Analysis of Knowles QTL data
ERM
2024-02-05
Last updated: 2024-02-05
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version df08393. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/variance_values by gene.png
Ignored: data/41588_2018_171_MOESM3_ESMeQTL_ST2_for paper.csv
Ignored: data/Arr_GWAS.txt
Ignored: data/Arr_geneset.RDS
Ignored: data/BC_cell_lines.csv
Ignored: data/BurridgeDOXTOX.RDS
Ignored: data/CADGWASgene_table.csv
Ignored: data/CAD_geneset.RDS
Ignored: data/CALIMA_Data/
Ignored: data/CMD04_75DRCviability.csv
Ignored: data/CMD04_87DRCviability.csv
Ignored: data/CMD05_75DRCviability.csv
Ignored: data/CMD05_87DRCviability.csv
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/Counts_RNA_ERMatthews.txt
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDEMresp_list.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG-GO/
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/DRC48hoursdata.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/EPIstorelist24.RDS
Ignored: data/Ep24counts.txt
Ignored: data/FC_necela.RDS
Ignored: data/FC_necela_names.RDS
Ignored: data/Full_LD_rep.csv
Ignored: data/GOIsig.csv
Ignored: data/GOplots.R
Ignored: data/GTEX_setsimple.csv
Ignored: data/GTEX_sig24.RDS
Ignored: data/GTEx_gene_list.csv
Ignored: data/HFGWASgene_table.csv
Ignored: data/HF_geneset.RDS
Ignored: data/Heart_Left_Ventricle.v8.egenes.txt
Ignored: data/Heatmap_mat.RDS
Ignored: data/Heatmap_sig.RDS
Ignored: data/Hf_GWAS.txt
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/Knowles_log2cpm_real.RDS
Ignored: data/Knowles_variation_data.RDS
Ignored: data/Knowles_variation_data_conc.RDS
Ignored: data/Knowlesvarlist.RDS
Ignored: data/LD50_05via.csv
Ignored: data/LDH48hoursdata.csv
Ignored: data/Mt24counts.txt
Ignored: data/NoRespDEG_final.csv
Ignored: data/RINsamplelist.txt
Ignored: data/RNA_seq_trial.RDS
Ignored: data/Schneider_GWAS.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/Sup_replicate_values.csv
Ignored: data/TMMnormed_x.RDS
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/TableS10.csv
Ignored: data/TableS11.csv
Ignored: data/TableS9.csv
Ignored: data/Top2_expression.RDS
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Var_test_list.RDS
Ignored: data/Var_test_list24.RDS
Ignored: data/Var_test_list24alt.RDS
Ignored: data/Var_test_list3.RDS
Ignored: data/Vargenes.RDS
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allfinal3hour.RDS
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/allmymatrix.RDS
Ignored: data/annotation_data_frame.RDS
Ignored: data/averageviabilitytable.RDS
Ignored: data/averageviabilitytable.csv
Ignored: data/avgLD50.RDS
Ignored: data/avg_LD50.RDS
Ignored: data/avg_via_table.csv
Ignored: data/backGL.txt
Ignored: data/burr_genes.RDS
Ignored: data/calcium_data.RDS
Ignored: data/clamp_summary.RDS
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/cormotifprobs.csv
Ignored: data/counts24hours.RDS
Ignored: data/cpmcount.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/crispr_genes.csv
Ignored: data/ctnnt_results.txt
Ignored: data/cvd_GWAS.txt
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/drug_noveh1.csv
Ignored: data/efit2.RDS
Ignored: data/efit2_final.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filcpm_counts.RDS
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/framefun24.RDS
Ignored: data/geneDoxonlyQTL.csv
Ignored: data/gene_corr_df.RDS
Ignored: data/gene_corr_frame.RDS
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/geneset_24.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/gtex/
Ignored: data/heartgenes.csv
Ignored: data/highly_var_genelist.RDS
Ignored: data/hsa_kegg_anno.RDS
Ignored: data/individualDRCfile.RDS
Ignored: data/individual_DRC48.RDS
Ignored: data/individual_LDH48.RDS
Ignored: data/indv_noveh1.csv
Ignored: data/kegglistDEG.RDS
Ignored: data/kegglistDEG24.RDS
Ignored: data/kegglistDEG3.RDS
Ignored: data/knowfig4.csv
Ignored: data/knowfig5.csv
Ignored: data/label_list.RDS
Ignored: data/ld50_table.csv
Ignored: data/mean_vardrug1.csv
Ignored: data/mean_varframe.csv
Ignored: data/mymatrix.RDS
Ignored: data/new_ld50avg.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/organized_drugframe.csv
Ignored: data/pca_all_anno.csv
Ignored: data/plan2plot.png
Ignored: data/plot_intv_list.RDS
Ignored: data/plot_list_DRC.RDS
Ignored: data/qval24hr.RDS
Ignored: data/qval3hr.RDS
Ignored: data/qvalueEPItemp.RDS
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sampsettrz.RDS
Ignored: data/schneider_closest_output.RDS
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/siglist_final.RDS
Ignored: data/siglist_old.RDS
Ignored: data/slope_table.csv
Ignored: data/supp10_24hlist.RDS
Ignored: data/supp10_3hlist.RDS
Ignored: data/supp_normLDH48.RDS
Ignored: data/supp_pca_all_anno.RDS
Ignored: data/supp_viadata.csv
Ignored: data/table3a.omar
Ignored: data/test_run_sample_list.txt
Ignored: data/testlist.txt
Ignored: data/toplistall.RDS
Ignored: data/trtonly_24h_genes.RDS
Ignored: data/trtonly_3h_genes.RDS
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Ignored: data/viability.RDS
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: .RDataTmp3
Untracked: 3hr all.pdf
Untracked: Code_files_list.csv
Untracked: Data_files_list.csv
Untracked: Doxorubicin_vehicle_3_24.csv
Untracked: Doxtoplist.csv
Untracked: EPIqvalue_analysis.Rmd
Untracked: Final.sup.pdf
Untracked: GWAS_list_of_interest.xlsx
Untracked: KEGGpathwaylist.R
Untracked: NA
Untracked: OmicNavigator_learn.R
Untracked: SNP_egenes_allfiles.RDS
Untracked: SNP_frame_pdf
Untracked: SNP_frame_pdf.pdf
Untracked: SigDoxtoplist.csv
Untracked: analysis/DRC_viability_check.Rmd
Untracked: analysis/New_code_dec-23.R
Untracked: analysis/cellcycle_kegg_genes.R
Untracked: analysis/ciFIT.R
Untracked: analysis/export_to_excel.R
Untracked: analysis/featureCountsPLAY.R
Untracked: cleanupfiles_script.R
Untracked: code/biomart_gene_names.R
Untracked: code/constantcode.R
Untracked: code/corMotifcustom.R
Untracked: code/cpm_boxplot.R
Untracked: code/extracting_ggplot_data.R
Untracked: code/movingfilesto_ppl.R
Untracked: code/pearson_extract_func.R
Untracked: code/pearson_tox_extract.R
Untracked: code/plot1C.fun.R
Untracked: code/spearman_extract_func.R
Untracked: code/venndiagramcolor_control.R
Untracked: cormotif_p.post.list_4.csv
Untracked: figS1024h.pdf
Untracked: final.pdf
Untracked: individual-legenddark2.png
Untracked: installed_old.rda
Untracked: listoftranscripts
Untracked: motif_ER.txt
Untracked: motif_LR.txt
Untracked: motif_NR.txt
Untracked: motif_TI.txt
Untracked: output/ABHD8_dif_values.RDS
Untracked: output/C3orf18_dif_values.RDS
Untracked: output/Cardiotox_dif_values.RDS
Untracked: output/DNR_DEGlist.csv
Untracked: output/DNRvenn.RDS
Untracked: output/DOX_DEGlist.csv
Untracked: output/DOX_de_goi.csv
Untracked: output/DOXvenn.RDS
Untracked: output/EEF1B2_dif_values.RDS
Untracked: output/EEIG1_dif_values.RDS
Untracked: output/EPI_DEGlist.csv
Untracked: output/EPIvenn.RDS
Untracked: output/ESGN_rds.RDS
Untracked: output/FC_necela.RDS
Untracked: output/FC_necela_names.RDS
Untracked: output/FRS2_dif_values.RDS
Untracked: output/Figures/
Untracked: output/GTEXv8_gene_median_tpm.RDS
Untracked: output/GTEXv8_gene_tpm_heart_left_ventricle.RDS
Untracked: output/HDDC2_dif_values.RDS
Untracked: output/HER2_gene.RDS
Untracked: output/KEGGcellcyclegenes.RDS
Untracked: output/Knowles_S13.csv
Untracked: output/Knowles_log2cpm.csv
Untracked: output/Knowles_supp13.csv
Untracked: output/LD50tox_table.RDS
Untracked: output/MTX_DEGlist.csv
Untracked: output/MTXvenn.RDS
Untracked: output/PEX16_dif_values.RDS
Untracked: output/RASIP1_dif_values.RDS
Untracked: output/RMI1_dif_values.RDS
Untracked: output/RSID_QTL_list_full.txt
Untracked: output/SETA_analysis_reyes.RDS
Untracked: output/SGWAS_top50_order.csv
Untracked: output/SLC27A1_dif_values.RDS
Untracked: output/SLC28A3_dif_values.RDS
Untracked: output/SNP_egenes_allfiles.RDS
Untracked: output/SNP_list_ID.RDS
Untracked: output/SNP_list_full.txt
Untracked: output/SNP_supp.RDS
Untracked: output/TGFBR3L_dif_values.RDS
Untracked: output/TNS2_dif_values.RDS
Untracked: output/TOP_50SNPreffile.csv
Untracked: output/TRZ_DEGlist.csv
Untracked: output/TableS8.csv
Untracked: output/Volcanoplot_10
Untracked: output/Volcanoplot_10.RDS
Untracked: output/ZNF740_dif_values.RDS
Untracked: output/allfinal_sup10.RDS
Untracked: output/counts_v8_heart_left_ventricle_gct.RDS
Untracked: output/crisprfoldchange.RDS
Untracked: output/endocytosisgenes.csv
Untracked: output/expre7k.csv
Untracked: output/expressed_egenes_by_RSID.csv
Untracked: output/gene_corr_fig9.RDS
Untracked: output/genes.RDS
Untracked: output/legend_b.RDS
Untracked: output/motif_ERrep.RDS
Untracked: output/motif_LRrep.RDS
Untracked: output/motif_NRrep.RDS
Untracked: output/motif_TI_rep.RDS
Untracked: output/near_genes_SNP1.RDS
Untracked: output/necela_list_test.RDS
Untracked: output/necela_val_genes.RDS
Untracked: output/output-old/
Untracked: output/rank24genes.csv
Untracked: output/rank3genes.csv
Untracked: output/sequencinginformationforsupp.csv
Untracked: output/sequencinginformationforsupp.prn
Untracked: output/sigVDA24.txt
Untracked: output/sigVDA3.txt
Untracked: output/sigVDX24.txt
Untracked: output/sigVDX3.txt
Untracked: output/sigVEP24.txt
Untracked: output/sigVEP3.txt
Untracked: output/sigVMT24.txt
Untracked: output/sigVMT3.txt
Untracked: output/sigVTR24.txt
Untracked: output/sigVTR3.txt
Untracked: output/supplementary_motif_list_GO.RDS
Untracked: output/test_biomart_run.RDS
Untracked: output/toptablebydrug.RDS
Untracked: output/trop_knowles_fun.csv
Untracked: output/tvl24hour.txt
Untracked: output/x_counts.RDS
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/DRC_analysis.Rmd
Modified: analysis/GOI_plots.Rmd
Modified: analysis/GTEx_genes.Rmd
Deleted: analysis/Knowles2019.Rmd
Modified: output/daplot.RDS
Modified: output/dxplot.RDS
Modified: output/epplot.RDS
Modified: output/mtplot.RDS
Modified: output/plan2plot.png
Modified: output/trplot.RDS
Modified: output/veplot.RDS
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Knowles2018.Rmd) and HTML
(docs/Knowles2018.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | df08393 | reneeisnowhere | 2024-02-05 | updates to scripts |
package loading
library(limma)
library(RColorBrewer)
library(tidyverse)
library(ggsignif)
library(cowplot)
library(ggpubr)
library(scales)
library(sjmisc)
library(kableExtra)
library(broom)
library(ComplexHeatmap)
library(ggVennDiagram)function load
cpm_boxplot <-
function(cpmcounts,
GOI,
brewer_palette,
fill_colors,
ylab) {
##GOI needs to be ENTREZID
df <- cpmcounts
df_plot <- df %>%
dplyr::filter(rownames(.) == GOI) %>%
pivot_longer(everything(),
names_to = "treatment",
values_to = "counts") %>%
separate(treatment, c("drug", "indv", "time")) %>%
mutate(time = factor(time, levels = c("3h", "24h"))) %>%
mutate(indv = factor(indv, levels = c(1, 2, 3, 4, 5, 6))) %>%
mutate(
drug = case_match(
drug,
"Da" ~ "DNR",
"Do" ~ "DOX",
"Ep" ~ "EPI",
"Mi" ~ "MTX",
"Tr" ~ "TRZ",
"Ve" ~ "VEH",
.default = drug
)
) %>%
mutate(drug = factor(drug, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH')))
plot <- ggplot2::ggplot(df_plot, aes(x = drug, y = counts)) +
geom_boxplot(position = "identity", aes(fill = drug)) +
geom_point(aes(
col = indv,
size = 1.5,
alpha = 0.5
)) +
guides(alpha = "none", size = "none") +
scale_color_brewer(palette = brewer_palette, guide = "none") +
scale_fill_manual(values = fill_colors) +
facet_wrap("time", nrow = 1, ncol = 2) +
theme_bw() +
ylab(ylab) +
xlab("") +
theme(
strip.background = element_rect(
fill = "white",
linetype = 1,
linewidth = 0.5
),
plot.title = element_text(
size = 10,
hjust = 0.5,
face = "bold"
),
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(linewidth = 1.0),
axis.line = element_line(linewidth = 1.0),
axis.text.x = element_blank(),
strip.text.x = element_text(margin = margin(2, 0, 2, 0, "pt"), face = "bold")
)
print(plot)
}
cpm_boxplot_time <-
function(cpmcounts,
timex,
GOI,
brewer_palette,
fill_colors,
ylab) {
##GOI needs to be ENTREZID
df <- cpmcounts
df_plot <- df %>%
dplyr::filter(rownames(.)==GOI) %>%
pivot_longer(everything(),
names_to = "treatment",
values_to = "counts") %>%
separate(treatment, c("drug","indv","time")) %>%
dplyr::filter(time == paste(timex)) %>%
mutate(time=factor(time, levels =c("3h", "24h"), labels=c("3 hours", "24 hours"))) %>%
mutate(indv=factor(indv, levels = c(1,2,3,4,5,6))) %>%
mutate(drug =case_match(drug, "Da"~"DNR",
"Do"~"DOX",
"Ep"~"EPI",
"Mi"~"MTX",
"Tr"~"TRZ",
"Ve"~"VEH", .default = drug)) %>%
mutate(drug=factor(drug, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH')))
plot <- ggplot2::ggplot(df_plot, aes(x=drug, y=counts))+
geom_boxplot(position="identity",aes(fill=drug))+
geom_point(aes(col=indv, size=1.5, alpha=0.5))+
guides(alpha= "none", size= "none")+
scale_color_brewer(palette = brewer_palette, guide = "none")+
scale_fill_manual(values=fill_colors)+
# try(facet_wrap("time", nrow=1, ncol=2))+
theme_bw()+
ylab(ylab)+
xlab("")+
theme(strip.background = element_rect(fill = "white",linetype=1, linewidth = 0.5),
plot.title = element_text(size=10,hjust = 0.5,face="bold"),
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(linewidth = 1.0),
axis.line = element_line(linewidth = 1.0),
axis.text.x = element_blank(),
strip.text.x = element_text(margin = margin(2,0,2,0, "pt"),face = "bold"))
print(plot)
}Knowles data
The code below is how I wrangled the knowles supplemental lists into entrezgene_ids to overlap with my expressed gene lists
Enrichment of Pairwise
A note on Knowles data sets: the intersection of the Knowles-4 and Knowles-5 sets yielded 104 genes in
backGL <- read.csv("data/backGL.txt")
# drug_palNoVeh <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031")
drug_palc <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031","#41B333")
time <- rep((rep(c("3h", "24h"), c(6,6))), 6)
time <- ordered(time, levels =c("3h", "24h"))
drug <- rep(c("DNR","DOX","EPI","MTX","TRZ", "Vehicle"),12)
mat_drug <- c("DNR","DOX","EPI","MTX")
indv <- as.factor(rep(c(1,2,3,4,5,6), c(12,12,12,12,12,12)))
labeltop <- (interaction(substring(drug, 0, 2), indv, time))
level_order2 <- c('75','87','77','79','78','71')
toplistall <- readRDS("data/toplistall.RDS")
knowles4 <-readRDS("output/knowles4.RDS")
knowles5 <-readRDS("output/knowles5.RDS")
DOXmeSNPs <- readRDS("output/DOXmeSNPs.RDS")
DOXreQTLs <- readRDS("output/DOXreQTLs.RDS")
DNRmeSNPs <- readRDS("output/DNRmeSNPs.RDS")
DNRreQTLs <- readRDS("output/DNRreQTLs.RDS")
EPImeSNPs <- readRDS("output/EPImeSNPs.RDS")
EPIreQTLs <- readRDS("output/EPIreQTLs.RDS")
MTXmeSNPs <- readRDS("output/MTXmeSNPs.RDS")
MTXreQTLs <- readRDS("output/MTXreQTLs.RDS")
cpmcounts <- readRDS("data/cpmcount.RDS")
toplist24hr <- toplistall %>%
mutate(id =case_match(id, "Da"~"DNR",
"Do"~"DOX",
"Ep"~"EPI",
"Mi"~"MTX",
"Tr"~"TRZ",
"Ve"~"VEH", .default = id)) %>%
mutate(id=factor(id, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH'))) %>%
mutate(time=factor(time, levels=c("3_hours","24_hours"))) %>%
filter(time=="24_hours")
toplist3hr <- toplistall %>%
mutate(id =case_match(id, "Da"~"DNR",
"Do"~"DOX",
"Ep"~"EPI",
"Mi"~"MTX",
"Tr"~"TRZ",
"Ve"~"VEH", .default = id)) %>%
mutate(id=factor(id, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH'))) %>%
mutate(id = as.factor(id)) %>%
mutate(time=factor(time, levels=c("3_hours","24_hours"))) %>%
filter(time=="3_hours")Determining the genetic basis of anthracycline-cardiotoxicity by
molecular response QTL mapping in induced cardiomyocytes
David A Knowles, Courtney K Burrows†, John D Blischak, Kristen M
Patterson, Daniel J Serie, Nadine Norton, Carole Ober, Jonathan K
Pritchard, Yoav Gilad
Knowles \(~~et~ al.~\) eLife 2018;7:e33480. DOI: https://doi.org/10.7554/eLife.33480
My first question was about transcriptional response at the 24 hour mark with my treatments. 3 hour RNA-seq had low numbers of DEGs,so my initial focus is at 24 hours. This time also happens to be when the Knowles paper collected their RNA-seq data.
Supplementary 4 contains a list of 518 SNPs within 1 Mb of TSS, which had a detectable marginal effect on expression (5% FDR). When converted from ensembl gene id to entrez gene id, my list of genes for this supplement = 417. I will call these meSNPS for marginal effect QTLs.
Supplementary 5 contains a list of 376 response eQTLs (reQTLs). These are variants that were associated with modulation of transcriptomic response to DOX treatment. After database name conversion, I have 273 genes.
meSNPdf <- toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
mutate(meSNP = if_else(ENTREZID %in% knowles4$entrezgene_id, "y" , "no")) %>%
dplyr::select(ENTREZID, id, time, meSNP, sigcount) %>%
group_by(id, time, meSNP, sigcount) %>%
tally() %>%
pivot_wider(
id_cols = c(id, time, sigcount),
names_from = meSNP,
names_glue = "meSNP_{meSNP}",
values_from = n
)
meSNPdf %>%
kable(., caption = "Number of genes from minimally expressed SNPs found in DEGs (sig and nonsig) broken down by drug,time, and significance") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped"),
font_size = 18
) %>%
scroll_box(width = "80%", height = "400px")| id | time | sigcount | meSNP_no | meSNP_y |
|---|---|---|---|---|
| DOX | 3_hours | notsig | 13665 | 400 |
| DOX | 3_hours | sig | 19 | NA |
| DOX | 24_hours | notsig | 7181 | 258 |
| DOX | 24_hours | sig | 6503 | 142 |
| EPI | 3_hours | notsig | 13477 | 397 |
| EPI | 3_hours | sig | 207 | 3 |
| EPI | 24_hours | notsig | 7484 | 272 |
| EPI | 24_hours | sig | 6200 | 128 |
| DNR | 3_hours | notsig | 13156 | 396 |
| DNR | 3_hours | sig | 528 | 4 |
| DNR | 24_hours | notsig | 6828 | 239 |
| DNR | 24_hours | sig | 6856 | 161 |
| MTX | 3_hours | notsig | 13611 | 398 |
| MTX | 3_hours | sig | 73 | 2 |
| MTX | 24_hours | notsig | 12585 | 384 |
| MTX | 24_hours | sig | 1099 | 16 |
| TRZ | 3_hours | notsig | 13684 | 400 |
| TRZ | 24_hours | notsig | 13684 | 400 |
reQTLdf <- toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
mutate(reQTL = if_else(ENTREZID %in% knowles5$entrezgene_id, "y" , "no")) %>%
dplyr::select(ENTREZID, id, time, reQTL, sigcount) %>%
group_by(id, time, reQTL, sigcount) %>%
tally() %>%
pivot_wider(
id_cols = c(id, time, sigcount),
names_from = reQTL,
names_glue = "reQTL_{reQTL}",
values_from = n
)
reQTLdf %>%
kable(., caption = "Number of genes from response eQTLS found in DEGs (sig and nonsig) broken down by drug,time, and significance") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped"),
font_size = 18
) %>%
scroll_box(width = "80%", height = "400px")| id | time | sigcount | reQTL_no | reQTL_y |
|---|---|---|---|---|
| DOX | 3_hours | notsig | 13794 | 271 |
| DOX | 3_hours | sig | 19 | NA |
| DOX | 24_hours | notsig | 7310 | 129 |
| DOX | 24_hours | sig | 6503 | 142 |
| EPI | 3_hours | notsig | 13606 | 268 |
| EPI | 3_hours | sig | 207 | 3 |
| EPI | 24_hours | notsig | 7616 | 140 |
| EPI | 24_hours | sig | 6197 | 131 |
| DNR | 3_hours | notsig | 13293 | 259 |
| DNR | 3_hours | sig | 520 | 12 |
| DNR | 24_hours | notsig | 6944 | 123 |
| DNR | 24_hours | sig | 6869 | 148 |
| MTX | 3_hours | notsig | 13739 | 270 |
| MTX | 3_hours | sig | 74 | 1 |
| MTX | 24_hours | notsig | 12725 | 244 |
| MTX | 24_hours | sig | 1088 | 27 |
| TRZ | 3_hours | notsig | 13813 | 271 |
| TRZ | 24_hours | notsig | 13813 | 271 |
dataframK_45 <- toplistall %>%
mutate(
id = case_match(
id,
"Da" ~ "DNR",
"Do" ~ "DOX",
"Ep" ~ "EPI",
"Mi" ~ "MTX",
"Tr" ~ "TRZ",
"Ve" ~ "VEH",
.default = id
)
) %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
mutate(K4 = if_else(ENTREZID %in% knowles4$entrezgene_id, 'y', 'no')) %>%
mutate(K5 = if_else(ENTREZID %in% knowles5$entrezgene_id, 'y', 'no')) %>%
filter(adj.P.Val < 0.05) %>%
group_by(time, id) %>%
dplyr::summarize(n = n(),
meSNP = sum(if_else(K4 == "y", 1, 0)),
reQTL = sum(if_else(K5 == "y", 1, 0))) %>%
as.data.frame()
dataframK_45 %>%
kable(., caption = "Summary of meSNPs and reQTLs found in my DEGs") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(font_size = 18) %>%
scroll_box(width = "80%", height = "400px")| time | id | n | meSNP | reQTL |
|---|---|---|---|---|
| 3_hours | DOX | 19 | 0 | 0 |
| 3_hours | EPI | 210 | 3 | 3 |
| 3_hours | DNR | 532 | 4 | 12 |
| 3_hours | MTX | 75 | 2 | 1 |
| 24_hours | DOX | 6645 | 142 | 142 |
| 24_hours | EPI | 6328 | 128 | 131 |
| 24_hours | DNR | 7017 | 161 | 148 |
| 24_hours | MTX | 1115 | 16 | 27 |
Visualization of proportions
toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
mutate(meSNP = if_else(ENTREZID %in% knowles4$entrezgene_id, "y", "no")) %>%
mutate(reQTL = if_else(ENTREZID %in% knowles5$entrezgene_id, "a", "b")) %>%
filter(time == "24_hours") %>%
group_by(id, sigcount, meSNP) %>%
summarize(K4count = n()) %>%
pivot_wider(
id_cols = c(id, sigcount),
names_from = c(meSNP),
values_from = K4count
) %>%
mutate(SNPprop = (y / (y + no) * 100)) %>%
ggplot(., aes(x = id, y = SNPprop)) +
geom_col() +
geom_text(aes(
x = id,
label = sprintf("%.2f", SNPprop),
vjust = -.2
)) +
facet_wrap( ~ sigcount) +
ggtitle("non-significant and significant enrichment proportions of Knowles meSNPs ")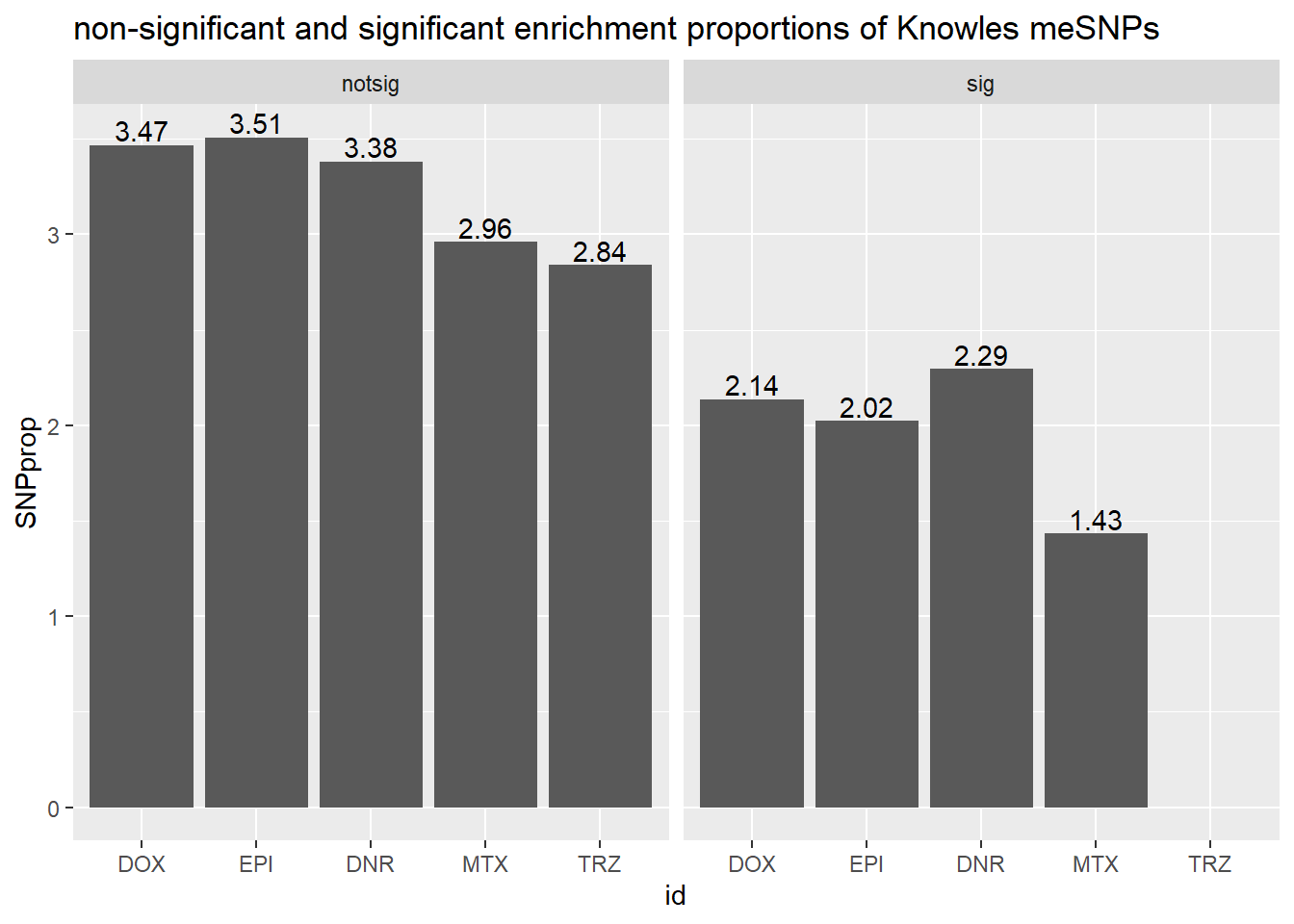
toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
mutate(meSNP = if_else(ENTREZID %in% knowles4$entrezgene_id, "y", "no")) %>%
mutate(reQTL = if_else(ENTREZID %in% knowles5$entrezgene_id, "a", "b")) %>%
filter(time == "24_hours") %>%
group_by(id, sigcount, reQTL) %>%
summarize(K5count = n()) %>%
pivot_wider(
id_cols = c(id, sigcount),
names_from = c(reQTL),
values_from = K5count
) %>%
mutate(QTLprop = (a / (a + b) * 100)) %>%
ggplot(., aes(x = id, y = QTLprop)) +
geom_col() +
geom_text(aes(
x = id,
label = sprintf("%.2f", QTLprop),
vjust = -.2
)) +
facet_wrap( ~ sigcount) +
ggtitle("non-significant and significant enrichment proportions of Knowles reQTLs ")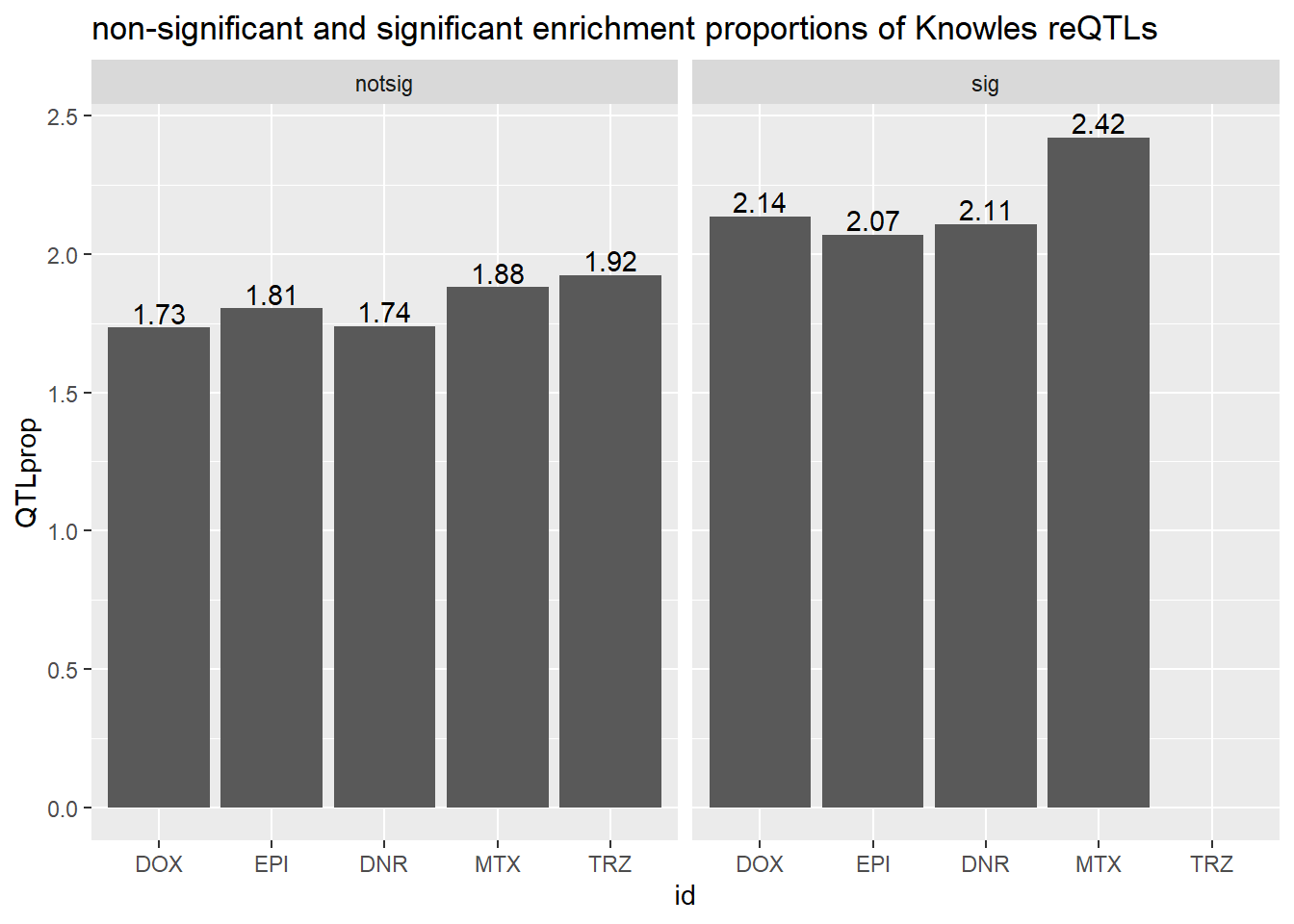
chi_squarek4 <- toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
dplyr::filter(id != "TRZ") %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
mutate(meSNP = if_else(ENTREZID %in% knowles4$entrezgene_id, "y", "no")) %>%
group_by(id, time) %>%
summarise(pvalue = chisq.test(meSNP, sigcount)$p.value)
chi_squarek5 <- toplistall %>%
mutate(id = factor(id, levels = c('DOX', 'EPI', 'DNR', 'MTX', 'TRZ', 'VEH'))) %>%
dplyr::filter(id != "TRZ") %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
mutate(reQTL = if_else(ENTREZID %in% knowles5$entrezgene_id, "a", "b")) %>%
group_by(id, time) %>%
summarise(pvalue = chisq.test(reQTL, sigcount)$p.value)
chi_squarek4 %>%
kable(., caption = "meSNPs (mininmally expressed QTLS) chi-squared pvalues") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "right",
bootstrap_options = c("striped"),
font_size = 18
) %>% scroll_box(width = "50%", height = "400px")| id | time | pvalue |
|---|---|---|
| DOX | 24_hours | 0.0000026 |
| DOX | 3_hours | 0.9563346 |
| EPI | 24_hours | 0.0000002 |
| EPI | 3_hours | 0.3023606 |
| DNR | 24_hours | 0.0001261 |
| DNR | 3_hours | 0.0047603 |
| MTX | 24_hours | 0.0043791 |
| MTX | 3_hours | 1.0000000 |
chi_squarek5 %>%
kable(., caption = "reQTLS (response eQTLS) chi-squared pvalues") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "right",
bootstrap_options = c("striped"),
font_size = 18
) %>% scroll_box(width = "50%", height = "400px")| id | time | pvalue |
|---|---|---|
| DOX | 24_hours | 0.0937665 |
| DOX | 3_hours | 1.0000000 |
| EPI | 24_hours | 0.2812223 |
| EPI | 3_hours | 0.7843268 |
| DNR | 24_hours | 0.1257321 |
| DNR | 3_hours | 0.6843822 |
| MTX | 24_hours | 0.2516965 |
| MTX | 3_hours | 1.0000000 |
We can “see” the proportions of SNPs are not enriched significantly
in the DE genes compared to the non-DE genes (chi square test).
We do not see significant enrichment of reQTLS in our DE gene sets over
the non-DE gene sets.
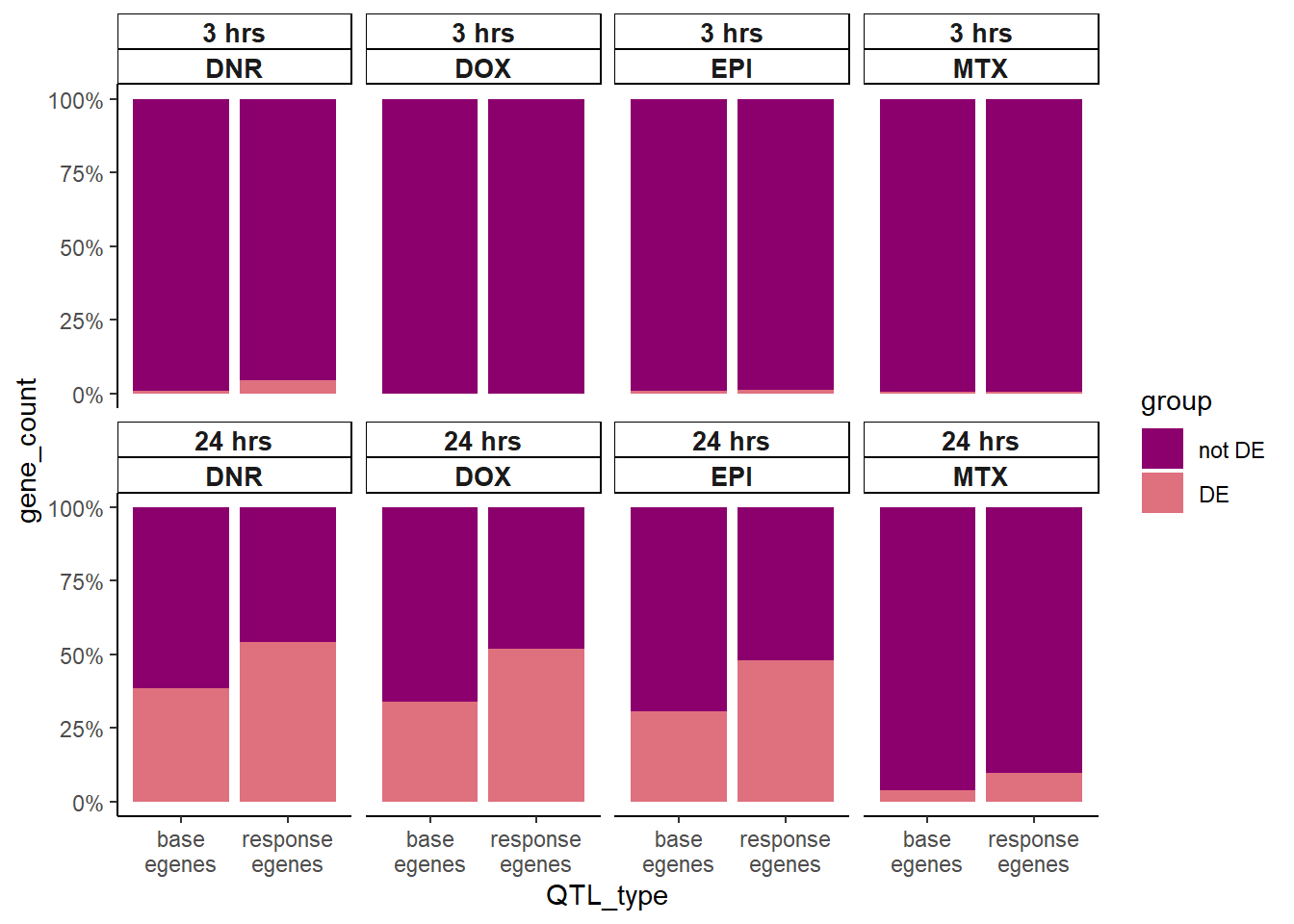
So what about enrichment of meSNPs in DE compared to reQTLs in significantly DE gene sets?
Knowles_count <-
toplistall %>%
filter(id != 'TRZ') %>%
mutate(id = as.factor(id)) %>%
mutate(time = factor(time, levels = c("3_hours", "24_hours"))) %>%
group_by(time, id) %>%
mutate(K4 = if_else(ENTREZID %in% knowles4$entrezgene_id, 1, 0)) %>%
mutate(K5 = if_else(ENTREZID %in% knowles5$entrezgene_id, 1, 0)) %>%
filter(adj.P.Val < 0.05) %>%
dplyr::summarize(n = n(),
K4 = sum(K4),
K5 = sum(K5)) %>%
as.tibble() %>%
dplyr::select(time, id, K4, K5) %>%
rename( 'K4_y'='K4', 'K5_y'='K5') %>%
mutate(time = case_match(time, '3_hours' ~ '3 hrs',
'24_hours' ~ '24 hrs', .default = time)) %>%
mutate(K4_n = 417 - K4_y, K5_n = 273 - K5_y) %>%
pivot_longer(!c(time, id), names_to = 'QTL', values_to = "gene_count") %>%
separate(QTL, into = c("QTL_type", 'group'), sep = '_') %>%
mutate(
QTL_type = case_match(
QTL_type,
'K4' ~ 'base\negenes',
'K5' ~ 'response\negenes',
.default = QTL_type
)
) %>%
mutate(time = factor(time, levels = c("3 hrs", "24 hrs"))) %>%
group_by(id, time, QTL_type) %>%
mutate(percent = gene_count / sum(gene_count) * 100) %>%
mutate(group = factor(
group,
levels = c("n", "y"),
labels = c("not DE", "DE")
))
ggplot(Knowles_count,
aes(
x = QTL_type,
y = gene_count,
group = group,
fill = group
)) +
geom_col(position = 'fill') +
# guides(fill="none")+
facet_wrap(time ~ id, nrow = 2, ncol = 4) +
theme_classic() +
scale_color_manual(values = drug_palc) +
scale_fill_manual(values = drug_palc) +
scale_y_continuous(labels = scales::percent) +
theme(
strip.text = element_text(size = 10, face = "bold"),
strip.text.x = element_text(margin = margin(2, 0, 2, 0, "pt")),
strip.background = element_rect (linetype = 1, linewidth = 0.5)
)
###This works
chi_frame <- Knowles_count %>%
pivot_wider(
id_cols = c(time, id, QTL_type),
names_from = group,
values_from = gene_count
)
testDNR3chi <- matrix(
c(4, 12, 413, 261),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
DNR_3chi <- chisq.test(testDNR3chi, correct = FALSE)$p.value
testDNR24chi <-
matrix(
c(161, 148, 256, 125),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
DNR_24chi <- chisq.test(testDNR24chi, correct = FALSE)$p.value
testDOX3chi <- matrix(
c(0, 0, 417, 273),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
DOX_3chi <- chisq.test(testDOX3chi, correct = FALSE)$p.value
testDOX24chi <-
matrix(
c(142, 142, 256, 125),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
DOX_24chi <- chisq.test(testDOX24chi, correct = FALSE)$p.value
testEPI3chi <- matrix(
c(3, 3, 414, 270),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
EPI_3chi <- chisq.test(testEPI3chi, correct = FALSE)$p.value
testEPI24chi <-
matrix(
c(128, 131, 289, 142),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
EPI_24chi <- chisq.test(testEPI24chi, correct = FALSE)$p.value
testMTX3chi <- matrix(
c(2, 1, 415, 272),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
MTX_3chi <- chisq.test(testMTX3chi, correct = FALSE)$p.value
testMTX24chi <-
matrix(
c(16, 27, 401, 246),
nrow = 2,
ncol = 2,
byrow = FALSE,
dimnames = list(c("k4", "k5"), c("y", "n"))
)
MTX_24chi <- chisq.test(testMTX24chi, correct = FALSE)$p.value
K4nK5chi <-
data.frame(
treatment = c(
'DNR_3',
'DNR_24',
'DOX_3',
'DOX_24',
'EPI_3',
'EPI_24',
'MTX_3',
'MTX_24'
),
chi_p.value = c(
DNR_3chi,
DNR_24chi,
DOX_3chi,
DOX_24chi,
EPI_3chi,
EPI_24chi,
MTX_3chi,
MTX_24chi
)
)
K4nK5chi %>%
separate(treatment, into = c('Drug', 'time')) %>%
pivot_wider(id_cols = Drug,
names_from = time,
values_from = chi_p.value) %>%
kable(., caption = "Chi Square p. values from chi-square test between proportions of sig-DE meQTLs and reQTLS by time and treatment") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| Drug | 3 | 24 |
|---|---|---|
| DNR | 0.0033591 | 0.0000557 |
| DOX | NaN | 0.0000077 |
| EPI | 0.5995866 | 0.0000045 |
| MTX | 0.8249205 | 0.0012981 |
Here we found that 24 hour reQTLs are significantly enriched in sig-DEgens compared to meQTLS, with DNR 3 hour treatment also showing significant enrichment.
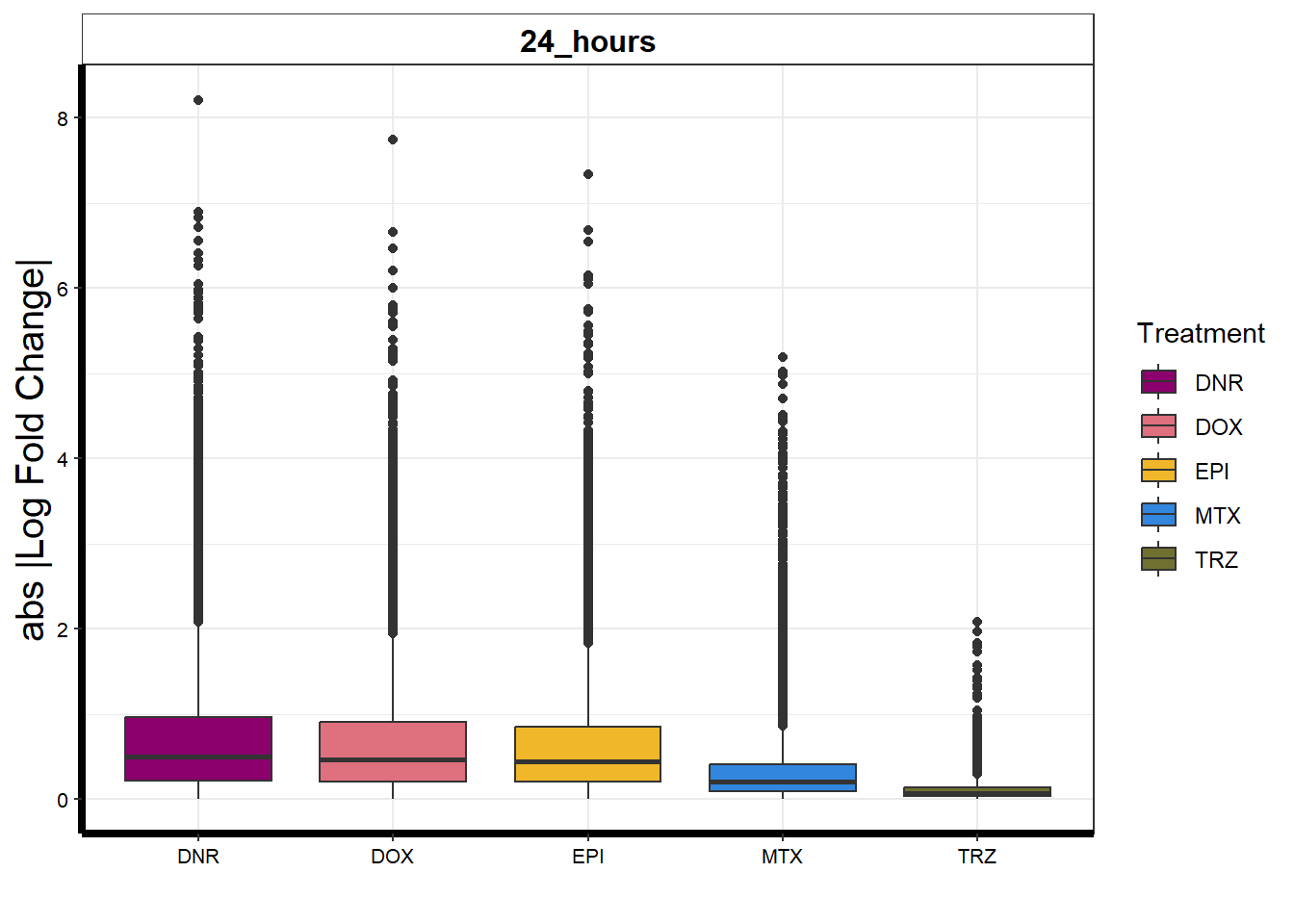
toplistall %>%
filter(time == "24_hours") %>%
group_by(time, id) %>%
mutate(sigcount = if_else(adj.P.Val < 0.05, 'sig', 'notsig')) %>%
ggplot(., aes(x = id, y = abs(logFC))) +
geom_boxplot(aes(fill = id)) +
ggpubr::fill_palette(palette = drug_palc) +
guides(fill = guide_legend(title = "Treatment")) +
facet_wrap(sigcount ~ time) +
theme_bw() +
xlab("") +
ylab("abs |Log Fold Change|") +
theme_bw() +
facet_wrap( ~ time) +
#ggtitle("All QTLs from all expressed genes (n=507)")+
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
# axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "transparent"),
axis.text = element_text(
size = 8,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 12,
color = "black",
face = "bold"
)
)
Dox-reQTLs
When I look ot the reQTLs across treatments at 24 hours, I see that dox has a total of 142 reQTLS. Because this gene set was specifically about DOX response eQTLs, I wanted to see if these reQTLs were only Dox specific, or had more in common with other treatments.
trtonly_24h_genes <- readRDS("data/trtonly_24h_genes.RDS")
list2env(trtonly_24h_genes, .GlobalEnv)<environment: R_GlobalEnv>siglist <- readRDS("data/siglist_final.RDS")
list2env(siglist, .GlobalEnv)<environment: R_GlobalEnv>reQTLcombine = list(DOXreQTLs$ENTREZID,
EPIreQTLs$ENTREZID,
DNRreQTLs$ENTREZID,
MTXreQTLs$ENTREZID)
length(unique(
c(
DNRreQTLs$ENTREZID,
DOXreQTLs$ENTREZID,
EPIreQTLs$ENTREZID,
MTXreQTLs$ENTREZID
)
))[1] 166print("number of unique genes expressed in pairwise DE set")[1] "number of unique genes expressed in pairwise DE set"QTLoverlaps <- VennDiagram::get.venn.partitions(reQTLcombine)
DoxonlyQTL <- QTLoverlaps$..values..[[14]]
ggVennDiagram::ggVennDiagram(
reQTLcombine,
category.names = c(
"DOX-142 \nsigDEG\n",
"EPI-131\nsigDEG\n",
"DNR-148\nsigDEG",
"MTX-27\nsigDEG"
),
label = "count"
) + scale_x_continuous(expand = expansion(mult = .2)) + scale_y_continuous(expand = expansion(mult = .1)) +
scale_fill_distiller(palette = "RdYlGn", direction = -1)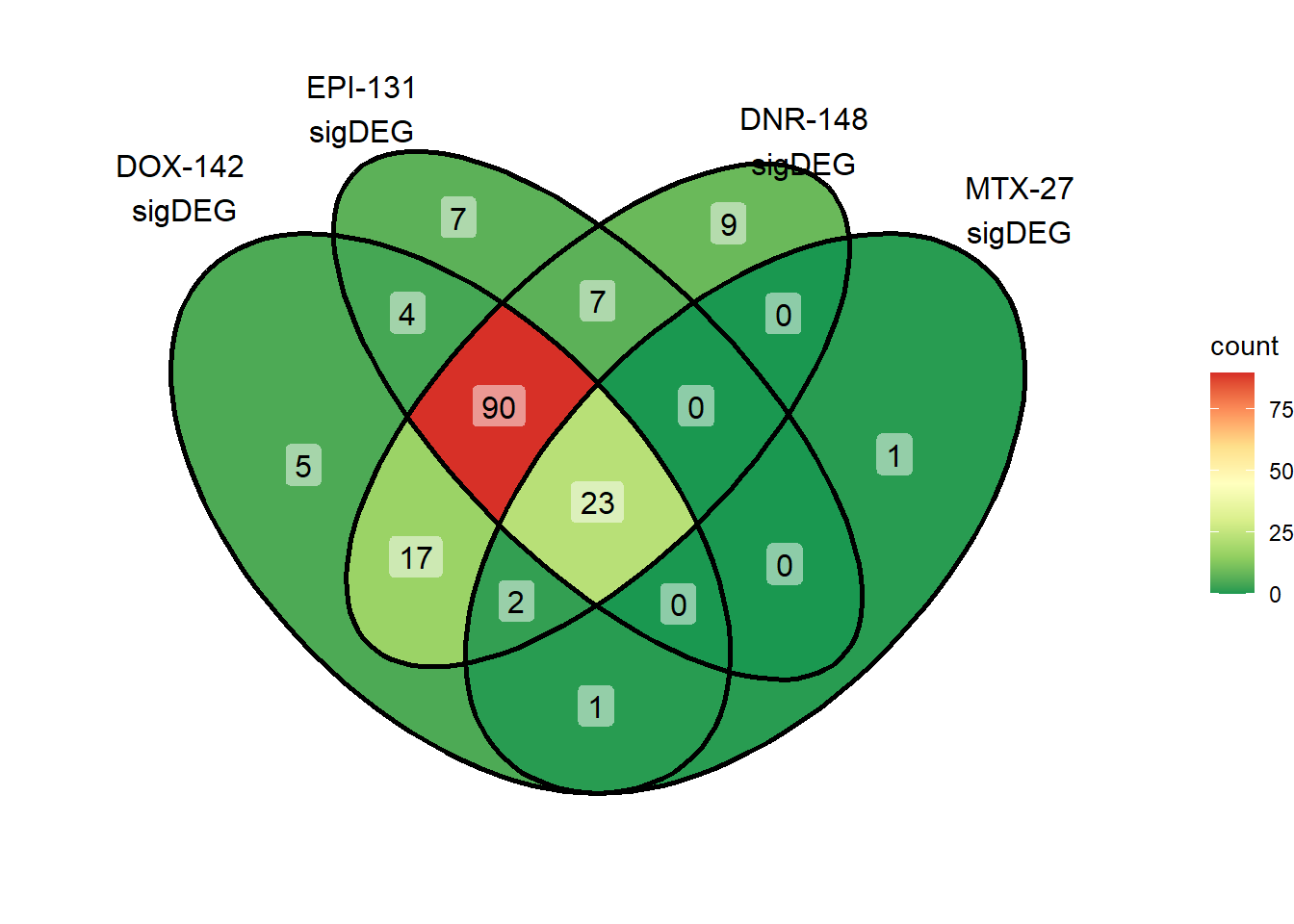 Here, I found 90 reQTLs that were related to only anthracyclines, with
23 reQTLS related to all Top2i drugs at 24 hours.
Here, I found 90 reQTLs that were related to only anthracyclines, with
23 reQTLS related to all Top2i drugs at 24 hours.
Venn of DOX egenes and all TOP2i DEGs
library(paletteer)
assignInNamespace(x="plot_venn", value=plot_venn, ns="ggVennDiagram")
reQTL_overlapDE24 <- list(DOXreQTLs$ENTREZID,sigVDA24$ENTREZID, sigVEP24$ENTREZID, sigVMT24$ENTREZID)
re_incommon <- c(DOXreQTLs$ENTREZID,sigVDA24$ENTREZID, sigVEP24$ENTREZID, sigVMT24$ENTREZID)
names(reQTL_overlapDE24) <- c("Dox_reQTLS", "DNR DEGs","EPI DEGs","MTX DEGs")
ggVennDiagram::ggVennDiagram(reQTL_overlapDE24,
category.names = c("DOX\negenes",
"DNR DEGs",
"EPI DEGs",
"MTX DEGs"),
show_intersect = FALSE,
set_color = "black",
category_size = c(6,6,6,6),
label = "count",
color = c("DOX\negenes" = "yellow","DNR DEGs" ="steelblue","EPI DEGs" = 'red', "MTX DEGs" = 'black') ,
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid", set_size = 4.5)+
scale_x_continuous(expand = expansion(mult = .3))+
scale_y_continuous(expand = expansion(mult = .2))+
# scale_color_paletteer_d(palette = "fishualize::Bodianus_pulchellus")+
# scale_colour_gradientn(colours = cols,
# values = rescale(c(0, 20, 75, 1000, 5000)),
# guide = "colorbar", limits=c(0, 100)) +
scale_fill_distiller(palette="Spectral", direction = -1, limits= c(1,200),oob=scales::squish)+
# scale_fill_manual(values = cbp1)+
labs(title = "24 hour DOX egenes in other DEG sets",
caption = paste("n =", length(unique(re_incommon)),"genes"))+
theme(plot.title = element_text(size = rel(1.6), hjust = 0.5, vjust =1))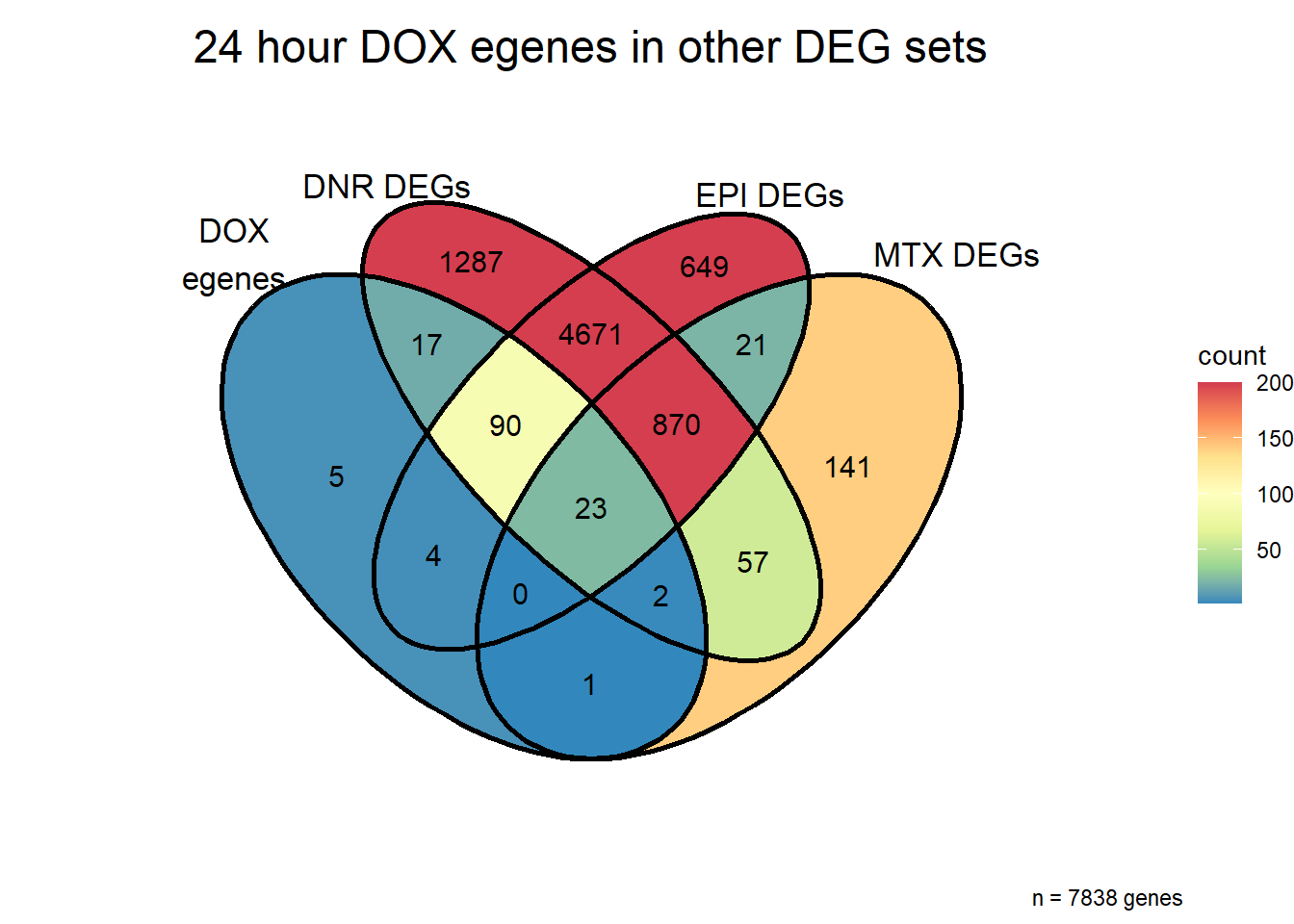
DEG_incommon <-c(sigVDA24$ENTREZID, sigVEP24$ENTREZID, sigVMT24$ENTREZID)
uniqueDEGs_incommon <- unique(DEG_incommon)
two_venn <- list( DOXreQTLs$ENTREZID,uniqueDEGs_incommon)
ggVennDiagram::ggVennDiagram(two_venn,
category.names = c("DOX egenes\nn = 142","union of TOP2i DEGs\n n = 7838"),
show_intersect = FALSE,
set_color = "black",
category_size = c(6,6),
label = "count",
# color = c("DOX\negenes" = "yellow","DNR DEGs" ="steelblue","EPI DEGs" = 'red', "MTX DEGs" = 'black') ,
#
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid", set_size = 4.5)+
scale_x_continuous(expand = expansion(mult = .3))+
scale_y_continuous(expand = expansion(mult = .1))+
scale_fill_distiller(palette="Spectral", direction = -1, limits= c(1,200),oob=scales::squish)#+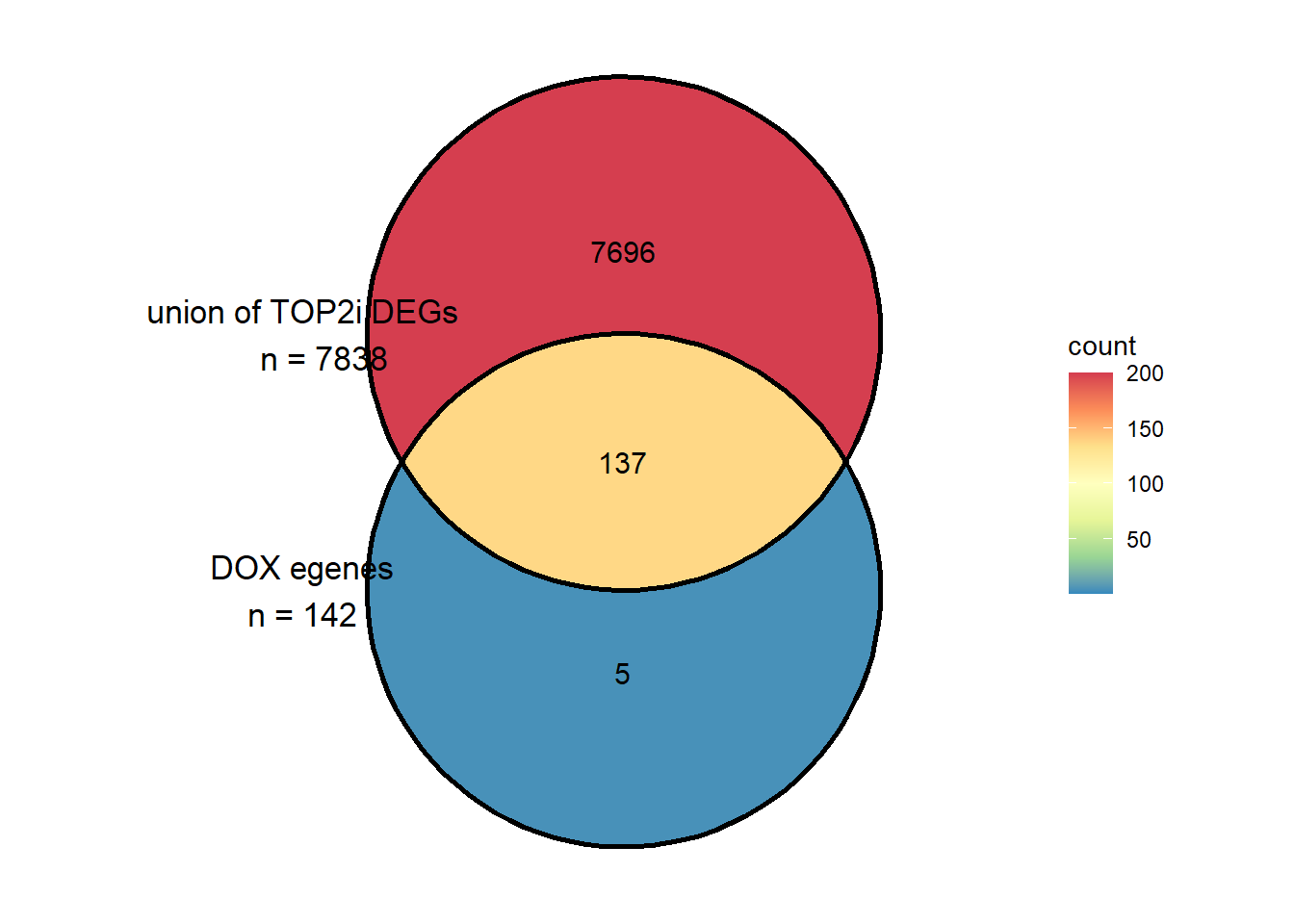
# labs(title = "24 hour DOX egenes in other DEG sets",
# caption = paste("n =", length(unique(re_incommon)),"genes"))+
# theme(plot.title = element_text(size = rel(1.6), hjust = 0.5, vjust =1))
ggsave("output/Figures/DOXegenesVENN.eps") DOXeQTL_table <- toplistall %>%
mutate(sigcount = if_else(adj.P.Val < 0.05,'sig','notsig'))%>%
mutate(DOXreQTLs=if_else(ENTREZID %in%DOXreQTLs$ENTREZID,"y","no")) %>%
dplyr::filter(time =="24_hours") %>%
dplyr::select(ENTREZID,id,DOXreQTLs,sigcount) %>%
group_by(id,DOXreQTLs,sigcount) %>%
tally() %>% as.data.frame() %>%
pivot_wider(.,id_cols = c(id,DOXreQTLs),names_from = sigcount,values_from = n) %>%
dplyr::select(id,DOXreQTLs,sig)
DOXeQTL_table%>%
kable(., caption= "All 24 hour sigDEGs broken down by drug and DOXreQTL status") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "80%", height = "400px")| id | DOXreQTLs | sig |
|---|---|---|
| DNR | no | 6885 |
| DNR | y | 132 |
| DOX | no | 6503 |
| DOX | y | 142 |
| EPI | no | 6211 |
| EPI | y | 117 |
| MTX | no | 1089 |
| MTX | y | 26 |
| TRZ | no | NA |
| TRZ | y | NA |
DOXeQTL_table %>%
add_row(id=c("Dox-specific DEGs","Dox-specific DEGs"),DOXreQTLs=c("no","y"), sig = c(62,1)) %>%
dplyr::filter(DOXreQTLs=="y") %>%
mutate(opp= 142-sig) %>%
filter(id !=c('TRZ','DOX')) %>%
rename("yes"=sig, "no"=opp) %>%
mutate(y_percent= paste0(sprintf("%2.1f", yes/(yes+no)*100), "%"),n_percent = paste0(sprintf("%2.1f", no/(yes+no)*100),"%")) %>%
pivot_longer(!c(id,DOXreQTLs,y_percent,n_percent), names_to="group", values_to = "count") %>%
mutate(id=factor(id, levels=c("DNR","EPI", "MTX","Dox-specific DEGs"))) %>%
ggplot(., aes(y=id,x=count,fill=group))+
geom_col(position='fill')+
theme_classic()+
scale_color_manual(values=c("red4","navy"))+
scale_fill_manual(values=c("#2297E6","red2"))+
ggtitle("DOXreQTLs overlaps")+
scale_y_discrete(limits=rev)+
geom_text(aes(y=id,x=1, label = n_percent,hjust=.8))+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.0),
axis.line = element_line(linewidth = 1.0),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 15, color = "black", face = "bold"))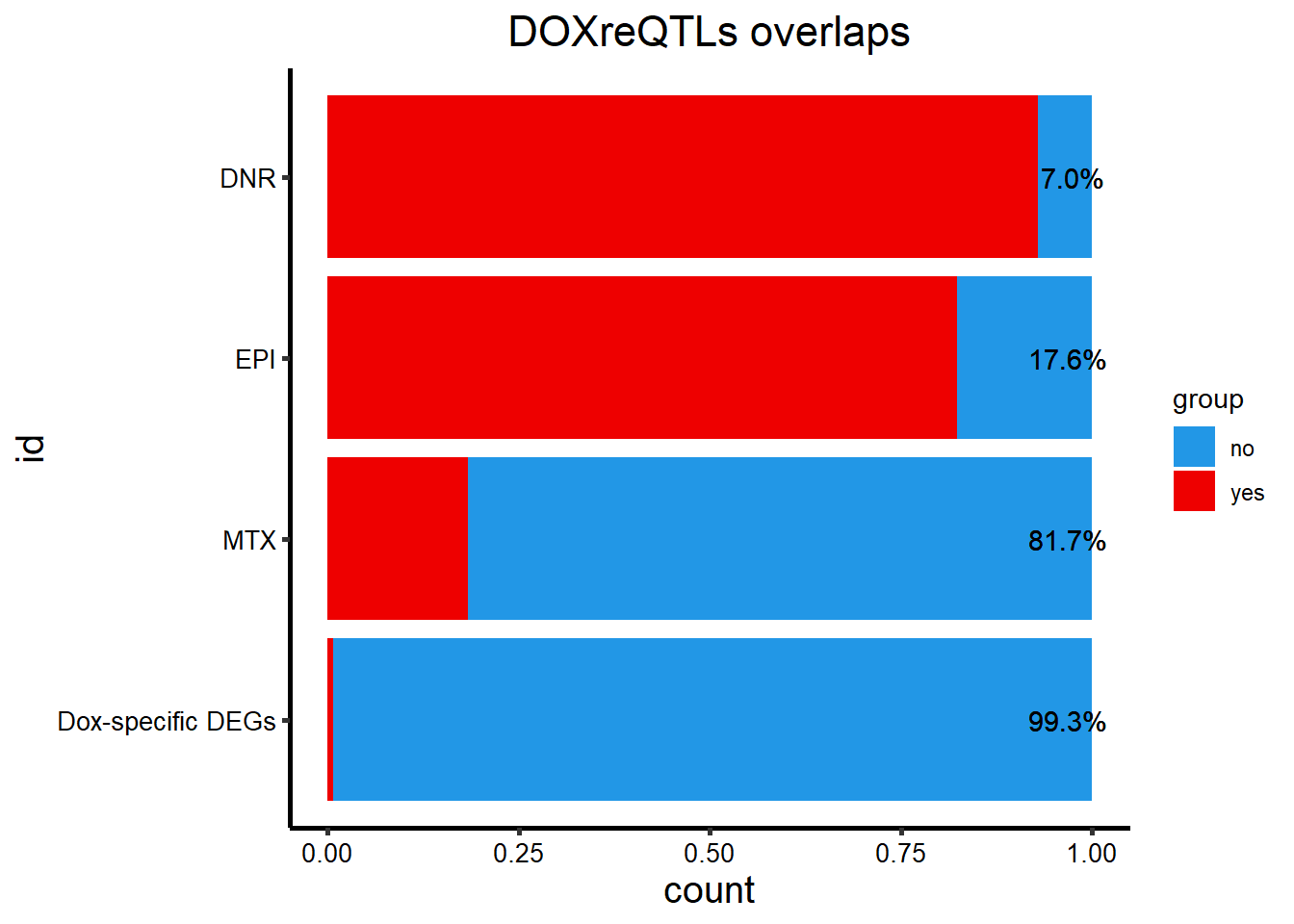
DOX specific-Dox dE-reqtl
# DoxonlyQTL <- as.numeric(QTLoverlaps$..values..[[14]])
# geneDoxonlyQTL <- getBM(attributes=my_attributes,filters ='entrezgene_id',
# values =DoxonlyQTL, mart = ensembl)
# write.csv(geneDoxonlyQTL,"data/geneDoxonlyQTL.csv")
geneDoxonlyQTL <- read.csv("data/geneDoxonlyQTL.csv",row.names = 1)
cpmcounts <- readRDS("data/cpmcount.RDS")
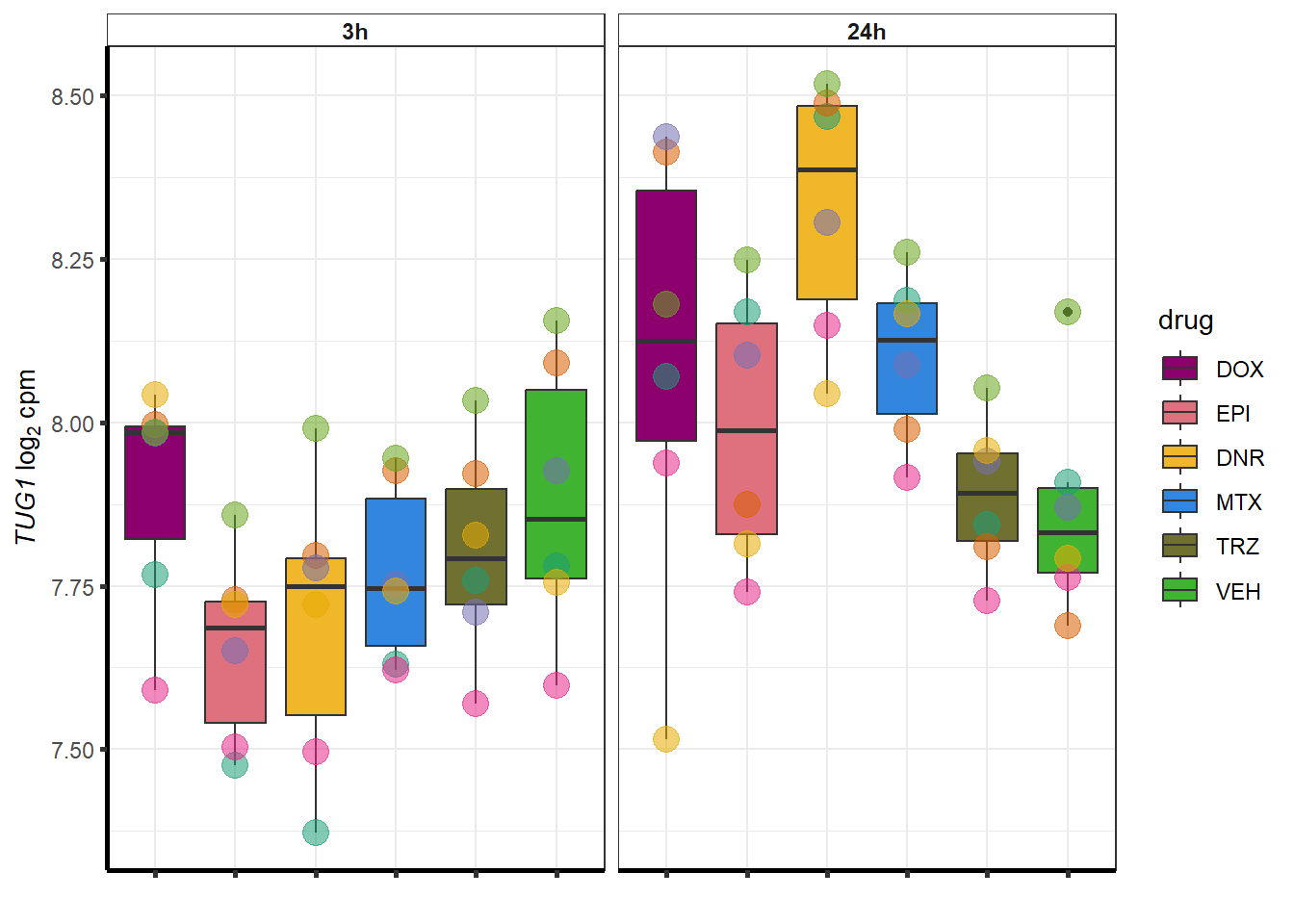
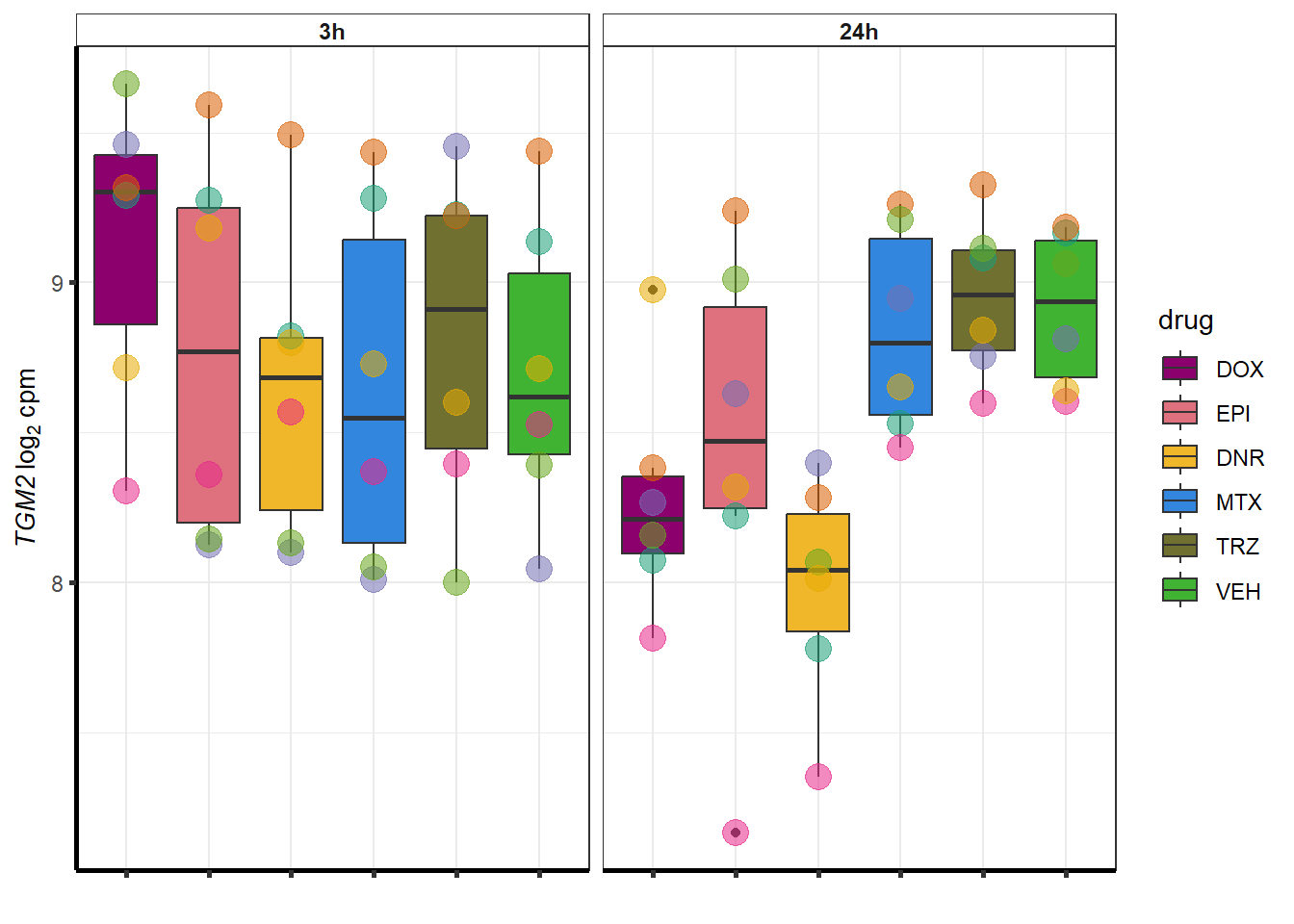
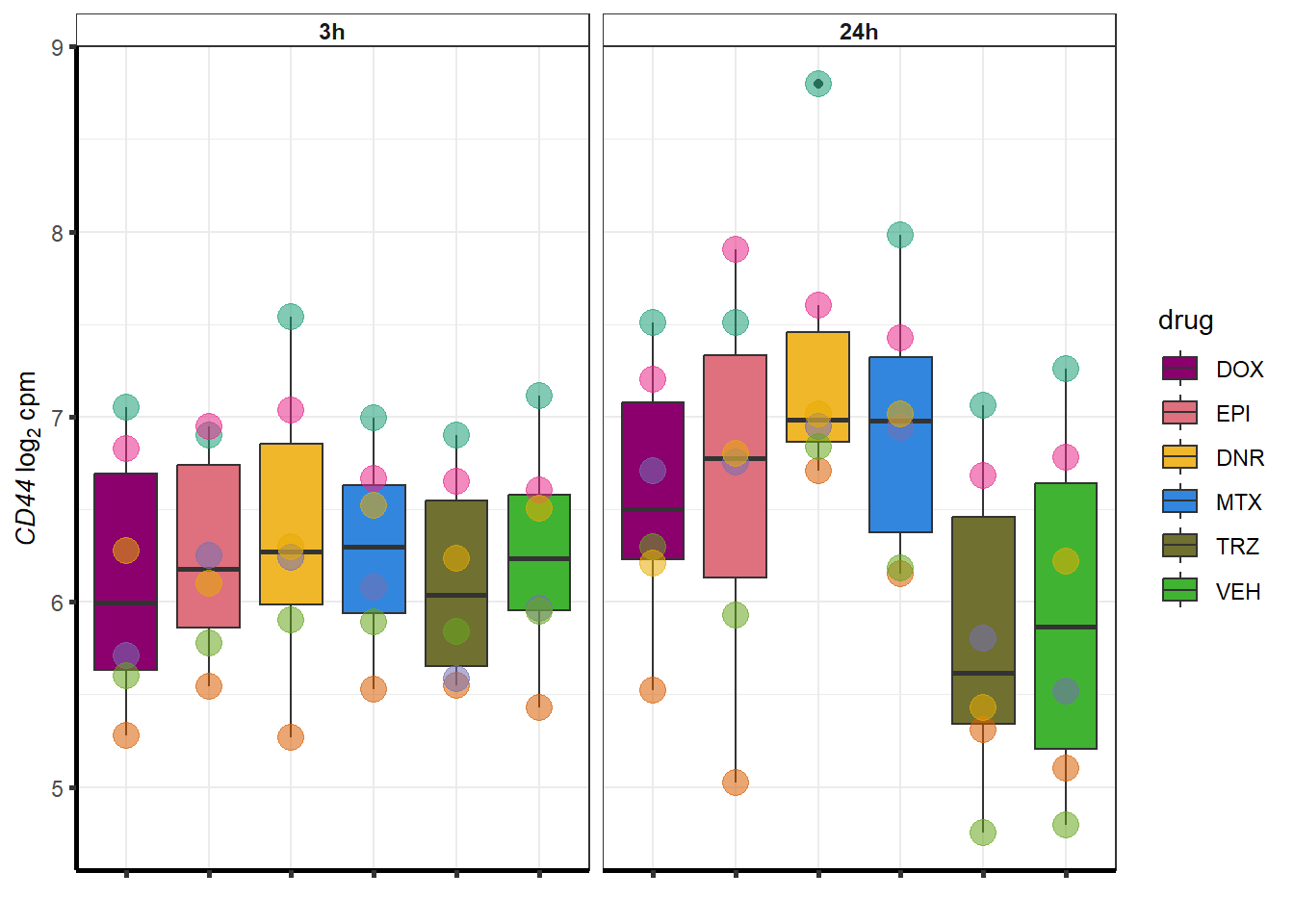
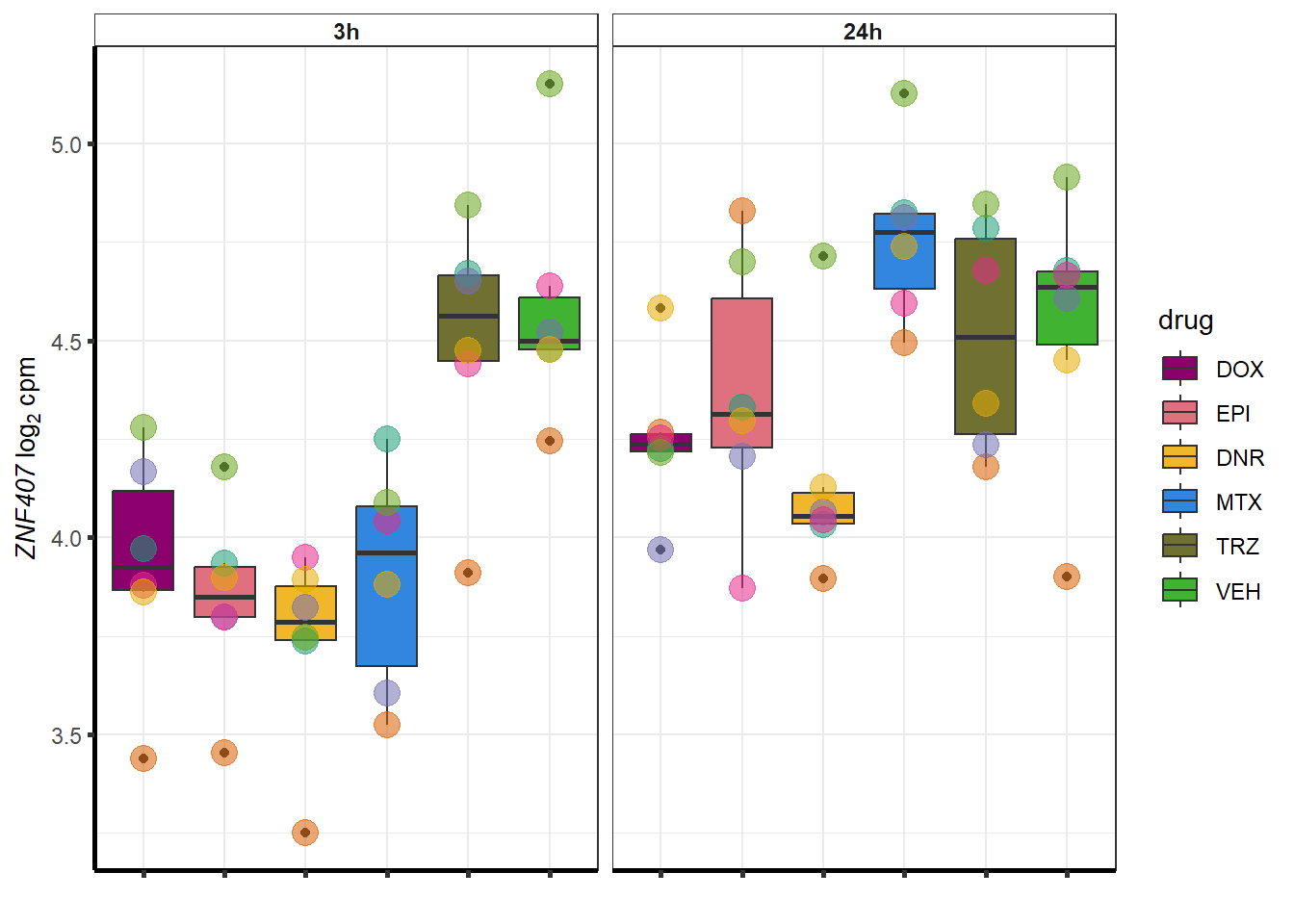
for (g in seq(from=1, to=length(geneDoxonlyQTL$entrezgene_id))){
a <- geneDoxonlyQTL$hgnc_symbol[g]
cpm_boxplot(cpmcounts,GOI=geneDoxonlyQTL[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}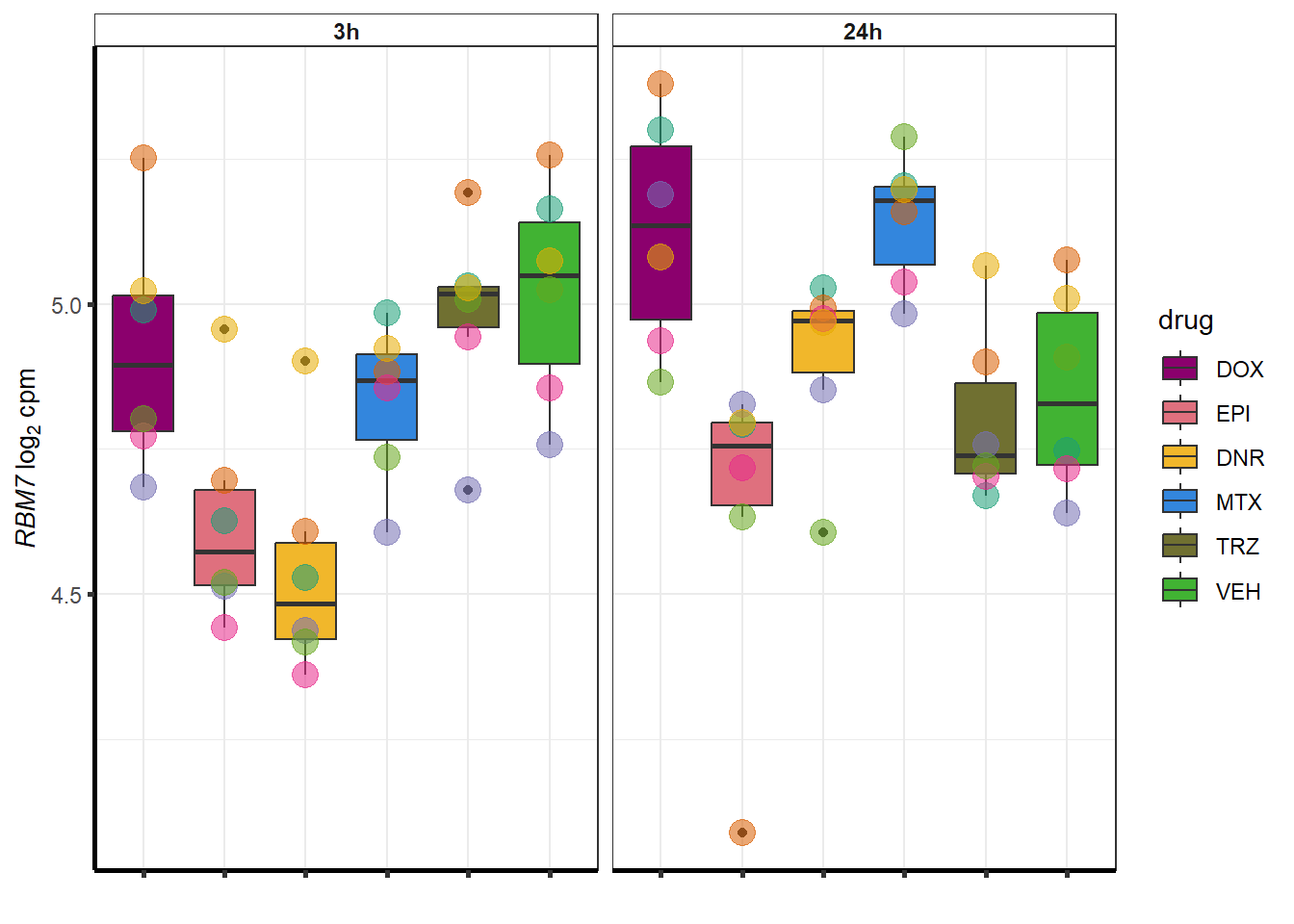
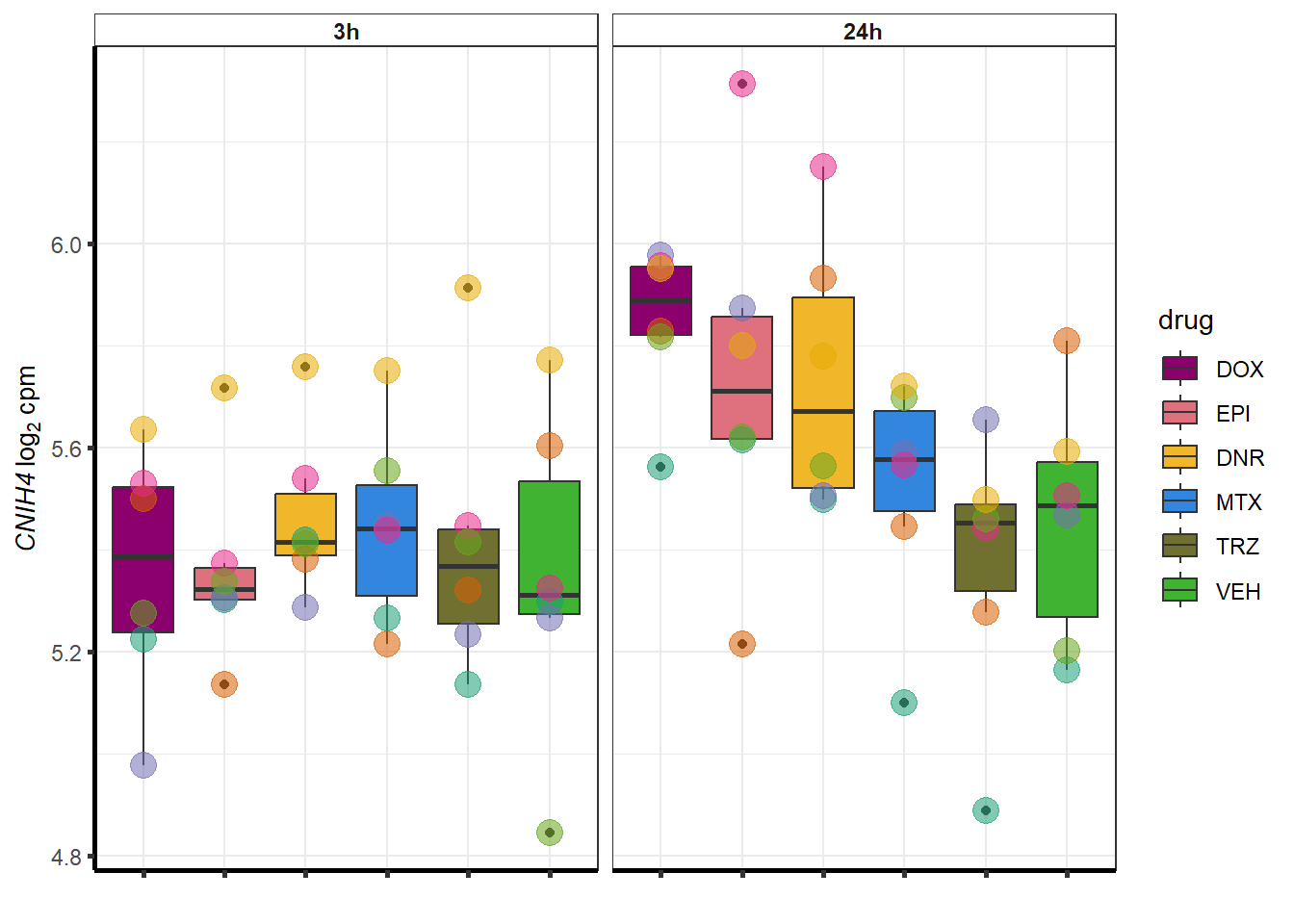
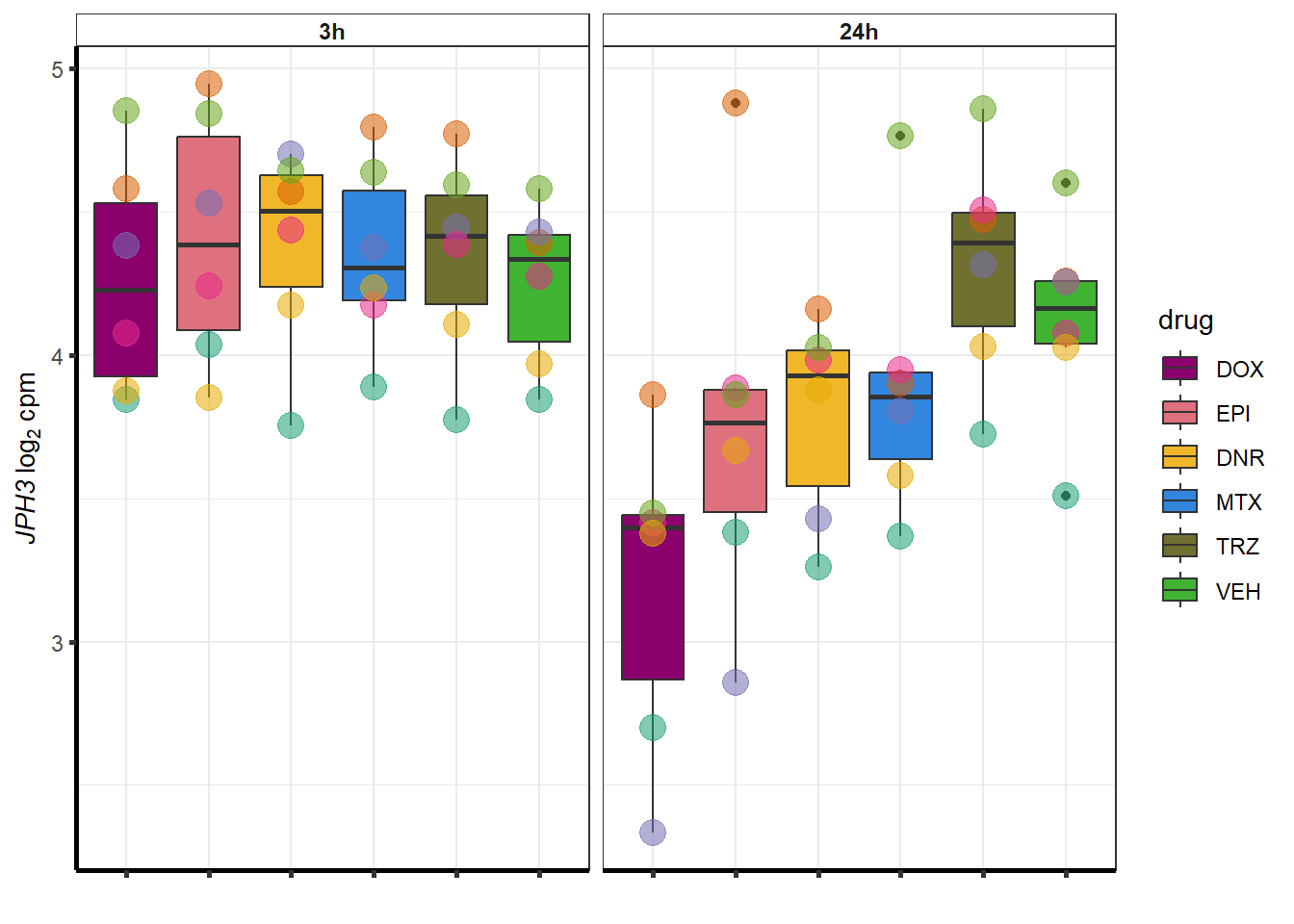
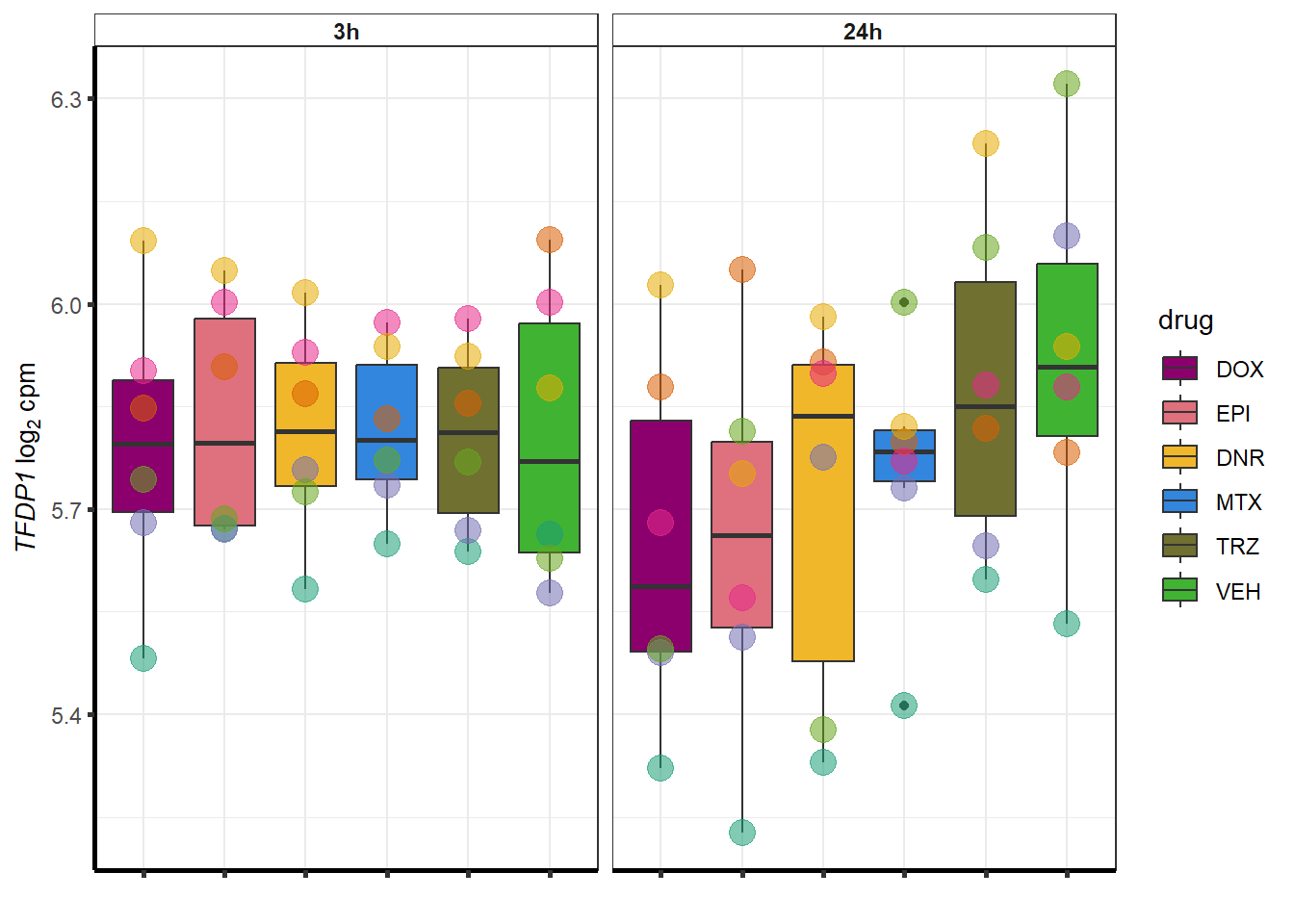
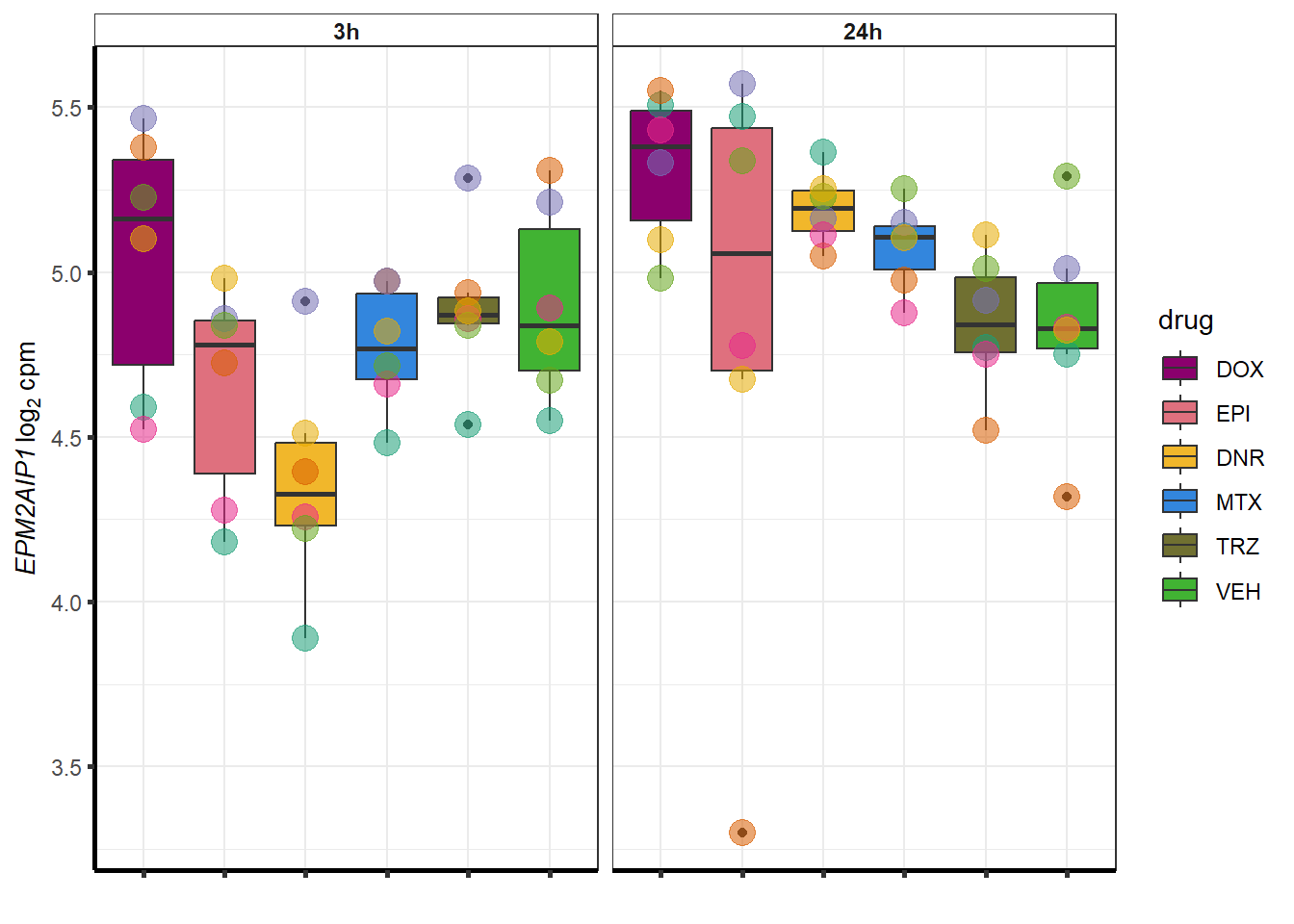 ### Dox specific DEG examination
### Dox specific DEG examination
#pull list
total24 <-list(sigVDA24$ENTREZID,sigVDX24$ENTREZID,sigVEP24$ENTREZID,sigVMT24$ENTREZID)
## do venn partition and pull doxspe genes
# total24 <- list(sigVDA24$SYMBOL,sigVDX24$SYMBOL,sigVEP24$SYMBOL,sigVMT24$SYMBOL)
trtonly_24h_genes <- readRDS("data/trtonly_24h_genes.RDS")
list2env(trtonly_24h_genes, envir = .GlobalEnv)<environment: R_GlobalEnv># venn_24h <- VennDiagram::get.venn.partitions(total24)
# DoxonlyDEG <- venn_24h$..values..[[14]]# 355
# EpionlyDEG <- venn_24h$..values..[[12]] #444
# DnronlyDEG <- venn_24h$..values..[[15]] #615
# MtxonlyDEG <- venn_24h$..values..[[8]] #122
# trtonly_24h_genes <- list(DnronlyDEG,DoxonlyDEG,EpionlyDEG, MtxonlyDEG)
# names(trtonly_24h_genes) <-c("DnronlyDEG","DoxonlyDEG","EpionlyDEG", "MtxonlyDEG")
# saveRDS(trtonly_24h_genes,"data/trtonly_24h_genes.RDS")
length(DoxonlyDEG)[1] 355intersect(DoxonlyDEG,DOXreQTLs$ENTREZID)[1] "57338" "29097" "7027" "10179" "9852" Dox24_lfc <- toplist24hr %>%
filter(ENTREZID %in% DoxonlyDEG) %>%
# group_by(id) %>%
# dplyr::filter(adj.P.Val<0.05) #%>%
mutate(logFC=logFC*(-1)) %>%
ggplot(., aes(x= id, y=logFC))+
geom_boxplot(aes(fill=id))+
theme_classic()+
fill_palette(palette = drug_palc)+
ggtitle("LogFC of Dox specific DEGs")+
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5,face = "bold"),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle = 20))
# strip.text.x = element_text(size = 12, color = "black", face = "italic"))
toplist24hr %>%
filter(ENTREZID %in% DoxonlyDEG) %>%
# filter(adj.P.Val<0.05) %>%
ggplot(., aes(x=adj.P.Val))+
# geom_histogram(aes(fill=id,position="dodge"))+
geom_density(aes(fill=id))+
# facet_wrap(~id)+
fill_palette(palette = drug_palc)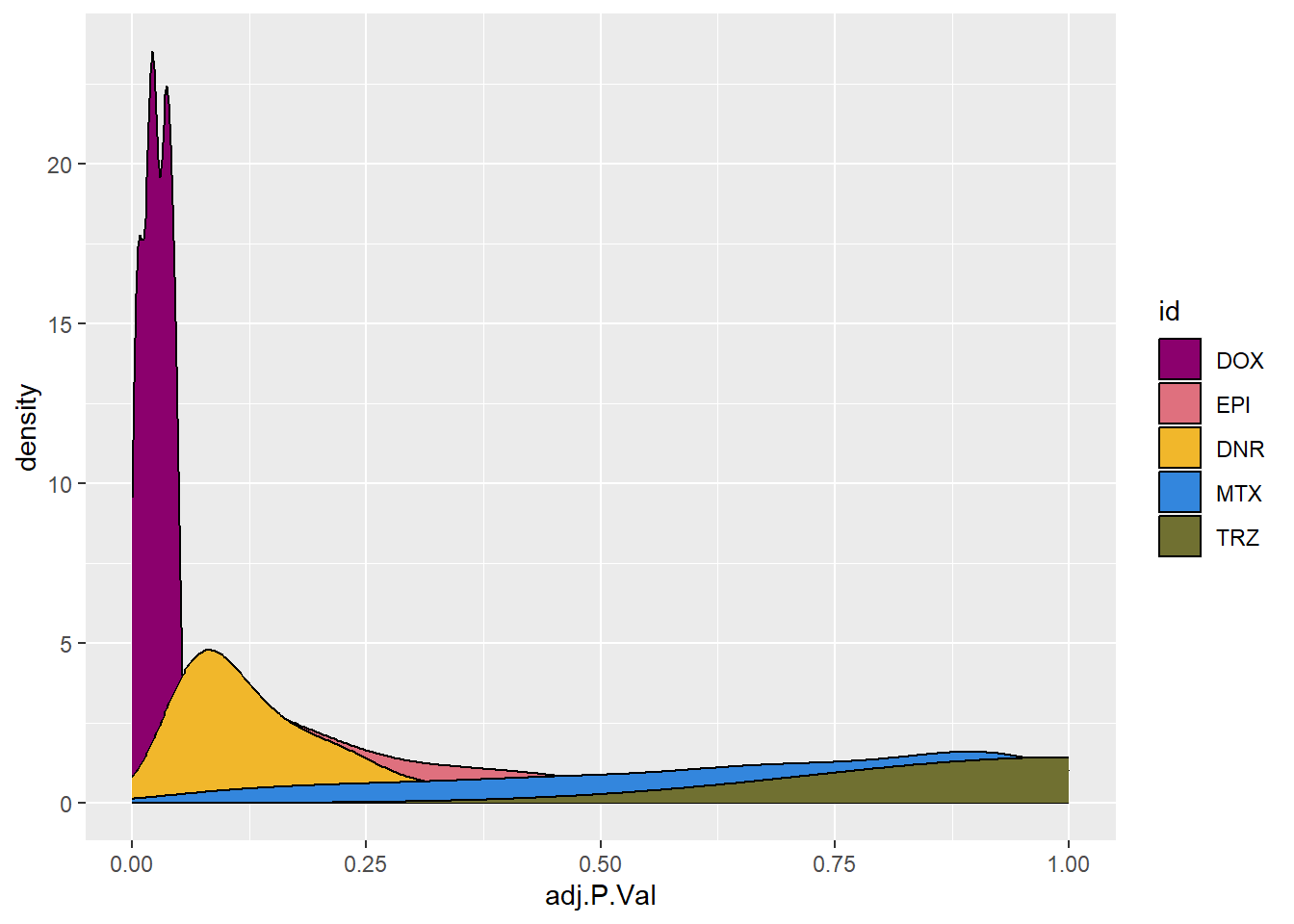
toplist24hr %>%
dplyr::filter(ENTREZID %in% DoxonlyDEG) %>%
# filter(adj.P.Val<0.5) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)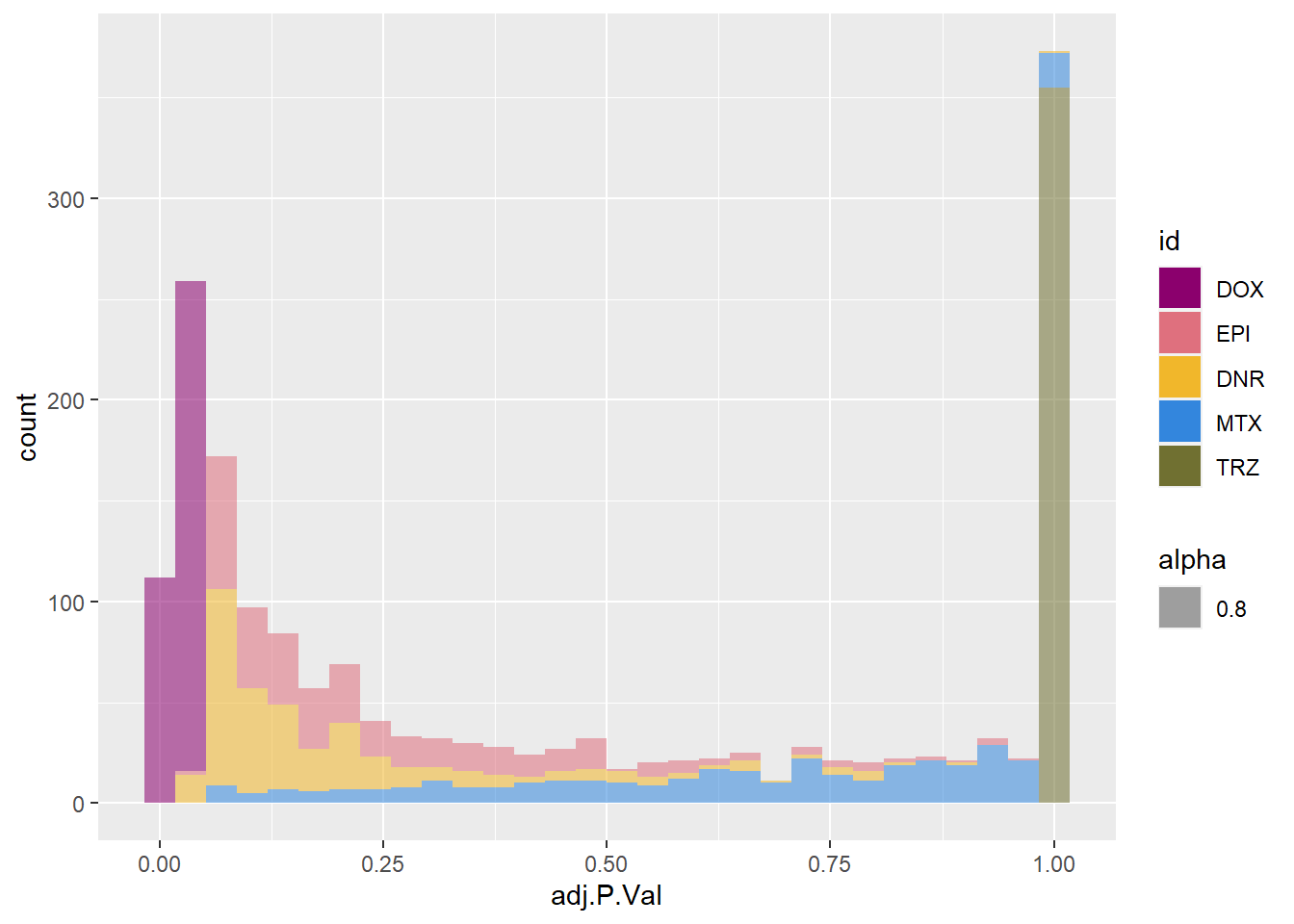
print(Dox24_lfc)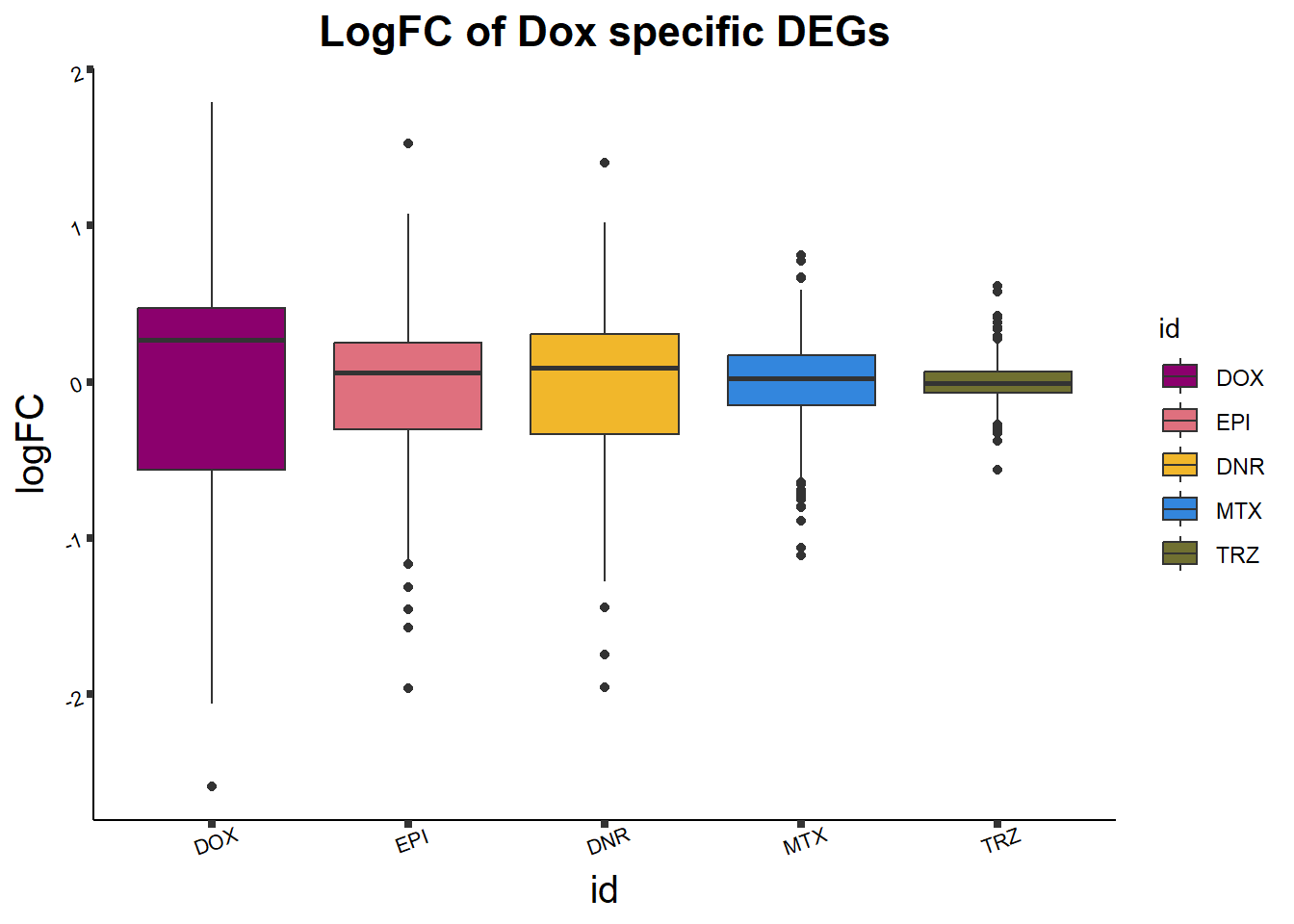
Plots of some genes:
Dox_eGene_sp <- intersect(DoxonlyDEG,DOXreQTLs$ENTREZID)
### making list
# #
# Doxonly_deg <- getBM(attributes=my_attributes,filters ='entrezgene_id',
# values =DoxonlyDEG, mart = ensembl)
# write.csv(Doxonly_deg,"output/Doxonly_deg.csv")
Doxonly_deg <- read.csv("output/Doxonly_deg.csv", row.names = 1)
set.seed(12345)
sampset <- Doxonly_deg %>%
distinct(entrezgene_id,.keep_all = TRUE) %>%
sample_n(.,12)
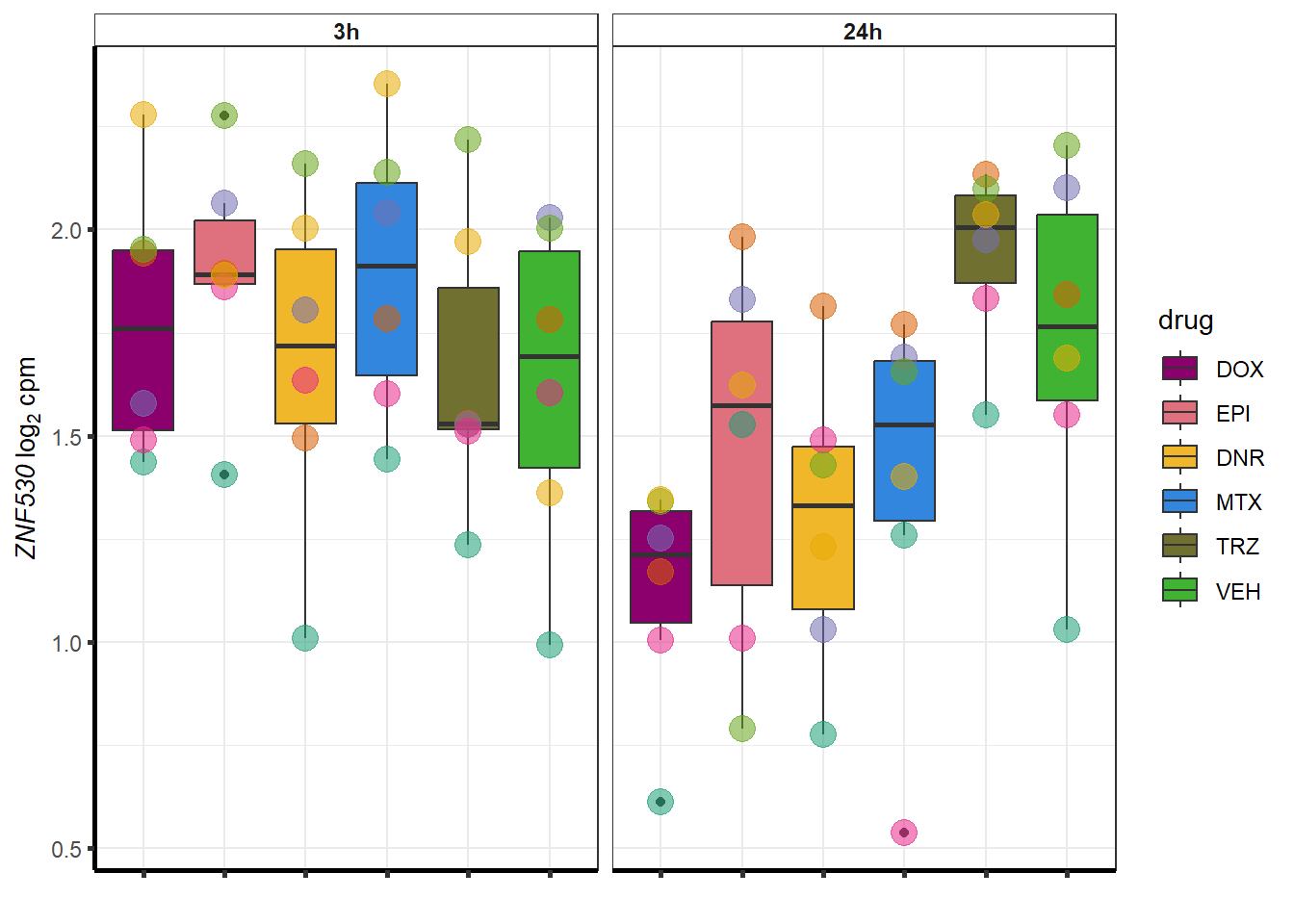
for (g in seq(from=1, to=length(sampset$entrezgene_id))){
a <- sampset$hgnc_symbol[g]
cpm_boxplot(cpmcounts,GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}
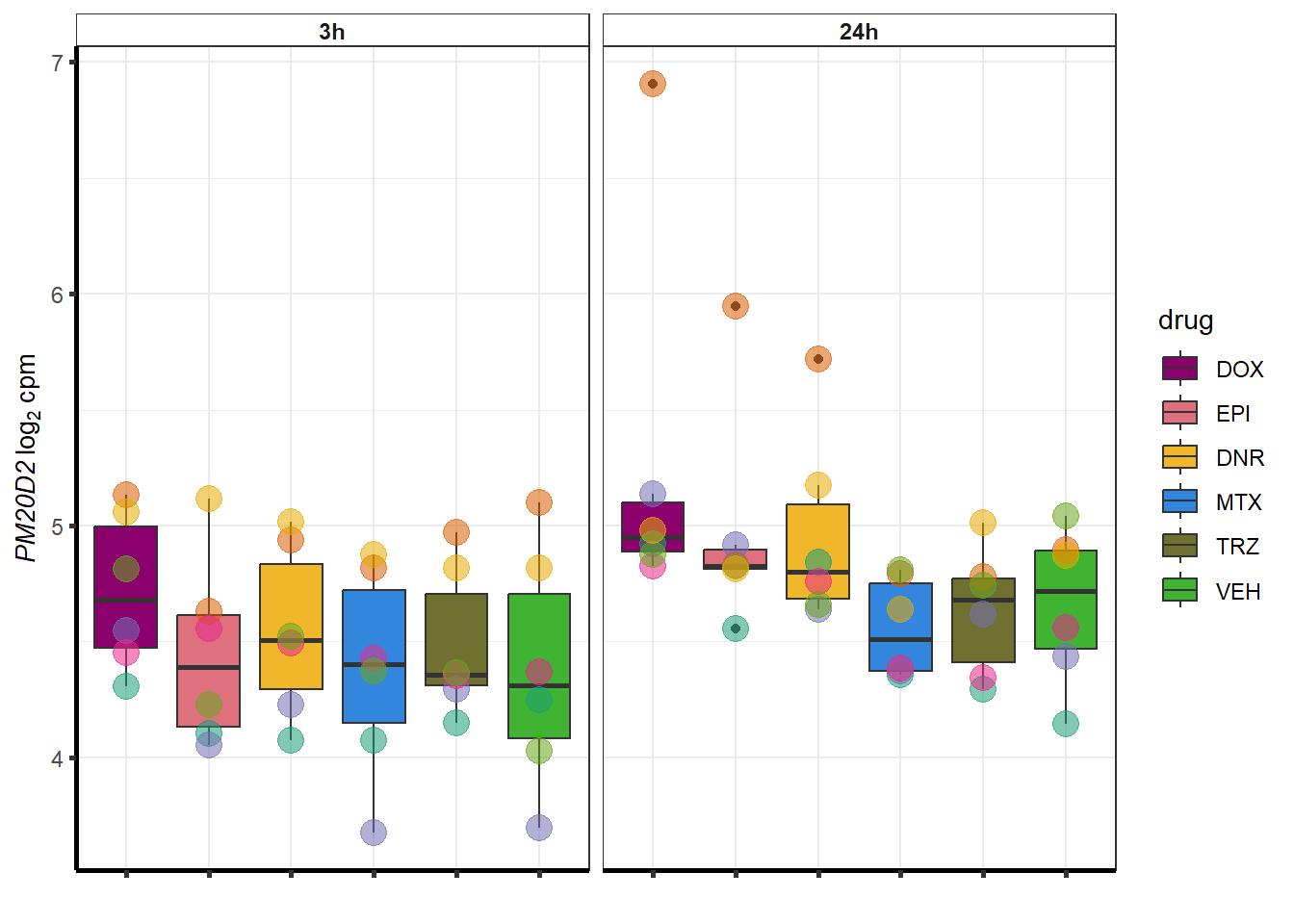
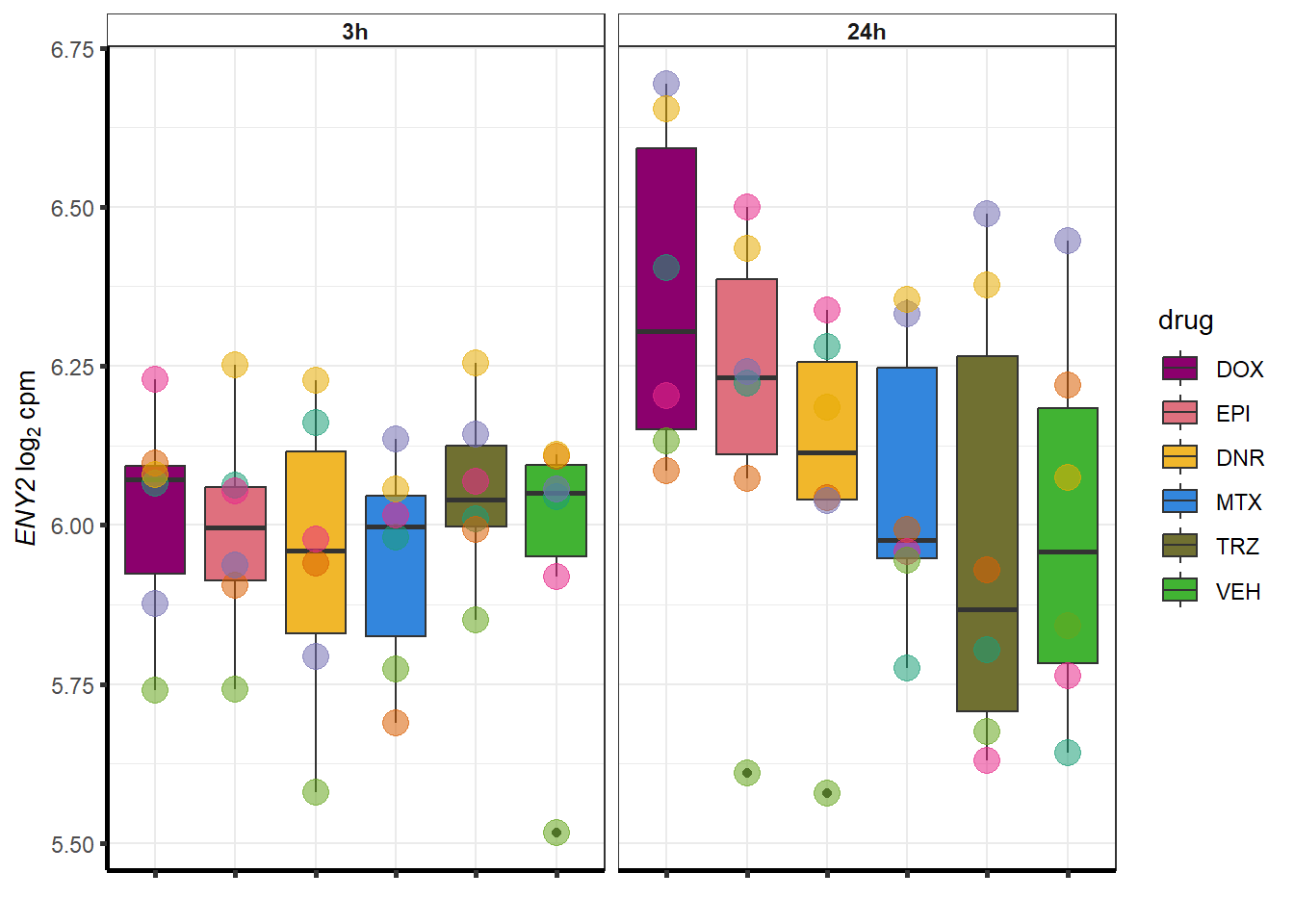
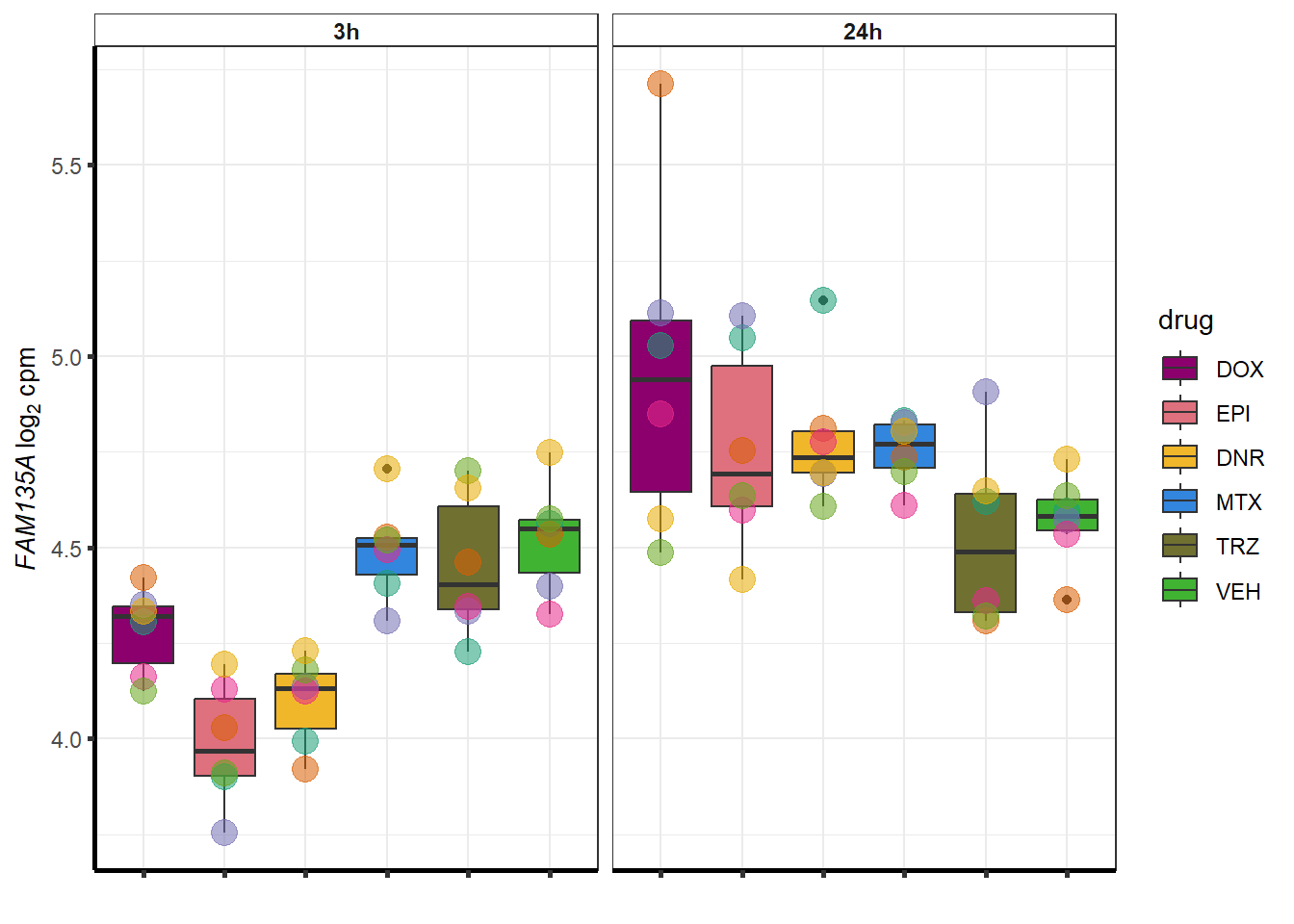
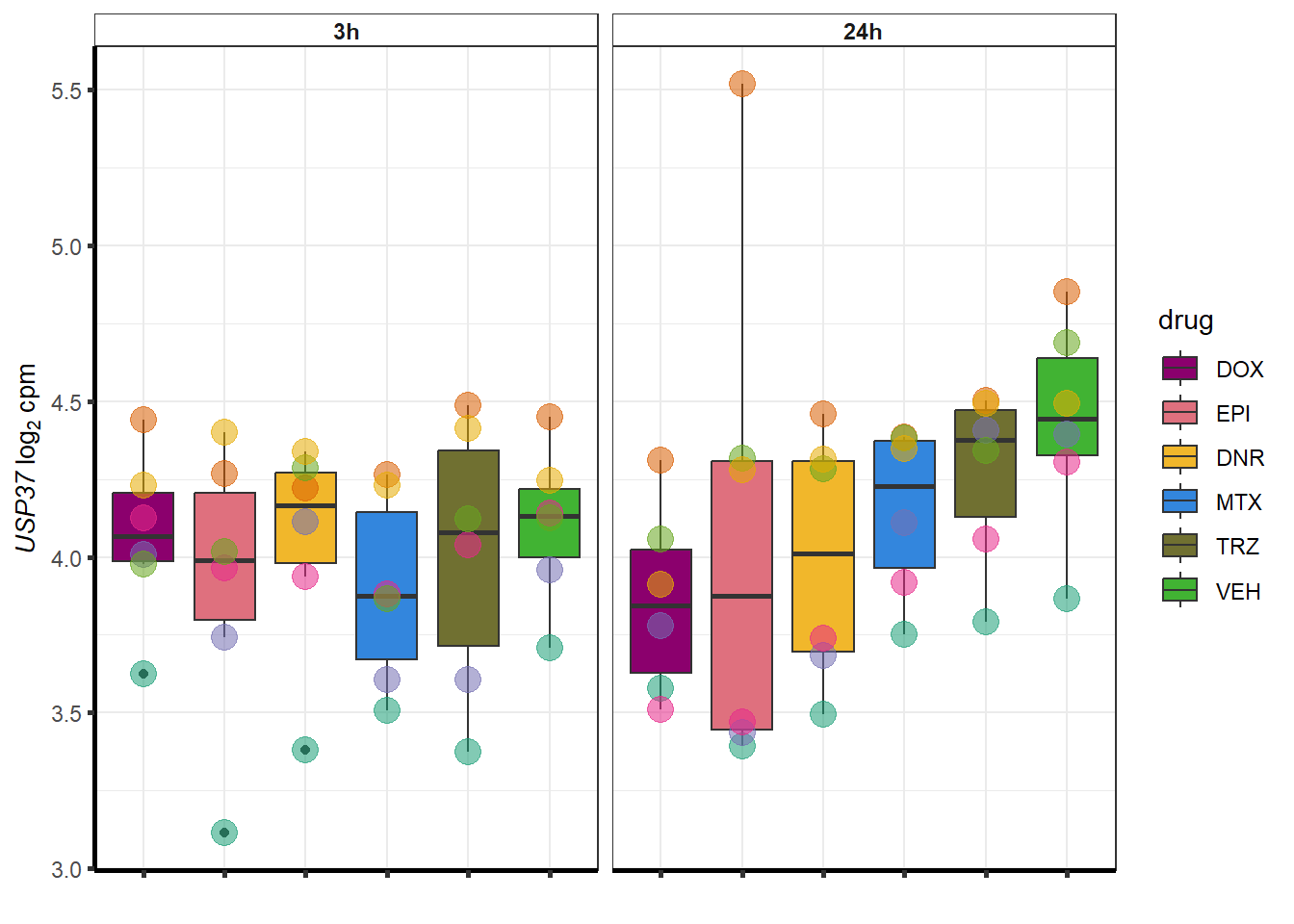
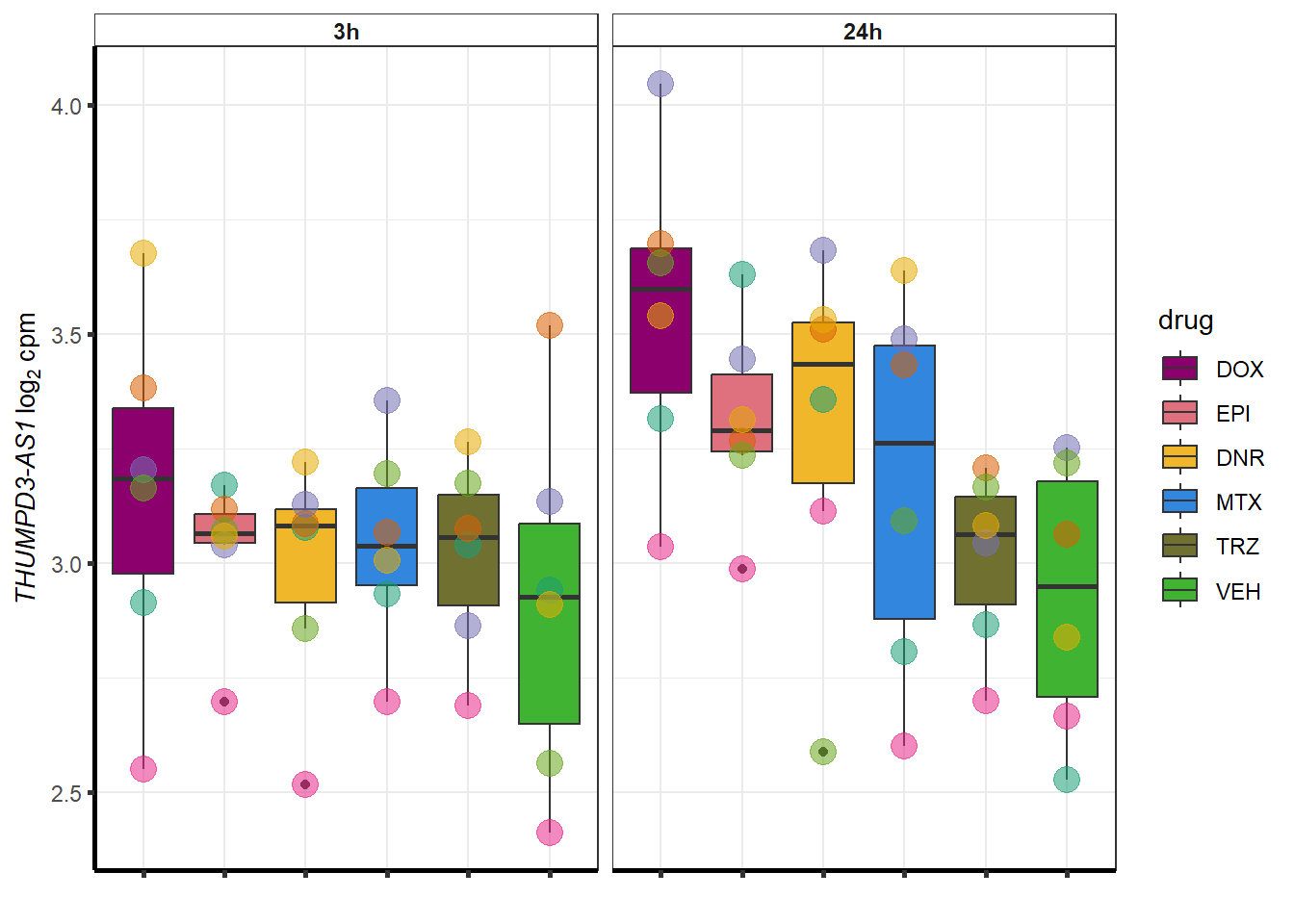
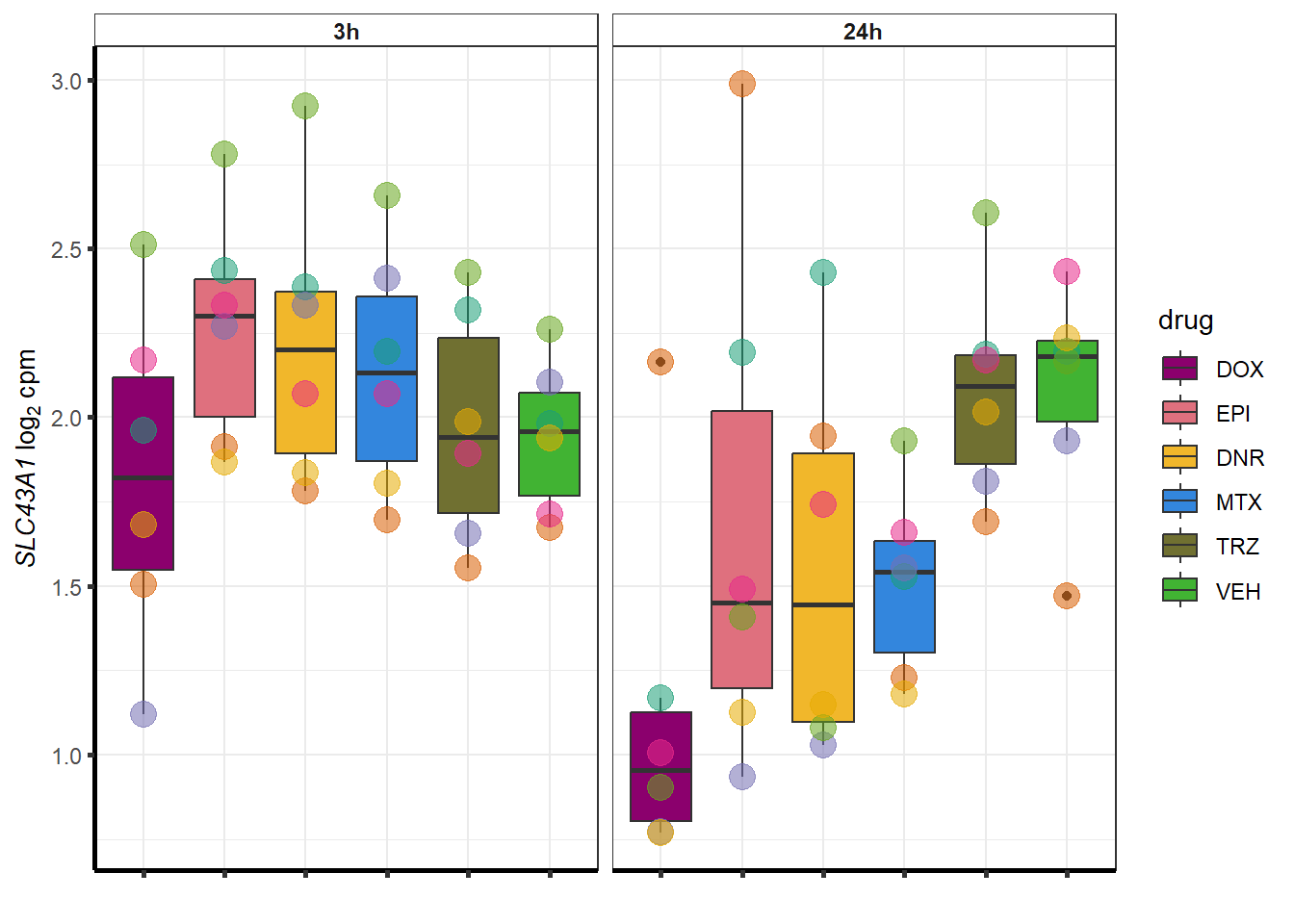
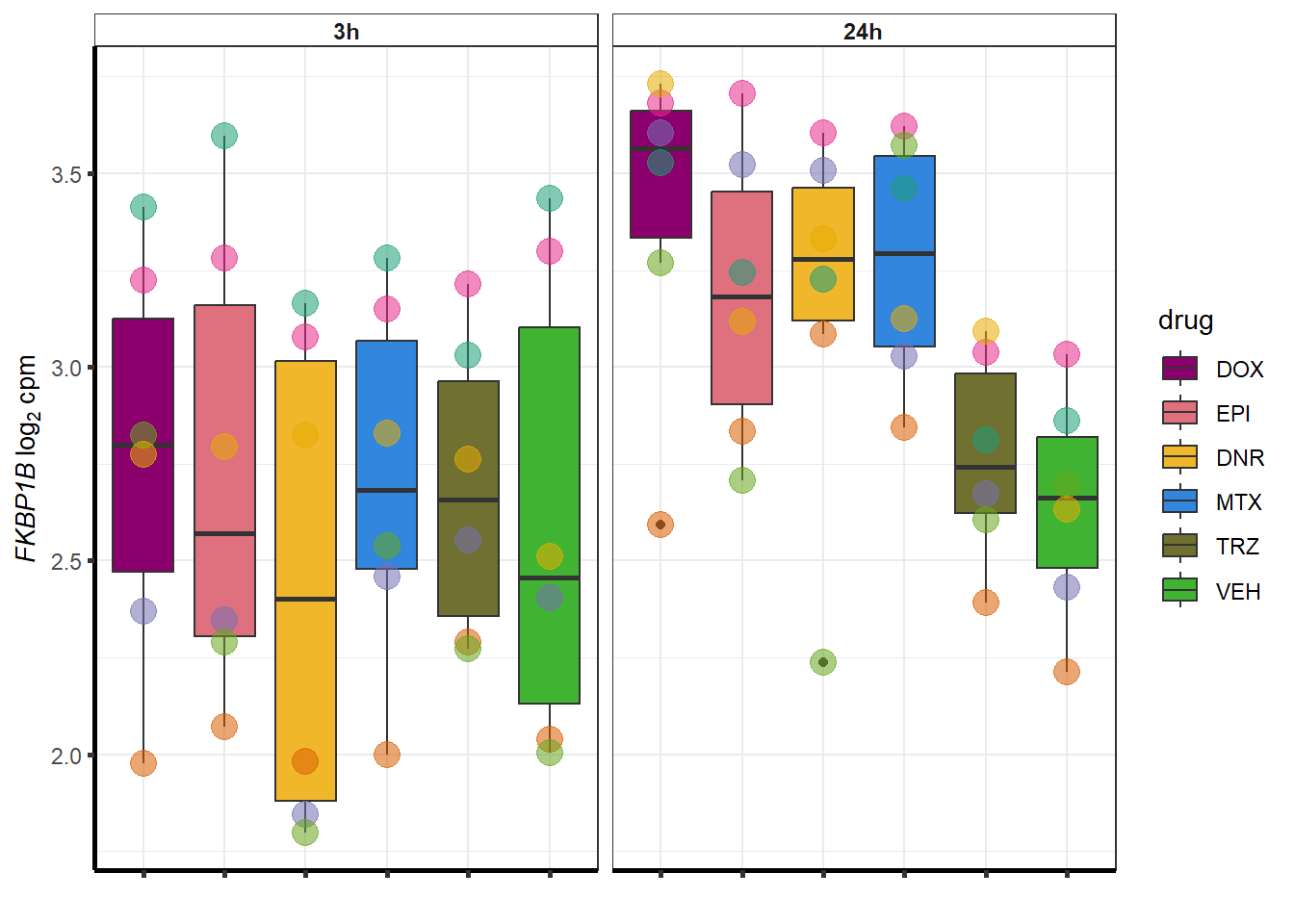
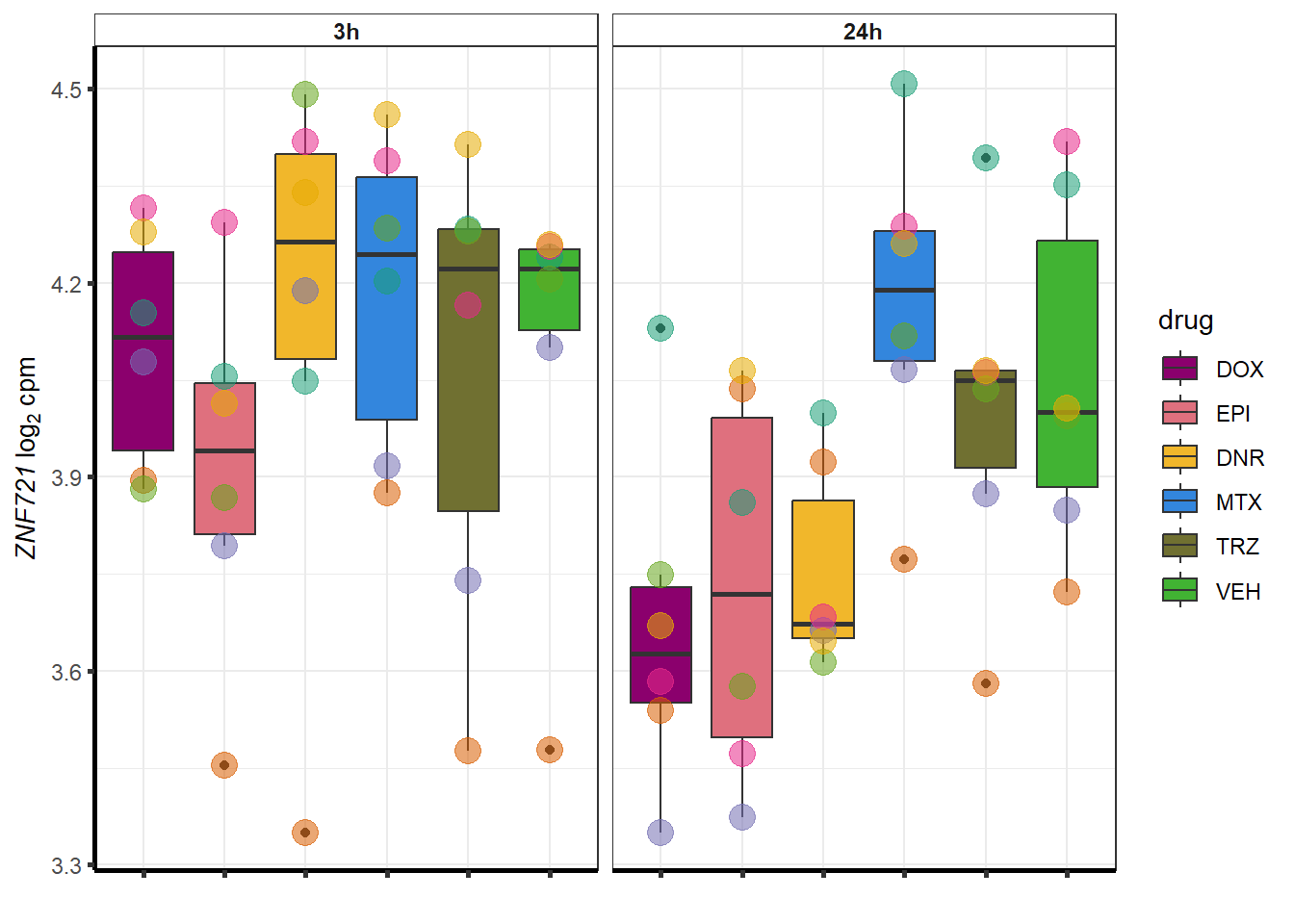
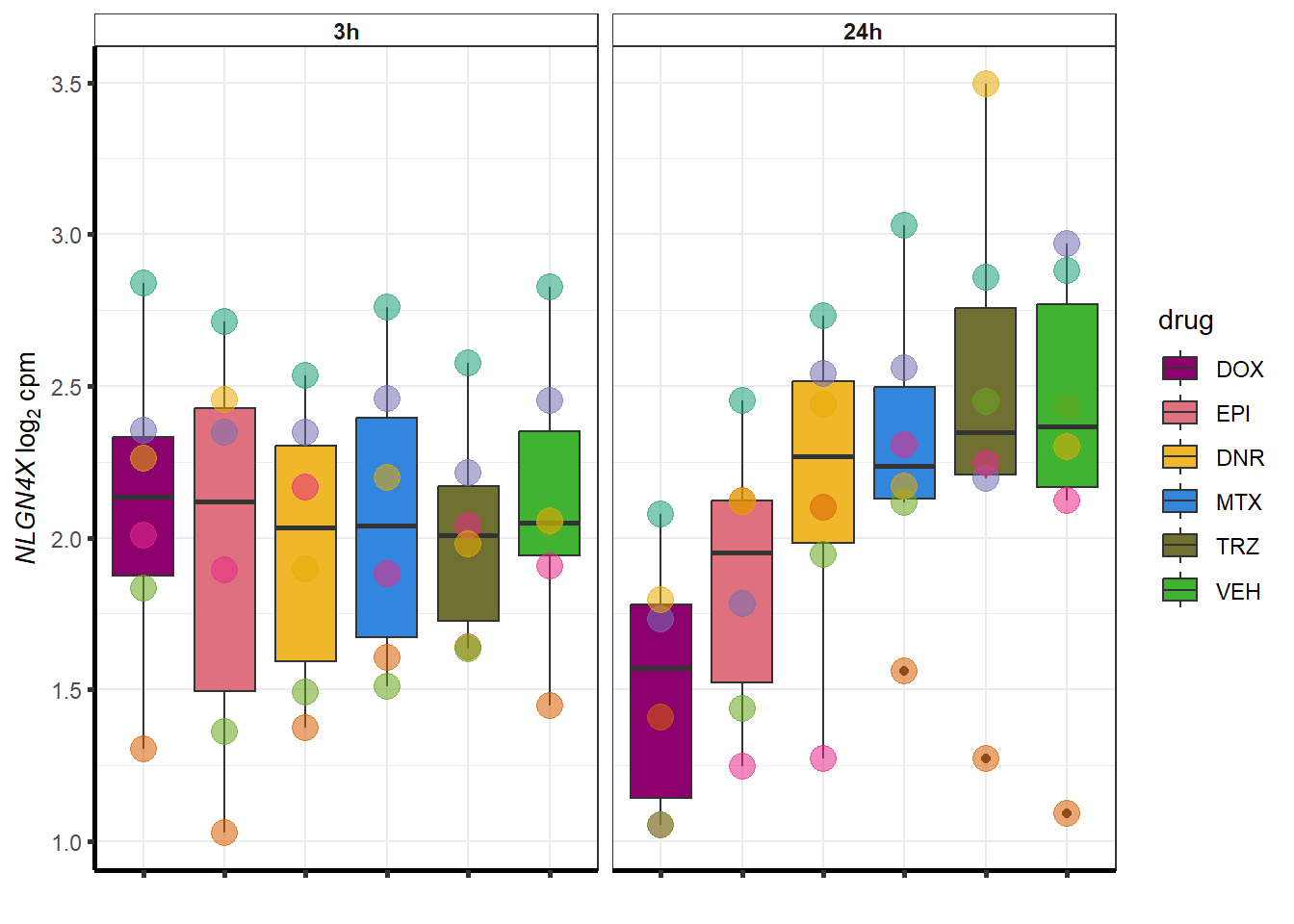
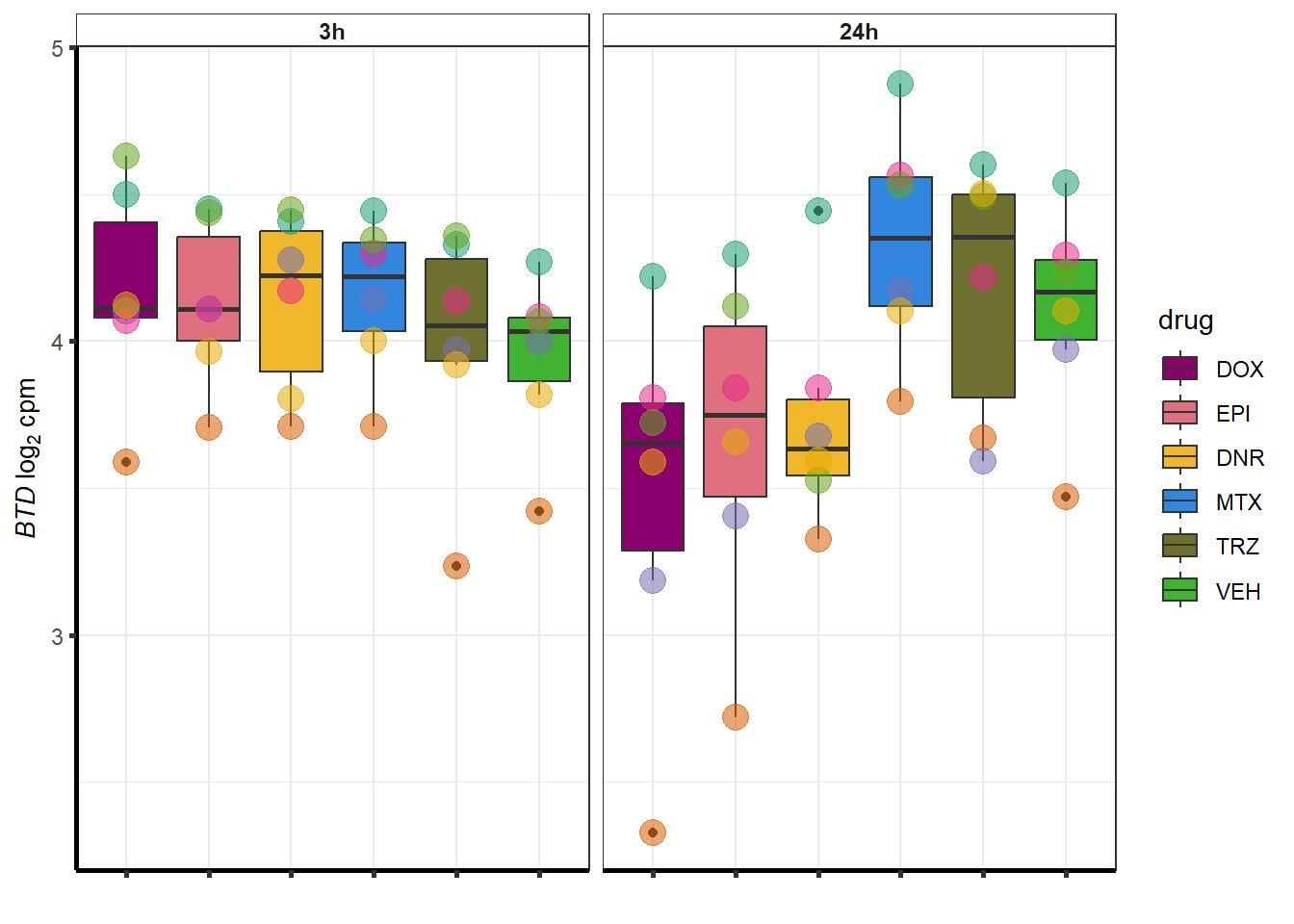
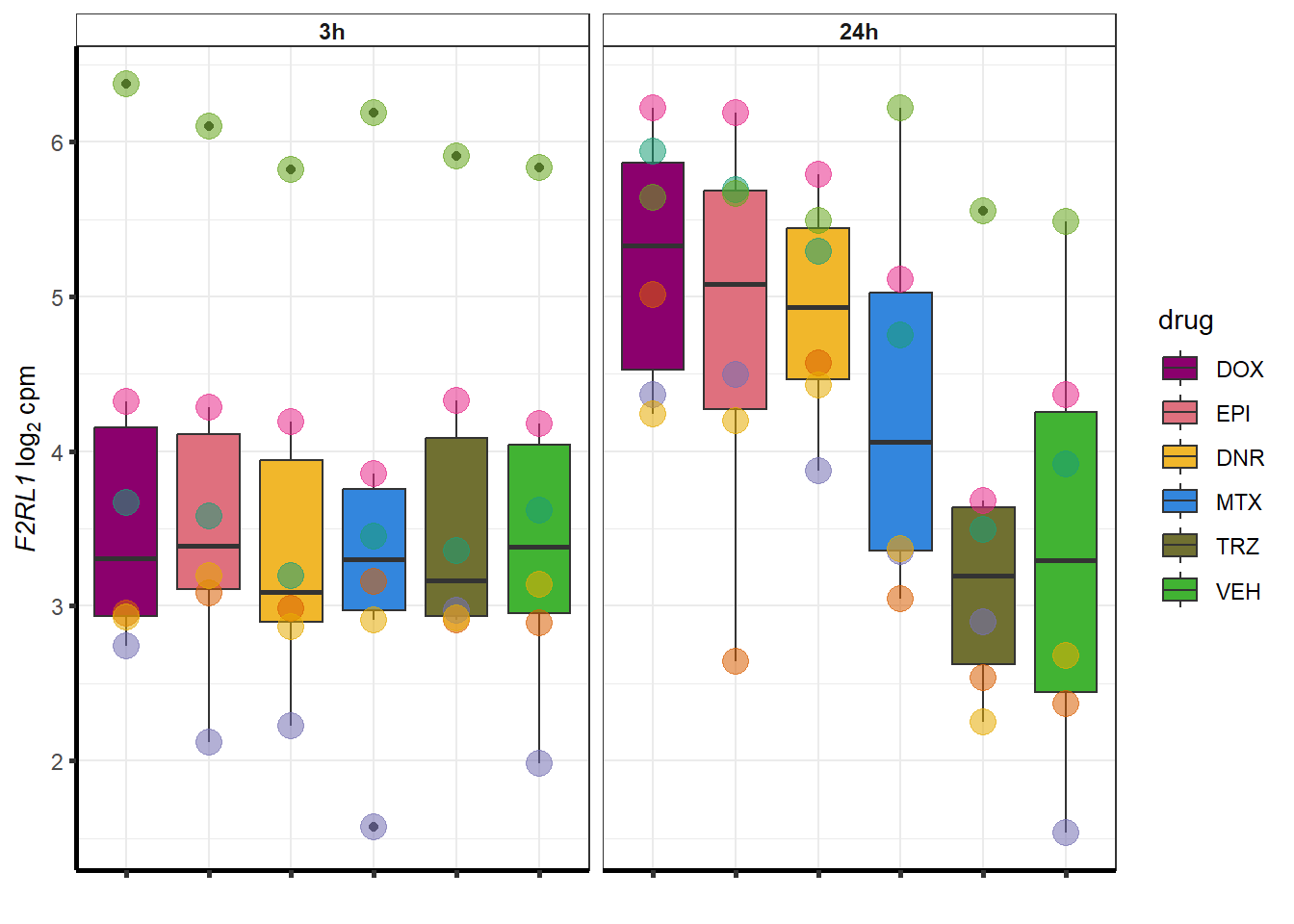
doxsponly <- Doxonly_deg %>%
distinct(entrezgene_id,.keep_all = TRUE) %>%
dplyr::filter(entrezgene_id %in%Dox_eGene_sp)
for (g in seq(from=1, to=length(doxsponly$entrezgene_id))){
a <- doxsponly$hgnc_symbol[g]
cpm_boxplot(cpmcounts,GOI=doxsponly[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
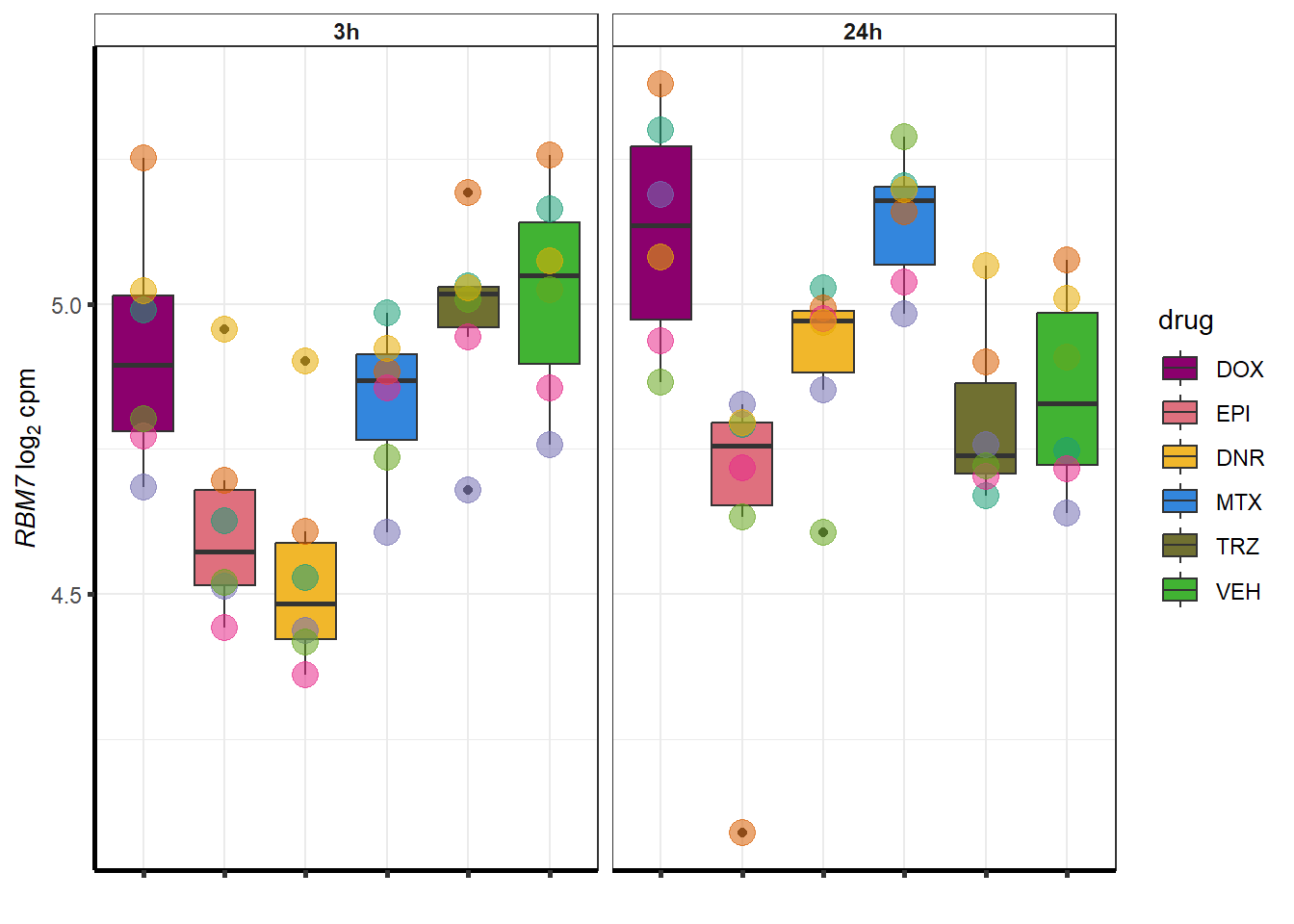
}
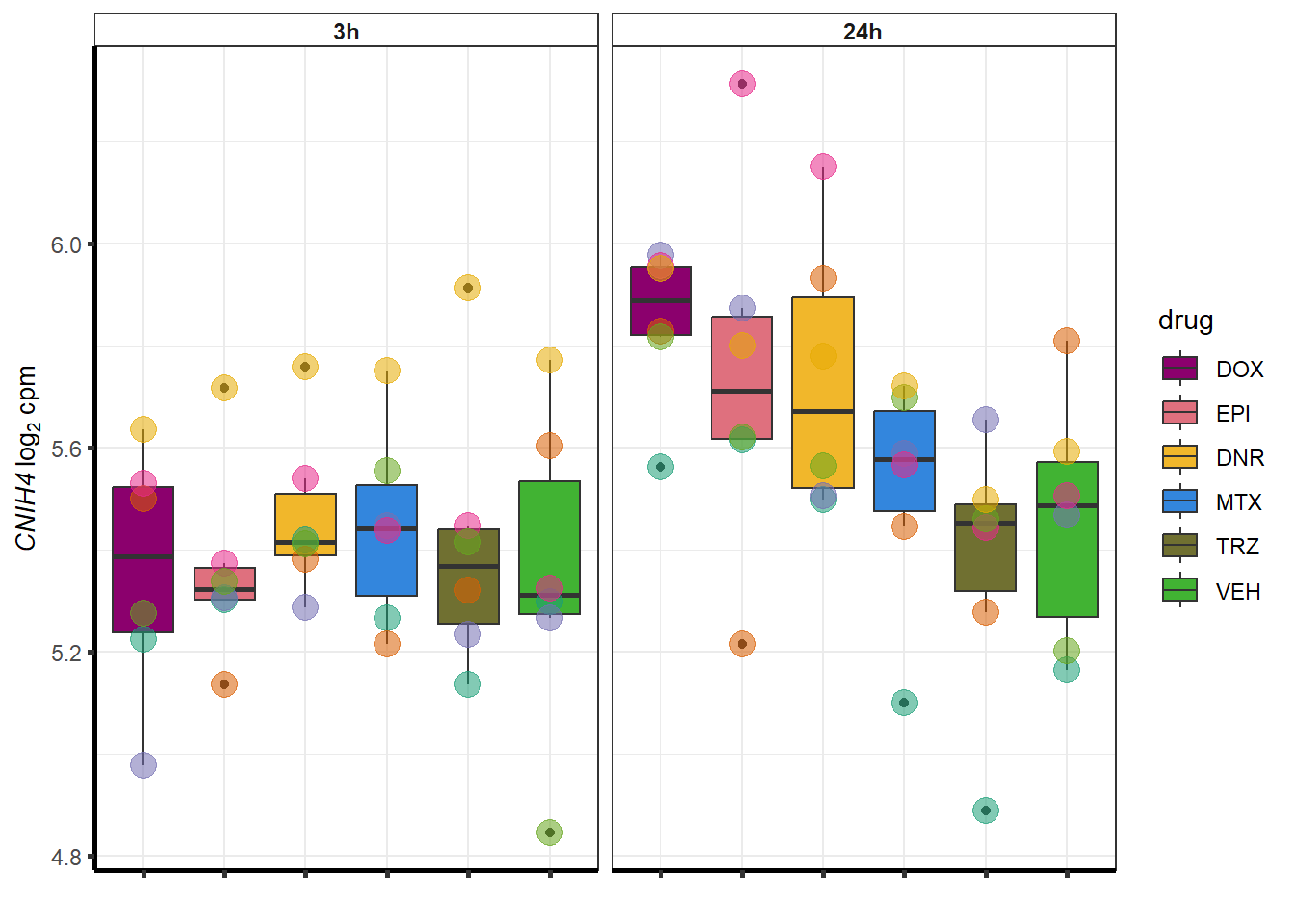
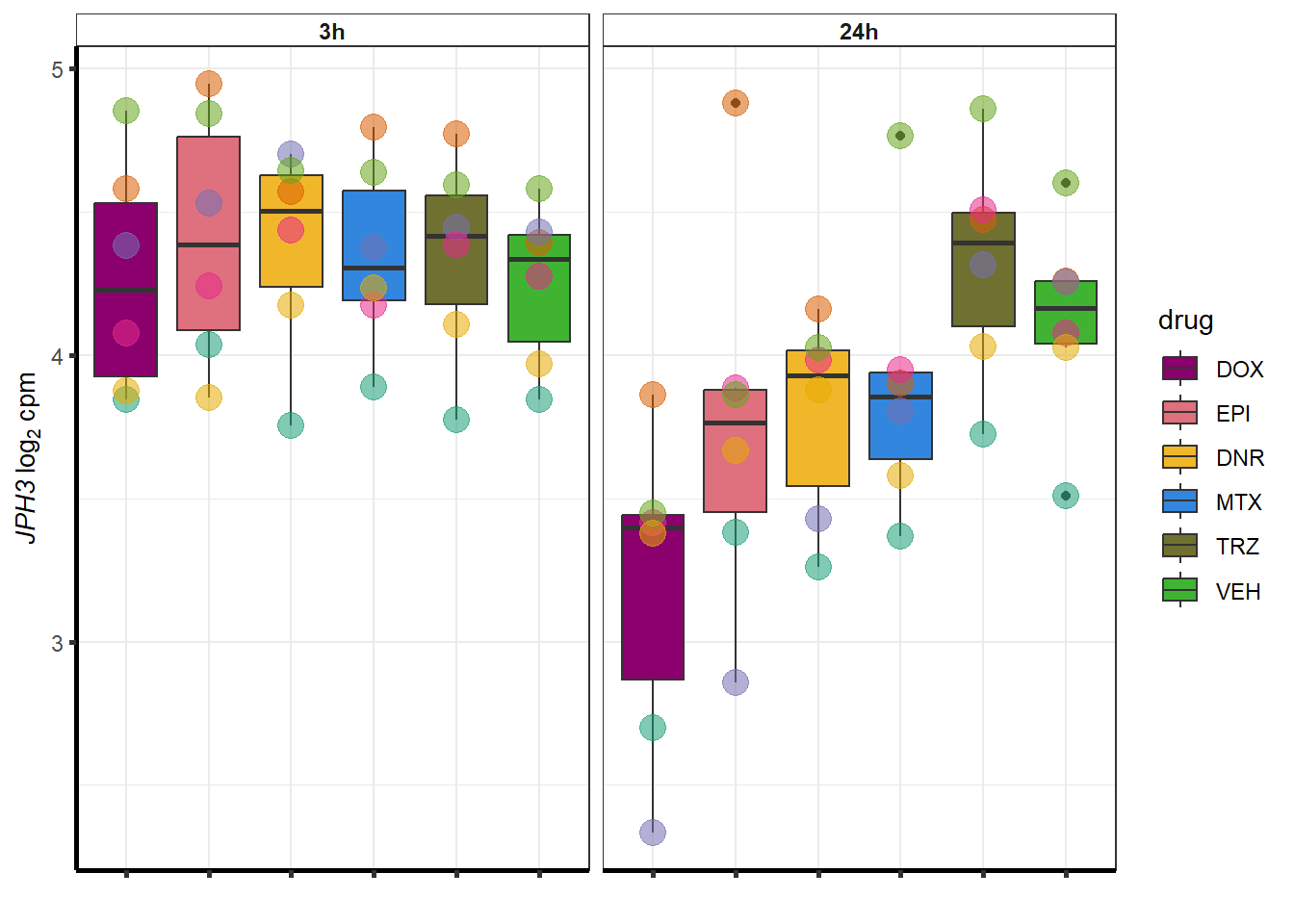
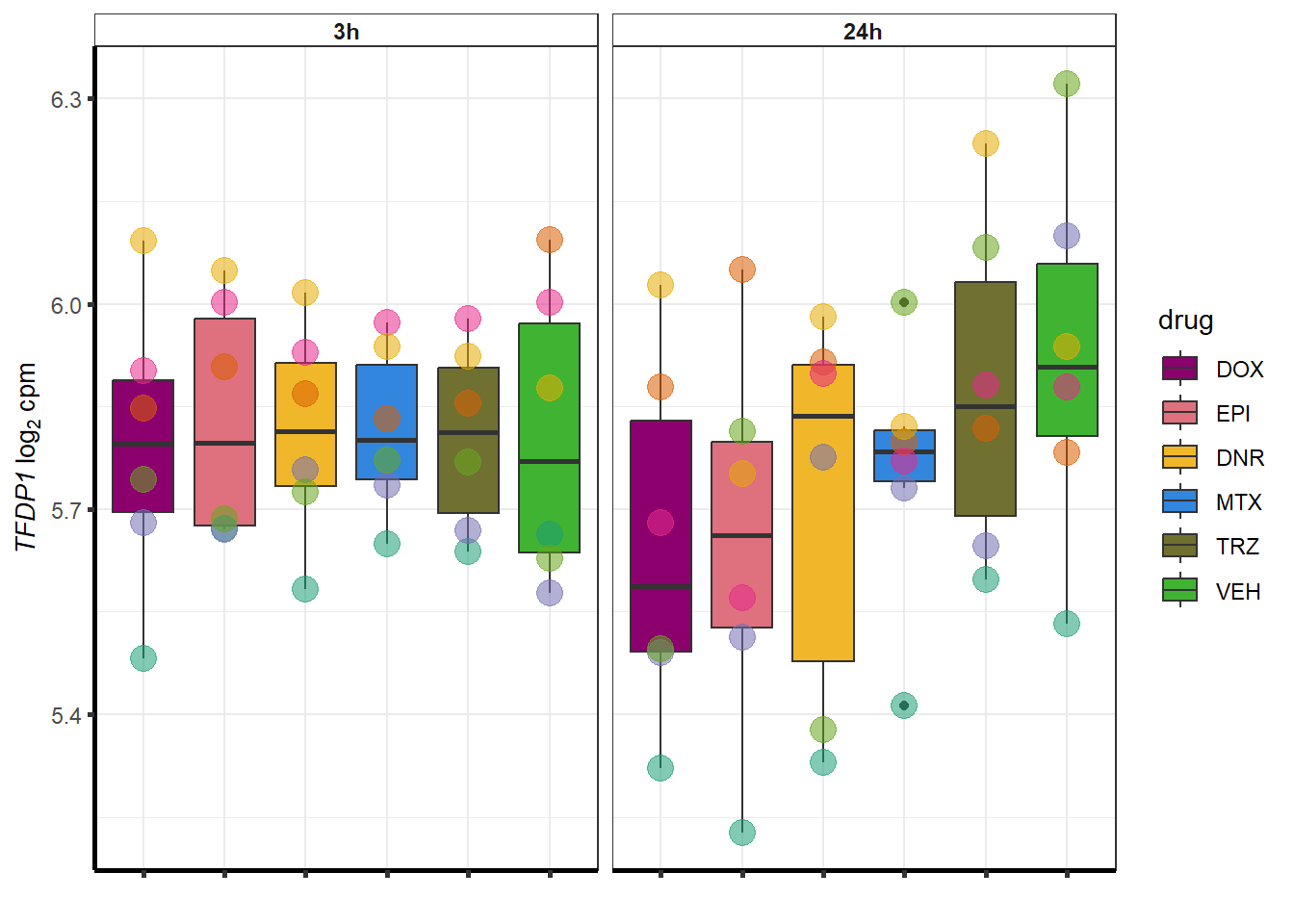
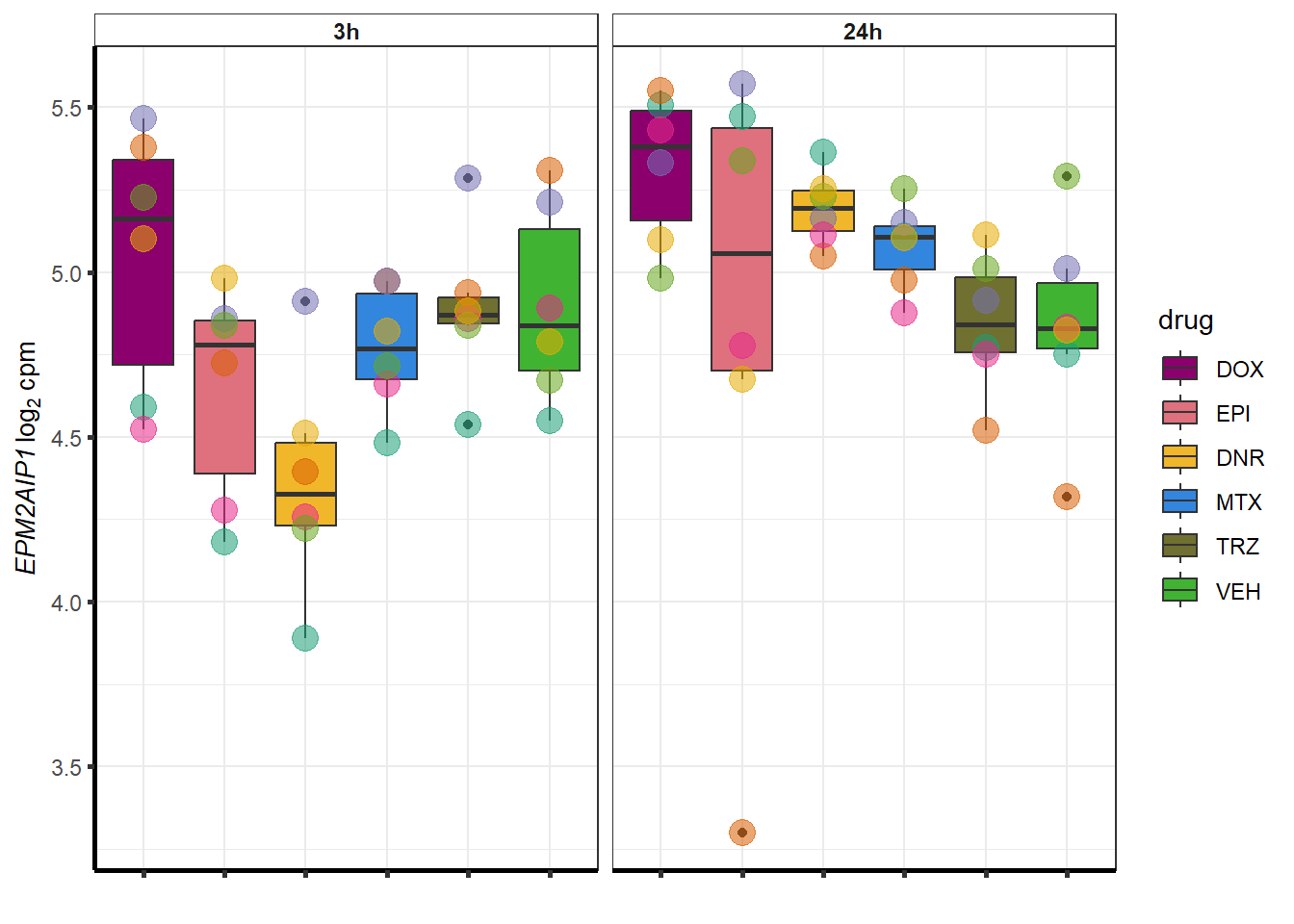
More investigation of 24 hour specific genes
DOX
DOXdeg_sp <- toplist24hr %>%
dplyr::filter(ENTREZID %in% DoxonlyDEG) %>%
dplyr::filter(adj.P.Val<0.01) %>%
filter(id=="DOX") %>%
dplyr::select(ENTREZID, SYMBOL)
DOXdeg_sp %>%
kable(., caption= "68 DOX specific genes") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box( height = "500px")| ENTREZID | SYMBOL | |
|---|---|---|
| 169200 | 169200 | TMEM64 |
| 1452 | 1452 | CSNK1A1 |
| 653082 | 653082 | ZDHHC11B |
| 202181 | 202181 | LOC202181 |
| 8501 | 8501 | SLC43A1 |
| 114882 | 114882 | OSBPL8 |
| 23108 | 23108 | RAP1GAP2 |
| 135154 | 135154 | SDHAF4 |
| 5000 | 5000 | ORC4 |
| 2130 | 2130 | EWSR1 |
| 122553 | 122553 | TRAPPC6B |
| 54165 | 54165 | DCUN1D1 |
| 57862 | 57862 | ZNF410 |
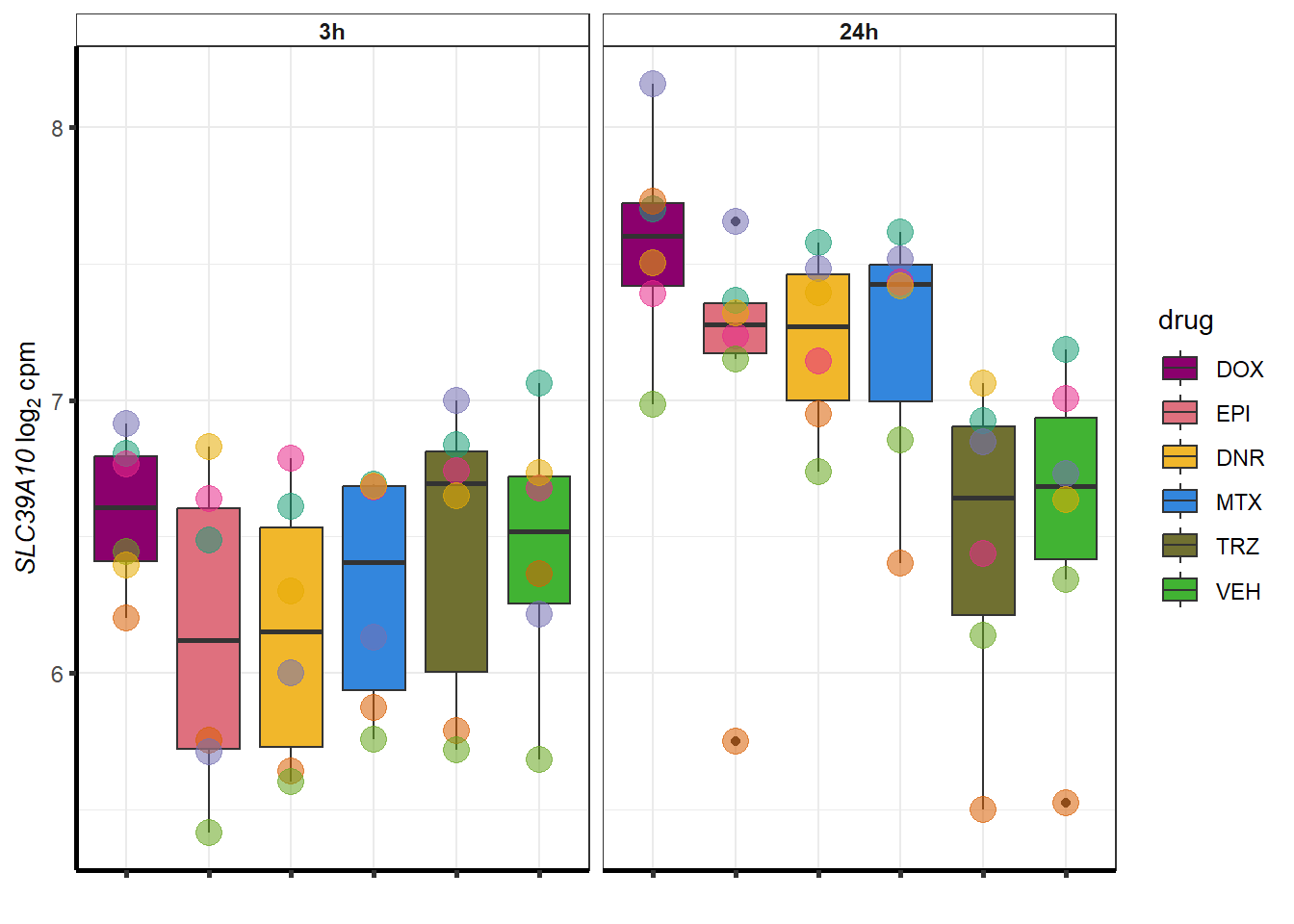
| 57181 | 57181 | SLC39A10 |
| 112858 | 112858 | TP53RK |
| 79065 | 79065 | ATG9A |
| 8515 | 8515 | ITGA10 |
| 25842 | 25842 | ASF1A |
| 284900 | 284900 | TTC28-AS1 |
| 2230 | 2230 | FDX1 |
| 9524 | 9524 | TECR |
| 256586 | 256586 | LYSMD2 |
| 57338 | 57338 | JPH3 |
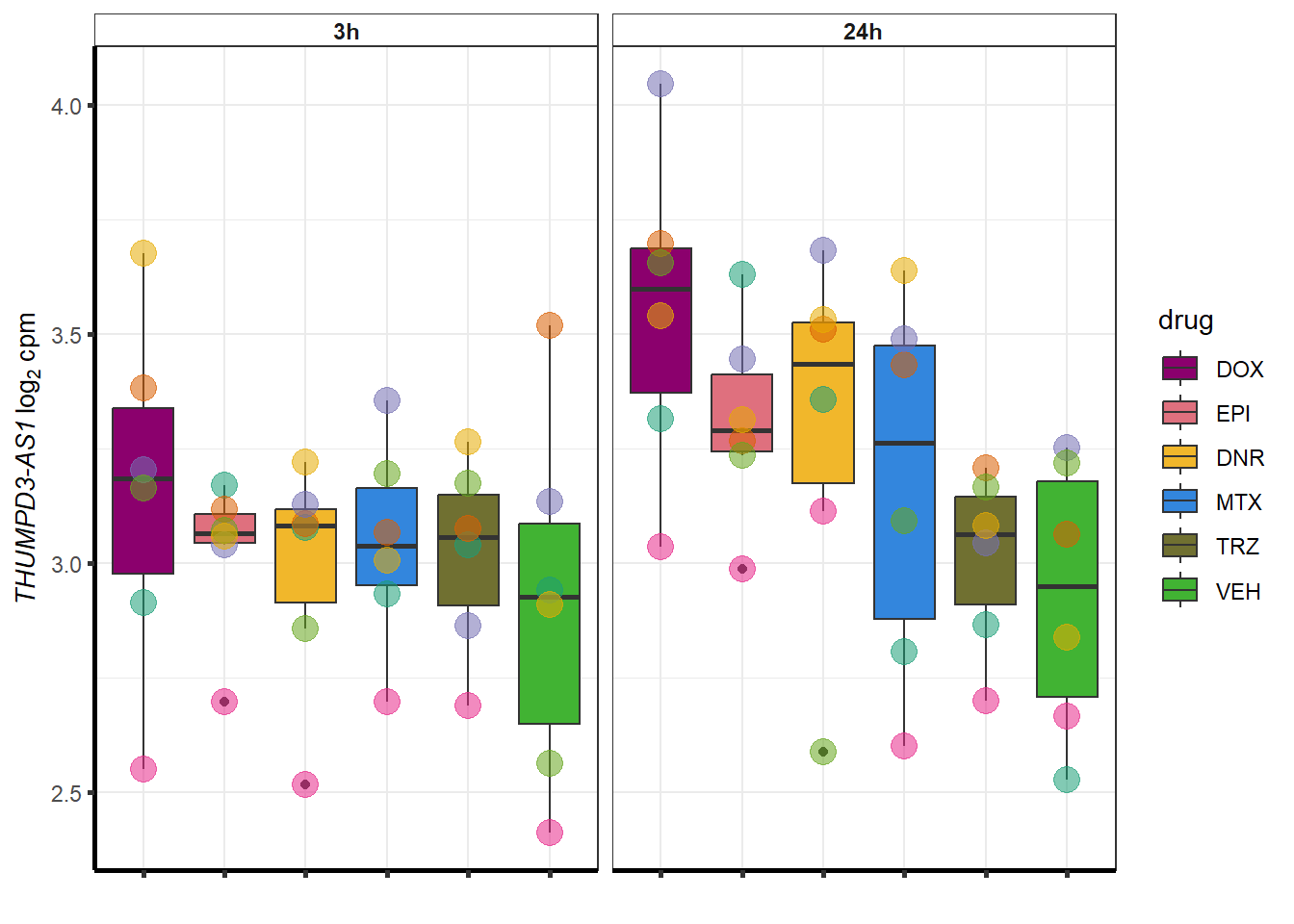
| 440944 | 440944 | THUMPD3-AS1 |
| 10190 | 10190 | TXNDC9 |
| 805 | 805 | CALM2 |
| 100131211 | 100131211 | NEMP2 |
| 4987 | 4987 | OPRL1 |
| 79624 | 79624 | ARMT1 |
| 23548 | 23548 | TTC33 |
| 27430 | 27430 | MAT2B |
| 81532 | 81532 | MOB2 |
| 158234 | 158234 | TRMT10B |
| 147 | 147 | ADRA1B |
| 26001 | 26001 | RNF167 |
| 57186 | 57186 | RALGAPA2 |
| 5431 | 5431 | POLR2B |
| 8763 | 8763 | CD164 |
| 55234 | 55234 | SMU1 |
| 84752 | 84752 | B3GNT9 |
| 375056 | 375056 | MIA3 |
| 7917 | 7917 | BAG6 |
| 54629 | 54629 | MINDY2 |
| 102723508 | 102723508 | KANTR |
| 26994 | 26994 | RNF11 |
| 29944 | 29944 | PNMA3 |
| 9446 | 9446 | GSTO1 |
| 7881 | 7881 | KCNAB1 |
| 6139 | 6139 | RPL17 |
| 100133331 | 100133331 | NA |
| 101927720 | 101927720 | ZNF793-AS1 |
| 6653 | 6653 | SORL1 |
| 5966 | 5966 | REL |
| 5411 | 5411 | PNN |
| 6498 | 6498 | SKIL |
| 8218 | 8218 | CLTCL1 |
| 83695 | 83695 | RHNO1 |
| 1838 | 1838 | DTNB |
| 347918 | 347918 | EP400P1 |
| 9894 | 9894 | TELO2 |
| 10973 | 10973 | ASCC3 |
| 5598 | 5598 | MAPK7 |
| 8241 | 8241 | RBM10 |
| 27229 | 27229 | TUBGCP4 |
| 2281 | 2281 | FKBP1B |
| 116068 | 116068 | LYSMD3 |
| 56998 | 56998 | CTNNBIP1 |
| 2551 | 2551 | GABPA |
intersect(DOXreQTLs$ENTREZID, DOXdeg_sp$ENTREZID)[1] "57338"#still jph3
##lfc
toplist24hr %>%
group_by(time,id) %>%
dplyr::filter(ENTREZID %in% DOXdeg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
ggplot(., aes(x= id, y=logFC))+
geom_boxplot(aes(fill=id))+
theme_classic()+
fill_palette(palette = drug_palc)+
ggtitle("LogFC of n = 68 Dox specific DEGs")+
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5,face = "bold"),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle = 20))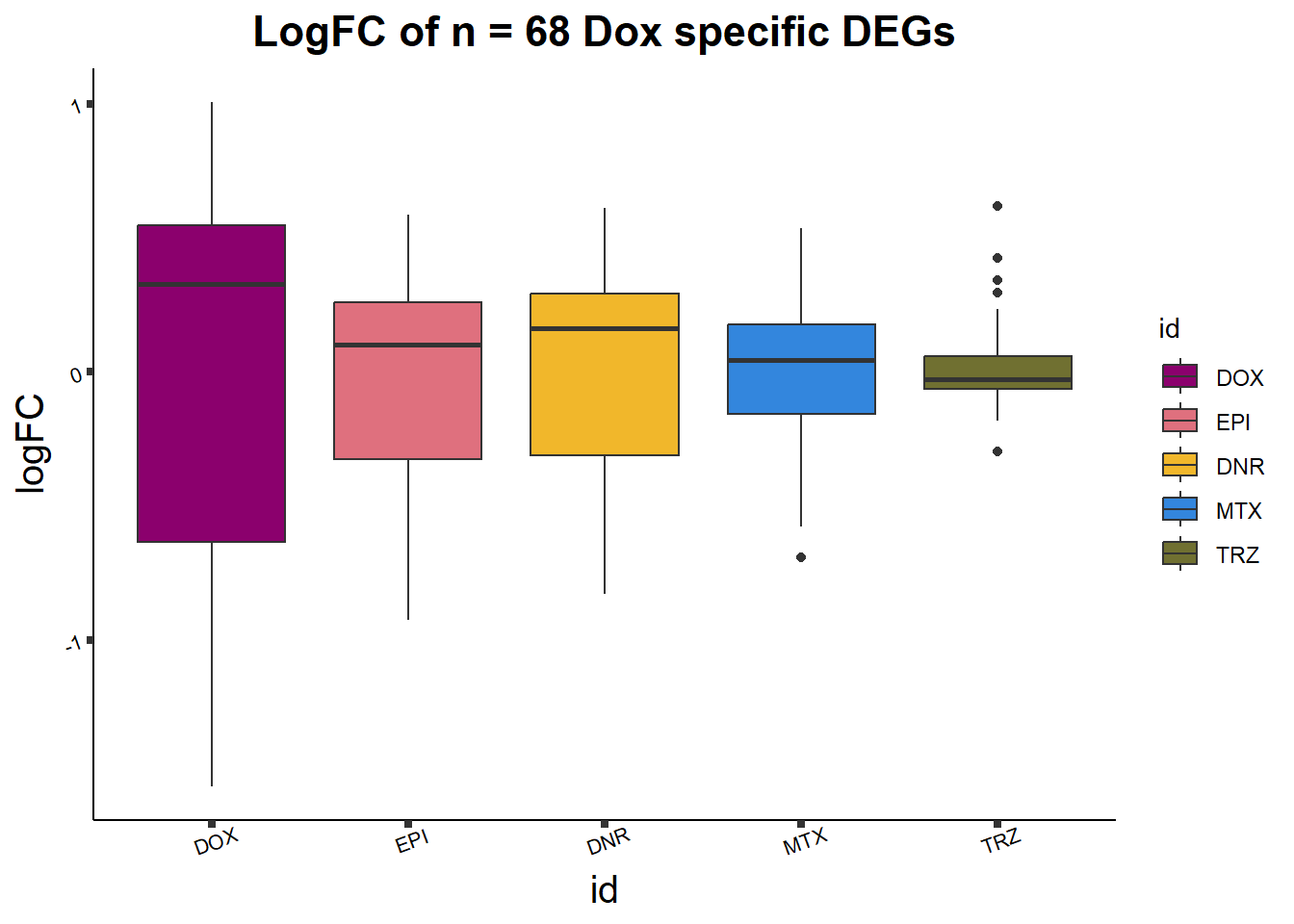
##histo check
toplist24hr %>%
dplyr::filter(ENTREZID %in% DOXdeg_sp$ENTREZID) %>%
dplyr::filter(adj.P.Val <0.05) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id))+
geom_vline(xintercept=0.01,linetype=2)+
# geom_histogram(aes(fill=id))+
facet_wrap(~id)+
ggtitle("all Dox specific DEG adj p. value <0.01")+
fill_palette(palette = drug_palc)+
theme_bw()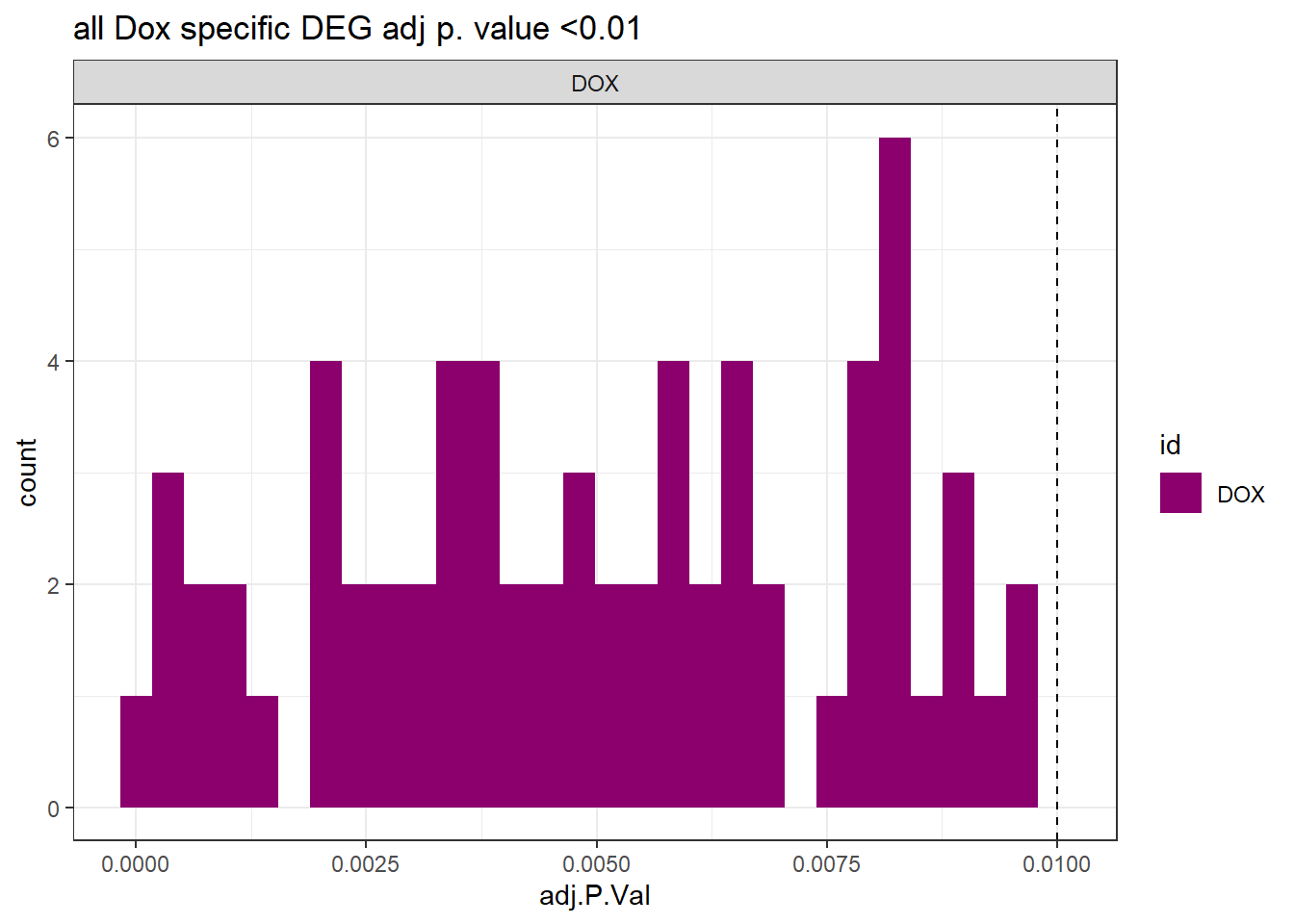
toplist24hr %>%
filter(ENTREZID %in% DoxonlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()
densityDOXsp <-
toplist24hr %>%
filter(ENTREZID %in% DoxonlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()
densityDOXsp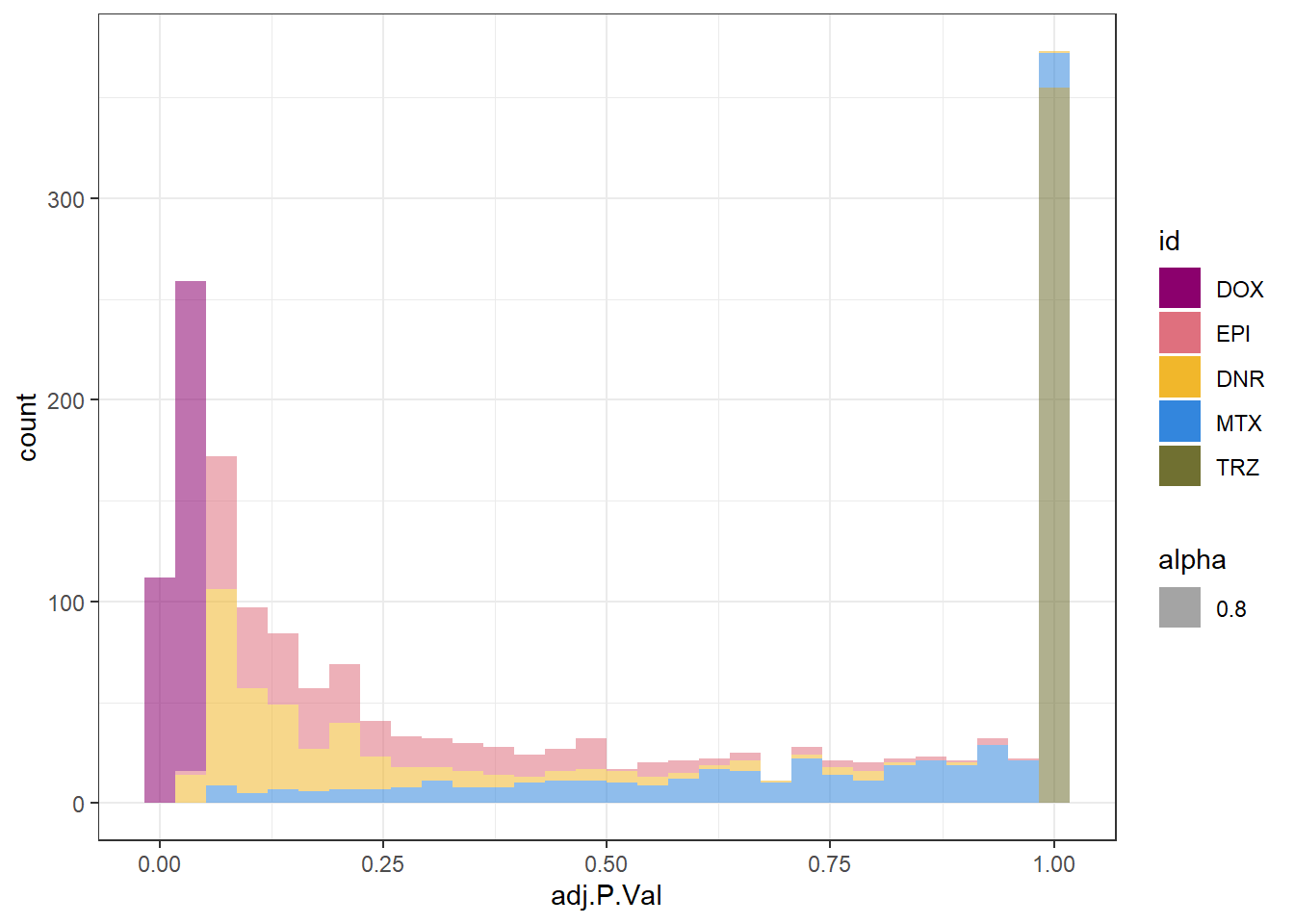
set.seed(12345)
sampset <- DOXdeg_sp %>%
sample_n(.,12)
for (g in seq(from=1, to=length(sampset$ENTREZID))){
a <- sampset$SYMBOL[g]
cpm_boxplot(cpmcounts,GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}
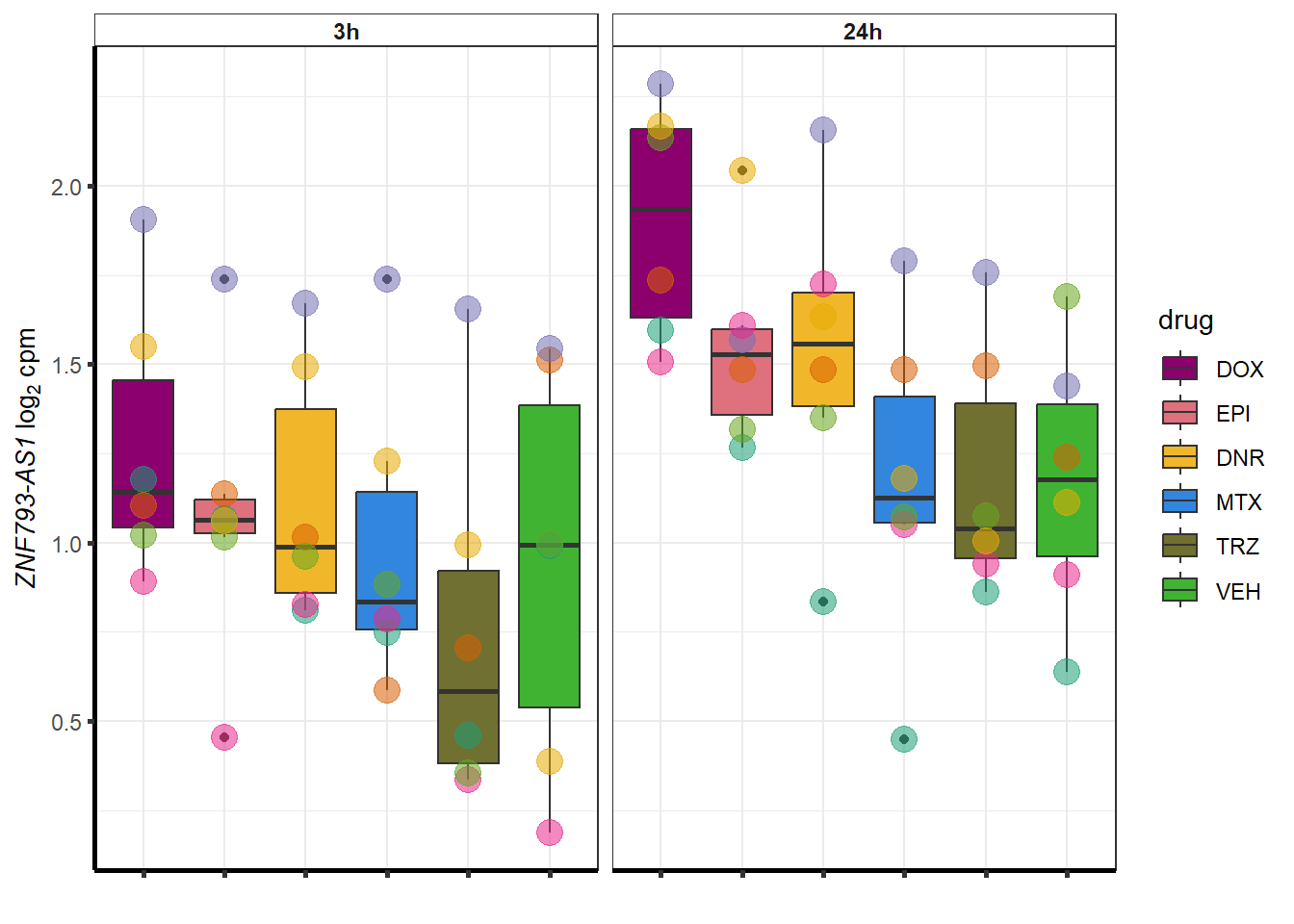
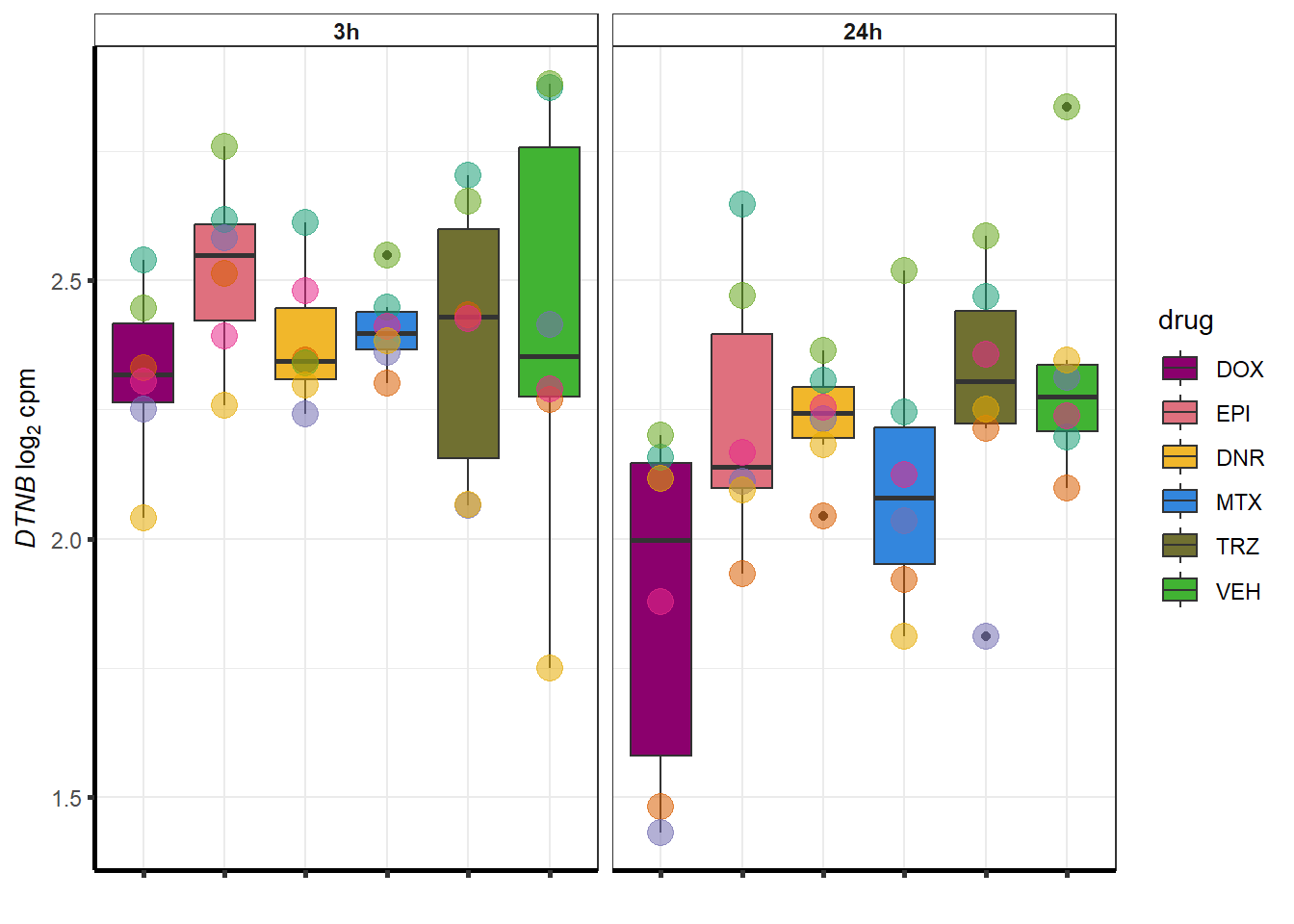
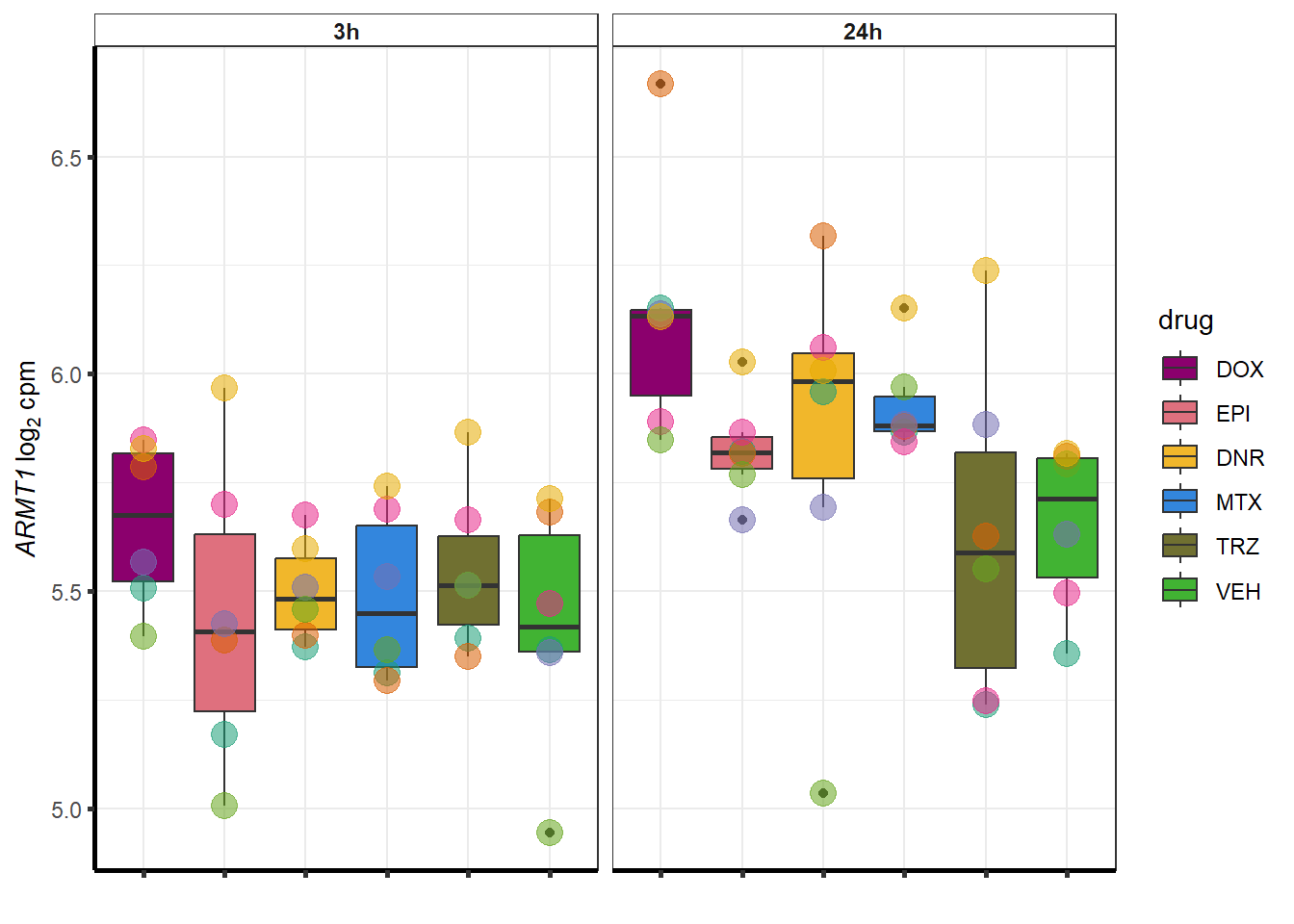
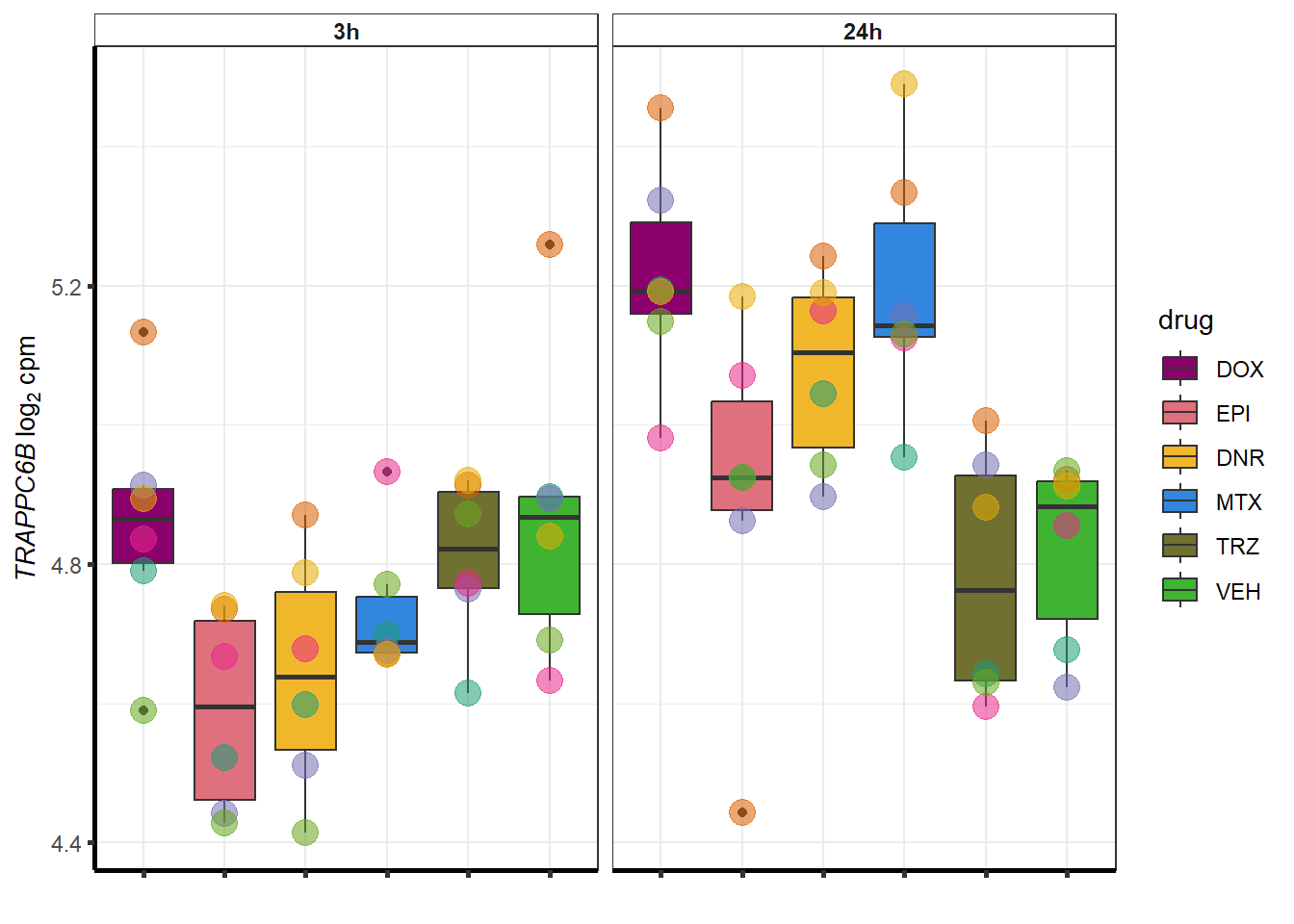
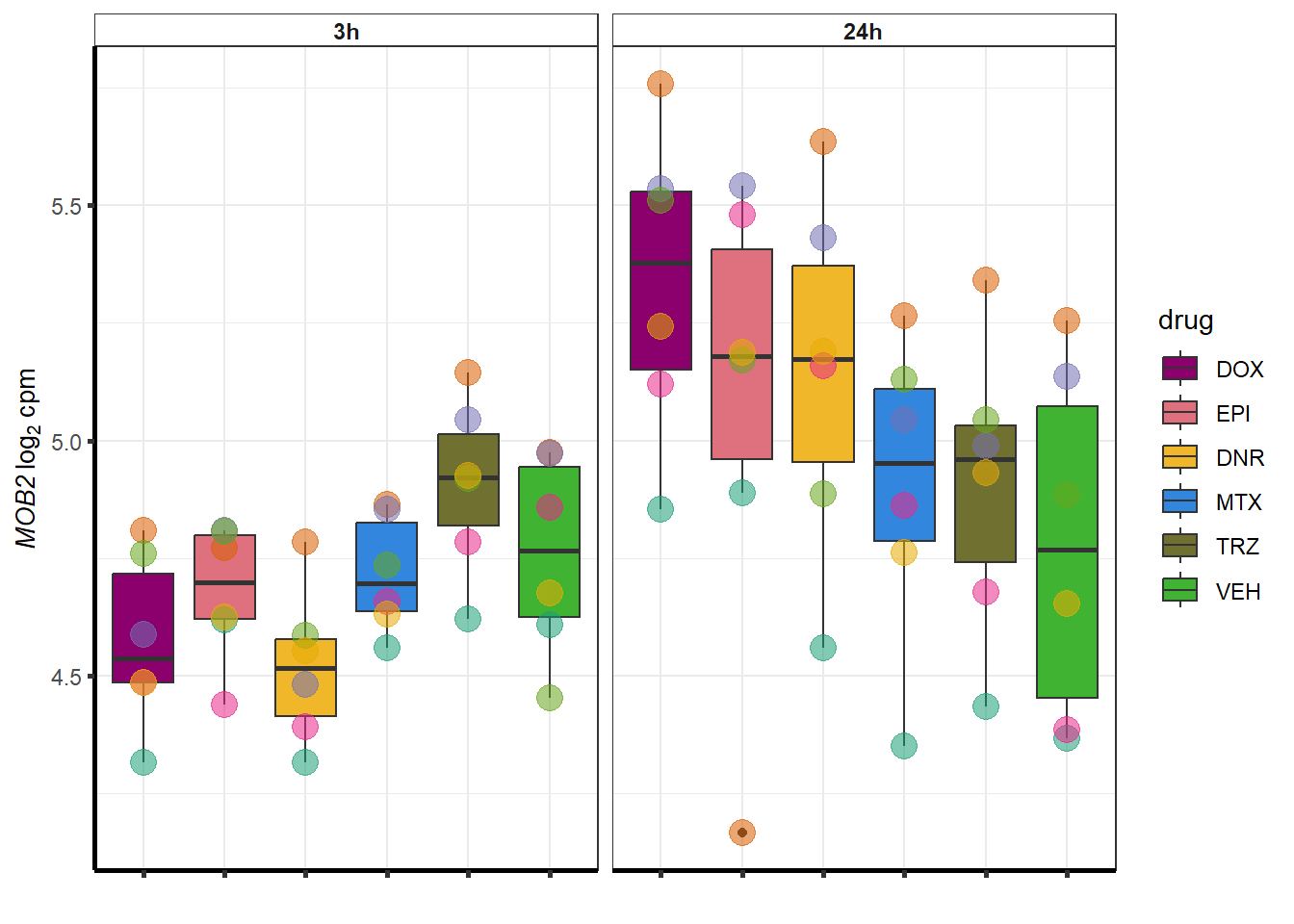
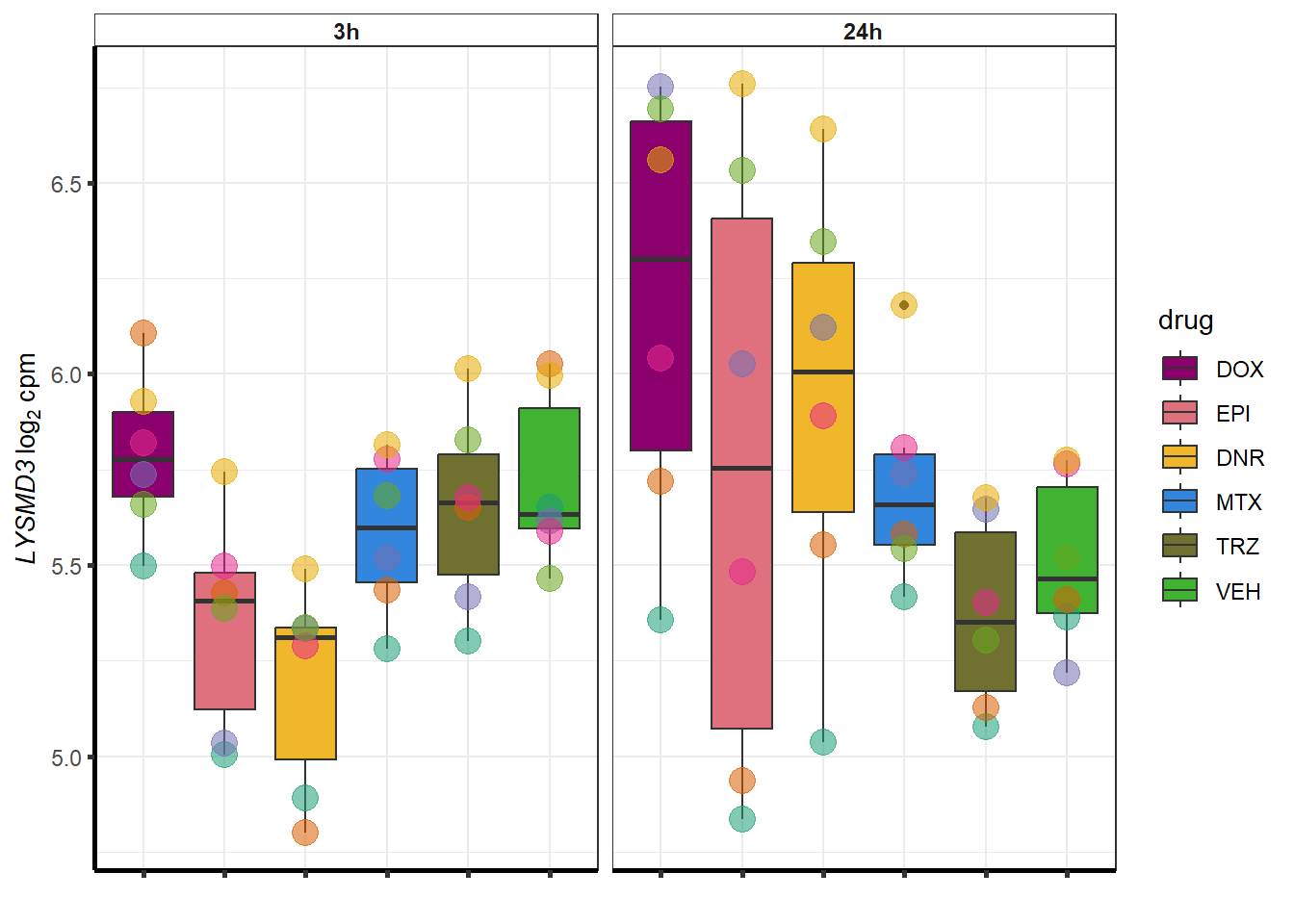
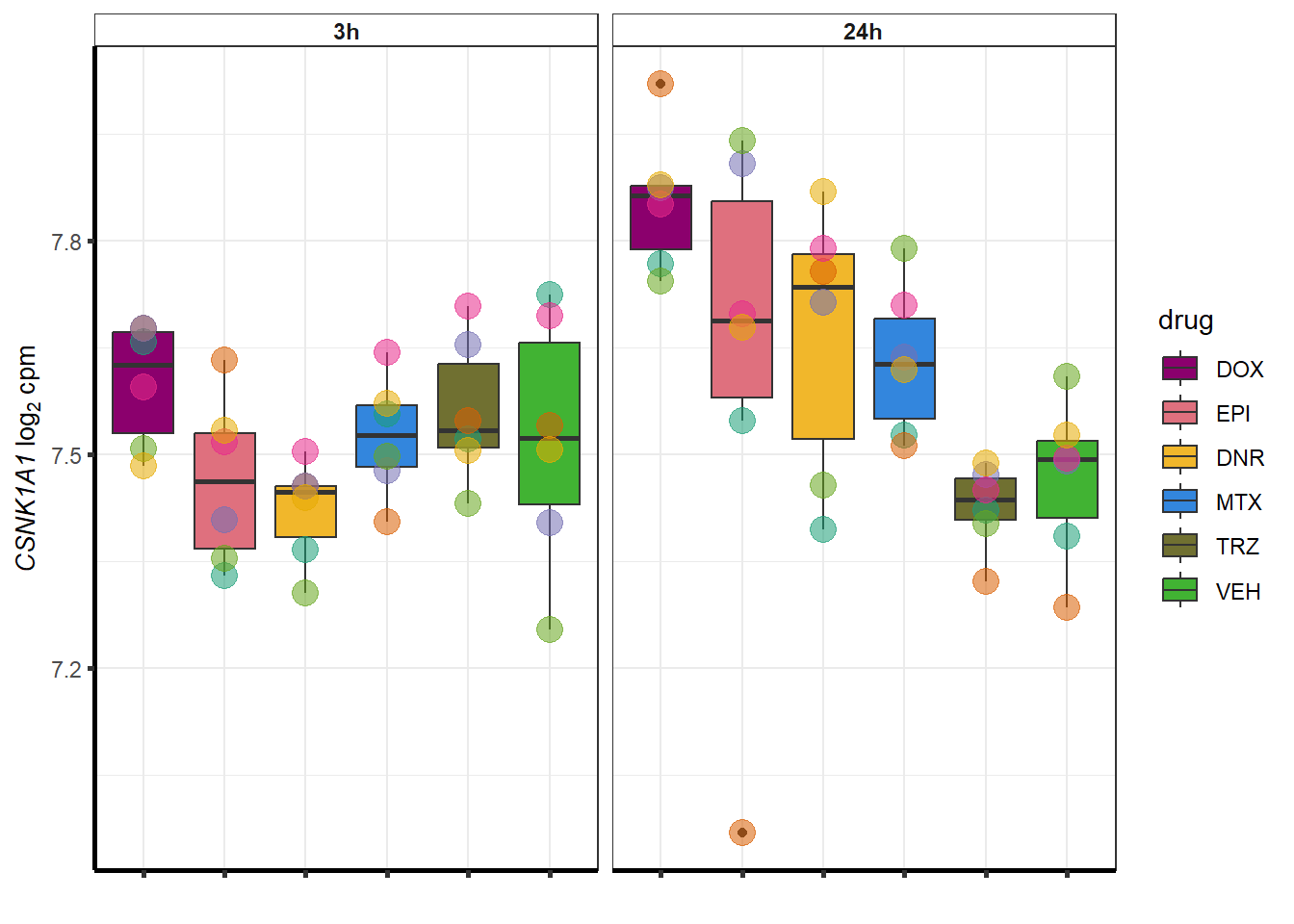
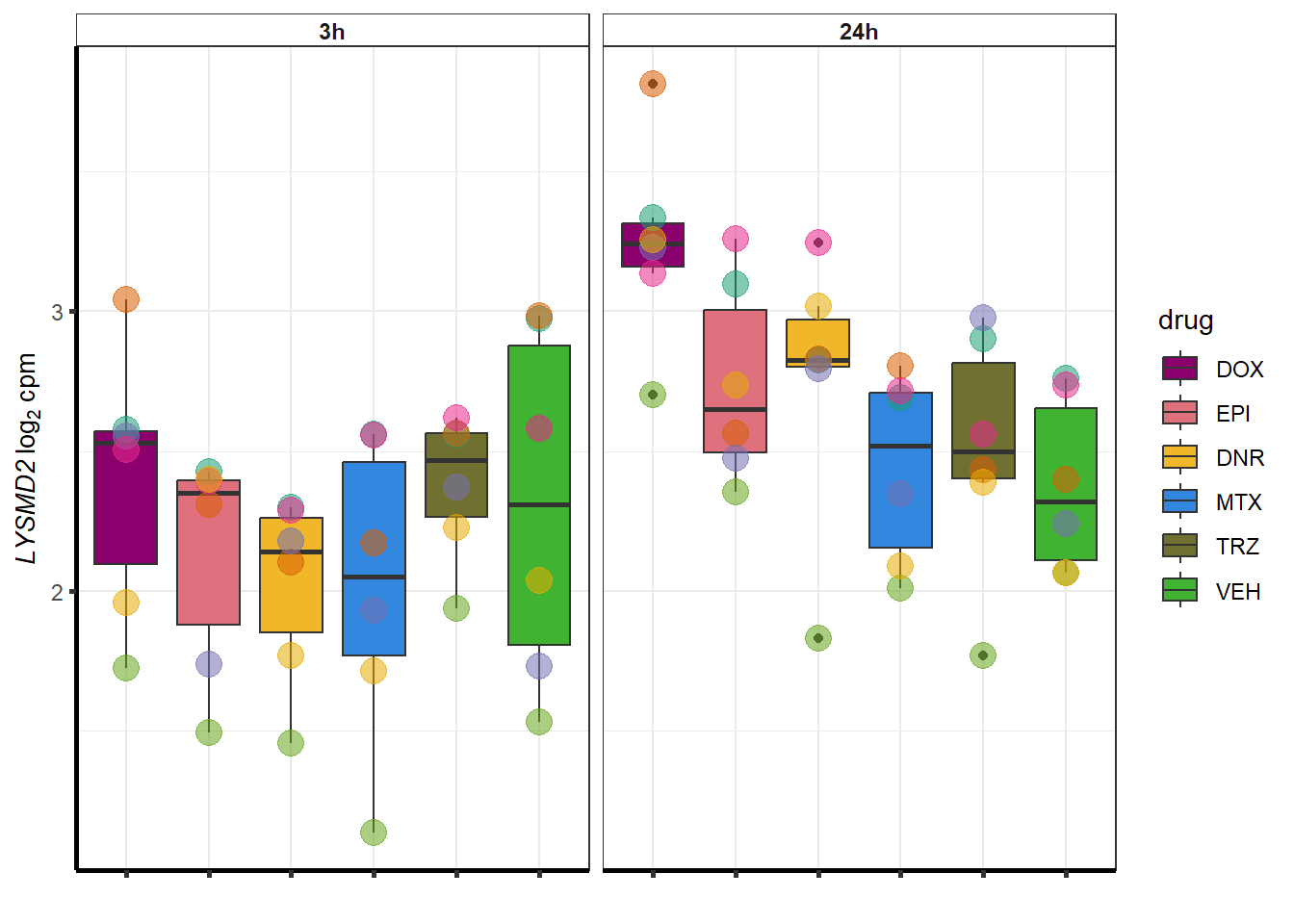
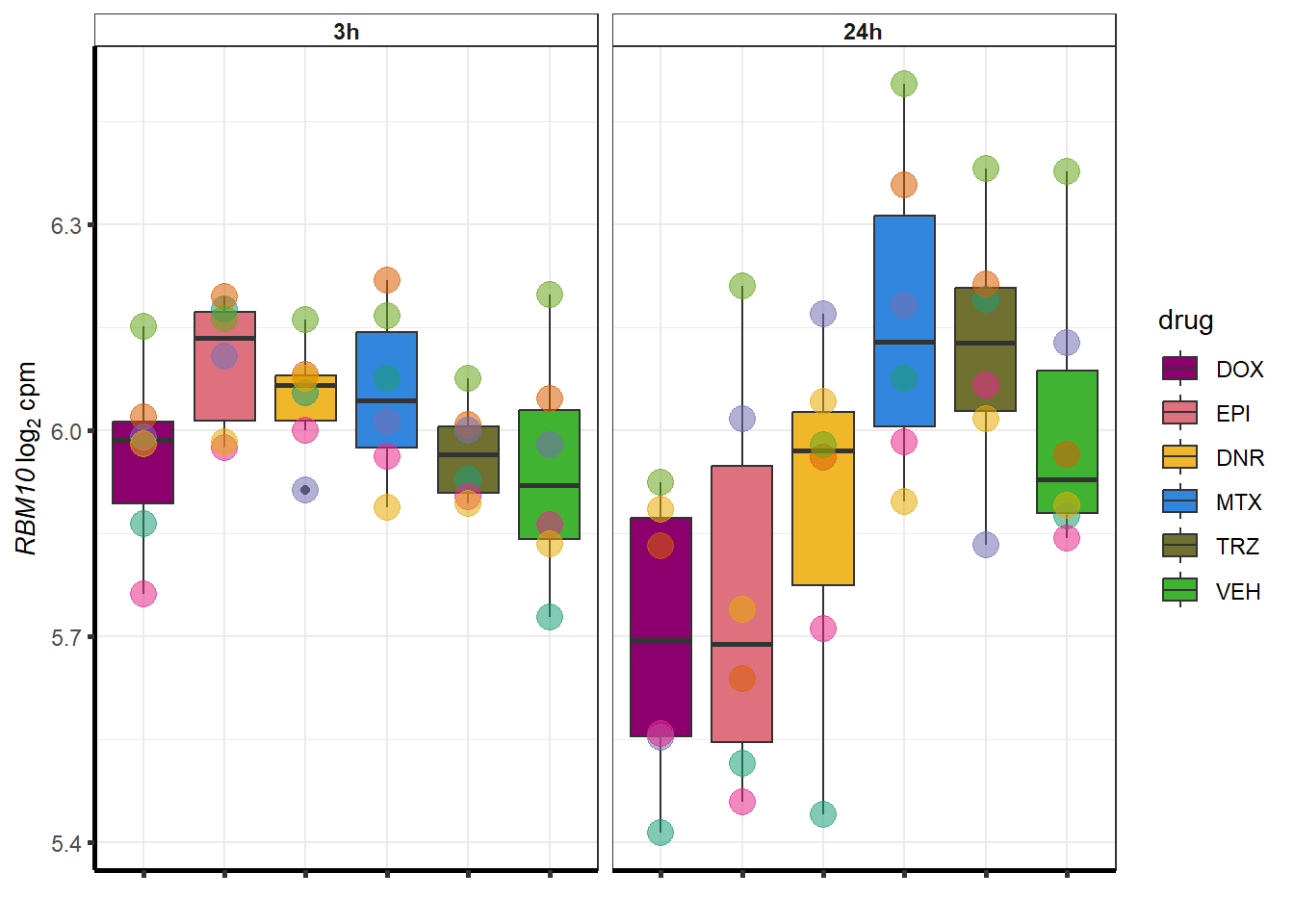
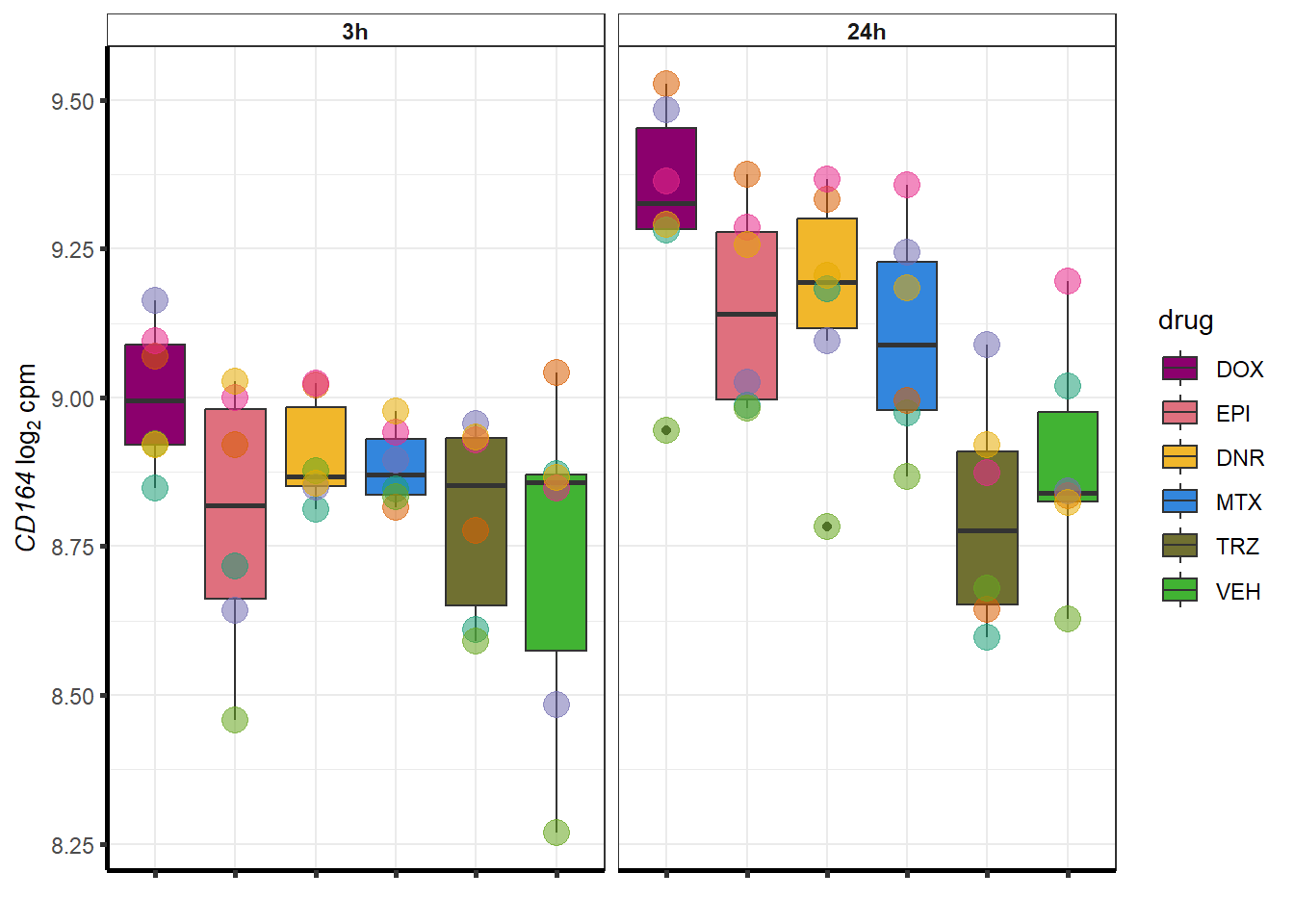
##most convincing of these is ZNF793-AS1 101927720MTX
MTXdeg_sp <- toplist24hr %>%
dplyr::filter(ENTREZID %in% MtxonlyDEG) %>%
dplyr::filter(adj.P.Val<0.01) %>%
filter(id=="MTX") %>%
dplyr::select(ENTREZID, SYMBOL)
MTXdeg_sp %>%
kable(., caption= "MTX specific genes") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box( height = "500px")| ENTREZID | SYMBOL | |
|---|---|---|
| 25894 | 25894 | PLEKHG4 |
| 253714 | 253714 | MMS22L |
| 126432 | 126432 | RINL |
| 4001 | 4001 | LMNB1 |
| 6240 | 6240 | RRM1 |
| 83879 | 83879 | CDCA7 |
| 25886 | 25886 | POC1A |
| 100128191 | 100128191 | TMPO-AS1 |
| 84892 | 84892 | POMGNT2 |
| 27346 | 27346 | TMEM97 |
| 63827 | 63827 | BCAN |
| 55723 | 55723 | ASF1B |
| 4173 | 4173 | MCM4 |
| 54853 | 54853 | WDR55 |
| 8317 | 8317 | CDC7 |
| 57699 | 57699 | CPNE5 |
| 655 | 655 | BMP7 |
| 9401 | 9401 | RECQL4 |
| 126382 | 126382 | NR2C2AP |
| 79109 | 79109 | MAPKAP1 |
| 441478 | 441478 | NRARP |
| 22950 | 22950 | SLC4A1AP |
| 100996573 | 100996573 | NA |
| 55780 | 55780 | ERMARD |
| 3148 | 3148 | HMGB2 |
| 3983 | 3983 | ABLIM1 |
| 9134 | 9134 | CCNE2 |
| 25981 | 25981 | DNAH1 |
| 9811 | 9811 | CTIF |
| 56970 | 56970 | ATXN7L3 |
| 79758 | 79758 | DHRS12 |
| 55147 | 55147 | RBM23 |
| 65057 | 65057 | ACD |
| 645954 | 645954 | SVIL2P |
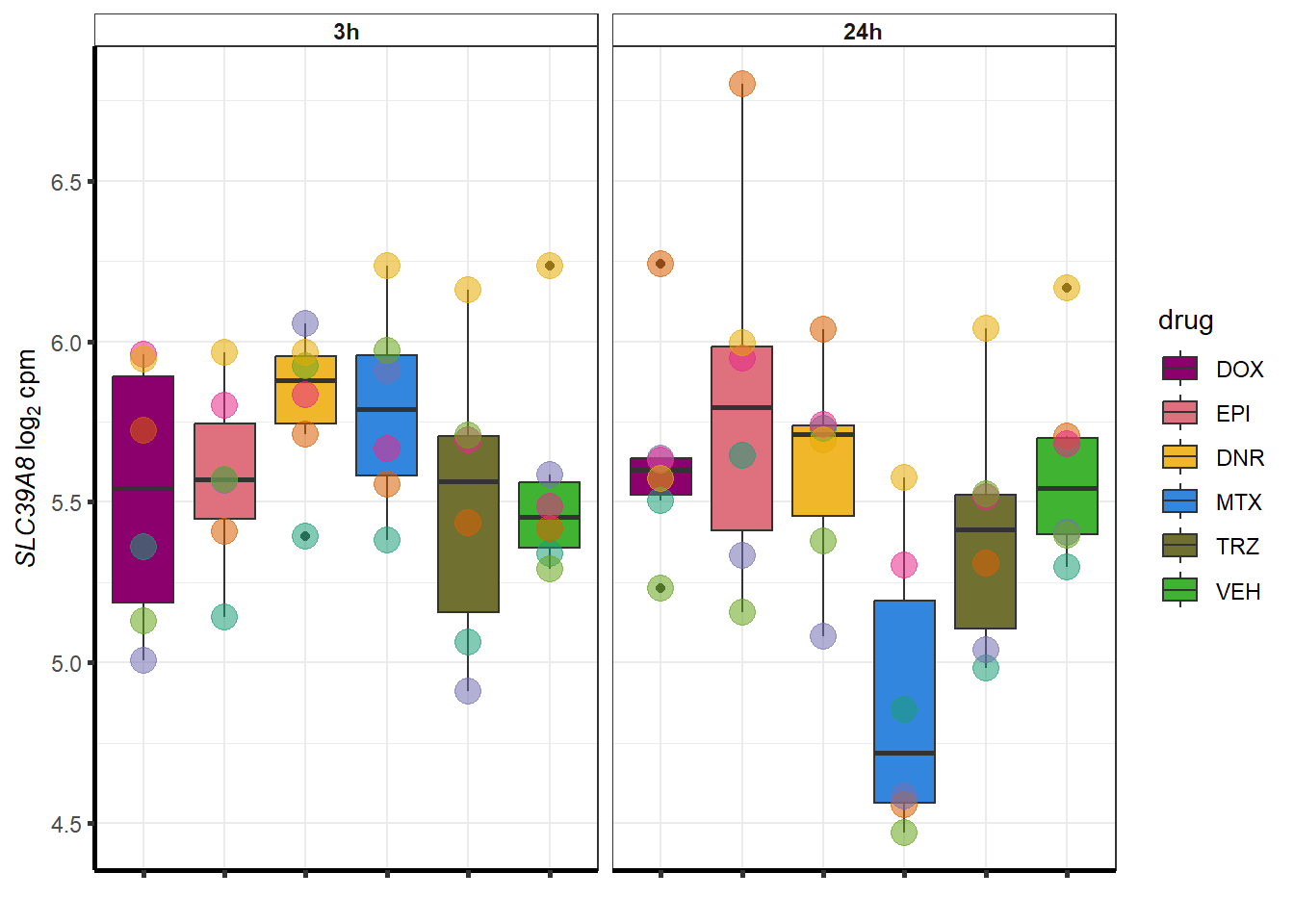
| 64116 | 64116 | SLC39A8 |
| 147645 | 147645 | VSIG10L |
| 23582 | 23582 | CCNDBP1 |
| 8321 | 8321 | FZD1 |
| 10432 | 10432 | RBM14 |
| 9820 | 9820 | CUL7 |
| 55706 | 55706 | NDC1 |
#intersect(DOXreQTLs$ENTREZID, MTXdeg_sp$ENTREZID)#none
##lfc
toplist24hr %>%
group_by(time,id) %>%
filter(ENTREZID %in% MtxonlyDEG) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment"=id)%>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
theme_classic()+
fill_palette(palette = drug_palc)+
ggtitle("LogFC of MTX specific DEGs (n = 48)")+
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5,face = "bold"),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle = 0))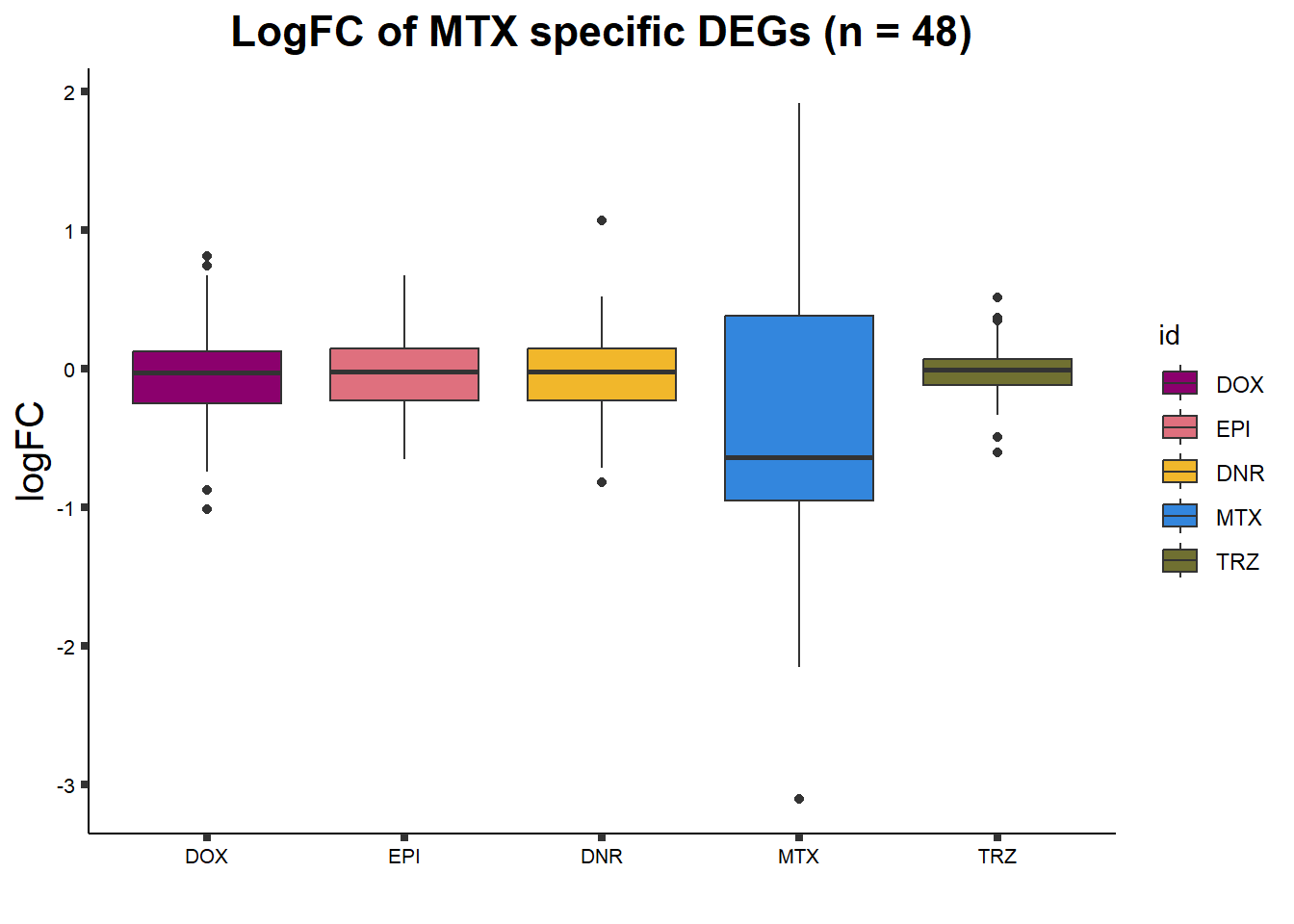
##histo check
toplist24hr %>%
dplyr::filter(ENTREZID %in% MTXdeg_sp$ENTREZID) %>%
dplyr::filter(adj.P.Val <0.05) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id))+
geom_vline(xintercept=0.01,linetype=2)+
# geom_histogram(aes(fill=id))+
facet_wrap(~id)+
ggtitle("all MTX specific DEG adj p. value <0.01")+
fill_palette(palette = drug_palc)+
theme_bw()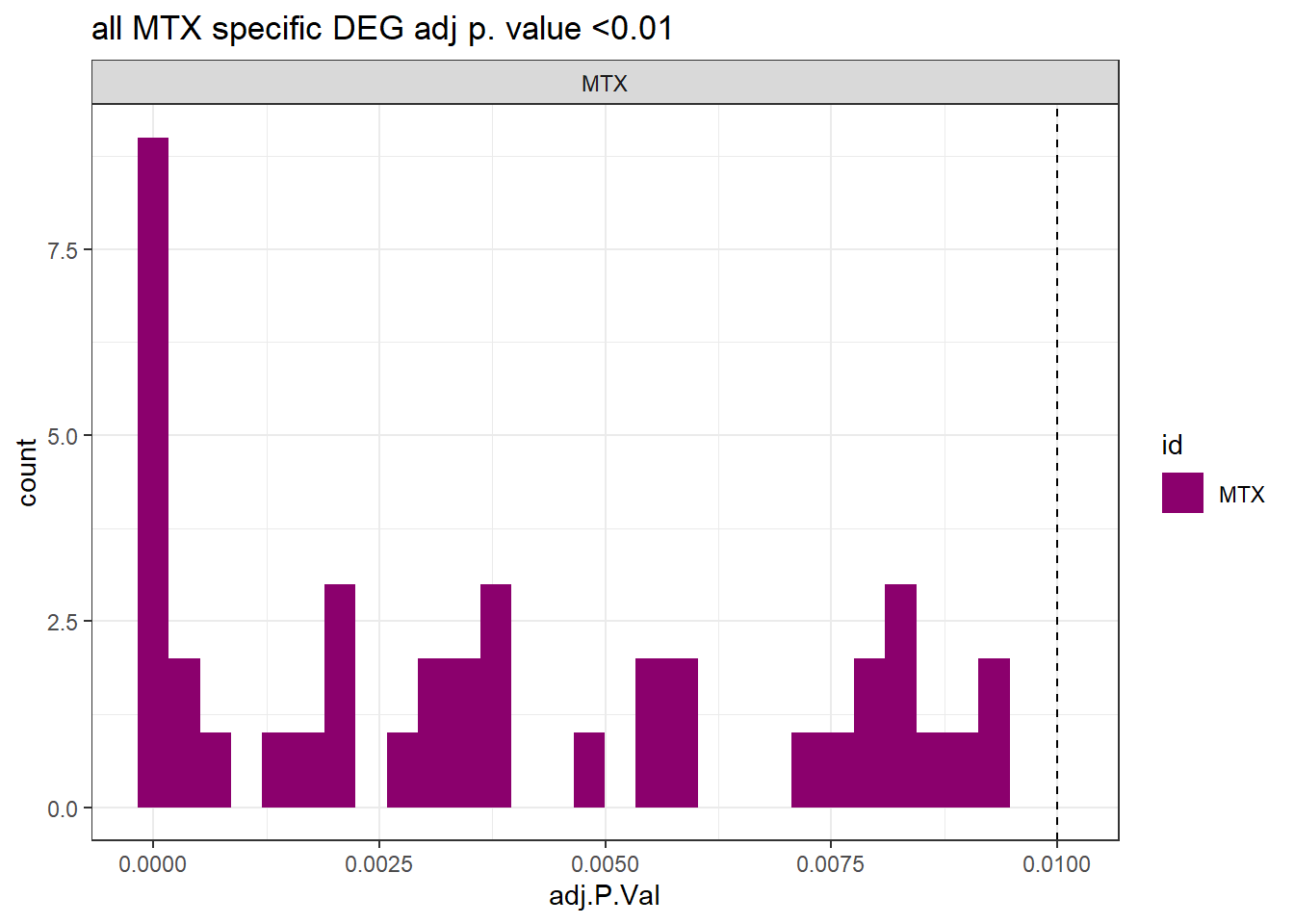
toplist24hr %>%
filter(ENTREZID %in% MtxonlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()
densityMTXsp <- toplist24hr %>%
filter(ENTREZID %in% MtxonlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()
densityMTXsp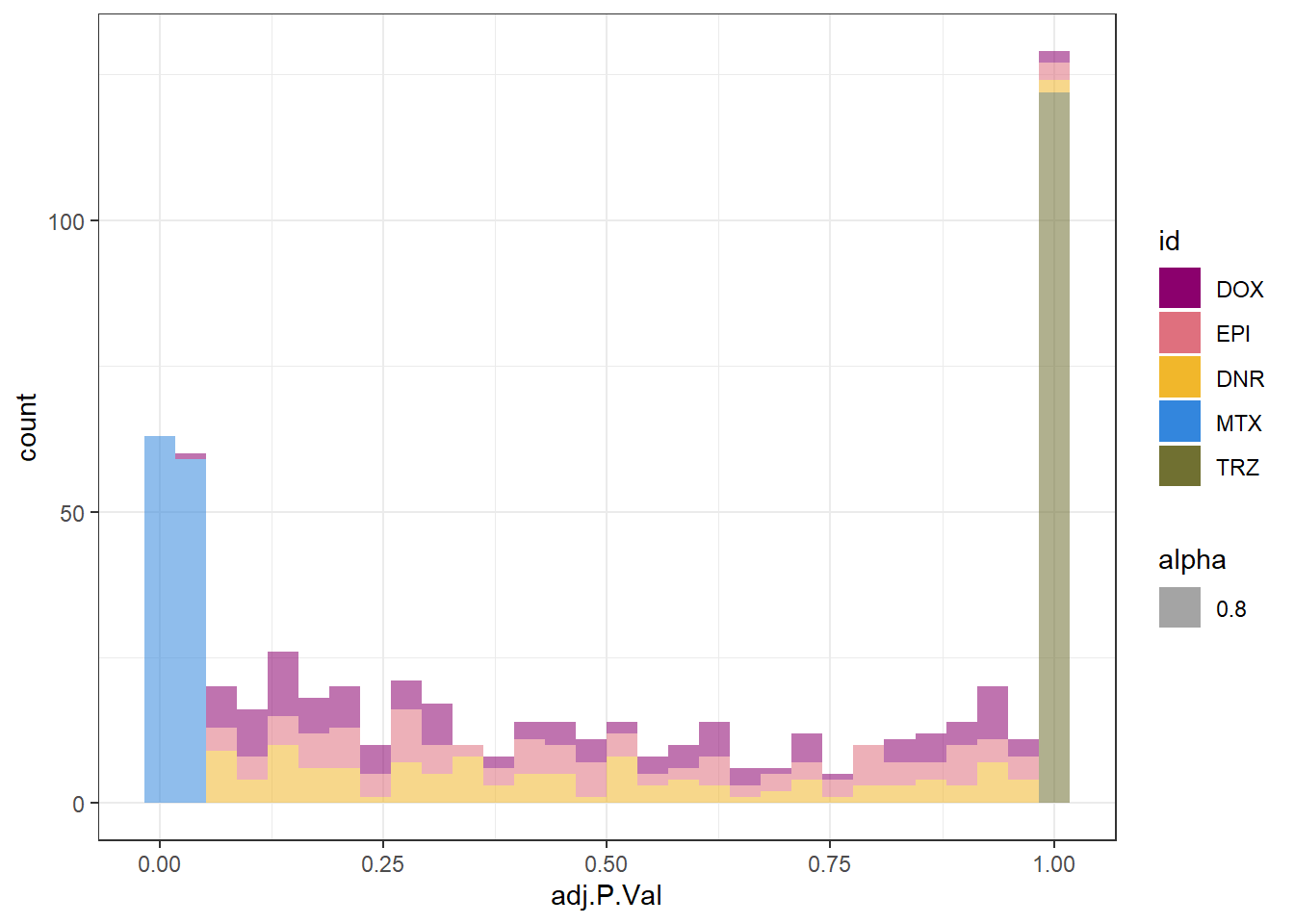
set.seed(12345)
sampset <- MTXdeg_sp %>%
sample_n(.,12)
for (g in seq(from=1, to=length(sampset$ENTREZID))){
a <- sampset$SYMBOL[g]
cpm_boxplot(cpmcounts,GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}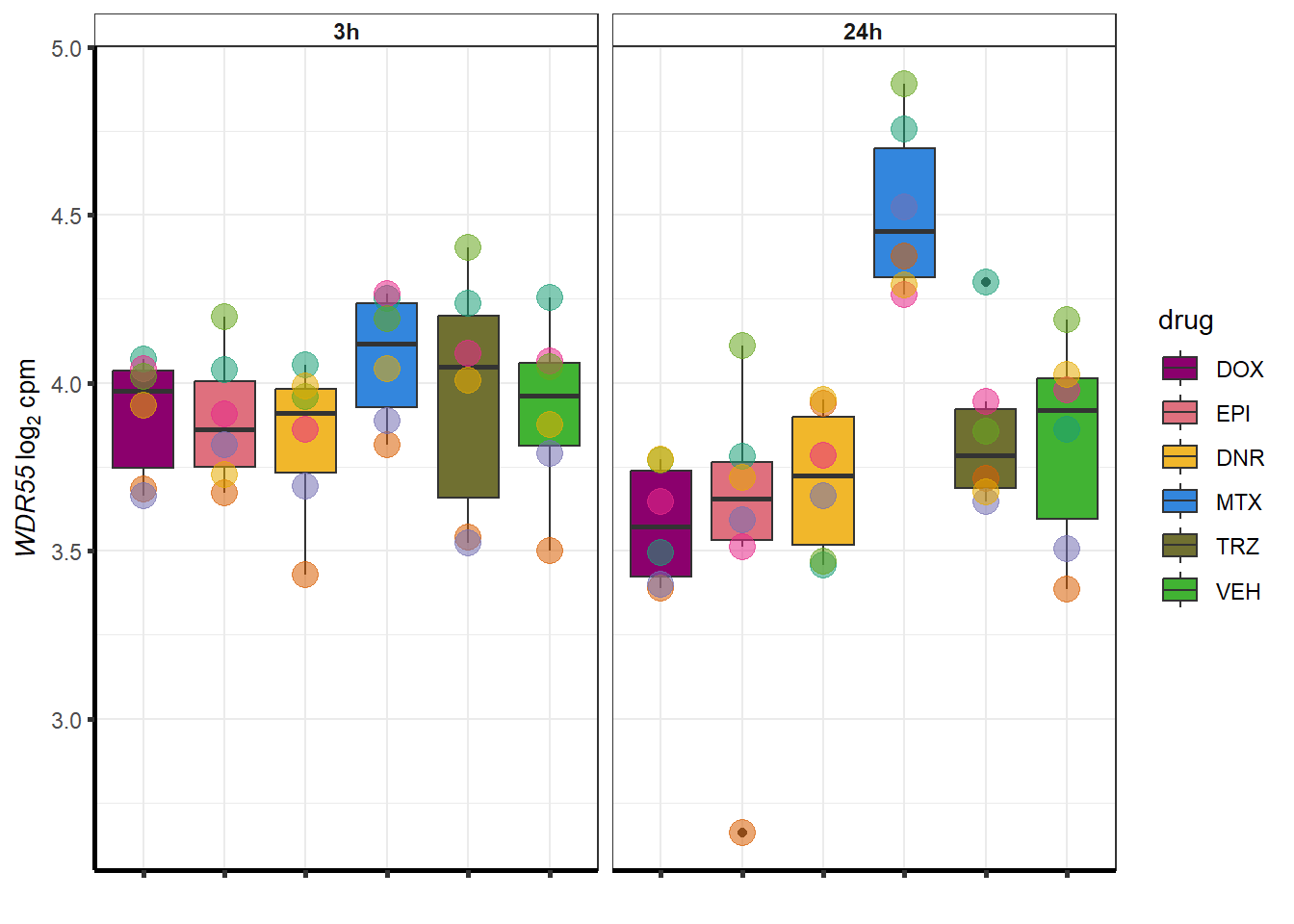
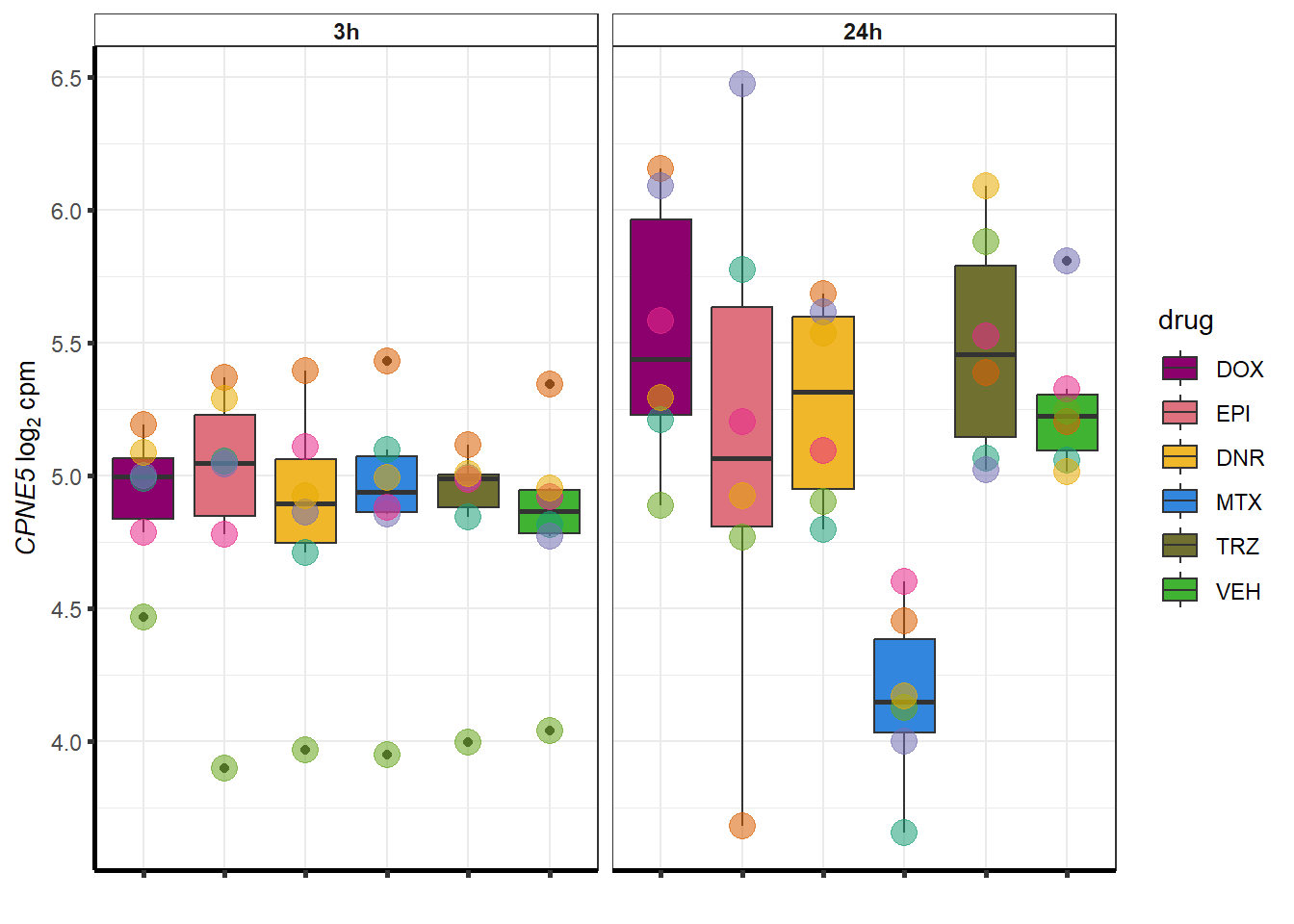
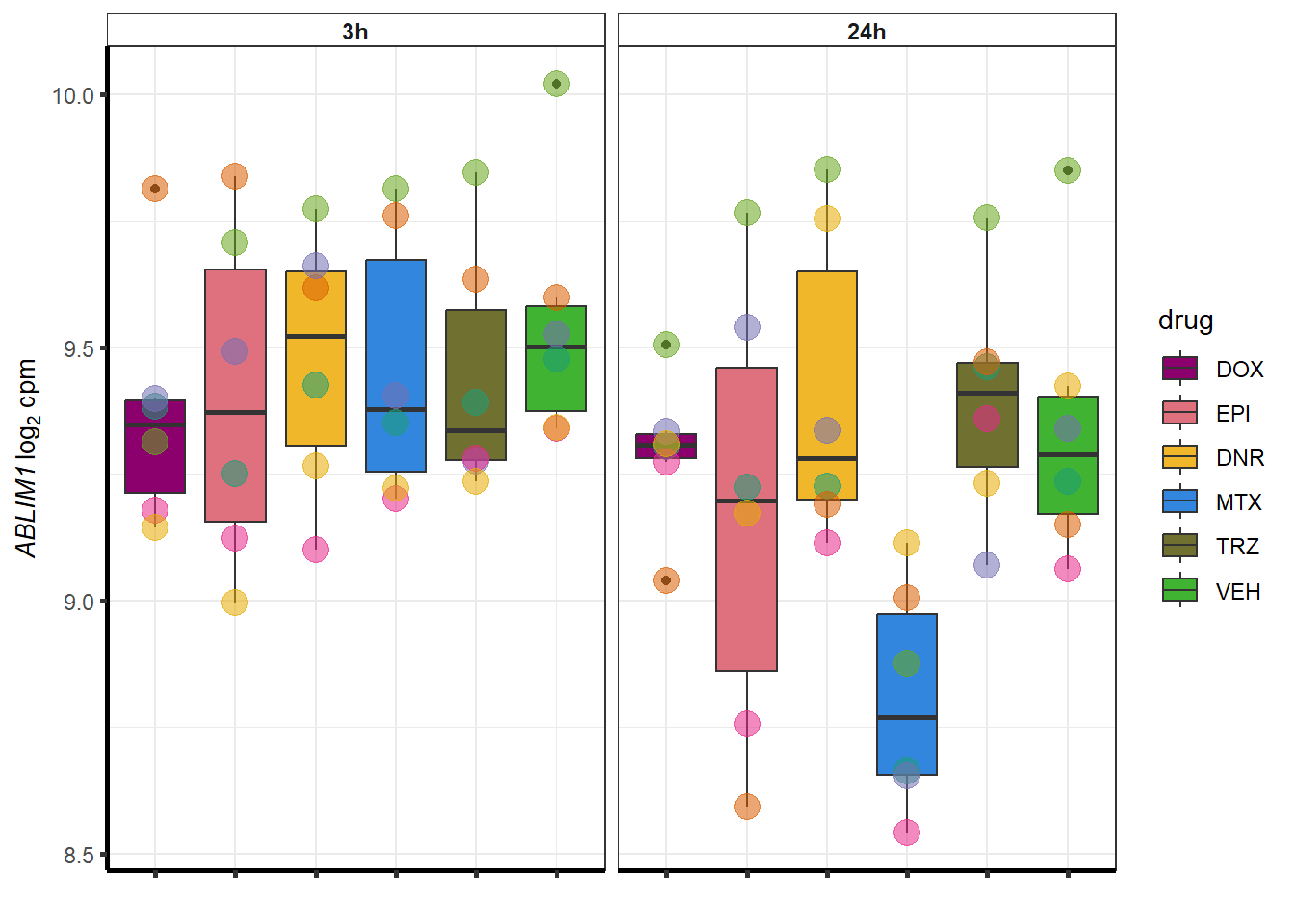
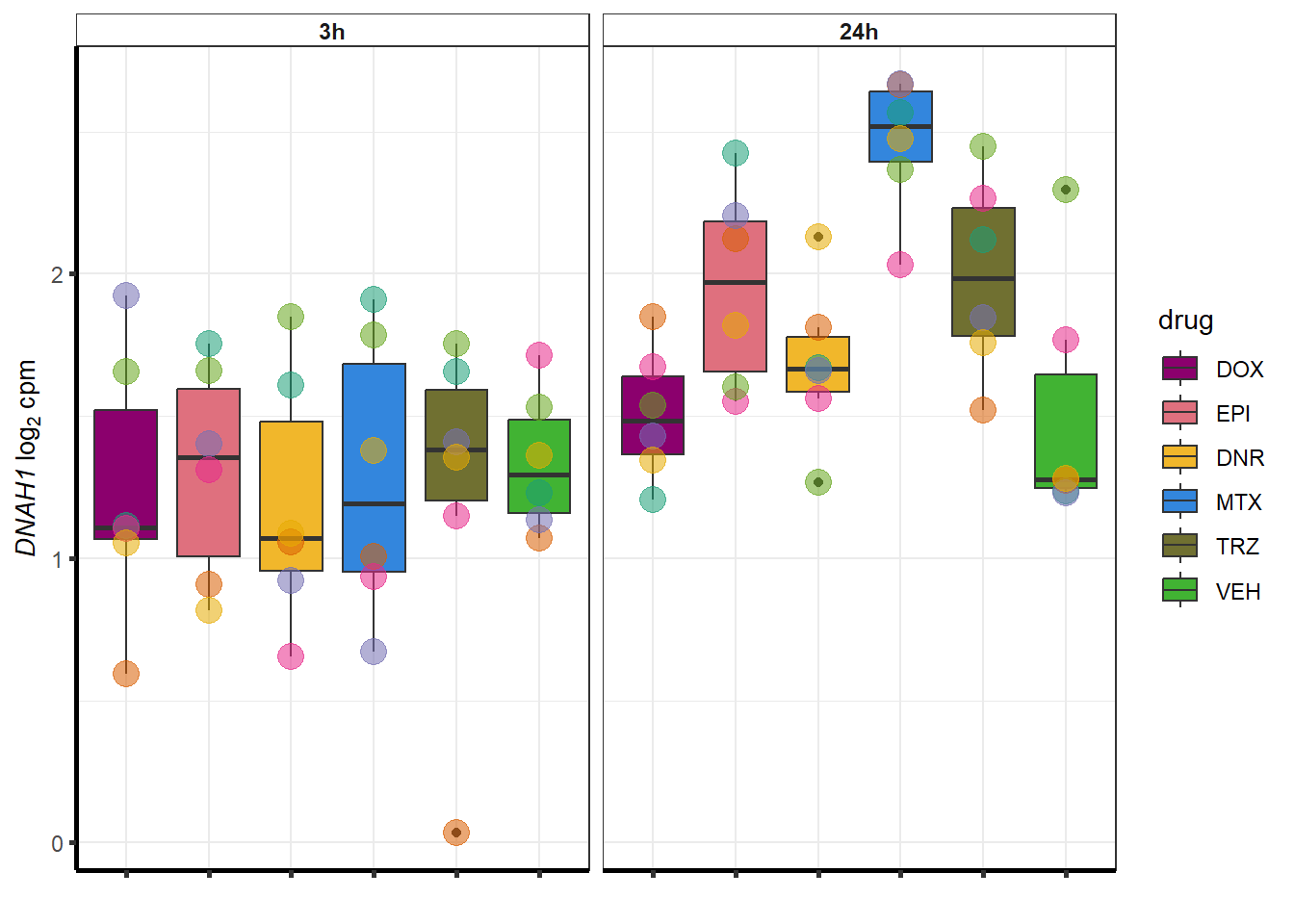
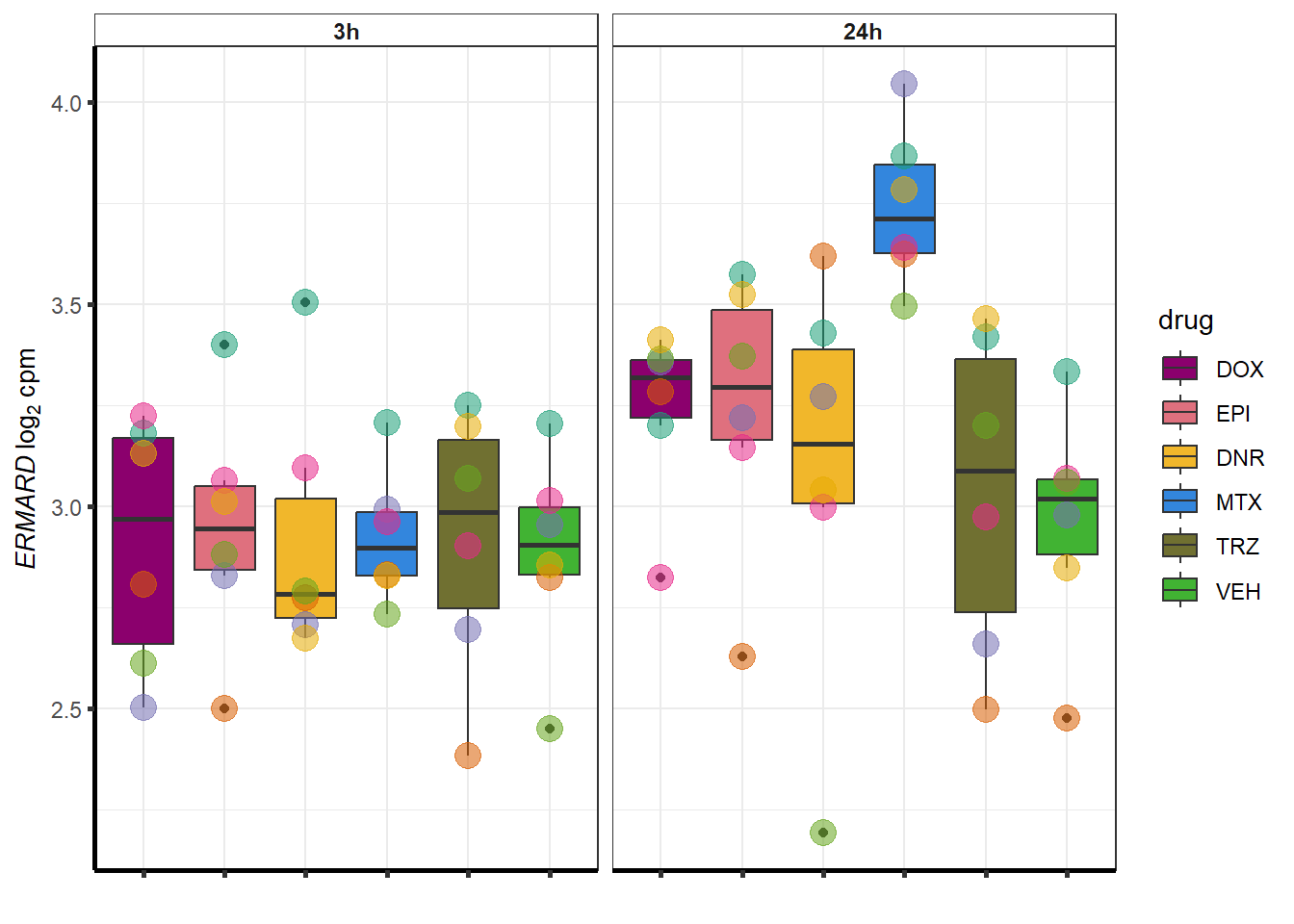
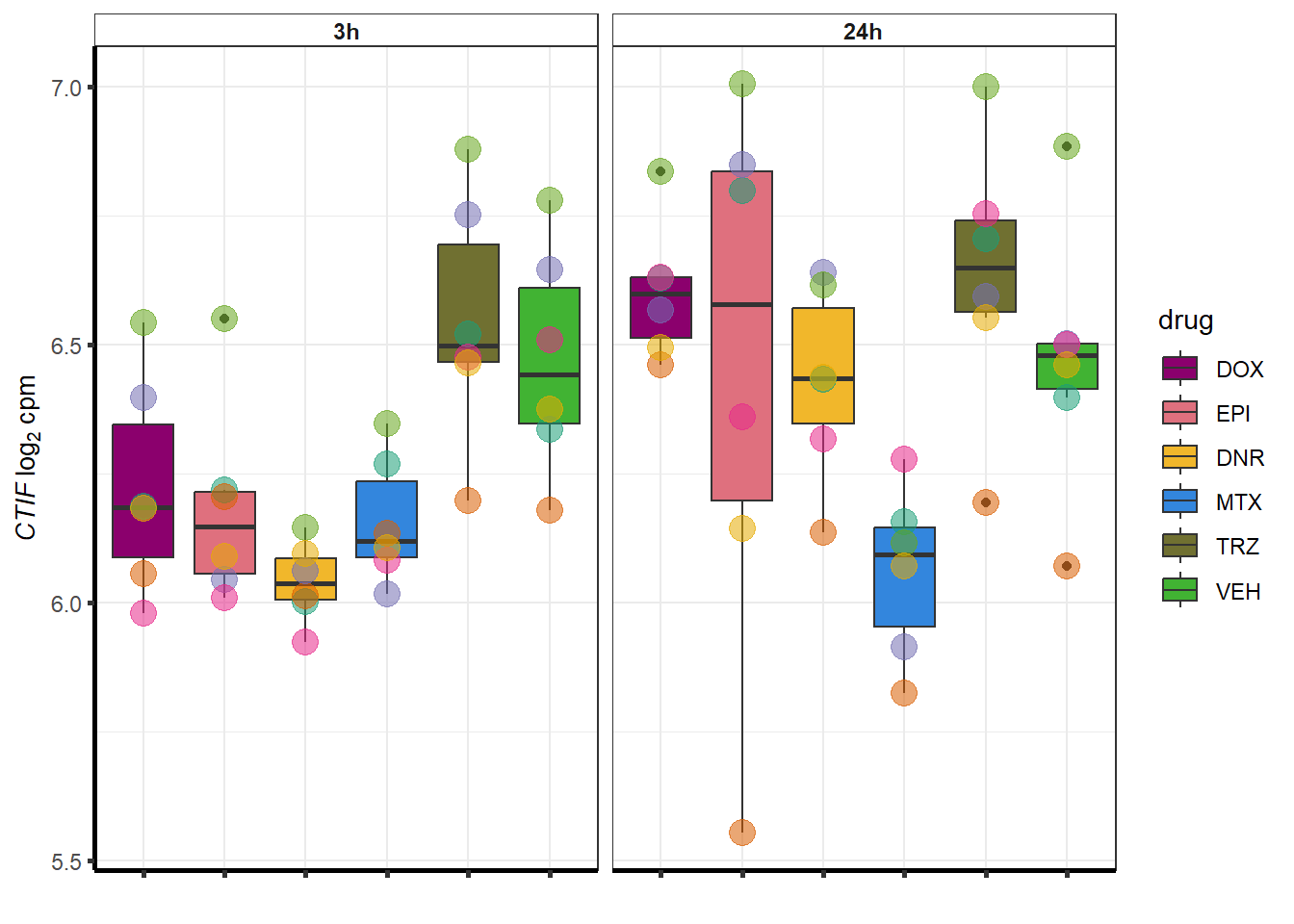
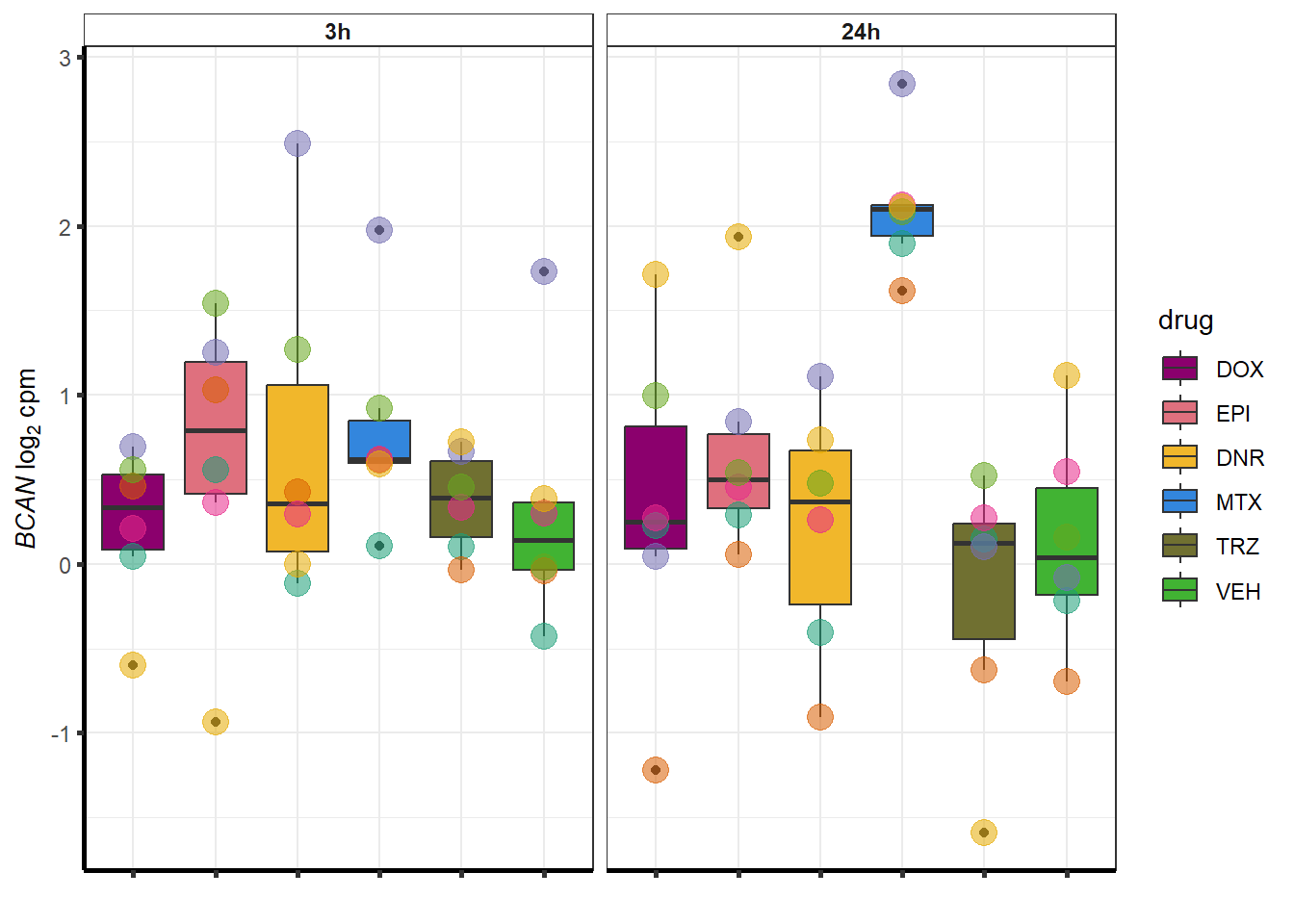
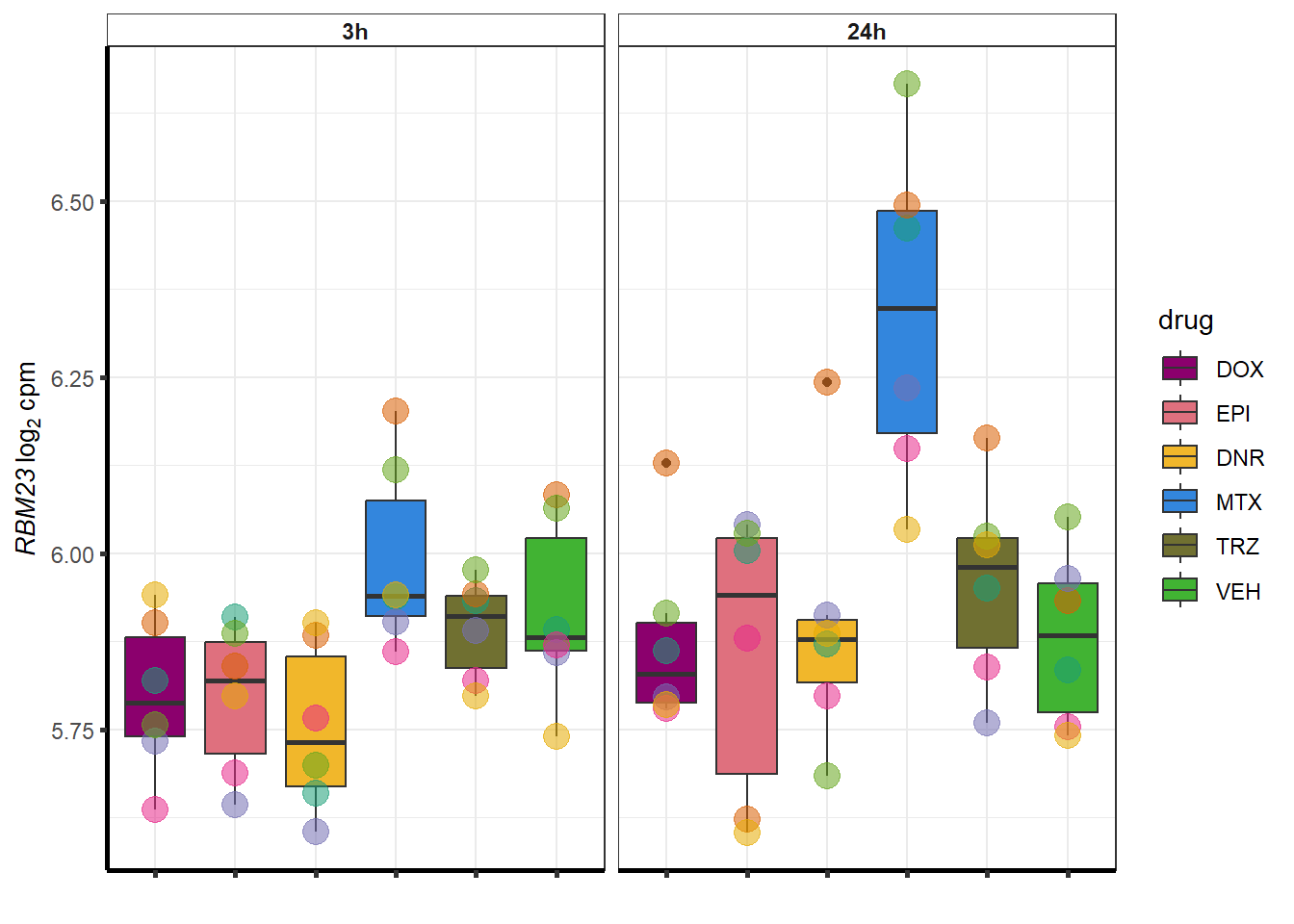
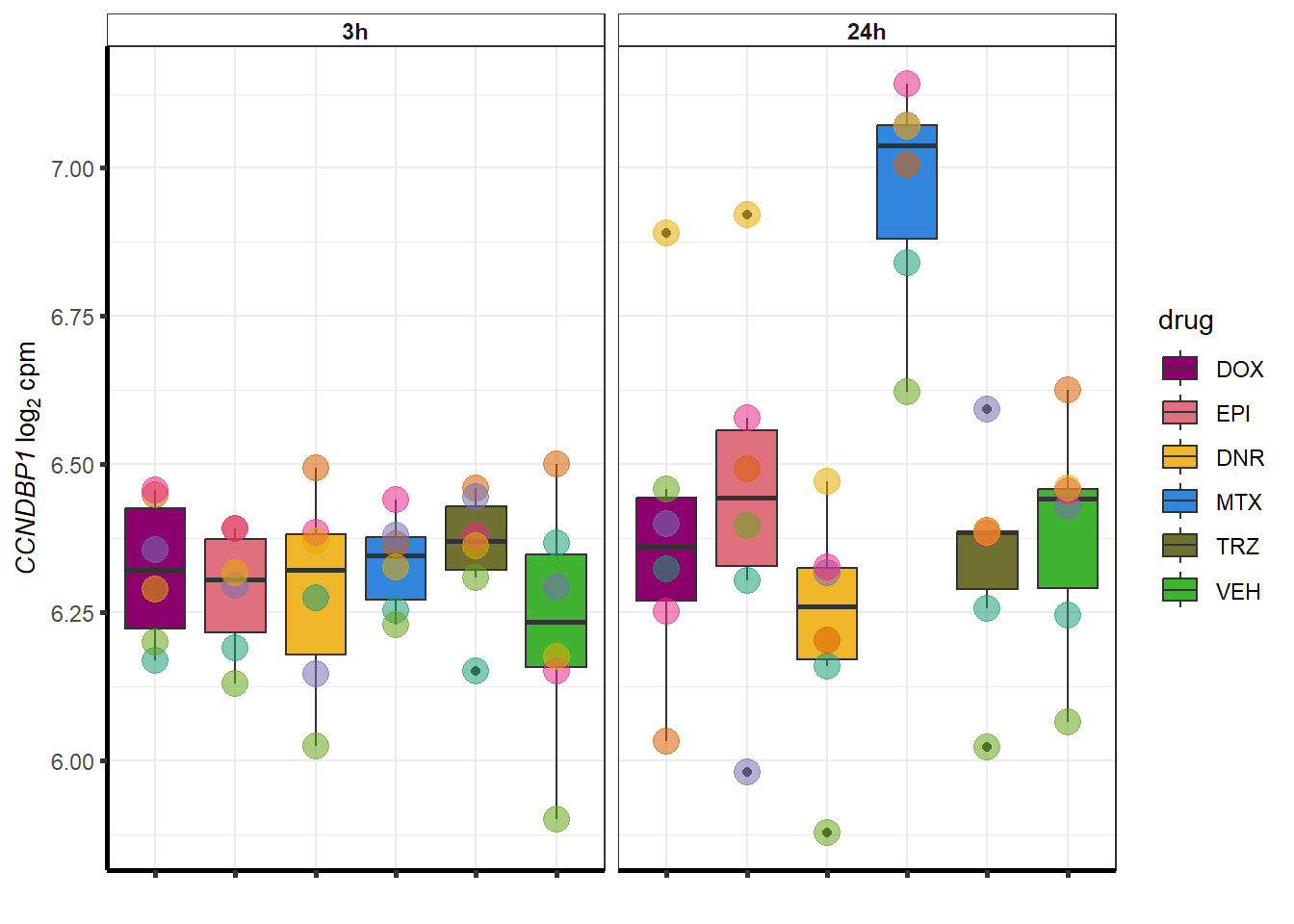
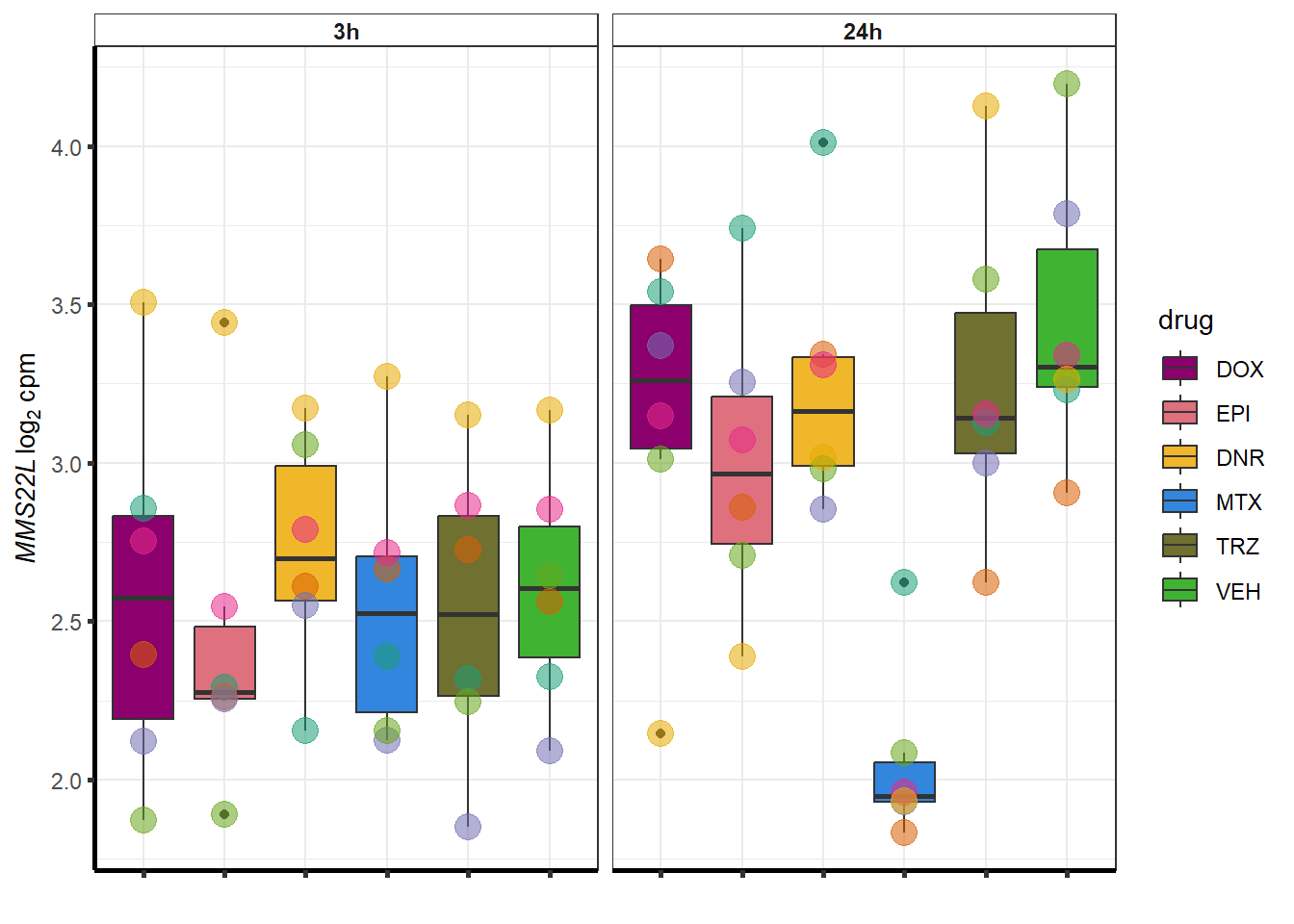
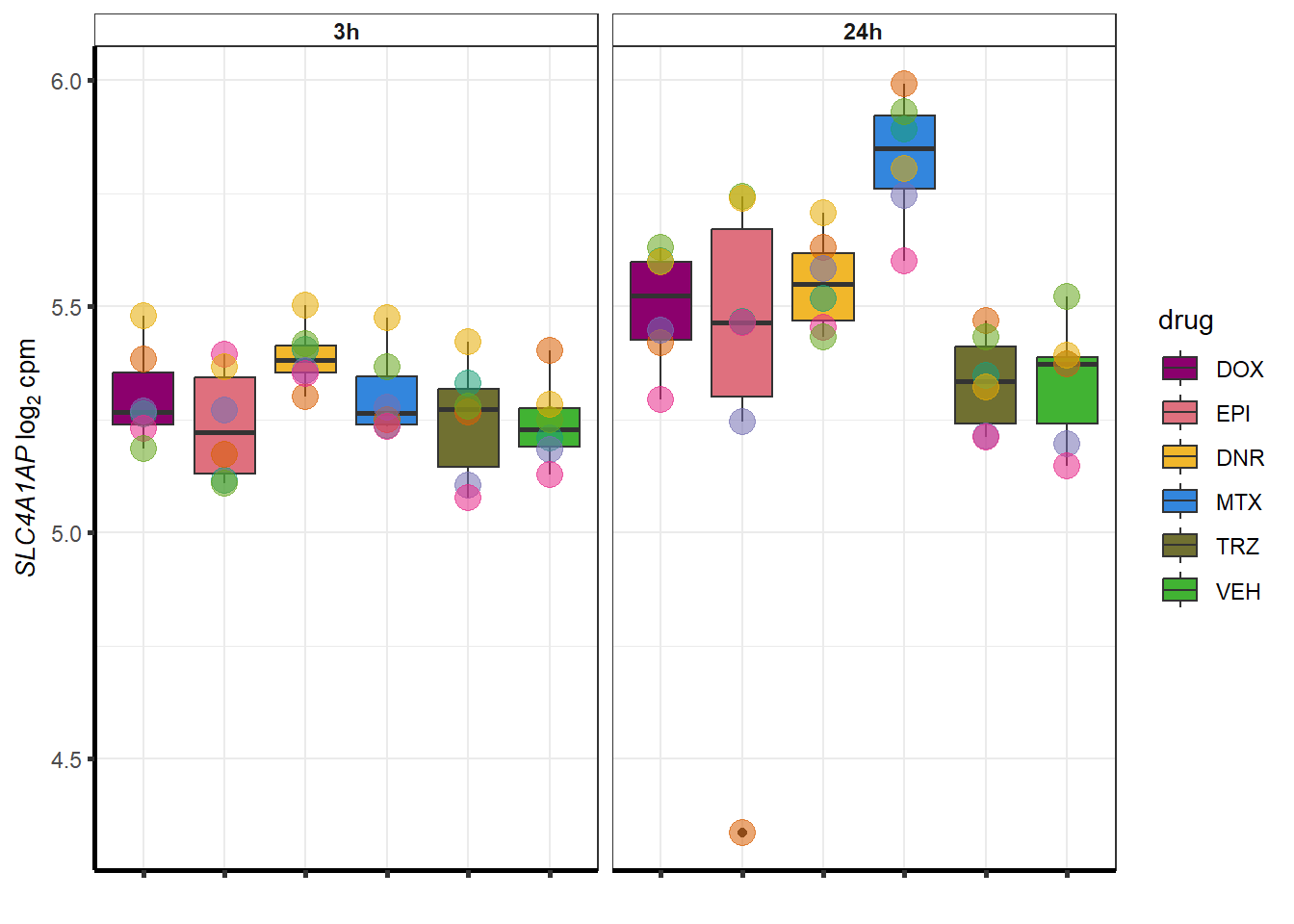
##most convincing of these is 54853 WDR55 DNR
DNRdeg_sp <- toplist24hr %>%
filter(adj.P.Val<0.01) %>%
filter(ENTREZID %in% DnronlyDEG) %>%
filter(id=="DNR") %>%
dplyr::select(ENTREZID, SYMBOL)
##lfc
toplist24hr %>%
filter(ENTREZID %in% DNRdeg_sp$ENTREZID) %>%
group_by(time,id) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
theme_classic()+
fill_palette(palette = drug_palc)+
ggtitle("LogFC of DNR specific DEGs (n = 112)")+
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5,face = "bold"),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle = 0))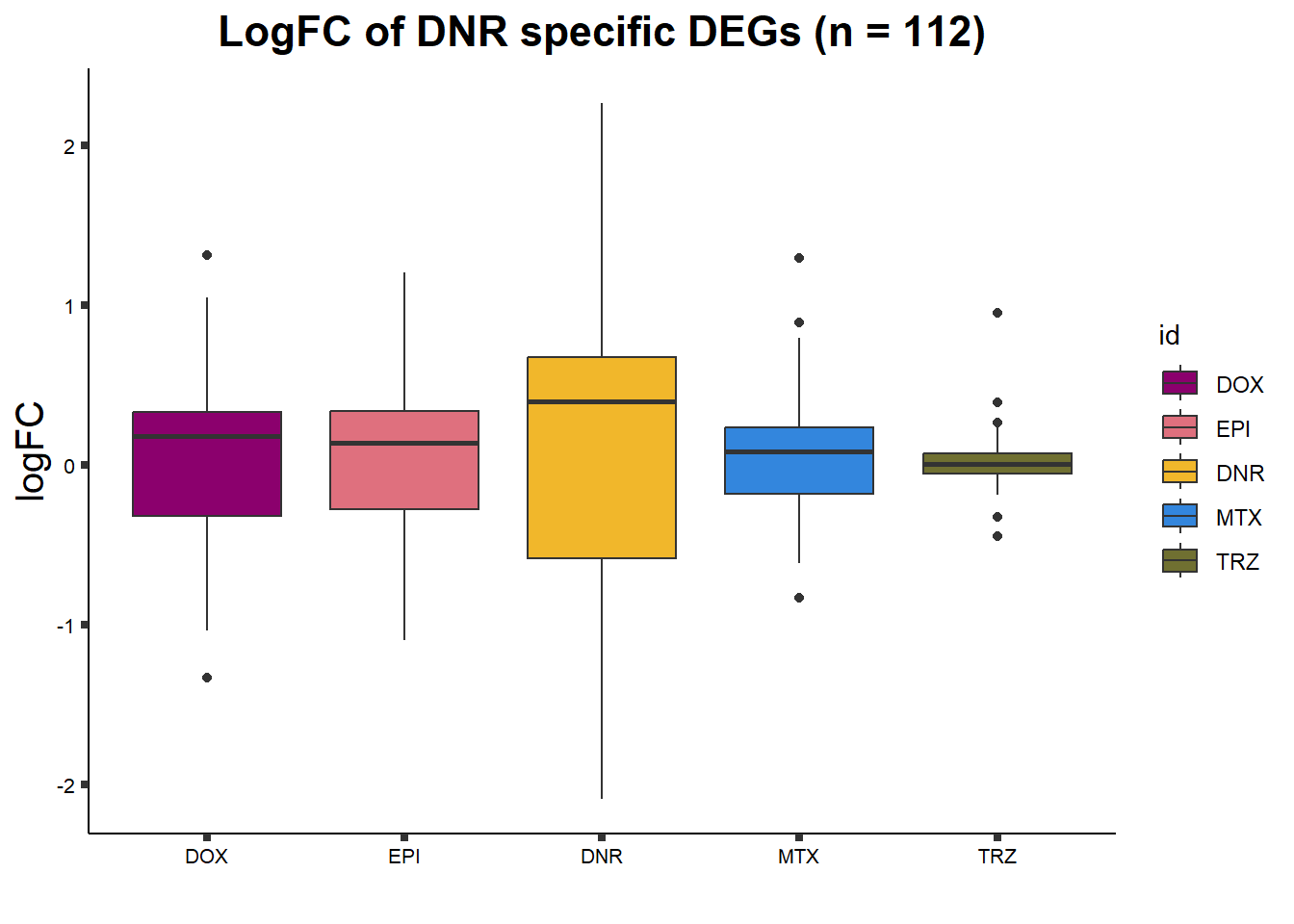
##histo check
toplist24hr %>%
dplyr::filter(ENTREZID %in% DNRdeg_sp$ENTREZID) %>%
dplyr::filter(adj.P.Val <0.05) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id))+
geom_vline(xintercept=0.01,linetype=2)+
# geom_histogram(aes(fill=id))+
facet_wrap(~id)+
ggtitle("all DNR specific DEG adj p. value <0.01")+
fill_palette(palette = drug_palc)+
theme_bw()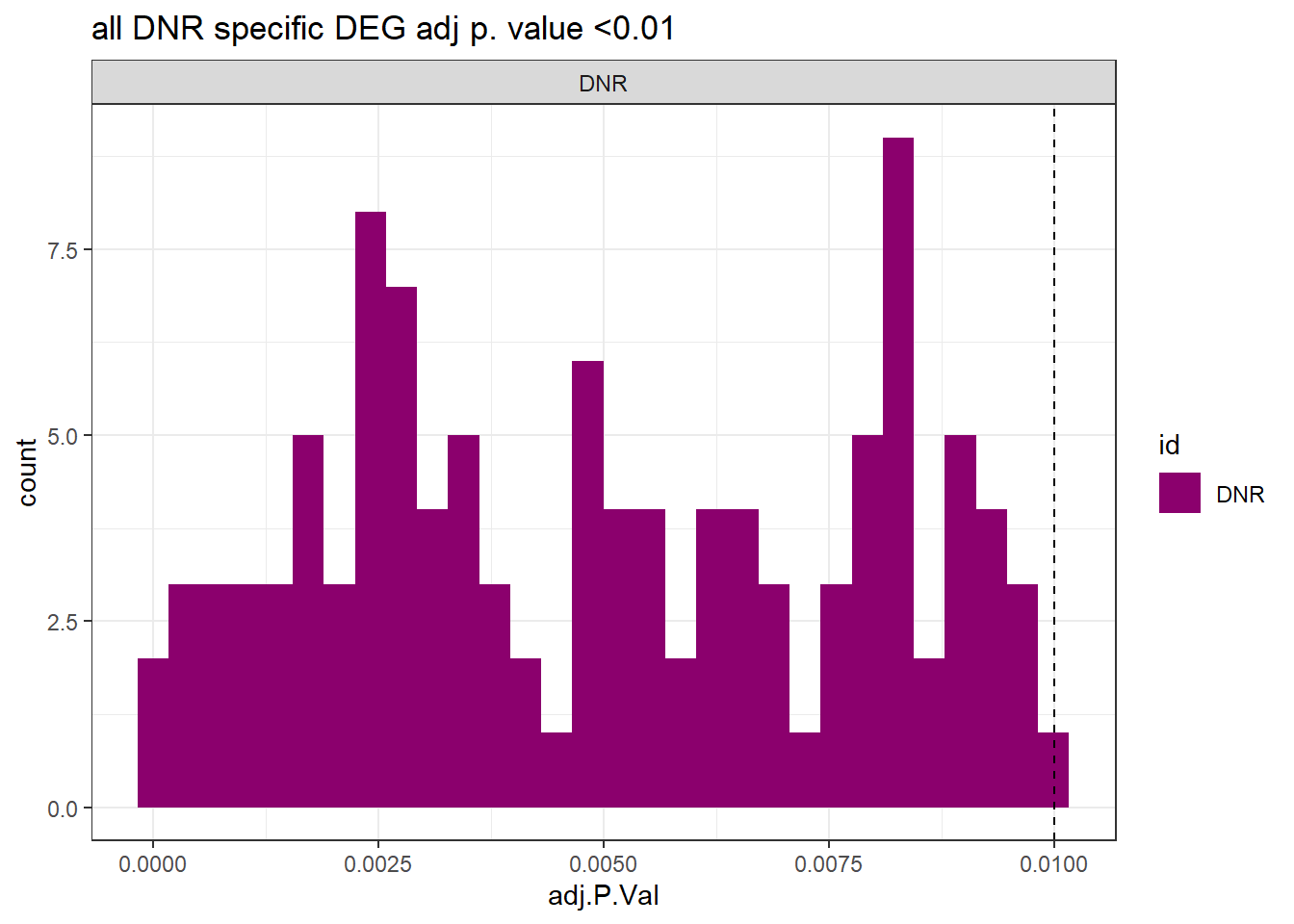
toplist24hr %>%
filter(ENTREZID %in% DnronlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()
set.seed(12345)
sampset <- DNRdeg_sp %>%
sample_n(.,12)
for (g in seq(from=1, to=length(sampset$ENTREZID))){
a <- sampset$SYMBOL[g]
cpm_boxplot(cpmcounts,GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}
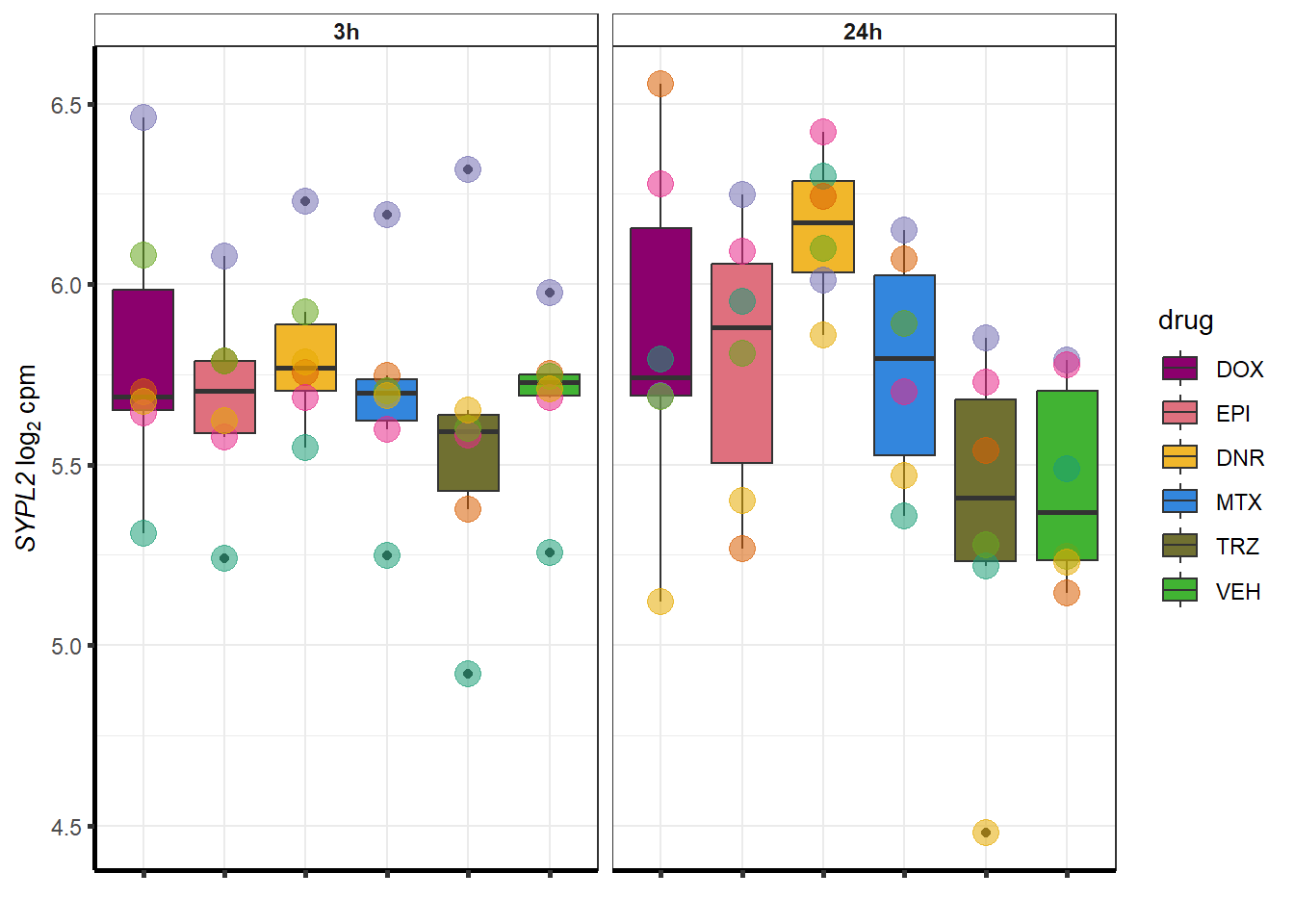
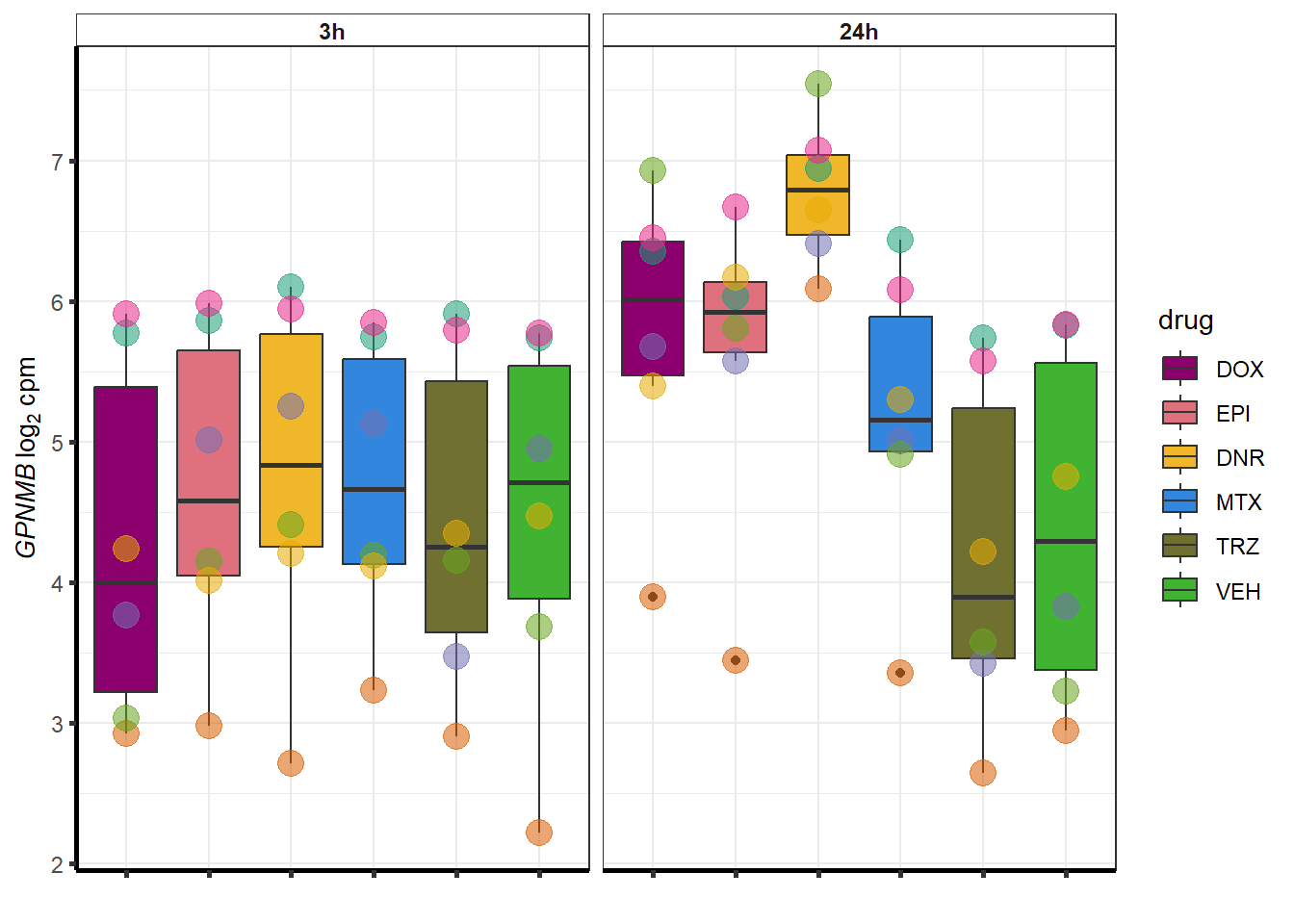
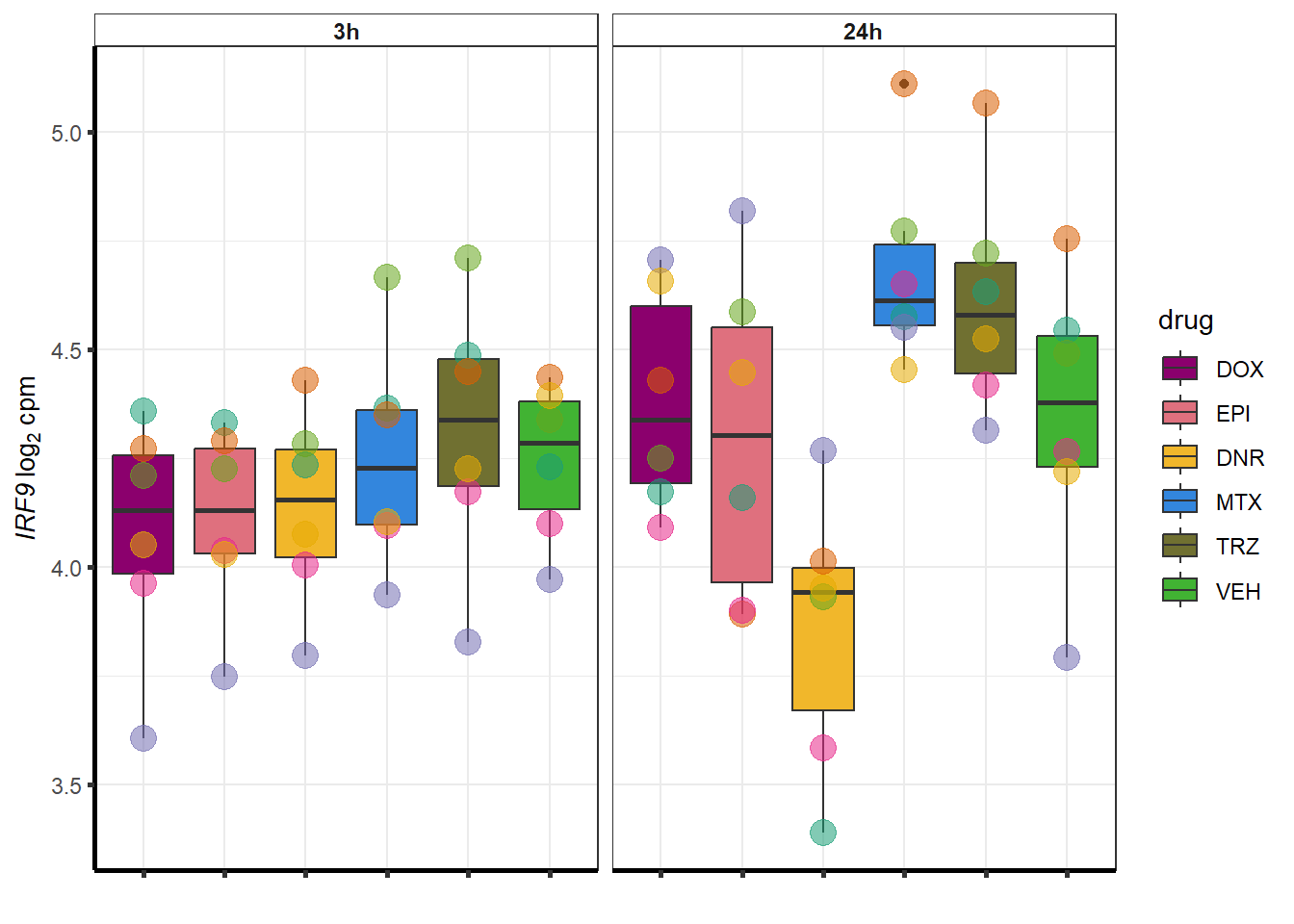
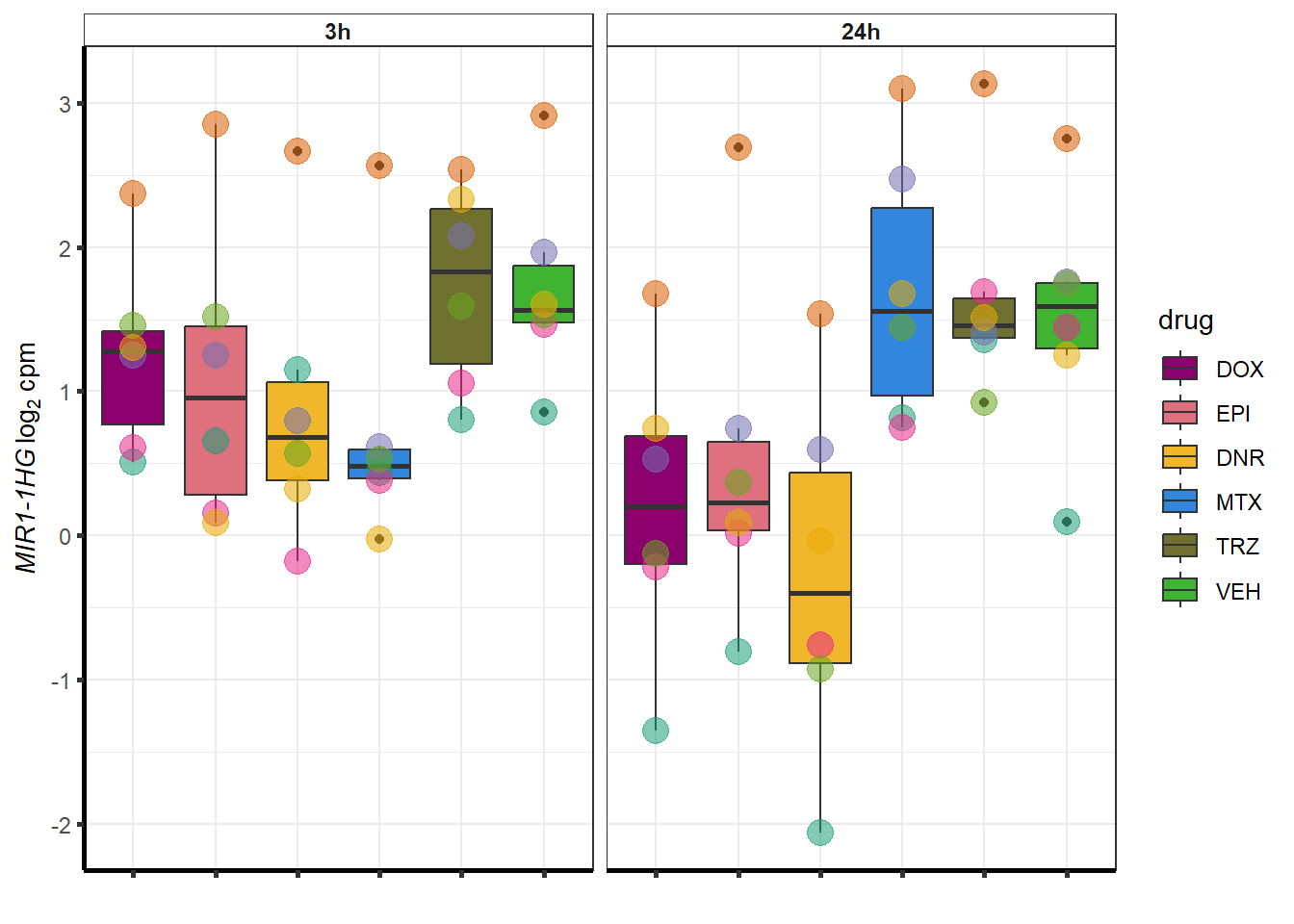
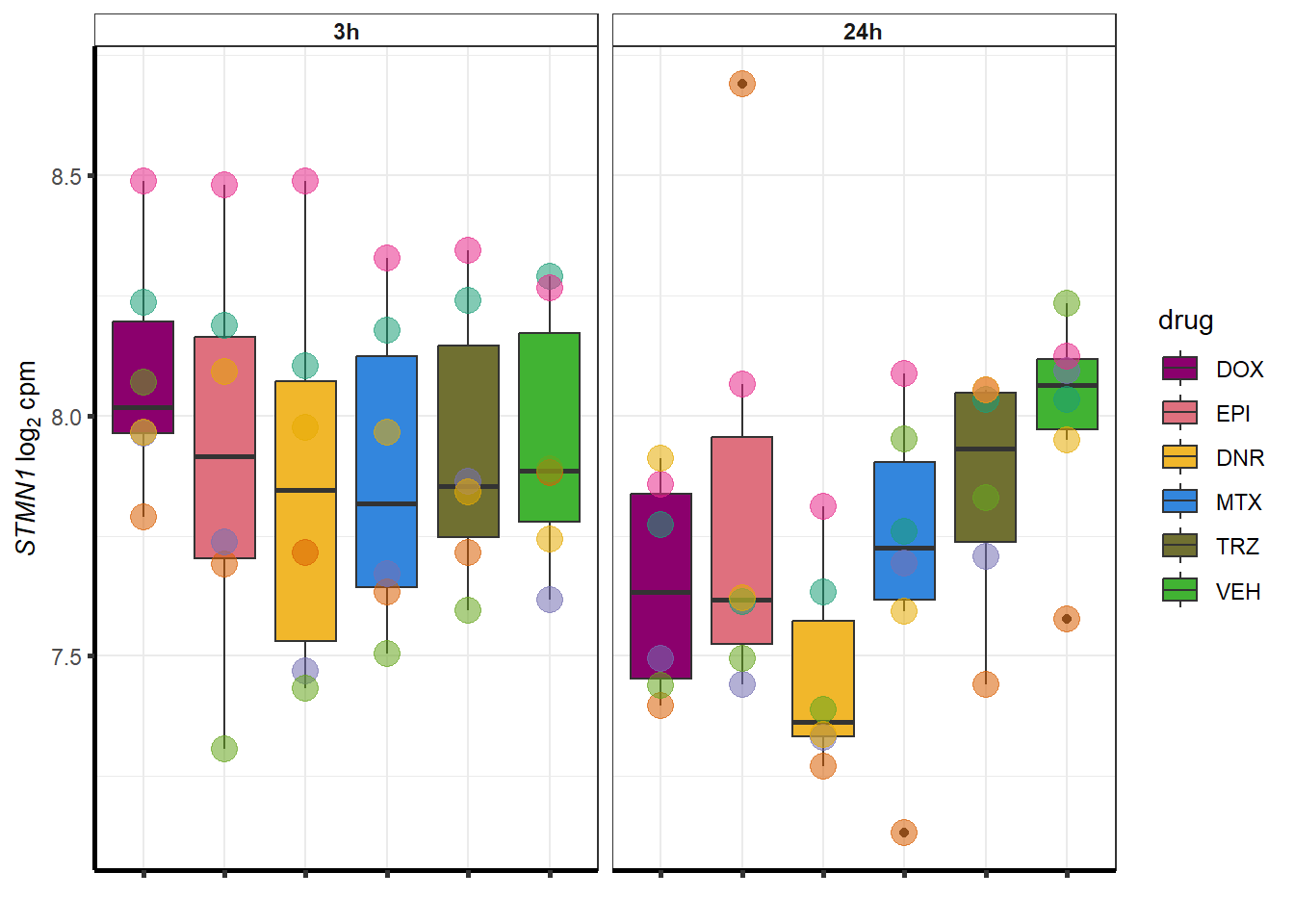
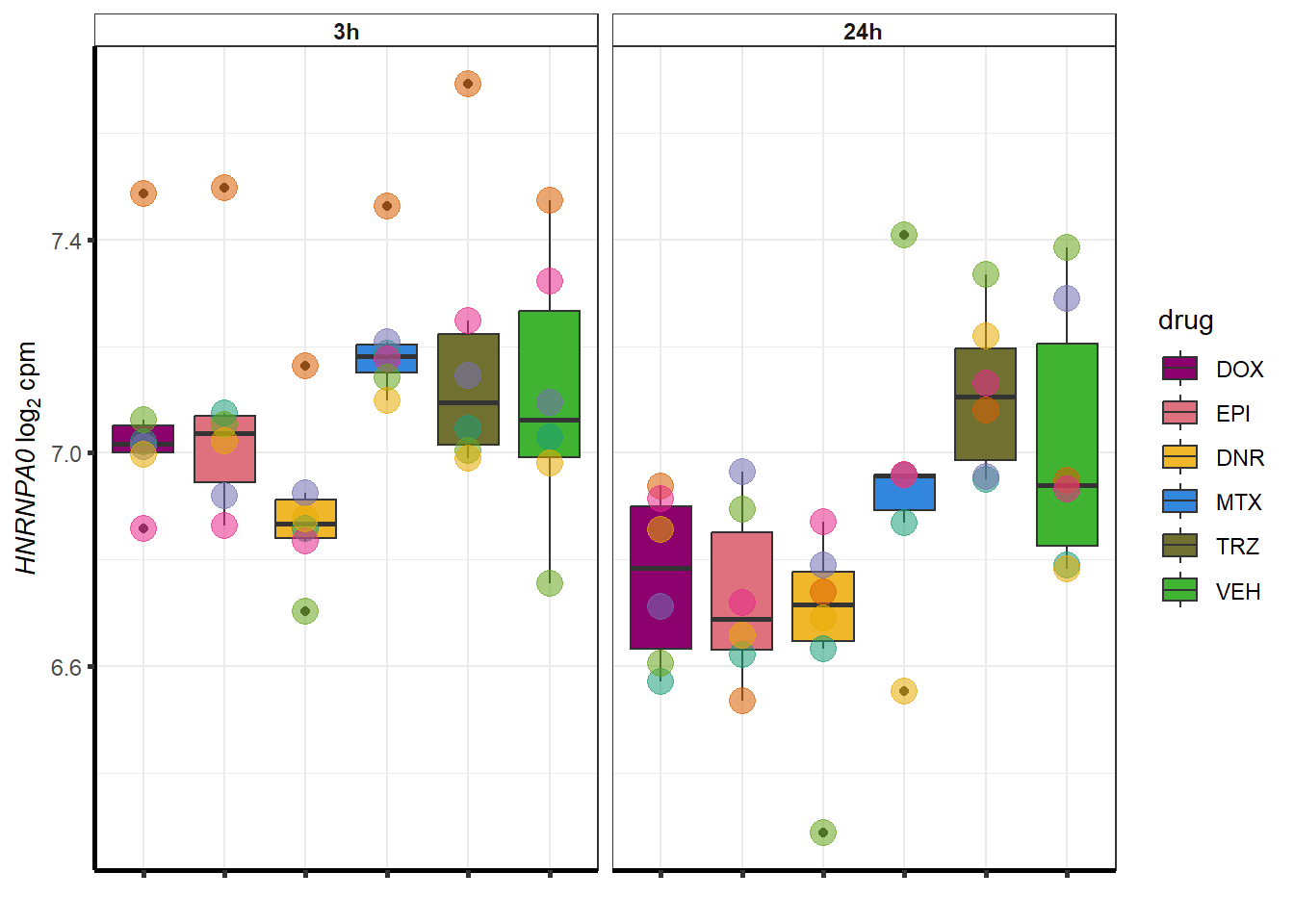
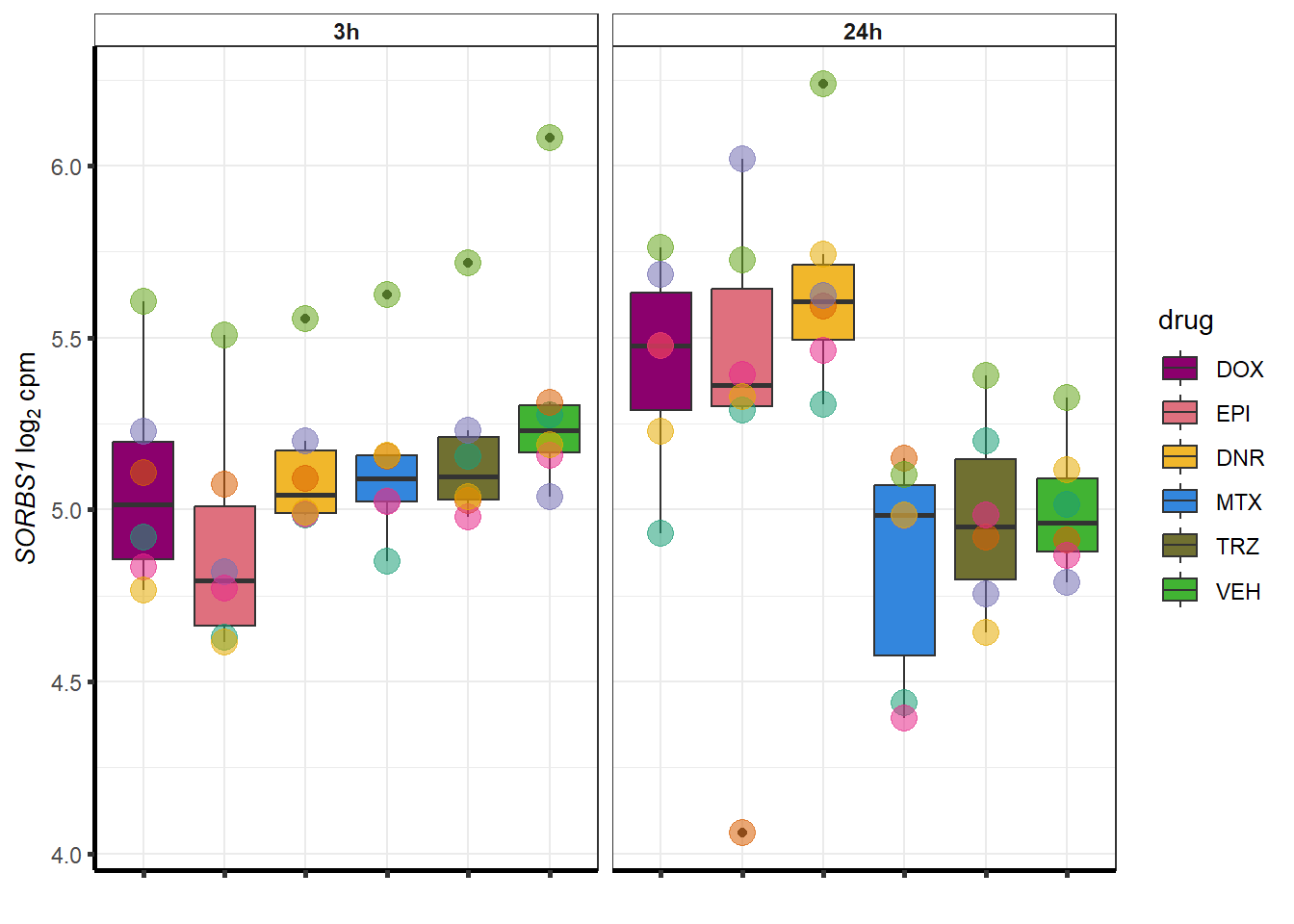
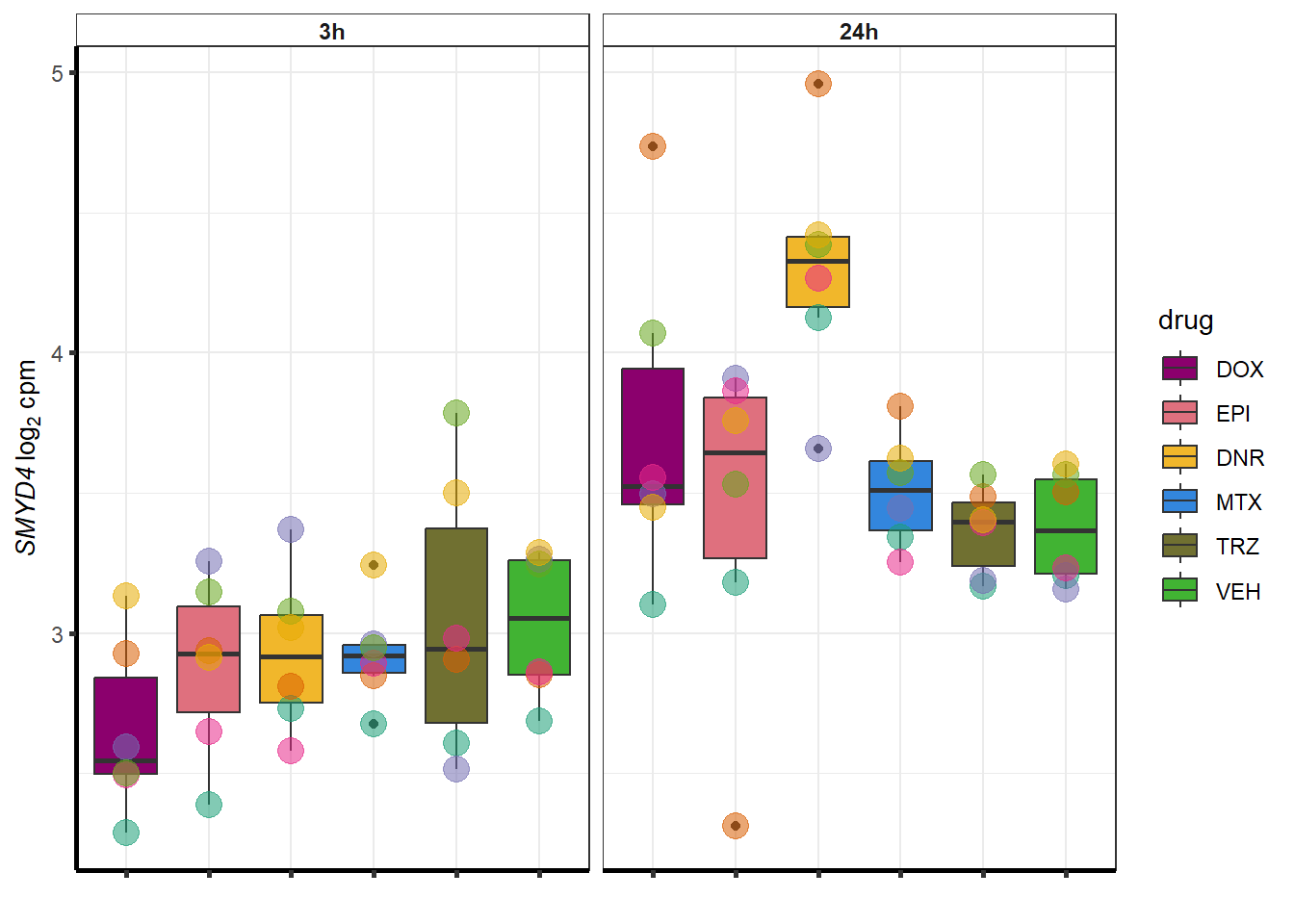
##most convincing of these is 114826 SMYD4EPI
EPIdeg_sp <- toplist24hr %>%
filter(adj.P.Val<0.01) %>%
filter(ENTREZID %in% EpionlyDEG) %>%
filter(id=="EPI") %>%
dplyr::select(ENTREZID, SYMBOL)
EPIdeg_sp %>%
kable(., caption= "EPI specific genes") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box( height = "500px")| ENTREZID | SYMBOL | |
|---|---|---|
| 9685 | 9685 | CLINT1 |
| 9373 | 9373 | PLAA |
| 55006 | 55006 | TRMT61B |
| 79798 | 79798 | ARMC5 |
| 79759 | 79759 | ZNF668 |
| 51434 | 51434 | ANAPC7 |
| 54508 | 54508 | EPB41L4A-DT |
| 92140 | 92140 | MTDH |
| 11097 | 11097 | NUP42 |
| 64781 | 64781 | CERK |
| 57587 | 57587 | CFAP97 |
| 57325 | 57325 | KAT14 |
| 201627 | 201627 | DENND6A |
| 55702 | 55702 | YJU2 |
| 51132 | 51132 | RLIM |
| 55105 | 55105 | GPATCH2 |
| 64863 | 64863 | METTL4 |
| 90864 | 90864 | SPSB3 |
| 8624 | 8624 | PSMG1 |
| 60561 | 60561 | RINT1 |
| 56252 | 56252 | YLPM1 |
| 55339 | 55339 | WDR33 |
| 23086 | 23086 | EXPH5 |
| 1938 | 1938 | EEF2 |
| 493812 | 493812 | HCG11 |
| 7110 | 7110 | TMF1 |
| 83852 | 83852 | SETDB2 |
| 5884 | 5884 | RAD17 |
| 64860 | 64860 | ARMCX5 |
| 220963 | 220963 | SLC16A9 |
| 22796 | 22796 | COG2 |
| 11232 | 11232 | POLG2 |
| 1385 | 1385 | CREB1 |
| 10111 | 10111 | RAD50 |
| 10342 | 10342 | TFG |
| 57609 | 57609 | DIP2B |
| 55727 | 55727 | BTBD7 |
| 2043 | 2043 | EPHA4 |
| 339210 | 339210 | C17orf67 |
| 9972 | 9972 | NUP153 |
| 285636 | 285636 | RIMOC1 |
| 54942 | 54942 | ABITRAM |
| 143282 | 143282 | FGFBP3 |
| 340359 | 340359 | KLHL38 |
| 339122 | 339122 | RAB43 |
| 79038 | 79038 | ZFYVE21 |
| 5533 | 5533 | PPP3CC |
| 2966 | 2966 | GTF2H2 |
| 105371932 | 105371932 | LOC105371932 |
| 55223 | 55223 | TRIM62 |
| 84967 | 84967 | LSM10 |
| 7592 | 7592 | ZNF41 |
| 11282 | 11282 | MGAT4B |
| 7869 | 7869 | SEMA3B |
| 65260 | 65260 | COA7 |
| 9967 | 9967 | THRAP3 |
| 10240 | 10240 | MRPS31 |
| 130507 | 130507 | UBR3 |
| 9829 | 9829 | DNAJC6 |
| 349136 | 349136 | WDR86 |
| 55676 | 55676 | SLC30A6 |
| 64844 | 64844 | MARCHF7 |
| 23122 | 23122 | CLASP2 |
| 26046 | 26046 | LTN1 |
| 138241 | 138241 | C9orf85 |
| 10021 | 10021 | HCN4 |
| 84859 | 84859 | LRCH3 |
| 85457 | 85457 | CIPC |
| 79713 | 79713 | IGFLR1 |
| 79657 | 79657 | RPAP3 |
| 5887 | 5887 | RAD23B |
| 8899 | 8899 | PRPF4B |
| 54904 | 54904 | NSD3 |
| 27246 | 27246 | RNF115 |
| 23064 | 23064 | SETX |
| 6897 | 6897 | TARS1 |
| 55937 | 55937 | APOM |
| 79230 | 79230 | ZNF557 |
| 11054 | 11054 | OGFR |
| 1032 | 1032 | CDKN2D |
| 55958 | 55958 | KLHL9 |
| 5048 | 5048 | PAFAH1B1 |
| 728229 | 728229 | TMEM191B |
| 80176 | 80176 | SPSB1 |
##lfc
toplist24hr %>%
filter(ENTREZID %in% EPIdeg_sp$ENTREZID) %>%
group_by(time,id) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
theme_classic()+
fill_palette(palette = drug_palc)+
ggtitle("LogFC of EPI specific DEGs (n = 84) ")+
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5,face = "bold"),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 8, color = "black", angle = 0))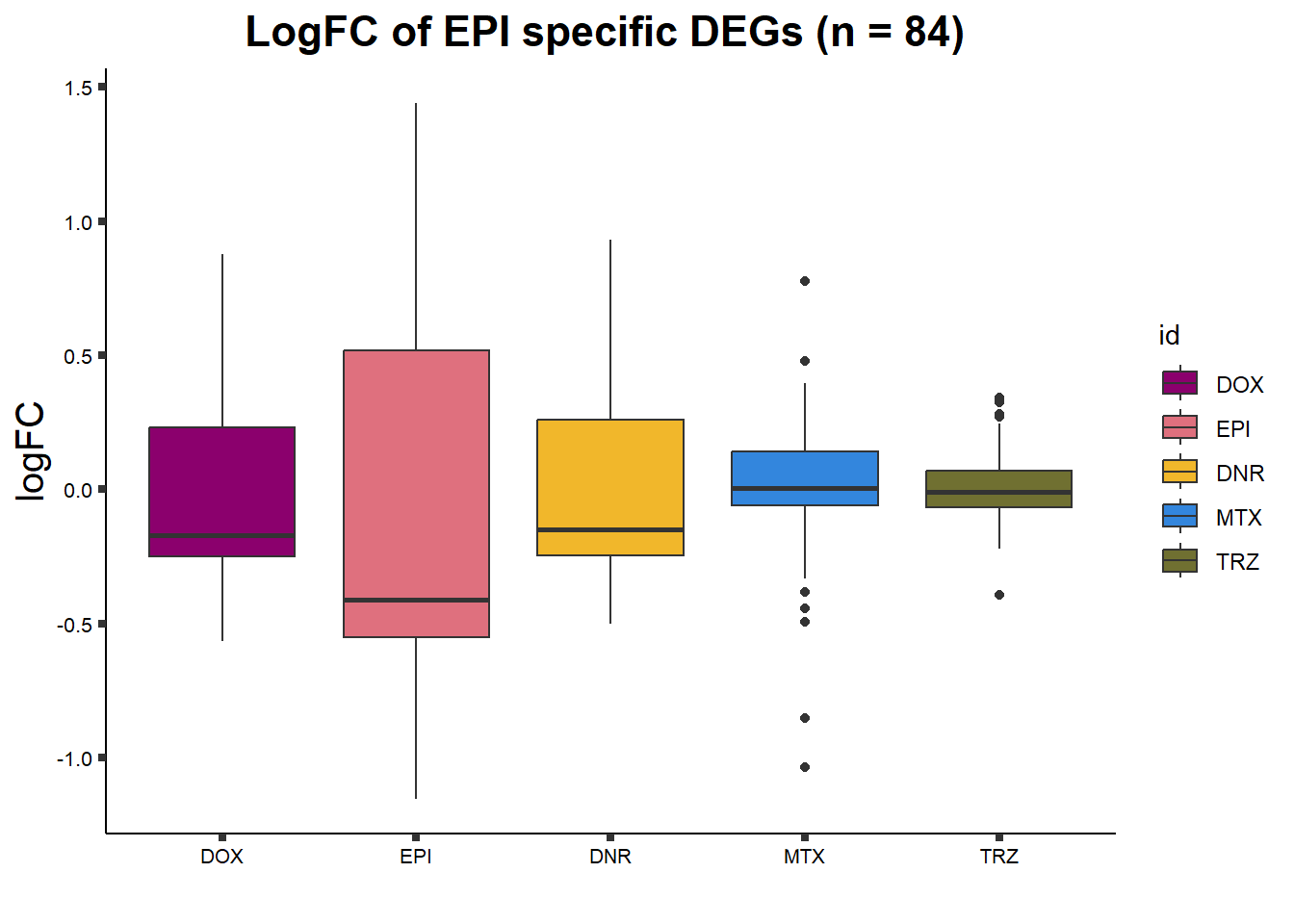
##histo check
toplist24hr %>%
dplyr::filter(ENTREZID %in% EPIdeg_sp$ENTREZID) %>%
dplyr::filter(adj.P.Val <0.05) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id))+
geom_vline(xintercept=0.01,linetype=2)+
# geom_histogram(aes(fill=id))+
facet_wrap(~id)+
ggtitle("all DNR specific DEG adj p. value <0.01")+
fill_palette(palette = drug_palc)+
theme_bw()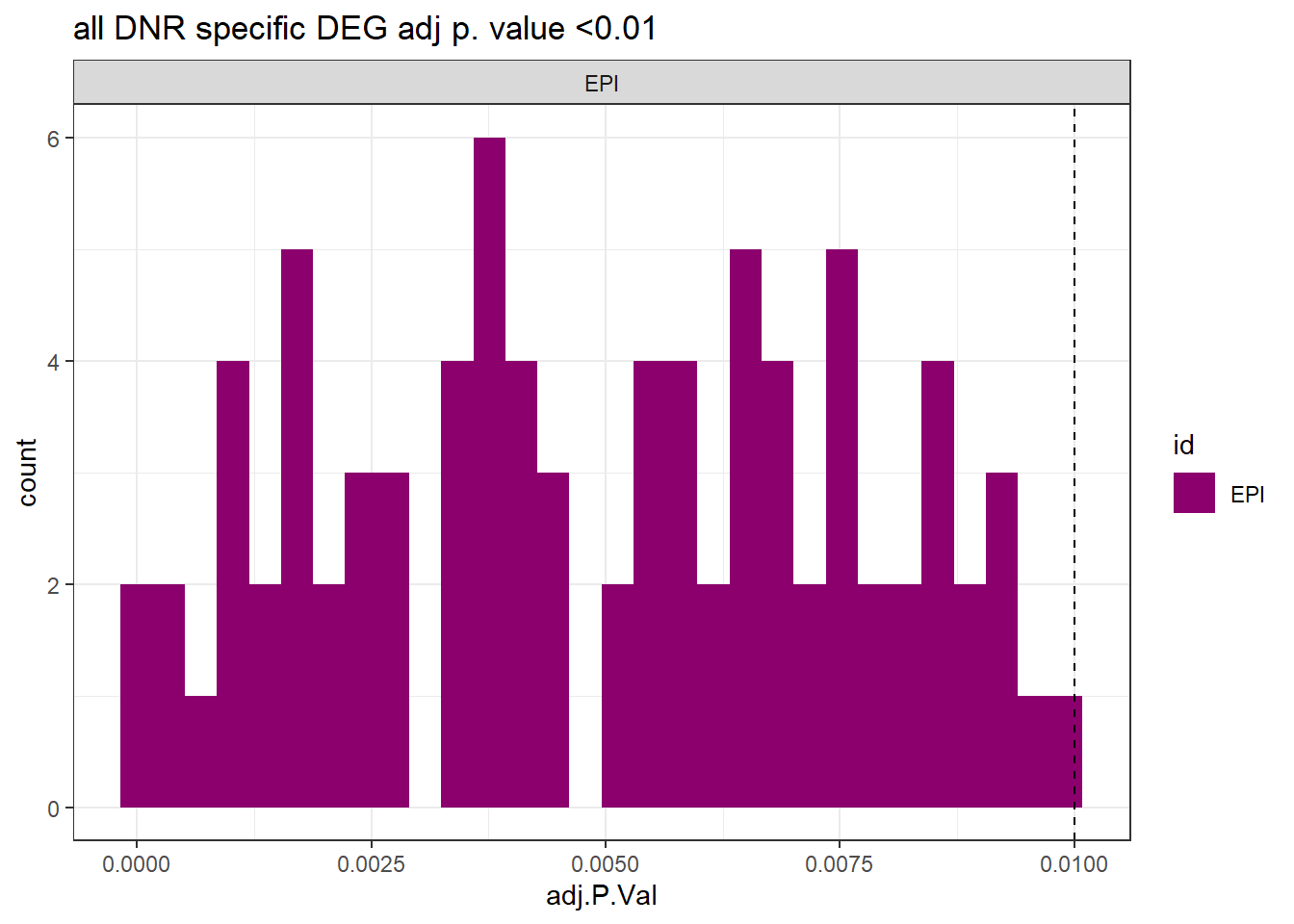
densityEPIsp <- toplist24hr %>%
filter(ENTREZID %in% EpionlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_histogram(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()
densityEPIsp
set.seed(12345)
sampset <- EPIdeg_sp %>%
sample_n(.,12)
for (g in seq(from=1, to=length(sampset$ENTREZID))){
a <- sampset$SYMBOL[g]
cpm_boxplot(cpmcounts,GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}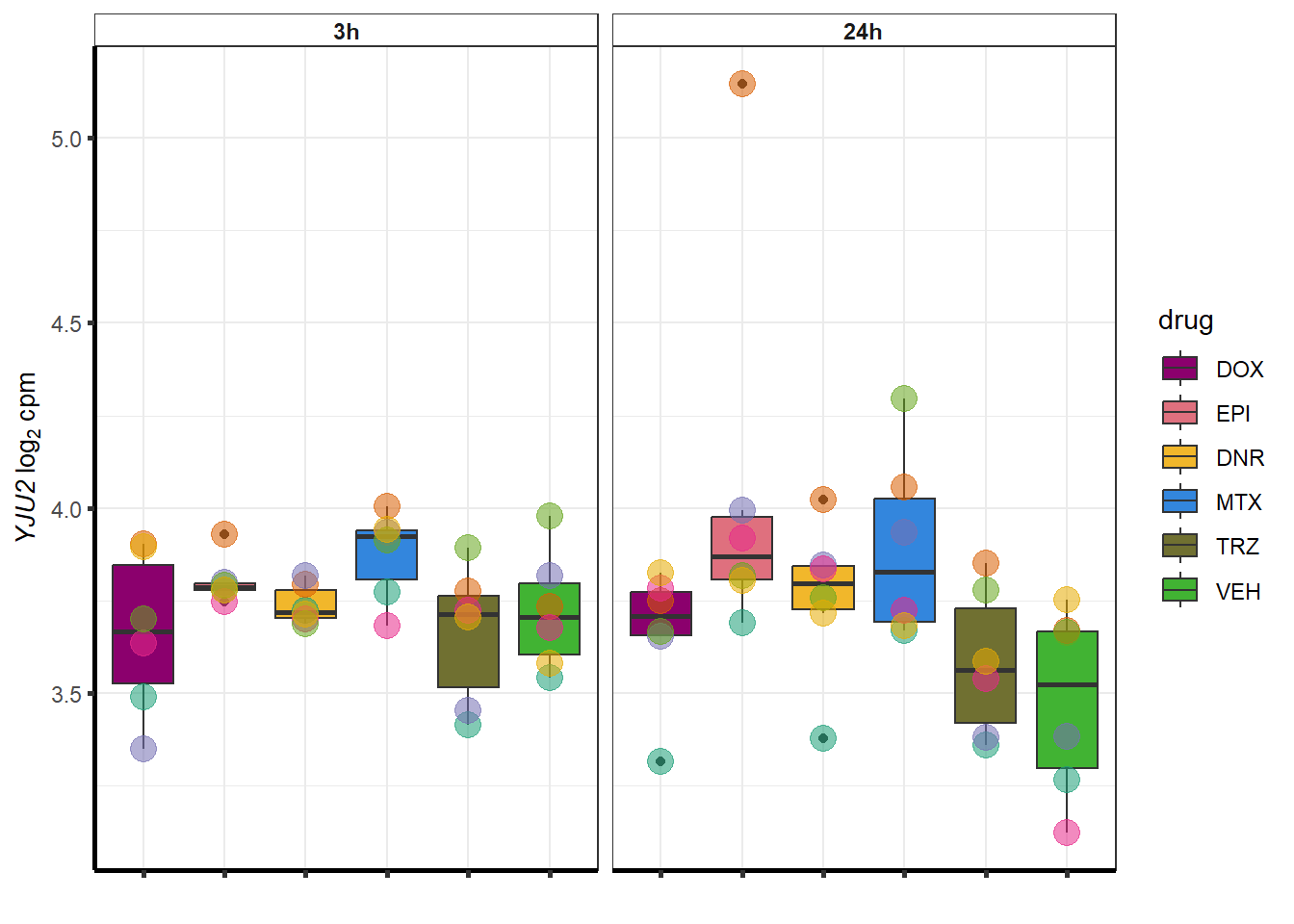
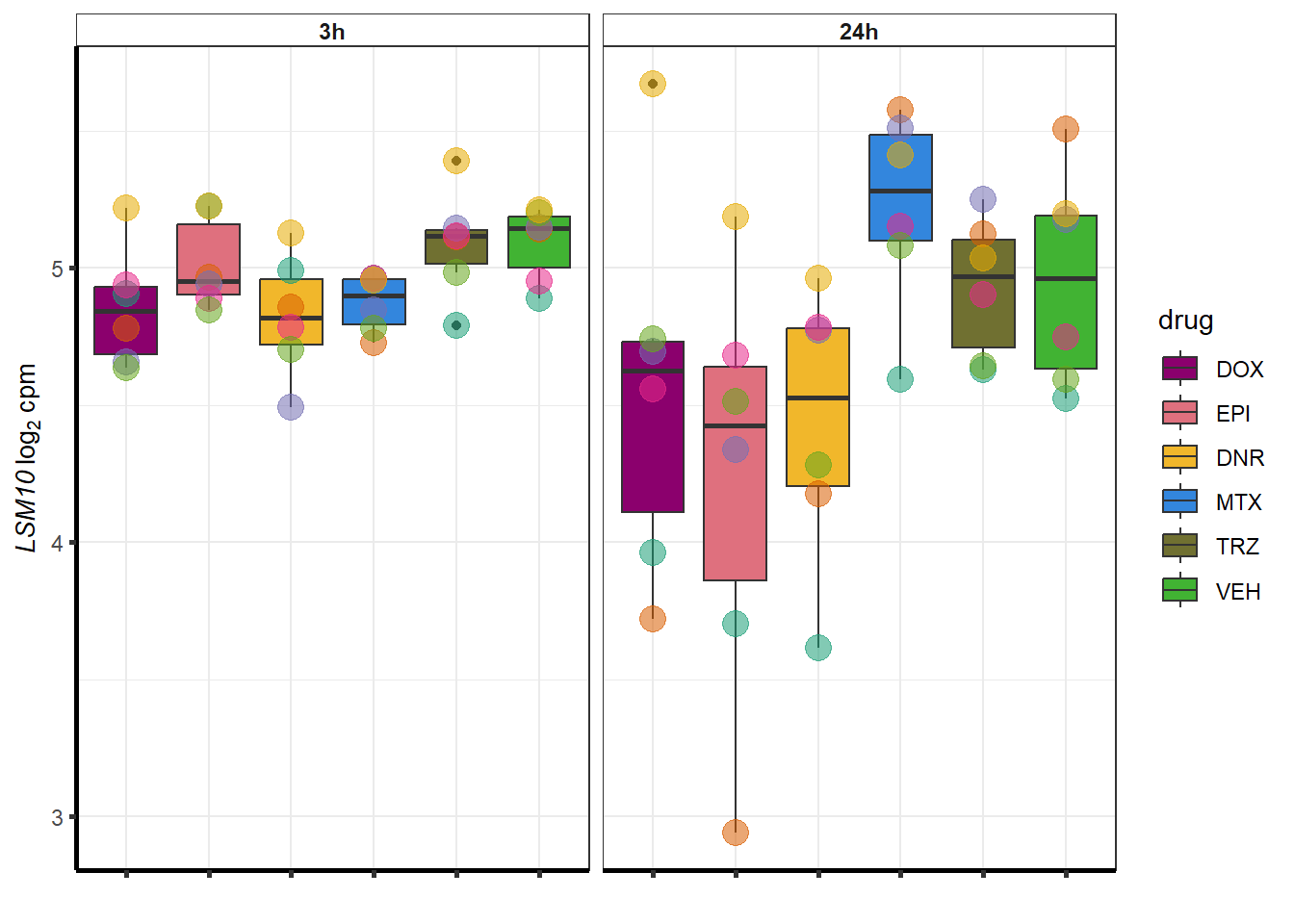
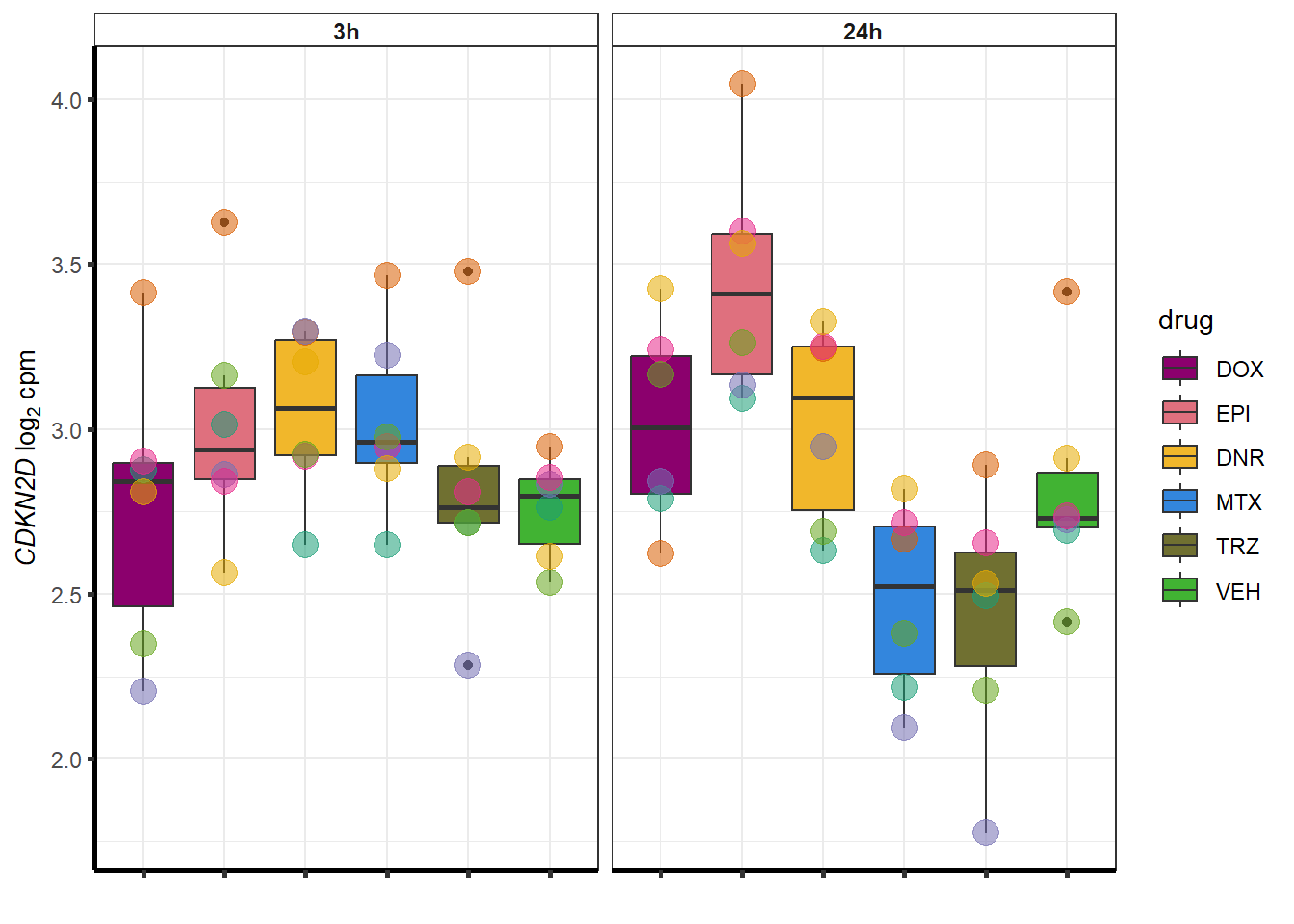
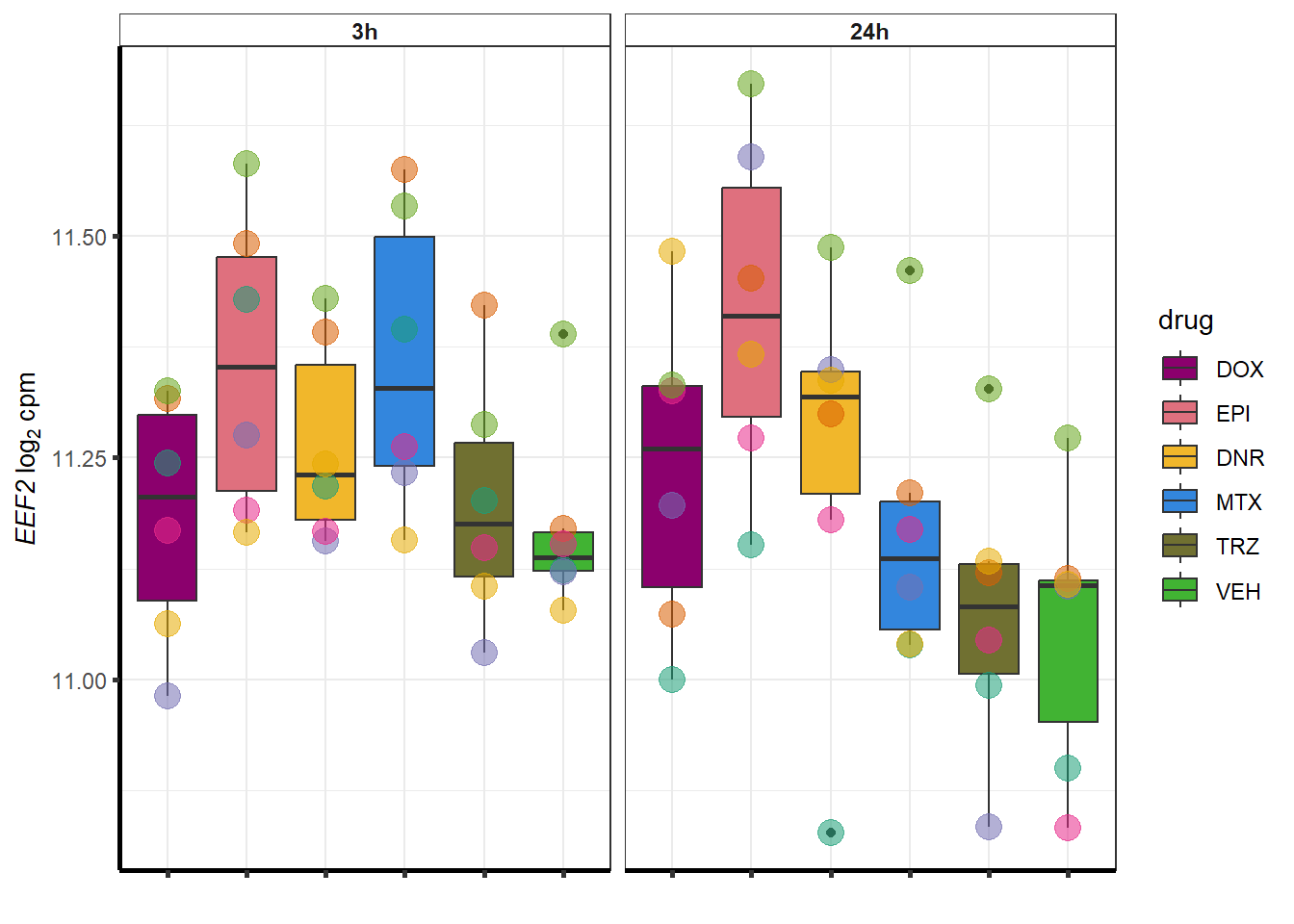
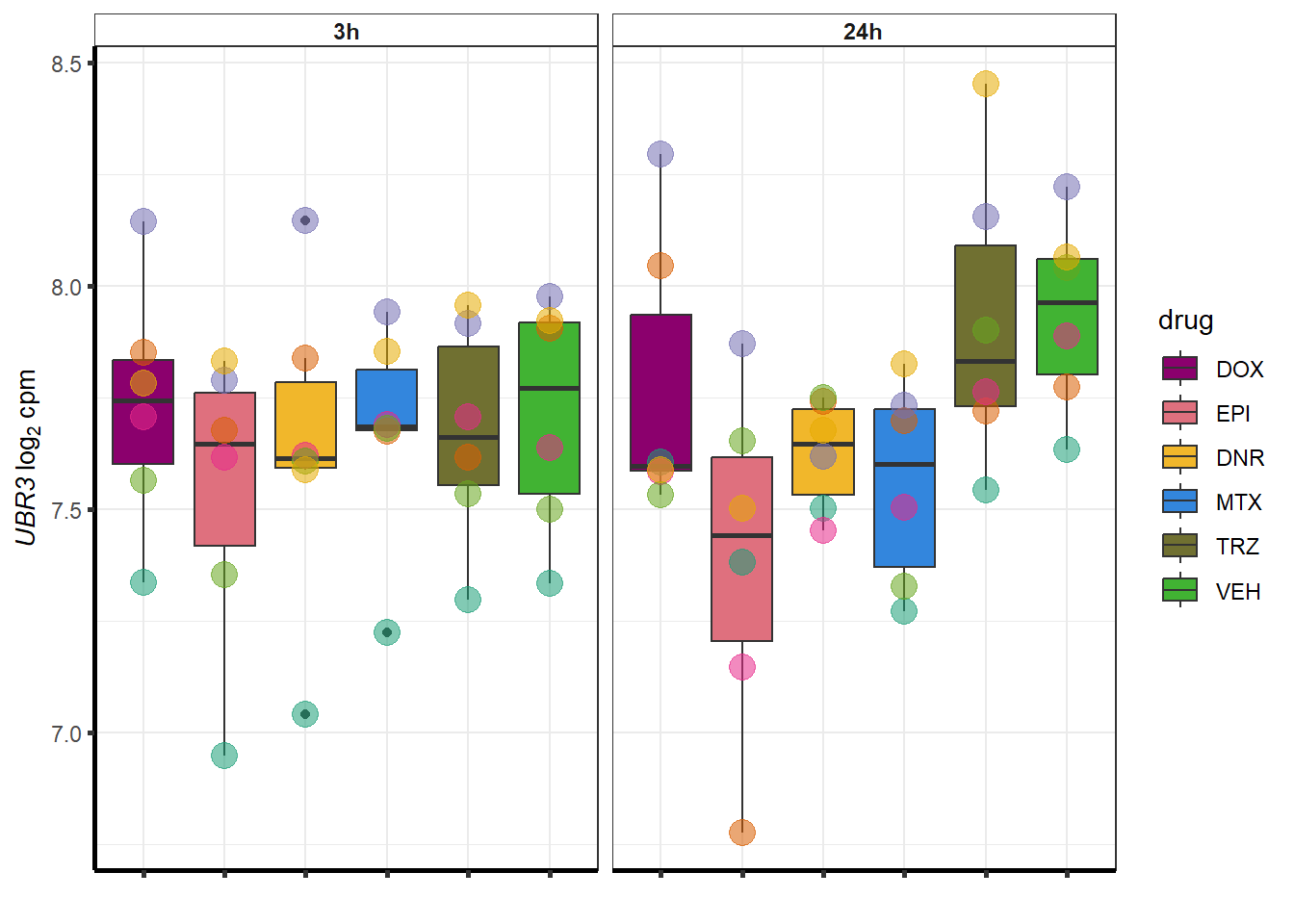
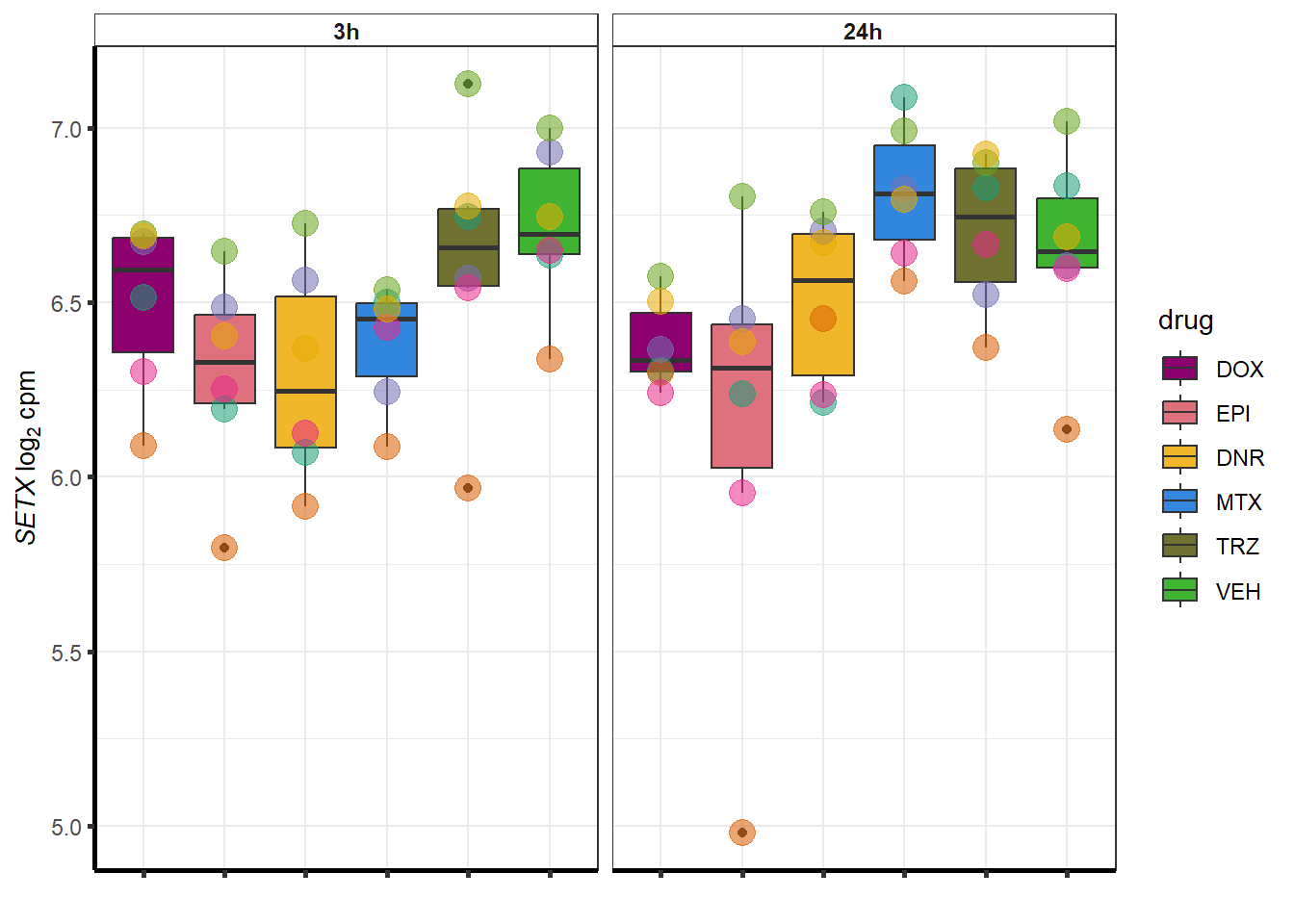
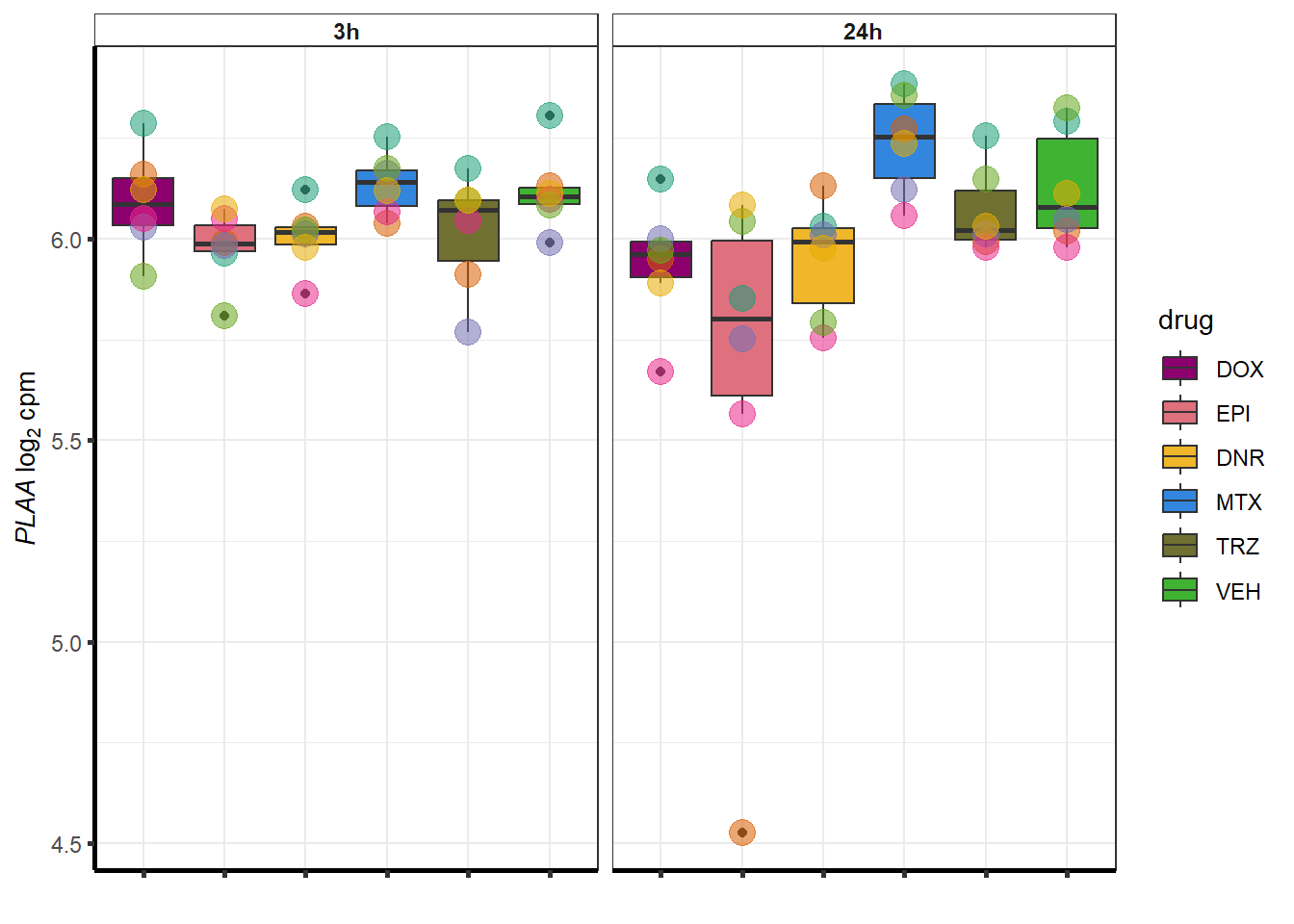
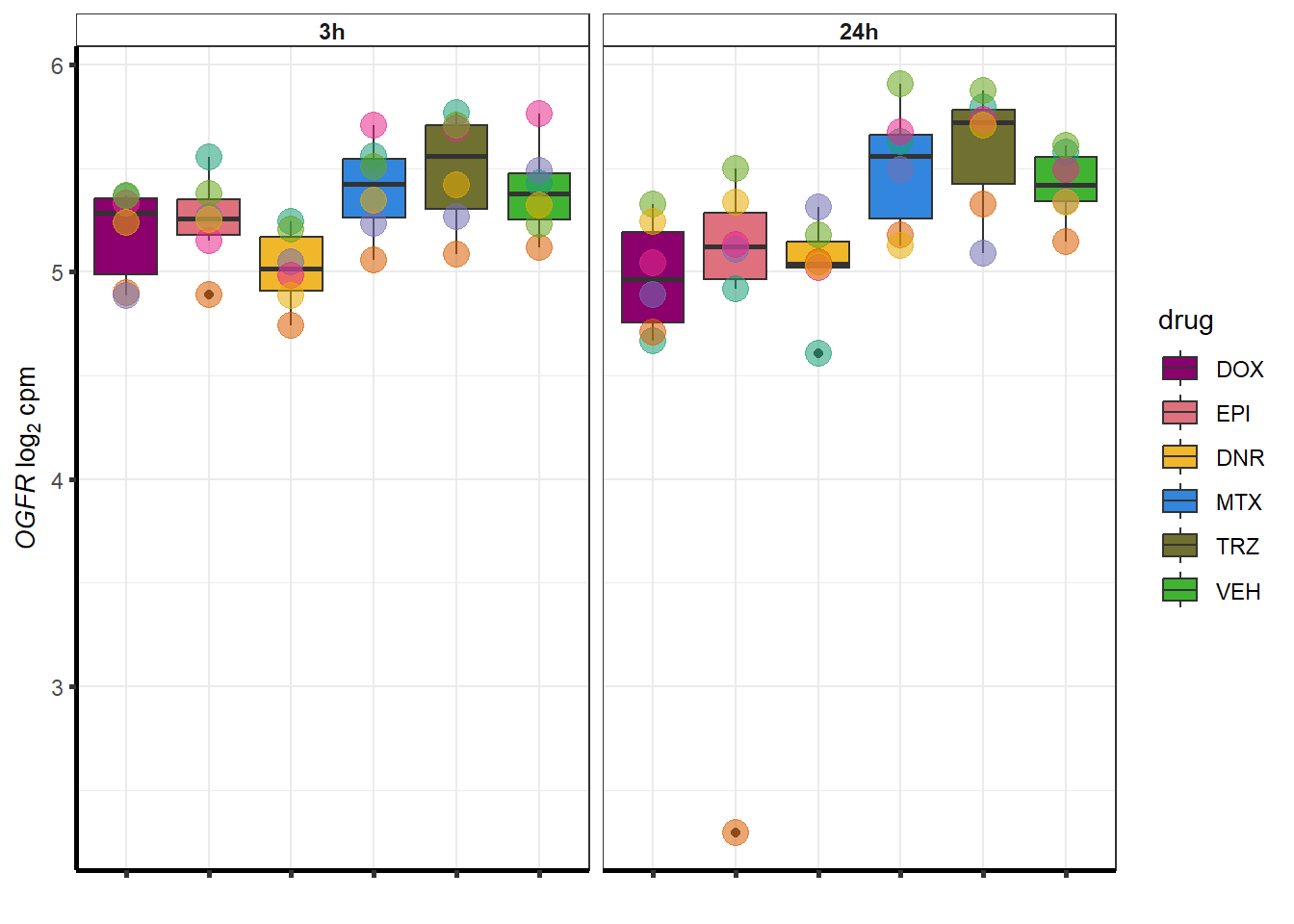
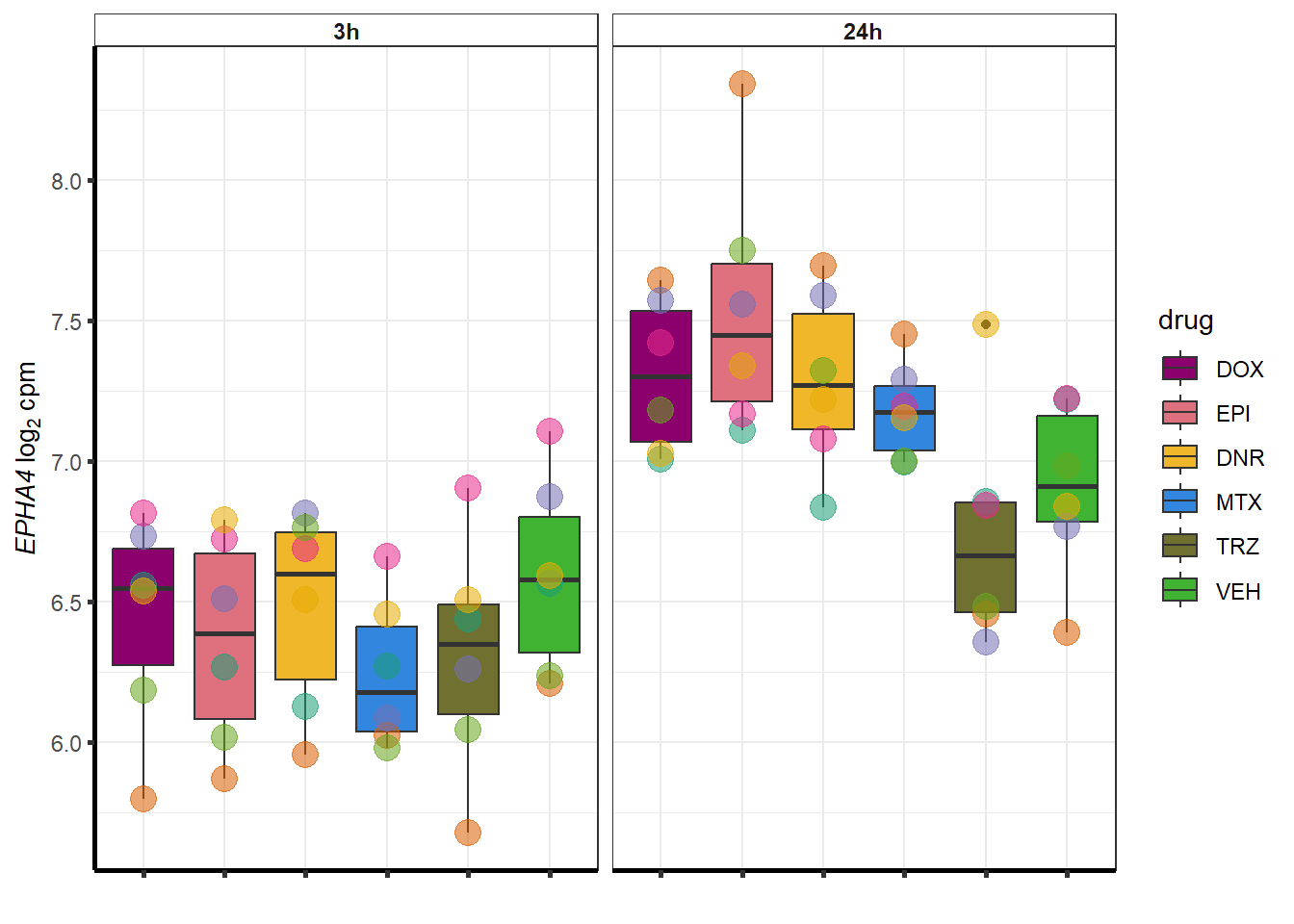
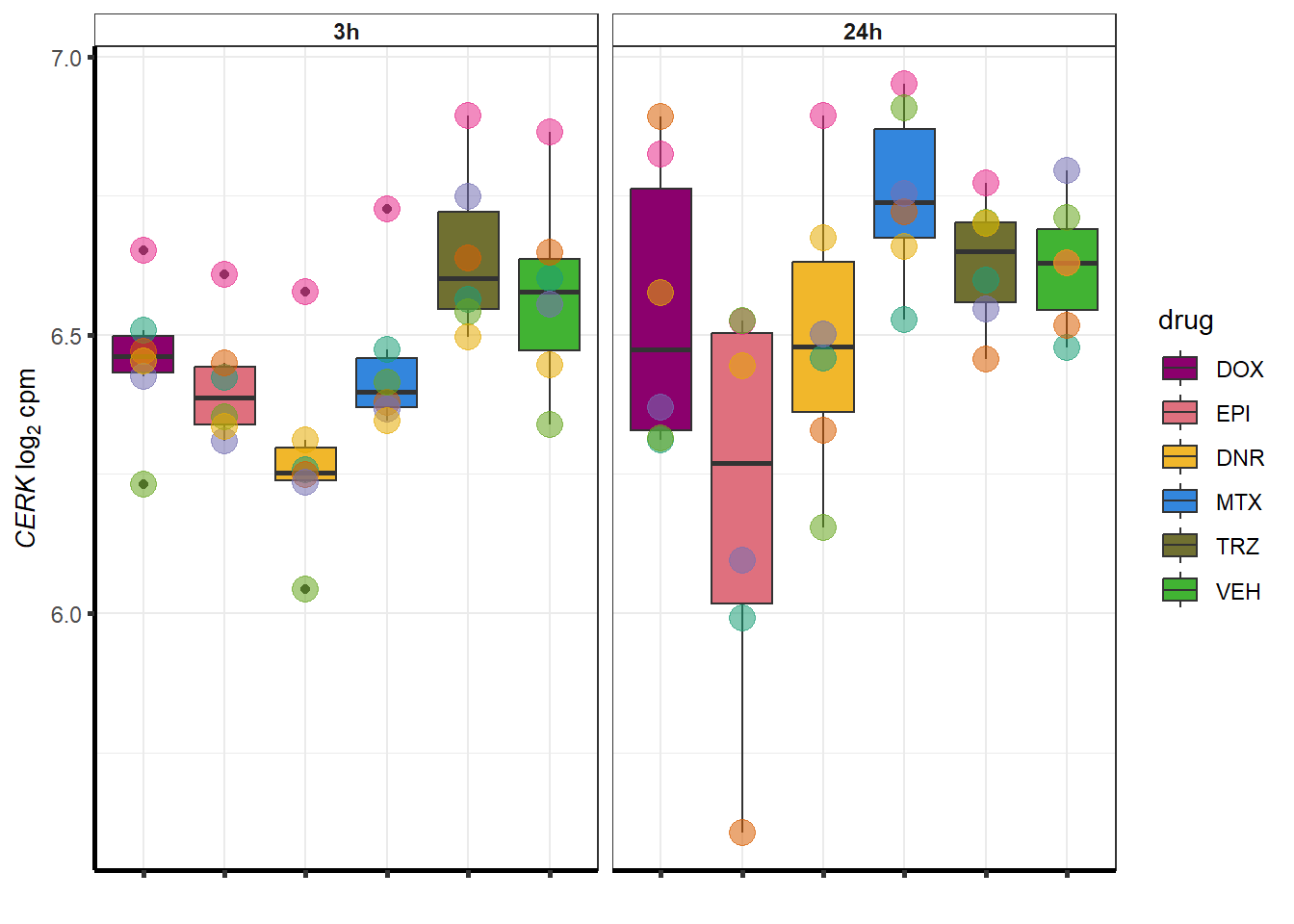
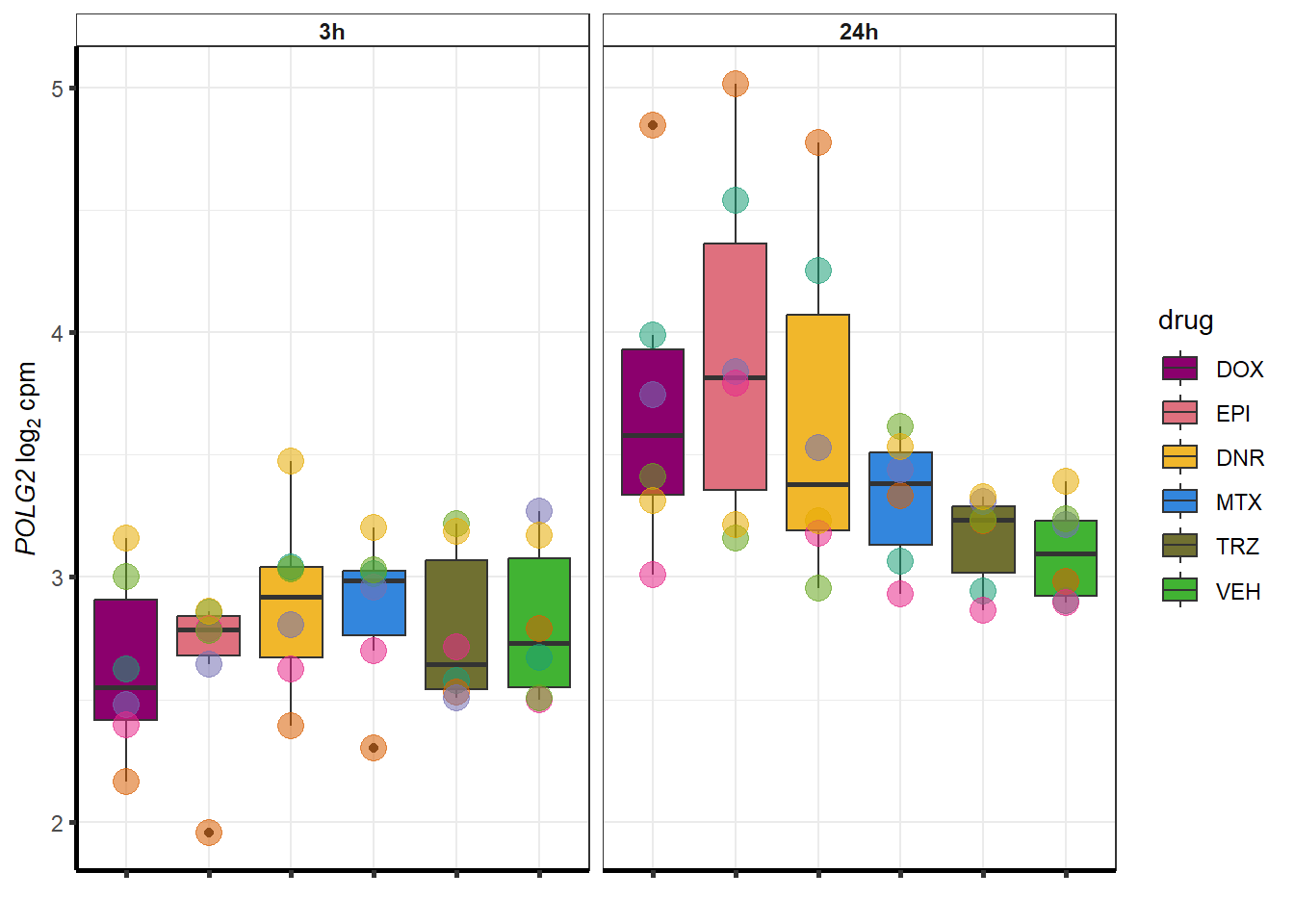
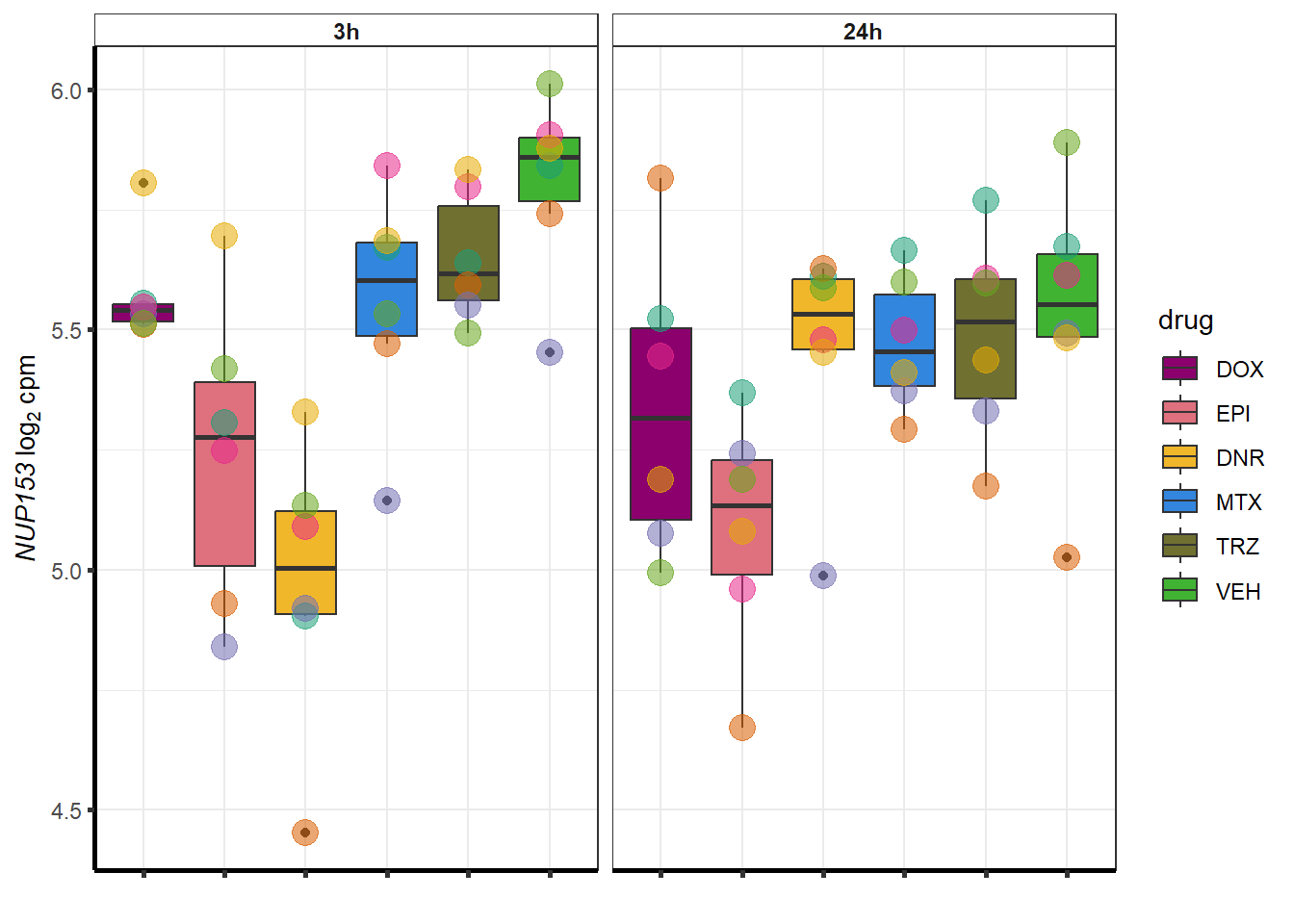
##most convincing of these is 220963 SLC16A9GO analysis 24h stringent adj.P.val <0.01 specific analysis
library(gprofiler2)
#
# DNRdeg_sp had NO enrichment
# EPIdeg_sp had NO enrichment
# MTXdeg_sep had 2, "DNA-templated DNA replication" "nuclear pore localization" "DNA replication"
# gostresMTXdeg_sp <- gost(query = c(MTXdeg_sp),
# organism = "hsapiens",
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP", "KEGG"))
# saveRDS(gostresMTXdeg_sp,"data/DEG-GO/gostresMTXdeg_sp.RDS")
# saveRDS(gostresDOXdeg_sp,"data/DEG-GO/gostresDOXdeg_sp.RDS")
DX_sp_DEGgostres <- readRDS("data/DEG-GO/gostresDOXdeg_sp.RDS")
MT_sp_DEGgostres <- readRDS("data/DEG-GO/gostresMTXdeg_sp.RDS")
DX_spgenes <- gostplot(DX_sp_DEGgostres, capped = FALSE, interactive = TRUE)
DX_spgenesMT_spgenes <- gostplot(MT_sp_DEGgostres, capped = FALSE, interactive = TRUE)
MT_spgenesDX_sp_DEGtable <- DX_sp_DEGgostres$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
MT_sp_DEGtable <- MT_sp_DEGgostres$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
DX_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = wrap_format(30))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('DOX specific gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))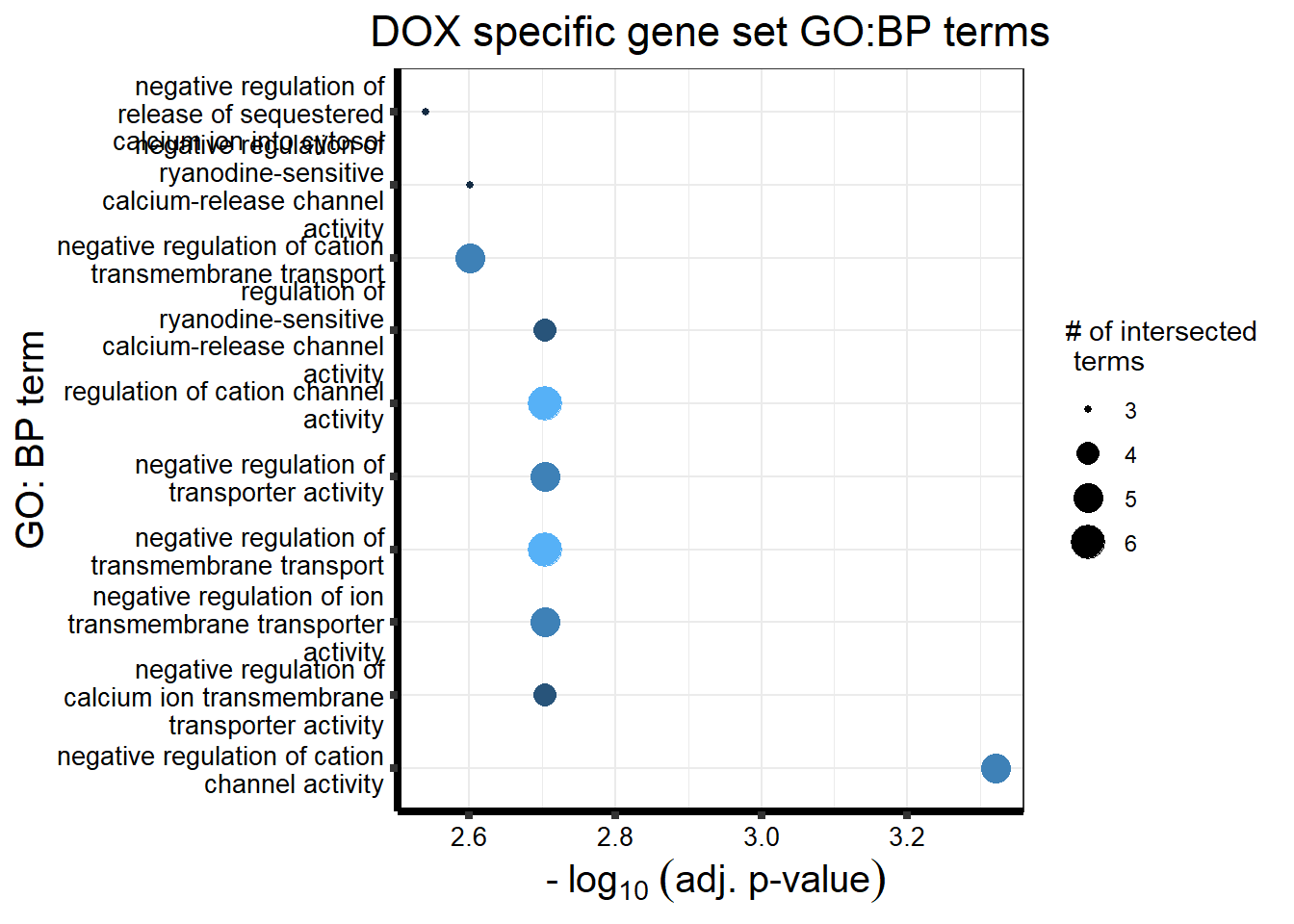
DX_sp_DEGtable %>%
mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Significant (adj. P value of <0.01) DOX specific genes (n = 68) and top 10 enriched GO terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped","hover")) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 5 | 30 | 4.78e-04 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 5 | 51 | 1.98e-03 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 4 | 27 | 1.98e-03 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 4 | 23 | 1.98e-03 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 5 | 58 | 1.98e-03 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 6 | 100 | 1.98e-03 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 6 | 96 | 1.98e-03 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 3 | 10 | 2.51e-03 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 5 | 64 | 2.51e-03 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 3 | 11 | 2.88e-03 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 5 | 71 | 2.95e-03 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 4 | 35 | 2.95e-03 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 5 | 70 | 2.95e-03 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 3 | 13 | 3.54e-03 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 4 | 45 | 7.02e-03 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 5 | 90 | 7.39e-03 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 9 | 391 | 9.58e-03 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 3 | 20 | 1.07e-02 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 4 | 60 | 1.72e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 3 | 25 | 1.91e-02 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 5 | 122 | 2.16e-02 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 6 | 190 | 2.16e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 3 | 27 | 2.16e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 3 | 28 | 2.16e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 3 | 28 | 2.16e-02 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 4 | 70 | 2.28e-02 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 6 | 199 | 2.34e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 3 | 31 | 2.62e-02 |
| GO:BP | GO:0032409 | regulation of transporter activity | 6 | 211 | 2.88e-02 |
| GO:BP | GO:0065009 | regulation of molecular function | 20 | 1939 | 2.88e-02 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 4 | 82 | 3.49e-02 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 4 | 83 | 3.54e-02 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 4 | 85 | 3.65e-02 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 4 | 85 | 3.65e-02 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 6 | 230 | 3.78e-02 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 2 | 9 | 3.78e-02 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 4 | 89 | 3.98e-02 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 4 | 90 | 4.04e-02 |
| GO:BP | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity | 2 | 10 | 4.35e-02 |
| GO:BP | GO:2001258 | negative regulation of cation channel activity | 4 | 30 | 1.88e-02 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 5 | 100 | 4.45e-02 |
| GO:BP | GO:0032413 | negative regulation of ion transmembrane transporter activity | 4 | 51 | 4.45e-02 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 5 | 96 | 4.45e-02 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 4 | 58 | 4.63e-02 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 3 | 23 | 4.63e-02 |
MT_sp_DEGtable %>%
mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Significant (adj. P value of <0.01) DOX specific genes (n = 41) and top 10 enriched GO terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped","hover")) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006261 | DNA-templated DNA replication | 6 | 148 | 4.77e-03 |
| GO:BP | GO:0051664 | nuclear pore localization | 2 | 4 | 3.13e-02 |
| GO:BP | GO:0006260 | DNA replication | 6 | 255 | 3.47e-02 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 6 | 148 | 4.77e-03 |
| GO:BP | GO:0051664 | nuclear pore localization | 2 | 4 | 3.13e-02 |
| GO:BP | GO:0006260 | DNA replication | 6 | 255 | 3.47e-02 |
Grid for supplement
densityDOXsp <- toplist24hr %>%
filter(ENTREZID %in% DoxonlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("DOX-only DEGs\n n = 355")+
guides(fill="none",alpha="none")+
theme(
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
densityDNRsp <- toplist24hr %>%
filter(ENTREZID %in% DnronlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("DNR-only DEGs\n n = 615")+
guides(fill="none",alpha="none")+
theme(
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
densityEPIsp <- toplist24hr %>%
filter(ENTREZID %in% EpionlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("EPI-only DEGs\n n = 444")+
guides(fill="none",alpha="none")+
theme(axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
densityMTXsp <- toplist24hr %>%
filter(ENTREZID %in% MtxonlyDEG) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("MTX-only DEGs\n n = 122")+
guides(fill="none",alpha="none")+
theme(axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0)) DoxBplot <- toplist24hr %>%
group_by(time,id) %>%
dplyr::filter(ENTREZID %in% DOXdeg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("DOX specific adj.P.V <0.01\n n = 68")+
theme_classic()+
fill_palette(palette = drug_palc)+
guides(fill= FALSE)+
theme(
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
DnrBplot <- toplist24hr %>%
group_by(time,id) %>%
dplyr::filter(ENTREZID %in% DNRdeg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("DNR-spec. adj.P.V <0.01\n n = 112")+
theme_classic()+
guides(fill= FALSE)+
fill_palette(palette = drug_palc)+
theme(
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
EpiBplot <- toplist24hr %>%
group_by(time,id) %>%
filter(ENTREZID %in% EPIdeg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("EPI-spec.adj.P.V <0.01\n n = 84")+
theme_classic()+
guides(fill= FALSE)+
fill_palette(palette = drug_palc)+
theme(axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
MtxBplot <- toplist24hr %>%
group_by(time,id) %>%
filter(ENTREZID %in% MtxonlyDEG) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("MTX-spec. adj.P.V <0.01\n n = 41")+
guides(fill= FALSE)+
theme_classic()+
fill_palette(palette = drug_palc)+
theme(axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
Doxgenesp_example <- cpm_boxplot_time(cpmcounts,"24h",GOI=101927720,"Dark2",drug_palc, ylab=bquote(~italic("ZNF793-AS1 ")~log[2]~"cpm "))+
theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))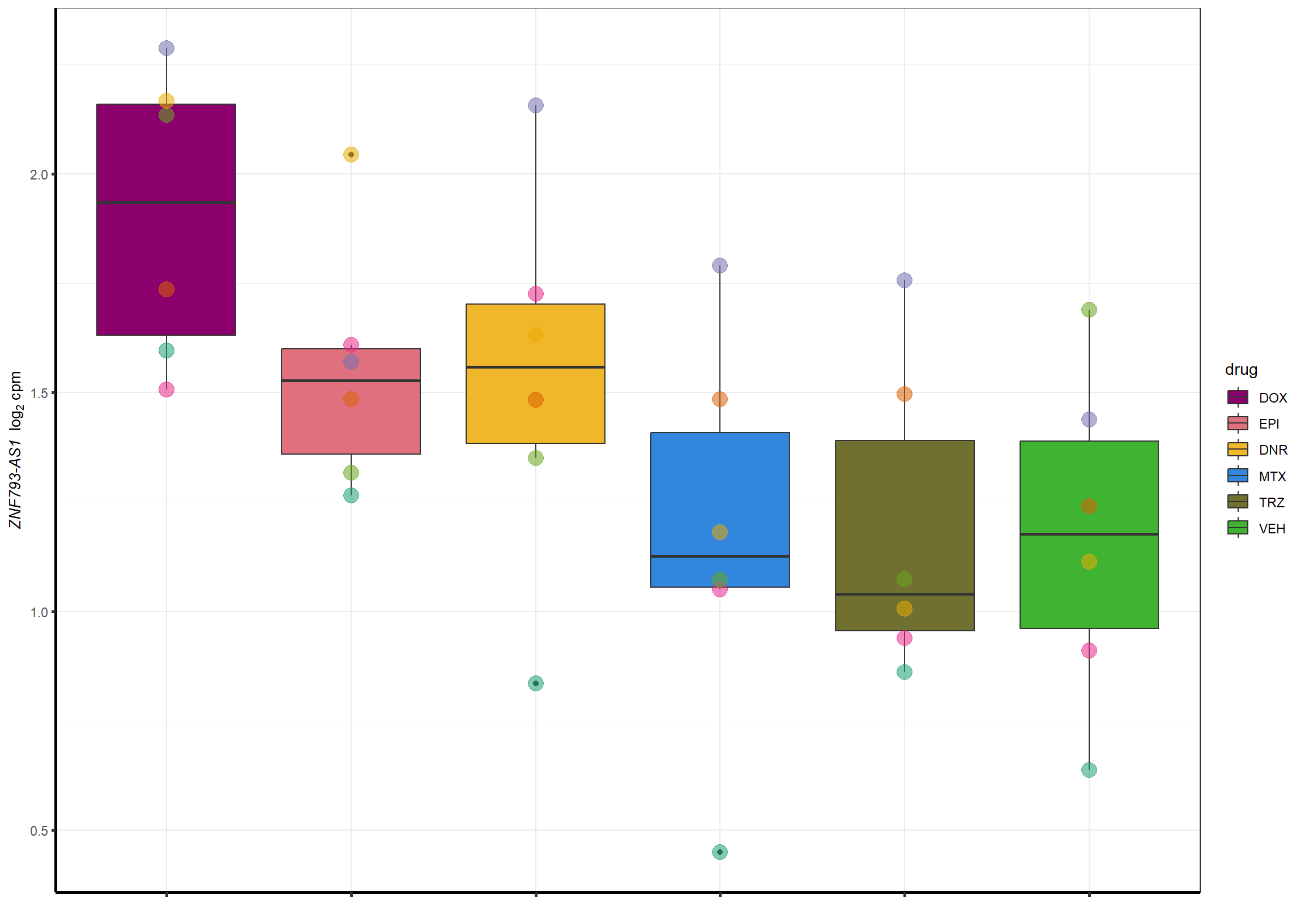
Dnrgenesp_example <- cpm_boxplot_time(cpmcounts,"24h",114826 ,"Dark2",drug_palc,
ylab=bquote(~italic("SMYD4")~log[2]~"cpm "))+
theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))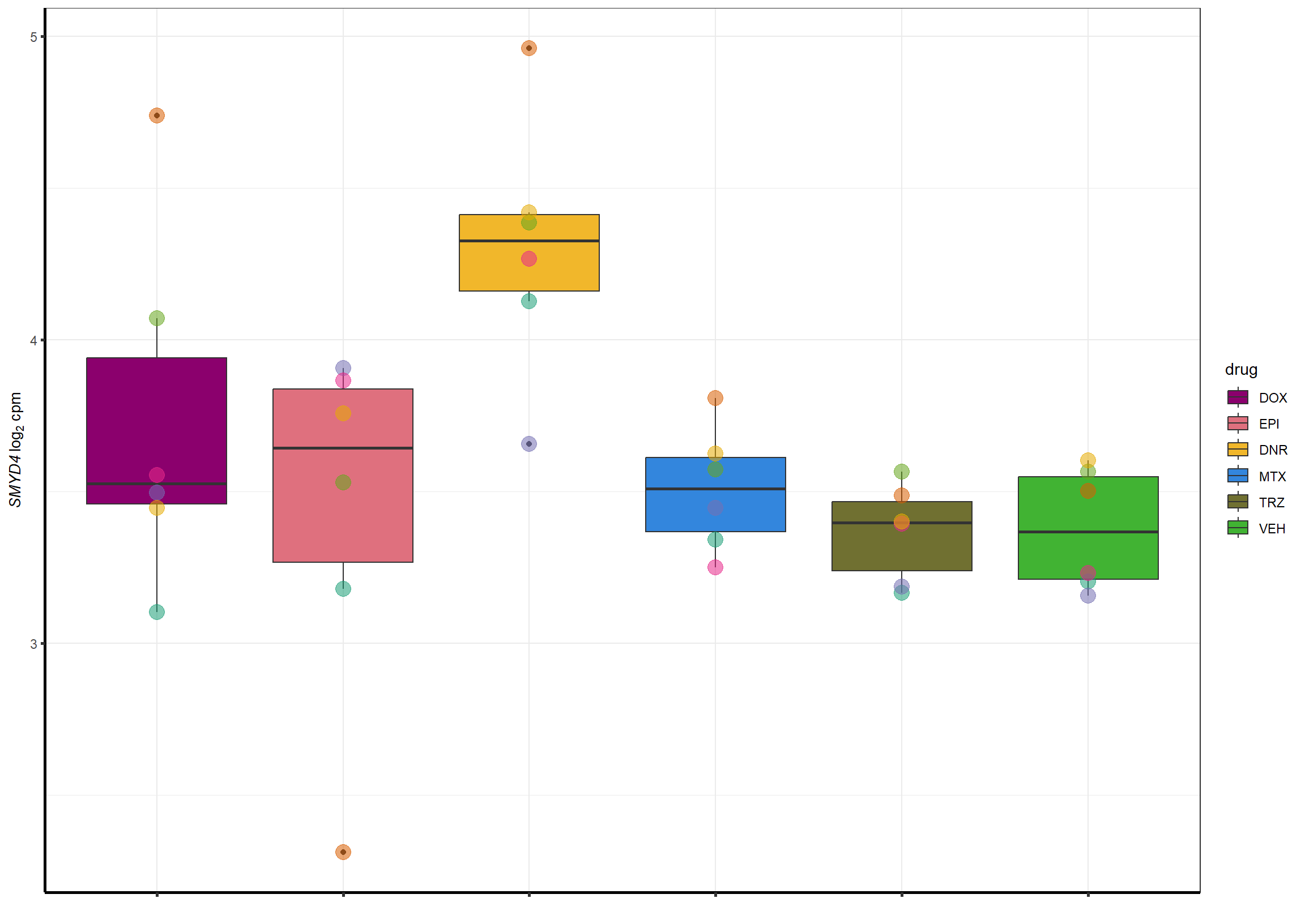
Epigenesp_example <- cpm_boxplot_time(cpmcounts,"24h",GOI=220963,"Dark2",drug_palc,
ylab=bquote(~italic("SLC16A9")~log[2]~"cpm "))+
theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))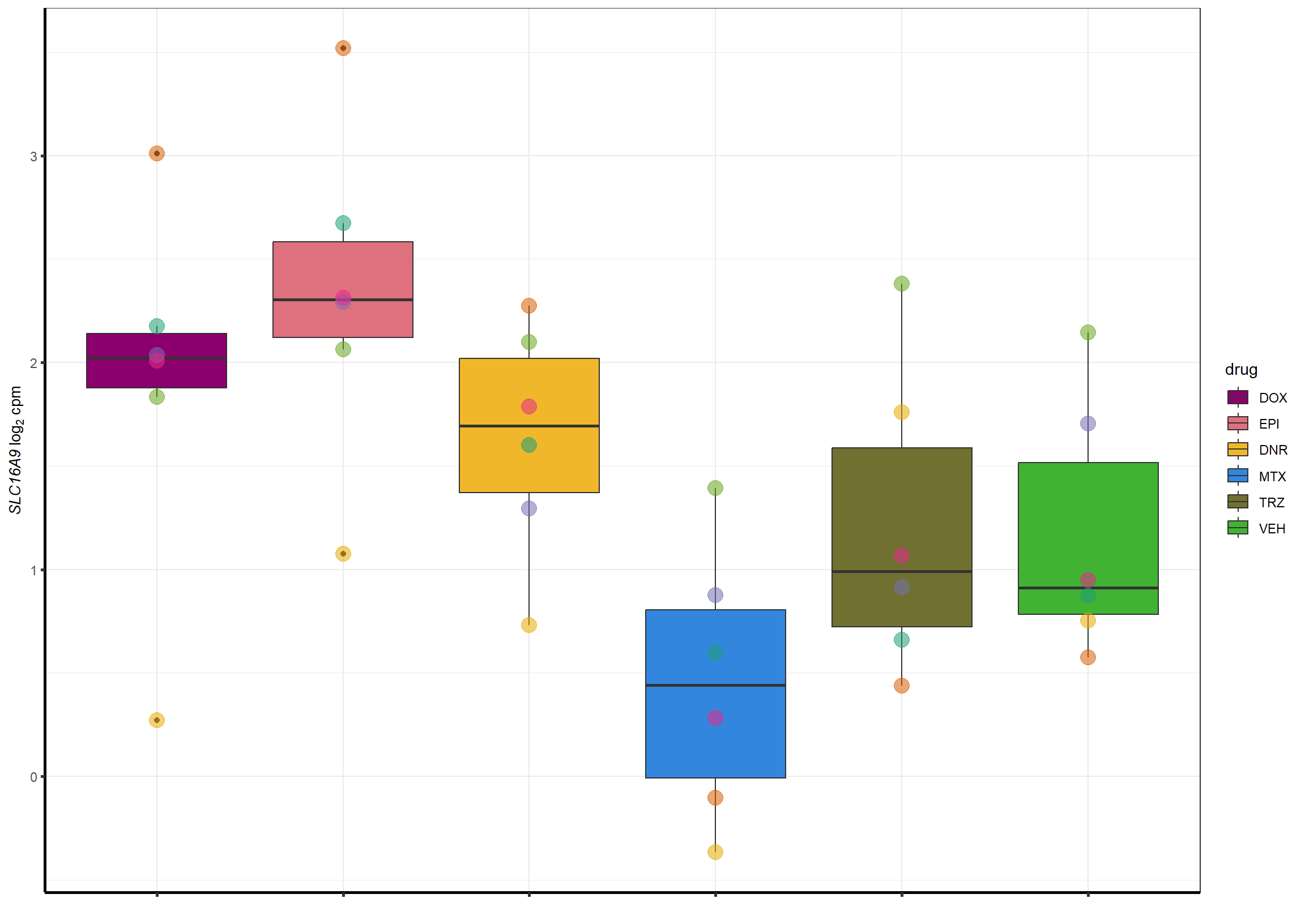
Mtxgenesp_example <- cpm_boxplot_time(cpmcounts,"24h",GOI=54853,"Dark2",drug_palc,
ylab=bquote(~italic("WDR55")~log[2]~"cpm "))+
theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))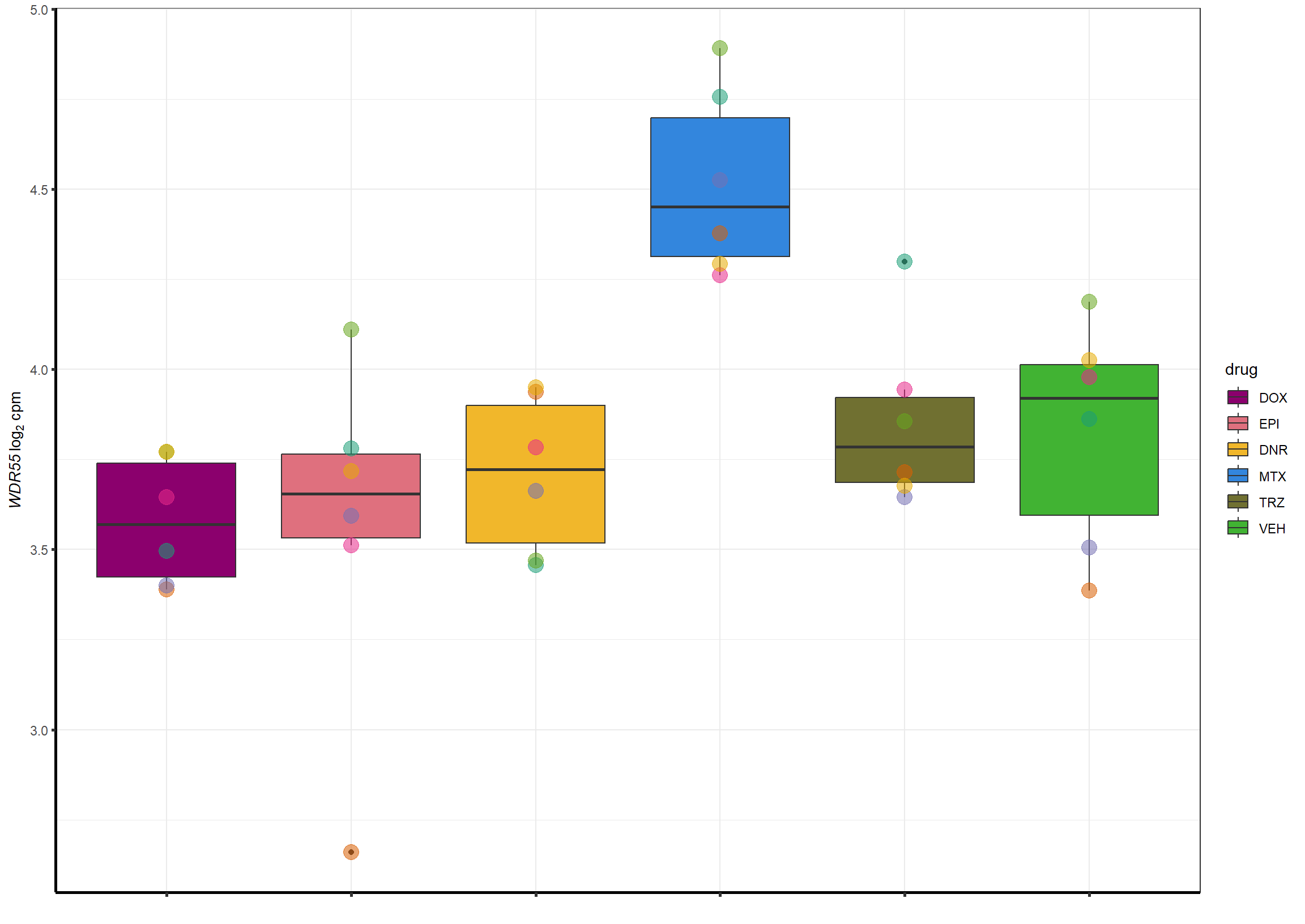
# supp10_24hlist <- list(densityDNRsp, DnrBplot, Dnrgenesp_example,densityDOXsp, DoxBplot, Doxgenesp_example,densityEPIsp, EpiBplot, Epigenesp_example,densityMTXsp, MtxBplot, Mtxgenesp_example)
#
# names(supp10_24hlist) <- c("densityDNRsp", "DnrBplot", "Dnrgenesp_example","densityDOXsp", "DoxBplot", "Doxgenesp_example","densityEPIsp", "EpiBplot", "Epigenesp_example","densityMTXsp", "MtxBplot", "Mtxgenesp_example")
mitopg <- plot_grid(densityMTXsp, MtxBplot, Mtxgenesp_example,
nrow = 1,
rel_heights = c(.8,2,1),
rel_widths=c(1.2,1,1.5),
scale=c(1,0.8,0.8))
Daunpg <- plot_grid(densityDNRsp, DnrBplot, Dnrgenesp_example, nrow = 1, rel_heights = c(.8,2,1), rel_widths=c(1.2,1,1.5),scale=c(1,0.8,0.8))
Doxopg <- plot_grid(densityDOXsp, DoxBplot, Doxgenesp_example, nrow = 1, rel_heights = c(.8,2,1), rel_widths=c(1.2,1,1.5),scale=c(1,0.8,0.8))
Epipg <- plot_grid(densityEPIsp, EpiBplot, Epigenesp_example, nrow = 1, rel_heights = c(.8,2,1), rel_widths=c(1.2,1,1.5),scale=c(1,0.8,0.8))
allfinal <- plot_grid(Doxopg,Epipg,Daunpg,mitopg,nrow=4)
allfinal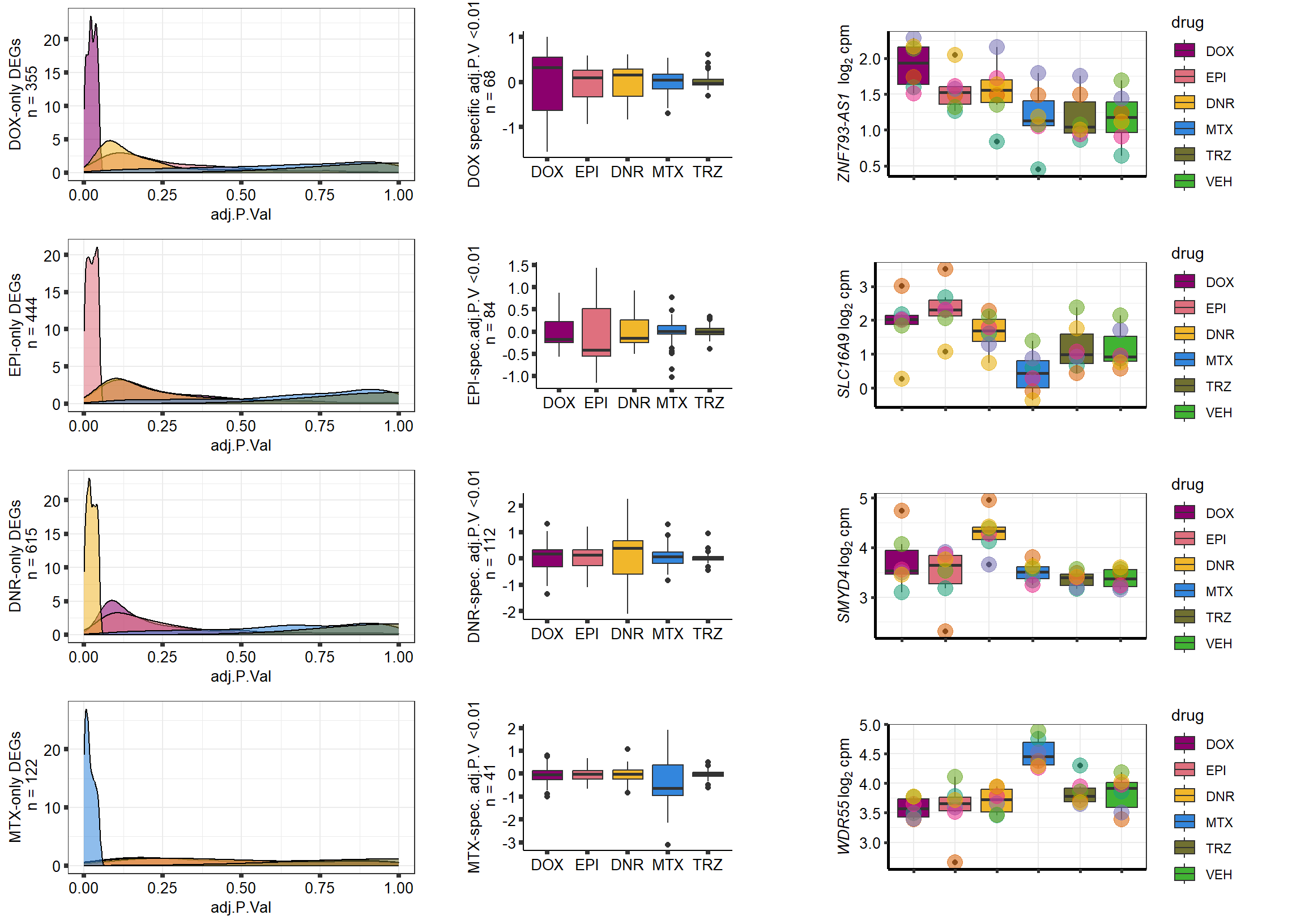
# saveRDS(supp10_24hlist,"data/supp10_24hlist.RDS")
# saveRDS(allfinal,"output/allfinal_sup10.RDS")3 hour update
siglist <- readRDS("data/siglist_final.RDS")
list2env(siglist,.GlobalEnv)<environment: R_GlobalEnv>total3h <-list(sigVDA3$ENTREZID,sigVDX3$ENTREZID,sigVEP3$ENTREZID,sigVMT3$ENTREZID)
## do venn partition and pull doxspe genes
# venn_3h <- VennDiagram::get.venn.partitions(total3h)
# Dox3onlyDEG <- venn_3h$..values..[[14]]## no genes
# Epi3onlyDEG <- venn_3h$..values..[[12]]## 18 genes
# Dnr3onlyDEG <- venn_3h$..values..[[15]]## 322
# Mtx3onlyDEG <- venn_3h$..values..[[8]] ## 16
# trtonly_3h_genes <- list(Dnr3onlyDEG,Dox3onlyDEG,Epi3onlyDEG,Mtx3onlyDEG)
# names(trtonly_3h_genes) <-c("Dnr3onlyDEG","Dox3onlyDEG","Epi3onlyDEG","Mtx3onlyDEG")
# saveRDS(trtonly_3h_genes,"data/trtonly_3h_genes.RDS")
trtonly_3h_genes <- readRDS("data/trtonly_3h_genes.RDS")
list2env(trtonly_3h_genes,.GlobalEnv)<environment: R_GlobalEnv>DOX3deg_sp <- toplist3hr %>%
dplyr::filter(ENTREZID %in% Dox3onlyDEG) %>%
dplyr::filter(adj.P.Val<0.01) %>%
filter(id=="DOX") %>%
dplyr::select(ENTREZID, SYMBOL)
EPI3deg_sp <- toplist3hr %>%
dplyr::filter(ENTREZID %in% Epi3onlyDEG) %>%
dplyr::filter(adj.P.Val<0.01) %>%
filter(id=="EPI") %>%
dplyr::select(ENTREZID, SYMBOL)
DNR3deg_sp <- toplist3hr %>%
dplyr::filter(ENTREZID %in% Dnr3onlyDEG) %>%
dplyr::filter(adj.P.Val<0.01) %>%
filter(id=="DNR") %>%
dplyr::select(ENTREZID, SYMBOL)
MTX3deg_sp <- toplist3hr %>%
dplyr::filter(ENTREZID %in% Mtx3onlyDEG) %>%
dplyr::filter(adj.P.Val<0.01) %>%
filter(id=="MTX") %>%
dplyr::select(ENTREZID, SYMBOL)
# set.seed(12345)
# sampset <- EPI3deg_sp %>%
# sample_n(.,2)
#
# for (g in seq(from=1, to=length(sampset$ENTREZID))){
# a <- sampset$SYMBOL[g]
# cpm_boxplot(cpmcounts,GOI=sampset[g,1],"Dark2",drug_palc,
# ylab=bquote(~italic(.(a))~log[2]~"cpm "))
# }
set.seed(12345)
sampset <- DNR3deg_sp %>%
sample_n(.,5)
print("DNR specfic")[1] "DNR specfic"for (g in seq(from=1, to=length(sampset$ENTREZID))){
a <- sampset$SYMBOL[g]
cpm_boxplot_time(cpmcounts,"3h",GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}
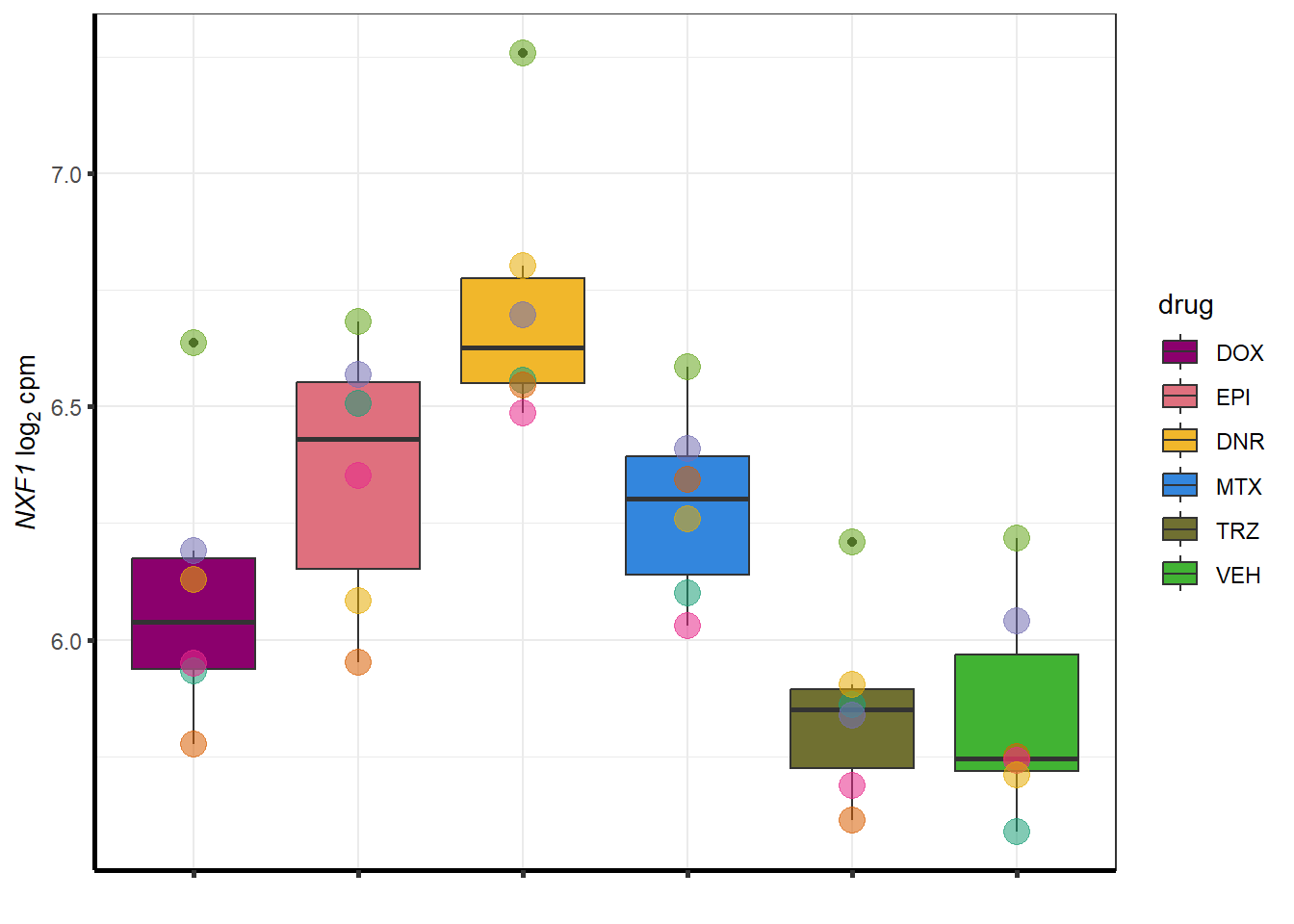
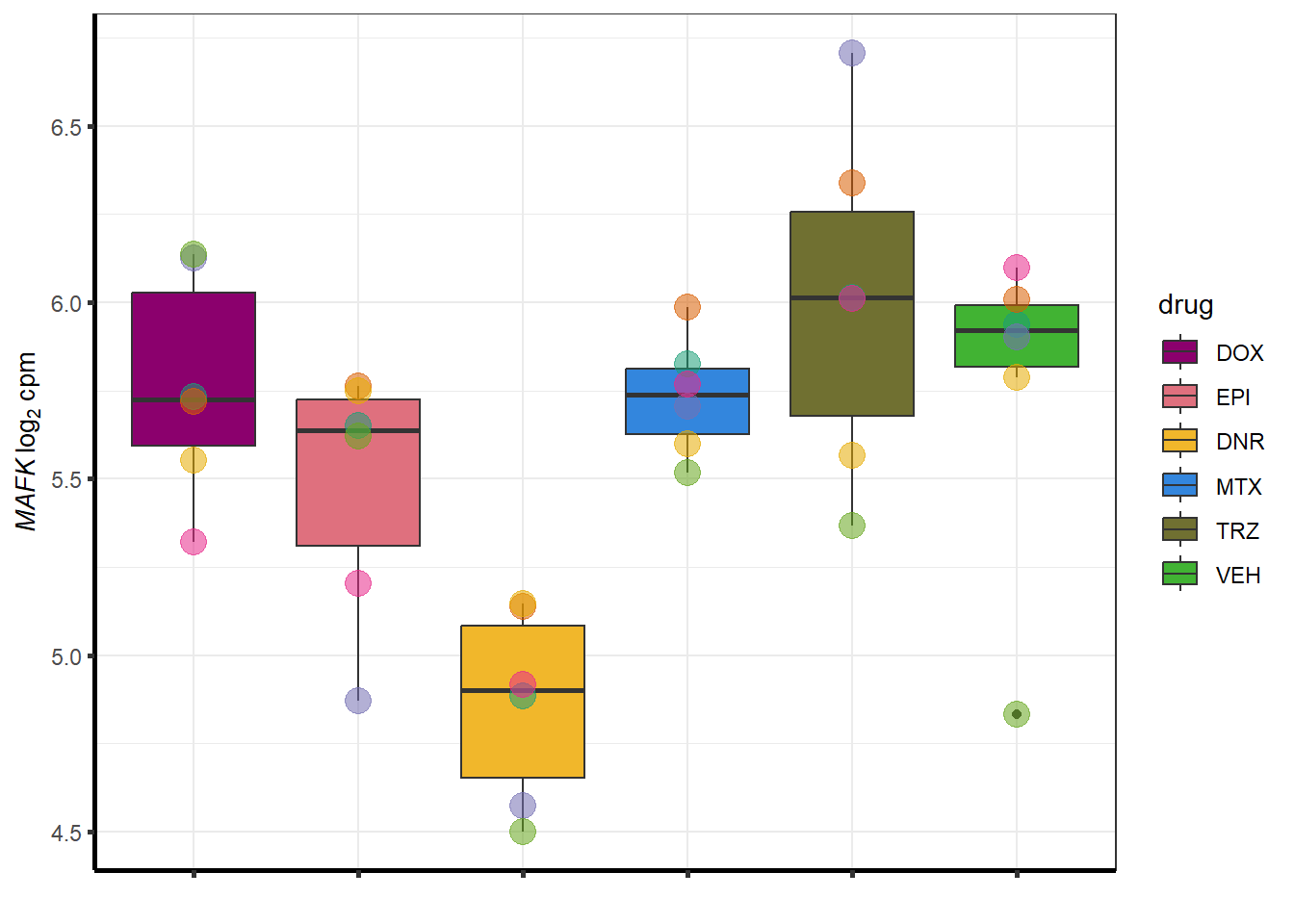
set.seed(12345)
sampset <- MTX3deg_sp %>%
sample_n(.,2)
print("MTX specfic")[1] "MTX specfic"for (g in seq(from=1, to=length(sampset$ENTREZID))){
a <- sampset$SYMBOL[g]
cpm_boxplot_time(cpmcounts,"3h",GOI=sampset[g,1],"Dark2",drug_palc,
ylab=bquote(~italic(.(a))~log[2]~"cpm "))
}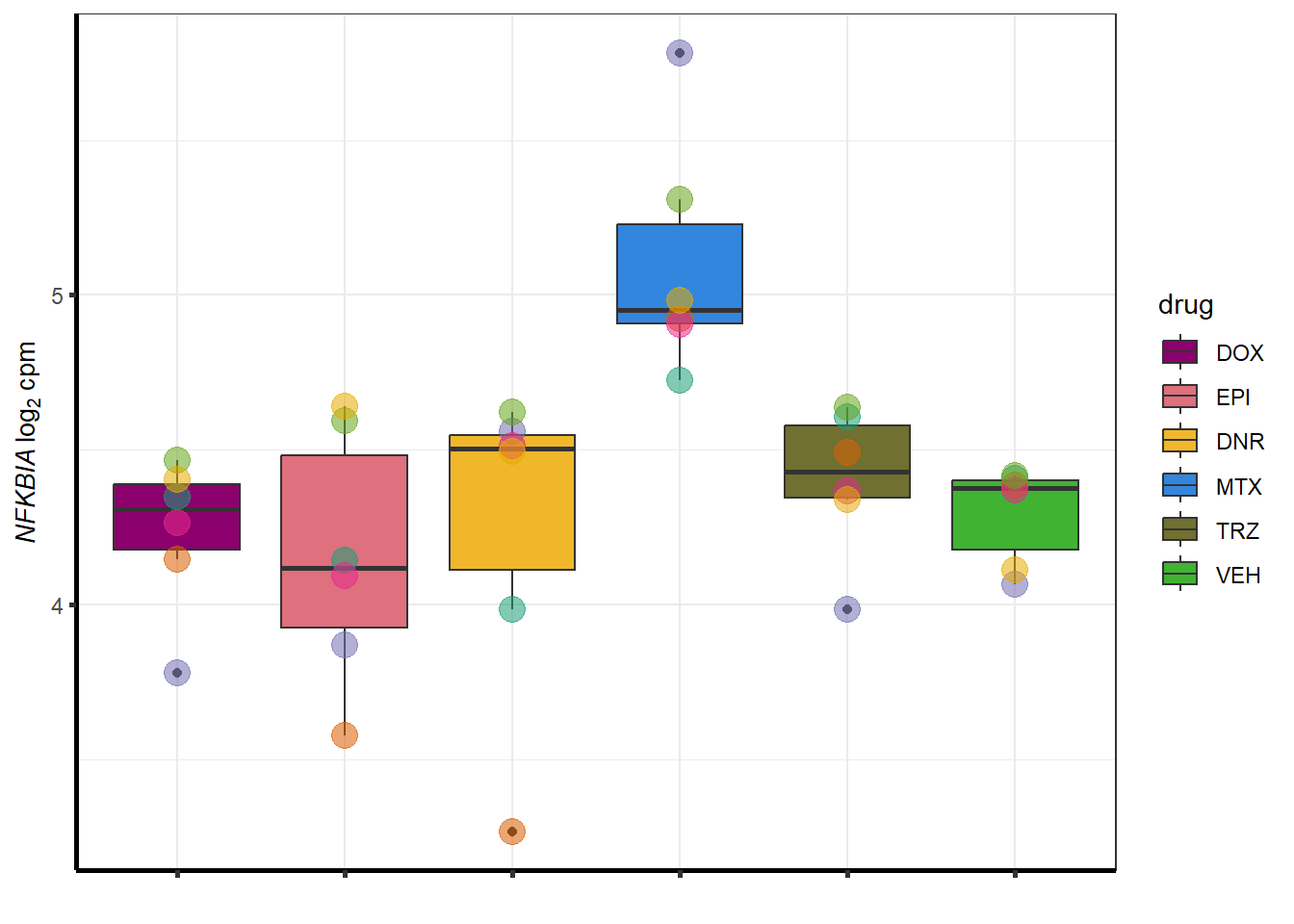
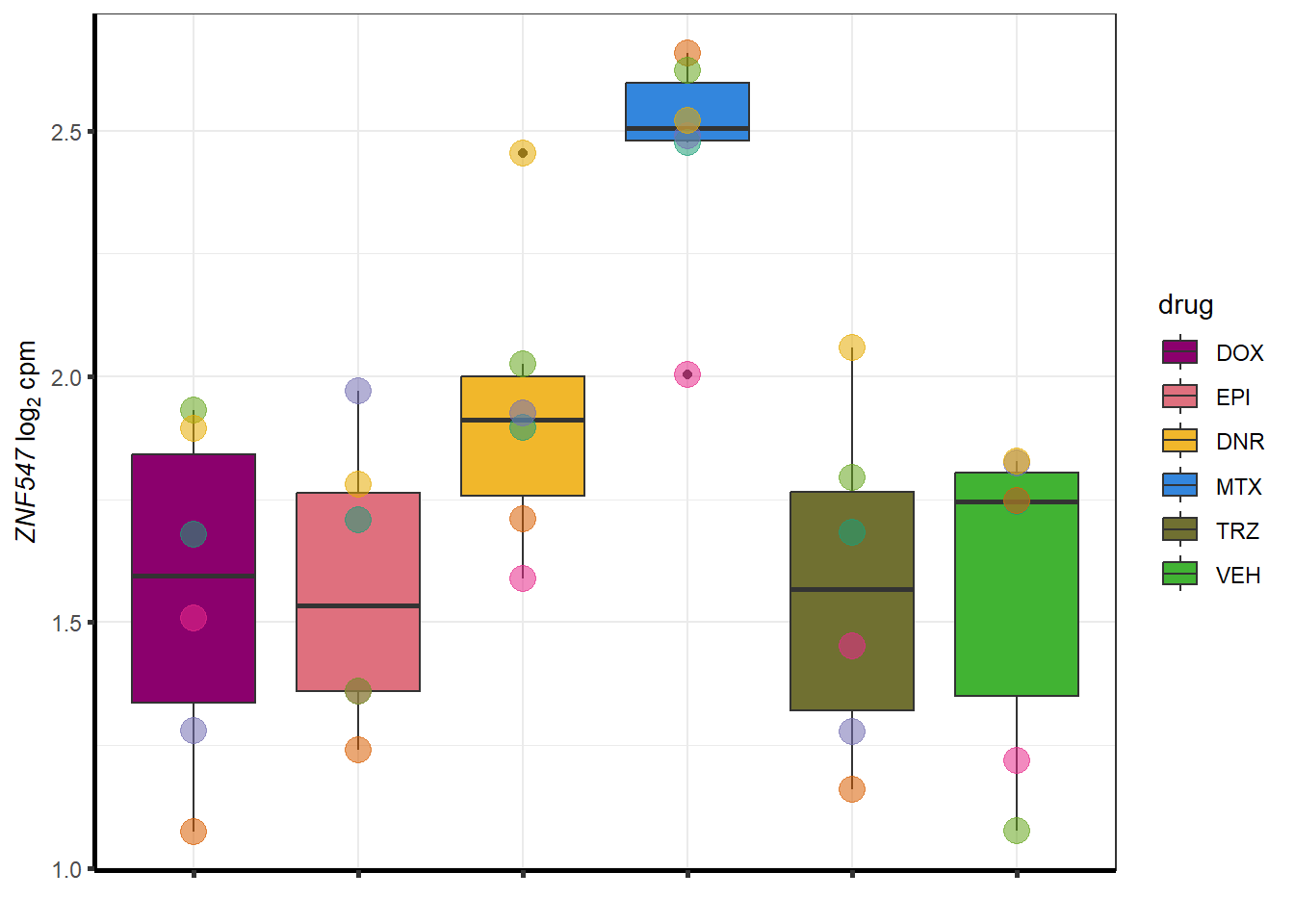
library(paletteer)
in_common3sp <- c(Dnr3onlyDEG,Dox3onlyDEG,Epi3onlyDEG,Mtx3onlyDEG)
ggVennDiagram::ggVennDiagram(trtonly_3h_genes,
category.names = c("DNR_sp\nn = 322 ",
"DOX_sp\n n = 0\n",
"EPI_sp\n n = 18\n",
" MTX_sp\n n = 16"),
show_intersect = FALSE,
set_color = "black",
category_size = c(6,6,6,6),
label = "count",
label_percent_digit = 1,
label_size = 4,
label_alpha = 0,
label_color = "black",
edge_lty = "solid", set_size = 4.5)+
scale_x_continuous(expand = expansion(mult = .3))+
scale_y_continuous(expand = expansion(mult = .2))+
scale_color_paletteer_d(palette = "fishualize::Bodianus_pulchellus")+
scale_fill_distiller(palette="Spectral", direction = -1)+
labs(title = "3 hour treatment specific genes p.adj. <0.05",
caption = paste("n =", length(unique(in_common3sp)),"genes"))+
theme(plot.title = element_text(size = rel(1.6), hjust = 0.5, vjust =1))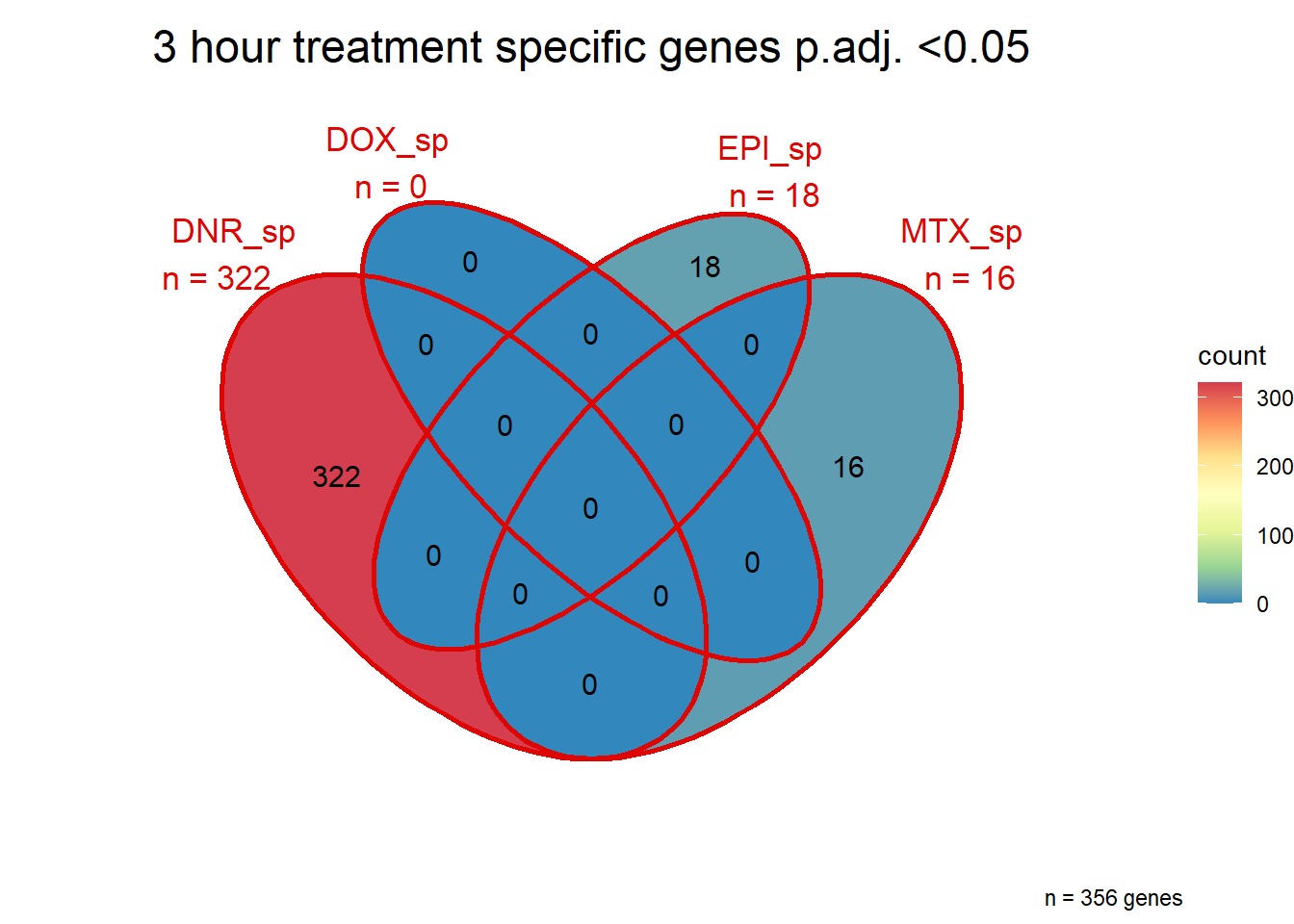
GO analysis 3h stringent adj.P.val <0.01 specific analysis
#
# DNRdeg_sp had NO enrichment
# EPIdeg_sp had NO enrichment
# MTXdeg_sep had 3, "DNA-templated DNA replication" "nuclear pore localization" "DNA replication"
# gostres3Mtxdeg_sp <- gost(query = MTX3deg_sp$ENTREZID,
# organism = "hsapiens",
# ordered_query = FALSE,
# domain_scope = "custom",
# significant = FALSE,
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = backGL$ENTREZID,
# sources=c("GO:BP", "KEGG"))
# saveRDS(gostres3Dnrdeg_sp,"data/DEG-GO/gostres3Dnrdeg_sp.RDS")
# saveRDS(gostres3Mtxdeg_sp,"data/DEG-GO/gostres3Mtxdeg_sp.RDS")
gostres3Dnrdeg_sp <- readRDS("data/DEG-GO/gostres3Dnrdeg_sp.RDS")
gostres3Mtxdeg_sp <- readRDS("data/DEG-GO/gostres3Mtxdeg_sp.RDS")
Dnr3_spgenes <- gostplot(gostres3Dnrdeg_sp, capped = FALSE, interactive = TRUE)
Dnr3_spgenesMtx3_spgenes <- gostplot(gostres3Mtxdeg_sp, capped = FALSE, interactive = TRUE)
Mtx3_spgenesMtx3_sp_DEGtable <- gostres3Mtxdeg_sp$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
Dnr3_sp_DEGtable <- gostres3Dnrdeg_sp$result %>%
dplyr::select(c(source, term_id,term_name,intersection_size,
term_size, p_value))
Dnr3_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = wrap_format(30))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('DNR 3 hour specific(stringent)\n gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))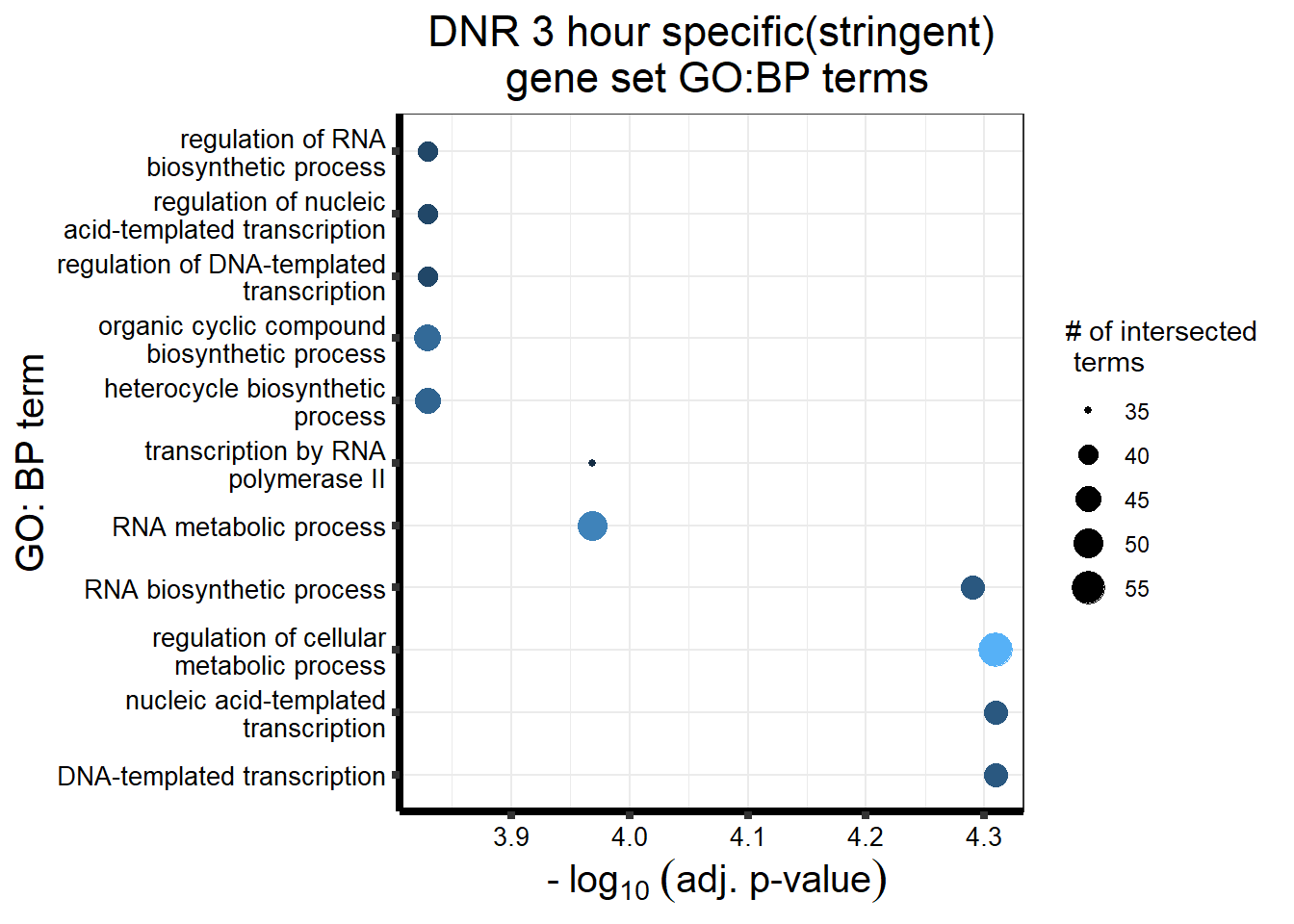
Dnr3_sp_DEGtable %>%
mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Stringent (adj. P value of <0.01) DNR 3 hour specific genes (n = 100) enriched GO and KEGG terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped","hover")) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006351 | DNA-templated transcription | 43 | 2683 | 4.90e-05 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 43 | 2684 | 4.90e-05 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 57 | 4174 | 4.90e-05 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 43 | 2714 | 5.13e-05 |
| GO:BP | GO:0016070 | RNA metabolic process | 50 | 3600 | 1.08e-04 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 35 | 1995 | 1.08e-04 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 40 | 2574 | 1.48e-04 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 45 | 3132 | 1.48e-04 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 46 | 3237 | 1.48e-04 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 40 | 2593 | 1.48e-04 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 40 | 2576 | 1.48e-04 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 33 | 1914 | 1.66e-04 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 50 | 3730 | 1.66e-04 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 42 | 2845 | 1.73e-04 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 44 | 3080 | 1.88e-04 |
| GO:BP | GO:0019222 | regulation of metabolic process | 60 | 5017 | 1.97e-04 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 52 | 4048 | 2.33e-04 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 43 | 3012 | 2.33e-04 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 44 | 3133 | 2.42e-04 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 44 | 3169 | 3.18e-04 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 43 | 3070 | 3.39e-04 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 43 | 3080 | 3.54e-04 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 55 | 4539 | 4.60e-04 |
| GO:BP | GO:0009058 | biosynthetic process | 55 | 4590 | 6.45e-04 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 53 | 4417 | 1.09e-03 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 53 | 4570 | 3.06e-03 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 52 | 4455 | 3.17e-03 |
| GO:BP | GO:0010467 | gene expression | 53 | 4587 | 3.19e-03 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 53 | 4596 | 3.27e-03 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 46 | 3750 | 3.48e-03 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 54 | 4747 | 3.48e-03 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 50 | 4245 | 3.48e-03 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 50 | 4292 | 4.63e-03 |
| GO:BP | GO:0010468 | regulation of gene expression | 44 | 3580 | 4.98e-03 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 52 | 4617 | 7.08e-03 |
| GO:BP | GO:0140467 | integrated stress response signaling | 4 | 35 | 7.50e-03 |
| GO:BP | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | 3 | 16 | 1.23e-02 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 53 | 4946 | 2.18e-02 |
| GO:BP | GO:0044237 | cellular metabolic process | 68 | 6996 | 2.19e-02 |
| GO:BP | GO:2000121 | regulation of removal of superoxide radicals | 2 | 6 | 4.47e-02 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 17 | 1017 | 5.42e-02 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 6 | 156 | 5.42e-02 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 17 | 1015 | 5.42e-02 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 17 | 1026 | 5.84e-02 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 5 | 108 | 5.87e-02 |
| GO:BP | GO:0050794 | regulation of cellular process | 70 | 7552 | 7.07e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 7 | 240 | 9.07e-02 |
| GO:BP | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress | 3 | 36 | 1.08e-01 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 18 | 1209 | 1.23e-01 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 18 | 1214 | 1.24e-01 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 13 | 740 | 1.24e-01 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 17 | 1119 | 1.25e-01 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 23 | 1735 | 1.25e-01 |
| GO:BP | GO:0065007 | biological regulation | 74 | 8320 | 1.35e-01 |
| GO:BP | GO:0043620 | regulation of DNA-templated transcription in response to stress | 3 | 41 | 1.38e-01 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 18 | 1239 | 1.40e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 72 | 8061 | 1.52e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 65 | 7069 | 1.63e-01 |
| GO:BP | GO:2000379 | positive regulation of reactive oxygen species metabolic process | 3 | 46 | 1.78e-01 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 18 | 1280 | 1.85e-01 |
| GO:BP | GO:0042636 | negative regulation of hair cycle | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0035998 | 7,8-dihydroneopterin 3’-triphosphate biosynthetic process | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0014861 | regulation of skeletal muscle contraction via regulation of action potential | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 4 | 99 | 1.86e-01 |
| GO:BP | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 1 | 1.86e-01 |
| GO:BP | GO:1990114 | RNA polymerase II core complex assembly | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | 2 | 18 | 1.86e-01 |
| GO:BP | GO:0002041 | intussusceptive angiogenesis | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 2 | 17 | 1.86e-01 |
| GO:BP | GO:0100001 | regulation of skeletal muscle contraction by action potential | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 2 | 15 | 1.86e-01 |
| GO:BP | GO:0071241 | cellular response to inorganic substance | 5 | 164 | 1.86e-01 |
| GO:BP | GO:2000761 | positive regulation of N-terminal peptidyl-lysine acetylation | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 7 | 293 | 1.86e-01 |
| GO:BP | GO:0062000 | positive regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0061999 | regulation of cardiac endothelial to mesenchymal transition | 1 | 1 | 1.86e-01 |
| GO:BP | GO:2000759 | regulation of N-terminal peptidyl-lysine acetylation | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 3 | 55 | 1.86e-01 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 2 | 17 | 1.86e-01 |
| GO:BP | GO:0072714 | response to selenite ion | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0072715 | cellular response to selenite ion | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0008152 | metabolic process | 74 | 8518 | 1.86e-01 |
| GO:BP | GO:1903788 | positive regulation of glutathione biosynthetic process | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 7 | 295 | 1.86e-01 |
| GO:BP | GO:1902217 | erythrocyte apoptotic process | 1 | 1 | 1.86e-01 |
| GO:BP | GO:1904574 | negative regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.86e-01 |
| GO:BP | GO:1904570 | negative regulation of selenocysteine incorporation | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0051169 | nuclear transport | 7 | 295 | 1.86e-01 |
| GO:BP | GO:1904572 | negative regulation of mRNA binding | 1 | 1 | 1.86e-01 |
| GO:BP | GO:1901327 | response to tacrolimus | 1 | 1 | 1.86e-01 |
| GO:BP | GO:1904573 | regulation of selenocysteine insertion sequence binding | 1 | 1 | 1.86e-01 |
| GO:BP | GO:1904569 | regulation of selenocysteine incorporation | 1 | 1 | 1.86e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 27 | 2293 | 1.86e-01 |
| GO:BP | GO:0090322 | regulation of superoxide metabolic process | 2 | 19 | 2.00e-01 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 2 | 19 | 2.00e-01 |
| GO:BP | GO:0071451 | cellular response to superoxide | 2 | 19 | 2.00e-01 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 3 | 59 | 2.16e-01 |
| GO:BP | GO:0003170 | heart valve development | 3 | 65 | 2.28e-01 |
| GO:BP | GO:1901376 | organic heteropentacyclic compound metabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 26 | 2270 | 2.28e-01 |
| GO:BP | GO:0003283 | atrial septum development | 2 | 23 | 2.28e-01 |
| GO:BP | GO:0035026 | leading edge cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0043385 | mycotoxin metabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 17 | 1315 | 2.28e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 17 | 1322 | 2.28e-01 |
| GO:BP | GO:1901377 | organic heteropentacyclic compound catabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 2 | 25 | 2.28e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 11 | 706 | 2.28e-01 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 4 | 121 | 2.28e-01 |
| GO:BP | GO:1903786 | regulation of glutathione biosynthetic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0043387 | mycotoxin catabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0035544 | negative regulation of SNARE complex assembly | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0045065 | cytotoxic T cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 2 | 25 | 2.28e-01 |
| GO:BP | GO:0009407 | toxin catabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:1905215 | negative regulation of RNA binding | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 22 | 1849 | 2.28e-01 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 2 | 21 | 2.28e-01 |
| GO:BP | GO:2000120 | positive regulation of sodium-dependent phosphate transport | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0006542 | glutamine biosynthetic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0072752 | cellular response to rapamycin | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0001174 | transcriptional start site selection at RNA polymerase II promoter | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0001173 | DNA-templated transcriptional start site selection | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0060689 | cell differentiation involved in salivary gland development | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0060819 | inactivation of X chromosome by genomic imprinting | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0071704 | organic substance metabolic process | 71 | 8169 | 2.28e-01 |
| GO:BP | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0000349 | generation of catalytic spliceosome for first transesterification step | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0000305 | response to oxygen radical | 2 | 22 | 2.28e-01 |
| GO:BP | GO:0000303 | response to superoxide | 2 | 21 | 2.28e-01 |
| GO:BP | GO:2001055 | positive regulation of mesenchymal cell apoptotic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0006979 | response to oxidative stress | 7 | 352 | 2.28e-01 |
| GO:BP | GO:0070982 | L-asparagine metabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0051066 | dihydrobiopterin metabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0001827 | inner cell mass cell fate commitment | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0090164 | asymmetric Golgi ribbon formation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:1990478 | response to ultrasound | 1 | 2 | 2.28e-01 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 3 | 67 | 2.28e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 68 | 7806 | 2.28e-01 |
| GO:BP | GO:0045585 | positive regulation of cytotoxic T cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0046223 | aflatoxin catabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0046222 | aflatoxin metabolic process | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0001828 | inner cell mass cellular morphogenesis | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0045583 | regulation of cytotoxic T cell differentiation | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0140074 | cardiac endothelial to mesenchymal transition | 1 | 2 | 2.28e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 17 | 1315 | 2.28e-01 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 5 | 195 | 2.28e-01 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 8 | 442 | 2.34e-01 |
| GO:BP | GO:0071772 | response to BMP | 4 | 131 | 2.34e-01 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 4 | 131 | 2.34e-01 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 2 | 26 | 2.34e-01 |
| GO:BP | GO:0036010 | protein localization to endosome | 2 | 26 | 2.34e-01 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 4 | 132 | 2.38e-01 |
| GO:BP | GO:0003171 | atrioventricular valve development | 2 | 27 | 2.46e-01 |
| GO:BP | GO:0010966 | regulation of phosphate transport | 1 | 3 | 2.51e-01 |
| GO:BP | GO:2000446 | regulation of macrophage migration inhibitory factor signaling pathway | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0014916 | regulation of lung blood pressure | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 4 | 135 | 2.51e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 11 | 742 | 2.51e-01 |
| GO:BP | GO:1904906 | positive regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 2.51e-01 |
| GO:BP | GO:1904904 | regulation of endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0006556 | S-adenosylmethionine biosynthetic process | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0008219 | cell death | 19 | 1596 | 2.51e-01 |
| GO:BP | GO:2000118 | regulation of sodium-dependent phosphate transport | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 24 | 2135 | 2.51e-01 |
| GO:BP | GO:0006405 | RNA export from nucleus | 3 | 78 | 2.51e-01 |
| GO:BP | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 19 | 1604 | 2.51e-01 |
| GO:BP | GO:1990737 | response to manganese-induced endoplasmic reticulum stress | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0010729 | positive regulation of hydrogen peroxide biosynthetic process | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 19 | 1564 | 2.51e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 65 | 7441 | 2.51e-01 |
| GO:BP | GO:1903300 | negative regulation of hexokinase activity | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 13 | 957 | 2.51e-01 |
| GO:BP | GO:0140468 | HRI-mediated signaling | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0035691 | macrophage migration inhibitory factor signaling pathway | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0003007 | heart morphogenesis | 5 | 216 | 2.51e-01 |
| GO:BP | GO:0090674 | endothelial cell-matrix adhesion via fibronectin | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 8 | 462 | 2.51e-01 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 2 | 29 | 2.51e-01 |
| GO:BP | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | 1 | 3 | 2.51e-01 |
| GO:BP | GO:1901355 | response to rapamycin | 1 | 3 | 2.51e-01 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 2 | 28 | 2.51e-01 |
| GO:BP | GO:0099578 | regulation of translation at postsynapse, modulating synaptic transmission | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0045776 | negative regulation of blood pressure | 2 | 30 | 2.51e-01 |
| GO:BP | GO:0031339 | negative regulation of vesicle fusion | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0043549 | regulation of kinase activity | 9 | 560 | 2.51e-01 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 23 | 2035 | 2.51e-01 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 3 | 80 | 2.51e-01 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 3 | 76 | 2.51e-01 |
| GO:BP | GO:0090076 | relaxation of skeletal muscle | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0051684 | maintenance of Golgi location | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0072740 | cellular response to anisomycin | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0033132 | negative regulation of glucokinase activity | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0060429 | epithelium development | 12 | 850 | 2.51e-01 |
| GO:BP | GO:0060591 | chondroblast differentiation | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 9 | 551 | 2.51e-01 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 6 | 294 | 2.51e-01 |
| GO:BP | GO:0071630 | nuclear protein quality control by the ubiquitin-proteasome system | 1 | 3 | 2.51e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 11 | 763 | 2.63e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 18 | 1507 | 2.64e-01 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 4 | 149 | 2.64e-01 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 3 | 84 | 2.64e-01 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 6 | 303 | 2.64e-01 |
| GO:BP | GO:0070848 | response to growth factor | 9 | 572 | 2.64e-01 |
| GO:BP | GO:0051168 | nuclear export | 4 | 150 | 2.68e-01 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 3 | 85 | 2.68e-01 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 2 | 34 | 2.71e-01 |
| GO:BP | GO:0045588 | positive regulation of gamma-delta T cell differentiation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0046645 | positive regulation of gamma-delta T cell activation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 5 | 230 | 2.71e-01 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 2 | 35 | 2.71e-01 |
| GO:BP | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0048732 | gland development | 6 | 318 | 2.71e-01 |
| GO:BP | GO:0072739 | response to anisomycin | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0009894 | regulation of catabolic process | 12 | 875 | 2.71e-01 |
| GO:BP | GO:0090427 | activation of meiosis | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0006408 | snRNA export from nucleus | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0090673 | endothelial cell-matrix adhesion | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0010726 | positive regulation of hydrogen peroxide metabolic process | 1 | 4 | 2.71e-01 |
| GO:BP | GO:1902037 | negative regulation of hematopoietic stem cell differentiation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:1902512 | positive regulation of apoptotic DNA fragmentation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 2 | 36 | 2.71e-01 |
| GO:BP | GO:1904029 | regulation of cyclin-dependent protein kinase activity | 3 | 88 | 2.71e-01 |
| GO:BP | GO:0003137 | Notch signaling pathway involved in heart induction | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0035470 | positive regulation of vascular wound healing | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0044341 | sodium-dependent phosphate transport | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0035480 | regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0035481 | positive regulation of Notch signaling pathway involved in heart induction | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0032447 | protein urmylation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0035880 | embryonic nail plate morphogenesis | 1 | 4 | 2.71e-01 |
| GO:BP | GO:1904798 | positive regulation of core promoter binding | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0035910 | ascending aorta morphogenesis | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0038159 | C-X-C chemokine receptor CXCR4 signaling pathway | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0018076 | N-terminal peptidyl-lysine acetylation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:1904886 | beta-catenin destruction complex disassembly | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0016477 | cell migration | 14 | 1081 | 2.71e-01 |
| GO:BP | GO:1905322 | positive regulation of mesenchymal stem cell migration | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0002143 | tRNA wobble position uridine thiolation | 1 | 4 | 2.71e-01 |
| GO:BP | GO:1905320 | regulation of mesenchymal stem cell migration | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0016480 | negative regulation of transcription by RNA polymerase III | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0003230 | cardiac atrium development | 2 | 34 | 2.71e-01 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 2 | 35 | 2.71e-01 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 2 | 34 | 2.71e-01 |
| GO:BP | GO:1905319 | mesenchymal stem cell migration | 1 | 4 | 2.71e-01 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 14 | 1107 | 2.74e-01 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 4 | 160 | 2.74e-01 |
| GO:BP | GO:0010941 | regulation of cell death | 15 | 1219 | 2.81e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 17 | 1441 | 2.83e-01 |
| GO:BP | GO:0006915 | apoptotic process | 17 | 1443 | 2.84e-01 |
| GO:BP | GO:0032879 | regulation of localization | 18 | 1555 | 2.84e-01 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 38 | 2.84e-01 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 2 | 38 | 2.84e-01 |
| GO:BP | GO:0008038 | neuron recognition | 2 | 39 | 2.86e-01 |
| GO:BP | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0060536 | cartilage morphogenesis | 1 | 5 | 2.86e-01 |
| GO:BP | GO:2000304 | positive regulation of ceramide biosynthetic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 2 | 39 | 2.86e-01 |
| GO:BP | GO:2001053 | regulation of mesenchymal cell apoptotic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0070309 | lens fiber cell morphogenesis | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1903202 | negative regulation of oxidative stress-induced cell death | 2 | 40 | 2.86e-01 |
| GO:BP | GO:0032057 | negative regulation of translational initiation in response to stress | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0045053 | protein retention in Golgi apparatus | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0034227 | tRNA thio-modification | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0010728 | regulation of hydrogen peroxide biosynthetic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 14 | 1137 | 2.86e-01 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1990089 | response to nerve growth factor | 2 | 41 | 2.86e-01 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 2 | 41 | 2.86e-01 |
| GO:BP | GO:1905166 | negative regulation of lysosomal protein catabolic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1903626 | positive regulation of DNA catabolic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0046654 | tetrahydrofolate biosynthetic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0035905 | ascending aorta development | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 29 | 2880 | 2.86e-01 |
| GO:BP | GO:0090650 | cellular response to oxygen-glucose deprivation | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 2 | 39 | 2.86e-01 |
| GO:BP | GO:0090154 | positive regulation of sphingolipid biosynthetic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1904351 | negative regulation of protein catabolic process in the vacuole | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1904796 | regulation of core promoter binding | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0090425 | acinar cell differentiation | 1 | 5 | 2.86e-01 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 2 | 40 | 2.86e-01 |
| GO:BP | GO:0071362 | cellular response to ether | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0001826 | inner cell mass cell differentiation | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0006528 | asparagine metabolic process | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 2 | 39 | 2.86e-01 |
| GO:BP | GO:0003289 | atrial septum primum morphogenesis | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0003284 | septum primum development | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation | 1 | 5 | 2.86e-01 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 5 | 251 | 2.88e-01 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 3 | 100 | 2.88e-01 |
| GO:BP | GO:0035295 | tube development | 11 | 821 | 2.88e-01 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 9 | 620 | 2.89e-01 |
| GO:BP | GO:0090132 | epithelium migration | 5 | 252 | 2.89e-01 |
| GO:BP | GO:0010631 | epithelial cell migration | 5 | 252 | 2.89e-01 |
| GO:BP | GO:0006801 | superoxide metabolic process | 2 | 42 | 2.91e-01 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 2 | 42 | 2.91e-01 |
| GO:BP | GO:0051030 | snRNA transport | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0140245 | regulation of translation at postsynapse | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0007023 | post-chaperonin tubulin folding pathway | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0140243 | regulation of translation at synapse | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0090303 | positive regulation of wound healing | 2 | 45 | 2.92e-01 |
| GO:BP | GO:0120183 | positive regulation of focal adhesion disassembly | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 20 | 1835 | 2.92e-01 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0045682 | regulation of epidermis development | 2 | 45 | 2.92e-01 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 4 | 179 | 2.92e-01 |
| GO:BP | GO:1902949 | positive regulation of tau-protein kinase activity | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0001568 | blood vessel development | 8 | 534 | 2.92e-01 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 2 | 43 | 2.92e-01 |
| GO:BP | GO:0090130 | tissue migration | 5 | 256 | 2.92e-01 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 2 | 45 | 2.92e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12 | 937 | 2.92e-01 |
| GO:BP | GO:0001649 | osteoblast differentiation | 4 | 179 | 2.92e-01 |
| GO:BP | GO:0032324 | molybdopterin cofactor biosynthetic process | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0019720 | Mo-molybdopterin cofactor metabolic process | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0051189 | prosthetic group metabolic process | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 42 | 4579 | 2.92e-01 |
| GO:BP | GO:0099562 | maintenance of postsynaptic density structure | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0003278 | apoptotic process involved in heart morphogenesis | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 3 | 104 | 2.92e-01 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 2 | 45 | 2.92e-01 |
| GO:BP | GO:0051049 | regulation of transport | 15 | 1267 | 2.92e-01 |
| GO:BP | GO:0060075 | regulation of resting membrane potential | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0120182 | regulation of focal adhesion disassembly | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0043545 | molybdopterin cofactor metabolic process | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 2 | 43 | 2.92e-01 |
| GO:BP | GO:0043009 | chordate embryonic development | 8 | 534 | 2.92e-01 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 1 | 6 | 2.92e-01 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0038146 | chemokine (C-X-C motif) ligand 12 signaling pathway | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 2 | 45 | 2.92e-01 |
| GO:BP | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | 1 | 6 | 2.92e-01 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 5 | 260 | 2.92e-01 |
| GO:BP | GO:0012501 | programmed cell death | 17 | 1479 | 2.92e-01 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 2 | 45 | 2.92e-01 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 5 | 265 | 2.97e-01 |
| GO:BP | GO:0003197 | endocardial cushion development | 2 | 46 | 2.97e-01 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 2 | 46 | 2.97e-01 |
| GO:BP | GO:0040011 | locomotion | 12 | 954 | 2.97e-01 |
| GO:BP | GO:0010823 | negative regulation of mitochondrion organization | 2 | 46 | 2.97e-01 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 3 | 109 | 3.02e-01 |
| GO:BP | GO:0030217 | T cell differentiation | 4 | 185 | 3.07e-01 |
| GO:BP | GO:0050708 | regulation of protein secretion | 4 | 185 | 3.07e-01 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0042492 | gamma-delta T cell differentiation | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0046543 | development of secondary female sexual characteristics | 1 | 7 | 3.08e-01 |
| GO:BP | GO:2000171 | negative regulation of dendrite development | 1 | 7 | 3.08e-01 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0048870 | cell motility | 14 | 1182 | 3.08e-01 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 3 | 111 | 3.08e-01 |
| GO:BP | GO:0009396 | folic acid-containing compound biosynthetic process | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0009404 | toxin metabolic process | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0006398 | mRNA 3’-end processing by stem-loop binding and cleavage | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0061312 | BMP signaling pathway involved in heart development | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 8 | 555 | 3.08e-01 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0090649 | response to oxygen-glucose deprivation | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 9 | 656 | 3.08e-01 |
| GO:BP | GO:0019083 | viral transcription | 2 | 48 | 3.08e-01 |
| GO:BP | GO:0045472 | response to ether | 1 | 7 | 3.08e-01 |
| GO:BP | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0003357 | noradrenergic neuron differentiation | 1 | 7 | 3.08e-01 |
| GO:BP | GO:1905461 | positive regulation of vascular associated smooth muscle cell apoptotic process | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0060548 | negative regulation of cell death | 10 | 756 | 3.08e-01 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 8 | 551 | 3.08e-01 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0033131 | regulation of glucokinase activity | 1 | 7 | 3.08e-01 |
| GO:BP | GO:0009299 | mRNA transcription | 2 | 50 | 3.14e-01 |
| GO:BP | GO:0001944 | vasculature development | 8 | 559 | 3.14e-01 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 2 | 50 | 3.14e-01 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 11 | 870 | 3.14e-01 |
| GO:BP | GO:0035239 | tube morphogenesis | 9 | 663 | 3.18e-01 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 7 | 464 | 3.19e-01 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 2 | 51 | 3.22e-01 |
| GO:BP | GO:1902510 | regulation of apoptotic DNA fragmentation | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0042416 | dopamine biosynthetic process | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0035878 | nail development | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0045607 | regulation of inner ear auditory receptor cell differentiation | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 1 | 8 | 3.24e-01 |
| GO:BP | GO:1902947 | regulation of tau-protein kinase activity | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0045631 | regulation of mechanoreceptor differentiation | 1 | 8 | 3.24e-01 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0034393 | positive regulation of smooth muscle cell apoptotic process | 1 | 8 | 3.24e-01 |
| GO:BP | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 6 | 373 | 3.24e-01 |
| GO:BP | GO:0036353 | histone H2A-K119 monoubiquitination | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0071287 | cellular response to manganese ion | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0090394 | negative regulation of excitatory postsynaptic potential | 1 | 8 | 3.24e-01 |
| GO:BP | GO:2000980 | regulation of inner ear receptor cell differentiation | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0051028 | mRNA transport | 3 | 117 | 3.24e-01 |
| GO:BP | GO:1903299 | regulation of hexokinase activity | 1 | 8 | 3.24e-01 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 2 | 53 | 3.24e-01 |
| GO:BP | GO:0001501 | skeletal system development | 6 | 377 | 3.25e-01 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 24 | 2412 | 3.32e-01 |
| GO:BP | GO:0097064 | ncRNA export from nucleus | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0034214 | protein hexamerization | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 10 | 787 | 3.32e-01 |
| GO:BP | GO:0060352 | cell adhesion molecule production | 1 | 9 | 3.32e-01 |
| GO:BP | GO:2001214 | positive regulation of vasculogenesis | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0007021 | tubulin complex assembly | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0006705 | mineralocorticoid biosynthetic process | 1 | 9 | 3.32e-01 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 2 | 55 | 3.32e-01 |
| GO:BP | GO:0030509 | BMP signaling pathway | 3 | 123 | 3.32e-01 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 7 | 479 | 3.32e-01 |
| GO:BP | GO:0045836 | positive regulation of meiotic nuclear division | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 2 | 54 | 3.32e-01 |
| GO:BP | GO:0051338 | regulation of transferase activity | 9 | 679 | 3.32e-01 |
| GO:BP | GO:0045136 | development of secondary sexual characteristics | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 2 | 54 | 3.32e-01 |
| GO:BP | GO:1905459 | regulation of vascular associated smooth muscle cell apoptotic process | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0003129 | heart induction | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0048757 | pigment granule maturation | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0031953 | negative regulation of protein autophosphorylation | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0032342 | aldosterone biosynthetic process | 1 | 9 | 3.32e-01 |
| GO:BP | GO:1905288 | vascular associated smooth muscle cell apoptotic process | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0061043 | regulation of vascular wound healing | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 13 | 1111 | 3.32e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 37 | 4045 | 3.32e-01 |
| GO:BP | GO:0002521 | leukocyte differentiation | 6 | 382 | 3.32e-01 |
| GO:BP | GO:0002407 | dendritic cell chemotaxis | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0035646 | endosome to melanosome transport | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 2 | 56 | 3.32e-01 |
| GO:BP | GO:0043485 | endosome to pigment granule transport | 1 | 9 | 3.32e-01 |
| GO:BP | GO:0098880 | maintenance of postsynaptic specialization structure | 1 | 9 | 3.32e-01 |
| GO:BP | GO:1903201 | regulation of oxidative stress-induced cell death | 2 | 56 | 3.32e-01 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 2 | 57 | 3.37e-01 |
| GO:BP | GO:0001525 | angiogenesis | 6 | 390 | 3.39e-01 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 3 | 128 | 3.45e-01 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 2 | 58 | 3.45e-01 |
| GO:BP | GO:0008544 | epidermis development | 4 | 210 | 3.46e-01 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 6 | 393 | 3.46e-01 |
| GO:BP | GO:0042481 | regulation of odontogenesis | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0001514 | selenocysteine incorporation | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0060174 | limb bud formation | 1 | 10 | 3.48e-01 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 3 | 129 | 3.48e-01 |
| GO:BP | GO:0033690 | positive regulation of osteoblast proliferation | 1 | 10 | 3.48e-01 |
| GO:BP | GO:1903624 | regulation of DNA catabolic process | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 1 | 10 | 3.48e-01 |
| GO:BP | GO:1902415 | regulation of mRNA binding | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 1 | 10 | 3.48e-01 |
| GO:BP | GO:2000826 | regulation of heart morphogenesis | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0006451 | translational readthrough | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0008212 | mineralocorticoid metabolic process | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 1 | 10 | 3.48e-01 |
| GO:BP | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0007423 | sensory organ development | 6 | 396 | 3.48e-01 |
| GO:BP | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0032341 | aldosterone metabolic process | 1 | 10 | 3.48e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 17 | 1602 | 3.48e-01 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 2 | 60 | 3.48e-01 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 6 | 399 | 3.49e-01 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 3 | 132 | 3.52e-01 |
| GO:BP | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | 11 | 3.56e-01 |
| GO:BP | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 1 | 11 | 3.56e-01 |
| GO:BP | GO:1902946 | protein localization to early endosome | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 37 | 4099 | 3.56e-01 |
| GO:BP | GO:0090280 | positive regulation of calcium ion import | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 3 | 133 | 3.56e-01 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0006396 | RNA processing | 11 | 929 | 3.56e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 6 | 407 | 3.56e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 3 | 135 | 3.56e-01 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 1 | 11 | 3.56e-01 |
| GO:BP | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0051549 | positive regulation of keratinocyte migration | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0043482 | cellular pigment accumulation | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0016567 | protein ubiquitination | 9 | 717 | 3.56e-01 |
| GO:BP | GO:0051657 | maintenance of organelle location | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0009713 | catechol-containing compound biosynthetic process | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 3 | 134 | 3.56e-01 |
| GO:BP | GO:0070307 | lens fiber cell development | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0001562 | response to protozoan | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 2 | 62 | 3.56e-01 |
| GO:BP | GO:0043476 | pigment accumulation | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 4 | 220 | 3.56e-01 |
| GO:BP | GO:0050665 | hydrogen peroxide biosynthetic process | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0042423 | catecholamine biosynthetic process | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0030007 | intracellular potassium ion homeostasis | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0043922 | negative regulation by host of viral transcription | 1 | 11 | 3.56e-01 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 1 | 11 | 3.56e-01 |
| GO:BP | GO:0065008 | regulation of biological quality | 21 | 2102 | 3.59e-01 |
| GO:BP | GO:0001503 | ossification | 5 | 313 | 3.59e-01 |
| GO:BP | GO:0010506 | regulation of autophagy | 5 | 313 | 3.59e-01 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 4 | 222 | 3.60e-01 |
| GO:BP | GO:0005977 | glycogen metabolic process | 2 | 64 | 3.60e-01 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 2 | 64 | 3.60e-01 |
| GO:BP | GO:0044042 | glucan metabolic process | 2 | 64 | 3.60e-01 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 14 | 1278 | 3.60e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 7 | 513 | 3.64e-01 |
| GO:BP | GO:0034309 | primary alcohol biosynthetic process | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0050884 | neuromuscular process controlling posture | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0071248 | cellular response to metal ion | 3 | 139 | 3.65e-01 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 1 | 12 | 3.65e-01 |
| GO:BP | GO:1903428 | positive regulation of reactive oxygen species biosynthetic process | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0007214 | gamma-aminobutyric acid signaling pathway | 1 | 12 | 3.65e-01 |
| GO:BP | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 1 | 12 | 3.65e-01 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 7 | 517 | 3.65e-01 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 2 | 65 | 3.65e-01 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0051098 | regulation of binding | 5 | 317 | 3.65e-01 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0010035 | response to inorganic substance | 6 | 414 | 3.65e-01 |
| GO:BP | GO:0003174 | mitral valve development | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 4 | 227 | 3.65e-01 |
| GO:BP | GO:0061029 | eyelid development in camera-type eye | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0050961 | detection of temperature stimulus involved in sensory perception | 1 | 12 | 3.65e-01 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 4 | 227 | 3.65e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 33 | 3625 | 3.67e-01 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 2 | 67 | 3.70e-01 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 2 | 67 | 3.70e-01 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0016048 | detection of temperature stimulus | 1 | 13 | 3.70e-01 |
| GO:BP | GO:1905214 | regulation of RNA binding | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 1 | 13 | 3.70e-01 |
| GO:BP | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death | 1 | 13 | 3.70e-01 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0006309 | apoptotic DNA fragmentation | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 6 | 424 | 3.70e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 2 | 69 | 3.70e-01 |
| GO:BP | GO:0003281 | ventricular septum development | 2 | 69 | 3.70e-01 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 3 | 144 | 3.70e-01 |
| GO:BP | GO:0051547 | regulation of keratinocyte migration | 1 | 13 | 3.70e-01 |
| GO:BP | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 1 | 13 | 3.70e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 6 | 421 | 3.70e-01 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 1 | 13 | 3.70e-01 |
| GO:BP | GO:2001212 | regulation of vasculogenesis | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0050951 | sensory perception of temperature stimulus | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0045820 | negative regulation of glycolytic process | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0046500 | S-adenosylmethionine metabolic process | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0050658 | RNA transport | 3 | 144 | 3.70e-01 |
| GO:BP | GO:1901524 | regulation of mitophagy | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 3 | 143 | 3.70e-01 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 3 | 143 | 3.70e-01 |
| GO:BP | GO:0042104 | positive regulation of activated T cell proliferation | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0048714 | positive regulation of oligodendrocyte differentiation | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0050657 | nucleic acid transport | 3 | 144 | 3.70e-01 |
| GO:BP | GO:1901032 | negative regulation of response to reactive oxygen species | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0042474 | middle ear morphogenesis | 1 | 13 | 3.70e-01 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 4 | 234 | 3.73e-01 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 2 | 70 | 3.74e-01 |
| GO:BP | GO:0051236 | establishment of RNA localization | 3 | 147 | 3.74e-01 |
| GO:BP | GO:0006813 | potassium ion transport | 3 | 147 | 3.74e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 6 | 429 | 3.77e-01 |
| GO:BP | GO:0003207 | cardiac chamber formation | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0006349 | regulation of gene expression by genomic imprinting | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 1 | 14 | 3.78e-01 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 4 | 236 | 3.78e-01 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0051000 | positive regulation of nitric-oxide synthase activity | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 3 | 149 | 3.78e-01 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 3 | 150 | 3.78e-01 |
| GO:BP | GO:0051645 | Golgi localization | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0010310 | regulation of hydrogen peroxide metabolic process | 1 | 14 | 3.78e-01 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0060021 | roof of mouth development | 2 | 71 | 3.78e-01 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 12 | 1088 | 3.78e-01 |
| GO:BP | GO:0071732 | cellular response to nitric oxide | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0003085 | negative regulation of systemic arterial blood pressure | 1 | 14 | 3.78e-01 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 2 | 73 | 3.82e-01 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 2 | 73 | 3.82e-01 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 2 | 73 | 3.82e-01 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 2 | 73 | 3.82e-01 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 2 | 73 | 3.82e-01 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 3 | 155 | 3.86e-01 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0060349 | bone morphogenesis | 2 | 74 | 3.86e-01 |
| GO:BP | GO:1903861 | positive regulation of dendrite extension | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0062149 | detection of stimulus involved in sensory perception of pain | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0016973 | poly(A)+ mRNA export from nucleus | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 12 | 1101 | 3.86e-01 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 3 | 155 | 3.86e-01 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0051546 | keratinocyte migration | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 6 | 440 | 3.86e-01 |
| GO:BP | GO:0036336 | dendritic cell migration | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0010042 | response to manganese ion | 1 | 15 | 3.86e-01 |
| GO:BP | GO:1902036 | regulation of hematopoietic stem cell differentiation | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0010942 | positive regulation of cell death | 6 | 441 | 3.86e-01 |
| GO:BP | GO:0036473 | cell death in response to oxidative stress | 2 | 75 | 3.86e-01 |
| GO:BP | GO:0043583 | ear development | 3 | 153 | 3.86e-01 |
| GO:BP | GO:0034390 | smooth muscle cell apoptotic process | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0034391 | regulation of smooth muscle cell apoptotic process | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0043217 | myelin maintenance | 1 | 15 | 3.86e-01 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 2 | 74 | 3.86e-01 |
| GO:BP | GO:0009790 | embryo development | 10 | 875 | 3.86e-01 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0043278 | response to morphine | 1 | 16 | 3.92e-01 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 1 | 16 | 3.92e-01 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0010226 | response to lithium ion | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0071731 | response to nitric oxide | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0042633 | hair cycle | 2 | 78 | 3.92e-01 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 10 | 881 | 3.92e-01 |
| GO:BP | GO:0042303 | molting cycle | 2 | 78 | 3.92e-01 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0003128 | heart field specification | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0072359 | circulatory system development | 10 | 884 | 3.92e-01 |
| GO:BP | GO:0009636 | response to toxic substance | 3 | 158 | 3.92e-01 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0050901 | leukocyte tethering or rolling | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 3 | 158 | 3.92e-01 |
| GO:BP | GO:0014072 | response to isoquinoline alkaloid | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 1 | 16 | 3.92e-01 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 1 | 16 | 3.92e-01 |
| GO:BP | GO:1905114 | cell surface receptor signaling pathway involved in cell-cell signaling | 6 | 448 | 3.92e-01 |
| GO:BP | GO:1990748 | cellular detoxification | 2 | 76 | 3.92e-01 |
| GO:BP | GO:0001701 | in utero embryonic development | 5 | 344 | 3.92e-01 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 1 | 16 | 3.92e-01 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 3 | 160 | 3.93e-01 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 3 | 160 | 3.93e-01 |
| GO:BP | GO:0048265 | response to pain | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0032456 | endocytic recycling | 2 | 80 | 3.98e-01 |
| GO:BP | GO:1903859 | regulation of dendrite extension | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0001759 | organ induction | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0061042 | vascular wound healing | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0060996 | dendritic spine development | 2 | 80 | 3.98e-01 |
| GO:BP | GO:0120255 | olefinic compound biosynthetic process | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0060713 | labyrinthine layer morphogenesis | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0031338 | regulation of vesicle fusion | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 8 | 667 | 3.98e-01 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 2 | 80 | 3.98e-01 |
| GO:BP | GO:0039535 | regulation of RIG-I signaling pathway | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0015693 | magnesium ion transport | 1 | 17 | 3.98e-01 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0006817 | phosphate ion transport | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0006700 | C21-steroid hormone biosynthetic process | 1 | 17 | 3.98e-01 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 10 | 898 | 4.02e-01 |
| GO:BP | GO:0001558 | regulation of cell growth | 5 | 357 | 4.05e-01 |
| GO:BP | GO:0030282 | bone mineralization | 2 | 82 | 4.07e-01 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 2 | 82 | 4.07e-01 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 5 | 358 | 4.07e-01 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 19 | 1974 | 4.07e-01 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0010575 | positive regulation of vascular endothelial growth factor production | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0007413 | axonal fasciculation | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 2 | 83 | 4.07e-01 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 2 | 83 | 4.07e-01 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 1 | 18 | 4.07e-01 |
| GO:BP | GO:1902883 | negative regulation of response to oxidative stress | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 12 | 1133 | 4.07e-01 |
| GO:BP | GO:0021522 | spinal cord motor neuron differentiation | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0071280 | cellular response to copper ion | 1 | 18 | 4.07e-01 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 1 | 18 | 4.07e-01 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 4 | 262 | 4.08e-01 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 4 | 262 | 4.08e-01 |
| GO:BP | GO:0009306 | protein secretion | 4 | 262 | 4.08e-01 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 4 | 266 | 4.15e-01 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0035162 | embryonic hemopoiesis | 1 | 19 | 4.15e-01 |
| GO:BP | GO:1903204 | negative regulation of oxidative stress-induced neuron death | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0002027 | regulation of heart rate | 2 | 86 | 4.15e-01 |
| GO:BP | GO:0010001 | glial cell differentiation | 3 | 170 | 4.15e-01 |
| GO:BP | GO:0035518 | histone H2A monoubiquitination | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0006448 | regulation of translational elongation | 1 | 19 | 4.15e-01 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0001774 | microglial cell activation | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0060341 | regulation of cellular localization | 9 | 798 | 4.15e-01 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 1 | 19 | 4.15e-01 |
| GO:BP | GO:1901071 | glucosamine-containing compound metabolic process | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 2 | 86 | 4.15e-01 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 4 | 265 | 4.15e-01 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 2 | 86 | 4.15e-01 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0006308 | DNA catabolic process | 1 | 19 | 4.15e-01 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 8 | 691 | 4.17e-01 |
| GO:BP | GO:0008037 | cell recognition | 2 | 87 | 4.21e-01 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0036260 | RNA capping | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0010574 | regulation of vascular endothelial growth factor production | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0034312 | diol biosynthetic process | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0150146 | cell junction disassembly | 1 | 20 | 4.26e-01 |
| GO:BP | GO:1903205 | regulation of hydrogen peroxide-induced cell death | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0050974 | detection of mechanical stimulus involved in sensory perception | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0022038 | corpus callosum development | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0030262 | apoptotic nuclear changes | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0032770 | positive regulation of monooxygenase activity | 1 | 20 | 4.26e-01 |
| GO:BP | GO:0043267 | negative regulation of potassium ion transport | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0010460 | positive regulation of heart rate | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 3 | 178 | 4.28e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 4 | 276 | 4.28e-01 |
| GO:BP | GO:0035640 | exploration behavior | 1 | 21 | 4.28e-01 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 2 | 90 | 4.28e-01 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 2 | 91 | 4.28e-01 |
| GO:BP | GO:0002269 | leukocyte activation involved in inflammatory response | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 5 | 376 | 4.28e-01 |
| GO:BP | GO:1900048 | positive regulation of hemostasis | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 1 | 21 | 4.28e-01 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0035268 | protein mannosylation | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 5 | 375 | 4.28e-01 |
| GO:BP | GO:0050773 | regulation of dendrite development | 2 | 90 | 4.28e-01 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 4 | 275 | 4.28e-01 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 2 | 91 | 4.28e-01 |
| GO:BP | GO:0006468 | protein phosphorylation | 12 | 1167 | 4.28e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 24 | 2646 | 4.28e-01 |
| GO:BP | GO:0046006 | regulation of activated T cell proliferation | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0030194 | positive regulation of blood coagulation | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0051291 | protein heterooligomerization | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0046885 | regulation of hormone biosynthetic process | 1 | 21 | 4.28e-01 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 6 | 485 | 4.28e-01 |
| GO:BP | GO:0098754 | detoxification | 2 | 91 | 4.28e-01 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 2 | 90 | 4.28e-01 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 4 | 277 | 4.31e-01 |
| GO:BP | GO:0048565 | digestive tract development | 2 | 92 | 4.34e-01 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 5 | 381 | 4.35e-01 |
| GO:BP | GO:0032880 | regulation of protein localization | 8 | 710 | 4.36e-01 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0030900 | forebrain development | 4 | 281 | 4.36e-01 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 1 | 22 | 4.36e-01 |
| GO:BP | GO:2000352 | negative regulation of endothelial cell apoptotic process | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0035493 | SNARE complex assembly | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0030182 | neuron differentiation | 11 | 1061 | 4.36e-01 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0006403 | RNA localization | 3 | 182 | 4.36e-01 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0043542 | endothelial cell migration | 3 | 182 | 4.36e-01 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0042634 | regulation of hair cycle | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis | 1 | 22 | 4.36e-01 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 2 | 94 | 4.37e-01 |
| GO:BP | GO:0001824 | blastocyst development | 2 | 94 | 4.37e-01 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 4 | 282 | 4.38e-01 |
| GO:BP | GO:0051259 | protein complex oligomerization | 3 | 185 | 4.41e-01 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0036474 | cell death in response to hydrogen peroxide | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0019080 | viral gene expression | 2 | 96 | 4.41e-01 |
| GO:BP | GO:0061900 | glial cell activation | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0009888 | tissue development | 14 | 1425 | 4.41e-01 |
| GO:BP | GO:0010573 | vascular endothelial growth factor production | 1 | 23 | 4.41e-01 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 2 | 96 | 4.41e-01 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0050820 | positive regulation of coagulation | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0042417 | dopamine metabolic process | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0061041 | regulation of wound healing | 2 | 95 | 4.41e-01 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 6 | 494 | 4.41e-01 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 1 | 23 | 4.41e-01 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 2 | 97 | 4.47e-01 |
| GO:BP | GO:0046777 | protein autophosphorylation | 3 | 188 | 4.48e-01 |
| GO:BP | GO:0055123 | digestive system development | 2 | 100 | 4.49e-01 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0033028 | myeloid cell apoptotic process | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0050821 | protein stabilization | 3 | 191 | 4.49e-01 |
| GO:BP | GO:0071276 | cellular response to cadmium ion | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0040008 | regulation of growth | 6 | 501 | 4.49e-01 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 12 | 1199 | 4.49e-01 |
| GO:BP | GO:0050798 | activated T cell proliferation | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0051788 | response to misfolded protein | 1 | 25 | 4.49e-01 |
| GO:BP | GO:1990776 | response to angiotensin | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0021515 | cell differentiation in spinal cord | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0048513 | animal organ development | 20 | 2189 | 4.49e-01 |
| GO:BP | GO:0021517 | ventral spinal cord development | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0006541 | glutamine metabolic process | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0042401 | biogenic amine biosynthetic process | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0034470 | ncRNA processing | 5 | 400 | 4.49e-01 |
| GO:BP | GO:1905606 | regulation of presynapse assembly | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0099174 | regulation of presynapse organization | 1 | 25 | 4.49e-01 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 2 | 101 | 4.49e-01 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 100 | 4.49e-01 |
| GO:BP | GO:0001841 | neural tube formation | 2 | 101 | 4.49e-01 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0090279 | regulation of calcium ion import | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 3 | 192 | 4.49e-01 |
| GO:BP | GO:0009309 | amine biosynthetic process | 1 | 25 | 4.49e-01 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0036075 | replacement ossification | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0003279 | cardiac septum development | 2 | 101 | 4.49e-01 |
| GO:BP | GO:0001958 | endochondral ossification | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0048729 | tissue morphogenesis | 6 | 504 | 4.49e-01 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0061311 | cell surface receptor signaling pathway involved in heart development | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0003272 | endocardial cushion formation | 1 | 24 | 4.49e-01 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 1 | 25 | 4.49e-01 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 2 | 101 | 4.49e-01 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 7 | 616 | 4.49e-01 |
| GO:BP | GO:1903203 | regulation of oxidative stress-induced neuron death | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0050999 | regulation of nitric-oxide synthase activity | 1 | 25 | 4.49e-01 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 2 | 100 | 4.49e-01 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 1 | 25 | 4.49e-01 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 2 | 98 | 4.49e-01 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 2 | 100 | 4.49e-01 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 3 | 194 | 4.50e-01 |
| GO:BP | GO:0035909 | aorta morphogenesis | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0030224 | monocyte differentiation | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0034311 | diol metabolic process | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0010092 | specification of animal organ identity | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0035994 | response to muscle stretch | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0050931 | pigment cell differentiation | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0031128 | developmental induction | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0006914 | autophagy | 6 | 513 | 4.54e-01 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 2 | 103 | 4.54e-01 |
| GO:BP | GO:0060674 | placenta blood vessel development | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 8 | 742 | 4.54e-01 |
| GO:BP | GO:0042398 | cellular modified amino acid biosynthetic process | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0051223 | regulation of protein transport | 5 | 404 | 4.54e-01 |
| GO:BP | GO:0007431 | salivary gland development | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0046326 | positive regulation of glucose import | 1 | 26 | 4.54e-01 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 6 | 513 | 4.54e-01 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 1 | 26 | 4.54e-01 |
| GO:BP | GO:1901361 | organic cyclic compound catabolic process | 5 | 406 | 4.57e-01 |
| GO:BP | GO:0015698 | inorganic anion transport | 2 | 104 | 4.59e-01 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 1 | 27 | 4.61e-01 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by RNA | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0071466 | cellular response to xenobiotic stimulus | 2 | 105 | 4.61e-01 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0033687 | osteoblast proliferation | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0097484 | dendrite extension | 1 | 27 | 4.61e-01 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 3 | 199 | 4.61e-01 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0010518 | positive regulation of phospholipase activity | 1 | 27 | 4.61e-01 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 3 | 199 | 4.61e-01 |
| GO:BP | GO:0061756 | leukocyte adhesion to vascular endothelial cell | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 1 | 27 | 4.61e-01 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0034063 | stress granule assembly | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0036475 | neuron death in response to oxidative stress | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0050905 | neuromuscular process | 2 | 107 | 4.63e-01 |
| GO:BP | GO:0033522 | histone H2A ubiquitination | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 1 | 28 | 4.63e-01 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0048483 | autonomic nervous system development | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0070098 | chemokine-mediated signaling pathway | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0060306 | regulation of membrane repolarization | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0009648 | photoperiodism | 1 | 28 | 4.63e-01 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0046189 | phenol-containing compound biosynthetic process | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0060325 | face morphogenesis | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0030879 | mammary gland development | 2 | 106 | 4.63e-01 |
| GO:BP | GO:0016358 | dendrite development | 3 | 202 | 4.63e-01 |
| GO:BP | GO:0021537 | telencephalon development | 3 | 201 | 4.63e-01 |
| GO:BP | GO:0008207 | C21-steroid hormone metabolic process | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0070873 | regulation of glycogen metabolic process | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 1 | 28 | 4.63e-01 |
| GO:BP | GO:0009611 | response to wounding | 5 | 415 | 4.63e-01 |
| GO:BP | GO:0120178 | steroid hormone biosynthetic process | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 5 | 418 | 4.63e-01 |
| GO:BP | GO:0016049 | cell growth | 5 | 418 | 4.63e-01 |
| GO:BP | GO:0060047 | heart contraction | 3 | 205 | 4.63e-01 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 2 | 110 | 4.63e-01 |
| GO:BP | GO:0007549 | dosage compensation | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0042743 | hydrogen peroxide metabolic process | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0045926 | negative regulation of growth | 3 | 206 | 4.63e-01 |
| GO:BP | GO:0060966 | regulation of gene silencing by RNA | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0048699 | generation of neurons | 11 | 1119 | 4.63e-01 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 2 | 110 | 4.63e-01 |
| GO:BP | GO:0060914 | heart formation | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 3 | 204 | 4.63e-01 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 2 | 110 | 4.63e-01 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 1 | 29 | 4.63e-01 |
| GO:BP | GO:1903426 | regulation of reactive oxygen species biosynthetic process | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 2 | 109 | 4.63e-01 |
| GO:BP | GO:0001889 | liver development | 2 | 110 | 4.63e-01 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0006730 | one-carbon metabolic process | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 2 | 108 | 4.63e-01 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0046688 | response to copper ion | 1 | 29 | 4.63e-01 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0000902 | cell morphogenesis | 8 | 761 | 4.63e-01 |
| GO:BP | GO:0090075 | relaxation of muscle | 1 | 29 | 4.63e-01 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 10 | 994 | 4.63e-01 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 10 | 993 | 4.63e-01 |
| GO:BP | GO:0007272 | ensheathment of neurons | 2 | 113 | 4.63e-01 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 3 | 213 | 4.63e-01 |
| GO:BP | GO:0008366 | axon ensheathment | 2 | 113 | 4.63e-01 |
| GO:BP | GO:0042110 | T cell activation | 4 | 313 | 4.63e-01 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0003009 | skeletal muscle contraction | 1 | 31 | 4.63e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 9 | 893 | 4.63e-01 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 1 | 30 | 4.63e-01 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 2 | 113 | 4.63e-01 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0048568 | embryonic organ development | 4 | 316 | 4.63e-01 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 5 | 428 | 4.63e-01 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 1 | 31 | 4.63e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 10 | 1014 | 4.63e-01 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 2 | 112 | 4.63e-01 |
| GO:BP | GO:0061448 | connective tissue development | 3 | 208 | 4.63e-01 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 6 | 539 | 4.63e-01 |
| GO:BP | GO:0023056 | positive regulation of signaling | 12 | 1256 | 4.63e-01 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0042552 | myelination | 2 | 111 | 4.63e-01 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 12 | 1255 | 4.63e-01 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0097106 | postsynaptic density organization | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0032350 | regulation of hormone metabolic process | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0035107 | appendage morphogenesis | 2 | 114 | 4.63e-01 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 1 | 31 | 4.63e-01 |
| GO:BP | GO:1901381 | positive regulation of potassium ion transmembrane transport | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 1 | 31 | 4.63e-01 |
| GO:BP | GO:2000758 | positive regulation of peptidyl-lysine acetylation | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0010390 | histone monoubiquitination | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0003231 | cardiac ventricle development | 2 | 114 | 4.63e-01 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 2 | 112 | 4.63e-01 |
| GO:BP | GO:0009712 | catechol-containing compound metabolic process | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0010828 | positive regulation of glucose transmembrane transport | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0001775 | cell activation | 7 | 655 | 4.63e-01 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0034405 | response to fluid shear stress | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0035108 | limb morphogenesis | 2 | 114 | 4.63e-01 |
| GO:BP | GO:0006584 | catecholamine metabolic process | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 2 | 113 | 4.63e-01 |
| GO:BP | GO:0051642 | centrosome localization | 1 | 31 | 4.63e-01 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0090504 | epiboly | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 2 | 114 | 4.63e-01 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 4 | 317 | 4.63e-01 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 1 | 30 | 4.63e-01 |
| GO:BP | GO:0051051 | negative regulation of transport | 4 | 318 | 4.63e-01 |
| GO:BP | GO:0031214 | biomineral tissue development | 2 | 111 | 4.63e-01 |
| GO:BP | GO:0016310 | phosphorylation | 13 | 1372 | 4.63e-01 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 1 | 31 | 4.63e-01 |
| GO:BP | GO:0010720 | positive regulation of cell development | 4 | 317 | 4.63e-01 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 2 | 115 | 4.64e-01 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 2 | 115 | 4.64e-01 |
| GO:BP | GO:0031929 | TOR signaling | 2 | 115 | 4.64e-01 |
| GO:BP | GO:0008217 | regulation of blood pressure | 2 | 115 | 4.64e-01 |
| GO:BP | GO:0042060 | wound healing | 4 | 320 | 4.64e-01 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 4 | 320 | 4.64e-01 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 7 | 660 | 4.65e-01 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 2 | 116 | 4.65e-01 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 2 | 116 | 4.65e-01 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 4.65e-01 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0031018 | endocrine pancreas development | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0010543 | regulation of platelet activation | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0003015 | heart process | 3 | 216 | 4.65e-01 |
| GO:BP | GO:0045823 | positive regulation of heart contraction | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0097502 | mannosylation | 1 | 32 | 4.65e-01 |
| GO:BP | GO:1903524 | positive regulation of blood circulation | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 1 | 32 | 4.65e-01 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 5 | 433 | 4.65e-01 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 2 | 116 | 4.65e-01 |
| GO:BP | GO:0051047 | positive regulation of secretion | 3 | 215 | 4.65e-01 |
| GO:BP | GO:0006402 | mRNA catabolic process | 3 | 216 | 4.65e-01 |
| GO:BP | GO:0016071 | mRNA metabolic process | 7 | 665 | 4.69e-01 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 2 | 118 | 4.72e-01 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 2 | 118 | 4.72e-01 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0003161 | cardiac conduction system development | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0150076 | neuroinflammatory response | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0060323 | head morphogenesis | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0006040 | amino sugar metabolic process | 1 | 33 | 4.74e-01 |
| GO:BP | GO:0007399 | nervous system development | 17 | 1904 | 4.76e-01 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 2 | 121 | 4.76e-01 |
| GO:BP | GO:0007283 | spermatogenesis | 4 | 330 | 4.76e-01 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 2 | 120 | 4.76e-01 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 20 | 2285 | 4.76e-01 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0045321 | leukocyte activation | 6 | 554 | 4.76e-01 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0060193 | positive regulation of lipase activity | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0019748 | secondary metabolic process | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0006364 | rRNA processing | 3 | 221 | 4.76e-01 |
| GO:BP | GO:0032768 | regulation of monooxygenase activity | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 2 | 120 | 4.76e-01 |
| GO:BP | GO:1903034 | regulation of response to wounding | 2 | 121 | 4.76e-01 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 3 | 221 | 4.76e-01 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 2 | 120 | 4.76e-01 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0035315 | hair cell differentiation | 1 | 34 | 4.76e-01 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0035272 | exocrine system development | 1 | 34 | 4.76e-01 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 4 | 331 | 4.78e-01 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 8 | 796 | 4.81e-01 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 2 | 123 | 4.82e-01 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 7 | 678 | 4.82e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 39 | 4794 | 4.82e-01 |
| GO:BP | GO:0034660 | ncRNA metabolic process | 6 | 561 | 4.82e-01 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0060711 | labyrinthine layer development | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0048538 | thymus development | 1 | 35 | 4.82e-01 |
| GO:BP | GO:0000209 | protein polyubiquitination | 3 | 226 | 4.85e-01 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 2 | 124 | 4.86e-01 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0043268 | positive regulation of potassium ion transport | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0050890 | cognition | 3 | 227 | 4.87e-01 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 22 | 2565 | 4.87e-01 |
| GO:BP | GO:1903409 | reactive oxygen species biosynthetic process | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0022008 | neurogenesis | 12 | 1294 | 4.87e-01 |
| GO:BP | GO:0048066 | developmental pigmentation | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0006397 | mRNA processing | 5 | 450 | 4.87e-01 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 1 | 36 | 4.87e-01 |
| GO:BP | GO:0010769 | regulation of cell morphogenesis involved in differentiation | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0097178 | ruffle assembly | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 2 | 126 | 4.89e-01 |
| GO:BP | GO:0001825 | blastocyst formation | 1 | 37 | 4.89e-01 |
| GO:BP | GO:1990868 | response to chemokine | 1 | 37 | 4.89e-01 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0008360 | regulation of cell shape | 2 | 126 | 4.89e-01 |
| GO:BP | GO:1990869 | cellular response to chemokine | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0045727 | positive regulation of translation | 2 | 126 | 4.89e-01 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0000423 | mitophagy | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0030832 | regulation of actin filament length | 2 | 127 | 4.89e-01 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0070509 | calcium ion import | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0003176 | aortic valve development | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0072175 | epithelial tube formation | 2 | 127 | 4.89e-01 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 1 | 37 | 4.89e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 22 | 2573 | 4.89e-01 |
| GO:BP | GO:0008033 | tRNA processing | 2 | 128 | 4.92e-01 |
| GO:BP | GO:0006986 | response to unfolded protein | 2 | 128 | 4.92e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 14 | 1555 | 4.92e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 18 | 2065 | 4.93e-01 |
| GO:BP | GO:0048232 | male gamete generation | 4 | 343 | 4.93e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 17 | 1939 | 4.95e-01 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 2 | 129 | 4.95e-01 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 1 | 38 | 4.96e-01 |
| GO:BP | GO:0045454 | cell redox homeostasis | 1 | 38 | 4.96e-01 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 3 | 234 | 4.96e-01 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 2 | 130 | 4.99e-01 |
| GO:BP | GO:0042063 | gliogenesis | 3 | 235 | 4.99e-01 |
| GO:BP | GO:0042311 | vasodilation | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 2 | 131 | 5.00e-01 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 2 | 131 | 5.00e-01 |
| GO:BP | GO:0043010 | camera-type eye development | 3 | 236 | 5.00e-01 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0099054 | presynapse assembly | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0006378 | mRNA polyadenylation | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0050906 | detection of stimulus involved in sensory perception | 1 | 39 | 5.00e-01 |
| GO:BP | GO:0001659 | temperature homeostasis | 2 | 132 | 5.02e-01 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 2 | 132 | 5.02e-01 |
| GO:BP | GO:0048839 | inner ear development | 2 | 132 | 5.02e-01 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 2 | 132 | 5.02e-01 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0042446 | hormone biosynthetic process | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0039531 | regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0045778 | positive regulation of ossification | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0014009 | glial cell proliferation | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0098739 | import across plasma membrane | 2 | 133 | 5.02e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 28 | 3386 | 5.02e-01 |
| GO:BP | GO:0034331 | cell junction maintenance | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 2 | 133 | 5.02e-01 |
| GO:BP | GO:0046649 | lymphocyte activation | 5 | 465 | 5.02e-01 |
| GO:BP | GO:0002376 | immune system process | 13 | 1447 | 5.02e-01 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 1 | 40 | 5.02e-01 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 134 | 5.03e-01 |
| GO:BP | GO:0030041 | actin filament polymerization | 2 | 134 | 5.03e-01 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 2 | 134 | 5.03e-01 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 2 | 134 | 5.03e-01 |
| GO:BP | GO:0046686 | response to cadmium ion | 1 | 41 | 5.03e-01 |
| GO:BP | GO:1905314 | semi-lunar valve development | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0050881 | musculoskeletal movement | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0035196 | miRNA processing | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0060612 | adipose tissue development | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0086009 | membrane repolarization | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0043631 | RNA polyadenylation | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0050879 | multicellular organismal movement | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0030520 | intracellular estrogen receptor signaling pathway | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 3 | 242 | 5.03e-01 |
| GO:BP | GO:0035329 | hippo signaling | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0046324 | regulation of glucose import | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 2 | 135 | 5.03e-01 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0006749 | glutathione metabolic process | 1 | 41 | 5.03e-01 |
| GO:BP | GO:0051260 | protein homooligomerization | 2 | 136 | 5.05e-01 |
| GO:BP | GO:0051726 | regulation of cell cycle | 9 | 956 | 5.05e-01 |
| GO:BP | GO:0090087 | regulation of peptide transport | 2 | 136 | 5.05e-01 |
| GO:BP | GO:0051289 | protein homotetramerization | 1 | 42 | 5.10e-01 |
| GO:BP | GO:0099172 | presynapse organization | 1 | 42 | 5.10e-01 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 1 | 42 | 5.10e-01 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 4 | 358 | 5.10e-01 |
| GO:BP | GO:0031103 | axon regeneration | 1 | 42 | 5.10e-01 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 1 | 42 | 5.10e-01 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 1 | 42 | 5.10e-01 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 2 | 138 | 5.12e-01 |
| GO:BP | GO:0035148 | tube formation | 2 | 138 | 5.12e-01 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 3 | 246 | 5.12e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 24 | 2890 | 5.16e-01 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 1 | 43 | 5.17e-01 |
| GO:BP | GO:0051353 | positive regulation of oxidoreductase activity | 1 | 43 | 5.17e-01 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 1 | 43 | 5.17e-01 |
| GO:BP | GO:0098900 | regulation of action potential | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0001961 | positive regulation of cytokine-mediated signaling pathway | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 3 | 249 | 5.20e-01 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 3 | 249 | 5.20e-01 |
| GO:BP | GO:0036211 | protein modification process | 24 | 2901 | 5.20e-01 |
| GO:BP | GO:0045123 | cellular extravasation | 1 | 44 | 5.20e-01 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0030073 | insulin secretion | 2 | 141 | 5.20e-01 |
| GO:BP | GO:0022029 | telencephalon cell migration | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0060485 | mesenchyme development | 3 | 249 | 5.20e-01 |
| GO:BP | GO:0048731 | system development | 24 | 2901 | 5.20e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 12 | 1345 | 5.20e-01 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 1 | 44 | 5.20e-01 |
| GO:BP | GO:0060173 | limb development | 2 | 142 | 5.22e-01 |
| GO:BP | GO:0048736 | appendage development | 2 | 142 | 5.22e-01 |
| GO:BP | GO:0031099 | regeneration | 2 | 142 | 5.22e-01 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 3 | 252 | 5.24e-01 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0031529 | ruffle organization | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 5 | 486 | 5.24e-01 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0060324 | face development | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0010171 | body morphogenesis | 1 | 45 | 5.24e-01 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 2 | 144 | 5.26e-01 |
| GO:BP | GO:0044248 | cellular catabolic process | 11 | 1229 | 5.26e-01 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 2 | 144 | 5.26e-01 |
| GO:BP | GO:0006417 | regulation of translation | 4 | 370 | 5.28e-01 |
| GO:BP | GO:0098609 | cell-cell adhesion | 6 | 609 | 5.28e-01 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 1 | 46 | 5.30e-01 |
| GO:BP | GO:0014823 | response to activity | 1 | 46 | 5.30e-01 |
| GO:BP | GO:0016574 | histone ubiquitination | 1 | 46 | 5.30e-01 |
| GO:BP | GO:0061462 | protein localization to lysosome | 1 | 46 | 5.30e-01 |
| GO:BP | GO:0021885 | forebrain cell migration | 1 | 46 | 5.30e-01 |
| GO:BP | GO:0006401 | RNA catabolic process | 3 | 256 | 5.30e-01 |
| GO:BP | GO:0016072 | rRNA metabolic process | 3 | 258 | 5.30e-01 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0008015 | blood circulation | 4 | 374 | 5.30e-01 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 8 | 859 | 5.30e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 2 | 147 | 5.30e-01 |
| GO:BP | GO:0031102 | neuron projection regeneration | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 4 | 372 | 5.30e-01 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0021915 | neural tube development | 2 | 146 | 5.30e-01 |
| GO:BP | GO:1903351 | cellular response to dopamine | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 1 | 47 | 5.30e-01 |
| GO:BP | GO:0042102 | positive regulation of T cell proliferation | 1 | 47 | 5.30e-01 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 3 | 258 | 5.30e-01 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 2 | 147 | 5.30e-01 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 1 | 48 | 5.34e-01 |
| GO:BP | GO:1903350 | response to dopamine | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0051235 | maintenance of location | 3 | 260 | 5.34e-01 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 33 | 4128 | 5.34e-01 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 3 | 260 | 5.34e-01 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0003205 | cardiac chamber development | 2 | 149 | 5.34e-01 |
| GO:BP | GO:0019827 | stem cell population maintenance | 2 | 149 | 5.34e-01 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0040007 | growth | 7 | 742 | 5.34e-01 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0019233 | sensory perception of pain | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 1 | 48 | 5.34e-01 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 4 | 378 | 5.34e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 10 | 1123 | 5.36e-01 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 1 | 49 | 5.36e-01 |
| GO:BP | GO:0098727 | maintenance of cell number | 2 | 151 | 5.36e-01 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 1 | 49 | 5.36e-01 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 2 | 150 | 5.36e-01 |
| GO:BP | GO:0006006 | glucose metabolic process | 2 | 151 | 5.36e-01 |
| GO:BP | GO:0001655 | urogenital system development | 1 | 49 | 5.36e-01 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 1 | 49 | 5.36e-01 |
| GO:BP | GO:0007005 | mitochondrion organization | 5 | 500 | 5.36e-01 |
| GO:BP | GO:0007596 | blood coagulation | 2 | 150 | 5.36e-01 |
| GO:BP | GO:0051099 | positive regulation of binding | 2 | 150 | 5.36e-01 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 1 | 49 | 5.36e-01 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 2 | 151 | 5.36e-01 |
| GO:BP | GO:0051216 | cartilage development | 2 | 151 | 5.36e-01 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 1 | 49 | 5.36e-01 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 2 | 151 | 5.36e-01 |
| GO:BP | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 153 | 5.37e-01 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 1 | 50 | 5.37e-01 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 4 | 382 | 5.37e-01 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 12 | 1380 | 5.37e-01 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0006941 | striated muscle contraction | 2 | 152 | 5.37e-01 |
| GO:BP | GO:1900046 | regulation of hemostasis | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 2 | 152 | 5.37e-01 |
| GO:BP | GO:0050865 | regulation of cell activation | 4 | 384 | 5.37e-01 |
| GO:BP | GO:1903317 | regulation of protein maturation | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0010038 | response to metal ion | 3 | 264 | 5.37e-01 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0035904 | aorta development | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0030193 | regulation of blood coagulation | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 1 | 50 | 5.37e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 5 | 504 | 5.37e-01 |
| GO:BP | GO:0007599 | hemostasis | 2 | 153 | 5.37e-01 |
| GO:BP | GO:0007416 | synapse assembly | 2 | 152 | 5.37e-01 |
| GO:BP | GO:0050817 | coagulation | 2 | 153 | 5.37e-01 |
| GO:BP | GO:0051170 | import into nucleus | 2 | 152 | 5.37e-01 |
| GO:BP | GO:0007507 | heart development | 5 | 506 | 5.40e-01 |
| GO:BP | GO:0045165 | cell fate commitment | 2 | 155 | 5.40e-01 |
| GO:BP | GO:1904888 | cranial skeletal system development | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 4 | 387 | 5.40e-01 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0099173 | postsynapse organization | 2 | 155 | 5.40e-01 |
| GO:BP | GO:1990928 | response to amino acid starvation | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0048645 | animal organ formation | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 2 | 154 | 5.40e-01 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 4 | 386 | 5.40e-01 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 1 | 51 | 5.40e-01 |
| GO:BP | GO:1902476 | chloride transmembrane transport | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 1 | 51 | 5.40e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 3 | 270 | 5.44e-01 |
| GO:BP | GO:0002064 | epithelial cell development | 2 | 156 | 5.44e-01 |
| GO:BP | GO:0006081 | cellular aldehyde metabolic process | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 1 | 52 | 5.44e-01 |
| GO:BP | GO:1902074 | response to salt | 3 | 271 | 5.44e-01 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0050877 | nervous system process | 6 | 634 | 5.44e-01 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0060191 | regulation of lipase activity | 1 | 52 | 5.44e-01 |
| GO:BP | GO:0007155 | cell adhesion | 9 | 1012 | 5.44e-01 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 2 | 157 | 5.44e-01 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 4 | 391 | 5.45e-01 |
| GO:BP | GO:0070482 | response to oxygen levels | 3 | 272 | 5.45e-01 |
| GO:BP | GO:0001654 | eye development | 3 | 272 | 5.45e-01 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 7 | 763 | 5.46e-01 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 1 | 53 | 5.46e-01 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 1 | 53 | 5.46e-01 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 1 | 53 | 5.46e-01 |
| GO:BP | GO:0031016 | pancreas development | 1 | 53 | 5.46e-01 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 2 | 158 | 5.46e-01 |
| GO:BP | GO:0150063 | visual system development | 3 | 273 | 5.46e-01 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 1 | 53 | 5.46e-01 |
| GO:BP | GO:0046323 | glucose import | 1 | 53 | 5.46e-01 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 3 | 273 | 5.46e-01 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 1 | 53 | 5.46e-01 |
| GO:BP | GO:0009615 | response to virus | 3 | 275 | 5.49e-01 |
| GO:BP | GO:0048880 | sensory system development | 3 | 275 | 5.49e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 10 | 1147 | 5.49e-01 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 2 | 160 | 5.50e-01 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 2 | 160 | 5.50e-01 |
| GO:BP | GO:0061035 | regulation of cartilage development | 1 | 54 | 5.50e-01 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 1 | 54 | 5.50e-01 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 1 | 54 | 5.50e-01 |
| GO:BP | GO:0042246 | tissue regeneration | 1 | 54 | 5.50e-01 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 1 | 54 | 5.50e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 2 | 161 | 5.52e-01 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 2 | 161 | 5.52e-01 |
| GO:BP | GO:0035195 | miRNA-mediated gene silencing | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0006611 | protein export from nucleus | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 1 | 55 | 5.54e-01 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 3 | 278 | 5.54e-01 |
| GO:BP | GO:0050818 | regulation of coagulation | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 1 | 55 | 5.54e-01 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 3 | 279 | 5.54e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 3 | 279 | 5.54e-01 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 4 | 400 | 5.55e-01 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 3 | 280 | 5.56e-01 |
| GO:BP | GO:0030163 | protein catabolic process | 8 | 901 | 5.56e-01 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 1 | 56 | 5.57e-01 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 1 | 56 | 5.57e-01 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 3 | 281 | 5.57e-01 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | 56 | 5.57e-01 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 1 | 56 | 5.57e-01 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 1 | 56 | 5.57e-01 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 1 | 56 | 5.57e-01 |
| GO:BP | GO:0046847 | filopodium assembly | 1 | 56 | 5.57e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 14 | 1681 | 5.60e-01 |
| GO:BP | GO:0007623 | circadian rhythm | 2 | 165 | 5.60e-01 |
| GO:BP | GO:0017148 | negative regulation of translation | 2 | 165 | 5.60e-01 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 1 | 57 | 5.60e-01 |
| GO:BP | GO:0098657 | import into cell | 2 | 166 | 5.60e-01 |
| GO:BP | GO:0044057 | regulation of system process | 4 | 405 | 5.60e-01 |
| GO:BP | GO:0061614 | miRNA transcription | 1 | 57 | 5.60e-01 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 1 | 57 | 5.60e-01 |
| GO:BP | GO:0042116 | macrophage activation | 1 | 57 | 5.60e-01 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 1 | 57 | 5.60e-01 |
| GO:BP | GO:0030072 | peptide hormone secretion | 2 | 166 | 5.60e-01 |
| GO:BP | GO:0006576 | biogenic amine metabolic process | 1 | 57 | 5.60e-01 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 1 | 57 | 5.60e-01 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 2 | 167 | 5.63e-01 |
| GO:BP | GO:1901215 | negative regulation of neuron death | 2 | 167 | 5.63e-01 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 2 | 168 | 5.64e-01 |
| GO:BP | GO:0048844 | artery morphogenesis | 1 | 58 | 5.64e-01 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 1 | 58 | 5.64e-01 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 1 | 58 | 5.64e-01 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 2 | 168 | 5.64e-01 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 1 | 58 | 5.64e-01 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 2 | 168 | 5.64e-01 |
| GO:BP | GO:0006821 | chloride transport | 1 | 58 | 5.64e-01 |
| GO:BP | GO:0033059 | cellular pigmentation | 1 | 58 | 5.64e-01 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 1 | 59 | 5.68e-01 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 3 | 288 | 5.68e-01 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 1 | 59 | 5.68e-01 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 1 | 59 | 5.68e-01 |
| GO:BP | GO:0043954 | cellular component maintenance | 1 | 59 | 5.68e-01 |
| GO:BP | GO:0021510 | spinal cord development | 1 | 59 | 5.68e-01 |
| GO:BP | GO:0032502 | developmental process | 35 | 4494 | 5.68e-01 |
| GO:BP | GO:0051651 | maintenance of location in cell | 2 | 170 | 5.69e-01 |
| GO:BP | GO:0060348 | bone development | 2 | 170 | 5.69e-01 |
| GO:BP | GO:0002790 | peptide secretion | 2 | 170 | 5.69e-01 |
| GO:BP | GO:0031647 | regulation of protein stability | 3 | 290 | 5.71e-01 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0051899 | membrane depolarization | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0035194 | ncRNA-mediated post-transcriptional gene silencing | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 1 | 60 | 5.71e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 2 | 172 | 5.72e-01 |
| GO:BP | GO:1901617 | organic hydroxy compound biosynthetic process | 2 | 172 | 5.72e-01 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 2 | 172 | 5.72e-01 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 2 | 172 | 5.72e-01 |
| GO:BP | GO:0008016 | regulation of heart contraction | 2 | 173 | 5.74e-01 |
| GO:BP | GO:0018958 | phenol-containing compound metabolic process | 1 | 61 | 5.74e-01 |
| GO:BP | GO:0034308 | primary alcohol metabolic process | 1 | 61 | 5.74e-01 |
| GO:BP | GO:0021954 | central nervous system neuron development | 1 | 61 | 5.74e-01 |
| GO:BP | GO:0030032 | lamellipodium assembly | 1 | 61 | 5.74e-01 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 1 | 61 | 5.74e-01 |
| GO:BP | GO:0042391 | regulation of membrane potential | 3 | 293 | 5.74e-01 |
| GO:BP | GO:0034394 | protein localization to cell surface | 1 | 61 | 5.74e-01 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 1 | 61 | 5.74e-01 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 1 | 62 | 5.76e-01 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 3 | 295 | 5.76e-01 |
| GO:BP | GO:0008380 | RNA splicing | 4 | 419 | 5.76e-01 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 1 | 62 | 5.76e-01 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 4 | 418 | 5.76e-01 |
| GO:BP | GO:0051046 | regulation of secretion | 4 | 418 | 5.76e-01 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 1 | 62 | 5.76e-01 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 1 | 62 | 5.76e-01 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 3 | 295 | 5.76e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 16 | 1974 | 5.76e-01 |
| GO:BP | GO:0071869 | response to catecholamine | 1 | 62 | 5.76e-01 |
| GO:BP | GO:0043588 | skin development | 2 | 175 | 5.76e-01 |
| GO:BP | GO:0071867 | response to monoamine | 1 | 62 | 5.76e-01 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 3 | 296 | 5.77e-01 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 5 | 547 | 5.80e-01 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 1 | 63 | 5.81e-01 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 1 | 63 | 5.81e-01 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 1 | 63 | 5.81e-01 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 1 | 63 | 5.81e-01 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 1 | 63 | 5.81e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 14 | 1719 | 5.82e-01 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 2 | 178 | 5.84e-01 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 1 | 64 | 5.84e-01 |
| GO:BP | GO:0051262 | protein tetramerization | 1 | 64 | 5.84e-01 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 3 | 300 | 5.84e-01 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 1 | 64 | 5.84e-01 |
| GO:BP | GO:0042594 | response to starvation | 2 | 178 | 5.84e-01 |
| GO:BP | GO:0048678 | response to axon injury | 1 | 64 | 5.84e-01 |
| GO:BP | GO:1901379 | regulation of potassium ion transmembrane transport | 1 | 64 | 5.84e-01 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 3 | 300 | 5.84e-01 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 4 | 425 | 5.84e-01 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 4 | 427 | 5.85e-01 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 3 | 302 | 5.85e-01 |
| GO:BP | GO:0015833 | peptide transport | 2 | 179 | 5.85e-01 |
| GO:BP | GO:0022411 | cellular component disassembly | 4 | 427 | 5.85e-01 |
| GO:BP | GO:0042254 | ribosome biogenesis | 3 | 301 | 5.85e-01 |
| GO:BP | GO:0006414 | translational elongation | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 1 | 65 | 5.85e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0008306 | associative learning | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0016441 | post-transcriptional gene silencing | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 1 | 65 | 5.85e-01 |
| GO:BP | GO:0065003 | protein-containing complex assembly | 11 | 1335 | 5.88e-01 |
| GO:BP | GO:0006954 | inflammatory response | 4 | 429 | 5.88e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 9 | 1075 | 5.90e-01 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 1 | 66 | 5.91e-01 |
| GO:BP | GO:0009582 | detection of abiotic stimulus | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0050807 | regulation of synapse organization | 2 | 184 | 5.91e-01 |
| GO:BP | GO:0019318 | hexose metabolic process | 2 | 185 | 5.91e-01 |
| GO:BP | GO:0006094 | gluconeogenesis | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0051851 | modulation by host of symbiont process | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 1 | 68 | 5.91e-01 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0051707 | response to other organism | 7 | 820 | 5.91e-01 |
| GO:BP | GO:0009581 | detection of external stimulus | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0001570 | vasculogenesis | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0001942 | hair follicle development | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0030001 | metal ion transport | 5 | 562 | 5.91e-01 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 2 | 185 | 5.91e-01 |
| GO:BP | GO:0051341 | regulation of oxidoreductase activity | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0097061 | dendritic spine organization | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0009308 | amine metabolic process | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0007422 | peripheral nervous system development | 1 | 67 | 5.91e-01 |
| GO:BP | GO:0006839 | mitochondrial transport | 2 | 185 | 5.91e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 7 | 820 | 5.91e-01 |
| GO:BP | GO:0010822 | positive regulation of mitochondrion organization | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 1 | 68 | 5.91e-01 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 3 | 307 | 5.91e-01 |
| GO:BP | GO:0043279 | response to alkaloid | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 1 | 68 | 5.91e-01 |
| GO:BP | GO:0090257 | regulation of muscle system process | 2 | 184 | 5.91e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 15 | 1869 | 5.91e-01 |
| GO:BP | GO:0022402 | cell cycle process | 9 | 1086 | 5.92e-01 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0090277 | positive regulation of peptide hormone secretion | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0022405 | hair cycle process | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0006399 | tRNA metabolic process | 2 | 186 | 5.92e-01 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 2 | 187 | 5.92e-01 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0022404 | molting cycle process | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 2 | 186 | 5.92e-01 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 2 | 187 | 5.92e-01 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 2 | 187 | 5.92e-01 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 12 | 1484 | 5.92e-01 |
| GO:BP | GO:0006486 | protein glycosylation | 2 | 187 | 5.92e-01 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0030901 | midbrain development | 1 | 69 | 5.92e-01 |
| GO:BP | GO:0030154 | cell differentiation | 23 | 2958 | 5.92e-01 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0097306 | cellular response to alcohol | 1 | 70 | 5.92e-01 |
| GO:BP | GO:1901216 | positive regulation of neuron death | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 24 | 3090 | 5.92e-01 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 2 | 189 | 5.92e-01 |
| GO:BP | GO:0006096 | glycolytic process | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 1 | 70 | 5.92e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 7 | 830 | 5.92e-01 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 2 | 189 | 5.92e-01 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 1 | 70 | 5.92e-01 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 1 | 70 | 5.92e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 10 | 1223 | 5.92e-01 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 1 | 71 | 5.95e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 8 | 963 | 5.95e-01 |
| GO:BP | GO:0098656 | monoatomic anion transmembrane transport | 1 | 71 | 5.95e-01 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 1 | 71 | 5.95e-01 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 1 | 71 | 5.95e-01 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 1 | 71 | 5.95e-01 |
| GO:BP | GO:0046620 | regulation of organ growth | 1 | 71 | 5.95e-01 |
| GO:BP | GO:0001892 | embryonic placenta development | 1 | 71 | 5.95e-01 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 3 | 316 | 5.95e-01 |
| GO:BP | GO:0006473 | protein acetylation | 2 | 190 | 5.95e-01 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 2 | 191 | 5.95e-01 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 2 | 191 | 5.95e-01 |
| GO:BP | GO:0043933 | protein-containing complex organization | 12 | 1496 | 5.97e-01 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 3 | 318 | 5.97e-01 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 6 | 706 | 5.97e-01 |
| GO:BP | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic | 1 | 72 | 5.97e-01 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 1 | 72 | 5.97e-01 |
| GO:BP | GO:0072665 | protein localization to vacuole | 1 | 72 | 5.97e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 15 | 1898 | 5.97e-01 |
| GO:BP | GO:0050671 | positive regulation of lymphocyte proliferation | 1 | 72 | 5.97e-01 |
| GO:BP | GO:0003013 | circulatory system process | 4 | 447 | 5.99e-01 |
| GO:BP | GO:0006446 | regulation of translational initiation | 1 | 73 | 6.00e-01 |
| GO:BP | GO:0042471 | ear morphogenesis | 1 | 73 | 6.00e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 2 | 194 | 6.00e-01 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 1 | 73 | 6.00e-01 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 1 | 73 | 6.00e-01 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 2 | 194 | 6.00e-01 |
| GO:BP | GO:0060993 | kidney morphogenesis | 1 | 73 | 6.00e-01 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 1 | 73 | 6.00e-01 |
| GO:BP | GO:0009987 | cellular process | 84 | 11336 | 6.01e-01 |
| GO:BP | GO:0014896 | muscle hypertrophy | 1 | 74 | 6.05e-01 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 1 | 74 | 6.05e-01 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 1 | 74 | 6.05e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 23 | 2984 | 6.05e-01 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 4 | 453 | 6.08e-01 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 75 | 6.08e-01 |
| GO:BP | GO:0051865 | protein autoubiquitination | 1 | 75 | 6.08e-01 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 1 | 75 | 6.08e-01 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 2 | 197 | 6.08e-01 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 1 | 75 | 6.08e-01 |
| GO:BP | GO:0048863 | stem cell differentiation | 2 | 197 | 6.08e-01 |
| GO:BP | GO:0030216 | keratinocyte differentiation | 1 | 75 | 6.08e-01 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 6 | 719 | 6.08e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 7 | 850 | 6.08e-01 |
| GO:BP | GO:0106027 | neuron projection organization | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 2 | 198 | 6.08e-01 |
| GO:BP | GO:0045927 | positive regulation of growth | 2 | 199 | 6.08e-01 |
| GO:BP | GO:0014812 | muscle cell migration | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 2 | 199 | 6.08e-01 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 1 | 76 | 6.08e-01 |
| GO:BP | GO:0006282 | regulation of DNA repair | 2 | 198 | 6.08e-01 |
| GO:BP | GO:1903522 | regulation of blood circulation | 2 | 198 | 6.08e-01 |
| GO:BP | GO:0051402 | neuron apoptotic process | 2 | 200 | 6.11e-01 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 1 | 77 | 6.13e-01 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 1 | 77 | 6.13e-01 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 1 | 77 | 6.13e-01 |
| GO:BP | GO:0007420 | brain development | 5 | 591 | 6.14e-01 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 1 | 78 | 6.16e-01 |
| GO:BP | GO:0007276 | gamete generation | 4 | 461 | 6.16e-01 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 1 | 78 | 6.16e-01 |
| GO:BP | GO:0097581 | lamellipodium organization | 1 | 78 | 6.16e-01 |
| GO:BP | GO:0070085 | glycosylation | 2 | 202 | 6.16e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 6.16e-01 |
| GO:BP | GO:0051702 | biological process involved in interaction with symbiont | 1 | 78 | 6.16e-01 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 6 | 726 | 6.16e-01 |
| GO:BP | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 203 | 6.17e-01 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 2 | 203 | 6.17e-01 |
| GO:BP | GO:0007049 | cell cycle | 12 | 1529 | 6.18e-01 |
| GO:BP | GO:0071824 | protein-DNA complex subunit organization | 2 | 204 | 6.19e-01 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 1 | 79 | 6.19e-01 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 1 | 79 | 6.19e-01 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 5 | 597 | 6.20e-01 |
| GO:BP | GO:0003012 | muscle system process | 3 | 334 | 6.20e-01 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 5 | 597 | 6.20e-01 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 5 | 598 | 6.21e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 11 | 1399 | 6.21e-01 |
| GO:BP | GO:1901342 | regulation of vasculature development | 2 | 206 | 6.21e-01 |
| GO:BP | GO:0006334 | nucleosome assembly | 1 | 80 | 6.21e-01 |
| GO:BP | GO:0045995 | regulation of embryonic development | 1 | 80 | 6.21e-01 |
| GO:BP | GO:0006513 | protein monoubiquitination | 1 | 80 | 6.21e-01 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 2 | 206 | 6.21e-01 |
| GO:BP | GO:0006672 | ceramide metabolic process | 1 | 80 | 6.21e-01 |
| GO:BP | GO:0010817 | regulation of hormone levels | 3 | 336 | 6.21e-01 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 4 | 468 | 6.22e-01 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 1 | 81 | 6.23e-01 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 2 | 207 | 6.23e-01 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 1 | 81 | 6.23e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 7 | 868 | 6.23e-01 |
| GO:BP | GO:0010586 | miRNA metabolic process | 1 | 81 | 6.23e-01 |
| GO:BP | GO:1904659 | glucose transmembrane transport | 1 | 81 | 6.23e-01 |
| GO:BP | GO:0007267 | cell-cell signaling | 9 | 1138 | 6.26e-01 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 1 | 82 | 6.26e-01 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 1 | 82 | 6.26e-01 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 1 | 82 | 6.26e-01 |
| GO:BP | GO:0019751 | polyol metabolic process | 1 | 82 | 6.26e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 5 | 605 | 6.26e-01 |
| GO:BP | GO:0050900 | leukocyte migration | 2 | 209 | 6.26e-01 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 82 | 6.26e-01 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 2 | 209 | 6.26e-01 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 3 | 340 | 6.26e-01 |
| GO:BP | GO:0014706 | striated muscle tissue development | 2 | 210 | 6.27e-01 |
| GO:BP | GO:0046879 | hormone secretion | 2 | 210 | 6.27e-01 |
| GO:BP | GO:0034341 | response to type II interferon | 1 | 83 | 6.28e-01 |
| GO:BP | GO:0030433 | ubiquitin-dependent ERAD pathway | 1 | 83 | 6.28e-01 |
| GO:BP | GO:0006820 | monoatomic anion transport | 1 | 83 | 6.28e-01 |
| GO:BP | GO:0009056 | catabolic process | 16 | 2088 | 6.28e-01 |
| GO:BP | GO:0060840 | artery development | 1 | 83 | 6.28e-01 |
| GO:BP | GO:0070665 | positive regulation of leukocyte proliferation | 1 | 83 | 6.28e-01 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 2 | 211 | 6.28e-01 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 2 | 211 | 6.28e-01 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 4 | 475 | 6.28e-01 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 3 | 344 | 6.28e-01 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0030278 | regulation of ossification | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0009408 | response to heat | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0016032 | viral process | 3 | 344 | 6.28e-01 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0120254 | olefinic compound metabolic process | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0021782 | glial cell development | 1 | 84 | 6.28e-01 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 1 | 84 | 6.28e-01 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 1 | 85 | 6.30e-01 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 1 | 85 | 6.30e-01 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 1 | 85 | 6.30e-01 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 3 | 345 | 6.30e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 37 | 4953 | 6.30e-01 |
| GO:BP | GO:0032526 | response to retinoic acid | 1 | 85 | 6.30e-01 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 2 | 214 | 6.30e-01 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 1 | 85 | 6.30e-01 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 1 | 85 | 6.30e-01 |
| GO:BP | GO:0034644 | cellular response to UV | 1 | 86 | 6.32e-01 |
| GO:BP | GO:0035270 | endocrine system development | 1 | 86 | 6.32e-01 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 5 | 615 | 6.32e-01 |
| GO:BP | GO:0009914 | hormone transport | 2 | 215 | 6.32e-01 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 3 | 348 | 6.32e-01 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 1 | 86 | 6.32e-01 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 1 | 86 | 6.32e-01 |
| GO:BP | GO:1901655 | cellular response to ketone | 1 | 86 | 6.32e-01 |
| GO:BP | GO:0042476 | odontogenesis | 1 | 86 | 6.32e-01 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 1 | 86 | 6.32e-01 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 3 | 348 | 6.32e-01 |
| GO:BP | GO:0050863 | regulation of T cell activation | 2 | 216 | 6.32e-01 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 1 | 87 | 6.35e-01 |
| GO:BP | GO:0098773 | skin epidermis development | 1 | 87 | 6.35e-01 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 1 | 87 | 6.35e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 6 | 755 | 6.38e-01 |
| GO:BP | GO:0061337 | cardiac conduction | 1 | 88 | 6.40e-01 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 1 | 88 | 6.40e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 2 | 219 | 6.40e-01 |
| GO:BP | GO:0033043 | regulation of organelle organization | 8 | 1028 | 6.41e-01 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0045445 | myoblast differentiation | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0006400 | tRNA modification | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0001843 | neural tube closure | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0043500 | muscle adaptation | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 2 | 222 | 6.41e-01 |
| GO:BP | GO:1901575 | organic substance catabolic process | 13 | 1712 | 6.41e-01 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 5 | 625 | 6.41e-01 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0031644 | regulation of nervous system process | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0046887 | positive regulation of hormone secretion | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0006520 | amino acid metabolic process | 2 | 222 | 6.41e-01 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0061912 | selective autophagy | 1 | 90 | 6.41e-01 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 1 | 89 | 6.41e-01 |
| GO:BP | GO:0051960 | regulation of nervous system development | 3 | 357 | 6.42e-01 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 2 | 223 | 6.43e-01 |
| GO:BP | GO:0060606 | tube closure | 1 | 91 | 6.44e-01 |
| GO:BP | GO:0031399 | regulation of protein modification process | 9 | 1173 | 6.44e-01 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 1 | 91 | 6.44e-01 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 1 | 91 | 6.44e-01 |
| GO:BP | GO:0030168 | platelet activation | 1 | 91 | 6.44e-01 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0061726 | mitochondrion disassembly | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 4 | 496 | 6.45e-01 |
| GO:BP | GO:0048144 | fibroblast proliferation | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 5 | 631 | 6.45e-01 |
| GO:BP | GO:0043473 | pigmentation | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0019953 | sexual reproduction | 5 | 631 | 6.45e-01 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 1 | 92 | 6.45e-01 |
| GO:BP | GO:0060322 | head development | 5 | 633 | 6.47e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 2 | 227 | 6.48e-01 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 1 | 93 | 6.48e-01 |
| GO:BP | GO:0006518 | peptide metabolic process | 6 | 771 | 6.48e-01 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 1 | 93 | 6.48e-01 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 1 | 93 | 6.48e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 2 | 227 | 6.48e-01 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 2 | 228 | 6.50e-01 |
| GO:BP | GO:0043543 | protein acylation | 2 | 228 | 6.50e-01 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 1 | 94 | 6.50e-01 |
| GO:BP | GO:0014020 | primary neural tube formation | 1 | 94 | 6.50e-01 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 5 | 637 | 6.50e-01 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 1 | 94 | 6.50e-01 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 2 | 229 | 6.51e-01 |
| GO:BP | GO:0006811 | monoatomic ion transport | 6 | 775 | 6.51e-01 |
| GO:BP | GO:0043604 | amide biosynthetic process | 6 | 775 | 6.51e-01 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 2 | 229 | 6.51e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 5 | 640 | 6.51e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 2 | 230 | 6.51e-01 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 1 | 95 | 6.51e-01 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 1 | 95 | 6.51e-01 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 1 | 95 | 6.51e-01 |
| GO:BP | GO:0043200 | response to amino acid | 1 | 95 | 6.51e-01 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 1 | 95 | 6.51e-01 |
| GO:BP | GO:0016197 | endosomal transport | 2 | 231 | 6.53e-01 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 15 | 2011 | 6.53e-01 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 2 | 232 | 6.54e-01 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 3 | 369 | 6.54e-01 |
| GO:BP | GO:0006575 | cellular modified amino acid metabolic process | 1 | 96 | 6.54e-01 |
| GO:BP | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 96 | 6.54e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 2 | 232 | 6.54e-01 |
| GO:BP | GO:0007059 | chromosome segregation | 3 | 369 | 6.54e-01 |
| GO:BP | GO:0031175 | neuron projection development | 6 | 780 | 6.54e-01 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 1 | 97 | 6.55e-01 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 1 | 97 | 6.55e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 6 | 782 | 6.55e-01 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 1 | 97 | 6.55e-01 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 1 | 97 | 6.55e-01 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 2 | 234 | 6.57e-01 |
| GO:BP | GO:0034728 | nucleosome organization | 1 | 98 | 6.59e-01 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 1 | 98 | 6.59e-01 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 1 | 98 | 6.59e-01 |
| GO:BP | GO:0042886 | amide transport | 2 | 235 | 6.59e-01 |
| GO:BP | GO:0008584 | male gonad development | 1 | 99 | 6.59e-01 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 1 | 99 | 6.59e-01 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 1 | 99 | 6.59e-01 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 1 | 99 | 6.59e-01 |
| GO:BP | GO:0035296 | regulation of tube diameter | 1 | 99 | 6.59e-01 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 1 | 99 | 6.59e-01 |
| GO:BP | GO:0090630 | activation of GTPase activity | 1 | 99 | 6.59e-01 |
| GO:BP | GO:0035150 | regulation of tube size | 1 | 99 | 6.59e-01 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 2 | 236 | 6.59e-01 |
| GO:BP | GO:0048511 | rhythmic process | 2 | 237 | 6.60e-01 |
| GO:BP | GO:0001666 | response to hypoxia | 2 | 237 | 6.60e-01 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 100 | 6.62e-01 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 1 | 100 | 6.62e-01 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 1 | 100 | 6.62e-01 |
| GO:BP | GO:0007417 | central nervous system development | 6 | 792 | 6.63e-01 |
| GO:BP | GO:0000904 | cell morphogenesis involved in differentiation | 4 | 516 | 6.64e-01 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1 | 101 | 6.64e-01 |
| GO:BP | GO:0021987 | cerebral cortex development | 1 | 101 | 6.64e-01 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 1 | 101 | 6.64e-01 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 1 | 101 | 6.64e-01 |
| GO:BP | GO:0051258 | protein polymerization | 2 | 241 | 6.68e-01 |
| GO:BP | GO:0071674 | mononuclear cell migration | 1 | 102 | 6.68e-01 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 1 | 102 | 6.68e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 2 | 242 | 6.70e-01 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 1 | 103 | 6.70e-01 |
| GO:BP | GO:0031047 | RNA-mediated gene silencing | 1 | 103 | 6.70e-01 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 1 | 103 | 6.70e-01 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 1 | 103 | 6.70e-01 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 1 | 104 | 6.74e-01 |
| GO:BP | GO:0003158 | endothelium development | 1 | 104 | 6.74e-01 |
| GO:BP | GO:0006906 | vesicle fusion | 1 | 104 | 6.74e-01 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 5 | 666 | 6.76e-01 |
| GO:BP | GO:0001101 | response to acid chemical | 1 | 105 | 6.76e-01 |
| GO:BP | GO:1901214 | regulation of neuron death | 2 | 246 | 6.76e-01 |
| GO:BP | GO:0006952 | defense response | 7 | 944 | 6.76e-01 |
| GO:BP | GO:0001508 | action potential | 1 | 105 | 6.76e-01 |
| GO:BP | GO:0009416 | response to light stimulus | 2 | 246 | 6.76e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 5 | 665 | 6.76e-01 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 1 | 105 | 6.76e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 6.78e-01 |
| GO:BP | GO:0090174 | organelle membrane fusion | 1 | 106 | 6.78e-01 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 1 | 106 | 6.78e-01 |
| GO:BP | GO:0036503 | ERAD pathway | 1 | 106 | 6.78e-01 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 4 | 529 | 6.78e-01 |
| GO:BP | GO:0006935 | chemotaxis | 3 | 389 | 6.78e-01 |
| GO:BP | GO:0006812 | monoatomic cation transport | 5 | 669 | 6.78e-01 |
| GO:BP | GO:0042330 | taxis | 3 | 389 | 6.78e-01 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 1 | 107 | 6.80e-01 |
| GO:BP | GO:0046718 | viral entry into host cell | 1 | 107 | 6.80e-01 |
| GO:BP | GO:0061024 | membrane organization | 5 | 671 | 6.80e-01 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 1 | 108 | 6.82e-01 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 1 | 108 | 6.82e-01 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 2 | 250 | 6.82e-01 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 1 | 108 | 6.82e-01 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 6 | 816 | 6.86e-01 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 1 | 109 | 6.86e-01 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 3 | 395 | 6.88e-01 |
| GO:BP | GO:0051606 | detection of stimulus | 1 | 110 | 6.88e-01 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 1 | 110 | 6.88e-01 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 1 | 110 | 6.88e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 10 | 1378 | 6.88e-01 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 4 | 538 | 6.89e-01 |
| GO:BP | GO:0051048 | negative regulation of secretion | 1 | 111 | 6.90e-01 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 1 | 111 | 6.90e-01 |
| GO:BP | GO:0008643 | carbohydrate transport | 1 | 111 | 6.90e-01 |
| GO:BP | GO:0030048 | actin filament-based movement | 1 | 112 | 6.92e-01 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 1 | 112 | 6.92e-01 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 1 | 112 | 6.92e-01 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 2 | 256 | 6.92e-01 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 4 | 541 | 6.92e-01 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 1 | 112 | 6.92e-01 |
| GO:BP | GO:0048592 | eye morphogenesis | 1 | 113 | 6.94e-01 |
| GO:BP | GO:0051592 | response to calcium ion | 1 | 113 | 6.94e-01 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 2 | 258 | 6.94e-01 |
| GO:BP | GO:0044409 | entry into host | 1 | 113 | 6.94e-01 |
| GO:BP | GO:0048638 | regulation of developmental growth | 2 | 258 | 6.94e-01 |
| GO:BP | GO:0006950 | response to stress | 20 | 2780 | 6.96e-01 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 1 | 114 | 6.97e-01 |
| GO:BP | GO:0046661 | male sex differentiation | 1 | 114 | 6.97e-01 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 1 | 114 | 6.97e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 1 | 115 | 6.99e-01 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 1 | 115 | 6.99e-01 |
| GO:BP | GO:0007612 | learning | 1 | 115 | 6.99e-01 |
| GO:BP | GO:0007568 | aging | 1 | 115 | 6.99e-01 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 115 | 6.99e-01 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 6 | 834 | 7.01e-01 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 1 | 116 | 7.01e-01 |
| GO:BP | GO:0051783 | regulation of nuclear division | 1 | 116 | 7.01e-01 |
| GO:BP | GO:0010033 | response to organic substance | 14 | 1957 | 7.01e-01 |
| GO:BP | GO:0006413 | translational initiation | 1 | 116 | 7.01e-01 |
| GO:BP | GO:0007009 | plasma membrane organization | 1 | 116 | 7.01e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 5 | 694 | 7.02e-01 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 1 | 117 | 7.02e-01 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 1 | 117 | 7.02e-01 |
| GO:BP | GO:0001890 | placenta development | 1 | 117 | 7.02e-01 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 1 | 117 | 7.02e-01 |
| GO:BP | GO:0048468 | cell development | 14 | 1963 | 7.04e-01 |
| GO:BP | GO:0006066 | alcohol metabolic process | 2 | 265 | 7.04e-01 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 1 | 118 | 7.05e-01 |
| GO:BP | GO:0000165 | MAPK cascade | 4 | 556 | 7.07e-01 |
| GO:BP | GO:0032504 | multicellular organism reproduction | 4 | 556 | 7.07e-01 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 2 | 267 | 7.07e-01 |
| GO:BP | GO:0006936 | muscle contraction | 2 | 267 | 7.07e-01 |
| GO:BP | GO:0042098 | T cell proliferation | 1 | 120 | 7.08e-01 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 1 | 120 | 7.08e-01 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 2 | 268 | 7.08e-01 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 2 | 268 | 7.08e-01 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 3 | 414 | 7.08e-01 |
| GO:BP | GO:0051225 | spindle assembly | 1 | 120 | 7.08e-01 |
| GO:BP | GO:0140013 | meiotic nuclear division | 1 | 120 | 7.08e-01 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 1 | 120 | 7.08e-01 |
| GO:BP | GO:0006816 | calcium ion transport | 2 | 269 | 7.09e-01 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 1 | 121 | 7.10e-01 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 1 | 121 | 7.10e-01 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 1 | 121 | 7.10e-01 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 1 | 121 | 7.10e-01 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 1 | 121 | 7.10e-01 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 4 | 563 | 7.11e-01 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 3 | 418 | 7.11e-01 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 122 | 7.11e-01 |
| GO:BP | GO:0050792 | regulation of viral process | 1 | 122 | 7.11e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 1 | 122 | 7.11e-01 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 1 | 122 | 7.11e-01 |
| GO:BP | GO:0043603 | amide metabolic process | 7 | 993 | 7.11e-01 |
| GO:BP | GO:0002253 | activation of immune response | 2 | 272 | 7.11e-01 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 1 | 123 | 7.12e-01 |
| GO:BP | GO:0003002 | regionalization | 2 | 273 | 7.12e-01 |
| GO:BP | GO:0051896 | regulation of protein kinase B signaling | 1 | 123 | 7.12e-01 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 1 | 123 | 7.12e-01 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 1 | 124 | 7.15e-01 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 1 | 124 | 7.15e-01 |
| GO:BP | GO:0032940 | secretion by cell | 4 | 569 | 7.17e-01 |
| GO:BP | GO:0019725 | cellular homeostasis | 4 | 569 | 7.17e-01 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 1 | 126 | 7.19e-01 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 1 | 126 | 7.19e-01 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 1 | 126 | 7.19e-01 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 1 | 126 | 7.19e-01 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 2 | 277 | 7.19e-01 |
| GO:BP | GO:1901565 | organonitrogen compound catabolic process | 8 | 1145 | 7.19e-01 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 1 | 127 | 7.21e-01 |
| GO:BP | GO:0007098 | centrosome cycle | 1 | 127 | 7.21e-01 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 1 | 127 | 7.21e-01 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 8 | 1149 | 7.21e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 15 | 2139 | 7.21e-01 |
| GO:BP | GO:2000241 | regulation of reproductive process | 1 | 128 | 7.24e-01 |
| GO:BP | GO:0007165 | signal transduction | 27 | 3818 | 7.25e-01 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 1 | 129 | 7.26e-01 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 1 | 129 | 7.26e-01 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 1 | 130 | 7.29e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 4 | 581 | 7.30e-01 |
| GO:BP | GO:0035265 | organ growth | 1 | 131 | 7.31e-01 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 1 | 131 | 7.31e-01 |
| GO:BP | GO:0070997 | neuron death | 2 | 286 | 7.34e-01 |
| GO:BP | GO:0048666 | neuron development | 6 | 877 | 7.35e-01 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 1 | 133 | 7.35e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 15 | 2161 | 7.35e-01 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 1 | 133 | 7.35e-01 |
| GO:BP | GO:0042445 | hormone metabolic process | 1 | 133 | 7.35e-01 |
| GO:BP | GO:0001764 | neuron migration | 1 | 133 | 7.35e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 6 | 877 | 7.35e-01 |
| GO:BP | GO:0046907 | intracellular transport | 10 | 1453 | 7.35e-01 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 3 | 439 | 7.36e-01 |
| GO:BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile | 2 | 289 | 7.36e-01 |
| GO:BP | GO:0000398 | mRNA splicing, via spliceosome | 2 | 289 | 7.36e-01 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 1 | 135 | 7.38e-01 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 1 | 135 | 7.38e-01 |
| GO:BP | GO:0048284 | organelle fusion | 1 | 135 | 7.38e-01 |
| GO:BP | GO:0006955 | immune response | 6 | 882 | 7.38e-01 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 1 | 135 | 7.38e-01 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 2 | 292 | 7.41e-01 |
| GO:BP | GO:0090501 | RNA phosphodiester bond hydrolysis | 1 | 136 | 7.41e-01 |
| GO:BP | GO:1903008 | organelle disassembly | 1 | 137 | 7.41e-01 |
| GO:BP | GO:0000375 | RNA splicing, via transesterification reactions | 2 | 293 | 7.41e-01 |
| GO:BP | GO:0009749 | response to glucose | 1 | 137 | 7.41e-01 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 1 | 137 | 7.41e-01 |
| GO:BP | GO:0061025 | membrane fusion | 1 | 137 | 7.41e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 2 | 293 | 7.41e-01 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 1 | 137 | 7.41e-01 |
| GO:BP | GO:0007030 | Golgi organization | 1 | 137 | 7.41e-01 |
| GO:BP | GO:0044000 | movement in host | 1 | 138 | 7.43e-01 |
| GO:BP | GO:0009746 | response to hexose | 1 | 138 | 7.43e-01 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 2 | 295 | 7.43e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 3 | 448 | 7.44e-01 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 1 | 139 | 7.44e-01 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 1 | 139 | 7.44e-01 |
| GO:BP | GO:0021543 | pallium development | 1 | 139 | 7.44e-01 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 1 | 139 | 7.44e-01 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 5 | 748 | 7.46e-01 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 1 | 140 | 7.46e-01 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 3 | 452 | 7.47e-01 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 1 | 141 | 7.47e-01 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 2 | 299 | 7.47e-01 |
| GO:BP | GO:0009411 | response to UV | 1 | 141 | 7.47e-01 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 1 | 141 | 7.47e-01 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 1 | 141 | 7.47e-01 |
| GO:BP | GO:0003008 | system process | 8 | 1189 | 7.47e-01 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 1 | 142 | 7.47e-01 |
| GO:BP | GO:0022900 | electron transport chain | 1 | 142 | 7.47e-01 |
| GO:BP | GO:0034284 | response to monosaccharide | 1 | 142 | 7.47e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 1 | 142 | 7.47e-01 |
| GO:BP | GO:1902075 | cellular response to salt | 1 | 142 | 7.47e-01 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 4 | 604 | 7.47e-01 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 2 | 301 | 7.47e-01 |
| GO:BP | GO:0051668 | localization within membrane | 4 | 605 | 7.47e-01 |
| GO:BP | GO:0034097 | response to cytokine | 4 | 607 | 7.50e-01 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 2 | 303 | 7.50e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 4 | 608 | 7.51e-01 |
| GO:BP | GO:0002520 | immune system development | 1 | 144 | 7.51e-01 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 6 | 906 | 7.52e-01 |
| GO:BP | GO:0032535 | regulation of cellular component size | 2 | 305 | 7.53e-01 |
| GO:BP | GO:0140352 | export from cell | 4 | 611 | 7.54e-01 |
| GO:BP | GO:0007389 | pattern specification process | 2 | 306 | 7.54e-01 |
| GO:BP | GO:0043491 | protein kinase B signaling | 1 | 146 | 7.55e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 7.55e-01 |
| GO:BP | GO:0030324 | lung development | 1 | 146 | 7.55e-01 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 1 | 147 | 7.56e-01 |
| GO:BP | GO:1901654 | response to ketone | 1 | 147 | 7.56e-01 |
| GO:BP | GO:0016236 | macroautophagy | 2 | 308 | 7.56e-01 |
| GO:BP | GO:1990138 | neuron projection extension | 1 | 148 | 7.59e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 2 | 310 | 7.59e-01 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 1 | 149 | 7.61e-01 |
| GO:BP | GO:0030323 | respiratory tube development | 1 | 150 | 7.63e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 7.65e-01 |
| GO:BP | GO:0015031 | protein transport | 9 | 1361 | 7.65e-01 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 1 | 151 | 7.65e-01 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 1 | 151 | 7.65e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 7.67e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 7 | 1074 | 7.68e-01 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 4 | 626 | 7.68e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 7.68e-01 |
| GO:BP | GO:0000725 | recombinational repair | 1 | 153 | 7.68e-01 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 2 | 317 | 7.68e-01 |
| GO:BP | GO:0022414 | reproductive process | 6 | 927 | 7.68e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 7 | 1076 | 7.69e-01 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 1 | 154 | 7.69e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 1 | 155 | 7.71e-01 |
| GO:BP | GO:0071478 | cellular response to radiation | 1 | 156 | 7.71e-01 |
| GO:BP | GO:0051641 | cellular localization | 19 | 2810 | 7.71e-01 |
| GO:BP | GO:0016050 | vesicle organization | 2 | 320 | 7.71e-01 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 1 | 156 | 7.71e-01 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | 155 | 7.71e-01 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 3 | 478 | 7.71e-01 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 5 | 783 | 7.71e-01 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 4 | 632 | 7.71e-01 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 1 | 155 | 7.71e-01 |
| GO:BP | GO:0023061 | signal release | 2 | 321 | 7.71e-01 |
| GO:BP | GO:0000003 | reproduction | 6 | 934 | 7.71e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 5 | 785 | 7.71e-01 |
| GO:BP | GO:0006974 | DNA damage response | 5 | 785 | 7.71e-01 |
| GO:BP | GO:0033002 | muscle cell proliferation | 1 | 157 | 7.72e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 3 | 482 | 7.73e-01 |
| GO:BP | GO:0008406 | gonad development | 1 | 158 | 7.73e-01 |
| GO:BP | GO:0042180 | cellular ketone metabolic process | 1 | 158 | 7.73e-01 |
| GO:BP | GO:0031348 | negative regulation of defense response | 1 | 159 | 7.75e-01 |
| GO:BP | GO:0097305 | response to alcohol | 1 | 159 | 7.75e-01 |
| GO:BP | GO:0043487 | regulation of RNA stability | 1 | 159 | 7.75e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 3 | 484 | 7.75e-01 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 2 | 326 | 7.76e-01 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 1 | 160 | 7.76e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 7.76e-01 |
| GO:BP | GO:0071702 | organic substance transport | 14 | 2116 | 7.76e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 29 | 4241 | 7.76e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 11 | 1684 | 7.76e-01 |
| GO:BP | GO:0009743 | response to carbohydrate | 1 | 161 | 7.76e-01 |
| GO:BP | GO:0045087 | innate immune response | 3 | 488 | 7.76e-01 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 1 | 161 | 7.76e-01 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 1 | 162 | 7.76e-01 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 3 | 486 | 7.76e-01 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 1 | 161 | 7.76e-01 |
| GO:BP | GO:0060326 | cell chemotaxis | 1 | 162 | 7.76e-01 |
| GO:BP | GO:0009451 | RNA modification | 1 | 162 | 7.76e-01 |
| GO:BP | GO:0016482 | cytosolic transport | 1 | 161 | 7.76e-01 |
| GO:BP | GO:0050896 | response to stimulus | 40 | 5770 | 7.77e-01 |
| GO:BP | GO:0046903 | secretion | 4 | 645 | 7.77e-01 |
| GO:BP | GO:0060541 | respiratory system development | 1 | 163 | 7.77e-01 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 1 | 163 | 7.77e-01 |
| GO:BP | GO:0006412 | translation | 4 | 646 | 7.77e-01 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 1 | 164 | 7.79e-01 |
| GO:BP | GO:0051179 | localization | 27 | 3969 | 7.79e-01 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 1 | 165 | 7.80e-01 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 1 | 165 | 7.80e-01 |
| GO:BP | GO:0031330 | negative regulation of cellular catabolic process | 1 | 165 | 7.80e-01 |
| GO:BP | GO:0042221 | response to chemical | 17 | 2559 | 7.80e-01 |
| GO:BP | GO:0008104 | protein localization | 14 | 2129 | 7.81e-01 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 1 | 166 | 7.81e-01 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 1 | 166 | 7.81e-01 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 1 | 166 | 7.81e-01 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 1 | 167 | 7.83e-01 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 5 | 809 | 7.83e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 2 | 337 | 7.83e-01 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 1 | 168 | 7.83e-01 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 1 | 168 | 7.83e-01 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 2 | 338 | 7.83e-01 |
| GO:BP | GO:0042593 | glucose homeostasis | 1 | 168 | 7.83e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 14 | 2138 | 7.83e-01 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 1 | 169 | 7.84e-01 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 1 | 169 | 7.84e-01 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 2 | 341 | 7.87e-01 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 2 | 342 | 7.88e-01 |
| GO:BP | GO:0060537 | muscle tissue development | 2 | 343 | 7.88e-01 |
| GO:BP | GO:0044085 | cellular component biogenesis | 18 | 2724 | 7.88e-01 |
| GO:BP | GO:0043393 | regulation of protein binding | 1 | 172 | 7.88e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 4 | 662 | 7.88e-01 |
| GO:BP | GO:0023052 | signaling | 28 | 4141 | 7.88e-01 |
| GO:BP | GO:0034329 | cell junction assembly | 2 | 344 | 7.89e-01 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 1 | 173 | 7.89e-01 |
| GO:BP | GO:0006810 | transport | 22 | 3298 | 7.89e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 16 | 2450 | 7.95e-01 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 3 | 512 | 7.96e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 5 | 827 | 7.97e-01 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 2 | 351 | 7.98e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 7.99e-01 |
| GO:BP | GO:0009314 | response to radiation | 2 | 352 | 7.99e-01 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 1 | 179 | 7.99e-01 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 1 | 179 | 7.99e-01 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 3 | 517 | 7.99e-01 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 1 | 179 | 7.99e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 2 | 353 | 7.99e-01 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 1 | 180 | 8.00e-01 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 2 | 357 | 8.03e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 1 | 182 | 8.03e-01 |
| GO:BP | GO:0048589 | developmental growth | 3 | 522 | 8.03e-01 |
| GO:BP | GO:0140694 | non-membrane-bounded organelle assembly | 2 | 357 | 8.03e-01 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 1 | 183 | 8.04e-01 |
| GO:BP | GO:0072657 | protein localization to membrane | 3 | 523 | 8.04e-01 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 1 | 183 | 8.04e-01 |
| GO:BP | GO:0006897 | endocytosis | 3 | 527 | 8.08e-01 |
| GO:BP | GO:0007411 | axon guidance | 1 | 185 | 8.08e-01 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 3 | 527 | 8.08e-01 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 2 | 362 | 8.08e-01 |
| GO:BP | GO:0097485 | neuron projection guidance | 1 | 186 | 8.08e-01 |
| GO:BP | GO:0045444 | fat cell differentiation | 1 | 187 | 8.10e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 8.10e-01 |
| GO:BP | GO:0051321 | meiotic cell cycle | 1 | 188 | 8.10e-01 |
| GO:BP | GO:0070661 | leukocyte proliferation | 1 | 188 | 8.10e-01 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 3 | 531 | 8.10e-01 |
| GO:BP | GO:0050808 | synapse organization | 2 | 365 | 8.10e-01 |
| GO:BP | GO:0007051 | spindle organization | 1 | 188 | 8.10e-01 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 3 | 532 | 8.10e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 6 | 1006 | 8.10e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 9 | 1462 | 8.11e-01 |
| GO:BP | GO:0006281 | DNA repair | 3 | 534 | 8.11e-01 |
| GO:BP | GO:0030162 | regulation of proteolysis | 3 | 535 | 8.12e-01 |
| GO:BP | GO:0019932 | second-messenger-mediated signaling | 1 | 191 | 8.13e-01 |
| GO:BP | GO:0007548 | sex differentiation | 1 | 191 | 8.13e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 2 | 369 | 8.13e-01 |
| GO:BP | GO:0006457 | protein folding | 1 | 192 | 8.14e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 1 | 192 | 8.14e-01 |
| GO:BP | GO:0048588 | developmental cell growth | 1 | 192 | 8.14e-01 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 4 | 701 | 8.15e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 3 | 540 | 8.15e-01 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 3 | 540 | 8.15e-01 |
| GO:BP | GO:0007015 | actin filament organization | 2 | 373 | 8.15e-01 |
| GO:BP | GO:0007154 | cell communication | 28 | 4226 | 8.17e-01 |
| GO:BP | GO:0051276 | chromosome organization | 3 | 543 | 8.17e-01 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 1 | 196 | 8.18e-01 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 3 | 544 | 8.18e-01 |
| GO:BP | GO:1901615 | organic hydroxy compound metabolic process | 2 | 376 | 8.18e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 2 | 377 | 8.19e-01 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 1 | 197 | 8.19e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 2 | 378 | 8.19e-01 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 1 | 198 | 8.20e-01 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 1 | 200 | 8.23e-01 |
| GO:BP | GO:0032990 | cell part morphogenesis | 3 | 551 | 8.23e-01 |
| GO:BP | GO:0006325 | chromatin organization | 3 | 551 | 8.23e-01 |
| GO:BP | GO:0140546 | defense response to symbiont | 1 | 202 | 8.25e-01 |
| GO:BP | GO:0007611 | learning or memory | 1 | 202 | 8.25e-01 |
| GO:BP | GO:0051607 | defense response to virus | 1 | 202 | 8.25e-01 |
| GO:BP | GO:0022618 | ribonucleoprotein complex assembly | 1 | 203 | 8.26e-01 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 1 | 204 | 8.27e-01 |
| GO:BP | GO:0008202 | steroid metabolic process | 1 | 204 | 8.27e-01 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 2 | 389 | 8.30e-01 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 7 | 1201 | 8.33e-01 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 1 | 209 | 8.35e-01 |
| GO:BP | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 1 | 211 | 8.36e-01 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 1 | 211 | 8.36e-01 |
| GO:BP | GO:0045333 | cellular respiration | 1 | 211 | 8.36e-01 |
| GO:BP | GO:0030100 | regulation of endocytosis | 1 | 211 | 8.36e-01 |
| GO:BP | GO:0021700 | developmental maturation | 1 | 211 | 8.36e-01 |
| GO:BP | GO:0071826 | ribonucleoprotein complex subunit organization | 1 | 211 | 8.36e-01 |
| GO:BP | GO:0008150 | biological_process | 86 | 12070 | 8.36e-01 |
| GO:BP | GO:0061564 | axon development | 2 | 398 | 8.36e-01 |
| GO:BP | GO:0048608 | reproductive structure development | 1 | 212 | 8.36e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 1 | 212 | 8.36e-01 |
| GO:BP | GO:0051234 | establishment of localization | 22 | 3433 | 8.36e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 2 | 398 | 8.36e-01 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 1 | 213 | 8.37e-01 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 1 | 214 | 8.38e-01 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 2 | 401 | 8.38e-01 |
| GO:BP | GO:0061458 | reproductive system development | 1 | 215 | 8.39e-01 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 2 | 403 | 8.40e-01 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 1 | 217 | 8.41e-01 |
| GO:BP | GO:0000819 | sister chromatid segregation | 1 | 217 | 8.41e-01 |
| GO:BP | GO:0002250 | adaptive immune response | 1 | 218 | 8.42e-01 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 2 | 408 | 8.44e-01 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 1 | 220 | 8.44e-01 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 1 | 222 | 8.46e-01 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 1 | 222 | 8.46e-01 |
| GO:BP | GO:0030030 | cell projection organization | 7 | 1233 | 8.46e-01 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 1 | 223 | 8.47e-01 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 1 | 224 | 8.48e-01 |
| GO:BP | GO:0016042 | lipid catabolic process | 1 | 225 | 8.49e-01 |
| GO:BP | GO:0033993 | response to lipid | 3 | 592 | 8.51e-01 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 1 | 228 | 8.53e-01 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 2 | 420 | 8.54e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 7 | 1248 | 8.54e-01 |
| GO:BP | GO:0007264 | small GTPase mediated signal transduction | 2 | 421 | 8.54e-01 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 1 | 230 | 8.54e-01 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 2 | 422 | 8.54e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 2 | 422 | 8.54e-01 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 1 | 232 | 8.56e-01 |
| GO:BP | GO:0030198 | extracellular matrix organization | 1 | 233 | 8.57e-01 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 1 | 233 | 8.57e-01 |
| GO:BP | GO:0043062 | extracellular structure organization | 1 | 234 | 8.57e-01 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 1 | 234 | 8.57e-01 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 1 | 235 | 8.58e-01 |
| GO:BP | GO:0016570 | histone modification | 2 | 433 | 8.63e-01 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 1 | 241 | 8.66e-01 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 2 | 438 | 8.68e-01 |
| GO:BP | GO:0001822 | kidney development | 1 | 244 | 8.68e-01 |
| GO:BP | GO:0019058 | viral life cycle | 1 | 244 | 8.68e-01 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 2 | 441 | 8.69e-01 |
| GO:BP | GO:0048545 | response to steroid hormone | 1 | 245 | 8.69e-01 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 1 | 247 | 8.71e-01 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 1 | 248 | 8.72e-01 |
| GO:BP | GO:0090150 | establishment of protein localization to membrane | 1 | 250 | 8.74e-01 |
| GO:BP | GO:0072001 | renal system development | 1 | 251 | 8.74e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 1 | 251 | 8.74e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 4 | 803 | 8.75e-01 |
| GO:BP | GO:0032989 | cellular component morphogenesis | 3 | 639 | 8.80e-01 |
| GO:BP | GO:0006310 | DNA recombination | 1 | 257 | 8.80e-01 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 1 | 257 | 8.80e-01 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 2 | 457 | 8.80e-01 |
| GO:BP | GO:0051604 | protein maturation | 1 | 259 | 8.81e-01 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 2 | 459 | 8.81e-01 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 1 | 259 | 8.81e-01 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 2 | 461 | 8.82e-01 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 2 | 463 | 8.83e-01 |
| GO:BP | GO:0006508 | proteolysis | 7 | 1318 | 8.84e-01 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 1 | 265 | 8.86e-01 |
| GO:BP | GO:0030031 | cell projection assembly | 2 | 469 | 8.87e-01 |
| GO:BP | GO:0055085 | transmembrane transport | 5 | 998 | 8.87e-01 |
| GO:BP | GO:0007600 | sensory perception | 1 | 269 | 8.88e-01 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 1 | 269 | 8.88e-01 |
| GO:BP | GO:0006302 | double-strand break repair | 1 | 271 | 8.90e-01 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 1 | 271 | 8.90e-01 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 1 | 273 | 8.91e-01 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 1 | 273 | 8.91e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 8.91e-01 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 1 | 274 | 8.91e-01 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 1 | 276 | 8.92e-01 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 1 | 276 | 8.92e-01 |
| GO:BP | GO:0007517 | muscle organ development | 1 | 277 | 8.93e-01 |
| GO:BP | GO:0048878 | chemical homeostasis | 3 | 676 | 8.98e-01 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 2 | 490 | 8.98e-01 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 2 | 490 | 8.98e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 10 | 1836 | 8.98e-01 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 1 | 285 | 8.99e-01 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 3 | 681 | 8.99e-01 |
| GO:BP | GO:0010256 | endomembrane system organization | 2 | 496 | 8.99e-01 |
| GO:BP | GO:1901653 | cellular response to peptide | 1 | 287 | 8.99e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 31 | 4955 | 8.99e-01 |
| GO:BP | GO:0022607 | cellular component assembly | 14 | 2466 | 8.99e-01 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 2 | 495 | 8.99e-01 |
| GO:BP | GO:0030029 | actin filament-based process | 3 | 687 | 9.01e-01 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 1 | 294 | 9.03e-01 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 2 | 506 | 9.05e-01 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 3 | 696 | 9.05e-01 |
| GO:BP | GO:0044281 | small molecule metabolic process | 7 | 1389 | 9.07e-01 |
| GO:BP | GO:0099536 | synaptic signaling | 2 | 511 | 9.07e-01 |
| GO:BP | GO:0006082 | organic acid metabolic process | 3 | 700 | 9.07e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 4 | 884 | 9.08e-01 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 1 | 308 | 9.12e-01 |
| GO:BP | GO:0009617 | response to bacterium | 1 | 321 | 9.21e-01 |
| GO:BP | GO:0043434 | response to peptide hormone | 1 | 321 | 9.21e-01 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 1 | 323 | 9.22e-01 |
| GO:BP | GO:0042692 | muscle cell differentiation | 1 | 324 | 9.22e-01 |
| GO:BP | GO:0061061 | muscle structure development | 2 | 546 | 9.25e-01 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 2 | 552 | 9.28e-01 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 1 | 336 | 9.29e-01 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 2 | 560 | 9.31e-01 |
| GO:BP | GO:0000280 | nuclear division | 1 | 344 | 9.33e-01 |
| GO:BP | GO:0051301 | cell division | 2 | 570 | 9.35e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 32 | 5274 | 9.36e-01 |
| GO:BP | GO:0034330 | cell junction organization | 2 | 575 | 9.37e-01 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 4 | 963 | 9.37e-01 |
| GO:BP | GO:0007409 | axonogenesis | 1 | 359 | 9.39e-01 |
| GO:BP | GO:0042592 | homeostatic process | 5 | 1159 | 9.42e-01 |
| GO:BP | GO:1901652 | response to peptide | 1 | 377 | 9.48e-01 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 2 | 612 | 9.50e-01 |
| GO:BP | GO:0048285 | organelle fission | 1 | 388 | 9.52e-01 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 1 | 391 | 9.52e-01 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 3 | 843 | 9.57e-01 |
| GO:BP | GO:0009725 | response to hormone | 2 | 642 | 9.58e-01 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 5 | 1238 | 9.60e-01 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 2 | 665 | 9.64e-01 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 1 | 433 | 9.65e-01 |
| GO:BP | GO:0007610 | behavior | 1 | 436 | 9.66e-01 |
| GO:BP | GO:0016043 | cellular component organization | 29 | 5076 | 9.68e-01 |
| GO:BP | GO:0001817 | regulation of cytokine production | 1 | 447 | 9.68e-01 |
| GO:BP | GO:0001816 | cytokine production | 1 | 451 | 9.69e-01 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 484 | 9.76e-01 |
| GO:BP | GO:0051640 | organelle localization | 1 | 503 | 9.79e-01 |
| GO:BP | GO:0007017 | microtubule-based process | 2 | 760 | 9.80e-01 |
| GO:BP | GO:0006629 | lipid metabolic process | 3 | 994 | 9.81e-01 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 2 | 784 | 9.82e-01 |
| GO:BP | GO:0007010 | cytoskeleton organization | 4 | 1226 | 9.83e-01 |
| GO:BP | GO:0070925 | organelle assembly | 2 | 817 | 9.85e-01 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 1 | 740 | 9.97e-01 |
| GO:BP | GO:0006996 | organelle organization | 11 | 3000 | 9.99e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 11 | 415 | 2.84e-02 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 4.85e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 2 | 70 | 6.29e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 2 | 117 | 6.29e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 2 | 87 | 6.29e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 2 | 99 | 6.29e-01 |
| KEGG | KEGG:03040 | Spliceosome | 3 | 132 | 6.29e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 6.29e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 3 | 86 | 6.29e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 6.29e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 3 | 145 | 6.29e-01 |
| KEGG | KEGG:05224 | Breast cancer | 3 | 117 | 6.29e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 1 | 84 | 6.29e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 2 | 115 | 6.29e-01 |
| KEGG | KEGG:03018 | RNA degradation | 1 | 74 | 6.29e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 1 | 75 | 6.29e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1 | 69 | 6.29e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 1 | 22 | 6.29e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 1 | 40 | 6.29e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 6.29e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 1 | 43 | 6.29e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 1 | 85 | 6.29e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 1 | 56 | 6.29e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 2 | 75 | 6.29e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 1 | 72 | 6.29e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 1 | 60 | 6.29e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 4 | 138 | 6.29e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 3 | 121 | 6.29e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 2 | 131 | 6.29e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 1 | 82 | 6.29e-01 |
| KEGG | KEGG:05142 | Chagas disease | 1 | 75 | 6.29e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 2 | 136 | 6.29e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 6.29e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 1 | 84 | 6.29e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 2 | 104 | 6.29e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 1 | 77 | 6.29e-01 |
| KEGG | KEGG:04924 | Renin secretion | 1 | 50 | 6.29e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 1 | 84 | 6.29e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 2 | 115 | 6.29e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 1 | 55 | 6.29e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 2 | 48 | 6.29e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 2 | 176 | 6.29e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 1 | 52 | 6.29e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 1 | 42 | 6.29e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 2 | 214 | 6.29e-01 |
| KEGG | KEGG:04210 | Apoptosis | 2 | 118 | 6.29e-01 |
| KEGG | KEGG:05133 | Pertussis | 1 | 45 | 6.29e-01 |
| KEGG | KEGG:05131 | Shigellosis | 2 | 210 | 6.29e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 2 | 150 | 6.29e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 1 | 78 | 6.29e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 2 | 112 | 6.29e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 1 | 58 | 6.29e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 2 | 35 | 6.29e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 2 | 124 | 6.29e-01 |
| KEGG | KEGG:04144 | Endocytosis | 3 | 232 | 6.29e-01 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 2 | 161 | 6.29e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 2 | 70 | 6.29e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 3 | 315 | 6.29e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 2 | 49 | 6.29e-01 |
| KEGG | KEGG:04360 | Axon guidance | 2 | 162 | 6.29e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 2 | 87 | 6.29e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 1 | 77 | 6.29e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 2 | 186 | 6.29e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 1 | 84 | 6.29e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 2 | 104 | 6.29e-01 |
| KEGG | KEGG:00000 | KEGG root term | 47 | 5558 | 6.29e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 6.29e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 6.29e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 2 | 142 | 6.29e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 1 | 38 | 6.29e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 6.29e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 1 | 48 | 6.29e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 6.29e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 1 | 19 | 6.29e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 1 | 28 | 6.29e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 6.29e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 1 | 65 | 6.29e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 1 | 25 | 6.29e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 3 | 172 | 6.29e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 6 | 409 | 6.29e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 2 | 97 | 6.29e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 3 | 180 | 6.29e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 6.29e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 1 | 20 | 6.29e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 2 | 134 | 6.29e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 2 | 98 | 6.29e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 2 | 76 | 6.29e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 1 | 64 | 6.29e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 2 | 83 | 6.29e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 2 | 110 | 6.29e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 55 | 6.29e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 12 | 6.29e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 1 | 78 | 6.29e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 2 | 165 | 6.29e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 1 | 79 | 6.29e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 1 | 49 | 6.29e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 1 | 56 | 6.29e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 2 | 114 | 6.29e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 3 | 153 | 6.29e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 1 | 91 | 6.48e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 2 | 226 | 6.49e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 6.55e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 1 | 98 | 6.56e-01 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 6.56e-01 |
| KEGG | KEGG:05164 | Influenza A | 1 | 103 | 6.59e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 1 | 103 | 6.59e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 2 | 240 | 6.59e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 3 | 385 | 6.59e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 1 | 110 | 6.76e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 6.82e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 6.91e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 1 | 121 | 6.97e-01 |
| KEGG | KEGG:05034 | Alcoholism | 1 | 122 | 6.97e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 1 | 128 | 7.11e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 1 | 135 | 7.15e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 1 | 133 | 7.15e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 1 | 134 | 7.15e-01 |
| KEGG | KEGG:04530 | Tight junction | 1 | 137 | 7.15e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 2 | 303 | 7.27e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 7.27e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 7.30e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 7.30e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 1 | 149 | 7.30e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 1 | 157 | 7.35e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 7.41e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 1 | 170 | 7.50e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 1 | 171 | 7.50e-01 |
| KEGG | KEGG:04510 | Focal adhesion | 1 | 177 | 7.55e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 1 | 179 | 7.55e-01 |
| KEGG | KEGG:05020 | Prion disease | 1 | 220 | 8.21e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 1 | 254 | 8.60e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 1 | 272 | 8.73e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 5 | 1146 | 9.28e-01 |
Dnr3_sp_DEGtable %>%
dplyr::filter(source=="KEGG") %>%
# dplyr::filter(p_value<0.05) %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=2 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
geom_vline(xintercept = (-log10(0.05)))+
scale_y_discrete(labels = wrap_format(30))+
scale_linetype(3)+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('DNR 3 hour specific(stringent)\n gene set KEGG terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("KEGG pathway")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))
Mtx3_sp_DEGtable %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
scale_y_discrete(labels = wrap_format(30))+
geom_vline(xintercept = (-log10(0.05)))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('MTX 3 hour specific(stringent)\n gene set GO:BP terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("GO: BP term")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))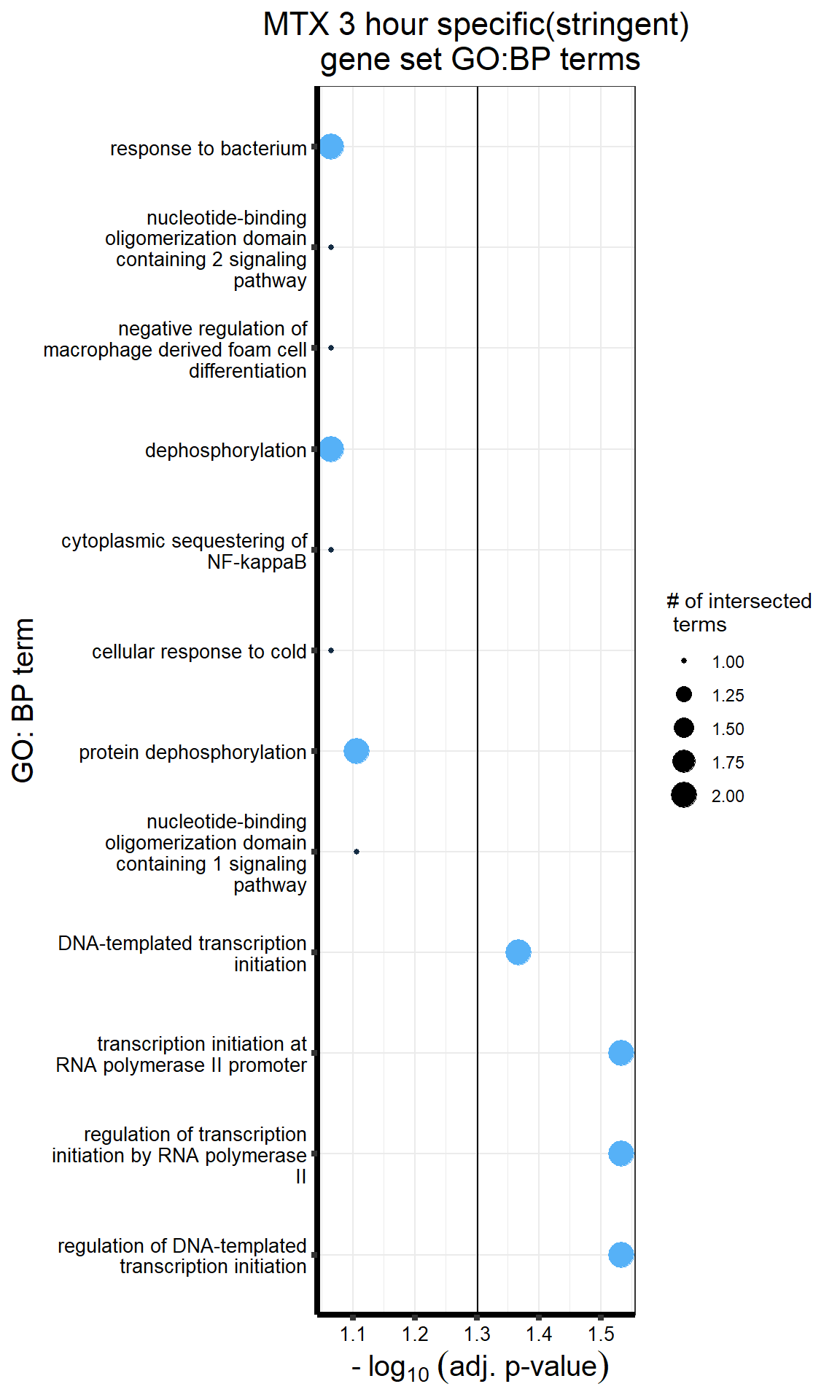
Mtx3_sp_DEGtable %>%
mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Stringent (adj. P value of <0.01) MTX 3 hour specific genes (n = 4) enriched GO and KEGG terms") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped","hover")) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 2 | 93 | 2.93e-02 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 2 | 69 | 2.93e-02 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 2 | 76 | 2.93e-02 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 2 | 130 | 4.29e-02 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 1 | 5 | 7.82e-02 |
| GO:BP | GO:0006470 | protein dephosphorylation | 2 | 214 | 7.82e-02 |
| GO:BP | GO:0016311 | dephosphorylation | 2 | 284 | 8.60e-02 |
| GO:BP | GO:0009617 | response to bacterium | 2 | 321 | 8.60e-02 |
| GO:BP | GO:0070417 | cellular response to cold | 1 | 8 | 8.60e-02 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 1 | 10 | 8.60e-02 |
| GO:BP | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | 1 | 11 | 8.60e-02 |
| GO:BP | GO:0007253 | cytoplasmic sequestering of NF-kappaB | 1 | 7 | 8.60e-02 |
| GO:BP | GO:0032495 | response to muramyl dipeptide | 1 | 15 | 8.81e-02 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 1 | 31 | 8.81e-02 |
| GO:BP | GO:0043330 | response to exogenous dsRNA | 1 | 27 | 8.81e-02 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 1 | 30 | 8.81e-02 |
| GO:BP | GO:0042994 | cytoplasmic sequestering of transcription factor | 1 | 15 | 8.81e-02 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 1 | 17 | 8.81e-02 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 1 | 19 | 8.81e-02 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 1 | 24 | 8.81e-02 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 1 | 22 | 8.81e-02 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 1 | 18 | 8.81e-02 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 1 | 30 | 8.81e-02 |
| GO:BP | GO:0050853 | B cell receptor signaling pathway | 1 | 32 | 8.81e-02 |
| GO:BP | GO:0010888 | negative regulation of lipid storage | 1 | 16 | 8.81e-02 |
| GO:BP | GO:0010875 | positive regulation of cholesterol efflux | 1 | 23 | 8.81e-02 |
| GO:BP | GO:0035994 | response to muscle stretch | 1 | 26 | 8.81e-02 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 1 | 31 | 8.81e-02 |
| GO:BP | GO:0051220 | cytoplasmic sequestering of protein | 1 | 23 | 8.81e-02 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 1 | 27 | 8.81e-02 |
| GO:BP | GO:0002752 | cell surface pattern recognition receptor signaling pathway | 1 | 27 | 8.81e-02 |
| GO:BP | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway | 1 | 18 | 8.81e-02 |
| GO:BP | GO:0090077 | foam cell differentiation | 1 | 22 | 8.81e-02 |
| GO:BP | GO:1905953 | negative regulation of lipid localization | 1 | 32 | 8.81e-02 |
| GO:BP | GO:0043331 | response to dsRNA | 1 | 33 | 8.82e-02 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 3 | 1914 | 8.85e-02 |
| GO:BP | GO:0031663 | lipopolysaccharide-mediated signaling pathway | 1 | 36 | 9.10e-02 |
| GO:BP | GO:0002220 | innate immune response activating cell surface receptor signaling pathway | 1 | 37 | 9.11e-02 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 3 | 1995 | 9.12e-02 |
| GO:BP | GO:0010883 | regulation of lipid storage | 1 | 39 | 9.12e-02 |
| GO:BP | GO:0033344 | cholesterol efflux | 1 | 40 | 9.12e-02 |
| GO:BP | GO:0009409 | response to cold | 1 | 41 | 9.13e-02 |
| GO:BP | GO:0006306 | DNA methylation | 1 | 46 | 9.15e-02 |
| GO:BP | GO:0032374 | regulation of cholesterol transport | 1 | 45 | 9.15e-02 |
| GO:BP | GO:0006305 | DNA alkylation | 1 | 46 | 9.15e-02 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 1 | 46 | 9.15e-02 |
| GO:BP | GO:0032371 | regulation of sterol transport | 1 | 45 | 9.15e-02 |
| GO:BP | GO:0032370 | positive regulation of lipid transport | 1 | 58 | 1.07e-01 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 1 | 66 | 1.07e-01 |
| GO:BP | GO:0031507 | heterochromatin formation | 1 | 69 | 1.07e-01 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 1 | 69 | 1.07e-01 |
| GO:BP | GO:0050729 | positive regulation of inflammatory response | 1 | 75 | 1.07e-01 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 1 | 75 | 1.07e-01 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 1 | 65 | 1.07e-01 |
| GO:BP | GO:0051707 | response to other organism | 2 | 820 | 1.07e-01 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 3 | 2576 | 1.07e-01 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 1 | 76 | 1.07e-01 |
| GO:BP | GO:1901222 | regulation of NIK/NF-kappaB signaling | 1 | 65 | 1.07e-01 |
| GO:BP | GO:0070828 | heterochromatin organization | 1 | 77 | 1.07e-01 |
| GO:BP | GO:0045638 | negative regulation of myeloid cell differentiation | 1 | 69 | 1.07e-01 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 1 | 59 | 1.07e-01 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 1 | 79 | 1.07e-01 |
| GO:BP | GO:0030301 | cholesterol transport | 1 | 80 | 1.07e-01 |
| GO:BP | GO:0019915 | lipid storage | 1 | 66 | 1.07e-01 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 2 | 820 | 1.07e-01 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 1 | 78 | 1.07e-01 |
| GO:BP | GO:0009607 | response to biotic stimulus | 2 | 850 | 1.07e-01 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 2 | 868 | 1.07e-01 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 3 | 2574 | 1.07e-01 |
| GO:BP | GO:0002753 | cytosolic pattern recognition receptor signaling pathway | 1 | 68 | 1.07e-01 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 3 | 2593 | 1.07e-01 |
| GO:BP | GO:0045185 | maintenance of protein location | 1 | 83 | 1.07e-01 |
| GO:BP | GO:0006304 | DNA modification | 1 | 89 | 1.09e-01 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 3 | 2684 | 1.09e-01 |
| GO:BP | GO:0015918 | sterol transport | 1 | 92 | 1.09e-01 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 3 | 2714 | 1.09e-01 |
| GO:BP | GO:0038061 | NIK/NF-kappaB signaling | 1 | 86 | 1.09e-01 |
| GO:BP | GO:0032368 | regulation of lipid transport | 1 | 92 | 1.09e-01 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 2 | 963 | 1.09e-01 |
| GO:BP | GO:0006351 | DNA-templated transcription | 3 | 2683 | 1.09e-01 |
| GO:BP | GO:0051101 | regulation of DNA binding | 1 | 104 | 1.19e-01 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 3 | 2845 | 1.19e-01 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 1 | 106 | 1.19e-01 |
| GO:BP | GO:0071222 | cellular response to lipopolysaccharide | 1 | 110 | 1.22e-01 |
| GO:BP | GO:1905952 | regulation of lipid localization | 1 | 112 | 1.22e-01 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 1 | 114 | 1.23e-01 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 1 | 115 | 1.23e-01 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 1 | 149 | 1.28e-01 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 1 | 138 | 1.28e-01 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 3 | 3012 | 1.28e-01 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 1 | 152 | 1.28e-01 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 2 | 1147 | 1.28e-01 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 1 | 153 | 1.28e-01 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 3 | 3133 | 1.28e-01 |
| GO:BP | GO:0009267 | cellular response to starvation | 1 | 152 | 1.28e-01 |
| GO:BP | GO:0051170 | import into nucleus | 1 | 152 | 1.28e-01 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 3 | 3169 | 1.28e-01 |
| GO:BP | GO:0071216 | cellular response to biotic stimulus | 1 | 138 | 1.28e-01 |
| GO:BP | GO:0009266 | response to temperature stimulus | 1 | 133 | 1.28e-01 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 3 | 3237 | 1.28e-01 |
| GO:BP | GO:0007219 | Notch signaling pathway | 1 | 146 | 1.28e-01 |
| GO:BP | GO:0006606 | protein import into nucleus | 1 | 147 | 1.28e-01 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 1 | 122 | 1.28e-01 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 1 | 151 | 1.28e-01 |
| GO:BP | GO:0002218 | activation of innate immune response | 1 | 142 | 1.28e-01 |
| GO:BP | GO:0051100 | negative regulation of binding | 1 | 135 | 1.28e-01 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 3 | 3070 | 1.28e-01 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 1 | 134 | 1.28e-01 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 3 | 3132 | 1.28e-01 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 3 | 3080 | 1.28e-01 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 3 | 3080 | 1.28e-01 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 1 | 155 | 1.28e-01 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 1 | 158 | 1.29e-01 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 1 | 161 | 1.30e-01 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 162 | 1.30e-01 |
| GO:BP | GO:0032496 | response to lipopolysaccharide | 1 | 178 | 1.37e-01 |
| GO:BP | GO:0042594 | response to starvation | 1 | 178 | 1.37e-01 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 1 | 178 | 1.37e-01 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 1 | 172 | 1.37e-01 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 1 | 174 | 1.37e-01 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 1 | 182 | 1.38e-01 |
| GO:BP | GO:0002237 | response to molecule of bacterial origin | 1 | 187 | 1.40e-01 |
| GO:BP | GO:0008283 | cell population proliferation | 2 | 1378 | 1.40e-01 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 1 | 192 | 1.42e-01 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 1 | 194 | 1.43e-01 |
| GO:BP | GO:0010468 | regulation of gene expression | 3 | 3580 | 1.49e-01 |
| GO:BP | GO:0016070 | RNA metabolic process | 3 | 3600 | 1.50e-01 |
| GO:BP | GO:0050727 | regulation of inflammatory response | 1 | 212 | 1.52e-01 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 1 | 219 | 1.56e-01 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 3 | 3750 | 1.56e-01 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 3 | 3730 | 1.56e-01 |
| GO:BP | GO:0045088 | regulation of innate immune response | 1 | 232 | 1.56e-01 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 1 | 227 | 1.56e-01 |
| GO:BP | GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 1 | 227 | 1.56e-01 |
| GO:BP | GO:0007049 | cell cycle | 2 | 1529 | 1.56e-01 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 1 | 230 | 1.56e-01 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 1 | 242 | 1.62e-01 |
| GO:BP | GO:0044237 | cellular metabolic process | 4 | 6996 | 1.62e-01 |
| GO:BP | GO:0031349 | positive regulation of defense response | 1 | 251 | 1.65e-01 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 4 | 7069 | 1.67e-01 |
| GO:BP | GO:0051235 | maintenance of location | 1 | 260 | 1.68e-01 |
| GO:BP | GO:0006869 | lipid transport | 1 | 272 | 1.70e-01 |
| GO:BP | GO:0033554 | cellular response to stress | 2 | 1681 | 1.70e-01 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 1 | 270 | 1.70e-01 |
| GO:BP | GO:0002253 | activation of immune response | 1 | 272 | 1.70e-01 |
| GO:BP | GO:0043414 | macromolecule methylation | 1 | 272 | 1.70e-01 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 1 | 274 | 1.70e-01 |
| GO:BP | GO:0034504 | protein localization to nucleus | 1 | 276 | 1.70e-01 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 1 | 279 | 1.71e-01 |
| GO:BP | GO:0009605 | response to external stimulus | 2 | 1719 | 1.71e-01 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 3 | 4048 | 1.74e-01 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 1 | 295 | 1.74e-01 |
| GO:BP | GO:0051169 | nuclear transport | 1 | 295 | 1.74e-01 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 1 | 293 | 1.74e-01 |
| GO:BP | GO:0010876 | lipid localization | 1 | 310 | 1.80e-01 |
| GO:BP | GO:0032259 | methylation | 1 | 313 | 1.80e-01 |
| GO:BP | GO:0051098 | regulation of binding | 1 | 317 | 1.80e-01 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 1 | 310 | 1.80e-01 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 4 | 7441 | 1.80e-01 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 3 | 4174 | 1.80e-01 |
| GO:BP | GO:0019538 | protein metabolic process | 3 | 4241 | 1.84e-01 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 3 | 4245 | 1.84e-01 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 1 | 337 | 1.85e-01 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 1 | 335 | 1.85e-01 |
| GO:BP | GO:0050794 | regulation of cellular process | 4 | 7552 | 1.85e-01 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 3 | 4292 | 1.85e-01 |
| GO:BP | GO:0009314 | response to radiation | 1 | 352 | 1.90e-01 |
| GO:BP | GO:0031667 | response to nutrient levels | 1 | 353 | 1.90e-01 |
| GO:BP | GO:1901652 | response to peptide | 1 | 377 | 1.96e-01 |
| GO:BP | GO:0006338 | chromatin remodeling | 1 | 378 | 1.96e-01 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 3 | 4455 | 1.96e-01 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 1 | 377 | 1.96e-01 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 3 | 4417 | 1.96e-01 |
| GO:BP | GO:0050778 | positive regulation of immune response | 1 | 369 | 1.96e-01 |
| GO:BP | GO:0044238 | primary metabolic process | 4 | 7806 | 1.99e-01 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 3 | 4539 | 2.01e-01 |
| GO:BP | GO:0071396 | cellular response to lipid | 1 | 398 | 2.01e-01 |
| GO:BP | GO:0035556 | intracellular signal transduction | 2 | 2065 | 2.01e-01 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 1 | 407 | 2.01e-01 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 3 | 4596 | 2.01e-01 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 3 | 4570 | 2.01e-01 |
| GO:BP | GO:0010467 | gene expression | 3 | 4587 | 2.01e-01 |
| GO:BP | GO:0009058 | biosynthetic process | 3 | 4590 | 2.01e-01 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 3 | 4617 | 2.02e-01 |
| GO:BP | GO:0006954 | inflammatory response | 1 | 429 | 2.02e-01 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 2 | 2139 | 2.02e-01 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 1 | 422 | 2.02e-01 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 1 | 429 | 2.02e-01 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 1 | 421 | 2.02e-01 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 1 | 418 | 2.02e-01 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 2 | 2161 | 2.03e-01 |
| GO:BP | GO:0050789 | regulation of biological process | 4 | 8061 | 2.06e-01 |
| GO:BP | GO:0031347 | regulation of defense response | 1 | 448 | 2.07e-01 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 3 | 4747 | 2.07e-01 |
| GO:BP | GO:0071704 | organic substance metabolic process | 4 | 8169 | 2.14e-01 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 1 | 482 | 2.19e-01 |
| GO:BP | GO:0050776 | regulation of immune response | 1 | 484 | 2.19e-01 |
| GO:BP | GO:0045087 | innate immune response | 1 | 488 | 2.19e-01 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 3 | 4955 | 2.23e-01 |
| GO:BP | GO:0044770 | cell cycle phase transition | 1 | 504 | 2.23e-01 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 3 | 4946 | 2.23e-01 |
| GO:BP | GO:0065007 | biological regulation | 4 | 8320 | 2.23e-01 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 1 | 513 | 2.24e-01 |
| GO:BP | GO:0019222 | regulation of metabolic process | 3 | 5017 | 2.28e-01 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 1 | 540 | 2.33e-01 |
| GO:BP | GO:0006325 | chromatin organization | 1 | 551 | 2.36e-01 |
| GO:BP | GO:0008152 | metabolic process | 4 | 8518 | 2.38e-01 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 1 | 581 | 2.46e-01 |
| GO:BP | GO:0033993 | response to lipid | 1 | 592 | 2.49e-01 |
| GO:BP | GO:0098542 | defense response to other organism | 1 | 608 | 2.52e-01 |
| GO:BP | GO:0060284 | regulation of cell development | 1 | 605 | 2.52e-01 |
| GO:BP | GO:0034097 | response to cytokine | 1 | 607 | 2.52e-01 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 1 | 640 | 2.63e-01 |
| GO:BP | GO:0030097 | hemopoiesis | 1 | 656 | 2.68e-01 |
| GO:BP | GO:0051050 | positive regulation of transport | 1 | 667 | 2.68e-01 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 1 | 665 | 2.68e-01 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 1 | 662 | 2.68e-01 |
| GO:BP | GO:0006950 | response to stress | 2 | 2780 | 2.75e-01 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 1 | 694 | 2.76e-01 |
| GO:BP | GO:0030334 | regulation of cell migration | 1 | 706 | 2.79e-01 |
| GO:BP | GO:2000145 | regulation of cell motility | 1 | 742 | 2.90e-01 |
| GO:BP | GO:0036211 | protein modification process | 2 | 2901 | 2.90e-01 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 1 | 755 | 2.92e-01 |
| GO:BP | GO:0040012 | regulation of locomotion | 1 | 763 | 2.94e-01 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 1 | 782 | 2.98e-01 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 1 | 785 | 2.98e-01 |
| GO:BP | GO:0006974 | DNA damage response | 1 | 785 | 2.98e-01 |
| GO:BP | GO:0006886 | intracellular protein transport | 1 | 803 | 3.03e-01 |
| GO:BP | GO:1901698 | response to nitrogen compound | 1 | 830 | 3.09e-01 |
| GO:BP | GO:0000278 | mitotic cell cycle | 1 | 827 | 3.09e-01 |
| GO:BP | GO:0043412 | macromolecule modification | 2 | 3090 | 3.10e-01 |
| GO:BP | GO:0033365 | protein localization to organelle | 1 | 859 | 3.16e-01 |
| GO:BP | GO:0002682 | regulation of immune system process | 1 | 877 | 3.20e-01 |
| GO:BP | GO:0006259 | DNA metabolic process | 1 | 884 | 3.20e-01 |
| GO:BP | GO:0006955 | immune response | 1 | 882 | 3.20e-01 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 1 | 893 | 3.22e-01 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1 | 937 | 3.35e-01 |
| GO:BP | GO:0006952 | defense response | 1 | 944 | 3.35e-01 |
| GO:BP | GO:0040011 | locomotion | 1 | 954 | 3.37e-01 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 1 | 1006 | 3.52e-01 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 1 | 1014 | 3.53e-01 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 1 | 1074 | 3.68e-01 |
| GO:BP | GO:0080134 | regulation of response to stress | 1 | 1075 | 3.68e-01 |
| GO:BP | GO:0023057 | negative regulation of signaling | 1 | 1076 | 3.68e-01 |
| GO:BP | GO:0022402 | cell cycle process | 1 | 1086 | 3.68e-01 |
| GO:BP | GO:0016477 | cell migration | 1 | 1081 | 3.68e-01 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2 | 3625 | 3.75e-01 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 1 | 1123 | 3.75e-01 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 1 | 1137 | 3.78e-01 |
| GO:BP | GO:0048870 | cell motility | 1 | 1182 | 3.89e-01 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1 | 1223 | 3.99e-01 |
| GO:BP | GO:0007165 | signal transduction | 2 | 3818 | 3.99e-01 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 1 | 1248 | 4.03e-01 |
| GO:BP | GO:0051049 | regulation of transport | 1 | 1267 | 4.07e-01 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1 | 1315 | 4.17e-01 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 1 | 1315 | 4.17e-01 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1 | 1322 | 4.17e-01 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1 | 1345 | 4.22e-01 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2 | 4045 | 4.23e-01 |
| GO:BP | GO:0015031 | protein transport | 1 | 1361 | 4.23e-01 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2 | 4099 | 4.29e-01 |
| GO:BP | GO:0071310 | cellular response to organic substance | 1 | 1399 | 4.29e-01 |
| GO:BP | GO:0023052 | signaling | 2 | 4141 | 4.33e-01 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1 | 1441 | 4.35e-01 |
| GO:BP | GO:0006915 | apoptotic process | 1 | 1443 | 4.35e-01 |
| GO:BP | GO:0046907 | intracellular transport | 1 | 1453 | 4.35e-01 |
| GO:BP | GO:0002376 | immune system process | 1 | 1447 | 4.35e-01 |
| GO:BP | GO:0045184 | establishment of protein localization | 1 | 1462 | 4.35e-01 |
| GO:BP | GO:0007154 | cell communication | 2 | 4226 | 4.36e-01 |
| GO:BP | GO:0012501 | programmed cell death | 1 | 1479 | 4.36e-01 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1 | 1507 | 4.42e-01 |
| GO:BP | GO:0032879 | regulation of localization | 1 | 1555 | 4.50e-01 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1 | 1555 | 4.50e-01 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1 | 1564 | 4.50e-01 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1 | 1602 | 4.55e-01 |
| GO:BP | GO:0008219 | cell death | 1 | 1596 | 4.55e-01 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1 | 1604 | 4.55e-01 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1 | 1684 | 4.72e-01 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2 | 4579 | 4.73e-01 |
| GO:BP | GO:0050793 | regulation of developmental process | 1 | 1835 | 5.00e-01 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1 | 1836 | 5.00e-01 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1 | 1869 | 5.05e-01 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1 | 1898 | 5.10e-01 |
| GO:BP | GO:0065009 | regulation of molecular function | 1 | 1939 | 5.16e-01 |
| GO:BP | GO:0048468 | cell development | 1 | 1963 | 5.16e-01 |
| GO:BP | GO:0010033 | response to organic substance | 1 | 1957 | 5.16e-01 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2 | 4953 | 5.16e-01 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1 | 1974 | 5.17e-01 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1 | 2035 | 5.27e-01 |
| GO:BP | GO:0009987 | cellular process | 4 | 11336 | 5.33e-01 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1 | 2138 | 5.38e-01 |
| GO:BP | GO:0008104 | protein localization | 1 | 2129 | 5.38e-01 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1 | 2135 | 5.38e-01 |
| GO:BP | GO:0071702 | organic substance transport | 1 | 2116 | 5.38e-01 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1 | 2270 | 5.61e-01 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1 | 2293 | 5.61e-01 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1 | 2285 | 5.61e-01 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1 | 2412 | 5.81e-01 |
| GO:BP | GO:0033036 | macromolecule localization | 1 | 2450 | 5.85e-01 |
| GO:BP | GO:0042221 | response to chemical | 1 | 2559 | 5.98e-01 |
| GO:BP | GO:0023051 | regulation of signaling | 1 | 2565 | 5.98e-01 |
| GO:BP | GO:0050896 | response to stimulus | 2 | 5770 | 5.98e-01 |
| GO:BP | GO:0010646 | regulation of cell communication | 1 | 2573 | 5.98e-01 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1 | 2646 | 6.08e-01 |
| GO:BP | GO:0051641 | cellular localization | 1 | 2810 | 6.32e-01 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1 | 2880 | 6.40e-01 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1 | 2890 | 6.40e-01 |
| GO:BP | GO:0008150 | biological_process | 4 | 12070 | 6.45e-01 |
| GO:BP | GO:0030154 | cell differentiation | 1 | 2958 | 6.46e-01 |
| GO:BP | GO:0048869 | cellular developmental process | 1 | 2984 | 6.47e-01 |
| GO:BP | GO:0006810 | transport | 1 | 3298 | 6.88e-01 |
| GO:BP | GO:0007275 | multicellular organism development | 1 | 3386 | 6.98e-01 |
| GO:BP | GO:0051234 | establishment of localization | 1 | 3433 | 7.01e-01 |
| GO:BP | GO:0051179 | localization | 1 | 3969 | 7.61e-01 |
| GO:BP | GO:0048856 | anatomical structure development | 1 | 4128 | 7.75e-01 |
| GO:BP | GO:0032502 | developmental process | 1 | 4494 | 8.07e-01 |
| GO:BP | GO:0032501 | multicellular organismal process | 1 | 4794 | 8.30e-01 |
| GO:BP | GO:0016043 | cellular component organization | 1 | 5076 | 8.49e-01 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1 | 5274 | 8.60e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 1 | 76 | 5.00e-02 |
| KEGG | KEGG:05135 | Yersinia infection | 1 | 112 | 5.00e-02 |
| KEGG | KEGG:05134 | Legionellosis | 1 | 39 | 5.00e-02 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 1 | 56 | 5.00e-02 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 1 | 53 | 5.00e-02 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 1 | 64 | 5.00e-02 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 1 | 113 | 5.00e-02 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 1 | 42 | 5.00e-02 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 1 | 56 | 5.00e-02 |
| KEGG | KEGG:04668 | TNF signaling pathway | 1 | 87 | 5.00e-02 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 1 | 58 | 5.00e-02 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 1 | 97 | 5.00e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 1 | 95 | 5.00e-02 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 1 | 104 | 5.00e-02 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 1 | 70 | 5.00e-02 |
| KEGG | KEGG:05140 | Leishmaniasis | 1 | 42 | 5.00e-02 |
| KEGG | KEGG:05145 | Toxoplasmosis | 1 | 82 | 5.00e-02 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 1 | 51 | 5.00e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 1 | 87 | 5.00e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 1 | 75 | 5.00e-02 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 1 | 69 | 5.00e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 1 | 86 | 5.00e-02 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 1 | 71 | 5.00e-02 |
| KEGG | KEGG:04115 | p53 signaling pathway | 1 | 65 | 5.00e-02 |
| KEGG | KEGG:05142 | Chagas disease | 1 | 75 | 5.00e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 2 | 415 | 5.00e-02 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 1 | 87 | 5.00e-02 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 1 | 60 | 5.00e-02 |
| KEGG | KEGG:05164 | Influenza A | 1 | 103 | 5.00e-02 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 1 | 117 | 5.00e-02 |
| KEGG | KEGG:05162 | Measles | 1 | 98 | 5.00e-02 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 1 | 49 | 5.00e-02 |
| KEGG | KEGG:05160 | Hepatitis C | 1 | 115 | 5.00e-02 |
| KEGG | KEGG:04210 | Apoptosis | 1 | 118 | 5.00e-02 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 1 | 110 | 5.00e-02 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 151 | 5.51e-02 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 1 | 153 | 5.51e-02 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 1 | 150 | 5.51e-02 |
| KEGG | KEGG:05161 | Hepatitis B | 1 | 136 | 5.51e-02 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 1 | 145 | 5.51e-02 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 1 | 153 | 5.51e-02 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 1 | 162 | 5.66e-02 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 1 | 165 | 5.66e-02 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 1 | 172 | 5.76e-02 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 1 | 176 | 5.76e-02 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 1 | 180 | 5.76e-02 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 1 | 186 | 5.82e-02 |
| KEGG | KEGG:05131 | Shigellosis | 1 | 210 | 6.41e-02 |
| KEGG | KEGG:05132 | Salmonella infection | 1 | 214 | 6.41e-02 |
| KEGG | KEGG:05200 | Pathways in cancer | 1 | 409 | 1.17e-01 |
| KEGG | KEGG:00000 | KEGG root term | 3 | 5558 | 1.90e-01 |
Mtx3_sp_DEGtable %>%
dplyr::filter(source=="KEGG") %>%
# dplyr::filter(p_value<0.05) %>%
dplyr::select(p_value,term_name,intersection_size) %>%
slice_min(., n=10 ,order_by=p_value) %>%
mutate(log_val = -log10(p_value)) %>%
# slice_max(., n=10,order_by = p_value) %>%
ggplot(., aes(x = log_val, y =reorder(term_name,p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
geom_vline(xintercept = (-log10(0.05)))+
scale_y_discrete(labels = wrap_format(30))+
guides(col="none", size=guide_legend(title = "# of intersected \n terms"))+
ggtitle('MTX 3 hour specific(stringent)\n gene set KEGG terms') +
xlab(expression(" -"~log[10]~("adj. p-value")))+
ylab("KEGG pathway")+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0),
strip.text.x = element_text(size = 12, color = "black", face = "bold"))
toplist3hr <- toplistall %>%
mutate(id =case_match(id, "Da"~"DNR",
"Do"~"DOX",
"Ep"~"EPI",
"Mi"~"MTX",
"Tr"~"TRZ",
"Ve"~"VEH", .default = id)) %>%
mutate(id=factor(id, levels = c('DOX','EPI','DNR','MTX','TRZ','VEH'))) %>%
mutate(id = as.factor(id)) %>%
mutate(time=factor(time, levels=c("3_hours","24_hours"))) %>%
filter(time=="3_hours")
densityDOX3sp <-
toplist3hr %>%
filter(ENTREZID %in% Dox3onlyDEG) %>%
# filter(adj.P.Val<0.5) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("DOX\n n = 0")+
guides(fill=FALSE,alpha=FALSE)+
theme(
axis.title = element_text(size = 14, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
densityDNR3sp <-
toplist3hr %>%
filter(ENTREZID %in% Dnr3onlyDEG) %>%
# filter(adj.P.Val<0.5) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
# geom_histogram(aes(y = ..density.., fill = id, alpha = 0.8), bins = 120)+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("DNR-only 3 hour DEGs\n n = 322")+
guides(fill=FALSE,alpha=FALSE)+
theme(
axis.title = element_text(size = 14, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
densityEPI3sp <-
toplist3hr %>%
filter(ENTREZID %in% Epi3onlyDEG) %>%
# filter(adj.P.Val<0.5) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("EPI-only DEGs\n n = 18")+
ylim(0,50)+
guides(fill=FALSE,alpha=FALSE)+
theme(axis.title = element_text(size = 14, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
densityMTX3sp <-
toplist3hr %>%
filter(ENTREZID %in% Mtx3onlyDEG) %>%
# filter(adj.P.Val<0.5) %>%
ggplot(., aes(x=adj.P.Val))+
geom_density(aes(fill=id, alpha= 0.8))+
fill_palette(palette = drug_palc)+
theme_bw()+
ylab("MTX-only DEGs\n n = 16")+
# ylim(0,0.5)+
guides(fill=FALSE,alpha=FALSE)+
theme(axis.title = element_text(size = 14, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))Dox3Bplot <- toplist3hr %>%
group_by(time,id) %>%
dplyr::filter(ENTREZID %in% DOX3deg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("DOX n = 0")+
theme_classic()+
fill_palette(palette = drug_palc)+
guides(fill= "none")+
theme(
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
Dnr3Bplot <-
toplist3hr %>%
group_by(time,id) %>%
dplyr::filter(ENTREZID %in% DNR3deg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("DNR-spec. adj.P.V <0.01\n n = 100")+
theme_classic()+
guides(fill= "none")+
fill_palette(palette = drug_palc)+
theme(
axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
Epi3Bplot <-
toplist3hr %>%
group_by(time,id) %>%
filter(ENTREZID %in% EPI3deg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("EPI-spec.adj.P.V <0.01\n n = 0")+
theme_classic()+
guides(fill= "none")+
fill_palette(palette = drug_palc)+
theme(axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))
Mtx3Bplot <-
toplist3hr %>%
group_by(time,id) %>%
filter(time=="3_hours") %>%
filter(ENTREZID %in% MTX3deg_sp$ENTREZID) %>%
mutate(logFC=logFC*(-1)) %>%
mutate("treatment" = id) %>%
ggplot(., aes(x= treatment, y=logFC))+
geom_boxplot(aes(fill=id))+
xlab(" ")+
ylab("MTX-spec. adj.P.V <0.01\n n = 4")+
guides(fill= "none")+
theme_classic()+
fill_palette(palette = drug_palc)+
theme(axis.title = element_text(size = 10, color = "black"),
axis.ticks = element_line(size = 1.5),
axis.text = element_text(size = 10, color = "black", angle = 0))Dox3genesp_example <- "none"#cpm_boxplot(cpmcounts,GOI=101927720,"Dark2",drug_palc,
# ylab=bquote(~italic("ZNF793-AS1 ")~log[2]~"cpm "))+
# theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))
Dnr3genesp_example <- cpm_boxplot_time(cpmcounts,"3h",7975 ,"Dark2",drug_palc,
ylab=bquote(~italic("MAFK")~log[2]~"cpm "))+
theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))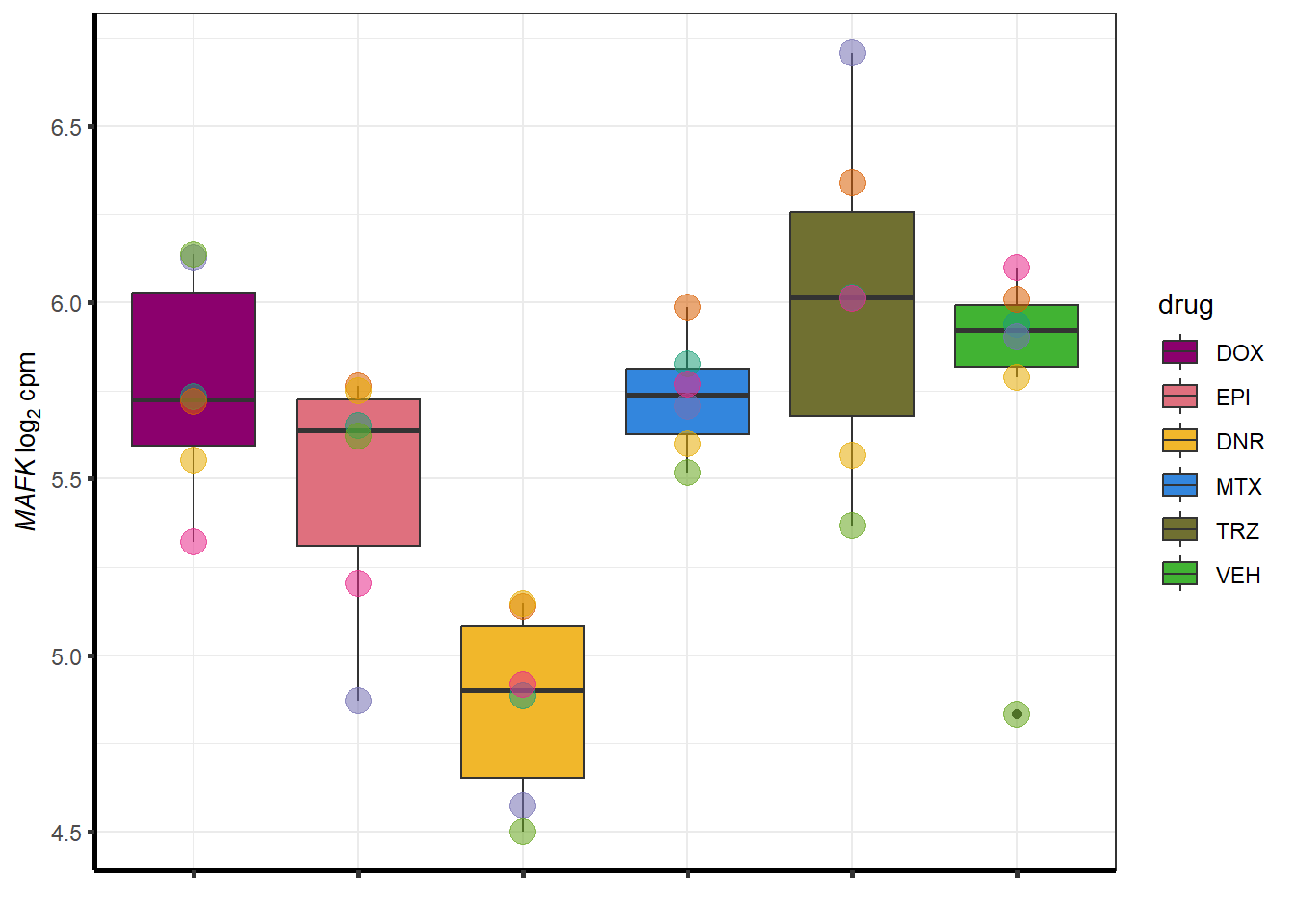
Epi3genesp_example <- "none"#cpm_boxplot(cpmcounts,GOI=79169,"Dark2",drug_palc,
# ylab=bquote(~italic("C1orf35")~log[2]~"cpm "))+
# theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))
Mtx3genesp_example <- cpm_boxplot_time(cpmcounts, "3h",GOI=284306,"Dark2",drug_palc,
ylab=bquote(~italic("ZNF547")~log[2]~"cpm "))+
theme(axis.title = element_text(size = 10, color = "black"), axis.text = element_text(size = 10, color = "black", angle = 0))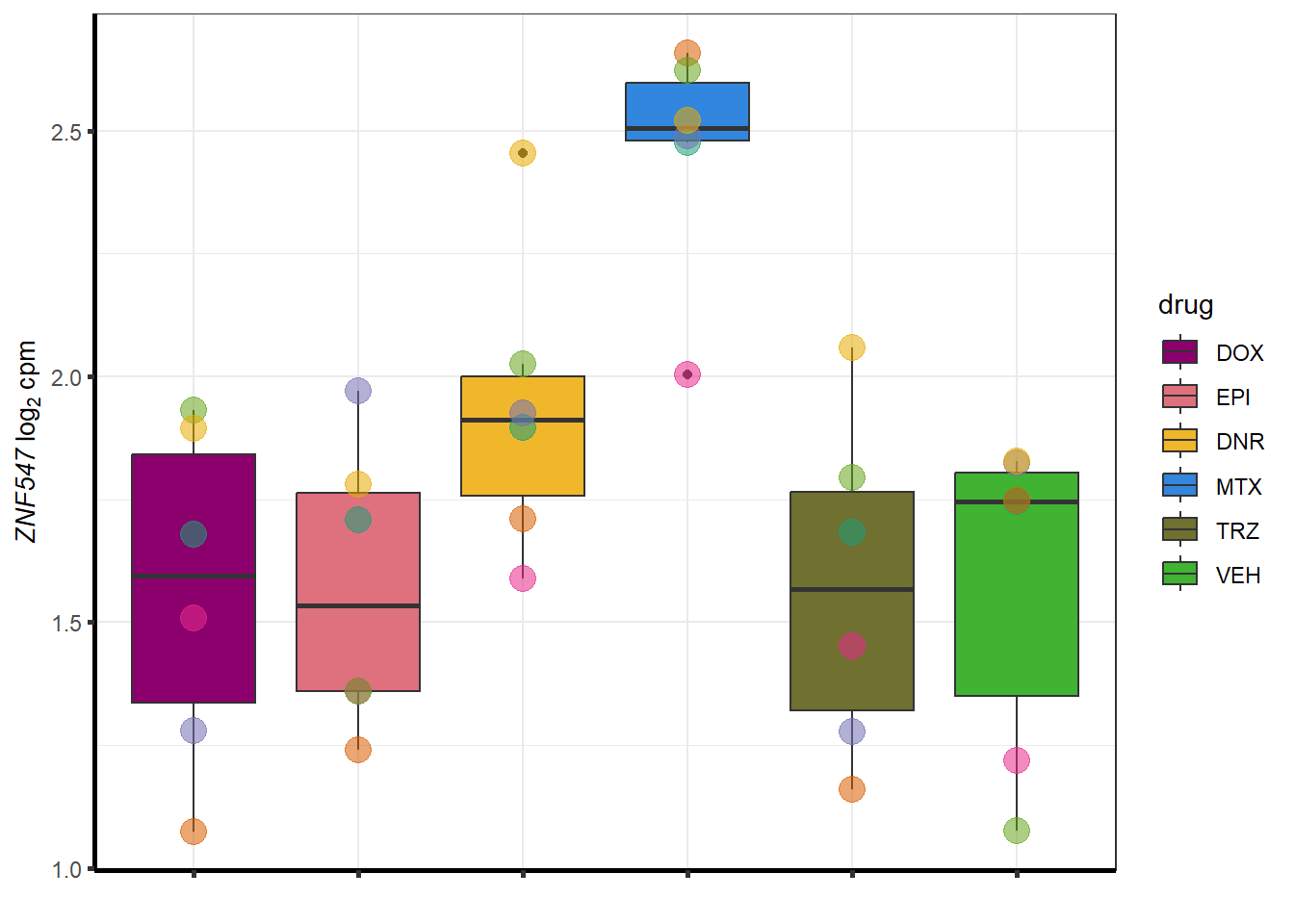
grid 3 hour
mito3pg <- plot_grid(densityMTX3sp, Mtx3Bplot, Mtx3genesp_example,
nrow = 1,
rel_heights = c(.8,2,1),
rel_widths=c(1.2,1,1.5),
scale=c(1,0.8,0.8))
Daun3pg <- plot_grid(densityDNR3sp, Dnr3Bplot, Dnr3genesp_example, nrow = 1, rel_heights = c(.8,2,1), rel_widths=c(1.2,1,1.5),scale=c(1,0.8,0.8))
Doxo3pg <- plot_grid(densityDOX3sp, Dox3Bplot, Dox3genesp_example, nrow = 1, rel_heights = c(.8,2,1), rel_widths=c(1.2,1,1.5),scale=c(1,0.8,0.8))
Epi3pg <- plot_grid(densityEPI3sp, Epi3Bplot, Epi3genesp_example, nrow = 1, rel_heights = c(.8,2,1), rel_widths=c(1.2,1,1.5),scale=c(1,0.8,0.8))
allfinal3hour <- plot_grid(Doxo3pg,Epi3pg,Daun3pg,mito3pg,nrow=4, rel_heights = c(0.5,1,1,1))
allfinal3hour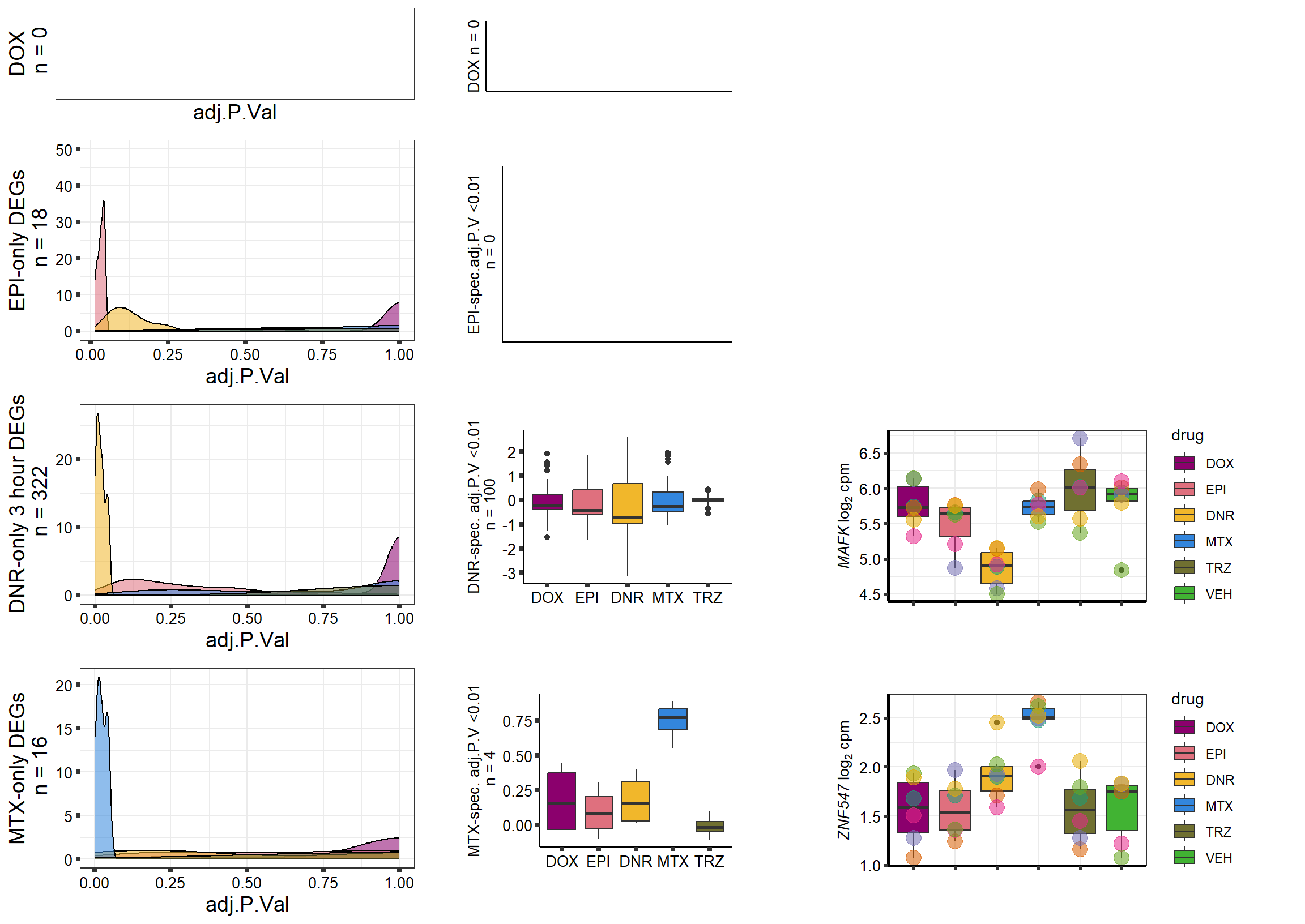
saveRDS(allfinal3hour,"data/allfinal3hour.RDS")
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] gprofiler2_0.2.2 paletteer_1.6.0 ggVennDiagram_1.5.0
[4] ComplexHeatmap_2.16.0 broom_1.0.5 kableExtra_1.3.4
[7] sjmisc_2.8.9 scales_1.3.0 ggpubr_0.6.0
[10] cowplot_1.1.1 ggsignif_0.6.4 lubridate_1.9.3
[13] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3
[16] purrr_1.0.2 readr_2.1.4 tidyr_1.3.0
[19] tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0
[22] RColorBrewer_1.1-3 limma_3.56.2 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] formatR_1.14 rematch2_2.1.2 rlang_1.1.2
[4] magrittr_2.0.3 clue_0.3-65 GetoptLong_1.0.5
[7] git2r_0.32.0 matrixStats_1.1.0 compiler_4.3.1
[10] getPass_0.2-2 png_0.1-8 systemfonts_1.0.5
[13] callr_3.7.3 vctrs_0.6.4 rvest_1.0.3
[16] pkgconfig_2.0.3 shape_1.4.6 crayon_1.5.2
[19] fastmap_1.1.1 ellipsis_0.3.2 backports_1.4.1
[22] labeling_0.4.3 utf8_1.2.4 promises_1.2.1
[25] rmarkdown_2.25 tzdb_0.4.0 ps_1.7.5
[28] ragg_1.2.7 xfun_0.41 cachem_1.0.8
[31] jsonlite_1.8.7 highr_0.10 later_1.3.1
[34] parallel_4.3.1 cluster_2.1.4 R6_2.5.1
[37] bslib_0.6.1 stringi_1.7.12 car_3.1-2
[40] jquerylib_0.1.4 Rcpp_1.0.11 iterators_1.0.14
[43] knitr_1.45 VennDiagram_1.7.3 IRanges_2.34.1
[46] httpuv_1.6.12 timechange_0.2.0 tidyselect_1.2.0
[49] rstudioapi_0.15.0 abind_1.4-5 yaml_2.3.7
[52] doParallel_1.0.17 codetools_0.2-19 sjlabelled_1.2.0
[55] processx_3.8.2 shiny_1.8.0 withr_3.0.0
[58] evaluate_0.23 lambda.r_1.2.4 futile.logger_1.4.3
[61] xml2_1.3.5 circlize_0.4.15 pillar_1.9.0
[64] carData_3.0-5 whisker_0.4.1 foreach_1.5.2
[67] stats4_4.3.1 insight_0.19.8 plotly_4.10.4
[70] generics_0.1.3 rprojroot_2.0.4 hms_1.1.3
[73] S4Vectors_0.38.2 munsell_0.5.0 xtable_1.8-4
[76] glue_1.6.2 lazyeval_0.2.2 tools_4.3.1
[79] data.table_1.14.8 webshot_0.5.5 fs_1.6.3
[82] crosstalk_1.2.1 colorspace_2.1-0 cli_3.6.1
[85] textshaping_0.3.7 futile.options_1.0.1 fansi_1.0.5
[88] viridisLite_0.4.2 svglite_2.1.2 gtable_0.3.4
[91] rstatix_0.7.2 sass_0.4.7 digest_0.6.33
[94] prismatic_1.1.1 BiocGenerics_0.46.0 htmlwidgets_1.6.4
[97] rjson_0.2.21 farver_2.1.1 htmltools_0.5.7
[100] lifecycle_1.0.4 httr_1.4.7 mime_0.12
[103] GlobalOptions_0.1.2