Comparisons with other data sets
ERM
2024-02-05
Last updated: 2024-02-05
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version df08393. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/variance_values by gene.png
Ignored: data/41588_2018_171_MOESM3_ESMeQTL_ST2_for paper.csv
Ignored: data/Arr_GWAS.txt
Ignored: data/Arr_geneset.RDS
Ignored: data/BC_cell_lines.csv
Ignored: data/BurridgeDOXTOX.RDS
Ignored: data/CADGWASgene_table.csv
Ignored: data/CAD_geneset.RDS
Ignored: data/CALIMA_Data/
Ignored: data/CMD04_75DRCviability.csv
Ignored: data/CMD04_87DRCviability.csv
Ignored: data/CMD05_75DRCviability.csv
Ignored: data/CMD05_87DRCviability.csv
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/Counts_RNA_ERMatthews.txt
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDEMresp_list.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG-GO/
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/DRC48hoursdata.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/EPIstorelist24.RDS
Ignored: data/Ep24counts.txt
Ignored: data/FC_necela.RDS
Ignored: data/FC_necela_names.RDS
Ignored: data/Full_LD_rep.csv
Ignored: data/GOIsig.csv
Ignored: data/GOplots.R
Ignored: data/GTEX_setsimple.csv
Ignored: data/GTEX_sig24.RDS
Ignored: data/GTEx_gene_list.csv
Ignored: data/HFGWASgene_table.csv
Ignored: data/HF_geneset.RDS
Ignored: data/Heart_Left_Ventricle.v8.egenes.txt
Ignored: data/Heatmap_mat.RDS
Ignored: data/Heatmap_sig.RDS
Ignored: data/Hf_GWAS.txt
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/Knowles_log2cpm_real.RDS
Ignored: data/Knowles_variation_data.RDS
Ignored: data/Knowles_variation_data_conc.RDS
Ignored: data/Knowlesvarlist.RDS
Ignored: data/LD50_05via.csv
Ignored: data/LDH48hoursdata.csv
Ignored: data/Mt24counts.txt
Ignored: data/NoRespDEG_final.csv
Ignored: data/RINsamplelist.txt
Ignored: data/RNA_seq_trial.RDS
Ignored: data/Schneider_GWAS.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/Sup_replicate_values.csv
Ignored: data/TMMnormed_x.RDS
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/TableS10.csv
Ignored: data/TableS11.csv
Ignored: data/TableS9.csv
Ignored: data/Top2_expression.RDS
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Var_test_list.RDS
Ignored: data/Var_test_list24.RDS
Ignored: data/Var_test_list24alt.RDS
Ignored: data/Var_test_list3.RDS
Ignored: data/Vargenes.RDS
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allfinal3hour.RDS
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/allmymatrix.RDS
Ignored: data/annotation_data_frame.RDS
Ignored: data/averageviabilitytable.RDS
Ignored: data/averageviabilitytable.csv
Ignored: data/avgLD50.RDS
Ignored: data/avg_LD50.RDS
Ignored: data/avg_via_table.csv
Ignored: data/backGL.txt
Ignored: data/burr_genes.RDS
Ignored: data/calcium_data.RDS
Ignored: data/clamp_summary.RDS
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/cormotifprobs.csv
Ignored: data/counts24hours.RDS
Ignored: data/cpmcount.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/crispr_genes.csv
Ignored: data/ctnnt_results.txt
Ignored: data/cvd_GWAS.txt
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/drug_noveh1.csv
Ignored: data/efit2.RDS
Ignored: data/efit2_final.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filcpm_counts.RDS
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/framefun24.RDS
Ignored: data/geneDoxonlyQTL.csv
Ignored: data/gene_corr_df.RDS
Ignored: data/gene_corr_frame.RDS
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/geneset_24.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/gtex/
Ignored: data/heartgenes.csv
Ignored: data/highly_var_genelist.RDS
Ignored: data/hsa_kegg_anno.RDS
Ignored: data/individualDRCfile.RDS
Ignored: data/individual_DRC48.RDS
Ignored: data/individual_LDH48.RDS
Ignored: data/indv_noveh1.csv
Ignored: data/kegglistDEG.RDS
Ignored: data/kegglistDEG24.RDS
Ignored: data/kegglistDEG3.RDS
Ignored: data/knowfig4.csv
Ignored: data/knowfig5.csv
Ignored: data/label_list.RDS
Ignored: data/ld50_table.csv
Ignored: data/mean_vardrug1.csv
Ignored: data/mean_varframe.csv
Ignored: data/mymatrix.RDS
Ignored: data/new_ld50avg.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/organized_drugframe.csv
Ignored: data/pca_all_anno.csv
Ignored: data/plan2plot.png
Ignored: data/plot_intv_list.RDS
Ignored: data/plot_list_DRC.RDS
Ignored: data/qval24hr.RDS
Ignored: data/qval3hr.RDS
Ignored: data/qvalueEPItemp.RDS
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sampsettrz.RDS
Ignored: data/schneider_closest_output.RDS
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/siglist_final.RDS
Ignored: data/siglist_old.RDS
Ignored: data/slope_table.csv
Ignored: data/supp10_24hlist.RDS
Ignored: data/supp10_3hlist.RDS
Ignored: data/supp_normLDH48.RDS
Ignored: data/supp_pca_all_anno.RDS
Ignored: data/supp_viadata.csv
Ignored: data/table3a.omar
Ignored: data/test_run_sample_list.txt
Ignored: data/testlist.txt
Ignored: data/toplistall.RDS
Ignored: data/trtonly_24h_genes.RDS
Ignored: data/trtonly_3h_genes.RDS
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Ignored: data/viability.RDS
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: .RDataTmp3
Untracked: 3hr all.pdf
Untracked: Code_files_list.csv
Untracked: Data_files_list.csv
Untracked: Doxorubicin_vehicle_3_24.csv
Untracked: Doxtoplist.csv
Untracked: EPIqvalue_analysis.Rmd
Untracked: Final.sup.pdf
Untracked: GWAS_list_of_interest.xlsx
Untracked: KEGGpathwaylist.R
Untracked: NA
Untracked: OmicNavigator_learn.R
Untracked: SNP_egenes_allfiles.RDS
Untracked: SNP_frame_pdf
Untracked: SNP_frame_pdf.pdf
Untracked: SigDoxtoplist.csv
Untracked: analysis/DRC_viability_check.Rmd
Untracked: analysis/New_code_dec-23.R
Untracked: analysis/cellcycle_kegg_genes.R
Untracked: analysis/ciFIT.R
Untracked: analysis/export_to_excel.R
Untracked: analysis/featureCountsPLAY.R
Untracked: cleanupfiles_script.R
Untracked: code/biomart_gene_names.R
Untracked: code/constantcode.R
Untracked: code/corMotifcustom.R
Untracked: code/cpm_boxplot.R
Untracked: code/extracting_ggplot_data.R
Untracked: code/movingfilesto_ppl.R
Untracked: code/pearson_extract_func.R
Untracked: code/pearson_tox_extract.R
Untracked: code/plot1C.fun.R
Untracked: code/spearman_extract_func.R
Untracked: code/venndiagramcolor_control.R
Untracked: cormotif_p.post.list_4.csv
Untracked: figS1024h.pdf
Untracked: final.pdf
Untracked: individual-legenddark2.png
Untracked: installed_old.rda
Untracked: listoftranscripts
Untracked: motif_ER.txt
Untracked: motif_LR.txt
Untracked: motif_NR.txt
Untracked: motif_TI.txt
Untracked: output/ABHD8_dif_values.RDS
Untracked: output/C3orf18_dif_values.RDS
Untracked: output/Cardiotox_dif_values.RDS
Untracked: output/DNR_DEGlist.csv
Untracked: output/DNRvenn.RDS
Untracked: output/DOX_DEGlist.csv
Untracked: output/DOX_de_goi.csv
Untracked: output/DOXvenn.RDS
Untracked: output/EEF1B2_dif_values.RDS
Untracked: output/EEIG1_dif_values.RDS
Untracked: output/EPI_DEGlist.csv
Untracked: output/EPIvenn.RDS
Untracked: output/ESGN_rds.RDS
Untracked: output/FC_necela.RDS
Untracked: output/FC_necela_names.RDS
Untracked: output/FRS2_dif_values.RDS
Untracked: output/Figures/
Untracked: output/GTEXv8_gene_median_tpm.RDS
Untracked: output/GTEXv8_gene_tpm_heart_left_ventricle.RDS
Untracked: output/HDDC2_dif_values.RDS
Untracked: output/HER2_gene.RDS
Untracked: output/KEGGcellcyclegenes.RDS
Untracked: output/Knowles_S13.csv
Untracked: output/Knowles_log2cpm.csv
Untracked: output/Knowles_supp13.csv
Untracked: output/LD50tox_table.RDS
Untracked: output/MTX_DEGlist.csv
Untracked: output/MTXvenn.RDS
Untracked: output/PEX16_dif_values.RDS
Untracked: output/RASIP1_dif_values.RDS
Untracked: output/RMI1_dif_values.RDS
Untracked: output/RSID_QTL_list_full.txt
Untracked: output/SETA_analysis_reyes.RDS
Untracked: output/SGWAS_top50_order.csv
Untracked: output/SLC27A1_dif_values.RDS
Untracked: output/SLC28A3_dif_values.RDS
Untracked: output/SNP_egenes_allfiles.RDS
Untracked: output/SNP_list_ID.RDS
Untracked: output/SNP_list_full.txt
Untracked: output/SNP_supp.RDS
Untracked: output/TGFBR3L_dif_values.RDS
Untracked: output/TNS2_dif_values.RDS
Untracked: output/TOP_50SNPreffile.csv
Untracked: output/TRZ_DEGlist.csv
Untracked: output/TableS8.csv
Untracked: output/Volcanoplot_10
Untracked: output/Volcanoplot_10.RDS
Untracked: output/ZNF740_dif_values.RDS
Untracked: output/allfinal_sup10.RDS
Untracked: output/counts_v8_heart_left_ventricle_gct.RDS
Untracked: output/crisprfoldchange.RDS
Untracked: output/endocytosisgenes.csv
Untracked: output/expre7k.csv
Untracked: output/expressed_egenes_by_RSID.csv
Untracked: output/gene_corr_fig9.RDS
Untracked: output/genes.RDS
Untracked: output/legend_b.RDS
Untracked: output/motif_ERrep.RDS
Untracked: output/motif_LRrep.RDS
Untracked: output/motif_NRrep.RDS
Untracked: output/motif_TI_rep.RDS
Untracked: output/near_genes_SNP1.RDS
Untracked: output/necela_list_test.RDS
Untracked: output/necela_val_genes.RDS
Untracked: output/output-old/
Untracked: output/rank24genes.csv
Untracked: output/rank3genes.csv
Untracked: output/sequencinginformationforsupp.csv
Untracked: output/sequencinginformationforsupp.prn
Untracked: output/sigVDA24.txt
Untracked: output/sigVDA3.txt
Untracked: output/sigVDX24.txt
Untracked: output/sigVDX3.txt
Untracked: output/sigVEP24.txt
Untracked: output/sigVEP3.txt
Untracked: output/sigVMT24.txt
Untracked: output/sigVMT3.txt
Untracked: output/sigVTR24.txt
Untracked: output/sigVTR3.txt
Untracked: output/supplementary_motif_list_GO.RDS
Untracked: output/test_biomart_run.RDS
Untracked: output/toptablebydrug.RDS
Untracked: output/trop_knowles_fun.csv
Untracked: output/tvl24hour.txt
Untracked: output/x_counts.RDS
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/DRC_analysis.Rmd
Modified: analysis/GOI_plots.Rmd
Modified: analysis/GTEx_genes.Rmd
Deleted: analysis/Knowles2019.Rmd
Modified: output/daplot.RDS
Modified: output/dxplot.RDS
Modified: output/epplot.RDS
Modified: output/mtplot.RDS
Modified: output/plan2plot.png
Modified: output/trplot.RDS
Modified: output/veplot.RDS
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/other_analysis.Rmd) and
HTML (docs/other_analysis.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | df08393 | reneeisnowhere | 2024-02-05 | updates to scripts |
| Rmd | 06800c9 | reneeisnowhere | 2023-07-26 | Commits to small changes and edits |
| html | 7758920 | reneeisnowhere | 2023-07-03 | Build site. |
| Rmd | 4126f95 | reneeisnowhere | 2023-07-03 | updated with final data |
| html | 771a192 | reneeisnowhere | 2023-06-29 | Build site. |
| Rmd | 112c968 | reneeisnowhere | 2023-06-29 | with final data |
| html | 6e4c867 | reneeisnowhere | 2023-06-21 | Build site. |
| Rmd | 028f18d | reneeisnowhere | 2023-06-21 | update on Fold Change plot |
| html | 4643600 | reneeisnowhere | 2023-06-16 | Build site. |
| Rmd | 751239e | reneeisnowhere | 2023-06-16 | updating and moving code |
| Rmd | 3d4ca64 | reneeisnowhere | 2023-06-16 | updates on Friday |
| html | 7ce5a2c | reneeisnowhere | 2023-06-15 | Build site. |
| Rmd | 9afd6a0 | reneeisnowhere | 2023-06-15 | fixing wflow error |
| html | 6b03af2 | reneeisnowhere | 2023-06-15 | Build site. |
| html | e02ca18 | reneeisnowhere | 2023-06-15 | Build site. |
| Rmd | 9ad6b91 | reneeisnowhere | 2023-06-15 | showing code adding pvalue text |
| html | 750ee45 | reneeisnowhere | 2023-06-15 | Build site. |
| Rmd | 637531c | reneeisnowhere | 2023-06-15 | moving out the knowles data |
| Rmd | f8f511a | reneeisnowhere | 2023-06-15 | updates and simplifications of code |
| Rmd | 7fc7ec7 | reneeisnowhere | 2023-06-14 | updating code |
| html | 4b6bd9b | reneeisnowhere | 2023-06-07 | Build site. |
| Rmd | 4b62a1e | reneeisnowhere | 2023-06-07 | updated numbers for grant |
| html | d64a0ae | reneeisnowhere | 2023-06-07 | Build site. |
| Rmd | 81f100c | reneeisnowhere | 2023-06-07 | add Dox reQTL grouping and AC shared numbers |
| html | 47f85a2 | reneeisnowhere | 2023-06-07 | Build site. |
| Rmd | 0ecede3 | reneeisnowhere | 2023-06-07 | data with CRispr set added and heatmap changes |
| html | 9a62d7c | reneeisnowhere | 2023-06-06 | Build site. |
| Rmd | 232d3b0 | reneeisnowhere | 2023-06-06 | Finally tested chisquare between knowles data |
| Rmd | 10bcf05 | reneeisnowhere | 2023-06-06 | updating the k4/k5 analysis of DEG |
| html | b4dd015 | reneeisnowhere | 2023-06-02 | Build site. |
| Rmd | 652d7e8 | reneeisnowhere | 2023-06-02 | updated heatmap Seoane Chisqure for cormotif |
| html | 5aeda27 | reneeisnowhere | 2023-06-02 | Build site. |
| Rmd | 6524ecd | reneeisnowhere | 2023-06-02 | Adding in heatmaps of chi values |
| html | 5dd9ddb | reneeisnowhere | 2023-06-02 | Build site. |
| Rmd | 8eaea47 | reneeisnowhere | 2023-06-02 | chi square updates |
| html | e4d118c | reneeisnowhere | 2023-06-01 | Build site. |
| Rmd | 573a477 | reneeisnowhere | 2023-06-01 | Updateing supplement 1 seoan chi results |
| html | cc3dfc3 | reneeisnowhere | 2023-06-01 | Build site. |
| Rmd | 522cce8 | reneeisnowhere | 2023-06-01 | Adding chisquare and other analysis |
| html | 4723cdd | reneeisnowhere | 2023-05-31 | Build site. |
| Rmd | 07a6e06 | reneeisnowhere | 2023-05-31 | adding in more data including Cormotif enrichment numbers |
| html | 6fd877b | reneeisnowhere | 2023-05-31 | Build site. |
| Rmd | b2ba055 | reneeisnowhere | 2023-05-31 | adding Seoane data with cormotif things |
| html | 4c0812e | reneeisnowhere | 2023-05-26 | Build site. |
| Rmd | c7e0fcc | reneeisnowhere | 2023-05-26 | adding in Gtex and chisquare values |
| html | e1bcef0 | reneeisnowhere | 2023-05-26 | Build site. |
| Rmd | 0f512c3 | reneeisnowhere | 2023-05-26 | adding in Gtex and chisquare values |
| Rmd | 1f8c483 | reneeisnowhere | 2023-05-26 | updating code with gtex and chisq |
| Rmd | 25d32da | reneeisnowhere | 2023-05-26 | Adding 3 hour and chisq test to populations |
| html | 5610749 | reneeisnowhere | 2023-05-22 | Build site. |
| Rmd | 889832a | reneeisnowhere | 2023-05-22 | add Seoane data again |
| html | 36cbdab | reneeisnowhere | 2023-05-22 | Build site. |
| Rmd | de54fd5 | reneeisnowhere | 2023-05-22 | add Seoane data |
| html | 7243a18 | reneeisnowhere | 2023-05-22 | Build site. |
| Rmd | e2b3215 | reneeisnowhere | 2023-05-22 | add Seoane data |
| html | c3481d8 | reneeisnowhere | 2023-05-22 | Build site. |
| Rmd | acbd0a8 | reneeisnowhere | 2023-05-22 | updates on GWAS enrichment |
| Rmd | e8c82ec | reneeisnowhere | 2023-05-18 | adding other_analysis and genes of interest log2cpm |
library(limma)
library(tidyverse)
library(ggsignif)
library(biomaRt)
library(RColorBrewer)
library(cowplot)
library(ggpubr)
library(scales)
library(sjmisc)
library(kableExtra)
library(broom)
library(ComplexHeatmap)Data set comparison
order:
ArrGWAS
HFGWAS
CADGWAS
Crispr sets
GWAS
ArrGWAS to 24 hour DEG genes p < 0.05
24 hour data set
# How I did the string split
# Arr_GWAS <- ArrGWAS[,13]
# names(Arr_GWAS) <- "genesplit"
# Arr_GWAS <- Arr_GWAS %>%
# separate_longer_delim(genesplit, delim = ",")
#write.csv(Arr_GWAS,"data/Arr_GWAS.txt")
arr_GWAS <- read.csv("data/Arr_GWAS.txt", row.names = 1)
Arr_geneset <- readRDS("data/Arr_geneset.RDS")
# Arr_geneset <- getBM(attributes=my_attributes,filters ='hgnc_symbol',
# values = arr_GWAS, mart = ensembl)
# #remove duplicates
# Arr_geneset <- Arr_geneset %>% distinct(entrezgene_id, .keep_all =TRUE)
# saveRDS(Arr_geneset,"data/Arr_geneset.RDS")
#Apply sorting
toplist24hr %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(ARR=if_else(ENTREZID %in%Arr_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,ARR) %>%
dplyr::summarize(ARRcount=n())%>%
pivot_wider(id_cols = c(id,sigcount), names_from=c(ARR), values_from=ARRcount) %>%
mutate(ARRprop=(y/(y+no)*100)) %>%
ggplot(., aes(x=id, y=ARRprop)) +
geom_col()+
geom_text(aes(x=id, label = sprintf("%.2f",ARRprop), vjust=-.2))+
#geom_text(aes(label = expression(paste0("number"~a,"out of",~b))))+
facet_wrap(~sigcount)+
ggtitle("24 hour non-significant and significant enrichment proporitions of Arrhythmia GWAS ")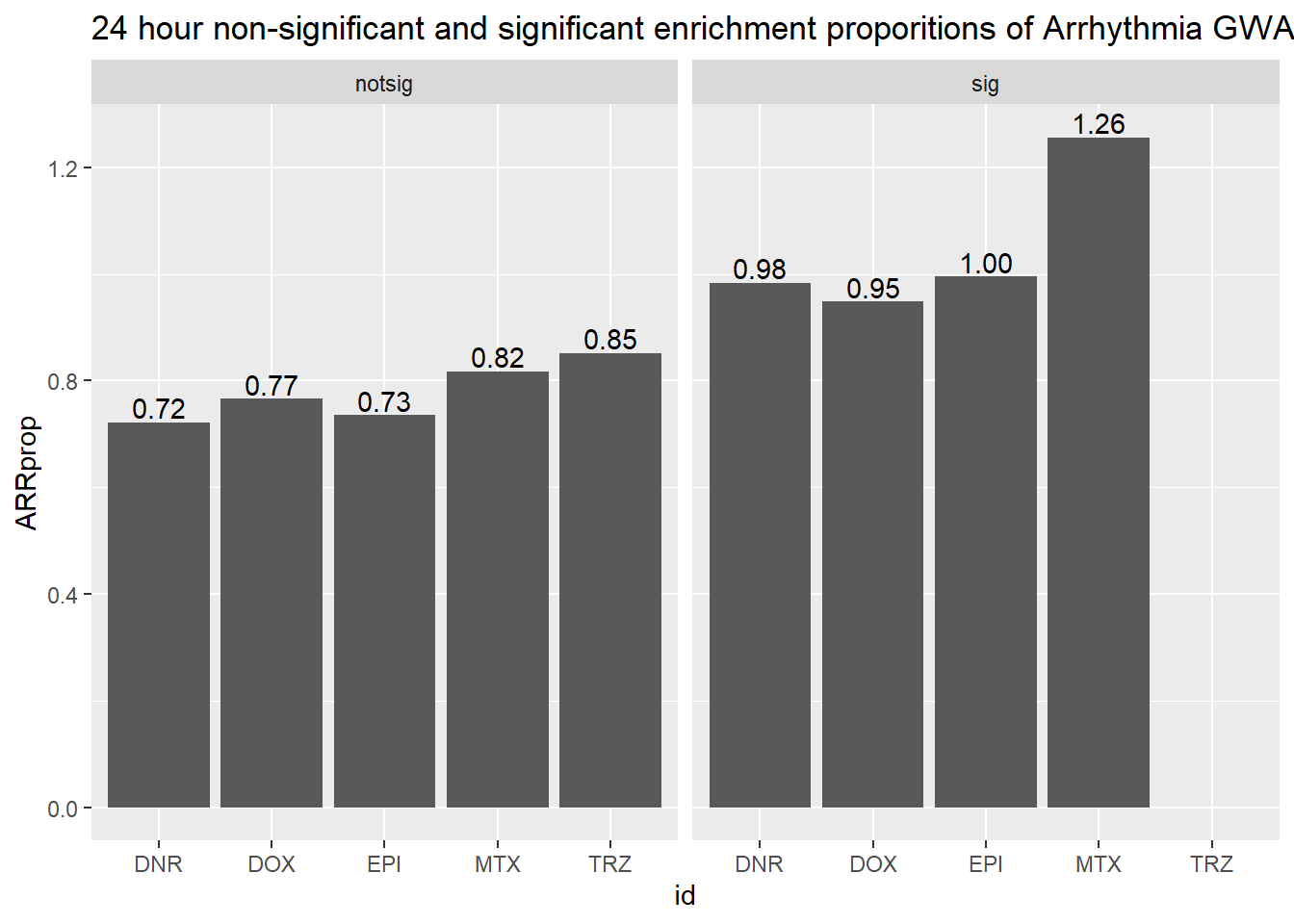
##make table of numbers:
dataframARR <- toplist24hr %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(ARR=if_else(ENTREZID %in%Arr_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,ARR) %>%
dplyr::summarize(ARRcount=n()) %>%
as.data.frame()
dataframARR %>%
kable(., caption= "Significant (adj. P value of <0.05) and non-sig gene counts in Arrhythmia 24 hour GWAS") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | sigcount | ARR | ARRcount |
|---|---|---|---|
| DNR | notsig | no | 7016 |
| DNR | notsig | y | 51 |
| DNR | sig | no | 6948 |
| DNR | sig | y | 69 |
| DOX | notsig | no | 7382 |
| DOX | notsig | y | 57 |
| DOX | sig | no | 6582 |
| DOX | sig | y | 63 |
| EPI | notsig | no | 7699 |
| EPI | notsig | y | 57 |
| EPI | sig | no | 6265 |
| EPI | sig | y | 63 |
| MTX | notsig | no | 12863 |
| MTX | notsig | y | 106 |
| MTX | sig | no | 1101 |
| MTX | sig | y | 14 |
| TRZ | notsig | no | 13964 |
| TRZ | notsig | y | 120 |
3 hour data set
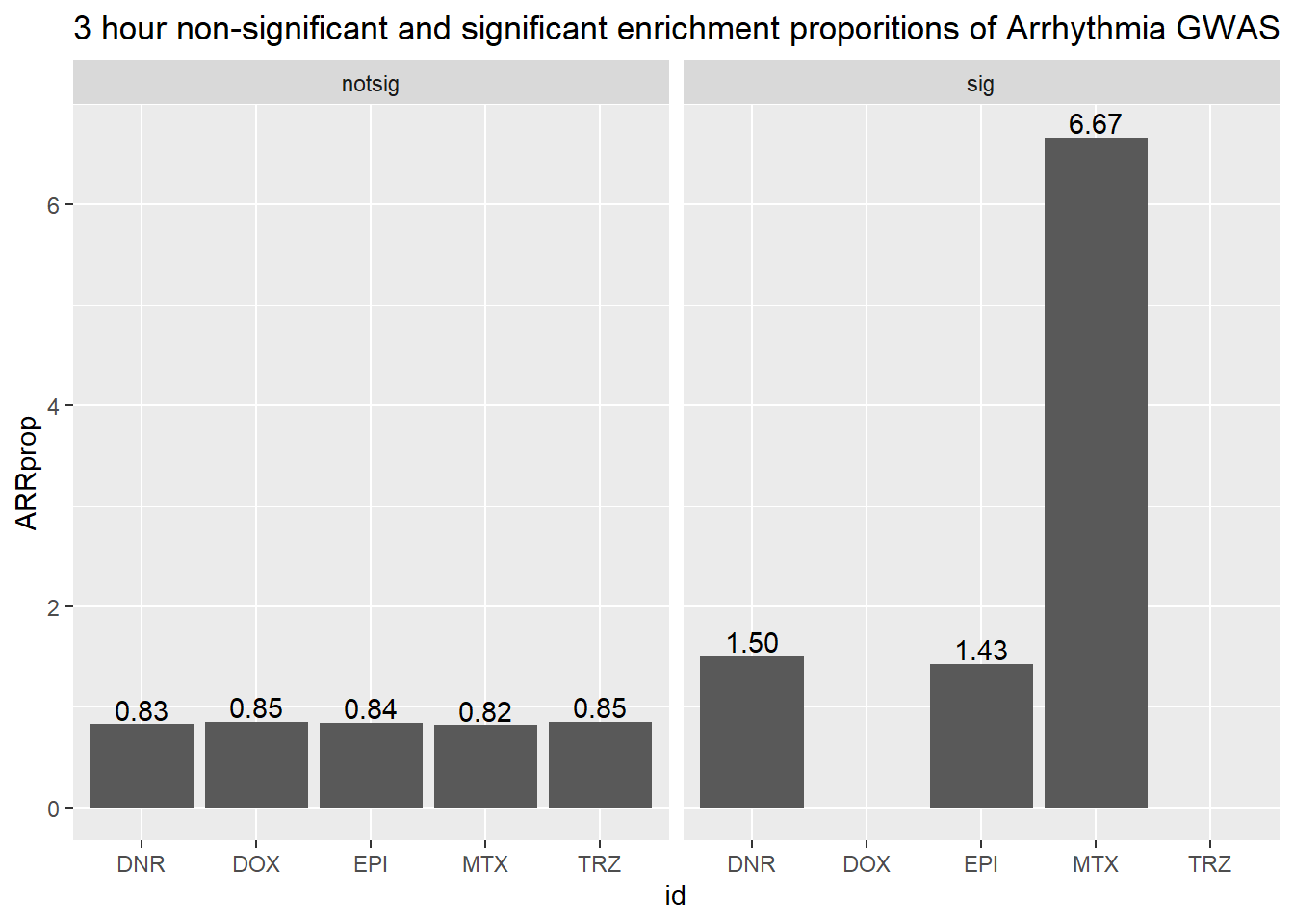
| id | sigcount | ARR | ARRcount |
|---|---|---|---|
| DNR | notsig | no | 13440 |
| DNR | notsig | y | 112 |
| DNR | sig | no | 524 |
| DNR | sig | y | 8 |
| DOX | notsig | no | 13945 |
| DOX | notsig | y | 120 |
| DOX | sig | no | 19 |
| EPI | notsig | no | 13757 |
| EPI | notsig | y | 117 |
| EPI | sig | no | 207 |
| EPI | sig | y | 3 |
| MTX | notsig | no | 13894 |
| MTX | notsig | y | 115 |
| MTX | sig | no | 70 |
| MTX | sig | y | 5 |
| TRZ | notsig | no | 13964 |
| TRZ | notsig | y | 120 |
chi square test ARR
chi_funarr <- toplistall %>%
dplyr::filter(id!="TRZ") %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(ARR=if_else(ENTREZID %in%Arr_geneset$entrezgene_id,"y","no")) %>%
group_by(id,time) %>%
dplyr::summarise(pvalue= chisq.test(ARR, sigcount)$p.value)
chi_funarr %>%
kable(., caption= "after performing chi square test between DEgenes, and non DE genes") %>%
kable_paper("striped") %>%
kable_styling(full_width = FALSE,font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | time | pvalue |
|---|---|---|
| DNR | 24_hours | 0.1101318 |
| DNR | 3_hours | 0.1536193 |
| DOX | 24_hours | 0.2799966 |
| DOX | 3_hours | 1.0000000 |
| EPI | 24_hours | 0.1136502 |
| EPI | 3_hours | 0.5908261 |
| MTX | 24_hours | 0.1744165 |
| MTX | 3_hours | 0.0000012 |
HFGWAS
24 hours HF
##just like ARrGWAS- imported the total csv, the took the "nearest" column and separated out the gene info
# test <- HFGWAS %>%
# select(nearest) %>%
# separate_wider_delim(nearest, delim = "[", names_sep = "", too_few = "align_start")
# test2 <- str_sub(test$nearest2,0,nchar(test$nearest2)-1)
# Hf_GWAS <- test2
#write.csv(Hf_GWAS, "data/Hf_GWAS.txt")
# HF_GWAS <- read.csv("data/Hf_GWAS.txt", row.names =1)
#
# HF_geneset <- getBM(attributes=my_attributes,filters ='hgnc_symbol',
# values = HF_GWAS, mart = ensembl)
# #remove duplicates
# HF_geneset <- HF_geneset %>% distinct(entrezgene_id, .keep_all =TRUE)
# saveRDS(HF_geneset,"data/HF_geneset.RDS")
HF_geneset <- readRDS("data/HF_geneset.RDS")
#Apply sorting
toplist24hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(HF=if_else(ENTREZID %in%HF_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,HF) %>%
dplyr::summarize(HFcount=n())%>%
pivot_wider(id_cols = c(id,sigcount), names_from=c(HF), values_from=HFcount) %>%
mutate(HFprop=(y/(y+no)*100)) %>%
ggplot(., aes(x=id, y=HFprop)) +
geom_col()+
geom_text(aes(x=id, label = sprintf("%.2f",HFprop), vjust=-.2))+
#geom_text(aes(label = expression(paste0("number"~a,"out of",~b))))+
facet_wrap(~sigcount)+
ggtitle("non-significant and significant enrichment proporitions of Heart Failure GWAS ")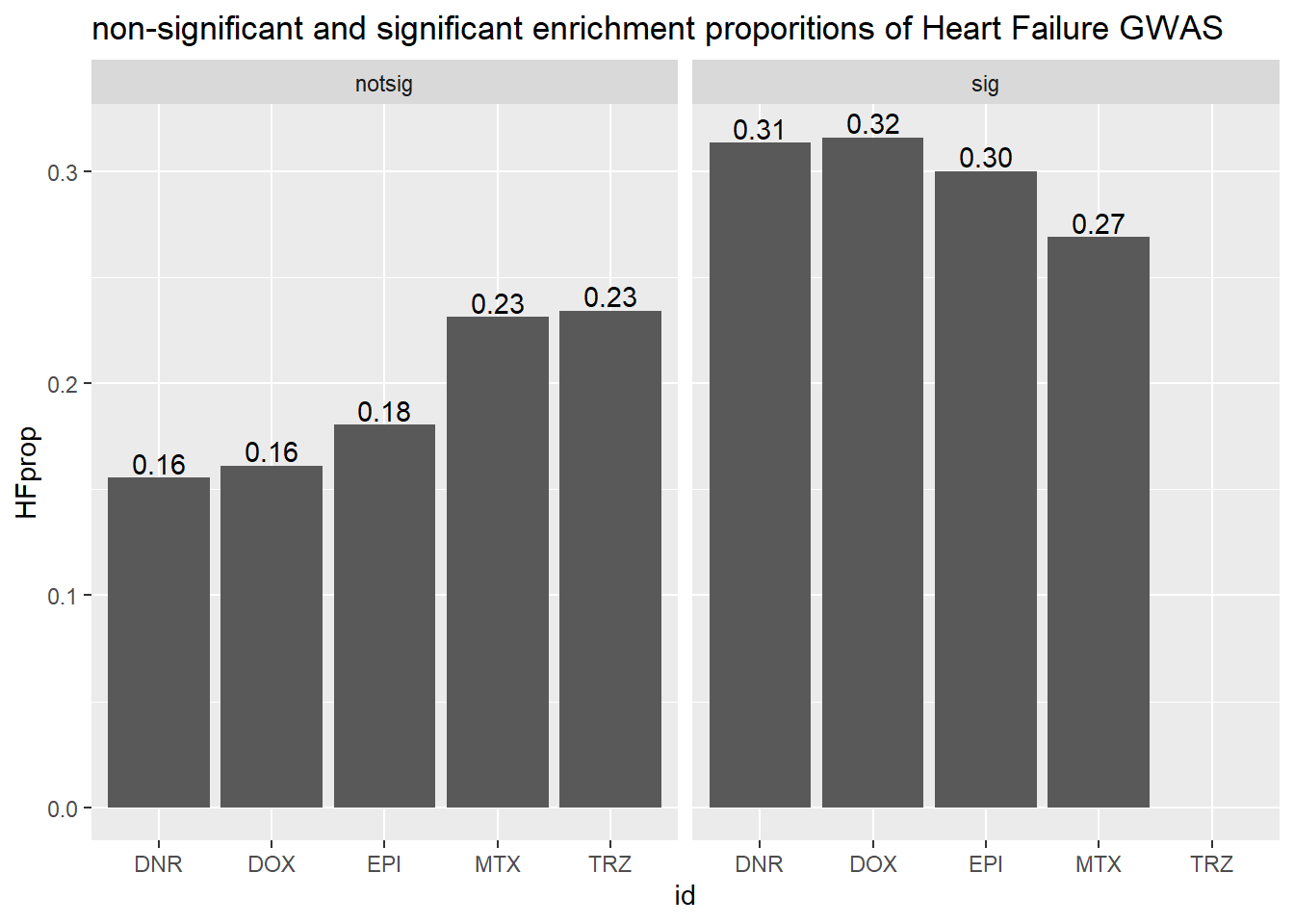
##make table of numbers:
dataframHF <- toplist24hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(HF=if_else(ENTREZID %in%HF_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,HF) %>%
dplyr::summarize(HFcount=n()) %>%
as.data.frame()
dataframHF %>% #mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Significant (adj. P value of <0.05) and non-sig gene counts in HFhythmia GWAS") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | sigcount | HF | HFcount |
|---|---|---|---|
| DNR | notsig | no | 7056 |
| DNR | notsig | y | 11 |
| DNR | sig | no | 6995 |
| DNR | sig | y | 22 |
| DOX | notsig | no | 7427 |
| DOX | notsig | y | 12 |
| DOX | sig | no | 6624 |
| DOX | sig | y | 21 |
| EPI | notsig | no | 7742 |
| EPI | notsig | y | 14 |
| EPI | sig | no | 6309 |
| EPI | sig | y | 19 |
| MTX | notsig | no | 12939 |
| MTX | notsig | y | 30 |
| MTX | sig | no | 1112 |
| MTX | sig | y | 3 |
| TRZ | notsig | no | 14051 |
| TRZ | notsig | y | 33 |
3 hours HF
#Apply sorting
toplist3hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(HF=if_else(ENTREZID %in%HF_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,HF) %>%
dplyr::summarize(HFcount=n())%>%
pivot_wider(id_cols = c(id,sigcount), names_from=c(HF), values_from=HFcount) %>%
mutate(HFprop=(y/(y+no)*100)) %>%
ggplot(., aes(x=id, y=HFprop)) +
geom_col()+
geom_text(aes(x=id, label = sprintf("%.2f",HFprop), vjust=-.2))+
#geom_text(aes(label = expression(paste0("number"~a,"out of",~b))))+
facet_wrap(~sigcount)+
ggtitle("non-significant and significant enrichment proportions of Heart Failure GWAS ")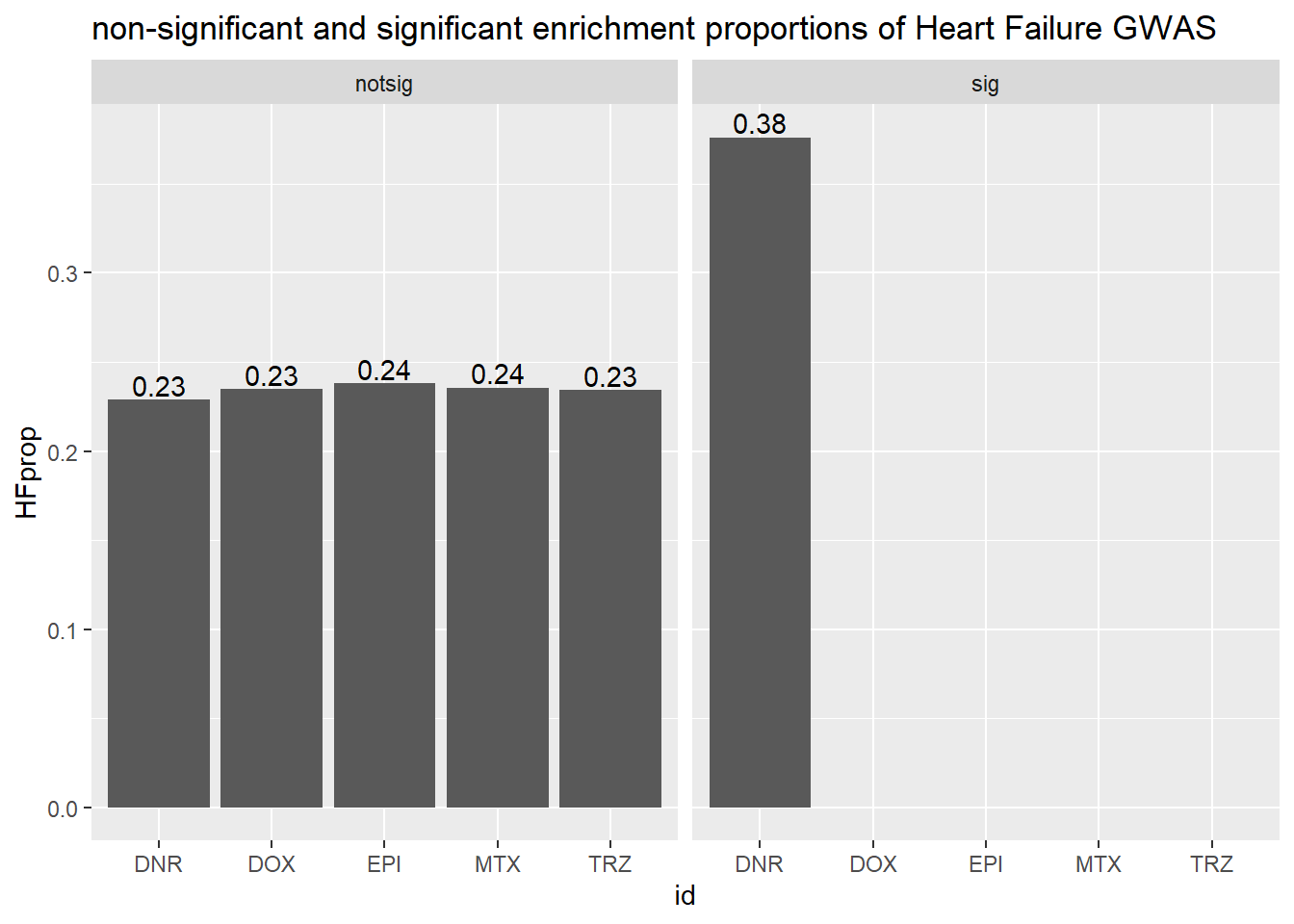
##make table of numbers:
dataframHF3 <- toplist3hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(HF=if_else(ENTREZID %in%HF_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,HF) %>%
dplyr::summarize(HFcount=n()) %>%
as.data.frame()
dataframHF3 %>%
kable(., caption= "Significant (adj. P value of <0.05) and non-sig gene counts in Three hour HFhythmia GWAS") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | sigcount | HF | HFcount |
|---|---|---|---|
| DNR | notsig | no | 13521 |
| DNR | notsig | y | 31 |
| DNR | sig | no | 530 |
| DNR | sig | y | 2 |
| DOX | notsig | no | 14032 |
| DOX | notsig | y | 33 |
| DOX | sig | no | 19 |
| EPI | notsig | no | 13841 |
| EPI | notsig | y | 33 |
| EPI | sig | no | 210 |
| MTX | notsig | no | 13976 |
| MTX | notsig | y | 33 |
| MTX | sig | no | 75 |
| TRZ | notsig | no | 14051 |
| TRZ | notsig | y | 33 |
chi square test HF
chi_funhf <- toplistall %>%
mutate(id = as.factor(id)) %>%
dplyr::filter(id!="TRZ") %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(HF=if_else(ENTREZID %in%HF_geneset$entrezgene_id,"y","no")) %>%
group_by(id,time) %>%
dplyr::summarise(pvalue= chisq.test(HF, sigcount)$p.value)
chi_funhf %>%
kable(., caption= "after performing chi square test between DEgenes, and non DE genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE,font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | time | pvalue |
|---|---|---|
| DNR | 24_hours | 0.0778586 |
| DNR | 3_hours | 0.8167557 |
| DOX | 24_hours | 0.0852093 |
| DOX | 3_hours | 1.0000000 |
| EPI | 24_hours | 0.1981312 |
| EPI | 3_hours | 1.0000000 |
| MTX | 24_hours | 1.0000000 |
| MTX | 3_hours | 1.0000000 |
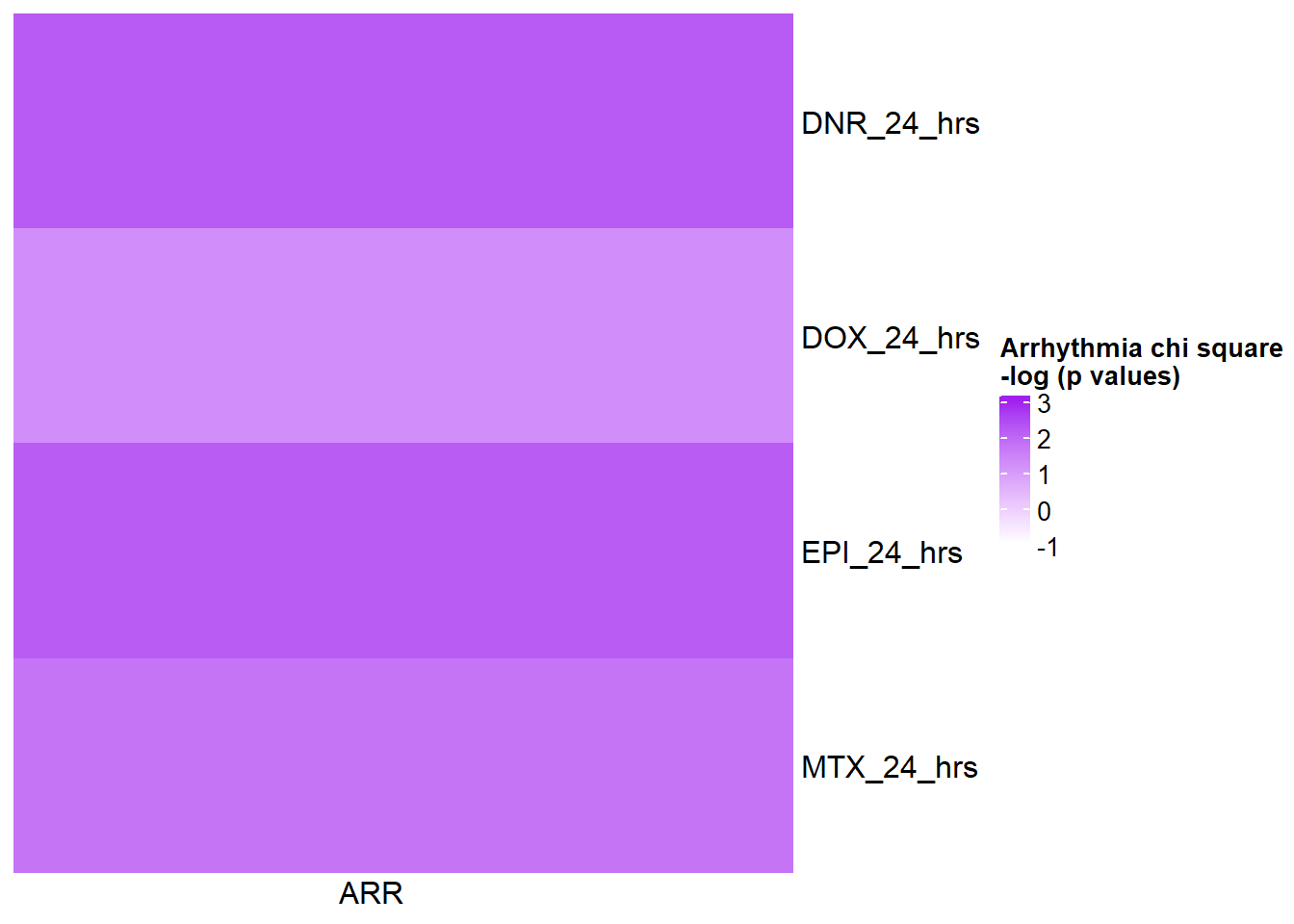
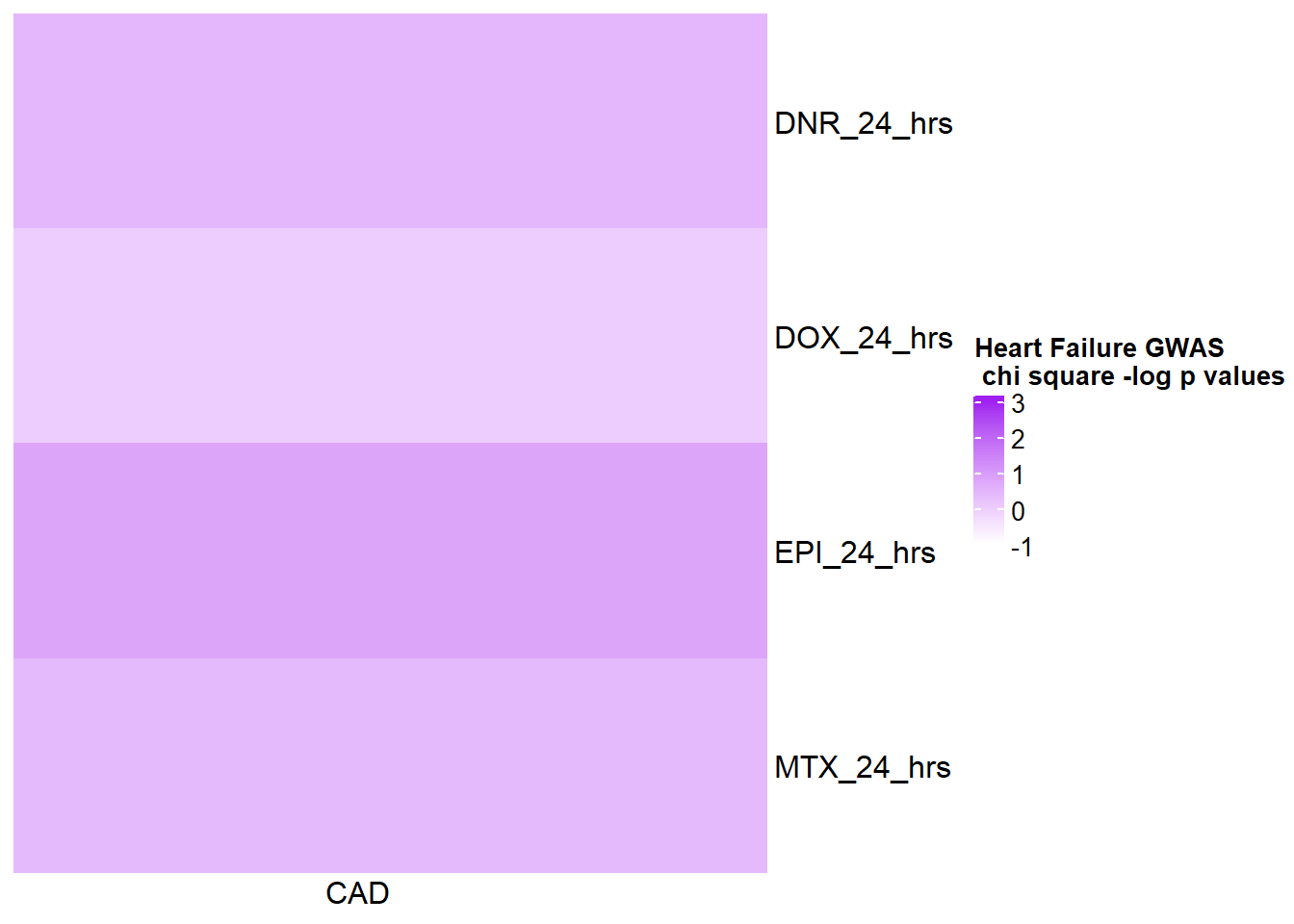
HFmat <- chi_funhf %>% ungroup() %>%
mutate(HF= log(pvalue)*(-1)) %>%
filter(time=="24_hours") %>%
# mutate(id =case_match( id, 'Daunorubicin'~'DNR',
# 'Doxorubicin'~'DOX',
# 'Epirubicin'~'EPI',
# 'Mitoxantrone'~'MTX',
# .default = id)) %>%
mutate(time=case_match(time,"24_hours"~"24_hrs",.default = time)) %>%
dplyr::select(id,time,HF) %>%
unite("term_name",id,time,sep="_") %>%
column_to_rownames('term_name')
col_fun5 = circlize::colorRamp2(c(0, 5), c("white", "purple"))
Heatmap( as.matrix(HFmat), name = "Heart Failure GWAS\n chi square -log p values",
cluster_rows = FALSE, cluster_columns = FALSE,
col=col_fun1,
column_names_rot = 0,
cell_fun = function(j, i, x, y, width, height, fill) {
if(HFmat[i, j] > -log(0.05))
grid.text("*", x, y, gp = gpar(fontsize = 20))
})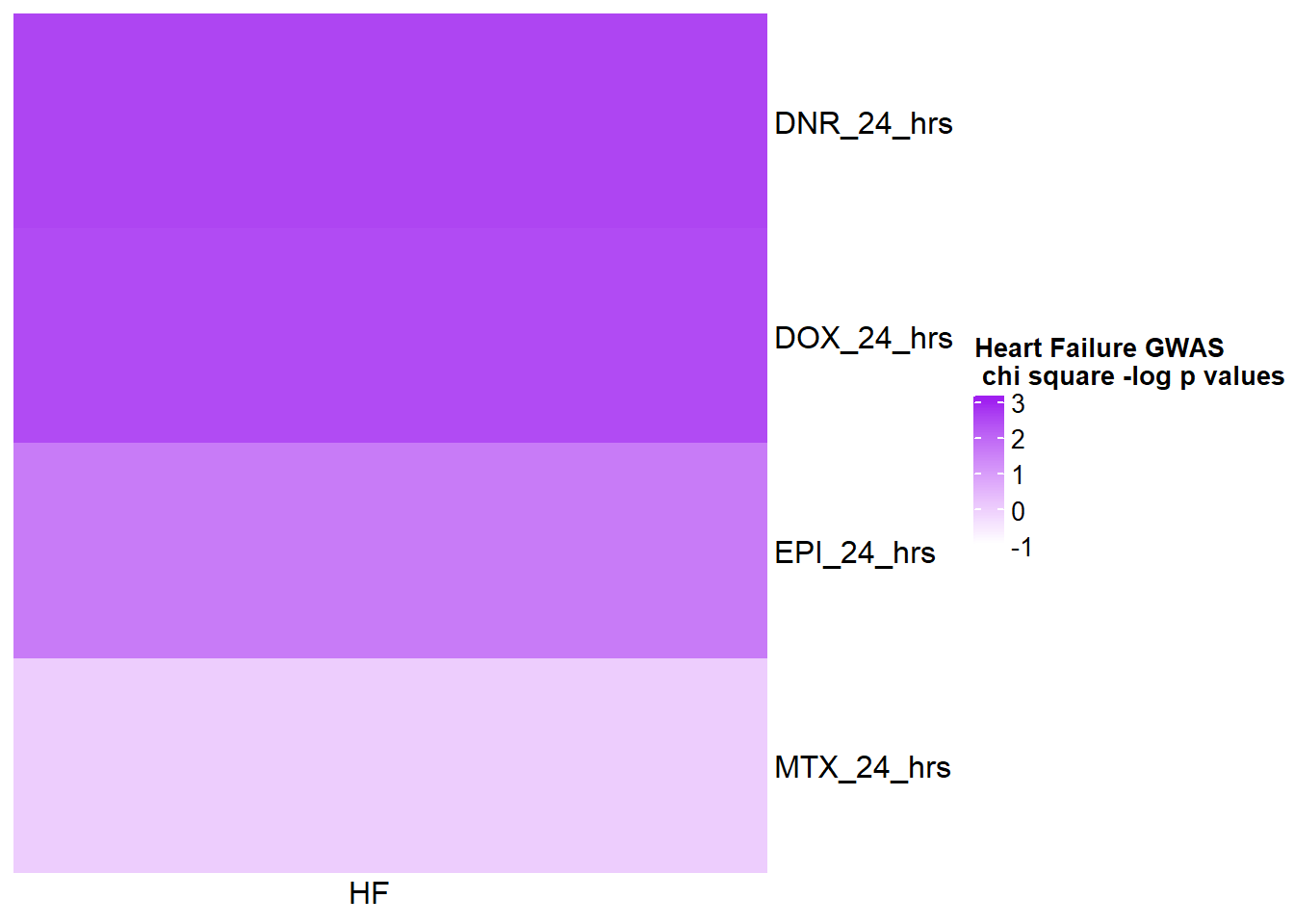
CAD GWAS
24 hour data set
# test <- CADGWAS %>%
# select(nearest) %>%
# separate_wider_delim(nearest, delim = "[", names_sep = "", too_few = "align_start")
# test2 <- str_sub(test$nearest2,0,nchar(test$nearest2)-1)
#
# test2[c(32,38,44,74,112,126,191,212)] <- c("TPCN1","C2orf43","FAM222A", "TDRD15" ,"AGPAT4","SVOP","SVOP","PLG")
#
# test2 [c(218,226,228,233,239,245,256,270,281,322,324,332,335,338,347,351,352,358)] <- c("HPCAL1", "KLHL29" , "COL4A3BP" , "ARAP1" ,
# "VEGFA", "TBPL1","SLC22A3" ,"C19orf38","LPA","VPS29","ATP2A2" ,"ATP2A2","KLHL29","GUCY1A3","KCNE2", "HOXB9","P2RY2" ,"CTC-236F12.4")
#
# CAD_GWAS <- test2
#write.csv(CAD_GWAS, "data/cvd_GWAS.txt")
CAD_GWAS <- read.csv("data/cvd_GWAS.txt", row.names =1)
# CAD_geneset <- getBM(attributes=my_attributes,filters ='hgnc_symbol',
# values = CAD_GWAS, mart = ensembl)
# #remove duplicates
# CAD_geneset <- CAD_geneset %>% distinct(entrezgene_id, .keep_all =TRUE)
#
# saveRDS(CAD_geneset,"data/CAD_geneset.RDS")
CAD_geneset <- readRDS("data/CAD_geneset.RDS")
#Apply sorting
toplist24hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(CAD=if_else(ENTREZID %in%CAD_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,CAD) %>%
dplyr::summarize(CADcount=n())%>%
pivot_wider(id_cols = c(id,sigcount), names_from=c(CAD), values_from=CADcount) %>%
mutate(CADprop=(y/(y+no)*100)) %>%
ggplot(., aes(x=id, y=CADprop)) +
geom_col()+
geom_text(aes(x=id, label = sprintf("%.2f",CADprop), vjust=-.2))+
#geom_text(aes(label = expression(paste0("number"~a,"out of",~b))))+
facet_wrap(~sigcount)+
ggtitle("non-significant and significant enrichment proporitions of CAD GWAS ")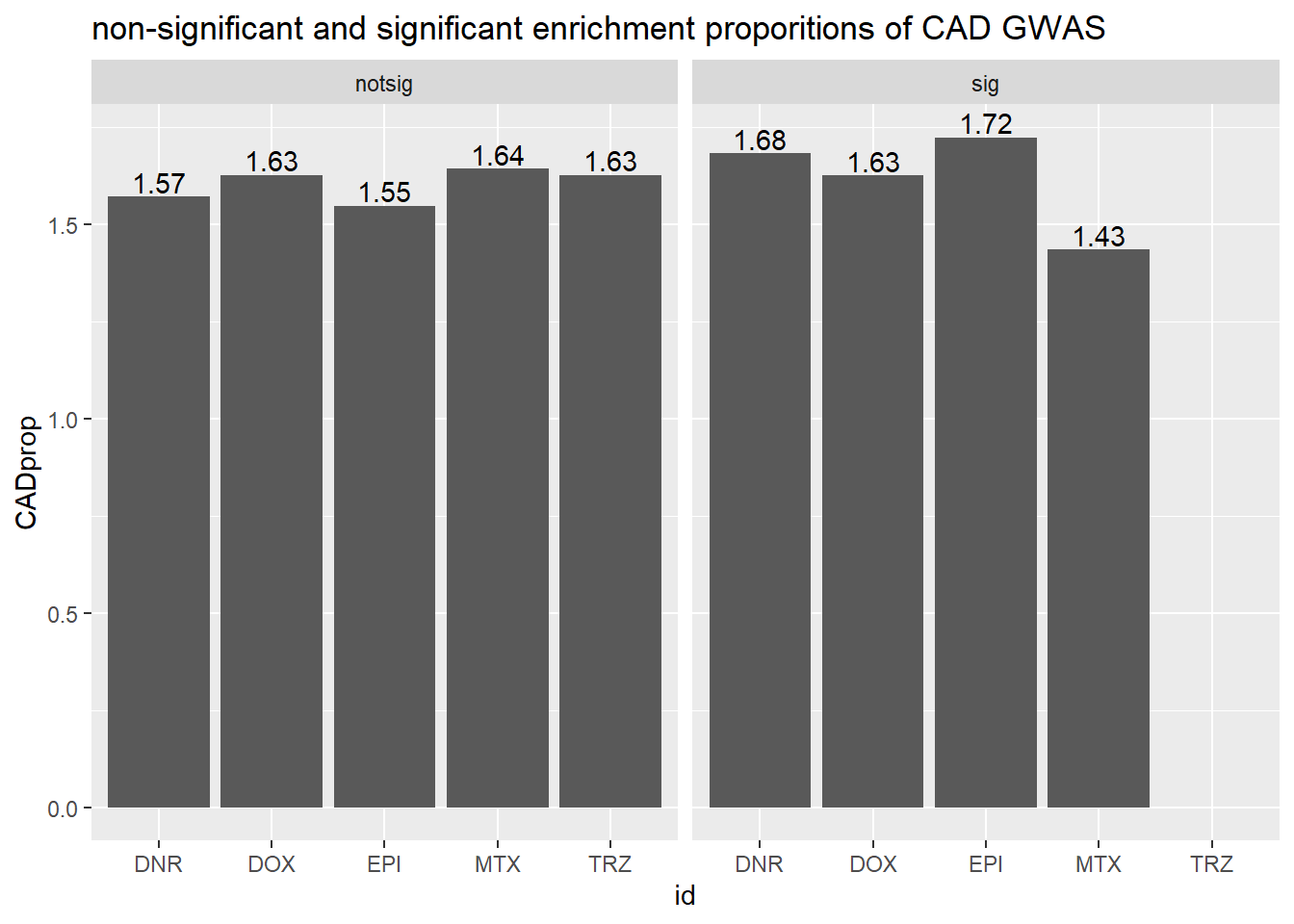
##make table of numbers:
dataframCAD <- toplist24hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(CAD=if_else(ENTREZID %in%CAD_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,CAD) %>%
dplyr::summarize(CADcount=n()) %>%
as.data.frame()
dataframCAD %>% #mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Significant (adj. P value of <0.05) and non-sig gene counts in CAD GWAS") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | sigcount | CAD | CADcount |
|---|---|---|---|
| DNR | notsig | no | 6956 |
| DNR | notsig | y | 111 |
| DNR | sig | no | 6899 |
| DNR | sig | y | 118 |
| DOX | notsig | no | 7318 |
| DOX | notsig | y | 121 |
| DOX | sig | no | 6537 |
| DOX | sig | y | 108 |
| EPI | notsig | no | 7636 |
| EPI | notsig | y | 120 |
| EPI | sig | no | 6219 |
| EPI | sig | y | 109 |
| MTX | notsig | no | 12756 |
| MTX | notsig | y | 213 |
| MTX | sig | no | 1099 |
| MTX | sig | y | 16 |
| TRZ | notsig | no | 13855 |
| TRZ | notsig | y | 229 |
3 hour data set
#Apply sorting
toplist3hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(CAD=if_else(ENTREZID %in%CAD_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,CAD) %>%
dplyr::summarize(CADcount=n())%>%
pivot_wider(id_cols = c(id,sigcount), names_from=c(CAD), values_from=CADcount) %>%
mutate(CADprop=(y/(y+no)*100)) %>%
ggplot(., aes(x=id, y=CADprop)) +
geom_col()+
geom_text(aes(x=id, label = sprintf("%.2f",CADprop), vjust=-.2))+
#geom_text(aes(label = expression(paste0("number"~a,"out of",~b))))+
facet_wrap(~sigcount)+
ggtitle("3 hour non-significant and significant enrichment proporitions of CAD GWAS ")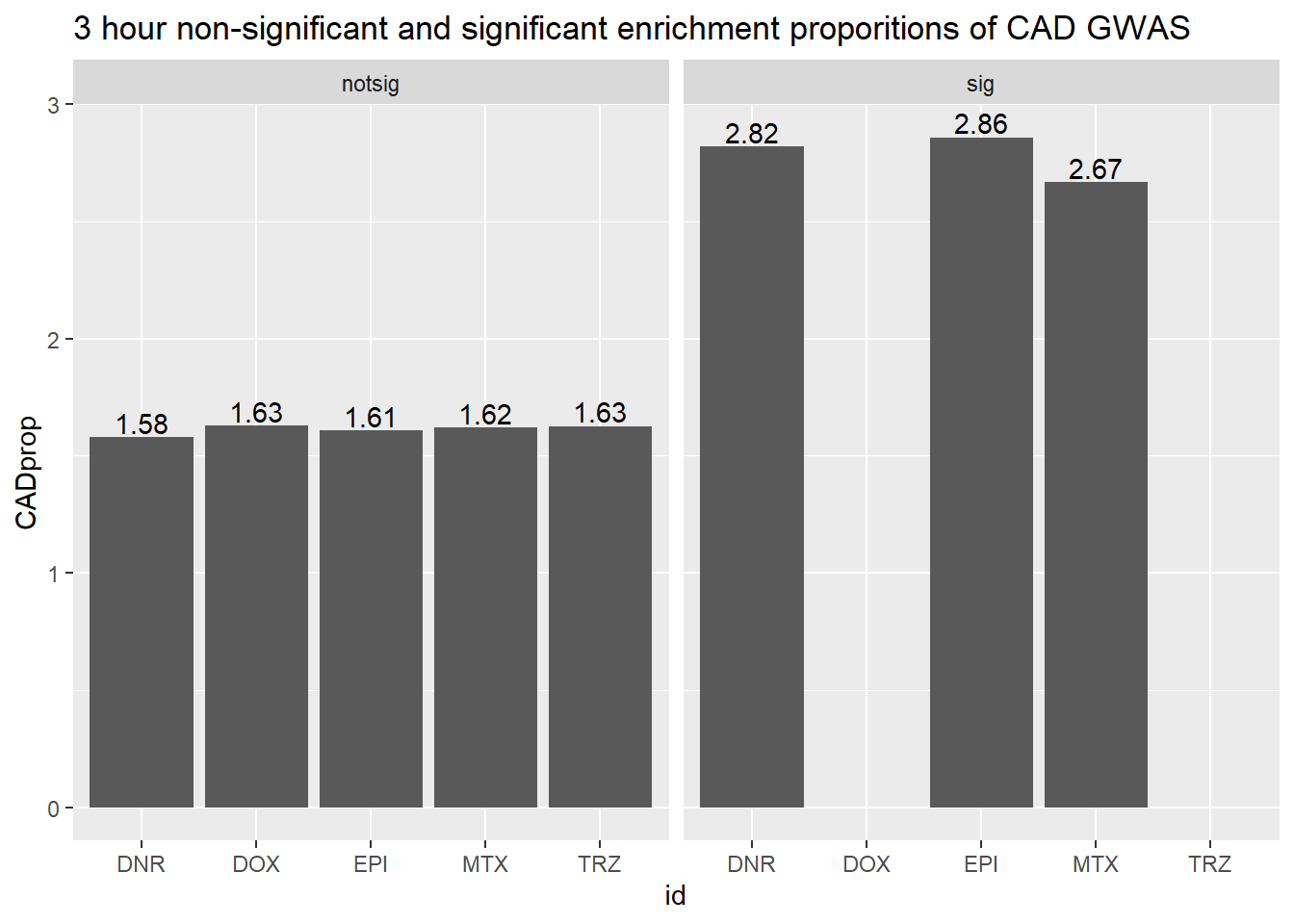
##make table of numbers:
dataframCAD3 <- toplist3hr %>%
mutate(id = as.factor(id)) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(CAD=if_else(ENTREZID %in%CAD_geneset$entrezgene_id,"y","no")) %>%
group_by(id,sigcount,CAD) %>%
dplyr::summarize(CADcount=n()) %>%
as.data.frame()
dataframCAD3 %>% #mutate_at(.vars = 6, .funs= scientific_format()) %>%
kable(., caption= "Significant (adj. P value of <0.05) and non-sig gene counts in 3 hour CAD GWAS") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | sigcount | CAD | CADcount |
|---|---|---|---|
| DNR | notsig | no | 13338 |
| DNR | notsig | y | 214 |
| DNR | sig | no | 517 |
| DNR | sig | y | 15 |
| DOX | notsig | no | 13836 |
| DOX | notsig | y | 229 |
| DOX | sig | no | 19 |
| EPI | notsig | no | 13651 |
| EPI | notsig | y | 223 |
| EPI | sig | no | 204 |
| EPI | sig | y | 6 |
| MTX | notsig | no | 13782 |
| MTX | notsig | y | 227 |
| MTX | sig | no | 73 |
| MTX | sig | y | 2 |
| TRZ | notsig | no | 13855 |
| TRZ | notsig | y | 229 |
chi square test CAD
chi_funCAD <- toplistall %>%
mutate(id = as.factor(id)) %>%
dplyr::filter(id!="TRZ") %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(CAD = if_else(ENTREZID %in%CAD_geneset$entrezgene_id,"y","no")) %>%
group_by(id,time) %>%
dplyr::summarise(pvalue = chisq.test(sigcount, CAD)$p.value)
chi_funCAD %>%
kable(., caption= "after performing chi square test between DEgenes, and non DE genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE,font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| id | time | pvalue |
|---|---|---|
| DNR | 24_hours | 0.6498845 |
| DNR | 3_hours | 0.0409172 |
| DOX | 24_hours | 1.0000000 |
| DOX | 3_hours | 1.0000000 |
| EPI | 24_hours | 0.4524570 |
| EPI | 3_hours | 0.2515982 |
| MTX | 24_hours | 0.6876234 |
| MTX | 3_hours | 0.7973241 |
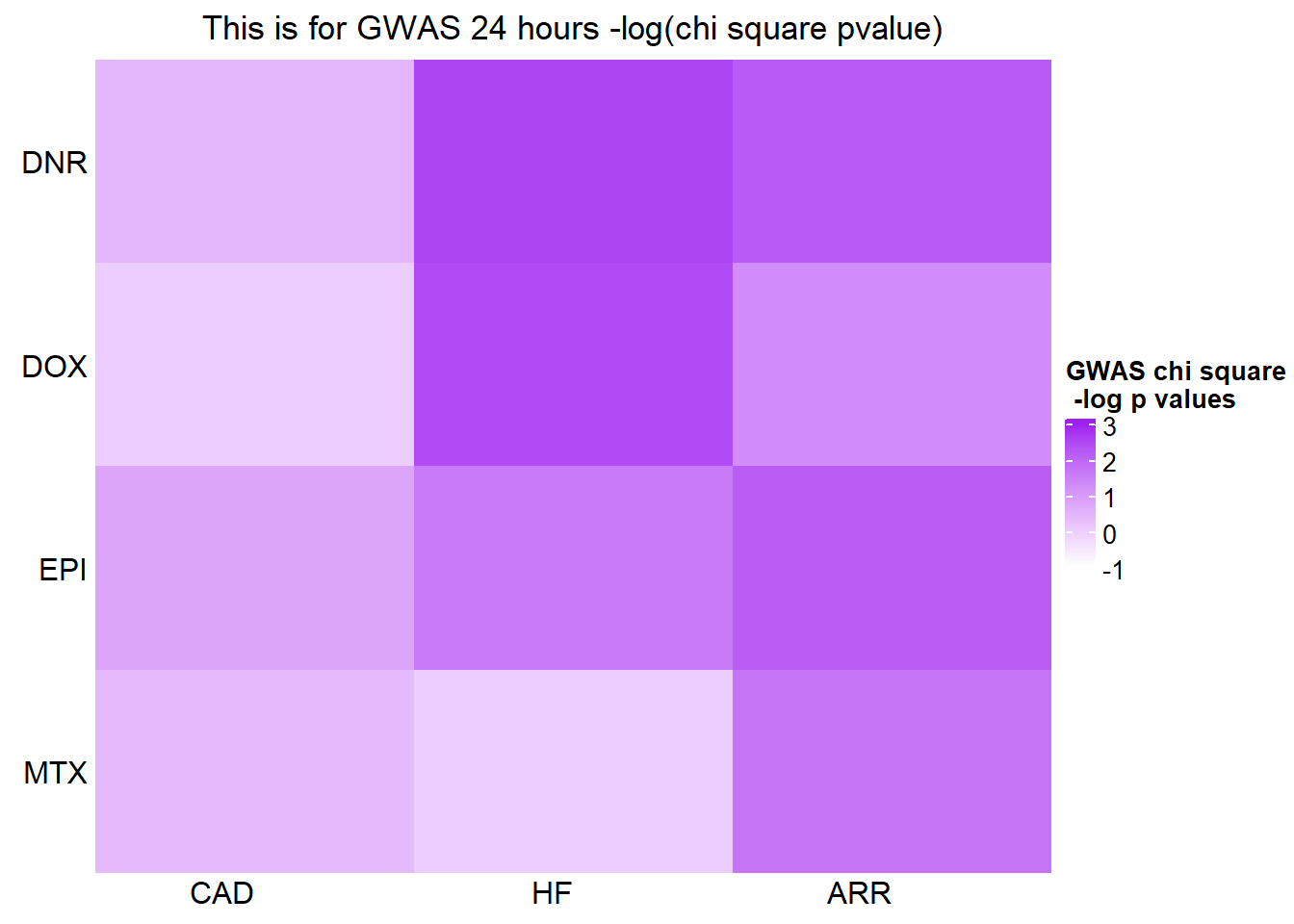
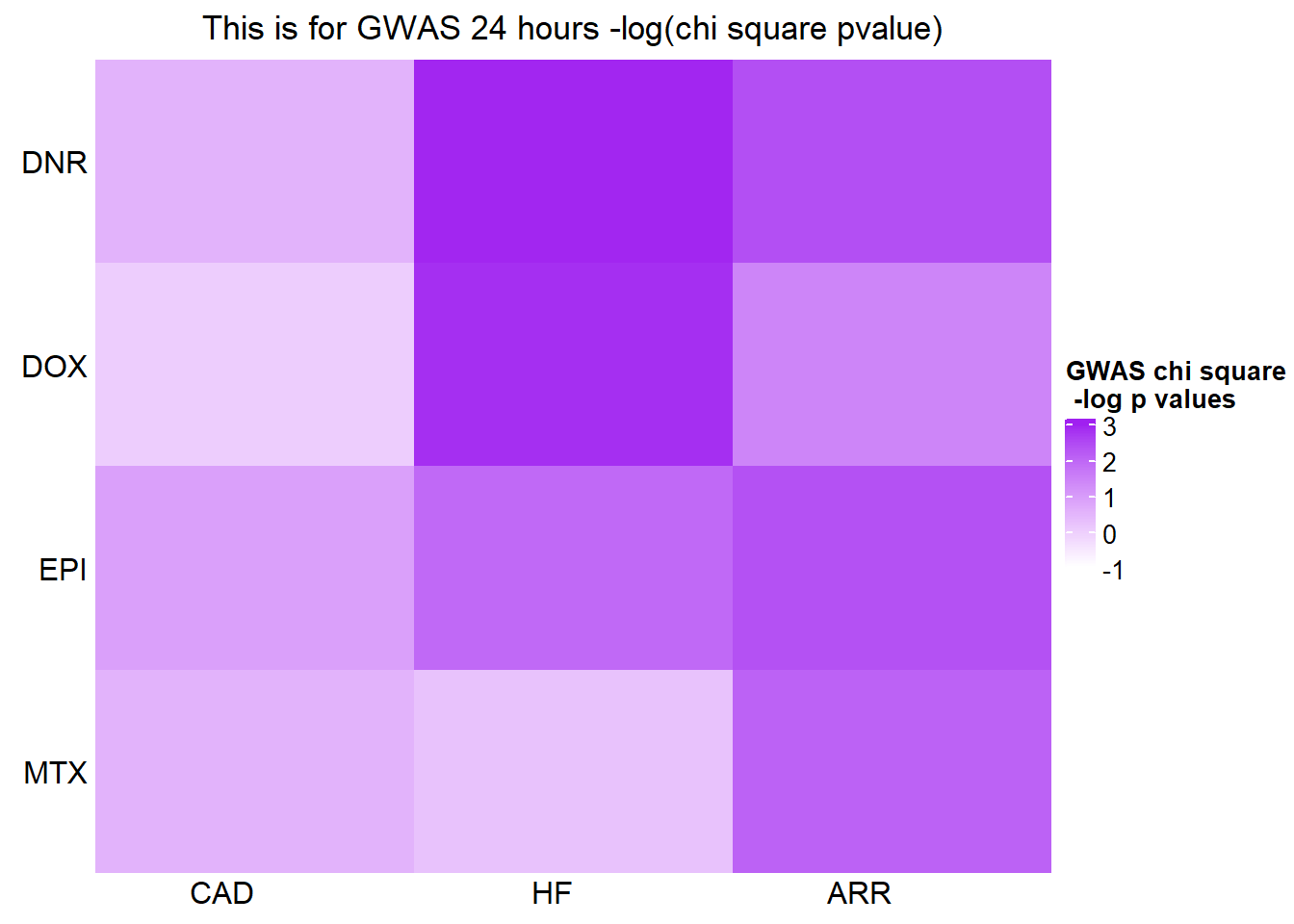
[1] "This is for GWAS 24 hours -log(chi square pvalue)"The star represents chi square p.value < 0.05.
GWAS heatmap
Genes in CVD and AC toxicity respond to Top2i
A. GWAS chi square results
toplist24hr <- toplistall %>%
filter(time=="24_hours")
chi_funarr <- toplist24hr %>%
mutate(id = as.factor(id)) %>%
dplyr::filter(id!="TRZ") %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(ARR=if_else(ENTREZID %in%Arr_geneset$entrezgene_id,"y","no")) %>%
group_by(id) %>%
summarise(pvalue= chisq.test(ARR, sigcount, correct =FALSE)$p.value)
chi_funhf <- toplist24hr %>%
mutate(id = as.factor(id)) %>%
dplyr::filter(id!="TRZ") %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(HF=if_else(ENTREZID %in%HF_geneset$entrezgene_id,"y","no")) %>%
group_by(id) %>%
summarise(pvalue= chisq.test(HF, sigcount, correct =FALSE)$p.value)
chi_funCAD <- toplist24hr %>%
mutate(id = as.factor(id)) %>%
dplyr::filter(id!="TRZ") %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(CAD=if_else(ENTREZID %in%CAD_geneset$entrezgene_id,"y","no")) %>%
group_by(id) %>%
summarise(pvalue= chisq.test(CAD, sigcount, correct =FALSE)$p.value)
HFmat <- chi_funhf %>% ungroup() %>%
mutate(HF= log(pvalue)*(-1)) %>%
dplyr::select(id,HF) %>%
column_to_rownames("id")
ARRmat <- chi_funarr %>% ungroup() %>%
mutate(ARR= log(pvalue)*(-1)) %>%
dplyr::select(id,ARR) %>%
column_to_rownames("id")
CADmat <- chi_funCAD %>% ungroup() %>%
mutate(CAD= -log(pvalue)) %>%
dplyr::select(id,CAD) %>%
column_to_rownames("id")
GWASmat <- bind_cols(CADmat$CAD,HFmat$HF,ARRmat$ARR)
colnames(GWASmat) <- c('CAD','HF','ARR')
rownames(GWASmat) <- c('DNR','DOX','EPI','MTX')
Heatmap(as.matrix(GWASmat), name = "GWAS chi square\n -log p values",
cluster_rows = FALSE,
column_title = 'This is for GWAS 24 hours -log(chi square pvalue)',
cluster_columns = FALSE,
row_names_side = 'left',
column_names_rot = 0, col_fun1,
cell_fun = function(j, i, x, y, width, height, fill) {
if(GWASmat[i, j] > -log(0.05))
grid.text("*", x, y, gp = gpar(fontsize = 20))
})
# GWAS_goi <- c('RARG', 'ITGB7', 'TNS2','ZNF740','SLC28A3','RMI1',
# 'FEDORA' ,'GDF5','FRS2','HDDC2','EEF1B2')
#
# library(biomaRt)
# ensembl <- useMart("ensembl", dataset="hsapiens_gene_ensembl")
# my_chr <- c(1:22, 'M', 'X', 'Y')
# my_attributes <- c('entrezgene_id', 'ensembl_gene_id', 'hgnc_symbol')
#
#
# GWAS_goi<- getBM(attributes=my_attributes,filters ='hgnc_symbol',
# values = GWAS_goi, mart = ensembl)
# GWAS_goi<-GWAS_goi %>% distinct(entrezgene_id,.keep_all = TRUE) %>% add_row(entrezgene_id='124903732',ensembl_gene_id='ENSG00000260788', hgnc_symbol="RP11-298D21.1
# ")
# write.csv(GWAS_goi,"output/GWAS_goi.csv")
GWAS_goi <- read.csv("output/GWAS_goi.csv")
##get the abs FC of all GOI
GWASabsFCsig <-
toplistall %>%
# mutate(absFC=abs(logFC)) %>%
mutate(id = as.factor(id)) %>%
filter(id !="Trastuzumab") %>%
mutate(time=factor(time, levels=c("3_hours","24_hours"))) %>%
filter(ENTREZID %in% GWAS_goi$entrezgene_id) %>%
filter(time =="24_hours") %>%
dplyr::select(ENTREZID ,time, id,logFC, adj.P.Val, SYMBOL) %>%
# mutate(id =case_match(id,
# 'Daunorubicin'~'DNR',
# 'Doxorubicin'~'DOX',
# 'Epirubicin'~'EPI',
# 'Mitoxantrone'~'MTX',
# .default = id)) %>%
pivot_wider(id_cols=id,
names_from = SYMBOL,
values_from =adj.P.Val)
gwas_sig_mat <- GWASabsFCsig %>%
column_to_rownames(var="id") %>%
as.matrix()
GWASabsFC <- toplistall %>%
# mutate(absFC=abs(logFC)) %>%
mutate(id = as.factor(id)) %>%
filter(id !="Trastuzumab") %>%
filter(time=="24_hours") %>%
mutate(logFC= logFC*(-1)) %>%
filter(ENTREZID %in% GWAS_goi$entrezgene_id) %>%
dplyr::select(SYMBOL ,time, id, logFC) %>%
# mutate(id =case_match(id,'Daunorubicin'~'DNR',
# 'Doxorubicin'~'DOX',
# 'Epirubicin'~'EPI',
# 'Mitoxantrone'~'MTX',
# .default = id)) %>%
pivot_wider(id_cols=id,
names_from = SYMBOL,
values_from = logFC) %>%
column_to_rownames(var="id") %>%
as.matrix()
Heatmap(GWASabsFC, name = "Fold change\nvalues",
cluster_rows = FALSE,
cluster_columns = FALSE,
row_names_side = "left",
column_title = "Fold change values of GWAS and TWAS genes",
column_title_side = "top",
column_title_gp = gpar(fontsize = 16, fontface = "bold"),
column_order= c('RARG',
'TNS2',
'ZNF740',
'SLC28A3',
'RMI1',
'EEF1B2',
'FRS2',
'HDDC2'),
column_names_rot = 0,
column_names_gp = gpar(fontsize = 12),
column_names_centered = TRUE,
cell_fun = function(j, i, x, y, width, height, fill) {
if(gwas_sig_mat[i, j] <0.05)
grid.text("*", x, y, gp = gpar(fontsize = 20))
})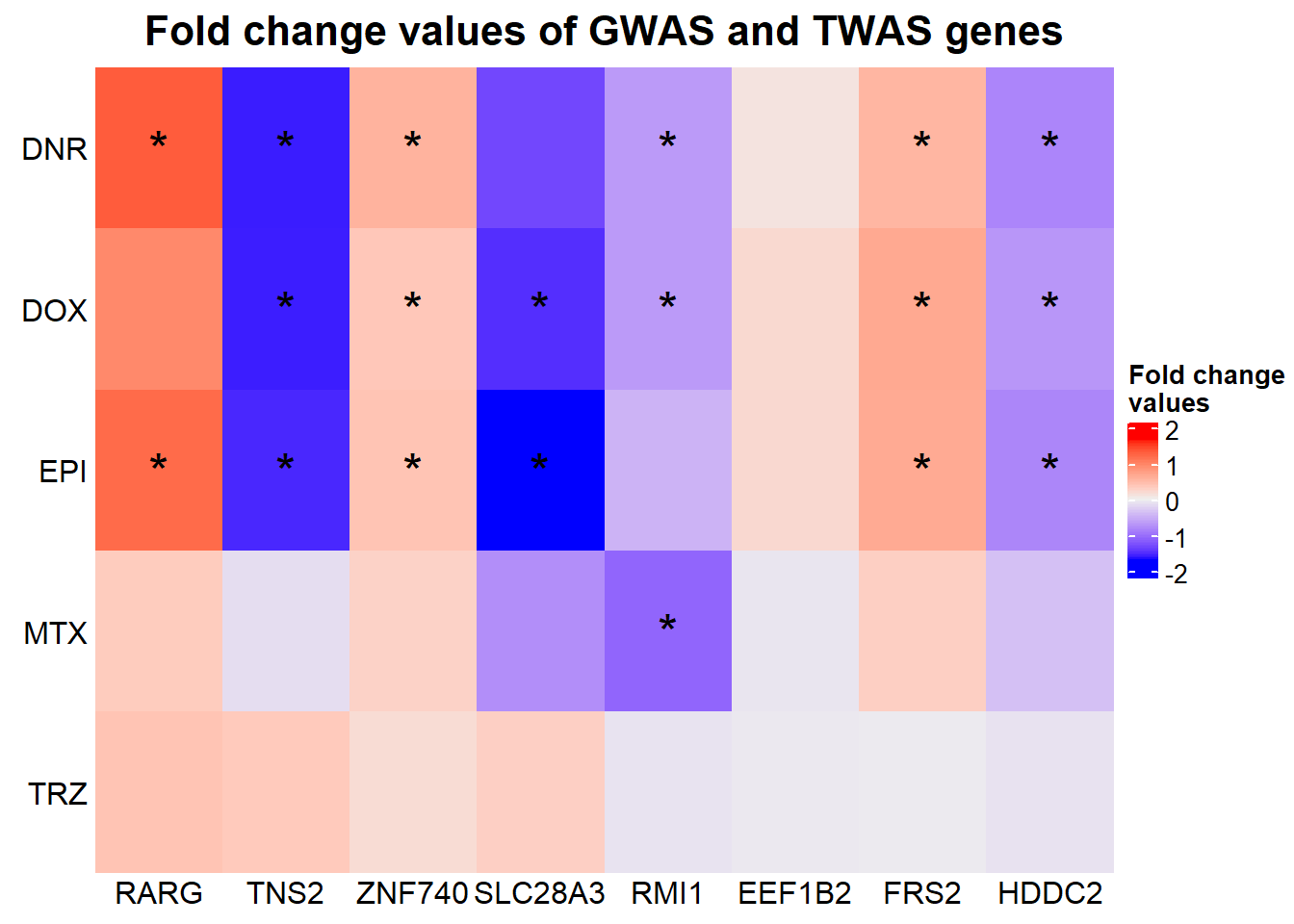
The stars represent all genes that have an adj. P. value of < 0.05 (significantly differentially expressed)
Crispr list
DEG_cormotif <- readRDS("data/DEG_cormotif.RDS")
list2env(DEG_cormotif,envir=.GlobalEnv)<environment: R_GlobalEnv># Crispr_list <- read_excel("C:/Users/renee/Downloads/41598_2021_92988_MOESM2_ESM.xlsx")
# View(Crispr_list)
# crispr_genes <- Crispr_list %>%
# dplyr::filter(p.value <0.05) %>%
# select(GeneName)
# crispr_genes <- getBM(attributes=my_attributes,filters ='hgnc_symbol',
# values =crispr_genes$GeneName, mart = ensembl)
# write.csv(crispr_genes,'data/crispr_genes.csv')
crispr_genes <- read.csv("data/crispr_genes.csv", row.names = 1)
print(" number of unique crispr_genes after conversion from hgnc symbol to entrezid")[1] " number of unique crispr_genes after conversion from hgnc symbol to entrezid"length(unique(crispr_genes$entrezgene_id))[1] 154crisprunique <- crispr_genes %>% distinct(entrezgene_id,.keep_all = TRUE)
Doxcrispall <- toplistall %>%
distinct(ENTREZID,.keep_all = TRUE) %>%
dplyr::select(ENTREZID,id,time)
crispmotifsummary <- Doxcrispall %>%
mutate(ER=if_else(ENTREZID %in% motif_ER,"y","no")) %>%
mutate(LR=if_else(ENTREZID %in% motif_LR,"y","no")) %>%
mutate(TI=if_else(ENTREZID %in% motif_TI,"y","no")) %>%
mutate(NR=if_else(ENTREZID %in% motif_NR,"y","no")) %>%
mutate(crisp = if_else(ENTREZID %in% crisprunique$entrezgene_id, "y", "no")) %>%
group_by(crisp,ER,TI,LR,NR) %>%
dplyr::summarize(n=n()) %>%
as.tibble %>%
pivot_wider(id_cols = c(crisp), names_from = c('ER', 'TI', 'LR', 'NR'), values_from= n) %>%
rename(.,c("crisp"=crisp,"none"= 2 , "ER" = 3 , "TI" = 4 , "LR" = 5 ,"NR" = 6))
cris_mat <- crispmotifsummary %>% dplyr::select(ER:NR) %>% as.matrix()
chicheck <- data.frame(one= c("LR","ER","TI"),two=rep("NR",3),p.value=c("","",""))
chicheck$p.value[1] <- chisq.test(cris_mat[,c('LR','NR')],correct = FALSE)$p.value
chicheck$p.value[2] <- chisq.test(cris_mat[,c('ER','NR')],correct = FALSE)$p.value
chicheck$p.value[3] <- chisq.test(cris_mat[,c('TI','NR')],correct = FALSE)$p.value
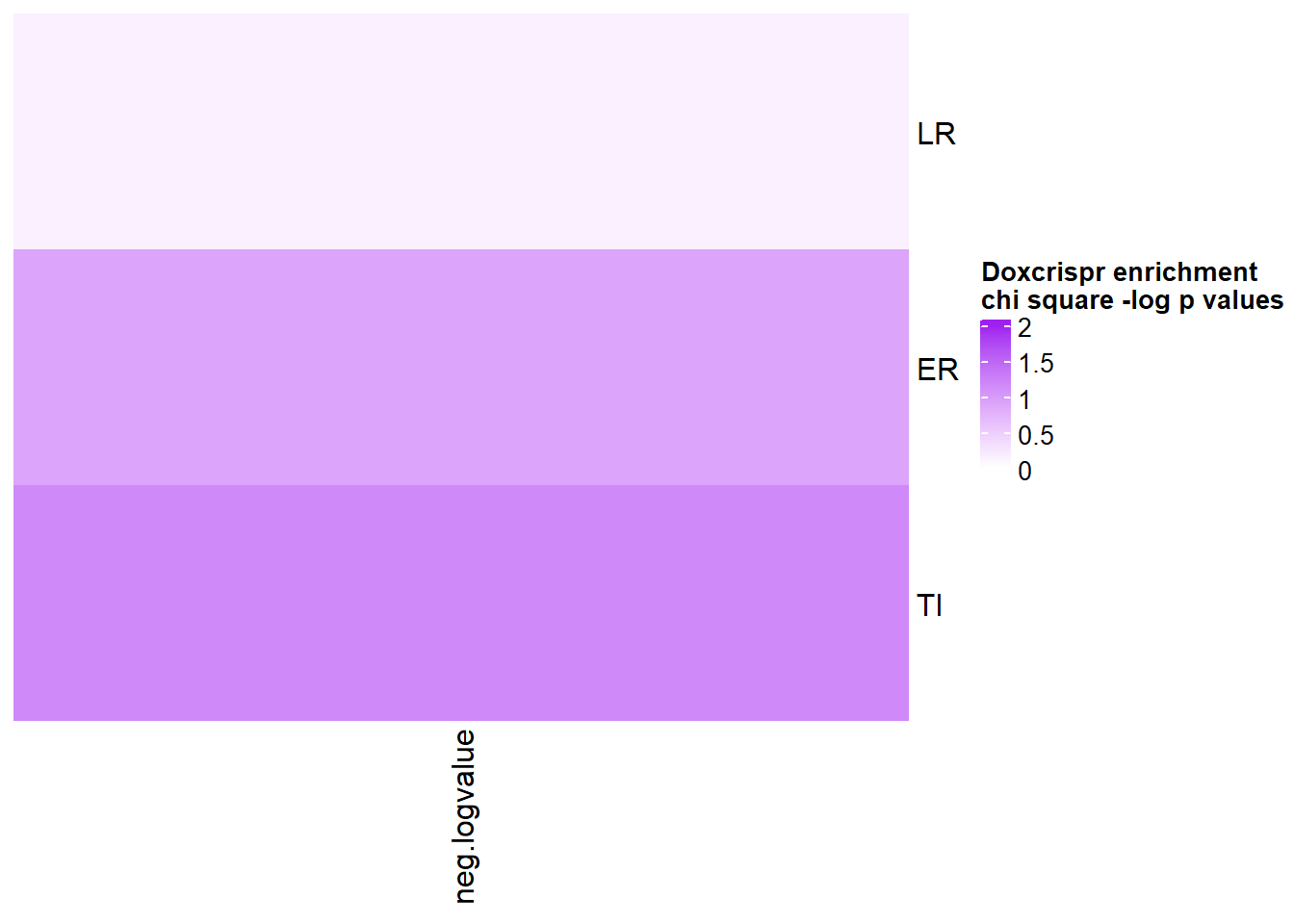
chicheck%>% kable(., caption= "chi square test p.values for encrichment of Doxcrispr gene sets in motif sets" )%>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| one | two | p.value |
|---|---|---|
| LR | NR | 0.861985991471947 |
| ER | NR | 0.402681880154749 |
| TI | NR | 0.309642007916355 |
chicheck_1 <- chicheck %>% mutate(p.value=as.numeric(p.value)) %>%
mutate(neg.logvalue=(-1*log(p.value))) %>% column_to_rownames('one') %>% dplyr::select(neg.logvalue) %>% as.matrix
col_fun = circlize::colorRamp2(c(0, 2), c("white", "purple"))
Heatmap( chicheck_1, name = "Doxcrispr enrichment \nchi square -log p values", cluster_rows = FALSE, cluster_columns = FALSE, col=col_fun,
cell_fun = function(j, i, x, y, width, height, fill) {
if(chicheck_1[i, j] > -log(0.05))
grid.text("*", x, y, gp = gpar(fontsize = 20))
})
| Version | Author | Date |
|---|---|---|
| 47f85a2 | reneeisnowhere | 2023-06-07 |
col_fun4 = circlize::colorRamp2(c(0, 5), c("white", "purple"))
pairwisecrispr <- toplistall %>%
filter(id!='TRZ') %>%
mutate(id = as.factor(id)) %>%
mutate(time=factor(time, levels=c("3_hours","24_hours"))) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(crisp = if_else(ENTREZID %in% crisprunique$entrezgene_id, "y", "no")) %>%
group_by(time, id) %>%
dplyr::summarise(pvalue= chisq.test(crisp, sigcount, correct=FALSE)$p.value)
crisprnumbers <- toplistall %>%
filter(id!='Trastuzumab') %>%
mutate(id = as.factor(id)) %>%
mutate(time=factor(time, levels=c("3_hours","24_hours"))) %>%
mutate(sigcount = if_else(adj.P.Val <0.05,'sig','notsig'))%>%
mutate(crisp = if_else(ENTREZID %in% crisprunique$entrezgene_id, "y", "no")) %>%
group_by(time, id,sigcount,crisp) %>%
dplyr::summarize(n=n()) %>%
as.tibble() #%>%
crisprnumbers %>% kable(., caption= "Summary of genes found in both sigDE and non sigDE by treatment" )%>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| time | id | sigcount | crisp | n |
|---|---|---|---|---|
| 3_hours | DNR | notsig | no | 13437 |
| 3_hours | DNR | notsig | y | 115 |
| 3_hours | DNR | sig | no | 530 |
| 3_hours | DNR | sig | y | 2 |
| 3_hours | DOX | notsig | no | 13948 |
| 3_hours | DOX | notsig | y | 117 |
| 3_hours | DOX | sig | no | 19 |
| 3_hours | EPI | notsig | no | 13758 |
| 3_hours | EPI | notsig | y | 116 |
| 3_hours | EPI | sig | no | 209 |
| 3_hours | EPI | sig | y | 1 |
| 3_hours | MTX | notsig | no | 13892 |
| 3_hours | MTX | notsig | y | 117 |
| 3_hours | MTX | sig | no | 75 |
| 3_hours | TRZ | notsig | no | 13967 |
| 3_hours | TRZ | notsig | y | 117 |
| 24_hours | DNR | notsig | no | 7016 |
| 24_hours | DNR | notsig | y | 51 |
| 24_hours | DNR | sig | no | 6951 |
| 24_hours | DNR | sig | y | 66 |
| 24_hours | DOX | notsig | no | 7385 |
| 24_hours | DOX | notsig | y | 54 |
| 24_hours | DOX | sig | no | 6582 |
| 24_hours | DOX | sig | y | 63 |
| 24_hours | EPI | notsig | no | 7700 |
| 24_hours | EPI | notsig | y | 56 |
| 24_hours | EPI | sig | no | 6267 |
| 24_hours | EPI | sig | y | 61 |
| 24_hours | MTX | notsig | no | 12861 |
| 24_hours | MTX | notsig | y | 108 |
| 24_hours | MTX | sig | no | 1106 |
| 24_hours | MTX | sig | y | 9 |
| 24_hours | TRZ | notsig | no | 13967 |
| 24_hours | TRZ | notsig | y | 117 |
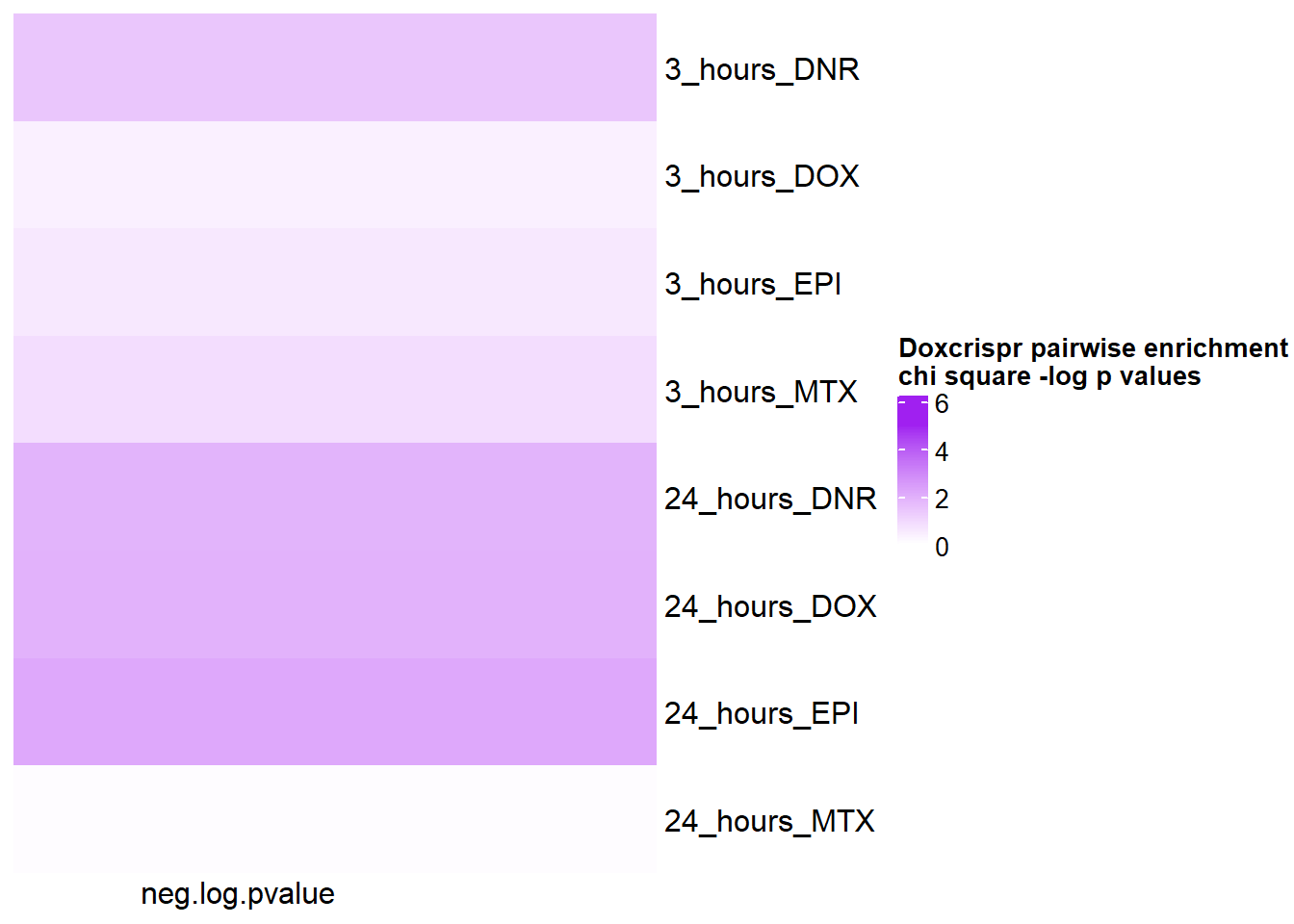
pairwisecrispr%>% kable(., caption= "Summary of chisqure values between numbers of sigDE and non sigDE by treatment" )%>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(full_width = FALSE, position = "left",bootstrap_options = c("striped"),font_size = 18) %>%
scroll_box(width = "60%", height = "400px")| time | id | pvalue |
|---|---|---|
| 3_hours | DNR | 0.2387271 |
| 3_hours | DOX | 0.6897318 |
| 3_hours | EPI | 0.5684613 |
| 3_hours | MTX | 0.4267580 |
| 24_hours | DNR | 0.1523968 |
| 24_hours | DOX | 0.1470074 |
| 24_hours | EPI | 0.1155814 |
| 24_hours | MTX | 0.9280448 |
crisp_pair_mat <- pairwisecrispr %>%
mutate(neg.log.pvalue= (-1*log(pvalue))) %>%
# mutate(time= case_match(time, '3_hours'~'3_hrs', '24_hours'~'24_hrs',.default = id)) %>%
# mutate(id =case_match( id, 'Daunorubicin'~'DNR', 'Doxorubicin'~'DOX' ,'Epirubicin'~'EPI' , 'Mitoxantrone' ~ 'MTX',.default = id)) %>%
unite('pairset',time,id ) %>%
column_to_rownames('pairset') %>% dplyr::select(neg.log.pvalue) %>% as.matrix()
Heatmap( crisp_pair_mat, name = "Doxcrispr pairwise enrichment \nchi square -log p values",
cluster_rows = FALSE,
cluster_columns = FALSE,
col=col_fun5, column_names_rot = 0,
cell_fun = function(j, i, x, y, width, height, fill) {
if(crisp_pair_mat[i, j] > -log(0.05))
grid.text("*", x, y, gp = gpar(fontsize = 20))
})
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ComplexHeatmap_2.16.0 broom_1.0.5 kableExtra_1.3.4
[4] sjmisc_2.8.9 scales_1.3.0 ggpubr_0.6.0
[7] cowplot_1.1.1 RColorBrewer_1.1-3 biomaRt_2.56.1
[10] ggsignif_0.6.4 lubridate_1.9.3 forcats_1.0.0
[13] stringr_1.5.0 dplyr_1.1.3 purrr_1.0.2
[16] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
[19] ggplot2_3.4.4 tidyverse_2.0.0 limma_3.56.2
[22] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] rstudioapi_0.15.0 jsonlite_1.8.7 shape_1.4.6
[4] magrittr_2.0.3 magick_2.8.1 farver_2.1.1
[7] rmarkdown_2.25 GlobalOptions_0.1.2 fs_1.6.3
[10] zlibbioc_1.46.0 vctrs_0.6.4 memoise_2.0.1
[13] RCurl_1.98-1.14 rstatix_0.7.2 webshot_0.5.5
[16] htmltools_0.5.7 progress_1.2.3 curl_5.2.0
[19] sass_0.4.7 bslib_0.6.1 cachem_1.0.8
[22] whisker_0.4.1 lifecycle_1.0.4 iterators_1.0.14
[25] pkgconfig_2.0.3 sjlabelled_1.2.0 R6_2.5.1
[28] fastmap_1.1.1 GenomeInfoDbData_1.2.10 clue_0.3-65
[31] digest_0.6.33 colorspace_2.1-0 AnnotationDbi_1.62.2
[34] S4Vectors_0.38.2 ps_1.7.5 rprojroot_2.0.4
[37] RSQLite_2.3.5 labeling_0.4.3 filelock_1.0.3
[40] fansi_1.0.5 timechange_0.2.0 httr_1.4.7
[43] abind_1.4-5 compiler_4.3.1 bit64_4.0.5
[46] withr_3.0.0 doParallel_1.0.17 backports_1.4.1
[49] carData_3.0-5 DBI_1.2.1 highr_0.10
[52] rappdirs_0.3.3 rjson_0.2.21 tools_4.3.1
[55] httpuv_1.6.12 glue_1.6.2 callr_3.7.3
[58] promises_1.2.1 getPass_0.2-2 cluster_2.1.4
[61] generics_0.1.3 gtable_0.3.4 tzdb_0.4.0
[64] hms_1.1.3 xml2_1.3.5 car_3.1-2
[67] utf8_1.2.4 XVector_0.40.0 BiocGenerics_0.46.0
[70] foreach_1.5.2 pillar_1.9.0 later_1.3.1
[73] circlize_0.4.15 BiocFileCache_2.8.0 bit_4.0.5
[76] tidyselect_1.2.0 Biostrings_2.68.1 knitr_1.45
[79] git2r_0.32.0 IRanges_2.34.1 svglite_2.1.2
[82] stats4_4.3.1 xfun_0.41 Biobase_2.60.0
[85] matrixStats_1.1.0 stringi_1.7.12 yaml_2.3.7
[88] evaluate_0.23 codetools_0.2-19 cli_3.6.1
[91] systemfonts_1.0.5 munsell_0.5.0 processx_3.8.2
[94] jquerylib_0.1.4 Rcpp_1.0.11 GenomeInfoDb_1.36.4
[97] dbplyr_2.4.0 png_0.1-8 XML_3.99-0.16.1
[100] parallel_4.3.1 blob_1.2.4 prettyunits_1.2.0
[103] bitops_1.0-7 viridisLite_0.4.2 insight_0.19.8
[106] crayon_1.5.2 GetoptLong_1.0.5 rlang_1.1.2
[109] KEGGREST_1.40.1 rvest_1.0.3