocyte differentiation | 2 | 6 | 6.30e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 6 | 199 | 6.30e-02 |
| GO:BP | GO:0060840 | artery development | 6 | 83 | 6.30e-02 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 6.30e-02 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 10 | 6.30e-02 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 1 | 8 | 6.30e-02 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 1 | 11 | 6.30e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 3 | 277 | 6.37e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 70 | 2880 | 6.47e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 5 | 67 | 6.47e-02 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 2 | 55 | 6.47e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 1 | 33 | 6.47e-02 |
| GO:BP | GO:0060430 | lung saccule development | 1 | 7 | 6.49e-02 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 1 | 3 | 6.49e-02 |
| GO:BP | GO:0048844 | artery morphogenesis | 5 | 58 | 6.64e-02 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 2 | 13 | 6.65e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 1 | 36 | 6.70e-02 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 1 | 36 | 6.82e-02 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 6.89e-02 |
| GO:BP | GO:0030222 | eosinophil differentiation | 1 | 2 | 6.89e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 1 | 39 | 7.13e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 5 | 69 | 7.13e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 3 | 300 | 7.24e-02 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 2 | 14 | 7.27e-02 |
| GO:BP | GO:0060212 | negative regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 3 | 7.41e-02 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 1 | 14 | 7.41e-02 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 4 | 69 | 7.66e-02 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 1 | 16 | 7.73e-02 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 1 | 16 | 7.73e-02 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 7.85e-02 |
| GO:BP | GO:0061780 | mitotic cohesin loading | 1 | 1 | 7.89e-02 |
| GO:BP | GO:0051102 | DNA ligation involved in DNA recombination | 1 | 1 | 7.89e-02 |
| GO:BP | GO:0075713 | establishment of integrated proviral latency | 1 | 1 | 7.89e-02 |
| GO:BP | GO:0032535 | regulation of cellular component size | 3 | 305 | 7.89e-02 |
| GO:BP | GO:0035261 | external genitalia morphogenesis | 1 | 1 | 7.89e-02 |
| GO:BP | GO:0034605 | cellular response to heat | 2 | 51 | 7.96e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 4 | 84 | 7.99e-02 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 4 | 124 | 7.99e-02 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 1 | 17 | 8.03e-02 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 8.03e-02 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 8.03e-02 |
| GO:BP | GO:0043041 | amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1 | 8.10e-02 |
| GO:BP | GO:1904322 | cellular response to forskolin | 1 | 11 | 8.10e-02 |
| GO:BP | GO:1904321 | response to forskolin | 1 | 11 | 8.10e-02 |
| GO:BP | GO:0015074 | DNA integration | 2 | 9 | 8.10e-02 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 56 | 2270 | 8.13e-02 |
| GO:BP | GO:0031056 | regulation of histone modification | 4 | 125 | 8.17e-02 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 2 | 439 | 8.17e-02 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 5 | 160 | 8.33e-02 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 8.37e-02 |
| GO:BP | GO:0043543 | protein acylation | 7 | 228 | 8.37e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 2 | 118 | 8.49e-02 |
| GO:BP | GO:2001145 | negative regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 8.49e-02 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 3 | 33 | 8.49e-02 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 5 | 72 | 8.49e-02 |
| GO:BP | GO:2001144 | regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 8.49e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 2 | 118 | 8.49e-02 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 8.49e-02 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 3 | 23 | 8.49e-02 |
| GO:BP | GO:0030854 | positive regulation of granulocyte differentiation | 1 | 5 | 8.49e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 3 | 344 | 8.49e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 1 | 56 | 8.49e-02 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 1 | 7 | 8.49e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 3 | 345 | 8.49e-02 |
| GO:BP | GO:0060324 | face development | 4 | 45 | 8.49e-02 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 8.63e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 4 | 620 | 8.63e-02 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 2 | 6 | 8.73e-02 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 1 | 64 | 8.75e-02 |
| GO:BP | GO:0007613 | memory | 2 | 92 | 8.75e-02 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 1 | 13 | 8.79e-02 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 8.79e-02 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 8.79e-02 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 6 | 65 | 8.79e-02 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 8.79e-02 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 8.79e-02 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 8.79e-02 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 1 | 23 | 8.79e-02 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 3 | 17 | 8.79e-02 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 8.79e-02 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 1 | 60 | 8.79e-02 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 3 | 45 | 8.79e-02 |
| GO:BP | GO:0043434 | response to peptide hormone | 2 | 321 | 8.89e-02 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 1 | 8 | 8.89e-02 |
| GO:BP | GO:0016573 | histone acetylation | 4 | 143 | 8.89e-02 |
| GO:BP | GO:0010720 | positive regulation of cell development | 3 | 317 | 8.91e-02 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 2 | 8 | 8.98e-02 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 2 | 26 | 8.99e-02 |
| GO:BP | GO:0030223 | neutrophil differentiation | 1 | 8 | 8.99e-02 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 1 | 15 | 8.99e-02 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 1 | 24 | 9.06e-02 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 3 | 27 | 9.09e-02 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 2 | 7 | 9.09e-02 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 9.09e-02 |
| GO:BP | GO:0006308 | DNA catabolic process | 3 | 19 | 9.10e-02 |
| GO:BP | GO:0033762 | response to glucagon | 1 | 13 | 9.16e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 2 | 152 | 9.18e-02 |
| GO:BP | GO:0045175 | basal protein localization | 1 | 1 | 9.18e-02 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 1 | 27 | 9.18e-02 |
| GO:BP | GO:0010165 | response to X-ray | 2 | 29 | 9.18e-02 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 6 | 69 | 9.18e-02 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 1 | 81 | 9.18e-02 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 9.18e-02 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 9.24e-02 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 2 | 22 | 9.28e-02 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 1 | 85 | 9.28e-02 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 4 | 148 | 9.28e-02 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 1 | 15 | 9.49e-02 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 1 | 23 | 9.51e-02 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 1 | 29 | 9.51e-02 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 4 | 150 | 9.52e-02 |
| GO:BP | GO:0097719 | neural tissue regeneration | 1 | 1 | 9.83e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 2 | 151 | 9.83e-02 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 2 | 13 | 9.87e-02 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 29 | 9.90e-02 |
| GO:BP | GO:0061010 | gallbladder development | 1 | 1 | 9.91e-02 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 1 | 18 | 9.91e-02 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 1 | 29 | 1.01e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 1.01e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 2 | 616 | 1.01e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 1 | 96 | 1.01e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 1.01e-01 |
| GO:BP | GO:0045672 | positive regulation of osteoclast differentiation | 1 | 11 | 1.01e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 3 | 418 | 1.01e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 3 | 20 | 1.01e-01 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 1 | 9 | 1.02e-01 |
| GO:BP | GO:1901652 | response to peptide | 2 | 377 | 1.02e-01 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 1.02e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 2 | 395 | 1.02e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 1 | 7 | 1.02e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 1.02e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 3 | 28 | 1.02e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 1.02e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 1 | 31 | 1.02e-01 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 1 | 18 | 1.02e-01 |
| GO:BP | GO:0060479 | lung cell differentiation | 1 | 18 | 1.02e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 1.02e-01 |
| GO:BP | GO:0035904 | aorta development | 4 | 50 | 1.03e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 1 | 103 | 1.04e-01 |
| GO:BP | GO:0010467 | gene expression | 27 | 4587 | 1.04e-01 |
| GO:BP | GO:0110064 | lncRNA catabolic process | 1 | 2 | 1.04e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 3 | 31 | 1.04e-01 |
| GO:BP | GO:0140744 | negative regulation of lncRNA transcription | 1 | 2 | 1.04e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 2 | 7 | 1.04e-01 |
| GO:BP | GO:0140742 | lncRNA transcription | 1 | 2 | 1.04e-01 |
| GO:BP | GO:0140743 | regulation of lncRNA transcription | 1 | 2 | 1.04e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 1 | 38 | 1.05e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 1 | 113 | 1.06e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 1 | 35 | 1.07e-01 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 1 | 11 | 1.08e-01 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 1 | 12 | 1.08e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 1.08e-01 |
| GO:BP | GO:0060010 | Sertoli cell fate commitment | 1 | 1 | 1.08e-01 |
| GO:BP | GO:1904791 | negative regulation of shelterin complex assembly | 1 | 1 | 1.08e-01 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 2 | 9 | 1.08e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 9 | 187 | 1.08e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 1 | 13 | 1.08e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 2 | 11 | 1.08e-01 |
| GO:BP | GO:0070898 | RNA polymerase III preinitiation complex assembly | 1 | 1 | 1.08e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 41 | 1.08e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 2 | 15 | 1.08e-01 |
| GO:BP | GO:0046599 | regulation of centriole replication | 2 | 21 | 1.08e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 1 | 41 | 1.08e-01 |
| GO:BP | GO:0007614 | short-term memory | 1 | 9 | 1.08e-01 |
| GO:BP | GO:0051693 | actin filament capping | 1 | 34 | 1.08e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 1 | 14 | 1.10e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 3 | 26 | 1.10e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 1.10e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 1.10e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 1 | 45 | 1.11e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 3 | 52 | 1.11e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 3 | 87 | 1.12e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 2 | 126 | 1.13e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 1.14e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 1.15e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 1 | 2 | 1.15e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 1.15e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 1.15e-01 |
| GO:BP | GO:0001543 | ovarian follicle rupture | 1 | 1 | 1.15e-01 |
| GO:BP | GO:0003174 | mitral valve development | 2 | 12 | 1.15e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 1.15e-01 |
| GO:BP | GO:0071921 | cohesin loading | 1 | 3 | 1.15e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 1.15e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 1.15e-01 |
| GO:BP | GO:0019043 | establishment of viral latency | 1 | 3 | 1.17e-01 |
| GO:BP | GO:0046832 | negative regulation of RNA export from nucleus | 1 | 2 | 1.17e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 4 | 135 | 1.17e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 1.17e-01 |
| GO:BP | GO:0009408 | response to heat | 2 | 84 | 1.17e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 2 | 159 | 1.17e-01 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 2 | 6 | 1.18e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 1 | 39 | 1.19e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 1 | 142 | 1.19e-01 |
| GO:BP | GO:0019482 | beta-alanine metabolic process | 1 | 2 | 1.22e-01 |
| GO:BP | GO:0007352 | zygotic specification of dorsal/ventral axis | 1 | 2 | 1.23e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 1.23e-01 |
| GO:BP | GO:0032200 | telomere organization | 1 | 166 | 1.23e-01 |
| GO:BP | GO:1902374 | regulation of rRNA catabolic process | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0048372 | lateral mesodermal cell fate commitment | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 13 | 1.23e-01 |
| GO:BP | GO:0021998 | neural plate mediolateral regionalization | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0048377 | lateral mesodermal cell fate specification | 1 | 1 | 1.23e-01 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 12 | 1.23e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 1 | 19 | 1.23e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 5 | 1076 | 1.23e-01 |
| GO:BP | GO:0048378 | regulation of lateral mesodermal cell fate specification | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 1 | 57 | 1.23e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 1 | 20 | 1.23e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 1.23e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | 56 | 1.23e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 5 | 1074 | 1.23e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 1.23e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 4 | 76 | 1.23e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 1 | 22 | 1.23e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 3 | 108 | 1.23e-01 |
| GO:BP | GO:0140200 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in regulation of heart rate | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 1.23e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 2 | 15 | 1.23e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 1 | 19 | 1.23e-01 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 1.23e-01 |
| GO:BP | GO:0061921 | peptidyl-lysine propionylation | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0097281 | immune complex formation | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0140067 | peptidyl-lysine butyrylation | 1 | 1 | 1.23e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 35 | 1.23e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 1 | 19 | 1.23e-01 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 2 | 30 | 1.23e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 3 | 23 | 1.23e-01 |
| GO:BP | GO:0021572 | rhombomere 6 development | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0021599 | abducens nerve formation | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 2 | 19 | 1.23e-01 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 1 | 5 | 1.23e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 1 | 56 | 1.23e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 3 | 33 | 1.23e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 2 | 149 | 1.23e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 2 | 199 | 1.23e-01 |
| GO:BP | GO:0051170 | import into nucleus | 1 | 152 | 1.23e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 4 | 76 | 1.23e-01 |
| GO:BP | GO:0021560 | abducens nerve development | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0086024 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in positive regulation of heart rate | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0019042 | viral latency | 1 | 4 | 1.23e-01 |
| GO:BP | GO:0021598 | abducens nerve morphogenesis | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0060428 | lung epithelium development | 1 | 30 | 1.23e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 5 | 101 | 1.23e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 178 | 1.23e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 1 | 52 | 1.23e-01 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 1.23e-01 |
| GO:BP | GO:1904412 | regulation of cardiac ventricle development | 1 | 1 | 1.23e-01 |
| GO:BP | GO:1904414 | positive regulation of cardiac ventricle development | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0035120 | post-embryonic appendage morphogenesis | 1 | 2 | 1.23e-01 |
| GO:BP | GO:0048338 | mesoderm structural organization | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 1.23e-01 |
| GO:BP | GO:1904155 | DN2 thymocyte differentiation | 1 | 1 | 1.23e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 5 | 103 | 1.23e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 4 | 75 | 1.23e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 1 | 53 | 1.23e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 2 | 47 | 1.23e-01 |
| GO:BP | GO:0035128 | post-embryonic forelimb morphogenesis | 1 | 2 | 1.23e-01 |
| GO:BP | GO:0036268 | swimming | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 1.23e-01 |
| GO:BP | GO:1905285 | fibrous ring of heart morphogenesis | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 7 | 192 | 1.23e-01 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 1.23e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 2 | 11 | 1.23e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 3 | 80 | 1.23e-01 |
| GO:BP | GO:0035127 | post-embryonic limb morphogenesis | 1 | 2 | 1.23e-01 |
| GO:BP | GO:0002383 | immune response in brain or nervous system | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 1.23e-01 |
| GO:BP | GO:0044357 | regulation of rRNA stability | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0048352 | paraxial mesoderm structural organization | 1 | 1 | 1.23e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 1 | 31 | 1.23e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 2 | 36 | 1.25e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 2 | 756 | 1.25e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 3 | 65 | 1.26e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 1 | 50 | 1.26e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 1.26e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 1 | 25 | 1.26e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 3 | 39 | 1.26e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 1 | 2 | 1.26e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 1 | 35 | 1.26e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 1.26e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 1 | 61 | 1.26e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 1.26e-01 |
| GO:BP | GO:0016570 | histone modification | 8 | 433 | 1.26e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 1.26e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 4 | 64 | 1.26e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 1 | 22 | 1.26e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 1 | 2 | 1.26e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 1 | 2 | 1.26e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 1 | 15 | 1.26e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 1.26e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 1.26e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 3 | 27 | 1.26e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 4 | 123 | 1.26e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 5 | 1123 | 1.26e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4 | 124 | 1.26e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 3 | 25 | 1.27e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 3 | 26 | 1.27e-01 |
| GO:BP | GO:0032240 | negative regulation of nucleobase-containing compound transport | 1 | 3 | 1.27e-01 |
| GO:BP | GO:0001823 | mesonephros development | 4 | 77 | 1.27e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 1 | 70 | 1.27e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 3 | 106 | 1.27e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 1.27e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 13 | 464 | 1.27e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 5 | 89 | 1.28e-01 |
| GO:BP | GO:0001756 | somitogenesis | 4 | 48 | 1.28e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 2 | 128 | 1.28e-01 |
| GO:BP | GO:0090084 | negative regulation of inclusion body assembly | 1 | 10 | 1.29e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 16 | 1173 | 1.29e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 2 | 15 | 1.30e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 1 | 4 | 1.31e-01 |
| GO:BP | GO:0051103 | DNA ligation involved in DNA repair | 1 | 5 | 1.31e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 1 | 71 | 1.31e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 1 | 4 | 1.31e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 1 | 76 | 1.31e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 4 | 53 | 1.31e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 3 | 53 | 1.31e-01 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 1 | 3 | 1.31e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 1.31e-01 |
| GO:BP | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 1 | 4 | 1.31e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 2 | 10 | 1.31e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 2 | 31 | 1.31e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 3 | 126 | 1.31e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 2 | 57 | 1.31e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 1 | 50 | 1.31e-01 |
| GO:BP | GO:0038018 | Wnt receptor catabolic process | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 1.31e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 1.31e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 1.31e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 1 | 209 | 1.31e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 1.32e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 1 | 26 | 1.32e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 2 | 906 | 1.33e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 1.33e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 1 | 50 | 1.35e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 1.35e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 1 | 2 | 1.35e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 2 | 13 | 1.35e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 2 | 14 | 1.35e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 1 | 49 | 1.35e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 3 | 581 | 1.36e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 1 | 78 | 1.36e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 1 | 38 | 1.36e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 1.36e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 6 | 165 | 1.37e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 4 | 131 | 1.38e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 1 | 78 | 1.38e-01 |
| GO:BP | GO:0071772 | response to BMP | 4 | 131 | 1.38e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 4 | 816 | 1.40e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 6 | 123 | 1.41e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 1 | 3 | 1.42e-01 |
| GO:BP | GO:0035601 | protein deacylation | 3 | 116 | 1.42e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 1.42e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 1.42e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 2 | 1028 | 1.42e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 1 | 3 | 1.42e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 1 | 3 | 1.42e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 1.42e-01 |
| GO:BP | GO:0061038 | uterus morphogenesis | 1 | 5 | 1.42e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 1.42e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 1 | 3 | 1.42e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 1 | 91 | 1.42e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 1.42e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 1.43e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 2 | 279 | 1.43e-01 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 1 | 6 | 1.43e-01 |
| GO:BP | GO:0009725 | response to hormone | 2 | 642 | 1.43e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 5 | 1137 | 1.43e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 2 | 150 | 1.43e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 1.43e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 1.43e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 4 | 957 | 1.43e-01 |
| GO:BP | GO:0021536 | diencephalon development | 1 | 41 | 1.43e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 1 | 4 | 1.43e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 2 | 18 | 1.43e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 2 | 17 | 1.43e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 1.43e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 1.44e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 3 | 43 | 1.44e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 1 | 276 | 1.44e-01 |
| GO:BP | GO:0097676 | histone H3-K36 dimethylation | 1 | 4 | 1.45e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 3 | 119 | 1.45e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 1.46e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 1 | 4 | 1.46e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 1 | 6 | 1.46e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 1 | 6 | 1.46e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 2 | 310 | 1.46e-01 |
| GO:BP | GO:0051169 | nuclear transport | 4 | 295 | 1.46e-01 |
| GO:BP | GO:0007611 | learning or memory | 2 | 202 | 1.46e-01 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 1 | 4 | 1.46e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 4 | 295 | 1.46e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 5 | 113 | 1.46e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 4 | 89 | 1.46e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 2 | 1006 | 1.46e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 3 | 43 | 1.47e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 4 | 138 | 1.48e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 2 | 55 | 1.48e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 1.50e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 1 | 36 | 1.50e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 3 | 44 | 1.50e-01 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 1 | 7 | 1.50e-01 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 1 | 5 | 1.50e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 1.50e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 12 | 358 | 1.51e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 1 | 7 | 1.51e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 1 | 5 | 1.51e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 1 | 34 | 1.52e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 1 | 274 | 1.52e-01 |
| GO:BP | GO:0140066 | peptidyl-lysine crotonylation | 1 | 2 | 1.52e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 3 | 81 | 1.52e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 3 | 2065 | 1.52e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 3 | 86 | 1.52e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 1 | 60 | 1.52e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 1 | 111 | 1.52e-01 |
| GO:BP | GO:1901301 | regulation of cargo loading into COPII-coated vesicle | 1 | 2 | 1.52e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 1 | 6 | 1.52e-01 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 1 | 16 | 1.52e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 4 | 89 | 1.52e-01 |
| GO:BP | GO:0035282 | segmentation | 5 | 75 | 1.52e-01 |
| GO:BP | GO:0006974 | DNA damage response | 15 | 785 | 1.52e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 3 | 45 | 1.53e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 1.53e-01 |
| GO:BP | GO:0097368 | establishment of Sertoli cell barrier | 1 | 3 | 1.53e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 5 | 109 | 1.53e-01 |
| GO:BP | GO:0060541 | respiratory system development | 7 | 163 | 1.53e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 1.53e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 2 | 17 | 1.53e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 1.53e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 1.53e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 1 | 105 | 1.53e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 1.53e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 1 | 4 | 1.53e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 1 | 4 | 1.53e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 3 | 100 | 1.53e-01 |
| GO:BP | GO:0061009 | common bile duct development | 1 | 4 | 1.53e-01 |
| GO:BP | GO:1903215 | negative regulation of protein targeting to mitochondrion | 1 | 3 | 1.53e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 3 | 82 | 1.54e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 1 | 105 | 1.54e-01 |
| GO:BP | GO:1901898 | negative regulation of relaxation of cardiac muscle | 1 | 2 | 1.54e-01 |
| GO:BP | GO:2000257 | regulation of protein activation cascade | 1 | 1 | 1.55e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 1.55e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 5 | 114 | 1.55e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 1.55e-01 |
| GO:BP | GO:0070171 | negative regulation of tooth mineralization | 1 | 2 | 1.55e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 1 | 108 | 1.55e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 2 | 25 | 1.55e-01 |
| GO:BP | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation | 1 | 1 | 1.55e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 1.55e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 1.55e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 1.55e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 1 | 70 | 1.55e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 338 | 1.55e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 1.55e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 3 | 41 | 1.55e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 2 | 33 | 1.55e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 1.55e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 4 | 1014 | 1.55e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 5 | 84 | 1.55e-01 |
| GO:BP | GO:0031175 | neuron projection development | 2 | 780 | 1.55e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 3 | 33 | 1.56e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 1 | 33 | 1.56e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 1 | 5 | 1.56e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 3 | 1.56e-01 |
| GO:BP | GO:0048511 | rhythmic process | 2 | 237 | 1.56e-01 |
| GO:BP | GO:0007398 | ectoderm development | 2 | 15 | 1.56e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 1.56e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 1 | 8 | 1.56e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 9 | 331 | 1.56e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 3 | 44 | 1.56e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 2 | 17 | 1.56e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 1 | 84 | 1.56e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 1 | 2 | 1.56e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 2 | 17 | 1.56e-01 |
| GO:BP | GO:0007099 | centriole replication | 2 | 38 | 1.56e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 4 | 92 | 1.56e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 2 | 133 | 1.56e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 1 | 5 | 1.56e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 4 | 93 | 1.56e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 2 | 191 | 1.56e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 2 | 21 | 1.56e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 1.56e-01 |
| GO:BP | GO:0050890 | cognition | 4 | 227 | 1.56e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 1 | 85 | 1.56e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 1 | 66 | 1.56e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 1 | 22 | 1.57e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 1 | 11 | 1.57e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 2 | 53 | 1.57e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 3 | 64 | 1.57e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 1 | 110 | 1.59e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 1 | 37 | 1.59e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 3 | 40 | 1.59e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 2 | 1075 | 1.59e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 1.60e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 5 | 104 | 1.60e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 1.61e-01 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 1 | 3 | 1.61e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 2 | 258 | 1.61e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 1 | 35 | 1.61e-01 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 1.61e-01 |
| GO:BP | GO:0006325 | chromatin organization | 15 | 551 | 1.61e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 391 | 1.61e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 4 | 81 | 1.61e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 2 | 1088 | 1.61e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 2 | 68 | 1.61e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 1 | 86 | 1.61e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 1.61e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 5 | 127 | 1.62e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 1 | 4 | 1.62e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 1.62e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 3 | 27 | 1.62e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 3 | 40 | 1.62e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 2 | 21 | 1.62e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 1 | 130 | 1.62e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 1.62e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 2 | 782 | 1.62e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 1 | 85 | 1.62e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 3 | 893 | 1.62e-01 |
| GO:BP | GO:0034773 | histone H4-K20 trimethylation | 1 | 5 | 1.62e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 3 | 605 | 1.63e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 2 | 1111 | 1.63e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 2 | 59 | 1.63e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 3 | 40 | 1.63e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 6 | 186 | 1.64e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 2 | 8 | 1.64e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 1 | 3 | 1.64e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 2 | 8 | 1.64e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 8 | 230 | 1.64e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 1 | 53 | 1.64e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 3 | 38 | 1.66e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 3 | 40 | 1.66e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 1 | 138 | 1.66e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 2 | 36 | 1.66e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 1 | 27 | 1.66e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 3 | 40 | 1.66e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 1.66e-01 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 1 | 7 | 1.66e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 1 | 126 | 1.67e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 1 | 52 | 1.67e-01 |
| GO:BP | GO:0001843 | neural tube closure | 4 | 90 | 1.67e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 1 | 140 | 1.67e-01 |
| GO:BP | GO:0070841 | inclusion body assembly | 1 | 22 | 1.67e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 1 | 8 | 1.67e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 3 | 679 | 1.67e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 1.67e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 1 | 6 | 1.67e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 5 | 128 | 1.67e-01 |
| GO:BP | GO:0007507 | heart development | 15 | 506 | 1.67e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 2 | 14 | 1.67e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 1 | 4 | 1.67e-01 |
| GO:BP | GO:0048703 | embryonic viscerocranium morphogenesis | 1 | 8 | 1.67e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 1 | 140 | 1.67e-01 |
| GO:BP | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion | 1 | 5 | 1.67e-01 |
| GO:BP | GO:0098534 | centriole assembly | 2 | 41 | 1.67e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 2 | 23 | 1.67e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 1 | 127 | 1.67e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 441 | 1.68e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 1 | 36 | 1.68e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 1 | 140 | 1.68e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 16 | 884 | 1.69e-01 |
| GO:BP | GO:0060606 | tube closure | 4 | 91 | 1.69e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 7 | 222 | 1.69e-01 |
| GO:BP | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1 | 3 | 1.69e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 1 | 147 | 1.69e-01 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 2 | 44 | 1.69e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 2 | 303 | 1.70e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 2 | 830 | 1.70e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 1 | 134 | 1.70e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 1 | 4 | 1.71e-01 |
| GO:BP | GO:0061053 | somite development | 4 | 63 | 1.71e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 2 | 16 | 1.71e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 1 | 6 | 1.71e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 438 | 1.71e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 1 | 3 | 1.71e-01 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 5 | 186 | 1.71e-01 |
| GO:BP | GO:0048666 | neuron development | 2 | 877 | 1.71e-01 |
| GO:BP | GO:0001966 | thigmotaxis | 1 | 2 | 1.71e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 5 | 129 | 1.71e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 2 | 41 | 1.71e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 2 | 21 | 1.71e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 3 | 52 | 1.71e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 1 | 151 | 1.72e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 1.72e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 1.72e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 1.72e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 1 | 102 | 1.72e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 2 | 1248 | 1.72e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 4 | 1167 | 1.72e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 5 | 1147 | 1.72e-01 |
| GO:BP | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 1 | 6 | 1.72e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 6 | 149 | 1.72e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 1 | 6 | 1.73e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 1.74e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 1 | 11 | 1.74e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 3 | 79 | 1.74e-01 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 1.74e-01 |
| GO:BP | GO:0048371 | lateral mesodermal cell differentiation | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0032915 | positive regulation of transforming growth factor beta2 production | 1 | 3 | 1.74e-01 |
| GO:BP | GO:0030879 | mammary gland development | 1 | 106 | 1.75e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 2 | 1219 | 1.76e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 1 | 90 | 1.76e-01 |
| GO:BP | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine | 1 | 2 | 1.77e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 3 | 39 | 1.78e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 504 | 1.78e-01 |
| GO:BP | GO:0035270 | endocrine system development | 1 | 86 | 1.78e-01 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 1.78e-01 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 1 | 2 | 1.81e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 1 | 7 | 1.81e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 1.81e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 1 | 12 | 1.81e-01 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 1.81e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 1 | 14 | 1.81e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 1 | 5 | 1.81e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 1 | 21 | 1.81e-01 |
| GO:BP | GO:0001822 | kidney development | 5 | 244 | 1.81e-01 |
| GO:BP | GO:0035112 | genitalia morphogenesis | 1 | 5 | 1.81e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 1 | 5 | 1.81e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 1 | 12 | 1.81e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 1.81e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 3 | 164 | 1.81e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 4 | 90 | 1.81e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 2 | 16 | 1.81e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 12 | 378 | 1.81e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 4 | 94 | 1.81e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 1.81e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 1 | 108 | 1.81e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 2 | 20 | 1.81e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 1.81e-01 |
| GO:BP | GO:0030324 | lung development | 6 | 146 | 1.82e-01 |
| GO:BP | GO:0016255 | attachment of GPI anchor to protein | 1 | 6 | 1.82e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 1 | 160 | 1.83e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 1 | 108 | 1.83e-01 |
| GO:BP | GO:0035284 | brain segmentation | 1 | 2 | 1.83e-01 |
| GO:BP | GO:0021571 | rhombomere 5 development | 1 | 2 | 1.83e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 2 | 1345 | 1.83e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 1 | 168 | 1.83e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 1 | 30 | 1.84e-01 |
| GO:BP | GO:0035781 | CD86 biosynthetic process | 1 | 2 | 1.84e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 1.84e-01 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 1 | 9 | 1.84e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 2 | 25 | 1.84e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 12 | 378 | 1.84e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 80 | 3625 | 1.84e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 12 | 2135 | 1.84e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 2 | 44 | 1.84e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 3 | 42 | 1.85e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 2 | 49 | 1.85e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 1 | 174 | 1.85e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 3 | 54 | 1.85e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 1 | 3 | 1.86e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 2 | 398 | 1.86e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 1.87e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 12 | 381 | 1.87e-01 |
| GO:BP | GO:0016571 | histone methylation | 5 | 117 | 1.88e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 9 | 4579 | 1.88e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 543 | 1.88e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 11 | 343 | 1.88e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 1 | 10 | 1.88e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 2 | 382 | 1.88e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 6 | 150 | 1.90e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 2 | 51 | 1.90e-01 |
| GO:BP | GO:0000012 | single strand break repair | 1 | 10 | 1.90e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 2 | 23 | 1.91e-01 |
| GO:BP | GO:0072001 | renal system development | 5 | 251 | 1.91e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 2 | 52 | 1.92e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 1.92e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 4 | 91 | 1.92e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 1 | 8 | 1.92e-01 |
| GO:BP | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 6 | 1.93e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 2 | 22 | 1.93e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 1 | 5 | 1.93e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 1 | 16 | 1.93e-01 |
| GO:BP | GO:0016180 | snRNA processing | 1 | 26 | 1.93e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 2 | 21 | 1.94e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 1 | 422 | 1.94e-01 |
| GO:BP | GO:0007517 | muscle organ development | 9 | 277 | 1.94e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 1.94e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 3 | 877 | 1.94e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 4 | 111 | 1.95e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 3 | 47 | 1.95e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 1.95e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 1.95e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 1 | 9 | 1.95e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 1 | 13 | 1.96e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 2 | 38 | 1.97e-01 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 1 | 4 | 1.97e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 2 | 22 | 1.97e-01 |
| GO:BP | GO:1900245 | positive regulation of MDA-5 signaling pathway | 1 | 2 | 1.97e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 4 | 112 | 1.97e-01 |
| GO:BP | GO:0009798 | axis specification | 4 | 63 | 1.97e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2565 | 1.97e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 4 | 94 | 1.97e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 1.97e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 4 | 94 | 1.97e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 1.97e-01 |
| GO:BP | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly | 1 | 7 | 1.97e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 3 | 50 | 1.97e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 1 | 35 | 1.97e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2573 | 1.97e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 1.98e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 2 | 41 | 1.98e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 3 | 33 | 1.99e-01 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 1 | 4 | 1.99e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 4 | 1101 | 1.99e-01 |
| GO:BP | GO:0001880 | Mullerian duct regression | 1 | 4 | 1.99e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 1.99e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 1.99e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 1 | 8 | 1.99e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 1 | 8 | 1.99e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 1.99e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 1 | 8 | 1.99e-01 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 1 | 4 | 1.99e-01 |
| GO:BP | GO:0048370 | lateral mesoderm formation | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 1 | 4 | 1.99e-01 |
| GO:BP | GO:0006915 | apoptotic process | 2 | 1443 | 1.99e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 1 | 9 | 1.99e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 1 | 9 | 1.99e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 1 | 69 | 1.99e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 626 | 1.99e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 2 | 34 | 1.99e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 1.99e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 1 | 144 | 1.99e-01 |
| GO:BP | GO:0048369 | lateral mesoderm morphogenesis | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 1.99e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 7 | 198 | 1.99e-01 |
| GO:BP | GO:0044154 | histone H3-K14 acetylation | 1 | 12 | 2.00e-01 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 1 | 5 | 2.00e-01 |
| GO:BP | GO:0001841 | neural tube formation | 4 | 101 | 2.00e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 2 | 24 | 2.01e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 2 | 27 | 2.01e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 1 | 214 | 2.02e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 2.02e-01 |
| GO:BP | GO:0014004 | microglia differentiation | 1 | 6 | 2.04e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 2 | 1681 | 2.04e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 2.04e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 26 | 7441 | 2.04e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 1 | 90 | 2.04e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 694 | 2.04e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 1 | 234 | 2.04e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 2 | 1061 | 2.04e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 8 | 2.04e-01 |
| GO:BP | GO:0012501 | programmed cell death | 2 | 1479 | 2.04e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 8 | 191 | 2.04e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 2.04e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 1 | 39 | 2.06e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 2 | 40 | 2.07e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 2.07e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 2 | 57 | 2.07e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 2 | 372 | 2.07e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 2.07e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 12 | 387 | 2.07e-01 |
| GO:BP | GO:0035283 | central nervous system segmentation | 1 | 2 | 2.08e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 6 | 4099 | 2.08e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 1 | 139 | 2.08e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 1 | 6 | 2.08e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 2 | 23 | 2.08e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 2.08e-01 |
| GO:BP | GO:0006266 | DNA ligation | 1 | 16 | 2.08e-01 |
| GO:BP | GO:0032868 | response to insulin | 1 | 215 | 2.08e-01 |
| GO:BP | GO:0021762 | substantia nigra development | 1 | 41 | 2.09e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 1 | 6 | 2.09e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 2.09e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 1 | 14 | 2.09e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 2.09e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 1 | 5 | 2.09e-01 |
| GO:BP | GO:0048468 | cell development | 4 | 1963 | 2.09e-01 |
| GO:BP | GO:0006996 | organelle organization | 3 | 3000 | 2.10e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 1 | 228 | 2.10e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 3 | 50 | 2.10e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 2 | 1107 | 2.10e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 1 | 14 | 2.10e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 1 | 7 | 2.10e-01 |
| GO:BP | GO:0007409 | axonogenesis | 2 | 359 | 2.10e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 2.10e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 5 | 109 | 2.11e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 5 | 265 | 2.11e-01 |
| GO:BP | GO:0050821 | protein stabilization | 1 | 191 | 2.11e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 1 | 8 | 2.11e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 1 | 8 | 2.11e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 2.11e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 19 | 875 | 2.11e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 15 | 534 | 2.11e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 2.12e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 6 | 179 | 2.12e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 3 | 63 | 2.12e-01 |
| GO:BP | GO:0048699 | generation of neurons | 2 | 1119 | 2.12e-01 |
| GO:BP | GO:0016310 | phosphorylation | 4 | 1372 | 2.12e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 3 | 47 | 2.12e-01 |
| GO:BP | GO:0051258 | protein polymerization | 1 | 241 | 2.13e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 2.13e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 3 | 55 | 2.13e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 1 | 9 | 2.13e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 1 | 9 | 2.13e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 1 | 40 | 2.14e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 4 | 1223 | 2.14e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 3 | 796 | 2.14e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 5 | 2.14e-01 |
| GO:BP | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 1 | 4 | 2.16e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 1 | 4 | 2.16e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 2.16e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 2.16e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 2.16e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 1 | 11 | 2.16e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 1 | 12 | 2.16e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 1 | 10 | 2.16e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 1 | 10 | 2.17e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 3 | 56 | 2.17e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 2.17e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 1 | 262 | 2.17e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 1 | 9 | 2.17e-01 |
| GO:BP | GO:0006621 | protein retention in ER lumen | 1 | 8 | 2.17e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 660 | 2.17e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 2 | 20 | 2.18e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 1 | 16 | 2.18e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 2 | 1201 | 2.18e-01 |
| GO:BP | GO:0003158 | endothelium development | 1 | 104 | 2.18e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 10 | 719 | 2.18e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 1 | 12 | 2.18e-01 |
| GO:BP | GO:0042661 | regulation of mesodermal cell fate specification | 1 | 4 | 2.18e-01 |
| GO:BP | GO:0008219 | cell death | 2 | 1596 | 2.18e-01 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 1 | 5 | 2.18e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 2.19e-01 |
| GO:BP | GO:0016076 | snRNA catabolic process | 1 | 5 | 2.20e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 678 | 2.20e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 1 | 241 | 2.20e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 3 | 57 | 2.20e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 3 | 57 | 2.20e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 1 | 7 | 2.20e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 1 | 164 | 2.20e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 1 | 16 | 2.21e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 1 | 247 | 2.21e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 1 | 7 | 2.21e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 2.21e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 1 | 10 | 2.21e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 2.21e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 1 | 19 | 2.21e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 827 | 2.21e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 1 | 88 | 2.21e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 2.21e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 1 | 6 | 2.21e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 1 | 210 | 2.21e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 2 | 71 | 2.21e-01 |
| GO:BP | GO:0065001 | specification of axis polarity | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 1 | 11 | 2.21e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 3 | 62 | 2.21e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 2 | 27 | 2.21e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 1 | 72 | 2.21e-01 |
| GO:BP | GO:0030030 | cell projection organization | 2 | 1233 | 2.21e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 3 | 60 | 2.21e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 1 | 16 | 2.21e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 189 | 2.21e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 4 | 120 | 2.21e-01 |
| GO:BP | GO:0090244 | Wnt signaling pathway involved in somitogenesis | 1 | 5 | 2.21e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 3 | 54 | 2.21e-01 |
| GO:BP | GO:0043627 | response to estrogen | 3 | 52 | 2.21e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 8 | 256 | 2.21e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 12 | 2293 | 2.21e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 6 | 2.24e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 2 | 28 | 2.24e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 2 | 24 | 2.24e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 1 | 15 | 2.24e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 1 | 12 | 2.25e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 1 | 259 | 2.26e-01 |
| GO:BP | GO:0021915 | neural tube development | 5 | 146 | 2.26e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 25 | 6996 | 2.27e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 4 | 121 | 2.27e-01 |
| GO:BP | GO:0061564 | axon development | 2 | 398 | 2.27e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 2 | 420 | 2.27e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 6 | 169 | 2.28e-01 |
| GO:BP | GO:0097305 | response to alcohol | 1 | 159 | 2.28e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 1 | 4 | 2.28e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 1 | 17 | 2.28e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 5 | 160 | 2.28e-01 |
| GO:BP | GO:0001708 | cell fate specification | 4 | 56 | 2.28e-01 |
| GO:BP | GO:0060633 | negative regulation of transcription initiation by RNA polymerase II | 1 | 6 | 2.28e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 1 | 168 | 2.28e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 5 | 1378 | 2.28e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 3 | 49 | 2.28e-01 |
| GO:BP | GO:0032197 | retrotransposition | 1 | 4 | 2.28e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 2.29e-01 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 1 | 8 | 2.29e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 2 | 99 | 2.29e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 2 | 61 | 2.29e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 3 | 69 | 2.29e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 1 | 284 | 2.29e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 2.30e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 1 | 6 | 2.30e-01 |
| GO:BP | GO:0006417 | regulation of translation | 2 | 370 | 2.30e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 1 | 8 | 2.30e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 1 | 12 | 2.30e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 1 | 6 | 2.30e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 2.30e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 4 | 165 | 2.31e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 3 | 280 | 2.31e-01 |
| GO:BP | GO:0048630 | skeletal muscle tissue growth | 1 | 8 | 2.32e-01 |
| GO:BP | GO:0048589 | developmental growth | 14 | 522 | 2.32e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 1 | 13 | 2.32e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 1 | 10 | 2.32e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 15 | 551 | 2.32e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 1 | 101 | 2.33e-01 |
| GO:BP | GO:0003170 | heart valve development | 3 | 65 | 2.33e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 2 | 25 | 2.33e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 1 | 14 | 2.33e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 1 | 57 | 2.33e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 5 | 2.33e-01 |
| GO:BP | GO:0060914 | heart formation | 2 | 29 | 2.34e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 2.35e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 211 | 2.37e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 5 | 149 | 2.37e-01 |
| GO:BP | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway | 1 | 9 | 2.37e-01 |
| GO:BP | GO:0003175 | tricuspid valve development | 1 | 6 | 2.37e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 2 | 400 | 2.37e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 1 | 299 | 2.37e-01 |
| GO:BP | GO:0060897 | neural plate regionalization | 1 | 6 | 2.37e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 1 | 318 | 2.37e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 4 | 2035 | 2.37e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 1 | 15 | 2.38e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 2 | 47 | 2.38e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 1 | 15 | 2.38e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 1 | 287 | 2.38e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 1 | 307 | 2.38e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 3 | 881 | 2.38e-01 |
| GO:BP | GO:0022008 | neurogenesis | 2 | 1294 | 2.38e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 3 | 232 | 2.38e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 2.38e-01 |
| GO:BP | GO:0003062 | regulation of heart rate by chemical signal | 1 | 6 | 2.38e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 2.38e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 2 | 1278 | 2.38e-01 |
| GO:BP | GO:0040008 | regulation of growth | 8 | 501 | 2.38e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 133 | 2.38e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 2.38e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 2.38e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 1 | 13 | 2.38e-01 |
| GO:BP | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | 1 | 5 | 2.38e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 3 | 50 | 2.38e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 2 | 68 | 2.39e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 26 | 7069 | 2.39e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 2.39e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | 7 | 2.39e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 3 | 65 | 2.39e-01 |
| GO:BP | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation | 1 | 5 | 2.40e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 1 | 15 | 2.40e-01 |
| GO:BP | GO:0090118 | receptor-mediated endocytosis involved in cholesterol transport | 1 | 5 | 2.40e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 1 | 326 | 2.41e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 84 | 2.41e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 2 | 1256 | 2.41e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 2 | 1255 | 2.41e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 1 | 4 | 2.41e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 2.41e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 2 | 30 | 2.41e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 1 | 956 | 2.42e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 1 | 199 | 2.42e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 12 | 406 | 2.42e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 2 | 35 | 2.42e-01 |
| GO:BP | GO:0006281 | DNA repair | 9 | 534 | 2.44e-01 |
| GO:BP | GO:2000143 | negative regulation of DNA-templated transcription initiation | 1 | 6 | 2.44e-01 |
| GO:BP | GO:0060065 | uterus development | 1 | 13 | 2.44e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 1 | 11 | 2.44e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 1 | 13 | 2.44e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 1 | 9 | 2.44e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 1 | 50 | 2.45e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 1 | 11 | 2.46e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 1 | 199 | 2.46e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 2 | 414 | 2.46e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 1 | 859 | 2.46e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 2.46e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 2.47e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 2.47e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 3 | 63 | 2.47e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 2 | 2011 | 2.47e-01 |
| GO:BP | GO:0043201 | response to leucine | 1 | 13 | 2.48e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 2.48e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 1 | 6 | 2.48e-01 |
| GO:BP | GO:0002457 | T cell antigen processing and presentation | 1 | 4 | 2.48e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 1 | 340 | 2.48e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 2.48e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 1 | 21 | 2.48e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 4 | 290 | 2.48e-01 |
| GO:BP | GO:0039533 | regulation of MDA-5 signaling pathway | 1 | 5 | 2.48e-01 |
| GO:BP | GO:0060463 | lung lobe morphogenesis | 1 | 4 | 2.49e-01 |
| GO:BP | GO:0060462 | lung lobe development | 1 | 4 | 2.49e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 1 | 7 | 2.49e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 1 | 22 | 2.49e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 1 | 21 | 2.49e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 1 | 9 | 2.49e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1 | 91 | 2.49e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 2.49e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 1 | 12 | 2.49e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 1 | 11 | 2.49e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 1 | 69 | 2.49e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 1 | 19 | 2.49e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 1 | 19 | 2.49e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 8 | 2.50e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 2.50e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 2.50e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 1 | 10 | 2.50e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 1 | 9 | 2.50e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 1 | 348 | 2.51e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 2.52e-01 |
| GO:BP | GO:1905770 | regulation of mesodermal cell differentiation | 1 | 6 | 2.53e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 2.53e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 2 | 27 | 2.53e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 1 | 215 | 2.53e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 215 | 2.54e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 2.55e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 1 | 4 | 2.55e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 1 | 5 | 2.55e-01 |
| GO:BP | GO:0035437 | maintenance of protein localization in endoplasmic reticulum | 1 | 11 | 2.55e-01 |
| GO:BP | GO:0001656 | metanephros development | 2 | 64 | 2.56e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 2 | 463 | 2.56e-01 |
| GO:BP | GO:0030901 | midbrain development | 2 | 69 | 2.56e-01 |
| GO:BP | GO:1901078 | negative regulation of relaxation of muscle | 1 | 4 | 2.56e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 6 | 2.56e-01 |
| GO:BP | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine | 1 | 5 | 2.56e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 2 | 32 | 2.56e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 4 | 116 | 2.56e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 2 | 267 | 2.57e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1086 | 2.57e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 2 | 424 | 2.57e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 1 | 10 | 2.57e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 3 | 67 | 2.57e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 97 | 2.58e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 2.58e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 1 | 12 | 2.58e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 1 | 17 | 2.58e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 2.58e-01 |
| GO:BP | GO:0001542 | ovulation from ovarian follicle | 1 | 8 | 2.58e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 1 | 222 | 2.58e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 7 | 227 | 2.58e-01 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 11 | 2.58e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 2.59e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 2.59e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 2.59e-01 |
| GO:BP | GO:0033993 | response to lipid | 2 | 592 | 2.60e-01 |
| GO:BP | GO:0007015 | actin filament organization | 1 | 373 | 2.60e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 2.60e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 250 | 2.60e-01 |
| GO:BP | GO:0007548 | sex differentiation | 7 | 191 | 2.60e-01 |
| GO:BP | GO:0071934 | thiamine transmembrane transport | 1 | 3 | 2.61e-01 |
| GO:BP | GO:0042723 | thiamine-containing compound metabolic process | 1 | 4 | 2.61e-01 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 2.61e-01 |
| GO:BP | GO:0007254 | JNK cascade | 1 | 138 | 2.61e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 2.61e-01 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 1 | 7 | 2.61e-01 |
| GO:BP | GO:0048736 | appendage development | 4 | 142 | 2.61e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 1 | 34 | 2.61e-01 |
| GO:BP | GO:0060173 | limb development | 4 | 142 | 2.61e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 3 | 120 | 2.61e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 1 | 8 | 2.62e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 1 | 18 | 2.62e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 1 | 229 | 2.62e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 2 | 706 | 2.62e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 1 | 23 | 2.63e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 3 | 82 | 2.63e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 4 | 112 | 2.63e-01 |
| GO:BP | GO:0001955 | blood vessel maturation | 1 | 6 | 2.63e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 1 | 16 | 2.63e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 19 | 665 | 2.63e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 1 | 18 | 2.63e-01 |
| GO:BP | GO:0021546 | rhombomere development | 1 | 4 | 2.64e-01 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 2 | 73 | 2.64e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 2 | 479 | 2.64e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 2 | 130 | 2.64e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 1 | 9 | 2.65e-01 |
| GO:BP | GO:0000303 | response to superoxide | 1 | 21 | 2.65e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 1 | 81 | 2.65e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 1 | 63 | 2.65e-01 |
| GO:BP | GO:0032815 | negative regulation of natural killer cell activation | 1 | 3 | 2.65e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 295 | 2.65e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 2.65e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 1 | 320 | 2.65e-01 |
| GO:BP | GO:0035295 | tube development | 2 | 821 | 2.65e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 2.65e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 2 | 61 | 2.65e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 2 | 656 | 2.66e-01 |
| GO:BP | GO:0003162 | atrioventricular node development | 1 | 8 | 2.66e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 2.66e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 1 | 15 | 2.66e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 3 | 58 | 2.66e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 1 | 5 | 2.66e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 2 | 31 | 2.66e-01 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 1 | 7 | 2.66e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 1 | 6 | 2.66e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 6 | 217 | 2.66e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 1 | 141 | 2.66e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 3 | 1399 | 2.66e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 1 | 8 | 2.66e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 2.66e-01 |
| GO:BP | GO:0016050 | vesicle organization | 1 | 320 | 2.66e-01 |
| GO:BP | GO:0019083 | viral transcription | 1 | 48 | 2.66e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 236 | 2.66e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 2.66e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 1 | 4 | 2.67e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 3 | 64 | 2.67e-01 |
| GO:BP | GO:0010038 | response to metal ion | 1 | 264 | 2.67e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 4 | 110 | 2.67e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 1 | 22 | 2.68e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 1 | 23 | 2.68e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 2 | 122 | 2.68e-01 |
| GO:BP | GO:0060419 | heart growth | 3 | 72 | 2.69e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 3 | 993 | 2.69e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 1 | 427 | 2.69e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 2 | 36 | 2.69e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 2.69e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 3 | 994 | 2.69e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 110 | 2.70e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 26 | 2.70e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 2 | 54 | 2.70e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 2 | 529 | 2.72e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 1 | 19 | 2.72e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 2.72e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 1 | 393 | 2.72e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 2.72e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 10 | 809 | 2.72e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 1 | 16 | 2.73e-01 |
| GO:BP | GO:0072359 | circulatory system development | 2 | 884 | 2.73e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 3 | 74 | 2.73e-01 |
| GO:BP | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration | 1 | 6 | 2.74e-01 |
| GO:BP | GO:0007420 | brain development | 3 | 591 | 2.74e-01 |
| GO:BP | GO:0007610 | behavior | 2 | 436 | 2.74e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 5 | 303 | 2.74e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 1 | 14 | 2.75e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 2 | 512 | 2.75e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 1 | 19 | 2.75e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 10 | 2.75e-01 |
| GO:BP | GO:0015888 | thiamine transport | 1 | 3 | 2.75e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 2 | 742 | 2.75e-01 |
| GO:BP | GO:0008585 | female gonad development | 3 | 77 | 2.75e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 1 | 14 | 2.75e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 1 | 183 | 2.75e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 1 | 399 | 2.75e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 3 | 64 | 2.75e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 1 | 28 | 2.76e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 2.76e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 1 | 15 | 2.76e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 12 | 963 | 2.76e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 2 | 541 | 2.76e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 2.76e-01 |
| GO:BP | GO:0007417 | central nervous system development | 7 | 792 | 2.76e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 2.76e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 2 | 42 | 2.76e-01 |
| GO:BP | GO:1900119 | positive regulation of execution phase of apoptosis | 1 | 7 | 2.77e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 3 | 55 | 2.78e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 2 | 516 | 2.78e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 1 | 7 | 2.79e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 1 | 16 | 2.80e-01 |
| GO:BP | GO:0030900 | forebrain development | 1 | 281 | 2.80e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 1 | 85 | 2.80e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 2 | 527 | 2.80e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 2.80e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 2 | 33 | 2.80e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 2.80e-01 |
| GO:BP | GO:0006353 | DNA-templated transcription termination | 1 | 22 | 2.80e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 2.80e-01 |
| GO:BP | GO:0008406 | gonad development | 5 | 158 | 2.80e-01 |
| GO:BP | GO:0009886 | post-embryonic animal morphogenesis | 1 | 10 | 2.80e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 1 | 179 | 2.80e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 1 | 459 | 2.80e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 1 | 8 | 2.80e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 2 | 531 | 2.81e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 2.81e-01 |
| GO:BP | GO:0090102 | cochlea development | 2 | 35 | 2.81e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 1 | 26 | 2.82e-01 |
| GO:BP | GO:0061061 | muscle structure development | 13 | 546 | 2.82e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 1 | 14 | 2.82e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 3 | 67 | 2.83e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 8 | 551 | 2.83e-01 |
| GO:BP | GO:0071637 | regulation of monocyte chemotactic protein-1 production | 1 | 7 | 2.83e-01 |
| GO:BP | GO:0071605 | monocyte chemotactic protein-1 production | 1 | 7 | 2.83e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 1 | 15 | 2.83e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 7 | 236 | 2.83e-01 |
| GO:BP | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 1 | 17 | 2.83e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 2.84e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 1 | 120 | 2.84e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 2.84e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 2 | 763 | 2.85e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 3 | 79 | 2.86e-01 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 1 | 21 | 2.86e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 5 | 357 | 2.86e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 2 | 27 | 2.87e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 1 | 30 | 2.87e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 1 | 11 | 2.87e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 1 | 15 | 2.87e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 1 | 15 | 2.87e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 1 | 15 | 2.87e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 1 | 11 | 2.87e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 4 | 127 | 2.87e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 1 | 30 | 2.87e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 1 | 296 | 2.87e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 2 | 551 | 2.88e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 1 | 279 | 2.88e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 1 | 5 | 2.88e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 1 | 461 | 2.88e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 5 | 161 | 2.88e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 5 | 1835 | 2.89e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 2.89e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 2 | 58 | 2.89e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 2.89e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 2.89e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 2.89e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 1 | 25 | 2.89e-01 |
| GO:BP | GO:0015884 | folic acid transport | 1 | 7 | 2.89e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 2.89e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 2 | 27 | 2.89e-01 |
| GO:BP | GO:0046794 | transport of virus | 1 | 19 | 2.89e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 1 | 44 | 2.90e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 1 | 15 | 2.90e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 1 | 441 | 2.90e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 1 | 15 | 2.90e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 1 | 469 | 2.90e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 1 | 22 | 2.90e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 6 | 212 | 2.90e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 5 | 1974 | 2.90e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 2 | 2285 | 2.91e-01 |
| GO:BP | GO:0048732 | gland development | 1 | 318 | 2.91e-01 |
| GO:BP | GO:0060322 | head development | 3 | 633 | 2.91e-01 |
| GO:BP | GO:0008213 | protein alkylation | 5 | 160 | 2.92e-01 |
| GO:BP | GO:0006479 | protein methylation | 5 | 160 | 2.92e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 1 | 494 | 2.93e-01 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 2 | 36 | 2.93e-01 |
| GO:BP | GO:0060429 | epithelium development | 2 | 850 | 2.93e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 13 | 504 | 2.93e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 2 | 38 | 2.94e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 1 | 197 | 2.94e-01 |
| GO:BP | GO:0061744 | motor behavior | 1 | 17 | 2.94e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 1 | 9 | 2.94e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 1 | 8 | 2.94e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 1 | 457 | 2.94e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 5 | 159 | 2.97e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 1 | 19 | 2.97e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 1 | 8 | 2.97e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 1 | 15 | 2.97e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 1 | 14 | 2.97e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 1 | 22 | 2.97e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 1 | 14 | 2.97e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 3 | 70 | 2.97e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 2 | 29 | 2.97e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 2.97e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 1 | 190 | 2.97e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 1 | 214 | 2.97e-01 |
| GO:BP | GO:0032736 | positive regulation of interleukin-13 production | 1 | 7 | 2.97e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 2 | 560 | 2.97e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 1 | 29 | 2.97e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 2.97e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 2.97e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 1 | 19 | 2.97e-01 |
| GO:BP | GO:0033962 | P-body assembly | 1 | 20 | 2.97e-01 |
| GO:BP | GO:0061458 | reproductive system development | 6 | 215 | 2.97e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 1 | 9 | 2.97e-01 |
| GO:BP | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine | 1 | 7 | 2.97e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 2 | 26 | 2.97e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 3 | 77 | 2.97e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 2.98e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 1 | 11 | 2.98e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 11 | 513 | 2.98e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 1 | 6 | 2.99e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1361 | 2.99e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 3 | 68 | 2.99e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 2.99e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 135 | 3.00e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 3.00e-01 |
| GO:BP | GO:0039530 | MDA-5 signaling pathway | 1 | 8 | 3.01e-01 |
| GO:BP | GO:0032682 | negative regulation of chemokine production | 1 | 12 | 3.01e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 5 | 271 | 3.01e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 4 | 146 | 3.01e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 1 | 15 | 3.01e-01 |
| GO:BP | GO:0007049 | cell cycle | 1 | 1529 | 3.01e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 1 | 17 | 3.01e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1453 | 3.01e-01 |
| GO:BP | GO:0021548 | pons development | 1 | 7 | 3.01e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 6 | 207 | 3.01e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 2 | 39 | 3.01e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 1 | 196 | 3.02e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 6 | 197 | 3.03e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 2 | 42 | 3.04e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 1 | 12 | 3.04e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 3.04e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 1 | 22 | 3.05e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 1 | 18 | 3.05e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 2 | 40 | 3.05e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 3 | 81 | 3.06e-01 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 1 | 15 | 3.06e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 3 | 189 | 3.06e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 1 | 14 | 3.06e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 14 | 482 | 3.06e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 4 | 147 | 3.06e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 5 | 144 | 3.06e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 1 | 14 | 3.06e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 1 | 485 | 3.06e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 3.06e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 1 | 348 | 3.06e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 1 | 9 | 3.06e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 1 | 15 | 3.06e-01 |
| GO:BP | GO:0021603 | cranial nerve formation | 1 | 4 | 3.06e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 1 | 50 | 3.06e-01 |
| GO:BP | GO:0070848 | response to growth factor | 8 | 572 | 3.06e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 1 | 15 | 3.06e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 3.06e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 1 | 18 | 3.06e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 3.06e-01 |
| GO:BP | GO:0006096 | glycolytic process | 3 | 70 | 3.06e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 3.06e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 1 | 21 | 3.06e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1462 | 3.08e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 3 | 86 | 3.08e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 3.09e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 6 | 222 | 3.09e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 1 | 36 | 3.10e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 7 | 251 | 3.10e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 1 | 106 | 3.11e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 1 | 22 | 3.11e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 1 | 16 | 3.11e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 3.11e-01 |
| GO:BP | GO:0035176 | social behavior | 1 | 34 | 3.11e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 3 | 71 | 3.12e-01 |
| GO:BP | GO:0021675 | nerve development | 3 | 57 | 3.12e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 2 | 74 | 3.13e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 3.13e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 1 | 18 | 3.13e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 1 | 24 | 3.14e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 3 | 86 | 3.15e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 3.15e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 1 | 9 | 3.15e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 1 | 14 | 3.15e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 1 | 9 | 3.15e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 2 | 42 | 3.16e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 1 | 517 | 3.16e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 1 | 11 | 3.16e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 2 | 32 | 3.16e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 1 | 8 | 3.16e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 1 | 16 | 3.17e-01 |
| GO:BP | GO:0040007 | growth | 18 | 742 | 3.17e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 1 | 34 | 3.17e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 2 | 54 | 3.17e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 3.17e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 1 | 27 | 3.17e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 1 | 52 | 3.17e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 1 | 234 | 3.18e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 5 | 155 | 3.18e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 1 | 384 | 3.18e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 336 | 3.20e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 1 | 19 | 3.20e-01 |
| GO:BP | GO:0035148 | tube formation | 4 | 138 | 3.20e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 3 | 68 | 3.20e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 4 | 111 | 3.20e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 2 | 42 | 3.21e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 1 | 604 | 3.21e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 1 | 26 | 3.22e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 1 | 24 | 3.22e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 1 | 612 | 3.22e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 4 | 132 | 3.22e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 1 | 24 | 3.22e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 1 | 21 | 3.22e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 1 | 31 | 3.22e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 26 | 1199 | 3.22e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 1 | 23 | 3.22e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 6 | 227 | 3.22e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 2 | 639 | 3.22e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 2 | 36 | 3.22e-01 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 1 | 10 | 3.23e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 2 | 74 | 3.23e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 3 | 80 | 3.23e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 1 | 10 | 3.23e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 12 | 3.23e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 1 | 21 | 3.23e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 1 | 14 | 3.24e-01 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 1 | 7 | 3.24e-01 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 1 | 10 | 3.24e-01 |
| GO:BP | GO:0006412 | translation | 1 | 646 | 3.24e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 1 | 16 | 3.24e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 1 | 382 | 3.24e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 3 | 78 | 3.24e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 2 | 46 | 3.24e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 2 | 40 | 3.24e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 1 | 37 | 3.26e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 3 | 91 | 3.26e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 3.26e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 3.26e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 1 | 13 | 3.26e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 1 | 35 | 3.26e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 8 | 3.26e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 1 | 68 | 3.26e-01 |
| GO:BP | GO:0070170 | regulation of tooth mineralization | 1 | 4 | 3.27e-01 |
| GO:BP | GO:0070266 | necroptotic process | 1 | 32 | 3.27e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 3.27e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 1 | 7 | 3.28e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 3.28e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 1 | 30 | 3.28e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 4 | 132 | 3.30e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 1 | 22 | 3.30e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 1 | 16 | 3.30e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 2 | 156 | 3.30e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 1 | 666 | 3.31e-01 |
| GO:BP | GO:0007399 | nervous system development | 2 | 1904 | 3.31e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 1 | 55 | 3.31e-01 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 1 | 5 | 3.31e-01 |
| GO:BP | GO:0034644 | cellular response to UV | 3 | 86 | 3.31e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 1 | 539 | 3.31e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 1 | 22 | 3.31e-01 |
| GO:BP | GO:0032528 | microvillus organization | 1 | 19 | 3.31e-01 |
| GO:BP | GO:0048806 | genitalia development | 1 | 28 | 3.31e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 2 | 40 | 3.32e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 1 | 12 | 3.32e-01 |
| GO:BP | GO:0060438 | trachea development | 1 | 13 | 3.32e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 1 | 37 | 3.32e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 1 | 15 | 3.32e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 1 | 15 | 3.33e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 4 | 149 | 3.33e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 152 | 3.33e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 3.33e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 3.33e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 3.33e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 1 | 10 | 3.35e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 1 | 29 | 3.35e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 3 | 155 | 3.35e-01 |
| GO:BP | GO:0048538 | thymus development | 2 | 35 | 3.35e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 2 | 45 | 3.35e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 7 | 185 | 3.35e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 1 | 11 | 3.35e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 1 | 496 | 3.35e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 3.35e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 3.35e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 1 | 14 | 3.35e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 3.35e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 4 | 272 | 3.35e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 3.35e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 3.35e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1684 | 3.37e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 1 | 14 | 3.37e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 1 | 31 | 3.37e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 2 | 44 | 3.37e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 2 | 57 | 3.39e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 1 | 687 | 3.39e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 3.39e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 1 | 665 | 3.39e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 2 | 34 | 3.40e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 418 | 3.40e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 2 | 54 | 3.40e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3 | 161 | 3.40e-01 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 1 | 18 | 3.40e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 1 | 16 | 3.40e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 1 | 29 | 3.40e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 3.40e-01 |
| GO:BP | GO:0048565 | digestive tract development | 2 | 92 | 3.40e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 1 | 22 | 3.40e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 15 | 3.40e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 5 | 216 | 3.40e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 1 | 96 | 3.40e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 3.41e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 1 | 13 | 3.41e-01 |
| GO:BP | GO:0000725 | recombinational repair | 4 | 153 | 3.42e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 1 | 16 | 3.42e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 1 | 168 | 3.42e-01 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 1 | 12 | 3.42e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 1 | 16 | 3.42e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 1 | 167 | 3.45e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 3.45e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 3.45e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 3.45e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 1 | 28 | 3.45e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 3.45e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 1 | 22 | 3.45e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 3.45e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 1 | 14 | 3.45e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 2 | 400 | 3.45e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 1 | 23 | 3.46e-01 |
| GO:BP | GO:0043038 | amino acid activation | 1 | 44 | 3.46e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 4 | 161 | 3.46e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 15 | 3.47e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 1 | 8 | 3.47e-01 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 1 | 13 | 3.47e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 2 | 631 | 3.47e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 1 | 39 | 3.48e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 1 | 46 | 3.48e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 7 | 265 | 3.48e-01 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 1 | 11 | 3.48e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 2 | 35 | 3.48e-01 |
| GO:BP | GO:0070914 | UV-damage excision repair | 1 | 13 | 3.48e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 1 | 15 | 3.48e-01 |
| GO:BP | GO:0002376 | immune system process | 2 | 1447 | 3.48e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 3 | 112 | 3.48e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 3.48e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 1 | 45 | 3.49e-01 |
| GO:BP | GO:0045190 | isotype switching | 1 | 39 | 3.49e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 1 | 48 | 3.49e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 1 | 39 | 3.49e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 1 | 39 | 3.49e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 2 | 1869 | 3.50e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 15 | 7552 | 3.51e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 3 | 113 | 3.51e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 3.51e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 3.51e-01 |
| GO:BP | GO:0035265 | organ growth | 4 | 131 | 3.51e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 26 | 7806 | 3.52e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 2 | 50 | 3.53e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 2 | 48 | 3.53e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 2 | 93 | 3.53e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 1 | 25 | 3.53e-01 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 1 | 15 | 3.53e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 1 | 771 | 3.53e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 1 | 39 | 3.53e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 1 | 41 | 3.53e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 1 | 10 | 3.53e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 2 | 75 | 3.53e-01 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 1 | 15 | 3.53e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 3.53e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 1 | 25 | 3.53e-01 |
| GO:BP | GO:0006284 | base-excision repair | 1 | 44 | 3.53e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 1 | 96 | 3.53e-01 |
| GO:BP | GO:0031099 | regeneration | 3 | 142 | 3.53e-01 |
| GO:BP | GO:0019985 | translesion synthesis | 1 | 24 | 3.53e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 2 | 63 | 3.53e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 2 | 2139 | 3.53e-01 |
| GO:BP | GO:0006950 | response to stress | 4 | 2780 | 3.53e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 1 | 37 | 3.53e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 3.53e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 3.53e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 1 | 19 | 3.53e-01 |
| GO:BP | GO:0040011 | locomotion | 2 | 954 | 3.56e-01 |
| GO:BP | GO:0016049 | cell growth | 5 | 418 | 3.56e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 1 | 775 | 3.56e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 2 | 2161 | 3.56e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 2 | 54 | 3.56e-01 |
| GO:BP | GO:0019080 | viral gene expression | 1 | 96 | 3.56e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 1 | 13 | 3.56e-01 |
| GO:BP | GO:0060384 | innervation | 1 | 22 | 3.56e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 1 | 161 | 3.56e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 1 | 14 | 3.56e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 3.56e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 1 | 12 | 3.56e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 1 | 15 | 3.57e-01 |
| GO:BP | GO:0010332 | response to gamma radiation | 1 | 50 | 3.57e-01 |
| GO:BP | GO:0010033 | response to organic substance | 2 | 1957 | 3.59e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 9 | 260 | 3.59e-01 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 1 | 17 | 3.59e-01 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 1 | 42 | 3.59e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 2 | 53 | 3.59e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 1 | 295 | 3.59e-01 |
| GO:BP | GO:0014029 | neural crest formation | 1 | 13 | 3.59e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 3.59e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 1 | 184 | 3.59e-01 |
| GO:BP | GO:0036211 | protein modification process | 5 | 2901 | 3.60e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 1 | 41 | 3.60e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 1 | 28 | 3.61e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 1 | 24 | 3.61e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 1 | 37 | 3.61e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 1 | 20 | 3.61e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 1 | 34 | 3.61e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 1 | 24 | 3.61e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 3.61e-01 |
| GO:BP | GO:0055123 | digestive system development | 2 | 100 | 3.61e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 2 | 52 | 3.61e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 2 | 55 | 3.61e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 609 | 3.62e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 1 | 282 | 3.62e-01 |
| GO:BP | GO:0110154 | RNA decapping | 1 | 17 | 3.63e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 1 | 187 | 3.64e-01 |
| GO:BP | GO:0045117 | azole transmembrane transport | 1 | 9 | 3.64e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 1 | 39 | 3.64e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 1 | 30 | 3.64e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 3.64e-01 |
| GO:BP | GO:0007498 | mesoderm development | 1 | 89 | 3.64e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 1 | 560 | 3.64e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 1 | 20 | 3.64e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 1 | 15 | 3.66e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 1 | 12 | 3.66e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 4 | 114 | 3.66e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 213 | 3.66e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 3.67e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 3 | 121 | 3.67e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2138 | 3.67e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 2 | 50 | 3.67e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 4 | 150 | 3.67e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 1 | 44 | 3.67e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 3.67e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 3.67e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 6 | 229 | 3.67e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 3 | 172 | 3.67e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 1 | 7 | 3.67e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2129 | 3.67e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 2 | 124 | 3.67e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 1 | 29 | 3.67e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 1 | 13 | 3.67e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 6 | 227 | 3.67e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 3.67e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 1 | 14 | 3.67e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 1 | 14 | 3.67e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 1 | 763 | 3.68e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 1 | 31 | 3.68e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 1 | 29 | 3.68e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 3.69e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 1 | 200 | 3.70e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 1 | 726 | 3.70e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 2 | 59 | 3.70e-01 |
| GO:BP | GO:0048873 | homeostasis of number of cells within a tissue | 1 | 24 | 3.70e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 1 | 26 | 3.70e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 1 | 47 | 3.71e-01 |
| GO:BP | GO:0090224 | regulation of spindle organization | 1 | 45 | 3.72e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 1 | 13 | 3.72e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 1 | 11 | 3.72e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 1 | 26 | 3.72e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 3 | 221 | 3.72e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 4 | 90 | 3.72e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 1 | 30 | 3.72e-01 |
| GO:BP | GO:0008584 | male gonad development | 3 | 99 | 3.72e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 3.72e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 3.72e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 1 | 26 | 3.72e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 1 | 22 | 3.72e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 3.73e-01 |
| GO:BP | GO:0030728 | ovulation | 1 | 14 | 3.73e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 3.73e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 17 | 3.73e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 2 | 2102 | 3.74e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2 | 2890 | 3.75e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 3 | 100 | 3.75e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 3.77e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 1 | 17 | 3.78e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 2 | 761 | 3.78e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 1 | 28 | 3.78e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 1 | 30 | 3.78e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 1 | 28 | 3.79e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 1 | 17 | 3.79e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 2 | 107 | 3.79e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 3.79e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 1 | 31 | 3.79e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 1 | 12 | 3.81e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 2 | 100 | 3.81e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 3 | 100 | 3.81e-01 |
| GO:BP | GO:0051168 | nuclear export | 3 | 150 | 3.81e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 1 | 127 | 3.81e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 3 | 121 | 3.81e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 3 | 1380 | 3.81e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 1 | 23 | 3.81e-01 |
| GO:BP | GO:0055070 | copper ion homeostasis | 1 | 18 | 3.81e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 4 | 120 | 3.81e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 2 | 135 | 3.81e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 1 | 25 | 3.82e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 3 | 119 | 3.82e-01 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 1 | 8 | 3.82e-01 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 1 | 9 | 3.83e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 3 | 80 | 3.84e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 1 | 78 | 3.84e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 468 | 3.84e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 1 | 31 | 3.84e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 2 | 57 | 3.84e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 1 | 52 | 3.84e-01 |
| GO:BP | GO:0016477 | cell migration | 2 | 1081 | 3.85e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 2 | 755 | 3.85e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 1 | 28 | 3.85e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 1 | 86 | 3.85e-01 |
| GO:BP | GO:0032940 | secretion by cell | 1 | 569 | 3.85e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 1 | 23 | 3.85e-01 |
| GO:BP | GO:0001709 | cell fate determination | 1 | 25 | 3.85e-01 |
| GO:BP | GO:0030902 | hindbrain development | 4 | 114 | 3.85e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 3 | 101 | 3.85e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 1 | 20 | 3.86e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2116 | 3.87e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 1 | 540 | 3.87e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 3 | 101 | 3.87e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 2 | 127 | 3.87e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 5 | 127 | 3.87e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 1 | 24 | 3.87e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 1 | 17 | 3.87e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 1 | 53 | 3.87e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 1 | 27 | 3.88e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 3 | 4045 | 3.88e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 1 | 179 | 3.89e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 2 | 67 | 3.89e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 1 | 41 | 3.90e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 1 | 37 | 3.90e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 2 | 31 | 3.91e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 1 | 31 | 3.91e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 2 | 88 | 3.91e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 3 | 121 | 3.92e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 1 | 22 | 3.92e-01 |
| GO:BP | GO:0030154 | cell differentiation | 4 | 2958 | 3.92e-01 |
| GO:BP | GO:0044409 | entry into host | 2 | 113 | 3.92e-01 |
| GO:BP | GO:0001503 | ossification | 3 | 313 | 3.92e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 1 | 17 | 3.93e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 1 | 15 | 3.93e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 1 | 35 | 3.94e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 1 | 21 | 3.94e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 1 | 15 | 3.94e-01 |
| GO:BP | GO:0031529 | ruffle organization | 1 | 45 | 3.95e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 2 | 107 | 3.95e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 4 | 2984 | 3.97e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 1 | 28 | 3.97e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 4 | 3090 | 3.97e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 3.97e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 3.97e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 3.98e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 1 | 30 | 3.98e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 3.98e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 1 | 28 | 3.99e-01 |
| GO:BP | GO:0007165 | signal transduction | 3 | 3818 | 3.99e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 1 | 41 | 3.99e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 5 | 308 | 3.99e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 3.99e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 1 | 55 | 4.00e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 1 | 55 | 4.00e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 1 | 26 | 4.00e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2450 | 4.00e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 4.01e-01 |
| GO:BP | GO:0140352 | export from cell | 1 | 611 | 4.01e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 3 | 156 | 4.01e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 4.01e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 1 | 12 | 4.01e-01 |
| GO:BP | GO:0051707 | response to other organism | 2 | 820 | 4.01e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 1 | 53 | 4.01e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 2 | 89 | 4.01e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 2 | 91 | 4.01e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 2 | 820 | 4.01e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 1 | 31 | 4.02e-01 |
| GO:BP | GO:0070509 | calcium ion import | 1 | 37 | 4.02e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 4.03e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 4.03e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 1 | 11 | 4.03e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 1 | 11 | 4.03e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 1 | 427 | 4.03e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 4.03e-01 |
| GO:BP | GO:1990840 | response to lectin | 1 | 11 | 4.03e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 1 | 22 | 4.03e-01 |
| GO:BP | GO:0048645 | animal organ formation | 2 | 51 | 4.03e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 1 | 993 | 4.06e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 1 | 22 | 4.06e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 1 | 22 | 4.07e-01 |
| GO:BP | GO:0032259 | methylation | 4 | 313 | 4.07e-01 |
| GO:BP | GO:0001525 | angiogenesis | 1 | 390 | 4.07e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 1 | 15 | 4.08e-01 |
| GO:BP | GO:0032570 | response to progesterone | 1 | 27 | 4.08e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 2 | 61 | 4.08e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 1 | 23 | 4.08e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 4.08e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 4.08e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 1 | 667 | 4.09e-01 |
| GO:BP | GO:0016233 | telomere capping | 1 | 36 | 4.09e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 4.10e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 2 | 850 | 4.10e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 1 | 61 | 4.11e-01 |
| GO:BP | GO:0001889 | liver development | 3 | 110 | 4.11e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 3 | 93 | 4.11e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 4.12e-01 |
| GO:BP | GO:0006301 | postreplication repair | 1 | 34 | 4.12e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 4.12e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 2 | 92 | 4.12e-01 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 1 | 41 | 4.13e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 4.13e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 1 | 16 | 4.13e-01 |
| GO:BP | GO:0030220 | platelet formation | 1 | 17 | 4.13e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 1 | 35 | 4.14e-01 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 1 | 37 | 4.14e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 4.15e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 1 | 16 | 4.15e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 1 | 15 | 4.15e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 2 | 151 | 4.15e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 1 | 246 | 4.15e-01 |
| GO:BP | GO:0021591 | ventricular system development | 1 | 29 | 4.16e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 4.17e-01 |
| GO:BP | GO:0046903 | secretion | 1 | 645 | 4.17e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 273 | 4.17e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 2 | 137 | 4.18e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 1 | 30 | 4.19e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 1 | 62 | 4.19e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 3 | 89 | 4.19e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 1 | 22 | 4.19e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 1 | 270 | 4.20e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 16 | 8061 | 4.21e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 1 | 48 | 4.22e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 27 | 4.22e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 1 | 27 | 4.22e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 2 | 70 | 4.22e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 2 | 70 | 4.22e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 2 | 834 | 4.23e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 1 | 29 | 4.23e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 4.23e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 1 | 19 | 4.23e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 4.23e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 1 | 34 | 4.23e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 1 | 26 | 4.23e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 4.23e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 1 | 48 | 4.25e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 1 | 70 | 4.25e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 3 | 101 | 4.25e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 2 | 50 | 4.25e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 1 | 19 | 4.25e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 1 | 19 | 4.25e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 3 | 186 | 4.25e-01 |
| GO:BP | GO:0007276 | gamete generation | 12 | 461 | 4.26e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 1 | 8 | 4.26e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 3 | 132 | 4.26e-01 |
| GO:BP | GO:0009790 | embryo development | 20 | 875 | 4.26e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 1 | 21 | 4.26e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 4.27e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 3 | 166 | 4.28e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 1 | 23 | 4.28e-01 |
| GO:BP | GO:0048870 | cell motility | 2 | 1182 | 4.29e-01 |
| GO:BP | GO:0006457 | protein folding | 2 | 192 | 4.29e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 4.29e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 4.29e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 1 | 42 | 4.29e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 1 | 14 | 4.30e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 4.30e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 1 | 14 | 4.30e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 1 | 28 | 4.30e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2810 | 4.30e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 1 | 19 | 4.31e-01 |
| GO:BP | GO:0001568 | blood vessel development | 13 | 534 | 4.32e-01 |
| GO:BP | GO:0007369 | gastrulation | 1 | 146 | 4.32e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 1 | 19 | 4.32e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 1 | 39 | 4.32e-01 |
| GO:BP | GO:0043583 | ear development | 5 | 153 | 4.32e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 2 | 146 | 4.33e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 1 | 51 | 4.33e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 3 | 194 | 4.33e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 4.33e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 1 | 42 | 4.33e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 4 | 785 | 4.34e-01 |
| GO:BP | GO:0007379 | segment specification | 1 | 14 | 4.34e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 1 | 486 | 4.34e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 1 | 30 | 4.34e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 1 | 31 | 4.34e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 4 | 198 | 4.35e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 1 | 16 | 4.36e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 3 | 67 | 4.38e-01 |
| GO:BP | GO:0035461 | vitamin transmembrane transport | 1 | 14 | 4.38e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 6 | 249 | 4.38e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 1 | 39 | 4.38e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 1 | 17 | 4.38e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 4 | 122 | 4.38e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 1 | 39 | 4.38e-01 |
| GO:BP | GO:0006304 | DNA modification | 1 | 89 | 4.38e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 2 | 963 | 4.38e-01 |
| GO:BP | GO:0009888 | tissue development | 2 | 1425 | 4.40e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 1 | 103 | 4.40e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 4.40e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 1 | 42 | 4.40e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 4.40e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1 | 26 | 4.40e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 1 | 462 | 4.40e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 1 | 260 | 4.40e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 1 | 28 | 4.40e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 1 | 51 | 4.40e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 2 | 99 | 4.40e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 4.40e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 1 | 28 | 4.40e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1 | 18 | 4.40e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 4.40e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 4 | 3386 | 4.40e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 1 | 60 | 4.40e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 4.40e-01 |
| GO:BP | GO:0023052 | signaling | 3 | 4141 | 4.40e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 4.40e-01 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 317 | 4.42e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 1 | 76 | 4.42e-01 |
| GO:BP | GO:0070997 | neuron death | 1 | 286 | 4.42e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 1 | 57 | 4.43e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 4.44e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 1 | 47 | 4.44e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 1 | 34 | 4.44e-01 |
| GO:BP | GO:0044000 | movement in host | 2 | 138 | 4.45e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 1 | 18 | 4.45e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 4.45e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 1 | 77 | 4.45e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 1 | 18 | 4.46e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 1 | 19 | 4.46e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 1 | 33 | 4.46e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 2 | 97 | 4.48e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 1 | 32 | 4.48e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 2 | 79 | 4.48e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 4.48e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 4.48e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 1 | 18 | 4.49e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 4.50e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 4.50e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 4.51e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 9 | 294 | 4.51e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 2 | 561 | 4.51e-01 |
| GO:BP | GO:0007154 | cell communication | 3 | 4226 | 4.52e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 1 | 110 | 4.53e-01 |
| GO:BP | GO:0007389 | pattern specification process | 9 | 306 | 4.53e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 2 | 114 | 4.53e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 2 | 114 | 4.53e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 15 | 4.54e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 1 | 28 | 4.54e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 1 | 28 | 4.54e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 1 | 16 | 4.54e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 1 | 43 | 4.55e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 4.57e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 1 | 43 | 4.57e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 1 | 35 | 4.57e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 2 | 67 | 4.57e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 2 | 117 | 4.58e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 1 | 107 | 4.58e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1 | 1226 | 4.59e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 1 | 277 | 4.60e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 3 | 99 | 4.61e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 1 | 66 | 4.62e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 1012 | 4.62e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 4.64e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 2 | 84 | 4.64e-01 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 1 | 23 | 4.64e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 17 | 598 | 4.64e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 3 | 73 | 4.64e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 1 | 26 | 4.66e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 1 | 50 | 4.66e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 4.66e-01 |
| GO:BP | GO:0002520 | immune system development | 2 | 144 | 4.66e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 1 | 50 | 4.66e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 2 | 110 | 4.66e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 1 | 58 | 4.66e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 4.67e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 1 | 45 | 4.67e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 1 | 41 | 4.67e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 1 | 45 | 4.67e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 1 | 24 | 4.67e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 4 | 105 | 4.67e-01 |
| GO:BP | GO:0031016 | pancreas development | 2 | 53 | 4.69e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 4.70e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 3 | 129 | 4.70e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 2 | 71 | 4.71e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 1 | 23 | 4.71e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 2 | 70 | 4.72e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 4.72e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 2 | 53 | 4.72e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 1 | 20 | 4.72e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 2 | 66 | 4.74e-01 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 1 | 21 | 4.74e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 1 | 38 | 4.74e-01 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 1 | 21 | 4.74e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 2 | 50 | 4.74e-01 |
| GO:BP | GO:0032835 | glomerulus development | 1 | 54 | 4.74e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 1 | 114 | 4.75e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 1 | 19 | 4.75e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 1 | 23 | 4.75e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 1 | 26 | 4.75e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 1 | 85 | 4.76e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 1 | 46 | 4.76e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 4.76e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 4.76e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 4.77e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 1 | 49 | 4.78e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 3 | 74 | 4.78e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 1 | 86 | 4.78e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 1 | 32 | 4.80e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 4.80e-01 |
| GO:BP | GO:0007492 | endoderm development | 2 | 65 | 4.80e-01 |
| GO:BP | GO:0001944 | vasculature development | 13 | 559 | 4.81e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 2 | 78 | 4.81e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 1 | 23 | 4.81e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 1 | 23 | 4.81e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 1 | 478 | 4.81e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 1 | 50 | 4.81e-01 |
| GO:BP | GO:0035188 | hatching | 1 | 23 | 4.81e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 2 | 160 | 4.82e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 4.82e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 1 | 39 | 4.82e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 1 | 39 | 4.82e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 1 | 86 | 4.83e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 1 | 28 | 4.83e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 13 | 538 | 4.83e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 2 | 100 | 4.83e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 1 | 28 | 4.84e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 1 | 317 | 4.86e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 1 | 70 | 4.86e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 2 | 160 | 4.86e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 344 | 4.88e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 1 | 92 | 4.89e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 1 | 56 | 4.89e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 1 | 20 | 4.89e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 1 | 20 | 4.89e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 2 | 80 | 4.90e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 3 | 98 | 4.90e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 1 | 30 | 4.90e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 1 | 19 | 4.90e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 2 | 146 | 4.90e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 1 | 45 | 4.90e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 1 | 38 | 4.90e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 1 | 16 | 4.90e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 1 | 375 | 4.90e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 2 | 154 | 4.90e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 4 | 344 | 4.90e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 1 | 376 | 4.92e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 1 | 37 | 4.92e-01 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 1 | 27 | 4.92e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 2 | 154 | 4.92e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 2 | 59 | 4.93e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 4.93e-01 |
| GO:BP | GO:0048731 | system development | 2 | 2901 | 4.94e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 4.94e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 1 | 26 | 4.95e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 1 | 24 | 4.95e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 4.96e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 2 | 157 | 4.96e-01 |
| GO:BP | GO:0006310 | DNA recombination | 4 | 257 | 4.96e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 4.97e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 2 | 71 | 4.97e-01 |
| GO:BP | GO:0048864 | stem cell development | 2 | 76 | 4.97e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 1 | 36 | 4.97e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 4.97e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 1 | 47 | 4.97e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 1 | 30 | 4.98e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 1 | 26 | 4.98e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 1 | 30 | 4.98e-01 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 1 | 50 | 4.99e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 1 | 615 | 5.00e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 5.02e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 1 | 1484 | 5.03e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 2 | 83 | 5.04e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 5.05e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 1 | 625 | 5.05e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 5.05e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 1 | 535 | 5.06e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 4 | 142 | 5.06e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 3 | 206 | 5.08e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 5.08e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 4 | 204 | 5.08e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 5.08e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 1 | 57 | 5.09e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 2 | 117 | 5.09e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 1 | 637 | 5.10e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 1 | 17 | 5.10e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 5.10e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 1 | 64 | 5.10e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 1 | 27 | 5.10e-01 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 1 | 42 | 5.10e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 2 | 79 | 5.10e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 1 | 25 | 5.10e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 5.10e-01 |
| GO:BP | GO:0042221 | response to chemical | 2 | 2559 | 5.11e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 5.13e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 994 | 5.13e-01 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 1 | 29 | 5.13e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 5.14e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 1 | 59 | 5.15e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 5.15e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 5.15e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 1 | 97 | 5.15e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 1 | 55 | 5.15e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 5.15e-01 |
| GO:BP | GO:0042594 | response to starvation | 3 | 178 | 5.15e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 5.15e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 5.15e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 18 | 742 | 5.15e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 2 | 55 | 5.15e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 6 | 278 | 5.15e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 2 | 556 | 5.15e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 1 | 35 | 5.16e-01 |
| GO:BP | GO:0007612 | learning | 1 | 115 | 5.16e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 1 | 24 | 5.16e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 1 | 22 | 5.17e-01 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 1 | 18 | 5.17e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 1 | 29 | 5.17e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 1 | 16 | 5.17e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 5.17e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 2 | 152 | 5.17e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 14 | 663 | 5.17e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 2 | 88 | 5.17e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 1 | 36 | 5.17e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 1 | 51 | 5.18e-01 |
| GO:BP | GO:0003407 | neural retina development | 1 | 48 | 5.18e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 1 | 61 | 5.18e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 1 | 70 | 5.18e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 5.18e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 5.18e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 5.18e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 1 | 53 | 5.18e-01 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 1 | 44 | 5.18e-01 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 1 | 44 | 5.18e-01 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 1 | 44 | 5.18e-01 |
| GO:BP | GO:0048839 | inner ear development | 4 | 132 | 5.19e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 3 | 206 | 5.20e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 5.20e-01 |
| GO:BP | GO:0030850 | prostate gland development | 1 | 35 | 5.20e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 2 | 268 | 5.20e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 2 | 268 | 5.20e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 5.20e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 1 | 62 | 5.20e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 5.21e-01 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 1 | 28 | 5.22e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 5.22e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 1 | 40 | 5.24e-01 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 1 | 556 | 5.25e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 5.25e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 5.25e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 1 | 69 | 5.25e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 3 | 129 | 5.25e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 5.25e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 5.25e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 3 | 155 | 5.25e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 1 | 140 | 5.27e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 5.27e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 5.29e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 1 | 31 | 5.29e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 1 | 34 | 5.29e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 4 | 124 | 5.29e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 5.29e-01 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 5.29e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 1 | 28 | 5.29e-01 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 1 | 46 | 5.29e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 90 | 5.29e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 1 | 69 | 5.29e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 1 | 26 | 5.29e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 5.29e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 1 | 22 | 5.30e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 1 | 38 | 5.30e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 2 | 251 | 5.30e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 1 | 78 | 5.31e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 1 | 100 | 5.31e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 1 | 20 | 5.31e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 1 | 104 | 5.32e-01 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 1 | 30 | 5.35e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 1 | 706 | 5.35e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 8 | 316 | 5.35e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 5.37e-01 |
| GO:BP | GO:0032743 | positive regulation of interleukin-2 production | 1 | 20 | 5.37e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 1 | 29 | 5.37e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 5.37e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 1 | 29 | 5.37e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2 | 114 | 5.37e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 1 | 40 | 5.37e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 2 | 204 | 5.37e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 1 | 51 | 5.39e-01 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 1 | 47 | 5.39e-01 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 1 | 47 | 5.39e-01 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 1 | 47 | 5.39e-01 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 1 | 47 | 5.39e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 2 | 166 | 5.39e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 3 | 1149 | 5.39e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 1 | 65 | 5.40e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 1 | 20 | 5.40e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 1 | 71 | 5.41e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 5.41e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 1 | 58 | 5.41e-01 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 1 | 30 | 5.42e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 5.42e-01 |
| GO:BP | GO:0032355 | response to estradiol | 2 | 83 | 5.42e-01 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 1 | 17 | 5.43e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 2 | 62 | 5.43e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 3 | 4953 | 5.43e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 5.43e-01 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 2 | 104 | 5.44e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 1 | 78 | 5.44e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 3 | 217 | 5.46e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 5.46e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 4 | 1898 | 5.46e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 2 | 103 | 5.47e-01 |
| GO:BP | GO:0001764 | neuron migration | 2 | 133 | 5.47e-01 |
| GO:BP | GO:0043588 | skin development | 1 | 175 | 5.48e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 5.48e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 1 | 63 | 5.49e-01 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 1 | 48 | 5.49e-01 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 1 | 48 | 5.49e-01 |
| GO:BP | GO:0006298 | mismatch repair | 1 | 29 | 5.50e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 1 | 60 | 5.50e-01 |
| GO:BP | GO:0006260 | DNA replication | 3 | 255 | 5.51e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 1 | 448 | 5.53e-01 |
| GO:BP | GO:0009411 | response to UV | 3 | 141 | 5.53e-01 |
| GO:BP | GO:0098751 | bone cell development | 1 | 26 | 5.53e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 1 | 60 | 5.53e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 3 | 1939 | 5.53e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 3 | 112 | 5.53e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 134 | 5.55e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 1 | 46 | 5.55e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 1 | 43 | 5.56e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 1 | 31 | 5.56e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 2 | 142 | 5.57e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 6 | 555 | 5.58e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 5.58e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 1 | 31 | 5.58e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1138 | 5.59e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 1 | 54 | 5.59e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 39 | 5.59e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 5.59e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 1 | 62 | 5.59e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 1 | 52 | 5.60e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 1 | 54 | 5.64e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 1 | 64 | 5.64e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 1 | 49 | 5.64e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 1 | 123 | 5.64e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 2 | 67 | 5.64e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 1 | 68 | 5.64e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 5.64e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 3 | 139 | 5.65e-01 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 1 | 52 | 5.65e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 2 | 90 | 5.65e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 1 | 71 | 5.65e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 5.65e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 1 | 61 | 5.65e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 4 | 717 | 5.65e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 2 | 1974 | 5.65e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 17 | 631 | 5.65e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 1 | 41 | 5.66e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 2 | 116 | 5.66e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 1 | 73 | 5.66e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 1 | 64 | 5.66e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 5.66e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 5.66e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 1 | 76 | 5.67e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 5.67e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 1 | 40 | 5.68e-01 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 1 | 54 | 5.68e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 5.68e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 1 | 91 | 5.68e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 1 | 21 | 5.69e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 10 | 330 | 5.69e-01 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1267 | 5.69e-01 |
| GO:BP | GO:0061912 | selective autophagy | 1 | 90 | 5.69e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 5.70e-01 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 1 | 31 | 5.70e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 1 | 30 | 5.70e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 1 | 39 | 5.70e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 5.70e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 5 | 496 | 5.71e-01 |
| GO:BP | GO:0065007 | biological regulation | 16 | 8320 | 5.72e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 2 | 108 | 5.72e-01 |
| GO:BP | GO:0016032 | viral process | 3 | 344 | 5.72e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 5.73e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 68 | 5.73e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 3 | 151 | 5.74e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 1 | 118 | 5.74e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 1 | 65 | 5.75e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 5.76e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 2 | 105 | 5.76e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 1 | 66 | 5.76e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 1 | 95 | 5.76e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 5.76e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 1 | 32 | 5.76e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 5.76e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 1 | 31 | 5.76e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 3 | 115 | 5.76e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 1 | 55 | 5.76e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 1 | 37 | 5.78e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 1 | 55 | 5.78e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 2 | 155 | 5.78e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 1 | 72 | 5.80e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 5.81e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 1 | 70 | 5.82e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 1 | 57 | 5.83e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 1 | 34 | 5.83e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 3 | 153 | 5.83e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 1 | 64 | 5.83e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 2 | 103 | 5.83e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 5.85e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 5.85e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 5.85e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 2 | 95 | 5.87e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 1 | 123 | 5.88e-01 |
| GO:BP | GO:1901998 | toxin transport | 1 | 37 | 5.89e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 3 | 318 | 5.89e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 2 | 166 | 5.89e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 5.89e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 1 | 53 | 5.89e-01 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 1 | 58 | 5.93e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 1 | 105 | 5.93e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 2 | 89 | 5.96e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 2 | 80 | 5.96e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 1 | 20 | 5.96e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 1 | 39 | 5.97e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 43 | 5.97e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 1 | 35 | 5.97e-01 |
| GO:BP | GO:0016197 | endosomal transport | 4 | 231 | 5.97e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 1 | 36 | 5.97e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 1 | 40 | 5.98e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 1 | 787 | 6.00e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 1 | 113 | 6.02e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 1 | 35 | 6.02e-01 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 1 | 57 | 6.03e-01 |
| GO:BP | GO:0048232 | male gamete generation | 10 | 343 | 6.04e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 2 | 104 | 6.04e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 2 | 173 | 6.05e-01 |
| GO:BP | GO:0007033 | vacuole organization | 2 | 197 | 6.05e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 5 | 418 | 6.05e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 6.05e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 3 | 139 | 6.05e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 1 | 121 | 6.05e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 870 | 6.05e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 1 | 62 | 6.07e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 6.07e-01 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 1 | 61 | 6.07e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 6.07e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 5 | 898 | 6.07e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 1 | 36 | 6.07e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 901 | 6.07e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 1 | 73 | 6.07e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 65 | 6.08e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 6.08e-01 |
| GO:BP | GO:0016043 | cellular component organization | 3 | 5076 | 6.08e-01 |
| GO:BP | GO:0021537 | telencephalon development | 3 | 201 | 6.08e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 1 | 38 | 6.08e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 2 | 98 | 6.08e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 5 | 257 | 6.09e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 6.10e-01 |
| GO:BP | GO:0007052 | mitotic spindle organization | 1 | 129 | 6.10e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 6.10e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 3 | 294 | 6.10e-01 |
| GO:BP | GO:0003002 | regionalization | 7 | 273 | 6.12e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 6.12e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 3 | 357 | 6.12e-01 |
| GO:BP | GO:0031648 | protein destabilization | 1 | 45 | 6.13e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 1 | 13 | 6.14e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 6.15e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 1 | 96 | 6.15e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 2 | 59 | 6.15e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 2 | 175 | 6.16e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 1 | 86 | 6.16e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 1 | 223 | 6.16e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 6.16e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 1 | 39 | 6.16e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 2 | 120 | 6.16e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 6.16e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 4 | 4128 | 6.18e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 1 | 43 | 6.18e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 8 | 418 | 6.19e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 81 | 6.19e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 1 | 88 | 6.19e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 1 | 76 | 6.19e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 1 | 37 | 6.19e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 2 | 170 | 6.20e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 3 | 1719 | 6.20e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 6.21e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 6.21e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 2 | 52 | 6.22e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 6.23e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 1 | 38 | 6.24e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 1 | 39 | 6.24e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 6.24e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 6.24e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 6.25e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 3 | 165 | 6.25e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 6.25e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 6.25e-01 |
| GO:BP | GO:0032609 | type II interferon production | 2 | 53 | 6.25e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 6.27e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 1 | 61 | 6.27e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 1 | 38 | 6.27e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 1 | 47 | 6.27e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 6.27e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 665 | 6.27e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 1 | 52 | 6.27e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 1 | 47 | 6.27e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 2 | 140 | 6.27e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 6.28e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3 | 5274 | 6.28e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 1 | 64 | 6.28e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 2 | 187 | 6.29e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 2 | 170 | 6.30e-01 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 2 | 240 | 6.30e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 2 | 116 | 6.30e-01 |
| GO:BP | GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | 1 | 30 | 6.32e-01 |
| GO:BP | GO:0001824 | blastocyst development | 2 | 94 | 6.32e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 5 | 386 | 6.33e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 6.33e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 1 | 179 | 6.33e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 6.34e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 85 | 6.36e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 547 | 6.36e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 1 | 80 | 6.36e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 6.36e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 1 | 129 | 6.37e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 1 | 45 | 6.37e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 1 | 47 | 6.37e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 1 | 35 | 6.37e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 28 | 6.37e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 6.37e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 2 | 868 | 6.38e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 1 | 41 | 6.38e-01 |
| GO:BP | GO:0009314 | response to radiation | 2 | 352 | 6.38e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 1 | 38 | 6.39e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 90 | 6.40e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 1 | 82 | 6.40e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 1 | 39 | 6.40e-01 |
| GO:BP | GO:0006497 | protein lipidation | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 3 | 116 | 6.44e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 6.44e-01 |
| GO:BP | GO:0009749 | response to glucose | 3 | 137 | 6.44e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 4 | 4241 | 6.44e-01 |
| GO:BP | GO:0051028 | mRNA transport | 1 | 117 | 6.45e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 6.45e-01 |
| GO:BP | GO:0015918 | sterol transport | 2 | 92 | 6.45e-01 |
| GO:BP | GO:0030282 | bone mineralization | 2 | 82 | 6.46e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 1 | 83 | 6.46e-01 |
| GO:BP | GO:0002252 | immune effector process | 3 | 318 | 6.48e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 1 | 48 | 6.48e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 6.48e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 2 | 152 | 6.48e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 1 | 111 | 6.48e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 1 | 51 | 6.48e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 1 | 42 | 6.48e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 1 | 51 | 6.49e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 1 | 252 | 6.50e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 1 | 108 | 6.50e-01 |
| GO:BP | GO:0043200 | response to amino acid | 1 | 95 | 6.52e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 1 | 64 | 6.52e-01 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 1 | 71 | 6.52e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 2 | 211 | 6.53e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 1 | 123 | 6.56e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 1 | 92 | 6.56e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 3 | 313 | 6.56e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 1 | 142 | 6.57e-01 |
| GO:BP | GO:0009746 | response to hexose | 3 | 138 | 6.58e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 2 | 112 | 6.58e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 6.58e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 6.58e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 1 | 41 | 6.59e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 1 | 168 | 6.60e-01 |
| GO:BP | GO:0120193 | tight junction organization | 1 | 52 | 6.60e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 1 | 60 | 6.62e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 4 | 182 | 6.62e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 6.63e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 92 | 6.64e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 597 | 6.64e-01 |
| GO:BP | GO:0007292 | female gamete generation | 2 | 103 | 6.64e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 597 | 6.64e-01 |
| GO:BP | GO:0051216 | cartilage development | 3 | 151 | 6.64e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 1 | 88 | 6.64e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 1 | 72 | 6.66e-01 |
| GO:BP | GO:0031929 | TOR signaling | 1 | 115 | 6.66e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 1 | 88 | 6.68e-01 |
| GO:BP | GO:0006997 | nucleus organization | 2 | 126 | 6.69e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 1 | 147 | 6.69e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 1 | 44 | 6.69e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 2 | 185 | 6.71e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 1 | 42 | 6.71e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 1 | 67 | 6.71e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 691 | 6.71e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 1 | 157 | 6.71e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 1 | 39 | 6.71e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 6.73e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 1 | 52 | 6.74e-01 |
| GO:BP | GO:0061448 | connective tissue development | 4 | 208 | 6.74e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 3 | 218 | 6.74e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 6.74e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 1 | 92 | 6.74e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 1 | 53 | 6.74e-01 |
| GO:BP | GO:0051304 | chromosome separation | 1 | 76 | 6.74e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 2 | 118 | 6.74e-01 |
| GO:BP | GO:0021766 | hippocampus development | 1 | 62 | 6.77e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 1 | 47 | 6.77e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 1 | 113 | 6.78e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 6.79e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 3 | 142 | 6.79e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 1 | 173 | 6.79e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1335 | 6.79e-01 |
| GO:BP | GO:0098754 | detoxification | 1 | 91 | 6.79e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 2 | 137 | 6.79e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 2 | 86 | 6.79e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 5 | 369 | 6.81e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 6.81e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 6.83e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 1 | 47 | 6.83e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 1 | 228 | 6.83e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 1 | 58 | 6.85e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 2 | 292 | 6.87e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 6.87e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 1 | 49 | 6.87e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 1 | 54 | 6.87e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 1 | 59 | 6.87e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 1 | 59 | 6.87e-01 |
| GO:BP | GO:0001890 | placenta development | 1 | 117 | 6.87e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 1 | 105 | 6.87e-01 |
| GO:BP | GO:0051604 | protein maturation | 5 | 259 | 6.88e-01 |
| GO:BP | GO:0072006 | nephron development | 1 | 117 | 6.89e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 1 | 118 | 6.89e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 6.90e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 2 | 130 | 6.90e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 1 | 53 | 6.90e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 1 | 40 | 6.91e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 6.93e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 9 | 440 | 6.93e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 6.93e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 2 | 88 | 6.96e-01 |
| GO:BP | GO:0071709 | membrane assembly | 1 | 53 | 6.96e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 2 | 165 | 6.97e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 6.97e-01 |
| GO:BP | GO:0016236 | macroautophagy | 2 | 308 | 6.97e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 1 | 46 | 6.97e-01 |
| GO:BP | GO:0042476 | odontogenesis | 2 | 86 | 6.97e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1496 | 6.97e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 1 | 31 | 6.98e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 1 | 31 | 6.98e-01 |
| GO:BP | GO:0032502 | developmental process | 4 | 4494 | 6.98e-01 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 1 | 81 | 6.98e-01 |
| GO:BP | GO:0007030 | Golgi organization | 2 | 137 | 6.98e-01 |
| GO:BP | GO:0001501 | skeletal system development | 5 | 377 | 6.99e-01 |
| GO:BP | GO:0051302 | regulation of cell division | 1 | 145 | 6.99e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 1 | 55 | 6.99e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 1 | 46 | 7.00e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 2 | 167 | 7.00e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 1 | 48 | 7.00e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 1 | 133 | 7.01e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 2 | 168 | 7.02e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 4 | 192 | 7.02e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 2 | 169 | 7.04e-01 |
| GO:BP | GO:0050896 | response to stimulus | 3 | 5770 | 7.05e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 7.05e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 1 | 69 | 7.06e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 5 | 324 | 7.07e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 103 | 7.08e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 2 | 102 | 7.08e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 1 | 64 | 7.09e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 7.09e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 676 | 7.10e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 1 | 160 | 7.10e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 1 | 185 | 7.10e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 1 | 144 | 7.10e-01 |
| GO:BP | GO:0050658 | RNA transport | 1 | 144 | 7.10e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 7.10e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 72 | 7.10e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 4 | 192 | 7.10e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 1 | 115 | 7.11e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 1 | 54 | 7.11e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 95 | 7.11e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 1 | 52 | 7.11e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 1 | 104 | 7.11e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 2 | 94 | 7.11e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 111 | 7.11e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 73 | 7.12e-01 |
| GO:BP | GO:0022414 | reproductive process | 22 | 927 | 7.12e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 2 | 188 | 7.13e-01 |
| GO:BP | GO:0051301 | cell division | 3 | 570 | 7.13e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 2 | 293 | 7.13e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 1 | 316 | 7.13e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 1 | 184 | 7.13e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 1 | 147 | 7.14e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 73 | 7.14e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 7.14e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 1 | 71 | 7.14e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 7.15e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 1 | 58 | 7.16e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 7 | 421 | 7.17e-01 |
| GO:BP | GO:0007051 | spindle organization | 1 | 188 | 7.18e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 2 | 209 | 7.19e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 1 | 51 | 7.20e-01 |
| GO:BP | GO:0000003 | reproduction | 2 | 934 | 7.20e-01 |
| GO:BP | GO:0000280 | nuclear division | 2 | 344 | 7.20e-01 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 1 | 90 | 7.20e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 7.20e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 7.22e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 7.23e-01 |
| GO:BP | GO:0060047 | heart contraction | 1 | 205 | 7.23e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 3 | 273 | 7.24e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 2 | 249 | 7.27e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 97 | 7.29e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 1 | 37 | 7.31e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 1 | 119 | 7.32e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 2 | 143 | 7.35e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 7.35e-01 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 7.36e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 1 | 76 | 7.37e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 8 | 710 | 7.37e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 1 | 67 | 7.37e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 9 | 798 | 7.37e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 198 | 7.38e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 1 | 83 | 7.40e-01 |
| GO:BP | GO:0003015 | heart process | 1 | 216 | 7.40e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 31 | 1712 | 7.47e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 7.47e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 1 | 71 | 7.47e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 2 | 197 | 7.47e-01 |
| GO:BP | GO:0055001 | muscle cell development | 1 | 165 | 7.47e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2 | 4955 | 7.47e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 1 | 96 | 7.48e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 7.48e-01 |
| GO:BP | GO:0000910 | cytokinesis | 2 | 164 | 7.48e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 2 | 78 | 7.48e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 7.48e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 1 | 116 | 7.48e-01 |
| GO:BP | GO:0006865 | amino acid transport | 1 | 102 | 7.49e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 5 | 232 | 7.49e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 1 | 76 | 7.51e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 5 | 252 | 7.52e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 1 | 98 | 7.52e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 7.52e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 7.52e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 1 | 36 | 7.53e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 7.54e-01 |
| GO:BP | GO:0001654 | eye development | 2 | 272 | 7.54e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 2 | 126 | 7.56e-01 |
| GO:BP | GO:0048469 | cell maturation | 1 | 108 | 7.57e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 1 | 69 | 7.57e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 1 | 136 | 7.57e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 1 | 161 | 7.58e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 1 | 57 | 7.58e-01 |
| GO:BP | GO:0150063 | visual system development | 2 | 273 | 7.58e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 1 | 113 | 7.58e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 65 | 7.59e-01 |
| GO:BP | GO:0021761 | limbic system development | 1 | 81 | 7.60e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 998 | 7.60e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 77 | 7.61e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 5 | 190 | 7.62e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 85 | 7.62e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 2 | 116 | 7.62e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 1 | 84 | 7.62e-01 |
| GO:BP | GO:0042063 | gliogenesis | 2 | 235 | 7.64e-01 |
| GO:BP | GO:0048880 | sensory system development | 2 | 275 | 7.64e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 1 | 94 | 7.65e-01 |
| GO:BP | GO:0048285 | organelle fission | 2 | 388 | 7.67e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 1 | 77 | 7.67e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 1 | 100 | 7.67e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 7.67e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 1 | 124 | 7.68e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 7.68e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 1 | 53 | 7.68e-01 |
| GO:BP | GO:0048477 | oogenesis | 1 | 64 | 7.70e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 1 | 88 | 7.70e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 2 | 194 | 7.70e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 7.72e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 1 | 43 | 7.72e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 1 | 81 | 7.72e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 1 | 49 | 7.73e-01 |
| GO:BP | GO:0006403 | RNA localization | 1 | 182 | 7.74e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 2 | 185 | 7.76e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 7.76e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 1 | 107 | 7.77e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 7.77e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 7.77e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 1 | 42 | 7.77e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 1 | 71 | 7.79e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 1 | 89 | 7.80e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 7.80e-01 |
| GO:BP | GO:0060271 | cilium assembly | 1 | 292 | 7.80e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 134 | 7.81e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 1 | 60 | 7.81e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 1 | 131 | 7.81e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 7.82e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 7.83e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 1 | 149 | 7.83e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 7.83e-01 |
| GO:BP | GO:0048284 | organelle fusion | 1 | 135 | 7.84e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 2 | 276 | 7.86e-01 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 1 | 74 | 7.87e-01 |
| GO:BP | GO:0045471 | response to ethanol | 1 | 73 | 7.88e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 1 | 140 | 7.88e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 1 | 98 | 7.89e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 1 | 67 | 7.90e-01 |
| GO:BP | GO:0006936 | muscle contraction | 2 | 267 | 7.90e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 2 | 407 | 7.90e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 1 | 74 | 7.91e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 2 | 245 | 7.91e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 1 | 75 | 7.91e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 2 | 288 | 7.91e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 1 | 222 | 7.91e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 1 | 161 | 7.91e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 2 | 91 | 7.92e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 2 | 91 | 7.92e-01 |
| GO:BP | GO:0051235 | maintenance of location | 4 | 260 | 7.93e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 3 | 116 | 7.93e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 2 | 225 | 7.95e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 2 | 111 | 7.97e-01 |
| GO:BP | GO:0030258 | lipid modification | 1 | 162 | 7.97e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 153 | 7.97e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 1 | 121 | 7.98e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 1 | 111 | 7.98e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 7.99e-01 |
| GO:BP | GO:0044782 | cilium organization | 1 | 313 | 8.00e-01 |
| GO:BP | GO:0022900 | electron transport chain | 1 | 142 | 8.01e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 8.01e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 1 | 110 | 8.01e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 4 | 353 | 8.02e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 1 | 91 | 8.02e-01 |
| GO:BP | GO:0051591 | response to cAMP | 1 | 66 | 8.02e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 4 | 246 | 8.03e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 8.03e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 2 | 94 | 8.04e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 2 | 94 | 8.04e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 1 | 194 | 8.06e-01 |
| GO:BP | GO:0032392 | DNA geometric change | 1 | 80 | 8.06e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 8.06e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 1 | 106 | 8.08e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 2 | 421 | 8.08e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 2 | 111 | 8.08e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 1 | 71 | 8.08e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 1 | 149 | 8.09e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 2 | 352 | 8.10e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 2 | 300 | 8.11e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 1 | 90 | 8.14e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 3 | 233 | 8.15e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 3 | 220 | 8.18e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 8.18e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 1 | 88 | 8.18e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 3 | 358 | 8.18e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 8.18e-01 |
| GO:BP | GO:0090398 | cellular senescence | 1 | 83 | 8.19e-01 |
| GO:BP | GO:0042552 | myelination | 1 | 111 | 8.20e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 2 | 429 | 8.20e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 71 | 8.20e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 1 | 98 | 8.22e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 1 | 113 | 8.23e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 1 | 113 | 8.23e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 1 | 156 | 8.25e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 1 | 66 | 8.26e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 3 | 500 | 8.26e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 1 | 89 | 8.28e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 1 | 49 | 8.28e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 1 | 62 | 8.28e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 1 | 60 | 8.28e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 1 | 60 | 8.28e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 2 | 252 | 8.28e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 1 | 74 | 8.28e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 1 | 101 | 8.30e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 94 | 8.30e-01 |
| GO:BP | GO:0071103 | DNA conformation change | 1 | 87 | 8.31e-01 |
| GO:BP | GO:0090132 | epithelium migration | 2 | 252 | 8.32e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 1 | 93 | 8.32e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 1 | 158 | 8.32e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 8.34e-01 |
| GO:BP | GO:1902074 | response to salt | 2 | 271 | 8.35e-01 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 8.35e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 1 | 86 | 8.36e-01 |
| GO:BP | GO:0071867 | response to monoamine | 1 | 62 | 8.36e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 1 | 62 | 8.36e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 2 | 428 | 8.36e-01 |
| GO:BP | GO:0090130 | tissue migration | 2 | 256 | 8.39e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 4 | 4794 | 8.43e-01 |
| GO:BP | GO:0070925 | organelle assembly | 5 | 817 | 8.44e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 3 | 285 | 8.44e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 1 | 172 | 8.44e-01 |
| GO:BP | GO:0007584 | response to nutrient | 1 | 109 | 8.47e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 1 | 143 | 8.47e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 5 | 552 | 8.47e-01 |
| GO:BP | GO:0007281 | germ cell development | 3 | 187 | 8.48e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 2 | 172 | 8.50e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 8.52e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 1 | 159 | 8.54e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 8.54e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 8.56e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 5 | 230 | 8.56e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 8.56e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 1 | 187 | 8.56e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 1 | 187 | 8.56e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 8.56e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 8.56e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 1 | 88 | 8.58e-01 |
| GO:BP | GO:0007040 | lysosome organization | 1 | 88 | 8.58e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 1 | 154 | 8.59e-01 |
| GO:BP | GO:0042592 | homeostatic process | 1 | 1159 | 8.59e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 3 | 200 | 8.61e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 73 | 8.63e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 1 | 544 | 8.63e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 1 | 157 | 8.64e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 1 | 189 | 8.65e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 1 | 97 | 8.65e-01 |
| GO:BP | GO:0042116 | macrophage activation | 1 | 57 | 8.66e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 1 | 69 | 8.69e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 1 | 88 | 8.71e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 1 | 86 | 8.72e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 96 | 8.75e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 4 | 251 | 8.75e-01 |
| GO:BP | GO:0009056 | catabolic process | 2 | 2088 | 8.75e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 2 | 271 | 8.75e-01 |
| GO:BP | GO:0003012 | muscle system process | 2 | 334 | 8.75e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 2 | 49 | 8.76e-01 |
| GO:BP | GO:0060348 | bone development | 2 | 170 | 8.78e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 1 | 258 | 8.79e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 1 | 513 | 8.81e-01 |
| GO:BP | GO:0006914 | autophagy | 1 | 513 | 8.81e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 2 | 53 | 8.82e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 1 | 226 | 8.84e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 1 | 170 | 8.84e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 1 | 175 | 8.84e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 1 | 293 | 8.86e-01 |
| GO:BP | GO:0007565 | female pregnancy | 1 | 122 | 8.86e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 5 | 242 | 8.88e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 8.88e-01 |
| GO:BP | GO:0021543 | pallium development | 1 | 139 | 8.89e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 1 | 1229 | 8.89e-01 |
| GO:BP | GO:0030534 | adult behavior | 1 | 92 | 8.89e-01 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 8.90e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 1 | 82 | 8.91e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 1 | 119 | 8.91e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 1 | 83 | 8.93e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 3 | 214 | 8.93e-01 |
| GO:BP | GO:0002253 | activation of immune response | 6 | 272 | 8.93e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 1 | 98 | 8.94e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 1 | 112 | 8.94e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 2 | 281 | 8.96e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 1 | 85 | 8.96e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 3 | 433 | 8.97e-01 |
| GO:BP | GO:0021700 | developmental maturation | 1 | 211 | 8.98e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 1 | 114 | 8.99e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 2 | 236 | 8.99e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 2 | 369 | 8.99e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 1 | 96 | 8.99e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 9.00e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 2 | 130 | 9.02e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 2 | 211 | 9.02e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 1 | 89 | 9.02e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 1 | 218 | 9.03e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 9.03e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 203 | 9.03e-01 |
| GO:BP | GO:0007423 | sensory organ development | 2 | 396 | 9.03e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 9.03e-01 |
| GO:BP | GO:0051648 | vesicle localization | 1 | 192 | 9.03e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 9.03e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 1 | 84 | 9.04e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 1 | 202 | 9.07e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 9.07e-01 |
| GO:BP | GO:0051607 | defense response to virus | 1 | 202 | 9.07e-01 |
| GO:BP | GO:0007338 | single fertilization | 1 | 70 | 9.07e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 9.07e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 1 | 131 | 9.08e-01 |
| GO:BP | GO:0006909 | phagocytosis | 3 | 148 | 9.08e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 1 | 182 | 9.08e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 1 | 234 | 9.09e-01 |
| GO:BP | GO:0006396 | RNA processing | 4 | 929 | 9.09e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 1 | 135 | 9.11e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 1 | 141 | 9.11e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 9.11e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 9.13e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 1 | 100 | 9.14e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 174 | 9.16e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 1 | 135 | 9.18e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 1 | 195 | 9.18e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 1 | 113 | 9.19e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 1 | 116 | 9.19e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 3 | 246 | 9.20e-01 |
| GO:BP | GO:0008015 | blood circulation | 1 | 374 | 9.22e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 9.25e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 5 | 532 | 9.29e-01 |
| GO:BP | GO:0008544 | epidermis development | 1 | 210 | 9.31e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 3 | 433 | 9.31e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 9.36e-01 |
| GO:BP | GO:0016485 | protein processing | 2 | 183 | 9.36e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 1 | 126 | 9.37e-01 |
| GO:BP | GO:0042886 | amide transport | 3 | 235 | 9.38e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 2 | 227 | 9.38e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 2 | 199 | 9.40e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 128 | 9.40e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 1 | 121 | 9.42e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 1 | 275 | 9.42e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 3 | 302 | 9.42e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 1 | 107 | 9.42e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 1 | 117 | 9.45e-01 |
| GO:BP | GO:0044057 | regulation of system process | 1 | 405 | 9.45e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 1 | 252 | 9.46e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 2 | 188 | 9.47e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 1 | 135 | 9.47e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 4 | 276 | 9.47e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 1 | 404 | 9.50e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 1 | 137 | 9.52e-01 |
| GO:BP | GO:0009566 | fertilization | 1 | 93 | 9.53e-01 |
| GO:BP | GO:0003013 | circulatory system process | 1 | 447 | 9.53e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 1 | 269 | 9.56e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 1 | 126 | 9.57e-01 |
| GO:BP | GO:0007286 | spermatid development | 1 | 105 | 9.58e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 1 | 138 | 9.60e-01 |
| GO:BP | GO:0009615 | response to virus | 1 | 275 | 9.62e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 1 | 216 | 9.63e-01 |
| GO:BP | GO:0007568 | aging | 1 | 115 | 9.63e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 115 | 9.63e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 1 | 203 | 9.63e-01 |
| GO:BP | GO:0015849 | organic acid transport | 1 | 217 | 9.63e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 843 | 9.63e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 1 | 122 | 9.64e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 1 | 75 | 9.64e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 1 | 206 | 9.64e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 1 | 116 | 9.64e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 7 | 369 | 9.65e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 4 | 357 | 9.67e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 1 | 194 | 9.68e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 9.68e-01 |
| GO:BP | GO:0030073 | insulin secretion | 1 | 141 | 9.68e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 7 | 447 | 9.68e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 1 | 115 | 9.68e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 9.69e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 4 | 362 | 9.70e-01 |
| GO:BP | GO:0001816 | cytokine production | 7 | 451 | 9.70e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 9 | 484 | 9.71e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 4 | 337 | 9.73e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 4 | 351 | 9.74e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 1 | 90 | 9.76e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 154 | 9.80e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 1 | 316 | 9.80e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 3 | 760 | 9.80e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 4 | 740 | 9.80e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 1 | 311 | 9.81e-01 |
| GO:BP | GO:0016358 | dendrite development | 1 | 202 | 9.84e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 9.85e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 9.85e-01 |
| GO:BP | GO:0015711 | organic anion transport | 1 | 264 | 9.85e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 9.85e-01 |
| GO:BP | GO:0009306 | protein secretion | 2 | 262 | 9.85e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 9.85e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 2 | 262 | 9.85e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 185 | 9.85e-01 |
| GO:BP | GO:0006869 | lipid transport | 1 | 272 | 9.85e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 1 | 179 | 9.86e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 2 | 151 | 9.86e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 2 | 266 | 9.87e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 9.89e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 1 | 166 | 9.89e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 5 | 448 | 9.89e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 1 | 373 | 9.90e-01 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 170 | 9.90e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 9.91e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 4 | 401 | 9.92e-01 |
| GO:BP | GO:0006364 | rRNA processing | 1 | 221 | 9.92e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 9.93e-01 |
| GO:BP | GO:0010876 | lipid localization | 3 | 310 | 9.93e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 9.93e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 4 | 408 | 9.93e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 2 | 162 | 9.93e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 1 | 425 | 9.94e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 9.94e-01 |
| GO:BP | GO:0015833 | peptide transport | 1 | 179 | 9.94e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 9.96e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 9.97e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 1 | 190 | 9.97e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 9.97e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 1 | 237 | 9.97e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 2 | 212 | 9.98e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 1 | 523 | 9.99e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 1 | 199 | 9.99e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 1 | 249 | 9.99e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 1 | 317 | 1.00e+00 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 378 | 1.00e+00 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 2 | 681 | 1.00e+00 |
| GO:BP | GO:0016072 | rRNA metabolic process | 1 | 258 | 1.00e+00 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 1 | 429 | 1.00e+00 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 1 | 506 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 1 | 248 | 1.00e+00 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 1.00e+00 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 1 | 295 | 1.00e+00 |
| GO:BP | GO:0042254 | ribosome biogenesis | 1 | 301 | 1.00e+00 |
| GO:BP | GO:0009611 | response to wounding | 1 | 415 | 1.00e+00 |
| GO:BP | GO:0044281 | small molecule metabolic process | 1 | 1389 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 230 | 1.00e+00 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 1.00e+00 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 1 | 371 | 1.00e+00 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 1 | 423 | 1.00e+00 |
| GO:BP | GO:0045087 | innate immune response | 5 | 488 | 1.00e+00 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 1 | 475 | 1.00e+00 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 1 | 311 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0006811 | monoatomic ion transport | 1 | 775 | 1.00e+00 |
| GO:BP | GO:0042742 | defense response to bacterium | 1 | 95 | 1.00e+00 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 1.00e+00 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 1 | 295 | 1.00e+00 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 213 | 1.00e+00 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 1 | 632 | 1.00e+00 |
| GO:BP | GO:0030001 | metal ion transport | 1 | 562 | 1.00e+00 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 1.00e+00 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 1 | 319 | 1.00e+00 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 1 | 784 | 1.00e+00 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 1 | 517 | 1.00e+00 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 1 | 487 | 1.00e+00 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 1.00e+00 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 1.00e+00 |
| GO:BP | GO:0051668 | localization within membrane | 1 | 605 | 1.00e+00 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 389 | 1.00e+00 |
| GO:BP | GO:0051640 | organelle localization | 1 | 503 | 1.00e+00 |
| GO:BP | GO:0061024 | membrane organization | 1 | 671 | 1.00e+00 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 422 | 1.00e+00 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 1.00e+00 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 1.00e+00 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 1 | 563 | 1.00e+00 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 10 | 1238 | 1.00e+00 |
| GO:BP | GO:0032409 | regulation of transporter activity | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 1 | 540 | 1.00e+00 |
| GO:BP | GO:0006954 | inflammatory response | 1 | 429 | 1.00e+00 |
| GO:BP | GO:0006897 | endocytosis | 4 | 527 | 1.00e+00 |
| GO:BP | GO:0070482 | response to oxygen levels | 1 | 272 | 1.00e+00 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0006812 | monoatomic cation transport | 1 | 669 | 1.00e+00 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 1 | 391 | 1.00e+00 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 1 | 390 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 29 | 415 | 4.08e-08 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 1 | 103 | 1.48e-01 |
| KEGG | KEGG:04924 | Renin secretion | 1 | 50 | 1.48e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 1.48e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 1.48e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 1 | 77 | 1.48e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 1 | 98 | 1.48e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 1.48e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 1 | 49 | 1.48e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 1.48e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 2 | 4 | 1.48e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 1 | 35 | 1.48e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 1 | 83 | 1.48e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 1 | 97 | 1.48e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 1 | 38 | 1.48e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 1.48e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 1.48e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 1 | 64 | 1.48e-01 |
| KEGG | KEGG:05016 | Huntington disease | 2 | 256 | 1.48e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 1 | 121 | 1.48e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 1 | 48 | 1.48e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 1.48e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 1 | 77 | 1.48e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 4 | 84 | 1.48e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 1.48e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 1 | 128 | 1.48e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 1.48e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 1 | 87 | 1.48e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 1.48e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 1.48e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 1.48e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 1 | 41 | 1.48e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 1.48e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 1 | 12 | 1.48e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 1 | 70 | 1.48e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 1.48e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 1 | 150 | 1.48e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 1 | 133 | 1.48e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 3 | 51 | 1.48e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 1 | 106 | 1.52e-01 |
| KEGG | KEGG:04146 | Peroxisome | 1 | 68 | 1.52e-01 |
| KEGG | KEGG:05034 | Alcoholism | 1 | 122 | 1.52e-01 |
| KEGG | KEGG:05224 | Breast cancer | 5 | 117 | 1.54e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 1 | 172 | 1.54e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 1.54e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 1 | 176 | 1.54e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 1.54e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 1 | 142 | 1.54e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 1 | 180 | 1.54e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 1 | 106 | 1.54e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 1.57e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 2 | 48 | 1.58e-01 |
| KEGG | KEGG:05020 | Prion disease | 1 | 220 | 1.76e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 1.80e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 4 | 145 | 1.80e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 2 | 124 | 1.80e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 5 | 138 | 1.94e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 1 | 272 | 1.94e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 2.03e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 2 | 57 | 2.24e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 2 | 49 | 2.52e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 3.01e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 4 | 117 | 3.01e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 1 | 25 | 3.31e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 2 | 56 | 3.54e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 2 | 84 | 3.78e-01 |
| KEGG | KEGG:05164 | Influenza A | 1 | 103 | 3.87e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 1 | 32 | 3.87e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 1 | 161 | 3.87e-01 |
| KEGG | KEGG:00000 | KEGG root term | 5 | 5558 | 4.02e-01 |
| KEGG | KEGG:04110 | Cell cycle | 2 | 151 | 4.28e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 4.33e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 4.53e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 4.80e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 3 | 134 | 5.65e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 1 | 65 | 5.65e-01 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 6.60e-01 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 6.60e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 6.60e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 6.60e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 1 | 99 | 6.60e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 1 | 44 | 6.60e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 1 | 35 | 6.60e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 1 | 75 | 6.60e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 6.60e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 6.67e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 7.28e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 2 | 128 | 7.34e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 2 | 164 | 7.56e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 2 | 104 | 7.68e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 1 | 65 | 7.68e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 7.68e-01 |
| KEGG | KEGG:04520 | Adherens junction | 1 | 67 | 7.72e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 5 | 409 | 7.72e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 1 | 135 | 7.72e-01 |
| KEGG | KEGG:03010 | Ribosome | 1 | 127 | 7.72e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 1 | 121 | 7.72e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 7.72e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 2 | 88 | 7.72e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 8.12e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 8.12e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 1 | 56 | 8.33e-01 |
| KEGG | KEGG:04530 | Tight junction | 1 | 137 | 8.33e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 8.66e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 1 | 89 | 8.69e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 3 | 149 | 8.71e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 8.77e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 1 | 112 | 8.77e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 8.77e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 1 | 109 | 8.92e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 1 | 97 | 8.92e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 1 | 115 | 8.92e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 1 | 114 | 9.02e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 9.22e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 1 | 179 | 9.28e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 2 | 315 | 9.28e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 1 | 240 | 9.36e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 1 | 171 | 9.80e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 9.88e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 1 | 98 | 9.88e-01 |
# EPgostres24 <- gost(query = sigVEP24$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EPgostres24,"data/DEG-GO/EPgostres24.RDS")
EPgostres24 <- readRDS("data/DEG-GO/EPgostres24.RDS")
# EPp24 <- gostplot(EPgostres24, capped = FALSE, interactive = TRUE)
# EPp24
EPtable124 <- EPgostres24$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
EPtable124 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('EPI 24 hour top 10 GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)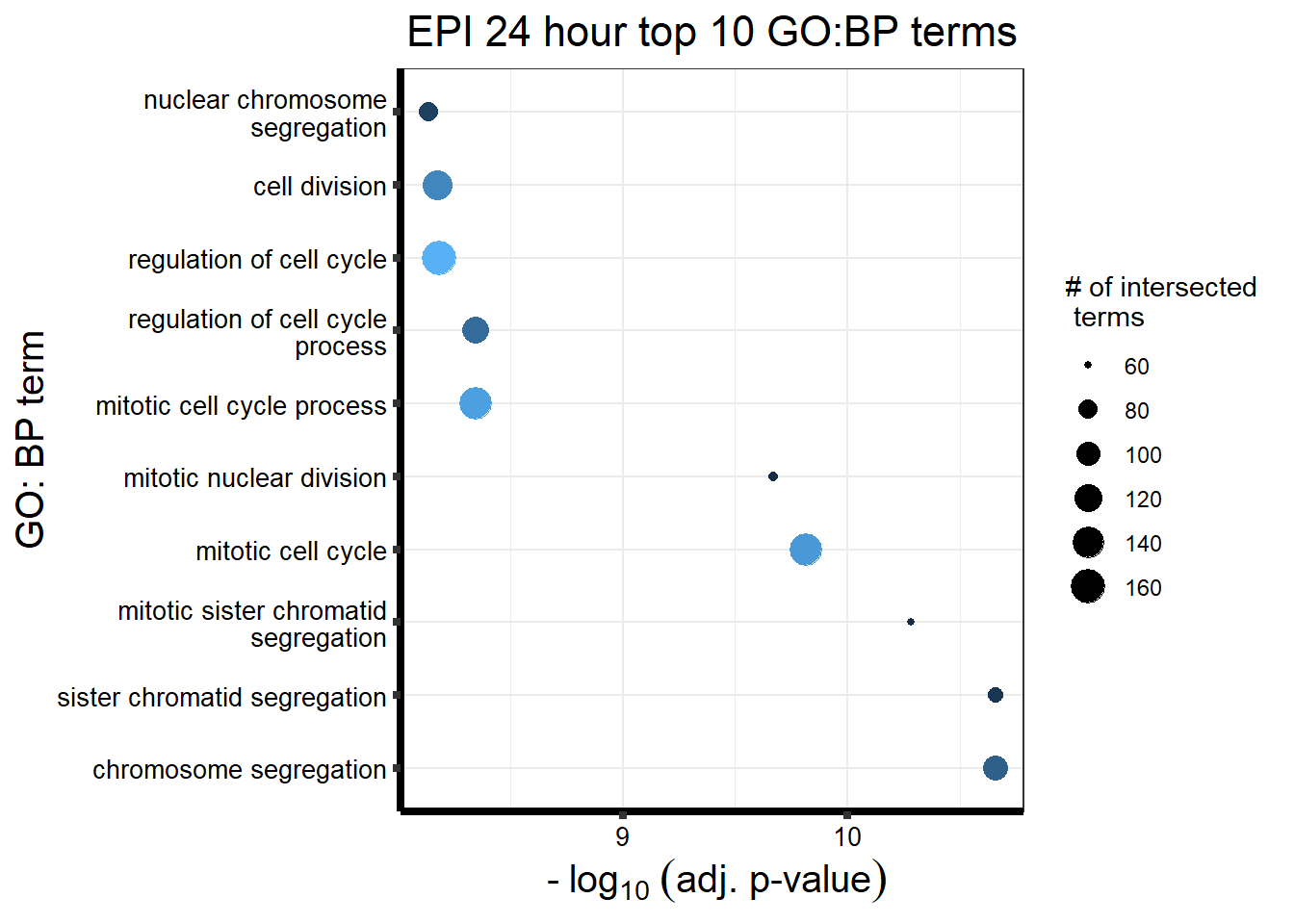
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
EPtable124 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "EPI 24 hour lists") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0007059 | chromosome segregation | 103 | 369 | 2.18e-11 |
| GO:BP | GO:0000819 | sister chromatid segregation | 69 | 217 | 2.18e-11 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 60 | 179 | 5.21e-11 |
| GO:BP | GO:0000278 | mitotic cell cycle | 145 | 827 | 1.52e-10 |
| GO:BP | GO:0140014 | mitotic nuclear division | 61 | 251 | 2.14e-10 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 150 | 694 | 4.52e-09 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 112 | 626 | 4.52e-09 |
| GO:BP | GO:0051726 | regulation of cell cycle | 162 | 956 | 6.60e-09 |
| GO:BP | GO:0051301 | cell division | 132 | 570 | 6.74e-09 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 79 | 273 | 7.35e-09 |
| GO:BP | GO:0022402 | cell cycle process | 175 | 1086 | 3.68e-08 |
| GO:BP | GO:0051276 | chromosome organization | 126 | 543 | 5.72e-08 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 45 | 139 | 8.54e-08 |
| GO:BP | GO:0000280 | nuclear division | 95 | 344 | 9.29e-08 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 58 | 183 | 1.14e-07 |
| GO:BP | GO:0007049 | cell cycle | 228 | 1529 | 1.22e-07 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 80 | 211 | 3.25e-07 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 70 | 338 | 4.00e-07 |
| GO:BP | GO:0007051 | spindle organization | 52 | 188 | 1.38e-06 |
| GO:BP | GO:0051225 | spindle assembly | 39 | 120 | 1.96e-06 |
| GO:BP | GO:0048285 | organelle fission | 97 | 388 | 3.56e-06 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 23 | 71 | 3.56e-06 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 20 | 42 | 4.67e-06 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 56 | 273 | 5.62e-06 |
| GO:BP | GO:0006974 | DNA damage response | 144 | 785 | 1.21e-05 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 36 | 121 | 1.24e-05 |
| GO:BP | GO:0007052 | mitotic spindle organization | 35 | 129 | 1.34e-05 |
| GO:BP | GO:0051304 | chromosome separation | 23 | 76 | 1.34e-05 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 36 | 157 | 1.34e-05 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 49 | 235 | 1.74e-05 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 16 | 44 | 2.53e-05 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 16 | 44 | 2.53e-05 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 16 | 44 | 2.53e-05 |
| GO:BP | GO:0006281 | DNA repair | 114 | 534 | 2.79e-05 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 16 | 45 | 3.38e-05 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 18 | 54 | 3.75e-05 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 16 | 46 | 4.47e-05 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 13 | 55 | 4.51e-05 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 111 | 441 | 4.75e-05 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 18 | 58 | 5.66e-05 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 17 | 52 | 5.72e-05 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 36 | 90 | 6.55e-05 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 16 | 47 | 6.69e-05 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 27 | 100 | 6.69e-05 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 16 | 47 | 6.69e-05 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 16 | 47 | 6.69e-05 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 16 | 47 | 6.69e-05 |
| GO:BP | GO:0051310 | metaphase plate congression | 14 | 69 | 9.19e-05 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 46 | 148 | 9.19e-05 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 22 | 88 | 9.69e-05 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 29 | 91 | 9.69e-05 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 18 | 61 | 9.69e-05 |
| GO:BP | GO:0006260 | DNA replication | 74 | 255 | 9.98e-05 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 16 | 48 | 1.05e-04 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 16 | 48 | 1.05e-04 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 56 | 209 | 1.06e-04 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 11 | 18 | 1.12e-04 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 39 | 129 | 1.16e-04 |
| GO:BP | GO:0006541 | glutamine metabolic process | 4 | 24 | 1.16e-04 |
| GO:BP | GO:0051783 | regulation of nuclear division | 29 | 116 | 1.17e-04 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 17 | 27 | 1.25e-04 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 67 | 391 | 1.26e-04 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 22 | 89 | 1.43e-04 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 18 | 57 | 1.53e-04 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 29 | 92 | 1.57e-04 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 22 | 79 | 1.60e-04 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 33 | 166 | 1.73e-04 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 34 | 99 | 1.91e-04 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 56 | 236 | 2.14e-04 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 24 | 47 | 2.20e-04 |
| GO:BP | GO:0051231 | spindle elongation | 8 | 12 | 2.29e-04 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 8 | 11 | 2.50e-04 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 31 | 318 | 2.70e-04 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 9 | 2.82e-04 |
| GO:BP | GO:0051255 | spindle midzone assembly | 9 | 12 | 2.87e-04 |
| GO:BP | GO:0006275 | regulation of DNA replication | 45 | 116 | 3.00e-04 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 93 | 552 | 3.00e-04 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 30 | 85 | 3.00e-04 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 29 | 308 | 3.15e-04 |
| GO:BP | GO:0044770 | cell cycle phase transition | 79 | 504 | 3.56e-04 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 14 | 75 | 3.78e-04 |
| GO:BP | GO:0006270 | DNA replication initiation | 19 | 35 | 3.97e-04 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 13 | 70 | 4.46e-04 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 36 | 178 | 4.75e-04 |
| GO:BP | GO:0033043 | regulation of organelle organization | 69 | 1028 | 6.05e-04 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 10 | 6.62e-04 |
| GO:BP | GO:0019985 | translesion synthesis | 11 | 24 | 6.69e-04 |
| GO:BP | GO:0050000 | chromosome localization | 16 | 82 | 6.96e-04 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 27 | 121 | 7.93e-04 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 78 | 418 | 8.52e-04 |
| GO:BP | GO:0006301 | postreplication repair | 13 | 34 | 8.52e-04 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 28 | 81 | 9.08e-04 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 29 | 92 | 9.48e-04 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 20 | 81 | 9.48e-04 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 16 | 36 | 9.96e-04 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 37 | 134 | 1.34e-03 |
| GO:BP | GO:0051383 | kinetochore organization | 10 | 23 | 1.47e-03 |
| GO:BP | GO:0010212 | response to ionizing radiation | 37 | 130 | 1.63e-03 |
| GO:BP | GO:0006259 | DNA metabolic process | 169 | 884 | 1.88e-03 |
| GO:BP | GO:0000910 | cytokinesis | 32 | 164 | 1.89e-03 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 12 | 21 | 1.97e-03 |
| GO:BP | GO:0006177 | GMP biosynthetic process | 3 | 13 | 2.00e-03 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 3 | 14 | 2.00e-03 |
| GO:BP | GO:0051302 | regulation of cell division | 19 | 145 | 2.03e-03 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 8 | 42 | 2.11e-03 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 12 | 28 | 2.63e-03 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 39 | 149 | 2.69e-03 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 13 | 15 | 2.97e-03 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 3.00e-03 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 19 | 50 | 3.27e-03 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 6 | 7 | 3.42e-03 |
| GO:BP | GO:0090224 | regulation of spindle organization | 8 | 45 | 3.79e-03 |
| GO:BP | GO:0046060 | dATP metabolic process | 2 | 3 | 4.37e-03 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 17 | 48 | 5.11e-03 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 4 | 60 | 5.13e-03 |
| GO:BP | GO:0009411 | response to UV | 29 | 141 | 5.34e-03 |
| GO:BP | GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 3 | 19 | 5.44e-03 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 7 | 8 | 5.62e-03 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 18 | 186 | 6.86e-03 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 22 | 104 | 6.86e-03 |
| GO:BP | GO:0044314 | protein K27-linked ubiquitination | 8 | 8 | 7.05e-03 |
| GO:BP | GO:0009127 | purine nucleoside monophosphate biosynthetic process | 3 | 21 | 7.05e-03 |
| GO:BP | GO:0036166 | phenotypic switching | 5 | 7 | 7.08e-03 |
| GO:BP | GO:0007098 | centrosome cycle | 26 | 127 | 7.18e-03 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 64 | 274 | 7.24e-03 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 29 | 103 | 7.40e-03 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 21 | 60 | 7.47e-03 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 13 | 45 | 7.67e-03 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 6 | 8 | 7.78e-03 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 23 | 74 | 8.41e-03 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 10 | 69 | 9.04e-03 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 8 | 11 | 1.02e-02 |
| GO:BP | GO:0007017 | microtubule-based process | 120 | 760 | 1.02e-02 |
| GO:BP | GO:0046033 | AMP metabolic process | 3 | 22 | 1.06e-02 |
| GO:BP | GO:0006284 | base-excision repair | 28 | 44 | 1.06e-02 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 56 | 357 | 1.09e-02 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 21 | 62 | 1.15e-02 |
| GO:BP | GO:0046037 | GMP metabolic process | 3 | 23 | 1.16e-02 |
| GO:BP | GO:0044209 | AMP salvage | 2 | 5 | 1.17e-02 |
| GO:BP | GO:0032263 | GMP salvage | 2 | 4 | 1.17e-02 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 27 | 137 | 1.17e-02 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 14 | 35 | 1.17e-02 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 1.21e-02 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 1.21e-02 |
| GO:BP | GO:1902969 | mitotic DNA replication | 10 | 14 | 1.22e-02 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 15 | 40 | 1.28e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 10 | 73 | 1.34e-02 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 9 | 18 | 1.38e-02 |
| GO:BP | GO:0006189 | ‘de novo’ IMP biosynthetic process | 5 | 6 | 1.53e-02 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 13 | 30 | 1.53e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 50 | 268 | 1.55e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 50 | 268 | 1.55e-02 |
| GO:BP | GO:0033554 | cellular response to stress | 230 | 1681 | 1.57e-02 |
| GO:BP | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process | 2 | 6 | 1.57e-02 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 4 | 11 | 1.57e-02 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 11 | 18 | 1.57e-02 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 15 | 37 | 1.57e-02 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 11 | 18 | 1.57e-02 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 9 | 14 | 1.62e-02 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 30 | 52 | 1.75e-02 |
| GO:BP | GO:0071478 | cellular response to radiation | 32 | 156 | 1.85e-02 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 3 | 27 | 1.99e-02 |
| GO:BP | GO:0007099 | centriole replication | 3 | 38 | 2.06e-02 |
| GO:BP | GO:0046053 | dAMP metabolic process | 2 | 5 | 2.09e-02 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 11 | 13 | 2.12e-02 |
| GO:BP | GO:1900262 | regulation of DNA-directed DNA polymerase activity | 11 | 13 | 2.12e-02 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 17 | 29 | 2.12e-02 |
| GO:BP | GO:0032392 | DNA geometric change | 23 | 80 | 2.17e-02 |
| GO:BP | GO:0042148 | strand invasion | 4 | 5 | 2.20e-02 |
| GO:BP | GO:0051299 | centrosome separation | 13 | 14 | 2.21e-02 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 10 | 12 | 2.21e-02 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 8 | 19 | 2.26e-02 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 11 | 20 | 2.37e-02 |
| GO:BP | GO:0010992 | ubiquitin recycling | 8 | 12 | 2.41e-02 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 5 | 7 | 2.41e-02 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 11 | 22 | 2.49e-02 |
| GO:BP | GO:0098534 | centriole assembly | 3 | 41 | 2.49e-02 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 61 | 175 | 2.54e-02 |
| GO:BP | GO:0106380 | purine ribonucleotide salvage | 2 | 7 | 2.54e-02 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 2 | 8 | 2.54e-02 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 2.56e-02 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 3 | 4 | 2.58e-02 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 2 | 16 | 2.62e-02 |
| GO:BP | GO:0034644 | cellular response to UV | 21 | 86 | 2.87e-02 |
| GO:BP | GO:0009314 | response to radiation | 59 | 352 | 2.92e-02 |
| GO:BP | GO:0034508 | centromere complex assembly | 9 | 28 | 2.97e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 15 | 82 | 3.00e-02 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 23 | 68 | 3.00e-02 |
| GO:BP | GO:0061642 | chemoattraction of axon | 2 | 2 | 3.04e-02 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 24 | 160 | 3.04e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 4 | 84 | 3.04e-02 |
| GO:BP | GO:0071103 | DNA conformation change | 24 | 87 | 3.04e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 31 | 240 | 3.13e-02 |
| GO:BP | GO:0033260 | nuclear DNA replication | 13 | 35 | 3.26e-02 |
| GO:BP | GO:0006302 | double-strand break repair | 79 | 271 | 3.29e-02 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 3 | 37 | 3.29e-02 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 18 | 30 | 3.29e-02 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 40 | 135 | 3.29e-02 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 41 | 604 | 3.39e-02 |
| GO:BP | GO:0070925 | organelle assembly | 118 | 817 | 3.48e-02 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 2 | 2 | 3.48e-02 |
| GO:BP | GO:0097293 | XMP biosynthetic process | 5 | 7 | 3.52e-02 |
| GO:BP | GO:0097292 | XMP metabolic process | 5 | 7 | 3.52e-02 |
| GO:BP | GO:0097294 | ‘de novo’ XMP biosynthetic process | 5 | 7 | 3.52e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 1 | 8 | 3.55e-02 |
| GO:BP | GO:0009170 | purine deoxyribonucleoside monophosphate metabolic process | 2 | 7 | 3.55e-02 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 3 | 39 | 3.57e-02 |
| GO:BP | GO:0140013 | meiotic nuclear division | 37 | 120 | 3.57e-02 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 6 | 7 | 3.61e-02 |
| GO:BP | GO:0051656 | establishment of organelle localization | 52 | 378 | 3.61e-02 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 6 | 10 | 3.66e-02 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 21 | 103 | 3.80e-02 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 12 | 13 | 3.84e-02 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 3.85e-02 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 6 | 12 | 3.86e-02 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 6 | 12 | 3.86e-02 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 7 | 13 | 4.00e-02 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 5 | 9 | 4.00e-02 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 2 | 11 | 4.00e-02 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 2 | 11 | 4.00e-02 |
| GO:BP | GO:0032261 | purine nucleotide salvage | 2 | 10 | 4.00e-02 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 2 | 11 | 4.00e-02 |
| GO:BP | GO:0051640 | organelle localization | 64 | 503 | 4.06e-02 |
| GO:BP | GO:1990227 | paranodal junction maintenance | 1 | 2 | 4.09e-02 |
| GO:BP | GO:0051298 | centrosome duplication | 7 | 68 | 4.40e-02 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 8 | 8 | 4.41e-02 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 47 | 149 | 4.52e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 4 | 7 | 4.60e-02 |
| GO:BP | GO:1990170 | stress response to cadmium ion | 2 | 2 | 4.70e-02 |
| GO:BP | GO:0071585 | detoxification of cadmium ion | 2 | 2 | 4.70e-02 |
| GO:BP | GO:0014029 | neural crest formation | 7 | 13 | 4.73e-02 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 9 | 270 | 4.79e-02 |
| GO:BP | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | 3 | 7 | 4.91e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 1 | 13 | 5.08e-02 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 9 | 161 | 5.11e-02 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 7 | 28 | 5.51e-02 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 5.52e-02 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 9 | 15 | 5.56e-02 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 6 | 9 | 5.60e-02 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 4 | 15 | 5.67e-02 |
| GO:BP | GO:0006979 | response to oxidative stress | 30 | 352 | 5.67e-02 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 1 | 5 | 5.67e-02 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 4 | 15 | 5.88e-02 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 9 | 14 | 5.94e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 13 | 35 | 5.94e-02 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 9 | 14 | 5.94e-02 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 9 | 14 | 5.94e-02 |
| GO:BP | GO:0031297 | replication fork processing | 26 | 42 | 5.94e-02 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 31 | 439 | 5.94e-02 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 5 | 13 | 5.94e-02 |
| GO:BP | GO:0010332 | response to gamma radiation | 26 | 50 | 5.94e-02 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 8 | 43 | 5.96e-02 |
| GO:BP | GO:0090677 | reversible differentiation | 3 | 5 | 5.96e-02 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 5 | 21 | 5.96e-02 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 6 | 194 | 5.97e-02 |
| GO:BP | GO:0031503 | protein-containing complex localization | 61 | 168 | 6.05e-02 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 14 | 33 | 6.14e-02 |
| GO:BP | GO:0022616 | DNA strand elongation | 22 | 35 | 6.17e-02 |
| GO:BP | GO:0061820 | telomeric D-loop disassembly | 5 | 8 | 6.19e-02 |
| GO:BP | GO:0051321 | meiotic cell cycle | 49 | 188 | 6.25e-02 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 2 | 14 | 6.39e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 2 | 15 | 6.39e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 2 | 15 | 6.39e-02 |
| GO:BP | GO:0035878 | nail development | 2 | 8 | 6.39e-02 |
| GO:BP | GO:0044208 | ‘de novo’ AMP biosynthetic process | 5 | 8 | 6.47e-02 |
| GO:BP | GO:0000725 | recombinational repair | 47 | 153 | 6.49e-02 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 7 | 11 | 6.49e-02 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 6.49e-02 |
| GO:BP | GO:0015691 | cadmium ion transport | 3 | 5 | 6.61e-02 |
| GO:BP | GO:0070574 | cadmium ion transmembrane transport | 3 | 5 | 6.61e-02 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 9 | 21 | 6.65e-02 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 3 | 50 | 6.65e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 13 | 36 | 6.75e-02 |
| GO:BP | GO:0006298 | mismatch repair | 18 | 29 | 6.75e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 13 | 36 | 6.75e-02 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 1 | 7 | 6.94e-02 |
| GO:BP | GO:0009151 | purine deoxyribonucleotide metabolic process | 2 | 12 | 6.94e-02 |
| GO:BP | GO:0010458 | exit from mitosis | 21 | 30 | 6.96e-02 |
| GO:BP | GO:0034502 | protein localization to chromosome | 11 | 110 | 7.01e-02 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 5 | 7.03e-02 |
| GO:BP | GO:1901491 | negative regulation of lymphangiogenesis | 3 | 3 | 7.03e-02 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 23 | 85 | 7.18e-02 |
| GO:BP | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 7.33e-02 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 2 | 3 | 7.33e-02 |
| GO:BP | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 7.33e-02 |
| GO:BP | GO:0072708 | response to sorbitol | 7 | 7 | 7.33e-02 |
| GO:BP | GO:0006175 | dATP biosynthetic process | 1 | 1 | 7.33e-02 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 6 | 19 | 7.49e-02 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 5 | 14 | 7.57e-02 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 7.57e-02 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 5 | 7.60e-02 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 4 | 151 | 7.62e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 4 | 7.62e-02 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 10 | 17 | 7.73e-02 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 13 | 41 | 7.78e-02 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 17 | 22 | 7.86e-02 |
| GO:BP | GO:0002076 | osteoblast development | 2 | 12 | 7.86e-02 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 3 | 9 | 7.86e-02 |
| GO:BP | GO:0086016 | AV node cell action potential | 3 | 9 | 7.86e-02 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 3 | 5 | 7.86e-02 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 5 | 42 | 7.88e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 11 | 21 | 8.00e-02 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 8 | 10 | 8.00e-02 |
| GO:BP | GO:0043173 | nucleotide salvage | 2 | 15 | 8.00e-02 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 4 | 4 | 8.08e-02 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 4 | 4 | 8.08e-02 |
| GO:BP | GO:0030261 | chromosome condensation | 10 | 33 | 8.08e-02 |
| GO:BP | GO:0046075 | dTTP metabolic process | 4 | 4 | 8.08e-02 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 6 | 65 | 8.18e-02 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 8.27e-02 |
| GO:BP | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process | 5 | 6 | 8.34e-02 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 13 | 28 | 8.34e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 22 | 293 | 8.46e-02 |
| GO:BP | GO:0035611 | protein branching point deglutamylation | 1 | 1 | 8.46e-02 |
| GO:BP | GO:0036438 | maintenance of lens transparency | 4 | 6 | 8.46e-02 |
| GO:BP | GO:0046599 | regulation of centriole replication | 2 | 21 | 8.50e-02 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 2 | 19 | 8.55e-02 |
| GO:BP | GO:0009092 | homoserine metabolic process | 3 | 5 | 8.56e-02 |
| GO:BP | GO:0019346 | transsulfuration | 3 | 5 | 8.56e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 4 | 123 | 8.66e-02 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 6 | 219 | 8.79e-02 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 8.81e-02 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 5 | 8 | 9.03e-02 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 23 | 88 | 9.03e-02 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 2 | 15 | 9.19e-02 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 8 | 40 | 9.19e-02 |
| GO:BP | GO:0097176 | epoxide metabolic process | 3 | 4 | 9.19e-02 |
| GO:BP | GO:0043928 | exonucleolytic catabolism of deadenylated mRNA | 4 | 13 | 9.26e-02 |
| GO:BP | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process | 10 | 17 | 9.26e-02 |
| GO:BP | GO:0032506 | cytokinetic process | 9 | 39 | 9.26e-02 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 7 | 57 | 9.26e-02 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 5 | 6 | 9.29e-02 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 9.31e-02 |
| GO:BP | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2 | 2 | 9.31e-02 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 4 | 9 | 9.41e-02 |
| GO:BP | GO:1990936 | vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 9.42e-02 |
| GO:BP | GO:1905174 | regulation of vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 9.42e-02 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 9.42e-02 |
| GO:BP | GO:0090678 | cell dedifferentiation involved in phenotypic switching | 2 | 2 | 9.42e-02 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 3 | 44 | 9.42e-02 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 3 | 4 | 9.42e-02 |
| GO:BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 4 | 18 | 9.55e-02 |
| GO:BP | GO:0051095 | regulation of helicase activity | 3 | 12 | 9.62e-02 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 9 | 41 | 9.72e-02 |
| GO:BP | GO:0099179 | regulation of synaptic membrane adhesion | 4 | 4 | 9.73e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 8 | 36 | 1.00e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 11 | 13 | 1.01e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 4 | 131 | 1.01e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 2 | 14 | 1.01e-01 |
| GO:BP | GO:0071772 | response to BMP | 4 | 131 | 1.01e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 1.01e-01 |
| GO:BP | GO:0051795 | positive regulation of timing of catagen | 1 | 3 | 1.01e-01 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 3 | 6 | 1.02e-01 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 3 | 6 | 1.02e-01 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 3 | 5 | 1.02e-01 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 2 | 16 | 1.02e-01 |
| GO:BP | GO:0035284 | brain segmentation | 2 | 2 | 1.03e-01 |
| GO:BP | GO:0021571 | rhombomere 5 development | 2 | 2 | 1.03e-01 |
| GO:BP | GO:0051305 | chromosome movement towards spindle pole | 3 | 7 | 1.03e-01 |
| GO:BP | GO:0051382 | kinetochore assembly | 7 | 18 | 1.03e-01 |
| GO:BP | GO:1903926 | cellular response to bisphenol A | 3 | 4 | 1.03e-01 |
| GO:BP | GO:1903925 | response to bisphenol A | 3 | 4 | 1.03e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 29 | 459 | 1.03e-01 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 6 | 19 | 1.03e-01 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 6 | 19 | 1.03e-01 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 6 | 19 | 1.03e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 16 | 28 | 1.03e-01 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 3 | 64 | 1.03e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 11 | 217 | 1.05e-01 |
| GO:BP | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process | 3 | 7 | 1.05e-01 |
| GO:BP | GO:0071046 | nuclear polyadenylation-dependent ncRNA catabolic process | 3 | 7 | 1.05e-01 |
| GO:BP | GO:0106354 | tRNA surveillance | 3 | 7 | 1.05e-01 |
| GO:BP | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process | 3 | 7 | 1.05e-01 |
| GO:BP | GO:0071029 | nuclear ncRNA surveillance | 3 | 7 | 1.05e-01 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 2 | 3 | 1.05e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 2 | 2 | 1.07e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 34 | 185 | 1.07e-01 |
| GO:BP | GO:0099068 | postsynapse assembly | 6 | 32 | 1.08e-01 |
| GO:BP | GO:0006312 | mitotic recombination | 14 | 22 | 1.08e-01 |
| GO:BP | GO:0000921 | septin ring assembly | 1 | 1 | 1.08e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 12 | 132 | 1.10e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 2 | 10 | 1.11e-01 |
| GO:BP | GO:0038101 | sequestering of nodal from receptor via nodal binding | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 1.12e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 3 | 20 | 1.13e-01 |
| GO:BP | GO:0060039 | pericardium development | 13 | 17 | 1.14e-01 |
| GO:BP | GO:0009439 | cyanate metabolic process | 2 | 2 | 1.14e-01 |
| GO:BP | GO:0009440 | cyanate catabolic process | 2 | 2 | 1.14e-01 |
| GO:BP | GO:0032223 | negative regulation of synaptic transmission, cholinergic | 1 | 1 | 1.15e-01 |
| GO:BP | GO:0046111 | xanthine biosynthetic process | 1 | 1 | 1.17e-01 |
| GO:BP | GO:0060407 | negative regulation of penile erection | 1 | 1 | 1.17e-01 |
| GO:BP | GO:0060169 | negative regulation of adenosine receptor signaling pathway | 1 | 1 | 1.17e-01 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 2 | 18 | 1.19e-01 |
| GO:BP | GO:0042637 | catagen | 1 | 4 | 1.19e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 3 | 51 | 1.19e-01 |
| GO:BP | GO:0071932 | replication fork reversal | 5 | 5 | 1.19e-01 |
| GO:BP | GO:0060364 | frontal suture morphogenesis | 1 | 4 | 1.19e-01 |
| GO:BP | GO:0051794 | regulation of timing of catagen | 1 | 4 | 1.19e-01 |
| GO:BP | GO:0090657 | telomeric loop disassembly | 5 | 9 | 1.22e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 6 | 35 | 1.23e-01 |
| GO:BP | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | 4 | 16 | 1.23e-01 |
| GO:BP | GO:0090222 | centrosome-templated microtubule nucleation | 1 | 2 | 1.23e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 10 | 14 | 1.23e-01 |
| GO:BP | GO:0086017 | Purkinje myocyte action potential | 2 | 4 | 1.23e-01 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 5 | 23 | 1.24e-01 |
| GO:BP | GO:0032060 | bleb assembly | 2 | 9 | 1.24e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 7 | 10 | 1.24e-01 |
| GO:BP | GO:0070266 | necroptotic process | 6 | 32 | 1.24e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 8 | 146 | 1.24e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 7 | 25 | 1.26e-01 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 6 | 13 | 1.28e-01 |
| GO:BP | GO:1900239 | regulation of phenotypic switching | 2 | 3 | 1.30e-01 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 1.31e-01 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 2 | 20 | 1.31e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 40 | 142 | 1.31e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 9 | 64 | 1.31e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 13 | 44 | 1.31e-01 |
| GO:BP | GO:0006543 | glutamine catabolic process | 1 | 3 | 1.31e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 8 | 9 | 1.32e-01 |
| GO:BP | GO:0048818 | positive regulation of hair follicle maturation | 2 | 5 | 1.32e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 6 | 76 | 1.32e-01 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 5 | 10 | 1.34e-01 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 4 | 41 | 1.34e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 6 | 25 | 1.34e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 37 | 206 | 1.34e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 7 | 20 | 1.35e-01 |
| GO:BP | GO:0051418 | microtubule nucleation by microtubule organizing center | 4 | 5 | 1.35e-01 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 14 | 28 | 1.36e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 18 | 25 | 1.37e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 36 | 1.37e-01 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 12 | 37 | 1.37e-01 |
| GO:BP | GO:1900121 | negative regulation of receptor binding | 1 | 8 | 1.38e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 4 | 25 | 1.38e-01 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 1.38e-01 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 3 | 8 | 1.38e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 2 | 6 | 1.38e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 2 | 59 | 1.38e-01 |
| GO:BP | GO:0043634 | polyadenylation-dependent ncRNA catabolic process | 3 | 8 | 1.38e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 7 | 30 | 1.38e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 7 | 9 | 1.39e-01 |
| GO:BP | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 2 | 1.40e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 5 | 48 | 1.40e-01 |
| GO:BP | GO:0044210 | ‘de novo’ CTP biosynthetic process | 1 | 2 | 1.40e-01 |
| GO:BP | GO:0009204 | deoxyribonucleoside triphosphate catabolic process | 5 | 5 | 1.40e-01 |
| GO:BP | GO:0009146 | purine nucleoside triphosphate catabolic process | 5 | 5 | 1.40e-01 |
| GO:BP | GO:0072719 | cellular response to cisplatin | 3 | 4 | 1.40e-01 |
| GO:BP | GO:0106381 | purine deoxyribonucleotide salvage | 1 | 3 | 1.40e-01 |
| GO:BP | GO:0106383 | dAMP salvage | 1 | 3 | 1.40e-01 |
| GO:BP | GO:0006170 | dAMP biosynthetic process | 1 | 3 | 1.40e-01 |
| GO:BP | GO:0009171 | purine deoxyribonucleoside monophosphate biosynthetic process | 1 | 3 | 1.40e-01 |
| GO:BP | GO:0001843 | neural tube closure | 22 | 90 | 1.40e-01 |
| GO:BP | GO:0045007 | depurination | 4 | 4 | 1.40e-01 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 4 | 4 | 1.40e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 4 | 32 | 1.44e-01 |
| GO:BP | GO:0106014 | regulation of inflammatory response to wounding | 2 | 4 | 1.44e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 6 | 14 | 1.44e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 2 | 5 | 1.44e-01 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 24 | 64 | 1.44e-01 |
| GO:BP | GO:0000722 | telomere maintenance via recombination | 9 | 14 | 1.45e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 5 | 31 | 1.45e-01 |
| GO:BP | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway | 3 | 3 | 1.45e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 6 | 11 | 1.45e-01 |
| GO:BP | GO:0043094 | cellular metabolic compound salvage | 2 | 25 | 1.45e-01 |
| GO:BP | GO:0070212 | protein poly-ADP-ribosylation | 2 | 6 | 1.45e-01 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 8 | 18 | 1.45e-01 |
| GO:BP | GO:1905364 | regulation of endosomal vesicle fusion | 1 | 1 | 1.46e-01 |
| GO:BP | GO:0021694 | cerebellar Purkinje cell layer formation | 1 | 12 | 1.46e-01 |
| GO:BP | GO:0035721 | intraciliary retrograde transport | 10 | 14 | 1.46e-01 |
| GO:BP | GO:0046521 | sphingoid catabolic process | 1 | 1 | 1.46e-01 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 2 | 5 | 1.48e-01 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 2 | 5 | 1.48e-01 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 2 | 5 | 1.48e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 1.50e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 2 | 17 | 1.50e-01 |
| GO:BP | GO:0032792 | negative regulation of CREB transcription factor activity | 1 | 4 | 1.50e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 1.50e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 2 | 2 | 1.50e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 15 | 45 | 1.50e-01 |
| GO:BP | GO:0035283 | central nervous system segmentation | 2 | 2 | 1.51e-01 |
| GO:BP | GO:0060606 | tube closure | 22 | 91 | 1.51e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 8 | 86 | 1.51e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 10 | 12 | 1.51e-01 |
| GO:BP | GO:0090215 | regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 4 | 4 | 1.51e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 2 | 64 | 1.52e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 34 | 112 | 1.52e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 1.53e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 1.53e-01 |
| GO:BP | GO:1903379 | regulation of mitotic chromosome condensation | 2 | 2 | 1.53e-01 |
| GO:BP | GO:1905213 | negative regulation of mitotic chromosome condensation | 2 | 2 | 1.53e-01 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 5 | 7 | 1.53e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 8 | 31 | 1.53e-01 |
| GO:BP | GO:1902340 | negative regulation of chromosome condensation | 2 | 2 | 1.53e-01 |
| GO:BP | GO:0021591 | ventricular system development | 13 | 29 | 1.54e-01 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 3 | 7 | 1.54e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 1.55e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 1.55e-01 |
| GO:BP | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1 | 8 | 1.56e-01 |
| GO:BP | GO:0045217 | cell-cell junction maintenance | 1 | 13 | 1.56e-01 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 8 | 21 | 1.56e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 6 | 9 | 1.56e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 4 | 222 | 1.57e-01 |
| GO:BP | GO:0090399 | replicative senescence | 2 | 13 | 1.58e-01 |
| GO:BP | GO:0034473 | U1 snRNA 3’-end processing | 2 | 3 | 1.58e-01 |
| GO:BP | GO:0034476 | U5 snRNA 3’-end processing | 2 | 3 | 1.58e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 2 | 4 | 1.59e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 42 | 96 | 1.59e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 6 | 82 | 1.60e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 1.60e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 3 | 5 | 1.60e-01 |
| GO:BP | GO:0098792 | xenophagy | 10 | 12 | 1.60e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 74 | 1226 | 1.60e-01 |
| GO:BP | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3’-5’ | 4 | 11 | 1.60e-01 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 5 | 19 | 1.60e-01 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 2 | 7 | 1.60e-01 |
| GO:BP | GO:1904172 | positive regulation of bleb assembly | 1 | 1 | 1.60e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 1.60e-01 |
| GO:BP | GO:1904170 | regulation of bleb assembly | 1 | 1 | 1.60e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 4 | 112 | 1.60e-01 |
| GO:BP | GO:0031106 | septin ring organization | 1 | 2 | 1.60e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 35 | 204 | 1.60e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 5 | 15 | 1.60e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 4 | 113 | 1.60e-01 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 18 | 33 | 1.60e-01 |
| GO:BP | GO:0097065 | anterior head development | 4 | 7 | 1.60e-01 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 7 | 9 | 1.60e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 2 | 34 | 1.63e-01 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 5 | 26 | 1.63e-01 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 1 | 16 | 1.63e-01 |
| GO:BP | GO:0060058 | positive regulation of apoptotic process involved in mammary gland involution | 2 | 3 | 1.63e-01 |
| GO:BP | GO:0060057 | apoptotic process involved in mammary gland involution | 2 | 3 | 1.63e-01 |
| GO:BP | GO:1900176 | negative regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0046713 | borate transport | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0000459 | exonucleolytic trimming involved in rRNA processing | 3 | 9 | 1.63e-01 |
| GO:BP | GO:1900146 | negative regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 9 | 1.63e-01 |
| GO:BP | GO:0043633 | polyadenylation-dependent RNA catabolic process | 3 | 9 | 1.63e-01 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 1.63e-01 |
| GO:BP | GO:0035445 | borate transmembrane transport | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 6 | 47 | 1.63e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 27 | 113 | 1.63e-01 |
| GO:BP | GO:0072201 | negative regulation of mesenchymal cell proliferation | 6 | 7 | 1.63e-01 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 10 | 25 | 1.63e-01 |
| GO:BP | GO:0009264 | deoxyribonucleotide catabolic process | 7 | 22 | 1.64e-01 |
| GO:BP | GO:0046386 | deoxyribose phosphate catabolic process | 7 | 22 | 1.64e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 5 | 28 | 1.64e-01 |
| GO:BP | GO:0001507 | acetylcholine catabolic process in synaptic cleft | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0046110 | xanthine metabolic process | 1 | 1 | 1.64e-01 |
| GO:BP | GO:0043103 | hypoxanthine salvage | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 1 | 5 | 1.64e-01 |
| GO:BP | GO:0070256 | negative regulation of mucus secretion | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 11 | 39 | 1.64e-01 |
| GO:BP | GO:0009217 | purine deoxyribonucleoside triphosphate catabolic process | 4 | 4 | 1.64e-01 |
| GO:BP | GO:0046061 | dATP catabolic process | 1 | 2 | 1.64e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 6 | 6 | 1.64e-01 |
| GO:BP | GO:0002636 | positive regulation of germinal center formation | 1 | 1 | 1.64e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 1.64e-01 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 5 | 7 | 1.65e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 5 | 17 | 1.67e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 4 | 20 | 1.67e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 5 | 40 | 1.67e-01 |
| GO:BP | GO:0098530 | positive regulation of strand invasion | 1 | 1 | 1.67e-01 |
| GO:BP | GO:0060542 | regulation of strand invasion | 1 | 1 | 1.67e-01 |
| GO:BP | GO:0035845 | photoreceptor cell outer segment organization | 3 | 10 | 1.67e-01 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 19 | 30 | 1.68e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 6 | 51 | 1.69e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 11 | 16 | 1.69e-01 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 15 | 33 | 1.69e-01 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 2 | 23 | 1.69e-01 |
| GO:BP | GO:0061912 | selective autophagy | 11 | 90 | 1.70e-01 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 3 | 7 | 1.71e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 2 | 3 | 1.71e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 44 | 103 | 1.71e-01 |
| GO:BP | GO:0008592 | regulation of Toll signaling pathway | 3 | 3 | 1.72e-01 |
| GO:BP | GO:0021756 | striatum development | 4 | 17 | 1.73e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 3 | 17 | 1.73e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 7 | 92 | 1.73e-01 |
| GO:BP | GO:0006188 | IMP biosynthetic process | 5 | 10 | 1.76e-01 |
| GO:BP | GO:0072308 | negative regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 1 | 1.76e-01 |
| GO:BP | GO:0044848 | biological phase | 1 | 9 | 1.79e-01 |
| GO:BP | GO:0060157 | urinary bladder development | 2 | 2 | 1.79e-01 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 1 | 8 | 1.79e-01 |
| GO:BP | GO:0009153 | purine deoxyribonucleotide biosynthetic process | 1 | 4 | 1.79e-01 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 4 | 16 | 1.79e-01 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 1.79e-01 |
| GO:BP | GO:0032901 | positive regulation of neurotrophin production | 1 | 1 | 1.79e-01 |
| GO:BP | GO:0072033 | renal vesicle formation | 6 | 8 | 1.79e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 2 | 19 | 1.79e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 8 | 12 | 1.79e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 8 | 9 | 1.79e-01 |
| GO:BP | GO:0045617 | negative regulation of keratinocyte differentiation | 1 | 3 | 1.79e-01 |
| GO:BP | GO:0048819 | regulation of hair follicle maturation | 1 | 9 | 1.79e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 9 | 76 | 1.79e-01 |
| GO:BP | GO:0044851 | hair cycle phase | 1 | 9 | 1.79e-01 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 2 | 8 | 1.79e-01 |
| GO:BP | GO:0032020 | ISG15-protein conjugation | 1 | 6 | 1.79e-01 |
| GO:BP | GO:0060677 | ureteric bud elongation | 7 | 7 | 1.79e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 3 | 5 | 1.81e-01 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 2 | 5 | 1.81e-01 |
| GO:BP | GO:0007144 | female meiosis I | 8 | 9 | 1.81e-01 |
| GO:BP | GO:0006310 | DNA recombination | 78 | 257 | 1.82e-01 |
| GO:BP | GO:0032458 | slow endocytic recycling | 2 | 2 | 1.82e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 6 | 43 | 1.83e-01 |
| GO:BP | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis | 3 | 4 | 1.83e-01 |
| GO:BP | GO:0010288 | response to lead ion | 3 | 18 | 1.84e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 9 | 17 | 1.86e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 4 | 121 | 1.86e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 5 | 7 | 1.88e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 6 | 39 | 1.88e-01 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 7 | 17 | 1.88e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 1 | 17 | 1.88e-01 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 7 | 17 | 1.88e-01 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 7 | 17 | 1.88e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 116 | 452 | 1.88e-01 |
| GO:BP | GO:0006266 | DNA ligation | 9 | 16 | 1.88e-01 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 5 | 6 | 1.88e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 3 | 26 | 1.88e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 1 | 99 | 1.88e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 4 | 28 | 1.91e-01 |
| GO:BP | GO:0060346 | bone trabecula formation | 1 | 9 | 1.91e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 1.91e-01 |
| GO:BP | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic | 8 | 19 | 1.92e-01 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 4 | 14 | 1.92e-01 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 4 | 4 | 1.92e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 16 | 121 | 1.92e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 110 | 189 | 1.92e-01 |
| GO:BP | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell communication | 2 | 6 | 1.92e-01 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 3 | 10 | 1.92e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 9 | 36 | 1.92e-01 |
| GO:BP | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling | 2 | 6 | 1.92e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 4 | 11 | 1.92e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 13 | 186 | 1.92e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 6 | 39 | 1.93e-01 |
| GO:BP | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | 5 | 5 | 1.93e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 14 | 35 | 1.93e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 14 | 77 | 1.93e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 3 | 116 | 1.93e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 1 | 41 | 1.93e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 1 | 22 | 1.93e-01 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 3 | 3 | 1.93e-01 |
| GO:BP | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | 2 | 4 | 1.94e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 1.94e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 3 | 22 | 1.94e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 5 | 47 | 1.95e-01 |
| GO:BP | GO:0021670 | lateral ventricle development | 6 | 11 | 1.95e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 5 | 11 | 1.95e-01 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 2 | 9 | 1.95e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 9 | 38 | 1.96e-01 |
| GO:BP | GO:0036245 | cellular response to menadione | 1 | 2 | 1.97e-01 |
| GO:BP | GO:0099606 | microtubule plus-end directed mitotic chromosome migration | 1 | 1 | 1.97e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 4 | 6 | 1.97e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 4 | 6 | 1.97e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 25 | 134 | 1.97e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 7 | 40 | 1.97e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 4 | 76 | 1.97e-01 |
| GO:BP | GO:0072709 | cellular response to sorbitol | 5 | 5 | 1.98e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 6 | 7 | 1.98e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 4 | 6 | 1.98e-01 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 4 | 191 | 1.98e-01 |
| GO:BP | GO:0033693 | neurofilament bundle assembly | 3 | 3 | 1.98e-01 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 3 | 3 | 1.98e-01 |
| GO:BP | GO:1900054 | positive regulation of retinoic acid biosynthetic process | 1 | 1 | 1.98e-01 |
| GO:BP | GO:0045828 | positive regulation of isoprenoid metabolic process | 1 | 1 | 1.98e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 1.98e-01 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 2 | 23 | 1.98e-01 |
| GO:BP | GO:0035609 | C-terminal protein deglutamylation | 1 | 2 | 1.98e-01 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 1 | 24 | 1.98e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 5 | 152 | 1.98e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 9 | 16 | 1.99e-01 |
| GO:BP | GO:0006581 | acetylcholine catabolic process | 1 | 3 | 2.00e-01 |
| GO:BP | GO:0043096 | purine nucleobase salvage | 1 | 3 | 2.01e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 24 | 144 | 2.01e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 21 | 86 | 2.01e-01 |
| GO:BP | GO:2000233 | negative regulation of rRNA processing | 3 | 3 | 2.01e-01 |
| GO:BP | GO:0008050 | female courtship behavior | 2 | 2 | 2.01e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 2 | 7 | 2.01e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 20 | 34 | 2.01e-01 |
| GO:BP | GO:0006157 | deoxyadenosine catabolic process | 1 | 2 | 2.01e-01 |
| GO:BP | GO:0046090 | deoxyadenosine metabolic process | 1 | 2 | 2.01e-01 |
| GO:BP | GO:0046101 | hypoxanthine biosynthetic process | 1 | 3 | 2.01e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 22 | 94 | 2.01e-01 |
| GO:BP | GO:0046103 | inosine biosynthetic process | 1 | 3 | 2.01e-01 |
| GO:BP | GO:0010165 | response to X-ray | 10 | 29 | 2.01e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 3 | 7 | 2.01e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 10 | 23 | 2.01e-01 |
| GO:BP | GO:0000423 | mitophagy | 6 | 37 | 2.01e-01 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 4 | 8 | 2.01e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 5 | 28 | 2.01e-01 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 2 | 11 | 2.01e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 3 | 22 | 2.01e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 3 | 10 | 2.02e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 27 | 116 | 2.03e-01 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 7 | 19 | 2.03e-01 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 2 | 17 | 2.03e-01 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 7 | 19 | 2.03e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 10 | 13 | 2.03e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 2 | 40 | 2.03e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 2.03e-01 |
| GO:BP | GO:0048642 | negative regulation of skeletal muscle tissue development | 1 | 3 | 2.03e-01 |
| GO:BP | GO:0051798 | positive regulation of hair follicle development | 1 | 9 | 2.03e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 8 | 88 | 2.04e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 8 | 147 | 2.04e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 2 | 214 | 2.04e-01 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 4 | 8 | 2.04e-01 |
| GO:BP | GO:0006273 | lagging strand elongation | 4 | 4 | 2.04e-01 |
| GO:BP | GO:0032077 | positive regulation of deoxyribonuclease activity | 3 | 5 | 2.04e-01 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 4 | 197 | 2.04e-01 |
| GO:BP | GO:0032202 | telomere assembly | 3 | 4 | 2.04e-01 |
| GO:BP | GO:0043622 | cortical microtubule organization | 4 | 4 | 2.04e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 8 | 64 | 2.04e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 5 | 19 | 2.05e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 5 | 19 | 2.05e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 11 | 29 | 2.05e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 10 | 13 | 2.05e-01 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 1 | 7 | 2.05e-01 |
| GO:BP | GO:0035904 | aorta development | 11 | 50 | 2.06e-01 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 19 | 30 | 2.06e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 2 | 17 | 2.06e-01 |
| GO:BP | GO:0061153 | trachea gland development | 2 | 2 | 2.06e-01 |
| GO:BP | GO:0061525 | hindgut development | 2 | 5 | 2.06e-01 |
| GO:BP | GO:0061152 | trachea submucosa development | 2 | 2 | 2.06e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 10 | 16 | 2.06e-01 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 2 | 13 | 2.06e-01 |
| GO:BP | GO:0090656 | t-circle formation | 2 | 13 | 2.06e-01 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 3 | 9 | 2.06e-01 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 2 | 13 | 2.06e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 5 | 69 | 2.06e-01 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 9 | 29 | 2.08e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 4 | 45 | 2.08e-01 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 2 | 5 | 2.08e-01 |
| GO:BP | GO:0035610 | protein side chain deglutamylation | 1 | 2 | 2.08e-01 |
| GO:BP | GO:1903995 | regulation of non-membrane spanning protein tyrosine kinase activity | 3 | 3 | 2.08e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 2.08e-01 |
| GO:BP | GO:0036164 | cell-abiotic substrate adhesion | 1 | 1 | 2.08e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 12 | 25 | 2.08e-01 |
| GO:BP | GO:0070194 | synaptonemal complex disassembly | 1 | 2 | 2.08e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 4 | 19 | 2.08e-01 |
| GO:BP | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus | 2 | 6 | 2.08e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 9 | 11 | 2.08e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 14 | 22 | 2.08e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 2.08e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 2 | 26 | 2.08e-01 |
| GO:BP | GO:1905198 | manchette assembly | 3 | 4 | 2.08e-01 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 13 | 46 | 2.08e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 2 | 2 | 2.08e-01 |
| GO:BP | GO:1902527 | positive regulation of protein monoubiquitination | 5 | 5 | 2.08e-01 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 2 | 6 | 2.08e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 16 | 23 | 2.08e-01 |
| GO:BP | GO:0097012 | response to granulocyte macrophage colony-stimulating factor | 2 | 6 | 2.08e-01 |
| GO:BP | GO:1904976 | cellular response to bleomycin | 2 | 2 | 2.08e-01 |
| GO:BP | GO:0003283 | atrial septum development | 3 | 23 | 2.08e-01 |
| GO:BP | GO:1904975 | response to bleomycin | 2 | 2 | 2.08e-01 |
| GO:BP | GO:0098754 | detoxification | 6 | 91 | 2.08e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 9 | 331 | 2.08e-01 |
| GO:BP | GO:0071047 | polyadenylation-dependent mRNA catabolic process | 2 | 4 | 2.08e-01 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 2 | 5 | 2.08e-01 |
| GO:BP | GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic process | 2 | 4 | 2.08e-01 |
| GO:BP | GO:1901339 | regulation of store-operated calcium channel activity | 4 | 8 | 2.08e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 9 | 107 | 2.08e-01 |
| GO:BP | GO:0048341 | paraxial mesoderm formation | 3 | 6 | 2.08e-01 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 1 | 12 | 2.08e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 6 | 10 | 2.08e-01 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 2 | 10 | 2.08e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 7 | 39 | 2.08e-01 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 2.08e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 2.08e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 15 | 39 | 2.08e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 2.08e-01 |
| GO:BP | GO:0016540 | protein autoprocessing | 7 | 16 | 2.08e-01 |
| GO:BP | GO:0046051 | UTP metabolic process | 9 | 10 | 2.08e-01 |
| GO:BP | GO:1904024 | negative regulation of glucose catabolic process to lactate via pyruvate | 2 | 3 | 2.09e-01 |
| GO:BP | GO:2000200 | regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | 2 | 5 | 2.09e-01 |
| GO:BP | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process | 5 | 15 | 2.09e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 4 | 132 | 2.09e-01 |
| GO:BP | GO:2000206 | regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.09e-01 |
| GO:BP | GO:2000202 | positive regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.09e-01 |
| GO:BP | GO:2000199 | positive regulation of ribonucleoprotein complex localization | 1 | 1 | 2.09e-01 |
| GO:BP | GO:2000208 | positive regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.09e-01 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 2 | 23 | 2.09e-01 |
| GO:BP | GO:0071505 | response to mycophenolic acid | 2 | 2 | 2.09e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 6 | 8 | 2.09e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 11 | 73 | 2.09e-01 |
| GO:BP | GO:0071506 | cellular response to mycophenolic acid | 2 | 2 | 2.09e-01 |
| GO:BP | GO:0032185 | septin cytoskeleton organization | 1 | 4 | 2.09e-01 |
| GO:BP | GO:1901490 | regulation of lymphangiogenesis | 3 | 5 | 2.10e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 1 | 6 | 2.10e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 1 | 6 | 2.10e-01 |
| GO:BP | GO:0044790 | suppression of viral release by host | 3 | 16 | 2.10e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 45 | 479 | 2.10e-01 |
| GO:BP | GO:0000017 | alpha-glucoside transport | 2 | 2 | 2.10e-01 |
| GO:BP | GO:0097694 | establishment of RNA localization to telomere | 3 | 3 | 2.11e-01 |
| GO:BP | GO:0097695 | establishment of protein-containing complex localization to telomere | 3 | 3 | 2.11e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 7 | 14 | 2.11e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 2.11e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 2.11e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 1 | 23 | 2.11e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 3 | 69 | 2.11e-01 |
| GO:BP | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 2 | 2.11e-01 |
| GO:BP | GO:0046398 | UDP-glucuronate metabolic process | 1 | 2 | 2.11e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 9 | 97 | 2.11e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 2 | 11 | 2.11e-01 |
| GO:BP | GO:0071504 | cellular response to heparin | 3 | 4 | 2.13e-01 |
| GO:BP | GO:0097688 | glutamate receptor clustering | 3 | 7 | 2.13e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 5 | 311 | 2.13e-01 |
| GO:BP | GO:0035582 | sequestering of BMP in extracellular matrix | 1 | 2 | 2.13e-01 |
| GO:BP | GO:0061009 | common bile duct development | 3 | 4 | 2.13e-01 |
| GO:BP | GO:0097113 | AMPA glutamate receptor clustering | 3 | 7 | 2.13e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 3 | 16 | 2.13e-01 |
| GO:BP | GO:1900108 | negative regulation of nodal signaling pathway | 1 | 2 | 2.13e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 8 | 425 | 2.13e-01 |
| GO:BP | GO:0072703 | cellular response to methyl methanesulfonate | 1 | 1 | 2.14e-01 |
| GO:BP | GO:0072702 | response to methyl methanesulfonate | 1 | 1 | 2.14e-01 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 4 | 5 | 2.14e-01 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 3 | 3 | 2.14e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 15 | 158 | 2.14e-01 |
| GO:BP | GO:0033212 | iron import into cell | 3 | 7 | 2.14e-01 |
| GO:BP | GO:0045008 | depyrimidination | 6 | 8 | 2.14e-01 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 12 | 29 | 2.15e-01 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 2 | 6 | 2.16e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 1 | 7 | 2.16e-01 |
| GO:BP | GO:0048820 | hair follicle maturation | 1 | 15 | 2.16e-01 |
| GO:BP | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential | 3 | 3 | 2.16e-01 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 2.17e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 10 | 14 | 2.17e-01 |
| GO:BP | GO:0003094 | glomerular filtration | 5 | 21 | 2.17e-01 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 11 | 19 | 2.18e-01 |
| GO:BP | GO:0046059 | dAMP catabolic process | 1 | 2 | 2.18e-01 |
| GO:BP | GO:0046100 | hypoxanthine metabolic process | 1 | 3 | 2.18e-01 |
| GO:BP | GO:0002906 | negative regulation of mature B cell apoptotic process | 1 | 3 | 2.18e-01 |
| GO:BP | GO:0002905 | regulation of mature B cell apoptotic process | 1 | 3 | 2.18e-01 |
| GO:BP | GO:0006154 | adenosine catabolic process | 1 | 3 | 2.18e-01 |
| GO:BP | GO:0002901 | mature B cell apoptotic process | 1 | 3 | 2.18e-01 |
| GO:BP | GO:0046124 | purine deoxyribonucleoside catabolic process | 1 | 2 | 2.18e-01 |
| GO:BP | GO:0060167 | regulation of adenosine receptor signaling pathway | 1 | 2 | 2.18e-01 |
| GO:BP | GO:0033197 | response to vitamin E | 2 | 9 | 2.18e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 4 | 21 | 2.19e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 9 | 77 | 2.19e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 5 | 69 | 2.19e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4 | 230 | 2.20e-01 |
| GO:BP | GO:0035608 | protein deglutamylation | 1 | 2 | 2.20e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 7 | 40 | 2.20e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 8 | 29 | 2.20e-01 |
| GO:BP | GO:0035825 | homologous recombination | 15 | 43 | 2.20e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 31 | 50 | 2.21e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 5 | 13 | 2.21e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 5 | 16 | 2.21e-01 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 3 | 8 | 2.21e-01 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 3 | 13 | 2.21e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 8 | 91 | 2.21e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 5 | 319 | 2.21e-01 |
| GO:BP | GO:0021685 | cerebellar granular layer structural organization | 1 | 1 | 2.21e-01 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 3 | 8 | 2.21e-01 |
| GO:BP | GO:0043174 | nucleoside salvage | 1 | 9 | 2.21e-01 |
| GO:BP | GO:0061146 | Peyer’s patch morphogenesis | 3 | 3 | 2.21e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 4 | 40 | 2.21e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 2 | 7 | 2.21e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 6 | 10 | 2.21e-01 |
| GO:BP | GO:0000301 | retrograde transport, vesicle recycling within Golgi | 7 | 9 | 2.21e-01 |
| GO:BP | GO:0090179 | planar cell polarity pathway involved in neural tube closure | 4 | 12 | 2.21e-01 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 1 | 10 | 2.22e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 5 | 9 | 2.23e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 4 | 11 | 2.24e-01 |
| GO:BP | GO:0032898 | neurotrophin production | 3 | 4 | 2.24e-01 |
| GO:BP | GO:0050893 | sensory processing | 4 | 4 | 2.24e-01 |
| GO:BP | GO:1903416 | response to glycoside | 3 | 6 | 2.25e-01 |
| GO:BP | GO:0016236 | macroautophagy | 105 | 308 | 2.30e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 2.30e-01 |
| GO:BP | GO:0055057 | neuroblast division | 5 | 12 | 2.30e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 2.30e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 2.30e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 10 | 19 | 2.30e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 2.30e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 4 | 28 | 2.31e-01 |
| GO:BP | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 2.31e-01 |
| GO:BP | GO:0046725 | negative regulation by virus of viral protein levels in host cell | 1 | 2 | 2.31e-01 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 2.31e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 2 | 48 | 2.31e-01 |
| GO:BP | GO:0060067 | cervix development | 2 | 2 | 2.31e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 3 | 17 | 2.31e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 5 | 41 | 2.32e-01 |
| GO:BP | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process | 4 | 6 | 2.33e-01 |
| GO:BP | GO:0006498 | N-terminal protein lipidation | 5 | 7 | 2.33e-01 |
| GO:BP | GO:0010225 | response to UV-C | 8 | 13 | 2.33e-01 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 5 | 6 | 2.33e-01 |
| GO:BP | GO:0060013 | righting reflex | 2 | 6 | 2.33e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 32 | 875 | 2.33e-01 |
| GO:BP | GO:0030903 | notochord development | 10 | 17 | 2.34e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 2.34e-01 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 1 | 9 | 2.35e-01 |
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 2.35e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 8 | 94 | 2.35e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 8 | 94 | 2.35e-01 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 5 | 10 | 2.35e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 5 | 6 | 2.35e-01 |
| GO:BP | GO:1901207 | regulation of heart looping | 2 | 2 | 2.35e-01 |
| GO:BP | GO:1903546 | protein localization to photoreceptor outer segment | 2 | 5 | 2.35e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 10 | 158 | 2.35e-01 |
| GO:BP | GO:0075071 | modulation by symbiont of host autophagy | 1 | 1 | 2.36e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 14 | 103 | 2.36e-01 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 10 | 38 | 2.36e-01 |
| GO:BP | GO:0140321 | suppression of host autophagy | 1 | 1 | 2.36e-01 |
| GO:BP | GO:0039521 | suppression by virus of host autophagy | 1 | 1 | 2.36e-01 |
| GO:BP | GO:1905672 | negative regulation of lysosome organization | 1 | 1 | 2.36e-01 |
| GO:BP | GO:0071275 | cellular response to aluminum ion | 1 | 1 | 2.36e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 2.36e-01 |
| GO:BP | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 2 | 2.36e-01 |
| GO:BP | GO:1900175 | regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 2 | 2.36e-01 |
| GO:BP | GO:1904550 | response to arachidonic acid | 1 | 1 | 2.36e-01 |
| GO:BP | GO:0097719 | neural tissue regeneration | 1 | 1 | 2.37e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 8 | 159 | 2.37e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 3 | 26 | 2.37e-01 |
| GO:BP | GO:0045760 | positive regulation of action potential | 2 | 4 | 2.37e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 6 | 27 | 2.37e-01 |
| GO:BP | GO:0051653 | spindle localization | 11 | 52 | 2.37e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 11 | 27 | 2.37e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 1 | 29 | 2.37e-01 |
| GO:BP | GO:1902608 | positive regulation of large conductance calcium-activated potassium channel activity | 1 | 2 | 2.37e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 11 | 49 | 2.37e-01 |
| GO:BP | GO:0090663 | galanin-activated signaling pathway | 1 | 1 | 2.37e-01 |
| GO:BP | GO:0060080 | inhibitory postsynaptic potential | 1 | 8 | 2.37e-01 |
| GO:BP | GO:0003360 | brainstem development | 2 | 7 | 2.37e-01 |
| GO:BP | GO:0015750 | pentose transmembrane transport | 2 | 2 | 2.37e-01 |
| GO:BP | GO:0015756 | fucose transmembrane transport | 2 | 2 | 2.37e-01 |
| GO:BP | GO:0031291 | Ran protein signal transduction | 2 | 3 | 2.37e-01 |
| GO:BP | GO:1903714 | isoleucine transmembrane transport | 1 | 1 | 2.37e-01 |
| GO:BP | GO:1903806 | L-isoleucine import across plasma membrane | 1 | 1 | 2.37e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 2 | 19 | 2.38e-01 |
| GO:BP | GO:1901859 | negative regulation of mitochondrial DNA metabolic process | 2 | 2 | 2.38e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 2.39e-01 |
| GO:BP | GO:0090031 | positive regulation of steroid hormone biosynthetic process | 5 | 5 | 2.39e-01 |
| GO:BP | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation | 2 | 5 | 2.39e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 10 | 34 | 2.39e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 2 | 5 | 2.39e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 2 | 5 | 2.39e-01 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 1 | 15 | 2.39e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 14 | 26 | 2.39e-01 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 2.39e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 10 | 68 | 2.39e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 7 | 47 | 2.39e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 3 | 13 | 2.40e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 6 | 11 | 2.41e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 1 | 226 | 2.41e-01 |
| GO:BP | GO:0046102 | inosine metabolic process | 1 | 4 | 2.41e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 5 | 8 | 2.41e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 5 | 8 | 2.41e-01 |
| GO:BP | GO:0009445 | putrescine metabolic process | 2 | 6 | 2.41e-01 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 7 | 11 | 2.41e-01 |
| GO:BP | GO:0006196 | AMP catabolic process | 1 | 3 | 2.41e-01 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 9 | 15 | 2.41e-01 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 12 | 16 | 2.41e-01 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 1 | 12 | 2.41e-01 |
| GO:BP | GO:0046122 | purine deoxyribonucleoside metabolic process | 1 | 3 | 2.41e-01 |
| GO:BP | GO:1990029 | vasomotion | 1 | 1 | 2.41e-01 |
| GO:BP | GO:0070997 | neuron death | 20 | 286 | 2.41e-01 |
| GO:BP | GO:0086044 | atrial cardiac muscle cell to AV node cell communication by electrical coupling | 1 | 1 | 2.41e-01 |
| GO:BP | GO:0086055 | Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling | 1 | 1 | 2.41e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 12 | 75 | 2.41e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 3 | 14 | 2.41e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 130 | 234 | 2.41e-01 |
| GO:BP | GO:0010652 | positive regulation of cell communication by chemical coupling | 1 | 1 | 2.41e-01 |
| GO:BP | GO:0009116 | nucleoside metabolic process | 2 | 42 | 2.41e-01 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 2 | 13 | 2.41e-01 |
| GO:BP | GO:0051152 | positive regulation of smooth muscle cell differentiation | 3 | 10 | 2.41e-01 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 2 | 13 | 2.41e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 1 | 33 | 2.41e-01 |
| GO:BP | GO:0098906 | regulation of Purkinje myocyte action potential | 1 | 1 | 2.41e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 3 | 19 | 2.41e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 2 | 22 | 2.41e-01 |
| GO:BP | GO:0010645 | regulation of cell communication by chemical coupling | 1 | 1 | 2.41e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 2 | 2 | 2.42e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 6 | 110 | 2.42e-01 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 1 | 16 | 2.42e-01 |
| GO:BP | GO:0021546 | rhombomere development | 2 | 4 | 2.42e-01 |
| GO:BP | GO:1904997 | regulation of leukocyte adhesion to arterial endothelial cell | 3 | 3 | 2.42e-01 |
| GO:BP | GO:0097205 | renal filtration | 5 | 23 | 2.42e-01 |
| GO:BP | GO:0071051 | polyadenylation-dependent snoRNA 3’-end processing | 4 | 7 | 2.42e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 3 | 440 | 2.42e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 2.42e-01 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 4 | 11 | 2.42e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 2 | 8 | 2.43e-01 |
| GO:BP | GO:1905462 | regulation of DNA duplex unwinding | 3 | 7 | 2.44e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 2.44e-01 |
| GO:BP | GO:0043111 | replication fork arrest | 2 | 2 | 2.44e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 2 | 53 | 2.44e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 4 | 48 | 2.44e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 2 | 35 | 2.45e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 4 | 42 | 2.45e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 5 | 74 | 2.45e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 32 | 101 | 2.45e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 8 | 161 | 2.45e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 13 | 55 | 2.45e-01 |
| GO:BP | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway | 4 | 5 | 2.45e-01 |
| GO:BP | GO:0021544 | subpallium development | 4 | 18 | 2.45e-01 |
| GO:BP | GO:0019606 | 2-oxobutyrate catabolic process | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 3 | 17 | 2.45e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 11 | 35 | 2.45e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 18 | 67 | 2.45e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 2.46e-01 |
| GO:BP | GO:1904979 | negative regulation of endosome organization | 2 | 2 | 2.46e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 15 | 378 | 2.46e-01 |
| GO:BP | GO:1905365 | regulation of intralumenal vesicle formation | 2 | 2 | 2.46e-01 |
| GO:BP | GO:1905366 | negative regulation of intralumenal vesicle formation | 2 | 2 | 2.46e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 6 | 45 | 2.46e-01 |
| GO:BP | GO:0019856 | pyrimidine nucleobase biosynthetic process | 1 | 8 | 2.46e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 2.46e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 4 | 22 | 2.47e-01 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 13 | 26 | 2.48e-01 |
| GO:BP | GO:0060840 | artery development | 16 | 83 | 2.48e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 4 | 28 | 2.48e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 2.48e-01 |
| GO:BP | GO:0019276 | UDP-N-acetylgalactosamine metabolic process | 1 | 2 | 2.48e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 3 | 80 | 2.48e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 11 | 119 | 2.48e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 18 | 138 | 2.48e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 4 | 6 | 2.48e-01 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 3 | 5 | 2.49e-01 |
| GO:BP | GO:0071307 | cellular response to vitamin K | 1 | 4 | 2.49e-01 |
| GO:BP | GO:0002138 | retinoic acid biosynthetic process | 2 | 8 | 2.49e-01 |
| GO:BP | GO:0043650 | dicarboxylic acid biosynthetic process | 1 | 9 | 2.49e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 2 | 284 | 2.49e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 15 | 381 | 2.49e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 7 | 56 | 2.49e-01 |
| GO:BP | GO:2000821 | regulation of grooming behavior | 2 | 3 | 2.49e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 3 | 18 | 2.49e-01 |
| GO:BP | GO:0006655 | phosphatidylglycerol biosynthetic process | 8 | 9 | 2.49e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 4 | 98 | 2.49e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 14 | 358 | 2.49e-01 |
| GO:BP | GO:0000303 | response to superoxide | 5 | 21 | 2.49e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 4 | 7 | 2.49e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 6 | 14 | 2.49e-01 |
| GO:BP | GO:0071468 | cellular response to acidic pH | 3 | 10 | 2.49e-01 |
| GO:BP | GO:0070166 | enamel mineralization | 1 | 11 | 2.49e-01 |
| GO:BP | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process | 3 | 3 | 2.49e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 10 | 23 | 2.49e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 11 | 173 | 2.49e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 5 | 45 | 2.49e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 5 | 69 | 2.49e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 4 | 13 | 2.49e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 5 | 28 | 2.49e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 24 | 56 | 2.49e-01 |
| GO:BP | GO:0090217 | negative regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 3 | 3 | 2.49e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 5 | 26 | 2.49e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 4 | 12 | 2.49e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 5 | 9 | 2.49e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 4 | 22 | 2.49e-01 |
| GO:BP | GO:0003336 | corneocyte desquamation | 1 | 1 | 2.49e-01 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 3 | 23 | 2.49e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 6 | 35 | 2.49e-01 |
| GO:BP | GO:0043157 | response to cation stress | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0042594 | response to starvation | 4 | 178 | 2.49e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 1 | 34 | 2.49e-01 |
| GO:BP | GO:0071580 | regulation of zinc ion transmembrane transport | 1 | 1 | 2.49e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 4 | 12 | 2.49e-01 |
| GO:BP | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process | 3 | 3 | 2.49e-01 |
| GO:BP | GO:0071581 | regulation of zinc ion transmembrane import | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0071582 | negative regulation of zinc ion transport | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0071583 | negative regulation of zinc ion transmembrane transport | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0071584 | negative regulation of zinc ion transmembrane import | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0072718 | response to cisplatin | 3 | 6 | 2.49e-01 |
| GO:BP | GO:0061349 | planar cell polarity pathway involved in cardiac right atrium morphogenesis | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0061350 | planar cell polarity pathway involved in cardiac muscle tissue morphogenesis | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0032200 | telomere organization | 43 | 166 | 2.49e-01 |
| GO:BP | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 3 | 3 | 2.49e-01 |
| GO:BP | GO:0051105 | regulation of DNA ligation | 4 | 6 | 2.49e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 28 | 127 | 2.49e-01 |
| GO:BP | GO:0061348 | planar cell polarity pathway involved in ventricular septum morphogenesis | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 3 | 11 | 2.49e-01 |
| GO:BP | GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0061346 | planar cell polarity pathway involved in heart morphogenesis | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 2.49e-01 |
| GO:BP | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration | 3 | 6 | 2.49e-01 |
| GO:BP | GO:0033082 | regulation of extrathymic T cell differentiation | 1 | 1 | 2.49e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 11 | 27 | 2.49e-01 |
| GO:BP | GO:1904934 | negative regulation of cell proliferation in midbrain | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0036517 | chemoattraction of serotonergic neuron axon | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 11 | 27 | 2.49e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 4 | 42 | 2.49e-01 |
| GO:BP | GO:0061354 | planar cell polarity pathway involved in pericardium morphogenesis | 1 | 1 | 2.49e-01 |
| GO:BP | GO:0051642 | centrosome localization | 21 | 31 | 2.50e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 21 | 31 | 2.50e-01 |
| GO:BP | GO:0046130 | purine ribonucleoside catabolic process | 1 | 5 | 2.50e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 5 | 36 | 2.50e-01 |
| GO:BP | GO:0046121 | deoxyribonucleoside catabolic process | 1 | 4 | 2.50e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 8 | 15 | 2.50e-01 |
| GO:BP | GO:0021599 | abducens nerve formation | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0021598 | abducens nerve morphogenesis | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0021572 | rhombomere 6 development | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0021560 | abducens nerve development | 1 | 1 | 2.50e-01 |
| GO:BP | GO:0009172 | purine deoxyribonucleoside monophosphate catabolic process | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 10 | 28 | 2.50e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 10 | 19 | 2.50e-01 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 10 | 19 | 2.51e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 3 | 69 | 2.51e-01 |
| GO:BP | GO:2000431 | regulation of cytokinesis, actomyosin contractile ring assembly | 1 | 1 | 2.51e-01 |
| GO:BP | GO:0060914 | heart formation | 12 | 29 | 2.52e-01 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 9 | 18 | 2.52e-01 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 5 | 7 | 2.52e-01 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 2.53e-01 |
| GO:BP | GO:0035641 | locomotory exploration behavior | 3 | 11 | 2.53e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 23 | 262 | 2.53e-01 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 3 | 12 | 2.53e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 2 | 14 | 2.53e-01 |
| GO:BP | GO:0072034 | renal vesicle induction | 3 | 3 | 2.53e-01 |
| GO:BP | GO:0097491 | sympathetic neuron projection guidance | 4 | 4 | 2.53e-01 |
| GO:BP | GO:0097490 | sympathetic neuron projection extension | 4 | 4 | 2.53e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 3 | 15 | 2.53e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 26 | 464 | 2.53e-01 |
| GO:BP | GO:0021768 | nucleus accumbens development | 1 | 1 | 2.54e-01 |
| GO:BP | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane | 1 | 3 | 2.57e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 9 | 12 | 2.57e-01 |
| GO:BP | GO:0045200 | establishment of neuroblast polarity | 1 | 3 | 2.57e-01 |
| GO:BP | GO:0045196 | establishment or maintenance of neuroblast polarity | 1 | 3 | 2.57e-01 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 13 | 16 | 2.57e-01 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 13 | 16 | 2.57e-01 |
| GO:BP | GO:1903423 | positive regulation of synaptic vesicle recycling | 6 | 6 | 2.57e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 4 | 24 | 2.57e-01 |
| GO:BP | GO:0009447 | putrescine catabolic process | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 3 | 3 | 2.57e-01 |
| GO:BP | GO:0032918 | spermidine acetylation | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0032917 | polyamine acetylation | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0072092 | ureteric bud invasion | 3 | 3 | 2.57e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 4 | 4 | 2.57e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 4 | 4 | 2.57e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 28 | 64 | 2.57e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 4 | 4 | 2.57e-01 |
| GO:BP | GO:1990646 | cellular response to prolactin | 2 | 2 | 2.57e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 4 | 265 | 2.57e-01 |
| GO:BP | GO:1905938 | positive regulation of germ cell proliferation | 2 | 2 | 2.57e-01 |
| GO:BP | GO:2000256 | positive regulation of male germ cell proliferation | 2 | 2 | 2.57e-01 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 4 | 12 | 2.58e-01 |
| GO:BP | GO:0036309 | protein localization to M-band | 2 | 2 | 2.58e-01 |
| GO:BP | GO:1901753 | leukotriene A4 biosynthetic process | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 9 | 279 | 2.58e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 11 | 21 | 2.58e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 16 | 868 | 2.58e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 2 | 17 | 2.58e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 27 | 46 | 2.58e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 1 | 23 | 2.58e-01 |
| GO:BP | GO:0046040 | IMP metabolic process | 1 | 15 | 2.58e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 3 | 67 | 2.58e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 18 | 27 | 2.58e-01 |
| GO:BP | GO:0071694 | maintenance of protein location in extracellular region | 1 | 4 | 2.58e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 2.58e-01 |
| GO:BP | GO:1903780 | negative regulation of cardiac conduction | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 3 | 23 | 2.58e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 2.58e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 3 | 31 | 2.58e-01 |
| GO:BP | GO:0010651 | negative regulation of cell communication by electrical coupling | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 2 | 2 | 2.58e-01 |
| GO:BP | GO:1901845 | negative regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 1 | 246 | 2.58e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 5 | 277 | 2.58e-01 |
| GO:BP | GO:0034755 | iron ion transmembrane transport | 4 | 16 | 2.58e-01 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 4 | 18 | 2.59e-01 |
| GO:BP | GO:0044806 | G-quadruplex DNA unwinding | 4 | 7 | 2.59e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 8 | 57 | 2.59e-01 |
| GO:BP | GO:0021773 | striatal medium spiny neuron differentiation | 2 | 3 | 2.59e-01 |
| GO:BP | GO:0072135 | kidney mesenchymal cell proliferation | 1 | 2 | 2.60e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 13 | 22 | 2.60e-01 |
| GO:BP | GO:0072136 | metanephric mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 2.60e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 4 | 33 | 2.60e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 5 | 22 | 2.60e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 3 | 21 | 2.60e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 2.60e-01 |
| GO:BP | GO:0071288 | cellular response to mercury ion | 2 | 2 | 2.60e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 4 | 240 | 2.60e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 2.61e-01 |
| GO:BP | GO:0016102 | diterpenoid biosynthetic process | 2 | 8 | 2.61e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 2 | 8 | 2.61e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 32 | 111 | 2.61e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 12 | 93 | 2.61e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 10 | 48 | 2.61e-01 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 8 | 13 | 2.61e-01 |
| GO:BP | GO:1900619 | acetate ester metabolic process | 1 | 3 | 2.61e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 2 | 20 | 2.61e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 2 | 20 | 2.61e-01 |
| GO:BP | GO:0009159 | deoxyribonucleoside monophosphate catabolic process | 2 | 8 | 2.61e-01 |
| GO:BP | GO:0008291 | acetylcholine metabolic process | 1 | 3 | 2.61e-01 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 2 | 4 | 2.61e-01 |
| GO:BP | GO:0019660 | glycolytic fermentation | 2 | 4 | 2.61e-01 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 2 | 4 | 2.61e-01 |
| GO:BP | GO:1904234 | positive regulation of aconitate hydratase activity | 2 | 2 | 2.61e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 17 | 65 | 2.61e-01 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 2 | 4 | 2.61e-01 |
| GO:BP | GO:1901003 | negative regulation of fermentation | 2 | 4 | 2.61e-01 |
| GO:BP | GO:1904232 | regulation of aconitate hydratase activity | 2 | 2 | 2.61e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 2.61e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 2.62e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 5 | 20 | 2.62e-01 |
| GO:BP | GO:0003273 | cell migration involved in endocardial cushion formation | 3 | 4 | 2.62e-01 |
| GO:BP | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway | 1 | 3 | 2.62e-01 |
| GO:BP | GO:0014034 | neural crest cell fate commitment | 3 | 4 | 2.62e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 6 | 59 | 2.62e-01 |
| GO:BP | GO:1905340 | regulation of protein localization to kinetochore | 3 | 4 | 2.62e-01 |
| GO:BP | GO:1905342 | positive regulation of protein localization to kinetochore | 3 | 4 | 2.62e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 4 | 241 | 2.62e-01 |
| GO:BP | GO:0071895 | odontoblast differentiation | 2 | 5 | 2.62e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 2.62e-01 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 6 | 29 | 2.62e-01 |
| GO:BP | GO:0001841 | neural tube formation | 22 | 101 | 2.62e-01 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 5 | 9 | 2.62e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 72 | 265 | 2.62e-01 |
| GO:BP | GO:0061485 | memory T cell proliferation | 1 | 1 | 2.62e-01 |
| GO:BP | GO:0060405 | regulation of penile erection | 1 | 4 | 2.62e-01 |
| GO:BP | GO:0032899 | regulation of neurotrophin production | 1 | 2 | 2.62e-01 |
| GO:BP | GO:0016078 | tRNA catabolic process | 3 | 14 | 2.63e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 2 | 12 | 2.63e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 25 | 393 | 2.63e-01 |
| GO:BP | GO:0014886 | transition between slow and fast fiber | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0097114 | NMDA glutamate receptor clustering | 2 | 5 | 2.64e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 3 | 30 | 2.64e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 4 | 50 | 2.64e-01 |
| GO:BP | GO:1901594 | response to capsazepine | 1 | 1 | 2.64e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 6 | 38 | 2.65e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 10 | 138 | 2.65e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 7 | 252 | 2.65e-01 |
| GO:BP | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity | 1 | 3 | 2.65e-01 |
| GO:BP | GO:0003032 | detection of oxygen | 2 | 5 | 2.65e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 4 | 72 | 2.66e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 3 | 20 | 2.66e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 7 | 48 | 2.66e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 5 | 29 | 2.66e-01 |
| GO:BP | GO:0030592 | DNA ADP-ribosylation | 2 | 3 | 2.66e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 18 | 28 | 2.66e-01 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 2 | 11 | 2.66e-01 |
| GO:BP | GO:0097186 | amelogenesis | 1 | 14 | 2.66e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 2.66e-01 |
| GO:BP | GO:0046168 | glycerol-3-phosphate catabolic process | 2 | 2 | 2.66e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 2.66e-01 |
| GO:BP | GO:0006265 | DNA topological change | 1 | 10 | 2.66e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 5 | 82 | 2.66e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 1 | 30 | 2.66e-01 |
| GO:BP | GO:2001170 | negative regulation of ATP biosynthetic process | 2 | 3 | 2.66e-01 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 3 | 4 | 2.66e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 10 | 71 | 2.66e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 4 | 18 | 2.67e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 2.68e-01 |
| GO:BP | GO:0006722 | triterpenoid metabolic process | 2 | 2 | 2.68e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 2.68e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 4 | 14 | 2.68e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 10 | 102 | 2.68e-01 |
| GO:BP | GO:2000521 | negative regulation of immunological synapse formation | 1 | 1 | 2.68e-01 |
| GO:BP | GO:2001189 | negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 1 | 2.68e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 6 | 28 | 2.68e-01 |
| GO:BP | GO:0060047 | heart contraction | 12 | 205 | 2.68e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 7 | 22 | 2.68e-01 |
| GO:BP | GO:0043201 | response to leucine | 3 | 13 | 2.69e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 15 | 387 | 2.70e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 4 | 8 | 2.70e-01 |
| GO:BP | GO:0090132 | epithelium migration | 7 | 252 | 2.70e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 2.70e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 2.70e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 14 | 187 | 2.70e-01 |
| GO:BP | GO:1900060 | negative regulation of ceramide biosynthetic process | 1 | 5 | 2.70e-01 |
| GO:BP | GO:1905463 | negative regulation of DNA duplex unwinding | 2 | 4 | 2.70e-01 |
| GO:BP | GO:1900107 | regulation of nodal signaling pathway | 1 | 3 | 2.70e-01 |
| GO:BP | GO:0006400 | tRNA modification | 34 | 90 | 2.70e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 2.70e-01 |
| GO:BP | GO:0031441 | negative regulation of mRNA 3’-end processing | 2 | 2 | 2.70e-01 |
| GO:BP | GO:0048854 | brain morphogenesis | 13 | 28 | 2.71e-01 |
| GO:BP | GO:0000296 | spermine transport | 1 | 3 | 2.71e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 30 | 300 | 2.71e-01 |
| GO:BP | GO:1904613 | cellular response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 2.71e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 12 | 58 | 2.71e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 7 | 56 | 2.71e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 2 | 32 | 2.71e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 5 | 89 | 2.71e-01 |
| GO:BP | GO:1904717 | regulation of AMPA glutamate receptor clustering | 2 | 4 | 2.72e-01 |
| GO:BP | GO:0106104 | regulation of glutamate receptor clustering | 2 | 4 | 2.72e-01 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 19 | 30 | 2.72e-01 |
| GO:BP | GO:0000425 | pexophagy | 3 | 6 | 2.72e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 16 | 23 | 2.72e-01 |
| GO:BP | GO:1903551 | regulation of extracellular exosome assembly | 3 | 6 | 2.72e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 2.72e-01 |
| GO:BP | GO:0006206 | pyrimidine nucleobase metabolic process | 3 | 13 | 2.73e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 7 | 17 | 2.73e-01 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 2 | 9 | 2.73e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 5 | 11 | 2.74e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 6 | 101 | 2.74e-01 |
| GO:BP | GO:0009174 | pyrimidine ribonucleoside monophosphate biosynthetic process | 4 | 9 | 2.74e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 3 | 4 | 2.74e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 7 | 50 | 2.74e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 7 | 50 | 2.74e-01 |
| GO:BP | GO:0032222 | regulation of synaptic transmission, cholinergic | 1 | 3 | 2.75e-01 |
| GO:BP | GO:0022614 | membrane to membrane docking | 3 | 5 | 2.75e-01 |
| GO:BP | GO:0009158 | ribonucleoside monophosphate catabolic process | 2 | 10 | 2.75e-01 |
| GO:BP | GO:0043063 | intercellular bridge organization | 1 | 1 | 2.76e-01 |
| GO:BP | GO:0006228 | UTP biosynthetic process | 5 | 8 | 2.76e-01 |
| GO:BP | GO:0002313 | mature B cell differentiation involved in immune response | 3 | 17 | 2.76e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 2.76e-01 |
| GO:BP | GO:0019240 | citrulline biosynthetic process | 4 | 4 | 2.76e-01 |
| GO:BP | GO:0006152 | purine nucleoside catabolic process | 1 | 6 | 2.76e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 10 | 140 | 2.76e-01 |
| GO:BP | GO:0070255 | regulation of mucus secretion | 1 | 6 | 2.76e-01 |
| GO:BP | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process | 2 | 5 | 2.76e-01 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 6 | 22 | 2.76e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 20 | 156 | 2.76e-01 |
| GO:BP | GO:0098735 | positive regulation of the force of heart contraction | 2 | 4 | 2.76e-01 |
| GO:BP | GO:0002634 | regulation of germinal center formation | 1 | 6 | 2.76e-01 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 3 | 2.76e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 3 | 76 | 2.76e-01 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 2.76e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 2.76e-01 |
| GO:BP | GO:0044878 | mitotic cytokinesis checkpoint signaling | 2 | 4 | 2.76e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 3 | 21 | 2.76e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 2 | 8 | 2.76e-01 |
| GO:BP | GO:0042245 | RNA repair | 2 | 2 | 2.76e-01 |
| GO:BP | GO:1904716 | positive regulation of chaperone-mediated autophagy | 1 | 1 | 2.76e-01 |
| GO:BP | GO:0030011 | maintenance of cell polarity | 3 | 16 | 2.76e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 20 | 326 | 2.76e-01 |
| GO:BP | GO:0046391 | 5-phosphoribose 1-diphosphate metabolic process | 2 | 5 | 2.76e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 2.76e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 13 | 21 | 2.76e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 2 | 18 | 2.76e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 14 | 45 | 2.76e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 3 | 3 | 2.76e-01 |
| GO:BP | GO:0098849 | cellular detoxification of cadmium ion | 1 | 1 | 2.76e-01 |
| GO:BP | GO:0018410 | C-terminal protein amino acid modification | 1 | 10 | 2.76e-01 |
| GO:BP | GO:0046073 | dTMP metabolic process | 3 | 7 | 2.76e-01 |
| GO:BP | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis | 1 | 1 | 2.76e-01 |
| GO:BP | GO:0140961 | cellular detoxification of metal ion | 1 | 1 | 2.76e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 2 | 11 | 2.76e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 11 | 114 | 2.76e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 7 | 53 | 2.76e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 2 | 35 | 2.77e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 26 | 81 | 2.77e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 8 | 186 | 2.78e-01 |
| GO:BP | GO:0090130 | tissue migration | 7 | 256 | 2.79e-01 |
| GO:BP | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 3 | 3 | 2.79e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 18 | 123 | 2.79e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 5 | 9 | 2.79e-01 |
| GO:BP | GO:0032938 | negative regulation of translation in response to oxidative stress | 1 | 1 | 2.79e-01 |
| GO:BP | GO:0043556 | regulation of translation in response to oxidative stress | 1 | 1 | 2.79e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 8 | 16 | 2.79e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 13 | 24 | 2.79e-01 |
| GO:BP | GO:0021693 | cerebellar Purkinje cell layer structural organization | 1 | 2 | 2.79e-01 |
| GO:BP | GO:0021698 | cerebellar cortex structural organization | 1 | 2 | 2.79e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 2.79e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 2.79e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 2.79e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 2.79e-01 |
| GO:BP | GO:0006244 | pyrimidine nucleotide catabolic process | 5 | 19 | 2.80e-01 |
| GO:BP | GO:1902729 | negative regulation of proteoglycan biosynthetic process | 1 | 1 | 2.81e-01 |
| GO:BP | GO:0032683 | negative regulation of connective tissue growth factor production | 1 | 1 | 2.81e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 4 | 14 | 2.81e-01 |
| GO:BP | GO:0060031 | mediolateral intercalation | 2 | 2 | 2.81e-01 |
| GO:BP | GO:0002157 | positive regulation of thyroid hormone mediated signaling pathway | 2 | 3 | 2.81e-01 |
| GO:BP | GO:0099526 | presynapse to nucleus signaling pathway | 1 | 2 | 2.81e-01 |
| GO:BP | GO:0060775 | planar cell polarity pathway involved in gastrula mediolateral intercalation | 2 | 2 | 2.81e-01 |
| GO:BP | GO:0090272 | negative regulation of fibroblast growth factor production | 2 | 2 | 2.81e-01 |
| GO:BP | GO:0098928 | presynaptic signal transduction | 1 | 2 | 2.81e-01 |
| GO:BP | GO:0007619 | courtship behavior | 2 | 3 | 2.81e-01 |
| GO:BP | GO:0036376 | sodium ion export across plasma membrane | 2 | 12 | 2.81e-01 |
| GO:BP | GO:0001100 | negative regulation of exit from mitosis | 1 | 1 | 2.82e-01 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 1 | 4 | 2.82e-01 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 3 | 5 | 2.82e-01 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 7 | 12 | 2.82e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 9 | 16 | 2.82e-01 |
| GO:BP | GO:0006432 | phenylalanyl-tRNA aminoacylation | 4 | 4 | 2.82e-01 |
| GO:BP | GO:0003431 | growth plate cartilage chondrocyte development | 3 | 5 | 2.82e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 2.83e-01 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 10 | 13 | 2.83e-01 |
| GO:BP | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery | 5 | 6 | 2.83e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 2.83e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 2.83e-01 |
| GO:BP | GO:0006404 | RNA import into nucleus | 4 | 4 | 2.83e-01 |
| GO:BP | GO:1900052 | regulation of retinoic acid biosynthetic process | 1 | 3 | 2.83e-01 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 4 | 26 | 2.83e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 100 | 2011 | 2.83e-01 |
| GO:BP | GO:0010459 | negative regulation of heart rate | 2 | 7 | 2.84e-01 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 2 | 9 | 2.84e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 2 | 12 | 2.84e-01 |
| GO:BP | GO:0021660 | rhombomere 3 formation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0021664 | rhombomere 5 morphogenesis | 1 | 1 | 2.84e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 2.84e-01 |
| GO:BP | GO:0021595 | rhombomere structural organization | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0110154 | RNA decapping | 2 | 17 | 2.84e-01 |
| GO:BP | GO:0021665 | rhombomere 5 structural organization | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0021666 | rhombomere 5 formation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 6 | 18 | 2.84e-01 |
| GO:BP | GO:1904747 | positive regulation of apoptotic process involved in development | 2 | 5 | 2.84e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 5 | 15 | 2.84e-01 |
| GO:BP | GO:0021659 | rhombomere 3 structural organization | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 8 | 13 | 2.84e-01 |
| GO:BP | GO:0021594 | rhombomere formation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0033078 | extrathymic T cell differentiation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:1902339 | positive regulation of apoptotic process involved in morphogenesis | 2 | 5 | 2.84e-01 |
| GO:BP | GO:1903952 | regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 4 | 5 | 2.84e-01 |
| GO:BP | GO:1903954 | positive regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 3 | 21 | 2.84e-01 |
| GO:BP | GO:0090398 | cellular senescence | 37 | 83 | 2.84e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 4 | 22 | 2.84e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 2.84e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 2 | 3 | 2.84e-01 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 10 | 21 | 2.85e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 3 | 159 | 2.85e-01 |
| GO:BP | GO:0008063 | Toll signaling pathway | 4 | 6 | 2.85e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 2 | 42 | 2.85e-01 |
| GO:BP | GO:0002314 | germinal center B cell differentiation | 1 | 3 | 2.85e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 2 | 3 | 2.85e-01 |
| GO:BP | GO:1905115 | regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 2.85e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 5 | 93 | 2.85e-01 |
| GO:BP | GO:1905116 | positive regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 2.85e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 32 | 81 | 2.85e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 2 | 42 | 2.85e-01 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 2 | 23 | 2.85e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 2 | 6 | 2.85e-01 |
| GO:BP | GO:0046340 | diacylglycerol catabolic process | 2 | 3 | 2.85e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 2.85e-01 |
| GO:BP | GO:0090155 | negative regulation of sphingolipid biosynthetic process | 1 | 6 | 2.85e-01 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 4 | 20 | 2.85e-01 |
| GO:BP | GO:1904394 | negative regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 1 | 2.85e-01 |
| GO:BP | GO:0048541 | Peyer’s patch development | 1 | 6 | 2.85e-01 |
| GO:BP | GO:0000255 | allantoin metabolic process | 1 | 4 | 2.85e-01 |
| GO:BP | GO:0090251 | protein localization involved in establishment of planar polarity | 2 | 2 | 2.85e-01 |
| GO:BP | GO:0048537 | mucosa-associated lymphoid tissue development | 1 | 6 | 2.85e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 5 | 92 | 2.85e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 18 | 31 | 2.85e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 2.85e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 39 | 1145 | 2.85e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 2.85e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 3 | 26 | 2.85e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 4 | 6 | 2.85e-01 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 5 | 20 | 2.85e-01 |
| GO:BP | GO:0018009 | N-terminal peptidyl-L-cysteine N-palmitoylation | 2 | 2 | 2.85e-01 |
| GO:BP | GO:1904636 | response to ionomycin | 1 | 3 | 2.85e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 7 | 293 | 2.85e-01 |
| GO:BP | GO:1904637 | cellular response to ionomycin | 1 | 3 | 2.85e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 2 | 16 | 2.85e-01 |
| GO:BP | GO:0008544 | epidermis development | 3 | 210 | 2.85e-01 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 20 | 67 | 2.85e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 2.85e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 19 | 71 | 2.85e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 4 | 30 | 2.85e-01 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 2 | 3 | 2.85e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 7 | 21 | 2.85e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 4 | 30 | 2.85e-01 |
| GO:BP | GO:0033566 | gamma-tubulin complex localization | 2 | 2 | 2.85e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 3 | 13 | 2.85e-01 |
| GO:BP | GO:1905663 | positive regulation of telomerase RNA reverse transcriptase activity | 2 | 2 | 2.86e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 4 | 39 | 2.86e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 6 | 16 | 2.86e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 32 | 73 | 2.86e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 8 | 30 | 2.87e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 49 | 560 | 2.88e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 5 | 14 | 2.88e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 6 | 21 | 2.89e-01 |
| GO:BP | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 3 | 2.89e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 7 | 38 | 2.89e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 2.89e-01 |
| GO:BP | GO:0086054 | bundle of His cell to Purkinje myocyte communication by electrical coupling | 2 | 2 | 2.89e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 6 | 7 | 2.89e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 2.89e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 6 | 7 | 2.89e-01 |
| GO:BP | GO:1902065 | response to L-glutamate | 8 | 11 | 2.89e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 2.89e-01 |
| GO:BP | GO:0032075 | positive regulation of nuclease activity | 3 | 7 | 2.90e-01 |
| GO:BP | GO:0035581 | sequestering of extracellular ligand from receptor | 1 | 7 | 2.90e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 28 | 64 | 2.90e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 10 | 19 | 2.90e-01 |
| GO:BP | GO:0035120 | post-embryonic appendage morphogenesis | 1 | 2 | 2.90e-01 |
| GO:BP | GO:0035127 | post-embryonic limb morphogenesis | 1 | 2 | 2.90e-01 |
| GO:BP | GO:0035128 | post-embryonic forelimb morphogenesis | 1 | 2 | 2.90e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 2 | 5 | 2.90e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 10 | 19 | 2.90e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 10 | 19 | 2.90e-01 |
| GO:BP | GO:1903281 | positive regulation of calcium:sodium antiporter activity | 1 | 2 | 2.90e-01 |
| GO:BP | GO:0044205 | ‘de novo’ UMP biosynthetic process | 2 | 3 | 2.90e-01 |
| GO:BP | GO:0086092 | regulation of the force of heart contraction by cardiac conduction | 1 | 2 | 2.90e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 7 | 137 | 2.90e-01 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 2 | 10 | 2.90e-01 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 1 | 12 | 2.90e-01 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 2 | 10 | 2.90e-01 |
| GO:BP | GO:0098532 | histone H3-K27 trimethylation | 1 | 4 | 2.90e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 2 | 41 | 2.90e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 25 | 399 | 2.92e-01 |
| GO:BP | GO:0033326 | cerebrospinal fluid secretion | 2 | 3 | 2.92e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 3 | 11 | 2.92e-01 |
| GO:BP | GO:0042551 | neuron maturation | 4 | 33 | 2.92e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 2.92e-01 |
| GO:BP | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 4 | 4 | 2.92e-01 |
| GO:BP | GO:0106015 | negative regulation of inflammatory response to wounding | 1 | 2 | 2.92e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 11 | 52 | 2.92e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 2 | 18 | 2.92e-01 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 13 | 21 | 2.93e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 19 | 30 | 2.93e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 7 | 51 | 2.93e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 12 | 113 | 2.93e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 7 | 90 | 2.93e-01 |
| GO:BP | GO:0046108 | uridine metabolic process | 1 | 1 | 2.93e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 2 | 45 | 2.93e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 11 | 14 | 2.93e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 11 | 21 | 2.93e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 2 | 8 | 2.93e-01 |
| GO:BP | GO:0006218 | uridine catabolic process | 1 | 1 | 2.93e-01 |
| GO:BP | GO:0042454 | ribonucleoside catabolic process | 3 | 12 | 2.93e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 6 | 14 | 2.93e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 14 | 22 | 2.93e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 22 | 36 | 2.93e-01 |
| GO:BP | GO:0032571 | response to vitamin K | 1 | 5 | 2.93e-01 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 2 | 62 | 2.95e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 4 | 155 | 2.95e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 2.95e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 13 | 155 | 2.95e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.95e-01 |
| GO:BP | GO:0016570 | histone modification | 1 | 433 | 2.95e-01 |
| GO:BP | GO:0097291 | renal phosphate ion absorption | 1 | 1 | 2.95e-01 |
| GO:BP | GO:0009155 | purine deoxyribonucleotide catabolic process | 1 | 7 | 2.95e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 2 | 8 | 2.95e-01 |
| GO:BP | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | 1 | 2 | 2.95e-01 |
| GO:BP | GO:0009169 | purine ribonucleoside monophosphate catabolic process | 1 | 7 | 2.95e-01 |
| GO:BP | GO:0070244 | negative regulation of thymocyte apoptotic process | 1 | 6 | 2.95e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 2.95e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 29 | 357 | 2.95e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 10 | 52 | 2.95e-01 |
| GO:BP | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation | 4 | 6 | 2.95e-01 |
| GO:BP | GO:0051126 | negative regulation of actin nucleation | 2 | 6 | 2.95e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 4 | 17 | 2.95e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 20 | 31 | 2.95e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 10 | 14 | 2.95e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 9 | 55 | 2.95e-01 |
| GO:BP | GO:0009128 | purine nucleoside monophosphate catabolic process | 1 | 7 | 2.95e-01 |
| GO:BP | GO:0000379 | tRNA-type intron splice site recognition and cleavage | 1 | 3 | 2.96e-01 |
| GO:BP | GO:1905647 | proline import across plasma membrane | 3 | 5 | 2.96e-01 |
| GO:BP | GO:0045769 | negative regulation of asymmetric cell division | 1 | 1 | 2.96e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 3 | 16 | 2.96e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 20 | 49 | 2.96e-01 |
| GO:BP | GO:0090071 | negative regulation of ribosome biogenesis | 3 | 4 | 2.96e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 6 | 8 | 2.96e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 6 | 16 | 2.96e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 2.96e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 5 | 30 | 2.96e-01 |
| GO:BP | GO:1904271 | L-proline import across plasma membrane | 3 | 5 | 2.96e-01 |
| GO:BP | GO:0006540 | glutamate decarboxylation to succinate | 1 | 1 | 2.96e-01 |
| GO:BP | GO:0018352 | protein-pyridoxal-5-phosphate linkage | 1 | 2 | 2.96e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 14 | 22 | 2.96e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 9 | 65 | 2.96e-01 |
| GO:BP | GO:0043490 | malate-aspartate shuttle | 2 | 4 | 2.96e-01 |
| GO:BP | GO:0019731 | antibacterial humoral response | 2 | 15 | 2.96e-01 |
| GO:BP | GO:0035148 | tube formation | 29 | 138 | 2.96e-01 |
| GO:BP | GO:1904555 | L-proline transmembrane transport | 3 | 5 | 2.96e-01 |
| GO:BP | GO:0051974 | negative regulation of telomerase activity | 5 | 11 | 2.96e-01 |
| GO:BP | GO:0098905 | regulation of bundle of His cell action potential | 1 | 2 | 2.96e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 5 | 57 | 2.96e-01 |
| GO:BP | GO:0003015 | heart process | 12 | 216 | 2.96e-01 |
| GO:BP | GO:0042946 | glucoside transport | 2 | 3 | 2.96e-01 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 4 | 20 | 2.96e-01 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 1 | 3 | 2.96e-01 |
| GO:BP | GO:0035623 | renal glucose absorption | 2 | 2 | 2.96e-01 |
| GO:BP | GO:0010643 | cell communication by chemical coupling | 1 | 2 | 2.96e-01 |
| GO:BP | GO:0035928 | rRNA import into mitochondrion | 2 | 3 | 2.96e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 2 | 12 | 2.96e-01 |
| GO:BP | GO:0003193 | pulmonary valve formation | 1 | 2 | 2.96e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 3 | 64 | 2.96e-01 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 4 | 28 | 2.96e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 8 | 14 | 2.97e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 18 | 64 | 2.97e-01 |
| GO:BP | GO:0021873 | forebrain neuroblast division | 2 | 3 | 2.97e-01 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 1 | 9 | 2.97e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 123 | 218 | 2.97e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 2 | 4 | 2.97e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 2 | 4 | 2.97e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 2 | 4 | 2.97e-01 |
| GO:BP | GO:0015761 | mannose transmembrane transport | 1 | 1 | 2.97e-01 |
| GO:BP | GO:0035513 | oxidative RNA demethylation | 3 | 5 | 2.97e-01 |
| GO:BP | GO:2000210 | positive regulation of anoikis | 1 | 6 | 2.97e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 8 | 15 | 2.97e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 2.97e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 17 | 160 | 2.97e-01 |
| GO:BP | GO:0035927 | RNA import into mitochondrion | 3 | 4 | 2.97e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 2.97e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 8 | 12 | 2.97e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 3 | 15 | 2.97e-01 |
| GO:BP | GO:0051835 | positive regulation of synapse structural plasticity | 2 | 2 | 2.98e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 2 | 32 | 2.98e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 3 | 22 | 2.98e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 73 | 278 | 2.99e-01 |
| GO:BP | GO:0007127 | meiosis I | 28 | 77 | 2.99e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 40 | 809 | 2.99e-01 |
| GO:BP | GO:0090244 | Wnt signaling pathway involved in somitogenesis | 2 | 5 | 2.99e-01 |
| GO:BP | GO:0071709 | membrane assembly | 4 | 53 | 2.99e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 5 | 16 | 2.99e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 3 | 75 | 2.99e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 23 | 50 | 3.00e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 2 | 2 | 3.00e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 5 | 103 | 3.00e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 22 | 80 | 3.00e-01 |
| GO:BP | GO:0034964 | box H/ACA RNA processing | 2 | 2 | 3.00e-01 |
| GO:BP | GO:1903444 | negative regulation of brown fat cell differentiation | 2 | 3 | 3.00e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 4 | 14 | 3.00e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 9 | 167 | 3.00e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 18 | 81 | 3.00e-01 |
| GO:BP | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation | 4 | 5 | 3.00e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 7 | 67 | 3.00e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 6 | 353 | 3.00e-01 |
| GO:BP | GO:0016557 | peroxisome membrane biogenesis | 3 | 3 | 3.00e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 10 | 22 | 3.00e-01 |
| GO:BP | GO:1901660 | calcium ion export | 3 | 3 | 3.00e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 7 | 60 | 3.00e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 2 | 6 | 3.00e-01 |
| GO:BP | GO:0021549 | cerebellum development | 1 | 78 | 3.00e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 3.00e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 5 | 343 | 3.00e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 3.00e-01 |
| GO:BP | GO:0000495 | box H/ACA RNA 3’-end processing | 2 | 2 | 3.00e-01 |
| GO:BP | GO:0046361 | 2-oxobutyrate metabolic process | 1 | 2 | 3.00e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 1 | 64 | 3.00e-01 |
| GO:BP | GO:0015680 | protein maturation by copper ion transfer | 2 | 2 | 3.00e-01 |
| GO:BP | GO:0032764 | negative regulation of mast cell cytokine production | 3 | 3 | 3.00e-01 |
| GO:BP | GO:0007616 | long-term memory | 8 | 31 | 3.00e-01 |
| GO:BP | GO:0035364 | thymine transport | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 3 | 5 | 3.00e-01 |
| GO:BP | GO:0009197 | pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 2 | 2 | 3.01e-01 |
| GO:BP | GO:0009196 | pyrimidine deoxyribonucleoside diphosphate metabolic process | 2 | 2 | 3.01e-01 |
| GO:BP | GO:1901862 | negative regulation of muscle tissue development | 3 | 9 | 3.01e-01 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 1 | 9 | 3.01e-01 |
| GO:BP | GO:0046338 | phosphatidylethanolamine catabolic process | 2 | 4 | 3.01e-01 |
| GO:BP | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | 4 | 4 | 3.01e-01 |
| GO:BP | GO:0046036 | CTP metabolic process | 1 | 14 | 3.01e-01 |
| GO:BP | GO:0033632 | regulation of cell-cell adhesion mediated by integrin | 1 | 6 | 3.01e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 7 | 42 | 3.01e-01 |
| GO:BP | GO:0048850 | hypophysis morphogenesis | 2 | 2 | 3.01e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 3.01e-01 |
| GO:BP | GO:0046072 | dTDP metabolic process | 2 | 2 | 3.01e-01 |
| GO:BP | GO:0006233 | dTDP biosynthetic process | 2 | 2 | 3.01e-01 |
| GO:BP | GO:0060945 | cardiac neuron differentiation | 1 | 1 | 3.01e-01 |
| GO:BP | GO:0062111 | zinc ion import into organelle | 2 | 7 | 3.01e-01 |
| GO:BP | GO:0006227 | dUDP biosynthetic process | 2 | 2 | 3.01e-01 |
| GO:BP | GO:1903723 | negative regulation of centriole elongation | 1 | 2 | 3.01e-01 |
| GO:BP | GO:0046085 | adenosine metabolic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0090129 | positive regulation of synapse maturation | 1 | 8 | 3.01e-01 |
| GO:BP | GO:0048742 | regulation of skeletal muscle fiber development | 1 | 8 | 3.01e-01 |
| GO:BP | GO:0006113 | fermentation | 2 | 5 | 3.01e-01 |
| GO:BP | GO:0043084 | penile erection | 1 | 6 | 3.01e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 2 | 59 | 3.01e-01 |
| GO:BP | GO:0033089 | positive regulation of T cell differentiation in thymus | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0046077 | dUDP metabolic process | 2 | 2 | 3.01e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 4 | 11 | 3.01e-01 |
| GO:BP | GO:0043465 | regulation of fermentation | 2 | 5 | 3.01e-01 |
| GO:BP | GO:0060419 | heart growth | 28 | 72 | 3.01e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 5 | 31 | 3.01e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 8 | 26 | 3.01e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 2 | 75 | 3.01e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 27 | 260 | 3.01e-01 |
| GO:BP | GO:0099576 | regulation of protein catabolic process at postsynapse, modulating synaptic transmission | 1 | 4 | 3.02e-01 |
| GO:BP | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning | 1 | 1 | 3.02e-01 |
| GO:BP | GO:0071503 | response to heparin | 3 | 5 | 3.02e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 3 | 14 | 3.02e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 9 | 22 | 3.02e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 3.02e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 3 | 129 | 3.02e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 2 | 57 | 3.02e-01 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 5 | 10 | 3.02e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 2 | 11 | 3.03e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 11 | 3.03e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 7 | 89 | 3.03e-01 |
| GO:BP | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway | 3 | 4 | 3.04e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 14 | 200 | 3.04e-01 |
| GO:BP | GO:1903553 | positive regulation of extracellular exosome assembly | 3 | 4 | 3.04e-01 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 1 | 9 | 3.04e-01 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 1 | 9 | 3.04e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 3 | 89 | 3.04e-01 |
| GO:BP | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 3.05e-01 |
| GO:BP | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0060789 | hair follicle placode formation | 3 | 5 | 3.05e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 2 | 19 | 3.05e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 7 | 9 | 3.05e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 3.05e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 32 | 68 | 3.05e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 4 | 8 | 3.05e-01 |
| GO:BP | GO:0046136 | positive regulation of vitamin metabolic process | 1 | 2 | 3.05e-01 |
| GO:BP | GO:2000537 | regulation of B cell chemotaxis | 1 | 1 | 3.05e-01 |
| GO:BP | GO:2000538 | positive regulation of B cell chemotaxis | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 3.05e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 4 | 303 | 3.05e-01 |
| GO:BP | GO:0003335 | corneocyte development | 1 | 2 | 3.05e-01 |
| GO:BP | GO:0090168 | Golgi reassembly | 4 | 5 | 3.05e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 2 | 42 | 3.05e-01 |
| GO:BP | GO:0060431 | primary lung bud formation | 1 | 2 | 3.05e-01 |
| GO:BP | GO:0046604 | positive regulation of mitotic centrosome separation | 3 | 3 | 3.05e-01 |
| GO:BP | GO:0071467 | cellular response to pH | 5 | 16 | 3.05e-01 |
| GO:BP | GO:0061757 | leukocyte adhesion to arterial endothelial cell | 3 | 4 | 3.05e-01 |
| GO:BP | GO:0019747 | regulation of isoprenoid metabolic process | 1 | 4 | 3.05e-01 |
| GO:BP | GO:2000810 | regulation of bicellular tight junction assembly | 8 | 15 | 3.05e-01 |
| GO:BP | GO:0098925 | retrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 4 | 6 | 3.05e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 1 | 5 | 3.05e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 2 | 21 | 3.05e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 3.05e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 19 | 162 | 3.05e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 1 | 5 | 3.05e-01 |
| GO:BP | GO:1903774 | positive regulation of viral budding via host ESCRT complex | 2 | 3 | 3.06e-01 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 6 | 30 | 3.06e-01 |
| GO:BP | GO:1990359 | stress response to zinc ion | 1 | 1 | 3.06e-01 |
| GO:BP | GO:0010312 | detoxification of zinc ion | 1 | 1 | 3.06e-01 |
| GO:BP | GO:0036486 | ventral trunk neural crest cell migration | 3 | 3 | 3.06e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 14 | 58 | 3.06e-01 |
| GO:BP | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 4 | 3.06e-01 |
| GO:BP | GO:1990535 | neuron projection maintenance | 4 | 9 | 3.07e-01 |
| GO:BP | GO:0060324 | face development | 4 | 45 | 3.07e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 7 | 14 | 3.07e-01 |
| GO:BP | GO:0032902 | nerve growth factor production | 2 | 2 | 3.07e-01 |
| GO:BP | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway | 3 | 3 | 3.08e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 4 | 30 | 3.08e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 3.08e-01 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 1 | 2 | 3.08e-01 |
| GO:BP | GO:1904439 | negative regulation of iron ion import across plasma membrane | 1 | 1 | 3.08e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 3.08e-01 |
| GO:BP | GO:1905438 | non-canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 3.08e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 13 | 30 | 3.08e-01 |
| GO:BP | GO:0048627 | myoblast development | 1 | 3 | 3.08e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 16 | 30 | 3.08e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 14 | 27 | 3.08e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 1 | 10 | 3.08e-01 |
| GO:BP | GO:0000729 | DNA double-strand break processing | 2 | 18 | 3.08e-01 |
| GO:BP | GO:0051885 | positive regulation of timing of anagen | 1 | 2 | 3.08e-01 |
| GO:BP | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis | 1 | 2 | 3.08e-01 |
| GO:BP | GO:1904933 | regulation of cell proliferation in midbrain | 1 | 1 | 3.08e-01 |
| GO:BP | GO:0036515 | serotonergic neuron axon guidance | 1 | 2 | 3.08e-01 |
| GO:BP | GO:0036518 | chemorepulsion of dopaminergic neuron axon | 1 | 2 | 3.08e-01 |
| GO:BP | GO:0003408 | optic cup formation involved in camera-type eye development | 1 | 1 | 3.08e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 2 | 7 | 3.08e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 4 | 15 | 3.08e-01 |
| GO:BP | GO:0106034 | protein maturation by [2Fe-2S] cluster transfer | 1 | 3 | 3.08e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 1 | 45 | 3.08e-01 |
| GO:BP | GO:0001560 | regulation of cell growth by extracellular stimulus | 2 | 3 | 3.08e-01 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 1 | 7 | 3.09e-01 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 7 | 41 | 3.09e-01 |
| GO:BP | GO:0032069 | regulation of nuclease activity | 4 | 17 | 3.09e-01 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 2 | 9 | 3.09e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 7 | 120 | 3.09e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 8 | 22 | 3.09e-01 |
| GO:BP | GO:0051793 | medium-chain fatty acid catabolic process | 2 | 3 | 3.09e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 3 | 11 | 3.09e-01 |
| GO:BP | GO:0160024 | Leydig cell proliferation | 2 | 2 | 3.09e-01 |
| GO:BP | GO:0045004 | DNA replication proofreading | 1 | 2 | 3.09e-01 |
| GO:BP | GO:0045951 | positive regulation of mitotic recombination | 1 | 1 | 3.09e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 2 | 7 | 3.09e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 4 | 5 | 3.09e-01 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 7 | 13 | 3.09e-01 |
| GO:BP | GO:0071578 | zinc ion import across plasma membrane | 2 | 6 | 3.09e-01 |
| GO:BP | GO:0009143 | nucleoside triphosphate catabolic process | 1 | 10 | 3.09e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 2 | 2 | 3.09e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 15 | 406 | 3.09e-01 |
| GO:BP | GO:0016077 | sno(s)RNA catabolic process | 2 | 3 | 3.09e-01 |
| GO:BP | GO:0002949 | tRNA threonylcarbamoyladenosine modification | 3 | 5 | 3.09e-01 |
| GO:BP | GO:0060453 | regulation of gastric acid secretion | 2 | 8 | 3.09e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 9 | 26 | 3.09e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 5 | 18 | 3.09e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 2 | 23 | 3.10e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 17 | 36 | 3.11e-01 |
| GO:BP | GO:0007382 | specification of segmental identity, maxillary segment | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0044751 | cellular response to human chorionic gonadotropin stimulus | 1 | 2 | 3.11e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 5 | 423 | 3.11e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 9 | 19 | 3.11e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 8 | 207 | 3.11e-01 |
| GO:BP | GO:1905871 | regulation of protein localization to cell leading edge | 1 | 2 | 3.11e-01 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 9 | 10 | 3.11e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 2 | 111 | 3.11e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 5 | 23 | 3.11e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 3 | 25 | 3.11e-01 |
| GO:BP | GO:0072423 | response to DNA damage checkpoint signaling | 3 | 8 | 3.11e-01 |
| GO:BP | GO:0072402 | response to DNA integrity checkpoint signaling | 3 | 8 | 3.11e-01 |
| GO:BP | GO:0031860 | telomeric 3’ overhang formation | 1 | 4 | 3.11e-01 |
| GO:BP | GO:0072396 | response to cell cycle checkpoint signaling | 3 | 8 | 3.11e-01 |
| GO:BP | GO:0009125 | nucleoside monophosphate catabolic process | 2 | 13 | 3.11e-01 |
| GO:BP | GO:0043584 | nose development | 2 | 10 | 3.11e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 37 | 84 | 3.11e-01 |
| GO:BP | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation | 2 | 2 | 3.11e-01 |
| GO:BP | GO:1905359 | protein localization to meiotic spindle | 1 | 1 | 3.11e-01 |
| GO:BP | GO:1903096 | protein localization to meiotic spindle midzone | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 23 | 420 | 3.11e-01 |
| GO:BP | GO:0071971 | extracellular exosome assembly | 3 | 6 | 3.11e-01 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 2 | 42 | 3.11e-01 |
| GO:BP | GO:0040009 | regulation of growth rate | 1 | 3 | 3.11e-01 |
| GO:BP | GO:0035973 | aggrephagy | 6 | 8 | 3.11e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 5 | 40 | 3.11e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 12 | 249 | 3.11e-01 |
| GO:BP | GO:1902145 | regulation of response to cell cycle checkpoint signaling | 1 | 1 | 3.11e-01 |
| GO:BP | GO:1902151 | regulation of response to DNA integrity checkpoint signaling | 1 | 1 | 3.11e-01 |
| GO:BP | GO:1902153 | regulation of response to DNA damage checkpoint signaling | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0032487 | regulation of Rap protein signal transduction | 1 | 3 | 3.11e-01 |
| GO:BP | GO:0033624 | negative regulation of integrin activation | 1 | 3 | 3.11e-01 |
| GO:BP | GO:0000706 | meiotic DNA double-strand break processing | 2 | 2 | 3.12e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 24 | 53 | 3.12e-01 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 6 | 9 | 3.12e-01 |
| GO:BP | GO:0006021 | inositol biosynthetic process | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 3.12e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 3 | 34 | 3.12e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 3 | 4 | 3.12e-01 |
| GO:BP | GO:0070295 | renal water absorption | 3 | 5 | 3.12e-01 |
| GO:BP | GO:0060611 | mammary gland fat development | 1 | 1 | 3.12e-01 |
| GO:BP | GO:1902228 | positive regulation of macrophage colony-stimulating factor signaling pathway | 1 | 1 | 3.12e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 3 | 128 | 3.12e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 3.12e-01 |
| GO:BP | GO:1904415 | regulation of xenophagy | 6 | 7 | 3.13e-01 |
| GO:BP | GO:1904417 | positive regulation of xenophagy | 6 | 7 | 3.13e-01 |
| GO:BP | GO:1903518 | positive regulation of single strand break repair | 2 | 2 | 3.13e-01 |
| GO:BP | GO:1904875 | regulation of DNA ligase activity | 2 | 2 | 3.13e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 3.13e-01 |
| GO:BP | GO:0015848 | spermidine transport | 1 | 3 | 3.13e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 2 | 11 | 3.14e-01 |
| GO:BP | GO:2000232 | regulation of rRNA processing | 7 | 16 | 3.14e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 5 | 29 | 3.15e-01 |
| GO:BP | GO:1900029 | positive regulation of ruffle assembly | 4 | 12 | 3.15e-01 |
| GO:BP | GO:0010224 | response to UV-B | 2 | 17 | 3.15e-01 |
| GO:BP | GO:0006598 | polyamine catabolic process | 1 | 4 | 3.15e-01 |
| GO:BP | GO:0061952 | midbody abscission | 4 | 17 | 3.16e-01 |
| GO:BP | GO:0035329 | hippo signaling | 23 | 41 | 3.16e-01 |
| GO:BP | GO:0001822 | kidney development | 14 | 244 | 3.17e-01 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 2 | 5 | 3.17e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 1 | 17 | 3.17e-01 |
| GO:BP | GO:1904999 | positive regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 3.17e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 4 | 44 | 3.17e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 3 | 179 | 3.17e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 1 | 17 | 3.17e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 6 | 113 | 3.17e-01 |
| GO:BP | GO:0072197 | ureter morphogenesis | 5 | 5 | 3.17e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 4 | 295 | 3.17e-01 |
| GO:BP | GO:0060449 | bud elongation involved in lung branching | 5 | 7 | 3.17e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 3.17e-01 |
| GO:BP | GO:0000712 | resolution of meiotic recombination intermediates | 1 | 13 | 3.17e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 17 | 61 | 3.18e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 3.18e-01 |
| GO:BP | GO:0070254 | mucus secretion | 1 | 8 | 3.18e-01 |
| GO:BP | GO:0015755 | fructose transmembrane transport | 3 | 5 | 3.18e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 3.18e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 2 | 42 | 3.18e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 3 | 31 | 3.18e-01 |
| GO:BP | GO:0009608 | response to symbiont | 1 | 2 | 3.18e-01 |
| GO:BP | GO:0009609 | response to symbiotic bacterium | 1 | 2 | 3.18e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 3.18e-01 |
| GO:BP | GO:0010868 | negative regulation of triglyceride biosynthetic process | 3 | 3 | 3.18e-01 |
| GO:BP | GO:0015791 | polyol transmembrane transport | 4 | 6 | 3.18e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 7 | 555 | 3.18e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 27 | 65 | 3.18e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 8 | 211 | 3.18e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 5 | 429 | 3.19e-01 |
| GO:BP | GO:0034199 | activation of protein kinase A activity | 3 | 3 | 3.19e-01 |
| GO:BP | GO:0051098 | regulation of binding | 2 | 317 | 3.19e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 15 | 37 | 3.19e-01 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 4 | 3.19e-01 |
| GO:BP | GO:0071476 | cellular hypotonic response | 3 | 8 | 3.19e-01 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 5 | 16 | 3.19e-01 |
| GO:BP | GO:0042727 | flavin-containing compound biosynthetic process | 2 | 2 | 3.19e-01 |
| GO:BP | GO:0002335 | mature B cell differentiation | 3 | 21 | 3.19e-01 |
| GO:BP | GO:1902954 | regulation of early endosome to recycling endosome transport | 1 | 2 | 3.19e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 4 | 172 | 3.19e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 5 | 18 | 3.19e-01 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 2 | 12 | 3.19e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 3.19e-01 |
| GO:BP | GO:0098760 | response to interleukin-7 | 2 | 12 | 3.19e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 10 | 140 | 3.19e-01 |
| GO:BP | GO:1900247 | regulation of cytoplasmic translational elongation | 7 | 10 | 3.19e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 18 | 131 | 3.20e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 3 | 70 | 3.20e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 1 | 69 | 3.20e-01 |
| GO:BP | GO:1905775 | negative regulation of DNA helicase activity | 1 | 2 | 3.20e-01 |
| GO:BP | GO:0003170 | heart valve development | 4 | 65 | 3.21e-01 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 4 | 14 | 3.21e-01 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 7 | 12 | 3.21e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 3 | 139 | 3.22e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 3.22e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 6 | 15 | 3.22e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 3 | 22 | 3.22e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 7 | 19 | 3.22e-01 |
| GO:BP | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 3 | 7 | 3.22e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 29 | 135 | 3.22e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 3.22e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 1 | 46 | 3.22e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 3.22e-01 |
| GO:BP | GO:0031990 | mRNA export from nucleus in response to heat stress | 2 | 2 | 3.22e-01 |
| GO:BP | GO:0036497 | eIF2alpha dephosphorylation in response to endoplasmic reticulum stress | 1 | 1 | 3.22e-01 |
| GO:BP | GO:1903916 | regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 3.22e-01 |
| GO:BP | GO:1903917 | positive regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 3.22e-01 |
| GO:BP | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation | 1 | 1 | 3.22e-01 |
| GO:BP | GO:0048369 | lateral mesoderm morphogenesis | 3 | 3 | 3.22e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 1 | 25 | 3.22e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 27 | 63 | 3.22e-01 |
| GO:BP | GO:0048370 | lateral mesoderm formation | 3 | 3 | 3.22e-01 |
| GO:BP | GO:0052648 | ribitol phosphate metabolic process | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0052647 | pentitol phosphate metabolic process | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0009231 | riboflavin biosynthetic process | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 2 | 5 | 3.23e-01 |
| GO:BP | GO:0046444 | FMN metabolic process | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 2 | 18 | 3.23e-01 |
| GO:BP | GO:1901563 | response to camptothecin | 4 | 5 | 3.23e-01 |
| GO:BP | GO:0002182 | cytoplasmic translational elongation | 8 | 12 | 3.23e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 14 | 228 | 3.23e-01 |
| GO:BP | GO:0036372 | opsin transport | 1 | 2 | 3.23e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 15 | 3.23e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 3 | 19 | 3.23e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 17 | 36 | 3.23e-01 |
| GO:BP | GO:0009398 | FMN biosynthetic process | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 6 | 377 | 3.23e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 56 | 414 | 3.23e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 7 | 114 | 3.23e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 4 | 58 | 3.23e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 18 | 104 | 3.23e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 11 | 156 | 3.23e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 11 | 23 | 3.23e-01 |
| GO:BP | GO:2000809 | positive regulation of synaptic vesicle clustering | 2 | 3 | 3.23e-01 |
| GO:BP | GO:0034402 | recruitment of 3’-end processing factors to RNA polymerase II holoenzyme complex | 1 | 2 | 3.24e-01 |
| GO:BP | GO:0010621 | negative regulation of transcription by transcription factor localization | 1 | 3 | 3.24e-01 |
| GO:BP | GO:0060322 | head development | 9 | 633 | 3.24e-01 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 3 | 7 | 3.24e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 4 | 94 | 3.24e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 3 | 9 | 3.24e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 3 | 9 | 3.24e-01 |
| GO:BP | GO:0010561 | negative regulation of glycoprotein biosynthetic process | 2 | 10 | 3.24e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 19 | 340 | 3.25e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 2 | 12 | 3.25e-01 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 5 | 6 | 3.25e-01 |
| GO:BP | GO:0098957 | anterograde axonal transport of mitochondrion | 1 | 5 | 3.25e-01 |
| GO:BP | GO:0001561 | fatty acid alpha-oxidation | 2 | 5 | 3.25e-01 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 1 | 3 | 3.25e-01 |
| GO:BP | GO:0002467 | germinal center formation | 1 | 10 | 3.25e-01 |
| GO:BP | GO:1902337 | regulation of apoptotic process involved in morphogenesis | 3 | 9 | 3.25e-01 |
| GO:BP | GO:1904748 | regulation of apoptotic process involved in development | 3 | 9 | 3.25e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 1 | 16 | 3.25e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 2 | 97 | 3.25e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 7 | 73 | 3.25e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 3.25e-01 |
| GO:BP | GO:0006824 | cobalt ion transport | 2 | 5 | 3.25e-01 |
| GO:BP | GO:1905176 | positive regulation of vascular associated smooth muscle cell dedifferentiation | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 3 | 82 | 3.25e-01 |
| GO:BP | GO:0072255 | metanephric glomerular mesangial cell development | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 3.25e-01 |
| GO:BP | GO:0001798 | positive regulation of type IIa hypersensitivity | 2 | 3 | 3.25e-01 |
| GO:BP | GO:0060995 | cell-cell signaling involved in kidney development | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0072204 | cell-cell signaling involved in metanephros development | 1 | 1 | 3.25e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 37 | 85 | 3.25e-01 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 6 | 36 | 3.25e-01 |
| GO:BP | GO:0061289 | Wnt signaling pathway involved in kidney development | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0090102 | cochlea development | 4 | 35 | 3.25e-01 |
| GO:BP | GO:0002894 | positive regulation of type II hypersensitivity | 2 | 3 | 3.25e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 3.25e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 3 | 94 | 3.25e-01 |
| GO:BP | GO:0061290 | canonical Wnt signaling pathway involved in metanephric kidney development | 1 | 1 | 3.25e-01 |
| GO:BP | GO:0090212 | negative regulation of establishment of blood-brain barrier | 1 | 1 | 3.26e-01 |
| GO:BP | GO:0060454 | positive regulation of gastric acid secretion | 1 | 2 | 3.26e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 10 | 140 | 3.26e-01 |
| GO:BP | GO:1903279 | regulation of calcium:sodium antiporter activity | 1 | 3 | 3.27e-01 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 3.28e-01 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 3.28e-01 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 3.28e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 2 | 2 | 3.28e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 6 | 10 | 3.28e-01 |
| GO:BP | GO:0060219 | camera-type eye photoreceptor cell differentiation | 10 | 18 | 3.28e-01 |
| GO:BP | GO:1900161 | regulation of phospholipid scramblase activity | 1 | 1 | 3.28e-01 |
| GO:BP | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development | 1 | 3 | 3.28e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 2 | 12 | 3.28e-01 |
| GO:BP | GO:1900163 | positive regulation of phospholipid scramblase activity | 1 | 1 | 3.28e-01 |
| GO:BP | GO:2000752 | regulation of glucosylceramide catabolic process | 1 | 1 | 3.28e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 7 | 14 | 3.28e-01 |
| GO:BP | GO:2000753 | positive regulation of glucosylceramide catabolic process | 1 | 1 | 3.28e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 6 | 18 | 3.28e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 1 | 19 | 3.28e-01 |
| GO:BP | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis | 3 | 3 | 3.28e-01 |
| GO:BP | GO:0044854 | plasma membrane raft assembly | 1 | 6 | 3.28e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 1 | 16 | 3.28e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 5 | 192 | 3.28e-01 |
| GO:BP | GO:0070455 | positive regulation of heme biosynthetic process | 3 | 3 | 3.29e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 3.29e-01 |
| GO:BP | GO:1901465 | positive regulation of tetrapyrrole biosynthetic process | 3 | 3 | 3.29e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 3.29e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 10 | 39 | 3.29e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 3 | 72 | 3.29e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 6 | 22 | 3.29e-01 |
| GO:BP | GO:0039689 | negative stranded viral RNA replication | 2 | 3 | 3.29e-01 |
| GO:BP | GO:0003271 | smoothened signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 3.29e-01 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 2 | 8 | 3.29e-01 |
| GO:BP | GO:1903943 | regulation of hepatocyte apoptotic process | 5 | 7 | 3.30e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 1 | 77 | 3.30e-01 |
| GO:BP | GO:0044068 | modulation by symbiont of host cellular process | 3 | 5 | 3.30e-01 |
| GO:BP | GO:0046074 | dTMP catabolic process | 1 | 3 | 3.30e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 3.30e-01 |
| GO:BP | GO:0044003 | modulation by symbiont of host process | 3 | 5 | 3.30e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 3 | 5 | 3.30e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 80 | 147 | 3.30e-01 |
| GO:BP | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 6 | 3.30e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 2 | 70 | 3.30e-01 |
| GO:BP | GO:0019054 | modulation by virus of host cellular process | 3 | 5 | 3.30e-01 |
| GO:BP | GO:0098749 | cerebellar neuron development | 1 | 3 | 3.30e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 5 | 60 | 3.30e-01 |
| GO:BP | GO:0003166 | bundle of His development | 5 | 6 | 3.30e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 3 | 5 | 3.30e-01 |
| GO:BP | GO:0019048 | modulation by virus of host process | 3 | 5 | 3.30e-01 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 1 | 2 | 3.30e-01 |
| GO:BP | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process | 1 | 8 | 3.30e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 6 | 8 | 3.30e-01 |
| GO:BP | GO:0006043 | glucosamine catabolic process | 2 | 2 | 3.30e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 7 | 19 | 3.30e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 2 | 69 | 3.30e-01 |
| GO:BP | GO:2000520 | regulation of immunological synapse formation | 1 | 2 | 3.30e-01 |
| GO:BP | GO:0014824 | artery smooth muscle contraction | 2 | 7 | 3.30e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 21 | 358 | 3.30e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 3.30e-01 |
| GO:BP | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain | 2 | 2 | 3.30e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 23 | 204 | 3.30e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 5 | 37 | 3.30e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 3 | 73 | 3.30e-01 |
| GO:BP | GO:0090594 | inflammatory response to wounding | 2 | 12 | 3.30e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 3 | 11 | 3.30e-01 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 2 | 13 | 3.31e-01 |
| GO:BP | GO:0097115 | neurexin clustering involved in presynaptic membrane assembly | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0048789 | cytoskeletal matrix organization at active zone | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 9 | 26 | 3.31e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 3.31e-01 |
| GO:BP | GO:0015675 | nickel cation transport | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0035444 | nickel cation transmembrane transport | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 3.31e-01 |
| GO:BP | GO:0015676 | vanadium ion transport | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0015692 | lead ion transport | 1 | 1 | 3.31e-01 |
| GO:BP | GO:0030202 | heparin metabolic process | 2 | 14 | 3.31e-01 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 2 | 5 | 3.32e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 9 | 92 | 3.32e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 9 | 92 | 3.32e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 228 | 3.32e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 2 | 28 | 3.32e-01 |
| GO:BP | GO:0071569 | protein ufmylation | 5 | 6 | 3.32e-01 |
| GO:BP | GO:1903837 | regulation of mRNA 3’-UTR binding | 1 | 1 | 3.32e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 8 | 14 | 3.32e-01 |
| GO:BP | GO:1903839 | positive regulation of mRNA 3’-UTR binding | 1 | 1 | 3.32e-01 |
| GO:BP | GO:0021915 | neural tube development | 30 | 146 | 3.32e-01 |
| GO:BP | GO:0001771 | immunological synapse formation | 2 | 7 | 3.32e-01 |
| GO:BP | GO:0046069 | cGMP catabolic process | 1 | 4 | 3.33e-01 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 3 | 5 | 3.33e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 3.33e-01 |
| GO:BP | GO:0070889 | platelet alpha granule organization | 1 | 2 | 3.33e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 3 | 73 | 3.34e-01 |
| GO:BP | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation | 2 | 3 | 3.34e-01 |
| GO:BP | GO:0060028 | convergent extension involved in axis elongation | 2 | 6 | 3.34e-01 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 1 | 4 | 3.34e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 2 | 8 | 3.34e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 1 | 52 | 3.35e-01 |
| GO:BP | GO:0038133 | ERBB2-ERBB3 signaling pathway | 2 | 3 | 3.35e-01 |
| GO:BP | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway | 5 | 6 | 3.35e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 3.35e-01 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 1 | 8 | 3.35e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 4 | 17 | 3.35e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 12 | 182 | 3.35e-01 |
| GO:BP | GO:1903966 | monounsaturated fatty acid biosynthetic process | 1 | 2 | 3.35e-01 |
| GO:BP | GO:0098904 | regulation of AV node cell action potential | 1 | 3 | 3.35e-01 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 6 | 3.35e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 3.35e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 2 | 55 | 3.35e-01 |
| GO:BP | GO:0000390 | spliceosomal complex disassembly | 2 | 3 | 3.35e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 3.35e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 43 | 756 | 3.35e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 1 | 13 | 3.35e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 6 | 10 | 3.35e-01 |
| GO:BP | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential | 2 | 3 | 3.35e-01 |
| GO:BP | GO:0043697 | cell dedifferentiation | 3 | 6 | 3.35e-01 |
| GO:BP | GO:0043696 | dedifferentiation | 3 | 6 | 3.35e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 3 | 51 | 3.35e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 2 | 4 | 3.35e-01 |
| GO:BP | GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 3.35e-01 |
| GO:BP | GO:0016114 | terpenoid biosynthetic process | 2 | 12 | 3.35e-01 |
| GO:BP | GO:0086048 | membrane depolarization during bundle of His cell action potential | 2 | 3 | 3.35e-01 |
| GO:BP | GO:0072257 | metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 3.35e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 4 | 21 | 3.35e-01 |
| GO:BP | GO:0070512 | positive regulation of histone H4-K20 methylation | 1 | 1 | 3.35e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 53 | 1712 | 3.35e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 3 | 40 | 3.35e-01 |
| GO:BP | GO:0070106 | interleukin-27-mediated signaling pathway | 1 | 4 | 3.35e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 100 | 3.35e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 2 | 23 | 3.35e-01 |
| GO:BP | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process | 2 | 2 | 3.36e-01 |
| GO:BP | GO:0097711 | ciliary basal body-plasma membrane docking | 2 | 2 | 3.36e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 3 | 22 | 3.37e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 13 | 108 | 3.37e-01 |
| GO:BP | GO:1904868 | telomerase catalytic core complex assembly | 2 | 2 | 3.37e-01 |
| GO:BP | GO:1904884 | positive regulation of telomerase catalytic core complex assembly | 2 | 2 | 3.37e-01 |
| GO:BP | GO:1904882 | regulation of telomerase catalytic core complex assembly | 2 | 2 | 3.37e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 31 | 58 | 3.37e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 6 | 69 | 3.37e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 3.38e-01 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 3 | 18 | 3.38e-01 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 1 | 10 | 3.38e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 3.38e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 3.38e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 3.38e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 13 | 30 | 3.38e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 3.38e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 5 | 20 | 3.38e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 22 | 72 | 3.38e-01 |
| GO:BP | GO:0009186 | deoxyribonucleoside diphosphate metabolic process | 3 | 4 | 3.38e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 1 | 9 | 3.38e-01 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 2 | 10 | 3.39e-01 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 2 | 10 | 3.39e-01 |
| GO:BP | GO:0072001 | renal system development | 14 | 251 | 3.39e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 3.39e-01 |
| GO:BP | GO:0043628 | regulatory ncRNA 3’-end processing | 3 | 19 | 3.39e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 3.39e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 5 | 69 | 3.39e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 2 | 57 | 3.39e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 5 | 16 | 3.39e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 21 | 124 | 3.39e-01 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 2 | 4 | 3.39e-01 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 2 | 4 | 3.39e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 21 | 45 | 3.39e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 3 | 13 | 3.39e-01 |
| GO:BP | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | 1 | 2 | 3.39e-01 |
| GO:BP | GO:0014839 | myoblast migration involved in skeletal muscle regeneration | 2 | 3 | 3.40e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 3.40e-01 |
| GO:BP | GO:0072553 | terminal button organization | 2 | 5 | 3.40e-01 |
| GO:BP | GO:0140450 | protein targeting to Golgi apparatus | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 3.40e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 7 | 18 | 3.40e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 3.40e-01 |
| GO:BP | GO:0110155 | NAD-cap decapping | 2 | 3 | 3.41e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 14 | 88 | 3.41e-01 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 2 | 2 | 3.41e-01 |
| GO:BP | GO:0150018 | basal dendrite development | 3 | 3 | 3.41e-01 |
| GO:BP | GO:0150019 | basal dendrite morphogenesis | 3 | 3 | 3.41e-01 |
| GO:BP | GO:1905931 | negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 2 | 3.41e-01 |
| GO:BP | GO:1905916 | negative regulation of cell differentiation involved in phenotypic switching | 1 | 2 | 3.41e-01 |
| GO:BP | GO:0150020 | basal dendrite arborization | 3 | 3 | 3.41e-01 |
| GO:BP | GO:0000212 | meiotic spindle organization | 5 | 14 | 3.41e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 2 | 30 | 3.41e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 4 | 23 | 3.41e-01 |
| GO:BP | GO:1900240 | negative regulation of phenotypic switching | 1 | 2 | 3.41e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 67 | 119 | 3.41e-01 |
| GO:BP | GO:0035351 | heme transmembrane transport | 1 | 2 | 3.41e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 2 | 40 | 3.41e-01 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 2 | 2 | 3.41e-01 |
| GO:BP | GO:0001696 | gastric acid secretion | 3 | 10 | 3.41e-01 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 5 | 7 | 3.41e-01 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 5 | 7 | 3.41e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 12 | 20 | 3.41e-01 |
| GO:BP | GO:0060243 | negative regulation of cell growth involved in contact inhibition | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 3 | 5 | 3.42e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 3.42e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 6 | 16 | 3.42e-01 |
| GO:BP | GO:0006499 | N-terminal protein myristoylation | 3 | 4 | 3.42e-01 |
| GO:BP | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process | 1 | 5 | 3.42e-01 |
| GO:BP | GO:0030034 | microvillar actin bundle assembly | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0046929 | negative regulation of neurotransmitter secretion | 4 | 6 | 3.42e-01 |
| GO:BP | GO:0008090 | retrograde axonal transport | 3 | 19 | 3.42e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 13 | 38 | 3.42e-01 |
| GO:BP | GO:0034058 | endosomal vesicle fusion | 1 | 10 | 3.42e-01 |
| GO:BP | GO:0018377 | protein myristoylation | 3 | 4 | 3.42e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 11 | 46 | 3.42e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 4 | 49 | 3.42e-01 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 4 | 8 | 3.42e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 11 | 86 | 3.42e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 10 | 20 | 3.42e-01 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 3 | 12 | 3.42e-01 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 3 | 30 | 3.42e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 3.42e-01 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 2 | 18 | 3.42e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 4 | 7 | 3.43e-01 |
| GO:BP | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway | 2 | 4 | 3.43e-01 |
| GO:BP | GO:2000066 | positive regulation of cortisol biosynthetic process | 1 | 1 | 3.44e-01 |
| GO:BP | GO:1903542 | negative regulation of exosomal secretion | 3 | 4 | 3.44e-01 |
| GO:BP | GO:2000180 | negative regulation of androgen biosynthetic process | 1 | 1 | 3.44e-01 |
| GO:BP | GO:0010738 | regulation of protein kinase A signaling | 1 | 13 | 3.44e-01 |
| GO:BP | GO:1903361 | protein localization to basolateral plasma membrane | 1 | 7 | 3.44e-01 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 5 | 17 | 3.44e-01 |
| GO:BP | GO:0031948 | positive regulation of glucocorticoid biosynthetic process | 1 | 1 | 3.44e-01 |
| GO:BP | GO:0031945 | positive regulation of glucocorticoid metabolic process | 1 | 1 | 3.44e-01 |
| GO:BP | GO:0015939 | pantothenate metabolic process | 2 | 2 | 3.44e-01 |
| GO:BP | GO:2000019 | negative regulation of male gonad development | 1 | 1 | 3.44e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 43 | 3.44e-01 |
| GO:BP | GO:0061369 | negative regulation of testicular blood vessel morphogenesis | 1 | 1 | 3.44e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 3 | 9 | 3.44e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 4 | 11 | 3.44e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 3.44e-01 |
| GO:BP | GO:0098930 | axonal transport | 6 | 60 | 3.44e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 3 | 26 | 3.45e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 3.45e-01 |
| GO:BP | GO:1905443 | regulation of clathrin coat assembly | 1 | 1 | 3.45e-01 |
| GO:BP | GO:1905445 | positive regulation of clathrin coat assembly | 1 | 1 | 3.45e-01 |
| GO:BP | GO:1900451 | positive regulation of glutamate receptor signaling pathway | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0035627 | ceramide transport | 2 | 15 | 3.45e-01 |
| GO:BP | GO:0033631 | cell-cell adhesion mediated by integrin | 1 | 12 | 3.45e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 10 | 15 | 3.45e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 2 | 138 | 3.45e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 6 | 94 | 3.45e-01 |
| GO:BP | GO:0097117 | guanylate kinase-associated protein clustering | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 1 | 8 | 3.45e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 2 | 60 | 3.45e-01 |
| GO:BP | GO:1904914 | negative regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0045794 | negative regulation of cell volume | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 1 | 45 | 3.45e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 1 | 717 | 3.45e-01 |
| GO:BP | GO:0071494 | cellular response to UV-C | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0006253 | dCTP catabolic process | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0050859 | negative regulation of B cell receptor signaling pathway | 1 | 3 | 3.45e-01 |
| GO:BP | GO:0046065 | dCTP metabolic process | 1 | 1 | 3.45e-01 |
| GO:BP | GO:2000836 | positive regulation of androgen secretion | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0033320 | UDP-D-xylose biosynthetic process | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 11 | 14 | 3.45e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 6 | 16 | 3.45e-01 |
| GO:BP | GO:0051309 | female meiosis chromosome separation | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0006362 | transcription elongation by RNA polymerase I | 3 | 4 | 3.45e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 3 | 34 | 3.45e-01 |
| GO:BP | GO:0035935 | androgen secretion | 1 | 1 | 3.45e-01 |
| GO:BP | GO:1904850 | negative regulation of establishment of protein localization to telomere | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 3 | 4 | 3.45e-01 |
| GO:BP | GO:1904913 | regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 3.45e-01 |
| GO:BP | GO:2000834 | regulation of androgen secretion | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 6 | 40 | 3.45e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 17 | 33 | 3.45e-01 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 4 | 21 | 3.45e-01 |
| GO:BP | GO:0048645 | animal organ formation | 4 | 51 | 3.45e-01 |
| GO:BP | GO:1904910 | regulation of establishment of RNA localization to telomere | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0033319 | UDP-D-xylose metabolic process | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0003064 | regulation of heart rate by hormone | 2 | 2 | 3.45e-01 |
| GO:BP | GO:1904792 | positive regulation of shelterin complex assembly | 1 | 1 | 3.45e-01 |
| GO:BP | GO:1904911 | negative regulation of establishment of RNA localization to telomere | 1 | 1 | 3.45e-01 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 1 | 10 | 3.45e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 2 | 11 | 3.45e-01 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 3 | 3 | 3.45e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 3.45e-01 |
| GO:BP | GO:0051792 | medium-chain fatty acid biosynthetic process | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0048864 | stem cell development | 19 | 76 | 3.45e-01 |
| GO:BP | GO:0060745 | mammary gland branching involved in pregnancy | 2 | 5 | 3.45e-01 |
| GO:BP | GO:0071579 | regulation of zinc ion transport | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 54 | 679 | 3.45e-01 |
| GO:BP | GO:0140882 | zinc export across plasma membrane | 1 | 3 | 3.45e-01 |
| GO:BP | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade | 2 | 5 | 3.45e-01 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 1 | 11 | 3.45e-01 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 14 | 24 | 3.45e-01 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 1 | 11 | 3.45e-01 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 1 | 17 | 3.45e-01 |
| GO:BP | GO:0060702 | negative regulation of endoribonuclease activity | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0009164 | nucleoside catabolic process | 3 | 15 | 3.45e-01 |
| GO:BP | GO:0060701 | negative regulation of ribonuclease activity | 1 | 2 | 3.45e-01 |
| GO:BP | GO:0006304 | DNA modification | 17 | 89 | 3.45e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 2 | 14 | 3.45e-01 |
| GO:BP | GO:0045054 | constitutive secretory pathway | 3 | 7 | 3.46e-01 |
| GO:BP | GO:0016233 | telomere capping | 10 | 36 | 3.46e-01 |
| GO:BP | GO:0036514 | dopaminergic neuron axon guidance | 1 | 3 | 3.46e-01 |
| GO:BP | GO:0014820 | tonic smooth muscle contraction | 2 | 8 | 3.46e-01 |
| GO:BP | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0086098 | angiotensin-activated signaling pathway involved in heart process | 2 | 3 | 3.46e-01 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 2 | 9 | 3.46e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation | 1 | 1 | 3.46e-01 |
| GO:BP | GO:1990637 | response to prolactin | 2 | 3 | 3.46e-01 |
| GO:BP | GO:0150037 | regulation of calcium-dependent activation of synaptic vesicle fusion | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 5 | 26 | 3.46e-01 |
| GO:BP | GO:0060065 | uterus development | 3 | 13 | 3.46e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 58 | 251 | 3.46e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 3 | 37 | 3.46e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 3.46e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 1 | 8 | 3.46e-01 |
| GO:BP | GO:1905026 | positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.46e-01 |
| GO:BP | GO:1905033 | positive regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 2 | 3.46e-01 |
| GO:BP | GO:1904938 | planar cell polarity pathway involved in axon guidance | 1 | 3 | 3.46e-01 |
| GO:BP | GO:2000620 | positive regulation of histone H4-K16 acetylation | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 17 | 299 | 3.46e-01 |
| GO:BP | GO:0003138 | primary heart field specification | 1 | 3 | 3.46e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 7 | 17 | 3.46e-01 |
| GO:BP | GO:0003213 | cardiac right atrium morphogenesis | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 3.46e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 1 | 54 | 3.46e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0051136 | regulation of NK T cell differentiation | 1 | 4 | 3.46e-01 |
| GO:BP | GO:1903997 | positive regulation of non-membrane spanning protein tyrosine kinase activity | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0003097 | renal water transport | 3 | 6 | 3.46e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 3.46e-01 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 1 | 4 | 3.46e-01 |
| GO:BP | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 1 | 2 | 3.46e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 13 | 26 | 3.46e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 1 | 48 | 3.46e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 9 | 21 | 3.46e-01 |
| GO:BP | GO:0021658 | rhombomere 3 morphogenesis | 1 | 1 | 3.46e-01 |
| GO:BP | GO:0035519 | protein K29-linked ubiquitination | 3 | 6 | 3.46e-01 |
| GO:BP | GO:0090557 | establishment of endothelial intestinal barrier | 5 | 10 | 3.46e-01 |
| GO:BP | GO:0014040 | positive regulation of Schwann cell differentiation | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0021593 | rhombomere morphogenesis | 1 | 1 | 3.46e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 2 | 4 | 3.46e-01 |
| GO:BP | GO:1902178 | fibroblast growth factor receptor apoptotic signaling pathway | 1 | 1 | 3.46e-01 |
| GO:BP | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0090735 | DNA repair complex assembly | 2 | 3 | 3.46e-01 |
| GO:BP | GO:0009194 | pyrimidine ribonucleoside diphosphate biosynthetic process | 2 | 4 | 3.46e-01 |
| GO:BP | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0006225 | UDP biosynthetic process | 2 | 4 | 3.46e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 4 | 506 | 3.46e-01 |
| GO:BP | GO:0042311 | vasodilation | 3 | 39 | 3.46e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 3.46e-01 |
| GO:BP | GO:1903910 | negative regulation of receptor clustering | 1 | 2 | 3.46e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 6 | 18 | 3.46e-01 |
| GO:BP | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | 1 | 4 | 3.46e-01 |
| GO:BP | GO:0000730 | DNA recombinase assembly | 2 | 3 | 3.46e-01 |
| GO:BP | GO:0009650 | UV protection | 3 | 11 | 3.46e-01 |
| GO:BP | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0016358 | dendrite development | 21 | 202 | 3.46e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 12 | 210 | 3.46e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 7 | 48 | 3.46e-01 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 2 | 2 | 3.46e-01 |
| GO:BP | GO:0097501 | stress response to metal ion | 3 | 11 | 3.46e-01 |
| GO:BP | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway | 2 | 6 | 3.46e-01 |
| GO:BP | GO:0061687 | detoxification of inorganic compound | 3 | 12 | 3.46e-01 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 7 | 15 | 3.46e-01 |
| GO:BP | GO:0046684 | response to pyrethroid | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 1 | 93 | 3.47e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 3.47e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 5 | 7 | 3.47e-01 |
| GO:BP | GO:1904628 | cellular response to phorbol 13-acetate 12-myristate | 1 | 4 | 3.47e-01 |
| GO:BP | GO:1904627 | response to phorbol 13-acetate 12-myristate | 1 | 4 | 3.47e-01 |
| GO:BP | GO:0032074 | negative regulation of nuclease activity | 2 | 6 | 3.47e-01 |
| GO:BP | GO:0061668 | mitochondrial ribosome assembly | 4 | 8 | 3.47e-01 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 1 | 3 | 3.47e-01 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 5 | 6 | 3.47e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 3.47e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 1 | 24 | 3.47e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 1 | 67 | 3.47e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 4 | 42 | 3.47e-01 |
| GO:BP | GO:0052150 | modulation by symbiont of host apoptotic process | 3 | 3 | 3.47e-01 |
| GO:BP | GO:0007032 | endosome organization | 22 | 92 | 3.47e-01 |
| GO:BP | GO:0039526 | modulation by virus of host apoptotic process | 3 | 3 | 3.47e-01 |
| GO:BP | GO:0035695 | mitophagy by induced vacuole formation | 1 | 1 | 3.47e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 6 | 11 | 3.47e-01 |
| GO:BP | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway | 2 | 2 | 3.47e-01 |
| GO:BP | GO:0086101 | endothelin receptor signaling pathway involved in heart process | 1 | 3 | 3.47e-01 |
| GO:BP | GO:0051257 | meiotic spindle midzone assembly | 1 | 2 | 3.47e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 73 | 133 | 3.47e-01 |
| GO:BP | GO:0052040 | modulation by symbiont of host programmed cell death | 3 | 3 | 3.47e-01 |
| GO:BP | GO:0042305 | specification of segmental identity, mandibular segment | 1 | 3 | 3.47e-01 |
| GO:BP | GO:0031175 | neuron projection development | 2 | 780 | 3.47e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 5 | 10 | 3.47e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 14 | 39 | 3.48e-01 |
| GO:BP | GO:0003162 | atrioventricular node development | 6 | 8 | 3.48e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 5 | 10 | 3.48e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 3 | 133 | 3.48e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 50 | 121 | 3.48e-01 |
| GO:BP | GO:0015793 | glycerol transmembrane transport | 3 | 4 | 3.48e-01 |
| GO:BP | GO:0016189 | synaptic vesicle to endosome fusion | 3 | 3 | 3.48e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 5 | 10 | 3.48e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 20 | 32 | 3.49e-01 |
| GO:BP | GO:2001188 | regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 2 | 3.49e-01 |
| GO:BP | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 2 | 2 | 3.49e-01 |
| GO:BP | GO:0090246 | convergent extension involved in somitogenesis | 1 | 1 | 3.49e-01 |
| GO:BP | GO:0044345 | stromal-epithelial cell signaling involved in prostate gland development | 1 | 1 | 3.49e-01 |
| GO:BP | GO:2000080 | negative regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 3.49e-01 |
| GO:BP | GO:0061053 | somite development | 10 | 63 | 3.49e-01 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 3.49e-01 |
| GO:BP | GO:1902038 | positive regulation of hematopoietic stem cell differentiation | 1 | 1 | 3.49e-01 |
| GO:BP | GO:0009838 | abscission | 2 | 6 | 3.49e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 11 | 64 | 3.49e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 2 | 65 | 3.49e-01 |
| GO:BP | GO:0032053 | ciliary basal body organization | 4 | 4 | 3.49e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 1 | 7 | 3.49e-01 |
| GO:BP | GO:0060166 | olfactory pit development | 1 | 1 | 3.50e-01 |
| GO:BP | GO:2000302 | positive regulation of synaptic vesicle exocytosis | 2 | 3 | 3.50e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 1 | 787 | 3.50e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 1 | 11 | 3.50e-01 |
| GO:BP | GO:0000917 | division septum assembly | 1 | 1 | 3.50e-01 |
| GO:BP | GO:0090529 | cell septum assembly | 1 | 1 | 3.50e-01 |
| GO:BP | GO:0030397 | membrane disassembly | 1 | 11 | 3.50e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 15 | 45 | 3.51e-01 |
| GO:BP | GO:0006421 | asparaginyl-tRNA aminoacylation | 3 | 3 | 3.51e-01 |
| GO:BP | GO:0008219 | cell death | 82 | 1596 | 3.51e-01 |
| GO:BP | GO:0032775 | DNA methylation on adenine | 2 | 2 | 3.52e-01 |
| GO:BP | GO:1904428 | negative regulation of tubulin deacetylation | 4 | 6 | 3.52e-01 |
| GO:BP | GO:1905774 | regulation of DNA helicase activity | 2 | 5 | 3.52e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 3.52e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 3 | 48 | 3.52e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 3.53e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 8 | 20 | 3.53e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 3.53e-01 |
| GO:BP | GO:0006833 | water transport | 6 | 10 | 3.53e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 5 | 51 | 3.53e-01 |
| GO:BP | GO:0009751 | response to salicylic acid | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 34 | 272 | 3.53e-01 |
| GO:BP | GO:0097576 | vacuole fusion | 1 | 12 | 3.53e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 3.53e-01 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 5 | 8 | 3.53e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 4 | 50 | 3.53e-01 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 5 | 12 | 3.53e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 2 | 14 | 3.53e-01 |
| GO:BP | GO:1903126 | negative regulation of centriole-centriole cohesion | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0003012 | muscle system process | 17 | 334 | 3.53e-01 |
| GO:BP | GO:0014740 | negative regulation of muscle hyperplasia | 1 | 1 | 3.53e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 49 | 89 | 3.54e-01 |
| GO:BP | GO:0035377 | transepithelial water transport | 2 | 2 | 3.54e-01 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 4 | 10 | 3.54e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 1 | 19 | 3.55e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 3.55e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 13 | 46 | 3.55e-01 |
| GO:BP | GO:0051106 | positive regulation of DNA ligation | 3 | 5 | 3.55e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 13 | 46 | 3.55e-01 |
| GO:BP | GO:1904631 | response to glucoside | 1 | 1 | 3.56e-01 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 5 | 16 | 3.56e-01 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 6 | 8 | 3.56e-01 |
| GO:BP | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response | 1 | 2 | 3.56e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 2 | 17 | 3.56e-01 |
| GO:BP | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential | 3 | 10 | 3.56e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 1 | 121 | 3.56e-01 |
| GO:BP | GO:0002540 | leukotriene production involved in inflammatory response | 1 | 2 | 3.56e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 7 | 10 | 3.57e-01 |
| GO:BP | GO:0046452 | dihydrofolate metabolic process | 1 | 3 | 3.57e-01 |
| GO:BP | GO:1902915 | negative regulation of protein polyubiquitination | 4 | 9 | 3.57e-01 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 11 | 15 | 3.58e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 3.58e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 14 | 194 | 3.58e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 1 | 2 | 3.58e-01 |
| GO:BP | GO:0070537 | histone H2A K63-linked deubiquitination | 2 | 8 | 3.58e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 5 | 6 | 3.60e-01 |
| GO:BP | GO:0061880 | regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 3.60e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 3.60e-01 |
| GO:BP | GO:1901656 | glycoside transport | 1 | 5 | 3.60e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 3.60e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 6 | 14 | 3.60e-01 |
| GO:BP | GO:0061764 | late endosome to lysosome transport via multivesicular body sorting pathway | 2 | 2 | 3.60e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 3.60e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 3 | 28 | 3.60e-01 |
| GO:BP | GO:0061881 | positive regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 3.60e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 5 | 6 | 3.60e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 6 | 14 | 3.60e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 2 | 14 | 3.60e-01 |
| GO:BP | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation | 2 | 2 | 3.60e-01 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 1 | 6 | 3.60e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 10 | 40 | 3.60e-01 |
| GO:BP | GO:1903566 | positive regulation of protein localization to cilium | 5 | 7 | 3.60e-01 |
| GO:BP | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development | 7 | 13 | 3.60e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 9 | 149 | 3.60e-01 |
| GO:BP | GO:0044722 | renal phosphate excretion | 1 | 2 | 3.60e-01 |
| GO:BP | GO:1903402 | regulation of renal phosphate excretion | 1 | 2 | 3.60e-01 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 2 | 10 | 3.60e-01 |
| GO:BP | GO:0032416 | negative regulation of sodium:proton antiporter activity | 1 | 1 | 3.60e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 26 | 48 | 3.60e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 4 | 14 | 3.60e-01 |
| GO:BP | GO:0042631 | cellular response to water deprivation | 1 | 1 | 3.61e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 5 | 487 | 3.61e-01 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 1 | 3 | 3.61e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 7 | 46 | 3.61e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 1 | 258 | 3.61e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 17 | 29 | 3.61e-01 |
| GO:BP | GO:0006272 | leading strand elongation | 2 | 4 | 3.61e-01 |
| GO:BP | GO:0043605 | amide catabolic process | 1 | 10 | 3.61e-01 |
| GO:BP | GO:0035601 | protein deacylation | 46 | 116 | 3.62e-01 |
| GO:BP | GO:0036359 | renal potassium excretion | 2 | 2 | 3.62e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 10 | 13 | 3.62e-01 |
| GO:BP | GO:0070384 | Harderian gland development | 2 | 4 | 3.62e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 3.62e-01 |
| GO:BP | GO:0060056 | mammary gland involution | 3 | 8 | 3.62e-01 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 5 | 26 | 3.62e-01 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 5 | 26 | 3.62e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 13 | 135 | 3.62e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 3.62e-01 |
| GO:BP | GO:0051660 | establishment of centrosome localization | 6 | 8 | 3.62e-01 |
| GO:BP | GO:0014904 | myotube cell development | 2 | 34 | 3.62e-01 |
| GO:BP | GO:0038018 | Wnt receptor catabolic process | 2 | 2 | 3.62e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 3 | 4 | 3.63e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 26 | 441 | 3.63e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 11 | 198 | 3.63e-01 |
| GO:BP | GO:0032049 | cardiolipin biosynthetic process | 6 | 7 | 3.63e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 2 | 23 | 3.63e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 6 | 9 | 3.63e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 6 | 9 | 3.63e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 3 | 111 | 3.63e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 2 | 31 | 3.63e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 7 | 54 | 3.64e-01 |
| GO:BP | GO:0006222 | UMP biosynthetic process | 2 | 8 | 3.64e-01 |
| GO:BP | GO:0048104 | establishment of body hair or bristle planar orientation | 2 | 3 | 3.64e-01 |
| GO:BP | GO:0048852 | diencephalon morphogenesis | 2 | 2 | 3.64e-01 |
| GO:BP | GO:0048105 | establishment of body hair planar orientation | 2 | 3 | 3.64e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 35 | 120 | 3.64e-01 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 3 | 4 | 3.64e-01 |
| GO:BP | GO:0006666 | 3-keto-sphinganine metabolic process | 1 | 1 | 3.64e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 4 | 54 | 3.64e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 9 | 41 | 3.64e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 5 | 10 | 3.64e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 1 | 70 | 3.64e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 3.64e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 3.64e-01 |
| GO:BP | GO:0003105 | negative regulation of glomerular filtration | 1 | 3 | 3.65e-01 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 5 | 8 | 3.65e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 3.65e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 3 | 11 | 3.65e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 4 | 58 | 3.65e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 132 | 3.65e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 2 | 20 | 3.65e-01 |
| GO:BP | GO:0036213 | contractile ring contraction | 1 | 1 | 3.66e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 2 | 6 | 3.66e-01 |
| GO:BP | GO:0000916 | actomyosin contractile ring contraction | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 4 | 25 | 3.66e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 3.66e-01 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 3.66e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 15 | 167 | 3.66e-01 |
| GO:BP | GO:0070293 | renal absorption | 7 | 24 | 3.66e-01 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 3.66e-01 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 3.66e-01 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 6 | 99 | 3.66e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 15 | 65 | 3.66e-01 |
| GO:BP | GO:2001178 | positive regulation of mediator complex assembly | 1 | 1 | 3.67e-01 |
| GO:BP | GO:2001176 | regulation of mediator complex assembly | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 2 | 20 | 3.67e-01 |
| GO:BP | GO:0097698 | telomere maintenance via base-excision repair | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0036034 | mediator complex assembly | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0120094 | negative regulation of peptidyl-lysine crotonylation | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0072239 | metanephric glomerulus vasculature development | 3 | 6 | 3.67e-01 |
| GO:BP | GO:0140131 | positive regulation of lymphocyte chemotaxis | 2 | 10 | 3.67e-01 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 10 | 34 | 3.67e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 13 | 35 | 3.67e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 3 | 13 | 3.67e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 3 | 13 | 3.67e-01 |
| GO:BP | GO:0120093 | regulation of peptidyl-lysine crotonylation | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0048290 | isotype switching to IgA isotypes | 1 | 4 | 3.67e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 1 | 29 | 3.67e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 2 | 8 | 3.67e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 3 | 27 | 3.67e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 7 | 44 | 3.67e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 11 | 133 | 3.67e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 3.67e-01 |
| GO:BP | GO:0001951 | intestinal D-glucose absorption | 2 | 3 | 3.67e-01 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 2 | 209 | 3.67e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 7 | 784 | 3.67e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 2 | 3 | 3.67e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 2 | 10 | 3.67e-01 |
| GO:BP | GO:1903019 | negative regulation of glycoprotein metabolic process | 2 | 12 | 3.67e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 65 | 119 | 3.67e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 8 | 216 | 3.67e-01 |
| GO:BP | GO:0060682 | primary ureteric bud growth | 1 | 1 | 3.67e-01 |
| GO:BP | GO:0032237 | activation of store-operated calcium channel activity | 2 | 5 | 3.67e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 3 | 4 | 3.67e-01 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 1 | 11 | 3.67e-01 |
| GO:BP | GO:0006386 | termination of RNA polymerase III transcription | 1 | 1 | 3.68e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 2 | 11 | 3.68e-01 |
| GO:BP | GO:0015854 | guanine transport | 1 | 2 | 3.68e-01 |
| GO:BP | GO:1902308 | regulation of peptidyl-serine dephosphorylation | 3 | 3 | 3.68e-01 |
| GO:BP | GO:0060083 | smooth muscle contraction involved in micturition | 1 | 2 | 3.68e-01 |
| GO:BP | GO:0061586 | positive regulation of transcription by transcription factor localization | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0035344 | hypoxanthine transport | 1 | 2 | 3.68e-01 |
| GO:BP | GO:0099156 | cell-cell signaling via exosome | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0006829 | zinc ion transport | 4 | 23 | 3.68e-01 |
| GO:BP | GO:0045110 | intermediate filament bundle assembly | 4 | 6 | 3.68e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 4 | 10 | 3.68e-01 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 2 | 6 | 3.68e-01 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 3 | 4 | 3.68e-01 |
| GO:BP | GO:0055014 | atrial cardiac muscle cell development | 3 | 3 | 3.68e-01 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 2 | 3 | 3.68e-01 |
| GO:BP | GO:0055011 | atrial cardiac muscle cell differentiation | 3 | 3 | 3.68e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 10 | 21 | 3.68e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 3.68e-01 |
| GO:BP | GO:0072230 | metanephric proximal straight tubule development | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0072232 | metanephric proximal convoluted tubule segment 2 development | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0072220 | metanephric descending thin limb development | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0097089 | methyl-branched fatty acid metabolic process | 1 | 3 | 3.68e-01 |
| GO:BP | GO:0072032 | proximal convoluted tubule segment 2 development | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0072022 | descending thin limb development | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0072020 | proximal straight tubule development | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 4 | 32 | 3.68e-01 |
| GO:BP | GO:0007517 | muscle organ development | 4 | 277 | 3.68e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 14 | 21 | 3.68e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 14 | 191 | 3.68e-01 |
| GO:BP | GO:1990743 | protein sialylation | 1 | 5 | 3.68e-01 |
| GO:BP | GO:1901628 | positive regulation of postsynaptic membrane organization | 1 | 2 | 3.68e-01 |
| GO:BP | GO:0035936 | testosterone secretion | 2 | 2 | 3.68e-01 |
| GO:BP | GO:2000845 | positive regulation of testosterone secretion | 2 | 2 | 3.68e-01 |
| GO:BP | GO:2000843 | regulation of testosterone secretion | 2 | 2 | 3.68e-01 |
| GO:BP | GO:2000310 | regulation of NMDA receptor activity | 5 | 19 | 3.68e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 2 | 7 | 3.68e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 50 | 494 | 3.68e-01 |
| GO:BP | GO:1904719 | positive regulation of AMPA glutamate receptor clustering | 1 | 2 | 3.68e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 2 | 33 | 3.68e-01 |
| GO:BP | GO:1904046 | negative regulation of vascular endothelial growth factor production | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 3 | 49 | 3.68e-01 |
| GO:BP | GO:0030954 | astral microtubule nucleation | 1 | 1 | 3.68e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 1 | 19 | 3.68e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 5 | 33 | 3.68e-01 |
| GO:BP | GO:0006936 | muscle contraction | 14 | 267 | 3.68e-01 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 2 | 5 | 3.68e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 8 | 149 | 3.68e-01 |
| GO:BP | GO:1903772 | regulation of viral budding via host ESCRT complex | 2 | 4 | 3.68e-01 |
| GO:BP | GO:0090069 | regulation of ribosome biogenesis | 3 | 5 | 3.68e-01 |
| GO:BP | GO:1903173 | fatty alcohol metabolic process | 2 | 2 | 3.68e-01 |
| GO:BP | GO:1900372 | negative regulation of purine nucleotide biosynthetic process | 2 | 3 | 3.68e-01 |
| GO:BP | GO:0006971 | hypotonic response | 3 | 10 | 3.68e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 5 | 16 | 3.68e-01 |
| GO:BP | GO:0033306 | phytol metabolic process | 2 | 2 | 3.68e-01 |
| GO:BP | GO:0001765 | membrane raft assembly | 1 | 9 | 3.68e-01 |
| GO:BP | GO:0035552 | oxidative single-stranded DNA demethylation | 2 | 3 | 3.68e-01 |
| GO:BP | GO:0062043 | positive regulation of cardiac epithelial to mesenchymal transition | 1 | 9 | 3.68e-01 |
| GO:BP | GO:0030809 | negative regulation of nucleotide biosynthetic process | 2 | 3 | 3.68e-01 |
| GO:BP | GO:0015823 | phenylalanine transport | 2 | 4 | 3.69e-01 |
| GO:BP | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process | 2 | 2 | 3.69e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 3 | 18 | 3.69e-01 |
| GO:BP | GO:0003096 | renal sodium ion transport | 2 | 10 | 3.69e-01 |
| GO:BP | GO:0140039 | cell-cell adhesion in response to extracellular stimulus | 1 | 2 | 3.69e-01 |
| GO:BP | GO:1904180 | negative regulation of membrane depolarization | 3 | 6 | 3.70e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 1 | 898 | 3.70e-01 |
| GO:BP | GO:1905936 | regulation of germ cell proliferation | 3 | 5 | 3.70e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 14 | 25 | 3.70e-01 |
| GO:BP | GO:0090149 | mitochondrial membrane fission | 1 | 2 | 3.70e-01 |
| GO:BP | GO:0003373 | dynamin family protein polymerization involved in membrane fission | 1 | 2 | 3.70e-01 |
| GO:BP | GO:0003374 | dynamin family protein polymerization involved in mitochondrial fission | 1 | 2 | 3.70e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 20 | 214 | 3.70e-01 |
| GO:BP | GO:0003192 | mitral valve formation | 3 | 3 | 3.71e-01 |
| GO:BP | GO:0007620 | copulation | 1 | 10 | 3.71e-01 |
| GO:BP | GO:0071864 | positive regulation of cell proliferation in bone marrow | 2 | 5 | 3.71e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 8 | 229 | 3.71e-01 |
| GO:BP | GO:0140018 | regulation of cytoplasmic translational fidelity | 1 | 1 | 3.71e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 6 | 19 | 3.71e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 38 | 357 | 3.71e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 2 | 19 | 3.72e-01 |
| GO:BP | GO:0030210 | heparin biosynthetic process | 1 | 9 | 3.72e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 2 | 19 | 3.72e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 4 | 8 | 3.72e-01 |
| GO:BP | GO:0007379 | segment specification | 2 | 14 | 3.72e-01 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 13 | 35 | 3.72e-01 |
| GO:BP | GO:0030091 | protein repair | 3 | 7 | 3.72e-01 |
| GO:BP | GO:0001705 | ectoderm formation | 3 | 3 | 3.72e-01 |
| GO:BP | GO:1903613 | regulation of protein tyrosine phosphatase activity | 1 | 4 | 3.72e-01 |
| GO:BP | GO:0097510 | base-excision repair, AP site formation via deaminated base removal | 1 | 1 | 3.72e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 2 | 218 | 3.72e-01 |
| GO:BP | GO:0031128 | developmental induction | 18 | 26 | 3.72e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 1 | 8 | 3.72e-01 |
| GO:BP | GO:0033387 | putrescine biosynthetic process from ornithine | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 5 | 18 | 3.73e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 11 | 70 | 3.73e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 11 | 70 | 3.73e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 1 | 27 | 3.73e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 5 | 18 | 3.73e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 7 | 15 | 3.73e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 5 | 56 | 3.74e-01 |
| GO:BP | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure | 2 | 14 | 3.74e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 5 | 52 | 3.74e-01 |
| GO:BP | GO:0070159 | mitochondrial threonyl-tRNA aminoacylation | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 4 | 23 | 3.74e-01 |
| GO:BP | GO:0001957 | intramembranous ossification | 1 | 7 | 3.74e-01 |
| GO:BP | GO:0036072 | direct ossification | 1 | 7 | 3.74e-01 |
| GO:BP | GO:0035499 | carnosine biosynthetic process | 1 | 1 | 3.76e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 14 | 39 | 3.76e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 13 | 127 | 3.77e-01 |
| GO:BP | GO:0002946 | tRNA C5-cytosine methylation | 3 | 3 | 3.77e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 4 | 12 | 3.77e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 22 | 73 | 3.77e-01 |
| GO:BP | GO:0006885 | regulation of pH | 1 | 74 | 3.77e-01 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 2 | 5 | 3.77e-01 |
| GO:BP | GO:0021700 | developmental maturation | 12 | 211 | 3.77e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 1 | 4 | 3.78e-01 |
| GO:BP | GO:0007495 | visceral mesoderm-endoderm interaction involved in midgut development | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0070934 | CRD-mediated mRNA stabilization | 7 | 11 | 3.78e-01 |
| GO:BP | GO:0010609 | mRNA localization resulting in post-transcriptional regulation of gene expression | 1 | 2 | 3.78e-01 |
| GO:BP | GO:0070291 | N-acylethanolamine metabolic process | 1 | 4 | 3.78e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 2 | 223 | 3.78e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 4 | 7 | 3.78e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0044313 | protein K6-linked deubiquitination | 2 | 3 | 3.78e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 5 | 54 | 3.78e-01 |
| GO:BP | GO:0003285 | septum secundum development | 3 | 3 | 3.78e-01 |
| GO:BP | GO:0099555 | trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 1 | 2 | 3.78e-01 |
| GO:BP | GO:0032815 | negative regulation of natural killer cell activation | 1 | 3 | 3.78e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 1 | 28 | 3.78e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 11 | 198 | 3.78e-01 |
| GO:BP | GO:0099554 | trans-synaptic signaling by soluble gas, modulating synaptic transmission | 1 | 2 | 3.78e-01 |
| GO:BP | GO:0098924 | retrograde trans-synaptic signaling by nitric oxide | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0098923 | retrograde trans-synaptic signaling by soluble gas | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0035694 | mitochondrial protein catabolic process | 2 | 6 | 3.78e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 3.78e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 3 | 3 | 3.78e-01 |
| GO:BP | GO:0032915 | positive regulation of transforming growth factor beta2 production | 2 | 3 | 3.78e-01 |
| GO:BP | GO:2001108 | positive regulation of Rho guanyl-nucleotide exchange factor activity | 2 | 2 | 3.78e-01 |
| GO:BP | GO:0046836 | glycolipid transport | 6 | 7 | 3.78e-01 |
| GO:BP | GO:1905099 | positive regulation of guanyl-nucleotide exchange factor activity | 2 | 2 | 3.78e-01 |
| GO:BP | GO:1901503 | ether biosynthetic process | 3 | 11 | 3.78e-01 |
| GO:BP | GO:0043276 | anoikis | 11 | 32 | 3.78e-01 |
| GO:BP | GO:0043200 | response to amino acid | 11 | 95 | 3.78e-01 |
| GO:BP | GO:0015881 | creatine transmembrane transport | 3 | 4 | 3.78e-01 |
| GO:BP | GO:1903964 | monounsaturated fatty acid metabolic process | 1 | 2 | 3.78e-01 |
| GO:BP | GO:0007420 | brain development | 8 | 591 | 3.78e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 2 | 24 | 3.78e-01 |
| GO:BP | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway | 2 | 3 | 3.78e-01 |
| GO:BP | GO:0001967 | suckling behavior | 8 | 10 | 3.78e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 5 | 32 | 3.78e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 11 | 19 | 3.79e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 3.79e-01 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 1 | 9 | 3.79e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 35 | 108 | 3.79e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 2 | 224 | 3.79e-01 |
| GO:BP | GO:0007000 | nucleolus organization | 3 | 17 | 3.79e-01 |
| GO:BP | GO:0061205 | paramesonephric duct development | 2 | 4 | 3.79e-01 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 1 | 4 | 3.79e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 10 | 16 | 3.79e-01 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 6 | 8 | 3.79e-01 |
| GO:BP | GO:0071955 | recycling endosome to Golgi transport | 1 | 2 | 3.79e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 2 | 4 | 3.79e-01 |
| GO:BP | GO:1904782 | negative regulation of NMDA glutamate receptor activity | 2 | 2 | 3.79e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 13 | 165 | 3.79e-01 |
| GO:BP | GO:0000056 | ribosomal small subunit export from nucleus | 4 | 8 | 3.80e-01 |
| GO:BP | GO:0010424 | DNA methylation on cytosine within a CG sequence | 1 | 1 | 3.80e-01 |
| GO:BP | GO:1904438 | regulation of iron ion import across plasma membrane | 1 | 1 | 3.80e-01 |
| GO:BP | GO:1905344 | prostaglandin catabolic process | 1 | 2 | 3.80e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 153 | 3.80e-01 |
| GO:BP | GO:0046755 | viral budding | 8 | 26 | 3.80e-01 |
| GO:BP | GO:0062232 | prostanoid catabolic process | 1 | 2 | 3.80e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 3.81e-01 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 3 | 23 | 3.81e-01 |
| GO:BP | GO:0046832 | negative regulation of RNA export from nucleus | 2 | 2 | 3.81e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 3 | 17 | 3.81e-01 |
| GO:BP | GO:0060049 | regulation of protein glycosylation | 2 | 10 | 3.81e-01 |
| GO:BP | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | 1 | 4 | 3.82e-01 |
| GO:BP | GO:1901321 | positive regulation of heart induction | 1 | 4 | 3.82e-01 |
| GO:BP | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | 1 | 3 | 3.82e-01 |
| GO:BP | GO:0060686 | negative regulation of prostatic bud formation | 1 | 4 | 3.82e-01 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 2 | 10 | 3.82e-01 |
| GO:BP | GO:0031915 | positive regulation of synaptic plasticity | 5 | 6 | 3.82e-01 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 4 | 14 | 3.82e-01 |
| GO:BP | GO:0006595 | polyamine metabolic process | 2 | 16 | 3.82e-01 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 1 | 13 | 3.82e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 1 | 13 | 3.82e-01 |
| GO:BP | GO:1903429 | regulation of cell maturation | 2 | 13 | 3.82e-01 |
| GO:BP | GO:0006424 | glutamyl-tRNA aminoacylation | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0014816 | skeletal muscle satellite cell differentiation | 5 | 6 | 3.83e-01 |
| GO:BP | GO:0006433 | prolyl-tRNA aminoacylation | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0035862 | dITP metabolic process | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0035863 | dITP catabolic process | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0032352 | positive regulation of hormone metabolic process | 4 | 11 | 3.83e-01 |
| GO:BP | GO:0090435 | protein localization to nuclear envelope | 1 | 14 | 3.83e-01 |
| GO:BP | GO:0055071 | manganese ion homeostasis | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0006248 | CMP catabolic process | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0097018 | renal albumin absorption | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 5 | 16 | 3.83e-01 |
| GO:BP | GO:0006203 | dGTP catabolic process | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 2 | 13 | 3.83e-01 |
| GO:BP | GO:0009790 | embryo development | 2 | 875 | 3.83e-01 |
| GO:BP | GO:0072011 | glomerular endothelium development | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0035287 | head segmentation | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0072739 | response to anisomycin | 1 | 4 | 3.83e-01 |
| GO:BP | GO:0035289 | posterior head segmentation | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0034478 | phosphatidylglycerol catabolic process | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 8 | 11 | 3.83e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 30 | 71 | 3.83e-01 |
| GO:BP | GO:0097497 | blood vessel endothelial cell delamination | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0015818 | isoleucine transport | 1 | 5 | 3.83e-01 |
| GO:BP | GO:0097492 | sympathetic neuron axon guidance | 1 | 4 | 3.83e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 1 | 7 | 3.83e-01 |
| GO:BP | GO:0010826 | negative regulation of centrosome duplication | 8 | 12 | 3.83e-01 |
| GO:BP | GO:1903210 | podocyte apoptotic process | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0046967 | cytosol to endoplasmic reticulum transport | 2 | 4 | 3.83e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 2 | 14 | 3.83e-01 |
| GO:BP | GO:1903804 | glycine import across plasma membrane | 1 | 4 | 3.83e-01 |
| GO:BP | GO:0044857 | plasma membrane raft organization | 1 | 9 | 3.83e-01 |
| GO:BP | GO:0030103 | vasopressin secretion | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0051593 | response to folic acid | 2 | 9 | 3.83e-01 |
| GO:BP | GO:0046050 | UMP catabolic process | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0051455 | monopolar spindle attachment to meiosis I kinetochore | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 3.83e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 2 | 32 | 3.83e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 1 | 16 | 3.83e-01 |
| GO:BP | GO:0050655 | dermatan sulfate proteoglycan metabolic process | 1 | 12 | 3.83e-01 |
| GO:BP | GO:1905222 | atrioventricular canal morphogenesis | 2 | 4 | 3.83e-01 |
| GO:BP | GO:0046606 | negative regulation of centrosome cycle | 8 | 12 | 3.83e-01 |
| GO:BP | GO:0048666 | neuron development | 2 | 877 | 3.83e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 3.83e-01 |
| GO:BP | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway | 2 | 2 | 3.83e-01 |
| GO:BP | GO:1905458 | positive regulation of lymphoid progenitor cell differentiation | 3 | 3 | 3.83e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 38 | 678 | 3.83e-01 |
| GO:BP | GO:0038129 | ERBB3 signaling pathway | 2 | 4 | 3.83e-01 |
| GO:BP | GO:0038135 | ERBB2-ERBB4 signaling pathway | 2 | 4 | 3.83e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 3.83e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 6 | 63 | 3.83e-01 |
| GO:BP | GO:0140042 | lipid droplet formation | 8 | 12 | 3.83e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 5 | 60 | 3.83e-01 |
| GO:BP | GO:1900063 | regulation of peroxisome organization | 3 | 4 | 3.83e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 57 | 96 | 3.83e-01 |
| GO:BP | GO:0007522 | visceral muscle development | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0062042 | regulation of cardiac epithelial to mesenchymal transition | 1 | 10 | 3.83e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 2 | 25 | 3.83e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 3.83e-01 |
| GO:BP | GO:0062026 | negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0062025 | regulation of SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0007380 | specification of segmental identity, head | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 2 | 18 | 3.83e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 4 | 30 | 3.83e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 3.83e-01 |
| GO:BP | GO:1903810 | L-histidine import across plasma membrane | 2 | 2 | 3.83e-01 |
| GO:BP | GO:1905420 | vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 3.83e-01 |
| GO:BP | GO:1903031 | regulation of microtubule plus-end binding | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0018872 | arsonoacetate metabolic process | 1 | 1 | 3.83e-01 |
| GO:BP | GO:1905070 | anterior visceral endoderm cell migration | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0090679 | cell differentiation involved in phenotypic switching | 1 | 3 | 3.83e-01 |
| GO:BP | GO:1905930 | regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 3 | 34 | 3.83e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 2 | 10 | 3.83e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 1 | 15 | 3.83e-01 |
| GO:BP | GO:1905915 | regulation of cell differentiation involved in phenotypic switching | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0001906 | cell killing | 4 | 69 | 3.83e-01 |
| GO:BP | GO:1903033 | positive regulation of microtubule plus-end binding | 2 | 2 | 3.83e-01 |
| GO:BP | GO:1905460 | negative regulation of vascular associated smooth muscle cell apoptotic process | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 15 | 52 | 3.83e-01 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 6 | 11 | 3.83e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 6 | 8 | 3.83e-01 |
| GO:BP | GO:0044154 | histone H3-K14 acetylation | 6 | 12 | 3.83e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 3 | 105 | 3.83e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 5 | 30 | 3.83e-01 |
| GO:BP | GO:0060232 | delamination | 2 | 5 | 3.83e-01 |
| GO:BP | GO:0055009 | atrial cardiac muscle tissue morphogenesis | 2 | 5 | 3.83e-01 |
| GO:BP | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 4 | 5 | 3.83e-01 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 5 | 7 | 3.83e-01 |
| GO:BP | GO:0048313 | Golgi inheritance | 4 | 15 | 3.83e-01 |
| GO:BP | GO:0061864 | basement membrane constituent secretion | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 12 | 105 | 3.83e-01 |
| GO:BP | GO:0061865 | polarized secretion of basement membrane proteins in epithelium | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0006500 | N-terminal protein palmitoylation | 2 | 3 | 3.83e-01 |
| GO:BP | GO:2000691 | negative regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0007072 | positive regulation of transcription involved in exit from mitosis | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 6 | 11 | 3.83e-01 |
| GO:BP | GO:0048308 | organelle inheritance | 4 | 15 | 3.83e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 2 | 17 | 3.83e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0006429 | leucyl-tRNA aminoacylation | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 3.83e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 1 | 130 | 3.83e-01 |
| GO:BP | GO:0016180 | snRNA processing | 4 | 26 | 3.83e-01 |
| GO:BP | GO:0006419 | alanyl-tRNA aminoacylation | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0006425 | glutaminyl-tRNA aminoacylation | 1 | 2 | 3.83e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 77 | 144 | 3.84e-01 |
| GO:BP | GO:0050658 | RNA transport | 77 | 144 | 3.84e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 2 | 12 | 3.84e-01 |
| GO:BP | GO:1901506 | regulation of acylglycerol transport | 1 | 2 | 3.84e-01 |
| GO:BP | GO:0032700 | negative regulation of interleukin-17 production | 1 | 9 | 3.84e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 3.84e-01 |
| GO:BP | GO:1905885 | positive regulation of triglyceride transport | 1 | 2 | 3.84e-01 |
| GO:BP | GO:0012501 | programmed cell death | 75 | 1479 | 3.84e-01 |
| GO:BP | GO:1905661 | regulation of telomerase RNA reverse transcriptase activity | 2 | 3 | 3.84e-01 |
| GO:BP | GO:1905883 | regulation of triglyceride transport | 1 | 2 | 3.84e-01 |
| GO:BP | GO:1901508 | positive regulation of acylglycerol transport | 1 | 2 | 3.84e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 2 | 60 | 3.84e-01 |
| GO:BP | GO:0015798 | myo-inositol transport | 2 | 3 | 3.84e-01 |
| GO:BP | GO:0060004 | reflex | 2 | 9 | 3.84e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 6 | 43 | 3.84e-01 |
| GO:BP | GO:0043602 | nitrate catabolic process | 1 | 1 | 3.84e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 2 | 35 | 3.84e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 9 | 80 | 3.84e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 1 | 4 | 3.84e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 2 | 4 | 3.84e-01 |
| GO:BP | GO:0072529 | pyrimidine-containing compound catabolic process | 6 | 29 | 3.84e-01 |
| GO:BP | GO:0070233 | negative regulation of T cell apoptotic process | 1 | 14 | 3.84e-01 |
| GO:BP | GO:0032601 | connective tissue growth factor production | 1 | 2 | 3.84e-01 |
| GO:BP | GO:2001045 | negative regulation of integrin-mediated signaling pathway | 1 | 3 | 3.84e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 36 | 663 | 3.84e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 2 | 870 | 3.84e-01 |
| GO:BP | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation | 1 | 1 | 3.84e-01 |
| GO:BP | GO:0032643 | regulation of connective tissue growth factor production | 1 | 2 | 3.84e-01 |
| GO:BP | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth | 1 | 2 | 3.84e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 2 | 5 | 3.84e-01 |
| GO:BP | GO:0032286 | central nervous system myelin maintenance | 3 | 3 | 3.84e-01 |
| GO:BP | GO:1903644 | regulation of chaperone-mediated protein folding | 3 | 3 | 3.84e-01 |
| GO:BP | GO:0046210 | nitric oxide catabolic process | 1 | 1 | 3.84e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 2 | 5 | 3.84e-01 |
| GO:BP | GO:0006915 | apoptotic process | 73 | 1443 | 3.84e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 8 | 75 | 3.84e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 2 | 418 | 3.84e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 5 | 37 | 3.84e-01 |
| GO:BP | GO:0110051 | metabolite repair | 1 | 1 | 3.84e-01 |
| GO:BP | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 3 | 3.85e-01 |
| GO:BP | GO:0006610 | ribosomal protein import into nucleus | 1 | 4 | 3.85e-01 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 10 | 29 | 3.85e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 2 | 32 | 3.85e-01 |
| GO:BP | GO:0046753 | non-lytic viral release | 1 | 14 | 3.85e-01 |
| GO:BP | GO:0016049 | cell growth | 32 | 418 | 3.85e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 2 | 6 | 3.85e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 3.86e-01 |
| GO:BP | GO:2001015 | negative regulation of skeletal muscle cell differentiation | 2 | 3 | 3.86e-01 |
| GO:BP | GO:0070221 | sulfide oxidation, using sulfide:quinone oxidoreductase | 1 | 3 | 3.86e-01 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 5 | 10 | 3.86e-01 |
| GO:BP | GO:0019418 | sulfide oxidation | 1 | 3 | 3.86e-01 |
| GO:BP | GO:0033750 | ribosome localization | 2 | 15 | 3.86e-01 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 2 | 15 | 3.86e-01 |
| GO:BP | GO:0009451 | RNA modification | 56 | 162 | 3.86e-01 |
| GO:BP | GO:0032349 | positive regulation of aldosterone biosynthetic process | 3 | 3 | 3.86e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 3.86e-01 |
| GO:BP | GO:0007174 | epidermal growth factor catabolic process | 1 | 2 | 3.86e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 3.86e-01 |
| GO:BP | GO:0032346 | positive regulation of aldosterone metabolic process | 3 | 3 | 3.86e-01 |
| GO:BP | GO:0010447 | response to acidic pH | 3 | 16 | 3.86e-01 |
| GO:BP | GO:0008216 | spermidine metabolic process | 1 | 7 | 3.86e-01 |
| GO:BP | GO:0003188 | heart valve formation | 10 | 16 | 3.86e-01 |
| GO:BP | GO:1905232 | cellular response to L-glutamate | 3 | 4 | 3.86e-01 |
| GO:BP | GO:0060383 | positive regulation of DNA strand elongation | 1 | 1 | 3.86e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 16 | 22 | 3.86e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 2 | 53 | 3.86e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 14 | 206 | 3.86e-01 |
| GO:BP | GO:0061743 | motor learning | 1 | 5 | 3.87e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 6 | 40 | 3.87e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 3 | 4 | 3.87e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 2 | 8 | 3.87e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 9 | 41 | 3.87e-01 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 3 | 24 | 3.87e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 14 | 85 | 3.87e-01 |
| GO:BP | GO:2000386 | positive regulation of ovarian follicle development | 2 | 2 | 3.87e-01 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 1 | 8 | 3.87e-01 |
| GO:BP | GO:0003219 | cardiac right ventricle formation | 3 | 4 | 3.87e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 5 | 33 | 3.87e-01 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 2 | 8 | 3.87e-01 |
| GO:BP | GO:0032070 | regulation of deoxyribonuclease activity | 3 | 9 | 3.87e-01 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 4 | 15 | 3.87e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 3 | 11 | 3.87e-01 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 4 | 15 | 3.88e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 6 | 11 | 3.88e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 3.88e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 3.88e-01 |
| GO:BP | GO:0046133 | pyrimidine ribonucleoside catabolic process | 2 | 7 | 3.90e-01 |
| GO:BP | GO:0046131 | pyrimidine ribonucleoside metabolic process | 2 | 7 | 3.90e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 17 | 183 | 3.90e-01 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 4 | 5 | 3.90e-01 |
| GO:BP | GO:0016080 | synaptic vesicle targeting | 3 | 3 | 3.90e-01 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 3 | 20 | 3.90e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 3 | 21 | 3.90e-01 |
| GO:BP | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells | 3 | 4 | 3.91e-01 |
| GO:BP | GO:0002892 | regulation of type II hypersensitivity | 2 | 3 | 3.91e-01 |
| GO:BP | GO:0060354 | negative regulation of cell adhesion molecule production | 3 | 5 | 3.91e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 20 | 151 | 3.91e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 9 | 24 | 3.91e-01 |
| GO:BP | GO:0001796 | regulation of type IIa hypersensitivity | 2 | 3 | 3.91e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 4 | 21 | 3.91e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 8 | 101 | 3.91e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 2 | 3 | 3.91e-01 |
| GO:BP | GO:0014038 | regulation of Schwann cell differentiation | 1 | 3 | 3.91e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 4 | 16 | 3.91e-01 |
| GO:BP | GO:0042415 | norepinephrine metabolic process | 2 | 7 | 3.91e-01 |
| GO:BP | GO:1905024 | regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.91e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 4 | 16 | 3.91e-01 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 9 | 33 | 3.91e-01 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 4 | 10 | 3.91e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 17 | 121 | 3.91e-01 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 9 | 33 | 3.91e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 3.91e-01 |
| GO:BP | GO:2000791 | negative regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 2 | 127 | 3.91e-01 |
| GO:BP | GO:0061502 | early endosome to recycling endosome transport | 1 | 3 | 3.91e-01 |
| GO:BP | GO:0019082 | viral protein processing | 2 | 31 | 3.91e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 11 | 246 | 3.91e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 2 | 2 | 3.91e-01 |
| GO:BP | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 3 | 39 | 3.91e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 9 | 53 | 3.92e-01 |
| GO:BP | GO:0016254 | preassembly of GPI anchor in ER membrane | 4 | 8 | 3.92e-01 |
| GO:BP | GO:0072347 | response to anesthetic | 1 | 1 | 3.92e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 4 | 371 | 3.92e-01 |
| GO:BP | GO:0032025 | response to cobalt ion | 3 | 10 | 3.92e-01 |
| GO:BP | GO:0071321 | cellular response to cGMP | 1 | 7 | 3.92e-01 |
| GO:BP | GO:0030238 | male sex determination | 4 | 8 | 3.93e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 25 | 188 | 3.93e-01 |
| GO:BP | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib | 2 | 4 | 3.93e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 7 | 13 | 3.93e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 2 | 21 | 3.93e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 6 | 40 | 3.94e-01 |
| GO:BP | GO:1990523 | bone regeneration | 1 | 2 | 3.94e-01 |
| GO:BP | GO:0030952 | establishment or maintenance of cytoskeleton polarity | 2 | 7 | 3.94e-01 |
| GO:BP | GO:0010722 | regulation of ferrochelatase activity | 1 | 1 | 3.94e-01 |
| GO:BP | GO:0070932 | histone H3 deacetylation | 5 | 12 | 3.94e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 4 | 36 | 3.94e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 41 | 147 | 3.94e-01 |
| GO:BP | GO:0070778 | L-aspartate transmembrane transport | 3 | 11 | 3.94e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 2 | 12 | 3.94e-01 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 3 | 6 | 3.94e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 3.94e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 6 | 34 | 3.94e-01 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 1 | 17 | 3.94e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 38 | 362 | 3.94e-01 |
| GO:BP | GO:2000639 | negative regulation of SREBP signaling pathway | 1 | 3 | 3.94e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 5 | 681 | 3.94e-01 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 2 | 18 | 3.94e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 3.95e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 3.95e-01 |
| GO:BP | GO:1903978 | regulation of microglial cell activation | 1 | 9 | 3.95e-01 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 4 | 22 | 3.95e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 1 | 21 | 3.95e-01 |
| GO:BP | GO:0090520 | sphingolipid mediated signaling pathway | 1 | 11 | 3.95e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 3 | 35 | 3.95e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 1 | 13 | 3.95e-01 |
| GO:BP | GO:0061739 | protein lipidation involved in autophagosome assembly | 2 | 3 | 3.95e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 3.95e-01 |
| GO:BP | GO:0006403 | RNA localization | 94 | 182 | 3.95e-01 |
| GO:BP | GO:0046898 | response to cycloheximide | 4 | 4 | 3.95e-01 |
| GO:BP | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | 2 | 2 | 3.96e-01 |
| GO:BP | GO:1900045 | negative regulation of protein K63-linked ubiquitination | 3 | 7 | 3.96e-01 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 2 | 14 | 3.96e-01 |
| GO:BP | GO:1903929 | primary palate development | 2 | 2 | 3.96e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 5 | 46 | 3.96e-01 |
| GO:BP | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | 1 | 3 | 3.96e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 1 | 7 | 3.97e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 3.97e-01 |
| GO:BP | GO:1990550 | mitochondrial alpha-ketoglutarate transmembrane transport | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 1 | 15 | 3.97e-01 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 2 | 18 | 3.97e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 21 | 551 | 3.97e-01 |
| GO:BP | GO:0002091 | negative regulation of receptor internalization | 3 | 14 | 3.97e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 9 | 23 | 3.97e-01 |
| GO:BP | GO:0034656 | nucleobase-containing small molecule catabolic process | 3 | 20 | 3.98e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 3 | 22 | 3.98e-01 |
| GO:BP | GO:0060429 | epithelium development | 2 | 850 | 3.98e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 4 | 14 | 3.98e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 2 | 2 | 3.98e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 13 | 1229 | 3.98e-01 |
| GO:BP | GO:1902685 | positive regulation of receptor localization to synapse | 3 | 3 | 3.98e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 25 | 282 | 3.98e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 7 | 31 | 3.99e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 38 | 186 | 3.99e-01 |
| GO:BP | GO:1904762 | positive regulation of myofibroblast differentiation | 1 | 1 | 3.99e-01 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 4 | 5 | 3.99e-01 |
| GO:BP | GO:0000098 | sulfur amino acid catabolic process | 2 | 7 | 3.99e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 13 | 19 | 3.99e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 3 | 7 | 3.99e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 6 | 39 | 3.99e-01 |
| GO:BP | GO:0062176 | R-loop processing | 4 | 6 | 4.00e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 10 | 265 | 4.00e-01 |
| GO:BP | GO:0031167 | rRNA methylation | 15 | 24 | 4.00e-01 |
| GO:BP | GO:0061072 | iris morphogenesis | 1 | 5 | 4.00e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 4 | 79 | 4.00e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 22 | 98 | 4.00e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 5 | 85 | 4.00e-01 |
| GO:BP | GO:0070676 | intralumenal vesicle formation | 2 | 4 | 4.00e-01 |
| GO:BP | GO:0033024 | mast cell apoptotic process | 2 | 3 | 4.00e-01 |
| GO:BP | GO:0033025 | regulation of mast cell apoptotic process | 2 | 3 | 4.00e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 4.00e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 2 | 7 | 4.00e-01 |
| GO:BP | GO:1905794 | response to puromycin | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0046886 | positive regulation of hormone biosynthetic process | 3 | 9 | 4.00e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 2 | 7 | 4.00e-01 |
| GO:BP | GO:1905795 | cellular response to puromycin | 1 | 1 | 4.00e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 3 | 12 | 4.00e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 8 | 23 | 4.01e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 4 | 4 | 4.01e-01 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 1 | 6 | 4.01e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 4.01e-01 |
| GO:BP | GO:0048641 | regulation of skeletal muscle tissue development | 1 | 20 | 4.02e-01 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 6 | 16 | 4.02e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 2 | 2 | 4.02e-01 |
| GO:BP | GO:0009173 | pyrimidine ribonucleoside monophosphate metabolic process | 4 | 12 | 4.02e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 4 | 14 | 4.02e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 12 | 84 | 4.02e-01 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 2 | 9 | 4.02e-01 |
| GO:BP | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis | 2 | 3 | 4.02e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 4.02e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 18 | 1239 | 4.03e-01 |
| GO:BP | GO:0072209 | metanephric mesangial cell differentiation | 1 | 1 | 4.03e-01 |
| GO:BP | GO:0072254 | metanephric glomerular mesangial cell differentiation | 1 | 1 | 4.03e-01 |
| GO:BP | GO:0098885 | modification of postsynaptic actin cytoskeleton | 3 | 10 | 4.03e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 22 | 74 | 4.03e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 1 | 4 | 4.03e-01 |
| GO:BP | GO:0035524 | proline transmembrane transport | 4 | 8 | 4.03e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 10 | 169 | 4.03e-01 |
| GO:BP | GO:0090210 | regulation of establishment of blood-brain barrier | 1 | 3 | 4.03e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 4 | 36 | 4.03e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 1 | 18 | 4.04e-01 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 3 | 8 | 4.04e-01 |
| GO:BP | GO:1901341 | positive regulation of store-operated calcium channel activity | 2 | 6 | 4.04e-01 |
| GO:BP | GO:0036445 | neuronal stem cell division | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0046600 | negative regulation of centriole replication | 4 | 7 | 4.05e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 5 | 13 | 4.05e-01 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 12 | 31 | 4.05e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 2 | 13 | 4.06e-01 |
| GO:BP | GO:0040008 | regulation of growth | 37 | 501 | 4.06e-01 |
| GO:BP | GO:0090400 | stress-induced premature senescence | 3 | 7 | 4.06e-01 |
| GO:BP | GO:0048703 | embryonic viscerocranium morphogenesis | 1 | 8 | 4.06e-01 |
| GO:BP | GO:0031120 | snRNA pseudouridine synthesis | 2 | 3 | 4.06e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 7 | 45 | 4.06e-01 |
| GO:BP | GO:0033979 | box H/ACA RNA metabolic process | 2 | 3 | 4.06e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 9 | 143 | 4.06e-01 |
| GO:BP | GO:0001759 | organ induction | 2 | 17 | 4.06e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 2 | 16 | 4.06e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 9 | 344 | 4.06e-01 |
| GO:BP | GO:1904274 | tricellular tight junction assembly | 1 | 1 | 4.06e-01 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 3 | 6 | 4.06e-01 |
| GO:BP | GO:0070342 | brown fat cell proliferation | 1 | 1 | 4.06e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 7 | 91 | 4.06e-01 |
| GO:BP | GO:2000755 | positive regulation of sphingomyelin catabolic process | 1 | 1 | 4.06e-01 |
| GO:BP | GO:0099627 | neurotransmitter receptor cycle | 1 | 1 | 4.06e-01 |
| GO:BP | GO:0099630 | postsynaptic neurotransmitter receptor cycle | 1 | 1 | 4.06e-01 |
| GO:BP | GO:2000754 | regulation of sphingomyelin catabolic process | 1 | 1 | 4.06e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 3 | 4 | 4.06e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 4 | 87 | 4.06e-01 |
| GO:BP | GO:0110113 | positive regulation of lipid transporter activity | 1 | 1 | 4.06e-01 |
| GO:BP | GO:0060574 | intestinal epithelial cell maturation | 2 | 3 | 4.06e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 7 | 47 | 4.06e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1 | 10 | 4.06e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 4.06e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 1 | 386 | 4.06e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 9 | 11 | 4.07e-01 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 13 | 21 | 4.07e-01 |
| GO:BP | GO:0009648 | photoperiodism | 18 | 28 | 4.08e-01 |
| GO:BP | GO:0006434 | seryl-tRNA aminoacylation | 2 | 2 | 4.08e-01 |
| GO:BP | GO:0044206 | UMP salvage | 1 | 4 | 4.08e-01 |
| GO:BP | GO:0032262 | pyrimidine nucleotide salvage | 1 | 4 | 4.08e-01 |
| GO:BP | GO:0006249 | dCMP catabolic process | 1 | 3 | 4.08e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 7 | 81 | 4.08e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 3 | 25 | 4.08e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 1 | 52 | 4.08e-01 |
| GO:BP | GO:0046079 | dUMP catabolic process | 1 | 4 | 4.08e-01 |
| GO:BP | GO:0046063 | dCMP metabolic process | 1 | 3 | 4.08e-01 |
| GO:BP | GO:0010138 | pyrimidine ribonucleotide salvage | 1 | 4 | 4.08e-01 |
| GO:BP | GO:0003241 | growth involved in heart morphogenesis | 3 | 4 | 4.08e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 5 | 696 | 4.08e-01 |
| GO:BP | GO:0009175 | pyrimidine ribonucleoside monophosphate catabolic process | 1 | 4 | 4.08e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 3 | 47 | 4.08e-01 |
| GO:BP | GO:0030576 | Cajal body organization | 2 | 4 | 4.08e-01 |
| GO:BP | GO:0032528 | microvillus organization | 4 | 19 | 4.08e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 4.09e-01 |
| GO:BP | GO:0036149 | phosphatidylinositol acyl-chain remodeling | 2 | 3 | 4.10e-01 |
| GO:BP | GO:0009611 | response to wounding | 2 | 415 | 4.10e-01 |
| GO:BP | GO:1903645 | negative regulation of chaperone-mediated protein folding | 2 | 2 | 4.10e-01 |
| GO:BP | GO:0042695 | thelarche | 1 | 5 | 4.10e-01 |
| GO:BP | GO:0030913 | paranodal junction assembly | 4 | 6 | 4.10e-01 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 1 | 5 | 4.10e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 4.10e-01 |
| GO:BP | GO:0060744 | mammary gland branching involved in thelarche | 1 | 5 | 4.10e-01 |
| GO:BP | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | 1 | 4 | 4.10e-01 |
| GO:BP | GO:0048022 | negative regulation of melanin biosynthetic process | 1 | 4 | 4.10e-01 |
| GO:BP | GO:0042640 | anagen | 1 | 5 | 4.10e-01 |
| GO:BP | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1 | 4 | 4.10e-01 |
| GO:BP | GO:0060751 | branch elongation involved in mammary gland duct branching | 1 | 4 | 4.10e-01 |
| GO:BP | GO:0051884 | regulation of timing of anagen | 1 | 5 | 4.10e-01 |
| GO:BP | GO:0097325 | melanocyte proliferation | 1 | 2 | 4.10e-01 |
| GO:BP | GO:0071863 | regulation of cell proliferation in bone marrow | 2 | 6 | 4.10e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 7 | 12 | 4.10e-01 |
| GO:BP | GO:0045105 | intermediate filament polymerization or depolymerization | 2 | 2 | 4.10e-01 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 2 | 2 | 4.10e-01 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 1 | 19 | 4.10e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 4.11e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 13 | 21 | 4.11e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 5 | 700 | 4.11e-01 |
| GO:BP | GO:0072223 | metanephric glomerular mesangium development | 2 | 3 | 4.12e-01 |
| GO:BP | GO:1903753 | negative regulation of p38MAPK cascade | 3 | 5 | 4.12e-01 |
| GO:BP | GO:1901647 | positive regulation of synoviocyte proliferation | 1 | 3 | 4.12e-01 |
| GO:BP | GO:0090125 | cell-cell adhesion involved in synapse maturation | 1 | 2 | 4.12e-01 |
| GO:BP | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | 5 | 10 | 4.12e-01 |
| GO:BP | GO:0002941 | synoviocyte proliferation | 1 | 3 | 4.12e-01 |
| GO:BP | GO:1901645 | regulation of synoviocyte proliferation | 1 | 3 | 4.12e-01 |
| GO:BP | GO:1903936 | cellular response to sodium arsenite | 1 | 5 | 4.12e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 3 | 54 | 4.12e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 5 | 7 | 4.12e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 4 | 36 | 4.12e-01 |
| GO:BP | GO:0036146 | cellular response to mycotoxin | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0072721 | cellular response to dithiothreitol | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 4 | 5 | 4.12e-01 |
| GO:BP | GO:1902990 | mitotic telomere maintenance via semi-conservative replication | 1 | 2 | 4.12e-01 |
| GO:BP | GO:0060035 | notochord cell development | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0060034 | notochord cell differentiation | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0035754 | B cell chemotaxis | 3 | 5 | 4.12e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 9 | 267 | 4.12e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 22 | 34 | 4.13e-01 |
| GO:BP | GO:0070483 | detection of hypoxia | 1 | 2 | 4.13e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 4 | 15 | 4.13e-01 |
| GO:BP | GO:0016926 | protein desumoylation | 1 | 9 | 4.13e-01 |
| GO:BP | GO:1904482 | cellular response to tetrahydrofolate | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 2 | 4 | 4.13e-01 |
| GO:BP | GO:1904481 | response to tetrahydrofolate | 1 | 1 | 4.13e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 2 | 71 | 4.13e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 1 | 27 | 4.13e-01 |
| GO:BP | GO:0106300 | protein-DNA covalent cross-linking repair | 2 | 7 | 4.13e-01 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 2 | 12 | 4.13e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 5 | 75 | 4.13e-01 |
| GO:BP | GO:0021979 | hypothalamus cell differentiation | 1 | 8 | 4.13e-01 |
| GO:BP | GO:0048865 | stem cell fate commitment | 3 | 5 | 4.13e-01 |
| GO:BP | GO:0060309 | elastin catabolic process | 2 | 2 | 4.13e-01 |
| GO:BP | GO:0051029 | rRNA transport | 2 | 5 | 4.13e-01 |
| GO:BP | GO:0040016 | embryonic cleavage | 2 | 7 | 4.13e-01 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 4.14e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 1 | 32 | 4.14e-01 |
| GO:BP | GO:0046710 | GDP metabolic process | 4 | 10 | 4.14e-01 |
| GO:BP | GO:0095500 | acetylcholine receptor signaling pathway | 1 | 18 | 4.14e-01 |
| GO:BP | GO:0010737 | protein kinase A signaling | 1 | 23 | 4.14e-01 |
| GO:BP | GO:1902047 | polyamine transmembrane transport | 1 | 10 | 4.14e-01 |
| GO:BP | GO:0009449 | gamma-aminobutyric acid biosynthetic process | 1 | 4 | 4.14e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 4.14e-01 |
| GO:BP | GO:0072414 | response to mitotic cell cycle checkpoint signaling | 1 | 1 | 4.14e-01 |
| GO:BP | GO:0072432 | response to G1 DNA damage checkpoint signaling | 1 | 1 | 4.14e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 3 | 78 | 4.14e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 4.14e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 9 | 160 | 4.14e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 1 | 5 | 4.14e-01 |
| GO:BP | GO:1902463 | protein localization to cell leading edge | 1 | 5 | 4.14e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 3 | 44 | 4.14e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 2 | 109 | 4.14e-01 |
| GO:BP | GO:0048102 | autophagic cell death | 1 | 6 | 4.14e-01 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 3 | 13 | 4.14e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 1 | 187 | 4.14e-01 |
| GO:BP | GO:0055001 | muscle cell development | 5 | 165 | 4.14e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 15 | 27 | 4.14e-01 |
| GO:BP | GO:1901329 | regulation of odontoblast differentiation | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 3 | 4 | 4.14e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 9 | 165 | 4.14e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 2 | 5 | 4.14e-01 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 4 | 23 | 4.14e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 47 | 213 | 4.14e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 5 | 7 | 4.14e-01 |
| GO:BP | GO:0019075 | virus maturation | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0070459 | prolactin secretion | 1 | 4 | 4.15e-01 |
| GO:BP | GO:1903569 | positive regulation of protein localization to ciliary membrane | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0018057 | peptidyl-lysine oxidation | 4 | 5 | 4.15e-01 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 2 | 8 | 4.15e-01 |
| GO:BP | GO:1904293 | negative regulation of ERAD pathway | 4 | 6 | 4.15e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 4.15e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 4.15e-01 |
| GO:BP | GO:1990953 | intramanchette transport | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 4 | 6 | 4.15e-01 |
| GO:BP | GO:1901623 | regulation of lymphocyte chemotaxis | 2 | 13 | 4.15e-01 |
| GO:BP | GO:0001880 | Mullerian duct regression | 2 | 4 | 4.15e-01 |
| GO:BP | GO:0033567 | DNA replication, Okazaki fragment processing | 2 | 2 | 4.16e-01 |
| GO:BP | GO:0044557 | relaxation of smooth muscle | 2 | 9 | 4.16e-01 |
| GO:BP | GO:0060856 | establishment of blood-brain barrier | 2 | 9 | 4.16e-01 |
| GO:BP | GO:0018215 | protein phosphopantetheinylation | 1 | 1 | 4.16e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 4 | 44 | 4.16e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 4 | 8 | 4.16e-01 |
| GO:BP | GO:0061002 | negative regulation of dendritic spine morphogenesis | 3 | 6 | 4.16e-01 |
| GO:BP | GO:0090672 | telomerase RNA localization | 7 | 19 | 4.16e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 8 | 51 | 4.16e-01 |
| GO:BP | GO:1904100 | positive regulation of protein O-linked glycosylation | 1 | 2 | 4.16e-01 |
| GO:BP | GO:1901165 | positive regulation of trophoblast cell migration | 1 | 6 | 4.16e-01 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 7 | 19 | 4.16e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 14 | 276 | 4.16e-01 |
| GO:BP | GO:1990564 | protein polyufmylation | 4 | 5 | 4.16e-01 |
| GO:BP | GO:1904098 | regulation of protein O-linked glycosylation | 1 | 2 | 4.16e-01 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 7 | 19 | 4.16e-01 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 7 | 19 | 4.16e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 12 | 41 | 4.16e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 11 | 29 | 4.16e-01 |
| GO:BP | GO:1990592 | protein K69-linked ufmylation | 4 | 5 | 4.16e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 90 | 4.16e-01 |
| GO:BP | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 3 | 6 | 4.16e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 29 | 237 | 4.16e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 1 | 7 | 4.16e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 4 | 37 | 4.16e-01 |
| GO:BP | GO:0032912 | negative regulation of transforming growth factor beta2 production | 2 | 2 | 4.16e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 6 | 24 | 4.16e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 1 | 7 | 4.16e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 1 | 63 | 4.16e-01 |
| GO:BP | GO:0042413 | carnitine catabolic process | 1 | 1 | 4.16e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 1 | 8 | 4.16e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 36 | 1199 | 4.16e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 4.16e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 4.16e-01 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 1 | 13 | 4.16e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 2 | 14 | 4.17e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 1 | 55 | 4.17e-01 |
| GO:BP | GO:0007271 | synaptic transmission, cholinergic | 1 | 15 | 4.17e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 6 | 16 | 4.17e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 1 | 5 | 4.17e-01 |
| GO:BP | GO:0035640 | exploration behavior | 3 | 21 | 4.17e-01 |
| GO:BP | GO:0008052 | sensory organ boundary specification | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 2 | 40 | 4.17e-01 |
| GO:BP | GO:0010160 | formation of animal organ boundary | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0061193 | taste bud development | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 17 | 740 | 4.17e-01 |
| GO:BP | GO:1901403 | positive regulation of tetrapyrrole metabolic process | 2 | 4 | 4.17e-01 |
| GO:BP | GO:0031119 | tRNA pseudouridine synthesis | 2 | 6 | 4.17e-01 |
| GO:BP | GO:0032876 | negative regulation of DNA endoreduplication | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0014901 | satellite cell activation involved in skeletal muscle regeneration | 2 | 3 | 4.18e-01 |
| GO:BP | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade | 4 | 16 | 4.18e-01 |
| GO:BP | GO:0106058 | positive regulation of calcineurin-mediated signaling | 4 | 16 | 4.18e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 34 | 61 | 4.19e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 5 | 203 | 4.19e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 4.19e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 4.19e-01 |
| GO:BP | GO:0033327 | Leydig cell differentiation | 7 | 9 | 4.19e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 4 | 390 | 4.19e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 4.19e-01 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 2 | 20 | 4.19e-01 |
| GO:BP | GO:0099172 | presynapse organization | 3 | 42 | 4.19e-01 |
| GO:BP | GO:0003129 | heart induction | 4 | 9 | 4.19e-01 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 25 | 4.20e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 2 | 43 | 4.20e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 11 | 34 | 4.20e-01 |
| GO:BP | GO:0071707 | immunoglobulin heavy chain V-D-J recombination | 2 | 2 | 4.20e-01 |
| GO:BP | GO:0007066 | female meiosis sister chromatid cohesion | 1 | 1 | 4.20e-01 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 3 | 11 | 4.20e-01 |
| GO:BP | GO:0003401 | axis elongation | 15 | 24 | 4.20e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 27 | 47 | 4.20e-01 |
| GO:BP | GO:0097531 | mast cell migration | 7 | 12 | 4.20e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 4 | 10 | 4.20e-01 |
| GO:BP | GO:0048874 | host-mediated regulation of intestinal microbiota composition | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0000707 | meiotic DNA recombinase assembly | 1 | 1 | 4.20e-01 |
| GO:BP | GO:0032238 | adenosine transport | 1 | 2 | 4.20e-01 |
| GO:BP | GO:1900244 | positive regulation of synaptic vesicle endocytosis | 4 | 4 | 4.20e-01 |
| GO:BP | GO:0042439 | ethanolamine-containing compound metabolic process | 1 | 5 | 4.20e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 4 | 9 | 4.20e-01 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 2 | 33 | 4.20e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 4 | 9 | 4.20e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 3 | 35 | 4.20e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 4.20e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 4.20e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 4.20e-01 |
| GO:BP | GO:0090038 | negative regulation of protein kinase C signaling | 2 | 4 | 4.20e-01 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 3 | 6 | 4.20e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 9 | 167 | 4.20e-01 |
| GO:BP | GO:0042552 | myelination | 2 | 111 | 4.20e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 24 | 39 | 4.20e-01 |
| GO:BP | GO:0044849 | estrous cycle | 2 | 13 | 4.21e-01 |
| GO:BP | GO:1901626 | regulation of postsynaptic membrane organization | 1 | 2 | 4.21e-01 |
| GO:BP | GO:0002383 | immune response in brain or nervous system | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0050808 | synapse organization | 24 | 365 | 4.21e-01 |
| GO:BP | GO:0097281 | immune complex formation | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0043588 | skin development | 2 | 175 | 4.21e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 10 | 18 | 4.21e-01 |
| GO:BP | GO:0061044 | negative regulation of vascular wound healing | 1 | 4 | 4.21e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 1 | 109 | 4.21e-01 |
| GO:BP | GO:1905145 | cellular response to acetylcholine | 1 | 20 | 4.21e-01 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 11 | 36 | 4.22e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 3 | 7 | 4.22e-01 |
| GO:BP | GO:1903434 | negative regulation of constitutive secretory pathway | 1 | 1 | 4.22e-01 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 11 | 36 | 4.22e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 3 | 7 | 4.22e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 32 | 73 | 4.22e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 18 | 33 | 4.22e-01 |
| GO:BP | GO:1901340 | negative regulation of store-operated calcium channel activity | 1 | 1 | 4.22e-01 |
| GO:BP | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | 7 | 10 | 4.22e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 2 | 16 | 4.22e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 4.22e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 5 | 136 | 4.22e-01 |
| GO:BP | GO:1903944 | negative regulation of hepatocyte apoptotic process | 4 | 6 | 4.22e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 2 | 6 | 4.23e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 13 | 33 | 4.23e-01 |
| GO:BP | GO:1990261 | pre-mRNA catabolic process | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 2 | 94 | 4.23e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 10 | 84 | 4.23e-01 |
| GO:BP | GO:0080163 | regulation of protein serine/threonine phosphatase activity | 2 | 2 | 4.23e-01 |
| GO:BP | GO:0001782 | B cell homeostasis | 1 | 24 | 4.23e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 29 | 317 | 4.23e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 4.23e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 27 | 77 | 4.23e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 2 | 113 | 4.23e-01 |
| GO:BP | GO:0035166 | post-embryonic hemopoiesis | 3 | 5 | 4.23e-01 |
| GO:BP | GO:0046380 | N-acetylneuraminate biosynthetic process | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 9 | 101 | 4.23e-01 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 4 | 4 | 4.23e-01 |
| GO:BP | GO:0140495 | migracytosis | 1 | 3 | 4.23e-01 |
| GO:BP | GO:2000493 | negative regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 4.23e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 2 | 113 | 4.23e-01 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 4.23e-01 |
| GO:BP | GO:1901658 | glycosyl compound catabolic process | 1 | 23 | 4.23e-01 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 1 | 3 | 4.23e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 11 | 137 | 4.23e-01 |
| GO:BP | GO:0022615 | protein to membrane docking | 2 | 5 | 4.23e-01 |
| GO:BP | GO:0034627 | ‘de novo’ NAD biosynthetic process | 2 | 2 | 4.23e-01 |
| GO:BP | GO:0160040 | mitocytosis | 1 | 3 | 4.23e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 3 | 122 | 4.23e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 4.23e-01 |
| GO:BP | GO:0106001 | intestinal hexose absorption | 2 | 3 | 4.23e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 5 | 34 | 4.23e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 2 | 6 | 4.23e-01 |
| GO:BP | GO:0090675 | intermicrovillar adhesion | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0000320 | re-entry into mitotic cell cycle | 2 | 2 | 4.23e-01 |
| GO:BP | GO:0044873 | lipoprotein localization to membrane | 1 | 1 | 4.23e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 5 | 66 | 4.23e-01 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 1 | 1 | 4.24e-01 |
| GO:BP | GO:0072303 | positive regulation of glomerular metanephric mesangial cell proliferation | 1 | 1 | 4.24e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 1 | 5 | 4.24e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 4.24e-01 |
| GO:BP | GO:0014806 | smooth muscle hyperplasia | 2 | 2 | 4.24e-01 |
| GO:BP | GO:0046135 | pyrimidine nucleoside catabolic process | 2 | 8 | 4.24e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 4.24e-01 |
| GO:BP | GO:0000389 | mRNA 3’-splice site recognition | 2 | 6 | 4.24e-01 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 2 | 15 | 4.24e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 3 | 141 | 4.24e-01 |
| GO:BP | GO:0030578 | PML body organization | 4 | 6 | 4.24e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 4.24e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 1 | 10 | 4.24e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 4.24e-01 |
| GO:BP | GO:0051097 | negative regulation of helicase activity | 1 | 5 | 4.24e-01 |
| GO:BP | GO:0006671 | phytosphingosine metabolic process | 1 | 1 | 4.24e-01 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 2 | 29 | 4.24e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 20 | 534 | 4.24e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 1 | 12 | 4.24e-01 |
| GO:BP | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 3 | 4.24e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 2 | 8 | 4.24e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 9 | 172 | 4.24e-01 |
| GO:BP | GO:0071602 | phytosphingosine biosynthetic process | 1 | 1 | 4.24e-01 |
| GO:BP | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 3 | 5 | 4.24e-01 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 4 | 6 | 4.24e-01 |
| GO:BP | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion | 3 | 5 | 4.24e-01 |
| GO:BP | GO:1904585 | response to putrescine | 1 | 1 | 4.25e-01 |
| GO:BP | GO:1904586 | cellular response to putrescine | 1 | 1 | 4.25e-01 |
| GO:BP | GO:1990828 | hepatocyte dedifferentiation | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 9 | 14 | 4.25e-01 |
| GO:BP | GO:0048853 | forebrain morphogenesis | 5 | 7 | 4.26e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 14 | 26 | 4.26e-01 |
| GO:BP | GO:1903516 | regulation of single strand break repair | 2 | 3 | 4.26e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 6 | 50 | 4.26e-01 |
| GO:BP | GO:2000225 | negative regulation of testosterone biosynthetic process | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 20 | 637 | 4.26e-01 |
| GO:BP | GO:0030237 | female sex determination | 1 | 1 | 4.26e-01 |
| GO:BP | GO:0033499 | galactose catabolic process via UDP-galactose | 2 | 4 | 4.26e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 17 | 112 | 4.26e-01 |
| GO:BP | GO:0055070 | copper ion homeostasis | 8 | 18 | 4.26e-01 |
| GO:BP | GO:1900222 | negative regulation of amyloid-beta clearance | 3 | 7 | 4.26e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 2 | 4 | 4.26e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 4 | 43 | 4.26e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 2 | 49 | 4.26e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 3 | 4 | 4.26e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 7 | 8 | 4.26e-01 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 7 | 18 | 4.27e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 12 | 55 | 4.27e-01 |
| GO:BP | GO:0045618 | positive regulation of keratinocyte differentiation | 10 | 12 | 4.27e-01 |
| GO:BP | GO:1904257 | zinc ion import into Golgi lumen | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0030393 | fructoselysine metabolic process | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 2 | 64 | 4.27e-01 |
| GO:BP | GO:0070086 | ubiquitin-dependent endocytosis | 2 | 4 | 4.27e-01 |
| GO:BP | GO:1902257 | negative regulation of apoptotic process involved in outflow tract morphogenesis | 2 | 2 | 4.28e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 7 | 81 | 4.28e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 1 | 16 | 4.28e-01 |
| GO:BP | GO:0051877 | pigment granule aggregation in cell center | 1 | 2 | 4.28e-01 |
| GO:BP | GO:0008272 | sulfate transport | 3 | 12 | 4.28e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 2 | 17 | 4.28e-01 |
| GO:BP | GO:0101030 | tRNA-guanine transglycosylation | 2 | 2 | 4.28e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 2 | 14 | 4.28e-01 |
| GO:BP | GO:0090148 | membrane fission | 6 | 44 | 4.28e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 7 | 112 | 4.28e-01 |
| GO:BP | GO:0008655 | pyrimidine-containing compound salvage | 1 | 5 | 4.28e-01 |
| GO:BP | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway | 1 | 4 | 4.29e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 11 | 31 | 4.29e-01 |
| GO:BP | GO:0060517 | epithelial cell proliferation involved in prostatic bud elongation | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0009213 | pyrimidine deoxyribonucleoside triphosphate catabolic process | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 4.29e-01 |
| GO:BP | GO:0070305 | response to cGMP | 1 | 7 | 4.29e-01 |
| GO:BP | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 1 | 37 | 4.29e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 3 | 3 | 4.29e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 4.29e-01 |
| GO:BP | GO:1990049 | retrograde neuronal dense core vesicle transport | 1 | 4 | 4.29e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 4 | 39 | 4.29e-01 |
| GO:BP | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 1 | 1 | 4.29e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 1 | 59 | 4.29e-01 |
| GO:BP | GO:1904534 | negative regulation of telomeric loop disassembly | 1 | 2 | 4.29e-01 |
| GO:BP | GO:1905838 | regulation of telomeric D-loop disassembly | 1 | 2 | 4.29e-01 |
| GO:BP | GO:1905839 | negative regulation of telomeric D-loop disassembly | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0030656 | regulation of vitamin metabolic process | 1 | 7 | 4.29e-01 |
| GO:BP | GO:0032960 | regulation of inositol trisphosphate biosynthetic process | 1 | 4 | 4.29e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 2 | 2 | 4.29e-01 |
| GO:BP | GO:0032213 | regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 1 | 40 | 4.29e-01 |
| GO:BP | GO:0032214 | negative regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 4.29e-01 |
| GO:BP | GO:1905778 | negative regulation of exonuclease activity | 1 | 2 | 4.29e-01 |
| GO:BP | GO:1905777 | regulation of exonuclease activity | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0048855 | adenohypophysis morphogenesis | 1 | 1 | 4.29e-01 |
| GO:BP | GO:1904780 | negative regulation of protein localization to centrosome | 2 | 2 | 4.30e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 4 | 4 | 4.30e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 4.30e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 6 | 97 | 4.30e-01 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 2 | 4.30e-01 |
| GO:BP | GO:2000744 | positive regulation of anterior head development | 2 | 2 | 4.30e-01 |
| GO:BP | GO:2000742 | regulation of anterior head development | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 3 | 6 | 4.30e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 4 | 28 | 4.30e-01 |
| GO:BP | GO:0099543 | trans-synaptic signaling by soluble gas | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0014028 | notochord formation | 2 | 3 | 4.30e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 1 | 5 | 4.30e-01 |
| GO:BP | GO:0003164 | His-Purkinje system development | 5 | 7 | 4.30e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 22 | 40 | 4.30e-01 |
| GO:BP | GO:0009203 | ribonucleoside triphosphate catabolic process | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0099548 | trans-synaptic signaling by nitric oxide | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0097601 | retina blood vessel maintenance | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0046041 | ITP metabolic process | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 5 | 424 | 4.30e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 10 | 19 | 4.30e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 4.30e-01 |
| GO:BP | GO:0071043 | CUT metabolic process | 2 | 5 | 4.30e-01 |
| GO:BP | GO:0071034 | CUT catabolic process | 2 | 5 | 4.30e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 3 | 14 | 4.30e-01 |
| GO:BP | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0021649 | vestibulocochlear nerve structural organization | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 4.30e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 4.30e-01 |
| GO:BP | GO:1903375 | facioacoustic ganglion development | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 20 | 103 | 4.30e-01 |
| GO:BP | GO:0071502 | cellular response to temperature stimulus | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0070544 | histone H3-K36 demethylation | 2 | 5 | 4.30e-01 |
| GO:BP | GO:0070509 | calcium ion import | 7 | 37 | 4.30e-01 |
| GO:BP | GO:0036337 | Fas signaling pathway | 2 | 5 | 4.30e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 8 | 12 | 4.30e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 2 | 8 | 4.30e-01 |
| GO:BP | GO:0018395 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0010808 | positive regulation of synaptic vesicle priming | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 4 | 106 | 4.30e-01 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 1 | 11 | 4.30e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 8 | 39 | 4.30e-01 |
| GO:BP | GO:0070079 | histone H4-R3 demethylation | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0070078 | histone H3-R2 demethylation | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0100012 | regulation of heart induction by canonical Wnt signaling pathway | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 2 | 504 | 4.30e-01 |
| GO:BP | GO:1902474 | positive regulation of protein localization to synapse | 1 | 6 | 4.30e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 28 | 46 | 4.30e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 3 | 84 | 4.30e-01 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 1 | 31 | 4.30e-01 |
| GO:BP | GO:1900006 | positive regulation of dendrite development | 3 | 14 | 4.30e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 4.30e-01 |
| GO:BP | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0061552 | ganglion morphogenesis | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0106101 | ER-dependent peroxisome localization | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 12 | 31 | 4.30e-01 |
| GO:BP | GO:1905144 | response to acetylcholine | 1 | 22 | 4.30e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 7 | 67 | 4.30e-01 |
| GO:BP | GO:0015846 | polyamine transport | 1 | 12 | 4.30e-01 |
| GO:BP | GO:2000348 | regulation of CD40 signaling pathway | 2 | 4 | 4.30e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 28 | 46 | 4.30e-01 |
| GO:BP | GO:0048304 | positive regulation of isotype switching to IgG isotypes | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 8 | 51 | 4.30e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 5 | 134 | 4.30e-01 |
| GO:BP | GO:0045578 | negative regulation of B cell differentiation | 3 | 6 | 4.30e-01 |
| GO:BP | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0060699 | regulation of endoribonuclease activity | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0000105 | histidine biosynthetic process | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 58 | 1111 | 4.30e-01 |
| GO:BP | GO:0036335 | intestinal stem cell homeostasis | 1 | 3 | 4.30e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 2 | 140 | 4.30e-01 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 5 | 13 | 4.30e-01 |
| GO:BP | GO:1904835 | dorsal root ganglion morphogenesis | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0060685 | regulation of prostatic bud formation | 1 | 5 | 4.30e-01 |
| GO:BP | GO:0006996 | organelle organization | 146 | 3000 | 4.30e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 2 | 1061 | 4.30e-01 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 14 | 32 | 4.30e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 2 | 28 | 4.30e-01 |
| GO:BP | GO:0050960 | detection of temperature stimulus involved in thermoception | 1 | 3 | 4.30e-01 |
| GO:BP | GO:0090381 | regulation of heart induction | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0046208 | spermine catabolic process | 2 | 2 | 4.30e-01 |
| GO:BP | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 3 | 4.30e-01 |
| GO:BP | GO:0032402 | melanosome transport | 5 | 19 | 4.30e-01 |
| GO:BP | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0032581 | ER-dependent peroxisome organization | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0034760 | negative regulation of iron ion transmembrane transport | 1 | 2 | 4.30e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 7 | 42 | 4.30e-01 |
| GO:BP | GO:0021891 | olfactory bulb interneuron development | 1 | 5 | 4.30e-01 |
| GO:BP | GO:0034757 | negative regulation of iron ion transport | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 18 | 35 | 4.30e-01 |
| GO:BP | GO:1905843 | regulation of cellular response to gamma radiation | 1 | 1 | 4.30e-01 |
| GO:BP | GO:1902296 | DNA strand elongation involved in cell cycle DNA replication | 1 | 1 | 4.30e-01 |
| GO:BP | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 4.30e-01 |
| GO:BP | GO:2000234 | positive regulation of rRNA processing | 1 | 10 | 4.30e-01 |
| GO:BP | GO:0000012 | single strand break repair | 5 | 10 | 4.30e-01 |
| GO:BP | GO:0051591 | response to cAMP | 4 | 66 | 4.30e-01 |
| GO:BP | GO:1904219 | positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0071839 | apoptotic process in bone marrow cell | 3 | 4 | 4.30e-01 |
| GO:BP | GO:1904222 | positive regulation of serine C-palmitoyltransferase activity | 1 | 2 | 4.30e-01 |
| GO:BP | GO:1902983 | DNA strand elongation involved in mitotic DNA replication | 1 | 1 | 4.30e-01 |
| GO:BP | GO:1902319 | DNA strand elongation involved in nuclear cell cycle DNA replication | 1 | 1 | 4.30e-01 |
| GO:BP | GO:2001228 | regulation of response to gamma radiation | 1 | 1 | 4.30e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 18 | 1280 | 4.30e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 4 | 55 | 4.30e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 1 | 9 | 4.30e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 15 | 1015 | 4.30e-01 |
| GO:BP | GO:0090309 | positive regulation of DNA methylation-dependent heterochromatin formation | 2 | 12 | 4.30e-01 |
| GO:BP | GO:0048672 | positive regulation of collateral sprouting | 5 | 10 | 4.30e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 4.30e-01 |
| GO:BP | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0090472 | dibasic protein processing | 1 | 1 | 4.31e-01 |
| GO:BP | GO:0002155 | regulation of thyroid hormone mediated signaling pathway | 2 | 5 | 4.31e-01 |
| GO:BP | GO:1901392 | regulation of transforming growth factor beta1 activation | 1 | 1 | 4.31e-01 |
| GO:BP | GO:1901394 | positive regulation of transforming growth factor beta1 activation | 1 | 1 | 4.31e-01 |
| GO:BP | GO:0061551 | trigeminal ganglion development | 3 | 3 | 4.31e-01 |
| GO:BP | GO:1903935 | response to sodium arsenite | 1 | 6 | 4.31e-01 |
| GO:BP | GO:2000346 | negative regulation of hepatocyte proliferation | 1 | 3 | 4.31e-01 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 2 | 30 | 4.31e-01 |
| GO:BP | GO:2001205 | negative regulation of osteoclast development | 1 | 2 | 4.31e-01 |
| GO:BP | GO:2001311 | lysobisphosphatidic acid metabolic process | 2 | 2 | 4.31e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 1 | 21 | 4.31e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 9 | 62 | 4.31e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 4 | 7 | 4.32e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 4.32e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 15 | 1017 | 4.32e-01 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 3 | 4 | 4.32e-01 |
| GO:BP | GO:1903996 | negative regulation of non-membrane spanning protein tyrosine kinase activity | 1 | 1 | 4.32e-01 |
| GO:BP | GO:2000615 | regulation of histone H3-K9 acetylation | 2 | 7 | 4.32e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 3 | 28 | 4.32e-01 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 4 | 33 | 4.32e-01 |
| GO:BP | GO:0090618 | DNA clamp unloading | 1 | 2 | 4.32e-01 |
| GO:BP | GO:0001787 | natural killer cell proliferation | 1 | 6 | 4.32e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 4.32e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 3 | 23 | 4.32e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 15 | 47 | 4.32e-01 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 3 | 19 | 4.32e-01 |
| GO:BP | GO:0021637 | trigeminal nerve structural organization | 3 | 4 | 4.32e-01 |
| GO:BP | GO:0021636 | trigeminal nerve morphogenesis | 3 | 4 | 4.32e-01 |
| GO:BP | GO:0030879 | mammary gland development | 1 | 106 | 4.33e-01 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 19 | 4.33e-01 |
| GO:BP | GO:0051028 | mRNA transport | 59 | 117 | 4.33e-01 |
| GO:BP | GO:0055123 | digestive system development | 10 | 100 | 4.33e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 3 | 31 | 4.33e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 36 | 660 | 4.33e-01 |
| GO:BP | GO:0060068 | vagina development | 3 | 8 | 4.33e-01 |
| GO:BP | GO:0071040 | nuclear polyadenylation-dependent antisense transcript catabolic process | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 4.33e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 3 | 53 | 4.33e-01 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 1 | 20 | 4.33e-01 |
| GO:BP | GO:0042868 | antisense RNA metabolic process | 1 | 1 | 4.33e-01 |
| GO:BP | GO:1903347 | negative regulation of bicellular tight junction assembly | 1 | 3 | 4.33e-01 |
| GO:BP | GO:0007566 | embryo implantation | 3 | 39 | 4.33e-01 |
| GO:BP | GO:0120222 | regulation of blastocyst development | 3 | 3 | 4.33e-01 |
| GO:BP | GO:0021592 | fourth ventricle development | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0021993 | initiation of neural tube closure | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0071041 | antisense RNA transcript catabolic process | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0018158 | protein oxidation | 1 | 12 | 4.33e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 4.33e-01 |
| GO:BP | GO:0019521 | D-gluconate metabolic process | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0019520 | aldonic acid metabolic process | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 1 | 5 | 4.33e-01 |
| GO:BP | GO:1903537 | meiotic cell cycle process involved in oocyte maturation | 1 | 5 | 4.33e-01 |
| GO:BP | GO:0046176 | aldonic acid catabolic process | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0046177 | D-gluconate catabolic process | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0015827 | tryptophan transport | 2 | 6 | 4.33e-01 |
| GO:BP | GO:0019322 | pentose biosynthetic process | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 25 | 71 | 4.33e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 4 | 86 | 4.33e-01 |
| GO:BP | GO:0034486 | vacuolar transmembrane transport | 2 | 3 | 4.33e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 1 | 24 | 4.33e-01 |
| GO:BP | GO:1902977 | mitotic DNA replication preinitiation complex assembly | 1 | 1 | 4.33e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 2 | 18 | 4.33e-01 |
| GO:BP | GO:1903608 | protein localization to cytoplasmic stress granule | 5 | 7 | 4.33e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 5 | 60 | 4.33e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 4 | 6 | 4.33e-01 |
| GO:BP | GO:0090403 | oxidative stress-induced premature senescence | 1 | 3 | 4.33e-01 |
| GO:BP | GO:0097043 | histone H3-K56 acetylation | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 4.34e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 16 | 30 | 4.34e-01 |
| GO:BP | GO:0007494 | midgut development | 3 | 7 | 4.34e-01 |
| GO:BP | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process | 1 | 7 | 4.34e-01 |
| GO:BP | GO:0010025 | wax biosynthetic process | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0070593 | dendrite self-avoidance | 1 | 11 | 4.34e-01 |
| GO:BP | GO:0010166 | wax metabolic process | 2 | 2 | 4.34e-01 |
| GO:BP | GO:2000041 | negative regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 4.34e-01 |
| GO:BP | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 4.34e-01 |
| GO:BP | GO:2000079 | regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 4.34e-01 |
| GO:BP | GO:0097722 | sperm motility | 2 | 61 | 4.34e-01 |
| GO:BP | GO:0035849 | nephric duct elongation | 1 | 1 | 4.34e-01 |
| GO:BP | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction | 2 | 2 | 4.34e-01 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.34e-01 |
| GO:BP | GO:0072105 | ureteric peristalsis | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0035847 | uterine epithelium development | 1 | 1 | 4.34e-01 |
| GO:BP | GO:0097379 | dorsal spinal cord interneuron posterior axon guidance | 1 | 1 | 4.34e-01 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 4 | 8 | 4.34e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 2 | 37 | 4.34e-01 |
| GO:BP | GO:0014849 | ureter smooth muscle contraction | 2 | 2 | 4.34e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 8 | 30 | 4.34e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 6 | 18 | 4.34e-01 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 3 | 17 | 4.34e-01 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 1 | 2 | 4.34e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 2 | 61 | 4.34e-01 |
| GO:BP | GO:0009446 | putrescine biosynthetic process | 1 | 4 | 4.34e-01 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 1 | 9 | 4.34e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 4.34e-01 |
| GO:BP | GO:0061303 | cornea development in camera-type eye | 1 | 4 | 4.34e-01 |
| GO:BP | GO:0061727 | methylglyoxal catabolic process to lactate | 4 | 5 | 4.34e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 4.34e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 2 | 1123 | 4.34e-01 |
| GO:BP | GO:0010967 | regulation of polyamine biosynthetic process | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0044343 | canonical Wnt signaling pathway involved in regulation of type B pancreatic cell proliferation | 1 | 1 | 4.34e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 1 | 35 | 4.34e-01 |
| GO:BP | GO:0051596 | methylglyoxal catabolic process | 4 | 5 | 4.34e-01 |
| GO:BP | GO:0032230 | positive regulation of synaptic transmission, GABAergic | 1 | 10 | 4.34e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 5 | 16 | 4.34e-01 |
| GO:BP | GO:1902775 | mitochondrial large ribosomal subunit assembly | 3 | 4 | 4.34e-01 |
| GO:BP | GO:0022013 | pallium cell proliferation in forebrain | 1 | 1 | 4.35e-01 |
| GO:BP | GO:0019860 | uracil metabolic process | 1 | 2 | 4.35e-01 |
| GO:BP | GO:0051666 | actin cortical patch localization | 1 | 2 | 4.36e-01 |
| GO:BP | GO:0008033 | tRNA processing | 30 | 128 | 4.36e-01 |
| GO:BP | GO:0019262 | N-acetylneuraminate catabolic process | 4 | 6 | 4.36e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 1 | 8 | 4.36e-01 |
| GO:BP | GO:1902034 | negative regulation of hematopoietic stem cell proliferation | 1 | 2 | 4.36e-01 |
| GO:BP | GO:0035702 | monocyte homeostasis | 1 | 2 | 4.36e-01 |
| GO:BP | GO:0061519 | macrophage homeostasis | 1 | 3 | 4.36e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 3 | 17 | 4.36e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 1 | 6 | 4.36e-01 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 4 | 8 | 4.37e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 1 | 74 | 4.37e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 1 | 63 | 4.37e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 4.37e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 5 | 57 | 4.37e-01 |
| GO:BP | GO:0071072 | negative regulation of phospholipid biosynthetic process | 4 | 6 | 4.37e-01 |
| GO:BP | GO:0009256 | 10-formyltetrahydrofolate metabolic process | 2 | 6 | 4.38e-01 |
| GO:BP | GO:0035265 | organ growth | 2 | 131 | 4.38e-01 |
| GO:BP | GO:0036216 | cellular response to stem cell factor stimulus | 3 | 4 | 4.38e-01 |
| GO:BP | GO:0038109 | Kit signaling pathway | 3 | 4 | 4.38e-01 |
| GO:BP | GO:1901211 | negative regulation of cardiac chamber formation | 1 | 1 | 4.38e-01 |
| GO:BP | GO:1901210 | regulation of cardiac chamber formation | 1 | 1 | 4.38e-01 |
| GO:BP | GO:1901208 | negative regulation of heart looping | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0036215 | response to stem cell factor | 3 | 4 | 4.38e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 1 | 25 | 4.38e-01 |
| GO:BP | GO:0014004 | microglia differentiation | 1 | 6 | 4.38e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 9 | 15 | 4.38e-01 |
| GO:BP | GO:0046049 | UMP metabolic process | 2 | 10 | 4.38e-01 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 5 | 16 | 4.38e-01 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 2 | 142 | 4.38e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 8 | 43 | 4.38e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0071838 | cell proliferation in bone marrow | 2 | 7 | 4.38e-01 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 3 | 13 | 4.38e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 2 | 33 | 4.38e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 1 | 8 | 4.38e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 4.38e-01 |
| GO:BP | GO:0007617 | mating behavior | 1 | 21 | 4.38e-01 |
| GO:BP | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 4 | 4.38e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 3 | 5 | 4.38e-01 |
| GO:BP | GO:0036302 | atrioventricular canal development | 8 | 12 | 4.38e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 12 | 18 | 4.38e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 5 | 25 | 4.38e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 4 | 20 | 4.38e-01 |
| GO:BP | GO:0014848 | urinary tract smooth muscle contraction | 3 | 6 | 4.38e-01 |
| GO:BP | GO:0014738 | regulation of muscle hyperplasia | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 4.39e-01 |
| GO:BP | GO:0035212 | cell competition in a multicellular organism | 1 | 1 | 4.39e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 3 | 21 | 4.39e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 33 | 87 | 4.39e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 3 | 63 | 4.39e-01 |
| GO:BP | GO:0098926 | postsynaptic signal transduction | 1 | 25 | 4.39e-01 |
| GO:BP | GO:0150066 | negative regulation of deacetylase activity | 2 | 2 | 4.39e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 2 | 21 | 4.39e-01 |
| GO:BP | GO:1901726 | negative regulation of histone deacetylase activity | 2 | 2 | 4.39e-01 |
| GO:BP | GO:0035864 | response to potassium ion | 3 | 12 | 4.39e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 3 | 23 | 4.39e-01 |
| GO:BP | GO:0070143 | mitochondrial alanyl-tRNA aminoacylation | 1 | 1 | 4.40e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 10 | 19 | 4.41e-01 |
| GO:BP | GO:0060754 | positive regulation of mast cell chemotaxis | 1 | 5 | 4.41e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 1 | 14 | 4.41e-01 |
| GO:BP | GO:0022406 | membrane docking | 36 | 84 | 4.41e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 4 | 39 | 4.41e-01 |
| GO:BP | GO:1904679 | myo-inositol import across plasma membrane | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0090184 | positive regulation of kidney development | 3 | 9 | 4.41e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 3 | 16 | 4.41e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 15 | 1026 | 4.41e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 17 | 1214 | 4.41e-01 |
| GO:BP | GO:1902908 | regulation of melanosome transport | 2 | 2 | 4.41e-01 |
| GO:BP | GO:0097338 | response to clozapine | 2 | 2 | 4.41e-01 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 4 | 7 | 4.41e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 2 | 26 | 4.41e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 2 | 13 | 4.41e-01 |
| GO:BP | GO:0070668 | positive regulation of mast cell proliferation | 2 | 4 | 4.41e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 2 | 20 | 4.41e-01 |
| GO:BP | GO:0042940 | D-amino acid transport | 2 | 5 | 4.41e-01 |
| GO:BP | GO:0090298 | negative regulation of mitochondrial DNA replication | 1 | 1 | 4.42e-01 |
| GO:BP | GO:0006288 | base-excision repair, DNA ligation | 1 | 1 | 4.42e-01 |
| GO:BP | GO:0048699 | generation of neurons | 2 | 1119 | 4.42e-01 |
| GO:BP | GO:0001944 | vasculature development | 30 | 559 | 4.42e-01 |
| GO:BP | GO:0048268 | clathrin coat assembly | 5 | 13 | 4.42e-01 |
| GO:BP | GO:0006041 | glucosamine metabolic process | 2 | 3 | 4.42e-01 |
| GO:BP | GO:0042074 | cell migration involved in gastrulation | 5 | 10 | 4.42e-01 |
| GO:BP | GO:0043038 | amino acid activation | 10 | 44 | 4.42e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 2 | 7 | 4.42e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 11 | 179 | 4.42e-01 |
| GO:BP | GO:0046078 | dUMP metabolic process | 1 | 6 | 4.42e-01 |
| GO:BP | GO:0046035 | CMP metabolic process | 1 | 6 | 4.42e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 2 | 7 | 4.42e-01 |
| GO:BP | GO:0009178 | pyrimidine deoxyribonucleoside monophosphate catabolic process | 1 | 5 | 4.42e-01 |
| GO:BP | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis | 3 | 4 | 4.43e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 15 | 106 | 4.43e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 2 | 9 | 4.43e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 4 | 14 | 4.43e-01 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 16 | 27 | 4.43e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 16 | 1119 | 4.44e-01 |
| GO:BP | GO:1900449 | regulation of glutamate receptor signaling pathway | 2 | 5 | 4.44e-01 |
| GO:BP | GO:0042942 | D-serine transport | 1 | 1 | 4.45e-01 |
| GO:BP | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development | 2 | 2 | 4.45e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 7 | 84 | 4.45e-01 |
| GO:BP | GO:1904322 | cellular response to forskolin | 2 | 11 | 4.45e-01 |
| GO:BP | GO:0050914 | sensory perception of salty taste | 1 | 1 | 4.45e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 114 | 4.45e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 114 | 4.45e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 4 | 13 | 4.45e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 10 | 46 | 4.45e-01 |
| GO:BP | GO:1904321 | response to forskolin | 2 | 11 | 4.45e-01 |
| GO:BP | GO:1900452 | regulation of long-term synaptic depression | 2 | 10 | 4.45e-01 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 7 | 19 | 4.45e-01 |
| GO:BP | GO:0033026 | negative regulation of mast cell apoptotic process | 1 | 1 | 4.45e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 2 | 551 | 4.45e-01 |
| GO:BP | GO:0048311 | mitochondrion distribution | 1 | 14 | 4.45e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 2 | 6 | 4.46e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 2 | 41 | 4.46e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 56 | 513 | 4.46e-01 |
| GO:BP | GO:0006914 | autophagy | 56 | 513 | 4.46e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 4.46e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 4.46e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 7 | 26 | 4.46e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 6 | 38 | 4.46e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 29 | 47 | 4.46e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 2 | 20 | 4.46e-01 |
| GO:BP | GO:0009133 | nucleoside diphosphate biosynthetic process | 6 | 11 | 4.46e-01 |
| GO:BP | GO:0001815 | positive regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 6 | 38 | 4.46e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 2 | 16 | 4.46e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 3 | 12 | 4.46e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 3 | 18 | 4.46e-01 |
| GO:BP | GO:0006046 | N-acetylglucosamine catabolic process | 3 | 3 | 4.46e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 4.47e-01 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 1 | 5 | 4.47e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 4.47e-01 |
| GO:BP | GO:1902747 | negative regulation of lens fiber cell differentiation | 3 | 5 | 4.47e-01 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 1 | 19 | 4.47e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion | 5 | 7 | 4.47e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 4.47e-01 |
| GO:BP | GO:0006207 | ‘de novo’ pyrimidine nucleobase biosynthetic process | 2 | 6 | 4.47e-01 |
| GO:BP | GO:1904527 | negative regulation of microtubule binding | 1 | 2 | 4.47e-01 |
| GO:BP | GO:1904441 | regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 4.47e-01 |
| GO:BP | GO:1990789 | thyroid gland epithelial cell proliferation | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0071283 | cellular response to iron(III) ion | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0009085 | lysine biosynthetic process | 2 | 2 | 4.47e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 3 | 4 | 4.47e-01 |
| GO:BP | GO:0042823 | pyridoxal phosphate biosynthetic process | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0050925 | negative regulation of negative chemotaxis | 2 | 2 | 4.47e-01 |
| GO:BP | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 1 | 10 | 4.47e-01 |
| GO:BP | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0021935 | cerebellar granule cell precursor tangential migration | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0019878 | lysine biosynthetic process via aminoadipic acid | 2 | 2 | 4.47e-01 |
| GO:BP | GO:0008614 | pyridoxine metabolic process | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0008615 | pyridoxine biosynthetic process | 1 | 2 | 4.47e-01 |
| GO:BP | GO:1904442 | negative regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0051607 | defense response to virus | 2 | 202 | 4.47e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 15 | 40 | 4.47e-01 |
| GO:BP | GO:1903697 | negative regulation of microvillus assembly | 1 | 1 | 4.47e-01 |
| GO:BP | GO:1903348 | positive regulation of bicellular tight junction assembly | 3 | 5 | 4.47e-01 |
| GO:BP | GO:0035176 | social behavior | 5 | 34 | 4.47e-01 |
| GO:BP | GO:0046688 | response to copper ion | 3 | 29 | 4.48e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 2 | 202 | 4.48e-01 |
| GO:BP | GO:1901463 | regulation of tetrapyrrole biosynthetic process | 5 | 7 | 4.48e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 50 | 427 | 4.48e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 9 | 72 | 4.48e-01 |
| GO:BP | GO:0070453 | regulation of heme biosynthetic process | 5 | 7 | 4.48e-01 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 5 | 7 | 4.48e-01 |
| GO:BP | GO:0009448 | gamma-aminobutyric acid metabolic process | 1 | 6 | 4.48e-01 |
| GO:BP | GO:0006538 | glutamate catabolic process | 1 | 7 | 4.48e-01 |
| GO:BP | GO:0055025 | positive regulation of cardiac muscle tissue development | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 20 | 35 | 4.48e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 8 | 29 | 4.49e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 32 | 130 | 4.49e-01 |
| GO:BP | GO:0006741 | NADP biosynthetic process | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 1 | 12 | 4.49e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 10 | 17 | 4.49e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 3 | 184 | 4.49e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 7 | 17 | 4.49e-01 |
| GO:BP | GO:0106016 | positive regulation of inflammatory response to wounding | 2 | 2 | 4.49e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 63 | 1219 | 4.49e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 3 | 49 | 4.49e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 5 | 56 | 4.49e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 20 | 39 | 4.50e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 2 | 187 | 4.50e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 5 | 96 | 4.50e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 2 | 4 | 4.50e-01 |
| GO:BP | GO:0010980 | positive regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0051168 | nuclear export | 9 | 150 | 4.50e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 1 | 10 | 4.50e-01 |
| GO:BP | GO:0048219 | inter-Golgi cisterna vesicle-mediated transport | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 2 | 187 | 4.50e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 4.50e-01 |
| GO:BP | GO:0042930 | enterobactin transport | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0010979 | regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0001788 | antibody-dependent cellular cytotoxicity | 2 | 2 | 4.50e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 3 | 24 | 4.50e-01 |
| GO:BP | GO:0051107 | negative regulation of DNA ligation | 1 | 1 | 4.50e-01 |
| GO:BP | GO:1904876 | negative regulation of DNA ligase activity | 1 | 1 | 4.50e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 4.50e-01 |
| GO:BP | GO:1900238 | regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 2 | 3 | 4.50e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 2 | 22 | 4.50e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 1 | 23 | 4.50e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 1 | 6 | 4.50e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 4 | 195 | 4.50e-01 |
| GO:BP | GO:0051394 | regulation of nerve growth factor receptor activity | 1 | 1 | 4.50e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 1 | 6 | 4.50e-01 |
| GO:BP | GO:0006883 | intracellular sodium ion homeostasis | 3 | 13 | 4.50e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 5 | 19 | 4.50e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 14 | 113 | 4.50e-01 |
| GO:BP | GO:0097484 | dendrite extension | 11 | 27 | 4.50e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 1 | 67 | 4.50e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 2 | 4 | 4.50e-01 |
| GO:BP | GO:0051645 | Golgi localization | 2 | 14 | 4.50e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 4.50e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 4.50e-01 |
| GO:BP | GO:1905371 | ceramide phosphoethanolamine metabolic process | 2 | 2 | 4.50e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 2 | 1201 | 4.50e-01 |
| GO:BP | GO:1905373 | ceramide phosphoethanolamine biosynthetic process | 2 | 2 | 4.50e-01 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 5 | 13 | 4.50e-01 |
| GO:BP | GO:1900826 | negative regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0031437 | regulation of mRNA cleavage | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 4.50e-01 |
| GO:BP | GO:0060310 | regulation of elastin catabolic process | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0060638 | mesenchymal-epithelial cell signaling | 1 | 4 | 4.50e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 4.50e-01 |
| GO:BP | GO:0060299 | negative regulation of sarcomere organization | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0007530 | sex determination | 5 | 12 | 4.50e-01 |
| GO:BP | GO:0043041 | amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0010711 | negative regulation of collagen catabolic process | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 2 | 4 | 4.50e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 2 | 4 | 4.50e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 2 | 14 | 4.50e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 4.50e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 5 | 20 | 4.50e-01 |
| GO:BP | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0060311 | negative regulation of elastin catabolic process | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 2 | 3 | 4.50e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 4.50e-01 |
| GO:BP | GO:2000591 | positive regulation of metanephric mesenchymal cell migration | 2 | 3 | 4.50e-01 |
| GO:BP | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 1 | 5 | 4.50e-01 |
| GO:BP | GO:0046068 | cGMP metabolic process | 2 | 11 | 4.50e-01 |
| GO:BP | GO:2000975 | positive regulation of pro-B cell differentiation | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0038009 | regulation of signal transduction by receptor internalization | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 1 | 24 | 4.50e-01 |
| GO:BP | GO:0031439 | positive regulation of mRNA cleavage | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 19 | 615 | 4.50e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 9 | 24 | 4.50e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 4.50e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 4.50e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 4.50e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 4.50e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 4.50e-01 |
| GO:BP | GO:1903232 | melanosome assembly | 2 | 19 | 4.50e-01 |
| GO:BP | GO:0090262 | regulation of transcription-coupled nucleotide-excision repair | 1 | 1 | 4.50e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 26 | 47 | 4.50e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 3 | 55 | 4.50e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 4.50e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 3 | 55 | 4.50e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 6 | 12 | 4.51e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 16 | 32 | 4.51e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 4.51e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 16 | 32 | 4.51e-01 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 5 | 35 | 4.51e-01 |
| GO:BP | GO:0150099 | neuron-glial cell signaling | 1 | 4 | 4.51e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 6 | 12 | 4.51e-01 |
| GO:BP | GO:0140066 | peptidyl-lysine crotonylation | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 8 | 18 | 4.51e-01 |
| GO:BP | GO:0035965 | cardiolipin acyl-chain remodeling | 2 | 4 | 4.51e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 1 | 17 | 4.51e-01 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 4 | 15 | 4.51e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 12 | 45 | 4.51e-01 |
| GO:BP | GO:0003158 | endothelium development | 7 | 104 | 4.51e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 4.51e-01 |
| GO:BP | GO:1903949 | positive regulation of atrial cardiac muscle cell action potential | 1 | 1 | 4.51e-01 |
| GO:BP | GO:1904199 | positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.51e-01 |
| GO:BP | GO:1990736 | regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 4.52e-01 |
| GO:BP | GO:0072584 | caveolin-mediated endocytosis | 5 | 9 | 4.52e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 11 | 14 | 4.52e-01 |
| GO:BP | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway | 2 | 4 | 4.52e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 4.52e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 5 | 90 | 4.52e-01 |
| GO:BP | GO:0098907 | regulation of SA node cell action potential | 5 | 7 | 4.52e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 12 | 19 | 4.52e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 4 | 10 | 4.52e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 4.52e-01 |
| GO:BP | GO:0032203 | telomere formation via telomerase | 1 | 1 | 4.52e-01 |
| GO:BP | GO:0017085 | response to insecticide | 1 | 4 | 4.52e-01 |
| GO:BP | GO:0051958 | methotrexate transport | 1 | 2 | 4.52e-01 |
| GO:BP | GO:0034337 | RNA folding | 1 | 1 | 4.52e-01 |
| GO:BP | GO:0140374 | antiviral innate immune response | 10 | 17 | 4.52e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 1 | 13 | 4.52e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 6 | 93 | 4.52e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 2 | 6 | 4.52e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 4 | 58 | 4.52e-01 |
| GO:BP | GO:1990683 | DNA double-strand break attachment to nuclear envelope | 2 | 2 | 4.53e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 4 | 9 | 4.53e-01 |
| GO:BP | GO:0003104 | positive regulation of glomerular filtration | 1 | 3 | 4.53e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 6 | 83 | 4.53e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 8 | 73 | 4.53e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 1 | 20 | 4.53e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 3 | 442 | 4.53e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 8 | 73 | 4.53e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 4.53e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 2 | 513 | 4.53e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 4 | 45 | 4.53e-01 |
| GO:BP | GO:0034499 | late endosome to Golgi transport | 2 | 5 | 4.53e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 4 | 8 | 4.53e-01 |
| GO:BP | GO:0070848 | response to growth factor | 2 | 572 | 4.53e-01 |
| GO:BP | GO:0019080 | viral gene expression | 4 | 96 | 4.53e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 4.53e-01 |
| GO:BP | GO:0032696 | negative regulation of interleukin-13 production | 2 | 3 | 4.53e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 6 | 17 | 4.53e-01 |
| GO:BP | GO:0032773 | positive regulation of tyrosinase activity | 2 | 2 | 4.53e-01 |
| GO:BP | GO:0015860 | purine nucleoside transmembrane transport | 1 | 3 | 4.53e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 9 | 23 | 4.53e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 1 | 9 | 4.53e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 2 | 69 | 4.53e-01 |
| GO:BP | GO:0009052 | pentose-phosphate shunt, non-oxidative branch | 4 | 6 | 4.53e-01 |
| GO:BP | GO:0032771 | regulation of tyrosinase activity | 2 | 2 | 4.53e-01 |
| GO:BP | GO:0097017 | renal protein absorption | 1 | 5 | 4.53e-01 |
| GO:BP | GO:0060003 | copper ion export | 2 | 2 | 4.53e-01 |
| GO:BP | GO:0003331 | positive regulation of extracellular matrix constituent secretion | 1 | 4 | 4.53e-01 |
| GO:BP | GO:0016255 | attachment of GPI anchor to protein | 4 | 6 | 4.53e-01 |
| GO:BP | GO:1990048 | anterograde neuronal dense core vesicle transport | 1 | 4 | 4.54e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 6 | 9 | 4.54e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 6 | 8 | 4.54e-01 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 1 | 3 | 4.54e-01 |
| GO:BP | GO:0036075 | replacement ossification | 4 | 25 | 4.54e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 4 | 25 | 4.54e-01 |
| GO:BP | GO:0016104 | triterpenoid biosynthetic process | 1 | 1 | 4.54e-01 |
| GO:BP | GO:0072229 | metanephric proximal convoluted tubule development | 1 | 2 | 4.54e-01 |
| GO:BP | GO:0072019 | proximal convoluted tubule development | 1 | 2 | 4.54e-01 |
| GO:BP | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development | 2 | 2 | 4.54e-01 |
| GO:BP | GO:0030030 | cell projection organization | 2 | 1233 | 4.54e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 5 | 98 | 4.54e-01 |
| GO:BP | GO:0018160 | peptidyl-pyrromethane cofactor linkage | 1 | 1 | 4.54e-01 |
| GO:BP | GO:1901678 | iron coordination entity transport | 4 | 11 | 4.55e-01 |
| GO:BP | GO:0060271 | cilium assembly | 41 | 292 | 4.55e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 3 | 32 | 4.55e-01 |
| GO:BP | GO:0039654 | fusion of virus membrane with host endosome membrane | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0006183 | GTP biosynthetic process | 4 | 9 | 4.55e-01 |
| GO:BP | GO:2000393 | negative regulation of lamellipodium morphogenesis | 2 | 2 | 4.55e-01 |
| GO:BP | GO:0017196 | N-terminal peptidyl-methionine acetylation | 3 | 8 | 4.55e-01 |
| GO:BP | GO:0061833 | protein localization to tricellular tight junction | 1 | 2 | 4.55e-01 |
| GO:BP | GO:0014832 | urinary bladder smooth muscle contraction | 2 | 4 | 4.55e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 2 | 25 | 4.55e-01 |
| GO:BP | GO:0009438 | methylglyoxal metabolic process | 5 | 7 | 4.55e-01 |
| GO:BP | GO:1904044 | response to aldosterone | 3 | 9 | 4.55e-01 |
| GO:BP | GO:1905285 | fibrous ring of heart morphogenesis | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 8 | 54 | 4.55e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 5 | 532 | 4.55e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 1 | 295 | 4.55e-01 |
| GO:BP | GO:0070673 | response to interleukin-18 | 4 | 5 | 4.55e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 3 | 191 | 4.55e-01 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 4 | 9 | 4.55e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 6 | 15 | 4.55e-01 |
| GO:BP | GO:0006680 | glucosylceramide catabolic process | 1 | 3 | 4.55e-01 |
| GO:BP | GO:0048378 | regulation of lateral mesodermal cell fate specification | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0021998 | neural plate mediolateral regionalization | 1 | 1 | 4.55e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 6 | 7 | 4.55e-01 |
| GO:BP | GO:0023021 | termination of signal transduction | 1 | 3 | 4.55e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 2 | 39 | 4.55e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 2 | 39 | 4.55e-01 |
| GO:BP | GO:0048377 | lateral mesodermal cell fate specification | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0014031 | mesenchymal cell development | 4 | 6 | 4.55e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 4 | 234 | 4.55e-01 |
| GO:BP | GO:0048372 | lateral mesodermal cell fate commitment | 1 | 1 | 4.55e-01 |
| GO:BP | GO:1904978 | regulation of endosome organization | 2 | 5 | 4.55e-01 |
| GO:BP | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 2 | 5 | 4.55e-01 |
| GO:BP | GO:0060697 | positive regulation of phospholipid catabolic process | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0048352 | paraxial mesoderm structural organization | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 5 | 38 | 4.55e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 2 | 84 | 4.55e-01 |
| GO:BP | GO:1904412 | regulation of cardiac ventricle development | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 3 | 8 | 4.55e-01 |
| GO:BP | GO:0045190 | isotype switching | 2 | 39 | 4.55e-01 |
| GO:BP | GO:1904414 | positive regulation of cardiac ventricle development | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0050923 | regulation of negative chemotaxis | 3 | 4 | 4.55e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 11 | 31 | 4.55e-01 |
| GO:BP | GO:0048338 | mesoderm structural organization | 1 | 1 | 4.55e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 7 | 85 | 4.55e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 5 | 5 | 4.55e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 5 | 13 | 4.55e-01 |
| GO:BP | GO:0060061 | Spemann organizer formation | 2 | 3 | 4.55e-01 |
| GO:BP | GO:0043970 | histone H3-K9 acetylation | 2 | 8 | 4.55e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 4 | 108 | 4.55e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 4.56e-01 |
| GO:BP | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER | 2 | 2 | 4.56e-01 |
| GO:BP | GO:0009222 | pyrimidine ribonucleotide catabolic process | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 4.56e-01 |
| GO:BP | GO:2000793 | cell proliferation involved in heart valve development | 3 | 4 | 4.56e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 56 | 1088 | 4.56e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 7 | 10 | 4.56e-01 |
| GO:BP | GO:0009131 | pyrimidine nucleoside monophosphate catabolic process | 1 | 6 | 4.56e-01 |
| GO:BP | GO:0051490 | negative regulation of filopodium assembly | 3 | 6 | 4.56e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 2 | 15 | 4.56e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 11 | 28 | 4.56e-01 |
| GO:BP | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process | 2 | 3 | 4.56e-01 |
| GO:BP | GO:1902261 | positive regulation of delayed rectifier potassium channel activity | 1 | 5 | 4.56e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 3 | 36 | 4.57e-01 |
| GO:BP | GO:0140466 | iron-sulfur cluster export from the mitochondrion | 1 | 1 | 4.57e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 4 | 13 | 4.57e-01 |
| GO:BP | GO:1903331 | positive regulation of iron-sulfur cluster assembly | 1 | 1 | 4.57e-01 |
| GO:BP | GO:1903329 | regulation of iron-sulfur cluster assembly | 1 | 1 | 4.57e-01 |
| GO:BP | GO:1902497 | iron-sulfur cluster transmembrane transport | 1 | 1 | 4.57e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 5 | 14 | 4.57e-01 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 2 | 14 | 4.58e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 4.58e-01 |
| GO:BP | GO:0007350 | blastoderm segmentation | 2 | 17 | 4.58e-01 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 4.58e-01 |
| GO:BP | GO:0001556 | oocyte maturation | 2 | 22 | 4.58e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 4.58e-01 |
| GO:BP | GO:0033599 | regulation of mammary gland epithelial cell proliferation | 1 | 14 | 4.59e-01 |
| GO:BP | GO:0021894 | cerebral cortex GABAergic interneuron development | 1 | 3 | 4.59e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 4.60e-01 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 7 | 29 | 4.60e-01 |
| GO:BP | GO:0009097 | isoleucine biosynthetic process | 1 | 2 | 4.61e-01 |
| GO:BP | GO:0009099 | valine biosynthetic process | 1 | 3 | 4.61e-01 |
| GO:BP | GO:0097332 | response to antipsychotic drug | 2 | 2 | 4.61e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 3 | 6 | 4.61e-01 |
| GO:BP | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 4.61e-01 |
| GO:BP | GO:0046149 | pigment catabolic process | 1 | 6 | 4.61e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 22 | 490 | 4.61e-01 |
| GO:BP | GO:0051648 | vesicle localization | 17 | 192 | 4.61e-01 |
| GO:BP | GO:0042167 | heme catabolic process | 1 | 6 | 4.61e-01 |
| GO:BP | GO:1900073 | regulation of neuromuscular synaptic transmission | 1 | 2 | 4.61e-01 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 3 | 66 | 4.61e-01 |
| GO:BP | GO:1900248 | negative regulation of cytoplasmic translational elongation | 2 | 2 | 4.61e-01 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 3 | 66 | 4.61e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 10 | 20 | 4.61e-01 |
| GO:BP | GO:1900075 | positive regulation of neuromuscular synaptic transmission | 1 | 2 | 4.61e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 22 | 490 | 4.61e-01 |
| GO:BP | GO:0072174 | metanephric tubule formation | 2 | 3 | 4.61e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 4.61e-01 |
| GO:BP | GO:0048371 | lateral mesodermal cell differentiation | 2 | 2 | 4.61e-01 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 7 | 27 | 4.61e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 3 | 26 | 4.61e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 4 | 47 | 4.61e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 2 | 24 | 4.62e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 4 | 19 | 4.62e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 4.62e-01 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 4.62e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 3 | 3 | 4.62e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 1 | 26 | 4.62e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 4.62e-01 |
| GO:BP | GO:0140820 | cytosol to Golgi apparatus transport | 1 | 4 | 4.62e-01 |
| GO:BP | GO:0035498 | carnosine metabolic process | 1 | 2 | 4.62e-01 |
| GO:BP | GO:0061734 | parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 4 | 6 | 4.62e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 2 | 86 | 4.62e-01 |
| GO:BP | GO:0070203 | regulation of establishment of protein localization to telomere | 4 | 11 | 4.62e-01 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 2 | 13 | 4.62e-01 |
| GO:BP | GO:0006527 | arginine catabolic process | 2 | 7 | 4.62e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 4 | 72 | 4.62e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 8 | 12 | 4.62e-01 |
| GO:BP | GO:0044057 | regulation of system process | 23 | 405 | 4.62e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 2 | 6 | 4.62e-01 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 2 | 3 | 4.62e-01 |
| GO:BP | GO:0006558 | L-phenylalanine metabolic process | 2 | 3 | 4.62e-01 |
| GO:BP | GO:1902222 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process | 2 | 3 | 4.62e-01 |
| GO:BP | GO:0006559 | L-phenylalanine catabolic process | 2 | 3 | 4.62e-01 |
| GO:BP | GO:0019068 | virion assembly | 9 | 35 | 4.62e-01 |
| GO:BP | GO:0048469 | cell maturation | 8 | 108 | 4.62e-01 |
| GO:BP | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | 1 | 8 | 4.62e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 4.62e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 3 | 29 | 4.62e-01 |
| GO:BP | GO:0038163 | thrombopoietin-mediated signaling pathway | 1 | 3 | 4.62e-01 |
| GO:BP | GO:0120187 | positive regulation of protein localization to chromatin | 1 | 3 | 4.62e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 5 | 34 | 4.62e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 4 | 10 | 4.62e-01 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 4.62e-01 |
| GO:BP | GO:0036089 | cleavage furrow formation | 2 | 7 | 4.62e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 2 | 18 | 4.62e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 1 | 111 | 4.62e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 18 | 4.62e-01 |
| GO:BP | GO:0030046 | parallel actin filament bundle assembly | 2 | 5 | 4.62e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 4 | 10 | 4.62e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 302 | 4.63e-01 |
| GO:BP | GO:0051694 | pointed-end actin filament capping | 2 | 6 | 4.63e-01 |
| GO:BP | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 1 | 3 | 4.63e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 6 | 35 | 4.63e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 4.63e-01 |
| GO:BP | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 5 | 4.63e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 3 | 19 | 4.64e-01 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 4.65e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 19 | 625 | 4.65e-01 |
| GO:BP | GO:0099551 | trans-synaptic signaling by neuropeptide, modulating synaptic transmission | 1 | 1 | 4.65e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 25 | 50 | 4.65e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 3 | 25 | 4.65e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 8 | 14 | 4.65e-01 |
| GO:BP | GO:0060708 | spongiotrophoblast differentiation | 2 | 2 | 4.65e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 2 | 70 | 4.65e-01 |
| GO:BP | GO:1905450 | negative regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 2 | 2 | 4.65e-01 |
| GO:BP | GO:0001525 | angiogenesis | 22 | 390 | 4.65e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 14 | 32 | 4.65e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 4.66e-01 |
| GO:BP | GO:0035781 | CD86 biosynthetic process | 2 | 2 | 4.66e-01 |
| GO:BP | GO:1905605 | positive regulation of blood-brain barrier permeability | 3 | 4 | 4.66e-01 |
| GO:BP | GO:0007292 | female gamete generation | 9 | 103 | 4.66e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 1 | 78 | 4.66e-01 |
| GO:BP | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0099163 | synaptic signaling by nitric oxide | 1 | 3 | 4.66e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0039536 | negative regulation of RIG-I signaling pathway | 2 | 7 | 4.66e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 3 | 5 | 4.67e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 1 | 10 | 4.67e-01 |
| GO:BP | GO:1905341 | negative regulation of protein localization to kinetochore | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 2 | 4 | 4.67e-01 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 2 | 4 | 4.67e-01 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 2 | 4 | 4.67e-01 |
| GO:BP | GO:0051350 | negative regulation of lyase activity | 3 | 15 | 4.67e-01 |
| GO:BP | GO:1903934 | positive regulation of DNA primase activity | 2 | 5 | 4.67e-01 |
| GO:BP | GO:1903932 | regulation of DNA primase activity | 2 | 5 | 4.67e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 3 | 82 | 4.67e-01 |
| GO:BP | GO:0030844 | positive regulation of intermediate filament depolymerization | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0030842 | regulation of intermediate filament depolymerization | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0045106 | intermediate filament depolymerization | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 3 | 34 | 4.67e-01 |
| GO:BP | GO:0045108 | regulation of intermediate filament polymerization or depolymerization | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0055111 | ingression involved in gastrulation with mouth forming second | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 7 | 29 | 4.67e-01 |
| GO:BP | GO:0006997 | nucleus organization | 7 | 126 | 4.67e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 4 | 57 | 4.67e-01 |
| GO:BP | GO:0080021 | response to benzoic acid | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 2 | 104 | 4.68e-01 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 2 | 20 | 4.68e-01 |
| GO:BP | GO:0051769 | regulation of nitric-oxide synthase biosynthetic process | 1 | 13 | 4.68e-01 |
| GO:BP | GO:0051767 | nitric-oxide synthase biosynthetic process | 1 | 13 | 4.68e-01 |
| GO:BP | GO:0019045 | latent virus replication | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0019046 | release from viral latency | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0110008 | ncRNA deadenylation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0034759 | regulation of iron ion transmembrane transport | 1 | 2 | 4.68e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 1 | 55 | 4.69e-01 |
| GO:BP | GO:0001947 | heart looping | 4 | 50 | 4.69e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 1 | 316 | 4.69e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 29 | 249 | 4.69e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 12 | 184 | 4.69e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 4 | 12 | 4.69e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 6 | 97 | 4.69e-01 |
| GO:BP | GO:0030208 | dermatan sulfate biosynthetic process | 2 | 5 | 4.69e-01 |
| GO:BP | GO:0010585 | glutamine secretion | 2 | 2 | 4.69e-01 |
| GO:BP | GO:0099171 | presynaptic modulation of chemical synaptic transmission | 2 | 26 | 4.69e-01 |
| GO:BP | GO:0071257 | cellular response to electrical stimulus | 2 | 8 | 4.69e-01 |
| GO:BP | GO:1902045 | negative regulation of Fas signaling pathway | 2 | 2 | 4.69e-01 |
| GO:BP | GO:0001889 | liver development | 16 | 110 | 4.70e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 9 | 26 | 4.70e-01 |
| GO:BP | GO:0009408 | response to heat | 5 | 84 | 4.70e-01 |
| GO:BP | GO:0033278 | cell proliferation in midbrain | 1 | 4 | 4.70e-01 |
| GO:BP | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process | 2 | 5 | 4.70e-01 |
| GO:BP | GO:0097466 | ubiquitin-dependent glycoprotein ERAD pathway | 2 | 5 | 4.70e-01 |
| GO:BP | GO:0051964 | negative regulation of synapse assembly | 1 | 6 | 4.70e-01 |
| GO:BP | GO:1905331 | negative regulation of morphogenesis of an epithelium | 1 | 6 | 4.70e-01 |
| GO:BP | GO:1904382 | mannose trimming involved in glycoprotein ERAD pathway | 2 | 5 | 4.70e-01 |
| GO:BP | GO:0002317 | plasma cell differentiation | 1 | 5 | 4.71e-01 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 8 | 12 | 4.71e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 4 | 22 | 4.71e-01 |
| GO:BP | GO:0048792 | spontaneous exocytosis of neurotransmitter | 1 | 1 | 4.71e-01 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 5 | 8 | 4.71e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 6 | 104 | 4.71e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 7 | 40 | 4.71e-01 |
| GO:BP | GO:0061000 | negative regulation of dendritic spine development | 5 | 10 | 4.71e-01 |
| GO:BP | GO:0097421 | liver regeneration | 3 | 23 | 4.72e-01 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 1 | 12 | 4.72e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 4.72e-01 |
| GO:BP | GO:0010820 | positive regulation of T cell chemotaxis | 3 | 7 | 4.72e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 2 | 15 | 4.72e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 4.72e-01 |
| GO:BP | GO:0019050 | suppression by virus of host apoptotic process | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0055129 | L-proline biosynthetic process | 3 | 5 | 4.72e-01 |
| GO:BP | GO:0033668 | suppression by symbiont of host apoptotic process | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0019388 | galactose catabolic process | 2 | 5 | 4.72e-01 |
| GO:BP | GO:0060180 | female mating behavior | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0045924 | regulation of female receptivity | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0033630 | positive regulation of cell adhesion mediated by integrin | 2 | 14 | 4.72e-01 |
| GO:BP | GO:0019051 | induction by virus of host apoptotic process | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0006391 | transcription initiation at mitochondrial promoter | 3 | 4 | 4.72e-01 |
| GO:BP | GO:1901166 | neural crest cell migration involved in autonomic nervous system development | 4 | 5 | 4.72e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 2 | 5 | 4.72e-01 |
| GO:BP | GO:0052151 | positive regulation by symbiont of host apoptotic process | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0006561 | proline biosynthetic process | 3 | 5 | 4.72e-01 |
| GO:BP | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway | 1 | 6 | 4.72e-01 |
| GO:BP | GO:0002154 | thyroid hormone mediated signaling pathway | 2 | 6 | 4.72e-01 |
| GO:BP | GO:0060613 | fat pad development | 1 | 6 | 4.72e-01 |
| GO:BP | GO:0052041 | suppression by symbiont of host programmed cell death | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 22 | 495 | 4.72e-01 |
| GO:BP | GO:0030220 | platelet formation | 2 | 17 | 4.72e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 40 | 106 | 4.72e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 2 | 106 | 4.72e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 88 | 164 | 4.72e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 31 | 665 | 4.72e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 4 | 40 | 4.72e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 30 | 400 | 4.72e-01 |
| GO:BP | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0052042 | induction by symbiont of host programmed cell death | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 5 | 20 | 4.72e-01 |
| GO:BP | GO:0098939 | dendritic transport of mitochondrion | 1 | 2 | 4.72e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 46 | 110 | 4.72e-01 |
| GO:BP | GO:0001907 | killing by symbiont of host cells | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 7 | 149 | 4.72e-01 |
| GO:BP | GO:0061709 | reticulophagy | 2 | 17 | 4.72e-01 |
| GO:BP | GO:1905395 | response to flavonoid | 2 | 2 | 4.72e-01 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 4 | 9 | 4.72e-01 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 1 | 14 | 4.72e-01 |
| GO:BP | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation | 1 | 6 | 4.72e-01 |
| GO:BP | GO:0021761 | limbic system development | 5 | 81 | 4.72e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 1 | 7 | 4.72e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 3 | 271 | 4.73e-01 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 1 | 5 | 4.73e-01 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0015824 | proline transport | 4 | 11 | 4.73e-01 |
| GO:BP | GO:1902389 | ceramide 1-phosphate transport | 1 | 7 | 4.73e-01 |
| GO:BP | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0097326 | melanocyte adhesion | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0019742 | pentacyclic triterpenoid metabolic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 6 | 85 | 4.73e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 3 | 39 | 4.73e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 2 | 19 | 4.73e-01 |
| GO:BP | GO:0060489 | planar dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0002481 | antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent | 2 | 2 | 4.73e-01 |
| GO:BP | GO:0036048 | protein desuccinylation | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0071528 | tRNA re-export from nucleus | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 5 | 17 | 4.73e-01 |
| GO:BP | GO:0071642 | positive regulation of macrophage inflammatory protein 1 alpha production | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 6 | 11 | 4.73e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 8 | 24 | 4.73e-01 |
| GO:BP | GO:0009056 | catabolic process | 59 | 2088 | 4.73e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 11 | 96 | 4.73e-01 |
| GO:BP | GO:0120072 | positive regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 2 | 33 | 4.73e-01 |
| GO:BP | GO:0015961 | diadenosine polyphosphate catabolic process | 4 | 6 | 4.73e-01 |
| GO:BP | GO:0120071 | regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 3 | 4 | 4.73e-01 |
| GO:BP | GO:0009597 | detection of virus | 2 | 5 | 4.73e-01 |
| GO:BP | GO:0006417 | regulation of translation | 4 | 370 | 4.73e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 2 | 15 | 4.73e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 2 | 15 | 4.73e-01 |
| GO:BP | GO:0035891 | exit from host cell | 1 | 28 | 4.73e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 39 | 140 | 4.73e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 2 | 67 | 4.73e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 24 | 281 | 4.73e-01 |
| GO:BP | GO:0014828 | distal stomach smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 8 | 256 | 4.73e-01 |
| GO:BP | GO:0120065 | pyloric antrum smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0071279 | cellular response to cobalt ion | 1 | 4 | 4.73e-01 |
| GO:BP | GO:0048736 | appendage development | 1 | 142 | 4.73e-01 |
| GO:BP | GO:0007417 | central nervous system development | 9 | 792 | 4.73e-01 |
| GO:BP | GO:1990575 | mitochondrial L-ornithine transmembrane transport | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 5 | 17 | 4.73e-01 |
| GO:BP | GO:0071625 | vocalization behavior | 2 | 13 | 4.73e-01 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 2 | 22 | 4.73e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 54 | 257 | 4.73e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 9 | 92 | 4.73e-01 |
| GO:BP | GO:0060490 | lateral sprouting involved in lung morphogenesis | 1 | 1 | 4.73e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 3 | 20 | 4.73e-01 |
| GO:BP | GO:0003174 | mitral valve development | 5 | 12 | 4.73e-01 |
| GO:BP | GO:0120063 | stomach smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0034263 | positive regulation of autophagy in response to ER overload | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0120064 | stomach pylorus smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway | 3 | 17 | 4.73e-01 |
| GO:BP | GO:0021896 | forebrain astrocyte differentiation | 2 | 2 | 4.73e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 5 | 10 | 4.73e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 6 | 11 | 4.73e-01 |
| GO:BP | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol | 2 | 3 | 4.73e-01 |
| GO:BP | GO:1902656 | calcium ion import into cytosol | 1 | 1 | 4.73e-01 |
| GO:BP | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 4 | 5 | 4.73e-01 |
| GO:BP | GO:1904343 | positive regulation of colon smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 4 | 16 | 4.73e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 1 | 20 | 4.73e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 5 | 10 | 4.73e-01 |
| GO:BP | GO:0098779 | positive regulation of mitophagy in response to mitochondrial depolarization | 6 | 10 | 4.73e-01 |
| GO:BP | GO:1904341 | regulation of colon smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 1 | 41 | 4.73e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 2 | 41 | 4.73e-01 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 5 | 11 | 4.73e-01 |
| GO:BP | GO:0006882 | intracellular zinc ion homeostasis | 4 | 28 | 4.73e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 4.73e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 1 | 30 | 4.73e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 2 | 13 | 4.73e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 9 | 62 | 4.73e-01 |
| GO:BP | GO:0051892 | negative regulation of cardioblast differentiation | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 1 | 7 | 4.73e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 4.73e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 2 | 15 | 4.73e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 4 | 59 | 4.73e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 6 | 90 | 4.73e-01 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0046057 | dADP catabolic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0018364 | peptidyl-glutamine methylation | 1 | 4 | 4.73e-01 |
| GO:BP | GO:0061159 | establishment of bipolar cell polarity involved in cell morphogenesis | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 2 | 72 | 4.73e-01 |
| GO:BP | GO:0007182 | common-partner SMAD protein phosphorylation | 1 | 6 | 4.73e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 2 | 74 | 4.73e-01 |
| GO:BP | GO:1990765 | colon smooth muscle contraction | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0071455 | cellular response to hyperoxia | 1 | 5 | 4.73e-01 |
| GO:BP | GO:0022008 | neurogenesis | 2 | 1294 | 4.73e-01 |
| GO:BP | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 7 | 17 | 4.73e-01 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 4 | 12 | 4.73e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 4.73e-01 |
| GO:BP | GO:0036049 | peptidyl-lysine desuccinylation | 1 | 2 | 4.73e-01 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0010038 | response to metal ion | 35 | 264 | 4.73e-01 |
| GO:BP | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 4.73e-01 |
| GO:BP | GO:0009192 | deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0039534 | negative regulation of MDA-5 signaling pathway | 2 | 3 | 4.73e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 1 | 34 | 4.73e-01 |
| GO:BP | GO:0061698 | protein deglutarylation | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 1 | 7 | 4.73e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 2 | 74 | 4.73e-01 |
| GO:BP | GO:0106230 | protein depropionylation | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 1 | 10 | 4.73e-01 |
| GO:BP | GO:0010831 | positive regulation of myotube differentiation | 5 | 10 | 4.73e-01 |
| GO:BP | GO:0000050 | urea cycle | 2 | 7 | 4.73e-01 |
| GO:BP | GO:0019076 | viral release from host cell | 1 | 28 | 4.73e-01 |
| GO:BP | GO:0046067 | dGDP catabolic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 4.73e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 4.73e-01 |
| GO:BP | GO:1900364 | negative regulation of mRNA polyadenylation | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 6 | 21 | 4.73e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 8 | 32 | 4.73e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 2 | 6 | 4.73e-01 |
| GO:BP | GO:0060712 | spongiotrophoblast layer development | 5 | 11 | 4.73e-01 |
| GO:BP | GO:0060173 | limb development | 1 | 142 | 4.73e-01 |
| GO:BP | GO:0021897 | forebrain astrocyte development | 2 | 2 | 4.73e-01 |
| GO:BP | GO:0009184 | purine deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 4.73e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 1 | 15 | 4.74e-01 |
| GO:BP | GO:1900365 | positive regulation of mRNA polyadenylation | 3 | 4 | 4.74e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 7 | 31 | 4.74e-01 |
| GO:BP | GO:0060763 | mammary duct terminal end bud growth | 1 | 3 | 4.74e-01 |
| GO:BP | GO:0071409 | cellular response to cycloheximide | 2 | 2 | 4.74e-01 |
| GO:BP | GO:1905303 | positive regulation of macropinocytosis | 2 | 2 | 4.74e-01 |
| GO:BP | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway | 1 | 2 | 4.74e-01 |
| GO:BP | GO:0048873 | homeostasis of number of cells within a tissue | 2 | 24 | 4.74e-01 |
| GO:BP | GO:0000160 | phosphorelay signal transduction system | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0035732 | nitric oxide storage | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 4 | 6 | 4.74e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 3 | 46 | 4.74e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 6 | 16 | 4.74e-01 |
| GO:BP | GO:1905608 | positive regulation of presynapse assembly | 1 | 2 | 4.74e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 8 | 47 | 4.75e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 25 | 462 | 4.75e-01 |
| GO:BP | GO:0051216 | cartilage development | 7 | 151 | 4.75e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 5 | 75 | 4.76e-01 |
| GO:BP | GO:0090345 | cellular organohalogen metabolic process | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0090346 | cellular organofluorine metabolic process | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 2 | 75 | 4.76e-01 |
| GO:BP | GO:0097211 | cellular response to gonadotropin-releasing hormone | 2 | 3 | 4.76e-01 |
| GO:BP | GO:0038130 | ERBB4 signaling pathway | 3 | 6 | 4.76e-01 |
| GO:BP | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis | 1 | 3 | 4.76e-01 |
| GO:BP | GO:0008355 | olfactory learning | 2 | 2 | 4.76e-01 |
| GO:BP | GO:2000224 | regulation of testosterone biosynthetic process | 1 | 2 | 4.76e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 8 | 58 | 4.76e-01 |
| GO:BP | GO:0061184 | positive regulation of dermatome development | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0009786 | regulation of asymmetric cell division | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 7 | 17 | 4.76e-01 |
| GO:BP | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination | 3 | 4 | 4.76e-01 |
| GO:BP | GO:1990983 | tRNA demethylation | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0002337 | B-1a B cell differentiation | 2 | 2 | 4.76e-01 |
| GO:BP | GO:0034514 | mitochondrial unfolded protein response | 2 | 2 | 4.76e-01 |
| GO:BP | GO:0006837 | serotonin transport | 1 | 11 | 4.76e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 11 | 179 | 4.76e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 6 | 12 | 4.76e-01 |
| GO:BP | GO:0040007 | growth | 53 | 742 | 4.76e-01 |
| GO:BP | GO:0090166 | Golgi disassembly | 2 | 6 | 4.76e-01 |
| GO:BP | GO:0042126 | nitrate metabolic process | 3 | 4 | 4.76e-01 |
| GO:BP | GO:0038203 | TORC2 signaling | 6 | 12 | 4.76e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 4.76e-01 |
| GO:BP | GO:0014041 | regulation of neuron maturation | 1 | 6 | 4.76e-01 |
| GO:BP | GO:0045837 | negative regulation of membrane potential | 2 | 9 | 4.76e-01 |
| GO:BP | GO:0010917 | negative regulation of mitochondrial membrane potential | 2 | 9 | 4.76e-01 |
| GO:BP | GO:0001778 | plasma membrane repair | 1 | 28 | 4.76e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 5 | 48 | 4.76e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 4.76e-01 |
| GO:BP | GO:0097067 | cellular response to thyroid hormone stimulus | 5 | 14 | 4.77e-01 |
| GO:BP | GO:0033504 | floor plate development | 3 | 8 | 4.77e-01 |
| GO:BP | GO:1905786 | positive regulation of anaphase-promoting complex-dependent catabolic process | 1 | 2 | 4.77e-01 |
| GO:BP | GO:0033023 | mast cell homeostasis | 2 | 4 | 4.77e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 1 | 6 | 4.77e-01 |
| GO:BP | GO:0015744 | succinate transport | 1 | 6 | 4.77e-01 |
| GO:BP | GO:0002931 | response to ischemia | 5 | 46 | 4.77e-01 |
| GO:BP | GO:0034729 | histone H3-K79 methylation | 2 | 2 | 4.77e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 5 | 52 | 4.77e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 4.78e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 1 | 27 | 4.78e-01 |
| GO:BP | GO:0090270 | regulation of fibroblast growth factor production | 1 | 4 | 4.78e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 4.78e-01 |
| GO:BP | GO:0150077 | regulation of neuroinflammatory response | 1 | 13 | 4.78e-01 |
| GO:BP | GO:0090269 | fibroblast growth factor production | 1 | 4 | 4.78e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 5 | 16 | 4.78e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 7 | 111 | 4.78e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 2 | 7 | 4.78e-01 |
| GO:BP | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome | 1 | 1 | 4.78e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 4 | 67 | 4.78e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 4.78e-01 |
| GO:BP | GO:0009957 | epidermal cell fate specification | 2 | 4 | 4.78e-01 |
| GO:BP | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis | 2 | 4 | 4.78e-01 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 2 | 13 | 4.78e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 2 | 3 | 4.78e-01 |
| GO:BP | GO:0060398 | regulation of growth hormone receptor signaling pathway | 2 | 5 | 4.78e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 4.78e-01 |
| GO:BP | GO:0001756 | somitogenesis | 7 | 48 | 4.78e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 21 | 706 | 4.78e-01 |
| GO:BP | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process | 1 | 3 | 4.78e-01 |
| GO:BP | GO:0060753 | regulation of mast cell chemotaxis | 1 | 7 | 4.78e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 2 | 4 | 4.78e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 4.78e-01 |
| GO:BP | GO:1903062 | regulation of reverse cholesterol transport | 1 | 1 | 4.78e-01 |
| GO:BP | GO:1903064 | positive regulation of reverse cholesterol transport | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0071317 | cellular response to isoquinoline alkaloid | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0071315 | cellular response to morphine | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 1 | 6 | 4.79e-01 |
| GO:BP | GO:0106217 | tRNA C3-cytosine methylation | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 7 | 15 | 4.79e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 2 | 6 | 4.79e-01 |
| GO:BP | GO:0002315 | marginal zone B cell differentiation | 1 | 6 | 4.79e-01 |
| GO:BP | GO:0001927 | exocyst assembly | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0060178 | regulation of exocyst localization | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0001928 | regulation of exocyst assembly | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 3 | 62 | 4.79e-01 |
| GO:BP | GO:1904117 | cellular response to vasopressin | 2 | 5 | 4.79e-01 |
| GO:BP | GO:1904116 | response to vasopressin | 2 | 5 | 4.79e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 1 | 272 | 4.79e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 6 | 26 | 4.79e-01 |
| GO:BP | GO:2000394 | positive regulation of lamellipodium morphogenesis | 3 | 6 | 4.79e-01 |
| GO:BP | GO:1904533 | regulation of telomeric loop disassembly | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 5 | 23 | 4.79e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 18 | 31 | 4.79e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 7 | 105 | 4.79e-01 |
| GO:BP | GO:0071313 | cellular response to caffeine | 2 | 10 | 4.79e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 3 | 14 | 4.79e-01 |
| GO:BP | GO:0061738 | late endosomal microautophagy | 2 | 3 | 4.79e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 3 | 5 | 4.80e-01 |
| GO:BP | GO:1904531 | positive regulation of actin filament binding | 1 | 2 | 4.80e-01 |
| GO:BP | GO:1904618 | positive regulation of actin binding | 1 | 2 | 4.80e-01 |
| GO:BP | GO:0002069 | columnar/cuboidal epithelial cell maturation | 3 | 9 | 4.80e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 3 | 5 | 4.80e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 56 | 486 | 4.80e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 16 | 30 | 4.80e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 2 | 17 | 4.80e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 8 | 1389 | 4.80e-01 |
| GO:BP | GO:0048630 | skeletal muscle tissue growth | 1 | 8 | 4.80e-01 |
| GO:BP | GO:0051589 | negative regulation of neurotransmitter transport | 1 | 8 | 4.80e-01 |
| GO:BP | GO:0036211 | protein modification process | 2 | 2901 | 4.80e-01 |
| GO:BP | GO:0120049 | snRNA (adenine-N6)-methylation | 2 | 2 | 4.81e-01 |
| GO:BP | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 3 | 4 | 4.81e-01 |
| GO:BP | GO:2000808 | negative regulation of synaptic vesicle clustering | 1 | 1 | 4.81e-01 |
| GO:BP | GO:1901026 | ripoptosome assembly involved in necroptotic process | 2 | 2 | 4.81e-01 |
| GO:BP | GO:0097343 | ripoptosome assembly | 2 | 2 | 4.81e-01 |
| GO:BP | GO:0090669 | telomerase RNA stabilization | 2 | 4 | 4.81e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 3 | 39 | 4.81e-01 |
| GO:BP | GO:0042407 | cristae formation | 2 | 18 | 4.81e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 2 | 11 | 4.81e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 4 | 29 | 4.82e-01 |
| GO:BP | GO:0051646 | mitochondrion localization | 4 | 47 | 4.82e-01 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 3 | 72 | 4.82e-01 |
| GO:BP | GO:0001508 | action potential | 7 | 105 | 4.82e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 4 | 23 | 4.82e-01 |
| GO:BP | GO:1904526 | regulation of microtubule binding | 2 | 10 | 4.82e-01 |
| GO:BP | GO:0035281 | pre-miRNA export from nucleus | 2 | 2 | 4.82e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 1 | 8 | 4.82e-01 |
| GO:BP | GO:0051697 | protein delipidation | 1 | 5 | 4.82e-01 |
| GO:BP | GO:1902576 | negative regulation of nuclear cell cycle DNA replication | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0051664 | nuclear pore localization | 2 | 4 | 4.82e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 3 | 61 | 4.82e-01 |
| GO:BP | GO:0061015 | snRNA import into nucleus | 2 | 2 | 4.82e-01 |
| GO:BP | GO:1903464 | negative regulation of mitotic cell cycle DNA replication | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0001568 | blood vessel development | 28 | 534 | 4.82e-01 |
| GO:BP | GO:1904998 | negative regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 2 | 26 | 4.82e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 4.82e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 2 | 52 | 4.82e-01 |
| GO:BP | GO:2001288 | positive regulation of caveolin-mediated endocytosis | 2 | 3 | 4.82e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 2 | 8 | 4.82e-01 |
| GO:BP | GO:0006787 | porphyrin-containing compound catabolic process | 1 | 7 | 4.82e-01 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 5 | 8 | 4.82e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0033015 | tetrapyrrole catabolic process | 1 | 7 | 4.82e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 8 | 80 | 4.82e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 4.82e-01 |
| GO:BP | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway | 3 | 6 | 4.83e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 2 | 8 | 4.83e-01 |
| GO:BP | GO:0036298 | recombinational interstrand cross-link repair | 1 | 2 | 4.83e-01 |
| GO:BP | GO:0070716 | mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication | 1 | 2 | 4.83e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 2 | 108 | 4.83e-01 |
| GO:BP | GO:0097070 | ductus arteriosus closure | 1 | 2 | 4.83e-01 |
| GO:BP | GO:1901634 | positive regulation of synaptic vesicle membrane organization | 1 | 4 | 4.83e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 3 | 5 | 4.83e-01 |
| GO:BP | GO:0060064 | Spemann organizer formation at the anterior end of the primitive streak | 1 | 1 | 4.83e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 4.83e-01 |
| GO:BP | GO:2001014 | regulation of skeletal muscle cell differentiation | 2 | 16 | 4.83e-01 |
| GO:BP | GO:0003307 | regulation of Wnt signaling pathway involved in heart development | 1 | 7 | 4.83e-01 |
| GO:BP | GO:0015855 | pyrimidine nucleobase transport | 1 | 3 | 4.83e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 28 | 57 | 4.83e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 2 | 6 | 4.83e-01 |
| GO:BP | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 4 | 4.83e-01 |
| GO:BP | GO:0120263 | positive regulation of heterochromatin organization | 2 | 15 | 4.83e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 4.83e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 3 | 5 | 4.83e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 154 | 4.83e-01 |
| GO:BP | GO:0009414 | response to water deprivation | 1 | 4 | 4.83e-01 |
| GO:BP | GO:0015853 | adenine transport | 1 | 5 | 4.83e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 2 | 20 | 4.83e-01 |
| GO:BP | GO:0099502 | calcium-dependent activation of synaptic vesicle fusion | 1 | 4 | 4.83e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 3 | 5 | 4.83e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 5 | 12 | 4.83e-01 |
| GO:BP | GO:0015862 | uridine transport | 1 | 4 | 4.83e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 13 | 4.83e-01 |
| GO:BP | GO:0031453 | positive regulation of heterochromatin formation | 2 | 15 | 4.83e-01 |
| GO:BP | GO:0015864 | pyrimidine nucleoside transport | 1 | 4 | 4.83e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 3 | 7 | 4.83e-01 |
| GO:BP | GO:0046048 | UDP metabolic process | 2 | 7 | 4.83e-01 |
| GO:BP | GO:0090118 | receptor-mediated endocytosis involved in cholesterol transport | 1 | 5 | 4.83e-01 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 2 | 7 | 4.83e-01 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 12 | 15 | 4.83e-01 |
| GO:BP | GO:1905474 | canonical Wnt signaling pathway involved in stem cell proliferation | 1 | 1 | 4.83e-01 |
| GO:BP | GO:0006560 | proline metabolic process | 4 | 7 | 4.83e-01 |
| GO:BP | GO:0021796 | cerebral cortex regionalization | 1 | 2 | 4.83e-01 |
| GO:BP | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process | 2 | 3 | 4.83e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 49 | 313 | 4.83e-01 |
| GO:BP | GO:0060929 | atrioventricular node cell fate commitment | 2 | 2 | 4.83e-01 |
| GO:BP | GO:0071361 | cellular response to ethanol | 1 | 9 | 4.84e-01 |
| GO:BP | GO:0099532 | synaptic vesicle endosomal processing | 5 | 7 | 4.84e-01 |
| GO:BP | GO:0018283 | iron incorporation into metallo-sulfur cluster | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0018282 | metal incorporation into metallo-sulfur cluster | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 12 | 187 | 4.84e-01 |
| GO:BP | GO:0051902 | negative regulation of mitochondrial depolarization | 1 | 5 | 4.84e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 10 | 25 | 4.84e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 5 | 41 | 4.84e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 2 | 12 | 4.84e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 7 | 12 | 4.84e-01 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 13 | 34 | 4.84e-01 |
| GO:BP | GO:0097474 | retinal cone cell apoptotic process | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 1 | 2 | 4.85e-01 |
| GO:BP | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 6 | 4.85e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | 11 | 4.86e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 4 | 53 | 4.86e-01 |
| GO:BP | GO:0072356 | chromosome passenger complex localization to kinetochore | 1 | 1 | 4.86e-01 |
| GO:BP | GO:0014829 | vascular associated smooth muscle contraction | 5 | 20 | 4.86e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 20 | 55 | 4.86e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 2 | 25 | 4.86e-01 |
| GO:BP | GO:2000565 | negative regulation of CD8-positive, alpha-beta T cell proliferation | 2 | 2 | 4.86e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 710 | 4.86e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 38 | 4.86e-01 |
| GO:BP | GO:1990428 | miRNA transport | 2 | 3 | 4.86e-01 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 3 | 11 | 4.86e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 5 | 10 | 4.86e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 9 | 21 | 4.86e-01 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 2 | 4 | 4.86e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 5 | 10 | 4.86e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 2 | 38 | 4.86e-01 |
| GO:BP | GO:0038138 | ERBB4-ERBB4 signaling pathway | 3 | 3 | 4.86e-01 |
| GO:BP | GO:0046548 | retinal rod cell development | 2 | 6 | 4.86e-01 |
| GO:BP | GO:0032058 | positive regulation of translational initiation in response to stress | 1 | 4 | 4.86e-01 |
| GO:BP | GO:0051415 | microtubule nucleation by interphase microtubule organizing center | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 6 | 27 | 4.87e-01 |
| GO:BP | GO:0032714 | negative regulation of interleukin-5 production | 2 | 2 | 4.87e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 2 | 21 | 4.87e-01 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 1 | 97 | 4.87e-01 |
| GO:BP | GO:2000254 | regulation of male germ cell proliferation | 1 | 4 | 4.87e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 69 | 127 | 4.87e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 9 | 53 | 4.87e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 2 | 4 | 4.87e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 6 | 27 | 4.87e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 2 | 16 | 4.87e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 2 | 16 | 4.87e-01 |
| GO:BP | GO:0007033 | vacuole organization | 6 | 197 | 4.87e-01 |
| GO:BP | GO:0019321 | pentose metabolic process | 2 | 9 | 4.87e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 8 | 24 | 4.88e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 4.88e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 27 | 901 | 4.88e-01 |
| GO:BP | GO:0071788 | endoplasmic reticulum tubular network maintenance | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0003195 | tricuspid valve formation | 2 | 2 | 4.88e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 8 | 197 | 4.88e-01 |
| GO:BP | GO:0098725 | symmetric cell division | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 29 | 170 | 4.88e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 8 | 151 | 4.88e-01 |
| GO:BP | GO:1904777 | negative regulation of protein localization to cell cortex | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 3 | 11 | 4.88e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 4.88e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 1 | 18 | 4.88e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 4.88e-01 |
| GO:BP | GO:0035022 | positive regulation of Rac protein signal transduction | 3 | 5 | 4.88e-01 |
| GO:BP | GO:1905216 | positive regulation of RNA binding | 2 | 9 | 4.88e-01 |
| GO:BP | GO:0032400 | melanosome localization | 5 | 22 | 4.88e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 2 | 12 | 4.88e-01 |
| GO:BP | GO:0001712 | ectodermal cell fate commitment | 2 | 2 | 4.88e-01 |
| GO:BP | GO:0001802 | type III hypersensitivity | 1 | 2 | 4.88e-01 |
| GO:BP | GO:0001805 | positive regulation of type III hypersensitivity | 1 | 2 | 4.88e-01 |
| GO:BP | GO:0001803 | regulation of type III hypersensitivity | 1 | 2 | 4.88e-01 |
| GO:BP | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 1 | 6 | 4.88e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 3 | 21 | 4.88e-01 |
| GO:BP | GO:0106088 | regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 4.88e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 1 | 33 | 4.88e-01 |
| GO:BP | GO:0106089 | negative regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 1 | 11 | 4.88e-01 |
| GO:BP | GO:0021943 | formation of radial glial scaffolds | 1 | 3 | 4.88e-01 |
| GO:BP | GO:0071873 | response to norepinephrine | 1 | 2 | 4.88e-01 |
| GO:BP | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway | 4 | 7 | 4.88e-01 |
| GO:BP | GO:0071874 | cellular response to norepinephrine stimulus | 1 | 2 | 4.88e-01 |
| GO:BP | GO:2001016 | positive regulation of skeletal muscle cell differentiation | 1 | 5 | 4.88e-01 |
| GO:BP | GO:0060385 | axonogenesis involved in innervation | 1 | 7 | 4.88e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 2 | 26 | 4.88e-01 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 4 | 11 | 4.88e-01 |
| GO:BP | GO:0060980 | cell migration involved in coronary vasculogenesis | 1 | 1 | 4.89e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 5 | 53 | 4.89e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 32 | 164 | 4.89e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 4 | 10 | 4.89e-01 |
| GO:BP | GO:1903781 | positive regulation of cardiac conduction | 1 | 1 | 4.89e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 2 | 3 | 4.89e-01 |
| GO:BP | GO:0044782 | cilium organization | 43 | 313 | 4.89e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 5 | 57 | 4.89e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 5 | 8 | 4.89e-01 |
| GO:BP | GO:0010587 | miRNA catabolic process | 7 | 10 | 4.89e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 2 | 7 | 4.89e-01 |
| GO:BP | GO:0010819 | regulation of T cell chemotaxis | 3 | 7 | 4.90e-01 |
| GO:BP | GO:0002023 | reduction of food intake in response to dietary excess | 1 | 2 | 4.90e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 24 | 185 | 4.90e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 4 | 5 | 4.90e-01 |
| GO:BP | GO:0042732 | D-xylose metabolic process | 2 | 2 | 4.90e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 1 | 17 | 4.91e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 4.91e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 10 | 156 | 4.91e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 3 | 2646 | 4.91e-01 |
| GO:BP | GO:0003275 | apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 4.91e-01 |
| GO:BP | GO:1902256 | regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 4.91e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 4 | 72 | 4.91e-01 |
| GO:BP | GO:0060442 | branching involved in prostate gland morphogenesis | 2 | 7 | 4.91e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 5 | 85 | 4.91e-01 |
| GO:BP | GO:0061364 | apoptotic process involved in luteolysis | 2 | 3 | 4.91e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 2 | 19 | 4.91e-01 |
| GO:BP | GO:0002101 | tRNA wobble cytosine modification | 2 | 2 | 4.91e-01 |
| GO:BP | GO:0017182 | peptidyl-diphthamide metabolic process | 4 | 7 | 4.92e-01 |
| GO:BP | GO:0017183 | peptidyl-diphthamide biosynthetic process from peptidyl-histidine | 4 | 7 | 4.92e-01 |
| GO:BP | GO:1905223 | epicardium morphogenesis | 1 | 2 | 4.92e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 5 | 12 | 4.92e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 5 | 12 | 4.92e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 8 | 21 | 4.92e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 16 | 52 | 4.92e-01 |
| GO:BP | GO:0009826 | unidimensional cell growth | 1 | 3 | 4.92e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 1 | 33 | 4.93e-01 |
| GO:BP | GO:0036255 | response to methylamine | 1 | 1 | 4.93e-01 |
| GO:BP | GO:1903442 | response to lipoic acid | 1 | 1 | 4.93e-01 |
| GO:BP | GO:0072126 | positive regulation of glomerular mesangial cell proliferation | 2 | 7 | 4.93e-01 |
| GO:BP | GO:0006547 | histidine metabolic process | 2 | 5 | 4.93e-01 |
| GO:BP | GO:0036093 | germ cell proliferation | 3 | 7 | 4.94e-01 |
| GO:BP | GO:0009229 | thiamine diphosphate biosynthetic process | 1 | 1 | 4.94e-01 |
| GO:BP | GO:0042724 | thiamine-containing compound biosynthetic process | 1 | 1 | 4.94e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 2 | 2 | 4.94e-01 |
| GO:BP | GO:0110135 | Norrin signaling pathway | 2 | 2 | 4.94e-01 |
| GO:BP | GO:1902866 | regulation of retina development in camera-type eye | 1 | 1 | 4.94e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 1 | 8 | 4.94e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 2 | 20 | 4.94e-01 |
| GO:BP | GO:2000589 | regulation of metanephric mesenchymal cell migration | 1 | 3 | 4.94e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 2 | 9 | 4.94e-01 |
| GO:BP | GO:0035789 | metanephric mesenchymal cell migration | 1 | 3 | 4.94e-01 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 4.94e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 4 | 10 | 4.94e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 8 | 18 | 4.94e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 3 | 19 | 4.94e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 17 | 61 | 4.94e-01 |
| GO:BP | GO:0007498 | mesoderm development | 2 | 89 | 4.95e-01 |
| GO:BP | GO:0031443 | fast-twitch skeletal muscle fiber contraction | 2 | 3 | 4.95e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 2 | 3 | 4.95e-01 |
| GO:BP | GO:0036371 | protein localization to T-tubule | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0050955 | thermoception | 1 | 3 | 4.95e-01 |
| GO:BP | GO:0070662 | mast cell proliferation | 3 | 6 | 4.95e-01 |
| GO:BP | GO:0070666 | regulation of mast cell proliferation | 3 | 6 | 4.95e-01 |
| GO:BP | GO:1903621 | protein localization to photoreceptor connecting cilium | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0032868 | response to insulin | 4 | 215 | 4.95e-01 |
| GO:BP | GO:0098657 | import into cell | 14 | 166 | 4.96e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 5 | 47 | 4.96e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 4 | 7 | 4.96e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 1 | 10 | 4.96e-01 |
| GO:BP | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process | 5 | 9 | 4.96e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 4 | 21 | 4.96e-01 |
| GO:BP | GO:0072189 | ureter development | 4 | 10 | 4.96e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 2 | 39 | 4.96e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 5 | 54 | 4.96e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 4.97e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 7 | 152 | 4.97e-01 |
| GO:BP | GO:0033564 | anterior/posterior axon guidance | 1 | 4 | 4.97e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 4.97e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 9 | 17 | 4.97e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 3 | 26 | 4.97e-01 |
| GO:BP | GO:0014900 | muscle hyperplasia | 1 | 2 | 4.97e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 4 | 378 | 4.97e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 4 | 9 | 4.97e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 14 | 4.98e-01 |
| GO:BP | GO:0003002 | regionalization | 3 | 273 | 4.98e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 4 | 24 | 4.98e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 1 | 13 | 4.98e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 4 | 24 | 4.98e-01 |
| GO:BP | GO:0062036 | sensory perception of hot stimulus | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0120168 | detection of hot stimulus involved in thermoception | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 1 | 4 | 4.98e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 3 | 18 | 4.98e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 9 | 70 | 4.98e-01 |
| GO:BP | GO:0002545 | chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0002445 | type II hypersensitivity | 2 | 3 | 4.98e-01 |
| GO:BP | GO:0002880 | regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 1 | 19 | 4.98e-01 |
| GO:BP | GO:0002882 | positive regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.98e-01 |
| GO:BP | GO:0015819 | lysine transport | 2 | 2 | 4.98e-01 |
| GO:BP | GO:0002885 | positive regulation of hypersensitivity | 2 | 5 | 4.98e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 9 | 100 | 4.98e-01 |
| GO:BP | GO:0001794 | type IIa hypersensitivity | 2 | 3 | 4.98e-01 |
| GO:BP | GO:0110112 | regulation of lipid transporter activity | 1 | 2 | 4.99e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 4 | 8 | 4.99e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 8 | 101 | 4.99e-01 |
| GO:BP | GO:0015838 | amino-acid betaine transport | 1 | 7 | 5.00e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 24 | 44 | 5.00e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 8 | 14 | 5.00e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 2 | 16 | 5.00e-01 |
| GO:BP | GO:0051548 | negative regulation of keratinocyte migration | 2 | 2 | 5.00e-01 |
| GO:BP | GO:0051823 | regulation of synapse structural plasticity | 2 | 5 | 5.00e-01 |
| GO:BP | GO:0034197 | triglyceride transport | 1 | 4 | 5.00e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 4 | 9 | 5.00e-01 |
| GO:BP | GO:0034196 | acylglycerol transport | 1 | 4 | 5.00e-01 |
| GO:BP | GO:1901390 | positive regulation of transforming growth factor beta activation | 2 | 3 | 5.01e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 6 | 55 | 5.01e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 5 | 25 | 5.01e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 5 | 5.01e-01 |
| GO:BP | GO:2000392 | regulation of lamellipodium morphogenesis | 4 | 10 | 5.01e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 5 | 25 | 5.01e-01 |
| GO:BP | GO:0003270 | Notch signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0060843 | venous endothelial cell differentiation | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 4 | 10 | 5.01e-01 |
| GO:BP | GO:0003182 | coronary sinus valve morphogenesis | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 2 | 14 | 5.01e-01 |
| GO:BP | GO:1902723 | negative regulation of skeletal muscle satellite cell proliferation | 2 | 2 | 5.01e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 9 | 97 | 5.01e-01 |
| GO:BP | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation | 2 | 2 | 5.01e-01 |
| GO:BP | GO:1903545 | cellular response to butyrate | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0061772 | xenobiotic transport across blood-nerve barrier | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0033027 | positive regulation of mast cell apoptotic process | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 7 | 21 | 5.01e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 2 | 40 | 5.01e-01 |
| GO:BP | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib | 2 | 5 | 5.01e-01 |
| GO:BP | GO:1903975 | regulation of glial cell migration | 2 | 12 | 5.01e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 1 | 18 | 5.01e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 10 | 20 | 5.01e-01 |
| GO:BP | GO:0003178 | coronary sinus valve development | 1 | 1 | 5.01e-01 |
| GO:BP | GO:0098711 | iron ion import across plasma membrane | 1 | 4 | 5.01e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 2 | 3090 | 5.01e-01 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 6 | 18 | 5.01e-01 |
| GO:BP | GO:1903765 | negative regulation of potassium ion export across plasma membrane | 1 | 1 | 5.02e-01 |
| GO:BP | GO:0032480 | negative regulation of type I interferon production | 9 | 23 | 5.02e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 16 | 31 | 5.02e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 3 | 11 | 5.02e-01 |
| GO:BP | GO:0007584 | response to nutrient | 3 | 109 | 5.02e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 40 | 198 | 5.02e-01 |
| GO:BP | GO:0070202 | regulation of establishment of protein localization to chromosome | 4 | 12 | 5.02e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 14 | 139 | 5.02e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 5.02e-01 |
| GO:BP | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | 1 | 5 | 5.02e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 2 | 29 | 5.02e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.02e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.02e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 5.02e-01 |
| GO:BP | GO:0001830 | trophectodermal cell fate commitment | 1 | 1 | 5.02e-01 |
| GO:BP | GO:0032534 | regulation of microvillus assembly | 2 | 6 | 5.02e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 22 | 511 | 5.02e-01 |
| GO:BP | GO:0098693 | regulation of synaptic vesicle cycle | 1 | 13 | 5.02e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.02e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 24 | 1209 | 5.02e-01 |
| GO:BP | GO:0007625 | grooming behavior | 2 | 10 | 5.02e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 5.02e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 14 | 142 | 5.02e-01 |
| GO:BP | GO:2000074 | regulation of type B pancreatic cell development | 2 | 8 | 5.02e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 5.02e-01 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 2 | 6 | 5.02e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 7 | 68 | 5.02e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 3 | 14 | 5.02e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 7 | 53 | 5.02e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 1 | 4 | 5.02e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 16 | 63 | 5.02e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 3 | 5 | 5.02e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 1 | 26 | 5.03e-01 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 4 | 5 | 5.03e-01 |
| GO:BP | GO:0006853 | carnitine shuttle | 1 | 3 | 5.03e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 5.03e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 3 | 30 | 5.03e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 5.03e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 5.03e-01 |
| GO:BP | GO:1905269 | positive regulation of chromatin organization | 2 | 15 | 5.03e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 2 | 10 | 5.03e-01 |
| GO:BP | GO:2001286 | regulation of caveolin-mediated endocytosis | 3 | 5 | 5.03e-01 |
| GO:BP | GO:0032808 | lacrimal gland development | 2 | 5 | 5.03e-01 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 4 | 5 | 5.04e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 2 | 26 | 5.04e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 1 | 179 | 5.04e-01 |
| GO:BP | GO:0002943 | tRNA dihydrouridine synthesis | 2 | 4 | 5.04e-01 |
| GO:BP | GO:0042746 | circadian sleep/wake cycle, wakefulness | 1 | 1 | 5.04e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 6 | 38 | 5.04e-01 |
| GO:BP | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 5.04e-01 |
| GO:BP | GO:1904426 | positive regulation of GTP binding | 2 | 3 | 5.04e-01 |
| GO:BP | GO:0031129 | inductive cell-cell signaling | 1 | 1 | 5.05e-01 |
| GO:BP | GO:1902558 | 5’-adenylyl sulfate transmembrane transport | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0046963 | 3’-phosphoadenosine 5’-phosphosulfate transport | 2 | 2 | 5.05e-01 |
| GO:BP | GO:1902559 | 3’-phospho-5’-adenylyl sulfate transmembrane transport | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 5.05e-01 |
| GO:BP | GO:0045792 | negative regulation of cell size | 3 | 9 | 5.05e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 14 | 27 | 5.05e-01 |
| GO:BP | GO:0060384 | innervation | 5 | 22 | 5.05e-01 |
| GO:BP | GO:0017055 | negative regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 2 | 5.05e-01 |
| GO:BP | GO:0045925 | positive regulation of female receptivity | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0000154 | rRNA modification | 19 | 35 | 5.05e-01 |
| GO:BP | GO:1901290 | succinyl-CoA biosynthetic process | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 9 | 79 | 5.05e-01 |
| GO:BP | GO:0021603 | cranial nerve formation | 1 | 4 | 5.05e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 3 | 42 | 5.05e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 2 | 8 | 5.05e-01 |
| GO:BP | GO:0051594 | detection of glucose | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0048565 | digestive tract development | 14 | 92 | 5.05e-01 |
| GO:BP | GO:1901142 | insulin metabolic process | 3 | 11 | 5.05e-01 |
| GO:BP | GO:0034287 | detection of monosaccharide stimulus | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0009732 | detection of hexose stimulus | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0061682 | seminal vesicle morphogenesis | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0045659 | negative regulation of neutrophil differentiation | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 2 | 146 | 5.05e-01 |
| GO:BP | GO:0000710 | meiotic mismatch repair | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 1 | 719 | 5.05e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 5 | 15 | 5.05e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 2 | 83 | 5.05e-01 |
| GO:BP | GO:0014022 | neural plate elongation | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0021854 | hypothalamus development | 1 | 15 | 5.05e-01 |
| GO:BP | GO:0009730 | detection of carbohydrate stimulus | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0019678 | propionate metabolic process, methylmalonyl pathway | 1 | 1 | 5.05e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 5 | 10 | 5.05e-01 |
| GO:BP | GO:0000023 | maltose metabolic process | 1 | 2 | 5.05e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 5.05e-01 |
| GO:BP | GO:0022007 | convergent extension involved in neural plate elongation | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0007207 | phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | 3 | 6 | 5.05e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 11 | 52 | 5.05e-01 |
| GO:BP | GO:0006307 | DNA dealkylation involved in DNA repair | 4 | 9 | 5.05e-01 |
| GO:BP | GO:0042819 | vitamin B6 biosynthetic process | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission | 2 | 3 | 5.05e-01 |
| GO:BP | GO:0019264 | glycine biosynthetic process from serine | 1 | 2 | 5.05e-01 |
| GO:BP | GO:2000625 | regulation of miRNA catabolic process | 4 | 5 | 5.05e-01 |
| GO:BP | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 1 | 31 | 5.05e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 16 | 122 | 5.05e-01 |
| GO:BP | GO:0019085 | early viral transcription | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 6 | 25 | 5.05e-01 |
| GO:BP | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity | 1 | 3 | 5.06e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 21 | 200 | 5.06e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 5.06e-01 |
| GO:BP | GO:0048477 | oogenesis | 6 | 64 | 5.06e-01 |
| GO:BP | GO:1904556 | L-tryptophan transmembrane transport | 1 | 2 | 5.06e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 1 | 14 | 5.06e-01 |
| GO:BP | GO:0033273 | response to vitamin | 3 | 61 | 5.06e-01 |
| GO:BP | GO:0032259 | methylation | 1 | 313 | 5.06e-01 |
| GO:BP | GO:0071921 | cohesin loading | 2 | 3 | 5.06e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 1 | 28 | 5.06e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 798 | 5.06e-01 |
| GO:BP | GO:0010836 | negative regulation of protein ADP-ribosylation | 2 | 4 | 5.06e-01 |
| GO:BP | GO:0032240 | negative regulation of nucleobase-containing compound transport | 2 | 3 | 5.06e-01 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 10 | 18 | 5.06e-01 |
| GO:BP | GO:1903677 | regulation of cap-independent translational initiation | 2 | 3 | 5.06e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 5.06e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 3 | 179 | 5.06e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 2 | 10 | 5.06e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 13 | 222 | 5.06e-01 |
| GO:BP | GO:0061944 | negative regulation of protein K48-linked ubiquitination | 1 | 2 | 5.06e-01 |
| GO:BP | GO:1903679 | positive regulation of cap-independent translational initiation | 2 | 3 | 5.06e-01 |
| GO:BP | GO:0070294 | renal sodium ion absorption | 1 | 9 | 5.06e-01 |
| GO:BP | GO:0035654 | clathrin-coated vesicle cargo loading, AP-3-mediated | 4 | 6 | 5.06e-01 |
| GO:BP | GO:1900186 | negative regulation of clathrin-dependent endocytosis | 3 | 4 | 5.06e-01 |
| GO:BP | GO:0035652 | clathrin-coated vesicle cargo loading | 4 | 6 | 5.06e-01 |
| GO:BP | GO:0006451 | translational readthrough | 2 | 10 | 5.06e-01 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 10 | 5.06e-01 |
| GO:BP | GO:0046754 | viral exocytosis | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0002505 | antigen processing and presentation of polysaccharide antigen via MHC class II | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0002747 | antigen processing and presentation following phagocytosis | 1 | 1 | 5.07e-01 |
| GO:BP | GO:1902766 | skeletal muscle satellite cell migration | 1 | 2 | 5.07e-01 |
| GO:BP | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation | 2 | 2 | 5.07e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 15 | 175 | 5.07e-01 |
| GO:BP | GO:0060312 | regulation of blood vessel remodeling | 2 | 4 | 5.07e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 5.07e-01 |
| GO:BP | GO:0032799 | low-density lipoprotein receptor particle metabolic process | 2 | 5 | 5.07e-01 |
| GO:BP | GO:0007423 | sensory organ development | 1 | 396 | 5.07e-01 |
| GO:BP | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process | 2 | 5 | 5.07e-01 |
| GO:BP | GO:2000250 | negative regulation of actin cytoskeleton reorganization | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 3 | 11 | 5.07e-01 |
| GO:BP | GO:0032802 | low-density lipoprotein particle receptor catabolic process | 2 | 5 | 5.07e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 1 | 64 | 5.07e-01 |
| GO:BP | GO:2000657 | negative regulation of apolipoprotein binding | 1 | 1 | 5.07e-01 |
| GO:BP | GO:2000656 | regulation of apolipoprotein binding | 1 | 1 | 5.07e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 5.07e-01 |
| GO:BP | GO:0032329 | serine transport | 2 | 10 | 5.07e-01 |
| GO:BP | GO:0098903 | regulation of membrane repolarization during action potential | 1 | 6 | 5.07e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 1 | 9 | 5.07e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 4 | 6 | 5.07e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 12 | 30 | 5.07e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 13 | 36 | 5.07e-01 |
| GO:BP | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 2 | 3 | 5.07e-01 |
| GO:BP | GO:1903567 | regulation of protein localization to ciliary membrane | 1 | 2 | 5.07e-01 |
| GO:BP | GO:0000821 | regulation of arginine metabolic process | 2 | 3 | 5.07e-01 |
| GO:BP | GO:0032967 | positive regulation of collagen biosynthetic process | 3 | 23 | 5.07e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 2 | 3 | 5.07e-01 |
| GO:BP | GO:0021889 | olfactory bulb interneuron differentiation | 6 | 7 | 5.07e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 42 | 834 | 5.07e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 5.07e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 5.07e-01 |
| GO:BP | GO:0140192 | regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 2 | 3 | 5.07e-01 |
| GO:BP | GO:0071787 | endoplasmic reticulum tubular network formation | 2 | 4 | 5.07e-01 |
| GO:BP | GO:0042539 | hypotonic salinity response | 2 | 2 | 5.08e-01 |
| GO:BP | GO:0060020 | Bergmann glial cell differentiation | 4 | 9 | 5.08e-01 |
| GO:BP | GO:0003014 | renal system process | 9 | 78 | 5.08e-01 |
| GO:BP | GO:0071477 | cellular hypotonic salinity response | 2 | 2 | 5.08e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 5.08e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 5 | 85 | 5.08e-01 |
| GO:BP | GO:0015707 | nitrite transport | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0060525 | prostate glandular acinus development | 4 | 5 | 5.08e-01 |
| GO:BP | GO:0071236 | cellular response to antibiotic | 1 | 11 | 5.08e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 6 | 64 | 5.08e-01 |
| GO:BP | GO:0070417 | cellular response to cold | 1 | 8 | 5.08e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 17 | 300 | 5.08e-01 |
| GO:BP | GO:0051031 | tRNA transport | 3 | 5 | 5.08e-01 |
| GO:BP | GO:0046697 | decidualization | 5 | 19 | 5.08e-01 |
| GO:BP | GO:0032532 | regulation of microvillus length | 2 | 4 | 5.08e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 7 | 57 | 5.08e-01 |
| GO:BP | GO:0060348 | bone development | 1 | 170 | 5.08e-01 |
| GO:BP | GO:0006003 | fructose 2,6-bisphosphate metabolic process | 1 | 4 | 5.08e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 2 | 11 | 5.08e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 398 | 5.08e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 2 | 117 | 5.08e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 5.08e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 3 | 33 | 5.08e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 5.08e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 5.08e-01 |
| GO:BP | GO:0070318 | positive regulation of G0 to G1 transition | 1 | 5 | 5.08e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 5.08e-01 |
| GO:BP | GO:0006600 | creatine metabolic process | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 6 | 20 | 5.08e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 5.08e-01 |
| GO:BP | GO:0006049 | UDP-N-acetylglucosamine catabolic process | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 12 | 76 | 5.08e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 6 | 12 | 5.08e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 5.08e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 7 | 90 | 5.09e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 3 | 115 | 5.09e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 9 | 13 | 5.09e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 6 | 39 | 5.09e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 21 | 227 | 5.09e-01 |
| GO:BP | GO:1904353 | regulation of telomere capping | 2 | 23 | 5.09e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 5 | 22 | 5.09e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 5 | 66 | 5.09e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 6 | 29 | 5.09e-01 |
| GO:BP | GO:0060932 | His-Purkinje system cell differentiation | 3 | 4 | 5.09e-01 |
| GO:BP | GO:0098789 | pre-mRNA cleavage required for polyadenylation | 4 | 12 | 5.09e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 5.09e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 1 | 6 | 5.09e-01 |
| GO:BP | GO:0090500 | endocardial cushion to mesenchymal transition | 3 | 4 | 5.09e-01 |
| GO:BP | GO:0060479 | lung cell differentiation | 11 | 18 | 5.09e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 1 | 27 | 5.09e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 3 | 70 | 5.09e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 1 | 7 | 5.09e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 4 | 14 | 5.09e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 2 | 21 | 5.09e-01 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 11 | 18 | 5.09e-01 |
| GO:BP | GO:0046070 | dGTP metabolic process | 2 | 3 | 5.09e-01 |
| GO:BP | GO:0001955 | blood vessel maturation | 2 | 6 | 5.09e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 17 | 96 | 5.10e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 7 | 42 | 5.10e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 6 | 10 | 5.10e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 6 | 18 | 5.10e-01 |
| GO:BP | GO:0007402 | ganglion mother cell fate determination | 1 | 1 | 5.10e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 7 | 67 | 5.10e-01 |
| GO:BP | GO:0021537 | telencephalon development | 15 | 201 | 5.10e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 11 | 161 | 5.10e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 2 | 7 | 5.10e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 2 | 78 | 5.10e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 4 | 7 | 5.10e-01 |
| GO:BP | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate | 2 | 4 | 5.10e-01 |
| GO:BP | GO:0051030 | snRNA transport | 4 | 6 | 5.10e-01 |
| GO:BP | GO:0006592 | ornithine biosynthetic process | 1 | 1 | 5.10e-01 |
| GO:BP | GO:0051601 | exocyst localization | 1 | 4 | 5.11e-01 |
| GO:BP | GO:1901911 | adenosine 5’-(hexahydrogen pentaphosphate) catabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:1901909 | diadenosine hexaphosphate catabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 5 | 40 | 5.11e-01 |
| GO:BP | GO:0071543 | diphosphoinositol polyphosphate metabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:1901908 | diadenosine hexaphosphate metabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:1901907 | diadenosine pentaphosphate catabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:1901906 | diadenosine pentaphosphate metabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:1901910 | adenosine 5’-(hexahydrogen pentaphosphate) metabolic process | 2 | 5 | 5.11e-01 |
| GO:BP | GO:0009268 | response to pH | 4 | 26 | 5.11e-01 |
| GO:BP | GO:0061945 | regulation of protein K48-linked ubiquitination | 2 | 4 | 5.11e-01 |
| GO:BP | GO:0016197 | endosomal transport | 19 | 231 | 5.11e-01 |
| GO:BP | GO:0035021 | negative regulation of Rac protein signal transduction | 2 | 4 | 5.11e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 3 | 10 | 5.11e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 17 | 5.11e-01 |
| GO:BP | GO:0042779 | tRNA 3’-trailer cleavage | 1 | 3 | 5.11e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 3 | 10 | 5.11e-01 |
| GO:BP | GO:0046717 | acid secretion | 4 | 25 | 5.11e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 5.11e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 2 | 43 | 5.11e-01 |
| GO:BP | GO:0072675 | osteoclast fusion | 1 | 5 | 5.11e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 9 | 160 | 5.12e-01 |
| GO:BP | GO:0035282 | segmentation | 10 | 75 | 5.12e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 2 | 3 | 5.12e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 5 | 51 | 5.12e-01 |
| GO:BP | GO:0035507 | regulation of myosin-light-chain-phosphatase activity | 1 | 5 | 5.12e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 2 | 3 | 5.12e-01 |
| GO:BP | GO:0034182 | regulation of maintenance of mitotic sister chromatid cohesion | 1 | 8 | 5.12e-01 |
| GO:BP | GO:0034091 | regulation of maintenance of sister chromatid cohesion | 1 | 8 | 5.12e-01 |
| GO:BP | GO:1904058 | positive regulation of sensory perception of pain | 2 | 3 | 5.12e-01 |
| GO:BP | GO:0060073 | micturition | 1 | 2 | 5.12e-01 |
| GO:BP | GO:1905673 | positive regulation of lysosome organization | 1 | 1 | 5.13e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 4 | 147 | 5.13e-01 |
| GO:BP | GO:1903676 | positive regulation of cap-dependent translational initiation | 1 | 1 | 5.13e-01 |
| GO:BP | GO:1903674 | regulation of cap-dependent translational initiation | 1 | 1 | 5.13e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 22 | 39 | 5.13e-01 |
| GO:BP | GO:0006553 | lysine metabolic process | 2 | 7 | 5.13e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 5.13e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 5.13e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 5 | 20 | 5.13e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 11 | 120 | 5.13e-01 |
| GO:BP | GO:0019408 | dolichol biosynthetic process | 1 | 2 | 5.13e-01 |
| GO:BP | GO:0072237 | metanephric proximal tubule development | 1 | 3 | 5.13e-01 |
| GO:BP | GO:0072014 | proximal tubule development | 2 | 7 | 5.13e-01 |
| GO:BP | GO:2000322 | regulation of glucocorticoid receptor signaling pathway | 1 | 7 | 5.13e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 1 | 19 | 5.13e-01 |
| GO:BP | GO:1901081 | negative regulation of relaxation of smooth muscle | 1 | 1 | 5.13e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 1 | 19 | 5.13e-01 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 3 | 5 | 5.13e-01 |
| GO:BP | GO:0009992 | intracellular water homeostasis | 2 | 3 | 5.13e-01 |
| GO:BP | GO:0003108 | negative regulation of the force of heart contraction by chemical signal | 1 | 1 | 5.13e-01 |
| GO:BP | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity | 1 | 3 | 5.13e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 9 | 13 | 5.13e-01 |
| GO:BP | GO:0035378 | carbon dioxide transmembrane transport | 1 | 1 | 5.13e-01 |
| GO:BP | GO:0046878 | positive regulation of saliva secretion | 1 | 2 | 5.13e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 5.14e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 8 | 88 | 5.14e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 28 | 59 | 5.15e-01 |
| GO:BP | GO:0009615 | response to virus | 2 | 275 | 5.15e-01 |
| GO:BP | GO:0006825 | copper ion transport | 2 | 13 | 5.15e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 2 | 22 | 5.15e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 2 | 15 | 5.15e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 3 | 232 | 5.15e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 4 | 6 | 5.15e-01 |
| GO:BP | GO:0070933 | histone H4 deacetylation | 3 | 7 | 5.15e-01 |
| GO:BP | GO:1903433 | regulation of constitutive secretory pathway | 1 | 2 | 5.15e-01 |
| GO:BP | GO:0001824 | blastocyst development | 3 | 94 | 5.15e-01 |
| GO:BP | GO:0007097 | nuclear migration | 2 | 23 | 5.15e-01 |
| GO:BP | GO:0036018 | cellular response to erythropoietin | 4 | 4 | 5.15e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 5 | 46 | 5.16e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 1 | 15 | 5.16e-01 |
| GO:BP | GO:1904877 | positive regulation of DNA ligase activity | 1 | 1 | 5.16e-01 |
| GO:BP | GO:0060681 | branch elongation involved in ureteric bud branching | 3 | 3 | 5.16e-01 |
| GO:BP | GO:0018206 | peptidyl-methionine modification | 4 | 13 | 5.16e-01 |
| GO:BP | GO:0003139 | secondary heart field specification | 1 | 10 | 5.16e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 2 | 15 | 5.16e-01 |
| GO:BP | GO:0002551 | mast cell chemotaxis | 2 | 9 | 5.16e-01 |
| GO:BP | GO:0097151 | positive regulation of inhibitory postsynaptic potential | 3 | 4 | 5.16e-01 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 1 | 8 | 5.17e-01 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 1 | 8 | 5.17e-01 |
| GO:BP | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing | 5 | 7 | 5.17e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 14 | 54 | 5.17e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 4 | 24 | 5.17e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 5.17e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 5.17e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 7 | 20 | 5.17e-01 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 1 | 17 | 5.17e-01 |
| GO:BP | GO:0015886 | heme transport | 5 | 9 | 5.17e-01 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 1 | 17 | 5.17e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0090135 | actin filament branching | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 5.17e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 2 | 6 | 5.17e-01 |
| GO:BP | GO:0019627 | urea metabolic process | 2 | 7 | 5.17e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 2 | 144 | 5.17e-01 |
| GO:BP | GO:0071941 | nitrogen cycle metabolic process | 2 | 7 | 5.17e-01 |
| GO:BP | GO:0060723 | regulation of cell proliferation involved in embryonic placenta development | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 3 | 29 | 5.17e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 14 | 37 | 5.17e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 6 | 47 | 5.17e-01 |
| GO:BP | GO:0060721 | regulation of spongiotrophoblast cell proliferation | 1 | 1 | 5.17e-01 |
| GO:BP | GO:0001731 | formation of translation preinitiation complex | 3 | 11 | 5.17e-01 |
| GO:BP | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine | 1 | 2 | 5.17e-01 |
| GO:BP | GO:0006768 | biotin metabolic process | 2 | 3 | 5.18e-01 |
| GO:BP | GO:0032691 | negative regulation of interleukin-1 beta production | 2 | 13 | 5.18e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 5 | 32 | 5.18e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 14 | 23 | 5.18e-01 |
| GO:BP | GO:0042136 | neurotransmitter biosynthetic process | 1 | 5 | 5.18e-01 |
| GO:BP | GO:0006045 | N-acetylglucosamine biosynthetic process | 1 | 2 | 5.18e-01 |
| GO:BP | GO:1901073 | glucosamine-containing compound biosynthetic process | 1 | 2 | 5.18e-01 |
| GO:BP | GO:0003010 | voluntary skeletal muscle contraction | 3 | 5 | 5.19e-01 |
| GO:BP | GO:0014721 | twitch skeletal muscle contraction | 3 | 5 | 5.19e-01 |
| GO:BP | GO:0001503 | ossification | 17 | 313 | 5.19e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 2 | 3 | 5.19e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 2 | 41 | 5.19e-01 |
| GO:BP | GO:0050894 | determination of affect | 1 | 1 | 5.19e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 3 | 21 | 5.19e-01 |
| GO:BP | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation | 1 | 1 | 5.19e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 2 | 28 | 5.19e-01 |
| GO:BP | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0035803 | egg coat formation | 2 | 2 | 5.20e-01 |
| GO:BP | GO:0097527 | necroptotic signaling pathway | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 31 | 74 | 5.20e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 28 | 496 | 5.20e-01 |
| GO:BP | GO:1903726 | negative regulation of phospholipid metabolic process | 3 | 7 | 5.20e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 2 | 4 | 5.20e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 12 | 47 | 5.20e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 12 | 34 | 5.20e-01 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 1 | 35 | 5.20e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 23 | 448 | 5.20e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 16 | 34 | 5.21e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 2 | 165 | 5.21e-01 |
| GO:BP | GO:0046443 | FAD metabolic process | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0072388 | flavin adenine dinucleotide biosynthetic process | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 5.21e-01 |
| GO:BP | GO:0044805 | late nucleophagy | 1 | 7 | 5.21e-01 |
| GO:BP | GO:0006747 | FAD biosynthetic process | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0006783 | heme biosynthetic process | 7 | 30 | 5.21e-01 |
| GO:BP | GO:0048671 | negative regulation of collateral sprouting | 4 | 9 | 5.21e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 3 | 22 | 5.21e-01 |
| GO:BP | GO:0031640 | killing of cells of another organism | 1 | 10 | 5.21e-01 |
| GO:BP | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 3 | 3 | 5.21e-01 |
| GO:BP | GO:1902803 | regulation of synaptic vesicle transport | 4 | 5 | 5.21e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 2 | 17 | 5.21e-01 |
| GO:BP | GO:0009888 | tissue development | 2 | 1425 | 5.21e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 2 | 117 | 5.21e-01 |
| GO:BP | GO:0001656 | metanephros development | 4 | 64 | 5.21e-01 |
| GO:BP | GO:1901163 | regulation of trophoblast cell migration | 1 | 12 | 5.21e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 5.21e-01 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 2 | 4 | 5.21e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 9 | 71 | 5.21e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 3 | 35 | 5.22e-01 |
| GO:BP | GO:0007497 | posterior midgut development | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0003175 | tricuspid valve development | 4 | 6 | 5.22e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 2 | 21 | 5.22e-01 |
| GO:BP | GO:0048265 | response to pain | 2 | 17 | 5.22e-01 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0034720 | histone H3-K4 demethylation | 1 | 5 | 5.22e-01 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 16 | 199 | 5.22e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 6 | 93 | 5.22e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 2 | 35 | 5.22e-01 |
| GO:BP | GO:1902888 | protein localization to astral microtubule | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 87 | 375 | 5.22e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 5.22e-01 |
| GO:BP | GO:0010714 | positive regulation of collagen metabolic process | 3 | 24 | 5.22e-01 |
| GO:BP | GO:0061183 | regulation of dermatome development | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0061713 | anterior neural tube closure | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 2 | 67 | 5.22e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 6 | 23 | 5.22e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 1 | 4 | 5.22e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 3 | 38 | 5.22e-01 |
| GO:BP | GO:0060897 | neural plate regionalization | 2 | 6 | 5.22e-01 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 4 | 14 | 5.22e-01 |
| GO:BP | GO:0061054 | dermatome development | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0035750 | protein localization to myelin sheath abaxonal region | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 2 | 16 | 5.22e-01 |
| GO:BP | GO:0006435 | threonyl-tRNA aminoacylation | 1 | 3 | 5.22e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 2 | 12 | 5.23e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 17 | 44 | 5.23e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 2 | 126 | 5.23e-01 |
| GO:BP | GO:0070836 | caveola assembly | 2 | 4 | 5.23e-01 |
| GO:BP | GO:0032071 | regulation of endodeoxyribonuclease activity | 2 | 7 | 5.23e-01 |
| GO:BP | GO:0098935 | dendritic transport | 7 | 13 | 5.23e-01 |
| GO:BP | GO:0036525 | protein deglycation | 1 | 2 | 5.23e-01 |
| GO:BP | GO:0030389 | fructosamine metabolic process | 1 | 1 | 5.23e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 8 | 341 | 5.23e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 5.23e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 5.23e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 2 | 665 | 5.23e-01 |
| GO:BP | GO:2000969 | positive regulation of AMPA receptor activity | 1 | 4 | 5.23e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 10 | 20 | 5.23e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 3 | 25 | 5.24e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 3 | 8 | 5.24e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 4 | 9 | 5.24e-01 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 1 | 16 | 5.24e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 8 | 342 | 5.24e-01 |
| GO:BP | GO:0019100 | male germ-line sex determination | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0060532 | bronchus cartilage development | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0003334 | keratinocyte development | 1 | 8 | 5.24e-01 |
| GO:BP | GO:0070460 | thyroid-stimulating hormone secretion | 1 | 2 | 5.24e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 4 | 14 | 5.24e-01 |
| GO:BP | GO:1904864 | negative regulation of beta-catenin-TCF complex assembly | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | 4 | 8 | 5.24e-01 |
| GO:BP | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 5.24e-01 |
| GO:BP | GO:1904863 | regulation of beta-catenin-TCF complex assembly | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 5 | 13 | 5.24e-01 |
| GO:BP | GO:0035622 | intrahepatic bile duct development | 1 | 2 | 5.24e-01 |
| GO:BP | GO:1902073 | positive regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0018992 | germ-line sex determination | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | 1 | 6 | 5.24e-01 |
| GO:BP | GO:0061145 | lung smooth muscle development | 1 | 2 | 5.24e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 5.24e-01 |
| GO:BP | GO:0140917 | zinc ion import into mitochondrion | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0140916 | zinc ion import into lysosome | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 5.24e-01 |
| GO:BP | GO:0061090 | positive regulation of sequestering of zinc ion | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0014036 | neural crest cell fate specification | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0140915 | zinc ion import into zymogen granule | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 1 | 216 | 5.24e-01 |
| GO:BP | GO:0015828 | tyrosine transport | 1 | 3 | 5.24e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 2 | 18 | 5.24e-01 |
| GO:BP | GO:0140914 | zinc ion import into secretory vesicle | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 2 | 18 | 5.24e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 3 | 51 | 5.24e-01 |
| GO:BP | GO:0060516 | primary prostatic bud elongation | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0007542 | primary sex determination, germ-line | 1 | 2 | 5.24e-01 |
| GO:BP | GO:1901585 | regulation of acid-sensing ion channel activity | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 6 | 10 | 5.25e-01 |
| GO:BP | GO:1905553 | regulation of blood vessel branching | 3 | 4 | 5.25e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 4 | 70 | 5.25e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 29 | 107 | 5.25e-01 |
| GO:BP | GO:0036115 | fatty-acyl-CoA catabolic process | 4 | 6 | 5.25e-01 |
| GO:BP | GO:0006771 | riboflavin metabolic process | 1 | 3 | 5.25e-01 |
| GO:BP | GO:0072359 | circulatory system development | 42 | 884 | 5.25e-01 |
| GO:BP | GO:1904800 | negative regulation of neuron remodeling | 1 | 1 | 5.25e-01 |
| GO:BP | GO:1904799 | regulation of neuron remodeling | 1 | 1 | 5.25e-01 |
| GO:BP | GO:2000173 | negative regulation of branching morphogenesis of a nerve | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 17 | 56 | 5.25e-01 |
| GO:BP | GO:0046533 | negative regulation of photoreceptor cell differentiation | 3 | 3 | 5.25e-01 |
| GO:BP | GO:0015910 | long-chain fatty acid import into peroxisome | 2 | 3 | 5.26e-01 |
| GO:BP | GO:0051001 | negative regulation of nitric-oxide synthase activity | 4 | 6 | 5.26e-01 |
| GO:BP | GO:0036153 | triglyceride acyl-chain remodeling | 1 | 1 | 5.26e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 13 | 227 | 5.26e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 3 | 4 | 5.26e-01 |
| GO:BP | GO:0072144 | glomerular mesangial cell development | 3 | 4 | 5.26e-01 |
| GO:BP | GO:0016559 | peroxisome fission | 5 | 9 | 5.26e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 1 | 19 | 5.26e-01 |
| GO:BP | GO:0003341 | cilium movement | 2 | 97 | 5.26e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 1 | 27 | 5.26e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 1 | 19 | 5.26e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 5.27e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 3 | 2880 | 5.27e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 27 | 121 | 5.27e-01 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 1 | 20 | 5.27e-01 |
| GO:BP | GO:2000454 | positive regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 5.27e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 4 | 544 | 5.27e-01 |
| GO:BP | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 3 | 8 | 5.27e-01 |
| GO:BP | GO:0032415 | regulation of sodium:proton antiporter activity | 1 | 4 | 5.28e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 10 | 26 | 5.28e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 63 | 287 | 5.28e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 422 | 5.28e-01 |
| GO:BP | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway | 2 | 3 | 5.28e-01 |
| GO:BP | GO:0036510 | trimming of terminal mannose on C branch | 1 | 3 | 5.28e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 3 | 30 | 5.28e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 1 | 25 | 5.28e-01 |
| GO:BP | GO:0007286 | spermatid development | 2 | 105 | 5.28e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 14 | 34 | 5.28e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 8 | 16 | 5.28e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 8 | 16 | 5.28e-01 |
| GO:BP | GO:0021892 | cerebral cortex GABAergic interneuron differentiation | 1 | 3 | 5.29e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 5 | 50 | 5.29e-01 |
| GO:BP | GO:0140212 | regulation of long-chain fatty acid import into cell | 4 | 7 | 5.29e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 10 | 41 | 5.29e-01 |
| GO:BP | GO:0001890 | placenta development | 3 | 117 | 5.29e-01 |
| GO:BP | GO:0010891 | negative regulation of sequestering of triglyceride | 3 | 5 | 5.29e-01 |
| GO:BP | GO:0060510 | type II pneumocyte differentiation | 1 | 5 | 5.29e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 2 | 126 | 5.29e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 1 | 906 | 5.29e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 6 | 107 | 5.29e-01 |
| GO:BP | GO:0036113 | very long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 1 | 57 | 5.29e-01 |
| GO:BP | GO:0048297 | negative regulation of isotype switching to IgA isotypes | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 1 | 31 | 5.29e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 12 | 24 | 5.29e-01 |
| GO:BP | GO:0018146 | keratan sulfate biosynthetic process | 6 | 13 | 5.29e-01 |
| GO:BP | GO:0009749 | response to glucose | 14 | 137 | 5.29e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 1 | 36 | 5.30e-01 |
| GO:BP | GO:0019043 | establishment of viral latency | 1 | 3 | 5.30e-01 |
| GO:BP | GO:0048245 | eosinophil chemotaxis | 1 | 5 | 5.30e-01 |
| GO:BP | GO:0090216 | positive regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 1 | 5.30e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 6 | 40 | 5.30e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 4 | 6 | 5.30e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 36 | 5.30e-01 |
| GO:BP | GO:0035821 | modulation of process of another organism | 1 | 5 | 5.30e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 3 | 15 | 5.30e-01 |
| GO:BP | GO:0033623 | regulation of integrin activation | 1 | 11 | 5.31e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 13 | 99 | 5.31e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 5 | 28 | 5.31e-01 |
| GO:BP | GO:0060134 | prepulse inhibition | 1 | 7 | 5.31e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 5.31e-01 |
| GO:BP | GO:1902374 | regulation of rRNA catabolic process | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0044357 | regulation of rRNA stability | 1 | 1 | 5.31e-01 |
| GO:BP | GO:1903636 | regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 5.31e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 5.31e-01 |
| GO:BP | GO:1903638 | positive regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 5.31e-01 |
| GO:BP | GO:0097210 | response to gonadotropin-releasing hormone | 2 | 4 | 5.31e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 7 | 13 | 5.31e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 3 | 28 | 5.31e-01 |
| GO:BP | GO:0035788 | cell migration involved in metanephros development | 1 | 4 | 5.31e-01 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 2 | 12 | 5.31e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 6 | 98 | 5.31e-01 |
| GO:BP | GO:0003128 | heart field specification | 5 | 16 | 5.31e-01 |
| GO:BP | GO:0010511 | regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 5.31e-01 |
| GO:BP | GO:0048284 | organelle fusion | 2 | 135 | 5.31e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 3 | 37 | 5.31e-01 |
| GO:BP | GO:0003306 | Wnt signaling pathway involved in heart development | 1 | 8 | 5.32e-01 |
| GO:BP | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity | 2 | 10 | 5.32e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 1 | 10 | 5.32e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 6 | 10 | 5.32e-01 |
| GO:BP | GO:1905809 | negative regulation of synapse organization | 1 | 8 | 5.32e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 3 | 16 | 5.32e-01 |
| GO:BP | GO:0061550 | cranial ganglion development | 3 | 4 | 5.32e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 5.32e-01 |
| GO:BP | GO:0036484 | trunk neural crest cell migration | 3 | 4 | 5.32e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 3 | 22 | 5.32e-01 |
| GO:BP | GO:0035290 | trunk segmentation | 3 | 4 | 5.32e-01 |
| GO:BP | GO:1900221 | regulation of amyloid-beta clearance | 4 | 13 | 5.32e-01 |
| GO:BP | GO:2000395 | regulation of ubiquitin-dependent endocytosis | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0060358 | negative regulation of leucine import | 1 | 2 | 5.32e-01 |
| GO:BP | GO:1905533 | negative regulation of leucine import across plasma membrane | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 18 | 35 | 5.32e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 5.32e-01 |
| GO:BP | GO:0061087 | positive regulation of histone H3-K27 methylation | 3 | 4 | 5.32e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 5.32e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 1 | 12 | 5.32e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 5.32e-01 |
| GO:BP | GO:0007389 | pattern specification process | 3 | 306 | 5.32e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 5.32e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 18 | 35 | 5.32e-01 |
| GO:BP | GO:0060357 | regulation of leucine import | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0061450 | trophoblast cell migration | 1 | 13 | 5.32e-01 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 2 | 5.32e-01 |
| GO:BP | GO:1904220 | regulation of serine C-palmitoyltransferase activity | 1 | 3 | 5.32e-01 |
| GO:BP | GO:0097428 | protein maturation by iron-sulfur cluster transfer | 1 | 17 | 5.32e-01 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 2 | 5.32e-01 |
| GO:BP | GO:2000397 | positive regulation of ubiquitin-dependent endocytosis | 1 | 2 | 5.32e-01 |
| GO:BP | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 6 | 5.32e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 3 | 15 | 5.33e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 5.33e-01 |
| GO:BP | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process | 1 | 2 | 5.33e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 6 | 25 | 5.33e-01 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 1 | 9 | 5.33e-01 |
| GO:BP | GO:0080170 | hydrogen peroxide transmembrane transport | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 5.33e-01 |
| GO:BP | GO:0090296 | regulation of mitochondrial DNA replication | 3 | 4 | 5.33e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 5.33e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 1 | 5 | 5.33e-01 |
| GO:BP | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 8 | 14 | 5.33e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 3 | 11 | 5.33e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 7 | 114 | 5.33e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 87 | 376 | 5.33e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 11 | 133 | 5.33e-01 |
| GO:BP | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway | 3 | 9 | 5.33e-01 |
| GO:BP | GO:0021785 | branchiomotor neuron axon guidance | 3 | 5 | 5.33e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 5 | 19 | 5.33e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 10 | 54 | 5.33e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 3 | 1719 | 5.34e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 5 | 33 | 5.34e-01 |
| GO:BP | GO:0033031 | positive regulation of neutrophil apoptotic process | 2 | 3 | 5.34e-01 |
| GO:BP | GO:0060992 | response to fungicide | 3 | 4 | 5.34e-01 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 6 | 17 | 5.34e-01 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 6 | 17 | 5.34e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 1 | 16 | 5.34e-01 |
| GO:BP | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 7 | 21 | 5.35e-01 |
| GO:BP | GO:2000419 | regulation of eosinophil extravasation | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 2 | 12 | 5.35e-01 |
| GO:BP | GO:2000504 | positive regulation of blood vessel remodeling | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 8 | 29 | 5.35e-01 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 1 | 14 | 5.35e-01 |
| GO:BP | GO:1990678 | histone H4-K16 deacetylation | 1 | 1 | 5.35e-01 |
| GO:BP | GO:2000420 | negative regulation of eosinophil extravasation | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 4 | 76 | 5.35e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 25 | 114 | 5.35e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 5.35e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 5.35e-01 |
| GO:BP | GO:0072682 | eosinophil extravasation | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0015816 | glycine transport | 1 | 9 | 5.35e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 2 | 56 | 5.35e-01 |
| GO:BP | GO:0008215 | spermine metabolic process | 3 | 5 | 5.35e-01 |
| GO:BP | GO:0006489 | dolichyl diphosphate biosynthetic process | 3 | 5 | 5.35e-01 |
| GO:BP | GO:0046465 | dolichyl diphosphate metabolic process | 3 | 5 | 5.35e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 3 | 6 | 5.35e-01 |
| GO:BP | GO:0035865 | cellular response to potassium ion | 2 | 9 | 5.35e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 2 | 9 | 5.35e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 6 | 60 | 5.36e-01 |
| GO:BP | GO:0045204 | MAPK export from nucleus | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0071037 | nuclear polyadenylation-dependent snRNA catabolic process | 1 | 2 | 5.36e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 2 | 19 | 5.36e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 4 | 11 | 5.36e-01 |
| GO:BP | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process | 1 | 2 | 5.36e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 42 | 78 | 5.36e-01 |
| GO:BP | GO:0071036 | nuclear polyadenylation-dependent snoRNA catabolic process | 1 | 2 | 5.36e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 1 | 26 | 5.36e-01 |
| GO:BP | GO:0045339 | farnesyl diphosphate catabolic process | 1 | 1 | 5.36e-01 |
| GO:BP | GO:1902247 | geranylgeranyl diphosphate catabolic process | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 1 | 16 | 5.36e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 10 | 5.36e-01 |
| GO:BP | GO:1990262 | anti-Mullerian hormone signaling pathway | 1 | 2 | 5.36e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 1 | 9 | 5.37e-01 |
| GO:BP | GO:0009051 | pentose-phosphate shunt, oxidative branch | 1 | 4 | 5.37e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 1 | 11 | 5.37e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 5.37e-01 |
| GO:BP | GO:0060683 | regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling | 1 | 1 | 5.37e-01 |
| GO:BP | GO:0006590 | thyroid hormone generation | 4 | 10 | 5.37e-01 |
| GO:BP | GO:0071865 | regulation of apoptotic process in bone marrow cell | 1 | 3 | 5.38e-01 |
| GO:BP | GO:0043248 | proteasome assembly | 1 | 13 | 5.38e-01 |
| GO:BP | GO:0071866 | negative regulation of apoptotic process in bone marrow cell | 1 | 3 | 5.38e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 5.38e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 1 | 23 | 5.38e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 5 | 7 | 5.38e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 5.38e-01 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 6 | 17 | 5.38e-01 |
| GO:BP | GO:0046337 | phosphatidylethanolamine metabolic process | 4 | 19 | 5.38e-01 |
| GO:BP | GO:0006497 | protein lipidation | 33 | 89 | 5.38e-01 |
| GO:BP | GO:0021561 | facial nerve development | 2 | 8 | 5.38e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 7 | 113 | 5.38e-01 |
| GO:BP | GO:1904210 | VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 5.38e-01 |
| GO:BP | GO:0019086 | late viral transcription | 3 | 4 | 5.38e-01 |
| GO:BP | GO:1904239 | regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.38e-01 |
| GO:BP | GO:1904240 | negative regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 2 | 8 | 5.38e-01 |
| GO:BP | GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 1 | 14 | 5.38e-01 |
| GO:BP | GO:0042983 | amyloid precursor protein biosynthetic process | 1 | 14 | 5.38e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 2 | 111 | 5.38e-01 |
| GO:BP | GO:0061448 | connective tissue development | 1 | 208 | 5.38e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 7 | 775 | 5.38e-01 |
| GO:BP | GO:0036306 | embryonic heart tube elongation | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0035922 | foramen ovale closure | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0030070 | insulin processing | 2 | 6 | 5.38e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 10 | 90 | 5.38e-01 |
| GO:BP | GO:0060577 | pulmonary vein morphogenesis | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 6 | 71 | 5.38e-01 |
| GO:BP | GO:0140214 | positive regulation of long-chain fatty acid import into cell | 2 | 2 | 5.38e-01 |
| GO:BP | GO:0007015 | actin filament organization | 17 | 373 | 5.38e-01 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 5 | 8 | 5.39e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 9 | 67 | 5.39e-01 |
| GO:BP | GO:0061171 | establishment of bipolar cell polarity | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0035971 | peptidyl-histidine dephosphorylation | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 14 | 70 | 5.39e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 9 | 31 | 5.39e-01 |
| GO:BP | GO:0099140 | presynaptic actin cytoskeleton organization | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0099187 | presynaptic cytoskeleton organization | 1 | 1 | 5.39e-01 |
| GO:BP | GO:2000984 | negative regulation of ATP citrate synthase activity | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 8 | 58 | 5.39e-01 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 4 | 8 | 5.39e-01 |
| GO:BP | GO:0070075 | tear secretion | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 12 | 31 | 5.39e-01 |
| GO:BP | GO:0097378 | dorsal spinal cord interneuron axon guidance | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1905065 | positive regulation of vascular associated smooth muscle cell differentiation | 4 | 6 | 5.39e-01 |
| GO:BP | GO:0042822 | pyridoxal phosphate metabolic process | 2 | 4 | 5.39e-01 |
| GO:BP | GO:2000252 | negative regulation of feeding behavior | 2 | 2 | 5.39e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 2 | 170 | 5.39e-01 |
| GO:BP | GO:1905719 | protein localization to perinuclear region of cytoplasm | 3 | 8 | 5.39e-01 |
| GO:BP | GO:0071243 | cellular response to arsenic-containing substance | 8 | 18 | 5.39e-01 |
| GO:BP | GO:2000727 | positive regulation of cardiac muscle cell differentiation | 1 | 6 | 5.39e-01 |
| GO:BP | GO:0009082 | branched-chain amino acid biosynthetic process | 1 | 4 | 5.39e-01 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 6 | 17 | 5.39e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 2 | 17 | 5.39e-01 |
| GO:BP | GO:0046084 | adenine biosynthetic process | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0006646 | phosphatidylethanolamine biosynthetic process | 6 | 8 | 5.39e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 1 | 10 | 5.39e-01 |
| GO:BP | GO:0035295 | tube development | 40 | 821 | 5.39e-01 |
| GO:BP | GO:1903301 | positive regulation of hexokinase activity | 1 | 5 | 5.39e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 5.39e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 4 | 22 | 5.39e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 8 | 31 | 5.39e-01 |
| GO:BP | GO:0043456 | regulation of pentose-phosphate shunt | 1 | 6 | 5.39e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 4 | 22 | 5.39e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 3 | 5 | 5.39e-01 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 1 | 9 | 5.39e-01 |
| GO:BP | GO:2000056 | regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0007604 | phototransduction, UV | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0120041 | positive regulation of macrophage proliferation | 1 | 4 | 5.39e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 10 | 28 | 5.39e-01 |
| GO:BP | GO:0035426 | extracellular matrix-cell signaling | 3 | 4 | 5.39e-01 |
| GO:BP | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0010021 | amylopectin biosynthetic process | 1 | 1 | 5.39e-01 |
| GO:BP | GO:2000896 | amylopectin metabolic process | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 3 | 36 | 5.39e-01 |
| GO:BP | GO:2000057 | negative regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 5.39e-01 |
| GO:BP | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity | 1 | 6 | 5.39e-01 |
| GO:BP | GO:0032105 | negative regulation of response to extracellular stimulus | 1 | 7 | 5.39e-01 |
| GO:BP | GO:0032099 | negative regulation of appetite | 1 | 7 | 5.39e-01 |
| GO:BP | GO:0032096 | negative regulation of response to food | 1 | 7 | 5.39e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 45 | 133 | 5.39e-01 |
| GO:BP | GO:1905007 | positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 5 | 5.39e-01 |
| GO:BP | GO:0022009 | central nervous system vasculogenesis | 1 | 4 | 5.39e-01 |
| GO:BP | GO:0006269 | DNA replication, synthesis of RNA primer | 2 | 7 | 5.39e-01 |
| GO:BP | GO:1902767 | isoprenoid biosynthetic process via mevalonate | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 8 | 52 | 5.39e-01 |
| GO:BP | GO:0033210 | leptin-mediated signaling pathway | 1 | 10 | 5.39e-01 |
| GO:BP | GO:0097503 | sialylation | 1 | 18 | 5.39e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 20 | 55 | 5.39e-01 |
| GO:BP | GO:0032108 | negative regulation of response to nutrient levels | 1 | 7 | 5.39e-01 |
| GO:BP | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0098694 | regulation of synaptic vesicle budding from presynaptic endocytic zone membrane | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1903972 | regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 5 | 50 | 5.39e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 2 | 30 | 5.39e-01 |
| GO:BP | GO:0042482 | positive regulation of odontogenesis | 1 | 5 | 5.39e-01 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 5 | 8 | 5.39e-01 |
| GO:BP | GO:1903969 | regulation of response to macrophage colony-stimulating factor | 1 | 3 | 5.39e-01 |
| GO:BP | GO:1903911 | positive regulation of receptor clustering | 1 | 6 | 5.39e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 44 | 100 | 5.39e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 3 | 32 | 5.39e-01 |
| GO:BP | GO:0062029 | positive regulation of stress granule assembly | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 2 | 35 | 5.39e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 5.40e-01 |
| GO:BP | GO:1903703 | enterocyte differentiation | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0014719 | skeletal muscle satellite cell activation | 2 | 5 | 5.41e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 57 | 1167 | 5.41e-01 |
| GO:BP | GO:0061956 | penetration of cumulus oophorus | 1 | 1 | 5.41e-01 |
| GO:BP | GO:1905667 | negative regulation of protein localization to endosome | 1 | 1 | 5.41e-01 |
| GO:BP | GO:0007521 | muscle cell fate determination | 1 | 2 | 5.41e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 5.41e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 24 | 5.41e-01 |
| GO:BP | GO:0010232 | vascular transport | 5 | 72 | 5.41e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 5 | 72 | 5.41e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 29 | 57 | 5.41e-01 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 6 | 11 | 5.41e-01 |
| GO:BP | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 1 | 2 | 5.41e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 5.41e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 1 | 96 | 5.41e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 2 | 763 | 5.41e-01 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 2 | 12 | 5.42e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 5.42e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 9 | 29 | 5.42e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 8 | 13 | 5.42e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 1 | 48 | 5.42e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 5 | 27 | 5.42e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 3 | 23 | 5.42e-01 |
| GO:BP | GO:0010818 | T cell chemotaxis | 4 | 12 | 5.42e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 1 | 28 | 5.42e-01 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 5 | 26 | 5.42e-01 |
| GO:BP | GO:0001661 | conditioned taste aversion | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 3 | 52 | 5.43e-01 |
| GO:BP | GO:0014805 | smooth muscle adaptation | 1 | 3 | 5.43e-01 |
| GO:BP | GO:0046204 | nor-spermidine metabolic process | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0016062 | adaptation of rhodopsin mediated signaling | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 4 | 14 | 5.44e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 1 | 71 | 5.44e-01 |
| GO:BP | GO:0032920 | putrescine acetylation | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0032919 | spermine acetylation | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 8 | 97 | 5.44e-01 |
| GO:BP | GO:1905776 | positive regulation of DNA helicase activity | 1 | 3 | 5.44e-01 |
| GO:BP | GO:1905464 | positive regulation of DNA duplex unwinding | 1 | 3 | 5.44e-01 |
| GO:BP | GO:0010886 | positive regulation of cholesterol storage | 2 | 5 | 5.44e-01 |
| GO:BP | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process | 5 | 16 | 5.44e-01 |
| GO:BP | GO:1905413 | regulation of dense core granule exocytosis | 2 | 2 | 5.44e-01 |
| GO:BP | GO:1904528 | positive regulation of microtubule binding | 3 | 6 | 5.44e-01 |
| GO:BP | GO:1901187 | regulation of ephrin receptor signaling pathway | 1 | 2 | 5.44e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 2 | 28 | 5.44e-01 |
| GO:BP | GO:0048468 | cell development | 15 | 1963 | 5.44e-01 |
| GO:BP | GO:0036285 | SAGA complex assembly | 1 | 1 | 5.44e-01 |
| GO:BP | GO:0098795 | global gene silencing by mRNA cleavage | 1 | 4 | 5.44e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 4 | 6 | 5.44e-01 |
| GO:BP | GO:0048312 | intracellular distribution of mitochondria | 1 | 5 | 5.44e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 2 | 16 | 5.44e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 3 | 197 | 5.44e-01 |
| GO:BP | GO:1903564 | regulation of protein localization to cilium | 3 | 11 | 5.45e-01 |
| GO:BP | GO:0097742 | de novo centriole assembly | 3 | 4 | 5.45e-01 |
| GO:BP | GO:0009757 | hexose mediated signaling | 1 | 5 | 5.45e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 30 | 547 | 5.45e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 2 | 755 | 5.45e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 1 | 16 | 5.45e-01 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 3 | 4 | 5.45e-01 |
| GO:BP | GO:0099519 | dense core granule cytoskeletal transport | 1 | 7 | 5.45e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 4 | 90 | 5.45e-01 |
| GO:BP | GO:0060353 | regulation of cell adhesion molecule production | 3 | 6 | 5.45e-01 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 5 | 15 | 5.45e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 2 | 25 | 5.45e-01 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 5 | 15 | 5.45e-01 |
| GO:BP | GO:1901950 | dense core granule transport | 1 | 7 | 5.45e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 9 | 18 | 5.45e-01 |
| GO:BP | GO:0032253 | dense core granule localization | 1 | 7 | 5.45e-01 |
| GO:BP | GO:0034498 | early endosome to Golgi transport | 5 | 9 | 5.45e-01 |
| GO:BP | GO:0010255 | glucose mediated signaling pathway | 1 | 5 | 5.45e-01 |
| GO:BP | GO:0010182 | sugar mediated signaling pathway | 1 | 5 | 5.45e-01 |
| GO:BP | GO:1900454 | positive regulation of long-term synaptic depression | 1 | 3 | 5.45e-01 |
| GO:BP | GO:0016477 | cell migration | 31 | 1081 | 5.45e-01 |
| GO:BP | GO:0032197 | retrotransposition | 2 | 4 | 5.45e-01 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 5 | 8 | 5.45e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 6 | 13 | 5.45e-01 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 9 | 11 | 5.45e-01 |
| GO:BP | GO:0007281 | germ cell development | 6 | 187 | 5.46e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 3 | 41 | 5.46e-01 |
| GO:BP | GO:0034756 | regulation of iron ion transport | 2 | 4 | 5.46e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 86 | 5.46e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 1 | 5 | 5.47e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 6 | 34 | 5.47e-01 |
| GO:BP | GO:1904529 | regulation of actin filament binding | 1 | 3 | 5.47e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 2 | 24 | 5.47e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 3 | 83 | 5.47e-01 |
| GO:BP | GO:0048103 | somatic stem cell division | 4 | 10 | 5.47e-01 |
| GO:BP | GO:1904645 | response to amyloid-beta | 10 | 32 | 5.47e-01 |
| GO:BP | GO:2000435 | negative regulation of protein neddylation | 2 | 4 | 5.47e-01 |
| GO:BP | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration | 1 | 1 | 5.47e-01 |
| GO:BP | GO:0002296 | T-helper 1 cell lineage commitment | 1 | 3 | 5.47e-01 |
| GO:BP | GO:0048560 | establishment of anatomical structure orientation | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0098787 | mRNA cleavage involved in mRNA processing | 4 | 13 | 5.47e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 76 | 273 | 5.47e-01 |
| GO:BP | GO:2000078 | positive regulation of type B pancreatic cell development | 1 | 2 | 5.47e-01 |
| GO:BP | GO:0001660 | fever generation | 1 | 5 | 5.47e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 9 | 16 | 5.48e-01 |
| GO:BP | GO:1901523 | icosanoid catabolic process | 1 | 2 | 5.48e-01 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 10 | 19 | 5.48e-01 |
| GO:BP | GO:0042753 | positive regulation of circadian rhythm | 2 | 10 | 5.48e-01 |
| GO:BP | GO:0046504 | glycerol ether biosynthetic process | 2 | 10 | 5.48e-01 |
| GO:BP | GO:0097384 | cellular lipid biosynthetic process | 2 | 10 | 5.48e-01 |
| GO:BP | GO:0008611 | ether lipid biosynthetic process | 2 | 10 | 5.48e-01 |
| GO:BP | GO:0043972 | histone H3-K23 acetylation | 1 | 1 | 5.48e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 9 | 26 | 5.48e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 2 | 4 | 5.48e-01 |
| GO:BP | GO:1902897 | regulation of postsynaptic density protein 95 clustering | 1 | 2 | 5.48e-01 |
| GO:BP | GO:0010915 | regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 5.48e-01 |
| GO:BP | GO:0060700 | regulation of ribonuclease activity | 1 | 5 | 5.48e-01 |
| GO:BP | GO:0010916 | negative regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 5.48e-01 |
| GO:BP | GO:0007386 | compartment pattern specification | 2 | 5 | 5.48e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 3 | 54 | 5.48e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 5 | 34 | 5.49e-01 |
| GO:BP | GO:0071472 | cellular response to salt stress | 5 | 11 | 5.49e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 17 | 27 | 5.49e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 27 | 51 | 5.49e-01 |
| GO:BP | GO:0006501 | C-terminal protein lipidation | 2 | 6 | 5.49e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 35 | 351 | 5.49e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 5.49e-01 |
| GO:BP | GO:0140708 | CAT tailing | 1 | 1 | 5.49e-01 |
| GO:BP | GO:1904045 | cellular response to aldosterone | 1 | 5 | 5.49e-01 |
| GO:BP | GO:0071415 | cellular response to purine-containing compound | 2 | 11 | 5.49e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 5.49e-01 |
| GO:BP | GO:0014906 | myotube cell development involved in skeletal muscle regeneration | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 12 | 31 | 5.49e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 8 | 19 | 5.49e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 8 | 19 | 5.49e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 12 | 5.49e-01 |
| GO:BP | GO:0021702 | cerebellar Purkinje cell differentiation | 7 | 10 | 5.49e-01 |
| GO:BP | GO:0051391 | tRNA acetylation | 1 | 1 | 5.49e-01 |
| GO:BP | GO:1904812 | rRNA acetylation involved in maturation of SSU-rRNA | 1 | 1 | 5.49e-01 |
| GO:BP | GO:1990884 | RNA acetylation | 1 | 1 | 5.49e-01 |
| GO:BP | GO:1990882 | rRNA acetylation | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 20 | 27 | 5.49e-01 |
| GO:BP | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity | 2 | 3 | 5.49e-01 |
| GO:BP | GO:0003330 | regulation of extracellular matrix constituent secretion | 4 | 7 | 5.49e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 3 | 36 | 5.50e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 3 | 8 | 5.50e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 2 | 17 | 5.50e-01 |
| GO:BP | GO:0097242 | amyloid-beta clearance | 7 | 27 | 5.50e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 2 | 15 | 5.50e-01 |
| GO:BP | GO:0051258 | protein polymerization | 2 | 241 | 5.50e-01 |
| GO:BP | GO:0001813 | regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 1 | 9 | 5.50e-01 |
| GO:BP | GO:0032759 | positive regulation of TRAIL production | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 5.50e-01 |
| GO:BP | GO:0032639 | TRAIL production | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0032679 | regulation of TRAIL production | 1 | 1 | 5.50e-01 |
| GO:BP | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 7 | 5.51e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 3 | 174 | 5.51e-01 |
| GO:BP | GO:0009190 | cyclic nucleotide biosynthetic process | 2 | 14 | 5.51e-01 |
| GO:BP | GO:1903712 | cysteine transmembrane transport | 1 | 3 | 5.51e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 32 | 140 | 5.51e-01 |
| GO:BP | GO:0042883 | cysteine transport | 1 | 3 | 5.51e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 2 | 15 | 5.51e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 3 | 63 | 5.51e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 5.51e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 2 | 16 | 5.51e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 4 | 64 | 5.51e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 31 | 540 | 5.51e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 6 | 47 | 5.51e-01 |
| GO:BP | GO:0030853 | negative regulation of granulocyte differentiation | 5 | 6 | 5.52e-01 |
| GO:BP | GO:0150093 | amyloid-beta clearance by transcytosis | 2 | 7 | 5.52e-01 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 2 | 6 | 5.52e-01 |
| GO:BP | GO:0090666 | scaRNA localization to Cajal body | 2 | 5 | 5.52e-01 |
| GO:BP | GO:0090188 | negative regulation of pancreatic juice secretion | 1 | 4 | 5.52e-01 |
| GO:BP | GO:0009188 | ribonucleoside diphosphate biosynthetic process | 4 | 8 | 5.52e-01 |
| GO:BP | GO:1904437 | positive regulation of transferrin receptor binding | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0030282 | bone mineralization | 10 | 82 | 5.52e-01 |
| GO:BP | GO:1904434 | positive regulation of ferrous iron binding | 1 | 2 | 5.52e-01 |
| GO:BP | GO:1904435 | regulation of transferrin receptor binding | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 5.52e-01 |
| GO:BP | GO:1905069 | allantois development | 2 | 4 | 5.52e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 5 | 8 | 5.52e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 5 | 8 | 5.52e-01 |
| GO:BP | GO:1904432 | regulation of ferrous iron binding | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 5 | 96 | 5.52e-01 |
| GO:BP | GO:0007568 | aging | 10 | 115 | 5.52e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 5.52e-01 |
| GO:BP | GO:1904229 | regulation of succinate dehydrogenase activity | 1 | 2 | 5.52e-01 |
| GO:BP | GO:1904231 | positive regulation of succinate dehydrogenase activity | 1 | 2 | 5.52e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 5.52e-01 |
| GO:BP | GO:0009746 | response to hexose | 14 | 138 | 5.52e-01 |
| GO:BP | GO:0009886 | post-embryonic animal morphogenesis | 1 | 10 | 5.53e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 5.53e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 8 | 145 | 5.53e-01 |
| GO:BP | GO:1990792 | cellular response to glial cell derived neurotrophic factor | 1 | 1 | 5.53e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 5.53e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 6 | 37 | 5.53e-01 |
| GO:BP | GO:1990790 | response to glial cell derived neurotrophic factor | 1 | 1 | 5.53e-01 |
| GO:BP | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate | 1 | 1 | 5.53e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 3 | 4 | 5.54e-01 |
| GO:BP | GO:0071374 | cellular response to parathyroid hormone stimulus | 2 | 7 | 5.54e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 4 | 612 | 5.54e-01 |
| GO:BP | GO:0060087 | relaxation of vascular associated smooth muscle | 1 | 7 | 5.54e-01 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 1 | 14 | 5.54e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 5 | 16 | 5.54e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 6 | 27 | 5.55e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 2 | 5 | 5.55e-01 |
| GO:BP | GO:0036369 | transcription factor catabolic process | 2 | 5 | 5.55e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 3 | 13 | 5.55e-01 |
| GO:BP | GO:0039530 | MDA-5 signaling pathway | 6 | 8 | 5.55e-01 |
| GO:BP | GO:0016310 | phosphorylation | 65 | 1372 | 5.55e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 4 | 62 | 5.55e-01 |
| GO:BP | GO:0032730 | positive regulation of interleukin-1 alpha production | 1 | 3 | 5.55e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 2 | 25 | 5.55e-01 |
| GO:BP | GO:0038002 | endocrine signaling | 1 | 1 | 5.55e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 3 | 12 | 5.56e-01 |
| GO:BP | GO:0035995 | detection of muscle stretch | 2 | 7 | 5.56e-01 |
| GO:BP | GO:0051597 | response to methylmercury | 2 | 5 | 5.56e-01 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 4 | 16 | 5.56e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 4 | 36 | 5.56e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 2 | 39 | 5.56e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 3 | 88 | 5.56e-01 |
| GO:BP | GO:1903945 | positive regulation of hepatocyte apoptotic process | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0098937 | anterograde dendritic transport | 5 | 9 | 5.56e-01 |
| GO:BP | GO:0009624 | response to nematode | 1 | 4 | 5.56e-01 |
| GO:BP | GO:0007040 | lysosome organization | 3 | 88 | 5.56e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 4 | 461 | 5.56e-01 |
| GO:BP | GO:1905178 | regulation of cardiac muscle tissue regeneration | 1 | 2 | 5.56e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 5.56e-01 |
| GO:BP | GO:0030157 | pancreatic juice secretion | 2 | 11 | 5.56e-01 |
| GO:BP | GO:1905179 | negative regulation of cardiac muscle tissue regeneration | 1 | 2 | 5.56e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 7 | 54 | 5.56e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 5.56e-01 |
| GO:BP | GO:0043045 | DNA methylation involved in embryo development | 1 | 7 | 5.56e-01 |
| GO:BP | GO:0002176 | male germ cell proliferation | 1 | 6 | 5.56e-01 |
| GO:BP | GO:0006591 | ornithine metabolic process | 1 | 6 | 5.56e-01 |
| GO:BP | GO:0034351 | negative regulation of glial cell apoptotic process | 2 | 8 | 5.56e-01 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 1 | 12 | 5.56e-01 |
| GO:BP | GO:0070564 | positive regulation of vitamin D receptor signaling pathway | 1 | 5 | 5.56e-01 |
| GO:BP | GO:0051352 | negative regulation of ligase activity | 1 | 2 | 5.57e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 6 | 42 | 5.57e-01 |
| GO:BP | GO:1904381 | Golgi apparatus mannose trimming | 2 | 3 | 5.57e-01 |
| GO:BP | GO:0042113 | B cell activation | 3 | 170 | 5.57e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 2 | 14 | 5.57e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 11 | 66 | 5.57e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 2 | 28 | 5.57e-01 |
| GO:BP | GO:0140251 | regulation protein catabolic process at presynapse | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0060313 | negative regulation of blood vessel remodeling | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 1 | 6 | 5.57e-01 |
| GO:BP | GO:0046707 | IDP metabolic process | 1 | 1 | 5.57e-01 |
| GO:BP | GO:0045606 | positive regulation of epidermal cell differentiation | 4 | 16 | 5.57e-01 |
| GO:BP | GO:1903382 | negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0060235 | lens induction in camera-type eye | 2 | 3 | 5.57e-01 |
| GO:BP | GO:1901143 | insulin catabolic process | 1 | 3 | 5.57e-01 |
| GO:BP | GO:1905281 | positive regulation of retrograde transport, endosome to Golgi | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0042726 | flavin-containing compound metabolic process | 1 | 4 | 5.57e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 1 | 37 | 5.57e-01 |
| GO:BP | GO:1903381 | regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 5.57e-01 |
| GO:BP | GO:1903800 | positive regulation of miRNA processing | 2 | 3 | 5.57e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 5.57e-01 |
| GO:BP | GO:0007500 | mesodermal cell fate determination | 2 | 2 | 5.57e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 3 | 78 | 5.57e-01 |
| GO:BP | GO:0010669 | epithelial structure maintenance | 3 | 15 | 5.57e-01 |
| GO:BP | GO:0046709 | IDP catabolic process | 1 | 1 | 5.57e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 2 | 9 | 5.57e-01 |
| GO:BP | GO:1902530 | positive regulation of protein linear polyubiquitination | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 3 | 40 | 5.57e-01 |
| GO:BP | GO:0060197 | cloacal septation | 2 | 2 | 5.57e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 2 | 10 | 5.57e-01 |
| GO:BP | GO:1904048 | regulation of spontaneous neurotransmitter secretion | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 3 | 53 | 5.57e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 5.57e-01 |
| GO:BP | GO:1902528 | regulation of protein linear polyubiquitination | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 8 | 280 | 5.57e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 5.57e-01 |
| GO:BP | GO:0072190 | ureter urothelium development | 2 | 3 | 5.57e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 3 | 52 | 5.57e-01 |
| GO:BP | GO:0040031 | snRNA modification | 3 | 7 | 5.57e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 19 | 1849 | 5.57e-01 |
| GO:BP | GO:1905335 | regulation of aggrephagy | 2 | 3 | 5.57e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 2 | 4 | 5.57e-01 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 4 | 10 | 5.57e-01 |
| GO:BP | GO:0051169 | nuclear transport | 13 | 295 | 5.57e-01 |
| GO:BP | GO:1905337 | positive regulation of aggrephagy | 2 | 3 | 5.57e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 13 | 295 | 5.57e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 3 | 191 | 5.58e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 3 | 20 | 5.58e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 4 | 30 | 5.58e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 19 | 203 | 5.58e-01 |
| GO:BP | GO:0061780 | mitotic cohesin loading | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 7 | 76 | 5.58e-01 |
| GO:BP | GO:0060050 | positive regulation of protein glycosylation | 1 | 6 | 5.58e-01 |
| GO:BP | GO:0035261 | external genitalia morphogenesis | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0006924 | activation-induced cell death of T cells | 1 | 8 | 5.58e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 5.58e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 5.58e-01 |
| GO:BP | GO:0080129 | proteasome core complex assembly | 1 | 1 | 5.58e-01 |
| GO:BP | GO:0019083 | viral transcription | 6 | 48 | 5.58e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 2 | 5 | 5.59e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 2 | 44 | 5.59e-01 |
| GO:BP | GO:1905704 | positive regulation of inhibitory synapse assembly | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0060916 | mesenchymal cell proliferation involved in lung development | 3 | 5 | 5.59e-01 |
| GO:BP | GO:0009195 | pyrimidine ribonucleoside diphosphate catabolic process | 2 | 3 | 5.59e-01 |
| GO:BP | GO:0009140 | pyrimidine nucleoside diphosphate catabolic process | 2 | 3 | 5.59e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 39 | 408 | 5.59e-01 |
| GO:BP | GO:0006256 | UDP catabolic process | 2 | 3 | 5.59e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 5 | 34 | 5.59e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 2 | 22 | 5.59e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 5.59e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 10 | 29 | 5.59e-01 |
| GO:BP | GO:0019254 | carnitine metabolic process, CoA-linked | 2 | 3 | 5.59e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 8 | 36 | 5.59e-01 |
| GO:BP | GO:0060374 | mast cell differentiation | 2 | 4 | 5.59e-01 |
| GO:BP | GO:1900038 | negative regulation of cellular response to hypoxia | 5 | 8 | 5.59e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 2 | 33 | 5.59e-01 |
| GO:BP | GO:1901475 | pyruvate transmembrane transport | 1 | 5 | 5.59e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 7 | 212 | 5.59e-01 |
| GO:BP | GO:0006848 | pyruvate transport | 1 | 5 | 5.59e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 6 | 111 | 5.59e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 2 | 2 | 5.59e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 4 | 12 | 5.59e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 4 | 12 | 5.59e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 4 | 12 | 5.59e-01 |
| GO:BP | GO:0016203 | muscle attachment | 1 | 1 | 5.59e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 1 | 33 | 5.59e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 1 | 23 | 5.59e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 5 | 8 | 5.60e-01 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 4 | 12 | 5.60e-01 |
| GO:BP | GO:0007565 | female pregnancy | 6 | 122 | 5.60e-01 |
| GO:BP | GO:0099044 | vesicle tethering to endoplasmic reticulum | 1 | 2 | 5.60e-01 |
| GO:BP | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly | 1 | 3 | 5.60e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 1 | 102 | 5.60e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 5 | 52 | 5.60e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 6 | 123 | 5.60e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 4 | 21 | 5.60e-01 |
| GO:BP | GO:0006294 | nucleotide-excision repair, preincision complex assembly | 1 | 1 | 5.60e-01 |
| GO:BP | GO:1905267 | endonucleolytic cleavage involved in tRNA processing | 2 | 3 | 5.60e-01 |
| GO:BP | GO:1901723 | negative regulation of cell proliferation involved in kidney development | 1 | 3 | 5.60e-01 |
| GO:BP | GO:0090384 | phagosome-lysosome docking | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 6 | 97 | 5.60e-01 |
| GO:BP | GO:0060126 | somatotropin secreting cell differentiation | 1 | 2 | 5.60e-01 |
| GO:BP | GO:0021675 | nerve development | 5 | 57 | 5.60e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 16 | 40 | 5.60e-01 |
| GO:BP | GO:1900079 | regulation of arginine biosynthetic process | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0044375 | regulation of peroxisome size | 2 | 3 | 5.60e-01 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 4 | 7 | 5.60e-01 |
| GO:BP | GO:0002100 | tRNA wobble adenosine to inosine editing | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0002192 | IRES-dependent translational initiation of linear mRNA | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0110017 | cap-independent translational initiation of linear mRNA | 1 | 1 | 5.60e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 2 | 25 | 5.60e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 2 | 4 | 5.60e-01 |
| GO:BP | GO:0032289 | central nervous system myelin formation | 1 | 4 | 5.60e-01 |
| GO:BP | GO:0000019 | regulation of mitotic recombination | 1 | 5 | 5.60e-01 |
| GO:BP | GO:0140361 | cyclic-GMP-AMP transmembrane import across plasma membrane | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0106048 | spermidine deacetylation | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0106047 | polyamine deacetylation | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 15 | 236 | 5.61e-01 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 1 | 11 | 5.61e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 11 | 30 | 5.61e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 2 | 335 | 5.61e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 4 | 121 | 5.61e-01 |
| GO:BP | GO:2000822 | regulation of behavioral fear response | 1 | 7 | 5.61e-01 |
| GO:BP | GO:1990961 | xenobiotic detoxification by transmembrane export across the plasma membrane | 5 | 7 | 5.61e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 7 | 20 | 5.61e-01 |
| GO:BP | GO:0035787 | cell migration involved in kidney development | 1 | 5 | 5.62e-01 |
| GO:BP | GO:0072008 | glomerular mesangial cell differentiation | 1 | 4 | 5.62e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 5 | 4241 | 5.62e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 4 | 4 | 5.62e-01 |
| GO:BP | GO:0006096 | glycolytic process | 2 | 70 | 5.62e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 2 | 631 | 5.62e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 5.62e-01 |
| GO:BP | GO:0072143 | mesangial cell development | 1 | 5 | 5.62e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 2 | 28 | 5.62e-01 |
| GO:BP | GO:0035385 | Roundabout signaling pathway | 3 | 5 | 5.62e-01 |
| GO:BP | GO:0072677 | eosinophil migration | 1 | 7 | 5.62e-01 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 5.62e-01 |
| GO:BP | GO:0031648 | protein destabilization | 1 | 45 | 5.63e-01 |
| GO:BP | GO:0030961 | peptidyl-arginine hydroxylation | 1 | 1 | 5.63e-01 |
| GO:BP | GO:0060092 | regulation of synaptic transmission, glycinergic | 1 | 1 | 5.63e-01 |
| GO:BP | GO:0016322 | neuron remodeling | 1 | 11 | 5.63e-01 |
| GO:BP | GO:0002071 | glandular epithelial cell maturation | 2 | 7 | 5.63e-01 |
| GO:BP | GO:0061817 | endoplasmic reticulum-plasma membrane tethering | 1 | 5 | 5.63e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 2 | 19 | 5.64e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 4 | 469 | 5.64e-01 |
| GO:BP | GO:0055024 | regulation of cardiac muscle tissue development | 3 | 3 | 5.64e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 4 | 37 | 5.65e-01 |
| GO:BP | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 1 | 3 | 5.65e-01 |
| GO:BP | GO:0001544 | initiation of primordial ovarian follicle growth | 1 | 1 | 5.65e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 4 | 12 | 5.65e-01 |
| GO:BP | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis | 1 | 3 | 5.65e-01 |
| GO:BP | GO:0042976 | activation of Janus kinase activity | 1 | 7 | 5.65e-01 |
| GO:BP | GO:0019449 | L-cysteine catabolic process to hypotaurine | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 3 | 7 | 5.66e-01 |
| GO:BP | GO:1990386 | mitotic cleavage furrow ingression | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0019452 | L-cysteine catabolic process to taurine | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 1 | 616 | 5.66e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 4 | 50 | 5.66e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 5 | 23 | 5.66e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 2 | 5 | 5.66e-01 |
| GO:BP | GO:0034230 | enkephalin processing | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 5.66e-01 |
| GO:BP | GO:0140647 | P450-containing electron transport chain | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 10 | 31 | 5.66e-01 |
| GO:BP | GO:0006171 | cAMP biosynthetic process | 2 | 8 | 5.67e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 5 | 17 | 5.67e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 58 | 881 | 5.67e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 2 | 30 | 5.67e-01 |
| GO:BP | GO:0052472 | modulation by host of symbiont transcription | 1 | 2 | 5.67e-01 |
| GO:BP | GO:0031642 | negative regulation of myelination | 5 | 8 | 5.67e-01 |
| GO:BP | GO:0090667 | cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 5.67e-01 |
| GO:BP | GO:1901837 | negative regulation of transcription of nucleolar large rRNA by RNA polymerase I | 2 | 4 | 5.67e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 3 | 18 | 5.67e-01 |
| GO:BP | GO:0090668 | endothelial cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 7 | 26 | 5.67e-01 |
| GO:BP | GO:0019391 | glucuronoside catabolic process | 1 | 2 | 5.67e-01 |
| GO:BP | GO:0019389 | glucuronoside metabolic process | 1 | 2 | 5.67e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 15 | 115 | 5.67e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 1 | 330 | 5.67e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 15 | 115 | 5.67e-01 |
| GO:BP | GO:0006678 | glucosylceramide metabolic process | 1 | 5 | 5.67e-01 |
| GO:BP | GO:0061091 | regulation of phospholipid translocation | 1 | 5 | 5.67e-01 |
| GO:BP | GO:0046477 | glycosylceramide catabolic process | 1 | 5 | 5.67e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 2 | 7 | 5.67e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 5.67e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 7 | 66 | 5.68e-01 |
| GO:BP | GO:0098828 | modulation of inhibitory postsynaptic potential | 2 | 4 | 5.68e-01 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 3 | 25 | 5.68e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 14 | 76 | 5.68e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 166 | 3625 | 5.68e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 9 | 15 | 5.68e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 1 | 535 | 5.68e-01 |
| GO:BP | GO:0140454 | protein aggregate center assembly | 1 | 1 | 5.68e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 1 | 31 | 5.69e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 5.69e-01 |
| GO:BP | GO:0090245 | axis elongation involved in somitogenesis | 3 | 4 | 5.69e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 2 | 742 | 5.69e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 3 | 9 | 5.69e-01 |
| GO:BP | GO:0110009 | formin-nucleated actin cable organization | 1 | 3 | 5.69e-01 |
| GO:BP | GO:0070649 | formin-nucleated actin cable assembly | 1 | 3 | 5.69e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 18 | 36 | 5.69e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 2 | 6 | 5.69e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 2 | 6 | 5.69e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 2 | 6 | 5.69e-01 |
| GO:BP | GO:0071351 | cellular response to interleukin-18 | 2 | 3 | 5.69e-01 |
| GO:BP | GO:0035655 | interleukin-18-mediated signaling pathway | 2 | 3 | 5.69e-01 |
| GO:BP | GO:1904141 | positive regulation of microglial cell migration | 1 | 3 | 5.69e-01 |
| GO:BP | GO:0120040 | regulation of macrophage proliferation | 1 | 4 | 5.69e-01 |
| GO:BP | GO:0038145 | macrophage colony-stimulating factor signaling pathway | 1 | 3 | 5.69e-01 |
| GO:BP | GO:1901740 | negative regulation of myoblast fusion | 2 | 2 | 5.69e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 2 | 9 | 5.70e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 11 | 31 | 5.70e-01 |
| GO:BP | GO:1902722 | positive regulation of prolactin secretion | 1 | 1 | 5.70e-01 |
| GO:BP | GO:0006426 | glycyl-tRNA aminoacylation | 1 | 1 | 5.70e-01 |
| GO:BP | GO:1901724 | positive regulation of cell proliferation involved in kidney development | 2 | 8 | 5.70e-01 |
| GO:BP | GO:0060718 | chorionic trophoblast cell differentiation | 2 | 5 | 5.70e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 14 | 27 | 5.70e-01 |
| GO:BP | GO:0045763 | negative regulation of cellular amino acid metabolic process | 2 | 2 | 5.70e-01 |
| GO:BP | GO:0046458 | hexadecanal metabolic process | 1 | 1 | 5.70e-01 |
| GO:BP | GO:0070150 | mitochondrial glycyl-tRNA aminoacylation | 1 | 1 | 5.70e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 2 | 12 | 5.70e-01 |
| GO:BP | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway | 2 | 5 | 5.70e-01 |
| GO:BP | GO:0061301 | cerebellum vasculature morphogenesis | 1 | 1 | 5.70e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 5.70e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 13 | 36 | 5.71e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 5 | 40 | 5.71e-01 |
| GO:BP | GO:0106134 | positive regulation of cardiac muscle cell contraction | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 3 | 33 | 5.71e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 12 | 5.71e-01 |
| GO:BP | GO:0097374 | sensory neuron axon guidance | 2 | 2 | 5.71e-01 |
| GO:BP | GO:0051458 | corticotropin secretion | 2 | 4 | 5.71e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 5 | 40 | 5.71e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 34 | 92 | 5.71e-01 |
| GO:BP | GO:0015760 | glucose-6-phosphate transport | 1 | 3 | 5.71e-01 |
| GO:BP | GO:1902068 | regulation of sphingolipid mediated signaling pathway | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0015712 | hexose phosphate transport | 1 | 3 | 5.71e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 13 | 26 | 5.71e-01 |
| GO:BP | GO:0106227 | peptidyl-lysine glutarylation | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0031000 | response to caffeine | 2 | 13 | 5.71e-01 |
| GO:BP | GO:0050915 | sensory perception of sour taste | 1 | 4 | 5.71e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 3 | 12 | 5.71e-01 |
| GO:BP | GO:0007612 | learning | 3 | 115 | 5.71e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 4 | 11 | 5.71e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 51 | 569 | 5.71e-01 |
| GO:BP | GO:0031034 | myosin filament assembly | 2 | 4 | 5.71e-01 |
| GO:BP | GO:0008015 | blood circulation | 16 | 374 | 5.72e-01 |
| GO:BP | GO:0001764 | neuron migration | 7 | 133 | 5.72e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 12 | 1484 | 5.72e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 5.72e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 6 | 107 | 5.73e-01 |
| GO:BP | GO:1901229 | regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 5.73e-01 |
| GO:BP | GO:1901231 | positive regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 5.73e-01 |
| GO:BP | GO:1901232 | regulation of convergent extension involved in axis elongation | 1 | 1 | 5.73e-01 |
| GO:BP | GO:1901233 | negative regulation of convergent extension involved in axis elongation | 1 | 1 | 5.73e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 40 | 81 | 5.73e-01 |
| GO:BP | GO:0014859 | negative regulation of skeletal muscle cell proliferation | 2 | 3 | 5.73e-01 |
| GO:BP | GO:0070586 | cell-cell adhesion involved in gastrulation | 2 | 7 | 5.73e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 3 | 13 | 5.73e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 1 | 11 | 5.73e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 5.73e-01 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 2 | 3 | 5.73e-01 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 2 | 3 | 5.73e-01 |
| GO:BP | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering | 1 | 9 | 5.73e-01 |
| GO:BP | GO:0034635 | glutathione transport | 1 | 9 | 5.73e-01 |
| GO:BP | GO:0090383 | phagosome acidification | 1 | 5 | 5.74e-01 |
| GO:BP | GO:0048232 | male gamete generation | 1 | 343 | 5.74e-01 |
| GO:BP | GO:0090209 | negative regulation of triglyceride metabolic process | 1 | 7 | 5.75e-01 |
| GO:BP | GO:1904003 | negative regulation of sebum secreting cell proliferation | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0072733 | response to staurosporine | 1 | 3 | 5.75e-01 |
| GO:BP | GO:0072734 | cellular response to staurosporine | 1 | 3 | 5.75e-01 |
| GO:BP | GO:2001204 | regulation of osteoclast development | 1 | 4 | 5.75e-01 |
| GO:BP | GO:0010739 | positive regulation of protein kinase A signaling | 2 | 6 | 5.75e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 1 | 3 | 5.75e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 10 | 52 | 5.75e-01 |
| GO:BP | GO:0048320 | axial mesoderm formation | 1 | 2 | 5.75e-01 |
| GO:BP | GO:0032692 | negative regulation of interleukin-1 production | 2 | 15 | 5.75e-01 |
| GO:BP | GO:0061744 | motor behavior | 1 | 17 | 5.75e-01 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 7 | 36 | 5.75e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 4 | 7 | 5.76e-01 |
| GO:BP | GO:0033962 | P-body assembly | 1 | 20 | 5.76e-01 |
| GO:BP | GO:0001545 | primary ovarian follicle growth | 1 | 1 | 5.76e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 21 | 236 | 5.76e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 2 | 3 | 5.76e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 3 | 30 | 5.76e-01 |
| GO:BP | GO:0009227 | nucleotide-sugar catabolic process | 2 | 3 | 5.76e-01 |
| GO:BP | GO:0006855 | xenobiotic transmembrane transport | 2 | 11 | 5.76e-01 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 4 | 12 | 5.76e-01 |
| GO:BP | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 6 | 5.76e-01 |
| GO:BP | GO:0061824 | cytosolic ciliogenesis | 1 | 1 | 5.77e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 38 | 401 | 5.77e-01 |
| GO:BP | GO:0002070 | epithelial cell maturation | 4 | 16 | 5.77e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 5.77e-01 |
| GO:BP | GO:0030581 | symbiont intracellular protein transport in host | 1 | 2 | 5.77e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 8 | 61 | 5.77e-01 |
| GO:BP | GO:0019060 | intracellular transport of viral protein in host cell | 1 | 2 | 5.77e-01 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 3 | 7 | 5.77e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 19 | 91 | 5.78e-01 |
| GO:BP | GO:0032570 | response to progesterone | 3 | 27 | 5.78e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 7 | 15 | 5.78e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 3 | 16 | 5.78e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 1 | 6 | 5.78e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 7 | 15 | 5.78e-01 |
| GO:BP | GO:0099178 | regulation of retrograde trans-synaptic signaling by endocanabinoid | 2 | 3 | 5.78e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 3 | 51 | 5.78e-01 |
| GO:BP | GO:0019079 | viral genome replication | 1 | 101 | 5.78e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 5.78e-01 |
| GO:BP | GO:0043649 | dicarboxylic acid catabolic process | 1 | 14 | 5.78e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 5 | 69 | 5.78e-01 |
| GO:BP | GO:2000609 | regulation of thyroid hormone generation | 1 | 2 | 5.78e-01 |
| GO:BP | GO:0015779 | glucuronoside transport | 1 | 1 | 5.78e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 2 | 51 | 5.78e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 5.78e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 1 | 16 | 5.79e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 5.79e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 5.79e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 26 | 101 | 5.79e-01 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 2 | 15 | 5.79e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 7 | 68 | 5.79e-01 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 4 | 11 | 5.79e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 3 | 13 | 5.79e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2 | 26 | 5.79e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 1 | 13 | 5.79e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 5.79e-01 |
| GO:BP | GO:0040010 | positive regulation of growth rate | 2 | 2 | 5.79e-01 |
| GO:BP | GO:0031929 | TOR signaling | 1 | 115 | 5.79e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 61 | 145 | 5.79e-01 |
| GO:BP | GO:0060820 | inactivation of X chromosome by heterochromatin formation | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 4 | 16 | 5.79e-01 |
| GO:BP | GO:0060026 | convergent extension | 2 | 14 | 5.79e-01 |
| GO:BP | GO:0070383 | DNA cytosine deamination | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0051791 | medium-chain fatty acid metabolic process | 2 | 9 | 5.79e-01 |
| GO:BP | GO:0018201 | peptidyl-glycine modification | 2 | 4 | 5.79e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 2 | 26 | 5.79e-01 |
| GO:BP | GO:0010610 | regulation of mRNA stability involved in response to stress | 2 | 3 | 5.79e-01 |
| GO:BP | GO:1901254 | positive regulation of intracellular transport of viral material | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0016237 | lysosomal microautophagy | 1 | 11 | 5.79e-01 |
| GO:BP | GO:1903512 | phytanic acid metabolic process | 1 | 2 | 5.79e-01 |
| GO:BP | GO:1903059 | regulation of protein lipidation | 1 | 11 | 5.79e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0106028 | neuron projection retraction | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 2 | 8 | 5.79e-01 |
| GO:BP | GO:0099540 | trans-synaptic signaling by neuropeptide | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 5 | 35 | 5.80e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 26 | 80 | 5.80e-01 |
| GO:BP | GO:0008584 | male gonad development | 3 | 99 | 5.80e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 9 | 23 | 5.80e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 9 | 23 | 5.80e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 9 | 23 | 5.80e-01 |
| GO:BP | GO:1904383 | response to sodium phosphate | 3 | 5 | 5.81e-01 |
| GO:BP | GO:0042168 | heme metabolic process | 9 | 40 | 5.81e-01 |
| GO:BP | GO:1904990 | regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1904992 | positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1905857 | positive regulation of pentose-phosphate shunt | 2 | 3 | 5.81e-01 |
| GO:BP | GO:0060687 | regulation of branching involved in prostate gland morphogenesis | 1 | 3 | 5.81e-01 |
| GO:BP | GO:1902595 | regulation of DNA replication origin binding | 1 | 2 | 5.81e-01 |
| GO:BP | GO:1902732 | positive regulation of chondrocyte proliferation | 2 | 5 | 5.81e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 5 | 13 | 5.81e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 10 | 22 | 5.81e-01 |
| GO:BP | GO:1903248 | regulation of citrulline biosynthetic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1903249 | negative regulation of citrulline biosynthetic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation | 1 | 5 | 5.81e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 5.81e-01 |
| GO:BP | GO:1903999 | negative regulation of eating behavior | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0033241 | regulation of amine catabolic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 3 | 39 | 5.81e-01 |
| GO:BP | GO:1902074 | response to salt | 29 | 271 | 5.81e-01 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 4 | 21 | 5.81e-01 |
| GO:BP | GO:0033242 | negative regulation of amine catabolic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0098736 | negative regulation of the force of heart contraction | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0009258 | 10-formyltetrahydrofolate catabolic process | 1 | 3 | 5.81e-01 |
| GO:BP | GO:0090219 | negative regulation of lipid kinase activity | 4 | 8 | 5.81e-01 |
| GO:BP | GO:1905799 | regulation of intraciliary retrograde transport | 1 | 2 | 5.81e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 3 | 8 | 5.81e-01 |
| GO:BP | GO:0070084 | protein initiator methionine removal | 2 | 4 | 5.81e-01 |
| GO:BP | GO:2000283 | negative regulation of amino acid biosynthetic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1900082 | negative regulation of arginine catabolic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 4 | 21 | 5.81e-01 |
| GO:BP | GO:1900081 | regulation of arginine catabolic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 58 | 118 | 5.81e-01 |
| GO:BP | GO:0043105 | negative regulation of GTP cyclohydrolase I activity | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0043095 | regulation of GTP cyclohydrolase I activity | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0030497 | fatty acid elongation | 7 | 12 | 5.82e-01 |
| GO:BP | GO:0042761 | very long-chain fatty acid biosynthetic process | 7 | 12 | 5.82e-01 |
| GO:BP | GO:1905706 | regulation of mitochondrial ATP synthesis coupled proton transport | 2 | 3 | 5.82e-01 |
| GO:BP | GO:1905273 | positive regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 5.82e-01 |
| GO:BP | GO:1905271 | regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0015916 | fatty-acyl-CoA transport | 2 | 2 | 5.82e-01 |
| GO:BP | GO:0019042 | viral latency | 1 | 4 | 5.82e-01 |
| GO:BP | GO:2000627 | positive regulation of miRNA catabolic process | 1 | 4 | 5.82e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 22 | 53 | 5.82e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 3 | 100 | 5.82e-01 |
| GO:BP | GO:1905072 | cardiac jelly development | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 29 | 1380 | 5.82e-01 |
| GO:BP | GO:1903365 | regulation of fear response | 1 | 8 | 5.82e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 1 | 6 | 5.82e-01 |
| GO:BP | GO:0042635 | positive regulation of hair cycle | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0006579 | amino-acid betaine catabolic process | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 4 | 8 | 5.82e-01 |
| GO:BP | GO:0007528 | neuromuscular junction development | 3 | 39 | 5.82e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 10 | 68 | 5.82e-01 |
| GO:BP | GO:0060764 | cell-cell signaling involved in mammary gland development | 1 | 1 | 5.82e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 13 | 41 | 5.82e-01 |
| GO:BP | GO:0060596 | mammary placode formation | 2 | 2 | 5.82e-01 |
| GO:BP | GO:0060527 | prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0060526 | prostate glandular acinus morphogenesis | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 12 | 90 | 5.82e-01 |
| GO:BP | GO:0060010 | Sertoli cell fate commitment | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0051573 | negative regulation of histone H3-K9 methylation | 1 | 5 | 5.82e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 58 | 500 | 5.82e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 5 | 40 | 5.82e-01 |
| GO:BP | GO:0072658 | maintenance of protein location in membrane | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 8 | 35 | 5.82e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 5 | 49 | 5.82e-01 |
| GO:BP | GO:0030900 | forebrain development | 2 | 281 | 5.82e-01 |
| GO:BP | GO:0072660 | maintenance of protein location in plasma membrane | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0032423 | regulation of mismatch repair | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0010650 | positive regulation of cell communication by electrical coupling | 2 | 3 | 5.82e-01 |
| GO:BP | GO:1901337 | thioester transport | 2 | 2 | 5.82e-01 |
| GO:BP | GO:0140060 | axon arborization | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 6 | 113 | 5.82e-01 |
| GO:BP | GO:0097039 | protein linear polyubiquitination | 2 | 5 | 5.82e-01 |
| GO:BP | GO:0003167 | atrioventricular bundle cell differentiation | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0090240 | positive regulation of histone H4 acetylation | 1 | 6 | 5.82e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 5 | 32 | 5.82e-01 |
| GO:BP | GO:0010159 | specification of animal organ position | 2 | 2 | 5.82e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 6 | 170 | 5.82e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 5.83e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 5.83e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 28 | 56 | 5.83e-01 |
| GO:BP | GO:0003430 | growth plate cartilage chondrocyte growth | 2 | 3 | 5.83e-01 |
| GO:BP | GO:0002280 | monocyte activation involved in immune response | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 5.83e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 5.83e-01 |
| GO:BP | GO:0051647 | nucleus localization | 2 | 28 | 5.83e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 5.83e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 32 | 70 | 5.83e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 5.83e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 4 | 17 | 5.83e-01 |
| GO:BP | GO:0045730 | respiratory burst | 4 | 21 | 5.83e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 5 | 55 | 5.84e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 6 | 101 | 5.84e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 5 | 8 | 5.84e-01 |
| GO:BP | GO:1903811 | L-asparagine import across plasma membrane | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 1 | 19 | 5.84e-01 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 3 | 44 | 5.84e-01 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 2 | 7 | 5.84e-01 |
| GO:BP | GO:0070408 | carbamoyl phosphate metabolic process | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0035624 | receptor transactivation | 1 | 3 | 5.84e-01 |
| GO:BP | GO:0070409 | carbamoyl phosphate biosynthetic process | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 4 | 9 | 5.84e-01 |
| GO:BP | GO:0090088 | regulation of oligopeptide transport | 1 | 2 | 5.84e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 4 | 11 | 5.84e-01 |
| GO:BP | GO:0045959 | negative regulation of complement activation, classical pathway | 2 | 4 | 5.84e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 13 | 2139 | 5.84e-01 |
| GO:BP | GO:1902951 | negative regulation of dendritic spine maintenance | 1 | 2 | 5.84e-01 |
| GO:BP | GO:2001150 | positive regulation of dipeptide transmembrane transport | 1 | 2 | 5.84e-01 |
| GO:BP | GO:2001148 | regulation of dipeptide transmembrane transport | 1 | 2 | 5.84e-01 |
| GO:BP | GO:1902938 | regulation of intracellular calcium activated chloride channel activity | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 6 | 12 | 5.84e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 1 | 1173 | 5.84e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 10 | 17 | 5.84e-01 |
| GO:BP | GO:2000880 | positive regulation of dipeptide transport | 1 | 2 | 5.84e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 5 | 22 | 5.84e-01 |
| GO:BP | GO:2000878 | positive regulation of oligopeptide transport | 1 | 2 | 5.84e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 6 | 9 | 5.84e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 1 | 8 | 5.84e-01 |
| GO:BP | GO:0090089 | regulation of dipeptide transport | 1 | 2 | 5.84e-01 |
| GO:BP | GO:0010700 | negative regulation of norepinephrine secretion | 1 | 2 | 5.84e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 12 | 252 | 5.84e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 5.84e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2780 | 5.84e-01 |
| GO:BP | GO:0032811 | negative regulation of epinephrine secretion | 1 | 2 | 5.84e-01 |
| GO:BP | GO:0046318 | negative regulation of glucosylceramide biosynthetic process | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0046317 | regulation of glucosylceramide biosynthetic process | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 5.84e-01 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 2 | 7 | 5.84e-01 |
| GO:BP | GO:1902426 | deactivation of mitotic spindle assembly checkpoint | 2 | 3 | 5.84e-01 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 3 | 5.84e-01 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 3 | 5.84e-01 |
| GO:BP | GO:0006968 | cellular defense response | 2 | 15 | 5.85e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 1 | 14 | 5.85e-01 |
| GO:BP | GO:0060558 | regulation of calcidiol 1-monooxygenase activity | 1 | 2 | 5.85e-01 |
| GO:BP | GO:0072674 | multinuclear osteoclast differentiation | 1 | 6 | 5.85e-01 |
| GO:BP | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 1 | 4 | 5.86e-01 |
| GO:BP | GO:0003057 | regulation of the force of heart contraction by chemical signal | 3 | 5 | 5.86e-01 |
| GO:BP | GO:1902216 | positive regulation of interleukin-4-mediated signaling pathway | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0042797 | tRNA transcription by RNA polymerase III | 4 | 7 | 5.86e-01 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 5 | 11 | 5.86e-01 |
| GO:BP | GO:0006863 | purine nucleobase transport | 1 | 7 | 5.86e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 5.86e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 2 | 36 | 5.86e-01 |
| GO:BP | GO:0006127 | glycerophosphate shuttle | 1 | 2 | 5.86e-01 |
| GO:BP | GO:0070236 | negative regulation of activation-induced cell death of T cells | 2 | 3 | 5.86e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 2 | 2 | 5.86e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 13 | 15 | 5.86e-01 |
| GO:BP | GO:0055089 | fatty acid homeostasis | 1 | 12 | 5.86e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 3 | 225 | 5.86e-01 |
| GO:BP | GO:0050812 | regulation of acyl-CoA biosynthetic process | 2 | 5 | 5.86e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 2 | 30 | 5.86e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 4 | 9 | 5.86e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 12 | 43 | 5.86e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 5.86e-01 |
| GO:BP | GO:0072319 | vesicle uncoating | 5 | 9 | 5.86e-01 |
| GO:BP | GO:0001554 | luteolysis | 3 | 7 | 5.86e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 9 | 80 | 5.86e-01 |
| GO:BP | GO:0002184 | cytoplasmic translational termination | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 6 | 23 | 5.86e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 20 | 50 | 5.86e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 5 | 50 | 5.86e-01 |
| GO:BP | GO:0050860 | negative regulation of T cell receptor signaling pathway | 1 | 17 | 5.86e-01 |
| GO:BP | GO:0061025 | membrane fusion | 2 | 137 | 5.86e-01 |
| GO:BP | GO:2001144 | regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 5.87e-01 |
| GO:BP | GO:2001145 | negative regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | 1 | 1 | 5.87e-01 |
| GO:BP | GO:0016094 | polyprenol biosynthetic process | 1 | 3 | 5.87e-01 |
| GO:BP | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus | 2 | 6 | 5.87e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 1 | 12 | 5.87e-01 |
| GO:BP | GO:0050930 | induction of positive chemotaxis | 1 | 12 | 5.87e-01 |
| GO:BP | GO:0045010 | actin nucleation | 3 | 50 | 5.87e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 7 | 36 | 5.87e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 3 | 21 | 5.88e-01 |
| GO:BP | GO:0090206 | negative regulation of cholesterol metabolic process | 3 | 5 | 5.88e-01 |
| GO:BP | GO:0106119 | negative regulation of sterol biosynthetic process | 3 | 5 | 5.88e-01 |
| GO:BP | GO:0045541 | negative regulation of cholesterol biosynthetic process | 3 | 5 | 5.88e-01 |
| GO:BP | GO:0021849 | neuroblast division in subventricular zone | 1 | 3 | 5.88e-01 |
| GO:BP | GO:1900143 | positive regulation of oligodendrocyte apoptotic process | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0001809 | positive regulation of type IV hypersensitivity | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 3 | 5 | 5.88e-01 |
| GO:BP | GO:0060022 | hard palate development | 4 | 5 | 5.88e-01 |
| GO:BP | GO:0016256 | N-glycan processing to lysosome | 1 | 2 | 5.88e-01 |
| GO:BP | GO:2000387 | regulation of antral ovarian follicle growth | 1 | 1 | 5.88e-01 |
| GO:BP | GO:2000388 | positive regulation of antral ovarian follicle growth | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0031117 | positive regulation of microtubule depolymerization | 1 | 4 | 5.88e-01 |
| GO:BP | GO:1900130 | regulation of lipid binding | 1 | 1 | 5.88e-01 |
| GO:BP | GO:1900131 | negative regulation of lipid binding | 1 | 1 | 5.88e-01 |
| GO:BP | GO:0042118 | endothelial cell activation | 5 | 7 | 5.88e-01 |
| GO:BP | GO:0034463 | 90S preribosome assembly | 1 | 1 | 5.89e-01 |
| GO:BP | GO:0006012 | galactose metabolic process | 1 | 9 | 5.89e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 9 | 15 | 5.89e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 22 | 105 | 5.90e-01 |
| GO:BP | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway | 1 | 3 | 5.90e-01 |
| GO:BP | GO:0003150 | muscular septum morphogenesis | 1 | 4 | 5.90e-01 |
| GO:BP | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development | 1 | 3 | 5.90e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 7 | 130 | 5.90e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 407 | 5.90e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 2 | 57 | 5.90e-01 |
| GO:BP | GO:2000490 | negative regulation of hepatic stellate cell activation | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0015727 | lactate transport | 1 | 5 | 5.90e-01 |
| GO:BP | GO:0035873 | lactate transmembrane transport | 1 | 5 | 5.90e-01 |
| GO:BP | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching | 1 | 2 | 5.90e-01 |
| GO:BP | GO:0072684 | mitochondrial tRNA 3’-trailer cleavage, endonucleolytic | 1 | 1 | 5.90e-01 |
| GO:BP | GO:0031340 | positive regulation of vesicle fusion | 1 | 9 | 5.90e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 1 | 39 | 5.91e-01 |
| GO:BP | GO:0033034 | positive regulation of myeloid cell apoptotic process | 3 | 6 | 5.91e-01 |
| GO:BP | GO:0009224 | CMP biosynthetic process | 1 | 3 | 5.91e-01 |
| GO:BP | GO:0007601 | visual perception | 1 | 96 | 5.91e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 5.91e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 5.91e-01 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 1 | 30 | 5.91e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 5.91e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 23 | 111 | 5.91e-01 |
| GO:BP | GO:1905090 | negative regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 5.91e-01 |
| GO:BP | GO:0016577 | histone demethylation | 5 | 21 | 5.91e-01 |
| GO:BP | GO:2000413 | regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 5.91e-01 |
| GO:BP | GO:0072681 | fibronectin-dependent thymocyte migration | 1 | 1 | 5.91e-01 |
| GO:BP | GO:2000415 | positive regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 5.91e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 5.91e-01 |
| GO:BP | GO:0072680 | extracellular matrix-dependent thymocyte migration | 1 | 1 | 5.91e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 15 | 31 | 5.91e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 14 | 142 | 5.92e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 5 | 35 | 5.92e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 2 | 1974 | 5.92e-01 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 5 | 25 | 5.92e-01 |
| GO:BP | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis | 1 | 1 | 5.92e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 5 | 16 | 5.92e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 34 | 166 | 5.92e-01 |
| GO:BP | GO:1904970 | brush border assembly | 1 | 2 | 5.92e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 2 | 53 | 5.92e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 3 | 70 | 5.92e-01 |
| GO:BP | GO:0072007 | mesangial cell differentiation | 1 | 5 | 5.92e-01 |
| GO:BP | GO:0021978 | telencephalon regionalization | 1 | 4 | 5.92e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 4 | 18 | 5.92e-01 |
| GO:BP | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process | 1 | 2 | 5.92e-01 |
| GO:BP | GO:0006656 | phosphatidylcholine biosynthetic process | 12 | 25 | 5.92e-01 |
| GO:BP | GO:0090156 | intracellular sphingolipid homeostasis | 1 | 5 | 5.92e-01 |
| GO:BP | GO:0010764 | negative regulation of fibroblast migration | 1 | 9 | 5.92e-01 |
| GO:BP | GO:0038001 | paracrine signaling | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0060939 | epicardium-derived cardiac fibroblast cell development | 2 | 3 | 5.92e-01 |
| GO:BP | GO:1902847 | regulation of neuronal signal transduction | 1 | 1 | 5.92e-01 |
| GO:BP | GO:1902998 | positive regulation of neurofibrillary tangle assembly | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 13 | 172 | 5.92e-01 |
| GO:BP | GO:0060938 | epicardium-derived cardiac fibroblast cell differentiation | 2 | 3 | 5.92e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 8 | 25 | 5.92e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 8 | 25 | 5.92e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 2 | 27 | 5.92e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 7 | 22 | 5.92e-01 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 2 | 6 | 5.92e-01 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 1 | 6 | 5.92e-01 |
| GO:BP | GO:0051890 | regulation of cardioblast differentiation | 2 | 5 | 5.92e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 5.92e-01 |
| GO:BP | GO:0098759 | cellular response to interleukin-8 | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0098758 | response to interleukin-8 | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0072262 | metanephric glomerular mesangial cell proliferation involved in metanephros development | 1 | 2 | 5.92e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 5.92e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 5.92e-01 |
| GO:BP | GO:1990108 | protein linear deubiquitination | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 25 | 103 | 5.92e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 4 | 24 | 5.92e-01 |
| GO:BP | GO:0007416 | synapse assembly | 9 | 152 | 5.92e-01 |
| GO:BP | GO:1903352 | L-ornithine transmembrane transport | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0030643 | intracellular phosphate ion homeostasis | 1 | 5 | 5.92e-01 |
| GO:BP | GO:2000182 | regulation of progesterone biosynthetic process | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0060086 | circadian temperature homeostasis | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 60 | 5.92e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 1 | 40 | 5.92e-01 |
| GO:BP | GO:0046056 | dADP metabolic process | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0046712 | GDP catabolic process | 1 | 2 | 5.92e-01 |
| GO:BP | GO:0046066 | dGDP metabolic process | 1 | 2 | 5.92e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 3 | 15 | 5.92e-01 |
| GO:BP | GO:0060301 | positive regulation of cytokine activity | 1 | 1 | 5.92e-01 |
| GO:BP | GO:1905040 | otic placode development | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0030263 | apoptotic chromosome condensation | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 27 | 59 | 5.92e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 13 | 2161 | 5.92e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 5.92e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 2 | 83 | 5.92e-01 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 1 | 14 | 5.92e-01 |
| GO:BP | GO:1902378 | VEGF-activated neuropilin signaling pathway involved in axon guidance | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration | 1 | 3 | 5.93e-01 |
| GO:BP | GO:0032613 | interleukin-10 production | 1 | 24 | 5.93e-01 |
| GO:BP | GO:0007548 | sex differentiation | 5 | 191 | 5.93e-01 |
| GO:BP | GO:0007231 | osmosensory signaling pathway | 3 | 3 | 5.93e-01 |
| GO:BP | GO:0061518 | microglial cell proliferation | 1 | 4 | 5.93e-01 |
| GO:BP | GO:1902834 | regulation of proline import across plasma membrane | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 2 | 48 | 5.93e-01 |
| GO:BP | GO:1902836 | positive regulation of proline import across plasma membrane | 1 | 2 | 5.93e-01 |
| GO:BP | GO:1904124 | microglial cell migration | 1 | 3 | 5.93e-01 |
| GO:BP | GO:1904139 | regulation of microglial cell migration | 1 | 3 | 5.93e-01 |
| GO:BP | GO:0021629 | olfactory nerve structural organization | 1 | 1 | 5.93e-01 |
| GO:BP | GO:2000764 | positive regulation of semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 1 | 27 | 5.93e-01 |
| GO:BP | GO:0042701 | progesterone secretion | 2 | 3 | 5.93e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 2 | 48 | 5.93e-01 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 1 | 24 | 5.93e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 5.93e-01 |
| GO:BP | GO:0014042 | positive regulation of neuron maturation | 1 | 3 | 5.93e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 3 | 40 | 5.93e-01 |
| GO:BP | GO:0030329 | prenylcysteine metabolic process | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0030328 | prenylcysteine catabolic process | 1 | 2 | 5.93e-01 |
| GO:BP | GO:2000763 | positive regulation of transcription from RNA polymerase II promoter involved in norepinephrine biosynthetic process | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0009415 | response to water | 1 | 9 | 5.93e-01 |
| GO:BP | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 3 | 5.93e-01 |
| GO:BP | GO:0070881 | regulation of proline transport | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0014724 | regulation of twitch skeletal muscle contraction | 2 | 3 | 5.93e-01 |
| GO:BP | GO:0034773 | histone H4-K20 trimethylation | 3 | 5 | 5.93e-01 |
| GO:BP | GO:0098529 | neuromuscular junction development, skeletal muscle fiber | 1 | 1 | 5.93e-01 |
| GO:BP | GO:1905735 | regulation of L-proline import across plasma membrane | 1 | 2 | 5.93e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 4 | 14 | 5.93e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 10 | 222 | 5.93e-01 |
| GO:BP | GO:1905737 | positive regulation of L-proline import across plasma membrane | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0021934 | hindbrain tangential cell migration | 1 | 3 | 5.93e-01 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 1 | 17 | 5.93e-01 |
| GO:BP | GO:0072300 | positive regulation of metanephric glomerulus development | 1 | 1 | 5.93e-01 |
| GO:BP | GO:2001262 | positive regulation of semaphorin-plexin signaling pathway | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0071527 | semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 5.93e-01 |
| GO:BP | GO:0072216 | positive regulation of metanephros development | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 5 | 16 | 5.93e-01 |
| GO:BP | GO:0048132 | female germ-line stem cell asymmetric division | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0006702 | androgen biosynthetic process | 2 | 4 | 5.93e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 18 | 36 | 5.93e-01 |
| GO:BP | GO:0072298 | regulation of metanephric glomerulus development | 1 | 1 | 5.93e-01 |
| GO:BP | GO:0003419 | growth plate cartilage chondrocyte proliferation | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0042816 | vitamin B6 metabolic process | 1 | 5 | 5.93e-01 |
| GO:BP | GO:0032835 | glomerulus development | 11 | 54 | 5.93e-01 |
| GO:BP | GO:0075506 | entry of viral genome into host nucleus through nuclear pore complex via importin | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0034447 | very-low-density lipoprotein particle clearance | 2 | 4 | 5.93e-01 |
| GO:BP | GO:1904797 | negative regulation of core promoter binding | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0031449 | regulation of slow-twitch skeletal muscle fiber contraction | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0048549 | positive regulation of pinocytosis | 4 | 7 | 5.94e-01 |
| GO:BP | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation | 1 | 4 | 5.94e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 3 | 11 | 5.94e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 2 | 55 | 5.94e-01 |
| GO:BP | GO:2000774 | positive regulation of cellular senescence | 3 | 9 | 5.94e-01 |
| GO:BP | GO:0006884 | cell volume homeostasis | 4 | 24 | 5.94e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 21 | 214 | 5.94e-01 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 4 | 23 | 5.94e-01 |
| GO:BP | GO:0070376 | regulation of ERK5 cascade | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0003358 | noradrenergic neuron development | 1 | 2 | 5.94e-01 |
| GO:BP | GO:0031118 | rRNA pseudouridine synthesis | 3 | 10 | 5.94e-01 |
| GO:BP | GO:1904851 | positive regulation of establishment of protein localization to telomere | 3 | 10 | 5.94e-01 |
| GO:BP | GO:0030321 | transepithelial chloride transport | 3 | 7 | 5.94e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 2 | 5 | 5.95e-01 |
| GO:BP | GO:0060696 | regulation of phospholipid catabolic process | 1 | 5 | 5.95e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 5.95e-01 |
| GO:BP | GO:0016031 | tRNA import into mitochondrion | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 1 | 12 | 5.95e-01 |
| GO:BP | GO:0010839 | negative regulation of keratinocyte proliferation | 2 | 19 | 5.95e-01 |
| GO:BP | GO:0043931 | ossification involved in bone maturation | 1 | 17 | 5.95e-01 |
| GO:BP | GO:0099514 | synaptic vesicle cytoskeletal transport | 2 | 16 | 5.96e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 5.96e-01 |
| GO:BP | GO:0099517 | synaptic vesicle transport along microtubule | 2 | 16 | 5.96e-01 |
| GO:BP | GO:0048636 | positive regulation of muscle organ development | 2 | 3 | 5.96e-01 |
| GO:BP | GO:0009409 | response to cold | 10 | 41 | 5.96e-01 |
| GO:BP | GO:0045844 | positive regulation of striated muscle tissue development | 2 | 3 | 5.96e-01 |
| GO:BP | GO:0048490 | anterograde synaptic vesicle transport | 2 | 16 | 5.96e-01 |
| GO:BP | GO:0030258 | lipid modification | 2 | 162 | 5.96e-01 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 1 | 6 | 5.96e-01 |
| GO:BP | GO:0106035 | protein maturation by [4Fe-4S] cluster transfer | 2 | 7 | 5.96e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 14 | 101 | 5.96e-01 |
| GO:BP | GO:0003250 | regulation of cell proliferation involved in heart valve morphogenesis | 2 | 3 | 5.96e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 3 | 86 | 5.96e-01 |
| GO:BP | GO:0003249 | cell proliferation involved in heart valve morphogenesis | 2 | 3 | 5.96e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 1 | 101 | 5.96e-01 |
| GO:BP | GO:0046849 | bone remodeling | 6 | 59 | 5.96e-01 |
| GO:BP | GO:0044828 | negative regulation by host of viral genome replication | 2 | 7 | 5.96e-01 |
| GO:BP | GO:0007399 | nervous system development | 2 | 1904 | 5.96e-01 |
| GO:BP | GO:0106091 | glial cell projection elongation | 2 | 2 | 5.96e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 5.96e-01 |
| GO:BP | GO:0017145 | stem cell division | 13 | 26 | 5.97e-01 |
| GO:BP | GO:1904181 | positive regulation of membrane depolarization | 3 | 8 | 5.97e-01 |
| GO:BP | GO:0016076 | snRNA catabolic process | 2 | 5 | 5.97e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 5.97e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 8 | 227 | 5.97e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 4 | 32 | 5.97e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 5.97e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 7 | 5.97e-01 |
| GO:BP | GO:0071139 | resolution of recombination intermediates | 3 | 6 | 5.97e-01 |
| GO:BP | GO:0033080 | immature T cell proliferation in thymus | 2 | 7 | 5.97e-01 |
| GO:BP | GO:0033079 | immature T cell proliferation | 2 | 7 | 5.97e-01 |
| GO:BP | GO:1904837 | beta-catenin-TCF complex assembly | 1 | 3 | 5.97e-01 |
| GO:BP | GO:0032287 | peripheral nervous system myelin maintenance | 3 | 8 | 5.97e-01 |
| GO:BP | GO:0032877 | positive regulation of DNA endoreduplication | 1 | 2 | 5.97e-01 |
| GO:BP | GO:0007538 | primary sex determination | 1 | 2 | 5.97e-01 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 2 | 9 | 5.97e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 2 | 111 | 5.97e-01 |
| GO:BP | GO:0042262 | DNA protection | 1 | 6 | 5.97e-01 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 3 | 14 | 5.97e-01 |
| GO:BP | GO:0032252 | secretory granule localization | 1 | 9 | 5.97e-01 |
| GO:BP | GO:0090022 | regulation of neutrophil chemotaxis | 1 | 19 | 5.98e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 6 | 67 | 5.98e-01 |
| GO:BP | GO:0060268 | negative regulation of respiratory burst | 1 | 4 | 5.98e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 4 | 20 | 5.98e-01 |
| GO:BP | GO:0009756 | carbohydrate mediated signaling | 1 | 6 | 5.98e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 3 | 26 | 5.98e-01 |
| GO:BP | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 1 | 6 | 5.98e-01 |
| GO:BP | GO:0003013 | circulatory system process | 18 | 447 | 5.99e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 6 | 102 | 5.99e-01 |
| GO:BP | GO:1901622 | positive regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0106020 | regulation of vesicle docking | 1 | 2 | 5.99e-01 |
| GO:BP | GO:0033169 | histone H3-K9 demethylation | 1 | 5 | 5.99e-01 |
| GO:BP | GO:0007624 | ultradian rhythm | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0032438 | melanosome organization | 4 | 36 | 6.00e-01 |
| GO:BP | GO:0042335 | cuticle development | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0060018 | astrocyte fate commitment | 2 | 2 | 6.00e-01 |
| GO:BP | GO:0060948 | cardiac vascular smooth muscle cell development | 2 | 3 | 6.00e-01 |
| GO:BP | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation | 2 | 3 | 6.00e-01 |
| GO:BP | GO:0072289 | metanephric nephron tubule formation | 2 | 2 | 6.00e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 4 | 45 | 6.00e-01 |
| GO:BP | GO:0016182 | synaptic vesicle budding from endosome | 1 | 1 | 6.00e-01 |
| GO:BP | GO:1904616 | regulation of actin binding | 1 | 3 | 6.01e-01 |
| GO:BP | GO:1990547 | mitochondrial phosphate ion transmembrane transport | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 6.01e-01 |
| GO:BP | GO:0072560 | type B pancreatic cell maturation | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0001946 | lymphangiogenesis | 3 | 12 | 6.01e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 6.01e-01 |
| GO:BP | GO:1905584 | outer hair cell apoptotic process | 1 | 2 | 6.01e-01 |
| GO:BP | GO:1905349 | ciliary transition zone assembly | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 3 | 11 | 6.01e-01 |
| GO:BP | GO:0016191 | synaptic vesicle uncoating | 4 | 7 | 6.01e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 2 | 14 | 6.02e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 3 | 70 | 6.02e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 5 | 50 | 6.02e-01 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 2 | 5 | 6.02e-01 |
| GO:BP | GO:0009629 | response to gravity | 2 | 5 | 6.02e-01 |
| GO:BP | GO:0006573 | valine metabolic process | 1 | 7 | 6.02e-01 |
| GO:BP | GO:0006549 | isoleucine metabolic process | 1 | 6 | 6.02e-01 |
| GO:BP | GO:0019441 | tryptophan catabolic process to kynurenine | 1 | 1 | 6.02e-01 |
| GO:BP | GO:0032610 | interleukin-1 alpha production | 1 | 4 | 6.02e-01 |
| GO:BP | GO:0032650 | regulation of interleukin-1 alpha production | 1 | 4 | 6.02e-01 |
| GO:BP | GO:0006072 | glycerol-3-phosphate metabolic process | 3 | 7 | 6.02e-01 |
| GO:BP | GO:0061789 | dense core granule priming | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 1 | 10 | 6.03e-01 |
| GO:BP | GO:0045829 | negative regulation of isotype switching | 2 | 4 | 6.03e-01 |
| GO:BP | GO:0009932 | cell tip growth | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0003058 | hormonal regulation of the force of heart contraction | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 44 | 486 | 6.03e-01 |
| GO:BP | GO:0034201 | response to oleic acid | 3 | 5 | 6.03e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 1 | 17 | 6.03e-01 |
| GO:BP | GO:0072429 | response to intra-S DNA damage checkpoint signaling | 1 | 4 | 6.03e-01 |
| GO:BP | GO:1900099 | negative regulation of plasma cell differentiation | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 6.03e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 6.03e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 6.03e-01 |
| GO:BP | GO:1903463 | regulation of mitotic cell cycle DNA replication | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 3 | 26 | 6.03e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 6.03e-01 |
| GO:BP | GO:1902268 | negative regulation of polyamine transmembrane transport | 1 | 3 | 6.03e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 2 | 11 | 6.03e-01 |
| GO:BP | GO:1902896 | terminal web assembly | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 2 | 9 | 6.03e-01 |
| GO:BP | GO:0036155 | acylglycerol acyl-chain remodeling | 2 | 2 | 6.03e-01 |
| GO:BP | GO:0036506 | maintenance of unfolded protein | 1 | 3 | 6.03e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 6.03e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 1 | 101 | 6.03e-01 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 3 | 7 | 6.03e-01 |
| GO:BP | GO:1904431 | positive regulation of t-circle formation | 1 | 3 | 6.03e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 5 | 30 | 6.03e-01 |
| GO:BP | GO:1904378 | maintenance of unfolded protein involved in ERAD pathway | 1 | 3 | 6.03e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 4 | 24 | 6.03e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 3 | 27 | 6.03e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 7 | 15 | 6.03e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 4 | 159 | 6.03e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 6 | 103 | 6.03e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 29 | 6.03e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 6 | 11 | 6.04e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 9 | 230 | 6.04e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 9 | 17 | 6.04e-01 |
| GO:BP | GO:1904447 | folate import across plasma membrane | 1 | 4 | 6.04e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 8 | 44 | 6.04e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 55 | 162 | 6.04e-01 |
| GO:BP | GO:0003242 | cardiac chamber ballooning | 1 | 1 | 6.04e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 4 | 155 | 6.04e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 6 | 118 | 6.04e-01 |
| GO:BP | GO:0061670 | evoked neurotransmitter secretion | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0060267 | positive regulation of respiratory burst | 2 | 3 | 6.05e-01 |
| GO:BP | GO:0048625 | myoblast fate commitment | 2 | 4 | 6.05e-01 |
| GO:BP | GO:2000806 | positive regulation of termination of RNA polymerase II transcription, poly(A)-coupled | 1 | 1 | 6.05e-01 |
| GO:BP | GO:2000458 | regulation of astrocyte chemotaxis | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1904595 | positive regulation of termination of RNA polymerase II transcription | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0035700 | astrocyte chemotaxis | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 1 | 28 | 6.05e-01 |
| GO:BP | GO:0035196 | miRNA processing | 17 | 41 | 6.05e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 3 | 12 | 6.05e-01 |
| GO:BP | GO:0099161 | regulation of presynaptic dense core granule exocytosis | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0010759 | positive regulation of macrophage chemotaxis | 3 | 9 | 6.05e-01 |
| GO:BP | GO:1904828 | positive regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0036145 | dendritic cell homeostasis | 1 | 2 | 6.05e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 12 | 48 | 6.05e-01 |
| GO:BP | GO:0060566 | positive regulation of termination of DNA-templated transcription | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1904826 | regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 6.05e-01 |
| GO:BP | GO:0015851 | nucleobase transport | 1 | 7 | 6.05e-01 |
| GO:BP | GO:0070842 | aggresome assembly | 3 | 5 | 6.05e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 4 | 8 | 6.05e-01 |
| GO:BP | GO:2000489 | regulation of hepatic stellate cell activation | 2 | 6 | 6.05e-01 |
| GO:BP | GO:0031284 | positive regulation of guanylate cyclase activity | 2 | 2 | 6.05e-01 |
| GO:BP | GO:0046619 | lens placode formation involved in camera-type eye formation | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0001743 | lens placode formation | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 5 | 84 | 6.05e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 2 | 252 | 6.05e-01 |
| GO:BP | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 8 | 6.05e-01 |
| GO:BP | GO:1901632 | regulation of synaptic vesicle membrane organization | 1 | 8 | 6.05e-01 |
| GO:BP | GO:0048732 | gland development | 3 | 318 | 6.05e-01 |
| GO:BP | GO:0048697 | positive regulation of collateral sprouting in absence of injury | 1 | 1 | 6.05e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 3 | 54 | 6.05e-01 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 2 | 19 | 6.05e-01 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 3 | 23 | 6.05e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 1 | 88 | 6.06e-01 |
| GO:BP | GO:1902725 | negative regulation of satellite cell differentiation | 2 | 2 | 6.06e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 1 | 31 | 6.06e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 2 | 60 | 6.06e-01 |
| GO:BP | GO:0035733 | hepatic stellate cell activation | 5 | 8 | 6.06e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 3 | 10 | 6.06e-01 |
| GO:BP | GO:0061055 | myotome development | 2 | 2 | 6.06e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0006564 | L-serine biosynthetic process | 1 | 6 | 6.06e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 4 | 687 | 6.06e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 3 | 34 | 6.06e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 1 | 15 | 6.06e-01 |
| GO:BP | GO:0002331 | pre-B cell allelic exclusion | 1 | 2 | 6.06e-01 |
| GO:BP | GO:1905532 | regulation of leucine import across plasma membrane | 1 | 3 | 6.06e-01 |
| GO:BP | GO:1903650 | negative regulation of cytoplasmic transport | 2 | 3 | 6.06e-01 |
| GO:BP | GO:0030185 | nitric oxide transport | 1 | 2 | 6.06e-01 |
| GO:BP | GO:0002524 | hypersensitivity | 3 | 7 | 6.06e-01 |
| GO:BP | GO:0098970 | postsynaptic neurotransmitter receptor diffusion trapping | 1 | 4 | 6.06e-01 |
| GO:BP | GO:0036364 | transforming growth factor beta1 activation | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0007621 | negative regulation of female receptivity | 1 | 1 | 6.06e-01 |
| GO:BP | GO:2000645 | negative regulation of receptor catabolic process | 1 | 3 | 6.06e-01 |
| GO:BP | GO:0098953 | receptor diffusion trapping | 1 | 4 | 6.06e-01 |
| GO:BP | GO:0099628 | neurotransmitter receptor diffusion trapping | 1 | 4 | 6.06e-01 |
| GO:BP | GO:1901006 | ubiquinone-6 biosynthetic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:1901004 | ubiquinone-6 metabolic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 4 | 43 | 6.06e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 3 | 55 | 6.06e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 4 | 78 | 6.06e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 6 | 37 | 6.07e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 3 | 43 | 6.07e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 6.07e-01 |
| GO:BP | GO:0097154 | GABAergic neuron differentiation | 1 | 5 | 6.07e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 17 | 1735 | 6.07e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 4 | 16 | 6.08e-01 |
| GO:BP | GO:0051796 | negative regulation of timing of catagen | 1 | 1 | 6.08e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 4 | 72 | 6.08e-01 |
| GO:BP | GO:1902910 | positive regulation of melanosome transport | 1 | 1 | 6.08e-01 |
| GO:BP | GO:0060901 | regulation of hair cycle by canonical Wnt signaling pathway | 1 | 1 | 6.08e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 2 | 8 | 6.08e-01 |
| GO:BP | GO:0050652 | dermatan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 1 | 6.08e-01 |
| GO:BP | GO:0071389 | cellular response to mineralocorticoid stimulus | 7 | 9 | 6.08e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 6.09e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 1 | 22 | 6.09e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 7 | 33 | 6.09e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 6.09e-01 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 9 | 17 | 6.09e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 6.09e-01 |
| GO:BP | GO:0006325 | chromatin organization | 8 | 551 | 6.09e-01 |
| GO:BP | GO:0034139 | regulation of toll-like receptor 3 signaling pathway | 2 | 8 | 6.10e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 4 | 6 | 6.10e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 4 | 6 | 6.10e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 6.10e-01 |
| GO:BP | GO:0032485 | regulation of Ral protein signal transduction | 2 | 3 | 6.10e-01 |
| GO:BP | GO:0060956 | endocardial cell differentiation | 2 | 4 | 6.10e-01 |
| GO:BP | GO:0003348 | cardiac endothelial cell differentiation | 2 | 4 | 6.10e-01 |
| GO:BP | GO:0014807 | regulation of somitogenesis | 2 | 3 | 6.10e-01 |
| GO:BP | GO:0032095 | regulation of response to food | 1 | 8 | 6.10e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 4 | 59 | 6.10e-01 |
| GO:BP | GO:0032782 | bile acid secretion | 1 | 5 | 6.11e-01 |
| GO:BP | GO:0042339 | keratan sulfate metabolic process | 6 | 15 | 6.11e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 7 | 15 | 6.11e-01 |
| GO:BP | GO:0140157 | ammonium import across plasma membrane | 2 | 3 | 6.11e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 7 | 21 | 6.11e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 9 | 19 | 6.11e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 3 | 9 | 6.11e-01 |
| GO:BP | GO:0061669 | spontaneous neurotransmitter secretion | 2 | 5 | 6.11e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 2 | 9 | 6.11e-01 |
| GO:BP | GO:0072097 | negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 9 | 154 | 6.11e-01 |
| GO:BP | GO:0034097 | response to cytokine | 33 | 607 | 6.11e-01 |
| GO:BP | GO:1904193 | negative regulation of cholangiocyte apoptotic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0050882 | voluntary musculoskeletal movement | 3 | 7 | 6.11e-01 |
| GO:BP | GO:0097370 | protein O-GlcNAcylation via threonine | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1903689 | regulation of wound healing, spreading of epidermal cells | 1 | 8 | 6.11e-01 |
| GO:BP | GO:0060503 | bud dilation involved in lung branching | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072101 | specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072099 | anterior/posterior pattern specification involved in ureteric bud development | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0070343 | white fat cell proliferation | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0003218 | cardiac left ventricle formation | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0015677 | copper ion import | 2 | 5 | 6.11e-01 |
| GO:BP | GO:0046794 | transport of virus | 8 | 19 | 6.11e-01 |
| GO:BP | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway | 3 | 7 | 6.11e-01 |
| GO:BP | GO:0072200 | negative regulation of mesenchymal cell proliferation involved in ureter development | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 4 | 37 | 6.11e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 6.11e-01 |
| GO:BP | GO:0061031 | endodermal digestive tract morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1902488 | cholangiocyte apoptotic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1904192 | regulation of cholangiocyte apoptotic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 2 | 33 | 6.11e-01 |
| GO:BP | GO:0061151 | BMP signaling pathway involved in renal system segmentation | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072192 | ureter epithelial cell differentiation | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 23 | 6.11e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 6.11e-01 |
| GO:BP | GO:0048680 | positive regulation of axon regeneration | 1 | 8 | 6.11e-01 |
| GO:BP | GO:0009210 | pyrimidine ribonucleoside triphosphate catabolic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0090194 | negative regulation of glomerulus development | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0009237 | siderophore metabolic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072100 | specification of ureteric bud anterior/posterior symmetry | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 8 | 161 | 6.11e-01 |
| GO:BP | GO:0030302 | deoxynucleotide transport | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072096 | negative regulation of branch elongation involved in ureteric bud branching | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0070350 | regulation of white fat cell proliferation | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0044528 | regulation of mitochondrial mRNA stability | 2 | 7 | 6.11e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 2 | 27 | 6.11e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 5 | 84 | 6.11e-01 |
| GO:BP | GO:2000035 | regulation of stem cell division | 5 | 9 | 6.11e-01 |
| GO:BP | GO:0048390 | intermediate mesoderm morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000007 | negative regulation of metanephric comma-shaped body morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000006 | regulation of metanephric comma-shaped body morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 1 | 9 | 6.11e-01 |
| GO:BP | GO:0048392 | intermediate mesodermal cell differentiation | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000005 | negative regulation of metanephric S-shaped body morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000004 | regulation of metanephric S-shaped body morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0035553 | oxidative single-stranded RNA demethylation | 1 | 3 | 6.11e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 9 | 23 | 6.11e-01 |
| GO:BP | GO:1990966 | ATP generation from poly-ADP-D-ribose | 1 | 4 | 6.11e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 5 | 10 | 6.11e-01 |
| GO:BP | GO:0006254 | CTP catabolic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1904862 | inhibitory synapse assembly | 6 | 13 | 6.11e-01 |
| GO:BP | GO:0071403 | cellular response to high density lipoprotein particle stimulus | 1 | 3 | 6.11e-01 |
| GO:BP | GO:0061149 | BMP signaling pathway involved in ureter morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0055018 | regulation of cardiac muscle fiber development | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0055020 | positive regulation of cardiac muscle fiber development | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0120077 | angiogenic sprout fusion | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0072537 | fibroblast activation | 1 | 11 | 6.11e-01 |
| GO:BP | GO:0019290 | siderophore biosynthetic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0120078 | cell adhesion involved in sprouting angiogenesis | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 1 | 13 | 6.11e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 3 | 56 | 6.11e-01 |
| GO:BP | GO:0046052 | UTP catabolic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0061150 | renal system segmentation | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0061155 | pulmonary artery endothelial tube morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1901964 | positive regulation of cell proliferation involved in outflow tract morphogenesis | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0051541 | elastin metabolic process | 2 | 5 | 6.11e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 60 | 123 | 6.11e-01 |
| GO:BP | GO:0071893 | BMP signaling pathway involved in nephric duct formation | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 1 | 33 | 6.11e-01 |
| GO:BP | GO:0048391 | intermediate mesoderm formation | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0016139 | glycoside catabolic process | 2 | 8 | 6.11e-01 |
| GO:BP | GO:0008626 | granzyme-mediated apoptotic signaling pathway | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 4 | 68 | 6.11e-01 |
| GO:BP | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0036017 | response to erythropoietin | 4 | 4 | 6.11e-01 |
| GO:BP | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis | 2 | 6 | 6.11e-01 |
| GO:BP | GO:0035846 | oviduct epithelium development | 1 | 2 | 6.12e-01 |
| GO:BP | GO:0044321 | response to leptin | 2 | 19 | 6.12e-01 |
| GO:BP | GO:0045819 | positive regulation of glycogen catabolic process | 1 | 1 | 6.12e-01 |
| GO:BP | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation | 1 | 3 | 6.12e-01 |
| GO:BP | GO:0097377 | spinal cord interneuron axon guidance | 1 | 1 | 6.12e-01 |
| GO:BP | GO:0097477 | lateral motor column neuron migration | 1 | 3 | 6.12e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 12 | 35 | 6.12e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 2 | 126 | 6.12e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 6.12e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 7 | 115 | 6.12e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 4 | 5 | 6.12e-01 |
| GO:BP | GO:1904746 | negative regulation of apoptotic process involved in development | 1 | 3 | 6.12e-01 |
| GO:BP | GO:0046083 | adenine metabolic process | 1 | 3 | 6.12e-01 |
| GO:BP | GO:1902031 | regulation of NADP metabolic process | 1 | 8 | 6.12e-01 |
| GO:BP | GO:0032596 | protein transport into membrane raft | 3 | 4 | 6.12e-01 |
| GO:BP | GO:0045046 | protein import into peroxisome membrane | 2 | 6 | 6.12e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 2 | 8 | 6.12e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 6 | 29 | 6.12e-01 |
| GO:BP | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly | 1 | 5 | 6.12e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 1 | 89 | 6.12e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 2 | 6 | 6.12e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 13 | 23 | 6.12e-01 |
| GO:BP | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis | 1 | 3 | 6.12e-01 |
| GO:BP | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 3 | 4 | 6.12e-01 |
| GO:BP | GO:1900235 | negative regulation of Kit signaling pathway | 1 | 2 | 6.12e-01 |
| GO:BP | GO:1900234 | regulation of Kit signaling pathway | 1 | 2 | 6.12e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 1 | 37 | 6.12e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 1 | 11 | 6.12e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 6.12e-01 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 2 | 7 | 6.13e-01 |
| GO:BP | GO:0019236 | response to pheromone | 1 | 1 | 6.13e-01 |
| GO:BP | GO:0006412 | translation | 5 | 646 | 6.13e-01 |
| GO:BP | GO:0010265 | SCF complex assembly | 1 | 2 | 6.13e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 5 | 112 | 6.13e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 8 | 993 | 6.13e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 1 | 24 | 6.13e-01 |
| GO:BP | GO:0043984 | histone H4-K16 acetylation | 7 | 14 | 6.13e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 59 | 796 | 6.13e-01 |
| GO:BP | GO:0097252 | oligodendrocyte apoptotic process | 1 | 4 | 6.13e-01 |
| GO:BP | GO:2001198 | regulation of dendritic cell differentiation | 1 | 8 | 6.13e-01 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 5 | 17 | 6.13e-01 |
| GO:BP | GO:0034696 | response to prostaglandin F | 1 | 2 | 6.13e-01 |
| GO:BP | GO:0098814 | spontaneous synaptic transmission | 3 | 10 | 6.13e-01 |
| GO:BP | GO:0030540 | female genitalia development | 1 | 14 | 6.13e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 1 | 10 | 6.13e-01 |
| GO:BP | GO:0006621 | protein retention in ER lumen | 3 | 8 | 6.13e-01 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 4 | 11 | 6.14e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 3 | 59 | 6.14e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 3 | 60 | 6.14e-01 |
| GO:BP | GO:0007276 | gamete generation | 1 | 461 | 6.14e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 13 | 41 | 6.15e-01 |
| GO:BP | GO:0007141 | male meiosis I | 9 | 15 | 6.15e-01 |
| GO:BP | GO:0042357 | thiamine diphosphate metabolic process | 1 | 2 | 6.15e-01 |
| GO:BP | GO:0002939 | tRNA N1-guanine methylation | 1 | 3 | 6.15e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 32 | 6.15e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 4 | 11 | 6.15e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 1 | 108 | 6.15e-01 |
| GO:BP | GO:1902317 | nuclear DNA replication termination | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 7 | 81 | 6.15e-01 |
| GO:BP | GO:0097198 | histone H3-K36 trimethylation | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0018095 | protein polyglutamylation | 1 | 9 | 6.15e-01 |
| GO:BP | GO:1902979 | mitotic DNA replication termination | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0044565 | dendritic cell proliferation | 2 | 3 | 6.15e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 7 | 129 | 6.15e-01 |
| GO:BP | GO:1902294 | cell cycle DNA replication termination | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0070977 | bone maturation | 1 | 19 | 6.15e-01 |
| GO:BP | GO:0006274 | DNA replication termination | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 3 | 8 | 6.15e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 3 | 8 | 6.15e-01 |
| GO:BP | GO:0042592 | homeostatic process | 10 | 1159 | 6.15e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 6.16e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 6.16e-01 |
| GO:BP | GO:1901834 | regulation of deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 1 | 6.16e-01 |
| GO:BP | GO:1904246 | negative regulation of polynucleotide adenylyltransferase activity | 1 | 1 | 6.16e-01 |
| GO:BP | GO:1901835 | positive regulation of deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 23 | 44 | 6.16e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 2 | 6 | 6.16e-01 |
| GO:BP | GO:0140235 | RNA polyadenylation at postsynapse | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 1 | 7 | 6.16e-01 |
| GO:BP | GO:0031442 | positive regulation of mRNA 3’-end processing | 5 | 10 | 6.16e-01 |
| GO:BP | GO:2000738 | positive regulation of stem cell differentiation | 5 | 17 | 6.16e-01 |
| GO:BP | GO:0060456 | positive regulation of digestive system process | 1 | 8 | 6.16e-01 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 2 | 4 | 6.16e-01 |
| GO:BP | GO:2000626 | negative regulation of miRNA catabolic process | 1 | 1 | 6.16e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 2 | 6 | 6.16e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 2 | 6 | 6.16e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 8 | 100 | 6.16e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 2 | 6 | 6.16e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 2 | 6 | 6.16e-01 |
| GO:BP | GO:0030488 | tRNA methylation | 20 | 42 | 6.16e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 2 | 16 | 6.16e-01 |
| GO:BP | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay | 1 | 3 | 6.16e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 2 | 1974 | 6.17e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 6 | 8 | 6.17e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 6.17e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 24 | 61 | 6.17e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 6.17e-01 |
| GO:BP | GO:0048278 | vesicle docking | 23 | 57 | 6.17e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 3 | 58 | 6.17e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 6.17e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 2 | 129 | 6.17e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 12 | 48 | 6.17e-01 |
| GO:BP | GO:0097264 | self proteolysis | 2 | 7 | 6.17e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 1 | 53 | 6.17e-01 |
| GO:BP | GO:2000644 | regulation of receptor catabolic process | 3 | 9 | 6.17e-01 |
| GO:BP | GO:1990504 | dense core granule exocytosis | 2 | 7 | 6.17e-01 |
| GO:BP | GO:0003100 | regulation of systemic arterial blood pressure by endothelin | 1 | 5 | 6.18e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 9 | 30 | 6.18e-01 |
| GO:BP | GO:0021535 | cell migration in hindbrain | 2 | 12 | 6.18e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 3 | 32 | 6.18e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 2 | 25 | 6.18e-01 |
| GO:BP | GO:0043048 | dolichyl monophosphate biosynthetic process | 1 | 1 | 6.18e-01 |
| GO:BP | GO:1990784 | response to dsDNA | 1 | 2 | 6.18e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 1 | 30 | 6.18e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 2 | 25 | 6.18e-01 |
| GO:BP | GO:1990786 | cellular response to dsDNA | 1 | 2 | 6.18e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 2 | 28 | 6.18e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 3 | 17 | 6.18e-01 |
| GO:BP | GO:0034350 | regulation of glial cell apoptotic process | 2 | 10 | 6.18e-01 |
| GO:BP | GO:0036071 | N-glycan fucosylation | 2 | 3 | 6.19e-01 |
| GO:BP | GO:0008406 | gonad development | 4 | 158 | 6.19e-01 |
| GO:BP | GO:0021501 | prechordal plate formation | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0021547 | midbrain-hindbrain boundary initiation | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0009207 | purine ribonucleoside triphosphate catabolic process | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0007518 | myoblast fate determination | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0098917 | retrograde trans-synaptic signaling | 1 | 7 | 6.19e-01 |
| GO:BP | GO:0006193 | ITP catabolic process | 1 | 1 | 6.19e-01 |
| GO:BP | GO:0006659 | phosphatidylserine biosynthetic process | 2 | 4 | 6.19e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 2 | 15 | 6.19e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 32 | 67 | 6.19e-01 |
| GO:BP | GO:0061198 | fungiform papilla formation | 1 | 3 | 6.19e-01 |
| GO:BP | GO:1903764 | regulation of potassium ion export across plasma membrane | 2 | 5 | 6.19e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 1 | 118 | 6.19e-01 |
| GO:BP | GO:0015822 | ornithine transport | 1 | 5 | 6.20e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 1 | 63 | 6.20e-01 |
| GO:BP | GO:0006685 | sphingomyelin catabolic process | 1 | 6 | 6.20e-01 |
| GO:BP | GO:0003356 | regulation of cilium beat frequency | 1 | 10 | 6.20e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 10 | 206 | 6.20e-01 |
| GO:BP | GO:0021508 | floor plate formation | 1 | 3 | 6.20e-01 |
| GO:BP | GO:0045063 | T-helper 1 cell differentiation | 7 | 14 | 6.21e-01 |
| GO:BP | GO:0072678 | T cell migration | 3 | 38 | 6.21e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0090152 | establishment of protein localization to mitochondrial membrane involved in mitochondrial fission | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0003169 | coronary vein morphogenesis | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 10 | 22 | 6.21e-01 |
| GO:BP | GO:0006014 | D-ribose metabolic process | 2 | 3 | 6.21e-01 |
| GO:BP | GO:0010904 | regulation of UDP-glucose catabolic process | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0051340 | regulation of ligase activity | 6 | 11 | 6.21e-01 |
| GO:BP | GO:0010905 | negative regulation of UDP-glucose catabolic process | 1 | 1 | 6.21e-01 |
| GO:BP | GO:1901304 | regulation of spermidine biosynthetic process | 1 | 1 | 6.21e-01 |
| GO:BP | GO:1990859 | cellular response to endothelin | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0016198 | axon choice point recognition | 2 | 5 | 6.21e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 2 | 12 | 6.21e-01 |
| GO:BP | GO:0003252 | negative regulation of cell proliferation involved in heart valve morphogenesis | 1 | 2 | 6.21e-01 |
| GO:BP | GO:1903849 | positive regulation of aorta morphogenesis | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0044027 | negative regulation of gene expression via CpG island methylation | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 1 | 52 | 6.21e-01 |
| GO:BP | GO:0071896 | protein localization to adherens junction | 2 | 6 | 6.21e-01 |
| GO:BP | GO:1903367 | positive regulation of fear response | 2 | 4 | 6.21e-01 |
| GO:BP | GO:0070317 | negative regulation of G0 to G1 transition | 1 | 5 | 6.21e-01 |
| GO:BP | GO:0051729 | germline cell cycle switching, mitotic to meiotic cell cycle | 2 | 2 | 6.21e-01 |
| GO:BP | GO:0048870 | cell motility | 30 | 1182 | 6.21e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 6.21e-01 |
| GO:BP | GO:1904692 | positive regulation of type B pancreatic cell proliferation | 1 | 2 | 6.21e-01 |
| GO:BP | GO:1903847 | regulation of aorta morphogenesis | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0034334 | adherens junction maintenance | 4 | 6 | 6.21e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 2 | 104 | 6.21e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 2 | 20 | 6.21e-01 |
| GO:BP | GO:1901307 | positive regulation of spermidine biosynthetic process | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0051728 | cell cycle switching, mitotic to meiotic cell cycle | 2 | 2 | 6.21e-01 |
| GO:BP | GO:0060184 | cell cycle switching | 2 | 2 | 6.21e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 6 | 133 | 6.21e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 2 | 144 | 6.21e-01 |
| GO:BP | GO:1904227 | negative regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 6.21e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 3 | 6.21e-01 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 2 | 6 | 6.21e-01 |
| GO:BP | GO:1904880 | response to hydrogen sulfide | 2 | 3 | 6.21e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 2 | 6 | 6.21e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 25 | 120 | 6.21e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 3 | 6.21e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 6 | 23 | 6.21e-01 |
| GO:BP | GO:0044381 | glucose import in response to insulin stimulus | 2 | 4 | 6.21e-01 |
| GO:BP | GO:0015734 | taurine transport | 2 | 6 | 6.21e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 6.21e-01 |
| GO:BP | GO:0061847 | response to cholecystokinin | 1 | 1 | 6.21e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 3 | 113 | 6.21e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 3 | 34 | 6.21e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 5 | 843 | 6.21e-01 |
| GO:BP | GO:1901485 | positive regulation of transcription factor catabolic process | 1 | 3 | 6.21e-01 |
| GO:BP | GO:0099074 | mitochondrion to lysosome transport | 2 | 3 | 6.21e-01 |
| GO:BP | GO:1901483 | regulation of transcription factor catabolic process | 1 | 3 | 6.21e-01 |
| GO:BP | GO:0072137 | condensed mesenchymal cell proliferation | 1 | 2 | 6.21e-01 |
| GO:BP | GO:1904425 | negative regulation of GTP binding | 2 | 3 | 6.21e-01 |
| GO:BP | GO:0045955 | negative regulation of calcium ion-dependent exocytosis | 3 | 6 | 6.21e-01 |
| GO:BP | GO:0071879 | positive regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0099075 | mitochondrion-derived vesicle mediated transport | 2 | 3 | 6.21e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 12 | 115 | 6.21e-01 |
| GO:BP | GO:0021793 | chemorepulsion of branchiomotor axon | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0002051 | osteoblast fate commitment | 2 | 2 | 6.21e-01 |
| GO:BP | GO:1904226 | regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0097500 | receptor localization to non-motile cilium | 3 | 4 | 6.21e-01 |
| GO:BP | GO:0003062 | regulation of heart rate by chemical signal | 1 | 6 | 6.21e-01 |
| GO:BP | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation | 1 | 4 | 6.21e-01 |
| GO:BP | GO:2000064 | regulation of cortisol biosynthetic process | 1 | 7 | 6.21e-01 |
| GO:BP | GO:0003065 | positive regulation of heart rate by epinephrine | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 6 | 37 | 6.21e-01 |
| GO:BP | GO:0048806 | genitalia development | 2 | 28 | 6.21e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 3 | 8 | 6.21e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 6 | 131 | 6.21e-01 |
| GO:BP | GO:1900022 | regulation of D-erythro-sphingosine kinase activity | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 26 | 54 | 6.21e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 1 | 111 | 6.21e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 4 | 10 | 6.21e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 4 | 10 | 6.21e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 6.21e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 13 | 37 | 6.21e-01 |
| GO:BP | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0046031 | ADP metabolic process | 5 | 7 | 6.21e-01 |
| GO:BP | GO:0002883 | regulation of hypersensitivity | 2 | 5 | 6.21e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 1 | 122 | 6.22e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 17 | 41 | 6.22e-01 |
| GO:BP | GO:1905313 | transforming growth factor beta receptor signaling pathway involved in heart development | 1 | 1 | 6.22e-01 |
| GO:BP | GO:0014006 | regulation of microglia differentiation | 1 | 1 | 6.22e-01 |
| GO:BP | GO:0014008 | positive regulation of microglia differentiation | 1 | 1 | 6.22e-01 |
| GO:BP | GO:0045141 | meiotic telomere clustering | 2 | 5 | 6.22e-01 |
| GO:BP | GO:0006734 | NADH metabolic process | 3 | 12 | 6.22e-01 |
| GO:BP | GO:0060318 | definitive erythrocyte differentiation | 1 | 4 | 6.22e-01 |
| GO:BP | GO:0009304 | tRNA transcription | 1 | 8 | 6.22e-01 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 1 | 144 | 6.22e-01 |
| GO:BP | GO:0021627 | olfactory nerve morphogenesis | 2 | 3 | 6.22e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 6.22e-01 |
| GO:BP | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | 3 | 9 | 6.23e-01 |
| GO:BP | GO:0120179 | adherens junction disassembly | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0002537 | nitric oxide production involved in inflammatory response | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0006982 | response to lipid hydroperoxide | 1 | 2 | 6.23e-01 |
| GO:BP | GO:0160025 | sensory perception of itch | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0160023 | sneeze reflex | 1 | 1 | 6.23e-01 |
| GO:BP | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly | 1 | 8 | 6.23e-01 |
| GO:BP | GO:0043461 | proton-transporting ATP synthase complex assembly | 1 | 8 | 6.23e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 5 | 64 | 6.23e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 25 | 52 | 6.24e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 3 | 21 | 6.24e-01 |
| GO:BP | GO:1905609 | positive regulation of smooth muscle cell-matrix adhesion | 1 | 1 | 6.24e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 7 | 97 | 6.24e-01 |
| GO:BP | GO:0033366 | protein localization to secretory granule | 1 | 1 | 6.24e-01 |
| GO:BP | GO:0043473 | pigmentation | 12 | 92 | 6.24e-01 |
| GO:BP | GO:1901098 | positive regulation of autophagosome maturation | 4 | 6 | 6.24e-01 |
| GO:BP | GO:0032713 | negative regulation of interleukin-4 production | 4 | 5 | 6.24e-01 |
| GO:BP | GO:0002519 | natural killer cell tolerance induction | 1 | 1 | 6.24e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 6.24e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 10 | 18 | 6.24e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 10 | 6.24e-01 |
| GO:BP | GO:0097155 | fasciculation of sensory neuron axon | 2 | 3 | 6.24e-01 |
| GO:BP | GO:0042921 | glucocorticoid receptor signaling pathway | 1 | 13 | 6.24e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 6.24e-01 |
| GO:BP | GO:0070158 | mitochondrial seryl-tRNA aminoacylation | 1 | 1 | 6.25e-01 |
| GO:BP | GO:0017014 | protein nitrosylation | 1 | 11 | 6.25e-01 |
| GO:BP | GO:0018119 | peptidyl-cysteine S-nitrosylation | 1 | 11 | 6.25e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 6.25e-01 |
| GO:BP | GO:0060486 | club cell differentiation | 2 | 3 | 6.25e-01 |
| GO:BP | GO:0006826 | iron ion transport | 4 | 40 | 6.25e-01 |
| GO:BP | GO:0006769 | nicotinamide metabolic process | 1 | 1 | 6.25e-01 |
| GO:BP | GO:0061034 | olfactory bulb mitral cell layer development | 1 | 2 | 6.25e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 2 | 100 | 6.26e-01 |
| GO:BP | GO:0021722 | superior olivary nucleus maturation | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0021586 | pons maturation | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0021718 | superior olivary nucleus development | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | 1 | 8 | 6.26e-01 |
| GO:BP | GO:0048319 | axial mesoderm morphogenesis | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 29 | 114 | 6.26e-01 |
| GO:BP | GO:0033183 | negative regulation of histone ubiquitination | 1 | 4 | 6.26e-01 |
| GO:BP | GO:0008585 | female gonad development | 20 | 77 | 6.26e-01 |
| GO:BP | GO:0060264 | regulation of respiratory burst involved in inflammatory response | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 3 | 5 | 6.26e-01 |
| GO:BP | GO:1901314 | regulation of histone H2A K63-linked ubiquitination | 1 | 4 | 6.26e-01 |
| GO:BP | GO:1901315 | negative regulation of histone H2A K63-linked ubiquitination | 1 | 4 | 6.26e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 6 | 13 | 6.26e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 5 | 10 | 6.26e-01 |
| GO:BP | GO:0038180 | nerve growth factor signaling pathway | 3 | 10 | 6.26e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 29 | 59 | 6.26e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0061107 | seminal vesicle development | 1 | 2 | 6.26e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 13 | 24 | 6.26e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 6.26e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 6.27e-01 |
| GO:BP | GO:1902804 | negative regulation of synaptic vesicle transport | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 7 | 131 | 6.27e-01 |
| GO:BP | GO:1901609 | negative regulation of vesicle transport along microtubule | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1903937 | response to acrylamide | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0031649 | heat generation | 1 | 11 | 6.27e-01 |
| GO:BP | GO:1903743 | negative regulation of anterograde synaptic vesicle transport | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1902684 | negative regulation of receptor localization to synapse | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 5 | 10 | 6.27e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 1 | 21 | 6.27e-01 |
| GO:BP | GO:0061061 | muscle structure development | 4 | 546 | 6.27e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 25 | 49 | 6.27e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 3 | 41 | 6.27e-01 |
| GO:BP | GO:0061085 | regulation of histone H3-K27 methylation | 1 | 4 | 6.27e-01 |
| GO:BP | GO:1902416 | positive regulation of mRNA binding | 1 | 8 | 6.27e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 17 | 4245 | 6.27e-01 |
| GO:BP | GO:2000340 | positive regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 3 | 6.28e-01 |
| GO:BP | GO:0038026 | reelin-mediated signaling pathway | 5 | 9 | 6.28e-01 |
| GO:BP | GO:0042428 | serotonin metabolic process | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 4 | 223 | 6.28e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 3 | 56 | 6.28e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 2 | 8 | 6.28e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 4 | 233 | 6.28e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 1 | 51 | 6.28e-01 |
| GO:BP | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity | 1 | 6 | 6.28e-01 |
| GO:BP | GO:1903394 | protein localization to kinetochore involved in kinetochore assembly | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0017126 | nucleologenesis | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 1 | 31 | 6.28e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 6.28e-01 |
| GO:BP | GO:0019368 | fatty acid elongation, unsaturated fatty acid | 3 | 6 | 6.28e-01 |
| GO:BP | GO:0060160 | negative regulation of dopamine receptor signaling pathway | 2 | 3 | 6.28e-01 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 6 | 17 | 6.28e-01 |
| GO:BP | GO:0046951 | ketone body biosynthetic process | 2 | 5 | 6.28e-01 |
| GO:BP | GO:0009972 | cytidine deamination | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 4 | 114 | 6.28e-01 |
| GO:BP | GO:0035963 | cellular response to interleukin-13 | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 6.28e-01 |
| GO:BP | GO:0016554 | cytidine to uridine editing | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0045344 | negative regulation of MHC class I biosynthetic process | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 1 | 36 | 6.28e-01 |
| GO:BP | GO:0035246 | peptidyl-arginine N-methylation | 3 | 4 | 6.28e-01 |
| GO:BP | GO:0006216 | cytidine catabolic process | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 3 | 54 | 6.28e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 3 | 62 | 6.28e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 6.28e-01 |
| GO:BP | GO:0046087 | cytidine metabolic process | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0019367 | fatty acid elongation, saturated fatty acid | 3 | 6 | 6.28e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 4 | 4 | 6.28e-01 |
| GO:BP | GO:0071348 | cellular response to interleukin-11 | 1 | 1 | 6.28e-01 |
| GO:BP | GO:1903741 | negative regulation of phosphatidate phosphatase activity | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid | 3 | 6 | 6.28e-01 |
| GO:BP | GO:1903730 | regulation of phosphatidate phosphatase activity | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0034625 | fatty acid elongation, monounsaturated fatty acid | 3 | 6 | 6.28e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 4 | 161 | 6.28e-01 |
| GO:BP | GO:0046851 | negative regulation of bone remodeling | 2 | 9 | 6.28e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 2 | 31 | 6.28e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 2 | 106 | 6.28e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 2 | 2 | 6.28e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 2 | 2 | 6.28e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 2 | 2 | 6.28e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 6.28e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 3 | 23 | 6.28e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 3 | 13 | 6.28e-01 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 1 | 46 | 6.28e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 1 | 46 | 6.28e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 2 | 30 | 6.28e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 2 | 30 | 6.28e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 9 | 17 | 6.28e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 8 | 71 | 6.28e-01 |
| GO:BP | GO:0051026 | chiasma assembly | 1 | 3 | 6.28e-01 |
| GO:BP | GO:1900126 | negative regulation of hyaluronan biosynthetic process | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0042063 | gliogenesis | 2 | 235 | 6.28e-01 |
| GO:BP | GO:0043980 | histone H2B-K12 acetylation | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0002175 | protein localization to paranode region of axon | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0072006 | nephron development | 5 | 117 | 6.28e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 4 | 22 | 6.28e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 3 | 10 | 6.28e-01 |
| GO:BP | GO:0006669 | sphinganine-1-phosphate biosynthetic process | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 1 | 29 | 6.28e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 8 | 26 | 6.28e-01 |
| GO:BP | GO:0015891 | siderophore transport | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0090117 | endosome to lysosome transport of low-density lipoprotein particle | 1 | 2 | 6.28e-01 |
| GO:BP | GO:2000802 | positive regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 6.28e-01 |
| GO:BP | GO:2000800 | regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 6.28e-01 |
| GO:BP | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis | 3 | 6 | 6.28e-01 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 1 | 4 | 6.28e-01 |
| GO:BP | GO:0032488 | Cdc42 protein signal transduction | 1 | 10 | 6.28e-01 |
| GO:BP | GO:0043977 | histone H2A-K5 acetylation | 1 | 1 | 6.28e-01 |
| GO:BP | GO:2000553 | positive regulation of T-helper 2 cell cytokine production | 1 | 3 | 6.28e-01 |
| GO:BP | GO:1903119 | protein localization to actin cytoskeleton | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0071107 | response to parathyroid hormone | 3 | 9 | 6.28e-01 |
| GO:BP | GO:0044379 | protein localization to actin cortical patch | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 5 | 10 | 6.28e-01 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 1 | 7 | 6.28e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 10 | 94 | 6.28e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 6.28e-01 |
| GO:BP | GO:1904370 | regulation of protein localization to actin cortical patch | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0140051 | positive regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 6.28e-01 |
| GO:BP | GO:1904372 | positive regulation of protein localization to actin cortical patch | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0140049 | regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 6.28e-01 |
| GO:BP | GO:1904515 | positive regulation of TORC2 signaling | 2 | 4 | 6.28e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 6.28e-01 |
| GO:BP | GO:1904373 | response to kainic acid | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0008065 | establishment of blood-nerve barrier | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0005997 | xylulose metabolic process | 1 | 1 | 6.28e-01 |
| GO:BP | GO:1990379 | lipid transport across blood-brain barrier | 2 | 6 | 6.28e-01 |
| GO:BP | GO:0070572 | positive regulation of neuron projection regeneration | 1 | 8 | 6.28e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 13 | 20 | 6.28e-01 |
| GO:BP | GO:0006563 | L-serine metabolic process | 2 | 10 | 6.28e-01 |
| GO:BP | GO:0016320 | endoplasmic reticulum membrane fusion | 1 | 2 | 6.28e-01 |
| GO:BP | GO:1905517 | macrophage migration | 2 | 34 | 6.28e-01 |
| GO:BP | GO:0072383 | plus-end-directed vesicle transport along microtubule | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0010636 | positive regulation of mitochondrial fusion | 1 | 2 | 6.28e-01 |
| GO:BP | GO:1902283 | negative regulation of primary amine oxidase activity | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 2 | 10 | 6.28e-01 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0033029 | regulation of neutrophil apoptotic process | 3 | 5 | 6.28e-01 |
| GO:BP | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | 3 | 6.28e-01 |
| GO:BP | GO:0010716 | negative regulation of extracellular matrix disassembly | 1 | 2 | 6.28e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 8 | 23 | 6.28e-01 |
| GO:BP | GO:0001971 | negative regulation of activation of membrane attack complex | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 4 | 14 | 6.29e-01 |
| GO:BP | GO:0021766 | hippocampus development | 3 | 62 | 6.29e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 4 | 23 | 6.29e-01 |
| GO:BP | GO:0002265 | astrocyte activation involved in immune response | 2 | 4 | 6.29e-01 |
| GO:BP | GO:1990120 | messenger ribonucleoprotein complex assembly | 1 | 3 | 6.30e-01 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 2 | 8 | 6.30e-01 |
| GO:BP | GO:0006880 | intracellular sequestering of iron ion | 1 | 3 | 6.30e-01 |
| GO:BP | GO:0014856 | skeletal muscle cell proliferation | 4 | 14 | 6.30e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 2 | 54 | 6.30e-01 |
| GO:BP | GO:0031623 | receptor internalization | 1 | 95 | 6.30e-01 |
| GO:BP | GO:1905301 | regulation of macropinocytosis | 2 | 3 | 6.30e-01 |
| GO:BP | GO:0055118 | negative regulation of cardiac muscle contraction | 1 | 4 | 6.30e-01 |
| GO:BP | GO:0009631 | cold acclimation | 2 | 2 | 6.30e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 12 | 25 | 6.30e-01 |
| GO:BP | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 1 | 6 | 6.30e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 3 | 14 | 6.30e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 3 | 14 | 6.30e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 4 | 86 | 6.30e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 5 | 21 | 6.30e-01 |
| GO:BP | GO:0061549 | sympathetic ganglion development | 5 | 6 | 6.30e-01 |
| GO:BP | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation | 1 | 3 | 6.30e-01 |
| GO:BP | GO:0019482 | beta-alanine metabolic process | 1 | 2 | 6.30e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 15 | 376 | 6.30e-01 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 1 | 56 | 6.30e-01 |
| GO:BP | GO:2000617 | positive regulation of histone H3-K9 acetylation | 1 | 6 | 6.30e-01 |
| GO:BP | GO:0045186 | zonula adherens assembly | 1 | 1 | 6.30e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 6 | 14 | 6.31e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 5 | 666 | 6.31e-01 |
| GO:BP | GO:0042045 | epithelial fluid transport | 2 | 7 | 6.31e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 6.31e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 8 | 19 | 6.31e-01 |
| GO:BP | GO:1904891 | positive regulation of excitatory synapse assembly | 1 | 2 | 6.31e-01 |
| GO:BP | GO:0045002 | double-strand break repair via single-strand annealing | 3 | 7 | 6.31e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 9 | 38 | 6.31e-01 |
| GO:BP | GO:0043686 | co-translational protein modification | 2 | 3 | 6.31e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 2 | 22 | 6.31e-01 |
| GO:BP | GO:1904184 | positive regulation of pyruvate dehydrogenase activity | 1 | 3 | 6.31e-01 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 1 | 5 | 6.31e-01 |
| GO:BP | GO:0019102 | male somatic sex determination | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 3 | 80 | 6.31e-01 |
| GO:BP | GO:0043973 | histone H3-K4 acetylation | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 2 | 48 | 6.31e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 5 | 9 | 6.31e-01 |
| GO:BP | GO:0060520 | activation of prostate induction by androgen receptor signaling pathway | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 2 | 22 | 6.31e-01 |
| GO:BP | GO:0060514 | prostate induction | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0045041 | protein import into mitochondrial intermembrane space | 2 | 3 | 6.31e-01 |
| GO:BP | GO:1902462 | positive regulation of mesenchymal stem cell proliferation | 3 | 4 | 6.31e-01 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 1 | 7 | 6.31e-01 |
| GO:BP | GO:1902460 | regulation of mesenchymal stem cell proliferation | 3 | 4 | 6.31e-01 |
| GO:BP | GO:0045843 | negative regulation of striated muscle tissue development | 4 | 5 | 6.31e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 1 | 6 | 6.31e-01 |
| GO:BP | GO:0060515 | prostate field specification | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0061890 | positive regulation of astrocyte activation | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0070102 | interleukin-6-mediated signaling pathway | 8 | 14 | 6.31e-01 |
| GO:BP | GO:0140706 | protein-containing complex localization to centriolar satellite | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0003011 | involuntary skeletal muscle contraction | 2 | 3 | 6.32e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 18 | 301 | 6.32e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 6.32e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 8 | 35 | 6.32e-01 |
| GO:BP | GO:0002158 | osteoclast proliferation | 3 | 7 | 6.32e-01 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 3 | 8 | 6.32e-01 |
| GO:BP | GO:0045793 | positive regulation of cell size | 2 | 11 | 6.32e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 8 | 98 | 6.32e-01 |
| GO:BP | GO:0016561 | protein import into peroxisome matrix, translocation | 2 | 3 | 6.32e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 3 | 114 | 6.33e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 5 | 83 | 6.33e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 7 | 190 | 6.33e-01 |
| GO:BP | GO:1904205 | negative regulation of skeletal muscle hypertrophy | 1 | 1 | 6.33e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 78 | 6.33e-01 |
| GO:BP | GO:0003251 | positive regulation of cell proliferation involved in heart valve morphogenesis | 1 | 1 | 6.34e-01 |
| GO:BP | GO:2000525 | positive regulation of T cell costimulation | 1 | 2 | 6.34e-01 |
| GO:BP | GO:2000634 | regulation of primary miRNA processing | 2 | 2 | 6.34e-01 |
| GO:BP | GO:1905305 | negative regulation of cardiac myofibril assembly | 1 | 1 | 6.34e-01 |
| GO:BP | GO:1905304 | regulation of cardiac myofibril assembly | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0061040 | female gonad morphogenesis | 1 | 1 | 6.34e-01 |
| GO:BP | GO:2000635 | negative regulation of primary miRNA processing | 2 | 2 | 6.34e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 3 | 12 | 6.34e-01 |
| GO:BP | GO:1905135 | biotin import across plasma membrane | 1 | 2 | 6.34e-01 |
| GO:BP | GO:0001579 | medium-chain fatty acid transport | 1 | 2 | 6.34e-01 |
| GO:BP | GO:1903790 | guanine nucleotide transmembrane transport | 1 | 5 | 6.34e-01 |
| GO:BP | GO:0140754 | reorganization of cellular membranes to establish viral sites of replication | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0070729 | cyclic nucleotide transport | 1 | 6 | 6.34e-01 |
| GO:BP | GO:0006182 | cGMP biosynthetic process | 1 | 6 | 6.34e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 1 | 10 | 6.34e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 4 | 78 | 6.34e-01 |
| GO:BP | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis | 3 | 5 | 6.35e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 2 | 104 | 6.35e-01 |
| GO:BP | GO:0019065 | receptor-mediated endocytosis of virus by host cell | 3 | 9 | 6.35e-01 |
| GO:BP | GO:0098751 | bone cell development | 2 | 26 | 6.35e-01 |
| GO:BP | GO:0002432 | granuloma formation | 1 | 1 | 6.35e-01 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 1 | 3 | 6.35e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 2 | 9 | 6.35e-01 |
| GO:BP | GO:0043278 | response to morphine | 2 | 16 | 6.35e-01 |
| GO:BP | GO:0110024 | positive regulation of cardiac muscle myoblast proliferation | 2 | 3 | 6.35e-01 |
| GO:BP | GO:0031144 | proteasome localization | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0045684 | positive regulation of epidermis development | 4 | 20 | 6.35e-01 |
| GO:BP | GO:0110021 | cardiac muscle myoblast proliferation | 2 | 3 | 6.35e-01 |
| GO:BP | GO:0110022 | regulation of cardiac muscle myoblast proliferation | 2 | 3 | 6.35e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 11 | 22 | 6.35e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 7 | 198 | 6.35e-01 |
| GO:BP | GO:0070911 | global genome nucleotide-excision repair | 1 | 2 | 6.35e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 3 | 26 | 6.35e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 4 | 26 | 6.35e-01 |
| GO:BP | GO:0006479 | protein methylation | 3 | 160 | 6.35e-01 |
| GO:BP | GO:0160007 | glutathione import into mitochondrion | 1 | 1 | 6.35e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 3 | 48 | 6.35e-01 |
| GO:BP | GO:2000723 | negative regulation of cardiac vascular smooth muscle cell differentiation | 1 | 1 | 6.35e-01 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 6.35e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 20 | 42 | 6.35e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 4 | 71 | 6.35e-01 |
| GO:BP | GO:0071318 | cellular response to ATP | 1 | 12 | 6.35e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 6.35e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 2 | 16 | 6.35e-01 |
| GO:BP | GO:0008213 | protein alkylation | 3 | 160 | 6.35e-01 |
| GO:BP | GO:0035094 | response to nicotine | 1 | 28 | 6.36e-01 |
| GO:BP | GO:1903334 | positive regulation of protein folding | 1 | 2 | 6.36e-01 |
| GO:BP | GO:0048210 | Golgi vesicle fusion to target membrane | 2 | 4 | 6.36e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 36 | 234 | 6.36e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 9 | 37 | 6.36e-01 |
| GO:BP | GO:0032930 | positive regulation of superoxide anion generation | 2 | 10 | 6.36e-01 |
| GO:BP | GO:0061140 | lung secretory cell differentiation | 1 | 5 | 6.36e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 3 | 64 | 6.36e-01 |
| GO:BP | GO:0051643 | endoplasmic reticulum localization | 1 | 8 | 6.36e-01 |
| GO:BP | GO:1903755 | positive regulation of SUMO transferase activity | 1 | 1 | 6.36e-01 |
| GO:BP | GO:1903182 | regulation of SUMO transferase activity | 1 | 1 | 6.36e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 8 | 31 | 6.36e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 2 | 12 | 6.36e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 8 | 18 | 6.36e-01 |
| GO:BP | GO:0016095 | polyprenol catabolic process | 1 | 2 | 6.36e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 1 | 4 | 6.36e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 1 | 6 | 6.36e-01 |
| GO:BP | GO:0031958 | corticosteroid receptor signaling pathway | 1 | 14 | 6.36e-01 |
| GO:BP | GO:0043113 | receptor clustering | 4 | 45 | 6.36e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 15 | 27 | 6.36e-01 |
| GO:BP | GO:1901080 | regulation of relaxation of smooth muscle | 1 | 1 | 6.36e-01 |
| GO:BP | GO:2000360 | negative regulation of binding of sperm to zona pellucida | 2 | 2 | 6.37e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 6 | 126 | 6.37e-01 |
| GO:BP | GO:0038167 | epidermal growth factor receptor signaling pathway via positive regulation of NF-kappaB transcription factor activity | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0038168 | epidermal growth factor receptor signaling pathway via I-kappaB kinase/NF-kappaB cascade | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 6.37e-01 |
| GO:BP | GO:0071377 | cellular response to glucagon stimulus | 1 | 4 | 6.37e-01 |
| GO:BP | GO:0000451 | rRNA 2’-O-methylation | 2 | 3 | 6.37e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 22 | 150 | 6.37e-01 |
| GO:BP | GO:0072236 | metanephric loop of Henle development | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0048799 | animal organ maturation | 1 | 20 | 6.37e-01 |
| GO:BP | GO:0048599 | oocyte development | 2 | 38 | 6.37e-01 |
| GO:BP | GO:0031393 | negative regulation of prostaglandin biosynthetic process | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 7 | 29 | 6.37e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 7 | 29 | 6.37e-01 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 5 | 31 | 6.37e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 14 | 179 | 6.37e-01 |
| GO:BP | GO:1902524 | positive regulation of protein K48-linked ubiquitination | 2 | 2 | 6.37e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 6.37e-01 |
| GO:BP | GO:0006742 | NADP catabolic process | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0019677 | NAD catabolic process | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 1 | 31 | 6.37e-01 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 6.37e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 3 | 52 | 6.38e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 3 | 106 | 6.38e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 1 | 27 | 6.38e-01 |
| GO:BP | GO:1904055 | negative regulation of cholangiocyte proliferation | 1 | 1 | 6.38e-01 |
| GO:BP | GO:0002940 | tRNA N2-guanine methylation | 1 | 4 | 6.39e-01 |
| GO:BP | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway | 3 | 8 | 6.39e-01 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 1 | 10 | 6.39e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 5 | 538 | 6.39e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 2 | 12 | 6.39e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 11 | 99 | 6.39e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 2 | 32 | 6.39e-01 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 1 | 10 | 6.39e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 11 | 99 | 6.39e-01 |
| GO:BP | GO:0070926 | regulation of ATP:ADP antiporter activity | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 2 | 56 | 6.39e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 4 | 7 | 6.39e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 2 | 56 | 6.39e-01 |
| GO:BP | GO:0007252 | I-kappaB phosphorylation | 3 | 14 | 6.39e-01 |
| GO:BP | GO:1905908 | positive regulation of amyloid fibril formation | 3 | 5 | 6.39e-01 |
| GO:BP | GO:1903893 | positive regulation of ATF6-mediated unfolded protein response | 1 | 1 | 6.39e-01 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 3 | 10 | 6.40e-01 |
| GO:BP | GO:0010133 | proline catabolic process to glutamate | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 2 | 3 | 6.40e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 2 | 3 | 6.40e-01 |
| GO:BP | GO:0032682 | negative regulation of chemokine production | 5 | 12 | 6.40e-01 |
| GO:BP | GO:0001834 | trophectodermal cell proliferation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:1905280 | negative regulation of retrograde transport, endosome to Golgi | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0051351 | positive regulation of ligase activity | 5 | 9 | 6.40e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 1 | 59 | 6.40e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 12 | 3070 | 6.41e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 1 | 104 | 6.41e-01 |
| GO:BP | GO:1900533 | palmitic acid metabolic process | 1 | 1 | 6.41e-01 |
| GO:BP | GO:1900535 | palmitic acid biosynthetic process | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0097035 | regulation of membrane lipid distribution | 2 | 43 | 6.41e-01 |
| GO:BP | GO:0036116 | long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 151 | 6.41e-01 |
| GO:BP | GO:0045887 | positive regulation of synaptic assembly at neuromuscular junction | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 25 | 321 | 6.41e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 3 | 16 | 6.41e-01 |
| GO:BP | GO:0060722 | cell proliferation involved in embryonic placenta development | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 8 | 19 | 6.41e-01 |
| GO:BP | GO:0060720 | spongiotrophoblast cell proliferation | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0090214 | spongiotrophoblast layer developmental growth | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 1 | 110 | 6.41e-01 |
| GO:BP | GO:0070267 | oncosis | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 8 | 19 | 6.41e-01 |
| GO:BP | GO:1903547 | regulation of growth hormone activity | 1 | 2 | 6.41e-01 |
| GO:BP | GO:0030035 | microspike assembly | 3 | 5 | 6.41e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 8 | 19 | 6.41e-01 |
| GO:BP | GO:0061326 | renal tubule development | 22 | 77 | 6.41e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 6.41e-01 |
| GO:BP | GO:0048632 | negative regulation of skeletal muscle tissue growth | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0051103 | DNA ligation involved in DNA repair | 2 | 5 | 6.41e-01 |
| GO:BP | GO:0071228 | cellular response to tumor cell | 2 | 4 | 6.41e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 6 | 147 | 6.41e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 15 | 31 | 6.41e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 1 | 86 | 6.41e-01 |
| GO:BP | GO:0030889 | negative regulation of B cell proliferation | 3 | 9 | 6.42e-01 |
| GO:BP | GO:0097350 | neutrophil clearance | 1 | 5 | 6.42e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 3 | 39 | 6.42e-01 |
| GO:BP | GO:0045226 | extracellular polysaccharide biosynthetic process | 2 | 2 | 6.42e-01 |
| GO:BP | GO:0060927 | cardiac pacemaker cell fate commitment | 2 | 3 | 6.42e-01 |
| GO:BP | GO:0001715 | ectodermal cell fate specification | 1 | 1 | 6.42e-01 |
| GO:BP | GO:0042665 | regulation of ectodermal cell fate specification | 1 | 1 | 6.42e-01 |
| GO:BP | GO:0046379 | extracellular polysaccharide metabolic process | 2 | 2 | 6.42e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 4 | 12 | 6.42e-01 |
| GO:BP | GO:0045006 | DNA deamination | 1 | 9 | 6.42e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 1 | 128 | 6.42e-01 |
| GO:BP | GO:1903333 | negative regulation of protein folding | 2 | 4 | 6.42e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 3 | 29 | 6.42e-01 |
| GO:BP | GO:0042666 | negative regulation of ectodermal cell fate specification | 1 | 1 | 6.42e-01 |
| GO:BP | GO:1903401 | L-lysine transmembrane transport | 1 | 6 | 6.42e-01 |
| GO:BP | GO:2000492 | regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 6.42e-01 |
| GO:BP | GO:0032965 | regulation of collagen biosynthetic process | 3 | 30 | 6.42e-01 |
| GO:BP | GO:1902022 | L-lysine transport | 1 | 6 | 6.42e-01 |
| GO:BP | GO:1905430 | cellular response to glycine | 1 | 2 | 6.42e-01 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 6 | 16 | 6.42e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 50 | 241 | 6.42e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 3 | 115 | 6.42e-01 |
| GO:BP | GO:0006422 | aspartyl-tRNA aminoacylation | 1 | 2 | 6.42e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 2 | 24 | 6.43e-01 |
| GO:BP | GO:0070476 | rRNA (guanine-N7)-methylation | 1 | 2 | 6.43e-01 |
| GO:BP | GO:2000643 | positive regulation of early endosome to late endosome transport | 4 | 7 | 6.43e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 1 | 4 | 6.43e-01 |
| GO:BP | GO:0070779 | D-aspartate import across plasma membrane | 1 | 4 | 6.43e-01 |
| GO:BP | GO:0070777 | D-aspartate transport | 1 | 4 | 6.43e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 6.43e-01 |
| GO:BP | GO:0097222 | mitochondrial mRNA polyadenylation | 1 | 1 | 6.43e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 1 | 4 | 6.43e-01 |
| GO:BP | GO:0090616 | mitochondrial mRNA 3’-end processing | 1 | 1 | 6.43e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 6.43e-01 |
| GO:BP | GO:0048242 | epinephrine secretion | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0014060 | regulation of epinephrine secretion | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 2 | 141 | 6.43e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 1 | 60 | 6.43e-01 |
| GO:BP | GO:0098921 | retrograde trans-synaptic signaling by endocannabinoid | 3 | 4 | 6.43e-01 |
| GO:BP | GO:0098920 | retrograde trans-synaptic signaling by lipid | 3 | 4 | 6.43e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 3 | 15 | 6.43e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 6.43e-01 |
| GO:BP | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | 1 | 4 | 6.43e-01 |
| GO:BP | GO:1904980 | positive regulation of endosome organization | 1 | 1 | 6.43e-01 |
| GO:BP | GO:0098886 | modification of dendritic spine | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0072387 | flavin adenine dinucleotide metabolic process | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 2 | 22 | 6.43e-01 |
| GO:BP | GO:0051004 | regulation of lipoprotein lipase activity | 3 | 11 | 6.43e-01 |
| GO:BP | GO:0031033 | myosin filament organization | 2 | 5 | 6.44e-01 |
| GO:BP | GO:1902498 | regulation of protein autoubiquitination | 3 | 5 | 6.44e-01 |
| GO:BP | GO:1904923 | regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 7 | 15 | 6.44e-01 |
| GO:BP | GO:0030854 | positive regulation of granulocyte differentiation | 3 | 5 | 6.44e-01 |
| GO:BP | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process | 3 | 3 | 6.44e-01 |
| GO:BP | GO:0062009 | secondary palate development | 3 | 20 | 6.44e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 76 | 421 | 6.44e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 6.44e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 2 | 93 | 6.44e-01 |
| GO:BP | GO:0120323 | lipid ubiquitination | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0120322 | lipid modification by small protein conjugation | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0019230 | proprioception | 1 | 3 | 6.44e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 6.44e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 20 | 41 | 6.44e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 13 | 6.45e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 11 | 99 | 6.45e-01 |
| GO:BP | GO:0035799 | ureter maturation | 1 | 1 | 6.45e-01 |
| GO:BP | GO:1905755 | protein localization to cytoplasmic microtubule | 1 | 2 | 6.45e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 6.45e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 6.45e-01 |
| GO:BP | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | 2 | 4 | 6.45e-01 |
| GO:BP | GO:1902889 | protein localization to spindle microtubule | 1 | 2 | 6.45e-01 |
| GO:BP | GO:0033133 | positive regulation of glucokinase activity | 3 | 4 | 6.45e-01 |
| GO:BP | GO:0016081 | synaptic vesicle docking | 2 | 9 | 6.45e-01 |
| GO:BP | GO:0045988 | negative regulation of striated muscle contraction | 3 | 7 | 6.45e-01 |
| GO:BP | GO:0007613 | memory | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium | 1 | 1 | 6.45e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 2 | 157 | 6.45e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 31 | 418 | 6.45e-01 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 6.45e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 6.45e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 2 | 99 | 6.45e-01 |
| GO:BP | GO:1904587 | response to glycoprotein | 2 | 5 | 6.45e-01 |
| GO:BP | GO:0023041 | neuronal signal transduction | 1 | 6 | 6.45e-01 |
| GO:BP | GO:1990264 | peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | 2 | 4 | 6.45e-01 |
| GO:BP | GO:0010724 | regulation of definitive erythrocyte differentiation | 2 | 2 | 6.45e-01 |
| GO:BP | GO:0044860 | protein localization to plasma membrane raft | 1 | 4 | 6.45e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 2 | 7 | 6.45e-01 |
| GO:BP | GO:0018106 | peptidyl-histidine phosphorylation | 1 | 1 | 6.45e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 2 | 266 | 6.45e-01 |
| GO:BP | GO:0035066 | positive regulation of histone acetylation | 12 | 25 | 6.45e-01 |
| GO:BP | GO:0019470 | 4-hydroxyproline catabolic process | 2 | 4 | 6.45e-01 |
| GO:BP | GO:1903742 | regulation of anterograde synaptic vesicle transport | 2 | 3 | 6.45e-01 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 6 | 13 | 6.45e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 13 | 37 | 6.45e-01 |
| GO:BP | GO:0019471 | 4-hydroxyproline metabolic process | 2 | 4 | 6.45e-01 |
| GO:BP | GO:0034436 | glycoprotein transport | 2 | 3 | 6.45e-01 |
| GO:BP | GO:0015962 | diadenosine triphosphate metabolic process | 1 | 1 | 6.45e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 55 | 125 | 6.45e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 6.45e-01 |
| GO:BP | GO:0015964 | diadenosine triphosphate catabolic process | 1 | 1 | 6.45e-01 |
| GO:BP | GO:1903292 | protein localization to Golgi membrane | 1 | 4 | 6.45e-01 |
| GO:BP | GO:0080120 | CAAX-box protein maturation | 1 | 3 | 6.45e-01 |
| GO:BP | GO:0048266 | behavioral response to pain | 1 | 11 | 6.45e-01 |
| GO:BP | GO:0051866 | general adaptation syndrome | 2 | 4 | 6.45e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 20 | 157 | 6.45e-01 |
| GO:BP | GO:0033353 | S-adenosylmethionine cycle | 2 | 7 | 6.45e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 1 | 129 | 6.45e-01 |
| GO:BP | GO:0006364 | rRNA processing | 12 | 221 | 6.45e-01 |
| GO:BP | GO:0060632 | regulation of microtubule-based movement | 1 | 31 | 6.45e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 2 | 32 | 6.45e-01 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 5 | 6.45e-01 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 2 | 4 | 6.45e-01 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 2 | 4 | 6.45e-01 |
| GO:BP | GO:0080058 | protein deglutathionylation | 1 | 1 | 6.45e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 32 | 65 | 6.46e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 7 | 18 | 6.46e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 8 | 123 | 6.46e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 6.46e-01 |
| GO:BP | GO:0030154 | cell differentiation | 3 | 2958 | 6.46e-01 |
| GO:BP | GO:0006409 | tRNA export from nucleus | 2 | 3 | 6.46e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 6 | 135 | 6.47e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 23 | 91 | 6.47e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 6 | 127 | 6.47e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 22 | 265 | 6.47e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 8 | 34 | 6.47e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 1 | 8 | 6.47e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 5 | 154 | 6.47e-01 |
| GO:BP | GO:0048496 | maintenance of animal organ identity | 3 | 4 | 6.48e-01 |
| GO:BP | GO:0070562 | regulation of vitamin D receptor signaling pathway | 1 | 9 | 6.48e-01 |
| GO:BP | GO:0060438 | trachea development | 3 | 13 | 6.48e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 69 | 1378 | 6.48e-01 |
| GO:BP | GO:0007158 | neuron cell-cell adhesion | 4 | 13 | 6.48e-01 |
| GO:BP | GO:0044058 | regulation of digestive system process | 2 | 22 | 6.48e-01 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 2 | 24 | 6.48e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 2 | 15 | 6.48e-01 |
| GO:BP | GO:0000493 | box H/ACA snoRNP assembly | 1 | 2 | 6.48e-01 |
| GO:BP | GO:0072193 | ureter smooth muscle cell differentiation | 1 | 2 | 6.48e-01 |
| GO:BP | GO:0072191 | ureter smooth muscle development | 1 | 2 | 6.48e-01 |
| GO:BP | GO:2000741 | positive regulation of mesenchymal stem cell differentiation | 1 | 4 | 6.48e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 1 | 3 | 6.48e-01 |
| GO:BP | GO:0060737 | prostate gland morphogenetic growth | 1 | 2 | 6.48e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 2 | 38 | 6.48e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 3 | 50 | 6.48e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 6.49e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 3 | 63 | 6.49e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 6 | 34 | 6.49e-01 |
| GO:BP | GO:0032979 | protein insertion into mitochondrial inner membrane from matrix | 2 | 4 | 6.49e-01 |
| GO:BP | GO:0032978 | protein insertion into membrane from inner side | 2 | 4 | 6.49e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 16 | 31 | 6.49e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 556 | 6.49e-01 |
| GO:BP | GO:0033087 | negative regulation of immature T cell proliferation | 1 | 3 | 6.49e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 5 | 14 | 6.49e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 3 | 51 | 6.49e-01 |
| GO:BP | GO:1990134 | epithelial cell apoptotic process involved in palatal shelf morphogenesis | 1 | 1 | 6.49e-01 |
| GO:BP | GO:0002536 | respiratory burst involved in inflammatory response | 1 | 4 | 6.49e-01 |
| GO:BP | GO:0033088 | negative regulation of immature T cell proliferation in thymus | 1 | 3 | 6.49e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 2 | 86 | 6.49e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 10 | 24 | 6.49e-01 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 5 | 11 | 6.49e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 2 | 7 | 6.49e-01 |
| GO:BP | GO:0032463 | negative regulation of protein homooligomerization | 1 | 2 | 6.49e-01 |
| GO:BP | GO:0032460 | negative regulation of protein oligomerization | 1 | 2 | 6.49e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 3 | 82 | 6.49e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 6.49e-01 |
| GO:BP | GO:1903826 | L-arginine transmembrane transport | 5 | 12 | 6.49e-01 |
| GO:BP | GO:0061517 | macrophage proliferation | 1 | 7 | 6.49e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 2 | 33 | 6.49e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 2 | 33 | 6.49e-01 |
| GO:BP | GO:0090314 | positive regulation of protein targeting to membrane | 4 | 22 | 6.49e-01 |
| GO:BP | GO:0036138 | peptidyl-histidine hydroxylation | 1 | 1 | 6.49e-01 |
| GO:BP | GO:0039533 | regulation of MDA-5 signaling pathway | 2 | 5 | 6.49e-01 |
| GO:BP | GO:1900373 | positive regulation of purine nucleotide biosynthetic process | 2 | 12 | 6.49e-01 |
| GO:BP | GO:1905243 | cellular response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 6.49e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 4 | 75 | 6.49e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 6.49e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 3 | 2984 | 6.49e-01 |
| GO:BP | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | 1 | 2 | 6.49e-01 |
| GO:BP | GO:0030810 | positive regulation of nucleotide biosynthetic process | 2 | 12 | 6.49e-01 |
| GO:BP | GO:0097104 | postsynaptic membrane assembly | 2 | 8 | 6.49e-01 |
| GO:BP | GO:0042487 | regulation of odontogenesis of dentin-containing tooth | 1 | 6 | 6.49e-01 |
| GO:BP | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway | 2 | 2 | 6.49e-01 |
| GO:BP | GO:0035589 | G protein-coupled purinergic nucleotide receptor signaling pathway | 3 | 4 | 6.49e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 1 | 146 | 6.49e-01 |
| GO:BP | GO:1900208 | regulation of cardiolipin metabolic process | 2 | 3 | 6.49e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 4 | 8 | 6.50e-01 |
| GO:BP | GO:1905209 | positive regulation of cardiocyte differentiation | 1 | 11 | 6.50e-01 |
| GO:BP | GO:1904253 | positive regulation of bile acid metabolic process | 2 | 3 | 6.50e-01 |
| GO:BP | GO:0006657 | CDP-choline pathway | 4 | 6 | 6.50e-01 |
| GO:BP | GO:0070859 | positive regulation of bile acid biosynthetic process | 2 | 3 | 6.50e-01 |
| GO:BP | GO:0106004 | tRNA (guanine-N7)-methylation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 6 | 56 | 6.50e-01 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 3 | 12 | 6.50e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 6.50e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 6.50e-01 |
| GO:BP | GO:1990442 | intrinsic apoptotic signaling pathway in response to nitrosative stress | 2 | 3 | 6.50e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 4 | 1898 | 6.50e-01 |
| GO:BP | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation | 2 | 5 | 6.50e-01 |
| GO:BP | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:2000452 | regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0034397 | telomere localization | 2 | 5 | 6.50e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 13 | 35 | 6.50e-01 |
| GO:BP | GO:0070922 | RISC complex assembly | 3 | 9 | 6.50e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 32 | 123 | 6.50e-01 |
| GO:BP | GO:0097340 | inhibition of cysteine-type endopeptidase activity | 2 | 5 | 6.50e-01 |
| GO:BP | GO:0097341 | zymogen inhibition | 2 | 5 | 6.50e-01 |
| GO:BP | GO:2001137 | positive regulation of endocytic recycling | 1 | 7 | 6.50e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 8 | 21 | 6.50e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 4 | 31 | 6.51e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 4 | 27 | 6.51e-01 |
| GO:BP | GO:0046321 | positive regulation of fatty acid oxidation | 5 | 15 | 6.51e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 28 | 58 | 6.51e-01 |
| GO:BP | GO:0071247 | cellular response to chromate | 1 | 1 | 6.52e-01 |
| GO:BP | GO:1901423 | response to benzene | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 4 | 155 | 6.52e-01 |
| GO:BP | GO:0021510 | spinal cord development | 3 | 59 | 6.52e-01 |
| GO:BP | GO:0048702 | embryonic neurocranium morphogenesis | 1 | 7 | 6.52e-01 |
| GO:BP | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0046687 | response to chromate | 1 | 1 | 6.52e-01 |
| GO:BP | GO:1904710 | positive regulation of granulosa cell apoptotic process | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0055113 | epiboly involved in gastrulation with mouth forming second | 1 | 1 | 6.52e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 2 | 23 | 6.52e-01 |
| GO:BP | GO:1902269 | positive regulation of polyamine transmembrane transport | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0097055 | agmatine biosynthetic process | 1 | 1 | 6.52e-01 |
| GO:BP | GO:1905396 | cellular response to flavonoid | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0098629 | trans-Golgi network membrane organization | 1 | 2 | 6.52e-01 |
| GO:BP | GO:1901608 | regulation of vesicle transport along microtubule | 3 | 4 | 6.52e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 17 | 307 | 6.52e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 2 | 155 | 6.52e-01 |
| GO:BP | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 4 | 13 | 6.52e-01 |
| GO:BP | GO:0090186 | regulation of pancreatic juice secretion | 1 | 6 | 6.52e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 4 | 9 | 6.52e-01 |
| GO:BP | GO:0015697 | quaternary ammonium group transport | 1 | 17 | 6.53e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 6.53e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 1 | 63 | 6.53e-01 |
| GO:BP | GO:2000804 | regulation of termination of RNA polymerase II transcription, poly(A)-coupled | 2 | 3 | 6.53e-01 |
| GO:BP | GO:0006574 | valine catabolic process | 2 | 4 | 6.53e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 6.53e-01 |
| GO:BP | GO:0007549 | dosage compensation | 11 | 29 | 6.53e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 3 | 12 | 6.53e-01 |
| GO:BP | GO:0051641 | cellular localization | 2 | 2810 | 6.53e-01 |
| GO:BP | GO:1904594 | regulation of termination of RNA polymerase II transcription | 2 | 3 | 6.53e-01 |
| GO:BP | GO:0006414 | translational elongation | 25 | 65 | 6.53e-01 |
| GO:BP | GO:0030846 | termination of RNA polymerase II transcription, poly(A)-coupled | 2 | 3 | 6.53e-01 |
| GO:BP | GO:0060755 | negative regulation of mast cell chemotaxis | 1 | 1 | 6.53e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 1 | 45 | 6.53e-01 |
| GO:BP | GO:0031508 | pericentric heterochromatin formation | 2 | 3 | 6.54e-01 |
| GO:BP | GO:1905555 | positive regulation of blood vessel branching | 2 | 3 | 6.54e-01 |
| GO:BP | GO:0038016 | insulin receptor internalization | 2 | 3 | 6.54e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 2 | 68 | 6.54e-01 |
| GO:BP | GO:1903385 | regulation of homophilic cell adhesion | 2 | 3 | 6.54e-01 |
| GO:BP | GO:0051451 | myoblast migration | 2 | 11 | 6.54e-01 |
| GO:BP | GO:0090107 | regulation of high-density lipoprotein particle assembly | 1 | 3 | 6.54e-01 |
| GO:BP | GO:0042758 | long-chain fatty acid catabolic process | 3 | 3 | 6.55e-01 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 4 | 11 | 6.55e-01 |
| GO:BP | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0032094 | response to food | 2 | 22 | 6.55e-01 |
| GO:BP | GO:0035227 | regulation of glutamate-cysteine ligase activity | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0035229 | positive regulation of glutamate-cysteine ligase activity | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0045338 | farnesyl diphosphate metabolic process | 3 | 5 | 6.55e-01 |
| GO:BP | GO:0140570 | extraction of mislocalized protein from mitochondrial outer membrane | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 5 | 18 | 6.55e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 1 | 35 | 6.55e-01 |
| GO:BP | GO:0038003 | G protein-coupled opioid receptor signaling pathway | 1 | 5 | 6.55e-01 |
| GO:BP | GO:0042839 | D-glucuronate metabolic process | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0060633 | negative regulation of transcription initiation by RNA polymerase II | 3 | 6 | 6.56e-01 |
| GO:BP | GO:0042840 | D-glucuronate catabolic process | 1 | 1 | 6.56e-01 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 4 | 7 | 6.56e-01 |
| GO:BP | GO:0015801 | aromatic amino acid transport | 2 | 11 | 6.56e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 6.56e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 1 | 17 | 6.56e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 6.56e-01 |
| GO:BP | GO:0007635 | chemosensory behavior | 1 | 10 | 6.56e-01 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 2 | 7 | 6.56e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 1 | 598 | 6.56e-01 |
| GO:BP | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation | 4 | 7 | 6.56e-01 |
| GO:BP | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron development | 4 | 7 | 6.56e-01 |
| GO:BP | GO:0060023 | soft palate development | 2 | 3 | 6.57e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 2 | 77 | 6.58e-01 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 1 | 4 | 6.58e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 6.58e-01 |
| GO:BP | GO:0015821 | methionine transport | 1 | 4 | 6.58e-01 |
| GO:BP | GO:2000311 | regulation of AMPA receptor activity | 2 | 15 | 6.58e-01 |
| GO:BP | GO:0032098 | regulation of appetite | 1 | 8 | 6.58e-01 |
| GO:BP | GO:0051586 | positive regulation of dopamine uptake involved in synaptic transmission | 2 | 2 | 6.58e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 2 | 39 | 6.58e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 1 | 38 | 6.58e-01 |
| GO:BP | GO:0051944 | positive regulation of catecholamine uptake involved in synaptic transmission | 2 | 2 | 6.58e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 1 | 33 | 6.58e-01 |
| GO:BP | GO:0021686 | cerebellar granular layer maturation | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0021933 | radial glia guided migration of cerebellar granule cell | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 2 | 19 | 6.58e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 6.58e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 11 | 40 | 6.58e-01 |
| GO:BP | GO:0035844 | cloaca development | 2 | 3 | 6.58e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 1 | 26 | 6.58e-01 |
| GO:BP | GO:0072095 | regulation of branch elongation involved in ureteric bud branching | 2 | 2 | 6.58e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 12 | 3132 | 6.58e-01 |
| GO:BP | GO:0055107 | Golgi to secretory granule transport | 1 | 1 | 6.58e-01 |
| GO:BP | GO:0007507 | heart development | 21 | 506 | 6.58e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 9 | 17 | 6.58e-01 |
| GO:BP | GO:0007588 | excretion | 2 | 18 | 6.58e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 1 | 26 | 6.58e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 2 | 5 | 6.58e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 6 | 126 | 6.58e-01 |
| GO:BP | GO:1902214 | regulation of interleukin-4-mediated signaling pathway | 1 | 2 | 6.58e-01 |
| GO:BP | GO:0072278 | metanephric comma-shaped body morphogenesis | 2 | 3 | 6.58e-01 |
| GO:BP | GO:0061325 | cell proliferation involved in outflow tract morphogenesis | 2 | 4 | 6.58e-01 |
| GO:BP | GO:0051170 | import into nucleus | 6 | 152 | 6.58e-01 |
| GO:BP | GO:0070070 | proton-transporting V-type ATPase complex assembly | 1 | 7 | 6.59e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 1 | 132 | 6.59e-01 |
| GO:BP | GO:0010815 | bradykinin catabolic process | 1 | 6 | 6.59e-01 |
| GO:BP | GO:1902380 | positive regulation of endoribonuclease activity | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0072386 | plus-end-directed organelle transport along microtubule | 1 | 7 | 6.59e-01 |
| GO:BP | GO:1904685 | positive regulation of metalloendopeptidase activity | 1 | 1 | 6.59e-01 |
| GO:BP | GO:2000344 | positive regulation of acrosome reaction | 1 | 6 | 6.59e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 7 | 199 | 6.59e-01 |
| GO:BP | GO:0000741 | karyogamy | 1 | 1 | 6.59e-01 |
| GO:BP | GO:2001037 | positive regulation of tongue muscle cell differentiation | 1 | 1 | 6.59e-01 |
| GO:BP | GO:2001035 | regulation of tongue muscle cell differentiation | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0007344 | pronuclear fusion | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0035981 | tongue muscle cell differentiation | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 3 | 17 | 6.60e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 10 | 25 | 6.60e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 10 | 25 | 6.60e-01 |
| GO:BP | GO:0043181 | vacuolar sequestering | 1 | 1 | 6.60e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 6 | 190 | 6.60e-01 |
| GO:BP | GO:0036250 | peroxisome transport along microtubule | 1 | 1 | 6.60e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 9 | 19 | 6.60e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 54 | 1137 | 6.61e-01 |
| GO:BP | GO:1902044 | regulation of Fas signaling pathway | 1 | 4 | 6.61e-01 |
| GO:BP | GO:2000417 | negative regulation of eosinophil migration | 1 | 1 | 6.61e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 2 | 88 | 6.61e-01 |
| GO:BP | GO:0006101 | citrate metabolic process | 2 | 5 | 6.61e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 27 | 228 | 6.61e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 9 | 38 | 6.61e-01 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 4 | 9 | 6.61e-01 |
| GO:BP | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production | 1 | 3 | 6.61e-01 |
| GO:BP | GO:2000338 | regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 6.61e-01 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 38 | 6.61e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 14 | 68 | 6.61e-01 |
| GO:BP | GO:0071608 | macrophage inflammatory protein-1 alpha production | 1 | 3 | 6.61e-01 |
| GO:BP | GO:1901860 | positive regulation of mitochondrial DNA metabolic process | 2 | 3 | 6.61e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 14 | 3750 | 6.61e-01 |
| GO:BP | GO:0021543 | pallium development | 9 | 139 | 6.61e-01 |
| GO:BP | GO:0072566 | chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 6.61e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 12 | 3133 | 6.61e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 4 | 7 | 6.61e-01 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 6 | 28 | 6.61e-01 |
| GO:BP | GO:0060545 | positive regulation of necroptotic process | 3 | 4 | 6.61e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 6.61e-01 |
| GO:BP | GO:0051295 | establishment of meiotic spindle localization | 1 | 4 | 6.61e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 10 | 42 | 6.61e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 1 | 28 | 6.61e-01 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 4 | 120 | 6.61e-01 |
| GO:BP | GO:0040038 | polar body extrusion after meiotic divisions | 1 | 5 | 6.61e-01 |
| GO:BP | GO:0021678 | third ventricle development | 1 | 4 | 6.61e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 20 | 6.61e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 3 | 7 | 6.62e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 2 | 69 | 6.62e-01 |
| GO:BP | GO:0016571 | histone methylation | 1 | 117 | 6.62e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 3 | 10 | 6.62e-01 |
| GO:BP | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development | 1 | 2 | 6.62e-01 |
| GO:BP | GO:0060672 | epithelial cell morphogenesis involved in placental branching | 1 | 2 | 6.62e-01 |
| GO:BP | GO:0033385 | geranylgeranyl diphosphate metabolic process | 1 | 2 | 6.62e-01 |
| GO:BP | GO:0007611 | learning or memory | 4 | 202 | 6.62e-01 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 3 | 5 | 6.62e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 6.62e-01 |
| GO:BP | GO:0034354 | ‘de novo’ NAD biosynthetic process from tryptophan | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 2 | 13 | 6.62e-01 |
| GO:BP | GO:0043586 | tongue development | 8 | 15 | 6.62e-01 |
| GO:BP | GO:0034516 | response to vitamin B6 | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0048203 | vesicle targeting, trans-Golgi to endosome | 1 | 2 | 6.62e-01 |
| GO:BP | GO:1903959 | regulation of monoatomic anion transmembrane transport | 5 | 6 | 6.62e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 1 | 28 | 6.62e-01 |
| GO:BP | GO:0035635 | entry of bacterium into host cell | 2 | 8 | 6.62e-01 |
| GO:BP | GO:1905572 | ganglioside GM1 transport to membrane | 1 | 1 | 6.62e-01 |
| GO:BP | GO:1903771 | positive regulation of beta-galactosidase activity | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 3 | 39 | 6.62e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 1 | 18 | 6.62e-01 |
| GO:BP | GO:0009234 | menaquinone biosynthetic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 5 | 138 | 6.62e-01 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 2 | 11 | 6.62e-01 |
| GO:BP | GO:0045658 | regulation of neutrophil differentiation | 2 | 2 | 6.62e-01 |
| GO:BP | GO:0098870 | action potential propagation | 2 | 5 | 6.62e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 7 | 560 | 6.62e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 1 | 14 | 6.62e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 6.62e-01 |
| GO:BP | GO:0042256 | cytosolic ribosome assembly | 1 | 5 | 6.62e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 2 | 15 | 6.62e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 6 | 116 | 6.62e-01 |
| GO:BP | GO:0048793 | pronephros development | 2 | 5 | 6.62e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 2 | 1107 | 6.62e-01 |
| GO:BP | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis | 1 | 3 | 6.62e-01 |
| GO:BP | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation | 1 | 5 | 6.62e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 2 | 13 | 6.62e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 1 | 18 | 6.62e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 2 | 39 | 6.62e-01 |
| GO:BP | GO:0008582 | regulation of synaptic assembly at neuromuscular junction | 3 | 4 | 6.62e-01 |
| GO:BP | GO:0098838 | folate transmembrane transport | 1 | 6 | 6.62e-01 |
| GO:BP | GO:1901175 | lycopene metabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 2 | 348 | 6.62e-01 |
| GO:BP | GO:0002318 | myeloid progenitor cell differentiation | 1 | 7 | 6.62e-01 |
| GO:BP | GO:0042371 | vitamin K biosynthetic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0006390 | mitochondrial transcription | 8 | 18 | 6.62e-01 |
| GO:BP | GO:1990401 | embryonic lung development | 1 | 1 | 6.62e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 4 | 169 | 6.62e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 13 | 3237 | 6.62e-01 |
| GO:BP | GO:0097476 | spinal cord motor neuron migration | 1 | 3 | 6.62e-01 |
| GO:BP | GO:0062172 | lutein catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0016110 | tetraterpenoid catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0062170 | lutein metabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0097376 | interneuron axon guidance | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0001408 | guanine nucleotide transport | 1 | 6 | 6.62e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 9 | 42 | 6.62e-01 |
| GO:BP | GO:0061819 | telomeric DNA-containing double minutes formation | 2 | 3 | 6.62e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 4 | 102 | 6.62e-01 |
| GO:BP | GO:1904693 | midbrain morphogenesis | 2 | 3 | 6.62e-01 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 5 | 8 | 6.62e-01 |
| GO:BP | GO:0019227 | neuronal action potential propagation | 2 | 5 | 6.62e-01 |
| GO:BP | GO:1902443 | negative regulation of ripoptosome assembly involved in necroptotic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:1902442 | regulation of ripoptosome assembly involved in necroptotic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 6.62e-01 |
| GO:BP | GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | 10 | 14 | 6.62e-01 |
| GO:BP | GO:0031651 | negative regulation of heat generation | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 8 | 165 | 6.62e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 30 | 893 | 6.62e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 1 | 4 | 6.62e-01 |
| GO:BP | GO:0016118 | carotenoid catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:1901826 | zeaxanthin catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0034975 | protein folding in endoplasmic reticulum | 1 | 10 | 6.62e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 7 | 161 | 6.62e-01 |
| GO:BP | GO:0016124 | xanthophyll catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 3 | 27 | 6.62e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 7 | 10 | 6.62e-01 |
| GO:BP | GO:1905765 | negative regulation of protection from non-homologous end joining at telomere | 2 | 3 | 6.62e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 2 | 2412 | 6.62e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 2 | 19 | 6.62e-01 |
| GO:BP | GO:0042538 | hyperosmotic salinity response | 4 | 8 | 6.62e-01 |
| GO:BP | GO:1905869 | negative regulation of 3’-UTR-mediated mRNA stabilization | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0032194 | ubiquinone biosynthetic process via 3,4-dihydroxy-5-polyprenylbenzoate | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 2 | 19 | 6.62e-01 |
| GO:BP | GO:1905764 | regulation of protection from non-homologous end joining at telomere | 2 | 3 | 6.62e-01 |
| GO:BP | GO:0099093 | calcium export from the mitochondrion | 1 | 2 | 6.62e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 2 | 19 | 6.62e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 52 | 1149 | 6.62e-01 |
| GO:BP | GO:0030916 | otic vesicle formation | 2 | 6 | 6.62e-01 |
| GO:BP | GO:1901176 | lycopene catabolic process | 1 | 1 | 6.62e-01 |
| GO:BP | GO:1903048 | regulation of acetylcholine-gated cation channel activity | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 2 | 20 | 6.62e-01 |
| GO:BP | GO:0060242 | contact inhibition | 1 | 8 | 6.62e-01 |
| GO:BP | GO:0006415 | translational termination | 1 | 17 | 6.62e-01 |
| GO:BP | GO:1903049 | negative regulation of acetylcholine-gated cation channel activity | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0035441 | cell migration involved in vasculogenesis | 2 | 4 | 6.62e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 1 | 126 | 6.62e-01 |
| GO:BP | GO:2000860 | positive regulation of aldosterone secretion | 2 | 3 | 6.62e-01 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 2 | 39 | 6.62e-01 |
| GO:BP | GO:2000857 | positive regulation of mineralocorticoid secretion | 2 | 3 | 6.62e-01 |
| GO:BP | GO:1901569 | fatty acid derivative catabolic process | 1 | 10 | 6.62e-01 |
| GO:BP | GO:0051780 | behavioral response to nutrient | 1 | 1 | 6.62e-01 |
| GO:BP | GO:0010713 | negative regulation of collagen metabolic process | 3 | 8 | 6.62e-01 |
| GO:BP | GO:0015728 | mevalonate transport | 1 | 1 | 6.62e-01 |
| GO:BP | GO:2000983 | regulation of ATP citrate synthase activity | 1 | 2 | 6.62e-01 |
| GO:BP | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | 1 | 13 | 6.63e-01 |
| GO:BP | GO:0010832 | negative regulation of myotube differentiation | 1 | 10 | 6.63e-01 |
| GO:BP | GO:0090660 | cerebrospinal fluid circulation | 1 | 9 | 6.63e-01 |
| GO:BP | GO:0097577 | sequestering of iron ion | 1 | 3 | 6.63e-01 |
| GO:BP | GO:1904580 | regulation of intracellular mRNA localization | 2 | 4 | 6.63e-01 |
| GO:BP | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process | 1 | 2 | 6.63e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 3 | 17 | 6.63e-01 |
| GO:BP | GO:0003168 | Purkinje myocyte differentiation | 1 | 1 | 6.63e-01 |
| GO:BP | GO:1904347 | regulation of small intestine smooth muscle contraction | 1 | 1 | 6.63e-01 |
| GO:BP | GO:1904349 | positive regulation of small intestine smooth muscle contraction | 1 | 1 | 6.63e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 6.63e-01 |
| GO:BP | GO:0150045 | regulation of synaptic signaling by nitric oxide | 1 | 1 | 6.63e-01 |
| GO:BP | GO:0060397 | growth hormone receptor signaling pathway via JAK-STAT | 3 | 9 | 6.64e-01 |
| GO:BP | GO:0097105 | presynaptic membrane assembly | 1 | 8 | 6.64e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 6.64e-01 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 8 | 16 | 6.64e-01 |
| GO:BP | GO:2001295 | malonyl-CoA biosynthetic process | 1 | 2 | 6.64e-01 |
| GO:BP | GO:0032625 | interleukin-21 production | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0032745 | positive regulation of interleukin-21 production | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 3 | 95 | 6.64e-01 |
| GO:BP | GO:0032665 | regulation of interleukin-21 production | 1 | 1 | 6.64e-01 |
| GO:BP | GO:0045337 | farnesyl diphosphate biosynthetic process | 1 | 3 | 6.64e-01 |
| GO:BP | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 2 | 5 | 6.65e-01 |
| GO:BP | GO:0050746 | regulation of lipoprotein metabolic process | 1 | 15 | 6.65e-01 |
| GO:BP | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0120158 | positive regulation of collagen catabolic process | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0006565 | L-serine catabolic process | 1 | 4 | 6.65e-01 |
| GO:BP | GO:1904197 | positive regulation of granulosa cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:1904897 | regulation of hepatic stellate cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:1904195 | regulation of granulosa cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 4 | 23 | 6.65e-01 |
| GO:BP | GO:0010039 | response to iron ion | 1 | 24 | 6.65e-01 |
| GO:BP | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | 1 | 10 | 6.65e-01 |
| GO:BP | GO:0048668 | collateral sprouting | 8 | 25 | 6.65e-01 |
| GO:BP | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 5 | 8 | 6.65e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 6.65e-01 |
| GO:BP | GO:0006572 | tyrosine catabolic process | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 15 | 37 | 6.65e-01 |
| GO:BP | GO:1904899 | positive regulation of hepatic stellate cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 1 | 45 | 6.65e-01 |
| GO:BP | GO:0007398 | ectoderm development | 1 | 15 | 6.65e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 2 | 9 | 6.65e-01 |
| GO:BP | GO:0030327 | prenylated protein catabolic process | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 6.65e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 9 | 74 | 6.65e-01 |
| GO:BP | GO:0036118 | hyaluranon cable assembly | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0014862 | regulation of skeletal muscle contraction by chemo-mechanical energy conversion | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0009589 | detection of UV | 1 | 2 | 6.65e-01 |
| GO:BP | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 6.65e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 2 | 2 | 6.65e-01 |
| GO:BP | GO:0014860 | neurotransmitter secretion involved in regulation of skeletal muscle contraction | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 6.65e-01 |
| GO:BP | GO:0014843 | growth factor dependent regulation of skeletal muscle satellite cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0014728 | regulation of the force of skeletal muscle contraction | 1 | 3 | 6.65e-01 |
| GO:BP | GO:1900104 | regulation of hyaluranon cable assembly | 1 | 3 | 6.65e-01 |
| GO:BP | GO:1900106 | positive regulation of hyaluranon cable assembly | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0031444 | slow-twitch skeletal muscle fiber contraction | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0015847 | putrescine transport | 2 | 2 | 6.65e-01 |
| GO:BP | GO:1990922 | hepatic stellate cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 5 | 25 | 6.65e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 20 | 79 | 6.65e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 3 | 29 | 6.65e-01 |
| GO:BP | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0060377 | negative regulation of mast cell differentiation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 4 | 7 | 6.65e-01 |
| GO:BP | GO:1902748 | positive regulation of lens fiber cell differentiation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:1990739 | granulosa cell proliferation | 1 | 1 | 6.65e-01 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 2 | 7 | 6.65e-01 |
| GO:BP | GO:0043474 | pigment metabolic process involved in pigmentation | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0060465 | pharynx development | 1 | 3 | 6.65e-01 |
| GO:BP | GO:0003406 | retinal pigment epithelium development | 2 | 4 | 6.65e-01 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 8 | 12 | 6.66e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 2 | 14 | 6.66e-01 |
| GO:BP | GO:0071045 | nuclear histone mRNA catabolic process | 1 | 1 | 6.66e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 16 | 323 | 6.66e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 11 | 189 | 6.66e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 3 | 12 | 6.66e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 3 | 12 | 6.66e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 3 | 320 | 6.66e-01 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 17 | 36 | 6.67e-01 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 1 | 57 | 6.67e-01 |
| GO:BP | GO:1901492 | positive regulation of lymphangiogenesis | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 6 | 80 | 6.67e-01 |
| GO:BP | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion | 2 | 3 | 6.67e-01 |
| GO:BP | GO:0036035 | osteoclast development | 1 | 10 | 6.68e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 1 | 13 | 6.68e-01 |
| GO:BP | GO:0016129 | phytosteroid biosynthetic process | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0016128 | phytosteroid metabolic process | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0016131 | brassinosteroid metabolic process | 1 | 1 | 6.68e-01 |
| GO:BP | GO:0016132 | brassinosteroid biosynthetic process | 1 | 1 | 6.68e-01 |
| GO:BP | GO:1903387 | positive regulation of homophilic cell adhesion | 1 | 1 | 6.68e-01 |
| GO:BP | GO:1905454 | negative regulation of myeloid progenitor cell differentiation | 1 | 1 | 6.68e-01 |
| GO:BP | GO:1905711 | response to phosphatidylethanolamine | 1 | 1 | 6.69e-01 |
| GO:BP | GO:0002127 | tRNA wobble base cytosine methylation | 1 | 1 | 6.69e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 57 | 149 | 6.69e-01 |
| GO:BP | GO:0036367 | light adaption | 1 | 3 | 6.69e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 3 | 57 | 6.69e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 2 | 3 | 6.69e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 6.69e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 1 | 24 | 6.69e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1 | 24 | 6.69e-01 |
| GO:BP | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 5 | 6.69e-01 |
| GO:BP | GO:2000353 | positive regulation of endothelial cell apoptotic process | 1 | 13 | 6.69e-01 |
| GO:BP | GO:0071305 | cellular response to vitamin D | 2 | 14 | 6.69e-01 |
| GO:BP | GO:0018117 | protein adenylylation | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0018175 | protein nucleotidylation | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 6.69e-01 |
| GO:BP | GO:2000491 | positive regulation of hepatic stellate cell activation | 1 | 4 | 6.69e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 1 | 18 | 6.70e-01 |
| GO:BP | GO:1901193 | regulation of formation of translation preinitiation complex | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle | 1 | 3 | 6.70e-01 |
| GO:BP | GO:1904689 | negative regulation of cytoplasmic translational initiation | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0009386 | translational attenuation | 1 | 2 | 6.70e-01 |
| GO:BP | GO:1901194 | negative regulation of formation of translation preinitiation complex | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 6.70e-01 |
| GO:BP | GO:0031591 | wybutosine biosynthetic process | 2 | 4 | 6.70e-01 |
| GO:BP | GO:0031590 | wybutosine metabolic process | 2 | 4 | 6.70e-01 |
| GO:BP | GO:0015782 | CMP-N-acetylneuraminate transmembrane transport | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0009651 | response to salt stress | 3 | 18 | 6.70e-01 |
| GO:BP | GO:1905770 | regulation of mesodermal cell differentiation | 3 | 6 | 6.71e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 5 | 8 | 6.71e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 1 | 53 | 6.71e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 5 | 8 | 6.71e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 3 | 12 | 6.71e-01 |
| GO:BP | GO:1901160 | primary amino compound metabolic process | 1 | 8 | 6.71e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 4 | 8 | 6.71e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 5 | 8 | 6.71e-01 |
| GO:BP | GO:0033505 | floor plate morphogenesis | 1 | 3 | 6.71e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 105 | 485 | 6.71e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 6.71e-01 |
| GO:BP | GO:1990046 | stress-induced mitochondrial fusion | 1 | 2 | 6.71e-01 |
| GO:BP | GO:1905603 | regulation of blood-brain barrier permeability | 3 | 6 | 6.71e-01 |
| GO:BP | GO:1901550 | regulation of endothelial cell development | 1 | 12 | 6.71e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 4 | 16 | 6.71e-01 |
| GO:BP | GO:1903140 | regulation of establishment of endothelial barrier | 1 | 12 | 6.71e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 2 | 38 | 6.71e-01 |
| GO:BP | GO:0015700 | arsenite transport | 1 | 1 | 6.71e-01 |
| GO:BP | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration | 3 | 6 | 6.72e-01 |
| GO:BP | GO:0071140 | resolution of mitotic recombination intermediates | 1 | 2 | 6.72e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 6.72e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 16 | 79 | 6.72e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 5 | 4955 | 6.72e-01 |
| GO:BP | GO:0022414 | reproductive process | 2 | 927 | 6.72e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 2 | 316 | 6.72e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 3 | 277 | 6.72e-01 |
| GO:BP | GO:0010748 | negative regulation of long-chain fatty acid import across plasma membrane | 1 | 4 | 6.72e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 3 | 41 | 6.72e-01 |
| GO:BP | GO:0046511 | sphinganine biosynthetic process | 1 | 2 | 6.72e-01 |
| GO:BP | GO:0140213 | negative regulation of long-chain fatty acid import into cell | 1 | 4 | 6.72e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 7 | 49 | 6.72e-01 |
| GO:BP | GO:1905323 | telomerase holoenzyme complex assembly | 2 | 6 | 6.72e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 13 | 149 | 6.72e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 1 | 10 | 6.72e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 9 | 1074 | 6.73e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 9 | 1076 | 6.73e-01 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 4 | 15 | 6.73e-01 |
| GO:BP | GO:0016573 | histone acetylation | 66 | 143 | 6.73e-01 |
| GO:BP | GO:0036148 | phosphatidylglycerol acyl-chain remodeling | 1 | 5 | 6.73e-01 |
| GO:BP | GO:0086072 | AV node cell-bundle of His cell adhesion involved in cell communication | 1 | 1 | 6.73e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 3 | 60 | 6.73e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 35 | 95 | 6.73e-01 |
| GO:BP | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration | 1 | 2 | 6.73e-01 |
| GO:BP | GO:0035780 | CD80 biosynthetic process | 1 | 1 | 6.73e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 9 | 30 | 6.73e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 2 | 40 | 6.74e-01 |
| GO:BP | GO:0051013 | microtubule severing | 3 | 7 | 6.74e-01 |
| GO:BP | GO:0060024 | rhythmic synaptic transmission | 1 | 3 | 6.74e-01 |
| GO:BP | GO:0019377 | glycolipid catabolic process | 3 | 12 | 6.74e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 1 | 3 | 6.74e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 1 | 3 | 6.74e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 4 | 38 | 6.74e-01 |
| GO:BP | GO:2000174 | regulation of pro-T cell differentiation | 1 | 1 | 6.74e-01 |
| GO:BP | GO:2000176 | positive regulation of pro-T cell differentiation | 1 | 1 | 6.74e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 1 | 72 | 6.74e-01 |
| GO:BP | GO:0051138 | positive regulation of NK T cell differentiation | 2 | 3 | 6.74e-01 |
| GO:BP | GO:0099105 | ion channel modulating, G protein-coupled receptor signaling pathway | 1 | 7 | 6.74e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 9 | 403 | 6.74e-01 |
| GO:BP | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | 3 | 10 | 6.74e-01 |
| GO:BP | GO:1902744 | negative regulation of lamellipodium organization | 3 | 4 | 6.74e-01 |
| GO:BP | GO:0000003 | reproduction | 2 | 934 | 6.74e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 3 | 62 | 6.74e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 6.74e-01 |
| GO:BP | GO:1900223 | positive regulation of amyloid-beta clearance | 1 | 5 | 6.74e-01 |
| GO:BP | GO:2000724 | positive regulation of cardiac vascular smooth muscle cell differentiation | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 30 | 597 | 6.75e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 30 | 597 | 6.75e-01 |
| GO:BP | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling | 2 | 8 | 6.75e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 6.75e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 10 | 67 | 6.75e-01 |
| GO:BP | GO:0072318 | clathrin coat disassembly | 4 | 8 | 6.75e-01 |
| GO:BP | GO:0043060 | meiotic metaphase I plate congression | 1 | 1 | 6.75e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 6.75e-01 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 10 | 18 | 6.75e-01 |
| GO:BP | GO:0051900 | regulation of mitochondrial depolarization | 5 | 15 | 6.75e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 27 | 150 | 6.75e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 17 | 33 | 6.75e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 4 | 131 | 6.75e-01 |
| GO:BP | GO:0051707 | response to other organism | 4 | 820 | 6.75e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 5 | 25 | 6.75e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 7 | 168 | 6.75e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 4 | 11 | 6.75e-01 |
| GO:BP | GO:1904925 | positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 1 | 14 | 6.75e-01 |
| GO:BP | GO:0048294 | negative regulation of isotype switching to IgE isotypes | 1 | 2 | 6.75e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 4 | 11 | 6.75e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 2 | 311 | 6.75e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 6 | 13 | 6.76e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 2 | 22 | 6.76e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 2 | 14 | 6.76e-01 |
| GO:BP | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 6 | 12 | 6.76e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0045453 | bone resorption | 4 | 40 | 6.76e-01 |
| GO:BP | GO:0044804 | autophagy of nucleus | 1 | 18 | 6.76e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 2 | 104 | 6.76e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 4 | 820 | 6.76e-01 |
| GO:BP | GO:0061520 | Langerhans cell differentiation | 2 | 3 | 6.76e-01 |
| GO:BP | GO:0021541 | ammon gyrus development | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 2 | 10 | 6.76e-01 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 2 | 10 | 6.76e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 1 | 18 | 6.76e-01 |
| GO:BP | GO:0052779 | amino disaccharide metabolic process | 2 | 9 | 6.76e-01 |
| GO:BP | GO:0097712 | vesicle targeting, trans-Golgi to periciliary membrane compartment | 1 | 1 | 6.76e-01 |
| GO:BP | GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 1 | 5 | 6.76e-01 |
| GO:BP | GO:0030205 | dermatan sulfate metabolic process | 2 | 9 | 6.76e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 1 | 5 | 6.76e-01 |
| GO:BP | GO:2000301 | negative regulation of synaptic vesicle exocytosis | 2 | 4 | 6.76e-01 |
| GO:BP | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 1 | 5 | 6.76e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 1 | 11 | 6.76e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 1 | 134 | 6.76e-01 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 9 | 17 | 6.76e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 3 | 25 | 6.76e-01 |
| GO:BP | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling | 1 | 12 | 6.76e-01 |
| GO:BP | GO:0006782 | protoporphyrinogen IX biosynthetic process | 2 | 9 | 6.76e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 11 | 276 | 6.76e-01 |
| GO:BP | GO:0090410 | malonate catabolic process | 1 | 1 | 6.76e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 2 | 12 | 6.76e-01 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 1 | 14 | 6.76e-01 |
| GO:BP | GO:0045907 | positive regulation of vasoconstriction | 1 | 15 | 6.76e-01 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 4 | 13 | 6.76e-01 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 1 | 4 | 6.76e-01 |
| GO:BP | GO:0140009 | L-aspartate import across plasma membrane | 1 | 5 | 6.76e-01 |
| GO:BP | GO:0051006 | positive regulation of lipoprotein lipase activity | 1 | 4 | 6.76e-01 |
| GO:BP | GO:0010898 | positive regulation of triglyceride catabolic process | 1 | 4 | 6.76e-01 |
| GO:BP | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0140141 | mitochondrial potassium ion transmembrane transport | 1 | 2 | 6.76e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 6 | 196 | 6.76e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 3 | 9 | 6.76e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 13 | 36 | 6.76e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 3 | 61 | 6.76e-01 |
| GO:BP | GO:0042116 | macrophage activation | 1 | 57 | 6.77e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 1 | 12 | 6.77e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 8 | 20 | 6.77e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 6.77e-01 |
| GO:BP | GO:0061470 | T follicular helper cell differentiation | 2 | 4 | 6.77e-01 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 2 | 16 | 6.77e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 6.77e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 6.77e-01 |
| GO:BP | GO:0003407 | neural retina development | 3 | 48 | 6.77e-01 |
| GO:BP | GO:0061197 | fungiform papilla morphogenesis | 2 | 4 | 6.77e-01 |
| GO:BP | GO:0048696 | regulation of collateral sprouting in absence of injury | 1 | 2 | 6.77e-01 |
| GO:BP | GO:0044338 | canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation | 1 | 3 | 6.77e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 4 | 58 | 6.77e-01 |
| GO:BP | GO:2000230 | negative regulation of pancreatic stellate cell proliferation | 1 | 1 | 6.77e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 8 | 83 | 6.77e-01 |
| GO:BP | GO:0010041 | response to iron(III) ion | 1 | 4 | 6.78e-01 |
| GO:BP | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis | 4 | 9 | 6.78e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 9 | 17 | 6.78e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 1 | 19 | 6.78e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 9 | 91 | 6.78e-01 |
| GO:BP | GO:0015825 | L-serine transport | 1 | 7 | 6.78e-01 |
| GO:BP | GO:0071360 | cellular response to exogenous dsRNA | 6 | 14 | 6.78e-01 |
| GO:BP | GO:0070781 | response to biotin | 1 | 1 | 6.78e-01 |
| GO:BP | GO:0070175 | positive regulation of enamel mineralization | 1 | 2 | 6.78e-01 |
| GO:BP | GO:2000870 | regulation of progesterone secretion | 1 | 1 | 6.78e-01 |
| GO:BP | GO:0034143 | regulation of toll-like receptor 4 signaling pathway | 5 | 15 | 6.78e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0021999 | neural plate anterior/posterior regionalization | 1 | 5 | 6.78e-01 |
| GO:BP | GO:1902688 | regulation of NAD metabolic process | 1 | 1 | 6.79e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 58 | 151 | 6.79e-01 |
| GO:BP | GO:0061622 | glycolytic process through glucose-1-phosphate | 1 | 2 | 6.79e-01 |
| GO:BP | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 3 | 9 | 6.79e-01 |
| GO:BP | GO:1990009 | retinal cell apoptotic process | 1 | 5 | 6.79e-01 |
| GO:BP | GO:0062196 | regulation of lysosome size | 2 | 8 | 6.79e-01 |
| GO:BP | GO:1904690 | positive regulation of cytoplasmic translational initiation | 2 | 5 | 6.79e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 6.79e-01 |
| GO:BP | GO:0072046 | establishment of planar polarity involved in nephron morphogenesis | 1 | 1 | 6.79e-01 |
| GO:BP | GO:0001781 | neutrophil apoptotic process | 3 | 5 | 6.79e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 1 | 12 | 6.79e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 1 | 12 | 6.79e-01 |
| GO:BP | GO:2000369 | regulation of clathrin-dependent endocytosis | 1 | 18 | 6.79e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 185 | 6.79e-01 |
| GO:BP | GO:0035962 | response to interleukin-13 | 1 | 1 | 6.79e-01 |
| GO:BP | GO:0015808 | L-alanine transport | 2 | 9 | 6.79e-01 |
| GO:BP | GO:0009110 | vitamin biosynthetic process | 2 | 14 | 6.80e-01 |
| GO:BP | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion | 4 | 5 | 6.80e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 2 | 9 | 6.81e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 5 | 52 | 6.81e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 5 | 49 | 6.81e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 7 | 91 | 6.81e-01 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 2 | 7 | 6.81e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 6.81e-01 |
| GO:BP | GO:0051409 | response to nitrosative stress | 5 | 10 | 6.81e-01 |
| GO:BP | GO:0097360 | chorionic trophoblast cell proliferation | 1 | 2 | 6.81e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 4 | 163 | 6.81e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 7 | 31 | 6.81e-01 |
| GO:BP | GO:1901382 | regulation of chorionic trophoblast cell proliferation | 1 | 2 | 6.81e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 1 | 12 | 6.81e-01 |
| GO:BP | GO:2000312 | regulation of kainate selective glutamate receptor activity | 1 | 3 | 6.81e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 2 | 29 | 6.81e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 8 | 43 | 6.82e-01 |
| GO:BP | GO:0006959 | humoral immune response | 2 | 80 | 6.82e-01 |
| GO:BP | GO:0002520 | immune system development | 5 | 144 | 6.82e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 19 | 46 | 6.82e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 4 | 29 | 6.82e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 7 | 114 | 6.82e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 6.83e-01 |
| GO:BP | GO:0038183 | bile acid signaling pathway | 1 | 3 | 6.83e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 3 | 82 | 6.83e-01 |
| GO:BP | GO:0070561 | vitamin D receptor signaling pathway | 1 | 10 | 6.83e-01 |
| GO:BP | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway | 2 | 10 | 6.83e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 2 | 42 | 6.83e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 5 | 23 | 6.83e-01 |
| GO:BP | GO:0098902 | regulation of membrane depolarization during action potential | 2 | 5 | 6.83e-01 |
| GO:BP | GO:0032928 | regulation of superoxide anion generation | 2 | 11 | 6.83e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 1 | 11 | 6.83e-01 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 4 | 20 | 6.83e-01 |
| GO:BP | GO:0015742 | alpha-ketoglutarate transport | 1 | 3 | 6.83e-01 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 1 | 6 | 6.83e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 6 | 54 | 6.83e-01 |
| GO:BP | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway | 1 | 7 | 6.84e-01 |
| GO:BP | GO:2000584 | negative regulation of platelet-derived growth factor receptor-alpha signaling pathway | 1 | 1 | 6.84e-01 |
| GO:BP | GO:0060245 | detection of cell density | 1 | 9 | 6.84e-01 |
| GO:BP | GO:0097090 | presynaptic membrane organization | 1 | 8 | 6.84e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 3 | 26 | 6.84e-01 |
| GO:BP | GO:0150094 | amyloid-beta clearance by cellular catabolic process | 1 | 7 | 6.84e-01 |
| GO:BP | GO:0006842 | tricarboxylic acid transport | 1 | 3 | 6.84e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 3 | 259 | 6.84e-01 |
| GO:BP | GO:0015746 | citrate transport | 1 | 3 | 6.84e-01 |
| GO:BP | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 3 | 6 | 6.84e-01 |
| GO:BP | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 3 | 6 | 6.84e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 2 | 7 | 6.84e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 3 | 4 | 6.84e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 5 | 14 | 6.84e-01 |
| GO:BP | GO:0033280 | response to vitamin D | 3 | 24 | 6.84e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 27 | 482 | 6.84e-01 |
| GO:BP | GO:0010710 | regulation of collagen catabolic process | 1 | 3 | 6.84e-01 |
| GO:BP | GO:0046032 | ADP catabolic process | 1 | 2 | 6.84e-01 |
| GO:BP | GO:0016032 | viral process | 4 | 344 | 6.85e-01 |
| GO:BP | GO:0010725 | regulation of primitive erythrocyte differentiation | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0035854 | eosinophil fate commitment | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0010522 | regulation of calcium ion transport into cytosol | 1 | 10 | 6.85e-01 |
| GO:BP | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination | 1 | 3 | 6.85e-01 |
| GO:BP | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 4 | 6.85e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 2 | 116 | 6.85e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 4 | 53 | 6.85e-01 |
| GO:BP | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0021932 | hindbrain radial glia guided cell migration | 1 | 8 | 6.85e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 3 | 1914 | 6.85e-01 |
| GO:BP | GO:0048635 | negative regulation of muscle organ development | 1 | 6 | 6.85e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 6.85e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 2 | 162 | 6.85e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 5 | 25 | 6.85e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 12 | 41 | 6.85e-01 |
| GO:BP | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0015844 | monoamine transport | 2 | 45 | 6.85e-01 |
| GO:BP | GO:0035585 | calcium-mediated signaling using extracellular calcium source | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 6.85e-01 |
| GO:BP | GO:0070235 | regulation of activation-induced cell death of T cells | 2 | 3 | 6.85e-01 |
| GO:BP | GO:0008039 | synaptic target recognition | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 4 | 25 | 6.85e-01 |
| GO:BP | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 1 | 7 | 6.85e-01 |
| GO:BP | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0061010 | gallbladder development | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0021828 | gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 3 | 6 | 6.86e-01 |
| GO:BP | GO:0021856 | hypothalamic tangential migration using cell-axon interactions | 3 | 6 | 6.86e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 3 | 135 | 6.86e-01 |
| GO:BP | GO:0021780 | glial cell fate specification | 3 | 3 | 6.86e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 38 | 116 | 6.86e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 4 | 12 | 6.86e-01 |
| GO:BP | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome | 6 | 13 | 6.86e-01 |
| GO:BP | GO:1905702 | regulation of inhibitory synapse assembly | 1 | 3 | 6.87e-01 |
| GO:BP | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 5 | 10 | 6.87e-01 |
| GO:BP | GO:0050890 | cognition | 11 | 227 | 6.87e-01 |
| GO:BP | GO:1904020 | regulation of G protein-coupled receptor internalization | 2 | 3 | 6.87e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 3 | 17 | 6.87e-01 |
| GO:BP | GO:0001510 | RNA methylation | 40 | 87 | 6.87e-01 |
| GO:BP | GO:1903818 | positive regulation of voltage-gated potassium channel activity | 1 | 11 | 6.87e-01 |
| GO:BP | GO:1901072 | glucosamine-containing compound catabolic process | 1 | 5 | 6.87e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 6.87e-01 |
| GO:BP | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 5 | 6.87e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 3 | 40 | 6.88e-01 |
| GO:BP | GO:1905551 | negative regulation of protein localization to endoplasmic reticulum | 1 | 2 | 6.88e-01 |
| GO:BP | GO:1904182 | regulation of pyruvate dehydrogenase activity | 1 | 4 | 6.88e-01 |
| GO:BP | GO:0110096 | cellular response to aldehyde | 1 | 10 | 6.88e-01 |
| GO:BP | GO:0099527 | postsynapse to nucleus signaling pathway | 2 | 6 | 6.88e-01 |
| GO:BP | GO:0008354 | germ cell migration | 2 | 6 | 6.88e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 6.88e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 1 | 88 | 6.88e-01 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 5 | 9 | 6.88e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 6.88e-01 |
| GO:BP | GO:0031064 | negative regulation of histone deacetylation | 2 | 2 | 6.88e-01 |
| GO:BP | GO:0098942 | retrograde trans-synaptic signaling by trans-synaptic protein complex | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0090193 | positive regulation of glomerulus development | 1 | 3 | 6.88e-01 |
| GO:BP | GO:0016340 | calcium-dependent cell-matrix adhesion | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 4 | 850 | 6.88e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 3 | 61 | 6.88e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 2 | 40 | 6.89e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 1 | 12 | 6.89e-01 |
| GO:BP | GO:0034651 | cortisol biosynthetic process | 1 | 8 | 6.89e-01 |
| GO:BP | GO:1902645 | tertiary alcohol biosynthetic process | 1 | 8 | 6.89e-01 |
| GO:BP | GO:0032353 | negative regulation of hormone biosynthetic process | 1 | 8 | 6.89e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 5 | 18 | 6.89e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 2 | 7 | 6.89e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 2 | 7 | 6.89e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 13 | 71 | 6.89e-01 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 4 | 40 | 6.89e-01 |
| GO:BP | GO:0016243 | regulation of autophagosome size | 1 | 2 | 6.89e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 1 | 8 | 6.89e-01 |
| GO:BP | GO:0050832 | defense response to fungus | 1 | 14 | 6.89e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 3 | 34 | 6.89e-01 |
| GO:BP | GO:0070375 | ERK5 cascade | 1 | 3 | 6.89e-01 |
| GO:BP | GO:1903939 | regulation of TORC2 signaling | 2 | 5 | 6.90e-01 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 2 | 5 | 6.90e-01 |
| GO:BP | GO:1903108 | regulation of mitochondrial transcription | 2 | 5 | 6.90e-01 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1 | 9 | 6.90e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 2 | 15 | 6.90e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 8 | 85 | 6.90e-01 |
| GO:BP | GO:0007614 | short-term memory | 5 | 9 | 6.90e-01 |
| GO:BP | GO:0033030 | negative regulation of neutrophil apoptotic process | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 7 | 135 | 6.90e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 7 | 17 | 6.90e-01 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 4 | 19 | 6.90e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 3 | 63 | 6.90e-01 |
| GO:BP | GO:0042791 | 5S class rRNA transcription by RNA polymerase III | 3 | 6 | 6.90e-01 |
| GO:BP | GO:0071918 | urea transmembrane transport | 1 | 2 | 6.90e-01 |
| GO:BP | GO:0042780 | tRNA 3’-end processing | 1 | 7 | 6.91e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 6.91e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 6.91e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 6.91e-01 |
| GO:BP | GO:0061966 | establishment of left/right asymmetry | 1 | 3 | 6.91e-01 |
| GO:BP | GO:0032196 | transposition | 4 | 17 | 6.91e-01 |
| GO:BP | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production | 1 | 3 | 6.91e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 5 | 57 | 6.91e-01 |
| GO:BP | GO:0021527 | spinal cord association neuron differentiation | 2 | 3 | 6.91e-01 |
| GO:BP | GO:0097752 | regulation of DNA stability | 2 | 3 | 6.91e-01 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 2 | 11 | 6.91e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 6.91e-01 |
| GO:BP | GO:0030539 | male genitalia development | 1 | 13 | 6.91e-01 |
| GO:BP | GO:1903238 | positive regulation of leukocyte tethering or rolling | 2 | 5 | 6.92e-01 |
| GO:BP | GO:1902174 | positive regulation of keratinocyte apoptotic process | 1 | 3 | 6.92e-01 |
| GO:BP | GO:0010795 | regulation of ubiquinone biosynthetic process | 1 | 2 | 6.92e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 1 | 18 | 6.92e-01 |
| GO:BP | GO:0019284 | L-methionine salvage from S-adenosylmethionine | 1 | 3 | 6.92e-01 |
| GO:BP | GO:0061386 | closure of optic fissure | 1 | 2 | 6.92e-01 |
| GO:BP | GO:2001111 | positive regulation of lens epithelial cell proliferation | 1 | 1 | 6.92e-01 |
| GO:BP | GO:2001109 | regulation of lens epithelial cell proliferation | 1 | 1 | 6.92e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 31 | 120 | 6.92e-01 |
| GO:BP | GO:0097166 | lens epithelial cell proliferation | 1 | 1 | 6.92e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 5 | 10 | 6.92e-01 |
| GO:BP | GO:1903651 | positive regulation of cytoplasmic transport | 6 | 11 | 6.92e-01 |
| GO:BP | GO:0071264 | positive regulation of translational initiation in response to starvation | 1 | 2 | 6.92e-01 |
| GO:BP | GO:0071262 | regulation of translational initiation in response to starvation | 1 | 2 | 6.92e-01 |
| GO:BP | GO:0010712 | regulation of collagen metabolic process | 3 | 34 | 6.92e-01 |
| GO:BP | GO:1990167 | protein K27-linked deubiquitination | 2 | 3 | 6.92e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 7 | 90 | 6.92e-01 |
| GO:BP | GO:0021750 | vestibular nucleus development | 1 | 1 | 6.92e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 7 | 29 | 6.92e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 3 | 50 | 6.92e-01 |
| GO:BP | GO:0097402 | neuroblast migration | 1 | 2 | 6.93e-01 |
| GO:BP | GO:0060935 | cardiac fibroblast cell differentiation | 2 | 4 | 6.93e-01 |
| GO:BP | GO:0000350 | generation of catalytic spliceosome for second transesterification step | 1 | 2 | 6.93e-01 |
| GO:BP | GO:0060936 | cardiac fibroblast cell development | 2 | 4 | 6.93e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 1 | 9 | 6.93e-01 |
| GO:BP | GO:0090238 | positive regulation of arachidonic acid secretion | 1 | 2 | 6.93e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 2 | 65 | 6.93e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 2 | 168 | 6.93e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 8 | 66 | 6.94e-01 |
| GO:BP | GO:0001654 | eye development | 15 | 272 | 6.94e-01 |
| GO:BP | GO:0046877 | regulation of saliva secretion | 1 | 3 | 6.94e-01 |
| GO:BP | GO:0045039 | protein insertion into mitochondrial inner membrane | 3 | 13 | 6.94e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 6 | 51 | 6.94e-01 |
| GO:BP | GO:0001542 | ovulation from ovarian follicle | 1 | 8 | 6.94e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 6 | 18 | 6.94e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 1 | 12 | 6.94e-01 |
| GO:BP | GO:0016485 | protein processing | 6 | 183 | 6.94e-01 |
| GO:BP | GO:0046348 | amino sugar catabolic process | 6 | 9 | 6.94e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 3 | 7 | 6.94e-01 |
| GO:BP | GO:0007270 | neuron-neuron synaptic transmission | 3 | 5 | 6.94e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 6.94e-01 |
| GO:BP | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway | 3 | 4 | 6.94e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 12 | 303 | 6.95e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 2 | 260 | 6.95e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 6.95e-01 |
| GO:BP | GO:0030449 | regulation of complement activation | 3 | 11 | 6.95e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 2 | 14 | 6.95e-01 |
| GO:BP | GO:0045653 | negative regulation of megakaryocyte differentiation | 1 | 17 | 6.95e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 6 | 41 | 6.95e-01 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 1 | 4 | 6.95e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 16 | 38 | 6.95e-01 |
| GO:BP | GO:1900864 | mitochondrial RNA modification | 2 | 9 | 6.95e-01 |
| GO:BP | GO:0070900 | mitochondrial tRNA modification | 2 | 9 | 6.95e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 3 | 73 | 6.95e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 4 | 47 | 6.95e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 3 | 7 | 6.95e-01 |
| GO:BP | GO:1905949 | negative regulation of calcium ion import across plasma membrane | 2 | 4 | 6.96e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 2 | 38 | 6.96e-01 |
| GO:BP | GO:0045986 | negative regulation of smooth muscle contraction | 1 | 12 | 6.96e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 5 | 7 | 6.96e-01 |
| GO:BP | GO:0031048 | small ncRNA-mediated heterochromatin formation | 1 | 2 | 6.96e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 6.96e-01 |
| GO:BP | GO:0072695 | regulation of DNA recombination at telomere | 1 | 2 | 6.96e-01 |
| GO:BP | GO:0048239 | negative regulation of DNA recombination at telomere | 1 | 2 | 6.96e-01 |
| GO:BP | GO:0001806 | type IV hypersensitivity | 2 | 2 | 6.97e-01 |
| GO:BP | GO:0010985 | negative regulation of lipoprotein particle clearance | 2 | 6 | 6.97e-01 |
| GO:BP | GO:0051461 | positive regulation of corticotropin secretion | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 6.97e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 2 | 52 | 6.97e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 2 | 189 | 6.97e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 8 | 24 | 6.97e-01 |
| GO:BP | GO:0032765 | positive regulation of mast cell cytokine production | 2 | 3 | 6.97e-01 |
| GO:BP | GO:0006562 | proline catabolic process | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 6.97e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 7 | 211 | 6.97e-01 |
| GO:BP | GO:1901509 | regulation of endothelial tube morphogenesis | 2 | 3 | 6.97e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 6.97e-01 |
| GO:BP | GO:0035279 | miRNA-mediated gene silencing by mRNA destabilization | 1 | 3 | 6.97e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 6.97e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 2 | 7 | 6.97e-01 |
| GO:BP | GO:0010526 | retrotransposon silencing | 1 | 3 | 6.97e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 1 | 168 | 6.97e-01 |
| GO:BP | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly | 1 | 6 | 6.97e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 14 | 45 | 6.97e-01 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 9 | 21 | 6.97e-01 |
| GO:BP | GO:0015074 | DNA integration | 4 | 9 | 6.98e-01 |
| GO:BP | GO:0031581 | hemidesmosome assembly | 2 | 6 | 6.98e-01 |
| GO:BP | GO:0016050 | vesicle organization | 3 | 320 | 6.99e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 1 | 169 | 6.99e-01 |
| GO:BP | GO:1900148 | negative regulation of Schwann cell migration | 1 | 3 | 6.99e-01 |
| GO:BP | GO:0035523 | protein K29-linked deubiquitination | 1 | 4 | 6.99e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 6.99e-01 |
| GO:BP | GO:0042117 | monocyte activation | 1 | 8 | 6.99e-01 |
| GO:BP | GO:0009450 | gamma-aminobutyric acid catabolic process | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0061528 | aspartate secretion | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0060717 | chorion development | 2 | 8 | 6.99e-01 |
| GO:BP | GO:1904448 | regulation of aspartate secretion | 1 | 2 | 6.99e-01 |
| GO:BP | GO:1904450 | positive regulation of aspartate secretion | 1 | 2 | 6.99e-01 |
| GO:BP | GO:1902259 | regulation of delayed rectifier potassium channel activity | 1 | 12 | 7.00e-01 |
| GO:BP | GO:0021986 | habenula development | 1 | 1 | 7.00e-01 |
| GO:BP | GO:0021538 | epithalamus development | 1 | 1 | 7.00e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 2 | 43 | 7.00e-01 |
| GO:BP | GO:0034104 | negative regulation of tissue remodeling | 2 | 13 | 7.00e-01 |
| GO:BP | GO:0015965 | diadenosine tetraphosphate metabolic process | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0015960 | diadenosine polyphosphate biosynthetic process | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0015966 | diadenosine tetraphosphate biosynthetic process | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0051930 | regulation of sensory perception of pain | 1 | 13 | 7.00e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 4 | 35 | 7.00e-01 |
| GO:BP | GO:0051931 | regulation of sensory perception | 1 | 13 | 7.00e-01 |
| GO:BP | GO:0061300 | cerebellum vasculature development | 1 | 2 | 7.00e-01 |
| GO:BP | GO:1905449 | regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 3 | 5 | 7.00e-01 |
| GO:BP | GO:0042670 | retinal cone cell differentiation | 3 | 6 | 7.00e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 3 | 73 | 7.00e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 3 | 75 | 7.00e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 7.00e-01 |
| GO:BP | GO:1905429 | response to glycine | 1 | 2 | 7.00e-01 |
| GO:BP | GO:1902082 | positive regulation of calcium ion import into sarcoplasmic reticulum | 1 | 1 | 7.00e-01 |
| GO:BP | GO:1903116 | positive regulation of actin filament-based movement | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0051659 | maintenance of mitochondrion location | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0031448 | positive regulation of fast-twitch skeletal muscle fiber contraction | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 4 | 16 | 7.01e-01 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 4 | 59 | 7.01e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 1 | 149 | 7.01e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 1 | 13 | 7.01e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 7.01e-01 |
| GO:BP | GO:0032536 | regulation of cell projection size | 2 | 8 | 7.01e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 3 | 20 | 7.01e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 7.01e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 7.01e-01 |
| GO:BP | GO:0061365 | positive regulation of triglyceride lipase activity | 1 | 4 | 7.01e-01 |
| GO:BP | GO:0048569 | post-embryonic animal organ development | 4 | 15 | 7.01e-01 |
| GO:BP | GO:0071888 | macrophage apoptotic process | 5 | 9 | 7.01e-01 |
| GO:BP | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway | 3 | 4 | 7.01e-01 |
| GO:BP | GO:0071281 | cellular response to iron ion | 4 | 6 | 7.01e-01 |
| GO:BP | GO:0033762 | response to glucagon | 2 | 13 | 7.01e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 2 | 10 | 7.01e-01 |
| GO:BP | GO:0035745 | T-helper 2 cell cytokine production | 1 | 3 | 7.01e-01 |
| GO:BP | GO:2000551 | regulation of T-helper 2 cell cytokine production | 1 | 3 | 7.01e-01 |
| GO:BP | GO:0014047 | glutamate secretion | 1 | 14 | 7.01e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 13 | 64 | 7.02e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 10 | 26 | 7.02e-01 |
| GO:BP | GO:0090248 | cell migration involved in somitogenic axis elongation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0002329 | pre-B cell differentiation | 1 | 4 | 7.02e-01 |
| GO:BP | GO:0090249 | regulation of cell migration involved in somitogenic axis elongation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0036378 | calcitriol biosynthetic process from calciol | 2 | 2 | 7.03e-01 |
| GO:BP | GO:0000055 | ribosomal large subunit export from nucleus | 2 | 8 | 7.03e-01 |
| GO:BP | GO:0046104 | thymidine metabolic process | 1 | 5 | 7.03e-01 |
| GO:BP | GO:1900114 | positive regulation of histone H3-K9 trimethylation | 1 | 2 | 7.03e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 7.03e-01 |
| GO:BP | GO:0098713 | leucine import across plasma membrane | 1 | 5 | 7.03e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 25 | 7.03e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 1 | 25 | 7.03e-01 |
| GO:BP | GO:0072377 | blood coagulation, common pathway | 1 | 1 | 7.03e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 30 | 691 | 7.03e-01 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 1 | 5 | 7.03e-01 |
| GO:BP | GO:1903801 | L-leucine import across plasma membrane | 1 | 5 | 7.03e-01 |
| GO:BP | GO:1903649 | regulation of cytoplasmic transport | 1 | 25 | 7.03e-01 |
| GO:BP | GO:0018344 | protein geranylgeranylation | 1 | 6 | 7.03e-01 |
| GO:BP | GO:0090385 | phagosome-lysosome fusion | 1 | 12 | 7.03e-01 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 2 | 9 | 7.04e-01 |
| GO:BP | GO:0042748 | circadian sleep/wake cycle, non-REM sleep | 2 | 3 | 7.04e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 3 | 13 | 7.04e-01 |
| GO:BP | GO:0001949 | sebaceous gland cell differentiation | 1 | 1 | 7.04e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 7.04e-01 |
| GO:BP | GO:0007493 | endodermal cell fate determination | 1 | 1 | 7.04e-01 |
| GO:BP | GO:1904002 | regulation of sebum secreting cell proliferation | 1 | 1 | 7.04e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 1 | 74 | 7.04e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 7.04e-01 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 1 | 19 | 7.04e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 8 | 316 | 7.05e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 3 | 76 | 7.05e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 3 | 76 | 7.05e-01 |
| GO:BP | GO:0150063 | visual system development | 15 | 273 | 7.05e-01 |
| GO:BP | GO:0030505 | inorganic diphosphate transport | 1 | 3 | 7.05e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 7.05e-01 |
| GO:BP | GO:0033182 | regulation of histone ubiquitination | 1 | 6 | 7.05e-01 |
| GO:BP | GO:0070535 | histone H2A K63-linked ubiquitination | 1 | 6 | 7.05e-01 |
| GO:BP | GO:0072177 | mesonephric duct development | 2 | 7 | 7.06e-01 |
| GO:BP | GO:0060741 | prostate gland stromal morphogenesis | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0060592 | mammary gland formation | 3 | 6 | 7.06e-01 |
| GO:BP | GO:0060330 | regulation of response to type II interferon | 1 | 12 | 7.06e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 2 | 74 | 7.06e-01 |
| GO:BP | GO:0060334 | regulation of type II interferon-mediated signaling pathway | 1 | 12 | 7.06e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 7.06e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 28 | 79 | 7.06e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 7.06e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 2 | 57 | 7.06e-01 |
| GO:BP | GO:0006548 | histidine catabolic process | 1 | 3 | 7.06e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 3 | 4 | 7.07e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 7.07e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 7.07e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 7.07e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 3 | 1995 | 7.07e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 5 | 29 | 7.07e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 8 | 1006 | 7.07e-01 |
| GO:BP | GO:0140029 | exocytic process | 1 | 69 | 7.08e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 2 | 60 | 7.08e-01 |
| GO:BP | GO:0021536 | diencephalon development | 1 | 41 | 7.08e-01 |
| GO:BP | GO:0043585 | nose morphogenesis | 1 | 3 | 7.08e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 1 | 13 | 7.08e-01 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 4 | 19 | 7.08e-01 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 2 | 14 | 7.08e-01 |
| GO:BP | GO:0002726 | positive regulation of T cell cytokine production | 4 | 14 | 7.08e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 2 | 10 | 7.08e-01 |
| GO:BP | GO:0097156 | fasciculation of motor neuron axon | 1 | 2 | 7.08e-01 |
| GO:BP | GO:0097202 | activation of cysteine-type endopeptidase activity | 5 | 16 | 7.08e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 1 | 30 | 7.08e-01 |
| GO:BP | GO:1900098 | regulation of plasma cell differentiation | 2 | 2 | 7.08e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 1 | 2 | 7.08e-01 |
| GO:BP | GO:0045342 | MHC class II biosynthetic process | 8 | 12 | 7.08e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 7.08e-01 |
| GO:BP | GO:0034285 | response to disaccharide | 2 | 4 | 7.08e-01 |
| GO:BP | GO:0140448 | signaling receptor ligand precursor processing | 2 | 29 | 7.08e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 209 | 7.08e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 1 | 1075 | 7.08e-01 |
| GO:BP | GO:0009744 | response to sucrose | 2 | 4 | 7.08e-01 |
| GO:BP | GO:0075509 | endocytosis involved in viral entry into host cell | 3 | 11 | 7.08e-01 |
| GO:BP | GO:0051901 | positive regulation of mitochondrial depolarization | 2 | 6 | 7.08e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 2 | 6 | 7.09e-01 |
| GO:BP | GO:0006901 | vesicle coating | 14 | 36 | 7.09e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 1 | 35 | 7.09e-01 |
| GO:BP | GO:1902001 | fatty acid transmembrane transport | 5 | 15 | 7.09e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 3 | 8 | 7.09e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 2 | 19 | 7.09e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 1 | 19 | 7.09e-01 |
| GO:BP | GO:0038134 | ERBB2-EGFR signaling pathway | 1 | 6 | 7.09e-01 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 1 | 10 | 7.09e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 1 | 17 | 7.09e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 11 | 859 | 7.09e-01 |
| GO:BP | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1 | 17 | 7.09e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 2 | 1147 | 7.09e-01 |
| GO:BP | GO:0070995 | NADPH oxidation | 2 | 3 | 7.09e-01 |
| GO:BP | GO:0021871 | forebrain regionalization | 1 | 8 | 7.09e-01 |
| GO:BP | GO:0006116 | NADH oxidation | 2 | 4 | 7.09e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 3 | 65 | 7.10e-01 |
| GO:BP | GO:1904429 | regulation of t-circle formation | 1 | 5 | 7.10e-01 |
| GO:BP | GO:0034383 | low-density lipoprotein particle clearance | 1 | 17 | 7.10e-01 |
| GO:BP | GO:1990927 | calcium ion regulated lysosome exocytosis | 1 | 3 | 7.10e-01 |
| GO:BP | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing | 1 | 6 | 7.10e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 7.10e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 7.10e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 7.10e-01 |
| GO:BP | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 1 | 2 | 7.10e-01 |
| GO:BP | GO:0045981 | positive regulation of nucleotide metabolic process | 2 | 16 | 7.10e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 1 | 14 | 7.10e-01 |
| GO:BP | GO:0071000 | response to magnetism | 1 | 2 | 7.10e-01 |
| GO:BP | GO:1901529 | positive regulation of anion channel activity | 2 | 2 | 7.10e-01 |
| GO:BP | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | 1 | 12 | 7.10e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 3 | 300 | 7.10e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 1 | 118 | 7.10e-01 |
| GO:BP | GO:0071922 | regulation of cohesin loading | 1 | 2 | 7.10e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 7.10e-01 |
| GO:BP | GO:0006627 | protein processing involved in protein targeting to mitochondrion | 2 | 6 | 7.10e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 12 | 59 | 7.10e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 6 | 31 | 7.10e-01 |
| GO:BP | GO:1900544 | positive regulation of purine nucleotide metabolic process | 2 | 16 | 7.10e-01 |
| GO:BP | GO:0051764 | actin crosslink formation | 2 | 10 | 7.10e-01 |
| GO:BP | GO:1903793 | positive regulation of monoatomic anion transport | 2 | 2 | 7.10e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 1 | 118 | 7.10e-01 |
| GO:BP | GO:1903961 | positive regulation of anion transmembrane transport | 2 | 2 | 7.10e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 12 | 59 | 7.10e-01 |
| GO:BP | GO:0035583 | sequestering of TGFbeta in extracellular matrix | 2 | 3 | 7.10e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 8 | 16 | 7.10e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 2 | 236 | 7.10e-01 |
| GO:BP | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation | 2 | 3 | 7.10e-01 |
| GO:BP | GO:0070126 | mitochondrial translational termination | 2 | 4 | 7.10e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 10 | 30 | 7.10e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 7.10e-01 |
| GO:BP | GO:0006427 | histidyl-tRNA aminoacylation | 1 | 2 | 7.10e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 7.10e-01 |
| GO:BP | GO:0033633 | negative regulation of cell-cell adhesion mediated by integrin | 1 | 2 | 7.10e-01 |
| GO:BP | GO:1903215 | negative regulation of protein targeting to mitochondrion | 3 | 3 | 7.10e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 3 | 10 | 7.10e-01 |
| GO:BP | GO:0043615 | astrocyte cell migration | 2 | 6 | 7.10e-01 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 1 | 11 | 7.11e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 3 | 18 | 7.11e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 1 | 9 | 7.11e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 2 | 44 | 7.11e-01 |
| GO:BP | GO:0034163 | regulation of toll-like receptor 9 signaling pathway | 3 | 6 | 7.11e-01 |
| GO:BP | GO:0060066 | oviduct development | 1 | 4 | 7.11e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 17 | 4539 | 7.11e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 17 | 153 | 7.11e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 110 | 517 | 7.11e-01 |
| GO:BP | GO:0001905 | activation of membrane attack complex | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0001969 | regulation of activation of membrane attack complex | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0006431 | methionyl-tRNA aminoacylation | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0002074 | extraocular skeletal muscle development | 1 | 2 | 7.11e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 7 | 15 | 7.11e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 5 | 771 | 7.12e-01 |
| GO:BP | GO:0101024 | mitotic nuclear membrane organization | 1 | 9 | 7.12e-01 |
| GO:BP | GO:0072109 | glomerular mesangium development | 6 | 13 | 7.12e-01 |
| GO:BP | GO:0007084 | mitotic nuclear membrane reassembly | 1 | 9 | 7.12e-01 |
| GO:BP | GO:0009180 | purine ribonucleoside diphosphate biosynthetic process | 2 | 4 | 7.12e-01 |
| GO:BP | GO:0006172 | ADP biosynthetic process | 2 | 4 | 7.12e-01 |
| GO:BP | GO:0042760 | very long-chain fatty acid catabolic process | 2 | 5 | 7.13e-01 |
| GO:BP | GO:0006482 | protein demethylation | 5 | 27 | 7.13e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 5 | 27 | 7.13e-01 |
| GO:BP | GO:0010626 | negative regulation of Schwann cell proliferation | 2 | 6 | 7.13e-01 |
| GO:BP | GO:0046618 | xenobiotic export from cell | 5 | 8 | 7.13e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 9 | 17 | 7.13e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 8 | 89 | 7.13e-01 |
| GO:BP | GO:1901652 | response to peptide | 28 | 377 | 7.14e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 68 | 148 | 7.14e-01 |
| GO:BP | GO:0061073 | ciliary body morphogenesis | 1 | 3 | 7.14e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 5 | 233 | 7.14e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 1 | 160 | 7.14e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 12 | 114 | 7.14e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 3 | 17 | 7.14e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 10 | 7.14e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 7.14e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 3 | 34 | 7.14e-01 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 1 | 17 | 7.14e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 3 | 36 | 7.15e-01 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 1 | 1 | 7.15e-01 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 1 | 1 | 7.15e-01 |
| GO:BP | GO:1905590 | fibronectin fibril organization | 1 | 1 | 7.15e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 73 | 160 | 7.15e-01 |
| GO:BP | GO:0015840 | urea transport | 1 | 2 | 7.15e-01 |
| GO:BP | GO:0010747 | positive regulation of long-chain fatty acid import across plasma membrane | 1 | 1 | 7.15e-01 |
| GO:BP | GO:0046479 | glycosphingolipid catabolic process | 1 | 10 | 7.15e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 14 | 89 | 7.15e-01 |
| GO:BP | GO:1901654 | response to ketone | 4 | 147 | 7.16e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 7.16e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 7.16e-01 |
| GO:BP | GO:0055062 | phosphate ion homeostasis | 1 | 8 | 7.16e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 5 | 234 | 7.16e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 11 | 740 | 7.16e-01 |
| GO:BP | GO:1903332 | regulation of protein folding | 3 | 8 | 7.16e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 3 | 28 | 7.16e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 5 | 13 | 7.16e-01 |
| GO:BP | GO:2000833 | positive regulation of steroid hormone secretion | 1 | 6 | 7.16e-01 |
| GO:BP | GO:1903713 | asparagine transmembrane transport | 1 | 1 | 7.16e-01 |
| GO:BP | GO:2000487 | positive regulation of glutamine transport | 1 | 2 | 7.16e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 7.16e-01 |
| GO:BP | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH | 1 | 1 | 7.16e-01 |
| GO:BP | GO:0051365 | cellular response to potassium ion starvation | 1 | 1 | 7.16e-01 |
| GO:BP | GO:0043029 | T cell homeostasis | 4 | 27 | 7.16e-01 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 1 | 7 | 7.16e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 7 | 37 | 7.17e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 1 | 25 | 7.17e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 1 | 12 | 7.17e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 9 | 194 | 7.17e-01 |
| GO:BP | GO:0006379 | mRNA cleavage | 5 | 19 | 7.17e-01 |
| GO:BP | GO:0048241 | epinephrine transport | 1 | 4 | 7.17e-01 |
| GO:BP | GO:0021615 | glossopharyngeal nerve morphogenesis | 2 | 2 | 7.18e-01 |
| GO:BP | GO:0015878 | biotin transport | 1 | 3 | 7.18e-01 |
| GO:BP | GO:0008306 | associative learning | 3 | 65 | 7.18e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 2 | 1278 | 7.18e-01 |
| GO:BP | GO:0019532 | oxalate transport | 1 | 5 | 7.18e-01 |
| GO:BP | GO:0015884 | folic acid transport | 1 | 7 | 7.18e-01 |
| GO:BP | GO:1903109 | positive regulation of mitochondrial transcription | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0060766 | negative regulation of androgen receptor signaling pathway | 5 | 12 | 7.18e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 146 | 7.18e-01 |
| GO:BP | GO:0051230 | spindle disassembly | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0051228 | mitotic spindle disassembly | 1 | 2 | 7.18e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 2 | 71 | 7.18e-01 |
| GO:BP | GO:0042308 | negative regulation of protein import into nucleus | 7 | 13 | 7.18e-01 |
| GO:BP | GO:0140719 | constitutive heterochromatin formation | 4 | 10 | 7.19e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 9 | 21 | 7.19e-01 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 5 | 19 | 7.19e-01 |
| GO:BP | GO:0023035 | CD40 signaling pathway | 2 | 11 | 7.19e-01 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 5 | 19 | 7.19e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 7.19e-01 |
| GO:BP | GO:0046449 | creatinine metabolic process | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 3 | 58 | 7.19e-01 |
| GO:BP | GO:0031282 | regulation of guanylate cyclase activity | 1 | 3 | 7.19e-01 |
| GO:BP | GO:0006554 | lysine catabolic process | 1 | 6 | 7.19e-01 |
| GO:BP | GO:2000359 | regulation of binding of sperm to zona pellucida | 2 | 2 | 7.19e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 3 | 48 | 7.19e-01 |
| GO:BP | GO:0006526 | arginine biosynthetic process | 2 | 4 | 7.19e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 2 | 49 | 7.19e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 11 | 31 | 7.19e-01 |
| GO:BP | GO:1904021 | negative regulation of G protein-coupled receptor internalization | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 5 | 234 | 7.19e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 1 | 11 | 7.19e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 1 | 11 | 7.19e-01 |
| GO:BP | GO:1990840 | response to lectin | 1 | 11 | 7.19e-01 |
| GO:BP | GO:0014841 | skeletal muscle satellite cell proliferation | 2 | 10 | 7.20e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 2 | 19 | 7.20e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 6 | 24 | 7.20e-01 |
| GO:BP | GO:0006102 | isocitrate metabolic process | 3 | 6 | 7.20e-01 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 1 | 8 | 7.20e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 7.20e-01 |
| GO:BP | GO:0048880 | sensory system development | 15 | 275 | 7.20e-01 |
| GO:BP | GO:0018125 | peptidyl-cysteine methylation | 1 | 2 | 7.21e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 3 | 13 | 7.21e-01 |
| GO:BP | GO:0061088 | regulation of sequestering of zinc ion | 2 | 4 | 7.21e-01 |
| GO:BP | GO:1903598 | positive regulation of gap junction assembly | 2 | 7 | 7.21e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 3 | 23 | 7.21e-01 |
| GO:BP | GO:0035526 | retrograde transport, plasma membrane to Golgi | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0016553 | base conversion or substitution editing | 1 | 10 | 7.22e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 9 | 22 | 7.22e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 7.22e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 7.23e-01 |
| GO:BP | GO:0062100 | positive regulation of programmed necrotic cell death | 1 | 5 | 7.23e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 5 | 26 | 7.23e-01 |
| GO:BP | GO:0034352 | positive regulation of glial cell apoptotic process | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0001823 | mesonephros development | 3 | 77 | 7.23e-01 |
| GO:BP | GO:0036150 | phosphatidylserine acyl-chain remodeling | 1 | 7 | 7.23e-01 |
| GO:BP | GO:0061868 | hepatic stellate cell migration | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0061869 | regulation of hepatic stellate cell migration | 1 | 2 | 7.23e-01 |
| GO:BP | GO:1900141 | regulation of oligodendrocyte apoptotic process | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0061870 | positive regulation of hepatic stellate cell migration | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0002457 | T cell antigen processing and presentation | 1 | 4 | 7.23e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 10 | 95 | 7.23e-01 |
| GO:BP | GO:2000368 | positive regulation of acrosomal vesicle exocytosis | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 16 | 51 | 7.23e-01 |
| GO:BP | GO:0002159 | desmosome assembly | 2 | 3 | 7.23e-01 |
| GO:BP | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 1 | 16 | 7.23e-01 |
| GO:BP | GO:0045016 | mitochondrial magnesium ion transmembrane transport | 1 | 2 | 7.23e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 9 | 20 | 7.23e-01 |
| GO:BP | GO:0061792 | secretory granule maturation | 1 | 3 | 7.23e-01 |
| GO:BP | GO:1990502 | dense core granule maturation | 1 | 3 | 7.23e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 150 | 7.23e-01 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 1 | 8 | 7.23e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 1 | 83 | 7.23e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 6 | 37 | 7.24e-01 |
| GO:BP | GO:0048861 | leukemia inhibitory factor signaling pathway | 2 | 6 | 7.24e-01 |
| GO:BP | GO:0097502 | mannosylation | 6 | 32 | 7.24e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 3 | 5 | 7.24e-01 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 1 | 8 | 7.24e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 12 | 324 | 7.25e-01 |
| GO:BP | GO:0032288 | myelin assembly | 4 | 19 | 7.25e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 1 | 22 | 7.25e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 3 | 17 | 7.25e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 1 | 25 | 7.25e-01 |
| GO:BP | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 2 | 7.25e-01 |
| GO:BP | GO:0034414 | tRNA 3’-trailer cleavage, endonucleolytic | 1 | 2 | 7.25e-01 |
| GO:BP | GO:0010668 | ectodermal cell differentiation | 3 | 5 | 7.25e-01 |
| GO:BP | GO:0090237 | regulation of arachidonic acid secretion | 1 | 2 | 7.26e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 14 | 29 | 7.26e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 3 | 14 | 7.26e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 3 | 14 | 7.26e-01 |
| GO:BP | GO:0046469 | platelet activating factor metabolic process | 1 | 5 | 7.26e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 1 | 60 | 7.26e-01 |
| GO:BP | GO:0045916 | negative regulation of complement activation | 2 | 8 | 7.26e-01 |
| GO:BP | GO:0048934 | peripheral nervous system neuron differentiation | 5 | 9 | 7.26e-01 |
| GO:BP | GO:0048935 | peripheral nervous system neuron development | 5 | 9 | 7.26e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 7.26e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 10 | 41 | 7.26e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 2 | 18 | 7.26e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 7.26e-01 |
| GO:BP | GO:0006050 | mannosamine metabolic process | 1 | 2 | 7.26e-01 |
| GO:BP | GO:0006051 | N-acetylmannosamine metabolic process | 1 | 2 | 7.26e-01 |
| GO:BP | GO:1905089 | regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 2 | 7.26e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 2 | 61 | 7.26e-01 |
| GO:BP | GO:0071672 | negative regulation of smooth muscle cell chemotaxis | 1 | 2 | 7.26e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 7.26e-01 |
| GO:BP | GO:0032743 | positive regulation of interleukin-2 production | 1 | 20 | 7.26e-01 |
| GO:BP | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 7.26e-01 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 4 | 8 | 7.27e-01 |
| GO:BP | GO:0021730 | trigeminal sensory nucleus development | 1 | 1 | 7.27e-01 |
| GO:BP | GO:0021740 | principal sensory nucleus of trigeminal nerve development | 1 | 1 | 7.27e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 8 | 15 | 7.27e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 7.28e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 7.28e-01 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 1 | 9 | 7.29e-01 |
| GO:BP | GO:0140447 | cytokine precursor processing | 1 | 1 | 7.29e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 4 | 116 | 7.29e-01 |
| GO:BP | GO:0036031 | recruitment of mRNA capping enzyme to RNA polymerase II holoenzyme complex | 1 | 1 | 7.29e-01 |
| GO:BP | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process | 3 | 5 | 7.29e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 7 | 15 | 7.30e-01 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 8 | 17 | 7.30e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 1 | 118 | 7.30e-01 |
| GO:BP | GO:0034772 | histone H4-K20 dimethylation | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 7.30e-01 |
| GO:BP | GO:0033625 | positive regulation of integrin activation | 2 | 6 | 7.30e-01 |
| GO:BP | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 1 | 4 | 7.30e-01 |
| GO:BP | GO:0007254 | JNK cascade | 4 | 138 | 7.30e-01 |
| GO:BP | GO:0010637 | negative regulation of mitochondrial fusion | 3 | 8 | 7.30e-01 |
| GO:BP | GO:0051604 | protein maturation | 8 | 259 | 7.30e-01 |
| GO:BP | GO:1902996 | regulation of neurofibrillary tangle assembly | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 2 | 5 | 7.30e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 4 | 65 | 7.30e-01 |
| GO:BP | GO:1990770 | small intestine smooth muscle contraction | 1 | 1 | 7.30e-01 |
| GO:BP | GO:0038162 | erythropoietin-mediated signaling pathway | 1 | 2 | 7.30e-01 |
| GO:BP | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis | 3 | 7 | 7.30e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 2 | 46 | 7.30e-01 |
| GO:BP | GO:0097102 | endothelial tip cell fate specification | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0061441 | renal artery morphogenesis | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0048859 | formation of anatomical boundary | 3 | 3 | 7.30e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0002478 | antigen processing and presentation of exogenous peptide antigen | 6 | 24 | 7.30e-01 |
| GO:BP | GO:1904386 | response to L-phenylalanine derivative | 3 | 5 | 7.30e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 7.30e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 5 | 11 | 7.30e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 3 | 78 | 7.30e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 2 | 60 | 7.30e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 2 | 60 | 7.30e-01 |
| GO:BP | GO:0035270 | endocrine system development | 5 | 86 | 7.30e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 4 | 605 | 7.31e-01 |
| GO:BP | GO:0099152 | regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane | 1 | 3 | 7.31e-01 |
| GO:BP | GO:0009620 | response to fungus | 1 | 22 | 7.31e-01 |
| GO:BP | GO:1901685 | glutathione derivative metabolic process | 1 | 3 | 7.31e-01 |
| GO:BP | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 1 | 107 | 7.31e-01 |
| GO:BP | GO:1901687 | glutathione derivative biosynthetic process | 1 | 3 | 7.31e-01 |
| GO:BP | GO:1903489 | positive regulation of lactation | 1 | 1 | 7.31e-01 |
| GO:BP | GO:0061032 | visceral serous pericardium development | 1 | 1 | 7.31e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 6 | 438 | 7.31e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 9 | 21 | 7.31e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 20 | 357 | 7.31e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 16 | 748 | 7.31e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 4 | 656 | 7.31e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 4 | 7.32e-01 |
| GO:BP | GO:0036466 | synaptic vesicle recycling via endosome | 2 | 4 | 7.32e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 17 | 4590 | 7.32e-01 |
| GO:BP | GO:0045213 | neurotransmitter receptor metabolic process | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0060857 | establishment of glial blood-brain barrier | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 2 | 188 | 7.32e-01 |
| GO:BP | GO:0071500 | cellular response to nitrosative stress | 3 | 6 | 7.32e-01 |
| GO:BP | GO:0045875 | negative regulation of sister chromatid cohesion | 1 | 5 | 7.32e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 7 | 15 | 7.32e-01 |
| GO:BP | GO:0090192 | regulation of glomerulus development | 1 | 4 | 7.32e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 4 | 65 | 7.32e-01 |
| GO:BP | GO:0072215 | regulation of metanephros development | 1 | 2 | 7.32e-01 |
| GO:BP | GO:1905524 | negative regulation of protein autoubiquitination | 1 | 2 | 7.32e-01 |
| GO:BP | GO:1903850 | regulation of cristae formation | 2 | 4 | 7.32e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 59 | 7.32e-01 |
| GO:BP | GO:0035272 | exocrine system development | 6 | 34 | 7.32e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 7.32e-01 |
| GO:BP | GO:1904688 | regulation of cytoplasmic translational initiation | 3 | 6 | 7.32e-01 |
| GO:BP | GO:0140249 | protein catabolic process at postsynapse | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 7 | 15 | 7.32e-01 |
| GO:BP | GO:0089700 | protein kinase D signaling | 2 | 4 | 7.32e-01 |
| GO:BP | GO:0140246 | protein catabolic process at synapse | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0032000 | positive regulation of fatty acid beta-oxidation | 3 | 9 | 7.32e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 7 | 16 | 7.32e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 7 | 16 | 7.32e-01 |
| GO:BP | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis | 1 | 2 | 7.32e-01 |
| GO:BP | GO:0071359 | cellular response to dsRNA | 7 | 18 | 7.32e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 1 | 9 | 7.32e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 4 | 963 | 7.32e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 1 | 9 | 7.32e-01 |
| GO:BP | GO:0034650 | cortisol metabolic process | 1 | 9 | 7.32e-01 |
| GO:BP | GO:0032351 | negative regulation of hormone metabolic process | 1 | 10 | 7.32e-01 |
| GO:BP | GO:0035771 | interleukin-4-mediated signaling pathway | 2 | 4 | 7.33e-01 |
| GO:BP | GO:1990773 | matrix metallopeptidase secretion | 1 | 5 | 7.33e-01 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 1 | 14 | 7.33e-01 |
| GO:BP | GO:1904464 | regulation of matrix metallopeptidase secretion | 1 | 5 | 7.33e-01 |
| GO:BP | GO:0002115 | store-operated calcium entry | 2 | 15 | 7.33e-01 |
| GO:BP | GO:0072156 | distal tubule morphogenesis | 1 | 3 | 7.33e-01 |
| GO:BP | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching | 2 | 4 | 7.34e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 3 | 7 | 7.34e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 3 | 15 | 7.34e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 3 | 9 | 7.34e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 6 | 13 | 7.34e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 2 | 8 | 7.34e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 2 | 50 | 7.34e-01 |
| GO:BP | GO:0070901 | mitochondrial tRNA methylation | 1 | 4 | 7.34e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 4 | 8 | 7.34e-01 |
| GO:BP | GO:0010273 | detoxification of copper ion | 1 | 8 | 7.35e-01 |
| GO:BP | GO:1990169 | stress response to copper ion | 1 | 8 | 7.35e-01 |
| GO:BP | GO:0035747 | natural killer cell chemotaxis | 1 | 3 | 7.35e-01 |
| GO:BP | GO:1903525 | regulation of membrane tubulation | 3 | 5 | 7.35e-01 |
| GO:BP | GO:2000143 | negative regulation of DNA-templated transcription initiation | 3 | 6 | 7.35e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 2 | 19 | 7.35e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 1 | 67 | 7.36e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 2 | 151 | 7.36e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 11 | 249 | 7.36e-01 |
| GO:BP | GO:0021540 | corpus callosum morphogenesis | 2 | 5 | 7.36e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 6 | 70 | 7.37e-01 |
| GO:BP | GO:0032736 | positive regulation of interleukin-13 production | 1 | 7 | 7.37e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 4 | 47 | 7.37e-01 |
| GO:BP | GO:1904491 | protein localization to ciliary transition zone | 2 | 7 | 7.37e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 9 | 192 | 7.37e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 6 | 19 | 7.38e-01 |
| GO:BP | GO:0061046 | regulation of branching involved in lung morphogenesis | 1 | 3 | 7.38e-01 |
| GO:BP | GO:0048866 | stem cell fate specification | 1 | 2 | 7.38e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 2 | 4 | 7.38e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 1 | 64 | 7.38e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 3 | 8 | 7.38e-01 |
| GO:BP | GO:0140250 | regulation protein catabolic process at synapse | 1 | 5 | 7.38e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 2 | 294 | 7.38e-01 |
| GO:BP | GO:0098908 | regulation of neuronal action potential | 2 | 3 | 7.38e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 1 | 3 | 7.38e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 1 | 3 | 7.38e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 5 | 19 | 7.38e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 8 | 517 | 7.38e-01 |
| GO:BP | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity | 1 | 4 | 7.39e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 1 | 1248 | 7.39e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 7.39e-01 |
| GO:BP | GO:0042351 | ‘de novo’ GDP-L-fucose biosynthetic process | 1 | 2 | 7.39e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 4 | 18 | 7.39e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 4 | 12 | 7.39e-01 |
| GO:BP | GO:0014043 | negative regulation of neuron maturation | 1 | 2 | 7.39e-01 |
| GO:BP | GO:1903860 | negative regulation of dendrite extension | 1 | 2 | 7.39e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 11 | 220 | 7.39e-01 |
| GO:BP | GO:0110104 | mRNA alternative polyadenylation | 1 | 5 | 7.39e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 4 | 276 | 7.39e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 7.39e-01 |
| GO:BP | GO:1905242 | response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 7.40e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 69 | 993 | 7.40e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 2 | 18 | 7.40e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 2 | 49 | 7.40e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 2 | 45 | 7.40e-01 |
| GO:BP | GO:0031016 | pancreas development | 3 | 53 | 7.40e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 4 | 180 | 7.41e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 7.41e-01 |
| GO:BP | GO:0038128 | ERBB2 signaling pathway | 3 | 14 | 7.41e-01 |
| GO:BP | GO:0071550 | death-inducing signaling complex assembly | 1 | 3 | 7.41e-01 |
| GO:BP | GO:2000409 | positive regulation of T cell extravasation | 1 | 2 | 7.41e-01 |
| GO:BP | GO:0035697 | CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.41e-01 |
| GO:BP | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.41e-01 |
| GO:BP | GO:0034381 | plasma lipoprotein particle clearance | 3 | 25 | 7.41e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 7.41e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 3 | 10 | 7.41e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 3 | 10 | 7.41e-01 |
| GO:BP | GO:0061458 | reproductive system development | 12 | 215 | 7.41e-01 |
| GO:BP | GO:0060541 | respiratory system development | 5 | 163 | 7.41e-01 |
| GO:BP | GO:0050996 | positive regulation of lipid catabolic process | 2 | 16 | 7.41e-01 |
| GO:BP | GO:1905772 | positive regulation of mesodermal cell differentiation | 1 | 1 | 7.41e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 12 | 43 | 7.42e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 91 | 1869 | 7.42e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 69 | 994 | 7.42e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 12 | 43 | 7.42e-01 |
| GO:BP | GO:2001279 | regulation of unsaturated fatty acid biosynthetic process | 3 | 7 | 7.42e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 1 | 6 | 7.42e-01 |
| GO:BP | GO:0071867 | response to monoamine | 2 | 62 | 7.42e-01 |
| GO:BP | GO:0035600 | tRNA methylthiolation | 1 | 2 | 7.42e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 2 | 62 | 7.42e-01 |
| GO:BP | GO:1990145 | maintenance of translational fidelity | 1 | 2 | 7.42e-01 |
| GO:BP | GO:0035752 | lysosomal lumen pH elevation | 2 | 3 | 7.42e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 8 | 68 | 7.42e-01 |
| GO:BP | GO:0071926 | endocannabinoid signaling pathway | 3 | 5 | 7.42e-01 |
| GO:BP | GO:0060629 | regulation of homologous chromosome segregation | 1 | 1 | 7.42e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 7.42e-01 |
| GO:BP | GO:0051582 | positive regulation of neurotransmitter uptake | 3 | 4 | 7.42e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 1 | 41 | 7.42e-01 |
| GO:BP | GO:0099525 | presynaptic dense core vesicle exocytosis | 1 | 4 | 7.42e-01 |
| GO:BP | GO:0048675 | axon extension | 4 | 110 | 7.43e-01 |
| GO:BP | GO:0009086 | methionine biosynthetic process | 1 | 10 | 7.43e-01 |
| GO:BP | GO:0021502 | neural fold elevation formation | 1 | 3 | 7.43e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 18 | 36 | 7.43e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 18 | 36 | 7.43e-01 |
| GO:BP | GO:0001575 | globoside metabolic process | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0001576 | globoside biosynthetic process | 1 | 2 | 7.43e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 3 | 14 | 7.43e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 7.43e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 7.43e-01 |
| GO:BP | GO:0002414 | immunoglobulin transcytosis in epithelial cells | 1 | 2 | 7.43e-01 |
| GO:BP | GO:1905424 | regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation | 2 | 8 | 7.43e-01 |
| GO:BP | GO:1901380 | negative regulation of potassium ion transmembrane transport | 2 | 18 | 7.43e-01 |
| GO:BP | GO:1905426 | positive regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0070315 | G1 to G0 transition involved in cell differentiation | 2 | 4 | 7.43e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 3 | 19 | 7.43e-01 |
| GO:BP | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0045629 | negative regulation of T-helper 2 cell differentiation | 1 | 3 | 7.44e-01 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 2 | 8 | 7.44e-01 |
| GO:BP | GO:0003084 | positive regulation of systemic arterial blood pressure | 1 | 7 | 7.44e-01 |
| GO:BP | GO:2000670 | positive regulation of dendritic cell apoptotic process | 1 | 2 | 7.44e-01 |
| GO:BP | GO:0007258 | JUN phosphorylation | 1 | 2 | 7.44e-01 |
| GO:BP | GO:0006545 | glycine biosynthetic process | 1 | 4 | 7.45e-01 |
| GO:BP | GO:0021511 | spinal cord patterning | 1 | 9 | 7.45e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 3 | 57 | 7.45e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 5 | 20 | 7.45e-01 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 1 | 12 | 7.45e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 22 | 344 | 7.45e-01 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 1 | 12 | 7.45e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 11 | 26 | 7.45e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 4 | 15 | 7.45e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 2 | 50 | 7.45e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 2 | 66 | 7.46e-01 |
| GO:BP | GO:1990258 | histone glutamine methylation | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 9 | 24 | 7.46e-01 |
| GO:BP | GO:1904567 | response to wortmannin | 1 | 2 | 7.46e-01 |
| GO:BP | GO:1904568 | cellular response to wortmannin | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0061086 | negative regulation of histone H3-K27 methylation | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0014075 | response to amine | 2 | 34 | 7.46e-01 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 2 | 8 | 7.46e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 3 | 47 | 7.46e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 7.46e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 52 | 294 | 7.46e-01 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 3 | 39 | 7.46e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 25 | 57 | 7.46e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 8 | 186 | 7.46e-01 |
| GO:BP | GO:0043382 | positive regulation of memory T cell differentiation | 1 | 3 | 7.46e-01 |
| GO:BP | GO:0034444 | regulation of plasma lipoprotein oxidation | 1 | 1 | 7.46e-01 |
| GO:BP | GO:0034445 | negative regulation of plasma lipoprotein oxidation | 1 | 1 | 7.46e-01 |
| GO:BP | GO:0046501 | protoporphyrinogen IX metabolic process | 2 | 11 | 7.46e-01 |
| GO:BP | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c | 1 | 7 | 7.46e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 2 | 15 | 7.46e-01 |
| GO:BP | GO:0019551 | glutamate catabolic process to 2-oxoglutarate | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0019550 | glutamate catabolic process to aspartate | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 1 | 27 | 7.46e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 2 | 83 | 7.46e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 1 | 27 | 7.46e-01 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 2 | 8 | 7.46e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 6 | 102 | 7.46e-01 |
| GO:BP | GO:0099040 | ceramide translocation | 1 | 2 | 7.46e-01 |
| GO:BP | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 5 | 34 | 7.47e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 7.47e-01 |
| GO:BP | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane | 1 | 19 | 7.47e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 7.47e-01 |
| GO:BP | GO:2001223 | negative regulation of neuron migration | 3 | 8 | 7.47e-01 |
| GO:BP | GO:0035349 | coenzyme A transmembrane transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0035350 | FAD transmembrane transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 3 | 28 | 7.47e-01 |
| GO:BP | GO:0015880 | coenzyme A transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:2000642 | negative regulation of early endosome to late endosome transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0080121 | AMP transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0015883 | FAD transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 2 | 11 | 7.47e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 2 | 21 | 7.47e-01 |
| GO:BP | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus | 2 | 8 | 7.47e-01 |
| GO:BP | GO:0071106 | adenosine 3’,5’-bisphosphate transmembrane transport | 1 | 2 | 7.47e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 29 | 55 | 7.47e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 22 | 345 | 7.47e-01 |
| GO:BP | GO:0030422 | siRNA processing | 1 | 7 | 7.47e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 14 | 62 | 7.47e-01 |
| GO:BP | GO:1905050 | positive regulation of metallopeptidase activity | 3 | 5 | 7.48e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 2 | 19 | 7.48e-01 |
| GO:BP | GO:0060399 | positive regulation of growth hormone receptor signaling pathway | 1 | 2 | 7.48e-01 |
| GO:BP | GO:1903044 | protein localization to membrane raft | 4 | 8 | 7.48e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 2 | 19 | 7.48e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 7 | 448 | 7.48e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 2 | 211 | 7.48e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 4 | 26 | 7.49e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 1 | 64 | 7.49e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 3 | 12 | 7.49e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 9 | 23 | 7.49e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 7.49e-01 |
| GO:BP | GO:0034115 | negative regulation of heterotypic cell-cell adhesion | 2 | 6 | 7.49e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 7.49e-01 |
| GO:BP | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation | 2 | 6 | 7.49e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 7.49e-01 |
| GO:BP | GO:0016560 | protein import into peroxisome matrix, docking | 2 | 3 | 7.49e-01 |
| GO:BP | GO:0019064 | fusion of virus membrane with host plasma membrane | 1 | 4 | 7.49e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 5 | 83 | 7.49e-01 |
| GO:BP | GO:0039663 | membrane fusion involved in viral entry into host cell | 1 | 4 | 7.49e-01 |
| GO:BP | GO:0006423 | cysteinyl-tRNA aminoacylation | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0009750 | response to fructose | 1 | 3 | 7.49e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 9 | 620 | 7.49e-01 |
| GO:BP | GO:0051012 | microtubule sliding | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 14 | 114 | 7.49e-01 |
| GO:BP | GO:0048817 | negative regulation of hair follicle maturation | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 9 | 191 | 7.49e-01 |
| GO:BP | GO:1902811 | positive regulation of skeletal muscle fiber differentiation | 1 | 2 | 7.49e-01 |
| GO:BP | GO:2001256 | regulation of store-operated calcium entry | 1 | 11 | 7.50e-01 |
| GO:BP | GO:0060152 | microtubule-based peroxisome localization | 1 | 2 | 7.50e-01 |
| GO:BP | GO:0060151 | peroxisome localization | 1 | 2 | 7.50e-01 |
| GO:BP | GO:0051459 | regulation of corticotropin secretion | 1 | 3 | 7.51e-01 |
| GO:BP | GO:0061302 | smooth muscle cell-matrix adhesion | 1 | 5 | 7.51e-01 |
| GO:BP | GO:0002475 | antigen processing and presentation via MHC class Ib | 2 | 8 | 7.51e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 1 | 35 | 7.51e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 14 | 43 | 7.51e-01 |
| GO:BP | GO:1903727 | positive regulation of phospholipid metabolic process | 1 | 9 | 7.51e-01 |
| GO:BP | GO:2001138 | regulation of phospholipid transport | 1 | 12 | 7.51e-01 |
| GO:BP | GO:0035483 | gastric emptying | 1 | 2 | 7.51e-01 |
| GO:BP | GO:1902370 | regulation of tRNA catabolic process | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 10 | 209 | 7.51e-01 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 4 | 17 | 7.51e-01 |
| GO:BP | GO:0036416 | tRNA stabilization | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0032484 | Ral protein signal transduction | 1 | 5 | 7.51e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 13 | 170 | 7.51e-01 |
| GO:BP | GO:1902371 | negative regulation of tRNA catabolic process | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0036415 | regulation of tRNA stability | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 4 | 13 | 7.51e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 1 | 25 | 7.51e-01 |
| GO:BP | GO:0043007 | maintenance of rDNA | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0061196 | fungiform papilla development | 2 | 5 | 7.51e-01 |
| GO:BP | GO:0042661 | regulation of mesodermal cell fate specification | 1 | 4 | 7.51e-01 |
| GO:BP | GO:0019058 | viral life cycle | 2 | 244 | 7.51e-01 |
| GO:BP | GO:1903326 | regulation of tRNA metabolic process | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 8 | 77 | 7.51e-01 |
| GO:BP | GO:0007340 | acrosome reaction | 3 | 20 | 7.51e-01 |
| GO:BP | GO:1903327 | negative regulation of tRNA metabolic process | 1 | 2 | 7.51e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 68 | 150 | 7.51e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 6 | 42 | 7.51e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 1 | 11 | 7.51e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 4 | 22 | 7.51e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 1 | 87 | 7.51e-01 |
| GO:BP | GO:1905523 | positive regulation of macrophage migration | 3 | 13 | 7.51e-01 |
| GO:BP | GO:0002544 | chronic inflammatory response | 1 | 6 | 7.51e-01 |
| GO:BP | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization | 1 | 4 | 7.52e-01 |
| GO:BP | GO:0010635 | regulation of mitochondrial fusion | 3 | 11 | 7.52e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 7.52e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 1 | 10 | 7.52e-01 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 1 | 17 | 7.52e-01 |
| GO:BP | GO:0099022 | vesicle tethering | 12 | 31 | 7.52e-01 |
| GO:BP | GO:0009566 | fertilization | 1 | 93 | 7.52e-01 |
| GO:BP | GO:0033383 | geranyl diphosphate metabolic process | 1 | 3 | 7.52e-01 |
| GO:BP | GO:0071395 | cellular response to jasmonic acid stimulus | 2 | 3 | 7.52e-01 |
| GO:BP | GO:0009753 | response to jasmonic acid | 2 | 3 | 7.52e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4048 | 7.52e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 2 | 5 | 7.52e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 1 | 19 | 7.53e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 2 | 14 | 7.53e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 10 | 92 | 7.53e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 1 | 71 | 7.53e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 1 | 71 | 7.53e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 3 | 14 | 7.53e-01 |
| GO:BP | GO:0018277 | protein deamination | 1 | 2 | 7.53e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1315 | 7.53e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 1 | 1315 | 7.53e-01 |
| GO:BP | GO:0043950 | positive regulation of cAMP-mediated signaling | 3 | 5 | 7.53e-01 |
| GO:BP | GO:0030002 | intracellular monoatomic anion homeostasis | 1 | 7 | 7.53e-01 |
| GO:BP | GO:0030644 | intracellular chloride ion homeostasis | 1 | 7 | 7.53e-01 |
| GO:BP | GO:0006953 | acute-phase response | 2 | 18 | 7.53e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 7.53e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 4 | 212 | 7.53e-01 |
| GO:BP | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 11 | 131 | 7.54e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 1 | 39 | 7.54e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 1 | 39 | 7.54e-01 |
| GO:BP | GO:0070391 | response to lipoteichoic acid | 2 | 6 | 7.54e-01 |
| GO:BP | GO:0071223 | cellular response to lipoteichoic acid | 2 | 6 | 7.54e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 8 | 14 | 7.54e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 8 | 17 | 7.54e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1 | 1322 | 7.54e-01 |
| GO:BP | GO:0071475 | cellular hyperosmotic salinity response | 2 | 6 | 7.54e-01 |
| GO:BP | GO:0051891 | positive regulation of cardioblast differentiation | 1 | 4 | 7.54e-01 |
| GO:BP | GO:0071712 | ER-associated misfolded protein catabolic process | 5 | 12 | 7.54e-01 |
| GO:BP | GO:0030974 | thiamine pyrophosphate transmembrane transport | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0021524 | visceral motor neuron differentiation | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0072098 | anterior/posterior pattern specification involved in kidney development | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0061047 | positive regulation of branching involved in lung morphogenesis | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0061227 | pattern specification involved in mesonephros development | 1 | 2 | 7.54e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 7.54e-01 |
| GO:BP | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0045626 | negative regulation of T-helper 1 cell differentiation | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0048389 | intermediate mesoderm development | 1 | 2 | 7.54e-01 |
| GO:BP | GO:1903721 | positive regulation of I-kappaB phosphorylation | 1 | 5 | 7.54e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 2 | 27 | 7.54e-01 |
| GO:BP | GO:0051562 | negative regulation of mitochondrial calcium ion concentration | 1 | 2 | 7.55e-01 |
| GO:BP | GO:0015789 | UDP-N-acetylgalactosamine transmembrane transport | 2 | 4 | 7.55e-01 |
| GO:BP | GO:0015787 | UDP-glucuronic acid transmembrane transport | 2 | 4 | 7.55e-01 |
| GO:BP | GO:0016556 | mRNA modification | 9 | 29 | 7.55e-01 |
| GO:BP | GO:0070257 | positive regulation of mucus secretion | 1 | 4 | 7.55e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 7.55e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 2 | 216 | 7.55e-01 |
| GO:BP | GO:0099500 | vesicle fusion to plasma membrane | 1 | 19 | 7.55e-01 |
| GO:BP | GO:2000110 | negative regulation of macrophage apoptotic process | 2 | 3 | 7.55e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 7.55e-01 |
| GO:BP | GO:0035437 | maintenance of protein localization in endoplasmic reticulum | 5 | 11 | 7.55e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 3 | 21 | 7.55e-01 |
| GO:BP | GO:0035502 | metanephric part of ureteric bud development | 1 | 3 | 7.55e-01 |
| GO:BP | GO:0072284 | metanephric S-shaped body morphogenesis | 1 | 4 | 7.55e-01 |
| GO:BP | GO:0071071 | regulation of phospholipid biosynthetic process | 7 | 15 | 7.55e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 3 | 12 | 7.55e-01 |
| GO:BP | GO:0015918 | sterol transport | 4 | 92 | 7.55e-01 |
| GO:BP | GO:1904338 | regulation of dopaminergic neuron differentiation | 1 | 7 | 7.55e-01 |
| GO:BP | GO:0044597 | daunorubicin metabolic process | 4 | 7 | 7.55e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 2 | 103 | 7.55e-01 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 3 | 27 | 7.56e-01 |
| GO:BP | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 2 | 7.56e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 1 | 67 | 7.56e-01 |
| GO:BP | GO:0071386 | cellular response to corticosterone stimulus | 1 | 1 | 7.56e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 2 | 11 | 7.56e-01 |
| GO:BP | GO:0014732 | skeletal muscle atrophy | 1 | 5 | 7.56e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 7.57e-01 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 1 | 12 | 7.57e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 5 | 15 | 7.57e-01 |
| GO:BP | GO:0071420 | cellular response to histamine | 3 | 4 | 7.57e-01 |
| GO:BP | GO:0016183 | synaptic vesicle coating | 2 | 4 | 7.58e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 16 | 28 | 7.58e-01 |
| GO:BP | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0006788 | heme oxidation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0006895 | Golgi to endosome transport | 6 | 17 | 7.58e-01 |
| GO:BP | GO:0031627 | telomeric loop formation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 31 | 105 | 7.58e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 7.58e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 2 | 45 | 7.58e-01 |
| GO:BP | GO:0070173 | regulation of enamel mineralization | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 2 | 122 | 7.59e-01 |
| GO:BP | GO:0048631 | regulation of skeletal muscle tissue growth | 2 | 7 | 7.59e-01 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 7.59e-01 |
| GO:BP | GO:0002890 | negative regulation of immunoglobulin mediated immune response | 4 | 9 | 7.59e-01 |
| GO:BP | GO:0002713 | negative regulation of B cell mediated immunity | 4 | 9 | 7.59e-01 |
| GO:BP | GO:0033239 | negative regulation of amine metabolic process | 2 | 2 | 7.59e-01 |
| GO:BP | GO:0010746 | regulation of long-chain fatty acid import across plasma membrane | 2 | 5 | 7.59e-01 |
| GO:BP | GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1 | 8 | 7.59e-01 |
| GO:BP | GO:0014010 | Schwann cell proliferation | 2 | 7 | 7.59e-01 |
| GO:BP | GO:0010624 | regulation of Schwann cell proliferation | 2 | 7 | 7.59e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 2 | 13 | 7.59e-01 |
| GO:BP | GO:0044793 | negative regulation by host of viral process | 2 | 10 | 7.59e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 4 | 27 | 7.59e-01 |
| GO:BP | GO:0046833 | positive regulation of RNA export from nucleus | 2 | 5 | 7.59e-01 |
| GO:BP | GO:0006772 | thiamine metabolic process | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0008300 | isoprenoid catabolic process | 3 | 5 | 7.59e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 10 | 93 | 7.59e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 7.59e-01 |
| GO:BP | GO:0033572 | transferrin transport | 4 | 8 | 7.59e-01 |
| GO:BP | GO:0140354 | lipid import into cell | 1 | 15 | 7.59e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 1 | 69 | 7.59e-01 |
| GO:BP | GO:0048731 | system development | 2 | 2901 | 7.59e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 3 | 213 | 7.59e-01 |
| GO:BP | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 1 | 9 | 7.59e-01 |
| GO:BP | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway | 1 | 3 | 7.59e-01 |
| GO:BP | GO:0038020 | insulin receptor recycling | 1 | 4 | 7.59e-01 |
| GO:BP | GO:1901334 | lactone metabolic process | 1 | 9 | 7.59e-01 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 1 | 12 | 7.59e-01 |
| GO:BP | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | 2 | 6 | 7.59e-01 |
| GO:BP | GO:1900363 | regulation of mRNA polyadenylation | 2 | 11 | 7.59e-01 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 1 | 31 | 7.59e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 1 | 124 | 7.59e-01 |
| GO:BP | GO:0016598 | protein arginylation | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0048073 | regulation of eye pigmentation | 1 | 2 | 7.59e-01 |
| GO:BP | GO:1904245 | regulation of polynucleotide adenylyltransferase activity | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 5 | 27 | 7.59e-01 |
| GO:BP | GO:0035247 | peptidyl-arginine omega-N-methylation | 2 | 3 | 7.59e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 2 | 24 | 7.60e-01 |
| GO:BP | GO:1990822 | basic amino acid transmembrane transport | 8 | 14 | 7.60e-01 |
| GO:BP | GO:0089709 | L-histidine transmembrane transport | 2 | 5 | 7.60e-01 |
| GO:BP | GO:0001974 | blood vessel remodeling | 3 | 32 | 7.60e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 2 | 29 | 7.60e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 7.60e-01 |
| GO:BP | GO:0033198 | response to ATP | 2 | 24 | 7.60e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 7.60e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 7.60e-01 |
| GO:BP | GO:0042048 | olfactory behavior | 3 | 7 | 7.60e-01 |
| GO:BP | GO:0046726 | positive regulation by virus of viral protein levels in host cell | 2 | 4 | 7.60e-01 |
| GO:BP | GO:0043508 | negative regulation of JUN kinase activity | 6 | 13 | 7.60e-01 |
| GO:BP | GO:0048342 | paraxial mesodermal cell differentiation | 1 | 1 | 7.60e-01 |
| GO:BP | GO:0048343 | paraxial mesodermal cell fate commitment | 1 | 1 | 7.60e-01 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 4 | 14 | 7.60e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 5 | 34 | 7.60e-01 |
| GO:BP | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 1 | 6 | 7.60e-01 |
| GO:BP | GO:0006204 | IMP catabolic process | 2 | 3 | 7.60e-01 |
| GO:BP | GO:0046054 | dGMP metabolic process | 2 | 2 | 7.60e-01 |
| GO:BP | GO:0044062 | regulation of excretion | 1 | 12 | 7.60e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 7 | 105 | 7.60e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 35 | 7.60e-01 |
| GO:BP | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0120127 | response to zinc ion starvation | 1 | 6 | 7.60e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 7.60e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 2 | 21 | 7.60e-01 |
| GO:BP | GO:1905550 | regulation of protein localization to endoplasmic reticulum | 2 | 4 | 7.60e-01 |
| GO:BP | GO:0034224 | cellular response to zinc ion starvation | 1 | 6 | 7.60e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 36 | 1939 | 7.61e-01 |
| GO:BP | GO:2000546 | positive regulation of endothelial cell chemotaxis to fibroblast growth factor | 2 | 3 | 7.61e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 7.61e-01 |
| GO:BP | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor | 2 | 3 | 7.61e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 676 | 7.61e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 2 | 7 | 7.61e-01 |
| GO:BP | GO:0035332 | positive regulation of hippo signaling | 4 | 6 | 7.61e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 3 | 104 | 7.61e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 3 | 104 | 7.61e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 12 | 221 | 7.61e-01 |
| GO:BP | GO:0090386 | phagosome maturation involved in apoptotic cell clearance | 1 | 2 | 7.61e-01 |
| GO:BP | GO:0090387 | phagolysosome assembly involved in apoptotic cell clearance | 1 | 2 | 7.61e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 2 | 162 | 7.61e-01 |
| GO:BP | GO:2000436 | positive regulation of protein neddylation | 1 | 5 | 7.61e-01 |
| GO:BP | GO:0016036 | cellular response to phosphate starvation | 1 | 2 | 7.61e-01 |
| GO:BP | GO:0010359 | regulation of anion channel activity | 4 | 5 | 7.61e-01 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 7.62e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 59 | 197 | 7.62e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 3 | 42 | 7.62e-01 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 6 | 20 | 7.62e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 3 | 70 | 7.62e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 8 | 41 | 7.63e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 2 | 43 | 7.63e-01 |
| GO:BP | GO:0099118 | microtubule-based protein transport | 1 | 14 | 7.63e-01 |
| GO:BP | GO:0098840 | protein transport along microtubule | 1 | 14 | 7.63e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 2 | 31 | 7.63e-01 |
| GO:BP | GO:0071800 | podosome assembly | 3 | 13 | 7.63e-01 |
| GO:BP | GO:2000467 | positive regulation of glycogen (starch) synthase activity | 2 | 4 | 7.63e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 1 | 2 | 7.63e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 26 | 50 | 7.63e-01 |
| GO:BP | GO:0002678 | positive regulation of chronic inflammatory response | 1 | 1 | 7.63e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 3 | 7 | 7.64e-01 |
| GO:BP | GO:0006701 | progesterone biosynthetic process | 1 | 5 | 7.64e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 4 | 64 | 7.64e-01 |
| GO:BP | GO:0006677 | glycosylceramide metabolic process | 1 | 13 | 7.64e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 8 | 540 | 7.64e-01 |
| GO:BP | GO:0007418 | ventral midline development | 1 | 4 | 7.64e-01 |
| GO:BP | GO:0007597 | blood coagulation, intrinsic pathway | 1 | 3 | 7.64e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 5 | 15 | 7.65e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 3 | 49 | 7.65e-01 |
| GO:BP | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation | 2 | 4 | 7.65e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 3 | 20 | 7.65e-01 |
| GO:BP | GO:0021644 | vagus nerve morphogenesis | 1 | 1 | 7.65e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 9 | 38 | 7.65e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 16 | 427 | 7.65e-01 |
| GO:BP | GO:0060155 | platelet dense granule organization | 1 | 23 | 7.66e-01 |
| GO:BP | GO:0002395 | immune response in nasopharyngeal-associated lymphoid tissue | 1 | 1 | 7.66e-01 |
| GO:BP | GO:0002387 | immune response in gut-associated lymphoid tissue | 1 | 1 | 7.66e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 4 | 154 | 7.66e-01 |
| GO:BP | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process | 2 | 4 | 7.66e-01 |
| GO:BP | GO:0006909 | phagocytosis | 2 | 148 | 7.66e-01 |
| GO:BP | GO:0140331 | aminophospholipid translocation | 2 | 6 | 7.66e-01 |
| GO:BP | GO:1900147 | regulation of Schwann cell migration | 1 | 4 | 7.66e-01 |
| GO:BP | GO:0006667 | sphinganine metabolic process | 1 | 2 | 7.66e-01 |
| GO:BP | GO:0070667 | negative regulation of mast cell proliferation | 1 | 2 | 7.66e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 7.66e-01 |
| GO:BP | GO:0033490 | cholesterol biosynthetic process via lathosterol | 2 | 4 | 7.66e-01 |
| GO:BP | GO:0033489 | cholesterol biosynthetic process via desmosterol | 2 | 4 | 7.66e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 17 | 46 | 7.66e-01 |
| GO:BP | GO:0046203 | spermidine catabolic process | 1 | 2 | 7.66e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 11 | 22 | 7.66e-01 |
| GO:BP | GO:0033206 | meiotic cytokinesis | 1 | 7 | 7.66e-01 |
| GO:BP | GO:0036135 | Schwann cell migration | 1 | 4 | 7.66e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 1 | 13 | 7.66e-01 |
| GO:BP | GO:0035685 | helper T cell diapedesis | 1 | 2 | 7.66e-01 |
| GO:BP | GO:1990839 | response to endothelin | 1 | 2 | 7.66e-01 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 3 | 9 | 7.66e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 2 | 32 | 7.67e-01 |
| GO:BP | GO:0006258 | UDP-glucose catabolic process | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 1 | 49 | 7.67e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 2 | 21 | 7.67e-01 |
| GO:BP | GO:0003332 | negative regulation of extracellular matrix constituent secretion | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0061723 | glycophagy | 2 | 4 | 7.67e-01 |
| GO:BP | GO:1903431 | positive regulation of cell maturation | 1 | 6 | 7.67e-01 |
| GO:BP | GO:2000077 | negative regulation of type B pancreatic cell development | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0036265 | RNA (guanine-N7)-methylation | 3 | 6 | 7.67e-01 |
| GO:BP | GO:2001027 | negative regulation of endothelial cell chemotaxis | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 7.67e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 1 | 28 | 7.68e-01 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 2 | 29 | 7.68e-01 |
| GO:BP | GO:0034462 | small-subunit processome assembly | 1 | 3 | 7.68e-01 |
| GO:BP | GO:1904430 | negative regulation of t-circle formation | 1 | 2 | 7.68e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 1 | 64 | 7.68e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 5 | 75 | 7.68e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 1 | 10 | 7.68e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 7.68e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 3 | 14 | 7.68e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 3 | 14 | 7.68e-01 |
| GO:BP | GO:0003400 | regulation of COPII vesicle coating | 2 | 3 | 7.68e-01 |
| GO:BP | GO:0006850 | mitochondrial pyruvate transmembrane transport | 1 | 2 | 7.68e-01 |
| GO:BP | GO:2000354 | regulation of ovarian follicle development | 2 | 5 | 7.68e-01 |
| GO:BP | GO:0048213 | Golgi vesicle prefusion complex stabilization | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 3 | 4 | 7.69e-01 |
| GO:BP | GO:0045654 | positive regulation of megakaryocyte differentiation | 3 | 9 | 7.69e-01 |
| GO:BP | GO:2000815 | regulation of mRNA stability involved in response to oxidative stress | 1 | 2 | 7.69e-01 |
| GO:BP | GO:0021855 | hypothalamus cell migration | 3 | 6 | 7.69e-01 |
| GO:BP | GO:0015770 | sucrose transport | 2 | 4 | 7.69e-01 |
| GO:BP | GO:0015766 | disaccharide transport | 2 | 4 | 7.69e-01 |
| GO:BP | GO:0043306 | positive regulation of mast cell degranulation | 4 | 10 | 7.69e-01 |
| GO:BP | GO:0033008 | positive regulation of mast cell activation involved in immune response | 4 | 10 | 7.69e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 1 | 12 | 7.69e-01 |
| GO:BP | GO:0015772 | oligosaccharide transport | 2 | 4 | 7.69e-01 |
| GO:BP | GO:0048521 | negative regulation of behavior | 2 | 5 | 7.69e-01 |
| GO:BP | GO:1903974 | positive regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 1 | 7.69e-01 |
| GO:BP | GO:1903971 | positive regulation of response to macrophage colony-stimulating factor | 1 | 1 | 7.69e-01 |
| GO:BP | GO:1905602 | positive regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 2 | 7.69e-01 |
| GO:BP | GO:0032707 | negative regulation of interleukin-23 production | 1 | 2 | 7.69e-01 |
| GO:BP | GO:0043587 | tongue morphogenesis | 3 | 7 | 7.69e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 2 | 317 | 7.70e-01 |
| GO:BP | GO:0046950 | cellular ketone body metabolic process | 2 | 7 | 7.70e-01 |
| GO:BP | GO:0090290 | positive regulation of osteoclast proliferation | 1 | 1 | 7.70e-01 |
| GO:BP | GO:0031529 | ruffle organization | 6 | 45 | 7.70e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 3 | 30 | 7.70e-01 |
| GO:BP | GO:0032849 | positive regulation of cellular pH reduction | 1 | 2 | 7.70e-01 |
| GO:BP | GO:1903942 | positive regulation of respiratory gaseous exchange | 1 | 1 | 7.70e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 10 | 22 | 7.70e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 7.70e-01 |
| GO:BP | GO:1903527 | positive regulation of membrane tubulation | 2 | 3 | 7.71e-01 |
| GO:BP | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 1 | 5 | 7.71e-01 |
| GO:BP | GO:1904829 | regulation of aortic smooth muscle cell differentiation | 1 | 2 | 7.71e-01 |
| GO:BP | GO:1904831 | positive regulation of aortic smooth muscle cell differentiation | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0035887 | aortic smooth muscle cell differentiation | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0010155 | regulation of proton transport | 1 | 18 | 7.71e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 7 | 16 | 7.71e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 7.71e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 4 | 64 | 7.71e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 6 | 478 | 7.71e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 1 | 93 | 7.71e-01 |
| GO:BP | GO:0031554 | regulation of termination of DNA-templated transcription | 2 | 4 | 7.71e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 8 | 22 | 7.71e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 10 | 23 | 7.71e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1 | 1441 | 7.71e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 3 | 13 | 7.71e-01 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 2 | 17 | 7.71e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 1 | 6 | 7.71e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 1 | 11 | 7.71e-01 |
| GO:BP | GO:0072717 | cellular response to actinomycin D | 1 | 4 | 7.71e-01 |
| GO:BP | GO:0046549 | retinal cone cell development | 2 | 5 | 7.71e-01 |
| GO:BP | GO:0015876 | acetyl-CoA transport | 1 | 1 | 7.71e-01 |
| GO:BP | GO:0045779 | negative regulation of bone resorption | 1 | 7 | 7.71e-01 |
| GO:BP | GO:0060931 | sinoatrial node cell development | 2 | 4 | 7.71e-01 |
| GO:BP | GO:0038065 | collagen-activated signaling pathway | 1 | 13 | 7.71e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 1 | 35 | 7.71e-01 |
| GO:BP | GO:0002838 | negative regulation of immune response to tumor cell | 2 | 2 | 7.71e-01 |
| GO:BP | GO:0060729 | intestinal epithelial structure maintenance | 1 | 4 | 7.71e-01 |
| GO:BP | GO:0002835 | negative regulation of response to tumor cell | 2 | 2 | 7.71e-01 |
| GO:BP | GO:0070948 | regulation of neutrophil mediated cytotoxicity | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0009136 | purine nucleoside diphosphate biosynthetic process | 2 | 5 | 7.72e-01 |
| GO:BP | GO:0045898 | regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 7 | 7.72e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 3 | 16 | 7.72e-01 |
| GO:BP | GO:0061017 | hepatoblast differentiation | 1 | 1 | 7.72e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 4 | 157 | 7.72e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 1 | 14 | 7.72e-01 |
| GO:BP | GO:0048014 | Tie signaling pathway | 2 | 3 | 7.73e-01 |
| GO:BP | GO:2000416 | regulation of eosinophil migration | 2 | 2 | 7.73e-01 |
| GO:BP | GO:0051598 | meiotic recombination checkpoint signaling | 1 | 2 | 7.73e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 2 | 10 | 7.73e-01 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 3 | 20 | 7.73e-01 |
| GO:BP | GO:0046167 | glycerol-3-phosphate biosynthetic process | 1 | 2 | 7.73e-01 |
| GO:BP | GO:0019541 | propionate metabolic process | 1 | 2 | 7.73e-01 |
| GO:BP | GO:0001555 | oocyte growth | 1 | 2 | 7.73e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 1 | 15 | 7.74e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 1 | 15 | 7.74e-01 |
| GO:BP | GO:0071461 | cellular response to redox state | 1 | 1 | 7.74e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 1 | 15 | 7.74e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 3 | 29 | 7.74e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 9 | 194 | 7.74e-01 |
| GO:BP | GO:1903226 | positive regulation of endodermal cell differentiation | 1 | 2 | 7.74e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 7.74e-01 |
| GO:BP | GO:0042749 | regulation of circadian sleep/wake cycle | 2 | 9 | 7.74e-01 |
| GO:BP | GO:0021551 | central nervous system morphogenesis | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0005984 | disaccharide metabolic process | 1 | 5 | 7.74e-01 |
| GO:BP | GO:0090241 | negative regulation of histone H4 acetylation | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 7 | 82 | 7.74e-01 |
| GO:BP | GO:1990044 | protein localization to lipid droplet | 2 | 4 | 7.75e-01 |
| GO:BP | GO:0030309 | poly-N-acetyllactosamine metabolic process | 3 | 6 | 7.75e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 21 | 50 | 7.75e-01 |
| GO:BP | GO:0001553 | luteinization | 4 | 10 | 7.75e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 5 | 250 | 7.75e-01 |
| GO:BP | GO:0001845 | phagolysosome assembly | 1 | 14 | 7.75e-01 |
| GO:BP | GO:0009635 | response to herbicide | 1 | 3 | 7.75e-01 |
| GO:BP | GO:0030311 | poly-N-acetyllactosamine biosynthetic process | 2 | 5 | 7.75e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 3 | 71 | 7.75e-01 |
| GO:BP | GO:0043335 | protein unfolding | 1 | 5 | 7.75e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4455 | 7.76e-01 |
| GO:BP | GO:0071105 | response to interleukin-11 | 1 | 1 | 7.76e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 3 | 20 | 7.76e-01 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 1 | 13 | 7.76e-01 |
| GO:BP | GO:0099011 | neuronal dense core vesicle exocytosis | 1 | 4 | 7.77e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 1 | 18 | 7.77e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 20 | 512 | 7.77e-01 |
| GO:BP | GO:2000510 | positive regulation of dendritic cell chemotaxis | 1 | 3 | 7.77e-01 |
| GO:BP | GO:1904022 | positive regulation of G protein-coupled receptor internalization | 1 | 1 | 7.77e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 3 | 4579 | 7.77e-01 |
| GO:BP | GO:0051412 | response to corticosterone | 2 | 10 | 7.77e-01 |
| GO:BP | GO:0006668 | sphinganine-1-phosphate metabolic process | 1 | 2 | 7.77e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 88 | 2065 | 7.77e-01 |
| GO:BP | GO:0036261 | 7-methylguanosine cap hypermethylation | 1 | 8 | 7.77e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 7.77e-01 |
| GO:BP | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.77e-01 |
| GO:BP | GO:1904237 | positive regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.77e-01 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 1 | 7 | 7.77e-01 |
| GO:BP | GO:0120193 | tight junction organization | 4 | 52 | 7.77e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 2 | 61 | 7.77e-01 |
| GO:BP | GO:0030241 | skeletal muscle myosin thick filament assembly | 1 | 3 | 7.77e-01 |
| GO:BP | GO:0071688 | striated muscle myosin thick filament assembly | 1 | 3 | 7.77e-01 |
| GO:BP | GO:0048769 | sarcomerogenesis | 1 | 3 | 7.77e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 6 | 13 | 7.78e-01 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 1 | 7 | 7.78e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 3 | 196 | 7.78e-01 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 1 | 10 | 7.78e-01 |
| GO:BP | GO:1902396 | protein localization to bicellular tight junction | 1 | 3 | 7.78e-01 |
| GO:BP | GO:0140291 | peptidyl-glutamate ADP-deribosylation | 1 | 3 | 7.78e-01 |
| GO:BP | GO:0006726 | eye pigment biosynthetic process | 1 | 1 | 7.78e-01 |
| GO:BP | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity | 1 | 2 | 7.78e-01 |
| GO:BP | GO:1903056 | regulation of melanosome organization | 1 | 2 | 7.78e-01 |
| GO:BP | GO:0043324 | pigment metabolic process involved in developmental pigmentation | 1 | 1 | 7.78e-01 |
| GO:BP | GO:0042441 | eye pigment metabolic process | 1 | 1 | 7.78e-01 |
| GO:BP | GO:0090170 | regulation of Golgi inheritance | 1 | 4 | 7.78e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 2 | 1399 | 7.78e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 2 | 24 | 7.78e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 7.78e-01 |
| GO:BP | GO:0070498 | interleukin-1-mediated signaling pathway | 1 | 20 | 7.78e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 4 | 27 | 7.78e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 2 | 16 | 7.78e-01 |
| GO:BP | GO:0042447 | hormone catabolic process | 1 | 7 | 7.78e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 7.79e-01 |
| GO:BP | GO:0033604 | negative regulation of catecholamine secretion | 1 | 6 | 7.79e-01 |
| GO:BP | GO:0002790 | peptide secretion | 8 | 170 | 7.79e-01 |
| GO:BP | GO:0032430 | positive regulation of phospholipase A2 activity | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0150003 | regulation of spontaneous synaptic transmission | 1 | 6 | 7.79e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 2 | 199 | 7.79e-01 |
| GO:BP | GO:0002934 | desmosome organization | 4 | 9 | 7.79e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 7.79e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 7.79e-01 |
| GO:BP | GO:0055095 | lipoprotein particle mediated signaling | 1 | 3 | 7.79e-01 |
| GO:BP | GO:0060478 | acrosomal vesicle exocytosis | 3 | 5 | 7.79e-01 |
| GO:BP | GO:0018342 | protein prenylation | 4 | 10 | 7.79e-01 |
| GO:BP | GO:0055096 | low-density lipoprotein particle mediated signaling | 1 | 3 | 7.79e-01 |
| GO:BP | GO:0015911 | long-chain fatty acid import across plasma membrane | 3 | 9 | 7.79e-01 |
| GO:BP | GO:0097354 | prenylation | 4 | 10 | 7.79e-01 |
| GO:BP | GO:0034371 | chylomicron remodeling | 1 | 1 | 7.79e-01 |
| GO:BP | GO:0070475 | rRNA base methylation | 1 | 8 | 7.79e-01 |
| GO:BP | GO:0002309 | T cell proliferation involved in immune response | 1 | 3 | 7.79e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 1 | 40 | 7.79e-01 |
| GO:BP | GO:0010896 | regulation of triglyceride catabolic process | 1 | 7 | 7.80e-01 |
| GO:BP | GO:1905035 | negative regulation of antifungal innate immune response | 1 | 2 | 7.80e-01 |
| GO:BP | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 7.80e-01 |
| GO:BP | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 7.80e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 1 | 62 | 7.80e-01 |
| GO:BP | GO:0043654 | recognition of apoptotic cell | 1 | 4 | 7.80e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 1 | 57 | 7.81e-01 |
| GO:BP | GO:0051234 | establishment of localization | 2 | 3433 | 7.81e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 2 | 57 | 7.81e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 7.81e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 7.81e-01 |
| GO:BP | GO:0014858 | positive regulation of skeletal muscle cell proliferation | 1 | 2 | 7.81e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 7.81e-01 |
| GO:BP | GO:0090086 | negative regulation of protein deubiquitination | 2 | 4 | 7.81e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 7.81e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 7.81e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 7.81e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 2 | 290 | 7.82e-01 |
| GO:BP | GO:0071691 | cardiac muscle thin filament assembly | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 2 | 234 | 7.82e-01 |
| GO:BP | GO:1903845 | negative regulation of cellular response to transforming growth factor beta stimulus | 2 | 2 | 7.82e-01 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 1 | 6 | 7.82e-01 |
| GO:BP | GO:0090258 | negative regulation of mitochondrial fission | 2 | 3 | 7.82e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 1 | 18 | 7.82e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 1 | 18 | 7.82e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 1 | 56 | 7.82e-01 |
| GO:BP | GO:0060300 | regulation of cytokine activity | 2 | 3 | 7.82e-01 |
| GO:BP | GO:1903093 | regulation of protein K48-linked deubiquitination | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0036316 | SREBP-SCAP complex retention in endoplasmic reticulum | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0070189 | kynurenine metabolic process | 1 | 6 | 7.82e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 7 | 85 | 7.82e-01 |
| GO:BP | GO:0045720 | negative regulation of integrin biosynthetic process | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 7.82e-01 |
| GO:BP | GO:0018993 | somatic sex determination | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 11 | 46 | 7.82e-01 |
| GO:BP | GO:0032914 | positive regulation of transforming growth factor beta1 production | 1 | 6 | 7.82e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0031509 | subtelomeric heterochromatin formation | 2 | 7 | 7.82e-01 |
| GO:BP | GO:0060174 | limb bud formation | 3 | 10 | 7.82e-01 |
| GO:BP | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 10 | 44 | 7.82e-01 |
| GO:BP | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0021825 | substrate-dependent cerebral cortex tangential migration | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway | 1 | 12 | 7.82e-01 |
| GO:BP | GO:1904506 | negative regulation of telomere maintenance in response to DNA damage | 2 | 3 | 7.82e-01 |
| GO:BP | GO:0032364 | intracellular oxygen homeostasis | 1 | 6 | 7.82e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 4 | 24 | 7.82e-01 |
| GO:BP | GO:0021827 | postnatal olfactory bulb interneuron migration | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 39 | 7.82e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 4 | 9 | 7.82e-01 |
| GO:BP | GO:0051709 | regulation of killing of cells of another organism | 2 | 3 | 7.82e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1 | 1507 | 7.82e-01 |
| GO:BP | GO:0033085 | negative regulation of T cell differentiation in thymus | 1 | 5 | 7.82e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 7 | 418 | 7.82e-01 |
| GO:BP | GO:0035551 | protein initiator methionine removal involved in protein maturation | 1 | 3 | 7.82e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 2 | 28 | 7.82e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 10 | 1101 | 7.82e-01 |
| GO:BP | GO:1905034 | regulation of antifungal innate immune response | 2 | 3 | 7.82e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 1 | 73 | 7.82e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 7.82e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4570 | 7.82e-01 |
| GO:BP | GO:0048561 | establishment of animal organ orientation | 1 | 1 | 7.82e-01 |
| GO:BP | GO:0021548 | pons development | 3 | 7 | 7.82e-01 |
| GO:BP | GO:1901610 | positive regulation of vesicle transport along microtubule | 2 | 3 | 7.82e-01 |
| GO:BP | GO:0032594 | protein transport within lipid bilayer | 3 | 6 | 7.82e-01 |
| GO:BP | GO:1904204 | regulation of skeletal muscle hypertrophy | 1 | 1 | 7.83e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 1 | 32 | 7.83e-01 |
| GO:BP | GO:0045346 | regulation of MHC class II biosynthetic process | 7 | 11 | 7.83e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 7.83e-01 |
| GO:BP | GO:1905162 | regulation of phagosome maturation | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0070171 | negative regulation of tooth mineralization | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 6 | 816 | 7.83e-01 |
| GO:BP | GO:2000523 | regulation of T cell costimulation | 1 | 2 | 7.83e-01 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 2 | 11 | 7.83e-01 |
| GO:BP | GO:0035262 | gonad morphogenesis | 1 | 1 | 7.83e-01 |
| GO:BP | GO:1903719 | regulation of I-kappaB phosphorylation | 1 | 6 | 7.83e-01 |
| GO:BP | GO:0072134 | nephrogenic mesenchyme morphogenesis | 1 | 2 | 7.83e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 2 | 8 | 7.83e-01 |
| GO:BP | GO:0030421 | defecation | 1 | 1 | 7.83e-01 |
| GO:BP | GO:1903054 | negative regulation of extracellular matrix organization | 2 | 6 | 7.83e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 2 | 7 | 7.83e-01 |
| GO:BP | GO:0097400 | interleukin-17-mediated signaling pathway | 2 | 8 | 7.83e-01 |
| GO:BP | GO:0097164 | ammonium ion metabolic process | 1 | 14 | 7.84e-01 |
| GO:BP | GO:0000266 | mitochondrial fission | 2 | 38 | 7.84e-01 |
| GO:BP | GO:1901896 | positive regulation of ATPase-coupled calcium transmembrane transporter activity | 2 | 6 | 7.84e-01 |
| GO:BP | GO:0032425 | positive regulation of mismatch repair | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0045176 | apical protein localization | 1 | 13 | 7.84e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 14 | 56 | 7.84e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4596 | 7.84e-01 |
| GO:BP | GO:0150032 | positive regulation of protein localization to lysosome | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 1 | 23 | 7.84e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 1 | 22 | 7.84e-01 |
| GO:BP | GO:0010912 | positive regulation of isomerase activity | 1 | 5 | 7.84e-01 |
| GO:BP | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process | 1 | 2 | 7.84e-01 |
| GO:BP | GO:0010911 | regulation of isomerase activity | 1 | 5 | 7.84e-01 |
| GO:BP | GO:0060437 | lung growth | 1 | 5 | 7.84e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 2 | 147 | 7.84e-01 |
| GO:BP | GO:0050821 | protein stabilization | 2 | 191 | 7.85e-01 |
| GO:BP | GO:0006954 | inflammatory response | 9 | 429 | 7.85e-01 |
| GO:BP | GO:0006983 | ER overload response | 3 | 12 | 7.85e-01 |
| GO:BP | GO:0006887 | exocytosis | 4 | 256 | 7.85e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 11 | 128 | 7.85e-01 |
| GO:BP | GO:0061027 | umbilical cord development | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0036304 | umbilical cord morphogenesis | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0038172 | interleukin-33-mediated signaling pathway | 1 | 1 | 7.86e-01 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 2 | 6 | 7.86e-01 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 1 | 22 | 7.86e-01 |
| GO:BP | GO:0140469 | GCN2-mediated signaling | 1 | 3 | 7.86e-01 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 4 | 7.86e-01 |
| GO:BP | GO:1900028 | negative regulation of ruffle assembly | 2 | 4 | 7.86e-01 |
| GO:BP | GO:0000973 | post-transcriptional tethering of RNA polymerase II gene DNA at nuclear periphery | 1 | 2 | 7.86e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 12 | 258 | 7.86e-01 |
| GO:BP | GO:0042355 | L-fucose catabolic process | 1 | 3 | 7.86e-01 |
| GO:BP | GO:0019317 | fucose catabolic process | 1 | 3 | 7.86e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 1 | 20 | 7.86e-01 |
| GO:BP | GO:0016199 | axon midline choice point recognition | 2 | 3 | 7.87e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 7.87e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 3 | 10 | 7.87e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 5 | 69 | 7.87e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 6 | 22 | 7.87e-01 |
| GO:BP | GO:0070172 | positive regulation of tooth mineralization | 1 | 2 | 7.87e-01 |
| GO:BP | GO:1905382 | positive regulation of snRNA transcription by RNA polymerase II | 1 | 3 | 7.87e-01 |
| GO:BP | GO:0072488 | ammonium transmembrane transport | 2 | 6 | 7.87e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 7.87e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 7 | 433 | 7.87e-01 |
| GO:BP | GO:0072070 | loop of Henle development | 1 | 8 | 7.87e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 3 | 19 | 7.87e-01 |
| GO:BP | GO:0000348 | mRNA branch site recognition | 1 | 1 | 7.87e-01 |
| GO:BP | GO:0071801 | regulation of podosome assembly | 2 | 8 | 7.88e-01 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 1 | 23 | 7.88e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 7.88e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 3 | 7 | 7.88e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 7.88e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 9 | 40 | 7.88e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 7.88e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 52 | 7.88e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 8 | 21 | 7.88e-01 |
| GO:BP | GO:0000413 | protein peptidyl-prolyl isomerization | 9 | 26 | 7.89e-01 |
| GO:BP | GO:1905651 | regulation of artery morphogenesis | 2 | 5 | 7.89e-01 |
| GO:BP | GO:1905653 | positive regulation of artery morphogenesis | 2 | 5 | 7.89e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 6 | 93 | 7.89e-01 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 4 | 10 | 7.89e-01 |
| GO:BP | GO:2001136 | negative regulation of endocytic recycling | 1 | 1 | 7.89e-01 |
| GO:BP | GO:0097638 | L-arginine import across plasma membrane | 2 | 4 | 7.89e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 2 | 158 | 7.89e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 5 | 37 | 7.89e-01 |
| GO:BP | GO:0015802 | basic amino acid transport | 6 | 17 | 7.89e-01 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 1 | 5 | 7.89e-01 |
| GO:BP | GO:0001502 | cartilage condensation | 3 | 12 | 7.89e-01 |
| GO:BP | GO:2000543 | positive regulation of gastrulation | 1 | 5 | 7.89e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 8 | 33 | 7.89e-01 |
| GO:BP | GO:0031017 | exocrine pancreas development | 1 | 4 | 7.90e-01 |
| GO:BP | GO:0048002 | antigen processing and presentation of peptide antigen | 3 | 41 | 7.90e-01 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 2 | 10 | 7.90e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 7.90e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 2 | 44 | 7.90e-01 |
| GO:BP | GO:0060455 | negative regulation of gastric acid secretion | 1 | 3 | 7.90e-01 |
| GO:BP | GO:2000987 | positive regulation of behavioral fear response | 1 | 3 | 7.90e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 1 | 35 | 7.90e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 7.91e-01 |
| GO:BP | GO:0051542 | elastin biosynthetic process | 1 | 2 | 7.91e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 22 | 310 | 7.91e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 3 | 11 | 7.91e-01 |
| GO:BP | GO:0045964 | positive regulation of dopamine metabolic process | 2 | 6 | 7.91e-01 |
| GO:BP | GO:0045915 | positive regulation of catecholamine metabolic process | 2 | 6 | 7.91e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 11 | 46 | 7.91e-01 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 2 | 22 | 7.91e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 23 | 539 | 7.92e-01 |
| GO:BP | GO:0050916 | sensory perception of sweet taste | 2 | 3 | 7.92e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 2 | 52 | 7.92e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 2 | 52 | 7.92e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 7.92e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 185 | 7.92e-01 |
| GO:BP | GO:0051639 | actin filament network formation | 1 | 7 | 7.92e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1 | 1564 | 7.92e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 7 | 168 | 7.92e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 3 | 39 | 7.92e-01 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 4 | 9 | 7.93e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 7.93e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 3 | 51 | 7.93e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 1 | 43 | 7.93e-01 |
| GO:BP | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell | 1 | 2 | 7.93e-01 |
| GO:BP | GO:0006413 | translational initiation | 8 | 116 | 7.93e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 6 | 126 | 7.93e-01 |
| GO:BP | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion | 1 | 3 | 7.93e-01 |
| GO:BP | GO:0038192 | gastric inhibitory peptide signaling pathway | 1 | 1 | 7.93e-01 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 7.93e-01 |
| GO:BP | GO:0006714 | sesquiterpenoid metabolic process | 1 | 2 | 7.93e-01 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 2 | 5 | 7.93e-01 |
| GO:BP | GO:0009798 | axis specification | 2 | 63 | 7.94e-01 |
| GO:BP | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway | 1 | 6 | 7.94e-01 |
| GO:BP | GO:0060011 | Sertoli cell proliferation | 1 | 4 | 7.95e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 7.95e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 3 | 10 | 7.95e-01 |
| GO:BP | GO:0061187 | regulation of rDNA heterochromatin formation | 1 | 2 | 7.96e-01 |
| GO:BP | GO:0061188 | negative regulation of rDNA heterochromatin formation | 1 | 2 | 7.96e-01 |
| GO:BP | GO:0048243 | norepinephrine secretion | 1 | 5 | 7.96e-01 |
| GO:BP | GO:0014061 | regulation of norepinephrine secretion | 1 | 5 | 7.96e-01 |
| GO:BP | GO:0090615 | mitochondrial mRNA processing | 1 | 2 | 7.96e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 5 | 58 | 7.96e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 4 | 16 | 7.96e-01 |
| GO:BP | GO:0044691 | tooth eruption | 2 | 5 | 7.96e-01 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 3 | 9 | 7.96e-01 |
| GO:BP | GO:2001171 | positive regulation of ATP biosynthetic process | 1 | 10 | 7.96e-01 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 40 | 7.96e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 7 | 162 | 7.96e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1602 | 7.96e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 7.97e-01 |
| GO:BP | GO:1990034 | calcium ion export across plasma membrane | 1 | 6 | 7.97e-01 |
| GO:BP | GO:0048669 | collateral sprouting in absence of injury | 1 | 4 | 7.97e-01 |
| GO:BP | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus | 2 | 7 | 7.97e-01 |
| GO:BP | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation | 1 | 4 | 7.97e-01 |
| GO:BP | GO:0003056 | regulation of vascular associated smooth muscle contraction | 1 | 8 | 7.97e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 2 | 104 | 7.97e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 6 | 24 | 7.97e-01 |
| GO:BP | GO:1902603 | carnitine transmembrane transport | 2 | 4 | 7.97e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4747 | 7.97e-01 |
| GO:BP | GO:0048289 | isotype switching to IgE isotypes | 2 | 5 | 7.97e-01 |
| GO:BP | GO:0048293 | regulation of isotype switching to IgE isotypes | 2 | 5 | 7.97e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 7.97e-01 |
| GO:BP | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | 3 | 11 | 7.98e-01 |
| GO:BP | GO:0048936 | peripheral nervous system neuron axonogenesis | 1 | 4 | 7.98e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 2 | 61 | 7.98e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 3 | 53 | 7.98e-01 |
| GO:BP | GO:0120045 | stereocilium maintenance | 1 | 2 | 7.98e-01 |
| GO:BP | GO:1903430 | negative regulation of cell maturation | 2 | 4 | 7.98e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 9 | 90 | 7.99e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 7.99e-01 |
| GO:BP | GO:0061885 | positive regulation of mini excitatory postsynaptic potential | 1 | 2 | 7.99e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 26 | 301 | 7.99e-01 |
| GO:BP | GO:0002032 | desensitization of G protein-coupled receptor signaling pathway by arrestin | 1 | 2 | 7.99e-01 |
| GO:BP | GO:1905258 | regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 7.99e-01 |
| GO:BP | GO:1905259 | negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 7.99e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 6 | 94 | 7.99e-01 |
| GO:BP | GO:0061884 | regulation of mini excitatory postsynaptic potential | 1 | 2 | 7.99e-01 |
| GO:BP | GO:0098816 | mini excitatory postsynaptic potential | 1 | 2 | 7.99e-01 |
| GO:BP | GO:0006704 | glucocorticoid biosynthetic process | 1 | 11 | 7.99e-01 |
| GO:BP | GO:0044830 | modulation by host of viral RNA genome replication | 2 | 6 | 7.99e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 4 | 113 | 7.99e-01 |
| GO:BP | GO:0021884 | forebrain neuron development | 1 | 16 | 7.99e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 1 | 21 | 7.99e-01 |
| GO:BP | GO:0032474 | otolith morphogenesis | 1 | 3 | 7.99e-01 |
| GO:BP | GO:0042220 | response to cocaine | 1 | 28 | 7.99e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 22 | 52 | 7.99e-01 |
| GO:BP | GO:0048318 | axial mesoderm development | 1 | 6 | 8.00e-01 |
| GO:BP | GO:1990791 | dorsal root ganglion development | 3 | 6 | 8.00e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 10 | 152 | 8.00e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 55 | 1138 | 8.00e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 1 | 30 | 8.00e-01 |
| GO:BP | GO:0007409 | axonogenesis | 6 | 359 | 8.00e-01 |
| GO:BP | GO:0071284 | cellular response to lead ion | 2 | 4 | 8.00e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1 | 1604 | 8.00e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 2 | 20 | 8.00e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 1 | 6 | 8.00e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 1 | 73 | 8.00e-01 |
| GO:BP | GO:0009306 | protein secretion | 8 | 262 | 8.01e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 4 | 258 | 8.01e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 4 | 8 | 8.01e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 3 | 9 | 8.01e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 3 | 10 | 8.01e-01 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 8.01e-01 |
| GO:BP | GO:0043378 | positive regulation of CD8-positive, alpha-beta T cell differentiation | 2 | 2 | 8.01e-01 |
| GO:BP | GO:0002841 | negative regulation of T cell mediated immune response to tumor cell | 1 | 1 | 8.01e-01 |
| GO:BP | GO:1903842 | response to arsenite ion | 1 | 4 | 8.01e-01 |
| GO:BP | GO:1903843 | cellular response to arsenite ion | 1 | 4 | 8.01e-01 |
| GO:BP | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway | 4 | 9 | 8.01e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 1 | 17 | 8.01e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 1 | 4 | 8.01e-01 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 2 | 12 | 8.01e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 8.01e-01 |
| GO:BP | GO:0045932 | negative regulation of muscle contraction | 2 | 20 | 8.01e-01 |
| GO:BP | GO:0001923 | B-1 B cell differentiation | 1 | 3 | 8.02e-01 |
| GO:BP | GO:0006393 | termination of mitochondrial transcription | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0035811 | negative regulation of urine volume | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 8.02e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 3 | 7 | 8.02e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 6 | 138 | 8.02e-01 |
| GO:BP | GO:0014842 | regulation of skeletal muscle satellite cell proliferation | 5 | 8 | 8.02e-01 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 1 | 10 | 8.02e-01 |
| GO:BP | GO:0043097 | pyrimidine nucleoside salvage | 1 | 2 | 8.02e-01 |
| GO:BP | GO:0099041 | vesicle tethering to Golgi | 2 | 5 | 8.02e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 8 | 262 | 8.03e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 8.03e-01 |
| GO:BP | GO:0070327 | thyroid hormone transport | 1 | 7 | 8.03e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 8.03e-01 |
| GO:BP | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 5 | 12 | 8.03e-01 |
| GO:BP | GO:0045989 | positive regulation of striated muscle contraction | 2 | 12 | 8.03e-01 |
| GO:BP | GO:1903729 | regulation of plasma membrane organization | 1 | 15 | 8.03e-01 |
| GO:BP | GO:0035768 | endothelial cell chemotaxis to fibroblast growth factor | 3 | 4 | 8.03e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 8 | 2684 | 8.03e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 4 | 12 | 8.03e-01 |
| GO:BP | GO:0035766 | cell chemotaxis to fibroblast growth factor | 3 | 4 | 8.03e-01 |
| GO:BP | GO:1904847 | regulation of cell chemotaxis to fibroblast growth factor | 3 | 4 | 8.03e-01 |
| GO:BP | GO:1990868 | response to chemokine | 3 | 37 | 8.03e-01 |
| GO:BP | GO:0070445 | regulation of oligodendrocyte progenitor proliferation | 1 | 5 | 8.03e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 6 | 76 | 8.03e-01 |
| GO:BP | GO:0090023 | positive regulation of neutrophil chemotaxis | 1 | 14 | 8.03e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 3 | 37 | 8.03e-01 |
| GO:BP | GO:0070444 | oligodendrocyte progenitor proliferation | 1 | 5 | 8.03e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 3 | 7 | 8.03e-01 |
| GO:BP | GO:1901252 | regulation of intracellular transport of viral material | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 5 | 22 | 8.03e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 3 | 8 | 8.03e-01 |
| GO:BP | GO:2000347 | positive regulation of hepatocyte proliferation | 4 | 9 | 8.03e-01 |
| GO:BP | GO:2000544 | regulation of endothelial cell chemotaxis to fibroblast growth factor | 3 | 4 | 8.03e-01 |
| GO:BP | GO:0051665 | membrane raft localization | 2 | 4 | 8.03e-01 |
| GO:BP | GO:0030850 | prostate gland development | 3 | 35 | 8.03e-01 |
| GO:BP | GO:0060426 | lung vasculature development | 1 | 3 | 8.03e-01 |
| GO:BP | GO:1900453 | negative regulation of long-term synaptic depression | 1 | 3 | 8.03e-01 |
| GO:BP | GO:0110077 | vesicle-mediated intercellular transport | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0014857 | regulation of skeletal muscle cell proliferation | 5 | 10 | 8.03e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 8 | 2683 | 8.03e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 20 | 527 | 8.03e-01 |
| GO:BP | GO:0019303 | D-ribose catabolic process | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration | 2 | 4 | 8.03e-01 |
| GO:BP | GO:0071698 | olfactory placode development | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0099538 | synaptic signaling via neuropeptide | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0071716 | leukotriene transport | 2 | 5 | 8.03e-01 |
| GO:BP | GO:0030910 | olfactory placode formation | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0071699 | olfactory placode morphogenesis | 1 | 2 | 8.03e-01 |
| GO:BP | GO:0003157 | endocardium development | 3 | 9 | 8.03e-01 |
| GO:BP | GO:1904542 | regulation of free ubiquitin chain polymerization | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 1 | 31 | 8.04e-01 |
| GO:BP | GO:0002579 | positive regulation of antigen processing and presentation | 2 | 3 | 8.04e-01 |
| GO:BP | GO:1903384 | negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0071907 | determination of digestive tract left/right asymmetry | 3 | 5 | 8.04e-01 |
| GO:BP | GO:1904544 | positive regulation of free ubiquitin chain polymerization | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 3 | 16 | 8.04e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 4 | 98 | 8.04e-01 |
| GO:BP | GO:0042264 | peptidyl-aspartic acid hydroxylation | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 4 | 173 | 8.04e-01 |
| GO:BP | GO:0036482 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 2 | 8.04e-01 |
| GO:BP | GO:1904881 | cellular response to hydrogen sulfide | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 1 | 73 | 8.04e-01 |
| GO:BP | GO:1903383 | regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4946 | 8.04e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 4 | 47 | 8.04e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 4 | 19 | 8.04e-01 |
| GO:BP | GO:0098508 | endothelial to hematopoietic transition | 1 | 2 | 8.04e-01 |
| GO:BP | GO:0048511 | rhythmic process | 13 | 237 | 8.04e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 4 | 4617 | 8.04e-01 |
| GO:BP | GO:0061737 | leukotriene signaling pathway | 1 | 1 | 8.05e-01 |
| GO:BP | GO:0046532 | regulation of photoreceptor cell differentiation | 3 | 4 | 8.05e-01 |
| GO:BP | GO:0021628 | olfactory nerve formation | 1 | 2 | 8.05e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 1 | 8 | 8.05e-01 |
| GO:BP | GO:0021563 | glossopharyngeal nerve development | 2 | 3 | 8.05e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 1 | 3 | 8.06e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 6 | 95 | 8.07e-01 |
| GO:BP | GO:2000065 | negative regulation of cortisol biosynthetic process | 1 | 4 | 8.07e-01 |
| GO:BP | GO:0003383 | apical constriction | 1 | 4 | 8.07e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 8.07e-01 |
| GO:BP | GO:0046541 | saliva secretion | 1 | 4 | 8.07e-01 |
| GO:BP | GO:0032345 | negative regulation of aldosterone metabolic process | 1 | 4 | 8.07e-01 |
| GO:BP | GO:0032348 | negative regulation of aldosterone biosynthetic process | 1 | 4 | 8.07e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 5 | 11 | 8.07e-01 |
| GO:BP | GO:0006555 | methionine metabolic process | 1 | 13 | 8.07e-01 |
| GO:BP | GO:0097168 | mesenchymal stem cell proliferation | 3 | 4 | 8.07e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 2 | 2574 | 8.07e-01 |
| GO:BP | GO:0048280 | vesicle fusion with Golgi apparatus | 1 | 8 | 8.07e-01 |
| GO:BP | GO:1902224 | ketone body metabolic process | 2 | 7 | 8.07e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 2 | 2576 | 8.07e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 4 | 10 | 8.07e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 8.07e-01 |
| GO:BP | GO:0010936 | negative regulation of macrophage cytokine production | 1 | 7 | 8.08e-01 |
| GO:BP | GO:0042541 | hemoglobin biosynthetic process | 6 | 13 | 8.08e-01 |
| GO:BP | GO:0008078 | mesodermal cell migration | 1 | 3 | 8.09e-01 |
| GO:BP | GO:0019464 | glycine decarboxylation via glycine cleavage system | 1 | 3 | 8.09e-01 |
| GO:BP | GO:0006382 | adenosine to inosine editing | 2 | 4 | 8.09e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 3 | 26 | 8.09e-01 |
| GO:BP | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment | 1 | 1 | 8.10e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 4 | 68 | 8.10e-01 |
| GO:BP | GO:1902362 | melanocyte apoptotic process | 1 | 1 | 8.10e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 15 | 27 | 8.10e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 1 | 92 | 8.10e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 1 | 92 | 8.10e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 1 | 92 | 8.10e-01 |
| GO:BP | GO:0097069 | cellular response to thyroxine stimulus | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 8.10e-01 |
| GO:BP | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0036228 | protein localization to nuclear inner membrane | 1 | 3 | 8.10e-01 |
| GO:BP | GO:1904387 | cellular response to L-phenylalanine derivative | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0000432 | positive regulation of transcription from RNA polymerase II promoter by glucose | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 9 | 269 | 8.10e-01 |
| GO:BP | GO:0140568 | extraction of mislocalized protein from membrane | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 10 | 55 | 8.10e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 2 | 11 | 8.10e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 2 | 2593 | 8.10e-01 |
| GO:BP | GO:1904396 | regulation of neuromuscular junction development | 3 | 5 | 8.10e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 3 | 10 | 8.10e-01 |
| GO:BP | GO:1903350 | response to dopamine | 4 | 48 | 8.10e-01 |
| GO:BP | GO:0021769 | orbitofrontal cortex development | 1 | 2 | 8.10e-01 |
| GO:BP | GO:2000485 | regulation of glutamine transport | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0019509 | L-methionine salvage from methylthioadenosine | 1 | 5 | 8.10e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 1 | 122 | 8.10e-01 |
| GO:BP | GO:1904959 | regulation of cytochrome-c oxidase activity | 3 | 7 | 8.10e-01 |
| GO:BP | GO:0051124 | synaptic assembly at neuromuscular junction | 3 | 6 | 8.10e-01 |
| GO:BP | GO:0043380 | regulation of memory T cell differentiation | 1 | 4 | 8.10e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 2 | 6 | 8.10e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 10 | 66 | 8.10e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 2 | 5 | 8.10e-01 |
| GO:BP | GO:0046940 | nucleoside monophosphate phosphorylation | 3 | 9 | 8.10e-01 |
| GO:BP | GO:0051410 | detoxification of nitrogen compound | 2 | 5 | 8.11e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 8 | 23 | 8.11e-01 |
| GO:BP | GO:1903580 | positive regulation of ATP metabolic process | 1 | 11 | 8.11e-01 |
| GO:BP | GO:0070458 | cellular detoxification of nitrogen compound | 2 | 5 | 8.11e-01 |
| GO:BP | GO:0006085 | acetyl-CoA biosynthetic process | 1 | 16 | 8.11e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 8 | 2714 | 8.11e-01 |
| GO:BP | GO:0097494 | regulation of vesicle size | 8 | 19 | 8.11e-01 |
| GO:BP | GO:0072672 | neutrophil extravasation | 1 | 7 | 8.11e-01 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 3 | 28 | 8.11e-01 |
| GO:BP | GO:0045348 | positive regulation of MHC class II biosynthetic process | 5 | 8 | 8.11e-01 |
| GO:BP | GO:0097473 | retinal rod cell apoptotic process | 1 | 2 | 8.11e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 8.11e-01 |
| GO:BP | GO:0006679 | glucosylceramide biosynthetic process | 1 | 2 | 8.11e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 20 | 531 | 8.11e-01 |
| GO:BP | GO:0071400 | cellular response to oleic acid | 1 | 3 | 8.11e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 3 | 31 | 8.11e-01 |
| GO:BP | GO:0007603 | phototransduction, visible light | 1 | 9 | 8.11e-01 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 8.11e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 2 | 90 | 8.11e-01 |
| GO:BP | GO:0016486 | peptide hormone processing | 5 | 27 | 8.12e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 7 | 25 | 8.12e-01 |
| GO:BP | GO:0003352 | regulation of cilium movement | 1 | 17 | 8.12e-01 |
| GO:BP | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | 3 | 8 | 8.12e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 3 | 86 | 8.12e-01 |
| GO:BP | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0097532 | stress response to acid chemical | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0043011 | myeloid dendritic cell differentiation | 1 | 10 | 8.12e-01 |
| GO:BP | GO:0097533 | cellular stress response to acid chemical | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 3 | 7 | 8.12e-01 |
| GO:BP | GO:0072338 | lactam metabolic process | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 4 | 295 | 8.12e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 3 | 86 | 8.12e-01 |
| GO:BP | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0070142 | synaptic vesicle budding | 3 | 8 | 8.12e-01 |
| GO:BP | GO:1902871 | positive regulation of amacrine cell differentiation | 1 | 1 | 8.12e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 1 | 7 | 8.12e-01 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 1 | 11 | 8.12e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 1 | 22 | 8.12e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 8.12e-01 |
| GO:BP | GO:0060684 | epithelial-mesenchymal cell signaling | 2 | 3 | 8.12e-01 |
| GO:BP | GO:0014891 | striated muscle atrophy | 1 | 6 | 8.12e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 1 | 129 | 8.12e-01 |
| GO:BP | GO:0050428 | 3’-phosphoadenosine 5’-phosphosulfate biosynthetic process | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 3 | 9 | 8.12e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 5 | 18 | 8.12e-01 |
| GO:BP | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0000103 | sulfate assimilation | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 5 | 18 | 8.12e-01 |
| GO:BP | GO:0070973 | protein localization to endoplasmic reticulum exit site | 2 | 6 | 8.12e-01 |
| GO:BP | GO:2000726 | negative regulation of cardiac muscle cell differentiation | 3 | 8 | 8.12e-01 |
| GO:BP | GO:0044861 | protein transport into plasma membrane raft | 1 | 1 | 8.12e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 2 | 50 | 8.13e-01 |
| GO:BP | GO:1905279 | regulation of retrograde transport, endosome to Golgi | 1 | 6 | 8.13e-01 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 8.13e-01 |
| GO:BP | GO:0002921 | negative regulation of humoral immune response | 2 | 10 | 8.13e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 8.13e-01 |
| GO:BP | GO:0036066 | protein O-linked fucosylation | 2 | 5 | 8.13e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 8 | 563 | 8.13e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 5 | 468 | 8.13e-01 |
| GO:BP | GO:0002086 | diaphragm contraction | 1 | 2 | 8.13e-01 |
| GO:BP | GO:0034477 | U6 snRNA 3’-end processing | 1 | 2 | 8.13e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 1 | 7 | 8.14e-01 |
| GO:BP | GO:0044721 | protein import into peroxisome matrix, substrate release | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0022010 | central nervous system myelination | 1 | 12 | 8.14e-01 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 1 | 18 | 8.14e-01 |
| GO:BP | GO:0046476 | glycosylceramide biosynthetic process | 3 | 6 | 8.14e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 4 | 289 | 8.14e-01 |
| GO:BP | GO:0032291 | axon ensheathment in central nervous system | 1 | 12 | 8.14e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 7 | 18 | 8.14e-01 |
| GO:BP | GO:0000961 | negative regulation of mitochondrial RNA catabolic process | 1 | 2 | 8.14e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 1 | 199 | 8.14e-01 |
| GO:BP | GO:2000632 | negative regulation of pre-miRNA processing | 1 | 2 | 8.15e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 2 | 5 | 8.15e-01 |
| GO:BP | GO:0042473 | outer ear morphogenesis | 3 | 5 | 8.15e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 6 | 382 | 8.15e-01 |
| GO:BP | GO:2000367 | regulation of acrosomal vesicle exocytosis | 1 | 3 | 8.15e-01 |
| GO:BP | GO:0001807 | regulation of type IV hypersensitivity | 1 | 1 | 8.15e-01 |
| GO:BP | GO:0046947 | hydroxylysine biosynthetic process | 1 | 2 | 8.15e-01 |
| GO:BP | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production | 1 | 6 | 8.15e-01 |
| GO:BP | GO:0071609 | chemokine (C-C motif) ligand 5 production | 1 | 6 | 8.15e-01 |
| GO:BP | GO:0046946 | hydroxylysine metabolic process | 1 | 2 | 8.15e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 7 | 16 | 8.15e-01 |
| GO:BP | GO:0060120 | inner ear receptor cell fate commitment | 2 | 4 | 8.15e-01 |
| GO:BP | GO:0009912 | auditory receptor cell fate commitment | 2 | 4 | 8.15e-01 |
| GO:BP | GO:0031392 | regulation of prostaglandin biosynthetic process | 2 | 6 | 8.15e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 2 | 222 | 8.16e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 1 | 100 | 8.16e-01 |
| GO:BP | GO:0021764 | amygdala development | 1 | 3 | 8.16e-01 |
| GO:BP | GO:1903236 | regulation of leukocyte tethering or rolling | 2 | 6 | 8.16e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 16 | 242 | 8.16e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 8.16e-01 |
| GO:BP | GO:0030647 | aminoglycoside antibiotic metabolic process | 4 | 7 | 8.16e-01 |
| GO:BP | GO:0030638 | polyketide metabolic process | 4 | 8 | 8.16e-01 |
| GO:BP | GO:0021559 | trigeminal nerve development | 1 | 5 | 8.16e-01 |
| GO:BP | GO:0044598 | doxorubicin metabolic process | 4 | 8 | 8.16e-01 |
| GO:BP | GO:0045992 | negative regulation of embryonic development | 1 | 2 | 8.16e-01 |
| GO:BP | GO:1905036 | positive regulation of antifungal innate immune response | 1 | 1 | 8.16e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 9 | 16 | 8.16e-01 |
| GO:BP | GO:0071376 | cellular response to corticotropin-releasing hormone stimulus | 1 | 3 | 8.16e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 1 | 31 | 8.16e-01 |
| GO:BP | GO:0043435 | response to corticotropin-releasing hormone | 1 | 3 | 8.16e-01 |
| GO:BP | GO:0045124 | regulation of bone resorption | 2 | 24 | 8.16e-01 |
| GO:BP | GO:0071529 | cementum mineralization | 1 | 3 | 8.16e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 6 | 14 | 8.16e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 8.16e-01 |
| GO:BP | GO:2000508 | regulation of dendritic cell chemotaxis | 1 | 3 | 8.17e-01 |
| GO:BP | GO:1903803 | L-glutamine import across plasma membrane | 2 | 4 | 8.17e-01 |
| GO:BP | GO:0032966 | negative regulation of collagen biosynthetic process | 2 | 7 | 8.17e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 10 | 52 | 8.17e-01 |
| GO:BP | GO:1903141 | negative regulation of establishment of endothelial barrier | 1 | 2 | 8.17e-01 |
| GO:BP | GO:1901551 | negative regulation of endothelial cell development | 1 | 2 | 8.17e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 4 | 9 | 8.17e-01 |
| GO:BP | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 3 | 7 | 8.18e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 41 | 143 | 8.18e-01 |
| GO:BP | GO:0033240 | positive regulation of amine metabolic process | 3 | 7 | 8.18e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 3 | 96 | 8.18e-01 |
| GO:BP | GO:0072535 | tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 2 | 80 | 8.18e-01 |
| GO:BP | GO:2000307 | regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 8.18e-01 |
| GO:BP | GO:2000412 | positive regulation of thymocyte migration | 1 | 1 | 8.18e-01 |
| GO:BP | GO:1903061 | positive regulation of protein lipidation | 1 | 4 | 8.18e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 3 | 97 | 8.18e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 1 | 14 | 8.18e-01 |
| GO:BP | GO:0050747 | positive regulation of lipoprotein metabolic process | 1 | 4 | 8.18e-01 |
| GO:BP | GO:0048538 | thymus development | 1 | 35 | 8.18e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 22 | 782 | 8.18e-01 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 3 | 16 | 8.18e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 7 | 28 | 8.18e-01 |
| GO:BP | GO:1901000 | regulation of response to salt stress | 1 | 4 | 8.18e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 2 | 382 | 8.18e-01 |
| GO:BP | GO:0016119 | carotene metabolic process | 1 | 1 | 8.18e-01 |
| GO:BP | GO:1900069 | regulation of cellular hyperosmotic salinity response | 1 | 4 | 8.18e-01 |
| GO:BP | GO:0016122 | xanthophyll metabolic process | 1 | 1 | 8.18e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 12 | 30 | 8.18e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 3 | 71 | 8.18e-01 |
| GO:BP | GO:0016121 | carotene catabolic process | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0046247 | terpene catabolic process | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 3 | 30 | 8.18e-01 |
| GO:BP | GO:0032836 | glomerular basement membrane development | 1 | 9 | 8.18e-01 |
| GO:BP | GO:0046039 | GTP metabolic process | 6 | 21 | 8.18e-01 |
| GO:BP | GO:0044000 | movement in host | 1 | 138 | 8.18e-01 |
| GO:BP | GO:1901825 | zeaxanthin metabolic process | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0048733 | sebaceous gland development | 1 | 6 | 8.18e-01 |
| GO:BP | GO:0090327 | negative regulation of locomotion involved in locomotory behavior | 1 | 1 | 8.18e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 8 | 199 | 8.18e-01 |
| GO:BP | GO:0016115 | terpenoid catabolic process | 2 | 4 | 8.18e-01 |
| GO:BP | GO:1902661 | positive regulation of glucose mediated signaling pathway | 1 | 3 | 8.19e-01 |
| GO:BP | GO:1902659 | regulation of glucose mediated signaling pathway | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0106005 | RNA 5’-cap (guanine-N7)-methylation | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 5 | 172 | 8.19e-01 |
| GO:BP | GO:0097028 | dendritic cell differentiation | 2 | 23 | 8.19e-01 |
| GO:BP | GO:1904251 | regulation of bile acid metabolic process | 1 | 9 | 8.19e-01 |
| GO:BP | GO:0035672 | oligopeptide transmembrane transport | 2 | 5 | 8.19e-01 |
| GO:BP | GO:0042745 | circadian sleep/wake cycle | 2 | 11 | 8.19e-01 |
| GO:BP | GO:0006857 | oligopeptide transport | 2 | 5 | 8.19e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 4 | 142 | 8.19e-01 |
| GO:BP | GO:2000674 | regulation of type B pancreatic cell apoptotic process | 1 | 6 | 8.19e-01 |
| GO:BP | GO:1902988 | neurofibrillary tangle assembly | 1 | 3 | 8.19e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 2 | 25 | 8.19e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 2 | 74 | 8.19e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 1 | 19 | 8.19e-01 |
| GO:BP | GO:0003342 | proepicardium development | 1 | 2 | 8.19e-01 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 2 | 11 | 8.19e-01 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 2 | 11 | 8.19e-01 |
| GO:BP | GO:0097101 | blood vessel endothelial cell fate specification | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 2 | 3386 | 8.19e-01 |
| GO:BP | GO:0097254 | renal tubular secretion | 1 | 15 | 8.19e-01 |
| GO:BP | GO:0033084 | regulation of immature T cell proliferation in thymus | 1 | 6 | 8.19e-01 |
| GO:BP | GO:0003350 | pulmonary myocardium development | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 2 | 11 | 8.19e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 8.19e-01 |
| GO:BP | GO:0060012 | synaptic transmission, glycinergic | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0060846 | blood vessel endothelial cell fate commitment | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0033083 | regulation of immature T cell proliferation | 1 | 6 | 8.19e-01 |
| GO:BP | GO:1902172 | regulation of keratinocyte apoptotic process | 1 | 5 | 8.19e-01 |
| GO:BP | GO:0003343 | septum transversum development | 1 | 2 | 8.19e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 4 | 116 | 8.19e-01 |
| GO:BP | GO:0097283 | keratinocyte apoptotic process | 1 | 5 | 8.19e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 39 | 292 | 8.19e-01 |
| GO:BP | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0000478 | endonucleolytic cleavage involved in rRNA processing | 7 | 17 | 8.19e-01 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5770 | 8.19e-01 |
| GO:BP | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 7 | 17 | 8.19e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1 | 72 | 8.19e-01 |
| GO:BP | GO:0021578 | hindbrain maturation | 1 | 4 | 8.19e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 14 | 50 | 8.19e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 3 | 187 | 8.19e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 5 | 116 | 8.19e-01 |
| GO:BP | GO:2000659 | regulation of interleukin-1-mediated signaling pathway | 2 | 4 | 8.19e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 3 | 6 | 8.19e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 3 | 187 | 8.19e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 9 | 111 | 8.20e-01 |
| GO:BP | GO:2000982 | positive regulation of inner ear receptor cell differentiation | 1 | 1 | 8.20e-01 |
| GO:BP | GO:1990418 | response to insulin-like growth factor stimulus | 1 | 2 | 8.20e-01 |
| GO:BP | GO:0097021 | lymphocyte migration into lymphoid organs | 1 | 6 | 8.20e-01 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 10 | 36 | 8.20e-01 |
| GO:BP | GO:1903744 | positive regulation of anterograde synaptic vesicle transport | 1 | 2 | 8.20e-01 |
| GO:BP | GO:0014852 | regulation of skeletal muscle contraction by neural stimulation via neuromuscular junction | 1 | 2 | 8.20e-01 |
| GO:BP | GO:1904811 | positive regulation of dense core granule transport | 1 | 1 | 8.20e-01 |
| GO:BP | GO:1904809 | regulation of dense core granule transport | 1 | 1 | 8.20e-01 |
| GO:BP | GO:1901951 | regulation of anterograde dense core granule transport | 1 | 1 | 8.20e-01 |
| GO:BP | GO:1901843 | positive regulation of high voltage-gated calcium channel activity | 1 | 4 | 8.20e-01 |
| GO:BP | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 1 | 10 | 8.20e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 3 | 26 | 8.20e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 19 | 40 | 8.20e-01 |
| GO:BP | GO:1900100 | positive regulation of plasma cell differentiation | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0030073 | insulin secretion | 6 | 141 | 8.20e-01 |
| GO:BP | GO:0071332 | cellular response to fructose stimulus | 1 | 2 | 8.20e-01 |
| GO:BP | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1 | 3 | 8.20e-01 |
| GO:BP | GO:0031133 | regulation of axon diameter | 1 | 4 | 8.20e-01 |
| GO:BP | GO:0006097 | glyoxylate cycle | 1 | 2 | 8.20e-01 |
| GO:BP | GO:0045633 | positive regulation of mechanoreceptor differentiation | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0042660 | positive regulation of cell fate specification | 1 | 1 | 8.20e-01 |
| GO:BP | GO:1905552 | positive regulation of protein localization to endoplasmic reticulum | 1 | 2 | 8.20e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 8.20e-01 |
| GO:BP | GO:1901953 | positive regulation of anterograde dense core granule transport | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0015833 | peptide transport | 8 | 179 | 8.20e-01 |
| GO:BP | GO:1902805 | positive regulation of synaptic vesicle transport | 1 | 2 | 8.20e-01 |
| GO:BP | GO:1903487 | regulation of lactation | 1 | 1 | 8.20e-01 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 1 | 7 | 8.20e-01 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 2 | 9 | 8.20e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 8 | 25 | 8.20e-01 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 8.20e-01 |
| GO:BP | GO:0045609 | positive regulation of inner ear auditory receptor cell differentiation | 1 | 1 | 8.20e-01 |
| GO:BP | GO:1902913 | positive regulation of neuroepithelial cell differentiation | 1 | 2 | 8.20e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 3 | 28 | 8.20e-01 |
| GO:BP | GO:0035964 | COPI-coated vesicle budding | 1 | 6 | 8.20e-01 |
| GO:BP | GO:0036260 | RNA capping | 2 | 20 | 8.20e-01 |
| GO:BP | GO:0048200 | Golgi transport vesicle coating | 1 | 6 | 8.20e-01 |
| GO:BP | GO:0048205 | COPI coating of Golgi vesicle | 1 | 6 | 8.20e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 8.21e-01 |
| GO:BP | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death | 1 | 3 | 8.21e-01 |
| GO:BP | GO:0007512 | adult heart development | 2 | 9 | 8.22e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 3 | 23 | 8.22e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 1 | 49 | 8.22e-01 |
| GO:BP | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly | 1 | 7 | 8.22e-01 |
| GO:BP | GO:0035482 | gastric motility | 1 | 3 | 8.22e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 5 | 29 | 8.22e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 8.22e-01 |
| GO:BP | GO:1901159 | xylulose 5-phosphate biosynthetic process | 2 | 4 | 8.22e-01 |
| GO:BP | GO:0035505 | positive regulation of myosin light chain kinase activity | 1 | 1 | 8.22e-01 |
| GO:BP | GO:0033602 | negative regulation of dopamine secretion | 2 | 4 | 8.22e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 6 | 75 | 8.22e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 1 | 185 | 8.22e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 2 | 28 | 8.22e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 3 | 7 | 8.22e-01 |
| GO:BP | GO:0032241 | positive regulation of nucleobase-containing compound transport | 2 | 6 | 8.22e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 2 | 31 | 8.22e-01 |
| GO:BP | GO:0038007 | netrin-activated signaling pathway | 4 | 8 | 8.22e-01 |
| GO:BP | GO:0051167 | xylulose 5-phosphate metabolic process | 2 | 4 | 8.22e-01 |
| GO:BP | GO:0006064 | glucuronate catabolic process | 2 | 4 | 8.22e-01 |
| GO:BP | GO:0035726 | common myeloid progenitor cell proliferation | 2 | 5 | 8.22e-01 |
| GO:BP | GO:0035504 | regulation of myosin light chain kinase activity | 1 | 1 | 8.22e-01 |
| GO:BP | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | 2 | 4 | 8.22e-01 |
| GO:BP | GO:0045218 | zonula adherens maintenance | 1 | 3 | 8.22e-01 |
| GO:BP | GO:1903393 | positive regulation of adherens junction organization | 1 | 2 | 8.23e-01 |
| GO:BP | GO:0045056 | transcytosis | 3 | 17 | 8.23e-01 |
| GO:BP | GO:0034983 | peptidyl-lysine deacetylation | 2 | 8 | 8.23e-01 |
| GO:BP | GO:0097240 | chromosome attachment to the nuclear envelope | 3 | 3 | 8.23e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 8 | 275 | 8.24e-01 |
| GO:BP | GO:0060177 | regulation of angiotensin metabolic process | 1 | 1 | 8.24e-01 |
| GO:BP | GO:1903597 | negative regulation of gap junction assembly | 1 | 1 | 8.24e-01 |
| GO:BP | GO:0048148 | behavioral response to cocaine | 4 | 9 | 8.24e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 13 | 62 | 8.24e-01 |
| GO:BP | GO:0043379 | memory T cell differentiation | 1 | 5 | 8.24e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 2 | 229 | 8.24e-01 |
| GO:BP | GO:0009067 | aspartate family amino acid biosynthetic process | 2 | 18 | 8.24e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 4 | 232 | 8.24e-01 |
| GO:BP | GO:2000016 | negative regulation of determination of dorsal identity | 1 | 1 | 8.24e-01 |
| GO:BP | GO:0071624 | positive regulation of granulocyte chemotaxis | 1 | 14 | 8.24e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 8.24e-01 |
| GO:BP | GO:0006438 | valyl-tRNA aminoacylation | 1 | 2 | 8.25e-01 |
| GO:BP | GO:1905912 | regulation of calcium ion export across plasma membrane | 1 | 4 | 8.25e-01 |
| GO:BP | GO:1905913 | negative regulation of calcium ion export across plasma membrane | 1 | 4 | 8.25e-01 |
| GO:BP | GO:0010533 | regulation of activation of Janus kinase activity | 1 | 3 | 8.26e-01 |
| GO:BP | GO:1903526 | negative regulation of membrane tubulation | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 5 | 15 | 8.26e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 8.26e-01 |
| GO:BP | GO:0042350 | GDP-L-fucose biosynthetic process | 1 | 3 | 8.26e-01 |
| GO:BP | GO:0060263 | regulation of respiratory burst | 1 | 10 | 8.26e-01 |
| GO:BP | GO:1900194 | negative regulation of oocyte maturation | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0031998 | regulation of fatty acid beta-oxidation | 5 | 16 | 8.26e-01 |
| GO:BP | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | 2 | 5 | 8.26e-01 |
| GO:BP | GO:0033227 | dsRNA transport | 1 | 2 | 8.26e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 4 | 161 | 8.27e-01 |
| GO:BP | GO:0034203 | glycolipid translocation | 1 | 1 | 8.27e-01 |
| GO:BP | GO:2000805 | negative regulation of termination of RNA polymerase II transcription, poly(A)-coupled | 1 | 2 | 8.27e-01 |
| GO:BP | GO:0060567 | negative regulation of termination of DNA-templated transcription | 1 | 2 | 8.27e-01 |
| GO:BP | GO:0120191 | negative regulation of termination of RNA polymerase II transcription | 1 | 2 | 8.27e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 1 | 20 | 8.27e-01 |
| GO:BP | GO:2000192 | negative regulation of fatty acid transport | 1 | 7 | 8.27e-01 |
| GO:BP | GO:0002859 | negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 1 | 8.27e-01 |
| GO:BP | GO:0038158 | granulocyte colony-stimulating factor signaling pathway | 1 | 2 | 8.27e-01 |
| GO:BP | GO:0097749 | membrane tubulation | 1 | 4 | 8.27e-01 |
| GO:BP | GO:0006011 | UDP-glucose metabolic process | 2 | 6 | 8.27e-01 |
| GO:BP | GO:0007411 | axon guidance | 7 | 185 | 8.27e-01 |
| GO:BP | GO:0002856 | negative regulation of natural killer cell mediated immune response to tumor cell | 1 | 1 | 8.27e-01 |
| GO:BP | GO:0009313 | oligosaccharide catabolic process | 1 | 11 | 8.27e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 8.27e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 3 | 10 | 8.27e-01 |
| GO:BP | GO:0010286 | heat acclimation | 2 | 5 | 8.27e-01 |
| GO:BP | GO:0070370 | cellular heat acclimation | 2 | 5 | 8.27e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 1 | 16 | 8.27e-01 |
| GO:BP | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | 1 | 5 | 8.27e-01 |
| GO:BP | GO:0051695 | actin filament uncapping | 1 | 2 | 8.28e-01 |
| GO:BP | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway | 1 | 1 | 8.28e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 40 | 8.28e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 10 | 26 | 8.28e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 3 | 47 | 8.28e-01 |
| GO:BP | GO:0051179 | localization | 2 | 3969 | 8.28e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 1 | 74 | 8.28e-01 |
| GO:BP | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway | 3 | 6 | 8.28e-01 |
| GO:BP | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 6 | 8.28e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 9 | 123 | 8.29e-01 |
| GO:BP | GO:0042989 | sequestering of actin monomers | 1 | 9 | 8.29e-01 |
| GO:BP | GO:0061888 | regulation of astrocyte activation | 2 | 3 | 8.29e-01 |
| GO:BP | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient | 1 | 7 | 8.29e-01 |
| GO:BP | GO:1990399 | epithelium regeneration | 1 | 2 | 8.29e-01 |
| GO:BP | GO:1905041 | regulation of epithelium regeneration | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 2 | 64 | 8.29e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 73 | 8.29e-01 |
| GO:BP | GO:0001709 | cell fate determination | 8 | 25 | 8.29e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 11 | 30 | 8.29e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 11 | 30 | 8.29e-01 |
| GO:BP | GO:1902626 | assembly of large subunit precursor of preribosome | 1 | 1 | 8.30e-01 |
| GO:BP | GO:1903766 | positive regulation of potassium ion export across plasma membrane | 1 | 3 | 8.30e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 5 | 155 | 8.30e-01 |
| GO:BP | GO:0021553 | olfactory nerve development | 3 | 5 | 8.30e-01 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 3 | 8 | 8.30e-01 |
| GO:BP | GO:0045292 | mRNA cis splicing, via spliceosome | 1 | 26 | 8.30e-01 |
| GO:BP | GO:0000960 | regulation of mitochondrial RNA catabolic process | 2 | 5 | 8.30e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 55 | 1345 | 8.30e-01 |
| GO:BP | GO:0051311 | meiotic metaphase plate congression | 1 | 2 | 8.30e-01 |
| GO:BP | GO:1904958 | positive regulation of midbrain dopaminergic neuron differentiation | 1 | 3 | 8.30e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 8.30e-01 |
| GO:BP | GO:0071380 | cellular response to prostaglandin E stimulus | 1 | 14 | 8.30e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 23 | 61 | 8.30e-01 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 5 | 31 | 8.30e-01 |
| GO:BP | GO:0060921 | sinoatrial node cell differentiation | 1 | 5 | 8.31e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 2 | 230 | 8.31e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 10 | 29 | 8.31e-01 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 1 | 3 | 8.31e-01 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 2 | 5 | 8.31e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 5 | 166 | 8.31e-01 |
| GO:BP | GO:0071352 | cellular response to interleukin-2 | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0002322 | B cell proliferation involved in immune response | 2 | 3 | 8.31e-01 |
| GO:BP | GO:0003099 | positive regulation of the force of heart contraction by chemical signal | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0035356 | intracellular triglyceride homeostasis | 2 | 5 | 8.31e-01 |
| GO:BP | GO:0038110 | interleukin-2-mediated signaling pathway | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0045672 | positive regulation of osteoclast differentiation | 2 | 11 | 8.31e-01 |
| GO:BP | GO:0001547 | antral ovarian follicle growth | 3 | 5 | 8.31e-01 |
| GO:BP | GO:0070602 | regulation of centromeric sister chromatid cohesion | 1 | 4 | 8.31e-01 |
| GO:BP | GO:0006651 | diacylglycerol biosynthetic process | 3 | 7 | 8.31e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 8.31e-01 |
| GO:BP | GO:0046813 | receptor-mediated virion attachment to host cell | 1 | 6 | 8.31e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 18 | 39 | 8.31e-01 |
| GO:BP | GO:2000251 | positive regulation of actin cytoskeleton reorganization | 2 | 14 | 8.31e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 8.31e-01 |
| GO:BP | GO:0090297 | positive regulation of mitochondrial DNA replication | 1 | 2 | 8.31e-01 |
| GO:BP | GO:0000494 | box C/D RNA 3’-end processing | 1 | 3 | 8.31e-01 |
| GO:BP | GO:0002729 | positive regulation of natural killer cell cytokine production | 1 | 4 | 8.31e-01 |
| GO:BP | GO:0034963 | box C/D RNA processing | 1 | 3 | 8.31e-01 |
| GO:BP | GO:0033967 | box C/D RNA metabolic process | 1 | 3 | 8.31e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 8.32e-01 |
| GO:BP | GO:0045976 | negative regulation of mitotic cell cycle, embryonic | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 1 | 215 | 8.32e-01 |
| GO:BP | GO:0045329 | carnitine biosynthetic process | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 6 | 39 | 8.32e-01 |
| GO:BP | GO:0002282 | microglial cell activation involved in immune response | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0006491 | N-glycan processing | 3 | 18 | 8.32e-01 |
| GO:BP | GO:0072343 | pancreatic stellate cell proliferation | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0006532 | aspartate biosynthetic process | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0006106 | fumarate metabolic process | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 2 | 19 | 8.32e-01 |
| GO:BP | GO:0071306 | cellular response to vitamin E | 1 | 2 | 8.32e-01 |
| GO:BP | GO:2000535 | regulation of entry of bacterium into host cell | 1 | 6 | 8.32e-01 |
| GO:BP | GO:0060694 | regulation of cholesterol transporter activity | 1 | 1 | 8.32e-01 |
| GO:BP | GO:1902564 | negative regulation of neutrophil activation | 1 | 2 | 8.32e-01 |
| GO:BP | GO:1903286 | regulation of potassium ion import | 2 | 8 | 8.32e-01 |
| GO:BP | GO:1901558 | response to metformin | 1 | 1 | 8.32e-01 |
| GO:BP | GO:2000229 | regulation of pancreatic stellate cell proliferation | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0017185 | peptidyl-lysine hydroxylation | 1 | 9 | 8.32e-01 |
| GO:BP | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress | 1 | 5 | 8.32e-01 |
| GO:BP | GO:1903595 | positive regulation of histamine secretion by mast cell | 1 | 4 | 8.32e-01 |
| GO:BP | GO:1903593 | regulation of histamine secretion by mast cell | 1 | 4 | 8.32e-01 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 1 | 63 | 8.32e-01 |
| GO:BP | GO:0034441 | plasma lipoprotein particle oxidation | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 1 | 27 | 8.32e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 6 | 53 | 8.32e-01 |
| GO:BP | GO:0038196 | type III interferon-mediated signaling pathway | 2 | 4 | 8.32e-01 |
| GO:BP | GO:0071358 | cellular response to type III interferon | 2 | 4 | 8.32e-01 |
| GO:BP | GO:1904597 | negative regulation of connective tissue replacement involved in inflammatory response wound healing | 1 | 1 | 8.32e-01 |
| GO:BP | GO:1904596 | regulation of connective tissue replacement involved in inflammatory response wound healing | 1 | 1 | 8.32e-01 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 5 | 8.32e-01 |
| GO:BP | GO:1905204 | negative regulation of connective tissue replacement | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0150007 | clathrin-dependent synaptic vesicle endocytosis | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0045627 | positive regulation of T-helper 1 cell differentiation | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0033091 | positive regulation of immature T cell proliferation | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0033092 | positive regulation of immature T cell proliferation in thymus | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 3 | 3580 | 8.32e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 3 | 26 | 8.32e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 44 | 1014 | 8.32e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 1 | 199 | 8.32e-01 |
| GO:BP | GO:0071316 | cellular response to nicotine | 1 | 4 | 8.32e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 2 | 295 | 8.32e-01 |
| GO:BP | GO:1905384 | regulation of protein localization to presynapse | 1 | 2 | 8.32e-01 |
| GO:BP | GO:1905386 | positive regulation of protein localization to presynapse | 1 | 2 | 8.32e-01 |
| GO:BP | GO:2000405 | negative regulation of T cell migration | 1 | 4 | 8.32e-01 |
| GO:BP | GO:1905793 | protein localization to pericentriolar material | 1 | 2 | 8.32e-01 |
| GO:BP | GO:0045643 | regulation of eosinophil differentiation | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0045645 | positive regulation of eosinophil differentiation | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0044070 | regulation of monoatomic anion transport | 7 | 12 | 8.32e-01 |
| GO:BP | GO:1905380 | regulation of snRNA transcription by RNA polymerase II | 1 | 4 | 8.32e-01 |
| GO:BP | GO:0035026 | leading edge cell differentiation | 1 | 2 | 8.33e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 8.33e-01 |
| GO:BP | GO:1904683 | regulation of metalloendopeptidase activity | 2 | 6 | 8.33e-01 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 1 | 11 | 8.34e-01 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 1 | 12 | 8.34e-01 |
| GO:BP | GO:0003176 | aortic valve development | 15 | 37 | 8.34e-01 |
| GO:BP | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 3 | 8.34e-01 |
| GO:BP | GO:0061548 | ganglion development | 7 | 12 | 8.34e-01 |
| GO:BP | GO:0060428 | lung epithelium development | 12 | 30 | 8.34e-01 |
| GO:BP | GO:0021798 | forebrain dorsal/ventral pattern formation | 1 | 2 | 8.35e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 5 | 194 | 8.35e-01 |
| GO:BP | GO:0022400 | regulation of rhodopsin mediated signaling pathway | 3 | 5 | 8.35e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 4 | 252 | 8.35e-01 |
| GO:BP | GO:0010989 | negative regulation of low-density lipoprotein particle clearance | 2 | 4 | 8.35e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 2 | 2845 | 8.36e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 15 | 2270 | 8.36e-01 |
| GO:BP | GO:0051693 | actin filament capping | 1 | 34 | 8.36e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 4 | 135 | 8.36e-01 |
| GO:BP | GO:0090325 | regulation of locomotion involved in locomotory behavior | 2 | 3 | 8.36e-01 |
| GO:BP | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development | 1 | 2 | 8.36e-01 |
| GO:BP | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate | 1 | 4 | 8.36e-01 |
| GO:BP | GO:0034138 | toll-like receptor 3 signaling pathway | 2 | 12 | 8.36e-01 |
| GO:BP | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | 1 | 5 | 8.37e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 8 | 112 | 8.37e-01 |
| GO:BP | GO:0045347 | negative regulation of MHC class II biosynthetic process | 2 | 3 | 8.37e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 8.37e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 8.37e-01 |
| GO:BP | GO:1905167 | positive regulation of lysosomal protein catabolic process | 1 | 4 | 8.37e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 8.37e-01 |
| GO:BP | GO:0019884 | antigen processing and presentation of exogenous antigen | 2 | 28 | 8.37e-01 |
| GO:BP | GO:0009812 | flavonoid metabolic process | 1 | 4 | 8.37e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 4 | 73 | 8.37e-01 |
| GO:BP | GO:0010501 | RNA secondary structure unwinding | 2 | 8 | 8.37e-01 |
| GO:BP | GO:0051235 | maintenance of location | 1 | 260 | 8.37e-01 |
| GO:BP | GO:0071344 | diphosphate metabolic process | 1 | 2 | 8.37e-01 |
| GO:BP | GO:0007431 | salivary gland development | 2 | 26 | 8.37e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 20 | 551 | 8.37e-01 |
| GO:BP | GO:0061762 | CAMKK-AMPK signaling cascade | 3 | 5 | 8.37e-01 |
| GO:BP | GO:1903371 | regulation of endoplasmic reticulum tubular network organization | 1 | 6 | 8.38e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 2 | 37 | 8.38e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 2 | 37 | 8.38e-01 |
| GO:BP | GO:0032328 | alanine transport | 2 | 14 | 8.38e-01 |
| GO:BP | GO:1900119 | positive regulation of execution phase of apoptosis | 1 | 7 | 8.38e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 8.38e-01 |
| GO:BP | GO:0019835 | cytolysis | 1 | 6 | 8.38e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 8.38e-01 |
| GO:BP | GO:0071492 | cellular response to UV-A | 1 | 8 | 8.39e-01 |
| GO:BP | GO:0015879 | carnitine transport | 2 | 4 | 8.39e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 1 | 16 | 8.39e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 1 | 17 | 8.39e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 3 | 12 | 8.40e-01 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 1 | 82 | 8.40e-01 |
| GO:BP | GO:0046959 | habituation | 1 | 5 | 8.40e-01 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 1 | 82 | 8.40e-01 |
| GO:BP | GO:0006689 | ganglioside catabolic process | 1 | 5 | 8.40e-01 |
| GO:BP | GO:0090715 | immunological memory formation process | 1 | 6 | 8.40e-01 |
| GO:BP | GO:0035621 | ER to Golgi ceramide transport | 1 | 3 | 8.40e-01 |
| GO:BP | GO:0042886 | amide transport | 8 | 235 | 8.40e-01 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 7 | 21 | 8.41e-01 |
| GO:BP | GO:0035442 | dipeptide transmembrane transport | 1 | 4 | 8.41e-01 |
| GO:BP | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0098967 | exocytic insertion of neurotransmitter receptor to postsynaptic membrane | 1 | 5 | 8.41e-01 |
| GO:BP | GO:0072125 | negative regulation of glomerular mesangial cell proliferation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 4 | 63 | 8.41e-01 |
| GO:BP | GO:1905312 | positive regulation of cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0035993 | deltoid tuberosity development | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0035992 | tendon formation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0035990 | tendon cell differentiation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0003130 | BMP signaling pathway involved in heart induction | 1 | 2 | 8.41e-01 |
| GO:BP | GO:1905310 | regulation of cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 3 | 8.41e-01 |
| GO:BP | GO:0003134 | endodermal-mesodermal cell signaling involved in heart induction | 1 | 2 | 8.41e-01 |
| GO:BP | GO:1901998 | toxin transport | 3 | 37 | 8.41e-01 |
| GO:BP | GO:0042938 | dipeptide transport | 1 | 4 | 8.41e-01 |
| GO:BP | GO:0003133 | endodermal-mesodermal cell signaling | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0097048 | dendritic cell apoptotic process | 2 | 7 | 8.41e-01 |
| GO:BP | GO:0072198 | mesenchymal cell proliferation involved in ureter development | 1 | 1 | 8.41e-01 |
| GO:BP | GO:2000668 | regulation of dendritic cell apoptotic process | 2 | 7 | 8.41e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 6 | 70 | 8.41e-01 |
| GO:BP | GO:0072199 | regulation of mesenchymal cell proliferation involved in ureter development | 1 | 1 | 8.41e-01 |
| GO:BP | GO:0030168 | platelet activation | 8 | 91 | 8.41e-01 |
| GO:BP | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 2 | 11 | 8.41e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 2 | 3 | 8.41e-01 |
| GO:BP | GO:1905749 | regulation of endosome to plasma membrane protein transport | 1 | 5 | 8.41e-01 |
| GO:BP | GO:0048069 | eye pigmentation | 2 | 4 | 8.41e-01 |
| GO:BP | GO:0060319 | primitive erythrocyte differentiation | 2 | 4 | 8.42e-01 |
| GO:BP | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 3 | 6 | 8.42e-01 |
| GO:BP | GO:0031179 | peptide modification | 1 | 4 | 8.42e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 15 | 44 | 8.42e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 11 | 28 | 8.42e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 1 | 154 | 8.42e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 10 | 151 | 8.42e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 11 | 994 | 8.43e-01 |
| GO:BP | GO:0021682 | nerve maturation | 1 | 2 | 8.43e-01 |
| GO:BP | GO:0034204 | lipid translocation | 1 | 38 | 8.43e-01 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 2 | 11 | 8.43e-01 |
| GO:BP | GO:0051867 | general adaptation syndrome, behavioral process | 1 | 3 | 8.43e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 4 | 19 | 8.43e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 1 | 33 | 8.43e-01 |
| GO:BP | GO:1904179 | positive regulation of adipose tissue development | 1 | 6 | 8.43e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 6 | 31 | 8.44e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 8.44e-01 |
| GO:BP | GO:0030277 | maintenance of gastrointestinal epithelium | 1 | 8 | 8.44e-01 |
| GO:BP | GO:0021781 | glial cell fate commitment | 4 | 5 | 8.44e-01 |
| GO:BP | GO:0006546 | glycine catabolic process | 1 | 3 | 8.45e-01 |
| GO:BP | GO:1900150 | regulation of defense response to fungus | 2 | 3 | 8.45e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 9 | 39 | 8.45e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 9 | 39 | 8.45e-01 |
| GO:BP | GO:1903596 | regulation of gap junction assembly | 2 | 8 | 8.45e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 2 | 11 | 8.45e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 5 | 66 | 8.45e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 1 | 10 | 8.45e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 2 | 17 | 8.45e-01 |
| GO:BP | GO:0030049 | muscle filament sliding | 1 | 10 | 8.45e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 4 | 139 | 8.45e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 3 | 348 | 8.46e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 14 | 395 | 8.46e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 4 | 13 | 8.46e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 4 | 155 | 8.46e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 8.46e-01 |
| GO:BP | GO:1903391 | regulation of adherens junction organization | 2 | 5 | 8.46e-01 |
| GO:BP | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production | 1 | 4 | 8.46e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 2 | 13 | 8.46e-01 |
| GO:BP | GO:0060693 | regulation of branching involved in salivary gland morphogenesis | 2 | 6 | 8.46e-01 |
| GO:BP | GO:0060457 | negative regulation of digestive system process | 1 | 7 | 8.46e-01 |
| GO:BP | GO:1905247 | positive regulation of aspartic-type peptidase activity | 2 | 8 | 8.46e-01 |
| GO:BP | GO:0022600 | digestive system process | 7 | 54 | 8.46e-01 |
| GO:BP | GO:0097068 | response to thyroxine | 2 | 4 | 8.46e-01 |
| GO:BP | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway | 1 | 4 | 8.46e-01 |
| GO:BP | GO:0033007 | negative regulation of mast cell activation involved in immune response | 4 | 4 | 8.46e-01 |
| GO:BP | GO:1904582 | positive regulation of intracellular mRNA localization | 1 | 3 | 8.46e-01 |
| GO:BP | GO:1990603 | dark adaptation | 1 | 3 | 8.47e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 2 | 110 | 8.47e-01 |
| GO:BP | GO:0042053 | regulation of dopamine metabolic process | 5 | 11 | 8.47e-01 |
| GO:BP | GO:0042069 | regulation of catecholamine metabolic process | 5 | 11 | 8.47e-01 |
| GO:BP | GO:0071485 | cellular response to absence of light | 1 | 3 | 8.47e-01 |
| GO:BP | GO:0009646 | response to absence of light | 1 | 3 | 8.47e-01 |
| GO:BP | GO:0021550 | medulla oblongata development | 1 | 2 | 8.47e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 8.47e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 4 | 7 | 8.47e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 8.47e-01 |
| GO:BP | GO:2001054 | negative regulation of mesenchymal cell apoptotic process | 1 | 3 | 8.48e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 20 | 62 | 8.49e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 10 | 465 | 8.49e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 3 | 22 | 8.49e-01 |
| GO:BP | GO:0003284 | septum primum development | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0042412 | taurine biosynthetic process | 1 | 2 | 8.49e-01 |
| GO:BP | GO:2000172 | regulation of branching morphogenesis of a nerve | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0015670 | carbon dioxide transport | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 5 | 11 | 8.49e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 11 | 51 | 8.49e-01 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 4 | 25 | 8.49e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 10 | 305 | 8.49e-01 |
| GO:BP | GO:0046439 | L-cysteine metabolic process | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0019448 | L-cysteine catabolic process | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0098528 | skeletal muscle fiber differentiation | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 3 | 4292 | 8.49e-01 |
| GO:BP | GO:0009093 | cysteine catabolic process | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 2 | 168 | 8.49e-01 |
| GO:BP | GO:0046305 | alkanesulfonate biosynthetic process | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 2 | 242 | 8.49e-01 |
| GO:BP | GO:0090131 | mesenchyme migration | 1 | 4 | 8.49e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 2 | 5 | 8.49e-01 |
| GO:BP | GO:0110095 | cellular detoxification of aldehyde | 1 | 5 | 8.49e-01 |
| GO:BP | GO:0051036 | regulation of endosome size | 2 | 13 | 8.49e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 8.50e-01 |
| GO:BP | GO:0006891 | intra-Golgi vesicle-mediated transport | 14 | 33 | 8.50e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 3 | 42 | 8.50e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 2 | 111 | 8.50e-01 |
| GO:BP | GO:0015917 | aminophospholipid transport | 2 | 8 | 8.50e-01 |
| GO:BP | GO:0070085 | glycosylation | 3 | 202 | 8.50e-01 |
| GO:BP | GO:0007608 | sensory perception of smell | 1 | 16 | 8.50e-01 |
| GO:BP | GO:2000407 | regulation of T cell extravasation | 1 | 3 | 8.50e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 4 | 5017 | 8.50e-01 |
| GO:BP | GO:0010753 | positive regulation of cGMP-mediated signaling | 1 | 3 | 8.50e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 8.50e-01 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 4 | 20 | 8.50e-01 |
| GO:BP | GO:0033523 | histone H2B ubiquitination | 2 | 10 | 8.50e-01 |
| GO:BP | GO:0034471 | ncRNA 5’-end processing | 2 | 5 | 8.50e-01 |
| GO:BP | GO:0000967 | rRNA 5’-end processing | 2 | 5 | 8.50e-01 |
| GO:BP | GO:0007632 | visual behavior | 9 | 41 | 8.50e-01 |
| GO:BP | GO:0080009 | mRNA methylation | 4 | 16 | 8.50e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 19 | 40 | 8.50e-01 |
| GO:BP | GO:0001922 | B-1 B cell homeostasis | 1 | 4 | 8.50e-01 |
| GO:BP | GO:0000472 | endonucleolytic cleavage to generate mature 5’-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 5 | 8.50e-01 |
| GO:BP | GO:0071671 | regulation of smooth muscle cell chemotaxis | 2 | 4 | 8.51e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 8.51e-01 |
| GO:BP | GO:1900272 | negative regulation of long-term synaptic potentiation | 1 | 8 | 8.51e-01 |
| GO:BP | GO:0021564 | vagus nerve development | 1 | 2 | 8.52e-01 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 1 | 7 | 8.52e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 26 | 631 | 8.52e-01 |
| GO:BP | GO:0031944 | negative regulation of glucocorticoid metabolic process | 1 | 5 | 8.52e-01 |
| GO:BP | GO:0031947 | negative regulation of glucocorticoid biosynthetic process | 1 | 5 | 8.52e-01 |
| GO:BP | GO:1904037 | positive regulation of epithelial cell apoptotic process | 1 | 26 | 8.52e-01 |
| GO:BP | GO:0019682 | glyceraldehyde-3-phosphate metabolic process | 2 | 5 | 8.52e-01 |
| GO:BP | GO:1903976 | negative regulation of glial cell migration | 1 | 5 | 8.53e-01 |
| GO:BP | GO:0098597 | observational learning | 1 | 5 | 8.53e-01 |
| GO:BP | GO:0061535 | glutamate secretion, neurotransmission | 1 | 5 | 8.53e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 12 | 33 | 8.53e-01 |
| GO:BP | GO:0010887 | negative regulation of cholesterol storage | 4 | 9 | 8.53e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 2 | 32 | 8.53e-01 |
| GO:BP | GO:0035684 | helper T cell extravasation | 1 | 3 | 8.53e-01 |
| GO:BP | GO:0002295 | T-helper cell lineage commitment | 1 | 7 | 8.53e-01 |
| GO:BP | GO:0072176 | nephric duct development | 5 | 12 | 8.53e-01 |
| GO:BP | GO:0070640 | vitamin D3 metabolic process | 2 | 2 | 8.53e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 4 | 65 | 8.53e-01 |
| GO:BP | GO:0060842 | arterial endothelial cell differentiation | 1 | 5 | 8.54e-01 |
| GO:BP | GO:0003160 | endocardium morphogenesis | 1 | 3 | 8.54e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 2 | 3012 | 8.54e-01 |
| GO:BP | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II | 1 | 17 | 8.54e-01 |
| GO:BP | GO:0006002 | fructose 6-phosphate metabolic process | 3 | 8 | 8.55e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 5 | 13 | 8.55e-01 |
| GO:BP | GO:0031204 | post-translational protein targeting to membrane, translocation | 1 | 7 | 8.55e-01 |
| GO:BP | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum | 1 | 2 | 8.55e-01 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 2 | 10 | 8.55e-01 |
| GO:BP | GO:0060215 | primitive hemopoiesis | 2 | 7 | 8.55e-01 |
| GO:BP | GO:2000124 | regulation of endocannabinoid signaling pathway | 1 | 3 | 8.55e-01 |
| GO:BP | GO:0045625 | regulation of T-helper 1 cell differentiation | 2 | 5 | 8.55e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 1 | 31 | 8.55e-01 |
| GO:BP | GO:0106077 | histone succinylation | 1 | 4 | 8.55e-01 |
| GO:BP | GO:0018335 | protein succinylation | 1 | 4 | 8.55e-01 |
| GO:BP | GO:0071929 | alpha-tubulin acetylation | 1 | 3 | 8.55e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 1 | 31 | 8.55e-01 |
| GO:BP | GO:0072752 | cellular response to rapamycin | 1 | 2 | 8.55e-01 |
| GO:BP | GO:1990402 | embryonic liver development | 1 | 3 | 8.56e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 8.56e-01 |
| GO:BP | GO:0072102 | glomerulus morphogenesis | 1 | 7 | 8.56e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 3 | 7 | 8.56e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 8.56e-01 |
| GO:BP | GO:0071073 | positive regulation of phospholipid biosynthetic process | 3 | 7 | 8.56e-01 |
| GO:BP | GO:0070430 | positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 2 | 8.56e-01 |
| GO:BP | GO:0046185 | aldehyde catabolic process | 6 | 12 | 8.56e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 8.56e-01 |
| GO:BP | GO:1905581 | positive regulation of low-density lipoprotein particle clearance | 1 | 2 | 8.56e-01 |
| GO:BP | GO:0032483 | regulation of Rab protein signal transduction | 2 | 7 | 8.56e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 5 | 8 | 8.56e-01 |
| GO:BP | GO:0016046 | detection of fungus | 1 | 1 | 8.56e-01 |
| GO:BP | GO:1904466 | positive regulation of matrix metallopeptidase secretion | 1 | 2 | 8.56e-01 |
| GO:BP | GO:0030050 | vesicle transport along actin filament | 7 | 13 | 8.56e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 3 | 9 | 8.56e-01 |
| GO:BP | GO:0007525 | somatic muscle development | 1 | 4 | 8.56e-01 |
| GO:BP | GO:0048295 | positive regulation of isotype switching to IgE isotypes | 1 | 3 | 8.56e-01 |
| GO:BP | GO:0090289 | regulation of osteoclast proliferation | 1 | 3 | 8.56e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 3 | 9 | 8.56e-01 |
| GO:BP | GO:0001732 | formation of cytoplasmic translation initiation complex | 5 | 16 | 8.56e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 3 | 9 | 8.56e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 6 | 24 | 8.56e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 8.56e-01 |
| GO:BP | GO:1900112 | regulation of histone H3-K9 trimethylation | 1 | 5 | 8.56e-01 |
| GO:BP | GO:0006865 | amino acid transport | 3 | 102 | 8.56e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 43 | 256 | 8.57e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 15 | 8.57e-01 |
| GO:BP | GO:0015820 | leucine transport | 1 | 10 | 8.57e-01 |
| GO:BP | GO:1901537 | positive regulation of DNA demethylation | 3 | 7 | 8.57e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 1 | 957 | 8.57e-01 |
| GO:BP | GO:0002727 | regulation of natural killer cell cytokine production | 1 | 4 | 8.57e-01 |
| GO:BP | GO:0002370 | natural killer cell cytokine production | 1 | 4 | 8.57e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 4 | 9 | 8.57e-01 |
| GO:BP | GO:0006949 | syncytium formation | 2 | 39 | 8.57e-01 |
| GO:BP | GO:0030043 | actin filament fragmentation | 1 | 4 | 8.57e-01 |
| GO:BP | GO:0070988 | demethylation | 8 | 52 | 8.57e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 13 | 84 | 8.57e-01 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 1 | 9 | 8.58e-01 |
| GO:BP | GO:1900195 | positive regulation of oocyte maturation | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0098743 | cell aggregation | 3 | 12 | 8.58e-01 |
| GO:BP | GO:0016056 | rhodopsin mediated signaling pathway | 4 | 6 | 8.59e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 14 | 344 | 8.59e-01 |
| GO:BP | GO:0032502 | developmental process | 3 | 4494 | 8.59e-01 |
| GO:BP | GO:0034653 | retinoic acid catabolic process | 1 | 1 | 8.59e-01 |
| GO:BP | GO:0016103 | diterpenoid catabolic process | 1 | 1 | 8.59e-01 |
| GO:BP | GO:1902809 | regulation of skeletal muscle fiber differentiation | 1 | 4 | 8.59e-01 |
| GO:BP | GO:0110091 | negative regulation of hippocampal neuron apoptotic process | 1 | 2 | 8.59e-01 |
| GO:BP | GO:1901033 | positive regulation of response to reactive oxygen species | 2 | 6 | 8.59e-01 |
| GO:BP | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death | 2 | 6 | 8.59e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 3 | 91 | 8.59e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 5 | 13 | 8.59e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 8.59e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 1 | 25 | 8.59e-01 |
| GO:BP | GO:0007610 | behavior | 2 | 436 | 8.60e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 2 | 76 | 8.60e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 2 | 67 | 8.60e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 1 | 43 | 8.60e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 4 | 37 | 8.60e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 7 | 14 | 8.60e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 2 | 114 | 8.60e-01 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 1 | 21 | 8.60e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 10 | 21 | 8.60e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 8.61e-01 |
| GO:BP | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine | 1 | 5 | 8.61e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 8.61e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 8.61e-01 |
| GO:BP | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration | 2 | 5 | 8.61e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 3 | 64 | 8.61e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 8.61e-01 |
| GO:BP | GO:0045723 | positive regulation of fatty acid biosynthetic process | 1 | 12 | 8.61e-01 |
| GO:BP | GO:0090091 | positive regulation of extracellular matrix disassembly | 3 | 6 | 8.61e-01 |
| GO:BP | GO:0071934 | thiamine transmembrane transport | 2 | 3 | 8.61e-01 |
| GO:BP | GO:0035336 | long-chain fatty-acyl-CoA metabolic process | 10 | 19 | 8.61e-01 |
| GO:BP | GO:0042723 | thiamine-containing compound metabolic process | 2 | 4 | 8.61e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 1 | 87 | 8.61e-01 |
| GO:BP | GO:0030222 | eosinophil differentiation | 2 | 2 | 8.61e-01 |
| GO:BP | GO:2000370 | positive regulation of clathrin-dependent endocytosis | 2 | 5 | 8.62e-01 |
| GO:BP | GO:0060440 | trachea formation | 2 | 7 | 8.62e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 25 | 49 | 8.63e-01 |
| GO:BP | GO:0060183 | apelin receptor signaling pathway | 1 | 1 | 8.63e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 8.63e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 6 | 110 | 8.63e-01 |
| GO:BP | GO:0043969 | histone H2B acetylation | 1 | 3 | 8.63e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 1 | 44 | 8.63e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 1 | 38 | 8.64e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 2 | 3080 | 8.64e-01 |
| GO:BP | GO:0015867 | ATP transport | 1 | 9 | 8.64e-01 |
| GO:BP | GO:0060742 | epithelial cell differentiation involved in prostate gland development | 4 | 8 | 8.64e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 2 | 3080 | 8.64e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 3 | 4417 | 8.64e-01 |
| GO:BP | GO:2000474 | regulation of opioid receptor signaling pathway | 1 | 2 | 8.64e-01 |
| GO:BP | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 3 | 8.64e-01 |
| GO:BP | GO:0031580 | membrane raft distribution | 1 | 3 | 8.64e-01 |
| GO:BP | GO:1990834 | response to odorant | 1 | 3 | 8.64e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 8.65e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 1 | 2 | 8.65e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 2 | 77 | 8.65e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 43 | 258 | 8.65e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 2 | 105 | 8.65e-01 |
| GO:BP | GO:1903998 | regulation of eating behavior | 1 | 1 | 8.65e-01 |
| GO:BP | GO:1902644 | tertiary alcohol metabolic process | 1 | 17 | 8.65e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 8.65e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 31 | 88 | 8.65e-01 |
| GO:BP | GO:0034030 | ribonucleoside bisphosphate biosynthetic process | 22 | 49 | 8.65e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 8.65e-01 |
| GO:BP | GO:0033866 | nucleoside bisphosphate biosynthetic process | 22 | 49 | 8.65e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 4 | 276 | 8.65e-01 |
| GO:BP | GO:0019375 | galactolipid biosynthetic process | 2 | 4 | 8.65e-01 |
| GO:BP | GO:0006682 | galactosylceramide biosynthetic process | 2 | 4 | 8.65e-01 |
| GO:BP | GO:0034033 | purine nucleoside bisphosphate biosynthetic process | 22 | 49 | 8.65e-01 |
| GO:BP | GO:1905203 | regulation of connective tissue replacement | 1 | 5 | 8.65e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 4 | 147 | 8.66e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 19 | 40 | 8.66e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 19 | 40 | 8.66e-01 |
| GO:BP | GO:1990127 | intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 8.66e-01 |
| GO:BP | GO:1902238 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 8.66e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 1 | 13 | 8.66e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 3 | 433 | 8.66e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 8.66e-01 |
| GO:BP | GO:1902499 | positive regulation of protein autoubiquitination | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 8 | 27 | 8.66e-01 |
| GO:BP | GO:0043305 | negative regulation of mast cell degranulation | 2 | 2 | 8.67e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 1 | 55 | 8.67e-01 |
| GO:BP | GO:0006776 | vitamin A metabolic process | 1 | 3 | 8.67e-01 |
| GO:BP | GO:0071224 | cellular response to peptidoglycan | 1 | 2 | 8.67e-01 |
| GO:BP | GO:0070671 | response to interleukin-12 | 1 | 6 | 8.67e-01 |
| GO:BP | GO:1904732 | regulation of electron transfer activity | 3 | 8 | 8.67e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 6 | 204 | 8.67e-01 |
| GO:BP | GO:0038165 | oncostatin-M-mediated signaling pathway | 1 | 4 | 8.67e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 2 | 75 | 8.68e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 8.68e-01 |
| GO:BP | GO:0048539 | bone marrow development | 1 | 7 | 8.68e-01 |
| GO:BP | GO:0002604 | regulation of dendritic cell antigen processing and presentation | 2 | 5 | 8.68e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 1 | 55 | 8.68e-01 |
| GO:BP | GO:0045726 | positive regulation of integrin biosynthetic process | 1 | 2 | 8.68e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 5 | 554 | 8.68e-01 |
| GO:BP | GO:0060433 | bronchus development | 1 | 6 | 8.68e-01 |
| GO:BP | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development | 1 | 2 | 8.68e-01 |
| GO:BP | GO:0032681 | regulation of lymphotoxin A production | 1 | 1 | 8.68e-01 |
| GO:BP | GO:0060736 | prostate gland growth | 1 | 7 | 8.68e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 3 | 122 | 8.68e-01 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 7 | 24 | 8.68e-01 |
| GO:BP | GO:0032761 | positive regulation of lymphotoxin A production | 1 | 1 | 8.68e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 8.68e-01 |
| GO:BP | GO:0032641 | lymphotoxin A production | 1 | 1 | 8.68e-01 |
| GO:BP | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline | 1 | 9 | 8.69e-01 |
| GO:BP | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0046016 | positive regulation of transcription by glucose | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0033869 | nucleoside bisphosphate catabolic process | 2 | 11 | 8.69e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 15 | 2285 | 8.69e-01 |
| GO:BP | GO:0045471 | response to ethanol | 2 | 73 | 8.69e-01 |
| GO:BP | GO:2000831 | regulation of steroid hormone secretion | 1 | 13 | 8.69e-01 |
| GO:BP | GO:0034031 | ribonucleoside bisphosphate catabolic process | 2 | 11 | 8.69e-01 |
| GO:BP | GO:0034034 | purine nucleoside bisphosphate catabolic process | 2 | 11 | 8.69e-01 |
| GO:BP | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0006867 | asparagine transport | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0008347 | glial cell migration | 2 | 47 | 8.69e-01 |
| GO:BP | GO:1902024 | L-histidine transport | 1 | 4 | 8.69e-01 |
| GO:BP | GO:0015817 | histidine transport | 1 | 4 | 8.69e-01 |
| GO:BP | GO:0018171 | peptidyl-cysteine oxidation | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0014734 | skeletal muscle hypertrophy | 1 | 1 | 8.69e-01 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 1 | 31 | 8.70e-01 |
| GO:BP | GO:0072076 | nephrogenic mesenchyme development | 1 | 3 | 8.70e-01 |
| GO:BP | GO:0032847 | regulation of cellular pH reduction | 1 | 5 | 8.70e-01 |
| GO:BP | GO:2001225 | regulation of chloride transport | 1 | 4 | 8.70e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 2 | 23 | 8.70e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 2 | 15 | 8.71e-01 |
| GO:BP | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway | 1 | 3 | 8.71e-01 |
| GO:BP | GO:0000480 | endonucleolytic cleavage in 5’-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 8 | 8.72e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 5 | 23 | 8.72e-01 |
| GO:BP | GO:0150031 | regulation of protein localization to lysosome | 1 | 3 | 8.72e-01 |
| GO:BP | GO:0030259 | lipid glycosylation | 1 | 6 | 8.72e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 3 | 95 | 8.72e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 3 | 14 | 8.72e-01 |
| GO:BP | GO:0060336 | negative regulation of type II interferon-mediated signaling pathway | 1 | 6 | 8.72e-01 |
| GO:BP | GO:0060331 | negative regulation of response to type II interferon | 1 | 6 | 8.72e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 3 | 211 | 8.72e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 2 | 18 | 8.72e-01 |
| GO:BP | GO:0014889 | muscle atrophy | 1 | 9 | 8.72e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 8.72e-01 |
| GO:BP | GO:0090382 | phagosome maturation | 1 | 24 | 8.73e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 8.73e-01 |
| GO:BP | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | 3 | 8.73e-01 |
| GO:BP | GO:0030001 | metal ion transport | 7 | 562 | 8.73e-01 |
| GO:BP | GO:0031645 | negative regulation of nervous system process | 2 | 9 | 8.73e-01 |
| GO:BP | GO:1902624 | positive regulation of neutrophil migration | 1 | 21 | 8.73e-01 |
| GO:BP | GO:0035723 | interleukin-15-mediated signaling pathway | 1 | 2 | 8.73e-01 |
| GO:BP | GO:0071350 | cellular response to interleukin-15 | 1 | 2 | 8.73e-01 |
| GO:BP | GO:0060463 | lung lobe morphogenesis | 1 | 4 | 8.73e-01 |
| GO:BP | GO:0060462 | lung lobe development | 1 | 4 | 8.73e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 2 | 608 | 8.73e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 7 | 40 | 8.73e-01 |
| GO:BP | GO:0031652 | positive regulation of heat generation | 2 | 4 | 8.74e-01 |
| GO:BP | GO:0070170 | regulation of tooth mineralization | 1 | 4 | 8.74e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 2 | 3169 | 8.74e-01 |
| GO:BP | GO:0042426 | choline catabolic process | 1 | 3 | 8.74e-01 |
| GO:BP | GO:0021942 | radial glia guided migration of Purkinje cell | 1 | 3 | 8.75e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 28 | 1223 | 8.75e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 2 | 54 | 8.75e-01 |
| GO:BP | GO:0040019 | positive regulation of embryonic development | 1 | 17 | 8.75e-01 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 9 | 23 | 8.75e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 3 | 36 | 8.75e-01 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 1 | 10 | 8.75e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 4 | 124 | 8.75e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 7 | 19 | 8.75e-01 |
| GO:BP | GO:0015949 | nucleobase-containing small molecule interconversion | 3 | 8 | 8.75e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 15 | 2035 | 8.75e-01 |
| GO:BP | GO:0032687 | negative regulation of interferon-alpha production | 1 | 2 | 8.75e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 2 | 15 | 8.75e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 3 | 11 | 8.75e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 2 | 248 | 8.75e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 4 | 39 | 8.75e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 6 | 69 | 8.75e-01 |
| GO:BP | GO:0099541 | trans-synaptic signaling by lipid | 3 | 6 | 8.75e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 8.75e-01 |
| GO:BP | GO:0099542 | trans-synaptic signaling by endocannabinoid | 3 | 6 | 8.75e-01 |
| GO:BP | GO:0021648 | vestibulocochlear nerve morphogenesis | 2 | 3 | 8.75e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 4 | 830 | 8.75e-01 |
| GO:BP | GO:0099641 | anterograde axonal protein transport | 3 | 9 | 8.75e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 8 | 23 | 8.75e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 3 | 141 | 8.76e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 1 | 90 | 8.76e-01 |
| GO:BP | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway | 1 | 2 | 8.76e-01 |
| GO:BP | GO:0009584 | detection of visible light | 2 | 20 | 8.76e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 8.76e-01 |
| GO:BP | GO:0002253 | activation of immune response | 2 | 272 | 8.76e-01 |
| GO:BP | GO:0051238 | sequestering of metal ion | 2 | 17 | 8.76e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 3 | 384 | 8.77e-01 |
| GO:BP | GO:0060452 | positive regulation of cardiac muscle contraction | 1 | 7 | 8.77e-01 |
| GO:BP | GO:0042414 | epinephrine metabolic process | 1 | 2 | 8.77e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 16 | 31 | 8.77e-01 |
| GO:BP | GO:0034342 | response to type III interferon | 2 | 5 | 8.78e-01 |
| GO:BP | GO:0045087 | innate immune response | 1 | 488 | 8.78e-01 |
| GO:BP | GO:1904398 | positive regulation of neuromuscular junction development | 1 | 2 | 8.78e-01 |
| GO:BP | GO:0071599 | otic vesicle development | 2 | 10 | 8.78e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 31 | 88 | 8.78e-01 |
| GO:BP | GO:0031054 | pre-miRNA processing | 3 | 14 | 8.78e-01 |
| GO:BP | GO:2000418 | positive regulation of eosinophil migration | 1 | 1 | 8.78e-01 |
| GO:BP | GO:2000410 | regulation of thymocyte migration | 1 | 1 | 8.78e-01 |
| GO:BP | GO:0042110 | T cell activation | 2 | 313 | 8.78e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 21 | 61 | 8.78e-01 |
| GO:BP | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone | 2 | 5 | 8.78e-01 |
| GO:BP | GO:1904783 | positive regulation of NMDA glutamate receptor activity | 2 | 4 | 8.78e-01 |
| GO:BP | GO:0050892 | intestinal absorption | 4 | 22 | 8.78e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 8 | 108 | 8.78e-01 |
| GO:BP | GO:1901727 | positive regulation of histone deacetylase activity | 2 | 5 | 8.78e-01 |
| GO:BP | GO:0061141 | lung ciliated cell differentiation | 1 | 1 | 8.78e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 32 | 76 | 8.79e-01 |
| GO:BP | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1 | 19 | 8.79e-01 |
| GO:BP | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia | 3 | 9 | 8.79e-01 |
| GO:BP | GO:0048007 | antigen processing and presentation, exogenous lipid antigen via MHC class Ib | 2 | 3 | 8.79e-01 |
| GO:BP | GO:0002438 | acute inflammatory response to antigenic stimulus | 3 | 10 | 8.79e-01 |
| GO:BP | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib | 2 | 3 | 8.79e-01 |
| GO:BP | GO:0018343 | protein farnesylation | 1 | 2 | 8.79e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 9 | 8.79e-01 |
| GO:BP | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure | 1 | 4 | 8.79e-01 |
| GO:BP | GO:0002001 | renin secretion into blood stream | 1 | 4 | 8.79e-01 |
| GO:BP | GO:2000646 | positive regulation of receptor catabolic process | 1 | 5 | 8.79e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 6 | 23 | 8.79e-01 |
| GO:BP | GO:1903392 | negative regulation of adherens junction organization | 1 | 3 | 8.79e-01 |
| GO:BP | GO:0051712 | positive regulation of killing of cells of another organism | 1 | 2 | 8.79e-01 |
| GO:BP | GO:0036016 | cellular response to interleukin-3 | 2 | 6 | 8.79e-01 |
| GO:BP | GO:0036015 | response to interleukin-3 | 2 | 6 | 8.79e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 12 | 51 | 8.79e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 8.79e-01 |
| GO:BP | GO:0032819 | positive regulation of natural killer cell proliferation | 1 | 4 | 8.79e-01 |
| GO:BP | GO:0071211 | protein targeting to vacuole involved in autophagy | 1 | 4 | 8.79e-01 |
| GO:BP | GO:0043102 | amino acid salvage | 1 | 5 | 8.79e-01 |
| GO:BP | GO:0071798 | response to prostaglandin D | 1 | 2 | 8.79e-01 |
| GO:BP | GO:0071265 | L-methionine biosynthetic process | 1 | 5 | 8.79e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 16 | 39 | 8.79e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 8.79e-01 |
| GO:BP | GO:0071267 | L-methionine salvage | 1 | 5 | 8.79e-01 |
| GO:BP | GO:0071799 | cellular response to prostaglandin D stimulus | 1 | 2 | 8.79e-01 |
| GO:BP | GO:0000958 | mitochondrial mRNA catabolic process | 1 | 3 | 8.79e-01 |
| GO:BP | GO:0060430 | lung saccule development | 2 | 7 | 8.79e-01 |
| GO:BP | GO:0000962 | positive regulation of mitochondrial RNA catabolic process | 1 | 3 | 8.79e-01 |
| GO:BP | GO:0014827 | intestine smooth muscle contraction | 1 | 3 | 8.79e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 1 | 4 | 8.79e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 5 | 20 | 8.79e-01 |
| GO:BP | GO:0060847 | endothelial cell fate specification | 1 | 3 | 8.79e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 1 | 3 | 8.79e-01 |
| GO:BP | GO:1904878 | negative regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 4 | 8.80e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 4 | 178 | 8.80e-01 |
| GO:BP | GO:0034105 | positive regulation of tissue remodeling | 1 | 4 | 8.80e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 8 | 632 | 8.80e-01 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 1 | 43 | 8.81e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 1 | 46 | 8.81e-01 |
| GO:BP | GO:0140650 | radial glia-guided pyramidal neuron migration | 2 | 4 | 8.81e-01 |
| GO:BP | GO:0006369 | termination of RNA polymerase II transcription | 1 | 8 | 8.81e-01 |
| GO:BP | GO:0071205 | protein localization to juxtaparanode region of axon | 1 | 3 | 8.81e-01 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 1 | 102 | 8.81e-01 |
| GO:BP | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 1 | 4 | 8.81e-01 |
| GO:BP | GO:1904844 | response to L-glutamine | 1 | 3 | 8.81e-01 |
| GO:BP | GO:0030728 | ovulation | 1 | 14 | 8.81e-01 |
| GO:BP | GO:0020027 | hemoglobin metabolic process | 6 | 16 | 8.81e-01 |
| GO:BP | GO:0050904 | diapedesis | 2 | 6 | 8.81e-01 |
| GO:BP | GO:0090713 | immunological memory process | 1 | 7 | 8.81e-01 |
| GO:BP | GO:1904772 | response to tetrachloromethane | 1 | 4 | 8.81e-01 |
| GO:BP | GO:0002246 | wound healing involved in inflammatory response | 3 | 6 | 8.81e-01 |
| GO:BP | GO:0090234 | regulation of kinetochore assembly | 1 | 3 | 8.82e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 4 | 142 | 8.82e-01 |
| GO:BP | GO:0060192 | negative regulation of lipase activity | 1 | 13 | 8.82e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 3 | 39 | 8.82e-01 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 3 | 19 | 8.82e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 5 | 62 | 8.82e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 8.82e-01 |
| GO:BP | GO:0032418 | lysosome localization | 5 | 62 | 8.82e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 8.82e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 1 | 9 | 8.82e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 8.82e-01 |
| GO:BP | GO:0002038 | positive regulation of L-glutamate import across plasma membrane | 1 | 3 | 8.82e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 1 | 58 | 8.82e-01 |
| GO:BP | GO:1904751 | positive regulation of protein localization to nucleolus | 1 | 3 | 8.82e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 1 | 18 | 8.82e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 25 | 404 | 8.82e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 6 | 31 | 8.83e-01 |
| GO:BP | GO:0097305 | response to alcohol | 3 | 159 | 8.83e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 1 | 336 | 8.83e-01 |
| GO:BP | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 5 | 21 | 8.83e-01 |
| GO:BP | GO:0018242 | protein O-linked glycosylation via serine | 1 | 7 | 8.84e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 70 | 1133 | 8.84e-01 |
| GO:BP | GO:0061092 | positive regulation of phospholipid translocation | 1 | 4 | 8.84e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 11 | 53 | 8.85e-01 |
| GO:BP | GO:0090208 | positive regulation of triglyceride metabolic process | 1 | 18 | 8.85e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 2 | 74 | 8.85e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 6 | 32 | 8.85e-01 |
| GO:BP | GO:0071379 | cellular response to prostaglandin stimulus | 1 | 16 | 8.85e-01 |
| GO:BP | GO:1900193 | regulation of oocyte maturation | 2 | 5 | 8.85e-01 |
| GO:BP | GO:0032119 | sequestering of zinc ion | 2 | 5 | 8.85e-01 |
| GO:BP | GO:0140367 | antibacterial innate immune response | 1 | 2 | 8.85e-01 |
| GO:BP | GO:0001773 | myeloid dendritic cell activation | 1 | 12 | 8.85e-01 |
| GO:BP | GO:1904669 | ATP export | 1 | 1 | 8.85e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 8.85e-01 |
| GO:BP | GO:0072017 | distal tubule development | 2 | 6 | 8.85e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 3 | 26 | 8.86e-01 |
| GO:BP | GO:0001574 | ganglioside biosynthetic process | 1 | 14 | 8.86e-01 |
| GO:BP | GO:0018026 | peptidyl-lysine monomethylation | 1 | 11 | 8.86e-01 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 1 | 31 | 8.86e-01 |
| GO:BP | GO:0044211 | CTP salvage | 1 | 3 | 8.86e-01 |
| GO:BP | GO:0043373 | CD4-positive, alpha-beta T cell lineage commitment | 1 | 9 | 8.86e-01 |
| GO:BP | GO:0000338 | protein deneddylation | 4 | 11 | 8.86e-01 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 9 | 22 | 8.86e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 7 | 15 | 8.86e-01 |
| GO:BP | GO:0010360 | negative regulation of anion channel activity | 1 | 2 | 8.86e-01 |
| GO:BP | GO:1903960 | negative regulation of anion transmembrane transport | 1 | 2 | 8.86e-01 |
| GO:BP | GO:0015669 | gas transport | 3 | 6 | 8.86e-01 |
| GO:BP | GO:0046368 | GDP-L-fucose metabolic process | 1 | 4 | 8.86e-01 |
| GO:BP | GO:1905664 | regulation of calcium ion import across plasma membrane | 1 | 8 | 8.86e-01 |
| GO:BP | GO:0072350 | tricarboxylic acid metabolic process | 5 | 13 | 8.86e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 28 | 8.86e-01 |
| GO:BP | GO:0048633 | positive regulation of skeletal muscle tissue growth | 1 | 6 | 8.86e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 8.86e-01 |
| GO:BP | GO:0010984 | regulation of lipoprotein particle clearance | 1 | 12 | 8.86e-01 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 9 | 27 | 8.86e-01 |
| GO:BP | GO:0061038 | uterus morphogenesis | 2 | 5 | 8.86e-01 |
| GO:BP | GO:0046784 | viral mRNA export from host cell nucleus | 2 | 6 | 8.87e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 2 | 77 | 8.87e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 7 | 45 | 8.87e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 9 | 23 | 8.87e-01 |
| GO:BP | GO:0032929 | negative regulation of superoxide anion generation | 1 | 1 | 8.87e-01 |
| GO:BP | GO:0045728 | respiratory burst after phagocytosis | 1 | 2 | 8.87e-01 |
| GO:BP | GO:0035188 | hatching | 9 | 23 | 8.87e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 9 | 23 | 8.87e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 8.87e-01 |
| GO:BP | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway | 2 | 5 | 8.88e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 6 | 40 | 8.88e-01 |
| GO:BP | GO:1903852 | positive regulation of cristae formation | 1 | 3 | 8.88e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 3 | 65 | 8.88e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 8.88e-01 |
| GO:BP | GO:0032226 | positive regulation of synaptic transmission, dopaminergic | 1 | 3 | 8.88e-01 |
| GO:BP | GO:0061182 | negative regulation of chondrocyte development | 1 | 2 | 8.88e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 6 | 963 | 8.88e-01 |
| GO:BP | GO:0010033 | response to organic substance | 2 | 1957 | 8.88e-01 |
| GO:BP | GO:0032695 | negative regulation of interleukin-12 production | 1 | 6 | 8.88e-01 |
| GO:BP | GO:0018094 | protein polyglycylation | 1 | 1 | 8.89e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 4 | 8 | 8.89e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 2 | 296 | 8.89e-01 |
| GO:BP | GO:0032275 | luteinizing hormone secretion | 3 | 6 | 8.89e-01 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 1 | 11 | 8.89e-01 |
| GO:BP | GO:0006353 | DNA-templated transcription termination | 2 | 22 | 8.89e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 1 | 50 | 8.89e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 1 | 279 | 8.90e-01 |
| GO:BP | GO:0030431 | sleep | 2 | 13 | 8.90e-01 |
| GO:BP | GO:0060484 | lung-associated mesenchyme development | 1 | 5 | 8.90e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 16 | 2102 | 8.90e-01 |
| GO:BP | GO:0044351 | macropinocytosis | 1 | 8 | 8.90e-01 |
| GO:BP | GO:0090032 | negative regulation of steroid hormone biosynthetic process | 1 | 6 | 8.90e-01 |
| GO:BP | GO:0045163 | clustering of voltage-gated potassium channels | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0098664 | G protein-coupled serotonin receptor signaling pathway | 1 | 8 | 8.90e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2 | 4099 | 8.90e-01 |
| GO:BP | GO:1904708 | regulation of granulosa cell apoptotic process | 1 | 2 | 8.90e-01 |
| GO:BP | GO:1904700 | granulosa cell apoptotic process | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 3 | 108 | 8.90e-01 |
| GO:BP | GO:0070141 | response to UV-A | 1 | 11 | 8.90e-01 |
| GO:BP | GO:1901162 | primary amino compound biosynthetic process | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0021626 | central nervous system maturation | 1 | 5 | 8.90e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 4 | 496 | 8.90e-01 |
| GO:BP | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 1 | 14 | 8.90e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 9 | 22 | 8.90e-01 |
| GO:BP | GO:0042908 | xenobiotic transport | 12 | 30 | 8.90e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 1 | 17 | 8.90e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 2 | 84 | 8.90e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 1 | 18 | 8.90e-01 |
| GO:BP | GO:0035669 | TRAM-dependent toll-like receptor 4 signaling pathway | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0000957 | mitochondrial RNA catabolic process | 2 | 6 | 8.90e-01 |
| GO:BP | GO:0014826 | vein smooth muscle contraction | 1 | 3 | 8.90e-01 |
| GO:BP | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0060585 | positive regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity | 3 | 12 | 8.91e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 8.91e-01 |
| GO:BP | GO:0019563 | glycerol catabolic process | 1 | 5 | 8.91e-01 |
| GO:BP | GO:0070669 | response to interleukin-2 | 1 | 3 | 8.91e-01 |
| GO:BP | GO:0034776 | response to histamine | 4 | 5 | 8.91e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 4 | 15 | 8.91e-01 |
| GO:BP | GO:1990569 | UDP-N-acetylglucosamine transmembrane transport | 1 | 6 | 8.91e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 3 | 13 | 8.91e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 20 | 2135 | 8.92e-01 |
| GO:BP | GO:0050975 | sensory perception of touch | 1 | 2 | 8.92e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 1 | 93 | 8.92e-01 |
| GO:BP | GO:0046968 | peptide antigen transport | 1 | 1 | 8.92e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 5 | 400 | 8.92e-01 |
| GO:BP | GO:0140300 | serine import into mitochondrion | 1 | 3 | 8.92e-01 |
| GO:BP | GO:0043353 | enucleate erythrocyte differentiation | 3 | 9 | 8.92e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 4 | 148 | 8.92e-01 |
| GO:BP | GO:0044467 | glial cell-derived neurotrophic factor production | 1 | 2 | 8.92e-01 |
| GO:BP | GO:1900168 | positive regulation of glial cell-derived neurotrophic factor production | 1 | 2 | 8.92e-01 |
| GO:BP | GO:0034375 | high-density lipoprotein particle remodeling | 2 | 11 | 8.92e-01 |
| GO:BP | GO:1900166 | regulation of glial cell-derived neurotrophic factor production | 1 | 2 | 8.92e-01 |
| GO:BP | GO:0099612 | protein localization to axon | 3 | 6 | 8.92e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 11 | 150 | 8.92e-01 |
| GO:BP | GO:1901336 | lactone biosynthetic process | 1 | 3 | 8.92e-01 |
| GO:BP | GO:0019853 | L-ascorbic acid biosynthetic process | 1 | 3 | 8.92e-01 |
| GO:BP | GO:2000380 | regulation of mesoderm development | 2 | 3 | 8.92e-01 |
| GO:BP | GO:0006533 | aspartate catabolic process | 1 | 2 | 8.92e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 17 | 78 | 8.92e-01 |
| GO:BP | GO:0099039 | sphingolipid translocation | 1 | 4 | 8.92e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 1 | 35 | 8.93e-01 |
| GO:BP | GO:2000503 | positive regulation of natural killer cell chemotaxis | 1 | 1 | 8.93e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 5 | 24 | 8.93e-01 |
| GO:BP | GO:0021776 | smoothened signaling pathway involved in spinal cord motor neuron cell fate specification | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 2 | 138 | 8.93e-01 |
| GO:BP | GO:0035054 | embryonic heart tube anterior/posterior pattern specification | 1 | 2 | 8.93e-01 |
| GO:BP | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification | 1 | 3 | 8.93e-01 |
| GO:BP | GO:2000864 | regulation of estradiol secretion | 1 | 2 | 8.93e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 1 | 39 | 8.93e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 2 | 29 | 8.93e-01 |
| GO:BP | GO:0035938 | estradiol secretion | 1 | 2 | 8.93e-01 |
| GO:BP | GO:0006570 | tyrosine metabolic process | 1 | 6 | 8.93e-01 |
| GO:BP | GO:0070358 | actin polymerization-dependent cell motility | 1 | 6 | 8.93e-01 |
| GO:BP | GO:0018243 | protein O-linked glycosylation via threonine | 3 | 7 | 8.93e-01 |
| GO:BP | GO:0035352 | NAD transmembrane transport | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 6 | 95 | 8.93e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 1 | 17 | 8.93e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 2 | 5 | 8.93e-01 |
| GO:BP | GO:0043415 | positive regulation of skeletal muscle tissue regeneration | 1 | 4 | 8.93e-01 |
| GO:BP | GO:0045117 | azole transmembrane transport | 4 | 9 | 8.93e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 5 | 19 | 8.93e-01 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 1 | 89 | 8.93e-01 |
| GO:BP | GO:1904694 | negative regulation of vascular associated smooth muscle contraction | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 5 | 9 | 8.93e-01 |
| GO:BP | GO:0021590 | cerebellum maturation | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0021699 | cerebellar cortex maturation | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0034344 | regulation of type III interferon production | 1 | 3 | 8.95e-01 |
| GO:BP | GO:1903406 | regulation of P-type sodium:potassium-exchanging transporter activity | 1 | 2 | 8.95e-01 |
| GO:BP | GO:0034343 | type III interferon production | 1 | 3 | 8.95e-01 |
| GO:BP | GO:1903408 | positive regulation of P-type sodium:potassium-exchanging transporter activity | 1 | 2 | 8.95e-01 |
| GO:BP | GO:1990074 | polyuridylation-dependent mRNA catabolic process | 1 | 3 | 8.95e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 1 | 40 | 8.95e-01 |
| GO:BP | GO:0048548 | regulation of pinocytosis | 4 | 11 | 8.95e-01 |
| GO:BP | GO:0006000 | fructose metabolic process | 1 | 10 | 8.95e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 67 | 561 | 8.95e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 8.95e-01 |
| GO:BP | GO:0016137 | glycoside metabolic process | 7 | 16 | 8.95e-01 |
| GO:BP | GO:0045333 | cellular respiration | 3 | 211 | 8.95e-01 |
| GO:BP | GO:0061889 | negative regulation of astrocyte activation | 1 | 2 | 8.95e-01 |
| GO:BP | GO:0035455 | response to interferon-alpha | 1 | 16 | 8.95e-01 |
| GO:BP | GO:0023061 | signal release | 9 | 321 | 8.96e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0051005 | negative regulation of lipoprotein lipase activity | 1 | 4 | 8.97e-01 |
| GO:BP | GO:2000631 | regulation of pre-miRNA processing | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0048712 | negative regulation of astrocyte differentiation | 2 | 9 | 8.97e-01 |
| GO:BP | GO:1905604 | negative regulation of blood-brain barrier permeability | 1 | 2 | 8.97e-01 |
| GO:BP | GO:1903565 | negative regulation of protein localization to cilium | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0045797 | positive regulation of intestinal cholesterol absorption | 1 | 1 | 8.97e-01 |
| GO:BP | GO:0015874 | norepinephrine transport | 1 | 9 | 8.97e-01 |
| GO:BP | GO:0090659 | walking behavior | 5 | 25 | 8.98e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 8.98e-01 |
| GO:BP | GO:0070100 | negative regulation of chemokine-mediated signaling pathway | 2 | 6 | 8.98e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 2 | 28 | 8.98e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 1 | 55 | 8.98e-01 |
| GO:BP | GO:0072378 | blood coagulation, fibrin clot formation | 1 | 7 | 8.98e-01 |
| GO:BP | GO:1902565 | positive regulation of neutrophil activation | 1 | 1 | 8.98e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 5 | 132 | 8.98e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 8 | 669 | 8.98e-01 |
| GO:BP | GO:2000381 | negative regulation of mesoderm development | 1 | 1 | 8.98e-01 |
| GO:BP | GO:0090160 | Golgi to lysosome transport | 4 | 10 | 8.98e-01 |
| GO:BP | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | 2 | 3 | 8.99e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 6 | 29 | 8.99e-01 |
| GO:BP | GO:1905383 | protein localization to presynapse | 4 | 11 | 8.99e-01 |
| GO:BP | GO:2000391 | positive regulation of neutrophil extravasation | 2 | 6 | 9.00e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 16 | 39 | 9.00e-01 |
| GO:BP | GO:1905806 | regulation of synapse pruning | 2 | 5 | 9.00e-01 |
| GO:BP | GO:0060304 | regulation of phosphatidylinositol dephosphorylation | 2 | 5 | 9.00e-01 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 1 | 13 | 9.00e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 10 | 20 | 9.00e-01 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 1 | 7 | 9.00e-01 |
| GO:BP | GO:0030901 | midbrain development | 2 | 69 | 9.00e-01 |
| GO:BP | GO:0022027 | interkinetic nuclear migration | 1 | 4 | 9.00e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 318 | 9.00e-01 |
| GO:BP | GO:0061209 | cell proliferation involved in mesonephros development | 1 | 3 | 9.01e-01 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 3 | 12 | 9.01e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 2 | 23 | 9.01e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 2 | 13 | 9.01e-01 |
| GO:BP | GO:0016116 | carotenoid metabolic process | 1 | 1 | 9.01e-01 |
| GO:BP | GO:0120253 | hydrocarbon catabolic process | 1 | 1 | 9.01e-01 |
| GO:BP | GO:0016108 | tetraterpenoid metabolic process | 1 | 1 | 9.01e-01 |
| GO:BP | GO:0042214 | terpene metabolic process | 1 | 1 | 9.01e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 9.01e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 6 | 43 | 9.01e-01 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 1 | 8 | 9.01e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 9.01e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 4 | 187 | 9.01e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 5 | 11 | 9.01e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 1 | 59 | 9.01e-01 |
| GO:BP | GO:0042867 | pyruvate catabolic process | 1 | 2 | 9.01e-01 |
| GO:BP | GO:0035879 | plasma membrane lactate transport | 1 | 3 | 9.01e-01 |
| GO:BP | GO:0050434 | positive regulation of viral transcription | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0015888 | thiamine transport | 2 | 3 | 9.02e-01 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 2 | 7 | 9.02e-01 |
| GO:BP | GO:0055005 | ventricular cardiac myofibril assembly | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0035701 | hematopoietic stem cell migration | 1 | 6 | 9.02e-01 |
| GO:BP | GO:1900625 | positive regulation of monocyte aggregation | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 5 | 55 | 9.02e-01 |
| GO:BP | GO:1900623 | regulation of monocyte aggregation | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0003165 | Purkinje myocyte development | 1 | 3 | 9.02e-01 |
| GO:BP | GO:0061439 | kidney vasculature morphogenesis | 2 | 7 | 9.02e-01 |
| GO:BP | GO:0061438 | renal system vasculature morphogenesis | 2 | 7 | 9.02e-01 |
| GO:BP | GO:2001293 | malonyl-CoA metabolic process | 1 | 6 | 9.03e-01 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 1 | 5 | 9.03e-01 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 1 | 11 | 9.03e-01 |
| GO:BP | GO:1901256 | regulation of macrophage colony-stimulating factor production | 1 | 2 | 9.03e-01 |
| GO:BP | GO:0036301 | macrophage colony-stimulating factor production | 1 | 2 | 9.03e-01 |
| GO:BP | GO:0008594 | photoreceptor cell morphogenesis | 1 | 4 | 9.03e-01 |
| GO:BP | GO:1901078 | negative regulation of relaxation of muscle | 1 | 4 | 9.03e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 5 | 159 | 9.03e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 1 | 36 | 9.03e-01 |
| GO:BP | GO:0071803 | positive regulation of podosome assembly | 1 | 6 | 9.03e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 9.04e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 9.04e-01 |
| GO:BP | GO:0043171 | peptide catabolic process | 1 | 16 | 9.04e-01 |
| GO:BP | GO:0060128 | corticotropin hormone secreting cell differentiation | 1 | 2 | 9.04e-01 |
| GO:BP | GO:1904749 | regulation of protein localization to nucleolus | 2 | 7 | 9.04e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 4 | 11 | 9.04e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 5 | 134 | 9.04e-01 |
| GO:BP | GO:0097676 | histone H3-K36 dimethylation | 1 | 4 | 9.04e-01 |
| GO:BP | GO:0022900 | electron transport chain | 2 | 142 | 9.04e-01 |
| GO:BP | GO:1902931 | negative regulation of alcohol biosynthetic process | 4 | 10 | 9.05e-01 |
| GO:BP | GO:0046903 | secretion | 3 | 645 | 9.05e-01 |
| GO:BP | GO:0060375 | regulation of mast cell differentiation | 1 | 2 | 9.05e-01 |
| GO:BP | GO:0015711 | organic anion transport | 2 | 264 | 9.05e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 1 | 19 | 9.05e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 2 | 22 | 9.05e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 9 | 63 | 9.05e-01 |
| GO:BP | GO:0007567 | parturition | 1 | 8 | 9.05e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 8 | 20 | 9.06e-01 |
| GO:BP | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | 7 | 14 | 9.06e-01 |
| GO:BP | GO:0045345 | positive regulation of MHC class I biosynthetic process | 1 | 2 | 9.07e-01 |
| GO:BP | GO:0015938 | coenzyme A catabolic process | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0044580 | butyryl-CoA catabolic process | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0010842 | retina layer formation | 6 | 16 | 9.07e-01 |
| GO:BP | GO:0006935 | chemotaxis | 12 | 389 | 9.07e-01 |
| GO:BP | GO:1905453 | regulation of myeloid progenitor cell differentiation | 1 | 1 | 9.07e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 2 | 43 | 9.07e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 2 | 43 | 9.07e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 9.07e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 2 | 4128 | 9.07e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 9.08e-01 |
| GO:BP | GO:0061753 | substrate localization to autophagosome | 1 | 3 | 9.08e-01 |
| GO:BP | GO:0030574 | collagen catabolic process | 6 | 26 | 9.08e-01 |
| GO:BP | GO:0002019 | regulation of renal output by angiotensin | 1 | 3 | 9.08e-01 |
| GO:BP | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation | 1 | 3 | 9.08e-01 |
| GO:BP | GO:0010814 | substance P catabolic process | 1 | 3 | 9.08e-01 |
| GO:BP | GO:0002092 | positive regulation of receptor internalization | 1 | 21 | 9.08e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 1 | 15 | 9.08e-01 |
| GO:BP | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine | 3 | 7 | 9.09e-01 |
| GO:BP | GO:0060161 | positive regulation of dopamine receptor signaling pathway | 1 | 3 | 9.09e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 5 | 136 | 9.09e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 9 | 27 | 9.09e-01 |
| GO:BP | GO:0042330 | taxis | 12 | 389 | 9.09e-01 |
| GO:BP | GO:0046958 | nonassociative learning | 1 | 6 | 9.09e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 3 | 9 | 9.09e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 21 | 2293 | 9.09e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 1 | 30 | 9.09e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 9.09e-01 |
| GO:BP | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 3 | 9.09e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 2 | 41 | 9.10e-01 |
| GO:BP | GO:0035929 | steroid hormone secretion | 1 | 18 | 9.10e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 44 | 192 | 9.10e-01 |
| GO:BP | GO:0035461 | vitamin transmembrane transport | 1 | 14 | 9.10e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 26 | 667 | 9.10e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 1 | 18 | 9.10e-01 |
| GO:BP | GO:0016043 | cellular component organization | 3 | 5076 | 9.10e-01 |
| GO:BP | GO:0009068 | aspartate family amino acid catabolic process | 1 | 12 | 9.10e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 2 | 13 | 9.10e-01 |
| GO:BP | GO:0046055 | dGMP catabolic process | 1 | 1 | 9.10e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 12 | 36 | 9.10e-01 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 1 | 5 | 9.10e-01 |
| GO:BP | GO:0043317 | regulation of cytotoxic T cell degranulation | 1 | 1 | 9.10e-01 |
| GO:BP | GO:0043318 | negative regulation of cytotoxic T cell degranulation | 1 | 1 | 9.10e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 4 | 373 | 9.10e-01 |
| GO:BP | GO:0008295 | spermidine biosynthetic process | 1 | 3 | 9.11e-01 |
| GO:BP | GO:0046294 | formaldehyde catabolic process | 1 | 2 | 9.11e-01 |
| GO:BP | GO:0015871 | choline transport | 2 | 6 | 9.11e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 1 | 49 | 9.11e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 2 | 7 | 9.11e-01 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 1 | 11 | 9.11e-01 |
| GO:BP | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport | 2 | 5 | 9.11e-01 |
| GO:BP | GO:0106071 | positive regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 3 | 9.11e-01 |
| GO:BP | GO:1903003 | positive regulation of protein deubiquitination | 1 | 6 | 9.12e-01 |
| GO:BP | GO:0006681 | galactosylceramide metabolic process | 2 | 5 | 9.12e-01 |
| GO:BP | GO:0019374 | galactolipid metabolic process | 2 | 5 | 9.12e-01 |
| GO:BP | GO:0002572 | pro-T cell differentiation | 1 | 1 | 9.12e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 1 | 2565 | 9.12e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 2 | 99 | 9.12e-01 |
| GO:BP | GO:0045990 | carbon catabolite regulation of transcription | 1 | 4 | 9.13e-01 |
| GO:BP | GO:0006452 | translational frameshifting | 1 | 3 | 9.13e-01 |
| GO:BP | GO:0031424 | keratinization | 1 | 13 | 9.13e-01 |
| GO:BP | GO:0045991 | carbon catabolite activation of transcription | 1 | 4 | 9.13e-01 |
| GO:BP | GO:0032790 | ribosome disassembly | 1 | 11 | 9.13e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 1 | 2573 | 9.13e-01 |
| GO:BP | GO:0044241 | lipid digestion | 1 | 10 | 9.13e-01 |
| GO:BP | GO:0001839 | neural plate morphogenesis | 1 | 4 | 9.13e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 2 | 30 | 9.14e-01 |
| GO:BP | GO:0032385 | positive regulation of intracellular cholesterol transport | 1 | 3 | 9.14e-01 |
| GO:BP | GO:0032382 | positive regulation of intracellular sterol transport | 1 | 3 | 9.14e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 9.14e-01 |
| GO:BP | GO:1905600 | regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 3 | 9.14e-01 |
| GO:BP | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 3 | 9.14e-01 |
| GO:BP | GO:0032497 | detection of lipopolysaccharide | 1 | 2 | 9.14e-01 |
| GO:BP | GO:0042420 | dopamine catabolic process | 1 | 5 | 9.14e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 1 | 165 | 9.14e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 9.14e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 9.14e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 10 | 19 | 9.14e-01 |
| GO:BP | GO:0007197 | adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway | 1 | 3 | 9.14e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 8 | 112 | 9.14e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 1 | 6 | 9.14e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 9.14e-01 |
| GO:BP | GO:0120188 | regulation of bile acid secretion | 1 | 2 | 9.14e-01 |
| GO:BP | GO:0099640 | axo-dendritic protein transport | 3 | 10 | 9.14e-01 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 1 | 4 | 9.14e-01 |
| GO:BP | GO:1904028 | positive regulation of collagen fibril organization | 1 | 4 | 9.14e-01 |
| GO:BP | GO:2000097 | regulation of smooth muscle cell-matrix adhesion | 1 | 3 | 9.14e-01 |
| GO:BP | GO:0032817 | regulation of natural killer cell proliferation | 1 | 4 | 9.14e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 7 | 218 | 9.15e-01 |
| GO:BP | GO:0006578 | amino-acid betaine biosynthetic process | 1 | 5 | 9.15e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 1 | 68 | 9.15e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 1 | 9 | 9.15e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 2 | 55 | 9.16e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 4 | 271 | 9.16e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 9.16e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 9 | 41 | 9.16e-01 |
| GO:BP | GO:0070165 | positive regulation of adiponectin secretion | 1 | 1 | 9.16e-01 |
| GO:BP | GO:0090292 | nuclear matrix anchoring at nuclear membrane | 1 | 3 | 9.16e-01 |
| GO:BP | GO:0038098 | sequestering of BMP from receptor via BMP binding | 1 | 2 | 9.16e-01 |
| GO:BP | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration | 1 | 3 | 9.16e-01 |
| GO:BP | GO:0043578 | nuclear matrix organization | 1 | 3 | 9.16e-01 |
| GO:BP | GO:0030534 | adult behavior | 3 | 92 | 9.17e-01 |
| GO:BP | GO:0019405 | alditol catabolic process | 1 | 6 | 9.17e-01 |
| GO:BP | GO:0006114 | glycerol biosynthetic process | 1 | 3 | 9.17e-01 |
| GO:BP | GO:0034695 | response to prostaglandin E | 1 | 17 | 9.17e-01 |
| GO:BP | GO:0019401 | alditol biosynthetic process | 1 | 3 | 9.17e-01 |
| GO:BP | GO:0014012 | peripheral nervous system axon regeneration | 1 | 6 | 9.17e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 1 | 45 | 9.17e-01 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 2 | 17 | 9.17e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 9.17e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 3 | 10 | 9.17e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 3 | 10 | 9.17e-01 |
| GO:BP | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity | 1 | 4 | 9.17e-01 |
| GO:BP | GO:0002638 | negative regulation of immunoglobulin production | 2 | 5 | 9.17e-01 |
| GO:BP | GO:0036065 | fucosylation | 1 | 10 | 9.17e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 9.17e-01 |
| GO:BP | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis | 1 | 4 | 9.17e-01 |
| GO:BP | GO:0071605 | monocyte chemotactic protein-1 production | 1 | 7 | 9.17e-01 |
| GO:BP | GO:0071637 | regulation of monocyte chemotactic protein-1 production | 1 | 7 | 9.17e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 20 | 42 | 9.18e-01 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 2 | 15 | 9.18e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 2 | 21 | 9.18e-01 |
| GO:BP | GO:2000855 | regulation of mineralocorticoid secretion | 2 | 4 | 9.18e-01 |
| GO:BP | GO:2000858 | regulation of aldosterone secretion | 2 | 4 | 9.18e-01 |
| GO:BP | GO:0006642 | triglyceride mobilization | 1 | 3 | 9.18e-01 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 2 | 15 | 9.18e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 9.18e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 5 | 78 | 9.18e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 9.18e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 3 | 33 | 9.18e-01 |
| GO:BP | GO:0002252 | immune effector process | 7 | 318 | 9.18e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 15 | 42 | 9.18e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 4 | 174 | 9.18e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 1 | 26 | 9.19e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 9.19e-01 |
| GO:BP | GO:0007210 | serotonin receptor signaling pathway | 1 | 8 | 9.19e-01 |
| GO:BP | GO:1901387 | positive regulation of voltage-gated calcium channel activity | 4 | 13 | 9.19e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 1 | 105 | 9.19e-01 |
| GO:BP | GO:1904352 | positive regulation of protein catabolic process in the vacuole | 1 | 6 | 9.19e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 1 | 5 | 9.19e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 3 | 447 | 9.20e-01 |
| GO:BP | GO:0090026 | positive regulation of monocyte chemotaxis | 1 | 7 | 9.20e-01 |
| GO:BP | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity | 1 | 3 | 9.20e-01 |
| GO:BP | GO:0002399 | MHC class II protein complex assembly | 2 | 4 | 9.20e-01 |
| GO:BP | GO:0002503 | peptide antigen assembly with MHC class II protein complex | 2 | 4 | 9.20e-01 |
| GO:BP | GO:0099515 | actin filament-based transport | 7 | 15 | 9.20e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 2 | 3730 | 9.21e-01 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 4 | 29 | 9.21e-01 |
| GO:BP | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain | 1 | 1 | 9.21e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 1 | 71 | 9.21e-01 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 1 | 5 | 9.21e-01 |
| GO:BP | GO:1903659 | regulation of complement-dependent cytotoxicity | 1 | 2 | 9.21e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 1 | 109 | 9.22e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 2 | 8 | 9.22e-01 |
| GO:BP | GO:0090261 | positive regulation of inclusion body assembly | 1 | 5 | 9.22e-01 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 10 | 20 | 9.22e-01 |
| GO:BP | GO:0002434 | immune complex clearance | 1 | 1 | 9.22e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 1 | 43 | 9.22e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3 | 5274 | 9.22e-01 |
| GO:BP | GO:0075525 | viral translational termination-reinitiation | 1 | 5 | 9.22e-01 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 1 | 9 | 9.22e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 3 | 56 | 9.22e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 11 | 166 | 9.22e-01 |
| GO:BP | GO:0019062 | virion attachment to host cell | 1 | 9 | 9.22e-01 |
| GO:BP | GO:0001816 | cytokine production | 3 | 451 | 9.22e-01 |
| GO:BP | GO:0010706 | ganglioside biosynthetic process via lactosylceramide | 1 | 5 | 9.22e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 7 | 45 | 9.22e-01 |
| GO:BP | GO:0034134 | toll-like receptor 2 signaling pathway | 2 | 11 | 9.23e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 21 | 639 | 9.23e-01 |
| GO:BP | GO:0060872 | semicircular canal development | 2 | 7 | 9.23e-01 |
| GO:BP | GO:0048808 | male genitalia morphogenesis | 1 | 2 | 9.24e-01 |
| GO:BP | GO:0090598 | male anatomical structure morphogenesis | 1 | 2 | 9.24e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 3 | 28 | 9.24e-01 |
| GO:BP | GO:1903577 | cellular response to L-arginine | 1 | 3 | 9.24e-01 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 1 | 5 | 9.24e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 3 | 7 | 9.24e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 3 | 11 | 9.24e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 3 | 11 | 9.24e-01 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 2 | 11 | 9.24e-01 |
| GO:BP | GO:0046440 | L-lysine metabolic process | 1 | 4 | 9.24e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 1 | 66 | 9.24e-01 |
| GO:BP | GO:0019477 | L-lysine catabolic process | 1 | 4 | 9.24e-01 |
| GO:BP | GO:0019474 | L-lysine catabolic process to acetyl-CoA | 1 | 4 | 9.24e-01 |
| GO:BP | GO:1900118 | negative regulation of execution phase of apoptosis | 3 | 6 | 9.24e-01 |
| GO:BP | GO:0044417 | translocation of molecules into host | 2 | 7 | 9.24e-01 |
| GO:BP | GO:1904177 | regulation of adipose tissue development | 1 | 8 | 9.24e-01 |
| GO:BP | GO:0006107 | oxaloacetate metabolic process | 2 | 7 | 9.24e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 2 | 6 | 9.25e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 1 | 6996 | 9.25e-01 |
| GO:BP | GO:0072131 | kidney mesenchyme morphogenesis | 1 | 4 | 9.25e-01 |
| GO:BP | GO:0072133 | metanephric mesenchyme morphogenesis | 1 | 4 | 9.25e-01 |
| GO:BP | GO:0033686 | positive regulation of luteinizing hormone secretion | 1 | 2 | 9.25e-01 |
| GO:BP | GO:0099004 | calmodulin dependent kinase signaling pathway | 3 | 6 | 9.25e-01 |
| GO:BP | GO:0045341 | MHC class I biosynthetic process | 2 | 3 | 9.25e-01 |
| GO:BP | GO:0045343 | regulation of MHC class I biosynthetic process | 2 | 3 | 9.25e-01 |
| GO:BP | GO:0071673 | positive regulation of smooth muscle cell chemotaxis | 1 | 2 | 9.25e-01 |
| GO:BP | GO:0061314 | Notch signaling involved in heart development | 3 | 9 | 9.25e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 2 | 13 | 9.25e-01 |
| GO:BP | GO:0001775 | cell activation | 5 | 655 | 9.25e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 5 | 92 | 9.25e-01 |
| GO:BP | GO:0001886 | endothelial cell morphogenesis | 1 | 10 | 9.26e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 9 | 44 | 9.27e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 9 | 28 | 9.27e-01 |
| GO:BP | GO:0090158 | endoplasmic reticulum membrane organization | 2 | 14 | 9.27e-01 |
| GO:BP | GO:1903979 | negative regulation of microglial cell activation | 1 | 5 | 9.28e-01 |
| GO:BP | GO:0006952 | defense response | 16 | 944 | 9.28e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 1 | 10 | 9.28e-01 |
| GO:BP | GO:0070672 | response to interleukin-15 | 1 | 3 | 9.28e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 1 | 7 | 9.28e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 5 | 13 | 9.28e-01 |
| GO:BP | GO:0051953 | negative regulation of amine transport | 1 | 15 | 9.28e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 12 | 28 | 9.28e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 1 | 49 | 9.28e-01 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 1 | 8 | 9.28e-01 |
| GO:BP | GO:0007217 | tachykinin receptor signaling pathway | 1 | 3 | 9.28e-01 |
| GO:BP | GO:1990384 | hyaloid vascular plexus regression | 2 | 3 | 9.29e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.29e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 9 | 52 | 9.29e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 9.29e-01 |
| GO:BP | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 3 | 10 | 9.29e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 80 | 726 | 9.29e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 21 | 62 | 9.29e-01 |
| GO:BP | GO:0002826 | negative regulation of T-helper 1 type immune response | 1 | 3 | 9.29e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 1 | 129 | 9.29e-01 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 1 | 5 | 9.30e-01 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 1 | 5 | 9.30e-01 |
| GO:BP | GO:2001245 | regulation of phosphatidylcholine biosynthetic process | 1 | 5 | 9.30e-01 |
| GO:BP | GO:1904054 | regulation of cholangiocyte proliferation | 1 | 2 | 9.30e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 3 | 53 | 9.30e-01 |
| GO:BP | GO:1903891 | regulation of ATF6-mediated unfolded protein response | 1 | 4 | 9.31e-01 |
| GO:BP | GO:0042756 | drinking behavior | 1 | 6 | 9.31e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 6 | 32 | 9.31e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 9.31e-01 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 4 | 14 | 9.31e-01 |
| GO:BP | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition | 1 | 3 | 9.32e-01 |
| GO:BP | GO:0071670 | smooth muscle cell chemotaxis | 1 | 6 | 9.32e-01 |
| GO:BP | GO:0010960 | magnesium ion homeostasis | 4 | 9 | 9.32e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 9.32e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 4 | 13 | 9.32e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 30 | 9.32e-01 |
| GO:BP | GO:1904340 | positive regulation of dopaminergic neuron differentiation | 1 | 3 | 9.33e-01 |
| GO:BP | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 3 | 9.33e-01 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 5 | 11 | 9.33e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 2 | 10 | 9.33e-01 |
| GO:BP | GO:1903288 | positive regulation of potassium ion import across plasma membrane | 2 | 7 | 9.33e-01 |
| GO:BP | GO:1905289 | regulation of CAMKK-AMPK signaling cascade | 1 | 2 | 9.33e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 12 | 118 | 9.33e-01 |
| GO:BP | GO:0071394 | cellular response to testosterone stimulus | 4 | 11 | 9.33e-01 |
| GO:BP | GO:0060406 | positive regulation of penile erection | 1 | 3 | 9.33e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 4 | 10 | 9.33e-01 |
| GO:BP | GO:0048254 | snoRNA localization | 1 | 5 | 9.34e-01 |
| GO:BP | GO:0019530 | taurine metabolic process | 2 | 6 | 9.34e-01 |
| GO:BP | GO:0090084 | negative regulation of inclusion body assembly | 4 | 10 | 9.34e-01 |
| GO:BP | GO:0019694 | alkanesulfonate metabolic process | 2 | 6 | 9.34e-01 |
| GO:BP | GO:0000964 | mitochondrial RNA 5’-end processing | 1 | 4 | 9.35e-01 |
| GO:BP | GO:0060347 | heart trabecula formation | 5 | 13 | 9.35e-01 |
| GO:BP | GO:1905155 | positive regulation of membrane invagination | 1 | 11 | 9.35e-01 |
| GO:BP | GO:0060100 | positive regulation of phagocytosis, engulfment | 1 | 11 | 9.35e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 4 | 8 | 9.35e-01 |
| GO:BP | GO:1903980 | positive regulation of microglial cell activation | 2 | 2 | 9.35e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 9.35e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 3 | 10 | 9.35e-01 |
| GO:BP | GO:0002468 | dendritic cell antigen processing and presentation | 2 | 6 | 9.35e-01 |
| GO:BP | GO:0046864 | isoprenoid transport | 1 | 3 | 9.35e-01 |
| GO:BP | GO:0046865 | terpenoid transport | 1 | 3 | 9.35e-01 |
| GO:BP | GO:0019614 | catechol-containing compound catabolic process | 1 | 5 | 9.35e-01 |
| GO:BP | GO:0009249 | protein lipoylation | 1 | 6 | 9.35e-01 |
| GO:BP | GO:0042424 | catecholamine catabolic process | 1 | 5 | 9.35e-01 |
| GO:BP | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 5 | 9.36e-01 |
| GO:BP | GO:0043132 | NAD transport | 1 | 3 | 9.36e-01 |
| GO:BP | GO:0034465 | response to carbon monoxide | 1 | 3 | 9.36e-01 |
| GO:BP | GO:0051956 | negative regulation of amino acid transport | 1 | 10 | 9.36e-01 |
| GO:BP | GO:0090230 | regulation of centromere complex assembly | 1 | 4 | 9.36e-01 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 1 | 30 | 9.36e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 12 | 119 | 9.36e-01 |
| GO:BP | GO:1990911 | response to psychosocial stress | 1 | 2 | 9.36e-01 |
| GO:BP | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway | 1 | 4 | 9.36e-01 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 1 | 11 | 9.36e-01 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 1 | 11 | 9.36e-01 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 2 | 10 | 9.36e-01 |
| GO:BP | GO:0002025 | norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure | 1 | 3 | 9.36e-01 |
| GO:BP | GO:0046884 | follicle-stimulating hormone secretion | 2 | 4 | 9.36e-01 |
| GO:BP | GO:2000389 | regulation of neutrophil extravasation | 2 | 6 | 9.36e-01 |
| GO:BP | GO:0023058 | adaptation of signaling pathway | 8 | 16 | 9.36e-01 |
| GO:BP | GO:0046879 | hormone secretion | 5 | 210 | 9.36e-01 |
| GO:BP | GO:0071603 | endothelial cell-cell adhesion | 1 | 4 | 9.37e-01 |
| GO:BP | GO:0010593 | negative regulation of lamellipodium assembly | 1 | 2 | 9.37e-01 |
| GO:BP | GO:0034372 | very-low-density lipoprotein particle remodeling | 2 | 4 | 9.37e-01 |
| GO:BP | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway | 4 | 9 | 9.37e-01 |
| GO:BP | GO:0048840 | otolith development | 1 | 6 | 9.37e-01 |
| GO:BP | GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 2 | 4 | 9.37e-01 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 1 | 23 | 9.37e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 2 | 44 | 9.38e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 9.38e-01 |
| GO:BP | GO:0032429 | regulation of phospholipase A2 activity | 1 | 8 | 9.38e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 11 | 27 | 9.38e-01 |
| GO:BP | GO:0097368 | establishment of Sertoli cell barrier | 1 | 3 | 9.38e-01 |
| GO:BP | GO:0038116 | chemokine (C-C motif) ligand 21 signaling pathway | 1 | 2 | 9.39e-01 |
| GO:BP | GO:2000439 | positive regulation of monocyte extravasation | 1 | 3 | 9.39e-01 |
| GO:BP | GO:0007196 | adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway | 1 | 1 | 9.39e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 2 | 4 | 9.39e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 9.40e-01 |
| GO:BP | GO:0060923 | cardiac muscle cell fate commitment | 3 | 7 | 9.40e-01 |
| GO:BP | GO:0042832 | defense response to protozoan | 1 | 10 | 9.40e-01 |
| GO:BP | GO:0019511 | peptidyl-proline hydroxylation | 1 | 14 | 9.40e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 3 | 10 | 9.41e-01 |
| GO:BP | GO:0019323 | pentose catabolic process | 1 | 2 | 9.41e-01 |
| GO:BP | GO:0021631 | optic nerve morphogenesis | 1 | 2 | 9.41e-01 |
| GO:BP | GO:1904273 | L-alanine import across plasma membrane | 1 | 4 | 9.41e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 2 | 13 | 9.41e-01 |
| GO:BP | GO:0009725 | response to hormone | 42 | 642 | 9.41e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 9 | 23 | 9.41e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 6 | 15 | 9.41e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 3 | 4794 | 9.41e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 6 | 21 | 9.42e-01 |
| GO:BP | GO:1900158 | negative regulation of bone mineralization involved in bone maturation | 1 | 2 | 9.42e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 2 | 17 | 9.42e-01 |
| GO:BP | GO:0007165 | signal transduction | 5 | 3818 | 9.42e-01 |
| GO:BP | GO:1905048 | regulation of metallopeptidase activity | 5 | 14 | 9.42e-01 |
| GO:BP | GO:0072179 | nephric duct formation | 1 | 1 | 9.42e-01 |
| GO:BP | GO:2000109 | regulation of macrophage apoptotic process | 2 | 6 | 9.42e-01 |
| GO:BP | GO:0060159 | regulation of dopamine receptor signaling pathway | 2 | 7 | 9.43e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 9.43e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 2 | 28 | 9.43e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 9.43e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 9.43e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 9.43e-01 |
| GO:BP | GO:1905906 | regulation of amyloid fibril formation | 4 | 14 | 9.43e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 5 | 10 | 9.44e-01 |
| GO:BP | GO:0006897 | endocytosis | 3 | 527 | 9.44e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 1 | 37 | 9.44e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 3 | 9 | 9.44e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 4 | 84 | 9.44e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 3 | 3600 | 9.44e-01 |
| GO:BP | GO:0061591 | calcium activated galactosylceramide scrambling | 1 | 2 | 9.44e-01 |
| GO:BP | GO:0061589 | calcium activated phosphatidylserine scrambling | 1 | 3 | 9.44e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 9.44e-01 |
| GO:BP | GO:0032693 | negative regulation of interleukin-10 production | 1 | 7 | 9.44e-01 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 5 | 38 | 9.44e-01 |
| GO:BP | GO:0042448 | progesterone metabolic process | 1 | 13 | 9.45e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 1 | 7 | 9.45e-01 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 1 | 9 | 9.45e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 9.45e-01 |
| GO:BP | GO:1900245 | positive regulation of MDA-5 signaling pathway | 1 | 2 | 9.45e-01 |
| GO:BP | GO:1903224 | regulation of endodermal cell differentiation | 1 | 6 | 9.45e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 28 | 457 | 9.45e-01 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 1 | 4 | 9.45e-01 |
| GO:BP | GO:0030252 | growth hormone secretion | 1 | 12 | 9.46e-01 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 1 | 5 | 9.46e-01 |
| GO:BP | GO:0042321 | negative regulation of circadian sleep/wake cycle, sleep | 1 | 2 | 9.46e-01 |
| GO:BP | GO:0006583 | melanin biosynthetic process from tyrosine | 1 | 1 | 9.46e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 3 | 8 | 9.46e-01 |
| GO:BP | GO:1902563 | regulation of neutrophil activation | 2 | 3 | 9.47e-01 |
| GO:BP | GO:2000501 | regulation of natural killer cell chemotaxis | 1 | 2 | 9.47e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 10 | 28 | 9.47e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 10 | 28 | 9.47e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1 | 2890 | 9.47e-01 |
| GO:BP | GO:0032494 | response to peptidoglycan | 2 | 5 | 9.47e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 2 | 7 | 9.47e-01 |
| GO:BP | GO:0015732 | prostaglandin transport | 1 | 10 | 9.47e-01 |
| GO:BP | GO:0006363 | termination of RNA polymerase I transcription | 1 | 4 | 9.47e-01 |
| GO:BP | GO:0140352 | export from cell | 24 | 611 | 9.47e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 7 | 31 | 9.47e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 12 | 61 | 9.48e-01 |
| GO:BP | GO:0009312 | oligosaccharide biosynthetic process | 2 | 15 | 9.48e-01 |
| GO:BP | GO:1905451 | positive regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 3 | 9.48e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 1 | 20 | 9.48e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 2 | 119 | 9.48e-01 |
| GO:BP | GO:1903278 | positive regulation of sodium ion export across plasma membrane | 1 | 4 | 9.48e-01 |
| GO:BP | GO:1903276 | regulation of sodium ion export across plasma membrane | 1 | 4 | 9.48e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 1 | 11 | 9.48e-01 |
| GO:BP | GO:0009583 | detection of light stimulus | 2 | 27 | 9.48e-01 |
| GO:BP | GO:0034162 | toll-like receptor 9 signaling pathway | 3 | 12 | 9.48e-01 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 3 | 45 | 9.49e-01 |
| GO:BP | GO:0070268 | cornification | 1 | 2 | 9.49e-01 |
| GO:BP | GO:0045650 | negative regulation of macrophage differentiation | 1 | 5 | 9.49e-01 |
| GO:BP | GO:0021877 | forebrain neuron fate commitment | 1 | 1 | 9.49e-01 |
| GO:BP | GO:0002676 | regulation of chronic inflammatory response | 1 | 2 | 9.49e-01 |
| GO:BP | GO:0005985 | sucrose metabolic process | 1 | 1 | 9.49e-01 |
| GO:BP | GO:0050917 | sensory perception of umami taste | 1 | 3 | 9.49e-01 |
| GO:BP | GO:0009914 | hormone transport | 5 | 215 | 9.49e-01 |
| GO:BP | GO:0019695 | choline metabolic process | 1 | 4 | 9.49e-01 |
| GO:BP | GO:0034694 | response to prostaglandin | 3 | 21 | 9.49e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 9.49e-01 |
| GO:BP | GO:0002692 | negative regulation of cellular extravasation | 1 | 5 | 9.49e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 2 | 317 | 9.49e-01 |
| GO:BP | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process | 1 | 4 | 9.50e-01 |
| GO:BP | GO:1901535 | regulation of DNA demethylation | 3 | 8 | 9.50e-01 |
| GO:BP | GO:1904480 | positive regulation of intestinal absorption | 1 | 1 | 9.50e-01 |
| GO:BP | GO:1904731 | positive regulation of intestinal lipid absorption | 1 | 1 | 9.50e-01 |
| GO:BP | GO:0034378 | chylomicron assembly | 1 | 3 | 9.50e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 5 | 33 | 9.50e-01 |
| GO:BP | GO:0032940 | secretion by cell | 22 | 569 | 9.50e-01 |
| GO:BP | GO:0060332 | positive regulation of response to type II interferon | 1 | 4 | 9.50e-01 |
| GO:BP | GO:0060335 | positive regulation of type II interferon-mediated signaling pathway | 1 | 4 | 9.50e-01 |
| GO:BP | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production | 2 | 5 | 9.50e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 13 | 29 | 9.50e-01 |
| GO:BP | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing | 1 | 5 | 9.50e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 2 | 38 | 9.50e-01 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 5 | 14 | 9.50e-01 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 1 | 14 | 9.50e-01 |
| GO:BP | GO:0048172 | regulation of short-term neuronal synaptic plasticity | 1 | 9 | 9.50e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 1 | 15 | 9.50e-01 |
| GO:BP | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | 5 | 13 | 9.51e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 3 | 9 | 9.51e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 5 | 389 | 9.51e-01 |
| GO:BP | GO:0014051 | gamma-aminobutyric acid secretion | 2 | 8 | 9.51e-01 |
| GO:BP | GO:0031650 | regulation of heat generation | 2 | 7 | 9.51e-01 |
| GO:BP | GO:0006556 | S-adenosylmethionine biosynthetic process | 1 | 3 | 9.51e-01 |
| GO:BP | GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1 | 10 | 9.51e-01 |
| GO:BP | GO:0050427 | 3’-phosphoadenosine 5’-phosphosulfate metabolic process | 1 | 10 | 9.51e-01 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 1 | 131 | 9.51e-01 |
| GO:BP | GO:0044650 | adhesion of symbiont to host cell | 1 | 9 | 9.51e-01 |
| GO:BP | GO:0016480 | negative regulation of transcription by RNA polymerase III | 1 | 4 | 9.51e-01 |
| GO:BP | GO:0050765 | negative regulation of phagocytosis | 2 | 15 | 9.51e-01 |
| GO:BP | GO:0090027 | negative regulation of monocyte chemotaxis | 1 | 4 | 9.51e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 2 | 369 | 9.51e-01 |
| GO:BP | GO:0001562 | response to protozoan | 1 | 11 | 9.51e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 8 | 18 | 9.51e-01 |
| GO:BP | GO:0044872 | lipoprotein localization | 1 | 15 | 9.51e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 11 | 998 | 9.51e-01 |
| GO:BP | GO:1905665 | positive regulation of calcium ion import across plasma membrane | 2 | 6 | 9.51e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 9 | 100 | 9.51e-01 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 11 | 38 | 9.51e-01 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 11 | 38 | 9.51e-01 |
| GO:BP | GO:0150147 | cell-cell junction disassembly | 1 | 4 | 9.52e-01 |
| GO:BP | GO:0044546 | NLRP3 inflammasome complex assembly | 1 | 23 | 9.52e-01 |
| GO:BP | GO:1990079 | cartilage homeostasis | 1 | 2 | 9.52e-01 |
| GO:BP | GO:0048563 | post-embryonic animal organ morphogenesis | 1 | 8 | 9.52e-01 |
| GO:BP | GO:0038171 | cannabinoid signaling pathway | 3 | 5 | 9.52e-01 |
| GO:BP | GO:0022401 | negative adaptation of signaling pathway | 7 | 15 | 9.53e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 9.53e-01 |
| GO:BP | GO:0046325 | negative regulation of glucose import | 4 | 10 | 9.53e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 1 | 42 | 9.53e-01 |
| GO:BP | GO:0042693 | muscle cell fate commitment | 2 | 11 | 9.53e-01 |
| GO:BP | GO:0061523 | cilium disassembly | 1 | 4 | 9.54e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 1 | 55 | 9.54e-01 |
| GO:BP | GO:0002674 | negative regulation of acute inflammatory response | 1 | 5 | 9.54e-01 |
| GO:BP | GO:0071549 | cellular response to dexamethasone stimulus | 1 | 24 | 9.54e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 14 | 43 | 9.54e-01 |
| GO:BP | GO:1904977 | lymphatic endothelial cell migration | 1 | 3 | 9.55e-01 |
| GO:BP | GO:0042360 | vitamin E metabolic process | 1 | 2 | 9.55e-01 |
| GO:BP | GO:0006104 | succinyl-CoA metabolic process | 1 | 9 | 9.55e-01 |
| GO:BP | GO:0035095 | behavioral response to nicotine | 1 | 3 | 9.56e-01 |
| GO:BP | GO:0019915 | lipid storage | 10 | 66 | 9.56e-01 |
| GO:BP | GO:0140507 | granzyme-mediated programmed cell death signaling pathway | 2 | 3 | 9.56e-01 |
| GO:BP | GO:0052782 | amino disaccharide catabolic process | 1 | 4 | 9.56e-01 |
| GO:BP | GO:0090045 | positive regulation of deacetylase activity | 2 | 7 | 9.56e-01 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 1 | 20 | 9.56e-01 |
| GO:BP | GO:0014850 | response to muscle activity | 1 | 17 | 9.56e-01 |
| GO:BP | GO:0032225 | regulation of synaptic transmission, dopaminergic | 2 | 5 | 9.56e-01 |
| GO:BP | GO:0033006 | regulation of mast cell activation involved in immune response | 10 | 21 | 9.56e-01 |
| GO:BP | GO:0060099 | regulation of phagocytosis, engulfment | 1 | 11 | 9.56e-01 |
| GO:BP | GO:0070841 | inclusion body assembly | 6 | 22 | 9.56e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 9.56e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 2 | 37 | 9.56e-01 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 9.56e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 1 | 16 | 9.56e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 33 | 4174 | 9.56e-01 |
| GO:BP | GO:0030209 | dermatan sulfate catabolic process | 1 | 4 | 9.56e-01 |
| GO:BP | GO:1905153 | regulation of membrane invagination | 1 | 11 | 9.56e-01 |
| GO:BP | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells | 1 | 4 | 9.56e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 3 | 12 | 9.56e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 4 | 160 | 9.56e-01 |
| GO:BP | GO:2001294 | malonyl-CoA catabolic process | 1 | 4 | 9.56e-01 |
| GO:BP | GO:1902858 | propionyl-CoA metabolic process | 1 | 4 | 9.56e-01 |
| GO:BP | GO:0061357 | positive regulation of Wnt protein secretion | 1 | 4 | 9.56e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 1 | 7441 | 9.56e-01 |
| GO:BP | GO:2000848 | positive regulation of corticosteroid hormone secretion | 2 | 4 | 9.56e-01 |
| GO:BP | GO:0035932 | aldosterone secretion | 2 | 5 | 9.56e-01 |
| GO:BP | GO:0035931 | mineralocorticoid secretion | 2 | 5 | 9.56e-01 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 1 | 14 | 9.56e-01 |
| GO:BP | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell | 1 | 3 | 9.56e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 9.56e-01 |
| GO:BP | GO:0035814 | negative regulation of renal sodium excretion | 1 | 2 | 9.57e-01 |
| GO:BP | GO:0010813 | neuropeptide catabolic process | 1 | 4 | 9.57e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 11 | 27 | 9.57e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 3 | 78 | 9.57e-01 |
| GO:BP | GO:2000015 | regulation of determination of dorsal identity | 1 | 3 | 9.57e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0072334 | UDP-galactose transmembrane transport | 1 | 4 | 9.57e-01 |
| GO:BP | GO:1903792 | negative regulation of monoatomic anion transport | 1 | 3 | 9.57e-01 |
| GO:BP | GO:0034587 | piRNA processing | 1 | 4 | 9.57e-01 |
| GO:BP | GO:0033033 | negative regulation of myeloid cell apoptotic process | 3 | 10 | 9.57e-01 |
| GO:BP | GO:1905245 | regulation of aspartic-type peptidase activity | 2 | 11 | 9.57e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.58e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 9.58e-01 |
| GO:BP | GO:0010988 | regulation of low-density lipoprotein particle clearance | 1 | 8 | 9.58e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 8 | 41 | 9.58e-01 |
| GO:BP | GO:0006030 | chitin metabolic process | 1 | 2 | 9.58e-01 |
| GO:BP | GO:0006032 | chitin catabolic process | 1 | 2 | 9.58e-01 |
| GO:BP | GO:0002693 | positive regulation of cellular extravasation | 5 | 13 | 9.58e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 2 | 106 | 9.58e-01 |
| GO:BP | GO:0045923 | positive regulation of fatty acid metabolic process | 3 | 26 | 9.58e-01 |
| GO:BP | GO:0043316 | cytotoxic T cell degranulation | 1 | 2 | 9.58e-01 |
| GO:BP | GO:0001706 | endoderm formation | 5 | 46 | 9.58e-01 |
| GO:BP | GO:1905150 | regulation of voltage-gated sodium channel activity | 2 | 6 | 9.58e-01 |
| GO:BP | GO:1904823 | purine nucleobase transmembrane transport | 1 | 3 | 9.58e-01 |
| GO:BP | GO:0046292 | formaldehyde metabolic process | 1 | 3 | 9.58e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 1 | 57 | 9.58e-01 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 2 | 11 | 9.58e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 9.58e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 5 | 29 | 9.58e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 2 | 72 | 9.58e-01 |
| GO:BP | GO:0007220 | Notch receptor processing | 2 | 9 | 9.58e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 6 | 96 | 9.58e-01 |
| GO:BP | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration | 1 | 7 | 9.58e-01 |
| GO:BP | GO:0010387 | COP9 signalosome assembly | 1 | 3 | 9.58e-01 |
| GO:BP | GO:1901164 | negative regulation of trophoblast cell migration | 1 | 4 | 9.58e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 4 | 27 | 9.59e-01 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 2 | 34 | 9.59e-01 |
| GO:BP | GO:0045113 | regulation of integrin biosynthetic process | 1 | 2 | 9.59e-01 |
| GO:BP | GO:0051454 | intracellular pH elevation | 2 | 4 | 9.59e-01 |
| GO:BP | GO:1903576 | response to L-arginine | 1 | 3 | 9.59e-01 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 6 | 27 | 9.59e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 1 | 81 | 9.59e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 9.59e-01 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 5 | 18 | 9.59e-01 |
| GO:BP | GO:0110088 | hippocampal neuron apoptotic process | 1 | 2 | 9.59e-01 |
| GO:BP | GO:2001280 | positive regulation of unsaturated fatty acid biosynthetic process | 1 | 4 | 9.59e-01 |
| GO:BP | GO:0110089 | regulation of hippocampal neuron apoptotic process | 1 | 2 | 9.59e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 1 | 20 | 9.59e-01 |
| GO:BP | GO:0007060 | male meiosis chromosome segregation | 1 | 1 | 9.59e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 5 | 33 | 9.59e-01 |
| GO:BP | GO:1905867 | epididymis development | 1 | 2 | 9.59e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 9.59e-01 |
| GO:BP | GO:0045950 | negative regulation of mitotic recombination | 1 | 2 | 9.59e-01 |
| GO:BP | GO:0036123 | histone H3-K9 dimethylation | 1 | 3 | 9.59e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 2 | 49 | 9.59e-01 |
| GO:BP | GO:0140632 | canonical inflammasome complex assembly | 1 | 23 | 9.59e-01 |
| GO:BP | GO:0065001 | specification of axis polarity | 1 | 2 | 9.59e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 9.59e-01 |
| GO:BP | GO:0048512 | circadian behavior | 3 | 30 | 9.60e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 9.60e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 4 | 10 | 9.60e-01 |
| GO:BP | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 4 | 15 | 9.60e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 11 | 47 | 9.60e-01 |
| GO:BP | GO:0021957 | corticospinal tract morphogenesis | 1 | 7 | 9.61e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 1 | 11 | 9.61e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 16 | 82 | 9.61e-01 |
| GO:BP | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process | 1 | 7 | 9.61e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 9.61e-01 |
| GO:BP | GO:0030322 | stabilization of membrane potential | 1 | 6 | 9.61e-01 |
| GO:BP | GO:0071354 | cellular response to interleukin-6 | 1 | 23 | 9.61e-01 |
| GO:BP | GO:1990962 | xenobiotic transport across blood-brain barrier | 1 | 3 | 9.61e-01 |
| GO:BP | GO:1903450 | regulation of G1 to G0 transition | 1 | 4 | 9.61e-01 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 3 | 22 | 9.61e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 5 | 22 | 9.61e-01 |
| GO:BP | GO:0021859 | pyramidal neuron differentiation | 4 | 11 | 9.61e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 1 | 47 | 9.61e-01 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 1 | 3 | 9.61e-01 |
| GO:BP | GO:1905516 | positive regulation of fertilization | 1 | 3 | 9.61e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 9.61e-01 |
| GO:BP | GO:0002840 | regulation of T cell mediated immune response to tumor cell | 2 | 5 | 9.61e-01 |
| GO:BP | GO:1990544 | mitochondrial ATP transmembrane transport | 1 | 4 | 9.61e-01 |
| GO:BP | GO:0140021 | mitochondrial ADP transmembrane transport | 1 | 4 | 9.61e-01 |
| GO:BP | GO:0043416 | regulation of skeletal muscle tissue regeneration | 1 | 5 | 9.61e-01 |
| GO:BP | GO:0007586 | digestion | 5 | 60 | 9.62e-01 |
| GO:BP | GO:0002827 | positive regulation of T-helper 1 type immune response | 2 | 5 | 9.62e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 1 | 7069 | 9.62e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 9.62e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 9.62e-01 |
| GO:BP | GO:1990705 | cholangiocyte proliferation | 1 | 3 | 9.63e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 9.63e-01 |
| GO:BP | GO:0001920 | negative regulation of receptor recycling | 1 | 3 | 9.63e-01 |
| GO:BP | GO:0051799 | negative regulation of hair follicle development | 1 | 5 | 9.63e-01 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 9.63e-01 |
| GO:BP | GO:0070103 | regulation of interleukin-6-mediated signaling pathway | 1 | 4 | 9.63e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 9.63e-01 |
| GO:BP | GO:0010467 | gene expression | 4 | 4587 | 9.63e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 5 | 11 | 9.63e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 5 | 11 | 9.63e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 5 | 11 | 9.63e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 1 | 30 | 9.63e-01 |
| GO:BP | GO:0034135 | regulation of toll-like receptor 2 signaling pathway | 1 | 7 | 9.63e-01 |
| GO:BP | GO:1903412 | response to bile acid | 1 | 3 | 9.63e-01 |
| GO:BP | GO:0006477 | protein sulfation | 2 | 6 | 9.64e-01 |
| GO:BP | GO:0035562 | negative regulation of chromatin binding | 1 | 8 | 9.64e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 1 | 27 | 9.64e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 3 | 44 | 9.64e-01 |
| GO:BP | GO:0090025 | regulation of monocyte chemotaxis | 1 | 12 | 9.64e-01 |
| GO:BP | GO:0014002 | astrocyte development | 14 | 27 | 9.64e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 3 | 484 | 9.64e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 16 | 47 | 9.64e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 11 | 25 | 9.64e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 1 | 285 | 9.64e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 1 | 108 | 9.64e-01 |
| GO:BP | GO:0034380 | high-density lipoprotein particle assembly | 1 | 10 | 9.64e-01 |
| GO:BP | GO:2000328 | regulation of T-helper 17 cell lineage commitment | 1 | 1 | 9.64e-01 |
| GO:BP | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway | 1 | 4 | 9.64e-01 |
| GO:BP | GO:0006105 | succinate metabolic process | 1 | 5 | 9.64e-01 |
| GO:BP | GO:0032264 | IMP salvage | 1 | 4 | 9.64e-01 |
| GO:BP | GO:0009996 | negative regulation of cell fate specification | 1 | 4 | 9.64e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 8 | 19 | 9.65e-01 |
| GO:BP | GO:0001916 | positive regulation of T cell mediated cytotoxicity | 2 | 13 | 9.65e-01 |
| GO:BP | GO:0046174 | polyol catabolic process | 1 | 7 | 9.65e-01 |
| GO:BP | GO:0046949 | fatty-acyl-CoA biosynthetic process | 9 | 20 | 9.65e-01 |
| GO:BP | GO:0000965 | mitochondrial RNA 3’-end processing | 1 | 5 | 9.65e-01 |
| GO:BP | GO:0036305 | ameloblast differentiation | 1 | 5 | 9.65e-01 |
| GO:BP | GO:0035989 | tendon development | 1 | 4 | 9.65e-01 |
| GO:BP | GO:0072104 | glomerular capillary formation | 1 | 5 | 9.65e-01 |
| GO:BP | GO:0006071 | glycerol metabolic process | 4 | 12 | 9.65e-01 |
| GO:BP | GO:0072103 | glomerulus vasculature morphogenesis | 1 | 5 | 9.65e-01 |
| GO:BP | GO:0044409 | entry into host | 19 | 113 | 9.67e-01 |
| GO:BP | GO:0044060 | regulation of endocrine process | 1 | 24 | 9.67e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 1 | 84 | 9.67e-01 |
| GO:BP | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity | 1 | 3 | 9.67e-01 |
| GO:BP | GO:0042026 | protein refolding | 9 | 26 | 9.67e-01 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 2 | 18 | 9.67e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 28 | 106 | 9.67e-01 |
| GO:BP | GO:0007309 | oocyte axis specification | 1 | 3 | 9.67e-01 |
| GO:BP | GO:0007308 | oocyte construction | 1 | 3 | 9.67e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 1 | 31 | 9.67e-01 |
| GO:BP | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 9.68e-01 |
| GO:BP | GO:0046487 | glyoxylate metabolic process | 2 | 5 | 9.68e-01 |
| GO:BP | GO:0043031 | negative regulation of macrophage activation | 2 | 9 | 9.68e-01 |
| GO:BP | GO:0060439 | trachea morphogenesis | 1 | 7 | 9.68e-01 |
| GO:BP | GO:0072385 | minus-end-directed organelle transport along microtubule | 1 | 5 | 9.68e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 17 | 96 | 9.68e-01 |
| GO:BP | GO:0009311 | oligosaccharide metabolic process | 4 | 43 | 9.68e-01 |
| GO:BP | GO:0023052 | signaling | 5 | 4141 | 9.68e-01 |
| GO:BP | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | 4 | 13 | 9.68e-01 |
| GO:BP | GO:1900425 | negative regulation of defense response to bacterium | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0032627 | interleukin-23 production | 2 | 6 | 9.68e-01 |
| GO:BP | GO:0032667 | regulation of interleukin-23 production | 2 | 6 | 9.68e-01 |
| GO:BP | GO:0050909 | sensory perception of taste | 1 | 13 | 9.68e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 5 | 121 | 9.68e-01 |
| GO:BP | GO:0097709 | connective tissue replacement | 1 | 8 | 9.68e-01 |
| GO:BP | GO:0097278 | complement-dependent cytotoxicity | 1 | 4 | 9.68e-01 |
| GO:BP | GO:2000437 | regulation of monocyte extravasation | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 2 | 3 | 9.68e-01 |
| GO:BP | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin | 1 | 1 | 9.68e-01 |
| GO:BP | GO:0071484 | cellular response to light intensity | 1 | 4 | 9.69e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 9.69e-01 |
| GO:BP | GO:0061377 | mammary gland lobule development | 2 | 13 | 9.69e-01 |
| GO:BP | GO:0060749 | mammary gland alveolus development | 2 | 13 | 9.69e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 5 | 13 | 9.69e-01 |
| GO:BP | GO:0051135 | positive regulation of NK T cell activation | 1 | 3 | 9.69e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 1 | 33 | 9.69e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 22 | 137 | 9.69e-01 |
| GO:BP | GO:0033004 | negative regulation of mast cell activation | 3 | 4 | 9.69e-01 |
| GO:BP | GO:0072679 | thymocyte migration | 1 | 1 | 9.69e-01 |
| GO:BP | GO:1904557 | L-alanine transmembrane transport | 1 | 4 | 9.69e-01 |
| GO:BP | GO:0001708 | cell fate specification | 1 | 56 | 9.69e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 2 | 20 | 9.69e-01 |
| GO:BP | GO:0018197 | peptidyl-aspartic acid modification | 1 | 5 | 9.70e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 2 | 5 | 9.70e-01 |
| GO:BP | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway | 1 | 5 | 9.70e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 9.70e-01 |
| GO:BP | GO:1905907 | negative regulation of amyloid fibril formation | 2 | 10 | 9.70e-01 |
| GO:BP | GO:0072656 | maintenance of protein location in mitochondrion | 1 | 5 | 9.70e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 9.70e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 9.70e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 9.71e-01 |
| GO:BP | GO:0070741 | response to interleukin-6 | 1 | 23 | 9.71e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 2 | 47 | 9.71e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 8 | 22 | 9.71e-01 |
| GO:BP | GO:0033299 | secretion of lysosomal enzymes | 1 | 7 | 9.71e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 9.71e-01 |
| GO:BP | GO:0061590 | calcium activated phosphatidylcholine scrambling | 1 | 3 | 9.71e-01 |
| GO:BP | GO:0002396 | MHC protein complex assembly | 2 | 8 | 9.71e-01 |
| GO:BP | GO:0002501 | peptide antigen assembly with MHC protein complex | 2 | 8 | 9.71e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 9.71e-01 |
| GO:BP | GO:0002858 | regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 5 | 23 | 9.71e-01 |
| GO:BP | GO:0042421 | norepinephrine biosynthetic process | 1 | 2 | 9.71e-01 |
| GO:BP | GO:0022417 | protein maturation by protein folding | 2 | 8 | 9.71e-01 |
| GO:BP | GO:0014052 | regulation of gamma-aminobutyric acid secretion | 1 | 5 | 9.71e-01 |
| GO:BP | GO:0070474 | positive regulation of uterine smooth muscle contraction | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0061024 | membrane organization | 3 | 671 | 9.72e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 8 | 775 | 9.72e-01 |
| GO:BP | GO:0007492 | endoderm development | 26 | 65 | 9.72e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 9.72e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 2 | 37 | 9.72e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 9.72e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 6 | 16 | 9.73e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2 | 4045 | 9.73e-01 |
| GO:BP | GO:0060435 | bronchiole development | 1 | 2 | 9.73e-01 |
| GO:BP | GO:0007154 | cell communication | 5 | 4226 | 9.73e-01 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0060732 | positive regulation of inositol phosphate biosynthetic process | 4 | 5 | 9.73e-01 |
| GO:BP | GO:0010986 | positive regulation of lipoprotein particle clearance | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0019336 | phenol-containing compound catabolic process | 1 | 6 | 9.73e-01 |
| GO:BP | GO:0032274 | gonadotropin secretion | 4 | 10 | 9.73e-01 |
| GO:BP | GO:0021523 | somatic motor neuron differentiation | 1 | 2 | 9.73e-01 |
| GO:BP | GO:1990787 | negative regulation of hh target transcription factor activity | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0007602 | phototransduction | 1 | 17 | 9.73e-01 |
| GO:BP | GO:0002716 | negative regulation of natural killer cell mediated immunity | 1 | 10 | 9.74e-01 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 5 | 28 | 9.74e-01 |
| GO:BP | GO:0002553 | histamine secretion by mast cell | 1 | 8 | 9.74e-01 |
| GO:BP | GO:0002349 | histamine production involved in inflammatory response | 1 | 8 | 9.74e-01 |
| GO:BP | GO:0002441 | histamine secretion involved in inflammatory response | 1 | 8 | 9.74e-01 |
| GO:BP | GO:0048752 | semicircular canal morphogenesis | 1 | 5 | 9.75e-01 |
| GO:BP | GO:0061975 | articular cartilage development | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0019673 | GDP-mannose metabolic process | 1 | 7 | 9.75e-01 |
| GO:BP | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 9.75e-01 |
| GO:BP | GO:0002034 | maintenance of blood vessel diameter homeostasis by renin-angiotensin | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0045576 | mast cell activation | 3 | 35 | 9.75e-01 |
| GO:BP | GO:0003072 | renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0048755 | branching morphogenesis of a nerve | 3 | 8 | 9.75e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 3 | 23 | 9.75e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 10 | 1238 | 9.75e-01 |
| GO:BP | GO:1903028 | positive regulation of opsonization | 1 | 1 | 9.75e-01 |
| GO:BP | GO:0044406 | adhesion of symbiont to host | 1 | 10 | 9.75e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 5 | 168 | 9.75e-01 |
| GO:BP | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome | 1 | 2 | 9.75e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 2 | 14 | 9.75e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 9.75e-01 |
| GO:BP | GO:0007155 | cell adhesion | 3 | 1012 | 9.75e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 2 | 288 | 9.75e-01 |
| GO:BP | GO:0009642 | response to light intensity | 1 | 11 | 9.75e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 2 | 19 | 9.75e-01 |
| GO:BP | GO:0140894 | endolysosomal toll-like receptor signaling pathway | 3 | 24 | 9.75e-01 |
| GO:BP | GO:0032277 | negative regulation of gonadotropin secretion | 1 | 4 | 9.75e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 1 | 68 | 9.76e-01 |
| GO:BP | GO:0033574 | response to testosterone | 2 | 29 | 9.76e-01 |
| GO:BP | GO:1903307 | positive regulation of regulated secretory pathway | 1 | 32 | 9.76e-01 |
| GO:BP | GO:0009233 | menaquinone metabolic process | 1 | 1 | 9.76e-01 |
| GO:BP | GO:0070099 | regulation of chemokine-mediated signaling pathway | 2 | 8 | 9.76e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 3 | 16 | 9.76e-01 |
| GO:BP | GO:0060986 | endocrine hormone secretion | 2 | 35 | 9.76e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 1 | 10 | 9.76e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 2 | 36 | 9.76e-01 |
| GO:BP | GO:0002829 | negative regulation of type 2 immune response | 1 | 7 | 9.76e-01 |
| GO:BP | GO:0010961 | intracellular magnesium ion homeostasis | 1 | 4 | 9.76e-01 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 9.76e-01 |
| GO:BP | GO:0001915 | negative regulation of T cell mediated cytotoxicity | 1 | 3 | 9.77e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 9 | 23 | 9.77e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 1 | 16 | 9.77e-01 |
| GO:BP | GO:1905684 | regulation of plasma membrane repair | 1 | 7 | 9.77e-01 |
| GO:BP | GO:0006531 | aspartate metabolic process | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0046907 | intracellular transport | 3 | 1453 | 9.77e-01 |
| GO:BP | GO:0097052 | L-kynurenine metabolic process | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0001990 | regulation of systemic arterial blood pressure by hormone | 1 | 22 | 9.77e-01 |
| GO:BP | GO:0070487 | monocyte aggregation | 1 | 4 | 9.77e-01 |
| GO:BP | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 9.77e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 1 | 51 | 9.77e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 1 | 7 | 9.77e-01 |
| GO:BP | GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | 6 | 14 | 9.78e-01 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 8 | 18 | 9.78e-01 |
| GO:BP | GO:0042368 | vitamin D biosynthetic process | 2 | 4 | 9.78e-01 |
| GO:BP | GO:2000402 | negative regulation of lymphocyte migration | 1 | 10 | 9.78e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 1 | 7806 | 9.78e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 9.78e-01 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 2 | 4 | 9.78e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 9 | 68 | 9.78e-01 |
| GO:BP | GO:0045112 | integrin biosynthetic process | 1 | 3 | 9.78e-01 |
| GO:BP | GO:0046668 | regulation of retinal cell programmed cell death | 1 | 2 | 9.78e-01 |
| GO:BP | GO:0043302 | positive regulation of leukocyte degranulation | 4 | 15 | 9.79e-01 |
| GO:BP | GO:0007341 | penetration of zona pellucida | 1 | 4 | 9.79e-01 |
| GO:BP | GO:0021800 | cerebral cortex tangential migration | 1 | 4 | 9.79e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 2 | 27 | 9.79e-01 |
| GO:BP | GO:0035469 | determination of pancreatic left/right asymmetry | 1 | 3 | 9.79e-01 |
| GO:BP | GO:0014905 | myoblast fusion involved in skeletal muscle regeneration | 1 | 3 | 9.79e-01 |
| GO:BP | GO:0033684 | regulation of luteinizing hormone secretion | 1 | 3 | 9.79e-01 |
| GO:BP | GO:0046881 | positive regulation of follicle-stimulating hormone secretion | 1 | 3 | 9.79e-01 |
| GO:BP | GO:0044788 | modulation by host of viral process | 2 | 40 | 9.79e-01 |
| GO:BP | GO:0002424 | T cell mediated immune response to tumor cell | 2 | 6 | 9.79e-01 |
| GO:BP | GO:0046880 | regulation of follicle-stimulating hormone secretion | 1 | 3 | 9.79e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 3 | 69 | 9.79e-01 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 4 | 12 | 9.79e-01 |
| GO:BP | GO:0001711 | endodermal cell fate commitment | 3 | 9 | 9.79e-01 |
| GO:BP | GO:0042221 | response to chemical | 2 | 2559 | 9.79e-01 |
| GO:BP | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 1 | 10 | 9.79e-01 |
| GO:BP | GO:0061356 | regulation of Wnt protein secretion | 1 | 4 | 9.79e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 2 | 220 | 9.80e-01 |
| GO:BP | GO:0002752 | cell surface pattern recognition receptor signaling pathway | 1 | 27 | 9.80e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 2 | 50 | 9.80e-01 |
| GO:BP | GO:1990051 | activation of protein kinase C activity | 1 | 3 | 9.80e-01 |
| GO:BP | GO:0045332 | phospholipid translocation | 1 | 33 | 9.80e-01 |
| GO:BP | GO:0002082 | regulation of oxidative phosphorylation | 1 | 19 | 9.80e-01 |
| GO:BP | GO:0009644 | response to high light intensity | 1 | 2 | 9.81e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 5 | 26 | 9.81e-01 |
| GO:BP | GO:0048535 | lymph node development | 4 | 11 | 9.83e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 3 | 70 | 9.83e-01 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 1 | 5 | 9.83e-01 |
| GO:BP | GO:0044648 | histone H3-K4 dimethylation | 1 | 6 | 9.83e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 2 | 29 | 9.83e-01 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 5 | 13 | 9.83e-01 |
| GO:BP | GO:0002855 | regulation of natural killer cell mediated immune response to tumor cell | 1 | 4 | 9.83e-01 |
| GO:BP | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 9.83e-01 |
| GO:BP | GO:0001840 | neural plate development | 1 | 7 | 9.83e-01 |
| GO:BP | GO:0019400 | alditol metabolic process | 4 | 14 | 9.84e-01 |
| GO:BP | GO:0007030 | Golgi organization | 4 | 137 | 9.84e-01 |
| GO:BP | GO:0120262 | negative regulation of heterochromatin organization | 1 | 4 | 9.84e-01 |
| GO:BP | GO:0042747 | circadian sleep/wake cycle, REM sleep | 1 | 2 | 9.84e-01 |
| GO:BP | GO:0032489 | regulation of Cdc42 protein signal transduction | 1 | 6 | 9.84e-01 |
| GO:BP | GO:0006710 | androgen catabolic process | 1 | 3 | 9.84e-01 |
| GO:BP | GO:0043321 | regulation of natural killer cell degranulation | 1 | 3 | 9.84e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 9.84e-01 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 2 | 9 | 9.84e-01 |
| GO:BP | GO:1905563 | negative regulation of vascular endothelial cell proliferation | 1 | 5 | 9.84e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 4 | 24 | 9.84e-01 |
| GO:BP | GO:0034398 | telomere tethering at nuclear periphery | 1 | 1 | 9.84e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 2 | 49 | 9.84e-01 |
| GO:BP | GO:0043323 | positive regulation of natural killer cell degranulation | 1 | 3 | 9.84e-01 |
| GO:BP | GO:2001026 | regulation of endothelial cell chemotaxis | 2 | 16 | 9.84e-01 |
| GO:BP | GO:0031452 | negative regulation of heterochromatin formation | 1 | 4 | 9.84e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 9.84e-01 |
| GO:BP | GO:0072540 | T-helper 17 cell lineage commitment | 2 | 3 | 9.84e-01 |
| GO:BP | GO:1905268 | negative regulation of chromatin organization | 1 | 4 | 9.84e-01 |
| GO:BP | GO:0072015 | podocyte development | 2 | 9 | 9.84e-01 |
| GO:BP | GO:1905599 | positive regulation of low-density lipoprotein receptor activity | 1 | 4 | 9.84e-01 |
| GO:BP | GO:0030473 | nuclear migration along microtubule | 1 | 5 | 9.84e-01 |
| GO:BP | GO:0070197 | meiotic attachment of telomere to nuclear envelope | 1 | 1 | 9.84e-01 |
| GO:BP | GO:0044821 | meiotic telomere tethering at nuclear periphery | 1 | 1 | 9.84e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 2 | 484 | 9.84e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 17 | 107 | 9.85e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 9 | 58 | 9.85e-01 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 1 | 6 | 9.85e-01 |
| GO:BP | GO:1903963 | arachidonate transport | 1 | 11 | 9.85e-01 |
| GO:BP | GO:0050482 | arachidonic acid secretion | 1 | 11 | 9.85e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 4 | 609 | 9.85e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 5 | 16 | 9.85e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 8 | 53 | 9.85e-01 |
| GO:BP | GO:0035457 | cellular response to interferon-alpha | 1 | 7 | 9.85e-01 |
| GO:BP | GO:0035025 | positive regulation of Rho protein signal transduction | 2 | 22 | 9.85e-01 |
| GO:BP | GO:0021819 | layer formation in cerebral cortex | 3 | 12 | 9.86e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 9.86e-01 |
| GO:BP | GO:0001878 | response to yeast | 1 | 3 | 9.86e-01 |
| GO:BP | GO:0071349 | cellular response to interleukin-12 | 1 | 5 | 9.86e-01 |
| GO:BP | GO:0006688 | glycosphingolipid biosynthetic process | 1 | 25 | 9.86e-01 |
| GO:BP | GO:0019371 | cyclooxygenase pathway | 1 | 7 | 9.86e-01 |
| GO:BP | GO:0032963 | collagen metabolic process | 3 | 74 | 9.86e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 9.86e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 8 | 139 | 9.86e-01 |
| GO:BP | GO:0035630 | bone mineralization involved in bone maturation | 3 | 6 | 9.86e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 2 | 25 | 9.86e-01 |
| GO:BP | GO:0140353 | lipid export from cell | 1 | 28 | 9.87e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 58 | 9.87e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 9.87e-01 |
| GO:BP | GO:0002513 | tolerance induction to self antigen | 1 | 4 | 9.87e-01 |
| GO:BP | GO:0015671 | oxygen transport | 1 | 4 | 9.87e-01 |
| GO:BP | GO:1903208 | negative regulation of hydrogen peroxide-induced neuron death | 1 | 5 | 9.87e-01 |
| GO:BP | GO:1903207 | regulation of hydrogen peroxide-induced neuron death | 1 | 5 | 9.87e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 9 | 59 | 9.87e-01 |
| GO:BP | GO:0006868 | glutamine transport | 2 | 8 | 9.87e-01 |
| GO:BP | GO:0036476 | neuron death in response to hydrogen peroxide | 1 | 5 | 9.87e-01 |
| GO:BP | GO:0072205 | metanephric collecting duct development | 2 | 5 | 9.87e-01 |
| GO:BP | GO:0046641 | positive regulation of alpha-beta T cell proliferation | 2 | 11 | 9.87e-01 |
| GO:BP | GO:0072683 | T cell extravasation | 1 | 8 | 9.87e-01 |
| GO:BP | GO:1900426 | positive regulation of defense response to bacterium | 1 | 6 | 9.87e-01 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 1 | 6 | 9.87e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 3 | 16 | 9.87e-01 |
| GO:BP | GO:0072592 | oxygen metabolic process | 1 | 4 | 9.87e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 3 | 58 | 9.88e-01 |
| GO:BP | GO:0061588 | calcium activated phospholipid scrambling | 1 | 4 | 9.88e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 1 | 211 | 9.88e-01 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 14 | 48 | 9.88e-01 |
| GO:BP | GO:0032276 | regulation of gonadotropin secretion | 2 | 6 | 9.88e-01 |
| GO:BP | GO:0045579 | positive regulation of B cell differentiation | 4 | 10 | 9.88e-01 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 5 | 16 | 9.88e-01 |
| GO:BP | GO:0042362 | fat-soluble vitamin biosynthetic process | 3 | 6 | 9.88e-01 |
| GO:BP | GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | 1 | 9 | 9.88e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 9.88e-01 |
| GO:BP | GO:0007369 | gastrulation | 26 | 146 | 9.88e-01 |
| GO:BP | GO:0021853 | cerebral cortex GABAergic interneuron migration | 1 | 2 | 9.89e-01 |
| GO:BP | GO:0002691 | regulation of cellular extravasation | 3 | 24 | 9.89e-01 |
| GO:BP | GO:0051935 | glutamate reuptake | 1 | 4 | 9.89e-01 |
| GO:BP | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning | 1 | 5 | 9.89e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 7 | 34 | 9.90e-01 |
| GO:BP | GO:0045656 | negative regulation of monocyte differentiation | 1 | 4 | 9.90e-01 |
| GO:BP | GO:0060179 | male mating behavior | 1 | 5 | 9.90e-01 |
| GO:BP | GO:0007168 | receptor guanylyl cyclase signaling pathway | 2 | 5 | 9.90e-01 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 2 | 23 | 9.90e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 2 | 16 | 9.90e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 9.91e-01 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 1 | 18 | 9.91e-01 |
| GO:BP | GO:0048263 | determination of dorsal identity | 2 | 5 | 9.91e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 14 | 88 | 9.91e-01 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 2 | 8 | 9.91e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 3 | 23 | 9.91e-01 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 4 | 96 | 9.91e-01 |
| GO:BP | GO:0002423 | natural killer cell mediated immune response to tumor cell | 1 | 4 | 9.91e-01 |
| GO:BP | GO:0070472 | regulation of uterine smooth muscle contraction | 1 | 5 | 9.92e-01 |
| GO:BP | GO:0001821 | histamine secretion | 1 | 9 | 9.92e-01 |
| GO:BP | GO:0022617 | extracellular matrix disassembly | 5 | 36 | 9.92e-01 |
| GO:BP | GO:0050817 | coagulation | 10 | 153 | 9.92e-01 |
| GO:BP | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb | 1 | 4 | 9.92e-01 |
| GO:BP | GO:0048143 | astrocyte activation | 6 | 12 | 9.92e-01 |
| GO:BP | GO:1903027 | regulation of opsonization | 1 | 1 | 9.92e-01 |
| GO:BP | GO:0046034 | ATP metabolic process | 7 | 116 | 9.92e-01 |
| GO:BP | GO:0060911 | cardiac cell fate commitment | 1 | 11 | 9.92e-01 |
| GO:BP | GO:0032278 | positive regulation of gonadotropin secretion | 1 | 4 | 9.92e-01 |
| GO:BP | GO:0007599 | hemostasis | 10 | 153 | 9.93e-01 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 2 | 9 | 9.93e-01 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 12 | 25 | 9.93e-01 |
| GO:BP | GO:1900424 | regulation of defense response to bacterium | 2 | 12 | 9.93e-01 |
| GO:BP | GO:0017038 | protein import | 1 | 7 | 9.93e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 2 | 10 | 9.93e-01 |
| GO:BP | GO:0034112 | positive regulation of homotypic cell-cell adhesion | 1 | 10 | 9.93e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 2 | 10 | 9.93e-01 |
| GO:BP | GO:0150079 | negative regulation of neuroinflammatory response | 1 | 6 | 9.93e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 1 | 130 | 9.93e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 10 | 1335 | 9.93e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 2 | 190 | 9.93e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 5 | 90 | 9.93e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 5 | 14 | 9.93e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 2 | 475 | 9.94e-01 |
| GO:BP | GO:0010496 | intercellular transport | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0051049 | regulation of transport | 47 | 1267 | 9.94e-01 |
| GO:BP | GO:0048643 | positive regulation of skeletal muscle tissue development | 2 | 14 | 9.94e-01 |
| GO:BP | GO:0061355 | Wnt protein secretion | 1 | 5 | 9.94e-01 |
| GO:BP | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process | 1 | 10 | 9.94e-01 |
| GO:BP | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | 1 | 11 | 9.94e-01 |
| GO:BP | GO:0007216 | G protein-coupled glutamate receptor signaling pathway | 1 | 5 | 9.94e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 26 | 64 | 9.94e-01 |
| GO:BP | GO:0002283 | neutrophil activation involved in immune response | 1 | 7 | 9.94e-01 |
| GO:BP | GO:1901289 | succinyl-CoA catabolic process | 1 | 6 | 9.94e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 2 | 19 | 9.94e-01 |
| GO:BP | GO:2000556 | positive regulation of T-helper 1 cell cytokine production | 1 | 2 | 9.94e-01 |
| GO:BP | GO:2000554 | regulation of T-helper 1 cell cytokine production | 1 | 2 | 9.94e-01 |
| GO:BP | GO:0035744 | T-helper 1 cell cytokine production | 1 | 2 | 9.94e-01 |
| GO:BP | GO:0052695 | cellular glucuronidation | 1 | 2 | 9.94e-01 |
| GO:BP | GO:0031638 | zymogen activation | 10 | 45 | 9.94e-01 |
| GO:BP | GO:0071910 | determination of liver left/right asymmetry | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 1 | 42 | 9.94e-01 |
| GO:BP | GO:1901317 | regulation of flagellated sperm motility | 1 | 6 | 9.94e-01 |
| GO:BP | GO:0006907 | pinocytosis | 1 | 18 | 9.94e-01 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 1 | 104 | 9.94e-01 |
| GO:BP | GO:2001140 | positive regulation of phospholipid transport | 2 | 11 | 9.94e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 10 | 35 | 9.94e-01 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 1 | 13 | 9.94e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 10 | 24 | 9.94e-01 |
| GO:BP | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0072044 | collecting duct development | 3 | 6 | 9.94e-01 |
| GO:BP | GO:0120197 | mucociliary clearance | 1 | 5 | 9.95e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 10 | 245 | 9.95e-01 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 1 | 8 | 9.95e-01 |
| GO:BP | GO:1902260 | negative regulation of delayed rectifier potassium channel activity | 1 | 5 | 9.95e-01 |
| GO:BP | GO:0042363 | fat-soluble vitamin catabolic process | 1 | 1 | 9.95e-01 |
| GO:BP | GO:0035112 | genitalia morphogenesis | 2 | 5 | 9.95e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 1 | 8 | 9.95e-01 |
| GO:BP | GO:0015721 | bile acid and bile salt transport | 2 | 11 | 9.95e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 9.95e-01 |
| GO:BP | GO:1905920 | positive regulation of CoA-transferase activity | 1 | 5 | 9.95e-01 |
| GO:BP | GO:1905918 | regulation of CoA-transferase activity | 1 | 5 | 9.95e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 7 | 39 | 9.95e-01 |
| GO:BP | GO:0032383 | regulation of intracellular cholesterol transport | 1 | 6 | 9.95e-01 |
| GO:BP | GO:0032380 | regulation of intracellular sterol transport | 1 | 6 | 9.95e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 9 | 36 | 9.95e-01 |
| GO:BP | GO:1904936 | interneuron migration | 1 | 6 | 9.96e-01 |
| GO:BP | GO:0042182 | ketone catabolic process | 4 | 9 | 9.96e-01 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 6 | 37 | 9.96e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 3 | 15 | 9.96e-01 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 1 | 14 | 9.96e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 2 | 20 | 9.96e-01 |
| GO:BP | GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | 1 | 30 | 9.96e-01 |
| GO:BP | GO:0009820 | alkaloid metabolic process | 1 | 2 | 9.96e-01 |
| GO:BP | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing | 1 | 4 | 9.96e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 3 | 24 | 9.96e-01 |
| GO:BP | GO:0015866 | ADP transport | 1 | 6 | 9.96e-01 |
| GO:BP | GO:0070091 | glucagon secretion | 1 | 3 | 9.96e-01 |
| GO:BP | GO:0070092 | regulation of glucagon secretion | 1 | 3 | 9.96e-01 |
| GO:BP | GO:0070471 | uterine smooth muscle contraction | 1 | 6 | 9.96e-01 |
| GO:BP | GO:1900157 | regulation of bone mineralization involved in bone maturation | 1 | 4 | 9.96e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 1 | 91 | 9.96e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 1 | 91 | 9.96e-01 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 6 | 137 | 9.96e-01 |
| GO:BP | GO:0051014 | actin filament severing | 2 | 13 | 9.96e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 2 | 30 | 9.97e-01 |
| GO:BP | GO:0043299 | leukocyte degranulation | 1 | 41 | 9.97e-01 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 3 | 9 | 9.97e-01 |
| GO:BP | GO:0046415 | urate metabolic process | 1 | 5 | 9.97e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 6 | 21 | 9.97e-01 |
| GO:BP | GO:0021521 | ventral spinal cord interneuron specification | 1 | 3 | 9.98e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 1 | 94 | 9.98e-01 |
| GO:BP | GO:0060573 | cell fate specification involved in pattern specification | 1 | 3 | 9.98e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 7 | 336 | 9.98e-01 |
| GO:BP | GO:0071676 | negative regulation of mononuclear cell migration | 2 | 16 | 9.98e-01 |
| GO:BP | GO:0072048 | renal system pattern specification | 1 | 6 | 9.98e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 1 | 94 | 9.98e-01 |
| GO:BP | GO:0048251 | elastic fiber assembly | 2 | 12 | 9.98e-01 |
| GO:BP | GO:0045058 | T cell selection | 1 | 22 | 9.98e-01 |
| GO:BP | GO:0001914 | regulation of T cell mediated cytotoxicity | 2 | 17 | 9.98e-01 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 4 | 18 | 9.98e-01 |
| GO:BP | GO:0021860 | pyramidal neuron development | 2 | 8 | 9.98e-01 |
| GO:BP | GO:0061004 | pattern specification involved in kidney development | 1 | 6 | 9.98e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 2 | 581 | 9.98e-01 |
| GO:BP | GO:0051957 | positive regulation of amino acid transport | 4 | 16 | 9.98e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 4 | 90 | 9.98e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 9 | 422 | 9.98e-01 |
| GO:BP | GO:0048262 | determination of dorsal/ventral asymmetry | 2 | 6 | 9.98e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 2 | 14 | 9.98e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 11 | 39 | 9.98e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 2 | 14 | 9.98e-01 |
| GO:BP | GO:0050886 | endocrine process | 3 | 57 | 9.98e-01 |
| GO:BP | GO:0007596 | blood coagulation | 9 | 150 | 9.99e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3 | 26 | 9.99e-01 |
| GO:BP | GO:0042373 | vitamin K metabolic process | 2 | 6 | 9.99e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 6 | 25 | 9.99e-01 |
| GO:BP | GO:1902523 | positive regulation of protein K63-linked ubiquitination | 1 | 5 | 9.99e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 6 | 191 | 9.99e-01 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 3 | 87 | 9.99e-01 |
| GO:BP | GO:0071286 | cellular response to magnesium ion | 1 | 3 | 9.99e-01 |
| GO:BP | GO:0031342 | negative regulation of cell killing | 1 | 11 | 9.99e-01 |
| GO:BP | GO:0150172 | regulation of phosphatidylcholine metabolic process | 1 | 10 | 9.99e-01 |
| GO:BP | GO:0120252 | hydrocarbon metabolic process | 1 | 2 | 9.99e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 3 | 523 | 1.00e+00 |
| GO:BP | GO:0097692 | histone H3-K4 monomethylation | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0043933 | protein-containing complex organization | 10 | 1496 | 1.00e+00 |
| GO:BP | GO:0000165 | MAPK cascade | 17 | 556 | 1.00e+00 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 1.00e+00 |
| GO:BP | GO:0048536 | spleen development | 2 | 28 | 1.00e+00 |
| GO:BP | GO:0016045 | detection of bacterium | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 1.00e+00 |
| GO:BP | GO:0021762 | substantia nigra development | 6 | 41 | 1.00e+00 |
| GO:BP | GO:0010455 | positive regulation of cell fate commitment | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 2 | 29 | 1.00e+00 |
| GO:BP | GO:0045026 | plasma membrane fusion | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0021520 | spinal cord motor neuron cell fate specification | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0006810 | transport | 11 | 3298 | 1.00e+00 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 4 | 30 | 1.00e+00 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 2 | 38 | 1.00e+00 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 3 | 27 | 1.00e+00 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 2 | 34 | 1.00e+00 |
| GO:BP | GO:0071702 | organic substance transport | 7 | 2116 | 1.00e+00 |
| GO:BP | GO:0015031 | protein transport | 8 | 1361 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 2 | 38 | 1.00e+00 |
| GO:BP | GO:0071705 | nitrogen compound transport | 9 | 1684 | 1.00e+00 |
| GO:BP | GO:0051917 | regulation of fibrinolysis | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0032604 | granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0046164 | alcohol catabolic process | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 2 | 20 | 1.00e+00 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 10 | 227 | 1.00e+00 |
| GO:BP | GO:0031223 | auditory behavior | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0035813 | regulation of renal sodium excretion | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 1.00e+00 |
| GO:BP | GO:0016127 | sterol catabolic process | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 1 | 115 | 1.00e+00 |
| GO:BP | GO:0035930 | corticosteroid hormone secretion | 2 | 12 | 1.00e+00 |
| GO:BP | GO:0065007 | biological regulation | 1 | 8320 | 1.00e+00 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 15 | 65 | 1.00e+00 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 13 | 55 | 1.00e+00 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 1 | 21 | 1.00e+00 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0035812 | renal sodium excretion | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0021554 | optic nerve development | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0032645 | regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0035815 | positive regulation of renal sodium excretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0006457 | protein folding | 4 | 192 | 1.00e+00 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 2 | 16 | 1.00e+00 |
| GO:BP | GO:0002682 | regulation of immune system process | 9 | 877 | 1.00e+00 |
| GO:BP | GO:0006869 | lipid transport | 7 | 272 | 1.00e+00 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 2 | 27 | 1.00e+00 |
| GO:BP | GO:0030200 | heparan sulfate proteoglycan catabolic process | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 1.00e+00 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0010572 | positive regulation of platelet activation | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0002839 | positive regulation of immune response to tumor cell | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0051923 | sulfation | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0001867 | complement activation, lectin pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0007342 | fusion of sperm to egg plasma membrane involved in single fertilization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0046666 | retinal cell programmed cell death | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0002689 | negative regulation of leukocyte chemotaxis | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0061760 | antifungal innate immune response | 4 | 7 | 1.00e+00 |
| GO:BP | GO:0051969 | regulation of transmission of nerve impulse | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0003081 | regulation of systemic arterial blood pressure by renin-angiotensin | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0030299 | intestinal cholesterol absorption | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0030300 | regulation of intestinal cholesterol absorption | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0045906 | negative regulation of vasoconstriction | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0032490 | detection of molecule of bacterial origin | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition | 1 | 17 | 1.00e+00 |
| GO:BP | GO:0016071 | mRNA metabolic process | 3 | 665 | 1.00e+00 |
| GO:BP | GO:0042359 | vitamin D metabolic process | 3 | 9 | 1.00e+00 |
| GO:BP | GO:0045838 | positive regulation of membrane potential | 2 | 12 | 1.00e+00 |
| GO:BP | GO:0050789 | regulation of biological process | 6 | 8061 | 1.00e+00 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 2 | 122 | 1.00e+00 |
| GO:BP | GO:0070486 | leukocyte aggregation | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0007338 | single fertilization | 3 | 70 | 1.00e+00 |
| GO:BP | GO:0070857 | regulation of bile acid biosynthetic process | 2 | 7 | 1.00e+00 |
| GO:BP | GO:0050794 | regulation of cellular process | 1 | 7552 | 1.00e+00 |
| GO:BP | GO:0019081 | viral translation | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0006108 | malate metabolic process | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0002643 | regulation of tolerance induction | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0035739 | CD4-positive, alpha-beta T cell proliferation | 1 | 12 | 1.00e+00 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 4 | 19 | 1.00e+00 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 7 | 21 | 1.00e+00 |
| GO:BP | GO:0006821 | chloride transport | 5 | 58 | 1.00e+00 |
| GO:BP | GO:0007339 | binding of sperm to zona pellucida | 2 | 20 | 1.00e+00 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 5 | 18 | 1.00e+00 |
| GO:BP | GO:0035696 | monocyte extravasation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0006612 | protein targeting to membrane | 1 | 118 | 1.00e+00 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 1.00e+00 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 2 | 35 | 1.00e+00 |
| GO:BP | GO:0071715 | icosanoid transport | 2 | 24 | 1.00e+00 |
| GO:BP | GO:0002837 | regulation of immune response to tumor cell | 4 | 10 | 1.00e+00 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002836 | positive regulation of response to tumor cell | 1 | 7 | 1.00e+00 |
| GO:BP | GO:2000846 | regulation of corticosteroid hormone secretion | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0002218 | activation of innate immune response | 2 | 142 | 1.00e+00 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 1 | 16 | 1.00e+00 |
| GO:BP | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 1.00e+00 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0002834 | regulation of response to tumor cell | 4 | 11 | 1.00e+00 |
| GO:BP | GO:0060124 | positive regulation of growth hormone secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 1 | 28 | 1.00e+00 |
| GO:BP | GO:0010876 | lipid localization | 8 | 310 | 1.00e+00 |
| GO:BP | GO:0042953 | lipoprotein transport | 1 | 14 | 1.00e+00 |
| GO:BP | GO:1903817 | negative regulation of voltage-gated potassium channel activity | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0060123 | regulation of growth hormone secretion | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 2 | 192 | 1.00e+00 |
| GO:BP | GO:0051918 | negative regulation of fibrinolysis | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 1 | 66 | 1.00e+00 |
| GO:BP | GO:0022030 | telencephalon glial cell migration | 3 | 22 | 1.00e+00 |
| GO:BP | GO:0022029 | telencephalon cell migration | 7 | 44 | 1.00e+00 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0070207 | protein homotrimerization | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0032715 | negative regulation of interleukin-6 production | 1 | 24 | 1.00e+00 |
| GO:BP | GO:1904478 | regulation of intestinal absorption | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0070206 | protein trimerization | 3 | 12 | 1.00e+00 |
| GO:BP | GO:0006956 | complement activation | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0021801 | cerebral cortex radial glia-guided migration | 3 | 22 | 1.00e+00 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 6 | 211 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.00e+00 |
| GO:BP | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 1 | 40 | 1.00e+00 |
| GO:BP | GO:0002922 | positive regulation of humoral immune response | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.00e+00 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0045291 | mRNA trans splicing, SL addition | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0098543 | detection of other organism | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0006706 | steroid catabolic process | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 1.00e+00 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 1 | 74 | 1.00e+00 |
| GO:BP | GO:0008228 | opsonization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0002507 | tolerance induction | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0031639 | plasminogen activation | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 2 | 16 | 1.00e+00 |
| GO:BP | GO:0001991 | regulation of systemic arterial blood pressure by circulatory renin-angiotensin | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 1.00e+00 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 2 | 24 | 1.00e+00 |
| GO:BP | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 1.00e+00 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0098883 | synapse pruning | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0051608 | histamine transport | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 7 | 19 | 1.00e+00 |
| GO:BP | GO:0043320 | natural killer cell degranulation | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 2 | 22 | 1.00e+00 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 4 | 28 | 1.00e+00 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 2 | 20 | 1.00e+00 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 250 | 1.00e+00 |
| GO:BP | GO:0042730 | fibrinolysis | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0035036 | sperm-egg recognition | 2 | 22 | 1.00e+00 |
| GO:BP | GO:0021830 | interneuron migration from the subpallium to the cortex | 1 | 3 | 1.00e+00 |
| GO:BP | GO:1904729 | regulation of intestinal lipid absorption | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0070269 | pyroptosis | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0021843 | substrate-independent telencephalic tangential interneuron migration | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 15 | 1.00e+00 |
| GO:BP | GO:0015849 | organic acid transport | 4 | 217 | 1.00e+00 |
| GO:BP | GO:0006376 | mRNA splice site selection | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0021885 | forebrain cell migration | 7 | 46 | 1.00e+00 |
| GO:BP | GO:0006605 | protein targeting | 4 | 307 | 1.00e+00 |
| GO:BP | GO:0021826 | substrate-independent telencephalic tangential migration | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0010918 | positive regulation of mitochondrial membrane potential | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0032310 | prostaglandin secretion | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0032309 | icosanoid secretion | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0032308 | positive regulation of prostaglandin secretion | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0032306 | regulation of prostaglandin secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0045956 | positive regulation of calcium ion-dependent exocytosis | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0042178 | xenobiotic catabolic process | 3 | 12 | 1.00e+00 |
| GO:BP | GO:0032026 | response to magnesium ion | 4 | 10 | 1.00e+00 |
| GO:BP | GO:0006910 | phagocytosis, recognition | 1 | 12 | 1.00e+00 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 1 | 39 | 1.00e+00 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 1.00e+00 |
| GO:BP | GO:0060579 | ventral spinal cord interneuron fate commitment | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0042711 | maternal behavior | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0060581 | cell fate commitment involved in pattern specification | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0098856 | intestinal lipid absorption | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 7 | 337 | 1.00e+00 |
| GO:BP | GO:0002418 | immune response to tumor cell | 4 | 12 | 1.00e+00 |
| GO:BP | GO:0010755 | regulation of plasminogen activation | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0000395 | mRNA 5’-splice site recognition | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 13 | 55 | 1.00e+00 |
| GO:BP | GO:0048664 | neuron fate determination | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0034384 | high-density lipoprotein particle clearance | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 2 | 23 | 1.00e+00 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 2 | 14 | 1.00e+00 |
| GO:BP | GO:0031349 | positive regulation of defense response | 4 | 251 | 1.00e+00 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 7 | 21 | 1.00e+00 |
| GO:BP | GO:0031341 | regulation of cell killing | 5 | 36 | 1.00e+00 |
| GO:BP | GO:0045088 | regulation of innate immune response | 4 | 232 | 1.00e+00 |
| GO:BP | GO:0050907 | detection of chemical stimulus involved in sensory perception | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 2 | 182 | 1.00e+00 |
| GO:BP | GO:2000318 | positive regulation of T-helper 17 type immune response | 3 | 7 | 1.00e+00 |
| GO:BP | GO:1901616 | organic hydroxy compound catabolic process | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0045109 | intermediate filament organization | 7 | 21 | 1.00e+00 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 5 | 30 | 1.00e+00 |
| GO:BP | GO:0002031 | G protein-coupled receptor internalization | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0048663 | neuron fate commitment | 1 | 29 | 1.00e+00 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 1.00e+00 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 1.00e+00 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0019373 | epoxygenase P450 pathway | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0034374 | low-density lipoprotein particle remodeling | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0045059 | positive thymic T cell selection | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 1 | 57 | 1.00e+00 |
| GO:BP | GO:0017121 | plasma membrane phospholipid scrambling | 1 | 13 | 1.00e+00 |
| GO:BP | GO:0045061 | thymic T cell selection | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0060272 | embryonic skeletal joint morphogenesis | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0034379 | very-low-density lipoprotein particle assembly | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0043300 | regulation of leukocyte degranulation | 10 | 27 | 1.00e+00 |
| GO:BP | GO:0048149 | behavioral response to ethanol | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 1 | 94 | 1.00e+00 |
| GO:BP | GO:0046942 | carboxylic acid transport | 4 | 216 | 1.00e+00 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 1 | 6 | 1.00e+00 |
| GO:BP | GO:2000321 | positive regulation of T-helper 17 cell differentiation | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 1.00e+00 |
| GO:BP | GO:0060746 | parental behavior | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0080154 | regulation of fertilization | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0002003 | angiotensin maturation | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0070162 | adiponectin secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 4 | 19 | 1.00e+00 |
| GO:BP | GO:0070163 | regulation of adiponectin secretion | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0002002 | regulation of angiotensin levels in blood | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0010757 | negative regulation of plasminogen activation | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0021794 | thalamus development | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0006707 | cholesterol catabolic process | 1 | 8 | 1.00e+00 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 5 | 51 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 2 | 11 | 1.00e+00 |
| GO:BP | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0043589 | skin morphogenesis | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0002717 | positive regulation of natural killer cell mediated immunity | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 1 | 4 | 1.00e+00 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 1.00e+00 |
| GO:BP | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0000353 | formation of quadruple SL/U4/U5/U6 snRNP | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0051668 | localization within membrane | 3 | 605 | 1.00e+00 |
| GO:BP | GO:0007638 | mechanosensory behavior | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 3 | 19 | 1.00e+00 |
| GO:BP | GO:0150078 | positive regulation of neuroinflammatory response | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0009111 | vitamin catabolic process | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0006013 | mannose metabolic process | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0000365 | mRNA trans splicing, via spliceosome | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0002016 | regulation of blood volume by renin-angiotensin | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| KEGG | KEGG:04115 | p53 signaling pathway | 10 | 65 | 5.66e-06 |
| KEGG | KEGG:03410 | Base excision repair | 26 | 33 | 1.87e-05 |
| KEGG | KEGG:04110 | Cell cycle | 72 | 151 | 6.40e-05 |
| KEGG | KEGG:03030 | DNA replication | 13 | 35 | 4.26e-04 |
| KEGG | KEGG:03430 | Mismatch repair | 17 | 22 | 1.78e-03 |
| KEGG | KEGG:03440 | Homologous recombination | 11 | 39 | 6.98e-03 |
| KEGG | KEGG:04217 | Necroptosis | 7 | 106 | 2.64e-02 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 2 | 61 | 2.64e-02 |
| KEGG | KEGG:00230 | Purine metabolism | 4 | 97 | 3.73e-02 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 2 | 110 | 3.73e-02 |
| KEGG | KEGG:05034 | Alcoholism | 2 | 122 | 3.73e-02 |
| KEGG | KEGG:01524 | Platinum drug resistance | 26 | 65 | 3.84e-02 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 15 | 48 | 6.53e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 6.83e-02 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 9 | 43 | 7.13e-02 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 7.55e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 28 | 84 | 7.55e-02 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 3 | 69 | 7.55e-02 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 6 | 86 | 7.55e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 4 | 75 | 7.55e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 1 | 57 | 7.55e-02 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 2 | 20 | 7.55e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 7.55e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 7.55e-02 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 7.55e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 7.55e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 8.80e-02 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 1.10e-01 |
| KEGG | KEGG:04210 | Apoptosis | 39 | 118 | 1.16e-01 |
| KEGG | KEGG:05224 | Breast cancer | 1 | 117 | 1.16e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 1.16e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 1.16e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 5 | 24 | 1.16e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 1.19e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 6 | 145 | 1.19e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 1 | 145 | 1.19e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 1.34e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 1.35e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 3 | 10 | 2.06e-01 |
| KEGG | KEGG:03018 | RNA degradation | 17 | 74 | 2.19e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 25 | 56 | 2.29e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 2 | 66 | 2.56e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 1 | 16 | 2.61e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 6 | 106 | 2.66e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 1 | 409 | 3.04e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 1 | 84 | 3.21e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 11 | 35 | 3.46e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 7 | 42 | 3.46e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 3 | 91 | 3.46e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 1 | 29 | 3.46e-01 |
| KEGG | KEGG:04530 | Tight junction | 4 | 137 | 3.46e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 2 | 91 | 3.46e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 4 | 157 | 3.46e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 20 | 70 | 3.46e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 3 | 8 | 3.51e-01 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 2 | 6 | 3.51e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 1 | 34 | 3.60e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 7 | 46 | 3.78e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 8 | 109 | 4.10e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 4.18e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 2 | 73 | 4.18e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 10 | 30 | 4.18e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 2 | 78 | 4.18e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 8 | 58 | 4.18e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 9 | 135 | 4.18e-01 |
| KEGG | KEGG:04540 | Gap junction | 8 | 73 | 4.27e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 7 | 272 | 4.27e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 4 | 55 | 4.27e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 4 | 19 | 4.40e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 7 | 107 | 4.59e-01 |
| KEGG | KEGG:04142 | Lysosome | 1 | 118 | 4.59e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 5 | 18 | 4.59e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 14 | 56 | 4.59e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 55 | 99 | 4.59e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 5 | 180 | 4.59e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 4.59e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 2 | 44 | 4.59e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 8 | 44 | 4.59e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 4 | 108 | 4.59e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 69 | 134 | 4.69e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 5 | 19 | 4.69e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 7 | 56 | 4.96e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 4 | 172 | 4.96e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 2 | 51 | 4.96e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 5.08e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 5.21e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 1 | 21 | 5.33e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 18 | 30 | 5.33e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 2 | 74 | 5.38e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 4 | 40 | 5.38e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 13 | 31 | 5.38e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 22 | 87 | 5.38e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 34 | 133 | 5.69e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 3 | 49 | 5.81e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 5.82e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 5.83e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 2 | 44 | 5.83e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 6 | 63 | 5.83e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 12 | 77 | 5.83e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 2 | 55 | 5.83e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 8 | 82 | 5.83e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 3 | 26 | 5.83e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 4 | 35 | 5.83e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 3 | 16 | 5.83e-01 |
| KEGG | KEGG:05164 | Influenza A | 3 | 103 | 5.83e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 5 | 46 | 5.83e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 10 | 114 | 5.83e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 4 | 35 | 6.03e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 1 | 41 | 6.03e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 2 | 214 | 6.03e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 7 | 97 | 6.03e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 6.11e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 11 | 17 | 6.11e-01 |
| KEGG | KEGG:05016 | Huntington disease | 9 | 256 | 6.11e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 24 | 85 | 6.14e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 12 | 6.14e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 32 | 95 | 6.14e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 3 | 46 | 6.23e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 8 | 1146 | 6.24e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 24 | 74 | 6.24e-01 |
| KEGG | KEGG:04520 | Adherens junction | 18 | 67 | 6.26e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 6 | 128 | 6.26e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 42 | 121 | 6.26e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 5 | 33 | 6.26e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 11 | 55 | 6.43e-01 |
| KEGG | KEGG:05142 | Chagas disease | 5 | 75 | 6.67e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 2 | 103 | 6.67e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 6.67e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 13 | 25 | 6.77e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 1 | 20 | 6.77e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 1 | 8 | 6.77e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 2 | 112 | 6.77e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 1 | 63 | 6.77e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 4 | 82 | 6.77e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 6.77e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 14 | 115 | 6.77e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 10 | 303 | 6.77e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 5 | 76 | 6.77e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 2 | 6 | 6.77e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 4 | 10 | 6.80e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 4 | 79 | 6.85e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 5 | 12 | 6.85e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 6.85e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 2 | 22 | 6.85e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 11 | 69 | 6.85e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 20 | 76 | 6.85e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 4 | 124 | 6.85e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 6.85e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 1 | 43 | 6.85e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 1 | 37 | 6.85e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 10 | 86 | 6.85e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 6.85e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 1 | 67 | 6.85e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 3 | 4 | 6.85e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 3 | 53 | 6.94e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 7 | 138 | 7.01e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 23 | 72 | 7.01e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 7.01e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 1 | 78 | 7.01e-01 |
| KEGG | KEGG:05162 | Measles | 2 | 98 | 7.06e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 1 | 49 | 7.09e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 7 | 38 | 7.18e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 1 | 83 | 7.19e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 12 | 385 | 7.19e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 1 | 61 | 7.19e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 2 | 15 | 7.19e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 1 | 37 | 7.22e-01 |
| KEGG | KEGG:04976 | Bile secretion | 1 | 42 | 7.23e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 8 | 170 | 7.23e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 1 | 17 | 7.24e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 2 | 151 | 7.24e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 7.24e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 3 | 15 | 7.24e-01 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 1 | 5 | 7.24e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 1 | 51 | 7.24e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 7.24e-01 |
| KEGG | KEGG:04146 | Peroxisome | 30 | 68 | 7.24e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 1 | 22 | 7.24e-01 |
| KEGG | KEGG:04144 | Endocytosis | 20 | 232 | 7.24e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 7.24e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 9 | 165 | 7.24e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 14 | 315 | 7.24e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 20 | 78 | 7.24e-01 |
| KEGG | KEGG:05131 | Shigellosis | 22 | 210 | 7.24e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 3 | 83 | 7.24e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 26 | 98 | 7.24e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 11 | 121 | 7.24e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 7 | 56 | 7.24e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 3 | 28 | 7.24e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 8 | 150 | 7.24e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 4 | 104 | 7.24e-01 |
| KEGG | KEGG:04611 | Platelet activation | 7 | 89 | 7.24e-01 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 7.24e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 2 | 52 | 7.24e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 2 | 106 | 7.24e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 10 | 41 | 7.24e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 7.24e-01 |
| KEGG | KEGG:00000 | KEGG root term | 2 | 5558 | 7.24e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 5 | 133 | 7.24e-01 |
| KEGG | KEGG:02010 | ABC transporters | 2 | 29 | 7.24e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 3 | 22 | 7.24e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 6 | 164 | 7.24e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 3 | 16 | 7.24e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 5 | 56 | 7.24e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 2 | 115 | 7.24e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 1 | 58 | 7.24e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 2 | 38 | 7.28e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 7.36e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 4 | 70 | 7.36e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 1 | 56 | 7.36e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 10 | 7.36e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 1 | 22 | 7.36e-01 |
| KEGG | KEGG:04145 | Phagosome | 10 | 99 | 7.36e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 20 | 76 | 7.42e-01 |
| KEGG | KEGG:04924 | Renin secretion | 2 | 50 | 7.42e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 1 | 28 | 7.42e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 7.46e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 3 | 23 | 7.46e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 9 | 20 | 7.48e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 1 | 22 | 7.58e-01 |
| KEGG | KEGG:04730 | Long-term depression | 5 | 42 | 7.59e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 2 | 145 | 7.62e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 7.73e-01 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 7.73e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 3 | 63 | 7.73e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 7 | 50 | 7.73e-01 |
| KEGG | KEGG:05133 | Pertussis | 2 | 45 | 7.73e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 14 | 48 | 7.85e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 3 | 24 | 7.90e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 13 | 32 | 7.90e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 15 | 87 | 7.90e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 1 | 14 | 7.90e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 4 | 13 | 7.90e-01 |
| KEGG | KEGG:03050 | Proteasome | 12 | 42 | 7.90e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 10 | 113 | 7.90e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 7.90e-01 |
| KEGG | KEGG:03040 | Spliceosome | 40 | 132 | 7.97e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 1 | 12 | 7.97e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 1 | 115 | 7.97e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 2 | 14 | 7.97e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 1 | 131 | 7.97e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 2 | 27 | 8.08e-01 |
| KEGG | KEGG:00232 | Caffeine metabolism | 1 | 1 | 8.08e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 1 | 110 | 8.08e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 6 | 25 | 8.09e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 8.09e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 8.09e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 8.19e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 5 | 65 | 8.21e-01 |
| KEGG | KEGG:00780 | Biotin metabolism | 1 | 3 | 8.25e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 5 | 176 | 8.25e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 1 | 98 | 8.46e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 3 | 112 | 8.46e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 1 | 106 | 8.46e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 1 | 69 | 8.48e-01 |
| KEGG | KEGG:03060 | Protein export | 5 | 23 | 8.48e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 3 | 99 | 8.60e-01 |
| KEGG | KEGG:04744 | Phototransduction | 1 | 12 | 8.66e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 8.66e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 2 | 28 | 8.66e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 4 | 103 | 8.66e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 3 | 49 | 8.66e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 22 | 84 | 8.71e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 11 | 49 | 8.75e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 11 | 24 | 8.76e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 8.94e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 6 | 94 | 9.11e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 9.11e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 9.11e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 3 | 38 | 9.11e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 1 | 11 | 9.11e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 4 | 32 | 9.11e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 6 | 14 | 9.11e-01 |
| KEGG | KEGG:05020 | Prion disease | 5 | 220 | 9.11e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 4 | 53 | 9.11e-01 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 1 | 12 | 9.11e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 2 | 161 | 9.17e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 3 | 9 | 9.18e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 1 | 40 | 9.18e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 1 | 3 | 9.18e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 1 | 7 | 9.18e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 9 | 50 | 9.19e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 1 | 77 | 9.19e-01 |
| KEGG | KEGG:03265 | Virion - Lyssavirus | 1 | 3 | 9.19e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 1 | 88 | 9.19e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 8 | 110 | 9.19e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 1 | 44 | 9.29e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 2 | 33 | 9.31e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 23 | 49 | 9.31e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 7 | 17 | 9.31e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 9.31e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 9.31e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 2 | 104 | 9.31e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 3 | 415 | 9.39e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 11 | 240 | 9.39e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 3 | 55 | 9.39e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 4 | 31 | 9.39e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 3 | 10 | 9.40e-01 |
| KEGG | KEGG:00511 | Other glycan degradation | 2 | 16 | 9.48e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 4 | 226 | 9.51e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 5 | 74 | 9.51e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 9.51e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 1 | 28 | 9.51e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 2 | 108 | 9.51e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 9.51e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 1 | 25 | 9.51e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 5 | 179 | 9.51e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 1 | 12 | 9.51e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 10 | 64 | 9.51e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 8 | 26 | 9.51e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 8 | 171 | 9.55e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 9.59e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 6 | 86 | 9.69e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 1 | 24 | 9.70e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 9 | 97 | 9.73e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 18 | 142 | 9.73e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 2 | 28 | 9.93e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 2 | 44 | 9.93e-01 |
| KEGG | KEGG:00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 1 | 2 | 9.95e-01 |
| KEGG | KEGG:04742 | Taste transduction | 2 | 22 | 9.95e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 2 | 33 | 9.95e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 3 | 89 | 1.00e+00 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 2 | 23 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 3 | 186 | 1.00e+00 |
| KEGG | KEGG:00531 | Glycosaminoglycan degradation | 1 | 16 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 13 | 36 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 1 | 19 | 1.00e+00 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 1.00e+00 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 1 | 23 | 1.00e+00 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 5 | 1.00e+00 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 1 | 13 | 1.00e+00 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 5 | 60 | 1.00e+00 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 3 | 43 | 1.00e+00 |
| KEGG | KEGG:05310 | Asthma | 1 | 3 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 2 | 40 | 1.00e+00 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 6 | 41 | 1.00e+00 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 1 | 19 | 1.00e+00 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 3 | 12 | 1.00e+00 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 1 | 31 | 1.00e+00 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 1 | 9 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 5 | 169 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 1 | 31 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 3 | 98 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 4 | 127 | 1.00e+00 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:05134 | Legionellosis | 4 | 39 | 1.00e+00 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 2 | 9 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:05146 | Amoebiasis | 4 | 64 | 1.00e+00 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 1 | 21 | 1.00e+00 |
##epi 24 specifi n = 533
epsp24 <- list24totvenn24$..values..[[12]]
##ep 3 hour above
#
# EPspgostres24sp <- gost(query = epsp24,
# organism = "hsapiens", significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EPspgostres,"data/DEG-GO/EPspgostres.RDS")
# saveRDS(EPgostres24sp,"data/DEG-GO/EPgostres24sp.RDS")
EPspgostres <- readRDS("data/DEG-GO/EPspgostres.RDS")
##no enrichment foundmtsp24 <- list24totvenn24$..values..[[8]]
# MTgostres24sp <- gost(query = mtsp24,
# organism = "hsapiens", significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
#
# saveRDS(MTgostres24sp,"data/DEG-GO/MTgostres24sp.RDS")
MTgostres24sp <- readRDS("data/DEG-GO/MTgostres24sp.RDS")
MTsptable124 <- MTgostres24sp$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
MTsptable124 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle("MTX specific 24 hour top GO:BP terms") +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)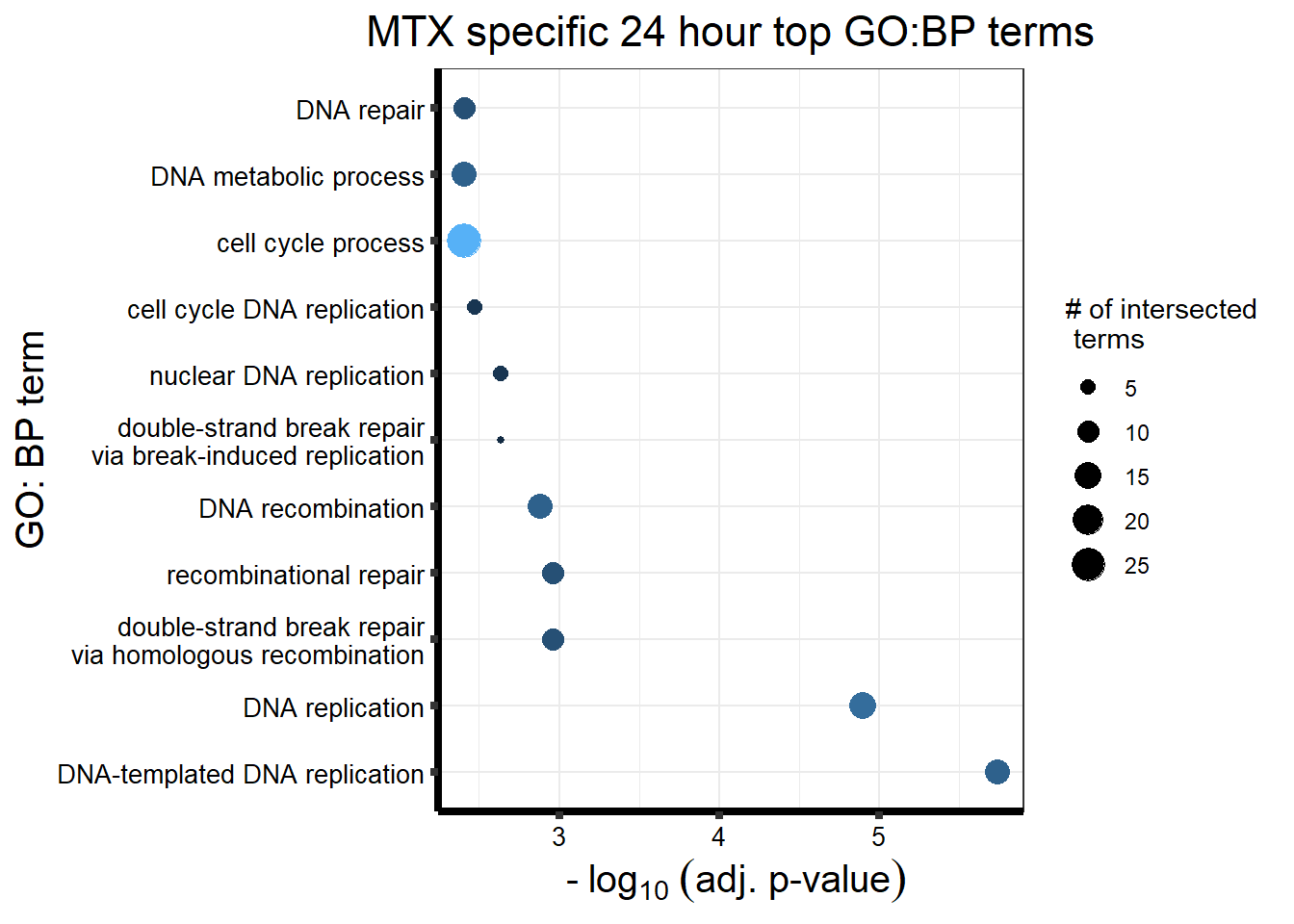
MTX-specific 24 hour genes show an enrichment for DNA repair. (n=188)
MTX DEGs (adj. p value < 0.05)
3 hour = 75 24 hour = 1115
# MTgostresstar <- gost(query = sigVMT3$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# numeric_ns="ENTREZGENE",
# user_threshold = 1,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("KEGG"))
# saveRDS(MTgostres,"data/DEG-GO/MTgostre.RDS")
MTgostres <- readRDS("data/DEG-GO/MTgostre.RDS")
# MTp <- gostplot(MTgostres, capped = FALSE, interactive = TRUE)
# MTp
MTtable1 <- MTgostres$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
MTtable1 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('MTX 3 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)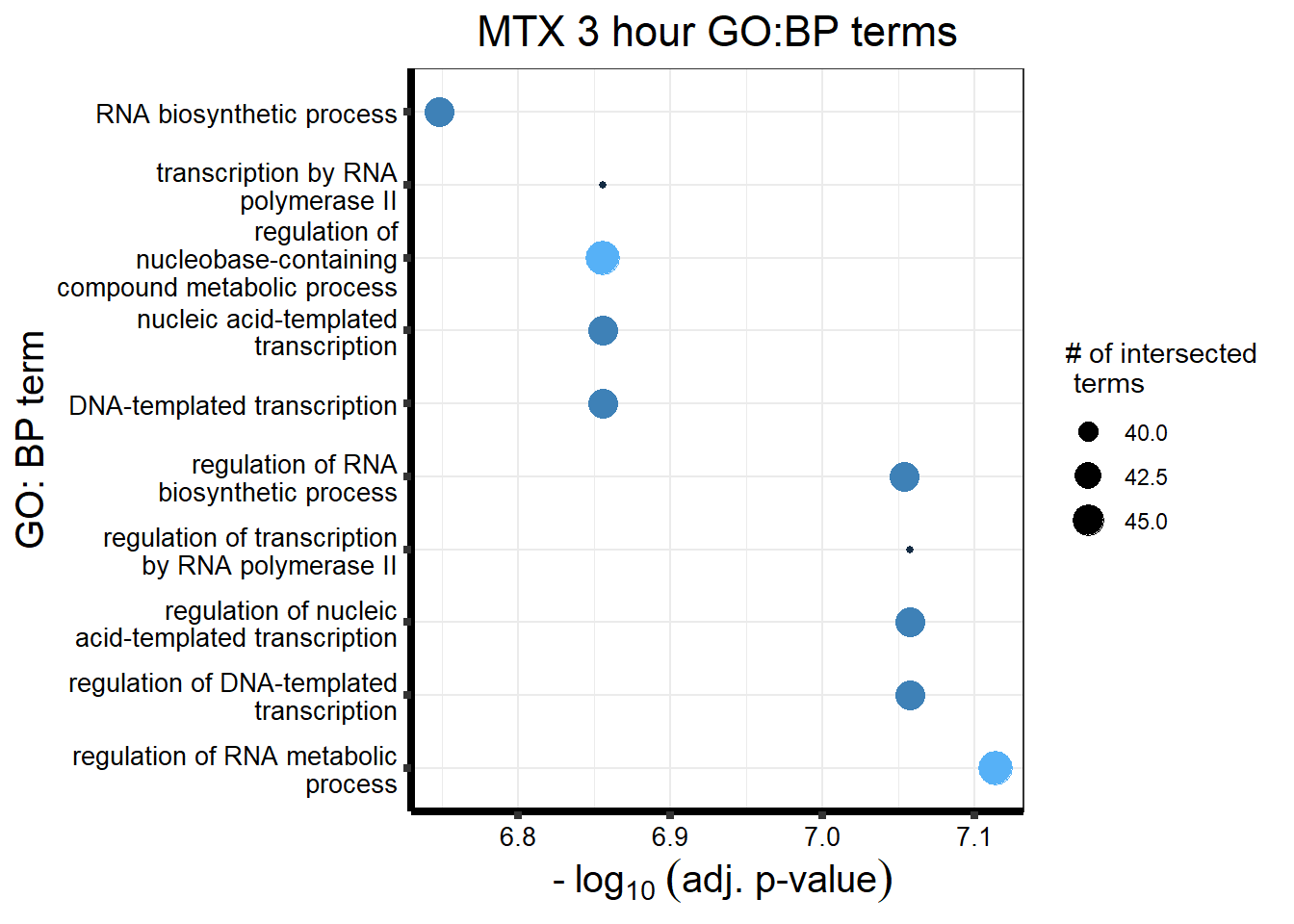
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
MTtable1 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "MTX 3 hour table of enrichment") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 47 | 2845 | 7.69e-08 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 38 | 1914 | 8.75e-08 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 44 | 2574 | 8.75e-08 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 44 | 2576 | 8.75e-08 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 44 | 2593 | 8.83e-08 |
| GO:BP | GO:0006351 | DNA-templated transcription | 44 | 2683 | 1.39e-07 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 44 | 2684 | 1.39e-07 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 38 | 1995 | 1.39e-07 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 47 | 3080 | 1.39e-07 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 44 | 2714 | 1.79e-07 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 45 | 3012 | 8.06e-07 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 45 | 3080 | 1.83e-06 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 21 | 740 | 2.54e-06 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 45 | 3169 | 3.98e-06 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 53 | 4292 | 5.04e-06 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 44 | 3070 | 5.04e-06 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 25 | 1119 | 5.16e-06 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 44 | 3132 | 7.93e-06 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 44 | 3133 | 8.19e-06 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 53 | 4417 | 1.35e-05 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 23 | 1015 | 1.52e-05 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 23 | 1017 | 1.52e-05 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 23 | 1026 | 1.69e-05 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 25 | 1209 | 1.69e-05 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 44 | 3237 | 1.91e-05 |
| GO:BP | GO:0010468 | regulation of gene expression | 47 | 3580 | 3.21e-05 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 50 | 4174 | 4.66e-05 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 24 | 1214 | 5.15e-05 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 24 | 1239 | 7.83e-05 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 46 | 3750 | 8.90e-05 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 6 | 67 | 1.08e-04 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 24 | 1280 | 1.24e-04 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 45 | 3730 | 1.80e-04 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 29 | 1849 | 2.74e-04 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 27 | 1735 | 6.00e-04 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 2 | 11 | 7.64e-04 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 6 | 103 | 9.43e-04 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 44 | 4245 | 9.65e-04 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 8 | 198 | 1.01e-03 |
| GO:BP | GO:0003279 | cardiac septum development | 6 | 101 | 1.01e-03 |
| GO:BP | GO:0014706 | striated muscle tissue development | 8 | 210 | 1.62e-03 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 26 | 4617 | 1.65e-03 |
| GO:BP | GO:0003281 | ventricular septum development | 5 | 69 | 1.84e-03 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 6 | 109 | 2.08e-03 |
| GO:BP | GO:0060419 | heart growth | 5 | 72 | 2.61e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 8 | 216 | 2.62e-03 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 2 | 23 | 2.93e-03 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 6 | 129 | 3.16e-03 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 20 | 4048 | 3.27e-03 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 3.27e-03 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 22 | 1441 | 3.42e-03 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 44 | 4539 | 4.72e-03 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 2 | 25 | 4.82e-03 |
| GO:BP | GO:0003162 | atrioventricular node development | 2 | 8 | 4.98e-03 |
| GO:BP | GO:0048589 | developmental growth | 12 | 522 | 5.35e-03 |
| GO:BP | GO:0035265 | organ growth | 6 | 131 | 5.54e-03 |
| GO:BP | GO:0003171 | atrioventricular valve development | 2 | 27 | 5.65e-03 |
| GO:BP | GO:0009058 | biosynthetic process | 44 | 4590 | 5.93e-03 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 3 | 23 | 5.93e-03 |
| GO:BP | GO:0019222 | regulation of metabolic process | 26 | 5017 | 6.01e-03 |
| GO:BP | GO:0016070 | RNA metabolic process | 19 | 3600 | 6.35e-03 |
| GO:BP | GO:0007507 | heart development | 11 | 506 | 7.47e-03 |
| GO:BP | GO:0060420 | regulation of heart growth | 4 | 50 | 7.52e-03 |
| GO:BP | GO:0003205 | cardiac chamber development | 6 | 149 | 7.67e-03 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 20 | 1315 | 7.97e-03 |
| GO:BP | GO:0060537 | muscle tissue development | 9 | 343 | 7.97e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 20 | 1315 | 7.97e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 20 | 1322 | 8.20e-03 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 20 | 4455 | 8.22e-03 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 2 | 33 | 8.40e-03 |
| GO:BP | GO:0048638 | regulation of developmental growth | 8 | 258 | 8.40e-03 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 10 | 2135 | 8.96e-03 |
| GO:BP | GO:0040007 | growth | 14 | 742 | 8.96e-03 |
| GO:BP | GO:0001657 | ureteric bud development | 2 | 75 | 9.42e-03 |
| GO:BP | GO:0072164 | mesonephric tubule development | 2 | 76 | 9.42e-03 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 2 | 76 | 9.42e-03 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 4 | 65 | 9.45e-03 |
| GO:BP | GO:0003176 | aortic valve development | 2 | 37 | 9.51e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 4 | 68 | 9.51e-03 |
| GO:BP | GO:0036302 | atrioventricular canal development | 2 | 12 | 9.51e-03 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 22 | 1602 | 9.51e-03 |
| GO:BP | GO:0001823 | mesonephros development | 2 | 77 | 9.51e-03 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 20 | 4570 | 1.01e-02 |
| GO:BP | GO:1905314 | semi-lunar valve development | 2 | 41 | 1.09e-02 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 3 | 26 | 1.11e-02 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 20 | 4596 | 1.11e-02 |
| GO:BP | GO:0007352 | zygotic specification of dorsal/ventral axis | 1 | 2 | 1.11e-02 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 1.11e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 4 | 62 | 1.13e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 5 | 114 | 1.16e-02 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 10 | 1.16e-02 |
| GO:BP | GO:1903676 | positive regulation of cap-dependent translational initiation | 1 | 1 | 1.17e-02 |
| GO:BP | GO:0009725 | response to hormone | 4 | 642 | 1.17e-02 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 3 | 27 | 1.17e-02 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 10 | 2293 | 1.17e-02 |
| GO:BP | GO:1903674 | regulation of cap-dependent translational initiation | 1 | 1 | 1.17e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 4 | 71 | 1.17e-02 |
| GO:BP | GO:0060976 | coronary vasculature development | 3 | 44 | 1.35e-02 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 11 | 482 | 1.46e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 4 | 77 | 1.47e-02 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 2 | 55 | 1.48e-02 |
| GO:BP | GO:0072073 | kidney epithelium development | 2 | 112 | 1.53e-02 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 20 | 4747 | 1.57e-02 |
| GO:BP | GO:0090721 | primary adaptive immune response involving T cells and B cells | 1 | 1 | 1.68e-02 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 13 | 665 | 1.68e-02 |
| GO:BP | GO:0090720 | primary adaptive immune response | 1 | 1 | 1.68e-02 |
| GO:BP | GO:0046620 | regulation of organ growth | 4 | 71 | 1.87e-02 |
| GO:BP | GO:0003170 | heart valve development | 2 | 65 | 2.00e-02 |
| GO:BP | GO:0034504 | protein localization to nucleus | 4 | 276 | 2.00e-02 |
| GO:BP | GO:0051170 | import into nucleus | 3 | 152 | 2.00e-02 |
| GO:BP | GO:0051169 | nuclear transport | 4 | 295 | 2.02e-02 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 4 | 295 | 2.02e-02 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 20 | 4946 | 2.04e-02 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 1 | 5 | 2.13e-02 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 3 | 33 | 2.25e-02 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 20 | 1507 | 2.26e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 3 | 40 | 2.28e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 2.30e-02 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 2 | 9 | 2.30e-02 |
| GO:BP | GO:0035264 | multicellular organism growth | 5 | 123 | 2.30e-02 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 2 | 17 | 2.31e-02 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 5 | 1107 | 2.31e-02 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 1 | 2 | 2.35e-02 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 1 | 6 | 2.35e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 15 | 937 | 2.39e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 3 | 84 | 2.48e-02 |
| GO:BP | GO:0010467 | gene expression | 21 | 4587 | 2.55e-02 |
| GO:BP | GO:1903679 | positive regulation of cap-independent translational initiation | 1 | 3 | 2.60e-02 |
| GO:BP | GO:0040008 | regulation of growth | 10 | 501 | 2.60e-02 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 2 | 76 | 2.60e-02 |
| GO:BP | GO:1903677 | regulation of cap-independent translational initiation | 1 | 3 | 2.60e-02 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 5 | 124 | 2.61e-02 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 3 | 43 | 2.81e-02 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 3 | 45 | 2.84e-02 |
| GO:BP | GO:0060840 | artery development | 4 | 83 | 2.85e-02 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 1 | 4 | 2.86e-02 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 18 | 2270 | 3.02e-02 |
| GO:BP | GO:0035054 | embryonic heart tube anterior/posterior pattern specification | 1 | 2 | 3.08e-02 |
| GO:BP | GO:0003285 | septum secundum development | 1 | 3 | 3.08e-02 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 2 | 13 | 3.08e-02 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 20 | 1564 | 3.08e-02 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 4 | 161 | 3.08e-02 |
| GO:BP | GO:0061061 | muscle structure development | 10 | 546 | 3.12e-02 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 1 | 4 | 3.13e-02 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 4 | 3.13e-02 |
| GO:BP | GO:1904003 | negative regulation of sebum secreting cell proliferation | 1 | 1 | 3.20e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 3 | 80 | 3.20e-02 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 3.26e-02 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 3.26e-02 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 2 | 14 | 3.36e-02 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 20 | 2412 | 3.36e-02 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 2 | 15 | 3.39e-02 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 6 | 233 | 3.43e-02 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 1 | 9 | 3.45e-02 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 1 | 11 | 3.45e-02 |
| GO:BP | GO:1904690 | positive regulation of cytoplasmic translational initiation | 1 | 5 | 3.52e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 2 | 33 | 3.52e-02 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 2 | 130 | 3.52e-02 |
| GO:BP | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 5 | 3.52e-02 |
| GO:BP | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress | 1 | 5 | 3.52e-02 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 3 | 56 | 3.52e-02 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 1 | 12 | 3.54e-02 |
| GO:BP | GO:0003161 | cardiac conduction system development | 2 | 33 | 3.54e-02 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 5 | 1278 | 3.54e-02 |
| GO:BP | GO:0003174 | mitral valve development | 1 | 12 | 3.54e-02 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 2 | 69 | 3.65e-02 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 20 | 1604 | 3.65e-02 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 2 | 69 | 3.65e-02 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 11 | 3625 | 3.66e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 3 | 52 | 3.67e-02 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 2 | 17 | 3.73e-02 |
| GO:BP | GO:0048565 | digestive tract development | 3 | 92 | 3.73e-02 |
| GO:BP | GO:0035904 | aorta development | 3 | 50 | 3.73e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 2 | 36 | 3.73e-02 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 2 | 36 | 3.76e-02 |
| GO:BP | GO:0001822 | kidney development | 2 | 244 | 3.76e-02 |
| GO:BP | GO:1904688 | regulation of cytoplasmic translational initiation | 1 | 6 | 3.76e-02 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 3.76e-02 |
| GO:BP | GO:0021542 | dentate gyrus development | 2 | 11 | 3.76e-02 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 2 | 34 | 3.76e-02 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 1 | 6 | 3.76e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 31 | 2880 | 3.76e-02 |
| GO:BP | GO:2000257 | regulation of protein activation cascade | 1 | 1 | 3.89e-02 |
| GO:BP | GO:0072001 | renal system development | 2 | 251 | 3.92e-02 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 3.92e-02 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 3.92e-02 |
| GO:BP | GO:0061865 | polarized secretion of basement membrane proteins in epithelium | 1 | 1 | 3.95e-02 |
| GO:BP | GO:1905070 | anterior visceral endoderm cell migration | 1 | 1 | 3.95e-02 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 3 | 54 | 3.95e-02 |
| GO:BP | GO:0061864 | basement membrane constituent secretion | 1 | 1 | 3.95e-02 |
| GO:BP | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.95e-02 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 2 | 76 | 3.95e-02 |
| GO:BP | GO:2000691 | negative regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.95e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 29 | 2646 | 3.96e-02 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 1 | 7 | 4.03e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 2 | 39 | 4.03e-02 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 1 | 7 | 4.03e-02 |
| GO:BP | GO:0055123 | digestive system development | 3 | 100 | 4.10e-02 |
| GO:BP | GO:0009749 | response to glucose | 3 | 137 | 4.12e-02 |
| GO:BP | GO:0021756 | striatum development | 2 | 17 | 4.20e-02 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 2 | 29 | 4.28e-02 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 4.31e-02 |
| GO:BP | GO:0009746 | response to hexose | 3 | 138 | 4.31e-02 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 1 | 6 | 4.31e-02 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 4.38e-02 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 4.38e-02 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 1 | 8 | 4.38e-02 |
| GO:BP | GO:0001949 | sebaceous gland cell differentiation | 1 | 1 | 4.42e-02 |
| GO:BP | GO:0032912 | negative regulation of transforming growth factor beta2 production | 1 | 2 | 4.42e-02 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 2 | 21 | 4.42e-02 |
| GO:BP | GO:0007493 | endodermal cell fate determination | 1 | 1 | 4.42e-02 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 1 | 19 | 4.42e-02 |
| GO:BP | GO:1904002 | regulation of sebum secreting cell proliferation | 1 | 1 | 4.42e-02 |
| GO:BP | GO:0072359 | circulatory system development | 13 | 884 | 4.53e-02 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 29 | 4.53e-02 |
| GO:BP | GO:0034284 | response to monosaccharide | 3 | 142 | 4.54e-02 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 13 | 748 | 4.58e-02 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 2 | 19 | 4.58e-02 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 2 | 19 | 4.58e-02 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 2 | 19 | 4.58e-02 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 4.58e-02 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 1 | 8 | 4.58e-02 |
| GO:BP | GO:0060290 | transdifferentiation | 1 | 2 | 4.58e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 3 | 123 | 4.60e-02 |
| GO:BP | GO:0043049 | otic placode formation | 1 | 1 | 4.67e-02 |
| GO:BP | GO:2000979 | positive regulation of forebrain neuron differentiation | 1 | 1 | 4.67e-02 |
| GO:BP | GO:0061114 | branching involved in pancreas morphogenesis | 1 | 1 | 4.67e-02 |
| GO:BP | GO:0071233 | cellular response to leucine | 1 | 10 | 4.67e-02 |
| GO:BP | GO:0002194 | hepatocyte cell migration | 1 | 1 | 4.67e-02 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 2 | 18 | 4.79e-02 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 2 | 20 | 4.79e-02 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 2 | 18 | 4.79e-02 |
| GO:BP | GO:0003177 | pulmonary valve development | 1 | 22 | 4.79e-02 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 4.81e-02 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 4.81e-02 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 10 | 4.81e-02 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 1 | 8 | 4.89e-02 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 3 | 72 | 4.89e-02 |
| GO:BP | GO:0071772 | response to BMP | 2 | 131 | 4.91e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 2 | 131 | 4.91e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 2 | 21 | 5.03e-02 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 2 | 34 | 5.04e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 5 | 206 | 5.06e-02 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 3 | 60 | 5.06e-02 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 5.06e-02 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 1 | 11 | 5.06e-02 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 1 | 13 | 5.11e-02 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 3 | 73 | 5.20e-02 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 15 | 5.20e-02 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 1 | 6 | 5.22e-02 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 2 | 21 | 5.22e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 4 | 1147 | 5.22e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 3 | 50 | 5.22e-02 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 5.22e-02 |
| GO:BP | GO:0110021 | cardiac muscle myoblast proliferation | 1 | 3 | 5.33e-02 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 5.33e-02 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 2 | 24 | 5.33e-02 |
| GO:BP | GO:1901390 | positive regulation of transforming growth factor beta activation | 1 | 3 | 5.33e-02 |
| GO:BP | GO:0021544 | subpallium development | 2 | 18 | 5.33e-02 |
| GO:BP | GO:0110024 | positive regulation of cardiac muscle myoblast proliferation | 1 | 3 | 5.33e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 3 | 74 | 5.33e-02 |
| GO:BP | GO:0110022 | regulation of cardiac muscle myoblast proliferation | 1 | 3 | 5.33e-02 |
| GO:BP | GO:0009743 | response to carbohydrate | 3 | 161 | 5.33e-02 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 4 | 1006 | 5.33e-02 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 4 | 100 | 5.35e-02 |
| GO:BP | GO:0048588 | developmental cell growth | 5 | 192 | 5.35e-02 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 5.35e-02 |
| GO:BP | GO:0007440 | foregut morphogenesis | 1 | 7 | 5.37e-02 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 5.37e-02 |
| GO:BP | GO:0043201 | response to leucine | 1 | 13 | 5.37e-02 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 5 | 5.37e-02 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 2 | 93 | 5.43e-02 |
| GO:BP | GO:0051892 | negative regulation of cardioblast differentiation | 1 | 1 | 5.55e-02 |
| GO:BP | GO:0061159 | establishment of bipolar cell polarity involved in cell morphogenesis | 1 | 1 | 5.55e-02 |
| GO:BP | GO:0043491 | protein kinase B signaling | 2 | 146 | 5.58e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 4 | 353 | 5.61e-02 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 2 | 234 | 5.72e-02 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 3 | 63 | 5.78e-02 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 1 | 18 | 5.79e-02 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 2 | 164 | 5.87e-02 |
| GO:BP | GO:0034405 | response to fluid shear stress | 1 | 31 | 5.87e-02 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 1 | 14 | 6.02e-02 |
| GO:BP | GO:0032868 | response to insulin | 2 | 215 | 6.06e-02 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 6.31e-02 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 1 | 16 | 6.31e-02 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 1 | 16 | 6.31e-02 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 6.31e-02 |
| GO:BP | GO:0023057 | negative regulation of signaling | 4 | 1076 | 6.34e-02 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 4 | 1074 | 6.34e-02 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 12 | 742 | 6.34e-02 |
| GO:BP | GO:0030279 | negative regulation of ossification | 1 | 25 | 6.34e-02 |
| GO:BP | GO:0060486 | club cell differentiation | 1 | 3 | 6.38e-02 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 4 | 158 | 6.38e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 6.38e-02 |
| GO:BP | GO:0032745 | positive regulation of interleukin-21 production | 1 | 1 | 6.41e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 2 | 56 | 6.41e-02 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 2 | 28 | 6.41e-02 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 1 | 16 | 6.41e-02 |
| GO:BP | GO:0032665 | regulation of interleukin-21 production | 1 | 1 | 6.41e-02 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 6.41e-02 |
| GO:BP | GO:0032625 | interleukin-21 production | 1 | 1 | 6.41e-02 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 4 | 377 | 6.41e-02 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 2 | 26 | 6.41e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 3 | 616 | 6.56e-02 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 6.57e-02 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 2 | 56 | 6.61e-02 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 1 | 17 | 6.63e-02 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 6.69e-02 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 1 | 26 | 6.69e-02 |
| GO:BP | GO:0061614 | miRNA transcription | 2 | 57 | 6.71e-02 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 6.75e-02 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 1 | 22 | 6.75e-02 |
| GO:BP | GO:0001743 | lens placode formation | 1 | 2 | 6.75e-02 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 6.75e-02 |
| GO:BP | GO:0046619 | lens placode formation involved in camera-type eye formation | 1 | 2 | 6.75e-02 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 6.75e-02 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 1 | 15 | 6.87e-02 |
| GO:BP | GO:0001568 | blood vessel development | 9 | 534 | 6.90e-02 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 2 | 60 | 6.90e-02 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 6.94e-02 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 2 | 47 | 6.94e-02 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 6.94e-02 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 6.94e-02 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 6.94e-02 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 6.94e-02 |
| GO:BP | GO:0080134 | regulation of response to stress | 4 | 1075 | 6.94e-02 |
| GO:BP | GO:0061171 | establishment of bipolar cell polarity | 1 | 1 | 6.94e-02 |
| GO:BP | GO:0070075 | tear secretion | 1 | 2 | 6.94e-02 |
| GO:BP | GO:0043627 | response to estrogen | 2 | 52 | 6.99e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 1 | 36 | 6.99e-02 |
| GO:BP | GO:0021537 | telencephalon development | 5 | 201 | 6.99e-02 |
| GO:BP | GO:0003188 | heart valve formation | 1 | 16 | 6.99e-02 |
| GO:BP | GO:0051602 | response to electrical stimulus | 2 | 26 | 6.99e-02 |
| GO:BP | GO:0042692 | muscle cell differentiation | 6 | 324 | 7.04e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 4 | 1088 | 7.04e-02 |
| GO:BP | GO:0042593 | glucose homeostasis | 2 | 168 | 7.08e-02 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 1 | 28 | 7.08e-02 |
| GO:BP | GO:0043414 | macromolecule methylation | 5 | 272 | 7.08e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 2 | 277 | 7.08e-02 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 7.08e-02 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 2 | 169 | 7.08e-02 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 7.08e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 4 | 1378 | 7.17e-02 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 3 | 816 | 7.17e-02 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 7.17e-02 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 1 | 24 | 7.25e-02 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 2 | 32 | 7.25e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 5 | 199 | 7.29e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 4 | 1111 | 7.29e-02 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 7.33e-02 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 7.33e-02 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 7.33e-02 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 2 | 31 | 7.35e-02 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 1 | 4 | 7.41e-02 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 7.41e-02 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 1 | 4 | 7.41e-02 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 7.41e-02 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 2 | 130 | 7.48e-02 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 7.65e-02 |
| GO:BP | GO:0060510 | type II pneumocyte differentiation | 1 | 5 | 7.65e-02 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 7.65e-02 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 1 | 3 | 7.65e-02 |
| GO:BP | GO:0070315 | G1 to G0 transition involved in cell differentiation | 1 | 4 | 7.65e-02 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 1 | 48 | 7.65e-02 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 2 | 67 | 7.65e-02 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 1 | 5 | 7.65e-02 |
| GO:BP | GO:0033189 | response to vitamin A | 1 | 12 | 7.65e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 2 | 300 | 7.67e-02 |
| GO:BP | GO:0055001 | muscle cell development | 4 | 165 | 7.70e-02 |
| GO:BP | GO:0001944 | vasculature development | 9 | 559 | 7.73e-02 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 1 | 29 | 7.82e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 230 | 7.92e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 3 | 796 | 7.93e-02 |
| GO:BP | GO:0098751 | bone cell development | 2 | 26 | 7.93e-02 |
| GO:BP | GO:0033554 | cellular response to stress | 6 | 1681 | 7.94e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 2 | 152 | 7.94e-02 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 1 | 50 | 7.95e-02 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 7.95e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 2 | 240 | 7.95e-02 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 1 | 24 | 7.97e-02 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 7.97e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 3 | 95 | 7.97e-02 |
| GO:BP | GO:0003230 | cardiac atrium development | 2 | 34 | 7.98e-02 |
| GO:BP | GO:0071698 | olfactory placode development | 1 | 2 | 8.02e-02 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 2 | 57 | 8.02e-02 |
| GO:BP | GO:1904252 | negative regulation of bile acid metabolic process | 1 | 1 | 8.02e-02 |
| GO:BP | GO:0055005 | ventricular cardiac myofibril assembly | 1 | 3 | 8.02e-02 |
| GO:BP | GO:0060838 | lymphatic endothelial cell fate commitment | 1 | 3 | 8.02e-02 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 1 | 10 | 8.02e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 8.02e-02 |
| GO:BP | GO:0070858 | negative regulation of bile acid biosynthetic process | 1 | 1 | 8.02e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 2 | 82 | 8.02e-02 |
| GO:BP | GO:0071699 | olfactory placode morphogenesis | 1 | 2 | 8.02e-02 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 2 | 51 | 8.02e-02 |
| GO:BP | GO:0070417 | cellular response to cold | 1 | 8 | 8.02e-02 |
| GO:BP | GO:0007492 | endoderm development | 2 | 65 | 8.02e-02 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 2 | 52 | 8.02e-02 |
| GO:BP | GO:0030910 | olfactory placode formation | 1 | 2 | 8.02e-02 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 2 | 42 | 8.07e-02 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 1 | 55 | 8.10e-02 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 1 | 27 | 8.13e-02 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 8.13e-02 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 1 | 38 | 8.16e-02 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 1 | 5 | 8.18e-02 |
| GO:BP | GO:0003283 | atrial septum development | 1 | 23 | 8.23e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 2 | 513 | 8.25e-02 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 4 | 1248 | 8.30e-02 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 1 | 37 | 8.53e-02 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 1 | 23 | 8.54e-02 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 8.61e-02 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 2 | 178 | 8.61e-02 |
| GO:BP | GO:0048729 | tissue morphogenesis | 8 | 504 | 8.61e-02 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 8.61e-02 |
| GO:BP | GO:0006606 | protein import into nucleus | 2 | 147 | 8.61e-02 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 2 | 187 | 8.61e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 8.68e-02 |
| GO:BP | GO:0010941 | regulation of cell death | 4 | 1219 | 8.68e-02 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 2 | 54 | 8.69e-02 |
| GO:BP | GO:0006305 | DNA alkylation | 2 | 46 | 8.69e-02 |
| GO:BP | GO:0006306 | DNA methylation | 2 | 46 | 8.69e-02 |
| GO:BP | GO:1905382 | positive regulation of snRNA transcription by RNA polymerase II | 1 | 3 | 8.70e-02 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 2 | 53 | 8.70e-02 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 85 | 8.71e-02 |
| GO:BP | GO:1901652 | response to peptide | 3 | 377 | 8.73e-02 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 1 | 2 | 8.77e-02 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 1 | 8 | 8.77e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 2 | 344 | 8.77e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 2 | 345 | 8.79e-02 |
| GO:BP | GO:0009880 | embryonic pattern specification | 1 | 50 | 8.89e-02 |
| GO:BP | GO:0006417 | regulation of translation | 2 | 370 | 8.89e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 2 | 81 | 8.89e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 3 | 881 | 8.89e-02 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 11 | 8.89e-02 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 1 | 15 | 8.89e-02 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 2 | 265 | 8.89e-02 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 9.00e-02 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 5 | 9.00e-02 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 3 | 88 | 9.00e-02 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 1 | 29 | 9.00e-02 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 1 | 6 | 9.00e-02 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 2 | 348 | 9.10e-02 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 1 | 17 | 9.15e-02 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 1 | 18 | 9.15e-02 |
| GO:BP | GO:0043434 | response to peptide hormone | 2 | 321 | 9.24e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 1 | 65 | 9.28e-02 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 1 | 7 | 9.28e-02 |
| GO:BP | GO:0060214 | endocardium formation | 1 | 1 | 9.28e-02 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 1 | 9 | 9.28e-02 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 1 | 9 | 9.28e-02 |
| GO:BP | GO:0060298 | positive regulation of sarcomere organization | 1 | 4 | 9.28e-02 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 9.28e-02 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 1 | 31 | 9.28e-02 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 1 | 4 | 9.28e-02 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 2 | 44 | 9.28e-02 |
| GO:BP | GO:0060430 | lung saccule development | 1 | 7 | 9.28e-02 |
| GO:BP | GO:0048645 | animal organ formation | 2 | 51 | 9.28e-02 |
| GO:BP | GO:0061113 | pancreas morphogenesis | 1 | 3 | 9.28e-02 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 1 | 8 | 9.28e-02 |
| GO:BP | GO:0006470 | protein dephosphorylation | 3 | 214 | 9.43e-02 |
| GO:BP | GO:0042594 | response to starvation | 2 | 178 | 9.43e-02 |
| GO:BP | GO:0048519 | negative regulation of biological process | 11 | 4045 | 9.43e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 2 | 293 | 9.43e-02 |
| GO:BP | GO:0001709 | cell fate determination | 2 | 25 | 9.55e-02 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 2 | 28 | 9.55e-02 |
| GO:BP | GO:0032259 | methylation | 5 | 313 | 9.57e-02 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 9.62e-02 |
| GO:BP | GO:0060348 | bone development | 4 | 170 | 9.62e-02 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 1 | 6 | 9.62e-02 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 1 | 55 | 9.62e-02 |
| GO:BP | GO:0051890 | regulation of cardioblast differentiation | 1 | 5 | 9.62e-02 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 1 | 38 | 9.62e-02 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 2 | 69 | 9.68e-02 |
| GO:BP | GO:0010506 | regulation of autophagy | 2 | 313 | 9.68e-02 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 1 | 86 | 9.68e-02 |
| GO:BP | GO:0140467 | integrated stress response signaling | 1 | 35 | 9.68e-02 |
| GO:BP | GO:1905380 | regulation of snRNA transcription by RNA polymerase II | 1 | 4 | 9.68e-02 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 1 | 20 | 9.68e-02 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 9.68e-02 |
| GO:BP | GO:0048733 | sebaceous gland development | 1 | 6 | 9.70e-02 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 9.70e-02 |
| GO:BP | GO:0009798 | axis specification | 1 | 63 | 9.74e-02 |
| GO:BP | GO:0061035 | regulation of cartilage development | 1 | 54 | 9.74e-02 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 1 | 48 | 9.78e-02 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 1 | 15 | 9.86e-02 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 9.86e-02 |
| GO:BP | GO:0035295 | tube development | 11 | 821 | 9.86e-02 |
| GO:BP | GO:0060485 | mesenchyme development | 5 | 249 | 9.86e-02 |
| GO:BP | GO:1901509 | regulation of endothelial tube morphogenesis | 1 | 3 | 9.86e-02 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 1 | 48 | 9.86e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 4 | 1223 | 9.86e-02 |
| GO:BP | GO:0006986 | response to unfolded protein | 3 | 128 | 9.86e-02 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 41 | 9.86e-02 |
| GO:BP | GO:0045165 | cell fate commitment | 3 | 155 | 9.86e-02 |
| GO:BP | GO:0051693 | actin filament capping | 2 | 34 | 9.86e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 1 | 34 | 9.92e-02 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 2 | 70 | 9.95e-02 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 1 | 41 | 9.95e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 9 | 868 | 9.95e-02 |
| GO:BP | GO:0010033 | response to organic substance | 5 | 1957 | 9.95e-02 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 2 | 424 | 9.96e-02 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 9.98e-02 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 9.98e-02 |
| GO:BP | GO:1990928 | response to amino acid starvation | 1 | 51 | 9.98e-02 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 1.00e-01 |
| GO:BP | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | 1 | 5 | 1.00e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 2 | 194 | 1.00e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 2 | 62 | 1.00e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 3 | 763 | 1.01e-01 |
| GO:BP | GO:0061140 | lung secretory cell differentiation | 1 | 5 | 1.01e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 1.01e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 2 | 307 | 1.01e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 2 | 418 | 1.01e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 1 | 45 | 1.01e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 1 | 23 | 1.01e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 1 | 51 | 1.01e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 1 | 6 | 1.01e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 1 | 7 | 1.02e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 1.02e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 1 | 4 | 1.02e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 3 | 756 | 1.02e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 3 | 993 | 1.03e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 3 | 994 | 1.03e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 303 | 1.03e-01 |
| GO:BP | GO:0008584 | male gonad development | 3 | 99 | 1.03e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 1.03e-01 |
| GO:BP | GO:0007389 | pattern specification process | 5 | 306 | 1.03e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 1.03e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 3 | 100 | 1.04e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 1 | 9 | 1.04e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 1 | 10 | 1.04e-01 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 1 | 3 | 1.04e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 1 | 10 | 1.04e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 2 | 69 | 1.04e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 1 | 9 | 1.04e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 1 | 10 | 1.04e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 2 | 52 | 1.04e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 1.04e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 2 | 76 | 1.05e-01 |
| GO:BP | GO:0036301 | macrophage colony-stimulating factor production | 1 | 2 | 1.05e-01 |
| GO:BP | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death | 1 | 6 | 1.05e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 100 | 1.05e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 1.05e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 2 | 393 | 1.05e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 1 | 58 | 1.05e-01 |
| GO:BP | GO:1901256 | regulation of macrophage colony-stimulating factor production | 1 | 2 | 1.05e-01 |
| GO:BP | GO:1901033 | positive regulation of response to reactive oxygen species | 1 | 6 | 1.05e-01 |
| GO:BP | GO:0061470 | T follicular helper cell differentiation | 1 | 4 | 1.05e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 1.05e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 1.05e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 1 | 23 | 1.05e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 1 | 39 | 1.05e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 1 | 84 | 1.05e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 1.05e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 1 | 9 | 1.07e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 1 | 39 | 1.07e-01 |
| GO:BP | GO:0006915 | apoptotic process | 4 | 1443 | 1.07e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 2 | 70 | 1.07e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 1 | 18 | 1.07e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 2 | 399 | 1.07e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 1 | 12 | 1.07e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 1.07e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 1.07e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 1 | 12 | 1.07e-01 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 1 | 6 | 1.07e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 2 | 70 | 1.08e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 2 | 70 | 1.08e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 1.08e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 2 | 340 | 1.09e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 1 | 26 | 1.09e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 1.09e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 1 | 19 | 1.09e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 1 | 5 | 1.10e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 1 | 6 | 1.10e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 1.10e-01 |
| GO:BP | GO:0007369 | gastrulation | 3 | 146 | 1.10e-01 |
| GO:BP | GO:0055009 | atrial cardiac muscle tissue morphogenesis | 1 | 5 | 1.10e-01 |
| GO:BP | GO:0003160 | endocardium morphogenesis | 1 | 3 | 1.10e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 2 | 219 | 1.10e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 1.10e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 2 | 96 | 1.10e-01 |
| GO:BP | GO:0040031 | snRNA modification | 1 | 7 | 1.10e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 1 | 4 | 1.12e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 1.12e-01 |
| GO:BP | GO:0012501 | programmed cell death | 4 | 1479 | 1.12e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 1.13e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 2 | 352 | 1.14e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 1 | 4 | 1.14e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 1 | 6 | 1.14e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 1 | 6 | 1.14e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 1 | 128 | 1.14e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 1 | 40 | 1.14e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 1.15e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 1 | 4 | 1.15e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 2 | 68 | 1.15e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 1 | 129 | 1.16e-01 |
| GO:BP | GO:0001711 | endodermal cell fate commitment | 1 | 9 | 1.16e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 1.16e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 1 | 11 | 1.16e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 2 | 37 | 1.16e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 1.17e-01 |
| GO:BP | GO:0003002 | regionalization | 2 | 273 | 1.17e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 1.17e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 1.18e-01 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 3 | 157 | 1.18e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 1.18e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 1 | 70 | 1.18e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 1.18e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 1 | 31 | 1.18e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 4 | 1399 | 1.19e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 3 | 1173 | 1.19e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 1.19e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 9 | 663 | 1.20e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 1 | 7 | 1.20e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 1 | 4 | 1.20e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 1.20e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 1 | 11 | 1.20e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 1 | 46 | 1.21e-01 |
| GO:BP | GO:0009790 | embryo development | 3 | 875 | 1.21e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 2 | 441 | 1.22e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 1 | 56 | 1.22e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 1 | 53 | 1.22e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 90 | 1.22e-01 |
| GO:BP | GO:0007517 | muscle organ development | 5 | 277 | 1.22e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 1 | 84 | 1.22e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 1 | 57 | 1.22e-01 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 1 | 6 | 1.22e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 2 | 57 | 1.23e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 3 | 113 | 1.23e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 2 | 316 | 1.24e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 1 | 65 | 1.24e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 1.24e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 1.24e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 2 | 91 | 1.25e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 3 | 114 | 1.25e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 2 | 457 | 1.25e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 3 | 1167 | 1.25e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 1 | 17 | 1.26e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 1.26e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 1.26e-01 |
| GO:BP | GO:0035562 | negative regulation of chromatin binding | 1 | 8 | 1.28e-01 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 1.28e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 1 | 5 | 1.28e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 1 | 61 | 1.28e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 1.29e-01 |
| GO:BP | GO:0060047 | heart contraction | 4 | 205 | 1.29e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 1 | 4 | 1.29e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 1 | 5 | 1.29e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 1.29e-01 |
| GO:BP | GO:0030916 | otic vesicle formation | 1 | 6 | 1.29e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 1.29e-01 |
| GO:BP | GO:0060836 | lymphatic endothelial cell differentiation | 1 | 5 | 1.29e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 2 | 50 | 1.29e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 1.29e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 1.29e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 1.29e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 1.29e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 3 | 468 | 1.29e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 1 | 75 | 1.29e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 1.30e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 1.30e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 1 | 9 | 1.30e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 1 | 71 | 1.30e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 3 | 118 | 1.30e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 2 | 56 | 1.30e-01 |
| GO:BP | GO:0008219 | cell death | 4 | 1596 | 1.30e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 1 | 64 | 1.31e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 7 | 462 | 1.31e-01 |
| GO:BP | GO:0061912 | selective autophagy | 1 | 90 | 1.32e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 1.32e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 1.33e-01 |
| GO:BP | GO:0061303 | cornea development in camera-type eye | 1 | 4 | 1.33e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 1 | 65 | 1.33e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 4 | 440 | 1.33e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 1 | 11 | 1.33e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 1 | 11 | 1.33e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 1 | 9 | 1.33e-01 |
| GO:BP | GO:0060429 | epithelium development | 3 | 850 | 1.33e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 1.33e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 1.33e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 1 | 13 | 1.34e-01 |
| GO:BP | GO:0016049 | cell growth | 2 | 418 | 1.34e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 79 | 1.35e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 1 | 15 | 1.35e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 1 | 13 | 1.35e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 1.35e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 1 | 71 | 1.35e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 1.35e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 1.35e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 2 | 485 | 1.35e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 1.35e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 1 | 9 | 1.36e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 3 | 127 | 1.36e-01 |
| GO:BP | GO:0001947 | heart looping | 1 | 50 | 1.36e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 3 | 284 | 1.36e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 1 | 9 | 1.36e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 2 | 227 | 1.36e-01 |
| GO:BP | GO:0042118 | endothelial cell activation | 1 | 7 | 1.37e-01 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 1 | 24 | 1.37e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 114 | 1.38e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 114 | 1.38e-01 |
| GO:BP | GO:0003015 | heart process | 4 | 216 | 1.38e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 1 | 16 | 1.38e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 1 | 14 | 1.38e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 1 | 11 | 1.38e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 2 | 270 | 1.38e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 1.38e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 2 | 88 | 1.38e-01 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 1 | 11 | 1.38e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 1 | 72 | 1.38e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 1 | 179 | 1.39e-01 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 1 | 11 | 1.40e-01 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 1 | 11 | 1.40e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 3 | 146 | 1.41e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 1 | 19 | 1.42e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 2 | 96 | 1.42e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 3 | 893 | 1.42e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 1 | 96 | 1.42e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 1 | 78 | 1.42e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 2 | 115 | 1.42e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 1.42e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 1.42e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 1 | 23 | 1.42e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 1.42e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 1 | 23 | 1.42e-01 |
| GO:BP | GO:0006304 | DNA modification | 2 | 89 | 1.42e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 2 | 517 | 1.42e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 2 | 115 | 1.42e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 81 | 1.42e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 1 | 23 | 1.42e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 1 | 56 | 1.42e-01 |
| GO:BP | GO:0030900 | forebrain development | 5 | 281 | 1.42e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 1 | 8 | 1.42e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 2 | 53 | 1.42e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 1 | 8 | 1.42e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 1 | 76 | 1.42e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 3 | 143 | 1.42e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 1 | 78 | 1.42e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 3 | 783 | 1.42e-01 |
| GO:BP | GO:0048664 | neuron fate determination | 1 | 4 | 1.42e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 1 | 67 | 1.42e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 1 | 12 | 1.42e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 1 | 20 | 1.42e-01 |
| GO:BP | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | 1 | 10 | 1.42e-01 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 1 | 8 | 1.42e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 1 | 52 | 1.42e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 1 | 10 | 1.42e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 3 | 113 | 1.42e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 1.43e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 2 | 40 | 1.44e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 2 | 51 | 1.44e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 1 | 12 | 1.44e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 84 | 1.44e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 1 | 12 | 1.44e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 1 | 70 | 1.44e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 1 | 179 | 1.44e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 1.45e-01 |
| GO:BP | GO:0016081 | synaptic vesicle docking | 1 | 9 | 1.45e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 2 | 111 | 1.45e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 2 | 604 | 1.45e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 1.46e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 85 | 1.46e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 1 | 12 | 1.46e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 2 | 67 | 1.47e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 2 | 178 | 1.47e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 1.47e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 2 | 40 | 1.47e-01 |
| GO:BP | GO:0060322 | head development | 8 | 633 | 1.47e-01 |
| GO:BP | GO:0009617 | response to bacterium | 3 | 321 | 1.47e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 1 | 82 | 1.47e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 2 | 513 | 1.47e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 1 | 10 | 1.47e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 3 | 149 | 1.47e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 1.47e-01 |
| GO:BP | GO:0006914 | autophagy | 2 | 513 | 1.47e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 2 | 57 | 1.47e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 1.47e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 1 | 20 | 1.47e-01 |
| GO:BP | GO:0070857 | regulation of bile acid biosynthetic process | 1 | 7 | 1.47e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 1 | 187 | 1.47e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 1 | 8 | 1.47e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 2 | 58 | 1.47e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 1.47e-01 |
| GO:BP | GO:0006412 | translation | 2 | 646 | 1.48e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 2 | 40 | 1.48e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 3 | 197 | 1.48e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 1.48e-01 |
| GO:BP | GO:0016310 | phosphorylation | 3 | 1372 | 1.48e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 2 | 40 | 1.48e-01 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 1 | 27 | 1.48e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 3 | 160 | 1.48e-01 |
| GO:BP | GO:0043200 | response to amino acid | 1 | 95 | 1.49e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 1 | 8 | 1.49e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 1 | 22 | 1.49e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 2 | 56 | 1.49e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 1 | 27 | 1.49e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 39 | 1.49e-01 |
| GO:BP | GO:0048736 | appendage development | 1 | 142 | 1.49e-01 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 1 | 18 | 1.49e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 3 | 160 | 1.49e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 1 | 14 | 1.49e-01 |
| GO:BP | GO:0060173 | limb development | 1 | 142 | 1.49e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 4 | 191 | 1.50e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 12 | 1014 | 1.50e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 1.50e-01 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 1 | 8 | 1.51e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 1 | 6 | 1.51e-01 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 1.51e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 1 | 69 | 1.51e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 2 | 86 | 1.51e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 1 | 17 | 1.51e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 92 | 1.51e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 1 | 10 | 1.51e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 2 | 187 | 1.52e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 1 | 222 | 1.52e-01 |
| GO:BP | GO:0010459 | negative regulation of heart rate | 1 | 7 | 1.52e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 2 | 666 | 1.52e-01 |
| GO:BP | GO:0031929 | TOR signaling | 1 | 115 | 1.52e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 1 | 74 | 1.52e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 6 | 2065 | 1.52e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 1.52e-01 |
| GO:BP | GO:0007417 | central nervous system development | 10 | 792 | 1.52e-01 |
| GO:BP | GO:0003157 | endocardium development | 1 | 9 | 1.52e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 2 | 494 | 1.52e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 2 | 61 | 1.52e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 1 | 13 | 1.53e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 1 | 14 | 1.53e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 1 | 227 | 1.53e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 1.53e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 1.53e-01 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 1 | 18 | 1.54e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 1.54e-01 |
| GO:BP | GO:0060479 | lung cell differentiation | 1 | 18 | 1.54e-01 |
| GO:BP | GO:0009409 | response to cold | 1 | 41 | 1.54e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 2 | 108 | 1.54e-01 |
| GO:BP | GO:0098754 | detoxification | 1 | 91 | 1.55e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 1 | 75 | 1.55e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 40 | 1.55e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 1.56e-01 |
| GO:BP | GO:0002752 | cell surface pattern recognition receptor signaling pathway | 1 | 27 | 1.57e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 1 | 27 | 1.57e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 1 | 27 | 1.57e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 1 | 105 | 1.57e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 12 | 1974 | 1.57e-01 |
| GO:BP | GO:0042117 | monocyte activation | 1 | 8 | 1.57e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 1 | 117 | 1.57e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 1 | 7 | 1.57e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 27 | 1.57e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 151 | 1.57e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 7 | 560 | 1.57e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 1 | 8 | 1.58e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 1 | 10 | 1.58e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 1 | 85 | 1.58e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 1 | 17 | 1.58e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 1 | 33 | 1.58e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 1 | 19 | 1.58e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 2 | 67 | 1.58e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 1 | 10 | 1.58e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 1 | 16 | 1.58e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 1 | 16 | 1.58e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 3 | 157 | 1.59e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 1 | 19 | 1.59e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 1 | 105 | 1.59e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 1.59e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 3 | 138 | 1.59e-01 |
| GO:BP | GO:0021543 | pallium development | 3 | 139 | 1.59e-01 |
| GO:BP | GO:0031016 | pancreas development | 2 | 53 | 1.60e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 1 | 133 | 1.60e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 1 | 105 | 1.60e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 1 | 12 | 1.60e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 1.60e-01 |
| GO:BP | GO:0001501 | skeletal system development | 6 | 377 | 1.61e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 2 | 57 | 1.61e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 1 | 15 | 1.61e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 1.61e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 2 | 42 | 1.61e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 2 | 78 | 1.61e-01 |
| GO:BP | GO:0030324 | lung development | 3 | 146 | 1.61e-01 |
| GO:BP | GO:0060766 | negative regulation of androgen receptor signaling pathway | 1 | 12 | 1.61e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 1.61e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 1 | 89 | 1.61e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 2 | 555 | 1.61e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 1 | 13 | 1.62e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 1 | 15 | 1.62e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 1.63e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 2 | 125 | 1.63e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 111 | 1.63e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 1 | 11 | 1.63e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 1 | 11 | 1.63e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 1 | 17 | 1.63e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 1 | 113 | 1.63e-01 |
| GO:BP | GO:1904251 | regulation of bile acid metabolic process | 1 | 9 | 1.63e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 1 | 11 | 1.63e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 1 | 59 | 1.63e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 3 | 152 | 1.63e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 2 | 123 | 1.64e-01 |
| GO:BP | GO:0021766 | hippocampus development | 2 | 62 | 1.64e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 2 | 101 | 1.64e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 1.64e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 1 | 13 | 1.64e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 3 | 108 | 1.64e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 1 | 36 | 1.65e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 3 | 150 | 1.65e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 10 | 1.65e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 1 | 22 | 1.65e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 1.65e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 3 | 267 | 1.65e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 1.65e-01 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 1 | 18 | 1.65e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 1.65e-01 |
| GO:BP | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development | 1 | 13 | 1.65e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 2 | 1137 | 1.65e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 1 | 10 | 1.65e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 1 | 151 | 1.66e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 1 | 23 | 1.66e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 2 | 57 | 1.67e-01 |
| GO:BP | GO:0043584 | nose development | 1 | 10 | 1.69e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 1 | 22 | 1.69e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 1 | 9 | 1.69e-01 |
| GO:BP | GO:0071599 | otic vesicle development | 1 | 10 | 1.69e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 1 | 110 | 1.70e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 1 | 126 | 1.70e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 1.70e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 2 | 127 | 1.70e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 2 | 551 | 1.70e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 1.71e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 1 | 65 | 1.72e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 3 | 293 | 1.72e-01 |
| GO:BP | GO:0007049 | cell cycle | 14 | 1529 | 1.72e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 1.72e-01 |
| GO:BP | GO:0036035 | osteoclast development | 1 | 10 | 1.72e-01 |
| GO:BP | GO:0007612 | learning | 2 | 115 | 1.72e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 1 | 23 | 1.72e-01 |
| GO:BP | GO:0035176 | social behavior | 1 | 34 | 1.72e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 2 | 131 | 1.73e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 2 | 771 | 1.73e-01 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 1 | 42 | 1.73e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 2 | 64 | 1.73e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 1.73e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 8 | 679 | 1.73e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 2 | 61 | 1.74e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 1 | 25 | 1.74e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 1 | 12 | 1.75e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 1 | 13 | 1.75e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 3 | 169 | 1.75e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 2 | 64 | 1.75e-01 |
| GO:BP | GO:0061448 | connective tissue development | 2 | 208 | 1.75e-01 |
| GO:BP | GO:0016573 | histone acetylation | 2 | 143 | 1.75e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 2 | 101 | 1.75e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 1 | 34 | 1.75e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 2 | 775 | 1.75e-01 |
| GO:BP | GO:0033993 | response to lipid | 3 | 592 | 1.75e-01 |
| GO:BP | GO:0006974 | DNA damage response | 8 | 785 | 1.76e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 2 | 561 | 1.76e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 1 | 14 | 1.76e-01 |
| GO:BP | GO:0008406 | gonad development | 3 | 158 | 1.76e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 1 | 14 | 1.76e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 1 | 18 | 1.77e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 1 | 165 | 1.77e-01 |
| GO:BP | GO:0070848 | response to growth factor | 2 | 572 | 1.77e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 1 | 144 | 1.77e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 1 | 23 | 1.79e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 2 | 50 | 1.79e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 2 | 90 | 1.79e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 1 | 21 | 1.80e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 15 | 6996 | 1.80e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 1 | 138 | 1.80e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 1 | 41 | 1.80e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 1 | 24 | 1.80e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 1.80e-01 |
| GO:BP | GO:0001946 | lymphangiogenesis | 1 | 12 | 1.80e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 1 | 31 | 1.80e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 1 | 19 | 1.80e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 1 | 19 | 1.80e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 2 | 142 | 1.80e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 1.80e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 151 | 1.80e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 1.80e-01 |
| GO:BP | GO:0090224 | regulation of spindle organization | 1 | 45 | 1.80e-01 |
| GO:BP | GO:0007052 | mitotic spindle organization | 2 | 129 | 1.80e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 1 | 281 | 1.80e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 6 | 1719 | 1.80e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 1 | 126 | 1.80e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 3 | 161 | 1.80e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 1 | 140 | 1.80e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 1.80e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 1 | 37 | 1.80e-01 |
| GO:BP | GO:1900424 | regulation of defense response to bacterium | 1 | 12 | 1.80e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 4 | 247 | 1.81e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 2 | 148 | 1.81e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 25 | 1.81e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1 | 26 | 1.81e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 1.81e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 1 | 127 | 1.81e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 1.82e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 1 | 69 | 1.82e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 1 | 112 | 1.82e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 1 | 140 | 1.82e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 4 | 278 | 1.82e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 2 | 150 | 1.83e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 1 | 22 | 1.83e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 1 | 28 | 1.83e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 1 | 140 | 1.83e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 2 | 103 | 1.84e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 1 | 149 | 1.84e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 2 | 809 | 1.85e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 1 | 147 | 1.85e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 89 | 1.85e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 1.85e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 1.85e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 18 | 1835 | 1.86e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 1 | 134 | 1.86e-01 |
| GO:BP | GO:0060541 | respiratory system development | 3 | 163 | 1.87e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 1 | 32 | 1.87e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 2 | 280 | 1.87e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 4 | 1869 | 1.87e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 1 | 19 | 1.87e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 1 | 186 | 1.87e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 1 | 97 | 1.87e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 1 | 123 | 1.87e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 1 | 155 | 1.87e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 4 | 859 | 1.87e-01 |
| GO:BP | GO:0030258 | lipid modification | 1 | 162 | 1.88e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 1 | 93 | 1.88e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 4 | 820 | 1.88e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 1 | 11 | 1.88e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 1 | 94 | 1.88e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 1.88e-01 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 1 | 19 | 1.88e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 1.88e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 3 | 182 | 1.88e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 1.88e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 2 | 158 | 1.88e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 1 | 19 | 1.88e-01 |
| GO:BP | GO:0001503 | ossification | 1 | 313 | 1.88e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 1.88e-01 |
| GO:BP | GO:0060428 | lung epithelium development | 1 | 30 | 1.88e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 3 | 203 | 1.88e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 1 | 151 | 1.88e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 2 | 144 | 1.88e-01 |
| GO:BP | GO:0051707 | response to other organism | 4 | 820 | 1.88e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 1 | 26 | 1.88e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 1 | 21 | 1.89e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 1 | 18 | 1.89e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 7 | 827 | 1.89e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 1 | 95 | 1.89e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 1 | 101 | 1.89e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 1.89e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 1.89e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 3 | 317 | 1.90e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 1 | 77 | 1.90e-01 |
| GO:BP | GO:0000303 | response to superoxide | 1 | 21 | 1.91e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 2 | 113 | 1.91e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 3 | 206 | 1.91e-01 |
| GO:BP | GO:0007420 | brain development | 7 | 591 | 1.91e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 7 | 691 | 1.91e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 2 | 160 | 1.91e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 1 | 58 | 1.91e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 1 | 102 | 1.92e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 2 | 94 | 1.92e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 1.92e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 1 | 103 | 1.92e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 3 | 173 | 1.93e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 7 | 694 | 1.93e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 1.93e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 1 | 33 | 1.93e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 5 | 2890 | 1.93e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 1 | 40 | 1.93e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 1.93e-01 |
| GO:BP | GO:0006950 | response to stress | 5 | 2780 | 1.93e-01 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 1 | 18 | 1.93e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 1 | 66 | 1.93e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 4 | 850 | 1.93e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 1 | 23 | 1.94e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 4 | 259 | 1.94e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 1 | 22 | 1.94e-01 |
| GO:BP | GO:0014002 | astrocyte development | 1 | 27 | 1.94e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 3 | 1345 | 1.96e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 1 | 23 | 1.96e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 1 | 156 | 1.96e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 1 | 105 | 1.97e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 1.97e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 1 | 19 | 1.97e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 185 | 1.97e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 1 | 19 | 1.97e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 1 | 170 | 1.97e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 2 | 84 | 1.97e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 2 | 130 | 1.97e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 1 | 38 | 1.98e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 3 | 195 | 1.98e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 2 | 782 | 1.98e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 2 | 687 | 1.98e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 1 | 22 | 1.98e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 1.98e-01 |
| GO:BP | GO:0006479 | protein methylation | 2 | 160 | 1.98e-01 |
| GO:BP | GO:0008213 | protein alkylation | 2 | 160 | 1.98e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 1.98e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 2 | 85 | 1.98e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 24 | 1.98e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 1 | 110 | 1.99e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 1.99e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 1 | 160 | 1.99e-01 |
| GO:BP | GO:0060914 | heart formation | 1 | 29 | 2.00e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 1 | 168 | 2.00e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 1 | 34 | 2.00e-01 |
| GO:BP | GO:0007584 | response to nutrient | 1 | 109 | 2.00e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 1 | 33 | 2.00e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 1 | 20 | 2.00e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 2.00e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 2.00e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 2 | 81 | 2.01e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 4 | 282 | 2.01e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 1 | 68 | 2.02e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 1 | 13 | 2.03e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 1 | 10 | 2.03e-01 |
| GO:BP | GO:0042221 | response to chemical | 5 | 2559 | 2.04e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 1 | 174 | 2.04e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 2.04e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 1 | 32 | 2.04e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 2.04e-01 |
| GO:BP | GO:0019083 | viral transcription | 1 | 48 | 2.04e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 2.04e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 2.05e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 1 | 123 | 2.05e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 1 | 28 | 2.06e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 1 | 26 | 2.07e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 1 | 17 | 2.07e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 2.07e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 5 | 441 | 2.08e-01 |
| GO:BP | GO:0009888 | tissue development | 2 | 1425 | 2.08e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 1 | 31 | 2.08e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 1 | 187 | 2.09e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 1 | 187 | 2.09e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 14 | 2.09e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 209 | 2.11e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 1 | 76 | 2.11e-01 |
| GO:BP | GO:0021761 | limbic system development | 2 | 81 | 2.11e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 2 | 127 | 2.12e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 5 | 4099 | 2.12e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 1 | 18 | 2.13e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 1 | 18 | 2.13e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 1 | 18 | 2.13e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 1 | 19 | 2.13e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 1 | 18 | 2.13e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 2 | 830 | 2.13e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 2 | 90 | 2.13e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 1 | 38 | 2.14e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 1 | 35 | 2.14e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 3 | 207 | 2.14e-01 |
| GO:BP | GO:0022402 | cell cycle process | 10 | 1086 | 2.14e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 1 | 152 | 2.14e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 1 | 21 | 2.14e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 2 | 660 | 2.15e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 5 | 418 | 2.15e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 1 | 21 | 2.15e-01 |
| GO:BP | GO:0007498 | mesoderm development | 2 | 89 | 2.16e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 1 | 17 | 2.16e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 4 | 310 | 2.16e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 1 | 80 | 2.16e-01 |
| GO:BP | GO:0001654 | eye development | 1 | 272 | 2.16e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 1 | 75 | 2.16e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 1 | 34 | 2.16e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 2 | 103 | 2.16e-01 |
| GO:BP | GO:0001706 | endoderm formation | 1 | 46 | 2.17e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 1 | 19 | 2.17e-01 |
| GO:BP | GO:0150063 | visual system development | 1 | 273 | 2.17e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 4 | 2285 | 2.19e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 1 | 16 | 2.19e-01 |
| GO:BP | GO:0048880 | sensory system development | 1 | 275 | 2.19e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 2.19e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 2 | 678 | 2.19e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 1 | 23 | 2.19e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 1 | 24 | 2.19e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 1 | 26 | 2.19e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 9 | 956 | 2.20e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 3 | 2011 | 2.20e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 4 | 963 | 2.20e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 4 | 2035 | 2.21e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 1 | 25 | 2.21e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 1 | 35 | 2.21e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 1 | 18 | 2.21e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 1 | 19 | 2.21e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 1 | 19 | 2.21e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 2.21e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 1 | 35 | 2.21e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 1 | 18 | 2.21e-01 |
| GO:BP | GO:1901998 | toxin transport | 1 | 37 | 2.21e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 1 | 38 | 2.21e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 3 | 184 | 2.22e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 2.22e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 1 | 144 | 2.22e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 2 | 993 | 2.23e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 2.24e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 2.24e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 2 | 64 | 2.25e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 1 | 222 | 2.25e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 1 | 24 | 2.25e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 1 | 54 | 2.25e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 2 | 32 | 2.25e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 1 | 22 | 2.26e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 1 | 33 | 2.26e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 2 | 55 | 2.26e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 2 | 182 | 2.26e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 1 | 19 | 2.26e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 1 | 33 | 2.26e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 1 | 165 | 2.26e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 1 | 31 | 2.26e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 7 | 4579 | 2.27e-01 |
| GO:BP | GO:0007548 | sex differentiation | 3 | 191 | 2.27e-01 |
| GO:BP | GO:0006473 | protein acetylation | 2 | 190 | 2.27e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 1 | 27 | 2.27e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 2.27e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 1 | 40 | 2.27e-01 |
| GO:BP | GO:0019079 | viral genome replication | 2 | 101 | 2.27e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 2 | 111 | 2.27e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 2 | 260 | 2.27e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 1 | 8 | 2.27e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 104 | 2.27e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 1 | 18 | 2.27e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 2 | 148 | 2.28e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 2 | 67 | 2.29e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 2 | 137 | 2.29e-01 |
| GO:BP | GO:0015918 | sterol transport | 1 | 92 | 2.30e-01 |
| GO:BP | GO:0001525 | angiogenesis | 4 | 390 | 2.33e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 2.33e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 1 | 42 | 2.33e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 2 | 121 | 2.33e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 2 | 108 | 2.34e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 1 | 17 | 2.34e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 1 | 86 | 2.34e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 1 | 25 | 2.34e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 2.34e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 1 | 33 | 2.34e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 1 | 27 | 2.35e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 1 | 43 | 2.35e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 1 | 121 | 2.35e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 3 | 479 | 2.35e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 1 | 43 | 2.35e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 2.35e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 1 | 39 | 2.35e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 2.35e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 1 | 277 | 2.35e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 1 | 27 | 2.36e-01 |
| GO:BP | GO:0003158 | endothelium development | 2 | 104 | 2.36e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 1 | 228 | 2.38e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 1 | 246 | 2.38e-01 |
| GO:BP | GO:0016236 | macroautophagy | 1 | 308 | 2.38e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 2 | 91 | 2.38e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 134 | 2.41e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 7 | 870 | 2.41e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 2 | 726 | 2.42e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 7 | 605 | 2.42e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 134 | 2.42e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 1 | 44 | 2.42e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 1 | 31 | 2.43e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 2 | 676 | 2.43e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 2 | 212 | 2.43e-01 |
| GO:BP | GO:0051258 | protein polymerization | 1 | 241 | 2.43e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 1 | 44 | 2.43e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 2 | 875 | 2.43e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 2 | 167 | 2.43e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 3 | 400 | 2.43e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 2 | 192 | 2.45e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 1 | 22 | 2.45e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 1 | 32 | 2.45e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 1 | 92 | 2.45e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 2.45e-01 |
| GO:BP | GO:0061458 | reproductive system development | 2 | 215 | 2.47e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 3 | 262 | 2.47e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 1 | 45 | 2.48e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 2 | 118 | 2.48e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 1 | 194 | 2.48e-01 |
| GO:BP | GO:0001708 | cell fate specification | 1 | 56 | 2.48e-01 |
| GO:BP | GO:0043588 | skin development | 2 | 175 | 2.48e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 1 | 225 | 2.48e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 1 | 14 | 2.48e-01 |
| GO:BP | GO:0070266 | necroptotic process | 1 | 32 | 2.49e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 1 | 41 | 2.49e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 1 | 42 | 2.49e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 3 | 229 | 2.51e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 3 | 198 | 2.51e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 2.52e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 2.53e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 2.53e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 1 | 31 | 2.53e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 2.53e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 1 | 16 | 2.54e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 2.54e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 1 | 241 | 2.54e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 1 | 75 | 2.57e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 1 | 38 | 2.57e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 1 | 41 | 2.57e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 2.57e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 1 | 149 | 2.59e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 189 | 2.59e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 2.60e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 2 | 127 | 2.60e-01 |
| GO:BP | GO:0007051 | spindle organization | 2 | 188 | 2.60e-01 |
| GO:BP | GO:0007423 | sensory organ development | 4 | 396 | 2.60e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 3 | 2139 | 2.61e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 5 | 504 | 2.62e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 4 | 358 | 2.62e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 1 | 23 | 2.62e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 2 | 227 | 2.62e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 1 | 47 | 2.63e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 2 | 156 | 2.63e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 1 | 271 | 2.63e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 1 | 41 | 2.64e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 1 | 25 | 2.64e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 2.64e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 2.64e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 3 | 2161 | 2.64e-01 |
| GO:BP | GO:0001889 | liver development | 2 | 110 | 2.66e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 1 | 54 | 2.66e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 1 | 22 | 2.66e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 2.66e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 1 | 45 | 2.67e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 1 | 36 | 2.67e-01 |
| GO:BP | GO:0007613 | memory | 1 | 92 | 2.67e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 2 | 127 | 2.68e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 1 | 54 | 2.69e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 2.69e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 6 | 626 | 2.70e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 4 | 279 | 2.71e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 1 | 36 | 2.71e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 2 | 112 | 2.71e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 2 | 132 | 2.71e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 2.72e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 2 | 126 | 2.72e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 3 | 234 | 2.73e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 1 | 85 | 2.73e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 1 | 28 | 2.73e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 2.73e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 1 | 29 | 2.74e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 2.74e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 133 | 2.74e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 2 | 1123 | 2.75e-01 |
| GO:BP | GO:0090659 | walking behavior | 1 | 25 | 2.75e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 1 | 39 | 2.75e-01 |
| GO:BP | GO:0043543 | protein acylation | 2 | 228 | 2.75e-01 |
| GO:BP | GO:0060324 | face development | 1 | 45 | 2.75e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 112 | 2.75e-01 |
| GO:BP | GO:0006284 | base-excision repair | 1 | 44 | 2.75e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 4 | 2565 | 2.75e-01 |
| GO:BP | GO:0048513 | animal organ development | 20 | 2189 | 2.75e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 2.75e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 1 | 303 | 2.75e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 1 | 39 | 2.76e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 1 | 57 | 2.76e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 2.76e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 4 | 2573 | 2.76e-01 |
| GO:BP | GO:0030261 | chromosome condensation | 1 | 33 | 2.77e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 1 | 21 | 2.77e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 1 | 34 | 2.80e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 211 | 2.82e-01 |
| GO:BP | GO:0033574 | response to testosterone | 1 | 29 | 2.82e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 2 | 232 | 2.82e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 1 | 299 | 2.82e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 1 | 318 | 2.83e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 1 | 40 | 2.83e-01 |
| GO:BP | GO:0035148 | tube formation | 2 | 138 | 2.84e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 2 | 179 | 2.84e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 1 | 287 | 2.84e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 357 | 2.85e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 2 | 114 | 2.85e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 305 | 2.85e-01 |
| GO:BP | GO:0051591 | response to cAMP | 1 | 66 | 2.85e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 2.85e-01 |
| GO:BP | GO:0003012 | muscle system process | 4 | 334 | 2.85e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 1 | 55 | 2.85e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 23 | 2.85e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 2.86e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 1 | 43 | 2.86e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 1 | 49 | 2.86e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 64 | 2.86e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 2.86e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 2.86e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 2.86e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 4 | 391 | 2.87e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 2 | 1974 | 2.88e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 2 | 200 | 2.88e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 1 | 35 | 2.88e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 2.88e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 2.88e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 3 | 252 | 2.88e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 1 | 326 | 2.89e-01 |
| GO:BP | GO:0007155 | cell adhesion | 2 | 1012 | 2.89e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 1 | 100 | 2.89e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 1 | 191 | 2.90e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 1 | 51 | 2.91e-01 |
| GO:BP | GO:0090132 | epithelium migration | 3 | 252 | 2.93e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 2.93e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 1 | 31 | 2.93e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 3 | 372 | 2.93e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 1 | 44 | 2.94e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 1 | 115 | 2.95e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 2.95e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 2.95e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 1 | 52 | 2.95e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 2 | 122 | 2.95e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 1 | 39 | 2.96e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 2.96e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 1 | 199 | 2.96e-01 |
| GO:BP | GO:0007611 | learning or memory | 2 | 202 | 2.96e-01 |
| GO:BP | GO:0019080 | viral gene expression | 1 | 96 | 2.96e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 3 | 279 | 2.96e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 1 | 26 | 2.96e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 2.97e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 15 | 7441 | 2.98e-01 |
| GO:BP | GO:0048511 | rhythmic process | 3 | 237 | 2.98e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 2.98e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 1 | 20 | 2.98e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 1 | 99 | 2.99e-01 |
| GO:BP | GO:0090130 | tissue migration | 3 | 256 | 2.99e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 1 | 386 | 2.99e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 3.00e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 1 | 43 | 3.00e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 1 | 64 | 3.01e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 1 | 62 | 3.01e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 4 | 335 | 3.01e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 1 | 211 | 3.04e-01 |
| GO:BP | GO:0021915 | neural tube development | 2 | 146 | 3.04e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 1 | 23 | 3.05e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 1 | 35 | 3.08e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 1 | 37 | 3.08e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 7 | 706 | 3.08e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 4 | 418 | 3.08e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 1 | 101 | 3.08e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 1 | 52 | 3.08e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 1 | 58 | 3.08e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 3 | 448 | 3.08e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 1 | 51 | 3.08e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 1 | 122 | 3.12e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 1 | 68 | 3.12e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 1 | 24 | 3.13e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 1 | 106 | 3.13e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 1 | 40 | 3.13e-01 |
| GO:BP | GO:0006936 | muscle contraction | 1 | 267 | 3.13e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 3.14e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 1 | 37 | 3.14e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 2 | 204 | 3.14e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 1 | 53 | 3.14e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 2 | 197 | 3.15e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 3.15e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 3.15e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 438 | 3.16e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 3.16e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 1 | 222 | 3.17e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 10 | 1133 | 3.18e-01 |
| GO:BP | GO:0051168 | nuclear export | 1 | 150 | 3.18e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 3.19e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 1 | 464 | 3.20e-01 |
| GO:BP | GO:0007015 | actin filament organization | 1 | 373 | 3.21e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 1 | 25 | 3.21e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 3.22e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 3.22e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 2 | 149 | 3.22e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 2 | 237 | 3.22e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 1 | 55 | 3.22e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 1 | 48 | 3.22e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 2 | 98 | 3.22e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 1 | 48 | 3.22e-01 |
| GO:BP | GO:0042551 | neuron maturation | 1 | 33 | 3.23e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 3 | 785 | 3.23e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 1 | 229 | 3.24e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 1 | 85 | 3.24e-01 |
| GO:BP | GO:0022008 | neurogenesis | 2 | 1294 | 3.24e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 3.24e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 1 | 53 | 3.24e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 1 | 33 | 3.24e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 1 | 500 | 3.26e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 3.27e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 1 | 46 | 3.28e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 1 | 53 | 3.28e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 2 | 151 | 3.28e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 2 | 251 | 3.28e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 1 | 116 | 3.28e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 65 | 3.28e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 295 | 3.29e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 1 | 320 | 3.30e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 1 | 155 | 3.30e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 3.30e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 1 | 49 | 3.32e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 1 | 27 | 3.33e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 77 | 3.34e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 1 | 433 | 3.36e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 2 | 249 | 3.37e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 2 | 160 | 3.37e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 3 | 212 | 3.38e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 1 | 425 | 3.39e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 2 | 89 | 3.39e-01 |
| GO:BP | GO:0051653 | spindle localization | 1 | 52 | 3.39e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 1 | 427 | 3.40e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 1 | 29 | 3.41e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 3.41e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 1 | 122 | 3.41e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 1 | 41 | 3.41e-01 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 1 | 70 | 3.41e-01 |
| GO:BP | GO:0050890 | cognition | 2 | 227 | 3.42e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 2 | 1484 | 3.42e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 2 | 459 | 3.43e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 6 | 884 | 3.43e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 1 | 52 | 3.43e-01 |
| GO:BP | GO:0009314 | response to radiation | 3 | 352 | 3.43e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 1 | 420 | 3.43e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 1 | 59 | 3.43e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 3.43e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 1 | 78 | 3.43e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 1 | 46 | 3.43e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 1 | 37 | 3.43e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 15 | 7069 | 3.43e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 1 | 146 | 3.44e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 1 | 439 | 3.46e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 1 | 71 | 3.46e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 1 | 28 | 3.47e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 1 | 34 | 3.47e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 3 | 274 | 3.47e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 1 | 61 | 3.47e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 1 | 89 | 3.47e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 3.47e-01 |
| GO:BP | GO:0048731 | system development | 24 | 2901 | 3.48e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 172 | 3.48e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 3.48e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 3.48e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 3 | 308 | 3.48e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 3.48e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 3.48e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 1 | 71 | 3.48e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 3.48e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 1 | 59 | 3.49e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 3 | 236 | 3.50e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 1 | 24 | 3.50e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 27 | 3.50e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 7 | 7552 | 3.51e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 3.52e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 7 | 742 | 3.52e-01 |
| GO:BP | GO:0051098 | regulation of binding | 3 | 317 | 3.52e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 1 | 96 | 3.54e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 1 | 40 | 3.54e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 3.54e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 1 | 93 | 3.55e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 1 | 92 | 3.55e-01 |
| GO:BP | GO:0046907 | intracellular transport | 4 | 1453 | 3.55e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 3.57e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 1 | 47 | 3.57e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 1 | 47 | 3.57e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 3.57e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 26 | 3.58e-01 |
| GO:BP | GO:0140014 | mitotic nuclear division | 2 | 251 | 3.59e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 3.62e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 3 | 265 | 3.62e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 1 | 89 | 3.62e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 3.62e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 2 | 246 | 3.62e-01 |
| GO:BP | GO:0031175 | neuron projection development | 5 | 780 | 3.62e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 2 | 137 | 3.62e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 3.62e-01 |
| GO:BP | GO:0048864 | stem cell development | 1 | 76 | 3.63e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 1 | 81 | 3.64e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 2 | 112 | 3.64e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 3.64e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 4 | 407 | 3.65e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 1 | 296 | 3.65e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 2 | 272 | 3.65e-01 |
| GO:BP | GO:0048278 | vesicle docking | 1 | 57 | 3.68e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 1 | 461 | 3.69e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 1 | 80 | 3.69e-01 |
| GO:BP | GO:0048839 | inner ear development | 2 | 132 | 3.69e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 1 | 33 | 3.70e-01 |
| GO:BP | GO:0035270 | endocrine system development | 1 | 86 | 3.71e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 2 | 1229 | 3.71e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 2 | 206 | 3.71e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 1 | 60 | 3.73e-01 |
| GO:BP | GO:0006325 | chromatin organization | 2 | 551 | 3.73e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 1 | 469 | 3.73e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 1 | 53 | 3.75e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 174 | 3.75e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 2 | 183 | 3.77e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 1 | 27 | 3.77e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 1 | 108 | 3.77e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 1 | 55 | 3.77e-01 |
| GO:BP | GO:0048468 | cell development | 3 | 1963 | 3.77e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 1 | 40 | 3.78e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 1 | 42 | 3.78e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 3.79e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 1 | 80 | 3.80e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 2 | 276 | 3.81e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 1 | 90 | 3.82e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 496 | 3.83e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 84 | 3.83e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 1 | 61 | 3.83e-01 |
| GO:BP | GO:0036211 | protein modification process | 3 | 2901 | 3.84e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 3.84e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 1 | 317 | 3.84e-01 |
| GO:BP | GO:0051235 | maintenance of location | 2 | 260 | 3.85e-01 |
| GO:BP | GO:0001843 | neural tube closure | 1 | 90 | 3.85e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 2 | 165 | 3.85e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 94 | 3.85e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 3 | 301 | 3.85e-01 |
| GO:BP | GO:0008015 | blood circulation | 4 | 374 | 3.86e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 3 | 295 | 3.86e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 1 | 75 | 3.86e-01 |
| GO:BP | GO:0060606 | tube closure | 1 | 91 | 3.87e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 7 | 763 | 3.87e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 4 | 552 | 3.87e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 1 | 66 | 3.88e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 3.89e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 3.90e-01 |
| GO:BP | GO:0007610 | behavior | 3 | 436 | 3.91e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 1 | 67 | 3.91e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 400 | 3.92e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 4 | 421 | 3.92e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 2 | 123 | 3.92e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 3.92e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 2 | 217 | 3.93e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 2 | 111 | 3.94e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 1 | 51 | 3.94e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 1 | 28 | 3.94e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 1 | 29 | 3.94e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 3 | 338 | 3.95e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 2 | 209 | 3.96e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 3.96e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 1 | 104 | 3.97e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 3.97e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 3 | 331 | 3.98e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 1 | 94 | 3.98e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 1 | 348 | 3.99e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 3 | 547 | 3.99e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 1 | 69 | 4.00e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 3 | 1898 | 4.00e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 2 | 191 | 4.03e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 1 | 100 | 4.03e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 1 | 72 | 4.03e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 1 | 67 | 4.04e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 4.05e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 1 | 114 | 4.05e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 1 | 98 | 4.05e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 2 | 192 | 4.06e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 2 | 138 | 4.06e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 4 | 429 | 4.09e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 2 | 336 | 4.10e-01 |
| GO:BP | GO:0042592 | homeostatic process | 2 | 1159 | 4.11e-01 |
| GO:BP | GO:0070997 | neuron death | 2 | 286 | 4.12e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 6 | 662 | 4.12e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 3 | 484 | 4.13e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 1 | 120 | 4.14e-01 |
| GO:BP | GO:0002253 | activation of immune response | 2 | 272 | 4.15e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 1 | 38 | 4.16e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 97 | 4.16e-01 |
| GO:BP | GO:0030154 | cell differentiation | 4 | 2958 | 4.17e-01 |
| GO:BP | GO:0001841 | neural tube formation | 1 | 101 | 4.18e-01 |
| GO:BP | GO:0043583 | ear development | 2 | 153 | 4.18e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 4.18e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 1 | 118 | 4.18e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 3 | 3090 | 4.18e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 4.18e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 1 | 52 | 4.18e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 1 | 384 | 4.19e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 1 | 93 | 4.19e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 96 | 4.19e-01 |
| GO:BP | GO:0001510 | RNA methylation | 1 | 87 | 4.19e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 4 | 2984 | 4.20e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 1 | 121 | 4.20e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 72 | 4.20e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 4.20e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 1 | 107 | 4.20e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 6 | 631 | 4.21e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 1 | 116 | 4.21e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 73 | 4.22e-01 |
| GO:BP | GO:0016571 | histone methylation | 1 | 117 | 4.22e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 1 | 82 | 4.22e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 1 | 73 | 4.22e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 1 | 74 | 4.23e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 1 | 612 | 4.23e-01 |
| GO:BP | GO:0044057 | regulation of system process | 1 | 405 | 4.23e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 1 | 83 | 4.23e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 1 | 104 | 4.23e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 73 | 4.23e-01 |
| GO:BP | GO:0009611 | response to wounding | 1 | 415 | 4.24e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 5 | 4953 | 4.24e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 597 | 4.25e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 2 | 1255 | 4.25e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 597 | 4.25e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 2 | 1256 | 4.25e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 4.26e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 1 | 85 | 4.26e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 4.26e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 1 | 123 | 4.26e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 2 | 337 | 4.26e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 2 | 217 | 4.28e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 1 | 710 | 4.28e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 1 | 39 | 4.28e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 1 | 54 | 4.29e-01 |
| GO:BP | GO:0140029 | exocytic process | 1 | 69 | 4.30e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 2 | 581 | 4.32e-01 |
| GO:BP | GO:0003013 | circulatory system process | 1 | 447 | 4.33e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 1 | 89 | 4.34e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 1 | 114 | 4.36e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 1 | 40 | 4.36e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 1 | 539 | 4.38e-01 |
| GO:BP | GO:0016032 | viral process | 3 | 344 | 4.38e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 1 | 64 | 4.40e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 2 | 214 | 4.40e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 4.41e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 4.41e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 1 | 48 | 4.44e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 1 | 48 | 4.44e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 110 | 4.45e-01 |
| GO:BP | GO:0030282 | bone mineralization | 1 | 82 | 4.46e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 1 | 60 | 4.46e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 1 | 97 | 4.46e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 4.46e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 11 | 1380 | 4.46e-01 |
| GO:BP | GO:0006396 | RNA processing | 4 | 929 | 4.46e-01 |
| GO:BP | GO:0007399 | nervous system development | 15 | 1904 | 4.48e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 4.48e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 1 | 76 | 4.49e-01 |
| GO:BP | GO:0002376 | immune system process | 4 | 1447 | 4.49e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 4.49e-01 |
| GO:BP | GO:0022406 | membrane docking | 1 | 84 | 4.51e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 4.51e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 681 | 4.51e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 1 | 665 | 4.51e-01 |
| GO:BP | GO:0006281 | DNA repair | 4 | 534 | 4.51e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 1 | 107 | 4.54e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 1 | 79 | 4.54e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 4.54e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 4.54e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 2 | 204 | 4.55e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 1 | 719 | 4.57e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 2 | 268 | 4.57e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 1 | 120 | 4.57e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 2 | 268 | 4.57e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 4.57e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 1 | 72 | 4.58e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 4.59e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 4.59e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 798 | 4.59e-01 |
| GO:BP | GO:0048666 | neuron development | 5 | 877 | 4.59e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 4.59e-01 |
| GO:BP | GO:0008585 | female gonad development | 1 | 77 | 4.59e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 1 | 116 | 4.60e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 1 | 74 | 4.60e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 6 | 1061 | 4.60e-01 |
| GO:BP | GO:0007165 | signal transduction | 5 | 3818 | 4.61e-01 |
| GO:BP | GO:0051225 | spindle assembly | 1 | 120 | 4.62e-01 |
| GO:BP | GO:0006955 | immune response | 3 | 882 | 4.62e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 4.62e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 1 | 475 | 4.62e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 4.62e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 5 | 877 | 4.63e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 1 | 135 | 4.63e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 4.63e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 2 | 236 | 4.64e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 1 | 66 | 4.64e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 4.64e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 740 | 4.66e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 1 | 88 | 4.66e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 4.66e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 4 | 3386 | 4.67e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 1 | 153 | 4.67e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 1 | 79 | 4.67e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 4.69e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 1 | 96 | 4.69e-01 |
| GO:BP | GO:0042116 | macrophage activation | 1 | 57 | 4.69e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 15 | 7806 | 4.69e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 2 | 232 | 4.70e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1 | 114 | 4.70e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 1 | 74 | 4.70e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 1 | 133 | 4.70e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 4.72e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 4.72e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 1 | 98 | 4.72e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 1 | 89 | 4.73e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 1 | 91 | 4.73e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 1 | 147 | 4.77e-01 |
| GO:BP | GO:0021510 | spinal cord development | 1 | 59 | 4.78e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 1 | 129 | 4.78e-01 |
| GO:BP | GO:0006869 | lipid transport | 1 | 272 | 4.78e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 1 | 90 | 4.78e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 1 | 71 | 4.78e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 3 | 357 | 4.81e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 1 | 92 | 4.83e-01 |
| GO:BP | GO:0048284 | organelle fusion | 1 | 135 | 4.83e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 1 | 56 | 4.83e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 1 | 86 | 4.83e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 1 | 56 | 4.83e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 1 | 447 | 4.87e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 4.88e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 1 | 88 | 4.88e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 609 | 4.89e-01 |
| GO:BP | GO:0001816 | cytokine production | 1 | 451 | 4.89e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 1 | 86 | 4.89e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 1 | 72 | 4.89e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 1 | 126 | 4.89e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 1 | 140 | 4.90e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 4.90e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 1 | 834 | 4.93e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 1 | 95 | 4.97e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 4.98e-01 |
| GO:BP | GO:0021549 | cerebellum development | 1 | 78 | 4.98e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 1 | 159 | 4.99e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 1 | 139 | 5.00e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 1 | 116 | 5.00e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 1 | 556 | 5.00e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 5.00e-01 |
| GO:BP | GO:0048699 | generation of neurons | 6 | 1119 | 5.01e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 1 | 108 | 5.01e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 1 | 230 | 5.01e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 5.01e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 6 | 755 | 5.02e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 1 | 91 | 5.02e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 1 | 101 | 5.04e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 1 | 161 | 5.04e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 1 | 99 | 5.04e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 4 | 534 | 5.04e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 1 | 77 | 5.04e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 1 | 135 | 5.05e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 1 | 86 | 5.06e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 5.06e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 5.06e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 1 | 119 | 5.06e-01 |
| GO:BP | GO:0010876 | lipid localization | 1 | 310 | 5.07e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 1 | 103 | 5.09e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 1 | 132 | 5.09e-01 |
| GO:BP | GO:0051641 | cellular localization | 7 | 2810 | 5.09e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 1 | 90 | 5.09e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 5.09e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 1 | 150 | 5.10e-01 |
| GO:BP | GO:0016570 | histone modification | 2 | 433 | 5.10e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 1 | 143 | 5.11e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 1 | 65 | 5.12e-01 |
| GO:BP | GO:0051604 | protein maturation | 1 | 259 | 5.13e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 115 | 5.13e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 5.15e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 1 | 242 | 5.16e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 2 | 165 | 5.16e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 7 | 1028 | 5.19e-01 |
| GO:BP | GO:0007409 | axonogenesis | 3 | 359 | 5.19e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 4 | 1836 | 5.20e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 5.21e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 2 | 227 | 5.23e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 1 | 170 | 5.23e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 1 | 102 | 5.23e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 1 | 69 | 5.23e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 1 | 97 | 5.23e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 4 | 551 | 5.25e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 2 | 249 | 5.28e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 5.28e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 97 | 5.29e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 1 | 140 | 5.30e-01 |
| GO:BP | GO:0023052 | signaling | 5 | 4141 | 5.30e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 2 | 369 | 5.33e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 5.34e-01 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 5.36e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 1 | 906 | 5.37e-01 |
| GO:BP | GO:0000280 | nuclear division | 2 | 344 | 5.37e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 7 | 963 | 5.37e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 5.38e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 1 | 111 | 5.38e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 5.38e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 5.38e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 1 | 106 | 5.38e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 1 | 170 | 5.38e-01 |
| GO:BP | GO:0019058 | viral life cycle | 2 | 244 | 5.39e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 2 | 154 | 5.39e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 4 | 516 | 5.40e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 1 | 185 | 5.40e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 1 | 112 | 5.41e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 1 | 302 | 5.42e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 5.43e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 1 | 86 | 5.44e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 1 | 98 | 5.44e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 2 | 185 | 5.44e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 2 | 157 | 5.44e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 1 | 59 | 5.44e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 5.44e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 2 | 375 | 5.45e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 2 | 236 | 5.45e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 1 | 203 | 5.45e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 159 | 5.45e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 1 | 104 | 5.45e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 2 | 376 | 5.45e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 14 | 1939 | 5.45e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 5.45e-01 |
| GO:BP | GO:0007154 | cell communication | 5 | 4226 | 5.45e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 5.46e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 5.47e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 156 | 5.48e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 5.49e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 1 | 88 | 5.52e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 2 | 316 | 5.54e-01 |
| GO:BP | GO:0007254 | JNK cascade | 1 | 138 | 5.54e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 5.56e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 1 | 104 | 5.58e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 1 | 104 | 5.58e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 1 | 140 | 5.59e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 1 | 199 | 5.60e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 5.60e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 994 | 5.61e-01 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 1 | 179 | 5.62e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 1 | 121 | 5.63e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 2 | 199 | 5.65e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 5.65e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 1 | 120 | 5.65e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 1 | 598 | 5.67e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 2 | 273 | 5.69e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 1 | 121 | 5.70e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 5.70e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 1 | 102 | 5.72e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 1 | 101 | 5.75e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 4 | 535 | 5.75e-01 |
| GO:BP | GO:0061564 | axon development | 3 | 398 | 5.76e-01 |
| GO:BP | GO:0008104 | protein localization | 5 | 2129 | 5.77e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 1 | 197 | 5.78e-01 |
| GO:BP | GO:0006959 | humoral immune response | 1 | 80 | 5.78e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 3 | 608 | 5.78e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 1 | 170 | 5.79e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 5 | 2138 | 5.80e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 1 | 216 | 5.80e-01 |
| GO:BP | GO:0048285 | organelle fission | 2 | 388 | 5.81e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 1 | 154 | 5.82e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 5.82e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 3 | 640 | 5.82e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 1 | 1149 | 5.84e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 5.85e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 1 | 157 | 5.87e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 1 | 91 | 5.89e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 1 | 91 | 5.89e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 1 | 194 | 5.89e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 998 | 5.92e-01 |
| GO:BP | GO:0016477 | cell migration | 8 | 1081 | 5.92e-01 |
| GO:BP | GO:0048870 | cell motility | 9 | 1182 | 5.96e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 5.96e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 2 | 344 | 5.96e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 1 | 94 | 5.99e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 1 | 94 | 5.99e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 142 | 5.99e-01 |
| GO:BP | GO:0009451 | RNA modification | 1 | 162 | 6.01e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 2 | 1335 | 6.04e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 3 | 656 | 6.05e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 7 | 957 | 6.06e-01 |
| GO:BP | GO:0009056 | catabolic process | 2 | 2088 | 6.10e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 1 | 116 | 6.13e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 1 | 105 | 6.13e-01 |
| GO:BP | GO:0030534 | adult behavior | 1 | 92 | 6.13e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 8 | 1226 | 6.13e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 6.13e-01 |
| GO:BP | GO:0042063 | gliogenesis | 1 | 235 | 6.16e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 6.16e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 2 | 300 | 6.21e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 4 | 760 | 6.22e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 7 | 8061 | 6.22e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 6.25e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 1 | 150 | 6.37e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 6.37e-01 |
| GO:BP | GO:0008544 | epidermis development | 1 | 210 | 6.37e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 2 | 1496 | 6.37e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 6.37e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 1 | 138 | 6.37e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 2 | 357 | 6.38e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1201 | 6.38e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 6.41e-01 |
| GO:BP | GO:0016358 | dendrite development | 1 | 202 | 6.42e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 1 | 256 | 6.42e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 6.43e-01 |
| GO:BP | GO:0050896 | response to stimulus | 5 | 5770 | 6.43e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1233 | 6.46e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 6.49e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 213 | 6.50e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 3 | 4241 | 6.51e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 2 | 4128 | 6.51e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 1 | 189 | 6.51e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 1 | 190 | 6.54e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 1 | 1389 | 6.55e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 128 | 6.60e-01 |
| GO:BP | GO:0006996 | organelle organization | 2 | 3000 | 6.60e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 6.60e-01 |
| GO:BP | GO:0016197 | endosomal transport | 1 | 231 | 6.61e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 1 | 228 | 6.63e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 1 | 196 | 6.65e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 6.66e-01 |
| GO:BP | GO:0006954 | inflammatory response | 1 | 429 | 6.66e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 6.66e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 3 | 463 | 6.67e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 2 | 448 | 6.67e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 1 | 200 | 6.67e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 6.67e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 6.67e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 4 | 639 | 6.69e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 6.72e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 6.72e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 6.72e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 138 | 6.73e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 1 | 540 | 6.73e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 6.75e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 6.75e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 1 | 163 | 6.76e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 1 | 166 | 6.76e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1 | 135 | 6.78e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 1 | 242 | 6.78e-01 |
| GO:BP | GO:0045087 | innate immune response | 2 | 488 | 6.81e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 1 | 111 | 6.82e-01 |
| GO:BP | GO:0048732 | gland development | 1 | 318 | 6.83e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 1 | 108 | 6.83e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 1 | 186 | 6.88e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 6.90e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 149 | 6.90e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 6.90e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 1 | 214 | 6.91e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 154 | 6.91e-01 |
| GO:BP | GO:0051276 | chromosome organization | 3 | 543 | 6.91e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 6.91e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 1 | 168 | 6.92e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 1 | 151 | 6.92e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 1 | 258 | 6.93e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 2 | 369 | 6.96e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 6.96e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 1 | 105 | 6.96e-01 |
| GO:BP | GO:0048469 | cell maturation | 1 | 108 | 6.99e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 1 | 292 | 7.00e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 1 | 218 | 7.00e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 7.03e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 7.04e-01 |
| GO:BP | GO:0015031 | protein transport | 2 | 1361 | 7.05e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 1 | 145 | 7.07e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 213 | 7.07e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 7.07e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 2 | 260 | 7.07e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 3 | 478 | 7.07e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 251 | 7.07e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 1 | 667 | 7.07e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 1 | 180 | 7.07e-01 |
| GO:BP | GO:0032502 | developmental process | 2 | 4494 | 7.07e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 7.07e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 7.10e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 1 | 234 | 7.10e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 7.11e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 7.12e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 5 | 2450 | 7.13e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 2 | 512 | 7.17e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 1 | 155 | 7.17e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 1 | 198 | 7.18e-01 |
| GO:BP | GO:0016485 | protein processing | 1 | 183 | 7.18e-01 |
| GO:BP | GO:0050821 | protein stabilization | 1 | 191 | 7.19e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 7.19e-01 |
| GO:BP | GO:0009566 | fertilization | 1 | 93 | 7.19e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 1 | 124 | 7.28e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 2 | 527 | 7.29e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 7.29e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 1 | 135 | 7.30e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 7.30e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 273 | 7.31e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 7.31e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 7.31e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 2 | 531 | 7.31e-01 |
| GO:BP | GO:0051301 | cell division | 1 | 570 | 7.31e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 2 | 194 | 7.33e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 2 | 427 | 7.35e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 1 | 172 | 7.39e-01 |
| GO:BP | GO:0016050 | vesicle organization | 1 | 320 | 7.40e-01 |
| GO:BP | GO:0065007 | biological regulation | 7 | 8320 | 7.40e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 2 | 551 | 7.45e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1138 | 7.46e-01 |
| GO:BP | GO:0002252 | immune effector process | 3 | 318 | 7.47e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 1 | 358 | 7.52e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 7.52e-01 |
| GO:BP | GO:0051668 | localization within membrane | 4 | 605 | 7.58e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 7.58e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 2 | 294 | 7.59e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 2 | 414 | 7.61e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 7.61e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 1 | 252 | 7.65e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 7.65e-01 |
| GO:BP | GO:0071702 | organic substance transport | 3 | 2116 | 7.68e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 236 | 7.69e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 3 | 4955 | 7.73e-01 |
| GO:BP | GO:0060271 | cilium assembly | 1 | 292 | 7.75e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 1 | 160 | 7.77e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 1 | 378 | 7.81e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 1 | 381 | 7.83e-01 |
| GO:BP | GO:0034330 | cell junction organization | 3 | 575 | 7.83e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 7.83e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 7.83e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 7.83e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 7.83e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 1 | 269 | 7.83e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 7.83e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 7.83e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 2 | 532 | 7.90e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 1 | 387 | 7.92e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 2 | 422 | 7.94e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 1 | 211 | 7.94e-01 |
| GO:BP | GO:0044782 | cilium organization | 1 | 313 | 7.97e-01 |
| GO:BP | GO:0051640 | organelle localization | 2 | 503 | 7.98e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 2 | 486 | 7.99e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 4 | 761 | 8.00e-01 |
| GO:BP | GO:0006952 | defense response | 3 | 944 | 8.05e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 1 | 406 | 8.12e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 8.13e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 1 | 192 | 8.14e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 2 | 1684 | 8.16e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 8.17e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 250 | 8.17e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 2 | 4794 | 8.18e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 1 | 245 | 8.20e-01 |
| GO:BP | GO:0061024 | membrane organization | 2 | 671 | 8.22e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 1 | 271 | 8.23e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 8.27e-01 |
| GO:BP | GO:0006909 | phagocytosis | 1 | 148 | 8.28e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 8.29e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 1 | 357 | 8.29e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 1 | 404 | 8.29e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 8.30e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 901 | 8.30e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 8.32e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 2 | 701 | 8.33e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 1 | 362 | 8.33e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 1 | 311 | 8.36e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 1 | 351 | 8.38e-01 |
| GO:BP | GO:0003008 | system process | 5 | 1189 | 8.39e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 3 | 523 | 8.41e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 8.41e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 1 | 294 | 8.41e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 1 | 290 | 8.43e-01 |
| GO:BP | GO:0006887 | exocytosis | 1 | 256 | 8.43e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 250 | 8.44e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 1 | 2102 | 8.50e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 3 | 620 | 8.50e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 665 | 8.50e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 1 | 95 | 8.52e-01 |
| GO:BP | GO:0021700 | developmental maturation | 1 | 211 | 8.52e-01 |
| GO:BP | GO:0050877 | nervous system process | 3 | 634 | 8.55e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 2 | 382 | 8.55e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 1 | 401 | 8.65e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 1 | 142 | 8.68e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 8.70e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 1 | 408 | 8.70e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 1 | 378 | 8.72e-01 |
| GO:BP | GO:0006810 | transport | 5 | 3298 | 8.82e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 1 | 190 | 8.83e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 1 | 330 | 8.91e-01 |
| GO:BP | GO:0048232 | male gamete generation | 1 | 343 | 8.98e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 3 | 1199 | 8.98e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 2 | 560 | 8.98e-01 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1267 | 9.00e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 6 | 1238 | 9.00e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 2 | 615 | 9.02e-01 |
| GO:BP | GO:0051234 | establishment of localization | 5 | 3433 | 9.02e-01 |
| GO:BP | GO:0006935 | chemotaxis | 1 | 389 | 9.04e-01 |
| GO:BP | GO:0042330 | taxis | 1 | 389 | 9.04e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 9.04e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 1 | 275 | 9.05e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 2 | 625 | 9.05e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 1 | 784 | 9.08e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 2 | 637 | 9.10e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 9.10e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 9.12e-01 |
| GO:BP | GO:0051179 | localization | 6 | 3969 | 9.13e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 1 | 293 | 9.13e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 9.13e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 9.14e-01 |
| GO:BP | GO:0032940 | secretion by cell | 2 | 569 | 9.17e-01 |
| GO:BP | GO:0016043 | cellular component organization | 3 | 5076 | 9.18e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 344 | 9.18e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 1 | 421 | 9.28e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 9.28e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 9.30e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3 | 5274 | 9.31e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 1 | 506 | 9.34e-01 |
| GO:BP | GO:0140352 | export from cell | 2 | 611 | 9.36e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 1 | 517 | 9.37e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 2 | 706 | 9.37e-01 |
| GO:BP | GO:0007276 | gamete generation | 1 | 461 | 9.38e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 9.38e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 1 | 540 | 9.43e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 2 | 717 | 9.44e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 9.46e-01 |
| GO:BP | GO:0030001 | metal ion transport | 1 | 562 | 9.50e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 1 | 376 | 9.50e-01 |
| GO:BP | GO:0046903 | secretion | 2 | 645 | 9.51e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 1 | 563 | 9.53e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 1 | 538 | 9.60e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 2 | 787 | 9.60e-01 |
| GO:BP | GO:0070925 | organelle assembly | 2 | 817 | 9.61e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 1 | 373 | 9.65e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 556 | 9.65e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 1 | 632 | 9.67e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 1 | 669 | 9.71e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 1 | 631 | 9.77e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 9.77e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 9.77e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 9.77e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 9.79e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 2 | 898 | 9.79e-01 |
| GO:BP | GO:0006897 | endocytosis | 1 | 527 | 9.85e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 1 | 775 | 9.85e-01 |
| GO:BP | GO:0006508 | proteolysis | 4 | 1318 | 9.88e-01 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| KEGG | KEGG:04115 | p53 signaling pathway | 4 | 65 | 1.41e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 8 | 415 | 2.58e-02 |
| KEGG | KEGG:04330 | Notch signaling pathway | 3 | 51 | 7.82e-02 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 2 | 70 | 8.97e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 2 | 87 | 8.97e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 3 | 75 | 8.97e-02 |
| KEGG | KEGG:04210 | Apoptosis | 3 | 118 | 1.84e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 1 | 84 | 2.04e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 2 | 150 | 2.04e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 2.04e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 2 | 53 | 2.13e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 1 | 49 | 2.48e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 2.48e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 2.48e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 2.48e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 2.48e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 2.48e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 2.48e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 2.48e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 5 | 409 | 2.48e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 2 | 124 | 2.48e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 1 | 58 | 2.48e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1 | 69 | 2.48e-01 |
| KEGG | KEGG:05134 | Legionellosis | 1 | 39 | 2.48e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 2.48e-01 |
| KEGG | KEGG:05142 | Chagas disease | 1 | 75 | 2.48e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 1 | 82 | 2.48e-01 |
| KEGG | KEGG:05162 | Measles | 2 | 98 | 2.48e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 2.48e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 1 | 112 | 2.48e-01 |
| KEGG | KEGG:04530 | Tight junction | 1 | 137 | 2.48e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 2.48e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 2 | 84 | 2.48e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 2.48e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 2 | 71 | 2.48e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 3 | 138 | 2.48e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 1 | 145 | 2.48e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 1 | 133 | 2.48e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 2.48e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 1 | 30 | 2.48e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 2.48e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 2.48e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 1 | 32 | 2.48e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 1 | 135 | 2.48e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 2.51e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 1 | 110 | 2.56e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 2.59e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 1 | 87 | 2.59e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 2.61e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 1 | 112 | 2.69e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 1 | 97 | 2.75e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 1 | 180 | 2.82e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 2.82e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 2 | 145 | 2.82e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 2.82e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 1 | 115 | 2.82e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 2 | 153 | 2.82e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 1 | 176 | 2.82e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 1 | 172 | 2.82e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 1 | 165 | 2.82e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 2.82e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 2.82e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 1 | 41 | 2.82e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 2.82e-01 |
| KEGG | KEGG:05164 | Influenza A | 1 | 103 | 2.82e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 1 | 65 | 2.82e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 2.82e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 1 | 57 | 2.82e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 2.82e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 1 | 186 | 2.82e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 2.82e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 2 | 134 | 2.82e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 2 | 128 | 2.82e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 1 | 44 | 2.82e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 2.82e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 2.87e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 1 | 75 | 2.90e-01 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 2.90e-01 |
| KEGG | KEGG:05131 | Shigellosis | 1 | 210 | 2.90e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 1 | 214 | 2.90e-01 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 2.90e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 2 | 171 | 3.54e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 1 | 104 | 3.63e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 1 | 74 | 3.63e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 1 | 109 | 4.34e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 1 | 106 | 4.35e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 2 | 149 | 4.40e-01 |
| KEGG | KEGG:05224 | Breast cancer | 1 | 117 | 4.55e-01 |
| KEGG | KEGG:00000 | KEGG root term | 5 | 5558 | 4.55e-01 |
| KEGG | KEGG:04110 | Cell cycle | 1 | 151 | 4.71e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 5.18e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 1 | 272 | 6.25e-01 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 6.27e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 1 | 240 | 6.80e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 7.04e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 9.92e-01 |
# ---------------------------------------------------------
#
# MTgostres24 <- gost(query = sigVMT24$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(MTgostres24,"data/DEG-GO/MTgostres24.RDS")
MTgostres24 <- readRDS("data/DEG-GO/MTgostres24.RDS")
# MTp24 <- gostplot(MTgostres24, capped = FALSE, interactive = TRUE)
# MTp24
MTtable124 <- MTgostres24$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
MTtable124 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('MTX 24 hour GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)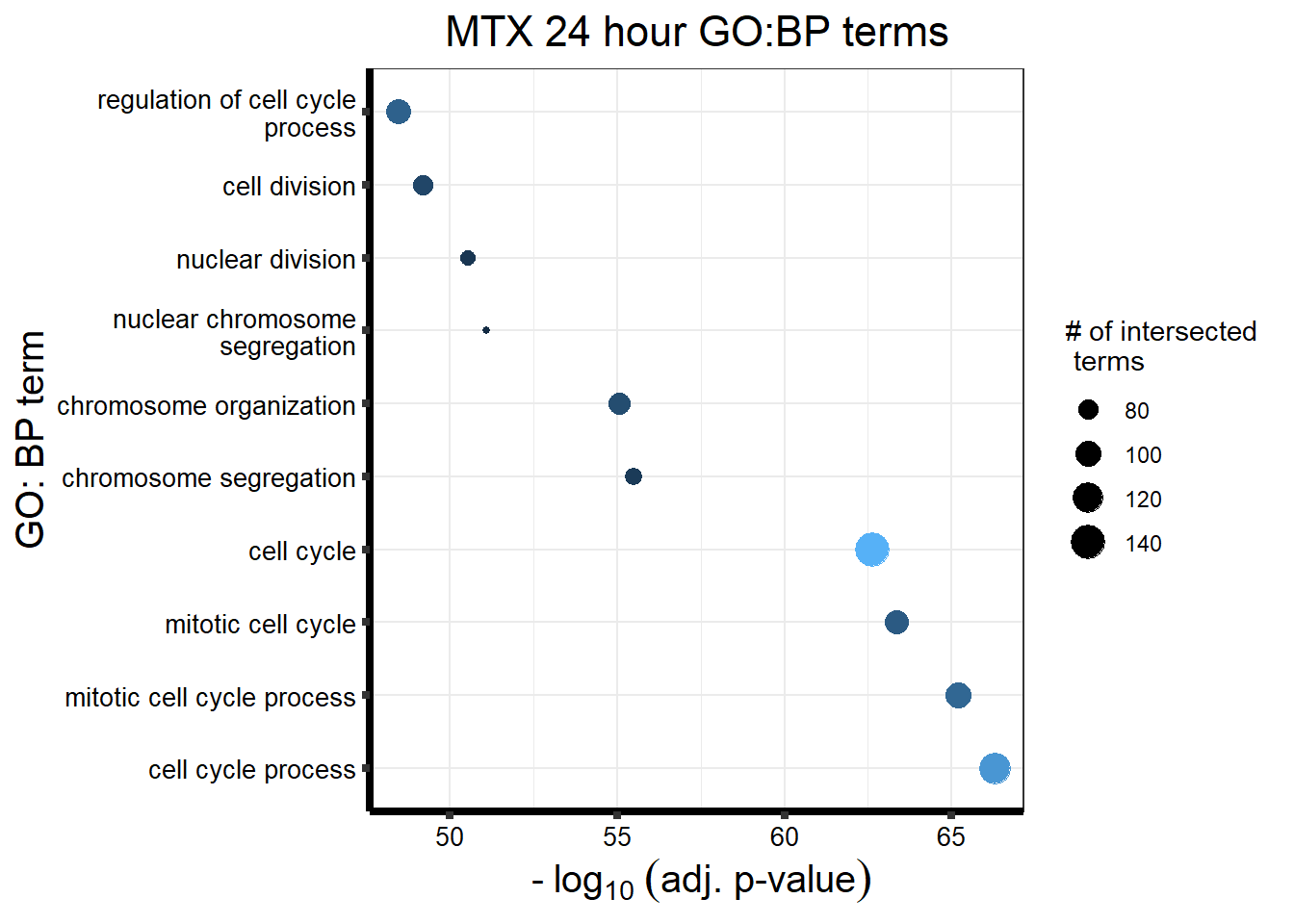
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
MTtable124 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "MTX 24hour enrichment of terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 126 | 1086 | 5.35e-67 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 100 | 694 | 6.64e-66 |
| GO:BP | GO:0000278 | mitotic cell cycle | 93 | 827 | 4.39e-64 |
| GO:BP | GO:0007049 | cell cycle | 140 | 1529 | 2.35e-63 |
| GO:BP | GO:0007059 | chromosome segregation | 74 | 369 | 3.09e-56 |
| GO:BP | GO:0051276 | chromosome organization | 85 | 543 | 8.59e-56 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 64 | 273 | 7.99e-52 |
| GO:BP | GO:0000280 | nuclear division | 71 | 344 | 2.85e-51 |
| GO:BP | GO:0051301 | cell division | 81 | 570 | 6.15e-50 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 97 | 626 | 3.22e-49 |
| GO:BP | GO:0140014 | mitotic nuclear division | 59 | 251 | 7.41e-49 |
| GO:BP | GO:0000819 | sister chromatid segregation | 55 | 217 | 8.59e-49 |
| GO:BP | GO:0048285 | organelle fission | 71 | 388 | 2.69e-48 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 51 | 179 | 6.31e-48 |
| GO:BP | GO:0051726 | regulation of cell cycle | 125 | 956 | 1.90e-42 |
| GO:BP | GO:0044770 | cell cycle phase transition | 78 | 504 | 7.72e-40 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 62 | 148 | 1.92e-38 |
| GO:BP | GO:0006260 | DNA replication | 79 | 255 | 3.02e-37 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 74 | 391 | 1.06e-35 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 52 | 183 | 8.48e-35 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 87 | 418 | 1.18e-34 |
| GO:BP | GO:0006259 | DNA metabolic process | 148 | 884 | 1.43e-32 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 74 | 441 | 7.44e-32 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 44 | 139 | 1.14e-31 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 54 | 235 | 4.46e-30 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 60 | 308 | 7.19e-30 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 34 | 157 | 9.90e-30 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 51 | 273 | 1.78e-29 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 33 | 121 | 3.66e-29 |
| GO:BP | GO:0006974 | DNA damage response | 125 | 785 | 4.29e-29 |
| GO:BP | GO:0007051 | spindle organization | 26 | 188 | 1.88e-28 |
| GO:BP | GO:0006281 | DNA repair | 99 | 534 | 3.77e-28 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 42 | 209 | 2.13e-27 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 51 | 552 | 7.55e-27 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 26 | 71 | 1.11e-26 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 30 | 100 | 1.86e-26 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 59 | 338 | 2.07e-26 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 55 | 211 | 2.51e-26 |
| GO:BP | GO:0007052 | mitotic spindle organization | 22 | 129 | 4.07e-26 |
| GO:BP | GO:0051304 | chromosome separation | 26 | 76 | 9.67e-26 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 22 | 91 | 1.45e-25 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 22 | 92 | 3.03e-25 |
| GO:BP | GO:0051783 | regulation of nuclear division | 31 | 116 | 7.39e-25 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 22 | 99 | 1.96e-24 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 21 | 88 | 3.23e-24 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 44 | 274 | 4.58e-24 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 21 | 89 | 6.55e-24 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 18 | 58 | 1.56e-23 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 18 | 61 | 4.14e-23 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 40 | 166 | 4.17e-23 |
| GO:BP | GO:0007017 | microtubule-based process | 55 | 760 | 1.30e-22 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 20 | 46 | 1.91e-22 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 27 | 236 | 2.81e-22 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 17 | 42 | 2.81e-22 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 16 | 44 | 3.61e-22 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 16 | 44 | 3.61e-22 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 16 | 44 | 3.61e-22 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 20 | 47 | 4.48e-22 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 20 | 47 | 4.48e-22 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 20 | 47 | 4.48e-22 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 20 | 47 | 4.48e-22 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 16 | 45 | 5.01e-22 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 27 | 54 | 5.33e-22 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 20 | 48 | 1.09e-21 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 20 | 48 | 1.09e-21 |
| GO:BP | GO:0006270 | DNA replication initiation | 23 | 35 | 1.44e-21 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 19 | 90 | 2.66e-21 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 20 | 52 | 4.03e-21 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 33 | 129 | 6.82e-21 |
| GO:BP | GO:0006996 | organelle organization | 101 | 3000 | 1.04e-20 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 27 | 57 | 2.45e-20 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 23 | 47 | 8.25e-20 |
| GO:BP | GO:0051225 | spindle assembly | 28 | 120 | 2.25e-19 |
| GO:BP | GO:0051310 | metaphase plate congression | 21 | 69 | 7.00e-19 |
| GO:BP | GO:0050000 | chromosome localization | 20 | 82 | 6.46e-18 |
| GO:BP | GO:0032392 | DNA geometric change | 28 | 80 | 7.41e-18 |
| GO:BP | GO:0071103 | DNA conformation change | 29 | 87 | 8.99e-18 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 33 | 149 | 1.04e-17 |
| GO:BP | GO:0051321 | meiotic cell cycle | 32 | 188 | 1.04e-17 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 27 | 74 | 1.08e-17 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 21 | 75 | 2.16e-17 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 31 | 134 | 3.44e-17 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 46 | 135 | 5.50e-17 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 18 | 55 | 8.78e-17 |
| GO:BP | GO:0140013 | meiotic nuclear division | 26 | 120 | 3.09e-16 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 22 | 357 | 4.36e-16 |
| GO:BP | GO:0033554 | cellular response to stress | 170 | 1681 | 4.56e-16 |
| GO:BP | GO:0033260 | nuclear DNA replication | 23 | 35 | 7.54e-16 |
| GO:BP | GO:0006275 | regulation of DNA replication | 29 | 116 | 1.59e-15 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 13 | 70 | 1.87e-15 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 24 | 40 | 1.95e-15 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 27 | 121 | 2.28e-15 |
| GO:BP | GO:0006310 | DNA recombination | 55 | 257 | 2.34e-15 |
| GO:BP | GO:0006302 | double-strand break repair | 49 | 271 | 2.45e-15 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 23 | 85 | 4.31e-15 |
| GO:BP | GO:0033043 | regulation of organelle organization | 59 | 1028 | 6.67e-15 |
| GO:BP | GO:0007010 | cytoskeleton organization | 36 | 1226 | 1.08e-14 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 32 | 178 | 1.44e-14 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 23 | 92 | 7.63e-14 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 21 | 81 | 2.58e-13 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 13 | 45 | 2.61e-13 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 18 | 81 | 3.11e-13 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 21 | 103 | 4.02e-13 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 19 | 86 | 6.12e-13 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 14 | 27 | 9.55e-13 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 23 | 103 | 1.02e-12 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 27 | 64 | 1.04e-12 |
| GO:BP | GO:0000910 | cytokinesis | 24 | 164 | 1.04e-12 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 13 | 22 | 1.12e-12 |
| GO:BP | GO:0000725 | recombinational repair | 36 | 153 | 1.41e-12 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 10 | 18 | 1.50e-12 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 31 | 81 | 1.89e-12 |
| GO:BP | GO:0051231 | spindle elongation | 8 | 12 | 2.09e-12 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 46 | 149 | 3.06e-12 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 67 | 452 | 3.92e-12 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 23 | 48 | 8.75e-12 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 11 | 28 | 1.01e-11 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 20 | 113 | 1.15e-11 |
| GO:BP | GO:0034508 | centromere complex assembly | 15 | 28 | 1.19e-11 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 20 | 318 | 1.39e-11 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 9 | 1.42e-11 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 17 | 79 | 1.55e-11 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 8 | 21 | 2.27e-11 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 14 | 104 | 2.56e-11 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 10 | 4.06e-11 |
| GO:BP | GO:0051302 | regulation of cell division | 22 | 145 | 6.81e-11 |
| GO:BP | GO:1902969 | mitotic DNA replication | 10 | 14 | 8.66e-11 |
| GO:BP | GO:0034502 | protein localization to chromosome | 31 | 110 | 1.06e-10 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 10 | 11 | 1.07e-10 |
| GO:BP | GO:0007127 | meiosis I | 28 | 77 | 1.32e-10 |
| GO:BP | GO:0051383 | kinetochore organization | 9 | 23 | 1.92e-10 |
| GO:BP | GO:0051255 | spindle midzone assembly | 8 | 12 | 2.28e-10 |
| GO:BP | GO:0007098 | centrosome cycle | 28 | 127 | 2.35e-10 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 36 | 175 | 2.53e-10 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 12 | 33 | 3.70e-10 |
| GO:BP | GO:1900262 | regulation of DNA-directed DNA polymerase activity | 10 | 13 | 4.19e-10 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 10 | 13 | 4.19e-10 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 16 | 42 | 1.45e-09 |
| GO:BP | GO:0051656 | establishment of organelle localization | 26 | 378 | 1.67e-09 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 7 | 14 | 1.84e-09 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 28 | 137 | 2.18e-09 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 7 | 19 | 2.38e-09 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 7 | 19 | 2.38e-09 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 7 | 19 | 2.38e-09 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 10 | 15 | 2.90e-09 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 9 | 15 | 3.58e-09 |
| GO:BP | GO:0070925 | organelle assembly | 23 | 817 | 3.62e-09 |
| GO:BP | GO:0030261 | chromosome condensation | 10 | 33 | 3.72e-09 |
| GO:BP | GO:0010212 | response to ionizing radiation | 21 | 130 | 5.24e-09 |
| GO:BP | GO:0090224 | regulation of spindle organization | 12 | 45 | 5.52e-09 |
| GO:BP | GO:0051640 | organelle localization | 21 | 503 | 6.81e-09 |
| GO:BP | GO:0051653 | spindle localization | 11 | 52 | 1.10e-08 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 17 | 85 | 1.82e-08 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 17 | 185 | 2.25e-08 |
| GO:BP | GO:0031297 | replication fork processing | 18 | 42 | 2.48e-08 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 17 | 88 | 2.74e-08 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 10 | 18 | 2.82e-08 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 10 | 18 | 2.82e-08 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 11 | 42 | 3.23e-08 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 11 | 52 | 4.80e-08 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 6 | 18 | 6.57e-08 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 36 | 6.85e-08 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 10 | 60 | 7.27e-08 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 17 | 204 | 7.36e-08 |
| GO:BP | GO:0051293 | establishment of spindle localization | 10 | 48 | 7.77e-08 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 6 | 19 | 7.84e-08 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 6 | 25 | 8.62e-08 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 34 | 2011 | 9.26e-08 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 10 | 62 | 9.64e-08 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 18 | 604 | 1.14e-07 |
| GO:BP | GO:0035825 | homologous recombination | 17 | 43 | 1.38e-07 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 7 | 8 | 1.48e-07 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 8 | 35 | 1.48e-07 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 9 | 50 | 1.72e-07 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 10 | 27 | 2.25e-07 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 6 | 12 | 2.75e-07 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 6 | 12 | 2.75e-07 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 15 | 36 | 4.59e-07 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 5 | 4.63e-07 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 5 | 7 | 4.63e-07 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 6 | 7 | 5.93e-07 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 29 | 161 | 6.20e-07 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 10 | 30 | 6.68e-07 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 29 | 207 | 6.71e-07 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 23 | 229 | 7.90e-07 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 11 | 68 | 1.25e-06 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 10 | 19 | 1.34e-06 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 14 | 43 | 1.47e-06 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 5 | 5 | 1.60e-06 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 8 | 36 | 1.60e-06 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 5 | 5 | 1.60e-06 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 5 | 5 | 1.60e-06 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 20 | 77 | 1.79e-06 |
| GO:BP | GO:0051781 | positive regulation of cell division | 12 | 64 | 1.83e-06 |
| GO:BP | GO:0051298 | centrosome duplication | 16 | 68 | 2.16e-06 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 6 | 11 | 2.46e-06 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 45 | 265 | 2.57e-06 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 7 | 8 | 3.67e-06 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 7 | 8 | 3.67e-06 |
| GO:BP | GO:0051382 | kinetochore assembly | 4 | 18 | 3.92e-06 |
| GO:BP | GO:0051338 | regulation of transferase activity | 45 | 679 | 4.56e-06 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 7 | 11 | 4.61e-06 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 5 | 8 | 4.76e-06 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 5 | 7 | 5.13e-06 |
| GO:BP | GO:0019953 | sexual reproduction | 38 | 631 | 5.73e-06 |
| GO:BP | GO:1903932 | regulation of DNA primase activity | 5 | 5 | 6.30e-06 |
| GO:BP | GO:1903934 | positive regulation of DNA primase activity | 5 | 5 | 6.30e-06 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 7 | 31 | 7.69e-06 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 6 | 13 | 8.23e-06 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 12 | 146 | 8.56e-06 |
| GO:BP | GO:0009314 | response to radiation | 45 | 352 | 8.57e-06 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 8 | 32 | 9.55e-06 |
| GO:BP | GO:0032506 | cytokinetic process | 8 | 39 | 9.75e-06 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 5 | 8 | 1.22e-05 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 5 | 8 | 1.22e-05 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 13 | 36 | 1.28e-05 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 6 | 25 | 1.28e-05 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 13 | 36 | 1.28e-05 |
| GO:BP | GO:0000212 | meiotic spindle organization | 4 | 14 | 1.29e-05 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 9 | 15 | 1.40e-05 |
| GO:BP | GO:0071478 | cellular response to radiation | 16 | 156 | 1.40e-05 |
| GO:BP | GO:0022616 | DNA strand elongation | 11 | 35 | 1.64e-05 |
| GO:BP | GO:0010332 | response to gamma radiation | 10 | 50 | 2.19e-05 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 7 | 37 | 2.80e-05 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 5 | 2.80e-05 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 25 | 112 | 2.96e-05 |
| GO:BP | GO:0007099 | centriole replication | 11 | 38 | 3.34e-05 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 5 | 6 | 3.94e-05 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 14 | 35 | 4.16e-05 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 14 | 217 | 4.66e-05 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 17 | 149 | 5.22e-05 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 4 | 4 | 5.81e-05 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 5 | 9 | 6.80e-05 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 12 | 51 | 7.04e-05 |
| GO:BP | GO:0098534 | centriole assembly | 11 | 41 | 8.59e-05 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 19 | 111 | 9.13e-05 |
| GO:BP | GO:0007292 | female gamete generation | 7 | 103 | 9.65e-05 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 3 | 5 | 9.92e-05 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 5 | 9 | 1.00e-04 |
| GO:BP | GO:0007144 | female meiosis I | 5 | 9 | 1.01e-04 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 18 | 169 | 1.05e-04 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 18 | 103 | 1.17e-04 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 13 | 34 | 1.24e-04 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 6 | 8 | 1.28e-04 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 9 | 45 | 1.32e-04 |
| GO:BP | GO:0007018 | microtubule-based movement | 11 | 300 | 1.35e-04 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 3 | 8 | 1.35e-04 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 9 | 30 | 1.41e-04 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 14 | 67 | 1.53e-04 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 1.73e-04 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 1.84e-04 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 9 | 30 | 1.84e-04 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 11 | 69 | 2.33e-04 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 3 | 11 | 2.37e-04 |
| GO:BP | GO:0009411 | response to UV | 25 | 141 | 2.71e-04 |
| GO:BP | GO:0006301 | postreplication repair | 10 | 34 | 2.93e-04 |
| GO:BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 7 | 18 | 3.18e-04 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 3 | 13 | 3.36e-04 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 4 | 5 | 3.53e-04 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 16 | 94 | 3.96e-04 |
| GO:BP | GO:0042148 | strand invasion | 5 | 5 | 4.44e-04 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 3 | 12 | 4.69e-04 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 33 | 268 | 4.69e-04 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 33 | 268 | 4.69e-04 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 8 | 52 | 4.80e-04 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 2 | 4 | 4.85e-04 |
| GO:BP | GO:0046599 | regulation of centriole replication | 9 | 21 | 5.25e-04 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 7 | 42 | 5.52e-04 |
| GO:BP | GO:0031399 | regulation of protein modification process | 17 | 1173 | 6.25e-04 |
| GO:BP | GO:0006513 | protein monoubiquitination | 8 | 80 | 6.28e-04 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 3 | 6.28e-04 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 6.28e-04 |
| GO:BP | GO:0035556 | intracellular signal transduction | 35 | 2065 | 6.34e-04 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 10 | 44 | 6.34e-04 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 3 | 4 | 6.59e-04 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 4 | 5 | 6.91e-04 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 10 | 101 | 7.24e-04 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 5 | 9 | 7.50e-04 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 5 | 7 | 7.51e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 8 | 119 | 7.53e-04 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 7 | 14 | 7.80e-04 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 7 | 14 | 7.80e-04 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 7 | 14 | 7.80e-04 |
| GO:BP | GO:0006541 | glutamine metabolic process | 7 | 24 | 7.80e-04 |
| GO:BP | GO:0071168 | protein localization to chromatin | 14 | 48 | 7.89e-04 |
| GO:BP | GO:0019985 | translesion synthesis | 8 | 24 | 8.20e-04 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 14 | 796 | 8.59e-04 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 5 | 18 | 8.66e-04 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 4 | 26 | 9.39e-04 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 3 | 6 | 9.94e-04 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 3 | 6 | 9.94e-04 |
| GO:BP | GO:0006282 | regulation of DNA repair | 29 | 198 | 1.00e-03 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 5 | 19 | 1.12e-03 |
| GO:BP | GO:1903925 | response to bisphenol A | 3 | 4 | 1.12e-03 |
| GO:BP | GO:1903926 | cellular response to bisphenol A | 3 | 4 | 1.12e-03 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 4 | 19 | 1.15e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 4 | 55 | 1.16e-03 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 8 | 19 | 1.23e-03 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 6 | 29 | 1.24e-03 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 4 | 6 | 1.30e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 18 | 1167 | 1.48e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 25 | 459 | 1.48e-03 |
| GO:BP | GO:0051305 | chromosome movement towards spindle pole | 2 | 7 | 1.49e-03 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 3 | 4 | 1.55e-03 |
| GO:BP | GO:0006312 | mitotic recombination | 9 | 22 | 1.59e-03 |
| GO:BP | GO:0022414 | reproductive process | 41 | 927 | 1.59e-03 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 3 | 7 | 1.63e-03 |
| GO:BP | GO:0000003 | reproduction | 41 | 934 | 1.80e-03 |
| GO:BP | GO:0010390 | histone monoubiquitination | 5 | 31 | 1.86e-03 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 15 | 400 | 1.86e-03 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 11 | 73 | 1.93e-03 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 3 | 23 | 1.93e-03 |
| GO:BP | GO:0051865 | protein autoubiquitination | 10 | 75 | 2.09e-03 |
| GO:BP | GO:0010165 | response to X-ray | 5 | 29 | 2.10e-03 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 5 | 30 | 2.14e-03 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 14 | 881 | 2.25e-03 |
| GO:BP | GO:0061820 | telomeric D-loop disassembly | 5 | 8 | 2.25e-03 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 3 | 4 | 2.36e-03 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 17 | 439 | 2.39e-03 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 2 | 4 | 2.40e-03 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 3 | 14 | 2.45e-03 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 3 | 8 | 2.60e-03 |
| GO:BP | GO:1905463 | negative regulation of DNA duplex unwinding | 2 | 4 | 2.64e-03 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 2.64e-03 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 2.64e-03 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 2.64e-03 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 2.67e-03 |
| GO:BP | GO:0016310 | phosphorylation | 19 | 1372 | 2.71e-03 |
| GO:BP | GO:0006269 | DNA replication, synthesis of RNA primer | 4 | 7 | 2.81e-03 |
| GO:BP | GO:0030010 | establishment of cell polarity | 9 | 134 | 2.88e-03 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 3 | 14 | 2.91e-03 |
| GO:BP | GO:0071139 | resolution of recombination intermediates | 4 | 6 | 3.12e-03 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 10 | 31 | 3.14e-03 |
| GO:BP | GO:0051782 | negative regulation of cell division | 4 | 15 | 3.51e-03 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 35 | 262 | 3.70e-03 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 29 | 479 | 3.75e-03 |
| GO:BP | GO:0090656 | t-circle formation | 6 | 13 | 3.94e-03 |
| GO:BP | GO:0072719 | cellular response to cisplatin | 3 | 4 | 3.94e-03 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 6 | 13 | 3.94e-03 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 6 | 13 | 3.94e-03 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 4 | 29 | 4.04e-03 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 3 | 31 | 4.35e-03 |
| GO:BP | GO:0006325 | chromatin organization | 44 | 551 | 4.63e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 19 | 121 | 4.65e-03 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 7 | 29 | 4.65e-03 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 3 | 10 | 4.68e-03 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 4 | 28 | 4.81e-03 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 4 | 11 | 4.91e-03 |
| GO:BP | GO:1905342 | positive regulation of protein localization to kinetochore | 2 | 4 | 4.96e-03 |
| GO:BP | GO:1905340 | regulation of protein localization to kinetochore | 2 | 4 | 4.96e-03 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 5 | 22 | 4.98e-03 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 5 | 14 | 5.28e-03 |
| GO:BP | GO:0000723 | telomere maintenance | 26 | 142 | 5.43e-03 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 5 | 11 | 5.43e-03 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 4 | 9 | 5.70e-03 |
| GO:BP | GO:0086016 | AV node cell action potential | 4 | 9 | 5.70e-03 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 5 | 38 | 6.01e-03 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 4 | 21 | 6.16e-03 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 6 | 25 | 6.40e-03 |
| GO:BP | GO:0006298 | mismatch repair | 6 | 29 | 6.47e-03 |
| GO:BP | GO:0090657 | telomeric loop disassembly | 5 | 9 | 6.50e-03 |
| GO:BP | GO:0034644 | cellular response to UV | 15 | 86 | 6.50e-03 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 4 | 12 | 6.65e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 14 | 993 | 7.01e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 14 | 994 | 7.05e-03 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 5 | 18 | 7.09e-03 |
| GO:BP | GO:0000921 | septin ring assembly | 1 | 1 | 7.22e-03 |
| GO:BP | GO:0032200 | telomere organization | 29 | 166 | 7.80e-03 |
| GO:BP | GO:1905462 | regulation of DNA duplex unwinding | 2 | 7 | 8.01e-03 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 14 | 186 | 8.27e-03 |
| GO:BP | GO:0006338 | chromatin remodeling | 26 | 378 | 8.40e-03 |
| GO:BP | GO:0070194 | synaptonemal complex disassembly | 1 | 2 | 8.55e-03 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 2 | 17 | 8.90e-03 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 4 | 10 | 8.92e-03 |
| GO:BP | GO:0044806 | G-quadruplex DNA unwinding | 4 | 7 | 8.92e-03 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 22 | 8.92e-03 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 10 | 35 | 9.26e-03 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 2 | 3 | 9.26e-03 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 4 | 13 | 9.63e-03 |
| GO:BP | GO:0051642 | centrosome localization | 4 | 31 | 9.75e-03 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 4 | 31 | 9.75e-03 |
| GO:BP | GO:0043549 | regulation of kinase activity | 31 | 560 | 9.76e-03 |
| GO:BP | GO:0007019 | microtubule depolymerization | 3 | 43 | 9.90e-03 |
| GO:BP | GO:0016043 | cellular component organization | 116 | 5076 | 1.05e-02 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 1.07e-02 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 19 | 282 | 1.08e-02 |
| GO:BP | GO:0003283 | atrial septum development | 4 | 23 | 1.09e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 1 | 8 | 1.09e-02 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 4 | 45 | 1.09e-02 |
| GO:BP | GO:0016574 | histone ubiquitination | 5 | 46 | 1.10e-02 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 3 | 4 | 1.19e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 3 | 11 | 1.22e-02 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 4 | 23 | 1.27e-02 |
| GO:BP | GO:0051664 | nuclear pore localization | 3 | 4 | 1.29e-02 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 4 | 10 | 1.29e-02 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 4 | 1.32e-02 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 1.32e-02 |
| GO:BP | GO:0031106 | septin ring organization | 1 | 2 | 1.33e-02 |
| GO:BP | GO:1904172 | positive regulation of bleb assembly | 1 | 1 | 1.33e-02 |
| GO:BP | GO:1904170 | regulation of bleb assembly | 1 | 1 | 1.33e-02 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 6 | 20 | 1.34e-02 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 2 | 8 | 1.38e-02 |
| GO:BP | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 2 | 1.38e-02 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 8 | 64 | 1.39e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 3 | 5 | 1.39e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 62 | 868 | 1.40e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 12 | 719 | 1.41e-02 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 12 | 63 | 1.42e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 10 | 206 | 1.42e-02 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 2 | 6 | 1.46e-02 |
| GO:BP | GO:0001556 | oocyte maturation | 6 | 22 | 1.48e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 18 | 1380 | 1.53e-02 |
| GO:BP | GO:0072718 | response to cisplatin | 3 | 6 | 1.54e-02 |
| GO:BP | GO:0006177 | GMP biosynthetic process | 5 | 13 | 1.55e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 10 | 496 | 1.55e-02 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 5 | 1.64e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 1 | 13 | 1.65e-02 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 5 | 18 | 1.70e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 14 | 97 | 1.76e-02 |
| GO:BP | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process | 1 | 2 | 1.76e-02 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 7 | 30 | 1.76e-02 |
| GO:BP | GO:0006334 | nucleosome assembly | 16 | 80 | 1.81e-02 |
| GO:BP | GO:0021987 | cerebral cortex development | 5 | 101 | 1.85e-02 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 5 | 29 | 1.87e-02 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 2 | 4 | 1.96e-02 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 4 | 7 | 1.98e-02 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 4 | 16 | 1.99e-02 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 3 | 11 | 2.02e-02 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 2 | 3 | 2.09e-02 |
| GO:BP | GO:0099606 | microtubule plus-end directed mitotic chromosome migration | 1 | 1 | 2.10e-02 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 5 | 11 | 2.11e-02 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 16 | 89 | 2.19e-02 |
| GO:BP | GO:1904975 | response to bleomycin | 2 | 2 | 2.19e-02 |
| GO:BP | GO:0010458 | exit from mitosis | 2 | 30 | 2.19e-02 |
| GO:BP | GO:1904976 | cellular response to bleomycin | 2 | 2 | 2.19e-02 |
| GO:BP | GO:0001578 | microtubule bundle formation | 4 | 79 | 2.24e-02 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 4 | 13 | 2.30e-02 |
| GO:BP | GO:0045004 | DNA replication proofreading | 2 | 2 | 2.34e-02 |
| GO:BP | GO:0033212 | iron import into cell | 4 | 7 | 2.35e-02 |
| GO:BP | GO:0071932 | replication fork reversal | 3 | 5 | 2.40e-02 |
| GO:BP | GO:0032185 | septin cytoskeleton organization | 1 | 4 | 2.40e-02 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 4 | 14 | 2.42e-02 |
| GO:BP | GO:0006566 | threonine metabolic process | 2 | 4 | 2.42e-02 |
| GO:BP | GO:0015691 | cadmium ion transport | 3 | 5 | 2.43e-02 |
| GO:BP | GO:0070574 | cadmium ion transmembrane transport | 3 | 5 | 2.43e-02 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 8 | 70 | 2.45e-02 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 1 | 7 | 2.49e-02 |
| GO:BP | GO:0006284 | base-excision repair | 7 | 44 | 2.52e-02 |
| GO:BP | GO:1903416 | response to glycoside | 2 | 6 | 2.54e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 10 | 69 | 2.54e-02 |
| GO:BP | GO:0051309 | female meiosis chromosome separation | 1 | 2 | 2.59e-02 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 4 | 15 | 2.59e-02 |
| GO:BP | GO:0008340 | determination of adult lifespan | 5 | 16 | 2.63e-02 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 7 | 252 | 2.67e-02 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 7 | 23 | 2.73e-02 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 5 | 52 | 2.73e-02 |
| GO:BP | GO:0045007 | depurination | 1 | 4 | 2.78e-02 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 6 | 2.81e-02 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 2 | 14 | 2.84e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 10 | 547 | 2.84e-02 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 6 | 48 | 2.85e-02 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 2 | 2.86e-02 |
| GO:BP | GO:0051299 | centrosome separation | 4 | 14 | 2.87e-02 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 2 | 6 | 2.92e-02 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 4 | 16 | 3.08e-02 |
| GO:BP | GO:0048239 | negative regulation of DNA recombination at telomere | 2 | 2 | 3.10e-02 |
| GO:BP | GO:0072695 | regulation of DNA recombination at telomere | 2 | 2 | 3.10e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 116 | 5274 | 3.14e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 9 | 35 | 3.14e-02 |
| GO:BP | GO:0019264 | glycine biosynthetic process from serine | 2 | 2 | 3.14e-02 |
| GO:BP | GO:0034728 | nucleosome organization | 17 | 98 | 3.14e-02 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 2 | 4 | 3.14e-02 |
| GO:BP | GO:0006265 | DNA topological change | 4 | 10 | 3.23e-02 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 8 | 24 | 3.26e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 9 | 82 | 3.26e-02 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 1 | 10 | 3.34e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 7 | 34 | 3.38e-02 |
| GO:BP | GO:1900060 | negative regulation of ceramide biosynthetic process | 3 | 5 | 3.38e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 6 | 26 | 3.40e-02 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 2 | 14 | 3.50e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 15 | 104 | 3.50e-02 |
| GO:BP | GO:0043687 | post-translational protein modification | 7 | 64 | 3.55e-02 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 5 | 17 | 3.55e-02 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 11 | 68 | 3.55e-02 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 5 | 17 | 3.55e-02 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 5 | 55 | 3.55e-02 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 5 | 17 | 3.55e-02 |
| GO:BP | GO:0030397 | membrane disassembly | 1 | 11 | 3.58e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 9 | 36 | 3.59e-02 |
| GO:BP | GO:2000348 | regulation of CD40 signaling pathway | 2 | 4 | 3.59e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 9 | 36 | 3.59e-02 |
| GO:BP | GO:1990928 | response to amino acid starvation | 6 | 51 | 3.64e-02 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 3.65e-02 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 19 | 3.67e-02 |
| GO:BP | GO:1905115 | regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 3.72e-02 |
| GO:BP | GO:0097742 | de novo centriole assembly | 2 | 4 | 3.72e-02 |
| GO:BP | GO:1905116 | positive regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 3.72e-02 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 2 | 4 | 3.72e-02 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 3.72e-02 |
| GO:BP | GO:0098904 | regulation of AV node cell action potential | 2 | 3 | 3.84e-02 |
| GO:BP | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis | 2 | 3 | 3.84e-02 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 5 | 15 | 3.91e-02 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 2 | 2 | 4.21e-02 |
| GO:BP | GO:0055071 | manganese ion homeostasis | 2 | 2 | 4.27e-02 |
| GO:BP | GO:2000241 | regulation of reproductive process | 11 | 128 | 4.27e-02 |
| GO:BP | GO:0021685 | cerebellar granular layer structural organization | 1 | 1 | 4.30e-02 |
| GO:BP | GO:0048290 | isotype switching to IgA isotypes | 2 | 4 | 4.33e-02 |
| GO:BP | GO:0090435 | protein localization to nuclear envelope | 1 | 14 | 4.37e-02 |
| GO:BP | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process | 1 | 5 | 4.37e-02 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 7 | 29 | 4.45e-02 |
| GO:BP | GO:0000722 | telomere maintenance via recombination | 5 | 14 | 4.45e-02 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 5 | 41 | 4.50e-02 |
| GO:BP | GO:0009127 | purine nucleoside monophosphate biosynthetic process | 6 | 21 | 4.50e-02 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 4.54e-02 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 4 | 35 | 4.61e-02 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 4 | 82 | 4.68e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 6 | 31 | 4.77e-02 |
| GO:BP | GO:1905777 | regulation of exonuclease activity | 2 | 2 | 4.87e-02 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 3 | 9 | 4.87e-02 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 10 | 597 | 4.87e-02 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 10 | 597 | 4.87e-02 |
| GO:BP | GO:0061952 | midbody abscission | 3 | 17 | 4.87e-02 |
| GO:BP | GO:1905839 | negative regulation of telomeric D-loop disassembly | 2 | 2 | 4.87e-02 |
| GO:BP | GO:1905778 | negative regulation of exonuclease activity | 2 | 2 | 4.87e-02 |
| GO:BP | GO:1905838 | regulation of telomeric D-loop disassembly | 2 | 2 | 4.87e-02 |
| GO:BP | GO:0006543 | glutamine catabolic process | 2 | 3 | 4.87e-02 |
| GO:BP | GO:0032214 | negative regulation of telomere maintenance via semi-conservative replication | 2 | 2 | 4.87e-02 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 1 | 1 | 4.87e-02 |
| GO:BP | GO:1904534 | negative regulation of telomeric loop disassembly | 2 | 2 | 4.87e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 20 | 240 | 4.87e-02 |
| GO:BP | GO:0006189 | ‘de novo’ IMP biosynthetic process | 3 | 6 | 4.87e-02 |
| GO:BP | GO:2000620 | positive regulation of histone H4-K16 acetylation | 2 | 2 | 4.87e-02 |
| GO:BP | GO:0032213 | regulation of telomere maintenance via semi-conservative replication | 2 | 2 | 4.87e-02 |
| GO:BP | GO:1905359 | protein localization to meiotic spindle | 1 | 1 | 4.87e-02 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 4 | 9 | 4.87e-02 |
| GO:BP | GO:1903096 | protein localization to meiotic spindle midzone | 1 | 1 | 4.87e-02 |
| GO:BP | GO:0003230 | cardiac atrium development | 4 | 34 | 4.87e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 23 | 293 | 4.89e-02 |
| GO:BP | GO:0051095 | regulation of helicase activity | 3 | 12 | 5.10e-02 |
| GO:BP | GO:0090166 | Golgi disassembly | 3 | 6 | 5.16e-02 |
| GO:BP | GO:0045769 | negative regulation of asymmetric cell division | 1 | 1 | 5.19e-02 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 4 | 19 | 5.27e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 5 | 27 | 5.32e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 4 | 77 | 5.34e-02 |
| GO:BP | GO:0032060 | bleb assembly | 1 | 9 | 5.34e-02 |
| GO:BP | GO:0046037 | GMP metabolic process | 6 | 23 | 5.47e-02 |
| GO:BP | GO:0006266 | DNA ligation | 2 | 16 | 5.51e-02 |
| GO:BP | GO:0090155 | negative regulation of sphingolipid biosynthetic process | 3 | 6 | 5.61e-02 |
| GO:BP | GO:1903126 | negative regulation of centriole-centriole cohesion | 1 | 1 | 5.61e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 60 | 616 | 5.70e-02 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 3 | 13 | 5.84e-02 |
| GO:BP | GO:0006997 | nucleus organization | 17 | 126 | 5.89e-02 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 2 | 23 | 5.92e-02 |
| GO:BP | GO:0090399 | replicative senescence | 5 | 13 | 5.96e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 2 | 8 | 5.99e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 13 | 114 | 5.99e-02 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 5 | 44 | 6.12e-02 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 5 | 17 | 6.19e-02 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 5 | 15 | 6.24e-02 |
| GO:BP | GO:0021591 | ventricular system development | 1 | 29 | 6.24e-02 |
| GO:BP | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing | 2 | 7 | 6.28e-02 |
| GO:BP | GO:0000712 | resolution of meiotic recombination intermediates | 4 | 13 | 6.33e-02 |
| GO:BP | GO:0002027 | regulation of heart rate | 10 | 86 | 6.47e-02 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 4 | 99 | 6.47e-02 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 4 | 11 | 6.48e-02 |
| GO:BP | GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 5 | 19 | 6.54e-02 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 4 | 191 | 6.62e-02 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 1 | 12 | 6.71e-02 |
| GO:BP | GO:0044314 | protein K27-linked ubiquitination | 3 | 8 | 6.79e-02 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 7 | 32 | 6.84e-02 |
| GO:BP | GO:0021543 | pallium development | 5 | 139 | 6.84e-02 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 9 | 37 | 6.84e-02 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 32 | 6.84e-02 |
| GO:BP | GO:1904353 | regulation of telomere capping | 2 | 23 | 6.84e-02 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 9 | 58 | 6.84e-02 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 6.99e-02 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 3 | 6 | 7.04e-02 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 7.14e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 7 | 39 | 7.23e-02 |
| GO:BP | GO:0071313 | cellular response to caffeine | 3 | 10 | 7.23e-02 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 16 | 120 | 7.24e-02 |
| GO:BP | GO:1903375 | facioacoustic ganglion development | 2 | 2 | 7.24e-02 |
| GO:BP | GO:1904835 | dorsal root ganglion morphogenesis | 2 | 2 | 7.24e-02 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 7 | 60 | 7.24e-02 |
| GO:BP | GO:0061552 | ganglion morphogenesis | 2 | 2 | 7.24e-02 |
| GO:BP | GO:0021649 | vestibulocochlear nerve structural organization | 2 | 2 | 7.24e-02 |
| GO:BP | GO:0097292 | XMP metabolic process | 3 | 7 | 7.25e-02 |
| GO:BP | GO:0097294 | ‘de novo’ XMP biosynthetic process | 3 | 7 | 7.25e-02 |
| GO:BP | GO:0097293 | XMP biosynthetic process | 3 | 7 | 7.25e-02 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 4 | 14 | 7.28e-02 |
| GO:BP | GO:0098942 | retrograde trans-synaptic signaling by trans-synaptic protein complex | 2 | 2 | 7.32e-02 |
| GO:BP | GO:0021698 | cerebellar cortex structural organization | 1 | 2 | 7.32e-02 |
| GO:BP | GO:0021693 | cerebellar Purkinje cell layer structural organization | 1 | 2 | 7.32e-02 |
| GO:BP | GO:1904631 | response to glucoside | 1 | 1 | 7.41e-02 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 1 | 23 | 7.47e-02 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 7.47e-02 |
| GO:BP | GO:1902977 | mitotic DNA replication preinitiation complex assembly | 1 | 1 | 7.64e-02 |
| GO:BP | GO:0033599 | regulation of mammary gland epithelial cell proliferation | 3 | 14 | 7.67e-02 |
| GO:BP | GO:1905341 | negative regulation of protein localization to kinetochore | 1 | 1 | 7.70e-02 |
| GO:BP | GO:0006273 | lagging strand elongation | 2 | 4 | 7.81e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 5 | 92 | 7.85e-02 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 27 | 7.90e-02 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 1 | 27 | 7.90e-02 |
| GO:BP | GO:0051657 | maintenance of organelle location | 2 | 11 | 7.93e-02 |
| GO:BP | GO:0071694 | maintenance of protein location in extracellular region | 2 | 4 | 8.06e-02 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 1 | 35 | 8.21e-02 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 4 | 12 | 8.28e-02 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 4 | 12 | 8.28e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 5 | 28 | 8.39e-02 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 1 | 2 | 8.50e-02 |
| GO:BP | GO:0000729 | DNA double-strand break processing | 3 | 18 | 8.51e-02 |
| GO:BP | GO:0019276 | UDP-N-acetylgalactosamine metabolic process | 2 | 2 | 8.54e-02 |
| GO:BP | GO:0072520 | seminiferous tubule development | 2 | 13 | 8.66e-02 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 4 | 14 | 8.67e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 16 | 101 | 8.71e-02 |
| GO:BP | GO:0051261 | protein depolymerization | 3 | 105 | 8.75e-02 |
| GO:BP | GO:0016233 | telomere capping | 8 | 36 | 8.76e-02 |
| GO:BP | GO:1904777 | negative regulation of protein localization to cell cortex | 1 | 1 | 8.82e-02 |
| GO:BP | GO:0098725 | symmetric cell division | 1 | 1 | 8.82e-02 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 3 | 108 | 8.85e-02 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 8.94e-02 |
| GO:BP | GO:0048313 | Golgi inheritance | 4 | 15 | 8.99e-02 |
| GO:BP | GO:0048308 | organelle inheritance | 4 | 15 | 8.99e-02 |
| GO:BP | GO:2001045 | negative regulation of integrin-mediated signaling pathway | 2 | 3 | 9.14e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 5 | 28 | 9.28e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 5 | 31 | 9.28e-02 |
| GO:BP | GO:0006272 | leading strand elongation | 2 | 4 | 9.31e-02 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 4 | 17 | 9.31e-02 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 4 | 8 | 9.32e-02 |
| GO:BP | GO:0006021 | inositol biosynthetic process | 2 | 3 | 9.49e-02 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 9.55e-02 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 5 | 13 | 9.62e-02 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 2 | 3 | 9.66e-02 |
| GO:BP | GO:1903516 | regulation of single strand break repair | 2 | 3 | 9.66e-02 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 3 | 10 | 9.66e-02 |
| GO:BP | GO:2001170 | negative regulation of ATP biosynthetic process | 2 | 3 | 9.66e-02 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 3 | 8 | 9.74e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 3 | 232 | 9.79e-02 |
| GO:BP | GO:0043111 | replication fork arrest | 2 | 2 | 9.79e-02 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 2 | 6 | 9.99e-02 |
| GO:BP | GO:0010826 | negative regulation of centrosome duplication | 4 | 12 | 1.00e-01 |
| GO:BP | GO:1905775 | negative regulation of DNA helicase activity | 1 | 2 | 1.00e-01 |
| GO:BP | GO:0046606 | negative regulation of centrosome cycle | 4 | 12 | 1.00e-01 |
| GO:BP | GO:0032069 | regulation of nuclease activity | 5 | 17 | 1.01e-01 |
| GO:BP | GO:0010224 | response to UV-B | 3 | 17 | 1.01e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 5 | 16 | 1.01e-01 |
| GO:BP | GO:0044208 | ‘de novo’ AMP biosynthetic process | 3 | 8 | 1.01e-01 |
| GO:BP | GO:1901723 | negative regulation of cell proliferation involved in kidney development | 2 | 3 | 1.01e-01 |
| GO:BP | GO:0032487 | regulation of Rap protein signal transduction | 1 | 3 | 1.01e-01 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 4 | 27 | 1.01e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 6 | 23 | 1.01e-01 |
| GO:BP | GO:0033624 | negative regulation of integrin activation | 1 | 3 | 1.01e-01 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 2 | 5 | 1.01e-01 |
| GO:BP | GO:0072092 | ureteric bud invasion | 2 | 3 | 1.01e-01 |
| GO:BP | GO:0030263 | apoptotic chromosome condensation | 1 | 4 | 1.01e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 2 | 31 | 1.01e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 2 | 31 | 1.01e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 6 | 30 | 1.02e-01 |
| GO:BP | GO:0030954 | astral microtubule nucleation | 1 | 1 | 1.03e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 17 | 1974 | 1.03e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 5 | 14 | 1.03e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 1 | 35 | 1.03e-01 |
| GO:BP | GO:1902480 | protein localization to mitotic spindle | 1 | 1 | 1.04e-01 |
| GO:BP | GO:0032202 | telomere assembly | 2 | 4 | 1.06e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 1 | 42 | 1.07e-01 |
| GO:BP | GO:1905786 | positive regulation of anaphase-promoting complex-dependent catabolic process | 1 | 2 | 1.07e-01 |
| GO:BP | GO:0098532 | histone H3-K27 trimethylation | 1 | 4 | 1.07e-01 |
| GO:BP | GO:0070318 | positive regulation of G0 to G1 transition | 2 | 5 | 1.08e-01 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 2 | 4 | 1.09e-01 |
| GO:BP | GO:0006545 | glycine biosynthetic process | 3 | 4 | 1.09e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 4 | 27 | 1.09e-01 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 2 | 6 | 1.09e-01 |
| GO:BP | GO:0044878 | mitotic cytokinesis checkpoint signaling | 1 | 4 | 1.10e-01 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 3 | 10 | 1.10e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 11 | 79 | 1.10e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 20 | 2139 | 1.10e-01 |
| GO:BP | GO:1904533 | regulation of telomeric loop disassembly | 2 | 2 | 1.10e-01 |
| GO:BP | GO:0070512 | positive regulation of histone H4-K20 methylation | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 5 | 34 | 1.12e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 2 | 717 | 1.14e-01 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 1.14e-01 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 1 | 2 | 1.14e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 8 | 76 | 1.14e-01 |
| GO:BP | GO:1905178 | regulation of cardiac muscle tissue regeneration | 1 | 2 | 1.14e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 2 | 33 | 1.14e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 9 | 631 | 1.14e-01 |
| GO:BP | GO:1905179 | negative regulation of cardiac muscle tissue regeneration | 1 | 2 | 1.14e-01 |
| GO:BP | GO:0060686 | negative regulation of prostatic bud formation | 2 | 4 | 1.15e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 5 | 19 | 1.15e-01 |
| GO:BP | GO:0031648 | protein destabilization | 1 | 45 | 1.15e-01 |
| GO:BP | GO:0000741 | karyogamy | 1 | 1 | 1.16e-01 |
| GO:BP | GO:0007344 | pronuclear fusion | 1 | 1 | 1.16e-01 |
| GO:BP | GO:0046600 | negative regulation of centriole replication | 3 | 7 | 1.16e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 1 | 15 | 1.17e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 3 | 126 | 1.17e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 7 | 59 | 1.17e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 5 | 31 | 1.17e-01 |
| GO:BP | GO:0060013 | righting reflex | 3 | 6 | 1.17e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 20 | 2161 | 1.18e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 1 | 4 | 1.18e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 2 | 5 | 1.18e-01 |
| GO:BP | GO:0098735 | positive regulation of the force of heart contraction | 2 | 4 | 1.18e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 3 | 133 | 1.18e-01 |
| GO:BP | GO:0044565 | dendritic cell proliferation | 2 | 3 | 1.18e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 6 | 33 | 1.18e-01 |
| GO:BP | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 2 | 3 | 1.18e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 16 | 104 | 1.19e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 7 | 37 | 1.20e-01 |
| GO:BP | GO:0071415 | cellular response to purine-containing compound | 3 | 11 | 1.21e-01 |
| GO:BP | GO:0031000 | response to caffeine | 4 | 13 | 1.22e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 10 | 64 | 1.23e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 29 | 352 | 1.23e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 2 | 7 | 1.24e-01 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 6 | 27 | 1.26e-01 |
| GO:BP | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell communication | 2 | 6 | 1.27e-01 |
| GO:BP | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling | 2 | 6 | 1.27e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 2 | 787 | 1.28e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 1.28e-01 |
| GO:BP | GO:1902595 | regulation of DNA replication origin binding | 1 | 2 | 1.28e-01 |
| GO:BP | GO:2000431 | regulation of cytokinesis, actomyosin contractile ring assembly | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1 | 4 | 1.29e-01 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 2 | 5 | 1.29e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 2 | 7 | 1.31e-01 |
| GO:BP | GO:1902319 | DNA strand elongation involved in nuclear cell cycle DNA replication | 1 | 1 | 1.31e-01 |
| GO:BP | GO:1902983 | DNA strand elongation involved in mitotic DNA replication | 1 | 1 | 1.31e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 5 | 35 | 1.31e-01 |
| GO:BP | GO:1902296 | DNA strand elongation involved in cell cycle DNA replication | 1 | 1 | 1.31e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 6 | 32 | 1.31e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 4 | 26 | 1.31e-01 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 4 | 8 | 1.31e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 17 | 1939 | 1.31e-01 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 1.32e-01 |
| GO:BP | GO:0035611 | protein branching point deglutamylation | 1 | 1 | 1.33e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 20 | 147 | 1.33e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 1 | 57 | 1.34e-01 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 1 | 4 | 1.35e-01 |
| GO:BP | GO:0009171 | purine deoxyribonucleoside monophosphate biosynthetic process | 2 | 3 | 1.35e-01 |
| GO:BP | GO:0106383 | dAMP salvage | 2 | 3 | 1.35e-01 |
| GO:BP | GO:0006170 | dAMP biosynthetic process | 2 | 3 | 1.35e-01 |
| GO:BP | GO:0090179 | planar cell polarity pathway involved in neural tube closure | 3 | 12 | 1.35e-01 |
| GO:BP | GO:1904439 | negative regulation of iron ion import across plasma membrane | 1 | 1 | 1.35e-01 |
| GO:BP | GO:0106381 | purine deoxyribonucleotide salvage | 2 | 3 | 1.35e-01 |
| GO:BP | GO:0035581 | sequestering of extracellular ligand from receptor | 2 | 7 | 1.36e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 29 | 344 | 1.38e-01 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 4 | 14 | 1.38e-01 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 1.38e-01 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 9 | 57 | 1.38e-01 |
| GO:BP | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination | 1 | 1 | 1.38e-01 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0046452 | dihydrofolate metabolic process | 2 | 3 | 1.39e-01 |
| GO:BP | GO:0032876 | negative regulation of DNA endoreduplication | 1 | 1 | 1.39e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 3 | 7 | 1.39e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 3 | 7 | 1.39e-01 |
| GO:BP | GO:0040016 | embryonic cleavage | 1 | 7 | 1.39e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 1 | 45 | 1.39e-01 |
| GO:BP | GO:0090398 | cellular senescence | 5 | 83 | 1.39e-01 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 2 | 7 | 1.39e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 5 | 33 | 1.39e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 3 | 73 | 1.39e-01 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 2 | 11 | 1.39e-01 |
| GO:BP | GO:0036164 | cell-abiotic substrate adhesion | 1 | 1 | 1.40e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 3 | 12 | 1.41e-01 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 7 | 30 | 1.41e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 4 | 23 | 1.41e-01 |
| GO:BP | GO:0006563 | L-serine metabolic process | 3 | 10 | 1.41e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 4 | 55 | 1.41e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 2 | 9 | 1.41e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 1 | 8 | 1.42e-01 |
| GO:BP | GO:0009838 | abscission | 1 | 6 | 1.43e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 1 | 16 | 1.43e-01 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 3 | 9 | 1.44e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 5 | 114 | 1.44e-01 |
| GO:BP | GO:1901660 | calcium ion export | 2 | 3 | 1.44e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 27 | 834 | 1.46e-01 |
| GO:BP | GO:0098711 | iron ion import across plasma membrane | 2 | 4 | 1.47e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 6 | 25 | 1.47e-01 |
| GO:BP | GO:0061757 | leukocyte adhesion to arterial endothelial cell | 2 | 4 | 1.47e-01 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 4 | 19 | 1.48e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 8 | 260 | 1.48e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 10 | 101 | 1.50e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 4 | 53 | 1.51e-01 |
| GO:BP | GO:0021629 | olfactory nerve structural organization | 1 | 1 | 1.51e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 4 | 16 | 1.51e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 2 | 23 | 1.51e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 4 | 28 | 1.52e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 4 | 11 | 1.52e-01 |
| GO:BP | GO:0038101 | sequestering of nodal from receptor via nodal binding | 1 | 1 | 1.52e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 1 | 6 | 1.52e-01 |
| GO:BP | GO:0097374 | sensory neuron axon guidance | 2 | 2 | 1.52e-01 |
| GO:BP | GO:0036486 | ventral trunk neural crest cell migration | 2 | 3 | 1.52e-01 |
| GO:BP | GO:0070537 | histone H2A K63-linked deubiquitination | 2 | 8 | 1.53e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 11 | 152 | 1.54e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 3 | 26 | 1.54e-01 |
| GO:BP | GO:0098906 | regulation of Purkinje myocyte action potential | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 2 | 898 | 1.54e-01 |
| GO:BP | GO:0046340 | diacylglycerol catabolic process | 2 | 3 | 1.54e-01 |
| GO:BP | GO:0010652 | positive regulation of cell communication by chemical coupling | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 5 | 21 | 1.54e-01 |
| GO:BP | GO:1990029 | vasomotion | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0010645 | regulation of cell communication by chemical coupling | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0072136 | metanephric mesenchymal cell proliferation involved in metanephros development | 2 | 2 | 1.54e-01 |
| GO:BP | GO:0086044 | atrial cardiac muscle cell to AV node cell communication by electrical coupling | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 5 | 427 | 1.54e-01 |
| GO:BP | GO:0086055 | Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling | 1 | 1 | 1.54e-01 |
| GO:BP | GO:0072135 | kidney mesenchymal cell proliferation | 2 | 2 | 1.54e-01 |
| GO:BP | GO:0097252 | oligodendrocyte apoptotic process | 2 | 4 | 1.54e-01 |
| GO:BP | GO:0097018 | renal albumin absorption | 2 | 3 | 1.54e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 9 | 67 | 1.55e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 3 | 13 | 1.55e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 2 | 4 | 1.55e-01 |
| GO:BP | GO:0060914 | heart formation | 5 | 29 | 1.55e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 3 | 76 | 1.56e-01 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 2 | 4 | 1.56e-01 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 2 | 5 | 1.56e-01 |
| GO:BP | GO:0006306 | DNA methylation | 5 | 46 | 1.57e-01 |
| GO:BP | GO:0006950 | response to stress | 174 | 2780 | 1.57e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 5 | 46 | 1.57e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 1 | 7 | 1.57e-01 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 1 | 27 | 1.57e-01 |
| GO:BP | GO:0036089 | cleavage furrow formation | 1 | 7 | 1.57e-01 |
| GO:BP | GO:0023035 | CD40 signaling pathway | 2 | 11 | 1.57e-01 |
| GO:BP | GO:0035695 | mitophagy by induced vacuole formation | 1 | 1 | 1.58e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 6 | 28 | 1.60e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 1 | 226 | 1.60e-01 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 2 | 7 | 1.61e-01 |
| GO:BP | GO:0006304 | DNA modification | 7 | 89 | 1.61e-01 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 4 | 21 | 1.61e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 3 | 11 | 1.61e-01 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 2 | 3 | 1.62e-01 |
| GO:BP | GO:0006565 | L-serine catabolic process | 2 | 4 | 1.62e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 3 | 1836 | 1.63e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 1.63e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 1.63e-01 |
| GO:BP | GO:0098930 | axonal transport | 1 | 60 | 1.64e-01 |
| GO:BP | GO:0009650 | UV protection | 3 | 11 | 1.65e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 2 | 45 | 1.65e-01 |
| GO:BP | GO:0009786 | regulation of asymmetric cell division | 1 | 1 | 1.66e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 7 | 45 | 1.66e-01 |
| GO:BP | GO:0001824 | blastocyst development | 11 | 94 | 1.66e-01 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 3 | 7 | 1.66e-01 |
| GO:BP | GO:0021873 | forebrain neuroblast division | 1 | 3 | 1.66e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 1 | 75 | 1.67e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 11 | 1123 | 1.67e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 4 | 17 | 1.68e-01 |
| GO:BP | GO:0045875 | negative regulation of sister chromatid cohesion | 1 | 5 | 1.68e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 4 | 16 | 1.69e-01 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 3 | 7 | 1.69e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 1.69e-01 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 4 | 32 | 1.69e-01 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 2 | 9 | 1.69e-01 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 2 | 9 | 1.69e-01 |
| GO:BP | GO:0008584 | male gonad development | 6 | 99 | 1.69e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 3 | 160 | 1.69e-01 |
| GO:BP | GO:0007276 | gamete generation | 19 | 461 | 1.70e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 2 | 5 | 1.70e-01 |
| GO:BP | GO:1901753 | leukotriene A4 biosynthetic process | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 3 | 7 | 1.71e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 14 | 149 | 1.71e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 1 | 31 | 1.71e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 4 | 16 | 1.71e-01 |
| GO:BP | GO:0008406 | gonad development | 8 | 158 | 1.71e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 22 | 3090 | 1.72e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 2 | 7 | 1.72e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 89 | 1.72e-01 |
| GO:BP | GO:0046338 | phosphatidylethanolamine catabolic process | 2 | 4 | 1.72e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 6 | 100 | 1.72e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 1.72e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 4 | 27 | 1.73e-01 |
| GO:BP | GO:0075506 | entry of viral genome into host nucleus through nuclear pore complex via importin | 1 | 2 | 1.74e-01 |
| GO:BP | GO:0036211 | protein modification process | 21 | 2901 | 1.75e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 1 | 3 | 1.76e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 2 | 24 | 1.76e-01 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 3 | 13 | 1.76e-01 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 3 | 13 | 1.76e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 3 | 14 | 1.76e-01 |
| GO:BP | GO:0002023 | reduction of food intake in response to dietary excess | 2 | 2 | 1.76e-01 |
| GO:BP | GO:0048599 | oocyte development | 6 | 38 | 1.77e-01 |
| GO:BP | GO:0071922 | regulation of cohesin loading | 1 | 2 | 1.77e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 5 | 427 | 1.78e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 1 | 12 | 1.78e-01 |
| GO:BP | GO:1904482 | cellular response to tetrahydrofolate | 1 | 1 | 1.78e-01 |
| GO:BP | GO:1904481 | response to tetrahydrofolate | 1 | 1 | 1.78e-01 |
| GO:BP | GO:0032901 | positive regulation of neurotrophin production | 1 | 1 | 1.78e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 4 | 20 | 1.78e-01 |
| GO:BP | GO:0090102 | cochlea development | 2 | 35 | 1.78e-01 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 1 | 2 | 1.78e-01 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 1 | 2 | 1.78e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 3 | 11 | 1.79e-01 |
| GO:BP | GO:0051097 | negative regulation of helicase activity | 1 | 5 | 1.79e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 6 | 69 | 1.79e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 2 | 49 | 1.79e-01 |
| GO:BP | GO:1905774 | regulation of DNA helicase activity | 1 | 5 | 1.79e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 11 | 906 | 1.80e-01 |
| GO:BP | GO:0030952 | establishment or maintenance of cytoskeleton polarity | 1 | 7 | 1.81e-01 |
| GO:BP | GO:0072347 | response to anesthetic | 1 | 1 | 1.83e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 1 | 73 | 1.83e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 8 | 161 | 1.84e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 7 | 70 | 1.85e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 11 | 88 | 1.85e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 2 | 12 | 1.86e-01 |
| GO:BP | GO:0039521 | suppression by virus of host autophagy | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0140321 | suppression of host autophagy | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0071275 | cellular response to aluminum ion | 1 | 1 | 1.86e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 5 | 54 | 1.86e-01 |
| GO:BP | GO:0075071 | modulation by symbiont of host autophagy | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0006188 | IMP biosynthetic process | 3 | 10 | 1.86e-01 |
| GO:BP | GO:1905672 | negative regulation of lysosome organization | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 3 | 17 | 1.88e-01 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 3 | 12 | 1.89e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 1 | 246 | 1.90e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 4 | 19 | 1.90e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 4 | 16 | 1.90e-01 |
| GO:BP | GO:0061511 | centriole elongation | 2 | 8 | 1.90e-01 |
| GO:BP | GO:0000730 | DNA recombinase assembly | 1 | 3 | 1.90e-01 |
| GO:BP | GO:0090735 | DNA repair complex assembly | 1 | 3 | 1.90e-01 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 2 | 5 | 1.91e-01 |
| GO:BP | GO:0046104 | thymidine metabolic process | 2 | 5 | 1.91e-01 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 7 | 34 | 1.91e-01 |
| GO:BP | GO:0032075 | positive regulation of nuclease activity | 2 | 7 | 1.92e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 1 | 103 | 1.92e-01 |
| GO:BP | GO:0034478 | phosphatidylglycerol catabolic process | 1 | 1 | 1.94e-01 |
| GO:BP | GO:0021537 | telencephalon development | 5 | 201 | 1.94e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 2 | 5 | 1.95e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1 | 101 | 1.96e-01 |
| GO:BP | GO:0034755 | iron ion transmembrane transport | 4 | 16 | 1.96e-01 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 1 | 3 | 1.97e-01 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 1 | 3 | 1.97e-01 |
| GO:BP | GO:1902426 | deactivation of mitotic spindle assembly checkpoint | 1 | 3 | 1.97e-01 |
| GO:BP | GO:0007548 | sex differentiation | 9 | 191 | 1.97e-01 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 3 | 23 | 1.97e-01 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 8 | 38 | 1.97e-01 |
| GO:BP | GO:0060685 | regulation of prostatic bud formation | 2 | 5 | 1.99e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 3 | 86 | 1.99e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 3 | 14 | 1.99e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 4 | 8 | 2.00e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 1 | 111 | 2.00e-01 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 2.01e-01 |
| GO:BP | GO:0030809 | negative regulation of nucleotide biosynthetic process | 2 | 3 | 2.01e-01 |
| GO:BP | GO:1900372 | negative regulation of purine nucleotide biosynthetic process | 2 | 3 | 2.01e-01 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 2 | 5 | 2.01e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 4 | 21 | 2.01e-01 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 2 | 5 | 2.01e-01 |
| GO:BP | GO:0033197 | response to vitamin E | 2 | 9 | 2.01e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 2 | 1017 | 2.01e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 2 | 1015 | 2.01e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 10 | 62 | 2.01e-01 |
| GO:BP | GO:0072429 | response to intra-S DNA damage checkpoint signaling | 1 | 4 | 2.01e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 12 | 100 | 2.01e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 1 | 2 | 2.03e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 2 | 234 | 2.04e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 5 | 29 | 2.04e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 2 | 1026 | 2.04e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 2 | 109 | 2.04e-01 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 2 | 10 | 2.05e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 7 | 35 | 2.05e-01 |
| GO:BP | GO:0006564 | L-serine biosynthetic process | 2 | 6 | 2.05e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 3 | 183 | 2.05e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 7 | 35 | 2.05e-01 |
| GO:BP | GO:1904438 | regulation of iron ion import across plasma membrane | 1 | 1 | 2.06e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 9 | 61 | 2.07e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 5 | 27 | 2.08e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 3 | 17 | 2.08e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 3 | 15 | 2.08e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 3 | 15 | 2.08e-01 |
| GO:BP | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 1 | 7 | 2.08e-01 |
| GO:BP | GO:0055001 | muscle cell development | 15 | 165 | 2.08e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 31 | 2466 | 2.09e-01 |
| GO:BP | GO:0090038 | negative regulation of protein kinase C signaling | 2 | 4 | 2.09e-01 |
| GO:BP | GO:0000706 | meiotic DNA double-strand break processing | 1 | 2 | 2.09e-01 |
| GO:BP | GO:0008272 | sulfate transport | 2 | 12 | 2.09e-01 |
| GO:BP | GO:0051257 | meiotic spindle midzone assembly | 1 | 2 | 2.10e-01 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 8 | 39 | 2.12e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 1 | 113 | 2.12e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 6 | 39 | 2.13e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 2 | 6 | 2.16e-01 |
| GO:BP | GO:0090129 | positive regulation of synapse maturation | 1 | 8 | 2.16e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 5 | 486 | 2.16e-01 |
| GO:BP | GO:0031915 | positive regulation of synaptic plasticity | 1 | 6 | 2.16e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 2 | 56 | 2.18e-01 |
| GO:BP | GO:0061485 | memory T cell proliferation | 1 | 1 | 2.18e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 1 | 8 | 2.18e-01 |
| GO:BP | GO:0003336 | corneocyte desquamation | 1 | 1 | 2.18e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 3 | 12 | 2.20e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 3 | 12 | 2.20e-01 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 1 | 8 | 2.20e-01 |
| GO:BP | GO:0051106 | positive regulation of DNA ligation | 1 | 5 | 2.20e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 1 | 17 | 2.21e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 1 | 127 | 2.21e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 2 | 7 | 2.21e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 1 | 117 | 2.22e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 17 | 144 | 2.22e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 3 | 15 | 2.24e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 3 | 11 | 2.25e-01 |
| GO:BP | GO:0051098 | regulation of binding | 6 | 317 | 2.26e-01 |
| GO:BP | GO:0032223 | negative regulation of synaptic transmission, cholinergic | 1 | 1 | 2.26e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 7 | 50 | 2.26e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 6 | 37 | 2.26e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 2 | 1119 | 2.26e-01 |
| GO:BP | GO:0035445 | borate transmembrane transport | 1 | 1 | 2.26e-01 |
| GO:BP | GO:0046713 | borate transport | 1 | 1 | 2.26e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 2 | 290 | 2.27e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 3 | 16 | 2.27e-01 |
| GO:BP | GO:0006824 | cobalt ion transport | 2 | 5 | 2.27e-01 |
| GO:BP | GO:0097490 | sympathetic neuron projection extension | 2 | 4 | 2.27e-01 |
| GO:BP | GO:0061551 | trigeminal ganglion development | 2 | 3 | 2.27e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 5 | 2.27e-01 |
| GO:BP | GO:0097491 | sympathetic neuron projection guidance | 2 | 4 | 2.27e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 2 | 21 | 2.27e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 3 | 12 | 2.28e-01 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 2 | 12 | 2.28e-01 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 1 | 3 | 2.28e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 1 | 6 | 2.29e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 5 | 421 | 2.29e-01 |
| GO:BP | GO:0071921 | cohesin loading | 1 | 3 | 2.29e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 4 | 19 | 2.29e-01 |
| GO:BP | GO:0060857 | establishment of glial blood-brain barrier | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0045213 | neurotransmitter receptor metabolic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0019054 | modulation by virus of host cellular process | 2 | 5 | 2.30e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 4 | 11 | 2.30e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 4 | 16 | 2.30e-01 |
| GO:BP | GO:0019048 | modulation by virus of host process | 2 | 5 | 2.30e-01 |
| GO:BP | GO:0044003 | modulation by symbiont of host process | 2 | 5 | 2.30e-01 |
| GO:BP | GO:0044068 | modulation by symbiont of host cellular process | 2 | 5 | 2.30e-01 |
| GO:BP | GO:0014038 | regulation of Schwann cell differentiation | 1 | 3 | 2.32e-01 |
| GO:BP | GO:1900146 | negative regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 1 | 2.32e-01 |
| GO:BP | GO:1900176 | negative regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 1 | 2.32e-01 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 6 | 64 | 2.32e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 3 | 8 | 2.32e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 2 | 10 | 2.32e-01 |
| GO:BP | GO:0048477 | oogenesis | 7 | 64 | 2.32e-01 |
| GO:BP | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | 2 | 8 | 2.33e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 1 | 19 | 2.33e-01 |
| GO:BP | GO:0021636 | trigeminal nerve morphogenesis | 2 | 4 | 2.33e-01 |
| GO:BP | GO:0097492 | sympathetic neuron axon guidance | 2 | 4 | 2.33e-01 |
| GO:BP | GO:0021637 | trigeminal nerve structural organization | 2 | 4 | 2.33e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 3 | 16 | 2.35e-01 |
| GO:BP | GO:0031291 | Ran protein signal transduction | 1 | 3 | 2.35e-01 |
| GO:BP | GO:0003193 | pulmonary valve formation | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0086054 | bundle of His cell to Purkinje myocyte communication by electrical coupling | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0098905 | regulation of bundle of His cell action potential | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0010643 | cell communication by chemical coupling | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 2 | 23 | 2.36e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 3 | 18 | 2.36e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 6 | 22 | 2.37e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 6 | 77 | 2.37e-01 |
| GO:BP | GO:0072703 | cellular response to methyl methanesulfonate | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 7 | 39 | 2.39e-01 |
| GO:BP | GO:0072702 | response to methyl methanesulfonate | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 1 | 210 | 2.39e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 4 | 10 | 2.39e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 5 | 46 | 2.41e-01 |
| GO:BP | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process | 2 | 6 | 2.41e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 4 | 16 | 2.42e-01 |
| GO:BP | GO:1900240 | negative regulation of phenotypic switching | 1 | 2 | 2.42e-01 |
| GO:BP | GO:1905916 | negative regulation of cell differentiation involved in phenotypic switching | 1 | 2 | 2.42e-01 |
| GO:BP | GO:1905931 | negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 2 | 2.42e-01 |
| GO:BP | GO:0051105 | regulation of DNA ligation | 1 | 6 | 2.44e-01 |
| GO:BP | GO:0071602 | phytosphingosine biosynthetic process | 1 | 1 | 2.44e-01 |
| GO:BP | GO:0006671 | phytosphingosine metabolic process | 1 | 1 | 2.44e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 2 | 1214 | 2.44e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 3 | 10 | 2.44e-01 |
| GO:BP | GO:0016570 | histone modification | 1 | 433 | 2.44e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 9 | 67 | 2.44e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 3 | 10 | 2.44e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 2 | 64 | 2.44e-01 |
| GO:BP | GO:0018057 | peptidyl-lysine oxidation | 2 | 5 | 2.44e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 2 | 1209 | 2.45e-01 |
| GO:BP | GO:0003094 | glomerular filtration | 5 | 21 | 2.45e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 9 | 55 | 2.45e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 9 | 55 | 2.45e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 5 | 40 | 2.45e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 3 | 30 | 2.45e-01 |
| GO:BP | GO:0048818 | positive regulation of hair follicle maturation | 2 | 5 | 2.46e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 2 | 63 | 2.46e-01 |
| GO:BP | GO:0015676 | vanadium ion transport | 1 | 1 | 2.46e-01 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 2 | 10 | 2.46e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 4 | 17 | 2.46e-01 |
| GO:BP | GO:0035444 | nickel cation transmembrane transport | 1 | 1 | 2.46e-01 |
| GO:BP | GO:0015692 | lead ion transport | 1 | 1 | 2.46e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 7 | 44 | 2.46e-01 |
| GO:BP | GO:0015675 | nickel cation transport | 1 | 1 | 2.46e-01 |
| GO:BP | GO:0000320 | re-entry into mitotic cell cycle | 1 | 2 | 2.46e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 25 | 494 | 2.47e-01 |
| GO:BP | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome | 1 | 1 | 2.48e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 2 | 22 | 2.48e-01 |
| GO:BP | GO:0071476 | cellular hypotonic response | 2 | 8 | 2.49e-01 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 3 | 10 | 2.49e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 2 | 1239 | 2.49e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 2 | 64 | 2.50e-01 |
| GO:BP | GO:2000615 | regulation of histone H3-K9 acetylation | 2 | 7 | 2.50e-01 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 2 | 8 | 2.50e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 2.50e-01 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0033567 | DNA replication, Okazaki fragment processing | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 2 | 6 | 2.50e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 2 | 6 | 2.50e-01 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 2 | 5 | 2.50e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 6 | 114 | 2.50e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 6 | 54 | 2.51e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 2.51e-01 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 1 | 13 | 2.51e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 3 | 28 | 2.51e-01 |
| GO:BP | GO:1904716 | positive regulation of chaperone-mediated autophagy | 1 | 1 | 2.51e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 8 | 165 | 2.51e-01 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 6 | 37 | 2.51e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 3 | 8 | 2.51e-01 |
| GO:BP | GO:0021561 | facial nerve development | 3 | 8 | 2.51e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 8 | 331 | 2.51e-01 |
| GO:BP | GO:0007000 | nucleolus organization | 4 | 17 | 2.51e-01 |
| GO:BP | GO:0099041 | vesicle tethering to Golgi | 2 | 5 | 2.51e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 4 | 84 | 2.52e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 1 | 48 | 2.52e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 2 | 26 | 2.52e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 2 | 7 | 2.54e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 1 | 12 | 2.54e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 6 | 79 | 2.54e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 13 | 1378 | 2.55e-01 |
| GO:BP | GO:0003285 | septum secundum development | 1 | 3 | 2.56e-01 |
| GO:BP | GO:1904999 | positive regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 2.56e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 1 | 2 | 2.56e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 2 | 1280 | 2.56e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 4 | 26 | 2.56e-01 |
| GO:BP | GO:0034337 | RNA folding | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0032203 | telomere formation via telomerase | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 4 | 11 | 2.57e-01 |
| GO:BP | GO:0000709 | meiotic joint molecule formation | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0000708 | meiotic strand invasion | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0010774 | meiotic strand invasion involved in reciprocal meiotic recombination | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 7 | 44 | 2.57e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 5 | 19 | 2.57e-01 |
| GO:BP | GO:1904232 | regulation of aconitate hydratase activity | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 1 | 94 | 2.57e-01 |
| GO:BP | GO:1904234 | positive regulation of aconitate hydratase activity | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 3 | 538 | 2.57e-01 |
| GO:BP | GO:0046521 | sphingoid catabolic process | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0034760 | negative regulation of iron ion transmembrane transport | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0061152 | trachea submucosa development | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0061153 | trachea gland development | 1 | 2 | 2.57e-01 |
| GO:BP | GO:1905364 | regulation of endosomal vesicle fusion | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0070342 | brown fat cell proliferation | 1 | 1 | 2.57e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 3 | 21 | 2.57e-01 |
| GO:BP | GO:0034757 | negative regulation of iron ion transport | 1 | 2 | 2.57e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 2.57e-01 |
| GO:BP | GO:0030026 | intracellular manganese ion homeostasis | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0015694 | mercury ion transport | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0060380 | regulation of single-stranded telomeric DNA binding | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0060381 | positive regulation of single-stranded telomeric DNA binding | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 2.59e-01 |
| GO:BP | GO:0036146 | cellular response to mycotoxin | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 9 | 65 | 2.59e-01 |
| GO:BP | GO:0097080 | plasma membrane selenite transport | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0072721 | cellular response to dithiothreitol | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 1 | 20 | 2.59e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 1 | 11 | 2.59e-01 |
| GO:BP | GO:1990540 | mitochondrial manganese ion transmembrane transport | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 1 | 4 | 2.59e-01 |
| GO:BP | GO:0016203 | muscle attachment | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 2 | 6 | 2.59e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 3 | 39 | 2.59e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 1 | 5 | 2.59e-01 |
| GO:BP | GO:0045190 | isotype switching | 3 | 39 | 2.59e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 3 | 39 | 2.59e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 3 | 214 | 2.59e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 3 | 12 | 2.59e-01 |
| GO:BP | GO:0110017 | cap-independent translational initiation of linear mRNA | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 2.59e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 2 | 26 | 2.59e-01 |
| GO:BP | GO:1900121 | negative regulation of receptor binding | 2 | 8 | 2.59e-01 |
| GO:BP | GO:0009153 | purine deoxyribonucleotide biosynthetic process | 2 | 4 | 2.59e-01 |
| GO:BP | GO:0002192 | IRES-dependent translational initiation of linear mRNA | 1 | 1 | 2.59e-01 |
| GO:BP | GO:0035519 | protein K29-linked ubiquitination | 2 | 6 | 2.59e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 2.60e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 2 | 9 | 2.60e-01 |
| GO:BP | GO:0090400 | stress-induced premature senescence | 1 | 7 | 2.62e-01 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 2 | 8 | 2.62e-01 |
| GO:BP | GO:1903394 | protein localization to kinetochore involved in kinetochore assembly | 1 | 1 | 2.62e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 4 | 200 | 2.62e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 4 | 40 | 2.62e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 11 | 68 | 2.62e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 3 | 8 | 2.62e-01 |
| GO:BP | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 1 | 3 | 2.62e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 3 | 21 | 2.63e-01 |
| GO:BP | GO:0007398 | ectoderm development | 4 | 15 | 2.63e-01 |
| GO:BP | GO:0061348 | planar cell polarity pathway involved in ventricular septum morphogenesis | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis | 1 | 1 | 2.63e-01 |
| GO:BP | GO:1904934 | negative regulation of cell proliferation in midbrain | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0061346 | planar cell polarity pathway involved in heart morphogenesis | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0036517 | chemoattraction of serotonergic neuron axon | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0061354 | planar cell polarity pathway involved in pericardium morphogenesis | 1 | 1 | 2.63e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 12 | 63 | 2.63e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 8 | 317 | 2.63e-01 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 2 | 9 | 2.63e-01 |
| GO:BP | GO:0036369 | transcription factor catabolic process | 2 | 5 | 2.63e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 3 | 14 | 2.63e-01 |
| GO:BP | GO:0061350 | planar cell polarity pathway involved in cardiac muscle tissue morphogenesis | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 2 | 5 | 2.63e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 9 | 65 | 2.63e-01 |
| GO:BP | GO:0061349 | planar cell polarity pathway involved in cardiac right atrium morphogenesis | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 2.63e-01 |
| GO:BP | GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 1 | 7 | 2.63e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 5 | 44 | 2.63e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 2.63e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 2 | 809 | 2.63e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 5 | 89 | 2.63e-01 |
| GO:BP | GO:1901862 | negative regulation of muscle tissue development | 2 | 9 | 2.64e-01 |
| GO:BP | GO:0099088 | axonal transport of messenger ribonucleoprotein complex | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0046065 | dCTP metabolic process | 1 | 1 | 2.65e-01 |
| GO:BP | GO:1903823 | telomere single strand break repair | 1 | 1 | 2.65e-01 |
| GO:BP | GO:1903517 | negative regulation of single strand break repair | 1 | 1 | 2.65e-01 |
| GO:BP | GO:1903824 | negative regulation of telomere single strand break repair | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 3 | 120 | 2.65e-01 |
| GO:BP | GO:0006253 | dCTP catabolic process | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 7 | 55 | 2.65e-01 |
| GO:BP | GO:0032207 | regulation of telomere maintenance via recombination | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0032208 | negative regulation of telomere maintenance via recombination | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 2.65e-01 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 1 | 179 | 2.65e-01 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 2.65e-01 |
| GO:BP | GO:1903770 | negative regulation of beta-galactosidase activity | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 2.65e-01 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 2.65e-01 |
| GO:BP | GO:1905331 | negative regulation of morphogenesis of an epithelium | 2 | 6 | 2.66e-01 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 2 | 7 | 2.66e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 29 | 3625 | 2.66e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 2 | 13 | 2.66e-01 |
| GO:BP | GO:0090240 | positive regulation of histone H4 acetylation | 2 | 6 | 2.66e-01 |
| GO:BP | GO:0001822 | kidney development | 1 | 244 | 2.67e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 9 | 60 | 2.69e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 18 | 247 | 2.69e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 3 | 556 | 2.69e-01 |
| GO:BP | GO:0051418 | microtubule nucleation by microtubule organizing center | 1 | 5 | 2.69e-01 |
| GO:BP | GO:0043970 | histone H3-K9 acetylation | 2 | 8 | 2.69e-01 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 1 | 11 | 2.70e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 1 | 192 | 2.70e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 17 | 133 | 2.71e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 11 | 194 | 2.71e-01 |
| GO:BP | GO:0097205 | renal filtration | 5 | 23 | 2.71e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 3 | 222 | 2.71e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 7 | 39 | 2.71e-01 |
| GO:BP | GO:0072001 | renal system development | 1 | 251 | 2.72e-01 |
| GO:BP | GO:0030157 | pancreatic juice secretion | 2 | 11 | 2.74e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 2 | 7 | 2.74e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 4 | 21 | 2.74e-01 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 4 | 19 | 2.74e-01 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 2 | 5 | 2.74e-01 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 2 | 7 | 2.74e-01 |
| GO:BP | GO:0035965 | cardiolipin acyl-chain remodeling | 2 | 4 | 2.74e-01 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 4 | 19 | 2.74e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 2 | 5 | 2.74e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 1 | 71 | 2.74e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 1 | 71 | 2.74e-01 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 2.75e-01 |
| GO:BP | GO:0071788 | endoplasmic reticulum tubular network maintenance | 1 | 1 | 2.76e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 12 | 219 | 2.76e-01 |
| GO:BP | GO:0021627 | olfactory nerve morphogenesis | 1 | 3 | 2.76e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 4 | 12 | 2.76e-01 |
| GO:BP | GO:0006567 | threonine catabolic process | 1 | 3 | 2.77e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 10 | 85 | 2.77e-01 |
| GO:BP | GO:0014075 | response to amine | 4 | 34 | 2.77e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 13 | 96 | 2.78e-01 |
| GO:BP | GO:0051026 | chiasma assembly | 1 | 3 | 2.78e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 1 | 162 | 2.79e-01 |
| GO:BP | GO:1901202 | negative regulation of extracellular matrix assembly | 1 | 1 | 2.79e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 59 | 1149 | 2.79e-01 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 1 | 29 | 2.81e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 3 | 77 | 2.81e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 10 | 89 | 2.81e-01 |
| GO:BP | GO:1903551 | regulation of extracellular exosome assembly | 2 | 6 | 2.81e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 1 | 197 | 2.81e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 3 | 41 | 2.82e-01 |
| GO:BP | GO:0035290 | trunk segmentation | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 3 | 11 | 2.82e-01 |
| GO:BP | GO:0030202 | heparin metabolic process | 3 | 14 | 2.82e-01 |
| GO:BP | GO:0033623 | regulation of integrin activation | 1 | 11 | 2.82e-01 |
| GO:BP | GO:0036484 | trunk neural crest cell migration | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0061550 | cranial ganglion development | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 6 | 33 | 2.82e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 6 | 25 | 2.82e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 6 | 25 | 2.82e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 12 | 2.82e-01 |
| GO:BP | GO:1901859 | negative regulation of mitochondrial DNA metabolic process | 1 | 2 | 2.83e-01 |
| GO:BP | GO:0007517 | muscle organ development | 1 | 277 | 2.84e-01 |
| GO:BP | GO:1990789 | thyroid gland epithelial cell proliferation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0031441 | negative regulation of mRNA 3’-end processing | 1 | 2 | 2.84e-01 |
| GO:BP | GO:1904441 | regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 10 | 70 | 2.84e-01 |
| GO:BP | GO:1904442 | negative regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 2.84e-01 |
| GO:BP | GO:1903697 | negative regulation of microvillus assembly | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0060243 | negative regulation of cell growth involved in contact inhibition | 1 | 1 | 2.86e-01 |
| GO:BP | GO:0070212 | protein poly-ADP-ribosylation | 2 | 6 | 2.86e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 4 | 135 | 2.86e-01 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 2 | 8 | 2.86e-01 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 2 | 8 | 2.86e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 2 | 39 | 2.87e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 7 | 59 | 2.87e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 3 | 18 | 2.87e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 11 | 71 | 2.87e-01 |
| GO:BP | GO:0032298 | positive regulation of DNA-templated DNA replication initiation | 1 | 2 | 2.87e-01 |
| GO:BP | GO:0072402 | response to DNA integrity checkpoint signaling | 1 | 8 | 2.87e-01 |
| GO:BP | GO:0072396 | response to cell cycle checkpoint signaling | 1 | 8 | 2.87e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 3 | 10 | 2.87e-01 |
| GO:BP | GO:1902729 | negative regulation of proteoglycan biosynthetic process | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0072423 | response to DNA damage checkpoint signaling | 1 | 8 | 2.87e-01 |
| GO:BP | GO:0030913 | paranodal junction assembly | 2 | 6 | 2.87e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 18 | 270 | 2.87e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 1 | 20 | 2.87e-01 |
| GO:BP | GO:0032683 | negative regulation of connective tissue growth factor production | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0044210 | ‘de novo’ CTP biosynthetic process | 1 | 2 | 2.88e-01 |
| GO:BP | GO:1900239 | regulation of phenotypic switching | 1 | 3 | 2.88e-01 |
| GO:BP | GO:1905460 | negative regulation of vascular associated smooth muscle cell apoptotic process | 1 | 2 | 2.88e-01 |
| GO:BP | GO:0090679 | cell differentiation involved in phenotypic switching | 1 | 3 | 2.88e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 2.88e-01 |
| GO:BP | GO:1905420 | vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 2.88e-01 |
| GO:BP | GO:1905915 | regulation of cell differentiation involved in phenotypic switching | 1 | 3 | 2.88e-01 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 6 | 36 | 2.88e-01 |
| GO:BP | GO:1905930 | regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 2.88e-01 |
| GO:BP | GO:0006971 | hypotonic response | 2 | 10 | 2.88e-01 |
| GO:BP | GO:0021785 | branchiomotor neuron axon guidance | 2 | 5 | 2.88e-01 |
| GO:BP | GO:0042413 | carnitine catabolic process | 1 | 1 | 2.89e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 5 | 26 | 2.89e-01 |
| GO:BP | GO:0055057 | neuroblast division | 1 | 12 | 2.89e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 4 | 89 | 2.89e-01 |
| GO:BP | GO:0006043 | glucosamine catabolic process | 1 | 2 | 2.89e-01 |
| GO:BP | GO:0098907 | regulation of SA node cell action potential | 2 | 7 | 2.89e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 2.91e-01 |
| GO:BP | GO:1902153 | regulation of response to DNA damage checkpoint signaling | 1 | 1 | 2.91e-01 |
| GO:BP | GO:0097017 | renal protein absorption | 2 | 5 | 2.91e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | 2 | 4 | 2.91e-01 |
| GO:BP | GO:1902151 | regulation of response to DNA integrity checkpoint signaling | 1 | 1 | 2.91e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 2.91e-01 |
| GO:BP | GO:1902145 | regulation of response to cell cycle checkpoint signaling | 1 | 1 | 2.91e-01 |
| GO:BP | GO:0034759 | regulation of iron ion transmembrane transport | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 3 | 13 | 2.91e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 19 | 259 | 2.91e-01 |
| GO:BP | GO:0046813 | receptor-mediated virion attachment to host cell | 2 | 6 | 2.91e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 2.91e-01 |
| GO:BP | GO:1904762 | positive regulation of myofibroblast differentiation | 1 | 1 | 2.91e-01 |
| GO:BP | GO:0000710 | meiotic mismatch repair | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 4 | 13 | 2.92e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 10 | 109 | 2.92e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 4 | 19 | 2.92e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 4 | 25 | 2.92e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 13 | 160 | 2.93e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 8 | 257 | 2.93e-01 |
| GO:BP | GO:0099172 | presynapse organization | 8 | 42 | 2.94e-01 |
| GO:BP | GO:0036298 | recombinational interstrand cross-link repair | 1 | 2 | 2.94e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 1 | 49 | 2.94e-01 |
| GO:BP | GO:0070716 | mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication | 1 | 2 | 2.94e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 4 | 51 | 2.94e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 1 | 22 | 2.94e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 4 | 47 | 2.94e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 4 | 22 | 2.94e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 2 | 10 | 2.94e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 1 | 3 | 2.94e-01 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 1 | 16 | 2.94e-01 |
| GO:BP | GO:1900142 | negative regulation of oligodendrocyte apoptotic process | 1 | 1 | 2.94e-01 |
| GO:BP | GO:2000533 | negative regulation of renal albumin absorption | 1 | 1 | 2.94e-01 |
| GO:BP | GO:0021768 | nucleus accumbens development | 1 | 1 | 2.94e-01 |
| GO:BP | GO:0035364 | thymine transport | 1 | 1 | 2.95e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 7 | 51 | 2.95e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 1 | 14 | 2.95e-01 |
| GO:BP | GO:0018158 | protein oxidation | 3 | 12 | 2.95e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 2.95e-01 |
| GO:BP | GO:0043157 | response to cation stress | 1 | 1 | 2.95e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 1 | 4 | 2.96e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 1 | 4 | 2.96e-01 |
| GO:BP | GO:1904116 | response to vasopressin | 1 | 5 | 2.96e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 1 | 4 | 2.96e-01 |
| GO:BP | GO:0007382 | specification of segmental identity, maxillary segment | 1 | 1 | 2.96e-01 |
| GO:BP | GO:0044751 | cellular response to human chorionic gonadotropin stimulus | 1 | 2 | 2.96e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 1 | 4 | 2.96e-01 |
| GO:BP | GO:0003170 | heart valve development | 6 | 65 | 2.96e-01 |
| GO:BP | GO:1904117 | cellular response to vasopressin | 1 | 5 | 2.96e-01 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 5 | 26 | 2.96e-01 |
| GO:BP | GO:1902990 | mitotic telomere maintenance via semi-conservative replication | 1 | 2 | 2.96e-01 |
| GO:BP | GO:1905871 | regulation of protein localization to cell leading edge | 1 | 2 | 2.96e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 14 | 123 | 2.96e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 3 | 13 | 2.97e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 1 | 28 | 2.97e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 2.97e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 41 | 534 | 2.97e-01 |
| GO:BP | GO:0019418 | sulfide oxidation | 1 | 3 | 2.97e-01 |
| GO:BP | GO:0060301 | positive regulation of cytokine activity | 1 | 1 | 2.97e-01 |
| GO:BP | GO:1905040 | otic placode development | 1 | 1 | 2.97e-01 |
| GO:BP | GO:0070221 | sulfide oxidation, using sulfide:quinone oxidoreductase | 1 | 3 | 2.97e-01 |
| GO:BP | GO:0072675 | osteoclast fusion | 2 | 5 | 2.97e-01 |
| GO:BP | GO:1902378 | VEGF-activated neuropilin signaling pathway involved in axon guidance | 1 | 1 | 2.97e-01 |
| GO:BP | GO:0042594 | response to starvation | 11 | 178 | 2.97e-01 |
| GO:BP | GO:0120263 | positive regulation of heterochromatin organization | 2 | 15 | 2.98e-01 |
| GO:BP | GO:0010668 | ectodermal cell differentiation | 2 | 5 | 2.98e-01 |
| GO:BP | GO:0031453 | positive regulation of heterochromatin formation | 2 | 15 | 2.98e-01 |
| GO:BP | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 2.98e-01 |
| GO:BP | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 2.98e-01 |
| GO:BP | GO:0006175 | dATP biosynthetic process | 1 | 1 | 2.98e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 13 | 156 | 2.98e-01 |
| GO:BP | GO:0010621 | negative regulation of transcription by transcription factor localization | 1 | 3 | 2.98e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 5 | 35 | 2.99e-01 |
| GO:BP | GO:0051598 | meiotic recombination checkpoint signaling | 1 | 2 | 2.99e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 3 | 20 | 3.00e-01 |
| GO:BP | GO:0003335 | corneocyte development | 1 | 2 | 3.00e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 3 | 16 | 3.02e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 31 | 2724 | 3.02e-01 |
| GO:BP | GO:0043201 | response to leucine | 2 | 13 | 3.02e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 3.02e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 1 | 15 | 3.02e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 3 | 47 | 3.02e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 1 | 343 | 3.02e-01 |
| GO:BP | GO:0002540 | leukotriene production involved in inflammatory response | 1 | 2 | 3.02e-01 |
| GO:BP | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response | 1 | 2 | 3.02e-01 |
| GO:BP | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 1 | 2 | 3.03e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 2 | 46 | 3.03e-01 |
| GO:BP | GO:1904914 | negative regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 3.03e-01 |
| GO:BP | GO:1904913 | regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 3.03e-01 |
| GO:BP | GO:1904911 | negative regulation of establishment of RNA localization to telomere | 1 | 1 | 3.03e-01 |
| GO:BP | GO:1904850 | negative regulation of establishment of protein localization to telomere | 1 | 1 | 3.03e-01 |
| GO:BP | GO:1904792 | positive regulation of shelterin complex assembly | 1 | 1 | 3.03e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 1 | 194 | 3.03e-01 |
| GO:BP | GO:0072717 | cellular response to actinomycin D | 1 | 4 | 3.03e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 2 | 46 | 3.03e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 2 | 6 | 3.03e-01 |
| GO:BP | GO:1904910 | regulation of establishment of RNA localization to telomere | 1 | 1 | 3.03e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 2 | 9 | 3.03e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 2 | 6 | 3.04e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 1 | 191 | 3.05e-01 |
| GO:BP | GO:0032072 | regulation of restriction endodeoxyribonuclease activity | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 1 | 14 | 3.05e-01 |
| GO:BP | GO:0036138 | peptidyl-histidine hydroxylation | 1 | 1 | 3.05e-01 |
| GO:BP | GO:0051641 | cellular localization | 3 | 2810 | 3.05e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 3.05e-01 |
| GO:BP | GO:0072134 | nephrogenic mesenchyme morphogenesis | 1 | 2 | 3.05e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 1 | 92 | 3.05e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 42 | 551 | 3.05e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 1 | 92 | 3.05e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 1 | 92 | 3.05e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 7 | 44 | 3.05e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 4 | 22 | 3.05e-01 |
| GO:BP | GO:1903429 | regulation of cell maturation | 1 | 13 | 3.06e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 3 | 11 | 3.06e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2 | 7 | 3.06e-01 |
| GO:BP | GO:0051573 | negative regulation of histone H3-K9 methylation | 1 | 5 | 3.06e-01 |
| GO:BP | GO:0001560 | regulation of cell growth by extracellular stimulus | 1 | 3 | 3.06e-01 |
| GO:BP | GO:0061087 | positive regulation of histone H3-K27 methylation | 1 | 4 | 3.06e-01 |
| GO:BP | GO:0032074 | negative regulation of nuclease activity | 2 | 6 | 3.07e-01 |
| GO:BP | GO:0001507 | acetylcholine catabolic process in synaptic cleft | 1 | 2 | 3.07e-01 |
| GO:BP | GO:0140009 | L-aspartate import across plasma membrane | 2 | 5 | 3.09e-01 |
| GO:BP | GO:0098530 | positive regulation of strand invasion | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0046317 | regulation of glucosylceramide biosynthetic process | 1 | 1 | 3.09e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 1 | 16 | 3.09e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 2 | 9 | 3.09e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 3 | 17 | 3.09e-01 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 3.09e-01 |
| GO:BP | GO:0060542 | regulation of strand invasion | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 4 | 19 | 3.09e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 1 | 279 | 3.09e-01 |
| GO:BP | GO:0046318 | negative regulation of glucosylceramide biosynthetic process | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 4 | 30 | 3.09e-01 |
| GO:BP | GO:0046033 | AMP metabolic process | 4 | 22 | 3.11e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 2 | 187 | 3.11e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 2 | 187 | 3.11e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 1 | 11 | 3.11e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 3 | 35 | 3.11e-01 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 4 | 21 | 3.11e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 3 | 6 | 3.12e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 3 | 6 | 3.12e-01 |
| GO:BP | GO:0097510 | base-excision repair, AP site formation via deaminated base removal | 1 | 1 | 3.12e-01 |
| GO:BP | GO:0009448 | gamma-aminobutyric acid metabolic process | 1 | 6 | 3.12e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 2 | 386 | 3.12e-01 |
| GO:BP | GO:0032899 | regulation of neurotrophin production | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 4 | 21 | 3.12e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 6 | 294 | 3.12e-01 |
| GO:BP | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 1 | 4 | 3.12e-01 |
| GO:BP | GO:0008065 | establishment of blood-nerve barrier | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 1 | 15 | 3.12e-01 |
| GO:BP | GO:0051660 | establishment of centrosome localization | 1 | 8 | 3.12e-01 |
| GO:BP | GO:0033566 | gamma-tubulin complex localization | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0031508 | pericentric heterochromatin formation | 1 | 3 | 3.12e-01 |
| GO:BP | GO:1905269 | positive regulation of chromatin organization | 2 | 15 | 3.16e-01 |
| GO:BP | GO:0035582 | sequestering of BMP in extracellular matrix | 1 | 2 | 3.16e-01 |
| GO:BP | GO:1900108 | negative regulation of nodal signaling pathway | 1 | 2 | 3.16e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 12 | 1345 | 3.16e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 1 | 5 | 3.16e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 22 | 151 | 3.16e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 6 | 41 | 3.16e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 3 | 19 | 3.16e-01 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 3.16e-01 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 3 | 15 | 3.16e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 3.16e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 1 | 13 | 3.16e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 4 | 16 | 3.16e-01 |
| GO:BP | GO:0035609 | C-terminal protein deglutamylation | 1 | 2 | 3.17e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 5 | 63 | 3.18e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 2 | 6 | 3.18e-01 |
| GO:BP | GO:0036438 | maintenance of lens transparency | 2 | 6 | 3.18e-01 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 1 | 6 | 3.19e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 5 | 3.19e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 4 | 22 | 3.19e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 3.19e-01 |
| GO:BP | GO:0003105 | negative regulation of glomerular filtration | 1 | 3 | 3.19e-01 |
| GO:BP | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation | 2 | 6 | 3.19e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 3.19e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 3.19e-01 |
| GO:BP | GO:0071578 | zinc ion import across plasma membrane | 2 | 6 | 3.19e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 4 | 70 | 3.19e-01 |
| GO:BP | GO:1903712 | cysteine transmembrane transport | 1 | 3 | 3.19e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 1 | 21 | 3.19e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 1 | 5 | 3.19e-01 |
| GO:BP | GO:0071288 | cellular response to mercury ion | 1 | 2 | 3.19e-01 |
| GO:BP | GO:0071971 | extracellular exosome assembly | 2 | 6 | 3.19e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 3.19e-01 |
| GO:BP | GO:0042883 | cysteine transport | 1 | 3 | 3.19e-01 |
| GO:BP | GO:0086017 | Purkinje myocyte action potential | 1 | 4 | 3.19e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 4 | 22 | 3.19e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 4 | 30 | 3.19e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 1 | 276 | 3.19e-01 |
| GO:BP | GO:0036333 | hepatocyte homeostasis | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0097552 | mitochondrial double-strand break repair via homologous recombination | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0097551 | mitochondrial double-strand break repair | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 2 | 7 | 3.20e-01 |
| GO:BP | GO:0099158 | regulation of recycling endosome localization within postsynapse | 1 | 1 | 3.21e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 2 | 11 | 3.22e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 5 | 51 | 3.22e-01 |
| GO:BP | GO:0097502 | mannosylation | 5 | 32 | 3.23e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 2 | 21 | 3.23e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 3 | 7 | 3.23e-01 |
| GO:BP | GO:0033762 | response to glucagon | 1 | 13 | 3.23e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 2 | 8 | 3.23e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 2 | 7 | 3.24e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 1 | 277 | 3.25e-01 |
| GO:BP | GO:1905091 | positive regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 3.26e-01 |
| GO:BP | GO:0110099 | negative regulation of calcium import into the mitochondrion | 1 | 1 | 3.26e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 2 | 1735 | 3.26e-01 |
| GO:BP | GO:0033082 | regulation of extrathymic T cell differentiation | 1 | 1 | 3.26e-01 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 1 | 14 | 3.27e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0000023 | maltose metabolic process | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 1 | 57 | 3.29e-01 |
| GO:BP | GO:1904613 | cellular response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 3.29e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 3 | 14 | 3.29e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 1 | 165 | 3.29e-01 |
| GO:BP | GO:0035754 | B cell chemotaxis | 2 | 5 | 3.29e-01 |
| GO:BP | GO:0051764 | actin crosslink formation | 1 | 10 | 3.29e-01 |
| GO:BP | GO:1902897 | regulation of postsynaptic density protein 95 clustering | 1 | 2 | 3.29e-01 |
| GO:BP | GO:2000210 | positive regulation of anoikis | 1 | 6 | 3.29e-01 |
| GO:BP | GO:0072033 | renal vesicle formation | 2 | 8 | 3.29e-01 |
| GO:BP | GO:0034589 | hydroxyproline transport | 1 | 1 | 3.29e-01 |
| GO:BP | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 4 | 19 | 3.29e-01 |
| GO:BP | GO:0031427 | response to methotrexate | 1 | 1 | 3.29e-01 |
| GO:BP | GO:0015826 | threonine transport | 1 | 1 | 3.29e-01 |
| GO:BP | GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 1 | 5 | 3.29e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 3.29e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 1 | 167 | 3.29e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 3.29e-01 |
| GO:BP | GO:1901166 | neural crest cell migration involved in autonomic nervous system development | 2 | 5 | 3.30e-01 |
| GO:BP | GO:0019694 | alkanesulfonate metabolic process | 1 | 6 | 3.30e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 1 | 21 | 3.30e-01 |
| GO:BP | GO:0021856 | hypothalamic tangential migration using cell-axon interactions | 2 | 6 | 3.30e-01 |
| GO:BP | GO:0021828 | gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 2 | 6 | 3.30e-01 |
| GO:BP | GO:0019530 | taurine metabolic process | 1 | 6 | 3.30e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 1 | 186 | 3.30e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 1 | 16 | 3.30e-01 |
| GO:BP | GO:0044321 | response to leptin | 1 | 19 | 3.30e-01 |
| GO:BP | GO:0048132 | female germ-line stem cell asymmetric division | 1 | 1 | 3.30e-01 |
| GO:BP | GO:0015728 | mevalonate transport | 1 | 1 | 3.30e-01 |
| GO:BP | GO:0051780 | behavioral response to nutrient | 1 | 1 | 3.30e-01 |
| GO:BP | GO:0035583 | sequestering of TGFbeta in extracellular matrix | 1 | 3 | 3.30e-01 |
| GO:BP | GO:0021979 | hypothalamus cell differentiation | 3 | 8 | 3.30e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 13 | 103 | 3.30e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 3.30e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 3 | 14 | 3.31e-01 |
| GO:BP | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential | 1 | 3 | 3.31e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 2 | 8 | 3.31e-01 |
| GO:BP | GO:2001113 | negative regulation of cellular response to hepatocyte growth factor stimulus | 1 | 1 | 3.32e-01 |
| GO:BP | GO:1902203 | negative regulation of hepatocyte growth factor receptor signaling pathway | 1 | 1 | 3.32e-01 |
| GO:BP | GO:0071140 | resolution of mitotic recombination intermediates | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 6 | 72 | 3.33e-01 |
| GO:BP | GO:0070285 | pigment cell development | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0099140 | presynaptic actin cytoskeleton organization | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0033241 | regulation of amine catabolic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 2 | 12 | 3.33e-01 |
| GO:BP | GO:1900082 | negative regulation of arginine catabolic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:1900081 | regulation of arginine catabolic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0099187 | presynaptic cytoskeleton organization | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 8 | 94 | 3.33e-01 |
| GO:BP | GO:0098736 | negative regulation of the force of heart contraction | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0060398 | regulation of growth hormone receptor signaling pathway | 1 | 5 | 3.33e-01 |
| GO:BP | GO:1903249 | negative regulation of citrulline biosynthetic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0033242 | negative regulation of amine catabolic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:2000283 | negative regulation of amino acid biosynthetic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:1903248 | regulation of citrulline biosynthetic process | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 2 | 7 | 3.33e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 5 | 36 | 3.33e-01 |
| GO:BP | GO:1900234 | regulation of Kit signaling pathway | 1 | 2 | 3.33e-01 |
| GO:BP | GO:1900235 | negative regulation of Kit signaling pathway | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 1 | 5 | 3.33e-01 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 2 | 8 | 3.34e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 1 | 21 | 3.34e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 4 | 418 | 3.34e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 2 | 11 | 3.35e-01 |
| GO:BP | GO:0060242 | contact inhibition | 2 | 8 | 3.36e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 1 | 300 | 3.36e-01 |
| GO:BP | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | 1 | 2 | 3.36e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 3.36e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 11 | 80 | 3.36e-01 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 2 | 6 | 3.37e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 1 | 300 | 3.37e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 2 | 10 | 3.37e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 3.37e-01 |
| GO:BP | GO:0036337 | Fas signaling pathway | 1 | 5 | 3.37e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 3 | 36 | 3.37e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 3 | 188 | 3.37e-01 |
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 3.37e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 2 | 21 | 3.37e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 6 | 41 | 3.38e-01 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 3 | 14 | 3.38e-01 |
| GO:BP | GO:0097045 | phosphatidylserine exposure on blood platelet | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 1 | 27 | 3.39e-01 |
| GO:BP | GO:0000019 | regulation of mitotic recombination | 2 | 5 | 3.39e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 5 | 51 | 3.40e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 1 | 295 | 3.40e-01 |
| GO:BP | GO:0043063 | intercellular bridge organization | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 2 | 21 | 3.40e-01 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 2 | 8 | 3.40e-01 |
| GO:BP | GO:1990968 | modulation by host of RNA binding by virus | 1 | 1 | 3.40e-01 |
| GO:BP | GO:1990969 | modulation by host of viral RNA-binding transcription factor activity | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 2 | 11 | 3.40e-01 |
| GO:BP | GO:0061983 | meiosis II cell cycle process | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0007135 | meiosis II | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0007147 | female meiosis II | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0072308 | negative regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 1 | 3.41e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 4 | 23 | 3.41e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 1 | 5 | 3.41e-01 |
| GO:BP | GO:0048864 | stem cell development | 11 | 76 | 3.42e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 1 | 6 | 3.42e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 5 | 72 | 3.42e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 1 | 2 | 3.42e-01 |
| GO:BP | GO:0046444 | FMN metabolic process | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0009398 | FMN biosynthetic process | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0099068 | postsynapse assembly | 5 | 32 | 3.42e-01 |
| GO:BP | GO:0009231 | riboflavin biosynthetic process | 1 | 1 | 3.42e-01 |
| GO:BP | GO:1904997 | regulation of leukocyte adhesion to arterial endothelial cell | 1 | 3 | 3.42e-01 |
| GO:BP | GO:0052647 | pentitol phosphate metabolic process | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 1 | 23 | 3.42e-01 |
| GO:BP | GO:0035610 | protein side chain deglutamylation | 1 | 2 | 3.42e-01 |
| GO:BP | GO:0052648 | ribitol phosphate metabolic process | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 5 | 615 | 3.42e-01 |
| GO:BP | GO:0006829 | zinc ion transport | 4 | 23 | 3.42e-01 |
| GO:BP | GO:0021793 | chemorepulsion of branchiomotor axon | 1 | 1 | 3.43e-01 |
| GO:BP | GO:0006041 | glucosamine metabolic process | 1 | 3 | 3.43e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | 24 | 3.44e-01 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 1 | 5 | 3.45e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 5 | 42 | 3.46e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 7 | 56 | 3.48e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 3.48e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 2 | 8 | 3.48e-01 |
| GO:BP | GO:1900175 | regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 2 | 3.48e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 6 | 30 | 3.48e-01 |
| GO:BP | GO:0098749 | cerebellar neuron development | 1 | 3 | 3.48e-01 |
| GO:BP | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | 1 | 4 | 3.48e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 9 | 55 | 3.48e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 1 | 5 | 3.48e-01 |
| GO:BP | GO:0046053 | dAMP metabolic process | 2 | 5 | 3.48e-01 |
| GO:BP | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 2 | 3.48e-01 |
| GO:BP | GO:2000617 | positive regulation of histone H3-K9 acetylation | 1 | 6 | 3.48e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 8 | 212 | 3.48e-01 |
| GO:BP | GO:1904744 | positive regulation of telomeric DNA binding | 1 | 2 | 3.48e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 3 | 17 | 3.48e-01 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 2 | 5 | 3.48e-01 |
| GO:BP | GO:0032071 | regulation of endodeoxyribonuclease activity | 2 | 7 | 3.48e-01 |
| GO:BP | GO:0106300 | protein-DNA covalent cross-linking repair | 1 | 7 | 3.48e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 2 | 1849 | 3.48e-01 |
| GO:BP | GO:0009749 | response to glucose | 19 | 137 | 3.48e-01 |
| GO:BP | GO:0050787 | detoxification of mercury ion | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0097752 | regulation of DNA stability | 1 | 3 | 3.49e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 14 | 131 | 3.49e-01 |
| GO:BP | GO:0003139 | secondary heart field specification | 2 | 10 | 3.49e-01 |
| GO:BP | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration | 2 | 6 | 3.49e-01 |
| GO:BP | GO:1905719 | protein localization to perinuclear region of cytoplasm | 2 | 8 | 3.49e-01 |
| GO:BP | GO:0016340 | calcium-dependent cell-matrix adhesion | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 14 | 102 | 3.49e-01 |
| GO:BP | GO:0021796 | cerebral cortex regionalization | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 1 | 17 | 3.49e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 17 | 168 | 3.49e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 2 | 6 | 3.50e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 5 | 625 | 3.51e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 2 | 9 | 3.51e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 5 | 51 | 3.51e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 7 | 64 | 3.51e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 1 | 17 | 3.51e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 1 | 17 | 3.51e-01 |
| GO:BP | GO:0048642 | negative regulation of skeletal muscle tissue development | 1 | 3 | 3.52e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 2 | 27 | 3.53e-01 |
| GO:BP | GO:0061289 | Wnt signaling pathway involved in kidney development | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 2 | 7 | 3.53e-01 |
| GO:BP | GO:0061290 | canonical Wnt signaling pathway involved in metanephric kidney development | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0072204 | cell-cell signaling involved in metanephros development | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0060995 | cell-cell signaling involved in kidney development | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0099022 | vesicle tethering | 5 | 31 | 3.53e-01 |
| GO:BP | GO:0042305 | specification of segmental identity, mandibular segment | 1 | 3 | 3.53e-01 |
| GO:BP | GO:0086101 | endothelin receptor signaling pathway involved in heart process | 1 | 3 | 3.53e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 3 | 40 | 3.53e-01 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 1 | 24 | 3.53e-01 |
| GO:BP | GO:0060775 | planar cell polarity pathway involved in gastrula mediolateral intercalation | 1 | 2 | 3.53e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0051885 | positive regulation of timing of anagen | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 5 | 32 | 3.53e-01 |
| GO:BP | GO:1990523 | bone regeneration | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0060031 | mediolateral intercalation | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0060067 | cervix development | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 2 | 8 | 3.53e-01 |
| GO:BP | GO:0061458 | reproductive system development | 8 | 215 | 3.53e-01 |
| GO:BP | GO:0006473 | protein acetylation | 13 | 190 | 3.53e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 2 | 6 | 3.53e-01 |
| GO:BP | GO:0061085 | regulation of histone H3-K27 methylation | 1 | 4 | 3.53e-01 |
| GO:BP | GO:1904933 | regulation of cell proliferation in midbrain | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0015880 | coenzyme A transport | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 4 | 104 | 3.53e-01 |
| GO:BP | GO:1905438 | non-canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0080121 | AMP transport | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 3 | 3.53e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 3 | 17 | 3.53e-01 |
| GO:BP | GO:0015883 | FAD transport | 1 | 2 | 3.53e-01 |
| GO:BP | GO:1904057 | negative regulation of sensory perception of pain | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0061642 | chemoattraction of axon | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0035349 | coenzyme A transmembrane transport | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0042074 | cell migration involved in gastrulation | 3 | 10 | 3.53e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 4 | 26 | 3.53e-01 |
| GO:BP | GO:0071106 | adenosine 3’,5’-bisphosphate transmembrane transport | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0048304 | positive regulation of isotype switching to IgG isotypes | 2 | 4 | 3.53e-01 |
| GO:BP | GO:0003408 | optic cup formation involved in camera-type eye development | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0035350 | FAD transmembrane transport | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0036518 | chemorepulsion of dopaminergic neuron axon | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0036515 | serotonergic neuron axon guidance | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 2 | 21 | 3.53e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 3 | 14 | 3.53e-01 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 1 | 4 | 3.53e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 3.53e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 3.53e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 1 | 4 | 3.53e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 58 | 2412 | 3.53e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0046040 | IMP metabolic process | 3 | 15 | 3.53e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 3 | 70 | 3.53e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 3 | 15 | 3.53e-01 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 3.53e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 1 | 31 | 3.54e-01 |
| GO:BP | GO:0051647 | nucleus localization | 2 | 28 | 3.54e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 3.54e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 4 | 27 | 3.54e-01 |
| GO:BP | GO:0090677 | reversible differentiation | 1 | 5 | 3.54e-01 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 3 | 10 | 3.54e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 4 | 22 | 3.55e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 5 | 34 | 3.55e-01 |
| GO:BP | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 2 | 6 | 3.55e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 1 | 344 | 3.55e-01 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 2 | 11 | 3.55e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 5 | 637 | 3.55e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 1 | 345 | 3.56e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 2 | 22 | 3.56e-01 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 3 | 16 | 3.56e-01 |
| GO:BP | GO:0060677 | ureteric bud elongation | 2 | 7 | 3.56e-01 |
| GO:BP | GO:1905936 | regulation of germ cell proliferation | 1 | 5 | 3.56e-01 |
| GO:BP | GO:0031627 | telomeric loop formation | 1 | 2 | 3.56e-01 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 3.56e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 3 | 55 | 3.56e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 3.57e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 3.57e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 3.57e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 1 | 2 | 3.57e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 3.57e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 2 | 8 | 3.57e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 2 | 8 | 3.57e-01 |
| GO:BP | GO:0009213 | pyrimidine deoxyribonucleoside triphosphate catabolic process | 1 | 2 | 3.57e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 2 | 8 | 3.57e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 3.57e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 6 | 54 | 3.57e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 4 | 26 | 3.60e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 7 | 39 | 3.60e-01 |
| GO:BP | GO:0070294 | renal sodium ion absorption | 2 | 9 | 3.61e-01 |
| GO:BP | GO:0046476 | glycosylceramide biosynthetic process | 2 | 6 | 3.61e-01 |
| GO:BP | GO:1900106 | positive regulation of hyaluranon cable assembly | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 1 | 54 | 3.61e-01 |
| GO:BP | GO:1900104 | regulation of hyaluranon cable assembly | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0071455 | cellular response to hyperoxia | 1 | 5 | 3.61e-01 |
| GO:BP | GO:1905312 | positive regulation of cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0001508 | action potential | 9 | 105 | 3.61e-01 |
| GO:BP | GO:1905310 | regulation of cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0021502 | neural fold elevation formation | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0072125 | negative regulation of glomerular mesangial cell proliferation | 1 | 2 | 3.61e-01 |
| GO:BP | GO:0072076 | nephrogenic mesenchyme development | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0036118 | hyaluranon cable assembly | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 4 | 14 | 3.61e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 4 | 31 | 3.61e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 2 | 14 | 3.61e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 2 | 8 | 3.61e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 6 | 34 | 3.61e-01 |
| GO:BP | GO:0060245 | detection of cell density | 2 | 9 | 3.61e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 4 | 24 | 3.61e-01 |
| GO:BP | GO:0060385 | axonogenesis involved in innervation | 2 | 7 | 3.61e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 1 | 9 | 3.61e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 4 | 24 | 3.61e-01 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 3.61e-01 |
| GO:BP | GO:0035608 | protein deglutamylation | 1 | 2 | 3.61e-01 |
| GO:BP | GO:0006581 | acetylcholine catabolic process | 1 | 3 | 3.62e-01 |
| GO:BP | GO:0048645 | animal organ formation | 6 | 51 | 3.63e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 2 | 9 | 3.65e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 1 | 19 | 3.65e-01 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 4 | 18 | 3.65e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 6 | 20 | 3.66e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 2 | 32 | 3.67e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 3 | 14 | 3.67e-01 |
| GO:BP | GO:0034756 | regulation of iron ion transport | 1 | 4 | 3.67e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 6 | 59 | 3.67e-01 |
| GO:BP | GO:0062232 | prostanoid catabolic process | 1 | 2 | 3.67e-01 |
| GO:BP | GO:1905344 | prostaglandin catabolic process | 1 | 2 | 3.67e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 1 | 2 | 3.67e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 3 | 9 | 3.68e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 1 | 23 | 3.68e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 5 | 33 | 3.68e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 1 | 4 | 3.68e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 3 | 18 | 3.68e-01 |
| GO:BP | GO:0009415 | response to water | 3 | 9 | 3.68e-01 |
| GO:BP | GO:0010886 | positive regulation of cholesterol storage | 2 | 5 | 3.68e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 5 | 41 | 3.69e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 2 | 10 | 3.69e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 5 | 45 | 3.69e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 1 | 18 | 3.69e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 3 | 859 | 3.69e-01 |
| GO:BP | GO:1905590 | fibronectin fibril organization | 1 | 1 | 3.70e-01 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 3 | 13 | 3.70e-01 |
| GO:BP | GO:1990791 | dorsal root ganglion development | 2 | 6 | 3.71e-01 |
| GO:BP | GO:0061061 | muscle structure development | 1 | 546 | 3.71e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 2 | 10 | 3.71e-01 |
| GO:BP | GO:0080163 | regulation of protein serine/threonine phosphatase activity | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0021648 | vestibulocochlear nerve morphogenesis | 2 | 3 | 3.71e-01 |
| GO:BP | GO:0009447 | putrescine catabolic process | 1 | 2 | 3.71e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 2 | 8 | 3.71e-01 |
| GO:BP | GO:0032918 | spermidine acetylation | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0021855 | hypothalamus cell migration | 2 | 6 | 3.71e-01 |
| GO:BP | GO:0032917 | polyamine acetylation | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0035585 | calcium-mediated signaling using extracellular calcium source | 1 | 2 | 3.71e-01 |
| GO:BP | GO:0042758 | long-chain fatty acid catabolic process | 2 | 3 | 3.73e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 2 | 6 | 3.75e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 1 | 19 | 3.76e-01 |
| GO:BP | GO:0051258 | protein polymerization | 3 | 241 | 3.76e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0106014 | regulation of inflammatory response to wounding | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0061044 | negative regulation of vascular wound healing | 1 | 4 | 3.76e-01 |
| GO:BP | GO:0090618 | DNA clamp unloading | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0046398 | UDP-glucuronate metabolic process | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 6 | 72 | 3.77e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 1 | 19 | 3.77e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 2 | 10 | 3.77e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 3.77e-01 |
| GO:BP | GO:0030900 | forebrain development | 5 | 281 | 3.77e-01 |
| GO:BP | GO:0009746 | response to hexose | 19 | 138 | 3.78e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 4 | 24 | 3.79e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 1 | 3 | 3.79e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 1 | 3 | 3.79e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 1 | 4 | 3.79e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 4 | 26 | 3.79e-01 |
| GO:BP | GO:1903444 | negative regulation of brown fat cell differentiation | 1 | 3 | 3.79e-01 |
| GO:BP | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 2 | 6 | 3.79e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 1 | 11 | 3.79e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 1 | 6 | 3.80e-01 |
| GO:BP | GO:0045907 | positive regulation of vasoconstriction | 2 | 15 | 3.80e-01 |
| GO:BP | GO:0018242 | protein O-linked glycosylation via serine | 2 | 7 | 3.81e-01 |
| GO:BP | GO:1990792 | cellular response to glial cell derived neurotrophic factor | 1 | 1 | 3.81e-01 |
| GO:BP | GO:1990790 | response to glial cell derived neurotrophic factor | 1 | 1 | 3.81e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 1 | 19 | 3.81e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 2 | 10 | 3.81e-01 |
| GO:BP | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production | 2 | 5 | 3.81e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 4 | 25 | 3.81e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 2 | 10 | 3.81e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 2 | 11 | 3.81e-01 |
| GO:BP | GO:0043029 | T cell homeostasis | 1 | 27 | 3.81e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 3 | 14 | 3.82e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 1 | 10 | 3.82e-01 |
| GO:BP | GO:0014854 | response to inactivity | 2 | 11 | 3.82e-01 |
| GO:BP | GO:0090675 | intermicrovillar adhesion | 1 | 1 | 3.82e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 35 | 3.82e-01 |
| GO:BP | GO:0006233 | dTDP biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0006227 | dUDP biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0009196 | pyrimidine deoxyribonucleoside diphosphate metabolic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0009197 | pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0030258 | lipid modification | 1 | 162 | 3.82e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 3 | 167 | 3.82e-01 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0046077 | dUDP metabolic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 3 | 167 | 3.82e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 1 | 5 | 3.82e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 2 | 27 | 3.82e-01 |
| GO:BP | GO:0042351 | ‘de novo’ GDP-L-fucose biosynthetic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0046072 | dTDP metabolic process | 1 | 2 | 3.82e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 3 | 21 | 3.83e-01 |
| GO:BP | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition | 1 | 3 | 3.83e-01 |
| GO:BP | GO:0014029 | neural crest formation | 3 | 13 | 3.83e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 1 | 21 | 3.85e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 2 | 8 | 3.85e-01 |
| GO:BP | GO:0033078 | extrathymic T cell differentiation | 1 | 1 | 3.86e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 3.86e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 8 | 108 | 3.86e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 1 | 4 | 3.87e-01 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 1 | 6 | 3.87e-01 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 2 | 16 | 3.87e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 5 | 24 | 3.87e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 1 | 21 | 3.87e-01 |
| GO:BP | GO:0001841 | neural tube formation | 8 | 101 | 3.87e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 6 | 140 | 3.88e-01 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 8 | 50 | 3.88e-01 |
| GO:BP | GO:0035694 | mitochondrial protein catabolic process | 1 | 6 | 3.88e-01 |
| GO:BP | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 3.88e-01 |
| GO:BP | GO:0006046 | N-acetylglucosamine catabolic process | 1 | 3 | 3.89e-01 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 2 | 7 | 3.89e-01 |
| GO:BP | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 3 | 3.89e-01 |
| GO:BP | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly | 1 | 2 | 3.89e-01 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 2 | 7 | 3.89e-01 |
| GO:BP | GO:2000809 | positive regulation of synaptic vesicle clustering | 1 | 3 | 3.89e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 3.89e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 3 | 22 | 3.89e-01 |
| GO:BP | GO:0032898 | neurotrophin production | 1 | 4 | 3.89e-01 |
| GO:BP | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 4 | 3.89e-01 |
| GO:BP | GO:0090188 | negative regulation of pancreatic juice secretion | 1 | 4 | 3.90e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 3.90e-01 |
| GO:BP | GO:0032020 | ISG15-protein conjugation | 1 | 6 | 3.91e-01 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 2 | 9 | 3.91e-01 |
| GO:BP | GO:0032730 | positive regulation of interleukin-1 alpha production | 1 | 3 | 3.91e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 3 | 11 | 3.91e-01 |
| GO:BP | GO:1903518 | positive regulation of single strand break repair | 1 | 2 | 3.91e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 1 | 10 | 3.92e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 3.92e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 4 | 25 | 3.92e-01 |
| GO:BP | GO:0048538 | thymus development | 1 | 35 | 3.92e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 1 | 156 | 3.92e-01 |
| GO:BP | GO:0007141 | male meiosis I | 2 | 15 | 3.92e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 12 | 88 | 3.92e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 1 | 252 | 3.92e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 7 | 68 | 3.93e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 11 | 83 | 3.93e-01 |
| GO:BP | GO:0036376 | sodium ion export across plasma membrane | 2 | 12 | 3.93e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 3.93e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 2 | 10 | 3.93e-01 |
| GO:BP | GO:1902462 | positive regulation of mesenchymal stem cell proliferation | 1 | 4 | 3.93e-01 |
| GO:BP | GO:1902460 | regulation of mesenchymal stem cell proliferation | 1 | 4 | 3.93e-01 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 4 | 29 | 3.93e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 3.93e-01 |
| GO:BP | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | 3 | 16 | 3.93e-01 |
| GO:BP | GO:1900195 | positive regulation of oocyte maturation | 1 | 3 | 3.93e-01 |
| GO:BP | GO:0090132 | epithelium migration | 1 | 252 | 3.94e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 9 | 81 | 3.94e-01 |
| GO:BP | GO:0036309 | protein localization to M-band | 1 | 2 | 3.94e-01 |
| GO:BP | GO:0048854 | brain morphogenesis | 5 | 28 | 3.94e-01 |
| GO:BP | GO:0036371 | protein localization to T-tubule | 1 | 2 | 3.94e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 1 | 39 | 3.95e-01 |
| GO:BP | GO:0009608 | response to symbiont | 1 | 2 | 3.95e-01 |
| GO:BP | GO:0070779 | D-aspartate import across plasma membrane | 1 | 4 | 3.95e-01 |
| GO:BP | GO:0009609 | response to symbiotic bacterium | 1 | 2 | 3.95e-01 |
| GO:BP | GO:0070777 | D-aspartate transport | 1 | 4 | 3.95e-01 |
| GO:BP | GO:0035344 | hypoxanthine transport | 1 | 2 | 3.95e-01 |
| GO:BP | GO:1900141 | regulation of oligodendrocyte apoptotic process | 1 | 2 | 3.95e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 2 | 14 | 3.95e-01 |
| GO:BP | GO:0001778 | plasma membrane repair | 2 | 28 | 3.95e-01 |
| GO:BP | GO:0015854 | guanine transport | 1 | 2 | 3.95e-01 |
| GO:BP | GO:0032764 | negative regulation of mast cell cytokine production | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0015744 | succinate transport | 2 | 6 | 3.95e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 2 | 6 | 3.95e-01 |
| GO:BP | GO:2000538 | positive regulation of B cell chemotaxis | 1 | 1 | 3.95e-01 |
| GO:BP | GO:0061364 | apoptotic process involved in luteolysis | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 3.95e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 1 | 4 | 3.95e-01 |
| GO:BP | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure | 2 | 14 | 3.95e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 7 | 52 | 3.95e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 1 | 64 | 3.95e-01 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 2 | 9 | 3.95e-01 |
| GO:BP | GO:2000537 | regulation of B cell chemotaxis | 1 | 1 | 3.95e-01 |
| GO:BP | GO:0090130 | tissue migration | 1 | 256 | 3.95e-01 |
| GO:BP | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 1 | 1 | 3.95e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 1 | 4 | 3.95e-01 |
| GO:BP | GO:2000532 | regulation of renal albumin absorption | 1 | 1 | 3.95e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 4 | 30 | 3.95e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 28 | 438 | 3.95e-01 |
| GO:BP | GO:0009744 | response to sucrose | 1 | 4 | 3.95e-01 |
| GO:BP | GO:0072011 | glomerular endothelium development | 1 | 2 | 3.95e-01 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 3 | 16 | 3.95e-01 |
| GO:BP | GO:0009414 | response to water deprivation | 2 | 4 | 3.95e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 1 | 9 | 3.95e-01 |
| GO:BP | GO:0009750 | response to fructose | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0035289 | posterior head segmentation | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0035287 | head segmentation | 1 | 3 | 3.95e-01 |
| GO:BP | GO:1903210 | podocyte apoptotic process | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0007380 | specification of segmental identity, head | 1 | 3 | 3.95e-01 |
| GO:BP | GO:0034285 | response to disaccharide | 1 | 4 | 3.95e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 1 | 9 | 3.95e-01 |
| GO:BP | GO:0070593 | dendrite self-avoidance | 1 | 11 | 3.95e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 4 | 49 | 3.96e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 2 | 22 | 3.96e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 3.96e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 1 | 8 | 3.96e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 3.96e-01 |
| GO:BP | GO:0140131 | positive regulation of lymphocyte chemotaxis | 3 | 10 | 3.96e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 3.96e-01 |
| GO:BP | GO:0048539 | bone marrow development | 2 | 7 | 3.96e-01 |
| GO:BP | GO:0044851 | hair cycle phase | 2 | 9 | 3.96e-01 |
| GO:BP | GO:0044848 | biological phase | 2 | 9 | 3.96e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 1 | 2 | 3.96e-01 |
| GO:BP | GO:0061441 | renal artery morphogenesis | 1 | 2 | 3.96e-01 |
| GO:BP | GO:0010912 | positive regulation of isomerase activity | 1 | 5 | 3.96e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 1 | 179 | 3.96e-01 |
| GO:BP | GO:0097102 | endothelial tip cell fate specification | 1 | 2 | 3.96e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 1 | 464 | 3.96e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 19 | 324 | 3.96e-01 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 1 | 3 | 3.96e-01 |
| GO:BP | GO:0031645 | negative regulation of nervous system process | 2 | 9 | 3.96e-01 |
| GO:BP | GO:0010911 | regulation of isomerase activity | 1 | 5 | 3.96e-01 |
| GO:BP | GO:0048819 | regulation of hair follicle maturation | 2 | 9 | 3.96e-01 |
| GO:BP | GO:0043007 | maintenance of rDNA | 1 | 2 | 3.96e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 6 | 70 | 3.96e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 1 | 5 | 3.96e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 6 | 70 | 3.96e-01 |
| GO:BP | GO:0002076 | osteoblast development | 3 | 12 | 3.97e-01 |
| GO:BP | GO:0034371 | chylomicron remodeling | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0000423 | mitophagy | 5 | 37 | 3.97e-01 |
| GO:BP | GO:0055095 | lipoprotein particle mediated signaling | 1 | 3 | 3.97e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 1 | 170 | 3.97e-01 |
| GO:BP | GO:0055096 | low-density lipoprotein particle mediated signaling | 1 | 3 | 3.97e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 2 | 7 | 3.97e-01 |
| GO:BP | GO:0071603 | endothelial cell-cell adhesion | 1 | 4 | 3.97e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 3.97e-01 |
| GO:BP | GO:0002520 | immune system development | 2 | 144 | 3.98e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 1 | 55 | 3.98e-01 |
| GO:BP | GO:0071874 | cellular response to norepinephrine stimulus | 1 | 2 | 3.98e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 10 | 1147 | 3.98e-01 |
| GO:BP | GO:0071873 | response to norepinephrine | 1 | 2 | 3.98e-01 |
| GO:BP | GO:0014832 | urinary bladder smooth muscle contraction | 2 | 4 | 3.98e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 3.98e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 5 | 15 | 3.99e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 2 | 16 | 3.99e-01 |
| GO:BP | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 1 | 3 | 3.99e-01 |
| GO:BP | GO:0016571 | histone methylation | 4 | 117 | 3.99e-01 |
| GO:BP | GO:0007420 | brain development | 1 | 591 | 3.99e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 2 | 12 | 3.99e-01 |
| GO:BP | GO:1904381 | Golgi apparatus mannose trimming | 1 | 3 | 3.99e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 3 | 32 | 4.00e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 2 | 39 | 4.00e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 4 | 39 | 4.01e-01 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 4 | 35 | 4.01e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 4.01e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 8 | 116 | 4.01e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 1 | 3 | 4.02e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 3 | 30 | 4.02e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 3 | 30 | 4.02e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 11 | 85 | 4.02e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 4.02e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 1 | 44 | 4.02e-01 |
| GO:BP | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | 1 | 4 | 4.03e-01 |
| GO:BP | GO:0046967 | cytosol to endoplasmic reticulum transport | 1 | 4 | 4.03e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 10 | 211 | 4.03e-01 |
| GO:BP | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 1 | 2 | 4.03e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 4.03e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 4.03e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 4.03e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 4.03e-01 |
| GO:BP | GO:0009386 | translational attenuation | 1 | 2 | 4.03e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 3 | 25 | 4.03e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 3 | 17 | 4.03e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 3 | 17 | 4.03e-01 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 5 | 36 | 4.04e-01 |
| GO:BP | GO:1902608 | positive regulation of large conductance calcium-activated potassium channel activity | 1 | 2 | 4.04e-01 |
| GO:BP | GO:0060789 | hair follicle placode formation | 1 | 5 | 4.04e-01 |
| GO:BP | GO:0090663 | galanin-activated signaling pathway | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 1 | 3 | 4.04e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 2 | 11 | 4.04e-01 |
| GO:BP | GO:0007040 | lysosome organization | 6 | 88 | 4.04e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 6 | 88 | 4.04e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 11 | 80 | 4.04e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 2 | 8 | 4.04e-01 |
| GO:BP | GO:0033668 | suppression by symbiont of host apoptotic process | 1 | 2 | 4.04e-01 |
| GO:BP | GO:0017055 | negative regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 2 | 4.04e-01 |
| GO:BP | GO:0019050 | suppression by virus of host apoptotic process | 1 | 2 | 4.04e-01 |
| GO:BP | GO:0072040 | negative regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis | 1 | 1 | 4.04e-01 |
| GO:BP | GO:1904673 | negative regulation of somatic stem cell population maintenance | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0052041 | suppression by symbiont of host programmed cell death | 1 | 2 | 4.04e-01 |
| GO:BP | GO:0072039 | regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0072131 | kidney mesenchyme morphogenesis | 1 | 4 | 4.04e-01 |
| GO:BP | GO:0072133 | metanephric mesenchyme morphogenesis | 1 | 4 | 4.04e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 8 | 67 | 4.04e-01 |
| GO:BP | GO:1905069 | allantois development | 1 | 4 | 4.04e-01 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 2 | 14 | 4.04e-01 |
| GO:BP | GO:0097527 | necroptotic signaling pathway | 1 | 3 | 4.04e-01 |
| GO:BP | GO:0042264 | peptidyl-aspartic acid hydroxylation | 1 | 2 | 4.04e-01 |
| GO:BP | GO:2000426 | negative regulation of apoptotic cell clearance | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0000425 | pexophagy | 1 | 6 | 4.04e-01 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 1 | 5 | 4.04e-01 |
| GO:BP | GO:0030210 | heparin biosynthetic process | 2 | 9 | 4.04e-01 |
| GO:BP | GO:1901145 | mesenchymal cell apoptotic process involved in nephron morphogenesis | 1 | 1 | 4.04e-01 |
| GO:BP | GO:0007431 | salivary gland development | 4 | 26 | 4.05e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 2 | 11 | 4.05e-01 |
| GO:BP | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis | 1 | 2 | 4.05e-01 |
| GO:BP | GO:1905704 | positive regulation of inhibitory synapse assembly | 1 | 2 | 4.05e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 4.05e-01 |
| GO:BP | GO:0036166 | phenotypic switching | 1 | 7 | 4.05e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 3 | 25 | 4.05e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 1 | 3 | 4.06e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 3 | 15 | 4.06e-01 |
| GO:BP | GO:0045617 | negative regulation of keratinocyte differentiation | 2 | 3 | 4.06e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 2 | 26 | 4.06e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 2 | 26 | 4.06e-01 |
| GO:BP | GO:0001843 | neural tube closure | 7 | 90 | 4.06e-01 |
| GO:BP | GO:1902268 | negative regulation of polyamine transmembrane transport | 1 | 3 | 4.06e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 3 | 16 | 4.06e-01 |
| GO:BP | GO:0032070 | regulation of deoxyribonuclease activity | 2 | 9 | 4.06e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 1 | 3 | 4.07e-01 |
| GO:BP | GO:0072674 | multinuclear osteoclast differentiation | 2 | 6 | 4.07e-01 |
| GO:BP | GO:1903173 | fatty alcohol metabolic process | 1 | 2 | 4.07e-01 |
| GO:BP | GO:0033306 | phytol metabolic process | 1 | 2 | 4.07e-01 |
| GO:BP | GO:0021884 | forebrain neuron development | 4 | 16 | 4.07e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 3 | 32 | 4.08e-01 |
| GO:BP | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron development | 2 | 7 | 4.08e-01 |
| GO:BP | GO:0060322 | head development | 1 | 633 | 4.08e-01 |
| GO:BP | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation | 2 | 7 | 4.08e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 1 | 23 | 4.08e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 8 | 61 | 4.08e-01 |
| GO:BP | GO:2000395 | regulation of ubiquitin-dependent endocytosis | 1 | 2 | 4.08e-01 |
| GO:BP | GO:2000397 | positive regulation of ubiquitin-dependent endocytosis | 1 | 2 | 4.08e-01 |
| GO:BP | GO:0060004 | reflex | 3 | 9 | 4.09e-01 |
| GO:BP | GO:1903109 | positive regulation of mitochondrial transcription | 1 | 2 | 4.09e-01 |
| GO:BP | GO:0044338 | canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation | 1 | 3 | 4.11e-01 |
| GO:BP | GO:0036520 | astrocyte-dopaminergic neuron signaling | 1 | 2 | 4.11e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 4 | 36 | 4.11e-01 |
| GO:BP | GO:0019062 | virion attachment to host cell | 2 | 9 | 4.11e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 2 | 8 | 4.11e-01 |
| GO:BP | GO:0036093 | germ cell proliferation | 1 | 7 | 4.12e-01 |
| GO:BP | GO:0070997 | neuron death | 4 | 286 | 4.13e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 1 | 303 | 4.13e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 5 | 32 | 4.13e-01 |
| GO:BP | GO:0021553 | olfactory nerve development | 1 | 5 | 4.14e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 3 | 15 | 4.14e-01 |
| GO:BP | GO:0035188 | hatching | 3 | 23 | 4.14e-01 |
| GO:BP | GO:0018277 | protein deamination | 1 | 2 | 4.14e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 3 | 23 | 4.14e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 3 | 23 | 4.14e-01 |
| GO:BP | GO:0098957 | anterograde axonal transport of mitochondrion | 1 | 5 | 4.14e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 2 | 478 | 4.14e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 2 | 271 | 4.14e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 3 | 19 | 4.14e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 4.15e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 4.15e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 4.15e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 4 | 18 | 4.15e-01 |
| GO:BP | GO:0060606 | tube closure | 7 | 91 | 4.15e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 2 | 18 | 4.15e-01 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 1 | 2 | 4.15e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 5 | 706 | 4.15e-01 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 3 | 18 | 4.15e-01 |
| GO:BP | GO:0003213 | cardiac right atrium morphogenesis | 1 | 2 | 4.16e-01 |
| GO:BP | GO:1900107 | regulation of nodal signaling pathway | 1 | 3 | 4.16e-01 |
| GO:BP | GO:0070889 | platelet alpha granule organization | 1 | 2 | 4.16e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 2 | 17 | 4.16e-01 |
| GO:BP | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation | 1 | 2 | 4.16e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 4 | 28 | 4.16e-01 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 3 | 22 | 4.16e-01 |
| GO:BP | GO:0036514 | dopaminergic neuron axon guidance | 1 | 3 | 4.16e-01 |
| GO:BP | GO:0003138 | primary heart field specification | 1 | 3 | 4.16e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 3 | 4.16e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 3 | 17 | 4.16e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 1 | 7 | 4.16e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 2 | 25 | 4.16e-01 |
| GO:BP | GO:1904938 | planar cell polarity pathway involved in axon guidance | 1 | 3 | 4.16e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 5 | 31 | 4.16e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 1 | 7 | 4.16e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 4.16e-01 |
| GO:BP | GO:0048850 | hypophysis morphogenesis | 1 | 2 | 4.16e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 2 | 20 | 4.17e-01 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 3 | 15 | 4.17e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 8 | 153 | 4.17e-01 |
| GO:BP | GO:0042407 | cristae formation | 3 | 18 | 4.18e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 54 | 2270 | 4.18e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 1 | 496 | 4.19e-01 |
| GO:BP | GO:0035904 | aorta development | 5 | 50 | 4.19e-01 |
| GO:BP | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation | 1 | 5 | 4.19e-01 |
| GO:BP | GO:0060764 | cell-cell signaling involved in mammary gland development | 1 | 1 | 4.19e-01 |
| GO:BP | GO:0044854 | plasma membrane raft assembly | 1 | 6 | 4.19e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 2 | 12 | 4.19e-01 |
| GO:BP | GO:1903553 | positive regulation of extracellular exosome assembly | 1 | 4 | 4.19e-01 |
| GO:BP | GO:0048627 | myoblast development | 1 | 3 | 4.19e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 6 | 47 | 4.19e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 5 | 71 | 4.19e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 2 | 6 | 4.20e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 2 | 11 | 4.20e-01 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 2 | 13 | 4.20e-01 |
| GO:BP | GO:1904430 | negative regulation of t-circle formation | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 1 | 5 | 4.20e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 4.20e-01 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 2 | 10 | 4.20e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 19 | 142 | 4.20e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 3 | 39 | 4.20e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 1 | 2 | 4.20e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 4.20e-01 |
| GO:BP | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process | 1 | 3 | 4.20e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 4.20e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 1 | 2 | 4.20e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 1 | 2 | 4.20e-01 |
| GO:BP | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | 1 | 7 | 4.20e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 4.20e-01 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 2 | 10 | 4.20e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 1 | 50 | 4.20e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 5 | 69 | 4.20e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 5 | 39 | 4.20e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 2 | 12 | 4.20e-01 |
| GO:BP | GO:0035329 | hippo signaling | 1 | 41 | 4.21e-01 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 4.21e-01 |
| GO:BP | GO:0035984 | cellular response to trichostatin A | 1 | 2 | 4.22e-01 |
| GO:BP | GO:0035983 | response to trichostatin A | 1 | 2 | 4.22e-01 |
| GO:BP | GO:0010587 | miRNA catabolic process | 2 | 10 | 4.22e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 1 | 53 | 4.22e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 6 | 44 | 4.22e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 3 | 22 | 4.23e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 3 | 26 | 4.23e-01 |
| GO:BP | GO:1903910 | negative regulation of receptor clustering | 1 | 2 | 4.23e-01 |
| GO:BP | GO:0007439 | ectodermal digestive tract development | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0048611 | embryonic ectodermal digestive tract development | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 16 | 160 | 4.24e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 4.24e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 4.24e-01 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 1 | 4 | 4.24e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 1 | 24 | 4.25e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 1 | 33 | 4.25e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 3 | 18 | 4.25e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 4.25e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 3 | 20 | 4.25e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 11 | 123 | 4.25e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 1 | 6 | 4.25e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 4.25e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 9 | 73 | 4.25e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 2 | 26 | 4.25e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 1 | 47 | 4.26e-01 |
| GO:BP | GO:0032650 | regulation of interleukin-1 alpha production | 1 | 4 | 4.27e-01 |
| GO:BP | GO:0032610 | interleukin-1 alpha production | 1 | 4 | 4.27e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2 | 12 | 4.27e-01 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 1 | 10 | 4.28e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 14 | 121 | 4.28e-01 |
| GO:BP | GO:0045210 | FasL biosynthetic process | 1 | 1 | 4.28e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 3 | 16 | 4.28e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 11 | 89 | 4.28e-01 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 1 | 2 | 4.28e-01 |
| GO:BP | GO:0110097 | regulation of calcium import into the mitochondrion | 1 | 2 | 4.28e-01 |
| GO:BP | GO:1905089 | regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 2 | 4.28e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 2 | 28 | 4.28e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 4.28e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 1 | 8 | 4.28e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 1 | 36 | 4.29e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 4 | 21 | 4.29e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 7 | 88 | 4.29e-01 |
| GO:BP | GO:0003307 | regulation of Wnt signaling pathway involved in heart development | 2 | 7 | 4.29e-01 |
| GO:BP | GO:1902463 | protein localization to cell leading edge | 1 | 5 | 4.29e-01 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 4 | 27 | 4.29e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 2 | 12 | 4.29e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 1 | 47 | 4.29e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 1 | 5 | 4.29e-01 |
| GO:BP | GO:0090148 | membrane fission | 6 | 44 | 4.29e-01 |
| GO:BP | GO:0046069 | cGMP catabolic process | 1 | 4 | 4.30e-01 |
| GO:BP | GO:0010232 | vascular transport | 4 | 72 | 4.30e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 4 | 72 | 4.30e-01 |
| GO:BP | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | 2 | 6 | 4.30e-01 |
| GO:BP | GO:0034514 | mitochondrial unfolded protein response | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 1 | 2 | 4.30e-01 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 2 | 14 | 4.31e-01 |
| GO:BP | GO:1905524 | negative regulation of protein autoubiquitination | 1 | 2 | 4.31e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 2 | 120 | 4.31e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 4 | 64 | 4.31e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 3 | 57 | 4.31e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 2 | 17 | 4.32e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 1 | 20 | 4.32e-01 |
| GO:BP | GO:0043984 | histone H4-K16 acetylation | 1 | 14 | 4.32e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 1 | 11 | 4.32e-01 |
| GO:BP | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 1 | 4 | 4.32e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 3 | 16 | 4.32e-01 |
| GO:BP | GO:0070203 | regulation of establishment of protein localization to telomere | 2 | 11 | 4.33e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 1 | 43 | 4.33e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 8 | 68 | 4.33e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 2 | 7 | 4.33e-01 |
| GO:BP | GO:1905608 | positive regulation of presynapse assembly | 1 | 2 | 4.33e-01 |
| GO:BP | GO:0003219 | cardiac right ventricle formation | 1 | 4 | 4.33e-01 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 1 | 9 | 4.33e-01 |
| GO:BP | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 11 | 96 | 4.33e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 3 | 23 | 4.34e-01 |
| GO:BP | GO:0010820 | positive regulation of T cell chemotaxis | 2 | 7 | 4.34e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 2 | 14 | 4.34e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 2 | 11 | 4.34e-01 |
| GO:BP | GO:1990227 | paranodal junction maintenance | 1 | 2 | 4.34e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 14 | 233 | 4.34e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 5 | 28 | 4.34e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 2 | 12 | 4.35e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 4 | 40 | 4.35e-01 |
| GO:BP | GO:0001575 | globoside metabolic process | 1 | 2 | 4.35e-01 |
| GO:BP | GO:0001576 | globoside biosynthetic process | 1 | 2 | 4.35e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 2 | 22 | 4.35e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 2 | 22 | 4.35e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 2 | 12 | 4.35e-01 |
| GO:BP | GO:2000344 | positive regulation of acrosome reaction | 1 | 6 | 4.35e-01 |
| GO:BP | GO:1905223 | epicardium morphogenesis | 1 | 2 | 4.35e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 4.35e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 1 | 4 | 4.35e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 2 | 57 | 4.35e-01 |
| GO:BP | GO:2001112 | regulation of cellular response to hepatocyte growth factor stimulus | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0003096 | renal sodium ion transport | 2 | 10 | 4.36e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 2 | 10 | 4.36e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 3 | 29 | 4.36e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 12 | 78 | 4.37e-01 |
| GO:BP | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process | 1 | 3 | 4.37e-01 |
| GO:BP | GO:0150173 | positive regulation of phosphatidylcholine metabolic process | 1 | 3 | 4.37e-01 |
| GO:BP | GO:0051234 | establishment of localization | 3 | 3433 | 4.37e-01 |
| GO:BP | GO:0016080 | synaptic vesicle targeting | 1 | 3 | 4.37e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 3 | 23 | 4.38e-01 |
| GO:BP | GO:0055129 | L-proline biosynthetic process | 1 | 5 | 4.38e-01 |
| GO:BP | GO:0006561 | proline biosynthetic process | 1 | 5 | 4.38e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 1 | 11 | 4.38e-01 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 2 | 10 | 4.38e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 6 | 47 | 4.39e-01 |
| GO:BP | GO:0046073 | dTMP metabolic process | 1 | 7 | 4.39e-01 |
| GO:BP | GO:0038109 | Kit signaling pathway | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 6 | 4.39e-01 |
| GO:BP | GO:0010225 | response to UV-C | 1 | 13 | 4.39e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 3 | 18 | 4.39e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 3 | 18 | 4.39e-01 |
| GO:BP | GO:0036215 | response to stem cell factor | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0036216 | cellular response to stem cell factor stimulus | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 4 | 39 | 4.39e-01 |
| GO:BP | GO:0009439 | cyanate metabolic process | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0009440 | cyanate catabolic process | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 1 | 3 | 4.40e-01 |
| GO:BP | GO:0120187 | positive regulation of protein localization to chromatin | 1 | 3 | 4.41e-01 |
| GO:BP | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly | 1 | 5 | 4.42e-01 |
| GO:BP | GO:1903800 | positive regulation of miRNA processing | 1 | 3 | 4.42e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 1 | 6 | 4.42e-01 |
| GO:BP | GO:0046710 | GDP metabolic process | 1 | 10 | 4.43e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 4.43e-01 |
| GO:BP | GO:0006924 | activation-induced cell death of T cells | 1 | 8 | 4.43e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 4.43e-01 |
| GO:BP | GO:0036150 | phosphatidylserine acyl-chain remodeling | 2 | 7 | 4.43e-01 |
| GO:BP | GO:1904425 | negative regulation of GTP binding | 1 | 3 | 4.43e-01 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 3 | 16 | 4.43e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 1 | 64 | 4.43e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 5 | 23 | 4.43e-01 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 3 | 16 | 4.43e-01 |
| GO:BP | GO:0070487 | monocyte aggregation | 1 | 4 | 4.44e-01 |
| GO:BP | GO:0060687 | regulation of branching involved in prostate gland morphogenesis | 1 | 3 | 4.44e-01 |
| GO:BP | GO:0072156 | distal tubule morphogenesis | 1 | 3 | 4.44e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 1 | 7 | 4.44e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 1 | 11 | 4.45e-01 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 2 | 9 | 4.45e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 1 | 13 | 4.45e-01 |
| GO:BP | GO:0097168 | mesenchymal stem cell proliferation | 1 | 4 | 4.45e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 3 | 14 | 4.45e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 2 | 18 | 4.45e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 3 | 17 | 4.45e-01 |
| GO:BP | GO:0002543 | activation of blood coagulation via clotting cascade | 1 | 2 | 4.45e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 16 | 131 | 4.45e-01 |
| GO:BP | GO:0021978 | telencephalon regionalization | 1 | 4 | 4.45e-01 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 2 | 13 | 4.45e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 4.45e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 5 | 4.45e-01 |
| GO:BP | GO:0042220 | response to cocaine | 1 | 28 | 4.46e-01 |
| GO:BP | GO:0061549 | sympathetic ganglion development | 2 | 6 | 4.46e-01 |
| GO:BP | GO:0042350 | GDP-L-fucose biosynthetic process | 1 | 3 | 4.46e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 3 | 48 | 4.46e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 4.46e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 1 | 535 | 4.47e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 1 | 358 | 4.47e-01 |
| GO:BP | GO:0051697 | protein delipidation | 1 | 5 | 4.47e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 2 | 38 | 4.47e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 2 | 24 | 4.47e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 4 | 36 | 4.47e-01 |
| GO:BP | GO:0043921 | modulation by host of viral transcription | 1 | 2 | 4.47e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 3 | 17 | 4.47e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 11 | 173 | 4.47e-01 |
| GO:BP | GO:0042940 | D-amino acid transport | 1 | 5 | 4.47e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 7 | 49 | 4.47e-01 |
| GO:BP | GO:0032643 | regulation of connective tissue growth factor production | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0032601 | connective tissue growth factor production | 1 | 2 | 4.47e-01 |
| GO:BP | GO:1902047 | polyamine transmembrane transport | 2 | 10 | 4.47e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 5 | 38 | 4.47e-01 |
| GO:BP | GO:0046725 | negative regulation by virus of viral protein levels in host cell | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 4.47e-01 |
| GO:BP | GO:0106091 | glial cell projection elongation | 1 | 2 | 4.48e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 2 | 12 | 4.48e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 5 | 41 | 4.48e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 2 | 12 | 4.48e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 1 | 37 | 4.49e-01 |
| GO:BP | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway | 1 | 3 | 4.49e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 4.49e-01 |
| GO:BP | GO:0006579 | amino-acid betaine catabolic process | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0021782 | glial cell development | 5 | 84 | 4.49e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 3 | 19 | 4.49e-01 |
| GO:BP | GO:0019254 | carnitine metabolic process, CoA-linked | 1 | 3 | 4.49e-01 |
| GO:BP | GO:0019262 | N-acetylneuraminate catabolic process | 1 | 6 | 4.49e-01 |
| GO:BP | GO:0051695 | actin filament uncapping | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 2 | 2135 | 4.49e-01 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 3 | 20 | 4.49e-01 |
| GO:BP | GO:0042727 | flavin-containing compound biosynthetic process | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 6 | 46 | 4.49e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 1 | 4 | 4.49e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 3 | 47 | 4.49e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 3 | 23 | 4.49e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 1 | 5 | 4.49e-01 |
| GO:BP | GO:0018009 | N-terminal peptidyl-L-cysteine N-palmitoylation | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 9 | 75 | 4.49e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 2 | 24 | 4.49e-01 |
| GO:BP | GO:0030576 | Cajal body organization | 1 | 4 | 4.49e-01 |
| GO:BP | GO:0021644 | vagus nerve morphogenesis | 1 | 1 | 4.49e-01 |
| GO:BP | GO:0009170 | purine deoxyribonucleoside monophosphate metabolic process | 2 | 7 | 4.49e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 3 | 20 | 4.49e-01 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 2 | 9 | 4.50e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 3 | 29 | 4.50e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 2 | 7 | 4.50e-01 |
| GO:BP | GO:0035973 | aggrephagy | 2 | 8 | 4.50e-01 |
| GO:BP | GO:0071787 | endoplasmic reticulum tubular network formation | 1 | 4 | 4.50e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 1 | 15 | 4.50e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 4.53e-01 |
| GO:BP | GO:0021756 | striatum development | 3 | 17 | 4.53e-01 |
| GO:BP | GO:1904024 | negative regulation of glucose catabolic process to lactate via pyruvate | 1 | 3 | 4.53e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 4.53e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 4.54e-01 |
| GO:BP | GO:0030592 | DNA ADP-ribosylation | 1 | 3 | 4.54e-01 |
| GO:BP | GO:0007417 | central nervous system development | 1 | 792 | 4.54e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 1 | 15 | 4.54e-01 |
| GO:BP | GO:0010819 | regulation of T cell chemotaxis | 2 | 7 | 4.54e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 1 | 248 | 4.54e-01 |
| GO:BP | GO:0000012 | single strand break repair | 2 | 10 | 4.54e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 2 | 11 | 4.54e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 2 | 8 | 4.54e-01 |
| GO:BP | GO:0030903 | notochord development | 3 | 17 | 4.54e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 1 | 8 | 4.55e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 2 | 34 | 4.55e-01 |
| GO:BP | GO:1904742 | regulation of telomeric DNA binding | 1 | 4 | 4.56e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 1 | 14 | 4.56e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 1 | 16 | 4.57e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 4 | 72 | 4.57e-01 |
| GO:BP | GO:0006000 | fructose metabolic process | 2 | 10 | 4.57e-01 |
| GO:BP | GO:0006784 | heme A biosynthetic process | 1 | 2 | 4.57e-01 |
| GO:BP | GO:0046160 | heme a metabolic process | 1 | 2 | 4.57e-01 |
| GO:BP | GO:0097065 | anterior head development | 1 | 7 | 4.57e-01 |
| GO:BP | GO:1990079 | cartilage homeostasis | 1 | 2 | 4.57e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 2 | 15 | 4.58e-01 |
| GO:BP | GO:0021682 | nerve maturation | 1 | 2 | 4.58e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 4 | 33 | 4.58e-01 |
| GO:BP | GO:0008585 | female gonad development | 3 | 77 | 4.58e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 3 | 23 | 4.59e-01 |
| GO:BP | GO:0003097 | renal water transport | 1 | 6 | 4.60e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 1 | 8 | 4.60e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 3 | 20 | 4.60e-01 |
| GO:BP | GO:0003104 | positive regulation of glomerular filtration | 1 | 3 | 4.60e-01 |
| GO:BP | GO:0060166 | olfactory pit development | 1 | 1 | 4.61e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 5 | 39 | 4.61e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 1 | 4 | 4.61e-01 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 1 | 4 | 4.61e-01 |
| GO:BP | GO:0032238 | adenosine transport | 1 | 2 | 4.62e-01 |
| GO:BP | GO:0070291 | N-acylethanolamine metabolic process | 1 | 4 | 4.62e-01 |
| GO:BP | GO:0003150 | muscular septum morphogenesis | 1 | 4 | 4.62e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 4 | 73 | 4.62e-01 |
| GO:BP | GO:0150098 | glial cell-neuron signaling | 1 | 2 | 4.62e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 8 | 69 | 4.62e-01 |
| GO:BP | GO:0070266 | necroptotic process | 4 | 32 | 4.62e-01 |
| GO:BP | GO:0070202 | regulation of establishment of protein localization to chromosome | 2 | 12 | 4.62e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 2 | 4 | 4.62e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 1 | 9 | 4.62e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 5 | 41 | 4.62e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 2 | 11 | 4.62e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 3 | 14 | 4.62e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 7 | 755 | 4.62e-01 |
| GO:BP | GO:0014904 | myotube cell development | 3 | 34 | 4.62e-01 |
| GO:BP | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | 2 | 10 | 4.62e-01 |
| GO:BP | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis | 1 | 4 | 4.62e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 2 | 2574 | 4.63e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 5 | 77 | 4.63e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 3 | 18 | 4.63e-01 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 3 | 82 | 4.63e-01 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 1 | 4 | 4.63e-01 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 3 | 82 | 4.63e-01 |
| GO:BP | GO:0014034 | neural crest cell fate commitment | 1 | 4 | 4.63e-01 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 4.63e-01 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 4.63e-01 |
| GO:BP | GO:1903537 | meiotic cell cycle process involved in oocyte maturation | 1 | 5 | 4.63e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 4 | 54 | 4.63e-01 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 1 | 4 | 4.63e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 2 | 2576 | 4.63e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 2 | 8 | 4.64e-01 |
| GO:BP | GO:0044650 | adhesion of symbiont to host cell | 2 | 9 | 4.64e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 1 | 12 | 4.64e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 1 | 4 | 4.64e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 6 | 43 | 4.64e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 16 | 151 | 4.65e-01 |
| GO:BP | GO:0150018 | basal dendrite development | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0097101 | blood vessel endothelial cell fate specification | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0150019 | basal dendrite morphogenesis | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0150020 | basal dendrite arborization | 1 | 3 | 4.65e-01 |
| GO:BP | GO:0060846 | blood vessel endothelial cell fate commitment | 1 | 3 | 4.65e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 1 | 2 | 4.65e-01 |
| GO:BP | GO:0000296 | spermine transport | 1 | 3 | 4.65e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 2 | 2593 | 4.65e-01 |
| GO:BP | GO:2000668 | regulation of dendritic cell apoptotic process | 2 | 7 | 4.65e-01 |
| GO:BP | GO:0097048 | dendritic cell apoptotic process | 2 | 7 | 4.65e-01 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 1 | 8 | 4.66e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 6 | 42 | 4.66e-01 |
| GO:BP | GO:0031099 | regeneration | 4 | 142 | 4.66e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 2 | 30 | 4.66e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 4 | 73 | 4.66e-01 |
| GO:BP | GO:0033504 | floor plate development | 1 | 8 | 4.66e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 4.66e-01 |
| GO:BP | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle | 1 | 3 | 4.66e-01 |
| GO:BP | GO:0048852 | diencephalon morphogenesis | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0046060 | dATP metabolic process | 1 | 3 | 4.66e-01 |
| GO:BP | GO:0035352 | NAD transmembrane transport | 1 | 3 | 4.66e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 2 | 17 | 4.66e-01 |
| GO:BP | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | 1 | 4 | 4.66e-01 |
| GO:BP | GO:0060157 | urinary bladder development | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 6 | 40 | 4.66e-01 |
| GO:BP | GO:1901321 | positive regulation of heart induction | 1 | 4 | 4.66e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | 7 | 4.66e-01 |
| GO:BP | GO:0090027 | negative regulation of monocyte chemotaxis | 1 | 4 | 4.66e-01 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 1 | 5 | 4.66e-01 |
| GO:BP | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | 1 | 3 | 4.66e-01 |
| GO:BP | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 1 | 4 | 4.66e-01 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 1 | 2 | 4.67e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 1 | 225 | 4.67e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 3 | 22 | 4.67e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 4.67e-01 |
| GO:BP | GO:0014848 | urinary tract smooth muscle contraction | 2 | 6 | 4.67e-01 |
| GO:BP | GO:0019731 | antibacterial humoral response | 4 | 15 | 4.67e-01 |
| GO:BP | GO:0043045 | DNA methylation involved in embryo development | 1 | 7 | 4.67e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 4.67e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 4.67e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 1 | 6 | 4.67e-01 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 3 | 20 | 4.67e-01 |
| GO:BP | GO:0006768 | biotin metabolic process | 1 | 3 | 4.67e-01 |
| GO:BP | GO:0060232 | delamination | 1 | 5 | 4.67e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 12 | 164 | 4.67e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 6 | 56 | 4.68e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 1 | 9 | 4.68e-01 |
| GO:BP | GO:0060856 | establishment of blood-brain barrier | 1 | 9 | 4.68e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 4 | 147 | 4.68e-01 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 1 | 11 | 4.69e-01 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 1 | 11 | 4.69e-01 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 4 | 122 | 4.69e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 1 | 5 | 4.70e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 7 | 60 | 4.70e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 1 | 5 | 4.70e-01 |
| GO:BP | GO:0045950 | negative regulation of mitotic recombination | 1 | 2 | 4.70e-01 |
| GO:BP | GO:0140719 | constitutive heterochromatin formation | 2 | 10 | 4.70e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 19 | 230 | 4.70e-01 |
| GO:BP | GO:1901896 | positive regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 6 | 4.70e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 1 | 7 | 4.70e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 1 | 3 | 4.71e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 18 | 194 | 4.71e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 4.71e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 4.71e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 4.71e-01 |
| GO:BP | GO:0032077 | positive regulation of deoxyribonuclease activity | 1 | 5 | 4.71e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 1 | 3 | 4.71e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 1 | 4 | 4.71e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 7 | 1145 | 4.71e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 4.71e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 11 | 158 | 4.71e-01 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 1 | 9 | 4.71e-01 |
| GO:BP | GO:0097694 | establishment of RNA localization to telomere | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 3 | 4953 | 4.71e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 34 | 357 | 4.71e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0097695 | establishment of protein-containing complex localization to telomere | 1 | 3 | 4.71e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 2 | 100 | 4.71e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 3 | 79 | 4.71e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 2 | 2293 | 4.71e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 2 | 8 | 4.71e-01 |
| GO:BP | GO:0048469 | cell maturation | 12 | 108 | 4.71e-01 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 4.71e-01 |
| GO:BP | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity | 1 | 3 | 4.71e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 2 | 8 | 4.71e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 3 | 4048 | 4.72e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 2 | 12 | 4.72e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 1 | 61 | 4.73e-01 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 3 | 21 | 4.73e-01 |
| GO:BP | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 3 | 36 | 4.73e-01 |
| GO:BP | GO:0032425 | positive regulation of mismatch repair | 1 | 2 | 4.73e-01 |
| GO:BP | GO:1904746 | negative regulation of apoptotic process involved in development | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0072038 | mesenchymal stem cell maintenance involved in nephron morphogenesis | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0036305 | ameloblast differentiation | 1 | 5 | 4.73e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 1 | 9 | 4.73e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 4 | 28 | 4.73e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 64 | 4.73e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 2 | 12 | 4.73e-01 |
| GO:BP | GO:0061523 | cilium disassembly | 1 | 4 | 4.73e-01 |
| GO:BP | GO:0039526 | modulation by virus of host apoptotic process | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0035711 | T-helper 1 cell activation | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 2 | 2684 | 4.73e-01 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 6 | 46 | 4.73e-01 |
| GO:BP | GO:0003032 | detection of oxygen | 1 | 5 | 4.73e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 2 | 2683 | 4.73e-01 |
| GO:BP | GO:0021915 | neural tube development | 11 | 146 | 4.73e-01 |
| GO:BP | GO:0052040 | modulation by symbiont of host programmed cell death | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0052150 | modulation by symbiont of host apoptotic process | 1 | 3 | 4.73e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 2 | 10 | 4.73e-01 |
| GO:BP | GO:0042853 | L-alanine catabolic process | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0042851 | L-alanine metabolic process | 1 | 2 | 4.73e-01 |
| GO:BP | GO:0010693 | negative regulation of alkaline phosphatase activity | 1 | 2 | 4.74e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 16 | 155 | 4.74e-01 |
| GO:BP | GO:0018410 | C-terminal protein amino acid modification | 1 | 10 | 4.75e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 1 | 420 | 4.75e-01 |
| GO:BP | GO:0006833 | water transport | 2 | 10 | 4.75e-01 |
| GO:BP | GO:0050859 | negative regulation of B cell receptor signaling pathway | 1 | 3 | 4.75e-01 |
| GO:BP | GO:1904044 | response to aldosterone | 2 | 9 | 4.76e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 1 | 7 | 4.76e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 1 | 114 | 4.76e-01 |
| GO:BP | GO:0035385 | Roundabout signaling pathway | 1 | 5 | 4.76e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 4.76e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 3 | 23 | 4.76e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 2 | 47 | 4.76e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 8 | 67 | 4.76e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 3 | 57 | 4.77e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 2 | 2714 | 4.77e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 6 | 28 | 4.77e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 3 | 18 | 4.77e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 2 | 30 | 4.77e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 1 | 295 | 4.77e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 4 | 28 | 4.77e-01 |
| GO:BP | GO:0032261 | purine nucleotide salvage | 2 | 10 | 4.78e-01 |
| GO:BP | GO:0031642 | negative regulation of myelination | 2 | 8 | 4.78e-01 |
| GO:BP | GO:0090186 | regulation of pancreatic juice secretion | 1 | 6 | 4.78e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 1 | 6 | 4.78e-01 |
| GO:BP | GO:0014824 | artery smooth muscle contraction | 2 | 7 | 4.79e-01 |
| GO:BP | GO:1903774 | positive regulation of viral budding via host ESCRT complex | 1 | 3 | 4.79e-01 |
| GO:BP | GO:0009631 | cold acclimation | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 2 | 19 | 4.79e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 4 | 20 | 4.79e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 2 | 19 | 4.79e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 2 | 21 | 4.79e-01 |
| GO:BP | GO:0006679 | glucosylceramide biosynthetic process | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 10 | 112 | 4.79e-01 |
| GO:BP | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine | 1 | 3 | 4.79e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 1 | 11 | 4.79e-01 |
| GO:BP | GO:0051126 | negative regulation of actin nucleation | 1 | 6 | 4.80e-01 |
| GO:BP | GO:1900029 | positive regulation of ruffle assembly | 1 | 12 | 4.80e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 2 | 710 | 4.80e-01 |
| GO:BP | GO:1901623 | regulation of lymphocyte chemotaxis | 3 | 13 | 4.80e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 10 | 82 | 4.81e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 5 | 26 | 4.81e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 5 | 72 | 4.81e-01 |
| GO:BP | GO:0016540 | protein autoprocessing | 3 | 16 | 4.81e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 3 | 22 | 4.81e-01 |
| GO:BP | GO:1905584 | outer hair cell apoptotic process | 1 | 2 | 4.81e-01 |
| GO:BP | GO:0098917 | retrograde trans-synaptic signaling | 2 | 7 | 4.81e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 2 | 10 | 4.81e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 13 | 158 | 4.81e-01 |
| GO:BP | GO:0051170 | import into nucleus | 4 | 152 | 4.81e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 5 | 62 | 4.82e-01 |
| GO:BP | GO:0048641 | regulation of skeletal muscle tissue development | 2 | 20 | 4.82e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 2 | 8 | 4.82e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 3 | 15 | 4.82e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 3 | 16 | 4.82e-01 |
| GO:BP | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization | 1 | 4 | 4.82e-01 |
| GO:BP | GO:0051798 | positive regulation of hair follicle development | 2 | 9 | 4.83e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 2 | 34 | 4.84e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 4 | 33 | 4.84e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 4.84e-01 |
| GO:BP | GO:0006598 | polyamine catabolic process | 1 | 4 | 4.84e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 4 | 25 | 4.84e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 4 | 25 | 4.84e-01 |
| GO:BP | GO:0097576 | vacuole fusion | 1 | 12 | 4.84e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 3 | 326 | 4.84e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 2 | 12 | 4.84e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 2 | 9 | 4.84e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 3 | 18 | 4.84e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 4 | 67 | 4.84e-01 |
| GO:BP | GO:0070070 | proton-transporting V-type ATPase complex assembly | 1 | 7 | 4.84e-01 |
| GO:BP | GO:0051974 | negative regulation of telomerase activity | 2 | 11 | 4.84e-01 |
| GO:BP | GO:0071236 | cellular response to antibiotic | 1 | 11 | 4.85e-01 |
| GO:BP | GO:0051795 | positive regulation of timing of catagen | 1 | 3 | 4.85e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 4.85e-01 |
| GO:BP | GO:0031581 | hemidesmosome assembly | 1 | 6 | 4.85e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 7 | 50 | 4.85e-01 |
| GO:BP | GO:0010609 | mRNA localization resulting in post-transcriptional regulation of gene expression | 1 | 2 | 4.85e-01 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 1 | 8 | 4.85e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 660 | 4.86e-01 |
| GO:BP | GO:0060840 | artery development | 10 | 83 | 4.86e-01 |
| GO:BP | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | 1 | 2 | 4.86e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 7 | 214 | 4.87e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 3 | 17 | 4.87e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 2 | 10 | 4.87e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 2 | 51 | 4.87e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 2 | 51 | 4.87e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 1 | 311 | 4.88e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 3 | 14 | 4.88e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 1 | 15 | 4.88e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 6 | 88 | 4.88e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 5 | 41 | 4.88e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 1 | 32 | 4.88e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 3 | 14 | 4.89e-01 |
| GO:BP | GO:0061024 | membrane organization | 1 | 671 | 4.90e-01 |
| GO:BP | GO:0090666 | scaRNA localization to Cajal body | 1 | 5 | 4.90e-01 |
| GO:BP | GO:0062043 | positive regulation of cardiac epithelial to mesenchymal transition | 1 | 9 | 4.90e-01 |
| GO:BP | GO:0001765 | membrane raft assembly | 1 | 9 | 4.90e-01 |
| GO:BP | GO:0008213 | protein alkylation | 1 | 160 | 4.90e-01 |
| GO:BP | GO:0110135 | Norrin signaling pathway | 1 | 2 | 4.90e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 1 | 23 | 4.90e-01 |
| GO:BP | GO:0006479 | protein methylation | 1 | 160 | 4.90e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 10 | 65 | 4.91e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 3 | 22 | 4.92e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 678 | 4.92e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 2 | 57 | 4.92e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 3 | 20 | 4.93e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 1 | 64 | 4.93e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 1 | 13 | 4.93e-01 |
| GO:BP | GO:0070383 | DNA cytosine deamination | 1 | 5 | 4.93e-01 |
| GO:BP | GO:0030091 | protein repair | 1 | 7 | 4.94e-01 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 1 | 8 | 4.94e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 1 | 319 | 4.94e-01 |
| GO:BP | GO:0070486 | leukocyte aggregation | 2 | 9 | 4.94e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 6 | 46 | 4.94e-01 |
| GO:BP | GO:0003174 | mitral valve development | 1 | 12 | 4.94e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 4.94e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 4 | 30 | 4.94e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 2 | 27 | 4.94e-01 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 1 | 10 | 4.94e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 2 | 2845 | 4.94e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 2 | 26 | 4.94e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 23 | 204 | 4.94e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 4.94e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 3 | 93 | 4.94e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 1 | 19 | 4.97e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 1 | 18 | 4.97e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 2 | 10 | 4.97e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 1 | 29 | 4.97e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 1 | 16 | 4.97e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 2 | 11 | 4.97e-01 |
| GO:BP | GO:0099152 | regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane | 1 | 3 | 4.97e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 1 | 52 | 4.97e-01 |
| GO:BP | GO:0009068 | aspartate family amino acid catabolic process | 1 | 12 | 4.97e-01 |
| GO:BP | GO:0015846 | polyamine transport | 2 | 12 | 4.98e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 4 | 39 | 4.98e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 3 | 19 | 4.98e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 3 | 34 | 4.98e-01 |
| GO:BP | GO:0042415 | norepinephrine metabolic process | 2 | 7 | 4.98e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 11 | 89 | 4.98e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 11 | 89 | 4.98e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 6 | 65 | 4.98e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.98e-01 |
| GO:BP | GO:0046368 | GDP-L-fucose metabolic process | 1 | 4 | 4.98e-01 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 1 | 4 | 4.98e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.98e-01 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 3 | 20 | 4.98e-01 |
| GO:BP | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process | 1 | 3 | 4.98e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.98e-01 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 1 | 4 | 4.98e-01 |
| GO:BP | GO:0046075 | dTTP metabolic process | 1 | 4 | 4.98e-01 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 1 | 5 | 4.98e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 3 | 23 | 4.98e-01 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 1 | 4 | 4.98e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 3 | 26 | 4.99e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 2 | 14 | 4.99e-01 |
| GO:BP | GO:0006362 | transcription elongation by RNA polymerase I | 1 | 4 | 4.99e-01 |
| GO:BP | GO:0001696 | gastric acid secretion | 1 | 10 | 4.99e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 3 | 67 | 4.99e-01 |
| GO:BP | GO:0032696 | negative regulation of interleukin-13 production | 1 | 3 | 5.00e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 2 | 13 | 5.00e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 2 | 11 | 5.00e-01 |
| GO:BP | GO:0051179 | localization | 3 | 3969 | 5.00e-01 |
| GO:BP | GO:0070317 | negative regulation of G0 to G1 transition | 1 | 5 | 5.00e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 4 | 40 | 5.00e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 20 | 161 | 5.00e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0060047 | heart contraction | 12 | 205 | 5.00e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0051136 | regulation of NK T cell differentiation | 1 | 4 | 5.00e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 3 | 20 | 5.00e-01 |
| GO:BP | GO:0019856 | pyrimidine nucleobase biosynthetic process | 1 | 8 | 5.00e-01 |
| GO:BP | GO:0007281 | germ cell development | 8 | 187 | 5.00e-01 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 1 | 7 | 5.00e-01 |
| GO:BP | GO:0009224 | CMP biosynthetic process | 1 | 3 | 5.00e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 5 | 65 | 5.00e-01 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 4 | 23 | 5.00e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 1 | 461 | 5.00e-01 |
| GO:BP | GO:0001503 | ossification | 1 | 313 | 5.00e-01 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 4 | 28 | 5.00e-01 |
| GO:BP | GO:0090309 | positive regulation of DNA methylation-dependent heterochromatin formation | 1 | 12 | 5.01e-01 |
| GO:BP | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation | 1 | 4 | 5.01e-01 |
| GO:BP | GO:0036335 | intestinal stem cell homeostasis | 1 | 3 | 5.02e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 2 | 11 | 5.03e-01 |
| GO:BP | GO:1901465 | positive regulation of tetrapyrrole biosynthetic process | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0070455 | positive regulation of heme biosynthetic process | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 5 | 73 | 5.06e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 5.06e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 1 | 6 | 5.06e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 6 | 86 | 5.06e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 12 | 113 | 5.06e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 4 | 29 | 5.06e-01 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 1 | 9 | 5.06e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 4 | 29 | 5.06e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 1 | 469 | 5.06e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 2 | 19 | 5.06e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 740 | 5.06e-01 |
| GO:BP | GO:0098718 | serine import across plasma membrane | 1 | 2 | 5.06e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 14 | 124 | 5.06e-01 |
| GO:BP | GO:1903812 | L-serine import across plasma membrane | 1 | 2 | 5.06e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 97 | 5.06e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 1 | 20 | 5.07e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 1 | 6 | 5.07e-01 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0019660 | glycolytic fermentation | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 1 | 4 | 5.07e-01 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 1 | 4 | 5.07e-01 |
| GO:BP | GO:1901003 | negative regulation of fermentation | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0071279 | cellular response to cobalt ion | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0035879 | plasma membrane lactate transport | 1 | 3 | 5.07e-01 |
| GO:BP | GO:0008291 | acetylcholine metabolic process | 1 | 3 | 5.07e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 3 | 42 | 5.07e-01 |
| GO:BP | GO:0051884 | regulation of timing of anagen | 1 | 5 | 5.07e-01 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 1 | 5 | 5.07e-01 |
| GO:BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 1 | 5 | 5.07e-01 |
| GO:BP | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 5.07e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 1 | 798 | 5.07e-01 |
| GO:BP | GO:0042640 | anagen | 1 | 5 | 5.07e-01 |
| GO:BP | GO:0042695 | thelarche | 1 | 5 | 5.07e-01 |
| GO:BP | GO:0042867 | pyruvate catabolic process | 1 | 2 | 5.07e-01 |
| GO:BP | GO:0006491 | N-glycan processing | 2 | 18 | 5.07e-01 |
| GO:BP | GO:0043132 | NAD transport | 1 | 3 | 5.07e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 3 | 18 | 5.07e-01 |
| GO:BP | GO:0060751 | branch elongation involved in mammary gland duct branching | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission | 1 | 3 | 5.07e-01 |
| GO:BP | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | 1 | 4 | 5.07e-01 |
| GO:BP | GO:1900619 | acetate ester metabolic process | 1 | 3 | 5.07e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 1 | 9 | 5.07e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 5.07e-01 |
| GO:BP | GO:0048022 | negative regulation of melanin biosynthetic process | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 14 | 138 | 5.07e-01 |
| GO:BP | GO:1990966 | ATP generation from poly-ADP-D-ribose | 1 | 4 | 5.07e-01 |
| GO:BP | GO:0097325 | melanocyte proliferation | 1 | 2 | 5.07e-01 |
| GO:BP | GO:0060744 | mammary gland branching involved in thelarche | 1 | 5 | 5.07e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 13 | 186 | 5.07e-01 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 1 | 17 | 5.07e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 7 | 58 | 5.08e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 2 | 12 | 5.08e-01 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 1 | 6 | 5.08e-01 |
| GO:BP | GO:0003128 | heart field specification | 2 | 16 | 5.08e-01 |
| GO:BP | GO:0060039 | pericardium development | 2 | 17 | 5.08e-01 |
| GO:BP | GO:0042631 | cellular response to water deprivation | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 2 | 10 | 5.08e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 5 | 665 | 5.08e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 5.09e-01 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 3 | 21 | 5.10e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 3 | 20 | 5.10e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 5.10e-01 |
| GO:BP | GO:0044857 | plasma membrane raft organization | 1 | 9 | 5.10e-01 |
| GO:BP | GO:0000821 | regulation of arginine metabolic process | 1 | 3 | 5.10e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 1 | 37 | 5.10e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 2 | 3012 | 5.10e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 1 | 3 | 5.10e-01 |
| GO:BP | GO:0014820 | tonic smooth muscle contraction | 2 | 8 | 5.10e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 2 | 5 | 5.10e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 1 | 12 | 5.10e-01 |
| GO:BP | GO:0045763 | negative regulation of cellular amino acid metabolic process | 1 | 2 | 5.10e-01 |
| GO:BP | GO:0140192 | regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 5.10e-01 |
| GO:BP | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 5.10e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 5.10e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 5.10e-01 |
| GO:BP | GO:0071896 | protein localization to adherens junction | 1 | 6 | 5.10e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 3 | 19 | 5.10e-01 |
| GO:BP | GO:0071307 | cellular response to vitamin K | 1 | 4 | 5.10e-01 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 2 | 4 | 5.10e-01 |
| GO:BP | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development | 1 | 2 | 5.10e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 1 | 875 | 5.10e-01 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 2 | 12 | 5.10e-01 |
| GO:BP | GO:0062042 | regulation of cardiac epithelial to mesenchymal transition | 1 | 10 | 5.10e-01 |
| GO:BP | GO:0042551 | neuron maturation | 3 | 33 | 5.10e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 1 | 3 | 5.10e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 5.10e-01 |
| GO:BP | GO:0070384 | Harderian gland development | 1 | 4 | 5.10e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 1 | 816 | 5.10e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 1 | 756 | 5.10e-01 |
| GO:BP | GO:0015860 | purine nucleoside transmembrane transport | 1 | 3 | 5.11e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 25 | 249 | 5.11e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 13 | 5.11e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 5.11e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 5.11e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 1 | 4 | 5.11e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 1 | 4 | 5.11e-01 |
| GO:BP | GO:0061009 | common bile duct development | 1 | 4 | 5.11e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 5.11e-01 |
| GO:BP | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process | 1 | 3 | 5.12e-01 |
| GO:BP | GO:0019068 | virion assembly | 4 | 35 | 5.12e-01 |
| GO:BP | GO:0090217 | negative regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 3 | 5.12e-01 |
| GO:BP | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process | 1 | 3 | 5.12e-01 |
| GO:BP | GO:1902527 | positive regulation of protein monoubiquitination | 1 | 5 | 5.12e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 5.12e-01 |
| GO:BP | GO:0019079 | viral genome replication | 1 | 101 | 5.12e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 3 | 4455 | 5.12e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 1 | 18 | 5.12e-01 |
| GO:BP | GO:0018095 | protein polyglutamylation | 2 | 9 | 5.13e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 9 | 67 | 5.13e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 2 | 12 | 5.13e-01 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 1 | 22 | 5.13e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 2 | 20 | 5.13e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 1 | 72 | 5.13e-01 |
| GO:BP | GO:0070778 | L-aspartate transmembrane transport | 2 | 11 | 5.13e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 2 | 37 | 5.13e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 4 | 16 | 5.13e-01 |
| GO:BP | GO:0048732 | gland development | 3 | 318 | 5.14e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 6 | 60 | 5.14e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 3 | 11 | 5.14e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 3 | 22 | 5.14e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 2 | 11 | 5.14e-01 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 4 | 24 | 5.14e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 3 | 23 | 5.15e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 5 | 74 | 5.15e-01 |
| GO:BP | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching | 1 | 4 | 5.15e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 1 | 3 | 5.15e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 1 | 4 | 5.15e-01 |
| GO:BP | GO:0060847 | endothelial cell fate specification | 1 | 3 | 5.15e-01 |
| GO:BP | GO:0009080 | pyruvate family amino acid catabolic process | 1 | 2 | 5.15e-01 |
| GO:BP | GO:0006522 | alanine metabolic process | 1 | 2 | 5.15e-01 |
| GO:BP | GO:0006524 | alanine catabolic process | 1 | 2 | 5.15e-01 |
| GO:BP | GO:0009078 | pyruvate family amino acid metabolic process | 1 | 2 | 5.15e-01 |
| GO:BP | GO:0060300 | regulation of cytokine activity | 1 | 3 | 5.15e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 4 | 30 | 5.15e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 1 | 12 | 5.16e-01 |
| GO:BP | GO:0007350 | blastoderm segmentation | 4 | 17 | 5.16e-01 |
| GO:BP | GO:1903766 | positive regulation of potassium ion export across plasma membrane | 1 | 3 | 5.17e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 901 | 5.18e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 1 | 18 | 5.20e-01 |
| GO:BP | GO:0006560 | proline metabolic process | 1 | 7 | 5.21e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 8 | 116 | 5.21e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 1 | 57 | 5.21e-01 |
| GO:BP | GO:0021559 | trigeminal nerve development | 2 | 5 | 5.21e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 2 | 3070 | 5.21e-01 |
| GO:BP | GO:0044868 | modulation by host of viral molecular function | 1 | 2 | 5.21e-01 |
| GO:BP | GO:0070459 | prolactin secretion | 1 | 4 | 5.21e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 2 | 3080 | 5.21e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 3 | 58 | 5.21e-01 |
| GO:BP | GO:1903613 | regulation of protein tyrosine phosphatase activity | 1 | 4 | 5.21e-01 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 2 | 15 | 5.21e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 5.21e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 2 | 3080 | 5.21e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 6 | 51 | 5.21e-01 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 2 | 15 | 5.21e-01 |
| GO:BP | GO:0048874 | host-mediated regulation of intestinal microbiota composition | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0042439 | ethanolamine-containing compound metabolic process | 1 | 5 | 5.22e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 1 | 12 | 5.22e-01 |
| GO:BP | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 2 | 21 | 5.22e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 5.22e-01 |
| GO:BP | GO:0003306 | Wnt signaling pathway involved in heart development | 2 | 8 | 5.22e-01 |
| GO:BP | GO:0036123 | histone H3-K9 dimethylation | 1 | 3 | 5.23e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 2 | 11 | 5.23e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 5.23e-01 |
| GO:BP | GO:0006500 | N-terminal protein palmitoylation | 1 | 3 | 5.24e-01 |
| GO:BP | GO:0000303 | response to superoxide | 2 | 21 | 5.25e-01 |
| GO:BP | GO:0014816 | skeletal muscle satellite cell differentiation | 1 | 6 | 5.25e-01 |
| GO:BP | GO:0046794 | transport of virus | 1 | 19 | 5.25e-01 |
| GO:BP | GO:1901503 | ether biosynthetic process | 1 | 11 | 5.25e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 5 | 58 | 5.25e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 3 | 23 | 5.25e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 1 | 371 | 5.25e-01 |
| GO:BP | GO:0071361 | cellular response to ethanol | 2 | 9 | 5.25e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 1 | 20 | 5.25e-01 |
| GO:BP | GO:0032714 | negative regulation of interleukin-5 production | 1 | 2 | 5.25e-01 |
| GO:BP | GO:0006595 | polyamine metabolic process | 2 | 16 | 5.25e-01 |
| GO:BP | GO:0006915 | apoptotic process | 10 | 1443 | 5.25e-01 |
| GO:BP | GO:0010737 | protein kinase A signaling | 4 | 23 | 5.26e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 3 | 348 | 5.26e-01 |
| GO:BP | GO:0021564 | vagus nerve development | 1 | 2 | 5.26e-01 |
| GO:BP | GO:0070189 | kynurenine metabolic process | 2 | 6 | 5.26e-01 |
| GO:BP | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process | 1 | 9 | 5.26e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 2 | 13 | 5.26e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 5.26e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 5 | 46 | 5.26e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 3 | 91 | 5.27e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 4 | 53 | 5.27e-01 |
| GO:BP | GO:0018119 | peptidyl-cysteine S-nitrosylation | 1 | 11 | 5.27e-01 |
| GO:BP | GO:0017014 | protein nitrosylation | 1 | 11 | 5.27e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 8 | 101 | 5.27e-01 |
| GO:BP | GO:1901998 | toxin transport | 4 | 37 | 5.28e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 2 | 10 | 5.28e-01 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 1 | 13 | 5.28e-01 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 2 | 20 | 5.28e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 3 | 14 | 5.28e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 1 | 13 | 5.28e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 2 | 3132 | 5.28e-01 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 2 | 13 | 5.29e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 1 | 78 | 5.29e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 3 | 4570 | 5.29e-01 |
| GO:BP | GO:2001200 | positive regulation of dendritic cell differentiation | 1 | 3 | 5.29e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 5.29e-01 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 2 | 11 | 5.29e-01 |
| GO:BP | GO:0032423 | regulation of mismatch repair | 1 | 3 | 5.29e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 2 | 3133 | 5.29e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 1 | 5 | 5.30e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 3 | 33 | 5.30e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 5.30e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 3 | 16 | 5.30e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 4 | 5.30e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 1 | 11 | 5.30e-01 |
| GO:BP | GO:0090156 | intracellular sphingolipid homeostasis | 1 | 5 | 5.31e-01 |
| GO:BP | GO:0006572 | tyrosine catabolic process | 1 | 2 | 5.31e-01 |
| GO:BP | GO:0043543 | protein acylation | 13 | 228 | 5.31e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 1 | 10 | 5.31e-01 |
| GO:BP | GO:0044406 | adhesion of symbiont to host | 2 | 10 | 5.31e-01 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 2 | 15 | 5.31e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 5.31e-01 |
| GO:BP | GO:1905702 | regulation of inhibitory synapse assembly | 1 | 3 | 5.31e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 3 | 17 | 5.32e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 1 | 14 | 5.32e-01 |
| GO:BP | GO:2001245 | regulation of phosphatidylcholine biosynthetic process | 1 | 5 | 5.32e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 11 | 93 | 5.32e-01 |
| GO:BP | GO:0051593 | response to folic acid | 1 | 9 | 5.33e-01 |
| GO:BP | GO:0016049 | cell growth | 38 | 418 | 5.33e-01 |
| GO:BP | GO:0032222 | regulation of synaptic transmission, cholinergic | 1 | 3 | 5.34e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 6 | 90 | 5.34e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 2 | 3169 | 5.34e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 3 | 37 | 5.34e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 3 | 4596 | 5.34e-01 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 1 | 8 | 5.34e-01 |
| GO:BP | GO:0071321 | cellular response to cGMP | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 6 | 185 | 5.34e-01 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 1 | 9 | 5.34e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 7 | 82 | 5.34e-01 |
| GO:BP | GO:0035995 | detection of muscle stretch | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0070444 | oligodendrocyte progenitor proliferation | 1 | 5 | 5.34e-01 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 2 | 15 | 5.34e-01 |
| GO:BP | GO:0070445 | regulation of oligodendrocyte progenitor proliferation | 1 | 5 | 5.34e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 1 | 7 | 5.34e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 1 | 21 | 5.34e-01 |
| GO:BP | GO:0022900 | electron transport chain | 4 | 142 | 5.35e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 7 | 56 | 5.36e-01 |
| GO:BP | GO:0055089 | fatty acid homeostasis | 1 | 12 | 5.37e-01 |
| GO:BP | GO:1903772 | regulation of viral budding via host ESCRT complex | 1 | 4 | 5.37e-01 |
| GO:BP | GO:1990044 | protein localization to lipid droplet | 1 | 4 | 5.37e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 2 | 13 | 5.38e-01 |
| GO:BP | GO:0046440 | L-lysine metabolic process | 1 | 4 | 5.38e-01 |
| GO:BP | GO:0019477 | L-lysine catabolic process | 1 | 4 | 5.38e-01 |
| GO:BP | GO:0019474 | L-lysine catabolic process to acetyl-CoA | 1 | 4 | 5.38e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 1 | 101 | 5.39e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 1 | 9 | 5.40e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 1 | 9 | 5.40e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 4 | 29 | 5.40e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 1 | 12 | 5.40e-01 |
| GO:BP | GO:0006688 | glycosphingolipid biosynthetic process | 4 | 25 | 5.40e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 1 | 390 | 5.40e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 3 | 52 | 5.40e-01 |
| GO:BP | GO:1903593 | regulation of histamine secretion by mast cell | 1 | 4 | 5.40e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 2 | 22 | 5.40e-01 |
| GO:BP | GO:1903595 | positive regulation of histamine secretion by mast cell | 1 | 4 | 5.40e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 2 | 40 | 5.41e-01 |
| GO:BP | GO:0035821 | modulation of process of another organism | 2 | 5 | 5.41e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 22 | 303 | 5.41e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 8 | 76 | 5.41e-01 |
| GO:BP | GO:0014829 | vascular associated smooth muscle contraction | 2 | 20 | 5.41e-01 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 2 | 12 | 5.41e-01 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 5 | 5.41e-01 |
| GO:BP | GO:0021766 | hippocampus development | 1 | 62 | 5.41e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 3 | 17 | 5.41e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 2 | 21 | 5.41e-01 |
| GO:BP | GO:0048678 | response to axon injury | 2 | 64 | 5.41e-01 |
| GO:BP | GO:0098935 | dendritic transport | 2 | 13 | 5.41e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0001554 | luteolysis | 1 | 7 | 5.41e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 5.41e-01 |
| GO:BP | GO:0100012 | regulation of heart induction by canonical Wnt signaling pathway | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 2 | 9 | 5.41e-01 |
| GO:BP | GO:0009186 | deoxyribonucleoside diphosphate metabolic process | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 5.41e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 5 | 69 | 5.41e-01 |
| GO:BP | GO:1903431 | positive regulation of cell maturation | 1 | 6 | 5.41e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 5.41e-01 |
| GO:BP | GO:1902474 | positive regulation of protein localization to synapse | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 2 | 39 | 5.41e-01 |
| GO:BP | GO:0042637 | catagen | 1 | 4 | 5.41e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | 40 | 5.41e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 4 | 25 | 5.41e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 2 | 39 | 5.41e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 3 | 30 | 5.41e-01 |
| GO:BP | GO:0002941 | synoviocyte proliferation | 1 | 3 | 5.41e-01 |
| GO:BP | GO:1901647 | positive regulation of synoviocyte proliferation | 1 | 3 | 5.41e-01 |
| GO:BP | GO:1901645 | regulation of synoviocyte proliferation | 1 | 3 | 5.41e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 2 | 3237 | 5.41e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 3 | 246 | 5.41e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 3 | 32 | 5.41e-01 |
| GO:BP | GO:1900193 | regulation of oocyte maturation | 1 | 5 | 5.41e-01 |
| GO:BP | GO:0021891 | olfactory bulb interneuron development | 1 | 5 | 5.41e-01 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0046753 | non-lytic viral release | 1 | 14 | 5.41e-01 |
| GO:BP | GO:0140021 | mitochondrial ADP transmembrane transport | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 5.41e-01 |
| GO:BP | GO:0002175 | protein localization to paranode region of axon | 1 | 3 | 5.41e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 3 | 30 | 5.41e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 3 | 23 | 5.41e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 2 | 10 | 5.41e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 12 | 93 | 5.41e-01 |
| GO:BP | GO:0060364 | frontal suture morphogenesis | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0090381 | regulation of heart induction | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0060028 | convergent extension involved in axis elongation | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | 1 | 3 | 5.41e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 1 | 10 | 5.41e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 3 | 39 | 5.41e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 1 | 6 | 5.41e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 5.41e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 2 | 22 | 5.41e-01 |
| GO:BP | GO:0051794 | regulation of timing of catagen | 1 | 4 | 5.41e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 8 | 76 | 5.41e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 12 | 118 | 5.41e-01 |
| GO:BP | GO:0050893 | sensory processing | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 1 | 10 | 5.41e-01 |
| GO:BP | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0048839 | inner ear development | 2 | 132 | 5.41e-01 |
| GO:BP | GO:0090168 | Golgi reassembly | 1 | 5 | 5.41e-01 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 1 | 9 | 5.41e-01 |
| GO:BP | GO:0043305 | negative regulation of mast cell degranulation | 1 | 2 | 5.41e-01 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0070100 | negative regulation of chemokine-mediated signaling pathway | 1 | 6 | 5.41e-01 |
| GO:BP | GO:1990544 | mitochondrial ATP transmembrane transport | 1 | 4 | 5.41e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 2 | 22 | 5.41e-01 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 1 | 5 | 5.41e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 1 | 6 | 5.42e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 12 | 198 | 5.42e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 9 | 69 | 5.42e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 2 | 10 | 5.43e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 5.44e-01 |
| GO:BP | GO:2000627 | positive regulation of miRNA catabolic process | 1 | 4 | 5.45e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 3 | 23 | 5.45e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 3 | 23 | 5.45e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 3 | 23 | 5.45e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 1 | 23 | 5.45e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 71 | 4099 | 5.45e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 1 | 5 | 5.47e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 1 | 5 | 5.47e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 4 | 25 | 5.47e-01 |
| GO:BP | GO:1901072 | glucosamine-containing compound catabolic process | 1 | 5 | 5.47e-01 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 2 | 7 | 5.47e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 3 | 20 | 5.47e-01 |
| GO:BP | GO:0003015 | heart process | 12 | 216 | 5.48e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 3 | 27 | 5.48e-01 |
| GO:BP | GO:0036149 | phosphatidylinositol acyl-chain remodeling | 1 | 3 | 5.48e-01 |
| GO:BP | GO:0071709 | membrane assembly | 5 | 53 | 5.48e-01 |
| GO:BP | GO:0046688 | response to copper ion | 2 | 29 | 5.48e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 1 | 612 | 5.48e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 2 | 40 | 5.48e-01 |
| GO:BP | GO:0009972 | cytidine deamination | 1 | 6 | 5.48e-01 |
| GO:BP | GO:0046087 | cytidine metabolic process | 1 | 6 | 5.48e-01 |
| GO:BP | GO:0016554 | cytidine to uridine editing | 1 | 6 | 5.48e-01 |
| GO:BP | GO:0006216 | cytidine catabolic process | 1 | 6 | 5.48e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 1 | 423 | 5.48e-01 |
| GO:BP | GO:0006113 | fermentation | 1 | 5 | 5.48e-01 |
| GO:BP | GO:0006003 | fructose 2,6-bisphosphate metabolic process | 1 | 4 | 5.48e-01 |
| GO:BP | GO:0009116 | nucleoside metabolic process | 5 | 42 | 5.48e-01 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 3 | 22 | 5.48e-01 |
| GO:BP | GO:0018344 | protein geranylgeranylation | 1 | 6 | 5.48e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 6 | 57 | 5.48e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 7 | 129 | 5.48e-01 |
| GO:BP | GO:0043465 | regulation of fermentation | 1 | 5 | 5.48e-01 |
| GO:BP | GO:0003188 | heart valve formation | 1 | 16 | 5.48e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 1 | 4 | 5.48e-01 |
| GO:BP | GO:0042262 | DNA protection | 1 | 6 | 5.48e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 5.48e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 2 | 25 | 5.48e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 5.49e-01 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 1 | 6 | 5.50e-01 |
| GO:BP | GO:1904772 | response to tetrachloromethane | 1 | 4 | 5.51e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 2 | 10 | 5.51e-01 |
| GO:BP | GO:0031860 | telomeric 3’ overhang formation | 1 | 4 | 5.51e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 3 | 19 | 5.51e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 963 | 5.52e-01 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 1 | 14 | 5.52e-01 |
| GO:BP | GO:0012501 | programmed cell death | 10 | 1479 | 5.53e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 3 | 4747 | 5.53e-01 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 1 | 17 | 5.54e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 1 | 429 | 5.55e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 2 | 18 | 5.55e-01 |
| GO:BP | GO:0048102 | autophagic cell death | 1 | 6 | 5.55e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 12 | 94 | 5.56e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 59 | 5.56e-01 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 1 | 5 | 5.56e-01 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 1 | 12 | 5.57e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1361 | 5.57e-01 |
| GO:BP | GO:0070293 | renal absorption | 3 | 24 | 5.57e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 336 | 5.57e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 2 | 9 | 5.57e-01 |
| GO:BP | GO:0097241 | hematopoietic stem cell migration to bone marrow | 1 | 5 | 5.58e-01 |
| GO:BP | GO:0035148 | tube formation | 9 | 138 | 5.58e-01 |
| GO:BP | GO:0039663 | membrane fusion involved in viral entry into host cell | 1 | 4 | 5.58e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 9 | 133 | 5.58e-01 |
| GO:BP | GO:0019064 | fusion of virus membrane with host plasma membrane | 1 | 4 | 5.58e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 1 | 11 | 5.58e-01 |
| GO:BP | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 1 | 3 | 5.58e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 8 | 1496 | 5.58e-01 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 2 | 14 | 5.58e-01 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 2 | 11 | 5.58e-01 |
| GO:BP | GO:0015853 | adenine transport | 1 | 5 | 5.59e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 2 | 13 | 5.59e-01 |
| GO:BP | GO:0015855 | pyrimidine nucleobase transport | 1 | 3 | 5.59e-01 |
| GO:BP | GO:0015862 | uridine transport | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0015864 | pyrimidine nucleoside transport | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 2 | 10 | 5.59e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 3 | 25 | 5.59e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 1 | 19 | 5.59e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 1 | 8 | 5.60e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 4 | 170 | 5.60e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 5.60e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 1 | 43 | 5.60e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 1 | 14 | 5.60e-01 |
| GO:BP | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery | 1 | 6 | 5.61e-01 |
| GO:BP | GO:0060384 | innervation | 3 | 22 | 5.62e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 11 | 107 | 5.62e-01 |
| GO:BP | GO:0048865 | stem cell fate commitment | 1 | 5 | 5.63e-01 |
| GO:BP | GO:0048820 | hair follicle maturation | 2 | 15 | 5.63e-01 |
| GO:BP | GO:0051601 | exocyst localization | 1 | 4 | 5.63e-01 |
| GO:BP | GO:0005984 | disaccharide metabolic process | 1 | 5 | 5.63e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 2 | 10 | 5.63e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 2 | 10 | 5.63e-01 |
| GO:BP | GO:0021871 | forebrain regionalization | 1 | 8 | 5.63e-01 |
| GO:BP | GO:0097340 | inhibition of cysteine-type endopeptidase activity | 1 | 5 | 5.63e-01 |
| GO:BP | GO:1901403 | positive regulation of tetrapyrrole metabolic process | 1 | 4 | 5.63e-01 |
| GO:BP | GO:0097341 | zymogen inhibition | 1 | 5 | 5.63e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 5.63e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 11 | 133 | 5.63e-01 |
| GO:BP | GO:1990504 | dense core granule exocytosis | 1 | 7 | 5.63e-01 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 1 | 7 | 5.63e-01 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 2 | 9 | 5.63e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 10 | 272 | 5.63e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 1 | 18 | 5.63e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 1 | 18 | 5.63e-01 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 5.64e-01 |
| GO:BP | GO:0031119 | tRNA pseudouridine synthesis | 1 | 6 | 5.65e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 5.65e-01 |
| GO:BP | GO:1904273 | L-alanine import across plasma membrane | 1 | 4 | 5.65e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 3 | 40 | 5.65e-01 |
| GO:BP | GO:0031103 | axon regeneration | 4 | 42 | 5.65e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 2 | 13 | 5.65e-01 |
| GO:BP | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis | 1 | 5 | 5.65e-01 |
| GO:BP | GO:0015848 | spermidine transport | 1 | 3 | 5.65e-01 |
| GO:BP | GO:0021544 | subpallium development | 3 | 18 | 5.65e-01 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 1 | 9 | 5.65e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 7 | 54 | 5.65e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 1 | 18 | 5.65e-01 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 1 | 5 | 5.65e-01 |
| GO:BP | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 1 | 5 | 5.65e-01 |
| GO:BP | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion | 1 | 5 | 5.65e-01 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 3 | 16 | 5.65e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 1 | 6 | 5.65e-01 |
| GO:BP | GO:1903371 | regulation of endoplasmic reticulum tubular network organization | 1 | 6 | 5.65e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 10 | 198 | 5.66e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 4 | 21 | 5.66e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 1 | 17 | 5.66e-01 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 2 | 17 | 5.66e-01 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 1 | 17 | 5.66e-01 |
| GO:BP | GO:0032263 | GMP salvage | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 3 | 20 | 5.66e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 3 | 20 | 5.66e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 4 | 62 | 5.66e-01 |
| GO:BP | GO:0044209 | AMP salvage | 1 | 5 | 5.66e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 5 | 47 | 5.66e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 4 | 23 | 5.66e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0035272 | exocrine system development | 4 | 34 | 5.66e-01 |
| GO:BP | GO:0021675 | nerve development | 9 | 57 | 5.66e-01 |
| GO:BP | GO:0045006 | DNA deamination | 1 | 9 | 5.67e-01 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 2 | 19 | 5.67e-01 |
| GO:BP | GO:0046337 | phosphatidylethanolamine metabolic process | 2 | 19 | 5.67e-01 |
| GO:BP | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process | 2 | 17 | 5.67e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 1 | 103 | 5.68e-01 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 1 | 11 | 5.69e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 3 | 4946 | 5.69e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 1 | 122 | 5.69e-01 |
| GO:BP | GO:1905050 | positive regulation of metallopeptidase activity | 1 | 5 | 5.69e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 20 | 187 | 5.69e-01 |
| GO:BP | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway | 1 | 3 | 5.70e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 1 | 8 | 5.70e-01 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 1 | 10 | 5.70e-01 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 4 | 26 | 5.70e-01 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 1 | 10 | 5.70e-01 |
| GO:BP | GO:0046755 | viral budding | 3 | 26 | 5.71e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 5.71e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 5 | 51 | 5.71e-01 |
| GO:BP | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport | 1 | 5 | 5.71e-01 |
| GO:BP | GO:0030002 | intracellular monoatomic anion homeostasis | 1 | 7 | 5.71e-01 |
| GO:BP | GO:0030644 | intracellular chloride ion homeostasis | 1 | 7 | 5.71e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 12 | 95 | 5.71e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 3 | 22 | 5.72e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 6 | 726 | 5.72e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 3 | 21 | 5.72e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 1 | 7 | 5.73e-01 |
| GO:BP | GO:0035066 | positive regulation of histone acetylation | 1 | 25 | 5.73e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 1 | 26 | 5.73e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 3 | 25 | 5.74e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 3 | 25 | 5.74e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 9 | 1133 | 5.74e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 5 | 55 | 5.74e-01 |
| GO:BP | GO:0019083 | viral transcription | 2 | 48 | 5.74e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 2 | 18 | 5.74e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 1 | 74 | 5.74e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 7 | 45 | 5.74e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 1 | 31 | 5.74e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 13 | 115 | 5.75e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 10 | 132 | 5.75e-01 |
| GO:BP | GO:0032355 | response to estradiol | 2 | 83 | 5.75e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 1 | 4 | 5.75e-01 |
| GO:BP | GO:0090215 | regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 4 | 5.75e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 1 | 687 | 5.75e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 11 | 170 | 5.76e-01 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 1 | 8 | 5.77e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 5 | 50 | 5.77e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 2 | 42 | 5.77e-01 |
| GO:BP | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 1 | 5 | 5.78e-01 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 2 | 33 | 5.79e-01 |
| GO:BP | GO:0001561 | fatty acid alpha-oxidation | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 5 | 45 | 5.79e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 4 | 97 | 5.79e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 1 | 8 | 5.79e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 5 | 43 | 5.79e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 7 | 56 | 5.79e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 2 | 87 | 5.79e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 1 | 24 | 5.79e-01 |
| GO:BP | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0072201 | negative regulation of mesenchymal cell proliferation | 1 | 7 | 5.79e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 5.79e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 1 | 7 | 5.79e-01 |
| GO:BP | GO:0032534 | regulation of microvillus assembly | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 5.79e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 7 | 53 | 5.79e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 1 | 15 | 5.79e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0060638 | mesenchymal-epithelial cell signaling | 1 | 4 | 5.79e-01 |
| GO:BP | GO:0046898 | response to cycloheximide | 1 | 4 | 5.79e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 5.79e-01 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 1 | 9 | 5.79e-01 |
| GO:BP | GO:2001054 | negative regulation of mesenchymal cell apoptotic process | 1 | 3 | 5.79e-01 |
| GO:BP | GO:0018197 | peptidyl-aspartic acid modification | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0071839 | apoptotic process in bone marrow cell | 1 | 4 | 5.79e-01 |
| GO:BP | GO:0097350 | neutrophil clearance | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 5 | 35 | 5.79e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 17 | 206 | 5.79e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 2 | 10 | 5.79e-01 |
| GO:BP | GO:0016573 | histone acetylation | 8 | 143 | 5.79e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 1 | 81 | 5.79e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 2 | 13 | 5.79e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 2 | 13 | 5.79e-01 |
| GO:BP | GO:1904970 | brush border assembly | 1 | 2 | 5.80e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 5 | 38 | 5.80e-01 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 3 | 23 | 5.80e-01 |
| GO:BP | GO:0061589 | calcium activated phosphatidylserine scrambling | 1 | 3 | 5.80e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 2 | 27 | 5.80e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 1 | 21 | 5.80e-01 |
| GO:BP | GO:0008038 | neuron recognition | 5 | 39 | 5.80e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 5 | 85 | 5.81e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 1 | 7 | 5.81e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 1 | 487 | 5.81e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 10 | 129 | 5.81e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 6 | 53 | 5.81e-01 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 2 | 12 | 5.81e-01 |
| GO:BP | GO:0009888 | tissue development | 1 | 1425 | 5.82e-01 |
| GO:BP | GO:0150099 | neuron-glial cell signaling | 1 | 4 | 5.82e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 2 | 11 | 5.82e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 13 | 137 | 5.82e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 3 | 55 | 5.82e-01 |
| GO:BP | GO:0035822 | gene conversion | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 2 | 11 | 5.82e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 1 | 110 | 5.82e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 5.82e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 2 | 11 | 5.82e-01 |
| GO:BP | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process | 1 | 6 | 5.82e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 5.82e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 4 | 33 | 5.82e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 3 | 28 | 5.82e-01 |
| GO:BP | GO:1904429 | regulation of t-circle formation | 1 | 5 | 5.82e-01 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 1 | 5 | 5.83e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 24 | 414 | 5.83e-01 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 1 | 5 | 5.83e-01 |
| GO:BP | GO:0046133 | pyrimidine ribonucleoside catabolic process | 1 | 7 | 5.83e-01 |
| GO:BP | GO:0006677 | glycosylceramide metabolic process | 2 | 13 | 5.83e-01 |
| GO:BP | GO:0046131 | pyrimidine ribonucleoside metabolic process | 1 | 7 | 5.83e-01 |
| GO:BP | GO:1903108 | regulation of mitochondrial transcription | 1 | 5 | 5.83e-01 |
| GO:BP | GO:0120188 | regulation of bile acid secretion | 1 | 2 | 5.83e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 5.83e-01 |
| GO:BP | GO:0070086 | ubiquitin-dependent endocytosis | 1 | 4 | 5.83e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 2 | 80 | 5.83e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 1 | 3 | 5.83e-01 |
| GO:BP | GO:0021761 | limbic system development | 11 | 81 | 5.83e-01 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 1 | 5 | 5.83e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 1 | 6 | 5.83e-01 |
| GO:BP | GO:0021549 | cerebellum development | 1 | 78 | 5.84e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 3 | 47 | 5.84e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 12 | 110 | 5.84e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 2 | 19 | 5.84e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 1 | 8 | 5.84e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1199 | 5.84e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 4 | 41 | 5.84e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 2 | 13 | 5.85e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 1 | 10 | 5.85e-01 |
| GO:BP | GO:1901523 | icosanoid catabolic process | 1 | 2 | 5.85e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 3 | 29 | 5.85e-01 |
| GO:BP | GO:0042044 | fluid transport | 2 | 17 | 5.86e-01 |
| GO:BP | GO:0090594 | inflammatory response to wounding | 1 | 12 | 5.86e-01 |
| GO:BP | GO:2000625 | regulation of miRNA catabolic process | 1 | 5 | 5.86e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 3 | 20 | 5.86e-01 |
| GO:BP | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing | 1 | 6 | 5.86e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 1 | 185 | 5.86e-01 |
| GO:BP | GO:0040007 | growth | 39 | 742 | 5.87e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 5.87e-01 |
| GO:BP | GO:0021615 | glossopharyngeal nerve morphogenesis | 1 | 2 | 5.87e-01 |
| GO:BP | GO:0050923 | regulation of negative chemotaxis | 1 | 4 | 5.87e-01 |
| GO:BP | GO:0070305 | response to cGMP | 1 | 7 | 5.87e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 3 | 28 | 5.87e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 5.88e-01 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 2 | 13 | 5.88e-01 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 2 | 14 | 5.88e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 1 | 19 | 5.88e-01 |
| GO:BP | GO:0043583 | ear development | 2 | 153 | 5.88e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 5 | 48 | 5.89e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 5.89e-01 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 1 | 8 | 5.90e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 1 | 10 | 5.90e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 1 | 64 | 5.91e-01 |
| GO:BP | GO:0051694 | pointed-end actin filament capping | 1 | 6 | 5.91e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 13 | 110 | 5.91e-01 |
| GO:BP | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 1 | 6 | 5.91e-01 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 2 | 10 | 5.91e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 5.91e-01 |
| GO:BP | GO:1903301 | positive regulation of hexokinase activity | 1 | 5 | 5.91e-01 |
| GO:BP | GO:0043456 | regulation of pentose-phosphate shunt | 1 | 6 | 5.91e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 18 | 173 | 5.91e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 4 | 29 | 5.92e-01 |
| GO:BP | GO:0060545 | positive regulation of necroptotic process | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | 1 | 2 | 5.92e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 3 | 15 | 5.93e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 2 | 15 | 5.93e-01 |
| GO:BP | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 4 | 5.93e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 3 | 15 | 5.93e-01 |
| GO:BP | GO:0060065 | uterus development | 2 | 13 | 5.93e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 97 | 5.93e-01 |
| GO:BP | GO:0006559 | L-phenylalanine catabolic process | 1 | 3 | 5.93e-01 |
| GO:BP | GO:0072656 | maintenance of protein location in mitochondrion | 1 | 5 | 5.93e-01 |
| GO:BP | GO:1902222 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process | 1 | 3 | 5.93e-01 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 1 | 3 | 5.93e-01 |
| GO:BP | GO:0006558 | L-phenylalanine metabolic process | 1 | 3 | 5.93e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 60 | 2646 | 5.93e-01 |
| GO:BP | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 6 | 5.94e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 5.95e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 1 | 20 | 5.95e-01 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 1 | 7 | 5.95e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 9 | 96 | 5.96e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 23 | 277 | 5.97e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 2 | 3730 | 5.97e-01 |
| GO:BP | GO:1900045 | negative regulation of protein K63-linked ubiquitination | 1 | 7 | 5.97e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 1 | 1088 | 5.97e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 2 | 38 | 5.98e-01 |
| GO:BP | GO:0051597 | response to methylmercury | 1 | 5 | 5.98e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 1 | 5 | 5.98e-01 |
| GO:BP | GO:0035426 | extracellular matrix-cell signaling | 1 | 4 | 5.98e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 1 | 8 | 5.98e-01 |
| GO:BP | GO:0071380 | cellular response to prostaglandin E stimulus | 2 | 14 | 5.98e-01 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 1 | 4 | 5.98e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 14 | 216 | 5.98e-01 |
| GO:BP | GO:0001787 | natural killer cell proliferation | 1 | 6 | 5.99e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 4 | 25 | 5.99e-01 |
| GO:BP | GO:2000491 | positive regulation of hepatic stellate cell activation | 1 | 4 | 5.99e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 5.99e-01 |
| GO:BP | GO:0032835 | glomerulus development | 6 | 54 | 5.99e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 1 | 14 | 5.99e-01 |
| GO:BP | GO:0060268 | negative regulation of respiratory burst | 1 | 4 | 5.99e-01 |
| GO:BP | GO:0043615 | astrocyte cell migration | 1 | 6 | 5.99e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 1 | 16 | 6.00e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 1 | 53 | 6.00e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 6.00e-01 |
| GO:BP | GO:0061912 | selective autophagy | 9 | 90 | 6.00e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 4 | 39 | 6.01e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 2 | 40 | 6.01e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 91 | 4292 | 6.02e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 2 | 14 | 6.02e-01 |
| GO:BP | GO:0090131 | mesenchyme migration | 1 | 4 | 6.02e-01 |
| GO:BP | GO:0032960 | regulation of inositol trisphosphate biosynthetic process | 1 | 4 | 6.02e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 3 | 24 | 6.02e-01 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 1 | 17 | 6.02e-01 |
| GO:BP | GO:2000328 | regulation of T-helper 17 cell lineage commitment | 1 | 1 | 6.02e-01 |
| GO:BP | GO:0060272 | embryonic skeletal joint morphogenesis | 1 | 7 | 6.02e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 2 | 8 | 6.02e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 6 | 39 | 6.02e-01 |
| GO:BP | GO:1904672 | regulation of somatic stem cell population maintenance | 1 | 6 | 6.02e-01 |
| GO:BP | GO:0031529 | ruffle organization | 2 | 45 | 6.02e-01 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 1 | 18 | 6.02e-01 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 1 | 10 | 6.02e-01 |
| GO:BP | GO:0060022 | hard palate development | 1 | 5 | 6.02e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 2 | 3750 | 6.02e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 3 | 33 | 6.02e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 1 | 1111 | 6.02e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 8 | 127 | 6.02e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 1 | 113 | 6.02e-01 |
| GO:BP | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration | 1 | 4 | 6.03e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 1 | 9 | 6.04e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 2 | 33 | 6.05e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 1 | 7 | 6.05e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 1 | 7 | 6.05e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 4 | 31 | 6.05e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 2 | 17 | 6.05e-01 |
| GO:BP | GO:1905749 | regulation of endosome to plasma membrane protein transport | 1 | 5 | 6.05e-01 |
| GO:BP | GO:0036466 | synaptic vesicle recycling via endosome | 1 | 4 | 6.05e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 5 | 58 | 6.05e-01 |
| GO:BP | GO:0021700 | developmental maturation | 2 | 211 | 6.06e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 1 | 90 | 6.06e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 2 | 46 | 6.06e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 1 | 8 | 6.06e-01 |
| GO:BP | GO:0032230 | positive regulation of synaptic transmission, GABAergic | 1 | 10 | 6.06e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 1 | 27 | 6.06e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 6.06e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 2 | 43 | 6.07e-01 |
| GO:BP | GO:0050896 | response to stimulus | 3 | 5770 | 6.07e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 12 | 114 | 6.07e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 12 | 114 | 6.07e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 6.07e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 6.07e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 1 | 55 | 6.07e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 1 | 14 | 6.08e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 2 | 16 | 6.08e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 2 | 15 | 6.08e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 4 | 45 | 6.08e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 15 | 6.08e-01 |
| GO:BP | GO:0001756 | somitogenesis | 1 | 48 | 6.08e-01 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 3 | 23 | 6.08e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 2 | 21 | 6.08e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 2 | 11 | 6.08e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 1 | 27 | 6.08e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 6.08e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 6 | 76 | 6.08e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 1 | 7 | 6.08e-01 |
| GO:BP | GO:0098664 | G protein-coupled serotonin receptor signaling pathway | 1 | 8 | 6.08e-01 |
| GO:BP | GO:0044557 | relaxation of smooth muscle | 1 | 9 | 6.08e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 5 | 48 | 6.08e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 6.08e-01 |
| GO:BP | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process | 1 | 6 | 6.08e-01 |
| GO:BP | GO:0048341 | paraxial mesoderm formation | 1 | 6 | 6.08e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 1 | 60 | 6.08e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 3 | 29 | 6.08e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 1 | 656 | 6.08e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 1 | 8 | 6.08e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 4 | 61 | 6.08e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 2 | 16 | 6.08e-01 |
| GO:BP | GO:0033278 | cell proliferation in midbrain | 1 | 4 | 6.08e-01 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 1 | 5 | 6.08e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 2 | 22 | 6.08e-01 |
| GO:BP | GO:0035865 | cellular response to potassium ion | 1 | 9 | 6.08e-01 |
| GO:BP | GO:0051964 | negative regulation of synapse assembly | 1 | 6 | 6.08e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 2 | 15 | 6.08e-01 |
| GO:BP | GO:0061525 | hindgut development | 1 | 5 | 6.08e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 2 | 20 | 6.08e-01 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 2 | 19 | 6.08e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 5 | 6.08e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 6 | 49 | 6.10e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 6.10e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 4 | 41 | 6.11e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 2 | 23 | 6.11e-01 |
| GO:BP | GO:0003334 | keratinocyte development | 1 | 8 | 6.11e-01 |
| GO:BP | GO:0070586 | cell-cell adhesion involved in gastrulation | 1 | 7 | 6.11e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 30 | 4045 | 6.11e-01 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 3 | 14 | 6.11e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 1 | 15 | 6.11e-01 |
| GO:BP | GO:0022615 | protein to membrane docking | 1 | 5 | 6.11e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 2 | 12 | 6.11e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 2 | 13 | 6.11e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 3 | 47 | 6.11e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 1 | 29 | 6.11e-01 |
| GO:BP | GO:0045541 | negative regulation of cholesterol biosynthetic process | 1 | 5 | 6.11e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 4 | 53 | 6.11e-01 |
| GO:BP | GO:0090206 | negative regulation of cholesterol metabolic process | 1 | 5 | 6.11e-01 |
| GO:BP | GO:0106119 | negative regulation of sterol biosynthetic process | 1 | 5 | 6.11e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 3 | 26 | 6.11e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 2 | 93 | 6.11e-01 |
| GO:BP | GO:0060049 | regulation of protein glycosylation | 1 | 10 | 6.11e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 3 | 65 | 6.11e-01 |
| GO:BP | GO:0033273 | response to vitamin | 4 | 61 | 6.11e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 6 | 59 | 6.11e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 4 | 35 | 6.11e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 9 | 123 | 6.11e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 3 | 26 | 6.11e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 2 | 17 | 6.11e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 3 | 19 | 6.11e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 4 | 53 | 6.12e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 6.12e-01 |
| GO:BP | GO:0097104 | postsynaptic membrane assembly | 1 | 8 | 6.12e-01 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 1 | 8 | 6.12e-01 |
| GO:BP | GO:0003356 | regulation of cilium beat frequency | 2 | 10 | 6.12e-01 |
| GO:BP | GO:0009151 | purine deoxyribonucleotide metabolic process | 2 | 12 | 6.13e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 2 | 1712 | 6.13e-01 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 3 | 31 | 6.13e-01 |
| GO:BP | GO:2000825 | positive regulation of androgen receptor activity | 1 | 5 | 6.13e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 4 | 59 | 6.13e-01 |
| GO:BP | GO:0003014 | renal system process | 5 | 78 | 6.13e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 3 | 26 | 6.13e-01 |
| GO:BP | GO:0046135 | pyrimidine nucleoside catabolic process | 1 | 8 | 6.13e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 1 | 5 | 6.13e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 10 | 6.13e-01 |
| GO:BP | GO:0032532 | regulation of microvillus length | 1 | 4 | 6.13e-01 |
| GO:BP | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | 1 | 5 | 6.13e-01 |
| GO:BP | GO:0035022 | positive regulation of Rac protein signal transduction | 1 | 5 | 6.13e-01 |
| GO:BP | GO:0060931 | sinoatrial node cell development | 1 | 4 | 6.13e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 6.13e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 2 | 25 | 6.13e-01 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 1 | 3 | 6.13e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1684 | 6.13e-01 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 1 | 11 | 6.13e-01 |
| GO:BP | GO:0042311 | vasodilation | 2 | 39 | 6.13e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 6.13e-01 |
| GO:BP | GO:1904557 | L-alanine transmembrane transport | 1 | 4 | 6.13e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 4 | 37 | 6.13e-01 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 4 | 35 | 6.13e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 1 | 5 | 6.13e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 4 | 22 | 6.13e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 3 | 22 | 6.13e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 3 | 29 | 6.13e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 1 | 12 | 6.13e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 3 | 34 | 6.13e-01 |
| GO:BP | GO:0032480 | negative regulation of type I interferon production | 1 | 23 | 6.13e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 1 | 10 | 6.13e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 3 | 25 | 6.13e-01 |
| GO:BP | GO:1903350 | response to dopamine | 3 | 48 | 6.14e-01 |
| GO:BP | GO:0044648 | histone H3-K4 dimethylation | 1 | 6 | 6.14e-01 |
| GO:BP | GO:2000370 | positive regulation of clathrin-dependent endocytosis | 1 | 5 | 6.14e-01 |
| GO:BP | GO:0003158 | endothelium development | 8 | 104 | 6.14e-01 |
| GO:BP | GO:0019673 | GDP-mannose metabolic process | 1 | 7 | 6.14e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 8 | 148 | 6.14e-01 |
| GO:BP | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 4 | 6.14e-01 |
| GO:BP | GO:0097305 | response to alcohol | 4 | 159 | 6.15e-01 |
| GO:BP | GO:0045002 | double-strand break repair via single-strand annealing | 1 | 7 | 6.15e-01 |
| GO:BP | GO:1905144 | response to acetylcholine | 2 | 22 | 6.16e-01 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 6.16e-01 |
| GO:BP | GO:0048311 | mitochondrion distribution | 1 | 14 | 6.16e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 2 | 17 | 6.16e-01 |
| GO:BP | GO:0007097 | nuclear migration | 1 | 23 | 6.16e-01 |
| GO:BP | GO:0060633 | negative regulation of transcription initiation by RNA polymerase II | 1 | 6 | 6.16e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 2 | 16 | 6.16e-01 |
| GO:BP | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway | 1 | 3 | 6.16e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 6.16e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 3 | 47 | 6.16e-01 |
| GO:BP | GO:0071895 | odontoblast differentiation | 1 | 5 | 6.17e-01 |
| GO:BP | GO:0009790 | embryo development | 16 | 875 | 6.17e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 3 | 22 | 6.17e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 1 | 5 | 6.17e-01 |
| GO:BP | GO:0043276 | anoikis | 1 | 32 | 6.17e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 1 | 11 | 6.17e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 5 | 39 | 6.17e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 2 | 37 | 6.17e-01 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 1 | 12 | 6.17e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 6.17e-01 |
| GO:BP | GO:1901509 | regulation of endothelial tube morphogenesis | 1 | 3 | 6.17e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 1 | 5 | 6.17e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 25 | 278 | 6.17e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 1 | 15 | 6.17e-01 |
| GO:BP | GO:0060442 | branching involved in prostate gland morphogenesis | 1 | 7 | 6.17e-01 |
| GO:BP | GO:0019441 | tryptophan catabolic process to kynurenine | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 4 | 51 | 6.18e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 2 | 17 | 6.18e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 3 | 28 | 6.18e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 3 | 41 | 6.18e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 1 | 25 | 6.18e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 18 | 6.18e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 1 | 28 | 6.18e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 1 | 31 | 6.18e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 2 | 25 | 6.18e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 2 | 25 | 6.18e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 1 | 23 | 6.18e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 25 | 307 | 6.18e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 1 | 442 | 6.18e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 6 | 44 | 6.18e-01 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 1 | 20 | 6.19e-01 |
| GO:BP | GO:0062009 | secondary palate development | 2 | 20 | 6.19e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 1 | 10 | 6.19e-01 |
| GO:BP | GO:0033239 | negative regulation of amine metabolic process | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0016236 | macroautophagy | 28 | 308 | 6.19e-01 |
| GO:BP | GO:0019240 | citrulline biosynthetic process | 1 | 4 | 6.19e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 1 | 13 | 6.19e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 3 | 38 | 6.19e-01 |
| GO:BP | GO:0006826 | iron ion transport | 5 | 40 | 6.19e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 5 | 50 | 6.19e-01 |
| GO:BP | GO:0070099 | regulation of chemokine-mediated signaling pathway | 1 | 8 | 6.19e-01 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 1 | 13 | 6.19e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 6.19e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 4 | 31 | 6.20e-01 |
| GO:BP | GO:0033007 | negative regulation of mast cell activation involved in immune response | 1 | 4 | 6.20e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 11 | 135 | 6.20e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 4 | 168 | 6.20e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 4 | 63 | 6.20e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 1 | 14 | 6.20e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 1 | 10 | 6.21e-01 |
| GO:BP | GO:0019346 | transsulfuration | 1 | 5 | 6.21e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 3 | 25 | 6.21e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 1 | 5 | 6.21e-01 |
| GO:BP | GO:0009092 | homoserine metabolic process | 1 | 5 | 6.21e-01 |
| GO:BP | GO:1900006 | positive regulation of dendrite development | 2 | 14 | 6.21e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 3 | 31 | 6.21e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 2 | 29 | 6.21e-01 |
| GO:BP | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process | 2 | 15 | 6.21e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 2 | 12 | 6.21e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 1 | 1219 | 6.21e-01 |
| GO:BP | GO:2000110 | negative regulation of macrophage apoptotic process | 1 | 3 | 6.21e-01 |
| GO:BP | GO:1904428 | negative regulation of tubulin deacetylation | 1 | 6 | 6.21e-01 |
| GO:BP | GO:0006678 | glucosylceramide metabolic process | 1 | 5 | 6.21e-01 |
| GO:BP | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion | 1 | 7 | 6.21e-01 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 1 | 7 | 6.21e-01 |
| GO:BP | GO:1903944 | negative regulation of hepatocyte apoptotic process | 1 | 6 | 6.21e-01 |
| GO:BP | GO:0021854 | hypothalamus development | 3 | 15 | 6.21e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 3 | 15 | 6.21e-01 |
| GO:BP | GO:0040009 | regulation of growth rate | 1 | 3 | 6.21e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 6.22e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 8 | 150 | 6.23e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 6.23e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 6.23e-01 |
| GO:BP | GO:0042748 | circadian sleep/wake cycle, non-REM sleep | 1 | 3 | 6.23e-01 |
| GO:BP | GO:0062100 | positive regulation of programmed necrotic cell death | 1 | 5 | 6.23e-01 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 1 | 23 | 6.23e-01 |
| GO:BP | GO:0045654 | positive regulation of megakaryocyte differentiation | 1 | 9 | 6.23e-01 |
| GO:BP | GO:0042552 | myelination | 11 | 111 | 6.24e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 5 | 84 | 6.25e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 1 | 26 | 6.25e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 6.25e-01 |
| GO:BP | GO:0060438 | trachea development | 1 | 13 | 6.26e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 6 | 62 | 6.26e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 1 | 9 | 6.26e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 8 | 284 | 6.26e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 2 | 16 | 6.26e-01 |
| GO:BP | GO:1904862 | inhibitory synapse assembly | 2 | 13 | 6.27e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 1 | 50 | 6.27e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 2 | 10 | 6.27e-01 |
| GO:BP | GO:0061590 | calcium activated phosphatidylcholine scrambling | 1 | 3 | 6.27e-01 |
| GO:BP | GO:0045794 | negative regulation of cell volume | 1 | 4 | 6.27e-01 |
| GO:BP | GO:0048742 | regulation of skeletal muscle fiber development | 1 | 8 | 6.27e-01 |
| GO:BP | GO:0030901 | midbrain development | 6 | 69 | 6.27e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 2 | 15 | 6.27e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 6.27e-01 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 1 | 17 | 6.28e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 6.28e-01 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 9 | 6.28e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 2 | 53 | 6.28e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 2 | 211 | 6.28e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 7 | 170 | 6.28e-01 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 1 | 22 | 6.28e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 1 | 9 | 6.28e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 1 | 22 | 6.28e-01 |
| GO:BP | GO:0008216 | spermidine metabolic process | 1 | 7 | 6.28e-01 |
| GO:BP | GO:0006682 | galactosylceramide biosynthetic process | 1 | 4 | 6.28e-01 |
| GO:BP | GO:0009445 | putrescine metabolic process | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 23 | 272 | 6.28e-01 |
| GO:BP | GO:0019375 | galactolipid biosynthetic process | 1 | 4 | 6.28e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 5 | 46 | 6.28e-01 |
| GO:BP | GO:0060264 | regulation of respiratory burst involved in inflammatory response | 1 | 3 | 6.28e-01 |
| GO:BP | GO:0032571 | response to vitamin K | 1 | 5 | 6.28e-01 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 1 | 21 | 6.28e-01 |
| GO:BP | GO:0032792 | negative regulation of CREB transcription factor activity | 1 | 4 | 6.28e-01 |
| GO:BP | GO:0052422 | modulation by host of symbiont catalytic activity | 1 | 3 | 6.28e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 6.28e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 6.28e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 5 | 46 | 6.28e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 6.28e-01 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 2 | 4 | 6.28e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 1 | 6 | 6.28e-01 |
| GO:BP | GO:0097012 | response to granulocyte macrophage colony-stimulating factor | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus | 1 | 6 | 6.29e-01 |
| GO:BP | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 5 | 6.29e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 5 | 47 | 6.29e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 8 | 80 | 6.29e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0015866 | ADP transport | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 17 | 222 | 6.29e-01 |
| GO:BP | GO:0010039 | response to iron ion | 2 | 24 | 6.29e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 7 | 67 | 6.29e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 1 | 3 | 6.29e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 1 | 8 | 6.29e-01 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 1 | 3 | 6.29e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 3 | 25 | 6.29e-01 |
| GO:BP | GO:0008219 | cell death | 8 | 1596 | 6.29e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 6.29e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 1 | 7 | 6.29e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 5 | 31 | 6.29e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 6.29e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 2 | 101 | 6.30e-01 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 1 | 13 | 6.30e-01 |
| GO:BP | GO:0006206 | pyrimidine nucleobase metabolic process | 1 | 13 | 6.30e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 6.30e-01 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 1 | 10 | 6.30e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 10 | 98 | 6.30e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 3 | 130 | 6.30e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 2 | 95 | 6.30e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 2 | 20 | 6.31e-01 |
| GO:BP | GO:0010818 | T cell chemotaxis | 2 | 12 | 6.31e-01 |
| GO:BP | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 3 | 6.31e-01 |
| GO:BP | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway | 1 | 5 | 6.31e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 2 | 12 | 6.32e-01 |
| GO:BP | GO:0016322 | neuron remodeling | 1 | 11 | 6.32e-01 |
| GO:BP | GO:0030035 | microspike assembly | 1 | 5 | 6.32e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 6.32e-01 |
| GO:BP | GO:0006771 | riboflavin metabolic process | 1 | 3 | 6.32e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 3 | 70 | 6.32e-01 |
| GO:BP | GO:1904748 | regulation of apoptotic process involved in development | 1 | 9 | 6.32e-01 |
| GO:BP | GO:1902337 | regulation of apoptotic process involved in morphogenesis | 1 | 9 | 6.32e-01 |
| GO:BP | GO:1904851 | positive regulation of establishment of protein localization to telomere | 1 | 10 | 6.32e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 2 | 14 | 6.32e-01 |
| GO:BP | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 6 | 6.32e-01 |
| GO:BP | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process | 1 | 8 | 6.32e-01 |
| GO:BP | GO:0008078 | mesodermal cell migration | 1 | 3 | 6.32e-01 |
| GO:BP | GO:0061744 | motor behavior | 2 | 17 | 6.32e-01 |
| GO:BP | GO:0021563 | glossopharyngeal nerve development | 1 | 3 | 6.33e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1 | 91 | 6.33e-01 |
| GO:BP | GO:0046348 | amino sugar catabolic process | 1 | 9 | 6.33e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 5 | 93 | 6.33e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 6.33e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 8 | 80 | 6.34e-01 |
| GO:BP | GO:0098754 | detoxification | 7 | 91 | 6.34e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 7 | 287 | 6.34e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 4 | 42 | 6.34e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 3 | 25 | 6.34e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 12 | 152 | 6.34e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 15 | 169 | 6.34e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 2 | 52 | 6.34e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 1 | 6 | 6.34e-01 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 6 | 84 | 6.34e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 4 | 47 | 6.34e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 6.35e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 2 | 189 | 6.35e-01 |
| GO:BP | GO:0070092 | regulation of glucagon secretion | 1 | 3 | 6.35e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 1 | 6 | 6.35e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 4 | 30 | 6.35e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 11 | 113 | 6.35e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 6.35e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 1 | 17 | 6.35e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 6.35e-01 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 1 | 15 | 6.35e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 1 | 8 | 6.35e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 11 | 113 | 6.35e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 3 | 9 | 6.35e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1904 | 6.35e-01 |
| GO:BP | GO:0001655 | urogenital system development | 4 | 49 | 6.35e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 1 | 21 | 6.35e-01 |
| GO:BP | GO:0070091 | glucagon secretion | 1 | 3 | 6.35e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 3 | 28 | 6.35e-01 |
| GO:BP | GO:0010832 | negative regulation of myotube differentiation | 1 | 10 | 6.35e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 1 | 7 | 6.35e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 4 | 23 | 6.35e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 4 | 57 | 6.35e-01 |
| GO:BP | GO:0016254 | preassembly of GPI anchor in ER membrane | 1 | 8 | 6.35e-01 |
| GO:BP | GO:0007210 | serotonin receptor signaling pathway | 1 | 8 | 6.35e-01 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 1 | 7 | 6.35e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 1 | 10 | 6.35e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 4 | 41 | 6.35e-01 |
| GO:BP | GO:0021548 | pons development | 1 | 7 | 6.35e-01 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 2 | 30 | 6.35e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 1 | 12 | 6.35e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 2 | 41 | 6.35e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 3 | 39 | 6.35e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 1 | 11 | 6.35e-01 |
| GO:BP | GO:0032259 | methylation | 1 | 313 | 6.35e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 6 | 90 | 6.35e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 3 | 17 | 6.36e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 6.36e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 6.36e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 3 | 27 | 6.37e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 1 | 336 | 6.37e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 1 | 87 | 6.37e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 6.38e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 6.38e-01 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 1 | 8 | 6.38e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 1 | 9 | 6.38e-01 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 2 | 26 | 6.38e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 2 | 48 | 6.39e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 1 | 25 | 6.39e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 1 | 32 | 6.39e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 8 | 113 | 6.40e-01 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 1 | 11 | 6.40e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 2 | 1335 | 6.41e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 4 | 27 | 6.41e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 1 | 43 | 6.42e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 1 | 30 | 6.42e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 2 | 33 | 6.42e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 2 | 16 | 6.42e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 2 | 16 | 6.42e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 1 | 12 | 6.42e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 2 | 27 | 6.42e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 1 | 9 | 6.42e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 5 | 57 | 6.42e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 5 | 50 | 6.42e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 1 | 6 | 6.42e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 3 | 28 | 6.43e-01 |
| GO:BP | GO:0097421 | liver regeneration | 3 | 23 | 6.43e-01 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 2 | 12 | 6.43e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 2 | 40 | 6.43e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 5 | 103 | 6.43e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 1 | 28 | 6.44e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 3 | 55 | 6.44e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 2 | 30 | 6.44e-01 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 1 | 24 | 6.44e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 2 | 23 | 6.45e-01 |
| GO:BP | GO:0046036 | CTP metabolic process | 1 | 14 | 6.45e-01 |
| GO:BP | GO:0007270 | neuron-neuron synaptic transmission | 1 | 5 | 6.45e-01 |
| GO:BP | GO:0061734 | parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 6 | 6.45e-01 |
| GO:BP | GO:0001974 | blood vessel remodeling | 2 | 32 | 6.45e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 1 | 13 | 6.45e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 4 | 47 | 6.45e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 6.45e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 3 | 34 | 6.45e-01 |
| GO:BP | GO:0032528 | microvillus organization | 2 | 19 | 6.45e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 3 | 34 | 6.46e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 6.46e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 8 | 74 | 6.46e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 2 | 4241 | 6.46e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 2 | 16 | 6.46e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 6 | 54 | 6.46e-01 |
| GO:BP | GO:0006882 | intracellular zinc ion homeostasis | 4 | 28 | 6.46e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 2 | 35 | 6.46e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 3 | 28 | 6.47e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 3 | 36 | 6.47e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 1 | 16 | 6.47e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 1 | 16 | 6.47e-01 |
| GO:BP | GO:0016553 | base conversion or substitution editing | 1 | 10 | 6.48e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 2 | 13 | 6.48e-01 |
| GO:BP | GO:2000143 | negative regulation of DNA-templated transcription initiation | 1 | 6 | 6.48e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 6.48e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 2 | 14 | 6.48e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 10 | 96 | 6.48e-01 |
| GO:BP | GO:0038203 | TORC2 signaling | 1 | 12 | 6.48e-01 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 1 | 6 | 6.48e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 4 | 136 | 6.48e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 4 | 60 | 6.48e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 1 | 23 | 6.48e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 4 | 64 | 6.48e-01 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 1 | 7 | 6.48e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 2 | 43 | 6.48e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 4 | 60 | 6.48e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 4 | 46 | 6.48e-01 |
| GO:BP | GO:1902498 | regulation of protein autoubiquitination | 1 | 5 | 6.49e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 6.49e-01 |
| GO:BP | GO:0090238 | positive regulation of arachidonic acid secretion | 1 | 2 | 6.49e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 7 | 69 | 6.49e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 2 | 3580 | 6.49e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 1 | 19 | 6.49e-01 |
| GO:BP | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 6 | 6.49e-01 |
| GO:BP | GO:0015811 | L-cystine transport | 1 | 3 | 6.49e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 2 | 4174 | 6.49e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 1 | 19 | 6.49e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 1 | 19 | 6.49e-01 |
| GO:BP | GO:0002837 | regulation of immune response to tumor cell | 2 | 10 | 6.49e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 2 | 37 | 6.49e-01 |
| GO:BP | GO:0007033 | vacuole organization | 9 | 197 | 6.49e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 2 | 15 | 6.49e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 1 | 30 | 6.50e-01 |
| GO:BP | GO:0060921 | sinoatrial node cell differentiation | 1 | 5 | 6.50e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 1 | 21 | 6.50e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 1 | 29 | 6.50e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 33 | 1602 | 6.50e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 1 | 29 | 6.50e-01 |
| GO:BP | GO:0043173 | nucleotide salvage | 2 | 15 | 6.51e-01 |
| GO:BP | GO:0051901 | positive regulation of mitochondrial depolarization | 1 | 6 | 6.51e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 4 | 89 | 6.51e-01 |
| GO:BP | GO:0031929 | TOR signaling | 10 | 115 | 6.51e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 12 | 105 | 6.51e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 2 | 4245 | 6.51e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 1 | 140 | 6.52e-01 |
| GO:BP | GO:0090025 | regulation of monocyte chemotaxis | 3 | 12 | 6.52e-01 |
| GO:BP | GO:0006848 | pyruvate transport | 1 | 5 | 6.53e-01 |
| GO:BP | GO:1901475 | pyruvate transmembrane transport | 1 | 5 | 6.53e-01 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 2 | 31 | 6.53e-01 |
| GO:BP | GO:0060068 | vagina development | 1 | 8 | 6.53e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 6.53e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 2 | 23 | 6.53e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 1 | 11 | 6.54e-01 |
| GO:BP | GO:0007568 | aging | 1 | 115 | 6.54e-01 |
| GO:BP | GO:0001771 | immunological synapse formation | 1 | 7 | 6.54e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 2 | 16 | 6.54e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 6.55e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 6.56e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 2 | 108 | 6.56e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 1 | 7 | 6.56e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 29 | 313 | 6.56e-01 |
| GO:BP | GO:1903943 | regulation of hepatocyte apoptotic process | 1 | 7 | 6.56e-01 |
| GO:BP | GO:0001889 | liver development | 1 | 110 | 6.56e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 2 | 17 | 6.56e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 2 | 36 | 6.56e-01 |
| GO:BP | GO:0006554 | lysine catabolic process | 1 | 6 | 6.56e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 4 | 64 | 6.56e-01 |
| GO:BP | GO:0030299 | intestinal cholesterol absorption | 1 | 8 | 6.56e-01 |
| GO:BP | GO:0097052 | L-kynurenine metabolic process | 1 | 4 | 6.56e-01 |
| GO:BP | GO:0061053 | somite development | 1 | 63 | 6.56e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 6 | 69 | 6.56e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 12 | 6.57e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 1 | 13 | 6.57e-01 |
| GO:BP | GO:0031640 | killing of cells of another organism | 2 | 10 | 6.57e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 2 | 21 | 6.57e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 2 | 161 | 6.57e-01 |
| GO:BP | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 1 | 6 | 6.58e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 1 | 6 | 6.58e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 13 | 6.58e-01 |
| GO:BP | GO:0002536 | respiratory burst involved in inflammatory response | 1 | 4 | 6.58e-01 |
| GO:BP | GO:0034058 | endosomal vesicle fusion | 1 | 10 | 6.58e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 1 | 7 | 6.58e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 6.58e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 1 | 13 | 6.58e-01 |
| GO:BP | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 1 | 5 | 6.58e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 6.58e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 1 | 16 | 6.58e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 5 | 39 | 6.59e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 2 | 42 | 6.59e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 4 | 24 | 6.59e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 1 | 130 | 6.59e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 1 | 10 | 6.59e-01 |
| GO:BP | GO:0042454 | ribonucleoside catabolic process | 1 | 12 | 6.59e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 1 | 9 | 6.60e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 1 | 9 | 6.60e-01 |
| GO:BP | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity | 1 | 3 | 6.60e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 6.60e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 6.60e-01 |
| GO:BP | GO:0110154 | RNA decapping | 1 | 17 | 6.60e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 6.60e-01 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 6.60e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 4 | 60 | 6.60e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 14 | 138 | 6.60e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 1 | 112 | 6.60e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 4 | 65 | 6.60e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 9 | 72 | 6.60e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 8 | 71 | 6.60e-01 |
| GO:BP | GO:0008090 | retrograde axonal transport | 1 | 19 | 6.60e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 5 | 36 | 6.60e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 1 | 9 | 6.61e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 2 | 299 | 6.61e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 6.61e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 2 | 33 | 6.61e-01 |
| GO:BP | GO:0019374 | galactolipid metabolic process | 1 | 5 | 6.61e-01 |
| GO:BP | GO:0048232 | male gamete generation | 10 | 343 | 6.61e-01 |
| GO:BP | GO:0006681 | galactosylceramide metabolic process | 1 | 5 | 6.61e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 6.61e-01 |
| GO:BP | GO:1990743 | protein sialylation | 1 | 5 | 6.61e-01 |
| GO:BP | GO:0071492 | cellular response to UV-A | 1 | 8 | 6.61e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 5 | 40 | 6.61e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 3 | 65 | 6.62e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 6.62e-01 |
| GO:BP | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | 1 | 5 | 6.62e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 2 | 14 | 6.62e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 3 | 27 | 6.63e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 6.63e-01 |
| GO:BP | GO:0090022 | regulation of neutrophil chemotaxis | 1 | 19 | 6.63e-01 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 2 | 16 | 6.63e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 1 | 81 | 6.63e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 6.63e-01 |
| GO:BP | GO:1903546 | protein localization to photoreceptor outer segment | 1 | 5 | 6.63e-01 |
| GO:BP | GO:0061438 | renal system vasculature morphogenesis | 1 | 7 | 6.63e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 1 | 7 | 6.63e-01 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 1 | 4 | 6.63e-01 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 1 | 8 | 6.63e-01 |
| GO:BP | GO:0061439 | kidney vasculature morphogenesis | 1 | 7 | 6.63e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 9 | 131 | 6.63e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 1 | 18 | 6.63e-01 |
| GO:BP | GO:0071867 | response to monoamine | 4 | 62 | 6.63e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 4 | 62 | 6.63e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 7 | 120 | 6.63e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 6.63e-01 |
| GO:BP | GO:0071772 | response to BMP | 9 | 131 | 6.63e-01 |
| GO:BP | GO:0032489 | regulation of Cdc42 protein signal transduction | 1 | 6 | 6.63e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 3 | 21 | 6.63e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 2 | 9 | 6.63e-01 |
| GO:BP | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | 1 | 13 | 6.63e-01 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 2 | 13 | 6.63e-01 |
| GO:BP | GO:0006858 | extracellular transport | 4 | 32 | 6.63e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 4 | 154 | 6.63e-01 |
| GO:BP | GO:0060613 | fat pad development | 1 | 6 | 6.63e-01 |
| GO:BP | GO:0060457 | negative regulation of digestive system process | 1 | 7 | 6.63e-01 |
| GO:BP | GO:0048675 | axon extension | 9 | 110 | 6.63e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 3 | 66 | 6.63e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 8 | 414 | 6.64e-01 |
| GO:BP | GO:0034091 | regulation of maintenance of sister chromatid cohesion | 1 | 8 | 6.64e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 1 | 6 | 6.64e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 6.64e-01 |
| GO:BP | GO:0034182 | regulation of maintenance of mitotic sister chromatid cohesion | 1 | 8 | 6.64e-01 |
| GO:BP | GO:0045653 | negative regulation of megakaryocyte differentiation | 1 | 17 | 6.64e-01 |
| GO:BP | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell | 1 | 3 | 6.64e-01 |
| GO:BP | GO:0061588 | calcium activated phospholipid scrambling | 1 | 4 | 6.64e-01 |
| GO:BP | GO:0061548 | ganglion development | 2 | 12 | 6.64e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 2 | 17 | 6.64e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 19 | 197 | 6.65e-01 |
| GO:BP | GO:0034372 | very-low-density lipoprotein particle remodeling | 1 | 4 | 6.65e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 3 | 92 | 6.65e-01 |
| GO:BP | GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 1 | 4 | 6.65e-01 |
| GO:BP | GO:0106380 | purine ribonucleotide salvage | 1 | 7 | 6.65e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 3 | 92 | 6.65e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 1 | 145 | 6.65e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 3 | 20 | 6.66e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 2 | 305 | 6.66e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 5 | 46 | 6.66e-01 |
| GO:BP | GO:2000489 | regulation of hepatic stellate cell activation | 1 | 6 | 6.66e-01 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 1 | 9 | 6.67e-01 |
| GO:BP | GO:0002834 | regulation of response to tumor cell | 2 | 11 | 6.67e-01 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 2 | 16 | 6.67e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 1 | 18 | 6.67e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 2 | 14 | 6.67e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 2 | 14 | 6.67e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 2 | 13 | 6.67e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 5 | 78 | 6.67e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 2 | 20 | 6.68e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 2 | 302 | 6.68e-01 |
| GO:BP | GO:0048643 | positive regulation of skeletal muscle tissue development | 2 | 14 | 6.68e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 10 | 6.68e-01 |
| GO:BP | GO:0048589 | developmental growth | 28 | 522 | 6.68e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 2 | 111 | 6.68e-01 |
| GO:BP | GO:0007625 | grooming behavior | 1 | 10 | 6.68e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 7 | 71 | 6.68e-01 |
| GO:BP | GO:0035627 | ceramide transport | 1 | 15 | 6.68e-01 |
| GO:BP | GO:0003360 | brainstem development | 1 | 7 | 6.68e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 15 | 184 | 6.68e-01 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 1 | 18 | 6.68e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 8 | 160 | 6.68e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 3 | 53 | 6.68e-01 |
| GO:BP | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | 1 | 8 | 6.69e-01 |
| GO:BP | GO:1902915 | negative regulation of protein polyubiquitination | 1 | 9 | 6.69e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 1 | 9 | 6.69e-01 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 1 | 16 | 6.69e-01 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 1 | 12 | 6.69e-01 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 3 | 27 | 6.69e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 5 | 71 | 6.69e-01 |
| GO:BP | GO:0035176 | social behavior | 1 | 34 | 6.70e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 3 | 38 | 6.70e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 1 | 12 | 6.70e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 6 | 65 | 6.70e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 8 | 132 | 6.70e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 6.70e-01 |
| GO:BP | GO:0101024 | mitotic nuclear membrane organization | 1 | 9 | 6.70e-01 |
| GO:BP | GO:0007084 | mitotic nuclear membrane reassembly | 1 | 9 | 6.70e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 6.70e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 3 | 53 | 6.70e-01 |
| GO:BP | GO:0031509 | subtelomeric heterochromatin formation | 1 | 7 | 6.70e-01 |
| GO:BP | GO:0010738 | regulation of protein kinase A signaling | 2 | 13 | 6.71e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 3 | 84 | 6.71e-01 |
| GO:BP | GO:0042138 | meiotic DNA double-strand break formation | 1 | 2 | 6.71e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 4 | 157 | 6.71e-01 |
| GO:BP | GO:0042726 | flavin-containing compound metabolic process | 1 | 4 | 6.71e-01 |
| GO:BP | GO:0007030 | Golgi organization | 1 | 137 | 6.71e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 1 | 12 | 6.71e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 1 | 12 | 6.71e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 3 | 24 | 6.71e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 3 | 149 | 6.71e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 4 | 93 | 6.72e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 1 | 9 | 6.72e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 2 | 30 | 6.72e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 4 | 92 | 6.72e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 5 | 92 | 6.72e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 1 | 9 | 6.72e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 4 | 81 | 6.72e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 2 | 256 | 6.72e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 6.72e-01 |
| GO:BP | GO:0003129 | heart induction | 1 | 9 | 6.72e-01 |
| GO:BP | GO:0021889 | olfactory bulb interneuron differentiation | 1 | 7 | 6.72e-01 |
| GO:BP | GO:0030879 | mammary gland development | 1 | 106 | 6.72e-01 |
| GO:BP | GO:0007494 | midgut development | 1 | 7 | 6.72e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 1 | 5 | 6.72e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 12 | 135 | 6.72e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 1 | 4 | 6.72e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 2 | 18 | 6.73e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 5 | 42 | 6.73e-01 |
| GO:BP | GO:0097692 | histone H3-K4 monomethylation | 1 | 8 | 6.73e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 3 | 241 | 6.73e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 2 | 258 | 6.74e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 3 | 28 | 6.74e-01 |
| GO:BP | GO:0035864 | response to potassium ion | 1 | 12 | 6.74e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 10 | 147 | 6.74e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 15 | 2285 | 6.74e-01 |
| GO:BP | GO:1902031 | regulation of NADP metabolic process | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0045898 | regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 7 | 6.74e-01 |
| GO:BP | GO:0002840 | regulation of T cell mediated immune response to tumor cell | 1 | 5 | 6.74e-01 |
| GO:BP | GO:0030154 | cell differentiation | 3 | 2958 | 6.74e-01 |
| GO:BP | GO:0071907 | determination of digestive tract left/right asymmetry | 1 | 5 | 6.74e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2116 | 6.74e-01 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 1 | 33 | 6.74e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 8 | 97 | 6.74e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 3 | 25 | 6.74e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 2 | 36 | 6.74e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 4 | 39 | 6.74e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 3 | 24 | 6.74e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 1 | 19 | 6.74e-01 |
| GO:BP | GO:0043650 | dicarboxylic acid biosynthetic process | 1 | 9 | 6.74e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 2 | 4417 | 6.74e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 3 | 85 | 6.74e-01 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 1 | 19 | 6.74e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 1 | 84 | 6.74e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 1 | 126 | 6.74e-01 |
| GO:BP | GO:0033033 | negative regulation of myeloid cell apoptotic process | 2 | 10 | 6.74e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 20 | 267 | 6.74e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 6.75e-01 |
| GO:BP | GO:0006885 | regulation of pH | 3 | 74 | 6.75e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 22 | 256 | 6.75e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 4 | 135 | 6.75e-01 |
| GO:BP | GO:0007340 | acrosome reaction | 1 | 20 | 6.75e-01 |
| GO:BP | GO:1903054 | negative regulation of extracellular matrix organization | 1 | 6 | 6.75e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 2 | 17 | 6.75e-01 |
| GO:BP | GO:0032700 | negative regulation of interleukin-17 production | 1 | 9 | 6.75e-01 |
| GO:BP | GO:0032049 | cardiolipin biosynthetic process | 1 | 7 | 6.75e-01 |
| GO:BP | GO:0090237 | regulation of arachidonic acid secretion | 1 | 2 | 6.75e-01 |
| GO:BP | GO:0072109 | glomerular mesangium development | 1 | 13 | 6.75e-01 |
| GO:BP | GO:0046469 | platelet activating factor metabolic process | 1 | 5 | 6.75e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 2 | 18 | 6.75e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 1 | 34 | 6.75e-01 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 2 | 17 | 6.75e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 7 | 109 | 6.75e-01 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 2 | 17 | 6.75e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 3 | 20 | 6.75e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 681 | 6.76e-01 |
| GO:BP | GO:0150172 | regulation of phosphatidylcholine metabolic process | 1 | 10 | 6.76e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 3 | 22 | 6.76e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 3 | 2984 | 6.76e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 1 | 9 | 6.76e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 6.76e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 12 | 150 | 6.76e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 63 | 2880 | 6.76e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 6 | 129 | 6.76e-01 |
| GO:BP | GO:0009056 | catabolic process | 2 | 2088 | 6.76e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 1 | 5 | 6.76e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 2 | 18 | 6.76e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 4 | 67 | 6.76e-01 |
| GO:BP | GO:0001845 | phagolysosome assembly | 1 | 14 | 6.76e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 2 | 18 | 6.76e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 6.76e-01 |
| GO:BP | GO:0045046 | protein import into peroxisome membrane | 1 | 6 | 6.76e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 9 | 1256 | 6.76e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 21 | 399 | 6.76e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 1 | 127 | 6.76e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 4 | 36 | 6.77e-01 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 3 | 20 | 6.77e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 9 | 1255 | 6.77e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 6.77e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 5 | 136 | 6.77e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 2 | 4539 | 6.78e-01 |
| GO:BP | GO:1901463 | regulation of tetrapyrrole biosynthetic process | 1 | 7 | 6.78e-01 |
| GO:BP | GO:0040008 | regulation of growth | 8 | 501 | 6.78e-01 |
| GO:BP | GO:0070453 | regulation of heme biosynthetic process | 1 | 7 | 6.78e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 4 | 40 | 6.78e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 1 | 12 | 6.79e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 3 | 151 | 6.79e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 3 | 36 | 6.79e-01 |
| GO:BP | GO:0019076 | viral release from host cell | 1 | 28 | 6.79e-01 |
| GO:BP | GO:0035891 | exit from host cell | 1 | 28 | 6.79e-01 |
| GO:BP | GO:0060267 | positive regulation of respiratory burst | 1 | 3 | 6.79e-01 |
| GO:BP | GO:0034351 | negative regulation of glial cell apoptotic process | 1 | 8 | 6.79e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 1 | 7 | 6.79e-01 |
| GO:BP | GO:0097354 | prenylation | 1 | 10 | 6.79e-01 |
| GO:BP | GO:0018342 | protein prenylation | 1 | 10 | 6.79e-01 |
| GO:BP | GO:0035701 | hematopoietic stem cell migration | 1 | 6 | 6.79e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 6.79e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 7 | 198 | 6.80e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 2 | 276 | 6.80e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 6.80e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 6.80e-01 |
| GO:BP | GO:0061762 | CAMKK-AMPK signaling cascade | 1 | 5 | 6.80e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 6.80e-01 |
| GO:BP | GO:0006863 | purine nucleobase transport | 1 | 7 | 6.80e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 6.80e-01 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 1 | 7 | 6.80e-01 |
| GO:BP | GO:0016198 | axon choice point recognition | 1 | 5 | 6.80e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 6.80e-01 |
| GO:BP | GO:0031204 | post-translational protein targeting to membrane, translocation | 1 | 7 | 6.80e-01 |
| GO:BP | GO:0006553 | lysine metabolic process | 1 | 7 | 6.81e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 2 | 13 | 6.81e-01 |
| GO:BP | GO:0071379 | cellular response to prostaglandin stimulus | 2 | 16 | 6.81e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 2 | 43 | 6.81e-01 |
| GO:BP | GO:0002315 | marginal zone B cell differentiation | 1 | 6 | 6.81e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 5 | 48 | 6.81e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 1 | 6 | 6.81e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 2 | 39 | 6.81e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 1 | 14 | 6.81e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 3 | 40 | 6.81e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 6 | 144 | 6.81e-01 |
| GO:BP | GO:0010288 | response to lead ion | 1 | 18 | 6.81e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 6.81e-01 |
| GO:BP | GO:0007032 | endosome organization | 8 | 92 | 6.81e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 6.81e-01 |
| GO:BP | GO:0035873 | lactate transmembrane transport | 1 | 5 | 6.81e-01 |
| GO:BP | GO:1904415 | regulation of xenophagy | 1 | 7 | 6.81e-01 |
| GO:BP | GO:1904417 | positive regulation of xenophagy | 1 | 7 | 6.81e-01 |
| GO:BP | GO:0015727 | lactate transport | 1 | 5 | 6.81e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 3 | 32 | 6.81e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 1 | 598 | 6.81e-01 |
| GO:BP | GO:0030011 | maintenance of cell polarity | 1 | 16 | 6.82e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 1 | 20 | 6.82e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 1 | 13 | 6.82e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 3 | 53 | 6.83e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 2 | 4590 | 6.83e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 1 | 28 | 6.83e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 2 | 36 | 6.83e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 3 | 60 | 6.83e-01 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 4 | 20 | 6.83e-01 |
| GO:BP | GO:0044241 | lipid digestion | 1 | 10 | 6.83e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 3 | 29 | 6.83e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 6.83e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 1 | 7 | 6.83e-01 |
| GO:BP | GO:0009648 | photoperiodism | 2 | 28 | 6.83e-01 |
| GO:BP | GO:1902065 | response to L-glutamate | 1 | 11 | 6.83e-01 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 1 | 48 | 6.83e-01 |
| GO:BP | GO:0045008 | depyrimidination | 1 | 8 | 6.83e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 1 | 784 | 6.83e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 5 | 52 | 6.83e-01 |
| GO:BP | GO:0021762 | substantia nigra development | 3 | 41 | 6.83e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 6.83e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 5 | 52 | 6.83e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 17 | 160 | 6.83e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 12 | 148 | 6.83e-01 |
| GO:BP | GO:0019082 | viral protein processing | 3 | 31 | 6.84e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 1 | 8 | 6.85e-01 |
| GO:BP | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 12 | 6.85e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 1 | 8 | 6.85e-01 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 1 | 13 | 6.85e-01 |
| GO:BP | GO:0051591 | response to cAMP | 4 | 66 | 6.86e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 2 | 36 | 6.86e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 10 | 100 | 6.86e-01 |
| GO:BP | GO:0034354 | ‘de novo’ NAD biosynthetic process from tryptophan | 1 | 1 | 6.86e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 5 | 121 | 6.86e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 2 | 14 | 6.86e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 15 | 182 | 6.86e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 1 | 23 | 6.86e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 2 | 47 | 6.86e-01 |
| GO:BP | GO:0006244 | pyrimidine nucleotide catabolic process | 2 | 19 | 6.86e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 1 | 7 | 6.86e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 6 | 6.86e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 1 | 7 | 6.86e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 1 | 5 | 6.86e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 2 | 16 | 6.86e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 1 | 9 | 6.86e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 3 | 25 | 6.87e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 3 | 37 | 6.87e-01 |
| GO:BP | GO:0043174 | nucleoside salvage | 1 | 9 | 6.87e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 4 | 84 | 6.87e-01 |
| GO:BP | GO:0007165 | signal transduction | 2 | 3818 | 6.87e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 2 | 42 | 6.87e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 2 | 22 | 6.87e-01 |
| GO:BP | GO:0006783 | heme biosynthetic process | 3 | 30 | 6.87e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 6.87e-01 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 1 | 5 | 6.87e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 2 | 135 | 6.87e-01 |
| GO:BP | GO:1905809 | negative regulation of synapse organization | 1 | 8 | 6.87e-01 |
| GO:BP | GO:0033004 | negative regulation of mast cell activation | 1 | 4 | 6.87e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 1 | 10 | 6.87e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 2 | 97 | 6.87e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 4 | 50 | 6.87e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 20 | 249 | 6.87e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 4 | 50 | 6.87e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 740 | 6.88e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 2 | 18 | 6.88e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 2 | 57 | 6.89e-01 |
| GO:BP | GO:0007286 | spermatid development | 2 | 105 | 6.89e-01 |
| GO:BP | GO:0002858 | regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 6.89e-01 |
| GO:BP | GO:0002729 | positive regulation of natural killer cell cytokine production | 1 | 4 | 6.89e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 1 | 28 | 6.89e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 2 | 65 | 6.90e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 2 | 31 | 6.91e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 9 | 179 | 6.92e-01 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 2 | 13 | 6.92e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 1 | 137 | 6.93e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 1 | 19 | 6.93e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 3 | 30 | 6.93e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 2 | 15 | 6.94e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 5 | 56 | 6.94e-01 |
| GO:BP | GO:0051169 | nuclear transport | 5 | 295 | 6.94e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 5 | 295 | 6.94e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 3 | 34 | 6.94e-01 |
| GO:BP | GO:0098856 | intestinal lipid absorption | 1 | 10 | 6.95e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 1 | 23 | 6.95e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 2 | 17 | 6.95e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 8 | 70 | 6.95e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 2 | 31 | 6.95e-01 |
| GO:BP | GO:0030889 | negative regulation of B cell proliferation | 1 | 9 | 6.95e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 2 | 340 | 6.95e-01 |
| GO:BP | GO:0032402 | melanosome transport | 2 | 19 | 6.95e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 1 | 13 | 6.95e-01 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 6.95e-01 |
| GO:BP | GO:0009143 | nucleoside triphosphate catabolic process | 1 | 10 | 6.95e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 2 | 17 | 6.95e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 7 | 74 | 6.95e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 6.95e-01 |
| GO:BP | GO:0045578 | negative regulation of B cell differentiation | 1 | 6 | 6.95e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 15 | 6.95e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 2 | 17 | 6.95e-01 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 1 | 12 | 6.95e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 6.95e-01 |
| GO:BP | GO:1903764 | regulation of potassium ion export across plasma membrane | 1 | 5 | 6.95e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 3 | 357 | 6.95e-01 |
| GO:BP | GO:2000535 | regulation of entry of bacterium into host cell | 1 | 6 | 6.96e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 3 | 67 | 6.96e-01 |
| GO:BP | GO:0032025 | response to cobalt ion | 1 | 10 | 6.96e-01 |
| GO:BP | GO:0071073 | positive regulation of phospholipid biosynthetic process | 1 | 7 | 6.97e-01 |
| GO:BP | GO:0034379 | very-low-density lipoprotein particle assembly | 1 | 8 | 6.97e-01 |
| GO:BP | GO:1904181 | positive regulation of membrane depolarization | 1 | 8 | 6.97e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 9 | 241 | 6.97e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 2 | 10 | 6.97e-01 |
| GO:BP | GO:0018243 | protein O-linked glycosylation via threonine | 1 | 7 | 6.97e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 1 | 44 | 6.97e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 5 | 32 | 6.98e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 10 | 164 | 6.98e-01 |
| GO:BP | GO:0046717 | acid secretion | 1 | 25 | 6.98e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 2 | 16 | 6.98e-01 |
| GO:BP | GO:0021860 | pyramidal neuron development | 1 | 8 | 6.98e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 5 | 52 | 6.99e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 10 | 6.99e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 9 | 165 | 6.99e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 4 | 67 | 6.99e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 9 | 82 | 6.99e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 2 | 14 | 6.99e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 2 | 107 | 6.99e-01 |
| GO:BP | GO:0099532 | synaptic vesicle endosomal processing | 1 | 7 | 6.99e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 2 | 24 | 6.99e-01 |
| GO:BP | GO:0002424 | T cell mediated immune response to tumor cell | 1 | 6 | 6.99e-01 |
| GO:BP | GO:1900118 | negative regulation of execution phase of apoptosis | 1 | 6 | 7.00e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 2 | 221 | 7.00e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 7.00e-01 |
| GO:BP | GO:0070106 | interleukin-27-mediated signaling pathway | 1 | 4 | 7.00e-01 |
| GO:BP | GO:0048468 | cell development | 2 | 1963 | 7.00e-01 |
| GO:BP | GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 7.00e-01 |
| GO:BP | GO:0072257 | metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 7.00e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 2 | 33 | 7.00e-01 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 1 | 102 | 7.00e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 19 | 237 | 7.01e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 1 | 843 | 7.01e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 17 | 377 | 7.01e-01 |
| GO:BP | GO:1901339 | regulation of store-operated calcium channel activity | 1 | 8 | 7.01e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 3 | 40 | 7.01e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 6 | 108 | 7.01e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 11 | 485 | 7.02e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 7 | 108 | 7.02e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 7.02e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 2 | 15 | 7.02e-01 |
| GO:BP | GO:0006498 | N-terminal protein lipidation | 1 | 7 | 7.02e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 7.02e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 7.02e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 1 | 15 | 7.03e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 4 | 85 | 7.03e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 1 | 8 | 7.03e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 7.03e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 6 | 61 | 7.03e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 7.03e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 3 | 34 | 7.03e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 1 | 14 | 7.03e-01 |
| GO:BP | GO:0002138 | retinoic acid biosynthetic process | 1 | 8 | 7.03e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 11 | 126 | 7.03e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 2 | 111 | 7.03e-01 |
| GO:BP | GO:0006627 | protein processing involved in protein targeting to mitochondrion | 1 | 6 | 7.03e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 3 | 27 | 7.03e-01 |
| GO:BP | GO:2000510 | positive regulation of dendritic cell chemotaxis | 1 | 3 | 7.03e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 7.03e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 6 | 125 | 7.03e-01 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 1 | 14 | 7.03e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 1 | 7 | 7.03e-01 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 1 | 5 | 7.03e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 1 | 22 | 7.03e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 16 | 353 | 7.04e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 2 | 18 | 7.04e-01 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 237 | 7.04e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 20 | 393 | 7.04e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 8 | 100 | 7.04e-01 |
| GO:BP | GO:0015851 | nucleobase transport | 1 | 7 | 7.04e-01 |
| GO:BP | GO:0009164 | nucleoside catabolic process | 1 | 15 | 7.04e-01 |
| GO:BP | GO:0046035 | CMP metabolic process | 1 | 6 | 7.04e-01 |
| GO:BP | GO:0097531 | mast cell migration | 1 | 12 | 7.04e-01 |
| GO:BP | GO:0001967 | suckling behavior | 1 | 10 | 7.04e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 7.04e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 4 | 33 | 7.04e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1453 | 7.05e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 2 | 273 | 7.05e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 2 | 13 | 7.05e-01 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 2 | 15 | 7.05e-01 |
| GO:BP | GO:0035282 | segmentation | 1 | 75 | 7.05e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 2 | 13 | 7.05e-01 |
| GO:BP | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 1 | 14 | 7.05e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 4 | 63 | 7.06e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 1 | 33 | 7.06e-01 |
| GO:BP | GO:0006656 | phosphatidylcholine biosynthetic process | 2 | 25 | 7.06e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 1 | 41 | 7.06e-01 |
| GO:BP | GO:0032288 | myelin assembly | 2 | 19 | 7.06e-01 |
| GO:BP | GO:0072529 | pyrimidine-containing compound catabolic process | 3 | 29 | 7.06e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 7.06e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 3 | 29 | 7.06e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 7.06e-01 |
| GO:BP | GO:0006810 | transport | 2 | 3298 | 7.07e-01 |
| GO:BP | GO:0046068 | cGMP metabolic process | 1 | 11 | 7.07e-01 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 1 | 8 | 7.08e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 1 | 5 | 7.08e-01 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 1 | 8 | 7.08e-01 |
| GO:BP | GO:0030539 | male genitalia development | 2 | 13 | 7.08e-01 |
| GO:BP | GO:0016477 | cell migration | 1 | 1081 | 7.08e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 1 | 60 | 7.08e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 7.09e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 2 | 24 | 7.09e-01 |
| GO:BP | GO:0015732 | prostaglandin transport | 1 | 10 | 7.09e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 2 | 26 | 7.09e-01 |
| GO:BP | GO:0002553 | histamine secretion by mast cell | 1 | 8 | 7.09e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 1 | 7 | 7.09e-01 |
| GO:BP | GO:0002441 | histamine secretion involved in inflammatory response | 1 | 8 | 7.09e-01 |
| GO:BP | GO:0002349 | histamine production involved in inflammatory response | 1 | 8 | 7.09e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 12 | 161 | 7.10e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 23 | 301 | 7.10e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 7.10e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 1 | 9 | 7.10e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 1 | 26 | 7.10e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 2 | 58 | 7.10e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 2 | 19 | 7.11e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 2 | 13 | 7.11e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 2 | 20 | 7.11e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 2 | 16 | 7.11e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 7.11e-01 |
| GO:BP | GO:0070141 | response to UV-A | 1 | 11 | 7.11e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 5 | 90 | 7.12e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 3 | 70 | 7.12e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 1 | 12 | 7.13e-01 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 1 | 26 | 7.13e-01 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 1 | 26 | 7.13e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 2 | 22 | 7.13e-01 |
| GO:BP | GO:0002689 | negative regulation of leukocyte chemotaxis | 1 | 10 | 7.13e-01 |
| GO:BP | GO:0010842 | retina layer formation | 1 | 16 | 7.13e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 1 | 21 | 7.13e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 5 | 57 | 7.13e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1201 | 7.14e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 1 | 166 | 7.14e-01 |
| GO:BP | GO:0099612 | protein localization to axon | 1 | 6 | 7.14e-01 |
| GO:BP | GO:0002418 | immune response to tumor cell | 2 | 12 | 7.15e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 23 | 441 | 7.15e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 1 | 21 | 7.15e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 8 | 121 | 7.15e-01 |
| GO:BP | GO:0006825 | copper ion transport | 1 | 13 | 7.15e-01 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 1 | 8 | 7.15e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 1 | 11 | 7.15e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 4 | 50 | 7.15e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 3 | 83 | 7.15e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 3 | 123 | 7.15e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 1 | 45 | 7.16e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 2 | 21 | 7.16e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 1 | 46 | 7.16e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 7.16e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 1 | 25 | 7.17e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 2 | 21 | 7.17e-01 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 1 | 9 | 7.17e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 2 | 19 | 7.17e-01 |
| GO:BP | GO:2001140 | positive regulation of phospholipid transport | 1 | 11 | 7.17e-01 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 1 | 8 | 7.19e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 1 | 14 | 7.19e-01 |
| GO:BP | GO:0007379 | segment specification | 1 | 14 | 7.19e-01 |
| GO:BP | GO:0060324 | face development | 4 | 45 | 7.19e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 1 | 7 | 7.19e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 4 | 28 | 7.19e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 2 | 52 | 7.19e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 7.19e-01 |
| GO:BP | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 7.19e-01 |
| GO:BP | GO:0002727 | regulation of natural killer cell cytokine production | 1 | 4 | 7.19e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 2 | 19 | 7.19e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 2 | 21 | 7.19e-01 |
| GO:BP | GO:0002370 | natural killer cell cytokine production | 1 | 4 | 7.19e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 2 | 14 | 7.19e-01 |
| GO:BP | GO:0034627 | ‘de novo’ NAD biosynthetic process | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0051001 | negative regulation of nitric-oxide synthase activity | 1 | 6 | 7.19e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 2 | 17 | 7.19e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 1 | 29 | 7.19e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 5 | 57 | 7.19e-01 |
| GO:BP | GO:0002855 | regulation of natural killer cell mediated immune response to tumor cell | 1 | 4 | 7.19e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 4 | 30 | 7.19e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 2 | 11 | 7.19e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 7.19e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 2 | 21 | 7.19e-01 |
| GO:BP | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 1 | 16 | 7.19e-01 |
| GO:BP | GO:0006651 | diacylglycerol biosynthetic process | 1 | 7 | 7.19e-01 |
| GO:BP | GO:0032693 | negative regulation of interleukin-10 production | 1 | 7 | 7.19e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 15 | 7.19e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1233 | 7.19e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 1 | 13 | 7.19e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 11 | 335 | 7.20e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 8 | 71 | 7.20e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 5 | 53 | 7.20e-01 |
| GO:BP | GO:0035733 | hepatic stellate cell activation | 1 | 8 | 7.20e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2 | 4955 | 7.20e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 35 | 7.20e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 7.20e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 1 | 35 | 7.20e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 1 | 9 | 7.21e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 2 | 49 | 7.21e-01 |
| GO:BP | GO:0045141 | meiotic telomere clustering | 1 | 5 | 7.21e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 3 | 28 | 7.21e-01 |
| GO:BP | GO:1901569 | fatty acid derivative catabolic process | 1 | 10 | 7.21e-01 |
| GO:BP | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade | 1 | 16 | 7.22e-01 |
| GO:BP | GO:0106058 | positive regulation of calcineurin-mediated signaling | 1 | 16 | 7.22e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 1 | 8 | 7.22e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 1 | 14 | 7.22e-01 |
| GO:BP | GO:0006171 | cAMP biosynthetic process | 1 | 8 | 7.23e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 1 | 14 | 7.23e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 1 | 14 | 7.23e-01 |
| GO:BP | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway | 1 | 7 | 7.23e-01 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 1 | 18 | 7.23e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 7.23e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 1 | 16 | 7.23e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 2 | 24 | 7.23e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 5 | 45 | 7.23e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 3 | 21 | 7.23e-01 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 1 | 24 | 7.23e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 1 | 9 | 7.23e-01 |
| GO:BP | GO:0051350 | negative regulation of lyase activity | 2 | 15 | 7.23e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 1 | 41 | 7.23e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 7.23e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 2 | 114 | 7.23e-01 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 1 | 10 | 7.23e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 3 | 98 | 7.23e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1 | 1914 | 7.23e-01 |
| GO:BP | GO:2000425 | regulation of apoptotic cell clearance | 1 | 7 | 7.23e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 1 | 116 | 7.23e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 7.23e-01 |
| GO:BP | GO:0051709 | regulation of killing of cells of another organism | 1 | 3 | 7.24e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 5 | 58 | 7.24e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 10 | 7.24e-01 |
| GO:BP | GO:0048853 | forebrain morphogenesis | 1 | 7 | 7.24e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 11 | 121 | 7.24e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 37 | 513 | 7.24e-01 |
| GO:BP | GO:0006914 | autophagy | 37 | 513 | 7.24e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 1 | 160 | 7.24e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 1 | 12 | 7.24e-01 |
| GO:BP | GO:0023052 | signaling | 2 | 4141 | 7.24e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 3 | 34 | 7.24e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 2 | 68 | 7.24e-01 |
| GO:BP | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 1 | 6 | 7.24e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 16 | 7.24e-01 |
| GO:BP | GO:0072017 | distal tubule development | 1 | 6 | 7.24e-01 |
| GO:BP | GO:0034350 | regulation of glial cell apoptotic process | 1 | 10 | 7.24e-01 |
| GO:BP | GO:0006655 | phosphatidylglycerol biosynthetic process | 1 | 9 | 7.24e-01 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 1 | 7 | 7.24e-01 |
| GO:BP | GO:0016102 | diterpenoid biosynthetic process | 1 | 8 | 7.24e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 62 | 7.24e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 7.24e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 2 | 40 | 7.24e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 6 | 139 | 7.25e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 3 | 30 | 7.25e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 2 | 20 | 7.26e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 1 | 9 | 7.26e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 7.26e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 2 | 20 | 7.26e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 1 | 26 | 7.27e-01 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 1 | 8 | 7.27e-01 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 2 | 18 | 7.27e-01 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 1 | 8 | 7.27e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 5 | 33 | 7.27e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 7.28e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 7.28e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 3 | 62 | 7.28e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 7 | 112 | 7.28e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 3 | 45 | 7.28e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 5 | 59 | 7.28e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 3 | 28 | 7.28e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 2 | 19 | 7.28e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 2 | 18 | 7.28e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 2 | 2102 | 7.28e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 1 | 9 | 7.28e-01 |
| GO:BP | GO:0046940 | nucleoside monophosphate phosphorylation | 1 | 9 | 7.28e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 9 | 96 | 7.28e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 1 | 24 | 7.28e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 9 | 75 | 7.28e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 7.28e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 7.28e-01 |
| GO:BP | GO:0051235 | maintenance of location | 14 | 260 | 7.28e-01 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 2 | 21 | 7.28e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 1 | 8 | 7.28e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 5 | 75 | 7.28e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 7.28e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 7.28e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 3 | 66 | 7.28e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 1 | 993 | 7.28e-01 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 2 | 21 | 7.28e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 1 | 8 | 7.28e-01 |
| GO:BP | GO:0010561 | negative regulation of glycoprotein biosynthetic process | 1 | 10 | 7.28e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 2 | 13 | 7.28e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 1 | 8 | 7.29e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 4 | 73 | 7.29e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 7.29e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 7.29e-01 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 4 | 24 | 7.30e-01 |
| GO:BP | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 1 | 16 | 7.30e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 7.30e-01 |
| GO:BP | GO:0099004 | calmodulin dependent kinase signaling pathway | 1 | 6 | 7.30e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 8 | 155 | 7.30e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 1 | 15 | 7.30e-01 |
| GO:BP | GO:2001138 | regulation of phospholipid transport | 1 | 12 | 7.30e-01 |
| GO:BP | GO:0060419 | heart growth | 2 | 72 | 7.30e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 9 | 122 | 7.30e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 1 | 10 | 7.31e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 2 | 18 | 7.31e-01 |
| GO:BP | GO:0007154 | cell communication | 2 | 4226 | 7.31e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 2 | 18 | 7.31e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 3 | 25 | 7.31e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 3 | 78 | 7.31e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 7 | 113 | 7.31e-01 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 1 | 13 | 7.32e-01 |
| GO:BP | GO:2001137 | positive regulation of endocytic recycling | 1 | 7 | 7.32e-01 |
| GO:BP | GO:2000310 | regulation of NMDA receptor activity | 2 | 19 | 7.32e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 7.32e-01 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 1 | 8 | 7.32e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 6 | 43 | 7.33e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 8 | 52 | 7.33e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 6 | 179 | 7.33e-01 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 1 | 11 | 7.33e-01 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 1 | 11 | 7.33e-01 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 1 | 11 | 7.33e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 1 | 14 | 7.33e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 12 | 7.33e-01 |
| GO:BP | GO:0071676 | negative regulation of mononuclear cell migration | 1 | 16 | 7.33e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 4 | 54 | 7.33e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1 | 1995 | 7.33e-01 |
| GO:BP | GO:0001881 | receptor recycling | 2 | 38 | 7.33e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 1 | 14 | 7.33e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 1 | 9 | 7.33e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 4 | 39 | 7.33e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 2 | 46 | 7.34e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 1 | 41 | 7.34e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 3 | 31 | 7.34e-01 |
| GO:BP | GO:0090026 | positive regulation of monocyte chemotaxis | 2 | 7 | 7.34e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 2 | 23 | 7.34e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 5 | 76 | 7.34e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 5 | 68 | 7.34e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 3 | 54 | 7.34e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 5 | 76 | 7.34e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 3 | 52 | 7.34e-01 |
| GO:BP | GO:0060429 | epithelium development | 1 | 850 | 7.36e-01 |
| GO:BP | GO:0015825 | L-serine transport | 1 | 7 | 7.36e-01 |
| GO:BP | GO:0098937 | anterograde dendritic transport | 1 | 9 | 7.36e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 3 | 31 | 7.36e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 3 | 41 | 7.36e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 1 | 8 | 7.37e-01 |
| GO:BP | GO:0050655 | dermatan sulfate proteoglycan metabolic process | 1 | 12 | 7.37e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 7 | 206 | 7.37e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 1 | 29 | 7.38e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 159 | 7.38e-01 |
| GO:BP | GO:0060346 | bone trabecula formation | 1 | 9 | 7.38e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 1 | 190 | 7.38e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 1 | 28 | 7.38e-01 |
| GO:BP | GO:0048870 | cell motility | 1 | 1182 | 7.38e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 7.39e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 7.39e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 7.39e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 1 | 43 | 7.39e-01 |
| GO:BP | GO:0032400 | melanosome localization | 2 | 22 | 7.39e-01 |
| GO:BP | GO:0006096 | glycolytic process | 6 | 70 | 7.39e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 3 | 40 | 7.39e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 7.39e-01 |
| GO:BP | GO:0006013 | mannose metabolic process | 1 | 9 | 7.39e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 7.39e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 2 | 30 | 7.39e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 1 | 288 | 7.39e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 1 | 14 | 7.39e-01 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 3 | 33 | 7.39e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 7.39e-01 |
| GO:BP | GO:0010739 | positive regulation of protein kinase A signaling | 1 | 6 | 7.39e-01 |
| GO:BP | GO:0046548 | retinal rod cell development | 1 | 6 | 7.39e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 7.39e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 2 | 184 | 7.39e-01 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 3 | 33 | 7.39e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 7.39e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 1 | 14 | 7.39e-01 |
| GO:BP | GO:0015867 | ATP transport | 1 | 9 | 7.39e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 1 | 43 | 7.39e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 3 | 63 | 7.39e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 7.39e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 2 | 30 | 7.39e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 7.39e-01 |
| GO:BP | GO:0034695 | response to prostaglandin E | 2 | 17 | 7.39e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 9 | 98 | 7.40e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 4 | 70 | 7.40e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 4 | 50 | 7.40e-01 |
| GO:BP | GO:0002423 | natural killer cell mediated immune response to tumor cell | 1 | 4 | 7.40e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 26 | 517 | 7.40e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 11 | 105 | 7.41e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 5 | 53 | 7.41e-01 |
| GO:BP | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation | 1 | 5 | 7.41e-01 |
| GO:BP | GO:0034397 | telomere localization | 1 | 5 | 7.41e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 5 | 61 | 7.41e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 1 | 151 | 7.41e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 1 | 50 | 7.41e-01 |
| GO:BP | GO:0007309 | oocyte axis specification | 1 | 3 | 7.42e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 2 | 323 | 7.42e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 1 | 159 | 7.42e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 1 | 37 | 7.42e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 2 | 52 | 7.42e-01 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 1 | 1 | 7.42e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 1 | 37 | 7.42e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 7 | 73 | 7.42e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 4 | 43 | 7.42e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 3 | 32 | 7.42e-01 |
| GO:BP | GO:0007308 | oocyte construction | 1 | 3 | 7.42e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 1 | 17 | 7.42e-01 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 1 | 30 | 7.42e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 1 | 8 | 7.42e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 1 | 17 | 7.42e-01 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 1 | 13 | 7.42e-01 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 1 | 1 | 7.42e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 2 | 66 | 7.42e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 7 | 86 | 7.42e-01 |
| GO:BP | GO:0032536 | regulation of cell projection size | 1 | 8 | 7.42e-01 |
| GO:BP | GO:0016358 | dendrite development | 2 | 202 | 7.42e-01 |
| GO:BP | GO:2000508 | regulation of dendritic cell chemotaxis | 1 | 3 | 7.42e-01 |
| GO:BP | GO:0035457 | cellular response to interferon-alpha | 1 | 7 | 7.42e-01 |
| GO:BP | GO:2001198 | regulation of dendritic cell differentiation | 1 | 8 | 7.43e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 1 | 142 | 7.43e-01 |
| GO:BP | GO:0034163 | regulation of toll-like receptor 9 signaling pathway | 1 | 6 | 7.43e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 1 | 9 | 7.43e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 2 | 23 | 7.43e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 1 | 19 | 7.43e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 16 | 2565 | 7.43e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 1 | 15 | 7.43e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 1 | 196 | 7.43e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 3 | 84 | 7.43e-01 |
| GO:BP | GO:1901380 | negative regulation of potassium ion transmembrane transport | 1 | 18 | 7.43e-01 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 1 | 11 | 7.43e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 7.43e-01 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 2 | 31 | 7.43e-01 |
| GO:BP | GO:0034656 | nucleobase-containing small molecule catabolic process | 1 | 20 | 7.43e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 1 | 37 | 7.43e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 1 | 17 | 7.44e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2901 | 7.44e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 2 | 19 | 7.44e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 1 | 16 | 7.44e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 2 | 58 | 7.44e-01 |
| GO:BP | GO:0072540 | T-helper 17 cell lineage commitment | 1 | 3 | 7.44e-01 |
| GO:BP | GO:0000098 | sulfur amino acid catabolic process | 1 | 7 | 7.45e-01 |
| GO:BP | GO:0048672 | positive regulation of collateral sprouting | 1 | 10 | 7.45e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 1 | 8 | 7.45e-01 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 1 | 9 | 7.45e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 7.45e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 2 | 4617 | 7.45e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 1 | 141 | 7.45e-01 |
| GO:BP | GO:0001764 | neuron migration | 14 | 133 | 7.45e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 3 | 42 | 7.45e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 2 | 26 | 7.45e-01 |
| GO:BP | GO:2000109 | regulation of macrophage apoptotic process | 1 | 6 | 7.45e-01 |
| GO:BP | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling | 1 | 8 | 7.46e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 2 | 23 | 7.46e-01 |
| GO:BP | GO:0021859 | pyramidal neuron differentiation | 1 | 11 | 7.46e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 1 | 228 | 7.46e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 2 | 187 | 7.46e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 16 | 2573 | 7.46e-01 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 1 | 9 | 7.46e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 2 | 104 | 7.46e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 34 | 425 | 7.46e-01 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 1 | 14 | 7.48e-01 |
| GO:BP | GO:0042428 | serotonin metabolic process | 1 | 6 | 7.48e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 1 | 17 | 7.49e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 7.49e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 5 | 35 | 7.49e-01 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 1 | 9 | 7.50e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 2 | 25 | 7.50e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 5 | 40 | 7.50e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 15 | 216 | 7.50e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 1 | 17 | 7.50e-01 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 7.50e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 1 | 42 | 7.51e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 1 | 11 | 7.51e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 3 | 64 | 7.51e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 4 | 34 | 7.51e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 1 | 28 | 7.51e-01 |
| GO:BP | GO:0032782 | bile acid secretion | 1 | 5 | 7.51e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 8 | 81 | 7.51e-01 |
| GO:BP | GO:0035635 | entry of bacterium into host cell | 1 | 8 | 7.52e-01 |
| GO:BP | GO:0003407 | neural retina development | 4 | 48 | 7.52e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 7.52e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 7.52e-01 |
| GO:BP | GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1 | 8 | 7.52e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 1 | 13 | 7.52e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 7.52e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 7.52e-01 |
| GO:BP | GO:0006570 | tyrosine metabolic process | 1 | 6 | 7.52e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 7.52e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 1 | 12 | 7.52e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 7.52e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 2 | 39 | 7.53e-01 |
| GO:BP | GO:0045063 | T-helper 1 cell differentiation | 2 | 14 | 7.53e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 2 | 22 | 7.53e-01 |
| GO:BP | GO:0060026 | convergent extension | 1 | 14 | 7.54e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 1 | 50 | 7.54e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 1 | 8 | 7.54e-01 |
| GO:BP | GO:0001821 | histamine secretion | 1 | 9 | 7.54e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 3 | 79 | 7.54e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 7.55e-01 |
| GO:BP | GO:0098779 | positive regulation of mitophagy in response to mitochondrial depolarization | 1 | 10 | 7.55e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 7.55e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 5 | 115 | 7.55e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 6 | 203 | 7.55e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 5 | 96 | 7.56e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 1 | 13 | 7.56e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 1 | 20 | 7.57e-01 |
| GO:BP | GO:0048712 | negative regulation of astrocyte differentiation | 1 | 9 | 7.57e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 1 | 29 | 7.57e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 4 | 44 | 7.57e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 7.57e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 7.57e-01 |
| GO:BP | GO:0006949 | syncytium formation | 1 | 39 | 7.57e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 1 | 31 | 7.57e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 5 | 31 | 7.57e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 3 | 99 | 7.57e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 7.57e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 7.57e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 4 | 49 | 7.57e-01 |
| GO:BP | GO:0001823 | mesonephros development | 5 | 77 | 7.58e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 18 | 168 | 7.58e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 7.59e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 7.59e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 2 | 3600 | 7.60e-01 |
| GO:BP | GO:0032836 | glomerular basement membrane development | 1 | 9 | 7.60e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 1 | 41 | 7.60e-01 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 7.60e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 3 | 77 | 7.60e-01 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 1 | 13 | 7.60e-01 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 5 | 62 | 7.60e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 1 | 9 | 7.61e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 1 | 9 | 7.61e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 1 | 18 | 7.61e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 2 | 26 | 7.61e-01 |
| GO:BP | GO:0006868 | glutamine transport | 1 | 8 | 7.61e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 7.61e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 5 | 330 | 7.61e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 1 | 27 | 7.61e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 1 | 10 | 7.61e-01 |
| GO:BP | GO:0032108 | negative regulation of response to nutrient levels | 1 | 7 | 7.61e-01 |
| GO:BP | GO:0033210 | leptin-mediated signaling pathway | 1 | 10 | 7.61e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 9 | 99 | 7.61e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 7.61e-01 |
| GO:BP | GO:0006002 | fructose 6-phosphate metabolic process | 1 | 8 | 7.61e-01 |
| GO:BP | GO:0032099 | negative regulation of appetite | 1 | 7 | 7.61e-01 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 1 | 18 | 7.61e-01 |
| GO:BP | GO:0032105 | negative regulation of response to extracellular stimulus | 1 | 7 | 7.61e-01 |
| GO:BP | GO:0032096 | negative regulation of response to food | 1 | 7 | 7.61e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 1 | 13 | 7.61e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 1 | 132 | 7.61e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 1 | 22 | 7.62e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 3 | 36 | 7.63e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 18 | 169 | 7.64e-01 |
| GO:BP | GO:0009133 | nucleoside diphosphate biosynthetic process | 1 | 11 | 7.64e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 2 | 54 | 7.64e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 1 | 28 | 7.64e-01 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 2 | 28 | 7.65e-01 |
| GO:BP | GO:0030073 | insulin secretion | 1 | 141 | 7.65e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 3 | 31 | 7.65e-01 |
| GO:BP | GO:1903978 | regulation of microglial cell activation | 1 | 9 | 7.65e-01 |
| GO:BP | GO:0090520 | sphingolipid mediated signaling pathway | 1 | 11 | 7.65e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 86 | 7.65e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 5 | 40 | 7.66e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 7.66e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 1 | 10 | 7.66e-01 |
| GO:BP | GO:0070233 | negative regulation of T cell apoptotic process | 1 | 14 | 7.66e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 1 | 134 | 7.66e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 7 | 127 | 7.66e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 2 | 38 | 7.66e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 7.66e-01 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 2 | 19 | 7.66e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 7.66e-01 |
| GO:BP | GO:1905048 | regulation of metallopeptidase activity | 1 | 14 | 7.66e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 1 | 15 | 7.66e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 5 | 53 | 7.66e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 7.66e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 2 | 52 | 7.66e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 1 | 43 | 7.66e-01 |
| GO:BP | GO:0031016 | pancreas development | 4 | 53 | 7.67e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 20 | 213 | 7.67e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 3 | 60 | 7.67e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 2 | 62 | 7.67e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 1 | 18 | 7.67e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 2 | 28 | 7.67e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 3 | 32 | 7.67e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 2 | 41 | 7.68e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 1 | 21 | 7.68e-01 |
| GO:BP | GO:1901658 | glycosyl compound catabolic process | 1 | 23 | 7.68e-01 |
| GO:BP | GO:0015886 | heme transport | 1 | 9 | 7.68e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 1 | 136 | 7.68e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 2 | 25 | 7.68e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 2 | 15 | 7.68e-01 |
| GO:BP | GO:1903727 | positive regulation of phospholipid metabolic process | 1 | 9 | 7.68e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 5 | 57 | 7.68e-01 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 3 | 26 | 7.68e-01 |
| GO:BP | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 1 | 9 | 7.68e-01 |
| GO:BP | GO:0060525 | prostate glandular acinus development | 1 | 5 | 7.68e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 7 | 113 | 7.68e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 1 | 10 | 7.69e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 1 | 11 | 7.69e-01 |
| GO:BP | GO:0061687 | detoxification of inorganic compound | 1 | 12 | 7.69e-01 |
| GO:BP | GO:0006527 | arginine catabolic process | 1 | 7 | 7.69e-01 |
| GO:BP | GO:0014850 | response to muscle activity | 1 | 17 | 7.69e-01 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 5 | 46 | 7.69e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 3 | 37 | 7.69e-01 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 2 | 8 | 7.69e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 1 | 10 | 7.69e-01 |
| GO:BP | GO:0006901 | vesicle coating | 2 | 36 | 7.69e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 3 | 25 | 7.69e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 2 | 9 | 7.69e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 1 | 422 | 7.69e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 2 | 23 | 7.69e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 23 | 362 | 7.69e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 5 | 111 | 7.69e-01 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 7.70e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 4 | 41 | 7.70e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 1 | 994 | 7.70e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 2 | 41 | 7.70e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 5 | 80 | 7.70e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 1 | 13 | 7.70e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 1 | 57 | 7.70e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 2 | 29 | 7.71e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 1 | 61 | 7.71e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 7.71e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 9 | 83 | 7.71e-01 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 1 | 18 | 7.71e-01 |
| GO:BP | GO:0090219 | negative regulation of lipid kinase activity | 1 | 8 | 7.71e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 1 | 10 | 7.71e-01 |
| GO:BP | GO:0008306 | associative learning | 3 | 65 | 7.71e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 2 | 114 | 7.71e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 2 | 64 | 7.72e-01 |
| GO:BP | GO:0038183 | bile acid signaling pathway | 1 | 3 | 7.72e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 2 | 39 | 7.72e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 2 | 39 | 7.72e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 1 | 18 | 7.72e-01 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 1 | 19 | 7.72e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 2 | 21 | 7.72e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 7.72e-01 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 1 | 19 | 7.72e-01 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 1 | 19 | 7.72e-01 |
| GO:BP | GO:0090672 | telomerase RNA localization | 1 | 19 | 7.72e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 1 | 106 | 7.72e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 3 | 52 | 7.72e-01 |
| GO:BP | GO:0046386 | deoxyribose phosphate catabolic process | 2 | 22 | 7.72e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 2 | 50 | 7.72e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 1 | 15 | 7.72e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 3 | 40 | 7.72e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 7.72e-01 |
| GO:BP | GO:0009264 | deoxyribonucleotide catabolic process | 2 | 22 | 7.72e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 1 | 24 | 7.73e-01 |
| GO:BP | GO:0001890 | placenta development | 7 | 117 | 7.73e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 2 | 395 | 7.73e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 9 | 150 | 7.73e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 7.73e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 1 | 17 | 7.73e-01 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 1 | 10 | 7.74e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 7.74e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 1 | 9 | 7.74e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 1 | 32 | 7.74e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 1 | 168 | 7.74e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 7.75e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 4 | 58 | 7.75e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 1 | 21 | 7.75e-01 |
| GO:BP | GO:0016032 | viral process | 1 | 344 | 7.76e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 1 | 137 | 7.76e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 7.76e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 10 | 292 | 7.76e-01 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 1 | 21 | 7.77e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 1 | 22 | 7.77e-01 |
| GO:BP | GO:0009174 | pyrimidine ribonucleoside monophosphate biosynthetic process | 1 | 9 | 7.77e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 1 | 41 | 7.77e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 7.77e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 7.77e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 1 | 23 | 7.78e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 2 | 17 | 7.78e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 2 | 24 | 7.78e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 2 | 28 | 7.78e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 3 | 47 | 7.78e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 7 | 73 | 7.79e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 7 | 73 | 7.79e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 4 | 49 | 7.79e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 7.80e-01 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 1 | 17 | 7.80e-01 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 1 | 5 | 7.80e-01 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 2 | 25 | 7.80e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 2 | 47 | 7.80e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 6 | 84 | 7.80e-01 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 1 | 10 | 7.80e-01 |
| GO:BP | GO:0090382 | phagosome maturation | 1 | 24 | 7.80e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 75 | 4579 | 7.80e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 7.81e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 14 | 172 | 7.81e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 2 | 24 | 7.81e-01 |
| GO:BP | GO:1903019 | negative regulation of glycoprotein metabolic process | 1 | 12 | 7.81e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 6 | 78 | 7.81e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 2 | 18 | 7.81e-01 |
| GO:BP | GO:0006855 | xenobiotic transmembrane transport | 1 | 11 | 7.82e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 2 | 20 | 7.82e-01 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 1 | 12 | 7.82e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 3 | 40 | 7.82e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 7.82e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 7.82e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 7.83e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 1 | 12 | 7.83e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 1 | 19 | 7.83e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 7.83e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 2 | 13 | 7.83e-01 |
| GO:BP | GO:0035524 | proline transmembrane transport | 1 | 8 | 7.83e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 1 | 10 | 7.83e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 3 | 34 | 7.84e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 2 | 30 | 7.85e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 3 | 41 | 7.85e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 21 | 191 | 7.86e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 7.86e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 7 | 223 | 7.86e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 4 | 74 | 7.86e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 2 | 24 | 7.86e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 8 | 240 | 7.86e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 2 | 29 | 7.86e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 5 | 48 | 7.86e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 1 | 12 | 7.86e-01 |
| GO:BP | GO:0006883 | intracellular sodium ion homeostasis | 1 | 13 | 7.86e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2 | 5017 | 7.86e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 1 | 14 | 7.86e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 4 | 50 | 7.88e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 1 | 1229 | 7.88e-01 |
| GO:BP | GO:0022600 | digestive system process | 3 | 54 | 7.89e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 2 | 59 | 7.89e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 2 | 55 | 7.89e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 7.89e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 2 | 26 | 7.89e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 1 | 16 | 7.89e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 1 | 10 | 7.89e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 4 | 67 | 7.89e-01 |
| GO:BP | GO:0021510 | spinal cord development | 1 | 59 | 7.90e-01 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 1 | 23 | 7.90e-01 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 1 | 131 | 7.90e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 1 | 16 | 7.90e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 1 | 16 | 7.90e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 1 | 17 | 7.91e-01 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 1 | 11 | 7.91e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 2 | 90 | 7.92e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 5 | 99 | 7.92e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 1 | 16 | 7.92e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 3 | 30 | 7.92e-01 |
| GO:BP | GO:0072006 | nephron development | 10 | 117 | 7.92e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 5 | 99 | 7.92e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 3 | 48 | 7.92e-01 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 7.92e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3386 | 7.92e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 1 | 15 | 7.92e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 1 | 16 | 7.92e-01 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 1 | 10 | 7.92e-01 |
| GO:BP | GO:0060742 | epithelial cell differentiation involved in prostate gland development | 1 | 8 | 7.92e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 2 | 14 | 7.93e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 1 | 35 | 7.93e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 2 | 22 | 7.93e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 7 | 224 | 7.93e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 2 | 29 | 7.93e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 10 | 147 | 7.93e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 1 | 10 | 7.93e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 3 | 76 | 7.94e-01 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 3 | 28 | 7.94e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 1 | 10 | 7.94e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 2 | 20 | 7.94e-01 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 1 | 29 | 7.94e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 2 | 52 | 7.94e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 1 | 26 | 7.94e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 1 | 13 | 7.95e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 7 | 199 | 7.95e-01 |
| GO:BP | GO:0019080 | viral gene expression | 2 | 96 | 7.96e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 9 | 124 | 7.96e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 3 | 69 | 7.96e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 7.96e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 7.96e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 5 | 99 | 7.96e-01 |
| GO:BP | GO:0098792 | xenophagy | 1 | 12 | 7.96e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 2 | 25 | 7.96e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 2 | 28 | 7.96e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 7.97e-01 |
| GO:BP | GO:0030540 | female genitalia development | 1 | 14 | 7.97e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 1 | 14 | 7.97e-01 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 3 | 25 | 7.97e-01 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 2 | 20 | 7.97e-01 |
| GO:BP | GO:0043931 | ossification involved in bone maturation | 2 | 17 | 7.98e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 1 | 13 | 7.98e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 2 | 77 | 7.98e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 5 | 153 | 7.98e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 2 | 21 | 7.98e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 6 | 65 | 7.98e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 1 | 23 | 7.98e-01 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 1 | 7 | 7.98e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 7.98e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 1 | 39 | 7.98e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 6 | 126 | 7.98e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 1 | 19 | 7.99e-01 |
| GO:BP | GO:0043584 | nose development | 1 | 10 | 7.99e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 2 | 49 | 8.00e-01 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 1 | 14 | 8.00e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 8.00e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 2 | 66 | 8.00e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 2 | 17 | 8.00e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 4 | 71 | 8.00e-01 |
| GO:BP | GO:0048806 | genitalia development | 3 | 28 | 8.00e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 1 | 43 | 8.00e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 8.00e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 1 | 228 | 8.00e-01 |
| GO:BP | GO:0044057 | regulation of system process | 17 | 405 | 8.00e-01 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 1 | 25 | 8.00e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 7 | 77 | 8.00e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 2 | 52 | 8.00e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 3 | 77 | 8.00e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 3 | 77 | 8.00e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 7 | 126 | 8.00e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 1 | 31 | 8.00e-01 |
| GO:BP | GO:0042976 | activation of Janus kinase activity | 1 | 7 | 8.00e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 2 | 26 | 8.01e-01 |
| GO:BP | GO:1904322 | cellular response to forskolin | 1 | 11 | 8.01e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 3 | 138 | 8.01e-01 |
| GO:BP | GO:1904321 | response to forskolin | 1 | 11 | 8.01e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 2 | 37 | 8.01e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 2 | 140 | 8.03e-01 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 1 | 10 | 8.03e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 3 | 111 | 8.03e-01 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 1 | 10 | 8.03e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 5 | 37 | 8.04e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 2 | 22 | 8.04e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 2 | 28 | 8.04e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 1 | 16 | 8.04e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 8 | 93 | 8.04e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 1 | 32 | 8.04e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 1 | 18 | 8.04e-01 |
| GO:BP | GO:0001656 | metanephros development | 6 | 64 | 8.05e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 1 | 9 | 8.06e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 4 | 35 | 8.06e-01 |
| GO:BP | GO:0015808 | L-alanine transport | 1 | 9 | 8.06e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 1 | 30 | 8.06e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 2 | 27 | 8.06e-01 |
| GO:BP | GO:0060173 | limb development | 12 | 142 | 8.06e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 3 | 50 | 8.06e-01 |
| GO:BP | GO:0048736 | appendage development | 12 | 142 | 8.06e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 8.06e-01 |
| GO:BP | GO:0032329 | serine transport | 1 | 10 | 8.06e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 2 | 17 | 8.06e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 5 | 146 | 8.06e-01 |
| GO:BP | GO:0032488 | Cdc42 protein signal transduction | 1 | 10 | 8.06e-01 |
| GO:BP | GO:0002931 | response to ischemia | 4 | 46 | 8.06e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 8.06e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 3 | 63 | 8.06e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 4 | 48 | 8.06e-01 |
| GO:BP | GO:0032418 | lysosome localization | 4 | 62 | 8.06e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 1 | 10 | 8.06e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 2 | 91 | 8.06e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 22 | 357 | 8.06e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 2 | 26 | 8.06e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 2 | 16 | 8.06e-01 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 1 | 18 | 8.06e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 4 | 62 | 8.06e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 2 | 22 | 8.06e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 1 | 23 | 8.06e-01 |
| GO:BP | GO:0072537 | fibroblast activation | 1 | 11 | 8.06e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 7 | 78 | 8.06e-01 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 1 | 13 | 8.06e-01 |
| GO:BP | GO:0007584 | response to nutrient | 6 | 109 | 8.06e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 12 | 230 | 8.06e-01 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 1 | 17 | 8.06e-01 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 1 | 7 | 8.06e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 9 | 160 | 8.07e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 1 | 30 | 8.07e-01 |
| GO:BP | GO:0002295 | T-helper cell lineage commitment | 1 | 7 | 8.07e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 2 | 68 | 8.08e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 1 | 39 | 8.08e-01 |
| GO:BP | GO:1901160 | primary amino compound metabolic process | 1 | 8 | 8.08e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 1 | 27 | 8.08e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 14 | 187 | 8.08e-01 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 2 | 20 | 8.08e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 20 | 258 | 8.08e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 1 | 13 | 8.08e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 1 | 500 | 8.08e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 8.08e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 8.08e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 1 | 41 | 8.08e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 14 | 187 | 8.08e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 1 | 20 | 8.08e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 4 | 118 | 8.08e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 8 | 227 | 8.08e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 3 | 140 | 8.08e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 8.08e-01 |
| GO:BP | GO:0032613 | interleukin-10 production | 2 | 24 | 8.09e-01 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 2 | 24 | 8.09e-01 |
| GO:BP | GO:0051014 | actin filament severing | 1 | 13 | 8.09e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 8.09e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 2 | 105 | 8.09e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 9 | 92 | 8.10e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 8.10e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 6 | 50 | 8.10e-01 |
| GO:BP | GO:0098760 | response to interleukin-7 | 1 | 12 | 8.10e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 2 | 13 | 8.10e-01 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 1 | 12 | 8.10e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 2 | 25 | 8.10e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 1 | 166 | 8.10e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 1 | 88 | 8.10e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 6 | 62 | 8.10e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 2 | 20 | 8.10e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 4 | 49 | 8.10e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 7 | 56 | 8.11e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 7 | 56 | 8.11e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 9 | 123 | 8.11e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 1 | 19 | 8.11e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 1 | 19 | 8.12e-01 |
| GO:BP | GO:0007507 | heart development | 26 | 506 | 8.12e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 2 | 46 | 8.12e-01 |
| GO:BP | GO:0006414 | translational elongation | 3 | 65 | 8.13e-01 |
| GO:BP | GO:0046323 | glucose import | 2 | 53 | 8.13e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 4 | 59 | 8.13e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 8.13e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 16 | 197 | 8.13e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 8.13e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 1 | 7 | 8.13e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 2 | 24 | 8.13e-01 |
| GO:BP | GO:0051608 | histamine transport | 1 | 11 | 8.13e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 1 | 88 | 8.13e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 2 | 4794 | 8.13e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 2 | 24 | 8.13e-01 |
| GO:BP | GO:1901652 | response to peptide | 14 | 377 | 8.13e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 1 | 19 | 8.13e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 3 | 6996 | 8.13e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 7 | 90 | 8.13e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 8.14e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 3 | 42 | 8.14e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 2 | 25 | 8.14e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 8.14e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 1 | 31 | 8.14e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 1 | 18 | 8.14e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 2 | 25 | 8.14e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 1 | 47 | 8.14e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 9 | 93 | 8.14e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 3 | 49 | 8.14e-01 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 170 | 8.14e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 29 | 8.14e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 1 | 11 | 8.14e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 4 | 104 | 8.14e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 4 | 61 | 8.14e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 1 | 13 | 8.14e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 1 | 10 | 8.14e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 3 | 321 | 8.14e-01 |
| GO:BP | GO:0016078 | tRNA catabolic process | 1 | 14 | 8.14e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 3 | 53 | 8.14e-01 |
| GO:BP | GO:0014002 | astrocyte development | 1 | 27 | 8.15e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 3 | 35 | 8.15e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 2 | 19 | 8.15e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 5 | 68 | 8.16e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 8 | 112 | 8.16e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 21 | 281 | 8.16e-01 |
| GO:BP | GO:0045010 | actin nucleation | 4 | 50 | 8.16e-01 |
| GO:BP | GO:0035265 | organ growth | 3 | 131 | 8.16e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 4 | 49 | 8.16e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 3 | 43 | 8.17e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 8.17e-01 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 1 | 11 | 8.17e-01 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 1 | 11 | 8.17e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 2 | 21 | 8.18e-01 |
| GO:BP | GO:0072678 | T cell migration | 3 | 38 | 8.18e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 1 | 33 | 8.18e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 13 | 8.18e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 2 | 19 | 8.18e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 8.18e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 8.18e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 1 | 14 | 8.18e-01 |
| GO:BP | GO:0035845 | photoreceptor cell outer segment organization | 1 | 10 | 8.18e-01 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 1 | 9 | 8.18e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 8.18e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 2 | 28 | 8.18e-01 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 2 | 20 | 8.18e-01 |
| GO:BP | GO:0070085 | glycosylation | 15 | 202 | 8.18e-01 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 2 | 12 | 8.18e-01 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 2 | 12 | 8.18e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 1 | 54 | 8.18e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 1 | 100 | 8.18e-01 |
| GO:BP | GO:0030252 | growth hormone secretion | 1 | 12 | 8.18e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 2 | 26 | 8.18e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 1 | 168 | 8.18e-01 |
| GO:BP | GO:1902931 | negative regulation of alcohol biosynthetic process | 1 | 10 | 8.18e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 1 | 280 | 8.18e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 2 | 375 | 8.19e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 1 | 27 | 8.19e-01 |
| GO:BP | GO:0002839 | positive regulation of immune response to tumor cell | 1 | 7 | 8.19e-01 |
| GO:BP | GO:0002836 | positive regulation of response to tumor cell | 1 | 7 | 8.19e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 5 | 71 | 8.19e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 3 | 40 | 8.20e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 2 | 376 | 8.21e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 8.23e-01 |
| GO:BP | GO:0015824 | proline transport | 1 | 11 | 8.23e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 7 | 71 | 8.23e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 8.23e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 1 | 34 | 8.23e-01 |
| GO:BP | GO:0071071 | regulation of phospholipid biosynthetic process | 1 | 15 | 8.24e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 2 | 25 | 8.24e-01 |
| GO:BP | GO:0140354 | lipid import into cell | 1 | 15 | 8.24e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 1 | 66 | 8.24e-01 |
| GO:BP | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | 1 | 14 | 8.24e-01 |
| GO:BP | GO:0007613 | memory | 3 | 92 | 8.24e-01 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 1 | 14 | 8.24e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 4 | 45 | 8.24e-01 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 2 | 14 | 8.24e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 2 | 19 | 8.24e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 2 | 19 | 8.24e-01 |
| GO:BP | GO:1901678 | iron coordination entity transport | 1 | 11 | 8.24e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 1 | 51 | 8.24e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 1 | 22 | 8.24e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 4 | 123 | 8.24e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 3 | 38 | 8.24e-01 |
| GO:BP | GO:0007338 | single fertilization | 2 | 70 | 8.25e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 8.25e-01 |
| GO:BP | GO:0007595 | lactation | 3 | 32 | 8.25e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 5 | 114 | 8.25e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 2 | 25 | 8.26e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 1 | 38 | 8.26e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 2 | 37 | 8.26e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 5 | 96 | 8.26e-01 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 8.26e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 6 | 72 | 8.26e-01 |
| GO:BP | GO:0009798 | axis specification | 4 | 63 | 8.26e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 2 | 28 | 8.26e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 11 | 156 | 8.26e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 3 | 36 | 8.26e-01 |
| GO:BP | GO:0018146 | keratan sulfate biosynthetic process | 1 | 13 | 8.26e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 2 | 45 | 8.26e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 2 | 64 | 8.26e-01 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 1 | 5 | 8.26e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 1 | 19 | 8.26e-01 |
| GO:BP | GO:0016114 | terpenoid biosynthetic process | 1 | 12 | 8.26e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 213 | 8.27e-01 |
| GO:BP | GO:0043306 | positive regulation of mast cell degranulation | 1 | 10 | 8.28e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 2 | 30 | 8.28e-01 |
| GO:BP | GO:0033008 | positive regulation of mast cell activation involved in immune response | 1 | 10 | 8.28e-01 |
| GO:BP | GO:0007416 | synapse assembly | 15 | 152 | 8.28e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 1 | 185 | 8.29e-01 |
| GO:BP | GO:0097484 | dendrite extension | 2 | 27 | 8.29e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 1 | 18 | 8.29e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 7 | 64 | 8.29e-01 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 1 | 10 | 8.30e-01 |
| GO:BP | GO:0043094 | cellular metabolic compound salvage | 2 | 25 | 8.30e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 30 | 424 | 8.30e-01 |
| GO:BP | GO:0095500 | acetylcholine receptor signaling pathway | 1 | 18 | 8.30e-01 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 1 | 10 | 8.30e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 1 | 53 | 8.30e-01 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 8.30e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 14 | 175 | 8.30e-01 |
| GO:BP | GO:0015833 | peptide transport | 1 | 179 | 8.31e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 10 | 143 | 8.31e-01 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 1 | 41 | 8.31e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 4 | 71 | 8.31e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 1 | 16 | 8.31e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 1 | 24 | 8.31e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 1 | 71 | 8.32e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 1 | 10 | 8.33e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 8.33e-01 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 38 | 8.34e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 3 | 21 | 8.34e-01 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 1 | 11 | 8.34e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 6 | 203 | 8.34e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 8 | 85 | 8.34e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 1 | 11 | 8.34e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 2 | 62 | 8.34e-01 |
| GO:BP | GO:0032606 | type I interferon production | 6 | 76 | 8.34e-01 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 1 | 9 | 8.34e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 6 | 76 | 8.34e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 2 | 35 | 8.34e-01 |
| GO:BP | GO:0120193 | tight junction organization | 1 | 52 | 8.34e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 1 | 25 | 8.34e-01 |
| GO:BP | GO:0071888 | macrophage apoptotic process | 1 | 9 | 8.34e-01 |
| GO:BP | GO:0043473 | pigmentation | 3 | 92 | 8.34e-01 |
| GO:BP | GO:0051028 | mRNA transport | 7 | 117 | 8.34e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 8.34e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 2 | 26 | 8.34e-01 |
| GO:BP | GO:0050892 | intestinal absorption | 1 | 22 | 8.34e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 8.34e-01 |
| GO:BP | GO:0070977 | bone maturation | 2 | 19 | 8.34e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 1 | 26 | 8.34e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 14 | 204 | 8.34e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 2 | 24 | 8.34e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 8 | 192 | 8.34e-01 |
| GO:BP | GO:1904925 | positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 1 | 14 | 8.35e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 2 | 31 | 8.36e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 8.36e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 5 | 60 | 8.36e-01 |
| GO:BP | GO:0034694 | response to prostaglandin | 2 | 21 | 8.36e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 1 | 513 | 8.37e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 6 | 85 | 8.37e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 2 | 19 | 8.37e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 2 | 24 | 8.37e-01 |
| GO:BP | GO:0002082 | regulation of oxidative phosphorylation | 1 | 19 | 8.37e-01 |
| GO:BP | GO:0007271 | synaptic transmission, cholinergic | 1 | 15 | 8.38e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 2 | 25 | 8.38e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 27 | 265 | 8.38e-01 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 1 | 13 | 8.38e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 8.38e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 8.38e-01 |
| GO:BP | GO:0030850 | prostate gland development | 2 | 35 | 8.38e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 2 | 38 | 8.38e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 8 | 117 | 8.38e-01 |
| GO:BP | GO:0042541 | hemoglobin biosynthetic process | 1 | 13 | 8.38e-01 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 1 | 5 | 8.39e-01 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 1 | 5 | 8.39e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 1 | 22 | 8.39e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 7 | 68 | 8.39e-01 |
| GO:BP | GO:0000266 | mitochondrial fission | 2 | 38 | 8.40e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 4 | 58 | 8.40e-01 |
| GO:BP | GO:0060263 | regulation of respiratory burst | 1 | 10 | 8.40e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 4 | 106 | 8.40e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 39 | 555 | 8.40e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 1 | 1389 | 8.40e-01 |
| GO:BP | GO:0043113 | receptor clustering | 2 | 45 | 8.40e-01 |
| GO:BP | GO:0033006 | regulation of mast cell activation involved in immune response | 2 | 21 | 8.40e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 1 | 58 | 8.40e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 3 | 39 | 8.40e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 1 | 11 | 8.40e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 4 | 38 | 8.40e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 8.40e-01 |
| GO:BP | GO:0042063 | gliogenesis | 7 | 235 | 8.40e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 3 | 27 | 8.40e-01 |
| GO:BP | GO:0043200 | response to amino acid | 3 | 95 | 8.41e-01 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 1 | 15 | 8.41e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 15 | 293 | 8.41e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 1 | 26 | 8.42e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 1 | 29 | 8.42e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 7 | 124 | 8.42e-01 |
| GO:BP | GO:0043373 | CD4-positive, alpha-beta T cell lineage commitment | 1 | 9 | 8.42e-01 |
| GO:BP | GO:0044793 | negative regulation by host of viral process | 1 | 10 | 8.42e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 1 | 13 | 8.42e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 1 | 10 | 8.42e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 1 | 42 | 8.43e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 2 | 83 | 8.43e-01 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 1 | 11 | 8.44e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 6 | 75 | 8.44e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 2 | 33 | 8.44e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1 | 10 | 8.44e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 1 | 12 | 8.44e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 3 | 4128 | 8.45e-01 |
| GO:BP | GO:1905145 | cellular response to acetylcholine | 1 | 20 | 8.45e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 9 | 111 | 8.45e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 3 | 35 | 8.45e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 3 | 38 | 8.45e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 8.45e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 25 | 8.45e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 1 | 15 | 8.45e-01 |
| GO:BP | GO:0042308 | negative regulation of protein import into nucleus | 1 | 13 | 8.45e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 8 | 91 | 8.45e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 2 | 25 | 8.45e-01 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 1 | 19 | 8.45e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 16 | 8.45e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 8.46e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 1 | 27 | 8.46e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 1 | 212 | 8.46e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 3 | 47 | 8.46e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 1 | 19 | 8.46e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 8 | 100 | 8.46e-01 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 1 | 10 | 8.46e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 1 | 12 | 8.46e-01 |
| GO:BP | GO:0051900 | regulation of mitochondrial depolarization | 1 | 15 | 8.46e-01 |
| GO:BP | GO:0007015 | actin filament organization | 8 | 373 | 8.46e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 1 | 10 | 8.46e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 1 | 19 | 8.46e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 1 | 10 | 8.46e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 8.46e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 11 | 143 | 8.46e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 1 | 17 | 8.46e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 10 | 102 | 8.47e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 1 | 13 | 8.47e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 2 | 30 | 8.48e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 1 | 66 | 8.48e-01 |
| GO:BP | GO:0070509 | calcium ion import | 2 | 37 | 8.48e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 1 | 24 | 8.48e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 1 | 17 | 8.48e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 1 | 10 | 8.48e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 3 | 64 | 8.48e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 2 | 317 | 8.48e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 2 | 20 | 8.48e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 1 | 10 | 8.48e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 3 | 35 | 8.48e-01 |
| GO:BP | GO:0001947 | heart looping | 1 | 50 | 8.48e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 8.48e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 1 | 15 | 8.48e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 1 | 15 | 8.48e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 1 | 15 | 8.48e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 1 | 19 | 8.48e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 1012 | 8.48e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 8 | 68 | 8.48e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 1 | 14 | 8.49e-01 |
| GO:BP | GO:0032095 | regulation of response to food | 1 | 8 | 8.49e-01 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 2 | 27 | 8.49e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 8.49e-01 |
| GO:BP | GO:0006936 | muscle contraction | 21 | 267 | 8.49e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 13 | 222 | 8.49e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 6 | 71 | 8.50e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 6 | 98 | 8.50e-01 |
| GO:BP | GO:1904923 | regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 1 | 15 | 8.50e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 5 | 73 | 8.51e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 8.51e-01 |
| GO:BP | GO:0021536 | diencephalon development | 4 | 41 | 8.51e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 8.51e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 10 | 159 | 8.51e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 2 | 62 | 8.52e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 1 | 14 | 8.52e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 3 | 49 | 8.52e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 6 | 117 | 8.52e-01 |
| GO:BP | GO:0001502 | cartilage condensation | 1 | 12 | 8.52e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 4 | 75 | 8.53e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 2 | 32 | 8.53e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 3 | 35 | 8.53e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 4 | 60 | 8.53e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 2 | 97 | 8.54e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 2 | 158 | 8.54e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 1 | 14 | 8.54e-01 |
| GO:BP | GO:0042339 | keratan sulfate metabolic process | 1 | 15 | 8.54e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 8.54e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 2 | 157 | 8.54e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 4 | 60 | 8.54e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 2 | 53 | 8.55e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 35 | 428 | 8.55e-01 |
| GO:BP | GO:0042168 | heme metabolic process | 3 | 40 | 8.55e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 1 | 16 | 8.55e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 5 | 87 | 8.55e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 6 | 86 | 8.55e-01 |
| GO:BP | GO:0051931 | regulation of sensory perception | 1 | 13 | 8.55e-01 |
| GO:BP | GO:0051930 | regulation of sensory perception of pain | 1 | 13 | 8.55e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 8.55e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 6 | 94 | 8.55e-01 |
| GO:BP | GO:0051648 | vesicle localization | 15 | 192 | 8.55e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 8.55e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 2 | 54 | 8.55e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 3 | 40 | 8.55e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 5 | 49 | 8.55e-01 |
| GO:BP | GO:1905155 | positive regulation of membrane invagination | 1 | 11 | 8.55e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 1 | 26 | 8.55e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 1 | 11 | 8.55e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 8.55e-01 |
| GO:BP | GO:0060100 | positive regulation of phagocytosis, engulfment | 1 | 11 | 8.55e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 6 | 61 | 8.55e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 3 | 41 | 8.56e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 4 | 52 | 8.56e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 12 | 154 | 8.56e-01 |
| GO:BP | GO:0042749 | regulation of circadian sleep/wake cycle | 1 | 9 | 8.57e-01 |
| GO:BP | GO:0035196 | miRNA processing | 2 | 41 | 8.57e-01 |
| GO:BP | GO:0051693 | actin filament capping | 2 | 34 | 8.57e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 8.58e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 8.58e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 8.58e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 9 | 8.58e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 2 | 145 | 8.58e-01 |
| GO:BP | GO:0021694 | cerebellar Purkinje cell layer formation | 1 | 12 | 8.58e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 3 | 35 | 8.58e-01 |
| GO:BP | GO:0001906 | cell killing | 6 | 69 | 8.58e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 2 | 27 | 8.58e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 6 | 84 | 8.58e-01 |
| GO:BP | GO:0001759 | organ induction | 1 | 17 | 8.58e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 8 | 154 | 8.58e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 2 | 22 | 8.58e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 8.58e-01 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 1 | 12 | 8.59e-01 |
| GO:BP | GO:0051646 | mitochondrion localization | 1 | 47 | 8.59e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 36 | 8.59e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 25 | 358 | 8.61e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 4 | 54 | 8.61e-01 |
| GO:BP | GO:0045576 | mast cell activation | 1 | 35 | 8.61e-01 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 1 | 28 | 8.61e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 2 | 33 | 8.61e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 6 | 89 | 8.61e-01 |
| GO:BP | GO:0035094 | response to nicotine | 1 | 28 | 8.61e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 1 | 13 | 8.61e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 8.61e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 8.62e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 2 | 35 | 8.62e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 3 | 42 | 8.62e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 12 | 90 | 8.63e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 2 | 68 | 8.63e-01 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 1 | 11 | 8.63e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 3 | 44 | 8.63e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 2 | 110 | 8.64e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 3 | 33 | 8.64e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 8.64e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 12 | 200 | 8.64e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 8.64e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 5 | 69 | 8.64e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 6 | 61 | 8.64e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 5 | 91 | 8.64e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 8.64e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 8.64e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 8.64e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 4 | 55 | 8.64e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 4 | 75 | 8.64e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 2 | 27 | 8.65e-01 |
| GO:BP | GO:0009173 | pyrimidine ribonucleoside monophosphate metabolic process | 1 | 12 | 8.65e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 8.65e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 2 | 101 | 8.65e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 3 | 47 | 8.67e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 2 | 46 | 8.67e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 4 | 99 | 8.67e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 8.67e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 1 | 211 | 8.67e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 1 | 11 | 8.67e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 2 | 20 | 8.67e-01 |
| GO:BP | GO:0006390 | mitochondrial transcription | 1 | 18 | 8.68e-01 |
| GO:BP | GO:0033280 | response to vitamin D | 1 | 24 | 8.68e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 4 | 71 | 8.68e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 1 | 14 | 8.68e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 1 | 18 | 8.68e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 1 | 23 | 8.69e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 1 | 23 | 8.69e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 1 | 127 | 8.69e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 3 | 39 | 8.69e-01 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 1 | 17 | 8.69e-01 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 1 | 10 | 8.69e-01 |
| GO:BP | GO:0030282 | bone mineralization | 6 | 82 | 8.69e-01 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 1 | 17 | 8.69e-01 |
| GO:BP | GO:0033198 | response to ATP | 2 | 24 | 8.69e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 8.69e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 4 | 43 | 8.69e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 3 | 25 | 8.69e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 8.69e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 6 | 97 | 8.69e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 2 | 12 | 8.70e-01 |
| GO:BP | GO:0007631 | feeding behavior | 1 | 52 | 8.70e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 13 | 227 | 8.70e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 1 | 20 | 8.70e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 2 | 475 | 8.70e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 1 | 32 | 8.70e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 1 | 50 | 8.70e-01 |
| GO:BP | GO:2000251 | positive regulation of actin cytoskeleton reorganization | 1 | 14 | 8.70e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 8.70e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 2 | 31 | 8.70e-01 |
| GO:BP | GO:0006837 | serotonin transport | 1 | 11 | 8.71e-01 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 2 | 29 | 8.71e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 1 | 33 | 8.71e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 1 | 33 | 8.71e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 2 | 35 | 8.71e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 251 | 8.71e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 3 | 91 | 8.72e-01 |
| GO:BP | GO:0061326 | renal tubule development | 5 | 77 | 8.72e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 1 | 14 | 8.72e-01 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 1 | 11 | 8.72e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 1 | 14 | 8.72e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 1 | 17 | 8.72e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 6 | 70 | 8.73e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 2 | 25 | 8.73e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 4 | 154 | 8.73e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 39 | 8.73e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 2 | 31 | 8.73e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 19 | 265 | 8.73e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 6 | 122 | 8.73e-01 |
| GO:BP | GO:0098657 | import into cell | 13 | 166 | 8.74e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 2 | 24 | 8.74e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 12 | 172 | 8.74e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 3 | 258 | 8.74e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 215 | 8.74e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 1 | 16 | 8.74e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 2 | 102 | 8.75e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 3 | 111 | 8.76e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 3 | 128 | 8.76e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 3 | 701 | 8.76e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 2 | 20 | 8.76e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 1 | 17 | 8.76e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 1 | 19 | 8.76e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 8.76e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 1 | 12 | 8.76e-01 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 1 | 21 | 8.76e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 1 | 32 | 8.76e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 2 | 60 | 8.76e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 1 | 63 | 8.76e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 1 | 12 | 8.76e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 2 | 103 | 8.77e-01 |
| GO:BP | GO:0044058 | regulation of digestive system process | 1 | 22 | 8.77e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 4 | 34 | 8.78e-01 |
| GO:BP | GO:0048799 | animal organ maturation | 2 | 20 | 8.78e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 8.78e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 2 | 19 | 8.78e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 2 | 19 | 8.78e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 1 | 22 | 8.79e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 11 | 143 | 8.79e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 1 | 20 | 8.79e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 4 | 63 | 8.79e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 7 | 105 | 8.79e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 8.79e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 8.79e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 2 | 20 | 8.79e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 2 | 31 | 8.79e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 4 | 157 | 8.79e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 8.80e-01 |
| GO:BP | GO:0006968 | cellular defense response | 1 | 15 | 8.81e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 8.81e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 8.81e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 5 | 51 | 8.81e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 20 | 8.81e-01 |
| GO:BP | GO:0044804 | autophagy of nucleus | 1 | 18 | 8.81e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 18 | 252 | 8.81e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 8.81e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 2 | 23 | 8.81e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 1 | 95 | 8.82e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 8.82e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 1 | 56 | 8.82e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 4 | 59 | 8.83e-01 |
| GO:BP | GO:0098926 | postsynaptic signal transduction | 1 | 25 | 8.83e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 8.83e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 1 | 18 | 8.83e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 3 | 70 | 8.83e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 1 | 18 | 8.83e-01 |
| GO:BP | GO:0060271 | cilium assembly | 7 | 292 | 8.83e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 1 | 17 | 8.84e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 9 | 149 | 8.84e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 2 | 25 | 8.84e-01 |
| GO:BP | GO:0002643 | regulation of tolerance induction | 1 | 10 | 8.84e-01 |
| GO:BP | GO:0034162 | toll-like receptor 9 signaling pathway | 1 | 12 | 8.84e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 3 | 129 | 8.84e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 17 | 8.84e-01 |
| GO:BP | GO:0007566 | embryo implantation | 1 | 39 | 8.85e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 3 | 95 | 8.85e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 7 | 65 | 8.85e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 1 | 46 | 8.85e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 1 | 15 | 8.86e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 8.86e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 3 | 43 | 8.86e-01 |
| GO:BP | GO:0035455 | response to interferon-alpha | 1 | 16 | 8.86e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 3 | 7441 | 8.86e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 2 | 67 | 8.86e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 5 | 66 | 8.86e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 7 | 93 | 8.86e-01 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 2 | 16 | 8.86e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 3 | 44 | 8.86e-01 |
| GO:BP | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1 | 8 | 8.86e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 8.86e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 1 | 15 | 8.86e-01 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 1 | 15 | 8.86e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 8.86e-01 |
| GO:BP | GO:1905153 | regulation of membrane invagination | 1 | 11 | 8.86e-01 |
| GO:BP | GO:0034204 | lipid translocation | 2 | 38 | 8.86e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 3 | 41 | 8.86e-01 |
| GO:BP | GO:0035336 | long-chain fatty-acyl-CoA metabolic process | 1 | 19 | 8.86e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 6 | 158 | 8.86e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 8.86e-01 |
| GO:BP | GO:0060099 | regulation of phagocytosis, engulfment | 1 | 11 | 8.86e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 2 | 83 | 8.86e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 1 | 16 | 8.86e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 8.86e-01 |
| GO:BP | GO:0044782 | cilium organization | 24 | 313 | 8.86e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 2 | 37 | 8.86e-01 |
| GO:BP | GO:0045217 | cell-cell junction maintenance | 1 | 13 | 8.86e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 3 | 58 | 8.86e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 8.86e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 10 | 155 | 8.86e-01 |
| GO:BP | GO:0006959 | humoral immune response | 2 | 80 | 8.86e-01 |
| GO:BP | GO:0097035 | regulation of membrane lipid distribution | 3 | 43 | 8.86e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 8.86e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 6 | 69 | 8.86e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 7 | 106 | 8.87e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 3 | 32 | 8.87e-01 |
| GO:BP | GO:0042745 | circadian sleep/wake cycle | 1 | 11 | 8.87e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 1 | 36 | 8.87e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 43 | 486 | 8.87e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 2 | 22 | 8.87e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 1 | 15 | 8.87e-01 |
| GO:BP | GO:0060632 | regulation of microtubule-based movement | 1 | 31 | 8.87e-01 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 1 | 74 | 8.87e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 32 | 418 | 8.87e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 62 | 1075 | 8.88e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 2 | 32 | 8.88e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 4 | 83 | 8.89e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 3 | 37 | 8.89e-01 |
| GO:BP | GO:1990868 | response to chemokine | 3 | 37 | 8.89e-01 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 1 | 18 | 8.90e-01 |
| GO:BP | GO:0032502 | developmental process | 1 | 4494 | 8.90e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 1 | 29 | 8.90e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 1 | 30 | 8.90e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 1 | 34 | 8.90e-01 |
| GO:BP | GO:0043605 | amide catabolic process | 1 | 10 | 8.90e-01 |
| GO:BP | GO:0098743 | cell aggregation | 1 | 12 | 8.90e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 2 | 39 | 8.90e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 2 | 32 | 8.90e-01 |
| GO:BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 1 | 16 | 8.90e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 13 | 155 | 8.90e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 13 | 236 | 8.91e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 1 | 14 | 8.91e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 97 | 8.92e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 2 | 31 | 8.92e-01 |
| GO:BP | GO:0009190 | cyclic nucleotide biosynthetic process | 1 | 14 | 8.92e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 51 | 7552 | 8.92e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 7 | 105 | 8.92e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 2 | 29 | 8.92e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 2 | 31 | 8.92e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 4 | 72 | 8.92e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 8.92e-01 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 1 | 16 | 8.92e-01 |
| GO:BP | GO:0048284 | organelle fusion | 2 | 135 | 8.92e-01 |
| GO:BP | GO:0007612 | learning | 3 | 115 | 8.92e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 4 | 45 | 8.92e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 3 | 49 | 8.92e-01 |
| GO:BP | GO:0071715 | icosanoid transport | 1 | 24 | 8.92e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 8.92e-01 |
| GO:BP | GO:1902074 | response to salt | 1 | 271 | 8.92e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 8.92e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 1 | 21 | 8.92e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 3 | 37 | 8.92e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 7 | 179 | 8.93e-01 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 1 | 19 | 8.93e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 1 | 12 | 8.93e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 4 | 69 | 8.93e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 38 | 8.93e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 2 | 28 | 8.94e-01 |
| GO:BP | GO:0071318 | cellular response to ATP | 1 | 12 | 8.94e-01 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 1 | 8 | 8.94e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 4 | 34 | 8.94e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 9 | 1555 | 8.94e-01 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 1 | 18 | 8.94e-01 |
| GO:BP | GO:0007586 | digestion | 3 | 60 | 8.94e-01 |
| GO:BP | GO:0042886 | amide transport | 1 | 235 | 8.94e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 1 | 39 | 8.94e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 4 | 62 | 8.94e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 1 | 37 | 8.94e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 1 | 18 | 8.94e-01 |
| GO:BP | GO:0003012 | muscle system process | 26 | 334 | 8.95e-01 |
| GO:BP | GO:0010467 | gene expression | 2 | 4587 | 8.95e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 8.96e-01 |
| GO:BP | GO:0043299 | leukocyte degranulation | 1 | 41 | 8.97e-01 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 4 | 45 | 8.97e-01 |
| GO:BP | GO:0009566 | fertilization | 2 | 93 | 8.97e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 10 | 191 | 8.97e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 11 | 236 | 8.97e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 2 | 29 | 8.97e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 8 | 122 | 8.97e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 24 | 8.98e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 7 | 234 | 8.99e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 5 | 76 | 8.99e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 3 | 33 | 9.00e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 2 | 36 | 9.00e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 2 | 35 | 9.00e-01 |
| GO:BP | GO:0006379 | mRNA cleavage | 1 | 19 | 9.00e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 1 | 43 | 9.00e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 1 | 29 | 9.00e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 5 | 67 | 9.00e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 1 | 29 | 9.00e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 2 | 16 | 9.00e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 1 | 21 | 9.00e-01 |
| GO:BP | GO:0009306 | protein secretion | 1 | 262 | 9.00e-01 |
| GO:BP | GO:0035270 | endocrine system development | 6 | 86 | 9.01e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 1 | 262 | 9.01e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 11 | 551 | 9.01e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 2 | 16 | 9.01e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 1 | 28 | 9.01e-01 |
| GO:BP | GO:0046949 | fatty-acyl-CoA biosynthetic process | 1 | 20 | 9.02e-01 |
| GO:BP | GO:0009408 | response to heat | 2 | 84 | 9.02e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 8 | 146 | 9.02e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 3 | 45 | 9.02e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 9.02e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 2 | 448 | 9.03e-01 |
| GO:BP | GO:0050765 | negative regulation of phagocytosis | 1 | 15 | 9.03e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 8 | 110 | 9.03e-01 |
| GO:BP | GO:0050808 | synapse organization | 34 | 365 | 9.03e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 3 | 7069 | 9.03e-01 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 1 | 16 | 9.03e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 9.03e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 9.03e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 1 | 14 | 9.04e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 7 | 98 | 9.05e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 1 | 23 | 9.05e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 3 | 46 | 9.05e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 1 | 26 | 9.05e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 35 | 782 | 9.05e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 7 | 69 | 9.05e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 1 | 19 | 9.05e-01 |
| GO:BP | GO:0020027 | hemoglobin metabolic process | 1 | 16 | 9.05e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 1 | 266 | 9.05e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 1 | 37 | 9.05e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 5 | 139 | 9.06e-01 |
| GO:BP | GO:0046697 | decidualization | 1 | 19 | 9.06e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 1 | 24 | 9.06e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 1 | 18 | 9.06e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 2 | 22 | 9.06e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 5 | 57 | 9.06e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 9.07e-01 |
| GO:BP | GO:0032328 | alanine transport | 1 | 14 | 9.07e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 3 | 40 | 9.07e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 12 | 227 | 9.07e-01 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 3 | 31 | 9.07e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 9.07e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 1 | 121 | 9.08e-01 |
| GO:BP | GO:0010839 | negative regulation of keratinocyte proliferation | 1 | 19 | 9.08e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 1 | 27 | 9.08e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 9.08e-01 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 3 | 44 | 9.08e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 9.08e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 2 | 30 | 9.08e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 3 | 57 | 9.08e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 3 | 116 | 9.08e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 2 | 19 | 9.08e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 2 | 20 | 9.08e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 9 | 223 | 9.08e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 2 | 40 | 9.08e-01 |
| GO:BP | GO:0032098 | regulation of appetite | 1 | 8 | 9.08e-01 |
| GO:BP | GO:2000369 | regulation of clathrin-dependent endocytosis | 1 | 18 | 9.08e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 3 | 46 | 9.08e-01 |
| GO:BP | GO:0150077 | regulation of neuroinflammatory response | 1 | 13 | 9.08e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 2 | 86 | 9.08e-01 |
| GO:BP | GO:0007423 | sensory organ development | 2 | 396 | 9.08e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 1 | 38 | 9.08e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 2 | 108 | 9.10e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 1 | 10 | 9.10e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 9.10e-01 |
| GO:BP | GO:2000353 | positive regulation of endothelial cell apoptotic process | 1 | 13 | 9.10e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 4 | 81 | 9.10e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 9.11e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 9.11e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 9.11e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 1 | 40 | 9.11e-01 |
| GO:BP | GO:0045471 | response to ethanol | 1 | 73 | 9.11e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 26 | 9.11e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 1 | 23 | 9.11e-01 |
| GO:BP | GO:0042113 | B cell activation | 4 | 170 | 9.11e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 3 | 66 | 9.12e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 1 | 43 | 9.14e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 9.14e-01 |
| GO:BP | GO:0007411 | axon guidance | 13 | 185 | 9.14e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 2 | 556 | 9.14e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 8 | 105 | 9.14e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 17 | 242 | 9.14e-01 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 1 | 9 | 9.14e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 2 | 132 | 9.14e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 1 | 42 | 9.14e-01 |
| GO:BP | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 1 | 10 | 9.15e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 4 | 43 | 9.15e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 8 | 139 | 9.15e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 4 | 43 | 9.15e-01 |
| GO:BP | GO:0097428 | protein maturation by iron-sulfur cluster transfer | 1 | 17 | 9.15e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 9.15e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 3 | 32 | 9.15e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 4 | 101 | 9.16e-01 |
| GO:BP | GO:0010038 | response to metal ion | 18 | 264 | 9.16e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 1 | 112 | 9.16e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 1 | 26 | 9.16e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 1 | 15 | 9.17e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 9.17e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 13 | 186 | 9.17e-01 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 1 | 21 | 9.17e-01 |
| GO:BP | GO:0070166 | enamel mineralization | 1 | 11 | 9.17e-01 |
| GO:BP | GO:0030220 | platelet formation | 1 | 17 | 9.17e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 26 | 378 | 9.17e-01 |
| GO:BP | GO:0097164 | ammonium ion metabolic process | 1 | 14 | 9.17e-01 |
| GO:BP | GO:0003352 | regulation of cilium movement | 2 | 17 | 9.17e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 10 | 165 | 9.18e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 2 | 106 | 9.19e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 9.19e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 9.19e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 1 | 25 | 9.19e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 17 | 893 | 9.19e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 8 | 190 | 9.19e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 4 | 65 | 9.19e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 2 | 33 | 9.19e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 2 | 106 | 9.19e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 16 | 9.19e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 7 | 90 | 9.19e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 1 | 15 | 9.19e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 1 | 23 | 9.19e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 2 | 51 | 9.19e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 1 | 16 | 9.19e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 2 | 88 | 9.19e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 1 | 51 | 9.20e-01 |
| GO:BP | GO:0032715 | negative regulation of interleukin-6 production | 2 | 24 | 9.20e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 3 | 38 | 9.20e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 12 | 214 | 9.21e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 2 | 40 | 9.21e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 1 | 42 | 9.21e-01 |
| GO:BP | GO:0097028 | dendritic cell differentiation | 2 | 23 | 9.22e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 42 | 9.22e-01 |
| GO:BP | GO:0051168 | nuclear export | 1 | 150 | 9.23e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 1 | 31 | 9.23e-01 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 1 | 39 | 9.23e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 9.23e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 1 | 18 | 9.23e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 5 | 55 | 9.23e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 60 | 9.23e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 8 | 172 | 9.23e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 2 | 60 | 9.23e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 9.23e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 8 | 147 | 9.23e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 16 | 2890 | 9.23e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 9.23e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 9.23e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 9.23e-01 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 4 | 22 | 9.24e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 1 | 23 | 9.24e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 1 | 19 | 9.24e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 1 | 63 | 9.24e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 1 | 19 | 9.24e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 1 | 19 | 9.24e-01 |
| GO:BP | GO:0032094 | response to food | 2 | 22 | 9.24e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 9.24e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 7 | 131 | 9.24e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 3 | 84 | 9.24e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 9.24e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 4 | 74 | 9.24e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 2 | 47 | 9.24e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 9.24e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 5 | 90 | 9.25e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 1 | 34 | 9.25e-01 |
| GO:BP | GO:0070848 | response to growth factor | 6 | 572 | 9.26e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 9.26e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 9.26e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 4 | 40 | 9.26e-01 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 9.26e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 1 | 60 | 9.27e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 9.27e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 2 | 41 | 9.27e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 1 | 15 | 9.28e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 23 | 408 | 9.28e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 2 | 43 | 9.28e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 2 | 55 | 9.28e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 2 | 86 | 9.28e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 6 | 99 | 9.28e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 22 | 316 | 9.29e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 5 | 457 | 9.29e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 2 | 120 | 9.29e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 2 | 27 | 9.29e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 3 | 87 | 9.30e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 1 | 59 | 9.30e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 2 | 234 | 9.31e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 4 | 73 | 9.32e-01 |
| GO:BP | GO:0001942 | hair follicle development | 3 | 67 | 9.32e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 9.32e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 3 | 48 | 9.32e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 3 | 48 | 9.32e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 1 | 21 | 9.32e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 1 | 19 | 9.32e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 1 | 19 | 9.32e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 9.32e-01 |
| GO:BP | GO:0140029 | exocytic process | 5 | 69 | 9.33e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 6 | 90 | 9.33e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 1 | 25 | 9.33e-01 |
| GO:BP | GO:0036075 | replacement ossification | 1 | 25 | 9.33e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 22 | 269 | 9.34e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 38 | 544 | 9.34e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 2 | 32 | 9.34e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 2 | 52 | 9.36e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 10 | 116 | 9.36e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 2 | 25 | 9.36e-01 |
| GO:BP | GO:0003013 | circulatory system process | 19 | 447 | 9.36e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 1 | 27 | 9.36e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 1 | 19 | 9.36e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 9.36e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 3 | 38 | 9.36e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 3 | 51 | 9.36e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 20 | 265 | 9.36e-01 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 1 | 20 | 9.36e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 1 | 26 | 9.36e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 1 | 27 | 9.36e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 1 | 27 | 9.36e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 2 | 27 | 9.36e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 1 | 28 | 9.36e-01 |
| GO:BP | GO:0031128 | developmental induction | 1 | 26 | 9.36e-01 |
| GO:BP | GO:0030431 | sleep | 1 | 13 | 9.36e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 9.36e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 9.36e-01 |
| GO:BP | GO:0055123 | digestive system development | 5 | 100 | 9.36e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 9.36e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 2 | 45 | 9.36e-01 |
| GO:BP | GO:0009651 | response to salt stress | 1 | 18 | 9.36e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 1 | 95 | 9.37e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 9.37e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 9.37e-01 |
| GO:BP | GO:0097503 | sialylation | 1 | 18 | 9.37e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 1 | 67 | 9.37e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 9.37e-01 |
| GO:BP | GO:0035601 | protein deacylation | 4 | 116 | 9.37e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 6 | 83 | 9.37e-01 |
| GO:BP | GO:0008015 | blood circulation | 16 | 374 | 9.37e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 4 | 40 | 9.37e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 6 | 68 | 9.37e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 5 | 74 | 9.37e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 3 | 60 | 9.38e-01 |
| GO:BP | GO:0008037 | cell recognition | 1 | 87 | 9.38e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 3 | 32 | 9.38e-01 |
| GO:BP | GO:0051216 | cartilage development | 11 | 151 | 9.39e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 9.41e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 9.41e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 2 | 44 | 9.41e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 2 | 71 | 9.41e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 9.41e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 2 | 115 | 9.41e-01 |
| GO:BP | GO:0009311 | oligosaccharide metabolic process | 1 | 43 | 9.41e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 4 | 115 | 9.41e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 8 | 199 | 9.41e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 2 | 54 | 9.42e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 40 | 9.42e-01 |
| GO:BP | GO:0022405 | hair cycle process | 3 | 69 | 9.42e-01 |
| GO:BP | GO:0022404 | molting cycle process | 3 | 69 | 9.42e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 3 | 58 | 9.43e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 3 | 605 | 9.43e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 1 | 20 | 9.43e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 1 | 30 | 9.43e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 4 | 40 | 9.43e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 9.43e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 1 | 19 | 9.43e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 1 | 14 | 9.43e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 4 | 40 | 9.43e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 1 | 36 | 9.44e-01 |
| GO:BP | GO:0006417 | regulation of translation | 21 | 370 | 9.44e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 1 | 44 | 9.44e-01 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 1 | 20 | 9.44e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 8 | 94 | 9.44e-01 |
| GO:BP | GO:0044000 | movement in host | 2 | 138 | 9.44e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 1 | 79 | 9.44e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 4 | 74 | 9.44e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 2 | 36 | 9.45e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 1 | 28 | 9.45e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 2 | 36 | 9.45e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 1 | 20 | 9.45e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 9.45e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 8 | 111 | 9.45e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 5 | 78 | 9.45e-01 |
| GO:BP | GO:0051769 | regulation of nitric-oxide synthase biosynthetic process | 1 | 13 | 9.45e-01 |
| GO:BP | GO:0051767 | nitric-oxide synthase biosynthetic process | 1 | 13 | 9.45e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 1 | 76 | 9.45e-01 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 19 | 9.45e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 9 | 163 | 9.45e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 1 | 45 | 9.45e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 5 | 59 | 9.45e-01 |
| GO:BP | GO:0043302 | positive regulation of leukocyte degranulation | 1 | 15 | 9.45e-01 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 1 | 19 | 9.45e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 1 | 107 | 9.45e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 9 | 133 | 9.46e-01 |
| GO:BP | GO:0070988 | demethylation | 1 | 52 | 9.46e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 1 | 38 | 9.46e-01 |
| GO:BP | GO:0009110 | vitamin biosynthetic process | 1 | 14 | 9.47e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 6 | 94 | 9.47e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 6 | 94 | 9.47e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 2 | 58 | 9.47e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 31 | 830 | 9.47e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 22 | 401 | 9.47e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 1 | 31 | 9.48e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 2 | 46 | 9.48e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 3 | 7806 | 9.48e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 2 | 119 | 9.48e-01 |
| GO:BP | GO:0006403 | RNA localization | 11 | 182 | 9.48e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 1 | 38 | 9.48e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 4 | 119 | 9.48e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 9.48e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 9.49e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 1 | 317 | 9.49e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 5 | 96 | 9.49e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 9.49e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 1 | 81 | 9.49e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 4 | 73 | 9.49e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 9.49e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 4 | 104 | 9.49e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 4 | 104 | 9.49e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 1 | 40 | 9.49e-01 |
| GO:BP | GO:0016197 | endosomal transport | 16 | 231 | 9.49e-01 |
| GO:BP | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | 1 | 13 | 9.49e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 1 | 18 | 9.49e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 9 | 134 | 9.49e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 9.49e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 1 | 115 | 9.50e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 1 | 115 | 9.50e-01 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 1 | 29 | 9.50e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 9.50e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 1 | 35 | 9.50e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 1 | 26 | 9.51e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 2 | 42 | 9.51e-01 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 1 | 21 | 9.51e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 1 | 37 | 9.52e-01 |
| GO:BP | GO:0048668 | collateral sprouting | 1 | 25 | 9.52e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 3 | 49 | 9.52e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 1 | 55 | 9.52e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 9.52e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 1 | 22 | 9.53e-01 |
| GO:BP | GO:0048565 | digestive tract development | 7 | 92 | 9.53e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 3 | 64 | 9.53e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 1 | 58 | 9.53e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 8 | 120 | 9.53e-01 |
| GO:BP | GO:0044409 | entry into host | 1 | 113 | 9.53e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 7 | 113 | 9.53e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 9.54e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 9.54e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 1 | 28 | 9.54e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 1 | 532 | 9.54e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 26 | 387 | 9.54e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 25 | 381 | 9.54e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 1 | 20 | 9.55e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 1 | 39 | 9.55e-01 |
| GO:BP | GO:0043300 | regulation of leukocyte degranulation | 2 | 27 | 9.55e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 1 | 19 | 9.55e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 12 | 275 | 9.56e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 13 | 506 | 9.56e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 1 | 37 | 9.57e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 2 | 53 | 9.57e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 2 | 130 | 9.57e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 1 | 22 | 9.57e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 9.57e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 5 | 61 | 9.57e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 2 | 50 | 9.58e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 3 | 90 | 9.58e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 13 | 174 | 9.58e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 7 | 155 | 9.58e-01 |
| GO:BP | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 9.58e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 2 | 80 | 9.58e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 34 | 663 | 9.58e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 7 | 75 | 9.58e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 1 | 76 | 9.58e-01 |
| GO:BP | GO:0007617 | mating behavior | 1 | 21 | 9.58e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 1 | 24 | 9.58e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 5 | 154 | 9.58e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 9.58e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 1 | 138 | 9.58e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 1 | 22 | 9.58e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 9.58e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 2 | 45 | 9.59e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 4 | 58 | 9.59e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 6 | 73 | 9.59e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 1 | 19 | 9.59e-01 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 5 | 191 | 9.59e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 7 | 144 | 9.59e-01 |
| GO:BP | GO:0050658 | RNA transport | 7 | 144 | 9.59e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 1 | 21 | 9.59e-01 |
| GO:BP | GO:0002313 | mature B cell differentiation involved in immune response | 1 | 17 | 9.59e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 3 | 61 | 9.59e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 2 | 33 | 9.59e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 4 | 70 | 9.60e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 3 | 54 | 9.60e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 5 | 64 | 9.60e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 2 | 102 | 9.60e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 7 | 107 | 9.60e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 1 | 15 | 9.60e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 1 | 107 | 9.60e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 3 | 107 | 9.60e-01 |
| GO:BP | GO:0042476 | odontogenesis | 3 | 86 | 9.60e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 1 | 53 | 9.60e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 2 | 34 | 9.60e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 4 | 59 | 9.60e-01 |
| GO:BP | GO:0097186 | amelogenesis | 1 | 14 | 9.61e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 17 | 233 | 9.61e-01 |
| GO:BP | GO:0022406 | membrane docking | 5 | 84 | 9.61e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 1 | 14 | 9.61e-01 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 1 | 29 | 9.61e-01 |
| GO:BP | GO:0001773 | myeloid dendritic cell activation | 1 | 12 | 9.61e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 9 | 69 | 9.61e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 2 | 55 | 9.61e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 9.62e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 1 | 16 | 9.62e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 17 | 234 | 9.62e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 1 | 18 | 9.62e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 1 | 20 | 9.62e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 18 | 9.62e-01 |
| GO:BP | GO:0030324 | lung development | 7 | 146 | 9.62e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 1 | 31 | 9.63e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 1 | 80 | 9.63e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 3 | 62 | 9.63e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 4 | 102 | 9.63e-01 |
| GO:BP | GO:0042116 | macrophage activation | 3 | 57 | 9.63e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 4 | 113 | 9.63e-01 |
| GO:BP | GO:0060541 | respiratory system development | 9 | 163 | 9.63e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 7 | 96 | 9.63e-01 |
| GO:BP | GO:0061564 | axon development | 27 | 398 | 9.63e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1 | 21 | 9.64e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 9.64e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 9.64e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 30 | 404 | 9.64e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 2 | 36 | 9.64e-01 |
| GO:BP | GO:0030534 | adult behavior | 2 | 92 | 9.64e-01 |
| GO:BP | GO:0007601 | visual perception | 1 | 96 | 9.64e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 1 | 21 | 9.64e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 9.64e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 1 | 37 | 9.65e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 1 | 72 | 9.65e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 3 | 39 | 9.65e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 1 | 37 | 9.65e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 3 | 56 | 9.65e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 9.65e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 9.65e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 1 | 20 | 9.65e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 1 | 48 | 9.65e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 17 | 234 | 9.65e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 1 | 12 | 9.66e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 8 | 211 | 9.66e-01 |
| GO:BP | GO:0033574 | response to testosterone | 1 | 29 | 9.66e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 19 | 351 | 9.66e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 1 | 58 | 9.66e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 1 | 30 | 9.66e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 4 | 63 | 9.66e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 1 | 19 | 9.66e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 1 | 19 | 9.66e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 1 | 31 | 9.66e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 1 | 97 | 9.66e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 3 | 38 | 9.66e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 2 | 28 | 9.66e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 2 | 61 | 9.66e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 1 | 34 | 9.66e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 1 | 51 | 9.66e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 2 | 118 | 9.66e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 1 | 32 | 9.66e-01 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 5 | 197 | 9.66e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 2 | 37 | 9.66e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 3 | 59 | 9.66e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 9.66e-01 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 1 | 22 | 9.66e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 1 | 81 | 9.66e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 1 | 19 | 9.66e-01 |
| GO:BP | GO:0009409 | response to cold | 2 | 41 | 9.66e-01 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 2 | 43 | 9.66e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 9.66e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 1 | 59 | 9.66e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 1 | 84 | 9.66e-01 |
| GO:BP | GO:0001525 | angiogenesis | 9 | 390 | 9.66e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 1 | 37 | 9.67e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 3 | 40 | 9.67e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 2 | 33 | 9.67e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 29 | 9.67e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 1 | 30 | 9.67e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 39 | 9.68e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 2 | 73 | 9.68e-01 |
| GO:BP | GO:0006887 | exocytosis | 11 | 256 | 9.68e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 7 | 150 | 9.68e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 1 | 37 | 9.68e-01 |
| GO:BP | GO:0007528 | neuromuscular junction development | 1 | 39 | 9.69e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 5 | 106 | 9.69e-01 |
| GO:BP | GO:0002507 | tolerance induction | 1 | 15 | 9.69e-01 |
| GO:BP | GO:0032692 | negative regulation of interleukin-1 production | 1 | 15 | 9.69e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 8 | 159 | 9.71e-01 |
| GO:BP | GO:0060219 | camera-type eye photoreceptor cell differentiation | 1 | 18 | 9.71e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 2 | 45 | 9.71e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 9.71e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 1 | 50 | 9.71e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 1 | 30 | 9.71e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 1 | 31 | 9.71e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 1 | 104 | 9.72e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 9.73e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 5 | 170 | 9.73e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 5 | 170 | 9.73e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 8 | 135 | 9.73e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 2 | 73 | 9.73e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 65 | 9.73e-01 |
| GO:BP | GO:0003341 | cilium movement | 12 | 97 | 9.74e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 3 | 42 | 9.74e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 8 | 640 | 9.74e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 2 | 54 | 9.74e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 3 | 316 | 9.74e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 1 | 14 | 9.74e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 9 | 151 | 9.74e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 9.74e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 3 | 61 | 9.74e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 2 | 153 | 9.74e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 10 | 172 | 9.74e-01 |
| GO:BP | GO:0015721 | bile acid and bile salt transport | 1 | 11 | 9.74e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 9.74e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 9.74e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 9.74e-01 |
| GO:BP | GO:0017121 | plasma membrane phospholipid scrambling | 1 | 13 | 9.74e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 2 | 45 | 9.74e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 29 | 344 | 9.75e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 9.75e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 9.75e-01 |
| GO:BP | GO:0050482 | arachidonic acid secretion | 1 | 11 | 9.75e-01 |
| GO:BP | GO:1903963 | arachidonate transport | 1 | 11 | 9.75e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 1 | 448 | 9.75e-01 |
| GO:BP | GO:0001709 | cell fate determination | 1 | 25 | 9.75e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 2 | 47 | 9.75e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 2 | 111 | 9.75e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 2 | 30 | 9.75e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 1 | 15 | 9.75e-01 |
| GO:BP | GO:0001654 | eye development | 11 | 272 | 9.75e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 3 | 39 | 9.75e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 6 | 85 | 9.75e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 2 | 83 | 9.76e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 5 | 91 | 9.76e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 3 | 51 | 9.76e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 2 | 49 | 9.76e-01 |
| GO:BP | GO:0015695 | organic cation transport | 2 | 34 | 9.76e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 5 | 147 | 9.76e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 2 | 44 | 9.76e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 1 | 57 | 9.77e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 1 | 53 | 9.77e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 1 | 382 | 9.77e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 14 | 295 | 9.77e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 2 | 67 | 9.77e-01 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 1 | 29 | 9.77e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 12 | 189 | 9.78e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 21 | 1507 | 9.78e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 1 | 30 | 9.78e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 9.78e-01 |
| GO:BP | GO:0048278 | vesicle docking | 3 | 57 | 9.78e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 12 | 1107 | 9.78e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 9.79e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 1 | 26 | 9.79e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 1 | 20 | 9.79e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 2 | 118 | 9.79e-01 |
| GO:BP | GO:0150063 | visual system development | 11 | 273 | 9.79e-01 |
| GO:BP | GO:0015918 | sterol transport | 2 | 92 | 9.80e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 4 | 78 | 9.80e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 2 | 26 | 9.80e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 3 | 49 | 9.80e-01 |
| GO:BP | GO:0002717 | positive regulation of natural killer cell mediated immunity | 1 | 11 | 9.80e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 2 | 119 | 9.80e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 1 | 26 | 9.80e-01 |
| GO:BP | GO:0007498 | mesoderm development | 4 | 89 | 9.80e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 2 | 119 | 9.80e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 4 | 74 | 9.80e-01 |
| GO:BP | GO:0045730 | respiratory burst | 1 | 21 | 9.80e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 23 | 512 | 9.80e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 8 | 504 | 9.81e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 1 | 28 | 9.81e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 2 | 130 | 9.81e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 10 | 142 | 9.81e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 30 | 9.81e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 1 | 25 | 9.82e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 9.82e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 3 | 1248 | 9.82e-01 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 1 | 20 | 9.82e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 2 | 59 | 9.82e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 1 | 28 | 9.82e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 2 | 59 | 9.82e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 9.82e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 1 | 25 | 9.82e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 9.82e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 1 | 28 | 9.82e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 1 | 199 | 9.82e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 3 | 195 | 9.82e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 1 | 40 | 9.82e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 9.82e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 1 | 52 | 9.82e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 9.82e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 9.82e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 1 | 22 | 9.82e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 29 | 9.82e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 9.82e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 3 | 142 | 9.82e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 2 | 43 | 9.82e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 4 | 185 | 9.82e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 1 | 52 | 9.83e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 1 | 30 | 9.83e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 1 | 26 | 9.83e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 1 | 62 | 9.83e-01 |
| GO:BP | GO:0050821 | protein stabilization | 1 | 191 | 9.83e-01 |
| GO:BP | GO:0001944 | vasculature development | 21 | 559 | 9.83e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 2 | 140 | 9.83e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 1 | 31 | 9.83e-01 |
| GO:BP | GO:0008347 | glial cell migration | 1 | 47 | 9.83e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 5 | 52 | 9.83e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 1 | 43 | 9.83e-01 |
| GO:BP | GO:0048880 | sensory system development | 11 | 275 | 9.83e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 7 | 116 | 9.83e-01 |
| GO:BP | GO:0002335 | mature B cell differentiation | 1 | 21 | 9.84e-01 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 1 | 38 | 9.84e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 9.84e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 9.84e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 3 | 957 | 9.84e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 1 | 166 | 9.84e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 5 | 68 | 9.85e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 9 | 180 | 9.85e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 3 | 161 | 9.85e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 1 | 20 | 9.85e-01 |
| GO:BP | GO:0003002 | regionalization | 1 | 273 | 9.85e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 3 | 49 | 9.85e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 8 | 138 | 9.85e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 32 | 9.85e-01 |
| GO:BP | GO:0032609 | type II interferon production | 5 | 53 | 9.85e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 2 | 57 | 9.85e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 6 | 84 | 9.85e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 1 | 27 | 9.85e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 1 | 27 | 9.85e-01 |
| GO:BP | GO:0045332 | phospholipid translocation | 1 | 33 | 9.85e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 2 | 33 | 9.86e-01 |
| GO:BP | GO:0006954 | inflammatory response | 1 | 429 | 9.86e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 1 | 94 | 9.86e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 14 | 218 | 9.86e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 1 | 34 | 9.86e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 9.86e-01 |
| GO:BP | GO:0042303 | molting cycle | 3 | 78 | 9.86e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 5 | 51 | 9.86e-01 |
| GO:BP | GO:0042633 | hair cycle | 3 | 78 | 9.86e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 9.86e-01 |
| GO:BP | GO:0032868 | response to insulin | 16 | 215 | 9.86e-01 |
| GO:BP | GO:0016050 | vesicle organization | 1 | 320 | 9.86e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 5 | 90 | 9.86e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 1 | 114 | 9.86e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 2 | 149 | 9.86e-01 |
| GO:BP | GO:0061448 | connective tissue development | 14 | 208 | 9.86e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2 | 56 | 9.86e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 2 | 35 | 9.86e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 3 | 70 | 9.87e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 1 | 418 | 9.87e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 4 | 69 | 9.87e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 4 | 179 | 9.87e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 1 | 71 | 9.87e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 1 | 23 | 9.87e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 9 | 74 | 9.88e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 9 | 74 | 9.88e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 10 | 462 | 9.88e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 2 | 52 | 9.88e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 2 | 185 | 9.89e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 4 | 59 | 9.89e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 9.89e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 1 | 26 | 9.89e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 1 | 57 | 9.89e-01 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 1 | 38 | 9.89e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 9.89e-01 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 1 | 38 | 9.89e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 26 | 406 | 9.89e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 2 | 82 | 9.89e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 18 | 252 | 9.89e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 1 | 30 | 9.89e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 2 | 1006 | 9.89e-01 |
| GO:BP | GO:0042592 | homeostatic process | 2 | 1159 | 9.89e-01 |
| GO:BP | GO:0017145 | stem cell division | 1 | 26 | 9.89e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 4 | 131 | 9.90e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 9.90e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 23 | 527 | 9.90e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 9.90e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 1 | 539 | 9.90e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 4 | 56 | 9.90e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 19 | 1399 | 9.90e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 25 | 676 | 9.90e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 1 | 43 | 9.90e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 12 | 218 | 9.90e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 9.90e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 2 | 161 | 9.90e-01 |
| GO:BP | GO:0035295 | tube development | 19 | 821 | 9.90e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 1 | 47 | 9.90e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 9.90e-01 |
| GO:BP | GO:0007409 | axonogenesis | 22 | 359 | 9.90e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 1 | 21 | 9.90e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 2 | 162 | 9.90e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 2 | 37 | 9.90e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 1 | 28 | 9.91e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 1 | 40 | 9.91e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 9.91e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 9.91e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 13 | 192 | 9.92e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 7 | 249 | 9.92e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 7 | 178 | 9.92e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 1 | 222 | 9.92e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 6 | 105 | 9.92e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 9.92e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 44 | 482 | 9.92e-01 |
| GO:BP | GO:0043038 | amino acid activation | 1 | 44 | 9.92e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 3 | 58 | 9.92e-01 |
| GO:BP | GO:0042908 | xenobiotic transport | 1 | 30 | 9.92e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 1 | 27 | 9.92e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 23 | 531 | 9.92e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 9.92e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 3 | 50 | 9.92e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 1 | 27 | 9.92e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 2 | 42 | 9.92e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 9 | 194 | 9.93e-01 |
| GO:BP | GO:0009611 | response to wounding | 16 | 415 | 9.93e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 2 | 92 | 9.93e-01 |
| GO:BP | GO:0140894 | endolysosomal toll-like receptor signaling pathway | 1 | 24 | 9.93e-01 |
| GO:BP | GO:0006413 | translational initiation | 3 | 116 | 9.93e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 9.93e-01 |
| GO:BP | GO:0031424 | keratinization | 1 | 13 | 9.93e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 9.93e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 9.93e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 9.93e-01 |
| GO:BP | GO:0007389 | pattern specification process | 1 | 306 | 9.93e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 6 | 132 | 9.93e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 21 | 1564 | 9.93e-01 |
| GO:BP | GO:0045058 | T cell selection | 1 | 22 | 9.94e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 2 | 199 | 9.94e-01 |
| GO:BP | GO:1901654 | response to ketone | 9 | 147 | 9.94e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 1 | 28 | 9.94e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 1 | 24 | 9.94e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 2 | 46 | 9.95e-01 |
| GO:BP | GO:0032602 | chemokine production | 2 | 46 | 9.95e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 1 | 229 | 9.95e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 5 | 296 | 9.95e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 9.95e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 9.95e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 4 | 112 | 9.95e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 1 | 40 | 9.95e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 2 | 53 | 9.95e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 2 | 63 | 9.95e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 4 | 86 | 9.95e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 1 | 52 | 9.95e-01 |
| GO:BP | GO:0007616 | long-term memory | 1 | 31 | 9.95e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 2 | 56 | 9.95e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 9.95e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 9.95e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 9.95e-01 |
| GO:BP | GO:0060348 | bone development | 2 | 170 | 9.95e-01 |
| GO:BP | GO:0001568 | blood vessel development | 25 | 534 | 9.95e-01 |
| GO:BP | GO:0006897 | endocytosis | 1 | 527 | 9.95e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 1 | 50 | 9.95e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 4 | 115 | 9.95e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 8 | 149 | 9.95e-01 |
| GO:BP | GO:0090659 | walking behavior | 1 | 25 | 9.95e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 2 | 38 | 9.95e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 1 | 19 | 9.95e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 2 | 87 | 9.95e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 9.95e-01 |
| GO:BP | GO:0006884 | cell volume homeostasis | 1 | 24 | 9.95e-01 |
| GO:BP | GO:0072359 | circulatory system development | 42 | 884 | 9.95e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 2 | 58 | 9.95e-01 |
| GO:BP | GO:1904645 | response to amyloid-beta | 1 | 32 | 9.96e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 9.96e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 28 | 376 | 9.96e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 28 | 9.96e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 1 | 114 | 9.97e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 1 | 23 | 9.97e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 2 | 40 | 9.97e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 2 | 65 | 9.97e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 9.97e-01 |
| GO:BP | GO:0051607 | defense response to virus | 4 | 202 | 9.97e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 2 | 38 | 9.97e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 1 | 95 | 9.97e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 9.97e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 2 | 209 | 9.97e-01 |
| GO:BP | GO:0098751 | bone cell development | 1 | 26 | 9.97e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 3 | 39 | 9.97e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 6 | 118 | 9.97e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 1 | 30 | 9.97e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 4 | 202 | 9.97e-01 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 1 | 19 | 9.97e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 20 | 463 | 9.97e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 1 | 20 | 9.97e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 3 | 73 | 9.97e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 2 | 1074 | 9.97e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 2 | 1076 | 9.97e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 4 | 194 | 9.97e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 9.98e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 23 | 311 | 9.98e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 3 | 44 | 9.99e-01 |
| GO:BP | GO:0006821 | chloride transport | 1 | 58 | 9.99e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 3 | 67 | 9.99e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 7 | 90 | 9.99e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 17 | 1322 | 9.99e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 3 | 40 | 9.99e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 2 | 52 | 9.99e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 6 | 91 | 1.00e+00 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 6 | 91 | 1.00e+00 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 1 | 39 | 1.00e+00 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 1 | 87 | 1.00e+00 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 5 | 422 | 1.00e+00 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 1.00e+00 |
| GO:BP | GO:0009791 | post-embryonic development | 1 | 74 | 1.00e+00 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 26 | 742 | 1.00e+00 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 21 | 1604 | 1.00e+00 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 11 | 116 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0010033 | response to organic substance | 21 | 1957 | 1.00e+00 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 9 | 468 | 1.00e+00 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 1.00e+00 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 1.00e+00 |
| GO:BP | GO:0010876 | lipid localization | 5 | 310 | 1.00e+00 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 1.00e+00 |
| GO:BP | GO:0015711 | organic anion transport | 4 | 264 | 1.00e+00 |
| GO:BP | GO:0015748 | organophosphate ester transport | 3 | 106 | 1.00e+00 |
| GO:BP | GO:0015844 | monoamine transport | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0015849 | organic acid transport | 3 | 217 | 1.00e+00 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 2 | 174 | 1.00e+00 |
| GO:BP | GO:0015914 | phospholipid transport | 2 | 75 | 1.00e+00 |
| GO:BP | GO:0016071 | mRNA metabolic process | 2 | 665 | 1.00e+00 |
| GO:BP | GO:0016072 | rRNA metabolic process | 5 | 258 | 1.00e+00 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 1.00e+00 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 1 | 1238 | 1.00e+00 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 2 | 65 | 1.00e+00 |
| GO:BP | GO:0009725 | response to hormone | 1 | 642 | 1.00e+00 |
| GO:BP | GO:0016485 | protein processing | 5 | 183 | 1.00e+00 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 12 | 1278 | 1.00e+00 |
| GO:BP | GO:0009620 | response to fungus | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 2 | 31 | 1.00e+00 |
| GO:BP | GO:0007608 | sensory perception of smell | 2 | 16 | 1.00e+00 |
| GO:BP | GO:0007610 | behavior | 3 | 436 | 1.00e+00 |
| GO:BP | GO:0008033 | tRNA processing | 1 | 128 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0008360 | regulation of cell shape | 5 | 126 | 1.00e+00 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 1.00e+00 |
| GO:BP | GO:0008544 | epidermis development | 17 | 210 | 1.00e+00 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 4 | 560 | 1.00e+00 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 1 | 142 | 1.00e+00 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1 | 1974 | 1.00e+00 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 1 | 97 | 1.00e+00 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 1 | 137 | 1.00e+00 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 1 | 96 | 1.00e+00 |
| GO:BP | GO:0009451 | RNA modification | 1 | 162 | 1.00e+00 |
| GO:BP | GO:0009581 | detection of external stimulus | 4 | 67 | 1.00e+00 |
| GO:BP | GO:0009583 | detection of light stimulus | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0009584 | detection of visible light | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0009605 | response to external stimulus | 2 | 1719 | 1.00e+00 |
| GO:BP | GO:0009607 | response to biotic stimulus | 1 | 850 | 1.00e+00 |
| GO:BP | GO:0009615 | response to virus | 5 | 275 | 1.00e+00 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 1.00e+00 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 2 | 144 | 1.00e+00 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 250 | 1.00e+00 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 6 | 274 | 1.00e+00 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 48 | 1.00e+00 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0031341 | regulation of cell killing | 2 | 36 | 1.00e+00 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0031349 | positive regulation of defense response | 14 | 251 | 1.00e+00 |
| GO:BP | GO:0031623 | receptor internalization | 2 | 95 | 1.00e+00 |
| GO:BP | GO:0031638 | zymogen activation | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0031960 | response to corticosteroid | 3 | 109 | 1.00e+00 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 1 | 56 | 1.00e+00 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 1.00e+00 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 18 | 337 | 1.00e+00 |
| GO:BP | GO:0031175 | neuron projection development | 1 | 780 | 1.00e+00 |
| GO:BP | GO:0032309 | icosanoid secretion | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0032526 | response to retinoic acid | 4 | 85 | 1.00e+00 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 1.00e+00 |
| GO:BP | GO:0032570 | response to progesterone | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 1.00e+00 |
| GO:BP | GO:0032635 | interleukin-6 production | 6 | 86 | 1.00e+00 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 1.00e+00 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 6 | 86 | 1.00e+00 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 2 | 55 | 1.00e+00 |
| GO:BP | GO:0032940 | secretion by cell | 1 | 569 | 1.00e+00 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 22 | 639 | 1.00e+00 |
| GO:BP | GO:0032990 | cell part morphogenesis | 23 | 551 | 1.00e+00 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 1.00e+00 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 1.00e+00 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 3 | 103 | 1.00e+00 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 227 | 1.00e+00 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 2 | 66 | 1.00e+00 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 1.00e+00 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 6 | 108 | 1.00e+00 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0022008 | neurogenesis | 3 | 1294 | 1.00e+00 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 13 | 295 | 1.00e+00 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 9 | 189 | 1.00e+00 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 28 | 691 | 1.00e+00 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 6 | 220 | 1.00e+00 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 1.00e+00 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 1.00e+00 |
| GO:BP | GO:0030001 | metal ion transport | 22 | 562 | 1.00e+00 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 3 | 260 | 1.00e+00 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 2 | 310 | 1.00e+00 |
| GO:BP | GO:0030100 | regulation of endocytosis | 8 | 211 | 1.00e+00 |
| GO:BP | GO:0030168 | platelet activation | 1 | 91 | 1.00e+00 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 5 | 140 | 1.00e+00 |
| GO:BP | GO:0030182 | neuron differentiation | 2 | 1061 | 1.00e+00 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 1 | 50 | 1.00e+00 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 7 | 75 | 1.00e+00 |
| GO:BP | GO:0030225 | macrophage differentiation | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0030316 | osteoclast differentiation | 3 | 66 | 1.00e+00 |
| GO:BP | GO:0030317 | flagellated sperm motility | 6 | 61 | 1.00e+00 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 1.00e+00 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 25 | 407 | 1.00e+00 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 1.00e+00 |
| GO:BP | GO:0033866 | nucleoside bisphosphate biosynthetic process | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0007599 | hemostasis | 11 | 153 | 1.00e+00 |
| GO:BP | GO:0007565 | female pregnancy | 6 | 122 | 1.00e+00 |
| GO:BP | GO:0002252 | immune effector process | 4 | 318 | 1.00e+00 |
| GO:BP | GO:0002253 | activation of immune response | 4 | 272 | 1.00e+00 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 1.00e+00 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 3 | 55 | 1.00e+00 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 4 | 70 | 1.00e+00 |
| GO:BP | GO:0002376 | immune system process | 2 | 1447 | 1.00e+00 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 4 | 151 | 1.00e+00 |
| GO:BP | GO:0002521 | leukocyte differentiation | 3 | 382 | 1.00e+00 |
| GO:BP | GO:0002526 | acute inflammatory response | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 2 | 151 | 1.00e+00 |
| GO:BP | GO:0002682 | regulation of immune system process | 1 | 877 | 1.00e+00 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 1.00e+00 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 9 | 289 | 1.00e+00 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 5 | 348 | 1.00e+00 |
| GO:BP | GO:0002697 | regulation of immune effector process | 2 | 190 | 1.00e+00 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 1.00e+00 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 4 | 70 | 1.00e+00 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 2 | 68 | 1.00e+00 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 4 | 230 | 1.00e+00 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 1 | 122 | 1.00e+00 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 2 | 77 | 1.00e+00 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 4 | 242 | 1.00e+00 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 4 | 162 | 1.00e+00 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 1 | 581 | 1.00e+00 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 1 | 70 | 1.00e+00 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 1.00e+00 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 3 | 115 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 1.00e+00 |
| GO:BP | GO:0000348 | mRNA branch site recognition | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 1.00e+00 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 1.00e+00 |
| GO:BP | GO:0000902 | cell morphogenesis | 27 | 761 | 1.00e+00 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 20 | 516 | 1.00e+00 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 1 | 50 | 1.00e+00 |
| GO:BP | GO:0001501 | skeletal system development | 24 | 377 | 1.00e+00 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 2 | 43 | 1.00e+00 |
| GO:BP | GO:0001704 | formation of primary germ layer | 2 | 96 | 1.00e+00 |
| GO:BP | GO:0001708 | cell fate specification | 1 | 56 | 1.00e+00 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 1.00e+00 |
| GO:BP | GO:0001816 | cytokine production | 2 | 451 | 1.00e+00 |
| GO:BP | GO:0001817 | regulation of cytokine production | 32 | 447 | 1.00e+00 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 12 | 165 | 1.00e+00 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 1 | 276 | 1.00e+00 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 1.00e+00 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 1.00e+00 |
| GO:BP | GO:0002218 | activation of innate immune response | 5 | 142 | 1.00e+00 |
| GO:BP | GO:0001706 | endoderm formation | 1 | 46 | 1.00e+00 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 1.00e+00 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 3 | 88 | 1.00e+00 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 9 | 192 | 1.00e+00 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 1.00e+00 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 3 | 103 | 1.00e+00 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 2 | 60 | 1.00e+00 |
| GO:BP | GO:0006909 | phagocytosis | 2 | 148 | 1.00e+00 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0006935 | chemotaxis | 1 | 389 | 1.00e+00 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 1.00e+00 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 1.00e+00 |
| GO:BP | GO:0006986 | response to unfolded protein | 3 | 128 | 1.00e+00 |
| GO:BP | GO:0007031 | peroxisome organization | 1 | 34 | 1.00e+00 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 11 | 234 | 1.00e+00 |
| GO:BP | GO:0006869 | lipid transport | 5 | 272 | 1.00e+00 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 4 | 1898 | 1.00e+00 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 1.00e+00 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 2 | 389 | 1.00e+00 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 9 | 118 | 1.00e+00 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 7 | 78 | 1.00e+00 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 3 | 40 | 1.00e+00 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0007254 | JNK cascade | 1 | 138 | 1.00e+00 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0007267 | cell-cell signaling | 4 | 1138 | 1.00e+00 |
| GO:BP | GO:0007369 | gastrulation | 4 | 146 | 1.00e+00 |
| GO:BP | GO:0007492 | endoderm development | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 2 | 763 | 1.00e+00 |
| GO:BP | GO:0006865 | amino acid transport | 1 | 102 | 1.00e+00 |
| GO:BP | GO:0006813 | potassium ion transport | 3 | 147 | 1.00e+00 |
| GO:BP | GO:0006812 | monoatomic cation transport | 23 | 669 | 1.00e+00 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 1.00e+00 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 10 | 433 | 1.00e+00 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 1.00e+00 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 4 | 209 | 1.00e+00 |
| GO:BP | GO:0006364 | rRNA processing | 3 | 221 | 1.00e+00 |
| GO:BP | GO:0006376 | mRNA splice site selection | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 1 | 54 | 1.00e+00 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 1.00e+00 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 1.00e+00 |
| GO:BP | GO:0006399 | tRNA metabolic process | 2 | 186 | 1.00e+00 |
| GO:BP | GO:0006400 | tRNA modification | 1 | 90 | 1.00e+00 |
| GO:BP | GO:0006405 | RNA export from nucleus | 1 | 78 | 1.00e+00 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 1 | 59 | 1.00e+00 |
| GO:BP | GO:0006412 | translation | 29 | 646 | 1.00e+00 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 1.00e+00 |
| GO:BP | GO:0006497 | protein lipidation | 3 | 89 | 1.00e+00 |
| GO:BP | GO:0006518 | peptide metabolic process | 15 | 771 | 1.00e+00 |
| GO:BP | GO:0006605 | protein targeting | 4 | 307 | 1.00e+00 |
| GO:BP | GO:0006612 | protein targeting to membrane | 2 | 118 | 1.00e+00 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 1 | 100 | 1.00e+00 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 1 | 56 | 1.00e+00 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 2 | 51 | 1.00e+00 |
| GO:BP | GO:0006811 | monoatomic ion transport | 29 | 775 | 1.00e+00 |
| GO:BP | GO:0007596 | blood coagulation | 7 | 150 | 1.00e+00 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 1.00e+00 |
| GO:BP | GO:0034030 | ribonucleoside bisphosphate biosynthetic process | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0034033 | purine nucleoside bisphosphate biosynthetic process | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0060326 | cell chemotaxis | 4 | 162 | 1.00e+00 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 3 | 51 | 1.00e+00 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 19 | 403 | 1.00e+00 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 1 | 48 | 1.00e+00 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 1.00e+00 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0061614 | miRNA transcription | 1 | 57 | 1.00e+00 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 6 | 246 | 1.00e+00 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 1 | 104 | 1.00e+00 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 3 | 68 | 1.00e+00 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 70 | 1.00e+00 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 1 | 191 | 1.00e+00 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 6 | 121 | 1.00e+00 |
| GO:BP | GO:0070555 | response to interleukin-1 | 3 | 83 | 1.00e+00 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 6 | 123 | 1.00e+00 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 21 | 1869 | 1.00e+00 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 1 | 78 | 1.00e+00 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 4 | 114 | 1.00e+00 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 4 | 110 | 1.00e+00 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 5 | 540 | 1.00e+00 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 1.00e+00 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 2 | 64 | 1.00e+00 |
| GO:BP | GO:0065007 | biological regulation | 2 | 8320 | 1.00e+00 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 3 | 52 | 1.00e+00 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 82 | 1.00e+00 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 2 | 44 | 1.00e+00 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 4 | 144 | 1.00e+00 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 1.00e+00 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 7 | 236 | 1.00e+00 |
| GO:BP | GO:0050890 | cognition | 1 | 227 | 1.00e+00 |
| GO:BP | GO:0050900 | leukocyte migration | 6 | 209 | 1.00e+00 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 3 | 39 | 1.00e+00 |
| GO:BP | GO:0051047 | positive regulation of secretion | 3 | 215 | 1.00e+00 |
| GO:BP | GO:0051048 | negative regulation of secretion | 4 | 111 | 1.00e+00 |
| GO:BP | GO:0051049 | regulation of transport | 2 | 1267 | 1.00e+00 |
| GO:BP | GO:0051050 | positive regulation of transport | 1 | 667 | 1.00e+00 |
| GO:BP | GO:0051051 | negative regulation of transport | 10 | 318 | 1.00e+00 |
| GO:BP | GO:0055085 | transmembrane transport | 24 | 998 | 1.00e+00 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 2 | 56 | 1.00e+00 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 3 | 2035 | 1.00e+00 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 4 | 1101 | 1.00e+00 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 9 | 748 | 1.00e+00 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 18 | 1441 | 1.00e+00 |
| GO:BP | GO:0051384 | response to glucocorticoid | 2 | 93 | 1.00e+00 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 1 | 47 | 1.00e+00 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 1 | 70 | 1.00e+00 |
| GO:BP | GO:0051604 | protein maturation | 5 | 259 | 1.00e+00 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 1.00e+00 |
| GO:BP | GO:0051668 | localization within membrane | 11 | 605 | 1.00e+00 |
| GO:BP | GO:0051707 | response to other organism | 1 | 820 | 1.00e+00 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 60 | 665 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 6 | 94 | 1.00e+00 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 1 | 129 | 1.00e+00 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 11 | 168 | 1.00e+00 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 3 | 82 | 1.00e+00 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 4 | 118 | 1.00e+00 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 1 | 56 | 1.00e+00 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 1.00e+00 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 1 | 55 | 1.00e+00 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 9 | 211 | 1.00e+00 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 5 | 157 | 1.00e+00 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 3 | 294 | 1.00e+00 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 1 | 35 | 1.00e+00 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 2 | 89 | 1.00e+00 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 15 | 1223 | 1.00e+00 |
| GO:BP | GO:1903307 | positive regulation of regulated secretory pathway | 1 | 32 | 1.00e+00 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 3 | 99 | 1.00e+00 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 3 | 199 | 1.00e+00 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 6 | 94 | 1.00e+00 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 2 | 57 | 1.00e+00 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 2 | 279 | 1.00e+00 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 4 | 118 | 1.00e+00 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 3 | 71 | 1.00e+00 |
| GO:BP | GO:1904037 | positive regulation of epithelial cell apoptotic process | 1 | 26 | 1.00e+00 |
| GO:BP | GO:1905517 | macrophage migration | 1 | 34 | 1.00e+00 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 1.00e+00 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 2 | 1014 | 1.00e+00 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 1 | 1315 | 1.00e+00 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 79 | 1484 | 1.00e+00 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 2 | 63 | 1.00e+00 |
| GO:BP | GO:1900046 | regulation of hemostasis | 1 | 50 | 1.00e+00 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 1.00e+00 |
| GO:BP | GO:0072337 | modified amino acid transport | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 2 | 72 | 1.00e+00 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 1 | 53 | 1.00e+00 |
| GO:BP | GO:0072657 | protein localization to membrane | 10 | 523 | 1.00e+00 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 2 | 40 | 1.00e+00 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 4 | 114 | 1.00e+00 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 4 | 250 | 1.00e+00 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 1 | 69 | 1.00e+00 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 1 | 120 | 1.00e+00 |
| GO:BP | GO:0097581 | lamellipodium organization | 3 | 78 | 1.00e+00 |
| GO:BP | GO:0097722 | sperm motility | 6 | 61 | 1.00e+00 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 1.00e+00 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 609 | 1.00e+00 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 2 | 48 | 1.00e+00 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 2 | 67 | 1.00e+00 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 20 | 540 | 1.00e+00 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 21 | 563 | 1.00e+00 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 20 | 517 | 1.00e+00 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 75 | 1.00e+00 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 2 | 40 | 1.00e+00 |
| GO:BP | GO:0140352 | export from cell | 1 | 611 | 1.00e+00 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 3 | 67 | 1.00e+00 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 6 | 124 | 1.00e+00 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 26 | 421 | 1.00e+00 |
| GO:BP | GO:0050865 | regulation of cell activation | 5 | 384 | 1.00e+00 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0042330 | taxis | 1 | 389 | 1.00e+00 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 1.00e+00 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 1.00e+00 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 12 | 666 | 1.00e+00 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 1 | 820 | 1.00e+00 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 16 | 373 | 1.00e+00 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 3 | 141 | 1.00e+00 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 4 | 82 | 1.00e+00 |
| GO:BP | GO:0043588 | skin development | 12 | 175 | 1.00e+00 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 1 | 73 | 1.00e+00 |
| GO:BP | GO:0043604 | amide biosynthetic process | 41 | 775 | 1.00e+00 |
| GO:BP | GO:0043631 | RNA polyadenylation | 1 | 41 | 1.00e+00 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 4 | 122 | 1.00e+00 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 1 | 963 | 1.00e+00 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 1 | 131 | 1.00e+00 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 1 | 135 | 1.00e+00 |
| GO:BP | GO:0044788 | modulation by host of viral process | 1 | 40 | 1.00e+00 |
| GO:BP | GO:0045047 | protein targeting to ER | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0045087 | innate immune response | 1 | 488 | 1.00e+00 |
| GO:BP | GO:0045088 | regulation of innate immune response | 10 | 232 | 1.00e+00 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 9 | 182 | 1.00e+00 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 1.00e+00 |
| GO:BP | GO:0045109 | intermediate filament organization | 2 | 21 | 1.00e+00 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 3 | 92 | 1.00e+00 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 1.00e+00 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 37 | 632 | 1.00e+00 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 1.00e+00 |
| GO:BP | GO:0034470 | ncRNA processing | 2 | 400 | 1.00e+00 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 3 | 561 | 1.00e+00 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 16 | 391 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 2 | 60 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 2 | 55 | 1.00e+00 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 3 | 64 | 1.00e+00 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 1.00e+00 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 5 | 111 | 1.00e+00 |
| GO:BP | GO:0042254 | ribosome biogenesis | 3 | 301 | 1.00e+00 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 1 | 38 | 1.00e+00 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 1 | 40 | 1.00e+00 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 1.00e+00 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 1.00e+00 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 4 | 429 | 1.00e+00 |
| GO:BP | GO:0042073 | intraciliary transport | 1 | 46 | 1.00e+00 |
| GO:BP | GO:0042098 | T cell proliferation | 5 | 120 | 1.00e+00 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 3 | 47 | 1.00e+00 |
| GO:BP | GO:0042110 | T cell activation | 2 | 313 | 1.00e+00 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 4 | 95 | 1.00e+00 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 3 | 88 | 1.00e+00 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 4 | 116 | 1.00e+00 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0045165 | cell fate commitment | 6 | 155 | 1.00e+00 |
| GO:BP | GO:0045292 | mRNA cis splicing, via spliceosome | 1 | 26 | 1.00e+00 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 1.00e+00 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 5 | 187 | 1.00e+00 |
| GO:BP | GO:0048568 | embryonic organ development | 11 | 316 | 1.00e+00 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 29 | 440 | 1.00e+00 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 19 | 870 | 1.00e+00 |
| GO:BP | GO:0048666 | neuron development | 1 | 877 | 1.00e+00 |
| GO:BP | GO:0048699 | generation of neurons | 3 | 1119 | 1.00e+00 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 1.00e+00 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 1.00e+00 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 1.00e+00 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 1.00e+00 |
| GO:BP | GO:0048545 | response to steroid hormone | 1 | 245 | 1.00e+00 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 5 | 105 | 1.00e+00 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 2 | 38 | 1.00e+00 |
| GO:BP | GO:0050776 | regulation of immune response | 2 | 484 | 1.00e+00 |
| GO:BP | GO:0050777 | negative regulation of immune response | 6 | 111 | 1.00e+00 |
| GO:BP | GO:0050778 | positive regulation of immune response | 2 | 369 | 1.00e+00 |
| GO:BP | GO:0050789 | regulation of biological process | 2 | 8061 | 1.00e+00 |
| GO:BP | GO:0050817 | coagulation | 8 | 153 | 1.00e+00 |
| GO:BP | GO:0050818 | regulation of coagulation | 2 | 55 | 1.00e+00 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 1 | 38 | 1.00e+00 |
| GO:BP | GO:0050832 | defense response to fungus | 1 | 14 | 1.00e+00 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 4 | 106 | 1.00e+00 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 1.00e+00 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 3 | 57 | 1.00e+00 |
| GO:BP | GO:0048536 | spleen development | 1 | 28 | 1.00e+00 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 1 | 42 | 1.00e+00 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 2 | 285 | 1.00e+00 |
| GO:BP | GO:0045453 | bone resorption | 1 | 40 | 1.00e+00 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 2 | 74 | 1.00e+00 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 2 | 1137 | 1.00e+00 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 2 | 620 | 1.00e+00 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 3 | 86 | 1.00e+00 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 9 | 152 | 1.00e+00 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 1 | 67 | 1.00e+00 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 2 | 38 | 1.00e+00 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 1 | 17 | 1.00e+00 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 5 | 104 | 1.00e+00 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 5 | 108 | 1.00e+00 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1315 | 1.00e+00 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 1.00e+00 |
| GO:BP | GO:0046034 | ATP metabolic process | 2 | 116 | 1.00e+00 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 2 | 55 | 1.00e+00 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 2 | 66 | 1.00e+00 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 2 | 45 | 1.00e+00 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 3 | 89 | 1.00e+00 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0046649 | lymphocyte activation | 6 | 465 | 1.00e+00 |
| GO:BP | GO:0046849 | bone remodeling | 3 | 59 | 1.00e+00 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 2 | 90 | 1.00e+00 |
| GO:BP | GO:0046903 | secretion | 1 | 645 | 1.00e+00 |
| GO:BP | GO:0046942 | carboxylic acid transport | 3 | 216 | 1.00e+00 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 1.00e+00 |
| GO:BP | GO:0050863 | regulation of T cell activation | 2 | 216 | 1.00e+00 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 14 | 320 | 1.00e+00 |
| KEGG | KEGG:04110 | Cell cycle | 51 | 151 | 1.99e-31 |
| KEGG | KEGG:03030 | DNA replication | 21 | 35 | 9.39e-18 |
| KEGG | KEGG:04115 | p53 signaling pathway | 19 | 65 | 3.73e-12 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 10 | 48 | 1.01e-07 |
| KEGG | KEGG:03440 | Homologous recombination | 14 | 39 | 1.06e-07 |
| KEGG | KEGG:03430 | Mismatch repair | 8 | 22 | 5.49e-06 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 12 | 86 | 1.17e-05 |
| KEGG | KEGG:04114 | Oocyte meiosis | 12 | 108 | 1.42e-05 |
| KEGG | KEGG:04218 | Cellular senescence | 15 | 145 | 7.56e-05 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 8 | 43 | 1.12e-04 |
| KEGG | KEGG:01524 | Platinum drug resistance | 7 | 65 | 3.57e-03 |
| KEGG | KEGG:03410 | Base excision repair | 9 | 33 | 4.23e-03 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 9 | 180 | 1.16e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 5 | 75 | 3.24e-02 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 17 | 172 | 3.51e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 5.26e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 3 | 109 | 5.88e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 1 | 57 | 7.20e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 7.20e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 7.20e-02 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 11 | 63 | 7.20e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 7.20e-02 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 7.20e-02 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 7.20e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 1 | 84 | 7.82e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 8.05e-02 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 4 | 61 | 9.02e-02 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 1 | 11 | 9.63e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 1.09e-01 |
| KEGG | KEGG:05224 | Breast cancer | 1 | 117 | 1.09e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 1.09e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 4 | 136 | 1.11e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 1 | 145 | 1.14e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 1.25e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 1 | 24 | 1.25e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 1.25e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 5 | 46 | 1.61e-01 |
| KEGG | KEGG:05034 | Alcoholism | 4 | 122 | 2.00e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 3 | 37 | 2.00e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 4 | 110 | 2.00e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 6 | 69 | 2.00e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 1 | 50 | 2.08e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 15 | 97 | 2.08e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 1 | 409 | 2.73e-01 |
| KEGG | KEGG:04146 | Peroxisome | 1 | 68 | 2.82e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 4 | 44 | 2.87e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 3 | 20 | 2.87e-01 |
| KEGG | KEGG:04210 | Apoptosis | 6 | 118 | 2.87e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 6 | 86 | 2.87e-01 |
| KEGG | KEGG:04217 | Necroptosis | 2 | 106 | 2.95e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 4 | 18 | 3.00e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 3 | 31 | 3.13e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 10 | 149 | 3.41e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 5 | 27 | 3.44e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 4 | 44 | 3.52e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 5 | 33 | 3.93e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 11 | 121 | 4.06e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 11 | 73 | 4.15e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 2 | 165 | 4.32e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 8 | 70 | 4.37e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 4 | 84 | 5.20e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 10 | 78 | 5.20e-01 |
| KEGG | KEGG:03265 | Virion - Lyssavirus | 1 | 3 | 5.20e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 1 | 30 | 5.38e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 2 | 17 | 6.33e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 4 | 85 | 6.75e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 3 | 25 | 6.96e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 3 | 19 | 6.96e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 7 | 49 | 6.98e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 4 | 24 | 7.13e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 10 | 76 | 7.45e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 6 | 60 | 7.45e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 4 | 44 | 7.52e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 8.08e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 8.39e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 3 | 10 | 8.58e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 2 | 38 | 8.59e-01 |
| KEGG | KEGG:04540 | Gap junction | 3 | 73 | 8.59e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 2 | 12 | 8.59e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 1 | 69 | 9.31e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 3 | 19 | 9.31e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 6 | 59 | 9.50e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 4 | 49 | 9.55e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 9.60e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 2 | 31 | 9.74e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 2 | 16 | 9.74e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 2 | 12 | 9.79e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 2 | 12 | 9.79e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 9.92e-01 |
| KEGG | KEGG:02010 | ABC transporters | 1 | 29 | 1.00e+00 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 5 | 108 | 1.00e+00 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 15 | 303 | 1.00e+00 |
| KEGG | KEGG:05012 | Parkinson disease | 12 | 226 | 1.00e+00 |
| KEGG | KEGG:03060 | Protein export | 1 | 23 | 1.00e+00 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 3 | 107 | 1.00e+00 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 10 | 157 | 1.00e+00 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 2 | 17 | 1.00e+00 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 1.00e+00 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 1.00e+00 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 1 | 22 | 1.00e+00 |
| KEGG | KEGG:05010 | Alzheimer disease | 19 | 315 | 1.00e+00 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 5 | 124 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 1 | 36 | 1.00e+00 |
| KEGG | KEGG:05134 | Legionellosis | 2 | 39 | 1.00e+00 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 3 | 240 | 1.00e+00 |
| KEGG | KEGG:05162 | Measles | 8 | 98 | 1.00e+00 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 3 | 145 | 1.00e+00 |
| KEGG | KEGG:01100 | Metabolic pathways | 1 | 1146 | 1.00e+00 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 5 | 77 | 1.00e+00 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 4 | 176 | 1.00e+00 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 1 | 35 | 1.00e+00 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 9 | 121 | 1.00e+00 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 3 | 23 | 1.00e+00 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 1 | 179 | 1.00e+00 |
| KEGG | KEGG:04140 | Autophagy - animal | 8 | 133 | 1.00e+00 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 5 | 133 | 1.00e+00 |
| KEGG | KEGG:04611 | Platelet activation | 1 | 89 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 6 | 98 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 1.00e+00 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 1.00e+00 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 1 | 164 | 1.00e+00 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 2 | 91 | 1.00e+00 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 2 | 103 | 1.00e+00 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 2 | 115 | 1.00e+00 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:05145 | Toxoplasmosis | 1 | 82 | 1.00e+00 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:04924 | Renin secretion | 2 | 50 | 1.00e+00 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 2 | 35 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 1 | 127 | 1.00e+00 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:04931 | Insulin resistance | 4 | 95 | 1.00e+00 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 1 | 131 | 1.00e+00 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 3 | 41 | 1.00e+00 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 6 | 50 | 1.00e+00 |
| KEGG | KEGG:05160 | Hepatitis C | 3 | 115 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 3 | 169 | 1.00e+00 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 1.00e+00 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 3 | 161 | 1.00e+00 |
| KEGG | KEGG:05416 | Viral myocarditis | 1 | 40 | 1.00e+00 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 2 | 74 | 1.00e+00 |
| KEGG | KEGG:04726 | Serotonergic synapse | 2 | 63 | 1.00e+00 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 3 | 43 | 1.00e+00 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 1.00e+00 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 3 | 46 | 1.00e+00 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 1.00e+00 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 1 | 37 | 1.00e+00 |
| KEGG | KEGG:05135 | Yersinia infection | 3 | 112 | 1.00e+00 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 3 | 272 | 1.00e+00 |
| KEGG | KEGG:05152 | Tuberculosis | 3 | 106 | 1.00e+00 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 6 | 110 | 1.00e+00 |
| KEGG | KEGG:04727 | GABAergic synapse | 3 | 55 | 1.00e+00 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 1 | 48 | 1.00e+00 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 5 | 117 | 1.00e+00 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 3 | 48 | 1.00e+00 |
| KEGG | KEGG:05016 | Huntington disease | 4 | 256 | 1.00e+00 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 1.00e+00 |
| KEGG | KEGG:04014 | Ras signaling pathway | 6 | 171 | 1.00e+00 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 4 | 56 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 4 | 415 | 1.00e+00 |
| KEGG | KEGG:04142 | Lysosome | 1 | 118 | 1.00e+00 |
| KEGG | KEGG:05143 | African trypanosomiasis | 1 | 16 | 1.00e+00 |
| KEGG | KEGG:05164 | Influenza A | 2 | 103 | 1.00e+00 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 2 | 20 | 1.00e+00 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 1 | 65 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 1.00e+00 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 1 | 56 | 1.00e+00 |
| KEGG | KEGG:04713 | Circadian entrainment | 4 | 69 | 1.00e+00 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 1 | 88 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 1 | 186 | 1.00e+00 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 1.00e+00 |
| KEGG | KEGG:04916 | Melanogenesis | 5 | 77 | 1.00e+00 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 3 | 142 | 1.00e+00 |
| KEGG | KEGG:00640 | Propanoate metabolism | 1 | 28 | 1.00e+00 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 2 | 97 | 1.00e+00 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 1 | 49 | 1.00e+00 |
| KEGG | KEGG:00650 | Butanoate metabolism | 1 | 17 | 1.00e+00 |
| KEGG | KEGG:04972 | Pancreatic secretion | 4 | 58 | 1.00e+00 |
| KEGG | KEGG:05032 | Morphine addiction | 7 | 56 | 1.00e+00 |
| KEGG | KEGG:05020 | Prion disease | 6 | 220 | 1.00e+00 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 1 | 86 | 1.00e+00 |
| KEGG | KEGG:04971 | Gastric acid secretion | 1 | 52 | 1.00e+00 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 4 | 106 | 1.00e+00 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 1 | 115 | 1.00e+00 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 1 | 6 | 1.00e+00 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 2 | 99 | 1.00e+00 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 1 | 110 | 1.00e+00 |
| KEGG | KEGG:03040 | Spliceosome | 6 | 132 | 1.00e+00 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 3 | 43 | 1.00e+00 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 1.00e+00 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 3 | 71 | 1.00e+00 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:04970 | Salivary secretion | 2 | 55 | 1.00e+00 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 4 | 51 | 1.00e+00 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 5 | 99 | 1.00e+00 |
| KEGG | KEGG:04720 | Long-term potentiation | 3 | 55 | 1.00e+00 |
| KEGG | KEGG:04714 | Thermogenesis | 9 | 195 | 1.00e+00 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 1 | 28 | 1.00e+00 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 1 | 31 | 1.00e+00 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 2 | 63 | 1.00e+00 |
| KEGG | KEGG:04710 | Circadian rhythm | 2 | 30 | 1.00e+00 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 3 | 55 | 1.00e+00 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 2 | 78 | 1.00e+00 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 1.00e+00 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 3 | 33 | 1.00e+00 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 1 | 103 | 1.00e+00 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 1.00e+00 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 1.00e+00 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 1.00e+00 |
| KEGG | KEGG:04740 | Olfactory transduction | 3 | 25 | 1.00e+00 |
| KEGG | KEGG:04976 | Bile secretion | 1 | 42 | 1.00e+00 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 1 | 46 | 1.00e+00 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 2 | 14 | 1.00e+00 |
| KEGG | KEGG:04742 | Taste transduction | 1 | 22 | 1.00e+00 |
| KEGG | KEGG:04978 | Mineral absorption | 4 | 42 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 1 | 19 | 1.00e+00 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 4 | 74 | 1.00e+00 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 12 | 135 | 1.00e+00 |
| KEGG | KEGG:04530 | Tight junction | 1 | 137 | 1.00e+00 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 6 | 98 | 1.00e+00 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 4 | 61 | 1.00e+00 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 2 | 29 | 1.00e+00 |
| KEGG | KEGG:05030 | Cocaine addiction | 2 | 35 | 1.00e+00 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 1 | 112 | 1.00e+00 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 3 | 51 | 1.00e+00 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 2 | 56 | 1.00e+00 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 1 | 7 | 1.00e+00 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 1 | 9 | 1.00e+00 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 4 | 83 | 1.00e+00 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 1.00e+00 |
| KEGG | KEGG:04520 | Adherens junction | 3 | 67 | 1.00e+00 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 3 | 79 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 1 | 89 | 1.00e+00 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 4 | 113 | 1.00e+00 |
| KEGG | KEGG:04725 | Cholinergic synapse | 3 | 83 | 1.00e+00 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 1 | 49 | 1.00e+00 |
| KEGG | KEGG:04744 | Phototransduction | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 1 | 4 | 1.00e+00 |
| KEGG | KEGG:05132 | Salmonella infection | 1 | 214 | 1.00e+00 |
| KEGG | KEGG:04144 | Endocytosis | 2 | 232 | 1.00e+00 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 3 | 72 | 1.00e+00 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 1 | 15 | 1.00e+00 |
| KEGG | KEGG:03020 | RNA polymerase | 1 | 34 | 1.00e+00 |
| KEGG | KEGG:04330 | Notch signaling pathway | 1 | 51 | 1.00e+00 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 1.00e+00 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 1 | 13 | 1.00e+00 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 3 | 84 | 1.00e+00 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 1 | 49 | 1.00e+00 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1 | 69 | 1.00e+00 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 1.00e+00 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 1 | 24 | 1.00e+00 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 2 | 44 | 1.00e+00 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 1.00e+00 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 1 | 104 | 1.00e+00 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 1 | 98 | 1.00e+00 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 1 | 14 | 1.00e+00 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 1 | 44 | 1.00e+00 |
| KEGG | KEGG:00920 | Sulfur metabolism | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:04929 | GnRH secretion | 1 | 53 | 1.00e+00 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 19 | 385 | 1.00e+00 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:04360 | Axon guidance | 8 | 162 | 1.00e+00 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 2 | 20 | 1.00e+00 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 1.00e+00 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 2 | 97 | 1.00e+00 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 2 | 23 | 1.00e+00 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 6 | 151 | 1.00e+00 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 1 | 38 | 1.00e+00 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 6 | 138 | 1.00e+00 |
| KEGG | KEGG:00052 | Galactose metabolism | 1 | 22 | 1.00e+00 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 3 | 43 | 1.00e+00 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 3 | 74 | 1.00e+00 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 7 | 91 | 1.00e+00 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 1.00e+00 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 4 | 67 | 1.00e+00 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 2 | 60 | 1.00e+00 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 2 | 66 | 1.00e+00 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 1 | 25 | 1.00e+00 |
| KEGG | KEGG:05142 | Chagas disease | 1 | 75 | 1.00e+00 |
| KEGG | KEGG:00071 | Fatty acid degradation | 2 | 32 | 1.00e+00 |
| KEGG | KEGG:03018 | RNA degradation | 2 | 74 | 1.00e+00 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 1.00e+00 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 9 | 106 | 1.00e+00 |
| KEGG | KEGG:04145 | Phagosome | 8 | 99 | 1.00e+00 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 3 | 58 | 1.00e+00 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 1.00e+00 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 1 | 22 | 1.00e+00 |
| KEGG | KEGG:04911 | Insulin secretion | 3 | 64 | 1.00e+00 |
| KEGG | KEGG:00830 | Retinol metabolism | 1 | 16 | 1.00e+00 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 4 | 76 | 1.00e+00 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 9 | 134 | 1.00e+00 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 1 | 87 | 1.00e+00 |
| KEGG | KEGG:03022 | Basal transcription factors | 1 | 40 | 1.00e+00 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 3 | 170 | 1.00e+00 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 2 | 94 | 1.00e+00 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 12 | 128 | 1.00e+00 |
| KEGG | KEGG:05131 | Shigellosis | 3 | 210 | 1.00e+00 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 3 | 41 | 1.00e+00 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 2 | 150 | 1.00e+00 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 6 | 84 | 1.00e+00 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 3 | 62 | 1.00e+00 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 1 | 114 | 1.00e+00 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 1.00e+00 |
Venn Diagrams
All DEGs adj. p value < 0.05, 24 hour
library(paletteer)
total24 <-
list(sigVDA24$ENTREZID,
sigVDX24$ENTREZID,
sigVEP24$ENTREZID,
sigVMT24$ENTREZID)
in_common24 <-
c(sigVDA24$ENTREZID,
sigVDX24$ENTREZID,
sigVEP24$ENTREZID,
sigVMT24$ENTREZID)
ggVennDiagram::ggVennDiagram(
total24,
category.names = c(
"DNR\nn = 7017 ",
"DOX\n n = 6645\n",
"EPI\n n = 6328\n",
" MTX\n n = 1115"
),
show_intersect = FALSE,
set_color = "black",
category_size = c(6, 6, 6, 6),
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid",
set_size = 4.5
) +
scale_x_continuous(expand = expansion(mult = .3)) +
scale_y_continuous(expand = expansion(mult = .2)) +
scale_color_paletteer_d(palette = "fishualize::Bodianus_pulchellus") +
scale_fill_distiller(palette = "Spectral", direction = -1) +
labs(title = "24 hour comparison of DE genes p.adj. <0.05",
caption = paste("n =", length(unique(in_common24)), "genes")) +
theme(plot.title = element_text(
size = rel(1.6),
hjust = 0.5,
vjust = 1
))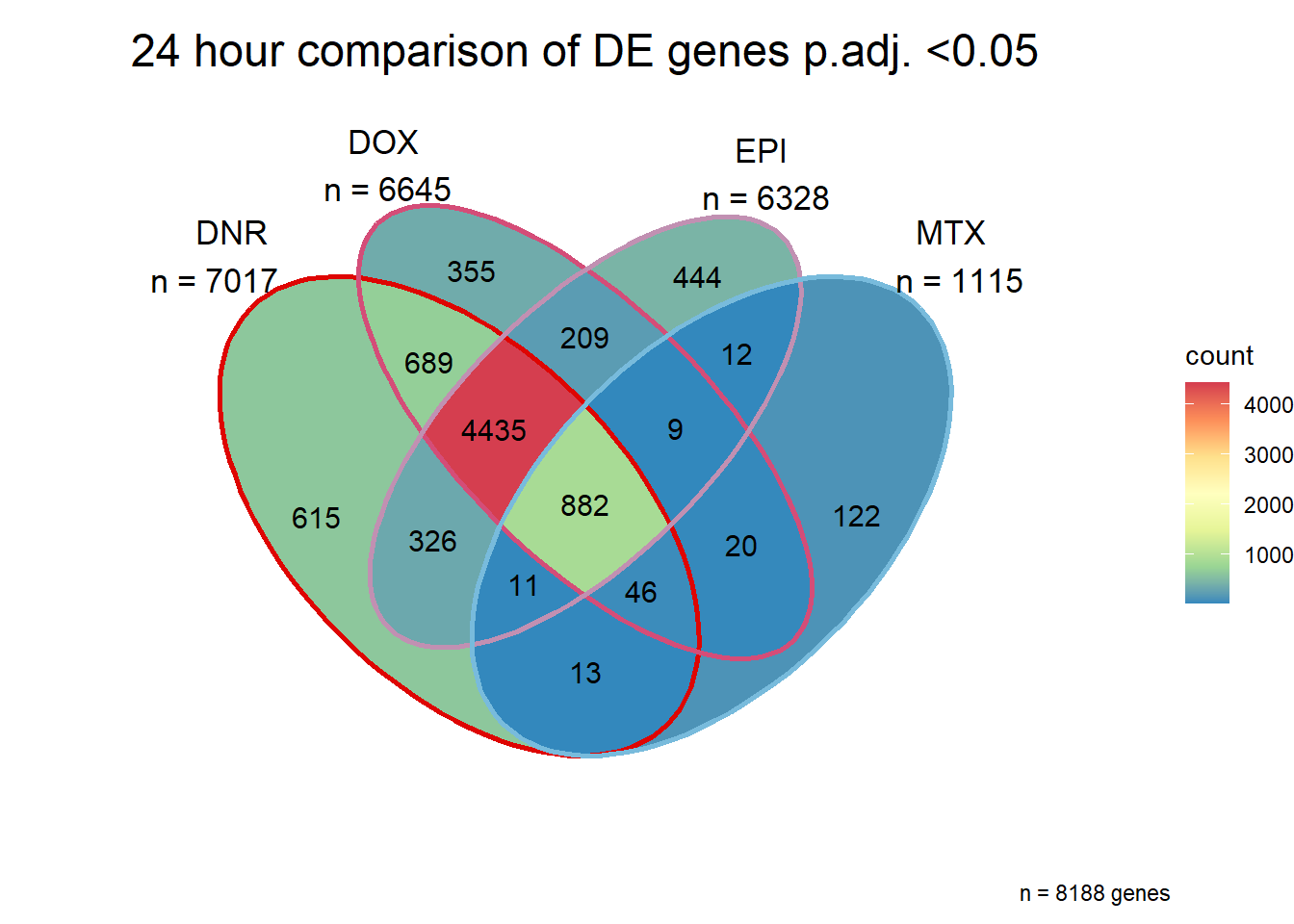
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
| ae53b54 | reneeisnowhere | 2023-07-04 |
| b2bec16 | reneeisnowhere | 2023-06-30 |
| b205acd | reneeisnowhere | 2023-06-29 |
| d9ebe68 | reneeisnowhere | 2023-06-01 |
| 00442f9 | reneeisnowhere | 2023-05-26 |
| 860985f | reneeisnowhere | 2023-05-19 |
| 8ad428c | reneeisnowhere | 2023-04-20 |
| 353fc4a | reneeisnowhere | 2023-04-17 |
All DEGs adj. p value < 0.05, 3 hour
# total 3 ----------------------------------------------------------------
drug_palNoVeh <- c("#8B006D",
"#DF707E",
"#F1B72B",
"#3386DD",
"#707031",
"#41B333")
library(paletteer)
total3 <-
list(sigVDA3$ENTREZID,
sigVDX3$ENTREZID,
sigVEP3$ENTREZID,
sigVMT3$ENTREZID)
totalin_common3 <-
c(sigVDA3$SYMBOL,
sigVDX3$SYMBOL,
sigVEP3$SYMBOL,
sigVMT3$SYMBOL)
ggVennDiagram::ggVennDiagram(
total3,
category.names = c("DNR\nn = 532 ",
"DOX\nn = 19\n",
"EPI\nn = 210\n",
"MTX\n n = 75"),
show_intersect = FALSE,
set_color = "black",
category_size = c(6, 6, 6, 6),
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid",
set_size = 4.5
) +
scale_x_continuous(expand = expansion(mult = .3)) +
scale_y_continuous(expand = expansion(mult = .2)) +
scale_color_paletteer_d(palette = "fishualize::Bodianus_pulchellus") +
scale_fill_distiller(palette = "Spectral",
direction = -1,
limits = c(0, 4500)) +
labs(title = "3 hour comparison of DE genes p.adj. <0.05",
caption = paste("n =", length(unique(totalin_common3)), "genes")) +
theme(plot.title = element_text(
size = rel(1.6),
hjust = 0.5,
vjust = 1
))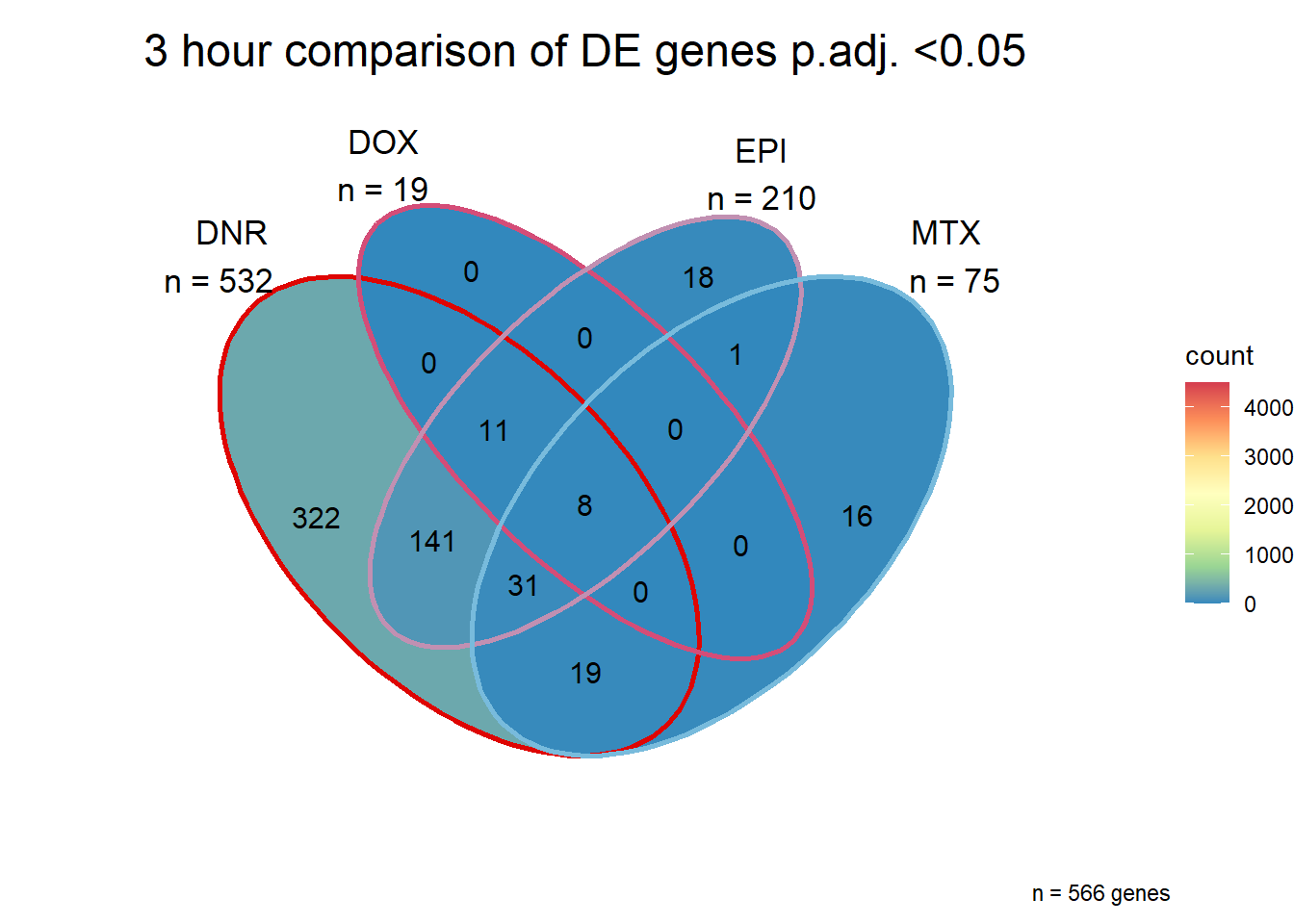
Because the 3 hour DEGs are low (expecially in DOX), we have chosen to focus on analyzing the 24 hour DEGs for GO: BP analysis ### Venn 3 and 24 hour sigDEGs by treatment (adj. p value <0.01) similarities
# Dauno comp --------------------------------------------------------------
Dauncomp <- list(sigVDA24$ENTREZID, sigVDA3$ENTREZID)
in_commonDa <- c(sigVDA24$ENTREZID, sigVDA3$ENTREZID)
length(unique(in_commonDa))[1] 7237# DNRvenn <-
ggVennDiagram(
Dauncomp,
category.names = c("DNR-24", "DNR-3"),
show_intersect = FALSE,
set_color = "black",
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid",
set_size =
) +
scale_x_continuous(expand = expansion(mult = .2)) +
scale_fill_gradient(low = "lightblue", high = "yellow") +
labs(title = "Comparison of DNR 3h v 24h", caption = paste("n =", length(unique(in_commonDa)), "genes")) +
theme(plot.title = element_text(size = rel(1), hjust = 0.5))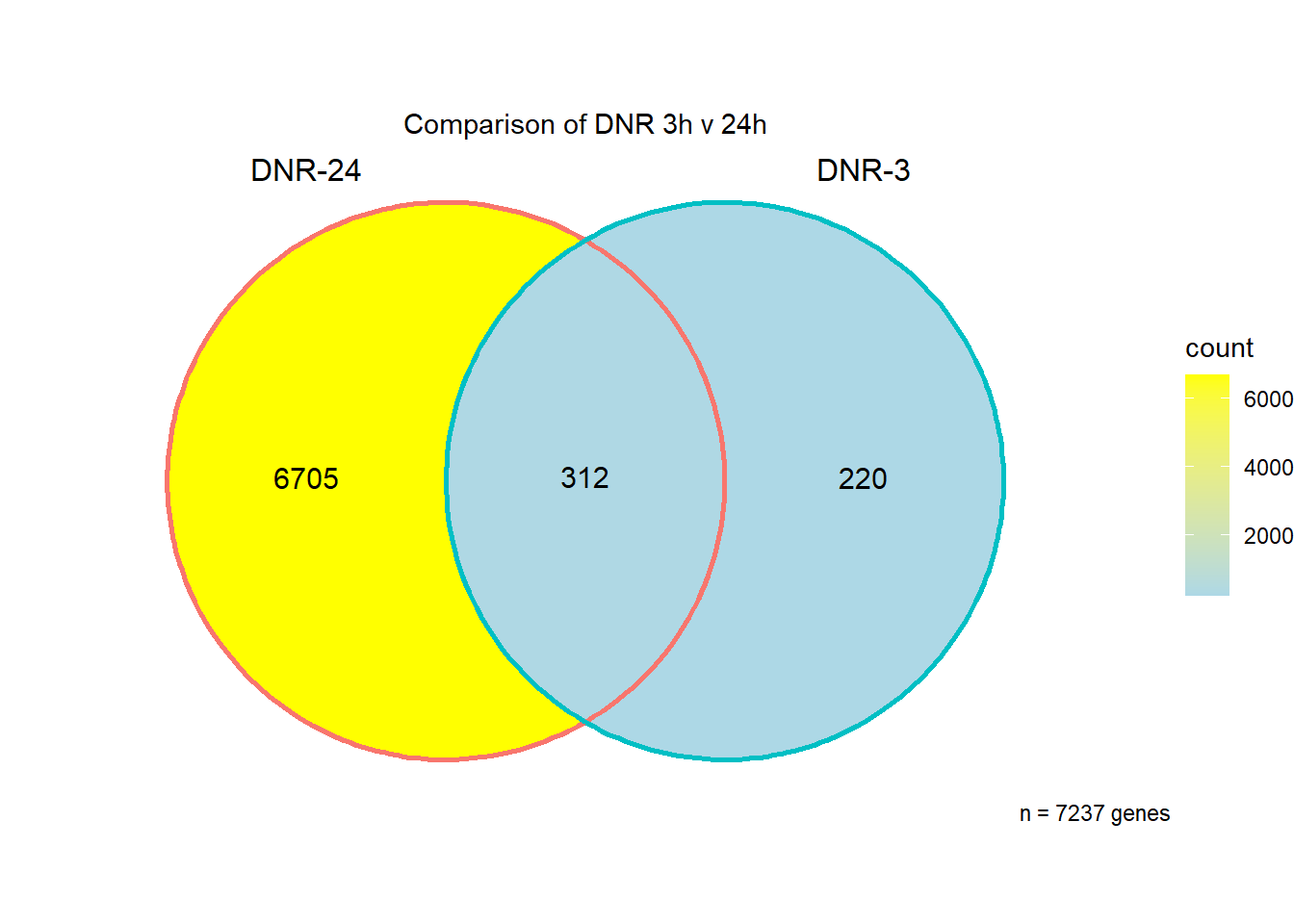
#Davenlist <- intersect(Dauncomp[[1]],Dauncomp[[2]])
# saveRDS(DNRvenn,"output/DNRvenn.RDS")
# Doxocomp ----------------------------------------------------------------
Doxcomp <- list(sigVDX24$ENTREZID, sigVDX3$ENTREZID)
in_commonDx <- c(sigVDX24$ENTREZID, sigVDX3$ENTREZID)
length(unique(in_commonDx))[1] 6656# DOXvenn <-
ggVennDiagram(
Doxcomp,
category.names = c("DOX-24", "DOX-3"),
show_intersect = FALSE,
set_color = "black",
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid",
set_size =
) +
scale_x_continuous(expand = expansion(mult = .2)) +
scale_fill_gradient(low = "light blue", high = "yellow") +
labs(title = "Comparison of Doxo 3h v 24h",
caption = paste("n =", length(unique(in_commonDx)), "genes")) +
theme(plot.title = element_text(size = rel(1), hjust = 0.5))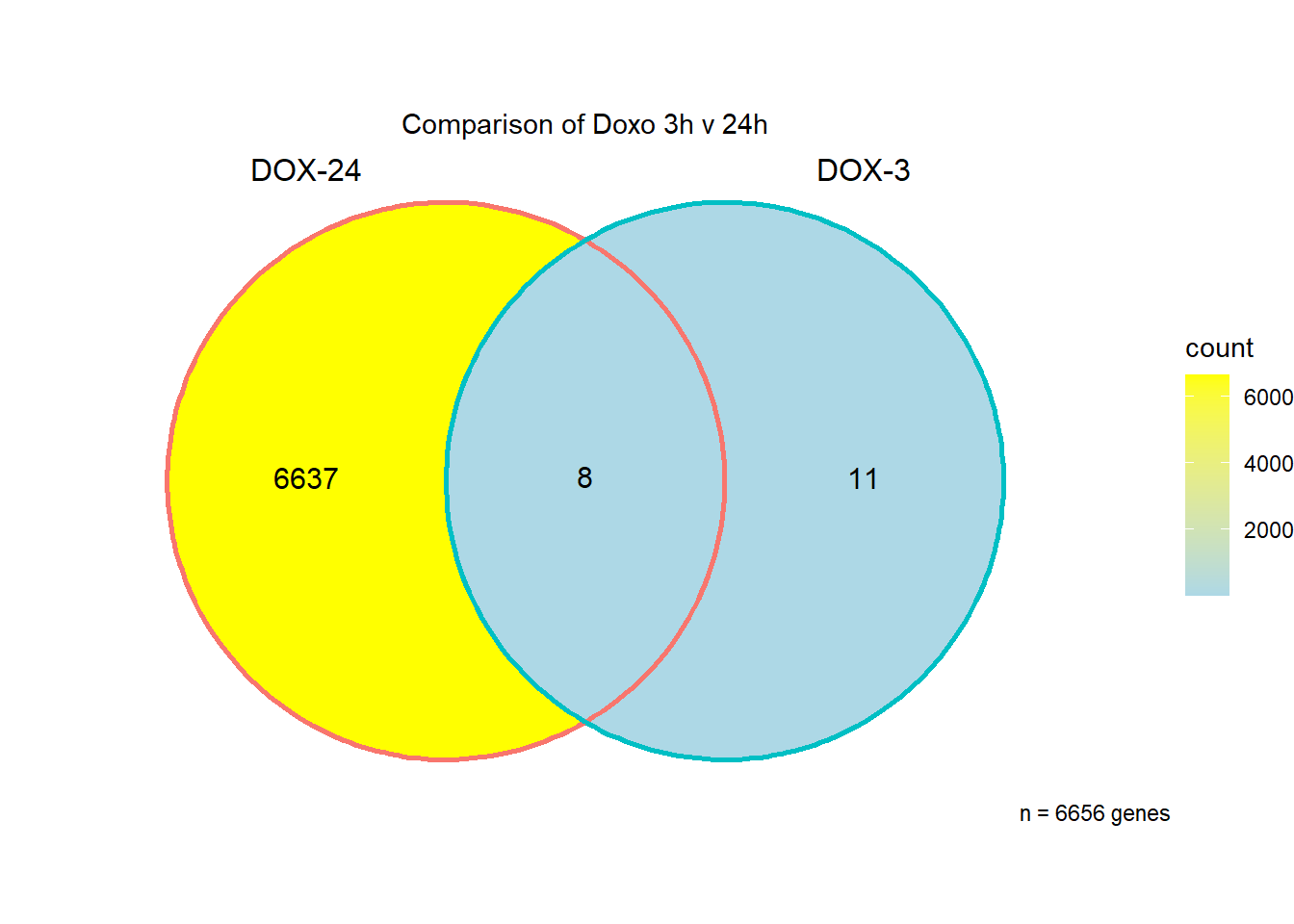
# saveRDS(DOXvenn,"output/DOXvenn.RDS")
# Dxvenlist <- intersect(Doxcomp[[1]],Doxcomp[[2]])
#length(intersect(Dxvenlist,Davenlist))## 7 of DX are in DA
# Epi Comp ----------------------------------------------------------------
Epicomp <- list(sigVEP24$ENTREZID, sigVEP3$ENTREZID)
in_commonEp <- c(sigVEP24$ENTREZID, sigVEP3$ENTREZID)
length(unique(in_commonEp))[1] 6416# EPIvenn <-
ggVennDiagram(
Epicomp,
category.names = c("EPI-24", "EPI-3"),
show_intersect = FALSE,
set_color = "black",
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid",
set_size =
) +
scale_x_continuous(expand = expansion(mult = .2)) +
scale_fill_gradient(low = "light blue", high = "yellow") +
labs(title = "Comparison of EPI 3h v 24h", caption = paste("n =", length(unique(in_commonEp)), "genes")) +
theme(plot.title = element_text(size = rel(1), hjust = 0.5))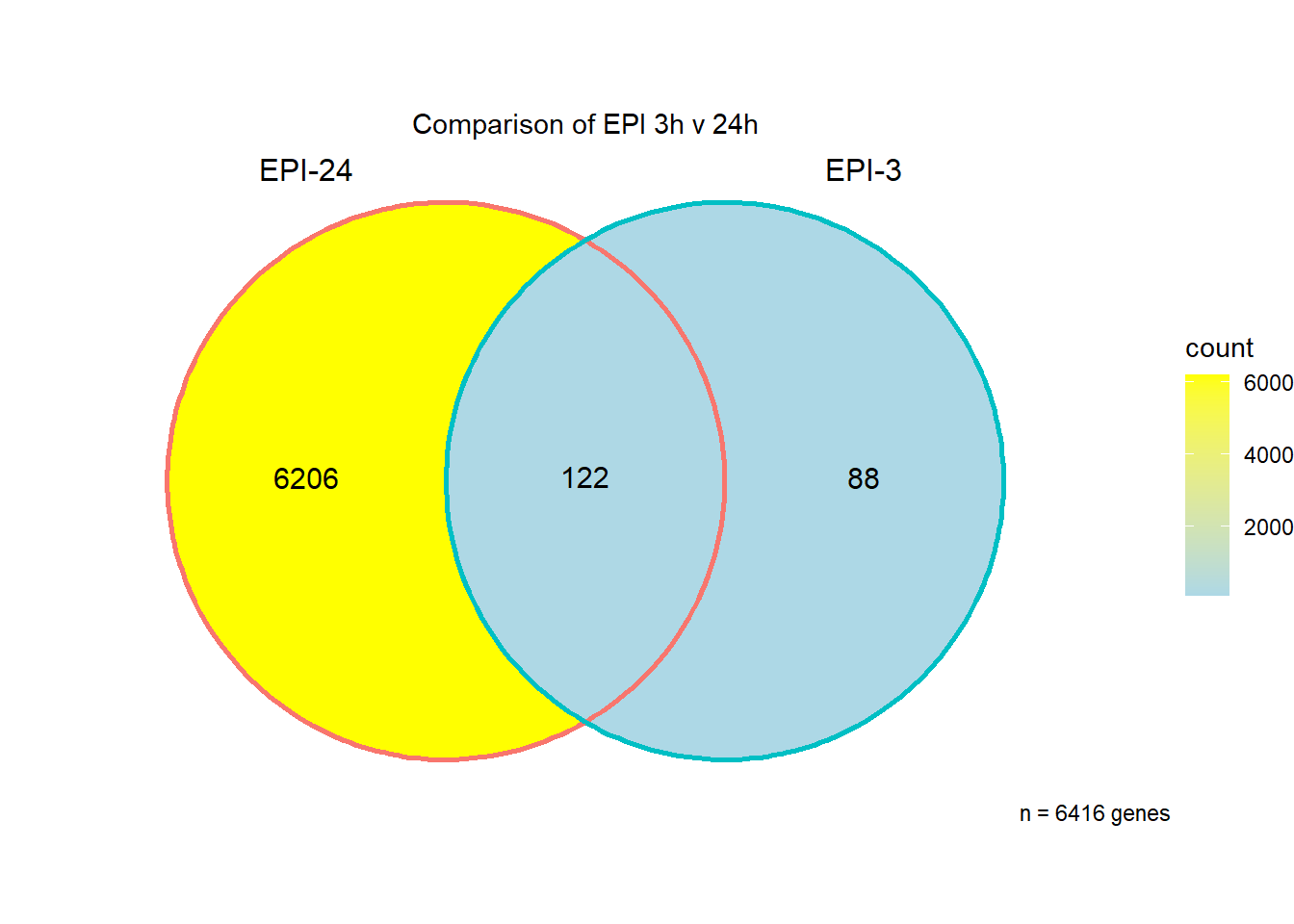
# saveRDS(EPIvenn,"output/EPIvenn.RDS")
#
# Mito comp ---------------------------------------------------------------
Mitocomp <- list(sigVMT24$ENTREZID, sigVMT3$ENTREZID)
in_commonMt <- c(sigVMT24$ENTREZID, sigVMT3$ENTREZID)
length(unique(in_commonMt))[1] 1174# MTXvenn<-
ggVennDiagram(
Mitocomp,
category.names = c("MTX-24", "MTX-3"),
show_intersect = FALSE,
set_color = "black",
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid",
set_size =
) +
scale_x_continuous(expand = expansion(mult = .2)) +
scale_fill_gradient(low = "light blue", high = "yellow") +
labs(title = "Comparison of Mito 3h v 24h",
caption = paste("n =", length(unique(in_commonMt)), "genes")) +
theme(plot.title = element_text(size = rel(1), hjust = 0.5))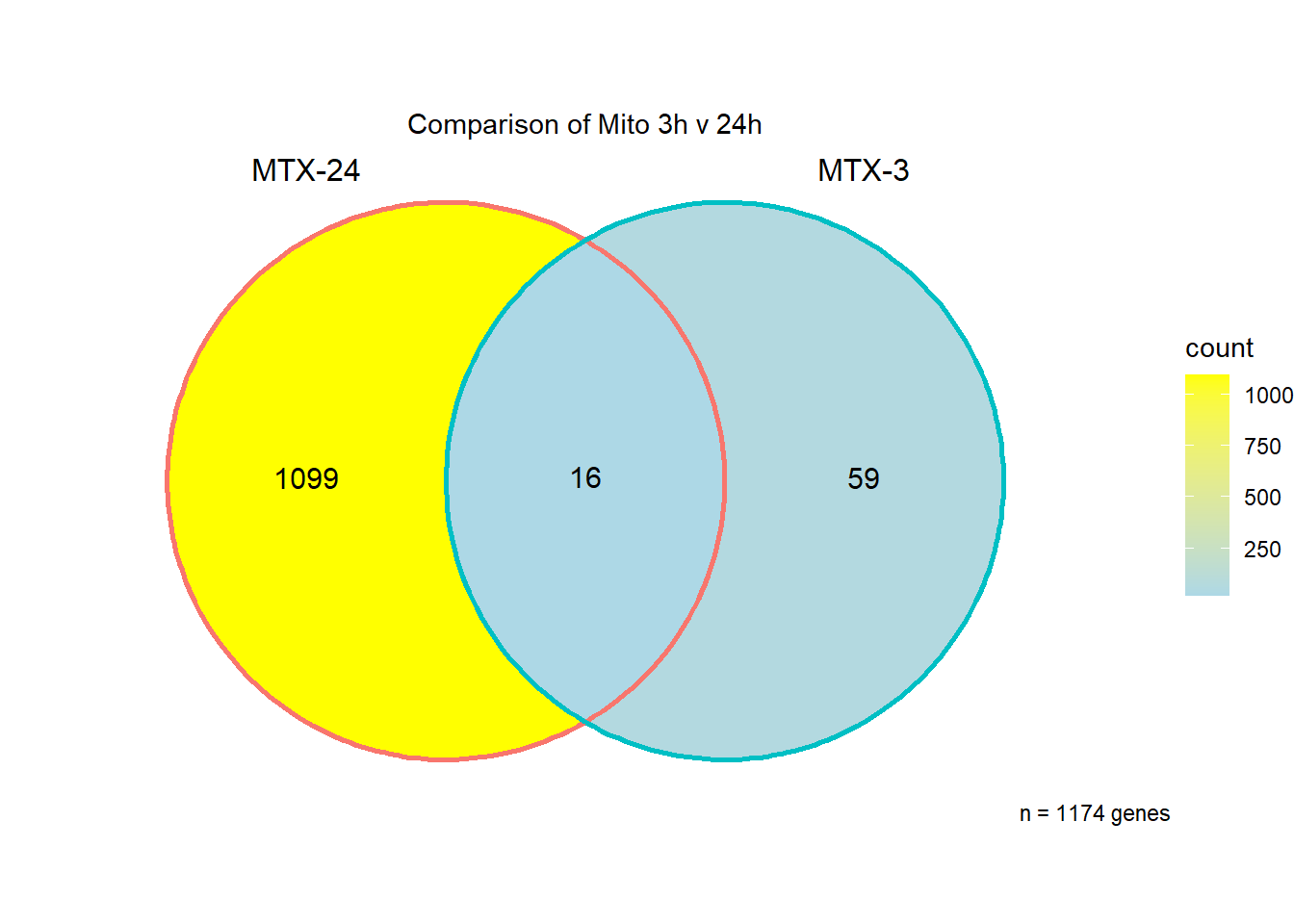
# saveRDS(MTXvenn,"output/MTXvenn.RDS")GO analysis on 24 hour Venn, adj. p value < 0.05
# > list24totvenn$..count..
# [1] 977 15 17 14 58 24 34 188 4178 225 287 489 697 342 616
# > list24totvenn$..values..[?]
##pairwise
DDEresp <- list24totvenn24$..values..[[9]]
DDEresp3 <- list3totvenn3$..values..[[9]]
#DDEresp1<- read.csv("data/DDEresp_list.csv",row.names = 1)
# write.csv(DDEresp,"data/DDEresp_list.csv")
DDEMresp <- list24totvenn24$..values..[[1]]
DDEMresp3 <- list3totvenn3$..values..[[1]]
DXsprespon <- list24totvenn24$..values..[[14]]
length(DXsprespon)[1] 355complete <-
c(
sigVDA24$ENTREZID,
sigVDX24$ENTREZID,
sigVEP24$ENTREZID,
sigVMT24$ENTREZID,
sigVDA3$ENTREZID,
sigVDX3$ENTREZID,
sigVEP3$ENTREZID,
sigVMT3$ENTREZID
)
complete <- as.data.frame(unique(complete))
colnames(complete) <- "ENTREZID" #9047 genes
print("Number of All genes that respond to some condition in DEG set, adj. P value < 0.05")[1] "Number of All genes that respond to some condition in DEG set, adj. P value < 0.05"length(complete$ENTREZID)[1] 8368#8368
NoRespDEG <- setdiff(backGL$ENTREZID , complete$ENTREZID) #
print("Number of No response genes in DEG set, adj. P value < 0.05")[1] "Number of No response genes in DEG set, adj. P value < 0.05"length(NoRespDEG)[1] 5716DDE response set
adj. P value < 0.05, intersection of DNR-DOX-EPI, excluding those in common with MTX, n = 4435 genes.
no significant enrichment found!
AC specific enrichment at 24 hours did not occur. The next step was Top2 inhibitior enrichment. This would be the intersection of all Top2bi drugs. This was 882 genes long. Below is that analysis.
##No enrichment for DDE only resp!
##overlap of DDEM + AC only gives:
DDEresp3All <- (union(DDEresp3, DDEMresp3))
# (just the overlap of DDE, i use the dataframe DDEresp3)
# DDEgostres3only <- gost(query = DDEresp3,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# #
#
# saveRDS(DDEgostres3,"data/DEG-GO/DDE_allgostres3.RDS")
DDEgostres3 <- readRDS("data/DEG-GO/DDE_allgostres3.RDS")
# saveRDS(DDEgostres3only,"data/DEG-GO/DDEgostres3only.RDS")
DDEgostres3only <- readRDS("data/DEG-GO/DDEgostres3only.RDS")
# saveRDS(DDEgostres24kegg,"data/DEG-GO/DDEgostres24kegg.RDS")
# DDEp3 <- gostplot(DDEgostres3, capped = FALSE, interactive = TRUE)
# DDEp3
# DDEp3only <- gostplot(DDEgostres3only, capped = FALSE, interactive = TRUE)
# DDEp3only
# kegg_term <- c("p53 signaling pathway","DNA replication","Cell cycle" ,"Base excision repair")
DDEtable3 <- DDEgostres3$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DDEtable3only <- DDEgostres3only$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DDEtable3only %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Anthracycline DEGs 3 hr GO:BP terms (not significantly enriched)') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)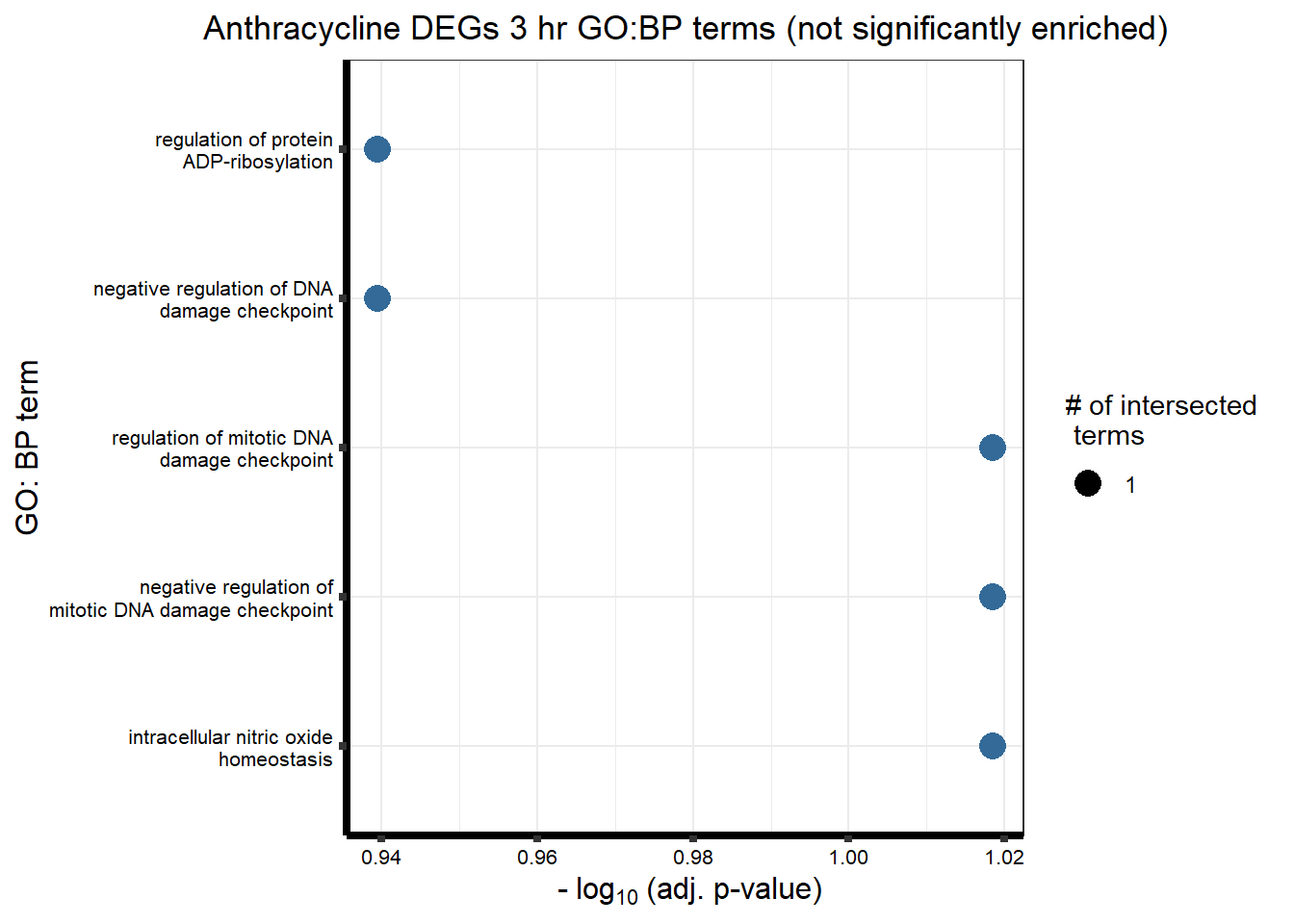
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEtable3only %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size, col = "red")) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Anthracycline DEGs 3 hr KEGG pathway terms ') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("KEGG term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)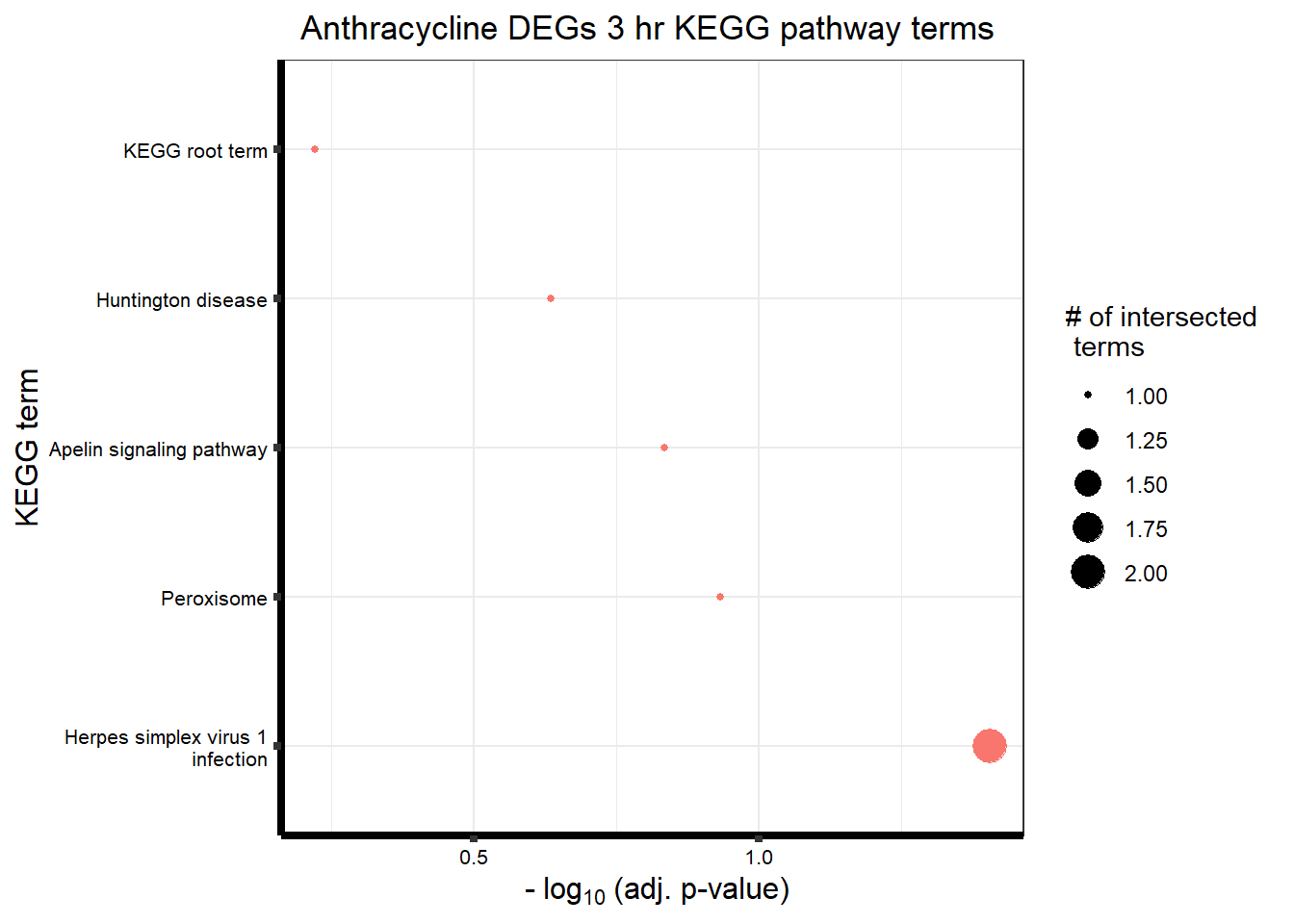
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEtable3 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "DDE all overlapped genes (including those that overlapped with MTX),n = 11+ n = 8") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 12 | 2845 | 6.92e-02 |
| GO:BP | GO:0090720 | primary adaptive immune response | 1 | 1 | 6.92e-02 |
| GO:BP | GO:0090721 | primary adaptive immune response involving T cells and B cells | 1 | 1 | 6.92e-02 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 6.92e-02 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 6.92e-02 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 1 | 9 | 6.92e-02 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 13 | 3080 | 6.92e-02 |
| GO:BP | GO:1904290 | negative regulation of mitotic DNA damage checkpoint | 1 | 1 | 6.92e-02 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 6.92e-02 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 1 | 3 | 6.92e-02 |
| GO:BP | GO:0007614 | short-term memory | 1 | 9 | 7.55e-02 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 12 | 4292 | 8.80e-02 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 13 | 8.80e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 8.80e-02 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 11 | 2576 | 8.80e-02 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 11 | 2574 | 8.80e-02 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 11 | 2593 | 8.80e-02 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 11 | 2684 | 9.18e-02 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 1 | 7 | 9.18e-02 |
| GO:BP | GO:0015074 | DNA integration | 1 | 9 | 9.18e-02 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 12 | 4417 | 9.18e-02 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 11 | 2714 | 9.18e-02 |
| GO:BP | GO:0006351 | DNA-templated transcription | 11 | 2683 | 9.18e-02 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 2 | 678 | 1.00e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 1 | 22 | 1.00e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 1.00e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 1 | 55 | 1.00e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 11 | 3012 | 1.00e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 1.00e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 1.00e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 11 | 3169 | 1.00e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 1 | 72 | 1.00e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 1.00e-01 |
| GO:BP | GO:0051170 | import into nucleus | 2 | 152 | 1.00e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 11 | 3080 | 1.00e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 11 | 4174 | 1.00e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 84 | 1.00e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 1.00e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 2 | 274 | 1.00e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 3 | 1074 | 1.00e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 11 | 3132 | 1.00e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 1.00e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 1 | 64 | 1.00e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 1 | 14 | 1.00e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 11 | 3070 | 1.00e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 11 | 3133 | 1.00e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 1 | 20 | 1.00e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 1.00e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 2 | 660 | 1.00e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 1 | 82 | 1.00e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 1 | 22 | 1.00e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 1.00e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 1.00e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 1.00e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 1 | 36 | 1.00e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 1 | 56 | 1.00e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 1 | 36 | 1.00e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 1 | 39 | 1.00e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 1.00e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 1 | 60 | 1.00e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 1 | 33 | 1.00e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 2 | 756 | 1.00e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 3 | 1076 | 1.00e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 5 | 1914 | 1.00e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 1 | 19 | 1.00e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 1.00e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 1.00e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 6 | 1995 | 1.00e-01 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 1.00e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 1.00e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 1 | 13 | 1.00e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 1.00e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 1 | 19 | 1.00e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1 | 91 | 1.00e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 16 | 4048 | 1.00e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 1 | 25 | 1.00e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 1 | 19 | 1.00e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 1.00e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 1.00e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 1.00e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 11 | 3237 | 1.00e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 97 | 1.03e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 1 | 26 | 1.04e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 1.04e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 12 | 4617 | 1.07e-01 |
| GO:BP | GO:0007613 | memory | 1 | 92 | 1.07e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 1.07e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 110 | 1.08e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 1 | 81 | 1.08e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 2 | 906 | 1.09e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 1 | 85 | 1.09e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 16 | 4455 | 1.09e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 1.13e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 1 | 120 | 1.13e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 1 | 21 | 1.17e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 1 | 131 | 1.17e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 1.17e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 1 | 120 | 1.17e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 1 | 36 | 1.17e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 1 | 34 | 1.17e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 1 | 96 | 1.17e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 1.17e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 1.17e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 135 | 1.17e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 1 | 44 | 1.17e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 1.18e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 1.18e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 1 | 103 | 1.18e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 1.18e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 16 | 4570 | 1.18e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 1.19e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 1 | 113 | 1.19e-01 |
| GO:BP | GO:0016180 | snRNA processing | 1 | 26 | 1.19e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 1.19e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 159 | 1.22e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 16 | 4596 | 1.22e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 1.22e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 1.22e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 1.25e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 1.25e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 152 | 1.25e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 1.25e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 1.26e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 1.26e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 1.26e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 1.26e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 1 | 167 | 1.26e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 1 | 52 | 1.26e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 2 | 1088 | 1.26e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 1.26e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 1 | 168 | 1.26e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 2 | 1111 | 1.27e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 1 | 62 | 1.27e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 12 | 5017 | 1.27e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 1 | 142 | 1.27e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 1.27e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 1 | 184 | 1.27e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 1.27e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 1 | 187 | 1.27e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 1 | 161 | 1.27e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 213 | 1.27e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 9 | 2646 | 1.27e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 189 | 1.27e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 10 | 3580 | 1.27e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 1.27e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 5 | 2412 | 1.27e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 1.27e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 1 | 55 | 1.27e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 2 | 400 | 1.27e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 1 | 200 | 1.28e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 16 | 4747 | 1.28e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 1.28e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 11 | 3730 | 1.28e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 1.28e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 1.28e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 1.28e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 1.30e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 14 | 3600 | 1.31e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 1.31e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 11 | 3750 | 1.31e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 178 | 1.31e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 2 | 1219 | 1.31e-01 |
| GO:BP | GO:0032200 | telomere organization | 1 | 166 | 1.31e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 1.31e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 1.31e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 251 | 1.35e-01 |
| GO:BP | GO:0051169 | nuclear transport | 2 | 295 | 1.37e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 1.37e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 258 | 1.37e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 1 | 246 | 1.37e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 1 | 277 | 1.37e-01 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 237 | 1.37e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 2 | 276 | 1.37e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 1.37e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 2 | 295 | 1.37e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 1.37e-01 |
| GO:BP | GO:0050890 | cognition | 1 | 227 | 1.37e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 273 | 1.37e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 1 | 270 | 1.38e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 16 | 4946 | 1.38e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 1.42e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 1 | 300 | 1.44e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 1 | 303 | 1.44e-01 |
| GO:BP | GO:0070997 | neuron death | 1 | 286 | 1.45e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 1.45e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 305 | 1.45e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 1.45e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 6 | 1602 | 1.45e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 1 | 124 | 1.45e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 1.45e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 1.45e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 1 | 260 | 1.45e-01 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 317 | 1.45e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 1.45e-01 |
| GO:BP | GO:0006915 | apoptotic process | 2 | 1443 | 1.45e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 1 | 209 | 1.45e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 1.46e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 1.46e-01 |
| GO:BP | GO:0012501 | programmed cell death | 2 | 1479 | 1.46e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 2 | 400 | 1.49e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 1 | 345 | 1.50e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 1 | 344 | 1.50e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 1.52e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 9 | 2880 | 1.52e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 1.55e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 357 | 1.55e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 1 | 85 | 1.55e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 344 | 1.57e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 1.57e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 1 | 317 | 1.57e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 1.57e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 1 | 376 | 1.57e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 1.57e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 1 | 375 | 1.57e-01 |
| GO:BP | GO:0008219 | cell death | 2 | 1596 | 1.57e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 359 | 1.58e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 1.58e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 1.59e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 1.60e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 2 | 1006 | 1.60e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 1.60e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 1.60e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 1.60e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 1.60e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 1 | 108 | 1.60e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 1 | 418 | 1.60e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 1.60e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 2 | 884 | 1.62e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 398 | 1.63e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 1 | 420 | 1.63e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 1.63e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 1 | 99 | 1.63e-01 |
| GO:BP | GO:0016049 | cell growth | 1 | 418 | 1.63e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 1.63e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 3 | 2285 | 1.64e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 1 | 106 | 1.70e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 1 | 126 | 1.73e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 1.73e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 338 | 1.73e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 1 | 414 | 1.73e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 2 | 631 | 1.73e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 1 | 448 | 1.73e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 5 | 1322 | 1.73e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 2 | 717 | 1.73e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 5 | 1315 | 1.73e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 5 | 1315 | 1.73e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 2 | 956 | 1.73e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 1.76e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 2 | 679 | 1.78e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 1.78e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 2 | 2011 | 1.78e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 1 | 149 | 1.79e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 1 | 151 | 1.80e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 1 | 122 | 1.80e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 1 | 479 | 1.80e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 1.80e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 501 | 1.80e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 1.80e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 1.80e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 1.81e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 1.81e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 2 | 787 | 1.81e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 1 | 512 | 1.81e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 391 | 1.81e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 1.81e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 1.81e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 1 | 96 | 1.81e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 1.81e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 1.81e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 1 | 527 | 1.82e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 1 | 531 | 1.82e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 11 | 4245 | 1.85e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 1 | 551 | 1.86e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 1.88e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 2 | 2065 | 1.88e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 7 | 2270 | 1.88e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 496 | 1.89e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 1 | 560 | 1.91e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2573 | 1.92e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 441 | 1.92e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 2 | 561 | 1.92e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2565 | 1.92e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 1.92e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 5 | 1441 | 1.92e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 2 | 719 | 1.92e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 1.92e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 1.92e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 1.93e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 1 | 439 | 1.94e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 438 | 1.94e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 2 | 1248 | 1.95e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 1.95e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 1.95e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 547 | 1.95e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 1 | 96 | 1.97e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 1 | 639 | 1.97e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 1.98e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 2.01e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 504 | 2.02e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 1 | 191 | 2.02e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 1 | 127 | 2.02e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 605 | 2.02e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 1 | 620 | 2.05e-01 |
| GO:BP | GO:0007498 | mesoderm development | 1 | 89 | 2.06e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 597 | 2.06e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 597 | 2.06e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 2 | 898 | 2.06e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 5 | 1507 | 2.06e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 691 | 2.08e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 1 | 538 | 2.12e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 3 | 859 | 2.18e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 2.19e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 543 | 2.19e-01 |
| GO:BP | GO:0040007 | growth | 1 | 742 | 2.19e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 556 | 2.19e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 2 | 834 | 2.20e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 2.22e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 1 | 809 | 2.22e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 2.23e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 5 | 1564 | 2.24e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 2 | 957 | 2.24e-01 |
| GO:BP | GO:0031175 | neuron projection development | 1 | 780 | 2.24e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 1 | 232 | 2.24e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 755 | 2.24e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 1 | 816 | 2.24e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 676 | 2.24e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 95 | 2.25e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 1 | 616 | 2.30e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 11 | 4539 | 2.31e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 1 | 164 | 2.32e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 626 | 2.35e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 1 | 796 | 2.35e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 5 | 1604 | 2.37e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 2.38e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 3 | 2890 | 2.40e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 11 | 4590 | 2.43e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 694 | 2.43e-01 |
| GO:BP | GO:0048666 | neuron development | 1 | 877 | 2.45e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 1 | 881 | 2.52e-01 |
| GO:BP | GO:0007369 | gastrulation | 1 | 146 | 2.55e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 963 | 2.57e-01 |
| GO:BP | GO:0006325 | chromatin organization | 2 | 551 | 2.59e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 785 | 2.72e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 2.72e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 1 | 994 | 2.77e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 1 | 993 | 2.77e-01 |
| GO:BP | GO:0007049 | cell cycle | 2 | 1529 | 2.77e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 2 | 1137 | 2.79e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 827 | 2.79e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 1 | 123 | 2.85e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 6 | 4579 | 2.85e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 1014 | 2.86e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 1 | 1061 | 2.87e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 4 | 3625 | 2.89e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 1 | 1123 | 2.92e-01 |
| GO:BP | GO:0048699 | generation of neurons | 1 | 1119 | 2.98e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 1 | 1173 | 2.99e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 1 | 1149 | 3.01e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 2 | 1147 | 3.01e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 3.01e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 3.01e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1201 | 3.01e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 3.01e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 3.01e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 1 | 1167 | 3.02e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 3.02e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 3.04e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1233 | 3.04e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 3.06e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 3.09e-01 |
| GO:BP | GO:0042592 | homeostatic process | 1 | 1159 | 3.19e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 1 | 615 | 3.19e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1138 | 3.21e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 1 | 625 | 3.21e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1294 | 3.22e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 1 | 637 | 3.24e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 3.25e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 1 | 267 | 3.25e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 3.25e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 1 | 1028 | 3.25e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 7 | 7552 | 3.29e-01 |
| GO:BP | GO:0016310 | phosphorylation | 1 | 1372 | 3.30e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1223 | 3.31e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 1 | 1380 | 3.37e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1086 | 3.43e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 4 | 1849 | 3.45e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 3.45e-01 |
| GO:BP | GO:0043588 | skin development | 1 | 175 | 3.46e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 1 | 706 | 3.47e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 2 | 1378 | 3.61e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 3.70e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 1 | 1075 | 3.73e-01 |
| GO:BP | GO:0015031 | protein transport | 2 | 1361 | 3.76e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 2 | 1898 | 3.76e-01 |
| GO:BP | GO:0046907 | intracellular transport | 3 | 1453 | 3.78e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 5 | 4099 | 3.82e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 206 | 3.83e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1 | 1735 | 3.83e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 3.88e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 3.94e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 3.94e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 1 | 280 | 3.99e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 3.99e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 4.00e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 2 | 901 | 4.07e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 10 | 7441 | 4.18e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 4.18e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1 | 1974 | 4.20e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1 | 1345 | 4.20e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 1 | 331 | 4.22e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1904 | 4.24e-01 |
| GO:BP | GO:0010467 | gene expression | 14 | 4587 | 4.26e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 1 | 1939 | 4.28e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1 | 1974 | 4.45e-01 |
| GO:BP | GO:0036211 | protein modification process | 3 | 2901 | 4.48e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 4 | 4045 | 4.48e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1 | 2139 | 4.48e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 4.48e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1 | 2161 | 4.51e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 1 | 1681 | 4.53e-01 |
| GO:BP | GO:0010033 | response to organic substance | 1 | 1957 | 4.53e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 2 | 665 | 4.57e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 10 | 7069 | 4.61e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1719 | 4.61e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 1 | 2102 | 4.69e-01 |
| GO:BP | GO:0007165 | signal transduction | 3 | 3818 | 4.76e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1 | 2035 | 4.76e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 2 | 1684 | 4.81e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 3 | 3090 | 4.83e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 4.96e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1199 | 4.98e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 5.01e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 2 | 1836 | 5.01e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 5.03e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 5.04e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 10 | 8061 | 5.04e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1 | 2135 | 5.07e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 2 | 1119 | 5.24e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 1 | 440 | 5.31e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1 | 2293 | 5.31e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 5.32e-01 |
| GO:BP | GO:0023052 | signaling | 3 | 4141 | 5.32e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 1 | 504 | 5.37e-01 |
| GO:BP | GO:0007154 | cell communication | 3 | 4226 | 5.48e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 2 | 1209 | 5.62e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2129 | 5.62e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2138 | 5.63e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2901 | 5.73e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 5.76e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 2 | 937 | 5.77e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 4 | 6996 | 5.79e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 5.84e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 10 | 7806 | 5.90e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2116 | 5.98e-01 |
| GO:BP | GO:0030154 | cell differentiation | 1 | 2958 | 6.03e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 6.03e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 1 | 2984 | 6.03e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 6.10e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2450 | 6.19e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 2 | 2466 | 6.23e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3386 | 6.32e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 6.43e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2 | 4953 | 6.43e-01 |
| GO:BP | GO:0065007 | biological regulation | 10 | 8320 | 6.53e-01 |
| GO:BP | GO:0016043 | cellular component organization | 5 | 5076 | 6.53e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 6.54e-01 |
| GO:BP | GO:0051641 | cellular localization | 4 | 2810 | 6.60e-01 |
| GO:BP | GO:0006996 | organelle organization | 1 | 3000 | 6.62e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 998 | 6.81e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 468 | 6.83e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 5 | 5274 | 6.83e-01 |
| GO:BP | GO:0006396 | RNA processing | 4 | 929 | 6.97e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4241 | 6.97e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 740 | 6.98e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2780 | 7.03e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2088 | 7.05e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 7.07e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 1 | 742 | 7.07e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 1 | 4128 | 7.28e-01 |
| GO:BP | GO:0009790 | embryo development | 1 | 875 | 7.28e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 1 | 875 | 7.29e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 870 | 7.36e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 1 | 4955 | 7.68e-01 |
| GO:BP | GO:0032502 | developmental process | 1 | 4494 | 7.71e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 7.72e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 7.84e-01 |
| GO:BP | GO:0050896 | response to stimulus | 2 | 5770 | 7.84e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1335 | 7.86e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 1 | 1015 | 7.97e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 1 | 1017 | 7.97e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 1 | 1026 | 7.99e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1496 | 8.02e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 868 | 8.28e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 8.28e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 8.28e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 8.28e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 8.28e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 8.28e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 1 | 4794 | 8.36e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1 | 1214 | 8.37e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 1 | 1239 | 8.44e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 8.48e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1 | 1280 | 8.51e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 8.53e-01 |
| GO:BP | GO:0009888 | tissue development | 1 | 1425 | 8.87e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 665 | 8.87e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 8.96e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 9.65e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 9.74e-01 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 2 | 415 | 3.94e-02 |
| KEGG | KEGG:04146 | Peroxisome | 1 | 68 | 1.17e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 1 | 106 | 1.46e-01 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 2.32e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 6.01e-01 |
DDEtable3only %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "DDE only the overlapped n= 11") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 1 | 3 | 9.58e-02 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 9.58e-02 |
| GO:BP | GO:1904290 | negative regulation of mitotic DNA damage checkpoint | 1 | 1 | 9.58e-02 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 1 | 9 | 1.15e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 1.15e-01 |
| GO:BP | GO:0007614 | short-term memory | 1 | 9 | 1.17e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 1.28e-01 |
| GO:BP | GO:0044849 | estrous cycle | 1 | 13 | 1.28e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 2 | 274 | 1.33e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 1 | 64 | 1.33e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 6 | 2714 | 1.33e-01 |
| GO:BP | GO:0007613 | memory | 1 | 92 | 1.33e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 1 | 21 | 1.33e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 1.33e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 2 | 660 | 1.33e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 2 | 678 | 1.33e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 1 | 120 | 1.33e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 1 | 22 | 1.33e-01 |
| GO:BP | GO:0048675 | axon extension | 1 | 110 | 1.33e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 1 | 36 | 1.33e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 1 | 81 | 1.33e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 1 | 85 | 1.33e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 1 | 96 | 1.33e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 1.33e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 1 | 113 | 1.33e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 1.33e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 1 | 30 | 1.33e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 1 | 82 | 1.33e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 1 | 84 | 1.33e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 4 | 4174 | 1.33e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 1 | 20 | 1.33e-01 |
| GO:BP | GO:0016180 | snRNA processing | 1 | 26 | 1.33e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 1 | 14 | 1.33e-01 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 1 | 7 | 1.33e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 1.33e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 1 | 33 | 1.33e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 1.33e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 1 | 36 | 1.33e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 1 | 56 | 1.33e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 1 | 36 | 1.33e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 1 | 39 | 1.33e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 1 | 60 | 1.33e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 1.33e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 3 | 1074 | 1.33e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 4 | 3080 | 1.33e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 1 | 22 | 1.33e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 1 | 55 | 1.33e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 3 | 1076 | 1.33e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 1 | 120 | 1.33e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 1.33e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 1 | 131 | 1.33e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 1 | 77 | 1.33e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 1.33e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 1.33e-01 |
| GO:BP | GO:0015074 | DNA integration | 1 | 9 | 1.33e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 1 | 103 | 1.33e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 6 | 2683 | 1.33e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 1.33e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 6 | 2574 | 1.33e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 2 | 756 | 1.33e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 5 | 1914 | 1.33e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 6 | 1995 | 1.33e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 1 | 97 | 1.33e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 1 | 45 | 1.33e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 1 | 72 | 1.33e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 1 | 19 | 1.33e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 1 | 19 | 1.33e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 1 | 19 | 1.33e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 1.33e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 8 | 4417 | 1.33e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 1.33e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 1 | 91 | 1.33e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 1.33e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 1 | 34 | 1.33e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 1.33e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 1 | 103 | 1.33e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 6 | 2593 | 1.33e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 135 | 1.33e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 2 | 906 | 1.33e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 1 | 44 | 1.33e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 1 | 25 | 1.33e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 6 | 2684 | 1.33e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 8 | 4292 | 1.33e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 1.33e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 1 | 13 | 1.33e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 1 | 26 | 1.33e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 6 | 2576 | 1.33e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 1.33e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 159 | 1.34e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 1.35e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 1 | 168 | 1.37e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 1 | 62 | 1.37e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 189 | 1.37e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 1.37e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 1 | 187 | 1.37e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 1 | 184 | 1.37e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 1.37e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 1.37e-01 |
| GO:BP | GO:0051170 | import into nucleus | 1 | 152 | 1.37e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 1 | 199 | 1.37e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 5 | 2412 | 1.37e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 6 | 2845 | 1.37e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 1 | 167 | 1.37e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 2 | 400 | 1.37e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 1 | 55 | 1.37e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 1 | 167 | 1.37e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 1.37e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 2 | 1111 | 1.37e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 1.37e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 1 | 213 | 1.37e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 1 | 52 | 1.37e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 1.37e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 1 | 200 | 1.37e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 1.37e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 2 | 1088 | 1.37e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 1.37e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 1 | 142 | 1.37e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 1.37e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 1 | 161 | 1.37e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 1.37e-01 |
| GO:BP | GO:0007416 | synapse assembly | 1 | 152 | 1.37e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 6 | 3012 | 1.41e-01 |
| GO:BP | GO:0032200 | telomere organization | 1 | 166 | 1.41e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 1.41e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 1.41e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 178 | 1.41e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 1.41e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 2 | 1219 | 1.41e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 1.41e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 1.46e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 6 | 3080 | 1.46e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 6 | 3070 | 1.46e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 1.46e-01 |
| GO:BP | GO:0050890 | cognition | 1 | 227 | 1.46e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 258 | 1.46e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 1 | 54 | 1.46e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 1 | 273 | 1.46e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 1 | 277 | 1.46e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 1 | 246 | 1.46e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 1 | 251 | 1.46e-01 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 237 | 1.46e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 1 | 270 | 1.47e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 1 | 300 | 1.50e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 1.50e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 1 | 260 | 1.50e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 1 | 236 | 1.50e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 1 | 303 | 1.50e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 1 | 305 | 1.50e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 6 | 3169 | 1.50e-01 |
| GO:BP | GO:0012501 | programmed cell death | 2 | 1479 | 1.50e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 1.50e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 1.50e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 1.50e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 6 | 3132 | 1.50e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 1.50e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 1 | 209 | 1.50e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 6 | 3133 | 1.50e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 1 | 124 | 1.50e-01 |
| GO:BP | GO:0070997 | neuron death | 1 | 286 | 1.50e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 8 | 4617 | 1.50e-01 |
| GO:BP | GO:0006915 | apoptotic process | 2 | 1443 | 1.50e-01 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 317 | 1.50e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 1.50e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 5 | 2646 | 1.51e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 1 | 345 | 1.55e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 1 | 344 | 1.55e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 6 | 3237 | 1.55e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 1.60e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 1 | 85 | 1.60e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 357 | 1.60e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 1 | 376 | 1.61e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 1 | 344 | 1.61e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 1 | 317 | 1.61e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 1.61e-01 |
| GO:BP | GO:0008219 | cell death | 2 | 1596 | 1.61e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 1.61e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 1.61e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 1 | 375 | 1.61e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 1.61e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 359 | 1.61e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 1 | 276 | 1.62e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 1 | 108 | 1.63e-01 |
| GO:BP | GO:0051169 | nuclear transport | 1 | 295 | 1.63e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 1 | 418 | 1.63e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 2 | 884 | 1.63e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 1 | 395 | 1.63e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 1 | 341 | 1.63e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 1 | 420 | 1.63e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 1 | 99 | 1.63e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 2 | 1006 | 1.63e-01 |
| GO:BP | GO:0061564 | axon development | 1 | 398 | 1.63e-01 |
| GO:BP | GO:0016049 | cell growth | 1 | 418 | 1.63e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 1 | 136 | 1.63e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 1 | 295 | 1.63e-01 |
| GO:BP | GO:0050808 | synapse organization | 1 | 365 | 1.63e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 1 | 342 | 1.63e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 1.64e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 3 | 2285 | 1.65e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 1 | 106 | 1.70e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 5 | 2880 | 1.73e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 1 | 126 | 1.73e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 1 | 414 | 1.74e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 2 | 631 | 1.74e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 2 | 717 | 1.74e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 1 | 448 | 1.75e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 1 | 338 | 1.75e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 2 | 956 | 1.75e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 1.77e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 4 | 5017 | 1.77e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 2 | 2011 | 1.78e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 1.78e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 2 | 679 | 1.78e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 1 | 149 | 1.79e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 1 | 122 | 1.80e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 1.80e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 1 | 151 | 1.80e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 1.80e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 501 | 1.80e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 1 | 479 | 1.80e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 1.81e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 2 | 787 | 1.81e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 391 | 1.81e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 1 | 512 | 1.81e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 1 | 529 | 1.81e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 1.81e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 4 | 4048 | 1.81e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 1 | 541 | 1.81e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 1.81e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 1 | 527 | 1.81e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 1 | 531 | 1.82e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 1 | 551 | 1.86e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 1 | 486 | 1.87e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 6 | 3730 | 1.87e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 2 | 2065 | 1.87e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 1 | 496 | 1.89e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 1 | 560 | 1.91e-01 |
| GO:BP | GO:0034330 | cell junction organization | 1 | 575 | 1.91e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 1 | 441 | 1.91e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 6 | 3750 | 1.91e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2565 | 1.91e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2573 | 1.91e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 2 | 719 | 1.91e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 1 | 490 | 1.91e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 1 | 490 | 1.91e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 1 | 495 | 1.92e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 1 | 438 | 1.93e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 1 | 439 | 1.93e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 1 | 511 | 1.94e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 1 | 452 | 1.94e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 2 | 1248 | 1.94e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 1 | 547 | 1.94e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 4 | 2270 | 1.94e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 1 | 639 | 1.95e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 1 | 569 | 1.96e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 504 | 2.01e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 1 | 191 | 2.01e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 605 | 2.01e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 1 | 620 | 2.04e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 4 | 4455 | 2.04e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 1 | 597 | 2.04e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 2 | 898 | 2.04e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 1 | 597 | 2.04e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 691 | 2.07e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 1 | 538 | 2.11e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 4 | 4570 | 2.17e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 2 | 803 | 2.17e-01 |
| GO:BP | GO:0040007 | growth | 1 | 742 | 2.17e-01 |
| GO:BP | GO:0051276 | chromosome organization | 1 | 543 | 2.17e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 556 | 2.17e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 2 | 834 | 2.18e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 1 | 809 | 2.20e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 4 | 4596 | 2.20e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 2.21e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 1 | 816 | 2.21e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 1 | 676 | 2.21e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 2 | 957 | 2.21e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 755 | 2.21e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 3 | 1602 | 2.21e-01 |
| GO:BP | GO:0031175 | neuron projection development | 1 | 780 | 2.21e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 1 | 232 | 2.21e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 2 | 859 | 2.26e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 1 | 616 | 2.26e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 1 | 626 | 2.32e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 1 | 796 | 2.32e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 4 | 4747 | 2.35e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 2.35e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 3 | 2890 | 2.37e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 694 | 2.41e-01 |
| GO:BP | GO:0048666 | neuron development | 1 | 877 | 2.42e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 2 | 3580 | 2.44e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 2 | 4245 | 2.46e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 4 | 4946 | 2.47e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 1 | 881 | 2.47e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 1 | 963 | 2.52e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 785 | 2.67e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 2.68e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 1 | 994 | 2.72e-01 |
| GO:BP | GO:0007049 | cell cycle | 2 | 1529 | 2.72e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 1 | 993 | 2.72e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 2 | 4539 | 2.72e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 2 | 1137 | 2.73e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 827 | 2.73e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 2 | 4590 | 2.76e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 6 | 4579 | 2.78e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 1014 | 2.79e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 1 | 1061 | 2.80e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 4 | 3625 | 2.82e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 1 | 1123 | 2.85e-01 |
| GO:BP | GO:0048699 | generation of neurons | 1 | 1119 | 2.91e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 1 | 1173 | 2.91e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 2.92e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1 | 1201 | 2.92e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 2.92e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 1 | 1149 | 2.92e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 2 | 1147 | 2.92e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 2.92e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 2.92e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 1 | 1167 | 2.93e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 2.93e-01 |
| GO:BP | GO:0030030 | cell projection organization | 1 | 1233 | 2.95e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 2.95e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 3.01e-01 |
| GO:BP | GO:0042592 | homeostatic process | 1 | 1159 | 3.10e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 1 | 615 | 3.10e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 1 | 1138 | 3.12e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 1 | 625 | 3.12e-01 |
| GO:BP | GO:0022008 | neurogenesis | 1 | 1294 | 3.12e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 1 | 637 | 3.15e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 3.16e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 3.16e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 1 | 1028 | 3.16e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 7 | 7552 | 3.19e-01 |
| GO:BP | GO:0016310 | phosphorylation | 1 | 1372 | 3.20e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1223 | 3.21e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 1 | 1380 | 3.27e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1086 | 3.32e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 1 | 706 | 3.39e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 2 | 3600 | 3.39e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 2 | 1378 | 3.51e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 3.60e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 1 | 1075 | 3.63e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 400 | 3.64e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 2 | 1898 | 3.65e-01 |
| GO:BP | GO:0015031 | protein transport | 2 | 1361 | 3.65e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 5 | 4099 | 3.70e-01 |
| GO:BP | GO:0046907 | intracellular transport | 2 | 1453 | 3.70e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1 | 1735 | 3.71e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 3.76e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 551 | 3.77e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 3.81e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 3.81e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 3.89e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 2 | 1315 | 3.90e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 2 | 1315 | 3.90e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 1 | 1849 | 3.90e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 2 | 1322 | 3.90e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 901 | 4.00e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 10 | 7441 | 4.01e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 4.01e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1 | 1974 | 4.02e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1 | 1345 | 4.03e-01 |
| GO:BP | GO:0007399 | nervous system development | 1 | 1904 | 4.07e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 1 | 1939 | 4.12e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 2 | 1441 | 4.22e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1 | 1974 | 4.27e-01 |
| GO:BP | GO:0036211 | protein modification process | 3 | 2901 | 4.30e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 4.30e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 4 | 4045 | 4.30e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1 | 2139 | 4.30e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1 | 2161 | 4.32e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 1 | 1681 | 4.34e-01 |
| GO:BP | GO:0010033 | response to organic substance | 1 | 1957 | 4.34e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 2 | 1507 | 4.37e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 1 | 561 | 4.40e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 1 | 1719 | 4.40e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 10 | 7069 | 4.40e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 1 | 2102 | 4.48e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 1 | 665 | 4.48e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 2 | 1564 | 4.52e-01 |
| GO:BP | GO:0007165 | signal transduction | 3 | 3818 | 4.52e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1 | 2035 | 4.52e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 2 | 1684 | 4.56e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 3 | 3090 | 4.58e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 2 | 1604 | 4.65e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 4.68e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1199 | 4.71e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 2 | 1836 | 4.75e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 10 | 8061 | 4.78e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1 | 2135 | 4.81e-01 |
| GO:BP | GO:0010467 | gene expression | 2 | 4587 | 4.90e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1 | 2293 | 5.05e-01 |
| GO:BP | GO:0023052 | signaling | 3 | 4141 | 5.08e-01 |
| GO:BP | GO:0007154 | cell communication | 3 | 4226 | 5.23e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2129 | 5.38e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2138 | 5.39e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2901 | 5.49e-01 |
| GO:BP | GO:0006508 | proteolysis | 1 | 1318 | 5.52e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 4 | 6996 | 5.55e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 5.60e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 10 | 7806 | 5.65e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2116 | 5.73e-01 |
| GO:BP | GO:0030154 | cell differentiation | 1 | 2958 | 5.77e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 1 | 2984 | 5.79e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2450 | 5.94e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 2 | 2466 | 5.98e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3386 | 6.07e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 6.17e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2 | 4953 | 6.17e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 6.18e-01 |
| GO:BP | GO:0016043 | cellular component organization | 5 | 5076 | 6.25e-01 |
| GO:BP | GO:0065007 | biological regulation | 10 | 8320 | 6.25e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 6.26e-01 |
| GO:BP | GO:0006996 | organelle organization | 1 | 3000 | 6.35e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2810 | 6.36e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 1 | 998 | 6.51e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 5 | 5274 | 6.54e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4241 | 6.67e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2780 | 6.76e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2088 | 6.78e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 1 | 4128 | 7.02e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 1 | 4955 | 7.46e-01 |
| GO:BP | GO:0032502 | developmental process | 1 | 4494 | 7.48e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 7.49e-01 |
| GO:BP | GO:0050896 | response to stimulus | 2 | 5770 | 7.60e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 7.60e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1335 | 7.62e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1496 | 7.82e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 8.08e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 1 | 4794 | 8.23e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 9.52e-01 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 2 | 415 | 3.94e-02 |
| KEGG | KEGG:04146 | Peroxisome | 1 | 68 | 1.17e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 1 | 106 | 1.46e-01 |
| KEGG | KEGG:05016 | Huntington disease | 1 | 256 | 2.32e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 6.01e-01 |
##No enrichment for DDE only resp!
##overlap of DDEM + AC only gives:
DDErespAll <-
(union(DDEresp, DDEMresp))## for the "24 hour all version (4435+882)
# DDEgostres24only <- gost(query = DDEresp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(DDEgostres24all,"data/DEG-GO/DDE_allgostres24.RDS")
DDEgostres24all <- readRDS("data/DEG-GO/DDE_allgostres24.RDS")
# saveRDS(DDEgostres24only,"data/DEG-GO/DDEgostres24only.RDS")
DDEgostres24only <- readRDS("data/DEG-GO/DDE_allgostres24.RDS")
# saveRDS(DDEgostres24kegg,"data/DEG-GO/DDEgostres24kegg.RDS")
# DDEp24all <- gostplot(DDEgostres24all, capped = FALSE, interactive = TRUE)
# DDEp24all
# DDEp24only <- gostplot(DDEgostres24only, capped = FALSE, interactive = TRUE)
# DDEp24only
kegg_term <-
c("p53 signaling pathway",
"DNA replication",
"Cell cycle" ,
"Base excision repair")
DDEtable24only <- DDEgostres24only$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DDEtable24all <- DDEgostres24all$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DDEtable24only %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
slice_max(., n = 10, order_by = log_val) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Anthracycline DEGs 24 hr GO:BP terms, n = 4435 genes') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)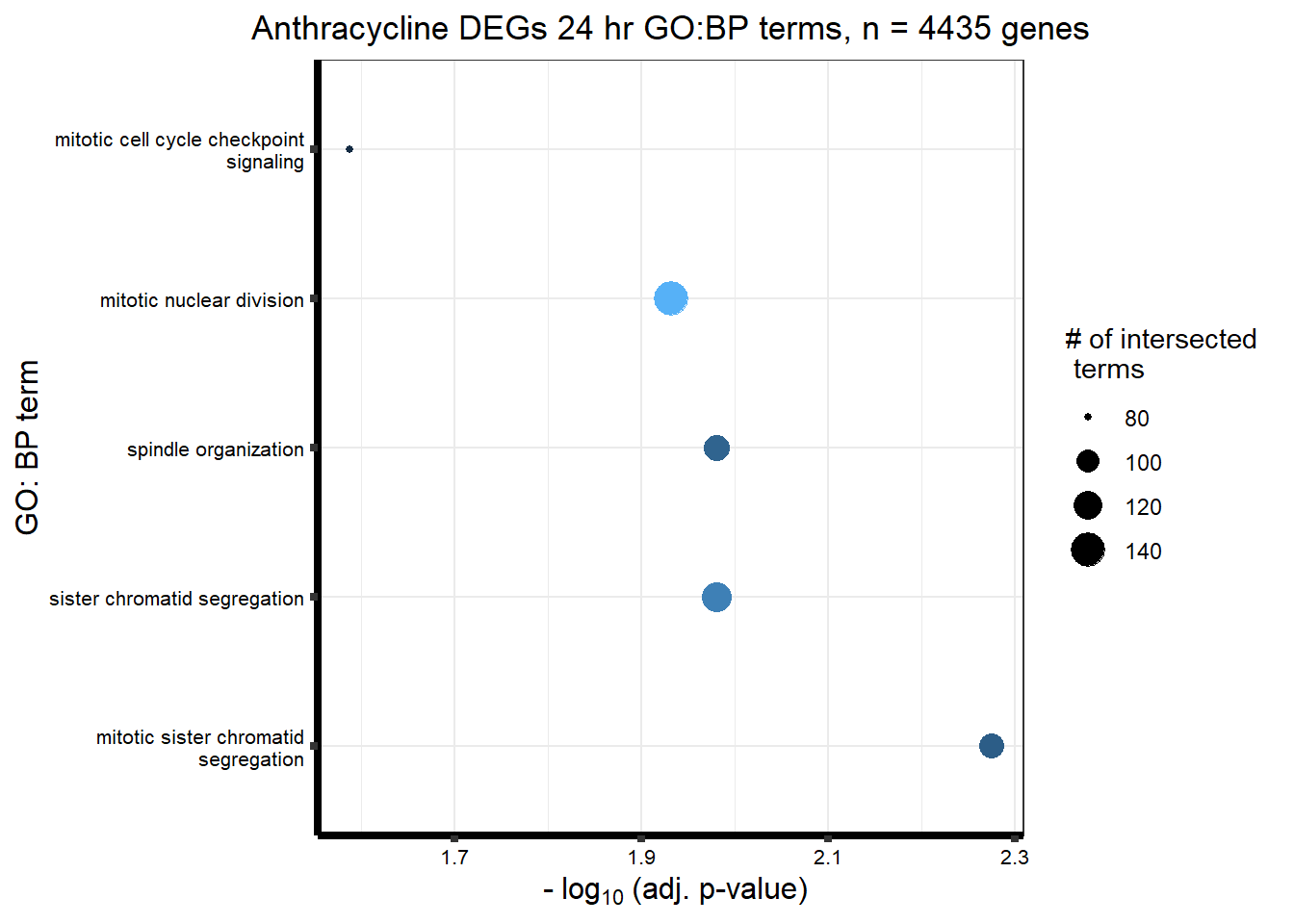
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEtable24only %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
filter(term_name %in% kegg_term) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size, col = "red")) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Anthracycline DEGs 24 hr KEGG pathway terms') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("KEGG term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)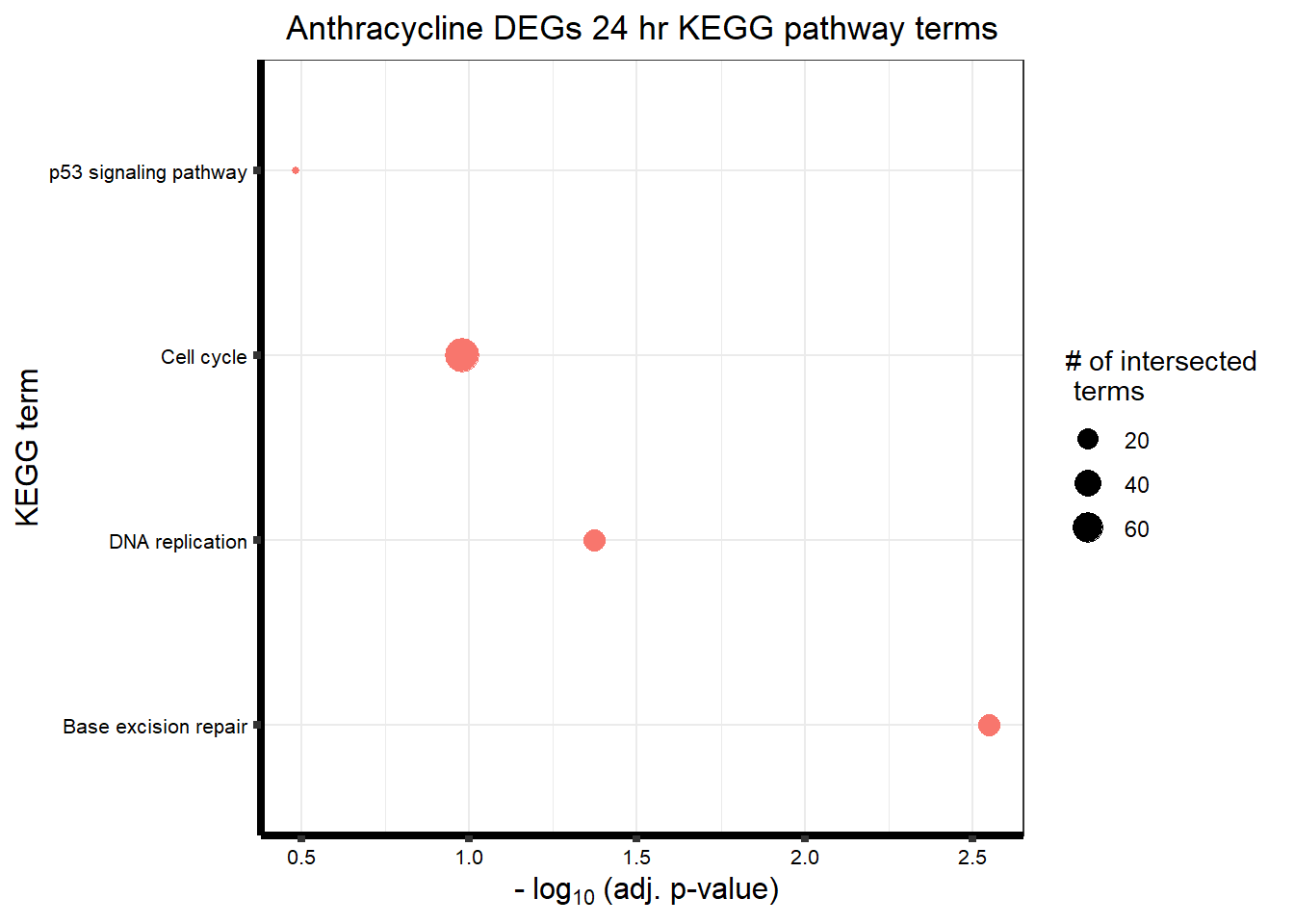
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEtable24only %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "24 hour DDE only enrichment values") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 105 | 179 | 5.31e-03 |
| GO:BP | GO:0000819 | sister chromatid segregation | 121 | 217 | 1.05e-02 |
| GO:BP | GO:0007051 | spindle organization | 108 | 188 | 1.05e-02 |
| GO:BP | GO:0140014 | mitotic nuclear division | 142 | 251 | 1.17e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 80 | 139 | 2.58e-02 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 57 | 90 | 3.34e-02 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 90 | 157 | 3.58e-02 |
| GO:BP | GO:0007052 | mitotic spindle organization | 74 | 129 | 6.44e-02 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 32 | 47 | 8.57e-02 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 30 | 44 | 8.57e-02 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 30 | 44 | 8.57e-02 |
| GO:BP | GO:0007059 | chromosome segregation | 196 | 369 | 8.57e-02 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 30 | 44 | 8.57e-02 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 338 | 694 | 8.57e-02 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 35 | 54 | 8.57e-02 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 31 | 46 | 8.57e-02 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 32 | 47 | 8.57e-02 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 32 | 47 | 8.57e-02 |
| GO:BP | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic | 3 | 41 | 9.91e-02 |
| GO:BP | GO:0060407 | negative regulation of penile erection | 1 | 1 | 9.91e-02 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 34 | 52 | 9.91e-02 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 97 | 183 | 9.91e-02 |
| GO:BP | GO:0060169 | negative regulation of adenosine receptor signaling pathway | 1 | 1 | 9.91e-02 |
| GO:BP | GO:0046111 | xanthine biosynthetic process | 1 | 1 | 9.91e-02 |
| GO:BP | GO:0036166 | phenotypic switching | 4 | 7 | 9.91e-02 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 30 | 45 | 9.91e-02 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 84 | 148 | 9.91e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 113 | 211 | 1.03e-01 |
| GO:BP | GO:0097114 | NMDA glutamate receptor clustering | 2 | 5 | 1.06e-01 |
| GO:BP | GO:0071585 | detoxification of cadmium ion | 2 | 2 | 1.10e-01 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 2 | 9 | 1.10e-01 |
| GO:BP | GO:0009264 | deoxyribonucleotide catabolic process | 2 | 22 | 1.10e-01 |
| GO:BP | GO:1990170 | stress response to cadmium ion | 2 | 2 | 1.10e-01 |
| GO:BP | GO:0046386 | deoxyribose phosphate catabolic process | 2 | 22 | 1.10e-01 |
| GO:BP | GO:0006260 | DNA replication | 135 | 255 | 1.15e-01 |
| GO:BP | GO:2000233 | negative regulation of rRNA processing | 3 | 3 | 1.17e-01 |
| GO:BP | GO:0046061 | dATP catabolic process | 1 | 2 | 1.17e-01 |
| GO:BP | GO:0070256 | negative regulation of mucus secretion | 1 | 2 | 1.17e-01 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 32 | 48 | 1.17e-01 |
| GO:BP | GO:0043928 | exonucleolytic catabolism of deadenylated mRNA | 2 | 13 | 1.17e-01 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 31 | 47 | 1.17e-01 |
| GO:BP | GO:0046110 | xanthine metabolic process | 1 | 1 | 1.17e-01 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 32 | 48 | 1.17e-01 |
| GO:BP | GO:0002636 | positive regulation of germinal center formation | 1 | 1 | 1.17e-01 |
| GO:BP | GO:0043103 | hypoxanthine salvage | 1 | 2 | 1.17e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 64 | 1.18e-01 |
| GO:BP | GO:0051310 | metaphase plate congression | 41 | 69 | 1.28e-01 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 34 | 55 | 1.28e-01 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 36 | 58 | 1.28e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 394 | 827 | 1.28e-01 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 43 | 71 | 1.30e-01 |
| GO:BP | GO:0043096 | purine nucleobase salvage | 1 | 3 | 1.37e-01 |
| GO:BP | GO:0097688 | glutamate receptor clustering | 3 | 7 | 1.37e-01 |
| GO:BP | GO:0046090 | deoxyadenosine metabolic process | 1 | 2 | 1.37e-01 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 2 | 36 | 1.37e-01 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 2 | 35 | 1.37e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 2 | 14 | 1.37e-01 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 2 | 36 | 1.37e-01 |
| GO:BP | GO:0097113 | AMPA glutamate receptor clustering | 3 | 7 | 1.37e-01 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 37 | 61 | 1.37e-01 |
| GO:BP | GO:0046103 | inosine biosynthetic process | 1 | 3 | 1.37e-01 |
| GO:BP | GO:0006157 | deoxyadenosine catabolic process | 1 | 2 | 1.37e-01 |
| GO:BP | GO:0046101 | hypoxanthine biosynthetic process | 1 | 3 | 1.37e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 2 | 2 | 1.37e-01 |
| GO:BP | GO:0046060 | dATP metabolic process | 1 | 3 | 1.37e-01 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 7 | 7 | 1.37e-01 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 27 | 42 | 1.37e-01 |
| GO:BP | GO:0002901 | mature B cell apoptotic process | 1 | 3 | 1.38e-01 |
| GO:BP | GO:0046059 | dAMP catabolic process | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0060167 | regulation of adenosine receptor signaling pathway | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0046124 | purine deoxyribonucleoside catabolic process | 1 | 2 | 1.38e-01 |
| GO:BP | GO:0009217 | purine deoxyribonucleoside triphosphate catabolic process | 1 | 4 | 1.38e-01 |
| GO:BP | GO:0002906 | negative regulation of mature B cell apoptotic process | 1 | 3 | 1.38e-01 |
| GO:BP | GO:0046100 | hypoxanthine metabolic process | 1 | 3 | 1.38e-01 |
| GO:BP | GO:0006154 | adenosine catabolic process | 1 | 3 | 1.38e-01 |
| GO:BP | GO:0002905 | regulation of mature B cell apoptotic process | 1 | 3 | 1.38e-01 |
| GO:BP | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic | 2 | 19 | 1.39e-01 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 12 | 15 | 1.44e-01 |
| GO:BP | GO:0051383 | kinetochore organization | 17 | 23 | 1.44e-01 |
| GO:BP | GO:0009146 | purine nucleoside triphosphate catabolic process | 1 | 5 | 1.48e-01 |
| GO:BP | GO:0032263 | GMP salvage | 1 | 4 | 1.48e-01 |
| GO:BP | GO:0046122 | purine deoxyribonucleoside metabolic process | 1 | 3 | 1.48e-01 |
| GO:BP | GO:0051304 | chromosome separation | 45 | 76 | 1.48e-01 |
| GO:BP | GO:0046102 | inosine metabolic process | 1 | 4 | 1.48e-01 |
| GO:BP | GO:0009204 | deoxyribonucleoside triphosphate catabolic process | 1 | 5 | 1.48e-01 |
| GO:BP | GO:0110154 | RNA decapping | 2 | 17 | 1.48e-01 |
| GO:BP | GO:0006196 | AMP catabolic process | 1 | 3 | 1.48e-01 |
| GO:BP | GO:0044209 | AMP salvage | 1 | 5 | 1.48e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 5 | 25 | 1.48e-01 |
| GO:BP | GO:0051255 | spindle midzone assembly | 11 | 12 | 1.49e-01 |
| GO:BP | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 2 | 2 | 1.49e-01 |
| GO:BP | GO:0035284 | brain segmentation | 2 | 2 | 1.52e-01 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 7 | 17 | 1.52e-01 |
| GO:BP | GO:0021571 | rhombomere 5 development | 2 | 2 | 1.52e-01 |
| GO:BP | GO:0098792 | xenophagy | 8 | 12 | 1.52e-01 |
| GO:BP | GO:0010459 | negative regulation of heart rate | 2 | 7 | 1.52e-01 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 36 | 57 | 1.52e-01 |
| GO:BP | GO:0046121 | deoxyribonucleoside catabolic process | 1 | 4 | 1.52e-01 |
| GO:BP | GO:0140467 | integrated stress response signaling | 9 | 35 | 1.52e-01 |
| GO:BP | GO:1990709 | presynaptic active zone organization | 5 | 9 | 1.52e-01 |
| GO:BP | GO:0035721 | intraciliary retrograde transport | 10 | 14 | 1.52e-01 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 15 | 20 | 1.52e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 66 | 121 | 1.52e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 104 | 234 | 1.52e-01 |
| GO:BP | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process | 1 | 6 | 1.52e-01 |
| GO:BP | GO:0046130 | purine ribonucleoside catabolic process | 1 | 5 | 1.52e-01 |
| GO:BP | GO:0009172 | purine deoxyribonucleoside monophosphate catabolic process | 1 | 3 | 1.52e-01 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 8 | 9 | 1.54e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 11 | 13 | 1.54e-01 |
| GO:BP | GO:0007032 | endosome organization | 12 | 92 | 1.54e-01 |
| GO:BP | GO:0060405 | regulation of penile erection | 1 | 4 | 1.61e-01 |
| GO:BP | GO:0046053 | dAMP metabolic process | 1 | 5 | 1.61e-01 |
| GO:BP | GO:0090071 | negative regulation of ribosome biogenesis | 3 | 4 | 1.61e-01 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 45 | 75 | 1.61e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 11 | 19 | 1.61e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 4 | 89 | 1.63e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 87 | 189 | 1.63e-01 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 8 | 9 | 1.64e-01 |
| GO:BP | GO:0050000 | chromosome localization | 48 | 82 | 1.66e-01 |
| GO:BP | GO:0045828 | positive regulation of isoprenoid metabolic process | 1 | 1 | 1.67e-01 |
| GO:BP | GO:1900054 | positive regulation of retinoic acid biosynthetic process | 1 | 1 | 1.67e-01 |
| GO:BP | GO:0070255 | regulation of mucus secretion | 1 | 6 | 1.67e-01 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 1 | 8 | 1.67e-01 |
| GO:BP | GO:0006152 | purine nucleoside catabolic process | 1 | 6 | 1.67e-01 |
| GO:BP | GO:0106380 | purine ribonucleotide salvage | 1 | 7 | 1.67e-01 |
| GO:BP | GO:0002634 | regulation of germinal center formation | 1 | 6 | 1.67e-01 |
| GO:BP | GO:0002314 | germinal center B cell differentiation | 1 | 3 | 1.69e-01 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 4 | 6 | 1.69e-01 |
| GO:BP | GO:1905174 | regulation of vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 1.69e-01 |
| GO:BP | GO:0015791 | polyol transmembrane transport | 4 | 6 | 1.69e-01 |
| GO:BP | GO:0000255 | allantoin metabolic process | 1 | 4 | 1.69e-01 |
| GO:BP | GO:0048541 | Peyer’s patch development | 1 | 6 | 1.69e-01 |
| GO:BP | GO:0031053 | primary miRNA processing | 11 | 16 | 1.69e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 3 | 28 | 1.69e-01 |
| GO:BP | GO:1990936 | vascular associated smooth muscle cell dedifferentiation | 2 | 2 | 1.69e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 7 | 80 | 1.69e-01 |
| GO:BP | GO:0090678 | cell dedifferentiation involved in phenotypic switching | 2 | 2 | 1.69e-01 |
| GO:BP | GO:0048537 | mucosa-associated lymphoid tissue development | 1 | 6 | 1.69e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 4 | 93 | 1.69e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 4 | 92 | 1.69e-01 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 16 | 22 | 1.69e-01 |
| GO:BP | GO:0009170 | purine deoxyribonucleoside monophosphate metabolic process | 1 | 7 | 1.75e-01 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 1 | 9 | 1.75e-01 |
| GO:BP | GO:0033632 | regulation of cell-cell adhesion mediated by integrin | 1 | 6 | 1.75e-01 |
| GO:BP | GO:0033089 | positive regulation of T cell differentiation in thymus | 1 | 5 | 1.75e-01 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 1 | 11 | 1.75e-01 |
| GO:BP | GO:0090222 | centrosome-templated microtubule nucleation | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0032261 | purine nucleotide salvage | 1 | 10 | 1.75e-01 |
| GO:BP | GO:0009128 | purine nucleoside monophosphate catabolic process | 1 | 7 | 1.75e-01 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 1 | 11 | 1.75e-01 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 1 | 10 | 1.75e-01 |
| GO:BP | GO:0034508 | centromere complex assembly | 19 | 28 | 1.75e-01 |
| GO:BP | GO:0070244 | negative regulation of thymocyte apoptotic process | 1 | 6 | 1.75e-01 |
| GO:BP | GO:0009155 | purine deoxyribonucleotide catabolic process | 1 | 7 | 1.75e-01 |
| GO:BP | GO:0046085 | adenosine metabolic process | 1 | 5 | 1.75e-01 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 5 | 6 | 1.75e-01 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 1 | 11 | 1.75e-01 |
| GO:BP | GO:0035283 | central nervous system segmentation | 2 | 2 | 1.75e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 3 | 15 | 1.75e-01 |
| GO:BP | GO:0009169 | purine ribonucleoside monophosphate catabolic process | 1 | 7 | 1.75e-01 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 10 | 39 | 1.75e-01 |
| GO:BP | GO:1901490 | regulation of lymphangiogenesis | 4 | 5 | 1.75e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 47 | 85 | 1.75e-01 |
| GO:BP | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3’-5’ | 5 | 11 | 1.75e-01 |
| GO:BP | GO:0043084 | penile erection | 1 | 6 | 1.75e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 2 | 69 | 1.75e-01 |
| GO:BP | GO:0051299 | centrosome separation | 11 | 14 | 1.75e-01 |
| GO:BP | GO:1901491 | negative regulation of lymphangiogenesis | 3 | 3 | 1.80e-01 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 4 | 28 | 1.84e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 5 | 165 | 1.84e-01 |
| GO:BP | GO:0009143 | nucleoside triphosphate catabolic process | 1 | 10 | 1.84e-01 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 49 | 91 | 1.85e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 12 | 381 | 1.85e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 1.93e-01 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 13 | 18 | 1.93e-01 |
| GO:BP | GO:1901845 | negative regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0006270 | DNA replication initiation | 23 | 35 | 1.93e-01 |
| GO:BP | GO:0035641 | locomotory exploration behavior | 3 | 11 | 1.93e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 3 | 36 | 1.93e-01 |
| GO:BP | GO:0009159 | deoxyribonucleoside monophosphate catabolic process | 1 | 8 | 1.93e-01 |
| GO:BP | GO:0009158 | ribonucleoside monophosphate catabolic process | 1 | 10 | 1.93e-01 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 13 | 18 | 1.93e-01 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 1 | 15 | 1.93e-01 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 1 | 15 | 1.93e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 2 | 74 | 1.93e-01 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 1 | 14 | 1.93e-01 |
| GO:BP | GO:0008592 | regulation of Toll signaling pathway | 3 | 3 | 1.93e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 1.93e-01 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 2 | 30 | 1.93e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 1.93e-01 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 1.93e-01 |
| GO:BP | GO:1903780 | negative regulation of cardiac conduction | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0070254 | mucus secretion | 1 | 8 | 1.93e-01 |
| GO:BP | GO:0002467 | germinal center formation | 1 | 10 | 1.93e-01 |
| GO:BP | GO:0006177 | GMP biosynthetic process | 1 | 13 | 1.93e-01 |
| GO:BP | GO:0036245 | cellular response to menadione | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 1.93e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 146 | 273 | 1.93e-01 |
| GO:BP | GO:0051225 | spindle assembly | 64 | 120 | 1.93e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 2 | 24 | 1.93e-01 |
| GO:BP | GO:0010651 | negative regulation of cell communication by electrical coupling | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 10 | 12 | 1.93e-01 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 18 | 27 | 1.93e-01 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 1 | 14 | 1.93e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 1 | 11 | 1.93e-01 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 2 | 12 | 1.95e-01 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 5 | 8 | 1.95e-01 |
| GO:BP | GO:0009151 | purine deoxyribonucleotide metabolic process | 1 | 12 | 1.97e-01 |
| GO:BP | GO:0044314 | protein K27-linked ubiquitination | 7 | 8 | 1.97e-01 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 13 | 18 | 1.97e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 1.97e-01 |
| GO:BP | GO:0090069 | regulation of ribosome biogenesis | 3 | 5 | 1.97e-01 |
| GO:BP | GO:0010458 | exit from mitosis | 20 | 30 | 1.97e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 1.97e-01 |
| GO:BP | GO:0060453 | regulation of gastric acid secretion | 2 | 8 | 1.97e-01 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 7 | 19 | 1.97e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 27 | 186 | 1.97e-01 |
| GO:BP | GO:0006400 | tRNA modification | 34 | 90 | 1.97e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 1.97e-01 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 4 | 11 | 1.97e-01 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 7 | 19 | 1.97e-01 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 1 | 10 | 1.97e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 7 | 16 | 1.99e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 6 | 9 | 1.99e-01 |
| GO:BP | GO:2000521 | negative regulation of immunological synapse formation | 1 | 1 | 1.99e-01 |
| GO:BP | GO:0003166 | bundle of His development | 5 | 6 | 1.99e-01 |
| GO:BP | GO:1904071 | presynaptic active zone assembly | 3 | 4 | 1.99e-01 |
| GO:BP | GO:2001189 | negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 1 | 1.99e-01 |
| GO:BP | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process | 13 | 17 | 1.99e-01 |
| GO:BP | GO:0033631 | cell-cell adhesion mediated by integrin | 1 | 12 | 1.99e-01 |
| GO:BP | GO:0006281 | DNA repair | 254 | 534 | 1.99e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 3 | 11 | 1.99e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 4 | 39 | 2.00e-01 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 1 | 18 | 2.01e-01 |
| GO:BP | GO:0043173 | nucleotide salvage | 1 | 15 | 2.01e-01 |
| GO:BP | GO:0071029 | nuclear ncRNA surveillance | 3 | 7 | 2.02e-01 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 1 | 19 | 2.02e-01 |
| GO:BP | GO:0097719 | neural tissue regeneration | 1 | 1 | 2.02e-01 |
| GO:BP | GO:0071046 | nuclear polyadenylation-dependent ncRNA catabolic process | 3 | 7 | 2.02e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 4 | 107 | 2.02e-01 |
| GO:BP | GO:0009125 | nucleoside monophosphate catabolic process | 1 | 13 | 2.02e-01 |
| GO:BP | GO:0032938 | negative regulation of translation in response to oxidative stress | 1 | 1 | 2.02e-01 |
| GO:BP | GO:0043556 | regulation of translation in response to oxidative stress | 1 | 1 | 2.02e-01 |
| GO:BP | GO:0001829 | trophectodermal cell differentiation | 1 | 15 | 2.02e-01 |
| GO:BP | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process | 3 | 7 | 2.02e-01 |
| GO:BP | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process | 3 | 7 | 2.02e-01 |
| GO:BP | GO:0106354 | tRNA surveillance | 3 | 7 | 2.02e-01 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 1 | 12 | 2.02e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 5 | 186 | 2.02e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 2.02e-01 |
| GO:BP | GO:0043605 | amide catabolic process | 1 | 10 | 2.02e-01 |
| GO:BP | GO:1902065 | response to L-glutamate | 6 | 11 | 2.02e-01 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 1 | 15 | 2.02e-01 |
| GO:BP | GO:0042454 | ribonucleoside catabolic process | 1 | 12 | 2.02e-01 |
| GO:BP | GO:0072201 | negative regulation of mesenchymal cell proliferation | 4 | 7 | 2.02e-01 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 9 | 12 | 2.02e-01 |
| GO:BP | GO:0044790 | suppression of viral release by host | 3 | 16 | 2.02e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 54 | 119 | 2.02e-01 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 5 | 29 | 2.02e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 4 | 65 | 2.02e-01 |
| GO:BP | GO:0002949 | tRNA threonylcarbamoyladenosine modification | 3 | 5 | 2.02e-01 |
| GO:BP | GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 1 | 19 | 2.02e-01 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 4 | 8 | 2.02e-01 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 53 | 99 | 2.03e-01 |
| GO:BP | GO:1900262 | regulation of DNA-directed DNA polymerase activity | 10 | 13 | 2.03e-01 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 10 | 13 | 2.03e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 28 | 50 | 2.04e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 4 | 75 | 2.04e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 2.04e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 3 | 120 | 2.04e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 3 | 19 | 2.04e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 25 | 45 | 2.04e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 5 | 358 | 2.04e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 1 | 17 | 2.04e-01 |
| GO:BP | GO:0048873 | homeostasis of number of cells within a tissue | 3 | 24 | 2.04e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 5 | 50 | 2.04e-01 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 14 | 21 | 2.04e-01 |
| GO:BP | GO:1901594 | response to capsazepine | 1 | 1 | 2.04e-01 |
| GO:BP | GO:0006284 | base-excision repair | 5 | 44 | 2.04e-01 |
| GO:BP | GO:0009127 | purine nucleoside monophosphate biosynthetic process | 1 | 21 | 2.04e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 1 | 59 | 2.04e-01 |
| GO:BP | GO:0007620 | copulation | 1 | 10 | 2.04e-01 |
| GO:BP | GO:2000232 | regulation of rRNA processing | 5 | 16 | 2.04e-01 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 1 | 16 | 2.04e-01 |
| GO:BP | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway | 3 | 3 | 2.06e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 44 | 81 | 2.06e-01 |
| GO:BP | GO:1900052 | regulation of retinoic acid biosynthetic process | 1 | 3 | 2.08e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 3 | 121 | 2.08e-01 |
| GO:BP | GO:2000821 | regulation of grooming behavior | 2 | 3 | 2.08e-01 |
| GO:BP | GO:0097065 | anterior head development | 6 | 7 | 2.08e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 4 | 29 | 2.08e-01 |
| GO:BP | GO:0097115 | neurexin clustering involved in presynaptic membrane assembly | 1 | 1 | 2.10e-01 |
| GO:BP | GO:0048789 | cytoskeletal matrix organization at active zone | 1 | 1 | 2.10e-01 |
| GO:BP | GO:0098928 | presynaptic signal transduction | 2 | 2 | 2.12e-01 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 1 | 18 | 2.12e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 8 | 406 | 2.12e-01 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 2 | 8 | 2.12e-01 |
| GO:BP | GO:0009164 | nucleoside catabolic process | 1 | 15 | 2.12e-01 |
| GO:BP | GO:0099526 | presynapse to nucleus signaling pathway | 2 | 2 | 2.12e-01 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 10 | 13 | 2.12e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 66 | 129 | 2.12e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 2.12e-01 |
| GO:BP | GO:0070233 | negative regulation of T cell apoptotic process | 1 | 14 | 2.12e-01 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 8 | 8 | 2.12e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 3 | 45 | 2.12e-01 |
| GO:BP | GO:0008063 | Toll signaling pathway | 4 | 6 | 2.12e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 4 | 70 | 2.12e-01 |
| GO:BP | GO:0051418 | microtubule nucleation by microtubule organizing center | 1 | 5 | 2.12e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 7 | 14 | 2.12e-01 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 2 | 15 | 2.12e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 2 | 17 | 2.12e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 97 | 218 | 2.13e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 85 | 166 | 2.15e-01 |
| GO:BP | GO:0046033 | AMP metabolic process | 1 | 22 | 2.15e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 2.15e-01 |
| GO:BP | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 1 | 3 | 2.15e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 6 | 9 | 2.15e-01 |
| GO:BP | GO:0060419 | heart growth | 36 | 72 | 2.16e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 2 | 18 | 2.16e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 3 | 42 | 2.16e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 8 | 16 | 2.16e-01 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 1 | 20 | 2.16e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 8 | 55 | 2.16e-01 |
| GO:BP | GO:0046037 | GMP metabolic process | 1 | 23 | 2.16e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 2 | 18 | 2.16e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 1 | 21 | 2.16e-01 |
| GO:BP | GO:0035601 | protein deacylation | 52 | 116 | 2.16e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 2 | 506 | 2.16e-01 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 49 | 92 | 2.17e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 2 | 39 | 2.17e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 4 | 25 | 2.18e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 4 | 64 | 2.19e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 2.19e-01 |
| GO:BP | GO:0032691 | negative regulation of interleukin-1 beta production | 2 | 13 | 2.19e-01 |
| GO:BP | GO:0000017 | alpha-glucoside transport | 2 | 2 | 2.19e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 4 | 106 | 2.19e-01 |
| GO:BP | GO:0019985 | translesion synthesis | 16 | 24 | 2.19e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 2 | 37 | 2.19e-01 |
| GO:BP | GO:0034656 | nucleobase-containing small molecule catabolic process | 1 | 20 | 2.19e-01 |
| GO:BP | GO:0045200 | establishment of neuroblast polarity | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 2 | 36 | 2.19e-01 |
| GO:BP | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0060431 | primary lung bud formation | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0043634 | polyadenylation-dependent ncRNA catabolic process | 3 | 8 | 2.19e-01 |
| GO:BP | GO:0045760 | positive regulation of action potential | 2 | 4 | 2.19e-01 |
| GO:BP | GO:0071307 | cellular response to vitamin K | 1 | 4 | 2.19e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 13 | 68 | 2.19e-01 |
| GO:BP | GO:0015750 | pentose transmembrane transport | 2 | 2 | 2.19e-01 |
| GO:BP | GO:0006974 | DNA damage response | 366 | 785 | 2.19e-01 |
| GO:BP | GO:0045196 | establishment or maintenance of neuroblast polarity | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 66 | 127 | 2.19e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 2.19e-01 |
| GO:BP | GO:0033081 | regulation of T cell differentiation in thymus | 1 | 16 | 2.19e-01 |
| GO:BP | GO:0019606 | 2-oxobutyrate catabolic process | 1 | 1 | 2.19e-01 |
| GO:BP | GO:0019747 | regulation of isoprenoid metabolic process | 1 | 4 | 2.19e-01 |
| GO:BP | GO:0060057 | apoptotic process involved in mammary gland involution | 2 | 3 | 2.19e-01 |
| GO:BP | GO:0046391 | 5-phosphoribose 1-diphosphate metabolic process | 2 | 5 | 2.19e-01 |
| GO:BP | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process | 2 | 5 | 2.19e-01 |
| GO:BP | GO:0060058 | positive regulation of apoptotic process involved in mammary gland involution | 2 | 3 | 2.19e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 18 | 1199 | 2.19e-01 |
| GO:BP | GO:0015793 | glycerol transmembrane transport | 2 | 4 | 2.19e-01 |
| GO:BP | GO:0015756 | fucose transmembrane transport | 2 | 2 | 2.19e-01 |
| GO:BP | GO:0071505 | response to mycophenolic acid | 2 | 2 | 2.19e-01 |
| GO:BP | GO:0043094 | cellular metabolic compound salvage | 1 | 25 | 2.19e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 3 | 35 | 2.19e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 2 | 40 | 2.19e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 5 | 378 | 2.19e-01 |
| GO:BP | GO:0046136 | positive regulation of vitamin metabolic process | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 1 | 17 | 2.19e-01 |
| GO:BP | GO:0071506 | cellular response to mycophenolic acid | 2 | 2 | 2.19e-01 |
| GO:BP | GO:0034473 | U1 snRNA 3’-end processing | 1 | 3 | 2.20e-01 |
| GO:BP | GO:1904717 | regulation of AMPA glutamate receptor clustering | 2 | 4 | 2.20e-01 |
| GO:BP | GO:0106104 | regulation of glutamate receptor clustering | 2 | 4 | 2.20e-01 |
| GO:BP | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 2 | 2 | 2.20e-01 |
| GO:BP | GO:0034476 | U5 snRNA 3’-end processing | 1 | 3 | 2.20e-01 |
| GO:BP | GO:0061146 | Peyer’s patch morphogenesis | 3 | 3 | 2.20e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 4 | 82 | 2.20e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 71 | 251 | 2.20e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 32 | 65 | 2.20e-01 |
| GO:BP | GO:0070278 | extracellular matrix constituent secretion | 4 | 12 | 2.20e-01 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 9 | 38 | 2.20e-01 |
| GO:BP | GO:0002313 | mature B cell differentiation involved in immune response | 1 | 17 | 2.20e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 34 | 168 | 2.21e-01 |
| GO:BP | GO:0007174 | epidermal growth factor catabolic process | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 9 | 12 | 2.21e-01 |
| GO:BP | GO:0071468 | cellular response to acidic pH | 2 | 10 | 2.21e-01 |
| GO:BP | GO:1901658 | glycosyl compound catabolic process | 1 | 23 | 2.21e-01 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 4 | 21 | 2.21e-01 |
| GO:BP | GO:1905366 | negative regulation of intralumenal vesicle formation | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 1 | 20 | 2.21e-01 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 9 | 12 | 2.21e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 9 | 86 | 2.21e-01 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 9 | 10 | 2.21e-01 |
| GO:BP | GO:1905365 | regulation of intralumenal vesicle formation | 1 | 2 | 2.21e-01 |
| GO:BP | GO:1904979 | negative regulation of endosome organization | 1 | 2 | 2.21e-01 |
| GO:BP | GO:0001782 | B cell homeostasis | 1 | 24 | 2.21e-01 |
| GO:BP | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation | 2 | 2 | 2.21e-01 |
| GO:BP | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation | 3 | 5 | 2.21e-01 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 1 | 27 | 2.21e-01 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 1 | 30 | 2.21e-01 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 9 | 11 | 2.21e-01 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 46 | 88 | 2.21e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 4 | 7 | 2.21e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 1 | 16 | 2.21e-01 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 5 | 240 | 2.22e-01 |
| GO:BP | GO:0045008 | depyrimidination | 1 | 8 | 2.23e-01 |
| GO:BP | GO:2000520 | regulation of immunological synapse formation | 1 | 2 | 2.23e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 6 | 147 | 2.23e-01 |
| GO:BP | GO:0006885 | regulation of pH | 4 | 74 | 2.24e-01 |
| GO:BP | GO:1900451 | positive regulation of glutamate receptor signaling pathway | 1 | 2 | 2.24e-01 |
| GO:BP | GO:0035740 | CD8-positive, alpha-beta T cell proliferation | 5 | 7 | 2.24e-01 |
| GO:BP | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation | 5 | 7 | 2.24e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 22 | 46 | 2.24e-01 |
| GO:BP | GO:0097117 | guanylate kinase-associated protein clustering | 1 | 2 | 2.24e-01 |
| GO:BP | GO:0099179 | regulation of synaptic membrane adhesion | 4 | 4 | 2.24e-01 |
| GO:BP | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential | 3 | 10 | 2.24e-01 |
| GO:BP | GO:2000311 | regulation of AMPA receptor activity | 2 | 15 | 2.24e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 2 | 15 | 2.24e-01 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 1 | 23 | 2.25e-01 |
| GO:BP | GO:0051835 | positive regulation of synapse structural plasticity | 2 | 2 | 2.26e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 2.26e-01 |
| GO:BP | GO:0035120 | post-embryonic appendage morphogenesis | 1 | 2 | 2.26e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 2.26e-01 |
| GO:BP | GO:0035127 | post-embryonic limb morphogenesis | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0035128 | post-embryonic forelimb morphogenesis | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 5 | 387 | 2.26e-01 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 3 | 13 | 2.26e-01 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 9 | 11 | 2.26e-01 |
| GO:BP | GO:0042245 | RNA repair | 2 | 2 | 2.26e-01 |
| GO:BP | GO:0021546 | rhombomere development | 2 | 4 | 2.27e-01 |
| GO:BP | GO:0090215 | regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 3 | 4 | 2.29e-01 |
| GO:BP | GO:1901165 | positive regulation of trophoblast cell migration | 2 | 6 | 2.29e-01 |
| GO:BP | GO:1903995 | regulation of non-membrane spanning protein tyrosine kinase activity | 3 | 3 | 2.29e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 2 | 113 | 2.29e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 1 | 23 | 2.29e-01 |
| GO:BP | GO:0007617 | mating behavior | 1 | 21 | 2.29e-01 |
| GO:BP | GO:0090272 | negative regulation of fibroblast growth factor production | 2 | 2 | 2.30e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 3 | 69 | 2.30e-01 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 28 | 47 | 2.30e-01 |
| GO:BP | GO:0002335 | mature B cell differentiation | 1 | 21 | 2.30e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 2.30e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process | 3 | 15 | 2.30e-01 |
| GO:BP | GO:0007274 | neuromuscular synaptic transmission | 2 | 14 | 2.30e-01 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 9 | 21 | 2.30e-01 |
| GO:BP | GO:1904550 | response to arachidonic acid | 1 | 1 | 2.30e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 4 | 22 | 2.30e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 2.30e-01 |
| GO:BP | GO:0045794 | negative regulation of cell volume | 2 | 4 | 2.30e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 6 | 52 | 2.30e-01 |
| GO:BP | GO:0002091 | negative regulation of receptor internalization | 5 | 14 | 2.30e-01 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 4 | 15 | 2.30e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 2.30e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 2.30e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 25 | 48 | 2.30e-01 |
| GO:BP | GO:0090244 | Wnt signaling pathway involved in somitogenesis | 2 | 5 | 2.30e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 3 | 136 | 2.31e-01 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 11 | 16 | 2.32e-01 |
| GO:BP | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | 2 | 7 | 2.32e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 10 | 24 | 2.32e-01 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 1 | 3 | 2.32e-01 |
| GO:BP | GO:0000280 | nuclear division | 190 | 344 | 2.32e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 2.32e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 2.33e-01 |
| GO:BP | GO:1902969 | mitotic DNA replication | 10 | 14 | 2.33e-01 |
| GO:BP | GO:0001561 | fatty acid alpha-oxidation | 2 | 5 | 2.33e-01 |
| GO:BP | GO:0043633 | polyadenylation-dependent RNA catabolic process | 3 | 9 | 2.33e-01 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 1 | 37 | 2.33e-01 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 5 | 46 | 2.33e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 5 | 40 | 2.33e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 3 | 27 | 2.33e-01 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 9 | 2.33e-01 |
| GO:BP | GO:0090212 | negative regulation of establishment of blood-brain barrier | 1 | 1 | 2.33e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 2 | 22 | 2.33e-01 |
| GO:BP | GO:0071047 | polyadenylation-dependent mRNA catabolic process | 1 | 4 | 2.33e-01 |
| GO:BP | GO:2001188 | regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 2 | 2.33e-01 |
| GO:BP | GO:0032692 | negative regulation of interleukin-1 production | 2 | 15 | 2.33e-01 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 4 | 9 | 2.33e-01 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 1 | 37 | 2.33e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 13 | 2.33e-01 |
| GO:BP | GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic process | 1 | 4 | 2.33e-01 |
| GO:BP | GO:0060080 | inhibitory postsynaptic potential | 1 | 8 | 2.33e-01 |
| GO:BP | GO:0000459 | exonucleolytic trimming involved in rRNA processing | 3 | 9 | 2.33e-01 |
| GO:BP | GO:0048341 | paraxial mesoderm formation | 4 | 6 | 2.33e-01 |
| GO:BP | GO:0060454 | positive regulation of gastric acid secretion | 1 | 2 | 2.33e-01 |
| GO:BP | GO:0001696 | gastric acid secretion | 2 | 10 | 2.33e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 11 | 23 | 2.33e-01 |
| GO:BP | GO:0099171 | presynaptic modulation of chemical synaptic transmission | 2 | 26 | 2.33e-01 |
| GO:BP | GO:0016557 | peroxisome membrane biogenesis | 3 | 3 | 2.34e-01 |
| GO:BP | GO:0071494 | cellular response to UV-C | 2 | 4 | 2.34e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 1 | 40 | 2.34e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 2 | 48 | 2.34e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 1 | 119 | 2.35e-01 |
| GO:BP | GO:0014839 | myoblast migration involved in skeletal muscle regeneration | 2 | 3 | 2.35e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 7 | 10 | 2.35e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 3 | 46 | 2.35e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 1 | 26 | 2.35e-01 |
| GO:BP | GO:1903379 | regulation of mitotic chromosome condensation | 2 | 2 | 2.35e-01 |
| GO:BP | GO:1905213 | negative regulation of mitotic chromosome condensation | 2 | 2 | 2.35e-01 |
| GO:BP | GO:1902340 | negative regulation of chromosome condensation | 2 | 2 | 2.35e-01 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 1 | 39 | 2.35e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 2.35e-01 |
| GO:BP | GO:0033693 | neurofilament bundle assembly | 3 | 3 | 2.35e-01 |
| GO:BP | GO:2001015 | negative regulation of skeletal muscle cell differentiation | 2 | 3 | 2.35e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 2 | 12 | 2.35e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 195 | 418 | 2.35e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 4 | 105 | 2.35e-01 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 7 | 8 | 2.35e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 7 | 9 | 2.35e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 2 | 425 | 2.35e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 1 | 26 | 2.35e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 4 | 40 | 2.35e-01 |
| GO:BP | GO:0090125 | cell-cell adhesion involved in synapse maturation | 1 | 2 | 2.35e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 14 | 2.35e-01 |
| GO:BP | GO:0051152 | positive regulation of smooth muscle cell differentiation | 2 | 10 | 2.35e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 10 | 14 | 2.35e-01 |
| GO:BP | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER | 2 | 2 | 2.35e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 1 | 32 | 2.37e-01 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 2 | 23 | 2.37e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 4 | 40 | 2.37e-01 |
| GO:BP | GO:0008033 | tRNA processing | 44 | 128 | 2.38e-01 |
| GO:BP | GO:0031584 | activation of phospholipase D activity | 2 | 4 | 2.38e-01 |
| GO:BP | GO:0031990 | mRNA export from nucleus in response to heat stress | 1 | 2 | 2.38e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 6 | 8 | 2.38e-01 |
| GO:BP | GO:0034402 | recruitment of 3’-end processing factors to RNA polymerase II holoenzyme complex | 1 | 2 | 2.38e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 7 | 9 | 2.38e-01 |
| GO:BP | GO:1902228 | positive regulation of macrophage colony-stimulating factor signaling pathway | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0060611 | mammary gland fat development | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0003164 | His-Purkinje system development | 5 | 7 | 2.39e-01 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 4 | 13 | 2.39e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 6 | 159 | 2.39e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 1 | 30 | 2.39e-01 |
| GO:BP | GO:0046051 | UTP metabolic process | 8 | 10 | 2.39e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 2.39e-01 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 3 | 18 | 2.39e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 3 | 14 | 2.39e-01 |
| GO:BP | GO:0039536 | negative regulation of RIG-I signaling pathway | 2 | 7 | 2.39e-01 |
| GO:BP | GO:0140374 | antiviral innate immune response | 13 | 17 | 2.39e-01 |
| GO:BP | GO:0003241 | growth involved in heart morphogenesis | 3 | 4 | 2.39e-01 |
| GO:BP | GO:0003273 | cell migration involved in endocardial cushion formation | 3 | 4 | 2.39e-01 |
| GO:BP | GO:0072709 | cellular response to sorbitol | 2 | 5 | 2.39e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 6 | 352 | 2.39e-01 |
| GO:BP | GO:0008050 | female courtship behavior | 2 | 2 | 2.41e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 2 | 21 | 2.42e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 1 | 45 | 2.42e-01 |
| GO:BP | GO:1900247 | regulation of cytoplasmic translational elongation | 7 | 10 | 2.42e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 1 | 27 | 2.42e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 2 | 137 | 2.42e-01 |
| GO:BP | GO:0032571 | response to vitamin K | 1 | 5 | 2.42e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 5 | 21 | 2.42e-01 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 23 | 36 | 2.42e-01 |
| GO:BP | GO:0006298 | mismatch repair | 2 | 29 | 2.42e-01 |
| GO:BP | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 4 | 4 | 2.42e-01 |
| GO:BP | GO:0006228 | UTP biosynthetic process | 4 | 8 | 2.42e-01 |
| GO:BP | GO:1902990 | mitotic telomere maintenance via semi-conservative replication | 1 | 2 | 2.42e-01 |
| GO:BP | GO:0050893 | sensory processing | 4 | 4 | 2.42e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 2 | 56 | 2.42e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 2 | 39 | 2.42e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 6 | 7 | 2.42e-01 |
| GO:BP | GO:1903613 | regulation of protein tyrosine phosphatase activity | 1 | 4 | 2.42e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 4 | 103 | 2.42e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 8 | 11 | 2.45e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 1 | 48 | 2.45e-01 |
| GO:BP | GO:0098925 | retrograde trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 1 | 1 | 2.45e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 3 | 37 | 2.45e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 4 | 15 | 2.45e-01 |
| GO:BP | GO:0006489 | dolichyl diphosphate biosynthetic process | 3 | 5 | 2.45e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 1 | 38 | 2.45e-01 |
| GO:BP | GO:0033387 | putrescine biosynthetic process from ornithine | 1 | 3 | 2.45e-01 |
| GO:BP | GO:0060449 | bud elongation involved in lung branching | 1 | 7 | 2.45e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 1 | 140 | 2.45e-01 |
| GO:BP | GO:0046465 | dolichyl diphosphate metabolic process | 3 | 5 | 2.45e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 6 | 161 | 2.45e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 10 | 28 | 2.45e-01 |
| GO:BP | GO:0061912 | selective autophagy | 42 | 90 | 2.45e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 1 | 33 | 2.49e-01 |
| GO:BP | GO:0046599 | regulation of centriole replication | 14 | 21 | 2.50e-01 |
| GO:BP | GO:0033326 | cerebrospinal fluid secretion | 2 | 3 | 2.51e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 5 | 9 | 2.52e-01 |
| GO:BP | GO:0022614 | membrane to membrane docking | 3 | 5 | 2.52e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 6 | 41 | 2.52e-01 |
| GO:BP | GO:0070914 | UV-damage excision repair | 9 | 13 | 2.52e-01 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 8 | 11 | 2.52e-01 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 9 | 2.52e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 3 | 6 | 2.52e-01 |
| GO:BP | GO:0016236 | macroautophagy | 83 | 308 | 2.52e-01 |
| GO:BP | GO:0009116 | nucleoside metabolic process | 1 | 42 | 2.52e-01 |
| GO:BP | GO:0106015 | negative regulation of inflammatory response to wounding | 1 | 2 | 2.52e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 8 | 11 | 2.52e-01 |
| GO:BP | GO:0045618 | positive regulation of keratinocyte differentiation | 10 | 12 | 2.52e-01 |
| GO:BP | GO:0071569 | protein ufmylation | 4 | 6 | 2.53e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 29 | 57 | 2.53e-01 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 3 | 10 | 2.54e-01 |
| GO:BP | GO:1990743 | protein sialylation | 1 | 5 | 2.54e-01 |
| GO:BP | GO:2000791 | negative regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 2.54e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 9 | 23 | 2.54e-01 |
| GO:BP | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development | 2 | 2 | 2.54e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 1 | 53 | 2.54e-01 |
| GO:BP | GO:1903954 | positive regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.54e-01 |
| GO:BP | GO:0070293 | renal absorption | 4 | 24 | 2.54e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 8 | 15 | 2.54e-01 |
| GO:BP | GO:1903952 | regulation of voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization | 1 | 1 | 2.54e-01 |
| GO:BP | GO:0046361 | 2-oxobutyrate metabolic process | 1 | 2 | 2.54e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 70 | 137 | 2.55e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 5 | 59 | 2.55e-01 |
| GO:BP | GO:0021591 | ventricular system development | 13 | 29 | 2.55e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 7 | 16 | 2.55e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 115 | 235 | 2.55e-01 |
| GO:BP | GO:0055059 | asymmetric neuroblast division | 1 | 6 | 2.55e-01 |
| GO:BP | GO:0080170 | hydrogen peroxide transmembrane transport | 1 | 1 | 2.55e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 1 | 3 | 2.55e-01 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 4 | 16 | 2.55e-01 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 2 | 26 | 2.55e-01 |
| GO:BP | GO:0045007 | depurination | 4 | 4 | 2.56e-01 |
| GO:BP | GO:1903714 | isoleucine transmembrane transport | 1 | 1 | 2.56e-01 |
| GO:BP | GO:1903806 | L-isoleucine import across plasma membrane | 1 | 1 | 2.56e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 1 | 42 | 2.57e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 11 | 38 | 2.57e-01 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 1 | 50 | 2.57e-01 |
| GO:BP | GO:0046688 | response to copper ion | 2 | 29 | 2.57e-01 |
| GO:BP | GO:0071581 | regulation of zinc ion transmembrane import | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0071582 | negative regulation of zinc ion transport | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0071584 | negative regulation of zinc ion transmembrane import | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0071583 | negative regulation of zinc ion transmembrane transport | 1 | 1 | 2.58e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 2 | 57 | 2.58e-01 |
| GO:BP | GO:0071580 | regulation of zinc ion transmembrane transport | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0003192 | mitral valve formation | 3 | 3 | 2.58e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 6 | 32 | 2.58e-01 |
| GO:BP | GO:0021670 | lateral ventricle development | 5 | 11 | 2.58e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 6 | 155 | 2.58e-01 |
| GO:BP | GO:0006244 | pyrimidine nucleotide catabolic process | 2 | 19 | 2.58e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 2.59e-01 |
| GO:BP | GO:0061687 | detoxification of inorganic compound | 3 | 12 | 2.59e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 9 | 569 | 2.59e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 2.59e-01 |
| GO:BP | GO:0097501 | stress response to metal ion | 3 | 11 | 2.59e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 19 | 30 | 2.59e-01 |
| GO:BP | GO:1903566 | positive regulation of protein localization to cilium | 5 | 7 | 2.59e-01 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 2 | 20 | 2.59e-01 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 8 | 11 | 2.60e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 3 | 48 | 2.60e-01 |
| GO:BP | GO:0021599 | abducens nerve formation | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 4 | 14 | 2.60e-01 |
| GO:BP | GO:0021560 | abducens nerve development | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0021598 | abducens nerve morphogenesis | 1 | 1 | 2.60e-01 |
| GO:BP | GO:0021572 | rhombomere 6 development | 1 | 1 | 2.60e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 4 | 110 | 2.60e-01 |
| GO:BP | GO:0140042 | lipid droplet formation | 8 | 12 | 2.60e-01 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 9 | 13 | 2.60e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 30 | 52 | 2.60e-01 |
| GO:BP | GO:0060083 | smooth muscle contraction involved in micturition | 1 | 2 | 2.60e-01 |
| GO:BP | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain | 1 | 2 | 2.60e-01 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 4 | 17 | 2.61e-01 |
| GO:BP | GO:0060219 | camera-type eye photoreceptor cell differentiation | 10 | 18 | 2.61e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 2 | 7 | 2.61e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 1 | 35 | 2.62e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 15 | 25 | 2.62e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 5 | 7 | 2.62e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 5 | 103 | 2.62e-01 |
| GO:BP | GO:0140450 | protein targeting to Golgi apparatus | 1 | 1 | 2.62e-01 |
| GO:BP | GO:0006275 | regulation of DNA replication | 61 | 116 | 2.62e-01 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 2 | 33 | 2.62e-01 |
| GO:BP | GO:1903542 | negative regulation of exosomal secretion | 1 | 4 | 2.62e-01 |
| GO:BP | GO:0070676 | intralumenal vesicle formation | 1 | 4 | 2.62e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 29 | 58 | 2.62e-01 |
| GO:BP | GO:0022406 | membrane docking | 3 | 84 | 2.62e-01 |
| GO:BP | GO:0035166 | post-embryonic hemopoiesis | 3 | 5 | 2.62e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 2.65e-01 |
| GO:BP | GO:0086092 | regulation of the force of heart contraction by cardiac conduction | 1 | 2 | 2.65e-01 |
| GO:BP | GO:2000809 | positive regulation of synaptic vesicle clustering | 1 | 3 | 2.65e-01 |
| GO:BP | GO:0010288 | response to lead ion | 2 | 18 | 2.65e-01 |
| GO:BP | GO:2000302 | positive regulation of synaptic vesicle exocytosis | 1 | 3 | 2.65e-01 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 2.65e-01 |
| GO:BP | GO:2000206 | regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 2 | 784 | 2.65e-01 |
| GO:BP | GO:1901656 | glycoside transport | 4 | 5 | 2.65e-01 |
| GO:BP | GO:2000202 | positive regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 5 | 22 | 2.65e-01 |
| GO:BP | GO:2000200 | regulation of ribosomal subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:2000199 | positive regulation of ribonucleoprotein complex localization | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0042946 | glucoside transport | 3 | 3 | 2.65e-01 |
| GO:BP | GO:0006272 | leading strand elongation | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0021773 | striatal medium spiny neuron differentiation | 1 | 3 | 2.65e-01 |
| GO:BP | GO:0044849 | estrous cycle | 2 | 13 | 2.65e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 3 | 28 | 2.65e-01 |
| GO:BP | GO:1903281 | positive regulation of calcium:sodium antiporter activity | 1 | 2 | 2.65e-01 |
| GO:BP | GO:1900073 | regulation of neuromuscular synaptic transmission | 1 | 2 | 2.65e-01 |
| GO:BP | GO:1900075 | positive regulation of neuromuscular synaptic transmission | 1 | 2 | 2.65e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 12 | 21 | 2.65e-01 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 2 | 19 | 2.65e-01 |
| GO:BP | GO:2000208 | positive regulation of ribosomal small subunit export from nucleus | 1 | 1 | 2.65e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 1 | 36 | 2.65e-01 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 46 | 89 | 2.65e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 28 | 61 | 2.65e-01 |
| GO:BP | GO:0001880 | Mullerian duct regression | 3 | 4 | 2.65e-01 |
| GO:BP | GO:0099555 | trans-synaptic signaling by nitric oxide, modulating synaptic transmission | 2 | 2 | 2.66e-01 |
| GO:BP | GO:0099554 | trans-synaptic signaling by soluble gas, modulating synaptic transmission | 2 | 2 | 2.66e-01 |
| GO:BP | GO:0036372 | opsin transport | 1 | 2 | 2.68e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 1 | 38 | 2.68e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 6 | 33 | 2.68e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 2 | 560 | 2.69e-01 |
| GO:BP | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning | 1 | 1 | 2.69e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 166 | 338 | 2.70e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 2.70e-01 |
| GO:BP | GO:0072719 | cellular response to cisplatin | 4 | 4 | 2.70e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 1 | 42 | 2.70e-01 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 2 | 21 | 2.70e-01 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 4 | 4 | 2.70e-01 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 14 | 33 | 2.70e-01 |
| GO:BP | GO:0046075 | dTTP metabolic process | 4 | 4 | 2.70e-01 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 4 | 4 | 2.70e-01 |
| GO:BP | GO:0046108 | uridine metabolic process | 1 | 1 | 2.70e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 15 | 93 | 2.70e-01 |
| GO:BP | GO:0006218 | uridine catabolic process | 1 | 1 | 2.70e-01 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 9 | 12 | 2.70e-01 |
| GO:BP | GO:0055070 | copper ion homeostasis | 4 | 18 | 2.70e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 7 | 41 | 2.71e-01 |
| GO:BP | GO:0032077 | positive regulation of deoxyribonuclease activity | 2 | 5 | 2.71e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 5 | 293 | 2.72e-01 |
| GO:BP | GO:0006222 | UMP biosynthetic process | 2 | 8 | 2.72e-01 |
| GO:BP | GO:1905443 | regulation of clathrin coat assembly | 1 | 1 | 2.72e-01 |
| GO:BP | GO:1905445 | positive regulation of clathrin coat assembly | 1 | 1 | 2.72e-01 |
| GO:BP | GO:0048703 | embryonic viscerocranium morphogenesis | 1 | 8 | 2.72e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 2.72e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 11 | 490 | 2.72e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 11 | 490 | 2.72e-01 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 1 | 3 | 2.72e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 9 | 13 | 2.72e-01 |
| GO:BP | GO:0060856 | establishment of blood-brain barrier | 2 | 9 | 2.72e-01 |
| GO:BP | GO:0071955 | recycling endosome to Golgi transport | 1 | 2 | 2.72e-01 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 1 | 4 | 2.72e-01 |
| GO:BP | GO:0002138 | retinoic acid biosynthetic process | 1 | 8 | 2.72e-01 |
| GO:BP | GO:0006421 | asparaginyl-tRNA aminoacylation | 3 | 3 | 2.72e-01 |
| GO:BP | GO:1990535 | neuron projection maintenance | 6 | 9 | 2.72e-01 |
| GO:BP | GO:0090042 | tubulin deacetylation | 9 | 23 | 2.72e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 4 | 9 | 2.72e-01 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 12 | 19 | 2.73e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 132 | 273 | 2.73e-01 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 56 | 100 | 2.73e-01 |
| GO:BP | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell | 1 | 3 | 2.74e-01 |
| GO:BP | GO:0021894 | cerebral cortex GABAergic interneuron development | 1 | 3 | 2.75e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 3 | 83 | 2.75e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 18 | 53 | 2.75e-01 |
| GO:BP | GO:0071467 | cellular response to pH | 2 | 16 | 2.75e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 27 | 1145 | 2.75e-01 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 1 | 64 | 2.75e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 2 | 532 | 2.75e-01 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 38 | 70 | 2.75e-01 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 5 | 16 | 2.75e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 5 | 6 | 2.75e-01 |
| GO:BP | GO:0098849 | cellular detoxification of cadmium ion | 1 | 1 | 2.75e-01 |
| GO:BP | GO:0033319 | UDP-D-xylose metabolic process | 1 | 1 | 2.75e-01 |
| GO:BP | GO:2000256 | positive regulation of male germ cell proliferation | 2 | 2 | 2.75e-01 |
| GO:BP | GO:1905938 | positive regulation of germ cell proliferation | 2 | 2 | 2.75e-01 |
| GO:BP | GO:1990646 | cellular response to prolactin | 2 | 2 | 2.75e-01 |
| GO:BP | GO:0006312 | mitotic recombination | 15 | 22 | 2.75e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 2 | 83 | 2.75e-01 |
| GO:BP | GO:0033320 | UDP-D-xylose biosynthetic process | 1 | 1 | 2.75e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 2.75e-01 |
| GO:BP | GO:0140961 | cellular detoxification of metal ion | 1 | 1 | 2.75e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 46 | 106 | 2.75e-01 |
| GO:BP | GO:1903361 | protein localization to basolateral plasma membrane | 1 | 7 | 2.75e-01 |
| GO:BP | GO:0090677 | reversible differentiation | 2 | 5 | 2.75e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 5 | 67 | 2.75e-01 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 1 | 62 | 2.75e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 3 | 21 | 2.77e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 11 | 495 | 2.77e-01 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 4 | 12 | 2.77e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 2.77e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 2 | 30 | 2.77e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 60 | 121 | 2.79e-01 |
| GO:BP | GO:0035623 | renal glucose absorption | 2 | 2 | 2.79e-01 |
| GO:BP | GO:1904978 | regulation of endosome organization | 1 | 5 | 2.79e-01 |
| GO:BP | GO:0022615 | protein to membrane docking | 1 | 5 | 2.79e-01 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 4 | 4 | 2.79e-01 |
| GO:BP | GO:0043622 | cortical microtubule organization | 3 | 4 | 2.80e-01 |
| GO:BP | GO:0016102 | diterpenoid biosynthetic process | 1 | 8 | 2.81e-01 |
| GO:BP | GO:0097490 | sympathetic neuron projection extension | 4 | 4 | 2.81e-01 |
| GO:BP | GO:0097491 | sympathetic neuron projection guidance | 4 | 4 | 2.81e-01 |
| GO:BP | GO:0042126 | nitrate metabolic process | 3 | 4 | 2.82e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 16 | 50 | 2.82e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 39 | 87 | 2.82e-01 |
| GO:BP | GO:0035640 | exploration behavior | 3 | 21 | 2.82e-01 |
| GO:BP | GO:0035927 | RNA import into mitochondrion | 3 | 4 | 2.84e-01 |
| GO:BP | GO:0016073 | snRNA metabolic process | 8 | 54 | 2.84e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 2 | 7 | 2.84e-01 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 2 | 15 | 2.84e-01 |
| GO:BP | GO:0099068 | postsynapse assembly | 2 | 32 | 2.84e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 1 | 26 | 2.84e-01 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 17 | 28 | 2.84e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 3 | 28 | 2.84e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 2.84e-01 |
| GO:BP | GO:0090210 | regulation of establishment of blood-brain barrier | 1 | 3 | 2.84e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 2.84e-01 |
| GO:BP | GO:1905198 | manchette assembly | 4 | 4 | 2.84e-01 |
| GO:BP | GO:0014886 | transition between slow and fast fiber | 1 | 1 | 2.84e-01 |
| GO:BP | GO:0045955 | negative regulation of calcium ion-dependent exocytosis | 4 | 6 | 2.84e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 7 | 10 | 2.84e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 3 | 94 | 2.85e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 1 | 57 | 2.86e-01 |
| GO:BP | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential | 2 | 3 | 2.87e-01 |
| GO:BP | GO:0021660 | rhombomere 3 formation | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 1 | 60 | 2.87e-01 |
| GO:BP | GO:0021664 | rhombomere 5 morphogenesis | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0036497 | eIF2alpha dephosphorylation in response to endoplasmic reticulum stress | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0021665 | rhombomere 5 structural organization | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.87e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 23 | 1712 | 2.87e-01 |
| GO:BP | GO:0106014 | regulation of inflammatory response to wounding | 1 | 4 | 2.87e-01 |
| GO:BP | GO:0070295 | renal water absorption | 3 | 5 | 2.87e-01 |
| GO:BP | GO:0021666 | rhombomere 5 formation | 1 | 1 | 2.87e-01 |
| GO:BP | GO:1903929 | primary palate development | 1 | 2 | 2.87e-01 |
| GO:BP | GO:0016080 | synaptic vesicle targeting | 1 | 3 | 2.87e-01 |
| GO:BP | GO:0021659 | rhombomere 3 structural organization | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 10 | 26 | 2.87e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 10 | 331 | 2.87e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 12 | 88 | 2.87e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 2 | 41 | 2.87e-01 |
| GO:BP | GO:0090031 | positive regulation of steroid hormone biosynthetic process | 3 | 5 | 2.87e-01 |
| GO:BP | GO:0021594 | rhombomere formation | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 45 | 79 | 2.87e-01 |
| GO:BP | GO:0032458 | slow endocytic recycling | 2 | 2 | 2.87e-01 |
| GO:BP | GO:0051877 | pigment granule aggregation in cell center | 1 | 2 | 2.87e-01 |
| GO:BP | GO:0021595 | rhombomere structural organization | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0016197 | endosomal transport | 26 | 231 | 2.87e-01 |
| GO:BP | GO:0009446 | putrescine biosynthetic process | 1 | 4 | 2.87e-01 |
| GO:BP | GO:1904046 | negative regulation of vascular endothelial growth factor production | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0006884 | cell volume homeostasis | 2 | 24 | 2.87e-01 |
| GO:BP | GO:0086048 | membrane depolarization during bundle of His cell action potential | 2 | 3 | 2.87e-01 |
| GO:BP | GO:0014041 | regulation of neuron maturation | 1 | 6 | 2.87e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 7 | 49 | 2.87e-01 |
| GO:BP | GO:1903917 | positive regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 2.87e-01 |
| GO:BP | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 2 | 18 | 2.87e-01 |
| GO:BP | GO:0060682 | primary ureteric bud growth | 1 | 1 | 2.87e-01 |
| GO:BP | GO:1903916 | regulation of endoplasmic reticulum stress-induced eIF2 alpha dephosphorylation | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 18 | 30 | 2.87e-01 |
| GO:BP | GO:0061642 | chemoattraction of axon | 1 | 2 | 2.87e-01 |
| GO:BP | GO:0009174 | pyrimidine ribonucleoside monophosphate biosynthetic process | 2 | 9 | 2.87e-01 |
| GO:BP | GO:0006655 | phosphatidylglycerol biosynthetic process | 7 | 9 | 2.87e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 4 | 147 | 2.87e-01 |
| GO:BP | GO:0030656 | regulation of vitamin metabolic process | 1 | 7 | 2.87e-01 |
| GO:BP | GO:0042746 | circadian sleep/wake cycle, wakefulness | 1 | 1 | 2.87e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 1 | 63 | 2.87e-01 |
| GO:BP | GO:1903546 | protein localization to photoreceptor outer segment | 3 | 5 | 2.87e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 4 | 90 | 2.88e-01 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 5 | 21 | 2.88e-01 |
| GO:BP | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway | 4 | 5 | 2.88e-01 |
| GO:BP | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I | 3 | 7 | 2.88e-01 |
| GO:BP | GO:0006189 | ‘de novo’ IMP biosynthetic process | 5 | 6 | 2.88e-01 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 28 | 48 | 2.88e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 5 | 6 | 2.88e-01 |
| GO:BP | GO:0018352 | protein-pyridoxal-5-phosphate linkage | 1 | 2 | 2.88e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 5 | 75 | 2.88e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 2 | 17 | 2.88e-01 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 3 | 11 | 2.88e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 2.88e-01 |
| GO:BP | GO:1905940 | negative regulation of gonad development | 2 | 2 | 2.88e-01 |
| GO:BP | GO:0006540 | glutamate decarboxylation to succinate | 1 | 1 | 2.88e-01 |
| GO:BP | GO:0051382 | kinetochore assembly | 12 | 18 | 2.88e-01 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 12 | 17 | 2.88e-01 |
| GO:BP | GO:0051648 | vesicle localization | 7 | 192 | 2.89e-01 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 1 | 3 | 2.89e-01 |
| GO:BP | GO:1902723 | negative regulation of skeletal muscle satellite cell proliferation | 2 | 2 | 2.89e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 5 | 29 | 2.89e-01 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 9 | 13 | 2.90e-01 |
| GO:BP | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth | 1 | 2 | 2.90e-01 |
| GO:BP | GO:0051301 | cell division | 274 | 570 | 2.90e-01 |
| GO:BP | GO:1900029 | positive regulation of ruffle assembly | 4 | 12 | 2.90e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 2 | 25 | 2.91e-01 |
| GO:BP | GO:0007616 | long-term memory | 7 | 31 | 2.91e-01 |
| GO:BP | GO:1901207 | regulation of heart looping | 2 | 2 | 2.91e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 2.92e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 2 | 35 | 2.92e-01 |
| GO:BP | GO:0010447 | response to acidic pH | 2 | 16 | 2.92e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 11 | 511 | 2.93e-01 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 5 | 11 | 2.93e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 18 | 875 | 2.93e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 2 | 59 | 2.93e-01 |
| GO:BP | GO:0150118 | negative regulation of cell-substrate junction organization | 6 | 14 | 2.93e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 6 | 39 | 2.93e-01 |
| GO:BP | GO:0051895 | negative regulation of focal adhesion assembly | 6 | 14 | 2.93e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 2.93e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 2.93e-01 |
| GO:BP | GO:0097338 | response to clozapine | 2 | 2 | 2.95e-01 |
| GO:BP | GO:0006424 | glutamyl-tRNA aminoacylation | 1 | 2 | 2.95e-01 |
| GO:BP | GO:0006433 | prolyl-tRNA aminoacylation | 1 | 2 | 2.95e-01 |
| GO:BP | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process | 1 | 5 | 2.97e-01 |
| GO:BP | GO:1902337 | regulation of apoptotic process involved in morphogenesis | 3 | 9 | 2.97e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 9 | 14 | 2.97e-01 |
| GO:BP | GO:1904748 | regulation of apoptotic process involved in development | 3 | 9 | 2.97e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 4 | 57 | 2.97e-01 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 3 | 5 | 2.97e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 9 | 14 | 2.97e-01 |
| GO:BP | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development | 3 | 5 | 2.97e-01 |
| GO:BP | GO:0035499 | carnosine biosynthetic process | 1 | 1 | 2.97e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 4 | 31 | 2.97e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 2 | 81 | 2.97e-01 |
| GO:BP | GO:1903279 | regulation of calcium:sodium antiporter activity | 1 | 3 | 2.97e-01 |
| GO:BP | GO:0097698 | telomere maintenance via base-excision repair | 1 | 1 | 2.97e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 4 | 23 | 2.98e-01 |
| GO:BP | GO:0099576 | regulation of protein catabolic process at postsynapse, modulating synaptic transmission | 1 | 4 | 2.98e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 2 | 3 | 2.98e-01 |
| GO:BP | GO:0003162 | atrioventricular node development | 5 | 8 | 2.98e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 1 | 252 | 2.98e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 10 | 35 | 2.98e-01 |
| GO:BP | GO:0035552 | oxidative single-stranded DNA demethylation | 2 | 3 | 2.99e-01 |
| GO:BP | GO:0140029 | exocytic process | 3 | 69 | 2.99e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 1 | 45 | 3.00e-01 |
| GO:BP | GO:0098924 | retrograde trans-synaptic signaling by nitric oxide | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 4 | 7 | 3.00e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 4 | 7 | 3.00e-01 |
| GO:BP | GO:0098923 | retrograde trans-synaptic signaling by soluble gas | 1 | 1 | 3.00e-01 |
| GO:BP | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | 1 | 5 | 3.00e-01 |
| GO:BP | GO:1900449 | regulation of glutamate receptor signaling pathway | 1 | 5 | 3.00e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 3 | 132 | 3.00e-01 |
| GO:BP | GO:0008614 | pyridoxine metabolic process | 1 | 2 | 3.00e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 3.00e-01 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 2 | 36 | 3.00e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 5 | 7 | 3.00e-01 |
| GO:BP | GO:0042823 | pyridoxal phosphate biosynthetic process | 1 | 2 | 3.00e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 4 | 14 | 3.00e-01 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 1 | 10 | 3.00e-01 |
| GO:BP | GO:0008615 | pyridoxine biosynthetic process | 1 | 2 | 3.00e-01 |
| GO:BP | GO:0106034 | protein maturation by [2Fe-2S] cluster transfer | 1 | 3 | 3.00e-01 |
| GO:BP | GO:0050960 | detection of temperature stimulus involved in thermoception | 1 | 3 | 3.00e-01 |
| GO:BP | GO:0060056 | mammary gland involution | 3 | 8 | 3.00e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 6 | 8 | 3.00e-01 |
| GO:BP | GO:0071502 | cellular response to temperature stimulus | 1 | 2 | 3.00e-01 |
| GO:BP | GO:0072034 | renal vesicle induction | 3 | 3 | 3.00e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 2 | 4 | 3.00e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 3.00e-01 |
| GO:BP | GO:0035513 | oxidative RNA demethylation | 3 | 5 | 3.00e-01 |
| GO:BP | GO:0033499 | galactose catabolic process via UDP-galactose | 2 | 4 | 3.00e-01 |
| GO:BP | GO:0060039 | pericardium development | 7 | 17 | 3.00e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 9 | 16 | 3.00e-01 |
| GO:BP | GO:1905222 | atrioventricular canal morphogenesis | 3 | 4 | 3.00e-01 |
| GO:BP | GO:0097428 | protein maturation by iron-sulfur cluster transfer | 3 | 17 | 3.01e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 3 | 97 | 3.02e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 4 | 16 | 3.04e-01 |
| GO:BP | GO:0034644 | cellular response to UV | 4 | 86 | 3.04e-01 |
| GO:BP | GO:0010038 | response to metal ion | 13 | 264 | 3.04e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 4 | 18 | 3.04e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 10 | 27 | 3.04e-01 |
| GO:BP | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | 3 | 16 | 3.04e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 5 | 22 | 3.04e-01 |
| GO:BP | GO:0046074 | dTMP catabolic process | 1 | 3 | 3.04e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 42 | 89 | 3.05e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 5 | 45 | 3.06e-01 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 9 | 18 | 3.06e-01 |
| GO:BP | GO:0043038 | amino acid activation | 7 | 44 | 3.06e-01 |
| GO:BP | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process | 5 | 6 | 3.06e-01 |
| GO:BP | GO:1902339 | positive regulation of apoptotic process involved in morphogenesis | 1 | 5 | 3.06e-01 |
| GO:BP | GO:0044205 | ‘de novo’ UMP biosynthetic process | 2 | 3 | 3.06e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 3 | 47 | 3.06e-01 |
| GO:BP | GO:1904747 | positive regulation of apoptotic process involved in development | 1 | 5 | 3.06e-01 |
| GO:BP | GO:0007182 | common-partner SMAD protein phosphorylation | 1 | 6 | 3.06e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 3.06e-01 |
| GO:BP | GO:0051231 | spindle elongation | 9 | 12 | 3.06e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 3.06e-01 |
| GO:BP | GO:0030578 | PML body organization | 1 | 6 | 3.06e-01 |
| GO:BP | GO:0045951 | positive regulation of mitotic recombination | 1 | 1 | 3.06e-01 |
| GO:BP | GO:2001014 | regulation of skeletal muscle cell differentiation | 4 | 16 | 3.06e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 13 | 464 | 3.06e-01 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 4 | 5 | 3.06e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 3.06e-01 |
| GO:BP | GO:0035329 | hippo signaling | 21 | 41 | 3.07e-01 |
| GO:BP | GO:0043584 | nose development | 1 | 10 | 3.07e-01 |
| GO:BP | GO:0032352 | positive regulation of hormone metabolic process | 1 | 11 | 3.07e-01 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 13 | 24 | 3.07e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 6 | 160 | 3.07e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 2 | 35 | 3.07e-01 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 4 | 41 | 3.07e-01 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 14 | 27 | 3.08e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 27 | 130 | 3.08e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 3 | 50 | 3.08e-01 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 2 | 9 | 3.08e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 3.08e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 57 | 213 | 3.08e-01 |
| GO:BP | GO:1905033 | positive regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 2 | 3.08e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 143 | 308 | 3.08e-01 |
| GO:BP | GO:0097089 | methyl-branched fatty acid metabolic process | 1 | 3 | 3.08e-01 |
| GO:BP | GO:0030034 | microvillar actin bundle assembly | 1 | 1 | 3.08e-01 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 3.08e-01 |
| GO:BP | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 1 | 2 | 3.08e-01 |
| GO:BP | GO:0090383 | phagosome acidification | 1 | 5 | 3.08e-01 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 17 | 30 | 3.08e-01 |
| GO:BP | GO:1905026 | positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.08e-01 |
| GO:BP | GO:1900452 | regulation of long-term synaptic depression | 2 | 10 | 3.08e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 2 | 22 | 3.08e-01 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 3.08e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 7 | 44 | 3.08e-01 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 6 | 8 | 3.08e-01 |
| GO:BP | GO:0001824 | blastocyst development | 1 | 94 | 3.08e-01 |
| GO:BP | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction | 2 | 2 | 3.09e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 3 | 20 | 3.09e-01 |
| GO:BP | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway | 2 | 6 | 3.09e-01 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 10 | 3.09e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 3 | 216 | 3.09e-01 |
| GO:BP | GO:2000565 | negative regulation of CD8-positive, alpha-beta T cell proliferation | 2 | 2 | 3.09e-01 |
| GO:BP | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter | 1 | 1 | 3.09e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 3 | 70 | 3.09e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 3 | 54 | 3.09e-01 |
| GO:BP | GO:0034964 | box H/ACA RNA processing | 2 | 2 | 3.09e-01 |
| GO:BP | GO:0010225 | response to UV-C | 4 | 13 | 3.09e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 3.09e-01 |
| GO:BP | GO:0000495 | box H/ACA RNA 3’-end processing | 2 | 2 | 3.09e-01 |
| GO:BP | GO:0009411 | response to UV | 3 | 141 | 3.10e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 17 | 32 | 3.10e-01 |
| GO:BP | GO:0022616 | DNA strand elongation | 2 | 35 | 3.10e-01 |
| GO:BP | GO:0055009 | atrial cardiac muscle tissue morphogenesis | 2 | 5 | 3.10e-01 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 3 | 25 | 3.10e-01 |
| GO:BP | GO:0032075 | positive regulation of nuclease activity | 1 | 7 | 3.10e-01 |
| GO:BP | GO:1903966 | monounsaturated fatty acid biosynthetic process | 1 | 2 | 3.11e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 1 | 11 | 3.11e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 8 | 72 | 3.11e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 13 | 901 | 3.11e-01 |
| GO:BP | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation | 1 | 1 | 3.11e-01 |
| GO:BP | GO:1902045 | negative regulation of Fas signaling pathway | 2 | 2 | 3.11e-01 |
| GO:BP | GO:0043490 | malate-aspartate shuttle | 2 | 4 | 3.11e-01 |
| GO:BP | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0051823 | regulation of synapse structural plasticity | 1 | 5 | 3.11e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 3 | 4 | 3.11e-01 |
| GO:BP | GO:0043602 | nitrate catabolic process | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0046210 | nitric oxide catabolic process | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0060945 | cardiac neuron differentiation | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0034199 | activation of protein kinase A activity | 3 | 3 | 3.11e-01 |
| GO:BP | GO:0006666 | 3-keto-sphinganine metabolic process | 1 | 1 | 3.11e-01 |
| GO:BP | GO:1990359 | stress response to zinc ion | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0010312 | detoxification of zinc ion | 1 | 1 | 3.11e-01 |
| GO:BP | GO:0031860 | telomeric 3’ overhang formation | 1 | 4 | 3.12e-01 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 1 | 56 | 3.12e-01 |
| GO:BP | GO:0021892 | cerebral cortex GABAergic interneuron differentiation | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 4 | 20 | 3.12e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 5 | 23 | 3.13e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 7 | 67 | 3.15e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 2 | 84 | 3.16e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 20 | 32 | 3.16e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 2 | 22 | 3.16e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 1 | 6 | 3.18e-01 |
| GO:BP | GO:0009056 | catabolic process | 26 | 2088 | 3.18e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 5 | 76 | 3.18e-01 |
| GO:BP | GO:0071393 | cellular response to progesterone stimulus | 1 | 5 | 3.18e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 3.18e-01 |
| GO:BP | GO:0010868 | negative regulation of triglyceride biosynthetic process | 3 | 3 | 3.18e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 3.18e-01 |
| GO:BP | GO:0046754 | viral exocytosis | 1 | 1 | 3.18e-01 |
| GO:BP | GO:0002505 | antigen processing and presentation of polysaccharide antigen via MHC class II | 1 | 1 | 3.18e-01 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 3.18e-01 |
| GO:BP | GO:0036072 | direct ossification | 1 | 7 | 3.18e-01 |
| GO:BP | GO:0071051 | polyadenylation-dependent snoRNA 3’-end processing | 5 | 7 | 3.18e-01 |
| GO:BP | GO:0002747 | antigen processing and presentation following phagocytosis | 1 | 1 | 3.18e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 35 | 71 | 3.18e-01 |
| GO:BP | GO:0000379 | tRNA-type intron splice site recognition and cleavage | 1 | 3 | 3.18e-01 |
| GO:BP | GO:0001771 | immunological synapse formation | 1 | 7 | 3.18e-01 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 3 | 28 | 3.18e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 3 | 2139 | 3.18e-01 |
| GO:BP | GO:0001957 | intramembranous ossification | 1 | 7 | 3.18e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 1 | 160 | 3.18e-01 |
| GO:BP | GO:0016114 | terpenoid biosynthetic process | 1 | 12 | 3.18e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 1 | 86 | 3.18e-01 |
| GO:BP | GO:0033484 | intracellular nitric oxide homeostasis | 2 | 3 | 3.18e-01 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 10 | 20 | 3.18e-01 |
| GO:BP | GO:0030592 | DNA ADP-ribosylation | 1 | 3 | 3.18e-01 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 10 | 18 | 3.18e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 5 | 114 | 3.18e-01 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 5 | 3.18e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 1 | 69 | 3.18e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 1 | 316 | 3.19e-01 |
| GO:BP | GO:0046755 | viral budding | 5 | 26 | 3.19e-01 |
| GO:BP | GO:0009451 | RNA modification | 53 | 162 | 3.19e-01 |
| GO:BP | GO:2000834 | regulation of androgen secretion | 1 | 1 | 3.19e-01 |
| GO:BP | GO:2000836 | positive regulation of androgen secretion | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0035935 | androgen secretion | 1 | 1 | 3.19e-01 |
| GO:BP | GO:0032902 | nerve growth factor production | 2 | 2 | 3.19e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 3 | 211 | 3.20e-01 |
| GO:BP | GO:0007522 | visceral muscle development | 1 | 1 | 3.20e-01 |
| GO:BP | GO:0045110 | intermediate filament bundle assembly | 5 | 6 | 3.20e-01 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 110 | 236 | 3.20e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 3 | 2161 | 3.20e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 1 | 311 | 3.20e-01 |
| GO:BP | GO:1901508 | positive regulation of acylglycerol transport | 1 | 2 | 3.20e-01 |
| GO:BP | GO:1901506 | regulation of acylglycerol transport | 1 | 2 | 3.20e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 5 | 234 | 3.20e-01 |
| GO:BP | GO:1905885 | positive regulation of triglyceride transport | 1 | 2 | 3.20e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 4 | 25 | 3.20e-01 |
| GO:BP | GO:1905883 | regulation of triglyceride transport | 1 | 2 | 3.20e-01 |
| GO:BP | GO:0061205 | paramesonephric duct development | 2 | 4 | 3.20e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 7 | 34 | 3.20e-01 |
| GO:BP | GO:0048565 | digestive tract development | 1 | 92 | 3.20e-01 |
| GO:BP | GO:0071504 | cellular response to heparin | 3 | 4 | 3.20e-01 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 18 | 30 | 3.20e-01 |
| GO:BP | GO:0003431 | growth plate cartilage chondrocyte development | 3 | 5 | 3.20e-01 |
| GO:BP | GO:1990261 | pre-mRNA catabolic process | 1 | 1 | 3.21e-01 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 23 | 33 | 3.21e-01 |
| GO:BP | GO:0097291 | renal phosphate ion absorption | 1 | 1 | 3.21e-01 |
| GO:BP | GO:0009092 | homoserine metabolic process | 2 | 5 | 3.21e-01 |
| GO:BP | GO:0019346 | transsulfuration | 2 | 5 | 3.21e-01 |
| GO:BP | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization | 2 | 4 | 3.21e-01 |
| GO:BP | GO:0015755 | fructose transmembrane transport | 3 | 5 | 3.22e-01 |
| GO:BP | GO:0032237 | activation of store-operated calcium channel activity | 2 | 5 | 3.22e-01 |
| GO:BP | GO:0001100 | negative regulation of exit from mitosis | 1 | 1 | 3.22e-01 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 2 | 23 | 3.22e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 1 | 11 | 3.22e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 10 | 55 | 3.22e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 3 | 40 | 3.22e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 2 | 740 | 3.22e-01 |
| GO:BP | GO:0055057 | neuroblast division | 1 | 12 | 3.22e-01 |
| GO:BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 12 | 18 | 3.22e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 3.22e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 3.22e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 3.22e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 3 | 28 | 3.22e-01 |
| GO:BP | GO:1901645 | regulation of synoviocyte proliferation | 1 | 3 | 3.22e-01 |
| GO:BP | GO:1901647 | positive regulation of synoviocyte proliferation | 1 | 3 | 3.22e-01 |
| GO:BP | GO:0072553 | terminal button organization | 1 | 5 | 3.22e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 3.22e-01 |
| GO:BP | GO:1900244 | positive regulation of synaptic vesicle endocytosis | 1 | 4 | 3.22e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 25 | 105 | 3.22e-01 |
| GO:BP | GO:0007296 | vitellogenesis | 2 | 4 | 3.22e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 19 | 50 | 3.22e-01 |
| GO:BP | GO:0014901 | satellite cell activation involved in skeletal muscle regeneration | 2 | 3 | 3.22e-01 |
| GO:BP | GO:0002941 | synoviocyte proliferation | 1 | 3 | 3.22e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 3.22e-01 |
| GO:BP | GO:0034498 | early endosome to Golgi transport | 3 | 9 | 3.23e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 63 | 287 | 3.23e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 6 | 14 | 3.23e-01 |
| GO:BP | GO:1904417 | positive regulation of xenophagy | 6 | 7 | 3.23e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 5 | 28 | 3.23e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 21 | 44 | 3.23e-01 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 6 | 7 | 3.23e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 7 | 46 | 3.23e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 8 | 24 | 3.23e-01 |
| GO:BP | GO:0090246 | convergent extension involved in somitogenesis | 1 | 1 | 3.23e-01 |
| GO:BP | GO:1904415 | regulation of xenophagy | 6 | 7 | 3.23e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 4 | 31 | 3.23e-01 |
| GO:BP | GO:2000969 | positive regulation of AMPA receptor activity | 1 | 4 | 3.23e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 3 | 74 | 3.23e-01 |
| GO:BP | GO:0007033 | vacuole organization | 5 | 197 | 3.23e-01 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 11 | 17 | 3.23e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 15 | 27 | 3.23e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 7 | 214 | 3.23e-01 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 11 | 17 | 3.23e-01 |
| GO:BP | GO:0099178 | regulation of retrograde trans-synaptic signaling by endocanabinoid | 2 | 3 | 3.23e-01 |
| GO:BP | GO:0002157 | positive regulation of thyroid hormone mediated signaling pathway | 2 | 3 | 3.23e-01 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 11 | 17 | 3.23e-01 |
| GO:BP | GO:0044345 | stromal-epithelial cell signaling involved in prostate gland development | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0007619 | courtship behavior | 2 | 3 | 3.23e-01 |
| GO:BP | GO:2000080 | negative regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 3.23e-01 |
| GO:BP | GO:0009445 | putrescine metabolic process | 1 | 6 | 3.23e-01 |
| GO:BP | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 1 | 17 | 3.23e-01 |
| GO:BP | GO:1902074 | response to salt | 13 | 271 | 3.24e-01 |
| GO:BP | GO:0072033 | renal vesicle formation | 6 | 8 | 3.24e-01 |
| GO:BP | GO:0061519 | macrophage homeostasis | 1 | 3 | 3.24e-01 |
| GO:BP | GO:0046049 | UMP metabolic process | 2 | 10 | 3.24e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 3.24e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 1 | 14 | 3.24e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 19 | 35 | 3.24e-01 |
| GO:BP | GO:0035702 | monocyte homeostasis | 1 | 2 | 3.24e-01 |
| GO:BP | GO:0014856 | skeletal muscle cell proliferation | 4 | 14 | 3.25e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 6 | 34 | 3.25e-01 |
| GO:BP | GO:0044154 | histone H3-K14 acetylation | 3 | 12 | 3.25e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 1 | 108 | 3.25e-01 |
| GO:BP | GO:0014004 | microglia differentiation | 1 | 6 | 3.25e-01 |
| GO:BP | GO:0001887 | selenium compound metabolic process | 2 | 2 | 3.25e-01 |
| GO:BP | GO:0038135 | ERBB2-ERBB4 signaling pathway | 3 | 4 | 3.25e-01 |
| GO:BP | GO:0043628 | regulatory ncRNA 3’-end processing | 10 | 19 | 3.25e-01 |
| GO:BP | GO:0001889 | liver development | 1 | 110 | 3.25e-01 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 3 | 13 | 3.25e-01 |
| GO:BP | GO:0055123 | digestive system development | 1 | 100 | 3.25e-01 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 1 | 11 | 3.25e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 10 | 27 | 3.26e-01 |
| GO:BP | GO:0072239 | metanephric glomerulus vasculature development | 3 | 6 | 3.26e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 4 | 12 | 3.26e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 1 | 112 | 3.26e-01 |
| GO:BP | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development | 5 | 13 | 3.26e-01 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 1 | 7 | 3.26e-01 |
| GO:BP | GO:1903975 | regulation of glial cell migration | 2 | 12 | 3.26e-01 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 1 | 8 | 3.26e-01 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 1 | 14 | 3.26e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 3.26e-01 |
| GO:BP | GO:0016078 | tRNA catabolic process | 1 | 14 | 3.26e-01 |
| GO:BP | GO:0000050 | urea cycle | 3 | 7 | 3.26e-01 |
| GO:BP | GO:0061743 | motor learning | 1 | 5 | 3.26e-01 |
| GO:BP | GO:1903723 | negative regulation of centriole elongation | 1 | 2 | 3.26e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 2 | 5 | 3.27e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 1 | 87 | 3.27e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 4 | 10 | 3.29e-01 |
| GO:BP | GO:0015680 | protein maturation by copper ion transfer | 2 | 2 | 3.29e-01 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 8 | 17 | 3.29e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 3.29e-01 |
| GO:BP | GO:1904875 | regulation of DNA ligase activity | 2 | 2 | 3.30e-01 |
| GO:BP | GO:0035845 | photoreceptor cell outer segment organization | 2 | 10 | 3.30e-01 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 2 | 6 | 3.30e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 5 | 127 | 3.30e-01 |
| GO:BP | GO:0070997 | neuron death | 9 | 286 | 3.30e-01 |
| GO:BP | GO:1904100 | positive regulation of protein O-linked glycosylation | 1 | 2 | 3.31e-01 |
| GO:BP | GO:0001890 | placenta development | 1 | 117 | 3.31e-01 |
| GO:BP | GO:1904098 | regulation of protein O-linked glycosylation | 1 | 2 | 3.31e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 1 | 30 | 3.31e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 3 | 8 | 3.31e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 3 | 8 | 3.31e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 9 | 14 | 3.31e-01 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 4 | 8 | 3.31e-01 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 26 | 50 | 3.31e-01 |
| GO:BP | GO:0014859 | negative regulation of skeletal muscle cell proliferation | 2 | 3 | 3.31e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 2 | 2 | 3.32e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 40 | 140 | 3.32e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 11 | 19 | 3.33e-01 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 1 | 8 | 3.33e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 5 | 88 | 3.33e-01 |
| GO:BP | GO:0039689 | negative stranded viral RNA replication | 2 | 3 | 3.33e-01 |
| GO:BP | GO:1990953 | intramanchette transport | 1 | 1 | 3.33e-01 |
| GO:BP | GO:0099543 | trans-synaptic signaling by soluble gas | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0099548 | trans-synaptic signaling by nitric oxide | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 9 | 29 | 3.33e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 4 | 9 | 3.33e-01 |
| GO:BP | GO:0070262 | peptidyl-serine dephosphorylation | 2 | 14 | 3.33e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 2 | 1229 | 3.33e-01 |
| GO:BP | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway | 2 | 4 | 3.33e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 2 | 10 | 3.33e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 3 | 40 | 3.33e-01 |
| GO:BP | GO:0015798 | myo-inositol transport | 2 | 3 | 3.33e-01 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 4 | 10 | 3.33e-01 |
| GO:BP | GO:0035351 | heme transmembrane transport | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 4 | 10 | 3.33e-01 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 7 | 9 | 3.33e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 1 | 94 | 3.33e-01 |
| GO:BP | GO:0034627 | ‘de novo’ NAD biosynthetic process | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0086016 | AV node cell action potential | 7 | 9 | 3.33e-01 |
| GO:BP | GO:0035377 | transepithelial water transport | 2 | 2 | 3.33e-01 |
| GO:BP | GO:0019388 | galactose catabolic process | 2 | 5 | 3.33e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 3.34e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 1 | 31 | 3.34e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 3.34e-01 |
| GO:BP | GO:0007099 | centriole replication | 22 | 38 | 3.34e-01 |
| GO:BP | GO:0042819 | vitamin B6 biosynthetic process | 1 | 3 | 3.35e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 3 | 114 | 3.35e-01 |
| GO:BP | GO:0046036 | CTP metabolic process | 9 | 14 | 3.36e-01 |
| GO:BP | GO:0160024 | Leydig cell proliferation | 2 | 2 | 3.36e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 40 | 77 | 3.36e-01 |
| GO:BP | GO:0000160 | phosphorelay signal transduction system | 1 | 1 | 3.36e-01 |
| GO:BP | GO:0048672 | positive regulation of collateral sprouting | 4 | 10 | 3.36e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 6 | 36 | 3.36e-01 |
| GO:BP | GO:2000639 | negative regulation of SREBP signaling pathway | 1 | 3 | 3.37e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 2 | 39 | 3.38e-01 |
| GO:BP | GO:0061002 | negative regulation of dendritic spine morphogenesis | 1 | 6 | 3.38e-01 |
| GO:BP | GO:0070159 | mitochondrial threonyl-tRNA aminoacylation | 1 | 1 | 3.38e-01 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 3 | 11 | 3.38e-01 |
| GO:BP | GO:0046886 | positive regulation of hormone biosynthetic process | 4 | 9 | 3.38e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 3.38e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 12 | 3.38e-01 |
| GO:BP | GO:1990592 | protein K69-linked ufmylation | 4 | 5 | 3.38e-01 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 2 | 12 | 3.38e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 7 | 13 | 3.38e-01 |
| GO:BP | GO:0046208 | spermine catabolic process | 2 | 2 | 3.38e-01 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 41 | 81 | 3.38e-01 |
| GO:BP | GO:0043113 | receptor clustering | 2 | 45 | 3.38e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 2 | 63 | 3.38e-01 |
| GO:BP | GO:0048245 | eosinophil chemotaxis | 1 | 5 | 3.38e-01 |
| GO:BP | GO:1990564 | protein polyufmylation | 4 | 5 | 3.38e-01 |
| GO:BP | GO:0072529 | pyrimidine-containing compound catabolic process | 1 | 29 | 3.38e-01 |
| GO:BP | GO:0006825 | copper ion transport | 1 | 13 | 3.38e-01 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 2 | 13 | 3.38e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 38 | 174 | 3.39e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 16 | 73 | 3.39e-01 |
| GO:BP | GO:1990049 | retrograde neuronal dense core vesicle transport | 1 | 4 | 3.39e-01 |
| GO:BP | GO:1903753 | negative regulation of p38MAPK cascade | 3 | 5 | 3.39e-01 |
| GO:BP | GO:0014849 | ureter smooth muscle contraction | 2 | 2 | 3.39e-01 |
| GO:BP | GO:0072105 | ureteric peristalsis | 2 | 2 | 3.39e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 6 | 357 | 3.39e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 3 | 28 | 3.39e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 3 | 59 | 3.39e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 206 | 3.39e-01 |
| GO:BP | GO:0015939 | pantothenate metabolic process | 2 | 2 | 3.39e-01 |
| GO:BP | GO:1904274 | tricellular tight junction assembly | 1 | 1 | 3.39e-01 |
| GO:BP | GO:0071318 | cellular response to ATP | 2 | 12 | 3.40e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 38 | 161 | 3.40e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 3.40e-01 |
| GO:BP | GO:0060134 | prepulse inhibition | 1 | 7 | 3.40e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 3.40e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 33 | 114 | 3.40e-01 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 38 | 73 | 3.40e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 6 | 22 | 3.40e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 3.40e-01 |
| GO:BP | GO:0140018 | regulation of cytoplasmic translational fidelity | 1 | 1 | 3.40e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 1 | 33 | 3.40e-01 |
| GO:BP | GO:0000301 | retrograde transport, vesicle recycling within Golgi | 5 | 9 | 3.40e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 6 | 175 | 3.40e-01 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 2 | 4 | 3.40e-01 |
| GO:BP | GO:0016180 | snRNA processing | 5 | 26 | 3.40e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 3.40e-01 |
| GO:BP | GO:0006386 | termination of RNA polymerase III transcription | 1 | 1 | 3.40e-01 |
| GO:BP | GO:0071025 | RNA surveillance | 1 | 16 | 3.40e-01 |
| GO:BP | GO:0072739 | response to anisomycin | 1 | 4 | 3.41e-01 |
| GO:BP | GO:1901163 | regulation of trophoblast cell migration | 2 | 12 | 3.41e-01 |
| GO:BP | GO:0009597 | detection of virus | 4 | 5 | 3.41e-01 |
| GO:BP | GO:1901628 | positive regulation of postsynaptic membrane organization | 1 | 2 | 3.41e-01 |
| GO:BP | GO:0033327 | Leydig cell differentiation | 6 | 9 | 3.41e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 3 | 57 | 3.41e-01 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 1 | 142 | 3.41e-01 |
| GO:BP | GO:1904719 | positive regulation of AMPA glutamate receptor clustering | 1 | 2 | 3.41e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 14 | 52 | 3.41e-01 |
| GO:BP | GO:0098735 | positive regulation of the force of heart contraction | 1 | 4 | 3.41e-01 |
| GO:BP | GO:0014832 | urinary bladder smooth muscle contraction | 1 | 4 | 3.42e-01 |
| GO:BP | GO:0032070 | regulation of deoxyribonuclease activity | 1 | 9 | 3.42e-01 |
| GO:BP | GO:0090149 | mitochondrial membrane fission | 1 | 2 | 3.42e-01 |
| GO:BP | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway | 3 | 4 | 3.42e-01 |
| GO:BP | GO:0003374 | dynamin family protein polymerization involved in mitochondrial fission | 1 | 2 | 3.42e-01 |
| GO:BP | GO:0015761 | mannose transmembrane transport | 1 | 1 | 3.42e-01 |
| GO:BP | GO:0003373 | dynamin family protein polymerization involved in membrane fission | 1 | 2 | 3.42e-01 |
| GO:BP | GO:0050955 | thermoception | 1 | 3 | 3.42e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 6 | 25 | 3.42e-01 |
| GO:BP | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway | 2 | 6 | 3.42e-01 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 7 | 12 | 3.43e-01 |
| GO:BP | GO:1905024 | regulation of membrane repolarization during ventricular cardiac muscle cell action potential | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 6 | 38 | 3.44e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 3.45e-01 |
| GO:BP | GO:0086017 | Purkinje myocyte action potential | 2 | 4 | 3.45e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 3 | 20 | 3.45e-01 |
| GO:BP | GO:2000822 | regulation of behavioral fear response | 1 | 7 | 3.45e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 3.45e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 3 | 14 | 3.45e-01 |
| GO:BP | GO:0034499 | late endosome to Golgi transport | 2 | 5 | 3.45e-01 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 2 | 16 | 3.45e-01 |
| GO:BP | GO:0061668 | mitochondrial ribosome assembly | 4 | 8 | 3.45e-01 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 24 | 43 | 3.46e-01 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 4 | 21 | 3.46e-01 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 7 | 29 | 3.46e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 6 | 362 | 3.46e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 2 | 21 | 3.47e-01 |
| GO:BP | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway | 2 | 3 | 3.47e-01 |
| GO:BP | GO:0014029 | neural crest formation | 9 | 13 | 3.48e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 8 | 139 | 3.48e-01 |
| GO:BP | GO:1904293 | negative regulation of ERAD pathway | 1 | 6 | 3.48e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 6 | 424 | 3.48e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 8 | 142 | 3.48e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 3.48e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 1 | 14 | 3.49e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 5 | 20 | 3.49e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 4 | 70 | 3.49e-01 |
| GO:BP | GO:0060173 | limb development | 2 | 142 | 3.49e-01 |
| GO:BP | GO:0048736 | appendage development | 2 | 142 | 3.49e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 2 | 19 | 3.49e-01 |
| GO:BP | GO:0071579 | regulation of zinc ion transport | 1 | 2 | 3.49e-01 |
| GO:BP | GO:1904800 | negative regulation of neuron remodeling | 1 | 1 | 3.49e-01 |
| GO:BP | GO:1903964 | monounsaturated fatty acid metabolic process | 1 | 2 | 3.49e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 4 | 17 | 3.49e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 8 | 173 | 3.49e-01 |
| GO:BP | GO:1904799 | regulation of neuron remodeling | 1 | 1 | 3.49e-01 |
| GO:BP | GO:0140882 | zinc export across plasma membrane | 1 | 3 | 3.49e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 2 | 19 | 3.49e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 1 | 118 | 3.49e-01 |
| GO:BP | GO:0006248 | CMP catabolic process | 1 | 3 | 3.49e-01 |
| GO:BP | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridinylation | 2 | 3 | 3.49e-01 |
| GO:BP | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation | 1 | 1 | 3.49e-01 |
| GO:BP | GO:0072107 | positive regulation of ureteric bud formation | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0009052 | pentose-phosphate shunt, non-oxidative branch | 4 | 6 | 3.49e-01 |
| GO:BP | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0046050 | UMP catabolic process | 1 | 3 | 3.49e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 3 | 15 | 3.49e-01 |
| GO:BP | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | 6 | 10 | 3.49e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 3 | 39 | 3.49e-01 |
| GO:BP | GO:0072106 | regulation of ureteric bud formation | 1 | 2 | 3.49e-01 |
| GO:BP | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process | 1 | 7 | 3.49e-01 |
| GO:BP | GO:0030324 | lung development | 2 | 146 | 3.49e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 1 | 118 | 3.49e-01 |
| GO:BP | GO:2000173 | negative regulation of branching morphogenesis of a nerve | 1 | 1 | 3.49e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 2 | 10 | 3.49e-01 |
| GO:BP | GO:0021658 | rhombomere 3 morphogenesis | 1 | 1 | 3.50e-01 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 1 | 7 | 3.50e-01 |
| GO:BP | GO:1903423 | positive regulation of synaptic vesicle recycling | 1 | 6 | 3.50e-01 |
| GO:BP | GO:0021593 | rhombomere morphogenesis | 1 | 1 | 3.50e-01 |
| GO:BP | GO:0014040 | positive regulation of Schwann cell differentiation | 1 | 2 | 3.50e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 27 | 51 | 3.50e-01 |
| GO:BP | GO:0009173 | pyrimidine ribonucleoside monophosphate metabolic process | 2 | 12 | 3.51e-01 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 3 | 3 | 3.51e-01 |
| GO:BP | GO:1901341 | positive regulation of store-operated calcium channel activity | 2 | 6 | 3.51e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 3.52e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 12 | 50 | 3.53e-01 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 2 | 20 | 3.53e-01 |
| GO:BP | GO:1903837 | regulation of mRNA 3’-UTR binding | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 2 | 12 | 3.53e-01 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 1 | 19 | 3.53e-01 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 3 | 33 | 3.53e-01 |
| GO:BP | GO:0031297 | replication fork processing | 2 | 42 | 3.53e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 1 | 159 | 3.53e-01 |
| GO:BP | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 2 | 5 | 3.53e-01 |
| GO:BP | GO:0002182 | cytoplasmic translational elongation | 7 | 12 | 3.53e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 7 | 91 | 3.53e-01 |
| GO:BP | GO:1903839 | positive regulation of mRNA 3’-UTR binding | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0140212 | regulation of long-chain fatty acid import into cell | 3 | 7 | 3.53e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 12 | 50 | 3.53e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 6 | 14 | 3.53e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 1 | 16 | 3.53e-01 |
| GO:BP | GO:0090529 | cell septum assembly | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 12 | 70 | 3.53e-01 |
| GO:BP | GO:0000917 | division septum assembly | 1 | 1 | 3.53e-01 |
| GO:BP | GO:0060763 | mammary duct terminal end bud growth | 1 | 3 | 3.54e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 205 | 441 | 3.54e-01 |
| GO:BP | GO:1900161 | regulation of phospholipid scramblase activity | 1 | 1 | 3.54e-01 |
| GO:BP | GO:1900163 | positive regulation of phospholipid scramblase activity | 1 | 1 | 3.54e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 3.54e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 3 | 256 | 3.54e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 2 | 55 | 3.54e-01 |
| GO:BP | GO:0043276 | anoikis | 10 | 32 | 3.54e-01 |
| GO:BP | GO:0061450 | trophoblast cell migration | 2 | 13 | 3.54e-01 |
| GO:BP | GO:1900239 | regulation of phenotypic switching | 1 | 3 | 3.54e-01 |
| GO:BP | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway | 1 | 2 | 3.54e-01 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 5 | 7 | 3.54e-01 |
| GO:BP | GO:1903365 | regulation of fear response | 1 | 8 | 3.54e-01 |
| GO:BP | GO:0097154 | GABAergic neuron differentiation | 1 | 5 | 3.54e-01 |
| GO:BP | GO:2000753 | positive regulation of glucosylceramide catabolic process | 1 | 1 | 3.54e-01 |
| GO:BP | GO:2001016 | positive regulation of skeletal muscle cell differentiation | 2 | 5 | 3.54e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 3.54e-01 |
| GO:BP | GO:0001560 | regulation of cell growth by extracellular stimulus | 1 | 3 | 3.54e-01 |
| GO:BP | GO:0060073 | micturition | 1 | 2 | 3.54e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 2 | 150 | 3.54e-01 |
| GO:BP | GO:2000752 | regulation of glucosylceramide catabolic process | 1 | 1 | 3.54e-01 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 8 | 12 | 3.54e-01 |
| GO:BP | GO:0006543 | glutamine catabolic process | 3 | 3 | 3.54e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 8 | 98 | 3.54e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 4 | 138 | 3.54e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 3 | 5 | 3.54e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 1 | 112 | 3.54e-01 |
| GO:BP | GO:0060047 | heart contraction | 9 | 205 | 3.54e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 84 | 178 | 3.54e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 5 | 22 | 3.55e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 9 | 64 | 3.55e-01 |
| GO:BP | GO:0009175 | pyrimidine ribonucleoside monophosphate catabolic process | 1 | 4 | 3.55e-01 |
| GO:BP | GO:1905371 | ceramide phosphoethanolamine metabolic process | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0097503 | sialylation | 1 | 18 | 3.55e-01 |
| GO:BP | GO:1904679 | myo-inositol import across plasma membrane | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 3.55e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 122 | 3.55e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 3 | 6 | 3.55e-01 |
| GO:BP | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 3 | 3.55e-01 |
| GO:BP | GO:0072677 | eosinophil migration | 1 | 7 | 3.55e-01 |
| GO:BP | GO:2000346 | negative regulation of hepatocyte proliferation | 1 | 3 | 3.55e-01 |
| GO:BP | GO:1904321 | response to forskolin | 3 | 11 | 3.55e-01 |
| GO:BP | GO:1904322 | cellular response to forskolin | 3 | 11 | 3.55e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 4 | 50 | 3.55e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 6 | 316 | 3.55e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 7 | 94 | 3.55e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 1 | 179 | 3.55e-01 |
| GO:BP | GO:1905373 | ceramide phosphoethanolamine biosynthetic process | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0032049 | cardiolipin biosynthetic process | 4 | 7 | 3.55e-01 |
| GO:BP | GO:0046079 | dUMP catabolic process | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 1 | 6 | 3.55e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 4 | 36 | 3.55e-01 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 2 | 66 | 3.55e-01 |
| GO:BP | GO:0097384 | cellular lipid biosynthetic process | 3 | 10 | 3.55e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 3 | 22 | 3.55e-01 |
| GO:BP | GO:2000386 | positive regulation of ovarian follicle development | 2 | 2 | 3.55e-01 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 3 | 8 | 3.55e-01 |
| GO:BP | GO:0072255 | metanephric glomerular mesangial cell development | 1 | 1 | 3.55e-01 |
| GO:BP | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation | 4 | 6 | 3.55e-01 |
| GO:BP | GO:0046168 | glycerol-3-phosphate catabolic process | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0000493 | box H/ACA snoRNP assembly | 1 | 2 | 3.55e-01 |
| GO:BP | GO:0046063 | dCMP metabolic process | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0072014 | proximal tubule development | 1 | 7 | 3.55e-01 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 46 | 82 | 3.55e-01 |
| GO:BP | GO:0086098 | angiotensin-activated signaling pathway involved in heart process | 2 | 3 | 3.55e-01 |
| GO:BP | GO:0032202 | telomere assembly | 2 | 4 | 3.55e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 33 | 219 | 3.55e-01 |
| GO:BP | GO:0045837 | negative regulation of membrane potential | 1 | 9 | 3.55e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 4 | 10 | 3.55e-01 |
| GO:BP | GO:2000393 | negative regulation of lamellipodium morphogenesis | 2 | 2 | 3.55e-01 |
| GO:BP | GO:1905176 | positive regulation of vascular associated smooth muscle cell dedifferentiation | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0000916 | actomyosin contractile ring contraction | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 3 | 6 | 3.55e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 3 | 48 | 3.55e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 6 | 21 | 3.55e-01 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 2 | 66 | 3.55e-01 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 12 | 19 | 3.55e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 6 | 19 | 3.55e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 3 | 12 | 3.55e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 6 | 19 | 3.55e-01 |
| GO:BP | GO:2001205 | negative regulation of osteoclast development | 1 | 2 | 3.55e-01 |
| GO:BP | GO:0006591 | ornithine metabolic process | 1 | 6 | 3.55e-01 |
| GO:BP | GO:0003330 | regulation of extracellular matrix constituent secretion | 4 | 7 | 3.55e-01 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 3 | 4 | 3.55e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 20 | 3.55e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 9 | 13 | 3.55e-01 |
| GO:BP | GO:2000742 | regulation of anterior head development | 2 | 2 | 3.55e-01 |
| GO:BP | GO:2000744 | positive regulation of anterior head development | 2 | 2 | 3.55e-01 |
| GO:BP | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process | 2 | 3 | 3.55e-01 |
| GO:BP | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process | 2 | 3 | 3.55e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 2 | 33 | 3.55e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 1 | 121 | 3.55e-01 |
| GO:BP | GO:1990048 | anterograde neuronal dense core vesicle transport | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 2 | 51 | 3.55e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 163 | 3.55e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 9 | 16 | 3.55e-01 |
| GO:BP | GO:0050860 | negative regulation of T cell receptor signaling pathway | 1 | 17 | 3.55e-01 |
| GO:BP | GO:0032262 | pyrimidine nucleotide salvage | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 15 | 22 | 3.55e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 3.55e-01 |
| GO:BP | GO:0009992 | intracellular water homeostasis | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0060754 | positive regulation of mast cell chemotaxis | 1 | 5 | 3.55e-01 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 12 | 19 | 3.55e-01 |
| GO:BP | GO:0014806 | smooth muscle hyperplasia | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0042822 | pyridoxal phosphate metabolic process | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 12 | 19 | 3.55e-01 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 2 | 40 | 3.55e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 11 | 36 | 3.55e-01 |
| GO:BP | GO:0010002 | cardioblast differentiation | 6 | 15 | 3.55e-01 |
| GO:BP | GO:1902389 | ceramide 1-phosphate transport | 2 | 7 | 3.55e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 1 | 104 | 3.55e-01 |
| GO:BP | GO:0010138 | pyrimidine ribonucleotide salvage | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 11 | 37 | 3.55e-01 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 1 | 23 | 3.55e-01 |
| GO:BP | GO:0014848 | urinary tract smooth muscle contraction | 1 | 6 | 3.55e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 4 | 28 | 3.55e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 8 | 41 | 3.55e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 3 | 10 | 3.55e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 2 | 17 | 3.55e-01 |
| GO:BP | GO:0006249 | dCMP catabolic process | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0006434 | seryl-tRNA aminoacylation | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 17 | 135 | 3.55e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 3.55e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 128 | 271 | 3.55e-01 |
| GO:BP | GO:0006225 | UDP biosynthetic process | 2 | 4 | 3.55e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 3 | 35 | 3.55e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 6 | 40 | 3.55e-01 |
| GO:BP | GO:0035928 | rRNA import into mitochondrion | 2 | 3 | 3.55e-01 |
| GO:BP | GO:0036438 | maintenance of lens transparency | 5 | 6 | 3.55e-01 |
| GO:BP | GO:1990637 | response to prolactin | 2 | 3 | 3.55e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 6 | 17 | 3.55e-01 |
| GO:BP | GO:1990550 | mitochondrial alpha-ketoglutarate transmembrane transport | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0036213 | contractile ring contraction | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 6 | 100 | 3.55e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 5 | 13 | 3.55e-01 |
| GO:BP | GO:0052040 | modulation by symbiont of host programmed cell death | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 51 | 214 | 3.55e-01 |
| GO:BP | GO:0150020 | basal dendrite arborization | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0150019 | basal dendrite morphogenesis | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0150018 | basal dendrite development | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 4 | 5 | 3.55e-01 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 1 | 11 | 3.55e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 15 | 1214 | 3.55e-01 |
| GO:BP | GO:0110155 | NAD-cap decapping | 2 | 3 | 3.55e-01 |
| GO:BP | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway | 2 | 4 | 3.55e-01 |
| GO:BP | GO:0051298 | centrosome duplication | 35 | 68 | 3.55e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 3 | 267 | 3.55e-01 |
| GO:BP | GO:0039526 | modulation by virus of host apoptotic process | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 3 | 30 | 3.55e-01 |
| GO:BP | GO:0010917 | negative regulation of mitochondrial membrane potential | 1 | 9 | 3.55e-01 |
| GO:BP | GO:0038133 | ERBB2-ERBB3 signaling pathway | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0035498 | carnosine metabolic process | 1 | 2 | 3.55e-01 |
| GO:BP | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway | 1 | 2 | 3.55e-01 |
| GO:BP | GO:0052150 | modulation by symbiont of host apoptotic process | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 1 | 113 | 3.55e-01 |
| GO:BP | GO:0060916 | mesenchymal cell proliferation involved in lung development | 3 | 5 | 3.55e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 8 | 108 | 3.55e-01 |
| GO:BP | GO:0023041 | neuronal signal transduction | 1 | 6 | 3.55e-01 |
| GO:BP | GO:0002520 | immune system development | 1 | 144 | 3.55e-01 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 4 | 28 | 3.55e-01 |
| GO:BP | GO:0032764 | negative regulation of mast cell cytokine production | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 7 | 58 | 3.55e-01 |
| GO:BP | GO:0009268 | response to pH | 2 | 26 | 3.55e-01 |
| GO:BP | GO:0043696 | dedifferentiation | 2 | 6 | 3.55e-01 |
| GO:BP | GO:0009301 | snRNA transcription | 3 | 18 | 3.55e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 4 | 42 | 3.55e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 11 | 46 | 3.55e-01 |
| GO:BP | GO:0032364 | intracellular oxygen homeostasis | 1 | 6 | 3.55e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 10 | 67 | 3.55e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 3 | 9 | 3.55e-01 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 22 | 40 | 3.55e-01 |
| GO:BP | GO:0009194 | pyrimidine ribonucleoside diphosphate biosynthetic process | 2 | 4 | 3.55e-01 |
| GO:BP | GO:0043697 | cell dedifferentiation | 2 | 6 | 3.55e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 3 | 9 | 3.55e-01 |
| GO:BP | GO:1902038 | positive regulation of hematopoietic stem cell differentiation | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0044057 | regulation of system process | 18 | 405 | 3.55e-01 |
| GO:BP | GO:1903997 | positive regulation of non-membrane spanning protein tyrosine kinase activity | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0070509 | calcium ion import | 3 | 37 | 3.55e-01 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 11 | 19 | 3.55e-01 |
| GO:BP | GO:0016075 | rRNA catabolic process | 1 | 19 | 3.55e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 3 | 30 | 3.55e-01 |
| GO:BP | GO:0032402 | melanosome transport | 4 | 19 | 3.55e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 1 | 111 | 3.55e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 128 | 3.55e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 11 | 46 | 3.55e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 5 | 73 | 3.55e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 5 | 36 | 3.55e-01 |
| GO:BP | GO:1903936 | cellular response to sodium arsenite | 1 | 5 | 3.55e-01 |
| GO:BP | GO:0046849 | bone remodeling | 2 | 59 | 3.55e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 7 | 89 | 3.55e-01 |
| GO:BP | GO:0032528 | microvillus organization | 4 | 19 | 3.55e-01 |
| GO:BP | GO:0032911 | negative regulation of transforming growth factor beta1 production | 4 | 5 | 3.55e-01 |
| GO:BP | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | 4 | 5 | 3.55e-01 |
| GO:BP | GO:0062025 | regulation of SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0008611 | ether lipid biosynthetic process | 3 | 10 | 3.55e-01 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 13 | 22 | 3.55e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 3.55e-01 |
| GO:BP | GO:0009751 | response to salicylic acid | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 7 | 94 | 3.55e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 11 | 47 | 3.55e-01 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 6 | 19 | 3.55e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 9 | 31 | 3.55e-01 |
| GO:BP | GO:0090217 | negative regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 2 | 3 | 3.55e-01 |
| GO:BP | GO:0060932 | His-Purkinje system cell differentiation | 3 | 4 | 3.55e-01 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 3 | 4 | 3.55e-01 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 3 | 4 | 3.55e-01 |
| GO:BP | GO:0046504 | glycerol ether biosynthetic process | 3 | 10 | 3.55e-01 |
| GO:BP | GO:2001311 | lysobisphosphatidic acid metabolic process | 2 | 2 | 3.55e-01 |
| GO:BP | GO:0099163 | synaptic signaling by nitric oxide | 1 | 3 | 3.55e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 5 | 7 | 3.55e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 4 | 140 | 3.55e-01 |
| GO:BP | GO:0048671 | negative regulation of collateral sprouting | 5 | 9 | 3.55e-01 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 4 | 19 | 3.55e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 17 | 57 | 3.55e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 25 | 3.55e-01 |
| GO:BP | GO:0048630 | skeletal muscle tissue growth | 1 | 8 | 3.55e-01 |
| GO:BP | GO:0061734 | parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 4 | 6 | 3.55e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 11 | 28 | 3.55e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 8 | 58 | 3.55e-01 |
| GO:BP | GO:0062026 | negative regulation of SCF-dependent proteasomal ubiquitin-dependent catabolic process | 1 | 1 | 3.55e-01 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 3 | 3 | 3.55e-01 |
| GO:BP | GO:0015823 | phenylalanine transport | 2 | 4 | 3.55e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 2 | 11 | 3.55e-01 |
| GO:BP | GO:0044206 | UMP salvage | 1 | 4 | 3.55e-01 |
| GO:BP | GO:0072092 | ureteric bud invasion | 3 | 3 | 3.56e-01 |
| GO:BP | GO:0010424 | DNA methylation on cytosine within a CG sequence | 1 | 1 | 3.56e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 1 | 19 | 3.57e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 2 | 24 | 3.57e-01 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 23 | 44 | 3.57e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 6 | 14 | 3.57e-01 |
| GO:BP | GO:0038018 | Wnt receptor catabolic process | 2 | 2 | 3.58e-01 |
| GO:BP | GO:0071625 | vocalization behavior | 2 | 13 | 3.58e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 5 | 124 | 3.58e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 3 | 26 | 3.58e-01 |
| GO:BP | GO:0061956 | penetration of cumulus oophorus | 1 | 1 | 3.59e-01 |
| GO:BP | GO:0106217 | tRNA C3-cytosine methylation | 1 | 2 | 3.59e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 4 | 26 | 3.59e-01 |
| GO:BP | GO:0098534 | centriole assembly | 1 | 41 | 3.59e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 4 | 13 | 3.59e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 5 | 67 | 3.59e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 24 | 3.59e-01 |
| GO:BP | GO:0008215 | spermine metabolic process | 3 | 5 | 3.59e-01 |
| GO:BP | GO:0099551 | trans-synaptic signaling by neuropeptide, modulating synaptic transmission | 1 | 1 | 3.59e-01 |
| GO:BP | GO:0045578 | negative regulation of B cell differentiation | 3 | 6 | 3.59e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 1 | 131 | 3.59e-01 |
| GO:BP | GO:0019377 | glycolipid catabolic process | 5 | 12 | 3.59e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 3 | 52 | 3.60e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 11 | 21 | 3.60e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 5 | 51 | 3.60e-01 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 3.60e-01 |
| GO:BP | GO:0048278 | vesicle docking | 2 | 57 | 3.60e-01 |
| GO:BP | GO:0061217 | regulation of mesonephros development | 1 | 2 | 3.60e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 1 | 17 | 3.60e-01 |
| GO:BP | GO:0036486 | ventral trunk neural crest cell migration | 3 | 3 | 3.60e-01 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 15 | 29 | 3.60e-01 |
| GO:BP | GO:0090385 | phagosome-lysosome fusion | 1 | 12 | 3.60e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 2 | 13 | 3.60e-01 |
| GO:BP | GO:0097104 | postsynaptic membrane assembly | 1 | 8 | 3.60e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 3 | 20 | 3.60e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 15 | 48 | 3.60e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 2 | 13 | 3.60e-01 |
| GO:BP | GO:0097105 | presynaptic membrane assembly | 1 | 8 | 3.60e-01 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 1 | 191 | 3.61e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 3.61e-01 |
| GO:BP | GO:0097601 | retina blood vessel maintenance | 1 | 1 | 3.61e-01 |
| GO:BP | GO:0006656 | phosphatidylcholine biosynthetic process | 7 | 25 | 3.61e-01 |
| GO:BP | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential | 1 | 3 | 3.61e-01 |
| GO:BP | GO:2001306 | lipoxin B4 biosynthetic process | 1 | 1 | 3.61e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 2 | 12 | 3.61e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 2 | 39 | 3.61e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 3.61e-01 |
| GO:BP | GO:0014841 | skeletal muscle satellite cell proliferation | 3 | 10 | 3.61e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 1 | 26 | 3.61e-01 |
| GO:BP | GO:1903429 | regulation of cell maturation | 1 | 13 | 3.61e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 1 | 184 | 3.61e-01 |
| GO:BP | GO:0060442 | branching involved in prostate gland morphogenesis | 2 | 7 | 3.61e-01 |
| GO:BP | GO:0000303 | response to superoxide | 2 | 21 | 3.61e-01 |
| GO:BP | GO:0043574 | peroxisomal transport | 10 | 22 | 3.61e-01 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 4 | 5 | 3.61e-01 |
| GO:BP | GO:0006391 | transcription initiation at mitochondrial promoter | 3 | 4 | 3.61e-01 |
| GO:BP | GO:1901465 | positive regulation of tetrapyrrole biosynthetic process | 3 | 3 | 3.61e-01 |
| GO:BP | GO:0071941 | nitrogen cycle metabolic process | 3 | 7 | 3.61e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 3 | 277 | 3.61e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 2 | 22 | 3.61e-01 |
| GO:BP | GO:0001660 | fever generation | 1 | 5 | 3.61e-01 |
| GO:BP | GO:0016189 | synaptic vesicle to endosome fusion | 1 | 3 | 3.61e-01 |
| GO:BP | GO:2000322 | regulation of glucocorticoid receptor signaling pathway | 2 | 7 | 3.61e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 2 | 145 | 3.61e-01 |
| GO:BP | GO:0070493 | thrombin-activated receptor signaling pathway | 1 | 9 | 3.61e-01 |
| GO:BP | GO:0019627 | urea metabolic process | 3 | 7 | 3.61e-01 |
| GO:BP | GO:0070455 | positive regulation of heme biosynthetic process | 3 | 3 | 3.61e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 2 | 84 | 3.61e-01 |
| GO:BP | GO:0002317 | plasma cell differentiation | 1 | 5 | 3.61e-01 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 3 | 15 | 3.61e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 4 | 280 | 3.61e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 1 | 132 | 3.61e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 3 | 143 | 3.61e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 1 | 21 | 3.61e-01 |
| GO:BP | GO:0044058 | regulation of digestive system process | 2 | 22 | 3.61e-01 |
| GO:BP | GO:1904782 | negative regulation of NMDA glutamate receptor activity | 2 | 2 | 3.62e-01 |
| GO:BP | GO:0090148 | membrane fission | 5 | 44 | 3.62e-01 |
| GO:BP | GO:0097292 | XMP metabolic process | 5 | 7 | 3.62e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 3.62e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 8 | 17 | 3.62e-01 |
| GO:BP | GO:0097294 | ‘de novo’ XMP biosynthetic process | 5 | 7 | 3.62e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 3.62e-01 |
| GO:BP | GO:1902527 | positive regulation of protein monoubiquitination | 4 | 5 | 3.62e-01 |
| GO:BP | GO:0097293 | XMP biosynthetic process | 5 | 7 | 3.62e-01 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 1 | 197 | 3.62e-01 |
| GO:BP | GO:0006722 | triterpenoid metabolic process | 2 | 2 | 3.62e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 4 | 41 | 3.62e-01 |
| GO:BP | GO:0072032 | proximal convoluted tubule segment 2 development | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0072022 | descending thin limb development | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0070212 | protein poly-ADP-ribosylation | 5 | 6 | 3.62e-01 |
| GO:BP | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus | 1 | 6 | 3.62e-01 |
| GO:BP | GO:0003057 | regulation of the force of heart contraction by chemical signal | 3 | 5 | 3.62e-01 |
| GO:BP | GO:0072232 | metanephric proximal convoluted tubule segment 2 development | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0150037 | regulation of calcium-dependent activation of synaptic vesicle fusion | 1 | 2 | 3.62e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 1 | 159 | 3.62e-01 |
| GO:BP | GO:0032200 | telomere organization | 9 | 166 | 3.62e-01 |
| GO:BP | GO:0072220 | metanephric descending thin limb development | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0051974 | negative regulation of telomerase activity | 1 | 11 | 3.62e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 4 | 20 | 3.62e-01 |
| GO:BP | GO:0019240 | citrulline biosynthetic process | 3 | 4 | 3.62e-01 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 1 | 13 | 3.62e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 17 | 49 | 3.62e-01 |
| GO:BP | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | 3 | 4 | 3.62e-01 |
| GO:BP | GO:0072230 | metanephric proximal straight tubule development | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 70 | 273 | 3.62e-01 |
| GO:BP | GO:0061864 | basement membrane constituent secretion | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 3.62e-01 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 1 | 7 | 3.62e-01 |
| GO:BP | GO:0061865 | polarized secretion of basement membrane proteins in epithelium | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 3.62e-01 |
| GO:BP | GO:0072020 | proximal straight tubule development | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0006915 | apoptotic process | 2 | 1443 | 3.62e-01 |
| GO:BP | GO:0033630 | positive regulation of cell adhesion mediated by integrin | 1 | 14 | 3.62e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 3 | 30 | 3.62e-01 |
| GO:BP | GO:2000504 | positive regulation of blood vessel remodeling | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 1 | 14 | 3.62e-01 |
| GO:BP | GO:1901660 | calcium ion export | 3 | 3 | 3.62e-01 |
| GO:BP | GO:0006833 | water transport | 2 | 10 | 3.62e-01 |
| GO:BP | GO:1903621 | protein localization to photoreceptor connecting cilium | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0097012 | response to granulocyte macrophage colony-stimulating factor | 1 | 6 | 3.62e-01 |
| GO:BP | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.62e-01 |
| GO:BP | GO:1905070 | anterior visceral endoderm cell migration | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0090346 | cellular organofluorine metabolic process | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0140039 | cell-cell adhesion in response to extracellular stimulus | 1 | 2 | 3.62e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 2 | 194 | 3.62e-01 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 1 | 8 | 3.62e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 1 | 144 | 3.62e-01 |
| GO:BP | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway | 3 | 17 | 3.62e-01 |
| GO:BP | GO:2000691 | negative regulation of cardiac muscle cell myoblast differentiation | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 2 | 43 | 3.62e-01 |
| GO:BP | GO:0046073 | dTMP metabolic process | 2 | 7 | 3.62e-01 |
| GO:BP | GO:0090345 | cellular organohalogen metabolic process | 1 | 1 | 3.62e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 4 | 19 | 3.62e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 3.62e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 80 | 194 | 3.62e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 9 | 85 | 3.62e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 3 | 8 | 3.62e-01 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 1 | 13 | 3.62e-01 |
| GO:BP | GO:1905065 | positive regulation of vascular associated smooth muscle cell differentiation | 3 | 6 | 3.62e-01 |
| GO:BP | GO:0036018 | cellular response to erythropoietin | 3 | 4 | 3.62e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 13 | 23 | 3.63e-01 |
| GO:BP | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 2 | 5 | 3.64e-01 |
| GO:BP | GO:0061833 | protein localization to tricellular tight junction | 1 | 2 | 3.64e-01 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 1 | 13 | 3.64e-01 |
| GO:BP | GO:0009190 | cyclic nucleotide biosynthetic process | 2 | 14 | 3.64e-01 |
| GO:BP | GO:0033962 | P-body assembly | 1 | 20 | 3.64e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 3 | 69 | 3.64e-01 |
| GO:BP | GO:0006498 | N-terminal protein lipidation | 5 | 7 | 3.64e-01 |
| GO:BP | GO:1901654 | response to ketone | 3 | 147 | 3.64e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 3 | 60 | 3.64e-01 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 3 | 14 | 3.64e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 2 | 22 | 3.64e-01 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 9 | 14 | 3.64e-01 |
| GO:BP | GO:0070574 | cadmium ion transmembrane transport | 4 | 5 | 3.64e-01 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 4 | 5 | 3.64e-01 |
| GO:BP | GO:0015691 | cadmium ion transport | 4 | 5 | 3.64e-01 |
| GO:BP | GO:0043041 | amino acid activation for nonribosomal peptide biosynthetic process | 1 | 1 | 3.64e-01 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 2 | 72 | 3.65e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 3.65e-01 |
| GO:BP | GO:0003015 | heart process | 9 | 216 | 3.65e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 1 | 198 | 3.65e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 3 | 69 | 3.65e-01 |
| GO:BP | GO:0097090 | presynaptic membrane organization | 1 | 8 | 3.65e-01 |
| GO:BP | GO:0051095 | regulation of helicase activity | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0019048 | modulation by virus of host process | 4 | 5 | 3.65e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 8 | 24 | 3.65e-01 |
| GO:BP | GO:0046684 | response to pyrethroid | 1 | 1 | 3.65e-01 |
| GO:BP | GO:0044003 | modulation by symbiont of host process | 4 | 5 | 3.65e-01 |
| GO:BP | GO:0046898 | response to cycloheximide | 4 | 4 | 3.65e-01 |
| GO:BP | GO:0044068 | modulation by symbiont of host cellular process | 4 | 5 | 3.65e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 4 | 6 | 3.65e-01 |
| GO:BP | GO:0019054 | modulation by virus of host cellular process | 4 | 5 | 3.65e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 4 | 6 | 3.65e-01 |
| GO:BP | GO:1903935 | response to sodium arsenite | 1 | 6 | 3.65e-01 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 1 | 8 | 3.65e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 22 | 42 | 3.65e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 3 | 25 | 3.65e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 52 | 241 | 3.65e-01 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 42 | 86 | 3.65e-01 |
| GO:BP | GO:1901626 | regulation of postsynaptic membrane organization | 1 | 2 | 3.65e-01 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 9 | 14 | 3.65e-01 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 9 | 14 | 3.65e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 4 | 74 | 3.65e-01 |
| GO:BP | GO:0051656 | establishment of organelle localization | 181 | 378 | 3.65e-01 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 9 | 14 | 3.65e-01 |
| GO:BP | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway | 1 | 4 | 3.66e-01 |
| GO:BP | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway | 2 | 6 | 3.66e-01 |
| GO:BP | GO:0006741 | NADP biosynthetic process | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0060157 | urinary bladder development | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0008655 | pyrimidine-containing compound salvage | 1 | 5 | 3.66e-01 |
| GO:BP | GO:1905667 | negative regulation of protein localization to endosome | 1 | 1 | 3.66e-01 |
| GO:BP | GO:0140495 | migracytosis | 1 | 3 | 3.66e-01 |
| GO:BP | GO:0160040 | mitocytosis | 1 | 3 | 3.66e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 65 | 185 | 3.66e-01 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 5 | 7 | 3.66e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 1 | 12 | 3.66e-01 |
| GO:BP | GO:0090117 | endosome to lysosome transport of low-density lipoprotein particle | 1 | 2 | 3.66e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 24 | 72 | 3.66e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 3.66e-01 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 4 | 8 | 3.66e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 2 | 26 | 3.66e-01 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 5 | 7 | 3.66e-01 |
| GO:BP | GO:0012501 | programmed cell death | 2 | 1479 | 3.66e-01 |
| GO:BP | GO:0038138 | ERBB4-ERBB4 signaling pathway | 3 | 3 | 3.66e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 11 | 427 | 3.66e-01 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 8 | 17 | 3.66e-01 |
| GO:BP | GO:0032506 | cytokinetic process | 20 | 39 | 3.67e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 2 | 994 | 3.67e-01 |
| GO:BP | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation | 1 | 5 | 3.68e-01 |
| GO:BP | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 1 | 4 | 3.68e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 8 | 24 | 3.68e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 12 | 31 | 3.68e-01 |
| GO:BP | GO:0035589 | G protein-coupled purinergic nucleotide receptor signaling pathway | 2 | 4 | 3.68e-01 |
| GO:BP | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 3 | 6 | 3.68e-01 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 1 | 6 | 3.68e-01 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 3 | 7 | 3.68e-01 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 24 | 45 | 3.68e-01 |
| GO:BP | GO:0072223 | metanephric glomerular mesangium development | 2 | 3 | 3.68e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 1 | 15 | 3.68e-01 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 25 | 3.68e-01 |
| GO:BP | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis | 2 | 3 | 3.68e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 6 | 192 | 3.68e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 2 | 5 | 3.68e-01 |
| GO:BP | GO:0001947 | heart looping | 3 | 50 | 3.68e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 2 | 39 | 3.68e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 3 | 31 | 3.68e-01 |
| GO:BP | GO:0060929 | atrioventricular node cell fate commitment | 2 | 2 | 3.68e-01 |
| GO:BP | GO:0042816 | vitamin B6 metabolic process | 1 | 5 | 3.68e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 3 | 6 | 3.68e-01 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 10 | 19 | 3.69e-01 |
| GO:BP | GO:0051640 | organelle localization | 233 | 503 | 3.69e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 7 | 27 | 3.69e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 2 | 21 | 3.69e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 7 | 27 | 3.69e-01 |
| GO:BP | GO:0030208 | dermatan sulfate biosynthetic process | 2 | 5 | 3.69e-01 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 10 | 15 | 3.69e-01 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 1 | 209 | 3.69e-01 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 10 | 19 | 3.69e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 3.69e-01 |
| GO:BP | GO:0034729 | histone H3-K79 methylation | 2 | 2 | 3.69e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 1 | 52 | 3.69e-01 |
| GO:BP | GO:0099156 | cell-cell signaling via exosome | 1 | 1 | 3.69e-01 |
| GO:BP | GO:0061586 | positive regulation of transcription by transcription factor localization | 1 | 1 | 3.69e-01 |
| GO:BP | GO:0046717 | acid secretion | 2 | 25 | 3.69e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 2 | 43 | 3.69e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 15 | 27 | 3.69e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 1 | 157 | 3.69e-01 |
| GO:BP | GO:2000392 | regulation of lamellipodium morphogenesis | 5 | 10 | 3.70e-01 |
| GO:BP | GO:0090224 | regulation of spindle organization | 24 | 45 | 3.70e-01 |
| GO:BP | GO:0032416 | negative regulation of sodium:proton antiporter activity | 1 | 1 | 3.70e-01 |
| GO:BP | GO:0044722 | renal phosphate excretion | 1 | 2 | 3.70e-01 |
| GO:BP | GO:0030238 | male sex determination | 5 | 8 | 3.70e-01 |
| GO:BP | GO:0060383 | positive regulation of DNA strand elongation | 1 | 1 | 3.70e-01 |
| GO:BP | GO:1903402 | regulation of renal phosphate excretion | 1 | 2 | 3.70e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 5 | 103 | 3.71e-01 |
| GO:BP | GO:1902178 | fibroblast growth factor receptor apoptotic signaling pathway | 1 | 1 | 3.71e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 6 | 13 | 3.71e-01 |
| GO:BP | GO:0071257 | cellular response to electrical stimulus | 4 | 8 | 3.71e-01 |
| GO:BP | GO:1904637 | cellular response to ionomycin | 1 | 3 | 3.72e-01 |
| GO:BP | GO:1904636 | response to ionomycin | 1 | 3 | 3.72e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 3.72e-01 |
| GO:BP | GO:0007625 | grooming behavior | 2 | 10 | 3.72e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 8 | 41 | 3.73e-01 |
| GO:BP | GO:1903764 | regulation of potassium ion export across plasma membrane | 3 | 5 | 3.73e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 38 | 71 | 3.73e-01 |
| GO:BP | GO:0003064 | regulation of heart rate by hormone | 1 | 2 | 3.73e-01 |
| GO:BP | GO:1901503 | ether biosynthetic process | 3 | 11 | 3.74e-01 |
| GO:BP | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification | 2 | 2 | 3.74e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 2 | 48 | 3.74e-01 |
| GO:BP | GO:0021569 | rhombomere 3 development | 1 | 2 | 3.74e-01 |
| GO:BP | GO:0019878 | lysine biosynthetic process via aminoadipic acid | 2 | 2 | 3.74e-01 |
| GO:BP | GO:0000707 | meiotic DNA recombinase assembly | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0009085 | lysine biosynthetic process | 2 | 2 | 3.74e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 2 | 48 | 3.74e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 1 | 13 | 3.74e-01 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 8 | 19 | 3.74e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 3 | 18 | 3.74e-01 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 15 | 36 | 3.74e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 18 | 40 | 3.74e-01 |
| GO:BP | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 2 | 2 | 3.74e-01 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 3 | 15 | 3.74e-01 |
| GO:BP | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification | 2 | 2 | 3.74e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 3.74e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 3.74e-01 |
| GO:BP | GO:0033603 | positive regulation of dopamine secretion | 1 | 4 | 3.74e-01 |
| GO:BP | GO:0014038 | regulation of Schwann cell differentiation | 1 | 3 | 3.74e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 2 | 57 | 3.74e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 1 | 218 | 3.74e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 2 | 8 | 3.74e-01 |
| GO:BP | GO:0007066 | female meiosis sister chromatid cohesion | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 4 | 20 | 3.74e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 3 | 33 | 3.74e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 3 | 44 | 3.74e-01 |
| GO:BP | GO:0031948 | positive regulation of glucocorticoid biosynthetic process | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 3.74e-01 |
| GO:BP | GO:2000079 | regulation of canonical Wnt signaling pathway involved in controlling type B pancreatic cell proliferation | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 4 | 18 | 3.74e-01 |
| GO:BP | GO:0002568 | somatic diversification of T cell receptor genes | 2 | 4 | 3.74e-01 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 1 | 9 | 3.74e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 1 | 28 | 3.74e-01 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 15 | 27 | 3.74e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 3 | 45 | 3.74e-01 |
| GO:BP | GO:2000066 | positive regulation of cortisol biosynthetic process | 1 | 1 | 3.74e-01 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 1 | 15 | 3.74e-01 |
| GO:BP | GO:0014031 | mesenchymal cell development | 4 | 6 | 3.74e-01 |
| GO:BP | GO:0044343 | canonical Wnt signaling pathway involved in regulation of type B pancreatic cell proliferation | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 5 | 166 | 3.74e-01 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 1 | 15 | 3.74e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 7 | 18 | 3.74e-01 |
| GO:BP | GO:2000180 | negative regulation of androgen biosynthetic process | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0003175 | tricuspid valve development | 4 | 6 | 3.74e-01 |
| GO:BP | GO:0031945 | positive regulation of glucocorticoid metabolic process | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0048792 | spontaneous exocytosis of neurotransmitter | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0033153 | T cell receptor V(D)J recombination | 2 | 4 | 3.74e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 6 | 81 | 3.74e-01 |
| GO:BP | GO:0006203 | dGTP catabolic process | 2 | 2 | 3.74e-01 |
| GO:BP | GO:0032329 | serine transport | 2 | 10 | 3.74e-01 |
| GO:BP | GO:2000019 | negative regulation of male gonad development | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 1 | 20 | 3.74e-01 |
| GO:BP | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells | 1 | 4 | 3.74e-01 |
| GO:BP | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity | 1 | 6 | 3.74e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 3.74e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 4 | 66 | 3.74e-01 |
| GO:BP | GO:0032366 | intracellular sterol transport | 4 | 30 | 3.74e-01 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 5 | 7 | 3.74e-01 |
| GO:BP | GO:0009449 | gamma-aminobutyric acid biosynthetic process | 1 | 4 | 3.74e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 2 | 75 | 3.74e-01 |
| GO:BP | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 3.74e-01 |
| GO:BP | GO:2000041 | negative regulation of planar cell polarity pathway involved in axis elongation | 1 | 2 | 3.74e-01 |
| GO:BP | GO:0061369 | negative regulation of testicular blood vessel morphogenesis | 1 | 1 | 3.74e-01 |
| GO:BP | GO:0002681 | somatic recombination of T cell receptor gene segments | 2 | 4 | 3.74e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 1 | 237 | 3.75e-01 |
| GO:BP | GO:0038130 | ERBB4 signaling pathway | 4 | 6 | 3.76e-01 |
| GO:BP | GO:0016358 | dendrite development | 26 | 202 | 3.76e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 1 | 227 | 3.76e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 1 | 55 | 3.76e-01 |
| GO:BP | GO:0098760 | response to interleukin-7 | 1 | 12 | 3.76e-01 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 1 | 12 | 3.76e-01 |
| GO:BP | GO:0120041 | positive regulation of macrophage proliferation | 1 | 4 | 3.76e-01 |
| GO:BP | GO:1903969 | regulation of response to macrophage colony-stimulating factor | 1 | 3 | 3.76e-01 |
| GO:BP | GO:0042482 | positive regulation of odontogenesis | 1 | 5 | 3.76e-01 |
| GO:BP | GO:1903972 | regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 3 | 3.76e-01 |
| GO:BP | GO:1900365 | positive regulation of mRNA polyadenylation | 3 | 4 | 3.76e-01 |
| GO:BP | GO:1903996 | negative regulation of non-membrane spanning protein tyrosine kinase activity | 1 | 1 | 3.77e-01 |
| GO:BP | GO:0071932 | replication fork reversal | 4 | 5 | 3.77e-01 |
| GO:BP | GO:0060753 | regulation of mast cell chemotaxis | 1 | 7 | 3.77e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 1 | 357 | 3.77e-01 |
| GO:BP | GO:1903645 | negative regulation of chaperone-mediated protein folding | 2 | 2 | 3.77e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 6 | 15 | 3.77e-01 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 2 | 46 | 3.77e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 2 | 8 | 3.77e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 6 | 401 | 3.77e-01 |
| GO:BP | GO:1902261 | positive regulation of delayed rectifier potassium channel activity | 1 | 5 | 3.77e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 6 | 43 | 3.77e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 6 | 21 | 3.78e-01 |
| GO:BP | GO:0006301 | postreplication repair | 18 | 34 | 3.78e-01 |
| GO:BP | GO:0062176 | R-loop processing | 4 | 6 | 3.78e-01 |
| GO:BP | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 2 | 17 | 3.78e-01 |
| GO:BP | GO:0097151 | positive regulation of inhibitory postsynaptic potential | 3 | 4 | 3.78e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 2 | 80 | 3.78e-01 |
| GO:BP | GO:0019290 | siderophore biosynthetic process | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0045054 | constitutive secretory pathway | 3 | 7 | 3.78e-01 |
| GO:BP | GO:0009237 | siderophore metabolic process | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 3 | 53 | 3.78e-01 |
| GO:BP | GO:0046035 | CMP metabolic process | 1 | 6 | 3.78e-01 |
| GO:BP | GO:0046078 | dUMP metabolic process | 1 | 6 | 3.78e-01 |
| GO:BP | GO:1903434 | negative regulation of constitutive secretory pathway | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0009178 | pyrimidine deoxyribonucleoside monophosphate catabolic process | 1 | 5 | 3.78e-01 |
| GO:BP | GO:1904394 | negative regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 1 | 3.78e-01 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 4 | 4 | 3.79e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 19 | 56 | 3.79e-01 |
| GO:BP | GO:0006482 | protein demethylation | 11 | 27 | 3.79e-01 |
| GO:BP | GO:0062111 | zinc ion import into organelle | 2 | 7 | 3.79e-01 |
| GO:BP | GO:0008214 | protein dealkylation | 11 | 27 | 3.79e-01 |
| GO:BP | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis | 2 | 3 | 3.79e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 7 | 39 | 3.79e-01 |
| GO:BP | GO:0070453 | regulation of heme biosynthetic process | 3 | 7 | 3.79e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 9 | 206 | 3.79e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 4 | 83 | 3.79e-01 |
| GO:BP | GO:1901463 | regulation of tetrapyrrole biosynthetic process | 3 | 7 | 3.79e-01 |
| GO:BP | GO:0090594 | inflammatory response to wounding | 1 | 12 | 3.79e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 23 | 81 | 3.79e-01 |
| GO:BP | GO:0009099 | valine biosynthetic process | 1 | 3 | 3.79e-01 |
| GO:BP | GO:0019082 | viral protein processing | 1 | 31 | 3.79e-01 |
| GO:BP | GO:0009097 | isoleucine biosynthetic process | 1 | 2 | 3.79e-01 |
| GO:BP | GO:0038180 | nerve growth factor signaling pathway | 3 | 10 | 3.79e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 3.79e-01 |
| GO:BP | GO:0010967 | regulation of polyamine biosynthetic process | 2 | 2 | 3.79e-01 |
| GO:BP | GO:0097396 | response to interleukin-17 | 1 | 13 | 3.79e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 1 | 154 | 3.79e-01 |
| GO:BP | GO:0051548 | negative regulation of keratinocyte migration | 2 | 2 | 3.80e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 264 | 552 | 3.80e-01 |
| GO:BP | GO:0032915 | positive regulation of transforming growth factor beta2 production | 2 | 3 | 3.80e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 2 | 65 | 3.80e-01 |
| GO:BP | GO:1904555 | L-proline transmembrane transport | 1 | 5 | 3.80e-01 |
| GO:BP | GO:1904271 | L-proline import across plasma membrane | 1 | 5 | 3.80e-01 |
| GO:BP | GO:1905647 | proline import across plasma membrane | 1 | 5 | 3.80e-01 |
| GO:BP | GO:0015818 | isoleucine transport | 1 | 5 | 3.80e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 4 | 69 | 3.80e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 1 | 249 | 3.80e-01 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 2 | 11 | 3.80e-01 |
| GO:BP | GO:1903804 | glycine import across plasma membrane | 1 | 4 | 3.80e-01 |
| GO:BP | GO:0060745 | mammary gland branching involved in pregnancy | 2 | 5 | 3.80e-01 |
| GO:BP | GO:0061000 | negative regulation of dendritic spine development | 1 | 10 | 3.80e-01 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 1 | 8 | 3.80e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 1 | 240 | 3.80e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 2 | 47 | 3.80e-01 |
| GO:BP | GO:0051594 | detection of glucose | 1 | 1 | 3.80e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 5 | 80 | 3.80e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 4 | 99 | 3.80e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 1 | 30 | 3.80e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 1 | 157 | 3.80e-01 |
| GO:BP | GO:1902775 | mitochondrial large ribosomal subunit assembly | 3 | 4 | 3.80e-01 |
| GO:BP | GO:0009730 | detection of carbohydrate stimulus | 1 | 1 | 3.80e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 1 | 228 | 3.80e-01 |
| GO:BP | GO:0048855 | adenohypophysis morphogenesis | 1 | 1 | 3.80e-01 |
| GO:BP | GO:1901339 | regulation of store-operated calcium channel activity | 2 | 8 | 3.80e-01 |
| GO:BP | GO:0034287 | detection of monosaccharide stimulus | 1 | 1 | 3.80e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 1 | 241 | 3.80e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 1 | 14 | 3.80e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 10 | 33 | 3.80e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 1 | 185 | 3.80e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 1 | 236 | 3.80e-01 |
| GO:BP | GO:0003174 | mitral valve development | 7 | 12 | 3.80e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 3 | 167 | 3.80e-01 |
| GO:BP | GO:0009732 | detection of hexose stimulus | 1 | 1 | 3.80e-01 |
| GO:BP | GO:1904222 | positive regulation of serine C-palmitoyltransferase activity | 1 | 2 | 3.81e-01 |
| GO:BP | GO:0009826 | unidimensional cell growth | 2 | 3 | 3.81e-01 |
| GO:BP | GO:1904219 | positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 3.81e-01 |
| GO:BP | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity | 1 | 2 | 3.81e-01 |
| GO:BP | GO:0008219 | cell death | 2 | 1596 | 3.81e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 6 | 11 | 3.81e-01 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 2 | 17 | 3.82e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 3.82e-01 |
| GO:BP | GO:0000469 | cleavage involved in rRNA processing | 1 | 29 | 3.82e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 14 | 262 | 3.82e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 4 | 39 | 3.82e-01 |
| GO:BP | GO:1902308 | regulation of peptidyl-serine dephosphorylation | 3 | 3 | 3.82e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 3.82e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 3.82e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 12 | 486 | 3.83e-01 |
| GO:BP | GO:0001951 | intestinal D-glucose absorption | 2 | 3 | 3.83e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 4 | 111 | 3.83e-01 |
| GO:BP | GO:0097324 | melanocyte migration | 2 | 3 | 3.83e-01 |
| GO:BP | GO:0009886 | post-embryonic animal morphogenesis | 1 | 10 | 3.83e-01 |
| GO:BP | GO:0019075 | virus maturation | 2 | 2 | 3.83e-01 |
| GO:BP | GO:0110051 | metabolite repair | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0042942 | D-serine transport | 1 | 1 | 3.83e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 108 | 209 | 3.83e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 16 | 44 | 3.83e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 7 | 43 | 3.83e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 7 | 172 | 3.83e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 12 | 23 | 3.83e-01 |
| GO:BP | GO:0034197 | triglyceride transport | 1 | 4 | 3.83e-01 |
| GO:BP | GO:0034196 | acylglycerol transport | 1 | 4 | 3.83e-01 |
| GO:BP | GO:0071243 | cellular response to arsenic-containing substance | 8 | 18 | 3.83e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 3.83e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 1 | 31 | 3.84e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 6 | 408 | 3.84e-01 |
| GO:BP | GO:1902685 | positive regulation of receptor localization to synapse | 2 | 3 | 3.84e-01 |
| GO:BP | GO:0001845 | phagolysosome assembly | 1 | 14 | 3.85e-01 |
| GO:BP | GO:0045105 | intermediate filament polymerization or depolymerization | 2 | 2 | 3.85e-01 |
| GO:BP | GO:0010561 | negative regulation of glycoprotein biosynthetic process | 1 | 10 | 3.85e-01 |
| GO:BP | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib | 3 | 4 | 3.85e-01 |
| GO:BP | GO:0006417 | regulation of translation | 5 | 370 | 3.85e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 3.85e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 3.85e-01 |
| GO:BP | GO:2000195 | negative regulation of female gonad development | 1 | 1 | 3.85e-01 |
| GO:BP | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis | 1 | 3 | 3.85e-01 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 5 | 8 | 3.85e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 5 | 10 | 3.85e-01 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 14 | 26 | 3.86e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 3 | 66 | 3.86e-01 |
| GO:BP | GO:0042732 | D-xylose metabolic process | 1 | 2 | 3.86e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 1 | 189 | 3.86e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 5 | 37 | 3.86e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 1 | 199 | 3.86e-01 |
| GO:BP | GO:0000056 | ribosomal small subunit export from nucleus | 5 | 8 | 3.86e-01 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 4 | 5 | 3.86e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 6 | 113 | 3.86e-01 |
| GO:BP | GO:0030497 | fatty acid elongation | 4 | 12 | 3.86e-01 |
| GO:BP | GO:0031291 | Ran protein signal transduction | 1 | 3 | 3.87e-01 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 2 | 44 | 3.87e-01 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 3 | 58 | 3.87e-01 |
| GO:BP | GO:0032400 | melanosome localization | 4 | 22 | 3.87e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 2 | 17 | 3.87e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 6 | 14 | 3.87e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 9 | 17 | 3.87e-01 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 5 | 36 | 3.87e-01 |
| GO:BP | GO:0051293 | establishment of spindle localization | 27 | 48 | 3.87e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 2 | 20 | 3.87e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 6 | 414 | 3.87e-01 |
| GO:BP | GO:0030011 | maintenance of cell polarity | 5 | 16 | 3.87e-01 |
| GO:BP | GO:0097070 | ductus arteriosus closure | 1 | 2 | 3.87e-01 |
| GO:BP | GO:2000310 | regulation of NMDA receptor activity | 2 | 19 | 3.87e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 15 | 1280 | 3.87e-01 |
| GO:BP | GO:2000493 | negative regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 3.87e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 3.87e-01 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 10 | 15 | 3.87e-01 |
| GO:BP | GO:0009440 | cyanate catabolic process | 1 | 2 | 3.87e-01 |
| GO:BP | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 6 | 3.87e-01 |
| GO:BP | GO:0061739 | protein lipidation involved in autophagosome assembly | 2 | 3 | 3.87e-01 |
| GO:BP | GO:0009439 | cyanate metabolic process | 1 | 2 | 3.87e-01 |
| GO:BP | GO:0060676 | ureteric bud formation | 1 | 5 | 3.87e-01 |
| GO:BP | GO:0006595 | polyamine metabolic process | 1 | 16 | 3.87e-01 |
| GO:BP | GO:0051276 | chromosome organization | 257 | 543 | 3.87e-01 |
| GO:BP | GO:0007231 | osmosensory signaling pathway | 3 | 3 | 3.87e-01 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 22 | 42 | 3.87e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 5 | 418 | 3.87e-01 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 4 | 4 | 3.87e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 5 | 18 | 3.87e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 5 | 18 | 3.87e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 2 | 62 | 3.87e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 15 | 31 | 3.87e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 2 | 73 | 3.87e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 6 | 149 | 3.88e-01 |
| GO:BP | GO:0010650 | positive regulation of cell communication by electrical coupling | 2 | 3 | 3.88e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 3 | 56 | 3.89e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 3 | 52 | 3.89e-01 |
| GO:BP | GO:0006646 | phosphatidylethanolamine biosynthetic process | 4 | 8 | 3.89e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 14 | 67 | 3.89e-01 |
| GO:BP | GO:0019856 | pyrimidine nucleobase biosynthetic process | 5 | 8 | 3.89e-01 |
| GO:BP | GO:0031120 | snRNA pseudouridine synthesis | 2 | 3 | 3.89e-01 |
| GO:BP | GO:0019227 | neuronal action potential propagation | 2 | 5 | 3.89e-01 |
| GO:BP | GO:0071317 | cellular response to isoquinoline alkaloid | 1 | 2 | 3.89e-01 |
| GO:BP | GO:0071315 | cellular response to morphine | 1 | 2 | 3.89e-01 |
| GO:BP | GO:0098870 | action potential propagation | 2 | 5 | 3.89e-01 |
| GO:BP | GO:0033979 | box H/ACA RNA metabolic process | 2 | 3 | 3.89e-01 |
| GO:BP | GO:0035907 | dorsal aorta development | 2 | 8 | 3.89e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 1 | 272 | 3.89e-01 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 12 | 21 | 3.89e-01 |
| GO:BP | GO:0031128 | developmental induction | 13 | 26 | 3.89e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 2 | 103 | 3.90e-01 |
| GO:BP | GO:0009222 | pyrimidine ribonucleotide catabolic process | 1 | 7 | 3.91e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 3.91e-01 |
| GO:BP | GO:0009131 | pyrimidine nucleoside monophosphate catabolic process | 1 | 6 | 3.91e-01 |
| GO:BP | GO:0034383 | low-density lipoprotein particle clearance | 1 | 17 | 3.91e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 1 | 18 | 3.91e-01 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 10 | 26 | 3.91e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 9 | 16 | 3.91e-01 |
| GO:BP | GO:0072414 | response to mitotic cell cycle checkpoint signaling | 1 | 1 | 3.91e-01 |
| GO:BP | GO:0072432 | response to G1 DNA damage checkpoint signaling | 1 | 1 | 3.91e-01 |
| GO:BP | GO:1905303 | positive regulation of macropinocytosis | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0101030 | tRNA-guanine transglycosylation | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0016049 | cell growth | 1 | 418 | 3.91e-01 |
| GO:BP | GO:0099630 | postsynaptic neurotransmitter receptor cycle | 1 | 1 | 3.91e-01 |
| GO:BP | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway | 2 | 2 | 3.91e-01 |
| GO:BP | GO:1905395 | response to flavonoid | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0099627 | neurotransmitter receptor cycle | 1 | 1 | 3.91e-01 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 1 | 23 | 3.91e-01 |
| GO:BP | GO:0001907 | killing by symbiont of host cells | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0051642 | centrosome localization | 16 | 31 | 3.91e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 2 | 61 | 3.91e-01 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 2 | 26 | 3.91e-01 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 2 | 18 | 3.91e-01 |
| GO:BP | GO:1902001 | fatty acid transmembrane transport | 5 | 15 | 3.91e-01 |
| GO:BP | GO:1903943 | regulation of hepatocyte apoptotic process | 4 | 7 | 3.91e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 6 | 21 | 3.91e-01 |
| GO:BP | GO:0006266 | DNA ligation | 9 | 16 | 3.91e-01 |
| GO:BP | GO:1900248 | negative regulation of cytoplasmic translational elongation | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0019051 | induction by virus of host apoptotic process | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0120040 | regulation of macrophage proliferation | 1 | 4 | 3.91e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 2 | 51 | 3.91e-01 |
| GO:BP | GO:0052151 | positive regulation by symbiont of host apoptotic process | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0052042 | induction by symbiont of host programmed cell death | 2 | 2 | 3.91e-01 |
| GO:BP | GO:0045106 | intermediate filament depolymerization | 1 | 1 | 3.91e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 3.91e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 16 | 31 | 3.91e-01 |
| GO:BP | GO:1904141 | positive regulation of microglial cell migration | 1 | 3 | 3.91e-01 |
| GO:BP | GO:0030844 | positive regulation of intermediate filament depolymerization | 1 | 1 | 3.91e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 3.91e-01 |
| GO:BP | GO:0044313 | protein K6-linked deubiquitination | 2 | 3 | 3.91e-01 |
| GO:BP | GO:0045108 | regulation of intermediate filament polymerization or depolymerization | 1 | 1 | 3.91e-01 |
| GO:BP | GO:0030842 | regulation of intermediate filament depolymerization | 1 | 1 | 3.91e-01 |
| GO:BP | GO:0046929 | negative regulation of neurotransmitter secretion | 3 | 6 | 3.91e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 2 | 57 | 3.91e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 21 | 64 | 3.91e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 1 | 15 | 3.91e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 3 | 112 | 3.91e-01 |
| GO:BP | GO:0061764 | late endosome to lysosome transport via multivesicular body sorting pathway | 2 | 2 | 3.91e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 16 | 56 | 3.91e-01 |
| GO:BP | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway | 2 | 4 | 3.91e-01 |
| GO:BP | GO:0061817 | endoplasmic reticulum-plasma membrane tethering | 1 | 5 | 3.91e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 2 | 14 | 3.91e-01 |
| GO:BP | GO:0038145 | macrophage colony-stimulating factor signaling pathway | 1 | 3 | 3.91e-01 |
| GO:BP | GO:0070842 | aggresome assembly | 3 | 5 | 3.91e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 3 | 4 | 3.91e-01 |
| GO:BP | GO:0051107 | negative regulation of DNA ligation | 1 | 1 | 3.92e-01 |
| GO:BP | GO:1904876 | negative regulation of DNA ligase activity | 1 | 1 | 3.92e-01 |
| GO:BP | GO:1902068 | regulation of sphingolipid mediated signaling pathway | 1 | 1 | 3.92e-01 |
| GO:BP | GO:1904428 | negative regulation of tubulin deacetylation | 3 | 6 | 3.92e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 1 | 265 | 3.92e-01 |
| GO:BP | GO:0090207 | regulation of triglyceride metabolic process | 3 | 29 | 3.93e-01 |
| GO:BP | GO:0035849 | nephric duct elongation | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 1 | 31 | 3.93e-01 |
| GO:BP | GO:0097379 | dorsal spinal cord interneuron posterior axon guidance | 1 | 1 | 3.93e-01 |
| GO:BP | GO:1905337 | positive regulation of aggrephagy | 2 | 3 | 3.93e-01 |
| GO:BP | GO:1905335 | regulation of aggrephagy | 2 | 3 | 3.93e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 7 | 86 | 3.93e-01 |
| GO:BP | GO:0097531 | mast cell migration | 5 | 12 | 3.93e-01 |
| GO:BP | GO:0071476 | cellular hypotonic response | 6 | 8 | 3.93e-01 |
| GO:BP | GO:0035847 | uterine epithelium development | 1 | 1 | 3.93e-01 |
| GO:BP | GO:0006936 | muscle contraction | 1 | 267 | 3.93e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 2 | 21 | 3.93e-01 |
| GO:BP | GO:0014028 | notochord formation | 2 | 3 | 3.93e-01 |
| GO:BP | GO:0061945 | regulation of protein K48-linked ubiquitination | 2 | 4 | 3.93e-01 |
| GO:BP | GO:0060456 | positive regulation of digestive system process | 1 | 8 | 3.93e-01 |
| GO:BP | GO:1901329 | regulation of odontoblast differentiation | 1 | 2 | 3.94e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 3.94e-01 |
| GO:BP | GO:1900063 | regulation of peroxisome organization | 1 | 4 | 3.94e-01 |
| GO:BP | GO:0061744 | motor behavior | 1 | 17 | 3.94e-01 |
| GO:BP | GO:0006404 | RNA import into nucleus | 3 | 4 | 3.94e-01 |
| GO:BP | GO:0035862 | dITP metabolic process | 1 | 1 | 3.94e-01 |
| GO:BP | GO:0035863 | dITP catabolic process | 1 | 1 | 3.94e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 19 | 60 | 3.94e-01 |
| GO:BP | GO:0120222 | regulation of blastocyst development | 2 | 3 | 3.94e-01 |
| GO:BP | GO:1904997 | regulation of leukocyte adhesion to arterial endothelial cell | 2 | 3 | 3.94e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 2 | 62 | 3.94e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 1 | 18 | 3.95e-01 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 1 | 12 | 3.95e-01 |
| GO:BP | GO:0036115 | fatty-acyl-CoA catabolic process | 4 | 6 | 3.95e-01 |
| GO:BP | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | 4 | 11 | 3.96e-01 |
| GO:BP | GO:0098693 | regulation of synaptic vesicle cycle | 7 | 13 | 3.96e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 3.96e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 39 | 270 | 3.96e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 3.96e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 3 | 843 | 3.96e-01 |
| GO:BP | GO:0014719 | skeletal muscle satellite cell activation | 2 | 5 | 3.96e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 4 | 56 | 3.96e-01 |
| GO:BP | GO:0051340 | regulation of ligase activity | 6 | 11 | 3.96e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 2 | 4 | 3.97e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 2 | 4 | 3.97e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 1 | 12 | 3.97e-01 |
| GO:BP | GO:0016577 | histone demethylation | 9 | 21 | 3.97e-01 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 16 | 30 | 3.97e-01 |
| GO:BP | GO:0032058 | positive regulation of translational initiation in response to stress | 1 | 4 | 3.97e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 3.97e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 3.97e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 2 | 13 | 3.97e-01 |
| GO:BP | GO:0034389 | lipid droplet organization | 14 | 30 | 3.97e-01 |
| GO:BP | GO:0031119 | tRNA pseudouridine synthesis | 1 | 6 | 3.97e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 15 | 49 | 3.97e-01 |
| GO:BP | GO:0042113 | B cell activation | 1 | 170 | 3.97e-01 |
| GO:BP | GO:0048511 | rhythmic process | 4 | 237 | 3.97e-01 |
| GO:BP | GO:0061072 | iris morphogenesis | 1 | 5 | 3.97e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 3 | 28 | 3.97e-01 |
| GO:BP | GO:0048285 | organelle fission | 199 | 388 | 3.97e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 2 | 30 | 3.97e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 2 | 4 | 3.97e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 3.97e-01 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 8 | 13 | 3.97e-01 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 1 | 5 | 3.97e-01 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 1 | 26 | 3.97e-01 |
| GO:BP | GO:0090398 | cellular senescence | 22 | 83 | 3.97e-01 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 5 | 35 | 3.97e-01 |
| GO:BP | GO:1903810 | L-histidine import across plasma membrane | 2 | 2 | 3.97e-01 |
| GO:BP | GO:0060064 | Spemann organizer formation at the anterior end of the primitive streak | 1 | 1 | 3.97e-01 |
| GO:BP | GO:1905474 | canonical Wnt signaling pathway involved in stem cell proliferation | 1 | 1 | 3.97e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 3.97e-01 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 4 | 27 | 3.97e-01 |
| GO:BP | GO:0051415 | microtubule nucleation by interphase microtubule organizing center | 1 | 1 | 3.97e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 2 | 30 | 3.97e-01 |
| GO:BP | GO:0006497 | protein lipidation | 30 | 89 | 3.97e-01 |
| GO:BP | GO:0070932 | histone H3 deacetylation | 6 | 12 | 3.98e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 3 | 104 | 3.98e-01 |
| GO:BP | GO:0097711 | ciliary basal body-plasma membrane docking | 2 | 2 | 3.98e-01 |
| GO:BP | GO:0042592 | homeostatic process | 2 | 1159 | 3.98e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 3 | 104 | 3.98e-01 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 1 | 16 | 3.98e-01 |
| GO:BP | GO:0033260 | nuclear DNA replication | 19 | 35 | 3.98e-01 |
| GO:BP | GO:0060035 | notochord cell development | 1 | 1 | 3.98e-01 |
| GO:BP | GO:0060034 | notochord cell differentiation | 1 | 1 | 3.98e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 2 | 53 | 3.98e-01 |
| GO:BP | GO:0002481 | antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent | 2 | 2 | 3.98e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 4 | 22 | 3.99e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 6 | 448 | 3.99e-01 |
| GO:BP | GO:1900006 | positive regulation of dendrite development | 5 | 14 | 3.99e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 14 | 637 | 3.99e-01 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 2 | 5 | 3.99e-01 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 2 | 5 | 3.99e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 3.99e-01 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 5 | 8 | 3.99e-01 |
| GO:BP | GO:0010808 | positive regulation of synaptic vesicle priming | 1 | 1 | 3.99e-01 |
| GO:BP | GO:0090038 | negative regulation of protein kinase C signaling | 2 | 4 | 3.99e-01 |
| GO:BP | GO:0006183 | GTP biosynthetic process | 4 | 9 | 4.00e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 5 | 20 | 4.00e-01 |
| GO:BP | GO:1904627 | response to phorbol 13-acetate 12-myristate | 2 | 4 | 4.00e-01 |
| GO:BP | GO:1904628 | cellular response to phorbol 13-acetate 12-myristate | 2 | 4 | 4.00e-01 |
| GO:BP | GO:0048370 | lateral mesoderm formation | 3 | 3 | 4.00e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 32 | 84 | 4.00e-01 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 4 | 10 | 4.00e-01 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 4 | 30 | 4.00e-01 |
| GO:BP | GO:0048369 | lateral mesoderm morphogenesis | 3 | 3 | 4.00e-01 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 9 | 19 | 4.00e-01 |
| GO:BP | GO:0010891 | negative regulation of sequestering of triglyceride | 1 | 5 | 4.00e-01 |
| GO:BP | GO:0032460 | negative regulation of protein oligomerization | 1 | 2 | 4.00e-01 |
| GO:BP | GO:0032463 | negative regulation of protein homooligomerization | 1 | 2 | 4.00e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 2 | 5 | 4.00e-01 |
| GO:BP | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 6 | 80 | 4.00e-01 |
| GO:BP | GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 1 | 14 | 4.00e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 7 | 12 | 4.00e-01 |
| GO:BP | GO:0042983 | amyloid precursor protein biosynthetic process | 1 | 14 | 4.00e-01 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 2 | 28 | 4.00e-01 |
| GO:BP | GO:0002551 | mast cell chemotaxis | 6 | 9 | 4.00e-01 |
| GO:BP | GO:0060087 | relaxation of vascular associated smooth muscle | 1 | 7 | 4.00e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 9 | 197 | 4.00e-01 |
| GO:BP | GO:0006307 | DNA dealkylation involved in DNA repair | 3 | 9 | 4.00e-01 |
| GO:BP | GO:0060708 | spongiotrophoblast differentiation | 2 | 2 | 4.00e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 236 | 4.00e-01 |
| GO:BP | GO:1901585 | regulation of acid-sensing ion channel activity | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 8 | 73 | 4.00e-01 |
| GO:BP | GO:0031649 | heat generation | 1 | 11 | 4.00e-01 |
| GO:BP | GO:0051940 | regulation of catecholamine uptake involved in synaptic transmission | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 8 | 164 | 4.00e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 1 | 211 | 4.00e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 3 | 10 | 4.00e-01 |
| GO:BP | GO:0021756 | striatum development | 1 | 17 | 4.00e-01 |
| GO:BP | GO:0090618 | DNA clamp unloading | 1 | 2 | 4.00e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 8 | 23 | 4.00e-01 |
| GO:BP | GO:0032053 | ciliary basal body organization | 1 | 4 | 4.00e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 4 | 12 | 4.00e-01 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 9 | 19 | 4.00e-01 |
| GO:BP | GO:0008052 | sensory organ boundary specification | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 4.00e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 3 | 29 | 4.00e-01 |
| GO:BP | GO:1901950 | dense core granule transport | 1 | 7 | 4.00e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 5 | 142 | 4.00e-01 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 1 | 11 | 4.00e-01 |
| GO:BP | GO:0015961 | diadenosine polyphosphate catabolic process | 4 | 6 | 4.00e-01 |
| GO:BP | GO:0003097 | renal water transport | 2 | 6 | 4.00e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 10 | 47 | 4.00e-01 |
| GO:BP | GO:0098917 | retrograde trans-synaptic signaling | 3 | 7 | 4.00e-01 |
| GO:BP | GO:0007158 | neuron cell-cell adhesion | 1 | 13 | 4.00e-01 |
| GO:BP | GO:0002545 | chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 18 | 4.00e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 1 | 295 | 4.00e-01 |
| GO:BP | GO:2000210 | positive regulation of anoikis | 3 | 6 | 4.00e-01 |
| GO:BP | GO:0021780 | glial cell fate specification | 3 | 3 | 4.00e-01 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 5 | 7 | 4.00e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 30 | 61 | 4.00e-01 |
| GO:BP | GO:0019068 | virion assembly | 5 | 35 | 4.00e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 13 | 106 | 4.00e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 4.00e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 2 | 56 | 4.00e-01 |
| GO:BP | GO:0001964 | startle response | 1 | 16 | 4.00e-01 |
| GO:BP | GO:0070544 | histone H3-K36 demethylation | 3 | 5 | 4.00e-01 |
| GO:BP | GO:0032069 | regulation of nuclease activity | 1 | 17 | 4.00e-01 |
| GO:BP | GO:1903232 | melanosome assembly | 2 | 19 | 4.00e-01 |
| GO:BP | GO:0071472 | cellular response to salt stress | 5 | 11 | 4.00e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 11 | 35 | 4.00e-01 |
| GO:BP | GO:0002880 | regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0051126 | negative regulation of actin nucleation | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0032230 | positive regulation of synaptic transmission, GABAergic | 1 | 10 | 4.00e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 4.00e-01 |
| GO:BP | GO:0035524 | proline transmembrane transport | 3 | 8 | 4.00e-01 |
| GO:BP | GO:0010522 | regulation of calcium ion transport into cytosol | 1 | 10 | 4.00e-01 |
| GO:BP | GO:0032253 | dense core granule localization | 1 | 7 | 4.00e-01 |
| GO:BP | GO:0006538 | glutamate catabolic process | 1 | 7 | 4.00e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 4.00e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 2 | 18 | 4.00e-01 |
| GO:BP | GO:0033026 | negative regulation of mast cell apoptotic process | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0002882 | positive regulation of chronic inflammatory response to non-antigenic stimulus | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0010160 | formation of animal organ boundary | 1 | 1 | 4.00e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 2 | 56 | 4.00e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 2 | 23 | 4.00e-01 |
| GO:BP | GO:0072197 | ureter morphogenesis | 3 | 5 | 4.00e-01 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 2 | 26 | 4.00e-01 |
| GO:BP | GO:0099519 | dense core granule cytoskeletal transport | 1 | 7 | 4.00e-01 |
| GO:BP | GO:0009448 | gamma-aminobutyric acid metabolic process | 1 | 6 | 4.00e-01 |
| GO:BP | GO:0039534 | negative regulation of MDA-5 signaling pathway | 1 | 3 | 4.00e-01 |
| GO:BP | GO:0061193 | taste bud development | 1 | 1 | 4.00e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 4.00e-01 |
| GO:BP | GO:0071503 | response to heparin | 3 | 5 | 4.00e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 1 | 7 | 4.00e-01 |
| GO:BP | GO:1900186 | negative regulation of clathrin-dependent endocytosis | 3 | 4 | 4.00e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 6 | 48 | 4.00e-01 |
| GO:BP | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 5 | 4.00e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 1 | 212 | 4.00e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 3 | 10 | 4.02e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 1 | 271 | 4.02e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 1 | 20 | 4.02e-01 |
| GO:BP | GO:0051666 | actin cortical patch localization | 1 | 2 | 4.02e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 4.02e-01 |
| GO:BP | GO:0009256 | 10-formyltetrahydrofolate metabolic process | 2 | 6 | 4.02e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 1 | 513 | 4.02e-01 |
| GO:BP | GO:1904124 | microglial cell migration | 1 | 3 | 4.02e-01 |
| GO:BP | GO:1904139 | regulation of microglial cell migration | 1 | 3 | 4.02e-01 |
| GO:BP | GO:0072303 | positive regulation of glomerular metanephric mesangial cell proliferation | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 1 | 18 | 4.02e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 4 | 344 | 4.02e-01 |
| GO:BP | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate | 1 | 1 | 4.02e-01 |
| GO:BP | GO:1903575 | cornified envelope assembly | 1 | 6 | 4.02e-01 |
| GO:BP | GO:0018215 | protein phosphopantetheinylation | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0070257 | positive regulation of mucus secretion | 1 | 4 | 4.02e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 2 | 64 | 4.02e-01 |
| GO:BP | GO:0061518 | microglial cell proliferation | 1 | 4 | 4.02e-01 |
| GO:BP | GO:0015707 | nitrite transport | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0003195 | tricuspid valve formation | 2 | 2 | 4.02e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 4 | 155 | 4.02e-01 |
| GO:BP | GO:0150066 | negative regulation of deacetylase activity | 2 | 2 | 4.02e-01 |
| GO:BP | GO:0060354 | negative regulation of cell adhesion molecule production | 1 | 5 | 4.02e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 65 | 162 | 4.02e-01 |
| GO:BP | GO:0110113 | positive regulation of lipid transporter activity | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0035176 | social behavior | 3 | 34 | 4.02e-01 |
| GO:BP | GO:0006362 | transcription elongation by RNA polymerase I | 2 | 4 | 4.02e-01 |
| GO:BP | GO:1902954 | regulation of early endosome to recycling endosome transport | 1 | 2 | 4.02e-01 |
| GO:BP | GO:0046149 | pigment catabolic process | 1 | 6 | 4.02e-01 |
| GO:BP | GO:0042167 | heme catabolic process | 1 | 6 | 4.02e-01 |
| GO:BP | GO:2000755 | positive regulation of sphingomyelin catabolic process | 1 | 1 | 4.02e-01 |
| GO:BP | GO:2000754 | regulation of sphingomyelin catabolic process | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 11 | 53 | 4.02e-01 |
| GO:BP | GO:1990927 | calcium ion regulated lysosome exocytosis | 1 | 3 | 4.02e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 6 | 103 | 4.02e-01 |
| GO:BP | GO:0006600 | creatine metabolic process | 1 | 3 | 4.02e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 5 | 167 | 4.02e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 3 | 41 | 4.02e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 1 | 37 | 4.02e-01 |
| GO:BP | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | 2 | 2 | 4.02e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 2 | 544 | 4.02e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 6 | 42 | 4.02e-01 |
| GO:BP | GO:0006896 | Golgi to vacuole transport | 1 | 19 | 4.02e-01 |
| GO:BP | GO:1901726 | negative regulation of histone deacetylase activity | 2 | 2 | 4.02e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 14 | 72 | 4.02e-01 |
| GO:BP | GO:0006914 | autophagy | 1 | 513 | 4.02e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 8 | 278 | 4.02e-01 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 1 | 1 | 4.02e-01 |
| GO:BP | GO:0046204 | nor-spermidine metabolic process | 1 | 1 | 4.03e-01 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 2 | 14 | 4.03e-01 |
| GO:BP | GO:0032020 | ISG15-protein conjugation | 4 | 6 | 4.03e-01 |
| GO:BP | GO:0032920 | putrescine acetylation | 1 | 1 | 4.03e-01 |
| GO:BP | GO:0032919 | spermine acetylation | 1 | 1 | 4.03e-01 |
| GO:BP | GO:0036017 | response to erythropoietin | 3 | 4 | 4.03e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 1 | 311 | 4.03e-01 |
| GO:BP | GO:1903347 | negative regulation of bicellular tight junction assembly | 1 | 3 | 4.03e-01 |
| GO:BP | GO:0098885 | modification of postsynaptic actin cytoskeleton | 4 | 10 | 4.03e-01 |
| GO:BP | GO:0071225 | cellular response to muramyl dipeptide | 4 | 7 | 4.04e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 2 | 21 | 4.04e-01 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 7 | 15 | 4.04e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 2 | 70 | 4.04e-01 |
| GO:BP | GO:1903019 | negative regulation of glycoprotein metabolic process | 1 | 12 | 4.04e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 2 | 54 | 4.04e-01 |
| GO:BP | GO:0032898 | neurotrophin production | 2 | 4 | 4.05e-01 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 3 | 16 | 4.05e-01 |
| GO:BP | GO:0062009 | secondary palate development | 5 | 20 | 4.05e-01 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 6 | 18 | 4.05e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 10 | 122 | 4.05e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 4 | 186 | 4.05e-01 |
| GO:BP | GO:0000423 | mitophagy | 17 | 37 | 4.05e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 3 | 39 | 4.05e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 2 | 32 | 4.05e-01 |
| GO:BP | GO:1904199 | positive regulation of regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.05e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 4.05e-01 |
| GO:BP | GO:1990736 | regulation of vascular associated smooth muscle cell membrane depolarization | 1 | 1 | 4.05e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 9 | 706 | 4.05e-01 |
| GO:BP | GO:0002894 | positive regulation of type II hypersensitivity | 2 | 3 | 4.05e-01 |
| GO:BP | GO:0001798 | positive regulation of type IIa hypersensitivity | 2 | 3 | 4.05e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 4.05e-01 |
| GO:BP | GO:1903949 | positive regulation of atrial cardiac muscle cell action potential | 1 | 1 | 4.05e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 4.05e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 4.05e-01 |
| GO:BP | GO:1905663 | positive regulation of telomerase RNA reverse transcriptase activity | 2 | 2 | 4.05e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 19 | 101 | 4.05e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 2 | 55 | 4.05e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 2 | 76 | 4.05e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 2 | 55 | 4.05e-01 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 2 | 14 | 4.05e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 1 | 209 | 4.05e-01 |
| GO:BP | GO:2000394 | positive regulation of lamellipodium morphogenesis | 3 | 6 | 4.05e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 1 | 319 | 4.06e-01 |
| GO:BP | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling | 2 | 6 | 4.06e-01 |
| GO:BP | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell communication | 2 | 6 | 4.06e-01 |
| GO:BP | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 1 | 4 | 4.06e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 1 | 199 | 4.07e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 6 | 318 | 4.07e-01 |
| GO:BP | GO:0003271 | smoothened signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 4.07e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 62 | 134 | 4.07e-01 |
| GO:BP | GO:0097211 | cellular response to gonadotropin-releasing hormone | 2 | 3 | 4.07e-01 |
| GO:BP | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 4.08e-01 |
| GO:BP | GO:0016926 | protein desumoylation | 2 | 9 | 4.08e-01 |
| GO:BP | GO:0072209 | metanephric mesangial cell differentiation | 1 | 1 | 4.08e-01 |
| GO:BP | GO:0072254 | metanephric glomerular mesangial cell differentiation | 1 | 1 | 4.08e-01 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 1 | 16 | 4.08e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 9 | 19 | 4.08e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 10 | 38 | 4.08e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 4.08e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 1 | 234 | 4.08e-01 |
| GO:BP | GO:0032912 | negative regulation of transforming growth factor beta2 production | 2 | 2 | 4.08e-01 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 7 | 25 | 4.08e-01 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 20 | 36 | 4.09e-01 |
| GO:BP | GO:0060050 | positive regulation of protein glycosylation | 1 | 6 | 4.09e-01 |
| GO:BP | GO:0098903 | regulation of membrane repolarization during action potential | 1 | 6 | 4.10e-01 |
| GO:BP | GO:0014740 | negative regulation of muscle hyperplasia | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0070483 | detection of hypoxia | 1 | 2 | 4.10e-01 |
| GO:BP | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis | 3 | 6 | 4.10e-01 |
| GO:BP | GO:1903553 | positive regulation of extracellular exosome assembly | 2 | 4 | 4.10e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 4.10e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 3 | 10 | 4.10e-01 |
| GO:BP | GO:0048266 | behavioral response to pain | 1 | 11 | 4.10e-01 |
| GO:BP | GO:0071409 | cellular response to cycloheximide | 2 | 2 | 4.10e-01 |
| GO:BP | GO:0030046 | parallel actin filament bundle assembly | 2 | 5 | 4.10e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 4 | 121 | 4.10e-01 |
| GO:BP | GO:0090216 | positive regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 1 | 4.10e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 9 | 31 | 4.10e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 1 | 10 | 4.10e-01 |
| GO:BP | GO:0097421 | liver regeneration | 4 | 23 | 4.10e-01 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 1 | 24 | 4.10e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 3 | 28 | 4.11e-01 |
| GO:BP | GO:1903765 | negative regulation of potassium ion export across plasma membrane | 1 | 1 | 4.11e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 6 | 90 | 4.11e-01 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 4.11e-01 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 3 | 5 | 4.11e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 3 | 249 | 4.11e-01 |
| GO:BP | GO:0032815 | negative regulation of natural killer cell activation | 1 | 3 | 4.11e-01 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 3 | 5 | 4.11e-01 |
| GO:BP | GO:1990683 | DNA double-strand break attachment to nuclear envelope | 2 | 2 | 4.11e-01 |
| GO:BP | GO:0001830 | trophectodermal cell fate commitment | 1 | 1 | 4.11e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | 40 | 4.11e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 3 | 34 | 4.12e-01 |
| GO:BP | GO:1903031 | regulation of microtubule plus-end binding | 2 | 2 | 4.12e-01 |
| GO:BP | GO:0071864 | positive regulation of cell proliferation in bone marrow | 2 | 5 | 4.12e-01 |
| GO:BP | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion | 3 | 7 | 4.12e-01 |
| GO:BP | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling | 2 | 8 | 4.12e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 8 | 21 | 4.12e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 2 | 33 | 4.12e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 4 | 27 | 4.12e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 1 | 24 | 4.12e-01 |
| GO:BP | GO:1990983 | tRNA demethylation | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 7 | 76 | 4.12e-01 |
| GO:BP | GO:0046710 | GDP metabolic process | 4 | 10 | 4.12e-01 |
| GO:BP | GO:0042753 | positive regulation of circadian rhythm | 1 | 10 | 4.12e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 6 | 10 | 4.12e-01 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 1 | 6 | 4.12e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 2 | 54 | 4.12e-01 |
| GO:BP | GO:0031284 | positive regulation of guanylate cyclase activity | 2 | 2 | 4.12e-01 |
| GO:BP | GO:0018377 | protein myristoylation | 3 | 4 | 4.12e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 31 | 92 | 4.12e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 3 | 17 | 4.12e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 6 | 90 | 4.12e-01 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 3 | 18 | 4.12e-01 |
| GO:BP | GO:0070934 | CRD-mediated mRNA stabilization | 6 | 11 | 4.12e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 35 | 90 | 4.12e-01 |
| GO:BP | GO:0006499 | N-terminal protein myristoylation | 3 | 4 | 4.12e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 4 | 10 | 4.12e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 57 | 133 | 4.12e-01 |
| GO:BP | GO:0046697 | decidualization | 5 | 19 | 4.12e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 3 | 19 | 4.12e-01 |
| GO:BP | GO:0043278 | response to morphine | 2 | 16 | 4.12e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 2 | 16 | 4.12e-01 |
| GO:BP | GO:0051455 | monopolar spindle attachment to meiosis I kinetochore | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0098754 | detoxification | 5 | 91 | 4.12e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 6 | 84 | 4.12e-01 |
| GO:BP | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 6 | 341 | 4.12e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 1 | 279 | 4.12e-01 |
| GO:BP | GO:0060479 | lung cell differentiation | 6 | 18 | 4.12e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 2 | 51 | 4.12e-01 |
| GO:BP | GO:1905278 | positive regulation of epithelial tube formation | 1 | 7 | 4.12e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 2 | 52 | 4.12e-01 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 53 | 113 | 4.12e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 2 | 423 | 4.12e-01 |
| GO:BP | GO:1905276 | regulation of epithelial tube formation | 1 | 7 | 4.12e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 7 | 29 | 4.12e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 4 | 10 | 4.12e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 2 | 102 | 4.12e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 11 | 43 | 4.12e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 32 | 162 | 4.12e-01 |
| GO:BP | GO:0120094 | negative regulation of peptidyl-lysine crotonylation | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0120093 | regulation of peptidyl-lysine crotonylation | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0034351 | negative regulation of glial cell apoptotic process | 2 | 8 | 4.12e-01 |
| GO:BP | GO:0006012 | galactose metabolic process | 2 | 9 | 4.12e-01 |
| GO:BP | GO:0033564 | anterior/posterior axon guidance | 2 | 4 | 4.12e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 22 | 152 | 4.12e-01 |
| GO:BP | GO:0060487 | lung epithelial cell differentiation | 6 | 18 | 4.12e-01 |
| GO:BP | GO:1905232 | cellular response to L-glutamate | 2 | 4 | 4.12e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 5 | 33 | 4.12e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 8 | 129 | 4.12e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 4 | 23 | 4.12e-01 |
| GO:BP | GO:1903033 | positive regulation of microtubule plus-end binding | 2 | 2 | 4.12e-01 |
| GO:BP | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development | 2 | 2 | 4.12e-01 |
| GO:BP | GO:0006610 | ribosomal protein import into nucleus | 1 | 4 | 4.12e-01 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 1 | 8 | 4.12e-01 |
| GO:BP | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 2 | 6 | 4.12e-01 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 1 | 4 | 4.12e-01 |
| GO:BP | GO:0098920 | retrograde trans-synaptic signaling by lipid | 2 | 4 | 4.12e-01 |
| GO:BP | GO:0098921 | retrograde trans-synaptic signaling by endocannabinoid | 2 | 4 | 4.12e-01 |
| GO:BP | GO:1903012 | positive regulation of bone development | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 501 | 4.12e-01 |
| GO:BP | GO:0044873 | lipoprotein localization to membrane | 1 | 1 | 4.12e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 6 | 342 | 4.12e-01 |
| GO:BP | GO:0006207 | ‘de novo’ pyrimidine nucleobase biosynthetic process | 3 | 6 | 4.13e-01 |
| GO:BP | GO:0006605 | protein targeting | 7 | 307 | 4.13e-01 |
| GO:BP | GO:0045730 | respiratory burst | 1 | 21 | 4.13e-01 |
| GO:BP | GO:0010165 | response to X-ray | 16 | 29 | 4.13e-01 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 5 | 9 | 4.13e-01 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 1 | 6 | 4.13e-01 |
| GO:BP | GO:0046031 | ADP metabolic process | 5 | 7 | 4.13e-01 |
| GO:BP | GO:1903064 | positive regulation of reverse cholesterol transport | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0003012 | muscle system process | 1 | 334 | 4.13e-01 |
| GO:BP | GO:1903062 | regulation of reverse cholesterol transport | 1 | 1 | 4.13e-01 |
| GO:BP | GO:0022402 | cell cycle process | 505 | 1086 | 4.13e-01 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 5 | 9 | 4.13e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 4 | 10 | 4.13e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 1 | 260 | 4.13e-01 |
| GO:BP | GO:1903223 | positive regulation of oxidative stress-induced neuron death | 1 | 5 | 4.13e-01 |
| GO:BP | GO:0071707 | immunoglobulin heavy chain V-D-J recombination | 2 | 2 | 4.13e-01 |
| GO:BP | GO:0007635 | chemosensory behavior | 1 | 10 | 4.13e-01 |
| GO:BP | GO:0032967 | positive regulation of collagen biosynthetic process | 4 | 23 | 4.13e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 18 | 40 | 4.13e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 2 | 9 | 4.13e-01 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 3 | 8 | 4.13e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 3 | 106 | 4.13e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 2 | 429 | 4.13e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 4.13e-01 |
| GO:BP | GO:0001705 | ectoderm formation | 2 | 3 | 4.13e-01 |
| GO:BP | GO:0042044 | fluid transport | 2 | 17 | 4.14e-01 |
| GO:BP | GO:0034486 | vacuolar transmembrane transport | 2 | 3 | 4.14e-01 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 7 | 13 | 4.14e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 2 | 26 | 4.14e-01 |
| GO:BP | GO:0038129 | ERBB3 signaling pathway | 3 | 4 | 4.14e-01 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 3 | 22 | 4.14e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 1 | 22 | 4.14e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 44 | 92 | 4.14e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 14 | 386 | 4.14e-01 |
| GO:BP | GO:0061709 | reticulophagy | 6 | 17 | 4.14e-01 |
| GO:BP | GO:0060318 | definitive erythrocyte differentiation | 3 | 4 | 4.14e-01 |
| GO:BP | GO:0035265 | organ growth | 40 | 131 | 4.15e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 4.15e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 4.15e-01 |
| GO:BP | GO:2001176 | regulation of mediator complex assembly | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0001927 | exocyst assembly | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0060178 | regulation of exocyst localization | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 1 | 279 | 4.15e-01 |
| GO:BP | GO:0001928 | regulation of exocyst assembly | 1 | 1 | 4.15e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 1 | 318 | 4.15e-01 |
| GO:BP | GO:0014816 | skeletal muscle satellite cell differentiation | 2 | 6 | 4.15e-01 |
| GO:BP | GO:0051793 | medium-chain fatty acid catabolic process | 2 | 3 | 4.15e-01 |
| GO:BP | GO:0090023 | positive regulation of neutrophil chemotaxis | 2 | 14 | 4.15e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 35 | 100 | 4.15e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 2 | 19 | 4.15e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 4.15e-01 |
| GO:BP | GO:0036034 | mediator complex assembly | 1 | 1 | 4.15e-01 |
| GO:BP | GO:2001178 | positive regulation of mediator complex assembly | 1 | 1 | 4.15e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 2 | 20 | 4.15e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 7 | 246 | 4.15e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 4.15e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 92 | 376 | 4.15e-01 |
| GO:BP | GO:0060028 | convergent extension involved in axis elongation | 4 | 6 | 4.15e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 2 | 1389 | 4.15e-01 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 3 | 53 | 4.15e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 1 | 16 | 4.15e-01 |
| GO:BP | GO:0048265 | response to pain | 2 | 17 | 4.15e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 10 | 16 | 4.16e-01 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 1 | 9 | 4.16e-01 |
| GO:BP | GO:0140454 | protein aggregate center assembly | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 4 | 30 | 4.17e-01 |
| GO:BP | GO:0071236 | cellular response to antibiotic | 1 | 11 | 4.17e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 226 | 504 | 4.17e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 11 | 36 | 4.17e-01 |
| GO:BP | GO:0006590 | thyroid hormone generation | 4 | 10 | 4.17e-01 |
| GO:BP | GO:0070417 | cellular response to cold | 1 | 8 | 4.17e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 3 | 115 | 4.17e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 3 | 170 | 4.17e-01 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 6 | 17 | 4.17e-01 |
| GO:BP | GO:1902257 | negative regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 2 | 4.17e-01 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 6 | 7 | 4.17e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 3 | 170 | 4.17e-01 |
| GO:BP | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway | 3 | 3 | 4.17e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 30 | 62 | 4.17e-01 |
| GO:BP | GO:0043048 | dolichyl monophosphate biosynthetic process | 1 | 1 | 4.17e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 1 | 9 | 4.17e-01 |
| GO:BP | GO:0003167 | atrioventricular bundle cell differentiation | 2 | 3 | 4.17e-01 |
| GO:BP | GO:0036359 | renal potassium excretion | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 2 | 14 | 4.17e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 9 | 16 | 4.17e-01 |
| GO:BP | GO:0098779 | positive regulation of mitophagy in response to mitochondrial depolarization | 5 | 10 | 4.17e-01 |
| GO:BP | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation | 2 | 2 | 4.17e-01 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 14 | 27 | 4.17e-01 |
| GO:BP | GO:0060596 | mammary placode formation | 2 | 2 | 4.17e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 2 | 12 | 4.17e-01 |
| GO:BP | GO:1905072 | cardiac jelly development | 2 | 3 | 4.17e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 1 | 26 | 4.17e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 4 | 6 | 4.17e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 8 | 123 | 4.17e-01 |
| GO:BP | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process | 3 | 6 | 4.18e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 1 | 222 | 4.18e-01 |
| GO:BP | GO:0048732 | gland development | 1 | 318 | 4.18e-01 |
| GO:BP | GO:0090251 | protein localization involved in establishment of planar polarity | 2 | 2 | 4.18e-01 |
| GO:BP | GO:2000727 | positive regulation of cardiac muscle cell differentiation | 1 | 6 | 4.19e-01 |
| GO:BP | GO:0019520 | aldonic acid metabolic process | 1 | 2 | 4.19e-01 |
| GO:BP | GO:0019521 | D-gluconate metabolic process | 1 | 2 | 4.19e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 5 | 19 | 4.19e-01 |
| GO:BP | GO:0046177 | D-gluconate catabolic process | 1 | 2 | 4.19e-01 |
| GO:BP | GO:0046176 | aldonic acid catabolic process | 1 | 2 | 4.19e-01 |
| GO:BP | GO:0019322 | pentose biosynthetic process | 1 | 2 | 4.19e-01 |
| GO:BP | GO:0009648 | photoperiodism | 11 | 28 | 4.19e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 3 | 200 | 4.19e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 3 | 51 | 4.19e-01 |
| GO:BP | GO:0051660 | establishment of centrosome localization | 3 | 8 | 4.19e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 1 | 19 | 4.21e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 4 | 60 | 4.21e-01 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 2 | 111 | 4.21e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 15 | 36 | 4.21e-01 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 4 | 29 | 4.21e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 4 | 4 | 4.21e-01 |
| GO:BP | GO:0098828 | modulation of inhibitory postsynaptic potential | 3 | 4 | 4.21e-01 |
| GO:BP | GO:0099540 | trans-synaptic signaling by neuropeptide | 1 | 1 | 4.21e-01 |
| GO:BP | GO:0016139 | glycoside catabolic process | 3 | 8 | 4.21e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 2 | 82 | 4.21e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 4.21e-01 |
| GO:BP | GO:0006787 | porphyrin-containing compound catabolic process | 1 | 7 | 4.21e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 68 | 149 | 4.21e-01 |
| GO:BP | GO:0033015 | tetrapyrrole catabolic process | 1 | 7 | 4.21e-01 |
| GO:BP | GO:0010700 | negative regulation of norepinephrine secretion | 2 | 2 | 4.21e-01 |
| GO:BP | GO:0006429 | leucyl-tRNA aminoacylation | 1 | 2 | 4.21e-01 |
| GO:BP | GO:0006425 | glutaminyl-tRNA aminoacylation | 1 | 2 | 4.21e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 4 | 706 | 4.22e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 2 | 22 | 4.22e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 5 | 41 | 4.23e-01 |
| GO:BP | GO:0072172 | mesonephric tubule formation | 1 | 6 | 4.23e-01 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 1 | 31 | 4.23e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 4 | 76 | 4.23e-01 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 1 | 13 | 4.23e-01 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 1 | 13 | 4.23e-01 |
| GO:BP | GO:0090656 | t-circle formation | 1 | 13 | 4.23e-01 |
| GO:BP | GO:0045606 | positive regulation of epidermal cell differentiation | 5 | 16 | 4.23e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 3 | 39 | 4.23e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 2 | 16 | 4.23e-01 |
| GO:BP | GO:0060980 | cell migration involved in coronary vasculogenesis | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 1 | 316 | 4.23e-01 |
| GO:BP | GO:0042749 | regulation of circadian sleep/wake cycle | 1 | 9 | 4.23e-01 |
| GO:BP | GO:0009203 | ribonucleoside triphosphate catabolic process | 2 | 2 | 4.23e-01 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 9 | 15 | 4.23e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 1 | 229 | 4.23e-01 |
| GO:BP | GO:0046041 | ITP metabolic process | 2 | 2 | 4.23e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 16 | 40 | 4.23e-01 |
| GO:BP | GO:0006853 | carnitine shuttle | 1 | 3 | 4.23e-01 |
| GO:BP | GO:0090269 | fibroblast growth factor production | 2 | 4 | 4.23e-01 |
| GO:BP | GO:1903781 | positive regulation of cardiac conduction | 1 | 1 | 4.23e-01 |
| GO:BP | GO:0090270 | regulation of fibroblast growth factor production | 2 | 4 | 4.23e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 3 | 165 | 4.23e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 4 | 310 | 4.23e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 12 | 120 | 4.24e-01 |
| GO:BP | GO:1901403 | positive regulation of tetrapyrrole metabolic process | 2 | 4 | 4.24e-01 |
| GO:BP | GO:0051958 | methotrexate transport | 1 | 2 | 4.24e-01 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 2 | 119 | 4.24e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 7 | 10 | 4.24e-01 |
| GO:BP | GO:0015911 | long-chain fatty acid import across plasma membrane | 3 | 9 | 4.24e-01 |
| GO:BP | GO:1990227 | paranodal junction maintenance | 2 | 2 | 4.24e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 5 | 68 | 4.24e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 4 | 67 | 4.24e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 4 | 16 | 4.25e-01 |
| GO:BP | GO:0044210 | ‘de novo’ CTP biosynthetic process | 2 | 2 | 4.25e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 1 | 289 | 4.25e-01 |
| GO:BP | GO:0046479 | glycosphingolipid catabolic process | 4 | 10 | 4.25e-01 |
| GO:BP | GO:0044208 | ‘de novo’ AMP biosynthetic process | 5 | 8 | 4.25e-01 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 2 | 30 | 4.25e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 14 | 1239 | 4.25e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 1 | 22 | 4.25e-01 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 14 | 26 | 4.25e-01 |
| GO:BP | GO:0036113 | very long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 8 | 149 | 4.25e-01 |
| GO:BP | GO:0051451 | myoblast migration | 2 | 11 | 4.25e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 2 | 62 | 4.25e-01 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 84 | 186 | 4.25e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 7 | 771 | 4.25e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 2 | 27 | 4.25e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 5 | 377 | 4.25e-01 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 9 | 20 | 4.25e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 8 | 12 | 4.25e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 4.25e-01 |
| GO:BP | GO:1904645 | response to amyloid-beta | 7 | 32 | 4.25e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 1 | 230 | 4.25e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 12 | 46 | 4.25e-01 |
| GO:BP | GO:2001304 | lipoxin B4 metabolic process | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0061303 | cornea development in camera-type eye | 1 | 4 | 4.25e-01 |
| GO:BP | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity | 1 | 10 | 4.25e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 1 | 2 | 4.25e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 23 | 52 | 4.25e-01 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 8 | 19 | 4.25e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 1 | 371 | 4.25e-01 |
| GO:BP | GO:0090262 | regulation of transcription-coupled nucleotide-excision repair | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 8 | 19 | 4.25e-01 |
| GO:BP | GO:0021544 | subpallium development | 1 | 18 | 4.25e-01 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 8 | 19 | 4.25e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 2 | 15 | 4.25e-01 |
| GO:BP | GO:0016050 | vesicle organization | 31 | 320 | 4.25e-01 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 3 | 4 | 4.25e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 1 | 41 | 4.25e-01 |
| GO:BP | GO:1901004 | ubiquinone-6 metabolic process | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0001507 | acetylcholine catabolic process in synaptic cleft | 2 | 2 | 4.25e-01 |
| GO:BP | GO:0036510 | trimming of terminal mannose on C branch | 1 | 3 | 4.25e-01 |
| GO:BP | GO:0034033 | purine nucleoside bisphosphate biosynthetic process | 11 | 49 | 4.25e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 4.25e-01 |
| GO:BP | GO:0033866 | nucleoside bisphosphate biosynthetic process | 11 | 49 | 4.25e-01 |
| GO:BP | GO:1901006 | ubiquinone-6 biosynthetic process | 1 | 1 | 4.25e-01 |
| GO:BP | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol | 1 | 3 | 4.25e-01 |
| GO:BP | GO:0034030 | ribonucleoside bisphosphate biosynthetic process | 11 | 49 | 4.25e-01 |
| GO:BP | GO:0045176 | apical protein localization | 1 | 13 | 4.25e-01 |
| GO:BP | GO:0035732 | nitric oxide storage | 1 | 1 | 4.25e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 6 | 418 | 4.25e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 5 | 134 | 4.25e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 1 | 11 | 4.25e-01 |
| GO:BP | GO:0061525 | hindgut development | 1 | 5 | 4.25e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 4.25e-01 |
| GO:BP | GO:1903348 | positive regulation of bicellular tight junction assembly | 3 | 5 | 4.25e-01 |
| GO:BP | GO:0070673 | response to interleukin-18 | 3 | 5 | 4.25e-01 |
| GO:BP | GO:0090557 | establishment of endothelial intestinal barrier | 3 | 10 | 4.25e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 1 | 295 | 4.25e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 4.25e-01 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 6 | 12 | 4.25e-01 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 1 | 7 | 4.26e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 1 | 320 | 4.26e-01 |
| GO:BP | GO:0090472 | dibasic protein processing | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 2 | 36 | 4.26e-01 |
| GO:BP | GO:1901394 | positive regulation of transforming growth factor beta1 activation | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1901392 | regulation of transforming growth factor beta1 activation | 1 | 1 | 4.26e-01 |
| GO:BP | GO:1903977 | positive regulation of glial cell migration | 1 | 5 | 4.26e-01 |
| GO:BP | GO:0010714 | positive regulation of collagen metabolic process | 4 | 24 | 4.27e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 2 | 36 | 4.27e-01 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 10 | 17 | 4.27e-01 |
| GO:BP | GO:0001946 | lymphangiogenesis | 5 | 12 | 4.27e-01 |
| GO:BP | GO:2000018 | regulation of male gonad development | 2 | 5 | 4.27e-01 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 10 | 17 | 4.27e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 5 | 4245 | 4.27e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 1 | 294 | 4.27e-01 |
| GO:BP | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway | 2 | 5 | 4.27e-01 |
| GO:BP | GO:0098939 | dendritic transport of mitochondrion | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 3 | 18 | 4.27e-01 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 1 | 16 | 4.27e-01 |
| GO:BP | GO:0051351 | positive regulation of ligase activity | 5 | 9 | 4.27e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 2 | 5 | 4.27e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 6 | 23 | 4.27e-01 |
| GO:BP | GO:1904613 | cellular response to 2,3,7,8-tetrachlorodibenzodioxine | 2 | 2 | 4.27e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 23 | 43 | 4.27e-01 |
| GO:BP | GO:0106001 | intestinal hexose absorption | 2 | 3 | 4.27e-01 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 1 | 10 | 4.27e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 6 | 121 | 4.27e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 4.27e-01 |
| GO:BP | GO:0006435 | threonyl-tRNA aminoacylation | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 12 | 21 | 4.27e-01 |
| GO:BP | GO:0034381 | plasma lipoprotein particle clearance | 1 | 25 | 4.27e-01 |
| GO:BP | GO:0009957 | epidermal cell fate specification | 2 | 4 | 4.27e-01 |
| GO:BP | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis | 2 | 4 | 4.27e-01 |
| GO:BP | GO:1903569 | positive regulation of protein localization to ciliary membrane | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0010182 | sugar mediated signaling pathway | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0010255 | glucose mediated signaling pathway | 1 | 5 | 4.27e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 8 | 28 | 4.27e-01 |
| GO:BP | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0009757 | hexose mediated signaling | 1 | 5 | 4.27e-01 |
| GO:BP | GO:1900454 | positive regulation of long-term synaptic depression | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 2 | 47 | 4.27e-01 |
| GO:BP | GO:0016104 | triterpenoid biosynthetic process | 1 | 1 | 4.27e-01 |
| GO:BP | GO:0051601 | exocyst localization | 2 | 4 | 4.27e-01 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 8 | 16 | 4.27e-01 |
| GO:BP | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery | 4 | 6 | 4.28e-01 |
| GO:BP | GO:0062196 | regulation of lysosome size | 4 | 8 | 4.28e-01 |
| GO:BP | GO:0070889 | platelet alpha granule organization | 2 | 2 | 4.28e-01 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 1 | 7 | 4.28e-01 |
| GO:BP | GO:0003419 | growth plate cartilage chondrocyte proliferation | 1 | 2 | 4.28e-01 |
| GO:BP | GO:0071578 | zinc ion import across plasma membrane | 1 | 6 | 4.28e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 6 | 70 | 4.28e-01 |
| GO:BP | GO:1902044 | regulation of Fas signaling pathway | 1 | 4 | 4.28e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 5 | 54 | 4.28e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 4 | 6 | 4.28e-01 |
| GO:BP | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis | 3 | 4 | 4.28e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 2 | 33 | 4.28e-01 |
| GO:BP | GO:0072229 | metanephric proximal convoluted tubule development | 1 | 2 | 4.28e-01 |
| GO:BP | GO:0072019 | proximal convoluted tubule development | 1 | 2 | 4.28e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 4.29e-01 |
| GO:BP | GO:1901254 | positive regulation of intracellular transport of viral material | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 4.29e-01 |
| GO:BP | GO:0061159 | establishment of bipolar cell polarity involved in cell morphogenesis | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0051892 | negative regulation of cardioblast differentiation | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0009082 | branched-chain amino acid biosynthetic process | 1 | 4 | 4.29e-01 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 1 | 16 | 4.29e-01 |
| GO:BP | GO:0000722 | telomere maintenance via recombination | 8 | 14 | 4.29e-01 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 2 | 33 | 4.29e-01 |
| GO:BP | GO:0090382 | phagosome maturation | 1 | 24 | 4.29e-01 |
| GO:BP | GO:0019045 | latent virus replication | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 4.29e-01 |
| GO:BP | GO:0019046 | release from viral latency | 1 | 2 | 4.29e-01 |
| GO:BP | GO:0015827 | tryptophan transport | 2 | 6 | 4.29e-01 |
| GO:BP | GO:0032252 | secretory granule localization | 1 | 9 | 4.29e-01 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 2 | 17 | 4.29e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 5 | 7 | 4.29e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 4.29e-01 |
| GO:BP | GO:0061772 | xenobiotic transport across blood-nerve barrier | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 11 | 30 | 4.29e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 14 | 73 | 4.29e-01 |
| GO:BP | GO:0017055 | negative regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 2 | 4.29e-01 |
| GO:BP | GO:1903545 | cellular response to butyrate | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0045925 | positive regulation of female receptivity | 1 | 1 | 4.29e-01 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 7 | 11 | 4.29e-01 |
| GO:BP | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization | 1 | 6 | 4.29e-01 |
| GO:BP | GO:0032480 | negative regulation of type I interferon production | 9 | 23 | 4.29e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 7 | 12 | 4.29e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 4.29e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 20 | 63 | 4.29e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 3 | 51 | 4.30e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 3 | 57 | 4.30e-01 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 20 | 36 | 4.30e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 4 | 33 | 4.30e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 3 | 69 | 4.30e-01 |
| GO:BP | GO:0001731 | formation of translation preinitiation complex | 3 | 11 | 4.30e-01 |
| GO:BP | GO:0006657 | CDP-choline pathway | 3 | 6 | 4.30e-01 |
| GO:BP | GO:0051643 | endoplasmic reticulum localization | 1 | 8 | 4.30e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 1 | 242 | 4.30e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 7 | 138 | 4.30e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 1 | 390 | 4.30e-01 |
| GO:BP | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0060775 | planar cell polarity pathway involved in gastrula mediolateral intercalation | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0060067 | cervix development | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0060031 | mediolateral intercalation | 2 | 2 | 4.30e-01 |
| GO:BP | GO:0051105 | regulation of DNA ligation | 4 | 6 | 4.30e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 91 | 375 | 4.30e-01 |
| GO:BP | GO:0048103 | somatic stem cell division | 4 | 10 | 4.30e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 15 | 96 | 4.31e-01 |
| GO:BP | GO:0006598 | polyamine catabolic process | 3 | 4 | 4.31e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 2 | 36 | 4.31e-01 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 2 | 13 | 4.31e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 10 | 23 | 4.31e-01 |
| GO:BP | GO:0033198 | response to ATP | 2 | 24 | 4.31e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 1 | 17 | 4.31e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 1 | 11 | 4.31e-01 |
| GO:BP | GO:1903444 | negative regulation of brown fat cell differentiation | 3 | 3 | 4.31e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 16 | 33 | 4.31e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 3 | 26 | 4.31e-01 |
| GO:BP | GO:0042779 | tRNA 3’-trailer cleavage | 1 | 3 | 4.31e-01 |
| GO:BP | GO:0006304 | DNA modification | 1 | 89 | 4.31e-01 |
| GO:BP | GO:0061511 | centriole elongation | 5 | 8 | 4.31e-01 |
| GO:BP | GO:0035754 | B cell chemotaxis | 2 | 5 | 4.32e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 1 | 21 | 4.32e-01 |
| GO:BP | GO:0036445 | neuronal stem cell division | 1 | 1 | 4.32e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 13 | 53 | 4.32e-01 |
| GO:BP | GO:0009447 | putrescine catabolic process | 2 | 2 | 4.32e-01 |
| GO:BP | GO:0032918 | spermidine acetylation | 2 | 2 | 4.32e-01 |
| GO:BP | GO:0032917 | polyamine acetylation | 2 | 2 | 4.32e-01 |
| GO:BP | GO:0035891 | exit from host cell | 1 | 28 | 4.33e-01 |
| GO:BP | GO:0019076 | viral release from host cell | 1 | 28 | 4.33e-01 |
| GO:BP | GO:0051257 | meiotic spindle midzone assembly | 2 | 2 | 4.33e-01 |
| GO:BP | GO:0072126 | positive regulation of glomerular mesangial cell proliferation | 2 | 7 | 4.33e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 2 | 25 | 4.33e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 1 | 12 | 4.33e-01 |
| GO:BP | GO:1901859 | negative regulation of mitochondrial DNA metabolic process | 2 | 2 | 4.33e-01 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 1 | 7 | 4.33e-01 |
| GO:BP | GO:0015881 | creatine transmembrane transport | 1 | 4 | 4.33e-01 |
| GO:BP | GO:0051596 | methylglyoxal catabolic process | 3 | 5 | 4.33e-01 |
| GO:BP | GO:0061727 | methylglyoxal catabolic process to lactate | 3 | 5 | 4.33e-01 |
| GO:BP | GO:2000553 | positive regulation of T-helper 2 cell cytokine production | 1 | 3 | 4.33e-01 |
| GO:BP | GO:0034263 | positive regulation of autophagy in response to ER overload | 1 | 1 | 4.33e-01 |
| GO:BP | GO:0006887 | exocytosis | 5 | 256 | 4.33e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 6 | 64 | 4.33e-01 |
| GO:BP | GO:0046131 | pyrimidine ribonucleoside metabolic process | 1 | 7 | 4.33e-01 |
| GO:BP | GO:0046133 | pyrimidine ribonucleoside catabolic process | 1 | 7 | 4.33e-01 |
| GO:BP | GO:0140448 | signaling receptor ligand precursor processing | 1 | 29 | 4.33e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 1 | 35 | 4.33e-01 |
| GO:BP | GO:0060947 | cardiac vascular smooth muscle cell differentiation | 1 | 8 | 4.33e-01 |
| GO:BP | GO:0008015 | blood circulation | 1 | 374 | 4.34e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 9 | 17 | 4.34e-01 |
| GO:BP | GO:0006419 | alanyl-tRNA aminoacylation | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0042551 | neuron maturation | 1 | 33 | 4.34e-01 |
| GO:BP | GO:0042727 | flavin-containing compound biosynthetic process | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0060897 | neural plate regionalization | 3 | 6 | 4.34e-01 |
| GO:BP | GO:0002885 | positive regulation of hypersensitivity | 3 | 5 | 4.34e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 5 | 183 | 4.34e-01 |
| GO:BP | GO:0071477 | cellular hypotonic salinity response | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0042539 | hypotonic salinity response | 2 | 2 | 4.34e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 13 | 133 | 4.34e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 17 | 37 | 4.34e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 6 | 14 | 4.35e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 14 | 615 | 4.35e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 4.35e-01 |
| GO:BP | GO:0090314 | positive regulation of protein targeting to membrane | 5 | 22 | 4.35e-01 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 2 | 21 | 4.35e-01 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 2 | 21 | 4.35e-01 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 2 | 6 | 4.36e-01 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 4 | 5 | 4.36e-01 |
| GO:BP | GO:0048645 | animal organ formation | 1 | 51 | 4.36e-01 |
| GO:BP | GO:0018160 | peptidyl-pyrromethane cofactor linkage | 1 | 1 | 4.36e-01 |
| GO:BP | GO:0003193 | pulmonary valve formation | 2 | 2 | 4.36e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 4.36e-01 |
| GO:BP | GO:0086054 | bundle of His cell to Purkinje myocyte communication by electrical coupling | 2 | 2 | 4.36e-01 |
| GO:BP | GO:1905517 | macrophage migration | 1 | 34 | 4.36e-01 |
| GO:BP | GO:0003283 | atrial septum development | 12 | 23 | 4.36e-01 |
| GO:BP | GO:0044557 | relaxation of smooth muscle | 1 | 9 | 4.36e-01 |
| GO:BP | GO:0010643 | cell communication by chemical coupling | 2 | 2 | 4.36e-01 |
| GO:BP | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration | 1 | 1 | 4.36e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 44 | 300 | 4.36e-01 |
| GO:BP | GO:0006547 | histidine metabolic process | 2 | 5 | 4.36e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 3 | 28 | 4.36e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 11 | 52 | 4.36e-01 |
| GO:BP | GO:0051890 | regulation of cardioblast differentiation | 2 | 5 | 4.36e-01 |
| GO:BP | GO:0007031 | peroxisome organization | 13 | 34 | 4.37e-01 |
| GO:BP | GO:2001037 | positive regulation of tongue muscle cell differentiation | 1 | 1 | 4.37e-01 |
| GO:BP | GO:2001035 | regulation of tongue muscle cell differentiation | 1 | 1 | 4.37e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 73 | 149 | 4.37e-01 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 10 | 18 | 4.37e-01 |
| GO:BP | GO:0002158 | osteoclast proliferation | 1 | 7 | 4.37e-01 |
| GO:BP | GO:0071374 | cellular response to parathyroid hormone stimulus | 1 | 7 | 4.37e-01 |
| GO:BP | GO:0035981 | tongue muscle cell differentiation | 1 | 1 | 4.37e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 1 | 30 | 4.37e-01 |
| GO:BP | GO:0042487 | regulation of odontogenesis of dentin-containing tooth | 1 | 6 | 4.37e-01 |
| GO:BP | GO:0061517 | macrophage proliferation | 1 | 7 | 4.37e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 1 | 26 | 4.37e-01 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 1 | 12 | 4.37e-01 |
| GO:BP | GO:0099532 | synaptic vesicle endosomal processing | 3 | 7 | 4.38e-01 |
| GO:BP | GO:2001204 | regulation of osteoclast development | 1 | 4 | 4.38e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 1 | 23 | 4.38e-01 |
| GO:BP | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process | 1 | 2 | 4.38e-01 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 4 | 6 | 4.38e-01 |
| GO:BP | GO:0001834 | trophectodermal cell proliferation | 1 | 2 | 4.38e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 18 | 1735 | 4.38e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 4.38e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 2 | 21 | 4.38e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 2 | 22 | 4.38e-01 |
| GO:BP | GO:1903898 | negative regulation of PERK-mediated unfolded protein response | 1 | 6 | 4.38e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 2 | 487 | 4.38e-01 |
| GO:BP | GO:0006171 | cAMP biosynthetic process | 2 | 8 | 4.38e-01 |
| GO:BP | GO:0080163 | regulation of protein serine/threonine phosphatase activity | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 6 | 9 | 4.38e-01 |
| GO:BP | GO:0051653 | spindle localization | 28 | 52 | 4.38e-01 |
| GO:BP | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0051216 | cartilage development | 4 | 151 | 4.38e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 4.38e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 1 | 276 | 4.38e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 8 | 46 | 4.38e-01 |
| GO:BP | GO:0031441 | negative regulation of mRNA 3’-end processing | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0016233 | telomere capping | 2 | 36 | 4.38e-01 |
| GO:BP | GO:0071624 | positive regulation of granulocyte chemotaxis | 2 | 14 | 4.38e-01 |
| GO:BP | GO:0042745 | circadian sleep/wake cycle | 1 | 11 | 4.38e-01 |
| GO:BP | GO:0014047 | glutamate secretion | 1 | 14 | 4.38e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 5 | 25 | 4.38e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 23 | 73 | 4.38e-01 |
| GO:BP | GO:0000105 | histidine biosynthetic process | 1 | 2 | 4.38e-01 |
| GO:BP | GO:0035803 | egg coat formation | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 8 | 65 | 4.38e-01 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 4 | 5 | 4.38e-01 |
| GO:BP | GO:0090209 | negative regulation of triglyceride metabolic process | 1 | 7 | 4.38e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 2 | 22 | 4.38e-01 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 5 | 7 | 4.38e-01 |
| GO:BP | GO:1902866 | regulation of retina development in camera-type eye | 1 | 1 | 4.38e-01 |
| GO:BP | GO:0033762 | response to glucagon | 2 | 13 | 4.38e-01 |
| GO:BP | GO:0030576 | Cajal body organization | 1 | 4 | 4.38e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 31 | 179 | 4.38e-01 |
| GO:BP | GO:1902722 | positive regulation of prolactin secretion | 1 | 1 | 4.38e-01 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 4 | 6 | 4.38e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 8 | 25 | 4.38e-01 |
| GO:BP | GO:1905775 | negative regulation of DNA helicase activity | 2 | 2 | 4.38e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 2 | 24 | 4.38e-01 |
| GO:BP | GO:0040011 | locomotion | 5 | 954 | 4.38e-01 |
| GO:BP | GO:0042148 | strand invasion | 4 | 5 | 4.38e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 4 | 5 | 4.38e-01 |
| GO:BP | GO:0071233 | cellular response to leucine | 6 | 10 | 4.38e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 4.38e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 6 | 34 | 4.38e-01 |
| GO:BP | GO:1904862 | inhibitory synapse assembly | 6 | 13 | 4.38e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 4 | 58 | 4.38e-01 |
| GO:BP | GO:2000810 | regulation of bicellular tight junction assembly | 6 | 15 | 4.38e-01 |
| GO:BP | GO:0035781 | CD86 biosynthetic process | 2 | 2 | 4.39e-01 |
| GO:BP | GO:0050930 | induction of positive chemotaxis | 1 | 12 | 4.39e-01 |
| GO:BP | GO:0060022 | hard palate development | 2 | 5 | 4.39e-01 |
| GO:BP | GO:0090118 | receptor-mediated endocytosis involved in cholesterol transport | 1 | 5 | 4.39e-01 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 17 | 31 | 4.39e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 2 | 2 | 4.39e-01 |
| GO:BP | GO:0051006 | positive regulation of lipoprotein lipase activity | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0061757 | leukocyte adhesion to arterial endothelial cell | 2 | 4 | 4.39e-01 |
| GO:BP | GO:0010898 | positive regulation of triglyceride catabolic process | 1 | 4 | 4.39e-01 |
| GO:BP | GO:0046380 | N-acetylneuraminate biosynthetic process | 1 | 1 | 4.40e-01 |
| GO:BP | GO:0014900 | muscle hyperplasia | 2 | 2 | 4.40e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 4 | 6 | 4.40e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 4.40e-01 |
| GO:BP | GO:0061152 | trachea submucosa development | 2 | 2 | 4.40e-01 |
| GO:BP | GO:0061153 | trachea gland development | 2 | 2 | 4.40e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 1 | 19 | 4.40e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 1 | 19 | 4.40e-01 |
| GO:BP | GO:0014043 | negative regulation of neuron maturation | 1 | 2 | 4.40e-01 |
| GO:BP | GO:1903860 | negative regulation of dendrite extension | 1 | 2 | 4.40e-01 |
| GO:BP | GO:0007495 | visceral mesoderm-endoderm interaction involved in midgut development | 1 | 1 | 4.40e-01 |
| GO:BP | GO:0036376 | sodium ion export across plasma membrane | 1 | 12 | 4.40e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 3 | 58 | 4.40e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 3 | 55 | 4.40e-01 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 4.40e-01 |
| GO:BP | GO:0019321 | pentose metabolic process | 2 | 9 | 4.40e-01 |
| GO:BP | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 2 | 2 | 4.40e-01 |
| GO:BP | GO:0140214 | positive regulation of long-chain fatty acid import into cell | 2 | 2 | 4.40e-01 |
| GO:BP | GO:0032930 | positive regulation of superoxide anion generation | 4 | 10 | 4.41e-01 |
| GO:BP | GO:1905454 | negative regulation of myeloid progenitor cell differentiation | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0106048 | spermidine deacetylation | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0106047 | polyamine deacetylation | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 4.41e-01 |
| GO:BP | GO:0030432 | peristalsis | 1 | 6 | 4.41e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 5 | 68 | 4.41e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 4.41e-01 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 2 | 17 | 4.41e-01 |
| GO:BP | GO:0036507 | protein demannosylation | 2 | 17 | 4.41e-01 |
| GO:BP | GO:0000320 | re-entry into mitotic cell cycle | 2 | 2 | 4.41e-01 |
| GO:BP | GO:0002253 | activation of immune response | 1 | 272 | 4.41e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 23 | 52 | 4.41e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 4 | 742 | 4.41e-01 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 3 | 10 | 4.41e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 4.41e-01 |
| GO:BP | GO:0051589 | negative regulation of neurotransmitter transport | 1 | 8 | 4.41e-01 |
| GO:BP | GO:0051583 | dopamine uptake involved in synaptic transmission | 1 | 8 | 4.41e-01 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 1 | 15 | 4.41e-01 |
| GO:BP | GO:0051934 | catecholamine uptake involved in synaptic transmission | 1 | 8 | 4.41e-01 |
| GO:BP | GO:0006273 | lagging strand elongation | 3 | 4 | 4.41e-01 |
| GO:BP | GO:0048219 | inter-Golgi cisterna vesicle-mediated transport | 1 | 1 | 4.41e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 3 | 155 | 4.42e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 1 | 23 | 4.42e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 4.42e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 2 | 35 | 4.42e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 4.42e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 4.42e-01 |
| GO:BP | GO:0071863 | regulation of cell proliferation in bone marrow | 2 | 6 | 4.42e-01 |
| GO:BP | GO:0000730 | DNA recombinase assembly | 3 | 3 | 4.42e-01 |
| GO:BP | GO:0090735 | DNA repair complex assembly | 3 | 3 | 4.42e-01 |
| GO:BP | GO:0016077 | sno(s)RNA catabolic process | 2 | 3 | 4.42e-01 |
| GO:BP | GO:0007610 | behavior | 6 | 436 | 4.42e-01 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 8 | 17 | 4.43e-01 |
| GO:BP | GO:0009408 | response to heat | 2 | 84 | 4.43e-01 |
| GO:BP | GO:0048382 | mesendoderm development | 4 | 5 | 4.43e-01 |
| GO:BP | GO:2000035 | regulation of stem cell division | 5 | 9 | 4.43e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 4.43e-01 |
| GO:BP | GO:0042256 | cytosolic ribosome assembly | 1 | 5 | 4.43e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 2 | 2 | 4.43e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 6 | 486 | 4.44e-01 |
| GO:BP | GO:1904618 | positive regulation of actin binding | 1 | 2 | 4.44e-01 |
| GO:BP | GO:1904531 | positive regulation of actin filament binding | 1 | 2 | 4.44e-01 |
| GO:BP | GO:0070145 | mitochondrial asparaginyl-tRNA aminoacylation | 1 | 1 | 4.44e-01 |
| GO:BP | GO:2001108 | positive regulation of Rho guanyl-nucleotide exchange factor activity | 1 | 2 | 4.44e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 2 | 25 | 4.44e-01 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 3 | 78 | 4.44e-01 |
| GO:BP | GO:1905099 | positive regulation of guanyl-nucleotide exchange factor activity | 1 | 2 | 4.44e-01 |
| GO:BP | GO:0006527 | arginine catabolic process | 1 | 7 | 4.44e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 3 | 70 | 4.44e-01 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 4 | 6 | 4.44e-01 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 4 | 6 | 4.44e-01 |
| GO:BP | GO:1903464 | negative regulation of mitotic cell cycle DNA replication | 1 | 1 | 4.44e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 3 | 56 | 4.44e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 1 | 62 | 4.44e-01 |
| GO:BP | GO:0070084 | protein initiator methionine removal | 2 | 4 | 4.44e-01 |
| GO:BP | GO:0043200 | response to amino acid | 11 | 95 | 4.44e-01 |
| GO:BP | GO:1902576 | negative regulation of nuclear cell cycle DNA replication | 1 | 1 | 4.44e-01 |
| GO:BP | GO:1904058 | positive regulation of sensory perception of pain | 2 | 3 | 4.44e-01 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 10 | 16 | 4.44e-01 |
| GO:BP | GO:2000808 | negative regulation of synaptic vesicle clustering | 1 | 1 | 4.44e-01 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 11 | 21 | 4.44e-01 |
| GO:BP | GO:1990547 | mitochondrial phosphate ion transmembrane transport | 1 | 1 | 4.44e-01 |
| GO:BP | GO:1903518 | positive regulation of single strand break repair | 2 | 2 | 4.44e-01 |
| GO:BP | GO:0060971 | embryonic heart tube left/right pattern formation | 3 | 5 | 4.44e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 1 | 41 | 4.44e-01 |
| GO:BP | GO:0036153 | triglyceride acyl-chain remodeling | 1 | 1 | 4.44e-01 |
| GO:BP | GO:0036309 | protein localization to M-band | 2 | 2 | 4.44e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 4 | 6 | 4.44e-01 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 5 | 11 | 4.44e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 13 | 77 | 4.44e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 4.44e-01 |
| GO:BP | GO:0003360 | brainstem development | 1 | 7 | 4.44e-01 |
| GO:BP | GO:0030237 | female sex determination | 1 | 1 | 4.44e-01 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 5 | 31 | 4.44e-01 |
| GO:BP | GO:1903689 | regulation of wound healing, spreading of epidermal cells | 1 | 8 | 4.44e-01 |
| GO:BP | GO:0120187 | positive regulation of protein localization to chromatin | 1 | 3 | 4.44e-01 |
| GO:BP | GO:1903959 | regulation of monoatomic anion transmembrane transport | 5 | 6 | 4.44e-01 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 10 | 16 | 4.44e-01 |
| GO:BP | GO:2000225 | negative regulation of testosterone biosynthetic process | 1 | 1 | 4.44e-01 |
| GO:BP | GO:2001223 | negative regulation of neuron migration | 3 | 8 | 4.44e-01 |
| GO:BP | GO:0007207 | phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway | 2 | 6 | 4.44e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 14 | 625 | 4.44e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 5 | 35 | 4.44e-01 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 1 | 3 | 4.44e-01 |
| GO:BP | GO:0090135 | actin filament branching | 1 | 1 | 4.44e-01 |
| GO:BP | GO:0070668 | positive regulation of mast cell proliferation | 2 | 4 | 4.44e-01 |
| GO:BP | GO:0019262 | N-acetylneuraminate catabolic process | 4 | 6 | 4.45e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 1 | 36 | 4.45e-01 |
| GO:BP | GO:2000234 | positive regulation of rRNA processing | 5 | 10 | 4.45e-01 |
| GO:BP | GO:0015824 | proline transport | 3 | 11 | 4.45e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 9 | 99 | 4.45e-01 |
| GO:BP | GO:0031443 | fast-twitch skeletal muscle fiber contraction | 1 | 3 | 4.45e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 32 | 96 | 4.45e-01 |
| GO:BP | GO:0097210 | response to gonadotropin-releasing hormone | 2 | 4 | 4.45e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 7 | 15 | 4.45e-01 |
| GO:BP | GO:0043174 | nucleoside salvage | 5 | 9 | 4.46e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 1 | 15 | 4.46e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 1 | 30 | 4.46e-01 |
| GO:BP | GO:0110021 | cardiac muscle myoblast proliferation | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 6 | 14 | 4.46e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 14 | 74 | 4.46e-01 |
| GO:BP | GO:0110022 | regulation of cardiac muscle myoblast proliferation | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0110024 | positive regulation of cardiac muscle myoblast proliferation | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 2 | 80 | 4.46e-01 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 6 | 12 | 4.46e-01 |
| GO:BP | GO:1903564 | regulation of protein localization to cilium | 5 | 11 | 4.46e-01 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 1 | 14 | 4.46e-01 |
| GO:BP | GO:1990575 | mitochondrial L-ornithine transmembrane transport | 1 | 2 | 4.46e-01 |
| GO:BP | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0010746 | regulation of long-chain fatty acid import across plasma membrane | 2 | 5 | 4.46e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 1 | 10 | 4.46e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 2 | 20 | 4.46e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 9 | 50 | 4.46e-01 |
| GO:BP | GO:0006680 | glucosylceramide catabolic process | 1 | 3 | 4.46e-01 |
| GO:BP | GO:1901290 | succinyl-CoA biosynthetic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 9 | 19 | 4.46e-01 |
| GO:BP | GO:0106089 | negative regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0030952 | establishment or maintenance of cytoskeleton polarity | 2 | 7 | 4.46e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 2 | 18 | 4.46e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 2 | 18 | 4.46e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 8 | 393 | 4.46e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 8 | 859 | 4.46e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 1 | 296 | 4.46e-01 |
| GO:BP | GO:1902497 | iron-sulfur cluster transmembrane transport | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0060697 | positive regulation of phospholipid catabolic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0030858 | positive regulation of epithelial cell differentiation | 14 | 37 | 4.46e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 11 | 37 | 4.46e-01 |
| GO:BP | GO:1903926 | cellular response to bisphenol A | 3 | 4 | 4.46e-01 |
| GO:BP | GO:0048268 | clathrin coat assembly | 5 | 13 | 4.46e-01 |
| GO:BP | GO:1904491 | protein localization to ciliary transition zone | 2 | 7 | 4.46e-01 |
| GO:BP | GO:1903925 | response to bisphenol A | 3 | 4 | 4.46e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 4.46e-01 |
| GO:BP | GO:2000584 | negative regulation of platelet-derived growth factor receptor-alpha signaling pathway | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 1 | 18 | 4.46e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 20 | 81 | 4.46e-01 |
| GO:BP | GO:0061880 | regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0061881 | positive regulation of anterograde axonal transport of mitochondrion | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0060353 | regulation of cell adhesion molecule production | 1 | 6 | 4.46e-01 |
| GO:BP | GO:0140466 | iron-sulfur cluster export from the mitochondrion | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1900130 | regulation of lipid binding | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0106088 | regulation of cell adhesion involved in sprouting angiogenesis | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0003219 | cardiac right ventricle formation | 3 | 4 | 4.46e-01 |
| GO:BP | GO:1901390 | positive regulation of transforming growth factor beta activation | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 2 | 39 | 4.46e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 4 | 16 | 4.46e-01 |
| GO:BP | GO:0023021 | termination of signal transduction | 1 | 3 | 4.46e-01 |
| GO:BP | GO:0006182 | cGMP biosynthetic process | 1 | 6 | 4.46e-01 |
| GO:BP | GO:1903331 | positive regulation of iron-sulfur cluster assembly | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1903329 | regulation of iron-sulfur cluster assembly | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 3 | 4 | 4.46e-01 |
| GO:BP | GO:0019678 | propionate metabolic process, methylmalonyl pathway | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1900222 | negative regulation of amyloid-beta clearance | 1 | 7 | 4.46e-01 |
| GO:BP | GO:1904998 | negative regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0018009 | N-terminal peptidyl-L-cysteine N-palmitoylation | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0055111 | ingression involved in gastrulation with mouth forming second | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 1 | 26 | 4.46e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 1 | 56 | 4.46e-01 |
| GO:BP | GO:0051155 | positive regulation of striated muscle cell differentiation | 2 | 31 | 4.46e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 4 | 97 | 4.46e-01 |
| GO:BP | GO:1900131 | negative regulation of lipid binding | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1903910 | negative regulation of receptor clustering | 1 | 2 | 4.46e-01 |
| GO:BP | GO:0003108 | negative regulation of the force of heart contraction by chemical signal | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 6 | 9 | 4.46e-01 |
| GO:BP | GO:1903551 | regulation of extracellular exosome assembly | 4 | 6 | 4.46e-01 |
| GO:BP | GO:0046067 | dGDP catabolic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 3 | 4 | 4.46e-01 |
| GO:BP | GO:0046600 | negative regulation of centriole replication | 3 | 7 | 4.46e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 5 | 816 | 4.46e-01 |
| GO:BP | GO:1903433 | regulation of constitutive secretory pathway | 1 | 2 | 4.46e-01 |
| GO:BP | GO:0045948 | positive regulation of translational initiation | 2 | 23 | 4.46e-01 |
| GO:BP | GO:0009184 | purine deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 9 | 23 | 4.46e-01 |
| GO:BP | GO:0009192 | deoxyribonucleoside diphosphate catabolic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0061713 | anterior neural tube closure | 1 | 2 | 4.46e-01 |
| GO:BP | GO:1905706 | regulation of mitochondrial ATP synthesis coupled proton transport | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0060510 | type II pneumocyte differentiation | 3 | 5 | 4.46e-01 |
| GO:BP | GO:0009314 | response to radiation | 4 | 352 | 4.46e-01 |
| GO:BP | GO:0017085 | response to insecticide | 1 | 4 | 4.46e-01 |
| GO:BP | GO:1901081 | negative regulation of relaxation of smooth muscle | 1 | 1 | 4.46e-01 |
| GO:BP | GO:1905271 | regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 1 | 10 | 4.46e-01 |
| GO:BP | GO:1905273 | positive regulation of proton-transporting ATP synthase activity, rotational mechanism | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0051694 | pointed-end actin filament capping | 2 | 6 | 4.46e-01 |
| GO:BP | GO:0001508 | action potential | 11 | 105 | 4.46e-01 |
| GO:BP | GO:0046057 | dADP catabolic process | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0046135 | pyrimidine nucleoside catabolic process | 1 | 8 | 4.46e-01 |
| GO:BP | GO:0006880 | intracellular sequestering of iron ion | 3 | 3 | 4.46e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 21 | 1849 | 4.46e-01 |
| GO:BP | GO:0045204 | MAPK export from nucleus | 1 | 3 | 4.46e-01 |
| GO:BP | GO:0006043 | glucosamine catabolic process | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 4 | 14 | 4.46e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 1 | 57 | 4.46e-01 |
| GO:BP | GO:0034201 | response to oleic acid | 3 | 5 | 4.46e-01 |
| GO:BP | GO:0052041 | suppression by symbiont of host programmed cell death | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0016254 | preassembly of GPI anchor in ER membrane | 2 | 8 | 4.46e-01 |
| GO:BP | GO:0033668 | suppression by symbiont of host apoptotic process | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 6 | 993 | 4.46e-01 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 1 | 22 | 4.46e-01 |
| GO:BP | GO:0019050 | suppression by virus of host apoptotic process | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 4.46e-01 |
| GO:BP | GO:1902725 | negative regulation of satellite cell differentiation | 1 | 2 | 4.46e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 2 | 114 | 4.46e-01 |
| GO:BP | GO:0002159 | desmosome assembly | 2 | 3 | 4.46e-01 |
| GO:BP | GO:0016182 | synaptic vesicle budding from endosome | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 2 | 2 | 4.46e-01 |
| GO:BP | GO:0038203 | TORC2 signaling | 5 | 12 | 4.46e-01 |
| GO:BP | GO:2000301 | negative regulation of synaptic vesicle exocytosis | 2 | 4 | 4.46e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 2 | 114 | 4.46e-01 |
| GO:BP | GO:0002155 | regulation of thyroid hormone mediated signaling pathway | 2 | 5 | 4.46e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 4.46e-01 |
| GO:BP | GO:0035963 | cellular response to interleukin-13 | 1 | 1 | 4.46e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 3 | 5 | 4.46e-01 |
| GO:BP | GO:0003188 | heart valve formation | 8 | 16 | 4.46e-01 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 3 | 20 | 4.46e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 2 | 16 | 4.47e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 62 | 116 | 4.47e-01 |
| GO:BP | GO:0007530 | sex determination | 6 | 12 | 4.47e-01 |
| GO:BP | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 2 | 4.47e-01 |
| GO:BP | GO:1905605 | positive regulation of blood-brain barrier permeability | 1 | 4 | 4.47e-01 |
| GO:BP | GO:0033605 | positive regulation of catecholamine secretion | 1 | 7 | 4.47e-01 |
| GO:BP | GO:1903911 | positive regulation of receptor clustering | 1 | 6 | 4.47e-01 |
| GO:BP | GO:0072718 | response to cisplatin | 4 | 6 | 4.47e-01 |
| GO:BP | GO:0050914 | sensory perception of salty taste | 1 | 1 | 4.47e-01 |
| GO:BP | GO:0045063 | T-helper 1 cell differentiation | 5 | 14 | 4.47e-01 |
| GO:BP | GO:0035655 | interleukin-18-mediated signaling pathway | 2 | 3 | 4.47e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 11 | 152 | 4.47e-01 |
| GO:BP | GO:0071351 | cellular response to interleukin-18 | 2 | 3 | 4.47e-01 |
| GO:BP | GO:0090669 | telomerase RNA stabilization | 2 | 4 | 4.47e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 1 | 23 | 4.47e-01 |
| GO:BP | GO:0007612 | learning | 3 | 115 | 4.48e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 4 | 358 | 4.48e-01 |
| GO:BP | GO:0048312 | intracellular distribution of mitochondria | 1 | 5 | 4.48e-01 |
| GO:BP | GO:0061365 | positive regulation of triglyceride lipase activity | 1 | 4 | 4.48e-01 |
| GO:BP | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade | 6 | 16 | 4.48e-01 |
| GO:BP | GO:0003104 | positive regulation of glomerular filtration | 1 | 3 | 4.48e-01 |
| GO:BP | GO:1903774 | positive regulation of viral budding via host ESCRT complex | 1 | 3 | 4.48e-01 |
| GO:BP | GO:0051029 | rRNA transport | 2 | 5 | 4.48e-01 |
| GO:BP | GO:0106058 | positive regulation of calcineurin-mediated signaling | 6 | 16 | 4.48e-01 |
| GO:BP | GO:0061738 | late endosomal microautophagy | 1 | 3 | 4.48e-01 |
| GO:BP | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 1 | 3 | 4.48e-01 |
| GO:BP | GO:2000591 | positive regulation of metanephric mesenchymal cell migration | 1 | 3 | 4.48e-01 |
| GO:BP | GO:0003013 | circulatory system process | 1 | 447 | 4.48e-01 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 4 | 15 | 4.48e-01 |
| GO:BP | GO:1900238 | regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway | 1 | 3 | 4.48e-01 |
| GO:BP | GO:0010610 | regulation of mRNA stability involved in response to stress | 2 | 3 | 4.48e-01 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 4 | 15 | 4.48e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 10 | 14 | 4.48e-01 |
| GO:BP | GO:0010759 | positive regulation of macrophage chemotaxis | 2 | 9 | 4.48e-01 |
| GO:BP | GO:0098657 | import into cell | 6 | 166 | 4.48e-01 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0035973 | aggrephagy | 5 | 8 | 4.48e-01 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0010359 | regulation of anion channel activity | 4 | 5 | 4.48e-01 |
| GO:BP | GO:0031915 | positive regulation of synaptic plasticity | 4 | 6 | 4.48e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 4 | 539 | 4.48e-01 |
| GO:BP | GO:0019367 | fatty acid elongation, saturated fatty acid | 2 | 6 | 4.48e-01 |
| GO:BP | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid | 2 | 6 | 4.48e-01 |
| GO:BP | GO:0033566 | gamma-tubulin complex localization | 2 | 2 | 4.48e-01 |
| GO:BP | GO:1904976 | cellular response to bleomycin | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 5 | 504 | 4.48e-01 |
| GO:BP | GO:0019368 | fatty acid elongation, unsaturated fatty acid | 2 | 6 | 4.48e-01 |
| GO:BP | GO:0006227 | dUDP biosynthetic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0086050 | membrane repolarization during bundle of His cell action potential | 1 | 1 | 4.48e-01 |
| GO:BP | GO:0006233 | dTDP biosynthetic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0009196 | pyrimidine deoxyribonucleoside diphosphate metabolic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:1904975 | response to bleomycin | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0086052 | membrane repolarization during SA node cell action potential | 1 | 1 | 4.48e-01 |
| GO:BP | GO:0034625 | fatty acid elongation, monounsaturated fatty acid | 2 | 6 | 4.48e-01 |
| GO:BP | GO:1904220 | regulation of serine C-palmitoyltransferase activity | 1 | 3 | 4.48e-01 |
| GO:BP | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0046077 | dUDP metabolic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0042136 | neurotransmitter biosynthetic process | 1 | 5 | 4.48e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 2 | 91 | 4.48e-01 |
| GO:BP | GO:0009197 | pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0046072 | dTDP metabolic process | 2 | 2 | 4.48e-01 |
| GO:BP | GO:0034350 | regulation of glial cell apoptotic process | 2 | 10 | 4.49e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 4.49e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 4.49e-01 |
| GO:BP | GO:0006294 | nucleotide-excision repair, preincision complex assembly | 1 | 1 | 4.49e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 1 | 24 | 4.50e-01 |
| GO:BP | GO:0036337 | Fas signaling pathway | 1 | 5 | 4.50e-01 |
| GO:BP | GO:0046967 | cytosol to endoplasmic reticulum transport | 1 | 4 | 4.50e-01 |
| GO:BP | GO:0006046 | N-acetylglucosamine catabolic process | 3 | 3 | 4.50e-01 |
| GO:BP | GO:0032581 | ER-dependent peroxisome organization | 1 | 1 | 4.50e-01 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 9 | 19 | 4.50e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 1 | 382 | 4.50e-01 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 7 | 39 | 4.50e-01 |
| GO:BP | GO:0106101 | ER-dependent peroxisome localization | 1 | 1 | 4.50e-01 |
| GO:BP | GO:1902034 | negative regulation of hematopoietic stem cell proliferation | 1 | 2 | 4.50e-01 |
| GO:BP | GO:0032485 | regulation of Ral protein signal transduction | 2 | 3 | 4.50e-01 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 5 | 7 | 4.51e-01 |
| GO:BP | GO:0016540 | protein autoprocessing | 3 | 16 | 4.51e-01 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 5 | 7 | 4.51e-01 |
| GO:BP | GO:0009790 | embryo development | 8 | 875 | 4.51e-01 |
| GO:BP | GO:0000710 | meiotic mismatch repair | 2 | 2 | 4.51e-01 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 2 | 2 | 4.51e-01 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 8 | 18 | 4.51e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 4.51e-01 |
| GO:BP | GO:1905609 | positive regulation of smooth muscle cell-matrix adhesion | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 4 | 185 | 4.51e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 1 | 26 | 4.51e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 16 | 321 | 4.51e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 3 | 8 | 4.51e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 2 | 44 | 4.51e-01 |
| GO:BP | GO:1904382 | mannose trimming involved in glycoprotein ERAD pathway | 3 | 5 | 4.51e-01 |
| GO:BP | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 13 | 47 | 4.51e-01 |
| GO:BP | GO:0017182 | peptidyl-diphthamide metabolic process | 4 | 7 | 4.51e-01 |
| GO:BP | GO:0017183 | peptidyl-diphthamide biosynthetic process from peptidyl-histidine | 4 | 7 | 4.51e-01 |
| GO:BP | GO:0000725 | recombinational repair | 74 | 153 | 4.51e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 1 | 3 | 4.51e-01 |
| GO:BP | GO:1901740 | negative regulation of myoblast fusion | 2 | 2 | 4.51e-01 |
| GO:BP | GO:0060081 | membrane hyperpolarization | 1 | 7 | 4.51e-01 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 1 | 7 | 4.51e-01 |
| GO:BP | GO:0051792 | medium-chain fatty acid biosynthetic process | 2 | 4 | 4.51e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 1 | 5 | 4.51e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 2 | 6 | 4.51e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 13 | 26 | 4.51e-01 |
| GO:BP | GO:0071918 | urea transmembrane transport | 1 | 2 | 4.51e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 4.51e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 4 | 763 | 4.51e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 8 | 70 | 4.51e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 2 | 19 | 4.51e-01 |
| GO:BP | GO:0021592 | fourth ventricle development | 1 | 1 | 4.51e-01 |
| GO:BP | GO:1902767 | isoprenoid biosynthetic process via mevalonate | 1 | 1 | 4.51e-01 |
| GO:BP | GO:1905463 | negative regulation of DNA duplex unwinding | 3 | 4 | 4.51e-01 |
| GO:BP | GO:0003010 | voluntary skeletal muscle contraction | 2 | 5 | 4.51e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 1 | 6 | 4.51e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 4 | 76 | 4.51e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 1 | 23 | 4.51e-01 |
| GO:BP | GO:0030903 | notochord development | 6 | 17 | 4.51e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 20 | 39 | 4.51e-01 |
| GO:BP | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process | 3 | 5 | 4.51e-01 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 3 | 9 | 4.51e-01 |
| GO:BP | GO:0033306 | phytol metabolic process | 2 | 2 | 4.51e-01 |
| GO:BP | GO:0014721 | twitch skeletal muscle contraction | 2 | 5 | 4.51e-01 |
| GO:BP | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0097466 | ubiquitin-dependent glycoprotein ERAD pathway | 3 | 5 | 4.51e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 3 | 16 | 4.51e-01 |
| GO:BP | GO:0021993 | initiation of neural tube closure | 1 | 1 | 4.51e-01 |
| GO:BP | GO:0097500 | receptor localization to non-motile cilium | 3 | 4 | 4.51e-01 |
| GO:BP | GO:1903173 | fatty alcohol metabolic process | 2 | 2 | 4.51e-01 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 4 | 8 | 4.52e-01 |
| GO:BP | GO:0010936 | negative regulation of macrophage cytokine production | 1 | 7 | 4.52e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 7 | 420 | 4.52e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 19 | 94 | 4.53e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 2 | 25 | 4.53e-01 |
| GO:BP | GO:0001843 | neural tube closure | 18 | 90 | 4.54e-01 |
| GO:BP | GO:0071481 | cellular response to X-ray | 4 | 10 | 4.54e-01 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 3 | 4 | 4.55e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 5 | 4539 | 4.55e-01 |
| GO:BP | GO:0021612 | facial nerve structural organization | 5 | 7 | 4.56e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 11 | 38 | 4.56e-01 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0033151 | V(D)J recombination | 6 | 16 | 4.56e-01 |
| GO:BP | GO:0040007 | growth | 1 | 742 | 4.56e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 2 | 5 | 4.56e-01 |
| GO:BP | GO:0046048 | UDP metabolic process | 1 | 7 | 4.56e-01 |
| GO:BP | GO:0038109 | Kit signaling pathway | 3 | 4 | 4.56e-01 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 1 | 8 | 4.56e-01 |
| GO:BP | GO:0001815 | positive regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 4.56e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 8 | 65 | 4.56e-01 |
| GO:BP | GO:0036216 | cellular response to stem cell factor stimulus | 3 | 4 | 4.56e-01 |
| GO:BP | GO:0036215 | response to stem cell factor | 3 | 4 | 4.56e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 4.56e-01 |
| GO:BP | GO:0021603 | cranial nerve formation | 1 | 4 | 4.56e-01 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 1 | 8 | 4.56e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 1 | 138 | 4.57e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 4 | 10 | 4.57e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 1 | 8 | 4.57e-01 |
| GO:BP | GO:0070076 | histone lysine demethylation | 8 | 20 | 4.57e-01 |
| GO:BP | GO:0042594 | response to starvation | 25 | 178 | 4.57e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 2 | 10 | 4.57e-01 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 5 | 7 | 4.57e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 1 | 4 | 4.58e-01 |
| GO:BP | GO:0006895 | Golgi to endosome transport | 7 | 17 | 4.58e-01 |
| GO:BP | GO:0000729 | DNA double-strand break processing | 1 | 18 | 4.58e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 4.58e-01 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 4.58e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 2 | 4 | 4.58e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 12 | 29 | 4.58e-01 |
| GO:BP | GO:1901906 | diadenosine pentaphosphate metabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:1901907 | diadenosine pentaphosphate catabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:1901908 | diadenosine hexaphosphate metabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:1901909 | diadenosine hexaphosphate catabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 4.58e-01 |
| GO:BP | GO:1901910 | adenosine 5’-(hexahydrogen pentaphosphate) metabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:1901911 | adenosine 5’-(hexahydrogen pentaphosphate) catabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:0048549 | positive regulation of pinocytosis | 4 | 7 | 4.58e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 4.58e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 8 | 17 | 4.58e-01 |
| GO:BP | GO:0006553 | lysine metabolic process | 2 | 7 | 4.58e-01 |
| GO:BP | GO:0001832 | blastocyst growth | 2 | 19 | 4.58e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 1 | 40 | 4.58e-01 |
| GO:BP | GO:1904835 | dorsal root ganglion morphogenesis | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0010831 | positive regulation of myotube differentiation | 4 | 10 | 4.58e-01 |
| GO:BP | GO:0060049 | regulation of protein glycosylation | 1 | 10 | 4.58e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 9 | 740 | 4.58e-01 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 2 | 9 | 4.58e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 3 | 23 | 4.58e-01 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 8 | 399 | 4.58e-01 |
| GO:BP | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis | 1 | 3 | 4.58e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 3 | 4 | 4.58e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 1 | 353 | 4.58e-01 |
| GO:BP | GO:1902488 | cholangiocyte apoptotic process | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 2 | 20 | 4.58e-01 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0009756 | carbohydrate mediated signaling | 1 | 6 | 4.58e-01 |
| GO:BP | GO:0048297 | negative regulation of isotype switching to IgA isotypes | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0021649 | vestibulocochlear nerve structural organization | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 3 | 28 | 4.58e-01 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 83 | 175 | 4.58e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 17 | 67 | 4.58e-01 |
| GO:BP | GO:0039533 | regulation of MDA-5 signaling pathway | 1 | 5 | 4.58e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 2 | 123 | 4.58e-01 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 3 | 20 | 4.58e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 18 | 43 | 4.58e-01 |
| GO:BP | GO:1904193 | negative regulation of cholangiocyte apoptotic process | 1 | 1 | 4.58e-01 |
| GO:BP | GO:1903375 | facioacoustic ganglion development | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0061682 | seminal vesicle morphogenesis | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0007072 | positive regulation of transcription involved in exit from mitosis | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 26 | 179 | 4.58e-01 |
| GO:BP | GO:0015910 | long-chain fatty acid import into peroxisome | 2 | 3 | 4.58e-01 |
| GO:BP | GO:1904192 | regulation of cholangiocyte apoptotic process | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0071543 | diphosphoinositol polyphosphate metabolic process | 2 | 5 | 4.58e-01 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 2 | 33 | 4.58e-01 |
| GO:BP | GO:0048668 | collateral sprouting | 9 | 25 | 4.58e-01 |
| GO:BP | GO:0061552 | ganglion morphogenesis | 2 | 2 | 4.58e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 2 | 41 | 4.58e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 4 | 30 | 4.58e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 8 | 147 | 4.58e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 2 | 100 | 4.58e-01 |
| GO:BP | GO:0060397 | growth hormone receptor signaling pathway via JAK-STAT | 3 | 9 | 4.58e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 2 | 39 | 4.58e-01 |
| GO:BP | GO:0060348 | bone development | 6 | 170 | 4.58e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 32 | 4.58e-01 |
| GO:BP | GO:0039654 | fusion of virus membrane with host endosome membrane | 1 | 1 | 4.58e-01 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 2 | 33 | 4.58e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 7 | 41 | 4.58e-01 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 6 | 31 | 4.58e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 4.58e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 1 | 13 | 4.58e-01 |
| GO:BP | GO:0032682 | negative regulation of chemokine production | 5 | 12 | 4.58e-01 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 2 | 26 | 4.58e-01 |
| GO:BP | GO:0010979 | regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 4.59e-01 |
| GO:BP | GO:0010980 | positive regulation of vitamin D 24-hydroxylase activity | 1 | 1 | 4.59e-01 |
| GO:BP | GO:0003358 | noradrenergic neuron development | 1 | 2 | 4.59e-01 |
| GO:BP | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process | 2 | 8 | 4.59e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 7 | 112 | 4.59e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 5 | 120 | 4.60e-01 |
| GO:BP | GO:0072337 | modified amino acid transport | 5 | 35 | 4.60e-01 |
| GO:BP | GO:0034514 | mitochondrial unfolded protein response | 2 | 2 | 4.60e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 36 | 77 | 4.60e-01 |
| GO:BP | GO:0072356 | chromosome passenger complex localization to kinetochore | 1 | 1 | 4.60e-01 |
| GO:BP | GO:0003275 | apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 4.60e-01 |
| GO:BP | GO:1902256 | regulation of apoptotic process involved in outflow tract morphogenesis | 1 | 3 | 4.60e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 4.60e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 4.60e-01 |
| GO:BP | GO:2000620 | positive regulation of histone H4-K16 acetylation | 2 | 2 | 4.61e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 11 | 42 | 4.62e-01 |
| GO:BP | GO:1901166 | neural crest cell migration involved in autonomic nervous system development | 4 | 5 | 4.62e-01 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 12 | 23 | 4.62e-01 |
| GO:BP | GO:0060061 | Spemann organizer formation | 2 | 3 | 4.62e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 3 | 37 | 4.62e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 5 | 403 | 4.62e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 1 | 348 | 4.62e-01 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 5 | 8 | 4.62e-01 |
| GO:BP | GO:0005981 | regulation of glycogen catabolic process | 2 | 4 | 4.62e-01 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 5 | 8 | 4.62e-01 |
| GO:BP | GO:0019236 | response to pheromone | 1 | 1 | 4.62e-01 |
| GO:BP | GO:2001228 | regulation of response to gamma radiation | 1 | 1 | 4.62e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 4.62e-01 |
| GO:BP | GO:0042117 | monocyte activation | 1 | 8 | 4.62e-01 |
| GO:BP | GO:0006188 | IMP biosynthetic process | 6 | 10 | 4.62e-01 |
| GO:BP | GO:0009188 | ribonucleoside diphosphate biosynthetic process | 5 | 8 | 4.62e-01 |
| GO:BP | GO:1905843 | regulation of cellular response to gamma radiation | 1 | 1 | 4.62e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 5 | 138 | 4.62e-01 |
| GO:BP | GO:1902888 | protein localization to astral microtubule | 1 | 1 | 4.62e-01 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 2 | 8 | 4.62e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 5 | 4590 | 4.62e-01 |
| GO:BP | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing | 5 | 7 | 4.62e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 1 | 31 | 4.62e-01 |
| GO:BP | GO:0045004 | DNA replication proofreading | 2 | 2 | 4.62e-01 |
| GO:BP | GO:0016255 | attachment of GPI anchor to protein | 3 | 6 | 4.62e-01 |
| GO:BP | GO:1905939 | regulation of gonad development | 4 | 8 | 4.62e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 1 | 30 | 4.62e-01 |
| GO:BP | GO:0051939 | gamma-aminobutyric acid import | 1 | 3 | 4.63e-01 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 11 | 22 | 4.63e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 15 | 37 | 4.63e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 3 | 93 | 4.63e-01 |
| GO:BP | GO:0051352 | negative regulation of ligase activity | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 10 | 33 | 4.63e-01 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 4 | 28 | 4.63e-01 |
| GO:BP | GO:0003331 | positive regulation of extracellular matrix constituent secretion | 1 | 4 | 4.63e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 10 | 113 | 4.63e-01 |
| GO:BP | GO:0060429 | epithelium development | 8 | 850 | 4.63e-01 |
| GO:BP | GO:0097067 | cellular response to thyroid hormone stimulus | 5 | 14 | 4.63e-01 |
| GO:BP | GO:0048864 | stem cell development | 1 | 76 | 4.63e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 11 | 72 | 4.63e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 74 | 161 | 4.63e-01 |
| GO:BP | GO:0009629 | response to gravity | 2 | 5 | 4.63e-01 |
| GO:BP | GO:1905342 | positive regulation of protein localization to kinetochore | 3 | 4 | 4.63e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 5 | 442 | 4.63e-01 |
| GO:BP | GO:0010212 | response to ionizing radiation | 58 | 130 | 4.63e-01 |
| GO:BP | GO:1905340 | regulation of protein localization to kinetochore | 3 | 4 | 4.63e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 1 | 43 | 4.63e-01 |
| GO:BP | GO:0045829 | negative regulation of isotype switching | 2 | 4 | 4.63e-01 |
| GO:BP | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0022013 | pallium cell proliferation in forebrain | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0061326 | renal tubule development | 11 | 77 | 4.63e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 3 | 136 | 4.63e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 2 | 132 | 4.63e-01 |
| GO:BP | GO:1904184 | positive regulation of pyruvate dehydrogenase activity | 1 | 3 | 4.63e-01 |
| GO:BP | GO:0006432 | phenylalanyl-tRNA aminoacylation | 2 | 4 | 4.63e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 1 | 8 | 4.63e-01 |
| GO:BP | GO:0048854 | brain morphogenesis | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 4 | 6 | 4.63e-01 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 1 | 6 | 4.63e-01 |
| GO:BP | GO:0097378 | dorsal spinal cord interneuron axon guidance | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0003129 | heart induction | 3 | 9 | 4.63e-01 |
| GO:BP | GO:0060723 | regulation of cell proliferation involved in embryonic placenta development | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0060721 | regulation of spongiotrophoblast cell proliferation | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0032775 | DNA methylation on adenine | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 4 | 143 | 4.63e-01 |
| GO:BP | GO:0060606 | tube closure | 18 | 91 | 4.63e-01 |
| GO:BP | GO:0018872 | arsonoacetate metabolic process | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0099183 | trans-synaptic signaling by BDNF, modulating synaptic transmission | 1 | 2 | 4.63e-01 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 3 | 4 | 4.63e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 4 | 90 | 4.63e-01 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 18 | 32 | 4.63e-01 |
| GO:BP | GO:1904373 | response to kainic acid | 1 | 1 | 4.63e-01 |
| GO:BP | GO:1904372 | positive regulation of protein localization to actin cortical patch | 1 | 1 | 4.63e-01 |
| GO:BP | GO:1904370 | regulation of protein localization to actin cortical patch | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0044379 | protein localization to actin cortical patch | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 6 | 36 | 4.63e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 1 | 534 | 4.63e-01 |
| GO:BP | GO:1903119 | protein localization to actin cytoskeleton | 1 | 1 | 4.63e-01 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 7 | 12 | 4.63e-01 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 7 | 16 | 4.63e-01 |
| GO:BP | GO:0035507 | regulation of myosin-light-chain-phosphatase activity | 1 | 5 | 4.63e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 24 | 109 | 4.64e-01 |
| GO:BP | GO:0045046 | protein import into peroxisome membrane | 3 | 6 | 4.64e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 1 | 11 | 4.65e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 1 | 7 | 4.65e-01 |
| GO:BP | GO:0033169 | histone H3-K9 demethylation | 1 | 5 | 4.65e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 8 | 13 | 4.65e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 10 | 22 | 4.65e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 3 | 13 | 4.65e-01 |
| GO:BP | GO:0090520 | sphingolipid mediated signaling pathway | 2 | 11 | 4.65e-01 |
| GO:BP | GO:0006549 | isoleucine metabolic process | 1 | 6 | 4.65e-01 |
| GO:BP | GO:0006573 | valine metabolic process | 1 | 7 | 4.65e-01 |
| GO:BP | GO:0070078 | histone H3-R2 demethylation | 1 | 1 | 4.65e-01 |
| GO:BP | GO:0070079 | histone H4-R3 demethylation | 1 | 1 | 4.65e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 2 | 605 | 4.65e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 10 | 665 | 4.65e-01 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 3 | 5 | 4.65e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 2 | 3 | 4.65e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 2 | 3 | 4.65e-01 |
| GO:BP | GO:0018395 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine | 1 | 1 | 4.65e-01 |
| GO:BP | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication | 1 | 1 | 4.66e-01 |
| GO:BP | GO:0050890 | cognition | 3 | 227 | 4.66e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 2 | 252 | 4.66e-01 |
| GO:BP | GO:0042761 | very long-chain fatty acid biosynthetic process | 5 | 12 | 4.66e-01 |
| GO:BP | GO:0010724 | regulation of definitive erythrocyte differentiation | 2 | 2 | 4.66e-01 |
| GO:BP | GO:0015840 | urea transport | 1 | 2 | 4.66e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 2 | 39 | 4.67e-01 |
| GO:BP | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway | 3 | 6 | 4.67e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 4.67e-01 |
| GO:BP | GO:0060687 | regulation of branching involved in prostate gland morphogenesis | 1 | 3 | 4.67e-01 |
| GO:BP | GO:0090245 | axis elongation involved in somitogenesis | 1 | 4 | 4.67e-01 |
| GO:BP | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation | 1 | 5 | 4.67e-01 |
| GO:BP | GO:0003242 | cardiac chamber ballooning | 1 | 1 | 4.67e-01 |
| GO:BP | GO:1902747 | negative regulation of lens fiber cell differentiation | 3 | 5 | 4.67e-01 |
| GO:BP | GO:0060927 | cardiac pacemaker cell fate commitment | 2 | 3 | 4.67e-01 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 4.67e-01 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 3 | 4.67e-01 |
| GO:BP | GO:0052651 | monoacylglycerol catabolic process | 3 | 6 | 4.67e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 4.67e-01 |
| GO:BP | GO:0002252 | immune effector process | 1 | 318 | 4.67e-01 |
| GO:BP | GO:0003270 | Notch signaling pathway involved in regulation of secondary heart field cardioblast proliferation | 1 | 1 | 4.67e-01 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 1 | 6 | 4.67e-01 |
| GO:BP | GO:0003182 | coronary sinus valve morphogenesis | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0060843 | venous endothelial cell differentiation | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0003178 | coronary sinus valve development | 1 | 1 | 4.67e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 8 | 18 | 4.68e-01 |
| GO:BP | GO:0046548 | retinal rod cell development | 2 | 6 | 4.68e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 5 | 21 | 4.68e-01 |
| GO:BP | GO:0071072 | negative regulation of phospholipid biosynthetic process | 2 | 6 | 4.68e-01 |
| GO:BP | GO:1990379 | lipid transport across blood-brain barrier | 2 | 6 | 4.68e-01 |
| GO:BP | GO:0043931 | ossification involved in bone maturation | 1 | 17 | 4.68e-01 |
| GO:BP | GO:2000657 | negative regulation of apolipoprotein binding | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 3 | 56 | 4.68e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 2 | 104 | 4.68e-01 |
| GO:BP | GO:2000656 | regulation of apolipoprotein binding | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 1 | 551 | 4.68e-01 |
| GO:BP | GO:0001967 | suckling behavior | 6 | 10 | 4.68e-01 |
| GO:BP | GO:2001303 | lipoxin A4 biosynthetic process | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0010332 | response to gamma radiation | 24 | 50 | 4.68e-01 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 2 | 15 | 4.68e-01 |
| GO:BP | GO:0001656 | metanephros development | 1 | 64 | 4.68e-01 |
| GO:BP | GO:0032570 | response to progesterone | 1 | 27 | 4.68e-01 |
| GO:BP | GO:1901211 | negative regulation of cardiac chamber formation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:1901210 | regulation of cardiac chamber formation | 1 | 1 | 4.68e-01 |
| GO:BP | GO:1901208 | negative regulation of heart looping | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0003065 | positive regulation of heart rate by epinephrine | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 4.68e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 4.68e-01 |
| GO:BP | GO:1905267 | endonucleolytic cleavage involved in tRNA processing | 2 | 3 | 4.68e-01 |
| GO:BP | GO:0036491 | regulation of translation initiation in response to endoplasmic reticulum stress | 1 | 8 | 4.68e-01 |
| GO:BP | GO:0071838 | cell proliferation in bone marrow | 2 | 7 | 4.68e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 1 | 9 | 4.68e-01 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 1 | 9 | 4.68e-01 |
| GO:BP | GO:2000369 | regulation of clathrin-dependent endocytosis | 1 | 18 | 4.68e-01 |
| GO:BP | GO:0048697 | positive regulation of collateral sprouting in absence of injury | 1 | 1 | 4.68e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 4 | 8 | 4.69e-01 |
| GO:BP | GO:0048560 | establishment of anatomical structure orientation | 1 | 2 | 4.70e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 4.70e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 4 | 6 | 4.70e-01 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 3 | 5 | 4.70e-01 |
| GO:BP | GO:0048290 | isotype switching to IgA isotypes | 4 | 4 | 4.70e-01 |
| GO:BP | GO:0090022 | regulation of neutrophil chemotaxis | 2 | 19 | 4.70e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 4 | 65 | 4.70e-01 |
| GO:BP | GO:0015918 | sterol transport | 2 | 92 | 4.70e-01 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 1 | 17 | 4.70e-01 |
| GO:BP | GO:0061025 | membrane fusion | 3 | 137 | 4.70e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 41 | 123 | 4.70e-01 |
| GO:BP | GO:0021637 | trigeminal nerve structural organization | 3 | 4 | 4.70e-01 |
| GO:BP | GO:0021636 | trigeminal nerve morphogenesis | 3 | 4 | 4.70e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 1 | 25 | 4.71e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 4.71e-01 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 4 | 7 | 4.71e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 9 | 187 | 4.71e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 4.71e-01 |
| GO:BP | GO:0032868 | response to insulin | 43 | 215 | 4.72e-01 |
| GO:BP | GO:0071107 | response to parathyroid hormone | 1 | 9 | 4.72e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 65 | 4.72e-01 |
| GO:BP | GO:0015838 | amino-acid betaine transport | 1 | 7 | 4.72e-01 |
| GO:BP | GO:0001796 | regulation of type IIa hypersensitivity | 2 | 3 | 4.72e-01 |
| GO:BP | GO:0046851 | negative regulation of bone remodeling | 2 | 9 | 4.72e-01 |
| GO:BP | GO:0002892 | regulation of type II hypersensitivity | 2 | 3 | 4.72e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 1 | 53 | 4.72e-01 |
| GO:BP | GO:0110008 | ncRNA deadenylation | 1 | 1 | 4.72e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 4.72e-01 |
| GO:BP | GO:0021702 | cerebellar Purkinje cell differentiation | 6 | 10 | 4.72e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 9 | 50 | 4.72e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 4 | 6 | 4.72e-01 |
| GO:BP | GO:0072423 | response to DNA damage checkpoint signaling | 2 | 8 | 4.72e-01 |
| GO:BP | GO:0048311 | mitochondrion distribution | 2 | 14 | 4.72e-01 |
| GO:BP | GO:1901634 | positive regulation of synaptic vesicle membrane organization | 1 | 4 | 4.72e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 2 | 106 | 4.72e-01 |
| GO:BP | GO:0072402 | response to DNA integrity checkpoint signaling | 2 | 8 | 4.72e-01 |
| GO:BP | GO:0072396 | response to cell cycle checkpoint signaling | 2 | 8 | 4.72e-01 |
| GO:BP | GO:0070143 | mitochondrial alanyl-tRNA aminoacylation | 1 | 1 | 4.72e-01 |
| GO:BP | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 4 | 4.72e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 7 | 158 | 4.72e-01 |
| GO:BP | GO:0099502 | calcium-dependent activation of synaptic vesicle fusion | 1 | 4 | 4.72e-01 |
| GO:BP | GO:0051013 | microtubule severing | 1 | 7 | 4.72e-01 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 2 | 89 | 4.72e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 5 | 38 | 4.72e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 1 | 384 | 4.72e-01 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 1 | 4 | 4.73e-01 |
| GO:BP | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0061470 | T follicular helper cell differentiation | 2 | 4 | 4.73e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 31 | 67 | 4.73e-01 |
| GO:BP | GO:0060309 | elastin catabolic process | 2 | 2 | 4.73e-01 |
| GO:BP | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle | 1 | 2 | 4.73e-01 |
| GO:BP | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 2 | 11 | 4.73e-01 |
| GO:BP | GO:0010710 | regulation of collagen catabolic process | 2 | 3 | 4.73e-01 |
| GO:BP | GO:1902951 | negative regulation of dendritic spine maintenance | 1 | 2 | 4.73e-01 |
| GO:BP | GO:1905090 | negative regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 4.73e-01 |
| GO:BP | GO:1902938 | regulation of intracellular calcium activated chloride channel activity | 1 | 1 | 4.73e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 4.73e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 5 | 9 | 4.73e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 9 | 68 | 4.73e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 1 | 22 | 4.73e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 24 | 52 | 4.73e-01 |
| GO:BP | GO:0051866 | general adaptation syndrome | 2 | 4 | 4.73e-01 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 3 | 4 | 4.74e-01 |
| GO:BP | GO:0045041 | protein import into mitochondrial intermembrane space | 2 | 3 | 4.74e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 19 | 150 | 4.74e-01 |
| GO:BP | GO:1904257 | zinc ion import into Golgi lumen | 1 | 3 | 4.74e-01 |
| GO:BP | GO:0072488 | ammonium transmembrane transport | 2 | 6 | 4.74e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 5 | 28 | 4.74e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 2 | 3 | 4.74e-01 |
| GO:BP | GO:2000194 | regulation of female gonad development | 1 | 4 | 4.74e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 5 | 422 | 4.74e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 4.74e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 4.74e-01 |
| GO:BP | GO:0031129 | inductive cell-cell signaling | 1 | 1 | 4.74e-01 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 16 | 25 | 4.74e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 1 | 448 | 4.74e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 4.74e-01 |
| GO:BP | GO:0060312 | regulation of blood vessel remodeling | 1 | 4 | 4.74e-01 |
| GO:BP | GO:0008306 | associative learning | 2 | 65 | 4.75e-01 |
| GO:BP | GO:0070266 | necroptotic process | 17 | 32 | 4.75e-01 |
| GO:BP | GO:0061171 | establishment of bipolar cell polarity | 1 | 1 | 4.75e-01 |
| GO:BP | GO:1902247 | geranylgeranyl diphosphate catabolic process | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0045339 | farnesyl diphosphate catabolic process | 1 | 1 | 4.75e-01 |
| GO:BP | GO:2000551 | regulation of T-helper 2 cell cytokine production | 1 | 3 | 4.75e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 1 | 369 | 4.75e-01 |
| GO:BP | GO:0035378 | carbon dioxide transmembrane transport | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity | 1 | 3 | 4.75e-01 |
| GO:BP | GO:0070075 | tear secretion | 1 | 2 | 4.75e-01 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 3 | 3 | 4.75e-01 |
| GO:BP | GO:0046878 | positive regulation of saliva secretion | 1 | 2 | 4.75e-01 |
| GO:BP | GO:0007402 | ganglion mother cell fate determination | 1 | 1 | 4.75e-01 |
| GO:BP | GO:0060374 | mast cell differentiation | 3 | 4 | 4.75e-01 |
| GO:BP | GO:0046070 | dGTP metabolic process | 2 | 3 | 4.75e-01 |
| GO:BP | GO:0072237 | metanephric proximal tubule development | 1 | 3 | 4.75e-01 |
| GO:BP | GO:0035745 | T-helper 2 cell cytokine production | 1 | 3 | 4.75e-01 |
| GO:BP | GO:0071288 | cellular response to mercury ion | 1 | 2 | 4.75e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 3 | 71 | 4.75e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 7 | 22 | 4.75e-01 |
| GO:BP | GO:0051305 | chromosome movement towards spindle pole | 1 | 7 | 4.75e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 5 | 45 | 4.75e-01 |
| GO:BP | GO:0050927 | positive regulation of positive chemotaxis | 2 | 20 | 4.75e-01 |
| GO:BP | GO:0006413 | translational initiation | 2 | 116 | 4.75e-01 |
| GO:BP | GO:0019043 | establishment of viral latency | 1 | 3 | 4.75e-01 |
| GO:BP | GO:0007270 | neuron-neuron synaptic transmission | 2 | 5 | 4.75e-01 |
| GO:BP | GO:0097484 | dendrite extension | 9 | 27 | 4.76e-01 |
| GO:BP | GO:0060515 | prostate field specification | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0060520 | activation of prostate induction by androgen receptor signaling pathway | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 1 | 23 | 4.76e-01 |
| GO:BP | GO:0060514 | prostate induction | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0070649 | formin-nucleated actin cable assembly | 2 | 3 | 4.76e-01 |
| GO:BP | GO:0110009 | formin-nucleated actin cable organization | 2 | 3 | 4.76e-01 |
| GO:BP | GO:0042921 | glucocorticoid receptor signaling pathway | 2 | 13 | 4.76e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 4.76e-01 |
| GO:BP | GO:0019102 | male somatic sex determination | 1 | 1 | 4.76e-01 |
| GO:BP | GO:0010836 | negative regulation of protein ADP-ribosylation | 2 | 4 | 4.76e-01 |
| GO:BP | GO:0032287 | peripheral nervous system myelin maintenance | 3 | 8 | 4.77e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 2 | 67 | 4.77e-01 |
| GO:BP | GO:1905285 | fibrous ring of heart morphogenesis | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0048338 | mesoderm structural organization | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0099172 | presynapse organization | 2 | 42 | 4.77e-01 |
| GO:BP | GO:0006865 | amino acid transport | 2 | 102 | 4.77e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 3 | 155 | 4.77e-01 |
| GO:BP | GO:0046753 | non-lytic viral release | 2 | 14 | 4.77e-01 |
| GO:BP | GO:0140157 | ammonium import across plasma membrane | 1 | 3 | 4.77e-01 |
| GO:BP | GO:0048352 | paraxial mesoderm structural organization | 1 | 1 | 4.77e-01 |
| GO:BP | GO:1904412 | regulation of cardiac ventricle development | 1 | 1 | 4.77e-01 |
| GO:BP | GO:1904414 | positive regulation of cardiac ventricle development | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0021998 | neural plate mediolateral regionalization | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0048372 | lateral mesodermal cell fate commitment | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 2 | 108 | 4.77e-01 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 5 | 18 | 4.77e-01 |
| GO:BP | GO:0048377 | lateral mesodermal cell fate specification | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 1 | 14 | 4.77e-01 |
| GO:BP | GO:0048378 | regulation of lateral mesodermal cell fate specification | 1 | 1 | 4.77e-01 |
| GO:BP | GO:0019482 | beta-alanine metabolic process | 1 | 2 | 4.77e-01 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 1 | 34 | 4.77e-01 |
| GO:BP | GO:0015816 | glycine transport | 2 | 9 | 4.77e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 4 | 12 | 4.77e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 28 | 60 | 4.77e-01 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 2 | 10 | 4.77e-01 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 6 | 36 | 4.77e-01 |
| GO:BP | GO:0071548 | response to dexamethasone | 1 | 36 | 4.78e-01 |
| GO:BP | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis | 1 | 1 | 4.78e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 5 | 39 | 4.78e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 5 | 54 | 4.78e-01 |
| GO:BP | GO:0019860 | uracil metabolic process | 1 | 2 | 4.78e-01 |
| GO:BP | GO:0060517 | epithelial cell proliferation involved in prostatic bud elongation | 1 | 1 | 4.78e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 1 | 16 | 4.78e-01 |
| GO:BP | GO:0035750 | protein localization to myelin sheath abaxonal region | 1 | 1 | 4.79e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 7 | 66 | 4.79e-01 |
| GO:BP | GO:0072144 | glomerular mesangial cell development | 2 | 4 | 4.79e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 38 | 127 | 4.79e-01 |
| GO:BP | GO:0007386 | compartment pattern specification | 2 | 5 | 4.79e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 1 | 50 | 4.79e-01 |
| GO:BP | GO:1903644 | regulation of chaperone-mediated protein folding | 2 | 3 | 4.79e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 4 | 265 | 4.79e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 3 | 246 | 4.79e-01 |
| GO:BP | GO:0000389 | mRNA 3’-splice site recognition | 1 | 6 | 4.79e-01 |
| GO:BP | GO:0050877 | nervous system process | 11 | 634 | 4.79e-01 |
| GO:BP | GO:0003058 | hormonal regulation of the force of heart contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0009932 | cell tip growth | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 3 | 25 | 4.79e-01 |
| GO:BP | GO:0061622 | glycolytic process through glucose-1-phosphate | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0060184 | cell cycle switching | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0120063 | stomach smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:1904343 | positive regulation of colon smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:1990262 | anti-Mullerian hormone signaling pathway | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0120072 | positive regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0120071 | regulation of pyloric antrum smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0120065 | pyloric antrum smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0120064 | stomach pylorus smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:1990765 | colon smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 7 | 10 | 4.79e-01 |
| GO:BP | GO:1904341 | regulation of colon smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 3 | 6 | 4.79e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 43 | 84 | 4.79e-01 |
| GO:BP | GO:0097326 | melanocyte adhesion | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0014828 | distal stomach smooth muscle contraction | 1 | 1 | 4.79e-01 |
| GO:BP | GO:0051728 | cell cycle switching, mitotic to meiotic cell cycle | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0051729 | germline cell cycle switching, mitotic to meiotic cell cycle | 1 | 2 | 4.79e-01 |
| GO:BP | GO:2000609 | regulation of thyroid hormone generation | 1 | 2 | 4.79e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 4.79e-01 |
| GO:BP | GO:0046084 | adenine biosynthetic process | 1 | 2 | 4.80e-01 |
| GO:BP | GO:0072301 | regulation of metanephric glomerular mesangial cell proliferation | 1 | 1 | 4.80e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 2 | 44 | 4.80e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 4 | 92 | 4.80e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 2 | 38 | 4.80e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 4 | 92 | 4.80e-01 |
| GO:BP | GO:0061502 | early endosome to recycling endosome transport | 1 | 3 | 4.80e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 37 | 114 | 4.80e-01 |
| GO:BP | GO:0021889 | olfactory bulb interneuron differentiation | 4 | 7 | 4.81e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 1 | 9 | 4.81e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 21 | 667 | 4.81e-01 |
| GO:BP | GO:0110112 | regulation of lipid transporter activity | 1 | 2 | 4.81e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 2 | 121 | 4.81e-01 |
| GO:BP | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 1 | 4 | 4.81e-01 |
| GO:BP | GO:1904024 | negative regulation of glucose catabolic process to lactate via pyruvate | 1 | 3 | 4.81e-01 |
| GO:BP | GO:0007097 | nuclear migration | 2 | 23 | 4.81e-01 |
| GO:BP | GO:0090179 | planar cell polarity pathway involved in neural tube closure | 2 | 12 | 4.82e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 8 | 12 | 4.82e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 19 | 40 | 4.82e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 12 | 62 | 4.82e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 2 | 31 | 4.82e-01 |
| GO:BP | GO:1903093 | regulation of protein K48-linked deubiquitination | 1 | 3 | 4.82e-01 |
| GO:BP | GO:0032965 | regulation of collagen biosynthetic process | 8 | 30 | 4.82e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 39 | 118 | 4.82e-01 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 4 | 12 | 4.82e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 1 | 11 | 4.82e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 4.82e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 2 | 68 | 4.82e-01 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 4 | 30 | 4.82e-01 |
| GO:BP | GO:0014860 | neurotransmitter secretion involved in regulation of skeletal muscle contraction | 1 | 1 | 4.82e-01 |
| GO:BP | GO:1901652 | response to peptide | 18 | 377 | 4.82e-01 |
| GO:BP | GO:0002069 | columnar/cuboidal epithelial cell maturation | 5 | 9 | 4.82e-01 |
| GO:BP | GO:0000390 | spliceosomal complex disassembly | 2 | 3 | 4.83e-01 |
| GO:BP | GO:0071348 | cellular response to interleukin-11 | 1 | 1 | 4.83e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 2 | 24 | 4.83e-01 |
| GO:BP | GO:2000312 | regulation of kainate selective glutamate receptor activity | 1 | 3 | 4.84e-01 |
| GO:BP | GO:0006206 | pyrimidine nucleobase metabolic process | 3 | 13 | 4.84e-01 |
| GO:BP | GO:0045986 | negative regulation of smooth muscle contraction | 1 | 12 | 4.84e-01 |
| GO:BP | GO:0032289 | central nervous system myelin formation | 1 | 4 | 4.84e-01 |
| GO:BP | GO:0017145 | stem cell division | 11 | 26 | 4.84e-01 |
| GO:BP | GO:0032916 | positive regulation of transforming growth factor beta3 production | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 4 | 15 | 4.84e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 12 | 58 | 4.84e-01 |
| GO:BP | GO:0021694 | cerebellar Purkinje cell layer formation | 7 | 12 | 4.84e-01 |
| GO:BP | GO:0070977 | bone maturation | 1 | 19 | 4.84e-01 |
| GO:BP | GO:0061767 | negative regulation of lung blood pressure | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0072734 | cellular response to staurosporine | 1 | 3 | 4.84e-01 |
| GO:BP | GO:1905704 | positive regulation of inhibitory synapse assembly | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0006883 | intracellular sodium ion homeostasis | 1 | 13 | 4.84e-01 |
| GO:BP | GO:0055113 | epiboly involved in gastrulation with mouth forming second | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0000019 | regulation of mitotic recombination | 1 | 5 | 4.84e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 4.84e-01 |
| GO:BP | GO:0072733 | response to staurosporine | 1 | 3 | 4.84e-01 |
| GO:BP | GO:0033504 | floor plate development | 1 | 8 | 4.84e-01 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 7 | 11 | 4.84e-01 |
| GO:BP | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 1 | 6 | 4.84e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 174 | 391 | 4.84e-01 |
| GO:BP | GO:0120158 | positive regulation of collagen catabolic process | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 2 | 70 | 4.84e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 1 | 23 | 4.84e-01 |
| GO:BP | GO:0035654 | clathrin-coated vesicle cargo loading, AP-3-mediated | 1 | 6 | 4.84e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 23 | 494 | 4.84e-01 |
| GO:BP | GO:0035652 | clathrin-coated vesicle cargo loading | 1 | 6 | 4.84e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 2 | 96 | 4.84e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 16 | 32 | 4.84e-01 |
| GO:BP | GO:1902624 | positive regulation of neutrophil migration | 2 | 21 | 4.84e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 1 | 39 | 4.84e-01 |
| GO:BP | GO:0030321 | transepithelial chloride transport | 1 | 7 | 4.84e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 2 | 30 | 4.84e-01 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 4 | 9 | 4.84e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 1 | 75 | 4.84e-01 |
| GO:BP | GO:0019408 | dolichol biosynthetic process | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 12 | 19 | 4.84e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 8 | 38 | 4.84e-01 |
| GO:BP | GO:0050926 | regulation of positive chemotaxis | 2 | 20 | 4.84e-01 |
| GO:BP | GO:1902811 | positive regulation of skeletal muscle fiber differentiation | 1 | 2 | 4.84e-01 |
| GO:BP | GO:0060180 | female mating behavior | 2 | 2 | 4.84e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 2 | 29 | 4.84e-01 |
| GO:BP | GO:2000064 | regulation of cortisol biosynthetic process | 2 | 7 | 4.84e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 2 | 6 | 4.84e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 1 | 34 | 4.84e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 4 | 7 | 4.84e-01 |
| GO:BP | GO:0045924 | regulation of female receptivity | 2 | 2 | 4.84e-01 |
| GO:BP | GO:0002154 | thyroid hormone mediated signaling pathway | 2 | 6 | 4.84e-01 |
| GO:BP | GO:0035962 | response to interleukin-13 | 1 | 1 | 4.84e-01 |
| GO:BP | GO:0045654 | positive regulation of megakaryocyte differentiation | 2 | 9 | 4.84e-01 |
| GO:BP | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport | 3 | 4 | 4.85e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 11 | 51 | 4.85e-01 |
| GO:BP | GO:0097043 | histone H3-K56 acetylation | 1 | 2 | 4.86e-01 |
| GO:BP | GO:0106035 | protein maturation by [4Fe-4S] cluster transfer | 1 | 7 | 4.86e-01 |
| GO:BP | GO:0097009 | energy homeostasis | 20 | 39 | 4.86e-01 |
| GO:BP | GO:0043461 | proton-transporting ATP synthase complex assembly | 1 | 8 | 4.86e-01 |
| GO:BP | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly | 1 | 8 | 4.86e-01 |
| GO:BP | GO:1904353 | regulation of telomere capping | 1 | 23 | 4.86e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 21 | 48 | 4.86e-01 |
| GO:BP | GO:0018206 | peptidyl-methionine modification | 4 | 13 | 4.87e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 1 | 14 | 4.87e-01 |
| GO:BP | GO:0060252 | positive regulation of glial cell proliferation | 5 | 14 | 4.87e-01 |
| GO:BP | GO:0043583 | ear development | 6 | 153 | 4.87e-01 |
| GO:BP | GO:0042996 | regulation of Golgi to plasma membrane protein transport | 5 | 9 | 4.87e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 3 | 225 | 4.87e-01 |
| GO:BP | GO:1900127 | positive regulation of hyaluronan biosynthetic process | 1 | 4 | 4.87e-01 |
| GO:BP | GO:0035789 | metanephric mesenchymal cell migration | 1 | 3 | 4.87e-01 |
| GO:BP | GO:2000057 | negative regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 4.87e-01 |
| GO:BP | GO:2000056 | regulation of Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 4.87e-01 |
| GO:BP | GO:0042930 | enterobactin transport | 1 | 1 | 4.87e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 15 | 4.87e-01 |
| GO:BP | GO:0120323 | lipid ubiquitination | 1 | 1 | 4.87e-01 |
| GO:BP | GO:1903772 | regulation of viral budding via host ESCRT complex | 1 | 4 | 4.87e-01 |
| GO:BP | GO:2000589 | regulation of metanephric mesenchymal cell migration | 1 | 3 | 4.87e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 6 | 39 | 4.87e-01 |
| GO:BP | GO:0072319 | vesicle uncoating | 2 | 9 | 4.87e-01 |
| GO:BP | GO:0044878 | mitotic cytokinesis checkpoint signaling | 1 | 4 | 4.87e-01 |
| GO:BP | GO:0060068 | vagina development | 2 | 8 | 4.87e-01 |
| GO:BP | GO:0044805 | late nucleophagy | 1 | 7 | 4.87e-01 |
| GO:BP | GO:0071895 | odontoblast differentiation | 1 | 5 | 4.87e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 5 | 61 | 4.87e-01 |
| GO:BP | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0042045 | epithelial fluid transport | 3 | 7 | 4.87e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 4.87e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 8 | 71 | 4.87e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 9 | 53 | 4.87e-01 |
| GO:BP | GO:0120322 | lipid modification by small protein conjugation | 1 | 1 | 4.87e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 28 | 197 | 4.87e-01 |
| GO:BP | GO:0048368 | lateral mesoderm development | 7 | 10 | 4.87e-01 |
| GO:BP | GO:1905464 | positive regulation of DNA duplex unwinding | 1 | 3 | 4.87e-01 |
| GO:BP | GO:1905776 | positive regulation of DNA helicase activity | 1 | 3 | 4.87e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 15 | 37 | 4.87e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 2 | 156 | 4.88e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 8 | 13 | 4.88e-01 |
| GO:BP | GO:0014738 | regulation of muscle hyperplasia | 1 | 1 | 4.88e-01 |
| GO:BP | GO:0006548 | histidine catabolic process | 1 | 3 | 4.88e-01 |
| GO:BP | GO:1901724 | positive regulation of cell proliferation involved in kidney development | 2 | 8 | 4.88e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 7 | 67 | 4.89e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 7 | 459 | 4.89e-01 |
| GO:BP | GO:0071377 | cellular response to glucagon stimulus | 1 | 4 | 4.89e-01 |
| GO:BP | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis | 2 | 4 | 4.89e-01 |
| GO:BP | GO:0007144 | female meiosis I | 7 | 9 | 4.89e-01 |
| GO:BP | GO:0061053 | somite development | 18 | 63 | 4.89e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 18 | 34 | 4.89e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 1 | 23 | 4.89e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 14 | 133 | 4.89e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 1 | 8 | 4.89e-01 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 3 | 9 | 4.89e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 5 | 10 | 4.89e-01 |
| GO:BP | GO:0006954 | inflammatory response | 1 | 429 | 4.89e-01 |
| GO:BP | GO:0060677 | ureteric bud elongation | 1 | 7 | 4.89e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 5 | 10 | 4.89e-01 |
| GO:BP | GO:0006592 | ornithine biosynthetic process | 1 | 1 | 4.89e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 47 | 103 | 4.90e-01 |
| GO:BP | GO:0014842 | regulation of skeletal muscle satellite cell proliferation | 2 | 8 | 4.90e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 3 | 133 | 4.90e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 10 | 42 | 4.90e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 4.90e-01 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 6 | 11 | 4.90e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 4 | 73 | 4.90e-01 |
| GO:BP | GO:0003062 | regulation of heart rate by chemical signal | 1 | 6 | 4.90e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 1 | 61 | 4.90e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 3 | 9 | 4.90e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 1 | 127 | 4.90e-01 |
| GO:BP | GO:0036149 | phosphatidylinositol acyl-chain remodeling | 1 | 3 | 4.90e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 4 | 73 | 4.90e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 3 | 9 | 4.90e-01 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 1 | 9 | 4.91e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 17 | 42 | 4.91e-01 |
| GO:BP | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 7 | 4.91e-01 |
| GO:BP | GO:1903387 | positive regulation of homophilic cell adhesion | 1 | 1 | 4.91e-01 |
| GO:BP | GO:0001661 | conditioned taste aversion | 1 | 2 | 4.91e-01 |
| GO:BP | GO:0090494 | dopamine uptake | 1 | 10 | 4.91e-01 |
| GO:BP | GO:0070933 | histone H4 deacetylation | 3 | 7 | 4.91e-01 |
| GO:BP | GO:0002070 | epithelial cell maturation | 8 | 16 | 4.91e-01 |
| GO:BP | GO:0098789 | pre-mRNA cleavage required for polyadenylation | 1 | 12 | 4.91e-01 |
| GO:BP | GO:0033625 | positive regulation of integrin activation | 3 | 6 | 4.91e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 13 | 135 | 4.91e-01 |
| GO:BP | GO:0071389 | cellular response to mineralocorticoid stimulus | 6 | 9 | 4.91e-01 |
| GO:BP | GO:1990961 | xenobiotic detoxification by transmembrane export across the plasma membrane | 4 | 7 | 4.91e-01 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 6 | 10 | 4.91e-01 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 1 | 30 | 4.91e-01 |
| GO:BP | GO:2000635 | negative regulation of primary miRNA processing | 2 | 2 | 4.91e-01 |
| GO:BP | GO:2000634 | regulation of primary miRNA processing | 2 | 2 | 4.91e-01 |
| GO:BP | GO:0031167 | rRNA methylation | 10 | 24 | 4.91e-01 |
| GO:BP | GO:0045684 | positive regulation of epidermis development | 5 | 20 | 4.91e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 73 | 4.91e-01 |
| GO:BP | GO:0072750 | cellular response to leptomycin B | 1 | 1 | 4.91e-01 |
| GO:BP | GO:1901862 | negative regulation of muscle tissue development | 6 | 9 | 4.91e-01 |
| GO:BP | GO:1901344 | response to leptomycin B | 1 | 1 | 4.91e-01 |
| GO:BP | GO:0140354 | lipid import into cell | 4 | 15 | 4.91e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 18 | 35 | 4.91e-01 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 12 | 23 | 4.91e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 1 | 27 | 4.91e-01 |
| GO:BP | GO:0031064 | negative regulation of histone deacetylation | 1 | 2 | 4.91e-01 |
| GO:BP | GO:0035247 | peptidyl-arginine omega-N-methylation | 1 | 3 | 4.91e-01 |
| GO:BP | GO:0034773 | histone H4-K20 trimethylation | 3 | 5 | 4.91e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 33 | 71 | 4.92e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 11 | 38 | 4.92e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 1 | 23 | 4.92e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 3 | 60 | 4.92e-01 |
| GO:BP | GO:0042311 | vasodilation | 2 | 39 | 4.92e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 3 | 45 | 4.92e-01 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 2 | 17 | 4.92e-01 |
| GO:BP | GO:0044860 | protein localization to plasma membrane raft | 1 | 4 | 4.92e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 7 | 45 | 4.92e-01 |
| GO:BP | GO:1905209 | positive regulation of cardiocyte differentiation | 1 | 11 | 4.93e-01 |
| GO:BP | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | 4 | 9 | 4.93e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 2 | 111 | 4.93e-01 |
| GO:BP | GO:0046068 | cGMP metabolic process | 1 | 11 | 4.93e-01 |
| GO:BP | GO:0007588 | excretion | 1 | 18 | 4.93e-01 |
| GO:BP | GO:0060526 | prostate glandular acinus morphogenesis | 1 | 3 | 4.93e-01 |
| GO:BP | GO:0060527 | prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis | 1 | 3 | 4.93e-01 |
| GO:BP | GO:0014034 | neural crest cell fate commitment | 1 | 4 | 4.93e-01 |
| GO:BP | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process | 4 | 9 | 4.93e-01 |
| GO:BP | GO:2000020 | positive regulation of male gonad development | 1 | 4 | 4.93e-01 |
| GO:BP | GO:0051781 | positive regulation of cell division | 2 | 64 | 4.94e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 2 | 40 | 4.94e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 21 | 39 | 4.94e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 2 | 11 | 4.94e-01 |
| GO:BP | GO:0006971 | hypotonic response | 6 | 10 | 4.94e-01 |
| GO:BP | GO:1904529 | regulation of actin filament binding | 1 | 3 | 4.94e-01 |
| GO:BP | GO:1904426 | positive regulation of GTP binding | 1 | 3 | 4.94e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 16 | 34 | 4.94e-01 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 1 | 2 | 4.94e-01 |
| GO:BP | GO:0140066 | peptidyl-lysine crotonylation | 1 | 2 | 4.94e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 1 | 78 | 4.94e-01 |
| GO:BP | GO:0006659 | phosphatidylserine biosynthetic process | 2 | 4 | 4.94e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 14 | 31 | 4.94e-01 |
| GO:BP | GO:0002357 | defense response to tumor cell | 1 | 6 | 4.94e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 2 | 23 | 4.94e-01 |
| GO:BP | GO:0010896 | regulation of triglyceride catabolic process | 1 | 7 | 4.94e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 4 | 154 | 4.94e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 5 | 40 | 4.94e-01 |
| GO:BP | GO:0002101 | tRNA wobble cytosine modification | 1 | 2 | 4.94e-01 |
| GO:BP | GO:0140647 | P450-containing electron transport chain | 1 | 2 | 4.94e-01 |
| GO:BP | GO:0043649 | dicarboxylic acid catabolic process | 1 | 14 | 4.95e-01 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 2 | 9 | 4.95e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 4.95e-01 |
| GO:BP | GO:1904227 | negative regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0098970 | postsynaptic neurotransmitter receptor diffusion trapping | 1 | 4 | 4.95e-01 |
| GO:BP | GO:0099628 | neurotransmitter receptor diffusion trapping | 1 | 4 | 4.95e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 3 | 10 | 4.95e-01 |
| GO:BP | GO:0098953 | receptor diffusion trapping | 1 | 4 | 4.95e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 2 | 23 | 4.95e-01 |
| GO:BP | GO:0031099 | regeneration | 11 | 142 | 4.95e-01 |
| GO:BP | GO:0031958 | corticosteroid receptor signaling pathway | 2 | 14 | 4.95e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 3 | 163 | 4.95e-01 |
| GO:BP | GO:2000224 | regulation of testosterone biosynthetic process | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 4 | 8 | 4.95e-01 |
| GO:BP | GO:1904226 | regulation of glycogen synthase activity, transferring glucose-1-phosphate | 1 | 1 | 4.95e-01 |
| GO:BP | GO:1905533 | negative regulation of leucine import across plasma membrane | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0071879 | positive regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 1 | 1 | 4.95e-01 |
| GO:BP | GO:1903944 | negative regulation of hepatocyte apoptotic process | 3 | 6 | 4.95e-01 |
| GO:BP | GO:0061184 | positive regulation of dermatome development | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 1 | 41 | 4.95e-01 |
| GO:BP | GO:0072137 | condensed mesenchymal cell proliferation | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0032346 | positive regulation of aldosterone metabolic process | 1 | 3 | 4.95e-01 |
| GO:BP | GO:0007049 | cell cycle | 706 | 1529 | 4.95e-01 |
| GO:BP | GO:0032349 | positive regulation of aldosterone biosynthetic process | 1 | 3 | 4.95e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 4.95e-01 |
| GO:BP | GO:0044027 | negative regulation of gene expression via CpG island methylation | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0060358 | negative regulation of leucine import | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0032736 | positive regulation of interleukin-13 production | 1 | 7 | 4.95e-01 |
| GO:BP | GO:0009624 | response to nematode | 1 | 4 | 4.95e-01 |
| GO:BP | GO:0060357 | regulation of leucine import | 1 | 2 | 4.95e-01 |
| GO:BP | GO:0010905 | negative regulation of UDP-glucose catabolic process | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 1 | 12 | 4.95e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 1 | 12 | 4.95e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 1 | 41 | 4.95e-01 |
| GO:BP | GO:0033604 | negative regulation of catecholamine secretion | 3 | 6 | 4.95e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 4 | 14 | 4.95e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 4.95e-01 |
| GO:BP | GO:0050894 | determination of affect | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0010904 | regulation of UDP-glucose catabolic process | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 1 | 29 | 4.95e-01 |
| GO:BP | GO:0030103 | vasopressin secretion | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0097497 | blood vessel endothelial cell delamination | 1 | 1 | 4.95e-01 |
| GO:BP | GO:0001955 | blood vessel maturation | 2 | 6 | 4.95e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 1 | 33 | 4.96e-01 |
| GO:BP | GO:0021510 | spinal cord development | 5 | 59 | 4.96e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 8 | 14 | 4.96e-01 |
| GO:BP | GO:0031439 | positive regulation of mRNA cleavage | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0031437 | regulation of mRNA cleavage | 1 | 1 | 4.96e-01 |
| GO:BP | GO:2000975 | positive regulation of pro-B cell differentiation | 1 | 1 | 4.96e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 22 | 51 | 4.96e-01 |
| GO:BP | GO:0019470 | 4-hydroxyproline catabolic process | 2 | 4 | 4.96e-01 |
| GO:BP | GO:0019471 | 4-hydroxyproline metabolic process | 2 | 4 | 4.96e-01 |
| GO:BP | GO:0055129 | L-proline biosynthetic process | 4 | 5 | 4.96e-01 |
| GO:BP | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 1 | 9 | 4.96e-01 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 6 | 30 | 4.96e-01 |
| GO:BP | GO:0016559 | peroxisome fission | 1 | 9 | 4.96e-01 |
| GO:BP | GO:0006561 | proline biosynthetic process | 4 | 5 | 4.96e-01 |
| GO:BP | GO:0015886 | heme transport | 1 | 9 | 4.96e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 2 | 6 | 4.96e-01 |
| GO:BP | GO:0030393 | fructoselysine metabolic process | 1 | 1 | 4.96e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 3 | 200 | 4.96e-01 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 6 | 9 | 4.96e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 10 | 198 | 4.96e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 1 | 465 | 4.96e-01 |
| GO:BP | GO:0061009 | common bile duct development | 2 | 4 | 4.96e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 4.96e-01 |
| GO:BP | GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1 | 8 | 4.96e-01 |
| GO:BP | GO:1905301 | regulation of macropinocytosis | 2 | 3 | 4.96e-01 |
| GO:BP | GO:0007015 | actin filament organization | 6 | 373 | 4.96e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 1 | 620 | 4.97e-01 |
| GO:BP | GO:0032928 | regulation of superoxide anion generation | 4 | 11 | 4.97e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 7 | 295 | 4.97e-01 |
| GO:BP | GO:0060271 | cilium assembly | 31 | 292 | 4.97e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 1 | 45 | 4.97e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 2 | 5 | 4.97e-01 |
| GO:BP | GO:0008090 | retrograde axonal transport | 1 | 19 | 4.97e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 5 | 85 | 4.98e-01 |
| GO:BP | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion | 1 | 1 | 4.99e-01 |
| GO:BP | GO:0002414 | immunoglobulin transcytosis in epithelial cells | 1 | 2 | 4.99e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 43 | 85 | 4.99e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 4.99e-01 |
| GO:BP | GO:0106020 | regulation of vesicle docking | 1 | 2 | 4.99e-01 |
| GO:BP | GO:1905458 | positive regulation of lymphoid progenitor cell differentiation | 2 | 3 | 4.99e-01 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 1 | 15 | 4.99e-01 |
| GO:BP | GO:0090493 | catecholamine uptake | 1 | 11 | 4.99e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 4.99e-01 |
| GO:BP | GO:0042538 | hyperosmotic salinity response | 4 | 8 | 4.99e-01 |
| GO:BP | GO:2000078 | positive regulation of type B pancreatic cell development | 2 | 2 | 4.99e-01 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 3 | 10 | 5.00e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 9 | 191 | 5.00e-01 |
| GO:BP | GO:0006660 | phosphatidylserine catabolic process | 2 | 3 | 5.00e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 3 | 21 | 5.00e-01 |
| GO:BP | GO:1990009 | retinal cell apoptotic process | 3 | 5 | 5.00e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 45 | 210 | 5.00e-01 |
| GO:BP | GO:1990523 | bone regeneration | 1 | 2 | 5.00e-01 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 2 | 103 | 5.01e-01 |
| GO:BP | GO:0070498 | interleukin-1-mediated signaling pathway | 4 | 20 | 5.01e-01 |
| GO:BP | GO:0060023 | soft palate development | 1 | 3 | 5.01e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 14 | 113 | 5.01e-01 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 1 | 16 | 5.01e-01 |
| GO:BP | GO:0008216 | spermidine metabolic process | 3 | 7 | 5.02e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 30 | 75 | 5.02e-01 |
| GO:BP | GO:0035196 | miRNA processing | 16 | 41 | 5.02e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 5.02e-01 |
| GO:BP | GO:0061302 | smooth muscle cell-matrix adhesion | 2 | 5 | 5.02e-01 |
| GO:BP | GO:0090107 | regulation of high-density lipoprotein particle assembly | 1 | 3 | 5.02e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 1 | 26 | 5.02e-01 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 1 | 25 | 5.02e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 57 | 133 | 5.03e-01 |
| GO:BP | GO:0010722 | regulation of ferrochelatase activity | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0060310 | regulation of elastin catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0010711 | negative regulation of collagen catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0060311 | negative regulation of elastin catabolic process | 1 | 1 | 5.03e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 1 | 4 | 5.03e-01 |
| GO:BP | GO:1904923 | regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 6 | 15 | 5.03e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 1 | 40 | 5.03e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 2 | 88 | 5.03e-01 |
| GO:BP | GO:0061944 | negative regulation of protein K48-linked ubiquitination | 1 | 2 | 5.03e-01 |
| GO:BP | GO:0098787 | mRNA cleavage involved in mRNA processing | 1 | 13 | 5.04e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 2 | 30 | 5.04e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 2 | 13 | 5.05e-01 |
| GO:BP | GO:0051302 | regulation of cell division | 15 | 145 | 5.05e-01 |
| GO:BP | GO:0014906 | myotube cell development involved in skeletal muscle regeneration | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 2 | 31 | 5.05e-01 |
| GO:BP | GO:0060992 | response to fungicide | 1 | 4 | 5.05e-01 |
| GO:BP | GO:0048799 | animal organ maturation | 1 | 20 | 5.05e-01 |
| GO:BP | GO:1904556 | L-tryptophan transmembrane transport | 1 | 2 | 5.05e-01 |
| GO:BP | GO:0007613 | memory | 15 | 92 | 5.05e-01 |
| GO:BP | GO:0070986 | left/right axis specification | 1 | 13 | 5.05e-01 |
| GO:BP | GO:0038003 | G protein-coupled opioid receptor signaling pathway | 1 | 5 | 5.05e-01 |
| GO:BP | GO:0003014 | renal system process | 6 | 78 | 5.05e-01 |
| GO:BP | GO:2000254 | regulation of male germ cell proliferation | 2 | 4 | 5.05e-01 |
| GO:BP | GO:0090672 | telomerase RNA localization | 5 | 19 | 5.05e-01 |
| GO:BP | GO:0051902 | negative regulation of mitochondrial depolarization | 2 | 5 | 5.05e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 5.05e-01 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 32 | 68 | 5.05e-01 |
| GO:BP | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process | 3 | 16 | 5.05e-01 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 1 | 9 | 5.05e-01 |
| GO:BP | GO:0007601 | visual perception | 2 | 96 | 5.05e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 1 | 48 | 5.05e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 9 | 16 | 5.05e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 1 | 660 | 5.05e-01 |
| GO:BP | GO:0048850 | hypophysis morphogenesis | 1 | 2 | 5.05e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 5 | 86 | 5.05e-01 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 5 | 19 | 5.05e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 2 | 8 | 5.05e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 1 | 90 | 5.05e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 5 | 7 | 5.05e-01 |
| GO:BP | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0010832 | negative regulation of myotube differentiation | 1 | 10 | 5.05e-01 |
| GO:BP | GO:0007141 | male meiosis I | 4 | 15 | 5.05e-01 |
| GO:BP | GO:2000172 | regulation of branching morphogenesis of a nerve | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0040016 | embryonic cleavage | 1 | 7 | 5.05e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 3 | 26 | 5.05e-01 |
| GO:BP | GO:0035864 | response to potassium ion | 2 | 12 | 5.05e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 7 | 23 | 5.05e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 2 | 23 | 5.05e-01 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 5 | 19 | 5.05e-01 |
| GO:BP | GO:0042335 | cuticle development | 1 | 2 | 5.05e-01 |
| GO:BP | GO:0040010 | positive regulation of growth rate | 2 | 2 | 5.05e-01 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 5 | 19 | 5.05e-01 |
| GO:BP | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 4 | 293 | 5.05e-01 |
| GO:BP | GO:0009051 | pentose-phosphate shunt, oxidative branch | 1 | 4 | 5.05e-01 |
| GO:BP | GO:0061820 | telomeric D-loop disassembly | 2 | 8 | 5.05e-01 |
| GO:BP | GO:0043435 | response to corticotropin-releasing hormone | 2 | 3 | 5.05e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 7 | 11 | 5.05e-01 |
| GO:BP | GO:0061448 | connective tissue development | 5 | 208 | 5.05e-01 |
| GO:BP | GO:0071376 | cellular response to corticotropin-releasing hormone stimulus | 2 | 3 | 5.05e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 19 | 141 | 5.05e-01 |
| GO:BP | GO:0000413 | protein peptidyl-prolyl isomerization | 6 | 26 | 5.05e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 155 | 357 | 5.05e-01 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 6 | 7 | 5.05e-01 |
| GO:BP | GO:1902073 | positive regulation of hypoxia-inducible factor-1alpha signaling pathway | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 15 | 27 | 5.05e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 1 | 14 | 5.05e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 1 | 6 | 5.05e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 4 | 7 | 5.05e-01 |
| GO:BP | GO:1904877 | positive regulation of DNA ligase activity | 1 | 1 | 5.05e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 2 | 30 | 5.05e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 5.05e-01 |
| GO:BP | GO:1904381 | Golgi apparatus mannose trimming | 2 | 3 | 5.05e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 5.05e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 5.05e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 1 | 691 | 5.05e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 5.05e-01 |
| GO:BP | GO:1904182 | regulation of pyruvate dehydrogenase activity | 1 | 4 | 5.05e-01 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 3 | 5 | 5.05e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 1 | 16 | 5.05e-01 |
| GO:BP | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 2 | 5 | 5.05e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 1 | 13 | 5.05e-01 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 1 | 39 | 5.05e-01 |
| GO:BP | GO:0010739 | positive regulation of protein kinase A signaling | 1 | 6 | 5.05e-01 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 1 | 13 | 5.05e-01 |
| GO:BP | GO:0099191 | trans-synaptic signaling by BDNF | 1 | 3 | 5.05e-01 |
| GO:BP | GO:0036035 | osteoclast development | 1 | 10 | 5.05e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 2 | 16 | 5.05e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 1 | 29 | 5.05e-01 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 14 | 30 | 5.05e-01 |
| GO:BP | GO:0032286 | central nervous system myelin maintenance | 2 | 3 | 5.06e-01 |
| GO:BP | GO:1904502 | regulation of lipophagy | 3 | 5 | 5.06e-01 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 3 | 5 | 5.06e-01 |
| GO:BP | GO:0106227 | peptidyl-lysine glutarylation | 1 | 1 | 5.06e-01 |
| GO:BP | GO:0006564 | L-serine biosynthetic process | 1 | 6 | 5.06e-01 |
| GO:BP | GO:0035553 | oxidative single-stranded RNA demethylation | 1 | 3 | 5.07e-01 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 5 | 8 | 5.07e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 285 | 626 | 5.07e-01 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 10 | 20 | 5.07e-01 |
| GO:BP | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation | 2 | 2 | 5.07e-01 |
| GO:BP | GO:0072079 | nephron tubule formation | 1 | 12 | 5.07e-01 |
| GO:BP | GO:0044782 | cilium organization | 33 | 313 | 5.07e-01 |
| GO:BP | GO:0070343 | white fat cell proliferation | 1 | 2 | 5.07e-01 |
| GO:BP | GO:1901529 | positive regulation of anion channel activity | 2 | 2 | 5.07e-01 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 1 | 7 | 5.07e-01 |
| GO:BP | GO:1903793 | positive regulation of monoatomic anion transport | 2 | 2 | 5.07e-01 |
| GO:BP | GO:0070350 | regulation of white fat cell proliferation | 1 | 2 | 5.07e-01 |
| GO:BP | GO:1903961 | positive regulation of anion transmembrane transport | 2 | 2 | 5.07e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 5.07e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 1 | 538 | 5.07e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 5.07e-01 |
| GO:BP | GO:0014854 | response to inactivity | 1 | 11 | 5.08e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 1 | 54 | 5.08e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 14 | 60 | 5.08e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 1 | 8 | 5.08e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 1 | 678 | 5.08e-01 |
| GO:BP | GO:0021561 | facial nerve development | 1 | 8 | 5.08e-01 |
| GO:BP | GO:0003218 | cardiac left ventricle formation | 1 | 2 | 5.08e-01 |
| GO:BP | GO:1990428 | miRNA transport | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 3 | 172 | 5.08e-01 |
| GO:BP | GO:0140914 | zinc ion import into secretory vesicle | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0140915 | zinc ion import into zymogen granule | 1 | 1 | 5.08e-01 |
| GO:BP | GO:1905661 | regulation of telomerase RNA reverse transcriptase activity | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 3 | 31 | 5.08e-01 |
| GO:BP | GO:0140916 | zinc ion import into lysosome | 1 | 1 | 5.08e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 21 | 46 | 5.08e-01 |
| GO:BP | GO:0042403 | thyroid hormone metabolic process | 4 | 11 | 5.08e-01 |
| GO:BP | GO:0140917 | zinc ion import into mitochondrion | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 2 | 8 | 5.08e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 7 | 54 | 5.08e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 2 | 97 | 5.08e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 21 | 46 | 5.08e-01 |
| GO:BP | GO:0006364 | rRNA processing | 2 | 221 | 5.08e-01 |
| GO:BP | GO:0051668 | localization within membrane | 9 | 605 | 5.08e-01 |
| GO:BP | GO:0061090 | positive regulation of sequestering of zinc ion | 1 | 1 | 5.08e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 1 | 31 | 5.08e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 1 | 40 | 5.08e-01 |
| GO:BP | GO:0140361 | cyclic-GMP-AMP transmembrane import across plasma membrane | 1 | 4 | 5.08e-01 |
| GO:BP | GO:2000510 | positive regulation of dendritic cell chemotaxis | 1 | 3 | 5.08e-01 |
| GO:BP | GO:0060020 | Bergmann glial cell differentiation | 4 | 9 | 5.08e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 3 | 53 | 5.08e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 1 | 40 | 5.09e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 1 | 484 | 5.09e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 3 | 85 | 5.09e-01 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 6 | 8 | 5.09e-01 |
| GO:BP | GO:0070236 | negative regulation of activation-induced cell death of T cells | 2 | 3 | 5.09e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 3 | 35 | 5.09e-01 |
| GO:BP | GO:1901304 | regulation of spermidine biosynthetic process | 1 | 1 | 5.09e-01 |
| GO:BP | GO:0055014 | atrial cardiac muscle cell development | 2 | 3 | 5.09e-01 |
| GO:BP | GO:1901307 | positive regulation of spermidine biosynthetic process | 1 | 1 | 5.09e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 3 | 85 | 5.09e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 2 | 3 | 5.09e-01 |
| GO:BP | GO:0055118 | negative regulation of cardiac muscle contraction | 1 | 4 | 5.09e-01 |
| GO:BP | GO:0055011 | atrial cardiac muscle cell differentiation | 2 | 3 | 5.09e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 5.09e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 5.09e-01 |
| GO:BP | GO:1902174 | positive regulation of keratinocyte apoptotic process | 1 | 3 | 5.09e-01 |
| GO:BP | GO:1903726 | negative regulation of phospholipid metabolic process | 2 | 7 | 5.09e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 11 | 42 | 5.09e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 5 | 299 | 5.10e-01 |
| GO:BP | GO:0043201 | response to leucine | 7 | 13 | 5.10e-01 |
| GO:BP | GO:0061824 | cytosolic ciliogenesis | 1 | 1 | 5.10e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 2 | 71 | 5.11e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 1 | 45 | 5.11e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 1 | 2 | 5.11e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 3 | 107 | 5.11e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 4 | 165 | 5.11e-01 |
| GO:BP | GO:1905941 | positive regulation of gonad development | 1 | 4 | 5.11e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 29 | 151 | 5.11e-01 |
| GO:BP | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 3 | 9 | 5.11e-01 |
| GO:BP | GO:0007506 | gonadal mesoderm development | 1 | 2 | 5.11e-01 |
| GO:BP | GO:0032415 | regulation of sodium:proton antiporter activity | 1 | 4 | 5.12e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 1 | 29 | 5.12e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 6 | 48 | 5.12e-01 |
| GO:BP | GO:0045342 | MHC class II biosynthetic process | 8 | 12 | 5.12e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 4 | 97 | 5.12e-01 |
| GO:BP | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | 6 | 10 | 5.13e-01 |
| GO:BP | GO:0003016 | respiratory system process | 1 | 30 | 5.13e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 6 | 11 | 5.13e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 8 | 20 | 5.13e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 3 | 42 | 5.14e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 2 | 5 | 5.14e-01 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 5 | 8 | 5.14e-01 |
| GO:BP | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly | 1 | 3 | 5.14e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 5.14e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 5.14e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 14 | 92 | 5.15e-01 |
| GO:BP | GO:0006501 | C-terminal protein lipidation | 2 | 6 | 5.15e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 5.15e-01 |
| GO:BP | GO:1905794 | response to puromycin | 1 | 1 | 5.15e-01 |
| GO:BP | GO:0043650 | dicarboxylic acid biosynthetic process | 2 | 9 | 5.15e-01 |
| GO:BP | GO:1905795 | cellular response to puromycin | 1 | 1 | 5.15e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 5.15e-01 |
| GO:BP | GO:0030643 | intracellular phosphate ion homeostasis | 3 | 5 | 5.15e-01 |
| GO:BP | GO:1905603 | regulation of blood-brain barrier permeability | 1 | 6 | 5.15e-01 |
| GO:BP | GO:1904239 | regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.15e-01 |
| GO:BP | GO:1904210 | VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.15e-01 |
| GO:BP | GO:1904240 | negative regulation of VCP-NPL4-UFD1 AAA ATPase complex assembly | 1 | 1 | 5.15e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 2 | 24 | 5.15e-01 |
| GO:BP | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process | 1 | 2 | 5.15e-01 |
| GO:BP | GO:0035733 | hepatic stellate cell activation | 5 | 8 | 5.15e-01 |
| GO:BP | GO:1903945 | positive regulation of hepatocyte apoptotic process | 1 | 1 | 5.16e-01 |
| GO:BP | GO:2000360 | negative regulation of binding of sperm to zona pellucida | 2 | 2 | 5.16e-01 |
| GO:BP | GO:0070409 | carbamoyl phosphate biosynthetic process | 1 | 1 | 5.16e-01 |
| GO:BP | GO:0070408 | carbamoyl phosphate metabolic process | 1 | 1 | 5.16e-01 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 9 | 17 | 5.16e-01 |
| GO:BP | GO:0015779 | glucuronoside transport | 1 | 1 | 5.16e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 1 | 38 | 5.16e-01 |
| GO:BP | GO:0046604 | positive regulation of mitotic centrosome separation | 2 | 3 | 5.16e-01 |
| GO:BP | GO:0097398 | cellular response to interleukin-17 | 4 | 12 | 5.16e-01 |
| GO:BP | GO:1902366 | regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.16e-01 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 10 | 22 | 5.16e-01 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 1 | 14 | 5.16e-01 |
| GO:BP | GO:1902359 | Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.16e-01 |
| GO:BP | GO:0035336 | long-chain fatty-acyl-CoA metabolic process | 5 | 19 | 5.16e-01 |
| GO:BP | GO:1902367 | negative regulation of Notch signaling pathway involved in somitogenesis | 1 | 2 | 5.16e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 15 | 170 | 5.16e-01 |
| GO:BP | GO:0071709 | membrane assembly | 3 | 53 | 5.16e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 1 | 656 | 5.16e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 5.16e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 3 | 6 | 5.17e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 3 | 6 | 5.17e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 3 | 6 | 5.17e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 3 | 6 | 5.17e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 3 | 6 | 5.17e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 5.17e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 3 | 56 | 5.17e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 1 | 556 | 5.17e-01 |
| GO:BP | GO:1990108 | protein linear deubiquitination | 1 | 3 | 5.17e-01 |
| GO:BP | GO:0019080 | viral gene expression | 1 | 96 | 5.17e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 4 | 167 | 5.17e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 6 | 11 | 5.17e-01 |
| GO:BP | GO:0002051 | osteoblast fate commitment | 2 | 2 | 5.17e-01 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 2 | 4 | 5.17e-01 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 6 | 15 | 5.17e-01 |
| GO:BP | GO:2000074 | regulation of type B pancreatic cell development | 3 | 8 | 5.17e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 1 | 609 | 5.17e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 5 | 42 | 5.17e-01 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 6 | 11 | 5.17e-01 |
| GO:BP | GO:0010747 | positive regulation of long-chain fatty acid import across plasma membrane | 1 | 1 | 5.18e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 3 | 13 | 5.18e-01 |
| GO:BP | GO:1903567 | regulation of protein localization to ciliary membrane | 1 | 2 | 5.18e-01 |
| GO:BP | GO:1902524 | positive regulation of protein K48-linked ubiquitination | 1 | 2 | 5.18e-01 |
| GO:BP | GO:0140820 | cytosol to Golgi apparatus transport | 1 | 4 | 5.18e-01 |
| GO:BP | GO:1905673 | positive regulation of lysosome organization | 1 | 1 | 5.18e-01 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 1 | 10 | 5.18e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 2 | 3 | 5.18e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 2 | 29 | 5.18e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 5.18e-01 |
| GO:BP | GO:0046903 | secretion | 1 | 645 | 5.18e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 1 | 631 | 5.18e-01 |
| GO:BP | GO:0072177 | mesonephric duct development | 2 | 7 | 5.18e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 13 | 27 | 5.18e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 3 | 16 | 5.18e-01 |
| GO:BP | GO:0030329 | prenylcysteine metabolic process | 1 | 2 | 5.18e-01 |
| GO:BP | GO:0043586 | tongue development | 5 | 15 | 5.18e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 1 | 47 | 5.18e-01 |
| GO:BP | GO:0030328 | prenylcysteine catabolic process | 1 | 2 | 5.18e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 2 | 27 | 5.18e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 2 | 13 | 5.18e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 2 | 8 | 5.18e-01 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 2 | 9 | 5.18e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 3 | 16 | 5.18e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 1 | 35 | 5.18e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 2 | 27 | 5.18e-01 |
| GO:BP | GO:0120168 | detection of hot stimulus involved in thermoception | 1 | 1 | 5.19e-01 |
| GO:BP | GO:1904045 | cellular response to aldosterone | 4 | 5 | 5.19e-01 |
| GO:BP | GO:0014805 | smooth muscle adaptation | 2 | 3 | 5.19e-01 |
| GO:BP | GO:0062036 | sensory perception of hot stimulus | 1 | 1 | 5.19e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 11 | 18 | 5.19e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 3 | 16 | 5.19e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 5 | 156 | 5.19e-01 |
| GO:BP | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | 5 | 8 | 5.20e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 3 | 60 | 5.20e-01 |
| GO:BP | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 5 | 5.20e-01 |
| GO:BP | GO:0021849 | neuroblast division in subventricular zone | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0052779 | amino disaccharide metabolic process | 2 | 9 | 5.20e-01 |
| GO:BP | GO:1902766 | skeletal muscle satellite cell migration | 1 | 2 | 5.20e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 6 | 11 | 5.20e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 5.20e-01 |
| GO:BP | GO:0097577 | sequestering of iron ion | 3 | 3 | 5.20e-01 |
| GO:BP | GO:0030205 | dermatan sulfate metabolic process | 2 | 9 | 5.20e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 5.20e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 1 | 78 | 5.20e-01 |
| GO:BP | GO:0072682 | eosinophil extravasation | 1 | 1 | 5.20e-01 |
| GO:BP | GO:1901187 | regulation of ephrin receptor signaling pathway | 1 | 2 | 5.20e-01 |
| GO:BP | GO:0006558 | L-phenylalanine metabolic process | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0019042 | viral latency | 1 | 4 | 5.20e-01 |
| GO:BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 3 | 5 | 5.20e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 2 | 161 | 5.20e-01 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 1 | 14 | 5.20e-01 |
| GO:BP | GO:0042695 | thelarche | 3 | 5 | 5.20e-01 |
| GO:BP | GO:0043972 | histone H3-K23 acetylation | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0090403 | oxidative stress-induced premature senescence | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 1 | 71 | 5.20e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 5.20e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 2 | 5 | 5.20e-01 |
| GO:BP | GO:0034447 | very-low-density lipoprotein particle clearance | 2 | 4 | 5.20e-01 |
| GO:BP | GO:1902222 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 1 | 12 | 5.20e-01 |
| GO:BP | GO:0006559 | L-phenylalanine catabolic process | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 4 | 228 | 5.20e-01 |
| GO:BP | GO:0097474 | retinal cone cell apoptotic process | 1 | 1 | 5.20e-01 |
| GO:BP | GO:2000419 | regulation of eosinophil extravasation | 1 | 1 | 5.20e-01 |
| GO:BP | GO:2000420 | negative regulation of eosinophil extravasation | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 6 | 15 | 5.20e-01 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 1 | 3 | 5.20e-01 |
| GO:BP | GO:0060744 | mammary gland branching involved in thelarche | 3 | 5 | 5.20e-01 |
| GO:BP | GO:0000425 | pexophagy | 2 | 6 | 5.20e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 3 | 24 | 5.20e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 5.20e-01 |
| GO:BP | GO:0002318 | myeloid progenitor cell differentiation | 3 | 7 | 5.20e-01 |
| GO:BP | GO:1903544 | response to butyrate | 1 | 2 | 5.20e-01 |
| GO:BP | GO:0009133 | nucleoside diphosphate biosynthetic process | 7 | 11 | 5.20e-01 |
| GO:BP | GO:1904528 | positive regulation of microtubule binding | 2 | 6 | 5.20e-01 |
| GO:BP | GO:0035212 | cell competition in a multicellular organism | 1 | 1 | 5.20e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 4 | 92 | 5.20e-01 |
| GO:BP | GO:1902416 | positive regulation of mRNA binding | 2 | 8 | 5.21e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 7 | 12 | 5.21e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 4 | 9 | 5.21e-01 |
| GO:BP | GO:0048680 | positive regulation of axon regeneration | 1 | 8 | 5.21e-01 |
| GO:BP | GO:0090668 | endothelial cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 5.21e-01 |
| GO:BP | GO:0106300 | protein-DNA covalent cross-linking repair | 1 | 7 | 5.21e-01 |
| GO:BP | GO:0090667 | cell chemotaxis to vascular endothelial growth factor | 1 | 1 | 5.21e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 3 | 44 | 5.21e-01 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 4 | 16 | 5.22e-01 |
| GO:BP | GO:2000644 | regulation of receptor catabolic process | 3 | 9 | 5.22e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 7 | 5.22e-01 |
| GO:BP | GO:0060948 | cardiac vascular smooth muscle cell development | 2 | 3 | 5.22e-01 |
| GO:BP | GO:2000454 | positive regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 2 | 20 | 5.22e-01 |
| GO:BP | GO:0070376 | regulation of ERK5 cascade | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0010795 | regulation of ubiquinone biosynthetic process | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 2 | 8 | 5.22e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 11 | 24 | 5.22e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 2 | 8 | 5.22e-01 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 1 | 4 | 5.22e-01 |
| GO:BP | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation | 2 | 3 | 5.22e-01 |
| GO:BP | GO:0035519 | protein K29-linked ubiquitination | 4 | 6 | 5.22e-01 |
| GO:BP | GO:0007611 | learning or memory | 24 | 202 | 5.22e-01 |
| GO:BP | GO:0003094 | glomerular filtration | 3 | 21 | 5.22e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 7 | 160 | 5.22e-01 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 1 | 4 | 5.22e-01 |
| GO:BP | GO:0010511 | regulation of phosphatidylinositol biosynthetic process | 1 | 2 | 5.22e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 2 | 5 | 5.22e-01 |
| GO:BP | GO:0035788 | cell migration involved in metanephros development | 1 | 4 | 5.22e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 2 | 1719 | 5.22e-01 |
| GO:BP | GO:1905857 | positive regulation of pentose-phosphate shunt | 2 | 3 | 5.22e-01 |
| GO:BP | GO:0001544 | initiation of primordial ovarian follicle growth | 1 | 1 | 5.22e-01 |
| GO:BP | GO:0099041 | vesicle tethering to Golgi | 1 | 5 | 5.22e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 5 | 7 | 5.22e-01 |
| GO:BP | GO:0033572 | transferrin transport | 4 | 8 | 5.22e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 1 | 14 | 5.23e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 5.23e-01 |
| GO:BP | GO:0042168 | heme metabolic process | 2 | 40 | 5.23e-01 |
| GO:BP | GO:0009950 | dorsal/ventral axis specification | 2 | 9 | 5.23e-01 |
| GO:BP | GO:0032633 | interleukin-4 production | 2 | 14 | 5.23e-01 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 2 | 14 | 5.23e-01 |
| GO:BP | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration | 1 | 3 | 5.23e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 1 | 55 | 5.23e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 2 | 14 | 5.23e-01 |
| GO:BP | GO:0035627 | ceramide transport | 2 | 15 | 5.24e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 1 | 52 | 5.24e-01 |
| GO:BP | GO:1905216 | positive regulation of RNA binding | 4 | 9 | 5.24e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 28 | 5.24e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 1 | 47 | 5.24e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 9 | 46 | 5.24e-01 |
| GO:BP | GO:0046477 | glycosylceramide catabolic process | 2 | 5 | 5.24e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 8 | 59 | 5.24e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 6 | 10 | 5.24e-01 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 1 | 7 | 5.24e-01 |
| GO:BP | GO:2000768 | positive regulation of nephron tubule epithelial cell differentiation | 1 | 3 | 5.24e-01 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 1 | 10 | 5.24e-01 |
| GO:BP | GO:0097377 | spinal cord interneuron axon guidance | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 3 | 214 | 5.24e-01 |
| GO:BP | GO:1904515 | positive regulation of TORC2 signaling | 2 | 4 | 5.24e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 6 | 23 | 5.24e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 9 | 523 | 5.24e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 1 | 726 | 5.24e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 2 | 47 | 5.24e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 15 | 63 | 5.24e-01 |
| GO:BP | GO:1905007 | positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 5 | 5.24e-01 |
| GO:BP | GO:0039530 | MDA-5 signaling pathway | 1 | 8 | 5.24e-01 |
| GO:BP | GO:0003032 | detection of oxygen | 1 | 5 | 5.24e-01 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 3 | 9 | 5.24e-01 |
| GO:BP | GO:0097477 | lateral motor column neuron migration | 1 | 3 | 5.24e-01 |
| GO:BP | GO:0002265 | astrocyte activation involved in immune response | 2 | 4 | 5.24e-01 |
| GO:BP | GO:0035846 | oviduct epithelium development | 1 | 2 | 5.24e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 2 | 107 | 5.24e-01 |
| GO:BP | GO:0061966 | establishment of left/right asymmetry | 1 | 3 | 5.24e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 5.24e-01 |
| GO:BP | GO:0099542 | trans-synaptic signaling by endocannabinoid | 2 | 6 | 5.24e-01 |
| GO:BP | GO:0022009 | central nervous system vasculogenesis | 1 | 4 | 5.24e-01 |
| GO:BP | GO:0031449 | regulation of slow-twitch skeletal muscle fiber contraction | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0099541 | trans-synaptic signaling by lipid | 2 | 6 | 5.24e-01 |
| GO:BP | GO:0070778 | L-aspartate transmembrane transport | 2 | 11 | 5.24e-01 |
| GO:BP | GO:1904181 | positive regulation of membrane depolarization | 2 | 8 | 5.24e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 5.24e-01 |
| GO:BP | GO:0019660 | glycolytic fermentation | 1 | 4 | 5.25e-01 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 1 | 4 | 5.25e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 1 | 756 | 5.25e-01 |
| GO:BP | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process | 4 | 14 | 5.25e-01 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 1 | 4 | 5.25e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1 | 47 | 5.25e-01 |
| GO:BP | GO:0060574 | intestinal epithelial cell maturation | 2 | 3 | 5.25e-01 |
| GO:BP | GO:2000490 | negative regulation of hepatic stellate cell activation | 1 | 2 | 5.25e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 14 | 62 | 5.25e-01 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 1 | 4 | 5.25e-01 |
| GO:BP | GO:1901003 | negative regulation of fermentation | 1 | 4 | 5.25e-01 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 7 | 15 | 5.25e-01 |
| GO:BP | GO:0033750 | ribosome localization | 7 | 15 | 5.25e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 1 | 59 | 5.25e-01 |
| GO:BP | GO:0006702 | androgen biosynthetic process | 2 | 4 | 5.25e-01 |
| GO:BP | GO:0045186 | zonula adherens assembly | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 2 | 68 | 5.25e-01 |
| GO:BP | GO:0072680 | extracellular matrix-dependent thymocyte migration | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0034436 | glycoprotein transport | 2 | 3 | 5.25e-01 |
| GO:BP | GO:1905462 | regulation of DNA duplex unwinding | 4 | 7 | 5.25e-01 |
| GO:BP | GO:2000413 | regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0072681 | fibronectin-dependent thymocyte migration | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0048371 | lateral mesodermal cell differentiation | 2 | 2 | 5.25e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 5.25e-01 |
| GO:BP | GO:0071403 | cellular response to high density lipoprotein particle stimulus | 2 | 3 | 5.25e-01 |
| GO:BP | GO:2000415 | positive regulation of fibronectin-dependent thymocyte migration | 1 | 1 | 5.25e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 5.25e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 2 | 12 | 5.26e-01 |
| GO:BP | GO:0071971 | extracellular exosome assembly | 4 | 6 | 5.26e-01 |
| GO:BP | GO:0001554 | luteolysis | 2 | 7 | 5.26e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 4 | 101 | 5.26e-01 |
| GO:BP | GO:0001809 | positive regulation of type IV hypersensitivity | 1 | 1 | 5.26e-01 |
| GO:BP | GO:1904693 | midbrain morphogenesis | 1 | 3 | 5.26e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 1 | 554 | 5.26e-01 |
| GO:BP | GO:0032802 | low-density lipoprotein particle receptor catabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0021501 | prechordal plate formation | 1 | 1 | 5.26e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 1 | 37 | 5.26e-01 |
| GO:BP | GO:0032799 | low-density lipoprotein receptor particle metabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0010166 | wax metabolic process | 1 | 2 | 5.26e-01 |
| GO:BP | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis | 3 | 9 | 5.26e-01 |
| GO:BP | GO:0010025 | wax biosynthetic process | 1 | 2 | 5.26e-01 |
| GO:BP | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process | 2 | 5 | 5.26e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 2 | 12 | 5.26e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 2 | 6 | 5.26e-01 |
| GO:BP | GO:2000387 | regulation of antral ovarian follicle growth | 1 | 1 | 5.26e-01 |
| GO:BP | GO:0016081 | synaptic vesicle docking | 2 | 9 | 5.26e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 2 | 8 | 5.26e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 7 | 12 | 5.26e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 5.26e-01 |
| GO:BP | GO:0070102 | interleukin-6-mediated signaling pathway | 4 | 14 | 5.26e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 4 | 32 | 5.26e-01 |
| GO:BP | GO:0021547 | midbrain-hindbrain boundary initiation | 1 | 1 | 5.26e-01 |
| GO:BP | GO:2000388 | positive regulation of antral ovarian follicle growth | 1 | 1 | 5.26e-01 |
| GO:BP | GO:0035246 | peptidyl-arginine N-methylation | 1 | 4 | 5.26e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 2 | 12 | 5.26e-01 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 1 | 11 | 5.26e-01 |
| GO:BP | GO:0032438 | melanosome organization | 4 | 36 | 5.26e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 5 | 11 | 5.26e-01 |
| GO:BP | GO:0006891 | intra-Golgi vesicle-mediated transport | 5 | 33 | 5.26e-01 |
| GO:BP | GO:2000670 | positive regulation of dendritic cell apoptotic process | 1 | 2 | 5.26e-01 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 4 | 11 | 5.26e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 7 | 19 | 5.26e-01 |
| GO:BP | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration | 1 | 2 | 5.26e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 13 | 159 | 5.26e-01 |
| GO:BP | GO:0035904 | aorta development | 5 | 50 | 5.26e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 11 | 105 | 5.26e-01 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 1 | 4 | 5.26e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 1 | 85 | 5.27e-01 |
| GO:BP | GO:0032978 | protein insertion into membrane from inner side | 2 | 4 | 5.27e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 1 | 123 | 5.27e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 1 | 71 | 5.27e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 15 | 27 | 5.27e-01 |
| GO:BP | GO:0030900 | forebrain development | 3 | 281 | 5.27e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 5.27e-01 |
| GO:BP | GO:0032979 | protein insertion into mitochondrial inner membrane from matrix | 2 | 4 | 5.27e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 7 | 63 | 5.27e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 47 | 469 | 5.27e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 5.27e-01 |
| GO:BP | GO:0045002 | double-strand break repair via single-strand annealing | 4 | 7 | 5.27e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 6 | 59 | 5.27e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 6 | 59 | 5.27e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 5.27e-01 |
| GO:BP | GO:0002383 | immune response in brain or nervous system | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0097281 | immune complex formation | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0016062 | adaptation of rhodopsin mediated signaling | 1 | 1 | 5.27e-01 |
| GO:BP | GO:1990859 | cellular response to endothelin | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0060486 | club cell differentiation | 2 | 3 | 5.27e-01 |
| GO:BP | GO:0061847 | response to cholecystokinin | 1 | 1 | 5.27e-01 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 7 | 36 | 5.27e-01 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 1 | 13 | 5.27e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 2 | 19 | 5.28e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 17 | 35 | 5.28e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 17 | 35 | 5.28e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 5.28e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 5 | 407 | 5.28e-01 |
| GO:BP | GO:0044806 | G-quadruplex DNA unwinding | 4 | 7 | 5.28e-01 |
| GO:BP | GO:0071041 | antisense RNA transcript catabolic process | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0060956 | endocardial cell differentiation | 2 | 4 | 5.28e-01 |
| GO:BP | GO:0070458 | cellular detoxification of nitrogen compound | 2 | 5 | 5.28e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 8 | 16 | 5.28e-01 |
| GO:BP | GO:1904685 | positive regulation of metalloendopeptidase activity | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0033079 | immature T cell proliferation | 2 | 7 | 5.28e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 3 | 24 | 5.28e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 10 | 85 | 5.28e-01 |
| GO:BP | GO:0042868 | antisense RNA metabolic process | 1 | 1 | 5.28e-01 |
| GO:BP | GO:1905774 | regulation of DNA helicase activity | 3 | 5 | 5.28e-01 |
| GO:BP | GO:0090330 | regulation of platelet aggregation | 3 | 21 | 5.28e-01 |
| GO:BP | GO:0048284 | organelle fusion | 2 | 135 | 5.28e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 6 | 11 | 5.28e-01 |
| GO:BP | GO:0072008 | glomerular mesangial cell differentiation | 2 | 4 | 5.28e-01 |
| GO:BP | GO:0048696 | regulation of collateral sprouting in absence of injury | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0071040 | nuclear polyadenylation-dependent antisense transcript catabolic process | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0009450 | gamma-aminobutyric acid catabolic process | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0072143 | mesangial cell development | 2 | 5 | 5.28e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 1 | 30 | 5.28e-01 |
| GO:BP | GO:0050808 | synapse organization | 7 | 365 | 5.28e-01 |
| GO:BP | GO:0031282 | regulation of guanylate cyclase activity | 2 | 3 | 5.28e-01 |
| GO:BP | GO:0021740 | principal sensory nucleus of trigeminal nerve development | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 1 | 19 | 5.28e-01 |
| GO:BP | GO:1904448 | regulation of aspartate secretion | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0048320 | axial mesoderm formation | 1 | 2 | 5.28e-01 |
| GO:BP | GO:1904450 | positive regulation of aspartate secretion | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 1 | 126 | 5.28e-01 |
| GO:BP | GO:0006953 | acute-phase response | 1 | 18 | 5.28e-01 |
| GO:BP | GO:0031118 | rRNA pseudouridine synthesis | 3 | 10 | 5.28e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 5 | 19 | 5.28e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 5 | 11 | 5.28e-01 |
| GO:BP | GO:0051097 | negative regulation of helicase activity | 3 | 5 | 5.28e-01 |
| GO:BP | GO:0033080 | immature T cell proliferation in thymus | 2 | 7 | 5.28e-01 |
| GO:BP | GO:1903209 | positive regulation of oxidative stress-induced cell death | 1 | 13 | 5.28e-01 |
| GO:BP | GO:0014807 | regulation of somitogenesis | 2 | 3 | 5.28e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 1 | 55 | 5.28e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 6 | 10 | 5.28e-01 |
| GO:BP | GO:0006848 | pyruvate transport | 1 | 5 | 5.28e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 1 | 13 | 5.28e-01 |
| GO:BP | GO:0032488 | Cdc42 protein signal transduction | 1 | 10 | 5.28e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 1 | 2 | 5.28e-01 |
| GO:BP | GO:0010712 | regulation of collagen metabolic process | 10 | 34 | 5.28e-01 |
| GO:BP | GO:1901475 | pyruvate transmembrane transport | 1 | 5 | 5.28e-01 |
| GO:BP | GO:0061528 | aspartate secretion | 1 | 2 | 5.28e-01 |
| GO:BP | GO:1990678 | histone H4-K16 deacetylation | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 9 | 46 | 5.28e-01 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 5 | 11 | 5.28e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 17 | 78 | 5.28e-01 |
| GO:BP | GO:0021730 | trigeminal sensory nucleus development | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 5.28e-01 |
| GO:BP | GO:1904826 | regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0044338 | canonical Wnt signaling pathway involved in mesenchymal stem cell differentiation | 1 | 3 | 5.28e-01 |
| GO:BP | GO:0003348 | cardiac endothelial cell differentiation | 2 | 4 | 5.28e-01 |
| GO:BP | GO:1904828 | positive regulation of hydrogen sulfide biosynthetic process | 1 | 1 | 5.28e-01 |
| GO:BP | GO:0030091 | protein repair | 1 | 7 | 5.28e-01 |
| GO:BP | GO:0001756 | somitogenesis | 14 | 48 | 5.28e-01 |
| GO:BP | GO:0051410 | detoxification of nitrogen compound | 2 | 5 | 5.28e-01 |
| GO:BP | GO:0019882 | antigen processing and presentation | 1 | 69 | 5.28e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 5 | 45 | 5.28e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 2 | 41 | 5.28e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 2 | 179 | 5.28e-01 |
| GO:BP | GO:1905765 | negative regulation of protection from non-homologous end joining at telomere | 2 | 3 | 5.28e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 6 | 21 | 5.28e-01 |
| GO:BP | GO:0000055 | ribosomal large subunit export from nucleus | 3 | 8 | 5.28e-01 |
| GO:BP | GO:0051490 | negative regulation of filopodium assembly | 3 | 6 | 5.28e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 4 | 29 | 5.28e-01 |
| GO:BP | GO:0061819 | telomeric DNA-containing double minutes formation | 2 | 3 | 5.28e-01 |
| GO:BP | GO:1905764 | regulation of protection from non-homologous end joining at telomere | 2 | 3 | 5.28e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 2 | 128 | 5.28e-01 |
| GO:BP | GO:0045603 | positive regulation of endothelial cell differentiation | 3 | 8 | 5.29e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 5.29e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 438 | 956 | 5.29e-01 |
| GO:BP | GO:0060938 | epicardium-derived cardiac fibroblast cell differentiation | 2 | 3 | 5.29e-01 |
| GO:BP | GO:0034104 | negative regulation of tissue remodeling | 2 | 13 | 5.29e-01 |
| GO:BP | GO:0106004 | tRNA (guanine-N7)-methylation | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 1 | 8 | 5.29e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 4 | 31 | 5.29e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 3 | 180 | 5.29e-01 |
| GO:BP | GO:0000154 | rRNA modification | 14 | 35 | 5.29e-01 |
| GO:BP | GO:0060939 | epicardium-derived cardiac fibroblast cell development | 2 | 3 | 5.29e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 5.29e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 9 | 893 | 5.29e-01 |
| GO:BP | GO:0007604 | phototransduction, UV | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 5 | 51 | 5.29e-01 |
| GO:BP | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation | 2 | 3 | 5.29e-01 |
| GO:BP | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | 2 | 5 | 5.29e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 3 | 10 | 5.29e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 2 | 2035 | 5.29e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 3 | 101 | 5.29e-01 |
| GO:BP | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib | 3 | 5 | 5.29e-01 |
| GO:BP | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production | 1 | 5 | 5.29e-01 |
| GO:BP | GO:0061789 | dense core granule priming | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 1 | 782 | 5.29e-01 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 4 | 15 | 5.29e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 5 | 151 | 5.29e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 8 | 5.29e-01 |
| GO:BP | GO:0042940 | D-amino acid transport | 1 | 5 | 5.29e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 8 | 441 | 5.29e-01 |
| GO:BP | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0051004 | regulation of lipoprotein lipase activity | 1 | 11 | 5.29e-01 |
| GO:BP | GO:0048308 | organelle inheritance | 8 | 15 | 5.29e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 5.29e-01 |
| GO:BP | GO:0032877 | positive regulation of DNA endoreduplication | 1 | 2 | 5.29e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 5 | 54 | 5.29e-01 |
| GO:BP | GO:0098529 | neuromuscular junction development, skeletal muscle fiber | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0048313 | Golgi inheritance | 8 | 15 | 5.29e-01 |
| GO:BP | GO:0006517 | protein deglycosylation | 2 | 25 | 5.29e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 12 | 116 | 5.29e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 18 | 55 | 5.29e-01 |
| GO:BP | GO:1901073 | glucosamine-containing compound biosynthetic process | 1 | 2 | 5.29e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 1 | 40 | 5.29e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 1 | 104 | 5.29e-01 |
| GO:BP | GO:0035694 | mitochondrial protein catabolic process | 1 | 6 | 5.29e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 5.29e-01 |
| GO:BP | GO:0007416 | synapse assembly | 5 | 152 | 5.29e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 15 | 31 | 5.29e-01 |
| GO:BP | GO:0006045 | N-acetylglucosamine biosynthetic process | 1 | 2 | 5.29e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 1 | 13 | 5.29e-01 |
| GO:BP | GO:0002100 | tRNA wobble adenosine to inosine editing | 1 | 1 | 5.29e-01 |
| GO:BP | GO:0016562 | protein import into peroxisome matrix, receptor recycling | 3 | 7 | 5.29e-01 |
| GO:BP | GO:1904797 | negative regulation of core promoter binding | 1 | 1 | 5.29e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 5 | 326 | 5.29e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 2 | 54 | 5.29e-01 |
| GO:BP | GO:0009409 | response to cold | 9 | 41 | 5.29e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 1 | 67 | 5.29e-01 |
| GO:BP | GO:1903577 | cellular response to L-arginine | 1 | 3 | 5.29e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 9 | 30 | 5.29e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 24 | 47 | 5.29e-01 |
| GO:BP | GO:0048569 | post-embryonic animal organ development | 4 | 15 | 5.30e-01 |
| GO:BP | GO:1905135 | biotin import across plasma membrane | 1 | 2 | 5.30e-01 |
| GO:BP | GO:1990386 | mitotic cleavage furrow ingression | 1 | 1 | 5.30e-01 |
| GO:BP | GO:0001579 | medium-chain fatty acid transport | 1 | 2 | 5.30e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 3 | 5 | 5.30e-01 |
| GO:BP | GO:1901678 | iron coordination entity transport | 1 | 11 | 5.30e-01 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 7 | 16 | 5.30e-01 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 3 | 5 | 5.30e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 2 | 18 | 5.30e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 1 | 581 | 5.30e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 2 | 115 | 5.30e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 9 | 32 | 5.30e-01 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 3 | 5 | 5.30e-01 |
| GO:BP | GO:0042918 | alkanesulfonate transport | 2 | 6 | 5.30e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 13 | 34 | 5.30e-01 |
| GO:BP | GO:0015734 | taurine transport | 2 | 6 | 5.30e-01 |
| GO:BP | GO:0036302 | atrioventricular canal development | 5 | 12 | 5.30e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 46 | 461 | 5.30e-01 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 3 | 5 | 5.30e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 10 | 830 | 5.30e-01 |
| GO:BP | GO:0032745 | positive regulation of interleukin-21 production | 1 | 1 | 5.30e-01 |
| GO:BP | GO:0032625 | interleukin-21 production | 1 | 1 | 5.30e-01 |
| GO:BP | GO:0032665 | regulation of interleukin-21 production | 1 | 1 | 5.30e-01 |
| GO:BP | GO:1903818 | positive regulation of voltage-gated potassium channel activity | 1 | 11 | 5.30e-01 |
| GO:BP | GO:0001841 | neural tube formation | 19 | 101 | 5.30e-01 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 1 | 17 | 5.30e-01 |
| GO:BP | GO:0014821 | phasic smooth muscle contraction | 1 | 12 | 5.30e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 19 | 70 | 5.30e-01 |
| GO:BP | GO:1904003 | negative regulation of sebum secreting cell proliferation | 1 | 1 | 5.30e-01 |
| GO:BP | GO:0003150 | muscular septum morphogenesis | 1 | 4 | 5.31e-01 |
| GO:BP | GO:0001822 | kidney development | 5 | 244 | 5.31e-01 |
| GO:BP | GO:0045453 | bone resorption | 1 | 40 | 5.31e-01 |
| GO:BP | GO:0051106 | positive regulation of DNA ligation | 3 | 5 | 5.31e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 3 | 8 | 5.31e-01 |
| GO:BP | GO:0016237 | lysosomal microautophagy | 1 | 11 | 5.31e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 25 | 67 | 5.31e-01 |
| GO:BP | GO:1903059 | regulation of protein lipidation | 1 | 11 | 5.31e-01 |
| GO:BP | GO:0007566 | embryo implantation | 1 | 39 | 5.31e-01 |
| GO:BP | GO:1903367 | positive regulation of fear response | 2 | 4 | 5.31e-01 |
| GO:BP | GO:0001788 | antibody-dependent cellular cytotoxicity | 2 | 2 | 5.31e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 3 | 23 | 5.31e-01 |
| GO:BP | GO:0051458 | corticotropin secretion | 2 | 4 | 5.31e-01 |
| GO:BP | GO:0035865 | cellular response to potassium ion | 1 | 9 | 5.32e-01 |
| GO:BP | GO:0007258 | JUN phosphorylation | 1 | 2 | 5.32e-01 |
| GO:BP | GO:1905523 | positive regulation of macrophage migration | 2 | 13 | 5.32e-01 |
| GO:BP | GO:0035295 | tube development | 1 | 821 | 5.32e-01 |
| GO:BP | GO:0009110 | vitamin biosynthetic process | 1 | 14 | 5.32e-01 |
| GO:BP | GO:0070572 | positive regulation of neuron projection regeneration | 1 | 8 | 5.32e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 7 | 884 | 5.32e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 22 | 117 | 5.33e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 1 | 21 | 5.33e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 5 | 268 | 5.33e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 5 | 268 | 5.33e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 5.33e-01 |
| GO:BP | GO:2000508 | regulation of dendritic cell chemotaxis | 1 | 3 | 5.33e-01 |
| GO:BP | GO:2000458 | regulation of astrocyte chemotaxis | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0036145 | dendritic cell homeostasis | 1 | 2 | 5.33e-01 |
| GO:BP | GO:0035700 | astrocyte chemotaxis | 1 | 1 | 5.33e-01 |
| GO:BP | GO:0060896 | neural plate pattern specification | 3 | 8 | 5.33e-01 |
| GO:BP | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus | 3 | 6 | 5.33e-01 |
| GO:BP | GO:0048753 | pigment granule organization | 4 | 37 | 5.33e-01 |
| GO:BP | GO:0006422 | aspartyl-tRNA aminoacylation | 1 | 2 | 5.33e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 7 | 26 | 5.33e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 1 | 13 | 5.33e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 2 | 57 | 5.33e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 6 | 23 | 5.34e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 4 | 102 | 5.34e-01 |
| GO:BP | GO:1903463 | regulation of mitotic cell cycle DNA replication | 1 | 2 | 5.34e-01 |
| GO:BP | GO:0002713 | negative regulation of B cell mediated immunity | 4 | 9 | 5.34e-01 |
| GO:BP | GO:0002890 | negative regulation of immunoglobulin mediated immune response | 4 | 9 | 5.34e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 10 | 64 | 5.34e-01 |
| GO:BP | GO:1900099 | negative regulation of plasma cell differentiation | 1 | 1 | 5.34e-01 |
| GO:BP | GO:1904616 | regulation of actin binding | 1 | 3 | 5.34e-01 |
| GO:BP | GO:1901337 | thioester transport | 2 | 2 | 5.34e-01 |
| GO:BP | GO:0015916 | fatty-acyl-CoA transport | 2 | 2 | 5.34e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 48 | 204 | 5.34e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 6 | 23 | 5.34e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 6 | 23 | 5.34e-01 |
| GO:BP | GO:0043248 | proteasome assembly | 1 | 13 | 5.34e-01 |
| GO:BP | GO:0033024 | mast cell apoptotic process | 1 | 3 | 5.34e-01 |
| GO:BP | GO:0033025 | regulation of mast cell apoptotic process | 1 | 3 | 5.34e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 3 | 307 | 5.34e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 1 | 53 | 5.34e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 5.34e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 1 | 52 | 5.34e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 2 | 54 | 5.34e-01 |
| GO:BP | GO:0044070 | regulation of monoatomic anion transport | 7 | 12 | 5.34e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 13 | 127 | 5.34e-01 |
| GO:BP | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion | 2 | 3 | 5.34e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 2 | 94 | 5.35e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 26 | 70 | 5.35e-01 |
| GO:BP | GO:0045725 | positive regulation of glycogen biosynthetic process | 1 | 12 | 5.35e-01 |
| GO:BP | GO:0022600 | digestive system process | 2 | 54 | 5.35e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 15 | 39 | 5.35e-01 |
| GO:BP | GO:0048490 | anterograde synaptic vesicle transport | 2 | 16 | 5.35e-01 |
| GO:BP | GO:0099517 | synaptic vesicle transport along microtubule | 2 | 16 | 5.35e-01 |
| GO:BP | GO:0099514 | synaptic vesicle cytoskeletal transport | 2 | 16 | 5.35e-01 |
| GO:BP | GO:0044528 | regulation of mitochondrial mRNA stability | 2 | 7 | 5.35e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 2 | 21 | 5.35e-01 |
| GO:BP | GO:1903307 | positive regulation of regulated secretory pathway | 1 | 32 | 5.35e-01 |
| GO:BP | GO:0051647 | nucleus localization | 2 | 28 | 5.35e-01 |
| GO:BP | GO:0046337 | phosphatidylethanolamine metabolic process | 3 | 19 | 5.35e-01 |
| GO:BP | GO:0060702 | negative regulation of endoribonuclease activity | 1 | 2 | 5.35e-01 |
| GO:BP | GO:0060701 | negative regulation of ribonuclease activity | 1 | 2 | 5.35e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 30 | 149 | 5.35e-01 |
| GO:BP | GO:0071865 | regulation of apoptotic process in bone marrow cell | 2 | 3 | 5.35e-01 |
| GO:BP | GO:0015712 | hexose phosphate transport | 1 | 3 | 5.35e-01 |
| GO:BP | GO:0015760 | glucose-6-phosphate transport | 1 | 3 | 5.35e-01 |
| GO:BP | GO:0071866 | negative regulation of apoptotic process in bone marrow cell | 2 | 3 | 5.35e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 1 | 105 | 5.35e-01 |
| GO:BP | GO:0061183 | regulation of dermatome development | 1 | 2 | 5.35e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 10 | 47 | 5.35e-01 |
| GO:BP | GO:0061054 | dermatome development | 1 | 2 | 5.35e-01 |
| GO:BP | GO:1905243 | cellular response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 5.35e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 9 | 22 | 5.35e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 5.35e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 3 | 93 | 5.35e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 15 | 59 | 5.36e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 4 | 52 | 5.36e-01 |
| GO:BP | GO:0016561 | protein import into peroxisome matrix, translocation | 2 | 3 | 5.36e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 3 | 18 | 5.36e-01 |
| GO:BP | GO:0048839 | inner ear development | 5 | 132 | 5.36e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 17 | 1869 | 5.36e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 11 | 40 | 5.36e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 2 | 58 | 5.36e-01 |
| GO:BP | GO:0071712 | ER-associated misfolded protein catabolic process | 1 | 12 | 5.36e-01 |
| GO:BP | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process | 1 | 2 | 5.36e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 2 | 149 | 5.36e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 4 | 103 | 5.36e-01 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 3 | 5 | 5.36e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 5.36e-01 |
| GO:BP | GO:0098759 | cellular response to interleukin-8 | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation | 2 | 3 | 5.36e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 1 | 108 | 5.36e-01 |
| GO:BP | GO:0061091 | regulation of phospholipid translocation | 1 | 5 | 5.36e-01 |
| GO:BP | GO:1902259 | regulation of delayed rectifier potassium channel activity | 1 | 12 | 5.36e-01 |
| GO:BP | GO:0099118 | microtubule-based protein transport | 1 | 14 | 5.36e-01 |
| GO:BP | GO:1900221 | regulation of amyloid-beta clearance | 1 | 13 | 5.36e-01 |
| GO:BP | GO:0009258 | 10-formyltetrahydrofolate catabolic process | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0072710 | response to hydroxyurea | 6 | 11 | 5.36e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 1 | 5 | 5.36e-01 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 2 | 10 | 5.36e-01 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 5 | 26 | 5.36e-01 |
| GO:BP | GO:0048853 | forebrain morphogenesis | 3 | 7 | 5.36e-01 |
| GO:BP | GO:0019742 | pentacyclic triterpenoid metabolic process | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0003008 | system process | 3 | 1189 | 5.36e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 5.36e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 6 | 83 | 5.36e-01 |
| GO:BP | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0106028 | neuron projection retraction | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 2 | 18 | 5.36e-01 |
| GO:BP | GO:0097623 | potassium ion export across plasma membrane | 4 | 10 | 5.36e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 6 | 13 | 5.36e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 1 | 25 | 5.36e-01 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 1 | 4 | 5.36e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 2 | 18 | 5.36e-01 |
| GO:BP | GO:0046606 | negative regulation of centrosome cycle | 6 | 12 | 5.36e-01 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 8 | 38 | 5.36e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 6 | 666 | 5.36e-01 |
| GO:BP | GO:0098758 | response to interleukin-8 | 1 | 1 | 5.36e-01 |
| GO:BP | GO:0072584 | caveolin-mediated endocytosis | 5 | 9 | 5.36e-01 |
| GO:BP | GO:0002296 | T-helper 1 cell lineage commitment | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0031529 | ruffle organization | 14 | 45 | 5.36e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 2 | 198 | 5.36e-01 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 8 | 38 | 5.36e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 9 | 21 | 5.36e-01 |
| GO:BP | GO:0060086 | circadian temperature homeostasis | 1 | 3 | 5.36e-01 |
| GO:BP | GO:2001288 | positive regulation of caveolin-mediated endocytosis | 2 | 3 | 5.36e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 1 | 5 | 5.36e-01 |
| GO:BP | GO:0006678 | glucosylceramide metabolic process | 1 | 5 | 5.36e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 4 | 5 | 5.36e-01 |
| GO:BP | GO:0098840 | protein transport along microtubule | 1 | 14 | 5.36e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 5.36e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 2 | 34 | 5.36e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 10 | 204 | 5.36e-01 |
| GO:BP | GO:0060160 | negative regulation of dopamine receptor signaling pathway | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 2 | 14 | 5.36e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 11 | 53 | 5.36e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 2 | 5 | 5.36e-01 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 15 | 67 | 5.36e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 2 | 5 | 5.36e-01 |
| GO:BP | GO:0001759 | organ induction | 1 | 17 | 5.36e-01 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 5 | 26 | 5.36e-01 |
| GO:BP | GO:0048104 | establishment of body hair or bristle planar orientation | 2 | 3 | 5.36e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 2 | 112 | 5.36e-01 |
| GO:BP | GO:0010826 | negative regulation of centrosome duplication | 6 | 12 | 5.36e-01 |
| GO:BP | GO:0048105 | establishment of body hair planar orientation | 2 | 3 | 5.36e-01 |
| GO:BP | GO:2000182 | regulation of progesterone biosynthetic process | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0007584 | response to nutrient | 1 | 109 | 5.36e-01 |
| GO:BP | GO:0072262 | metanephric glomerular mesangial cell proliferation involved in metanephros development | 1 | 2 | 5.36e-01 |
| GO:BP | GO:0046083 | adenine metabolic process | 1 | 3 | 5.36e-01 |
| GO:BP | GO:0090166 | Golgi disassembly | 1 | 6 | 5.37e-01 |
| GO:BP | GO:0045338 | farnesyl diphosphate metabolic process | 2 | 5 | 5.37e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 12 | 23 | 5.37e-01 |
| GO:BP | GO:1904447 | folate import across plasma membrane | 1 | 4 | 5.37e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 5.37e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 1 | 73 | 5.38e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 5.38e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 12 | 43 | 5.38e-01 |
| GO:BP | GO:0050925 | negative regulation of negative chemotaxis | 1 | 2 | 5.38e-01 |
| GO:BP | GO:0120078 | cell adhesion involved in sprouting angiogenesis | 1 | 2 | 5.38e-01 |
| GO:BP | GO:1903352 | L-ornithine transmembrane transport | 1 | 4 | 5.38e-01 |
| GO:BP | GO:0046443 | FAD metabolic process | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0072388 | flavin adenine dinucleotide biosynthetic process | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0120077 | angiogenic sprout fusion | 1 | 2 | 5.38e-01 |
| GO:BP | GO:0097205 | renal filtration | 3 | 23 | 5.38e-01 |
| GO:BP | GO:0006747 | FAD biosynthetic process | 1 | 1 | 5.38e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 10 | 15 | 5.38e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 1 | 43 | 5.38e-01 |
| GO:BP | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 4 | 5.39e-01 |
| GO:BP | GO:0014008 | positive regulation of microglia differentiation | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 2 | 14 | 5.39e-01 |
| GO:BP | GO:1905313 | transforming growth factor beta receptor signaling pathway involved in heart development | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 3 | 215 | 5.39e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 11 | 31 | 5.39e-01 |
| GO:BP | GO:0014006 | regulation of microglia differentiation | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1901080 | regulation of relaxation of smooth muscle | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1901233 | negative regulation of convergent extension involved in axis elongation | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1904891 | positive regulation of excitatory synapse assembly | 1 | 2 | 5.39e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 10 | 21 | 5.39e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 8 | 104 | 5.39e-01 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 1 | 9 | 5.39e-01 |
| GO:BP | GO:1901232 | regulation of convergent extension involved in axis elongation | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1901229 | regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 5.39e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 5.39e-01 |
| GO:BP | GO:1901231 | positive regulation of non-canonical Wnt signaling pathway via JNK cascade | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0015828 | tyrosine transport | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0070460 | thyroid-stimulating hormone secretion | 1 | 2 | 5.39e-01 |
| GO:BP | GO:2001302 | lipoxin A4 metabolic process | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 15 | 32 | 5.39e-01 |
| GO:BP | GO:0006004 | fucose metabolic process | 2 | 10 | 5.39e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 10 | 43 | 5.39e-01 |
| GO:BP | GO:0090168 | Golgi reassembly | 3 | 5 | 5.39e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 2 | 46 | 5.39e-01 |
| GO:BP | GO:0032765 | positive regulation of mast cell cytokine production | 2 | 3 | 5.39e-01 |
| GO:BP | GO:0046066 | dGDP metabolic process | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0046056 | dADP metabolic process | 1 | 1 | 5.39e-01 |
| GO:BP | GO:0046712 | GDP catabolic process | 1 | 2 | 5.39e-01 |
| GO:BP | GO:0019452 | L-cysteine catabolic process to taurine | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 868 | 5.40e-01 |
| GO:BP | GO:0019449 | L-cysteine catabolic process to hypotaurine | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0042635 | positive regulation of hair cycle | 1 | 1 | 5.40e-01 |
| GO:BP | GO:0046533 | negative regulation of photoreceptor cell differentiation | 3 | 3 | 5.40e-01 |
| GO:BP | GO:0072174 | metanephric tubule formation | 3 | 3 | 5.40e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 9 | 144 | 5.40e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 2 | 97 | 5.40e-01 |
| GO:BP | GO:1990264 | peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | 2 | 4 | 5.41e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 3 | 5 | 5.41e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 2 | 154 | 5.41e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 1 | 74 | 5.41e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 7 | 14 | 5.41e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 1 | 45 | 5.42e-01 |
| GO:BP | GO:0007165 | signal transduction | 3 | 3818 | 5.42e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 1 | 34 | 5.42e-01 |
| GO:BP | GO:0016094 | polyprenol biosynthetic process | 1 | 3 | 5.42e-01 |
| GO:BP | GO:0071043 | CUT metabolic process | 1 | 5 | 5.42e-01 |
| GO:BP | GO:1905936 | regulation of germ cell proliferation | 2 | 5 | 5.42e-01 |
| GO:BP | GO:1904180 | negative regulation of membrane depolarization | 2 | 6 | 5.42e-01 |
| GO:BP | GO:0071034 | CUT catabolic process | 1 | 5 | 5.42e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 5 | 43 | 5.42e-01 |
| GO:BP | GO:0048666 | neuron development | 8 | 877 | 5.42e-01 |
| GO:BP | GO:0001775 | cell activation | 1 | 655 | 5.42e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 3 | 4 | 5.42e-01 |
| GO:BP | GO:0048477 | oogenesis | 2 | 64 | 5.42e-01 |
| GO:BP | GO:1904117 | cellular response to vasopressin | 1 | 5 | 5.42e-01 |
| GO:BP | GO:0021943 | formation of radial glial scaffolds | 1 | 3 | 5.42e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 1 | 8 | 5.42e-01 |
| GO:BP | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway | 2 | 3 | 5.42e-01 |
| GO:BP | GO:1904116 | response to vasopressin | 1 | 5 | 5.42e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 1 | 8 | 5.42e-01 |
| GO:BP | GO:0034696 | response to prostaglandin F | 1 | 2 | 5.42e-01 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 6 | 13 | 5.42e-01 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 1 | 11 | 5.42e-01 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 2 | 3 | 5.42e-01 |
| GO:BP | GO:0006621 | protein retention in ER lumen | 4 | 8 | 5.44e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 1 | 13 | 5.44e-01 |
| GO:BP | GO:0006735 | NADH regeneration | 1 | 15 | 5.44e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 2 | 10 | 5.44e-01 |
| GO:BP | GO:0070875 | positive regulation of glycogen metabolic process | 1 | 13 | 5.44e-01 |
| GO:BP | GO:0015742 | alpha-ketoglutarate transport | 1 | 3 | 5.44e-01 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 1 | 15 | 5.44e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 2 | 26 | 5.44e-01 |
| GO:BP | GO:0046321 | positive regulation of fatty acid oxidation | 3 | 15 | 5.44e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 5.44e-01 |
| GO:BP | GO:0061621 | canonical glycolysis | 1 | 15 | 5.44e-01 |
| GO:BP | GO:0051031 | tRNA transport | 3 | 5 | 5.44e-01 |
| GO:BP | GO:0031117 | positive regulation of microtubule depolymerization | 2 | 4 | 5.44e-01 |
| GO:BP | GO:0009838 | abscission | 1 | 6 | 5.44e-01 |
| GO:BP | GO:0014857 | regulation of skeletal muscle cell proliferation | 2 | 10 | 5.44e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 26 | 62 | 5.45e-01 |
| GO:BP | GO:0032025 | response to cobalt ion | 2 | 10 | 5.45e-01 |
| GO:BP | GO:0051900 | regulation of mitochondrial depolarization | 4 | 15 | 5.45e-01 |
| GO:BP | GO:0002184 | cytoplasmic translational termination | 1 | 1 | 5.45e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 17 | 39 | 5.45e-01 |
| GO:BP | GO:0061088 | regulation of sequestering of zinc ion | 2 | 4 | 5.45e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 10 | 88 | 5.45e-01 |
| GO:BP | GO:0035787 | cell migration involved in kidney development | 1 | 5 | 5.45e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 2 | 258 | 5.45e-01 |
| GO:BP | GO:0048852 | diencephalon morphogenesis | 1 | 2 | 5.45e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 1 | 84 | 5.45e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 870 | 5.45e-01 |
| GO:BP | GO:0046039 | GTP metabolic process | 4 | 21 | 5.46e-01 |
| GO:BP | GO:1901142 | insulin metabolic process | 3 | 11 | 5.46e-01 |
| GO:BP | GO:0072001 | renal system development | 5 | 251 | 5.46e-01 |
| GO:BP | GO:0034635 | glutathione transport | 1 | 9 | 5.46e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 14 | 54 | 5.46e-01 |
| GO:BP | GO:0006581 | acetylcholine catabolic process | 2 | 3 | 5.47e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 12 | 45 | 5.47e-01 |
| GO:BP | GO:0021513 | spinal cord dorsal/ventral patterning | 1 | 7 | 5.47e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 3 | 63 | 5.47e-01 |
| GO:BP | GO:0048642 | negative regulation of skeletal muscle tissue development | 2 | 3 | 5.47e-01 |
| GO:BP | GO:0071839 | apoptotic process in bone marrow cell | 3 | 4 | 5.47e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 2 | 12 | 5.47e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 5 | 257 | 5.47e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 2 | 20 | 5.47e-01 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 9 | 18 | 5.48e-01 |
| GO:BP | GO:0080058 | protein deglutathionylation | 1 | 1 | 5.48e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 5 | 421 | 5.48e-01 |
| GO:BP | GO:0060377 | negative regulation of mast cell differentiation | 1 | 1 | 5.48e-01 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 9 | 19 | 5.48e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 12 | 1209 | 5.48e-01 |
| GO:BP | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 6 | 5.48e-01 |
| GO:BP | GO:0003100 | regulation of systemic arterial blood pressure by endothelin | 2 | 5 | 5.48e-01 |
| GO:BP | GO:0008347 | glial cell migration | 2 | 47 | 5.48e-01 |
| GO:BP | GO:0003168 | Purkinje myocyte differentiation | 1 | 1 | 5.48e-01 |
| GO:BP | GO:0070267 | oncosis | 1 | 1 | 5.48e-01 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 6 | 11 | 5.48e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 3 | 34 | 5.48e-01 |
| GO:BP | GO:0061031 | endodermal digestive tract morphogenesis | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 55 | 561 | 5.49e-01 |
| GO:BP | GO:0007624 | ultradian rhythm | 1 | 2 | 5.49e-01 |
| GO:BP | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity | 1 | 3 | 5.49e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 7 | 17 | 5.49e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 9 | 32 | 5.49e-01 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 5 | 11 | 5.49e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 5.49e-01 |
| GO:BP | GO:0030185 | nitric oxide transport | 1 | 2 | 5.49e-01 |
| GO:BP | GO:0009650 | UV protection | 1 | 11 | 5.49e-01 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 8 | 15 | 5.49e-01 |
| GO:BP | GO:1903182 | regulation of SUMO transferase activity | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0006426 | glycyl-tRNA aminoacylation | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 4 | 18 | 5.49e-01 |
| GO:BP | GO:0070150 | mitochondrial glycyl-tRNA aminoacylation | 1 | 1 | 5.49e-01 |
| GO:BP | GO:1903755 | positive regulation of SUMO transferase activity | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0021686 | cerebellar granular layer maturation | 1 | 1 | 5.49e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 2 | 2 | 5.49e-01 |
| GO:BP | GO:0072684 | mitochondrial tRNA 3’-trailer cleavage, endonucleolytic | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0021933 | radial glia guided migration of cerebellar granule cell | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 2 | 3 | 5.49e-01 |
| GO:BP | GO:0046949 | fatty-acyl-CoA biosynthetic process | 5 | 20 | 5.49e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 3 | 555 | 5.49e-01 |
| GO:BP | GO:0033058 | directional locomotion | 2 | 3 | 5.49e-01 |
| GO:BP | GO:0080021 | response to benzoic acid | 1 | 1 | 5.49e-01 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 5 | 16 | 5.50e-01 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 7 | 29 | 5.50e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 25 | 118 | 5.50e-01 |
| GO:BP | GO:1990134 | epithelial cell apoptotic process involved in palatal shelf morphogenesis | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0000350 | generation of catalytic spliceosome for second transesterification step | 1 | 2 | 5.50e-01 |
| GO:BP | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine | 2 | 2 | 5.50e-01 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 3 | 21 | 5.50e-01 |
| GO:BP | GO:0045337 | farnesyl diphosphate biosynthetic process | 1 | 3 | 5.50e-01 |
| GO:BP | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway | 2 | 3 | 5.50e-01 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 2 | 9 | 5.50e-01 |
| GO:BP | GO:0030961 | peptidyl-arginine hydroxylation | 1 | 1 | 5.50e-01 |
| GO:BP | GO:0001963 | synaptic transmission, dopaminergic | 1 | 16 | 5.50e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 5 | 512 | 5.51e-01 |
| GO:BP | GO:1900114 | positive regulation of histone H3-K9 trimethylation | 1 | 2 | 5.51e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 1 | 6 | 5.52e-01 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 1 | 3 | 5.52e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 5 | 21 | 5.52e-01 |
| GO:BP | GO:0045672 | positive regulation of osteoclast differentiation | 1 | 11 | 5.52e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 34 | 95 | 5.52e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 4 | 7 | 5.52e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 29 | 5.52e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 22 | 1681 | 5.52e-01 |
| GO:BP | GO:0009180 | purine ribonucleoside diphosphate biosynthetic process | 3 | 4 | 5.53e-01 |
| GO:BP | GO:0051891 | positive regulation of cardioblast differentiation | 2 | 4 | 5.53e-01 |
| GO:BP | GO:1904831 | positive regulation of aortic smooth muscle cell differentiation | 1 | 2 | 5.53e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 2 | 14 | 5.53e-01 |
| GO:BP | GO:1904829 | regulation of aortic smooth muscle cell differentiation | 1 | 2 | 5.53e-01 |
| GO:BP | GO:0035873 | lactate transmembrane transport | 1 | 5 | 5.53e-01 |
| GO:BP | GO:0015727 | lactate transport | 1 | 5 | 5.53e-01 |
| GO:BP | GO:0018106 | peptidyl-histidine phosphorylation | 1 | 1 | 5.53e-01 |
| GO:BP | GO:0010609 | mRNA localization resulting in post-transcriptional regulation of gene expression | 2 | 2 | 5.53e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 5.53e-01 |
| GO:BP | GO:0006172 | ADP biosynthetic process | 3 | 4 | 5.53e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 5.53e-01 |
| GO:BP | GO:0035887 | aortic smooth muscle cell differentiation | 1 | 2 | 5.53e-01 |
| GO:BP | GO:0000296 | spermine transport | 2 | 3 | 5.53e-01 |
| GO:BP | GO:0007518 | myoblast fate determination | 1 | 1 | 5.53e-01 |
| GO:BP | GO:0045932 | negative regulation of muscle contraction | 1 | 20 | 5.53e-01 |
| GO:BP | GO:0006689 | ganglioside catabolic process | 2 | 5 | 5.53e-01 |
| GO:BP | GO:0043508 | negative regulation of JUN kinase activity | 4 | 13 | 5.54e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 4 | 13 | 5.54e-01 |
| GO:BP | GO:2001198 | regulation of dendritic cell differentiation | 1 | 8 | 5.54e-01 |
| GO:BP | GO:0051931 | regulation of sensory perception | 1 | 13 | 5.54e-01 |
| GO:BP | GO:0051930 | regulation of sensory perception of pain | 1 | 13 | 5.54e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 2 | 38 | 5.54e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 5.54e-01 |
| GO:BP | GO:0070158 | mitochondrial seryl-tRNA aminoacylation | 1 | 1 | 5.54e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 11 | 33 | 5.54e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 1 | 58 | 5.54e-01 |
| GO:BP | GO:0046709 | IDP catabolic process | 1 | 1 | 5.54e-01 |
| GO:BP | GO:0046707 | IDP metabolic process | 1 | 1 | 5.54e-01 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 11 | 19 | 5.55e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 1 | 57 | 5.55e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 2 | 17 | 5.55e-01 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 5.55e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 3 | 23 | 5.55e-01 |
| GO:BP | GO:0031175 | neuron projection development | 7 | 780 | 5.55e-01 |
| GO:BP | GO:0016031 | tRNA import into mitochondrion | 1 | 1 | 5.55e-01 |
| GO:BP | GO:0072189 | ureter development | 5 | 10 | 5.55e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 3 | 77 | 5.55e-01 |
| GO:BP | GO:0002372 | myeloid dendritic cell cytokine production | 1 | 6 | 5.55e-01 |
| GO:BP | GO:0009186 | deoxyribonucleoside diphosphate metabolic process | 3 | 4 | 5.55e-01 |
| GO:BP | GO:0050821 | protein stabilization | 4 | 191 | 5.55e-01 |
| GO:BP | GO:0002735 | positive regulation of myeloid dendritic cell cytokine production | 1 | 6 | 5.55e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 3 | 45 | 5.55e-01 |
| GO:BP | GO:0001545 | primary ovarian follicle growth | 1 | 1 | 5.55e-01 |
| GO:BP | GO:0002733 | regulation of myeloid dendritic cell cytokine production | 1 | 6 | 5.55e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 1 | 17 | 5.55e-01 |
| GO:BP | GO:0002537 | nitric oxide production involved in inflammatory response | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0043095 | regulation of GTP cyclohydrolase I activity | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0043105 | negative regulation of GTP cyclohydrolase I activity | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 1 | 61 | 5.56e-01 |
| GO:BP | GO:1903010 | regulation of bone development | 1 | 6 | 5.56e-01 |
| GO:BP | GO:0006379 | mRNA cleavage | 1 | 19 | 5.56e-01 |
| GO:BP | GO:0090657 | telomeric loop disassembly | 2 | 9 | 5.56e-01 |
| GO:BP | GO:0033689 | negative regulation of osteoblast proliferation | 5 | 10 | 5.56e-01 |
| GO:BP | GO:0032759 | positive regulation of TRAIL production | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0001813 | regulation of antibody-dependent cellular cytotoxicity | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 9 | 70 | 5.56e-01 |
| GO:BP | GO:0032639 | TRAIL production | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 8 | 1074 | 5.56e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 9 | 70 | 5.56e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 8 | 242 | 5.56e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 2 | 15 | 5.56e-01 |
| GO:BP | GO:0032679 | regulation of TRAIL production | 1 | 1 | 5.56e-01 |
| GO:BP | GO:1901837 | negative regulation of transcription of nucleolar large rRNA by RNA polymerase I | 2 | 4 | 5.56e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 1 | 98 | 5.56e-01 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 4 | 8 | 5.56e-01 |
| GO:BP | GO:0099044 | vesicle tethering to endoplasmic reticulum | 1 | 2 | 5.56e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 14 | 45 | 5.56e-01 |
| GO:BP | GO:0030810 | positive regulation of nucleotide biosynthetic process | 3 | 12 | 5.56e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 1 | 14 | 5.56e-01 |
| GO:BP | GO:1905532 | regulation of leucine import across plasma membrane | 1 | 3 | 5.56e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 6 | 30 | 5.56e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 5 | 17 | 5.56e-01 |
| GO:BP | GO:1904055 | negative regulation of cholangiocyte proliferation | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 4 | 16 | 5.56e-01 |
| GO:BP | GO:1900373 | positive regulation of purine nucleotide biosynthetic process | 3 | 12 | 5.56e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 1 | 195 | 5.56e-01 |
| GO:BP | GO:0031642 | negative regulation of myelination | 5 | 8 | 5.56e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 1 | 12 | 5.56e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 34 | 145 | 5.56e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 3 | 47 | 5.56e-01 |
| GO:BP | GO:1903636 | regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 5.56e-01 |
| GO:BP | GO:1903638 | positive regulation of protein insertion into mitochondrial outer membrane | 1 | 1 | 5.56e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 4 | 31 | 5.56e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 2 | 58 | 5.56e-01 |
| GO:BP | GO:0003213 | cardiac right atrium morphogenesis | 2 | 2 | 5.57e-01 |
| GO:BP | GO:0003138 | primary heart field specification | 2 | 3 | 5.57e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 2 | 90 | 5.57e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 3 | 6 | 5.57e-01 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 2 | 3 | 5.57e-01 |
| GO:BP | GO:0043465 | regulation of fermentation | 1 | 5 | 5.57e-01 |
| GO:BP | GO:0006113 | fermentation | 1 | 5 | 5.57e-01 |
| GO:BP | GO:1904692 | positive regulation of type B pancreatic cell proliferation | 1 | 2 | 5.57e-01 |
| GO:BP | GO:2000492 | regulation of interleukin-18-mediated signaling pathway | 1 | 1 | 5.57e-01 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 33 | 74 | 5.57e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 4 | 422 | 5.57e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 27 | 61 | 5.57e-01 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 3 | 9 | 5.57e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 4 | 351 | 5.57e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 5 | 28 | 5.57e-01 |
| GO:BP | GO:0030854 | positive regulation of granulocyte differentiation | 3 | 5 | 5.57e-01 |
| GO:BP | GO:0060197 | cloacal septation | 1 | 2 | 5.57e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 14 | 30 | 5.57e-01 |
| GO:BP | GO:0036155 | acylglycerol acyl-chain remodeling | 2 | 2 | 5.58e-01 |
| GO:BP | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0061107 | seminal vesicle development | 1 | 2 | 5.58e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 3 | 5 | 5.58e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 1 | 389 | 5.59e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 5 | 20 | 5.59e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 1 | 1006 | 5.59e-01 |
| GO:BP | GO:1903527 | positive regulation of membrane tubulation | 2 | 3 | 5.59e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 7 | 38 | 5.59e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 1 | 681 | 5.59e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 5 | 340 | 5.59e-01 |
| GO:BP | GO:0034230 | enkephalin processing | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis | 2 | 6 | 5.59e-01 |
| GO:BP | GO:0032423 | regulation of mismatch repair | 2 | 3 | 5.59e-01 |
| GO:BP | GO:0006627 | protein processing involved in protein targeting to mitochondrion | 2 | 6 | 5.59e-01 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 1 | 10 | 5.59e-01 |
| GO:BP | GO:0043973 | histone H3-K4 acetylation | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0106230 | protein depropionylation | 1 | 2 | 5.59e-01 |
| GO:BP | GO:1903442 | response to lipoic acid | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0048022 | negative regulation of melanin biosynthetic process | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0061698 | protein deglutarylation | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate | 1 | 10 | 5.59e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 9 | 21 | 5.59e-01 |
| GO:BP | GO:0036255 | response to methylamine | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 11 | 1138 | 5.59e-01 |
| GO:BP | GO:0061301 | cerebellum vasculature morphogenesis | 1 | 1 | 5.59e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 7 | 68 | 5.59e-01 |
| GO:BP | GO:0046951 | ketone body biosynthetic process | 2 | 5 | 5.59e-01 |
| GO:BP | GO:0070926 | regulation of ATP:ADP antiporter activity | 1 | 1 | 5.59e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 2 | 28 | 5.59e-01 |
| GO:BP | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | 1 | 4 | 5.59e-01 |
| GO:BP | GO:0036048 | protein desuccinylation | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0009438 | methylglyoxal metabolic process | 3 | 7 | 5.59e-01 |
| GO:BP | GO:0070989 | oxidative demethylation | 4 | 10 | 5.59e-01 |
| GO:BP | GO:0007213 | G protein-coupled acetylcholine receptor signaling pathway | 4 | 14 | 5.59e-01 |
| GO:BP | GO:0036049 | peptidyl-lysine desuccinylation | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0019682 | glyceraldehyde-3-phosphate metabolic process | 2 | 5 | 5.59e-01 |
| GO:BP | GO:0006769 | nicotinamide metabolic process | 1 | 1 | 5.59e-01 |
| GO:BP | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0030195 | negative regulation of blood coagulation | 4 | 34 | 5.59e-01 |
| GO:BP | GO:0099093 | calcium export from the mitochondrion | 1 | 2 | 5.59e-01 |
| GO:BP | GO:0033353 | S-adenosylmethionine cycle | 2 | 7 | 5.59e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 6 | 11 | 5.59e-01 |
| GO:BP | GO:0034058 | endosomal vesicle fusion | 5 | 10 | 5.60e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 11 | 40 | 5.60e-01 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 47 | 103 | 5.60e-01 |
| GO:BP | GO:0032713 | negative regulation of interleukin-4 production | 3 | 5 | 5.60e-01 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 3 | 4 | 5.60e-01 |
| GO:BP | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate | 2 | 4 | 5.60e-01 |
| GO:BP | GO:0060577 | pulmonary vein morphogenesis | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 9 | 49 | 5.60e-01 |
| GO:BP | GO:0036306 | embryonic heart tube elongation | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0035922 | foramen ovale closure | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0140706 | protein-containing complex localization to centriolar satellite | 1 | 1 | 5.60e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 8 | 31 | 5.60e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 2 | 16 | 5.61e-01 |
| GO:BP | GO:0046040 | IMP metabolic process | 8 | 15 | 5.61e-01 |
| GO:BP | GO:0060755 | negative regulation of mast cell chemotaxis | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 6 | 29 | 5.61e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 1 | 134 | 5.61e-01 |
| GO:BP | GO:0016103 | diterpenoid catabolic process | 1 | 1 | 5.61e-01 |
| GO:BP | GO:1902809 | regulation of skeletal muscle fiber differentiation | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0034653 | retinoic acid catabolic process | 1 | 1 | 5.61e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 32 | 90 | 5.61e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 8 | 604 | 5.61e-01 |
| GO:BP | GO:0070564 | positive regulation of vitamin D receptor signaling pathway | 1 | 5 | 5.61e-01 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 1 | 23 | 5.61e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 1 | 76 | 5.62e-01 |
| GO:BP | GO:0035148 | tube formation | 26 | 138 | 5.62e-01 |
| GO:BP | GO:0097370 | protein O-GlcNAcylation via threonine | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0017196 | N-terminal peptidyl-methionine acetylation | 2 | 8 | 5.62e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 6 | 12 | 5.62e-01 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 4 | 12 | 5.62e-01 |
| GO:BP | GO:0051168 | nuclear export | 5 | 150 | 5.62e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 2 | 18 | 5.62e-01 |
| GO:BP | GO:0062232 | prostanoid catabolic process | 2 | 2 | 5.62e-01 |
| GO:BP | GO:0071557 | histone H3-K27 demethylation | 1 | 2 | 5.62e-01 |
| GO:BP | GO:0045988 | negative regulation of striated muscle contraction | 2 | 7 | 5.62e-01 |
| GO:BP | GO:1905344 | prostaglandin catabolic process | 2 | 2 | 5.62e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 12 | 484 | 5.62e-01 |
| GO:BP | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration | 4 | 6 | 5.62e-01 |
| GO:BP | GO:0120179 | adherens junction disassembly | 1 | 1 | 5.62e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 1 | 696 | 5.62e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 7 | 20 | 5.62e-01 |
| GO:BP | GO:0031340 | positive regulation of vesicle fusion | 1 | 9 | 5.62e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 4 | 429 | 5.62e-01 |
| GO:BP | GO:1904746 | negative regulation of apoptotic process involved in development | 1 | 3 | 5.62e-01 |
| GO:BP | GO:1902338 | negative regulation of apoptotic process involved in morphogenesis | 1 | 3 | 5.62e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 11 | 19 | 5.62e-01 |
| GO:BP | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway | 1 | 7 | 5.63e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 2 | 74 | 5.63e-01 |
| GO:BP | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process | 2 | 3 | 5.63e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 3 | 11 | 5.63e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 14 | 31 | 5.63e-01 |
| GO:BP | GO:0038009 | regulation of signal transduction by receptor internalization | 1 | 1 | 5.64e-01 |
| GO:BP | GO:0055024 | regulation of cardiac muscle tissue development | 1 | 3 | 5.64e-01 |
| GO:BP | GO:1900826 | negative regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 1 | 5.64e-01 |
| GO:BP | GO:0048861 | leukemia inhibitory factor signaling pathway | 2 | 6 | 5.64e-01 |
| GO:BP | GO:0051394 | regulation of nerve growth factor receptor activity | 1 | 1 | 5.64e-01 |
| GO:BP | GO:0060299 | negative regulation of sarcomere organization | 1 | 1 | 5.64e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 1 | 700 | 5.64e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 5 | 527 | 5.64e-01 |
| GO:BP | GO:0061140 | lung secretory cell differentiation | 1 | 5 | 5.64e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 6 | 439 | 5.64e-01 |
| GO:BP | GO:0010820 | positive regulation of T cell chemotaxis | 1 | 7 | 5.65e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 1 | 4 | 5.65e-01 |
| GO:BP | GO:1904882 | regulation of telomerase catalytic core complex assembly | 1 | 2 | 5.65e-01 |
| GO:BP | GO:0031034 | myosin filament assembly | 1 | 4 | 5.65e-01 |
| GO:BP | GO:0001501 | skeletal system development | 11 | 377 | 5.65e-01 |
| GO:BP | GO:0007379 | segment specification | 1 | 14 | 5.65e-01 |
| GO:BP | GO:1904884 | positive regulation of telomerase catalytic core complex assembly | 1 | 2 | 5.65e-01 |
| GO:BP | GO:0071635 | negative regulation of transforming growth factor beta production | 5 | 10 | 5.65e-01 |
| GO:BP | GO:1904868 | telomerase catalytic core complex assembly | 1 | 2 | 5.65e-01 |
| GO:BP | GO:0080120 | CAAX-box protein maturation | 1 | 3 | 5.65e-01 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 7 | 12 | 5.65e-01 |
| GO:BP | GO:0050915 | sensory perception of sour taste | 1 | 4 | 5.65e-01 |
| GO:BP | GO:1905755 | protein localization to cytoplasmic microtubule | 1 | 2 | 5.66e-01 |
| GO:BP | GO:1902889 | protein localization to spindle microtubule | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0019276 | UDP-N-acetylgalactosamine metabolic process | 2 | 2 | 5.66e-01 |
| GO:BP | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway | 1 | 7 | 5.66e-01 |
| GO:BP | GO:0010764 | negative regulation of fibroblast migration | 1 | 9 | 5.66e-01 |
| GO:BP | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | 1 | 8 | 5.66e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 10 | 1484 | 5.66e-01 |
| GO:BP | GO:0034105 | positive regulation of tissue remodeling | 1 | 4 | 5.66e-01 |
| GO:BP | GO:0006574 | valine catabolic process | 2 | 4 | 5.66e-01 |
| GO:BP | GO:2001279 | regulation of unsaturated fatty acid biosynthetic process | 3 | 7 | 5.66e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 4 | 285 | 5.66e-01 |
| GO:BP | GO:0042780 | tRNA 3’-end processing | 1 | 7 | 5.66e-01 |
| GO:BP | GO:0060696 | regulation of phospholipid catabolic process | 1 | 5 | 5.66e-01 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 15 | 32 | 5.66e-01 |
| GO:BP | GO:0090387 | phagolysosome assembly involved in apoptotic cell clearance | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 6 | 123 | 5.66e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 5 | 250 | 5.66e-01 |
| GO:BP | GO:1903333 | negative regulation of protein folding | 2 | 4 | 5.66e-01 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 5.66e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 31 | 234 | 5.66e-01 |
| GO:BP | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0090386 | phagosome maturation involved in apoptotic cell clearance | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 1 | 957 | 5.66e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 2 | 9 | 5.66e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 5.66e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 6 | 64 | 5.66e-01 |
| GO:BP | GO:0071528 | tRNA re-export from nucleus | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0035780 | CD80 biosynthetic process | 1 | 1 | 5.66e-01 |
| GO:BP | GO:0035282 | segmentation | 5 | 75 | 5.66e-01 |
| GO:BP | GO:0033280 | response to vitamin D | 3 | 24 | 5.66e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 3 | 8 | 5.66e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 5.66e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 6 | 111 | 5.66e-01 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 1 | 13 | 5.66e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 2 | 8 | 5.66e-01 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 1 | 17 | 5.66e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 3 | 290 | 5.66e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 1 | 15 | 5.66e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 4 | 10 | 5.66e-01 |
| GO:BP | GO:0021511 | spinal cord patterning | 1 | 9 | 5.67e-01 |
| GO:BP | GO:0015822 | ornithine transport | 1 | 5 | 5.67e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 8 | 612 | 5.67e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 40 | 5.67e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 1 | 18 | 5.67e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 5 | 531 | 5.67e-01 |
| GO:BP | GO:0032811 | negative regulation of epinephrine secretion | 1 | 2 | 5.67e-01 |
| GO:BP | GO:0060092 | regulation of synaptic transmission, glycinergic | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 10 | 19 | 5.67e-01 |
| GO:BP | GO:0035624 | receptor transactivation | 1 | 3 | 5.67e-01 |
| GO:BP | GO:0097376 | interneuron axon guidance | 1 | 1 | 5.67e-01 |
| GO:BP | GO:0072278 | metanephric comma-shaped body morphogenesis | 1 | 3 | 5.67e-01 |
| GO:BP | GO:0097476 | spinal cord motor neuron migration | 1 | 3 | 5.67e-01 |
| GO:BP | GO:0036303 | lymph vessel morphogenesis | 5 | 17 | 5.67e-01 |
| GO:BP | GO:0099538 | synaptic signaling via neuropeptide | 1 | 2 | 5.67e-01 |
| GO:BP | GO:1903576 | response to L-arginine | 1 | 3 | 5.67e-01 |
| GO:BP | GO:0018364 | peptidyl-glutamine methylation | 1 | 4 | 5.67e-01 |
| GO:BP | GO:0042554 | superoxide anion generation | 6 | 20 | 5.67e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 6 | 84 | 5.67e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 11 | 1119 | 5.67e-01 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 2 | 10 | 5.68e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 2 | 150 | 5.68e-01 |
| GO:BP | GO:0090214 | spongiotrophoblast layer developmental growth | 1 | 2 | 5.68e-01 |
| GO:BP | GO:0060722 | cell proliferation involved in embryonic placenta development | 1 | 2 | 5.68e-01 |
| GO:BP | GO:0060720 | spongiotrophoblast cell proliferation | 1 | 2 | 5.68e-01 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 2 | 10 | 5.68e-01 |
| GO:BP | GO:1903547 | regulation of growth hormone activity | 1 | 2 | 5.68e-01 |
| GO:BP | GO:0007565 | female pregnancy | 3 | 122 | 5.68e-01 |
| GO:BP | GO:0015825 | L-serine transport | 1 | 7 | 5.68e-01 |
| GO:BP | GO:0010839 | negative regulation of keratinocyte proliferation | 1 | 19 | 5.68e-01 |
| GO:BP | GO:0002432 | granuloma formation | 1 | 1 | 5.68e-01 |
| GO:BP | GO:0036484 | trunk neural crest cell migration | 3 | 4 | 5.69e-01 |
| GO:BP | GO:1900047 | negative regulation of hemostasis | 4 | 34 | 5.69e-01 |
| GO:BP | GO:0035290 | trunk segmentation | 3 | 4 | 5.69e-01 |
| GO:BP | GO:0033624 | negative regulation of integrin activation | 2 | 3 | 5.69e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 1 | 67 | 5.69e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 3 | 158 | 5.69e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 45 | 284 | 5.69e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 1 | 20 | 5.69e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 2 | 3 | 5.69e-01 |
| GO:BP | GO:1904232 | regulation of aconitate hydratase activity | 2 | 2 | 5.70e-01 |
| GO:BP | GO:1904234 | positive regulation of aconitate hydratase activity | 2 | 2 | 5.70e-01 |
| GO:BP | GO:0097694 | establishment of RNA localization to telomere | 2 | 3 | 5.71e-01 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 1 | 5 | 5.71e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 6 | 12 | 5.71e-01 |
| GO:BP | GO:0090208 | positive regulation of triglyceride metabolic process | 1 | 18 | 5.71e-01 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 2 | 60 | 5.71e-01 |
| GO:BP | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation | 1 | 2 | 5.71e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 6 | 12 | 5.71e-01 |
| GO:BP | GO:0097695 | establishment of protein-containing complex localization to telomere | 2 | 3 | 5.71e-01 |
| GO:BP | GO:0090102 | cochlea development | 5 | 35 | 5.71e-01 |
| GO:BP | GO:0033567 | DNA replication, Okazaki fragment processing | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0060914 | heart formation | 7 | 29 | 5.71e-01 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 6 | 27 | 5.71e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 1 | 50 | 5.71e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 21 | 710 | 5.71e-01 |
| GO:BP | GO:0033599 | regulation of mammary gland epithelial cell proliferation | 8 | 14 | 5.71e-01 |
| GO:BP | GO:0072658 | maintenance of protein location in membrane | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0072660 | maintenance of protein location in plasma membrane | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 5.71e-01 |
| GO:BP | GO:0031048 | small ncRNA-mediated heterochromatin formation | 1 | 2 | 5.71e-01 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 2 | 37 | 5.71e-01 |
| GO:BP | GO:1903790 | guanine nucleotide transmembrane transport | 1 | 5 | 5.71e-01 |
| GO:BP | GO:1901423 | response to benzene | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0051646 | mitochondrion localization | 5 | 47 | 5.71e-01 |
| GO:BP | GO:1904710 | positive regulation of granulosa cell apoptotic process | 1 | 1 | 5.71e-01 |
| GO:BP | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0140060 | axon arborization | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0006554 | lysine catabolic process | 1 | 6 | 5.71e-01 |
| GO:BP | GO:0002732 | positive regulation of dendritic cell cytokine production | 3 | 8 | 5.71e-01 |
| GO:BP | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0002943 | tRNA dihydrouridine synthesis | 2 | 4 | 5.71e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 11 | 40 | 5.71e-01 |
| GO:BP | GO:0048865 | stem cell fate commitment | 1 | 5 | 5.71e-01 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 12 | 25 | 5.71e-01 |
| GO:BP | GO:0070729 | cyclic nucleotide transport | 1 | 6 | 5.71e-01 |
| GO:BP | GO:0060346 | bone trabecula formation | 1 | 9 | 5.71e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 2 | 111 | 5.71e-01 |
| GO:BP | GO:0021785 | branchiomotor neuron axon guidance | 3 | 5 | 5.71e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 3 | 22 | 5.71e-01 |
| GO:BP | GO:0051412 | response to corticosterone | 5 | 10 | 5.71e-01 |
| GO:BP | GO:1903847 | regulation of aorta morphogenesis | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0071247 | cellular response to chromate | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0046458 | hexadecanal metabolic process | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 2 | 53 | 5.71e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 11 | 96 | 5.71e-01 |
| GO:BP | GO:0035227 | regulation of glutamate-cysteine ligase activity | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway | 1 | 1 | 5.71e-01 |
| GO:BP | GO:1905396 | cellular response to flavonoid | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 3 | 42 | 5.71e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 5 | 8 | 5.71e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 8 | 41 | 5.71e-01 |
| GO:BP | GO:0019391 | glucuronoside catabolic process | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0035229 | positive regulation of glutamate-cysteine ligase activity | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 6 | 22 | 5.71e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 11 | 40 | 5.71e-01 |
| GO:BP | GO:0003252 | negative regulation of cell proliferation involved in heart valve morphogenesis | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0046687 | response to chromate | 1 | 1 | 5.71e-01 |
| GO:BP | GO:0003169 | coronary vein morphogenesis | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 4 | 69 | 5.71e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 2 | 73 | 5.71e-01 |
| GO:BP | GO:0019389 | glucuronoside metabolic process | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0098749 | cerebellar neuron development | 2 | 3 | 5.71e-01 |
| GO:BP | GO:0060525 | prostate glandular acinus development | 1 | 5 | 5.71e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 1 | 90 | 5.71e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 19 | 2102 | 5.71e-01 |
| GO:BP | GO:1903849 | positive regulation of aorta morphogenesis | 1 | 2 | 5.71e-01 |
| GO:BP | GO:0007409 | axonogenesis | 2 | 359 | 5.71e-01 |
| GO:BP | GO:0006269 | DNA replication, synthesis of RNA primer | 4 | 7 | 5.71e-01 |
| GO:BP | GO:0072383 | plus-end-directed vesicle transport along microtubule | 2 | 6 | 5.71e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 1 | 55 | 5.72e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 5.72e-01 |
| GO:BP | GO:0048148 | behavioral response to cocaine | 3 | 9 | 5.72e-01 |
| GO:BP | GO:0072007 | mesangial cell differentiation | 2 | 5 | 5.72e-01 |
| GO:BP | GO:0035426 | extracellular matrix-cell signaling | 3 | 4 | 5.72e-01 |
| GO:BP | GO:1900125 | regulation of hyaluronan biosynthetic process | 1 | 6 | 5.72e-01 |
| GO:BP | GO:0038001 | paracrine signaling | 1 | 3 | 5.72e-01 |
| GO:BP | GO:0031442 | positive regulation of mRNA 3’-end processing | 4 | 10 | 5.72e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 1 | 64 | 5.72e-01 |
| GO:BP | GO:0051409 | response to nitrosative stress | 4 | 10 | 5.72e-01 |
| GO:BP | GO:2001286 | regulation of caveolin-mediated endocytosis | 3 | 5 | 5.72e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 1 | 1076 | 5.72e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 1 | 3 | 5.72e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 11 | 64 | 5.72e-01 |
| GO:BP | GO:0008300 | isoprenoid catabolic process | 2 | 5 | 5.73e-01 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 5 | 8 | 5.73e-01 |
| GO:BP | GO:0007521 | muscle cell fate determination | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0060789 | hair follicle placode formation | 2 | 5 | 5.73e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 2 | 194 | 5.73e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 1 | 3 | 5.73e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 5.73e-01 |
| GO:BP | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission | 2 | 3 | 5.73e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 41 | 89 | 5.73e-01 |
| GO:BP | GO:1900143 | positive regulation of oligodendrocyte apoptotic process | 1 | 1 | 5.73e-01 |
| GO:BP | GO:0060126 | somatotropin secreting cell differentiation | 1 | 2 | 5.73e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 9 | 43 | 5.73e-01 |
| GO:BP | GO:0016320 | endoplasmic reticulum membrane fusion | 1 | 2 | 5.73e-01 |
| GO:BP | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation | 1 | 6 | 5.73e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 5 | 25 | 5.73e-01 |
| GO:BP | GO:0030070 | insulin processing | 2 | 6 | 5.73e-01 |
| GO:BP | GO:0010526 | retrotransposon silencing | 1 | 3 | 5.73e-01 |
| GO:BP | GO:0035279 | miRNA-mediated gene silencing by mRNA destabilization | 1 | 3 | 5.73e-01 |
| GO:BP | GO:1904587 | response to glycoprotein | 1 | 5 | 5.73e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 4 | 16 | 5.73e-01 |
| GO:BP | GO:0018057 | peptidyl-lysine oxidation | 3 | 5 | 5.73e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 11 | 77 | 5.73e-01 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 7 | 19 | 5.74e-01 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 8 | 13 | 5.74e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 1 | 20 | 5.74e-01 |
| GO:BP | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine | 2 | 5 | 5.74e-01 |
| GO:BP | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process | 1 | 3 | 5.74e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 5 | 28 | 5.74e-01 |
| GO:BP | GO:0036075 | replacement ossification | 3 | 25 | 5.74e-01 |
| GO:BP | GO:0071500 | cellular response to nitrosative stress | 2 | 6 | 5.74e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 3 | 25 | 5.74e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 1 | 50 | 5.74e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 4 | 18 | 5.74e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 2 | 4 | 5.74e-01 |
| GO:BP | GO:1903939 | regulation of TORC2 signaling | 2 | 5 | 5.74e-01 |
| GO:BP | GO:0048319 | axial mesoderm morphogenesis | 1 | 3 | 5.74e-01 |
| GO:BP | GO:0042797 | tRNA transcription by RNA polymerase III | 1 | 7 | 5.74e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 29 | 1137 | 5.74e-01 |
| GO:BP | GO:0010819 | regulation of T cell chemotaxis | 1 | 7 | 5.74e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 5.74e-01 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 1 | 17 | 5.74e-01 |
| GO:BP | GO:0051258 | protein polymerization | 1 | 241 | 5.74e-01 |
| GO:BP | GO:1990786 | cellular response to dsDNA | 1 | 2 | 5.74e-01 |
| GO:BP | GO:1990784 | response to dsDNA | 1 | 2 | 5.74e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 5.74e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 5.74e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 10 | 39 | 5.74e-01 |
| GO:BP | GO:0009234 | menaquinone biosynthetic process | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 1 | 15 | 5.74e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 5.74e-01 |
| GO:BP | GO:0006560 | proline metabolic process | 3 | 7 | 5.74e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 3 | 48 | 5.74e-01 |
| GO:BP | GO:0097742 | de novo centriole assembly | 3 | 4 | 5.74e-01 |
| GO:BP | GO:0032194 | ubiquinone biosynthetic process via 3,4-dihydroxy-5-polyprenylbenzoate | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0097332 | response to antipsychotic drug | 1 | 2 | 5.74e-01 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 3 | 4 | 5.74e-01 |
| GO:BP | GO:2001170 | negative regulation of ATP biosynthetic process | 2 | 3 | 5.74e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 2 | 25 | 5.74e-01 |
| GO:BP | GO:1903516 | regulation of single strand break repair | 2 | 3 | 5.74e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 5.74e-01 |
| GO:BP | GO:0042371 | vitamin K biosynthetic process | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0043111 | replication fork arrest | 2 | 2 | 5.74e-01 |
| GO:BP | GO:1903811 | L-asparagine import across plasma membrane | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0060718 | chorionic trophoblast cell differentiation | 2 | 5 | 5.74e-01 |
| GO:BP | GO:2000645 | negative regulation of receptor catabolic process | 1 | 3 | 5.74e-01 |
| GO:BP | GO:0036364 | transforming growth factor beta1 activation | 1 | 1 | 5.74e-01 |
| GO:BP | GO:0045056 | transcytosis | 5 | 17 | 5.75e-01 |
| GO:BP | GO:0006829 | zinc ion transport | 3 | 23 | 5.75e-01 |
| GO:BP | GO:0031393 | negative regulation of prostaglandin biosynthetic process | 1 | 2 | 5.75e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 9 | 34 | 5.75e-01 |
| GO:BP | GO:2000833 | positive regulation of steroid hormone secretion | 1 | 6 | 5.75e-01 |
| GO:BP | GO:0070836 | caveola assembly | 2 | 4 | 5.75e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 24 | 55 | 5.75e-01 |
| GO:BP | GO:0016124 | xanthophyll catabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 3 | 18 | 5.75e-01 |
| GO:BP | GO:0030581 | symbiont intracellular protein transport in host | 1 | 2 | 5.75e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 4 | 39 | 5.75e-01 |
| GO:BP | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic active zone membrane | 1 | 8 | 5.75e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 4 | 7 | 5.75e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 10 | 803 | 5.75e-01 |
| GO:BP | GO:0016110 | tetraterpenoid catabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0019060 | intracellular transport of viral protein in host cell | 1 | 2 | 5.75e-01 |
| GO:BP | GO:1903703 | enterocyte differentiation | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 2 | 24 | 5.75e-01 |
| GO:BP | GO:1901175 | lycopene metabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0000098 | sulfur amino acid catabolic process | 1 | 7 | 5.75e-01 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 3 | 5 | 5.75e-01 |
| GO:BP | GO:0045344 | negative regulation of MHC class I biosynthetic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0016118 | carotenoid catabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 1 | 55 | 5.75e-01 |
| GO:BP | GO:1901826 | zeaxanthin catabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 5 | 32 | 5.75e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 4 | 7 | 5.75e-01 |
| GO:BP | GO:0010915 | regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 5.75e-01 |
| GO:BP | GO:0010916 | negative regulation of very-low-density lipoprotein particle clearance | 1 | 2 | 5.75e-01 |
| GO:BP | GO:1901176 | lycopene catabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0062172 | lutein catabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:1901632 | regulation of synaptic vesicle membrane organization | 1 | 8 | 5.75e-01 |
| GO:BP | GO:0071283 | cellular response to iron(III) ion | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 2 | 137 | 5.75e-01 |
| GO:BP | GO:1903430 | negative regulation of cell maturation | 1 | 4 | 5.75e-01 |
| GO:BP | GO:0062170 | lutein metabolic process | 1 | 1 | 5.75e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 8 | 19 | 5.75e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 5.76e-01 |
| GO:BP | GO:0071321 | cellular response to cGMP | 2 | 7 | 5.76e-01 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 17 | 37 | 5.76e-01 |
| GO:BP | GO:1905089 | regulation of parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization | 1 | 2 | 5.77e-01 |
| GO:BP | GO:0031652 | positive regulation of heat generation | 2 | 4 | 5.77e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 4 | 150 | 5.77e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 96 | 5.77e-01 |
| GO:BP | GO:0006500 | N-terminal protein palmitoylation | 2 | 3 | 5.78e-01 |
| GO:BP | GO:0045047 | protein targeting to ER | 3 | 49 | 5.78e-01 |
| GO:BP | GO:0021999 | neural plate anterior/posterior regionalization | 2 | 5 | 5.78e-01 |
| GO:BP | GO:2000359 | regulation of binding of sperm to zona pellucida | 2 | 2 | 5.78e-01 |
| GO:BP | GO:0001806 | type IV hypersensitivity | 2 | 2 | 5.78e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 13 | 91 | 5.78e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 1014 | 5.78e-01 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 1 | 6 | 5.78e-01 |
| GO:BP | GO:0015677 | copper ion import | 1 | 5 | 5.78e-01 |
| GO:BP | GO:0061034 | olfactory bulb mitral cell layer development | 1 | 2 | 5.78e-01 |
| GO:BP | GO:0030327 | prenylated protein catabolic process | 1 | 3 | 5.78e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 1 | 2 | 5.78e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 1 | 9 | 5.78e-01 |
| GO:BP | GO:0023052 | signaling | 3 | 4141 | 5.78e-01 |
| GO:BP | GO:1901492 | positive regulation of lymphangiogenesis | 1 | 2 | 5.79e-01 |
| GO:BP | GO:1904205 | negative regulation of skeletal muscle hypertrophy | 1 | 1 | 5.79e-01 |
| GO:BP | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis | 1 | 3 | 5.79e-01 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 1 | 22 | 5.79e-01 |
| GO:BP | GO:2000643 | positive regulation of early endosome to late endosome transport | 3 | 7 | 5.79e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 36 | 116 | 5.79e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 9 | 1111 | 5.79e-01 |
| GO:BP | GO:0070925 | organelle assembly | 7 | 817 | 5.79e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 2 | 2 | 5.79e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 2 | 2 | 5.79e-01 |
| GO:BP | GO:0043473 | pigmentation | 9 | 92 | 5.79e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 2 | 2 | 5.79e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 9 | 51 | 5.79e-01 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 6 | 23 | 5.79e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 2 | 12 | 5.79e-01 |
| GO:BP | GO:0099527 | postsynapse to nucleus signaling pathway | 1 | 6 | 5.79e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2 | 3625 | 5.79e-01 |
| GO:BP | GO:0046511 | sphinganine biosynthetic process | 1 | 2 | 5.79e-01 |
| GO:BP | GO:0036148 | phosphatidylglycerol acyl-chain remodeling | 1 | 5 | 5.79e-01 |
| GO:BP | GO:0033385 | geranylgeranyl diphosphate metabolic process | 1 | 2 | 5.79e-01 |
| GO:BP | GO:0072236 | metanephric loop of Henle development | 1 | 3 | 5.79e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 1 | 11 | 5.79e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 2 | 22 | 5.79e-01 |
| GO:BP | GO:1900453 | negative regulation of long-term synaptic depression | 1 | 3 | 5.80e-01 |
| GO:BP | GO:1901252 | regulation of intracellular transport of viral material | 1 | 2 | 5.80e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 5.80e-01 |
| GO:BP | GO:1903608 | protein localization to cytoplasmic stress granule | 1 | 7 | 5.80e-01 |
| GO:BP | GO:0002726 | positive regulation of T cell cytokine production | 1 | 14 | 5.80e-01 |
| GO:BP | GO:0097350 | neutrophil clearance | 1 | 5 | 5.80e-01 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 5.80e-01 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 3 | 6 | 5.80e-01 |
| GO:BP | GO:0098930 | axonal transport | 3 | 60 | 5.80e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 6 | 49 | 5.80e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 6 | 11 | 5.80e-01 |
| GO:BP | GO:0032782 | bile acid secretion | 1 | 5 | 5.80e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 10 | 1015 | 5.80e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 13 | 62 | 5.80e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 1 | 55 | 5.80e-01 |
| GO:BP | GO:1902645 | tertiary alcohol biosynthetic process | 2 | 8 | 5.80e-01 |
| GO:BP | GO:0034651 | cortisol biosynthetic process | 2 | 8 | 5.80e-01 |
| GO:BP | GO:0021502 | neural fold elevation formation | 1 | 3 | 5.80e-01 |
| GO:BP | GO:0002883 | regulation of hypersensitivity | 3 | 5 | 5.80e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 2 | 14 | 5.80e-01 |
| GO:BP | GO:0003056 | regulation of vascular associated smooth muscle contraction | 3 | 8 | 5.80e-01 |
| GO:BP | GO:0032353 | negative regulation of hormone biosynthetic process | 2 | 8 | 5.80e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 3 | 174 | 5.80e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 5 | 551 | 5.80e-01 |
| GO:BP | GO:0002115 | store-operated calcium entry | 2 | 15 | 5.80e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 5.80e-01 |
| GO:BP | GO:0032197 | retrotransposition | 2 | 4 | 5.80e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 7 | 41 | 5.80e-01 |
| GO:BP | GO:0000266 | mitochondrial fission | 4 | 38 | 5.80e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 11 | 56 | 5.80e-01 |
| GO:BP | GO:0048280 | vesicle fusion with Golgi apparatus | 1 | 8 | 5.80e-01 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 5 | 10 | 5.80e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 10 | 1017 | 5.80e-01 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 5 | 10 | 5.80e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 21 | 58 | 5.80e-01 |
| GO:BP | GO:1901340 | negative regulation of store-operated calcium channel activity | 1 | 1 | 5.80e-01 |
| GO:BP | GO:0060633 | negative regulation of transcription initiation by RNA polymerase II | 1 | 6 | 5.81e-01 |
| GO:BP | GO:0045720 | negative regulation of integrin biosynthetic process | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 2 | 19 | 5.81e-01 |
| GO:BP | GO:0018993 | somatic sex determination | 1 | 1 | 5.81e-01 |
| GO:BP | GO:1900364 | negative regulation of mRNA polyadenylation | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0006041 | glucosamine metabolic process | 2 | 3 | 5.81e-01 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 3 | 7 | 5.81e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 13 | 482 | 5.81e-01 |
| GO:BP | GO:1903044 | protein localization to membrane raft | 1 | 8 | 5.81e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 2 | 7 | 5.81e-01 |
| GO:BP | GO:0002678 | positive regulation of chronic inflammatory response | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 1 | 19 | 5.81e-01 |
| GO:BP | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 5.81e-01 |
| GO:BP | GO:0150045 | regulation of synaptic signaling by nitric oxide | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0033023 | mast cell homeostasis | 1 | 4 | 5.81e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 1 | 1075 | 5.81e-01 |
| GO:BP | GO:2000124 | regulation of endocannabinoid signaling pathway | 1 | 3 | 5.81e-01 |
| GO:BP | GO:0021541 | ammon gyrus development | 1 | 1 | 5.81e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 6 | 60 | 5.81e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 14 | 64 | 5.81e-01 |
| GO:BP | GO:0140192 | regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 5.81e-01 |
| GO:BP | GO:0071475 | cellular hyperosmotic salinity response | 2 | 6 | 5.82e-01 |
| GO:BP | GO:1904429 | regulation of t-circle formation | 2 | 5 | 5.82e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 13 | 33 | 5.82e-01 |
| GO:BP | GO:0071305 | cellular response to vitamin D | 2 | 14 | 5.82e-01 |
| GO:BP | GO:0007542 | primary sex determination, germ-line | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0019100 | male germ-line sex determination | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0097494 | regulation of vesicle size | 7 | 19 | 5.82e-01 |
| GO:BP | GO:0014036 | neural crest cell fate specification | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 2 | 75 | 5.82e-01 |
| GO:BP | GO:0035622 | intrahepatic bile duct development | 1 | 2 | 5.82e-01 |
| GO:BP | GO:1904864 | negative regulation of beta-catenin-TCF complex assembly | 1 | 2 | 5.82e-01 |
| GO:BP | GO:1904863 | regulation of beta-catenin-TCF complex assembly | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0106381 | purine deoxyribonucleotide salvage | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0060532 | bronchus cartilage development | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0006170 | dAMP biosynthetic process | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0009171 | purine deoxyribonucleoside monophosphate biosynthetic process | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0060516 | primary prostatic bud elongation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0044357 | regulation of rRNA stability | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0061145 | lung smooth muscle development | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 5.82e-01 |
| GO:BP | GO:0106383 | dAMP salvage | 2 | 3 | 5.82e-01 |
| GO:BP | GO:1902374 | regulation of rRNA catabolic process | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 2 | 47 | 5.82e-01 |
| GO:BP | GO:0018992 | germ-line sex determination | 1 | 2 | 5.82e-01 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 2 | 5 | 5.82e-01 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 4 | 45 | 5.82e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 2 | 16 | 5.82e-01 |
| GO:BP | GO:0048200 | Golgi transport vesicle coating | 1 | 6 | 5.82e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 4 | 32 | 5.82e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 32 | 68 | 5.82e-01 |
| GO:BP | GO:0033088 | negative regulation of immature T cell proliferation in thymus | 1 | 3 | 5.82e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 6 | 15 | 5.82e-01 |
| GO:BP | GO:2000723 | negative regulation of cardiac vascular smooth muscle cell differentiation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 19 | 44 | 5.82e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 1 | 63 | 5.82e-01 |
| GO:BP | GO:1902216 | positive regulation of interleukin-4-mediated signaling pathway | 1 | 1 | 5.82e-01 |
| GO:BP | GO:2000800 | regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0043977 | histone H2A-K5 acetylation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0002071 | glandular epithelial cell maturation | 4 | 7 | 5.82e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 5 | 18 | 5.82e-01 |
| GO:BP | GO:0043980 | histone H2B-K12 acetylation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 14 | 46 | 5.82e-01 |
| GO:BP | GO:0048205 | COPI coating of Golgi vesicle | 1 | 6 | 5.82e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 5 | 47 | 5.82e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 1 | 1088 | 5.82e-01 |
| GO:BP | GO:0097502 | mannosylation | 4 | 32 | 5.82e-01 |
| GO:BP | GO:0140049 | regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 5.82e-01 |
| GO:BP | GO:2000802 | positive regulation of endocardial cushion to mesenchymal transition involved in heart valve formation | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0140051 | positive regulation of endocardial cushion to mesenchymal transition | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0097039 | protein linear polyubiquitination | 2 | 5 | 5.82e-01 |
| GO:BP | GO:0033087 | negative regulation of immature T cell proliferation | 1 | 3 | 5.82e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 10 | 809 | 5.82e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 1 | 47 | 5.82e-01 |
| GO:BP | GO:0006669 | sphinganine-1-phosphate biosynthetic process | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0035964 | COPI-coated vesicle budding | 1 | 6 | 5.82e-01 |
| GO:BP | GO:0045887 | positive regulation of synaptic assembly at neuromuscular junction | 1 | 1 | 5.82e-01 |
| GO:BP | GO:0019254 | carnitine metabolic process, CoA-linked | 2 | 3 | 5.82e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 5 | 10 | 5.83e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 5 | 10 | 5.83e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 3 | 43 | 5.83e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 4 | 34 | 5.83e-01 |
| GO:BP | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 2 | 3 | 5.83e-01 |
| GO:BP | GO:0016477 | cell migration | 3 | 1081 | 5.83e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 2 | 13 | 5.83e-01 |
| GO:BP | GO:0042724 | thiamine-containing compound biosynthetic process | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0016076 | snRNA catabolic process | 2 | 5 | 5.83e-01 |
| GO:BP | GO:0090400 | stress-induced premature senescence | 2 | 7 | 5.83e-01 |
| GO:BP | GO:0021537 | telencephalon development | 2 | 201 | 5.83e-01 |
| GO:BP | GO:0009229 | thiamine diphosphate biosynthetic process | 1 | 1 | 5.83e-01 |
| GO:BP | GO:0060840 | artery development | 7 | 83 | 5.83e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 1 | 31 | 5.83e-01 |
| GO:BP | GO:0014862 | regulation of skeletal muscle contraction by chemo-mechanical energy conversion | 1 | 3 | 5.83e-01 |
| GO:BP | GO:0002309 | T cell proliferation involved in immune response | 1 | 3 | 5.83e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 1 | 26 | 5.83e-01 |
| GO:BP | GO:2000467 | positive regulation of glycogen (starch) synthase activity | 2 | 4 | 5.83e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 5.83e-01 |
| GO:BP | GO:0042670 | retinal cone cell differentiation | 3 | 6 | 5.83e-01 |
| GO:BP | GO:0031444 | slow-twitch skeletal muscle fiber contraction | 1 | 2 | 5.83e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 5.83e-01 |
| GO:BP | GO:0014728 | regulation of the force of skeletal muscle contraction | 1 | 3 | 5.83e-01 |
| GO:BP | GO:0061890 | positive regulation of astrocyte activation | 1 | 1 | 5.84e-01 |
| GO:BP | GO:0045346 | regulation of MHC class II biosynthetic process | 7 | 11 | 5.84e-01 |
| GO:BP | GO:0006116 | NADH oxidation | 2 | 4 | 5.84e-01 |
| GO:BP | GO:0098967 | exocytic insertion of neurotransmitter receptor to postsynaptic membrane | 1 | 5 | 5.85e-01 |
| GO:BP | GO:1990828 | hepatocyte dedifferentiation | 1 | 1 | 5.85e-01 |
| GO:BP | GO:1904586 | cellular response to putrescine | 1 | 1 | 5.85e-01 |
| GO:BP | GO:1904585 | response to putrescine | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 7 | 81 | 5.85e-01 |
| GO:BP | GO:0060558 | regulation of calcidiol 1-monooxygenase activity | 1 | 2 | 5.85e-01 |
| GO:BP | GO:0006072 | glycerol-3-phosphate metabolic process | 4 | 7 | 5.85e-01 |
| GO:BP | GO:0007154 | cell communication | 3 | 4226 | 5.85e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 8 | 258 | 5.85e-01 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 8 | 23 | 5.85e-01 |
| GO:BP | GO:0036285 | SAGA complex assembly | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0002445 | type II hypersensitivity | 2 | 3 | 5.85e-01 |
| GO:BP | GO:0032071 | regulation of endodeoxyribonuclease activity | 1 | 7 | 5.85e-01 |
| GO:BP | GO:0001794 | type IIa hypersensitivity | 2 | 3 | 5.85e-01 |
| GO:BP | GO:0060065 | uterus development | 1 | 13 | 5.85e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 3 | 34 | 5.85e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 5.85e-01 |
| GO:BP | GO:1903937 | response to acrylamide | 1 | 1 | 5.85e-01 |
| GO:BP | GO:0006685 | sphingomyelin catabolic process | 1 | 6 | 5.85e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 4 | 5 | 5.85e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 1 | 15 | 5.85e-01 |
| GO:BP | GO:0071360 | cellular response to exogenous dsRNA | 5 | 14 | 5.85e-01 |
| GO:BP | GO:0042354 | L-fucose metabolic process | 2 | 4 | 5.86e-01 |
| GO:BP | GO:1904251 | regulation of bile acid metabolic process | 1 | 9 | 5.86e-01 |
| GO:BP | GO:0051796 | negative regulation of timing of catagen | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 8 | 16 | 5.86e-01 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 5 | 14 | 5.86e-01 |
| GO:BP | GO:0120127 | response to zinc ion starvation | 3 | 6 | 5.86e-01 |
| GO:BP | GO:0060901 | regulation of hair cycle by canonical Wnt signaling pathway | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0032808 | lacrimal gland development | 1 | 5 | 5.86e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 6 | 15 | 5.86e-01 |
| GO:BP | GO:0003400 | regulation of COPII vesicle coating | 2 | 3 | 5.86e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 2 | 49 | 5.86e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 2 | 301 | 5.86e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 6 | 51 | 5.86e-01 |
| GO:BP | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 3 | 5.86e-01 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 1 | 8 | 5.86e-01 |
| GO:BP | GO:0034224 | cellular response to zinc ion starvation | 3 | 6 | 5.86e-01 |
| GO:BP | GO:0050655 | dermatan sulfate proteoglycan metabolic process | 2 | 12 | 5.86e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 5 | 452 | 5.86e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 1 | 35 | 5.86e-01 |
| GO:BP | GO:1902910 | positive regulation of melanosome transport | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0014852 | regulation of skeletal muscle contraction by neural stimulation via neuromuscular junction | 1 | 2 | 5.86e-01 |
| GO:BP | GO:0060010 | Sertoli cell fate commitment | 1 | 1 | 5.86e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 7 | 45 | 5.86e-01 |
| GO:BP | GO:1903401 | L-lysine transmembrane transport | 1 | 6 | 5.86e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 9 | 37 | 5.86e-01 |
| GO:BP | GO:1902022 | L-lysine transport | 1 | 6 | 5.86e-01 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 2 | 17 | 5.86e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 25 | 51 | 5.86e-01 |
| GO:BP | GO:0006541 | glutamine metabolic process | 12 | 24 | 5.87e-01 |
| GO:BP | GO:0071105 | response to interleukin-11 | 1 | 1 | 5.87e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 2 | 19 | 5.87e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 10 | 1026 | 5.87e-01 |
| GO:BP | GO:0035261 | external genitalia morphogenesis | 1 | 1 | 5.87e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 1 | 22 | 5.87e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 1 | 70 | 5.87e-01 |
| GO:BP | GO:0061780 | mitotic cohesin loading | 1 | 1 | 5.87e-01 |
| GO:BP | GO:0072046 | establishment of planar polarity involved in nephron morphogenesis | 1 | 1 | 5.87e-01 |
| GO:BP | GO:2000435 | negative regulation of protein neddylation | 1 | 4 | 5.88e-01 |
| GO:BP | GO:1905702 | regulation of inhibitory synapse assembly | 1 | 3 | 5.88e-01 |
| GO:BP | GO:2000436 | positive regulation of protein neddylation | 1 | 5 | 5.88e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 3 | 29 | 5.88e-01 |
| GO:BP | GO:0010033 | response to organic substance | 20 | 1957 | 5.88e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 5.88e-01 |
| GO:BP | GO:1903598 | positive regulation of gap junction assembly | 2 | 7 | 5.88e-01 |
| GO:BP | GO:0034463 | 90S preribosome assembly | 1 | 1 | 5.89e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 1 | 877 | 5.89e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 2 | 7 | 5.89e-01 |
| GO:BP | GO:1990922 | hepatic stellate cell proliferation | 1 | 1 | 5.89e-01 |
| GO:BP | GO:2000763 | positive regulation of transcription from RNA polymerase II promoter involved in norepinephrine biosynthetic process | 1 | 1 | 5.89e-01 |
| GO:BP | GO:1904899 | positive regulation of hepatic stellate cell proliferation | 1 | 1 | 5.89e-01 |
| GO:BP | GO:2000764 | positive regulation of semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 5.89e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 4 | 44 | 5.89e-01 |
| GO:BP | GO:1904897 | regulation of hepatic stellate cell proliferation | 1 | 1 | 5.89e-01 |
| GO:BP | GO:0071527 | semaphorin-plexin signaling pathway involved in outflow tract morphogenesis | 1 | 1 | 5.89e-01 |
| GO:BP | GO:2001262 | positive regulation of semaphorin-plexin signaling pathway | 1 | 1 | 5.89e-01 |
| GO:BP | GO:1904425 | negative regulation of GTP binding | 2 | 3 | 5.89e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 3 | 33 | 5.90e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 1 | 8 | 5.90e-01 |
| GO:BP | GO:0010637 | negative regulation of mitochondrial fusion | 3 | 8 | 5.90e-01 |
| GO:BP | GO:0033189 | response to vitamin A | 2 | 12 | 5.90e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 3 | 29 | 5.90e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 2 | 222 | 5.90e-01 |
| GO:BP | GO:0032695 | negative regulation of interleukin-12 production | 1 | 6 | 5.90e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 1 | 22 | 5.91e-01 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 4 | 17 | 5.91e-01 |
| GO:BP | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0016095 | polyprenol catabolic process | 1 | 2 | 5.91e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 12 | 27 | 5.91e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 14 | 137 | 5.91e-01 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 1 | 12 | 5.92e-01 |
| GO:BP | GO:1904990 | regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 12 | 26 | 5.92e-01 |
| GO:BP | GO:1904925 | positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization | 5 | 14 | 5.92e-01 |
| GO:BP | GO:0009812 | flavonoid metabolic process | 1 | 4 | 5.92e-01 |
| GO:BP | GO:0022414 | reproductive process | 1 | 927 | 5.92e-01 |
| GO:BP | GO:1904992 | positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 1 | 5.92e-01 |
| GO:BP | GO:0035437 | maintenance of protein localization in endoplasmic reticulum | 5 | 11 | 5.92e-01 |
| GO:BP | GO:0006096 | glycolytic process | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 5 | 11 | 5.92e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 4 | 38 | 5.92e-01 |
| GO:BP | GO:0071926 | endocannabinoid signaling pathway | 2 | 5 | 5.92e-01 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 9 | 19 | 5.92e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 1 | 59 | 5.92e-01 |
| GO:BP | GO:0035771 | interleukin-4-mediated signaling pathway | 2 | 4 | 5.92e-01 |
| GO:BP | GO:0010738 | regulation of protein kinase A signaling | 1 | 13 | 5.92e-01 |
| GO:BP | GO:2000738 | positive regulation of stem cell differentiation | 5 | 17 | 5.92e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 1 | 23 | 5.92e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 11 | 26 | 5.92e-01 |
| GO:BP | GO:0018146 | keratan sulfate biosynthetic process | 4 | 13 | 5.92e-01 |
| GO:BP | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion | 1 | 3 | 5.92e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 1 | 25 | 5.92e-01 |
| GO:BP | GO:0061364 | apoptotic process involved in luteolysis | 2 | 3 | 5.93e-01 |
| GO:BP | GO:0032000 | positive regulation of fatty acid beta-oxidation | 2 | 9 | 5.93e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 5.93e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 1 | 111 | 5.93e-01 |
| GO:BP | GO:0010133 | proline catabolic process to glutamate | 1 | 2 | 5.93e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 1 | 86 | 5.93e-01 |
| GO:BP | GO:0007155 | cell adhesion | 1 | 1012 | 5.93e-01 |
| GO:BP | GO:0000003 | reproduction | 1 | 934 | 5.93e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 2 | 170 | 5.93e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 5 | 14 | 5.93e-01 |
| GO:BP | GO:0003002 | regionalization | 1 | 273 | 5.93e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 4 | 199 | 5.93e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 2 | 16 | 5.93e-01 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 5.93e-01 |
| GO:BP | GO:0044565 | dendritic cell proliferation | 2 | 3 | 5.94e-01 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 1 | 22 | 5.94e-01 |
| GO:BP | GO:0032534 | regulation of microvillus assembly | 1 | 6 | 5.94e-01 |
| GO:BP | GO:0050996 | positive regulation of lipid catabolic process | 4 | 16 | 5.94e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 5.94e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1 | 1133 | 5.94e-01 |
| GO:BP | GO:0010265 | SCF complex assembly | 1 | 2 | 5.94e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 1 | 24 | 5.94e-01 |
| GO:BP | GO:1903826 | L-arginine transmembrane transport | 6 | 12 | 5.94e-01 |
| GO:BP | GO:0008585 | female gonad development | 11 | 77 | 5.94e-01 |
| GO:BP | GO:1904812 | rRNA acetylation involved in maturation of SSU-rRNA | 1 | 1 | 5.94e-01 |
| GO:BP | GO:1900533 | palmitic acid metabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:1900535 | palmitic acid biosynthetic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 22 | 114 | 5.94e-01 |
| GO:BP | GO:0071359 | cellular response to dsRNA | 6 | 18 | 5.94e-01 |
| GO:BP | GO:0098838 | folate transmembrane transport | 1 | 6 | 5.94e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 5 | 10 | 5.94e-01 |
| GO:BP | GO:0006254 | CTP catabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0120193 | tight junction organization | 10 | 52 | 5.94e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 4 | 135 | 5.94e-01 |
| GO:BP | GO:0070175 | positive regulation of enamel mineralization | 1 | 2 | 5.94e-01 |
| GO:BP | GO:0006782 | protoporphyrinogen IX biosynthetic process | 2 | 9 | 5.94e-01 |
| GO:BP | GO:0019915 | lipid storage | 2 | 66 | 5.94e-01 |
| GO:BP | GO:0097712 | vesicle targeting, trans-Golgi to periciliary membrane compartment | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 1 | 13 | 5.94e-01 |
| GO:BP | GO:0030889 | negative regulation of B cell proliferation | 2 | 9 | 5.94e-01 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 4 | 8 | 5.94e-01 |
| GO:BP | GO:0071921 | cohesin loading | 2 | 3 | 5.94e-01 |
| GO:BP | GO:1904431 | positive regulation of t-circle formation | 1 | 3 | 5.94e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 1 | 39 | 5.94e-01 |
| GO:BP | GO:0051391 | tRNA acetylation | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle | 1 | 3 | 5.94e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 5.94e-01 |
| GO:BP | GO:0098935 | dendritic transport | 6 | 13 | 5.94e-01 |
| GO:BP | GO:0051001 | negative regulation of nitric-oxide synthase activity | 1 | 6 | 5.94e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 1 | 39 | 5.94e-01 |
| GO:BP | GO:0042118 | endothelial cell activation | 3 | 7 | 5.94e-01 |
| GO:BP | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway | 5 | 9 | 5.94e-01 |
| GO:BP | GO:1990884 | RNA acetylation | 1 | 1 | 5.94e-01 |
| GO:BP | GO:1990882 | rRNA acetylation | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0021508 | floor plate formation | 1 | 3 | 5.94e-01 |
| GO:BP | GO:1990822 | basic amino acid transmembrane transport | 7 | 14 | 5.94e-01 |
| GO:BP | GO:0090410 | malonate catabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0040009 | regulation of growth rate | 3 | 3 | 5.94e-01 |
| GO:BP | GO:1902896 | terminal web assembly | 1 | 2 | 5.94e-01 |
| GO:BP | GO:0046052 | UTP catabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 1 | 12 | 5.94e-01 |
| GO:BP | GO:0021891 | olfactory bulb interneuron development | 3 | 5 | 5.94e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 6 | 131 | 5.94e-01 |
| GO:BP | GO:0036116 | long-chain fatty-acyl-CoA catabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0072429 | response to intra-S DNA damage checkpoint signaling | 1 | 4 | 5.94e-01 |
| GO:BP | GO:0035912 | dorsal aorta morphogenesis | 1 | 6 | 5.94e-01 |
| GO:BP | GO:0015962 | diadenosine triphosphate metabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0015964 | diadenosine triphosphate catabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 2 | 236 | 5.94e-01 |
| GO:BP | GO:0043060 | meiotic metaphase I plate congression | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 2 | 7 | 5.94e-01 |
| GO:BP | GO:1903301 | positive regulation of hexokinase activity | 3 | 5 | 5.94e-01 |
| GO:BP | GO:0015802 | basic amino acid transport | 8 | 17 | 5.94e-01 |
| GO:BP | GO:1903730 | regulation of phosphatidate phosphatase activity | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0000706 | meiotic DNA double-strand break processing | 1 | 2 | 5.94e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 14 | 64 | 5.94e-01 |
| GO:BP | GO:0001408 | guanine nucleotide transport | 1 | 6 | 5.94e-01 |
| GO:BP | GO:1903741 | negative regulation of phosphatidate phosphatase activity | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0008626 | granzyme-mediated apoptotic signaling pathway | 1 | 1 | 5.94e-01 |
| GO:BP | GO:0009210 | pyrimidine ribonucleoside triphosphate catabolic process | 1 | 1 | 5.94e-01 |
| GO:BP | GO:1901143 | insulin catabolic process | 1 | 3 | 5.94e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 65 | 5.94e-01 |
| GO:BP | GO:0045190 | isotype switching | 1 | 39 | 5.94e-01 |
| GO:BP | GO:0045292 | mRNA cis splicing, via spliceosome | 1 | 26 | 5.94e-01 |
| GO:BP | GO:0043456 | regulation of pentose-phosphate shunt | 3 | 6 | 5.94e-01 |
| GO:BP | GO:0098904 | regulation of AV node cell action potential | 2 | 3 | 5.94e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 2 | 40 | 5.94e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 6 | 64 | 5.94e-01 |
| GO:BP | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis | 2 | 3 | 5.94e-01 |
| GO:BP | GO:0044854 | plasma membrane raft assembly | 3 | 6 | 5.94e-01 |
| GO:BP | GO:0009304 | tRNA transcription | 1 | 8 | 5.94e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 4 | 38 | 5.95e-01 |
| GO:BP | GO:0097283 | keratinocyte apoptotic process | 1 | 5 | 5.95e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 2 | 11 | 5.95e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 2 | 38 | 5.95e-01 |
| GO:BP | GO:1902172 | regulation of keratinocyte apoptotic process | 1 | 5 | 5.95e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 1 | 39 | 5.95e-01 |
| GO:BP | GO:0006288 | base-excision repair, DNA ligation | 1 | 1 | 5.95e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 2 | 20 | 5.95e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 2 | 9 | 5.95e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 12 | 55 | 5.95e-01 |
| GO:BP | GO:0090298 | negative regulation of mitochondrial DNA replication | 1 | 1 | 5.95e-01 |
| GO:BP | GO:0097264 | self proteolysis | 2 | 7 | 5.95e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 2 | 227 | 5.95e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 172 | 5.95e-01 |
| GO:BP | GO:0036250 | peroxisome transport along microtubule | 1 | 1 | 5.95e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 2 | 18 | 5.96e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 5.96e-01 |
| GO:BP | GO:0097374 | sensory neuron axon guidance | 2 | 2 | 5.96e-01 |
| GO:BP | GO:0015878 | biotin transport | 1 | 3 | 5.96e-01 |
| GO:BP | GO:0090384 | phagosome-lysosome docking | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 21 | 91 | 5.96e-01 |
| GO:BP | GO:1900079 | regulation of arginine biosynthetic process | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 5.96e-01 |
| GO:BP | GO:0006768 | biotin metabolic process | 2 | 3 | 5.96e-01 |
| GO:BP | GO:0034755 | iron ion transmembrane transport | 1 | 16 | 5.96e-01 |
| GO:BP | GO:1903393 | positive regulation of adherens junction organization | 1 | 2 | 5.96e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 1 | 82 | 5.96e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 7 | 31 | 5.96e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 5 | 14 | 5.96e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 5.96e-01 |
| GO:BP | GO:1905602 | positive regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 2 | 5.96e-01 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 5.96e-01 |
| GO:BP | GO:0032960 | regulation of inositol trisphosphate biosynthetic process | 3 | 4 | 5.96e-01 |
| GO:BP | GO:0060490 | lateral sprouting involved in lung morphogenesis | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0070235 | regulation of activation-induced cell death of T cells | 2 | 3 | 5.96e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 2 | 28 | 5.96e-01 |
| GO:BP | GO:0060489 | planar dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 9 | 28 | 5.96e-01 |
| GO:BP | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis | 1 | 1 | 5.96e-01 |
| GO:BP | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0034110 | regulation of homotypic cell-cell adhesion | 3 | 26 | 5.96e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 5.96e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 10 | 64 | 5.96e-01 |
| GO:BP | GO:1901550 | regulation of endothelial cell development | 1 | 12 | 5.96e-01 |
| GO:BP | GO:0048589 | developmental growth | 24 | 522 | 5.96e-01 |
| GO:BP | GO:1903512 | phytanic acid metabolic process | 1 | 2 | 5.96e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 1 | 40 | 5.96e-01 |
| GO:BP | GO:1903140 | regulation of establishment of endothelial barrier | 1 | 12 | 5.96e-01 |
| GO:BP | GO:0015787 | UDP-glucuronic acid transmembrane transport | 2 | 4 | 5.96e-01 |
| GO:BP | GO:0015789 | UDP-N-acetylgalactosamine transmembrane transport | 2 | 4 | 5.96e-01 |
| GO:BP | GO:1905453 | regulation of myeloid progenitor cell differentiation | 1 | 1 | 5.96e-01 |
| GO:BP | GO:0032425 | positive regulation of mismatch repair | 1 | 2 | 5.96e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 1 | 25 | 5.96e-01 |
| GO:BP | GO:0051664 | nuclear pore localization | 1 | 4 | 5.96e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 25 | 5.96e-01 |
| GO:BP | GO:0044321 | response to leptin | 1 | 19 | 5.96e-01 |
| GO:BP | GO:0033031 | positive regulation of neutrophil apoptotic process | 1 | 3 | 5.96e-01 |
| GO:BP | GO:0071691 | cardiac muscle thin filament assembly | 1 | 3 | 5.96e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 6 | 29 | 5.97e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 5.97e-01 |
| GO:BP | GO:0002280 | monocyte activation involved in immune response | 1 | 1 | 5.97e-01 |
| GO:BP | GO:0061564 | axon development | 2 | 398 | 5.97e-01 |
| GO:BP | GO:0034097 | response to cytokine | 6 | 607 | 5.98e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 1 | 34 | 5.98e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 4 | 16 | 5.98e-01 |
| GO:BP | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation | 1 | 6 | 5.98e-01 |
| GO:BP | GO:0034115 | negative regulation of heterotypic cell-cell adhesion | 1 | 6 | 5.98e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 3 | 51 | 5.98e-01 |
| GO:BP | GO:1903048 | regulation of acetylcholine-gated cation channel activity | 1 | 1 | 5.98e-01 |
| GO:BP | GO:1903049 | negative regulation of acetylcholine-gated cation channel activity | 1 | 1 | 5.98e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 13 | 43 | 5.98e-01 |
| GO:BP | GO:0071228 | cellular response to tumor cell | 2 | 4 | 5.98e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 13 | 43 | 5.98e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 13 | 5.99e-01 |
| GO:BP | GO:0021627 | olfactory nerve morphogenesis | 2 | 3 | 5.99e-01 |
| GO:BP | GO:1905258 | regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 5.99e-01 |
| GO:BP | GO:0110135 | Norrin signaling pathway | 2 | 2 | 5.99e-01 |
| GO:BP | GO:1905259 | negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway | 1 | 2 | 5.99e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 2 | 61 | 5.99e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 3 | 22 | 5.99e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 2 | 12 | 5.99e-01 |
| GO:BP | GO:1904338 | regulation of dopaminergic neuron differentiation | 1 | 7 | 5.99e-01 |
| GO:BP | GO:0070375 | ERK5 cascade | 1 | 3 | 5.99e-01 |
| GO:BP | GO:0032596 | protein transport into membrane raft | 2 | 4 | 5.99e-01 |
| GO:BP | GO:0002934 | desmosome organization | 3 | 9 | 5.99e-01 |
| GO:BP | GO:0021538 | epithalamus development | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 1 | 105 | 5.99e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 13 | 50 | 5.99e-01 |
| GO:BP | GO:0048820 | hair follicle maturation | 1 | 15 | 5.99e-01 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 1 | 20 | 5.99e-01 |
| GO:BP | GO:0046618 | xenobiotic export from cell | 4 | 8 | 5.99e-01 |
| GO:BP | GO:0021986 | habenula development | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0006193 | ITP catabolic process | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 3 | 46 | 5.99e-01 |
| GO:BP | GO:0009207 | purine ribonucleoside triphosphate catabolic process | 1 | 1 | 5.99e-01 |
| GO:BP | GO:0031033 | myosin filament organization | 1 | 5 | 5.99e-01 |
| GO:BP | GO:0032525 | somite rostral/caudal axis specification | 2 | 7 | 6.00e-01 |
| GO:BP | GO:0016128 | phytosteroid metabolic process | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0016129 | phytosteroid biosynthetic process | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0016132 | brassinosteroid biosynthetic process | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0016131 | brassinosteroid metabolic process | 1 | 1 | 6.00e-01 |
| GO:BP | GO:1903041 | regulation of chondrocyte hypertrophy | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 1 | 9 | 6.00e-01 |
| GO:BP | GO:0048294 | negative regulation of isotype switching to IgE isotypes | 1 | 2 | 6.00e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 19 | 52 | 6.00e-01 |
| GO:BP | GO:0032484 | Ral protein signal transduction | 2 | 5 | 6.00e-01 |
| GO:BP | GO:1901334 | lactone metabolic process | 1 | 9 | 6.00e-01 |
| GO:BP | GO:0030302 | deoxynucleotide transport | 1 | 1 | 6.00e-01 |
| GO:BP | GO:1903043 | positive regulation of chondrocyte hypertrophy | 1 | 1 | 6.00e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 34 | 79 | 6.00e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 6 | 34 | 6.00e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 9 | 59 | 6.00e-01 |
| GO:BP | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 5 | 16 | 6.00e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 1 | 63 | 6.00e-01 |
| GO:BP | GO:0040038 | polar body extrusion after meiotic divisions | 1 | 5 | 6.00e-01 |
| GO:BP | GO:0051295 | establishment of meiotic spindle localization | 1 | 4 | 6.00e-01 |
| GO:BP | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development | 2 | 2 | 6.00e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 8 | 115 | 6.00e-01 |
| GO:BP | GO:0002579 | positive regulation of antigen processing and presentation | 2 | 3 | 6.00e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 2 | 41 | 6.00e-01 |
| GO:BP | GO:0150093 | amyloid-beta clearance by transcytosis | 2 | 7 | 6.00e-01 |
| GO:BP | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 4 | 7 | 6.00e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 8 | 21 | 6.01e-01 |
| GO:BP | GO:0060024 | rhythmic synaptic transmission | 1 | 3 | 6.01e-01 |
| GO:BP | GO:0035611 | protein branching point deglutamylation | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 1 | 15 | 6.01e-01 |
| GO:BP | GO:0150032 | positive regulation of protein localization to lysosome | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 1 | 1248 | 6.01e-01 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 6.01e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 8 | 98 | 6.01e-01 |
| GO:BP | GO:0006412 | translation | 3 | 646 | 6.01e-01 |
| GO:BP | GO:1902656 | calcium ion import into cytosol | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0007040 | lysosome organization | 1 | 88 | 6.01e-01 |
| GO:BP | GO:0006824 | cobalt ion transport | 1 | 5 | 6.01e-01 |
| GO:BP | GO:0051764 | actin crosslink formation | 4 | 10 | 6.01e-01 |
| GO:BP | GO:0015821 | methionine transport | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0032223 | negative regulation of synaptic transmission, cholinergic | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 1 | 252 | 6.01e-01 |
| GO:BP | GO:0046794 | transport of virus | 5 | 19 | 6.01e-01 |
| GO:BP | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 8 | 46 | 6.01e-01 |
| GO:BP | GO:1905242 | response to 3,3’,5-triiodo-L-thyronine | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0003285 | septum secundum development | 2 | 3 | 6.01e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 7 | 24 | 6.01e-01 |
| GO:BP | GO:0006258 | UDP-glucose catabolic process | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0072308 | negative regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0060066 | oviduct development | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0040031 | snRNA modification | 2 | 7 | 6.01e-01 |
| GO:BP | GO:0006175 | dATP biosynthetic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 6.01e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 11 | 1223 | 6.01e-01 |
| GO:BP | GO:0014059 | regulation of dopamine secretion | 1 | 22 | 6.01e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 1 | 8 | 6.01e-01 |
| GO:BP | GO:0071672 | negative regulation of smooth muscle cell chemotaxis | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0000012 | single strand break repair | 4 | 10 | 6.01e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 1 | 74 | 6.01e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 3 | 13 | 6.01e-01 |
| GO:BP | GO:0072537 | fibroblast activation | 6 | 11 | 6.01e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 1 | 88 | 6.01e-01 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 2 | 8 | 6.01e-01 |
| GO:BP | GO:2000077 | negative regulation of type B pancreatic cell development | 1 | 2 | 6.01e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 1 | 19 | 6.01e-01 |
| GO:BP | GO:0038101 | sequestering of nodal from receptor via nodal binding | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 4 | 26 | 6.01e-01 |
| GO:BP | GO:0071642 | positive regulation of macrophage inflammatory protein 1 alpha production | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0045763 | negative regulation of cellular amino acid metabolic process | 2 | 2 | 6.01e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 2 | 3 | 6.01e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 2 | 15 | 6.01e-01 |
| GO:BP | GO:0000821 | regulation of arginine metabolic process | 2 | 3 | 6.01e-01 |
| GO:BP | GO:0032901 | positive regulation of neurotrophin production | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 2 | 16 | 6.01e-01 |
| GO:BP | GO:1990167 | protein K27-linked deubiquitination | 2 | 3 | 6.01e-01 |
| GO:BP | GO:0021915 | neural tube development | 27 | 146 | 6.01e-01 |
| GO:BP | GO:1990046 | stress-induced mitochondrial fusion | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0007512 | adult heart development | 1 | 9 | 6.01e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 6.01e-01 |
| GO:BP | GO:1905364 | regulation of endosomal vesicle fusion | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0034975 | protein folding in endoplasmic reticulum | 1 | 10 | 6.01e-01 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 3 | 29 | 6.01e-01 |
| GO:BP | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0045792 | negative regulation of cell size | 2 | 9 | 6.01e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 8 | 17 | 6.01e-01 |
| GO:BP | GO:0034772 | histone H4-K20 dimethylation | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0007252 | I-kappaB phosphorylation | 2 | 14 | 6.01e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 3 | 131 | 6.01e-01 |
| GO:BP | GO:0000921 | septin ring assembly | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0015808 | L-alanine transport | 2 | 9 | 6.01e-01 |
| GO:BP | GO:0046521 | sphingoid catabolic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0010748 | negative regulation of long-chain fatty acid import across plasma membrane | 1 | 4 | 6.01e-01 |
| GO:BP | GO:1901321 | positive regulation of heart induction | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0140213 | negative regulation of long-chain fatty acid import into cell | 1 | 4 | 6.01e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 3 | 21 | 6.01e-01 |
| GO:BP | GO:0021700 | developmental maturation | 3 | 211 | 6.01e-01 |
| GO:BP | GO:0031627 | telomeric loop formation | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0035844 | cloaca development | 1 | 3 | 6.01e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 5 | 59 | 6.01e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 6.01e-01 |
| GO:BP | GO:0090132 | epithelium migration | 1 | 252 | 6.01e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 3 | 11 | 6.01e-01 |
| GO:BP | GO:1900378 | positive regulation of secondary metabolite biosynthetic process | 1 | 7 | 6.01e-01 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 3 | 10 | 6.01e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 6.01e-01 |
| GO:BP | GO:0048023 | positive regulation of melanin biosynthetic process | 1 | 7 | 6.01e-01 |
| GO:BP | GO:0030389 | fructosamine metabolic process | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 16 | 124 | 6.01e-01 |
| GO:BP | GO:0036525 | protein deglycation | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 2 | 87 | 6.01e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 5 | 14 | 6.01e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 1 | 126 | 6.01e-01 |
| GO:BP | GO:0035445 | borate transmembrane transport | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0060712 | spongiotrophoblast layer development | 3 | 11 | 6.01e-01 |
| GO:BP | GO:0140719 | constitutive heterochromatin formation | 4 | 10 | 6.01e-01 |
| GO:BP | GO:0046713 | borate transport | 1 | 1 | 6.01e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 3 | 11 | 6.01e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 6.01e-01 |
| GO:BP | GO:0007500 | mesodermal cell fate determination | 1 | 2 | 6.01e-01 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 4 | 55 | 6.01e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 8 | 108 | 6.01e-01 |
| GO:BP | GO:0071420 | cellular response to histamine | 3 | 4 | 6.02e-01 |
| GO:BP | GO:1902530 | positive regulation of protein linear polyubiquitination | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0070988 | demethylation | 3 | 52 | 6.02e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 135 | 274 | 6.02e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 1 | 7 | 6.02e-01 |
| GO:BP | GO:0015697 | quaternary ammonium group transport | 1 | 17 | 6.02e-01 |
| GO:BP | GO:0010635 | regulation of mitochondrial fusion | 4 | 11 | 6.02e-01 |
| GO:BP | GO:0007389 | pattern specification process | 12 | 306 | 6.02e-01 |
| GO:BP | GO:2000143 | negative regulation of DNA-templated transcription initiation | 1 | 6 | 6.02e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 1 | 102 | 6.02e-01 |
| GO:BP | GO:0055107 | Golgi to secretory granule transport | 1 | 1 | 6.02e-01 |
| GO:BP | GO:1902528 | regulation of protein linear polyubiquitination | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 5 | 294 | 6.02e-01 |
| GO:BP | GO:1903381 | regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 5 | 17 | 6.02e-01 |
| GO:BP | GO:1903332 | regulation of protein folding | 3 | 8 | 6.02e-01 |
| GO:BP | GO:1905281 | positive regulation of retrograde transport, endosome to Golgi | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 12 | 23 | 6.02e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 1 | 1219 | 6.02e-01 |
| GO:BP | GO:0140251 | regulation protein catabolic process at presynapse | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0006783 | heme biosynthetic process | 6 | 30 | 6.02e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 1 | 56 | 6.02e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 1 | 65 | 6.02e-01 |
| GO:BP | GO:0007586 | digestion | 2 | 60 | 6.02e-01 |
| GO:BP | GO:1905664 | regulation of calcium ion import across plasma membrane | 2 | 8 | 6.02e-01 |
| GO:BP | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway | 3 | 4 | 6.02e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 2 | 11 | 6.02e-01 |
| GO:BP | GO:1904048 | regulation of spontaneous neurotransmitter secretion | 1 | 2 | 6.02e-01 |
| GO:BP | GO:0072703 | cellular response to methyl methanesulfonate | 1 | 1 | 6.02e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 13 | 93 | 6.02e-01 |
| GO:BP | GO:0072702 | response to methyl methanesulfonate | 1 | 1 | 6.02e-01 |
| GO:BP | GO:1903382 | negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 2 | 6.02e-01 |
| GO:BP | GO:1901753 | leukotriene A4 biosynthetic process | 1 | 1 | 6.02e-01 |
| GO:BP | GO:0006526 | arginine biosynthetic process | 1 | 4 | 6.02e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 7 | 29 | 6.03e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 3 | 45 | 6.03e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 3 | 190 | 6.03e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 1 | 26 | 6.03e-01 |
| GO:BP | GO:0014046 | dopamine secretion | 1 | 23 | 6.03e-01 |
| GO:BP | GO:0001805 | positive regulation of type III hypersensitivity | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 2 | 16 | 6.03e-01 |
| GO:BP | GO:0061348 | planar cell polarity pathway involved in ventricular septum morphogenesis | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 2 | 9 | 6.03e-01 |
| GO:BP | GO:1902153 | regulation of response to DNA damage checkpoint signaling | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 2 | 55 | 6.03e-01 |
| GO:BP | GO:1904716 | positive regulation of chaperone-mediated autophagy | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 6 | 24 | 6.03e-01 |
| GO:BP | GO:0060018 | astrocyte fate commitment | 2 | 2 | 6.03e-01 |
| GO:BP | GO:0021768 | nucleus accumbens development | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0015766 | disaccharide transport | 2 | 4 | 6.03e-01 |
| GO:BP | GO:0090130 | tissue migration | 1 | 256 | 6.03e-01 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 46 | 104 | 6.03e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 2 | 249 | 6.03e-01 |
| GO:BP | GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0003096 | renal sodium ion transport | 1 | 10 | 6.03e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 7 | 665 | 6.03e-01 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 2 | 30 | 6.03e-01 |
| GO:BP | GO:0036164 | cell-abiotic substrate adhesion | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 1 | 43 | 6.03e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 20 | 99 | 6.03e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 4 | 7 | 6.03e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 1 | 9 | 6.03e-01 |
| GO:BP | GO:1902151 | regulation of response to DNA integrity checkpoint signaling | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0045626 | negative regulation of T-helper 1 cell differentiation | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0061346 | planar cell polarity pathway involved in heart morphogenesis | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0061350 | planar cell polarity pathway involved in cardiac muscle tissue morphogenesis | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0001803 | regulation of type III hypersensitivity | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0061354 | planar cell polarity pathway involved in pericardium morphogenesis | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0015770 | sucrose transport | 2 | 4 | 6.03e-01 |
| GO:BP | GO:0036517 | chemoattraction of serotonergic neuron axon | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0072289 | metanephric nephron tubule formation | 2 | 2 | 6.03e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 5 | 11 | 6.03e-01 |
| GO:BP | GO:0038128 | ERBB2 signaling pathway | 4 | 14 | 6.03e-01 |
| GO:BP | GO:0001802 | type III hypersensitivity | 1 | 2 | 6.03e-01 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 2 | 11 | 6.03e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 1 | 41 | 6.03e-01 |
| GO:BP | GO:0015772 | oligosaccharide transport | 2 | 4 | 6.03e-01 |
| GO:BP | GO:0098530 | positive regulation of strand invasion | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0060542 | regulation of strand invasion | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0044828 | negative regulation by host of viral genome replication | 1 | 7 | 6.03e-01 |
| GO:BP | GO:0097028 | dendritic cell differentiation | 2 | 23 | 6.03e-01 |
| GO:BP | GO:1904934 | negative regulation of cell proliferation in midbrain | 1 | 1 | 6.03e-01 |
| GO:BP | GO:1902145 | regulation of response to cell cycle checkpoint signaling | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0061349 | planar cell polarity pathway involved in cardiac right atrium morphogenesis | 1 | 1 | 6.03e-01 |
| GO:BP | GO:0045923 | positive regulation of fatty acid metabolic process | 5 | 26 | 6.03e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 2 | 14 | 6.03e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 2 | 63 | 6.04e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 1 | 57 | 6.04e-01 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 18 | 42 | 6.04e-01 |
| GO:BP | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process | 1 | 2 | 6.04e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 15 | 197 | 6.04e-01 |
| GO:BP | GO:0018282 | metal incorporation into metallo-sulfur cluster | 1 | 2 | 6.04e-01 |
| GO:BP | GO:0042473 | outer ear morphogenesis | 3 | 5 | 6.04e-01 |
| GO:BP | GO:0018283 | iron incorporation into metallo-sulfur cluster | 1 | 2 | 6.04e-01 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 1 | 43 | 6.04e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 8 | 49 | 6.04e-01 |
| GO:BP | GO:0008406 | gonad development | 1 | 158 | 6.04e-01 |
| GO:BP | GO:0060313 | negative regulation of blood vessel remodeling | 1 | 2 | 6.04e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1 | 1147 | 6.04e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 2 | 59 | 6.04e-01 |
| GO:BP | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation | 1 | 4 | 6.04e-01 |
| GO:BP | GO:0048669 | collateral sprouting in absence of injury | 1 | 4 | 6.04e-01 |
| GO:BP | GO:0043157 | response to cation stress | 1 | 1 | 6.04e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 6.05e-01 |
| GO:BP | GO:2000793 | cell proliferation involved in heart valve development | 2 | 4 | 6.05e-01 |
| GO:BP | GO:0016137 | glycoside metabolic process | 4 | 16 | 6.05e-01 |
| GO:BP | GO:0099606 | microtubule plus-end directed mitotic chromosome migration | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 1 | 2 | 6.05e-01 |
| GO:BP | GO:0090500 | endocardial cushion to mesenchymal transition | 2 | 4 | 6.05e-01 |
| GO:BP | GO:0003336 | corneocyte desquamation | 1 | 1 | 6.05e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 3 | 11 | 6.05e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 8 | 24 | 6.05e-01 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 1 | 9 | 6.05e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 6 | 513 | 6.06e-01 |
| GO:BP | GO:0006882 | intracellular zinc ion homeostasis | 2 | 28 | 6.06e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 1 | 7 | 6.06e-01 |
| GO:BP | GO:0046444 | FMN metabolic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0009398 | FMN biosynthetic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0002524 | hypersensitivity | 4 | 7 | 6.06e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1 | 2011 | 6.06e-01 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 18 | 42 | 6.06e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 6 | 14 | 6.06e-01 |
| GO:BP | GO:0052648 | ribitol phosphate metabolic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0052647 | pentitol phosphate metabolic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0021781 | glial cell fate commitment | 3 | 5 | 6.06e-01 |
| GO:BP | GO:0060003 | copper ion export | 1 | 2 | 6.06e-01 |
| GO:BP | GO:0032988 | ribonucleoprotein complex disassembly | 3 | 8 | 6.06e-01 |
| GO:BP | GO:2000431 | regulation of cytokinesis, actomyosin contractile ring assembly | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 6.06e-01 |
| GO:BP | GO:0009231 | riboflavin biosynthetic process | 1 | 1 | 6.06e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 10 | 88 | 6.06e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 1 | 18 | 6.06e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 3 | 165 | 6.06e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 5 | 6 | 6.06e-01 |
| GO:BP | GO:0009888 | tissue development | 10 | 1425 | 6.06e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 6.06e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 6.06e-01 |
| GO:BP | GO:0052472 | modulation by host of symbiont transcription | 1 | 2 | 6.06e-01 |
| GO:BP | GO:0046782 | regulation of viral transcription | 1 | 19 | 6.06e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 4 | 382 | 6.06e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 3 | 28 | 6.06e-01 |
| GO:BP | GO:0097202 | activation of cysteine-type endopeptidase activity | 5 | 16 | 6.06e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 5 | 184 | 6.06e-01 |
| GO:BP | GO:0048870 | cell motility | 1 | 1182 | 6.06e-01 |
| GO:BP | GO:0061485 | memory T cell proliferation | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 1 | 6 | 6.07e-01 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 1 | 21 | 6.07e-01 |
| GO:BP | GO:0062043 | positive regulation of cardiac epithelial to mesenchymal transition | 4 | 9 | 6.07e-01 |
| GO:BP | GO:0001765 | membrane raft assembly | 4 | 9 | 6.07e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 6.07e-01 |
| GO:BP | GO:0086044 | atrial cardiac muscle cell to AV node cell communication by electrical coupling | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 1 | 17 | 6.07e-01 |
| GO:BP | GO:0101024 | mitotic nuclear membrane organization | 1 | 9 | 6.07e-01 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 2 | 9 | 6.07e-01 |
| GO:BP | GO:0031590 | wybutosine metabolic process | 2 | 4 | 6.07e-01 |
| GO:BP | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation | 2 | 4 | 6.07e-01 |
| GO:BP | GO:0031591 | wybutosine biosynthetic process | 2 | 4 | 6.07e-01 |
| GO:BP | GO:0098886 | modification of dendritic spine | 1 | 2 | 6.07e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 1 | 161 | 6.07e-01 |
| GO:BP | GO:1990029 | vasomotion | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 6.07e-01 |
| GO:BP | GO:0007084 | mitotic nuclear membrane reassembly | 1 | 9 | 6.07e-01 |
| GO:BP | GO:0010652 | positive regulation of cell communication by chemical coupling | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0046065 | dCTP metabolic process | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0098906 | regulation of Purkinje myocyte action potential | 1 | 1 | 6.07e-01 |
| GO:BP | GO:1903416 | response to glycoside | 3 | 6 | 6.07e-01 |
| GO:BP | GO:0006253 | dCTP catabolic process | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0086055 | Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 1 | 21 | 6.07e-01 |
| GO:BP | GO:0010645 | regulation of cell communication by chemical coupling | 1 | 1 | 6.07e-01 |
| GO:BP | GO:0080009 | mRNA methylation | 1 | 16 | 6.08e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 2 | 165 | 6.08e-01 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 1 | 12 | 6.09e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 8 | 25 | 6.09e-01 |
| GO:BP | GO:0050746 | regulation of lipoprotein metabolic process | 1 | 15 | 6.09e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 8 | 25 | 6.09e-01 |
| GO:BP | GO:0045659 | negative regulation of neutrophil differentiation | 1 | 2 | 6.09e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 2 | 107 | 6.09e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 3 | 188 | 6.09e-01 |
| GO:BP | GO:0021934 | hindbrain tangential cell migration | 2 | 3 | 6.09e-01 |
| GO:BP | GO:1990169 | stress response to copper ion | 1 | 8 | 6.09e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 17 | 53 | 6.09e-01 |
| GO:BP | GO:0030505 | inorganic diphosphate transport | 1 | 3 | 6.09e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 9 | 29 | 6.09e-01 |
| GO:BP | GO:0010273 | detoxification of copper ion | 1 | 8 | 6.09e-01 |
| GO:BP | GO:1905069 | allantois development | 2 | 4 | 6.09e-01 |
| GO:BP | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron development | 4 | 7 | 6.09e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 1 | 33 | 6.09e-01 |
| GO:BP | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation | 4 | 7 | 6.09e-01 |
| GO:BP | GO:0000910 | cytokinesis | 73 | 164 | 6.09e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1 | 1345 | 6.09e-01 |
| GO:BP | GO:0099563 | modification of synaptic structure | 6 | 24 | 6.09e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 17 | 135 | 6.09e-01 |
| GO:BP | GO:0045769 | negative regulation of asymmetric cell division | 1 | 1 | 6.10e-01 |
| GO:BP | GO:0035695 | mitophagy by induced vacuole formation | 1 | 1 | 6.10e-01 |
| GO:BP | GO:0009753 | response to jasmonic acid | 2 | 3 | 6.10e-01 |
| GO:BP | GO:0071395 | cellular response to jasmonic acid stimulus | 2 | 3 | 6.10e-01 |
| GO:BP | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response | 1 | 5 | 6.10e-01 |
| GO:BP | GO:0006563 | L-serine metabolic process | 1 | 10 | 6.10e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 5 | 19 | 6.11e-01 |
| GO:BP | GO:0043331 | response to dsRNA | 13 | 33 | 6.11e-01 |
| GO:BP | GO:0032683 | negative regulation of connective tissue growth factor production | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1902729 | negative regulation of proteoglycan biosynthetic process | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0042760 | very long-chain fatty acid catabolic process | 2 | 5 | 6.11e-01 |
| GO:BP | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | 5 | 12 | 6.11e-01 |
| GO:BP | GO:0070586 | cell-cell adhesion involved in gastrulation | 1 | 7 | 6.11e-01 |
| GO:BP | GO:0030049 | muscle filament sliding | 3 | 10 | 6.11e-01 |
| GO:BP | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 7 | 10 | 6.11e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 6.11e-01 |
| GO:BP | GO:0002526 | acute inflammatory response | 2 | 49 | 6.11e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 6.11e-01 |
| GO:BP | GO:0001712 | ectodermal cell fate commitment | 1 | 2 | 6.11e-01 |
| GO:BP | GO:1904439 | negative regulation of iron ion import across plasma membrane | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1904910 | regulation of establishment of RNA localization to telomere | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1904911 | negative regulation of establishment of RNA localization to telomere | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1904913 | regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1904914 | negative regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 43 | 94 | 6.11e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 6 | 40 | 6.11e-01 |
| GO:BP | GO:0075071 | modulation by symbiont of host autophagy | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 11 | 27 | 6.11e-01 |
| GO:BP | GO:0021685 | cerebellar granular layer structural organization | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1 | 1255 | 6.11e-01 |
| GO:BP | GO:0035936 | testosterone secretion | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 1 | 71 | 6.11e-01 |
| GO:BP | GO:0061952 | midbody abscission | 2 | 17 | 6.11e-01 |
| GO:BP | GO:0045039 | protein insertion into mitochondrial inner membrane | 2 | 13 | 6.11e-01 |
| GO:BP | GO:1905672 | negative regulation of lysosome organization | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0140321 | suppression of host autophagy | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0043063 | intercellular bridge organization | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0036146 | cellular response to mycotoxin | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0045475 | locomotor rhythm | 3 | 13 | 6.11e-01 |
| GO:BP | GO:1903721 | positive regulation of I-kappaB phosphorylation | 1 | 5 | 6.11e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 10 | 6.11e-01 |
| GO:BP | GO:0039521 | suppression by virus of host autophagy | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 4 | 49 | 6.11e-01 |
| GO:BP | GO:0009136 | purine nucleoside diphosphate biosynthetic process | 3 | 5 | 6.11e-01 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 3 | 20 | 6.11e-01 |
| GO:BP | GO:1904792 | positive regulation of shelterin complex assembly | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1 | 1256 | 6.11e-01 |
| GO:BP | GO:0002176 | male germ cell proliferation | 2 | 6 | 6.11e-01 |
| GO:BP | GO:0035364 | thymine transport | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000845 | positive regulation of testosterone secretion | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0160025 | sensory perception of itch | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 10 | 34 | 6.11e-01 |
| GO:BP | GO:0160023 | sneeze reflex | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072386 | plus-end-directed organelle transport along microtubule | 2 | 7 | 6.11e-01 |
| GO:BP | GO:0071275 | cellular response to aluminum ion | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0072721 | cellular response to dithiothreitol | 1 | 1 | 6.11e-01 |
| GO:BP | GO:2000843 | regulation of testosterone secretion | 1 | 2 | 6.11e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 6.11e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 1 | 18 | 6.11e-01 |
| GO:BP | GO:0007028 | cytoplasm organization | 2 | 8 | 6.11e-01 |
| GO:BP | GO:0021527 | spinal cord association neuron differentiation | 2 | 3 | 6.11e-01 |
| GO:BP | GO:0035441 | cell migration involved in vasculogenesis | 1 | 4 | 6.11e-01 |
| GO:BP | GO:1904850 | negative regulation of establishment of protein localization to telomere | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 7 | 13 | 6.11e-01 |
| GO:BP | GO:1904631 | response to glucoside | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0097510 | base-excision repair, AP site formation via deaminated base removal | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway | 1 | 2 | 6.11e-01 |
| GO:BP | GO:1903096 | protein localization to meiotic spindle midzone | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 3 | 5 | 6.11e-01 |
| GO:BP | GO:0051769 | regulation of nitric-oxide synthase biosynthetic process | 8 | 13 | 6.11e-01 |
| GO:BP | GO:0051767 | nitric-oxide synthase biosynthetic process | 8 | 13 | 6.11e-01 |
| GO:BP | GO:0035669 | TRAM-dependent toll-like receptor 4 signaling pathway | 1 | 2 | 6.11e-01 |
| GO:BP | GO:1905359 | protein localization to meiotic spindle | 1 | 1 | 6.11e-01 |
| GO:BP | GO:1903867 | extraembryonic membrane development | 3 | 11 | 6.11e-01 |
| GO:BP | GO:1905115 | regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 13 | 31 | 6.11e-01 |
| GO:BP | GO:0048014 | Tie signaling pathway | 2 | 3 | 6.11e-01 |
| GO:BP | GO:0006101 | citrate metabolic process | 1 | 5 | 6.11e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 10 | 88 | 6.11e-01 |
| GO:BP | GO:1905116 | positive regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 1 | 1 | 6.11e-01 |
| GO:BP | GO:0090435 | protein localization to nuclear envelope | 6 | 14 | 6.11e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 1 | 8 | 6.12e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 6.12e-01 |
| GO:BP | GO:1903649 | regulation of cytoplasmic transport | 6 | 25 | 6.12e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 1 | 8 | 6.12e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 11 | 51 | 6.12e-01 |
| GO:BP | GO:1903126 | negative regulation of centriole-centriole cohesion | 1 | 1 | 6.13e-01 |
| GO:BP | GO:0006935 | chemotaxis | 2 | 389 | 6.13e-01 |
| GO:BP | GO:0046203 | spermidine catabolic process | 1 | 2 | 6.13e-01 |
| GO:BP | GO:0000212 | meiotic spindle organization | 7 | 14 | 6.13e-01 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1267 | 6.13e-01 |
| GO:BP | GO:0072347 | response to anesthetic | 1 | 1 | 6.13e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 6.13e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 3 | 36 | 6.14e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 8 | 33 | 6.14e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 6.14e-01 |
| GO:BP | GO:0003406 | retinal pigment epithelium development | 1 | 4 | 6.14e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 59 | 147 | 6.14e-01 |
| GO:BP | GO:0030913 | paranodal junction assembly | 3 | 6 | 6.14e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 1 | 31 | 6.14e-01 |
| GO:BP | GO:0060766 | negative regulation of androgen receptor signaling pathway | 3 | 12 | 6.14e-01 |
| GO:BP | GO:0140131 | positive regulation of lymphocyte chemotaxis | 1 | 10 | 6.14e-01 |
| GO:BP | GO:0032876 | negative regulation of DNA endoreduplication | 1 | 1 | 6.14e-01 |
| GO:BP | GO:0042330 | taxis | 2 | 389 | 6.14e-01 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 1 | 14 | 6.14e-01 |
| GO:BP | GO:0071394 | cellular response to testosterone stimulus | 3 | 11 | 6.14e-01 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 1 | 15 | 6.14e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 16 | 6.14e-01 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 4 | 15 | 6.14e-01 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 2 | 28 | 6.15e-01 |
| GO:BP | GO:0007497 | posterior midgut development | 1 | 2 | 6.15e-01 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 2 | 28 | 6.15e-01 |
| GO:BP | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity | 1 | 3 | 6.15e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 2 | 86 | 6.15e-01 |
| GO:BP | GO:1904044 | response to aldosterone | 6 | 9 | 6.15e-01 |
| GO:BP | GO:0097240 | chromosome attachment to the nuclear envelope | 3 | 3 | 6.15e-01 |
| GO:BP | GO:0060319 | primitive erythrocyte differentiation | 2 | 4 | 6.15e-01 |
| GO:BP | GO:0061017 | hepatoblast differentiation | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0045591 | positive regulation of regulatory T cell differentiation | 5 | 14 | 6.15e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 3 | 7 | 6.15e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 1 | 10 | 6.15e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 127 | 313 | 6.15e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 6.15e-01 |
| GO:BP | GO:0048002 | antigen processing and presentation of peptide antigen | 10 | 41 | 6.15e-01 |
| GO:BP | GO:1904762 | positive regulation of myofibroblast differentiation | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 6.15e-01 |
| GO:BP | GO:0008354 | germ cell migration | 3 | 6 | 6.15e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 9 | 281 | 6.15e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 6 | 26 | 6.15e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 6.15e-01 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 19 | 38 | 6.15e-01 |
| GO:BP | GO:1904506 | negative regulation of telomere maintenance in response to DNA damage | 2 | 3 | 6.15e-01 |
| GO:BP | GO:0001971 | negative regulation of activation of membrane attack complex | 1 | 1 | 6.15e-01 |
| GO:BP | GO:0034330 | cell junction organization | 9 | 575 | 6.15e-01 |
| GO:BP | GO:0010886 | positive regulation of cholesterol storage | 1 | 5 | 6.15e-01 |
| GO:BP | GO:0032474 | otolith morphogenesis | 1 | 3 | 6.15e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 2 | 7 | 6.15e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 2 | 44 | 6.15e-01 |
| GO:BP | GO:0071071 | regulation of phospholipid biosynthetic process | 7 | 15 | 6.15e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 5 | 101 | 6.15e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 3 | 135 | 6.15e-01 |
| GO:BP | GO:1901223 | negative regulation of NIK/NF-kappaB signaling | 3 | 21 | 6.15e-01 |
| GO:BP | GO:0042552 | myelination | 11 | 111 | 6.16e-01 |
| GO:BP | GO:1901687 | glutathione derivative biosynthetic process | 1 | 3 | 6.16e-01 |
| GO:BP | GO:1901685 | glutathione derivative metabolic process | 1 | 3 | 6.16e-01 |
| GO:BP | GO:0060936 | cardiac fibroblast cell development | 2 | 4 | 6.16e-01 |
| GO:BP | GO:0060935 | cardiac fibroblast cell differentiation | 2 | 4 | 6.16e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 3 | 199 | 6.16e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 1 | 17 | 6.16e-01 |
| GO:BP | GO:1901509 | regulation of endothelial tube morphogenesis | 2 | 3 | 6.16e-01 |
| GO:BP | GO:0060243 | negative regulation of cell growth involved in contact inhibition | 1 | 1 | 6.16e-01 |
| GO:BP | GO:1900544 | positive regulation of purine nucleotide metabolic process | 3 | 16 | 6.16e-01 |
| GO:BP | GO:0045981 | positive regulation of nucleotide metabolic process | 3 | 16 | 6.16e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 11 | 79 | 6.16e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 6 | 676 | 6.16e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 3 | 46 | 6.16e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 23 | 282 | 6.16e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 6 | 187 | 6.16e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 1 | 26 | 6.16e-01 |
| GO:BP | GO:0018125 | peptidyl-cysteine methylation | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0048242 | epinephrine secretion | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0048073 | regulation of eye pigmentation | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0014060 | regulation of epinephrine secretion | 1 | 2 | 6.17e-01 |
| GO:BP | GO:1904378 | maintenance of unfolded protein involved in ERAD pathway | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0036506 | maintenance of unfolded protein | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 5 | 21 | 6.17e-01 |
| GO:BP | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 1 | 5 | 6.17e-01 |
| GO:BP | GO:0002519 | natural killer cell tolerance induction | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 2 | 31 | 6.17e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 4 | 52 | 6.17e-01 |
| GO:BP | GO:0006742 | NADP catabolic process | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0019677 | NAD catabolic process | 1 | 3 | 6.17e-01 |
| GO:BP | GO:0044804 | autophagy of nucleus | 1 | 18 | 6.17e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 3 | 9 | 6.17e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 3 | 9 | 6.17e-01 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0031179 | peptide modification | 1 | 4 | 6.17e-01 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 6.17e-01 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 3 | 3070 | 6.17e-01 |
| GO:BP | GO:0016256 | N-glycan processing to lysosome | 1 | 2 | 6.17e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 3 | 9 | 6.17e-01 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 1 | 20 | 6.17e-01 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 6.17e-01 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0045723 | positive regulation of fatty acid biosynthetic process | 3 | 12 | 6.17e-01 |
| GO:BP | GO:0034337 | RNA folding | 1 | 1 | 6.17e-01 |
| GO:BP | GO:0032203 | telomere formation via telomerase | 1 | 1 | 6.17e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 5 | 21 | 6.17e-01 |
| GO:BP | GO:0090184 | positive regulation of kidney development | 3 | 9 | 6.17e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 5 | 187 | 6.17e-01 |
| GO:BP | GO:0046836 | glycolipid transport | 1 | 7 | 6.18e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 1 | 24 | 6.18e-01 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 5 | 10 | 6.18e-01 |
| GO:BP | GO:0009912 | auditory receptor cell fate commitment | 2 | 4 | 6.18e-01 |
| GO:BP | GO:1904482 | cellular response to tetrahydrofolate | 1 | 1 | 6.18e-01 |
| GO:BP | GO:1904481 | response to tetrahydrofolate | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0060120 | inner ear receptor cell fate commitment | 2 | 4 | 6.18e-01 |
| GO:BP | GO:1904526 | regulation of microtubule binding | 4 | 10 | 6.18e-01 |
| GO:BP | GO:0036015 | response to interleukin-3 | 2 | 6 | 6.18e-01 |
| GO:BP | GO:0036016 | cellular response to interleukin-3 | 2 | 6 | 6.18e-01 |
| GO:BP | GO:0035444 | nickel cation transmembrane transport | 1 | 1 | 6.18e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 7 | 38 | 6.18e-01 |
| GO:BP | GO:0030954 | astral microtubule nucleation | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0015692 | lead ion transport | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0015676 | vanadium ion transport | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 6.18e-01 |
| GO:BP | GO:0015675 | nickel cation transport | 1 | 1 | 6.18e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 3 | 57 | 6.18e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 7 | 303 | 6.18e-01 |
| GO:BP | GO:0071787 | endoplasmic reticulum tubular network formation | 1 | 4 | 6.18e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 3 | 10 | 6.19e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 7 | 35 | 6.19e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 2 | 9 | 6.19e-01 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 4 | 8 | 6.19e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 3 | 269 | 6.19e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 11 | 160 | 6.19e-01 |
| GO:BP | GO:0060741 | prostate gland stromal morphogenesis | 1 | 2 | 6.19e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 1 | 72 | 6.20e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 2 | 16 | 6.20e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 2 | 17 | 6.20e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 10 | 95 | 6.20e-01 |
| GO:BP | GO:0006050 | mannosamine metabolic process | 1 | 2 | 6.20e-01 |
| GO:BP | GO:0006051 | N-acetylmannosamine metabolic process | 1 | 2 | 6.20e-01 |
| GO:BP | GO:1902319 | DNA strand elongation involved in nuclear cell cycle DNA replication | 1 | 1 | 6.21e-01 |
| GO:BP | GO:1902296 | DNA strand elongation involved in cell cycle DNA replication | 1 | 1 | 6.21e-01 |
| GO:BP | GO:1902983 | DNA strand elongation involved in mitotic DNA replication | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0106016 | positive regulation of inflammatory response to wounding | 1 | 2 | 6.21e-01 |
| GO:BP | GO:1903334 | positive regulation of protein folding | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 1 | 16 | 6.21e-01 |
| GO:BP | GO:0098937 | anterograde dendritic transport | 4 | 9 | 6.21e-01 |
| GO:BP | GO:0019551 | glutamate catabolic process to 2-oxoglutarate | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0019550 | glutamate catabolic process to aspartate | 1 | 2 | 6.21e-01 |
| GO:BP | GO:0042413 | carnitine catabolic process | 1 | 1 | 6.21e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 6 | 775 | 6.21e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 2 | 10 | 6.21e-01 |
| GO:BP | GO:0030449 | regulation of complement activation | 1 | 11 | 6.21e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 1 | 64 | 6.21e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 8 | 18 | 6.22e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 4 | 211 | 6.22e-01 |
| GO:BP | GO:0060192 | negative regulation of lipase activity | 1 | 13 | 6.22e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 10 | 35 | 6.22e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 1 | 73 | 6.22e-01 |
| GO:BP | GO:0015801 | aromatic amino acid transport | 2 | 11 | 6.22e-01 |
| GO:BP | GO:0051562 | negative regulation of mitochondrial calcium ion concentration | 1 | 2 | 6.22e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 6 | 40 | 6.22e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 10 | 125 | 6.22e-01 |
| GO:BP | GO:0032392 | DNA geometric change | 34 | 80 | 6.23e-01 |
| GO:BP | GO:0046877 | regulation of saliva secretion | 1 | 3 | 6.24e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 9 | 116 | 6.24e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 2 | 49 | 6.24e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 6.24e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 6.24e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 38 | 400 | 6.24e-01 |
| GO:BP | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway | 1 | 10 | 6.24e-01 |
| GO:BP | GO:0070512 | positive regulation of histone H4-K20 methylation | 1 | 1 | 6.24e-01 |
| GO:BP | GO:0046597 | negative regulation of viral entry into host cell | 1 | 14 | 6.24e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 3 | 5 | 6.24e-01 |
| GO:BP | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process | 2 | 4 | 6.25e-01 |
| GO:BP | GO:0051954 | positive regulation of amine transport | 1 | 26 | 6.25e-01 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 4 | 8 | 6.25e-01 |
| GO:BP | GO:0050812 | regulation of acyl-CoA biosynthetic process | 2 | 5 | 6.25e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 1 | 73 | 6.25e-01 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 1 | 10 | 6.25e-01 |
| GO:BP | GO:0032345 | negative regulation of aldosterone metabolic process | 1 | 4 | 6.25e-01 |
| GO:BP | GO:2000417 | negative regulation of eosinophil migration | 1 | 1 | 6.25e-01 |
| GO:BP | GO:0060428 | lung epithelium development | 6 | 30 | 6.25e-01 |
| GO:BP | GO:2000065 | negative regulation of cortisol biosynthetic process | 1 | 4 | 6.25e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 1 | 48 | 6.25e-01 |
| GO:BP | GO:0032348 | negative regulation of aldosterone biosynthetic process | 1 | 4 | 6.25e-01 |
| GO:BP | GO:1900028 | negative regulation of ruffle assembly | 2 | 4 | 6.26e-01 |
| GO:BP | GO:1903766 | positive regulation of potassium ion export across plasma membrane | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 9 | 34 | 6.26e-01 |
| GO:BP | GO:0007621 | negative regulation of female receptivity | 1 | 1 | 6.26e-01 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 2 | 4 | 6.26e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 3 | 34 | 6.26e-01 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 2 | 4 | 6.26e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 6 | 51 | 6.26e-01 |
| GO:BP | GO:0019532 | oxalate transport | 1 | 5 | 6.26e-01 |
| GO:BP | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade | 1 | 5 | 6.26e-01 |
| GO:BP | GO:0061370 | testosterone biosynthetic process | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 1 | 4 | 6.26e-01 |
| GO:BP | GO:0061198 | fungiform papilla formation | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 100 | 6.26e-01 |
| GO:BP | GO:0021873 | forebrain neuroblast division | 1 | 3 | 6.26e-01 |
| GO:BP | GO:0008065 | establishment of blood-nerve barrier | 1 | 3 | 6.26e-01 |
| GO:BP | GO:1903651 | positive regulation of cytoplasmic transport | 3 | 11 | 6.26e-01 |
| GO:BP | GO:0045898 | regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | 7 | 6.26e-01 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 1 | 4 | 6.26e-01 |
| GO:BP | GO:0007528 | neuromuscular junction development | 5 | 39 | 6.26e-01 |
| GO:BP | GO:1902977 | mitotic DNA replication preinitiation complex assembly | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 9 | 457 | 6.27e-01 |
| GO:BP | GO:0006535 | cysteine biosynthetic process from serine | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0048755 | branching morphogenesis of a nerve | 1 | 8 | 6.27e-01 |
| GO:BP | GO:0007494 | midgut development | 3 | 7 | 6.27e-01 |
| GO:BP | GO:1904442 | negative regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0006671 | phytosphingosine metabolic process | 1 | 1 | 6.27e-01 |
| GO:BP | GO:2000416 | regulation of eosinophil migration | 2 | 2 | 6.27e-01 |
| GO:BP | GO:1904441 | regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0035825 | homologous recombination | 1 | 43 | 6.27e-01 |
| GO:BP | GO:1903697 | negative regulation of microvillus assembly | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 43 | 192 | 6.27e-01 |
| GO:BP | GO:0071602 | phytosphingosine biosynthetic process | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0051953 | negative regulation of amine transport | 4 | 15 | 6.27e-01 |
| GO:BP | GO:0016560 | protein import into peroxisome matrix, docking | 2 | 3 | 6.27e-01 |
| GO:BP | GO:1990789 | thyroid gland epithelial cell proliferation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 3 | 9 | 6.27e-01 |
| GO:BP | GO:0098725 | symmetric cell division | 1 | 1 | 6.27e-01 |
| GO:BP | GO:2000452 | regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:1904777 | negative regulation of protein localization to cell cortex | 1 | 1 | 6.27e-01 |
| GO:BP | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 4 | 35 | 6.27e-01 |
| GO:BP | GO:0035698 | CD8-positive, alpha-beta cytotoxic T cell extravasation | 1 | 1 | 6.27e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 3 | 3132 | 6.27e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 4 | 463 | 6.27e-01 |
| GO:BP | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0050428 | 3’-phosphoadenosine 5’-phosphosulfate biosynthetic process | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0000103 | sulfate assimilation | 1 | 2 | 6.27e-01 |
| GO:BP | GO:0099187 | presynaptic cytoskeleton organization | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0099140 | presynaptic actin cytoskeleton organization | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0019284 | L-methionine salvage from S-adenosylmethionine | 1 | 3 | 6.28e-01 |
| GO:BP | GO:0035752 | lysosomal lumen pH elevation | 2 | 3 | 6.28e-01 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 1 | 9 | 6.28e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 9 | 56 | 6.28e-01 |
| GO:BP | GO:1904464 | regulation of matrix metallopeptidase secretion | 1 | 5 | 6.28e-01 |
| GO:BP | GO:1990773 | matrix metallopeptidase secretion | 1 | 5 | 6.28e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 9 | 18 | 6.28e-01 |
| GO:BP | GO:0018201 | peptidyl-glycine modification | 2 | 4 | 6.28e-01 |
| GO:BP | GO:0070911 | global genome nucleotide-excision repair | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 5 | 761 | 6.28e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 2 | 203 | 6.28e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 11 | 113 | 6.28e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 11 | 113 | 6.28e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 19 | 50 | 6.28e-01 |
| GO:BP | GO:0002331 | pre-B cell allelic exclusion | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 6.28e-01 |
| GO:BP | GO:0050882 | voluntary musculoskeletal movement | 3 | 7 | 6.28e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 3 | 3133 | 6.28e-01 |
| GO:BP | GO:1905040 | otic placode development | 1 | 1 | 6.28e-01 |
| GO:BP | GO:1902378 | VEGF-activated neuropilin signaling pathway involved in axon guidance | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0090660 | cerebrospinal fluid circulation | 1 | 9 | 6.28e-01 |
| GO:BP | GO:0060301 | positive regulation of cytokine activity | 1 | 1 | 6.28e-01 |
| GO:BP | GO:0032351 | negative regulation of hormone metabolic process | 2 | 10 | 6.28e-01 |
| GO:BP | GO:0071344 | diphosphate metabolic process | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0035281 | pre-miRNA export from nucleus | 1 | 2 | 6.28e-01 |
| GO:BP | GO:1902744 | negative regulation of lamellipodium organization | 2 | 4 | 6.28e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 6.28e-01 |
| GO:BP | GO:0034650 | cortisol metabolic process | 2 | 9 | 6.28e-01 |
| GO:BP | GO:0061015 | snRNA import into nucleus | 1 | 2 | 6.28e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 2 | 222 | 6.28e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 2 | 9 | 6.28e-01 |
| GO:BP | GO:1905341 | negative regulation of protein localization to kinetochore | 1 | 1 | 6.28e-01 |
| GO:BP | GO:2000354 | regulation of ovarian follicle development | 2 | 5 | 6.28e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 1 | 29 | 6.28e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 10 | 18 | 6.28e-01 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 13 | 29 | 6.28e-01 |
| GO:BP | GO:0038134 | ERBB2-EGFR signaling pathway | 1 | 6 | 6.29e-01 |
| GO:BP | GO:0099040 | ceramide translocation | 1 | 2 | 6.29e-01 |
| GO:BP | GO:0071788 | endoplasmic reticulum tubular network maintenance | 1 | 1 | 6.29e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 1 | 14 | 6.30e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 7 | 61 | 6.30e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 1 | 105 | 6.30e-01 |
| GO:BP | GO:0062042 | regulation of cardiac epithelial to mesenchymal transition | 1 | 10 | 6.31e-01 |
| GO:BP | GO:0021750 | vestibular nucleus development | 1 | 1 | 6.31e-01 |
| GO:BP | GO:0042476 | odontogenesis | 3 | 86 | 6.31e-01 |
| GO:BP | GO:0070666 | regulation of mast cell proliferation | 2 | 6 | 6.31e-01 |
| GO:BP | GO:0043378 | positive regulation of CD8-positive, alpha-beta T cell differentiation | 2 | 2 | 6.31e-01 |
| GO:BP | GO:0070662 | mast cell proliferation | 2 | 6 | 6.31e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 2 | 10 | 6.31e-01 |
| GO:BP | GO:0014724 | regulation of twitch skeletal muscle contraction | 1 | 3 | 6.31e-01 |
| GO:BP | GO:0001654 | eye development | 2 | 272 | 6.31e-01 |
| GO:BP | GO:1990840 | response to lectin | 1 | 11 | 6.31e-01 |
| GO:BP | GO:1990858 | cellular response to lectin | 1 | 11 | 6.31e-01 |
| GO:BP | GO:0008037 | cell recognition | 1 | 87 | 6.31e-01 |
| GO:BP | GO:0002223 | stimulatory C-type lectin receptor signaling pathway | 1 | 11 | 6.31e-01 |
| GO:BP | GO:0035502 | metanephric part of ureteric bud development | 1 | 3 | 6.32e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 1 | 4 | 6.32e-01 |
| GO:BP | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0072284 | metanephric S-shaped body morphogenesis | 1 | 4 | 6.32e-01 |
| GO:BP | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process | 3 | 5 | 6.32e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 4 | 23 | 6.32e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 7 | 30 | 6.32e-01 |
| GO:BP | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | 1 | 10 | 6.32e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 6.32e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 6.32e-01 |
| GO:BP | GO:0110017 | cap-independent translational initiation of linear mRNA | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 1 | 10 | 6.32e-01 |
| GO:BP | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum | 1 | 10 | 6.32e-01 |
| GO:BP | GO:0002192 | IRES-dependent translational initiation of linear mRNA | 1 | 1 | 6.32e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 2 | 11 | 6.32e-01 |
| GO:BP | GO:0060215 | primitive hemopoiesis | 3 | 7 | 6.32e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 5 | 8 | 6.32e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 4 | 75 | 6.32e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 2 | 206 | 6.32e-01 |
| GO:BP | GO:2000474 | regulation of opioid receptor signaling pathway | 1 | 2 | 6.33e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 8 | 687 | 6.33e-01 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 3 | 5 | 6.33e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 6.33e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 6.33e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 1 | 14 | 6.33e-01 |
| GO:BP | GO:0010713 | negative regulation of collagen metabolic process | 3 | 8 | 6.33e-01 |
| GO:BP | GO:0006403 | RNA localization | 71 | 182 | 6.33e-01 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 9 | 50 | 6.34e-01 |
| GO:BP | GO:2000659 | regulation of interleukin-1-mediated signaling pathway | 1 | 4 | 6.34e-01 |
| GO:BP | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 6.34e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 1 | 29 | 6.34e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 6.34e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 1 | 13 | 6.34e-01 |
| GO:BP | GO:0036265 | RNA (guanine-N7)-methylation | 3 | 6 | 6.35e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 6.35e-01 |
| GO:BP | GO:1905949 | negative regulation of calcium ion import across plasma membrane | 1 | 4 | 6.35e-01 |
| GO:BP | GO:1905550 | regulation of protein localization to endoplasmic reticulum | 2 | 4 | 6.35e-01 |
| GO:BP | GO:0021793 | chemorepulsion of branchiomotor axon | 1 | 1 | 6.35e-01 |
| GO:BP | GO:0150063 | visual system development | 2 | 273 | 6.35e-01 |
| GO:BP | GO:0021629 | olfactory nerve structural organization | 1 | 1 | 6.35e-01 |
| GO:BP | GO:0006002 | fructose 6-phosphate metabolic process | 2 | 8 | 6.35e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 6.35e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 6.35e-01 |
| GO:BP | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing | 3 | 6 | 6.35e-01 |
| GO:BP | GO:0038163 | thrombopoietin-mediated signaling pathway | 1 | 3 | 6.35e-01 |
| GO:BP | GO:0060013 | righting reflex | 4 | 6 | 6.35e-01 |
| GO:BP | GO:0017126 | nucleologenesis | 1 | 2 | 6.35e-01 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 1 | 22 | 6.35e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 1 | 71 | 6.35e-01 |
| GO:BP | GO:0038116 | chemokine (C-C motif) ligand 21 signaling pathway | 1 | 2 | 6.35e-01 |
| GO:BP | GO:0071386 | cellular response to corticosterone stimulus | 1 | 1 | 6.35e-01 |
| GO:BP | GO:0048210 | Golgi vesicle fusion to target membrane | 2 | 4 | 6.35e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 1 | 94 | 6.35e-01 |
| GO:BP | GO:0016203 | muscle attachment | 1 | 1 | 6.35e-01 |
| GO:BP | GO:2000368 | positive regulation of acrosomal vesicle exocytosis | 1 | 2 | 6.36e-01 |
| GO:BP | GO:0042063 | gliogenesis | 2 | 235 | 6.36e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 1 | 4 | 6.36e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 1 | 25 | 6.36e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 5 | 69 | 6.36e-01 |
| GO:BP | GO:2000252 | negative regulation of feeding behavior | 2 | 2 | 6.36e-01 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 1 | 120 | 6.36e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 4 | 44 | 6.36e-01 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 8 | 39 | 6.36e-01 |
| GO:BP | GO:0048625 | myoblast fate commitment | 2 | 4 | 6.37e-01 |
| GO:BP | GO:0015884 | folic acid transport | 1 | 7 | 6.37e-01 |
| GO:BP | GO:0042262 | DNA protection | 3 | 6 | 6.37e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 14 | 31 | 6.37e-01 |
| GO:BP | GO:0045348 | positive regulation of MHC class II biosynthetic process | 5 | 8 | 6.37e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 8 | 93 | 6.37e-01 |
| GO:BP | GO:0048496 | maintenance of animal organ identity | 1 | 4 | 6.37e-01 |
| GO:BP | GO:0003250 | regulation of cell proliferation involved in heart valve morphogenesis | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0048132 | female germ-line stem cell asymmetric division | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity | 3 | 4 | 6.37e-01 |
| GO:BP | GO:0003249 | cell proliferation involved in heart valve morphogenesis | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0035685 | helper T cell diapedesis | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 6 | 138 | 6.37e-01 |
| GO:BP | GO:0070268 | cornification | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0097473 | retinal rod cell apoptotic process | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 17 | 78 | 6.37e-01 |
| GO:BP | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering | 4 | 9 | 6.37e-01 |
| GO:BP | GO:2000283 | negative regulation of amino acid biosynthetic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0036367 | light adaption | 1 | 3 | 6.37e-01 |
| GO:BP | GO:0042637 | catagen | 2 | 4 | 6.37e-01 |
| GO:BP | GO:0060364 | frontal suture morphogenesis | 2 | 4 | 6.37e-01 |
| GO:BP | GO:0051794 | regulation of timing of catagen | 2 | 4 | 6.37e-01 |
| GO:BP | GO:1903719 | regulation of I-kappaB phosphorylation | 1 | 6 | 6.37e-01 |
| GO:BP | GO:1903394 | protein localization to kinetochore involved in kinetochore assembly | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0033241 | regulation of amine catabolic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0033242 | negative regulation of amine catabolic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:1901098 | positive regulation of autophagosome maturation | 3 | 6 | 6.37e-01 |
| GO:BP | GO:0036138 | peptidyl-histidine hydroxylation | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 22 | 798 | 6.37e-01 |
| GO:BP | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development | 1 | 2 | 6.37e-01 |
| GO:BP | GO:1990839 | response to endothelin | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0007344 | pronuclear fusion | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0098528 | skeletal muscle fiber differentiation | 1 | 5 | 6.37e-01 |
| GO:BP | GO:0000741 | karyogamy | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0045124 | regulation of bone resorption | 5 | 24 | 6.37e-01 |
| GO:BP | GO:0034720 | histone H3-K4 demethylation | 1 | 5 | 6.37e-01 |
| GO:BP | GO:0098736 | negative regulation of the force of heart contraction | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 1 | 11 | 6.37e-01 |
| GO:BP | GO:0060672 | epithelial cell morphogenesis involved in placental branching | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0032288 | myelin assembly | 6 | 19 | 6.37e-01 |
| GO:BP | GO:1903248 | regulation of citrulline biosynthetic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0021932 | hindbrain radial glia guided cell migration | 3 | 8 | 6.37e-01 |
| GO:BP | GO:1900082 | negative regulation of arginine catabolic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:1900081 | regulation of arginine catabolic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:1903249 | negative regulation of citrulline biosynthetic process | 1 | 1 | 6.37e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 10 | 30 | 6.37e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 1 | 93 | 6.37e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 1 | 24 | 6.37e-01 |
| GO:BP | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0006788 | heme oxidation | 1 | 2 | 6.37e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 12 | 25 | 6.37e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 9 | 229 | 6.37e-01 |
| GO:BP | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway | 2 | 7 | 6.37e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 35 | 6.37e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 6.37e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 9 | 19 | 6.38e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 1 | 78 | 6.38e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 7 | 28 | 6.38e-01 |
| GO:BP | GO:0030879 | mammary gland development | 3 | 106 | 6.38e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 61 | 479 | 6.38e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 4 | 17 | 6.38e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 4 | 17 | 6.38e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 5 | 21 | 6.38e-01 |
| GO:BP | GO:0071313 | cellular response to caffeine | 5 | 10 | 6.38e-01 |
| GO:BP | GO:1903999 | negative regulation of eating behavior | 1 | 1 | 6.38e-01 |
| GO:BP | GO:0048880 | sensory system development | 2 | 275 | 6.38e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 1 | 1378 | 6.38e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 1 | 68 | 6.38e-01 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 1 | 9 | 6.38e-01 |
| GO:BP | GO:1905799 | regulation of intraciliary retrograde transport | 1 | 2 | 6.38e-01 |
| GO:BP | GO:1904567 | response to wortmannin | 1 | 2 | 6.38e-01 |
| GO:BP | GO:1904568 | cellular response to wortmannin | 1 | 2 | 6.38e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 5 | 12 | 6.39e-01 |
| GO:BP | GO:0048702 | embryonic neurocranium morphogenesis | 1 | 7 | 6.39e-01 |
| GO:BP | GO:0006085 | acetyl-CoA biosynthetic process | 1 | 16 | 6.39e-01 |
| GO:BP | GO:0061377 | mammary gland lobule development | 5 | 13 | 6.39e-01 |
| GO:BP | GO:0098713 | leucine import across plasma membrane | 1 | 5 | 6.39e-01 |
| GO:BP | GO:1903801 | L-leucine import across plasma membrane | 1 | 5 | 6.39e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 2 | 12 | 6.39e-01 |
| GO:BP | GO:0060749 | mammary gland alveolus development | 5 | 13 | 6.39e-01 |
| GO:BP | GO:0070922 | RISC complex assembly | 3 | 9 | 6.40e-01 |
| GO:BP | GO:0006562 | proline catabolic process | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0006423 | cysteinyl-tRNA aminoacylation | 1 | 2 | 6.40e-01 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 1 | 6 | 6.40e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 6 | 82 | 6.40e-01 |
| GO:BP | GO:0071037 | nuclear polyadenylation-dependent snRNA catabolic process | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0071039 | nuclear polyadenylation-dependent CUT catabolic process | 1 | 2 | 6.40e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 3 | 112 | 6.40e-01 |
| GO:BP | GO:0045959 | negative regulation of complement activation, classical pathway | 2 | 4 | 6.40e-01 |
| GO:BP | GO:0071036 | nuclear polyadenylation-dependent snoRNA catabolic process | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 7 | 276 | 6.40e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 4 | 22 | 6.40e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 7 | 33 | 6.40e-01 |
| GO:BP | GO:0098751 | bone cell development | 1 | 26 | 6.40e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 6 | 540 | 6.40e-01 |
| GO:BP | GO:0021578 | hindbrain maturation | 2 | 4 | 6.40e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 6.40e-01 |
| GO:BP | GO:0061141 | lung ciliated cell differentiation | 1 | 1 | 6.40e-01 |
| GO:BP | GO:0002478 | antigen processing and presentation of exogenous peptide antigen | 2 | 24 | 6.40e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 2 | 30 | 6.40e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 2 | 69 | 6.40e-01 |
| GO:BP | GO:0002673 | regulation of acute inflammatory response | 1 | 21 | 6.40e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 3 | 17 | 6.40e-01 |
| GO:BP | GO:0006127 | glycerophosphate shuttle | 1 | 2 | 6.40e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 3 | 211 | 6.40e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 4 | 9 | 6.40e-01 |
| GO:BP | GO:0001881 | receptor recycling | 8 | 38 | 6.40e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 1 | 62 | 6.40e-01 |
| GO:BP | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1 | 4 | 6.40e-01 |
| GO:BP | GO:0033366 | protein localization to secretory granule | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0033505 | floor plate morphogenesis | 1 | 3 | 6.41e-01 |
| GO:BP | GO:0035854 | eosinophil fate commitment | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0010725 | regulation of primitive erythrocyte differentiation | 1 | 1 | 6.41e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 4 | 19 | 6.41e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 7 | 6.41e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 2 | 115 | 6.41e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 2 | 115 | 6.41e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 2 | 20 | 6.41e-01 |
| GO:BP | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling | 1 | 12 | 6.41e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 8 | 27 | 6.42e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 1 | 1167 | 6.42e-01 |
| GO:BP | GO:0031929 | TOR signaling | 40 | 115 | 6.42e-01 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 3 | 12 | 6.42e-01 |
| GO:BP | GO:0140246 | protein catabolic process at synapse | 1 | 2 | 6.42e-01 |
| GO:BP | GO:0002369 | T cell cytokine production | 1 | 18 | 6.42e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 3 | 73 | 6.42e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 1 | 4 | 6.42e-01 |
| GO:BP | GO:0140249 | protein catabolic process at postsynapse | 1 | 2 | 6.42e-01 |
| GO:BP | GO:0099525 | presynaptic dense core vesicle exocytosis | 1 | 4 | 6.42e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 6.42e-01 |
| GO:BP | GO:0046069 | cGMP catabolic process | 2 | 4 | 6.42e-01 |
| GO:BP | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis | 1 | 2 | 6.42e-01 |
| GO:BP | GO:0001510 | RNA methylation | 27 | 87 | 6.42e-01 |
| GO:BP | GO:1900363 | regulation of mRNA polyadenylation | 4 | 11 | 6.42e-01 |
| GO:BP | GO:2001137 | positive regulation of endocytic recycling | 1 | 7 | 6.42e-01 |
| GO:BP | GO:0002724 | regulation of T cell cytokine production | 1 | 18 | 6.42e-01 |
| GO:BP | GO:0051238 | sequestering of metal ion | 7 | 17 | 6.42e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 8 | 260 | 6.42e-01 |
| GO:BP | GO:0043011 | myeloid dendritic cell differentiation | 1 | 10 | 6.42e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 2 | 8 | 6.42e-01 |
| GO:BP | GO:0072109 | glomerular mesangium development | 2 | 13 | 6.42e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 6.42e-01 |
| GO:BP | GO:0033240 | positive regulation of amine metabolic process | 3 | 7 | 6.42e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 2 | 30 | 6.42e-01 |
| GO:BP | GO:0098902 | regulation of membrane depolarization during action potential | 2 | 5 | 6.42e-01 |
| GO:BP | GO:0002337 | B-1a B cell differentiation | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 4 | 25 | 6.43e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 3 | 39 | 6.43e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 6.43e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 3 | 496 | 6.43e-01 |
| GO:BP | GO:0034143 | regulation of toll-like receptor 4 signaling pathway | 3 | 15 | 6.43e-01 |
| GO:BP | GO:0009589 | detection of UV | 1 | 2 | 6.43e-01 |
| GO:BP | GO:0060841 | venous blood vessel development | 5 | 13 | 6.44e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 6 | 25 | 6.44e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 6 | 25 | 6.44e-01 |
| GO:BP | GO:0042339 | keratan sulfate metabolic process | 4 | 15 | 6.44e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 81 | 6.44e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 6 | 37 | 6.44e-01 |
| GO:BP | GO:1905772 | positive regulation of mesodermal cell differentiation | 1 | 1 | 6.44e-01 |
| GO:BP | GO:0045793 | positive regulation of cell size | 1 | 11 | 6.44e-01 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 3 | 13 | 6.44e-01 |
| GO:BP | GO:0048469 | cell maturation | 1 | 108 | 6.44e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 6.44e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 2 | 78 | 6.44e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 2 | 54 | 6.44e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 2 | 276 | 6.44e-01 |
| GO:BP | GO:1905691 | lipid droplet disassembly | 1 | 7 | 6.44e-01 |
| GO:BP | GO:0036090 | cleavage furrow ingression | 1 | 2 | 6.45e-01 |
| GO:BP | GO:0034462 | small-subunit processome assembly | 1 | 3 | 6.45e-01 |
| GO:BP | GO:1904430 | negative regulation of t-circle formation | 1 | 2 | 6.45e-01 |
| GO:BP | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste | 1 | 1 | 6.45e-01 |
| GO:BP | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway | 1 | 2 | 6.45e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 71 | 560 | 6.45e-01 |
| GO:BP | GO:0046501 | protoporphyrinogen IX metabolic process | 2 | 11 | 6.45e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 8 | 36 | 6.45e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 4 | 78 | 6.45e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 2 | 40 | 6.45e-01 |
| GO:BP | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | 2 | 4 | 6.45e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 3 | 3237 | 6.45e-01 |
| GO:BP | GO:0097242 | amyloid-beta clearance | 7 | 27 | 6.45e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 1 | 129 | 6.45e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 2 | 11 | 6.45e-01 |
| GO:BP | GO:0008340 | determination of adult lifespan | 9 | 16 | 6.45e-01 |
| GO:BP | GO:0034333 | adherens junction assembly | 5 | 13 | 6.45e-01 |
| GO:BP | GO:0036093 | germ cell proliferation | 2 | 7 | 6.45e-01 |
| GO:BP | GO:0070562 | regulation of vitamin D receptor signaling pathway | 1 | 9 | 6.45e-01 |
| GO:BP | GO:0090219 | negative regulation of lipid kinase activity | 2 | 8 | 6.45e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 2 | 114 | 6.45e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 1 | 13 | 6.45e-01 |
| GO:BP | GO:0098908 | regulation of neuronal action potential | 2 | 3 | 6.45e-01 |
| GO:BP | GO:0007548 | sex differentiation | 1 | 191 | 6.45e-01 |
| GO:BP | GO:0001949 | sebaceous gland cell differentiation | 1 | 1 | 6.46e-01 |
| GO:BP | GO:1904002 | regulation of sebum secreting cell proliferation | 1 | 1 | 6.46e-01 |
| GO:BP | GO:0007493 | endodermal cell fate determination | 1 | 1 | 6.46e-01 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 1 | 8 | 6.46e-01 |
| GO:BP | GO:0051036 | regulation of endosome size | 4 | 13 | 6.47e-01 |
| GO:BP | GO:0099105 | ion channel modulating, G protein-coupled receptor signaling pathway | 1 | 7 | 6.47e-01 |
| GO:BP | GO:0021828 | gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 3 | 6 | 6.47e-01 |
| GO:BP | GO:0033383 | geranyl diphosphate metabolic process | 1 | 3 | 6.47e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 4 | 485 | 6.47e-01 |
| GO:BP | GO:0006667 | sphinganine metabolic process | 1 | 2 | 6.47e-01 |
| GO:BP | GO:0021856 | hypothalamic tangential migration using cell-axon interactions | 3 | 6 | 6.47e-01 |
| GO:BP | GO:0060670 | branching involved in labyrinthine layer morphogenesis | 3 | 10 | 6.47e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 14 | 1226 | 6.47e-01 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 2 | 6 | 6.47e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 5 | 639 | 6.47e-01 |
| GO:BP | GO:0007000 | nucleolus organization | 1 | 17 | 6.47e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 9 | 25 | 6.48e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 6.48e-01 |
| GO:BP | GO:0009067 | aspartate family amino acid biosynthetic process | 7 | 18 | 6.48e-01 |
| GO:BP | GO:0034502 | protein localization to chromosome | 1 | 110 | 6.48e-01 |
| GO:BP | GO:0021535 | cell migration in hindbrain | 5 | 12 | 6.49e-01 |
| GO:BP | GO:0072377 | blood coagulation, common pathway | 1 | 1 | 6.49e-01 |
| GO:BP | GO:0070384 | Harderian gland development | 2 | 4 | 6.49e-01 |
| GO:BP | GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 3 | 5 | 6.49e-01 |
| GO:BP | GO:0060426 | lung vasculature development | 1 | 3 | 6.49e-01 |
| GO:BP | GO:0072257 | metanephric nephron tubule epithelial cell differentiation | 3 | 5 | 6.49e-01 |
| GO:BP | GO:0010818 | T cell chemotaxis | 4 | 12 | 6.49e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 4 | 8 | 6.49e-01 |
| GO:BP | GO:0060026 | convergent extension | 1 | 14 | 6.50e-01 |
| GO:BP | GO:0070305 | response to cGMP | 2 | 7 | 6.50e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 13 | 94 | 6.50e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 1 | 22 | 6.50e-01 |
| GO:BP | GO:0005997 | xylulose metabolic process | 1 | 1 | 6.50e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 1 | 222 | 6.50e-01 |
| GO:BP | GO:0010621 | negative regulation of transcription by transcription factor localization | 2 | 3 | 6.50e-01 |
| GO:BP | GO:0033623 | regulation of integrin activation | 3 | 11 | 6.50e-01 |
| GO:BP | GO:1905424 | regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 6.50e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 7 | 91 | 6.50e-01 |
| GO:BP | GO:0045726 | positive regulation of integrin biosynthetic process | 1 | 2 | 6.50e-01 |
| GO:BP | GO:0051867 | general adaptation syndrome, behavioral process | 1 | 3 | 6.50e-01 |
| GO:BP | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development | 1 | 2 | 6.50e-01 |
| GO:BP | GO:1905426 | positive regulation of Wnt-mediated midbrain dopaminergic neuron differentiation | 1 | 2 | 6.50e-01 |
| GO:BP | GO:2000815 | regulation of mRNA stability involved in response to oxidative stress | 1 | 2 | 6.51e-01 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 2 | 6 | 6.51e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 3 | 40 | 6.51e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 6.51e-01 |
| GO:BP | GO:0045629 | negative regulation of T-helper 2 cell differentiation | 1 | 3 | 6.51e-01 |
| GO:BP | GO:0051901 | positive regulation of mitochondrial depolarization | 1 | 6 | 6.51e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 4 | 16 | 6.51e-01 |
| GO:BP | GO:0060152 | microtubule-based peroxisome localization | 1 | 2 | 6.51e-01 |
| GO:BP | GO:0060151 | peroxisome localization | 1 | 2 | 6.51e-01 |
| GO:BP | GO:0010753 | positive regulation of cGMP-mediated signaling | 1 | 3 | 6.52e-01 |
| GO:BP | GO:2000525 | positive regulation of T cell costimulation | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 6.52e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 1 | 120 | 6.52e-01 |
| GO:BP | GO:0002931 | response to ischemia | 6 | 46 | 6.52e-01 |
| GO:BP | GO:2001136 | negative regulation of endocytic recycling | 1 | 1 | 6.52e-01 |
| GO:BP | GO:0016036 | cellular response to phosphate starvation | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 16 | 97 | 6.52e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 3 | 4953 | 6.52e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 4 | 27 | 6.52e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 6.52e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 2 | 17 | 6.52e-01 |
| GO:BP | GO:0072190 | ureter urothelium development | 1 | 3 | 6.52e-01 |
| GO:BP | GO:0003430 | growth plate cartilage chondrocyte growth | 1 | 3 | 6.52e-01 |
| GO:BP | GO:0040019 | positive regulation of embryonic development | 3 | 17 | 6.52e-01 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 9 | 21 | 6.52e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 6.52e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 6 | 18 | 6.52e-01 |
| GO:BP | GO:0022008 | neurogenesis | 20 | 1294 | 6.52e-01 |
| GO:BP | GO:1902214 | regulation of interleukin-4-mediated signaling pathway | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0090240 | positive regulation of histone H4 acetylation | 3 | 6 | 6.52e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 1 | 31 | 6.52e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 29 | 69 | 6.52e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 42 | 101 | 6.52e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 12 | 26 | 6.52e-01 |
| GO:BP | GO:0072560 | type B pancreatic cell maturation | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0007341 | penetration of zona pellucida | 1 | 4 | 6.52e-01 |
| GO:BP | GO:0060592 | mammary gland formation | 2 | 6 | 6.52e-01 |
| GO:BP | GO:0060699 | regulation of endoribonuclease activity | 1 | 4 | 6.52e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 6.52e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 84 | 6.52e-01 |
| GO:BP | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation | 3 | 4 | 6.52e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 14 | 90 | 6.52e-01 |
| GO:BP | GO:1904837 | beta-catenin-TCF complex assembly | 1 | 3 | 6.52e-01 |
| GO:BP | GO:0090309 | positive regulation of DNA methylation-dependent heterochromatin formation | 1 | 12 | 6.52e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 2 | 49 | 6.52e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 11 | 42 | 6.52e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 3 | 421 | 6.52e-01 |
| GO:BP | GO:0007538 | primary sex determination | 1 | 2 | 6.52e-01 |
| GO:BP | GO:0046833 | positive regulation of RNA export from nucleus | 2 | 5 | 6.52e-01 |
| GO:BP | GO:0051707 | response to other organism | 8 | 820 | 6.52e-01 |
| GO:BP | GO:0036261 | 7-methylguanosine cap hypermethylation | 2 | 8 | 6.52e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 2 | 68 | 6.52e-01 |
| GO:BP | GO:0070173 | regulation of enamel mineralization | 1 | 2 | 6.53e-01 |
| GO:BP | GO:0071139 | resolution of recombination intermediates | 3 | 6 | 6.53e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 2 | 97 | 6.53e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 1 | 10 | 6.53e-01 |
| GO:BP | GO:0035551 | protein initiator methionine removal involved in protein maturation | 1 | 3 | 6.53e-01 |
| GO:BP | GO:0042116 | macrophage activation | 1 | 57 | 6.53e-01 |
| GO:BP | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity | 2 | 6 | 6.53e-01 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 12 | 31 | 6.53e-01 |
| GO:BP | GO:1905430 | cellular response to glycine | 1 | 2 | 6.53e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 8 | 820 | 6.54e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 4 | 12 | 6.54e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 4 | 27 | 6.54e-01 |
| GO:BP | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration | 2 | 5 | 6.54e-01 |
| GO:BP | GO:0003099 | positive regulation of the force of heart contraction by chemical signal | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0006097 | glyoxylate cycle | 1 | 2 | 6.54e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 2 | 178 | 6.54e-01 |
| GO:BP | GO:0090158 | endoplasmic reticulum membrane organization | 3 | 14 | 6.54e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 2 | 19 | 6.54e-01 |
| GO:BP | GO:0042762 | regulation of sulfur metabolic process | 4 | 12 | 6.54e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 5 | 344 | 6.54e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 4 | 35 | 6.54e-01 |
| GO:BP | GO:0007507 | heart development | 11 | 506 | 6.55e-01 |
| GO:BP | GO:0046813 | receptor-mediated virion attachment to host cell | 1 | 6 | 6.55e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 52 | 144 | 6.55e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 7 | 74 | 6.55e-01 |
| GO:BP | GO:0050658 | RNA transport | 52 | 144 | 6.55e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 16 | 6.55e-01 |
| GO:BP | GO:0016183 | synaptic vesicle coating | 2 | 4 | 6.55e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 2 | 47 | 6.55e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 5 | 763 | 6.55e-01 |
| GO:BP | GO:1904347 | regulation of small intestine smooth muscle contraction | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0048632 | negative regulation of skeletal muscle tissue growth | 1 | 1 | 6.55e-01 |
| GO:BP | GO:1904349 | positive regulation of small intestine smooth muscle contraction | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0002939 | tRNA N1-guanine methylation | 1 | 3 | 6.55e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 6 | 23 | 6.55e-01 |
| GO:BP | GO:0034516 | response to vitamin B6 | 1 | 1 | 6.55e-01 |
| GO:BP | GO:0019541 | propionate metabolic process | 1 | 2 | 6.56e-01 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 3 | 7 | 6.56e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 2 | 140 | 6.56e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 3 | 15 | 6.56e-01 |
| GO:BP | GO:0008355 | olfactory learning | 1 | 2 | 6.56e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 1 | 6 | 6.56e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 2 | 10 | 6.56e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 6.56e-01 |
| GO:BP | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 2 | 6.56e-01 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 3 | 6 | 6.56e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 9 | 31 | 6.56e-01 |
| GO:BP | GO:0051782 | negative regulation of cell division | 7 | 15 | 6.56e-01 |
| GO:BP | GO:0032908 | regulation of transforming growth factor beta1 production | 4 | 10 | 6.57e-01 |
| GO:BP | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c | 1 | 7 | 6.57e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 47 | 149 | 6.57e-01 |
| GO:BP | GO:0002457 | T cell antigen processing and presentation | 2 | 4 | 6.57e-01 |
| GO:BP | GO:0045010 | actin nucleation | 1 | 50 | 6.57e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 5 | 31 | 6.57e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 2 | 245 | 6.57e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 6 | 49 | 6.57e-01 |
| GO:BP | GO:0030282 | bone mineralization | 9 | 82 | 6.57e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 4 | 8 | 6.57e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2 | 4045 | 6.58e-01 |
| GO:BP | GO:0019509 | L-methionine salvage from methylthioadenosine | 2 | 5 | 6.58e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 85 | 6.58e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 6 | 16 | 6.58e-01 |
| GO:BP | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway | 3 | 9 | 6.58e-01 |
| GO:BP | GO:0071716 | leukotriene transport | 2 | 5 | 6.58e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 6.58e-01 |
| GO:BP | GO:1901623 | regulation of lymphocyte chemotaxis | 1 | 13 | 6.58e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 2 | 126 | 6.58e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 6.58e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 4 | 27 | 6.58e-01 |
| GO:BP | GO:0035600 | tRNA methylthiolation | 1 | 2 | 6.58e-01 |
| GO:BP | GO:0009725 | response to hormone | 11 | 642 | 6.58e-01 |
| GO:BP | GO:0033133 | positive regulation of glucokinase activity | 2 | 4 | 6.58e-01 |
| GO:BP | GO:0050923 | regulation of negative chemotaxis | 1 | 4 | 6.58e-01 |
| GO:BP | GO:1990145 | maintenance of translational fidelity | 1 | 2 | 6.58e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 1 | 9 | 6.58e-01 |
| GO:BP | GO:0016056 | rhodopsin mediated signaling pathway | 3 | 6 | 6.58e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 7 | 61 | 6.58e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 11 | 1028 | 6.58e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 48 | 151 | 6.58e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 8 | 22 | 6.58e-01 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 1 | 9 | 6.58e-01 |
| GO:BP | GO:1903901 | negative regulation of viral life cycle | 1 | 17 | 6.58e-01 |
| GO:BP | GO:0072387 | flavin adenine dinucleotide metabolic process | 1 | 2 | 6.58e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 1 | 20 | 6.59e-01 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 1 | 16 | 6.59e-01 |
| GO:BP | GO:0042092 | type 2 immune response | 1 | 19 | 6.59e-01 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 1 | 17 | 6.59e-01 |
| GO:BP | GO:0035272 | exocrine system development | 3 | 34 | 6.59e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 8 | 14 | 6.59e-01 |
| GO:BP | GO:0006102 | isocitrate metabolic process | 2 | 6 | 6.59e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 10 | 24 | 6.59e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 3 | 372 | 6.59e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 10 | 24 | 6.59e-01 |
| GO:BP | GO:0070881 | regulation of proline transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0032907 | transforming growth factor beta3 production | 1 | 3 | 6.59e-01 |
| GO:BP | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion | 2 | 5 | 6.59e-01 |
| GO:BP | GO:0048203 | vesicle targeting, trans-Golgi to endosome | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0032910 | regulation of transforming growth factor beta3 production | 1 | 3 | 6.59e-01 |
| GO:BP | GO:2000878 | positive regulation of oligopeptide transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:2000880 | positive regulation of dipeptide transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | 2 | 6 | 6.59e-01 |
| GO:BP | GO:0051591 | response to cAMP | 7 | 66 | 6.59e-01 |
| GO:BP | GO:0046338 | phosphatidylethanolamine catabolic process | 2 | 4 | 6.59e-01 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 3 | 10 | 6.59e-01 |
| GO:BP | GO:0021551 | central nervous system morphogenesis | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 3 | 22 | 6.59e-01 |
| GO:BP | GO:2001148 | regulation of dipeptide transmembrane transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:1902836 | positive regulation of proline import across plasma membrane | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0060613 | fat pad development | 3 | 6 | 6.59e-01 |
| GO:BP | GO:0090088 | regulation of oligopeptide transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0038192 | gastric inhibitory peptide signaling pathway | 1 | 1 | 6.59e-01 |
| GO:BP | GO:0016115 | terpenoid catabolic process | 2 | 4 | 6.59e-01 |
| GO:BP | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 2 | 4 | 6.59e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 2 | 140 | 6.59e-01 |
| GO:BP | GO:0097155 | fasciculation of sensory neuron axon | 1 | 3 | 6.59e-01 |
| GO:BP | GO:0090249 | regulation of cell migration involved in somitogenic axis elongation | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0090248 | cell migration involved in somitogenic axis elongation | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway | 3 | 8 | 6.59e-01 |
| GO:BP | GO:0009617 | response to bacterium | 4 | 321 | 6.59e-01 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 4 | 10 | 6.59e-01 |
| GO:BP | GO:0001547 | antral ovarian follicle growth | 3 | 5 | 6.59e-01 |
| GO:BP | GO:2001150 | positive regulation of dipeptide transmembrane transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0110096 | cellular response to aldehyde | 6 | 10 | 6.59e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 3 | 8 | 6.59e-01 |
| GO:BP | GO:1905735 | regulation of L-proline import across plasma membrane | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 2 | 8 | 6.59e-01 |
| GO:BP | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 3 | 6.59e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 2 | 101 | 6.59e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 1 | 7 | 6.59e-01 |
| GO:BP | GO:1905737 | positive regulation of L-proline import across plasma membrane | 1 | 2 | 6.59e-01 |
| GO:BP | GO:1902834 | regulation of proline import across plasma membrane | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0090089 | regulation of dipeptide transport | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 1 | 212 | 6.59e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 6 | 13 | 6.59e-01 |
| GO:BP | GO:1900126 | negative regulation of hyaluronan biosynthetic process | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0031650 | regulation of heat generation | 2 | 7 | 6.59e-01 |
| GO:BP | GO:0002604 | regulation of dendritic cell antigen processing and presentation | 2 | 5 | 6.59e-01 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 2 | 7 | 6.59e-01 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 6 | 17 | 6.59e-01 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 1 | 5 | 6.59e-01 |
| GO:BP | GO:0043587 | tongue morphogenesis | 2 | 7 | 6.59e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 7 | 25 | 6.59e-01 |
| GO:BP | GO:0062029 | positive regulation of stress granule assembly | 1 | 2 | 6.59e-01 |
| GO:BP | GO:0061073 | ciliary body morphogenesis | 1 | 3 | 6.59e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 1 | 128 | 6.59e-01 |
| GO:BP | GO:0036089 | cleavage furrow formation | 1 | 7 | 6.59e-01 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 3 | 16 | 6.59e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 9 | 199 | 6.60e-01 |
| GO:BP | GO:1901072 | glucosamine-containing compound catabolic process | 3 | 5 | 6.60e-01 |
| GO:BP | GO:0048213 | Golgi vesicle prefusion complex stabilization | 1 | 2 | 6.60e-01 |
| GO:BP | GO:0031054 | pre-miRNA processing | 5 | 14 | 6.60e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 2 | 50 | 6.60e-01 |
| GO:BP | GO:0001743 | lens placode formation | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 1 | 25 | 6.61e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 2 | 122 | 6.61e-01 |
| GO:BP | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | 3 | 4 | 6.61e-01 |
| GO:BP | GO:0071380 | cellular response to prostaglandin E stimulus | 1 | 14 | 6.61e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 10 | 53 | 6.61e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 2 | 3 | 6.61e-01 |
| GO:BP | GO:0046619 | lens placode formation involved in camera-type eye formation | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0030540 | female genitalia development | 2 | 14 | 6.61e-01 |
| GO:BP | GO:0061458 | reproductive system development | 1 | 215 | 6.61e-01 |
| GO:BP | GO:0006703 | estrogen biosynthetic process | 2 | 6 | 6.61e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 2 | 3 | 6.61e-01 |
| GO:BP | GO:0051138 | positive regulation of NK T cell differentiation | 1 | 3 | 6.61e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 3 | 6 | 6.61e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 6.61e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 3 | 6 | 6.61e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 1 | 7 | 6.61e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 1 | 49 | 6.61e-01 |
| GO:BP | GO:0001778 | plasma membrane repair | 2 | 28 | 6.61e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 3 | 6 | 6.61e-01 |
| GO:BP | GO:0071415 | cellular response to purine-containing compound | 6 | 11 | 6.61e-01 |
| GO:BP | GO:1904527 | negative regulation of microtubule binding | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0021935 | cerebellar granule cell precursor tangential migration | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0060398 | regulation of growth hormone receptor signaling pathway | 3 | 5 | 6.61e-01 |
| GO:BP | GO:0009313 | oligosaccharide catabolic process | 3 | 11 | 6.61e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 15 | 73 | 6.61e-01 |
| GO:BP | GO:0048133 | male germ-line stem cell asymmetric division | 1 | 1 | 6.61e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 8 | 16 | 6.61e-01 |
| GO:BP | GO:2000543 | positive regulation of gastrulation | 1 | 5 | 6.61e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 6.61e-01 |
| GO:BP | GO:0051230 | spindle disassembly | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0051228 | mitotic spindle disassembly | 1 | 2 | 6.61e-01 |
| GO:BP | GO:0046340 | diacylglycerol catabolic process | 2 | 3 | 6.61e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 3 | 17 | 6.61e-01 |
| GO:BP | GO:0002281 | macrophage activation involved in immune response | 5 | 8 | 6.61e-01 |
| GO:BP | GO:0015872 | dopamine transport | 1 | 29 | 6.61e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 1 | 20 | 6.61e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 3 | 158 | 6.62e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 8 | 25 | 6.62e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 8 | 25 | 6.62e-01 |
| GO:BP | GO:0048172 | regulation of short-term neuronal synaptic plasticity | 1 | 9 | 6.62e-01 |
| GO:BP | GO:0003128 | heart field specification | 3 | 16 | 6.62e-01 |
| GO:BP | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly | 1 | 7 | 6.62e-01 |
| GO:BP | GO:0046449 | creatinine metabolic process | 1 | 2 | 6.62e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 2 | 16 | 6.62e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 34 | 277 | 6.63e-01 |
| GO:BP | GO:0007603 | phototransduction, visible light | 4 | 9 | 6.63e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 92 | 258 | 6.63e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 9 | 28 | 6.64e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 5 | 54 | 6.64e-01 |
| GO:BP | GO:0032940 | secretion by cell | 5 | 569 | 6.64e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 3 | 9 | 6.64e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 1 | 204 | 6.64e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 1 | 129 | 6.64e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 12 | 56 | 6.65e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 6.65e-01 |
| GO:BP | GO:0007292 | female gamete generation | 2 | 103 | 6.65e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 2 | 60 | 6.65e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 7 | 16 | 6.65e-01 |
| GO:BP | GO:0003157 | endocardium development | 3 | 9 | 6.65e-01 |
| GO:BP | GO:0015891 | siderophore transport | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0002544 | chronic inflammatory response | 1 | 6 | 6.65e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 33 | 96 | 6.65e-01 |
| GO:BP | GO:0051461 | positive regulation of corticotropin secretion | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0006427 | histidyl-tRNA aminoacylation | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0048699 | generation of neurons | 15 | 1119 | 6.65e-01 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 1 | 11 | 6.65e-01 |
| GO:BP | GO:1901841 | regulation of high voltage-gated calcium channel activity | 1 | 12 | 6.65e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 2 | 9 | 6.65e-01 |
| GO:BP | GO:0021675 | nerve development | 3 | 57 | 6.65e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 2 | 14 | 6.65e-01 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 2 | 18 | 6.65e-01 |
| GO:BP | GO:0045843 | negative regulation of striated muscle tissue development | 3 | 5 | 6.65e-01 |
| GO:BP | GO:0007420 | brain development | 4 | 591 | 6.65e-01 |
| GO:BP | GO:0015812 | gamma-aminobutyric acid transport | 1 | 11 | 6.65e-01 |
| GO:BP | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering | 1 | 2 | 6.65e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 6.65e-01 |
| GO:BP | GO:0030311 | poly-N-acetyllactosamine biosynthetic process | 2 | 5 | 6.65e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 10 | 1602 | 6.66e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 2 | 4 | 6.66e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 2 | 29 | 6.66e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 5 | 13 | 6.66e-01 |
| GO:BP | GO:0030213 | hyaluronan biosynthetic process | 3 | 10 | 6.66e-01 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 5 | 63 | 6.66e-01 |
| GO:BP | GO:1902732 | positive regulation of chondrocyte proliferation | 2 | 5 | 6.66e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 4 | 33 | 6.66e-01 |
| GO:BP | GO:0055025 | positive regulation of cardiac muscle tissue development | 1 | 2 | 6.66e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 4 | 40 | 6.66e-01 |
| GO:BP | GO:1903141 | negative regulation of establishment of endothelial barrier | 1 | 2 | 6.66e-01 |
| GO:BP | GO:1901551 | negative regulation of endothelial cell development | 1 | 2 | 6.66e-01 |
| GO:BP | GO:0060304 | regulation of phosphatidylinositol dephosphorylation | 2 | 5 | 6.66e-01 |
| GO:BP | GO:1905349 | ciliary transition zone assembly | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 4 | 9 | 6.67e-01 |
| GO:BP | GO:2000353 | positive regulation of endothelial cell apoptotic process | 1 | 13 | 6.67e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 6.67e-01 |
| GO:BP | GO:0003350 | pulmonary myocardium development | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0003343 | septum transversum development | 1 | 2 | 6.67e-01 |
| GO:BP | GO:0003342 | proepicardium development | 1 | 2 | 6.67e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 4 | 19 | 6.67e-01 |
| GO:BP | GO:0044857 | plasma membrane raft organization | 4 | 9 | 6.67e-01 |
| GO:BP | GO:0031144 | proteasome localization | 1 | 2 | 6.68e-01 |
| GO:BP | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development | 1 | 2 | 6.68e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 1 | 147 | 6.68e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 5 | 29 | 6.68e-01 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 5 | 9 | 6.69e-01 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 5 | 9 | 6.69e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 4 | 8 | 6.69e-01 |
| GO:BP | GO:0015965 | diadenosine tetraphosphate metabolic process | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0015960 | diadenosine polyphosphate biosynthetic process | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0015966 | diadenosine tetraphosphate biosynthetic process | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 5 | 30 | 6.69e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 5 | 70 | 6.69e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 1 | 93 | 6.69e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 12 | 222 | 6.69e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 6.69e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 6.69e-01 |
| GO:BP | GO:0071455 | cellular response to hyperoxia | 1 | 5 | 6.69e-01 |
| GO:BP | GO:0021941 | negative regulation of cerebellar granule cell precursor proliferation | 1 | 2 | 6.69e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 2 | 3386 | 6.69e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 1 | 36 | 6.69e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 1 | 36 | 6.69e-01 |
| GO:BP | GO:0034414 | tRNA 3’-trailer cleavage, endonucleolytic | 1 | 2 | 6.69e-01 |
| GO:BP | GO:1903385 | regulation of homophilic cell adhesion | 1 | 3 | 6.69e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 2 | 12 | 6.69e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 11 | 129 | 6.69e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 2 | 4 | 6.69e-01 |
| GO:BP | GO:0007417 | central nervous system development | 6 | 792 | 6.69e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 2 | 4 | 6.69e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 2 | 4 | 6.69e-01 |
| GO:BP | GO:1905581 | positive regulation of low-density lipoprotein particle clearance | 1 | 2 | 6.70e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 4 | 17 | 6.70e-01 |
| GO:BP | GO:1902688 | regulation of NAD metabolic process | 1 | 1 | 6.70e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 8 | 850 | 6.70e-01 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 2 | 3 | 6.70e-01 |
| GO:BP | GO:0018117 | protein adenylylation | 1 | 2 | 6.70e-01 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 2 | 3 | 6.70e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 6.70e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 1 | 192 | 6.70e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 10 | 25 | 6.70e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 1 | 17 | 6.70e-01 |
| GO:BP | GO:0031947 | negative regulation of glucocorticoid biosynthetic process | 1 | 5 | 6.70e-01 |
| GO:BP | GO:0035747 | natural killer cell chemotaxis | 2 | 3 | 6.70e-01 |
| GO:BP | GO:1904383 | response to sodium phosphate | 3 | 5 | 6.70e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 6 | 54 | 6.70e-01 |
| GO:BP | GO:0031944 | negative regulation of glucocorticoid metabolic process | 1 | 5 | 6.70e-01 |
| GO:BP | GO:0018175 | protein nucleotidylation | 1 | 2 | 6.70e-01 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 2 | 13 | 6.70e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 1 | 19 | 6.70e-01 |
| GO:BP | GO:1900112 | regulation of histone H3-K9 trimethylation | 1 | 5 | 6.71e-01 |
| GO:BP | GO:0071284 | cellular response to lead ion | 1 | 4 | 6.71e-01 |
| GO:BP | GO:0061669 | spontaneous neurotransmitter secretion | 2 | 5 | 6.72e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 20 | 172 | 6.72e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 2 | 187 | 6.72e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 6.72e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 3 | 233 | 6.72e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 6.72e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 14 | 72 | 6.72e-01 |
| GO:BP | GO:0060334 | regulation of type II interferon-mediated signaling pathway | 2 | 12 | 6.72e-01 |
| GO:BP | GO:0060330 | regulation of response to type II interferon | 2 | 12 | 6.72e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 1 | 71 | 6.73e-01 |
| GO:BP | GO:1904028 | positive regulation of collagen fibril organization | 1 | 4 | 6.73e-01 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 1 | 4 | 6.73e-01 |
| GO:BP | GO:2000097 | regulation of smooth muscle cell-matrix adhesion | 1 | 3 | 6.73e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 3 | 234 | 6.74e-01 |
| GO:BP | GO:0021678 | third ventricle development | 1 | 4 | 6.74e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 1 | 121 | 6.74e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 2 | 151 | 6.74e-01 |
| GO:BP | GO:1990442 | intrinsic apoptotic signaling pathway in response to nitrosative stress | 1 | 3 | 6.74e-01 |
| GO:BP | GO:0006901 | vesicle coating | 15 | 36 | 6.74e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 4 | 17 | 6.74e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 6.74e-01 |
| GO:BP | GO:0006204 | IMP catabolic process | 2 | 3 | 6.75e-01 |
| GO:BP | GO:0046054 | dGMP metabolic process | 2 | 2 | 6.75e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 6.75e-01 |
| GO:BP | GO:0016310 | phosphorylation | 1 | 1372 | 6.75e-01 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 1 | 9 | 6.75e-01 |
| GO:BP | GO:0099011 | neuronal dense core vesicle exocytosis | 1 | 4 | 6.75e-01 |
| GO:BP | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development | 1 | 3 | 6.75e-01 |
| GO:BP | GO:0046032 | ADP catabolic process | 1 | 2 | 6.75e-01 |
| GO:BP | GO:0038020 | insulin receptor recycling | 1 | 4 | 6.75e-01 |
| GO:BP | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway | 1 | 3 | 6.75e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 2 | 221 | 6.75e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 2 | 55 | 6.76e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 3 | 295 | 6.76e-01 |
| GO:BP | GO:1904694 | negative regulation of vascular associated smooth muscle contraction | 1 | 3 | 6.76e-01 |
| GO:BP | GO:0015949 | nucleobase-containing small molecule interconversion | 3 | 8 | 6.76e-01 |
| GO:BP | GO:0051169 | nuclear transport | 3 | 295 | 6.76e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 6.76e-01 |
| GO:BP | GO:0071888 | macrophage apoptotic process | 1 | 9 | 6.77e-01 |
| GO:BP | GO:1903971 | positive regulation of response to macrophage colony-stimulating factor | 1 | 1 | 6.77e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 2 | 41 | 6.77e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 3 | 234 | 6.77e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 6.77e-01 |
| GO:BP | GO:0036071 | N-glycan fucosylation | 1 | 3 | 6.77e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 1 | 40 | 6.77e-01 |
| GO:BP | GO:0150031 | regulation of protein localization to lysosome | 1 | 3 | 6.77e-01 |
| GO:BP | GO:0032707 | negative regulation of interleukin-23 production | 1 | 2 | 6.77e-01 |
| GO:BP | GO:0030853 | negative regulation of granulocyte differentiation | 3 | 6 | 6.77e-01 |
| GO:BP | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II | 1 | 17 | 6.77e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 6 | 15 | 6.77e-01 |
| GO:BP | GO:1903974 | positive regulation of cellular response to macrophage colony-stimulating factor stimulus | 1 | 1 | 6.77e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 1 | 57 | 6.77e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 6 | 15 | 6.77e-01 |
| GO:BP | GO:0016322 | neuron remodeling | 1 | 11 | 6.77e-01 |
| GO:BP | GO:0046549 | retinal cone cell development | 2 | 5 | 6.77e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 2 | 516 | 6.77e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 6.77e-01 |
| GO:BP | GO:0007354 | zygotic determination of anterior/posterior axis, embryo | 1 | 2 | 6.77e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 2 | 135 | 6.77e-01 |
| GO:BP | GO:0030450 | regulation of complement activation, classical pathway | 2 | 4 | 6.77e-01 |
| GO:BP | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline | 3 | 9 | 6.77e-01 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 9 | 23 | 6.77e-01 |
| GO:BP | GO:0097252 | oligodendrocyte apoptotic process | 2 | 4 | 6.77e-01 |
| GO:BP | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation | 1 | 2 | 6.77e-01 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 1 | 4 | 6.78e-01 |
| GO:BP | GO:0034375 | high-density lipoprotein particle remodeling | 2 | 11 | 6.78e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 2 | 12 | 6.78e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 3 | 440 | 6.78e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 7 | 1061 | 6.78e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 4 | 517 | 6.78e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 2 | 4 | 6.78e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 2 | 3 | 6.78e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 2 | 3 | 6.78e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 2 | 3 | 6.78e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 2 | 3 | 6.78e-01 |
| GO:BP | GO:1905551 | negative regulation of protein localization to endoplasmic reticulum | 1 | 2 | 6.78e-01 |
| GO:BP | GO:0009227 | nucleotide-sugar catabolic process | 1 | 3 | 6.78e-01 |
| GO:BP | GO:2000674 | regulation of type B pancreatic cell apoptotic process | 2 | 6 | 6.78e-01 |
| GO:BP | GO:0051541 | elastin metabolic process | 2 | 5 | 6.78e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 1 | 15 | 6.79e-01 |
| GO:BP | GO:0055062 | phosphate ion homeostasis | 1 | 8 | 6.79e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 3 | 99 | 6.79e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 3 | 99 | 6.79e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 9 | 1441 | 6.79e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 13 | 120 | 6.79e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 3 | 427 | 6.79e-01 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 1 | 13 | 6.79e-01 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 4 | 5 | 6.80e-01 |
| GO:BP | GO:0010669 | epithelial structure maintenance | 1 | 15 | 6.80e-01 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 6 | 16 | 6.80e-01 |
| GO:BP | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | 1 | 4 | 6.80e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 14 | 71 | 6.80e-01 |
| GO:BP | GO:1990120 | messenger ribonucleoprotein complex assembly | 1 | 3 | 6.80e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 2 | 170 | 6.80e-01 |
| GO:BP | GO:0061549 | sympathetic ganglion development | 4 | 6 | 6.80e-01 |
| GO:BP | GO:0035995 | detection of muscle stretch | 3 | 7 | 6.80e-01 |
| GO:BP | GO:0097400 | interleukin-17-mediated signaling pathway | 2 | 8 | 6.80e-01 |
| GO:BP | GO:0090381 | regulation of heart induction | 1 | 4 | 6.80e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 6.80e-01 |
| GO:BP | GO:0100012 | regulation of heart induction by canonical Wnt signaling pathway | 1 | 4 | 6.80e-01 |
| GO:BP | GO:0070086 | ubiquitin-dependent endocytosis | 1 | 4 | 6.80e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 5 | 153 | 6.81e-01 |
| GO:BP | GO:0072176 | nephric duct development | 2 | 12 | 6.81e-01 |
| GO:BP | GO:1901537 | positive regulation of DNA demethylation | 1 | 7 | 6.81e-01 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 5 | 15 | 6.81e-01 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 1 | 5 | 6.81e-01 |
| GO:BP | GO:0036066 | protein O-linked fucosylation | 2 | 5 | 6.81e-01 |
| GO:BP | GO:0048859 | formation of anatomical boundary | 1 | 3 | 6.81e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 4 | 168 | 6.81e-01 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 6 | 11 | 6.81e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 2 | 70 | 6.82e-01 |
| GO:BP | GO:0009744 | response to sucrose | 3 | 4 | 6.82e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 2 | 70 | 6.82e-01 |
| GO:BP | GO:0034285 | response to disaccharide | 3 | 4 | 6.82e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 7 | 22 | 6.82e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 7 | 17 | 6.82e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 2 | 98 | 6.82e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 2 | 12 | 6.82e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 2 | 10 | 6.82e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 15 | 64 | 6.82e-01 |
| GO:BP | GO:0035583 | sequestering of TGFbeta in extracellular matrix | 2 | 3 | 6.82e-01 |
| GO:BP | GO:0070172 | positive regulation of tooth mineralization | 1 | 2 | 6.82e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 1 | 1380 | 6.82e-01 |
| GO:BP | GO:0010636 | positive regulation of mitochondrial fusion | 1 | 2 | 6.82e-01 |
| GO:BP | GO:1900098 | regulation of plasma cell differentiation | 2 | 2 | 6.82e-01 |
| GO:BP | GO:0061300 | cerebellum vasculature development | 1 | 2 | 6.82e-01 |
| GO:BP | GO:1905041 | regulation of epithelium regeneration | 1 | 2 | 6.82e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 1 | 8 | 6.82e-01 |
| GO:BP | GO:0046950 | cellular ketone body metabolic process | 2 | 7 | 6.82e-01 |
| GO:BP | GO:0060406 | positive regulation of penile erection | 1 | 3 | 6.82e-01 |
| GO:BP | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | 1 | 2 | 6.82e-01 |
| GO:BP | GO:1990399 | epithelium regeneration | 1 | 2 | 6.82e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 28 | 663 | 6.82e-01 |
| GO:BP | GO:1902283 | negative regulation of primary amine oxidase activity | 1 | 1 | 6.82e-01 |
| GO:BP | GO:0045347 | negative regulation of MHC class II biosynthetic process | 1 | 3 | 6.82e-01 |
| GO:BP | GO:1901026 | ripoptosome assembly involved in necroptotic process | 1 | 2 | 6.82e-01 |
| GO:BP | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine | 2 | 7 | 6.82e-01 |
| GO:BP | GO:0045016 | mitochondrial magnesium ion transmembrane transport | 1 | 2 | 6.82e-01 |
| GO:BP | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation | 1 | 5 | 6.82e-01 |
| GO:BP | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process | 2 | 3 | 6.82e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 1 | 8 | 6.82e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 4 | 25 | 6.82e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 2 | 4 | 6.82e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 1 | 41 | 6.82e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 6.82e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 4 | 25 | 6.82e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 5 | 701 | 6.82e-01 |
| GO:BP | GO:0097343 | ripoptosome assembly | 1 | 2 | 6.82e-01 |
| GO:BP | GO:0033008 | positive regulation of mast cell activation involved in immune response | 5 | 10 | 6.82e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 5 | 15 | 6.82e-01 |
| GO:BP | GO:1901000 | regulation of response to salt stress | 1 | 4 | 6.82e-01 |
| GO:BP | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | 3 | 6.82e-01 |
| GO:BP | GO:0099074 | mitochondrion to lysosome transport | 1 | 3 | 6.82e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 4 | 23 | 6.82e-01 |
| GO:BP | GO:0099075 | mitochondrion-derived vesicle mediated transport | 1 | 3 | 6.82e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 3 | 99 | 6.82e-01 |
| GO:BP | GO:0043306 | positive regulation of mast cell degranulation | 5 | 10 | 6.82e-01 |
| GO:BP | GO:1900069 | regulation of cellular hyperosmotic salinity response | 1 | 4 | 6.82e-01 |
| GO:BP | GO:0060923 | cardiac muscle cell fate commitment | 3 | 7 | 6.82e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 2 | 47 | 6.82e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 8 | 17 | 6.83e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 1 | 250 | 6.83e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 3 | 5 | 6.83e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 6.83e-01 |
| GO:BP | GO:0050433 | regulation of catecholamine secretion | 1 | 29 | 6.83e-01 |
| GO:BP | GO:1904683 | regulation of metalloendopeptidase activity | 2 | 6 | 6.83e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 6.83e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 6 | 27 | 6.83e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 4 | 19 | 6.83e-01 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 2 | 4 | 6.83e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 36 | 164 | 6.83e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 13 | 57 | 6.83e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 2 | 3 | 6.84e-01 |
| GO:BP | GO:0000451 | rRNA 2’-O-methylation | 1 | 3 | 6.84e-01 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 2 | 10 | 6.84e-01 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 2 | 4 | 6.84e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 2 | 21 | 6.84e-01 |
| GO:BP | GO:0034034 | purine nucleoside bisphosphate catabolic process | 4 | 11 | 6.84e-01 |
| GO:BP | GO:0034031 | ribonucleoside bisphosphate catabolic process | 4 | 11 | 6.84e-01 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 3 | 13 | 6.84e-01 |
| GO:BP | GO:0033869 | nucleoside bisphosphate catabolic process | 4 | 11 | 6.84e-01 |
| GO:BP | GO:0070561 | vitamin D receptor signaling pathway | 1 | 10 | 6.85e-01 |
| GO:BP | GO:0038183 | bile acid signaling pathway | 1 | 3 | 6.85e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 15 | 38 | 6.85e-01 |
| GO:BP | GO:0036118 | hyaluranon cable assembly | 1 | 3 | 6.85e-01 |
| GO:BP | GO:0046379 | extracellular polysaccharide metabolic process | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0045163 | clustering of voltage-gated potassium channels | 1 | 2 | 6.85e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 4 | 7552 | 6.85e-01 |
| GO:BP | GO:1900104 | regulation of hyaluranon cable assembly | 1 | 3 | 6.85e-01 |
| GO:BP | GO:0045226 | extracellular polysaccharide biosynthetic process | 1 | 2 | 6.85e-01 |
| GO:BP | GO:1900106 | positive regulation of hyaluranon cable assembly | 1 | 3 | 6.85e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 6 | 36 | 6.85e-01 |
| GO:BP | GO:0030850 | prostate gland development | 2 | 35 | 6.85e-01 |
| GO:BP | GO:0021722 | superior olivary nucleus maturation | 1 | 1 | 6.85e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 2 | 199 | 6.85e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 19 | 62 | 6.85e-01 |
| GO:BP | GO:0021718 | superior olivary nucleus development | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0021586 | pons maturation | 1 | 1 | 6.85e-01 |
| GO:BP | GO:0033029 | regulation of neutrophil apoptotic process | 1 | 5 | 6.85e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 10 | 55 | 6.85e-01 |
| GO:BP | GO:0140889 | DNA replication-dependent chromatin disassembly | 1 | 3 | 6.86e-01 |
| GO:BP | GO:0006612 | protein targeting to membrane | 3 | 118 | 6.86e-01 |
| GO:BP | GO:0008039 | synaptic target recognition | 1 | 2 | 6.86e-01 |
| GO:BP | GO:1904231 | positive regulation of succinate dehydrogenase activity | 1 | 2 | 6.86e-01 |
| GO:BP | GO:1904229 | regulation of succinate dehydrogenase activity | 1 | 2 | 6.86e-01 |
| GO:BP | GO:0010716 | negative regulation of extracellular matrix disassembly | 1 | 2 | 6.86e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 1 | 77 | 6.86e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 5 | 21 | 6.86e-01 |
| GO:BP | GO:0019835 | cytolysis | 1 | 6 | 6.86e-01 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 6.86e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 6 | 76 | 6.86e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 1 | 28 | 6.86e-01 |
| GO:BP | GO:0033602 | negative regulation of dopamine secretion | 1 | 4 | 6.86e-01 |
| GO:BP | GO:0070459 | prolactin secretion | 1 | 4 | 6.86e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 1 | 83 | 6.86e-01 |
| GO:BP | GO:0009749 | response to glucose | 1 | 137 | 6.86e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 2 | 4 | 6.86e-01 |
| GO:BP | GO:0015867 | ATP transport | 1 | 9 | 6.86e-01 |
| GO:BP | GO:0099022 | vesicle tethering | 5 | 31 | 6.87e-01 |
| GO:BP | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process | 2 | 3 | 6.87e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 6 | 38 | 6.87e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 14 | 157 | 6.87e-01 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 2 | 4 | 6.87e-01 |
| GO:BP | GO:0070995 | NADPH oxidation | 2 | 3 | 6.87e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 6.87e-01 |
| GO:BP | GO:1902950 | regulation of dendritic spine maintenance | 1 | 7 | 6.87e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 2 | 65 | 6.87e-01 |
| GO:BP | GO:0016191 | synaptic vesicle uncoating | 1 | 7 | 6.87e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 1 | 12 | 6.87e-01 |
| GO:BP | GO:1900272 | negative regulation of long-term synaptic potentiation | 1 | 8 | 6.87e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 8 | 529 | 6.87e-01 |
| GO:BP | GO:0019083 | viral transcription | 6 | 48 | 6.87e-01 |
| GO:BP | GO:0036228 | protein localization to nuclear inner membrane | 1 | 3 | 6.88e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 1 | 3 | 6.88e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 1 | 3 | 6.88e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 1 | 98 | 6.88e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 2 | 92 | 6.88e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 2 | 29 | 6.88e-01 |
| GO:BP | GO:0031648 | protein destabilization | 1 | 45 | 6.88e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 6.88e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 6.88e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 1 | 33 | 6.88e-01 |
| GO:BP | GO:0022010 | central nervous system myelination | 1 | 12 | 6.88e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 5 | 20 | 6.88e-01 |
| GO:BP | GO:0032291 | axon ensheathment in central nervous system | 1 | 12 | 6.88e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 2 | 112 | 6.88e-01 |
| GO:BP | GO:0031392 | regulation of prostaglandin biosynthetic process | 2 | 6 | 6.88e-01 |
| GO:BP | GO:0036316 | SREBP-SCAP complex retention in endoplasmic reticulum | 1 | 2 | 6.88e-01 |
| GO:BP | GO:1903525 | regulation of membrane tubulation | 2 | 5 | 6.88e-01 |
| GO:BP | GO:0006701 | progesterone biosynthetic process | 1 | 5 | 6.88e-01 |
| GO:BP | GO:0016486 | peptide hormone processing | 8 | 27 | 6.89e-01 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 2 | 10 | 6.89e-01 |
| GO:BP | GO:0042415 | norepinephrine metabolic process | 5 | 7 | 6.89e-01 |
| GO:BP | GO:0010232 | vascular transport | 5 | 72 | 6.89e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 5 | 72 | 6.89e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 8 | 55 | 6.89e-01 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 6.89e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 1 | 122 | 6.89e-01 |
| GO:BP | GO:0038172 | interleukin-33-mediated signaling pathway | 1 | 1 | 6.89e-01 |
| GO:BP | GO:0033206 | meiotic cytokinesis | 1 | 7 | 6.89e-01 |
| GO:BP | GO:2000632 | negative regulation of pre-miRNA processing | 1 | 2 | 6.89e-01 |
| GO:BP | GO:0061188 | negative regulation of rDNA heterochromatin formation | 1 | 2 | 6.89e-01 |
| GO:BP | GO:0061187 | regulation of rDNA heterochromatin formation | 1 | 2 | 6.89e-01 |
| GO:BP | GO:0045728 | respiratory burst after phagocytosis | 1 | 2 | 6.89e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 6.89e-01 |
| GO:BP | GO:0060155 | platelet dense granule organization | 1 | 23 | 6.89e-01 |
| GO:BP | GO:0099152 | regulation of neurotransmitter receptor transport, endosome to postsynaptic membrane | 1 | 3 | 6.89e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 6 | 17 | 6.89e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 1 | 7 | 6.89e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 3 | 24 | 6.89e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 3 | 24 | 6.89e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 10 | 906 | 6.89e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 4 | 146 | 6.90e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 1 | 33 | 6.90e-01 |
| GO:BP | GO:1901843 | positive regulation of high voltage-gated calcium channel activity | 1 | 4 | 6.90e-01 |
| GO:BP | GO:0019448 | L-cysteine catabolic process | 2 | 3 | 6.90e-01 |
| GO:BP | GO:0046439 | L-cysteine metabolic process | 2 | 3 | 6.90e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 4 | 8 | 6.90e-01 |
| GO:BP | GO:0050432 | catecholamine secretion | 1 | 31 | 6.90e-01 |
| GO:BP | GO:0060820 | inactivation of X chromosome by heterochromatin formation | 1 | 1 | 6.90e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 1 | 42 | 6.90e-01 |
| GO:BP | GO:0009093 | cysteine catabolic process | 2 | 3 | 6.90e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 3 | 20 | 6.90e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 6.90e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 16 | 92 | 6.90e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 2 | 7 | 6.90e-01 |
| GO:BP | GO:0019065 | receptor-mediated endocytosis of virus by host cell | 2 | 9 | 6.90e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 16 | 92 | 6.90e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 16 | 92 | 6.90e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 13 | 61 | 6.90e-01 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 7 | 17 | 6.90e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 6.90e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 9 | 317 | 6.90e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 1 | 179 | 6.90e-01 |
| GO:BP | GO:0045658 | regulation of neutrophil differentiation | 1 | 2 | 6.90e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 3 | 36 | 6.90e-01 |
| GO:BP | GO:0045141 | meiotic telomere clustering | 4 | 5 | 6.90e-01 |
| GO:BP | GO:0048241 | epinephrine transport | 1 | 4 | 6.91e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 3 | 373 | 6.91e-01 |
| GO:BP | GO:0048793 | pronephros development | 1 | 5 | 6.91e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 1 | 82 | 6.91e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 1 | 5 | 6.91e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 2 | 207 | 6.91e-01 |
| GO:BP | GO:0090234 | regulation of kinetochore assembly | 1 | 3 | 6.91e-01 |
| GO:BP | GO:0009746 | response to hexose | 1 | 138 | 6.91e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 5 | 80 | 6.91e-01 |
| GO:BP | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 3 | 6 | 6.91e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 2 | 36 | 6.91e-01 |
| GO:BP | GO:0016571 | histone methylation | 8 | 117 | 6.91e-01 |
| GO:BP | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death | 1 | 3 | 6.91e-01 |
| GO:BP | GO:0044381 | glucose import in response to insulin stimulus | 1 | 4 | 6.92e-01 |
| GO:BP | GO:0046931 | pore complex assembly | 6 | 19 | 6.92e-01 |
| GO:BP | GO:0032119 | sequestering of zinc ion | 2 | 5 | 6.92e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 6.92e-01 |
| GO:BP | GO:0035802 | adrenal cortex formation | 1 | 1 | 6.92e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 2 | 19 | 6.93e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 2 | 19 | 6.93e-01 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 5 | 18 | 6.93e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 6 | 10 | 6.93e-01 |
| GO:BP | GO:0001503 | ossification | 3 | 313 | 6.93e-01 |
| GO:BP | GO:1903286 | regulation of potassium ion import | 1 | 8 | 6.93e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 1 | 162 | 6.93e-01 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 2 | 5 | 6.93e-01 |
| GO:BP | GO:0002322 | B cell proliferation involved in immune response | 2 | 3 | 6.93e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 1 | 83 | 6.93e-01 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 3 | 18 | 6.94e-01 |
| GO:BP | GO:0031269 | pseudopodium assembly | 4 | 14 | 6.94e-01 |
| GO:BP | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 5 | 6.94e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 2 | 15 | 6.94e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 6.94e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 8 | 14 | 6.94e-01 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 5 | 13 | 6.95e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 1 | 26 | 6.95e-01 |
| GO:BP | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway | 2 | 5 | 6.95e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 6 | 38 | 6.95e-01 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 2 | 9 | 6.95e-01 |
| GO:BP | GO:0046348 | amino sugar catabolic process | 4 | 9 | 6.95e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 1 | 101 | 6.95e-01 |
| GO:BP | GO:0061086 | negative regulation of histone H3-K27 methylation | 1 | 2 | 6.95e-01 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 3 | 6.95e-01 |
| GO:BP | GO:0051607 | defense response to virus | 7 | 202 | 6.95e-01 |
| GO:BP | GO:1902426 | deactivation of mitotic spindle assembly checkpoint | 2 | 3 | 6.95e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 6.95e-01 |
| GO:BP | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching | 2 | 4 | 6.95e-01 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 3 | 6.95e-01 |
| GO:BP | GO:0051593 | response to folic acid | 1 | 9 | 6.95e-01 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 3 | 7 | 6.95e-01 |
| GO:BP | GO:0006508 | proteolysis | 13 | 1318 | 6.95e-01 |
| GO:BP | GO:0098814 | spontaneous synaptic transmission | 3 | 10 | 6.95e-01 |
| GO:BP | GO:1905450 | negative regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 2 | 6.95e-01 |
| GO:BP | GO:0070166 | enamel mineralization | 2 | 11 | 6.95e-01 |
| GO:BP | GO:0009086 | methionine biosynthetic process | 4 | 10 | 6.95e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 11 | 40 | 6.95e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 1 | 109 | 6.96e-01 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 1 | 34 | 6.96e-01 |
| GO:BP | GO:0016032 | viral process | 2 | 344 | 6.96e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 1 | 4 | 6.96e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 1 | 6 | 6.96e-01 |
| GO:BP | GO:0032696 | negative regulation of interleukin-13 production | 1 | 3 | 6.96e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 2 | 40 | 6.96e-01 |
| GO:BP | GO:0033085 | negative regulation of T cell differentiation in thymus | 1 | 5 | 6.96e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 3 | 68 | 6.96e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 8 | 75 | 6.97e-01 |
| GO:BP | GO:0006997 | nucleus organization | 1 | 126 | 6.97e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 1 | 66 | 6.97e-01 |
| GO:BP | GO:0038065 | collagen-activated signaling pathway | 1 | 13 | 6.97e-01 |
| GO:BP | GO:0045779 | negative regulation of bone resorption | 1 | 7 | 6.97e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 13 | 30 | 6.97e-01 |
| GO:BP | GO:0046963 | 3’-phosphoadenosine 5’-phosphosulfate transport | 1 | 2 | 6.97e-01 |
| GO:BP | GO:1902558 | 5’-adenylyl sulfate transmembrane transport | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 1 | 13 | 6.97e-01 |
| GO:BP | GO:1905382 | positive regulation of snRNA transcription by RNA polymerase II | 1 | 3 | 6.97e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 1 | 85 | 6.97e-01 |
| GO:BP | GO:1902559 | 3’-phospho-5’-adenylyl sulfate transmembrane transport | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0060322 | head development | 4 | 633 | 6.97e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 6.97e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 7 | 202 | 6.97e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 6.97e-01 |
| GO:BP | GO:1904387 | cellular response to L-phenylalanine derivative | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0097069 | cellular response to thyroxine stimulus | 1 | 2 | 6.97e-01 |
| GO:BP | GO:0003332 | negative regulation of extracellular matrix constituent secretion | 1 | 2 | 6.98e-01 |
| GO:BP | GO:2001027 | negative regulation of endothelial cell chemotaxis | 1 | 2 | 6.98e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 6.98e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 2 | 57 | 6.98e-01 |
| GO:BP | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent | 1 | 2 | 6.98e-01 |
| GO:BP | GO:0070391 | response to lipoteichoic acid | 3 | 6 | 6.98e-01 |
| GO:BP | GO:0071223 | cellular response to lipoteichoic acid | 3 | 6 | 6.98e-01 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 4 | 12 | 6.98e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 6 | 79 | 6.98e-01 |
| GO:BP | GO:0090170 | regulation of Golgi inheritance | 1 | 4 | 6.98e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 4 | 348 | 6.98e-01 |
| GO:BP | GO:0014061 | regulation of norepinephrine secretion | 2 | 5 | 6.99e-01 |
| GO:BP | GO:0048243 | norepinephrine secretion | 2 | 5 | 6.99e-01 |
| GO:BP | GO:0008584 | male gonad development | 5 | 99 | 6.99e-01 |
| GO:BP | GO:0007197 | adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway | 1 | 3 | 6.99e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 5 | 80 | 6.99e-01 |
| GO:BP | GO:0032483 | regulation of Rab protein signal transduction | 1 | 7 | 6.99e-01 |
| GO:BP | GO:0051098 | regulation of binding | 36 | 317 | 6.99e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 2 | 16 | 6.99e-01 |
| GO:BP | GO:0014042 | positive regulation of neuron maturation | 1 | 3 | 6.99e-01 |
| GO:BP | GO:0072298 | regulation of metanephric glomerulus development | 1 | 1 | 6.99e-01 |
| GO:BP | GO:0072216 | positive regulation of metanephros development | 1 | 1 | 6.99e-01 |
| GO:BP | GO:0072300 | positive regulation of metanephric glomerulus development | 1 | 1 | 6.99e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 2 | 69 | 6.99e-01 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 1 | 6 | 6.99e-01 |
| GO:BP | GO:0001905 | activation of membrane attack complex | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0061868 | hepatic stellate cell migration | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 6.99e-01 |
| GO:BP | GO:0010815 | bradykinin catabolic process | 1 | 6 | 6.99e-01 |
| GO:BP | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 6.99e-01 |
| GO:BP | GO:1990034 | calcium ion export across plasma membrane | 3 | 6 | 6.99e-01 |
| GO:BP | GO:0001969 | regulation of activation of membrane attack complex | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0031453 | positive regulation of heterochromatin formation | 1 | 15 | 6.99e-01 |
| GO:BP | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 1 | 142 | 6.99e-01 |
| GO:BP | GO:0061869 | regulation of hepatic stellate cell migration | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0061870 | positive regulation of hepatic stellate cell migration | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0060347 | heart trabecula formation | 5 | 13 | 6.99e-01 |
| GO:BP | GO:0033633 | negative regulation of cell-cell adhesion mediated by integrin | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0002074 | extraocular skeletal muscle development | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0072006 | nephron development | 2 | 117 | 6.99e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 2 | 23 | 6.99e-01 |
| GO:BP | GO:0061551 | trigeminal ganglion development | 2 | 3 | 6.99e-01 |
| GO:BP | GO:1905770 | regulation of mesodermal cell differentiation | 2 | 6 | 6.99e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 2 | 22 | 6.99e-01 |
| GO:BP | GO:0009611 | response to wounding | 3 | 415 | 6.99e-01 |
| GO:BP | GO:0120263 | positive regulation of heterochromatin organization | 1 | 15 | 6.99e-01 |
| GO:BP | GO:0043474 | pigment metabolic process involved in pigmentation | 1 | 3 | 6.99e-01 |
| GO:BP | GO:0010159 | specification of animal organ position | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 3 | 18 | 6.99e-01 |
| GO:BP | GO:0009306 | protein secretion | 9 | 262 | 6.99e-01 |
| GO:BP | GO:1900141 | regulation of oligodendrocyte apoptotic process | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0034352 | positive regulation of glial cell apoptotic process | 1 | 2 | 6.99e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 6.99e-01 |
| GO:BP | GO:0060465 | pharynx development | 1 | 3 | 6.99e-01 |
| GO:BP | GO:0010876 | lipid localization | 7 | 310 | 6.99e-01 |
| GO:BP | GO:1902463 | protein localization to cell leading edge | 1 | 5 | 6.99e-01 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 1 | 35 | 6.99e-01 |
| GO:BP | GO:0050916 | sensory perception of sweet taste | 2 | 3 | 7.00e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 7.00e-01 |
| GO:BP | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production | 1 | 3 | 7.00e-01 |
| GO:BP | GO:0061024 | membrane organization | 8 | 671 | 7.00e-01 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 4 | 8 | 7.00e-01 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 6 | 13 | 7.00e-01 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 6 | 13 | 7.00e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 4 | 31 | 7.00e-01 |
| GO:BP | GO:0003084 | positive regulation of systemic arterial blood pressure | 1 | 7 | 7.00e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 4 | 28 | 7.00e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 67 | 343 | 7.00e-01 |
| GO:BP | GO:0075525 | viral translational termination-reinitiation | 1 | 5 | 7.00e-01 |
| GO:BP | GO:0030422 | siRNA processing | 1 | 7 | 7.00e-01 |
| GO:BP | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 2 | 3 | 7.00e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 7.00e-01 |
| GO:BP | GO:1905206 | positive regulation of hydrogen peroxide-induced cell death | 2 | 6 | 7.00e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 7.00e-01 |
| GO:BP | GO:0032703 | negative regulation of interleukin-2 production | 4 | 10 | 7.00e-01 |
| GO:BP | GO:0044793 | negative regulation by host of viral process | 1 | 10 | 7.00e-01 |
| GO:BP | GO:1903596 | regulation of gap junction assembly | 2 | 8 | 7.00e-01 |
| GO:BP | GO:0044691 | tooth eruption | 2 | 5 | 7.00e-01 |
| GO:BP | GO:1901033 | positive regulation of response to reactive oxygen species | 2 | 6 | 7.00e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 9 | 262 | 7.00e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 3 | 11 | 7.00e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 3 | 11 | 7.00e-01 |
| GO:BP | GO:0071694 | maintenance of protein location in extracellular region | 3 | 4 | 7.00e-01 |
| GO:BP | GO:0071103 | DNA conformation change | 1 | 87 | 7.00e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 303 | 7.00e-01 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 4 | 19 | 7.00e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 7.00e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 8 | 53 | 7.00e-01 |
| GO:BP | GO:0030050 | vesicle transport along actin filament | 6 | 13 | 7.00e-01 |
| GO:BP | GO:0032715 | negative regulation of interleukin-6 production | 1 | 24 | 7.00e-01 |
| GO:BP | GO:0015819 | lysine transport | 1 | 2 | 7.00e-01 |
| GO:BP | GO:0060717 | chorion development | 2 | 8 | 7.01e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 1 | 265 | 7.01e-01 |
| GO:BP | GO:0046907 | intracellular transport | 16 | 1453 | 7.01e-01 |
| GO:BP | GO:2000487 | positive regulation of glutamine transport | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0010585 | glutamine secretion | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 5 | 100 | 7.01e-01 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 3 | 9 | 7.01e-01 |
| GO:BP | GO:0051365 | cellular response to potassium ion starvation | 1 | 1 | 7.01e-01 |
| GO:BP | GO:1903713 | asparagine transmembrane transport | 1 | 1 | 7.01e-01 |
| GO:BP | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH | 1 | 1 | 7.01e-01 |
| GO:BP | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 3 | 7.01e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 3 | 124 | 7.01e-01 |
| GO:BP | GO:0097492 | sympathetic neuron axon guidance | 2 | 4 | 7.01e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 1 | 109 | 7.01e-01 |
| GO:BP | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.01e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 8 | 541 | 7.01e-01 |
| GO:BP | GO:0071400 | cellular response to oleic acid | 1 | 3 | 7.01e-01 |
| GO:BP | GO:0021615 | glossopharyngeal nerve morphogenesis | 2 | 2 | 7.01e-01 |
| GO:BP | GO:1904237 | positive regulation of substrate-dependent cell migration, cell attachment to substrate | 1 | 3 | 7.01e-01 |
| GO:BP | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell | 1 | 2 | 7.01e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 1 | 20 | 7.01e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 1 | 89 | 7.01e-01 |
| GO:BP | GO:2000831 | regulation of steroid hormone secretion | 1 | 13 | 7.02e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 8 | 76 | 7.02e-01 |
| GO:BP | GO:0010737 | protein kinase A signaling | 1 | 23 | 7.02e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 8 | 76 | 7.02e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 2 | 25 | 7.02e-01 |
| GO:BP | GO:0019085 | early viral transcription | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 2 | 28 | 7.02e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 4 | 126 | 7.02e-01 |
| GO:BP | GO:0071352 | cellular response to interleukin-2 | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0002940 | tRNA N2-guanine methylation | 1 | 4 | 7.02e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 14 | 32 | 7.02e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 7 | 44 | 7.02e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 1 | 20 | 7.02e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 3 | 11 | 7.02e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 1 | 45 | 7.02e-01 |
| GO:BP | GO:1903727 | positive regulation of phospholipid metabolic process | 1 | 9 | 7.02e-01 |
| GO:BP | GO:0038110 | interleukin-2-mediated signaling pathway | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane | 1 | 19 | 7.02e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 7.02e-01 |
| GO:BP | GO:2001138 | regulation of phospholipid transport | 1 | 12 | 7.02e-01 |
| GO:BP | GO:1901825 | zeaxanthin metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0046247 | terpene catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0016119 | carotene metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:0016122 | xanthophyll metabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1900223 | positive regulation of amyloid-beta clearance | 1 | 5 | 7.02e-01 |
| GO:BP | GO:1904432 | regulation of ferrous iron binding | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1904434 | positive regulation of ferrous iron binding | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0016121 | carotene catabolic process | 1 | 1 | 7.02e-01 |
| GO:BP | GO:1904435 | regulation of transferrin receptor binding | 1 | 2 | 7.02e-01 |
| GO:BP | GO:1904437 | positive regulation of transferrin receptor binding | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0090241 | negative regulation of histone H4 acetylation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0006431 | methionyl-tRNA aminoacylation | 1 | 2 | 7.02e-01 |
| GO:BP | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification | 1 | 9 | 7.02e-01 |
| GO:BP | GO:0003176 | aortic valve development | 13 | 37 | 7.03e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 4 | 19 | 7.03e-01 |
| GO:BP | GO:0016199 | axon midline choice point recognition | 1 | 3 | 7.03e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 5 | 54 | 7.03e-01 |
| GO:BP | GO:1904781 | positive regulation of protein localization to centrosome | 1 | 7 | 7.03e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 2 | 12 | 7.04e-01 |
| GO:BP | GO:0048561 | establishment of animal organ orientation | 1 | 1 | 7.04e-01 |
| GO:BP | GO:0030258 | lipid modification | 21 | 162 | 7.04e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 2 | 59 | 7.04e-01 |
| GO:BP | GO:0002756 | MyD88-independent toll-like receptor signaling pathway | 4 | 8 | 7.04e-01 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 2 | 7 | 7.04e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 3 | 7 | 7.04e-01 |
| GO:BP | GO:0036466 | synaptic vesicle recycling via endosome | 2 | 4 | 7.04e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 2 | 19 | 7.05e-01 |
| GO:BP | GO:0006532 | aspartate biosynthetic process | 1 | 2 | 7.05e-01 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 4 | 12 | 7.05e-01 |
| GO:BP | GO:0006106 | fumarate metabolic process | 1 | 3 | 7.05e-01 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 1 | 146 | 7.05e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 7.05e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1 | 2065 | 7.05e-01 |
| GO:BP | GO:0140352 | export from cell | 5 | 611 | 7.06e-01 |
| GO:BP | GO:0010587 | miRNA catabolic process | 5 | 10 | 7.06e-01 |
| GO:BP | GO:1990834 | response to odorant | 1 | 3 | 7.06e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 3 | 20 | 7.06e-01 |
| GO:BP | GO:0010985 | negative regulation of lipoprotein particle clearance | 2 | 6 | 7.06e-01 |
| GO:BP | GO:1904204 | regulation of skeletal muscle hypertrophy | 1 | 1 | 7.06e-01 |
| GO:BP | GO:0070315 | G1 to G0 transition involved in cell differentiation | 2 | 4 | 7.06e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 1 | 26 | 7.06e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 4 | 7.06e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 2 | 165 | 7.06e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 5 | 12 | 7.06e-01 |
| GO:BP | GO:0006310 | DNA recombination | 124 | 257 | 7.06e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 3 | 14 | 7.06e-01 |
| GO:BP | GO:1990258 | histone glutamine methylation | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 2 | 2 | 7.06e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 2 | 2 | 7.06e-01 |
| GO:BP | GO:0030309 | poly-N-acetyllactosamine metabolic process | 2 | 6 | 7.06e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 2 | 2 | 7.06e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 1 | 220 | 7.06e-01 |
| GO:BP | GO:0070476 | rRNA (guanine-N7)-methylation | 1 | 2 | 7.06e-01 |
| GO:BP | GO:0042053 | regulation of dopamine metabolic process | 4 | 11 | 7.06e-01 |
| GO:BP | GO:0072070 | loop of Henle development | 1 | 8 | 7.06e-01 |
| GO:BP | GO:0140447 | cytokine precursor processing | 1 | 1 | 7.06e-01 |
| GO:BP | GO:0042069 | regulation of catecholamine metabolic process | 4 | 11 | 7.06e-01 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 6 | 14 | 7.06e-01 |
| GO:BP | GO:0021540 | corpus callosum morphogenesis | 1 | 5 | 7.07e-01 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 1 | 11 | 7.07e-01 |
| GO:BP | GO:0044830 | modulation by host of viral RNA genome replication | 1 | 6 | 7.07e-01 |
| GO:BP | GO:1904780 | negative regulation of protein localization to centrosome | 1 | 2 | 7.07e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 4 | 18 | 7.07e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 4 | 18 | 7.07e-01 |
| GO:BP | GO:0051964 | negative regulation of synapse assembly | 2 | 6 | 7.08e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 36 | 300 | 7.08e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 3 | 13 | 7.08e-01 |
| GO:BP | GO:1905035 | negative regulation of antifungal innate immune response | 1 | 2 | 7.08e-01 |
| GO:BP | GO:2000546 | positive regulation of endothelial cell chemotaxis to fibroblast growth factor | 1 | 3 | 7.08e-01 |
| GO:BP | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor | 1 | 3 | 7.08e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 2 | 35 | 7.08e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 3 | 213 | 7.08e-01 |
| GO:BP | GO:0051103 | DNA ligation involved in DNA repair | 2 | 5 | 7.08e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 20 | 55 | 7.08e-01 |
| GO:BP | GO:0019884 | antigen processing and presentation of exogenous antigen | 2 | 28 | 7.08e-01 |
| GO:BP | GO:0000961 | negative regulation of mitochondrial RNA catabolic process | 1 | 2 | 7.08e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 3 | 21 | 7.08e-01 |
| GO:BP | GO:0090032 | negative regulation of steroid hormone biosynthetic process | 1 | 6 | 7.08e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 2 | 8 | 7.09e-01 |
| GO:BP | GO:0042074 | cell migration involved in gastrulation | 3 | 10 | 7.09e-01 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 3 | 17 | 7.09e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 3 | 151 | 7.09e-01 |
| GO:BP | GO:0043585 | nose morphogenesis | 1 | 3 | 7.09e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 7.09e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 1 | 24 | 7.09e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1 | 1974 | 7.09e-01 |
| GO:BP | GO:0001781 | neutrophil apoptotic process | 1 | 5 | 7.09e-01 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 5 | 11 | 7.10e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 1 | 131 | 7.10e-01 |
| GO:BP | GO:0048808 | male genitalia morphogenesis | 1 | 2 | 7.10e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 1 | 6 | 7.10e-01 |
| GO:BP | GO:0090598 | male anatomical structure morphogenesis | 1 | 2 | 7.10e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 3 | 3750 | 7.10e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 1 | 15 | 7.10e-01 |
| GO:BP | GO:0150099 | neuron-glial cell signaling | 1 | 4 | 7.10e-01 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 4 | 13 | 7.11e-01 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 2 | 29 | 7.12e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 2 | 10 | 7.12e-01 |
| GO:BP | GO:0071550 | death-inducing signaling complex assembly | 1 | 3 | 7.12e-01 |
| GO:BP | GO:0032594 | protein transport within lipid bilayer | 2 | 6 | 7.12e-01 |
| GO:BP | GO:0015074 | DNA integration | 2 | 9 | 7.12e-01 |
| GO:BP | GO:0035697 | CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.12e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 5 | 179 | 7.12e-01 |
| GO:BP | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation | 1 | 1 | 7.12e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 2 | 18 | 7.12e-01 |
| GO:BP | GO:0006363 | termination of RNA polymerase I transcription | 1 | 4 | 7.12e-01 |
| GO:BP | GO:0032966 | negative regulation of collagen biosynthetic process | 2 | 7 | 7.12e-01 |
| GO:BP | GO:2000409 | positive regulation of T cell extravasation | 1 | 2 | 7.12e-01 |
| GO:BP | GO:0035385 | Roundabout signaling pathway | 1 | 5 | 7.12e-01 |
| GO:BP | GO:0061055 | myotome development | 1 | 2 | 7.12e-01 |
| GO:BP | GO:1903489 | positive regulation of lactation | 1 | 1 | 7.12e-01 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 1 | 13 | 7.12e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 18 | 42 | 7.12e-01 |
| GO:BP | GO:0051791 | medium-chain fatty acid metabolic process | 2 | 9 | 7.12e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 30 | 216 | 7.12e-01 |
| GO:BP | GO:0099500 | vesicle fusion to plasma membrane | 1 | 19 | 7.12e-01 |
| GO:BP | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity | 4 | 12 | 7.12e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 5 | 142 | 7.13e-01 |
| GO:BP | GO:0046832 | negative regulation of RNA export from nucleus | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0072193 | ureter smooth muscle cell differentiation | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0072191 | ureter smooth muscle development | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 1 | 9 | 7.13e-01 |
| GO:BP | GO:0060737 | prostate gland morphogenetic growth | 1 | 2 | 7.13e-01 |
| GO:BP | GO:2000741 | positive regulation of mesenchymal stem cell differentiation | 1 | 4 | 7.13e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 1 | 9 | 7.13e-01 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 6 | 23 | 7.13e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 4 | 23 | 7.13e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 35 | 7.13e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 5 | 16 | 7.13e-01 |
| GO:BP | GO:0035581 | sequestering of extracellular ligand from receptor | 4 | 7 | 7.13e-01 |
| GO:BP | GO:0051235 | maintenance of location | 2 | 260 | 7.13e-01 |
| GO:BP | GO:0035483 | gastric emptying | 1 | 2 | 7.13e-01 |
| GO:BP | GO:1904880 | response to hydrogen sulfide | 1 | 3 | 7.13e-01 |
| GO:BP | GO:1905269 | positive regulation of chromatin organization | 1 | 15 | 7.13e-01 |
| GO:BP | GO:0090084 | negative regulation of inclusion body assembly | 1 | 10 | 7.13e-01 |
| GO:BP | GO:0051459 | regulation of corticotropin secretion | 1 | 3 | 7.13e-01 |
| GO:BP | GO:0006668 | sphinganine-1-phosphate metabolic process | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 3 | 8 | 7.13e-01 |
| GO:BP | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0061027 | umbilical cord development | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0036304 | umbilical cord morphogenesis | 1 | 2 | 7.13e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 32 | 148 | 7.13e-01 |
| GO:BP | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway | 1 | 6 | 7.13e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 2 | 7 | 7.13e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 8 | 275 | 7.13e-01 |
| GO:BP | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 4 | 7.13e-01 |
| GO:BP | GO:0051639 | actin filament network formation | 1 | 7 | 7.13e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 3 | 19 | 7.13e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 1 | 20 | 7.13e-01 |
| GO:BP | GO:1905429 | response to glycine | 1 | 2 | 7.13e-01 |
| GO:BP | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 7.13e-01 |
| GO:BP | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0014826 | vein smooth muscle contraction | 1 | 3 | 7.13e-01 |
| GO:BP | GO:0097402 | neuroblast migration | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 9 | 115 | 7.13e-01 |
| GO:BP | GO:0030397 | membrane disassembly | 4 | 11 | 7.13e-01 |
| GO:BP | GO:0060585 | positive regulation of prostaglandin-endoperoxide synthase activity | 1 | 2 | 7.13e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 7.13e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 2 | 40 | 7.14e-01 |
| GO:BP | GO:0001709 | cell fate determination | 7 | 25 | 7.14e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 6 | 14 | 7.14e-01 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 5 | 12 | 7.14e-01 |
| GO:BP | GO:0042357 | thiamine diphosphate metabolic process | 1 | 2 | 7.14e-01 |
| GO:BP | GO:0014732 | skeletal muscle atrophy | 2 | 5 | 7.14e-01 |
| GO:BP | GO:0072318 | clathrin coat disassembly | 1 | 8 | 7.14e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 9 | 266 | 7.14e-01 |
| GO:BP | GO:2001256 | regulation of store-operated calcium entry | 1 | 11 | 7.14e-01 |
| GO:BP | GO:2000489 | regulation of hepatic stellate cell activation | 3 | 6 | 7.15e-01 |
| GO:BP | GO:0007440 | foregut morphogenesis | 2 | 7 | 7.15e-01 |
| GO:BP | GO:0038162 | erythropoietin-mediated signaling pathway | 1 | 2 | 7.15e-01 |
| GO:BP | GO:1990770 | small intestine smooth muscle contraction | 1 | 1 | 7.15e-01 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 11 | 29 | 7.15e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 3 | 17 | 7.15e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 7.15e-01 |
| GO:BP | GO:0071379 | cellular response to prostaglandin stimulus | 1 | 16 | 7.16e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 1 | 45 | 7.16e-01 |
| GO:BP | GO:0060177 | regulation of angiotensin metabolic process | 1 | 1 | 7.16e-01 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 2 | 11 | 7.16e-01 |
| GO:BP | GO:0051028 | mRNA transport | 1 | 117 | 7.16e-01 |
| GO:BP | GO:0038026 | reelin-mediated signaling pathway | 3 | 9 | 7.16e-01 |
| GO:BP | GO:1903597 | negative regulation of gap junction assembly | 1 | 1 | 7.16e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 3 | 7 | 7.16e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 27 | 7.16e-01 |
| GO:BP | GO:0051005 | negative regulation of lipoprotein lipase activity | 2 | 4 | 7.16e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 10 | 29 | 7.16e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 17 | 118 | 7.16e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 2 | 90 | 7.16e-01 |
| GO:BP | GO:0002387 | immune response in gut-associated lymphoid tissue | 1 | 1 | 7.16e-01 |
| GO:BP | GO:0018410 | C-terminal protein amino acid modification | 2 | 10 | 7.16e-01 |
| GO:BP | GO:0044000 | movement in host | 1 | 138 | 7.16e-01 |
| GO:BP | GO:0002395 | immune response in nasopharyngeal-associated lymphoid tissue | 1 | 1 | 7.16e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 3 | 68 | 7.16e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 8 | 28 | 7.17e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 2 | 62 | 7.17e-01 |
| GO:BP | GO:0006677 | glycosylceramide metabolic process | 1 | 13 | 7.17e-01 |
| GO:BP | GO:1904851 | positive regulation of establishment of protein localization to telomere | 2 | 10 | 7.17e-01 |
| GO:BP | GO:0006983 | ER overload response | 3 | 12 | 7.18e-01 |
| GO:BP | GO:0001773 | myeloid dendritic cell activation | 1 | 12 | 7.18e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 11 | 404 | 7.18e-01 |
| GO:BP | GO:0048817 | negative regulation of hair follicle maturation | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 1 | 11 | 7.18e-01 |
| GO:BP | GO:2000307 | regulation of tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 7.18e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 2 | 7 | 7.18e-01 |
| GO:BP | GO:2000412 | positive regulation of thymocyte migration | 1 | 1 | 7.18e-01 |
| GO:BP | GO:1902908 | regulation of melanosome transport | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0032771 | regulation of tyrosinase activity | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0021628 | olfactory nerve formation | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0044062 | regulation of excretion | 1 | 12 | 7.18e-01 |
| GO:BP | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 4 | 16 | 7.18e-01 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 1 | 12 | 7.18e-01 |
| GO:BP | GO:1902224 | ketone body metabolic process | 2 | 7 | 7.18e-01 |
| GO:BP | GO:0072535 | tumor necrosis factor (ligand) superfamily member 11 production | 1 | 1 | 7.18e-01 |
| GO:BP | GO:0070781 | response to biotin | 1 | 1 | 7.18e-01 |
| GO:BP | GO:0032773 | positive regulation of tyrosinase activity | 1 | 2 | 7.18e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 1 | 71 | 7.18e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 3 | 6 | 7.18e-01 |
| GO:BP | GO:0032241 | positive regulation of nucleobase-containing compound transport | 2 | 6 | 7.18e-01 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 3 | 17 | 7.19e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 5 | 78 | 7.19e-01 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 3 | 17 | 7.19e-01 |
| GO:BP | GO:2001295 | malonyl-CoA biosynthetic process | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 3 | 12 | 7.19e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 7.19e-01 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 1 | 149 | 7.19e-01 |
| GO:BP | GO:0071907 | determination of digestive tract left/right asymmetry | 3 | 5 | 7.19e-01 |
| GO:BP | GO:0051350 | negative regulation of lyase activity | 3 | 15 | 7.19e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 39 | 7.19e-01 |
| GO:BP | GO:0060462 | lung lobe development | 1 | 4 | 7.19e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 3 | 28 | 7.19e-01 |
| GO:BP | GO:0001807 | regulation of type IV hypersensitivity | 1 | 1 | 7.19e-01 |
| GO:BP | GO:0022400 | regulation of rhodopsin mediated signaling pathway | 2 | 5 | 7.19e-01 |
| GO:BP | GO:0097532 | stress response to acid chemical | 1 | 2 | 7.19e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 2 | 46 | 7.19e-01 |
| GO:BP | GO:0061010 | gallbladder development | 1 | 1 | 7.19e-01 |
| GO:BP | GO:2000367 | regulation of acrosomal vesicle exocytosis | 1 | 3 | 7.19e-01 |
| GO:BP | GO:0097533 | cellular stress response to acid chemical | 1 | 2 | 7.19e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 7 | 18 | 7.19e-01 |
| GO:BP | GO:2000774 | positive regulation of cellular senescence | 2 | 9 | 7.19e-01 |
| GO:BP | GO:0060463 | lung lobe morphogenesis | 1 | 4 | 7.19e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 2 | 13 | 7.20e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 7.20e-01 |
| GO:BP | GO:2000668 | regulation of dendritic cell apoptotic process | 1 | 7 | 7.20e-01 |
| GO:BP | GO:0097048 | dendritic cell apoptotic process | 1 | 7 | 7.20e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 5 | 22 | 7.20e-01 |
| GO:BP | GO:0048538 | thymus development | 1 | 35 | 7.20e-01 |
| GO:BP | GO:1990074 | polyuridylation-dependent mRNA catabolic process | 1 | 3 | 7.20e-01 |
| GO:BP | GO:1904021 | negative regulation of G protein-coupled receptor internalization | 1 | 2 | 7.20e-01 |
| GO:BP | GO:0030157 | pancreatic juice secretion | 2 | 11 | 7.21e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 2 | 302 | 7.21e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 3 | 16 | 7.21e-01 |
| GO:BP | GO:0043970 | histone H3-K9 acetylation | 4 | 8 | 7.21e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 5 | 41 | 7.21e-01 |
| GO:BP | GO:0009615 | response to virus | 9 | 275 | 7.21e-01 |
| GO:BP | GO:0071603 | endothelial cell-cell adhesion | 1 | 4 | 7.21e-01 |
| GO:BP | GO:0033007 | negative regulation of mast cell activation involved in immune response | 4 | 4 | 7.21e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 4 | 64 | 7.21e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 2 | 155 | 7.21e-01 |
| GO:BP | GO:0061032 | visceral serous pericardium development | 1 | 1 | 7.21e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 28 | 203 | 7.21e-01 |
| GO:BP | GO:1905449 | regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 2 | 5 | 7.21e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 9 | 68 | 7.21e-01 |
| GO:BP | GO:0002730 | regulation of dendritic cell cytokine production | 3 | 10 | 7.22e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 7.22e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 9 | 67 | 7.22e-01 |
| GO:BP | GO:0048635 | negative regulation of muscle organ development | 3 | 6 | 7.22e-01 |
| GO:BP | GO:0002371 | dendritic cell cytokine production | 3 | 10 | 7.22e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 7 | 27 | 7.22e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 15 | 73 | 7.22e-01 |
| GO:BP | GO:0002475 | antigen processing and presentation via MHC class Ib | 4 | 8 | 7.22e-01 |
| GO:BP | GO:1904179 | positive regulation of adipose tissue development | 1 | 6 | 7.22e-01 |
| GO:BP | GO:0070948 | regulation of neutrophil mediated cytotoxicity | 1 | 2 | 7.22e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 3 | 10 | 7.22e-01 |
| GO:BP | GO:0035684 | helper T cell extravasation | 1 | 3 | 7.22e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 1 | 15 | 7.22e-01 |
| GO:BP | GO:0032060 | bleb assembly | 4 | 9 | 7.22e-01 |
| GO:BP | GO:0070171 | negative regulation of tooth mineralization | 1 | 2 | 7.23e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 2 | 3 | 7.23e-01 |
| GO:BP | GO:0098907 | regulation of SA node cell action potential | 3 | 7 | 7.23e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 2 | 168 | 7.23e-01 |
| GO:BP | GO:0009249 | protein lipoylation | 1 | 6 | 7.23e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 6 | 29 | 7.23e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 1 | 52 | 7.23e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 17 | 119 | 7.23e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 1 | 52 | 7.23e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 2 | 29 | 7.23e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 4 | 6996 | 7.23e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 7 | 22 | 7.24e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 2 | 155 | 7.24e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 5 | 256 | 7.24e-01 |
| GO:BP | GO:0007431 | salivary gland development | 2 | 26 | 7.24e-01 |
| GO:BP | GO:0042541 | hemoglobin biosynthetic process | 4 | 13 | 7.24e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 10 | 52 | 7.24e-01 |
| GO:BP | GO:0032714 | negative regulation of interleukin-5 production | 1 | 2 | 7.24e-01 |
| GO:BP | GO:0035523 | protein K29-linked deubiquitination | 1 | 4 | 7.24e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 1 | 4 | 7.24e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 4 | 15 | 7.24e-01 |
| GO:BP | GO:0046541 | saliva secretion | 1 | 4 | 7.24e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 13 | 111 | 7.24e-01 |
| GO:BP | GO:0021855 | hypothalamus cell migration | 3 | 6 | 7.24e-01 |
| GO:BP | GO:1990791 | dorsal root ganglion development | 3 | 6 | 7.24e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 2 | 216 | 7.24e-01 |
| GO:BP | GO:1904386 | response to L-phenylalanine derivative | 2 | 5 | 7.24e-01 |
| GO:BP | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 3 | 7.24e-01 |
| GO:BP | GO:0014858 | positive regulation of skeletal muscle cell proliferation | 1 | 2 | 7.24e-01 |
| GO:BP | GO:0042886 | amide transport | 2 | 235 | 7.24e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 6 | 27 | 7.24e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 13 | 84 | 7.24e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 3 | 45 | 7.24e-01 |
| GO:BP | GO:0002711 | positive regulation of T cell mediated immunity | 1 | 29 | 7.24e-01 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 7 | 18 | 7.24e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 3 | 233 | 7.24e-01 |
| GO:BP | GO:0051030 | snRNA transport | 2 | 6 | 7.24e-01 |
| GO:BP | GO:0038016 | insulin receptor internalization | 1 | 3 | 7.24e-01 |
| GO:BP | GO:0010922 | positive regulation of phosphatase activity | 1 | 26 | 7.24e-01 |
| GO:BP | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 3 | 13 | 7.24e-01 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 4 | 22 | 7.25e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1 | 1898 | 7.25e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 1 | 60 | 7.25e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 2 | 59 | 7.25e-01 |
| GO:BP | GO:1905050 | positive regulation of metallopeptidase activity | 1 | 5 | 7.25e-01 |
| GO:BP | GO:0022900 | electron transport chain | 1 | 142 | 7.25e-01 |
| GO:BP | GO:0006704 | glucocorticoid biosynthetic process | 2 | 11 | 7.25e-01 |
| GO:BP | GO:0031268 | pseudopodium organization | 4 | 14 | 7.25e-01 |
| GO:BP | GO:0015849 | organic acid transport | 2 | 217 | 7.25e-01 |
| GO:BP | GO:0030488 | tRNA methylation | 12 | 42 | 7.25e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 1 | 161 | 7.25e-01 |
| GO:BP | GO:0044375 | regulation of peroxisome size | 1 | 3 | 7.25e-01 |
| GO:BP | GO:0032056 | positive regulation of translation in response to stress | 1 | 8 | 7.25e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 24 | 107 | 7.25e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 2 | 18 | 7.25e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 3 | 27 | 7.25e-01 |
| GO:BP | GO:0110091 | negative regulation of hippocampal neuron apoptotic process | 1 | 2 | 7.25e-01 |
| GO:BP | GO:0044788 | modulation by host of viral process | 5 | 40 | 7.25e-01 |
| GO:BP | GO:0048318 | axial mesoderm development | 1 | 6 | 7.25e-01 |
| GO:BP | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration | 1 | 6 | 7.25e-01 |
| GO:BP | GO:0002841 | negative regulation of T cell mediated immune response to tumor cell | 1 | 1 | 7.25e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 2 | 5 | 7.25e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 2 | 5 | 7.25e-01 |
| GO:BP | GO:1905331 | negative regulation of morphogenesis of an epithelium | 3 | 6 | 7.26e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 7.26e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 5 | 258 | 7.26e-01 |
| GO:BP | GO:0007418 | ventral midline development | 1 | 4 | 7.26e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 2 | 22 | 7.26e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 7.26e-01 |
| GO:BP | GO:0051311 | meiotic metaphase plate congression | 1 | 2 | 7.26e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 7 | 64 | 7.26e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 9 | 44 | 7.26e-01 |
| GO:BP | GO:0051956 | negative regulation of amino acid transport | 2 | 10 | 7.26e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 3 | 22 | 7.26e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 2 | 15 | 7.26e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 1 | 190 | 7.26e-01 |
| GO:BP | GO:0097527 | necroptotic signaling pathway | 3 | 3 | 7.26e-01 |
| GO:BP | GO:0046294 | formaldehyde catabolic process | 1 | 2 | 7.27e-01 |
| GO:BP | GO:0010155 | regulation of proton transport | 1 | 18 | 7.27e-01 |
| GO:BP | GO:0035811 | negative regulation of urine volume | 1 | 2 | 7.27e-01 |
| GO:BP | GO:0001945 | lymph vessel development | 5 | 22 | 7.27e-01 |
| GO:BP | GO:0043968 | histone H2A acetylation | 1 | 22 | 7.27e-01 |
| GO:BP | GO:0016485 | protein processing | 1 | 183 | 7.27e-01 |
| GO:BP | GO:0006970 | response to osmotic stress | 29 | 67 | 7.27e-01 |
| GO:BP | GO:0060478 | acrosomal vesicle exocytosis | 1 | 5 | 7.27e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 5 | 31 | 7.27e-01 |
| GO:BP | GO:0090399 | replicative senescence | 6 | 13 | 7.27e-01 |
| GO:BP | GO:0039663 | membrane fusion involved in viral entry into host cell | 1 | 4 | 7.27e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 1 | 23 | 7.27e-01 |
| GO:BP | GO:0019064 | fusion of virus membrane with host plasma membrane | 1 | 4 | 7.27e-01 |
| GO:BP | GO:0150094 | amyloid-beta clearance by cellular catabolic process | 1 | 7 | 7.27e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 11 | 23 | 7.27e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 3 | 39 | 7.27e-01 |
| GO:BP | GO:0015876 | acetyl-CoA transport | 1 | 1 | 7.27e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 2 | 97 | 7.27e-01 |
| GO:BP | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 2 | 4 | 7.27e-01 |
| GO:BP | GO:0051170 | import into nucleus | 1 | 152 | 7.27e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 2 | 15 | 7.27e-01 |
| GO:BP | GO:0043097 | pyrimidine nucleoside salvage | 1 | 2 | 7.27e-01 |
| GO:BP | GO:0045590 | negative regulation of regulatory T cell differentiation | 1 | 3 | 7.27e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 1 | 14 | 7.27e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 1 | 25 | 7.27e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 8 | 963 | 7.27e-01 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 3 | 12 | 7.27e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 3 | 43 | 7.27e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 9 | 344 | 7.27e-01 |
| GO:BP | GO:0006855 | xenobiotic transmembrane transport | 1 | 11 | 7.27e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 6 | 292 | 7.27e-01 |
| GO:BP | GO:0044597 | daunorubicin metabolic process | 3 | 7 | 7.28e-01 |
| GO:BP | GO:0045916 | negative regulation of complement activation | 3 | 8 | 7.28e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 6 | 24 | 7.28e-01 |
| GO:BP | GO:0060384 | innervation | 1 | 22 | 7.28e-01 |
| GO:BP | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 2 | 5 | 7.28e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 1 | 28 | 7.28e-01 |
| GO:BP | GO:0061386 | closure of optic fissure | 1 | 2 | 7.28e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 1 | 160 | 7.28e-01 |
| GO:BP | GO:0097166 | lens epithelial cell proliferation | 1 | 1 | 7.28e-01 |
| GO:BP | GO:2001111 | positive regulation of lens epithelial cell proliferation | 1 | 1 | 7.28e-01 |
| GO:BP | GO:2001109 | regulation of lens epithelial cell proliferation | 1 | 1 | 7.28e-01 |
| GO:BP | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion | 2 | 5 | 7.28e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 3 | 91 | 7.28e-01 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 2 | 5 | 7.28e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 7.28e-01 |
| GO:BP | GO:0071461 | cellular response to redox state | 1 | 1 | 7.28e-01 |
| GO:BP | GO:2000642 | negative regulation of early endosome to late endosome transport | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 2 | 38 | 7.29e-01 |
| GO:BP | GO:0044721 | protein import into peroxisome matrix, substrate release | 1 | 2 | 7.29e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 2 | 15 | 7.29e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 9 | 345 | 7.29e-01 |
| GO:BP | GO:0001823 | mesonephros development | 8 | 77 | 7.30e-01 |
| GO:BP | GO:0036031 | recruitment of mRNA capping enzyme to RNA polymerase II holoenzyme complex | 1 | 1 | 7.30e-01 |
| GO:BP | GO:0061357 | positive regulation of Wnt protein secretion | 1 | 4 | 7.30e-01 |
| GO:BP | GO:0016243 | regulation of autophagosome size | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 4 | 17 | 7.30e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 6 | 169 | 7.30e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 2 | 15 | 7.30e-01 |
| GO:BP | GO:0032385 | positive regulation of intracellular cholesterol transport | 1 | 3 | 7.30e-01 |
| GO:BP | GO:0032382 | positive regulation of intracellular sterol transport | 1 | 3 | 7.30e-01 |
| GO:BP | GO:1905600 | regulation of receptor-mediated endocytosis involved in cholesterol transport | 1 | 3 | 7.30e-01 |
| GO:BP | GO:0033083 | regulation of immature T cell proliferation | 1 | 6 | 7.30e-01 |
| GO:BP | GO:0033084 | regulation of immature T cell proliferation in thymus | 1 | 6 | 7.30e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 2 | 92 | 7.30e-01 |
| GO:BP | GO:0022007 | convergent extension involved in neural plate elongation | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1905386 | positive regulation of protein localization to presynapse | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 2 | 53 | 7.30e-01 |
| GO:BP | GO:0014022 | neural plate elongation | 1 | 2 | 7.30e-01 |
| GO:BP | GO:1905384 | regulation of protein localization to presynapse | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0006003 | fructose 2,6-bisphosphate metabolic process | 2 | 4 | 7.30e-01 |
| GO:BP | GO:1901256 | regulation of macrophage colony-stimulating factor production | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0036301 | macrophage colony-stimulating factor production | 1 | 2 | 7.30e-01 |
| GO:BP | GO:0021904 | dorsal/ventral neural tube patterning | 1 | 21 | 7.30e-01 |
| GO:BP | GO:0002032 | desensitization of G protein-coupled receptor signaling pathway by arrestin | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0061884 | regulation of mini excitatory postsynaptic potential | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0060742 | epithelial cell differentiation involved in prostate gland development | 2 | 8 | 7.31e-01 |
| GO:BP | GO:0060324 | face development | 18 | 45 | 7.31e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 71 | 7.31e-01 |
| GO:BP | GO:0061885 | positive regulation of mini excitatory postsynaptic potential | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0098816 | mini excitatory postsynaptic potential | 1 | 2 | 7.31e-01 |
| GO:BP | GO:0036335 | intestinal stem cell homeostasis | 2 | 3 | 7.31e-01 |
| GO:BP | GO:0007517 | muscle organ development | 36 | 277 | 7.31e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 2 | 85 | 7.31e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 1 | 39 | 7.31e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 1 | 5 | 7.31e-01 |
| GO:BP | GO:0033034 | positive regulation of myeloid cell apoptotic process | 1 | 6 | 7.31e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 12 | 41 | 7.31e-01 |
| GO:BP | GO:0061197 | fungiform papilla morphogenesis | 1 | 4 | 7.31e-01 |
| GO:BP | GO:0043950 | positive regulation of cAMP-mediated signaling | 4 | 5 | 7.31e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 7.31e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 1 | 39 | 7.31e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 5 | 189 | 7.31e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 11 | 139 | 7.31e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 10 | 62 | 7.31e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 7.31e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 1 | 143 | 7.31e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 3 | 100 | 7.31e-01 |
| GO:BP | GO:0089709 | L-histidine transmembrane transport | 2 | 5 | 7.31e-01 |
| GO:BP | GO:0048818 | positive regulation of hair follicle maturation | 2 | 5 | 7.31e-01 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 2 | 5 | 7.31e-01 |
| GO:BP | GO:1903850 | regulation of cristae formation | 1 | 4 | 7.31e-01 |
| GO:BP | GO:0060751 | branch elongation involved in mammary gland duct branching | 2 | 4 | 7.31e-01 |
| GO:BP | GO:0097325 | melanocyte proliferation | 2 | 2 | 7.31e-01 |
| GO:BP | GO:0014829 | vascular associated smooth muscle contraction | 9 | 20 | 7.32e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 18 | 742 | 7.32e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 2 | 17 | 7.32e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 2 | 93 | 7.32e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 2 | 33 | 7.32e-01 |
| GO:BP | GO:0002707 | negative regulation of lymphocyte mediated immunity | 8 | 24 | 7.32e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 6 | 29 | 7.33e-01 |
| GO:BP | GO:1904037 | positive regulation of epithelial cell apoptotic process | 4 | 26 | 7.33e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1 | 2285 | 7.33e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 6 | 51 | 7.33e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 4 | 110 | 7.33e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 1 | 52 | 7.33e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 6 | 29 | 7.33e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 7.33e-01 |
| GO:BP | GO:0007568 | aging | 5 | 115 | 7.33e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 7 | 72 | 7.33e-01 |
| GO:BP | GO:0048745 | smooth muscle tissue development | 4 | 19 | 7.33e-01 |
| GO:BP | GO:0033993 | response to lipid | 4 | 592 | 7.33e-01 |
| GO:BP | GO:0072350 | tricarboxylic acid metabolic process | 4 | 13 | 7.33e-01 |
| GO:BP | GO:0030322 | stabilization of membrane potential | 1 | 6 | 7.33e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 12 | 71 | 7.33e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 2 | 84 | 7.33e-01 |
| GO:BP | GO:0046167 | glycerol-3-phosphate biosynthetic process | 1 | 2 | 7.33e-01 |
| GO:BP | GO:0032792 | negative regulation of CREB transcription factor activity | 1 | 4 | 7.33e-01 |
| GO:BP | GO:0071801 | regulation of podosome assembly | 2 | 8 | 7.33e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 12 | 71 | 7.33e-01 |
| GO:BP | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus | 3 | 8 | 7.33e-01 |
| GO:BP | GO:1905552 | positive regulation of protein localization to endoplasmic reticulum | 1 | 2 | 7.33e-01 |
| GO:BP | GO:0009651 | response to salt stress | 6 | 18 | 7.33e-01 |
| GO:BP | GO:1903292 | protein localization to Golgi membrane | 1 | 4 | 7.33e-01 |
| GO:BP | GO:0032914 | positive regulation of transforming growth factor beta1 production | 1 | 6 | 7.33e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 5 | 40 | 7.33e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 6 | 19 | 7.33e-01 |
| GO:BP | GO:1904958 | positive regulation of midbrain dopaminergic neuron differentiation | 1 | 3 | 7.33e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 1 | 144 | 7.33e-01 |
| GO:BP | GO:0002329 | pre-B cell differentiation | 1 | 4 | 7.33e-01 |
| GO:BP | GO:1901535 | regulation of DNA demethylation | 1 | 8 | 7.33e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 5 | 21 | 7.33e-01 |
| GO:BP | GO:0070344 | regulation of fat cell proliferation | 1 | 9 | 7.33e-01 |
| GO:BP | GO:0060174 | limb bud formation | 2 | 10 | 7.33e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 8 | 19 | 7.33e-01 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 2 | 11 | 7.33e-01 |
| GO:BP | GO:0042221 | response to chemical | 22 | 2559 | 7.33e-01 |
| GO:BP | GO:1902761 | positive regulation of chondrocyte development | 1 | 1 | 7.33e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 2 | 4 | 7.33e-01 |
| GO:BP | GO:0046532 | regulation of photoreceptor cell differentiation | 2 | 4 | 7.33e-01 |
| GO:BP | GO:0060736 | prostate gland growth | 2 | 7 | 7.33e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 8 | 19 | 7.33e-01 |
| GO:BP | GO:0030974 | thiamine pyrophosphate transmembrane transport | 1 | 1 | 7.33e-01 |
| GO:BP | GO:0035768 | endothelial cell chemotaxis to fibroblast growth factor | 2 | 4 | 7.34e-01 |
| GO:BP | GO:0032700 | negative regulation of interleukin-17 production | 4 | 9 | 7.34e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 4 | 23 | 7.34e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 1 | 11 | 7.34e-01 |
| GO:BP | GO:0035766 | cell chemotaxis to fibroblast growth factor | 2 | 4 | 7.34e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 4 | 52 | 7.34e-01 |
| GO:BP | GO:0034163 | regulation of toll-like receptor 9 signaling pathway | 1 | 6 | 7.34e-01 |
| GO:BP | GO:0034983 | peptidyl-lysine deacetylation | 2 | 8 | 7.34e-01 |
| GO:BP | GO:1904847 | regulation of cell chemotaxis to fibroblast growth factor | 2 | 4 | 7.34e-01 |
| GO:BP | GO:2000544 | regulation of endothelial cell chemotaxis to fibroblast growth factor | 2 | 4 | 7.34e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 7.34e-01 |
| GO:BP | GO:0038196 | type III interferon-mediated signaling pathway | 1 | 4 | 7.34e-01 |
| GO:BP | GO:0071358 | cellular response to type III interferon | 1 | 4 | 7.34e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 2 | 52 | 7.34e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 5 | 37 | 7.34e-01 |
| GO:BP | GO:1990569 | UDP-N-acetylglucosamine transmembrane transport | 2 | 6 | 7.34e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 2 | 23 | 7.34e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 2 | 17 | 7.35e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 7.35e-01 |
| GO:BP | GO:0035066 | positive regulation of histone acetylation | 9 | 25 | 7.35e-01 |
| GO:BP | GO:0006734 | NADH metabolic process | 2 | 12 | 7.35e-01 |
| GO:BP | GO:0098926 | postsynaptic signal transduction | 5 | 25 | 7.35e-01 |
| GO:BP | GO:0009631 | cold acclimation | 1 | 2 | 7.35e-01 |
| GO:BP | GO:0048548 | regulation of pinocytosis | 4 | 11 | 7.35e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 4 | 9 | 7.35e-01 |
| GO:BP | GO:0006771 | riboflavin metabolic process | 2 | 3 | 7.35e-01 |
| GO:BP | GO:0006850 | mitochondrial pyruvate transmembrane transport | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0051937 | catecholamine transport | 1 | 38 | 7.36e-01 |
| GO:BP | GO:0097021 | lymphocyte migration into lymphoid organs | 2 | 6 | 7.36e-01 |
| GO:BP | GO:1900146 | negative regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 1 | 7.36e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 3 | 10 | 7.36e-01 |
| GO:BP | GO:0033227 | dsRNA transport | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0071671 | regulation of smooth muscle cell chemotaxis | 1 | 4 | 7.36e-01 |
| GO:BP | GO:1900176 | negative regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 1 | 7.36e-01 |
| GO:BP | GO:0009068 | aspartate family amino acid catabolic process | 1 | 12 | 7.36e-01 |
| GO:BP | GO:0043380 | regulation of memory T cell differentiation | 2 | 4 | 7.36e-01 |
| GO:BP | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 5 | 7.36e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 7.36e-01 |
| GO:BP | GO:0048935 | peripheral nervous system neuron development | 1 | 9 | 7.36e-01 |
| GO:BP | GO:0046725 | negative regulation by virus of viral protein levels in host cell | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 1 | 60 | 7.36e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 1 | 60 | 7.36e-01 |
| GO:BP | GO:0106071 | positive regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 1 | 3 | 7.36e-01 |
| GO:BP | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 7.36e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 19 | 47 | 7.36e-01 |
| GO:BP | GO:0048934 | peripheral nervous system neuron differentiation | 1 | 9 | 7.36e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 5 | 60 | 7.36e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 5 | 12 | 7.36e-01 |
| GO:BP | GO:0032613 | interleukin-10 production | 1 | 24 | 7.36e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 29 | 80 | 7.36e-01 |
| GO:BP | GO:0070291 | N-acylethanolamine metabolic process | 2 | 4 | 7.36e-01 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 1 | 24 | 7.36e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 4 | 24 | 7.36e-01 |
| GO:BP | GO:1901380 | negative regulation of potassium ion transmembrane transport | 1 | 18 | 7.36e-01 |
| GO:BP | GO:0070294 | renal sodium ion absorption | 4 | 9 | 7.36e-01 |
| GO:BP | GO:1902644 | tertiary alcohol metabolic process | 3 | 17 | 7.37e-01 |
| GO:BP | GO:0042363 | fat-soluble vitamin catabolic process | 1 | 1 | 7.37e-01 |
| GO:BP | GO:0031106 | septin ring organization | 1 | 2 | 7.37e-01 |
| GO:BP | GO:1904172 | positive regulation of bleb assembly | 1 | 1 | 7.37e-01 |
| GO:BP | GO:1904170 | regulation of bleb assembly | 1 | 1 | 7.37e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 7 | 35 | 7.37e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 3 | 16 | 7.37e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 3 | 117 | 7.37e-01 |
| GO:BP | GO:2000192 | negative regulation of fatty acid transport | 1 | 7 | 7.37e-01 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 5 | 25 | 7.37e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 4 | 8 | 7.37e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 4 | 8 | 7.37e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 4 | 8 | 7.37e-01 |
| GO:BP | GO:0042048 | olfactory behavior | 3 | 7 | 7.37e-01 |
| GO:BP | GO:0060872 | semicircular canal development | 2 | 7 | 7.37e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 17 | 50 | 7.37e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 15 | 616 | 7.37e-01 |
| GO:BP | GO:0060484 | lung-associated mesenchyme development | 1 | 5 | 7.37e-01 |
| GO:BP | GO:0033082 | regulation of extrathymic T cell differentiation | 1 | 1 | 7.37e-01 |
| GO:BP | GO:0042426 | choline catabolic process | 1 | 3 | 7.38e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 2 | 11 | 7.38e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 38 | 500 | 7.39e-01 |
| GO:BP | GO:0046462 | monoacylglycerol metabolic process | 2 | 8 | 7.39e-01 |
| GO:BP | GO:0033197 | response to vitamin E | 4 | 9 | 7.39e-01 |
| GO:BP | GO:0070342 | brown fat cell proliferation | 1 | 1 | 7.39e-01 |
| GO:BP | GO:0090663 | galanin-activated signaling pathway | 1 | 1 | 7.39e-01 |
| GO:BP | GO:1902608 | positive regulation of large conductance calcium-activated potassium channel activity | 1 | 2 | 7.39e-01 |
| GO:BP | GO:0095500 | acetylcholine receptor signaling pathway | 4 | 18 | 7.40e-01 |
| GO:BP | GO:0030261 | chromosome condensation | 17 | 33 | 7.40e-01 |
| GO:BP | GO:0110077 | vesicle-mediated intercellular transport | 1 | 2 | 7.40e-01 |
| GO:BP | GO:0001842 | neural fold formation | 1 | 5 | 7.40e-01 |
| GO:BP | GO:0070194 | synaptonemal complex disassembly | 1 | 2 | 7.40e-01 |
| GO:BP | GO:0070358 | actin polymerization-dependent cell motility | 1 | 6 | 7.40e-01 |
| GO:BP | GO:0097341 | zymogen inhibition | 1 | 5 | 7.40e-01 |
| GO:BP | GO:0097340 | inhibition of cysteine-type endopeptidase activity | 1 | 5 | 7.40e-01 |
| GO:BP | GO:0002468 | dendritic cell antigen processing and presentation | 2 | 6 | 7.40e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 1 | 25 | 7.40e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 1 | 12 | 7.40e-01 |
| GO:BP | GO:2000627 | positive regulation of miRNA catabolic process | 2 | 4 | 7.40e-01 |
| GO:BP | GO:0035938 | estradiol secretion | 1 | 2 | 7.40e-01 |
| GO:BP | GO:2000864 | regulation of estradiol secretion | 1 | 2 | 7.40e-01 |
| GO:BP | GO:0098957 | anterograde axonal transport of mitochondrion | 1 | 5 | 7.41e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 1 | 154 | 7.41e-01 |
| GO:BP | GO:1904999 | positive regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 7.41e-01 |
| GO:BP | GO:0070475 | rRNA base methylation | 1 | 8 | 7.41e-01 |
| GO:BP | GO:0045915 | positive regulation of catecholamine metabolic process | 2 | 6 | 7.42e-01 |
| GO:BP | GO:0045964 | positive regulation of dopamine metabolic process | 2 | 6 | 7.42e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 2 | 5 | 7.42e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 13 | 28 | 7.42e-01 |
| GO:BP | GO:0098795 | global gene silencing by mRNA cleavage | 1 | 4 | 7.42e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 6 | 7.43e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 7.43e-01 |
| GO:BP | GO:1905380 | regulation of snRNA transcription by RNA polymerase II | 1 | 4 | 7.43e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 1 | 74 | 7.43e-01 |
| GO:BP | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation | 4 | 5 | 7.43e-01 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 7.43e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 1 | 4 | 7.43e-01 |
| GO:BP | GO:1905653 | positive regulation of artery morphogenesis | 1 | 5 | 7.43e-01 |
| GO:BP | GO:1905651 | regulation of artery morphogenesis | 1 | 5 | 7.43e-01 |
| GO:BP | GO:0046398 | UDP-glucuronate metabolic process | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0034397 | telomere localization | 4 | 5 | 7.43e-01 |
| GO:BP | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 7.43e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 7.43e-01 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 5 | 11 | 7.43e-01 |
| GO:BP | GO:0061290 | canonical Wnt signaling pathway involved in metanephric kidney development | 1 | 1 | 7.43e-01 |
| GO:BP | GO:0061289 | Wnt signaling pathway involved in kidney development | 1 | 1 | 7.43e-01 |
| GO:BP | GO:0072204 | cell-cell signaling involved in metanephros development | 1 | 1 | 7.43e-01 |
| GO:BP | GO:0071867 | response to monoamine | 1 | 62 | 7.43e-01 |
| GO:BP | GO:0060995 | cell-cell signaling involved in kidney development | 1 | 1 | 7.43e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 1 | 62 | 7.43e-01 |
| GO:BP | GO:1990402 | embryonic liver development | 1 | 3 | 7.43e-01 |
| GO:BP | GO:0016556 | mRNA modification | 1 | 29 | 7.43e-01 |
| GO:BP | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway | 2 | 7 | 7.43e-01 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 2 | 12 | 7.43e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 5 | 13 | 7.43e-01 |
| GO:BP | GO:0003408 | optic cup formation involved in camera-type eye development | 1 | 1 | 7.43e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 4 | 10 | 7.43e-01 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 10 | 27 | 7.43e-01 |
| GO:BP | GO:0048293 | regulation of isotype switching to IgE isotypes | 2 | 5 | 7.43e-01 |
| GO:BP | GO:1905438 | non-canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 7.43e-01 |
| GO:BP | GO:1904933 | regulation of cell proliferation in midbrain | 1 | 1 | 7.43e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 12 | 53 | 7.43e-01 |
| GO:BP | GO:0036518 | chemorepulsion of dopaminergic neuron axon | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0036515 | serotonergic neuron axon guidance | 1 | 2 | 7.43e-01 |
| GO:BP | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0051885 | positive regulation of timing of anagen | 1 | 2 | 7.43e-01 |
| GO:BP | GO:0048289 | isotype switching to IgE isotypes | 2 | 5 | 7.43e-01 |
| GO:BP | GO:0070973 | protein localization to endoplasmic reticulum exit site | 1 | 6 | 7.44e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 3 | 14 | 7.44e-01 |
| GO:BP | GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 1 | 4 | 7.44e-01 |
| GO:BP | GO:0034372 | very-low-density lipoprotein particle remodeling | 1 | 4 | 7.44e-01 |
| GO:BP | GO:0002638 | negative regulation of immunoglobulin production | 2 | 5 | 7.44e-01 |
| GO:BP | GO:0032641 | lymphotoxin A production | 1 | 1 | 7.44e-01 |
| GO:BP | GO:0032761 | positive regulation of lymphotoxin A production | 1 | 1 | 7.44e-01 |
| GO:BP | GO:0032681 | regulation of lymphotoxin A production | 1 | 1 | 7.44e-01 |
| GO:BP | GO:0002327 | immature B cell differentiation | 3 | 11 | 7.44e-01 |
| GO:BP | GO:0075509 | endocytosis involved in viral entry into host cell | 2 | 11 | 7.44e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 2 | 37 | 7.44e-01 |
| GO:BP | GO:0035929 | steroid hormone secretion | 1 | 18 | 7.44e-01 |
| GO:BP | GO:0034478 | phosphatidylglycerol catabolic process | 1 | 1 | 7.44e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 3 | 468 | 7.44e-01 |
| GO:BP | GO:0018119 | peptidyl-cysteine S-nitrosylation | 1 | 11 | 7.44e-01 |
| GO:BP | GO:0017014 | protein nitrosylation | 1 | 11 | 7.44e-01 |
| GO:BP | GO:2000537 | regulation of B cell chemotaxis | 1 | 1 | 7.45e-01 |
| GO:BP | GO:2000538 | positive regulation of B cell chemotaxis | 1 | 1 | 7.45e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 27 | 140 | 7.45e-01 |
| GO:BP | GO:0060729 | intestinal epithelial structure maintenance | 2 | 4 | 7.46e-01 |
| GO:BP | GO:0003335 | corneocyte development | 1 | 2 | 7.46e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 1 | 18 | 7.46e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 5 | 43 | 7.46e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 4 | 49 | 7.46e-01 |
| GO:BP | GO:0051582 | positive regulation of neurotransmitter uptake | 2 | 4 | 7.46e-01 |
| GO:BP | GO:0051136 | regulation of NK T cell differentiation | 1 | 4 | 7.47e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 3 | 19 | 7.47e-01 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 1 | 7 | 7.48e-01 |
| GO:BP | GO:0008272 | sulfate transport | 1 | 12 | 7.48e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 7.48e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 7.48e-01 |
| GO:BP | GO:0031272 | regulation of pseudopodium assembly | 3 | 11 | 7.49e-01 |
| GO:BP | GO:0031274 | positive regulation of pseudopodium assembly | 3 | 11 | 7.49e-01 |
| GO:BP | GO:0032328 | alanine transport | 2 | 14 | 7.49e-01 |
| GO:BP | GO:0006982 | response to lipid hydroperoxide | 1 | 2 | 7.49e-01 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 2 | 11 | 7.49e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 5 | 111 | 7.49e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 1 | 31 | 7.49e-01 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 2 | 11 | 7.49e-01 |
| GO:BP | GO:1990502 | dense core granule maturation | 1 | 3 | 7.49e-01 |
| GO:BP | GO:2001171 | positive regulation of ATP biosynthetic process | 1 | 10 | 7.49e-01 |
| GO:BP | GO:1990504 | dense core granule exocytosis | 1 | 7 | 7.49e-01 |
| GO:BP | GO:1905413 | regulation of dense core granule exocytosis | 1 | 2 | 7.49e-01 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 2 | 11 | 7.49e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 2 | 16 | 7.49e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 9 | 53 | 7.49e-01 |
| GO:BP | GO:0060842 | arterial endothelial cell differentiation | 2 | 5 | 7.49e-01 |
| GO:BP | GO:0072338 | lactam metabolic process | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 2 | 16 | 7.49e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 7 | 52 | 7.49e-01 |
| GO:BP | GO:0061792 | secretory granule maturation | 1 | 3 | 7.49e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 8 | 29 | 7.49e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 7 | 187 | 7.49e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 1 | 16 | 7.49e-01 |
| GO:BP | GO:0008582 | regulation of synaptic assembly at neuromuscular junction | 2 | 4 | 7.49e-01 |
| GO:BP | GO:0019062 | virion attachment to host cell | 1 | 9 | 7.49e-01 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 1 | 104 | 7.49e-01 |
| GO:BP | GO:0035021 | negative regulation of Rac protein signal transduction | 1 | 4 | 7.49e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 7 | 187 | 7.49e-01 |
| GO:BP | GO:0070203 | regulation of establishment of protein localization to telomere | 2 | 11 | 7.49e-01 |
| GO:BP | GO:0009213 | pyrimidine deoxyribonucleoside triphosphate catabolic process | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0098905 | regulation of bundle of His cell action potential | 1 | 2 | 7.49e-01 |
| GO:BP | GO:0035332 | positive regulation of hippo signaling | 3 | 6 | 7.50e-01 |
| GO:BP | GO:0043373 | CD4-positive, alpha-beta T cell lineage commitment | 3 | 9 | 7.50e-01 |
| GO:BP | GO:0046185 | aldehyde catabolic process | 4 | 12 | 7.50e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 3 | 9 | 7.50e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 2 | 166 | 7.50e-01 |
| GO:BP | GO:1900425 | negative regulation of defense response to bacterium | 1 | 4 | 7.50e-01 |
| GO:BP | GO:0032929 | negative regulation of superoxide anion generation | 1 | 1 | 7.51e-01 |
| GO:BP | GO:0038165 | oncostatin-M-mediated signaling pathway | 1 | 4 | 7.51e-01 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 1 | 11 | 7.51e-01 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 52 | 81 | 7.51e-01 |
| GO:BP | GO:0060455 | negative regulation of gastric acid secretion | 1 | 3 | 7.51e-01 |
| GO:BP | GO:0021979 | hypothalamus cell differentiation | 5 | 8 | 7.51e-01 |
| GO:BP | GO:0034695 | response to prostaglandin E | 1 | 17 | 7.51e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 8 | 33 | 7.51e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 2 | 4 | 7.51e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 2 | 4 | 7.51e-01 |
| GO:BP | GO:2000987 | positive regulation of behavioral fear response | 1 | 3 | 7.51e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 7.51e-01 |
| GO:BP | GO:0036369 | transcription factor catabolic process | 2 | 5 | 7.52e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 4 | 13 | 7.52e-01 |
| GO:BP | GO:0090230 | regulation of centromere complex assembly | 1 | 4 | 7.52e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 2 | 5 | 7.52e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 5 | 166 | 7.52e-01 |
| GO:BP | GO:0007382 | specification of segmental identity, maxillary segment | 1 | 1 | 7.52e-01 |
| GO:BP | GO:1905871 | regulation of protein localization to cell leading edge | 1 | 2 | 7.52e-01 |
| GO:BP | GO:0044751 | cellular response to human chorionic gonadotropin stimulus | 1 | 2 | 7.52e-01 |
| GO:BP | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential | 1 | 6 | 7.52e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 1 | 5 | 7.52e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 7.52e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 7.52e-01 |
| GO:BP | GO:0001542 | ovulation from ovarian follicle | 1 | 8 | 7.52e-01 |
| GO:BP | GO:0045844 | positive regulation of striated muscle tissue development | 1 | 3 | 7.52e-01 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 1 | 45 | 7.52e-01 |
| GO:BP | GO:0048636 | positive regulation of muscle organ development | 1 | 3 | 7.52e-01 |
| GO:BP | GO:0031509 | subtelomeric heterochromatin formation | 2 | 7 | 7.52e-01 |
| GO:BP | GO:0061087 | positive regulation of histone H3-K27 methylation | 2 | 4 | 7.52e-01 |
| GO:BP | GO:1900060 | negative regulation of ceramide biosynthetic process | 2 | 5 | 7.52e-01 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 6 | 14 | 7.52e-01 |
| GO:BP | GO:0106077 | histone succinylation | 1 | 4 | 7.52e-01 |
| GO:BP | GO:0018335 | protein succinylation | 1 | 4 | 7.52e-01 |
| GO:BP | GO:0071929 | alpha-tubulin acetylation | 1 | 3 | 7.52e-01 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 2 | 37 | 7.52e-01 |
| GO:BP | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | 2 | 3 | 7.53e-01 |
| GO:BP | GO:0060375 | regulation of mast cell differentiation | 1 | 2 | 7.53e-01 |
| GO:BP | GO:0043982 | histone H4-K8 acetylation | 5 | 16 | 7.53e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 2 | 27 | 7.53e-01 |
| GO:BP | GO:0043981 | histone H4-K5 acetylation | 5 | 16 | 7.53e-01 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 2 | 4 | 7.53e-01 |
| GO:BP | GO:0000338 | protein deneddylation | 2 | 11 | 7.53e-01 |
| GO:BP | GO:1902460 | regulation of mesenchymal stem cell proliferation | 1 | 4 | 7.53e-01 |
| GO:BP | GO:0003165 | Purkinje myocyte development | 1 | 3 | 7.53e-01 |
| GO:BP | GO:1902462 | positive regulation of mesenchymal stem cell proliferation | 1 | 4 | 7.53e-01 |
| GO:BP | GO:1903729 | regulation of plasma membrane organization | 1 | 15 | 7.53e-01 |
| GO:BP | GO:0055005 | ventricular cardiac myofibril assembly | 1 | 3 | 7.53e-01 |
| GO:BP | GO:1903054 | negative regulation of extracellular matrix organization | 2 | 6 | 7.53e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 1 | 130 | 7.53e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 11 | 29 | 7.53e-01 |
| GO:BP | GO:1902897 | regulation of postsynaptic density protein 95 clustering | 1 | 2 | 7.53e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 10 | 400 | 7.53e-01 |
| GO:BP | GO:0021693 | cerebellar Purkinje cell layer structural organization | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 3 | 7 | 7.54e-01 |
| GO:BP | GO:0021698 | cerebellar cortex structural organization | 1 | 2 | 7.54e-01 |
| GO:BP | GO:1900372 | negative regulation of purine nucleotide biosynthetic process | 2 | 3 | 7.54e-01 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 3 | 17 | 7.54e-01 |
| GO:BP | GO:0031998 | regulation of fatty acid beta-oxidation | 6 | 16 | 7.54e-01 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 2 | 5 | 7.54e-01 |
| GO:BP | GO:0030809 | negative regulation of nucleotide biosynthetic process | 2 | 3 | 7.54e-01 |
| GO:BP | GO:0006390 | mitochondrial transcription | 5 | 18 | 7.54e-01 |
| GO:BP | GO:1905838 | regulation of telomeric D-loop disassembly | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0070126 | mitochondrial translational termination | 1 | 4 | 7.54e-01 |
| GO:BP | GO:0021590 | cerebellum maturation | 1 | 3 | 7.54e-01 |
| GO:BP | GO:1905778 | negative regulation of exonuclease activity | 1 | 2 | 7.54e-01 |
| GO:BP | GO:1905777 | regulation of exonuclease activity | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | 1 | 11 | 7.54e-01 |
| GO:BP | GO:0015854 | guanine transport | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0003158 | endothelium development | 15 | 104 | 7.54e-01 |
| GO:BP | GO:0032214 | negative regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0061975 | articular cartilage development | 1 | 3 | 7.54e-01 |
| GO:BP | GO:0035344 | hypoxanthine transport | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0055071 | manganese ion homeostasis | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0032213 | regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0003139 | secondary heart field specification | 4 | 10 | 7.54e-01 |
| GO:BP | GO:1904534 | negative regulation of telomeric loop disassembly | 1 | 2 | 7.54e-01 |
| GO:BP | GO:1905839 | negative regulation of telomeric D-loop disassembly | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0021699 | cerebellar cortex maturation | 1 | 3 | 7.54e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 7.54e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 48 | 418 | 7.54e-01 |
| GO:BP | GO:1904438 | regulation of iron ion import across plasma membrane | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 7 | 1107 | 7.54e-01 |
| GO:BP | GO:1903942 | positive regulation of respiratory gaseous exchange | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0090290 | positive regulation of osteoclast proliferation | 1 | 1 | 7.54e-01 |
| GO:BP | GO:0032606 | type I interferon production | 20 | 76 | 7.55e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 20 | 76 | 7.55e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 11 | 428 | 7.55e-01 |
| GO:BP | GO:0043335 | protein unfolding | 1 | 5 | 7.55e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 4 | 106 | 7.55e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 1 | 27 | 7.55e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 7 | 323 | 7.55e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 7.55e-01 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 1 | 2 | 7.55e-01 |
| GO:BP | GO:1903226 | positive regulation of endodermal cell differentiation | 1 | 2 | 7.55e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 3 | 20 | 7.55e-01 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 1 | 2 | 7.55e-01 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 1 | 17 | 7.56e-01 |
| GO:BP | GO:0043627 | response to estrogen | 1 | 52 | 7.56e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 10 | 1399 | 7.56e-01 |
| GO:BP | GO:0001915 | negative regulation of T cell mediated cytotoxicity | 1 | 3 | 7.56e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 4 | 13 | 7.56e-01 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 2 | 36 | 7.56e-01 |
| GO:BP | GO:2000251 | positive regulation of actin cytoskeleton reorganization | 6 | 14 | 7.56e-01 |
| GO:BP | GO:0045113 | regulation of integrin biosynthetic process | 1 | 2 | 7.56e-01 |
| GO:BP | GO:0038171 | cannabinoid signaling pathway | 2 | 5 | 7.56e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 5 | 53 | 7.56e-01 |
| GO:BP | GO:0008295 | spermidine biosynthetic process | 1 | 3 | 7.57e-01 |
| GO:BP | GO:0043029 | T cell homeostasis | 3 | 27 | 7.57e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 9 | 44 | 7.57e-01 |
| GO:BP | GO:2001293 | malonyl-CoA metabolic process | 2 | 6 | 7.57e-01 |
| GO:BP | GO:0010706 | ganglioside biosynthetic process via lactosylceramide | 1 | 5 | 7.57e-01 |
| GO:BP | GO:1904059 | regulation of locomotor rhythm | 1 | 3 | 7.57e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 2 | 13 | 7.57e-01 |
| GO:BP | GO:1905608 | positive regulation of presynapse assembly | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0002676 | regulation of chronic inflammatory response | 1 | 2 | 7.58e-01 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 1 | 24 | 7.58e-01 |
| GO:BP | GO:0042661 | regulation of mesodermal cell fate specification | 1 | 4 | 7.58e-01 |
| GO:BP | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine | 1 | 3 | 7.58e-01 |
| GO:BP | GO:0051309 | female meiosis chromosome separation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 755 | 7.58e-01 |
| GO:BP | GO:0018026 | peptidyl-lysine monomethylation | 1 | 11 | 7.58e-01 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 7.58e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 2 | 26 | 7.58e-01 |
| GO:BP | GO:0036371 | protein localization to T-tubule | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0032418 | lysosome localization | 7 | 62 | 7.58e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 1 | 111 | 7.58e-01 |
| GO:BP | GO:0061038 | uterus morphogenesis | 2 | 5 | 7.58e-01 |
| GO:BP | GO:0072678 | T cell migration | 2 | 38 | 7.58e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 2 | 5 | 7.58e-01 |
| GO:BP | GO:0032532 | regulation of microvillus length | 1 | 4 | 7.58e-01 |
| GO:BP | GO:0035799 | ureter maturation | 1 | 1 | 7.58e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 5 | 21 | 7.58e-01 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 2 | 7 | 7.58e-01 |
| GO:BP | GO:0018171 | peptidyl-cysteine oxidation | 1 | 3 | 7.58e-01 |
| GO:BP | GO:0090666 | scaRNA localization to Cajal body | 2 | 5 | 7.58e-01 |
| GO:BP | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity | 3 | 12 | 7.58e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 13 | 32 | 7.58e-01 |
| GO:BP | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G protein-coupled signaling pathway | 1 | 12 | 7.58e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 1 | 1939 | 7.58e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 13 | 7.58e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 4 | 116 | 7.58e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 6 | 18 | 7.58e-01 |
| GO:BP | GO:0032905 | transforming growth factor beta1 production | 2 | 11 | 7.58e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 4 | 16 | 7.58e-01 |
| GO:BP | GO:0020027 | hemoglobin metabolic process | 5 | 16 | 7.58e-01 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 3 | 23 | 7.58e-01 |
| GO:BP | GO:1900194 | negative regulation of oocyte maturation | 1 | 2 | 7.58e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 7.58e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 2 | 9 | 7.58e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 7 | 62 | 7.58e-01 |
| GO:BP | GO:0050904 | diapedesis | 2 | 6 | 7.59e-01 |
| GO:BP | GO:1905931 | negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0048840 | otolith development | 1 | 6 | 7.59e-01 |
| GO:BP | GO:0021626 | central nervous system maturation | 2 | 5 | 7.59e-01 |
| GO:BP | GO:1905223 | epicardium morphogenesis | 1 | 2 | 7.59e-01 |
| GO:BP | GO:0010533 | regulation of activation of Janus kinase activity | 1 | 3 | 7.59e-01 |
| GO:BP | GO:1900240 | negative regulation of phenotypic switching | 1 | 2 | 7.59e-01 |
| GO:BP | GO:1905916 | negative regulation of cell differentiation involved in phenotypic switching | 1 | 2 | 7.59e-01 |
| GO:BP | GO:1901998 | toxin transport | 1 | 37 | 7.60e-01 |
| GO:BP | GO:0035726 | common myeloid progenitor cell proliferation | 1 | 5 | 7.60e-01 |
| GO:BP | GO:0007423 | sensory organ development | 2 | 396 | 7.60e-01 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 3 | 11 | 7.60e-01 |
| GO:BP | GO:0006414 | translational elongation | 6 | 65 | 7.60e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 1 | 23 | 7.60e-01 |
| GO:BP | GO:0007597 | blood coagulation, intrinsic pathway | 1 | 3 | 7.60e-01 |
| GO:BP | GO:0090675 | intermicrovillar adhesion | 1 | 1 | 7.60e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 3 | 16 | 7.60e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 7.60e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 7.60e-01 |
| GO:BP | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 1 | 6 | 7.60e-01 |
| GO:BP | GO:0070430 | positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0016046 | detection of fungus | 1 | 1 | 7.60e-01 |
| GO:BP | GO:1904466 | positive regulation of matrix metallopeptidase secretion | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0036298 | recombinational interstrand cross-link repair | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0070716 | mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication | 1 | 2 | 7.60e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 1 | 72 | 7.60e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 4 | 11 | 7.60e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 3 | 33 | 7.60e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 1 | 6 | 7.60e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 3 | 7 | 7.61e-01 |
| GO:BP | GO:0048069 | eye pigmentation | 1 | 4 | 7.61e-01 |
| GO:BP | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 7.62e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 4 | 28 | 7.62e-01 |
| GO:BP | GO:0050747 | positive regulation of lipoprotein metabolic process | 1 | 4 | 7.62e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 1 | 7 | 7.62e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 1 | 7 | 7.62e-01 |
| GO:BP | GO:1903061 | positive regulation of protein lipidation | 1 | 4 | 7.62e-01 |
| GO:BP | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction | 1 | 1 | 7.62e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 2 | 130 | 7.62e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 17 | 41 | 7.62e-01 |
| GO:BP | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate | 1 | 4 | 7.62e-01 |
| GO:BP | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development | 1 | 2 | 7.62e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 7.62e-01 |
| GO:BP | GO:1902498 | regulation of protein autoubiquitination | 2 | 5 | 7.62e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 7.62e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 7.62e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 7.62e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 7.62e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 1 | 8 | 7.62e-01 |
| GO:BP | GO:0002092 | positive regulation of receptor internalization | 1 | 21 | 7.62e-01 |
| GO:BP | GO:1902871 | positive regulation of amacrine cell differentiation | 1 | 1 | 7.62e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 2 | 19 | 7.62e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 2 | 32 | 7.62e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 2 | 66 | 7.62e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 7 | 18 | 7.62e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 1 | 173 | 7.63e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 5 | 8 | 7.63e-01 |
| GO:BP | GO:0019264 | glycine biosynthetic process from serine | 1 | 2 | 7.63e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 6 | 775 | 7.63e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 1 | 35 | 7.63e-01 |
| GO:BP | GO:0031133 | regulation of axon diameter | 1 | 4 | 7.63e-01 |
| GO:BP | GO:0070669 | response to interleukin-2 | 1 | 3 | 7.63e-01 |
| GO:BP | GO:0043379 | memory T cell differentiation | 2 | 5 | 7.63e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 5 | 67 | 7.63e-01 |
| GO:BP | GO:0034342 | response to type III interferon | 1 | 5 | 7.63e-01 |
| GO:BP | GO:0009153 | purine deoxyribonucleotide biosynthetic process | 2 | 4 | 7.63e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 1 | 58 | 7.63e-01 |
| GO:BP | GO:1903580 | positive regulation of ATP metabolic process | 1 | 11 | 7.63e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 3 | 152 | 7.64e-01 |
| GO:BP | GO:0031508 | pericentric heterochromatin formation | 1 | 3 | 7.64e-01 |
| GO:BP | GO:1905179 | negative regulation of cardiac muscle tissue regeneration | 1 | 2 | 7.64e-01 |
| GO:BP | GO:1905178 | regulation of cardiac muscle tissue regeneration | 1 | 2 | 7.64e-01 |
| GO:BP | GO:0003307 | regulation of Wnt signaling pathway involved in heart development | 1 | 7 | 7.64e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 7.64e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 1 | 2565 | 7.64e-01 |
| GO:BP | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 1 | 7 | 7.64e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 2 | 169 | 7.64e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 5 | 7.64e-01 |
| GO:BP | GO:0060399 | positive regulation of growth hormone receptor signaling pathway | 1 | 2 | 7.65e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 1 | 2573 | 7.65e-01 |
| GO:BP | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination | 1 | 1 | 7.65e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 1 | 30 | 7.65e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 2 | 4128 | 7.65e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 15 | 55 | 7.65e-01 |
| GO:BP | GO:0034139 | regulation of toll-like receptor 3 signaling pathway | 1 | 8 | 7.65e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 13 | 42 | 7.65e-01 |
| GO:BP | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway | 1 | 2 | 7.65e-01 |
| GO:BP | GO:0072359 | circulatory system development | 35 | 884 | 7.65e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 6 | 25 | 7.65e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 2 | 7 | 7.65e-01 |
| GO:BP | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis | 1 | 4 | 7.65e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 9 | 66 | 7.65e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 9 | 40 | 7.65e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 8 | 61 | 7.65e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 2 | 86 | 7.65e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 1 | 30 | 7.65e-01 |
| GO:BP | GO:0006533 | aspartate catabolic process | 1 | 2 | 7.65e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 1 | 28 | 7.65e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 2 | 86 | 7.65e-01 |
| GO:BP | GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 2 | 5 | 7.65e-01 |
| GO:BP | GO:0070170 | regulation of tooth mineralization | 1 | 4 | 7.65e-01 |
| GO:BP | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 2 | 5 | 7.65e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 7.65e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 13 | 67 | 7.66e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 7.66e-01 |
| GO:BP | GO:0009386 | translational attenuation | 1 | 2 | 7.66e-01 |
| GO:BP | GO:2000395 | regulation of ubiquitin-dependent endocytosis | 1 | 2 | 7.66e-01 |
| GO:BP | GO:0015844 | monoamine transport | 1 | 45 | 7.66e-01 |
| GO:BP | GO:2000397 | positive regulation of ubiquitin-dependent endocytosis | 1 | 2 | 7.66e-01 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 5 | 7.66e-01 |
| GO:BP | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 20 | 2135 | 7.67e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 1 | 5 | 7.67e-01 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 4 | 15 | 7.67e-01 |
| GO:BP | GO:0071874 | cellular response to norepinephrine stimulus | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0071873 | response to norepinephrine | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 80 | 679 | 7.67e-01 |
| GO:BP | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | 3 | 8 | 7.67e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 2 | 21 | 7.67e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 1 | 15 | 7.67e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 2 | 47 | 7.67e-01 |
| GO:BP | GO:0021524 | visceral motor neuron differentiation | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 7.67e-01 |
| GO:BP | GO:0015670 | carbon dioxide transport | 1 | 2 | 7.67e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 7.67e-01 |
| GO:BP | GO:1900234 | regulation of Kit signaling pathway | 1 | 2 | 7.67e-01 |
| GO:BP | GO:1903960 | negative regulation of anion transmembrane transport | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 15 | 96 | 7.67e-01 |
| GO:BP | GO:0060438 | trachea development | 3 | 13 | 7.67e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 3 | 2684 | 7.67e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 7.67e-01 |
| GO:BP | GO:0006014 | D-ribose metabolic process | 1 | 3 | 7.67e-01 |
| GO:BP | GO:1900235 | negative regulation of Kit signaling pathway | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 3 | 7 | 7.67e-01 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 4 | 110 | 7.67e-01 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 3 | 2683 | 7.67e-01 |
| GO:BP | GO:2001225 | regulation of chloride transport | 2 | 4 | 7.67e-01 |
| GO:BP | GO:0010360 | negative regulation of anion channel activity | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 2 | 13 | 7.67e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 11 | 26 | 7.67e-01 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 2 | 7.67e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 3 | 19 | 7.67e-01 |
| GO:BP | GO:0009111 | vitamin catabolic process | 1 | 3 | 7.68e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 1 | 10 | 7.68e-01 |
| GO:BP | GO:0034334 | adherens junction maintenance | 2 | 6 | 7.68e-01 |
| GO:BP | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | 1 | 5 | 7.68e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 2 | 295 | 7.68e-01 |
| GO:BP | GO:0001732 | formation of cytoplasmic translation initiation complex | 2 | 16 | 7.68e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 1 | 114 | 7.68e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 2 | 69 | 7.68e-01 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 4 | 15 | 7.68e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 4 | 3730 | 7.68e-01 |
| GO:BP | GO:0032074 | negative regulation of nuclease activity | 1 | 6 | 7.68e-01 |
| GO:BP | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 3 | 10 | 7.68e-01 |
| GO:BP | GO:0072695 | regulation of DNA recombination at telomere | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0048239 | negative regulation of DNA recombination at telomere | 1 | 2 | 7.68e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 11 | 717 | 7.68e-01 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 3 | 8 | 7.68e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 5 | 17 | 7.68e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 1 | 34 | 7.68e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 10 | 7.69e-01 |
| GO:BP | GO:2000405 | negative regulation of T cell migration | 1 | 4 | 7.69e-01 |
| GO:BP | GO:0061196 | fungiform papilla development | 1 | 5 | 7.69e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 1 | 66 | 7.69e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 1 | 12 | 7.69e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 3 | 17 | 7.69e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 8 | 1278 | 7.69e-01 |
| GO:BP | GO:0019343 | cysteine biosynthetic process via cystathionine | 1 | 2 | 7.70e-01 |
| GO:BP | GO:1905145 | cellular response to acetylcholine | 4 | 20 | 7.70e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 7.70e-01 |
| GO:BP | GO:0006565 | L-serine catabolic process | 2 | 4 | 7.70e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 4 | 16 | 7.70e-01 |
| GO:BP | GO:0099515 | actin filament-based transport | 6 | 15 | 7.70e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 6 | 43 | 7.71e-01 |
| GO:BP | GO:1990790 | response to glial cell derived neurotrophic factor | 1 | 1 | 7.71e-01 |
| GO:BP | GO:1990792 | cellular response to glial cell derived neurotrophic factor | 1 | 1 | 7.71e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 1 | 49 | 7.71e-01 |
| GO:BP | GO:0032689 | negative regulation of type II interferon production | 1 | 16 | 7.71e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 1 | 13 | 7.71e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 2 | 79 | 7.71e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 4 | 551 | 7.71e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 6 | 26 | 7.71e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 7.71e-01 |
| GO:BP | GO:0071140 | resolution of mitotic recombination intermediates | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 2 | 18 | 7.71e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 2 | 99 | 7.71e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 6 | 230 | 7.71e-01 |
| GO:BP | GO:0035350 | FAD transmembrane transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0071106 | adenosine 3’,5’-bisphosphate transmembrane transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0035349 | coenzyme A transmembrane transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 2 | 10 | 7.71e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 21 | 139 | 7.71e-01 |
| GO:BP | GO:0080121 | AMP transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 1 | 1 | 7.71e-01 |
| GO:BP | GO:0034035 | purine ribonucleoside bisphosphate metabolic process | 1 | 10 | 7.71e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 2 | 10 | 7.71e-01 |
| GO:BP | GO:0050427 | 3’-phosphoadenosine 5’-phosphosulfate metabolic process | 1 | 10 | 7.71e-01 |
| GO:BP | GO:0097186 | amelogenesis | 2 | 14 | 7.71e-01 |
| GO:BP | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination | 1 | 3 | 7.71e-01 |
| GO:BP | GO:0015883 | FAD transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 4 | 9 | 7.71e-01 |
| GO:BP | GO:0015880 | coenzyme A transport | 1 | 2 | 7.71e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 2 | 17 | 7.71e-01 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 1 | 5 | 7.71e-01 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 1 | 30 | 7.71e-01 |
| GO:BP | GO:0030220 | platelet formation | 2 | 17 | 7.71e-01 |
| GO:BP | GO:0001974 | blood vessel remodeling | 4 | 32 | 7.71e-01 |
| GO:BP | GO:1905323 | telomerase holoenzyme complex assembly | 1 | 6 | 7.71e-01 |
| GO:BP | GO:0097102 | endothelial tip cell fate specification | 1 | 2 | 7.72e-01 |
| GO:BP | GO:1904772 | response to tetrachloromethane | 1 | 4 | 7.72e-01 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 5 | 14 | 7.72e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 10 | 25 | 7.72e-01 |
| GO:BP | GO:1904844 | response to L-glutamine | 1 | 3 | 7.72e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 2 | 3 | 7.72e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 5 | 26 | 7.72e-01 |
| GO:BP | GO:0050896 | response to stimulus | 3 | 5770 | 7.72e-01 |
| GO:BP | GO:2000491 | positive regulation of hepatic stellate cell activation | 2 | 4 | 7.72e-01 |
| GO:BP | GO:0061441 | renal artery morphogenesis | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 3 | 2714 | 7.72e-01 |
| GO:BP | GO:1905590 | fibronectin fibril organization | 1 | 1 | 7.72e-01 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 3 | 5 | 7.72e-01 |
| GO:BP | GO:0051597 | response to methylmercury | 1 | 5 | 7.72e-01 |
| GO:BP | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation | 1 | 3 | 7.72e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 1 | 45 | 7.72e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 2 | 110 | 7.72e-01 |
| GO:BP | GO:1902595 | regulation of DNA replication origin binding | 1 | 2 | 7.72e-01 |
| GO:BP | GO:0097254 | renal tubular secretion | 1 | 15 | 7.72e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 3 | 22 | 7.72e-01 |
| GO:BP | GO:2000631 | regulation of pre-miRNA processing | 1 | 3 | 7.72e-01 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 5 | 137 | 7.73e-01 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 4 | 9 | 7.73e-01 |
| GO:BP | GO:0099039 | sphingolipid translocation | 1 | 4 | 7.73e-01 |
| GO:BP | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport | 2 | 5 | 7.73e-01 |
| GO:BP | GO:0046726 | positive regulation by virus of viral protein levels in host cell | 1 | 4 | 7.73e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 2 | 68 | 7.73e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 2 | 70 | 7.73e-01 |
| GO:BP | GO:0044598 | doxorubicin metabolic process | 3 | 8 | 7.73e-01 |
| GO:BP | GO:0030638 | polyketide metabolic process | 3 | 8 | 7.73e-01 |
| GO:BP | GO:0030647 | aminoglycoside antibiotic metabolic process | 3 | 7 | 7.73e-01 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 3 | 9 | 7.73e-01 |
| GO:BP | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis | 2 | 5 | 7.73e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 3 | 47 | 7.74e-01 |
| GO:BP | GO:0021550 | medulla oblongata development | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0016198 | axon choice point recognition | 1 | 5 | 7.74e-01 |
| GO:BP | GO:0071922 | regulation of cohesin loading | 1 | 2 | 7.74e-01 |
| GO:BP | GO:0006409 | tRNA export from nucleus | 1 | 3 | 7.74e-01 |
| GO:BP | GO:1900119 | positive regulation of execution phase of apoptosis | 1 | 7 | 7.74e-01 |
| GO:BP | GO:1901485 | positive regulation of transcription factor catabolic process | 1 | 3 | 7.74e-01 |
| GO:BP | GO:0071265 | L-methionine biosynthetic process | 2 | 5 | 7.74e-01 |
| GO:BP | GO:1901483 | regulation of transcription factor catabolic process | 1 | 3 | 7.74e-01 |
| GO:BP | GO:0071267 | L-methionine salvage | 2 | 5 | 7.74e-01 |
| GO:BP | GO:0043102 | amino acid salvage | 2 | 5 | 7.74e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 6 | 29 | 7.74e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 1 | 104 | 7.74e-01 |
| GO:BP | GO:1901078 | negative regulation of relaxation of muscle | 1 | 4 | 7.75e-01 |
| GO:BP | GO:0045218 | zonula adherens maintenance | 1 | 3 | 7.75e-01 |
| GO:BP | GO:0075506 | entry of viral genome into host nucleus through nuclear pore complex via importin | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0048733 | sebaceous gland development | 1 | 6 | 7.75e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0007549 | dosage compensation | 3 | 29 | 7.75e-01 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 1 | 8 | 7.75e-01 |
| GO:BP | GO:0051712 | positive regulation of killing of cells of another organism | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 2 | 73 | 7.75e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 7.75e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 6 | 26 | 7.75e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 2 | 25 | 7.75e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 5 | 7 | 7.75e-01 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 3 | 12 | 7.75e-01 |
| GO:BP | GO:2000503 | positive regulation of natural killer cell chemotaxis | 1 | 1 | 7.75e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 7.75e-01 |
| GO:BP | GO:0002826 | negative regulation of T-helper 1 type immune response | 1 | 3 | 7.75e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 9 | 196 | 7.75e-01 |
| GO:BP | GO:0070202 | regulation of establishment of protein localization to chromosome | 2 | 12 | 7.75e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 2 | 12 | 7.75e-01 |
| GO:BP | GO:0061356 | regulation of Wnt protein secretion | 1 | 4 | 7.75e-01 |
| GO:BP | GO:1903934 | positive regulation of DNA primase activity | 2 | 5 | 7.75e-01 |
| GO:BP | GO:0035801 | adrenal cortex development | 1 | 2 | 7.75e-01 |
| GO:BP | GO:1903391 | regulation of adherens junction organization | 1 | 5 | 7.75e-01 |
| GO:BP | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway | 2 | 4 | 7.75e-01 |
| GO:BP | GO:0001706 | endoderm formation | 2 | 46 | 7.75e-01 |
| GO:BP | GO:1903932 | regulation of DNA primase activity | 2 | 5 | 7.75e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 2 | 12 | 7.75e-01 |
| GO:BP | GO:1902031 | regulation of NADP metabolic process | 3 | 8 | 7.75e-01 |
| GO:BP | GO:0001575 | globoside metabolic process | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0001576 | globoside biosynthetic process | 1 | 2 | 7.75e-01 |
| GO:BP | GO:0031115 | negative regulation of microtubule polymerization | 2 | 13 | 7.75e-01 |
| GO:BP | GO:0034380 | high-density lipoprotein particle assembly | 1 | 10 | 7.75e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 2 | 21 | 7.76e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 5 | 114 | 7.76e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 2 | 8 | 7.76e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 3 | 7 | 7.76e-01 |
| GO:BP | GO:0032743 | positive regulation of interleukin-2 production | 2 | 20 | 7.76e-01 |
| GO:BP | GO:0060931 | sinoatrial node cell development | 2 | 4 | 7.76e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 4 | 14 | 7.76e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 3 | 305 | 7.76e-01 |
| GO:BP | GO:2000410 | regulation of thymocyte migration | 1 | 1 | 7.76e-01 |
| GO:BP | GO:0090292 | nuclear matrix anchoring at nuclear membrane | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 2 | 293 | 7.76e-01 |
| GO:BP | GO:2000418 | positive regulation of eosinophil migration | 1 | 1 | 7.76e-01 |
| GO:BP | GO:0043578 | nuclear matrix organization | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0030030 | cell projection organization | 9 | 1233 | 7.76e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 7 | 48 | 7.76e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 1 | 56 | 7.76e-01 |
| GO:BP | GO:0048007 | antigen processing and presentation, exogenous lipid antigen via MHC class Ib | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib | 1 | 3 | 7.76e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 4 | 7.76e-01 |
| GO:BP | GO:0106119 | negative regulation of sterol biosynthetic process | 2 | 5 | 7.76e-01 |
| GO:BP | GO:1901860 | positive regulation of mitochondrial DNA metabolic process | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0050892 | intestinal absorption | 2 | 22 | 7.76e-01 |
| GO:BP | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 2 | 12 | 7.76e-01 |
| GO:BP | GO:0043007 | maintenance of rDNA | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 2 | 44 | 7.76e-01 |
| GO:BP | GO:0048819 | regulation of hair follicle maturation | 3 | 9 | 7.76e-01 |
| GO:BP | GO:0030916 | otic vesicle formation | 2 | 6 | 7.76e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 3 | 7 | 7.76e-01 |
| GO:BP | GO:0045541 | negative regulation of cholesterol biosynthetic process | 2 | 5 | 7.76e-01 |
| GO:BP | GO:0110095 | cellular detoxification of aldehyde | 2 | 5 | 7.76e-01 |
| GO:BP | GO:0044848 | biological phase | 3 | 9 | 7.76e-01 |
| GO:BP | GO:0044851 | hair cycle phase | 3 | 9 | 7.76e-01 |
| GO:BP | GO:0045213 | neurotransmitter receptor metabolic process | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0051695 | actin filament uncapping | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0060764 | cell-cell signaling involved in mammary gland development | 1 | 1 | 7.76e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 6 | 15 | 7.76e-01 |
| GO:BP | GO:0006842 | tricarboxylic acid transport | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 3 | 8 | 7.76e-01 |
| GO:BP | GO:0006071 | glycerol metabolic process | 1 | 12 | 7.76e-01 |
| GO:BP | GO:0021942 | radial glia guided migration of Purkinje cell | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0061550 | cranial ganglion development | 2 | 4 | 7.76e-01 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 3 | 20 | 7.76e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 1 | 10 | 7.76e-01 |
| GO:BP | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity | 3 | 11 | 7.76e-01 |
| GO:BP | GO:0098942 | retrograde trans-synaptic signaling by trans-synaptic protein complex | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0060857 | establishment of glial blood-brain barrier | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0015746 | citrate transport | 1 | 3 | 7.76e-01 |
| GO:BP | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway | 1 | 1 | 7.76e-01 |
| GO:BP | GO:0021644 | vagus nerve morphogenesis | 1 | 1 | 7.76e-01 |
| GO:BP | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0090206 | negative regulation of cholesterol metabolic process | 2 | 5 | 7.76e-01 |
| GO:BP | GO:0016340 | calcium-dependent cell-matrix adhesion | 1 | 2 | 7.76e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 5 | 17 | 7.77e-01 |
| GO:BP | GO:0018277 | protein deamination | 1 | 2 | 7.77e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 3 | 28 | 7.78e-01 |
| GO:BP | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration | 1 | 7 | 7.78e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 2 | 73 | 7.78e-01 |
| GO:BP | GO:0042908 | xenobiotic transport | 9 | 30 | 7.78e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 11 | 1564 | 7.78e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 5 | 226 | 7.78e-01 |
| GO:BP | GO:1904708 | regulation of granulosa cell apoptotic process | 1 | 2 | 7.78e-01 |
| GO:BP | GO:1904700 | granulosa cell apoptotic process | 1 | 2 | 7.78e-01 |
| GO:BP | GO:0015871 | choline transport | 2 | 6 | 7.78e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 336 | 7.78e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 3 | 6 | 7.78e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 15 | 40 | 7.79e-01 |
| GO:BP | GO:0061181 | regulation of chondrocyte development | 2 | 4 | 7.79e-01 |
| GO:BP | GO:0097360 | chorionic trophoblast cell proliferation | 1 | 2 | 7.79e-01 |
| GO:BP | GO:1901382 | regulation of chorionic trophoblast cell proliferation | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0042264 | peptidyl-aspartic acid hydroxylation | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0003160 | endocardium morphogenesis | 1 | 3 | 7.79e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0006714 | sesquiterpenoid metabolic process | 1 | 2 | 7.79e-01 |
| GO:BP | GO:0060159 | regulation of dopamine receptor signaling pathway | 1 | 7 | 7.79e-01 |
| GO:BP | GO:0046968 | peptide antigen transport | 1 | 1 | 7.79e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 3 | 41 | 7.79e-01 |
| GO:BP | GO:2001280 | positive regulation of unsaturated fatty acid biosynthetic process | 1 | 4 | 7.79e-01 |
| GO:BP | GO:0006415 | translational termination | 4 | 17 | 7.79e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 1 | 11 | 7.80e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 7.80e-01 |
| GO:BP | GO:0021563 | glossopharyngeal nerve development | 2 | 3 | 7.80e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 2 | 34 | 7.80e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 6 | 26 | 7.80e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 5 | 112 | 7.81e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 5 | 834 | 7.81e-01 |
| GO:BP | GO:1900208 | regulation of cardiolipin metabolic process | 1 | 3 | 7.81e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 2 | 5 | 7.81e-01 |
| GO:BP | GO:0048675 | axon extension | 20 | 110 | 7.81e-01 |
| GO:BP | GO:0022027 | interkinetic nuclear migration | 1 | 4 | 7.81e-01 |
| GO:BP | GO:0035482 | gastric motility | 1 | 3 | 7.81e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 1 | 53 | 7.81e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 1 | 17 | 7.81e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 14 | 80 | 7.82e-01 |
| GO:BP | GO:0015711 | organic anion transport | 2 | 264 | 7.82e-01 |
| GO:BP | GO:0006555 | methionine metabolic process | 1 | 13 | 7.82e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 7.82e-01 |
| GO:BP | GO:0014052 | regulation of gamma-aminobutyric acid secretion | 1 | 5 | 7.82e-01 |
| GO:BP | GO:0030035 | microspike assembly | 2 | 5 | 7.82e-01 |
| GO:BP | GO:0070474 | positive regulation of uterine smooth muscle contraction | 1 | 4 | 7.82e-01 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 38 | 7.82e-01 |
| GO:BP | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation | 1 | 2 | 7.82e-01 |
| GO:BP | GO:0044650 | adhesion of symbiont to host cell | 1 | 9 | 7.82e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 1 | 10 | 7.82e-01 |
| GO:BP | GO:0046959 | habituation | 1 | 5 | 7.82e-01 |
| GO:BP | GO:0015820 | leucine transport | 1 | 10 | 7.82e-01 |
| GO:BP | GO:0001655 | urogenital system development | 2 | 49 | 7.82e-01 |
| GO:BP | GO:2000340 | positive regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 3 | 7.83e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 5 | 74 | 7.83e-01 |
| GO:BP | GO:0017185 | peptidyl-lysine hydroxylation | 2 | 9 | 7.83e-01 |
| GO:BP | GO:0060629 | regulation of homologous chromosome segregation | 1 | 1 | 7.83e-01 |
| GO:BP | GO:0002920 | regulation of humoral immune response | 1 | 18 | 7.84e-01 |
| GO:BP | GO:0030001 | metal ion transport | 2 | 562 | 7.84e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 6 | 31 | 7.84e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 1 | 28 | 7.84e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 1 | 170 | 7.84e-01 |
| GO:BP | GO:0110104 | mRNA alternative polyadenylation | 1 | 5 | 7.84e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 14 | 209 | 7.85e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 10 | 114 | 7.85e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 2 | 8 | 7.85e-01 |
| GO:BP | GO:0002295 | T-helper cell lineage commitment | 2 | 7 | 7.85e-01 |
| GO:BP | GO:1904580 | regulation of intracellular mRNA localization | 1 | 4 | 7.85e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 3 | 37 | 7.85e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 7.85e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 7.85e-01 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 3 | 11 | 7.85e-01 |
| GO:BP | GO:1901097 | negative regulation of autophagosome maturation | 1 | 5 | 7.85e-01 |
| GO:BP | GO:0071285 | cellular response to lithium ion | 1 | 8 | 7.85e-01 |
| GO:BP | GO:2000485 | regulation of glutamine transport | 1 | 2 | 7.85e-01 |
| GO:BP | GO:0001553 | luteinization | 3 | 10 | 7.86e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 3 | 19 | 7.86e-01 |
| GO:BP | GO:0046292 | formaldehyde metabolic process | 1 | 3 | 7.86e-01 |
| GO:BP | GO:0045625 | regulation of T-helper 1 cell differentiation | 2 | 5 | 7.86e-01 |
| GO:BP | GO:1903003 | positive regulation of protein deubiquitination | 1 | 6 | 7.86e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 1 | 182 | 7.86e-01 |
| GO:BP | GO:0030043 | actin filament fragmentation | 1 | 4 | 7.86e-01 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 10 | 29 | 7.87e-01 |
| GO:BP | GO:0033239 | negative regulation of amine metabolic process | 2 | 2 | 7.87e-01 |
| GO:BP | GO:0002038 | positive regulation of L-glutamate import across plasma membrane | 1 | 3 | 7.87e-01 |
| GO:BP | GO:0090715 | immunological memory formation process | 2 | 6 | 7.87e-01 |
| GO:BP | GO:0048631 | regulation of skeletal muscle tissue growth | 2 | 7 | 7.87e-01 |
| GO:BP | GO:0002315 | marginal zone B cell differentiation | 1 | 6 | 7.87e-01 |
| GO:BP | GO:0071205 | protein localization to juxtaparanode region of axon | 1 | 3 | 7.87e-01 |
| GO:BP | GO:0002175 | protein localization to paranode region of axon | 1 | 3 | 7.87e-01 |
| GO:BP | GO:0010960 | magnesium ion homeostasis | 2 | 9 | 7.87e-01 |
| GO:BP | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly | 1 | 6 | 7.87e-01 |
| GO:BP | GO:2000291 | regulation of myoblast proliferation | 2 | 11 | 7.88e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 7.88e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 2 | 96 | 7.88e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 2 | 37 | 7.88e-01 |
| GO:BP | GO:0140250 | regulation protein catabolic process at synapse | 1 | 5 | 7.88e-01 |
| GO:BP | GO:0140331 | aminophospholipid translocation | 2 | 6 | 7.88e-01 |
| GO:BP | GO:0016116 | carotenoid metabolic process | 1 | 1 | 7.88e-01 |
| GO:BP | GO:0016108 | tetraterpenoid metabolic process | 1 | 1 | 7.88e-01 |
| GO:BP | GO:0042214 | terpene metabolic process | 1 | 1 | 7.88e-01 |
| GO:BP | GO:0120253 | hydrocarbon catabolic process | 1 | 1 | 7.88e-01 |
| GO:BP | GO:0002946 | tRNA C5-cytosine methylation | 1 | 3 | 7.88e-01 |
| GO:BP | GO:0021553 | olfactory nerve development | 3 | 5 | 7.88e-01 |
| GO:BP | GO:0048936 | peripheral nervous system neuron axonogenesis | 1 | 4 | 7.89e-01 |
| GO:BP | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient | 1 | 7 | 7.89e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 2 | 5 | 7.89e-01 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 2 | 9 | 7.89e-01 |
| GO:BP | GO:0035635 | entry of bacterium into host cell | 2 | 8 | 7.89e-01 |
| GO:BP | GO:0003011 | involuntary skeletal muscle contraction | 1 | 3 | 7.89e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 1 | 376 | 7.90e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 1 | 21 | 7.90e-01 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 10 | 28 | 7.90e-01 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 1 | 6 | 7.90e-01 |
| GO:BP | GO:0006826 | iron ion transport | 1 | 40 | 7.90e-01 |
| GO:BP | GO:0140291 | peptidyl-glutamate ADP-deribosylation | 1 | 3 | 7.90e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 1 | 39 | 7.91e-01 |
| GO:BP | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 2 | 5 | 7.91e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 11 | 39 | 7.91e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 13 | 41 | 7.91e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 12 | 28 | 7.91e-01 |
| GO:BP | GO:0002246 | wound healing involved in inflammatory response | 2 | 6 | 7.91e-01 |
| GO:BP | GO:0070848 | response to growth factor | 4 | 572 | 7.91e-01 |
| GO:BP | GO:0051604 | protein maturation | 1 | 259 | 7.91e-01 |
| GO:BP | GO:0035621 | ER to Golgi ceramide transport | 1 | 3 | 7.91e-01 |
| GO:BP | GO:0014734 | skeletal muscle hypertrophy | 1 | 1 | 7.91e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 2 | 211 | 7.92e-01 |
| GO:BP | GO:0033967 | box C/D RNA metabolic process | 1 | 3 | 7.92e-01 |
| GO:BP | GO:0000494 | box C/D RNA 3’-end processing | 1 | 3 | 7.92e-01 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 1 | 97 | 7.92e-01 |
| GO:BP | GO:0034963 | box C/D RNA processing | 1 | 3 | 7.92e-01 |
| GO:BP | GO:0035094 | response to nicotine | 2 | 28 | 7.92e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 7.92e-01 |
| GO:BP | GO:2000646 | positive regulation of receptor catabolic process | 1 | 5 | 7.92e-01 |
| GO:BP | GO:0006869 | lipid transport | 5 | 272 | 7.92e-01 |
| GO:BP | GO:0045345 | positive regulation of MHC class I biosynthetic process | 1 | 2 | 7.92e-01 |
| GO:BP | GO:0019230 | proprioception | 1 | 3 | 7.93e-01 |
| GO:BP | GO:1902565 | positive regulation of neutrophil activation | 1 | 1 | 7.93e-01 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 1 | 24 | 7.93e-01 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 2 | 18 | 7.93e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 7.93e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 7.93e-01 |
| GO:BP | GO:1902915 | negative regulation of protein polyubiquitination | 1 | 9 | 7.93e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 13 | 124 | 7.94e-01 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 7 | 24 | 7.94e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 2 | 98 | 7.94e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 8 | 26 | 7.94e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 1 | 25 | 7.94e-01 |
| GO:BP | GO:1990127 | intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 7.94e-01 |
| GO:BP | GO:1902238 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator | 1 | 2 | 7.94e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 2 | 115 | 7.95e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 7.95e-01 |
| GO:BP | GO:0002864 | regulation of acute inflammatory response to antigenic stimulus | 3 | 7 | 7.95e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 2 | 57 | 7.95e-01 |
| GO:BP | GO:0043171 | peptide catabolic process | 2 | 16 | 7.95e-01 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 1 | 7 | 7.95e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 2 | 12 | 7.96e-01 |
| GO:BP | GO:0097638 | L-arginine import across plasma membrane | 2 | 4 | 7.96e-01 |
| GO:BP | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway | 1 | 3 | 7.96e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 1 | 42 | 7.96e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 6 | 32 | 7.96e-01 |
| GO:BP | GO:0060685 | regulation of prostatic bud formation | 1 | 5 | 7.96e-01 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 37 | 64 | 7.96e-01 |
| GO:BP | GO:0045112 | integrin biosynthetic process | 1 | 3 | 7.96e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 2 | 11 | 7.96e-01 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 5 | 144 | 7.96e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 2 | 19 | 7.96e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 2 | 14 | 7.97e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 2 | 14 | 7.97e-01 |
| GO:BP | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade | 2 | 4 | 7.97e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 2 | 34 | 7.97e-01 |
| GO:BP | GO:0034344 | regulation of type III interferon production | 1 | 3 | 7.97e-01 |
| GO:BP | GO:0034343 | type III interferon production | 1 | 3 | 7.97e-01 |
| GO:BP | GO:1902474 | positive regulation of protein localization to synapse | 2 | 6 | 7.97e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 11 | 31 | 7.97e-01 |
| GO:BP | GO:0043415 | positive regulation of skeletal muscle tissue regeneration | 1 | 4 | 7.97e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 7.97e-01 |
| GO:BP | GO:0099159 | regulation of modification of postsynaptic structure | 1 | 6 | 7.97e-01 |
| GO:BP | GO:0032536 | regulation of cell projection size | 2 | 8 | 7.97e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 1 | 49 | 7.97e-01 |
| GO:BP | GO:2000523 | regulation of T cell costimulation | 1 | 2 | 7.97e-01 |
| GO:BP | GO:1905749 | regulation of endosome to plasma membrane protein transport | 1 | 5 | 7.97e-01 |
| GO:BP | GO:1903487 | regulation of lactation | 1 | 1 | 7.97e-01 |
| GO:BP | GO:1990418 | response to insulin-like growth factor stimulus | 1 | 2 | 7.97e-01 |
| GO:BP | GO:1900100 | positive regulation of plasma cell differentiation | 1 | 1 | 7.97e-01 |
| GO:BP | GO:0006858 | extracellular transport | 5 | 32 | 7.97e-01 |
| GO:BP | GO:0051952 | regulation of amine transport | 1 | 55 | 7.97e-01 |
| GO:BP | GO:0071332 | cellular response to fructose stimulus | 1 | 2 | 7.97e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 10 | 39 | 7.97e-01 |
| GO:BP | GO:0051944 | positive regulation of catecholamine uptake involved in synaptic transmission | 1 | 2 | 7.98e-01 |
| GO:BP | GO:0051586 | positive regulation of dopamine uptake involved in synaptic transmission | 1 | 2 | 7.98e-01 |
| GO:BP | GO:1905451 | positive regulation of Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 3 | 7.98e-01 |
| GO:BP | GO:0060100 | positive regulation of phagocytosis, engulfment | 3 | 11 | 7.98e-01 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 1 | 156 | 7.98e-01 |
| GO:BP | GO:1905155 | positive regulation of membrane invagination | 3 | 11 | 7.98e-01 |
| GO:BP | GO:0032240 | negative regulation of nucleobase-containing compound transport | 1 | 3 | 7.98e-01 |
| GO:BP | GO:0042832 | defense response to protozoan | 3 | 10 | 7.98e-01 |
| GO:BP | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway | 1 | 4 | 7.98e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2 | 4579 | 7.98e-01 |
| GO:BP | GO:0048866 | stem cell fate specification | 1 | 2 | 7.98e-01 |
| GO:BP | GO:0061046 | regulation of branching involved in lung morphogenesis | 1 | 3 | 7.98e-01 |
| GO:BP | GO:0043969 | histone H2B acetylation | 1 | 3 | 7.99e-01 |
| GO:BP | GO:1901314 | regulation of histone H2A K63-linked ubiquitination | 1 | 4 | 7.99e-01 |
| GO:BP | GO:1901315 | negative regulation of histone H2A K63-linked ubiquitination | 1 | 4 | 7.99e-01 |
| GO:BP | GO:0033183 | negative regulation of histone ubiquitination | 1 | 4 | 7.99e-01 |
| GO:BP | GO:0071670 | smooth muscle cell chemotaxis | 1 | 6 | 7.99e-01 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 1 | 16 | 7.99e-01 |
| GO:BP | GO:1904398 | positive regulation of neuromuscular junction development | 1 | 2 | 7.99e-01 |
| GO:BP | GO:1903843 | cellular response to arsenite ion | 1 | 4 | 7.99e-01 |
| GO:BP | GO:1903842 | response to arsenite ion | 1 | 4 | 7.99e-01 |
| GO:BP | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | 3 | 7.99e-01 |
| GO:BP | GO:0060232 | delamination | 2 | 5 | 7.99e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 2 | 13 | 7.99e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 8 | 395 | 7.99e-01 |
| GO:BP | GO:0035356 | intracellular triglyceride homeostasis | 2 | 5 | 7.99e-01 |
| GO:BP | GO:0043543 | protein acylation | 24 | 228 | 7.99e-01 |
| GO:BP | GO:0007596 | blood coagulation | 1 | 150 | 7.99e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 3 | 117 | 7.99e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 8 | 15 | 7.99e-01 |
| GO:BP | GO:0019400 | alditol metabolic process | 1 | 14 | 7.99e-01 |
| GO:BP | GO:0015938 | coenzyme A catabolic process | 1 | 3 | 7.99e-01 |
| GO:BP | GO:0014891 | striated muscle atrophy | 2 | 6 | 7.99e-01 |
| GO:BP | GO:0060011 | Sertoli cell proliferation | 1 | 4 | 7.99e-01 |
| GO:BP | GO:0019317 | fucose catabolic process | 1 | 3 | 7.99e-01 |
| GO:BP | GO:1905665 | positive regulation of calcium ion import across plasma membrane | 1 | 6 | 7.99e-01 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 2 | 24 | 7.99e-01 |
| GO:BP | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway | 1 | 3 | 7.99e-01 |
| GO:BP | GO:0042355 | L-fucose catabolic process | 1 | 3 | 7.99e-01 |
| GO:BP | GO:0044580 | butyryl-CoA catabolic process | 1 | 3 | 7.99e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 2 | 9 | 7.99e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 1 | 58 | 7.99e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 1 | 82 | 8.00e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 14 | 42 | 8.00e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 94 | 438 | 8.00e-01 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 10 | 28 | 8.00e-01 |
| GO:BP | GO:1905719 | protein localization to perinuclear region of cytoplasm | 3 | 8 | 8.00e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 4 | 191 | 8.01e-01 |
| GO:BP | GO:0046784 | viral mRNA export from host cell nucleus | 1 | 6 | 8.01e-01 |
| GO:BP | GO:1903845 | negative regulation of cellular response to transforming growth factor beta stimulus | 1 | 2 | 8.01e-01 |
| GO:BP | GO:1901569 | fatty acid derivative catabolic process | 6 | 10 | 8.01e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 9 | 37 | 8.01e-01 |
| GO:BP | GO:0042726 | flavin-containing compound metabolic process | 2 | 4 | 8.01e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 2 | 19 | 8.01e-01 |
| GO:BP | GO:0071529 | cementum mineralization | 1 | 3 | 8.01e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 1 | 14 | 8.01e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 4 | 39 | 8.01e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 9 | 148 | 8.01e-01 |
| GO:BP | GO:0030902 | hindbrain development | 3 | 114 | 8.02e-01 |
| GO:BP | GO:0002019 | regulation of renal output by angiotensin | 1 | 3 | 8.02e-01 |
| GO:BP | GO:1904959 | regulation of cytochrome-c oxidase activity | 2 | 7 | 8.02e-01 |
| GO:BP | GO:0010814 | substance P catabolic process | 1 | 3 | 8.02e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 2 | 22 | 8.02e-01 |
| GO:BP | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation | 1 | 3 | 8.02e-01 |
| GO:BP | GO:0043984 | histone H4-K16 acetylation | 5 | 14 | 8.02e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 5 | 185 | 8.02e-01 |
| GO:BP | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway | 1 | 3 | 8.02e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 8.02e-01 |
| GO:BP | GO:0030168 | platelet activation | 8 | 91 | 8.02e-01 |
| GO:BP | GO:0061156 | pulmonary artery morphogenesis | 2 | 7 | 8.02e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 5 | 324 | 8.02e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 4 | 8 | 8.02e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 8 | 70 | 8.02e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 2 | 20 | 8.02e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 9 | 71 | 8.02e-01 |
| GO:BP | GO:1904253 | positive regulation of bile acid metabolic process | 1 | 3 | 8.03e-01 |
| GO:BP | GO:0070859 | positive regulation of bile acid biosynthetic process | 1 | 3 | 8.03e-01 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 3 | 40 | 8.03e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 1 | 17 | 8.03e-01 |
| GO:BP | GO:0035063 | nuclear speck organization | 1 | 3 | 8.03e-01 |
| GO:BP | GO:0072672 | neutrophil extravasation | 1 | 7 | 8.03e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 6 | 227 | 8.03e-01 |
| GO:BP | GO:0007340 | acrosome reaction | 3 | 20 | 8.03e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 1 | 131 | 8.04e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 2 | 6 | 8.04e-01 |
| GO:BP | GO:0071772 | response to BMP | 1 | 131 | 8.04e-01 |
| GO:BP | GO:0031580 | membrane raft distribution | 1 | 3 | 8.04e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2901 | 8.04e-01 |
| GO:BP | GO:0001574 | ganglioside biosynthetic process | 1 | 14 | 8.04e-01 |
| GO:BP | GO:0046034 | ATP metabolic process | 4 | 116 | 8.04e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2 | 4955 | 8.04e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 11 | 1604 | 8.04e-01 |
| GO:BP | GO:0018158 | protein oxidation | 5 | 12 | 8.04e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 230 | 8.04e-01 |
| GO:BP | GO:0050817 | coagulation | 1 | 153 | 8.04e-01 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 3 | 11 | 8.04e-01 |
| GO:BP | GO:0090129 | positive regulation of synapse maturation | 3 | 8 | 8.04e-01 |
| GO:BP | GO:0007398 | ectoderm development | 7 | 15 | 8.04e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 2 | 83 | 8.05e-01 |
| GO:BP | GO:0060437 | lung growth | 1 | 5 | 8.05e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 6 | 36 | 8.05e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 1 | 53 | 8.05e-01 |
| GO:BP | GO:0045006 | DNA deamination | 4 | 9 | 8.05e-01 |
| GO:BP | GO:0072675 | osteoclast fusion | 1 | 5 | 8.05e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 7 | 26 | 8.05e-01 |
| GO:BP | GO:0061520 | Langerhans cell differentiation | 1 | 3 | 8.05e-01 |
| GO:BP | GO:0007217 | tachykinin receptor signaling pathway | 1 | 3 | 8.05e-01 |
| GO:BP | GO:0042758 | long-chain fatty acid catabolic process | 1 | 3 | 8.05e-01 |
| GO:BP | GO:0007599 | hemostasis | 1 | 153 | 8.05e-01 |
| GO:BP | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process | 1 | 4 | 8.05e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1 | 2890 | 8.05e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2780 | 8.05e-01 |
| GO:BP | GO:0071073 | positive regulation of phospholipid biosynthetic process | 3 | 7 | 8.05e-01 |
| GO:BP | GO:0045577 | regulation of B cell differentiation | 4 | 20 | 8.05e-01 |
| GO:BP | GO:2000625 | regulation of miRNA catabolic process | 2 | 5 | 8.05e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 8.05e-01 |
| GO:BP | GO:0007525 | somatic muscle development | 1 | 4 | 8.05e-01 |
| GO:BP | GO:2000347 | positive regulation of hepatocyte proliferation | 3 | 9 | 8.05e-01 |
| GO:BP | GO:0070327 | thyroid hormone transport | 1 | 7 | 8.06e-01 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 1 | 22 | 8.06e-01 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 2 | 19 | 8.06e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 2 | 15 | 8.06e-01 |
| GO:BP | GO:0090193 | positive regulation of glomerulus development | 1 | 3 | 8.06e-01 |
| GO:BP | GO:0010694 | positive regulation of alkaline phosphatase activity | 1 | 6 | 8.06e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 4 | 12 | 8.06e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 1 | 6 | 8.06e-01 |
| GO:BP | GO:0046305 | alkanesulfonate biosynthetic process | 1 | 2 | 8.07e-01 |
| GO:BP | GO:0042412 | taurine biosynthetic process | 1 | 2 | 8.07e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 4 | 21 | 8.07e-01 |
| GO:BP | GO:0032502 | developmental process | 2 | 4494 | 8.07e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 6 | 11 | 8.08e-01 |
| GO:BP | GO:0006353 | DNA-templated transcription termination | 1 | 22 | 8.08e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 5 | 196 | 8.08e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 1 | 22 | 8.08e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 1 | 22 | 8.08e-01 |
| GO:BP | GO:1904177 | regulation of adipose tissue development | 1 | 8 | 8.08e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 4 | 19 | 8.08e-01 |
| GO:BP | GO:0035672 | oligopeptide transmembrane transport | 2 | 5 | 8.08e-01 |
| GO:BP | GO:0006857 | oligopeptide transport | 2 | 5 | 8.08e-01 |
| GO:BP | GO:1902499 | positive regulation of protein autoubiquitination | 1 | 2 | 8.09e-01 |
| GO:BP | GO:0034694 | response to prostaglandin | 1 | 21 | 8.09e-01 |
| GO:BP | GO:0060242 | contact inhibition | 3 | 8 | 8.09e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 2 | 113 | 8.09e-01 |
| GO:BP | GO:1900016 | negative regulation of cytokine production involved in inflammatory response | 7 | 13 | 8.09e-01 |
| GO:BP | GO:0019694 | alkanesulfonate metabolic process | 2 | 6 | 8.09e-01 |
| GO:BP | GO:0019530 | taurine metabolic process | 2 | 6 | 8.09e-01 |
| GO:BP | GO:0071354 | cellular response to interleukin-6 | 5 | 23 | 8.09e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 24 | 114 | 8.09e-01 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 4 | 10 | 8.10e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 3 | 37 | 8.10e-01 |
| GO:BP | GO:0001502 | cartilage condensation | 3 | 12 | 8.10e-01 |
| GO:BP | GO:0060012 | synaptic transmission, glycinergic | 1 | 2 | 8.10e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 2 | 33 | 8.11e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 2 | 190 | 8.11e-01 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 2 | 19 | 8.11e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 8.11e-01 |
| GO:BP | GO:0060534 | trachea cartilage development | 2 | 4 | 8.11e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 9 | 150 | 8.11e-01 |
| GO:BP | GO:0000712 | resolution of meiotic recombination intermediates | 6 | 13 | 8.11e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 2 | 5 | 8.11e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 6 | 68 | 8.11e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 6 | 21 | 8.11e-01 |
| GO:BP | GO:0002082 | regulation of oxidative phosphorylation | 2 | 19 | 8.11e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 5 | 19 | 8.11e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 5 | 19 | 8.11e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 1 | 217 | 8.11e-01 |
| GO:BP | GO:0060179 | male mating behavior | 1 | 5 | 8.11e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 4 | 13 | 8.11e-01 |
| GO:BP | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 1 | 12 | 8.12e-01 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 8.12e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 5 | 96 | 8.12e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 4 | 95 | 8.12e-01 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 1 | 104 | 8.12e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 4 | 198 | 8.12e-01 |
| GO:BP | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity | 2 | 6 | 8.12e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 4 | 19 | 8.12e-01 |
| GO:BP | GO:0007614 | short-term memory | 1 | 9 | 8.12e-01 |
| GO:BP | GO:1905604 | negative regulation of blood-brain barrier permeability | 1 | 2 | 8.12e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 5 | 15 | 8.12e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 1 | 760 | 8.12e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 8 | 19 | 8.12e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 3 | 47 | 8.12e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 5 | 12 | 8.12e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 2 | 18 | 8.12e-01 |
| GO:BP | GO:0022417 | protein maturation by protein folding | 2 | 8 | 8.12e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 4 | 51 | 8.12e-01 |
| GO:BP | GO:1905144 | response to acetylcholine | 4 | 22 | 8.13e-01 |
| GO:BP | GO:0023061 | signal release | 3 | 321 | 8.13e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 70 | 120 | 8.13e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 3 | 116 | 8.13e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 1 | 75 | 8.13e-01 |
| GO:BP | GO:0071168 | protein localization to chromatin | 2 | 48 | 8.14e-01 |
| GO:BP | GO:0043654 | recognition of apoptotic cell | 1 | 4 | 8.14e-01 |
| GO:BP | GO:0002709 | regulation of T cell mediated immunity | 1 | 39 | 8.14e-01 |
| GO:BP | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning | 2 | 14 | 8.14e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 77 | 135 | 8.14e-01 |
| GO:BP | GO:0150007 | clathrin-dependent synaptic vesicle endocytosis | 1 | 2 | 8.14e-01 |
| GO:BP | GO:0035988 | chondrocyte proliferation | 2 | 18 | 8.14e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 8.15e-01 |
| GO:BP | GO:0042447 | hormone catabolic process | 1 | 7 | 8.15e-01 |
| GO:BP | GO:1903215 | negative regulation of protein targeting to mitochondrion | 1 | 3 | 8.15e-01 |
| GO:BP | GO:0060235 | lens induction in camera-type eye | 1 | 3 | 8.15e-01 |
| GO:BP | GO:1902564 | negative regulation of neutrophil activation | 1 | 2 | 8.15e-01 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 3 | 9 | 8.15e-01 |
| GO:BP | GO:0002282 | microglial cell activation involved in immune response | 1 | 1 | 8.15e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 2 | 126 | 8.15e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 3 | 13 | 8.15e-01 |
| GO:BP | GO:0044211 | CTP salvage | 1 | 3 | 8.15e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 1 | 8 | 8.15e-01 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 16 | 38 | 8.15e-01 |
| GO:BP | GO:0006256 | UDP catabolic process | 1 | 3 | 8.16e-01 |
| GO:BP | GO:0009195 | pyrimidine ribonucleoside diphosphate catabolic process | 1 | 3 | 8.16e-01 |
| GO:BP | GO:0009140 | pyrimidine nucleoside diphosphate catabolic process | 1 | 3 | 8.16e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 3 | 160 | 8.16e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 2 | 5 | 8.17e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 4 | 178 | 8.17e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 2 | 17 | 8.17e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 8.17e-01 |
| GO:BP | GO:1903650 | negative regulation of cytoplasmic transport | 1 | 3 | 8.18e-01 |
| GO:BP | GO:0060440 | trachea formation | 1 | 7 | 8.18e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 6 | 27 | 8.18e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 1 | 22 | 8.18e-01 |
| GO:BP | GO:0090155 | negative regulation of sphingolipid biosynthetic process | 2 | 6 | 8.18e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 2 | 7 | 8.18e-01 |
| GO:BP | GO:0015837 | amine transport | 1 | 56 | 8.18e-01 |
| GO:BP | GO:0106005 | RNA 5’-cap (guanine-N7)-methylation | 1 | 2 | 8.18e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 2 | 1995 | 8.18e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 1 | 17 | 8.18e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 2 | 31 | 8.18e-01 |
| GO:BP | GO:0061355 | Wnt protein secretion | 1 | 5 | 8.18e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 1 | 38 | 8.18e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 2 | 11 | 8.18e-01 |
| GO:BP | GO:1905162 | regulation of phagosome maturation | 1 | 2 | 8.18e-01 |
| GO:BP | GO:0010986 | positive regulation of lipoprotein particle clearance | 1 | 4 | 8.19e-01 |
| GO:BP | GO:0070841 | inclusion body assembly | 4 | 22 | 8.19e-01 |
| GO:BP | GO:0032835 | glomerulus development | 5 | 54 | 8.19e-01 |
| GO:BP | GO:0071688 | striated muscle myosin thick filament assembly | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0030241 | skeletal muscle myosin thick filament assembly | 1 | 3 | 8.19e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 2 | 6 | 8.19e-01 |
| GO:BP | GO:0048769 | sarcomerogenesis | 1 | 3 | 8.19e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 3 | 21 | 8.20e-01 |
| GO:BP | GO:0060545 | positive regulation of necroptotic process | 2 | 4 | 8.21e-01 |
| GO:BP | GO:0009584 | detection of visible light | 1 | 20 | 8.21e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 4 | 10 | 8.21e-01 |
| GO:BP | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing | 1 | 5 | 8.21e-01 |
| GO:BP | GO:0010668 | ectodermal cell differentiation | 2 | 5 | 8.21e-01 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 4 | 11 | 8.21e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 5 | 90 | 8.22e-01 |
| GO:BP | GO:0016573 | histone acetylation | 45 | 143 | 8.22e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 8 | 26 | 8.22e-01 |
| GO:BP | GO:0046940 | nucleoside monophosphate phosphorylation | 4 | 9 | 8.22e-01 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 13 | 37 | 8.23e-01 |
| GO:BP | GO:0070602 | regulation of centromeric sister chromatid cohesion | 1 | 4 | 8.23e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 2 | 127 | 8.23e-01 |
| GO:BP | GO:0019477 | L-lysine catabolic process | 1 | 4 | 8.23e-01 |
| GO:BP | GO:0019474 | L-lysine catabolic process to acetyl-CoA | 1 | 4 | 8.23e-01 |
| GO:BP | GO:0046440 | L-lysine metabolic process | 1 | 4 | 8.23e-01 |
| GO:BP | GO:2000407 | regulation of T cell extravasation | 1 | 3 | 8.23e-01 |
| GO:BP | GO:0042976 | activation of Janus kinase activity | 3 | 7 | 8.23e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 4 | 23 | 8.23e-01 |
| GO:BP | GO:0043615 | astrocyte cell migration | 1 | 6 | 8.24e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 10 | 28 | 8.24e-01 |
| GO:BP | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure | 1 | 14 | 8.25e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 4 | 65 | 8.25e-01 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 3 | 14 | 8.25e-01 |
| GO:BP | GO:0043983 | histone H4-K12 acetylation | 2 | 7 | 8.25e-01 |
| GO:BP | GO:0007127 | meiosis I | 48 | 77 | 8.26e-01 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 2 | 14 | 8.26e-01 |
| GO:BP | GO:0048295 | positive regulation of isotype switching to IgE isotypes | 1 | 3 | 8.26e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 5 | 32 | 8.26e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 1 | 102 | 8.26e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 2 | 123 | 8.26e-01 |
| GO:BP | GO:0051795 | positive regulation of timing of catagen | 1 | 3 | 8.26e-01 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 8.26e-01 |
| GO:BP | GO:2000615 | regulation of histone H3-K9 acetylation | 3 | 7 | 8.27e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 3 | 59 | 8.27e-01 |
| GO:BP | GO:0046865 | terpenoid transport | 1 | 3 | 8.27e-01 |
| GO:BP | GO:0046864 | isoprenoid transport | 1 | 3 | 8.27e-01 |
| GO:BP | GO:0042439 | ethanolamine-containing compound metabolic process | 1 | 5 | 8.27e-01 |
| GO:BP | GO:0007019 | microtubule depolymerization | 16 | 43 | 8.27e-01 |
| GO:BP | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 3 | 8.27e-01 |
| GO:BP | GO:0032497 | detection of lipopolysaccharide | 1 | 2 | 8.27e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 4 | 28 | 8.27e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 9 | 66 | 8.27e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 1 | 8 | 8.27e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 2 | 3 | 8.27e-01 |
| GO:BP | GO:0060700 | regulation of ribonuclease activity | 1 | 5 | 8.28e-01 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 2 | 14 | 8.28e-01 |
| GO:BP | GO:0002729 | positive regulation of natural killer cell cytokine production | 1 | 4 | 8.28e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 8.28e-01 |
| GO:BP | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity | 2 | 10 | 8.28e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 5 | 49 | 8.28e-01 |
| GO:BP | GO:1904054 | regulation of cholangiocyte proliferation | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0051709 | regulation of killing of cells of another organism | 1 | 3 | 8.29e-01 |
| GO:BP | GO:2000338 | regulation of chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 8.29e-01 |
| GO:BP | GO:0030154 | cell differentiation | 1 | 2958 | 8.29e-01 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 2 | 6 | 8.29e-01 |
| GO:BP | GO:0072136 | metanephric mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0044060 | regulation of endocrine process | 1 | 24 | 8.29e-01 |
| GO:BP | GO:0072135 | kidney mesenchymal cell proliferation | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 3 | 27 | 8.29e-01 |
| GO:BP | GO:0071608 | macrophage inflammatory protein-1 alpha production | 1 | 3 | 8.29e-01 |
| GO:BP | GO:0001562 | response to protozoan | 2 | 11 | 8.29e-01 |
| GO:BP | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production | 1 | 3 | 8.29e-01 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 2 | 6 | 8.29e-01 |
| GO:BP | GO:0045329 | carnitine biosynthetic process | 1 | 3 | 8.29e-01 |
| GO:BP | GO:0072566 | chemokine (C-X-C motif) ligand 1 production | 1 | 4 | 8.29e-01 |
| GO:BP | GO:0035461 | vitamin transmembrane transport | 1 | 14 | 8.29e-01 |
| GO:BP | GO:0006011 | UDP-glucose metabolic process | 1 | 6 | 8.29e-01 |
| GO:BP | GO:0010501 | RNA secondary structure unwinding | 2 | 8 | 8.29e-01 |
| GO:BP | GO:0032899 | regulation of neurotrophin production | 1 | 2 | 8.29e-01 |
| GO:BP | GO:0071800 | podosome assembly | 3 | 13 | 8.29e-01 |
| GO:BP | GO:0007600 | sensory perception | 3 | 269 | 8.29e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 7 | 106 | 8.29e-01 |
| GO:BP | GO:0097168 | mesenchymal stem cell proliferation | 1 | 4 | 8.29e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 1 | 17 | 8.30e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 1 | 16 | 8.30e-01 |
| GO:BP | GO:2000348 | regulation of CD40 signaling pathway | 2 | 4 | 8.30e-01 |
| GO:BP | GO:0002710 | negative regulation of T cell mediated immunity | 2 | 8 | 8.30e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 26 | 86 | 8.30e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 10 | 26 | 8.30e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 1 | 2984 | 8.30e-01 |
| GO:BP | GO:0001525 | angiogenesis | 2 | 390 | 8.30e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 6 | 1201 | 8.30e-01 |
| GO:BP | GO:0033078 | extrathymic T cell differentiation | 1 | 1 | 8.30e-01 |
| GO:BP | GO:0003306 | Wnt signaling pathway involved in heart development | 1 | 8 | 8.30e-01 |
| GO:BP | GO:0006924 | activation-induced cell death of T cells | 3 | 8 | 8.30e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 10 | 135 | 8.31e-01 |
| GO:BP | GO:0019695 | choline metabolic process | 1 | 4 | 8.31e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 8.31e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 2 | 6 | 8.31e-01 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 2 | 8.31e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 2 | 6 | 8.31e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 3 | 126 | 8.31e-01 |
| GO:BP | GO:0033622 | integrin activation | 3 | 18 | 8.31e-01 |
| GO:BP | GO:0042631 | cellular response to water deprivation | 1 | 1 | 8.32e-01 |
| GO:BP | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway | 2 | 4 | 8.32e-01 |
| GO:BP | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity | 1 | 3 | 8.32e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 1 | 25 | 8.32e-01 |
| GO:BP | GO:0090026 | positive regulation of monocyte chemotaxis | 1 | 7 | 8.32e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 17 | 1123 | 8.32e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 5 | 34 | 8.32e-01 |
| GO:BP | GO:0044872 | lipoprotein localization | 2 | 15 | 8.33e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 2 | 14 | 8.33e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 2 | 4 | 8.33e-01 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 19 | 8.33e-01 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 1 | 19 | 8.33e-01 |
| GO:BP | GO:0042701 | progesterone secretion | 1 | 3 | 8.33e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 8.33e-01 |
| GO:BP | GO:0150003 | regulation of spontaneous synaptic transmission | 1 | 6 | 8.33e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 2 | 5 | 8.33e-01 |
| GO:BP | GO:1902803 | regulation of synaptic vesicle transport | 1 | 5 | 8.33e-01 |
| GO:BP | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response | 1 | 2 | 8.33e-01 |
| GO:BP | GO:0002540 | leukotriene production involved in inflammatory response | 1 | 2 | 8.33e-01 |
| GO:BP | GO:0055001 | muscle cell development | 38 | 165 | 8.34e-01 |
| GO:BP | GO:0031623 | receptor internalization | 13 | 95 | 8.34e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 6 | 32 | 8.34e-01 |
| GO:BP | GO:0019249 | lactate biosynthetic process | 1 | 3 | 8.34e-01 |
| GO:BP | GO:0044406 | adhesion of symbiont to host | 1 | 10 | 8.34e-01 |
| GO:BP | GO:0031342 | negative regulation of cell killing | 3 | 11 | 8.34e-01 |
| GO:BP | GO:0006909 | phagocytosis | 1 | 148 | 8.34e-01 |
| GO:BP | GO:0044417 | translocation of molecules into host | 1 | 7 | 8.34e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 2 | 108 | 8.35e-01 |
| GO:BP | GO:0021523 | somatic motor neuron differentiation | 1 | 2 | 8.35e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 8 | 535 | 8.35e-01 |
| GO:BP | GO:0006651 | diacylglycerol biosynthetic process | 3 | 7 | 8.35e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 9 | 37 | 8.35e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 8.35e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 7 | 17 | 8.35e-01 |
| GO:BP | GO:0006751 | glutathione catabolic process | 1 | 5 | 8.35e-01 |
| GO:BP | GO:0042407 | cristae formation | 6 | 18 | 8.35e-01 |
| GO:BP | GO:0060166 | olfactory pit development | 1 | 1 | 8.36e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 3 | 9 | 8.36e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 1 | 223 | 8.36e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 7 | 48 | 8.36e-01 |
| GO:BP | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation | 1 | 2 | 8.37e-01 |
| GO:BP | GO:0036514 | dopaminergic neuron axon guidance | 1 | 3 | 8.37e-01 |
| GO:BP | GO:1904938 | planar cell polarity pathway involved in axon guidance | 1 | 3 | 8.37e-01 |
| GO:BP | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis | 1 | 7 | 8.37e-01 |
| GO:BP | GO:0060385 | axonogenesis involved in innervation | 3 | 7 | 8.37e-01 |
| GO:BP | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | 1 | 4 | 8.37e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 4 | 12 | 8.37e-01 |
| GO:BP | GO:2001245 | regulation of phosphatidylcholine biosynthetic process | 1 | 5 | 8.37e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 4 | 12 | 8.37e-01 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 6 | 15 | 8.37e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 1 | 25 | 8.38e-01 |
| GO:BP | GO:0051665 | membrane raft localization | 1 | 4 | 8.38e-01 |
| GO:BP | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration | 1 | 4 | 8.38e-01 |
| GO:BP | GO:0070472 | regulation of uterine smooth muscle contraction | 1 | 5 | 8.38e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 2 | 44 | 8.38e-01 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 4 | 24 | 8.38e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 9 | 67 | 8.39e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 13 | 64 | 8.39e-01 |
| GO:BP | GO:0002921 | negative regulation of humoral immune response | 3 | 10 | 8.39e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 18 | 134 | 8.39e-01 |
| GO:BP | GO:0006021 | inositol biosynthetic process | 1 | 3 | 8.39e-01 |
| GO:BP | GO:0035965 | cardiolipin acyl-chain remodeling | 2 | 4 | 8.39e-01 |
| GO:BP | GO:0035425 | autocrine signaling | 1 | 3 | 8.39e-01 |
| GO:BP | GO:0071316 | cellular response to nicotine | 2 | 4 | 8.39e-01 |
| GO:BP | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential | 1 | 4 | 8.39e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 3 | 23 | 8.39e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 2 | 34 | 8.39e-01 |
| GO:BP | GO:0045343 | regulation of MHC class I biosynthetic process | 2 | 3 | 8.39e-01 |
| GO:BP | GO:0070142 | synaptic vesicle budding | 1 | 8 | 8.39e-01 |
| GO:BP | GO:0045341 | MHC class I biosynthetic process | 2 | 3 | 8.39e-01 |
| GO:BP | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | 1 | 8 | 8.39e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 1 | 265 | 8.41e-01 |
| GO:BP | GO:1903595 | positive regulation of histamine secretion by mast cell | 1 | 4 | 8.41e-01 |
| GO:BP | GO:0031000 | response to caffeine | 5 | 13 | 8.41e-01 |
| GO:BP | GO:1903593 | regulation of histamine secretion by mast cell | 1 | 4 | 8.41e-01 |
| GO:BP | GO:0071934 | thiamine transmembrane transport | 2 | 3 | 8.41e-01 |
| GO:BP | GO:0042723 | thiamine-containing compound metabolic process | 2 | 4 | 8.41e-01 |
| GO:BP | GO:0030222 | eosinophil differentiation | 1 | 2 | 8.41e-01 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 5 | 29 | 8.41e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1 | 2646 | 8.41e-01 |
| GO:BP | GO:0021559 | trigeminal nerve development | 1 | 5 | 8.41e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 6 | 15 | 8.42e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 2 | 5 | 8.42e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 1 | 13 | 8.42e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 4 | 17 | 8.42e-01 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 2 | 36 | 8.42e-01 |
| GO:BP | GO:0002790 | peptide secretion | 1 | 170 | 8.42e-01 |
| GO:BP | GO:0030534 | adult behavior | 2 | 92 | 8.42e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 1 | 194 | 8.42e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 11 | 787 | 8.42e-01 |
| GO:BP | GO:0042693 | muscle cell fate commitment | 4 | 11 | 8.42e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 26 | 1974 | 8.42e-01 |
| GO:BP | GO:0044241 | lipid digestion | 1 | 10 | 8.42e-01 |
| GO:BP | GO:1905908 | positive regulation of amyloid fibril formation | 1 | 5 | 8.42e-01 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 1 | 27 | 8.42e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 13 | 157 | 8.42e-01 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 2 | 6 | 8.42e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 8.43e-01 |
| GO:BP | GO:0019511 | peptidyl-proline hydroxylation | 3 | 14 | 8.43e-01 |
| GO:BP | GO:0031017 | exocrine pancreas development | 1 | 4 | 8.43e-01 |
| GO:BP | GO:0060268 | negative regulation of respiratory burst | 2 | 4 | 8.43e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 3 | 106 | 8.43e-01 |
| GO:BP | GO:0001923 | B-1 B cell differentiation | 1 | 3 | 8.43e-01 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 1 | 27 | 8.43e-01 |
| GO:BP | GO:0009798 | axis specification | 8 | 63 | 8.43e-01 |
| GO:BP | GO:0061061 | muscle structure development | 40 | 546 | 8.44e-01 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 3 | 10 | 8.44e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 2 | 32 | 8.44e-01 |
| GO:BP | GO:0014824 | artery smooth muscle contraction | 3 | 7 | 8.44e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 2 | 3 | 8.44e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 2 | 3 | 8.44e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 2 | 6 | 8.44e-01 |
| GO:BP | GO:0060452 | positive regulation of cardiac muscle contraction | 1 | 7 | 8.44e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 20 | 8.44e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 8.44e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 7 | 998 | 8.44e-01 |
| GO:BP | GO:0030210 | heparin biosynthetic process | 4 | 9 | 8.44e-01 |
| GO:BP | GO:0042305 | specification of segmental identity, mandibular segment | 1 | 3 | 8.44e-01 |
| GO:BP | GO:0086101 | endothelin receptor signaling pathway involved in heart process | 1 | 3 | 8.44e-01 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 2 | 8 | 8.45e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 1 | 6 | 8.45e-01 |
| GO:BP | GO:0008291 | acetylcholine metabolic process | 2 | 3 | 8.45e-01 |
| GO:BP | GO:1900619 | acetate ester metabolic process | 2 | 3 | 8.45e-01 |
| GO:BP | GO:2000501 | regulation of natural killer cell chemotaxis | 1 | 2 | 8.45e-01 |
| GO:BP | GO:0070106 | interleukin-27-mediated signaling pathway | 2 | 4 | 8.45e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 1 | 36 | 8.45e-01 |
| GO:BP | GO:0032623 | interleukin-2 production | 3 | 31 | 8.45e-01 |
| GO:BP | GO:2001045 | negative regulation of integrin-mediated signaling pathway | 1 | 3 | 8.45e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 1 | 191 | 8.45e-01 |
| GO:BP | GO:0032687 | negative regulation of interferon-alpha production | 1 | 2 | 8.45e-01 |
| GO:BP | GO:0060290 | transdifferentiation | 1 | 2 | 8.45e-01 |
| GO:BP | GO:0032663 | regulation of interleukin-2 production | 3 | 31 | 8.45e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 3 | 16 | 8.45e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 20 | 2293 | 8.45e-01 |
| GO:BP | GO:0032601 | connective tissue growth factor production | 1 | 2 | 8.45e-01 |
| GO:BP | GO:0032643 | regulation of connective tissue growth factor production | 1 | 2 | 8.45e-01 |
| GO:BP | GO:0090160 | Golgi to lysosome transport | 3 | 10 | 8.45e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 8.45e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 1 | 14 | 8.46e-01 |
| GO:BP | GO:0097749 | membrane tubulation | 1 | 4 | 8.46e-01 |
| GO:BP | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 2 | 5 | 8.46e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 4 | 21 | 8.46e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 8.46e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 5 | 13 | 8.46e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 8.46e-01 |
| GO:BP | GO:1902916 | positive regulation of protein polyubiquitination | 1 | 13 | 8.46e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 8.46e-01 |
| GO:BP | GO:0010989 | negative regulation of low-density lipoprotein particle clearance | 1 | 4 | 8.46e-01 |
| GO:BP | GO:0036211 | protein modification process | 1 | 2901 | 8.46e-01 |
| GO:BP | GO:0034757 | negative regulation of iron ion transport | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0034760 | negative regulation of iron ion transmembrane transport | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0061591 | calcium activated galactosylceramide scrambling | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 6 | 39 | 8.46e-01 |
| GO:BP | GO:0061589 | calcium activated phosphatidylserine scrambling | 1 | 3 | 8.46e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 1 | 3 | 8.46e-01 |
| GO:BP | GO:0032487 | regulation of Rap protein signal transduction | 1 | 3 | 8.46e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 2 | 669 | 8.46e-01 |
| GO:BP | GO:1905383 | protein localization to presynapse | 3 | 11 | 8.46e-01 |
| GO:BP | GO:0032238 | adenosine transport | 1 | 2 | 8.46e-01 |
| GO:BP | GO:0032094 | response to food | 1 | 22 | 8.47e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 1 | 10 | 8.47e-01 |
| GO:BP | GO:0021766 | hippocampus development | 11 | 62 | 8.47e-01 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 3 | 48 | 8.47e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 4 | 67 | 8.47e-01 |
| GO:BP | GO:0060921 | sinoatrial node cell differentiation | 2 | 5 | 8.47e-01 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 1 | 10 | 8.47e-01 |
| GO:BP | GO:1904533 | regulation of telomeric loop disassembly | 1 | 2 | 8.47e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 2 | 88 | 8.47e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 4 | 8061 | 8.47e-01 |
| GO:BP | GO:0002716 | negative regulation of natural killer cell mediated immunity | 2 | 10 | 8.47e-01 |
| GO:BP | GO:0090289 | regulation of osteoclast proliferation | 1 | 3 | 8.47e-01 |
| GO:BP | GO:0034776 | response to histamine | 3 | 5 | 8.47e-01 |
| GO:BP | GO:0010984 | regulation of lipoprotein particle clearance | 3 | 12 | 8.47e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 1 | 40 | 8.48e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 2 | 39 | 8.48e-01 |
| GO:BP | GO:0048599 | oocyte development | 3 | 38 | 8.48e-01 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 1 | 2 | 8.48e-01 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 8.48e-01 |
| GO:BP | GO:0071896 | protein localization to adherens junction | 2 | 6 | 8.48e-01 |
| GO:BP | GO:0033006 | regulation of mast cell activation involved in immune response | 9 | 21 | 8.48e-01 |
| GO:BP | GO:1905809 | negative regulation of synapse organization | 2 | 8 | 8.48e-01 |
| GO:BP | GO:0051167 | xylulose 5-phosphate metabolic process | 1 | 4 | 8.48e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 1 | 28 | 8.48e-01 |
| GO:BP | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 2 | 10 | 8.48e-01 |
| GO:BP | GO:0019640 | glucuronate catabolic process to xylulose 5-phosphate | 1 | 4 | 8.48e-01 |
| GO:BP | GO:0090086 | negative regulation of protein deubiquitination | 1 | 4 | 8.48e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 1 | 72 | 8.48e-01 |
| GO:BP | GO:0006064 | glucuronate catabolic process | 1 | 4 | 8.48e-01 |
| GO:BP | GO:1901159 | xylulose 5-phosphate biosynthetic process | 1 | 4 | 8.48e-01 |
| GO:BP | GO:1904340 | positive regulation of dopaminergic neuron differentiation | 1 | 3 | 8.48e-01 |
| GO:BP | GO:1905167 | positive regulation of lysosomal protein catabolic process | 1 | 4 | 8.49e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 1 | 4 | 8.49e-01 |
| GO:BP | GO:1903978 | regulation of microglial cell activation | 3 | 9 | 8.49e-01 |
| GO:BP | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 22 | 45 | 8.49e-01 |
| GO:BP | GO:1903712 | cysteine transmembrane transport | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0042883 | cysteine transport | 1 | 3 | 8.49e-01 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 1 | 4 | 8.49e-01 |
| GO:BP | GO:0031581 | hemidesmosome assembly | 2 | 6 | 8.49e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 7 | 26 | 8.49e-01 |
| GO:BP | GO:0050859 | negative regulation of B cell receptor signaling pathway | 2 | 3 | 8.49e-01 |
| GO:BP | GO:0008544 | epidermis development | 6 | 210 | 8.49e-01 |
| GO:BP | GO:0110088 | hippocampal neuron apoptotic process | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0110089 | regulation of hippocampal neuron apoptotic process | 1 | 2 | 8.49e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 2 | 398 | 8.49e-01 |
| GO:BP | GO:1900038 | negative regulation of cellular response to hypoxia | 2 | 8 | 8.50e-01 |
| GO:BP | GO:0043353 | enucleate erythrocyte differentiation | 2 | 9 | 8.50e-01 |
| GO:BP | GO:0046055 | dGMP catabolic process | 1 | 1 | 8.50e-01 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 1 | 24 | 8.50e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 4 | 14 | 8.50e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 1 | 478 | 8.50e-01 |
| GO:BP | GO:0033574 | response to testosterone | 1 | 29 | 8.50e-01 |
| GO:BP | GO:1905786 | positive regulation of anaphase-promoting complex-dependent catabolic process | 1 | 2 | 8.50e-01 |
| GO:BP | GO:0090192 | regulation of glomerulus development | 1 | 4 | 8.50e-01 |
| GO:BP | GO:0072215 | regulation of metanephros development | 1 | 2 | 8.50e-01 |
| GO:BP | GO:0046452 | dihydrofolate metabolic process | 1 | 3 | 8.50e-01 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 8.50e-01 |
| GO:BP | GO:0008213 | protein alkylation | 12 | 160 | 8.50e-01 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 3 | 6 | 8.50e-01 |
| GO:BP | GO:0006479 | protein methylation | 12 | 160 | 8.50e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 2 | 57 | 8.51e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 7 | 20 | 8.51e-01 |
| GO:BP | GO:0048752 | semicircular canal morphogenesis | 1 | 5 | 8.51e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 29 | 8.51e-01 |
| GO:BP | GO:1905915 | regulation of cell differentiation involved in phenotypic switching | 1 | 3 | 8.51e-01 |
| GO:BP | GO:1905930 | regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 8.51e-01 |
| GO:BP | GO:1904732 | regulation of electron transfer activity | 2 | 8 | 8.51e-01 |
| GO:BP | GO:1905460 | negative regulation of vascular associated smooth muscle cell apoptotic process | 1 | 2 | 8.51e-01 |
| GO:BP | GO:1905420 | vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0090679 | cell differentiation involved in phenotypic switching | 1 | 3 | 8.51e-01 |
| GO:BP | GO:0035617 | stress granule disassembly | 1 | 5 | 8.52e-01 |
| GO:BP | GO:1904396 | regulation of neuromuscular junction development | 2 | 5 | 8.52e-01 |
| GO:BP | GO:0051124 | synaptic assembly at neuromuscular junction | 2 | 6 | 8.52e-01 |
| GO:BP | GO:0002674 | negative regulation of acute inflammatory response | 1 | 5 | 8.52e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 2 | 27 | 8.52e-01 |
| GO:BP | GO:0061314 | Notch signaling involved in heart development | 3 | 9 | 8.52e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 3 | 22 | 8.52e-01 |
| GO:BP | GO:1902268 | negative regulation of polyamine transmembrane transport | 1 | 3 | 8.53e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 2 | 90 | 8.53e-01 |
| GO:BP | GO:1900045 | negative regulation of protein K63-linked ubiquitination | 2 | 7 | 8.54e-01 |
| GO:BP | GO:0010626 | negative regulation of Schwann cell proliferation | 2 | 6 | 8.54e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 227 | 8.54e-01 |
| GO:BP | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway | 2 | 5 | 8.54e-01 |
| GO:BP | GO:0002370 | natural killer cell cytokine production | 1 | 4 | 8.54e-01 |
| GO:BP | GO:0002727 | regulation of natural killer cell cytokine production | 1 | 4 | 8.54e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 8.54e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 1 | 23 | 8.54e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 8.54e-01 |
| GO:BP | GO:0032836 | glomerular basement membrane development | 1 | 9 | 8.54e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 1 | 116 | 8.54e-01 |
| GO:BP | GO:0002827 | positive regulation of T-helper 1 type immune response | 3 | 5 | 8.54e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 1 | 18 | 8.54e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 1 | 9 | 8.54e-01 |
| GO:BP | GO:0006104 | succinyl-CoA metabolic process | 1 | 9 | 8.54e-01 |
| GO:BP | GO:1905912 | regulation of calcium ion export across plasma membrane | 1 | 4 | 8.54e-01 |
| GO:BP | GO:0006867 | asparagine transport | 1 | 3 | 8.54e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 8.54e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 8.54e-01 |
| GO:BP | GO:1902024 | L-histidine transport | 1 | 4 | 8.54e-01 |
| GO:BP | GO:1905913 | negative regulation of calcium ion export across plasma membrane | 1 | 4 | 8.54e-01 |
| GO:BP | GO:0015817 | histidine transport | 1 | 4 | 8.54e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 3 | 75 | 8.54e-01 |
| GO:BP | GO:0007030 | Golgi organization | 2 | 137 | 8.54e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 1 | 35 | 8.54e-01 |
| GO:BP | GO:1905584 | outer hair cell apoptotic process | 1 | 2 | 8.54e-01 |
| GO:BP | GO:0007369 | gastrulation | 6 | 146 | 8.54e-01 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 2 | 11 | 8.54e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 12 | 40 | 8.54e-01 |
| GO:BP | GO:0015833 | peptide transport | 1 | 179 | 8.55e-01 |
| GO:BP | GO:0032655 | regulation of interleukin-12 production | 1 | 29 | 8.55e-01 |
| GO:BP | GO:1903980 | positive regulation of microglial cell activation | 2 | 2 | 8.55e-01 |
| GO:BP | GO:0032615 | interleukin-12 production | 1 | 29 | 8.55e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 2 | 35 | 8.55e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 2 | 35 | 8.55e-01 |
| GO:BP | GO:0019086 | late viral transcription | 1 | 4 | 8.55e-01 |
| GO:BP | GO:0003334 | keratinocyte development | 1 | 8 | 8.55e-01 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 1 | 31 | 8.55e-01 |
| GO:BP | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis | 1 | 7 | 8.55e-01 |
| GO:BP | GO:0009635 | response to herbicide | 1 | 3 | 8.55e-01 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 8 | 22 | 8.55e-01 |
| GO:BP | GO:0090713 | immunological memory process | 2 | 7 | 8.55e-01 |
| GO:BP | GO:0000478 | endonucleolytic cleavage involved in rRNA processing | 2 | 17 | 8.55e-01 |
| GO:BP | GO:0019371 | cyclooxygenase pathway | 1 | 7 | 8.55e-01 |
| GO:BP | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 2 | 17 | 8.55e-01 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 4 | 84 | 8.55e-01 |
| GO:BP | GO:0010041 | response to iron(III) ion | 1 | 4 | 8.56e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 3 | 25 | 8.56e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 18 | 1836 | 8.56e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 3 | 14 | 8.57e-01 |
| GO:BP | GO:0045989 | positive regulation of striated muscle contraction | 2 | 12 | 8.57e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 3 | 14 | 8.57e-01 |
| GO:BP | GO:0070741 | response to interleukin-6 | 5 | 23 | 8.57e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 3 | 9 | 8.57e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 3 | 9 | 8.57e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 3 | 24 | 8.57e-01 |
| GO:BP | GO:0060099 | regulation of phagocytosis, engulfment | 3 | 11 | 8.57e-01 |
| GO:BP | GO:1905153 | regulation of membrane invagination | 3 | 11 | 8.57e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 8.57e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 8.58e-01 |
| GO:BP | GO:0009224 | CMP biosynthetic process | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0071799 | cellular response to prostaglandin D stimulus | 1 | 2 | 8.58e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 4 | 18 | 8.58e-01 |
| GO:BP | GO:0071798 | response to prostaglandin D | 1 | 2 | 8.58e-01 |
| GO:BP | GO:0003170 | heart valve development | 23 | 65 | 8.58e-01 |
| GO:BP | GO:0007271 | synaptic transmission, cholinergic | 9 | 15 | 8.58e-01 |
| GO:BP | GO:0006579 | amino-acid betaine catabolic process | 1 | 1 | 8.58e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 11 | 45 | 8.58e-01 |
| GO:BP | GO:0070471 | uterine smooth muscle contraction | 1 | 6 | 8.58e-01 |
| GO:BP | GO:0003407 | neural retina development | 5 | 48 | 8.58e-01 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 2 | 8 | 8.58e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 8.58e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 8.58e-01 |
| GO:BP | GO:0019563 | glycerol catabolic process | 1 | 5 | 8.58e-01 |
| GO:BP | GO:0060245 | detection of cell density | 3 | 9 | 8.58e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 8.58e-01 |
| GO:BP | GO:0033210 | leptin-mediated signaling pathway | 1 | 10 | 8.58e-01 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 2 | 14 | 8.58e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 17 | 33 | 8.58e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 17 | 33 | 8.58e-01 |
| GO:BP | GO:0006107 | oxaloacetate metabolic process | 2 | 7 | 8.59e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 4 | 44 | 8.59e-01 |
| GO:BP | GO:0048874 | host-mediated regulation of intestinal microbiota composition | 2 | 2 | 8.59e-01 |
| GO:BP | GO:0032849 | positive regulation of cellular pH reduction | 1 | 2 | 8.59e-01 |
| GO:BP | GO:0002693 | positive regulation of cellular extravasation | 1 | 13 | 8.59e-01 |
| GO:BP | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure | 1 | 4 | 8.59e-01 |
| GO:BP | GO:0002001 | renin secretion into blood stream | 1 | 4 | 8.59e-01 |
| GO:BP | GO:0015669 | gas transport | 1 | 6 | 8.59e-01 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 1 | 5 | 8.59e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 5 | 27 | 8.60e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 1 | 58 | 8.60e-01 |
| GO:BP | GO:0033092 | positive regulation of immature T cell proliferation in thymus | 1 | 2 | 8.60e-01 |
| GO:BP | GO:1903792 | negative regulation of monoatomic anion transport | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 2 | 7 | 8.60e-01 |
| GO:BP | GO:0019401 | alditol biosynthetic process | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0140650 | radial glia-guided pyramidal neuron migration | 1 | 4 | 8.60e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 14 | 40 | 8.60e-01 |
| GO:BP | GO:0006114 | glycerol biosynthetic process | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0033091 | positive regulation of immature T cell proliferation | 1 | 2 | 8.60e-01 |
| GO:BP | GO:0045627 | positive regulation of T-helper 1 cell differentiation | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 17 | 462 | 8.60e-01 |
| GO:BP | GO:0000023 | maltose metabolic process | 1 | 2 | 8.60e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 1 | 3090 | 8.60e-01 |
| GO:BP | GO:0097305 | response to alcohol | 7 | 159 | 8.60e-01 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 20 | 36 | 8.60e-01 |
| GO:BP | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination | 1 | 4 | 8.60e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 4 | 12 | 8.60e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 4 | 12 | 8.60e-01 |
| GO:BP | GO:0015847 | putrescine transport | 1 | 2 | 8.60e-01 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 20 | 36 | 8.60e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 8.60e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1 | 2880 | 8.60e-01 |
| GO:BP | GO:0002577 | regulation of antigen processing and presentation | 2 | 8 | 8.60e-01 |
| GO:BP | GO:0106091 | glial cell projection elongation | 1 | 2 | 8.60e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 2 | 51 | 8.60e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 4 | 632 | 8.60e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 6 | 15 | 8.60e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 6 | 1149 | 8.60e-01 |
| GO:BP | GO:0042748 | circadian sleep/wake cycle, non-REM sleep | 1 | 3 | 8.60e-01 |
| GO:BP | GO:0030259 | lipid glycosylation | 1 | 6 | 8.60e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 4 | 70 | 8.60e-01 |
| GO:BP | GO:0006306 | DNA methylation | 4 | 46 | 8.61e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 4 | 46 | 8.61e-01 |
| GO:BP | GO:0046958 | nonassociative learning | 1 | 6 | 8.61e-01 |
| GO:BP | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production | 1 | 3 | 8.61e-01 |
| GO:BP | GO:0043418 | homocysteine catabolic process | 1 | 3 | 8.62e-01 |
| GO:BP | GO:0019344 | cysteine biosynthetic process | 1 | 3 | 8.62e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 6 | 30 | 8.62e-01 |
| GO:BP | GO:0015874 | norepinephrine transport | 2 | 9 | 8.62e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 4 | 10 | 8.62e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 8.62e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 8.62e-01 |
| GO:BP | GO:0007492 | endoderm development | 20 | 65 | 8.62e-01 |
| GO:BP | GO:1905048 | regulation of metallopeptidase activity | 4 | 14 | 8.63e-01 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 1 | 7 | 8.63e-01 |
| GO:BP | GO:0030574 | collagen catabolic process | 7 | 26 | 8.63e-01 |
| GO:BP | GO:0001708 | cell fate specification | 16 | 56 | 8.63e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 9 | 19 | 8.63e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 4 | 122 | 8.63e-01 |
| GO:BP | GO:0033212 | iron import into cell | 3 | 7 | 8.63e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 6 | 14 | 8.63e-01 |
| GO:BP | GO:0006583 | melanin biosynthetic process from tyrosine | 1 | 1 | 8.63e-01 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 1 | 8 | 8.64e-01 |
| GO:BP | GO:0060331 | negative regulation of response to type II interferon | 1 | 6 | 8.65e-01 |
| GO:BP | GO:1901387 | positive regulation of voltage-gated calcium channel activity | 3 | 13 | 8.65e-01 |
| GO:BP | GO:0042360 | vitamin E metabolic process | 1 | 2 | 8.65e-01 |
| GO:BP | GO:0060846 | blood vessel endothelial cell fate commitment | 1 | 3 | 8.65e-01 |
| GO:BP | GO:0097101 | blood vessel endothelial cell fate specification | 1 | 3 | 8.65e-01 |
| GO:BP | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment | 1 | 1 | 8.65e-01 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 8 | 20 | 8.65e-01 |
| GO:BP | GO:0060336 | negative regulation of type II interferon-mediated signaling pathway | 1 | 6 | 8.65e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 1 | 2 | 8.65e-01 |
| GO:BP | GO:0097368 | establishment of Sertoli cell barrier | 1 | 3 | 8.65e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 13 | 65 | 8.65e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 6 | 118 | 8.65e-01 |
| GO:BP | GO:1905279 | regulation of retrograde transport, endosome to Golgi | 1 | 6 | 8.65e-01 |
| GO:BP | GO:0045117 | azole transmembrane transport | 4 | 9 | 8.65e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 8.65e-01 |
| GO:BP | GO:0071350 | cellular response to interleukin-15 | 1 | 2 | 8.65e-01 |
| GO:BP | GO:0035723 | interleukin-15-mediated signaling pathway | 1 | 2 | 8.65e-01 |
| GO:BP | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition | 1 | 3 | 8.65e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 1 | 41 | 8.65e-01 |
| GO:BP | GO:1990962 | xenobiotic transport across blood-brain barrier | 1 | 3 | 8.65e-01 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 8 | 23 | 8.65e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 8.65e-01 |
| GO:BP | GO:0070445 | regulation of oligodendrocyte progenitor proliferation | 2 | 5 | 8.65e-01 |
| GO:BP | GO:0070444 | oligodendrocyte progenitor proliferation | 2 | 5 | 8.65e-01 |
| GO:BP | GO:2001294 | malonyl-CoA catabolic process | 1 | 4 | 8.65e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 2 | 9 | 8.65e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 6 | 19 | 8.65e-01 |
| GO:BP | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity | 1 | 3 | 8.65e-01 |
| GO:BP | GO:0035092 | sperm DNA condensation | 1 | 7 | 8.65e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 3 | 9 | 8.65e-01 |
| GO:BP | GO:1902858 | propionyl-CoA metabolic process | 1 | 4 | 8.65e-01 |
| GO:BP | GO:0006369 | termination of RNA polymerase II transcription | 2 | 8 | 8.65e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 3 | 8 | 8.65e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 4 | 19 | 8.65e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 4 | 414 | 8.65e-01 |
| GO:BP | GO:0009750 | response to fructose | 2 | 3 | 8.65e-01 |
| GO:BP | GO:0006772 | thiamine metabolic process | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0048563 | post-embryonic animal organ morphogenesis | 1 | 8 | 8.66e-01 |
| GO:BP | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway | 2 | 15 | 8.66e-01 |
| GO:BP | GO:0035814 | negative regulation of renal sodium excretion | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 8.66e-01 |
| GO:BP | GO:0072156 | distal tubule morphogenesis | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 8.66e-01 |
| GO:BP | GO:0032096 | negative regulation of response to food | 3 | 7 | 8.66e-01 |
| GO:BP | GO:0032099 | negative regulation of appetite | 3 | 7 | 8.66e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0035744 | T-helper 1 cell cytokine production | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0060681 | branch elongation involved in ureteric bud branching | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0034162 | toll-like receptor 9 signaling pathway | 1 | 12 | 8.66e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 8.66e-01 |
| GO:BP | GO:2000554 | regulation of T-helper 1 cell cytokine production | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0008104 | protein localization | 5 | 2129 | 8.66e-01 |
| GO:BP | GO:0010813 | neuropeptide catabolic process | 1 | 4 | 8.66e-01 |
| GO:BP | GO:0014827 | intestine smooth muscle contraction | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0034756 | regulation of iron ion transport | 2 | 4 | 8.66e-01 |
| GO:BP | GO:0055095 | lipoprotein particle mediated signaling | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0055096 | low-density lipoprotein particle mediated signaling | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0032108 | negative regulation of response to nutrient levels | 3 | 7 | 8.66e-01 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 2 | 11 | 8.66e-01 |
| GO:BP | GO:0010988 | regulation of low-density lipoprotein particle clearance | 2 | 8 | 8.66e-01 |
| GO:BP | GO:2000556 | positive regulation of T-helper 1 cell cytokine production | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 1 | 1335 | 8.66e-01 |
| GO:BP | GO:0032105 | negative regulation of response to extracellular stimulus | 3 | 7 | 8.66e-01 |
| GO:BP | GO:2000344 | positive regulation of acrosome reaction | 2 | 6 | 8.66e-01 |
| GO:BP | GO:0034371 | chylomicron remodeling | 1 | 1 | 8.66e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 8 | 27 | 8.66e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 3 | 23 | 8.66e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 1 | 288 | 8.66e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 8.66e-01 |
| GO:BP | GO:0009312 | oligosaccharide biosynthetic process | 3 | 15 | 8.66e-01 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 1 | 28 | 8.66e-01 |
| GO:BP | GO:0006032 | chitin catabolic process | 1 | 2 | 8.66e-01 |
| GO:BP | GO:0006030 | chitin metabolic process | 1 | 2 | 8.66e-01 |
| GO:BP | GO:1903800 | positive regulation of miRNA processing | 1 | 3 | 8.66e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 8.66e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 8.66e-01 |
| GO:BP | GO:0014012 | peripheral nervous system axon regeneration | 1 | 6 | 8.66e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 2 | 168 | 8.66e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 5 | 2138 | 8.67e-01 |
| GO:BP | GO:0032627 | interleukin-23 production | 2 | 6 | 8.67e-01 |
| GO:BP | GO:1904273 | L-alanine import across plasma membrane | 1 | 4 | 8.67e-01 |
| GO:BP | GO:0023058 | adaptation of signaling pathway | 3 | 16 | 8.67e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 2 | 30 | 8.67e-01 |
| GO:BP | GO:0030728 | ovulation | 2 | 14 | 8.67e-01 |
| GO:BP | GO:0036378 | calcitriol biosynthetic process from calciol | 1 | 2 | 8.67e-01 |
| GO:BP | GO:0032667 | regulation of interleukin-23 production | 2 | 6 | 8.67e-01 |
| GO:BP | GO:0002840 | regulation of T cell mediated immune response to tumor cell | 2 | 5 | 8.67e-01 |
| GO:BP | GO:0032963 | collagen metabolic process | 6 | 74 | 8.67e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 4 | 11 | 8.67e-01 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 3 | 11 | 8.67e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 4 | 19 | 8.67e-01 |
| GO:BP | GO:0051598 | meiotic recombination checkpoint signaling | 1 | 2 | 8.67e-01 |
| GO:BP | GO:1900424 | regulation of defense response to bacterium | 3 | 12 | 8.67e-01 |
| GO:BP | GO:2000535 | regulation of entry of bacterium into host cell | 2 | 6 | 8.67e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 2 | 13 | 8.67e-01 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 3 | 11 | 8.67e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 2 | 75 | 8.67e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 3 | 9 | 8.67e-01 |
| GO:BP | GO:1905203 | regulation of connective tissue replacement | 1 | 5 | 8.67e-01 |
| GO:BP | GO:0002752 | cell surface pattern recognition receptor signaling pathway | 1 | 27 | 8.67e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 3 | 252 | 8.67e-01 |
| GO:BP | GO:0021564 | vagus nerve development | 1 | 2 | 8.67e-01 |
| GO:BP | GO:0098743 | cell aggregation | 1 | 12 | 8.67e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 551 | 8.67e-01 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 11 | 8.67e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 7 | 23 | 8.67e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 8.67e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 3 | 12 | 8.67e-01 |
| GO:BP | GO:0035188 | hatching | 7 | 23 | 8.67e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 26 | 154 | 8.67e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 7 | 23 | 8.67e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 1 | 3 | 8.68e-01 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 11 | 30 | 8.68e-01 |
| GO:BP | GO:0003383 | apical constriction | 1 | 4 | 8.68e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 9 | 51 | 8.68e-01 |
| GO:BP | GO:0072683 | T cell extravasation | 2 | 8 | 8.68e-01 |
| GO:BP | GO:1905867 | epididymis development | 1 | 2 | 8.68e-01 |
| GO:BP | GO:0043382 | positive regulation of memory T cell differentiation | 1 | 3 | 8.68e-01 |
| GO:BP | GO:0061085 | regulation of histone H3-K27 methylation | 2 | 4 | 8.69e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 13 | 337 | 8.69e-01 |
| GO:BP | GO:1904020 | regulation of G protein-coupled receptor internalization | 1 | 3 | 8.69e-01 |
| GO:BP | GO:0008211 | glucocorticoid metabolic process | 2 | 18 | 8.69e-01 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 1 | 11 | 8.69e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 9 | 153 | 8.69e-01 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 1 | 19 | 8.69e-01 |
| GO:BP | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation | 2 | 8 | 8.69e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 8 | 28 | 8.69e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 8.69e-01 |
| GO:BP | GO:0072015 | podocyte development | 1 | 9 | 8.69e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 2 | 121 | 8.69e-01 |
| GO:BP | GO:0061325 | cell proliferation involved in outflow tract morphogenesis | 1 | 4 | 8.69e-01 |
| GO:BP | GO:0051935 | glutamate reuptake | 1 | 4 | 8.70e-01 |
| GO:BP | GO:0140353 | lipid export from cell | 1 | 28 | 8.70e-01 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 4 | 13 | 8.70e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 10 | 58 | 8.70e-01 |
| GO:BP | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | 2 | 13 | 8.70e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 7 | 14 | 8.70e-01 |
| GO:BP | GO:0048521 | negative regulation of behavior | 2 | 5 | 8.70e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 6 | 61 | 8.70e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 1 | 3 | 8.70e-01 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 5 | 21 | 8.70e-01 |
| GO:BP | GO:0062100 | positive regulation of programmed necrotic cell death | 2 | 5 | 8.71e-01 |
| GO:BP | GO:0030539 | male genitalia development | 7 | 13 | 8.71e-01 |
| GO:BP | GO:0071803 | positive regulation of podosome assembly | 1 | 6 | 8.71e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 45 | 101 | 8.71e-01 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 5 | 27 | 8.71e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 1 | 28 | 8.72e-01 |
| GO:BP | GO:0045656 | negative regulation of monocyte differentiation | 1 | 4 | 8.72e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 8.72e-01 |
| GO:BP | GO:0032819 | positive regulation of natural killer cell proliferation | 1 | 4 | 8.73e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 5 | 39 | 8.73e-01 |
| GO:BP | GO:0097035 | regulation of membrane lipid distribution | 2 | 43 | 8.73e-01 |
| GO:BP | GO:0051454 | intracellular pH elevation | 2 | 4 | 8.73e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 1 | 19 | 8.73e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 2 | 7 | 8.73e-01 |
| GO:BP | GO:0006821 | chloride transport | 4 | 58 | 8.73e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 1 | 19 | 8.73e-01 |
| GO:BP | GO:0014051 | gamma-aminobutyric acid secretion | 1 | 8 | 8.73e-01 |
| GO:BP | GO:0035469 | determination of pancreatic left/right asymmetry | 1 | 3 | 8.73e-01 |
| GO:BP | GO:1990705 | cholangiocyte proliferation | 1 | 3 | 8.73e-01 |
| GO:BP | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis | 1 | 4 | 8.73e-01 |
| GO:BP | GO:0071281 | cellular response to iron ion | 2 | 6 | 8.73e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 8 | 46 | 8.73e-01 |
| GO:BP | GO:0032225 | regulation of synaptic transmission, dopaminergic | 1 | 5 | 8.73e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 8.73e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 3 | 23 | 8.73e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 1 | 15 | 8.73e-01 |
| GO:BP | GO:0015917 | aminophospholipid transport | 2 | 8 | 8.74e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 1 | 30 | 8.74e-01 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 2 | 12 | 8.75e-01 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 2 | 7 | 8.75e-01 |
| GO:BP | GO:0002835 | negative regulation of response to tumor cell | 1 | 2 | 8.75e-01 |
| GO:BP | GO:0002838 | negative regulation of immune response to tumor cell | 1 | 2 | 8.75e-01 |
| GO:BP | GO:0089700 | protein kinase D signaling | 1 | 4 | 8.75e-01 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 5 | 15 | 8.75e-01 |
| GO:BP | GO:1902931 | negative regulation of alcohol biosynthetic process | 1 | 10 | 8.75e-01 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 1 | 5 | 8.75e-01 |
| GO:BP | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain | 1 | 1 | 8.75e-01 |
| GO:BP | GO:0006642 | triglyceride mobilization | 1 | 3 | 8.75e-01 |
| GO:BP | GO:0009311 | oligosaccharide metabolic process | 8 | 43 | 8.75e-01 |
| GO:BP | GO:0045950 | negative regulation of mitotic recombination | 1 | 2 | 8.75e-01 |
| GO:BP | GO:0015031 | protein transport | 13 | 1361 | 8.75e-01 |
| GO:BP | GO:0007060 | male meiosis chromosome segregation | 1 | 1 | 8.75e-01 |
| GO:BP | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway | 2 | 6 | 8.75e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 11 | 60 | 8.76e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 3 | 55 | 8.77e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 2 | 105 | 8.77e-01 |
| GO:BP | GO:0001944 | vasculature development | 20 | 559 | 8.77e-01 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 5 | 9 | 8.77e-01 |
| GO:BP | GO:1903238 | positive regulation of leukocyte tethering or rolling | 1 | 5 | 8.77e-01 |
| GO:BP | GO:0035815 | positive regulation of renal sodium excretion | 1 | 7 | 8.77e-01 |
| GO:BP | GO:0021549 | cerebellum development | 4 | 78 | 8.77e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 2 | 18 | 8.77e-01 |
| GO:BP | GO:0070103 | regulation of interleukin-6-mediated signaling pathway | 1 | 4 | 8.77e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 1 | 20 | 8.77e-01 |
| GO:BP | GO:0019464 | glycine decarboxylation via glycine cleavage system | 1 | 3 | 8.77e-01 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 4 | 10 | 8.78e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 2 | 129 | 8.78e-01 |
| GO:BP | GO:0021884 | forebrain neuron development | 1 | 16 | 8.78e-01 |
| GO:BP | GO:0042448 | progesterone metabolic process | 1 | 13 | 8.78e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 18 | 57 | 8.78e-01 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 14 | 36 | 8.78e-01 |
| GO:BP | GO:0090296 | regulation of mitochondrial DNA replication | 1 | 4 | 8.79e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 13 | 50 | 8.79e-01 |
| GO:BP | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome | 3 | 13 | 8.79e-01 |
| GO:BP | GO:1901164 | negative regulation of trophoblast cell migration | 1 | 4 | 8.79e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 1 | 1496 | 8.79e-01 |
| GO:BP | GO:0014820 | tonic smooth muscle contraction | 3 | 8 | 8.79e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 1 | 108 | 8.79e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 3 | 15 | 8.79e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 1 | 31 | 8.79e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 1 | 24 | 8.79e-01 |
| GO:BP | GO:0009583 | detection of light stimulus | 1 | 27 | 8.79e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 2 | 47 | 8.79e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 1 | 58 | 8.79e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 3 | 147 | 8.79e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 3 | 22 | 8.80e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 3 | 28 | 8.80e-01 |
| GO:BP | GO:0032380 | regulation of intracellular sterol transport | 1 | 6 | 8.80e-01 |
| GO:BP | GO:0032383 | regulation of intracellular cholesterol transport | 1 | 6 | 8.80e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 1 | 232 | 8.80e-01 |
| GO:BP | GO:1903431 | positive regulation of cell maturation | 1 | 6 | 8.80e-01 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 6 | 38 | 8.80e-01 |
| GO:BP | GO:1900193 | regulation of oocyte maturation | 1 | 5 | 8.80e-01 |
| GO:BP | GO:0015888 | thiamine transport | 2 | 3 | 8.80e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 2 | 14 | 8.80e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 2 | 14 | 8.80e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 7 | 39 | 8.80e-01 |
| GO:BP | GO:0035455 | response to interferon-alpha | 1 | 16 | 8.80e-01 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 14 | 31 | 8.80e-01 |
| GO:BP | GO:1990911 | response to psychosocial stress | 1 | 2 | 8.81e-01 |
| GO:BP | GO:0002025 | norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure | 1 | 3 | 8.81e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 7 | 1507 | 8.81e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 2 | 3580 | 8.81e-01 |
| GO:BP | GO:1900864 | mitochondrial RNA modification | 1 | 9 | 8.81e-01 |
| GO:BP | GO:0015846 | polyamine transport | 4 | 12 | 8.81e-01 |
| GO:BP | GO:0150147 | cell-cell junction disassembly | 1 | 4 | 8.81e-01 |
| GO:BP | GO:0001568 | blood vessel development | 19 | 534 | 8.81e-01 |
| GO:BP | GO:0070900 | mitochondrial tRNA modification | 1 | 9 | 8.81e-01 |
| GO:BP | GO:0051135 | positive regulation of NK T cell activation | 1 | 3 | 8.81e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 4 | 25 | 8.82e-01 |
| GO:BP | GO:0034332 | adherens junction organization | 14 | 41 | 8.82e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 2 | 23 | 8.82e-01 |
| GO:BP | GO:0001578 | microtubule bundle formation | 3 | 79 | 8.83e-01 |
| GO:BP | GO:0001764 | neuron migration | 2 | 133 | 8.83e-01 |
| GO:BP | GO:0038007 | netrin-activated signaling pathway | 2 | 8 | 8.83e-01 |
| GO:BP | GO:0060911 | cardiac cell fate commitment | 3 | 11 | 8.83e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 3 | 11 | 8.83e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 1 | 9 | 8.83e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 2 | 4 | 8.83e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 15 | 185 | 8.84e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 8.84e-01 |
| GO:BP | GO:0019405 | alditol catabolic process | 1 | 6 | 8.84e-01 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 7 | 23 | 8.84e-01 |
| GO:BP | GO:0046325 | negative regulation of glucose import | 3 | 10 | 8.84e-01 |
| GO:BP | GO:0043305 | negative regulation of mast cell degranulation | 2 | 2 | 8.85e-01 |
| GO:BP | GO:0033490 | cholesterol biosynthetic process via lathosterol | 1 | 4 | 8.85e-01 |
| GO:BP | GO:0033489 | cholesterol biosynthetic process via desmosterol | 1 | 4 | 8.85e-01 |
| GO:BP | GO:1902731 | negative regulation of chondrocyte proliferation | 1 | 3 | 8.85e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 5 | 16 | 8.85e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 2 | 110 | 8.85e-01 |
| GO:BP | GO:0045906 | negative regulation of vasoconstriction | 1 | 6 | 8.85e-01 |
| GO:BP | GO:0018095 | protein polyglutamylation | 4 | 9 | 8.85e-01 |
| GO:BP | GO:0033033 | negative regulation of myeloid cell apoptotic process | 1 | 10 | 8.85e-01 |
| GO:BP | GO:0043416 | regulation of skeletal muscle tissue regeneration | 1 | 5 | 8.85e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 4 | 90 | 8.86e-01 |
| GO:BP | GO:0018243 | protein O-linked glycosylation via threonine | 3 | 7 | 8.86e-01 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 1 | 20 | 8.86e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 8.86e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 6 | 68 | 8.87e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 1 | 10 | 8.87e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 2 | 4794 | 8.87e-01 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 1 | 9 | 8.87e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 5 | 14 | 8.87e-01 |
| GO:BP | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin | 2 | 6 | 8.87e-01 |
| GO:BP | GO:1900108 | negative regulation of nodal signaling pathway | 1 | 2 | 8.87e-01 |
| GO:BP | GO:0035582 | sequestering of BMP in extracellular matrix | 1 | 2 | 8.87e-01 |
| GO:BP | GO:0070535 | histone H2A K63-linked ubiquitination | 1 | 6 | 8.88e-01 |
| GO:BP | GO:0033182 | regulation of histone ubiquitination | 1 | 6 | 8.88e-01 |
| GO:BP | GO:0042026 | protein refolding | 1 | 26 | 8.88e-01 |
| GO:BP | GO:0060264 | regulation of respiratory burst involved in inflammatory response | 1 | 3 | 8.88e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 8.88e-01 |
| GO:BP | GO:0001920 | negative regulation of receptor recycling | 1 | 3 | 8.88e-01 |
| GO:BP | GO:0097052 | L-kynurenine metabolic process | 1 | 4 | 8.88e-01 |
| GO:BP | GO:0006531 | aspartate metabolic process | 1 | 4 | 8.88e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 3 | 7 | 8.88e-01 |
| GO:BP | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 3 | 8.88e-01 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 1 | 20 | 8.88e-01 |
| GO:BP | GO:0042303 | molting cycle | 5 | 78 | 8.88e-01 |
| GO:BP | GO:0042633 | hair cycle | 5 | 78 | 8.88e-01 |
| GO:BP | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone | 1 | 5 | 8.88e-01 |
| GO:BP | GO:0014010 | Schwann cell proliferation | 2 | 7 | 8.88e-01 |
| GO:BP | GO:0010624 | regulation of Schwann cell proliferation | 2 | 7 | 8.88e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 1 | 21 | 8.88e-01 |
| GO:BP | GO:0030901 | midbrain development | 2 | 69 | 8.88e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 13 | 116 | 8.88e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 2 | 86 | 8.88e-01 |
| GO:BP | GO:0032185 | septin cytoskeleton organization | 1 | 4 | 8.88e-01 |
| GO:BP | GO:0007281 | germ cell development | 2 | 187 | 8.88e-01 |
| GO:BP | GO:0007168 | receptor guanylyl cyclase signaling pathway | 1 | 5 | 8.88e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 4 | 44 | 8.89e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 12 | 898 | 8.89e-01 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 1 | 2 | 8.89e-01 |
| GO:BP | GO:0070197 | meiotic attachment of telomere to nuclear envelope | 1 | 1 | 8.89e-01 |
| GO:BP | GO:0071549 | cellular response to dexamethasone stimulus | 1 | 24 | 8.89e-01 |
| GO:BP | GO:0034398 | telomere tethering at nuclear periphery | 1 | 1 | 8.89e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 6 | 44 | 8.89e-01 |
| GO:BP | GO:0044821 | meiotic telomere tethering at nuclear periphery | 1 | 1 | 8.89e-01 |
| GO:BP | GO:0030473 | nuclear migration along microtubule | 1 | 5 | 8.89e-01 |
| GO:BP | GO:0035701 | hematopoietic stem cell migration | 1 | 6 | 8.89e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 1 | 19 | 8.89e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 4 | 31 | 8.89e-01 |
| GO:BP | GO:0061590 | calcium activated phosphatidylcholine scrambling | 1 | 3 | 8.89e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 8.89e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 1 | 10 | 8.89e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 6 | 1315 | 8.89e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 6 | 1315 | 8.89e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 3 | 12 | 8.89e-01 |
| GO:BP | GO:0006897 | endocytosis | 3 | 527 | 8.89e-01 |
| GO:BP | GO:0071224 | cellular response to peptidoglycan | 1 | 2 | 8.89e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 10 | 8.89e-01 |
| GO:BP | GO:0070671 | response to interleukin-12 | 1 | 6 | 8.89e-01 |
| GO:BP | GO:0034182 | regulation of maintenance of mitotic sister chromatid cohesion | 2 | 8 | 8.89e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 4 | 15 | 8.89e-01 |
| GO:BP | GO:0048304 | positive regulation of isotype switching to IgG isotypes | 3 | 4 | 8.89e-01 |
| GO:BP | GO:0034091 | regulation of maintenance of sister chromatid cohesion | 2 | 8 | 8.89e-01 |
| GO:BP | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | 3 | 6 | 8.89e-01 |
| GO:BP | GO:1904783 | positive regulation of NMDA glutamate receptor activity | 1 | 4 | 8.90e-01 |
| GO:BP | GO:0021631 | optic nerve morphogenesis | 1 | 2 | 8.90e-01 |
| GO:BP | GO:0090025 | regulation of monocyte chemotaxis | 1 | 12 | 8.90e-01 |
| GO:BP | GO:0007254 | JNK cascade | 1 | 138 | 8.90e-01 |
| GO:BP | GO:0042119 | neutrophil activation | 5 | 16 | 8.90e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 8.90e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 5 | 172 | 8.91e-01 |
| GO:BP | GO:0048627 | myoblast development | 1 | 3 | 8.91e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 6 | 1322 | 8.91e-01 |
| GO:BP | GO:0060267 | positive regulation of respiratory burst | 1 | 3 | 8.92e-01 |
| GO:BP | GO:0140367 | antibacterial innate immune response | 1 | 2 | 8.93e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 4 | 26 | 8.93e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 3 | 15 | 8.93e-01 |
| GO:BP | GO:0001710 | mesodermal cell fate commitment | 2 | 9 | 8.93e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 12 | 2270 | 8.93e-01 |
| GO:BP | GO:1902563 | regulation of neutrophil activation | 2 | 3 | 8.93e-01 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 4 | 10 | 8.93e-01 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 4 | 10 | 8.93e-01 |
| GO:BP | GO:0097068 | response to thyroxine | 1 | 4 | 8.93e-01 |
| GO:BP | GO:0044351 | macropinocytosis | 2 | 8 | 8.93e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 11 | 25 | 8.93e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 9 | 66 | 8.93e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 1 | 13 | 8.93e-01 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 22 | 34 | 8.93e-01 |
| GO:BP | GO:0045617 | negative regulation of keratinocyte differentiation | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0060435 | bronchiole development | 1 | 2 | 8.93e-01 |
| GO:BP | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0072674 | multinuclear osteoclast differentiation | 1 | 6 | 8.93e-01 |
| GO:BP | GO:0071279 | cellular response to cobalt ion | 1 | 4 | 8.93e-01 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 4 | 17 | 8.93e-01 |
| GO:BP | GO:0014889 | muscle atrophy | 2 | 9 | 8.93e-01 |
| GO:BP | GO:0060686 | negative regulation of prostatic bud formation | 1 | 4 | 8.93e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 3 | 259 | 8.93e-01 |
| GO:BP | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | 1 | 3 | 8.93e-01 |
| GO:BP | GO:0010692 | regulation of alkaline phosphatase activity | 1 | 8 | 8.94e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 3 | 11 | 8.94e-01 |
| GO:BP | GO:0008594 | photoreceptor cell morphogenesis | 1 | 4 | 8.94e-01 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 1 | 6 | 8.94e-01 |
| GO:BP | GO:1990051 | activation of protein kinase C activity | 1 | 3 | 8.94e-01 |
| GO:BP | GO:0021782 | glial cell development | 6 | 84 | 8.94e-01 |
| GO:BP | GO:1905244 | regulation of modification of synaptic structure | 1 | 10 | 8.94e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 17 | 122 | 8.94e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 11 | 496 | 8.95e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 2 | 27 | 8.95e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 1 | 17 | 8.95e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 7 | 27 | 8.95e-01 |
| GO:BP | GO:0033004 | negative regulation of mast cell activation | 4 | 4 | 8.95e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 3 | 26 | 8.95e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 1 | 4 | 8.95e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 2 | 14 | 8.95e-01 |
| GO:BP | GO:0021648 | vestibulocochlear nerve morphogenesis | 2 | 3 | 8.95e-01 |
| GO:BP | GO:0006996 | organelle organization | 16 | 3000 | 8.96e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 8.96e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 8.96e-01 |
| GO:BP | GO:0022617 | extracellular matrix disassembly | 5 | 36 | 8.96e-01 |
| GO:BP | GO:0140052 | cellular response to oxidised low-density lipoprotein particle stimulus | 2 | 7 | 8.96e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1 | 4099 | 8.96e-01 |
| GO:BP | GO:0032494 | response to peptidoglycan | 2 | 5 | 8.96e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 8.97e-01 |
| GO:BP | GO:0090188 | negative regulation of pancreatic juice secretion | 1 | 4 | 8.97e-01 |
| GO:BP | GO:0019418 | sulfide oxidation | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0036065 | fucosylation | 4 | 10 | 8.97e-01 |
| GO:BP | GO:0009233 | menaquinone metabolic process | 1 | 1 | 8.97e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 14 | 107 | 8.97e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 2 | 7 | 8.97e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 8.97e-01 |
| GO:BP | GO:0070221 | sulfide oxidation, using sulfide:quinone oxidoreductase | 1 | 3 | 8.97e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 31 | 89 | 8.97e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 1 | 336 | 8.97e-01 |
| GO:BP | GO:0019079 | viral genome replication | 16 | 101 | 8.97e-01 |
| GO:BP | GO:1903803 | L-glutamine import across plasma membrane | 1 | 4 | 8.97e-01 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 1 | 11 | 8.97e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 8.97e-01 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 1 | 11 | 8.97e-01 |
| GO:BP | GO:0072378 | blood coagulation, fibrin clot formation | 1 | 7 | 8.97e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 2 | 12 | 8.97e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 6 | 16 | 8.97e-01 |
| GO:BP | GO:0003105 | negative regulation of glomerular filtration | 1 | 3 | 8.98e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 4 | 78 | 8.99e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 8.99e-01 |
| GO:BP | GO:0010961 | intracellular magnesium ion homeostasis | 1 | 4 | 8.99e-01 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 3 | 12 | 8.99e-01 |
| GO:BP | GO:0072679 | thymocyte migration | 1 | 1 | 8.99e-01 |
| GO:BP | GO:2000391 | positive regulation of neutrophil extravasation | 1 | 6 | 8.99e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 2 | 16 | 8.99e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 4 | 63 | 8.99e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 3 | 785 | 8.99e-01 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 4 | 18 | 8.99e-01 |
| GO:BP | GO:0042756 | drinking behavior | 1 | 6 | 9.00e-01 |
| GO:BP | GO:1903210 | podocyte apoptotic process | 1 | 3 | 9.00e-01 |
| GO:BP | GO:0035289 | posterior head segmentation | 1 | 3 | 9.00e-01 |
| GO:BP | GO:0035287 | head segmentation | 1 | 3 | 9.00e-01 |
| GO:BP | GO:0072011 | glomerular endothelium development | 1 | 2 | 9.00e-01 |
| GO:BP | GO:0097018 | renal albumin absorption | 1 | 3 | 9.00e-01 |
| GO:BP | GO:0061092 | positive regulation of phospholipid translocation | 1 | 4 | 9.00e-01 |
| GO:BP | GO:0007380 | specification of segmental identity, head | 1 | 3 | 9.00e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 9 | 100 | 9.00e-01 |
| GO:BP | GO:0097576 | vacuole fusion | 3 | 12 | 9.01e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 3 | 10 | 9.01e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 11 | 300 | 9.01e-01 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 5 | 14 | 9.01e-01 |
| GO:BP | GO:0014904 | myotube cell development | 8 | 34 | 9.01e-01 |
| GO:BP | GO:0060732 | positive regulation of inositol phosphate biosynthetic process | 1 | 5 | 9.01e-01 |
| GO:BP | GO:0032264 | IMP salvage | 1 | 4 | 9.01e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 9.01e-01 |
| GO:BP | GO:0033299 | secretion of lysosomal enzymes | 1 | 7 | 9.01e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 1 | 232 | 9.01e-01 |
| GO:BP | GO:0021543 | pallium development | 23 | 139 | 9.01e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 1 | 141 | 9.01e-01 |
| GO:BP | GO:0072385 | minus-end-directed organelle transport along microtubule | 1 | 5 | 9.02e-01 |
| GO:BP | GO:0060986 | endocrine hormone secretion | 1 | 35 | 9.02e-01 |
| GO:BP | GO:0048254 | snoRNA localization | 1 | 5 | 9.02e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 4 | 51 | 9.02e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 2 | 5 | 9.03e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 8 | 31 | 9.03e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 2 | 24 | 9.03e-01 |
| GO:BP | GO:0007399 | nervous system development | 36 | 1904 | 9.03e-01 |
| GO:BP | GO:0002424 | T cell mediated immune response to tumor cell | 2 | 6 | 9.03e-01 |
| GO:BP | GO:0015860 | purine nucleoside transmembrane transport | 1 | 3 | 9.03e-01 |
| GO:BP | GO:0034759 | regulation of iron ion transmembrane transport | 1 | 2 | 9.03e-01 |
| GO:BP | GO:0001878 | response to yeast | 1 | 3 | 9.03e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 1 | 10 | 9.03e-01 |
| GO:BP | GO:0055089 | fatty acid homeostasis | 4 | 12 | 9.03e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 3 | 100 | 9.04e-01 |
| GO:BP | GO:1900121 | negative regulation of receptor binding | 3 | 8 | 9.04e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 17 | 47 | 9.04e-01 |
| GO:BP | GO:0000960 | regulation of mitochondrial RNA catabolic process | 1 | 5 | 9.04e-01 |
| GO:BP | GO:0021798 | forebrain dorsal/ventral pattern formation | 1 | 2 | 9.04e-01 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 9.04e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 9.04e-01 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 1 | 2 | 9.04e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 9.04e-01 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 9.04e-01 |
| GO:BP | GO:0065007 | biological regulation | 4 | 8320 | 9.04e-01 |
| GO:BP | GO:1905034 | regulation of antifungal innate immune response | 1 | 3 | 9.04e-01 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 1 | 2 | 9.04e-01 |
| GO:BP | GO:2000318 | positive regulation of T-helper 17 type immune response | 2 | 7 | 9.04e-01 |
| GO:BP | GO:0035630 | bone mineralization involved in bone maturation | 2 | 6 | 9.04e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 30 | 9.04e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 2 | 2574 | 9.04e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 7 | 30 | 9.04e-01 |
| GO:BP | GO:0061548 | ganglion development | 5 | 12 | 9.04e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 2 | 2576 | 9.04e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 6 | 81 | 9.05e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 9.05e-01 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 1 | 31 | 9.05e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 9.05e-01 |
| GO:BP | GO:0072540 | T-helper 17 cell lineage commitment | 1 | 3 | 9.05e-01 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 9.05e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 3 | 9 | 9.05e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 4 | 9 | 9.05e-01 |
| GO:BP | GO:0072717 | cellular response to actinomycin D | 1 | 4 | 9.05e-01 |
| GO:BP | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 2 | 14 | 9.05e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 9.06e-01 |
| GO:BP | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway | 1 | 4 | 9.06e-01 |
| GO:BP | GO:0042182 | ketone catabolic process | 3 | 9 | 9.06e-01 |
| GO:BP | GO:0048251 | elastic fiber assembly | 1 | 12 | 9.06e-01 |
| GO:BP | GO:0030202 | heparin metabolic process | 5 | 14 | 9.06e-01 |
| GO:BP | GO:0021877 | forebrain neuron fate commitment | 1 | 1 | 9.06e-01 |
| GO:BP | GO:0045650 | negative regulation of macrophage differentiation | 1 | 5 | 9.06e-01 |
| GO:BP | GO:0048633 | positive regulation of skeletal muscle tissue growth | 1 | 6 | 9.06e-01 |
| GO:BP | GO:1903288 | positive regulation of potassium ion import across plasma membrane | 1 | 7 | 9.06e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 2 | 2593 | 9.06e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 1 | 77 | 9.06e-01 |
| GO:BP | GO:0046884 | follicle-stimulating hormone secretion | 1 | 4 | 9.07e-01 |
| GO:BP | GO:1990966 | ATP generation from poly-ADP-D-ribose | 1 | 4 | 9.07e-01 |
| GO:BP | GO:2001054 | negative regulation of mesenchymal cell apoptotic process | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 1 | 50 | 9.07e-01 |
| GO:BP | GO:0048665 | neuron fate specification | 4 | 14 | 9.07e-01 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 1 | 11 | 9.07e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 9 | 30 | 9.07e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 4 | 142 | 9.07e-01 |
| GO:BP | GO:1904751 | positive regulation of protein localization to nucleolus | 1 | 3 | 9.07e-01 |
| GO:BP | GO:0021548 | pons development | 2 | 7 | 9.07e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 9 | 30 | 9.07e-01 |
| GO:BP | GO:1903659 | regulation of complement-dependent cytotoxicity | 1 | 2 | 9.07e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 1 | 126 | 9.07e-01 |
| GO:BP | GO:0007276 | gamete generation | 5 | 461 | 9.07e-01 |
| GO:BP | GO:0035812 | renal sodium excretion | 2 | 11 | 9.08e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 2 | 8 | 9.08e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 4 | 937 | 9.08e-01 |
| GO:BP | GO:0006382 | adenosine to inosine editing | 1 | 4 | 9.08e-01 |
| GO:BP | GO:0034471 | ncRNA 5’-end processing | 1 | 5 | 9.08e-01 |
| GO:BP | GO:0000967 | rRNA 5’-end processing | 1 | 5 | 9.08e-01 |
| GO:BP | GO:0000472 | endonucleolytic cleavage to generate mature 5’-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 5 | 9.08e-01 |
| GO:BP | GO:1990384 | hyaloid vascular plexus regression | 1 | 3 | 9.08e-01 |
| GO:BP | GO:0036492 | eiF2alpha phosphorylation in response to endoplasmic reticulum stress | 1 | 7 | 9.08e-01 |
| GO:BP | GO:0032817 | regulation of natural killer cell proliferation | 1 | 4 | 9.09e-01 |
| GO:BP | GO:0032277 | negative regulation of gonadotropin secretion | 1 | 4 | 9.09e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 13 | 35 | 9.09e-01 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 1 | 7 | 9.09e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 9.09e-01 |
| GO:BP | GO:0071910 | determination of liver left/right asymmetry | 1 | 4 | 9.09e-01 |
| GO:BP | GO:0099641 | anterograde axonal protein transport | 2 | 9 | 9.09e-01 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 1 | 15 | 9.09e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 9.09e-01 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 1 | 15 | 9.09e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 1 | 11 | 9.09e-01 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 1 | 19 | 9.09e-01 |
| GO:BP | GO:0007567 | parturition | 1 | 8 | 9.09e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 9.09e-01 |
| GO:BP | GO:0061888 | regulation of astrocyte activation | 1 | 3 | 9.09e-01 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 9.09e-01 |
| GO:BP | GO:0050917 | sensory perception of umami taste | 1 | 3 | 9.09e-01 |
| GO:BP | GO:0006578 | amino-acid betaine biosynthetic process | 1 | 5 | 9.09e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 9.09e-01 |
| GO:BP | GO:0035821 | modulation of process of another organism | 1 | 5 | 9.09e-01 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 1 | 42 | 9.09e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 9.09e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 1 | 598 | 9.09e-01 |
| GO:BP | GO:0010912 | positive regulation of isomerase activity | 1 | 5 | 9.10e-01 |
| GO:BP | GO:0010911 | regulation of isomerase activity | 1 | 5 | 9.10e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 9.10e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 1 | 13 | 9.10e-01 |
| GO:BP | GO:0035585 | calcium-mediated signaling using extracellular calcium source | 1 | 2 | 9.10e-01 |
| GO:BP | GO:0036260 | RNA capping | 5 | 20 | 9.10e-01 |
| GO:BP | GO:0032816 | positive regulation of natural killer cell activation | 4 | 16 | 9.10e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 7 | 1238 | 9.10e-01 |
| GO:BP | GO:0060439 | trachea morphogenesis | 1 | 7 | 9.10e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 9.10e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 2 | 6 | 9.10e-01 |
| GO:BP | GO:1904557 | L-alanine transmembrane transport | 1 | 4 | 9.10e-01 |
| GO:BP | GO:0022037 | metencephalon development | 4 | 84 | 9.11e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 2 | 447 | 9.11e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 9.11e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 1 | 68 | 9.11e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 1 | 3 | 9.11e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 11 | 63 | 9.11e-01 |
| GO:BP | GO:0014905 | myoblast fusion involved in skeletal muscle regeneration | 1 | 3 | 9.11e-01 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 2 | 12 | 9.11e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 2 | 110 | 9.12e-01 |
| GO:BP | GO:1905247 | positive regulation of aspartic-type peptidase activity | 1 | 8 | 9.12e-01 |
| GO:BP | GO:0002536 | respiratory burst involved in inflammatory response | 1 | 4 | 9.12e-01 |
| GO:BP | GO:1903979 | negative regulation of microglial cell activation | 1 | 5 | 9.12e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 13 | 26 | 9.12e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 13 | 26 | 9.12e-01 |
| GO:BP | GO:0045087 | innate immune response | 3 | 488 | 9.12e-01 |
| GO:BP | GO:0034138 | toll-like receptor 3 signaling pathway | 1 | 12 | 9.12e-01 |
| GO:BP | GO:0007286 | spermatid development | 10 | 105 | 9.12e-01 |
| GO:BP | GO:0006546 | glycine catabolic process | 1 | 3 | 9.12e-01 |
| GO:BP | GO:0007220 | Notch receptor processing | 2 | 9 | 9.13e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 2 | 11 | 9.13e-01 |
| GO:BP | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | 1 | 4 | 9.13e-01 |
| GO:BP | GO:0001816 | cytokine production | 2 | 451 | 9.13e-01 |
| GO:BP | GO:0071676 | negative regulation of mononuclear cell migration | 2 | 16 | 9.13e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 7 | 12 | 9.13e-01 |
| GO:BP | GO:0002829 | negative regulation of type 2 immune response | 1 | 7 | 9.13e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 1 | 4 | 9.14e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 9.14e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 2 | 87 | 9.14e-01 |
| GO:BP | GO:0045653 | negative regulation of megakaryocyte differentiation | 1 | 17 | 9.15e-01 |
| GO:BP | GO:0034135 | regulation of toll-like receptor 2 signaling pathway | 1 | 7 | 9.15e-01 |
| GO:BP | GO:0065001 | specification of axis polarity | 1 | 2 | 9.15e-01 |
| GO:BP | GO:0002692 | negative regulation of cellular extravasation | 1 | 5 | 9.16e-01 |
| GO:BP | GO:0060332 | positive regulation of response to type II interferon | 1 | 4 | 9.16e-01 |
| GO:BP | GO:0060335 | positive regulation of type II interferon-mediated signaling pathway | 1 | 4 | 9.16e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 1 | 16 | 9.16e-01 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 1 | 2 | 9.16e-01 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 1 | 2 | 9.16e-01 |
| GO:BP | GO:0030431 | sleep | 1 | 13 | 9.16e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 2 | 46 | 9.16e-01 |
| GO:BP | GO:0060161 | positive regulation of dopamine receptor signaling pathway | 1 | 3 | 9.16e-01 |
| GO:BP | GO:0043588 | skin development | 4 | 175 | 9.16e-01 |
| GO:BP | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 1 | 6 | 9.16e-01 |
| GO:BP | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 1 | 6 | 9.16e-01 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 1 | 5 | 9.16e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 11 | 60 | 9.17e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 2 | 13 | 9.17e-01 |
| GO:BP | GO:0045217 | cell-cell junction maintenance | 4 | 13 | 9.17e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 3 | 79 | 9.17e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 3 | 6 | 9.17e-01 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 1 | 3 | 9.17e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 9.17e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 9.17e-01 |
| GO:BP | GO:0032602 | chemokine production | 9 | 46 | 9.17e-01 |
| GO:BP | GO:0072102 | glomerulus morphogenesis | 1 | 7 | 9.17e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 9 | 46 | 9.17e-01 |
| GO:BP | GO:1903412 | response to bile acid | 1 | 3 | 9.18e-01 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 1 | 8 | 9.18e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 2 | 42 | 9.18e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 4 | 32 | 9.18e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 5 | 12 | 9.18e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 6 | 76 | 9.18e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 1 | 11 | 9.18e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 4 | 25 | 9.18e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 6 | 52 | 9.18e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 13 | 1462 | 9.18e-01 |
| GO:BP | GO:0035352 | NAD transmembrane transport | 1 | 3 | 9.18e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 8 | 76 | 9.18e-01 |
| GO:BP | GO:0050765 | negative regulation of phagocytosis | 5 | 15 | 9.18e-01 |
| GO:BP | GO:0021854 | hypothalamus development | 2 | 15 | 9.18e-01 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 2 | 96 | 9.18e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 4 | 71 | 9.18e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 1 | 3 | 9.18e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 4 | 21 | 9.18e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 1 | 4 | 9.18e-01 |
| GO:BP | GO:0090027 | negative regulation of monocyte chemotaxis | 1 | 4 | 9.18e-01 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 4 | 14 | 9.18e-01 |
| GO:BP | GO:0060300 | regulation of cytokine activity | 1 | 3 | 9.18e-01 |
| GO:BP | GO:0060847 | endothelial cell fate specification | 1 | 3 | 9.18e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 5 | 33 | 9.19e-01 |
| GO:BP | GO:0120262 | negative regulation of heterochromatin organization | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0070672 | response to interleukin-15 | 1 | 3 | 9.19e-01 |
| GO:BP | GO:0044409 | entry into host | 18 | 113 | 9.19e-01 |
| GO:BP | GO:1905268 | negative regulation of chromatin organization | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0031452 | negative regulation of heterochromatin formation | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0032825 | positive regulation of natural killer cell differentiation | 1 | 7 | 9.19e-01 |
| GO:BP | GO:0030073 | insulin secretion | 15 | 141 | 9.19e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 5 | 47 | 9.19e-01 |
| GO:BP | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization | 1 | 4 | 9.19e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 3 | 115 | 9.19e-01 |
| GO:BP | GO:0010887 | negative regulation of cholesterol storage | 2 | 9 | 9.19e-01 |
| GO:BP | GO:0098532 | histone H3-K27 trimethylation | 2 | 4 | 9.20e-01 |
| GO:BP | GO:0016043 | cellular component organization | 1 | 5076 | 9.20e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 2 | 59 | 9.20e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 18 | 2845 | 9.20e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 5 | 22 | 9.20e-01 |
| GO:BP | GO:0010387 | COP9 signalosome assembly | 1 | 3 | 9.20e-01 |
| GO:BP | GO:0070141 | response to UV-A | 2 | 11 | 9.20e-01 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 2 | 97 | 9.20e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 1 | 88 | 9.21e-01 |
| GO:BP | GO:0002513 | tolerance induction to self antigen | 1 | 4 | 9.21e-01 |
| GO:BP | GO:0021871 | forebrain regionalization | 1 | 8 | 9.21e-01 |
| GO:BP | GO:0034587 | piRNA processing | 1 | 4 | 9.21e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 1 | 8 | 9.21e-01 |
| GO:BP | GO:2000617 | positive regulation of histone H3-K9 acetylation | 2 | 6 | 9.21e-01 |
| GO:BP | GO:0034112 | positive regulation of homotypic cell-cell adhesion | 1 | 10 | 9.21e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 1 | 10 | 9.21e-01 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 1 | 4 | 9.21e-01 |
| GO:BP | GO:0070537 | histone H2A K63-linked deubiquitination | 2 | 8 | 9.21e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 7 | 20 | 9.21e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 4 | 13 | 9.21e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 7 | 86 | 9.21e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 3 | 563 | 9.21e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 1 | 27 | 9.21e-01 |
| GO:BP | GO:0070383 | DNA cytosine deamination | 2 | 5 | 9.21e-01 |
| GO:BP | GO:0042938 | dipeptide transport | 1 | 4 | 9.21e-01 |
| GO:BP | GO:0014850 | response to muscle activity | 1 | 17 | 9.21e-01 |
| GO:BP | GO:0035442 | dipeptide transmembrane transport | 1 | 4 | 9.21e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 5 | 30 | 9.22e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 9.22e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 5 | 30 | 9.22e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 1 | 64 | 9.22e-01 |
| GO:BP | GO:2000389 | regulation of neutrophil extravasation | 1 | 6 | 9.22e-01 |
| GO:BP | GO:0002675 | positive regulation of acute inflammatory response | 4 | 11 | 9.22e-01 |
| GO:BP | GO:0002438 | acute inflammatory response to antigenic stimulus | 4 | 10 | 9.22e-01 |
| GO:BP | GO:0061588 | calcium activated phospholipid scrambling | 1 | 4 | 9.22e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 4 | 665 | 9.22e-01 |
| GO:BP | GO:0010286 | heat acclimation | 1 | 5 | 9.22e-01 |
| GO:BP | GO:0070370 | cellular heat acclimation | 1 | 5 | 9.22e-01 |
| GO:BP | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 1 | 4 | 9.23e-01 |
| GO:BP | GO:0060433 | bronchus development | 1 | 6 | 9.23e-01 |
| GO:BP | GO:0070365 | hepatocyte differentiation | 2 | 12 | 9.23e-01 |
| GO:BP | GO:0021682 | nerve maturation | 1 | 2 | 9.23e-01 |
| GO:BP | GO:1903350 | response to dopamine | 5 | 48 | 9.23e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 2 | 113 | 9.24e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 9.24e-01 |
| GO:BP | GO:0021819 | layer formation in cerebral cortex | 3 | 12 | 9.24e-01 |
| GO:BP | GO:0097752 | regulation of DNA stability | 1 | 3 | 9.24e-01 |
| GO:BP | GO:0032790 | ribosome disassembly | 1 | 11 | 9.24e-01 |
| GO:BP | GO:0007350 | blastoderm segmentation | 2 | 17 | 9.24e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 9.24e-01 |
| GO:BP | GO:0051969 | regulation of transmission of nerve impulse | 1 | 6 | 9.24e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 11 | 2412 | 9.24e-01 |
| GO:BP | GO:0043302 | positive regulation of leukocyte degranulation | 2 | 15 | 9.25e-01 |
| GO:BP | GO:1904352 | positive regulation of protein catabolic process in the vacuole | 1 | 6 | 9.25e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 9.25e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 9.25e-01 |
| GO:BP | GO:0072017 | distal tubule development | 1 | 6 | 9.25e-01 |
| GO:BP | GO:0021859 | pyramidal neuron differentiation | 1 | 11 | 9.25e-01 |
| GO:BP | GO:0048535 | lymph node development | 3 | 11 | 9.26e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 9.26e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 10 | 185 | 9.26e-01 |
| GO:BP | GO:0072376 | protein activation cascade | 1 | 9 | 9.26e-01 |
| GO:BP | GO:0042373 | vitamin K metabolic process | 2 | 6 | 9.26e-01 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 1 | 7 | 9.26e-01 |
| GO:BP | GO:0035609 | C-terminal protein deglutamylation | 1 | 2 | 9.26e-01 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 4 | 19 | 9.26e-01 |
| GO:BP | GO:0031645 | negative regulation of nervous system process | 5 | 9 | 9.27e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 6 | 39 | 9.27e-01 |
| GO:BP | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 2 | 9.27e-01 |
| GO:BP | GO:1900175 | regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 2 | 9.27e-01 |
| GO:BP | GO:0097354 | prenylation | 2 | 10 | 9.27e-01 |
| GO:BP | GO:0018342 | protein prenylation | 2 | 10 | 9.27e-01 |
| GO:BP | GO:1903998 | regulation of eating behavior | 1 | 1 | 9.27e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1 | 5274 | 9.27e-01 |
| GO:BP | GO:0021801 | cerebral cortex radial glia-guided migration | 6 | 22 | 9.28e-01 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 6 | 20 | 9.28e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 9.28e-01 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 1 | 21 | 9.28e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 9.28e-01 |
| GO:BP | GO:0022030 | telencephalon glial cell migration | 6 | 22 | 9.28e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 2 | 7 | 9.28e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 9.28e-01 |
| GO:BP | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 4 | 9.28e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 9.28e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 9.28e-01 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 2 | 9 | 9.28e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 22 | 123 | 9.28e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 2 | 79 | 9.28e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 9.28e-01 |
| GO:BP | GO:0042420 | dopamine catabolic process | 1 | 5 | 9.29e-01 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 1 | 30 | 9.29e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 6 | 15 | 9.29e-01 |
| GO:BP | GO:2001026 | regulation of endothelial cell chemotaxis | 2 | 16 | 9.29e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 3 | 88 | 9.29e-01 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 1 | 2 | 9.29e-01 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 6 | 9.29e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 5 | 182 | 9.29e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 13 | 39 | 9.30e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 13 | 39 | 9.30e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 18 | 63 | 9.30e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 3 | 11 | 9.30e-01 |
| GO:BP | GO:0002036 | regulation of L-glutamate import across plasma membrane | 1 | 6 | 9.30e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 2 | 102 | 9.30e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 6 | 84 | 9.30e-01 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 12 | 55 | 9.30e-01 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 12 | 55 | 9.30e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 1 | 19 | 9.30e-01 |
| GO:BP | GO:1903224 | regulation of endodermal cell differentiation | 1 | 6 | 9.30e-01 |
| GO:BP | GO:0015848 | spermidine transport | 1 | 3 | 9.30e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 3 | 63 | 9.31e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 16 | 89 | 9.31e-01 |
| GO:BP | GO:0061044 | negative regulation of vascular wound healing | 1 | 4 | 9.31e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 1 | 37 | 9.31e-01 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 5 | 15 | 9.31e-01 |
| GO:BP | GO:0009608 | response to symbiont | 1 | 2 | 9.32e-01 |
| GO:BP | GO:0009609 | response to symbiotic bacterium | 1 | 2 | 9.32e-01 |
| GO:BP | GO:0051697 | protein delipidation | 1 | 5 | 9.32e-01 |
| GO:BP | GO:0015671 | oxygen transport | 1 | 4 | 9.32e-01 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 2 | 6 | 9.32e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 9.32e-01 |
| GO:BP | GO:0021761 | limbic system development | 13 | 81 | 9.32e-01 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 3 | 27 | 9.32e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 9.32e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 19 | 104 | 9.32e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 2 | 105 | 9.32e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 3 | 24 | 9.32e-01 |
| GO:BP | GO:0019441 | tryptophan catabolic process to kynurenine | 1 | 1 | 9.33e-01 |
| GO:BP | GO:0030277 | maintenance of gastrointestinal epithelium | 2 | 8 | 9.33e-01 |
| GO:BP | GO:0051884 | regulation of timing of anagen | 1 | 5 | 9.33e-01 |
| GO:BP | GO:0009414 | response to water deprivation | 2 | 4 | 9.33e-01 |
| GO:BP | GO:0042640 | anagen | 1 | 5 | 9.33e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 2 | 118 | 9.33e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 1 | 188 | 9.33e-01 |
| GO:BP | GO:0048663 | neuron fate commitment | 2 | 29 | 9.34e-01 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 1 | 4 | 9.34e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 4 | 15 | 9.34e-01 |
| GO:BP | GO:0010842 | retina layer formation | 1 | 16 | 9.34e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 3 | 11 | 9.34e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 13 | 1684 | 9.34e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 8 | 37 | 9.34e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 9.34e-01 |
| GO:BP | GO:1901723 | negative regulation of cell proliferation involved in kidney development | 1 | 3 | 9.35e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 2 | 60 | 9.35e-01 |
| GO:BP | GO:0051014 | actin filament severing | 1 | 13 | 9.35e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 7 | 556 | 9.35e-01 |
| GO:BP | GO:1900150 | regulation of defense response to fungus | 1 | 3 | 9.35e-01 |
| GO:BP | GO:0000957 | mitochondrial RNA catabolic process | 1 | 6 | 9.35e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 2 | 69 | 9.35e-01 |
| GO:BP | GO:0099640 | axo-dendritic protein transport | 2 | 10 | 9.35e-01 |
| GO:BP | GO:0035112 | genitalia morphogenesis | 1 | 5 | 9.36e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 9.36e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 9.36e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 9.36e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 2 | 14 | 9.38e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 8 | 36 | 9.38e-01 |
| GO:BP | GO:0048712 | negative regulation of astrocyte differentiation | 1 | 9 | 9.38e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 5 | 2450 | 9.38e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 1 | 25 | 9.40e-01 |
| GO:BP | GO:0071384 | cellular response to corticosteroid stimulus | 2 | 47 | 9.40e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 19 | 3012 | 9.40e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 4 | 160 | 9.40e-01 |
| GO:BP | GO:0009786 | regulation of asymmetric cell division | 1 | 1 | 9.40e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 4 | 8 | 9.40e-01 |
| GO:BP | GO:1901289 | succinyl-CoA catabolic process | 1 | 6 | 9.40e-01 |
| GO:BP | GO:0046487 | glyoxylate metabolic process | 1 | 5 | 9.40e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 19 | 3080 | 9.41e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4241 | 9.41e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 2 | 47 | 9.41e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0006968 | cellular defense response | 2 | 15 | 9.41e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 4 | 8 | 9.41e-01 |
| GO:BP | GO:0001711 | endodermal cell fate commitment | 2 | 9 | 9.41e-01 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 1 | 5 | 9.41e-01 |
| GO:BP | GO:0072044 | collecting duct development | 2 | 6 | 9.41e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 19 | 3080 | 9.41e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 10 | 50 | 9.41e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 7 | 547 | 9.41e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 2 | 6 | 9.41e-01 |
| GO:BP | GO:0015855 | pyrimidine nucleobase transport | 1 | 3 | 9.41e-01 |
| GO:BP | GO:0015853 | adenine transport | 1 | 5 | 9.41e-01 |
| GO:BP | GO:0098711 | iron ion import across plasma membrane | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0015864 | pyrimidine nucleoside transport | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 9 | 719 | 9.41e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 5 | 9.41e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 5 | 100 | 9.41e-01 |
| GO:BP | GO:0046174 | polyol catabolic process | 1 | 7 | 9.41e-01 |
| GO:BP | GO:1903236 | regulation of leukocyte tethering or rolling | 1 | 6 | 9.41e-01 |
| GO:BP | GO:0015862 | uridine transport | 1 | 4 | 9.41e-01 |
| GO:BP | GO:0002825 | regulation of T-helper 1 type immune response | 4 | 10 | 9.41e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 2 | 9 | 9.41e-01 |
| GO:BP | GO:0061762 | CAMKK-AMPK signaling cascade | 1 | 5 | 9.41e-01 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 1 | 15 | 9.41e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 3 | 20 | 9.41e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 9.41e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 9.41e-01 |
| GO:BP | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin | 1 | 1 | 9.42e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 3 | 23 | 9.42e-01 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 3 | 13 | 9.42e-01 |
| GO:BP | GO:0036230 | granulocyte activation | 1 | 19 | 9.42e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 8 | 37 | 9.42e-01 |
| GO:BP | GO:2000402 | negative regulation of lymphocyte migration | 1 | 10 | 9.42e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 3 | 49 | 9.42e-01 |
| GO:BP | GO:0070640 | vitamin D3 metabolic process | 1 | 2 | 9.42e-01 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 2 | 8 | 9.42e-01 |
| GO:BP | GO:0051799 | negative regulation of hair follicle development | 1 | 5 | 9.42e-01 |
| GO:BP | GO:0006688 | glycosphingolipid biosynthetic process | 1 | 25 | 9.43e-01 |
| GO:BP | GO:0007216 | G protein-coupled glutamate receptor signaling pathway | 1 | 5 | 9.43e-01 |
| GO:BP | GO:0016570 | histone modification | 1 | 433 | 9.43e-01 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 1 | 5 | 9.43e-01 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 1 | 35 | 9.43e-01 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 6 | 14 | 9.43e-01 |
| GO:BP | GO:0060297 | regulation of sarcomere organization | 1 | 7 | 9.43e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 3 | 7 | 9.43e-01 |
| GO:BP | GO:0070779 | D-aspartate import across plasma membrane | 1 | 4 | 9.43e-01 |
| GO:BP | GO:0070777 | D-aspartate transport | 1 | 4 | 9.43e-01 |
| GO:BP | GO:0042428 | serotonin metabolic process | 1 | 6 | 9.43e-01 |
| GO:BP | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 9.43e-01 |
| GO:BP | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II | 1 | 15 | 9.43e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 6 | 41 | 9.43e-01 |
| GO:BP | GO:0048806 | genitalia development | 2 | 28 | 9.43e-01 |
| GO:BP | GO:0060795 | cell fate commitment involved in formation of primary germ layer | 5 | 18 | 9.43e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 9.43e-01 |
| GO:BP | GO:0090156 | intracellular sphingolipid homeostasis | 1 | 5 | 9.43e-01 |
| GO:BP | GO:0031638 | zymogen activation | 11 | 45 | 9.44e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 3 | 42 | 9.44e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 9.44e-01 |
| GO:BP | GO:0042989 | sequestering of actin monomers | 1 | 9 | 9.44e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 1 | 18 | 9.44e-01 |
| GO:BP | GO:0032847 | regulation of cellular pH reduction | 1 | 5 | 9.44e-01 |
| GO:BP | GO:0034134 | toll-like receptor 2 signaling pathway | 2 | 11 | 9.44e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 2 | 132 | 9.44e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 28 | 156 | 9.44e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 59 | 9.44e-01 |
| GO:BP | GO:0032275 | luteinizing hormone secretion | 1 | 6 | 9.44e-01 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 3 | 9 | 9.44e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 9.45e-01 |
| GO:BP | GO:0007289 | spermatid nucleus differentiation | 1 | 16 | 9.45e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 9.45e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 20 | 3169 | 9.45e-01 |
| GO:BP | GO:0071605 | monocyte chemotactic protein-1 production | 1 | 7 | 9.45e-01 |
| GO:BP | GO:0071637 | regulation of monocyte chemotactic protein-1 production | 1 | 7 | 9.45e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 1 | 60 | 9.45e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 1 | 46 | 9.45e-01 |
| GO:BP | GO:2000726 | negative regulation of cardiac muscle cell differentiation | 1 | 8 | 9.45e-01 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 1 | 5 | 9.45e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 1 | 10 | 9.45e-01 |
| GO:BP | GO:1900118 | negative regulation of execution phase of apoptosis | 2 | 6 | 9.45e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 1 | 10 | 9.45e-01 |
| GO:BP | GO:0022401 | negative adaptation of signaling pathway | 2 | 15 | 9.45e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 12 | 1914 | 9.45e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 3 | 330 | 9.45e-01 |
| GO:BP | GO:0048143 | astrocyte activation | 3 | 12 | 9.45e-01 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 2 | 35 | 9.45e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 2 | 13 | 9.45e-01 |
| GO:BP | GO:0001787 | natural killer cell proliferation | 1 | 6 | 9.45e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 1 | 5 | 9.46e-01 |
| GO:BP | GO:1902603 | carnitine transmembrane transport | 1 | 4 | 9.46e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 9.46e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 2 | 54 | 9.46e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 1 | 10 | 9.46e-01 |
| GO:BP | GO:1905564 | positive regulation of vascular endothelial cell proliferation | 1 | 15 | 9.47e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 2 | 134 | 9.47e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 3 | 33 | 9.47e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 5 | 23 | 9.48e-01 |
| GO:BP | GO:0042424 | catecholamine catabolic process | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence | 1 | 10 | 9.48e-01 |
| GO:BP | GO:0019614 | catechol-containing compound catabolic process | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 3 | 16 | 9.48e-01 |
| GO:BP | GO:0051641 | cellular localization | 16 | 2810 | 9.48e-01 |
| GO:BP | GO:0050907 | detection of chemical stimulus involved in sensory perception | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0032095 | regulation of response to food | 1 | 8 | 9.48e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 9 | 78 | 9.48e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 9.48e-01 |
| GO:BP | GO:0035457 | cellular response to interferon-alpha | 1 | 7 | 9.48e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 9.48e-01 |
| GO:BP | GO:1905906 | regulation of amyloid fibril formation | 3 | 14 | 9.48e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 1 | 20 | 9.48e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 1 | 10 | 9.48e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 1 | 20 | 9.48e-01 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 1 | 66 | 9.48e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 1 | 13 | 9.48e-01 |
| GO:BP | GO:0030263 | apoptotic chromosome condensation | 1 | 4 | 9.48e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 2 | 136 | 9.48e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 2 | 111 | 9.48e-01 |
| GO:BP | GO:0099612 | protein localization to axon | 1 | 6 | 9.48e-01 |
| GO:BP | GO:0007501 | mesodermal cell fate specification | 1 | 5 | 9.48e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 3 | 7 | 9.49e-01 |
| GO:BP | GO:0008380 | RNA splicing | 2 | 419 | 9.49e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 4 | 4617 | 9.49e-01 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 5 | 16 | 9.49e-01 |
| GO:BP | GO:0014823 | response to activity | 3 | 46 | 9.50e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 13 | 44 | 9.50e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 2 | 25 | 9.50e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 12 | 36 | 9.50e-01 |
| GO:BP | GO:1901523 | icosanoid catabolic process | 1 | 2 | 9.50e-01 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 1 | 5 | 9.50e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 2 | 91 | 9.50e-01 |
| GO:BP | GO:0006776 | vitamin A metabolic process | 1 | 3 | 9.50e-01 |
| GO:BP | GO:0046104 | thymidine metabolic process | 1 | 5 | 9.50e-01 |
| GO:BP | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 5 | 9.50e-01 |
| GO:BP | GO:0001839 | neural plate morphogenesis | 1 | 4 | 9.51e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 1 | 20 | 9.51e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 9.51e-01 |
| GO:BP | GO:0035025 | positive regulation of Rho protein signal transduction | 1 | 22 | 9.51e-01 |
| GO:BP | GO:0060430 | lung saccule development | 1 | 7 | 9.51e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 2 | 37 | 9.51e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 2 | 37 | 9.51e-01 |
| GO:BP | GO:0045875 | negative regulation of sister chromatid cohesion | 1 | 5 | 9.52e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 5 | 21 | 9.52e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 12 | 517 | 9.52e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 2 | 18 | 9.52e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 3 | 12 | 9.52e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 1 | 37 | 9.52e-01 |
| GO:BP | GO:0035813 | regulation of renal sodium excretion | 1 | 10 | 9.52e-01 |
| GO:BP | GO:0009642 | response to light intensity | 1 | 11 | 9.52e-01 |
| GO:BP | GO:0060263 | regulation of respiratory burst | 2 | 10 | 9.52e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 9.52e-01 |
| GO:BP | GO:2000380 | regulation of mesoderm development | 1 | 3 | 9.52e-01 |
| GO:BP | GO:0043031 | negative regulation of macrophage activation | 2 | 9 | 9.52e-01 |
| GO:BP | GO:0042791 | 5S class rRNA transcription by RNA polymerase III | 1 | 6 | 9.53e-01 |
| GO:BP | GO:0048232 | male gamete generation | 3 | 343 | 9.53e-01 |
| GO:BP | GO:0043132 | NAD transport | 1 | 3 | 9.53e-01 |
| GO:BP | GO:0007602 | phototransduction | 1 | 17 | 9.53e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 9.53e-01 |
| GO:BP | GO:0090131 | mesenchyme migration | 1 | 4 | 9.53e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 9.54e-01 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 10 | 25 | 9.54e-01 |
| GO:BP | GO:0140894 | endolysosomal toll-like receptor signaling pathway | 2 | 24 | 9.54e-01 |
| GO:BP | GO:0007339 | binding of sperm to zona pellucida | 6 | 20 | 9.54e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 3 | 4455 | 9.54e-01 |
| GO:BP | GO:0071286 | cellular response to magnesium ion | 1 | 3 | 9.54e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 5 | 192 | 9.54e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 9 | 25 | 9.54e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 2 | 50 | 9.54e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 9 | 25 | 9.54e-01 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 12 | 36 | 9.54e-01 |
| GO:BP | GO:0045956 | positive regulation of calcium ion-dependent exocytosis | 2 | 11 | 9.54e-01 |
| GO:BP | GO:0035610 | protein side chain deglutamylation | 1 | 2 | 9.54e-01 |
| GO:BP | GO:0001976 | nervous system process involved in regulation of systemic arterial blood pressure | 1 | 9 | 9.54e-01 |
| GO:BP | GO:0120252 | hydrocarbon metabolic process | 1 | 2 | 9.54e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 2 | 191 | 9.54e-01 |
| GO:BP | GO:0006397 | mRNA processing | 2 | 450 | 9.54e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 10 | 9.54e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 9.54e-01 |
| GO:BP | GO:0016045 | detection of bacterium | 1 | 8 | 9.54e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 15 | 43 | 9.54e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 7 | 111 | 9.55e-01 |
| GO:BP | GO:0036123 | histone H3-K9 dimethylation | 1 | 3 | 9.55e-01 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 20 | 94 | 9.55e-01 |
| GO:BP | GO:0070593 | dendrite self-avoidance | 1 | 11 | 9.55e-01 |
| GO:BP | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway | 1 | 4 | 9.55e-01 |
| GO:BP | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production | 2 | 5 | 9.55e-01 |
| GO:BP | GO:0045332 | phospholipid translocation | 1 | 33 | 9.55e-01 |
| GO:BP | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production | 1 | 6 | 9.55e-01 |
| GO:BP | GO:0071609 | chemokine (C-C motif) ligand 5 production | 1 | 6 | 9.55e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 3 | 168 | 9.55e-01 |
| GO:BP | GO:0019081 | viral translation | 1 | 18 | 9.55e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 4 | 12 | 9.55e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 2 | 77 | 9.55e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 3 | 13 | 9.55e-01 |
| GO:BP | GO:0060457 | negative regulation of digestive system process | 1 | 7 | 9.56e-01 |
| GO:BP | GO:0050909 | sensory perception of taste | 1 | 13 | 9.56e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 2 | 95 | 9.56e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 2 | 8 | 9.56e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 3 | 169 | 9.56e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 3 | 11 | 9.56e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 5 | 22 | 9.56e-01 |
| GO:BP | GO:0006958 | complement activation, classical pathway | 1 | 16 | 9.56e-01 |
| GO:BP | GO:0042953 | lipoprotein transport | 1 | 14 | 9.56e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 3 | 4292 | 9.56e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 9.56e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 1 | 32 | 9.56e-01 |
| GO:BP | GO:0035095 | behavioral response to nicotine | 1 | 3 | 9.56e-01 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 1 | 5 | 9.56e-01 |
| GO:BP | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | 1 | 2 | 9.56e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 2 | 10 | 9.56e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 2 | 18 | 9.56e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 15 | 64 | 9.56e-01 |
| GO:BP | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | 1 | 13 | 9.56e-01 |
| GO:BP | GO:0097278 | complement-dependent cytotoxicity | 1 | 4 | 9.57e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 4 | 45 | 9.57e-01 |
| GO:BP | GO:0048557 | embryonic digestive tract morphogenesis | 1 | 8 | 9.57e-01 |
| GO:BP | GO:0009820 | alkaloid metabolic process | 1 | 2 | 9.57e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 1 | 79 | 9.57e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 4 | 45 | 9.57e-01 |
| GO:BP | GO:2000110 | negative regulation of macrophage apoptotic process | 1 | 3 | 9.57e-01 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 15 | 9.57e-01 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 3 | 30 | 9.57e-01 |
| GO:BP | GO:0045579 | positive regulation of B cell differentiation | 3 | 10 | 9.57e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 3 | 16 | 9.57e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 12 | 34 | 9.57e-01 |
| GO:BP | GO:0070318 | positive regulation of G0 to G1 transition | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 13 | 30 | 9.57e-01 |
| GO:BP | GO:0042308 | negative regulation of protein import into nucleus | 2 | 13 | 9.57e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 19 | 272 | 9.57e-01 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 3 | 22 | 9.57e-01 |
| GO:BP | GO:0018197 | peptidyl-aspartic acid modification | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 9.57e-01 |
| GO:BP | GO:0042220 | response to cocaine | 1 | 28 | 9.57e-01 |
| GO:BP | GO:0051933 | amino acid neurotransmitter reuptake | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 3 | 22 | 9.57e-01 |
| GO:BP | GO:0048536 | spleen development | 1 | 28 | 9.57e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 1 | 18 | 9.57e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 1 | 9 | 9.57e-01 |
| GO:BP | GO:0032026 | response to magnesium ion | 2 | 10 | 9.57e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 9.57e-01 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 1 | 27 | 9.57e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 6 | 631 | 9.57e-01 |
| GO:BP | GO:0071599 | otic vesicle development | 2 | 10 | 9.57e-01 |
| GO:BP | GO:1901317 | regulation of flagellated sperm motility | 1 | 6 | 9.57e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 5 | 28 | 9.57e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 9 | 9.57e-01 |
| GO:BP | GO:0098664 | G protein-coupled serotonin receptor signaling pathway | 1 | 8 | 9.57e-01 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 7 | 39 | 9.57e-01 |
| GO:BP | GO:0002023 | reduction of food intake in response to dietary excess | 1 | 2 | 9.57e-01 |
| GO:BP | GO:0150172 | regulation of phosphatidylcholine metabolic process | 1 | 10 | 9.58e-01 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 1 | 6 | 9.58e-01 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 2 | 9 | 9.58e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 9.58e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 2 | 11 | 9.59e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 3 | 4570 | 9.59e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 2 | 15 | 9.59e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 1 | 433 | 9.59e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 9.59e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 5 | 38 | 9.59e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 14 | 151 | 9.59e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 61 | 9.59e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 2 | 27 | 9.59e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 61 | 9.59e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 8 | 27 | 9.60e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 9.60e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 9.60e-01 |
| GO:BP | GO:1905150 | regulation of voltage-gated sodium channel activity | 1 | 6 | 9.60e-01 |
| GO:BP | GO:0007196 | adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway | 1 | 1 | 9.60e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 2 | 17 | 9.60e-01 |
| GO:BP | GO:0010224 | response to UV-B | 5 | 17 | 9.60e-01 |
| GO:BP | GO:0032274 | gonadotropin secretion | 2 | 10 | 9.60e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 2 | 15 | 9.60e-01 |
| GO:BP | GO:0006949 | syncytium formation | 2 | 39 | 9.60e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 4 | 87 | 9.60e-01 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 8 | 35 | 9.60e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 3 | 4596 | 9.61e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 4 | 10 | 9.61e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 3 | 13 | 9.61e-01 |
| GO:BP | GO:0051180 | vitamin transport | 2 | 28 | 9.61e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 1 | 13 | 9.61e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 17 | 166 | 9.61e-01 |
| GO:BP | GO:0010039 | response to iron ion | 5 | 24 | 9.61e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 9 | 36 | 9.61e-01 |
| GO:BP | GO:0051693 | actin filament capping | 2 | 34 | 9.61e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 2 | 2 | 9.61e-01 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 1 | 8 | 9.61e-01 |
| GO:BP | GO:1903537 | meiotic cell cycle process involved in oocyte maturation | 1 | 5 | 9.61e-01 |
| GO:BP | GO:0006956 | complement activation | 2 | 23 | 9.62e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 4 | 24 | 9.62e-01 |
| GO:BP | GO:0015879 | carnitine transport | 1 | 4 | 9.62e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 1 | 142 | 9.62e-01 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 1 | 31 | 9.62e-01 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 1 | 19 | 9.62e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 9.62e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 9.62e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 9.62e-01 |
| GO:BP | GO:0045907 | positive regulation of vasoconstriction | 2 | 15 | 9.62e-01 |
| GO:BP | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation | 1 | 7 | 9.62e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 9.62e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 9.62e-01 |
| GO:BP | GO:0042421 | norepinephrine biosynthetic process | 1 | 2 | 9.62e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 9.62e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 1 | 19 | 9.62e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 1 | 64 | 9.62e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 7 | 23 | 9.62e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 9.62e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 3 | 4417 | 9.62e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 4 | 146 | 9.63e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 5 | 24 | 9.63e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 9.63e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 4 | 83 | 9.63e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 1 | 28 | 9.63e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 9.63e-01 |
| GO:BP | GO:0007498 | mesoderm development | 12 | 89 | 9.63e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 12 | 540 | 9.63e-01 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 1 | 13 | 9.63e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 7 | 30 | 9.63e-01 |
| GO:BP | GO:0006265 | DNA topological change | 2 | 10 | 9.63e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 1 | 93 | 9.63e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 2 | 8 | 9.63e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 3 | 75 | 9.63e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 2 | 9 | 9.63e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 4 | 8 | 9.63e-01 |
| GO:BP | GO:0060004 | reflex | 5 | 9 | 9.63e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 2 | 53 | 9.63e-01 |
| GO:BP | GO:1902260 | negative regulation of delayed rectifier potassium channel activity | 1 | 5 | 9.64e-01 |
| GO:BP | GO:0070314 | G1 to G0 transition | 3 | 15 | 9.64e-01 |
| GO:BP | GO:0140009 | L-aspartate import across plasma membrane | 1 | 5 | 9.64e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 7 | 251 | 9.64e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 5 | 123 | 9.65e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 2 | 211 | 9.65e-01 |
| GO:BP | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | 1 | 2 | 9.65e-01 |
| GO:BP | GO:0002385 | mucosal immune response | 1 | 11 | 9.65e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 12 | 597 | 9.65e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 12 | 597 | 9.65e-01 |
| GO:BP | GO:0070317 | negative regulation of G0 to G1 transition | 1 | 5 | 9.65e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 5 | 22 | 9.65e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 1 | 94 | 9.65e-01 |
| GO:BP | GO:0072205 | metanephric collecting duct development | 1 | 5 | 9.65e-01 |
| GO:BP | GO:0021536 | diencephalon development | 3 | 41 | 9.65e-01 |
| GO:BP | GO:0140507 | granzyme-mediated programmed cell death signaling pathway | 1 | 3 | 9.65e-01 |
| GO:BP | GO:0097709 | connective tissue replacement | 1 | 8 | 9.65e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 3 | 9 | 9.65e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 3 | 58 | 9.66e-01 |
| GO:BP | GO:1904970 | brush border assembly | 1 | 2 | 9.66e-01 |
| GO:BP | GO:0034204 | lipid translocation | 1 | 38 | 9.66e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 9.66e-01 |
| GO:BP | GO:0016554 | cytidine to uridine editing | 1 | 6 | 9.66e-01 |
| GO:BP | GO:0009972 | cytidine deamination | 1 | 6 | 9.66e-01 |
| GO:BP | GO:0046087 | cytidine metabolic process | 1 | 6 | 9.66e-01 |
| GO:BP | GO:0006216 | cytidine catabolic process | 1 | 6 | 9.66e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 9.67e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 9.67e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 13 | 46 | 9.67e-01 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 9.67e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 1 | 95 | 9.67e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 9.67e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2 | 5017 | 9.67e-01 |
| GO:BP | GO:0099004 | calmodulin dependent kinase signaling pathway | 1 | 6 | 9.67e-01 |
| GO:BP | GO:0010496 | intercellular transport | 1 | 4 | 9.67e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 2 | 29 | 9.67e-01 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 9.67e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 2 | 28 | 9.67e-01 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 2 | 27 | 9.67e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 8 | 109 | 9.67e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 3 | 59 | 9.67e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 2 | 20 | 9.68e-01 |
| GO:BP | GO:0006572 | tyrosine catabolic process | 1 | 2 | 9.68e-01 |
| GO:BP | GO:0150078 | positive regulation of neuroinflammatory response | 1 | 4 | 9.68e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 17 | 49 | 9.68e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 1 | 69 | 9.68e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 3 | 9 | 9.68e-01 |
| GO:BP | GO:2000321 | positive regulation of T-helper 17 cell differentiation | 1 | 3 | 9.68e-01 |
| GO:BP | GO:1905245 | regulation of aspartic-type peptidase activity | 1 | 11 | 9.68e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 3 | 4747 | 9.68e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 18 | 155 | 9.68e-01 |
| GO:BP | GO:0070189 | kynurenine metabolic process | 1 | 6 | 9.68e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 5 | 20 | 9.68e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 8 | 32 | 9.69e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 1 | 4 | 9.69e-01 |
| GO:BP | GO:0003356 | regulation of cilium beat frequency | 4 | 10 | 9.70e-01 |
| GO:BP | GO:0051573 | negative regulation of histone H3-K9 methylation | 1 | 5 | 9.70e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 18 | 55 | 9.70e-01 |
| GO:BP | GO:0071492 | cellular response to UV-A | 2 | 8 | 9.70e-01 |
| GO:BP | GO:0042362 | fat-soluble vitamin biosynthetic process | 2 | 6 | 9.70e-01 |
| GO:BP | GO:0035608 | protein deglutamylation | 1 | 2 | 9.70e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 1 | 49 | 9.70e-01 |
| GO:BP | GO:0097164 | ammonium ion metabolic process | 2 | 14 | 9.71e-01 |
| GO:BP | GO:1901160 | primary amino compound metabolic process | 1 | 8 | 9.71e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 12 | 9.71e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 8 | 16 | 9.71e-01 |
| GO:BP | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia | 1 | 9 | 9.71e-01 |
| GO:BP | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0002399 | MHC class II protein complex assembly | 2 | 4 | 9.71e-01 |
| GO:BP | GO:0002283 | neutrophil activation involved in immune response | 1 | 7 | 9.71e-01 |
| GO:BP | GO:0098543 | detection of other organism | 1 | 9 | 9.71e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 1 | 993 | 9.71e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 4 | 98 | 9.71e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 9.71e-01 |
| GO:BP | GO:1990544 | mitochondrial ATP transmembrane transport | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0032276 | regulation of gonadotropin secretion | 1 | 6 | 9.71e-01 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 1 | 11 | 9.71e-01 |
| GO:BP | GO:0140021 | mitochondrial ADP transmembrane transport | 1 | 4 | 9.71e-01 |
| GO:BP | GO:0045576 | mast cell activation | 3 | 35 | 9.71e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 13 | 46 | 9.71e-01 |
| GO:BP | GO:1903028 | positive regulation of opsonization | 1 | 1 | 9.71e-01 |
| GO:BP | GO:0018242 | protein O-linked glycosylation via serine | 2 | 7 | 9.71e-01 |
| GO:BP | GO:0006449 | regulation of translational termination | 1 | 10 | 9.71e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 9.71e-01 |
| GO:BP | GO:0002503 | peptide antigen assembly with MHC class II protein complex | 2 | 4 | 9.71e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 1 | 994 | 9.71e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 4 | 51 | 9.71e-01 |
| GO:BP | GO:0006491 | N-glycan processing | 2 | 18 | 9.71e-01 |
| GO:BP | GO:0002553 | histamine secretion by mast cell | 1 | 8 | 9.71e-01 |
| GO:BP | GO:0002349 | histamine production involved in inflammatory response | 1 | 8 | 9.71e-01 |
| GO:BP | GO:0002441 | histamine secretion involved in inflammatory response | 1 | 8 | 9.71e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 9.71e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 9.72e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 1 | 11 | 9.72e-01 |
| GO:BP | GO:0042178 | xenobiotic catabolic process | 1 | 12 | 9.72e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 3 | 10 | 9.72e-01 |
| GO:BP | GO:1903371 | regulation of endoplasmic reticulum tubular network organization | 1 | 6 | 9.72e-01 |
| GO:BP | GO:0001556 | oocyte maturation | 8 | 22 | 9.72e-01 |
| GO:BP | GO:0000480 | endonucleolytic cleavage in 5’-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 8 | 9.72e-01 |
| GO:BP | GO:0050886 | endocrine process | 1 | 57 | 9.72e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 9.73e-01 |
| GO:BP | GO:1905920 | positive regulation of CoA-transferase activity | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 2 | 37 | 9.73e-01 |
| GO:BP | GO:1901896 | positive regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 6 | 9.73e-01 |
| GO:BP | GO:1905918 | regulation of CoA-transferase activity | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 1 | 7806 | 9.73e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 3 | 4946 | 9.73e-01 |
| GO:BP | GO:0060638 | mesenchymal-epithelial cell signaling | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0034354 | ‘de novo’ NAD biosynthetic process from tryptophan | 1 | 1 | 9.73e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 9.73e-01 |
| GO:BP | GO:0007210 | serotonin receptor signaling pathway | 1 | 8 | 9.73e-01 |
| GO:BP | GO:0120197 | mucociliary clearance | 1 | 5 | 9.73e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 1 | 248 | 9.73e-01 |
| GO:BP | GO:0002691 | regulation of cellular extravasation | 5 | 24 | 9.74e-01 |
| GO:BP | GO:0006775 | fat-soluble vitamin metabolic process | 1 | 20 | 9.74e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 1 | 20 | 9.74e-01 |
| GO:BP | GO:0035022 | positive regulation of Rac protein signal transduction | 1 | 5 | 9.74e-01 |
| GO:BP | GO:0008078 | mesodermal cell migration | 1 | 3 | 9.74e-01 |
| GO:BP | GO:0006000 | fructose metabolic process | 3 | 10 | 9.75e-01 |
| GO:BP | GO:0016553 | base conversion or substitution editing | 2 | 10 | 9.75e-01 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 1 | 13 | 9.75e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 9.75e-01 |
| GO:BP | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 3 | 8 | 9.75e-01 |
| GO:BP | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0090186 | regulation of pancreatic juice secretion | 1 | 6 | 9.75e-01 |
| GO:BP | GO:0021554 | optic nerve development | 2 | 7 | 9.75e-01 |
| GO:BP | GO:0090091 | positive regulation of extracellular matrix disassembly | 1 | 6 | 9.75e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 2 | 80 | 9.75e-01 |
| GO:BP | GO:0051798 | positive regulation of hair follicle development | 2 | 9 | 9.75e-01 |
| GO:BP | GO:0014002 | astrocyte development | 8 | 27 | 9.75e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 3 | 7 | 9.75e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 1 | 50 | 9.75e-01 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 2 | 29 | 9.75e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 4 | 93 | 9.75e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 1 | 74 | 9.75e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 1 | 74 | 9.75e-01 |
| GO:BP | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity | 1 | 3 | 9.75e-01 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 5 | 14 | 9.75e-01 |
| GO:BP | GO:0021796 | cerebral cortex regionalization | 1 | 2 | 9.75e-01 |
| GO:BP | GO:1900426 | positive regulation of defense response to bacterium | 1 | 6 | 9.75e-01 |
| GO:BP | GO:0010572 | positive regulation of platelet activation | 1 | 3 | 9.75e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 1 | 101 | 9.75e-01 |
| GO:BP | GO:0006907 | pinocytosis | 4 | 18 | 9.75e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 9.75e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 8 | 34 | 9.75e-01 |
| GO:BP | GO:0090238 | positive regulation of arachidonic acid secretion | 1 | 2 | 9.76e-01 |
| GO:BP | GO:0032098 | regulation of appetite | 1 | 8 | 9.76e-01 |
| GO:BP | GO:0097017 | renal protein absorption | 1 | 5 | 9.76e-01 |
| GO:BP | GO:0051957 | positive regulation of amino acid transport | 4 | 16 | 9.76e-01 |
| GO:BP | GO:0090659 | walking behavior | 7 | 25 | 9.76e-01 |
| GO:BP | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway | 1 | 6 | 9.76e-01 |
| GO:BP | GO:0032730 | positive regulation of interleukin-1 alpha production | 1 | 3 | 9.76e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 14 | 119 | 9.76e-01 |
| GO:BP | GO:0002029 | desensitization of G protein-coupled receptor signaling pathway | 4 | 14 | 9.76e-01 |
| GO:BP | GO:0019336 | phenol-containing compound catabolic process | 1 | 6 | 9.77e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 9.77e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 3 | 21 | 9.78e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 3 | 12 | 9.78e-01 |
| GO:BP | GO:0071385 | cellular response to glucocorticoid stimulus | 9 | 40 | 9.78e-01 |
| GO:BP | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway | 2 | 9 | 9.78e-01 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 9.78e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 2 | 12 | 9.78e-01 |
| GO:BP | GO:0018344 | protein geranylgeranylation | 1 | 6 | 9.78e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 2 | 3600 | 9.78e-01 |
| GO:BP | GO:0002076 | osteoblast development | 4 | 12 | 9.78e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 8 | 57 | 9.78e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 2 | 168 | 9.78e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 2 | 30 | 9.79e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 3 | 317 | 9.79e-01 |
| GO:BP | GO:0080154 | regulation of fertilization | 1 | 7 | 9.79e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 20 | 57 | 9.79e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 4 | 35 | 9.79e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 5 | 21 | 9.79e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 9.79e-01 |
| GO:BP | GO:0002759 | regulation of antimicrobial humoral response | 1 | 3 | 9.80e-01 |
| GO:BP | GO:1903817 | negative regulation of voltage-gated potassium channel activity | 1 | 6 | 9.80e-01 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 1 | 87 | 9.80e-01 |
| GO:BP | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 9.80e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 15 | 796 | 9.80e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 9.80e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 9.80e-01 |
| GO:BP | GO:0042755 | eating behavior | 2 | 16 | 9.80e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 9.80e-01 |
| GO:BP | GO:0048742 | regulation of skeletal muscle fiber development | 2 | 8 | 9.81e-01 |
| GO:BP | GO:0033523 | histone H2B ubiquitination | 1 | 10 | 9.81e-01 |
| GO:BP | GO:0045471 | response to ethanol | 4 | 73 | 9.81e-01 |
| GO:BP | GO:0006545 | glycine biosynthetic process | 1 | 4 | 9.81e-01 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 1 | 23 | 9.81e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 9.81e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 1 | 1173 | 9.82e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 1 | 30 | 9.82e-01 |
| GO:BP | GO:1904936 | interneuron migration | 1 | 6 | 9.82e-01 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 2 | 4 | 9.82e-01 |
| GO:BP | GO:1900107 | regulation of nodal signaling pathway | 1 | 3 | 9.82e-01 |
| GO:BP | GO:0150079 | negative regulation of neuroinflammatory response | 1 | 6 | 9.82e-01 |
| GO:BP | GO:1904749 | regulation of protein localization to nucleolus | 1 | 7 | 9.82e-01 |
| GO:BP | GO:0032222 | regulation of synaptic transmission, cholinergic | 1 | 3 | 9.82e-01 |
| GO:BP | GO:0051026 | chiasma assembly | 1 | 3 | 9.82e-01 |
| GO:BP | GO:0010467 | gene expression | 3 | 4587 | 9.82e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 2 | 31 | 9.83e-01 |
| GO:BP | GO:0045838 | positive regulation of membrane potential | 2 | 12 | 9.83e-01 |
| GO:BP | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly | 1 | 5 | 9.83e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 1 | 16 | 9.83e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 7 | 65 | 9.83e-01 |
| GO:BP | GO:0002016 | regulation of blood volume by renin-angiotensin | 1 | 5 | 9.84e-01 |
| GO:BP | GO:0002003 | angiotensin maturation | 1 | 5 | 9.84e-01 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 1 | 7 | 9.84e-01 |
| GO:BP | GO:0048102 | autophagic cell death | 1 | 6 | 9.84e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 1 | 4174 | 9.84e-01 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 1 | 21 | 9.84e-01 |
| GO:BP | GO:0031424 | keratinization | 2 | 13 | 9.84e-01 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 1 | 21 | 9.84e-01 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 6 | 8 | 9.84e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 2 | 38 | 9.84e-01 |
| GO:BP | GO:1902047 | polyamine transmembrane transport | 2 | 10 | 9.84e-01 |
| GO:BP | GO:0021514 | ventral spinal cord interneuron differentiation | 1 | 5 | 9.84e-01 |
| GO:BP | GO:0023035 | CD40 signaling pathway | 3 | 11 | 9.84e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 2 | 31 | 9.84e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 2 | 4 | 9.84e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 5 | 29 | 9.84e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 7 | 43 | 9.84e-01 |
| GO:BP | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 6 | 9.84e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 2 | 33 | 9.84e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 9 | 65 | 9.84e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 2 | 3 | 9.84e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 2 | 13 | 9.84e-01 |
| GO:BP | GO:0003341 | cilium movement | 3 | 97 | 9.84e-01 |
| GO:BP | GO:0033278 | cell proliferation in midbrain | 1 | 4 | 9.85e-01 |
| GO:BP | GO:2000328 | regulation of T-helper 17 cell lineage commitment | 1 | 1 | 9.85e-01 |
| GO:BP | GO:0070100 | negative regulation of chemokine-mediated signaling pathway | 1 | 6 | 9.85e-01 |
| GO:BP | GO:0014075 | response to amine | 6 | 34 | 9.85e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 1 | 194 | 9.85e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 5 | 101 | 9.85e-01 |
| GO:BP | GO:0009415 | response to water | 3 | 9 | 9.86e-01 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 1 | 9 | 9.86e-01 |
| GO:BP | GO:0015744 | succinate transport | 1 | 6 | 9.86e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 9.86e-01 |
| GO:BP | GO:0006868 | glutamine transport | 1 | 8 | 9.86e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 1 | 6 | 9.86e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 14 | 55 | 9.86e-01 |
| GO:BP | GO:0051918 | negative regulation of fibrinolysis | 1 | 8 | 9.87e-01 |
| GO:BP | GO:0035562 | negative regulation of chromatin binding | 1 | 8 | 9.87e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 9.87e-01 |
| GO:BP | GO:0046469 | platelet activating factor metabolic process | 1 | 5 | 9.87e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 9.87e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 1 | 156 | 9.87e-01 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 6 | 23 | 9.87e-01 |
| GO:BP | GO:0090237 | regulation of arachidonic acid secretion | 1 | 2 | 9.87e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 1 | 130 | 9.87e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 9.87e-01 |
| GO:BP | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell | 1 | 3 | 9.87e-01 |
| GO:BP | GO:0071702 | organic substance transport | 11 | 2116 | 9.87e-01 |
| GO:BP | GO:0045058 | T cell selection | 1 | 22 | 9.87e-01 |
| GO:BP | GO:0052695 | cellular glucuronidation | 1 | 2 | 9.88e-01 |
| GO:BP | GO:1903027 | regulation of opsonization | 1 | 1 | 9.88e-01 |
| GO:BP | GO:0044546 | NLRP3 inflammasome complex assembly | 1 | 23 | 9.88e-01 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 1 | 5 | 9.88e-01 |
| GO:BP | GO:0001821 | histamine secretion | 1 | 9 | 9.88e-01 |
| GO:BP | GO:0090527 | actin filament reorganization | 1 | 10 | 9.89e-01 |
| GO:BP | GO:0003044 | regulation of systemic arterial blood pressure mediated by a chemical signal | 4 | 30 | 9.89e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 2 | 17 | 9.89e-01 |
| GO:BP | GO:0098581 | detection of external biotic stimulus | 1 | 11 | 9.90e-01 |
| GO:BP | GO:0051955 | regulation of amino acid transport | 2 | 28 | 9.90e-01 |
| GO:BP | GO:0035739 | CD4-positive, alpha-beta T cell proliferation | 2 | 12 | 9.90e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 2 | 8 | 9.90e-01 |
| GO:BP | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation | 2 | 12 | 9.90e-01 |
| GO:BP | GO:0046879 | hormone secretion | 5 | 210 | 9.90e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 2 | 168 | 9.90e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 2 | 4048 | 9.90e-01 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 11 | 57 | 9.91e-01 |
| GO:BP | GO:0032490 | detection of molecule of bacterial origin | 1 | 4 | 9.91e-01 |
| GO:BP | GO:0002002 | regulation of angiotensin levels in blood | 1 | 6 | 9.91e-01 |
| GO:BP | GO:0010918 | positive regulation of mitochondrial membrane potential | 1 | 9 | 9.91e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 2 | 45 | 9.92e-01 |
| GO:BP | GO:0046164 | alcohol catabolic process | 1 | 23 | 9.92e-01 |
| GO:BP | GO:0042368 | vitamin D biosynthetic process | 1 | 4 | 9.92e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 2 | 11 | 9.92e-01 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 1 | 5 | 9.92e-01 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 1 | 12 | 9.92e-01 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 1 | 12 | 9.92e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 2 | 12 | 9.93e-01 |
| GO:BP | GO:0140632 | canonical inflammasome complex assembly | 1 | 23 | 9.93e-01 |
| GO:BP | GO:0001914 | regulation of T cell mediated cytotoxicity | 2 | 17 | 9.93e-01 |
| GO:BP | GO:0002839 | positive regulation of immune response to tumor cell | 1 | 7 | 9.93e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 2 | 9 | 9.93e-01 |
| GO:BP | GO:0002836 | positive regulation of response to tumor cell | 1 | 7 | 9.93e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 6 | 105 | 9.93e-01 |
| GO:BP | GO:0032693 | negative regulation of interleukin-10 production | 1 | 7 | 9.93e-01 |
| GO:BP | GO:0001886 | endothelial cell morphogenesis | 1 | 10 | 9.93e-01 |
| GO:BP | GO:0032645 | regulation of granulocyte macrophage colony-stimulating factor production | 1 | 5 | 9.93e-01 |
| GO:BP | GO:0032259 | methylation | 20 | 313 | 9.93e-01 |
| GO:BP | GO:0001990 | regulation of systemic arterial blood pressure by hormone | 1 | 22 | 9.93e-01 |
| GO:BP | GO:0032604 | granulocyte macrophage colony-stimulating factor production | 1 | 5 | 9.93e-01 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 1 | 1 | 9.93e-01 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 1 | 1 | 9.93e-01 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 2 | 20 | 9.93e-01 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 3 | 17 | 9.93e-01 |
| GO:BP | GO:0046641 | positive regulation of alpha-beta T cell proliferation | 2 | 11 | 9.93e-01 |
| GO:BP | GO:0001916 | positive regulation of T cell mediated cytotoxicity | 4 | 13 | 9.93e-01 |
| GO:BP | GO:0032735 | positive regulation of interleukin-12 production | 6 | 19 | 9.94e-01 |
| GO:BP | GO:0009914 | hormone transport | 5 | 215 | 9.94e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 9.94e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 4 | 78 | 9.94e-01 |
| GO:BP | GO:0061438 | renal system vasculature morphogenesis | 1 | 7 | 9.94e-01 |
| GO:BP | GO:0032489 | regulation of Cdc42 protein signal transduction | 1 | 6 | 9.94e-01 |
| GO:BP | GO:0061439 | kidney vasculature morphogenesis | 1 | 7 | 9.94e-01 |
| GO:BP | GO:0007338 | single fertilization | 2 | 70 | 9.94e-01 |
| GO:BP | GO:0001906 | cell killing | 2 | 69 | 9.94e-01 |
| GO:BP | GO:0031204 | post-translational protein targeting to membrane, translocation | 1 | 7 | 9.94e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 11 | 34 | 9.94e-01 |
| GO:BP | GO:1905907 | negative regulation of amyloid fibril formation | 1 | 10 | 9.94e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 5 | 108 | 9.94e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 7 | 35 | 9.94e-01 |
| GO:BP | GO:0021762 | substantia nigra development | 2 | 41 | 9.94e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 13 | 36 | 9.94e-01 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 1 | 12 | 9.94e-01 |
| GO:BP | GO:0150077 | regulation of neuroinflammatory response | 3 | 13 | 9.94e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 3 | 64 | 9.94e-01 |
| GO:BP | GO:0032650 | regulation of interleukin-1 alpha production | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 10 | 52 | 9.94e-01 |
| GO:BP | GO:0043320 | natural killer cell degranulation | 1 | 6 | 9.94e-01 |
| GO:BP | GO:0032610 | interleukin-1 alpha production | 1 | 4 | 9.94e-01 |
| GO:BP | GO:0001840 | neural plate development | 1 | 7 | 9.94e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 17 | 881 | 9.95e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 2 | 74 | 9.95e-01 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 2 | 24 | 9.95e-01 |
| GO:BP | GO:0006863 | purine nucleobase transport | 1 | 7 | 9.95e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 9.95e-01 |
| GO:BP | GO:0048539 | bone marrow development | 1 | 7 | 9.95e-01 |
| GO:BP | GO:0002858 | regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 9.95e-01 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 1 | 17 | 9.95e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 2 | 6 | 9.95e-01 |
| GO:BP | GO:0001991 | regulation of systemic arterial blood pressure by circulatory renin-angiotensin | 1 | 10 | 9.95e-01 |
| GO:BP | GO:0017121 | plasma membrane phospholipid scrambling | 2 | 13 | 9.95e-01 |
| GO:BP | GO:0036150 | phosphatidylserine acyl-chain remodeling | 1 | 7 | 9.95e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 2 | 23 | 9.95e-01 |
| GO:BP | GO:0002501 | peptide antigen assembly with MHC protein complex | 2 | 8 | 9.95e-01 |
| GO:BP | GO:0002396 | MHC protein complex assembly | 2 | 8 | 9.95e-01 |
| GO:BP | GO:0035270 | endocrine system development | 5 | 86 | 9.95e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 9.95e-01 |
| GO:BP | GO:0032429 | regulation of phospholipase A2 activity | 1 | 8 | 9.95e-01 |
| GO:BP | GO:0070857 | regulation of bile acid biosynthetic process | 1 | 7 | 9.95e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 3 | 11 | 9.96e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 2 | 15 | 9.96e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 2 | 50 | 9.96e-01 |
| GO:BP | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 9.96e-01 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 3 | 18 | 9.96e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 2 | 27 | 9.96e-01 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 3 | 16 | 9.97e-01 |
| GO:BP | GO:0070269 | pyroptosis | 1 | 11 | 9.97e-01 |
| GO:BP | GO:0015721 | bile acid and bile salt transport | 3 | 11 | 9.97e-01 |
| GO:BP | GO:0002689 | negative regulation of leukocyte chemotaxis | 2 | 10 | 9.98e-01 |
| GO:BP | GO:0070070 | proton-transporting V-type ATPase complex assembly | 1 | 7 | 9.98e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 7 | 42 | 9.99e-01 |
| GO:BP | GO:0061760 | antifungal innate immune response | 3 | 7 | 9.99e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 3 | 22 | 9.99e-01 |
| GO:BP | GO:0050766 | positive regulation of phagocytosis | 3 | 38 | 9.99e-01 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 9.99e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 2 | 32 | 9.99e-01 |
| GO:BP | GO:0035036 | sperm-egg recognition | 6 | 22 | 9.99e-01 |
| GO:BP | GO:0015866 | ADP transport | 1 | 6 | 9.99e-01 |
| GO:BP | GO:0006063 | uronic acid metabolic process | 1 | 6 | 9.99e-01 |
| GO:BP | GO:0019585 | glucuronate metabolic process | 1 | 6 | 9.99e-01 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 1 | 7 | 9.99e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 13 | 1.00e+00 |
| GO:BP | GO:0015851 | nucleobase transport | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0002725 | negative regulation of T cell cytokine production | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 14 | 86 | 1.00e+00 |
| GO:BP | GO:0099024 | plasma membrane invagination | 1 | 42 | 1.00e+00 |
| GO:BP | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 1 | 2 | 1.00e+00 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 3 | 72 | 1.00e+00 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 14 | 88 | 1.00e+00 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 22 | 94 | 1.00e+00 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 9 | 1.00e+00 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 4 | 33 | 1.00e+00 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0051608 | histamine transport | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0019373 | epoxygenase P450 pathway | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0045109 | intermediate filament organization | 5 | 21 | 1.00e+00 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0006910 | phagocytosis, recognition | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 1.00e+00 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 2 | 276 | 1.00e+00 |
| GO:BP | GO:0006957 | complement activation, alternative pathway | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0006959 | humoral immune response | 1 | 80 | 1.00e+00 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 2 | 57 | 1.00e+00 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 1 | 23 | 1.00e+00 |
| GO:BP | GO:0071361 | cellular response to ethanol | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 5 | 963 | 1.00e+00 |
| GO:BP | GO:0043045 | DNA methylation involved in embryo development | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 3 | 104 | 1.00e+00 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.00e+00 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 1.00e+00 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 3 | 7069 | 1.00e+00 |
| GO:BP | GO:0045061 | thymic T cell selection | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 1.00e+00 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 20 | 113 | 1.00e+00 |
| GO:BP | GO:0043300 | regulation of leukocyte degranulation | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0007308 | oocyte construction | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 3 | 17 | 1.00e+00 |
| GO:BP | GO:0098856 | intestinal lipid absorption | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0000365 | mRNA trans splicing, via spliceosome | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0000353 | formation of quadruple SL/U4/U5/U6 snRNP | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0000348 | mRNA branch site recognition | 1 | 1 | 1.00e+00 |
| GO:BP | GO:0010755 | regulation of plasminogen activation | 1 | 16 | 1.00e+00 |
| GO:BP | GO:2000109 | regulation of macrophage apoptotic process | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.00e+00 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.00e+00 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 2 | 73 | 1.00e+00 |
| GO:BP | GO:0009566 | fertilization | 9 | 93 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0009620 | response to fungus | 5 | 22 | 1.00e+00 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 7 | 131 | 1.00e+00 |
| GO:BP | GO:1990868 | response to chemokine | 1 | 37 | 1.00e+00 |
| GO:BP | GO:1990869 | cellular response to chemokine | 1 | 37 | 1.00e+00 |
| GO:BP | GO:0010324 | membrane invagination | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0000395 | mRNA 5’-splice site recognition | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 2 | 13 | 1.00e+00 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 1 | 102 | 1.00e+00 |
| GO:BP | GO:0048857 | neural nucleus development | 3 | 50 | 1.00e+00 |
| GO:BP | GO:0007309 | oocyte axis specification | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0007342 | fusion of sperm to egg plasma membrane involved in single fertilization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0048641 | regulation of skeletal muscle tissue development | 4 | 20 | 1.00e+00 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0043299 | leukocyte degranulation | 2 | 41 | 1.00e+00 |
| GO:BP | GO:0006837 | serotonin transport | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0034374 | low-density lipoprotein particle remodeling | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0045026 | plasma membrane fusion | 2 | 5 | 1.00e+00 |
| GO:BP | GO:0070091 | glucagon secretion | 1 | 3 | 1.00e+00 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 5 | 40 | 1.00e+00 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0070092 | regulation of glucagon secretion | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 1 | 28 | 1.00e+00 |
| GO:BP | GO:0070099 | regulation of chemokine-mediated signaling pathway | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0007608 | sensory perception of smell | 3 | 16 | 1.00e+00 |
| GO:BP | GO:0048643 | positive regulation of skeletal muscle tissue development | 1 | 14 | 1.00e+00 |
| GO:BP | GO:0007631 | feeding behavior | 2 | 52 | 1.00e+00 |
| GO:BP | GO:0098586 | cellular response to virus | 11 | 40 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0050482 | arachidonic acid secretion | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0008228 | opsonization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 45 | 475 | 1.00e+00 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 2 | 26 | 1.00e+00 |
| GO:BP | GO:0015732 | prostaglandin transport | 2 | 10 | 1.00e+00 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 1.00e+00 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 5 | 30 | 1.00e+00 |
| GO:BP | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0002418 | immune response to tumor cell | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 21 | 91 | 1.00e+00 |
| GO:BP | GO:0002834 | regulation of response to tumor cell | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0002837 | regulation of immune response to tumor cell | 3 | 10 | 1.00e+00 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 4 | 32 | 1.00e+00 |
| GO:BP | GO:0031640 | killing of cells of another organism | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0031639 | plasminogen activation | 1 | 19 | 1.00e+00 |
| GO:BP | GO:0002323 | natural killer cell activation involved in immune response | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0002855 | regulation of natural killer cell mediated immune response to tumor cell | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0050913 | sensory perception of bitter taste | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 2 | 23 | 1.00e+00 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 12 | 48 | 1.00e+00 |
| GO:BP | GO:0031341 | regulation of cell killing | 6 | 36 | 1.00e+00 |
| GO:BP | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste | 2 | 3 | 1.00e+00 |
| GO:BP | GO:0002922 | positive regulation of humoral immune response | 2 | 4 | 1.00e+00 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 2 | 18 | 1.00e+00 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 1 | 18 | 1.00e+00 |
| GO:BP | GO:1901616 | organic hydroxy compound catabolic process | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0003081 | regulation of systemic arterial blood pressure by renin-angiotensin | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 1.00e+00 |
| GO:BP | GO:0031016 | pancreas development | 1 | 53 | 1.00e+00 |
| GO:BP | GO:0002423 | natural killer cell mediated immune response to tumor cell | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 3 | 17 | 1.00e+00 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 1 | 21 | 1.00e+00 |
| GO:BP | GO:1903963 | arachidonate transport | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0032637 | interleukin-8 production | 3 | 43 | 1.00e+00 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 21 | 91 | 1.00e+00 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 2 | 18 | 1.00e+00 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0002507 | tolerance induction | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 3 | 43 | 1.00e+00 |
| GO:BP | GO:0032310 | prostaglandin secretion | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0032309 | icosanoid secretion | 1 | 18 | 1.00e+00 |
| GO:BP | GO:0032308 | positive regulation of prostaglandin secretion | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0032306 | regulation of prostaglandin secretion | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 2 | 8 | 1.00e+00 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 8 | 60 | 1.00e+00 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 3 | 14 | 1.00e+00 |
| GO:BP | GO:0002717 | positive regulation of natural killer cell mediated immunity | 1 | 11 | 1.00e+00 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 1 | 82 | 1.00e+00 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 1 | 14 | 1.00e+00 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 10 | 54 | 1.00e+00 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0006570 | tyrosine metabolic process | 2 | 6 | 1.00e+00 |
| GO:BP | GO:0050832 | defense response to fungus | 3 | 14 | 1.00e+00 |
| GO:BP | GO:0060746 | parental behavior | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 1.00e+00 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 4 | 20 | 1.00e+00 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 1.00e+00 |
| GO:BP | GO:0060632 | regulation of microtubule-based movement | 2 | 31 | 1.00e+00 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 1 | 33 | 1.00e+00 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0042711 | maternal behavior | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0042730 | fibrinolysis | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0019731 | antibacterial humoral response | 1 | 15 | 1.00e+00 |
| GO:BP | GO:0042742 | defense response to bacterium | 1 | 95 | 1.00e+00 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 9 | 74 | 1.00e+00 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 2 | 7441 | 1.00e+00 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 1 | 82 | 1.00e+00 |
| GO:BP | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition | 1 | 17 | 1.00e+00 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 4 | 43 | 1.00e+00 |
| GO:BP | GO:0006396 | RNA processing | 3 | 929 | 1.00e+00 |
| GO:BP | GO:0021860 | pyramidal neuron development | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0042359 | vitamin D metabolic process | 2 | 9 | 1.00e+00 |
| GO:BP | GO:0030644 | intracellular chloride ion homeostasis | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0003352 | regulation of cilium movement | 1 | 17 | 1.00e+00 |
| GO:BP | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0005984 | disaccharide metabolic process | 1 | 5 | 1.00e+00 |
| GO:BP | GO:0030299 | intestinal cholesterol absorption | 1 | 8 | 1.00e+00 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 2 | 31 | 1.00e+00 |
| GO:BP | GO:2001140 | positive regulation of phospholipid transport | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0030252 | growth hormone secretion | 1 | 12 | 1.00e+00 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 2 | 11 | 1.00e+00 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 1 | 8 | 1.00e+00 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 22 | 94 | 1.00e+00 |
| GO:BP | GO:0030002 | intracellular monoatomic anion homeostasis | 1 | 7 | 1.00e+00 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 1 | 122 | 1.00e+00 |
| GO:BP | GO:0051917 | regulation of fibrinolysis | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 1 | 16 | 1.00e+00 |
| GO:BP | GO:0045291 | mRNA trans splicing, SL addition | 1 | 6 | 1.00e+00 |
| GO:BP | GO:0071715 | icosanoid transport | 1 | 24 | 1.00e+00 |
| GO:BP | GO:0021978 | telencephalon regionalization | 1 | 4 | 1.00e+00 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 2 | 32 | 1.00e+00 |
| GO:BP | GO:0006376 | mRNA splice site selection | 2 | 31 | 1.00e+00 |
| GO:BP | GO:0002031 | G protein-coupled receptor internalization | 1 | 10 | 1.00e+00 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 1 | 28 | 1.00e+00 |
| KEGG | KEGG:03410 | Base excision repair | 25 | 33 | 2.82e-03 |
| KEGG | KEGG:03030 | DNA replication | 24 | 35 | 4.23e-02 |
| KEGG | KEGG:03430 | Mismatch repair | 17 | 22 | 4.23e-02 |
| KEGG | KEGG:04110 | Cell cycle | 78 | 151 | 1.05e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 21 | 87 | 1.91e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 2 | 66 | 2.40e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 23 | 56 | 3.30e-01 |
| KEGG | KEGG:04530 | Tight junction | 2 | 137 | 3.30e-01 |
| KEGG | KEGG:04144 | Endocytosis | 4 | 232 | 3.30e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 1 | 55 | 3.30e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 4 | 35 | 3.30e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 1 | 65 | 3.30e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 1 | 91 | 3.30e-01 |
| KEGG | KEGG:03018 | RNA degradation | 2 | 74 | 3.30e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 3.30e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 3 | 10 | 3.30e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 1 | 69 | 3.30e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 8 | 19 | 3.30e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 25 | 43 | 3.30e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 3.30e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 2 | 34 | 3.30e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 16 | 77 | 3.30e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 3.30e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 2 | 44 | 3.30e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 1 | 121 | 3.33e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 2 | 103 | 3.33e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 3.33e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 3.33e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 6 | 24 | 3.33e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 1 | 106 | 3.33e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 1 | 97 | 3.33e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 3.33e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 10 | 65 | 3.33e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 1 | 115 | 3.33e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 1 | 73 | 3.33e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 5 | 58 | 3.33e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 1 | 109 | 3.33e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 2 | 1146 | 3.33e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 11 | 49 | 3.33e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 24 | 56 | 3.33e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 1 | 131 | 3.33e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 3.33e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 9 | 115 | 3.33e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 17 | 44 | 3.33e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 3.33e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 2 | 6 | 3.33e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 1 | 97 | 3.33e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 6 | 164 | 3.44e-01 |
| KEGG | KEGG:04210 | Apoptosis | 16 | 118 | 3.44e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 3 | 35 | 3.44e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 7 | 107 | 3.44e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 11 | 82 | 3.44e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 21 | 39 | 3.50e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 9 | 32 | 3.68e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 12 | 84 | 3.70e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 17 | 74 | 3.70e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 15 | 84 | 3.70e-01 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 1 | 6 | 3.70e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 1 | 195 | 3.80e-01 |
| KEGG | KEGG:05016 | Huntington disease | 2 | 256 | 3.80e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 4 | 91 | 3.80e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 16 | 133 | 3.80e-01 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 1 | 5 | 3.81e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 1 | 22 | 4.04e-01 |
| KEGG | KEGG:04360 | Axon guidance | 2 | 162 | 4.04e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 2 | 22 | 4.23e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 11 | 56 | 4.23e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 19 | 98 | 4.23e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 13 | 86 | 4.23e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 18 | 145 | 4.23e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 4.23e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 4 | 114 | 4.37e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 12 | 70 | 4.37e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 4 | 72 | 4.37e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 4 | 85 | 4.37e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 4 | 110 | 4.37e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 4.37e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 25 | 214 | 4.37e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 3 | 46 | 4.37e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 13 | 25 | 4.37e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 6 | 240 | 4.37e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 2 | 49 | 4.55e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 1 | 16 | 4.58e-01 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 3 | 61 | 4.58e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 3 | 46 | 4.59e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 4 | 75 | 4.59e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 2 | 171 | 4.66e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 59 | 134 | 4.67e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 2 | 46 | 4.67e-01 |
| KEGG | KEGG:04217 | Necroptosis | 6 | 106 | 4.67e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 3 | 35 | 4.67e-01 |
| KEGG | KEGG:04142 | Lysosome | 1 | 118 | 4.67e-01 |
| KEGG | KEGG:00000 | KEGG root term | 4 | 5558 | 4.67e-01 |
| KEGG | KEGG:04611 | Platelet activation | 6 | 89 | 4.67e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 1 | 40 | 4.67e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 16 | 135 | 4.67e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 3 | 56 | 4.67e-01 |
| KEGG | KEGG:05164 | Influenza A | 5 | 103 | 4.67e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 4.67e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 2 | 4 | 4.67e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 18 | 110 | 4.74e-01 |
| KEGG | KEGG:04730 | Long-term depression | 12 | 42 | 4.74e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 12 | 57 | 4.74e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 3 | 8 | 4.74e-01 |
| KEGG | KEGG:04540 | Gap junction | 8 | 73 | 4.74e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 24 | 48 | 4.75e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 1 | 28 | 4.75e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 9 | 56 | 4.76e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 3 | 43 | 4.76e-01 |
| KEGG | KEGG:04520 | Adherens junction | 15 | 67 | 4.76e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 2 | 22 | 4.76e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 2 | 37 | 4.76e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 2 | 22 | 4.76e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 1 | 112 | 4.76e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 12 | 76 | 4.76e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 2 | 18 | 4.76e-01 |
| KEGG | KEGG:05224 | Breast cancer | 15 | 117 | 4.76e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 7 | 128 | 4.76e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 1 | 8 | 4.76e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 9 | 110 | 4.76e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 14 | 69 | 4.82e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 2 | 23 | 4.85e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 2 | 19 | 4.85e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 8 | 70 | 4.85e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 1 | 38 | 4.85e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 2 | 58 | 4.85e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 4.85e-01 |
| KEGG | KEGG:05034 | Alcoholism | 26 | 122 | 4.85e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 2 | 61 | 4.85e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 10 | 4.90e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 3 | 44 | 4.90e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 3 | 41 | 4.94e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 29 | 104 | 4.94e-01 |
| KEGG | KEGG:02010 | ABC transporters | 12 | 29 | 4.94e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 2 | 134 | 4.95e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 2 | 128 | 4.95e-01 |
| KEGG | KEGG:04145 | Phagosome | 2 | 99 | 4.98e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 3 | 49 | 5.01e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 2 | 53 | 5.12e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 4 | 104 | 5.12e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 4 | 37 | 5.43e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 3 | 75 | 5.45e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 9 | 83 | 5.45e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 10 | 25 | 5.45e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 9 | 17 | 5.45e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 3 | 18 | 5.45e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 3 | 55 | 5.45e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 4 | 24 | 5.45e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 17 | 121 | 5.46e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 1 | 272 | 5.46e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 2 | 38 | 5.46e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 2 | 179 | 5.46e-01 |
| KEGG | KEGG:00232 | Caffeine metabolism | 1 | 1 | 5.46e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 4 | 409 | 5.46e-01 |
| KEGG | KEGG:04146 | Peroxisome | 33 | 68 | 5.48e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 15 | 136 | 5.48e-01 |
| KEGG | KEGG:05131 | Shigellosis | 22 | 210 | 5.48e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 1 | 17 | 5.48e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 2 | 161 | 5.50e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 19 | 138 | 5.50e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 13 | 31 | 5.61e-01 |
| KEGG | KEGG:05142 | Chagas disease | 4 | 75 | 5.72e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 1 | 63 | 5.76e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 9 | 35 | 5.80e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 1 | 157 | 5.80e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 10 | 76 | 5.89e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 9 | 50 | 5.90e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 2 | 40 | 5.90e-01 |
| KEGG | KEGG:03050 | Proteasome | 7 | 42 | 5.90e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 8 | 78 | 5.90e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 1 | 112 | 5.90e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 3 | 4 | 5.90e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 15 | 87 | 6.03e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 6 | 48 | 6.03e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 3 | 170 | 6.07e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 35 | 99 | 6.07e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 21 | 133 | 6.09e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 3 | 52 | 6.12e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 6.12e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 21 | 51 | 6.14e-01 |
| KEGG | KEGG:04924 | Renin secretion | 5 | 50 | 6.14e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 4 | 49 | 6.14e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 1 | 29 | 6.17e-01 |
| KEGG | KEGG:05214 | Glioma | 7 | 67 | 6.21e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 4 | 74 | 6.41e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 13 | 117 | 6.41e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 1 | 15 | 6.43e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 2 | 33 | 6.47e-01 |
| KEGG | KEGG:03040 | Spliceosome | 31 | 132 | 6.47e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 9 | 64 | 6.47e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 6 | 17 | 6.52e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 6 | 24 | 6.56e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 3 | 51 | 6.60e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 6 | 113 | 6.63e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 6.64e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 5 | 24 | 6.68e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 1 | 3 | 6.70e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 8 | 87 | 6.78e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 3 | 56 | 6.78e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 21 | 180 | 6.78e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 3 | 30 | 6.78e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 6 | 65 | 6.78e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 1 | 12 | 6.78e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 3 | 55 | 6.78e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 3 | 76 | 6.78e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 2 | 74 | 6.78e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 1 | 303 | 6.78e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 6.78e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 3 | 15 | 6.78e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 6.83e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 1 | 165 | 6.83e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 3 | 145 | 6.86e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 7 | 176 | 6.86e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 5 | 33 | 6.86e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 7.01e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 3 | 43 | 7.10e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 2 | 20 | 7.22e-01 |
| KEGG | KEGG:00511 | Other glycan degradation | 3 | 16 | 7.22e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 3 | 13 | 7.22e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 4 | 69 | 7.22e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 7.32e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 9 | 106 | 7.32e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 24 | 172 | 7.32e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 5 | 16 | 7.39e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 5 | 10 | 7.39e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 3 | 86 | 7.39e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 3 | 21 | 7.39e-01 |
| KEGG | KEGG:04976 | Bile secretion | 2 | 42 | 7.39e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 1 | 67 | 7.39e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 23 | 153 | 7.39e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 1 | 385 | 7.41e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 13 | 142 | 7.41e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 7.44e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 12 | 150 | 7.45e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 5 | 82 | 7.45e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 5 | 14 | 7.45e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 4 | 12 | 7.45e-01 |
| KEGG | KEGG:05133 | Pertussis | 1 | 45 | 7.50e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 6 | 62 | 7.50e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 8 | 20 | 7.50e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 3 | 415 | 7.59e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 2 | 44 | 7.61e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 4 | 31 | 7.79e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 12 | 97 | 7.79e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 23 | 49 | 7.79e-01 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 7.79e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 4 | 117 | 7.79e-01 |
| KEGG | KEGG:05218 | Melanoma | 11 | 58 | 7.79e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 2 | 28 | 7.79e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 1 | 83 | 7.79e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 1 | 28 | 7.79e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 2 | 86 | 7.79e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 8 | 44 | 7.89e-01 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 1 | 12 | 7.90e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 2 | 50 | 7.90e-01 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 3 | 23 | 7.90e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 8 | 67 | 7.99e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 3 | 79 | 7.99e-01 |
| KEGG | KEGG:03060 | Protein export | 4 | 23 | 7.99e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 1 | 11 | 8.00e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 2 | 153 | 8.04e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 16 | 84 | 8.04e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 25 | 145 | 8.04e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 8.04e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 1 | 16 | 8.17e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 8.17e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 2 | 186 | 8.17e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 8 | 55 | 8.19e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 8.19e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 17 | 115 | 8.19e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 1 | 31 | 8.27e-01 |
| KEGG | KEGG:00780 | Biotin metabolism | 1 | 3 | 8.33e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 3 | 103 | 8.52e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 7 | 71 | 8.52e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 1 | 315 | 8.52e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 1 | 32 | 8.52e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 5 | 77 | 8.53e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 2 | 12 | 8.53e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 2 | 9 | 8.53e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 8.53e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 2 | 108 | 8.53e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 6 | 75 | 8.59e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 1 | 226 | 8.60e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 2 | 94 | 8.62e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 6 | 63 | 8.64e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 8.73e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 4 | 41 | 8.77e-01 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 2 | 169 | 8.82e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 1 | 20 | 8.82e-01 |
| KEGG | KEGG:00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 1 | 2 | 8.82e-01 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 8.84e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 3 | 99 | 8.87e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 5 | 49 | 8.89e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 3 | 28 | 9.05e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 2 | 27 | 9.05e-01 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 2 | 26 | 9.05e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 2 | 78 | 9.05e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 9.17e-01 |
| KEGG | KEGG:05134 | Legionellosis | 3 | 39 | 9.17e-01 |
| KEGG | KEGG:04742 | Taste transduction | 1 | 22 | 9.19e-01 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 1 | 19 | 9.32e-01 |
| KEGG | KEGG:05162 | Measles | 2 | 98 | 9.32e-01 |
| KEGG | KEGG:01200 | Carbon metabolism | 2 | 98 | 9.32e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 2 | 41 | 9.41e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 2 | 124 | 9.45e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 2 | 26 | 9.45e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 4 | 63 | 9.45e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 1 | 108 | 9.45e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 1 | 38 | 9.45e-01 |
| KEGG | KEGG:03265 | Virion - Lyssavirus | 1 | 3 | 9.49e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 3 | 53 | 9.52e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 4 | 60 | 9.71e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 3 | 14 | 9.77e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 9.86e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 2 | 106 | 9.86e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 9.86e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 2 | 25 | 9.86e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 4 | 43 | 9.86e-01 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 1 | 9 | 9.86e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 1 | 10 | 9.99e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 2 | 33 | 9.99e-01 |
| KEGG | KEGG:05020 | Prion disease | 1 | 220 | 1.00e+00 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:05310 | Asthma | 1 | 3 | 1.00e+00 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 1.00e+00 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 4 | 89 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 3 | 31 | 1.00e+00 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 4 | 19 | 1.00e+00 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 1 | 9 | 1.00e+00 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 2 | 98 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 2 | 12 | 1.00e+00 |
| KEGG | KEGG:00310 | Lysine degradation | 12 | 59 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 15 | 127 | 1.00e+00 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 3 | 7 | 1.00e+00 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 1 | 26 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 4 | 36 | 1.00e+00 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 1 | 23 | 1.00e+00 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 5 | 1.00e+00 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 3 | 13 | 1.00e+00 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 4 | 12 | 1.00e+00 |
| KEGG | KEGG:04740 | Olfactory transduction | 1 | 25 | 1.00e+00 |
| KEGG | KEGG:04744 | Phototransduction | 3 | 12 | 1.00e+00 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 1 | 69 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 1.00e+00 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 3 | 12 | 1.00e+00 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 11 | 88 | 1.00e+00 |
of the AC genes that overlapped without Mito (n= 4435 and all AC gens plus those which overlapped with Mito (n= 882, the significantly enriched GO: BP terms are the same. This may support the idea that the specific AC set can tell us more about mechanism in AC induced cardiotoxicity. ## DDEM 24 hour response of 8
# DDEMgostres3 <- gost(query = DDEMresp3,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
#
# saveRDS(DDEMgostres3,"data/DEG-GO/DDEMgostres3.RDS")
DDEMgostres3 <- readRDS("data/DEG-GO/DDEMgostres3.RDS")
# DDEMp3 <- gostplot(DDEMgostres3, capped = FALSE, interactive = TRUE)
# DDEMp3
DDEMtable3 <- DDEMgostres3$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DDEMtable3 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Top2i DEGs (n=8) 3 hr GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)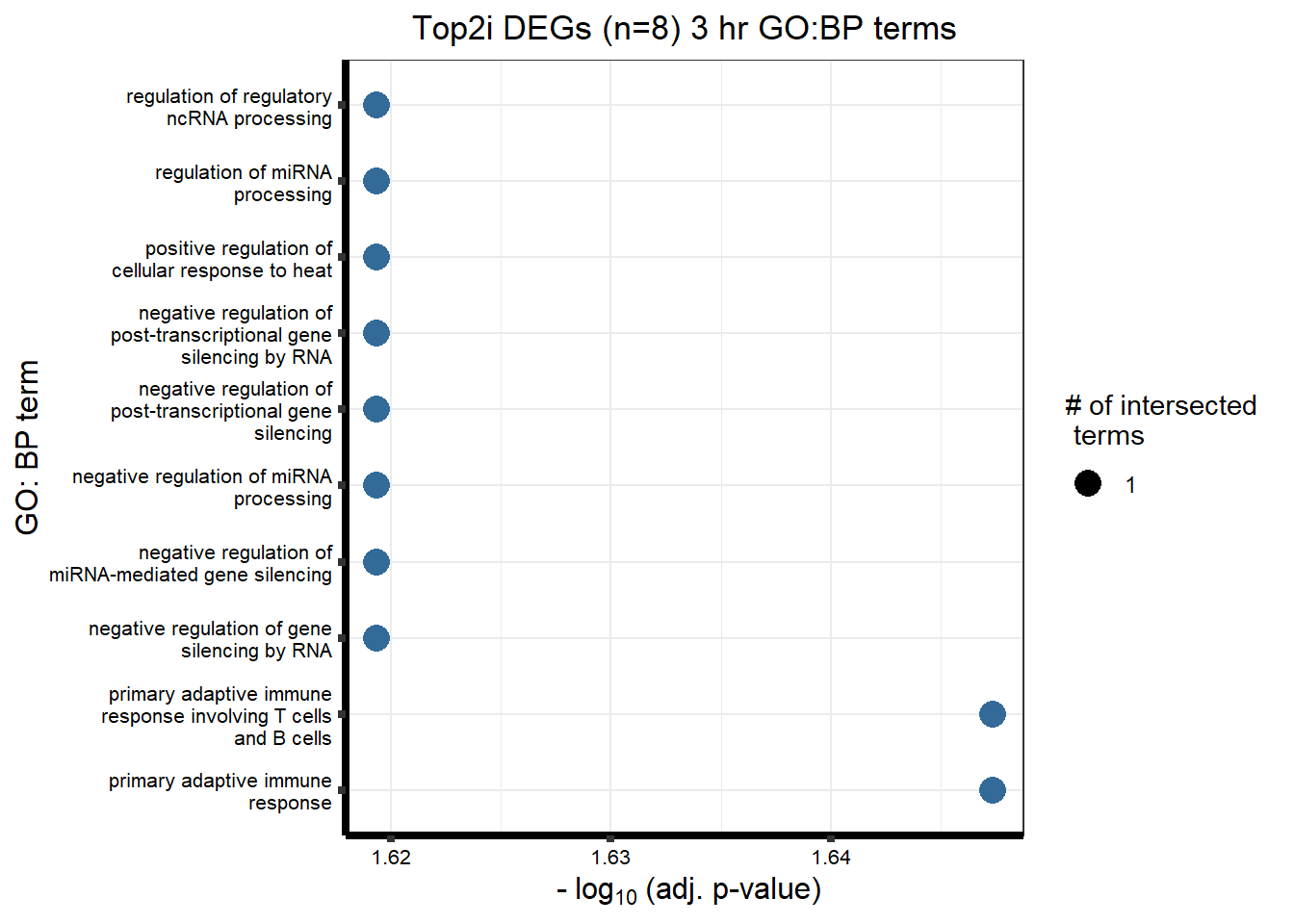
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEMtable3 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "Top2 inhibitor enrichment list, n=8") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0090721 | primary adaptive immune response involving T cells and B cells | 1 | 1 | 2.25e-02 |
| GO:BP | GO:0090720 | primary adaptive immune response | 1 | 1 | 2.25e-02 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 2.40e-02 |
| GO:BP | GO:1903799 | negative regulation of miRNA processing | 1 | 6 | 2.40e-02 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 1 | 11 | 2.40e-02 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 1 | 11 | 2.40e-02 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 1 | 10 | 2.40e-02 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 1 | 11 | 2.40e-02 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 2.40e-02 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 2.40e-02 |
| GO:BP | GO:0043589 | skin morphogenesis | 1 | 10 | 3.27e-02 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 3.60e-02 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 3.60e-02 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 3.60e-02 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 3.60e-02 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 4.22e-02 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 5.51e-02 |
| GO:BP | GO:0016070 | RNA metabolic process | 8 | 3600 | 5.51e-02 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 5.53e-02 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 5.93e-02 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 4 | 2845 | 5.93e-02 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 4 | 3080 | 6.94e-02 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 1 | 96 | 6.94e-02 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 6.94e-02 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 8 | 4048 | 7.01e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 7.01e-02 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 7.75e-02 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 1 | 127 | 7.75e-02 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 7.75e-02 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 1 | 164 | 9.11e-02 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 8 | 4455 | 9.97e-02 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 3 | 1849 | 1.10e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 1 | 96 | 1.10e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 8 | 4596 | 1.10e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 8 | 4570 | 1.10e-01 |
| GO:BP | GO:0007498 | mesoderm development | 1 | 89 | 1.12e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 1 | 267 | 1.24e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 8 | 4747 | 1.28e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 1.32e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 1.32e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 1 | 400 | 1.32e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 4 | 3580 | 1.32e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 4 | 4292 | 1.32e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 4 | 4174 | 1.32e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 8 | 4946 | 1.32e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 2 | 1119 | 1.32e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 5 | 2593 | 1.32e-01 |
| GO:BP | GO:0007369 | gastrulation | 1 | 146 | 1.32e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 5 | 2576 | 1.32e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 4 | 4417 | 1.32e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 5 | 2684 | 1.32e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 5 | 2683 | 1.32e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 5 | 2574 | 1.32e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 2 | 1209 | 1.35e-01 |
| GO:BP | GO:0010467 | gene expression | 8 | 4587 | 1.35e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 5 | 2714 | 1.35e-01 |
| GO:BP | GO:0051170 | import into nucleus | 1 | 152 | 1.35e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 1 | 53 | 1.36e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 1.42e-01 |
| GO:BP | GO:0043588 | skin development | 1 | 175 | 1.47e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 1.47e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 1.47e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 1.47e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 1 | 561 | 1.49e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 3 | 2135 | 1.52e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 1.54e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 5 | 3012 | 1.57e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 1.66e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 3 | 2293 | 1.66e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 5 | 3080 | 1.66e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 5 | 3070 | 1.66e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 1 | 95 | 1.72e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 5 | 3169 | 1.72e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 5 | 3133 | 1.72e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 5 | 3132 | 1.72e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 4 | 4617 | 1.72e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 3 | 1315 | 1.76e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 5 | 3237 | 1.76e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 3 | 1315 | 1.76e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 3 | 1322 | 1.76e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 2 | 1735 | 1.76e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 1 | 295 | 1.76e-01 |
| GO:BP | GO:0051169 | nuclear transport | 1 | 295 | 1.76e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 161 | 1.88e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 1 | 468 | 1.89e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 1.99e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 4 | 5017 | 1.99e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 3 | 1441 | 1.99e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 1 | 440 | 2.00e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 1 | 123 | 2.00e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 1 | 276 | 2.00e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 1 | 504 | 2.02e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 2.02e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 3 | 1507 | 2.03e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 1 | 280 | 2.04e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 3 | 1564 | 2.18e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 1 | 331 | 2.18e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 3 | 1602 | 2.22e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 3 | 1604 | 2.28e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 5 | 3730 | 2.28e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 2.33e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 5 | 3750 | 2.34e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 1 | 165 | 2.36e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 2.44e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 2.44e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 2.44e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 4 | 2646 | 2.45e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 1 | 206 | 2.58e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 2.58e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 2.61e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 1 | 742 | 2.77e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 2.77e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 1 | 665 | 2.86e-01 |
| GO:BP | GO:0009790 | embryo development | 1 | 875 | 2.89e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 4 | 2880 | 2.91e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 1 | 870 | 2.91e-01 |
| GO:BP | GO:0048511 | rhythmic process | 1 | 237 | 2.92e-01 |
| GO:BP | GO:0006396 | RNA processing | 3 | 929 | 2.92e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 1 | 258 | 3.00e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 3 | 1914 | 3.04e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 5 | 4245 | 3.10e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 551 | 3.12e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 3 | 1995 | 3.26e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 1 | 740 | 3.27e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 3 | 4539 | 3.60e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 3 | 4045 | 3.62e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 3 | 4590 | 3.65e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 3 | 2270 | 3.93e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 1 | 816 | 4.06e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 1 | 1015 | 4.06e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 8 | 6996 | 4.06e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 1 | 1017 | 4.06e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 1 | 1026 | 4.06e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 1 | 875 | 4.06e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 1 | 901 | 4.08e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 3 | 2412 | 4.08e-01 |
| GO:BP | GO:0009888 | tissue development | 1 | 1425 | 4.08e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 4.13e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2 | 3625 | 4.13e-01 |
| GO:BP | GO:0051641 | cellular localization | 2 | 2810 | 4.25e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 1 | 859 | 4.37e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1 | 1214 | 4.37e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 1 | 1239 | 4.41e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 1 | 616 | 4.41e-01 |
| GO:BP | GO:0040008 | regulation of growth | 1 | 501 | 4.45e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1 | 1280 | 4.46e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 4.53e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1453 | 4.80e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 1 | 1145 | 4.80e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1 | 1199 | 4.80e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1 | 1974 | 4.86e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 4.98e-01 |
| GO:BP | GO:0048513 | animal organ development | 1 | 2189 | 5.43e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 8 | 7441 | 5.47e-01 |
| GO:BP | GO:0007049 | cell cycle | 1 | 1529 | 5.68e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 5.68e-01 |
| GO:BP | GO:0040007 | growth | 1 | 742 | 5.68e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 1 | 665 | 5.68e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 4 | 7552 | 5.73e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 8 | 7069 | 5.86e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 1 | 868 | 5.86e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1 | 4579 | 5.89e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 6.10e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 6.11e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 6.11e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 1 | 1712 | 6.14e-01 |
| GO:BP | GO:0030154 | cell differentiation | 1 | 2958 | 6.52e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 1 | 2984 | 6.52e-01 |
| GO:BP | GO:0051179 | localization | 2 | 3969 | 6.56e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 1 | 1075 | 6.56e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1 | 1974 | 6.63e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3386 | 6.74e-01 |
| GO:BP | GO:0009056 | catabolic process | 1 | 2088 | 6.75e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 8 | 7806 | 6.88e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2138 | 6.88e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2129 | 6.88e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1 | 1147 | 6.96e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2450 | 7.54e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 1 | 1681 | 7.54e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 1 | 1378 | 7.60e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 1 | 4128 | 7.66e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 3 | 4099 | 7.94e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 4 | 8061 | 7.94e-01 |
| GO:BP | GO:0032502 | developmental process | 1 | 4494 | 8.00e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 8.00e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 8.00e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 8.12e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 8.69e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 1 | 4794 | 8.73e-01 |
| GO:BP | GO:0065007 | biological regulation | 4 | 8320 | 8.73e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1 | 2035 | 9.25e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 1 | 4241 | 9.26e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1 | 2890 | 9.61e-01 |
| GO:BP | GO:0006950 | response to stress | 1 | 2780 | 9.61e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 1 | 4955 | 9.66e-01 |
| GO:BP | GO:0048731 | system development | 1 | 2901 | 9.76e-01 |
| GO:BP | GO:0050896 | response to stimulus | 1 | 5770 | 9.98e-01 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1 | 5274 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 1.00e+00 |
| GO:BP | GO:0016043 | cellular component organization | 1 | 5076 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
DDEM 24 hour response of 882
# DDEMgostres24 <- gost(query = DDEMresp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
#
# saveRDS(DDEMgostres24,"data/DEG-GO/DDEMgostres24.RDS")
DDEMgostres24 <- readRDS("data/DEG-GO/DDEMgostres24.RDS")
# DDEMp24 <- gostplot(DDEMgostres24, capped = FALSE, interactive = TRUE)
# DDEMp24
DDEMtable24 <- DDEMgostres24$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DDEMtable24 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Top2i DEGs (n=832) 24 hr GO:BP terms') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)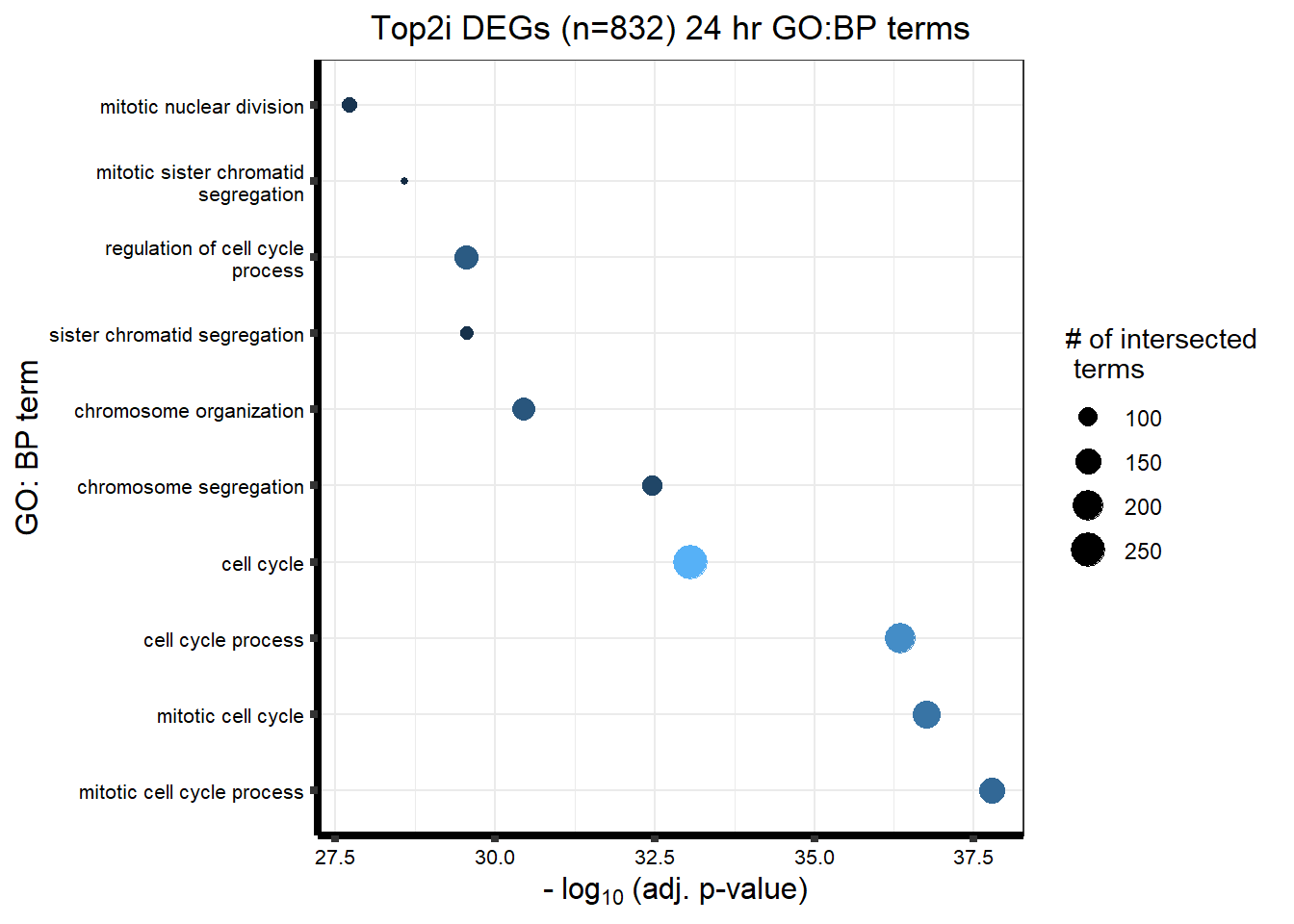
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEMtable24 %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size, col = "red")) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Top2 inhibitor DEGs (n=832) 24 hr KEGG terms') +
xlab(expression(" -" ~ log[10] ~ "(adj. p-value)")) +
ylab("KEGG term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)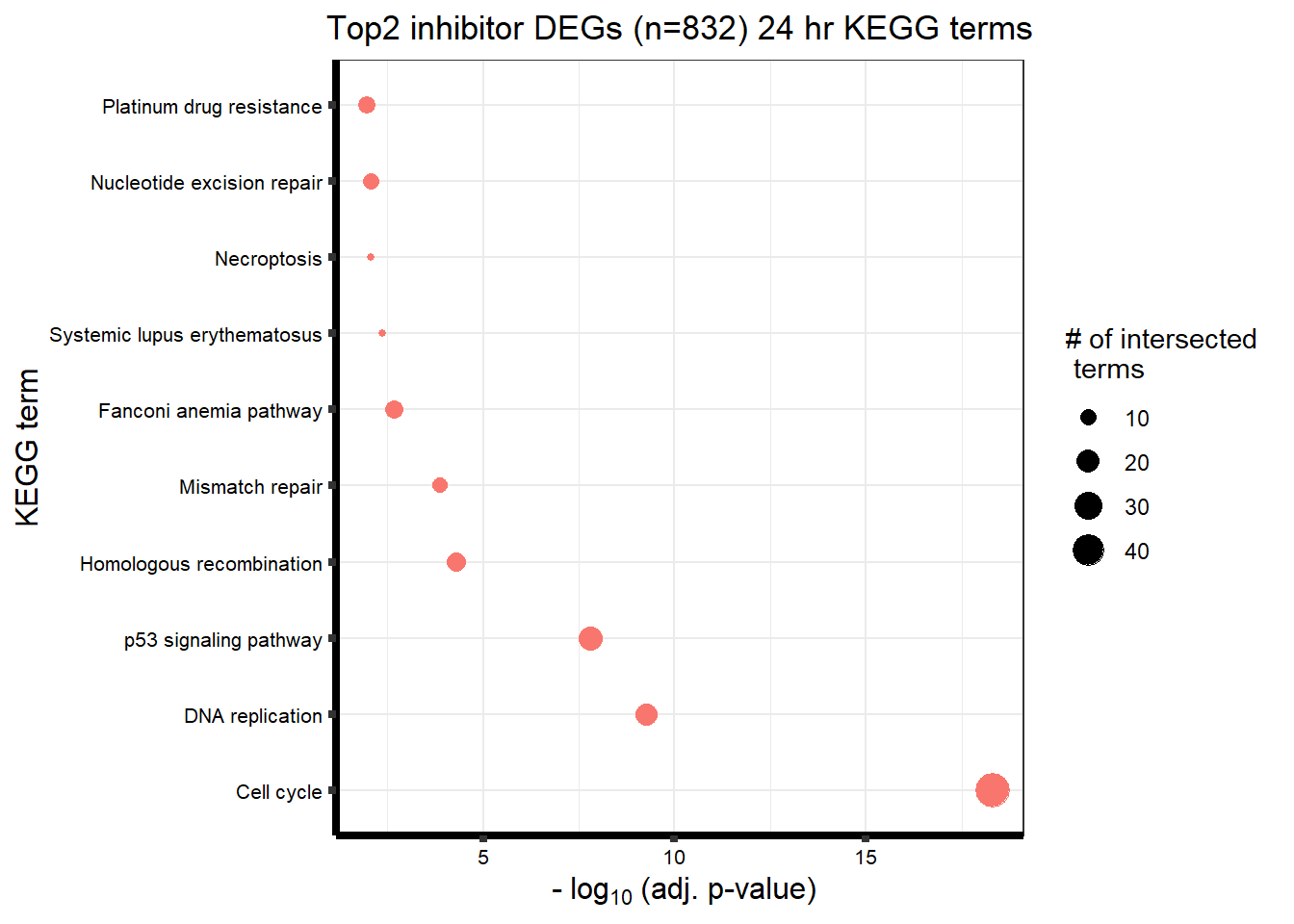
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DDEMtable24 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "Top2 inhibitor enrichment list, n=832") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:1903047 | mitotic cell cycle process | 154 | 694 | 1.65e-38 |
| GO:BP | GO:0000278 | mitotic cell cycle | 170 | 827 | 1.74e-37 |
| GO:BP | GO:0022402 | cell cycle process | 204 | 1086 | 4.54e-37 |
| GO:BP | GO:0007049 | cell cycle | 250 | 1529 | 8.71e-34 |
| GO:BP | GO:0007059 | chromosome segregation | 104 | 369 | 3.53e-33 |
| GO:BP | GO:0051276 | chromosome organization | 128 | 543 | 3.57e-31 |
| GO:BP | GO:0000819 | sister chromatid segregation | 73 | 217 | 2.74e-30 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 135 | 626 | 2.87e-30 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 63 | 179 | 2.61e-29 |
| GO:BP | GO:0140014 | mitotic nuclear division | 78 | 251 | 1.90e-28 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 83 | 273 | 1.34e-27 |
| GO:BP | GO:0000280 | nuclear division | 96 | 344 | 2.53e-26 |
| GO:BP | GO:0051301 | cell division | 122 | 570 | 3.37e-26 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 62 | 183 | 3.56e-26 |
| GO:BP | GO:0051726 | regulation of cell cycle | 168 | 956 | 1.16e-25 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 53 | 139 | 2.61e-25 |
| GO:BP | GO:0048285 | organelle fission | 92 | 388 | 4.89e-23 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 48 | 121 | 4.89e-23 |
| GO:BP | GO:0044770 | cell cycle phase transition | 104 | 504 | 2.35e-22 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 90 | 418 | 1.51e-21 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 36 | 71 | 1.73e-21 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 51 | 148 | 1.06e-20 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 87 | 391 | 1.06e-20 |
| GO:BP | GO:0006260 | DNA replication | 68 | 255 | 1.33e-20 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 70 | 273 | 1.74e-20 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 93 | 441 | 2.22e-20 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 64 | 235 | 2.58e-20 |
| GO:BP | GO:0051304 | chromosome separation | 35 | 76 | 3.11e-20 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 59 | 211 | 1.99e-19 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 72 | 308 | 2.61e-19 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 76 | 338 | 3.13e-19 |
| GO:BP | GO:0006974 | DNA damage response | 129 | 785 | 1.51e-18 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 30 | 58 | 1.69e-18 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 27 | 47 | 5.59e-18 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 27 | 47 | 5.59e-18 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 27 | 47 | 5.59e-18 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 38 | 99 | 6.30e-18 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 28 | 52 | 8.54e-18 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 30 | 61 | 8.54e-18 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 40 | 100 | 9.35e-18 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 28 | 54 | 1.53e-17 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 27 | 48 | 1.84e-17 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 27 | 48 | 1.84e-17 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 26 | 46 | 2.08e-17 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 34 | 91 | 4.42e-17 |
| GO:BP | GO:0006281 | DNA repair | 97 | 534 | 6.42e-17 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 26 | 47 | 7.68e-17 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 25 | 44 | 8.27e-17 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 25 | 44 | 8.27e-17 |
| GO:BP | GO:0006259 | DNA metabolic process | 139 | 884 | 8.27e-17 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 25 | 44 | 8.27e-17 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 58 | 209 | 8.64e-17 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 33 | 88 | 1.16e-16 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 34 | 92 | 1.16e-16 |
| GO:BP | GO:0051783 | regulation of nuclear division | 42 | 116 | 1.20e-16 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 48 | 166 | 1.37e-16 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 25 | 45 | 1.47e-16 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 56 | 236 | 2.16e-16 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 33 | 89 | 3.29e-16 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 28 | 57 | 5.71e-16 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 32 | 90 | 8.49e-15 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 39 | 129 | 9.34e-15 |
| GO:BP | GO:0007051 | spindle organization | 48 | 188 | 1.28e-14 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 43 | 157 | 4.27e-14 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 21 | 42 | 8.62e-14 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 63 | 274 | 2.47e-13 |
| GO:BP | GO:0007052 | mitotic spindle organization | 37 | 129 | 4.74e-13 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 91 | 552 | 5.50e-12 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 28 | 85 | 1.48e-11 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 32 | 121 | 3.24e-11 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 37 | 149 | 6.57e-11 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 41 | 178 | 7.70e-11 |
| GO:BP | GO:0051225 | spindle assembly | 26 | 120 | 1.52e-10 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 34 | 134 | 2.85e-10 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 15 | 27 | 4.22e-10 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 20 | 47 | 4.39e-10 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 40 | 135 | 7.97e-10 |
| GO:BP | GO:0051310 | metaphase plate congression | 22 | 69 | 8.25e-10 |
| GO:BP | GO:0006270 | DNA replication initiation | 17 | 35 | 1.12e-09 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 18 | 42 | 1.36e-09 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 25 | 81 | 1.36e-09 |
| GO:BP | GO:0032508 | DNA duplex unwinding | 24 | 74 | 1.81e-09 |
| GO:BP | GO:0006275 | regulation of DNA replication | 31 | 116 | 2.21e-09 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 11 | 18 | 2.40e-09 |
| GO:BP | GO:0050000 | chromosome localization | 24 | 82 | 2.63e-09 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 23 | 75 | 2.96e-09 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 14 | 28 | 3.10e-09 |
| GO:BP | GO:0007080 | mitotic metaphase plate congression | 19 | 55 | 3.74e-09 |
| GO:BP | GO:0034502 | protein localization to chromosome | 29 | 110 | 4.87e-09 |
| GO:BP | GO:0032392 | DNA geometric change | 24 | 80 | 9.15e-09 |
| GO:BP | GO:0071103 | DNA conformation change | 25 | 87 | 1.19e-08 |
| GO:BP | GO:0051383 | kinetochore organization | 13 | 23 | 1.43e-08 |
| GO:BP | GO:0051231 | spindle elongation | 9 | 12 | 1.48e-08 |
| GO:BP | GO:0051321 | meiotic cell cycle | 42 | 188 | 1.49e-08 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 21 | 79 | 1.53e-08 |
| GO:BP | GO:0033554 | cellular response to stress | 187 | 1681 | 1.74e-08 |
| GO:BP | GO:0140013 | meiotic nuclear division | 33 | 120 | 2.34e-08 |
| GO:BP | GO:0034508 | centromere complex assembly | 14 | 28 | 2.47e-08 |
| GO:BP | GO:0007017 | microtubule-based process | 106 | 760 | 2.47e-08 |
| GO:BP | GO:0033043 | regulation of organelle organization | 122 | 1028 | 2.97e-08 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 17 | 45 | 3.15e-08 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 9 | 3.27e-08 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 25 | 92 | 3.96e-08 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 11 | 18 | 5.57e-08 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 11 | 18 | 5.57e-08 |
| GO:BP | GO:0010212 | response to ionizing radiation | 26 | 130 | 5.58e-08 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 21 | 81 | 6.30e-08 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 16 | 40 | 8.32e-08 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 10 | 8.88e-08 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 26 | 103 | 9.79e-08 |
| GO:BP | GO:0033260 | nuclear DNA replication | 15 | 35 | 1.01e-07 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 51 | 318 | 1.13e-07 |
| GO:BP | GO:0006268 | DNA unwinding involved in DNA replication | 11 | 22 | 1.70e-07 |
| GO:BP | GO:0006310 | DNA recombination | 50 | 257 | 1.73e-07 |
| GO:BP | GO:0000910 | cytokinesis | 31 | 164 | 2.13e-07 |
| GO:BP | GO:0006302 | double-strand break repair | 47 | 271 | 2.57e-07 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 32 | 149 | 2.60e-07 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 10 | 15 | 3.09e-07 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 17 | 48 | 4.06e-07 |
| GO:BP | GO:0051255 | spindle midzone assembly | 8 | 12 | 4.54e-07 |
| GO:BP | GO:0000725 | recombinational repair | 32 | 153 | 4.54e-07 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 67 | 452 | 5.51e-07 |
| GO:BP | GO:0007098 | centrosome cycle | 28 | 127 | 5.75e-07 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 54 | 357 | 5.94e-07 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 16 | 70 | 7.29e-07 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 10 | 21 | 1.10e-06 |
| GO:BP | GO:0006996 | organelle organization | 290 | 3000 | 1.25e-06 |
| GO:BP | GO:0071478 | cellular response to radiation | 28 | 156 | 1.53e-06 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 24 | 113 | 2.22e-06 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 15 | 43 | 2.52e-06 |
| GO:BP | GO:0006541 | glutamine metabolic process | 4 | 24 | 2.99e-06 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 20 | 86 | 3.24e-06 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 20 | 77 | 3.86e-06 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 28 | 137 | 4.13e-06 |
| GO:BP | GO:0009314 | response to radiation | 47 | 352 | 4.83e-06 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 27 | 81 | 5.14e-06 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 20 | 64 | 5.51e-06 |
| GO:BP | GO:0051302 | regulation of cell division | 26 | 145 | 6.04e-06 |
| GO:BP | GO:0010332 | response to gamma radiation | 13 | 50 | 6.38e-06 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 29 | 161 | 7.87e-06 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 8.21e-06 |
| GO:BP | GO:1900262 | regulation of DNA-directed DNA polymerase activity | 8 | 13 | 8.32e-06 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 8 | 13 | 8.32e-06 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 15 | 50 | 8.91e-06 |
| GO:BP | GO:1902969 | mitotic DNA replication | 8 | 14 | 9.07e-06 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 38 | 185 | 9.12e-06 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 21 | 104 | 1.04e-05 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 23 | 103 | 1.41e-05 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 9 | 19 | 1.46e-05 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 7 | 12 | 1.46e-05 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 9 | 19 | 1.46e-05 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 9 | 19 | 1.46e-05 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 7 | 12 | 1.46e-05 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 39 | 204 | 2.59e-05 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 7 | 11 | 2.69e-05 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 8 | 15 | 2.86e-05 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 16 | 60 | 2.96e-05 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 15 | 68 | 3.95e-05 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 6 | 11 | 4.14e-05 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 15 | 52 | 4.45e-05 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 12 | 33 | 4.46e-05 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 16 | 62 | 4.52e-05 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 8 | 19 | 4.74e-05 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 31 | 175 | 5.59e-05 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 35 | 268 | 6.53e-05 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 35 | 268 | 6.53e-05 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 8 | 18 | 7.29e-05 |
| GO:BP | GO:0007127 | meiosis I | 24 | 77 | 8.44e-05 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 5 | 7 | 9.53e-05 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 8 | 19 | 9.58e-05 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 6 | 11 | 9.97e-05 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 5 | 1.03e-04 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 10 | 36 | 1.03e-04 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 5 | 8 | 1.33e-04 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 5 | 8 | 1.33e-04 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 7 | 14 | 1.36e-04 |
| GO:BP | GO:0090224 | regulation of spindle organization | 11 | 45 | 1.42e-04 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 11 | 35 | 1.52e-04 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 10 | 42 | 1.55e-04 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 4 | 60 | 1.96e-04 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 7 | 15 | 2.10e-04 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 5 | 7 | 2.18e-04 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 6 | 9 | 2.34e-04 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 6 | 13 | 2.34e-04 |
| GO:BP | GO:0031297 | replication fork processing | 12 | 42 | 2.50e-04 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 5 | 9 | 2.64e-04 |
| GO:BP | GO:0051382 | kinetochore assembly | 8 | 18 | 2.73e-04 |
| GO:BP | GO:0006513 | protein monoubiquitination | 14 | 80 | 2.73e-04 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 8 | 29 | 3.20e-04 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 6 | 7 | 3.20e-04 |
| GO:BP | GO:0030261 | chromosome condensation | 12 | 33 | 3.49e-04 |
| GO:BP | GO:0042148 | strand invasion | 4 | 5 | 5.18e-04 |
| GO:BP | GO:0009411 | response to UV | 19 | 141 | 5.25e-04 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 5 | 9 | 5.32e-04 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 5.99e-04 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 15 | 69 | 6.25e-04 |
| GO:BP | GO:0034644 | cellular response to UV | 14 | 86 | 6.92e-04 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 19 | 94 | 7.00e-04 |
| GO:BP | GO:0051298 | centrosome duplication | 15 | 68 | 8.66e-04 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 5 | 8 | 8.70e-04 |
| GO:BP | GO:0035825 | homologous recombination | 12 | 43 | 8.92e-04 |
| GO:BP | GO:0051656 | establishment of organelle localization | 43 | 378 | 1.12e-03 |
| GO:BP | GO:0046599 | regulation of centriole replication | 7 | 21 | 1.13e-03 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 5 | 8 | 1.41e-03 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 5 | 8 | 1.41e-03 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 10 | 44 | 1.82e-03 |
| GO:BP | GO:0071168 | protein localization to chromatin | 6 | 48 | 1.84e-03 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 6 | 14 | 1.98e-03 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 9 | 30 | 1.98e-03 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 6 | 14 | 1.98e-03 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 6 | 14 | 1.98e-03 |
| GO:BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 7 | 18 | 1.98e-03 |
| GO:BP | GO:0007010 | cytoskeleton organization | 128 | 1226 | 2.03e-03 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 9 | 37 | 2.07e-03 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 21 | 112 | 2.16e-03 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 8 | 32 | 2.18e-03 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 5 | 17 | 2.20e-03 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 8 | 25 | 2.33e-03 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 9 | 25 | 2.45e-03 |
| GO:BP | GO:0006301 | postreplication repair | 8 | 34 | 2.76e-03 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 12 | 73 | 3.45e-03 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 11 | 186 | 3.55e-03 |
| GO:BP | GO:1903926 | cellular response to bisphenol A | 3 | 4 | 3.61e-03 |
| GO:BP | GO:1903925 | response to bisphenol A | 3 | 4 | 3.61e-03 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 65 | 604 | 3.90e-03 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 3 | 5 | 4.12e-03 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 3 | 5 | 4.12e-03 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 3 | 5 | 4.12e-03 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 36 | 4.23e-03 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 4.59e-03 |
| GO:BP | GO:0007099 | centriole replication | 10 | 38 | 4.59e-03 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 5 | 11 | 4.63e-03 |
| GO:BP | GO:0006298 | mismatch repair | 8 | 29 | 5.12e-03 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 8 | 27 | 5.19e-03 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 10 | 51 | 5.45e-03 |
| GO:BP | GO:0051653 | spindle localization | 11 | 52 | 5.45e-03 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 12 | 45 | 5.81e-03 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 4 | 151 | 6.20e-03 |
| GO:BP | GO:0032506 | cytokinetic process | 8 | 39 | 6.77e-03 |
| GO:BP | GO:0022616 | DNA strand elongation | 9 | 35 | 6.77e-03 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 11 | 160 | 6.77e-03 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 5 | 11 | 6.85e-03 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 5 | 12 | 6.89e-03 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 69 | 868 | 7.14e-03 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 46 | 439 | 7.19e-03 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 7 | 19 | 7.21e-03 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 8 | 30 | 7.28e-03 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 35 | 265 | 7.50e-03 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 22 | 7.54e-03 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 3 | 4 | 7.56e-03 |
| GO:BP | GO:0072719 | cellular response to cisplatin | 3 | 4 | 8.01e-03 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 16 | 85 | 8.23e-03 |
| GO:BP | GO:0019985 | translesion synthesis | 6 | 24 | 8.23e-03 |
| GO:BP | GO:0099068 | postsynapse assembly | 6 | 32 | 8.38e-03 |
| GO:BP | GO:0070925 | organelle assembly | 85 | 817 | 8.53e-03 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 8 | 30 | 8.53e-03 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 8.53e-03 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 8 | 30 | 8.75e-03 |
| GO:BP | GO:0006177 | GMP biosynthetic process | 2 | 13 | 8.75e-03 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 2 | 14 | 8.75e-03 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 2 | 16 | 8.86e-03 |
| GO:BP | GO:0098534 | centriole assembly | 10 | 41 | 8.97e-03 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 4 | 7 | 9.61e-03 |
| GO:BP | GO:0051640 | organelle localization | 51 | 503 | 9.63e-03 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 4 | 8 | 9.87e-03 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 11 | 217 | 1.01e-02 |
| GO:BP | GO:0051293 | establishment of spindle localization | 10 | 48 | 1.04e-02 |
| GO:BP | GO:0006312 | mitotic recombination | 7 | 22 | 1.04e-02 |
| GO:BP | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 6 | 18 | 1.04e-02 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 16 | 88 | 1.04e-02 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 14 | 103 | 1.07e-02 |
| GO:BP | GO:1990227 | paranodal junction maintenance | 1 | 2 | 1.08e-02 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 34 | 262 | 1.13e-02 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 5 | 1.15e-02 |
| GO:BP | GO:1905340 | regulation of protein localization to kinetochore | 3 | 4 | 1.18e-02 |
| GO:BP | GO:1905342 | positive regulation of protein localization to kinetochore | 3 | 4 | 1.18e-02 |
| GO:BP | GO:0007144 | female meiosis I | 5 | 9 | 1.19e-02 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 8 | 29 | 1.22e-02 |
| GO:BP | GO:0006189 | ‘de novo’ IMP biosynthetic process | 3 | 6 | 1.23e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 7 | 25 | 1.24e-02 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 6 | 26 | 1.29e-02 |
| GO:BP | GO:0051782 | negative regulation of cell division | 5 | 15 | 1.29e-02 |
| GO:BP | GO:0000212 | meiotic spindle organization | 5 | 14 | 1.30e-02 |
| GO:BP | GO:0003283 | atrial septum development | 5 | 23 | 1.34e-02 |
| GO:BP | GO:0042772 | DNA damage response, signal transduction resulting in transcription | 6 | 19 | 1.35e-02 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 3 | 4 | 1.36e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 1 | 8 | 1.37e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 6 | 36 | 1.40e-02 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 3 | 1.40e-02 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 1.40e-02 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 7 | 23 | 1.41e-02 |
| GO:BP | GO:0048368 | lateral mesoderm development | 2 | 10 | 1.43e-02 |
| GO:BP | GO:0006979 | response to oxidative stress | 17 | 352 | 1.49e-02 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 1.53e-02 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 14 | 99 | 1.54e-02 |
| GO:BP | GO:0009168 | purine ribonucleoside monophosphate biosynthetic process | 2 | 19 | 1.59e-02 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 11 | 36 | 1.77e-02 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 11 | 36 | 1.77e-02 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 8 | 36 | 1.79e-02 |
| GO:BP | GO:0006520 | amino acid metabolic process | 4 | 222 | 1.87e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 4 | 10 | 1.89e-02 |
| GO:BP | GO:0009127 | purine nucleoside monophosphate biosynthetic process | 2 | 21 | 1.89e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 4 | 13 | 1.89e-02 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 4 | 5 | 1.91e-02 |
| GO:BP | GO:0097294 | ‘de novo’ XMP biosynthetic process | 3 | 7 | 1.91e-02 |
| GO:BP | GO:0097293 | XMP biosynthetic process | 3 | 7 | 1.91e-02 |
| GO:BP | GO:0097292 | XMP metabolic process | 3 | 7 | 1.91e-02 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 3 | 6 | 1.92e-02 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 3 | 6 | 1.92e-02 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 12 | 100 | 1.96e-02 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 6 | 20 | 1.99e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 10 | 82 | 2.00e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 9 | 152 | 2.04e-02 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 13 | 270 | 2.05e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 3 | 4 | 2.14e-02 |
| GO:BP | GO:0046037 | GMP metabolic process | 3 | 23 | 2.16e-02 |
| GO:BP | GO:0044210 | ‘de novo’ CTP biosynthetic process | 1 | 2 | 2.17e-02 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 6 | 31 | 2.22e-02 |
| GO:BP | GO:0010825 | positive regulation of centrosome duplication | 1 | 3 | 2.31e-02 |
| GO:BP | GO:0019276 | UDP-N-acetylgalactosamine metabolic process | 2 | 2 | 2.41e-02 |
| GO:BP | GO:0046033 | AMP metabolic process | 2 | 22 | 2.49e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 8 | 35 | 2.50e-02 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 7 | 30 | 2.50e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 4 | 11 | 2.56e-02 |
| GO:BP | GO:0035611 | protein branching point deglutamylation | 1 | 1 | 2.57e-02 |
| GO:BP | GO:0002076 | osteoblast development | 2 | 12 | 2.57e-02 |
| GO:BP | GO:0086016 | AV node cell action potential | 4 | 9 | 2.66e-02 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 4 | 9 | 2.66e-02 |
| GO:BP | GO:0051795 | positive regulation of timing of catagen | 1 | 3 | 2.67e-02 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 4 | 13 | 2.67e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 4 | 84 | 2.67e-02 |
| GO:BP | GO:0002227 | innate immune response in mucosa | 4 | 8 | 2.67e-02 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 2.67e-02 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 4 | 76 | 2.73e-02 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 7 | 37 | 2.73e-02 |
| GO:BP | GO:0044208 | ‘de novo’ AMP biosynthetic process | 3 | 8 | 2.73e-02 |
| GO:BP | GO:0048818 | positive regulation of hair follicle maturation | 2 | 5 | 2.73e-02 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 2.76e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 8 | 36 | 2.77e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 8 | 36 | 2.77e-02 |
| GO:BP | GO:0072718 | response to cisplatin | 3 | 6 | 2.77e-02 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 26 | 207 | 2.84e-02 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 2 | 14 | 2.89e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 3 | 7 | 2.92e-02 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 5 | 12 | 2.92e-02 |
| GO:BP | GO:0035973 | aggrephagy | 2 | 8 | 2.92e-02 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 3 | 5 | 2.95e-02 |
| GO:BP | GO:0045870 | positive regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 2 | 3.02e-02 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 11 | 35 | 3.03e-02 |
| GO:BP | GO:1902010 | negative regulation of translation in response to endoplasmic reticulum stress | 1 | 1 | 3.09e-02 |
| GO:BP | GO:0060013 | righting reflex | 3 | 6 | 3.13e-02 |
| GO:BP | GO:0051781 | positive regulation of cell division | 11 | 64 | 3.24e-02 |
| GO:BP | GO:0042637 | catagen | 1 | 4 | 3.31e-02 |
| GO:BP | GO:0051794 | regulation of timing of catagen | 1 | 4 | 3.31e-02 |
| GO:BP | GO:0060364 | frontal suture morphogenesis | 1 | 4 | 3.31e-02 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 3.31e-02 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 12 | 67 | 3.36e-02 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 11 | 34 | 3.36e-02 |
| GO:BP | GO:1904975 | response to bleomycin | 2 | 2 | 3.38e-02 |
| GO:BP | GO:1904976 | cellular response to bleomycin | 2 | 2 | 3.38e-02 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 1 | 5 | 3.40e-02 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 5 | 18 | 3.45e-02 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 4 | 11 | 3.45e-02 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 5 | 15 | 3.48e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 3 | 5 | 3.50e-02 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 6 | 30 | 3.57e-02 |
| GO:BP | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 3.59e-02 |
| GO:BP | GO:0009216 | purine deoxyribonucleoside triphosphate biosynthetic process | 1 | 1 | 3.59e-02 |
| GO:BP | GO:0006175 | dATP biosynthetic process | 1 | 1 | 3.59e-02 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 5 | 17 | 3.64e-02 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 5 | 17 | 3.64e-02 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 7 | 29 | 3.64e-02 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 2 | 27 | 3.64e-02 |
| GO:BP | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling | 2 | 2 | 3.64e-02 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 3 | 8 | 3.64e-02 |
| GO:BP | GO:0006334 | nucleosome assembly | 11 | 80 | 3.64e-02 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 5 | 17 | 3.64e-02 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 3.64e-02 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 14 | 146 | 3.68e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 6 | 26 | 3.68e-02 |
| GO:BP | GO:0097106 | postsynaptic density organization | 5 | 30 | 3.69e-02 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 9 | 103 | 3.77e-02 |
| GO:BP | GO:0048532 | anatomical structure arrangement | 5 | 13 | 3.80e-02 |
| GO:BP | GO:0034728 | nucleosome organization | 12 | 98 | 3.88e-02 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 5 | 23 | 3.90e-02 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 6 | 16 | 3.90e-02 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 3.93e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 11 | 97 | 3.97e-02 |
| GO:BP | GO:0046338 | phosphatidylethanolamine catabolic process | 2 | 4 | 3.97e-02 |
| GO:BP | GO:0044806 | G-quadruplex DNA unwinding | 3 | 7 | 4.07e-02 |
| GO:BP | GO:0044314 | protein K27-linked ubiquitination | 3 | 8 | 4.07e-02 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 4 | 23 | 4.07e-02 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 10 | 158 | 4.09e-02 |
| GO:BP | GO:0030010 | establishment of cell polarity | 15 | 134 | 4.09e-02 |
| GO:BP | GO:0051338 | regulation of transferase activity | 64 | 679 | 4.11e-02 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 10 | 38 | 4.14e-02 |
| GO:BP | GO:0090399 | replicative senescence | 4 | 13 | 4.16e-02 |
| GO:BP | GO:0038101 | sequestering of nodal from receptor via nodal binding | 1 | 1 | 4.19e-02 |
| GO:BP | GO:1903722 | regulation of centriole elongation | 1 | 7 | 4.30e-02 |
| GO:BP | GO:0021604 | cranial nerve structural organization | 4 | 8 | 4.35e-02 |
| GO:BP | GO:0006970 | response to osmotic stress | 6 | 67 | 4.35e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 7 | 34 | 4.35e-02 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 2 | 8 | 4.36e-02 |
| GO:BP | GO:0032792 | negative regulation of CREB transcription factor activity | 1 | 4 | 4.36e-02 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 4.36e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 22 | 240 | 4.36e-02 |
| GO:BP | GO:2001170 | negative regulation of ATP biosynthetic process | 2 | 3 | 4.53e-02 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 4.56e-02 |
| GO:BP | GO:0043111 | replication fork arrest | 2 | 2 | 4.56e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 17 | 101 | 4.56e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 6 | 92 | 4.61e-02 |
| GO:BP | GO:0002072 | optic cup morphogenesis involved in camera-type eye development | 2 | 4 | 4.62e-02 |
| GO:BP | GO:0021649 | vestibulocochlear nerve structural organization | 2 | 2 | 4.74e-02 |
| GO:BP | GO:1903375 | facioacoustic ganglion development | 2 | 2 | 4.74e-02 |
| GO:BP | GO:0061511 | centriole elongation | 1 | 8 | 4.74e-02 |
| GO:BP | GO:0032223 | negative regulation of synaptic transmission, cholinergic | 1 | 1 | 4.74e-02 |
| GO:BP | GO:1904835 | dorsal root ganglion morphogenesis | 2 | 2 | 4.74e-02 |
| GO:BP | GO:0061552 | ganglion morphogenesis | 2 | 2 | 4.74e-02 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 2 | 13 | 4.85e-02 |
| GO:BP | GO:0046340 | diacylglycerol catabolic process | 2 | 3 | 4.86e-02 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 4.86e-02 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 4.86e-02 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 28 | 229 | 4.86e-02 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 4.86e-02 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 4.86e-02 |
| GO:BP | GO:0072308 | negative regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 1 | 4.95e-02 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 14 | 89 | 5.04e-02 |
| GO:BP | GO:0006282 | regulation of DNA repair | 24 | 198 | 5.07e-02 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 2 | 37 | 5.07e-02 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 6 | 681 | 5.07e-02 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 11 | 68 | 5.08e-02 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 17 | 121 | 5.08e-02 |
| GO:BP | GO:0006543 | glutamine catabolic process | 2 | 3 | 5.09e-02 |
| GO:BP | GO:0021694 | cerebellar Purkinje cell layer formation | 1 | 12 | 5.09e-02 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 10 | 156 | 5.11e-02 |
| GO:BP | GO:2000620 | positive regulation of histone H4-K16 acetylation | 2 | 2 | 5.11e-02 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 5 | 17 | 5.15e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 9 | 132 | 5.25e-02 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 5.26e-02 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 7 | 32 | 5.26e-02 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 32 | 5.26e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 25 | 293 | 5.26e-02 |
| GO:BP | GO:0071139 | resolution of recombination intermediates | 3 | 6 | 5.26e-02 |
| GO:BP | GO:0045004 | DNA replication proofreading | 2 | 2 | 5.31e-02 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 2 | 39 | 5.35e-02 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 17 | 377 | 5.36e-02 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 8 | 31 | 5.36e-02 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 3 | 10 | 5.37e-02 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 3 | 23 | 5.37e-02 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 3 | 22 | 5.40e-02 |
| GO:BP | GO:0042594 | response to starvation | 9 | 178 | 5.40e-02 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 6 | 696 | 5.40e-02 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 14 | 149 | 5.40e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 2 | 123 | 5.41e-02 |
| GO:BP | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | 1 | 8 | 5.46e-02 |
| GO:BP | GO:0045217 | cell-cell junction maintenance | 1 | 13 | 5.46e-02 |
| GO:BP | GO:0051100 | negative regulation of binding | 2 | 135 | 5.46e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 16 | 111 | 5.46e-02 |
| GO:BP | GO:0032901 | positive regulation of neurotrophin production | 1 | 1 | 5.46e-02 |
| GO:BP | GO:0006188 | IMP biosynthetic process | 3 | 10 | 5.47e-02 |
| GO:BP | GO:0006082 | organic acid metabolic process | 6 | 700 | 5.47e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 16 | 459 | 5.48e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 4 | 101 | 5.48e-02 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 4 | 26 | 5.49e-02 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 2 | 11 | 5.50e-02 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 5 | 155 | 5.50e-02 |
| GO:BP | GO:0032020 | ISG15-protein conjugation | 1 | 6 | 5.59e-02 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 105 | 2011 | 5.59e-02 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 1 | 8 | 5.59e-02 |
| GO:BP | GO:0044848 | biological phase | 1 | 9 | 5.59e-02 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 1 | 9 | 5.59e-02 |
| GO:BP | GO:0045617 | negative regulation of keratinocyte differentiation | 1 | 3 | 5.59e-02 |
| GO:BP | GO:0044851 | hair cycle phase | 1 | 9 | 5.59e-02 |
| GO:BP | GO:0048819 | regulation of hair follicle maturation | 1 | 9 | 5.59e-02 |
| GO:BP | GO:0019856 | pyrimidine nucleobase biosynthetic process | 1 | 8 | 5.78e-02 |
| GO:BP | GO:0000209 | protein polyubiquitination | 4 | 226 | 5.82e-02 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 1 | 16 | 5.84e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 2 | 131 | 5.84e-02 |
| GO:BP | GO:0071772 | response to BMP | 2 | 131 | 5.84e-02 |
| GO:BP | GO:0140131 | positive regulation of lymphocyte chemotaxis | 3 | 10 | 5.93e-02 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 5 | 29 | 6.00e-02 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 2 | 59 | 6.02e-02 |
| GO:BP | GO:0090037 | positive regulation of protein kinase C signaling | 3 | 7 | 6.03e-02 |
| GO:BP | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 3 | 23 | 6.04e-02 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 1 | 10 | 6.04e-02 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 3 | 5 | 6.04e-02 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 3 | 440 | 6.04e-02 |
| GO:BP | GO:0060346 | bone trabecula formation | 1 | 9 | 6.04e-02 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 3 | 11 | 6.06e-02 |
| GO:BP | GO:0000921 | septin ring assembly | 1 | 1 | 6.11e-02 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 4 | 38 | 6.11e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 11 | 104 | 6.12e-02 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 4 | 40 | 6.13e-02 |
| GO:BP | GO:0106381 | purine deoxyribonucleotide salvage | 2 | 3 | 6.14e-02 |
| GO:BP | GO:0106383 | dAMP salvage | 2 | 3 | 6.14e-02 |
| GO:BP | GO:0009171 | purine deoxyribonucleoside monophosphate biosynthetic process | 2 | 3 | 6.14e-02 |
| GO:BP | GO:0006170 | dAMP biosynthetic process | 2 | 3 | 6.14e-02 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 2 | 116 | 6.14e-02 |
| GO:BP | GO:0002385 | mucosal immune response | 4 | 11 | 6.14e-02 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 10 | 219 | 6.14e-02 |
| GO:BP | GO:0046521 | sphingoid catabolic process | 1 | 1 | 6.16e-02 |
| GO:BP | GO:1905364 | regulation of endosomal vesicle fusion | 1 | 1 | 6.16e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 16 | 353 | 6.24e-02 |
| GO:BP | GO:0019372 | lipoxygenase pathway | 2 | 5 | 6.25e-02 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 5 | 17 | 6.26e-02 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 6.39e-02 |
| GO:BP | GO:0033212 | iron import into cell | 3 | 7 | 6.41e-02 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 4 | 13 | 6.46e-02 |
| GO:BP | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 2 | 3 | 6.46e-02 |
| GO:BP | GO:0090656 | t-circle formation | 4 | 13 | 6.46e-02 |
| GO:BP | GO:0097009 | energy homeostasis | 4 | 39 | 6.46e-02 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 4 | 13 | 6.46e-02 |
| GO:BP | GO:1900176 | negative regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 1 | 6.49e-02 |
| GO:BP | GO:1900146 | negative regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 1 | 6.49e-02 |
| GO:BP | GO:0036438 | maintenance of lens transparency | 2 | 6 | 6.49e-02 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 2 | 3 | 6.53e-02 |
| GO:BP | GO:0035904 | aorta development | 3 | 50 | 6.72e-02 |
| GO:BP | GO:0003230 | cardiac atrium development | 5 | 34 | 6.72e-02 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 5 | 48 | 6.72e-02 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 3 | 8 | 6.72e-02 |
| GO:BP | GO:0061820 | telomeric D-loop disassembly | 3 | 8 | 6.72e-02 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 4 | 14 | 6.72e-02 |
| GO:BP | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity | 1 | 5 | 6.73e-02 |
| GO:BP | GO:0051798 | positive regulation of hair follicle development | 1 | 9 | 6.73e-02 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 2 | 64 | 6.80e-02 |
| GO:BP | GO:0016567 | protein ubiquitination | 7 | 717 | 6.90e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 6 | 27 | 6.95e-02 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 2 | 4 | 6.97e-02 |
| GO:BP | GO:2000403 | positive regulation of lymphocyte migration | 4 | 24 | 7.00e-02 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 4 | 14 | 7.00e-02 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 4 | 21 | 7.00e-02 |
| GO:BP | GO:0035445 | borate transmembrane transport | 1 | 1 | 7.00e-02 |
| GO:BP | GO:0046713 | borate transport | 1 | 1 | 7.00e-02 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 3 | 12 | 7.05e-02 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 3 | 12 | 7.05e-02 |
| GO:BP | GO:0060117 | auditory receptor cell development | 1 | 14 | 7.08e-02 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 7.08e-02 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 5 | 172 | 7.08e-02 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 1 | 12 | 7.08e-02 |
| GO:BP | GO:0006284 | base-excision repair | 5 | 44 | 7.08e-02 |
| GO:BP | GO:0001507 | acetylcholine catabolic process in synaptic cleft | 1 | 2 | 7.25e-02 |
| GO:BP | GO:0046060 | dATP metabolic process | 1 | 3 | 7.28e-02 |
| GO:BP | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate | 4 | 15 | 7.28e-02 |
| GO:BP | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling | 2 | 3 | 7.28e-02 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 1 | 22 | 7.28e-02 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 3 | 9 | 7.35e-02 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 7.36e-02 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 4 | 14 | 7.40e-02 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 3 | 27 | 7.41e-02 |
| GO:BP | GO:0007292 | female gamete generation | 19 | 103 | 7.45e-02 |
| GO:BP | GO:0045683 | negative regulation of epidermis development | 1 | 6 | 7.45e-02 |
| GO:BP | GO:0045605 | negative regulation of epidermal cell differentiation | 1 | 6 | 7.45e-02 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 7.46e-02 |
| GO:BP | GO:0002027 | regulation of heart rate | 13 | 86 | 7.49e-02 |
| GO:BP | GO:0035609 | C-terminal protein deglutamylation | 1 | 2 | 7.50e-02 |
| GO:BP | GO:0072183 | negative regulation of nephron tubule epithelial cell differentiation | 1 | 2 | 7.55e-02 |
| GO:BP | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 2 | 7.55e-02 |
| GO:BP | GO:0046725 | negative regulation by virus of viral protein levels in host cell | 1 | 2 | 7.55e-02 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 9 | 194 | 7.58e-02 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 1 | 24 | 7.61e-02 |
| GO:BP | GO:0030879 | mammary gland development | 2 | 106 | 7.62e-02 |
| GO:BP | GO:1900121 | negative regulation of receptor binding | 1 | 8 | 7.64e-02 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 4 | 21 | 7.64e-02 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 9 | 58 | 7.64e-02 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 2 | 50 | 7.64e-02 |
| GO:BP | GO:0032055 | negative regulation of translation in response to stress | 1 | 4 | 7.64e-02 |
| GO:BP | GO:1990928 | response to amino acid starvation | 4 | 51 | 7.66e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 5 | 77 | 7.67e-02 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 4 | 13 | 7.67e-02 |
| GO:BP | GO:0048820 | hair follicle maturation | 1 | 15 | 7.69e-02 |
| GO:BP | GO:0007019 | microtubule depolymerization | 6 | 43 | 7.69e-02 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 1 | 5 | 7.69e-02 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 1 | 7 | 7.69e-02 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 6 | 103 | 7.75e-02 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 7 | 55 | 7.79e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 7 | 39 | 7.82e-02 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 9 | 64 | 7.85e-02 |
| GO:BP | GO:0003094 | glomerular filtration | 4 | 21 | 7.90e-02 |
| GO:BP | GO:0006338 | chromatin remodeling | 26 | 378 | 7.90e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 21 | 560 | 7.90e-02 |
| GO:BP | GO:0043542 | endothelial cell migration | 9 | 182 | 7.90e-02 |
| GO:BP | GO:0001556 | oocyte maturation | 6 | 22 | 7.90e-02 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 7 | 42 | 7.90e-02 |
| GO:BP | GO:1902855 | regulation of non-motile cilium assembly | 2 | 10 | 7.95e-02 |
| GO:BP | GO:0000722 | telomere maintenance via recombination | 4 | 14 | 8.10e-02 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 4 | 14 | 8.12e-02 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 3 | 8 | 8.13e-02 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 1 | 12 | 8.15e-02 |
| GO:BP | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate | 4 | 16 | 8.15e-02 |
| GO:BP | GO:0060900 | embryonic camera-type eye formation | 2 | 8 | 8.19e-02 |
| GO:BP | GO:0072702 | response to methyl methanesulfonate | 1 | 1 | 8.20e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 7 | 31 | 8.20e-02 |
| GO:BP | GO:0072703 | cellular response to methyl methanesulfonate | 1 | 1 | 8.20e-02 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 8.24e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 26 | 547 | 8.24e-02 |
| GO:BP | GO:0044565 | dendritic cell proliferation | 2 | 3 | 8.28e-02 |
| GO:BP | GO:0035610 | protein side chain deglutamylation | 1 | 2 | 8.33e-02 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 8.33e-02 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 8.33e-02 |
| GO:BP | GO:0032060 | bleb assembly | 2 | 9 | 8.34e-02 |
| GO:BP | GO:0072092 | ureteric bud invasion | 2 | 3 | 8.38e-02 |
| GO:BP | GO:0048681 | negative regulation of axon regeneration | 1 | 11 | 8.45e-02 |
| GO:BP | GO:0006325 | chromatin organization | 34 | 551 | 8.47e-02 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 2 | 65 | 8.48e-02 |
| GO:BP | GO:0006206 | pyrimidine nucleobase metabolic process | 1 | 13 | 8.48e-02 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 1 | 13 | 8.48e-02 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 5 | 19 | 8.53e-02 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 7 | 37 | 8.58e-02 |
| GO:BP | GO:1901623 | regulation of lymphocyte chemotaxis | 3 | 13 | 8.62e-02 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 3 | 13 | 8.65e-02 |
| GO:BP | GO:0090129 | positive regulation of synapse maturation | 1 | 8 | 8.65e-02 |
| GO:BP | GO:1900060 | negative regulation of ceramide biosynthetic process | 2 | 5 | 8.71e-02 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 11 | 119 | 8.75e-02 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 1 | 5 | 8.75e-02 |
| GO:BP | GO:1904502 | regulation of lipophagy | 1 | 5 | 8.75e-02 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 1 | 30 | 8.78e-02 |
| GO:BP | GO:0060122 | inner ear receptor cell stereocilium organization | 1 | 23 | 8.78e-02 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 8.78e-02 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 12 | 425 | 8.78e-02 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 1 | 12 | 8.80e-02 |
| GO:BP | GO:0070245 | positive regulation of thymocyte apoptotic process | 2 | 5 | 8.80e-02 |
| GO:BP | GO:0008340 | determination of adult lifespan | 5 | 16 | 8.80e-02 |
| GO:BP | GO:0046036 | CTP metabolic process | 1 | 14 | 8.80e-02 |
| GO:BP | GO:0098904 | regulation of AV node cell action potential | 2 | 3 | 8.80e-02 |
| GO:BP | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis | 2 | 3 | 8.80e-02 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 2 | 4 | 8.81e-02 |
| GO:BP | GO:0097094 | craniofacial suture morphogenesis | 1 | 15 | 8.88e-02 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 8.88e-02 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 5 | 65 | 8.88e-02 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 3 | 15 | 8.88e-02 |
| GO:BP | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 2 | 26 | 8.93e-02 |
| GO:BP | GO:0021612 | facial nerve structural organization | 3 | 7 | 8.93e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 6 | 31 | 8.96e-02 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 2 | 13 | 8.96e-02 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 2 | 13 | 8.96e-02 |
| GO:BP | GO:0097374 | sensory neuron axon guidance | 2 | 2 | 8.99e-02 |
| GO:BP | GO:0036486 | ventral trunk neural crest cell migration | 2 | 3 | 8.99e-02 |
| GO:BP | GO:1901753 | leukotriene A4 biosynthetic process | 1 | 1 | 9.00e-02 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 7 | 787 | 9.03e-02 |
| GO:BP | GO:0035608 | protein deglutamylation | 1 | 2 | 9.09e-02 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 2 | 5 | 9.10e-02 |
| GO:BP | GO:0030809 | negative regulation of nucleotide biosynthetic process | 2 | 3 | 9.10e-02 |
| GO:BP | GO:0097205 | renal filtration | 4 | 23 | 9.10e-02 |
| GO:BP | GO:0006581 | acetylcholine catabolic process | 1 | 3 | 9.10e-02 |
| GO:BP | GO:1903579 | negative regulation of ATP metabolic process | 2 | 5 | 9.10e-02 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 9.10e-02 |
| GO:BP | GO:1900372 | negative regulation of purine nucleotide biosynthetic process | 2 | 3 | 9.10e-02 |
| GO:BP | GO:0051797 | regulation of hair follicle development | 1 | 16 | 9.10e-02 |
| GO:BP | GO:0031106 | septin ring organization | 1 | 2 | 9.11e-02 |
| GO:BP | GO:1904172 | positive regulation of bleb assembly | 1 | 1 | 9.11e-02 |
| GO:BP | GO:1905463 | negative regulation of DNA duplex unwinding | 2 | 4 | 9.11e-02 |
| GO:BP | GO:1903416 | response to glycoside | 2 | 6 | 9.11e-02 |
| GO:BP | GO:0071233 | cellular response to leucine | 2 | 10 | 9.11e-02 |
| GO:BP | GO:0019731 | antibacterial humoral response | 5 | 15 | 9.11e-02 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 4 | 11 | 9.11e-02 |
| GO:BP | GO:1904170 | regulation of bleb assembly | 1 | 1 | 9.11e-02 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 3 | 29 | 9.11e-02 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 4 | 260 | 9.13e-02 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 6 | 97 | 9.17e-02 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 3 | 16 | 9.24e-02 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 1 | 16 | 9.27e-02 |
| GO:BP | GO:0009749 | response to glucose | 11 | 137 | 9.27e-02 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 1 | 18 | 9.27e-02 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 1 | 17 | 9.27e-02 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 2 | 71 | 9.27e-02 |
| GO:BP | GO:0003170 | heart valve development | 3 | 65 | 9.35e-02 |
| GO:BP | GO:1901186 | positive regulation of ERBB signaling pathway | 2 | 28 | 9.35e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 3 | 69 | 9.35e-02 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 9 | 63 | 9.35e-02 |
| GO:BP | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | 1 | 11 | 9.35e-02 |
| GO:BP | GO:0001578 | microtubule bundle formation | 3 | 79 | 9.35e-02 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 6 | 19 | 9.37e-02 |
| GO:BP | GO:0072135 | kidney mesenchymal cell proliferation | 1 | 2 | 9.37e-02 |
| GO:BP | GO:0072136 | metanephric mesenchymal cell proliferation involved in metanephros development | 1 | 2 | 9.37e-02 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 2 | 22 | 9.46e-02 |
| GO:BP | GO:0070166 | enamel mineralization | 1 | 11 | 9.52e-02 |
| GO:BP | GO:0090657 | telomeric loop disassembly | 3 | 9 | 9.52e-02 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 1 | 36 | 9.52e-02 |
| GO:BP | GO:0051299 | centrosome separation | 3 | 14 | 9.52e-02 |
| GO:BP | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process | 1 | 17 | 9.56e-02 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 6 | 33 | 9.61e-02 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 6 | 22 | 9.61e-02 |
| GO:BP | GO:0032263 | GMP salvage | 1 | 4 | 9.61e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 4 | 21 | 9.61e-02 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 2 | 23 | 9.61e-02 |
| GO:BP | GO:0009153 | purine deoxyribonucleotide biosynthetic process | 1 | 4 | 9.61e-02 |
| GO:BP | GO:0044209 | AMP salvage | 1 | 5 | 9.61e-02 |
| GO:BP | GO:0035582 | sequestering of BMP in extracellular matrix | 1 | 2 | 9.65e-02 |
| GO:BP | GO:1900108 | negative regulation of nodal signaling pathway | 1 | 2 | 9.65e-02 |
| GO:BP | GO:0001843 | neural tube closure | 5 | 90 | 9.67e-02 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 3 | 59 | 9.74e-02 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 1 | 20 | 9.74e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 30 | 881 | 9.74e-02 |
| GO:BP | GO:0048642 | negative regulation of skeletal muscle tissue development | 1 | 3 | 9.74e-02 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 9.76e-02 |
| GO:BP | GO:0097065 | anterior head development | 1 | 7 | 9.83e-02 |
| GO:BP | GO:1900036 | positive regulation of cellular response to heat | 1 | 1 | 9.86e-02 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 1 | 38 | 9.87e-02 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 4 | 23 | 9.87e-02 |
| GO:BP | GO:0071476 | cellular hypotonic response | 2 | 8 | 9.92e-02 |
| GO:BP | GO:0033082 | regulation of extrathymic T cell differentiation | 1 | 1 | 9.92e-02 |
| GO:BP | GO:0060606 | tube closure | 5 | 91 | 9.92e-02 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 2 | 31 | 9.93e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 6 | 28 | 9.97e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 230 | 9.98e-02 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 1 | 29 | 9.98e-02 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 3 | 191 | 1.01e-01 |
| GO:BP | GO:1902145 | regulation of response to cell cycle checkpoint signaling | 1 | 1 | 1.01e-01 |
| GO:BP | GO:1902153 | regulation of response to DNA damage checkpoint signaling | 1 | 1 | 1.01e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 1 | 40 | 1.01e-01 |
| GO:BP | GO:1902151 | regulation of response to DNA integrity checkpoint signaling | 1 | 1 | 1.01e-01 |
| GO:BP | GO:0014029 | neural crest formation | 3 | 13 | 1.02e-01 |
| GO:BP | GO:0021768 | nucleus accumbens development | 1 | 1 | 1.03e-01 |
| GO:BP | GO:0009746 | response to hexose | 11 | 138 | 1.03e-01 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 1 | 23 | 1.03e-01 |
| GO:BP | GO:0070459 | prolactin secretion | 1 | 4 | 1.03e-01 |
| GO:BP | GO:0000423 | mitophagy | 4 | 37 | 1.03e-01 |
| GO:BP | GO:1901660 | calcium ion export | 2 | 3 | 1.03e-01 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 6 | 36 | 1.03e-01 |
| GO:BP | GO:0098530 | positive regulation of strand invasion | 1 | 1 | 1.03e-01 |
| GO:BP | GO:0060576 | intestinal epithelial cell development | 2 | 11 | 1.03e-01 |
| GO:BP | GO:0060542 | regulation of strand invasion | 1 | 1 | 1.03e-01 |
| GO:BP | GO:0008544 | epidermis development | 2 | 210 | 1.03e-01 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 4 | 11 | 1.03e-01 |
| GO:BP | GO:0060119 | inner ear receptor cell development | 1 | 33 | 1.03e-01 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 4 | 16 | 1.03e-01 |
| GO:BP | GO:0032899 | regulation of neurotrophin production | 1 | 2 | 1.03e-01 |
| GO:BP | GO:0061724 | lipophagy | 1 | 7 | 1.03e-01 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 8 | 55 | 1.04e-01 |
| GO:BP | GO:0015701 | bicarbonate transport | 2 | 19 | 1.04e-01 |
| GO:BP | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | 1 | 8 | 1.04e-01 |
| GO:BP | GO:0060343 | trabecula formation | 1 | 23 | 1.04e-01 |
| GO:BP | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process | 1 | 6 | 1.05e-01 |
| GO:BP | GO:0036517 | chemoattraction of serotonergic neuron axon | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0036164 | cell-abiotic substrate adhesion | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0021695 | cerebellar cortex development | 1 | 44 | 1.05e-01 |
| GO:BP | GO:1904934 | negative regulation of cell proliferation in midbrain | 1 | 1 | 1.05e-01 |
| GO:BP | GO:1904613 | cellular response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 1.05e-01 |
| GO:BP | GO:0090155 | negative regulation of sphingolipid biosynthetic process | 2 | 6 | 1.05e-01 |
| GO:BP | GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0090179 | planar cell polarity pathway involved in neural tube closure | 2 | 12 | 1.05e-01 |
| GO:BP | GO:1904716 | positive regulation of chaperone-mediated autophagy | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0061354 | planar cell polarity pathway involved in pericardium morphogenesis | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0061350 | planar cell polarity pathway involved in cardiac muscle tissue morphogenesis | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0061349 | planar cell polarity pathway involved in cardiac right atrium morphogenesis | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0061348 | planar cell polarity pathway involved in ventricular septum morphogenesis | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0021756 | striatum development | 3 | 17 | 1.05e-01 |
| GO:BP | GO:0061347 | planar cell polarity pathway involved in outflow tract morphogenesis | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0061346 | planar cell polarity pathway involved in heart morphogenesis | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development | 1 | 1 | 1.05e-01 |
| GO:BP | GO:0010458 | exit from mitosis | 6 | 30 | 1.05e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 5 | 464 | 1.05e-01 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 3 | 197 | 1.05e-01 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 4 | 64 | 1.05e-01 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 1 | 18 | 1.05e-01 |
| GO:BP | GO:0070814 | hydrogen sulfide biosynthetic process | 2 | 4 | 1.06e-01 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 4 | 72 | 1.06e-01 |
| GO:BP | GO:0006950 | response to stress | 64 | 2780 | 1.06e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 1.06e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 8 | 156 | 1.06e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 5 | 479 | 1.06e-01 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 6 | 28 | 1.07e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 1.07e-01 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 1 | 22 | 1.07e-01 |
| GO:BP | GO:1900120 | regulation of receptor binding | 1 | 17 | 1.07e-01 |
| GO:BP | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration | 2 | 6 | 1.07e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 7 | 121 | 1.07e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 1.07e-01 |
| GO:BP | GO:1901862 | negative regulation of muscle tissue development | 2 | 9 | 1.07e-01 |
| GO:BP | GO:0097186 | amelogenesis | 1 | 14 | 1.07e-01 |
| GO:BP | GO:0015691 | cadmium ion transport | 2 | 5 | 1.07e-01 |
| GO:BP | GO:1900175 | regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 2 | 1.07e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 4 | 47 | 1.07e-01 |
| GO:BP | GO:0070574 | cadmium ion transmembrane transport | 2 | 5 | 1.07e-01 |
| GO:BP | GO:0060393 | regulation of pathway-restricted SMAD protein phosphorylation | 6 | 50 | 1.07e-01 |
| GO:BP | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry | 1 | 2 | 1.07e-01 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 4 | 73 | 1.07e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 2 | 13 | 1.07e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 1 | 34 | 1.07e-01 |
| GO:BP | GO:0001779 | natural killer cell differentiation | 2 | 18 | 1.07e-01 |
| GO:BP | GO:0033194 | response to hydroperoxide | 2 | 12 | 1.07e-01 |
| GO:BP | GO:2000697 | negative regulation of epithelial cell differentiation involved in kidney development | 1 | 3 | 1.07e-01 |
| GO:BP | GO:0010752 | regulation of cGMP-mediated signaling | 2 | 8 | 1.08e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 1.08e-01 |
| GO:BP | GO:0003416 | endochondral bone growth | 1 | 23 | 1.08e-01 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 4 | 73 | 1.09e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 5 | 94 | 1.09e-01 |
| GO:BP | GO:0010990 | regulation of SMAD protein complex assembly | 2 | 6 | 1.10e-01 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 2 | 5 | 1.10e-01 |
| GO:BP | GO:0032466 | negative regulation of cytokinesis | 2 | 7 | 1.10e-01 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 3 | 10 | 1.10e-01 |
| GO:BP | GO:0070342 | brown fat cell proliferation | 1 | 1 | 1.11e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 5 | 89 | 1.12e-01 |
| GO:BP | GO:0044068 | modulation by symbiont of host cellular process | 2 | 5 | 1.12e-01 |
| GO:BP | GO:0019048 | modulation by virus of host process | 2 | 5 | 1.12e-01 |
| GO:BP | GO:0043157 | response to cation stress | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0019054 | modulation by virus of host cellular process | 2 | 5 | 1.12e-01 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 2 | 9 | 1.12e-01 |
| GO:BP | GO:1902608 | positive regulation of large conductance calcium-activated potassium channel activity | 1 | 2 | 1.12e-01 |
| GO:BP | GO:0090663 | galanin-activated signaling pathway | 1 | 1 | 1.12e-01 |
| GO:BP | GO:0044003 | modulation by symbiont of host process | 2 | 5 | 1.12e-01 |
| GO:BP | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure | 2 | 13 | 1.13e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 18 | 206 | 1.13e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 1.13e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 2 | 34 | 1.13e-01 |
| GO:BP | GO:0140290 | peptidyl-serine ADP-deribosylation | 1 | 1 | 1.14e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 2 | 265 | 1.14e-01 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 1 | 7 | 1.14e-01 |
| GO:BP | GO:0046053 | dAMP metabolic process | 1 | 5 | 1.14e-01 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 3 | 94 | 1.14e-01 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 14 | 169 | 1.15e-01 |
| GO:BP | GO:0098868 | bone growth | 1 | 24 | 1.15e-01 |
| GO:BP | GO:0034505 | tooth mineralization | 1 | 17 | 1.15e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 3 | 277 | 1.15e-01 |
| GO:BP | GO:0099606 | microtubule plus-end directed mitotic chromosome migration | 1 | 1 | 1.15e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 1.15e-01 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 1 | 18 | 1.15e-01 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 7 | 29 | 1.15e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 5 | 39 | 1.15e-01 |
| GO:BP | GO:0051591 | response to cAMP | 5 | 66 | 1.15e-01 |
| GO:BP | GO:0003336 | corneocyte desquamation | 1 | 1 | 1.15e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 6 | 110 | 1.16e-01 |
| GO:BP | GO:0002063 | chondrocyte development | 1 | 24 | 1.16e-01 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 4 | 38 | 1.16e-01 |
| GO:BP | GO:0097120 | receptor localization to synapse | 5 | 55 | 1.16e-01 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 1 | 17 | 1.16e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 1.16e-01 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 1 | 28 | 1.16e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 11 | 142 | 1.16e-01 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 3 | 35 | 1.16e-01 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 3 | 12 | 1.17e-01 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 12 | 120 | 1.17e-01 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 3 | 12 | 1.17e-01 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 2 | 4 | 1.17e-01 |
| GO:BP | GO:0070194 | synaptonemal complex disassembly | 1 | 2 | 1.17e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 22 | 246 | 1.18e-01 |
| GO:BP | GO:0051095 | regulation of helicase activity | 3 | 12 | 1.19e-01 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 2 | 5 | 1.19e-01 |
| GO:BP | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 3 | 1.19e-01 |
| GO:BP | GO:0006971 | hypotonic response | 2 | 10 | 1.19e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 2 | 6 | 1.19e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 1.20e-01 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 1 | 23 | 1.20e-01 |
| GO:BP | GO:0006269 | DNA replication, synthesis of RNA primer | 2 | 7 | 1.20e-01 |
| GO:BP | GO:0070889 | platelet alpha granule organization | 1 | 2 | 1.20e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 12 | 88 | 1.21e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 1.21e-01 |
| GO:BP | GO:0006287 | base-excision repair, gap-filling | 2 | 14 | 1.21e-01 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 3 | 12 | 1.21e-01 |
| GO:BP | GO:0033078 | extrathymic T cell differentiation | 1 | 1 | 1.21e-01 |
| GO:BP | GO:0052647 | pentitol phosphate metabolic process | 1 | 1 | 1.21e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 1.21e-01 |
| GO:BP | GO:2000431 | regulation of cytokinesis, actomyosin contractile ring assembly | 1 | 1 | 1.21e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 5 | 76 | 1.21e-01 |
| GO:BP | GO:1990654 | sebum secreting cell proliferation | 1 | 2 | 1.21e-01 |
| GO:BP | GO:0009231 | riboflavin biosynthetic process | 1 | 1 | 1.21e-01 |
| GO:BP | GO:0009398 | FMN biosynthetic process | 1 | 1 | 1.21e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 10 | 206 | 1.21e-01 |
| GO:BP | GO:0106380 | purine ribonucleotide salvage | 1 | 7 | 1.21e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 23 | 496 | 1.21e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 1.21e-01 |
| GO:BP | GO:0090185 | negative regulation of kidney development | 1 | 3 | 1.21e-01 |
| GO:BP | GO:0090177 | establishment of planar polarity involved in neural tube closure | 2 | 14 | 1.21e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 1.21e-01 |
| GO:BP | GO:0072162 | metanephric mesenchymal cell differentiation | 1 | 3 | 1.21e-01 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 4 | 39 | 1.21e-01 |
| GO:BP | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 3 | 1.21e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 2 | 278 | 1.21e-01 |
| GO:BP | GO:0046444 | FMN metabolic process | 1 | 1 | 1.21e-01 |
| GO:BP | GO:0060322 | head development | 18 | 633 | 1.21e-01 |
| GO:BP | GO:0052648 | ribitol phosphate metabolic process | 1 | 1 | 1.21e-01 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 5 | 23 | 1.21e-01 |
| GO:BP | GO:0043951 | negative regulation of cAMP-mediated signaling | 3 | 10 | 1.22e-01 |
| GO:BP | GO:0014829 | vascular associated smooth muscle contraction | 3 | 20 | 1.22e-01 |
| GO:BP | GO:0046040 | IMP metabolic process | 3 | 15 | 1.22e-01 |
| GO:BP | GO:0000723 | telomere maintenance | 17 | 142 | 1.23e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 1.23e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 1.23e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 1.23e-01 |
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 1.23e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 4 | 249 | 1.23e-01 |
| GO:BP | GO:0070266 | necroptotic process | 3 | 32 | 1.23e-01 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 1 | 25 | 1.24e-01 |
| GO:BP | GO:0043201 | response to leucine | 2 | 13 | 1.24e-01 |
| GO:BP | GO:0097490 | sympathetic neuron projection extension | 2 | 4 | 1.24e-01 |
| GO:BP | GO:0097491 | sympathetic neuron projection guidance | 2 | 4 | 1.24e-01 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 5 | 1.24e-01 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 2 | 18 | 1.24e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 1 | 41 | 1.24e-01 |
| GO:BP | GO:0061551 | trigeminal ganglion development | 2 | 3 | 1.24e-01 |
| GO:BP | GO:0002021 | response to dietary excess | 3 | 22 | 1.24e-01 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 4 | 70 | 1.24e-01 |
| GO:BP | GO:0021610 | facial nerve morphogenesis | 3 | 8 | 1.24e-01 |
| GO:BP | GO:0021561 | facial nerve development | 3 | 8 | 1.24e-01 |
| GO:BP | GO:0045616 | regulation of keratinocyte differentiation | 1 | 25 | 1.24e-01 |
| GO:BP | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 3 | 1.25e-01 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 1 | 28 | 1.26e-01 |
| GO:BP | GO:0043200 | response to amino acid | 2 | 95 | 1.26e-01 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 1 | 25 | 1.26e-01 |
| GO:BP | GO:0061485 | memory T cell proliferation | 1 | 1 | 1.27e-01 |
| GO:BP | GO:0014824 | artery smooth muscle contraction | 2 | 7 | 1.27e-01 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 4 | 12 | 1.27e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 4 | 317 | 1.27e-01 |
| GO:BP | GO:1901751 | leukotriene A4 metabolic process | 1 | 2 | 1.27e-01 |
| GO:BP | GO:0086100 | endothelin receptor signaling pathway | 2 | 9 | 1.27e-01 |
| GO:BP | GO:1904999 | positive regulation of leukocyte adhesion to arterial endothelial cell | 1 | 1 | 1.27e-01 |
| GO:BP | GO:0006470 | protein dephosphorylation | 2 | 214 | 1.27e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 6 | 76 | 1.27e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 1 | 21 | 1.27e-01 |
| GO:BP | GO:0018410 | C-terminal protein amino acid modification | 1 | 10 | 1.27e-01 |
| GO:BP | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry | 1 | 3 | 1.27e-01 |
| GO:BP | GO:0071694 | maintenance of protein location in extracellular region | 1 | 4 | 1.27e-01 |
| GO:BP | GO:1900164 | nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry | 1 | 3 | 1.27e-01 |
| GO:BP | GO:0090128 | regulation of synapse maturation | 1 | 15 | 1.27e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 4 | 25 | 1.27e-01 |
| GO:BP | GO:0010224 | response to UV-B | 2 | 17 | 1.27e-01 |
| GO:BP | GO:0043687 | post-translational protein modification | 9 | 64 | 1.27e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 18 | 147 | 1.27e-01 |
| GO:BP | GO:0001841 | neural tube formation | 5 | 101 | 1.27e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 4 | 25 | 1.27e-01 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 1 | 26 | 1.27e-01 |
| GO:BP | GO:0001895 | retina homeostasis | 1 | 44 | 1.27e-01 |
| GO:BP | GO:0061912 | selective autophagy | 4 | 90 | 1.28e-01 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 1 | 11 | 1.28e-01 |
| GO:BP | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation | 5 | 37 | 1.28e-01 |
| GO:BP | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 6 | 55 | 1.28e-01 |
| GO:BP | GO:0043174 | nucleoside salvage | 1 | 9 | 1.28e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 6 | 127 | 1.28e-01 |
| GO:BP | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine | 1 | 2 | 1.28e-01 |
| GO:BP | GO:0086055 | Purkinje myocyte to ventricular cardiac muscle cell communication by electrical coupling | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0007420 | brain development | 17 | 591 | 1.28e-01 |
| GO:BP | GO:0006253 | dCTP catabolic process | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0010652 | positive regulation of cell communication by chemical coupling | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0010645 | regulation of cell communication by chemical coupling | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0086044 | atrial cardiac muscle cell to AV node cell communication by electrical coupling | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0098906 | regulation of Purkinje myocyte action potential | 1 | 1 | 1.28e-01 |
| GO:BP | GO:1990029 | vasomotion | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0097492 | sympathetic neuron axon guidance | 2 | 4 | 1.28e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 2 | 20 | 1.28e-01 |
| GO:BP | GO:0071313 | cellular response to caffeine | 3 | 10 | 1.28e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 7 | 45 | 1.28e-01 |
| GO:BP | GO:0032185 | septin cytoskeleton organization | 1 | 4 | 1.28e-01 |
| GO:BP | GO:0021636 | trigeminal nerve morphogenesis | 2 | 4 | 1.28e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 12 | 420 | 1.28e-01 |
| GO:BP | GO:0046065 | dCTP metabolic process | 1 | 1 | 1.28e-01 |
| GO:BP | GO:0021637 | trigeminal nerve structural organization | 2 | 4 | 1.28e-01 |
| GO:BP | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway | 1 | 3 | 1.28e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 1 | 36 | 1.28e-01 |
| GO:BP | GO:0051305 | chromosome movement towards spindle pole | 2 | 7 | 1.28e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 20 | 276 | 1.28e-01 |
| GO:BP | GO:0070212 | protein poly-ADP-ribosylation | 2 | 6 | 1.28e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 4 | 149 | 1.28e-01 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 4 | 21 | 1.28e-01 |
| GO:BP | GO:0043650 | dicarboxylic acid biosynthetic process | 1 | 9 | 1.29e-01 |
| GO:BP | GO:0055001 | muscle cell development | 19 | 165 | 1.29e-01 |
| GO:BP | GO:0051098 | regulation of binding | 2 | 317 | 1.30e-01 |
| GO:BP | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity | 1 | 4 | 1.30e-01 |
| GO:BP | GO:0032898 | neurotrophin production | 1 | 4 | 1.30e-01 |
| GO:BP | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development | 2 | 6 | 1.30e-01 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 4 | 23 | 1.30e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 3 | 192 | 1.30e-01 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 2 | 9 | 1.31e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 20 | 809 | 1.31e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 7 | 898 | 1.31e-01 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 4 | 41 | 1.31e-01 |
| GO:BP | GO:0003323 | type B pancreatic cell development | 3 | 19 | 1.31e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 2 | 13 | 1.31e-01 |
| GO:BP | GO:2000348 | regulation of CD40 signaling pathway | 2 | 4 | 1.31e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 1.32e-01 |
| GO:BP | GO:0099172 | presynapse organization | 5 | 42 | 1.32e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 6 | 133 | 1.32e-01 |
| GO:BP | GO:0019346 | transsulfuration | 2 | 5 | 1.32e-01 |
| GO:BP | GO:0018904 | ether metabolic process | 3 | 20 | 1.32e-01 |
| GO:BP | GO:0016310 | phosphorylation | 61 | 1372 | 1.32e-01 |
| GO:BP | GO:0009092 | homoserine metabolic process | 2 | 5 | 1.32e-01 |
| GO:BP | GO:0070813 | hydrogen sulfide metabolic process | 2 | 5 | 1.32e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 6 | 112 | 1.32e-01 |
| GO:BP | GO:0006565 | L-serine catabolic process | 2 | 4 | 1.32e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 303 | 1.32e-01 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 3 | 129 | 1.32e-01 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 1 | 33 | 1.32e-01 |
| GO:BP | GO:0046337 | phosphatidylethanolamine metabolic process | 3 | 19 | 1.32e-01 |
| GO:BP | GO:0042631 | cellular response to water deprivation | 1 | 1 | 1.32e-01 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 3 | 28 | 1.32e-01 |
| GO:BP | GO:0006272 | leading strand elongation | 2 | 4 | 1.32e-01 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 2 | 14 | 1.32e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 10 | 279 | 1.32e-01 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 2 | 19 | 1.32e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 9 | 187 | 1.33e-01 |
| GO:BP | GO:0009170 | purine deoxyribonucleoside monophosphate metabolic process | 1 | 7 | 1.33e-01 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 3 | 17 | 1.33e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 10 | 252 | 1.33e-01 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 2 | 32 | 1.33e-01 |
| GO:BP | GO:1900107 | regulation of nodal signaling pathway | 1 | 3 | 1.33e-01 |
| GO:BP | GO:2000406 | positive regulation of T cell migration | 3 | 20 | 1.33e-01 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 2 | 16 | 1.33e-01 |
| GO:BP | GO:0003140 | determination of left/right asymmetry in lateral mesoderm | 1 | 4 | 1.33e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 6 | 113 | 1.33e-01 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 25 | 1026 | 1.33e-01 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 6 | 35 | 1.33e-01 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 6 | 35 | 1.33e-01 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 5 | 23 | 1.33e-01 |
| GO:BP | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity | 1 | 3 | 1.34e-01 |
| GO:BP | GO:0046398 | UDP-glucuronate metabolic process | 1 | 2 | 1.34e-01 |
| GO:BP | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 2 | 1.34e-01 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 4 | 1.34e-01 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 1 | 20 | 1.34e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 3 | 19 | 1.34e-01 |
| GO:BP | GO:0070231 | T cell apoptotic process | 4 | 39 | 1.34e-01 |
| GO:BP | GO:0042249 | establishment of planar polarity of embryonic epithelium | 2 | 15 | 1.34e-01 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 2 | 2 | 1.34e-01 |
| GO:BP | GO:0008291 | acetylcholine metabolic process | 1 | 3 | 1.35e-01 |
| GO:BP | GO:1900619 | acetate ester metabolic process | 1 | 3 | 1.35e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 9 | 67 | 1.35e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 5 | 82 | 1.36e-01 |
| GO:BP | GO:0014820 | tonic smooth muscle contraction | 2 | 8 | 1.36e-01 |
| GO:BP | GO:0060040 | retinal bipolar neuron differentiation | 1 | 2 | 1.36e-01 |
| GO:BP | GO:0045636 | positive regulation of melanocyte differentiation | 1 | 4 | 1.36e-01 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 3 | 22 | 1.36e-01 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 3 | 10 | 1.37e-01 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 2 | 3 | 1.37e-01 |
| GO:BP | GO:0090132 | epithelium migration | 10 | 252 | 1.37e-01 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 1 | 43 | 1.37e-01 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 1 | 39 | 1.37e-01 |
| GO:BP | GO:0045769 | negative regulation of asymmetric cell division | 1 | 1 | 1.38e-01 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 3 | 240 | 1.38e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 2 | 105 | 1.38e-01 |
| GO:BP | GO:0035695 | mitophagy by induced vacuole formation | 1 | 1 | 1.38e-01 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 3 | 22 | 1.39e-01 |
| GO:BP | GO:0021683 | cerebellar granular layer morphogenesis | 2 | 8 | 1.39e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 8 | 172 | 1.39e-01 |
| GO:BP | GO:0003339 | regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 4 | 1.39e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 3 | 513 | 1.39e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 5 | 80 | 1.39e-01 |
| GO:BP | GO:0010754 | negative regulation of cGMP-mediated signaling | 2 | 4 | 1.39e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 5 | 82 | 1.39e-01 |
| GO:BP | GO:0007018 | microtubule-based movement | 17 | 300 | 1.39e-01 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 1 | 11 | 1.39e-01 |
| GO:BP | GO:0060995 | cell-cell signaling involved in kidney development | 1 | 1 | 1.39e-01 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 1 | 11 | 1.39e-01 |
| GO:BP | GO:0070106 | interleukin-27-mediated signaling pathway | 1 | 4 | 1.39e-01 |
| GO:BP | GO:0061290 | canonical Wnt signaling pathway involved in metanephric kidney development | 1 | 1 | 1.39e-01 |
| GO:BP | GO:0061289 | Wnt signaling pathway involved in kidney development | 1 | 1 | 1.39e-01 |
| GO:BP | GO:0060840 | artery development | 3 | 83 | 1.39e-01 |
| GO:BP | GO:0072204 | cell-cell signaling involved in metanephros development | 1 | 1 | 1.39e-01 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 3 | 241 | 1.39e-01 |
| GO:BP | GO:0032200 | telomere organization | 19 | 166 | 1.39e-01 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 2 | 14 | 1.39e-01 |
| GO:BP | GO:0032261 | purine nucleotide salvage | 1 | 10 | 1.39e-01 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 4 | 51 | 1.39e-01 |
| GO:BP | GO:0072307 | regulation of metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 1.39e-01 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 1 | 11 | 1.39e-01 |
| GO:BP | GO:0072257 | metanephric nephron tubule epithelial cell differentiation | 1 | 5 | 1.39e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 7 | 45 | 1.40e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 1.40e-01 |
| GO:BP | GO:0090036 | regulation of protein kinase C signaling | 4 | 14 | 1.40e-01 |
| GO:BP | GO:0032683 | negative regulation of connective tissue growth factor production | 1 | 1 | 1.40e-01 |
| GO:BP | GO:1903551 | regulation of extracellular exosome assembly | 2 | 6 | 1.40e-01 |
| GO:BP | GO:0098957 | anterograde axonal transport of mitochondrion | 1 | 5 | 1.40e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 3 | 40 | 1.40e-01 |
| GO:BP | GO:1902729 | negative regulation of proteoglycan biosynthetic process | 1 | 1 | 1.40e-01 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 2 | 17 | 1.40e-01 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 13 | 487 | 1.40e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 22 | 597 | 1.40e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 22 | 597 | 1.40e-01 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 1 | 10 | 1.40e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 6 | 132 | 1.41e-01 |
| GO:BP | GO:0051642 | centrosome localization | 4 | 31 | 1.41e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 4 | 31 | 1.41e-01 |
| GO:BP | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation | 1 | 3 | 1.41e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 11 | 184 | 1.41e-01 |
| GO:BP | GO:0046069 | cGMP catabolic process | 1 | 4 | 1.41e-01 |
| GO:BP | GO:0075071 | modulation by symbiont of host autophagy | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0097510 | base-excision repair, AP site formation via deaminated base removal | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0060067 | cervix development | 1 | 2 | 1.41e-01 |
| GO:BP | GO:1905438 | non-canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0010820 | positive regulation of T cell chemotaxis | 2 | 7 | 1.41e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 2 | 54 | 1.41e-01 |
| GO:BP | GO:0021685 | cerebellar granular layer structural organization | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0072721 | cellular response to dithiothreitol | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0000296 | spermine transport | 1 | 3 | 1.41e-01 |
| GO:BP | GO:0021549 | cerebellum development | 1 | 78 | 1.41e-01 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 1 | 43 | 1.41e-01 |
| GO:BP | GO:0060004 | reflex | 3 | 9 | 1.41e-01 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 7 | 52 | 1.41e-01 |
| GO:BP | GO:0021544 | subpallium development | 3 | 18 | 1.41e-01 |
| GO:BP | GO:1905672 | negative regulation of lysosome organization | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0003408 | optic cup formation involved in camera-type eye development | 1 | 1 | 1.41e-01 |
| GO:BP | GO:1905719 | protein localization to perinuclear region of cytoplasm | 2 | 8 | 1.41e-01 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 1 | 59 | 1.41e-01 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 2 | 30 | 1.41e-01 |
| GO:BP | GO:0071275 | cellular response to aluminum ion | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0060031 | mediolateral intercalation | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0090130 | tissue migration | 10 | 256 | 1.41e-01 |
| GO:BP | GO:0071932 | replication fork reversal | 2 | 5 | 1.41e-01 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 4 | 26 | 1.41e-01 |
| GO:BP | GO:0060775 | planar cell polarity pathway involved in gastrula mediolateral intercalation | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0036146 | cellular response to mycotoxin | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0120316 | sperm flagellum assembly | 2 | 26 | 1.41e-01 |
| GO:BP | GO:0010609 | mRNA localization resulting in post-transcriptional regulation of gene expression | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0061642 | chemoattraction of axon | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0140321 | suppression of host autophagy | 1 | 1 | 1.41e-01 |
| GO:BP | GO:0035418 | protein localization to synapse | 5 | 68 | 1.41e-01 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 3 | 11 | 1.41e-01 |
| GO:BP | GO:0032222 | regulation of synaptic transmission, cholinergic | 1 | 3 | 1.41e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 12 | 393 | 1.41e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 1 | 46 | 1.41e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 1 | 43 | 1.41e-01 |
| GO:BP | GO:0036515 | serotonergic neuron axon guidance | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0036518 | chemorepulsion of dopaminergic neuron axon | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0039521 | suppression by virus of host autophagy | 1 | 1 | 1.41e-01 |
| GO:BP | GO:1904933 | regulation of cell proliferation in midbrain | 1 | 1 | 1.41e-01 |
| GO:BP | GO:1900543 | negative regulation of purine nucleotide metabolic process | 2 | 6 | 1.41e-01 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 3 | 17 | 1.41e-01 |
| GO:BP | GO:0060324 | face development | 4 | 45 | 1.41e-01 |
| GO:BP | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0051885 | positive regulation of timing of anagen | 1 | 2 | 1.41e-01 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 3 | 139 | 1.41e-01 |
| GO:BP | GO:0036258 | multivesicular body assembly | 5 | 32 | 1.41e-01 |
| GO:BP | GO:2001151 | regulation of renal water transport | 1 | 1 | 1.42e-01 |
| GO:BP | GO:2001153 | positive regulation of renal water transport | 1 | 1 | 1.42e-01 |
| GO:BP | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation | 1 | 5 | 1.42e-01 |
| GO:BP | GO:1904439 | negative regulation of iron ion import across plasma membrane | 1 | 1 | 1.43e-01 |
| GO:BP | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 2 | 18 | 1.43e-01 |
| GO:BP | GO:0009790 | embryo development | 3 | 875 | 1.43e-01 |
| GO:BP | GO:0035364 | thymine transport | 1 | 1 | 1.43e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 14 | 101 | 1.43e-01 |
| GO:BP | GO:1903934 | positive regulation of DNA primase activity | 2 | 5 | 1.43e-01 |
| GO:BP | GO:1903932 | regulation of DNA primase activity | 2 | 5 | 1.43e-01 |
| GO:BP | GO:0036376 | sodium ion export across plasma membrane | 1 | 12 | 1.44e-01 |
| GO:BP | GO:0043063 | intercellular bridge organization | 1 | 1 | 1.44e-01 |
| GO:BP | GO:0007288 | sperm axoneme assembly | 1 | 20 | 1.44e-01 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 2 | 4 | 1.45e-01 |
| GO:BP | GO:0035581 | sequestering of extracellular ligand from receptor | 1 | 7 | 1.45e-01 |
| GO:BP | GO:0009187 | cyclic nucleotide metabolic process | 2 | 31 | 1.45e-01 |
| GO:BP | GO:1904913 | regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 1.45e-01 |
| GO:BP | GO:1904910 | regulation of establishment of RNA localization to telomere | 1 | 1 | 1.45e-01 |
| GO:BP | GO:1904911 | negative regulation of establishment of RNA localization to telomere | 1 | 1 | 1.45e-01 |
| GO:BP | GO:0045634 | regulation of melanocyte differentiation | 1 | 5 | 1.45e-01 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 3 | 11 | 1.45e-01 |
| GO:BP | GO:1904792 | positive regulation of shelterin complex assembly | 1 | 1 | 1.45e-01 |
| GO:BP | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process | 2 | 6 | 1.45e-01 |
| GO:BP | GO:1904914 | negative regulation of establishment of protein-containing complex localization to telomere | 1 | 1 | 1.45e-01 |
| GO:BP | GO:1904850 | negative regulation of establishment of protein localization to telomere | 1 | 1 | 1.45e-01 |
| GO:BP | GO:0035519 | protein K29-linked ubiquitination | 2 | 6 | 1.45e-01 |
| GO:BP | GO:0046719 | regulation by virus of viral protein levels in host cell | 1 | 8 | 1.45e-01 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 2 | 41 | 1.45e-01 |
| GO:BP | GO:2001012 | mesenchymal cell differentiation involved in renal system development | 1 | 5 | 1.45e-01 |
| GO:BP | GO:0072161 | mesenchymal cell differentiation involved in kidney development | 1 | 5 | 1.45e-01 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 3 | 39 | 1.46e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 1 | 50 | 1.46e-01 |
| GO:BP | GO:1904631 | response to glucoside | 1 | 1 | 1.46e-01 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 4 | 44 | 1.46e-01 |
| GO:BP | GO:0033197 | response to vitamin E | 2 | 9 | 1.46e-01 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 8 | 64 | 1.47e-01 |
| GO:BP | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization | 3 | 16 | 1.47e-01 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 1 | 34 | 1.47e-01 |
| GO:BP | GO:0098735 | positive regulation of the force of heart contraction | 2 | 4 | 1.47e-01 |
| GO:BP | GO:0022037 | metencephalon development | 1 | 84 | 1.47e-01 |
| GO:BP | GO:0002540 | leukotriene production involved in inflammatory response | 1 | 2 | 1.47e-01 |
| GO:BP | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response | 1 | 2 | 1.47e-01 |
| GO:BP | GO:0030903 | notochord development | 2 | 17 | 1.47e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 62 | 631 | 1.47e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 1 | 52 | 1.47e-01 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 4 | 106 | 1.47e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 1.47e-01 |
| GO:BP | GO:1905962 | glutamatergic neuron differentiation | 1 | 2 | 1.47e-01 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 7 | 311 | 1.47e-01 |
| GO:BP | GO:0071539 | protein localization to centrosome | 5 | 31 | 1.47e-01 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 3 | 11 | 1.47e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 3 | 870 | 1.47e-01 |
| GO:BP | GO:1905116 | positive regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 1.48e-01 |
| GO:BP | GO:1905115 | regulation of lateral attachment of mitotic spindle microtubules to kinetochore | 1 | 1 | 1.48e-01 |
| GO:BP | GO:0034478 | phosphatidylglycerol catabolic process | 1 | 1 | 1.48e-01 |
| GO:BP | GO:0010819 | regulation of T cell chemotaxis | 2 | 7 | 1.48e-01 |
| GO:BP | GO:0036257 | multivesicular body organization | 5 | 33 | 1.48e-01 |
| GO:BP | GO:0001967 | suckling behavior | 1 | 10 | 1.48e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 3 | 28 | 1.48e-01 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 1 | 24 | 1.48e-01 |
| GO:BP | GO:1903096 | protein localization to meiotic spindle midzone | 1 | 1 | 1.48e-01 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 1 | 1 | 1.48e-01 |
| GO:BP | GO:1905359 | protein localization to meiotic spindle | 1 | 1 | 1.48e-01 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 6 | 102 | 1.49e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 11 | 89 | 1.49e-01 |
| GO:BP | GO:2000537 | regulation of B cell chemotaxis | 1 | 1 | 1.49e-01 |
| GO:BP | GO:0036484 | trunk neural crest cell migration | 2 | 4 | 1.49e-01 |
| GO:BP | GO:2000538 | positive regulation of B cell chemotaxis | 1 | 1 | 1.49e-01 |
| GO:BP | GO:0035290 | trunk segmentation | 2 | 4 | 1.49e-01 |
| GO:BP | GO:0035965 | cardiolipin acyl-chain remodeling | 2 | 4 | 1.49e-01 |
| GO:BP | GO:0061550 | cranial ganglion development | 2 | 4 | 1.49e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 1.49e-01 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 4 | 26 | 1.49e-01 |
| GO:BP | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 5 | 1.50e-01 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 3 | 8 | 1.50e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 11 | 187 | 1.50e-01 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 1 | 25 | 1.51e-01 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 1 | 9 | 1.51e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 9 | 222 | 1.52e-01 |
| GO:BP | GO:1903126 | negative regulation of centriole-centriole cohesion | 1 | 1 | 1.52e-01 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 7 | 56 | 1.52e-01 |
| GO:BP | GO:0048742 | regulation of skeletal muscle fiber development | 1 | 8 | 1.52e-01 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 3 | 12 | 1.52e-01 |
| GO:BP | GO:0090175 | regulation of establishment of planar polarity | 4 | 54 | 1.52e-01 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 2 | 6 | 1.52e-01 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 1 | 46 | 1.52e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 1.53e-01 |
| GO:BP | GO:0050688 | regulation of defense response to virus | 2 | 63 | 1.53e-01 |
| GO:BP | GO:0032918 | spermidine acetylation | 1 | 2 | 1.53e-01 |
| GO:BP | GO:0032917 | polyamine acetylation | 1 | 2 | 1.53e-01 |
| GO:BP | GO:0009447 | putrescine catabolic process | 1 | 2 | 1.53e-01 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 4 | 34 | 1.53e-01 |
| GO:BP | GO:0072243 | metanephric nephron epithelium development | 2 | 15 | 1.55e-01 |
| GO:BP | GO:0072170 | metanephric tubule development | 2 | 15 | 1.55e-01 |
| GO:BP | GO:0072347 | response to anesthetic | 1 | 1 | 1.55e-01 |
| GO:BP | GO:0003335 | corneocyte development | 1 | 2 | 1.55e-01 |
| GO:BP | GO:0042262 | DNA protection | 2 | 6 | 1.55e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 1.55e-01 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 2 | 40 | 1.55e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 5 | 343 | 1.55e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 1.56e-01 |
| GO:BP | GO:1901859 | negative regulation of mitochondrial DNA metabolic process | 1 | 2 | 1.56e-01 |
| GO:BP | GO:0042415 | norepinephrine metabolic process | 2 | 7 | 1.56e-01 |
| GO:BP | GO:0051257 | meiotic spindle midzone assembly | 1 | 2 | 1.56e-01 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 7 | 319 | 1.56e-01 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 1 | 4 | 1.56e-01 |
| GO:BP | GO:2000401 | regulation of lymphocyte migration | 4 | 41 | 1.56e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 1 | 45 | 1.56e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 12 | 399 | 1.56e-01 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 2 | 111 | 1.56e-01 |
| GO:BP | GO:1905462 | regulation of DNA duplex unwinding | 2 | 7 | 1.57e-01 |
| GO:BP | GO:0032876 | negative regulation of DNA endoreduplication | 1 | 1 | 1.57e-01 |
| GO:BP | GO:0050942 | positive regulation of pigment cell differentiation | 1 | 5 | 1.57e-01 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 2 | 12 | 1.57e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 1.57e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 11 | 282 | 1.57e-01 |
| GO:BP | GO:0021785 | branchiomotor neuron axon guidance | 2 | 5 | 1.58e-01 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 1 | 15 | 1.58e-01 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 1 | 14 | 1.58e-01 |
| GO:BP | GO:0045980 | negative regulation of nucleotide metabolic process | 2 | 6 | 1.58e-01 |
| GO:BP | GO:0060429 | epithelium development | 3 | 850 | 1.58e-01 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 1 | 15 | 1.58e-01 |
| GO:BP | GO:0060914 | heart formation | 5 | 29 | 1.58e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 2 | 17 | 1.58e-01 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 1 | 9 | 1.58e-01 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 1 | 9 | 1.58e-01 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 2 | 9 | 1.58e-01 |
| GO:BP | GO:1902857 | positive regulation of non-motile cilium assembly | 1 | 7 | 1.58e-01 |
| GO:BP | GO:0071695 | anatomical structure maturation | 1 | 48 | 1.58e-01 |
| GO:BP | GO:0060074 | synapse maturation | 1 | 25 | 1.58e-01 |
| GO:BP | GO:0016236 | macroautophagy | 9 | 308 | 1.58e-01 |
| GO:BP | GO:1901861 | regulation of muscle tissue development | 3 | 35 | 1.58e-01 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 2 | 6 | 1.59e-01 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 2 | 60 | 1.59e-01 |
| GO:BP | GO:0039694 | viral RNA genome replication | 2 | 27 | 1.59e-01 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 2 | 7 | 1.59e-01 |
| GO:BP | GO:0072282 | metanephric nephron tubule morphogenesis | 2 | 5 | 1.59e-01 |
| GO:BP | GO:0009240 | isopentenyl diphosphate biosynthetic process | 1 | 5 | 1.60e-01 |
| GO:BP | GO:0046490 | isopentenyl diphosphate metabolic process | 1 | 5 | 1.60e-01 |
| GO:BP | GO:0016311 | dephosphorylation | 2 | 284 | 1.60e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 7 | 60 | 1.60e-01 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 24 | 1015 | 1.60e-01 |
| GO:BP | GO:0071971 | extracellular exosome assembly | 2 | 6 | 1.60e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 9 | 70 | 1.60e-01 |
| GO:BP | GO:0010165 | response to X-ray | 5 | 29 | 1.60e-01 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 2 | 16 | 1.60e-01 |
| GO:BP | GO:0032620 | interleukin-17 production | 2 | 20 | 1.60e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 1 | 67 | 1.60e-01 |
| GO:BP | GO:0032660 | regulation of interleukin-17 production | 2 | 20 | 1.60e-01 |
| GO:BP | GO:0006198 | cAMP catabolic process | 1 | 9 | 1.60e-01 |
| GO:BP | GO:0071895 | odontoblast differentiation | 1 | 5 | 1.60e-01 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 2 | 69 | 1.60e-01 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 2 | 42 | 1.60e-01 |
| GO:BP | GO:0003307 | regulation of Wnt signaling pathway involved in heart development | 2 | 7 | 1.60e-01 |
| GO:BP | GO:0021979 | hypothalamus cell differentiation | 3 | 8 | 1.60e-01 |
| GO:BP | GO:1904762 | positive regulation of myofibroblast differentiation | 1 | 1 | 1.60e-01 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 24 | 1017 | 1.61e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 9 | 227 | 1.61e-01 |
| GO:BP | GO:0051136 | regulation of NK T cell differentiation | 1 | 4 | 1.61e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 2 | 40 | 1.61e-01 |
| GO:BP | GO:0018057 | peptidyl-lysine oxidation | 2 | 5 | 1.61e-01 |
| GO:BP | GO:0090521 | podocyte cell migration | 1 | 7 | 1.61e-01 |
| GO:BP | GO:0006915 | apoptotic process | 6 | 1443 | 1.62e-01 |
| GO:BP | GO:0001832 | blastocyst growth | 4 | 19 | 1.62e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 2 | 25 | 1.62e-01 |
| GO:BP | GO:0072203 | cell proliferation involved in metanephros development | 1 | 8 | 1.62e-01 |
| GO:BP | GO:0051972 | regulation of telomerase activity | 5 | 45 | 1.62e-01 |
| GO:BP | GO:0009151 | purine deoxyribonucleotide metabolic process | 1 | 12 | 1.62e-01 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 1 | 33 | 1.62e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 3 | 40 | 1.62e-01 |
| GO:BP | GO:0042727 | flavin-containing compound biosynthetic process | 1 | 2 | 1.62e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 1 | 57 | 1.62e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 13 | 441 | 1.62e-01 |
| GO:BP | GO:1905408 | negative regulation of creatine transmembrane transporter activity | 1 | 2 | 1.62e-01 |
| GO:BP | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation | 1 | 4 | 1.62e-01 |
| GO:BP | GO:1905407 | regulation of creatine transmembrane transporter activity | 1 | 2 | 1.62e-01 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 7 | 52 | 1.63e-01 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 3 | 64 | 1.63e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 1.63e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 9 | 331 | 1.63e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 11 | 161 | 1.63e-01 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 1 | 16 | 1.63e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 1.63e-01 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 1 | 15 | 1.64e-01 |
| GO:BP | GO:0008219 | cell death | 41 | 1596 | 1.64e-01 |
| GO:BP | GO:1904024 | negative regulation of glucose catabolic process to lactate via pyruvate | 1 | 3 | 1.64e-01 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 9 | 79 | 1.64e-01 |
| GO:BP | GO:0038066 | p38MAPK cascade | 2 | 43 | 1.64e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 4 | 400 | 1.64e-01 |
| GO:BP | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | 1 | 4 | 1.64e-01 |
| GO:BP | GO:0060243 | negative regulation of cell growth involved in contact inhibition | 1 | 1 | 1.65e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 5 | 70 | 1.65e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 1.65e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 1.65e-01 |
| GO:BP | GO:0097742 | de novo centriole assembly | 2 | 4 | 1.65e-01 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 2 | 4 | 1.65e-01 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 2 | 12 | 1.65e-01 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 5 | 33 | 1.65e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 42 | 616 | 1.65e-01 |
| GO:BP | GO:0060166 | olfactory pit development | 1 | 1 | 1.65e-01 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 4 | 15 | 1.65e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 7 | 293 | 1.66e-01 |
| GO:BP | GO:0021700 | developmental maturation | 2 | 211 | 1.66e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 5 | 111 | 1.66e-01 |
| GO:BP | GO:0045023 | G0 to G1 transition | 4 | 35 | 1.66e-01 |
| GO:BP | GO:0050932 | regulation of pigment cell differentiation | 1 | 6 | 1.66e-01 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 1 | 31 | 1.66e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 8 | 1123 | 1.66e-01 |
| GO:BP | GO:0060547 | negative regulation of necrotic cell death | 2 | 21 | 1.67e-01 |
| GO:BP | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress | 1 | 7 | 1.67e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 157 | 2065 | 1.67e-01 |
| GO:BP | GO:1903553 | positive regulation of extracellular exosome assembly | 1 | 4 | 1.67e-01 |
| GO:BP | GO:0048627 | myoblast development | 1 | 3 | 1.67e-01 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 2 | 4 | 1.67e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 84 | 1380 | 1.68e-01 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 1 | 45 | 1.68e-01 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 3 | 30 | 1.68e-01 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 2 | 7 | 1.68e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 9 | 114 | 1.68e-01 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 3 | 30 | 1.68e-01 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 2 | 5 | 1.68e-01 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 1 | 20 | 1.69e-01 |
| GO:BP | GO:1904997 | regulation of leukocyte adhesion to arterial endothelial cell | 1 | 3 | 1.69e-01 |
| GO:BP | GO:2001301 | lipoxin biosynthetic process | 1 | 2 | 1.69e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 5 | 41 | 1.69e-01 |
| GO:BP | GO:0051126 | negative regulation of actin nucleation | 1 | 6 | 1.69e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 10 | 326 | 1.69e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 2 | 144 | 1.69e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 2 | 26 | 1.69e-01 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 2 | 21 | 1.69e-01 |
| GO:BP | GO:0071373 | cellular response to luteinizing hormone stimulus | 2 | 5 | 1.69e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 4 | 14 | 1.69e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 1.70e-01 |
| GO:BP | GO:0006914 | autophagy | 14 | 513 | 1.70e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 14 | 513 | 1.70e-01 |
| GO:BP | GO:0006972 | hyperosmotic response | 2 | 19 | 1.70e-01 |
| GO:BP | GO:1905934 | negative regulation of cell fate determination | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0032203 | telomere formation via telomerase | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0048850 | hypophysis morphogenesis | 1 | 2 | 1.70e-01 |
| GO:BP | GO:0003138 | primary heart field specification | 1 | 3 | 1.70e-01 |
| GO:BP | GO:0006265 | DNA topological change | 1 | 10 | 1.70e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 7 | 204 | 1.70e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 5 | 116 | 1.70e-01 |
| GO:BP | GO:2000226 | regulation of pancreatic A cell differentiation | 1 | 1 | 1.70e-01 |
| GO:BP | GO:2000227 | negative regulation of pancreatic A cell differentiation | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 2 | 18 | 1.70e-01 |
| GO:BP | GO:0030902 | hindbrain development | 1 | 114 | 1.70e-01 |
| GO:BP | GO:0061106 | negative regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0061105 | regulation of stomach neuroendocrine cell differentiation | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0012501 | programmed cell death | 6 | 1479 | 1.70e-01 |
| GO:BP | GO:0036514 | dopaminergic neuron axon guidance | 1 | 3 | 1.70e-01 |
| GO:BP | GO:0009213 | pyrimidine deoxyribonucleoside triphosphate catabolic process | 1 | 2 | 1.70e-01 |
| GO:BP | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation | 1 | 2 | 1.70e-01 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 1 | 3 | 1.70e-01 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 5 | 46 | 1.70e-01 |
| GO:BP | GO:0042135 | neurotransmitter catabolic process | 1 | 10 | 1.70e-01 |
| GO:BP | GO:0098754 | detoxification | 5 | 91 | 1.70e-01 |
| GO:BP | GO:0045977 | positive regulation of mitotic cell cycle, embryonic | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 2 | 12 | 1.70e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 4 | 38 | 1.70e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 3 | 51 | 1.70e-01 |
| GO:BP | GO:0034337 | RNA folding | 1 | 1 | 1.70e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 9 | 203 | 1.70e-01 |
| GO:BP | GO:1904938 | planar cell polarity pathway involved in axon guidance | 1 | 3 | 1.70e-01 |
| GO:BP | GO:0003213 | cardiac right atrium morphogenesis | 1 | 2 | 1.70e-01 |
| GO:BP | GO:0043173 | nucleotide salvage | 1 | 15 | 1.70e-01 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 2 | 66 | 1.70e-01 |
| GO:BP | GO:0034638 | phosphatidylcholine catabolic process | 2 | 9 | 1.70e-01 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 3 | 48 | 1.70e-01 |
| GO:BP | GO:0009435 | NAD biosynthetic process | 2 | 17 | 1.70e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 9 | 135 | 1.70e-01 |
| GO:BP | GO:0071415 | cellular response to purine-containing compound | 3 | 11 | 1.70e-01 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 2 | 25 | 1.71e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 3 | 17 | 1.71e-01 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 2 | 13 | 1.71e-01 |
| GO:BP | GO:0086054 | bundle of His cell to Purkinje myocyte communication by electrical coupling | 1 | 2 | 1.71e-01 |
| GO:BP | GO:0003193 | pulmonary valve formation | 1 | 2 | 1.71e-01 |
| GO:BP | GO:0007605 | sensory perception of sound | 1 | 98 | 1.71e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 5 | 87 | 1.71e-01 |
| GO:BP | GO:0010643 | cell communication by chemical coupling | 1 | 2 | 1.71e-01 |
| GO:BP | GO:0098905 | regulation of bundle of His cell action potential | 1 | 2 | 1.71e-01 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 2 | 17 | 1.71e-01 |
| GO:BP | GO:0061952 | midbody abscission | 3 | 17 | 1.71e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 2 | 418 | 1.71e-01 |
| GO:BP | GO:0016570 | histone modification | 1 | 433 | 1.71e-01 |
| GO:BP | GO:1904481 | response to tetrahydrofolate | 1 | 1 | 1.72e-01 |
| GO:BP | GO:1904482 | cellular response to tetrahydrofolate | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 11 | 494 | 1.72e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 10 | 68 | 1.72e-01 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 2 | 26 | 1.72e-01 |
| GO:BP | GO:0007595 | lactation | 1 | 32 | 1.72e-01 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 3 | 13 | 1.72e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 6 | 89 | 1.72e-01 |
| GO:BP | GO:0014032 | neural crest cell development | 9 | 71 | 1.72e-01 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 1 | 3 | 1.72e-01 |
| GO:BP | GO:0007250 | activation of NF-kappaB-inducing kinase activity | 1 | 13 | 1.72e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 2 | 442 | 1.72e-01 |
| GO:BP | GO:0015692 | lead ion transport | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0019755 | one-carbon compound transport | 2 | 23 | 1.72e-01 |
| GO:BP | GO:0015676 | vanadium ion transport | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0015675 | nickel cation transport | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0035444 | nickel cation transmembrane transport | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 2 | 7 | 1.72e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 2 | 13 | 1.72e-01 |
| GO:BP | GO:0030954 | astral microtubule nucleation | 1 | 1 | 1.72e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 16 | 200 | 1.72e-01 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 1 | 9 | 1.72e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 7 | 92 | 1.72e-01 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 1 | 17 | 1.72e-01 |
| GO:BP | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling | 2 | 6 | 1.72e-01 |
| GO:BP | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell communication | 2 | 6 | 1.72e-01 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 1 | 5 | 1.72e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 2 | 75 | 1.72e-01 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 2 | 45 | 1.72e-01 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 1 | 19 | 1.72e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 38 | 796 | 1.72e-01 |
| GO:BP | GO:0048087 | positive regulation of developmental pigmentation | 1 | 7 | 1.72e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 7 | 158 | 1.73e-01 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 4 | 113 | 1.73e-01 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 1 | 8 | 1.73e-01 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 3 | 14 | 1.73e-01 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 4 | 27 | 1.73e-01 |
| GO:BP | GO:0072207 | metanephric epithelium development | 2 | 16 | 1.73e-01 |
| GO:BP | GO:0071677 | positive regulation of mononuclear cell migration | 4 | 39 | 1.74e-01 |
| GO:BP | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 4 | 1.74e-01 |
| GO:BP | GO:0016477 | cell migration | 35 | 1081 | 1.74e-01 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 2 | 27 | 1.74e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 4 | 28 | 1.74e-01 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 4 | 31 | 1.75e-01 |
| GO:BP | GO:0009439 | cyanate metabolic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 12 | 358 | 1.75e-01 |
| GO:BP | GO:0048596 | embryonic camera-type eye morphogenesis | 2 | 18 | 1.75e-01 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 11 | 114 | 1.75e-01 |
| GO:BP | GO:0009440 | cyanate catabolic process | 1 | 2 | 1.75e-01 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 3 | 11 | 1.75e-01 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 1 | 23 | 1.75e-01 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 2 | 42 | 1.76e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 3 | 47 | 1.76e-01 |
| GO:BP | GO:0021828 | gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 2 | 6 | 1.76e-01 |
| GO:BP | GO:0021856 | hypothalamic tangential migration using cell-axon interactions | 2 | 6 | 1.76e-01 |
| GO:BP | GO:1901166 | neural crest cell migration involved in autonomic nervous system development | 2 | 5 | 1.76e-01 |
| GO:BP | GO:0071462 | cellular response to water stimulus | 1 | 4 | 1.76e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 1.76e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 4 | 101 | 1.76e-01 |
| GO:BP | GO:0015848 | spermidine transport | 1 | 3 | 1.76e-01 |
| GO:BP | GO:0061098 | positive regulation of protein tyrosine kinase activity | 1 | 33 | 1.78e-01 |
| GO:BP | GO:0000425 | pexophagy | 1 | 6 | 1.78e-01 |
| GO:BP | GO:1902296 | DNA strand elongation involved in cell cycle DNA replication | 1 | 1 | 1.78e-01 |
| GO:BP | GO:1902319 | DNA strand elongation involved in nuclear cell cycle DNA replication | 1 | 1 | 1.78e-01 |
| GO:BP | GO:1902983 | DNA strand elongation involved in mitotic DNA replication | 1 | 1 | 1.78e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 15 | 191 | 1.78e-01 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 9 | 71 | 1.78e-01 |
| GO:BP | GO:0003309 | type B pancreatic cell differentiation | 3 | 22 | 1.78e-01 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 2 | 23 | 1.79e-01 |
| GO:BP | GO:0008089 | anterograde axonal transport | 3 | 45 | 1.79e-01 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 2 | 48 | 1.80e-01 |
| GO:BP | GO:0042413 | carnitine catabolic process | 1 | 1 | 1.80e-01 |
| GO:BP | GO:2000273 | positive regulation of signaling receptor activity | 1 | 33 | 1.80e-01 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 4 | 194 | 1.80e-01 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 3 | 23 | 1.80e-01 |
| GO:BP | GO:0034755 | iron ion transmembrane transport | 3 | 16 | 1.80e-01 |
| GO:BP | GO:0051974 | negative regulation of telomerase activity | 2 | 11 | 1.81e-01 |
| GO:BP | GO:0042267 | natural killer cell mediated cytotoxicity | 2 | 34 | 1.81e-01 |
| GO:BP | GO:0099623 | regulation of cardiac muscle cell membrane repolarization | 3 | 20 | 1.81e-01 |
| GO:BP | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction | 2 | 8 | 1.81e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 1.81e-01 |
| GO:BP | GO:0072182 | regulation of nephron tubule epithelial cell differentiation | 1 | 10 | 1.81e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 3 | 46 | 1.81e-01 |
| GO:BP | GO:0070945 | neutrophil-mediated killing of gram-negative bacterium | 1 | 2 | 1.81e-01 |
| GO:BP | GO:0019363 | pyridine nucleotide biosynthetic process | 2 | 19 | 1.82e-01 |
| GO:BP | GO:0001906 | cell killing | 4 | 69 | 1.82e-01 |
| GO:BP | GO:0019359 | nicotinamide nucleotide biosynthetic process | 2 | 19 | 1.82e-01 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 2 | 8 | 1.82e-01 |
| GO:BP | GO:1905775 | negative regulation of DNA helicase activity | 1 | 2 | 1.82e-01 |
| GO:BP | GO:0002931 | response to ischemia | 3 | 46 | 1.82e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 1.82e-01 |
| GO:BP | GO:0007382 | specification of segmental identity, maxillary segment | 1 | 1 | 1.82e-01 |
| GO:BP | GO:0044751 | cellular response to human chorionic gonadotropin stimulus | 1 | 2 | 1.82e-01 |
| GO:BP | GO:1905871 | regulation of protein localization to cell leading edge | 1 | 2 | 1.82e-01 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 11 | 423 | 1.82e-01 |
| GO:BP | GO:0071321 | cellular response to cGMP | 1 | 7 | 1.82e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 1.83e-01 |
| GO:BP | GO:0021681 | cerebellar granular layer development | 2 | 11 | 1.83e-01 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 1 | 24 | 1.83e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 9 | 179 | 1.84e-01 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 3 | 14 | 1.85e-01 |
| GO:BP | GO:0106014 | regulation of inflammatory response to wounding | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 2 | 14 | 1.85e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 1.85e-01 |
| GO:BP | GO:0097176 | epoxide metabolic process | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 1.85e-01 |
| GO:BP | GO:0061044 | negative regulation of vascular wound healing | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion | 1 | 5 | 1.85e-01 |
| GO:BP | GO:0009611 | response to wounding | 2 | 415 | 1.85e-01 |
| GO:BP | GO:0071279 | cellular response to cobalt ion | 1 | 4 | 1.85e-01 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 1 | 4 | 1.85e-01 |
| GO:BP | GO:1901003 | negative regulation of fermentation | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0061643 | chemorepulsion of axon | 2 | 6 | 1.85e-01 |
| GO:BP | GO:0061757 | leukocyte adhesion to arterial endothelial cell | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0035082 | axoneme assembly | 2 | 52 | 1.85e-01 |
| GO:BP | GO:0045719 | negative regulation of glycogen biosynthetic process | 2 | 7 | 1.85e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 7 | 44 | 1.85e-01 |
| GO:BP | GO:0050893 | sensory processing | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway | 1 | 3 | 1.85e-01 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 1 | 67 | 1.85e-01 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 1 | 7 | 1.85e-01 |
| GO:BP | GO:0080163 | regulation of protein serine/threonine phosphatase activity | 1 | 2 | 1.85e-01 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 1 | 5 | 1.85e-01 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 6 | 44 | 1.85e-01 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion | 1 | 5 | 1.85e-01 |
| GO:BP | GO:0006021 | inositol biosynthetic process | 1 | 3 | 1.85e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 10 | 265 | 1.85e-01 |
| GO:BP | GO:0019660 | glycolytic fermentation | 1 | 4 | 1.85e-01 |
| GO:BP | GO:0031000 | response to caffeine | 3 | 13 | 1.86e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 10 | 295 | 1.86e-01 |
| GO:BP | GO:0070512 | positive regulation of histone H4-K20 methylation | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 1 | 16 | 1.86e-01 |
| GO:BP | GO:0090240 | positive regulation of histone H4 acetylation | 2 | 6 | 1.86e-01 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 2 | 11 | 1.86e-01 |
| GO:BP | GO:0045876 | positive regulation of sister chromatid cohesion | 2 | 8 | 1.86e-01 |
| GO:BP | GO:0070537 | histone H2A K63-linked deubiquitination | 2 | 8 | 1.86e-01 |
| GO:BP | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis | 1 | 4 | 1.86e-01 |
| GO:BP | GO:0106049 | regulation of cellular response to osmotic stress | 1 | 10 | 1.86e-01 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 2 | 7 | 1.86e-01 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 5 | 108 | 1.87e-01 |
| GO:BP | GO:0002228 | natural killer cell mediated immunity | 2 | 34 | 1.87e-01 |
| GO:BP | GO:0042407 | cristae formation | 2 | 18 | 1.87e-01 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 2 | 17 | 1.87e-01 |
| GO:BP | GO:0070384 | Harderian gland development | 1 | 4 | 1.87e-01 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 2 | 23 | 1.87e-01 |
| GO:BP | GO:0031441 | negative regulation of mRNA 3’-end processing | 1 | 2 | 1.88e-01 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 2 | 14 | 1.88e-01 |
| GO:BP | GO:0043570 | maintenance of DNA repeat elements | 2 | 6 | 1.88e-01 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 1 | 8 | 1.88e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 3 | 179 | 1.89e-01 |
| GO:BP | GO:0038092 | nodal signaling pathway | 1 | 8 | 1.89e-01 |
| GO:BP | GO:0006536 | glutamate metabolic process | 1 | 25 | 1.89e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 30 | 993 | 1.89e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 3 | 47 | 1.89e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 2 | 127 | 1.90e-01 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 3 | 21 | 1.90e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 6 | 144 | 1.90e-01 |
| GO:BP | GO:0043550 | regulation of lipid kinase activity | 3 | 43 | 1.90e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 30 | 994 | 1.90e-01 |
| GO:BP | GO:0048839 | inner ear development | 1 | 132 | 1.90e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 1 | 74 | 1.90e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 22 | 875 | 1.90e-01 |
| GO:BP | GO:0007498 | mesoderm development | 2 | 89 | 1.90e-01 |
| GO:BP | GO:0003139 | secondary heart field specification | 2 | 10 | 1.90e-01 |
| GO:BP | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation | 1 | 6 | 1.90e-01 |
| GO:BP | GO:0045216 | cell-cell junction organization | 1 | 153 | 1.90e-01 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 3 | 14 | 1.90e-01 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 2 | 6 | 1.90e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 4 | 47 | 1.90e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 1.90e-01 |
| GO:BP | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 1 | 5 | 1.90e-01 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 3 | 72 | 1.91e-01 |
| GO:BP | GO:0007000 | nucleolus organization | 3 | 17 | 1.91e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 14 | 376 | 1.91e-01 |
| GO:BP | GO:1902897 | regulation of postsynaptic density protein 95 clustering | 1 | 2 | 1.91e-01 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 2 | 26 | 1.93e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 20 | 313 | 1.93e-01 |
| GO:BP | GO:0071602 | phytosphingosine biosynthetic process | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0021698 | cerebellar cortex structural organization | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0048852 | diencephalon morphogenesis | 1 | 2 | 1.93e-01 |
| GO:BP | GO:2000002 | negative regulation of DNA damage checkpoint | 1 | 5 | 1.93e-01 |
| GO:BP | GO:1902977 | mitotic DNA replication preinitiation complex assembly | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 25 | 1119 | 1.93e-01 |
| GO:BP | GO:0072075 | metanephric mesenchyme development | 1 | 10 | 1.93e-01 |
| GO:BP | GO:0010621 | negative regulation of transcription by transcription factor localization | 1 | 3 | 1.93e-01 |
| GO:BP | GO:0009608 | response to symbiont | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0006671 | phytosphingosine metabolic process | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0009609 | response to symbiotic bacterium | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 6 | 143 | 1.93e-01 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 2 | 10 | 1.93e-01 |
| GO:BP | GO:0018158 | protein oxidation | 3 | 12 | 1.93e-01 |
| GO:BP | GO:0021693 | cerebellar Purkinje cell layer structural organization | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 5 | 88 | 1.93e-01 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 3 | 16 | 1.93e-01 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 2 | 15 | 1.93e-01 |
| GO:BP | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis | 1 | 4 | 1.93e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 27 | 1239 | 1.93e-01 |
| GO:BP | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | 1 | 3 | 1.93e-01 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 2 | 53 | 1.93e-01 |
| GO:BP | GO:0060686 | negative regulation of prostatic bud formation | 1 | 4 | 1.93e-01 |
| GO:BP | GO:0036211 | protein modification process | 13 | 2901 | 1.93e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 10 | 85 | 1.93e-01 |
| GO:BP | GO:0090082 | positive regulation of heart induction by negative regulation of canonical Wnt signaling pathway | 1 | 4 | 1.93e-01 |
| GO:BP | GO:1901321 | positive regulation of heart induction | 1 | 4 | 1.93e-01 |
| GO:BP | GO:0072720 | response to dithiothreitol | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0035902 | response to immobilization stress | 2 | 16 | 1.93e-01 |
| GO:BP | GO:0060157 | urinary bladder development | 1 | 2 | 1.93e-01 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 11 | 429 | 1.93e-01 |
| GO:BP | GO:1904442 | negative regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 1.93e-01 |
| GO:BP | GO:1903697 | negative regulation of microvillus assembly | 1 | 1 | 1.93e-01 |
| GO:BP | GO:1904441 | regulation of thyroid gland epithelial cell proliferation | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0072160 | nephron tubule epithelial cell differentiation | 1 | 12 | 1.93e-01 |
| GO:BP | GO:0098801 | regulation of renal system process | 3 | 21 | 1.93e-01 |
| GO:BP | GO:0006535 | cysteine biosynthetic process from serine | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 3 | 57 | 1.93e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 7 | 57 | 1.93e-01 |
| GO:BP | GO:1990789 | thyroid gland epithelial cell proliferation | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0051697 | protein delipidation | 1 | 5 | 1.93e-01 |
| GO:BP | GO:1905446 | regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 6 | 1.93e-01 |
| GO:BP | GO:0050830 | defense response to Gram-positive bacterium | 2 | 33 | 1.93e-01 |
| GO:BP | GO:0035883 | enteroendocrine cell differentiation | 4 | 24 | 1.93e-01 |
| GO:BP | GO:0098725 | symmetric cell division | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0030202 | heparin metabolic process | 2 | 14 | 1.93e-01 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 2 | 30 | 1.93e-01 |
| GO:BP | GO:1904777 | negative regulation of protein localization to cell cortex | 1 | 1 | 1.93e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 1 | 48 | 1.94e-01 |
| GO:BP | GO:1904438 | regulation of iron ion import across plasma membrane | 1 | 1 | 1.94e-01 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 5 | 140 | 1.94e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 6 | 160 | 1.94e-01 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 4 | 62 | 1.95e-01 |
| GO:BP | GO:0015854 | guanine transport | 1 | 2 | 1.95e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 6 | 81 | 1.95e-01 |
| GO:BP | GO:0035344 | hypoxanthine transport | 1 | 2 | 1.95e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 1.95e-01 |
| GO:BP | GO:0031642 | negative regulation of myelination | 2 | 8 | 1.95e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 3 | 40 | 1.95e-01 |
| GO:BP | GO:0099140 | presynaptic actin cytoskeleton organization | 1 | 1 | 1.95e-01 |
| GO:BP | GO:0021591 | ventricular system development | 2 | 29 | 1.95e-01 |
| GO:BP | GO:0048290 | isotype switching to IgA isotypes | 2 | 4 | 1.95e-01 |
| GO:BP | GO:0099187 | presynaptic cytoskeleton organization | 1 | 1 | 1.95e-01 |
| GO:BP | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0016540 | protein autoprocessing | 3 | 16 | 1.96e-01 |
| GO:BP | GO:0055071 | manganese ion homeostasis | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0071573 | shelterin complex assembly | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0061152 | trachea submucosa development | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0006999 | nuclear pore organization | 2 | 14 | 1.96e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 6 | 165 | 1.96e-01 |
| GO:BP | GO:1905778 | negative regulation of exonuclease activity | 1 | 2 | 1.96e-01 |
| GO:BP | GO:1905777 | regulation of exonuclease activity | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0032213 | regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 6 | 138 | 1.96e-01 |
| GO:BP | GO:0032214 | negative regulation of telomere maintenance via semi-conservative replication | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 4 | 27 | 1.96e-01 |
| GO:BP | GO:0050953 | sensory perception of light stimulus | 1 | 97 | 1.96e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 22 | 719 | 1.96e-01 |
| GO:BP | GO:1904534 | negative regulation of telomeric loop disassembly | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 6 | 76 | 1.96e-01 |
| GO:BP | GO:2000404 | regulation of T cell migration | 3 | 28 | 1.96e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 7 | 44 | 1.96e-01 |
| GO:BP | GO:1905838 | regulation of telomeric D-loop disassembly | 1 | 2 | 1.96e-01 |
| GO:BP | GO:1904790 | regulation of shelterin complex assembly | 1 | 2 | 1.96e-01 |
| GO:BP | GO:0072525 | pyridine-containing compound biosynthetic process | 2 | 22 | 1.96e-01 |
| GO:BP | GO:0061153 | trachea gland development | 1 | 2 | 1.96e-01 |
| GO:BP | GO:1905839 | negative regulation of telomeric D-loop disassembly | 1 | 2 | 1.96e-01 |
| GO:BP | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation | 2 | 6 | 1.96e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 2 | 53 | 1.96e-01 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 1 | 18 | 1.96e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 2 | 11 | 1.96e-01 |
| GO:BP | GO:0042430 | indole-containing compound metabolic process | 2 | 8 | 1.97e-01 |
| GO:BP | GO:0010886 | positive regulation of cholesterol storage | 2 | 5 | 1.97e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 37 | 1145 | 1.97e-01 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 5 | 140 | 1.97e-01 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 1 | 13 | 1.97e-01 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 2 | 11 | 1.97e-01 |
| GO:BP | GO:2000767 | positive regulation of cytoplasmic translation | 1 | 13 | 1.97e-01 |
| GO:BP | GO:1902378 | VEGF-activated neuropilin signaling pathway involved in axon guidance | 1 | 1 | 1.97e-01 |
| GO:BP | GO:0034058 | endosomal vesicle fusion | 1 | 10 | 1.97e-01 |
| GO:BP | GO:0006828 | manganese ion transport | 2 | 7 | 1.97e-01 |
| GO:BP | GO:1905040 | otic placode development | 1 | 1 | 1.97e-01 |
| GO:BP | GO:0071421 | manganese ion transmembrane transport | 2 | 7 | 1.97e-01 |
| GO:BP | GO:0060301 | positive regulation of cytokine activity | 1 | 1 | 1.97e-01 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 1 | 25 | 1.97e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 4 | 141 | 1.97e-01 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 1 | 25 | 1.97e-01 |
| GO:BP | GO:0098907 | regulation of SA node cell action potential | 2 | 7 | 1.97e-01 |
| GO:BP | GO:0033292 | T-tubule organization | 2 | 8 | 1.98e-01 |
| GO:BP | GO:1905341 | negative regulation of protein localization to kinetochore | 1 | 1 | 1.98e-01 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 8 | 77 | 1.98e-01 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 1 | 7 | 1.98e-01 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 3 | 15 | 1.99e-01 |
| GO:BP | GO:0007183 | SMAD protein complex assembly | 2 | 12 | 1.99e-01 |
| GO:BP | GO:0043583 | ear development | 1 | 153 | 1.99e-01 |
| GO:BP | GO:2001300 | lipoxin metabolic process | 1 | 3 | 1.99e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 2 | 504 | 1.99e-01 |
| GO:BP | GO:0021884 | forebrain neuron development | 4 | 16 | 2.00e-01 |
| GO:BP | GO:1905344 | prostaglandin catabolic process | 1 | 2 | 2.00e-01 |
| GO:BP | GO:0062232 | prostanoid catabolic process | 1 | 2 | 2.00e-01 |
| GO:BP | GO:0001865 | NK T cell differentiation | 1 | 7 | 2.00e-01 |
| GO:BP | GO:0071788 | endoplasmic reticulum tubular network maintenance | 1 | 1 | 2.00e-01 |
| GO:BP | GO:1903035 | negative regulation of response to wounding | 1 | 64 | 2.00e-01 |
| GO:BP | GO:0009414 | response to water deprivation | 1 | 4 | 2.00e-01 |
| GO:BP | GO:0019730 | antimicrobial humoral response | 2 | 28 | 2.00e-01 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 1 | 90 | 2.01e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 32 | 1219 | 2.01e-01 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 6 | 52 | 2.01e-01 |
| GO:BP | GO:0031175 | neuron projection development | 3 | 780 | 2.01e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 3 | 763 | 2.01e-01 |
| GO:BP | GO:0070265 | necrotic cell death | 3 | 53 | 2.01e-01 |
| GO:BP | GO:0035148 | tube formation | 2 | 138 | 2.01e-01 |
| GO:BP | GO:0021648 | vestibulocochlear nerve morphogenesis | 2 | 3 | 2.01e-01 |
| GO:BP | GO:0042633 | hair cycle | 1 | 78 | 2.01e-01 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 1 | 2 | 2.01e-01 |
| GO:BP | GO:0042303 | molting cycle | 1 | 78 | 2.01e-01 |
| GO:BP | GO:1990791 | dorsal root ganglion development | 2 | 6 | 2.01e-01 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 3 | 16 | 2.01e-01 |
| GO:BP | GO:0021855 | hypothalamus cell migration | 2 | 6 | 2.01e-01 |
| GO:BP | GO:0000320 | re-entry into mitotic cell cycle | 1 | 2 | 2.01e-01 |
| GO:BP | GO:0032874 | positive regulation of stress-activated MAPK cascade | 5 | 96 | 2.01e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 104 | 2.02e-01 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 1 | 20 | 2.02e-01 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 1 | 12 | 2.02e-01 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 1 | 2 | 2.02e-01 |
| GO:BP | GO:0070305 | response to cGMP | 1 | 7 | 2.02e-01 |
| GO:BP | GO:0001822 | kidney development | 12 | 244 | 2.02e-01 |
| GO:BP | GO:0002068 | glandular epithelial cell development | 3 | 28 | 2.02e-01 |
| GO:BP | GO:0047484 | regulation of response to osmotic stress | 1 | 12 | 2.03e-01 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 2.03e-01 |
| GO:BP | GO:0006003 | fructose 2,6-bisphosphate metabolic process | 1 | 4 | 2.03e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 3 | 101 | 2.03e-01 |
| GO:BP | GO:0006113 | fermentation | 1 | 5 | 2.03e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 5 | 114 | 2.03e-01 |
| GO:BP | GO:0003306 | Wnt signaling pathway involved in heart development | 2 | 8 | 2.03e-01 |
| GO:BP | GO:0043465 | regulation of fermentation | 1 | 5 | 2.03e-01 |
| GO:BP | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process | 1 | 3 | 2.03e-01 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 2.03e-01 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 2 | 32 | 2.03e-01 |
| GO:BP | GO:0048864 | stem cell development | 9 | 76 | 2.03e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 2.03e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 2.03e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 2 | 58 | 2.03e-01 |
| GO:BP | GO:0032355 | response to estradiol | 1 | 83 | 2.03e-01 |
| GO:BP | GO:0070831 | basement membrane assembly | 3 | 13 | 2.03e-01 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 6 | 2.03e-01 |
| GO:BP | GO:0090183 | regulation of kidney development | 2 | 22 | 2.03e-01 |
| GO:BP | GO:2000210 | positive regulation of anoikis | 1 | 6 | 2.03e-01 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 3 | 26 | 2.03e-01 |
| GO:BP | GO:0036335 | intestinal stem cell homeostasis | 1 | 3 | 2.04e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 51 | 1167 | 2.04e-01 |
| GO:BP | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome | 1 | 1 | 2.04e-01 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 1 | 17 | 2.04e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 1 | 109 | 2.04e-01 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 1 | 17 | 2.04e-01 |
| GO:BP | GO:0021761 | limbic system development | 4 | 81 | 2.04e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 17 | 551 | 2.04e-01 |
| GO:BP | GO:0051141 | negative regulation of NK T cell proliferation | 1 | 1 | 2.04e-01 |
| GO:BP | GO:2000615 | regulation of histone H3-K9 acetylation | 2 | 7 | 2.04e-01 |
| GO:BP | GO:0030900 | forebrain development | 15 | 281 | 2.05e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 9 | 222 | 2.05e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 133 | 2.05e-01 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 2 | 159 | 2.05e-01 |
| GO:BP | GO:0002192 | IRES-dependent translational initiation of linear mRNA | 1 | 1 | 2.05e-01 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 7 | 113 | 2.05e-01 |
| GO:BP | GO:0050859 | negative regulation of B cell receptor signaling pathway | 1 | 3 | 2.05e-01 |
| GO:BP | GO:0110017 | cap-independent translational initiation of linear mRNA | 1 | 1 | 2.05e-01 |
| GO:BP | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate | 2 | 12 | 2.05e-01 |
| GO:BP | GO:0009070 | serine family amino acid biosynthetic process | 3 | 16 | 2.05e-01 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 3 | 56 | 2.05e-01 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 3 | 52 | 2.05e-01 |
| GO:BP | GO:0006598 | polyamine catabolic process | 1 | 4 | 2.05e-01 |
| GO:BP | GO:0051607 | defense response to virus | 2 | 202 | 2.05e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 1 | 99 | 2.05e-01 |
| GO:BP | GO:0022612 | gland morphogenesis | 1 | 98 | 2.05e-01 |
| GO:BP | GO:0038202 | TORC1 signaling | 3 | 58 | 2.06e-01 |
| GO:BP | GO:0014044 | Schwann cell development | 1 | 28 | 2.06e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 2 | 202 | 2.06e-01 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 2 | 6 | 2.06e-01 |
| GO:BP | GO:0070874 | negative regulation of glycogen metabolic process | 2 | 7 | 2.06e-01 |
| GO:BP | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 5 | 98 | 2.06e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 6 | 28 | 2.06e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 6 | 84 | 2.06e-01 |
| GO:BP | GO:0009650 | UV protection | 2 | 11 | 2.07e-01 |
| GO:BP | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 1 | 10 | 2.07e-01 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 1 | 14 | 2.07e-01 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 2 | 25 | 2.07e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 7 | 1484 | 2.07e-01 |
| GO:BP | GO:0070203 | regulation of establishment of protein localization to telomere | 2 | 11 | 2.07e-01 |
| GO:BP | GO:0019441 | tryptophan catabolic process to kynurenine | 1 | 1 | 2.07e-01 |
| GO:BP | GO:1904161 | DNA synthesis involved in UV-damage excision repair | 1 | 2 | 2.08e-01 |
| GO:BP | GO:0001778 | plasma membrane repair | 2 | 28 | 2.08e-01 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 1 | 4 | 2.08e-01 |
| GO:BP | GO:1903444 | negative regulation of brown fat cell differentiation | 1 | 3 | 2.08e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 5 | 197 | 2.08e-01 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 4 | 22 | 2.08e-01 |
| GO:BP | GO:0072210 | metanephric nephron development | 5 | 29 | 2.09e-01 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 5 | 71 | 2.10e-01 |
| GO:BP | GO:0098532 | histone H3-K27 trimethylation | 1 | 4 | 2.10e-01 |
| GO:BP | GO:0046683 | response to organophosphorus | 5 | 96 | 2.10e-01 |
| GO:BP | GO:0048102 | autophagic cell death | 1 | 6 | 2.10e-01 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 1 | 69 | 2.10e-01 |
| GO:BP | GO:1905608 | positive regulation of presynapse assembly | 1 | 2 | 2.10e-01 |
| GO:BP | GO:0044458 | motile cilium assembly | 1 | 45 | 2.10e-01 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 6 | 295 | 2.10e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 26 | 1214 | 2.10e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 11 | 198 | 2.10e-01 |
| GO:BP | GO:0060744 | mammary gland branching involved in thelarche | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway | 1 | 4 | 2.10e-01 |
| GO:BP | GO:0070328 | triglyceride homeostasis | 1 | 17 | 2.10e-01 |
| GO:BP | GO:0048022 | negative regulation of melanin biosynthetic process | 1 | 4 | 2.10e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 69 | 1173 | 2.10e-01 |
| GO:BP | GO:0035050 | embryonic heart tube development | 9 | 69 | 2.10e-01 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 1 | 170 | 2.10e-01 |
| GO:BP | GO:0072529 | pyrimidine-containing compound catabolic process | 3 | 29 | 2.10e-01 |
| GO:BP | GO:0097325 | melanocyte proliferation | 1 | 2 | 2.10e-01 |
| GO:BP | GO:0048732 | gland development | 2 | 318 | 2.10e-01 |
| GO:BP | GO:0055090 | acylglycerol homeostasis | 1 | 17 | 2.10e-01 |
| GO:BP | GO:0048854 | brain morphogenesis | 4 | 28 | 2.10e-01 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 2 | 32 | 2.10e-01 |
| GO:BP | GO:0051309 | female meiosis chromosome separation | 1 | 2 | 2.10e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 21 | 756 | 2.10e-01 |
| GO:BP | GO:0036159 | inner dynein arm assembly | 1 | 8 | 2.10e-01 |
| GO:BP | GO:1902047 | polyamine transmembrane transport | 2 | 10 | 2.10e-01 |
| GO:BP | GO:0021629 | olfactory nerve structural organization | 1 | 1 | 2.10e-01 |
| GO:BP | GO:0042758 | long-chain fatty acid catabolic process | 2 | 3 | 2.10e-01 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 3 | 9 | 2.10e-01 |
| GO:BP | GO:0050808 | synapse organization | 5 | 365 | 2.10e-01 |
| GO:BP | GO:0016203 | muscle attachment | 1 | 1 | 2.10e-01 |
| GO:BP | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | 1 | 4 | 2.10e-01 |
| GO:BP | GO:0001894 | tissue homeostasis | 1 | 170 | 2.10e-01 |
| GO:BP | GO:0042695 | thelarche | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0001909 | leukocyte mediated cytotoxicity | 3 | 54 | 2.10e-01 |
| GO:BP | GO:0021793 | chemorepulsion of branchiomotor axon | 1 | 1 | 2.10e-01 |
| GO:BP | GO:0042640 | anagen | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 25 | 1088 | 2.10e-01 |
| GO:BP | GO:0032464 | positive regulation of protein homooligomerization | 1 | 2 | 2.10e-01 |
| GO:BP | GO:0043094 | cellular metabolic compound salvage | 1 | 25 | 2.10e-01 |
| GO:BP | GO:0070318 | positive regulation of G0 to G1 transition | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0060751 | branch elongation involved in mammary gland duct branching | 1 | 4 | 2.10e-01 |
| GO:BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0021537 | telencephalon development | 11 | 201 | 2.10e-01 |
| GO:BP | GO:0021936 | regulation of cerebellar granule cell precursor proliferation | 1 | 8 | 2.10e-01 |
| GO:BP | GO:0051884 | regulation of timing of anagen | 1 | 5 | 2.10e-01 |
| GO:BP | GO:0072074 | kidney mesenchyme development | 1 | 13 | 2.11e-01 |
| GO:BP | GO:0048741 | skeletal muscle fiber development | 2 | 30 | 2.11e-01 |
| GO:BP | GO:0072087 | renal vesicle development | 1 | 15 | 2.11e-01 |
| GO:BP | GO:0032230 | positive regulation of synaptic transmission, GABAergic | 1 | 10 | 2.11e-01 |
| GO:BP | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | 5 | 40 | 2.11e-01 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 1 | 8 | 2.11e-01 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 5 | 41 | 2.12e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 2.12e-01 |
| GO:BP | GO:1901856 | negative regulation of cellular respiration | 2 | 8 | 2.12e-01 |
| GO:BP | GO:0055006 | cardiac cell development | 9 | 77 | 2.12e-01 |
| GO:BP | GO:1904291 | positive regulation of mitotic DNA damage checkpoint | 1 | 2 | 2.12e-01 |
| GO:BP | GO:0021915 | neural tube development | 2 | 146 | 2.12e-01 |
| GO:BP | GO:0035234 | ectopic germ cell programmed cell death | 1 | 11 | 2.12e-01 |
| GO:BP | GO:1902414 | protein localization to cell junction | 6 | 93 | 2.12e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 3 | 165 | 2.12e-01 |
| GO:BP | GO:0072033 | renal vesicle formation | 2 | 8 | 2.13e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 2.13e-01 |
| GO:BP | GO:0031503 | protein-containing complex localization | 10 | 168 | 2.13e-01 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 3 | 17 | 2.13e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 6 | 92 | 2.13e-01 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 1 | 19 | 2.13e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 8 | 151 | 2.13e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 6 | 92 | 2.13e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 19 | 532 | 2.13e-01 |
| GO:BP | GO:0034699 | response to luteinizing hormone | 2 | 7 | 2.13e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 4 | 26 | 2.13e-01 |
| GO:BP | GO:0071314 | cellular response to cocaine | 2 | 7 | 2.13e-01 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 3 | 22 | 2.13e-01 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 1 | 11 | 2.13e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 18 | 663 | 2.13e-01 |
| GO:BP | GO:0036337 | Fas signaling pathway | 1 | 5 | 2.14e-01 |
| GO:BP | GO:0007344 | pronuclear fusion | 1 | 1 | 2.14e-01 |
| GO:BP | GO:0000741 | karyogamy | 1 | 1 | 2.14e-01 |
| GO:BP | GO:1903518 | positive regulation of single strand break repair | 1 | 2 | 2.14e-01 |
| GO:BP | GO:0036309 | protein localization to M-band | 1 | 2 | 2.14e-01 |
| GO:BP | GO:0048599 | oocyte development | 7 | 38 | 2.14e-01 |
| GO:BP | GO:0036371 | protein localization to T-tubule | 1 | 2 | 2.14e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 3 | 167 | 2.14e-01 |
| GO:BP | GO:0086101 | endothelin receptor signaling pathway involved in heart process | 1 | 3 | 2.14e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 25 | 834 | 2.14e-01 |
| GO:BP | GO:0042305 | specification of segmental identity, mandibular segment | 1 | 3 | 2.14e-01 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 1 | 20 | 2.14e-01 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 4 | 34 | 2.15e-01 |
| GO:BP | GO:1900081 | regulation of arginine catabolic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0120305 | regulation of pigmentation | 1 | 11 | 2.15e-01 |
| GO:BP | GO:0000098 | sulfur amino acid catabolic process | 2 | 7 | 2.15e-01 |
| GO:BP | GO:1900082 | negative regulation of arginine catabolic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 2.15e-01 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 1 | 102 | 2.15e-01 |
| GO:BP | GO:0036138 | peptidyl-histidine hydroxylation | 1 | 1 | 2.15e-01 |
| GO:BP | GO:1905916 | negative regulation of cell differentiation involved in phenotypic switching | 1 | 2 | 2.15e-01 |
| GO:BP | GO:2000283 | negative regulation of amino acid biosynthetic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0048070 | regulation of developmental pigmentation | 1 | 11 | 2.15e-01 |
| GO:BP | GO:0042074 | cell migration involved in gastrulation | 3 | 10 | 2.15e-01 |
| GO:BP | GO:1905223 | epicardium morphogenesis | 1 | 2 | 2.15e-01 |
| GO:BP | GO:0048132 | female germ-line stem cell asymmetric division | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 2 | 31 | 2.15e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 4 | 36 | 2.15e-01 |
| GO:BP | GO:1905931 | negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 2 | 2.15e-01 |
| GO:BP | GO:0048311 | mitochondrion distribution | 1 | 14 | 2.15e-01 |
| GO:BP | GO:1903394 | protein localization to kinetochore involved in kinetochore assembly | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 1 | 12 | 2.15e-01 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 3 | 22 | 2.15e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 26 | 1209 | 2.15e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 6 | 85 | 2.15e-01 |
| GO:BP | GO:0033241 | regulation of amine catabolic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 7 | 214 | 2.15e-01 |
| GO:BP | GO:0098736 | negative regulation of the force of heart contraction | 1 | 1 | 2.15e-01 |
| GO:BP | GO:1903248 | regulation of citrulline biosynthetic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:1900240 | negative regulation of phenotypic switching | 1 | 2 | 2.15e-01 |
| GO:BP | GO:1903249 | negative regulation of citrulline biosynthetic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0070848 | response to growth factor | 2 | 572 | 2.15e-01 |
| GO:BP | GO:0000712 | resolution of meiotic recombination intermediates | 1 | 13 | 2.15e-01 |
| GO:BP | GO:0033242 | negative regulation of amine catabolic process | 1 | 1 | 2.15e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 29 | 1111 | 2.15e-01 |
| GO:BP | GO:0006991 | response to sterol depletion | 3 | 16 | 2.15e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 1 | 178 | 2.15e-01 |
| GO:BP | GO:0090335 | regulation of brown fat cell differentiation | 2 | 17 | 2.15e-01 |
| GO:BP | GO:0009415 | response to water | 3 | 9 | 2.15e-01 |
| GO:BP | GO:0090188 | negative regulation of pancreatic juice secretion | 1 | 4 | 2.15e-01 |
| GO:BP | GO:0003341 | cilium movement | 5 | 97 | 2.15e-01 |
| GO:BP | GO:0090435 | protein localization to nuclear envelope | 2 | 14 | 2.15e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 12 | 257 | 2.15e-01 |
| GO:BP | GO:0003158 | endothelium development | 6 | 104 | 2.16e-01 |
| GO:BP | GO:0090166 | Golgi disassembly | 2 | 6 | 2.16e-01 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 2 | 11 | 2.16e-01 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 1 | 19 | 2.17e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 3 | 49 | 2.17e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 1 | 76 | 2.17e-01 |
| GO:BP | GO:0046834 | lipid phosphorylation | 3 | 16 | 2.17e-01 |
| GO:BP | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2 | 7 | 2.17e-01 |
| GO:BP | GO:0010738 | regulation of protein kinase A signaling | 1 | 13 | 2.17e-01 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 2 | 93 | 2.17e-01 |
| GO:BP | GO:1903301 | positive regulation of hexokinase activity | 1 | 5 | 2.17e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 2.17e-01 |
| GO:BP | GO:0043456 | regulation of pentose-phosphate shunt | 1 | 6 | 2.17e-01 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 2 | 187 | 2.17e-01 |
| GO:BP | GO:0048477 | oogenesis | 11 | 64 | 2.17e-01 |
| GO:BP | GO:0016054 | organic acid catabolic process | 2 | 187 | 2.17e-01 |
| GO:BP | GO:0072001 | renal system development | 12 | 251 | 2.17e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 27 | 1280 | 2.17e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 6 | 146 | 2.17e-01 |
| GO:BP | GO:0097012 | response to granulocyte macrophage colony-stimulating factor | 1 | 6 | 2.17e-01 |
| GO:BP | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus | 1 | 6 | 2.17e-01 |
| GO:BP | GO:0071447 | cellular response to hydroperoxide | 1 | 7 | 2.17e-01 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 4 | 70 | 2.18e-01 |
| GO:BP | GO:0090675 | intermicrovillar adhesion | 1 | 1 | 2.18e-01 |
| GO:BP | GO:0001736 | establishment of planar polarity | 4 | 70 | 2.18e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 3 | 40 | 2.18e-01 |
| GO:BP | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process | 2 | 15 | 2.19e-01 |
| GO:BP | GO:0051769 | regulation of nitric-oxide synthase biosynthetic process | 1 | 13 | 2.19e-01 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 1 | 10 | 2.19e-01 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 2 | 12 | 2.19e-01 |
| GO:BP | GO:0051767 | nitric-oxide synthase biosynthetic process | 1 | 13 | 2.19e-01 |
| GO:BP | GO:0046464 | acylglycerol catabolic process | 3 | 25 | 2.19e-01 |
| GO:BP | GO:0046461 | neutral lipid catabolic process | 3 | 25 | 2.19e-01 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 2 | 10 | 2.19e-01 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 2 | 28 | 2.19e-01 |
| GO:BP | GO:0097084 | vascular associated smooth muscle cell development | 2 | 11 | 2.19e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 2 | 10 | 2.19e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 1 | 114 | 2.19e-01 |
| GO:BP | GO:2001045 | negative regulation of integrin-mediated signaling pathway | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 2.19e-01 |
| GO:BP | GO:0033599 | regulation of mammary gland epithelial cell proliferation | 3 | 14 | 2.19e-01 |
| GO:BP | GO:0036298 | recombinational interstrand cross-link repair | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 1 | 69 | 2.19e-01 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 2 | 25 | 2.19e-01 |
| GO:BP | GO:0032643 | regulation of connective tissue growth factor production | 1 | 2 | 2.19e-01 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 1 | 17 | 2.19e-01 |
| GO:BP | GO:0015709 | thiosulfate transport | 1 | 4 | 2.19e-01 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 2 | 10 | 2.19e-01 |
| GO:BP | GO:0040009 | regulation of growth rate | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 4 | 85 | 2.19e-01 |
| GO:BP | GO:0019418 | sulfide oxidation | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0070716 | mismatch repair involved in maintenance of fidelity involved in DNA-dependent DNA replication | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 1 | 187 | 2.19e-01 |
| GO:BP | GO:0070221 | sulfide oxidation, using sulfide:quinone oxidoreductase | 1 | 3 | 2.19e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 1 | 114 | 2.19e-01 |
| GO:BP | GO:0070202 | regulation of establishment of protein localization to chromosome | 2 | 12 | 2.19e-01 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 1 | 17 | 2.19e-01 |
| GO:BP | GO:0032601 | connective tissue growth factor production | 1 | 2 | 2.19e-01 |
| GO:BP | GO:0070200 | establishment of protein localization to telomere | 3 | 16 | 2.19e-01 |
| GO:BP | GO:0043970 | histone H3-K9 acetylation | 2 | 8 | 2.20e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 1 | 101 | 2.20e-01 |
| GO:BP | GO:0060707 | trophoblast giant cell differentiation | 1 | 10 | 2.20e-01 |
| GO:BP | GO:0060294 | cilium movement involved in cell motility | 3 | 69 | 2.20e-01 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 3 | 22 | 2.20e-01 |
| GO:BP | GO:0032823 | regulation of natural killer cell differentiation | 1 | 8 | 2.20e-01 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 3 | 41 | 2.21e-01 |
| GO:BP | GO:0031032 | actomyosin structure organization | 9 | 186 | 2.21e-01 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 3 | 66 | 2.21e-01 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 3 | 66 | 2.21e-01 |
| GO:BP | GO:0018009 | N-terminal peptidyl-L-cysteine N-palmitoylation | 1 | 2 | 2.21e-01 |
| GO:BP | GO:1903905 | positive regulation of establishment of T cell polarity | 1 | 4 | 2.22e-01 |
| GO:BP | GO:0070593 | dendrite self-avoidance | 1 | 11 | 2.22e-01 |
| GO:BP | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron development | 2 | 7 | 2.22e-01 |
| GO:BP | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation | 2 | 7 | 2.22e-01 |
| GO:BP | GO:0002232 | leukocyte chemotaxis involved in inflammatory response | 1 | 4 | 2.22e-01 |
| GO:BP | GO:1904044 | response to aldosterone | 2 | 9 | 2.23e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 7 | 1389 | 2.23e-01 |
| GO:BP | GO:0034091 | regulation of maintenance of sister chromatid cohesion | 1 | 8 | 2.23e-01 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 1 | 121 | 2.23e-01 |
| GO:BP | GO:0070199 | establishment of protein localization to chromosome | 4 | 25 | 2.23e-01 |
| GO:BP | GO:0034182 | regulation of maintenance of mitotic sister chromatid cohesion | 1 | 8 | 2.23e-01 |
| GO:BP | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly | 1 | 2 | 2.23e-01 |
| GO:BP | GO:2000809 | positive regulation of synaptic vesicle clustering | 1 | 3 | 2.23e-01 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 2 | 18 | 2.23e-01 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 1 | 10 | 2.23e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 5 | 424 | 2.24e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 3 | 186 | 2.25e-01 |
| GO:BP | GO:0010818 | T cell chemotaxis | 2 | 12 | 2.25e-01 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 3 | 14 | 2.26e-01 |
| GO:BP | GO:0048313 | Golgi inheritance | 3 | 15 | 2.26e-01 |
| GO:BP | GO:0033624 | negative regulation of integrin activation | 1 | 3 | 2.26e-01 |
| GO:BP | GO:0032487 | regulation of Rap protein signal transduction | 1 | 3 | 2.26e-01 |
| GO:BP | GO:0048308 | organelle inheritance | 3 | 15 | 2.26e-01 |
| GO:BP | GO:0061102 | stomach neuroendocrine cell differentiation | 1 | 1 | 2.26e-01 |
| GO:BP | GO:0032379 | positive regulation of intracellular lipid transport | 1 | 4 | 2.26e-01 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 1 | 65 | 2.26e-01 |
| GO:BP | GO:0021558 | trochlear nerve development | 1 | 1 | 2.26e-01 |
| GO:BP | GO:0021891 | olfactory bulb interneuron development | 1 | 5 | 2.26e-01 |
| GO:BP | GO:1902474 | positive regulation of protein localization to synapse | 1 | 6 | 2.26e-01 |
| GO:BP | GO:0060028 | convergent extension involved in axis elongation | 1 | 6 | 2.26e-01 |
| GO:BP | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade | 1 | 4 | 2.26e-01 |
| GO:BP | GO:0070734 | histone H3-K27 methylation | 2 | 9 | 2.26e-01 |
| GO:BP | GO:0042551 | neuron maturation | 3 | 33 | 2.26e-01 |
| GO:BP | GO:1905933 | regulation of cell fate determination | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0072141 | renal interstitial fibroblast development | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 4 | 42 | 2.26e-01 |
| GO:BP | GO:0060685 | regulation of prostatic bud formation | 1 | 5 | 2.26e-01 |
| GO:BP | GO:0003402 | planar cell polarity pathway involved in axis elongation | 1 | 6 | 2.26e-01 |
| GO:BP | GO:0090381 | regulation of heart induction | 1 | 4 | 2.26e-01 |
| GO:BP | GO:0010046 | response to mycotoxin | 1 | 3 | 2.26e-01 |
| GO:BP | GO:0021870 | Cajal-Retzius cell differentiation | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0100012 | regulation of heart induction by canonical Wnt signaling pathway | 1 | 4 | 2.26e-01 |
| GO:BP | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway | 1 | 18 | 2.26e-01 |
| GO:BP | GO:0072071 | kidney interstitial fibroblast differentiation | 1 | 2 | 2.26e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 5 | 123 | 2.26e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 2 | 14 | 2.27e-01 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 2 | 179 | 2.27e-01 |
| GO:BP | GO:1901723 | negative regulation of cell proliferation involved in kidney development | 1 | 3 | 2.27e-01 |
| GO:BP | GO:2000466 | negative regulation of glycogen (starch) synthase activity | 1 | 3 | 2.27e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 27 | 1229 | 2.27e-01 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 2 | 19 | 2.27e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 3 | 906 | 2.27e-01 |
| GO:BP | GO:0021675 | nerve development | 9 | 57 | 2.28e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 7 | 3090 | 2.28e-01 |
| GO:BP | GO:0035265 | organ growth | 1 | 131 | 2.28e-01 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 1 | 31 | 2.28e-01 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 2 | 6 | 2.28e-01 |
| GO:BP | GO:0044854 | plasma membrane raft assembly | 1 | 6 | 2.28e-01 |
| GO:BP | GO:0032238 | adenosine transport | 1 | 2 | 2.28e-01 |
| GO:BP | GO:1902958 | positive regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 3 | 2.28e-01 |
| GO:BP | GO:0034760 | negative regulation of iron ion transmembrane transport | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0060385 | axonogenesis involved in innervation | 2 | 7 | 2.28e-01 |
| GO:BP | GO:0003105 | negative regulation of glomerular filtration | 1 | 3 | 2.28e-01 |
| GO:BP | GO:0034757 | negative regulation of iron ion transport | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0086017 | Purkinje myocyte action potential | 1 | 4 | 2.28e-01 |
| GO:BP | GO:0046015 | regulation of transcription by glucose | 1 | 6 | 2.28e-01 |
| GO:BP | GO:1904232 | regulation of aconitate hydratase activity | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 2 | 160 | 2.28e-01 |
| GO:BP | GO:1904234 | positive regulation of aconitate hydratase activity | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0042462 | eye photoreceptor cell development | 2 | 19 | 2.28e-01 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 2 | 11 | 2.28e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 2 | 71 | 2.28e-01 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 2 | 8 | 2.28e-01 |
| GO:BP | GO:0019264 | glycine biosynthetic process from serine | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 1 | 10 | 2.29e-01 |
| GO:BP | GO:0002023 | reduction of food intake in response to dietary excess | 1 | 2 | 2.30e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 4 | 121 | 2.30e-01 |
| GO:BP | GO:0014904 | myotube cell development | 2 | 34 | 2.30e-01 |
| GO:BP | GO:0046058 | cAMP metabolic process | 1 | 19 | 2.30e-01 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 2 | 28 | 2.30e-01 |
| GO:BP | GO:0071455 | cellular response to hyperoxia | 1 | 5 | 2.30e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 1 | 77 | 2.31e-01 |
| GO:BP | GO:1903546 | protein localization to photoreceptor outer segment | 1 | 5 | 2.31e-01 |
| GO:BP | GO:0060613 | fat pad development | 1 | 6 | 2.31e-01 |
| GO:BP | GO:0021543 | pallium development | 17 | 139 | 2.32e-01 |
| GO:BP | GO:0070944 | neutrophil-mediated killing of bacterium | 1 | 3 | 2.32e-01 |
| GO:BP | GO:0097695 | establishment of protein-containing complex localization to telomere | 1 | 3 | 2.32e-01 |
| GO:BP | GO:1904533 | regulation of telomeric loop disassembly | 1 | 2 | 2.32e-01 |
| GO:BP | GO:0097694 | establishment of RNA localization to telomere | 1 | 3 | 2.32e-01 |
| GO:BP | GO:0006939 | smooth muscle contraction | 6 | 75 | 2.32e-01 |
| GO:BP | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 2 | 9 | 2.32e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 4 | 91 | 2.33e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 2.33e-01 |
| GO:BP | GO:1905178 | regulation of cardiac muscle tissue regeneration | 1 | 2 | 2.34e-01 |
| GO:BP | GO:1905179 | negative regulation of cardiac muscle tissue regeneration | 1 | 2 | 2.34e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 8 | 155 | 2.34e-01 |
| GO:BP | GO:0048666 | neuron development | 3 | 877 | 2.34e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 10 | 183 | 2.34e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 2 | 223 | 2.34e-01 |
| GO:BP | GO:0060677 | ureteric bud elongation | 2 | 7 | 2.34e-01 |
| GO:BP | GO:0042311 | vasodilation | 3 | 39 | 2.35e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 2.35e-01 |
| GO:BP | GO:0009888 | tissue development | 41 | 1425 | 2.35e-01 |
| GO:BP | GO:0006043 | glucosamine catabolic process | 1 | 2 | 2.35e-01 |
| GO:BP | GO:0006473 | protein acetylation | 1 | 190 | 2.35e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 1 | 111 | 2.35e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 7 | 75 | 2.35e-01 |
| GO:BP | GO:0033668 | suppression by symbiont of host apoptotic process | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 3 | 19 | 2.36e-01 |
| GO:BP | GO:0052041 | suppression by symbiont of host programmed cell death | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0098749 | cerebellar neuron development | 1 | 3 | 2.36e-01 |
| GO:BP | GO:0098930 | axonal transport | 3 | 60 | 2.36e-01 |
| GO:BP | GO:0019050 | suppression by virus of host apoptotic process | 1 | 2 | 2.36e-01 |
| GO:BP | GO:0051121 | hepoxilin metabolic process | 1 | 5 | 2.36e-01 |
| GO:BP | GO:0036006 | cellular response to macrophage colony-stimulating factor stimulus | 1 | 6 | 2.36e-01 |
| GO:BP | GO:0051122 | hepoxilin biosynthetic process | 1 | 5 | 2.36e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 2 | 224 | 2.36e-01 |
| GO:BP | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure | 2 | 14 | 2.36e-01 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 2 | 7 | 2.36e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 1 | 138 | 2.37e-01 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 8 | 2.37e-01 |
| GO:BP | GO:0061436 | establishment of skin barrier | 2 | 18 | 2.37e-01 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 2 | 49 | 2.37e-01 |
| GO:BP | GO:0016260 | selenocysteine biosynthetic process | 1 | 2 | 2.37e-01 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 3 | 71 | 2.37e-01 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 2 | 20 | 2.37e-01 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 7 | 49 | 2.38e-01 |
| GO:BP | GO:0007589 | body fluid secretion | 1 | 60 | 2.38e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 2 | 742 | 2.38e-01 |
| GO:BP | GO:0008090 | retrograde axonal transport | 1 | 19 | 2.38e-01 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 1 | 20 | 2.38e-01 |
| GO:BP | GO:1990182 | exosomal secretion | 3 | 18 | 2.38e-01 |
| GO:BP | GO:0070383 | DNA cytosine deamination | 1 | 5 | 2.38e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 3 | 42 | 2.38e-01 |
| GO:BP | GO:0032727 | positive regulation of interferon-alpha production | 1 | 17 | 2.38e-01 |
| GO:BP | GO:0035458 | cellular response to interferon-beta | 1 | 15 | 2.38e-01 |
| GO:BP | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination | 1 | 1 | 2.38e-01 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 3 | 27 | 2.38e-01 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 1 | 2 | 2.38e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 407 | 2.38e-01 |
| GO:BP | GO:0015846 | polyamine transport | 2 | 12 | 2.38e-01 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 2.38e-01 |
| GO:BP | GO:0006771 | riboflavin metabolic process | 1 | 3 | 2.38e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 6 | 99 | 2.38e-01 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 1 | 11 | 2.38e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 6 | 99 | 2.38e-01 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 2 | 13 | 2.38e-01 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 4 | 19 | 2.39e-01 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 2 | 69 | 2.39e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 5 | 2139 | 2.39e-01 |
| GO:BP | GO:0034354 | ‘de novo’ NAD biosynthetic process from tryptophan | 1 | 1 | 2.39e-01 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 2 | 16 | 2.39e-01 |
| GO:BP | GO:0048870 | cell motility | 37 | 1182 | 2.39e-01 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 2 | 16 | 2.39e-01 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 2 | 22 | 2.39e-01 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 4 | 19 | 2.39e-01 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 6 | 73 | 2.39e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 7 | 111 | 2.40e-01 |
| GO:BP | GO:1900045 | negative regulation of protein K63-linked ubiquitination | 1 | 7 | 2.40e-01 |
| GO:BP | GO:0033628 | regulation of cell adhesion mediated by integrin | 2 | 36 | 2.40e-01 |
| GO:BP | GO:0032960 | regulation of inositol trisphosphate biosynthetic process | 1 | 4 | 2.40e-01 |
| GO:BP | GO:0048565 | digestive tract development | 4 | 92 | 2.40e-01 |
| GO:BP | GO:1904355 | positive regulation of telomere capping | 2 | 14 | 2.40e-01 |
| GO:BP | GO:0030397 | membrane disassembly | 1 | 11 | 2.40e-01 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 1 | 9 | 2.40e-01 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 1 | 42 | 2.40e-01 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 9 | 80 | 2.40e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 3 | 115 | 2.40e-01 |
| GO:BP | GO:0010571 | positive regulation of nuclear cell cycle DNA replication | 1 | 6 | 2.40e-01 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 2 | 19 | 2.40e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 3 | 31 | 2.40e-01 |
| GO:BP | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process | 1 | 6 | 2.40e-01 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 3 | 72 | 2.40e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 9 | 108 | 2.40e-01 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 2 | 20 | 2.40e-01 |
| GO:BP | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process | 1 | 8 | 2.40e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 6 | 99 | 2.40e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 2.41e-01 |
| GO:BP | GO:1900019 | regulation of protein kinase C activity | 1 | 6 | 2.41e-01 |
| GO:BP | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0048247 | lymphocyte chemotaxis | 3 | 24 | 2.41e-01 |
| GO:BP | GO:1903210 | podocyte apoptotic process | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 1 | 35 | 2.41e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 1 | 80 | 2.41e-01 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 2 | 20 | 2.41e-01 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 2 | 20 | 2.41e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 13 | 170 | 2.41e-01 |
| GO:BP | GO:2000397 | positive regulation of ubiquitin-dependent endocytosis | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 2 | 21 | 2.41e-01 |
| GO:BP | GO:0010992 | ubiquitin recycling | 2 | 12 | 2.41e-01 |
| GO:BP | GO:0051725 | protein de-ADP-ribosylation | 1 | 6 | 2.41e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 2.41e-01 |
| GO:BP | GO:0046710 | GDP metabolic process | 1 | 10 | 2.41e-01 |
| GO:BP | GO:0030997 | regulation of centriole-centriole cohesion | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0072201 | negative regulation of mesenchymal cell proliferation | 1 | 7 | 2.41e-01 |
| GO:BP | GO:0007442 | hindgut morphogenesis | 1 | 4 | 2.41e-01 |
| GO:BP | GO:0021924 | cell proliferation in external granule layer | 1 | 12 | 2.41e-01 |
| GO:BP | GO:0007380 | specification of segmental identity, head | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 3 | 28 | 2.41e-01 |
| GO:BP | GO:0070234 | positive regulation of T cell apoptotic process | 2 | 8 | 2.41e-01 |
| GO:BP | GO:0035287 | head segmentation | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 1 | 143 | 2.41e-01 |
| GO:BP | GO:0021930 | cerebellar granule cell precursor proliferation | 1 | 12 | 2.41e-01 |
| GO:BP | GO:0035289 | posterior head segmentation | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0090520 | sphingolipid mediated signaling pathway | 1 | 11 | 2.41e-01 |
| GO:BP | GO:0010991 | negative regulation of SMAD protein complex assembly | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0060173 | limb development | 1 | 142 | 2.41e-01 |
| GO:BP | GO:0051694 | pointed-end actin filament capping | 1 | 6 | 2.41e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 2.41e-01 |
| GO:BP | GO:2000395 | regulation of ubiquitin-dependent endocytosis | 1 | 2 | 2.41e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 16 | 232 | 2.41e-01 |
| GO:BP | GO:0009994 | oocyte differentiation | 7 | 39 | 2.41e-01 |
| GO:BP | GO:0001539 | cilium or flagellum-dependent cell motility | 3 | 74 | 2.41e-01 |
| GO:BP | GO:0070943 | neutrophil-mediated killing of symbiont cell | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0060638 | mesenchymal-epithelial cell signaling | 1 | 4 | 2.41e-01 |
| GO:BP | GO:0006304 | DNA modification | 7 | 89 | 2.41e-01 |
| GO:BP | GO:0008088 | axo-dendritic transport | 4 | 73 | 2.41e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 5 | 2161 | 2.41e-01 |
| GO:BP | GO:0048736 | appendage development | 1 | 142 | 2.41e-01 |
| GO:BP | GO:1903978 | regulation of microglial cell activation | 1 | 9 | 2.41e-01 |
| GO:BP | GO:1900020 | positive regulation of protein kinase C activity | 1 | 6 | 2.41e-01 |
| GO:BP | GO:0140058 | neuron projection arborization | 2 | 29 | 2.41e-01 |
| GO:BP | GO:0021534 | cell proliferation in hindbrain | 1 | 12 | 2.41e-01 |
| GO:BP | GO:0009386 | translational attenuation | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0060285 | cilium-dependent cell motility | 3 | 74 | 2.41e-01 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 2 | 22 | 2.41e-01 |
| GO:BP | GO:0072011 | glomerular endothelium development | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0097018 | renal albumin absorption | 1 | 3 | 2.41e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 7 | 228 | 2.41e-01 |
| GO:BP | GO:0010737 | protein kinase A signaling | 4 | 23 | 2.41e-01 |
| GO:BP | GO:0050872 | white fat cell differentiation | 1 | 15 | 2.41e-01 |
| GO:BP | GO:0007617 | mating behavior | 1 | 21 | 2.41e-01 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 6 | 65 | 2.41e-01 |
| GO:BP | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 1 | 2 | 2.41e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 2.41e-01 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 2 | 107 | 2.41e-01 |
| GO:BP | GO:1902356 | oxaloacetate(2-) transmembrane transport | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 3 | 133 | 2.41e-01 |
| GO:BP | GO:0015743 | malate transport | 1 | 5 | 2.41e-01 |
| GO:BP | GO:0071423 | malate transmembrane transport | 1 | 5 | 2.41e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 10 | 126 | 2.41e-01 |
| GO:BP | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 2 | 16 | 2.42e-01 |
| GO:BP | GO:0071874 | cellular response to norepinephrine stimulus | 1 | 2 | 2.42e-01 |
| GO:BP | GO:0071873 | response to norepinephrine | 1 | 2 | 2.42e-01 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 1 | 2 | 2.42e-01 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 1 | 2 | 2.42e-01 |
| GO:BP | GO:0014074 | response to purine-containing compound | 5 | 108 | 2.42e-01 |
| GO:BP | GO:0060136 | embryonic process involved in female pregnancy | 1 | 6 | 2.42e-01 |
| GO:BP | GO:0042133 | neurotransmitter metabolic process | 1 | 18 | 2.42e-01 |
| GO:BP | GO:0070291 | N-acylethanolamine metabolic process | 1 | 4 | 2.42e-01 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 2 | 11 | 2.42e-01 |
| GO:BP | GO:0060271 | cilium assembly | 5 | 292 | 2.42e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 3 | 20 | 2.42e-01 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 3 | 21 | 2.42e-01 |
| GO:BP | GO:0007194 | negative regulation of adenylate cyclase activity | 2 | 13 | 2.42e-01 |
| GO:BP | GO:0071288 | cellular response to mercury ion | 1 | 2 | 2.43e-01 |
| GO:BP | GO:0042883 | cysteine transport | 1 | 3 | 2.43e-01 |
| GO:BP | GO:1903712 | cysteine transmembrane transport | 1 | 3 | 2.43e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 5 | 83 | 2.43e-01 |
| GO:BP | GO:0060232 | delamination | 1 | 5 | 2.43e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 17 | 740 | 2.43e-01 |
| GO:BP | GO:0060065 | uterus development | 2 | 13 | 2.43e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 2.44e-01 |
| GO:BP | GO:0070341 | fat cell proliferation | 1 | 10 | 2.44e-01 |
| GO:BP | GO:0045653 | negative regulation of megakaryocyte differentiation | 2 | 17 | 2.44e-01 |
| GO:BP | GO:0061549 | sympathetic ganglion development | 2 | 6 | 2.44e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 4 | 35 | 2.44e-01 |
| GO:BP | GO:0060398 | regulation of growth hormone receptor signaling pathway | 1 | 5 | 2.44e-01 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 2 | 60 | 2.44e-01 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 1 | 6 | 2.44e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 3 | 555 | 2.44e-01 |
| GO:BP | GO:0098780 | response to mitochondrial depolarisation | 2 | 21 | 2.44e-01 |
| GO:BP | GO:0072423 | response to DNA damage checkpoint signaling | 1 | 8 | 2.44e-01 |
| GO:BP | GO:1902306 | negative regulation of sodium ion transmembrane transport | 2 | 10 | 2.44e-01 |
| GO:BP | GO:0072402 | response to DNA integrity checkpoint signaling | 1 | 8 | 2.44e-01 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 2 | 21 | 2.44e-01 |
| GO:BP | GO:0072396 | response to cell cycle checkpoint signaling | 1 | 8 | 2.44e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 5 | 131 | 2.44e-01 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 3 | 47 | 2.45e-01 |
| GO:BP | GO:0014832 | urinary bladder smooth muscle contraction | 2 | 4 | 2.45e-01 |
| GO:BP | GO:0033566 | gamma-tubulin complex localization | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission | 1 | 3 | 2.45e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 3 | 18 | 2.45e-01 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 1 | 19 | 2.45e-01 |
| GO:BP | GO:0046072 | dTDP metabolic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 3 | 18 | 2.45e-01 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0009196 | pyrimidine deoxyribonucleoside diphosphate metabolic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0006233 | dTDP biosynthetic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 2 | 20 | 2.45e-01 |
| GO:BP | GO:0010623 | programmed cell death involved in cell development | 1 | 13 | 2.45e-01 |
| GO:BP | GO:0009197 | pyrimidine deoxyribonucleoside diphosphate biosynthetic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:1900235 | negative regulation of Kit signaling pathway | 1 | 2 | 2.45e-01 |
| GO:BP | GO:1900234 | regulation of Kit signaling pathway | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0006227 | dUDP biosynthetic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0046077 | dUDP metabolic process | 1 | 2 | 2.45e-01 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 5 | 35 | 2.45e-01 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 6 | 154 | 2.45e-01 |
| GO:BP | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing | 2 | 7 | 2.45e-01 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 2 | 46 | 2.45e-01 |
| GO:BP | GO:0044242 | cellular lipid catabolic process | 9 | 170 | 2.45e-01 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 2 | 46 | 2.45e-01 |
| GO:BP | GO:0060333 | type II interferon-mediated signaling pathway | 1 | 21 | 2.46e-01 |
| GO:BP | GO:0060019 | radial glial cell differentiation | 2 | 12 | 2.46e-01 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 1 | 15 | 2.46e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 19 | 4955 | 2.46e-01 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 6 | 29 | 2.46e-01 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 3 | 18 | 2.47e-01 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 5 | 83 | 2.47e-01 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 1 | 144 | 2.47e-01 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 1 | 31 | 2.48e-01 |
| GO:BP | GO:0007350 | blastoderm segmentation | 4 | 17 | 2.48e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 6 | 86 | 2.48e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 6 | 80 | 2.48e-01 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 1 | 154 | 2.48e-01 |
| GO:BP | GO:1905786 | positive regulation of anaphase-promoting complex-dependent catabolic process | 1 | 2 | 2.48e-01 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 4 | 26 | 2.48e-01 |
| GO:BP | GO:0046456 | icosanoid biosynthetic process | 2 | 29 | 2.48e-01 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 1 | 3 | 2.49e-01 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 6 | 60 | 2.49e-01 |
| GO:BP | GO:0090102 | cochlea development | 3 | 35 | 2.49e-01 |
| GO:BP | GO:0048239 | negative regulation of DNA recombination at telomere | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0072695 | regulation of DNA recombination at telomere | 1 | 2 | 2.49e-01 |
| GO:BP | GO:0042310 | vasoconstriction | 4 | 51 | 2.49e-01 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 1 | 7 | 2.50e-01 |
| GO:BP | GO:1904289 | regulation of mitotic DNA damage checkpoint | 1 | 3 | 2.50e-01 |
| GO:BP | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 23 | 2.50e-01 |
| GO:BP | GO:0095500 | acetylcholine receptor signaling pathway | 1 | 18 | 2.50e-01 |
| GO:BP | GO:0048641 | regulation of skeletal muscle tissue development | 1 | 20 | 2.50e-01 |
| GO:BP | GO:0090148 | membrane fission | 5 | 44 | 2.50e-01 |
| GO:BP | GO:0034627 | ‘de novo’ NAD biosynthetic process | 1 | 2 | 2.50e-01 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 2 | 5 | 2.50e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 1 | 93 | 2.50e-01 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 3 | 33 | 2.50e-01 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 2 | 8 | 2.51e-01 |
| GO:BP | GO:0046452 | dihydrofolate metabolic process | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 1 | 11 | 2.51e-01 |
| GO:BP | GO:2000193 | positive regulation of fatty acid transport | 2 | 11 | 2.51e-01 |
| GO:BP | GO:0000710 | meiotic mismatch repair | 1 | 2 | 2.51e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 128 | 2.51e-01 |
| GO:BP | GO:0043543 | protein acylation | 1 | 228 | 2.52e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 18 | 173 | 2.52e-01 |
| GO:BP | GO:0051593 | response to folic acid | 2 | 9 | 2.52e-01 |
| GO:BP | GO:0035295 | tube development | 21 | 821 | 2.52e-01 |
| GO:BP | GO:0009615 | response to virus | 2 | 275 | 2.52e-01 |
| GO:BP | GO:1900015 | regulation of cytokine production involved in inflammatory response | 2 | 33 | 2.52e-01 |
| GO:BP | GO:0009116 | nucleoside metabolic process | 2 | 42 | 2.52e-01 |
| GO:BP | GO:0002534 | cytokine production involved in inflammatory response | 2 | 33 | 2.52e-01 |
| GO:BP | GO:1903516 | regulation of single strand break repair | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0030592 | DNA ADP-ribosylation | 1 | 3 | 2.53e-01 |
| GO:BP | GO:0051216 | cartilage development | 1 | 151 | 2.53e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 2 | 14 | 2.53e-01 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 6 | 157 | 2.53e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 5 | 151 | 2.53e-01 |
| GO:BP | GO:0007398 | ectoderm development | 3 | 15 | 2.54e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 16 | 234 | 2.54e-01 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 1 | 23 | 2.54e-01 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 4 | 28 | 2.54e-01 |
| GO:BP | GO:0033629 | negative regulation of cell adhesion mediated by integrin | 2 | 10 | 2.54e-01 |
| GO:BP | GO:0019343 | cysteine biosynthetic process via cystathionine | 1 | 2 | 2.54e-01 |
| GO:BP | GO:0007271 | synaptic transmission, cholinergic | 1 | 15 | 2.54e-01 |
| GO:BP | GO:2000310 | regulation of NMDA receptor activity | 2 | 19 | 2.54e-01 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 1 | 9 | 2.54e-01 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 2 | 10 | 2.54e-01 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 2 | 10 | 2.54e-01 |
| GO:BP | GO:1904714 | regulation of chaperone-mediated autophagy | 1 | 8 | 2.54e-01 |
| GO:BP | GO:0061525 | hindgut development | 1 | 5 | 2.55e-01 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 4 | 30 | 2.55e-01 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 1 | 7 | 2.55e-01 |
| GO:BP | GO:0019674 | NAD metabolic process | 2 | 33 | 2.55e-01 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 9 | 116 | 2.55e-01 |
| GO:BP | GO:0048341 | paraxial mesoderm formation | 1 | 6 | 2.55e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 2 | 21 | 2.55e-01 |
| GO:BP | GO:1905915 | regulation of cell differentiation involved in phenotypic switching | 1 | 3 | 2.55e-01 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 9 | 371 | 2.55e-01 |
| GO:BP | GO:0090679 | cell differentiation involved in phenotypic switching | 1 | 3 | 2.55e-01 |
| GO:BP | GO:1900239 | regulation of phenotypic switching | 1 | 3 | 2.55e-01 |
| GO:BP | GO:0046068 | cGMP metabolic process | 1 | 11 | 2.55e-01 |
| GO:BP | GO:1905460 | negative regulation of vascular associated smooth muscle cell apoptotic process | 1 | 2 | 2.55e-01 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 1 | 4 | 2.55e-01 |
| GO:BP | GO:0046606 | negative regulation of centrosome cycle | 2 | 12 | 2.55e-01 |
| GO:BP | GO:0016050 | vesicle organization | 6 | 320 | 2.55e-01 |
| GO:BP | GO:1905331 | negative regulation of morphogenesis of an epithelium | 1 | 6 | 2.55e-01 |
| GO:BP | GO:0033278 | cell proliferation in midbrain | 1 | 4 | 2.55e-01 |
| GO:BP | GO:1905930 | regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 2.55e-01 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 1 | 8 | 2.55e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 2.55e-01 |
| GO:BP | GO:0042726 | flavin-containing compound metabolic process | 1 | 4 | 2.55e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 1 | 121 | 2.55e-01 |
| GO:BP | GO:0010826 | negative regulation of centrosome duplication | 2 | 12 | 2.55e-01 |
| GO:BP | GO:0051964 | negative regulation of synapse assembly | 1 | 6 | 2.55e-01 |
| GO:BP | GO:1905420 | vascular associated smooth muscle cell differentiation involved in phenotypic switching | 1 | 3 | 2.55e-01 |
| GO:BP | GO:1990792 | cellular response to glial cell derived neurotrophic factor | 1 | 1 | 2.55e-01 |
| GO:BP | GO:0070942 | neutrophil mediated cytotoxicity | 1 | 4 | 2.55e-01 |
| GO:BP | GO:1990790 | response to glial cell derived neurotrophic factor | 1 | 1 | 2.55e-01 |
| GO:BP | GO:0021602 | cranial nerve morphogenesis | 4 | 16 | 2.55e-01 |
| GO:BP | GO:0006244 | pyrimidine nucleotide catabolic process | 2 | 19 | 2.55e-01 |
| GO:BP | GO:0033559 | unsaturated fatty acid metabolic process | 3 | 66 | 2.56e-01 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 1 | 12 | 2.56e-01 |
| GO:BP | GO:0071140 | resolution of mitotic recombination intermediates | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0019228 | neuronal action potential | 1 | 23 | 2.57e-01 |
| GO:BP | GO:0034759 | regulation of iron ion transmembrane transport | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0048304 | positive regulation of isotype switching to IgG isotypes | 1 | 4 | 2.57e-01 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 2 | 8 | 2.57e-01 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 1 | 10 | 2.57e-01 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 2 | 8 | 2.57e-01 |
| GO:BP | GO:1905145 | cellular response to acetylcholine | 1 | 20 | 2.57e-01 |
| GO:BP | GO:0033306 | phytol metabolic process | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 1 | 10 | 2.57e-01 |
| GO:BP | GO:1903173 | fatty alcohol metabolic process | 1 | 2 | 2.57e-01 |
| GO:BP | GO:0021854 | hypothalamus development | 1 | 15 | 2.57e-01 |
| GO:BP | GO:1902031 | regulation of NADP metabolic process | 1 | 8 | 2.57e-01 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 1 | 13 | 2.57e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 13 | 172 | 2.57e-01 |
| GO:BP | GO:0015860 | purine nucleoside transmembrane transport | 1 | 3 | 2.57e-01 |
| GO:BP | GO:0032647 | regulation of interferon-alpha production | 1 | 19 | 2.58e-01 |
| GO:BP | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response | 1 | 1 | 2.58e-01 |
| GO:BP | GO:0032607 | interferon-alpha production | 1 | 19 | 2.58e-01 |
| GO:BP | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity | 1 | 26 | 2.58e-01 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 1 | 27 | 2.58e-01 |
| GO:BP | GO:0006936 | muscle contraction | 5 | 267 | 2.58e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 13 | 462 | 2.59e-01 |
| GO:BP | GO:0007417 | central nervous system development | 10 | 792 | 2.59e-01 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 5 | 64 | 2.59e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 8 | 166 | 2.59e-01 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 3 | 39 | 2.59e-01 |
| GO:BP | GO:0006273 | lagging strand elongation | 1 | 4 | 2.59e-01 |
| GO:BP | GO:0007416 | synapse assembly | 8 | 152 | 2.59e-01 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 1 | 10 | 2.59e-01 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 2 | 12 | 2.59e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 1 | 198 | 2.59e-01 |
| GO:BP | GO:0071106 | adenosine 3’,5’-bisphosphate transmembrane transport | 1 | 2 | 2.59e-01 |
| GO:BP | GO:0019098 | reproductive behavior | 1 | 26 | 2.59e-01 |
| GO:BP | GO:0035350 | FAD transmembrane transport | 1 | 2 | 2.59e-01 |
| GO:BP | GO:0015883 | FAD transport | 1 | 2 | 2.59e-01 |
| GO:BP | GO:0035349 | coenzyme A transmembrane transport | 1 | 2 | 2.59e-01 |
| GO:BP | GO:0015880 | coenzyme A transport | 1 | 2 | 2.59e-01 |
| GO:BP | GO:0097527 | necroptotic signaling pathway | 1 | 3 | 2.59e-01 |
| GO:BP | GO:0080121 | AMP transport | 1 | 2 | 2.59e-01 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 2 | 74 | 2.59e-01 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 8 | 81 | 2.60e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 1 | 127 | 2.60e-01 |
| GO:BP | GO:0051339 | regulation of lyase activity | 4 | 39 | 2.60e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 2 | 246 | 2.60e-01 |
| GO:BP | GO:2000627 | positive regulation of miRNA catabolic process | 1 | 4 | 2.60e-01 |
| GO:BP | GO:0002052 | positive regulation of neuroblast proliferation | 4 | 19 | 2.60e-01 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 8 | 96 | 2.60e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 3 | 37 | 2.60e-01 |
| GO:BP | GO:0022412 | cellular process involved in reproduction in multicellular organism | 26 | 252 | 2.60e-01 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 4 | 35 | 2.60e-01 |
| GO:BP | GO:0031280 | negative regulation of cyclase activity | 2 | 13 | 2.60e-01 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 2 | 15 | 2.60e-01 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 2 | 15 | 2.60e-01 |
| GO:BP | GO:0031291 | Ran protein signal transduction | 1 | 3 | 2.60e-01 |
| GO:BP | GO:1902268 | negative regulation of polyamine transmembrane transport | 1 | 3 | 2.61e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 3 | 16 | 2.61e-01 |
| GO:BP | GO:0006829 | zinc ion transport | 3 | 23 | 2.61e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 3 | 266 | 2.61e-01 |
| GO:BP | GO:0016233 | telomere capping | 3 | 36 | 2.61e-01 |
| GO:BP | GO:0015729 | oxaloacetate transport | 1 | 5 | 2.61e-01 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 1 | 26 | 2.61e-01 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 1 | 26 | 2.61e-01 |
| GO:BP | GO:0036005 | response to macrophage colony-stimulating factor | 1 | 8 | 2.61e-01 |
| GO:BP | GO:0009395 | phospholipid catabolic process | 3 | 37 | 2.61e-01 |
| GO:BP | GO:2000773 | negative regulation of cellular senescence | 3 | 19 | 2.61e-01 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 1 | 14 | 2.61e-01 |
| GO:BP | GO:0045007 | depurination | 1 | 4 | 2.61e-01 |
| GO:BP | GO:0099639 | neurotransmitter receptor transport, endosome to plasma membrane | 2 | 10 | 2.61e-01 |
| GO:BP | GO:0097102 | endothelial tip cell fate specification | 1 | 2 | 2.61e-01 |
| GO:BP | GO:0008584 | male gonad development | 4 | 99 | 2.61e-01 |
| GO:BP | GO:1905590 | fibronectin fibril organization | 1 | 1 | 2.61e-01 |
| GO:BP | GO:0038190 | VEGF-activated neuropilin signaling pathway | 1 | 2 | 2.61e-01 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 2 | 15 | 2.61e-01 |
| GO:BP | GO:0061441 | renal artery morphogenesis | 1 | 2 | 2.61e-01 |
| GO:BP | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane | 2 | 10 | 2.61e-01 |
| GO:BP | GO:0071361 | cellular response to ethanol | 1 | 9 | 2.62e-01 |
| GO:BP | GO:0000730 | DNA recombinase assembly | 1 | 3 | 2.62e-01 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 8 | 83 | 2.62e-01 |
| GO:BP | GO:0090735 | DNA repair complex assembly | 1 | 3 | 2.62e-01 |
| GO:BP | GO:0051610 | serotonin uptake | 1 | 8 | 2.62e-01 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 2 | 209 | 2.62e-01 |
| GO:BP | GO:2000378 | negative regulation of reactive oxygen species metabolic process | 5 | 34 | 2.62e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 3 | 31 | 2.62e-01 |
| GO:BP | GO:1902595 | regulation of DNA replication origin binding | 1 | 2 | 2.62e-01 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 1 | 6 | 2.62e-01 |
| GO:BP | GO:0006500 | N-terminal protein palmitoylation | 1 | 3 | 2.62e-01 |
| GO:BP | GO:0042436 | indole-containing compound catabolic process | 1 | 1 | 2.62e-01 |
| GO:BP | GO:0070294 | renal sodium ion absorption | 2 | 9 | 2.62e-01 |
| GO:BP | GO:0032700 | negative regulation of interleukin-17 production | 1 | 9 | 2.62e-01 |
| GO:BP | GO:0006569 | tryptophan catabolic process | 1 | 1 | 2.62e-01 |
| GO:BP | GO:0051097 | negative regulation of helicase activity | 1 | 5 | 2.63e-01 |
| GO:BP | GO:1905774 | regulation of DNA helicase activity | 1 | 5 | 2.63e-01 |
| GO:BP | GO:1901503 | ether biosynthetic process | 1 | 11 | 2.63e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 1 | 316 | 2.63e-01 |
| GO:BP | GO:0140115 | export across plasma membrane | 1 | 46 | 2.63e-01 |
| GO:BP | GO:0010232 | vascular transport | 5 | 72 | 2.63e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 1 | 155 | 2.63e-01 |
| GO:BP | GO:0043651 | linoleic acid metabolic process | 2 | 12 | 2.63e-01 |
| GO:BP | GO:0055123 | digestive system development | 4 | 100 | 2.63e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 2 | 665 | 2.63e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 4 | 100 | 2.63e-01 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 5 | 72 | 2.63e-01 |
| GO:BP | GO:0044752 | response to human chorionic gonadotropin | 1 | 5 | 2.64e-01 |
| GO:BP | GO:1902463 | protein localization to cell leading edge | 1 | 5 | 2.64e-01 |
| GO:BP | GO:0034086 | maintenance of sister chromatid cohesion | 1 | 12 | 2.64e-01 |
| GO:BP | GO:0035456 | response to interferon-beta | 1 | 22 | 2.64e-01 |
| GO:BP | GO:0002230 | positive regulation of defense response to virus by host | 1 | 23 | 2.64e-01 |
| GO:BP | GO:0034088 | maintenance of mitotic sister chromatid cohesion | 1 | 12 | 2.64e-01 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 1 | 18 | 2.64e-01 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 2 | 6 | 2.64e-01 |
| GO:BP | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex | 1 | 5 | 2.65e-01 |
| GO:BP | GO:0009972 | cytidine deamination | 1 | 6 | 2.65e-01 |
| GO:BP | GO:0010259 | multicellular organism aging | 2 | 16 | 2.65e-01 |
| GO:BP | GO:0044878 | mitotic cytokinesis checkpoint signaling | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 7 | 68 | 2.65e-01 |
| GO:BP | GO:0060348 | bone development | 1 | 170 | 2.65e-01 |
| GO:BP | GO:0046087 | cytidine metabolic process | 1 | 6 | 2.65e-01 |
| GO:BP | GO:0030318 | melanocyte differentiation | 1 | 16 | 2.65e-01 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 1 | 17 | 2.65e-01 |
| GO:BP | GO:0070125 | mitochondrial translational elongation | 1 | 4 | 2.65e-01 |
| GO:BP | GO:0006216 | cytidine catabolic process | 1 | 6 | 2.65e-01 |
| GO:BP | GO:0016554 | cytidine to uridine editing | 1 | 6 | 2.65e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 2 | 35 | 2.65e-01 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 2.65e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 2 | 8 | 2.65e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 27 | 2.65e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 1 | 210 | 2.65e-01 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 1 | 2 | 2.65e-01 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 3 | 35 | 2.65e-01 |
| GO:BP | GO:0050774 | negative regulation of dendrite morphogenesis | 1 | 7 | 2.65e-01 |
| GO:BP | GO:0045602 | negative regulation of endothelial cell differentiation | 1 | 7 | 2.65e-01 |
| GO:BP | GO:0006814 | sodium ion transport | 4 | 160 | 2.65e-01 |
| GO:BP | GO:0035329 | hippo signaling | 1 | 41 | 2.65e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 2 | 8 | 2.65e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 3 | 27 | 2.65e-01 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 1 | 2 | 2.65e-01 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 1 | 2 | 2.65e-01 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 5 | 48 | 2.65e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 2 | 8 | 2.65e-01 |
| GO:BP | GO:1905144 | response to acetylcholine | 1 | 22 | 2.65e-01 |
| GO:BP | GO:1902870 | negative regulation of amacrine cell differentiation | 1 | 2 | 2.66e-01 |
| GO:BP | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | 1 | 6 | 2.66e-01 |
| GO:BP | GO:0021873 | forebrain neuroblast division | 1 | 3 | 2.66e-01 |
| GO:BP | GO:0009786 | regulation of asymmetric cell division | 1 | 1 | 2.66e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 2 | 69 | 2.66e-01 |
| GO:BP | GO:2000974 | negative regulation of pro-B cell differentiation | 1 | 2 | 2.66e-01 |
| GO:BP | GO:0071922 | regulation of cohesin loading | 1 | 2 | 2.66e-01 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 1 | 25 | 2.66e-01 |
| GO:BP | GO:0046548 | retinal rod cell development | 1 | 6 | 2.66e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 3 | 23 | 2.66e-01 |
| GO:BP | GO:0046331 | lateral inhibition | 1 | 3 | 2.66e-01 |
| GO:BP | GO:0045168 | cell-cell signaling involved in cell fate commitment | 1 | 3 | 2.66e-01 |
| GO:BP | GO:0071763 | nuclear membrane organization | 2 | 42 | 2.66e-01 |
| GO:BP | GO:2000978 | negative regulation of forebrain neuron differentiation | 1 | 2 | 2.66e-01 |
| GO:BP | GO:0021555 | midbrain-hindbrain boundary morphogenesis | 1 | 2 | 2.66e-01 |
| GO:BP | GO:0060164 | regulation of timing of neuron differentiation | 1 | 1 | 2.66e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 11 | 1712 | 2.66e-01 |
| GO:BP | GO:1905584 | outer hair cell apoptotic process | 1 | 2 | 2.66e-01 |
| GO:BP | GO:0035754 | B cell chemotaxis | 1 | 5 | 2.66e-01 |
| GO:BP | GO:0060027 | convergent extension involved in gastrulation | 1 | 8 | 2.66e-01 |
| GO:BP | GO:0007172 | signal complex assembly | 1 | 7 | 2.66e-01 |
| GO:BP | GO:1902035 | positive regulation of hematopoietic stem cell proliferation | 1 | 7 | 2.66e-01 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 3 | 50 | 2.67e-01 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 3 | 50 | 2.67e-01 |
| GO:BP | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 1 | 6 | 2.67e-01 |
| GO:BP | GO:0032025 | response to cobalt ion | 1 | 10 | 2.67e-01 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 2 | 9 | 2.68e-01 |
| GO:BP | GO:0006534 | cysteine metabolic process | 2 | 11 | 2.68e-01 |
| GO:BP | GO:0006563 | L-serine metabolic process | 2 | 10 | 2.68e-01 |
| GO:BP | GO:0019369 | arachidonic acid metabolic process | 2 | 26 | 2.68e-01 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 1 | 6 | 2.68e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 5 | 45 | 2.68e-01 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 2 | 218 | 2.68e-01 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 4 | 39 | 2.68e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 4 | 32 | 2.69e-01 |
| GO:BP | GO:0044782 | cilium organization | 5 | 313 | 2.69e-01 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 2 | 52 | 2.69e-01 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 8 | 85 | 2.69e-01 |
| GO:BP | GO:0062043 | positive regulation of cardiac epithelial to mesenchymal transition | 1 | 9 | 2.69e-01 |
| GO:BP | GO:0001765 | membrane raft assembly | 1 | 9 | 2.69e-01 |
| GO:BP | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization | 1 | 6 | 2.69e-01 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 2 | 25 | 2.70e-01 |
| GO:BP | GO:0075506 | entry of viral genome into host nucleus through nuclear pore complex via importin | 1 | 2 | 2.70e-01 |
| GO:BP | GO:0018242 | protein O-linked glycosylation via serine | 2 | 7 | 2.70e-01 |
| GO:BP | GO:0043588 | skin development | 2 | 175 | 2.70e-01 |
| GO:BP | GO:2000640 | positive regulation of SREBP signaling pathway | 1 | 2 | 2.70e-01 |
| GO:BP | GO:0051134 | negative regulation of NK T cell activation | 1 | 1 | 2.70e-01 |
| GO:BP | GO:0098935 | dendritic transport | 2 | 13 | 2.70e-01 |
| GO:BP | GO:0042552 | myelination | 2 | 111 | 2.70e-01 |
| GO:BP | GO:0002138 | retinoic acid biosynthetic process | 1 | 8 | 2.70e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 2.71e-01 |
| GO:BP | GO:0033864 | positive regulation of NAD(P)H oxidase activity | 1 | 6 | 2.71e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 3 | 78 | 2.71e-01 |
| GO:BP | GO:0045190 | isotype switching | 6 | 39 | 2.71e-01 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 3 | 20 | 2.71e-01 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 6 | 39 | 2.71e-01 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 6 | 39 | 2.71e-01 |
| GO:BP | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential | 1 | 3 | 2.71e-01 |
| GO:BP | GO:0090186 | regulation of pancreatic juice secretion | 1 | 6 | 2.72e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 1 | 6 | 2.72e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 6 | 341 | 2.72e-01 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 4 | 75 | 2.72e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 4 | 105 | 2.72e-01 |
| GO:BP | GO:0009445 | putrescine metabolic process | 1 | 6 | 2.72e-01 |
| GO:BP | GO:1902730 | positive regulation of proteoglycan biosynthetic process | 1 | 4 | 2.72e-01 |
| GO:BP | GO:0008216 | spermidine metabolic process | 1 | 7 | 2.72e-01 |
| GO:BP | GO:0098708 | glucose import across plasma membrane | 1 | 5 | 2.72e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 156 | 2.72e-01 |
| GO:BP | GO:0015757 | galactose transmembrane transport | 1 | 4 | 2.72e-01 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 3 | 18 | 2.72e-01 |
| GO:BP | GO:0070230 | positive regulation of lymphocyte apoptotic process | 2 | 10 | 2.73e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 6 | 342 | 2.73e-01 |
| GO:BP | GO:0034330 | cell junction organization | 6 | 575 | 2.73e-01 |
| GO:BP | GO:0031099 | regeneration | 1 | 142 | 2.73e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 18 | 678 | 2.73e-01 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 1 | 28 | 2.73e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 2 | 113 | 2.73e-01 |
| GO:BP | GO:0043116 | negative regulation of vascular permeability | 1 | 16 | 2.73e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 2 | 113 | 2.73e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 2 | 529 | 2.73e-01 |
| GO:BP | GO:0001575 | globoside metabolic process | 1 | 2 | 2.74e-01 |
| GO:BP | GO:0001576 | globoside biosynthetic process | 1 | 2 | 2.74e-01 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 2.74e-01 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 2.74e-01 |
| GO:BP | GO:0045006 | DNA deamination | 1 | 9 | 2.74e-01 |
| GO:BP | GO:1990708 | conditioned place preference | 1 | 2 | 2.74e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 2 | 34 | 2.75e-01 |
| GO:BP | GO:0098926 | postsynaptic signal transduction | 1 | 25 | 2.76e-01 |
| GO:BP | GO:0010269 | response to selenium ion | 1 | 8 | 2.76e-01 |
| GO:BP | GO:1902915 | negative regulation of protein polyubiquitination | 1 | 9 | 2.76e-01 |
| GO:BP | GO:0150099 | neuron-glial cell signaling | 1 | 4 | 2.76e-01 |
| GO:BP | GO:0072717 | cellular response to actinomycin D | 1 | 4 | 2.76e-01 |
| GO:BP | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway | 1 | 8 | 2.76e-01 |
| GO:BP | GO:0010939 | regulation of necrotic cell death | 2 | 38 | 2.76e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 2.76e-01 |
| GO:BP | GO:0006041 | glucosamine metabolic process | 1 | 3 | 2.76e-01 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 1 | 8 | 2.76e-01 |
| GO:BP | GO:0043149 | stress fiber assembly | 5 | 94 | 2.76e-01 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 5 | 94 | 2.76e-01 |
| GO:BP | GO:0030091 | protein repair | 1 | 7 | 2.76e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 1 | 216 | 2.76e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 2.76e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 10 | 302 | 2.76e-01 |
| GO:BP | GO:0035821 | modulation of process of another organism | 2 | 5 | 2.77e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 3 | 25 | 2.77e-01 |
| GO:BP | GO:0039526 | modulation by virus of host apoptotic process | 1 | 3 | 2.77e-01 |
| GO:BP | GO:0052150 | modulation by symbiont of host apoptotic process | 1 | 3 | 2.77e-01 |
| GO:BP | GO:0052040 | modulation by symbiont of host programmed cell death | 1 | 3 | 2.77e-01 |
| GO:BP | GO:0010044 | response to aluminum ion | 1 | 5 | 2.77e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 6 | 121 | 2.77e-01 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 1 | 30 | 2.77e-01 |
| GO:BP | GO:0006833 | water transport | 2 | 10 | 2.77e-01 |
| GO:BP | GO:0099550 | trans-synaptic signaling, modulating synaptic transmission | 2 | 10 | 2.77e-01 |
| GO:BP | GO:0032099 | negative regulation of appetite | 1 | 7 | 2.77e-01 |
| GO:BP | GO:0051350 | negative regulation of lyase activity | 2 | 15 | 2.77e-01 |
| GO:BP | GO:0032108 | negative regulation of response to nutrient levels | 1 | 7 | 2.77e-01 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 5 | 82 | 2.77e-01 |
| GO:BP | GO:0021784 | postganglionic parasympathetic fiber development | 1 | 2 | 2.77e-01 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 1 | 34 | 2.77e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 1 | 4 | 2.77e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 1 | 4 | 2.77e-01 |
| GO:BP | GO:0021644 | vagus nerve morphogenesis | 1 | 1 | 2.77e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 1 | 4 | 2.77e-01 |
| GO:BP | GO:0032875 | regulation of DNA endoreduplication | 1 | 4 | 2.77e-01 |
| GO:BP | GO:0032096 | negative regulation of response to food | 1 | 7 | 2.77e-01 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 5 | 23 | 2.77e-01 |
| GO:BP | GO:0016259 | selenocysteine metabolic process | 1 | 2 | 2.77e-01 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 1 | 60 | 2.77e-01 |
| GO:BP | GO:0061299 | retina vasculature morphogenesis in camera-type eye | 2 | 7 | 2.77e-01 |
| GO:BP | GO:0032105 | negative regulation of response to extracellular stimulus | 1 | 7 | 2.77e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 2 | 206 | 2.77e-01 |
| GO:BP | GO:0090239 | regulation of histone H4 acetylation | 2 | 10 | 2.77e-01 |
| GO:BP | GO:0034514 | mitochondrial unfolded protein response | 1 | 2 | 2.77e-01 |
| GO:BP | GO:0033210 | leptin-mediated signaling pathway | 1 | 10 | 2.77e-01 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 3 | 25 | 2.77e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 2 | 541 | 2.78e-01 |
| GO:BP | GO:0048806 | genitalia development | 2 | 28 | 2.78e-01 |
| GO:BP | GO:0006690 | icosanoid metabolic process | 3 | 60 | 2.78e-01 |
| GO:BP | GO:0001764 | neuron migration | 7 | 133 | 2.78e-01 |
| GO:BP | GO:0014870 | response to muscle inactivity | 2 | 9 | 2.78e-01 |
| GO:BP | GO:0045213 | neurotransmitter receptor metabolic process | 1 | 2 | 2.78e-01 |
| GO:BP | GO:0060857 | establishment of glial blood-brain barrier | 1 | 2 | 2.78e-01 |
| GO:BP | GO:0007284 | spermatogonial cell division | 1 | 3 | 2.78e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 2.79e-01 |
| GO:BP | GO:0051664 | nuclear pore localization | 1 | 4 | 2.79e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 2 | 122 | 2.79e-01 |
| GO:BP | GO:1900740 | positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 2.79e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 2 | 755 | 2.79e-01 |
| GO:BP | GO:0032462 | regulation of protein homooligomerization | 1 | 4 | 2.79e-01 |
| GO:BP | GO:0046457 | prostanoid biosynthetic process | 1 | 19 | 2.79e-01 |
| GO:BP | GO:0048874 | host-mediated regulation of intestinal microbiota composition | 1 | 2 | 2.79e-01 |
| GO:BP | GO:0032461 | positive regulation of protein oligomerization | 1 | 4 | 2.79e-01 |
| GO:BP | GO:0042439 | ethanolamine-containing compound metabolic process | 1 | 5 | 2.79e-01 |
| GO:BP | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 3 | 2.79e-01 |
| GO:BP | GO:0060764 | cell-cell signaling involved in mammary gland development | 1 | 1 | 2.79e-01 |
| GO:BP | GO:0001516 | prostaglandin biosynthetic process | 1 | 19 | 2.79e-01 |
| GO:BP | GO:0060068 | vagina development | 1 | 8 | 2.79e-01 |
| GO:BP | GO:0060513 | prostatic bud formation | 1 | 8 | 2.79e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 2.79e-01 |
| GO:BP | GO:0016340 | calcium-dependent cell-matrix adhesion | 1 | 2 | 2.79e-01 |
| GO:BP | GO:0098942 | retrograde trans-synaptic signaling by trans-synaptic protein complex | 1 | 2 | 2.79e-01 |
| GO:BP | GO:2000625 | regulation of miRNA catabolic process | 1 | 5 | 2.80e-01 |
| GO:BP | GO:0043007 | maintenance of rDNA | 1 | 2 | 2.80e-01 |
| GO:BP | GO:0055089 | fatty acid homeostasis | 1 | 12 | 2.80e-01 |
| GO:BP | GO:0060047 | heart contraction | 19 | 205 | 2.80e-01 |
| GO:BP | GO:0051695 | actin filament uncapping | 1 | 2 | 2.80e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 1 | 5 | 2.80e-01 |
| GO:BP | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway | 1 | 1 | 2.80e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 1 | 5 | 2.80e-01 |
| GO:BP | GO:0009224 | CMP biosynthetic process | 1 | 3 | 2.80e-01 |
| GO:BP | GO:0003417 | growth plate cartilage development | 1 | 15 | 2.80e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 2 | 15 | 2.80e-01 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 6 | 51 | 2.80e-01 |
| GO:BP | GO:0002315 | marginal zone B cell differentiation | 1 | 6 | 2.80e-01 |
| GO:BP | GO:0032959 | inositol trisphosphate biosynthetic process | 1 | 6 | 2.80e-01 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 2 | 14 | 2.80e-01 |
| GO:BP | GO:0015853 | adenine transport | 1 | 5 | 2.80e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 2 | 11 | 2.80e-01 |
| GO:BP | GO:0016102 | diterpenoid biosynthetic process | 1 | 8 | 2.80e-01 |
| GO:BP | GO:0015862 | uridine transport | 1 | 4 | 2.80e-01 |
| GO:BP | GO:0007566 | embryo implantation | 1 | 39 | 2.80e-01 |
| GO:BP | GO:0018277 | protein deamination | 1 | 2 | 2.80e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 5 | 115 | 2.80e-01 |
| GO:BP | GO:0030210 | heparin biosynthetic process | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0044857 | plasma membrane raft organization | 1 | 9 | 2.80e-01 |
| GO:BP | GO:0098711 | iron ion import across plasma membrane | 1 | 4 | 2.80e-01 |
| GO:BP | GO:0015864 | pyrimidine nucleoside transport | 1 | 4 | 2.80e-01 |
| GO:BP | GO:0015855 | pyrimidine nucleobase transport | 1 | 3 | 2.80e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 5 | 115 | 2.80e-01 |
| GO:BP | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone | 1 | 5 | 2.80e-01 |
| GO:BP | GO:0006579 | amino-acid betaine catabolic process | 1 | 1 | 2.80e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 1 | 258 | 2.80e-01 |
| GO:BP | GO:0030317 | flagellated sperm motility | 1 | 61 | 2.80e-01 |
| GO:BP | GO:0019254 | carnitine metabolic process, CoA-linked | 1 | 3 | 2.80e-01 |
| GO:BP | GO:0044572 | [4Fe-4S] cluster assembly | 1 | 5 | 2.80e-01 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 4 | 37 | 2.80e-01 |
| GO:BP | GO:0007517 | muscle organ development | 3 | 277 | 2.80e-01 |
| GO:BP | GO:0062042 | regulation of cardiac epithelial to mesenchymal transition | 1 | 10 | 2.80e-01 |
| GO:BP | GO:0097049 | motor neuron apoptotic process | 3 | 18 | 2.80e-01 |
| GO:BP | GO:0097722 | sperm motility | 1 | 61 | 2.80e-01 |
| GO:BP | GO:2000320 | negative regulation of T-helper 17 cell differentiation | 2 | 6 | 2.81e-01 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 4 | 29 | 2.81e-01 |
| GO:BP | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity | 1 | 30 | 2.81e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 2 | 86 | 2.81e-01 |
| GO:BP | GO:0060049 | regulation of protein glycosylation | 1 | 10 | 2.81e-01 |
| GO:BP | GO:0046131 | pyrimidine ribonucleoside metabolic process | 1 | 7 | 2.81e-01 |
| GO:BP | GO:0046133 | pyrimidine ribonucleoside catabolic process | 1 | 7 | 2.81e-01 |
| GO:BP | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 2.81e-01 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 1 | 68 | 2.81e-01 |
| GO:BP | GO:1903751 | negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 1 | 3 | 2.81e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 3 | 22 | 2.81e-01 |
| GO:BP | GO:0090618 | DNA clamp unloading | 1 | 2 | 2.81e-01 |
| GO:BP | GO:0080182 | histone H3-K4 trimethylation | 1 | 16 | 2.81e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 3 | 29 | 2.81e-01 |
| GO:BP | GO:0001754 | eye photoreceptor cell differentiation | 2 | 30 | 2.82e-01 |
| GO:BP | GO:0030101 | natural killer cell activation | 3 | 42 | 2.82e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 5 | 274 | 2.82e-01 |
| GO:BP | GO:0035176 | social behavior | 2 | 34 | 2.82e-01 |
| GO:BP | GO:0006924 | activation-induced cell death of T cells | 1 | 8 | 2.82e-01 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 2 | 13 | 2.82e-01 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 2 | 17 | 2.82e-01 |
| GO:BP | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 8 | 2.82e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 11 | 112 | 2.82e-01 |
| GO:BP | GO:1990966 | ATP generation from poly-ADP-D-ribose | 1 | 4 | 2.82e-01 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 7 | 53 | 2.82e-01 |
| GO:BP | GO:0030219 | megakaryocyte differentiation | 2 | 54 | 2.82e-01 |
| GO:BP | GO:1903774 | positive regulation of viral budding via host ESCRT complex | 1 | 3 | 2.82e-01 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 4 | 70 | 2.82e-01 |
| GO:BP | GO:0060662 | salivary gland cavitation | 1 | 3 | 2.82e-01 |
| GO:BP | GO:0060605 | tube lumen cavitation | 1 | 3 | 2.82e-01 |
| GO:BP | GO:0070997 | neuron death | 19 | 286 | 2.82e-01 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 1 | 63 | 2.82e-01 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 1 | 74 | 2.82e-01 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 2 | 27 | 2.82e-01 |
| GO:BP | GO:0061448 | connective tissue development | 1 | 208 | 2.82e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 1 | 130 | 2.82e-01 |
| GO:BP | GO:0021562 | vestibulocochlear nerve development | 2 | 4 | 2.82e-01 |
| GO:BP | GO:0051973 | positive regulation of telomerase activity | 3 | 31 | 2.82e-01 |
| GO:BP | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway | 1 | 7 | 2.82e-01 |
| GO:BP | GO:0014034 | neural crest cell fate commitment | 1 | 4 | 2.82e-01 |
| GO:BP | GO:1903537 | meiotic cell cycle process involved in oocyte maturation | 1 | 5 | 2.82e-01 |
| GO:BP | GO:0032740 | positive regulation of interleukin-17 production | 1 | 12 | 2.82e-01 |
| GO:BP | GO:0001503 | ossification | 10 | 313 | 2.83e-01 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 1 | 52 | 2.83e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 8 | 540 | 2.83e-01 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 8 | 89 | 2.83e-01 |
| GO:BP | GO:0006595 | polyamine metabolic process | 2 | 16 | 2.83e-01 |
| GO:BP | GO:0003128 | heart field specification | 2 | 16 | 2.83e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 3 | 506 | 2.83e-01 |
| GO:BP | GO:0050667 | homocysteine metabolic process | 2 | 11 | 2.83e-01 |
| GO:BP | GO:0006568 | tryptophan metabolic process | 1 | 3 | 2.83e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 6 | 137 | 2.83e-01 |
| GO:BP | GO:0006437 | tyrosyl-tRNA aminoacylation | 1 | 2 | 2.83e-01 |
| GO:BP | GO:0009069 | serine family amino acid metabolic process | 4 | 30 | 2.83e-01 |
| GO:BP | GO:0071169 | establishment of protein localization to chromatin | 1 | 8 | 2.83e-01 |
| GO:BP | GO:0042264 | peptidyl-aspartic acid hydroxylation | 1 | 2 | 2.83e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 2 | 16 | 2.83e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 4 | 615 | 2.83e-01 |
| GO:BP | GO:0001568 | blood vessel development | 14 | 534 | 2.83e-01 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 2 | 11 | 2.83e-01 |
| GO:BP | GO:0048634 | regulation of muscle organ development | 1 | 14 | 2.83e-01 |
| GO:BP | GO:0001881 | receptor recycling | 1 | 38 | 2.83e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 2 | 23 | 2.83e-01 |
| GO:BP | GO:0003290 | atrial septum secundum morphogenesis | 1 | 2 | 2.83e-01 |
| GO:BP | GO:0023035 | CD40 signaling pathway | 2 | 11 | 2.83e-01 |
| GO:BP | GO:0003219 | cardiac right ventricle formation | 1 | 4 | 2.84e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 4 | 84 | 2.84e-01 |
| GO:BP | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 1 | 6 | 2.84e-01 |
| GO:BP | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | 7 | 2.84e-01 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 1 | 8 | 2.84e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 9 | 299 | 2.84e-01 |
| GO:BP | GO:0060009 | Sertoli cell development | 1 | 10 | 2.84e-01 |
| GO:BP | GO:0042461 | photoreceptor cell development | 2 | 30 | 2.84e-01 |
| GO:BP | GO:0051106 | positive regulation of DNA ligation | 1 | 5 | 2.85e-01 |
| GO:BP | GO:0010457 | centriole-centriole cohesion | 2 | 14 | 2.85e-01 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 1 | 6 | 2.85e-01 |
| GO:BP | GO:0019068 | virion assembly | 4 | 35 | 2.85e-01 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 1 | 6 | 2.85e-01 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 1 | 12 | 2.85e-01 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 9 | 390 | 2.85e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 4 | 99 | 2.85e-01 |
| GO:BP | GO:0032377 | regulation of intracellular lipid transport | 1 | 7 | 2.85e-01 |
| GO:BP | GO:0072678 | T cell migration | 3 | 38 | 2.86e-01 |
| GO:BP | GO:0006020 | inositol metabolic process | 1 | 8 | 2.86e-01 |
| GO:BP | GO:0035694 | mitochondrial protein catabolic process | 1 | 6 | 2.86e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 3 | 1061 | 2.86e-01 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 2 | 35 | 2.87e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 6 | 93 | 2.87e-01 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 1 | 13 | 2.87e-01 |
| GO:BP | GO:1901661 | quinone metabolic process | 1 | 32 | 2.87e-01 |
| GO:BP | GO:0030539 | male genitalia development | 2 | 13 | 2.87e-01 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 2 | 10 | 2.87e-01 |
| GO:BP | GO:0009074 | aromatic amino acid family catabolic process | 2 | 4 | 2.87e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 5 | 461 | 2.88e-01 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 3 | 91 | 2.88e-01 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 3 | 42 | 2.88e-01 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 2 | 17 | 2.88e-01 |
| GO:BP | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity | 1 | 4 | 2.88e-01 |
| GO:BP | GO:0001525 | angiogenesis | 12 | 390 | 2.89e-01 |
| GO:BP | GO:0032764 | negative regulation of mast cell cytokine production | 1 | 3 | 2.89e-01 |
| GO:BP | GO:0007494 | midgut development | 1 | 7 | 2.89e-01 |
| GO:BP | GO:0048570 | notochord morphogenesis | 1 | 9 | 2.89e-01 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 1 | 9 | 2.89e-01 |
| GO:BP | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin | 1 | 8 | 2.89e-01 |
| GO:BP | GO:0003129 | heart induction | 1 | 9 | 2.89e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 4 | 110 | 2.89e-01 |
| GO:BP | GO:0090009 | primitive streak formation | 1 | 5 | 2.89e-01 |
| GO:BP | GO:0021889 | olfactory bulb interneuron differentiation | 1 | 7 | 2.89e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 4 | 625 | 2.89e-01 |
| GO:BP | GO:0014848 | urinary tract smooth muscle contraction | 2 | 6 | 2.90e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 5 | 101 | 2.90e-01 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 2 | 34 | 2.90e-01 |
| GO:BP | GO:0021796 | cerebral cortex regionalization | 1 | 2 | 2.90e-01 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 1 | 11 | 2.90e-01 |
| GO:BP | GO:0050655 | dermatan sulfate proteoglycan metabolic process | 1 | 12 | 2.90e-01 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 1 | 11 | 2.90e-01 |
| GO:BP | GO:0044282 | small molecule catabolic process | 2 | 271 | 2.90e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 4 | 30 | 2.90e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 1 | 463 | 2.90e-01 |
| GO:BP | GO:0006096 | glycolytic process | 1 | 70 | 2.91e-01 |
| GO:BP | GO:0000023 | maltose metabolic process | 1 | 2 | 2.91e-01 |
| GO:BP | GO:0030282 | bone mineralization | 4 | 82 | 2.91e-01 |
| GO:BP | GO:0070528 | protein kinase C signaling | 3 | 24 | 2.91e-01 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 1 | 6 | 2.92e-01 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | 24 | 2.92e-01 |
| GO:BP | GO:0003014 | renal system process | 6 | 78 | 2.92e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 3 | 76 | 2.92e-01 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 4 | 64 | 2.92e-01 |
| GO:BP | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 1 | 15 | 2.93e-01 |
| GO:BP | GO:0140271 | hexose import across plasma membrane | 1 | 5 | 2.93e-01 |
| GO:BP | GO:0015744 | succinate transport | 1 | 6 | 2.93e-01 |
| GO:BP | GO:0098704 | carbohydrate import across plasma membrane | 1 | 5 | 2.93e-01 |
| GO:BP | GO:0071422 | succinate transmembrane transport | 1 | 6 | 2.93e-01 |
| GO:BP | GO:0072520 | seminiferous tubule development | 2 | 13 | 2.93e-01 |
| GO:BP | GO:0007406 | negative regulation of neuroblast proliferation | 1 | 9 | 2.93e-01 |
| GO:BP | GO:0032228 | regulation of synaptic transmission, GABAergic | 1 | 23 | 2.93e-01 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 1 | 33 | 2.93e-01 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 2 | 35 | 2.94e-01 |
| GO:BP | GO:0007423 | sensory organ development | 1 | 396 | 2.94e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 3 | 53 | 2.94e-01 |
| GO:BP | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 3 | 2.94e-01 |
| GO:BP | GO:0060632 | regulation of microtubule-based movement | 2 | 31 | 2.94e-01 |
| GO:BP | GO:0106091 | glial cell projection elongation | 1 | 2 | 2.94e-01 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 3 | 101 | 2.94e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 2 | 47 | 2.94e-01 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 2 | 36 | 2.94e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 2.94e-01 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 1 | 65 | 2.94e-01 |
| GO:BP | GO:0048302 | regulation of isotype switching to IgG isotypes | 1 | 7 | 2.94e-01 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 4 | 56 | 2.94e-01 |
| GO:BP | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery | 1 | 6 | 2.94e-01 |
| GO:BP | GO:0001508 | action potential | 12 | 105 | 2.94e-01 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 4 | 31 | 2.95e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 6 | 94 | 2.95e-01 |
| GO:BP | GO:0021559 | trigeminal nerve development | 2 | 5 | 2.95e-01 |
| GO:BP | GO:0070189 | kynurenine metabolic process | 1 | 6 | 2.96e-01 |
| GO:BP | GO:0040014 | regulation of multicellular organism growth | 3 | 53 | 2.96e-01 |
| GO:BP | GO:0090594 | inflammatory response to wounding | 1 | 12 | 2.97e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 4 | 637 | 2.98e-01 |
| GO:BP | GO:0070286 | axonemal dynein complex assembly | 1 | 20 | 2.98e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 2.98e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 5 | 469 | 2.98e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 5 | 65 | 2.98e-01 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 1 | 6 | 2.98e-01 |
| GO:BP | GO:0007548 | sex differentiation | 6 | 191 | 2.98e-01 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 4 | 98 | 2.98e-01 |
| GO:BP | GO:0050919 | negative chemotaxis | 5 | 42 | 2.98e-01 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 3 | 21 | 2.98e-01 |
| GO:BP | GO:0050691 | regulation of defense response to virus by host | 1 | 30 | 2.98e-01 |
| GO:BP | GO:0045987 | positive regulation of smooth muscle contraction | 1 | 17 | 2.98e-01 |
| GO:BP | GO:0010040 | response to iron(II) ion | 1 | 5 | 2.98e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 2 | 109 | 2.98e-01 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 1 | 10 | 2.98e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 9 | 517 | 2.98e-01 |
| GO:BP | GO:0097252 | oligodendrocyte apoptotic process | 1 | 4 | 2.99e-01 |
| GO:BP | GO:0006266 | DNA ligation | 2 | 16 | 3.00e-01 |
| GO:BP | GO:0046135 | pyrimidine nucleoside catabolic process | 1 | 8 | 3.00e-01 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 4 | 3.00e-01 |
| GO:BP | GO:0090238 | positive regulation of arachidonic acid secretion | 1 | 2 | 3.00e-01 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 4 | 35 | 3.00e-01 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 2 | 11 | 3.00e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 3.00e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 3.00e-01 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 2 | 12 | 3.00e-01 |
| GO:BP | GO:0043418 | homocysteine catabolic process | 1 | 3 | 3.00e-01 |
| GO:BP | GO:0086015 | SA node cell action potential | 2 | 12 | 3.00e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 20 | 1719 | 3.00e-01 |
| GO:BP | GO:0060706 | cell differentiation involved in embryonic placenta development | 1 | 20 | 3.00e-01 |
| GO:BP | GO:0019344 | cysteine biosynthetic process | 1 | 3 | 3.00e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 3 | 1107 | 3.01e-01 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 1 | 31 | 3.01e-01 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 2 | 11 | 3.01e-01 |
| GO:BP | GO:0009794 | regulation of mitotic cell cycle, embryonic | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0021557 | oculomotor nerve development | 1 | 2 | 3.01e-01 |
| GO:BP | GO:0070779 | D-aspartate import across plasma membrane | 1 | 4 | 3.01e-01 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 1 | 3 | 3.01e-01 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 1 | 3 | 3.01e-01 |
| GO:BP | GO:1905457 | negative regulation of lymphoid progenitor cell differentiation | 1 | 3 | 3.01e-01 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 1 | 35 | 3.01e-01 |
| GO:BP | GO:0045448 | mitotic cell cycle, embryonic | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0055129 | L-proline biosynthetic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0070777 | D-aspartate transport | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0090461 | intracellular glutamate homeostasis | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 3 | 24 | 3.01e-01 |
| GO:BP | GO:0050829 | defense response to Gram-negative bacterium | 2 | 20 | 3.01e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 1 | 39 | 3.01e-01 |
| GO:BP | GO:0006561 | proline biosynthetic process | 1 | 5 | 3.01e-01 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 1 | 3 | 3.01e-01 |
| GO:BP | GO:0071242 | cellular response to ammonium ion | 1 | 4 | 3.01e-01 |
| GO:BP | GO:0042572 | retinol metabolic process | 3 | 24 | 3.02e-01 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 1 | 6 | 3.02e-01 |
| GO:BP | GO:0036369 | transcription factor catabolic process | 1 | 5 | 3.02e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 6 | 95 | 3.02e-01 |
| GO:BP | GO:0072716 | response to actinomycin D | 1 | 5 | 3.02e-01 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 2 | 13 | 3.02e-01 |
| GO:BP | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation | 1 | 8 | 3.02e-01 |
| GO:BP | GO:1904425 | negative regulation of GTP binding | 1 | 3 | 3.02e-01 |
| GO:BP | GO:0030576 | Cajal body organization | 1 | 4 | 3.02e-01 |
| GO:BP | GO:0030641 | regulation of cellular pH | 1 | 70 | 3.02e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 1 | 10 | 3.02e-01 |
| GO:BP | GO:2000317 | negative regulation of T-helper 17 type immune response | 2 | 7 | 3.02e-01 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 1 | 10 | 3.02e-01 |
| GO:BP | GO:0010454 | negative regulation of cell fate commitment | 2 | 6 | 3.02e-01 |
| GO:BP | GO:0032202 | telomere assembly | 1 | 4 | 3.02e-01 |
| GO:BP | GO:1905809 | negative regulation of synapse organization | 1 | 8 | 3.02e-01 |
| GO:BP | GO:0001885 | endothelial cell development | 2 | 53 | 3.02e-01 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 1 | 107 | 3.02e-01 |
| GO:BP | GO:0090315 | negative regulation of protein targeting to membrane | 1 | 5 | 3.02e-01 |
| GO:BP | GO:2001169 | regulation of ATP biosynthetic process | 2 | 17 | 3.02e-01 |
| GO:BP | GO:0097576 | vacuole fusion | 1 | 12 | 3.02e-01 |
| GO:BP | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | 2 | 10 | 3.02e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 1 | 18 | 3.02e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 2 | 33 | 3.03e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 1 | 67 | 3.03e-01 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 2 | 11 | 3.03e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 3.03e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 5 | 56 | 3.03e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 3.04e-01 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 1 | 33 | 3.04e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 1 | 37 | 3.04e-01 |
| GO:BP | GO:0097017 | renal protein absorption | 1 | 5 | 3.04e-01 |
| GO:BP | GO:0048699 | generation of neurons | 3 | 1119 | 3.04e-01 |
| GO:BP | GO:0048291 | isotype switching to IgG isotypes | 1 | 8 | 3.04e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 4 | 53 | 3.04e-01 |
| GO:BP | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | 1 | 7 | 3.04e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2 | 12 | 3.05e-01 |
| GO:BP | GO:0042754 | negative regulation of circadian rhythm | 1 | 10 | 3.05e-01 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 2 | 13 | 3.05e-01 |
| GO:BP | GO:0042023 | DNA endoreduplication | 1 | 5 | 3.05e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 3.05e-01 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 1 | 4 | 3.05e-01 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 1 | 4 | 3.05e-01 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 6 | 41 | 3.05e-01 |
| GO:BP | GO:0061087 | positive regulation of histone H3-K27 methylation | 1 | 4 | 3.05e-01 |
| GO:BP | GO:0009264 | deoxyribonucleotide catabolic process | 2 | 22 | 3.05e-01 |
| GO:BP | GO:0090156 | intracellular sphingolipid homeostasis | 1 | 5 | 3.05e-01 |
| GO:BP | GO:0046386 | deoxyribose phosphate catabolic process | 2 | 22 | 3.05e-01 |
| GO:BP | GO:2000253 | positive regulation of feeding behavior | 2 | 4 | 3.06e-01 |
| GO:BP | GO:0001523 | retinoid metabolic process | 4 | 40 | 3.06e-01 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 3 | 72 | 3.06e-01 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 4 | 68 | 3.07e-01 |
| GO:BP | GO:0042501 | serine phosphorylation of STAT protein | 1 | 9 | 3.07e-01 |
| GO:BP | GO:0051261 | protein depolymerization | 7 | 105 | 3.07e-01 |
| GO:BP | GO:0035585 | calcium-mediated signaling using extracellular calcium source | 1 | 2 | 3.07e-01 |
| GO:BP | GO:0006586 | indolalkylamine metabolic process | 1 | 3 | 3.07e-01 |
| GO:BP | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 6 | 3.07e-01 |
| GO:BP | GO:0008272 | sulfate transport | 2 | 12 | 3.07e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 6 | 73 | 3.07e-01 |
| GO:BP | GO:0031640 | killing of cells of another organism | 1 | 10 | 3.08e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 1 | 516 | 3.08e-01 |
| GO:BP | GO:0060789 | hair follicle placode formation | 1 | 5 | 3.08e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 5 | 168 | 3.08e-01 |
| GO:BP | GO:0090370 | negative regulation of cholesterol efflux | 1 | 3 | 3.08e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 108 | 1939 | 3.08e-01 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 1 | 9 | 3.08e-01 |
| GO:BP | GO:0035845 | photoreceptor cell outer segment organization | 1 | 10 | 3.08e-01 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 1 | 14 | 3.08e-01 |
| GO:BP | GO:0071353 | cellular response to interleukin-4 | 2 | 21 | 3.08e-01 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 2 | 15 | 3.08e-01 |
| GO:BP | GO:0046475 | glycerophospholipid catabolic process | 2 | 21 | 3.09e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 3.09e-01 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 1 | 7 | 3.10e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 1 | 26 | 3.10e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 4 | 113 | 3.11e-01 |
| GO:BP | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 1 | 36 | 3.11e-01 |
| GO:BP | GO:0061364 | apoptotic process involved in luteolysis | 1 | 3 | 3.11e-01 |
| GO:BP | GO:0007281 | germ cell development | 6 | 187 | 3.12e-01 |
| GO:BP | GO:0097101 | blood vessel endothelial cell fate specification | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0060846 | blood vessel endothelial cell fate commitment | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0150020 | basal dendrite arborization | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0150019 | basal dendrite morphogenesis | 1 | 3 | 3.12e-01 |
| GO:BP | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment | 1 | 1 | 3.12e-01 |
| GO:BP | GO:1902336 | positive regulation of retinal ganglion cell axon guidance | 1 | 2 | 3.12e-01 |
| GO:BP | GO:0150018 | basal dendrite development | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0009063 | amino acid catabolic process | 1 | 78 | 3.12e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 3 | 544 | 3.12e-01 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 4 | 111 | 3.12e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 1 | 5 | 3.12e-01 |
| GO:BP | GO:0070292 | N-acylphosphatidylethanolamine metabolic process | 1 | 5 | 3.12e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 6 | 195 | 3.12e-01 |
| GO:BP | GO:0051105 | regulation of DNA ligation | 1 | 6 | 3.12e-01 |
| GO:BP | GO:0006046 | N-acetylglucosamine catabolic process | 1 | 3 | 3.12e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 3.12e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 63 | 1345 | 3.12e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 8 | 438 | 3.12e-01 |
| GO:BP | GO:0007405 | neuroblast proliferation | 5 | 51 | 3.12e-01 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 1 | 26 | 3.12e-01 |
| GO:BP | GO:0007507 | heart development | 6 | 506 | 3.12e-01 |
| GO:BP | GO:0006417 | regulation of translation | 2 | 370 | 3.12e-01 |
| GO:BP | GO:0071320 | cellular response to cAMP | 3 | 41 | 3.13e-01 |
| GO:BP | GO:0043949 | regulation of cAMP-mediated signaling | 3 | 17 | 3.13e-01 |
| GO:BP | GO:0035583 | sequestering of TGFbeta in extracellular matrix | 1 | 3 | 3.13e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 3 | 90 | 3.13e-01 |
| GO:BP | GO:0032730 | positive regulation of interleukin-1 alpha production | 1 | 3 | 3.13e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 6 | 620 | 3.13e-01 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 1 | 35 | 3.14e-01 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 2 | 31 | 3.14e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 79 | 3.15e-01 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 1 | 5 | 3.15e-01 |
| GO:BP | GO:0010225 | response to UV-C | 1 | 13 | 3.15e-01 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 1 | 17 | 3.15e-01 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 1 | 17 | 3.15e-01 |
| GO:BP | GO:0021684 | cerebellar granular layer formation | 1 | 6 | 3.16e-01 |
| GO:BP | GO:0021707 | cerebellar granule cell differentiation | 1 | 6 | 3.16e-01 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 2 | 27 | 3.16e-01 |
| GO:BP | GO:0010749 | regulation of nitric oxide mediated signal transduction | 2 | 10 | 3.16e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 2 | 34 | 3.16e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 4 | 32 | 3.16e-01 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 5 | 32 | 3.16e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 3.16e-01 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 1 | 39 | 3.17e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 3 | 1201 | 3.17e-01 |
| GO:BP | GO:0009838 | abscission | 1 | 6 | 3.17e-01 |
| GO:BP | GO:0006837 | serotonin transport | 1 | 11 | 3.17e-01 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 7 | 57 | 3.17e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 3.17e-01 |
| GO:BP | GO:0044557 | relaxation of smooth muscle | 1 | 9 | 3.17e-01 |
| GO:BP | GO:0150077 | regulation of neuroinflammatory response | 1 | 13 | 3.17e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 4 | 28 | 3.17e-01 |
| GO:BP | GO:0000706 | meiotic DNA double-strand break processing | 1 | 2 | 3.17e-01 |
| GO:BP | GO:2000344 | positive regulation of acrosome reaction | 1 | 6 | 3.17e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 3 | 55 | 3.17e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 2 | 662 | 3.17e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 2 | 70 | 3.17e-01 |
| GO:BP | GO:2000191 | regulation of fatty acid transport | 2 | 19 | 3.17e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 2 | 901 | 3.18e-01 |
| GO:BP | GO:0031645 | negative regulation of nervous system process | 2 | 9 | 3.18e-01 |
| GO:BP | GO:0051898 | negative regulation of protein kinase B signaling | 3 | 36 | 3.18e-01 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 1 | 19 | 3.18e-01 |
| GO:BP | GO:0007631 | feeding behavior | 3 | 52 | 3.18e-01 |
| GO:BP | GO:0006885 | regulation of pH | 1 | 74 | 3.18e-01 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 13 | 3.18e-01 |
| GO:BP | GO:0090677 | reversible differentiation | 1 | 5 | 3.18e-01 |
| GO:BP | GO:0090116 | C-5 methylation of cytosine | 1 | 3 | 3.18e-01 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 3 | 108 | 3.18e-01 |
| GO:BP | GO:0071709 | membrane assembly | 3 | 53 | 3.18e-01 |
| GO:BP | GO:0045717 | negative regulation of fatty acid biosynthetic process | 2 | 14 | 3.18e-01 |
| GO:BP | GO:0090217 | negative regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 3 | 3.19e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 2 | 13 | 3.19e-01 |
| GO:BP | GO:0090237 | regulation of arachidonic acid secretion | 1 | 2 | 3.19e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 2 | 13 | 3.19e-01 |
| GO:BP | GO:0071921 | cohesin loading | 1 | 3 | 3.19e-01 |
| GO:BP | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process | 1 | 3 | 3.19e-01 |
| GO:BP | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway | 1 | 4 | 3.19e-01 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 4 | 40 | 3.19e-01 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 1 | 13 | 3.19e-01 |
| GO:BP | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process | 1 | 3 | 3.19e-01 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 1 | 105 | 3.19e-01 |
| GO:BP | GO:1902527 | positive regulation of protein monoubiquitination | 1 | 5 | 3.19e-01 |
| GO:BP | GO:0046469 | platelet activating factor metabolic process | 1 | 5 | 3.19e-01 |
| GO:BP | GO:0032049 | cardiolipin biosynthetic process | 1 | 7 | 3.19e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 2 | 1462 | 3.19e-01 |
| GO:BP | GO:0060384 | innervation | 3 | 22 | 3.19e-01 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 1 | 62 | 3.19e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 2 | 48 | 3.19e-01 |
| GO:BP | GO:0001944 | vasculature development | 7 | 559 | 3.19e-01 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 1 | 27 | 3.19e-01 |
| GO:BP | GO:0006826 | iron ion transport | 4 | 40 | 3.19e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 17 | 500 | 3.20e-01 |
| GO:BP | GO:2000807 | regulation of synaptic vesicle clustering | 1 | 7 | 3.20e-01 |
| GO:BP | GO:0070445 | regulation of oligodendrocyte progenitor proliferation | 1 | 5 | 3.20e-01 |
| GO:BP | GO:0035995 | detection of muscle stretch | 1 | 7 | 3.20e-01 |
| GO:BP | GO:0070444 | oligodendrocyte progenitor proliferation | 1 | 5 | 3.20e-01 |
| GO:BP | GO:0072675 | osteoclast fusion | 1 | 5 | 3.20e-01 |
| GO:BP | GO:0016101 | diterpenoid metabolic process | 4 | 43 | 3.21e-01 |
| GO:BP | GO:0055096 | low-density lipoprotein particle mediated signaling | 1 | 3 | 3.21e-01 |
| GO:BP | GO:0055095 | lipoprotein particle mediated signaling | 1 | 3 | 3.21e-01 |
| GO:BP | GO:0034371 | chylomicron remodeling | 1 | 1 | 3.21e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 3.21e-01 |
| GO:BP | GO:0010587 | miRNA catabolic process | 2 | 10 | 3.22e-01 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 1 | 4 | 3.22e-01 |
| GO:BP | GO:0019370 | leukotriene biosynthetic process | 1 | 7 | 3.22e-01 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 7 | 42 | 3.22e-01 |
| GO:BP | GO:0043030 | regulation of macrophage activation | 2 | 31 | 3.23e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 8 | 4241 | 3.23e-01 |
| GO:BP | GO:0071696 | ectodermal placode development | 2 | 11 | 3.23e-01 |
| GO:BP | GO:0019433 | triglyceride catabolic process | 2 | 17 | 3.23e-01 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 2 | 11 | 3.23e-01 |
| GO:BP | GO:0021747 | cochlear nucleus development | 1 | 3 | 3.23e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 9 | 540 | 3.23e-01 |
| GO:BP | GO:0014075 | response to amine | 3 | 34 | 3.23e-01 |
| GO:BP | GO:0060042 | retina morphogenesis in camera-type eye | 2 | 40 | 3.23e-01 |
| GO:BP | GO:0034205 | amyloid-beta formation | 1 | 41 | 3.23e-01 |
| GO:BP | GO:0072676 | lymphocyte migration | 4 | 59 | 3.23e-01 |
| GO:BP | GO:0042428 | serotonin metabolic process | 1 | 6 | 3.23e-01 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 1 | 4 | 3.23e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 3.23e-01 |
| GO:BP | GO:0003096 | renal sodium ion transport | 2 | 10 | 3.24e-01 |
| GO:BP | GO:1903772 | regulation of viral budding via host ESCRT complex | 1 | 4 | 3.24e-01 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 3 | 72 | 3.24e-01 |
| GO:BP | GO:0043377 | negative regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 3 | 3.24e-01 |
| GO:BP | GO:0072156 | distal tubule morphogenesis | 1 | 3 | 3.24e-01 |
| GO:BP | GO:0043584 | nose development | 1 | 10 | 3.24e-01 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 1 | 40 | 3.24e-01 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 2 | 29 | 3.24e-01 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 4 | 175 | 3.24e-01 |
| GO:BP | GO:0071711 | basement membrane organization | 3 | 33 | 3.24e-01 |
| GO:BP | GO:2001260 | regulation of semaphorin-plexin signaling pathway | 1 | 3 | 3.24e-01 |
| GO:BP | GO:0046471 | phosphatidylglycerol metabolic process | 2 | 23 | 3.24e-01 |
| GO:BP | GO:0016553 | base conversion or substitution editing | 1 | 10 | 3.24e-01 |
| GO:BP | GO:0030030 | cell projection organization | 3 | 1233 | 3.24e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 4 | 213 | 3.24e-01 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 3 | 35 | 3.24e-01 |
| GO:BP | GO:0050855 | regulation of B cell receptor signaling pathway | 1 | 11 | 3.25e-01 |
| GO:BP | GO:0048853 | forebrain morphogenesis | 1 | 7 | 3.25e-01 |
| GO:BP | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter | 1 | 4 | 3.25e-01 |
| GO:BP | GO:1902033 | regulation of hematopoietic stem cell proliferation | 1 | 12 | 3.25e-01 |
| GO:BP | GO:0046967 | cytosol to endoplasmic reticulum transport | 1 | 4 | 3.25e-01 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 1 | 5 | 3.25e-01 |
| GO:BP | GO:0009744 | response to sucrose | 1 | 4 | 3.25e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 5 | 117 | 3.25e-01 |
| GO:BP | GO:0006997 | nucleus organization | 8 | 126 | 3.25e-01 |
| GO:BP | GO:0035563 | positive regulation of chromatin binding | 2 | 12 | 3.25e-01 |
| GO:BP | GO:0009750 | response to fructose | 1 | 3 | 3.25e-01 |
| GO:BP | GO:0034285 | response to disaccharide | 1 | 4 | 3.25e-01 |
| GO:BP | GO:0030011 | maintenance of cell polarity | 1 | 16 | 3.25e-01 |
| GO:BP | GO:1903800 | positive regulation of miRNA processing | 1 | 3 | 3.26e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 3.26e-01 |
| GO:BP | GO:0030220 | platelet formation | 1 | 17 | 3.26e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 4 | 84 | 3.26e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 4 | 485 | 3.26e-01 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 1 | 22 | 3.26e-01 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 1 | 41 | 3.26e-01 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 4 | 71 | 3.26e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 9 | 168 | 3.26e-01 |
| GO:BP | GO:0006768 | biotin metabolic process | 1 | 3 | 3.26e-01 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 1 | 3 | 3.27e-01 |
| GO:BP | GO:0016358 | dendrite development | 5 | 202 | 3.27e-01 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 2 | 57 | 3.27e-01 |
| GO:BP | GO:0090168 | Golgi reassembly | 1 | 5 | 3.27e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 2 | 117 | 3.27e-01 |
| GO:BP | GO:0034756 | regulation of iron ion transport | 1 | 4 | 3.27e-01 |
| GO:BP | GO:0002532 | production of molecular mediator involved in inflammatory response | 2 | 49 | 3.28e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 4 | 227 | 3.28e-01 |
| GO:BP | GO:0003015 | heart process | 19 | 216 | 3.28e-01 |
| GO:BP | GO:0009566 | fertilization | 2 | 93 | 3.28e-01 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 1 | 16 | 3.28e-01 |
| GO:BP | GO:1901642 | nucleoside transmembrane transport | 1 | 6 | 3.28e-01 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 1 | 16 | 3.28e-01 |
| GO:BP | GO:0140009 | L-aspartate import across plasma membrane | 1 | 5 | 3.28e-01 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 1 | 4 | 3.28e-01 |
| GO:BP | GO:0060359 | response to ammonium ion | 1 | 5 | 3.28e-01 |
| GO:BP | GO:0032095 | regulation of response to food | 1 | 8 | 3.28e-01 |
| GO:BP | GO:0003334 | keratinocyte development | 1 | 8 | 3.28e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 2 | 330 | 3.29e-01 |
| GO:BP | GO:0032048 | cardiolipin metabolic process | 2 | 13 | 3.29e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 9 | 169 | 3.29e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 2 | 39 | 3.29e-01 |
| GO:BP | GO:1900044 | regulation of protein K63-linked ubiquitination | 1 | 13 | 3.29e-01 |
| GO:BP | GO:0006362 | transcription elongation by RNA polymerase I | 1 | 4 | 3.29e-01 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 1 | 39 | 3.29e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 3.29e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 5 | 490 | 3.29e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 5 | 490 | 3.29e-01 |
| GO:BP | GO:1904353 | regulation of telomere capping | 2 | 23 | 3.29e-01 |
| GO:BP | GO:0032927 | positive regulation of activin receptor signaling pathway | 1 | 7 | 3.29e-01 |
| GO:BP | GO:0001560 | regulation of cell growth by extracellular stimulus | 1 | 3 | 3.30e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 3 | 21 | 3.30e-01 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 1 | 89 | 3.30e-01 |
| GO:BP | GO:0051598 | meiotic recombination checkpoint signaling | 1 | 2 | 3.30e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 2 | 75 | 3.30e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 3 | 27 | 3.30e-01 |
| GO:BP | GO:1901523 | icosanoid catabolic process | 1 | 2 | 3.31e-01 |
| GO:BP | GO:0002084 | protein depalmitoylation | 1 | 10 | 3.31e-01 |
| GO:BP | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 49 | 798 | 3.31e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 2 | 16 | 3.31e-01 |
| GO:BP | GO:0038109 | Kit signaling pathway | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 10 | 323 | 3.31e-01 |
| GO:BP | GO:0030644 | intracellular chloride ion homeostasis | 1 | 7 | 3.31e-01 |
| GO:BP | GO:1901465 | positive regulation of tetrapyrrole biosynthetic process | 1 | 3 | 3.31e-01 |
| GO:BP | GO:0030002 | intracellular monoatomic anion homeostasis | 1 | 7 | 3.31e-01 |
| GO:BP | GO:0042454 | ribonucleoside catabolic process | 1 | 12 | 3.31e-01 |
| GO:BP | GO:0033860 | regulation of NAD(P)H oxidase activity | 1 | 6 | 3.31e-01 |
| GO:BP | GO:0036216 | cellular response to stem cell factor stimulus | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0046075 | dTTP metabolic process | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0051572 | negative regulation of histone H3-K4 methylation | 1 | 4 | 3.31e-01 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 1 | 5 | 3.31e-01 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 3 | 26 | 3.31e-01 |
| GO:BP | GO:0032074 | negative regulation of nuclease activity | 1 | 6 | 3.31e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 76 | 3625 | 3.31e-01 |
| GO:BP | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process | 1 | 3 | 3.31e-01 |
| GO:BP | GO:0070670 | response to interleukin-4 | 2 | 22 | 3.31e-01 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 3 | 21 | 3.31e-01 |
| GO:BP | GO:0032957 | inositol trisphosphate metabolic process | 1 | 10 | 3.31e-01 |
| GO:BP | GO:0070455 | positive regulation of heme biosynthetic process | 1 | 3 | 3.31e-01 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 1 | 96 | 3.31e-01 |
| GO:BP | GO:0016125 | sterol metabolic process | 3 | 111 | 3.31e-01 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 1 | 16 | 3.31e-01 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 2 | 13 | 3.31e-01 |
| GO:BP | GO:0036215 | response to stem cell factor | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 1 | 4 | 3.31e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 11 | 123 | 3.31e-01 |
| GO:BP | GO:0003407 | neural retina development | 5 | 48 | 3.31e-01 |
| GO:BP | GO:1903614 | negative regulation of protein tyrosine phosphatase activity | 1 | 3 | 3.31e-01 |
| GO:BP | GO:0035855 | megakaryocyte development | 1 | 16 | 3.31e-01 |
| GO:BP | GO:2000179 | positive regulation of neural precursor cell proliferation | 4 | 34 | 3.31e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 1 | 93 | 3.31e-01 |
| GO:BP | GO:0021564 | vagus nerve development | 1 | 2 | 3.31e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 44 | 710 | 3.32e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 2 | 17 | 3.32e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 3.32e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 1 | 386 | 3.32e-01 |
| GO:BP | GO:0007286 | spermatid development | 1 | 105 | 3.32e-01 |
| GO:BP | GO:0021627 | olfactory nerve morphogenesis | 1 | 3 | 3.33e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 5 | 495 | 3.33e-01 |
| GO:BP | GO:2000977 | regulation of forebrain neuron differentiation | 1 | 4 | 3.33e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 4 | 1974 | 3.33e-01 |
| GO:BP | GO:0062094 | stomach development | 1 | 4 | 3.33e-01 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 1 | 4 | 3.33e-01 |
| GO:BP | GO:0061009 | common bile duct development | 1 | 4 | 3.33e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 3.33e-01 |
| GO:BP | GO:0110135 | Norrin signaling pathway | 1 | 2 | 3.33e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 2 | 30 | 3.34e-01 |
| GO:BP | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | 1 | 5 | 3.34e-01 |
| GO:BP | GO:0048021 | regulation of melanin biosynthetic process | 1 | 12 | 3.34e-01 |
| GO:BP | GO:0061458 | reproductive system development | 11 | 215 | 3.34e-01 |
| GO:BP | GO:1900376 | regulation of secondary metabolite biosynthetic process | 1 | 12 | 3.34e-01 |
| GO:BP | GO:0043455 | regulation of secondary metabolic process | 1 | 12 | 3.34e-01 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 1 | 6 | 3.34e-01 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 1 | 14 | 3.34e-01 |
| GO:BP | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange | 1 | 5 | 3.34e-01 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 2 | 25 | 3.34e-01 |
| GO:BP | GO:0016476 | regulation of embryonic cell shape | 1 | 3 | 3.34e-01 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 4 | 138 | 3.34e-01 |
| GO:BP | GO:0090666 | scaRNA localization to Cajal body | 1 | 5 | 3.34e-01 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 1 | 6 | 3.34e-01 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 4 | 47 | 3.34e-01 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 1 | 13 | 3.35e-01 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 1 | 13 | 3.35e-01 |
| GO:BP | GO:1901339 | regulation of store-operated calcium channel activity | 1 | 8 | 3.35e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 23 | 344 | 3.35e-01 |
| GO:BP | GO:0002523 | leukocyte migration involved in inflammatory response | 1 | 9 | 3.36e-01 |
| GO:BP | GO:0048232 | male gamete generation | 2 | 343 | 3.36e-01 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 1 | 9 | 3.36e-01 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 1 | 8 | 3.36e-01 |
| GO:BP | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle | 1 | 20 | 3.36e-01 |
| GO:BP | GO:0019395 | fatty acid oxidation | 1 | 93 | 3.36e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 5 | 167 | 3.36e-01 |
| GO:BP | GO:0003356 | regulation of cilium beat frequency | 1 | 10 | 3.36e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 3.37e-01 |
| GO:BP | GO:0045010 | actin nucleation | 2 | 50 | 3.37e-01 |
| GO:BP | GO:2000003 | positive regulation of DNA damage checkpoint | 1 | 6 | 3.37e-01 |
| GO:BP | GO:0010520 | regulation of reciprocal meiotic recombination | 1 | 2 | 3.37e-01 |
| GO:BP | GO:0010845 | positive regulation of reciprocal meiotic recombination | 1 | 2 | 3.37e-01 |
| GO:BP | GO:0001787 | natural killer cell proliferation | 1 | 6 | 3.37e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 3 | 184 | 3.37e-01 |
| GO:BP | GO:0001893 | maternal placenta development | 1 | 28 | 3.37e-01 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 2 | 58 | 3.37e-01 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 2 | 10 | 3.37e-01 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 4 | 32 | 3.37e-01 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 1 | 17 | 3.37e-01 |
| GO:BP | GO:0045686 | negative regulation of glial cell differentiation | 3 | 15 | 3.37e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 8 | 281 | 3.37e-01 |
| GO:BP | GO:0036089 | cleavage furrow formation | 1 | 7 | 3.38e-01 |
| GO:BP | GO:0048515 | spermatid differentiation | 1 | 111 | 3.38e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 5 | 60 | 3.38e-01 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 1 | 15 | 3.38e-01 |
| GO:BP | GO:0036344 | platelet morphogenesis | 1 | 19 | 3.38e-01 |
| GO:BP | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development | 1 | 2 | 3.38e-01 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 1 | 12 | 3.38e-01 |
| GO:BP | GO:0070317 | negative regulation of G0 to G1 transition | 1 | 5 | 3.39e-01 |
| GO:BP | GO:0042044 | fluid transport | 1 | 17 | 3.39e-01 |
| GO:BP | GO:0051897 | positive regulation of protein kinase B signaling | 3 | 79 | 3.39e-01 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 1 | 7 | 3.39e-01 |
| GO:BP | GO:0051349 | positive regulation of lyase activity | 4 | 23 | 3.39e-01 |
| GO:BP | GO:2000535 | regulation of entry of bacterium into host cell | 1 | 6 | 3.39e-01 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 4 | 126 | 3.40e-01 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 2 | 40 | 3.40e-01 |
| GO:BP | GO:0048469 | cell maturation | 7 | 108 | 3.40e-01 |
| GO:BP | GO:0045188 | regulation of circadian sleep/wake cycle, non-REM sleep | 1 | 2 | 3.40e-01 |
| GO:BP | GO:0060545 | positive regulation of necroptotic process | 1 | 4 | 3.40e-01 |
| GO:BP | GO:0006998 | nuclear envelope organization | 2 | 57 | 3.40e-01 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 1 | 15 | 3.40e-01 |
| GO:BP | GO:0014038 | regulation of Schwann cell differentiation | 1 | 3 | 3.40e-01 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 1 | 6 | 3.40e-01 |
| GO:BP | GO:0045069 | regulation of viral genome replication | 2 | 64 | 3.40e-01 |
| GO:BP | GO:0003032 | detection of oxygen | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0051418 | microtubule nucleation by microtubule organizing center | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0051597 | response to methylmercury | 1 | 5 | 3.40e-01 |
| GO:BP | GO:0030913 | paranodal junction assembly | 1 | 6 | 3.40e-01 |
| GO:BP | GO:0072224 | metanephric glomerulus development | 1 | 10 | 3.40e-01 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 2 | 40 | 3.41e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 7 | 1076 | 3.41e-01 |
| GO:BP | GO:0061512 | protein localization to cilium | 2 | 61 | 3.41e-01 |
| GO:BP | GO:0045008 | depyrimidination | 1 | 8 | 3.41e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 7 | 1074 | 3.41e-01 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 6 | 47 | 3.42e-01 |
| GO:BP | GO:0046439 | L-cysteine metabolic process | 1 | 3 | 3.42e-01 |
| GO:BP | GO:0019448 | L-cysteine catabolic process | 1 | 3 | 3.42e-01 |
| GO:BP | GO:0009093 | cysteine catabolic process | 1 | 3 | 3.42e-01 |
| GO:BP | GO:0016114 | terpenoid biosynthetic process | 1 | 12 | 3.42e-01 |
| GO:BP | GO:0046755 | viral budding | 3 | 26 | 3.42e-01 |
| GO:BP | GO:0010668 | ectodermal cell differentiation | 1 | 5 | 3.42e-01 |
| GO:BP | GO:0098937 | anterograde dendritic transport | 1 | 9 | 3.42e-01 |
| GO:BP | GO:0046514 | ceramide catabolic process | 1 | 16 | 3.42e-01 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 1 | 16 | 3.42e-01 |
| GO:BP | GO:0061572 | actin filament bundle organization | 4 | 140 | 3.42e-01 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 1 | 15 | 3.43e-01 |
| GO:BP | GO:0019226 | transmission of nerve impulse | 1 | 40 | 3.43e-01 |
| GO:BP | GO:0014854 | response to inactivity | 2 | 11 | 3.43e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 4 | 706 | 3.43e-01 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 1 | 8 | 3.43e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 3 | 87 | 3.43e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 3 | 29 | 3.43e-01 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 1 | 3 | 3.43e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 3 | 33 | 3.43e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 4 | 102 | 3.43e-01 |
| GO:BP | GO:0003285 | septum secundum development | 1 | 3 | 3.43e-01 |
| GO:BP | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 1 | 7 | 3.44e-01 |
| GO:BP | GO:2000282 | regulation of cellular amino acid biosynthetic process | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0034440 | lipid oxidation | 1 | 94 | 3.44e-01 |
| GO:BP | GO:0060026 | convergent extension | 1 | 14 | 3.44e-01 |
| GO:BP | GO:0045763 | negative regulation of cellular amino acid metabolic process | 1 | 2 | 3.44e-01 |
| GO:BP | GO:0000821 | regulation of arginine metabolic process | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 13 | 422 | 3.44e-01 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 1 | 11 | 3.44e-01 |
| GO:BP | GO:0018344 | protein geranylgeranylation | 1 | 6 | 3.44e-01 |
| GO:BP | GO:0032776 | DNA methylation on cytosine | 1 | 4 | 3.44e-01 |
| GO:BP | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0140192 | regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 3 | 3.44e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 2 | 126 | 3.44e-01 |
| GO:BP | GO:0051646 | mitochondrion localization | 1 | 47 | 3.44e-01 |
| GO:BP | GO:0070193 | synaptonemal complex organization | 1 | 13 | 3.44e-01 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 3 | 28 | 3.44e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 9 | 109 | 3.44e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 3 | 41 | 3.44e-01 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 6 | 51 | 3.44e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 6 | 105 | 3.44e-01 |
| GO:BP | GO:0046530 | photoreceptor cell differentiation | 2 | 40 | 3.44e-01 |
| GO:BP | GO:0044320 | cellular response to leptin stimulus | 1 | 15 | 3.44e-01 |
| GO:BP | GO:0046753 | non-lytic viral release | 1 | 14 | 3.44e-01 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 1 | 6 | 3.45e-01 |
| GO:BP | GO:0044091 | membrane biogenesis | 3 | 58 | 3.45e-01 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 2 | 44 | 3.45e-01 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 6 | 58 | 3.45e-01 |
| GO:BP | GO:0032402 | melanosome transport | 2 | 19 | 3.45e-01 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 2 | 40 | 3.45e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 7 | 96 | 3.45e-01 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 6 | 190 | 3.46e-01 |
| GO:BP | GO:0046898 | response to cycloheximide | 1 | 4 | 3.46e-01 |
| GO:BP | GO:0071839 | apoptotic process in bone marrow cell | 1 | 4 | 3.46e-01 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 5 | 27 | 3.46e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 5 | 511 | 3.46e-01 |
| GO:BP | GO:0006692 | prostanoid metabolic process | 1 | 32 | 3.46e-01 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 3 | 40 | 3.46e-01 |
| GO:BP | GO:0006636 | unsaturated fatty acid biosynthetic process | 1 | 34 | 3.46e-01 |
| GO:BP | GO:0006693 | prostaglandin metabolic process | 1 | 32 | 3.46e-01 |
| GO:BP | GO:0003002 | regionalization | 1 | 273 | 3.47e-01 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 4 | 81 | 3.47e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 10 | 227 | 3.47e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 1 | 48 | 3.47e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 4 | 127 | 3.47e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 3 | 1147 | 3.47e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 1 | 67 | 3.47e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 1 | 12 | 3.47e-01 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 5 | 46 | 3.48e-01 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 1 | 20 | 3.48e-01 |
| GO:BP | GO:0061564 | axon development | 4 | 398 | 3.49e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 16 | 448 | 3.49e-01 |
| GO:BP | GO:1901741 | positive regulation of myoblast fusion | 1 | 9 | 3.49e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 4 | 236 | 3.49e-01 |
| GO:BP | GO:1904970 | brush border assembly | 1 | 2 | 3.50e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 3.50e-01 |
| GO:BP | GO:0042060 | wound healing | 1 | 320 | 3.50e-01 |
| GO:BP | GO:0032650 | regulation of interleukin-1 alpha production | 1 | 4 | 3.50e-01 |
| GO:BP | GO:0051873 | killing by host of symbiont cells | 1 | 5 | 3.50e-01 |
| GO:BP | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell | 1 | 13 | 3.50e-01 |
| GO:BP | GO:0034097 | response to cytokine | 8 | 607 | 3.50e-01 |
| GO:BP | GO:0032610 | interleukin-1 alpha production | 1 | 4 | 3.50e-01 |
| GO:BP | GO:0071578 | zinc ion import across plasma membrane | 1 | 6 | 3.50e-01 |
| GO:BP | GO:0006882 | intracellular zinc ion homeostasis | 3 | 28 | 3.50e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 3 | 775 | 3.50e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 2 | 21 | 3.50e-01 |
| GO:BP | GO:0035352 | NAD transmembrane transport | 1 | 3 | 3.50e-01 |
| GO:BP | GO:0018095 | protein polyglutamylation | 2 | 9 | 3.50e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 4 | 517 | 3.50e-01 |
| GO:BP | GO:0042574 | retinal metabolic process | 1 | 11 | 3.51e-01 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 1 | 45 | 3.51e-01 |
| GO:BP | GO:0042940 | D-amino acid transport | 1 | 5 | 3.51e-01 |
| GO:BP | GO:0033700 | phospholipid efflux | 1 | 9 | 3.51e-01 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 2 | 47 | 3.51e-01 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 1 | 6 | 3.52e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 9 | 203 | 3.52e-01 |
| GO:BP | GO:0006655 | phosphatidylglycerol biosynthetic process | 1 | 9 | 3.52e-01 |
| GO:BP | GO:0016578 | histone deubiquitination | 4 | 46 | 3.52e-01 |
| GO:BP | GO:0051967 | negative regulation of synaptic transmission, glutamatergic | 1 | 7 | 3.52e-01 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 3 | 51 | 3.52e-01 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 1 | 9 | 3.52e-01 |
| GO:BP | GO:0046785 | microtubule polymerization | 1 | 84 | 3.52e-01 |
| GO:BP | GO:0003012 | muscle system process | 5 | 334 | 3.53e-01 |
| GO:BP | GO:0042976 | activation of Janus kinase activity | 1 | 7 | 3.53e-01 |
| GO:BP | GO:1901722 | regulation of cell proliferation involved in kidney development | 1 | 11 | 3.53e-01 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 1 | 28 | 3.53e-01 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 7 | 82 | 3.53e-01 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 1 | 8 | 3.53e-01 |
| GO:BP | GO:0022008 | neurogenesis | 3 | 1294 | 3.53e-01 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 3 | 44 | 3.53e-01 |
| GO:BP | GO:0009071 | serine family amino acid catabolic process | 2 | 10 | 3.53e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 1 | 428 | 3.53e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 6 | 115 | 3.53e-01 |
| GO:BP | GO:0030263 | apoptotic chromosome condensation | 1 | 4 | 3.53e-01 |
| GO:BP | GO:0019626 | short-chain fatty acid catabolic process | 1 | 6 | 3.53e-01 |
| GO:BP | GO:0060847 | endothelial cell fate specification | 1 | 3 | 3.54e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 12 | 414 | 3.54e-01 |
| GO:BP | GO:0043112 | receptor metabolic process | 1 | 62 | 3.54e-01 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 3 | 49 | 3.54e-01 |
| GO:BP | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching | 1 | 4 | 3.54e-01 |
| GO:BP | GO:1904948 | midbrain dopaminergic neuron differentiation | 1 | 12 | 3.54e-01 |
| GO:BP | GO:0016198 | axon choice point recognition | 1 | 5 | 3.54e-01 |
| GO:BP | GO:1902669 | positive regulation of axon guidance | 1 | 3 | 3.54e-01 |
| GO:BP | GO:0038189 | neuropilin signaling pathway | 1 | 4 | 3.54e-01 |
| GO:BP | GO:0060300 | regulation of cytokine activity | 1 | 3 | 3.54e-01 |
| GO:BP | GO:0000348 | mRNA branch site recognition | 1 | 1 | 3.54e-01 |
| GO:BP | GO:2000209 | regulation of anoikis | 1 | 23 | 3.54e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 2 | 67 | 3.54e-01 |
| GO:BP | GO:0006699 | bile acid biosynthetic process | 1 | 25 | 3.54e-01 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 2 | 85 | 3.54e-01 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 1 | 17 | 3.55e-01 |
| GO:BP | GO:0001947 | heart looping | 2 | 50 | 3.55e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 14 | 486 | 3.56e-01 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 1 | 3 | 3.56e-01 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 1 | 19 | 3.56e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 1 | 36 | 3.56e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 3.56e-01 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 1 | 3 | 3.56e-01 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 16 | 3.56e-01 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 2 | 34 | 3.56e-01 |
| GO:BP | GO:0061437 | renal system vasculature development | 3 | 24 | 3.56e-01 |
| GO:BP | GO:0061440 | kidney vasculature development | 3 | 24 | 3.56e-01 |
| GO:BP | GO:1902426 | deactivation of mitotic spindle assembly checkpoint | 1 | 3 | 3.56e-01 |
| GO:BP | GO:0061085 | regulation of histone H3-K27 methylation | 1 | 4 | 3.56e-01 |
| GO:BP | GO:1902884 | positive regulation of response to oxidative stress | 1 | 9 | 3.56e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 3 | 1278 | 3.56e-01 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 3 | 37 | 3.56e-01 |
| GO:BP | GO:0052646 | alditol phosphate metabolic process | 1 | 10 | 3.56e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 3.57e-01 |
| GO:BP | GO:0070086 | ubiquitin-dependent endocytosis | 1 | 4 | 3.57e-01 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 2 | 15 | 3.57e-01 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 5 | 37 | 3.57e-01 |
| GO:BP | GO:0030157 | pancreatic juice secretion | 1 | 11 | 3.57e-01 |
| GO:BP | GO:0035865 | cellular response to potassium ion | 1 | 9 | 3.57e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 1 | 189 | 3.57e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 9 | 340 | 3.58e-01 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 1 | 13 | 3.58e-01 |
| GO:BP | GO:0034122 | negative regulation of toll-like receptor signaling pathway | 1 | 13 | 3.58e-01 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 3 | 27 | 3.58e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 3.58e-01 |
| GO:BP | GO:0051904 | pigment granule transport | 2 | 19 | 3.58e-01 |
| GO:BP | GO:0021542 | dentate gyrus development | 1 | 11 | 3.58e-01 |
| GO:BP | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 1 | 5 | 3.58e-01 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 1 | 10 | 3.58e-01 |
| GO:BP | GO:0021772 | olfactory bulb development | 2 | 19 | 3.58e-01 |
| GO:BP | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | 2 | 23 | 3.58e-01 |
| GO:BP | GO:2000373 | positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity | 1 | 5 | 3.58e-01 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 2 | 20 | 3.58e-01 |
| GO:BP | GO:0048865 | stem cell fate commitment | 1 | 5 | 3.58e-01 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 2 | 22 | 3.58e-01 |
| GO:BP | GO:1903373 | positive regulation of endoplasmic reticulum tubular network organization | 1 | 4 | 3.58e-01 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 2 | 12 | 3.58e-01 |
| GO:BP | GO:0048255 | mRNA stabilization | 1 | 50 | 3.59e-01 |
| GO:BP | GO:1901160 | primary amino compound metabolic process | 1 | 8 | 3.59e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 6 | 110 | 3.59e-01 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 1 | 53 | 3.59e-01 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 2 | 23 | 3.59e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 2 | 19 | 3.59e-01 |
| GO:BP | GO:2001267 | regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | 1 | 13 | 3.59e-01 |
| GO:BP | GO:0031929 | TOR signaling | 1 | 115 | 3.59e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 3.59e-01 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 1 | 16 | 3.59e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 2 | 82 | 3.59e-01 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 2 | 10 | 3.59e-01 |
| GO:BP | GO:0008213 | protein alkylation | 4 | 160 | 3.59e-01 |
| GO:BP | GO:0006479 | protein methylation | 4 | 160 | 3.59e-01 |
| GO:BP | GO:0060046 | regulation of acrosome reaction | 1 | 8 | 3.59e-01 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 6 | 50 | 3.59e-01 |
| GO:BP | GO:1901897 | regulation of relaxation of cardiac muscle | 1 | 5 | 3.59e-01 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 1 | 26 | 3.59e-01 |
| GO:BP | GO:0000045 | autophagosome assembly | 2 | 103 | 3.59e-01 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 3 | 137 | 3.60e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 2 | 49 | 3.60e-01 |
| GO:BP | GO:0099111 | microtubule-based transport | 4 | 191 | 3.61e-01 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 1 | 53 | 3.61e-01 |
| GO:BP | GO:0006000 | fructose metabolic process | 2 | 10 | 3.61e-01 |
| GO:BP | GO:0043984 | histone H4-K16 acetylation | 2 | 14 | 3.61e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 2 | 34 | 3.61e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 3.61e-01 |
| GO:BP | GO:0048525 | negative regulation of viral process | 2 | 64 | 3.61e-01 |
| GO:BP | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation | 1 | 13 | 3.61e-01 |
| GO:BP | GO:0090215 | regulation of 1-phosphatidylinositol-4-phosphate 5-kinase activity | 1 | 4 | 3.62e-01 |
| GO:BP | GO:0051648 | vesicle localization | 4 | 192 | 3.62e-01 |
| GO:BP | GO:0072049 | comma-shaped body morphogenesis | 1 | 4 | 3.62e-01 |
| GO:BP | GO:1902869 | regulation of amacrine cell differentiation | 1 | 3 | 3.62e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 3.62e-01 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 2 | 32 | 3.63e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 3 | 179 | 3.63e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 3.63e-01 |
| GO:BP | GO:0071787 | endoplasmic reticulum tubular network formation | 1 | 4 | 3.63e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 1 | 9 | 3.63e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 2 | 28 | 3.63e-01 |
| GO:BP | GO:0010919 | regulation of inositol phosphate biosynthetic process | 1 | 6 | 3.63e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 2 | 36 | 3.63e-01 |
| GO:BP | GO:0007389 | pattern specification process | 1 | 306 | 3.63e-01 |
| GO:BP | GO:0009143 | nucleoside triphosphate catabolic process | 1 | 10 | 3.63e-01 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 2 | 12 | 3.63e-01 |
| GO:BP | GO:0016264 | gap junction assembly | 2 | 14 | 3.63e-01 |
| GO:BP | GO:2000465 | regulation of glycogen (starch) synthase activity | 1 | 8 | 3.64e-01 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 2 | 15 | 3.64e-01 |
| GO:BP | GO:0003097 | renal water transport | 1 | 6 | 3.64e-01 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 6 | 196 | 3.65e-01 |
| GO:BP | GO:0042748 | circadian sleep/wake cycle, non-REM sleep | 1 | 3 | 3.65e-01 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 1 | 105 | 3.65e-01 |
| GO:BP | GO:0062100 | positive regulation of programmed necrotic cell death | 1 | 5 | 3.65e-01 |
| GO:BP | GO:0050890 | cognition | 6 | 227 | 3.65e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 6 | 608 | 3.65e-01 |
| GO:BP | GO:0051932 | synaptic transmission, GABAergic | 1 | 36 | 3.65e-01 |
| GO:BP | GO:0032077 | positive regulation of deoxyribonuclease activity | 1 | 5 | 3.65e-01 |
| GO:BP | GO:0032148 | activation of protein kinase B activity | 2 | 23 | 3.65e-01 |
| GO:BP | GO:0061647 | histone H3-K9 modification | 3 | 23 | 3.65e-01 |
| GO:BP | GO:0045933 | positive regulation of muscle contraction | 1 | 32 | 3.65e-01 |
| GO:BP | GO:0071681 | cellular response to indole-3-methanol | 1 | 5 | 3.65e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 3.65e-01 |
| GO:BP | GO:0048208 | COPII vesicle coating | 1 | 24 | 3.65e-01 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 1 | 24 | 3.65e-01 |
| GO:BP | GO:0055002 | striated muscle cell development | 2 | 67 | 3.65e-01 |
| GO:BP | GO:0090398 | cellular senescence | 3 | 83 | 3.66e-01 |
| GO:BP | GO:0046104 | thymidine metabolic process | 1 | 5 | 3.66e-01 |
| GO:BP | GO:0009164 | nucleoside catabolic process | 1 | 15 | 3.66e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 4 | 194 | 3.66e-01 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 1 | 5 | 3.66e-01 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 1 | 20 | 3.66e-01 |
| GO:BP | GO:0009186 | deoxyribonucleoside diphosphate metabolic process | 1 | 4 | 3.66e-01 |
| GO:BP | GO:0003174 | mitral valve development | 1 | 12 | 3.66e-01 |
| GO:BP | GO:0016080 | synaptic vesicle targeting | 1 | 3 | 3.66e-01 |
| GO:BP | GO:0015858 | nucleoside transport | 1 | 8 | 3.66e-01 |
| GO:BP | GO:0006863 | purine nucleobase transport | 1 | 7 | 3.66e-01 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 1 | 104 | 3.66e-01 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 1 | 4 | 3.66e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 3 | 39 | 3.67e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 2 | 90 | 3.67e-01 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 3 | 64 | 3.68e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 4 | 134 | 3.69e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 5 | 357 | 3.69e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 3.69e-01 |
| GO:BP | GO:0048675 | axon extension | 10 | 110 | 3.69e-01 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 5 | 44 | 3.69e-01 |
| GO:BP | GO:0002941 | synoviocyte proliferation | 1 | 3 | 3.69e-01 |
| GO:BP | GO:1901647 | positive regulation of synoviocyte proliferation | 1 | 3 | 3.69e-01 |
| GO:BP | GO:1901645 | regulation of synoviocyte proliferation | 1 | 3 | 3.69e-01 |
| GO:BP | GO:0002190 | cap-independent translational initiation | 1 | 4 | 3.69e-01 |
| GO:BP | GO:0002690 | positive regulation of leukocyte chemotaxis | 3 | 51 | 3.69e-01 |
| GO:BP | GO:0097167 | circadian regulation of translation | 1 | 4 | 3.69e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 1 | 761 | 3.69e-01 |
| GO:BP | GO:0036166 | phenotypic switching | 1 | 7 | 3.69e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 29 | 1149 | 3.69e-01 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 1 | 166 | 3.69e-01 |
| GO:BP | GO:0006560 | proline metabolic process | 1 | 7 | 3.69e-01 |
| GO:BP | GO:0006721 | terpenoid metabolic process | 4 | 50 | 3.69e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 13 | 551 | 3.69e-01 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 1 | 6 | 3.70e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 3.70e-01 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 1 | 57 | 3.70e-01 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 2 | 20 | 3.70e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 2 | 33 | 3.70e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 2 | 63 | 3.70e-01 |
| GO:BP | GO:0032098 | regulation of appetite | 1 | 8 | 3.70e-01 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 1 | 12 | 3.70e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 3 | 1974 | 3.71e-01 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 1 | 44 | 3.71e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 9 | 563 | 3.72e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 5 | 128 | 3.72e-01 |
| GO:BP | GO:0060039 | pericardium development | 1 | 17 | 3.73e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 3.73e-01 |
| GO:BP | GO:0060602 | branch elongation of an epithelium | 1 | 14 | 3.73e-01 |
| GO:BP | GO:1904862 | inhibitory synapse assembly | 1 | 13 | 3.73e-01 |
| GO:BP | GO:0030540 | female genitalia development | 1 | 14 | 3.73e-01 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 7 | 62 | 3.73e-01 |
| GO:BP | GO:0048645 | animal organ formation | 2 | 51 | 3.73e-01 |
| GO:BP | GO:1990504 | dense core granule exocytosis | 1 | 7 | 3.73e-01 |
| GO:BP | GO:0036149 | phosphatidylinositol acyl-chain remodeling | 1 | 3 | 3.73e-01 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 2 | 63 | 3.74e-01 |
| GO:BP | GO:0080144 | intracellular amino acid homeostasis | 1 | 7 | 3.74e-01 |
| GO:BP | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation | 1 | 3 | 3.74e-01 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 1 | 6 | 3.74e-01 |
| GO:BP | GO:0045087 | innate immune response | 5 | 488 | 3.74e-01 |
| GO:BP | GO:0035627 | ceramide transport | 1 | 15 | 3.74e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 14 | 660 | 3.74e-01 |
| GO:BP | GO:0030952 | establishment or maintenance of cytoskeleton polarity | 1 | 7 | 3.74e-01 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 3 | 90 | 3.74e-01 |
| GO:BP | GO:0048484 | enteric nervous system development | 2 | 7 | 3.74e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 3.74e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 3 | 36 | 3.74e-01 |
| GO:BP | GO:0010996 | response to auditory stimulus | 1 | 16 | 3.74e-01 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 4 | 41 | 3.74e-01 |
| GO:BP | GO:0014902 | myotube differentiation | 3 | 88 | 3.74e-01 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 8 | 72 | 3.74e-01 |
| GO:BP | GO:0021988 | olfactory lobe development | 2 | 21 | 3.74e-01 |
| GO:BP | GO:0048524 | positive regulation of viral process | 1 | 55 | 3.74e-01 |
| GO:BP | GO:0006720 | isoprenoid metabolic process | 4 | 85 | 3.75e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 2 | 17 | 3.75e-01 |
| GO:BP | GO:0019262 | N-acetylneuraminate catabolic process | 1 | 6 | 3.75e-01 |
| GO:BP | GO:0032532 | regulation of microvillus length | 1 | 4 | 3.75e-01 |
| GO:BP | GO:0015740 | C4-dicarboxylate transport | 2 | 24 | 3.75e-01 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 1 | 52 | 3.75e-01 |
| GO:BP | GO:0060059 | embryonic retina morphogenesis in camera-type eye | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 3 | 67 | 3.76e-01 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 4 | 40 | 3.76e-01 |
| GO:BP | GO:0097104 | postsynaptic membrane assembly | 1 | 8 | 3.76e-01 |
| GO:BP | GO:0042438 | melanin biosynthetic process | 2 | 17 | 3.76e-01 |
| GO:BP | GO:0006572 | tyrosine catabolic process | 1 | 2 | 3.76e-01 |
| GO:BP | GO:0045875 | negative regulation of sister chromatid cohesion | 1 | 5 | 3.76e-01 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 5 | 63 | 3.76e-01 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 2 | 39 | 3.77e-01 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 3 | 37 | 3.77e-01 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 2 | 25 | 3.77e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 11 | 167 | 3.78e-01 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 2 | 14 | 3.78e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 3 | 23 | 3.78e-01 |
| GO:BP | GO:1903533 | regulation of protein targeting | 4 | 66 | 3.78e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 4 | 230 | 3.78e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 5 | 362 | 3.78e-01 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 1 | 20 | 3.78e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 3 | 25 | 3.78e-01 |
| GO:BP | GO:1901403 | positive regulation of tetrapyrrole metabolic process | 1 | 4 | 3.78e-01 |
| GO:BP | GO:1905037 | autophagosome organization | 2 | 110 | 3.79e-01 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 8 | 124 | 3.79e-01 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 1 | 97 | 3.79e-01 |
| GO:BP | GO:0043473 | pigmentation | 6 | 92 | 3.79e-01 |
| GO:BP | GO:0044321 | response to leptin | 1 | 19 | 3.79e-01 |
| GO:BP | GO:0050923 | regulation of negative chemotaxis | 1 | 4 | 3.79e-01 |
| GO:BP | GO:0021615 | glossopharyngeal nerve morphogenesis | 1 | 2 | 3.79e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 4 | 113 | 3.79e-01 |
| GO:BP | GO:0048468 | cell development | 3 | 1963 | 3.79e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 6 | 115 | 3.79e-01 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 2 | 16 | 3.79e-01 |
| GO:BP | GO:0001767 | establishment of lymphocyte polarity | 2 | 11 | 3.79e-01 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 2 | 15 | 3.79e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 3 | 303 | 3.80e-01 |
| GO:BP | GO:0051481 | negative regulation of cytosolic calcium ion concentration | 1 | 9 | 3.80e-01 |
| GO:BP | GO:0010528 | regulation of transposition | 1 | 14 | 3.80e-01 |
| GO:BP | GO:0032868 | response to insulin | 2 | 215 | 3.80e-01 |
| GO:BP | GO:0008065 | establishment of blood-nerve barrier | 1 | 3 | 3.80e-01 |
| GO:BP | GO:0010529 | negative regulation of transposition | 1 | 14 | 3.80e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 9 | 97 | 3.80e-01 |
| GO:BP | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion | 1 | 4 | 3.80e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 4 | 30 | 3.80e-01 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 2 | 11 | 3.81e-01 |
| GO:BP | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway | 1 | 4 | 3.81e-01 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 2 | 93 | 3.81e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 1 | 86 | 3.81e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 2 | 18 | 3.81e-01 |
| GO:BP | GO:0035635 | entry of bacterium into host cell | 1 | 8 | 3.81e-01 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 1 | 9 | 3.81e-01 |
| GO:BP | GO:0006740 | NADPH regeneration | 1 | 22 | 3.81e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 3.82e-01 |
| GO:BP | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 1 | 7 | 3.82e-01 |
| GO:BP | GO:0036342 | post-anal tail morphogenesis | 1 | 13 | 3.82e-01 |
| GO:BP | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | 1 | 17 | 3.82e-01 |
| GO:BP | GO:0021682 | nerve maturation | 1 | 2 | 3.82e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 3.82e-01 |
| GO:BP | GO:0006498 | N-terminal protein lipidation | 1 | 7 | 3.82e-01 |
| GO:BP | GO:0051450 | myoblast proliferation | 1 | 17 | 3.82e-01 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 4 | 145 | 3.82e-01 |
| GO:BP | GO:0090038 | negative regulation of protein kinase C signaling | 1 | 4 | 3.82e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 17 | 427 | 3.82e-01 |
| GO:BP | GO:0043489 | RNA stabilization | 1 | 59 | 3.82e-01 |
| GO:BP | GO:1903020 | positive regulation of glycoprotein metabolic process | 2 | 17 | 3.82e-01 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 1 | 30 | 3.82e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 3.82e-01 |
| GO:BP | GO:0032400 | melanosome localization | 2 | 22 | 3.82e-01 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 11 | 113 | 3.83e-01 |
| GO:BP | GO:0060337 | type I interferon-mediated signaling pathway | 1 | 51 | 3.83e-01 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 3 | 27 | 3.83e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 15 | 421 | 3.83e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 5 | 129 | 3.83e-01 |
| GO:BP | GO:0007032 | endosome organization | 8 | 92 | 3.83e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 2 | 22 | 3.83e-01 |
| GO:BP | GO:0042759 | long-chain fatty acid biosynthetic process | 1 | 14 | 3.83e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 11 | 300 | 3.83e-01 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 1 | 14 | 3.83e-01 |
| GO:BP | GO:0071514 | genomic imprinting | 1 | 9 | 3.83e-01 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 2 | 17 | 3.83e-01 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 2 | 18 | 3.83e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 4 | 123 | 3.83e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 3 | 86 | 3.83e-01 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 5 | 64 | 3.83e-01 |
| GO:BP | GO:0043129 | surfactant homeostasis | 1 | 12 | 3.83e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 3 | 36 | 3.84e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 19 | 1849 | 3.84e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 3 | 30 | 3.84e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 3 | 30 | 3.84e-01 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 1 | 12 | 3.84e-01 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 1 | 13 | 3.84e-01 |
| GO:BP | GO:0072674 | multinuclear osteoclast differentiation | 1 | 6 | 3.84e-01 |
| GO:BP | GO:0032835 | glomerulus development | 4 | 54 | 3.84e-01 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 1 | 57 | 3.84e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 7 | 69 | 3.84e-01 |
| GO:BP | GO:0015851 | nucleobase transport | 1 | 7 | 3.85e-01 |
| GO:BP | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response | 1 | 3 | 3.85e-01 |
| GO:BP | GO:0003360 | brainstem development | 1 | 7 | 3.85e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 3.85e-01 |
| GO:BP | GO:0032696 | negative regulation of interleukin-13 production | 1 | 3 | 3.85e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 3.85e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 3.86e-01 |
| GO:BP | GO:0001755 | neural crest cell migration | 5 | 49 | 3.86e-01 |
| GO:BP | GO:0021978 | telencephalon regionalization | 1 | 4 | 3.86e-01 |
| GO:BP | GO:0090022 | regulation of neutrophil chemotaxis | 1 | 19 | 3.86e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 6 | 893 | 3.86e-01 |
| GO:BP | GO:2000491 | positive regulation of hepatic stellate cell activation | 1 | 4 | 3.86e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 15 | 375 | 3.86e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 2 | 107 | 3.86e-01 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 2 | 27 | 3.86e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 3.86e-01 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 2 | 59 | 3.86e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 3.86e-01 |
| GO:BP | GO:0051641 | cellular localization | 49 | 2810 | 3.86e-01 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 1 | 5 | 3.86e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 5 | 142 | 3.86e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 3 | 41 | 3.86e-01 |
| GO:BP | GO:0097752 | regulation of DNA stability | 1 | 3 | 3.86e-01 |
| GO:BP | GO:0001501 | skeletal system development | 1 | 377 | 3.86e-01 |
| GO:BP | GO:0071357 | cellular response to type I interferon | 1 | 52 | 3.86e-01 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 2 | 21 | 3.87e-01 |
| GO:BP | GO:0036150 | phosphatidylserine acyl-chain remodeling | 1 | 7 | 3.87e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 2 | 1399 | 3.87e-01 |
| GO:BP | GO:0060965 | negative regulation of miRNA-mediated gene silencing | 2 | 10 | 3.87e-01 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 1 | 10 | 3.87e-01 |
| GO:BP | GO:0006582 | melanin metabolic process | 2 | 18 | 3.87e-01 |
| GO:BP | GO:0043132 | NAD transport | 1 | 3 | 3.87e-01 |
| GO:BP | GO:0051901 | positive regulation of mitochondrial depolarization | 1 | 6 | 3.87e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 3 | 97 | 3.88e-01 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 3 | 97 | 3.88e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 18 | 198 | 3.88e-01 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 2 | 86 | 3.89e-01 |
| GO:BP | GO:0030917 | midbrain-hindbrain boundary development | 1 | 3 | 3.89e-01 |
| GO:BP | GO:0021861 | forebrain radial glial cell differentiation | 1 | 6 | 3.89e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 2 | 18 | 3.89e-01 |
| GO:BP | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | 1 | 9 | 3.89e-01 |
| GO:BP | GO:0090131 | mesenchyme migration | 1 | 4 | 3.90e-01 |
| GO:BP | GO:0060976 | coronary vasculature development | 1 | 44 | 3.90e-01 |
| GO:BP | GO:0045141 | meiotic telomere clustering | 1 | 5 | 3.90e-01 |
| GO:BP | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin | 1 | 16 | 3.90e-01 |
| GO:BP | GO:0034089 | establishment of meiotic sister chromatid cohesion | 1 | 4 | 3.90e-01 |
| GO:BP | GO:0000729 | DNA double-strand break processing | 2 | 18 | 3.90e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 5 | 88 | 3.90e-01 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 2 | 26 | 3.90e-01 |
| GO:BP | GO:1990036 | calcium ion import into sarcoplasmic reticulum | 1 | 4 | 3.90e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 15 | 376 | 3.90e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 1 | 55 | 3.90e-01 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 1 | 15 | 3.90e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 3 | 94 | 3.90e-01 |
| GO:BP | GO:0031062 | positive regulation of histone methylation | 4 | 36 | 3.90e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 35 | 534 | 3.90e-01 |
| GO:BP | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 4 | 3.90e-01 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 5 | 24 | 3.90e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 1 | 221 | 3.90e-01 |
| GO:BP | GO:0042402 | cellular biogenic amine catabolic process | 1 | 13 | 3.90e-01 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 1 | 33 | 3.90e-01 |
| GO:BP | GO:0072073 | kidney epithelium development | 3 | 112 | 3.91e-01 |
| GO:BP | GO:0072359 | circulatory system development | 9 | 884 | 3.91e-01 |
| GO:BP | GO:0051414 | response to cortisol | 1 | 5 | 3.91e-01 |
| GO:BP | GO:0002446 | neutrophil mediated immunity | 1 | 17 | 3.91e-01 |
| GO:BP | GO:0035385 | Roundabout signaling pathway | 1 | 5 | 3.91e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 3.91e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 3.91e-01 |
| GO:BP | GO:0051707 | response to other organism | 11 | 820 | 3.91e-01 |
| GO:BP | GO:0042078 | germ-line stem cell division | 1 | 2 | 3.92e-01 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 1 | 2 | 3.92e-01 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 1 | 2 | 3.92e-01 |
| GO:BP | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis | 1 | 5 | 3.92e-01 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 2 | 21 | 3.92e-01 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 1 | 50 | 3.92e-01 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 1 | 8 | 3.92e-01 |
| GO:BP | GO:0030502 | negative regulation of bone mineralization | 1 | 10 | 3.92e-01 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 3 | 33 | 3.92e-01 |
| GO:BP | GO:0043562 | cellular response to nitrogen levels | 1 | 9 | 3.92e-01 |
| GO:BP | GO:0006995 | cellular response to nitrogen starvation | 1 | 9 | 3.92e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 7 | 72 | 3.92e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 2 | 46 | 3.92e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 11 | 820 | 3.93e-01 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 8 | 190 | 3.93e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 2 | 170 | 3.93e-01 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 2 | 14 | 3.93e-01 |
| GO:BP | GO:0051875 | pigment granule localization | 2 | 22 | 3.93e-01 |
| GO:BP | GO:0036109 | alpha-linolenic acid metabolic process | 1 | 9 | 3.93e-01 |
| GO:BP | GO:0046073 | dTMP metabolic process | 1 | 7 | 3.94e-01 |
| GO:BP | GO:0006545 | glycine biosynthetic process | 1 | 4 | 3.94e-01 |
| GO:BP | GO:0001975 | response to amphetamine | 2 | 22 | 3.94e-01 |
| GO:BP | GO:0001771 | immunological synapse formation | 1 | 7 | 3.94e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 3 | 90 | 3.94e-01 |
| GO:BP | GO:0055088 | lipid homeostasis | 1 | 115 | 3.94e-01 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 3 | 18 | 3.95e-01 |
| GO:BP | GO:1905936 | regulation of germ cell proliferation | 1 | 5 | 3.95e-01 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 9 | 103 | 3.95e-01 |
| GO:BP | GO:1904994 | regulation of leukocyte adhesion to vascular endothelial cell | 1 | 16 | 3.95e-01 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 1 | 12 | 3.95e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 10 | 197 | 3.95e-01 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 2 | 18 | 3.95e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 1 | 414 | 3.96e-01 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 3 | 19 | 3.96e-01 |
| GO:BP | GO:0055093 | response to hyperoxia | 1 | 18 | 3.96e-01 |
| GO:BP | GO:0006007 | glucose catabolic process | 1 | 22 | 3.96e-01 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 3 | 25 | 3.96e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 3.96e-01 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 5 | 114 | 3.96e-01 |
| GO:BP | GO:0006824 | cobalt ion transport | 1 | 5 | 3.97e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 1 | 272 | 3.97e-01 |
| GO:BP | GO:0072028 | nephron morphogenesis | 2 | 62 | 3.97e-01 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 2 | 11 | 3.97e-01 |
| GO:BP | GO:0051573 | negative regulation of histone H3-K9 methylation | 1 | 5 | 3.97e-01 |
| GO:BP | GO:0071680 | response to indole-3-methanol | 1 | 6 | 3.97e-01 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 2 | 16 | 3.97e-01 |
| GO:BP | GO:0031065 | positive regulation of histone deacetylation | 1 | 22 | 3.97e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 3 | 154 | 3.97e-01 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 2 | 60 | 3.97e-01 |
| GO:BP | GO:1903018 | regulation of glycoprotein metabolic process | 3 | 36 | 3.97e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 3 | 1898 | 3.97e-01 |
| GO:BP | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway | 1 | 20 | 3.98e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 15 | 378 | 3.98e-01 |
| GO:BP | GO:0034656 | nucleobase-containing small molecule catabolic process | 1 | 20 | 3.98e-01 |
| GO:BP | GO:0071492 | cellular response to UV-A | 1 | 8 | 3.98e-01 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 1 | 10 | 3.98e-01 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 1 | 10 | 3.98e-01 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 1 | 60 | 3.98e-01 |
| GO:BP | GO:0035270 | endocrine system development | 5 | 86 | 3.98e-01 |
| GO:BP | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process | 1 | 5 | 3.98e-01 |
| GO:BP | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process | 1 | 6 | 3.98e-01 |
| GO:BP | GO:2000108 | positive regulation of leukocyte apoptotic process | 2 | 18 | 3.98e-01 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 3 | 118 | 3.98e-01 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 2 | 88 | 3.99e-01 |
| GO:BP | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 9 | 3.99e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 3 | 73 | 3.99e-01 |
| GO:BP | GO:0044550 | secondary metabolite biosynthetic process | 2 | 18 | 3.99e-01 |
| GO:BP | GO:0097352 | autophagosome maturation | 4 | 55 | 4.00e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 27 | 324 | 4.00e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 4 | 116 | 4.00e-01 |
| GO:BP | GO:0034340 | response to type I interferon | 1 | 57 | 4.00e-01 |
| GO:BP | GO:0007276 | gamete generation | 2 | 461 | 4.00e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 2 | 433 | 4.00e-01 |
| GO:BP | GO:0009310 | amine catabolic process | 1 | 14 | 4.01e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 2 | 1199 | 4.01e-01 |
| GO:BP | GO:0044571 | [2Fe-2S] cluster assembly | 1 | 11 | 4.01e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 1 | 783 | 4.02e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 1 | 372 | 4.02e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 4 | 265 | 4.02e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 8 | 120 | 4.02e-01 |
| GO:BP | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 1 | 16 | 4.02e-01 |
| GO:BP | GO:0060931 | sinoatrial node cell development | 1 | 4 | 4.02e-01 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 1 | 9 | 4.02e-01 |
| GO:BP | GO:0051235 | maintenance of location | 16 | 260 | 4.02e-01 |
| GO:BP | GO:0061548 | ganglion development | 2 | 12 | 4.02e-01 |
| GO:BP | GO:0060972 | left/right pattern formation | 3 | 101 | 4.03e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 7 | 155 | 4.03e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 4 | 130 | 4.03e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 2 | 30 | 4.03e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 4.03e-01 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 1 | 12 | 4.03e-01 |
| GO:BP | GO:0010561 | negative regulation of glycoprotein biosynthetic process | 1 | 10 | 4.03e-01 |
| GO:BP | GO:0071871 | response to epinephrine | 2 | 12 | 4.03e-01 |
| GO:BP | GO:0031915 | positive regulation of synaptic plasticity | 1 | 6 | 4.03e-01 |
| GO:BP | GO:0086103 | G protein-coupled receptor signaling pathway involved in heart process | 2 | 13 | 4.03e-01 |
| GO:BP | GO:0017014 | protein nitrosylation | 2 | 11 | 4.03e-01 |
| GO:BP | GO:0018119 | peptidyl-cysteine S-nitrosylation | 2 | 11 | 4.03e-01 |
| GO:BP | GO:0006959 | humoral immune response | 2 | 80 | 4.03e-01 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 3 | 37 | 4.03e-01 |
| GO:BP | GO:0042116 | macrophage activation | 3 | 57 | 4.03e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 3 | 197 | 4.03e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 3 | 41 | 4.03e-01 |
| GO:BP | GO:0036123 | histone H3-K9 dimethylation | 1 | 3 | 4.03e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 4.03e-01 |
| GO:BP | GO:0021536 | diencephalon development | 2 | 41 | 4.03e-01 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 2 | 19 | 4.04e-01 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 1 | 9 | 4.04e-01 |
| GO:BP | GO:0060457 | negative regulation of digestive system process | 1 | 7 | 4.04e-01 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 1 | 8 | 4.04e-01 |
| GO:BP | GO:0110156 | methylguanosine-cap decapping | 1 | 14 | 4.04e-01 |
| GO:BP | GO:0001783 | B cell apoptotic process | 2 | 21 | 4.04e-01 |
| GO:BP | GO:0032196 | transposition | 1 | 17 | 4.04e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 30 | 486 | 4.04e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 4 | 69 | 4.04e-01 |
| GO:BP | GO:1990868 | response to chemokine | 3 | 37 | 4.04e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 3 | 37 | 4.04e-01 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 2 | 31 | 4.04e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 2 | 43 | 4.04e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 4 | 61 | 4.04e-01 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 2 | 30 | 4.05e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 2 | 38 | 4.05e-01 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 1 | 12 | 4.05e-01 |
| GO:BP | GO:0045652 | regulation of megakaryocyte differentiation | 2 | 33 | 4.07e-01 |
| GO:BP | GO:0032069 | regulation of nuclease activity | 2 | 17 | 4.07e-01 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 1 | 21 | 4.07e-01 |
| GO:BP | GO:0090220 | chromosome localization to nuclear envelope involved in homologous chromosome segregation | 1 | 5 | 4.07e-01 |
| GO:BP | GO:0034397 | telomere localization | 1 | 5 | 4.07e-01 |
| GO:BP | GO:0060967 | negative regulation of gene silencing by RNA | 2 | 11 | 4.07e-01 |
| GO:BP | GO:0043252 | sodium-independent organic anion transport | 1 | 4 | 4.07e-01 |
| GO:BP | GO:0060149 | negative regulation of post-transcriptional gene silencing | 2 | 11 | 4.07e-01 |
| GO:BP | GO:0051026 | chiasma assembly | 1 | 3 | 4.07e-01 |
| GO:BP | GO:0032717 | negative regulation of interleukin-8 production | 1 | 12 | 4.07e-01 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 1 | 20 | 4.07e-01 |
| GO:BP | GO:1900369 | negative regulation of post-transcriptional gene silencing by RNA | 2 | 11 | 4.07e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 3 | 37 | 4.07e-01 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 1 | 31 | 4.08e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 34 | 569 | 4.08e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 4 | 155 | 4.08e-01 |
| GO:BP | GO:0008206 | bile acid metabolic process | 1 | 34 | 4.08e-01 |
| GO:BP | GO:0042744 | hydrogen peroxide catabolic process | 1 | 12 | 4.09e-01 |
| GO:BP | GO:0042159 | lipoprotein catabolic process | 1 | 14 | 4.09e-01 |
| GO:BP | GO:0045541 | negative regulation of cholesterol biosynthetic process | 1 | 5 | 4.09e-01 |
| GO:BP | GO:0090206 | negative regulation of cholesterol metabolic process | 1 | 5 | 4.09e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 4 | 40 | 4.09e-01 |
| GO:BP | GO:0106119 | negative regulation of sterol biosynthetic process | 1 | 5 | 4.09e-01 |
| GO:BP | GO:0002891 | positive regulation of immunoglobulin mediated immune response | 4 | 26 | 4.09e-01 |
| GO:BP | GO:0002714 | positive regulation of B cell mediated immunity | 4 | 26 | 4.09e-01 |
| GO:BP | GO:1903376 | regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | 1 | 8 | 4.10e-01 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 8 | 4.10e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 2 | 40 | 4.10e-01 |
| GO:BP | GO:0032714 | negative regulation of interleukin-5 production | 1 | 2 | 4.10e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 2 | 1075 | 4.10e-01 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 2 | 14 | 4.10e-01 |
| GO:BP | GO:0032733 | positive regulation of interleukin-10 production | 1 | 16 | 4.11e-01 |
| GO:BP | GO:0030258 | lipid modification | 2 | 162 | 4.11e-01 |
| GO:BP | GO:0043276 | anoikis | 1 | 32 | 4.11e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 6 | 1006 | 4.12e-01 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 1 | 14 | 4.12e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 4.12e-01 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 1 | 42 | 4.12e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 21 | 272 | 4.12e-01 |
| GO:BP | GO:0031648 | protein destabilization | 3 | 45 | 4.12e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 1 | 55 | 4.12e-01 |
| GO:BP | GO:0031641 | regulation of myelination | 3 | 36 | 4.13e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 7 | 850 | 4.14e-01 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 1 | 69 | 4.14e-01 |
| GO:BP | GO:0006691 | leukotriene metabolic process | 1 | 12 | 4.14e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 5 | 116 | 4.14e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 4.14e-01 |
| GO:BP | GO:0010559 | regulation of glycoprotein biosynthetic process | 2 | 32 | 4.14e-01 |
| GO:BP | GO:2000010 | positive regulation of protein localization to cell surface | 1 | 16 | 4.14e-01 |
| GO:BP | GO:0072050 | S-shaped body morphogenesis | 1 | 5 | 4.14e-01 |
| GO:BP | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | 3 | 21 | 4.14e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 1 | 104 | 4.14e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 10 | 212 | 4.14e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 3 | 39 | 4.14e-01 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 3 | 19 | 4.15e-01 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 3 | 102 | 4.15e-01 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 1 | 35 | 4.15e-01 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 2 | 63 | 4.15e-01 |
| GO:BP | GO:1902358 | sulfate transmembrane transport | 1 | 11 | 4.15e-01 |
| GO:BP | GO:0060907 | positive regulation of macrophage cytokine production | 1 | 18 | 4.16e-01 |
| GO:BP | GO:0032534 | regulation of microvillus assembly | 1 | 6 | 4.16e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 6 | 147 | 4.16e-01 |
| GO:BP | GO:0035864 | response to potassium ion | 1 | 12 | 4.16e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 2 | 113 | 4.16e-01 |
| GO:BP | GO:0007611 | learning or memory | 5 | 202 | 4.16e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 3 | 58 | 4.16e-01 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 3 | 58 | 4.16e-01 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 3 | 72 | 4.16e-01 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 3 | 64 | 4.16e-01 |
| GO:BP | GO:0035810 | positive regulation of urine volume | 1 | 10 | 4.16e-01 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 1 | 22 | 4.17e-01 |
| GO:BP | GO:0034453 | microtubule anchoring | 1 | 25 | 4.17e-01 |
| GO:BP | GO:0007565 | female pregnancy | 2 | 122 | 4.17e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 2 | 20 | 4.17e-01 |
| GO:BP | GO:1901658 | glycosyl compound catabolic process | 1 | 23 | 4.17e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 1 | 19 | 4.17e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 3 | 55 | 4.17e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 1 | 19 | 4.17e-01 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 2 | 100 | 4.17e-01 |
| GO:BP | GO:0045623 | negative regulation of T-helper cell differentiation | 2 | 11 | 4.17e-01 |
| GO:BP | GO:0009799 | specification of symmetry | 3 | 103 | 4.17e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 3 | 1378 | 4.18e-01 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 11 | 118 | 4.18e-01 |
| GO:BP | GO:0042073 | intraciliary transport | 1 | 46 | 4.18e-01 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 2 | 81 | 4.18e-01 |
| GO:BP | GO:0016322 | neuron remodeling | 1 | 11 | 4.18e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 3 | 50 | 4.18e-01 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 2 | 40 | 4.18e-01 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 1 | 38 | 4.19e-01 |
| GO:BP | GO:0002687 | positive regulation of leukocyte migration | 4 | 84 | 4.19e-01 |
| GO:BP | GO:0016925 | protein sumoylation | 4 | 59 | 4.19e-01 |
| GO:BP | GO:0003188 | heart valve formation | 1 | 16 | 4.19e-01 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 2 | 53 | 4.19e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 7 | 73 | 4.19e-01 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 1 | 54 | 4.19e-01 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 3 | 26 | 4.19e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 1 | 165 | 4.19e-01 |
| GO:BP | GO:0031579 | membrane raft organization | 1 | 19 | 4.19e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 1 | 118 | 4.19e-01 |
| GO:BP | GO:0009056 | catabolic process | 11 | 2088 | 4.19e-01 |
| GO:BP | GO:0010970 | transport along microtubule | 4 | 162 | 4.19e-01 |
| GO:BP | GO:0086036 | regulation of cardiac muscle cell membrane potential | 1 | 7 | 4.19e-01 |
| GO:BP | GO:0016240 | autophagosome membrane docking | 1 | 7 | 4.19e-01 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 11 | 129 | 4.19e-01 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 1 | 7 | 4.19e-01 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 1 | 7 | 4.19e-01 |
| GO:BP | GO:0060268 | negative regulation of respiratory burst | 1 | 4 | 4.19e-01 |
| GO:BP | GO:0021563 | glossopharyngeal nerve development | 1 | 3 | 4.19e-01 |
| GO:BP | GO:0032892 | positive regulation of organic acid transport | 2 | 27 | 4.19e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 2 | 34 | 4.20e-01 |
| GO:BP | GO:0001656 | metanephros development | 1 | 64 | 4.20e-01 |
| GO:BP | GO:0032836 | glomerular basement membrane development | 1 | 9 | 4.20e-01 |
| GO:BP | GO:0048589 | developmental growth | 1 | 522 | 4.20e-01 |
| GO:BP | GO:0006901 | vesicle coating | 3 | 36 | 4.21e-01 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 1 | 12 | 4.21e-01 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 1 | 5 | 4.21e-01 |
| GO:BP | GO:0060242 | contact inhibition | 1 | 8 | 4.21e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 6 | 71 | 4.21e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 2 | 19 | 4.22e-01 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 1 | 4 | 4.22e-01 |
| GO:BP | GO:0061304 | retinal blood vessel morphogenesis | 1 | 5 | 4.22e-01 |
| GO:BP | GO:0035426 | extracellular matrix-cell signaling | 1 | 4 | 4.22e-01 |
| GO:BP | GO:0003013 | circulatory system process | 15 | 447 | 4.22e-01 |
| GO:BP | GO:0070229 | negative regulation of lymphocyte apoptotic process | 2 | 23 | 4.22e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 4.22e-01 |
| GO:BP | GO:0090313 | regulation of protein targeting to membrane | 2 | 27 | 4.22e-01 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 2 | 22 | 4.22e-01 |
| GO:BP | GO:2000271 | positive regulation of fibroblast apoptotic process | 1 | 9 | 4.22e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 6 | 147 | 4.22e-01 |
| GO:BP | GO:0006403 | RNA localization | 7 | 182 | 4.22e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 2 | 357 | 4.22e-01 |
| GO:BP | GO:0140021 | mitochondrial ADP transmembrane transport | 1 | 4 | 4.22e-01 |
| GO:BP | GO:0055057 | neuroblast division | 1 | 12 | 4.22e-01 |
| GO:BP | GO:1990544 | mitochondrial ATP transmembrane transport | 1 | 4 | 4.22e-01 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.22e-01 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 1 | 16 | 4.22e-01 |
| GO:BP | GO:0001759 | organ induction | 1 | 17 | 4.22e-01 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.22e-01 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 1 | 5 | 4.22e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 8 | 957 | 4.22e-01 |
| GO:BP | GO:0080111 | DNA demethylation | 1 | 23 | 4.22e-01 |
| GO:BP | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly | 1 | 5 | 4.22e-01 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 7 | 133 | 4.22e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 5 | 102 | 4.23e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 3 | 107 | 4.23e-01 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 8 | 199 | 4.23e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 3 | 52 | 4.23e-01 |
| GO:BP | GO:0045721 | negative regulation of gluconeogenesis | 1 | 12 | 4.23e-01 |
| GO:BP | GO:0030240 | skeletal muscle thin filament assembly | 1 | 6 | 4.23e-01 |
| GO:BP | GO:0030035 | microspike assembly | 1 | 5 | 4.24e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 1 | 71 | 4.24e-01 |
| GO:BP | GO:0043966 | histone H3 acetylation | 5 | 59 | 4.25e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 1 | 151 | 4.25e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 2 | 61 | 4.25e-01 |
| GO:BP | GO:0007015 | actin filament organization | 8 | 373 | 4.25e-01 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 1 | 25 | 4.26e-01 |
| GO:BP | GO:2000110 | negative regulation of macrophage apoptotic process | 1 | 3 | 4.26e-01 |
| GO:BP | GO:0022414 | reproductive process | 3 | 927 | 4.26e-01 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 4 | 77 | 4.26e-01 |
| GO:BP | GO:0090400 | stress-induced premature senescence | 1 | 7 | 4.26e-01 |
| GO:BP | GO:0032075 | positive regulation of nuclease activity | 1 | 7 | 4.27e-01 |
| GO:BP | GO:0008104 | protein localization | 2 | 2129 | 4.27e-01 |
| GO:BP | GO:0034114 | regulation of heterotypic cell-cell adhesion | 1 | 13 | 4.27e-01 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 1 | 34 | 4.27e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 5 | 233 | 4.27e-01 |
| GO:BP | GO:0046600 | negative regulation of centriole replication | 1 | 7 | 4.27e-01 |
| GO:BP | GO:0090259 | regulation of retinal ganglion cell axon guidance | 1 | 3 | 4.27e-01 |
| GO:BP | GO:0021545 | cranial nerve development | 5 | 31 | 4.27e-01 |
| GO:BP | GO:0043305 | negative regulation of mast cell degranulation | 1 | 2 | 4.27e-01 |
| GO:BP | GO:0032762 | mast cell cytokine production | 1 | 6 | 4.27e-01 |
| GO:BP | GO:0032763 | regulation of mast cell cytokine production | 1 | 6 | 4.27e-01 |
| GO:BP | GO:0098751 | bone cell development | 1 | 26 | 4.27e-01 |
| GO:BP | GO:0008015 | blood circulation | 13 | 374 | 4.27e-01 |
| GO:BP | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 1 | 40 | 4.27e-01 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 2 | 45 | 4.27e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2 | 2138 | 4.27e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 4.27e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 4.27e-01 |
| GO:BP | GO:0032259 | methylation | 1 | 313 | 4.27e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 35 | 4.27e-01 |
| GO:BP | GO:1902074 | response to salt | 8 | 271 | 4.28e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 16 | 2880 | 4.28e-01 |
| GO:BP | GO:0000003 | reproduction | 3 | 934 | 4.29e-01 |
| GO:BP | GO:1902460 | regulation of mesenchymal stem cell proliferation | 1 | 4 | 4.29e-01 |
| GO:BP | GO:1902462 | positive regulation of mesenchymal stem cell proliferation | 1 | 4 | 4.29e-01 |
| GO:BP | GO:0006558 | L-phenylalanine metabolic process | 1 | 3 | 4.29e-01 |
| GO:BP | GO:1902600 | proton transmembrane transport | 2 | 111 | 4.29e-01 |
| GO:BP | GO:1902222 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process | 1 | 3 | 4.29e-01 |
| GO:BP | GO:1902221 | erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process | 1 | 3 | 4.29e-01 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 1 | 18 | 4.29e-01 |
| GO:BP | GO:0006559 | L-phenylalanine catabolic process | 1 | 3 | 4.29e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 2 | 39 | 4.29e-01 |
| GO:BP | GO:0042592 | homeostatic process | 30 | 1159 | 4.29e-01 |
| GO:BP | GO:0010878 | cholesterol storage | 2 | 16 | 4.29e-01 |
| GO:BP | GO:0032107 | regulation of response to nutrient levels | 1 | 19 | 4.30e-01 |
| GO:BP | GO:0032104 | regulation of response to extracellular stimulus | 1 | 19 | 4.30e-01 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 1 | 23 | 4.30e-01 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 1 | 13 | 4.30e-01 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 1 | 23 | 4.30e-01 |
| GO:BP | GO:0062009 | secondary palate development | 1 | 20 | 4.30e-01 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 1 | 5 | 4.30e-01 |
| GO:BP | GO:1904181 | positive regulation of membrane depolarization | 1 | 8 | 4.30e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 6 | 246 | 4.30e-01 |
| GO:BP | GO:0006903 | vesicle targeting | 5 | 61 | 4.31e-01 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 1 | 24 | 4.32e-01 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 3 | 25 | 4.32e-01 |
| GO:BP | GO:0042219 | cellular modified amino acid catabolic process | 2 | 16 | 4.32e-01 |
| GO:BP | GO:0046689 | response to mercury ion | 1 | 7 | 4.32e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 11 | 249 | 4.32e-01 |
| GO:BP | GO:1903371 | regulation of endoplasmic reticulum tubular network organization | 1 | 6 | 4.32e-01 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 2 | 33 | 4.32e-01 |
| GO:BP | GO:0072080 | nephron tubule development | 1 | 73 | 4.32e-01 |
| GO:BP | GO:0060135 | maternal process involved in female pregnancy | 1 | 41 | 4.32e-01 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 2 | 20 | 4.32e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 2 | 433 | 4.32e-01 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 1 | 29 | 4.33e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 1 | 25 | 4.33e-01 |
| GO:BP | GO:0036075 | replacement ossification | 1 | 25 | 4.33e-01 |
| GO:BP | GO:0018197 | peptidyl-aspartic acid modification | 1 | 5 | 4.33e-01 |
| GO:BP | GO:0007033 | vacuole organization | 4 | 197 | 4.33e-01 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 6 | 55 | 4.34e-01 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 1 | 53 | 4.34e-01 |
| GO:BP | GO:0060037 | pharyngeal system development | 2 | 23 | 4.34e-01 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 6 | 55 | 4.34e-01 |
| GO:BP | GO:0007613 | memory | 5 | 92 | 4.34e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 2 | 18 | 4.34e-01 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 1 | 5 | 4.34e-01 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 4 | 35 | 4.34e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 4.34e-01 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 4.34e-01 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 1 | 67 | 4.34e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 4.34e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 3 | 24 | 4.34e-01 |
| GO:BP | GO:1901725 | regulation of histone deacetylase activity | 1 | 8 | 4.34e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 5 | 74 | 4.34e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 1 | 10 | 4.34e-01 |
| GO:BP | GO:0050832 | defense response to fungus | 1 | 14 | 4.34e-01 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 2 | 16 | 4.34e-01 |
| GO:BP | GO:0033239 | negative regulation of amine metabolic process | 1 | 2 | 4.35e-01 |
| GO:BP | GO:0071371 | cellular response to gonadotropin stimulus | 2 | 13 | 4.35e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 4.35e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 4.35e-01 |
| GO:BP | GO:0019240 | citrulline biosynthetic process | 1 | 4 | 4.35e-01 |
| GO:BP | GO:0043045 | DNA methylation involved in embryo development | 1 | 7 | 4.35e-01 |
| GO:BP | GO:0034392 | negative regulation of smooth muscle cell apoptotic process | 1 | 6 | 4.35e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 3 | 62 | 4.35e-01 |
| GO:BP | GO:0055064 | chloride ion homeostasis | 1 | 13 | 4.35e-01 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 1 | 16 | 4.35e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 4 | 269 | 4.35e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 3 | 200 | 4.35e-01 |
| GO:BP | GO:0042537 | benzene-containing compound metabolic process | 1 | 14 | 4.36e-01 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 4.36e-01 |
| GO:BP | GO:1990403 | embryonic brain development | 1 | 13 | 4.36e-01 |
| GO:BP | GO:0060219 | camera-type eye photoreceptor cell differentiation | 1 | 18 | 4.36e-01 |
| GO:BP | GO:0003266 | regulation of secondary heart field cardioblast proliferation | 1 | 8 | 4.36e-01 |
| GO:BP | GO:0046035 | CMP metabolic process | 1 | 6 | 4.37e-01 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 1 | 5 | 4.37e-01 |
| GO:BP | GO:0060921 | sinoatrial node cell differentiation | 1 | 5 | 4.37e-01 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 1 | 20 | 4.37e-01 |
| GO:BP | GO:0001702 | gastrulation with mouth forming second | 1 | 21 | 4.37e-01 |
| GO:BP | GO:0140888 | interferon-mediated signaling pathway | 1 | 67 | 4.37e-01 |
| GO:BP | GO:0043032 | positive regulation of macrophage activation | 1 | 12 | 4.37e-01 |
| GO:BP | GO:0048240 | sperm capacitation | 1 | 7 | 4.37e-01 |
| GO:BP | GO:0032606 | type I interferon production | 1 | 76 | 4.37e-01 |
| GO:BP | GO:0051660 | establishment of centrosome localization | 1 | 8 | 4.37e-01 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 1 | 76 | 4.37e-01 |
| GO:BP | GO:0060788 | ectodermal placode formation | 1 | 10 | 4.37e-01 |
| GO:BP | GO:0071697 | ectodermal placode morphogenesis | 1 | 10 | 4.37e-01 |
| GO:BP | GO:0040016 | embryonic cleavage | 1 | 7 | 4.37e-01 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 5 | 58 | 4.38e-01 |
| GO:BP | GO:0019082 | viral protein processing | 3 | 31 | 4.38e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 30 | 1735 | 4.39e-01 |
| GO:BP | GO:1904894 | positive regulation of receptor signaling pathway via STAT | 3 | 22 | 4.40e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 8 | 60 | 4.40e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 8 | 60 | 4.40e-01 |
| GO:BP | GO:0061326 | renal tubule development | 1 | 77 | 4.40e-01 |
| GO:BP | GO:0110154 | RNA decapping | 1 | 17 | 4.40e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 1 | 236 | 4.41e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 4 | 161 | 4.41e-01 |
| GO:BP | GO:1903672 | positive regulation of sprouting angiogenesis | 1 | 14 | 4.41e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 2 | 99 | 4.41e-01 |
| GO:BP | GO:0061298 | retina vasculature development in camera-type eye | 2 | 14 | 4.41e-01 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 1 | 18 | 4.41e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 2 | 28 | 4.41e-01 |
| GO:BP | GO:1901896 | positive regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 6 | 4.41e-01 |
| GO:BP | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 10 | 4.41e-01 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 1 | 18 | 4.41e-01 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 1 | 18 | 4.41e-01 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 1 | 12 | 4.41e-01 |
| GO:BP | GO:0051170 | import into nucleus | 6 | 152 | 4.42e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 11 | 344 | 4.42e-01 |
| GO:BP | GO:0060245 | detection of cell density | 1 | 9 | 4.42e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 4.42e-01 |
| GO:BP | GO:0071896 | protein localization to adherens junction | 1 | 6 | 4.42e-01 |
| GO:BP | GO:1901569 | fatty acid derivative catabolic process | 1 | 10 | 4.43e-01 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 5 | 57 | 4.43e-01 |
| GO:BP | GO:0035999 | tetrahydrofolate interconversion | 1 | 9 | 4.43e-01 |
| GO:BP | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation | 1 | 6 | 4.43e-01 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 2 | 20 | 4.43e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 10 | 188 | 4.43e-01 |
| GO:BP | GO:1902019 | regulation of cilium-dependent cell motility | 1 | 12 | 4.43e-01 |
| GO:BP | GO:0060295 | regulation of cilium movement involved in cell motility | 1 | 12 | 4.43e-01 |
| GO:BP | GO:0000303 | response to superoxide | 1 | 21 | 4.43e-01 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 3 | 78 | 4.43e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 1 | 256 | 4.43e-01 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 4 | 54 | 4.44e-01 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 1 | 18 | 4.44e-01 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 1 | 18 | 4.44e-01 |
| GO:BP | GO:0000041 | transition metal ion transport | 5 | 71 | 4.45e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 1 | 258 | 4.45e-01 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 1 | 23 | 4.45e-01 |
| GO:BP | GO:0009110 | vitamin biosynthetic process | 1 | 14 | 4.45e-01 |
| GO:BP | GO:0010912 | positive regulation of isomerase activity | 1 | 5 | 4.45e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 5 | 401 | 4.45e-01 |
| GO:BP | GO:0010911 | regulation of isomerase activity | 1 | 5 | 4.45e-01 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 1 | 84 | 4.45e-01 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 2 | 47 | 4.45e-01 |
| GO:BP | GO:0070141 | response to UV-A | 1 | 11 | 4.45e-01 |
| GO:BP | GO:0090312 | positive regulation of protein deacetylation | 1 | 28 | 4.46e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 4 | 4579 | 4.46e-01 |
| GO:BP | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 1 | 14 | 4.46e-01 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 1 | 7 | 4.46e-01 |
| GO:BP | GO:0030204 | chondroitin sulfate metabolic process | 1 | 28 | 4.46e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 4 | 160 | 4.46e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 4.46e-01 |
| GO:BP | GO:1903019 | negative regulation of glycoprotein metabolic process | 1 | 12 | 4.47e-01 |
| GO:BP | GO:0072498 | embryonic skeletal joint development | 1 | 10 | 4.47e-01 |
| GO:BP | GO:0001676 | long-chain fatty acid metabolic process | 2 | 65 | 4.47e-01 |
| GO:BP | GO:0006552 | leucine catabolic process | 1 | 6 | 4.47e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 2 | 538 | 4.47e-01 |
| GO:BP | GO:0031581 | hemidesmosome assembly | 1 | 6 | 4.47e-01 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 2 | 78 | 4.47e-01 |
| GO:BP | GO:0030223 | neutrophil differentiation | 1 | 8 | 4.47e-01 |
| GO:BP | GO:0021766 | hippocampus development | 2 | 62 | 4.47e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 5 | 212 | 4.47e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 1 | 301 | 4.47e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 3 | 191 | 4.48e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 3 | 535 | 4.48e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 3 | 48 | 4.49e-01 |
| GO:BP | GO:1901857 | positive regulation of cellular respiration | 1 | 11 | 4.49e-01 |
| GO:BP | GO:0001953 | negative regulation of cell-matrix adhesion | 1 | 26 | 4.49e-01 |
| GO:BP | GO:1904117 | cellular response to vasopressin | 1 | 5 | 4.49e-01 |
| GO:BP | GO:1904116 | response to vasopressin | 1 | 5 | 4.49e-01 |
| GO:BP | GO:0034121 | regulation of toll-like receptor signaling pathway | 1 | 19 | 4.49e-01 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 1 | 142 | 4.49e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 4.50e-01 |
| GO:BP | GO:0006855 | xenobiotic transmembrane transport | 1 | 11 | 4.50e-01 |
| GO:BP | GO:0070778 | L-aspartate transmembrane transport | 1 | 11 | 4.50e-01 |
| GO:BP | GO:0035510 | DNA dealkylation | 1 | 28 | 4.50e-01 |
| GO:BP | GO:0060264 | regulation of respiratory burst involved in inflammatory response | 1 | 3 | 4.51e-01 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 3 | 40 | 4.51e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 2 | 37 | 4.51e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 3 | 25 | 4.51e-01 |
| GO:BP | GO:2000489 | regulation of hepatic stellate cell activation | 1 | 6 | 4.51e-01 |
| GO:BP | GO:0009306 | protein secretion | 2 | 262 | 4.52e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 9 | 164 | 4.52e-01 |
| GO:BP | GO:0044703 | multi-organism reproductive process | 2 | 131 | 4.52e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 6 | 237 | 4.52e-01 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 3 | 114 | 4.52e-01 |
| GO:BP | GO:0097164 | ammonium ion metabolic process | 1 | 14 | 4.52e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 1 | 22 | 4.53e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 2 | 262 | 4.53e-01 |
| GO:BP | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process | 1 | 9 | 4.53e-01 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 1 | 24 | 4.53e-01 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 1 | 15 | 4.53e-01 |
| GO:BP | GO:0014816 | skeletal muscle satellite cell differentiation | 1 | 6 | 4.53e-01 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 1 | 21 | 4.53e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 1 | 26 | 4.53e-01 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 1 | 8 | 4.54e-01 |
| GO:BP | GO:0007612 | learning | 3 | 115 | 4.54e-01 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 1 | 38 | 4.54e-01 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 1 | 19 | 4.54e-01 |
| GO:BP | GO:0001561 | fatty acid alpha-oxidation | 1 | 5 | 4.54e-01 |
| GO:BP | GO:0032071 | regulation of endodeoxyribonuclease activity | 1 | 7 | 4.54e-01 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 1 | 14 | 4.54e-01 |
| GO:BP | GO:0040008 | regulation of growth | 11 | 501 | 4.54e-01 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 3 | 62 | 4.54e-01 |
| GO:BP | GO:0051574 | positive regulation of histone H3-K9 methylation | 1 | 5 | 4.54e-01 |
| GO:BP | GO:2000617 | positive regulation of histone H3-K9 acetylation | 1 | 6 | 4.54e-01 |
| GO:BP | GO:1900118 | negative regulation of execution phase of apoptosis | 1 | 6 | 4.55e-01 |
| GO:BP | GO:1902622 | regulation of neutrophil migration | 1 | 28 | 4.55e-01 |
| GO:BP | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 1 | 6 | 4.55e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 5 | 68 | 4.57e-01 |
| GO:BP | GO:0055081 | monoatomic anion homeostasis | 1 | 14 | 4.57e-01 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 1 | 6 | 4.57e-01 |
| GO:BP | GO:0001504 | neurotransmitter uptake | 2 | 29 | 4.57e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 3 | 75 | 4.57e-01 |
| GO:BP | GO:1903613 | regulation of protein tyrosine phosphatase activity | 1 | 4 | 4.57e-01 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 4 | 48 | 4.57e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 5 | 408 | 4.57e-01 |
| GO:BP | GO:0040018 | positive regulation of multicellular organism growth | 1 | 22 | 4.57e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 29 | 4.57e-01 |
| GO:BP | GO:0008406 | gonad development | 7 | 158 | 4.57e-01 |
| GO:BP | GO:0002312 | B cell activation involved in immune response | 7 | 57 | 4.57e-01 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 1 | 60 | 4.58e-01 |
| GO:BP | GO:0007568 | aging | 6 | 115 | 4.58e-01 |
| GO:BP | GO:0001554 | luteolysis | 1 | 7 | 4.59e-01 |
| GO:BP | GO:0097354 | prenylation | 1 | 10 | 4.59e-01 |
| GO:BP | GO:0070100 | negative regulation of chemokine-mediated signaling pathway | 1 | 6 | 4.59e-01 |
| GO:BP | GO:0018342 | protein prenylation | 1 | 10 | 4.59e-01 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 2 | 31 | 4.59e-01 |
| GO:BP | GO:0001657 | ureteric bud development | 2 | 75 | 4.59e-01 |
| GO:BP | GO:0050877 | nervous system process | 1 | 634 | 4.59e-01 |
| GO:BP | GO:0061061 | muscle structure development | 10 | 546 | 4.59e-01 |
| GO:BP | GO:0003310 | pancreatic A cell differentiation | 1 | 2 | 4.59e-01 |
| GO:BP | GO:2000328 | regulation of T-helper 17 cell lineage commitment | 1 | 1 | 4.59e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 6 | 47 | 4.59e-01 |
| GO:BP | GO:0030001 | metal ion transport | 8 | 562 | 4.59e-01 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 1 | 6 | 4.59e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 5 | 59 | 4.59e-01 |
| GO:BP | GO:1900029 | positive regulation of ruffle assembly | 1 | 12 | 4.59e-01 |
| GO:BP | GO:0034968 | histone lysine methylation | 2 | 89 | 4.59e-01 |
| GO:BP | GO:0090281 | negative regulation of calcium ion import | 1 | 6 | 4.59e-01 |
| GO:BP | GO:0003264 | regulation of cardioblast proliferation | 1 | 9 | 4.59e-01 |
| GO:BP | GO:0003263 | cardioblast proliferation | 1 | 9 | 4.59e-01 |
| GO:BP | GO:0008078 | mesodermal cell migration | 1 | 3 | 4.59e-01 |
| GO:BP | GO:0035022 | positive regulation of Rac protein signal transduction | 1 | 5 | 4.59e-01 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 1 | 17 | 4.59e-01 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 1 | 15 | 4.60e-01 |
| GO:BP | GO:0007431 | salivary gland development | 3 | 26 | 4.60e-01 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 1 | 18 | 4.61e-01 |
| GO:BP | GO:0003401 | axis elongation | 1 | 24 | 4.61e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 1 | 20 | 4.61e-01 |
| GO:BP | GO:0019079 | viral genome replication | 2 | 101 | 4.61e-01 |
| GO:BP | GO:1904851 | positive regulation of establishment of protein localization to telomere | 1 | 10 | 4.61e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 13 | 2412 | 4.61e-01 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 3 | 30 | 4.61e-01 |
| GO:BP | GO:0045191 | regulation of isotype switching | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 4 | 179 | 4.61e-01 |
| GO:BP | GO:0072708 | response to sorbitol | 1 | 7 | 4.61e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 9 | 632 | 4.61e-01 |
| GO:BP | GO:0031016 | pancreas development | 2 | 53 | 4.61e-01 |
| GO:BP | GO:0072164 | mesonephric tubule development | 2 | 76 | 4.61e-01 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 2 | 76 | 4.61e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 7 | 1248 | 4.61e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 1 | 11 | 4.61e-01 |
| GO:BP | GO:0097340 | inhibition of cysteine-type endopeptidase activity | 1 | 5 | 4.61e-01 |
| GO:BP | GO:0097341 | zymogen inhibition | 1 | 5 | 4.61e-01 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 5 | 56 | 4.61e-01 |
| GO:BP | GO:0050995 | negative regulation of lipid catabolic process | 2 | 14 | 4.61e-01 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 1 | 14 | 4.62e-01 |
| GO:BP | GO:0006883 | intracellular sodium ion homeostasis | 1 | 13 | 4.62e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 4 | 91 | 4.62e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 4.62e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 3 | 993 | 4.62e-01 |
| GO:BP | GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 3 | 4.64e-01 |
| GO:BP | GO:0015886 | heme transport | 1 | 9 | 4.64e-01 |
| GO:BP | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway | 1 | 5 | 4.64e-01 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 1 | 71 | 4.64e-01 |
| GO:BP | GO:0042742 | defense response to bacterium | 2 | 95 | 4.64e-01 |
| GO:BP | GO:0071907 | determination of digestive tract left/right asymmetry | 1 | 5 | 4.64e-01 |
| GO:BP | GO:0071867 | response to monoamine | 8 | 62 | 4.64e-01 |
| GO:BP | GO:1990743 | protein sialylation | 1 | 5 | 4.64e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 8 | 62 | 4.64e-01 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 2 | 17 | 4.65e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 2 | 556 | 4.65e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 1 | 124 | 4.65e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 6 | 90 | 4.65e-01 |
| GO:BP | GO:0097530 | granulocyte migration | 3 | 67 | 4.65e-01 |
| GO:BP | GO:0072531 | pyrimidine-containing compound transmembrane transport | 1 | 9 | 4.65e-01 |
| GO:BP | GO:0009072 | aromatic amino acid metabolic process | 1 | 12 | 4.65e-01 |
| GO:BP | GO:0033504 | floor plate development | 1 | 8 | 4.65e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 1 | 12 | 4.65e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 10 | 294 | 4.65e-01 |
| GO:BP | GO:0043985 | histone H4-R3 methylation | 1 | 6 | 4.66e-01 |
| GO:BP | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development | 1 | 6 | 4.66e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 10 | 2646 | 4.66e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 5 | 84 | 4.67e-01 |
| GO:BP | GO:0036093 | germ cell proliferation | 1 | 7 | 4.67e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 2 | 28 | 4.67e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 4 | 135 | 4.67e-01 |
| GO:BP | GO:0000101 | sulfur amino acid transport | 1 | 10 | 4.68e-01 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 5 | 53 | 4.68e-01 |
| GO:BP | GO:1901889 | negative regulation of cell junction assembly | 1 | 24 | 4.68e-01 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 4.68e-01 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 1 | 16 | 4.68e-01 |
| GO:BP | GO:0031623 | receptor internalization | 1 | 95 | 4.68e-01 |
| GO:BP | GO:0032528 | microvillus organization | 2 | 19 | 4.69e-01 |
| GO:BP | GO:0044706 | multi-multicellular organism process | 2 | 135 | 4.69e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 1 | 185 | 4.69e-01 |
| GO:BP | GO:0019076 | viral release from host cell | 1 | 28 | 4.69e-01 |
| GO:BP | GO:0035891 | exit from host cell | 1 | 28 | 4.69e-01 |
| GO:BP | GO:0071236 | cellular response to antibiotic | 1 | 11 | 4.69e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 3 | 204 | 4.69e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 2 | 122 | 4.70e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 4 | 161 | 4.70e-01 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 3 | 31 | 4.70e-01 |
| GO:BP | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 1 | 7 | 4.70e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 3 | 784 | 4.70e-01 |
| GO:BP | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 1 | 15 | 4.70e-01 |
| GO:BP | GO:0032611 | interleukin-1 beta production | 2 | 48 | 4.70e-01 |
| GO:BP | GO:0032651 | regulation of interleukin-1 beta production | 2 | 48 | 4.70e-01 |
| GO:BP | GO:0032354 | response to follicle-stimulating hormone | 1 | 13 | 4.70e-01 |
| GO:BP | GO:2000319 | regulation of T-helper 17 cell differentiation | 2 | 10 | 4.70e-01 |
| GO:BP | GO:1900371 | regulation of purine nucleotide biosynthetic process | 2 | 25 | 4.71e-01 |
| GO:BP | GO:0030808 | regulation of nucleotide biosynthetic process | 2 | 25 | 4.71e-01 |
| GO:BP | GO:0032409 | regulation of transporter activity | 8 | 211 | 4.71e-01 |
| GO:BP | GO:0009620 | response to fungus | 1 | 22 | 4.71e-01 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 1 | 19 | 4.71e-01 |
| GO:BP | GO:0009408 | response to heat | 3 | 84 | 4.71e-01 |
| GO:BP | GO:0009651 | response to salt stress | 1 | 18 | 4.71e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 3 | 214 | 4.71e-01 |
| GO:BP | GO:0042573 | retinoic acid metabolic process | 1 | 12 | 4.71e-01 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 1 | 38 | 4.71e-01 |
| GO:BP | GO:0106300 | protein-DNA covalent cross-linking repair | 1 | 7 | 4.71e-01 |
| GO:BP | GO:0072009 | nephron epithelium development | 1 | 86 | 4.71e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 7 | 963 | 4.71e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 13 | 358 | 4.72e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 39 | 859 | 4.72e-01 |
| GO:BP | GO:0043457 | regulation of cellular respiration | 3 | 42 | 4.73e-01 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 1 | 41 | 4.73e-01 |
| GO:BP | GO:0034770 | histone H4-K20 methylation | 1 | 6 | 4.73e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 5 | 85 | 4.73e-01 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 10 | 170 | 4.73e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 3 | 68 | 4.74e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 3 | 49 | 4.74e-01 |
| GO:BP | GO:0051599 | response to hydrostatic pressure | 1 | 5 | 4.75e-01 |
| GO:BP | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 1 | 5 | 4.75e-01 |
| GO:BP | GO:0001974 | blood vessel remodeling | 3 | 32 | 4.75e-01 |
| GO:BP | GO:0090309 | positive regulation of DNA methylation-dependent heterochromatin formation | 1 | 12 | 4.75e-01 |
| GO:BP | GO:0072006 | nephron development | 10 | 117 | 4.75e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 10 | 136 | 4.76e-01 |
| GO:BP | GO:1901072 | glucosamine-containing compound catabolic process | 1 | 5 | 4.76e-01 |
| GO:BP | GO:0001823 | mesonephros development | 2 | 77 | 4.76e-01 |
| GO:BP | GO:0016049 | cell growth | 2 | 418 | 4.76e-01 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 3 | 26 | 4.77e-01 |
| GO:BP | GO:0070070 | proton-transporting V-type ATPase complex assembly | 1 | 7 | 4.77e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 1 | 45 | 4.77e-01 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 2 | 19 | 4.77e-01 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 1 | 7 | 4.78e-01 |
| GO:BP | GO:0051140 | regulation of NK T cell proliferation | 1 | 3 | 4.78e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 4 | 56 | 4.78e-01 |
| GO:BP | GO:0007035 | vacuolar acidification | 3 | 30 | 4.78e-01 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 1 | 31 | 4.78e-01 |
| GO:BP | GO:0072575 | epithelial cell proliferation involved in liver morphogenesis | 2 | 18 | 4.78e-01 |
| GO:BP | GO:0072574 | hepatocyte proliferation | 2 | 18 | 4.78e-01 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 13 | 4.78e-01 |
| GO:BP | GO:0071622 | regulation of granulocyte chemotaxis | 1 | 27 | 4.78e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 1 | 317 | 4.78e-01 |
| GO:BP | GO:0030203 | glycosaminoglycan metabolic process | 6 | 87 | 4.78e-01 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 2 | 18 | 4.78e-01 |
| GO:BP | GO:0150065 | regulation of deacetylase activity | 1 | 10 | 4.78e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 1 | 14 | 4.78e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 2 | 598 | 4.78e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 4.78e-01 |
| GO:BP | GO:0055078 | sodium ion homeostasis | 2 | 26 | 4.78e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 1 | 14 | 4.78e-01 |
| GO:BP | GO:0051709 | regulation of killing of cells of another organism | 1 | 3 | 4.78e-01 |
| GO:BP | GO:0032375 | negative regulation of cholesterol transport | 1 | 5 | 4.78e-01 |
| GO:BP | GO:0032372 | negative regulation of sterol transport | 1 | 5 | 4.78e-01 |
| GO:BP | GO:0019080 | viral gene expression | 2 | 96 | 4.78e-01 |
| GO:BP | GO:0035561 | regulation of chromatin binding | 2 | 20 | 4.79e-01 |
| GO:BP | GO:0051168 | nuclear export | 5 | 150 | 4.79e-01 |
| GO:BP | GO:0032958 | inositol phosphate biosynthetic process | 1 | 19 | 4.79e-01 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 20 | 4.79e-01 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 1 | 69 | 4.79e-01 |
| GO:BP | GO:0010758 | regulation of macrophage chemotaxis | 1 | 14 | 4.79e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 3 | 27 | 4.79e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 3 | 44 | 4.80e-01 |
| GO:BP | GO:0072348 | sulfur compound transport | 3 | 39 | 4.80e-01 |
| GO:BP | GO:0002536 | respiratory burst involved in inflammatory response | 1 | 4 | 4.80e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 5 | 166 | 4.80e-01 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 2 | 55 | 4.80e-01 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 1 | 7 | 4.80e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 4 | 169 | 4.80e-01 |
| GO:BP | GO:0021903 | rostrocaudal neural tube patterning | 1 | 5 | 4.80e-01 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 3 | 24 | 4.80e-01 |
| GO:BP | GO:0051567 | histone H3-K9 methylation | 2 | 18 | 4.80e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 5 | 120 | 4.80e-01 |
| GO:BP | GO:0051385 | response to mineralocorticoid | 2 | 25 | 4.80e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 19 | 225 | 4.80e-01 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 1 | 31 | 4.80e-01 |
| GO:BP | GO:0002903 | negative regulation of B cell apoptotic process | 1 | 11 | 4.80e-01 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 1 | 16 | 4.80e-01 |
| GO:BP | GO:0032755 | positive regulation of interleukin-6 production | 2 | 56 | 4.80e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 16 | 292 | 4.81e-01 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 1 | 29 | 4.81e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 1 | 25 | 4.81e-01 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 1 | 9 | 4.82e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 3 | 41 | 4.82e-01 |
| GO:BP | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway | 1 | 7 | 4.82e-01 |
| GO:BP | GO:0006171 | cAMP biosynthetic process | 1 | 8 | 4.82e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 3 | 348 | 4.82e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 4 | 172 | 4.82e-01 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 1 | 12 | 4.83e-01 |
| GO:BP | GO:0036158 | outer dynein arm assembly | 2 | 11 | 4.83e-01 |
| GO:BP | GO:0010039 | response to iron ion | 2 | 24 | 4.84e-01 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 1 | 7 | 4.84e-01 |
| GO:BP | GO:0070586 | cell-cell adhesion involved in gastrulation | 1 | 7 | 4.84e-01 |
| GO:BP | GO:1901380 | negative regulation of potassium ion transmembrane transport | 1 | 18 | 4.84e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 12 | 152 | 4.86e-01 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 1 | 46 | 4.86e-01 |
| GO:BP | GO:0033623 | regulation of integrin activation | 1 | 11 | 4.86e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 14 | 381 | 4.87e-01 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 1 | 38 | 4.87e-01 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 1 | 11 | 4.87e-01 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 1 | 9 | 4.88e-01 |
| GO:BP | GO:0000012 | single strand break repair | 1 | 10 | 4.88e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 4.89e-01 |
| GO:BP | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation | 2 | 14 | 4.89e-01 |
| GO:BP | GO:0006508 | proteolysis | 2 | 1318 | 4.89e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 1 | 27 | 4.89e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 8 | 1137 | 4.89e-01 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 1 | 26 | 4.90e-01 |
| GO:BP | GO:0061439 | kidney vasculature morphogenesis | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0060839 | endothelial cell fate commitment | 1 | 7 | 4.90e-01 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 1 | 28 | 4.90e-01 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 1 | 8 | 4.90e-01 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 2 | 119 | 4.90e-01 |
| GO:BP | GO:0061438 | renal system vasculature morphogenesis | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 2 | 26 | 4.90e-01 |
| GO:BP | GO:1900122 | positive regulation of receptor binding | 1 | 7 | 4.90e-01 |
| GO:BP | GO:0032489 | regulation of Cdc42 protein signal transduction | 1 | 6 | 4.90e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 12 | 222 | 4.90e-01 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 2 | 23 | 4.90e-01 |
| GO:BP | GO:1903510 | mucopolysaccharide metabolic process | 5 | 74 | 4.91e-01 |
| GO:BP | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion | 1 | 7 | 4.91e-01 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 1 | 35 | 4.91e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 2 | 2450 | 4.91e-01 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 1 | 107 | 4.91e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 5 | 771 | 4.91e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 9 | 669 | 4.91e-01 |
| GO:BP | GO:0006968 | cellular defense response | 2 | 15 | 4.91e-01 |
| GO:BP | GO:1901463 | regulation of tetrapyrrole biosynthetic process | 1 | 7 | 4.91e-01 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 1 | 9 | 4.91e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 2 | 114 | 4.91e-01 |
| GO:BP | GO:0070453 | regulation of heme biosynthetic process | 1 | 7 | 4.91e-01 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 1 | 70 | 4.91e-01 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 2 | 53 | 4.91e-01 |
| GO:BP | GO:0030953 | astral microtubule organization | 1 | 11 | 4.91e-01 |
| GO:BP | GO:0032613 | interleukin-10 production | 1 | 24 | 4.92e-01 |
| GO:BP | GO:0032653 | regulation of interleukin-10 production | 1 | 24 | 4.92e-01 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 1 | 5 | 4.92e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 2 | 18 | 4.92e-01 |
| GO:BP | GO:0002861 | regulation of inflammatory response to antigenic stimulus | 1 | 24 | 4.92e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 1 | 357 | 4.93e-01 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 4 | 49 | 4.93e-01 |
| GO:BP | GO:1904415 | regulation of xenophagy | 1 | 7 | 4.93e-01 |
| GO:BP | GO:0072576 | liver morphogenesis | 2 | 19 | 4.93e-01 |
| GO:BP | GO:1904417 | positive regulation of xenophagy | 1 | 7 | 4.93e-01 |
| GO:BP | GO:0051258 | protein polymerization | 5 | 241 | 4.93e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 2 | 199 | 4.93e-01 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 5 | 63 | 4.93e-01 |
| GO:BP | GO:0002790 | peptide secretion | 5 | 170 | 4.93e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 3 | 28 | 4.94e-01 |
| GO:BP | GO:0031204 | post-translational protein targeting to membrane, translocation | 1 | 7 | 4.94e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 3 | 126 | 4.94e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 4 | 73 | 4.94e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 4 | 73 | 4.94e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 2 | 43 | 4.94e-01 |
| GO:BP | GO:0006412 | translation | 2 | 646 | 4.95e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 4.95e-01 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 1 | 33 | 4.95e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 1 | 20 | 4.95e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 4.95e-01 |
| GO:BP | GO:0032094 | response to food | 1 | 22 | 4.95e-01 |
| GO:BP | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response | 1 | 13 | 4.95e-01 |
| GO:BP | GO:0002181 | cytoplasmic translation | 3 | 156 | 4.96e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 4.96e-01 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 8 | 96 | 4.96e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 41 | 3237 | 4.97e-01 |
| GO:BP | GO:0006465 | signal peptide processing | 1 | 13 | 4.97e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 1 | 209 | 4.97e-01 |
| GO:BP | GO:0003177 | pulmonary valve development | 1 | 22 | 4.97e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 6 | 131 | 4.97e-01 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 3 | 1914 | 4.98e-01 |
| GO:BP | GO:0046849 | bone remodeling | 2 | 59 | 4.98e-01 |
| GO:BP | GO:0032305 | positive regulation of icosanoid secretion | 1 | 8 | 4.98e-01 |
| GO:BP | GO:0051571 | positive regulation of histone H3-K4 methylation | 2 | 20 | 4.98e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 2 | 1869 | 4.98e-01 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 2 | 54 | 4.98e-01 |
| GO:BP | GO:0045601 | regulation of endothelial cell differentiation | 1 | 27 | 4.98e-01 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 1 | 12 | 4.98e-01 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 1 | 11 | 4.98e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 1 | 234 | 4.99e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 3 | 32 | 4.99e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 16 | 1835 | 4.99e-01 |
| GO:BP | GO:0003351 | epithelial cilium movement involved in extracellular fluid movement | 1 | 29 | 4.99e-01 |
| GO:BP | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell | 1 | 3 | 4.99e-01 |
| GO:BP | GO:0042063 | gliogenesis | 2 | 235 | 5.00e-01 |
| GO:BP | GO:0019083 | viral transcription | 1 | 48 | 5.00e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 5.00e-01 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 2 | 17 | 5.01e-01 |
| GO:BP | GO:0040007 | growth | 1 | 742 | 5.01e-01 |
| GO:BP | GO:0008356 | asymmetric cell division | 1 | 14 | 5.01e-01 |
| GO:BP | GO:0140962 | multicellular organismal-level chemical homeostasis | 3 | 34 | 5.01e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 5.01e-01 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 1 | 94 | 5.01e-01 |
| GO:BP | GO:1901998 | toxin transport | 3 | 37 | 5.01e-01 |
| GO:BP | GO:0010739 | positive regulation of protein kinase A signaling | 1 | 6 | 5.01e-01 |
| GO:BP | GO:0051568 | histone H3-K4 methylation | 1 | 57 | 5.01e-01 |
| GO:BP | GO:0032536 | regulation of cell projection size | 1 | 8 | 5.01e-01 |
| GO:BP | GO:0031128 | developmental induction | 1 | 26 | 5.01e-01 |
| GO:BP | GO:0007040 | lysosome organization | 2 | 88 | 5.01e-01 |
| GO:BP | GO:0035881 | amacrine cell differentiation | 1 | 3 | 5.01e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 1 | 100 | 5.01e-01 |
| GO:BP | GO:0001188 | RNA polymerase I preinitiation complex assembly | 1 | 9 | 5.01e-01 |
| GO:BP | GO:0080171 | lytic vacuole organization | 2 | 88 | 5.01e-01 |
| GO:BP | GO:0097502 | mannosylation | 2 | 32 | 5.01e-01 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 5 | 131 | 5.01e-01 |
| GO:BP | GO:0070233 | negative regulation of T cell apoptotic process | 1 | 14 | 5.01e-01 |
| GO:BP | GO:0031063 | regulation of histone deacetylation | 1 | 36 | 5.01e-01 |
| GO:BP | GO:0002313 | mature B cell differentiation involved in immune response | 1 | 17 | 5.01e-01 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 2 | 51 | 5.01e-01 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 1 | 8 | 5.01e-01 |
| GO:BP | GO:0010934 | macrophage cytokine production | 1 | 27 | 5.01e-01 |
| GO:BP | GO:0032637 | interleukin-8 production | 2 | 43 | 5.01e-01 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 1 | 27 | 5.01e-01 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 1 | 12 | 5.01e-01 |
| GO:BP | GO:0060292 | long-term synaptic depression | 1 | 21 | 5.01e-01 |
| GO:BP | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 13 | 5.01e-01 |
| GO:BP | GO:0032677 | regulation of interleukin-8 production | 2 | 43 | 5.01e-01 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 2 | 101 | 5.01e-01 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 1 | 12 | 5.01e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 1 | 26 | 5.01e-01 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 2 | 51 | 5.01e-01 |
| GO:BP | GO:2000107 | negative regulation of leukocyte apoptotic process | 3 | 33 | 5.01e-01 |
| GO:BP | GO:0030183 | B cell differentiation | 3 | 94 | 5.01e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 5 | 418 | 5.01e-01 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 10 | 104 | 5.01e-01 |
| GO:BP | GO:1901570 | fatty acid derivative biosynthetic process | 1 | 33 | 5.01e-01 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 1 | 15 | 5.02e-01 |
| GO:BP | GO:0097168 | mesenchymal stem cell proliferation | 1 | 4 | 5.02e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 3 | 843 | 5.02e-01 |
| GO:BP | GO:0010829 | negative regulation of glucose transmembrane transport | 1 | 14 | 5.02e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 1 | 50 | 5.03e-01 |
| GO:BP | GO:0046459 | short-chain fatty acid metabolic process | 1 | 10 | 5.03e-01 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 4 | 92 | 5.03e-01 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 1 | 10 | 5.03e-01 |
| GO:BP | GO:0032070 | regulation of deoxyribonuclease activity | 1 | 9 | 5.04e-01 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 10 | 5.04e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 2 | 25 | 5.04e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 3 | 116 | 5.04e-01 |
| GO:BP | GO:0035065 | regulation of histone acetylation | 4 | 47 | 5.04e-01 |
| GO:BP | GO:0033007 | negative regulation of mast cell activation involved in immune response | 1 | 4 | 5.05e-01 |
| GO:BP | GO:0007379 | segment specification | 1 | 14 | 5.05e-01 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 1 | 10 | 5.05e-01 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 1 | 114 | 5.05e-01 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 3 | 23 | 5.05e-01 |
| GO:BP | GO:0034087 | establishment of mitotic sister chromatid cohesion | 1 | 7 | 5.07e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 2 | 1014 | 5.07e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 3 | 29 | 5.07e-01 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 1 | 9 | 5.07e-01 |
| GO:BP | GO:0060267 | positive regulation of respiratory burst | 1 | 3 | 5.07e-01 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 3 | 52 | 5.08e-01 |
| GO:BP | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation | 1 | 5 | 5.08e-01 |
| GO:BP | GO:0043652 | engulfment of apoptotic cell | 1 | 11 | 5.08e-01 |
| GO:BP | GO:0035733 | hepatic stellate cell activation | 1 | 8 | 5.08e-01 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 1 | 20 | 5.08e-01 |
| GO:BP | GO:0009174 | pyrimidine ribonucleoside monophosphate biosynthetic process | 1 | 9 | 5.08e-01 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 4 | 93 | 5.08e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 2 | 666 | 5.09e-01 |
| GO:BP | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1 | 21 | 5.09e-01 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 5 | 61 | 5.09e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 49 | 2270 | 5.09e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 2 | 24 | 5.09e-01 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0000820 | regulation of glutamine family amino acid metabolic process | 1 | 5 | 5.10e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 2 | 35 | 5.10e-01 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0006651 | diacylglycerol biosynthetic process | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0010033 | response to organic substance | 2 | 1957 | 5.10e-01 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 1 | 6 | 5.10e-01 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 2 | 29 | 5.10e-01 |
| GO:BP | GO:1905063 | regulation of vascular associated smooth muscle cell differentiation | 1 | 14 | 5.10e-01 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 1 | 7 | 5.10e-01 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 1 | 7 | 5.10e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 3 | 32 | 5.10e-01 |
| GO:BP | GO:0015866 | ADP transport | 1 | 6 | 5.10e-01 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 1 | 14 | 5.11e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 6 | 236 | 5.12e-01 |
| GO:BP | GO:0003352 | regulation of cilium movement | 1 | 17 | 5.12e-01 |
| GO:BP | GO:0006551 | leucine metabolic process | 1 | 8 | 5.13e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 5.13e-01 |
| GO:BP | GO:0014866 | skeletal myofibril assembly | 1 | 9 | 5.13e-01 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 1 | 8 | 5.14e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 8 | 209 | 5.14e-01 |
| GO:BP | GO:0031848 | protection from non-homologous end joining at telomere | 1 | 10 | 5.15e-01 |
| GO:BP | GO:0021871 | forebrain regionalization | 1 | 8 | 5.15e-01 |
| GO:BP | GO:0010884 | positive regulation of lipid storage | 2 | 18 | 5.16e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 2 | 21 | 5.16e-01 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 10 | 5.16e-01 |
| GO:BP | GO:0032780 | negative regulation of ATP-dependent activity | 1 | 18 | 5.16e-01 |
| GO:BP | GO:0006858 | extracellular transport | 1 | 32 | 5.16e-01 |
| GO:BP | GO:0019320 | hexose catabolic process | 1 | 32 | 5.16e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 7 | 276 | 5.16e-01 |
| GO:BP | GO:2000345 | regulation of hepatocyte proliferation | 1 | 13 | 5.17e-01 |
| GO:BP | GO:0071493 | cellular response to UV-B | 1 | 11 | 5.17e-01 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 2 | 23 | 5.17e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 1 | 57 | 5.18e-01 |
| GO:BP | GO:0072112 | podocyte differentiation | 1 | 17 | 5.18e-01 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 1 | 8 | 5.18e-01 |
| GO:BP | GO:0061318 | renal filtration cell differentiation | 1 | 17 | 5.18e-01 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 2 | 104 | 5.18e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 5 | 132 | 5.18e-01 |
| GO:BP | GO:0098760 | response to interleukin-7 | 1 | 12 | 5.18e-01 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 3 | 48 | 5.18e-01 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 1 | 12 | 5.18e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 5 | 61 | 5.18e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 5.18e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 5.19e-01 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 2 | 19 | 5.19e-01 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 2 | 19 | 5.19e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 2 | 66 | 5.19e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 3 | 75 | 5.19e-01 |
| GO:BP | GO:0032303 | regulation of icosanoid secretion | 1 | 9 | 5.20e-01 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 1 | 9 | 5.20e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 6 | 61 | 5.20e-01 |
| GO:BP | GO:0035264 | multicellular organism growth | 4 | 123 | 5.20e-01 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 2 | 17 | 5.20e-01 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 2 | 17 | 5.20e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 5.20e-01 |
| GO:BP | GO:1901678 | iron coordination entity transport | 1 | 11 | 5.20e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 5 | 65 | 5.20e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 2 | 25 | 5.20e-01 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 1 | 11 | 5.20e-01 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 3 | 1995 | 5.20e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 5.20e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 1 | 161 | 5.20e-01 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 1 | 21 | 5.21e-01 |
| GO:BP | GO:0097048 | dendritic cell apoptotic process | 1 | 7 | 5.21e-01 |
| GO:BP | GO:2000668 | regulation of dendritic cell apoptotic process | 1 | 7 | 5.21e-01 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 1 | 13 | 5.22e-01 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 2 | 39 | 5.22e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 6 | 67 | 5.22e-01 |
| GO:BP | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity | 1 | 10 | 5.22e-01 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 1 | 39 | 5.22e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 3 | 135 | 5.22e-01 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 2 | 29 | 5.22e-01 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 1 | 8 | 5.23e-01 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 1 | 8 | 5.23e-01 |
| GO:BP | GO:0050764 | regulation of phagocytosis | 1 | 57 | 5.23e-01 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 1 | 18 | 5.23e-01 |
| GO:BP | GO:0035094 | response to nicotine | 1 | 28 | 5.24e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 8 | 560 | 5.24e-01 |
| GO:BP | GO:0045494 | photoreceptor cell maintenance | 1 | 27 | 5.24e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 1 | 71 | 5.24e-01 |
| GO:BP | GO:0030073 | insulin secretion | 5 | 141 | 5.24e-01 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 1 | 19 | 5.24e-01 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 1 | 8 | 5.25e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 1 | 17 | 5.25e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 4 | 95 | 5.25e-01 |
| GO:BP | GO:0060438 | trachea development | 1 | 13 | 5.25e-01 |
| GO:BP | GO:0021553 | olfactory nerve development | 1 | 5 | 5.26e-01 |
| GO:BP | GO:0045071 | negative regulation of viral genome replication | 2 | 37 | 5.26e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 5.26e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 5.27e-01 |
| GO:BP | GO:0048539 | bone marrow development | 1 | 7 | 5.27e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 6 | 4292 | 5.27e-01 |
| GO:BP | GO:0045589 | regulation of regulatory T cell differentiation | 2 | 21 | 5.28e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 4 | 134 | 5.28e-01 |
| GO:BP | GO:0006739 | NADP metabolic process | 1 | 40 | 5.28e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 9 | 267 | 5.28e-01 |
| GO:BP | GO:0031453 | positive regulation of heterochromatin formation | 1 | 15 | 5.29e-01 |
| GO:BP | GO:0120263 | positive regulation of heterochromatin organization | 1 | 15 | 5.29e-01 |
| GO:BP | GO:0072311 | glomerular epithelial cell differentiation | 1 | 17 | 5.29e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 3 | 68 | 5.29e-01 |
| GO:BP | GO:0035067 | negative regulation of histone acetylation | 1 | 9 | 5.29e-01 |
| GO:BP | GO:0044057 | regulation of system process | 5 | 405 | 5.29e-01 |
| GO:BP | GO:0031061 | negative regulation of histone methylation | 1 | 9 | 5.29e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 14 | 387 | 5.29e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 3 | 54 | 5.29e-01 |
| GO:BP | GO:0061909 | autophagosome-lysosome fusion | 1 | 8 | 5.29e-01 |
| GO:BP | GO:0060416 | response to growth hormone | 1 | 25 | 5.30e-01 |
| GO:BP | GO:1905634 | regulation of protein localization to chromatin | 1 | 8 | 5.30e-01 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 4 | 127 | 5.30e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 1 | 126 | 5.30e-01 |
| GO:BP | GO:2001137 | positive regulation of endocytic recycling | 1 | 7 | 5.30e-01 |
| GO:BP | GO:0042220 | response to cocaine | 2 | 28 | 5.30e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 4 | 605 | 5.30e-01 |
| GO:BP | GO:0033363 | secretory granule organization | 1 | 50 | 5.31e-01 |
| GO:BP | GO:0060926 | cardiac pacemaker cell development | 1 | 8 | 5.31e-01 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 1 | 9 | 5.31e-01 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 1 | 14 | 5.31e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 5.31e-01 |
| GO:BP | GO:0098712 | L-glutamate import across plasma membrane | 1 | 11 | 5.31e-01 |
| GO:BP | GO:1901568 | fatty acid derivative metabolic process | 2 | 48 | 5.32e-01 |
| GO:BP | GO:0002729 | positive regulation of natural killer cell cytokine production | 1 | 4 | 5.32e-01 |
| GO:BP | GO:0002858 | regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 5.32e-01 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 2 | 24 | 5.32e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 5 | 185 | 5.32e-01 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 5 | 192 | 5.32e-01 |
| GO:BP | GO:0048821 | erythrocyte development | 3 | 30 | 5.32e-01 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 1 | 12 | 5.32e-01 |
| GO:BP | GO:0032652 | regulation of interleukin-1 production | 2 | 56 | 5.33e-01 |
| GO:BP | GO:0032612 | interleukin-1 production | 2 | 56 | 5.33e-01 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 1 | 19 | 5.33e-01 |
| GO:BP | GO:0006641 | triglyceride metabolic process | 2 | 68 | 5.33e-01 |
| GO:BP | GO:0071715 | icosanoid transport | 2 | 24 | 5.33e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 4 | 136 | 5.33e-01 |
| GO:BP | GO:0010890 | positive regulation of sequestering of triglyceride | 1 | 7 | 5.33e-01 |
| GO:BP | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 5 | 5.33e-01 |
| GO:BP | GO:0015695 | organic cation transport | 1 | 34 | 5.34e-01 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 1 | 22 | 5.34e-01 |
| GO:BP | GO:0007399 | nervous system development | 16 | 1904 | 5.34e-01 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 1 | 35 | 5.34e-01 |
| GO:BP | GO:0048026 | positive regulation of mRNA splicing, via spliceosome | 1 | 21 | 5.35e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 7 | 427 | 5.35e-01 |
| GO:BP | GO:0009791 | post-embryonic development | 1 | 74 | 5.35e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 1 | 448 | 5.35e-01 |
| GO:BP | GO:0030212 | hyaluronan metabolic process | 2 | 21 | 5.36e-01 |
| GO:BP | GO:1901077 | regulation of relaxation of muscle | 1 | 8 | 5.36e-01 |
| GO:BP | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | 1 | 12 | 5.36e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 5.36e-01 |
| GO:BP | GO:0042755 | eating behavior | 1 | 16 | 5.36e-01 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 1 | 54 | 5.36e-01 |
| GO:BP | GO:0043615 | astrocyte cell migration | 1 | 6 | 5.36e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 7 | 4617 | 5.37e-01 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 1 | 23 | 5.37e-01 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 2 | 19 | 5.37e-01 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 1 | 10 | 5.37e-01 |
| GO:BP | GO:1903429 | regulation of cell maturation | 1 | 13 | 5.37e-01 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 2 | 24 | 5.37e-01 |
| GO:BP | GO:0015833 | peptide transport | 5 | 179 | 5.37e-01 |
| GO:BP | GO:0001866 | NK T cell proliferation | 1 | 4 | 5.37e-01 |
| GO:BP | GO:0010940 | positive regulation of necrotic cell death | 1 | 13 | 5.37e-01 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 1 | 7 | 5.37e-01 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 2 | 95 | 5.37e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 1 | 25 | 5.38e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 2 | 321 | 5.38e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 6 | 162 | 5.39e-01 |
| GO:BP | GO:0009617 | response to bacterium | 3 | 321 | 5.39e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 5.39e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 5.39e-01 |
| GO:BP | GO:0070091 | glucagon secretion | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0070092 | regulation of glucagon secretion | 1 | 3 | 5.39e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 4 | 179 | 5.40e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 5 | 54 | 5.40e-01 |
| GO:BP | GO:0010761 | fibroblast migration | 2 | 52 | 5.40e-01 |
| GO:BP | GO:0031529 | ruffle organization | 2 | 45 | 5.40e-01 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 1 | 12 | 5.41e-01 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 1 | 14 | 5.41e-01 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 1 | 11 | 5.41e-01 |
| GO:BP | GO:0016032 | viral process | 6 | 344 | 5.42e-01 |
| GO:BP | GO:0035809 | regulation of urine volume | 1 | 14 | 5.42e-01 |
| GO:BP | GO:0007584 | response to nutrient | 1 | 109 | 5.42e-01 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 2 | 52 | 5.42e-01 |
| GO:BP | GO:0072530 | purine-containing compound transmembrane transport | 2 | 20 | 5.43e-01 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 1 | 8 | 5.43e-01 |
| GO:BP | GO:0002639 | positive regulation of immunoglobulin production | 1 | 30 | 5.43e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 5.43e-01 |
| GO:BP | GO:0070099 | regulation of chemokine-mediated signaling pathway | 1 | 8 | 5.43e-01 |
| GO:BP | GO:0045671 | negative regulation of osteoclast differentiation | 2 | 20 | 5.43e-01 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 1 | 29 | 5.43e-01 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 4 | 55 | 5.44e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 5.44e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 1 | 748 | 5.44e-01 |
| GO:BP | GO:0010288 | response to lead ion | 1 | 18 | 5.44e-01 |
| GO:BP | GO:0006022 | aminoglycan metabolic process | 6 | 93 | 5.44e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 4 | 139 | 5.44e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 2 | 25 | 5.44e-01 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 1 | 23 | 5.44e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 2 | 20 | 5.44e-01 |
| GO:BP | GO:0000052 | citrulline metabolic process | 1 | 7 | 5.44e-01 |
| GO:BP | GO:0050854 | regulation of antigen receptor-mediated signaling pathway | 1 | 40 | 5.44e-01 |
| GO:BP | GO:0140059 | dendrite arborization | 1 | 9 | 5.44e-01 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 3 | 60 | 5.44e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 2 | 25 | 5.44e-01 |
| GO:BP | GO:1905269 | positive regulation of chromatin organization | 1 | 15 | 5.44e-01 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 1 | 11 | 5.44e-01 |
| GO:BP | GO:0051001 | negative regulation of nitric-oxide synthase activity | 1 | 6 | 5.44e-01 |
| GO:BP | GO:0051151 | negative regulation of smooth muscle cell differentiation | 1 | 14 | 5.44e-01 |
| GO:BP | GO:0030279 | negative regulation of ossification | 1 | 25 | 5.44e-01 |
| GO:BP | GO:0090218 | positive regulation of lipid kinase activity | 1 | 25 | 5.44e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 7 | 98 | 5.44e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 2 | 1133 | 5.44e-01 |
| GO:BP | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process | 1 | 5 | 5.44e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 3 | 85 | 5.44e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 5 | 90 | 5.45e-01 |
| GO:BP | GO:0042304 | regulation of fatty acid biosynthetic process | 3 | 30 | 5.45e-01 |
| GO:BP | GO:0050658 | RNA transport | 5 | 144 | 5.45e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 5 | 144 | 5.45e-01 |
| GO:BP | GO:0046655 | folic acid metabolic process | 1 | 14 | 5.45e-01 |
| GO:BP | GO:0021983 | pituitary gland development | 1 | 22 | 5.45e-01 |
| GO:BP | GO:0071542 | dopaminergic neuron differentiation | 1 | 20 | 5.45e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 12 | 164 | 5.46e-01 |
| GO:BP | GO:0035588 | G protein-coupled purinergic receptor signaling pathway | 1 | 8 | 5.46e-01 |
| GO:BP | GO:0001973 | G protein-coupled adenosine receptor signaling pathway | 1 | 8 | 5.46e-01 |
| GO:BP | GO:0007610 | behavior | 9 | 436 | 5.46e-01 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 1 | 11 | 5.47e-01 |
| GO:BP | GO:0010560 | positive regulation of glycoprotein biosynthetic process | 1 | 15 | 5.47e-01 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 3 | 49 | 5.48e-01 |
| GO:BP | GO:0014014 | negative regulation of gliogenesis | 3 | 26 | 5.48e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 8 | 124 | 5.48e-01 |
| GO:BP | GO:0033198 | response to ATP | 1 | 24 | 5.49e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 9 | 251 | 5.49e-01 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 4 | 120 | 5.50e-01 |
| GO:BP | GO:0030154 | cell differentiation | 21 | 2958 | 5.50e-01 |
| GO:BP | GO:0098657 | import into cell | 8 | 166 | 5.51e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 3 | 132 | 5.51e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 3 | 80 | 5.51e-01 |
| GO:BP | GO:0051693 | actin filament capping | 1 | 34 | 5.51e-01 |
| GO:BP | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity | 1 | 3 | 5.51e-01 |
| GO:BP | GO:0002335 | mature B cell differentiation | 1 | 21 | 5.51e-01 |
| GO:BP | GO:0018146 | keratan sulfate biosynthetic process | 1 | 13 | 5.52e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 11 | 665 | 5.52e-01 |
| GO:BP | GO:0007190 | activation of adenylate cyclase activity | 2 | 13 | 5.52e-01 |
| GO:BP | GO:0007340 | acrosome reaction | 1 | 20 | 5.52e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 2 | 26 | 5.52e-01 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 2 | 26 | 5.52e-01 |
| GO:BP | GO:0046940 | nucleoside monophosphate phosphorylation | 1 | 9 | 5.52e-01 |
| GO:BP | GO:0099022 | vesicle tethering | 2 | 31 | 5.52e-01 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 1 | 19 | 5.52e-01 |
| GO:BP | GO:0000578 | embryonic axis specification | 1 | 26 | 5.52e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 5 | 70 | 5.52e-01 |
| GO:BP | GO:0009437 | carnitine metabolic process | 1 | 11 | 5.52e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 2 | 67 | 5.52e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 3 | 159 | 5.53e-01 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 5 | 57 | 5.54e-01 |
| GO:BP | GO:0007254 | JNK cascade | 3 | 138 | 5.55e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 2 | 34 | 5.55e-01 |
| GO:BP | GO:0005984 | disaccharide metabolic process | 1 | 5 | 5.55e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 4 | 120 | 5.55e-01 |
| GO:BP | GO:0030878 | thyroid gland development | 2 | 17 | 5.56e-01 |
| GO:BP | GO:0090219 | negative regulation of lipid kinase activity | 1 | 8 | 5.57e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 6 | 4417 | 5.57e-01 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 1 | 26 | 5.57e-01 |
| GO:BP | GO:0060920 | cardiac pacemaker cell differentiation | 1 | 9 | 5.59e-01 |
| GO:BP | GO:0051452 | intracellular pH reduction | 4 | 45 | 5.59e-01 |
| GO:BP | GO:0006887 | exocytosis | 9 | 256 | 5.59e-01 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 1 | 13 | 5.59e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 4 | 294 | 5.60e-01 |
| GO:BP | GO:0007601 | visual perception | 1 | 96 | 5.60e-01 |
| GO:BP | GO:0048513 | animal organ development | 2 | 2189 | 5.60e-01 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 3 | 35 | 5.60e-01 |
| GO:BP | GO:0015908 | fatty acid transport | 3 | 58 | 5.61e-01 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 2 | 29 | 5.61e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 4 | 276 | 5.61e-01 |
| GO:BP | GO:0072010 | glomerular epithelium development | 1 | 18 | 5.61e-01 |
| GO:BP | GO:0061744 | motor behavior | 1 | 17 | 5.61e-01 |
| GO:BP | GO:0036124 | histone H3-K9 trimethylation | 1 | 9 | 5.61e-01 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 5.61e-01 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 5.61e-01 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 1 | 19 | 5.61e-01 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 1 | 19 | 5.61e-01 |
| GO:BP | GO:0045066 | regulatory T cell differentiation | 2 | 22 | 5.61e-01 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 1 | 38 | 5.61e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 2 | 64 | 5.61e-01 |
| GO:BP | GO:0008053 | mitochondrial fusion | 1 | 28 | 5.61e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 2 | 28 | 5.61e-01 |
| GO:BP | GO:0016571 | histone methylation | 2 | 117 | 5.61e-01 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 2 | 23 | 5.61e-01 |
| GO:BP | GO:0099601 | regulation of neurotransmitter receptor activity | 2 | 40 | 5.61e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 21 | 2984 | 5.61e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 4 | 295 | 5.61e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 1 | 22 | 5.61e-01 |
| GO:BP | GO:0031060 | regulation of histone methylation | 4 | 47 | 5.61e-01 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 2 | 41 | 5.61e-01 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 2 | 23 | 5.61e-01 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 2 | 23 | 5.61e-01 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 1 | 73 | 5.61e-01 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 2 | 23 | 5.61e-01 |
| GO:BP | GO:0002370 | natural killer cell cytokine production | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0002727 | regulation of natural killer cell cytokine production | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0002855 | regulation of natural killer cell mediated immune response to tumor cell | 1 | 4 | 5.61e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 1 | 41 | 5.61e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 26 | 478 | 5.61e-01 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 1 | 140 | 5.61e-01 |
| GO:BP | GO:0097050 | type B pancreatic cell apoptotic process | 1 | 7 | 5.62e-01 |
| GO:BP | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 5 | 59 | 5.62e-01 |
| GO:BP | GO:2000109 | regulation of macrophage apoptotic process | 1 | 6 | 5.62e-01 |
| GO:BP | GO:0007020 | microtubule nucleation | 2 | 39 | 5.62e-01 |
| GO:BP | GO:0001913 | T cell mediated cytotoxicity | 1 | 22 | 5.62e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 24 | 351 | 5.62e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 3 | 42 | 5.62e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 3 | 277 | 5.63e-01 |
| GO:BP | GO:0051028 | mRNA transport | 4 | 117 | 5.63e-01 |
| GO:BP | GO:0003008 | system process | 1 | 1189 | 5.63e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 5.63e-01 |
| GO:BP | GO:0002902 | regulation of B cell apoptotic process | 1 | 15 | 5.64e-01 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 1 | 26 | 5.64e-01 |
| GO:BP | GO:0051602 | response to electrical stimulus | 1 | 26 | 5.64e-01 |
| GO:BP | GO:0007309 | oocyte axis specification | 1 | 3 | 5.64e-01 |
| GO:BP | GO:0007308 | oocyte construction | 1 | 3 | 5.64e-01 |
| GO:BP | GO:0034969 | histone arginine methylation | 1 | 8 | 5.64e-01 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 2 | 54 | 5.64e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 4 | 3070 | 5.64e-01 |
| GO:BP | GO:0015732 | prostaglandin transport | 1 | 10 | 5.64e-01 |
| GO:BP | GO:0140352 | export from cell | 4 | 611 | 5.64e-01 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 2 | 60 | 5.65e-01 |
| GO:BP | GO:1904321 | response to forskolin | 1 | 11 | 5.65e-01 |
| GO:BP | GO:1904322 | cellular response to forskolin | 1 | 11 | 5.65e-01 |
| GO:BP | GO:0140253 | cell-cell fusion | 3 | 37 | 5.68e-01 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 3 | 37 | 5.68e-01 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 1 | 9 | 5.68e-01 |
| GO:BP | GO:0006364 | rRNA processing | 3 | 221 | 5.68e-01 |
| GO:BP | GO:0060837 | blood vessel endothelial cell differentiation | 1 | 10 | 5.68e-01 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 1 | 15 | 5.68e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 5 | 168 | 5.69e-01 |
| GO:BP | GO:0035521 | monoubiquitinated histone deubiquitination | 2 | 30 | 5.69e-01 |
| GO:BP | GO:0035522 | monoubiquitinated histone H2A deubiquitination | 2 | 30 | 5.69e-01 |
| GO:BP | GO:0048731 | system development | 3 | 2901 | 5.69e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 1 | 28 | 5.69e-01 |
| GO:BP | GO:0030252 | growth hormone secretion | 1 | 12 | 5.69e-01 |
| GO:BP | GO:0046639 | negative regulation of alpha-beta T cell differentiation | 2 | 17 | 5.69e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 2 | 104 | 5.69e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 27 | 5.70e-01 |
| GO:BP | GO:0030325 | adrenal gland development | 1 | 22 | 5.71e-01 |
| GO:BP | GO:0048246 | macrophage chemotaxis | 1 | 21 | 5.71e-01 |
| GO:BP | GO:0009880 | embryonic pattern specification | 5 | 50 | 5.71e-01 |
| GO:BP | GO:0009235 | cobalamin metabolic process | 1 | 8 | 5.71e-01 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 1 | 14 | 5.72e-01 |
| GO:BP | GO:0045907 | positive regulation of vasoconstriction | 1 | 15 | 5.72e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 4 | 782 | 5.73e-01 |
| GO:BP | GO:0032543 | mitochondrial translation | 1 | 130 | 5.73e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 5.73e-01 |
| GO:BP | GO:0015810 | aspartate transmembrane transport | 1 | 16 | 5.74e-01 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 1 | 37 | 5.74e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 10 | 656 | 5.74e-01 |
| GO:BP | GO:0061101 | neuroendocrine cell differentiation | 1 | 9 | 5.74e-01 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 1 | 13 | 5.74e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 1 | 147 | 5.74e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 12 | 998 | 5.74e-01 |
| GO:BP | GO:0043029 | T cell homeostasis | 1 | 27 | 5.74e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 6 | 307 | 5.74e-01 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 4 | 121 | 5.74e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 4 | 4099 | 5.74e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 2 | 53 | 5.74e-01 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 1 | 9 | 5.75e-01 |
| GO:BP | GO:0006952 | defense response | 12 | 944 | 5.75e-01 |
| GO:BP | GO:0000096 | sulfur amino acid metabolic process | 2 | 27 | 5.75e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 1 | 149 | 5.75e-01 |
| GO:BP | GO:0018243 | protein O-linked glycosylation via threonine | 1 | 7 | 5.75e-01 |
| GO:BP | GO:0030575 | nuclear body organization | 1 | 14 | 5.75e-01 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 1 | 29 | 5.75e-01 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 1 | 21 | 5.76e-01 |
| GO:BP | GO:0061157 | mRNA destabilization | 3 | 89 | 5.76e-01 |
| GO:BP | GO:0046348 | amino sugar catabolic process | 1 | 9 | 5.76e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 5.76e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 5.76e-01 |
| GO:BP | GO:0003091 | renal water homeostasis | 1 | 13 | 5.76e-01 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 1 | 27 | 5.77e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 8 | 97 | 5.78e-01 |
| GO:BP | GO:0050821 | protein stabilization | 1 | 191 | 5.78e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 4 | 3132 | 5.79e-01 |
| GO:BP | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | 2 | 19 | 5.79e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 2 | 35 | 5.79e-01 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 1 | 71 | 5.79e-01 |
| GO:BP | GO:0035383 | thioester metabolic process | 1 | 71 | 5.79e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 5 | 76 | 5.79e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 13 | 5.79e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 5 | 3750 | 5.79e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 3 | 247 | 5.79e-01 |
| GO:BP | GO:0042308 | negative regulation of protein import into nucleus | 1 | 13 | 5.80e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 4 | 3133 | 5.81e-01 |
| GO:BP | GO:0045116 | protein neddylation | 1 | 26 | 5.81e-01 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 1 | 38 | 5.81e-01 |
| GO:BP | GO:0030850 | prostate gland development | 1 | 35 | 5.81e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 6 | 150 | 5.81e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 1 | 25 | 5.81e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 3 | 97 | 5.81e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 2 | 74 | 5.82e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 1 | 701 | 5.83e-01 |
| GO:BP | GO:0051900 | regulation of mitochondrial depolarization | 1 | 15 | 5.83e-01 |
| GO:BP | GO:0042339 | keratan sulfate metabolic process | 1 | 15 | 5.83e-01 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 1 | 58 | 5.84e-01 |
| GO:BP | GO:1905521 | regulation of macrophage migration | 1 | 24 | 5.84e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 2 | 83 | 5.84e-01 |
| GO:BP | GO:2000249 | regulation of actin cytoskeleton reorganization | 1 | 32 | 5.84e-01 |
| GO:BP | GO:0016024 | CDP-diacylglycerol biosynthetic process | 1 | 12 | 5.84e-01 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 2 | 27 | 5.84e-01 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 2 | 28 | 5.84e-01 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 1 | 31 | 5.84e-01 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 1 | 12 | 5.85e-01 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 1 | 15 | 5.86e-01 |
| GO:BP | GO:0006825 | copper ion transport | 1 | 13 | 5.86e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 1 | 31 | 5.87e-01 |
| GO:BP | GO:0098917 | retrograde trans-synaptic signaling | 1 | 7 | 5.87e-01 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 1 | 10 | 5.87e-01 |
| GO:BP | GO:0007625 | grooming behavior | 1 | 10 | 5.87e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 3 | 131 | 5.88e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 5.88e-01 |
| GO:BP | GO:0001890 | placenta development | 6 | 117 | 5.88e-01 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 3 | 47 | 5.88e-01 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 1 | 16 | 5.88e-01 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 4 | 105 | 5.88e-01 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 2 | 27 | 5.88e-01 |
| GO:BP | GO:0030214 | hyaluronan catabolic process | 1 | 11 | 5.89e-01 |
| GO:BP | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus | 1 | 18 | 5.89e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 1 | 17 | 5.89e-01 |
| GO:BP | GO:0006337 | nucleosome disassembly | 1 | 16 | 5.89e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 6 | 69 | 5.90e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 5.90e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 3 | 75 | 5.90e-01 |
| GO:BP | GO:0021548 | pons development | 1 | 7 | 5.90e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 2 | 1101 | 5.90e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 4 | 78 | 5.90e-01 |
| GO:BP | GO:0075733 | intracellular transport of virus | 1 | 8 | 5.90e-01 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 5 | 61 | 5.90e-01 |
| GO:BP | GO:0090308 | regulation of DNA methylation-dependent heterochromatin formation | 1 | 16 | 5.90e-01 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 3 | 66 | 5.90e-01 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 1 | 19 | 5.91e-01 |
| GO:BP | GO:0051133 | regulation of NK T cell activation | 1 | 4 | 5.91e-01 |
| GO:BP | GO:0021984 | adenohypophysis development | 1 | 9 | 5.91e-01 |
| GO:BP | GO:0002423 | natural killer cell mediated immune response to tumor cell | 1 | 4 | 5.91e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 15 | 691 | 5.91e-01 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 1 | 12 | 5.91e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 2 | 82 | 5.92e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 2 | 963 | 5.92e-01 |
| GO:BP | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 1 | 11 | 5.93e-01 |
| GO:BP | GO:1902667 | regulation of axon guidance | 1 | 6 | 5.93e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 5.93e-01 |
| GO:BP | GO:0033004 | negative regulation of mast cell activation | 1 | 4 | 5.93e-01 |
| GO:BP | GO:0043301 | negative regulation of leukocyte degranulation | 1 | 5 | 5.93e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 3 | 165 | 5.93e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 2 | 28 | 5.93e-01 |
| GO:BP | GO:0060525 | prostate glandular acinus development | 1 | 5 | 5.93e-01 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 5 | 96 | 5.93e-01 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 2 | 33 | 5.93e-01 |
| GO:BP | GO:0055094 | response to lipoprotein particle | 2 | 20 | 5.93e-01 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 2 | 26 | 5.93e-01 |
| GO:BP | GO:0033622 | integrin activation | 1 | 18 | 5.93e-01 |
| GO:BP | GO:0002712 | regulation of B cell mediated immunity | 1 | 39 | 5.93e-01 |
| GO:BP | GO:0045922 | negative regulation of fatty acid metabolic process | 2 | 25 | 5.93e-01 |
| GO:BP | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development | 1 | 9 | 5.93e-01 |
| GO:BP | GO:0002889 | regulation of immunoglobulin mediated immune response | 1 | 39 | 5.93e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 2 | 23 | 5.93e-01 |
| GO:BP | GO:0034698 | response to gonadotropin | 2 | 19 | 5.93e-01 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 1 | 37 | 5.93e-01 |
| GO:BP | GO:0009133 | nucleoside diphosphate biosynthetic process | 1 | 11 | 5.93e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 1 | 159 | 5.93e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 3 | 290 | 5.94e-01 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 2 | 113 | 5.94e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 1 | 50 | 5.94e-01 |
| GO:BP | GO:0007520 | myoblast fusion | 1 | 30 | 5.94e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 2 | 134 | 5.94e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 1 | 28 | 5.95e-01 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 1 | 53 | 5.95e-01 |
| GO:BP | GO:0046470 | phosphatidylcholine metabolic process | 2 | 53 | 5.95e-01 |
| GO:BP | GO:0042749 | regulation of circadian sleep/wake cycle | 1 | 9 | 5.97e-01 |
| GO:BP | GO:0055015 | ventricular cardiac muscle cell development | 1 | 10 | 5.97e-01 |
| GO:BP | GO:0002449 | lymphocyte mediated immunity | 2 | 142 | 5.97e-01 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 3 | 93 | 5.98e-01 |
| GO:BP | GO:0050779 | RNA destabilization | 3 | 92 | 5.98e-01 |
| GO:BP | GO:0007202 | activation of phospholipase C activity | 1 | 10 | 5.99e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 2 | 68 | 5.99e-01 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 2 | 23 | 5.99e-01 |
| GO:BP | GO:0032801 | receptor catabolic process | 1 | 24 | 5.99e-01 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 1 | 19 | 5.99e-01 |
| GO:BP | GO:0001505 | regulation of neurotransmitter levels | 1 | 149 | 5.99e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 2 | 1256 | 5.99e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 2 | 1255 | 6.00e-01 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 1 | 67 | 6.00e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 1 | 161 | 6.00e-01 |
| GO:BP | GO:0002702 | positive regulation of production of molecular mediator of immune response | 2 | 77 | 6.01e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 2 | 21 | 6.01e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 1 | 51 | 6.01e-01 |
| GO:BP | GO:0061045 | negative regulation of wound healing | 1 | 51 | 6.01e-01 |
| GO:BP | GO:0030889 | negative regulation of B cell proliferation | 1 | 9 | 6.01e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 10 | 6.01e-01 |
| GO:BP | GO:0006949 | syncytium formation | 3 | 39 | 6.01e-01 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 1 | 21 | 6.01e-01 |
| GO:BP | GO:0032732 | positive regulation of interleukin-1 production | 2 | 35 | 6.02e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 2 | 89 | 6.02e-01 |
| GO:BP | GO:0002377 | immunoglobulin production | 7 | 70 | 6.03e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 5 | 114 | 6.03e-01 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 4 | 49 | 6.03e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 6.04e-01 |
| GO:BP | GO:0006527 | arginine catabolic process | 1 | 7 | 6.04e-01 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 1 | 10 | 6.04e-01 |
| GO:BP | GO:0061515 | myeloid cell development | 1 | 57 | 6.04e-01 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 1 | 10 | 6.04e-01 |
| GO:BP | GO:0046341 | CDP-diacylglycerol metabolic process | 1 | 13 | 6.04e-01 |
| GO:BP | GO:0072537 | fibroblast activation | 1 | 11 | 6.04e-01 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 2 | 276 | 6.04e-01 |
| GO:BP | GO:0007205 | protein kinase C-activating G protein-coupled receptor signaling pathway | 2 | 19 | 6.04e-01 |
| GO:BP | GO:0009303 | rRNA transcription | 2 | 37 | 6.04e-01 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 1 | 14 | 6.04e-01 |
| GO:BP | GO:0007141 | male meiosis I | 2 | 15 | 6.04e-01 |
| GO:BP | GO:1990266 | neutrophil migration | 2 | 52 | 6.05e-01 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 28 | 6.05e-01 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 1 | 9 | 6.05e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 2 | 29 | 6.05e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 14 | 406 | 6.05e-01 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 2 | 63 | 6.06e-01 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 1 | 17 | 6.06e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 6.06e-01 |
| GO:BP | GO:0009173 | pyrimidine ribonucleoside monophosphate metabolic process | 1 | 12 | 6.06e-01 |
| GO:BP | GO:2000672 | negative regulation of motor neuron apoptotic process | 1 | 10 | 6.06e-01 |
| GO:BP | GO:1902065 | response to L-glutamate | 1 | 11 | 6.06e-01 |
| GO:BP | GO:0032890 | regulation of organic acid transport | 2 | 49 | 6.06e-01 |
| GO:BP | GO:0003163 | sinoatrial node development | 1 | 10 | 6.06e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 2 | 158 | 6.07e-01 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 1 | 31 | 6.07e-01 |
| GO:BP | GO:0051169 | nuclear transport | 8 | 295 | 6.07e-01 |
| GO:BP | GO:0070293 | renal absorption | 1 | 24 | 6.07e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 7 | 90 | 6.07e-01 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 4 | 49 | 6.07e-01 |
| GO:BP | GO:0010043 | response to zinc ion | 2 | 31 | 6.07e-01 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 1 | 31 | 6.07e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 8 | 295 | 6.07e-01 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 1 | 10 | 6.07e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 4 | 142 | 6.08e-01 |
| GO:BP | GO:0070988 | demethylation | 1 | 52 | 6.09e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 3 | 259 | 6.09e-01 |
| GO:BP | GO:0060856 | establishment of blood-brain barrier | 1 | 9 | 6.09e-01 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 1 | 17 | 6.09e-01 |
| GO:BP | GO:0034605 | cellular response to heat | 1 | 51 | 6.09e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 6.09e-01 |
| GO:BP | GO:1901652 | response to peptide | 2 | 377 | 6.09e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 4 | 3012 | 6.09e-01 |
| GO:BP | GO:0034370 | triglyceride-rich lipoprotein particle remodeling | 1 | 4 | 6.09e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 38 | 6.09e-01 |
| GO:BP | GO:0034372 | very-low-density lipoprotein particle remodeling | 1 | 4 | 6.09e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 3 | 300 | 6.09e-01 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 1 | 10 | 6.09e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 5 | 47 | 6.09e-01 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 1 | 15 | 6.09e-01 |
| GO:BP | GO:2000757 | negative regulation of peptidyl-lysine acetylation | 1 | 13 | 6.10e-01 |
| GO:BP | GO:0051570 | regulation of histone H3-K9 methylation | 1 | 11 | 6.10e-01 |
| GO:BP | GO:0032941 | secretion by tissue | 1 | 25 | 6.10e-01 |
| GO:BP | GO:0097531 | mast cell migration | 1 | 12 | 6.10e-01 |
| GO:BP | GO:0098664 | G protein-coupled serotonin receptor signaling pathway | 2 | 8 | 6.10e-01 |
| GO:BP | GO:0006570 | tyrosine metabolic process | 1 | 6 | 6.11e-01 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 4 | 50 | 6.11e-01 |
| GO:BP | GO:0110011 | regulation of basement membrane organization | 1 | 11 | 6.11e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 7 | 153 | 6.11e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 2 | 1223 | 6.12e-01 |
| GO:BP | GO:1900117 | regulation of execution phase of apoptosis | 1 | 13 | 6.12e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 31 | 6.12e-01 |
| GO:BP | GO:0030516 | regulation of axon extension | 6 | 84 | 6.12e-01 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 1 | 17 | 6.13e-01 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 1 | 30 | 6.13e-01 |
| GO:BP | GO:0006544 | glycine metabolic process | 1 | 10 | 6.13e-01 |
| GO:BP | GO:0048012 | hepatocyte growth factor receptor signaling pathway | 1 | 10 | 6.13e-01 |
| GO:BP | GO:0050482 | arachidonic acid secretion | 1 | 11 | 6.13e-01 |
| GO:BP | GO:0072017 | distal tubule development | 1 | 6 | 6.13e-01 |
| GO:BP | GO:1903963 | arachidonate transport | 1 | 11 | 6.13e-01 |
| GO:BP | GO:0072539 | T-helper 17 cell differentiation | 2 | 14 | 6.13e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 4 | 57 | 6.13e-01 |
| GO:BP | GO:0046903 | secretion | 4 | 645 | 6.13e-01 |
| GO:BP | GO:0001824 | blastocyst development | 7 | 94 | 6.13e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 8 | 100 | 6.13e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 6 | 133 | 6.14e-01 |
| GO:BP | GO:0060259 | regulation of feeding behavior | 2 | 9 | 6.14e-01 |
| GO:BP | GO:0033344 | cholesterol efflux | 2 | 40 | 6.14e-01 |
| GO:BP | GO:0048857 | neural nucleus development | 4 | 50 | 6.14e-01 |
| GO:BP | GO:0032986 | protein-DNA complex disassembly | 1 | 18 | 6.14e-01 |
| GO:BP | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway | 1 | 10 | 6.15e-01 |
| GO:BP | GO:0014002 | astrocyte development | 1 | 27 | 6.15e-01 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 1 | 174 | 6.15e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 2 | 21 | 6.15e-01 |
| GO:BP | GO:0016197 | endosomal transport | 2 | 231 | 6.15e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 4 | 830 | 6.16e-01 |
| GO:BP | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 1 | 13 | 6.16e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 11 | 395 | 6.16e-01 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | 24 | 6.16e-01 |
| GO:BP | GO:0015844 | monoamine transport | 1 | 45 | 6.16e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 6.16e-01 |
| GO:BP | GO:1903350 | response to dopamine | 5 | 48 | 6.16e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 2 | 676 | 6.16e-01 |
| GO:BP | GO:0098792 | xenophagy | 1 | 12 | 6.16e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 1 | 34 | 6.17e-01 |
| GO:BP | GO:0015711 | organic anion transport | 1 | 264 | 6.17e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 31 | 2593 | 6.17e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 1 | 165 | 6.17e-01 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 1 | 23 | 6.17e-01 |
| GO:BP | GO:0060742 | epithelial cell differentiation involved in prostate gland development | 1 | 8 | 6.17e-01 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 1 | 10 | 6.17e-01 |
| GO:BP | GO:0035272 | exocrine system development | 3 | 34 | 6.17e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 4 | 2890 | 6.18e-01 |
| GO:BP | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry | 1 | 10 | 6.18e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 1 | 16 | 6.18e-01 |
| GO:BP | GO:0001654 | eye development | 6 | 272 | 6.18e-01 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 1 | 45 | 6.18e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 7 | 213 | 6.18e-01 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 1 | 18 | 6.18e-01 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 2 | 28 | 6.19e-01 |
| GO:BP | GO:0106058 | positive regulation of calcineurin-mediated signaling | 1 | 16 | 6.19e-01 |
| GO:BP | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade | 1 | 16 | 6.19e-01 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 2 | 21 | 6.19e-01 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 1 | 19 | 6.20e-01 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 1 | 19 | 6.20e-01 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 1 | 43 | 6.20e-01 |
| GO:BP | GO:0090672 | telomerase RNA localization | 1 | 19 | 6.20e-01 |
| GO:BP | GO:0015918 | sterol transport | 4 | 92 | 6.20e-01 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 1 | 19 | 6.20e-01 |
| GO:BP | GO:2000272 | negative regulation of signaling receptor activity | 3 | 33 | 6.20e-01 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 6 | 91 | 6.21e-01 |
| GO:BP | GO:0032365 | intracellular lipid transport | 1 | 42 | 6.21e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 1 | 58 | 6.21e-01 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 2 | 30 | 6.23e-01 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 1 | 21 | 6.23e-01 |
| GO:BP | GO:2001222 | regulation of neuron migration | 1 | 38 | 6.23e-01 |
| GO:BP | GO:0006577 | amino-acid betaine metabolic process | 1 | 13 | 6.24e-01 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 2 | 31 | 6.24e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 7 | 5017 | 6.24e-01 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 1 | 104 | 6.24e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 1 | 70 | 6.26e-01 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 1 | 11 | 6.26e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 6.26e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 1 | 44 | 6.26e-01 |
| GO:BP | GO:0046879 | hormone secretion | 1 | 210 | 6.26e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 6.27e-01 |
| GO:BP | GO:0150063 | visual system development | 6 | 273 | 6.27e-01 |
| GO:BP | GO:2000316 | regulation of T-helper 17 type immune response | 2 | 16 | 6.27e-01 |
| GO:BP | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process | 1 | 12 | 6.27e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 1 | 90 | 6.27e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 8 | 317 | 6.27e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 1 | 17 | 6.27e-01 |
| GO:BP | GO:0048286 | lung alveolus development | 1 | 39 | 6.28e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 2 | 120 | 6.28e-01 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 4 | 44 | 6.28e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 4 | 3080 | 6.28e-01 |
| GO:BP | GO:0032693 | negative regulation of interleukin-10 production | 1 | 7 | 6.28e-01 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 1 | 122 | 6.28e-01 |
| GO:BP | GO:0021860 | pyramidal neuron development | 1 | 8 | 6.28e-01 |
| GO:BP | GO:0006954 | inflammatory response | 8 | 429 | 6.28e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 2 | 40 | 6.28e-01 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 2 | 19 | 6.28e-01 |
| GO:BP | GO:0044058 | regulation of digestive system process | 1 | 22 | 6.28e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 6.28e-01 |
| GO:BP | GO:0048520 | positive regulation of behavior | 2 | 13 | 6.28e-01 |
| GO:BP | GO:0035066 | positive regulation of histone acetylation | 2 | 25 | 6.28e-01 |
| GO:BP | GO:0051569 | regulation of histone H3-K4 methylation | 2 | 26 | 6.28e-01 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 1 | 20 | 6.28e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 2 | 100 | 6.29e-01 |
| GO:BP | GO:0017157 | regulation of exocytosis | 2 | 147 | 6.29e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 1 | 66 | 6.30e-01 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 1 | 162 | 6.30e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 8 | 160 | 6.30e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 5 | 61 | 6.30e-01 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 1 | 29 | 6.31e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 6.32e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 1 | 30 | 6.32e-01 |
| GO:BP | GO:1901894 | regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | 9 | 6.32e-01 |
| GO:BP | GO:0051775 | response to redox state | 1 | 12 | 6.32e-01 |
| GO:BP | GO:0072540 | T-helper 17 cell lineage commitment | 1 | 3 | 6.32e-01 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 1 | 23 | 6.33e-01 |
| GO:BP | GO:0097475 | motor neuron migration | 1 | 10 | 6.33e-01 |
| GO:BP | GO:0071294 | cellular response to zinc ion | 1 | 15 | 6.33e-01 |
| GO:BP | GO:0042745 | circadian sleep/wake cycle | 1 | 11 | 6.33e-01 |
| GO:BP | GO:0007097 | nuclear migration | 1 | 23 | 6.33e-01 |
| GO:BP | GO:0071402 | cellular response to lipoprotein particle stimulus | 2 | 23 | 6.34e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 3 | 64 | 6.34e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 32 | 2714 | 6.34e-01 |
| GO:BP | GO:0032288 | myelin assembly | 1 | 19 | 6.34e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 8 | 256 | 6.34e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 1 | 76 | 6.34e-01 |
| GO:BP | GO:0051132 | NK T cell activation | 1 | 5 | 6.34e-01 |
| GO:BP | GO:0009948 | anterior/posterior axis specification | 1 | 32 | 6.34e-01 |
| GO:BP | GO:0045471 | response to ethanol | 2 | 73 | 6.34e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 1 | 34 | 6.34e-01 |
| GO:BP | GO:0009914 | hormone transport | 1 | 215 | 6.35e-01 |
| GO:BP | GO:0002437 | inflammatory response to antigenic stimulus | 1 | 36 | 6.35e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 3 | 2285 | 6.35e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 2 | 48 | 6.36e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 32 | 726 | 6.37e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 2 | 31 | 6.37e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 3 | 92 | 6.37e-01 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 1 | 12 | 6.38e-01 |
| GO:BP | GO:1902931 | negative regulation of alcohol biosynthetic process | 1 | 10 | 6.38e-01 |
| GO:BP | GO:0048284 | organelle fusion | 1 | 135 | 6.38e-01 |
| GO:BP | GO:0048880 | sensory system development | 6 | 275 | 6.38e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 1 | 31 | 6.38e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 6.38e-01 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 1 | 15 | 6.38e-01 |
| GO:BP | GO:0070509 | calcium ion import | 2 | 37 | 6.38e-01 |
| GO:BP | GO:1903798 | regulation of miRNA processing | 1 | 10 | 6.39e-01 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 1 | 19 | 6.39e-01 |
| GO:BP | GO:0032731 | positive regulation of interleukin-1 beta production | 1 | 30 | 6.39e-01 |
| GO:BP | GO:0002637 | regulation of immunoglobulin production | 1 | 43 | 6.40e-01 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 2 | 49 | 6.41e-01 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 1 | 79 | 6.41e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 6.41e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 6.41e-01 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 1 | 8 | 6.41e-01 |
| GO:BP | GO:0032634 | interleukin-5 production | 1 | 8 | 6.41e-01 |
| GO:BP | GO:0002679 | respiratory burst involved in defense response | 1 | 7 | 6.41e-01 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 3 | 46 | 6.41e-01 |
| GO:BP | GO:0030042 | actin filament depolymerization | 1 | 50 | 6.42e-01 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 1 | 17 | 6.42e-01 |
| GO:BP | GO:0051647 | nucleus localization | 2 | 28 | 6.42e-01 |
| GO:BP | GO:0006414 | translational elongation | 2 | 65 | 6.44e-01 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 1 | 54 | 6.44e-01 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 2 | 25 | 6.44e-01 |
| GO:BP | GO:0010842 | retina layer formation | 1 | 16 | 6.44e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 6.44e-01 |
| GO:BP | GO:0007263 | nitric oxide mediated signal transduction | 2 | 20 | 6.44e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 6 | 194 | 6.44e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 7 | 249 | 6.45e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 2 | 468 | 6.46e-01 |
| GO:BP | GO:0015867 | ATP transport | 1 | 9 | 6.46e-01 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 2 | 36 | 6.46e-01 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 1 | 26 | 6.46e-01 |
| GO:BP | GO:0033865 | nucleoside bisphosphate metabolic process | 1 | 92 | 6.46e-01 |
| GO:BP | GO:0034032 | purine nucleoside bisphosphate metabolic process | 1 | 92 | 6.46e-01 |
| GO:BP | GO:0007218 | neuropeptide signaling pathway | 1 | 36 | 6.46e-01 |
| GO:BP | GO:0033875 | ribonucleoside bisphosphate metabolic process | 1 | 92 | 6.46e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 4 | 310 | 6.47e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 42 | 6.48e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 4 | 223 | 6.48e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 2 | 64 | 6.48e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 7 | 320 | 6.49e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 4 | 3169 | 6.49e-01 |
| GO:BP | GO:1903715 | regulation of aerobic respiration | 1 | 28 | 6.49e-01 |
| GO:BP | GO:0000097 | sulfur amino acid biosynthetic process | 1 | 14 | 6.49e-01 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 1 | 40 | 6.49e-01 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 1 | 12 | 6.49e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 3 | 70 | 6.51e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 6.51e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 2 | 128 | 6.51e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 6.51e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 3 | 258 | 6.52e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 6.52e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 2 | 57 | 6.53e-01 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 1 | 37 | 6.53e-01 |
| GO:BP | GO:0010038 | response to metal ion | 6 | 264 | 6.53e-01 |
| GO:BP | GO:0072148 | epithelial cell fate commitment | 1 | 9 | 6.53e-01 |
| GO:BP | GO:0032488 | Cdc42 protein signal transduction | 1 | 10 | 6.53e-01 |
| GO:BP | GO:0006521 | regulation of cellular amino acid metabolic process | 1 | 7 | 6.53e-01 |
| GO:BP | GO:0032769 | negative regulation of monooxygenase activity | 1 | 8 | 6.53e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 3 | 211 | 6.54e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 6.54e-01 |
| GO:BP | GO:0032757 | positive regulation of interleukin-8 production | 1 | 31 | 6.54e-01 |
| GO:BP | GO:0005513 | detection of calcium ion | 1 | 12 | 6.54e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 10 | 4245 | 6.54e-01 |
| GO:BP | GO:1904424 | regulation of GTP binding | 1 | 16 | 6.54e-01 |
| GO:BP | GO:0030033 | microvillus assembly | 1 | 15 | 6.54e-01 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 1 | 38 | 6.54e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 2 | 64 | 6.54e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 4 | 108 | 6.54e-01 |
| GO:BP | GO:0008045 | motor neuron axon guidance | 2 | 25 | 6.54e-01 |
| GO:BP | GO:0032656 | regulation of interleukin-13 production | 1 | 10 | 6.55e-01 |
| GO:BP | GO:0032616 | interleukin-13 production | 1 | 10 | 6.55e-01 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 1 | 29 | 6.55e-01 |
| GO:BP | GO:0002224 | toll-like receptor signaling pathway | 1 | 43 | 6.55e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 6.56e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 7 | 1507 | 6.56e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 6.56e-01 |
| GO:BP | GO:0061053 | somite development | 3 | 63 | 6.56e-01 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 1 | 15 | 6.57e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 2 | 43 | 6.57e-01 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 1 | 17 | 6.57e-01 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 1 | 60 | 6.57e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 6.57e-01 |
| GO:BP | GO:0046697 | decidualization | 1 | 19 | 6.57e-01 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 1 | 29 | 6.57e-01 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 1 | 29 | 6.57e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 1 | 185 | 6.57e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 1 | 135 | 6.57e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 1 | 49 | 6.57e-01 |
| GO:BP | GO:0035459 | vesicle cargo loading | 2 | 27 | 6.59e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 1 | 12 | 6.60e-01 |
| GO:BP | GO:0030836 | positive regulation of actin filament depolymerization | 1 | 10 | 6.60e-01 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 1 | 10 | 6.60e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 20 | 287 | 6.60e-01 |
| GO:BP | GO:0014003 | oligodendrocyte development | 1 | 27 | 6.60e-01 |
| GO:BP | GO:0070920 | regulation of regulatory ncRNA processing | 1 | 11 | 6.60e-01 |
| GO:BP | GO:0048712 | negative regulation of astrocyte differentiation | 1 | 9 | 6.60e-01 |
| GO:BP | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules | 2 | 28 | 6.60e-01 |
| GO:BP | GO:0050819 | negative regulation of coagulation | 1 | 38 | 6.61e-01 |
| GO:BP | GO:0032675 | regulation of interleukin-6 production | 2 | 86 | 6.62e-01 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 5 | 199 | 6.62e-01 |
| GO:BP | GO:0032635 | interleukin-6 production | 2 | 86 | 6.62e-01 |
| GO:BP | GO:0050918 | positive chemotaxis | 1 | 40 | 6.62e-01 |
| GO:BP | GO:0071888 | macrophage apoptotic process | 1 | 9 | 6.62e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 2 | 280 | 6.62e-01 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 1 | 39 | 6.62e-01 |
| GO:BP | GO:0051290 | protein heterotetramerization | 1 | 12 | 6.63e-01 |
| GO:BP | GO:2001140 | positive regulation of phospholipid transport | 1 | 11 | 6.63e-01 |
| GO:BP | GO:0042886 | amide transport | 1 | 235 | 6.64e-01 |
| GO:BP | GO:0050891 | multicellular organismal-level water homeostasis | 1 | 16 | 6.64e-01 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 1 | 22 | 6.64e-01 |
| GO:BP | GO:0050908 | detection of light stimulus involved in visual perception | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0050962 | detection of light stimulus involved in sensory perception | 1 | 9 | 6.65e-01 |
| GO:BP | GO:0031445 | regulation of heterochromatin formation | 1 | 22 | 6.65e-01 |
| GO:BP | GO:0120261 | regulation of heterochromatin organization | 1 | 22 | 6.65e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 6.65e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 6.65e-01 |
| GO:BP | GO:0031056 | regulation of histone modification | 5 | 125 | 6.65e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 9 | 775 | 6.65e-01 |
| GO:BP | GO:0007616 | long-term memory | 1 | 31 | 6.65e-01 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 2 | 29 | 6.65e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 5 | 80 | 6.65e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 3 | 275 | 6.65e-01 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 1 | 11 | 6.66e-01 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 1 | 24 | 6.66e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 6.66e-01 |
| GO:BP | GO:0045063 | T-helper 1 cell differentiation | 1 | 14 | 6.66e-01 |
| GO:BP | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling | 2 | 55 | 6.66e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1 | 4953 | 6.66e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 3 | 91 | 6.67e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 4 | 168 | 6.67e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1 | 4048 | 6.67e-01 |
| GO:BP | GO:0035372 | protein localization to microtubule | 1 | 18 | 6.67e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 3 | 3386 | 6.67e-01 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 1 | 13 | 6.68e-01 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 1 | 40 | 6.68e-01 |
| GO:BP | GO:0048643 | positive regulation of skeletal muscle tissue development | 1 | 14 | 6.68e-01 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 2 | 35 | 6.68e-01 |
| GO:BP | GO:0030239 | myofibril assembly | 1 | 66 | 6.68e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 1 | 58 | 6.69e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 22 | 404 | 6.69e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1 | 47 | 6.70e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 6.70e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 3 | 2574 | 6.70e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 3 | 2576 | 6.70e-01 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 1 | 12 | 6.71e-01 |
| GO:BP | GO:0070584 | mitochondrion morphogenesis | 1 | 21 | 6.71e-01 |
| GO:BP | GO:0007409 | axonogenesis | 2 | 359 | 6.72e-01 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 5 | 64 | 6.72e-01 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 2 | 25 | 6.73e-01 |
| GO:BP | GO:0042541 | hemoglobin biosynthetic process | 1 | 13 | 6.73e-01 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 1 | 13 | 6.73e-01 |
| GO:BP | GO:1905517 | macrophage migration | 1 | 34 | 6.73e-01 |
| GO:BP | GO:0051764 | actin crosslink formation | 1 | 10 | 6.74e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 5 | 305 | 6.74e-01 |
| GO:BP | GO:0043249 | erythrocyte maturation | 1 | 12 | 6.74e-01 |
| GO:BP | GO:0046007 | negative regulation of activated T cell proliferation | 1 | 8 | 6.74e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 5 | 173 | 6.74e-01 |
| GO:BP | GO:0001756 | somitogenesis | 1 | 48 | 6.75e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 6.75e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 4 | 258 | 6.75e-01 |
| GO:BP | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 1 | 9 | 6.75e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 9 | 139 | 6.75e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 11 | 539 | 6.75e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 1 | 216 | 6.75e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 3 | 260 | 6.76e-01 |
| GO:BP | GO:0006346 | DNA methylation-dependent heterochromatin formation | 1 | 22 | 6.76e-01 |
| GO:BP | GO:0007210 | serotonin receptor signaling pathway | 2 | 8 | 6.76e-01 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 1 | 15 | 6.76e-01 |
| GO:BP | GO:0097421 | liver regeneration | 1 | 23 | 6.77e-01 |
| GO:BP | GO:0009190 | cyclic nucleotide biosynthetic process | 1 | 14 | 6.77e-01 |
| GO:BP | GO:0010863 | positive regulation of phospholipase C activity | 1 | 18 | 6.77e-01 |
| GO:BP | GO:0052372 | modulation by symbiont of entry into host | 2 | 38 | 6.77e-01 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 1 | 14 | 6.78e-01 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 1 | 15 | 6.78e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 10 | 612 | 6.78e-01 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 2 | 45 | 6.79e-01 |
| GO:BP | GO:0032371 | regulation of sterol transport | 2 | 45 | 6.79e-01 |
| GO:BP | GO:0032722 | positive regulation of chemokine production | 1 | 32 | 6.79e-01 |
| GO:BP | GO:0006783 | heme biosynthetic process | 2 | 30 | 6.79e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 23 | 2102 | 6.80e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 1 | 39 | 6.80e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 1 | 15 | 6.80e-01 |
| GO:BP | GO:0034204 | lipid translocation | 1 | 38 | 6.80e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 6.80e-01 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 1 | 45 | 6.81e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 8 | 816 | 6.81e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 4 | 189 | 6.82e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 6.82e-01 |
| GO:BP | GO:0048041 | focal adhesion assembly | 5 | 81 | 6.83e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 2 | 133 | 6.83e-01 |
| GO:BP | GO:0002839 | positive regulation of immune response to tumor cell | 1 | 7 | 6.83e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 4 | 232 | 6.83e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 5 | 4174 | 6.83e-01 |
| GO:BP | GO:2001138 | regulation of phospholipid transport | 1 | 12 | 6.83e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 5 | 3580 | 6.83e-01 |
| GO:BP | GO:0002836 | positive regulation of response to tumor cell | 1 | 7 | 6.83e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 1 | 67 | 6.83e-01 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 3 | 39 | 6.84e-01 |
| GO:BP | GO:0015813 | L-glutamate transmembrane transport | 1 | 19 | 6.85e-01 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 1 | 52 | 6.85e-01 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 1 | 52 | 6.85e-01 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 1 | 29 | 6.85e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 2 | 106 | 6.86e-01 |
| GO:BP | GO:1990849 | vacuolar localization | 2 | 62 | 6.87e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 6.87e-01 |
| GO:BP | GO:0032418 | lysosome localization | 2 | 62 | 6.87e-01 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 3 | 41 | 6.87e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 3 | 344 | 6.88e-01 |
| GO:BP | GO:0033273 | response to vitamin | 1 | 61 | 6.88e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 4 | 90 | 6.89e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 6 | 1322 | 6.90e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 3 | 345 | 6.90e-01 |
| GO:BP | GO:0032369 | negative regulation of lipid transport | 1 | 20 | 6.90e-01 |
| GO:BP | GO:0072567 | chemokine (C-X-C motif) ligand 2 production | 1 | 10 | 6.90e-01 |
| GO:BP | GO:2000341 | regulation of chemokine (C-X-C motif) ligand 2 production | 1 | 10 | 6.90e-01 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 1 | 13 | 6.90e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 1 | 199 | 6.90e-01 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 2 | 122 | 6.90e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 29 | 2683 | 6.91e-01 |
| GO:BP | GO:1901984 | negative regulation of protein acetylation | 1 | 16 | 6.91e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 10 | 148 | 6.91e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 29 | 2684 | 6.91e-01 |
| GO:BP | GO:0042440 | pigment metabolic process | 3 | 60 | 6.91e-01 |
| GO:BP | GO:0001707 | mesoderm formation | 1 | 53 | 6.91e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 1 | 54 | 6.91e-01 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 1 | 20 | 6.92e-01 |
| GO:BP | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation | 2 | 21 | 6.92e-01 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 6.92e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 6.92e-01 |
| GO:BP | GO:0021859 | pyramidal neuron differentiation | 1 | 11 | 6.92e-01 |
| GO:BP | GO:2000671 | regulation of motor neuron apoptotic process | 1 | 12 | 6.92e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 7 | 1564 | 6.92e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 3 | 53 | 6.93e-01 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 1 | 63 | 6.93e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 3 | 122 | 6.93e-01 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 5 | 85 | 6.93e-01 |
| GO:BP | GO:0042221 | response to chemical | 2 | 2559 | 6.93e-01 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 2 | 35 | 6.93e-01 |
| GO:BP | GO:0060263 | regulation of respiratory burst | 1 | 10 | 6.93e-01 |
| GO:BP | GO:0043374 | CD8-positive, alpha-beta T cell differentiation | 1 | 8 | 6.94e-01 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 1 | 52 | 6.94e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 11 | 687 | 6.95e-01 |
| GO:BP | GO:0060541 | respiratory system development | 3 | 163 | 6.95e-01 |
| GO:BP | GO:0007411 | axon guidance | 5 | 185 | 6.95e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 4 | 1315 | 6.95e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 4 | 1315 | 6.95e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 1 | 14 | 6.97e-01 |
| GO:BP | GO:1904406 | negative regulation of nitric oxide metabolic process | 1 | 10 | 6.97e-01 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 1 | 26 | 6.97e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 1 | 91 | 6.97e-01 |
| GO:BP | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 1 | 10 | 6.97e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1 | 4455 | 6.97e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 5 | 186 | 6.98e-01 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 1 | 14 | 6.99e-01 |
| GO:BP | GO:0071397 | cellular response to cholesterol | 1 | 11 | 6.99e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 1 | 16 | 6.99e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 1 | 16 | 6.99e-01 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 1 | 55 | 6.99e-01 |
| GO:BP | GO:0034661 | ncRNA catabolic process | 1 | 41 | 7.00e-01 |
| GO:BP | GO:0030431 | sleep | 1 | 13 | 7.00e-01 |
| GO:BP | GO:0031100 | animal organ regeneration | 4 | 47 | 7.00e-01 |
| GO:BP | GO:0045453 | bone resorption | 1 | 40 | 7.00e-01 |
| GO:BP | GO:0050896 | response to stimulus | 3 | 5770 | 7.00e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 5 | 180 | 7.00e-01 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 7 | 106 | 7.01e-01 |
| GO:BP | GO:0001696 | gastric acid secretion | 1 | 10 | 7.02e-01 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 1 | 45 | 7.02e-01 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 2 | 24 | 7.02e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 2 | 74 | 7.03e-01 |
| GO:BP | GO:0032729 | positive regulation of type II interferon production | 1 | 36 | 7.03e-01 |
| GO:BP | GO:0000245 | spliceosomal complex assembly | 1 | 73 | 7.04e-01 |
| GO:BP | GO:0097035 | regulation of membrane lipid distribution | 1 | 43 | 7.05e-01 |
| GO:BP | GO:0051234 | establishment of localization | 2 | 3433 | 7.05e-01 |
| GO:BP | GO:0002295 | T-helper cell lineage commitment | 1 | 7 | 7.05e-01 |
| GO:BP | GO:0002443 | leukocyte mediated immunity | 2 | 194 | 7.05e-01 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 3 | 60 | 7.05e-01 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 1 | 17 | 7.05e-01 |
| GO:BP | GO:0035729 | cellular response to hepatocyte growth factor stimulus | 1 | 13 | 7.05e-01 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 2 | 61 | 7.05e-01 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 3 | 105 | 7.06e-01 |
| GO:BP | GO:0032940 | secretion by cell | 3 | 569 | 7.06e-01 |
| GO:BP | GO:0010721 | negative regulation of cell development | 15 | 191 | 7.08e-01 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 1 | 89 | 7.08e-01 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 1 | 18 | 7.08e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 1 | 4570 | 7.08e-01 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 1 | 29 | 7.08e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 7 | 496 | 7.09e-01 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 1 | 19 | 7.09e-01 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 1 | 19 | 7.09e-01 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 1 | 19 | 7.09e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 4 | 382 | 7.09e-01 |
| GO:BP | GO:0008306 | associative learning | 3 | 65 | 7.09e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 1 | 242 | 7.09e-01 |
| GO:BP | GO:0046688 | response to copper ion | 1 | 29 | 7.09e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 7.09e-01 |
| GO:BP | GO:0006491 | N-glycan processing | 1 | 18 | 7.09e-01 |
| GO:BP | GO:0035633 | maintenance of blood-brain barrier | 2 | 26 | 7.10e-01 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 1 | 15 | 7.10e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 1 | 4596 | 7.11e-01 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 9 | 311 | 7.11e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 2 | 31 | 7.11e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 4 | 72 | 7.11e-01 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 3 | 64 | 7.11e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 14 | 1138 | 7.11e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 4 | 373 | 7.11e-01 |
| GO:BP | GO:0007528 | neuromuscular junction development | 1 | 39 | 7.11e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 3 | 61 | 7.12e-01 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 1 | 78 | 7.13e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 1 | 110 | 7.14e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 3 | 2565 | 7.15e-01 |
| GO:BP | GO:0005980 | glycogen catabolic process | 1 | 14 | 7.15e-01 |
| GO:BP | GO:0002720 | positive regulation of cytokine production involved in immune response | 1 | 49 | 7.15e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 2 | 150 | 7.15e-01 |
| GO:BP | GO:2001239 | regulation of extrinsic apoptotic signaling pathway in absence of ligand | 2 | 35 | 7.16e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 1 | 15 | 7.16e-01 |
| GO:BP | GO:0050687 | negative regulation of defense response to virus | 1 | 30 | 7.16e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 3 | 2573 | 7.16e-01 |
| GO:BP | GO:0010839 | negative regulation of keratinocyte proliferation | 1 | 19 | 7.18e-01 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 1 | 14 | 7.18e-01 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 1 | 14 | 7.18e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 16 | 241 | 7.18e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 7 | 1604 | 7.18e-01 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 1 | 28 | 7.18e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 1 | 215 | 7.18e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 1 | 76 | 7.18e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 2 | 73 | 7.18e-01 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 1 | 43 | 7.18e-01 |
| GO:BP | GO:0007628 | adult walking behavior | 1 | 23 | 7.19e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 16 | 2035 | 7.20e-01 |
| GO:BP | GO:1901863 | positive regulation of muscle tissue development | 1 | 18 | 7.20e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 7.20e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 1 | 25 | 7.20e-01 |
| GO:BP | GO:0030534 | adult behavior | 4 | 92 | 7.20e-01 |
| GO:BP | GO:0045822 | negative regulation of heart contraction | 1 | 17 | 7.20e-01 |
| GO:BP | GO:0070633 | transepithelial transport | 1 | 24 | 7.22e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 1 | 51 | 7.22e-01 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 1 | 21 | 7.22e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 1 | 228 | 7.22e-01 |
| GO:BP | GO:0050862 | positive regulation of T cell receptor signaling pathway | 1 | 11 | 7.22e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 56 | 631 | 7.23e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 2 | 1555 | 7.23e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 7.23e-01 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 4 | 43 | 7.24e-01 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 5 | 74 | 7.24e-01 |
| GO:BP | GO:0010744 | positive regulation of macrophage derived foam cell differentiation | 1 | 8 | 7.24e-01 |
| GO:BP | GO:0030730 | sequestering of triglyceride | 1 | 13 | 7.24e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 1 | 28 | 7.24e-01 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 1 | 12 | 7.25e-01 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 7 | 122 | 7.25e-01 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 6 | 97 | 7.25e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 1 | 4747 | 7.25e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 3 | 2845 | 7.26e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 7.26e-01 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 1 | 24 | 7.28e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 3 | 80 | 7.29e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 10 | 475 | 7.29e-01 |
| GO:BP | GO:0046642 | negative regulation of alpha-beta T cell proliferation | 1 | 9 | 7.30e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 2 | 382 | 7.30e-01 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 2 | 33 | 7.31e-01 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 2 | 33 | 7.31e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 1 | 16 | 7.31e-01 |
| GO:BP | GO:0043903 | regulation of biological process involved in symbiotic interaction | 2 | 40 | 7.31e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 1 | 42 | 7.31e-01 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 6 | 106 | 7.33e-01 |
| GO:BP | GO:0032309 | icosanoid secretion | 1 | 18 | 7.34e-01 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 1 | 14 | 7.34e-01 |
| GO:BP | GO:0009251 | glucan catabolic process | 1 | 14 | 7.34e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 1 | 172 | 7.34e-01 |
| GO:BP | GO:1903523 | negative regulation of blood circulation | 1 | 18 | 7.34e-01 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 1 | 19 | 7.34e-01 |
| GO:BP | GO:0002762 | negative regulation of myeloid leukocyte differentiation | 2 | 33 | 7.34e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 1 | 64 | 7.34e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 3 | 301 | 7.34e-01 |
| GO:BP | GO:0060033 | anatomical structure regression | 1 | 11 | 7.35e-01 |
| GO:BP | GO:0006909 | phagocytosis | 2 | 148 | 7.36e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 2 | 279 | 7.36e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 1 | 4946 | 7.36e-01 |
| GO:BP | GO:0016575 | histone deacetylation | 1 | 87 | 7.37e-01 |
| GO:BP | GO:0090659 | walking behavior | 1 | 25 | 7.37e-01 |
| GO:BP | GO:0030901 | midbrain development | 3 | 69 | 7.37e-01 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 1 | 20 | 7.37e-01 |
| GO:BP | GO:0035728 | response to hepatocyte growth factor | 1 | 15 | 7.37e-01 |
| GO:BP | GO:2000251 | positive regulation of actin cytoskeleton reorganization | 1 | 14 | 7.37e-01 |
| GO:BP | GO:0010470 | regulation of gastrulation | 1 | 13 | 7.37e-01 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 2 | 33 | 7.37e-01 |
| GO:BP | GO:0048538 | thymus development | 2 | 35 | 7.38e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 4 | 147 | 7.38e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 1 | 44 | 7.38e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 2 | 39 | 7.38e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 2 | 39 | 7.38e-01 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 1 | 42 | 7.38e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 10 | 4539 | 7.39e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 3 | 112 | 7.39e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 3 | 155 | 7.39e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 1 | 28 | 7.39e-01 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 1 | 15 | 7.40e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 2 | 62 | 7.46e-01 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 1 | 34 | 7.46e-01 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 1 | 23 | 7.46e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 2 | 174 | 7.47e-01 |
| GO:BP | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation | 1 | 13 | 7.47e-01 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 2 | 39 | 7.47e-01 |
| GO:BP | GO:0042908 | xenobiotic transport | 1 | 30 | 7.47e-01 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 3 | 18 | 7.47e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 1 | 55 | 7.47e-01 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 7 | 123 | 7.47e-01 |
| GO:BP | GO:0070830 | bicellular tight junction assembly | 1 | 43 | 7.47e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 1 | 46 | 7.47e-01 |
| GO:BP | GO:0035601 | protein deacylation | 2 | 116 | 7.48e-01 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 1 | 59 | 7.48e-01 |
| GO:BP | GO:0002824 | positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 62 | 7.48e-01 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 1 | 58 | 7.48e-01 |
| GO:BP | GO:0051938 | L-glutamate import | 1 | 25 | 7.48e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 4 | 1441 | 7.49e-01 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 2 | 32 | 7.49e-01 |
| GO:BP | GO:0001541 | ovarian follicle development | 2 | 41 | 7.50e-01 |
| GO:BP | GO:0045622 | regulation of T-helper cell differentiation | 2 | 20 | 7.51e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 1 | 56 | 7.51e-01 |
| GO:BP | GO:0002700 | regulation of production of molecular mediator of immune response | 2 | 108 | 7.51e-01 |
| GO:BP | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process | 1 | 21 | 7.51e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 1 | 78 | 7.51e-01 |
| GO:BP | GO:0043373 | CD4-positive, alpha-beta T cell lineage commitment | 1 | 9 | 7.52e-01 |
| GO:BP | GO:0097066 | response to thyroid hormone | 1 | 23 | 7.52e-01 |
| GO:BP | GO:0045744 | negative regulation of G protein-coupled receptor signaling pathway | 3 | 38 | 7.52e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 7.52e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 6 | 248 | 7.52e-01 |
| GO:BP | GO:0023061 | signal release | 1 | 321 | 7.52e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 1 | 26 | 7.52e-01 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 1 | 21 | 7.53e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 1 | 111 | 7.53e-01 |
| GO:BP | GO:0016485 | protein processing | 2 | 183 | 7.54e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 8 | 447 | 7.54e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 2 | 29 | 7.56e-01 |
| GO:BP | GO:0006027 | glycosaminoglycan catabolic process | 1 | 17 | 7.56e-01 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 1 | 39 | 7.56e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 10 | 4590 | 7.56e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 3 | 80 | 7.58e-01 |
| GO:BP | GO:1903306 | negative regulation of regulated secretory pathway | 1 | 14 | 7.59e-01 |
| GO:BP | GO:1901679 | nucleotide transmembrane transport | 1 | 18 | 7.60e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 1 | 55 | 7.60e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 7.60e-01 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 3 | 70 | 7.60e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 4 | 3730 | 7.60e-01 |
| GO:BP | GO:0033033 | negative regulation of myeloid cell apoptotic process | 1 | 10 | 7.60e-01 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 1 | 140 | 7.60e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 7.61e-01 |
| GO:BP | GO:0033762 | response to glucagon | 1 | 13 | 7.62e-01 |
| GO:BP | GO:0051180 | vitamin transport | 1 | 28 | 7.62e-01 |
| GO:BP | GO:0098732 | macromolecule deacylation | 2 | 119 | 7.62e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 1 | 71 | 7.62e-01 |
| GO:BP | GO:0001816 | cytokine production | 8 | 451 | 7.62e-01 |
| GO:BP | GO:0050848 | regulation of calcium-mediated signaling | 1 | 52 | 7.62e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 7.62e-01 |
| GO:BP | GO:0048771 | tissue remodeling | 2 | 114 | 7.63e-01 |
| GO:BP | GO:0019348 | dolichol metabolic process | 1 | 22 | 7.64e-01 |
| GO:BP | GO:0002821 | positive regulation of adaptive immune response | 1 | 66 | 7.65e-01 |
| GO:BP | GO:0020027 | hemoglobin metabolic process | 1 | 16 | 7.65e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 1 | 132 | 7.65e-01 |
| GO:BP | GO:0031670 | cellular response to nutrient | 2 | 31 | 7.65e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 7.65e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 7.67e-01 |
| GO:BP | GO:0002708 | positive regulation of lymphocyte mediated immunity | 1 | 58 | 7.68e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 4 | 250 | 7.68e-01 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 1 | 14 | 7.68e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 1 | 95 | 7.68e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 7.68e-01 |
| GO:BP | GO:0032642 | regulation of chemokine production | 1 | 46 | 7.68e-01 |
| GO:BP | GO:0035282 | segmentation | 2 | 75 | 7.68e-01 |
| GO:BP | GO:0032602 | chemokine production | 1 | 46 | 7.68e-01 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 1 | 20 | 7.69e-01 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 1 | 16 | 7.69e-01 |
| GO:BP | GO:0120192 | tight junction assembly | 1 | 47 | 7.69e-01 |
| GO:BP | GO:0018345 | protein palmitoylation | 1 | 30 | 7.69e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 8 | 150 | 7.69e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 1 | 68 | 7.70e-01 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 1 | 38 | 7.70e-01 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 2 | 45 | 7.70e-01 |
| GO:BP | GO:0098586 | cellular response to virus | 1 | 40 | 7.70e-01 |
| GO:BP | GO:0007626 | locomotory behavior | 2 | 138 | 7.70e-01 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 4 | 72 | 7.70e-01 |
| GO:BP | GO:0031643 | positive regulation of myelination | 1 | 12 | 7.70e-01 |
| GO:BP | GO:0007165 | signal transduction | 5 | 3818 | 7.70e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 1 | 59 | 7.71e-01 |
| GO:BP | GO:0001662 | behavioral fear response | 1 | 29 | 7.71e-01 |
| GO:BP | GO:0043967 | histone H4 acetylation | 3 | 57 | 7.72e-01 |
| GO:BP | GO:1901881 | positive regulation of protein depolymerization | 1 | 15 | 7.72e-01 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 1 | 36 | 7.73e-01 |
| GO:BP | GO:0031638 | zymogen activation | 1 | 45 | 7.73e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 1 | 27 | 7.73e-01 |
| GO:BP | GO:0045333 | cellular respiration | 3 | 211 | 7.73e-01 |
| GO:BP | GO:0051179 | localization | 2 | 3969 | 7.73e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 3 | 3080 | 7.73e-01 |
| GO:BP | GO:1990000 | amyloid fibril formation | 1 | 35 | 7.73e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 2 | 418 | 7.74e-01 |
| GO:BP | GO:0002837 | regulation of immune response to tumor cell | 1 | 10 | 7.74e-01 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 1 | 17 | 7.74e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 1 | 16 | 7.74e-01 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 1 | 21 | 7.74e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 7.75e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 2 | 53 | 7.77e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 2 | 41 | 7.77e-01 |
| GO:BP | GO:0009798 | axis specification | 1 | 63 | 7.77e-01 |
| GO:BP | GO:0010923 | negative regulation of phosphatase activity | 2 | 30 | 7.77e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 7.77e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 4 | 163 | 7.77e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 2 | 67 | 7.77e-01 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 1 | 38 | 7.77e-01 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 1 | 37 | 7.77e-01 |
| GO:BP | GO:0002209 | behavioral defense response | 1 | 30 | 7.78e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 7.78e-01 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 1 | 11 | 7.78e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 3 | 64 | 7.79e-01 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 1 | 15 | 7.79e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 1 | 20 | 7.79e-01 |
| GO:BP | GO:0015718 | monocarboxylic acid transport | 3 | 90 | 7.80e-01 |
| GO:BP | GO:0043297 | apical junction assembly | 1 | 51 | 7.80e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 7.80e-01 |
| GO:BP | GO:0001502 | cartilage condensation | 1 | 12 | 7.80e-01 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 2 | 24 | 7.80e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 7.80e-01 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 1 | 29 | 7.80e-01 |
| GO:BP | GO:0046660 | female sex differentiation | 6 | 91 | 7.80e-01 |
| GO:BP | GO:0048016 | inositol phosphate-mediated signaling | 3 | 45 | 7.80e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 4 | 84 | 7.81e-01 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 4 | 84 | 7.81e-01 |
| GO:BP | GO:0043277 | apoptotic cell clearance | 1 | 35 | 7.82e-01 |
| GO:BP | GO:0031424 | keratinization | 1 | 13 | 7.82e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 5 | 96 | 7.82e-01 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 2 | 82 | 7.83e-01 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 2 | 82 | 7.83e-01 |
| GO:BP | GO:0001675 | acrosome assembly | 1 | 16 | 7.84e-01 |
| GO:BP | GO:0043303 | mast cell degranulation | 2 | 30 | 7.85e-01 |
| GO:BP | GO:0036315 | cellular response to sterol | 1 | 14 | 7.85e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 2 | 56 | 7.85e-01 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 1 | 51 | 7.85e-01 |
| GO:BP | GO:0016579 | protein deubiquitination | 7 | 129 | 7.85e-01 |
| GO:BP | GO:0002520 | immune system development | 11 | 144 | 7.85e-01 |
| GO:BP | GO:0035584 | calcium-mediated signaling using intracellular calcium source | 1 | 14 | 7.86e-01 |
| GO:BP | GO:0008585 | female gonad development | 5 | 77 | 7.86e-01 |
| GO:BP | GO:0016093 | polyprenol metabolic process | 1 | 23 | 7.86e-01 |
| GO:BP | GO:0042113 | B cell activation | 4 | 170 | 7.87e-01 |
| GO:BP | GO:0071391 | cellular response to estrogen stimulus | 1 | 15 | 7.87e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 7 | 288 | 7.88e-01 |
| GO:BP | GO:0032760 | positive regulation of tumor necrosis factor production | 1 | 55 | 7.88e-01 |
| GO:BP | GO:0002834 | regulation of response to tumor cell | 1 | 11 | 7.89e-01 |
| GO:BP | GO:0051384 | response to glucocorticoid | 1 | 93 | 7.89e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 2 | 55 | 7.90e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 1 | 336 | 7.90e-01 |
| GO:BP | GO:0009595 | detection of biotic stimulus | 1 | 20 | 7.90e-01 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 1 | 11 | 7.90e-01 |
| GO:BP | GO:0048511 | rhythmic process | 3 | 237 | 7.91e-01 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 2 | 33 | 7.91e-01 |
| GO:BP | GO:0120193 | tight junction organization | 1 | 52 | 7.91e-01 |
| GO:BP | GO:0050869 | negative regulation of B cell activation | 1 | 19 | 7.91e-01 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 3 | 54 | 7.92e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 4 | 403 | 7.92e-01 |
| GO:BP | GO:0006476 | protein deacetylation | 1 | 106 | 7.92e-01 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 2 | 36 | 7.92e-01 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 2 | 36 | 7.92e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 2 | 141 | 7.92e-01 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 1 | 19 | 7.92e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 5 | 112 | 7.92e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 1 | 46 | 7.92e-01 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 1 | 49 | 7.93e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 14 | 994 | 7.93e-01 |
| GO:BP | GO:0021762 | substantia nigra development | 2 | 41 | 7.94e-01 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 1 | 21 | 7.94e-01 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 1 | 42 | 7.94e-01 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 1 | 17 | 7.95e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 2 | 6996 | 7.95e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 1 | 90 | 7.96e-01 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 1 | 23 | 7.96e-01 |
| GO:BP | GO:0001835 | blastocyst hatching | 1 | 23 | 7.96e-01 |
| GO:BP | GO:0035188 | hatching | 1 | 23 | 7.96e-01 |
| GO:BP | GO:0060251 | regulation of glial cell proliferation | 1 | 26 | 7.96e-01 |
| GO:BP | GO:0043576 | regulation of respiratory gaseous exchange | 1 | 15 | 7.96e-01 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 1 | 29 | 7.97e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 1 | 22 | 7.97e-01 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 1 | 47 | 7.97e-01 |
| GO:BP | GO:0046636 | negative regulation of alpha-beta T cell activation | 2 | 25 | 7.98e-01 |
| GO:BP | GO:0002279 | mast cell activation involved in immune response | 2 | 31 | 7.98e-01 |
| GO:BP | GO:1903557 | positive regulation of tumor necrosis factor superfamily cytokine production | 1 | 57 | 7.98e-01 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 1 | 23 | 7.98e-01 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 1 | 46 | 7.98e-01 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 2 | 32 | 7.99e-01 |
| GO:BP | GO:0071295 | cellular response to vitamin | 1 | 22 | 7.99e-01 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 4 | 83 | 8.00e-01 |
| GO:BP | GO:0046794 | transport of virus | 1 | 19 | 8.00e-01 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 1 | 81 | 8.01e-01 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 2 | 35 | 8.01e-01 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 1 | 50 | 8.02e-01 |
| GO:BP | GO:0033005 | positive regulation of mast cell activation | 1 | 13 | 8.02e-01 |
| GO:BP | GO:0006525 | arginine metabolic process | 1 | 13 | 8.02e-01 |
| GO:BP | GO:0050849 | negative regulation of calcium-mediated signaling | 1 | 15 | 8.02e-01 |
| GO:BP | GO:0097503 | sialylation | 1 | 18 | 8.03e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 2 | 28 | 8.03e-01 |
| GO:BP | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling | 2 | 78 | 8.04e-01 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 1 | 21 | 8.04e-01 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 8 | 316 | 8.04e-01 |
| GO:BP | GO:0032873 | negative regulation of stress-activated MAPK cascade | 2 | 46 | 8.04e-01 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 2 | 46 | 8.04e-01 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 2 | 47 | 8.04e-01 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 3 | 40 | 8.05e-01 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 2 | 33 | 8.05e-01 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 3 | 118 | 8.06e-01 |
| GO:BP | GO:0050850 | positive regulation of calcium-mediated signaling | 1 | 22 | 8.06e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1602 | 8.09e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 8.09e-01 |
| GO:BP | GO:0009988 | cell-cell recognition | 1 | 32 | 8.09e-01 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 2 | 32 | 8.10e-01 |
| GO:BP | GO:0031295 | T cell costimulation | 1 | 22 | 8.10e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 11 | 154 | 8.10e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 8.10e-01 |
| GO:BP | GO:0042596 | fear response | 1 | 32 | 8.10e-01 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 5 | 79 | 8.10e-01 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 5 | 99 | 8.11e-01 |
| GO:BP | GO:0008209 | androgen metabolic process | 1 | 16 | 8.11e-01 |
| GO:BP | GO:0030851 | granulocyte differentiation | 1 | 27 | 8.12e-01 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 1 | 69 | 8.12e-01 |
| GO:BP | GO:0002456 | T cell mediated immunity | 1 | 53 | 8.12e-01 |
| GO:BP | GO:0006026 | aminoglycan catabolic process | 1 | 19 | 8.13e-01 |
| GO:BP | GO:0009060 | aerobic respiration | 2 | 168 | 8.14e-01 |
| GO:BP | GO:0002705 | positive regulation of leukocyte mediated immunity | 1 | 68 | 8.14e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 3 | 220 | 8.14e-01 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 1 | 58 | 8.14e-01 |
| GO:BP | GO:0007130 | synaptonemal complex assembly | 1 | 10 | 8.14e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 13 | 1238 | 8.14e-01 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 4 | 66 | 8.14e-01 |
| GO:BP | GO:0007608 | sensory perception of smell | 1 | 16 | 8.15e-01 |
| GO:BP | GO:0045214 | sarcomere organization | 2 | 43 | 8.15e-01 |
| GO:BP | GO:0002440 | production of molecular mediator of immune response | 2 | 135 | 8.16e-01 |
| GO:BP | GO:0010894 | negative regulation of steroid biosynthetic process | 1 | 18 | 8.16e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 2 | 785 | 8.16e-01 |
| GO:BP | GO:0015868 | purine ribonucleotide transport | 1 | 19 | 8.16e-01 |
| GO:BP | GO:0035307 | positive regulation of protein dephosphorylation | 2 | 36 | 8.16e-01 |
| GO:BP | GO:0043113 | receptor clustering | 1 | 45 | 8.17e-01 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 1 | 39 | 8.18e-01 |
| GO:BP | GO:0035590 | purinergic nucleotide receptor signaling pathway | 1 | 16 | 8.18e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 1 | 151 | 8.18e-01 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 1 | 22 | 8.19e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 4 | 271 | 8.19e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 1 | 211 | 8.19e-01 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 3 | 40 | 8.19e-01 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 7 | 252 | 8.20e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 2 | 512 | 8.22e-01 |
| GO:BP | GO:0015909 | long-chain fatty acid transport | 1 | 38 | 8.22e-01 |
| GO:BP | GO:0014850 | response to muscle activity | 1 | 17 | 8.22e-01 |
| GO:BP | GO:0031294 | lymphocyte costimulation | 1 | 23 | 8.22e-01 |
| GO:BP | GO:0031960 | response to corticosteroid | 2 | 109 | 8.22e-01 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 2 | 62 | 8.22e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 8.23e-01 |
| GO:BP | GO:0071806 | protein transmembrane transport | 1 | 62 | 8.23e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 3 | 7069 | 8.23e-01 |
| GO:BP | GO:0045581 | negative regulation of T cell differentiation | 2 | 27 | 8.23e-01 |
| GO:BP | GO:0001889 | liver development | 3 | 110 | 8.23e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 2 | 106 | 8.23e-01 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 8.23e-01 |
| GO:BP | GO:0050795 | regulation of behavior | 2 | 39 | 8.23e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 1 | 37 | 8.23e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 8.23e-01 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 5 | 234 | 8.23e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 3 | 273 | 8.24e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 2 | 41 | 8.24e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 8.24e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 8.24e-01 |
| GO:BP | GO:0048708 | astrocyte differentiation | 1 | 58 | 8.24e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 3 | 4128 | 8.24e-01 |
| GO:BP | GO:0010883 | regulation of lipid storage | 2 | 39 | 8.24e-01 |
| GO:BP | GO:0006656 | phosphatidylcholine biosynthetic process | 1 | 25 | 8.24e-01 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 3 | 40 | 8.25e-01 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 3 | 40 | 8.25e-01 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 3 | 38 | 8.25e-01 |
| GO:BP | GO:0001708 | cell fate specification | 1 | 56 | 8.25e-01 |
| GO:BP | GO:0030324 | lung development | 2 | 146 | 8.25e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 2 | 111 | 8.25e-01 |
| GO:BP | GO:0002418 | immune response to tumor cell | 1 | 12 | 8.25e-01 |
| GO:BP | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | 25 | 8.25e-01 |
| GO:BP | GO:0048512 | circadian behavior | 1 | 30 | 8.25e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 12 | 204 | 8.25e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 1 | 34 | 8.25e-01 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 1 | 50 | 8.25e-01 |
| GO:BP | GO:0033003 | regulation of mast cell activation | 2 | 25 | 8.25e-01 |
| GO:BP | GO:1901099 | negative regulation of signal transduction in absence of ligand | 1 | 25 | 8.25e-01 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 1 | 15 | 8.25e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 8.25e-01 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 1 | 20 | 8.25e-01 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 2 | 38 | 8.26e-01 |
| GO:BP | GO:0002718 | regulation of cytokine production involved in immune response | 1 | 70 | 8.26e-01 |
| GO:BP | GO:0098743 | cell aggregation | 1 | 12 | 8.26e-01 |
| GO:BP | GO:0002367 | cytokine production involved in immune response | 1 | 70 | 8.26e-01 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 3 | 123 | 8.26e-01 |
| GO:BP | GO:0046677 | response to antibiotic | 1 | 43 | 8.27e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 2 | 62 | 8.28e-01 |
| GO:BP | GO:0070527 | platelet aggregation | 1 | 52 | 8.28e-01 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 3 | 52 | 8.28e-01 |
| GO:BP | GO:0032649 | regulation of type II interferon production | 1 | 52 | 8.28e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 2 | 527 | 8.28e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 3 | 336 | 8.28e-01 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 1 | 38 | 8.29e-01 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 1 | 45 | 8.29e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 8 | 740 | 8.30e-01 |
| GO:BP | GO:0072089 | stem cell proliferation | 1 | 84 | 8.30e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 3 | 112 | 8.30e-01 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 1 | 23 | 8.30e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 2 | 285 | 8.30e-01 |
| GO:BP | GO:0023052 | signaling | 5 | 4141 | 8.30e-01 |
| GO:BP | GO:0006935 | chemotaxis | 11 | 389 | 8.30e-01 |
| GO:BP | GO:0032609 | type II interferon production | 1 | 53 | 8.30e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 22 | 2135 | 8.30e-01 |
| GO:BP | GO:0017156 | calcium-ion regulated exocytosis | 2 | 46 | 8.30e-01 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 1 | 21 | 8.30e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 1 | 63 | 8.30e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 2 | 531 | 8.30e-01 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 1 | 43 | 8.30e-01 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 2 | 51 | 8.30e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 3 | 50 | 8.30e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 2 | 150 | 8.30e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 3 | 252 | 8.31e-01 |
| GO:BP | GO:0070723 | response to cholesterol | 1 | 16 | 8.31e-01 |
| GO:BP | GO:0042330 | taxis | 11 | 389 | 8.32e-01 |
| GO:BP | GO:0002719 | negative regulation of cytokine production involved in immune response | 1 | 16 | 8.32e-01 |
| GO:BP | GO:0097305 | response to alcohol | 9 | 159 | 8.32e-01 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 1 | 63 | 8.32e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 4 | 72 | 8.34e-01 |
| GO:BP | GO:0045940 | positive regulation of steroid metabolic process | 1 | 20 | 8.34e-01 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 1 | 37 | 8.35e-01 |
| GO:BP | GO:0042168 | heme metabolic process | 2 | 40 | 8.35e-01 |
| GO:BP | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway | 1 | 16 | 8.35e-01 |
| GO:BP | GO:0002285 | lymphocyte activation involved in immune response | 2 | 111 | 8.35e-01 |
| GO:BP | GO:0009725 | response to hormone | 1 | 642 | 8.35e-01 |
| GO:BP | GO:0006925 | inflammatory cell apoptotic process | 1 | 15 | 8.35e-01 |
| GO:BP | GO:0007622 | rhythmic behavior | 1 | 32 | 8.35e-01 |
| GO:BP | GO:1904645 | response to amyloid-beta | 1 | 32 | 8.35e-01 |
| GO:BP | GO:0021532 | neural tube patterning | 1 | 31 | 8.36e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 1 | 129 | 8.36e-01 |
| GO:BP | GO:0030890 | positive regulation of B cell proliferation | 2 | 26 | 8.36e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 1 | 65 | 8.37e-01 |
| GO:BP | GO:0010951 | negative regulation of endopeptidase activity | 1 | 102 | 8.37e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 1 | 369 | 8.38e-01 |
| GO:BP | GO:0098781 | ncRNA transcription | 2 | 61 | 8.38e-01 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 1 | 21 | 8.39e-01 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 1 | 39 | 8.39e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 8.39e-01 |
| GO:BP | GO:0033006 | regulation of mast cell activation involved in immune response | 1 | 21 | 8.40e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 2 | 81 | 8.41e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 2 | 551 | 8.41e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 1 | 155 | 8.42e-01 |
| GO:BP | GO:0007154 | cell communication | 5 | 4226 | 8.42e-01 |
| GO:BP | GO:0001704 | formation of primary germ layer | 1 | 96 | 8.42e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 11 | 192 | 8.43e-01 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 4 | 75 | 8.43e-01 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 1 | 31 | 8.43e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 2 | 157 | 8.43e-01 |
| GO:BP | GO:0015914 | phospholipid transport | 1 | 75 | 8.43e-01 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 1 | 31 | 8.43e-01 |
| GO:BP | GO:0045939 | negative regulation of steroid metabolic process | 1 | 19 | 8.43e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 2 | 58 | 8.44e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 3 | 108 | 8.44e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 8.45e-01 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 3 | 165 | 8.45e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 2 | 121 | 8.45e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 1 | 59 | 8.46e-01 |
| GO:BP | GO:0032515 | negative regulation of phosphoprotein phosphatase activity | 1 | 21 | 8.46e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 1 | 34 | 8.46e-01 |
| GO:BP | GO:0002360 | T cell lineage commitment | 1 | 15 | 8.46e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 8.47e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 6 | 185 | 8.47e-01 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 1 | 87 | 8.47e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 8.48e-01 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 1 | 59 | 8.48e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 8.49e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 8.49e-01 |
| GO:BP | GO:0014888 | striated muscle adaptation | 2 | 40 | 8.49e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 2 | 41 | 8.49e-01 |
| GO:BP | GO:0036037 | CD8-positive, alpha-beta T cell activation | 1 | 15 | 8.50e-01 |
| GO:BP | GO:0031532 | actin cytoskeleton reorganization | 1 | 96 | 8.51e-01 |
| GO:BP | GO:0097720 | calcineurin-mediated signaling | 1 | 38 | 8.51e-01 |
| GO:BP | GO:0050871 | positive regulation of B cell activation | 1 | 53 | 8.51e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 10 | 556 | 8.51e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 8.51e-01 |
| GO:BP | GO:0007041 | lysosomal transport | 1 | 115 | 8.51e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 15 | 316 | 8.52e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 8.53e-01 |
| GO:BP | GO:0009451 | RNA modification | 1 | 162 | 8.54e-01 |
| GO:BP | GO:0045576 | mast cell activation | 1 | 35 | 8.54e-01 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 1 | 31 | 8.54e-01 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 1 | 22 | 8.54e-01 |
| GO:BP | GO:0016573 | histone acetylation | 7 | 143 | 8.54e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 7 | 143 | 8.55e-01 |
| GO:BP | GO:0007606 | sensory perception of chemical stimulus | 1 | 31 | 8.55e-01 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 2 | 52 | 8.55e-01 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 2 | 27 | 8.55e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 2 | 104 | 8.56e-01 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 1 | 49 | 8.56e-01 |
| GO:BP | GO:0070613 | regulation of protein processing | 2 | 47 | 8.56e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 2 | 45 | 8.56e-01 |
| GO:BP | GO:0051503 | adenine nucleotide transport | 1 | 23 | 8.57e-01 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 3 | 105 | 8.57e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 1 | 185 | 8.57e-01 |
| GO:BP | GO:0006836 | neurotransmitter transport | 1 | 138 | 8.57e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 2 | 138 | 8.57e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 23 | 2293 | 8.57e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 5 | 149 | 8.57e-01 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 7 | 147 | 8.58e-01 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 2 | 38 | 8.58e-01 |
| GO:BP | GO:0044000 | movement in host | 9 | 138 | 8.58e-01 |
| GO:BP | GO:0001709 | cell fate determination | 1 | 25 | 8.59e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 2 | 7441 | 8.59e-01 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 2 | 43 | 8.60e-01 |
| GO:BP | GO:0032608 | interferon-beta production | 2 | 43 | 8.60e-01 |
| GO:BP | GO:0007030 | Golgi organization | 4 | 137 | 8.60e-01 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 1 | 24 | 8.60e-01 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 1 | 32 | 8.61e-01 |
| GO:BP | GO:0034368 | protein-lipid complex remodeling | 1 | 14 | 8.62e-01 |
| GO:BP | GO:0034369 | plasma lipoprotein particle remodeling | 1 | 14 | 8.62e-01 |
| GO:BP | GO:0042255 | ribosome assembly | 2 | 56 | 8.62e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1 | 114 | 8.63e-01 |
| GO:BP | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 1 | 10 | 8.64e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 1 | 37 | 8.65e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 1 | 73 | 8.65e-01 |
| GO:BP | GO:0009311 | oligosaccharide metabolic process | 2 | 43 | 8.65e-01 |
| GO:BP | GO:0045662 | negative regulation of myoblast differentiation | 1 | 19 | 8.66e-01 |
| GO:BP | GO:0002699 | positive regulation of immune effector process | 2 | 130 | 8.66e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 2 | 108 | 8.66e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 3 | 191 | 8.66e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 5 | 295 | 8.67e-01 |
| GO:BP | GO:0045920 | negative regulation of exocytosis | 1 | 26 | 8.67e-01 |
| GO:BP | GO:0002823 | negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 26 | 8.68e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 8.68e-01 |
| GO:BP | GO:0051604 | protein maturation | 2 | 259 | 8.68e-01 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 3 | 42 | 8.68e-01 |
| GO:BP | GO:0046596 | regulation of viral entry into host cell | 1 | 32 | 8.69e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 1 | 199 | 8.69e-01 |
| GO:BP | GO:0032480 | negative regulation of type I interferon production | 1 | 23 | 8.69e-01 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 1 | 30 | 8.69e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 5 | 151 | 8.69e-01 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 2 | 39 | 8.69e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 1 | 35 | 8.69e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 74 | 4045 | 8.70e-01 |
| GO:BP | GO:0015804 | neutral amino acid transport | 1 | 43 | 8.70e-01 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 8.70e-01 |
| GO:BP | GO:0001706 | endoderm formation | 1 | 46 | 8.70e-01 |
| GO:BP | GO:0015865 | purine nucleotide transport | 1 | 23 | 8.71e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 1 | 56 | 8.71e-01 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 1 | 96 | 8.71e-01 |
| GO:BP | GO:0002706 | regulation of lymphocyte mediated immunity | 1 | 83 | 8.71e-01 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 3 | 74 | 8.71e-01 |
| GO:BP | GO:0002252 | immune effector process | 2 | 318 | 8.71e-01 |
| GO:BP | GO:0016043 | cellular component organization | 3 | 5076 | 8.73e-01 |
| GO:BP | GO:0045103 | intermediate filament-based process | 2 | 40 | 8.73e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 8.73e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 8.73e-01 |
| GO:BP | GO:0030225 | macrophage differentiation | 1 | 35 | 8.73e-01 |
| GO:BP | GO:0008210 | estrogen metabolic process | 1 | 16 | 8.73e-01 |
| GO:BP | GO:0022600 | digestive system process | 1 | 54 | 8.74e-01 |
| GO:BP | GO:0042088 | T-helper 1 type immune response | 1 | 26 | 8.74e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 2 | 42 | 8.74e-01 |
| GO:BP | GO:0048278 | vesicle docking | 1 | 57 | 8.75e-01 |
| GO:BP | GO:0051354 | negative regulation of oxidoreductase activity | 1 | 14 | 8.75e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 3 | 65 | 8.75e-01 |
| GO:BP | GO:0002822 | regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 1 | 97 | 8.75e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 8.75e-01 |
| GO:BP | GO:0032502 | developmental process | 3 | 4494 | 8.76e-01 |
| GO:BP | GO:0036314 | response to sterol | 1 | 19 | 8.76e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 8.76e-01 |
| GO:BP | GO:0060441 | epithelial tube branching involved in lung morphogenesis | 1 | 20 | 8.76e-01 |
| GO:BP | GO:0006766 | vitamin metabolic process | 1 | 73 | 8.77e-01 |
| GO:BP | GO:0034367 | protein-containing complex remodeling | 1 | 16 | 8.77e-01 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 1 | 24 | 8.77e-01 |
| GO:BP | GO:0008542 | visual learning | 1 | 37 | 8.77e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 8.78e-01 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 2 | 44 | 8.78e-01 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 1 | 31 | 8.79e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 2 | 50 | 8.79e-01 |
| GO:BP | GO:0017158 | regulation of calcium ion-dependent exocytosis | 1 | 29 | 8.80e-01 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 2 | 44 | 8.80e-01 |
| GO:BP | GO:0018393 | internal peptidyl-lysine acetylation | 7 | 148 | 8.80e-01 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 2 | 119 | 8.80e-01 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 2 | 100 | 8.80e-01 |
| GO:BP | GO:0018126 | protein hydroxylation | 1 | 26 | 8.80e-01 |
| GO:BP | GO:0015800 | acidic amino acid transport | 1 | 39 | 8.80e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 2 | 133 | 8.82e-01 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 2 | 70 | 8.82e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 1 | 26 | 8.83e-01 |
| GO:BP | GO:0045620 | negative regulation of lymphocyte differentiation | 2 | 33 | 8.84e-01 |
| GO:BP | GO:0050777 | negative regulation of immune response | 1 | 111 | 8.84e-01 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 4 | 65 | 8.84e-01 |
| GO:BP | GO:0007599 | hemostasis | 1 | 153 | 8.87e-01 |
| GO:BP | GO:0006475 | internal protein amino acid acetylation | 7 | 150 | 8.88e-01 |
| GO:BP | GO:0043372 | positive regulation of CD4-positive, alpha-beta T cell differentiation | 1 | 16 | 8.88e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3 | 5274 | 8.89e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 2 | 639 | 8.89e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 1 | 52 | 8.90e-01 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 1 | 65 | 8.90e-01 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 1 | 53 | 8.91e-01 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 2 | 59 | 8.94e-01 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 2 | 59 | 8.94e-01 |
| GO:BP | GO:0043299 | leukocyte degranulation | 1 | 41 | 8.94e-01 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 1 | 104 | 8.94e-01 |
| GO:BP | GO:0045730 | respiratory burst | 1 | 21 | 8.94e-01 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 1 | 33 | 8.95e-01 |
| GO:BP | GO:0002820 | negative regulation of adaptive immune response | 2 | 27 | 8.96e-01 |
| GO:BP | GO:0002819 | regulation of adaptive immune response | 1 | 106 | 8.96e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 3 | 211 | 8.97e-01 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 1 | 30 | 8.97e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 6 | 139 | 8.97e-01 |
| GO:BP | GO:0014065 | phosphatidylinositol 3-kinase signaling | 2 | 100 | 8.98e-01 |
| GO:BP | GO:0051205 | protein insertion into membrane | 2 | 62 | 8.98e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 4 | 68 | 9.00e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 1 | 27 | 9.00e-01 |
| GO:BP | GO:0044409 | entry into host | 7 | 113 | 9.00e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 1 | 52 | 9.01e-01 |
| GO:BP | GO:0070542 | response to fatty acid | 2 | 43 | 9.01e-01 |
| GO:BP | GO:0009584 | detection of visible light | 1 | 20 | 9.01e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 3 | 400 | 9.01e-01 |
| GO:BP | GO:1903707 | negative regulation of hemopoiesis | 4 | 69 | 9.01e-01 |
| GO:BP | GO:0007632 | visual behavior | 1 | 41 | 9.01e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 9.01e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 9.02e-01 |
| GO:BP | GO:0006688 | glycosphingolipid biosynthetic process | 1 | 25 | 9.02e-01 |
| GO:BP | GO:0043368 | positive T cell selection | 1 | 16 | 9.02e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 1 | 22 | 9.02e-01 |
| GO:BP | GO:0021510 | spinal cord development | 1 | 59 | 9.02e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 9.02e-01 |
| GO:BP | GO:0002717 | positive regulation of natural killer cell mediated immunity | 1 | 11 | 9.03e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 1 | 39 | 9.03e-01 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 1 | 20 | 9.03e-01 |
| GO:BP | GO:0032640 | tumor necrosis factor production | 1 | 91 | 9.04e-01 |
| GO:BP | GO:0140029 | exocytic process | 3 | 69 | 9.04e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 1 | 457 | 9.04e-01 |
| GO:BP | GO:0032680 | regulation of tumor necrosis factor production | 1 | 91 | 9.04e-01 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 3 | 46 | 9.04e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 7 | 218 | 9.04e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 1 | 27 | 9.04e-01 |
| GO:BP | GO:0046034 | ATP metabolic process | 3 | 116 | 9.04e-01 |
| GO:BP | GO:0002701 | negative regulation of production of molecular mediator of immune response | 1 | 21 | 9.05e-01 |
| GO:BP | GO:0007034 | vacuolar transport | 6 | 149 | 9.05e-01 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 2 | 47 | 9.05e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 1 | 216 | 9.05e-01 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 2 | 57 | 9.05e-01 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 1 | 63 | 9.06e-01 |
| GO:BP | GO:0016064 | immunoglobulin mediated immune response | 1 | 76 | 9.07e-01 |
| GO:BP | GO:0006862 | nucleotide transport | 1 | 28 | 9.07e-01 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 1 | 27 | 9.07e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 9.07e-01 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 3 | 46 | 9.07e-01 |
| GO:BP | GO:0035308 | negative regulation of protein dephosphorylation | 1 | 28 | 9.07e-01 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 1 | 88 | 9.07e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 2 | 348 | 9.08e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 1 | 45 | 9.08e-01 |
| GO:BP | GO:0071706 | tumor necrosis factor superfamily cytokine production | 1 | 94 | 9.10e-01 |
| GO:BP | GO:1903555 | regulation of tumor necrosis factor superfamily cytokine production | 1 | 94 | 9.10e-01 |
| GO:BP | GO:0050864 | regulation of B cell activation | 1 | 77 | 9.10e-01 |
| GO:BP | GO:0019724 | B cell mediated immunity | 1 | 77 | 9.10e-01 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 2 | 48 | 9.10e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 1 | 78 | 9.10e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 2 | 667 | 9.10e-01 |
| GO:BP | GO:0006869 | lipid transport | 6 | 272 | 9.10e-01 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 3 | 50 | 9.10e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 1 | 140 | 9.10e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 1 | 41 | 9.11e-01 |
| GO:BP | GO:0017145 | stem cell division | 1 | 26 | 9.11e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 2 | 220 | 9.11e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 1 | 56 | 9.11e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 2 | 182 | 9.11e-01 |
| GO:BP | GO:0022406 | membrane docking | 3 | 84 | 9.12e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 1 | 115 | 9.13e-01 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 1 | 42 | 9.13e-01 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 1 | 31 | 9.14e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 1 | 126 | 9.14e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 1 | 57 | 9.14e-01 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 1 | 29 | 9.14e-01 |
| GO:BP | GO:0097484 | dendrite extension | 1 | 27 | 9.15e-01 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 2 | 53 | 9.15e-01 |
| GO:BP | GO:0099024 | plasma membrane invagination | 2 | 42 | 9.16e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 1 | 2466 | 9.16e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 2 | 7806 | 9.16e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 2 | 95 | 9.17e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 7 | 160 | 9.17e-01 |
| GO:BP | GO:0006605 | protein targeting | 1 | 307 | 9.17e-01 |
| GO:BP | GO:0045109 | intermediate filament organization | 1 | 21 | 9.19e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 2 | 120 | 9.20e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 1 | 68 | 9.21e-01 |
| GO:BP | GO:0002686 | negative regulation of leukocyte migration | 1 | 30 | 9.21e-01 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 1 | 131 | 9.24e-01 |
| GO:BP | GO:0015748 | organophosphate ester transport | 1 | 106 | 9.24e-01 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 1 | 30 | 9.24e-01 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 3 | 58 | 9.24e-01 |
| GO:BP | GO:0033238 | regulation of amine metabolic process | 1 | 19 | 9.24e-01 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 2 | 88 | 9.24e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 1 | 87 | 9.24e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 3 | 251 | 9.25e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 9.25e-01 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 3 | 189 | 9.25e-01 |
| GO:BP | GO:0043300 | regulation of leukocyte degranulation | 1 | 27 | 9.25e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 2 | 151 | 9.25e-01 |
| GO:BP | GO:0032715 | negative regulation of interleukin-6 production | 1 | 24 | 9.25e-01 |
| GO:BP | GO:0032720 | negative regulation of tumor necrosis factor production | 1 | 32 | 9.25e-01 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 3 | 145 | 9.25e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 1 | 69 | 9.25e-01 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 3 | 289 | 9.25e-01 |
| GO:BP | GO:1902475 | L-alpha-amino acid transmembrane transport | 1 | 55 | 9.25e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 2 | 162 | 9.26e-01 |
| GO:BP | GO:0007338 | single fertilization | 1 | 70 | 9.26e-01 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 5 | 104 | 9.26e-01 |
| GO:BP | GO:0099643 | signal release from synapse | 5 | 104 | 9.26e-01 |
| GO:BP | GO:0055092 | sterol homeostasis | 3 | 59 | 9.26e-01 |
| GO:BP | GO:0006897 | endocytosis | 2 | 527 | 9.26e-01 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 1 | 21 | 9.26e-01 |
| GO:BP | GO:0007586 | digestion | 1 | 60 | 9.28e-01 |
| GO:BP | GO:0002698 | negative regulation of immune effector process | 4 | 59 | 9.28e-01 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 1 | 38 | 9.28e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 3 | 230 | 9.28e-01 |
| GO:BP | GO:0010324 | membrane invagination | 2 | 49 | 9.28e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 9.28e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 1 | 70 | 9.28e-01 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 1 | 74 | 9.28e-01 |
| GO:BP | GO:0002275 | myeloid cell activation involved in immune response | 1 | 48 | 9.28e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 2 | 384 | 9.28e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 7 | 1335 | 9.28e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 1 | 2724 | 9.28e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 1 | 122 | 9.29e-01 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 3 | 51 | 9.29e-01 |
| GO:BP | GO:1903556 | negative regulation of tumor necrosis factor superfamily cytokine production | 1 | 33 | 9.29e-01 |
| GO:BP | GO:0008361 | regulation of cell size | 1 | 159 | 9.29e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 9.29e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 2 | 168 | 9.30e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 2 | 192 | 9.30e-01 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 1 | 40 | 9.30e-01 |
| GO:BP | GO:0030199 | collagen fibril organization | 2 | 56 | 9.31e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 2 | 211 | 9.31e-01 |
| GO:BP | GO:0045055 | regulated exocytosis | 3 | 155 | 9.32e-01 |
| GO:BP | GO:0002703 | regulation of leukocyte mediated immunity | 1 | 118 | 9.32e-01 |
| GO:BP | GO:0002347 | response to tumor cell | 1 | 22 | 9.32e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 9.32e-01 |
| GO:BP | GO:0007369 | gastrulation | 1 | 146 | 9.32e-01 |
| GO:BP | GO:2000516 | positive regulation of CD4-positive, alpha-beta T cell activation | 1 | 20 | 9.33e-01 |
| GO:BP | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | 1 | 23 | 9.34e-01 |
| GO:BP | GO:0022900 | electron transport chain | 3 | 142 | 9.34e-01 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 1 | 38 | 9.34e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 2 | 45 | 9.34e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 6 | 107 | 9.34e-01 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 36 | 9.34e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 3 | 222 | 9.35e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 9.35e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 3 | 554 | 9.36e-01 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 1 | 32 | 9.36e-01 |
| GO:BP | GO:0046717 | acid secretion | 1 | 25 | 9.37e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 1 | 25 | 9.37e-01 |
| GO:BP | GO:0042100 | B cell proliferation | 2 | 45 | 9.38e-01 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 8 | 211 | 9.38e-01 |
| GO:BP | GO:0007155 | cell adhesion | 17 | 1012 | 9.38e-01 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 1 | 137 | 9.39e-01 |
| GO:BP | GO:0046907 | intracellular transport | 17 | 1453 | 9.40e-01 |
| GO:BP | GO:0002366 | leukocyte activation involved in immune response | 2 | 154 | 9.40e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 2 | 162 | 9.41e-01 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 2 | 64 | 9.42e-01 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 1 | 116 | 9.42e-01 |
| GO:BP | GO:0071827 | plasma lipoprotein particle organization | 1 | 27 | 9.42e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 3 | 242 | 9.43e-01 |
| GO:BP | GO:0002263 | cell activation involved in immune response | 2 | 157 | 9.43e-01 |
| GO:BP | GO:0048015 | phosphatidylinositol-mediated signaling | 2 | 123 | 9.44e-01 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 4 | 67 | 9.44e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 3 | 229 | 9.44e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 9.45e-01 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 2 | 61 | 9.45e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 1 | 4794 | 9.46e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 5 | 8061 | 9.47e-01 |
| GO:BP | GO:0007492 | endoderm development | 1 | 65 | 9.47e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 9.47e-01 |
| GO:BP | GO:0050817 | coagulation | 4 | 153 | 9.47e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 6 | 337 | 9.47e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 4 | 7552 | 9.47e-01 |
| GO:BP | GO:0046640 | regulation of alpha-beta T cell proliferation | 1 | 23 | 9.47e-01 |
| GO:BP | GO:0008344 | adult locomotory behavior | 1 | 60 | 9.47e-01 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 9 | 218 | 9.47e-01 |
| GO:BP | GO:0015807 | L-amino acid transport | 1 | 65 | 9.47e-01 |
| GO:BP | GO:0009583 | detection of light stimulus | 1 | 27 | 9.47e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 1 | 52 | 9.47e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 1 | 142 | 9.47e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 9.47e-01 |
| GO:BP | GO:0048017 | inositol lipid-mediated signaling | 2 | 126 | 9.47e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 9.48e-01 |
| GO:BP | GO:0010876 | lipid localization | 6 | 310 | 9.48e-01 |
| GO:BP | GO:2000514 | regulation of CD4-positive, alpha-beta T cell activation | 2 | 39 | 9.49e-01 |
| GO:BP | GO:0030168 | platelet activation | 1 | 91 | 9.49e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 2 | 144 | 9.49e-01 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 2 | 70 | 9.49e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 9.49e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 8 | 137 | 9.49e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 2 | 199 | 9.50e-01 |
| GO:BP | GO:0045058 | T cell selection | 1 | 22 | 9.50e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 7 | 1496 | 9.50e-01 |
| GO:BP | GO:0071825 | protein-lipid complex subunit organization | 1 | 29 | 9.50e-01 |
| GO:BP | GO:1901385 | regulation of voltage-gated calcium channel activity | 1 | 29 | 9.50e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 2 | 188 | 9.50e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 2 | 234 | 9.50e-01 |
| GO:BP | GO:0016266 | O-glycan processing | 1 | 30 | 9.50e-01 |
| GO:BP | GO:0046942 | carboxylic acid transport | 4 | 216 | 9.50e-01 |
| GO:BP | GO:0002886 | regulation of myeloid leukocyte mediated immunity | 1 | 34 | 9.51e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 3 | 70 | 9.51e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 1 | 57 | 9.51e-01 |
| GO:BP | GO:0015849 | organic acid transport | 4 | 217 | 9.51e-01 |
| GO:BP | GO:0022602 | ovulation cycle process | 1 | 37 | 9.51e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 9.53e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 2 | 61 | 9.55e-01 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 1 | 47 | 9.55e-01 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 1 | 40 | 9.55e-01 |
| GO:BP | GO:0014823 | response to activity | 2 | 46 | 9.55e-01 |
| GO:BP | GO:0046633 | alpha-beta T cell proliferation | 1 | 24 | 9.55e-01 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 1 | 118 | 9.56e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 4 | 98 | 9.56e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 2 | 232 | 9.56e-01 |
| GO:BP | GO:0042269 | regulation of natural killer cell mediated cytotoxicity | 1 | 18 | 9.56e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 3 | 465 | 9.56e-01 |
| GO:BP | GO:0050856 | regulation of T cell receptor signaling pathway | 1 | 30 | 9.56e-01 |
| GO:BP | GO:0002460 | adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 2 | 161 | 9.56e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 1 | 100 | 9.56e-01 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 1 | 116 | 9.56e-01 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 1 | 119 | 9.56e-01 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 1 | 34 | 9.56e-01 |
| GO:BP | GO:1903305 | regulation of regulated secretory pathway | 3 | 89 | 9.58e-01 |
| GO:BP | GO:0002697 | regulation of immune effector process | 2 | 190 | 9.59e-01 |
| GO:BP | GO:0030316 | osteoclast differentiation | 2 | 66 | 9.59e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 5 | 369 | 9.59e-01 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 1 | 27 | 9.60e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 1 | 90 | 9.60e-01 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 1 | 37 | 9.60e-01 |
| GO:BP | GO:0002253 | activation of immune response | 3 | 272 | 9.60e-01 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 3 | 88 | 9.60e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 9.60e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 3 | 296 | 9.61e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 1 | 36 | 9.61e-01 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 1 | 36 | 9.61e-01 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 9.61e-01 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 1 | 52 | 9.61e-01 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 4 | 74 | 9.61e-01 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 1 | 65 | 9.61e-01 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 3 | 88 | 9.62e-01 |
| GO:BP | GO:0002704 | negative regulation of leukocyte mediated immunity | 1 | 28 | 9.62e-01 |
| GO:BP | GO:0043627 | response to estrogen | 2 | 52 | 9.62e-01 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 1 | 71 | 9.63e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 10 | 318 | 9.63e-01 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 2 | 65 | 9.63e-01 |
| GO:BP | GO:0042110 | T cell activation | 1 | 313 | 9.63e-01 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 1 | 43 | 9.63e-01 |
| GO:BP | GO:0015031 | protein transport | 34 | 1361 | 9.63e-01 |
| GO:BP | GO:0009409 | response to cold | 1 | 41 | 9.63e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 1 | 245 | 9.63e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 1 | 51 | 9.63e-01 |
| GO:BP | GO:0001775 | cell activation | 3 | 655 | 9.63e-01 |
| GO:BP | GO:0061024 | membrane organization | 4 | 671 | 9.63e-01 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 1 | 43 | 9.64e-01 |
| GO:BP | GO:0002715 | regulation of natural killer cell mediated immunity | 1 | 18 | 9.64e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 1 | 56 | 9.64e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 1 | 103 | 9.64e-01 |
| GO:BP | GO:0006911 | phagocytosis, engulfment | 1 | 33 | 9.65e-01 |
| GO:BP | GO:0045332 | phospholipid translocation | 1 | 33 | 9.65e-01 |
| GO:BP | GO:0043038 | amino acid activation | 1 | 44 | 9.66e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 5 | 166 | 9.68e-01 |
| GO:BP | GO:0006810 | transport | 27 | 3298 | 9.68e-01 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 3 | 64 | 9.69e-01 |
| GO:BP | GO:0060425 | lung morphogenesis | 1 | 38 | 9.70e-01 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 1 | 44 | 9.70e-01 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 3 | 55 | 9.70e-01 |
| GO:BP | GO:0070555 | response to interleukin-1 | 3 | 83 | 9.73e-01 |
| GO:BP | GO:0008347 | glial cell migration | 1 | 47 | 9.75e-01 |
| GO:BP | GO:0010467 | gene expression | 1 | 4587 | 9.75e-01 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 1 | 77 | 9.76e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 3 | 561 | 9.77e-01 |
| GO:BP | GO:0006497 | protein lipidation | 1 | 89 | 9.77e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 6 | 484 | 9.77e-01 |
| GO:BP | GO:0065007 | biological regulation | 5 | 8320 | 9.78e-01 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 1 | 45 | 9.78e-01 |
| GO:BP | GO:0006821 | chloride transport | 1 | 58 | 9.78e-01 |
| GO:BP | GO:0060419 | heart growth | 2 | 72 | 9.78e-01 |
| GO:BP | GO:0002548 | monocyte chemotaxis | 1 | 22 | 9.78e-01 |
| GO:BP | GO:0001912 | positive regulation of leukocyte mediated cytotoxicity | 1 | 20 | 9.79e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 9.79e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 9.79e-01 |
| GO:BP | GO:0071702 | organic substance transport | 12 | 2116 | 9.79e-01 |
| GO:BP | GO:0007596 | blood coagulation | 3 | 150 | 9.80e-01 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 1 | 92 | 9.81e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 2 | 3600 | 9.81e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 8 | 609 | 9.81e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 2 | 78 | 9.83e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 10 | 877 | 9.84e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 1 | 41 | 9.85e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 2 | 523 | 9.85e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 4 | 389 | 9.85e-01 |
| GO:BP | GO:0006805 | xenobiotic metabolic process | 1 | 56 | 9.85e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 1 | 74 | 9.85e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 48 | 9.86e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 9.87e-01 |
| GO:BP | GO:0060420 | regulation of heart growth | 1 | 50 | 9.87e-01 |
| GO:BP | GO:0042698 | ovulation cycle | 1 | 54 | 9.87e-01 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 4 | 90 | 9.87e-01 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 1 | 45 | 9.88e-01 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 1 | 36 | 9.88e-01 |
| GO:BP | GO:0031341 | regulation of cell killing | 1 | 36 | 9.88e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 9.89e-01 |
| GO:BP | GO:0031343 | positive regulation of cell killing | 1 | 23 | 9.89e-01 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 5 | 105 | 9.90e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 9.90e-01 |
| GO:BP | GO:0051049 | regulation of transport | 9 | 1267 | 9.91e-01 |
| GO:BP | GO:0006865 | amino acid transport | 1 | 102 | 9.92e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 1 | 86 | 9.92e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 12 | 422 | 9.92e-01 |
| GO:BP | GO:0051668 | localization within membrane | 2 | 605 | 9.93e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 9.93e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 1 | 136 | 9.93e-01 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 1 | 57 | 9.93e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 9.93e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 9.94e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 1 | 86 | 9.94e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 9.94e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 9.94e-01 |
| GO:BP | GO:0006396 | RNA processing | 1 | 929 | 9.95e-01 |
| GO:BP | GO:0045292 | mRNA cis splicing, via spliceosome | 1 | 26 | 9.95e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 9.96e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 1 | 289 | 9.96e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 1 | 289 | 9.96e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 1 | 293 | 9.96e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 4 | 1684 | 9.96e-01 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 5 | 124 | 9.97e-01 |
| GO:BP | GO:0001910 | regulation of leukocyte mediated cytotoxicity | 1 | 30 | 9.98e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 9.99e-01 |
| GO:BP | GO:0008380 | RNA splicing | 1 | 419 | 9.99e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 1 | 453 | 9.99e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 9.99e-01 |
| GO:BP | GO:0006397 | mRNA processing | 1 | 450 | 9.99e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 9.99e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 9.99e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 3 | 227 | 9.99e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 2 | 665 | 1.00e+00 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 5 | 581 | 1.00e+00 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 1.00e+00 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 1 | 118 | 1.00e+00 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 1 | 118 | 1.00e+00 |
| GO:BP | GO:0000387 | spliceosomal snRNP assembly | 1 | 45 | 1.00e+00 |
| GO:BP | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | 1 | 22 | 1.00e+00 |
| GO:BP | GO:0008150 | biological_process | 1 | 12070 | 1.00e+00 |
| GO:BP | GO:0008152 | metabolic process | 1 | 8518 | 1.00e+00 |
| GO:BP | GO:0070972 | protein localization to endoplasmic reticulum | 1 | 78 | 1.00e+00 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 4 | 250 | 1.00e+00 |
| GO:BP | GO:0072599 | establishment of protein localization to endoplasmic reticulum | 2 | 53 | 1.00e+00 |
| GO:BP | GO:0006613 | cotranslational protein targeting to membrane | 1 | 35 | 1.00e+00 |
| GO:BP | GO:0000244 | spliceosomal tri-snRNP complex assembly | 1 | 20 | 1.00e+00 |
| GO:BP | GO:0006612 | protein targeting to membrane | 2 | 118 | 1.00e+00 |
| GO:BP | GO:0006399 | tRNA metabolic process | 1 | 186 | 1.00e+00 |
| GO:BP | GO:0006376 | mRNA splice site selection | 1 | 31 | 1.00e+00 |
| GO:BP | GO:0009987 | cellular process | 1 | 11336 | 1.00e+00 |
| GO:BP | GO:0045047 | protein targeting to ER | 1 | 49 | 1.00e+00 |
| GO:BP | GO:0009593 | detection of chemical stimulus | 1 | 27 | 1.00e+00 |
| GO:BP | GO:0071704 | organic substance metabolic process | 1 | 8169 | 1.00e+00 |
| GO:BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1 | 30 | 1.00e+00 |
| KEGG | KEGG:04110 | Cell cycle | 48 | 151 | 4.85e-19 |
| KEGG | KEGG:03030 | DNA replication | 17 | 35 | 5.41e-10 |
| KEGG | KEGG:04115 | p53 signaling pathway | 22 | 65 | 1.60e-08 |
| KEGG | KEGG:03440 | Homologous recombination | 13 | 39 | 5.01e-05 |
| KEGG | KEGG:03430 | Mismatch repair | 9 | 22 | 1.35e-04 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 12 | 48 | 2.12e-03 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 5 | 61 | 4.29e-03 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 10 | 43 | 8.56e-03 |
| KEGG | KEGG:04217 | Necroptosis | 5 | 106 | 8.56e-03 |
| KEGG | KEGG:01524 | Platinum drug resistance | 11 | 65 | 1.07e-02 |
| KEGG | KEGG:05034 | Alcoholism | 6 | 122 | 1.29e-02 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 5 | 110 | 1.29e-02 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 2 | 20 | 1.29e-02 |
| KEGG | KEGG:03410 | Base excision repair | 6 | 33 | 1.42e-02 |
| KEGG | KEGG:04114 | Oocyte meiosis | 18 | 108 | 1.67e-02 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 15 | 86 | 1.77e-02 |
| KEGG | KEGG:05216 | Thyroid cancer | 1 | 35 | 4.76e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 6.92e-02 |
| KEGG | KEGG:05218 | Melanoma | 1 | 58 | 6.92e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 6.92e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 1 | 67 | 6.92e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 1 | 75 | 6.92e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 1 | 57 | 6.92e-02 |
| KEGG | KEGG:05214 | Glioma | 1 | 67 | 6.92e-02 |
| KEGG | KEGG:00230 | Purine metabolism | 4 | 97 | 7.45e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 1 | 84 | 7.45e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 7.45e-02 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 8.41e-02 |
| KEGG | KEGG:05224 | Breast cancer | 1 | 117 | 1.04e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 1.04e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 1 | 117 | 1.04e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 2 | 69 | 1.07e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 1.07e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 1 | 145 | 1.07e-01 |
| KEGG | KEGG:04210 | Apoptosis | 4 | 118 | 1.13e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 1.13e-01 |
| KEGG | KEGG:04540 | Gap junction | 3 | 73 | 1.13e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 1 | 128 | 1.13e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 10 | 63 | 1.13e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 16 | 145 | 1.99e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 3 | 10 | 2.00e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 1 | 16 | 2.03e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 1 | 46 | 2.04e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 4 | 30 | 2.19e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 4 | 26 | 2.20e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 13 | 172 | 2.46e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 1 | 409 | 2.46e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 2 | 55 | 2.72e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 1 | 29 | 2.91e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 1 | 84 | 3.31e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 4 | 214 | 3.32e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 17 | 180 | 3.34e-01 |
| KEGG | KEGG:04145 | Phagosome | 3 | 99 | 3.62e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 3.64e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 1 | 115 | 4.12e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 9 | 73 | 4.16e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 7 | 56 | 4.40e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 1 | 62 | 4.48e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 2 | 44 | 4.48e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 5 | 303 | 4.54e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 3 | 25 | 4.54e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 7 | 71 | 4.54e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 4 | 109 | 4.54e-01 |
| KEGG | KEGG:05164 | Influenza A | 2 | 103 | 4.58e-01 |
| KEGG | KEGG:04360 | Axon guidance | 1 | 162 | 4.58e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 9 | 78 | 4.58e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 3 | 150 | 4.66e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 2 | 44 | 4.66e-01 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 1 | 6 | 5.09e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 2 | 7 | 5.09e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 2 | 16 | 5.14e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 2 | 106 | 5.17e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 5.17e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 4 | 76 | 5.17e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 2 | 74 | 5.17e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 1 | 21 | 5.17e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 9 | 385 | 5.17e-01 |
| KEGG | KEGG:04976 | Bile secretion | 1 | 42 | 5.17e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 3 | 157 | 5.32e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 1 | 58 | 5.32e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 5.32e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 1 | 19 | 5.65e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 2 | 18 | 5.81e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 2 | 11 | 5.83e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 2 | 31 | 5.86e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 1 | 10 | 6.03e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 6.03e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 1 | 20 | 6.03e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 1 | 41 | 6.03e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 2 | 27 | 6.03e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 4 | 135 | 6.03e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 1 | 21 | 6.09e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 2 | 87 | 6.09e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 12 | 6.09e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 1 | 56 | 6.09e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 2 | 106 | 6.09e-01 |
| KEGG | KEGG:03265 | Virion - Lyssavirus | 1 | 3 | 6.09e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 3 | 33 | 6.09e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 6.12e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 6.12e-01 |
| KEGG | KEGG:05020 | Prion disease | 3 | 220 | 6.19e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 4 | 134 | 6.47e-01 |
| KEGG | KEGG:04146 | Peroxisome | 6 | 68 | 6.47e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 1 | 63 | 6.47e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 3 | 70 | 6.52e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 1 | 35 | 6.59e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 2 | 82 | 6.64e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 1 | 49 | 6.64e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 4 | 170 | 6.65e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1 | 69 | 6.65e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 6.65e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 5 | 83 | 6.65e-01 |
| KEGG | KEGG:04530 | Tight junction | 2 | 137 | 6.65e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 6.65e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 1 | 91 | 6.65e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 1 | 14 | 6.65e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 6.65e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 6.65e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 2 | 133 | 6.65e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 2 | 38 | 6.65e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 4 | 49 | 6.65e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 1 | 67 | 6.65e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 1 | 83 | 6.65e-01 |
| KEGG | KEGG:05016 | Huntington disease | 3 | 256 | 6.65e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 2 | 28 | 6.65e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 6.65e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 1 | 43 | 6.65e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 2 | 128 | 6.71e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 1 | 14 | 6.71e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 3 | 37 | 6.71e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 1 | 40 | 6.71e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 2 | 24 | 6.71e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 6.86e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 4 | 165 | 6.88e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 1 | 12 | 6.89e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 1 | 61 | 6.89e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 2 | 107 | 6.89e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 2 | 24 | 6.89e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 1 | 98 | 6.89e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 1 | 112 | 6.89e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 2 | 114 | 6.89e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 5 | 77 | 6.93e-01 |
| KEGG | KEGG:04924 | Renin secretion | 4 | 50 | 6.93e-01 |
| KEGG | KEGG:03018 | RNA degradation | 3 | 74 | 6.93e-01 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 6.93e-01 |
| KEGG | KEGG:05162 | Measles | 4 | 98 | 6.93e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 6.93e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 10 | 6.93e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 2 | 151 | 6.93e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 6.95e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 1 | 51 | 6.95e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 1 | 28 | 6.95e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 1 | 58 | 6.95e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 3 | 315 | 6.95e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 6.95e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 1 | 115 | 6.95e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 6.95e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 5 | 272 | 6.95e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 10 | 6.95e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 1 | 15 | 6.95e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 6.95e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 5 | 50 | 6.95e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 4 | 1146 | 6.95e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 1 | 20 | 6.95e-01 |
| KEGG | KEGG:03040 | Spliceosome | 4 | 132 | 6.95e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 1 | 23 | 6.95e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 3 | 38 | 6.95e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 4 | 56 | 6.95e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 2 | 133 | 6.95e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 4 | 124 | 7.12e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 69 | 7.12e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 6 | 69 | 7.12e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 1 | 88 | 7.12e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 7.26e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 1 | 98 | 7.41e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 1 | 46 | 7.41e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 2 | 226 | 7.41e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 3 | 50 | 7.41e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 11 | 7.41e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 7.41e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 7.41e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 7.41e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 3 | 41 | 7.41e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 3 | 415 | 7.41e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 2 | 79 | 7.41e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 1 | 14 | 7.46e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 1 | 51 | 7.48e-01 |
| KEGG | KEGG:02010 | ABC transporters | 1 | 29 | 7.51e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 4 | 49 | 7.57e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 4 | 138 | 7.57e-01 |
| KEGG | KEGG:04744 | Phototransduction | 1 | 12 | 7.57e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 4 | 76 | 7.57e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 1 | 17 | 7.57e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 1 | 59 | 7.57e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 1 | 110 | 7.57e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 3 | 97 | 7.58e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 2 | 104 | 7.70e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 7.72e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 1 | 16 | 7.76e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 2 | 66 | 7.76e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 1 | 19 | 7.76e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 3 | 99 | 7.76e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 3 | 37 | 7.76e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 7.76e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 2 | 31 | 7.87e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 1 | 44 | 7.88e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 4 | 77 | 8.02e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 1 | 106 | 8.10e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 7 | 121 | 8.10e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 3 | 48 | 8.10e-01 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 1 | 9 | 8.10e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 12 | 8.15e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 2 | 23 | 8.31e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 2 | 22 | 8.31e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 1 | 4 | 8.31e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1 | 5558 | 8.31e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 1 | 28 | 8.31e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 1 | 17 | 8.40e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 1 | 65 | 8.42e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 1 | 30 | 8.42e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 1 | 63 | 8.42e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 4 | 60 | 8.42e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 2 | 49 | 8.42e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 2 | 161 | 8.49e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 10 | 8.49e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 4 | 85 | 8.57e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 3 | 64 | 8.60e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 4 | 86 | 8.70e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 2 | 53 | 8.88e-01 |
| KEGG | KEGG:03060 | Protein export | 1 | 23 | 9.02e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 2 | 55 | 9.03e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 2 | 164 | 9.06e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 2 | 108 | 9.08e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 2 | 48 | 9.08e-01 |
| KEGG | KEGG:05142 | Chagas disease | 1 | 75 | 9.08e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 5 | 115 | 9.10e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 3 | 52 | 9.12e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 2 | 25 | 9.12e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 1 | 33 | 9.21e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 3 | 72 | 9.21e-01 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 1 | 13 | 9.21e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 1 | 22 | 9.21e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 2 | 43 | 9.21e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 9.21e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 3 | 121 | 9.24e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 1 | 34 | 9.27e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 2 | 94 | 9.27e-01 |
| KEGG | KEGG:04520 | Adherens junction | 1 | 67 | 9.41e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 1 | 240 | 9.43e-01 |
| KEGG | KEGG:04144 | Endocytosis | 3 | 232 | 9.43e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 2 | 176 | 9.51e-01 |
| KEGG | KEGG:05131 | Shigellosis | 1 | 210 | 9.52e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 1 | 35 | 9.57e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 1 | 32 | 9.57e-01 |
| KEGG | KEGG:04742 | Taste transduction | 2 | 22 | 9.57e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 4 | 91 | 9.57e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 2 | 78 | 9.57e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 1 | 24 | 9.58e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 3 | 84 | 9.62e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 1 | 112 | 9.62e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 2 | 40 | 9.63e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 1 | 46 | 9.63e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 1 | 131 | 9.64e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 5 | 195 | 9.64e-01 |
| KEGG | KEGG:05134 | Legionellosis | 1 | 39 | 9.67e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 2 | 103 | 9.67e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 9.67e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 6 | 142 | 9.77e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 3 | 103 | 9.86e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 1.00e+00 |
| KEGG | KEGG:04931 | Insulin resistance | 3 | 95 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 3 | 169 | 1.00e+00 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 64 | 1.00e+00 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 1 | 49 | 1.00e+00 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 1 | 44 | 1.00e+00 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 1.00e+00 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 1 | 99 | 1.00e+00 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 2 | 179 | 1.00e+00 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 2 | 84 | 1.00e+00 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 1.00e+00 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 2 | 110 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 1 | 40 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 1 | 75 | 1.00e+00 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 1.00e+00 |
| KEGG | KEGG:04970 | Salivary secretion | 1 | 55 | 1.00e+00 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 5 | 254 | 1.00e+00 |
| KEGG | KEGG:04142 | Lysosome | 1 | 118 | 1.00e+00 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 1 | 44 | 1.00e+00 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 1 | 74 | 1.00e+00 |
| KEGG | KEGG:04014 | Ras signaling pathway | 2 | 171 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 2 | 98 | 1.00e+00 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 1 | 86 | 1.00e+00 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 1 | 97 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 4 | 127 | 1.00e+00 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 1 | 74 | 1.00e+00 |
| KEGG | KEGG:04611 | Platelet activation | 1 | 89 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 2 | 89 | 1.00e+00 |
What I will do is take the terms from the top three KEGG pathways and look at count changes or at FC values. Updated later.
DXspecific response (only the subset unique to Dox)
# DXspgostres24 <- gost(query = DXsprespon,
# organism = "hsapiens", significant = FALSE,
# ordered_query = TRUE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
#
# saveRDS(DXspgostres24,"data/DEG-GO/DXspgostres24.RDS")
DXspgostres24 <- readRDS("data/DEG-GO/DXspgostres24.RDS")
# DXspp24 <- gostplot(DXspgostres24, capped = FALSE, interactive = TRUE)
# DXspp24
DXsptable24 <- DXspgostres24$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
DXsptable24 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = scales::label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle(paste0("DOX specific DEGs (n = ", length(DXsprespon), ") 24 hr GO:BP terms")) +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)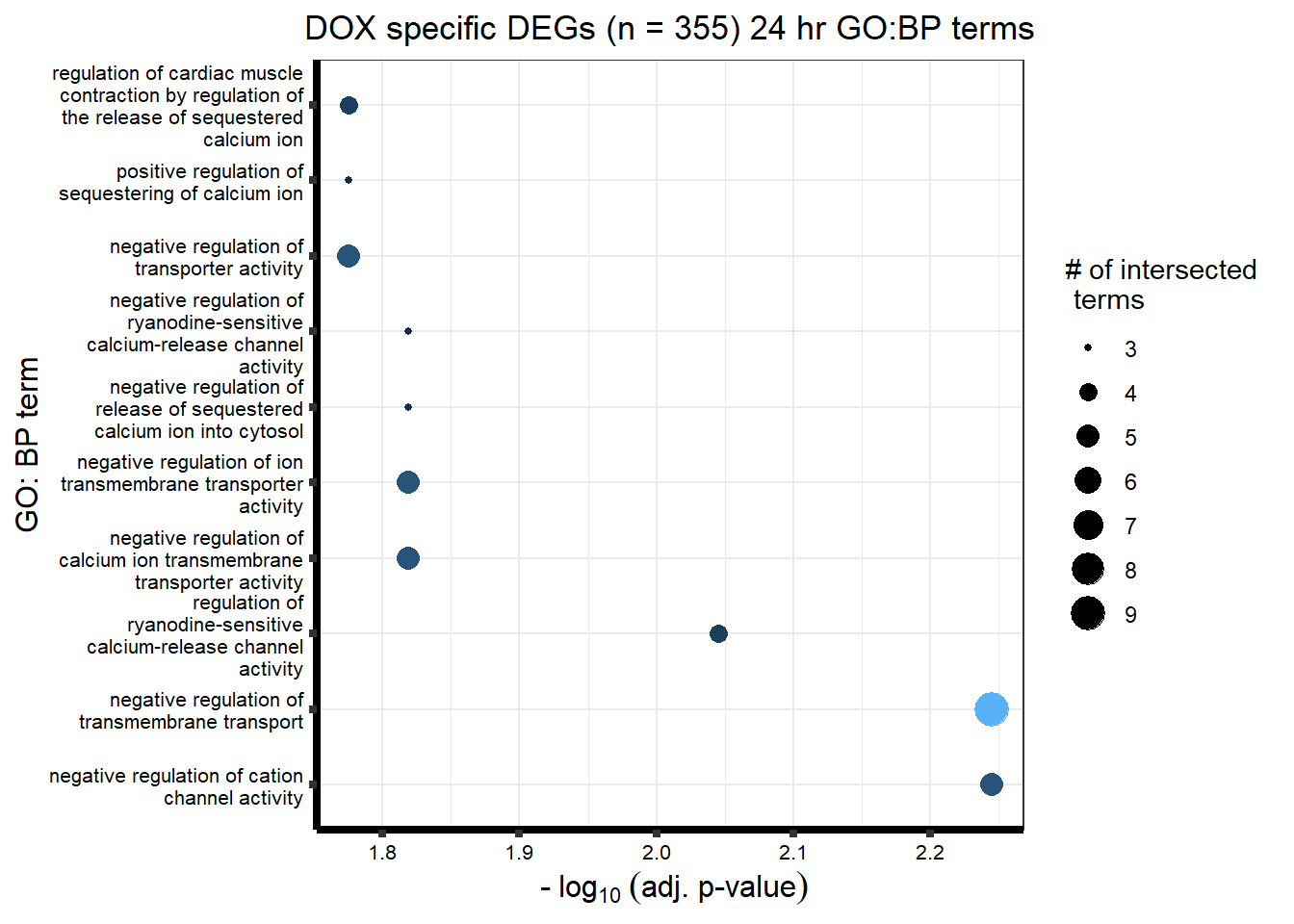
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
DXsptable24 %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 5 | 30 | 5.69e-03 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 9 | 96 | 5.69e-03 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 4 | 23 | 9.01e-03 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 3 | 10 | 1.52e-02 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 5 | 51 | 1.52e-02 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 5 | 27 | 1.52e-02 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 3 | 11 | 1.52e-02 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 4 | 20 | 1.68e-02 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 5 | 58 | 1.68e-02 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 3 | 13 | 1.68e-02 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 6 | 100 | 1.79e-02 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 5 | 64 | 1.94e-02 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 5 | 70 | 2.31e-02 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 4 | 35 | 2.31e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 4 | 25 | 2.31e-02 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 5 | 71 | 2.31e-02 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 2 | 114 | 2.48e-02 |
| GO:BP | GO:0043311 | positive regulation of eosinophil degranulation | 2 | 2 | 2.70e-02 |
| GO:BP | GO:1902568 | positive regulation of eosinophil activation | 2 | 2 | 2.70e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 4 | 27 | 2.88e-02 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 2 | 140 | 3.10e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 4 | 28 | 4.22e-02 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 4 | 45 | 4.22e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 4 | 28 | 4.52e-02 |
| GO:BP | GO:1903785 | L-valine transmembrane transport | 1 | 1 | 4.99e-02 |
As above, I will pull the specific genes and look at logCPM of these genes soon to examine if the change is seen in the other AC genes at 24 hours.
No Response genes n=5727
#
# gostresNoResp <- gost(query = NoRespDEG,
# organism = "hsapiens", significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP","KEGG"))
#
#saveRDS(gostresNoResp,"data/DEG-GO/gostresNoResp.RDS")
gostresNoResp <- readRDS("data/DEG-GO/gostresNoResp.RDS")
# Noresp24 <- gostplot(gostresNoResp, capped = FALSE, interactive = TRUE)
# Noresp24
Noresp24table <- gostresNoResp$result %>%
dplyr::select(c(
source,
term_id,
term_name,
intersection_size,
term_size,
p_value
))
Noresp24table %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle(paste0("No response genes(n = ", length(NoRespDEG), ") GO:BP terms")) +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("GO: BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)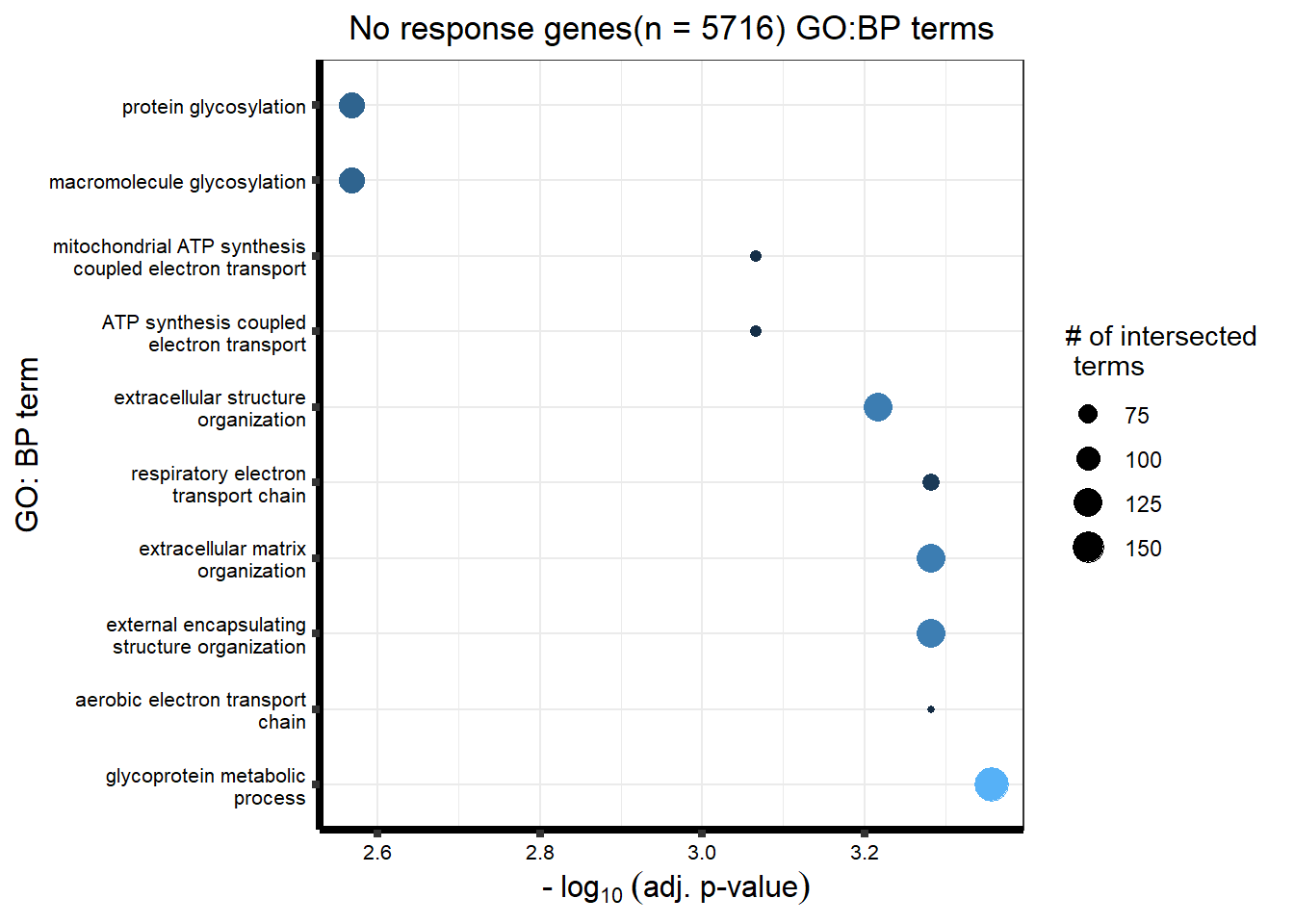
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
Noresp24table %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 10 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, p_value),
col = intersection_size
)) +
geom_point(aes(size = intersection_size, col = "red")) +
scale_y_discrete(labels = label_wrap(30)) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle(paste0("No response genes(n = ", length(NoRespDEG), ") 24 hr KEGG terms")) +
xlab(expression(" -" ~ log[10] ~ ("adj. p-value"))) +
ylab("KEGG term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.2), hjust = 0.5),
axis.title = element_text(size = 12, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 15,
color = "black",
face = "bold"
)
)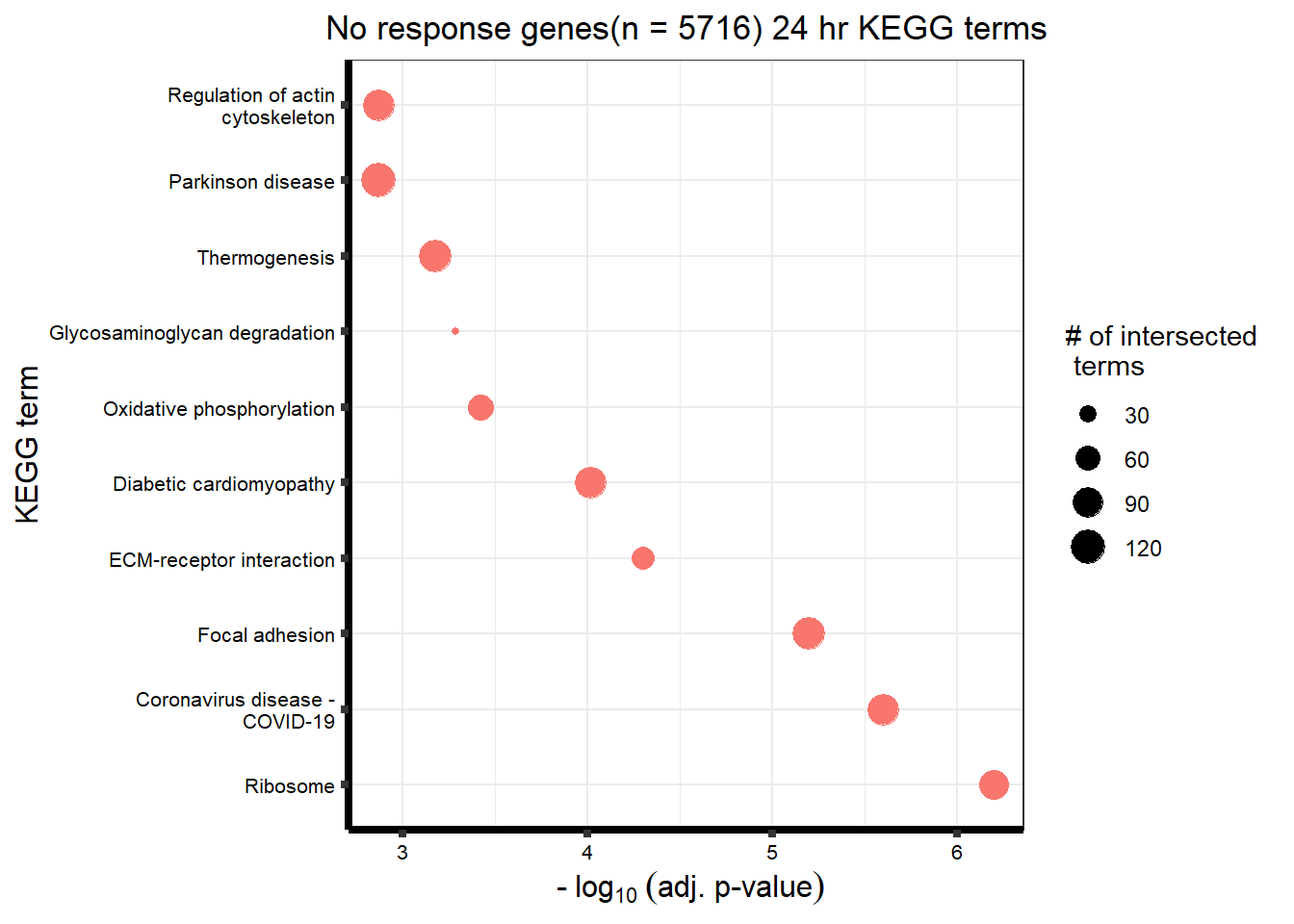
Noresp24table %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0009100 | glycoprotein metabolic process | 174 | 317 | 4.39e-04 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 67 | 103 | 5.22e-04 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 131 | 231 | 5.22e-04 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 52 | 74 | 5.22e-04 |
| GO:BP | GO:0030198 | extracellular matrix organization | 130 | 230 | 5.22e-04 |
| GO:BP | GO:0043062 | extracellular structure organization | 130 | 231 | 6.06e-04 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 55 | 82 | 8.58e-04 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 55 | 82 | 8.58e-04 |
| GO:BP | GO:0006486 | protein glycosylation | 107 | 189 | 2.71e-03 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 107 | 189 | 2.71e-03 |
| GO:BP | GO:0070085 | glycosylation | 114 | 204 | 2.76e-03 |
| GO:BP | GO:0006812 | monoatomic cation transport | 323 | 668 | 3.62e-03 |
| GO:BP | GO:0006811 | monoatomic ion transport | 367 | 774 | 7.37e-03 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 135 | 254 | 9.85e-03 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 78 | 134 | 1.06e-02 |
| GO:BP | GO:0002181 | cytoplasmic translation | 88 | 155 | 1.12e-02 |
| GO:BP | GO:0022900 | electron transport chain | 81 | 141 | 1.26e-02 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 99 | 179 | 1.35e-02 |
| GO:BP | GO:0009060 | aerobic respiration | 99 | 180 | 1.73e-02 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 54 | 89 | 3.50e-02 |
| GO:BP | GO:0030001 | metal ion transport | 269 | 563 | 3.59e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 455 | 996 | 3.59e-02 |
| GO:BP | GO:0045333 | cellular respiration | 117 | 222 | 3.59e-02 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 83 | 149 | 3.59e-02 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 259 | 541 | 3.75e-02 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 413 | 899 | 3.99e-02 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 298 | 633 | 4.66e-02 |
| GO:BP | GO:0034645 | cellular macromolecule biosynthetic process | 468 | 1030 | 4.66e-02 |
| KEGG | KEGG:03010 | Ribosome | 84 | 127 | 6.40e-07 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 100 | 162 | 2.53e-06 |
| KEGG | KEGG:04510 | Focal adhesion | 105 | 175 | 6.34e-06 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 48 | 69 | 4.97e-05 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 98 | 169 | 9.70e-05 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 65 | 106 | 3.78e-04 |
| KEGG | KEGG:00531 | Glycosaminoglycan degradation | 15 | 16 | 5.15e-04 |
| KEGG | KEGG:04714 | Thermogenesis | 107 | 195 | 6.63e-04 |
| KEGG | KEGG:05012 | Parkinson disease | 120 | 226 | 1.34e-03 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 98 | 179 | 1.34e-03 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 101 | 186 | 1.46e-03 |
| KEGG | KEGG:05020 | Prion disease | 114 | 220 | 6.11e-03 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 129 | 254 | 6.95e-03 |
| KEGG | KEGG:05146 | Amoebiasis | 39 | 64 | 1.33e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 85 | 161 | 1.36e-02 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 71 | 131 | 1.36e-02 |
| KEGG | KEGG:00000 | KEGG root term | 2295 | 5534 | 2.43e-02 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 50 | 89 | 2.63e-02 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 87 | 170 | 3.21e-02 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 39 | 67 | 3.21e-02 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 44 | 78 | 3.82e-02 |
##GO:BP full figure 24 DEG analysis
DDEdeg <- DDEtable24only %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DDEMdeg <- DDEMtable24 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DAsp3deg <- DAsptable3 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DXsp24deg <- DXsptable24 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
# EPsp24deg <- EPsptable124 %>%
# dplyr::filter(source=="GO:BP") %>%
# dplyr::select(p_value,term_name,intersection_size) %>%
# slice_min(., n=10 ,order_by=p_value) %>%
# mutate(log_val = -log10(p_value)) %>%
# mutate(order_val=rev(as.numeric(rownames(.))))
MTsp24deg <- MTsptable124 %>%
dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DAdeg <- DAtable124 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DXdeg <- DXtable124 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
EPdeg <- EPtable124 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
MTdeg <- MTtable124 %>% dplyr::filter(source == "GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
slice_min(., n = 5 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
GODEGlong <- list(
"DXdeg" = DXdeg,
"DXsp24deg" = DXsp24deg,
"DAdeg" = DAdeg,
"EPdeg" = EPdeg,
"MTdeg" = MTdeg,
"MTsp24deg" = MTsp24deg,
"DDEdeg" = DDEdeg,
"DDEMdeg" = DDEMdeg
)
GODEGlongtable <- data.table::rbindlist(GODEGlong, idcol = "deg")
p <- GODEGlongtable %>%
mutate(deg = fct_inorder(deg)) %>%
ggplot(., aes(
x = log_val,
y = reorder(term_name, order_val, desc = TRUE),
col = deg
)) +
geom_point(aes(size = intersection_size, col = deg)) +
scale_y_discrete(labels = label_wrap(40)) +
scale_color_manual(
values = c(
"DXdeg" = "#8B006D",
"DXsp24deg" = "#b0028a",
"DAdeg" = "#DF707E",
"EPdeg" = "#F1B72B",
"MTdeg" = "#33a1dd",
"MTsp24deg" = "#33a1dd",
"DDEdeg" = "red",
"DDEMdeg" = "blue",
alpha = 0.8
)
) +
facet_grid(deg ~ ., scales = "free_y" , labeller = label_bquote(.(as.expression(eval(
parse(text = paste0('facet_deg', '$`', deg, '`'))
))))) +
guides(col = "none",
size = guide_legend(title = "# of intersected \n terms")) +
ggtitle('Top enriched GO:BP terms for each 24 hour DEG list') +
xlab("-log 10 (adj. p-value)") +
ylab("GO:BP term") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white")
)
facet_deg = (
c(
DXdeg = expression(atop("DOX 24 hr", ~ italic("n = 6645"))),
DXsp24deg = expression(atop("DOX-specific 24 hr", ~ italic("n = 355"))),
DAdeg = expression(atop("DNR 24 hr", ~ italic("n = 7017"))),
EPdeg = expression(atop("EPI 24 hr", ~ italic("n = 6328"))),
MTdeg = expression(atop("MTX 24 hr", ~ italic("n = 1115"))),
MTsp24deg = expression(atop("MTX-specific 24 hr", ~ italic("n = 122"))),
DDEdeg = expression(atop("AC 24 hr", ~ italic("n = 4435,"))),
DDEMdeg = expression(atop("TOP2i 24 hr", ~ italic("n = 882")))
)
)
g <- ggplot_gtable(ggplot_build(p))
stripr <- which(grepl('strip-r', g$layout$name))
fills <-
c(
"#8B006D",
"#b0028a",
"#DF707E",
"#F1B72B",
"#3386DD",
"#33a1dd",
"red",
"blue",
alpha = 0.8
)
k <- 1
for (i in stripr) {
j <- which(grepl('rect', g$grobs[[i]]$grobs[[1]]$childrenOrder))
g$grobs[[i]]$grobs[[1]]$children[[j]]$gp$fill <- fills[k]
k <- k + 1
}
print(grid::grid.draw(g))NULLgrid::grid.draw(g)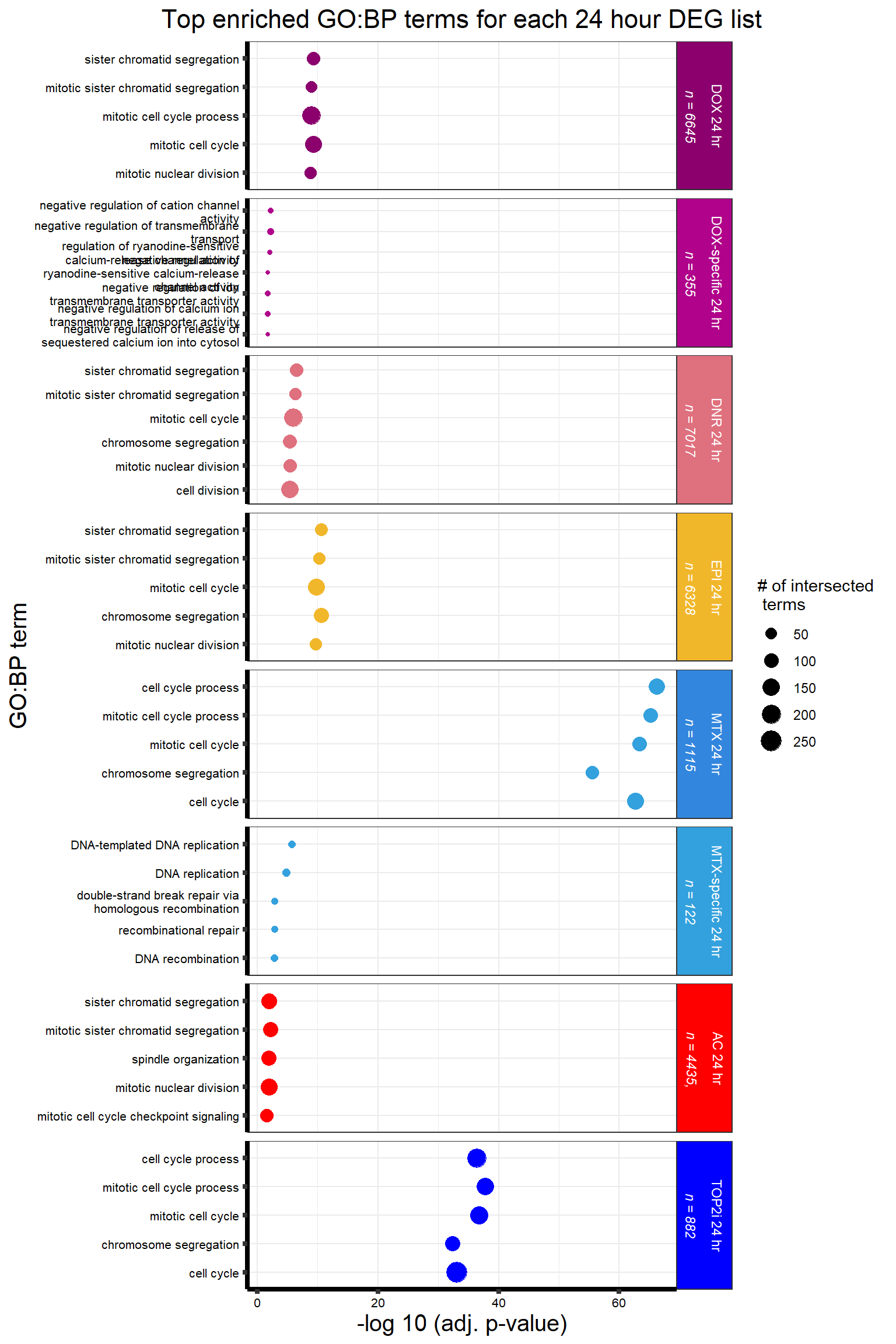
Visualization of FC correlation
efit2 <- readRDS(file = "data/efit2_final.RDS")
FCmatrix <- subset(efit2$coefficients)
is.matrix(subset(FCmatrix))[1] TRUEcolnames(FCmatrix) <-
c(
"DNR-3h",
"DOX-3h",
"EPI-3h",
"MTX-3h",
"TRZ-3h",
"DNR-24h",
"DOX-24h",
"EPI-24h",
"MTX-24h",
"TRZ-24h"
)
mat_col <- data.frame(time = c(rep("3 hours", 5), rep("24 hours", 5)))
rownames(mat_col) <- colnames(FCmatrix)
mat_colors <- list(time = c("#C25757", "#A77E69"))
names(mat_colors$time) <- unique(mat_col$time)
corrFC <- cor(FCmatrix)
ComplexHeatmap::pheatmap(
corrFC,
display_numbers = TRUE,
number_format = "%.2f",
main = ("Correlation of FC values across all expressed genes, n=14084"),
annotation_col = mat_col,
annotation_colors = mat_colors
)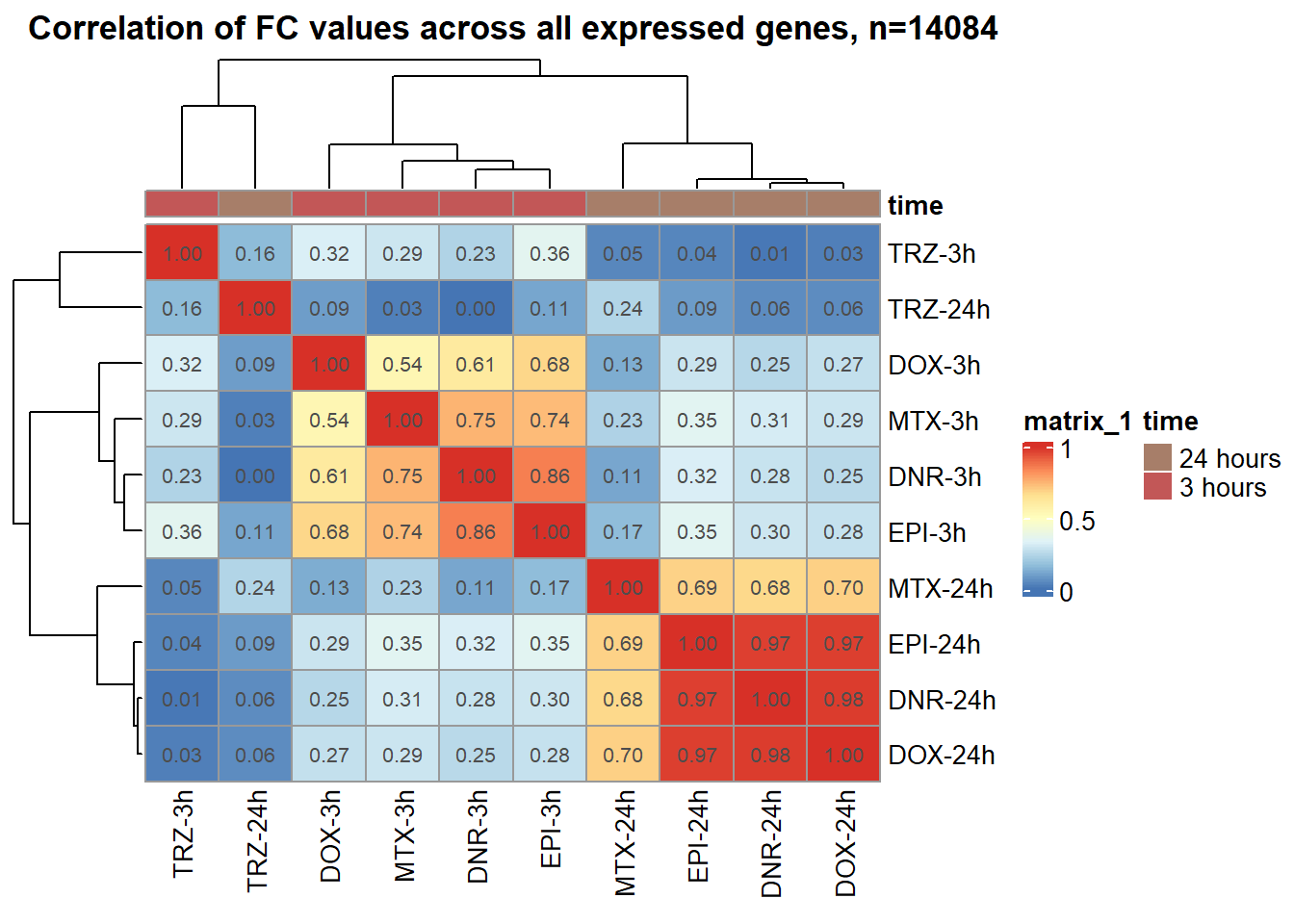
KEGG pathway intersection of pairwise DEG sets
note: grey box means these terms were not found to be enriched in the data set terms. I will try to change the levels to pull out the pvalue of the term for each (6-30-23)
kegglistDEG <- readRDS("data/kegglistDEG24.RDS")
list2env(kegglistDEG, envir = .GlobalEnv)<environment: R_GlobalEnv>kegg_term24 <-
data.frame(
term = c(
"p53 signaling pathway",
"DNA replication",
"Cell cycle" ,
"Base excision repair"
),
termid = c("KEGG:04115", "KEGG:03030", "KEGG:04110", "KEGG:03410")
)
DDEdegk <- DDEtable24only %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term24$termid) %>%
# slice_min(., n=4 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DDEMdegk <- DDEMtable24 %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term24$termid) %>%
# slice_min(., n=3 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DAdegk <- DAtable124 %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term24$termid) %>%
slice_min(., n = 3 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DXdegk <- DXtable124 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term24$termid) %>%
# slice_min(., n=3 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
EPdegk <- EPtable124 %>% dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term24$termid) %>%
# slice_min(., n=3 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
MTdegk <- MTtable124 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term24$termid) %>%
# slice_min(., n=3 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
library(ComplexHeatmap)
keggDEGlong <- list(
"DOX" = DXdegk,
"EPI" = EPdegk,
"DNR" = DAdegk,
"MTX" = MTdegk,
"ACs" = DDEdegk,
"TOP2i" = DDEMdegk
)
keggtable <- data.table::rbindlist(keggDEGlong, idcol = "deg")
kegg_sig_mat <- keggtable %>%
dplyr::select(deg, p_value, term_name) %>%
pivot_wider(
id_cols = everything(),
names_from = "term_name",
values_from = "p_value",
values_fill = list(p_value = 1)
) %>%
column_to_rownames('deg') %>%
as.matrix()#
kegg_mat <- keggtable %>%
dplyr::select(deg, log_val, term_name) %>%
pivot_wider(id_cols = everything(),
names_from = "term_name",
values_from = "log_val") %>%
column_to_rownames('deg') %>%
as.matrix()#
Heatmap(
kegg_mat,
column_title = "KEGG Pathway -log p values",
name = "-log (p value)",
cluster_rows = FALSE,
cluster_columns = FALSE,
cell_fun = function(j, i, x, y, width, height, fill) {
if (kegg_sig_mat[i, j] < 0.05)
grid.text("*", x, y, gp = gpar(fontsize = 20))
}
)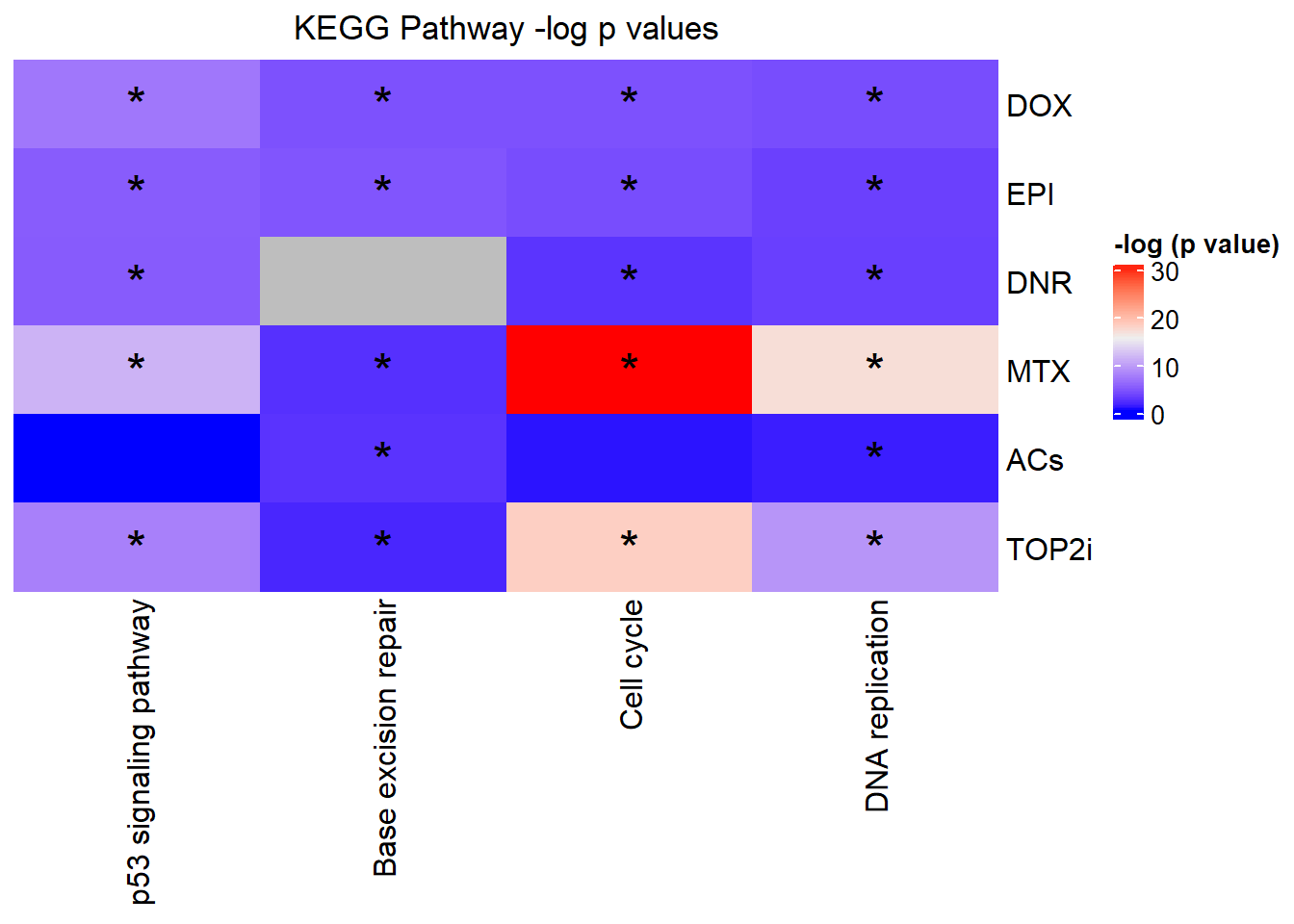
kegg_term3 <-
data.frame(
term = c("p53 signaling pathway", "Herpes simplex virus 1 infection"),
termid = c("KEGG:04115", "KEGG:05168")
)
DDEdeg3k <- DDEtable3only %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term3$termid) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DDEMdeg3k <- DDEMtable3 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term3$termid) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DAdeg3k <- DAtable3 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term3$termid) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
DXdeg3k <- DXtable1 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term3$termid) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
EPdeg3k <- EPtable1 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term3$termid) %>%
slice_min(., n = 3 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
MTdeg3k <- MTtable1 %>%
dplyr::filter(source == "KEGG") %>%
dplyr::select(p_value, term_id, term_name, intersection_size) %>%
filter(term_id %in% kegg_term3$termid) %>%
slice_min(., n = 3 , order_by = p_value) %>%
mutate(log_val = -log10(p_value)) %>%
mutate(order_val = rev(as.numeric(rownames(.))))
library(ComplexHeatmap)
keggDEGlong3 <- list(
"DOX" = DXdeg3k,
"EPI" = EPdeg3k,
"DNR" = DAdeg3k,
"MTX" = MTdeg3k,
"ACs" = DDEdeg3k,
"TOP2i" = DDEMdeg3k
)
keggtable3 <- data.table::rbindlist(keggDEGlong3, idcol = "deg")
kegg_sig_mat3 <- keggtable3 %>%
dplyr::select(deg, p_value, term_name) %>%
add_row(deg = "TOP2i",
p_value = 1,
term_name = "Herpes simplex virus 1 infection") %>%
pivot_wider(
id_cols = everything(),
names_from = "term_name",
values_from = "p_value",
values_fill = list(p_value = 1)
) %>%
column_to_rownames('deg') %>%
as.matrix()#
kegg_mat3 <- keggtable3 %>%
dplyr::select(deg, log_val, term_name) %>%
add_row(deg = "TOP2i",
log_val = NA,
term_name = "Herpes simplex virus 1 infection") %>%
pivot_wider(id_cols = everything(),
names_from = "term_name",
values_from = "log_val") %>%
column_to_rownames('deg') %>%
as.matrix()#
Heatmap(
kegg_mat3,
column_title = "KEGG Pathway -log p values",
name = "-log (p value)",
cluster_rows = FALSE,
cluster_columns = FALSE,
na_col = "grey",
cell_fun = function(j, i, x, y, width, height, fill) {
if (kegg_sig_mat3[i, j] < 0.05)
grid.text("*", x, y, gp = gpar(fontsize = 20))
}
)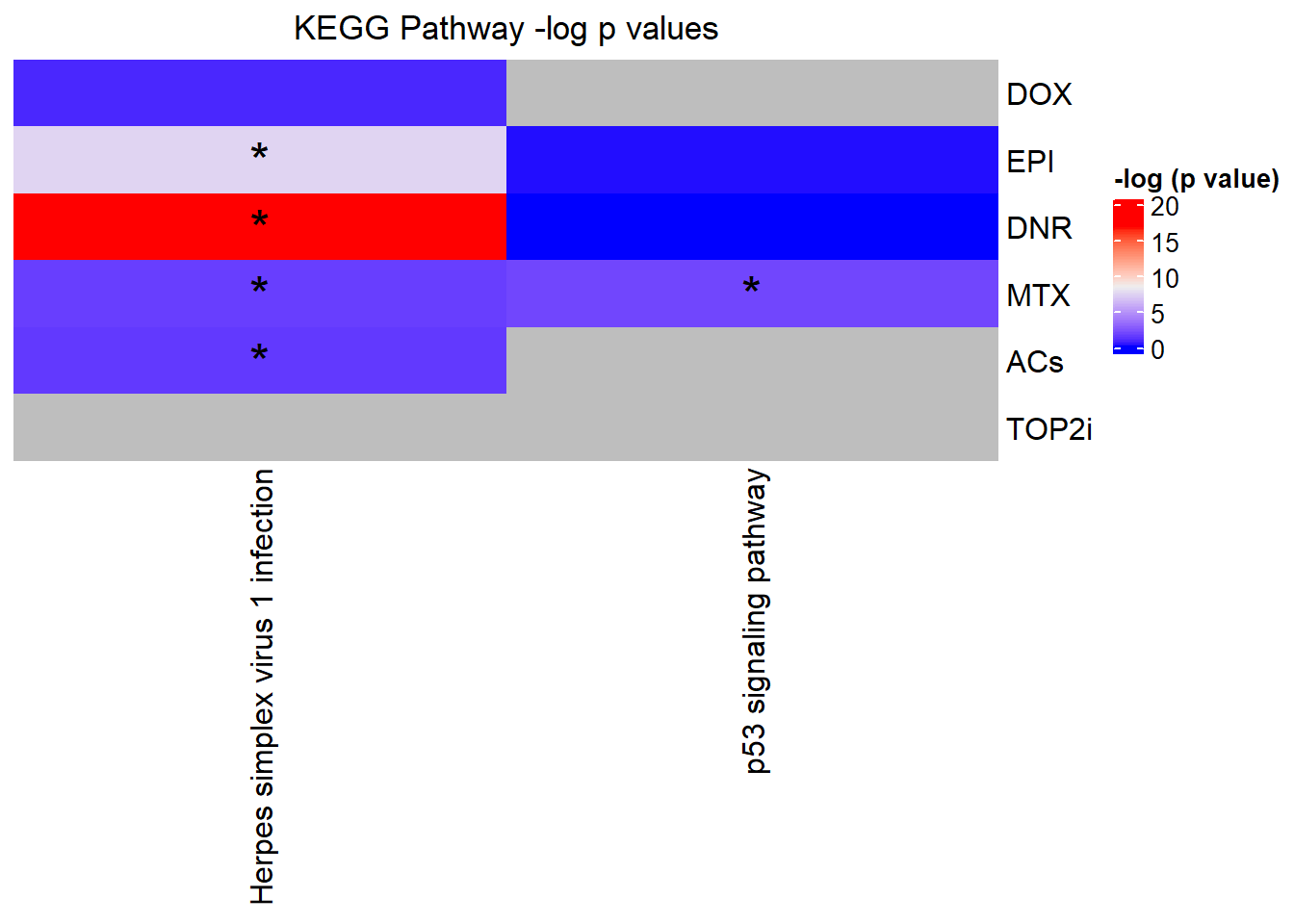
| Version | Author | Date |
|---|---|---|
| bcc7711 | reneeisnowhere | 2023-09-27 |
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ComplexHeatmap_2.16.0 paletteer_1.5.0 pheatmap_1.0.12
[4] limma_3.56.2 ggsignif_0.6.4 zoo_1.8-12
[7] rstatix_0.7.2 ggpubr_0.6.0 cowplot_1.1.1
[10] ggVennDiagram_1.2.3 scales_1.2.1 kableExtra_1.3.4
[13] gridExtra_2.3 BiocGenerics_0.46.0 gprofiler2_0.2.2
[16] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0
[19] dplyr_1.1.3 purrr_1.0.2 readr_2.1.4
[22] tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.3
[25] tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RColorBrewer_1.1-3 shape_1.4.6 rstudioapi_0.15.0
[4] jsonlite_1.8.7 magrittr_2.0.3 magick_2.7.5
[7] farver_2.1.1 rmarkdown_2.24 GlobalOptions_0.1.2
[10] fs_1.6.3 vctrs_0.6.3 webshot_0.5.5
[13] htmltools_0.5.6 lambda.r_1.2.4 broom_1.0.5
[16] sass_0.4.7 KernSmooth_2.23-22 bslib_0.5.1
[19] htmlwidgets_1.6.2 futile.options_1.0.1 plotly_4.10.2
[22] cachem_1.0.8 whisker_0.4.1 lifecycle_1.0.3
[25] iterators_1.0.14 pkgconfig_2.0.3 R6_2.5.1
[28] fastmap_1.1.1 clue_0.3-64 digest_0.6.33
[31] colorspace_2.1-0 rematch2_2.1.2 S4Vectors_0.38.1
[34] ps_1.7.5 rprojroot_2.0.3 prismatic_1.1.1
[37] labeling_0.4.3 fansi_1.0.4 timechange_0.2.0
[40] httr_1.4.7 abind_1.4-5 compiler_4.3.1
[43] proxy_0.4-27 bit64_4.0.5 withr_2.5.0
[46] doParallel_1.0.17 backports_1.4.1 carData_3.0-5
[49] DBI_1.1.3 highr_0.10 rjson_0.2.21
[52] classInt_0.4-10 tools_4.3.1 units_0.8-4
[55] httpuv_1.6.11 glue_1.6.2 VennDiagram_1.7.3
[58] callr_3.7.3 promises_1.2.1 sf_1.0-14
[61] getPass_0.2-2 cluster_2.1.4 generics_0.1.3
[64] gtable_0.3.4 tzdb_0.4.0 class_7.3-22
[67] data.table_1.14.8 hms_1.1.3 xml2_1.3.5
[70] car_3.1-2 utf8_1.2.3 foreach_1.5.2
[73] pillar_1.9.0 vroom_1.6.3 later_1.3.1
[76] circlize_0.4.15 lattice_0.21-8 bit_4.0.5
[79] tidyselect_1.2.0 knitr_1.44 git2r_0.32.0
[82] IRanges_2.34.1 svglite_2.1.1 stats4_4.3.1
[85] futile.logger_1.4.3 xfun_0.40 matrixStats_1.0.0
[88] stringi_1.7.12 lazyeval_0.2.2 yaml_2.3.7
[91] evaluate_0.21 codetools_0.2-19 RVenn_1.1.0
[94] cli_3.6.1 systemfonts_1.0.4 munsell_0.5.0
[97] processx_3.8.2 jquerylib_0.1.4 Rcpp_1.0.11
[100] png_0.1-8 parallel_4.3.1 viridisLite_0.4.2
[103] e1071_1.7-13 crayon_1.5.2 GetoptLong_1.0.5
[106] rlang_1.1.1 rvest_1.0.3 formatR_1.14