Cormotif_analysis
Renee Matthews
2025-05-06
Last updated: 2025-05-15
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d49fd7c. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/TFmapnames.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/datasave.RDS
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summarized_peaks_dataframe.txt
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Untracked files:
Untracked: RNA_seq_integration.Rmd
Untracked: analysis/Diagnosis-tmm.Rmd
Untracked: analysis/Expressed_RNA_associations.Rmd
Untracked: analysis/H3K27ac_integration_noM.Rmd
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/SVA.Rmd
Untracked: analysis/Tan2020.Rmd
Untracked: analysis/making_master_peaks_list.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/just_for_Fun.R
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: setup.RData
Unstaged changes:
Modified: ATAC_learning.Rproj
Modified: analysis/AF_HF_SNPs.Rmd
Modified: analysis/Cardiotox_SNPs.Rmd
Modified: analysis/Enhancer_enrichment.Rmd
Modified: analysis/H3K27ac_cormotif.Rmd
Modified: analysis/Jaspar_motif.Rmd
Modified: analysis/Jaspar_motif_ff.Rmd
Modified: analysis/RNA_seq_integration.Rmd
Modified: analysis/TSS_and_CUG.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Cormotif_analysis.Rmd) and
HTML (docs/Cormotif_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d49fd7c | reneeisnowhere | 2025-05-15 | rastr updates and spelling |
| html | 5e6e462 | reneeisnowhere | 2025-05-07 | Build site. |
| Rmd | d969893 | reneeisnowhere | 2025-05-07 | updating new pages |
| Rmd | 2db35c7 | reneeisnowhere | 2025-05-07 | updates to analysis |
| html | 8af2b68 | reneeisnowhere | 2025-05-06 | Build site. |
| Rmd | 02654a0 | reneeisnowhere | 2025-05-06 | updates to analysis |
| html | ac1a2a6 | reneeisnowhere | 2025-05-06 | Build site. |
| Rmd | 43a493f | reneeisnowhere | 2025-05-06 | updates to analysis |
library(tidyverse)
library(kableExtra)
library(broom)
library(RColorBrewer)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
library(Cormotif)
library(BiocParallel)
library(ggpubr)
library(devtools)
library(eulerr)
library(genomation)
library(ggsignif)
library(plyranges)
library(ggrepel)
library(ComplexHeatmap)
library(cowplot)
library(smplot2)## Fit limma model using code as it is found in the original cormotif code. It has
## only been modified to add names to the matrix of t values, as well as the
## limma fits
limmafit.default <- function(exprs,groupid,compid) {
limmafits <- list()
compnum <- nrow(compid)
genenum <- nrow(exprs)
limmat <- matrix(0,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- rownames(exprs)
colnames(limmat) <- rownames(compid)
names(limmas2) <- rownames(compid)
names(limmadf) <- rownames(compid)
names(limmav0) <- rownames(compid)
names(limmag1num) <- rownames(compid)
names(limmag2num) <- rownames(compid)
for(i in 1:compnum) {
selid1 <- which(groupid == compid[i,1])
selid2 <- which(groupid == compid[i,2])
eset <- new("ExpressionSet", exprs=cbind(exprs[,selid1],exprs[,selid2]))
g1num <- length(selid1)
g2num <- length(selid2)
designmat <- cbind(base=rep(1,(g1num+g2num)), delta=c(rep(0,g1num),rep(1,g2num)))
fit <- lmFit(eset,designmat)
fit <- eBayes(fit)
limmat[,i] <- fit$t[,2]
limmas2[i] <- fit$s2.prior
limmadf[i] <- fit$df.prior
limmav0[i] <- fit$var.prior[2]
limmag1num[i] <- g1num
limmag2num[i] <- g2num
limmafits[[i]] <- fit
# log odds
# w<-sqrt(1+fit$var.prior[2]/(1/g1num+1/g2num))
# log(0.99)+dt(fit$t[1,2],g1num+g2num-2+fit$df.prior,log=TRUE)-log(0.01)-dt(fit$t[1,2]/w, g1num+g2num-2+fit$df.prior, log=TRUE)+log(w)
}
names(limmafits) <- rownames(compid)
limmacompnum<-nrow(compid)
result<-list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
limmafit.counts <-
function (exprs, groupid, compid, norm.factor.method = "TMM", voom.normalize.method = "none")
{
limmafits <- list()
compnum <- nrow(compid)
genenum <- nrow(exprs)
limmat <- matrix(NA,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- rownames(exprs)
colnames(limmat) <- rownames(compid)
names(limmas2) <- rownames(compid)
names(limmadf) <- rownames(compid)
names(limmav0) <- rownames(compid)
names(limmag1num) <- rownames(compid)
names(limmag2num) <- rownames(compid)
for (i in 1:compnum) {
message(paste("Running limma for comparision",i,"/",compnum))
selid1 <- which(groupid == compid[i, 1])
selid2 <- which(groupid == compid[i, 2])
# make a new count data frame
counts <- cbind(exprs[, selid1], exprs[, selid2])
# remove NAs
not.nas <- which(apply(counts, 1, function(x) !any(is.na(x))) == TRUE)
# runn voom/limma
d <- DGEList(counts[not.nas,])
d <- calcNormFactors(d, method = norm.factor.method)
g1num <- length(selid1)
g2num <- length(selid2)
designmat <- cbind(base = rep(1, (g1num + g2num)), delta = c(rep(0,
g1num), rep(1, g2num)))
y <- voom(d, designmat, normalize.method = voom.normalize.method)
fit <- lmFit(y, designmat)
fit <- eBayes(fit)
limmafits[[i]] <- fit
limmat[not.nas, i] <- fit$t[, 2]
limmas2[i] <- fit$s2.prior
limmadf[i] <- fit$df.prior
limmav0[i] <- fit$var.prior[2]
limmag1num[i] <- g1num
limmag2num[i] <- g2num
}
limmacompnum <- nrow(compid)
names(limmafits) <- rownames(compid)
result <- list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
limmafit.list <-
function (fitlist, cmp.idx=2)
{
compnum <- length(fitlist)
genes <- c()
for (i in 1:compnum) genes <- unique(c(genes, rownames(fitlist[[i]])))
genenum <- length(genes)
limmat <- matrix(NA,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- genes
colnames(limmat) <- names(fitlist)
names(limmas2) <- names(fitlist)
names(limmadf) <- names(fitlist)
names(limmav0) <- names(fitlist)
names(limmag1num) <- names(fitlist)
names(limmag2num) <- names(fitlist)
for (i in 1:compnum) {
this.t <- fitlist[[i]]$t[,cmp.idx]
limmat[names(this.t),i] <- this.t
limmas2[i] <- fitlist[[i]]$s2.prior
limmadf[i] <- fitlist[[i]]$df.prior
limmav0[i] <- fitlist[[i]]$var.prior[cmp.idx]
limmag1num[i] <- sum(fitlist[[i]]$design[,cmp.idx]==0)
limmag2num[i] <- sum(fitlist[[i]]$design[,cmp.idx]==1)
}
limmacompnum <- compnum
result <- list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
## Rank genes based on statistics
generank<-function(x) {
xcol<-ncol(x)
xrow<-nrow(x)
result<-matrix(0,xrow,xcol)
z<-(1:1:xrow)
for(i in 1:xcol) {
y<-sort(x[,i],decreasing=TRUE,na.last=TRUE)
result[,i]<-match(x[,i],y)
result[,i]<-order(result[,i])
}
result
}
## Log-likelihood for moderated t under H0
modt.f0.loglike<-function(x,df) {
a<-dt(x, df, log=TRUE)
result<-as.vector(a)
flag<-which(is.na(result)==TRUE)
result[flag]<-0
result
}
## Log-likelihood for moderated t under H1
## param=c(df,g1num,g2num,v0)
modt.f1.loglike<-function(x,param) {
df<-param[1]
g1num<-param[2]
g2num<-param[3]
v0<-param[4]
w<-sqrt(1+v0/(1/g1num+1/g2num))
dt(x/w, df, log=TRUE)-log(w)
a<-dt(x/w, df, log=TRUE)-log(w)
result<-as.vector(a)
flag<-which(is.na(result)==TRUE)
result[flag]<-0
result
}
## Correlation Motif Fit
cmfit.X<-function(x, type, K=1, tol=1e-3, max.iter=100) {
## initialize
xrow <- nrow(x)
xcol <- ncol(x)
loglike0 <- list()
loglike1 <- list()
p <- rep(1, K)/K
q <- matrix(runif(K * xcol), K, xcol)
q[1, ] <- rep(0.01, xcol)
for (i in 1:xcol) {
f0 <- type[[i]][[1]]
f0param <- type[[i]][[2]]
f1 <- type[[i]][[3]]
f1param <- type[[i]][[4]]
loglike0[[i]] <- f0(x[, i], f0param)
loglike1[[i]] <- f1(x[, i], f1param)
}
condlike <- list()
for (i in 1:xcol) {
condlike[[i]] <- matrix(0, xrow, K)
}
loglike.old <- -1e+10
for (i.iter in 1:max.iter) {
if ((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations for K=",
K, sep = ""))
}
err <- tol + 1
clustlike <- matrix(0, xrow, K)
#templike <- matrix(0, xrow, 2)
templike1 <- rep(0, xrow)
templike2 <- rep(0, xrow)
for (j in 1:K) {
for (i in 1:xcol) {
templike1 <- log(q[j, i]) + loglike1[[i]]
templike2 <- log(1 - q[j, i]) + loglike0[[i]]
tempmax <- Rfast::Pmax(templike1, templike2)
templike1 <- exp(templike1 - tempmax)
templike2 <- exp(templike2 - tempmax)
tempsum <- templike1 + templike2
clustlike[, j] <- clustlike[, j] + tempmax +
log(tempsum)
condlike[[i]][, j] <- templike1/tempsum
}
clustlike[, j] <- clustlike[, j] + log(p[j])
}
#tempmax <- apply(clustlike, 1, max)
tempmax <- Rfast::rowMaxs(clustlike, value=TRUE)
for (j in 1:K) {
clustlike[, j] <- exp(clustlike[, j] - tempmax)
}
#tempsum <- apply(clustlike, 1, sum)
tempsum <- Rfast::rowsums(clustlike)
for (j in 1:K) {
clustlike[, j] <- clustlike[, j]/tempsum
}
#p.new <- (apply(clustlike, 2, sum) + 1)/(xrow + K)
p.new <- (Rfast::colsums(clustlike) + 1)/(xrow + K)
q.new <- matrix(0, K, xcol)
for (j in 1:K) {
clustpsum <- sum(clustlike[, j])
for (i in 1:xcol) {
q.new[j, i] <- (sum(clustlike[, j] * condlike[[i]][,
j]) + 1)/(clustpsum + 2)
}
}
err.p <- max(abs(p.new - p)/p)
err.q <- max(abs(q.new - q)/q)
err <- max(err.p, err.q)
loglike.new <- (sum(tempmax + log(tempsum)) + sum(log(p.new)) +
sum(log(q.new) + log(1 - q.new)))/xrow
p <- p.new
q <- q.new
loglike.old <- loglike.new
if (err < tol) {
break
}
}
clustlike <- matrix(0, xrow, K)
for (j in 1:K) {
for (i in 1:xcol) {
templike1 <- log(q[j, i]) + loglike1[[i]]
templike2 <- log(1 - q[j, i]) + loglike0[[i]]
tempmax <- Rfast::Pmax(templike1, templike2)
templike1 <- exp(templike1 - tempmax)
templike2 <- exp(templike2 - tempmax)
tempsum <- templike1 + templike2
clustlike[, j] <- clustlike[, j] + tempmax + log(tempsum)
condlike[[i]][, j] <- templike1/tempsum
}
clustlike[, j] <- clustlike[, j] + log(p[j])
}
#tempmax <- apply(clustlike, 1, max)
tempmax <- Rfast::rowMaxs(clustlike, value=TRUE)
for (j in 1:K) {
clustlike[, j] <- exp(clustlike[, j] - tempmax)
}
#tempsum <- apply(clustlike, 1, sum)
tempsum <- Rfast::rowsums(clustlike)
for (j in 1:K) {
clustlike[, j] <- clustlike[, j]/tempsum
}
p.post <- matrix(0, xrow, xcol)
for (j in 1:K) {
for (i in 1:xcol) {
p.post[, i] <- p.post[, i] + clustlike[, j] * condlike[[i]][,
j]
}
}
loglike.old <- loglike.old - (sum(log(p)) + sum(log(q) +
log(1 - q)))/xrow
loglike.old <- loglike.old * xrow
result <- list(p.post = p.post, motif.prior = p, motif.q = q,
loglike = loglike.old, clustlike=clustlike, condlike=condlike)
}
## Fit using (0,0,...,0) and (1,1,...,1)
cmfitall<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
p<-0.01
## compute loglikelihood
L0<-matrix(0,xrow,1)
L1<-matrix(0,xrow,1)
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
L0<-L0+loglike0[[i]]
L1<-L1+loglike1[[i]]
}
## EM algorithm to get MLE of p and q
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p)+L0
clustlike[,2]<-log(p)+L1
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## update motif occurrence rate
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(sum(clustlike[,2])+1)/(xrow+2)
## evaluate convergence
err<-abs(p.new-p)/p
## evaluate whether the log.likelihood increases
loglike.new<-(sum(tempmax+log(tempsum))+log(p.new)+log(1-p.new))/xrow
loglike.old<-loglike.new
p<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p)+L0
clustlike[,2]<-log(p)+L1
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post<-matrix(0,xrow,xcol)
for(i in 1:xcol) {
p.post[,i]<-clustlike[,2]
}
## return
#calculate back loglikelihood
loglike.old<-loglike.old-(log(p)+log(1-p))/xrow
loglike.old<-loglike.old*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.old)
}
## Fit each dataset separately
cmfitsep<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
p<-0.01*rep(1,xcol)
loglike.final<-rep(0,xcol)
## compute loglikelihood
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
}
p.post<-matrix(0,xrow,xcol)
## EM algorithm to get MLE of p
for(coli in 1:xcol) {
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p[coli])+loglike0[[coli]]
clustlike[,2]<-log(p[coli])+loglike1[[coli]]
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## evaluate whether the log.likelihood increases
loglike.new<-sum(tempmax+log(tempsum))/xrow
## update motif occurrence rate
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(sum(clustlike[,2]))/(xrow)
## evaluate convergence
err<-abs(p.new-p[coli])/p[coli]
loglike.old<-loglike.new
p[coli]<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p[coli])+loglike0[[coli]]
clustlike[,2]<-log(p[coli])+loglike1[[coli]]
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post[,coli]<-clustlike[,2]
loglike.final[coli]<-loglike.old
}
## return
loglike.final<-loglike.final*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.final)
}
## Fit the full model
cmfitfull<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
K<-2^xcol
p<-rep(1,K)/K
pattern<-rep(0,xcol)
patid<-matrix(0,K,xcol)
## compute loglikelihood
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
}
L<-matrix(0,xrow,K)
for(i in 1:K)
{
patid[i,]<-pattern
for(j in 1:xcol) {
if(pattern[j] < 0.5) {
L[,i]<-L[,i]+loglike0[[j]]
} else {
L[,i]<-L[,i]+loglike1[[j]]
}
}
if(i < K) {
pattern[xcol]<-pattern[xcol]+1
j<-xcol
while(pattern[j] > 1) {
pattern[j]<-0
j<-j-1
pattern[j]<-pattern[j]+1
}
}
}
## EM algorithm to get MLE of p and q
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,K)
for(j in 1:K) {
clustlike[,j]<-log(p[j])+L[,j]
}
tempmax<-apply(clustlike,1,max)
for(j in 1:K) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## update motif occurrence rate
for(j in 1:K) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(apply(clustlike,2,sum)+1)/(xrow+K)
## evaluate convergence
err<-max(abs(p.new-p)/p)
## evaluate whether the log.likelihood increases
loglike.new<-(sum(tempmax+log(tempsum))+sum(log(p.new)))/xrow
loglike.old<-loglike.new
p<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,K)
for(j in 1:K) {
clustlike[,j]<-log(p[j])+L[,j]
}
tempmax<-apply(clustlike,1,max)
for(j in 1:K) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:K) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post<-matrix(0,xrow,xcol)
for(j in 1:K) {
for(i in 1:xcol) {
if(patid[j,i] > 0.5) {
p.post[,i]<-p.post[,i]+clustlike[,j]
}
}
}
## return
#calculate back loglikelihood
loglike.old<-loglike.old-sum(log(p))/xrow
loglike.old<-loglike.old*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.old)
}
generatetype<-function(limfitted)
{
jtype<-list()
df<-limfitted$g1num+limfitted$g2num-2+limfitted$df0
for(j in 1:limfitted$compnum)
{
jtype[[j]]<-list(f0=modt.f0.loglike, f0.param=df[j], f1=modt.f1.loglike, f1.param=c(df[j],limfitted$g1num[j],limfitted$g2num[j],limfitted$v0[j]))
}
jtype
}
cormotiffit <- function(exprs, groupid=NULL, compid=NULL, K=1, tol=1e-3,
max.iter=100, BIC=TRUE, norm.factor.method="TMM",
voom.normalize.method = "none", runtype=c("logCPM","counts","limmafits"), each=3)
{
# first I want to do some typechecking. Input can be either a normalized
# matrix, a count matrix, or a list of limma fits. Dispatch the correct
# limmafit accordingly.
# todo: add some typechecking here
limfitted <- list()
if (runtype=="counts") {
limfitted <- limmafit.counts(exprs,groupid,compid, norm.factor.method, voom.normalize.method)
} else if (runtype=="logCPM") {
limfitted <- limmafit.default(exprs,groupid,compid)
} else if (runtype=="limmafits") {
limfitted <- limmafit.list(exprs)
} else {
stop("runtype must be one of 'logCPM', 'counts', or 'limmafits'")
}
jtype<-generatetype(limfitted)
fitresult<-list()
ks <- rep(K, each = each)
fitresult <- bplapply(1:length(ks), function(i, x, type, ks, tol, max.iter) {
cmfit.X(x, type, K = ks[i], tol = tol, max.iter = max.iter)
}, x=limfitted$t, type=jtype, ks=ks, tol=tol, max.iter=max.iter)
best.fitresults <- list()
for (i in 1:length(K)) {
w.k <- which(ks==K[i])
this.bic <- c()
for (j in w.k) this.bic[j] <- -2 * fitresult[[j]]$loglike + (K[i] - 1 + K[i] * limfitted$compnum) * log(dim(limfitted$t)[1])
w.min <- which(this.bic == min(this.bic, na.rm = TRUE))[1]
best.fitresults[[i]] <- fitresult[[w.min]]
}
fitresult <- best.fitresults
bic <- rep(0, length(K))
aic <- rep(0, length(K))
loglike <- rep(0, length(K))
for (i in 1:length(K)) loglike[i] <- fitresult[[i]]$loglike
for (i in 1:length(K)) bic[i] <- -2 * fitresult[[i]]$loglike + (K[i] - 1 + K[i] * limfitted$compnum) * log(dim(limfitted$t)[1])
for (i in 1:length(K)) aic[i] <- -2 * fitresult[[i]]$loglike + 2 * (K[i] - 1 + K[i] * limfitted$compnum)
if(BIC==TRUE) {
bestflag=which(bic==min(bic))
}
else {
bestflag=which(aic==min(aic))
}
result<-list(bestmotif=fitresult[[bestflag]],bic=cbind(K,bic),
aic=cbind(K,aic),loglike=cbind(K,loglike), allmotifs=fitresult)
}
cormotiffitall<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitall(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
cormotiffitsep<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitsep(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
cormotiffitfull<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitfull(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
plotIC<-function(fitted_cormotif)
{
oldpar<-par(mfrow=c(1,2))
plot(fitted_cormotif$bic[,1], fitted_cormotif$bic[,2], type="b",xlab="Motif Number", ylab="BIC", main="BIC")
plot(fitted_cormotif$aic[,1], fitted_cormotif$aic[,2], type="b",xlab="Motif Number", ylab="AIC", main="AIC")
}
plotMotif<-function(fitted_cormotif,title="")
{
layout(matrix(1:2,ncol=2))
u<-1:dim(fitted_cormotif$bestmotif$motif.q)[2]
v<-1:dim(fitted_cormotif$bestmotif$motif.q)[1]
image(u,v,t(fitted_cormotif$bestmotif$motif.q),
col=gray(seq(from=1,to=0,by=-0.1)),xlab="Study",yaxt = "n",
ylab="Corr. Motifs",main=paste(title,"pattern",sep=" "))
axis(2,at=1:length(v))
for(i in 1:(length(u)+1))
{
abline(v=(i-0.5))
}
for(i in 1:(length(v)+1))
{
abline(h=(i-0.5))
}
Ng=10000
if(is.null(fitted_cormotif$bestmotif$p.post)!=TRUE)
Ng=nrow(fitted_cormotif$bestmotif$p.post)
genecount=floor(fitted_cormotif$bestmotif$motif.p*Ng)
NK=nrow(fitted_cormotif$bestmotif$motif.q)
plot(0,0.7,pch=".",xlim=c(0,1.2),ylim=c(0.75,NK+0.25),
frame.plot=FALSE,axes=FALSE,xlab="No. of genes",ylab="", main=paste(title,"frequency",sep=" "))
segments(0,0.7,fitted_cormotif$bestmotif$motif.p[1],0.7)
rect(0,1:NK-0.3,fitted_cormotif$bestmotif$motif.p,1:NK+0.3,
col="dark grey")
mtext(1:NK,at=1:NK,side=2,cex=0.8)
text(fitted_cormotif$bestmotif$motif.p+0.15,1:NK,
labels=floor(fitted_cormotif$bestmotif$motif.p*Ng))
}
plotMotifnew<-function(fitted_cormotif,title="")
{
layout(matrix(1:2,ncol=2))
u<-1:dim(fitted_cormotif$motif.q)[2]
v<-1:dim(fitted_cormotif$motif.q)[1]
image(u,v,t(fitted_cormotif$motif.q),
col=gray(seq(from=1,to=0,by=-0.1)),xlab="Experiment",yaxt = "n",
ylab="Corr. Motifs",main=paste(title,"pattern",sep=" "))
axis(2,at=1:length(v))
for(i in 1:(length(u)+1))
{
abline(v=(i-0.5))
}
for(i in 1:(length(v)+1))
{
abline(h=(i-0.5))
}
Ng=10000
if(is.null(fitted_cormotif$p.post)!=TRUE)
Ng=nrow(fitted_cormotif$p.post)
genecount=floor(fitted_cormotif$motif.p*Ng)
NK=nrow(fitted_cormotif$motif.q)
plot(0,0.7,pch=".",xlim=c(0,1.2),ylim=c(0.75,NK+0.25),
frame.plot=FALSE,axes=FALSE,xlab="No. of regions",ylab="", main=paste(title,"frequency",sep=" "))
segments(0,0.7,fitted_cormotif$motif.p[1],0.7)
rect(0,1:NK-0.3,fitted_cormotif$motif.p,1:NK+0.3,
col="dark grey")
mtext(1:NK,at=1:NK,side=2,cex=0.8)
text(fitted_cormotif$motif.p+0.15,1:NK,
labels=floor(fitted_cormotif$motif.p*Ng))
}Loading counts matrix and making filtered matrix
drug_pal <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031","#41B333")
raw_counts <- read_delim("data/Final_four_data/re_analysis/Raw_unfiltered_counts.tsv",delim="\t") %>%
column_to_rownames("Peakid") %>%
as.matrix()
lcpm <- cpm(raw_counts, log= TRUE)
### for determining the basic cutoffs
filt_raw_counts <- raw_counts[rowMeans(lcpm)> 0,]
filt_raw_counts_noY <- filt_raw_counts[!grepl("chrY",rownames(filt_raw_counts)),]
dim(filt_raw_counts_noY)[1] 155557 48Number of filtered regions without the y chromosome = 155557 regions
making the metadata form
annotation_mat <- data.frame(timeset=colnames(filt_raw_counts_noY)) %>%
mutate(sample = timeset) %>%
separate(timeset, into = c("indv","trt","time"), sep= "_") %>%
mutate(time = factor(time, levels = c("3h", "24h"))) %>%
mutate(trt = factor(trt, levels = c("DOX","EPI", "DNR", "MTX", "TRZ", "VEH"))) %>%
mutate(indv=factor(indv, levels = c("A","B","C","D"))) %>%
mutate(trt_time=paste0(trt,"_",time))Preparing dge object
group <- c( rep(c(1,2,3,4,5,6,7,8,9,10,11,12),4))
group <- factor(group, levels =c("1","2","3","4","5","6","7","8","9","10","11","12"))
dge <- DGEList.data.frame(counts = filt_raw_counts_noY, group = group, genes = row.names(filt_raw_counts_noY))
dge <- calcNormFactors(dge)Adding group and comparison matrices
group_fac <- group
groupid <- as.numeric(group_fac)
compid <- data.frame(c1= c(2,4,6,8,10,1,3,5,7,9), c2 = c( 12,12,12,12,12,11,11,11,11,11))
compid c1 c2
1 2 12
2 4 12
3 6 12
4 8 12
5 10 12
6 1 11
7 3 11
8 5 11
9 7 11
10 9 11y_TMM_cpm <- cpm(dge, log = TRUE)
colnames(y_TMM_cpm) [1] "D_DNR_24h" "D_DNR_3h" "D_DOX_24h" "D_DOX_3h" "D_EPI_24h" "D_EPI_3h"
[7] "D_MTX_24h" "D_MTX_3h" "D_TRZ_24h" "D_TRZ_3h" "D_VEH_24h" "D_VEH_3h"
[13] "A_DNR_24h" "A_DNR_3h" "A_DOX_24h" "A_DOX_3h" "A_EPI_24h" "A_EPI_3h"
[19] "A_MTX_24h" "A_MTX_3h" "A_TRZ_24h" "A_TRZ_3h" "A_VEH_24h" "A_VEH_3h"
[25] "B_DNR_24h" "B_DNR_3h" "B_DOX_24h" "B_DOX_3h" "B_EPI_24h" "B_EPI_3h"
[31] "B_MTX_24h" "B_MTX_3h" "B_TRZ_24h" "B_TRZ_3h" "B_VEH_24h" "B_VEH_3h"
[37] "C_DNR_24h" "C_DNR_3h" "C_DOX_24h" "C_DOX_3h" "C_EPI_24h" "C_EPI_3h"
[43] "C_MTX_24h" "C_MTX_3h" "C_TRZ_24h" "C_TRZ_3h" "C_VEH_24h" "C_VEH_3h" Now that factors are grouped, We will call the cormotiffit function
to look at K=1:8.
This will be saved as an RDS for future use and lack of reruning
set.seed(31415)
cormotif_initial_norm <- cormotiffit(exprs = y_TMM_cpm, groupid = groupid, compid = compid, K=1:8, max.iter = 500, runtype = "logCPM")
saveRDS(cormotif_initial_norm,"data/Final_four_data/re_analysis/Cormotif_norm_initial.RDS")cormotif_initial_norm <- readRDS("data/Final_four_data/re_analysis/Cormotif_norm_initial.RDS")
plotMotif(cormotif_initial_norm)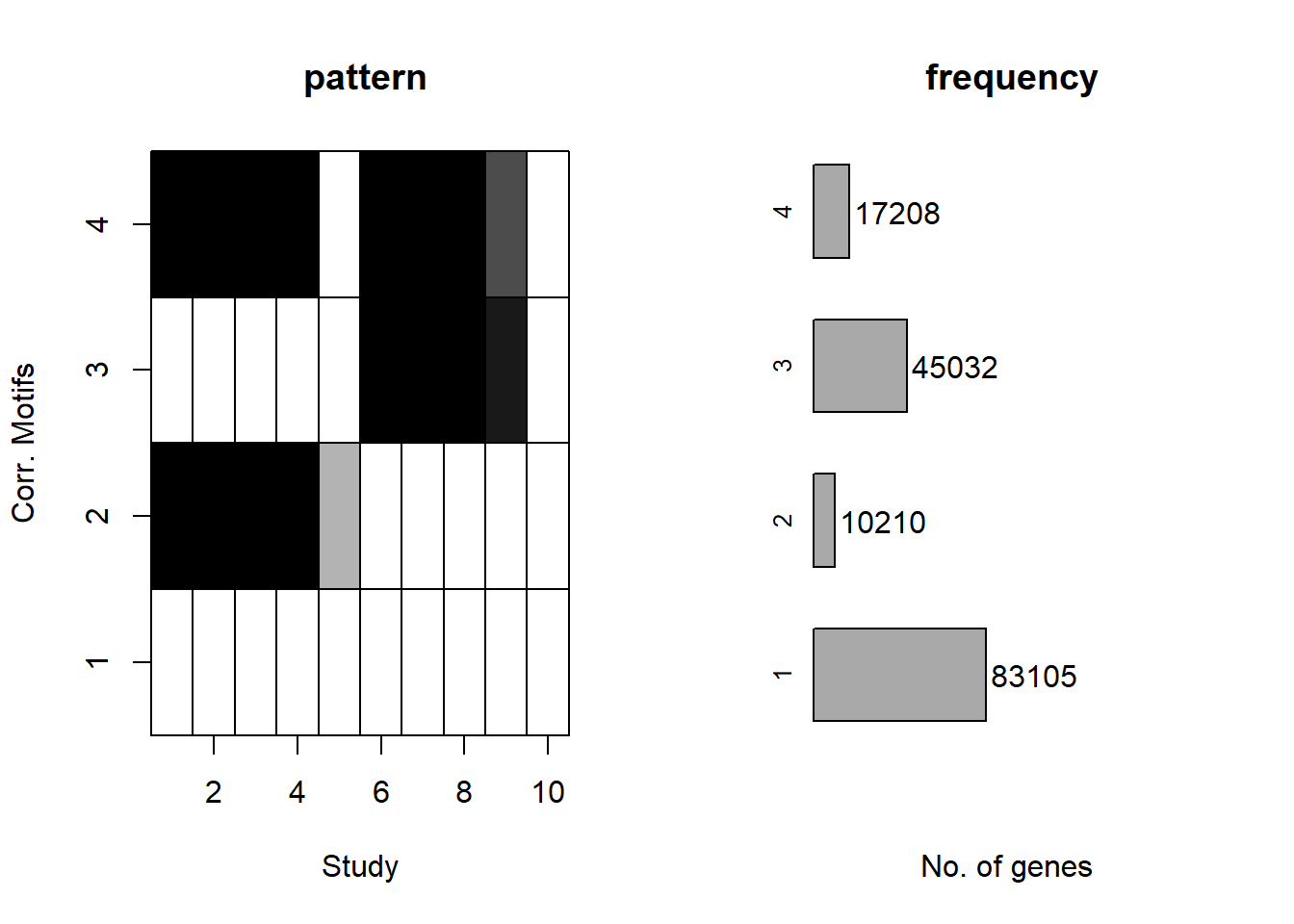
| Version | Author | Date |
|---|---|---|
| ac1a2a6 | reneeisnowhere | 2025-05-06 |
plotIC(cormotif_initial_norm)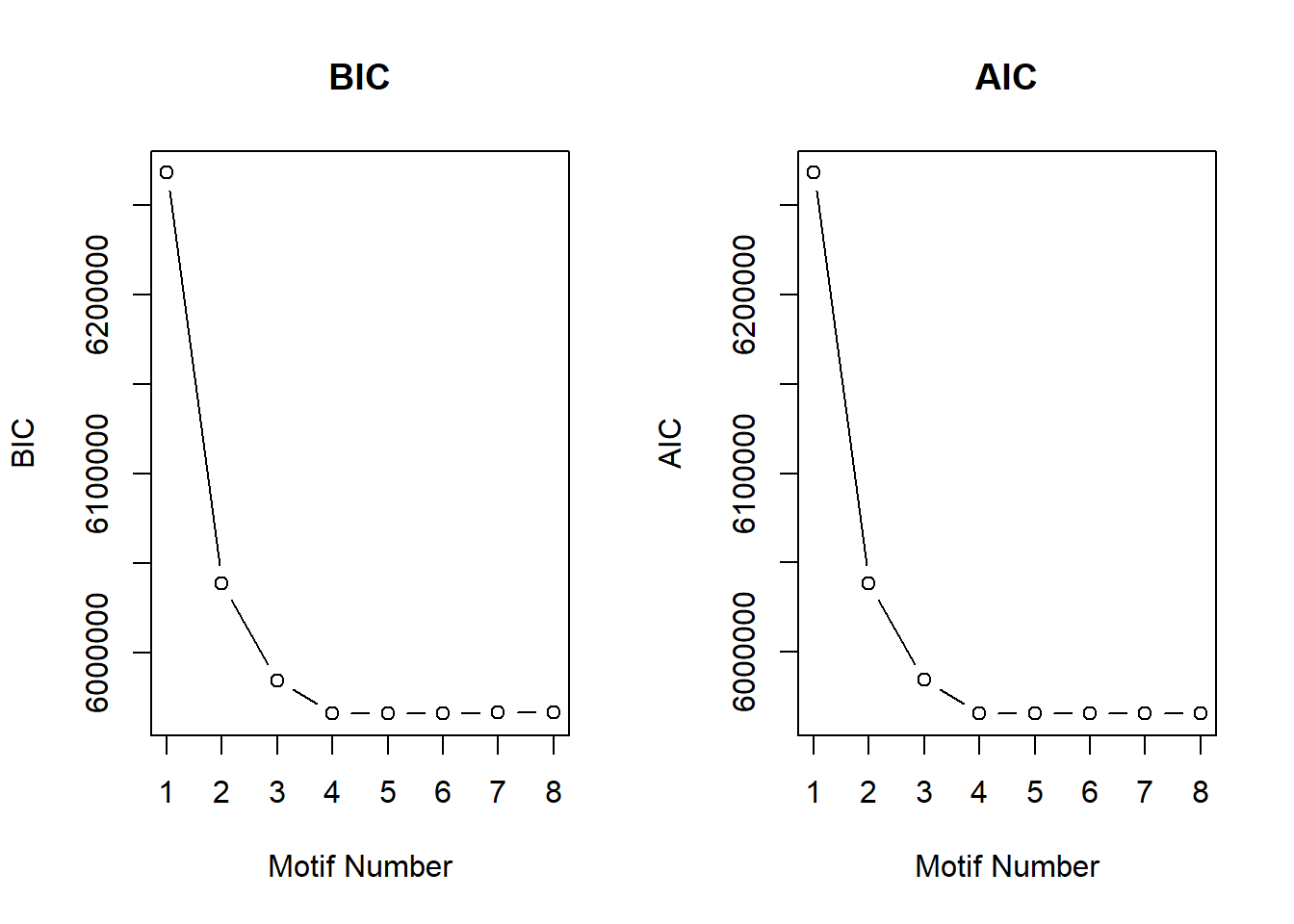
| Version | Author | Date |
|---|---|---|
| ac1a2a6 | reneeisnowhere | 2025-05-06 |
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('center',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
motif_prob <- cormotif_initial_norm$bestmotif$clustlike
rownames(motif_prob) <- rownames(y_TMM_cpm)
# saveRDS(motif_prob,"data/Final_four_data/re_analysis/motif_prob_norm.RDS")
| Version | Author | Date |
|---|---|---|
| ac1a2a6 | reneeisnowhere | 2025-05-06 |
Four motifs were found, now to generate the lists of regions that belong to each motif.
# motif_prob <- readRDS("data/Final_four_data/re_analysis/motif_prob_norm.RDS")
background_peaks <- motif_prob %>%
as.data.frame() %>%
rownames_to_column("Peakid") %>%
dplyr::select(Peakid) %>%
separate(Peakid, into=c("chr","start","end"),remove = FALSE)
NR_ff <- motif_prob %>%
as.data.frame() %>%
dplyr::filter(V1>.5 & V2<.5 & V3 <.5& V4<0.5) %>%
rownames_to_column("Peakid") %>%
dplyr::select(Peakid) %>%
separate(Peakid, into=c("chr","start","end"),remove = FALSE)
EAR_ff <- motif_prob %>%
as.data.frame() %>%
dplyr::filter(V1<.5 & V2>.5 & V3 <.5& V4<0.5) %>%
rownames_to_column("Peakid") %>%
dplyr::select(Peakid) %>%
separate(Peakid, into=c("chr","start","end"),remove = FALSE)
LR_ff <- motif_prob %>%
as.data.frame() %>%
dplyr::filter(V1<.5 & V2<.5 & V3 >.5& V4<0.5) %>%
rownames_to_column("Peakid") %>%
dplyr::select(Peakid) %>%
separate(Peakid, into=c("chr","start","end"),remove = FALSE)
ESR_ff <- motif_prob %>%
as.data.frame() %>%
dplyr::filter(V1<.5 & V2<.5 & V3 <.5& V4>0.5) %>%
rownames_to_column("Peakid") %>%
dplyr::select(Peakid) %>%
separate(Peakid, into=c("chr","start","end"),remove = FALSE)-EAR group has 7974 peaks.
-ESR group has 15128 peaks.
-LR group has 44227 peaks.
-NR group has 85154 peaks.
–Total number of peaks is 155557 –Total number of peaks in clusters is
152483
-percent of peaks classified into clusters: 98.0238755
-number of peaks not classified: 3074
Now to break into opening and closing motifs by group
median_24_lfc <- read_csv("data/Final_four_data/re_analysis/median_24_lfc_norm.csv")
median_3_lfc <- read_csv("data/Final_four_data/re_analysis/median_3_lfc_norm.csv")
open_3med <- median_3_lfc %>%
dplyr::filter(med_3h_lfc > 0)
close_3med <- median_3_lfc %>%
dplyr::filter(med_3h_lfc < 0)
open_24med <- median_24_lfc %>%
dplyr::filter(med_24h_lfc > 0)
close_24med <- median_24_lfc %>%
dplyr::filter(med_24h_lfc < 0)
medA <- median_3_lfc %>%
left_join(median_24_lfc, by=c("peak"="peak")) %>%
dplyr::filter(med_3h_lfc > 0 & med_24h_lfc>0)
medB <- median_3_lfc %>%
left_join(median_24_lfc, by=c("peak"="peak")) %>%
dplyr::filter(med_3h_lfc < 0 & med_24h_lfc < 0)
medC <- median_3_lfc %>%
left_join(median_24_lfc, by=c("peak"="peak")) %>%
dplyr::filter(med_3h_lfc > 0& med_24h_lfc <0)
medD <- median_3_lfc %>%
left_join(median_24_lfc, by=c("peak"="peak"))%>%
dplyr::filter(med_3h_lfc < 0 & med_24h_lfc > 0)
EAR_open <- EAR_ff %>%
dplyr::filter(Peakid %in% open_3med$peak)
EAR_open_gr <- EAR_open %>% GRanges()
EAR_close <- EAR_ff %>%
dplyr::filter(Peakid %in% close_3med$peak)
EAR_close_gr <- EAR_close %>% GRanges()
LR_open <- LR_ff %>%
dplyr::filter(Peakid %in% open_24med$peak)
LR_open_gr <- LR_open %>% GRanges()
LR_close <- LR_ff %>%
dplyr::filter(Peakid %in% close_24med$peak)
LR_close_gr <- LR_close %>% GRanges()
NR_gr <- NR_ff %>%
GRanges()
ESR_open <- ESR_ff %>%
dplyr::filter(Peakid %in% medA$peak)
ESR_open_gr <- ESR_open %>% GRanges()
ESR_close <- ESR_ff %>%
dplyr::filter(Peakid %in% medB$peak)
ESR_close_gr <- ESR_close %>% GRanges()
ESR_opcl <- ESR_ff %>%
dplyr::filter(Peakid %in% medC$peak)
ESR_opcl_gr <- ESR_opcl %>% GRanges()
ESR_clop <- ESR_ff %>%
dplyr::filter(Peakid %in% medD$peak)
ESR_clop_gr <- ESR_clop %>% GRanges()
NR <- NR_ff
NR_gr <- NR %>% GRanges()
background_peaks_gr <- background_peaks %>% GRanges
Motif_list <- list("EAR_open"=EAR_open,
"EAR_close"= EAR_close,
"ESR_open"=ESR_open,
"ESR_close"=ESR_close,
"ESR_opcl"=ESR_opcl,
"ESR_clop"=ESR_clop,
"LR_open"=LR_open,
"LR_close"=LR_close,
"NR"=NR_ff,
"all_regions"=background_peaks)
txdb <- TxDb.Hsapiens.UCSC.hg38.knownGeneExamining the Genomic features around each response motif region using Chipseeker
##make the Granges list
Motif_list_gr <- list("EAR_open"=EAR_open_gr,
"EAR_close"= EAR_close_gr,
"ESR_open"=ESR_open_gr,
"ESR_close"=ESR_close_gr,
"ESR_opcl"=ESR_opcl_gr,
"ESR_clop"=ESR_clop_gr,
"LR_open"=LR_open_gr,
"LR_close"=LR_close_gr,
"NR"=NR_gr,
"all_regions"=background_peaks_gr)
peakAnnoList<- lapply(Motif_list_gr, annotatePeak, tssRegion =c(-2000,2000), TxDb= txdb)>> preparing features information... 2025-05-15 11:33:35 AM
>> identifying nearest features... 2025-05-15 11:33:36 AM
>> calculating distance from peak to TSS... 2025-05-15 11:33:37 AM
>> assigning genomic annotation... 2025-05-15 11:33:37 AM
>> assigning chromosome lengths 2025-05-15 11:33:56 AM
>> done... 2025-05-15 11:33:56 AM
>> preparing features information... 2025-05-15 11:33:56 AM
>> identifying nearest features... 2025-05-15 11:33:56 AM
>> calculating distance from peak to TSS... 2025-05-15 11:33:57 AM
>> assigning genomic annotation... 2025-05-15 11:33:57 AM
>> assigning chromosome lengths 2025-05-15 11:34:00 AM
>> done... 2025-05-15 11:34:00 AM
>> preparing features information... 2025-05-15 11:34:00 AM
>> identifying nearest features... 2025-05-15 11:34:00 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:00 AM
>> assigning genomic annotation... 2025-05-15 11:34:00 AM
>> assigning chromosome lengths 2025-05-15 11:34:03 AM
>> done... 2025-05-15 11:34:03 AM
>> preparing features information... 2025-05-15 11:34:03 AM
>> identifying nearest features... 2025-05-15 11:34:03 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:04 AM
>> assigning genomic annotation... 2025-05-15 11:34:04 AM
>> assigning chromosome lengths 2025-05-15 11:34:07 AM
>> done... 2025-05-15 11:34:07 AM
>> preparing features information... 2025-05-15 11:34:07 AM
>> identifying nearest features... 2025-05-15 11:34:07 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:07 AM
>> assigning genomic annotation... 2025-05-15 11:34:07 AM
>> assigning chromosome lengths 2025-05-15 11:34:10 AM
>> done... 2025-05-15 11:34:10 AM
>> preparing features information... 2025-05-15 11:34:10 AM
>> identifying nearest features... 2025-05-15 11:34:10 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:10 AM
>> assigning genomic annotation... 2025-05-15 11:34:10 AM
>> assigning chromosome lengths 2025-05-15 11:34:14 AM
>> done... 2025-05-15 11:34:14 AM
>> preparing features information... 2025-05-15 11:34:14 AM
>> identifying nearest features... 2025-05-15 11:34:14 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:14 AM
>> assigning genomic annotation... 2025-05-15 11:34:14 AM
>> assigning chromosome lengths 2025-05-15 11:34:17 AM
>> done... 2025-05-15 11:34:17 AM
>> preparing features information... 2025-05-15 11:34:18 AM
>> identifying nearest features... 2025-05-15 11:34:18 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:18 AM
>> assigning genomic annotation... 2025-05-15 11:34:18 AM
>> assigning chromosome lengths 2025-05-15 11:34:21 AM
>> done... 2025-05-15 11:34:21 AM
>> preparing features information... 2025-05-15 11:34:21 AM
>> identifying nearest features... 2025-05-15 11:34:21 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:22 AM
>> assigning genomic annotation... 2025-05-15 11:34:22 AM
>> assigning chromosome lengths 2025-05-15 11:34:26 AM
>> done... 2025-05-15 11:34:26 AM
>> preparing features information... 2025-05-15 11:34:26 AM
>> identifying nearest features... 2025-05-15 11:34:26 AM
>> calculating distance from peak to TSS... 2025-05-15 11:34:27 AM
>> assigning genomic annotation... 2025-05-15 11:34:27 AM
>> assigning chromosome lengths 2025-05-15 11:34:31 AM
>> done... 2025-05-15 11:34:31 AM plotAnnoBar(peakAnnoList[c(1,3,7,5,2,4,8,6,9)])+
ggtitle ("Genomic Feature Distribution, All groups")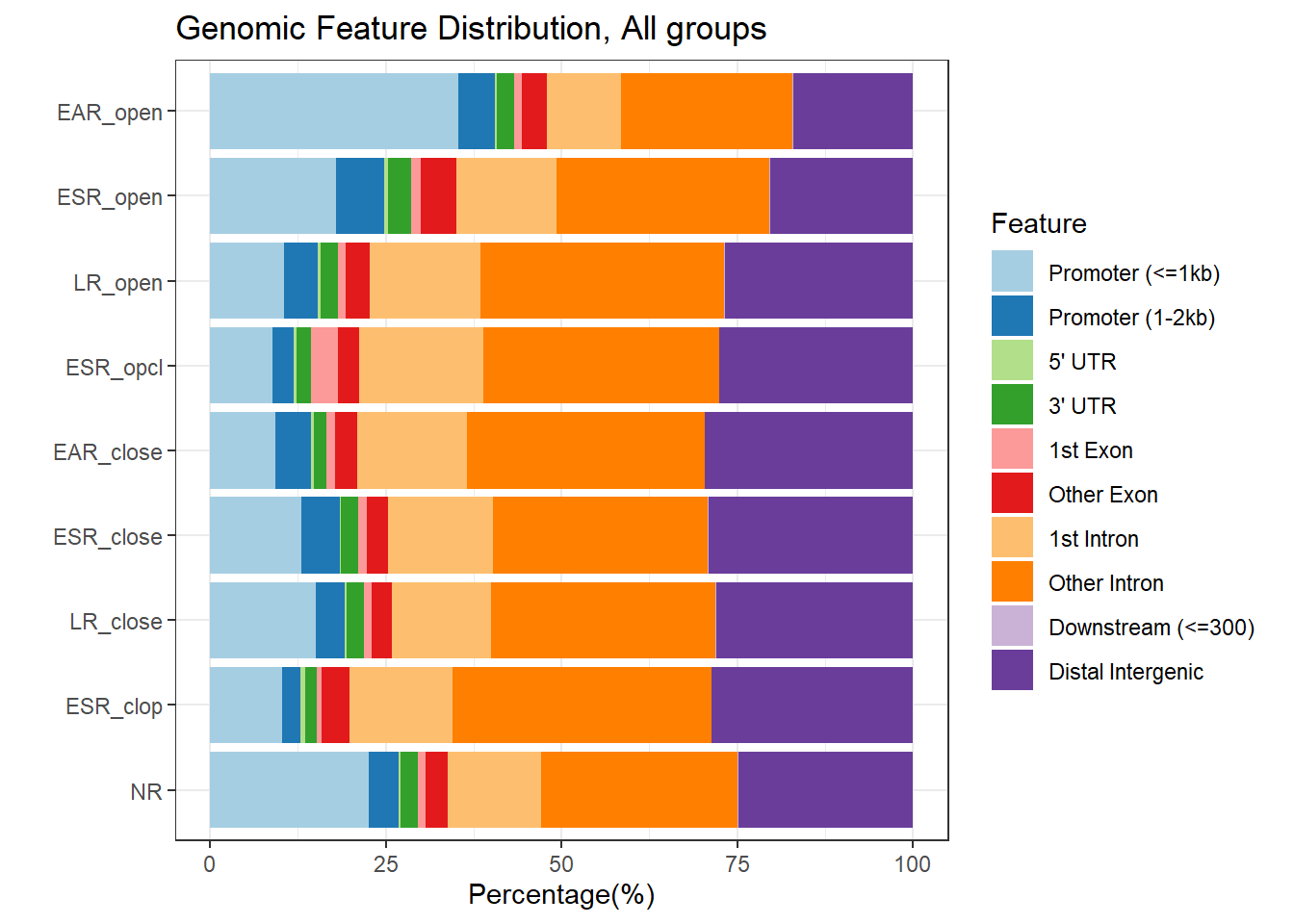
| Version | Author | Date |
|---|---|---|
| ac1a2a6 | reneeisnowhere | 2025-05-06 |
# saveRDS(Motif_list_gr, "data/Final_four_data/re_analysis/Motif_list_granges.RDS")Looking at the average logFC across clusters:
save_list <- readRDS("data/Final_four_data/re_analysis/Toptable_results.RDS")
all_results <- bind_rows(save_list, .id = "group")
mean_lfc <-
all_results %>%
separate(., group, into=c("trt","time")) %>%
dplyr::rename("Peakid"=genes) %>%
dplyr::select(Peakid,trt,time,logFC) %>%
mutate(EAR_open = if_else(Peakid %in% EAR_open$Peakid, "y", "no")) %>%
mutate(EAR_close = if_else(Peakid %in% EAR_close$Peakid, "y", "no")) %>%
mutate(ESR_open = if_else(Peakid %in% ESR_open$Peakid, "y", "no")) %>%
mutate(ESR_close= if_else(Peakid %in% ESR_close$Peakid, "y", "no")) %>%
mutate(ESR_opcl= if_else(Peakid %in% ESR_opcl$Peakid, "y", "no")) %>%
mutate(ESR_clop= if_else(Peakid %in% ESR_clop$Peakid, "y", "no")) %>%
mutate(LR_open= if_else(Peakid %in% LR_open$Peakid, "y", "no")) %>%
mutate(LR_close= if_else(Peakid %in% LR_close$Peakid, "y", "no")) %>%
mutate(NR = if_else(Peakid %in% NR_ff$Peakid, "y", "no"))%>%
mutate(trt = factor(trt,levels=c("DOX","EPI","DNR","MTX","TRZ","VEH"))) %>%
group_by(trt, time) %>%
mutate(absFC = (logFC)) %>% ##I took away abs(logFC) here but did not change code below
dplyr::select(trt, time, absFC, EAR_open:NR) %>%
dplyr::summarize(
EAR_op = mean(absFC[EAR_open == "y"]),
EAR_cl = mean(absFC[EAR_close == "y"]),
ESR_op = mean(absFC[ESR_open == "y"]),
ESR_cl = mean(absFC[ESR_close == "y"]),
ESR_opcl = mean(absFC[ESR_opcl == "y"]),
ESR_clop = mean(absFC[ESR_clop == "y"]),
LR_op = mean(absFC[LR_open == "y"]),
LR_cl = mean(absFC[LR_close == "y"]),
NR = mean(absFC[NR == "y"])
) %>%
as.data.frame()
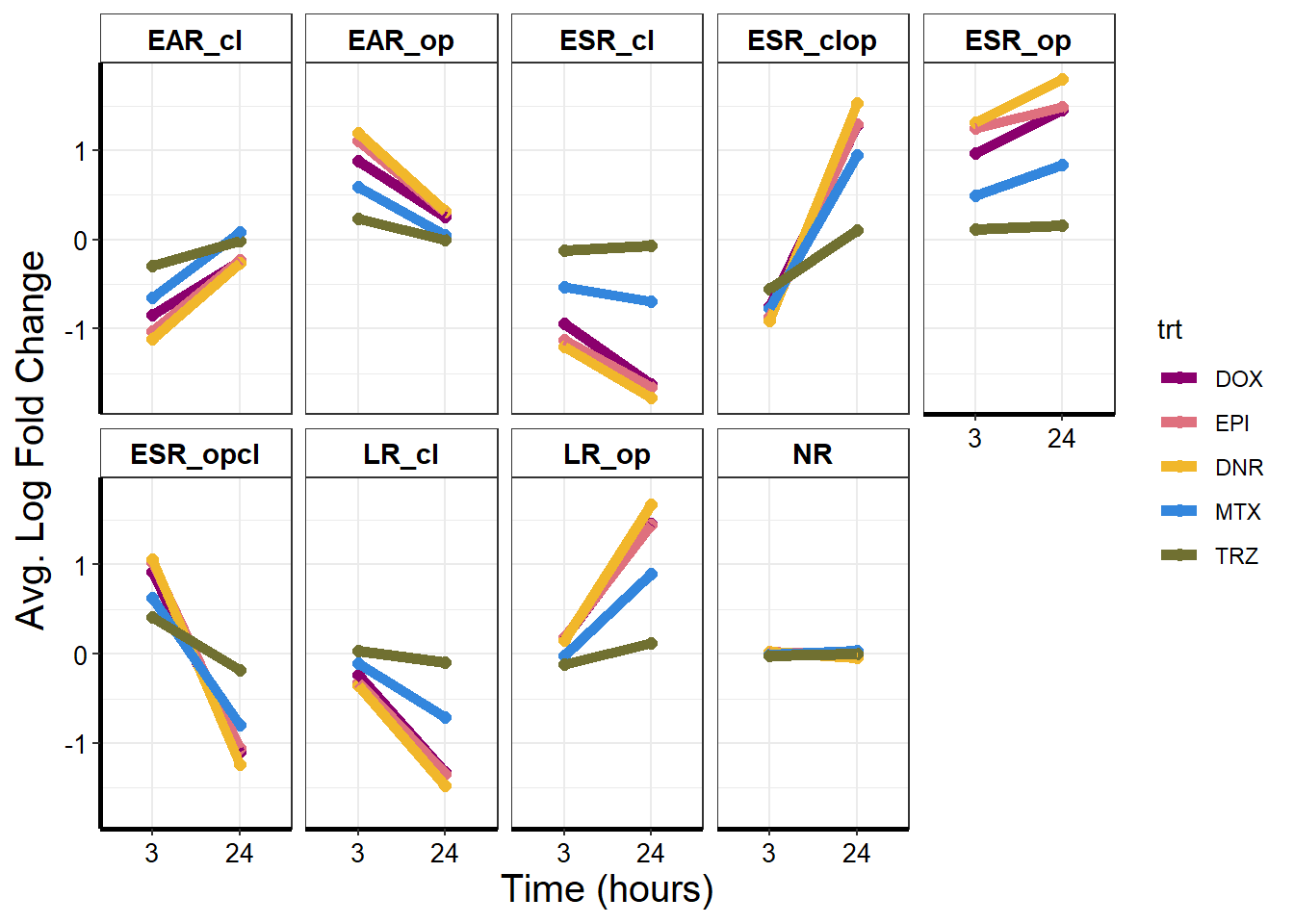
mean_lfc %>%
ungroup() %>%
pivot_longer(!c(trt, time), names_to = "Motif",
values_to = "meanLFC") %>%
mutate(time=factor(time, levels = c("3","24"))) %>%
ggplot(., aes(x = time,y = meanLFC,col = trt,
group = trt
)) +
geom_point(size = 2) +
geom_line(linewidth = 2) +
ggpubr::fill_palette(drug_pal) +
# guides(fill=guide_legend(title = "Treatment"))+
facet_wrap( ~ Motif, nrow = 2) +
theme_bw() +
xlab("Time (hours)") +
scale_color_manual(values = drug_pal) +
ylab(" Avg. Log Fold Change") +
theme_bw() +
theme(
plot.title = element_text(size = rel(1.0), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.0),
strip.background = element_rect(fill = "transparent"),
axis.text = element_text(
size = 10,
color = "black",
angle = 0
),
strip.text.x = element_text(
size = 11,
color = "black",
face = "bold"
)
)
# Folder with input BED files
output_dir <- "data/Final_four_data/re_analysis/motif_beds_centered"
# Create output folder if needed
dir.create(output_dir, showWarnings = FALSE)
# Loop through each BED file
for (name in names(Motif_list_gr)) {
gr <- Motif_list_gr[[name]]
# Recenter each region to 200 bp around its midpoint
gr_centered <- resize(gr, width = 200, fix = "center")
# Export to BED (auto converts to 0-based)
export(gr_centered, con = file.path(output_dir, paste0(name, "_centered.bed")), format = "BED")
}
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] smplot2_0.2.5
[2] cowplot_1.1.3
[3] ComplexHeatmap_2.22.0
[4] ggrepel_0.9.6
[5] plyranges_1.26.0
[6] ggsignif_0.6.4
[7] genomation_1.38.0
[8] eulerr_7.0.2
[9] devtools_2.4.5
[10] usethis_3.1.0
[11] ggpubr_0.6.0
[12] BiocParallel_1.40.0
[13] Cormotif_1.52.0
[14] affy_1.84.0
[15] scales_1.3.0
[16] VennDiagram_1.7.3
[17] futile.logger_1.4.3
[18] gridExtra_2.3
[19] ggfortify_0.4.17
[20] edgeR_4.4.2
[21] limma_3.62.2
[22] rtracklayer_1.66.0
[23] org.Hs.eg.db_3.20.0
[24] TxDb.Hsapiens.UCSC.hg38.knownGene_3.20.0
[25] GenomicFeatures_1.58.0
[26] AnnotationDbi_1.68.0
[27] Biobase_2.66.0
[28] GenomicRanges_1.58.0
[29] GenomeInfoDb_1.42.3
[30] IRanges_2.40.1
[31] S4Vectors_0.44.0
[32] BiocGenerics_0.52.0
[33] ChIPseeker_1.42.1
[34] RColorBrewer_1.1-3
[35] broom_1.0.7
[36] kableExtra_1.4.0
[37] lubridate_1.9.4
[38] forcats_1.0.0
[39] stringr_1.5.1
[40] dplyr_1.1.4
[41] purrr_1.0.4
[42] readr_2.1.5
[43] tidyr_1.3.1
[44] tibble_3.2.1
[45] ggplot2_3.5.1
[46] tidyverse_2.0.0
[47] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.5
[2] matrixStats_1.5.0
[3] bitops_1.0-9
[4] enrichplot_1.26.6
[5] doParallel_1.0.17
[6] httr_1.4.7
[7] profvis_0.4.0
[8] tools_4.4.2
[9] backports_1.5.0
[10] R6_2.6.1
[11] lazyeval_0.2.2
[12] GetoptLong_1.0.5
[13] urlchecker_1.0.1
[14] withr_3.0.2
[15] preprocessCore_1.68.0
[16] cli_3.6.4
[17] formatR_1.14
[18] labeling_0.4.3
[19] sass_0.4.9
[20] Rsamtools_2.22.0
[21] systemfonts_1.2.1
[22] yulab.utils_0.2.0
[23] foreign_0.8-88
[24] DOSE_4.0.0
[25] svglite_2.1.3
[26] R.utils_2.13.0
[27] sessioninfo_1.2.3
[28] plotrix_3.8-4
[29] BSgenome_1.74.0
[30] pwr_1.3-0
[31] impute_1.80.0
[32] rstudioapi_0.17.1
[33] RSQLite_2.3.9
[34] shape_1.4.6.1
[35] generics_0.1.3
[36] gridGraphics_0.5-1
[37] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[38] BiocIO_1.16.0
[39] vroom_1.6.5
[40] gtools_3.9.5
[41] car_3.1-3
[42] GO.db_3.20.0
[43] Matrix_1.7-3
[44] abind_1.4-8
[45] R.methodsS3_1.8.2
[46] lifecycle_1.0.4
[47] whisker_0.4.1
[48] yaml_2.3.10
[49] carData_3.0-5
[50] SummarizedExperiment_1.36.0
[51] gplots_3.2.0
[52] qvalue_2.38.0
[53] SparseArray_1.6.2
[54] blob_1.2.4
[55] promises_1.3.2
[56] crayon_1.5.3
[57] miniUI_0.1.1.1
[58] ggtangle_0.0.6
[59] lattice_0.22-6
[60] KEGGREST_1.46.0
[61] pillar_1.10.1
[62] knitr_1.49
[63] fgsea_1.32.2
[64] rjson_0.2.23
[65] boot_1.3-31
[66] codetools_0.2-20
[67] fastmatch_1.1-6
[68] glue_1.8.0
[69] getPass_0.2-4
[70] ggfun_0.1.8
[71] data.table_1.17.0
[72] remotes_2.5.0
[73] vctrs_0.6.5
[74] png_0.1-8
[75] treeio_1.30.0
[76] gtable_0.3.6
[77] cachem_1.1.0
[78] xfun_0.51
[79] S4Arrays_1.6.0
[80] mime_0.12
[81] iterators_1.0.14
[82] statmod_1.5.0
[83] ellipsis_0.3.2
[84] nlme_3.1-167
[85] ggtree_3.14.0
[86] bit64_4.6.0-1
[87] rprojroot_2.0.4
[88] bslib_0.9.0
[89] affyio_1.76.0
[90] rpart_4.1.24
[91] KernSmooth_2.23-26
[92] Hmisc_5.2-2
[93] colorspace_2.1-1
[94] DBI_1.2.3
[95] nnet_7.3-20
[96] seqPattern_1.38.0
[97] tidyselect_1.2.1
[98] processx_3.8.6
[99] bit_4.6.0
[100] compiler_4.4.2
[101] curl_6.2.1
[102] git2r_0.35.0
[103] htmlTable_2.4.3
[104] xml2_1.3.7
[105] DelayedArray_0.32.0
[106] checkmate_2.3.2
[107] caTools_1.18.3
[108] callr_3.7.6
[109] digest_0.6.37
[110] rmarkdown_2.29
[111] XVector_0.46.0
[112] base64enc_0.1-3
[113] htmltools_0.5.8.1
[114] pkgconfig_2.0.3
[115] MatrixGenerics_1.18.1
[116] fastmap_1.2.0
[117] GlobalOptions_0.1.2
[118] rlang_1.1.5
[119] htmlwidgets_1.6.4
[120] UCSC.utils_1.2.0
[121] shiny_1.10.0
[122] farver_2.1.2
[123] jquerylib_0.1.4
[124] zoo_1.8-13
[125] jsonlite_1.9.1
[126] GOSemSim_2.32.0
[127] R.oo_1.27.0
[128] RCurl_1.98-1.16
[129] magrittr_2.0.3
[130] Formula_1.2-5
[131] GenomeInfoDbData_1.2.13
[132] ggplotify_0.1.2
[133] patchwork_1.3.0
[134] munsell_0.5.1
[135] Rcpp_1.0.14
[136] ape_5.8-1
[137] stringi_1.8.4
[138] zlibbioc_1.52.0
[139] plyr_1.8.9
[140] pkgbuild_1.4.6
[141] parallel_4.4.2
[142] Biostrings_2.74.1
[143] splines_4.4.2
[144] circlize_0.4.16
[145] hms_1.1.3
[146] locfit_1.5-9.12
[147] ps_1.9.0
[148] igraph_2.1.4
[149] reshape2_1.4.4
[150] pkgload_1.4.0
[151] futile.options_1.0.1
[152] XML_3.99-0.18
[153] evaluate_1.0.3
[154] lambda.r_1.2.4
[155] BiocManager_1.30.25
[156] foreach_1.5.2
[157] tzdb_0.4.0
[158] httpuv_1.6.15
[159] clue_0.3-66
[160] gridBase_0.4-7
[161] xtable_1.8-4
[162] restfulr_0.0.15
[163] tidytree_0.4.6
[164] rstatix_0.7.2
[165] later_1.4.1
[166] viridisLite_0.4.2
[167] aplot_0.2.5
[168] memoise_2.0.1
[169] GenomicAlignments_1.42.0
[170] cluster_2.1.8.1
[171] timechange_0.3.0