Enhancer_enrichment
Renee Matthews
2025-05-07
Last updated: 2025-05-15
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b62ef0b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/H3K27ac_integration_noM.Rmd
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/TFmapnames.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/datasave.RDS
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summarized_peaks_dataframe.txt
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Untracked files:
Untracked: RNA_seq_integration.Rmd
Untracked: analysis/.gitignore
Untracked: analysis/Diagnosis-tmm.Rmd
Untracked: analysis/Expressed_RNA_associations.Rmd
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/SVA.Rmd
Untracked: analysis/Tan2020.Rmd
Untracked: analysis/making_master_peaks_list.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/just_for_Fun.R
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: setup.RData
Unstaged changes:
Modified: ATAC_learning.Rproj
Modified: analysis/AF_HF_SNPs.Rmd
Modified: analysis/Cardiotox_SNPs.Rmd
Modified: analysis/H3K27ac_cormotif.Rmd
Modified: analysis/Jaspar_motif.Rmd
Modified: analysis/Jaspar_motif_ff.Rmd
Modified: analysis/RNA_seq_integration.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Enhancer_enrichment.Rmd)
and HTML (docs/Enhancer_enrichment.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b62ef0b | reneeisnowhere | 2025-05-15 | updates and verification of runs |
| html | 5e6e462 | reneeisnowhere | 2025-05-07 | Build site. |
| Rmd | 2db35c7 | reneeisnowhere | 2025-05-07 | updates to analysis |
library(tidyverse)
library(kableExtra)
library(broom)
library(RColorBrewer)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
library(BiocParallel)
library(ggpubr)
library(devtools)
library(biomaRt)
library(eulerr)
library(smplot2)
library(genomation)
library(ggsignif)
library(plyranges)
library(ggrepel)
library(epitools)
library(circlize)Collapsed_peaks <- read_delim("data/Final_four_data/collapsed_new_peaks.txt",
delim = "\t",
escape_double = FALSE,
trim_ws = TRUE)
Motif_list_gr <- readRDS("data/Final_four_data/re_analysis/Motif_list_granges.RDS")
##order specific
df_list <- plyr::llply(Motif_list_gr, as.data.frame)
### no change motif_list_gr names so they do not overwrite the dataframes
names(Motif_list_gr) <- paste0(names(Motif_list_gr), "_gr")
list2env(Motif_list_gr,envir= .GlobalEnv)<environment: R_GlobalEnv>list2env(df_list,envir= .GlobalEnv)<environment: R_GlobalEnv>mrc_lookup <- bind_rows(
(EAR_open %>% dplyr::select(Peakid) %>% mutate(mrc = "EAR_open")),
(EAR_close %>% dplyr::select(Peakid) %>%mutate(mrc = "EAR_close")),
(ESR_open %>% dplyr::select(Peakid) %>%mutate(mrc = "ESR_open")),
(ESR_close %>% dplyr::select(Peakid) %>%mutate(mrc = "ESR_close")),
(ESR_opcl %>% dplyr::select(Peakid) %>%mutate(mrc = "ESR_opcl")),
(ESR_clop %>% dplyr::select(Peakid) %>%mutate(mrc = "ESR_clop")),
(LR_open %>% dplyr::select(Peakid) %>%mutate(mrc = "LR_open")),
(LR_close %>% dplyr::select(Peakid) %>%mutate(mrc = "LR_close")),
(NR %>% dplyr::select(Peakid) %>%mutate(mrc = "NR"))
) %>%
distinct(Peakid, mrc)
final_peaks <- Collapsed_peaks %>%
dplyr::filter(Peakid %in% mcols(all_regions_gr)$Peakid) %>%
left_join(.,(mrc_lookup %>%
dplyr::select(Peakid, mrc)), by = "Peakid") %>%
GRanges()Enhancers and Response clusters
First: obtained a list of cis Regulatory Elements from Encode Screen [(https://screen.encodeproject.org/#)]
cREs_HLV_46F <- genomation::readBed("data/enhancerdata/ENCFF867HAD_ENCFF152PBB_ENCFF352YYH_ENCFF252IVK.7group.bed")
# cREs_HLV_53F <- genomation::readBed("data/enhancerdata/ENCFF417JSF_ENCFF651XRK_ENCFF320IPT_ENCFF440RUS.7group.bed")
NR_cREs <- join_overlap_intersect(NR_gr,cREs_HLV_46F)
LR_open_cREs <- join_overlap_intersect(LR_open_gr,cREs_HLV_46F)
LR_close_cREs <- join_overlap_intersect(LR_close_gr,cREs_HLV_46F)
ESR_open_cREs <- join_overlap_intersect(ESR_open_gr,cREs_HLV_46F)
ESR_close_cREs <- join_overlap_intersect(ESR_close_gr,cREs_HLV_46F)
ESR_opcl_cREs <- join_overlap_intersect(ESR_opcl_gr, cREs_HLV_46F)
ESR_clop_cREs <- join_overlap_intersect(ESR_clop_gr, cREs_HLV_46F)
EAR_open_cREs <- join_overlap_intersect(EAR_open_gr,cREs_HLV_46F)
EAR_close_cREs <- join_overlap_intersect(EAR_close_gr,cREs_HLV_46F)
### These unique peaks are cREs that contain all types of cREs such as
# [1] "Low-DNase" "DNase-only" "CTCF-only,CTCF-bound"
# [4] "PLS,CTCF-bound" "PLS" "dELS"
# [7] "pELS" "DNase-H3K4me3" "DNase-H3K4me3,CTCF-bound"
# [10] "dELS,CTCF-bound" "pELS,CTCF-bound" #### NOT the exact ones I am interested in.
uni_EAR_open <- EAR_open_cREs %>% as.data.frame() %>% distinct(Peakid)
uni_EAR_close <- EAR_close_cREs %>% as.data.frame() %>% distinct(Peakid)
uni_ESR_open <- ESR_open_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_close <- ESR_close_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_opcl <- ESR_opcl_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_clop <- ESR_clop_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_LR_open <- LR_open_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_LR_close <- LR_close_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_NR <- NR_cREs%>% as.data.frame() %>% distinct(Peakid)
Whole_peaks <- join_overlap_intersect(final_peaks, cREs_HLV_46F)
Whole_peaks %>%
as.data.frame() %>%
left_join(.,mrc_lookup, by = c("Peakid","mrc"))%>%
group_by(blockCount, mrc) %>%
tally %>%
pivot_wider(., id_cols = mrc, names_from = blockCount, values_from = n) %>%
dplyr::select(mrc, PLS:'pELS,CTCF-bound') %>%
kable(., caption="Breakdown of peaks overlapping cREs") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 14)| mrc | PLS | PLS,CTCF-bound | dELS | dELS,CTCF-bound | pELS | pELS,CTCF-bound |
|---|---|---|---|---|---|---|
| EAR_close | 13 | 5 | 264 | 12 | 64 | 6 |
| EAR_open | 1185 | 219 | 215 | 60 | 1205 | 193 |
| ESR_close | 202 | 31 | 1276 | 174 | 399 | 75 |
| ESR_opcl | 1 | NA | 16 | 1 | 7 | 1 |
| ESR_open | 120 | 25 | 197 | 39 | 255 | 47 |
| LR_close | 1251 | 315 | 2190 | 441 | 1443 | 312 |
| LR_open | 125 | 17 | 877 | 78 | 280 | 47 |
| NR | 10961 | 3057 | 6511 | 1685 | 12260 | 2852 |
| NA | 128 | 34 | 246 | 52 | 151 | 39 |
| ESR_clop | NA | NA | 17 | NA | 5 | NA |
keep_cRE_names <- c("CTCF-only,CTCF-bound" ,"PLS,CTCF-bound","PLS","dELS,CTCF-bound", "pELS","pELS,CTCF-bound","dELS")
is_cRE <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount %in% keep_cRE_names) %>%
distinct(Peakid,blockCount)
is_CTCF <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "CTCF-only,CTCF-bound") %>%
distinct(Peakid,blockCount)
is_dELS <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "dELS,CTCF-bound"|blockCount == "dELS") %>%
distinct(Peakid,blockCount)
is_pELS <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "pELS,CTCF-bound"|blockCount == "pELS") %>%
distinct(Peakid,blockCount)
is_PLS <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "PLS,CTCF-bound"|blockCount == "PLS") %>%
distinct(Peakid,blockCount)
CRE_summary <-final_peaks %>%
as.data.frame() %>%
mutate(cRE_status=if_else(Peakid %in% is_cRE$Peakid,"cRE_peak","not_cRE_peak")) %>%
mutate(CTCF_status=if_else(Peakid %in% is_CTCF$Peakid,"CTCF_peak","not_CTCF_peak")) %>%
mutate(dELS_status=if_else(Peakid %in% is_dELS$Peakid,"dELS_peak","not_dELS_peak")) %>%
mutate(pELS_status=if_else(Peakid %in% is_pELS$Peakid,"pELS_peak","not_pELS_peak")) %>%
mutate(PLS_status=if_else(Peakid %in% is_PLS$Peakid,"PLS_peak","not_PLS_peak")) %>%
dplyr::select(Peakid:PLS_status)cRE_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(cRE_status, mrc) %>%
tally %>%
mutate(mrc=factor(mrc, levels = c("EAR_open","ESR_open","LR_open","ESR_opcl", "EAR_close","ESR_close","LR_close","ESR_clop", "NR"))) %>%
pivot_wider(id_cols = mrc, names_from = cRE_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
cRE_mat cRE_peak not_cRE_peak
EAR_close 322 2753
EAR_open 1518 3381
ESR_clop 18 696
ESR_close 1846 6088
ESR_opcl 24 179
ESR_open 489 5788
LR_close 4411 14199
LR_open 1206 24411
NR 22087 63067CTCF_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(CTCF_status, mrc) %>%
tally %>%
mutate(mrc=factor(mrc, levels = c("EAR_open","ESR_open","LR_open","ESR_opcl", "EAR_close","ESR_close","LR_close","ESR_clop", "NR"))) %>%
pivot_wider(id_cols = mrc, names_from = CTCF_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
CTCF_mat CTCF_peak not_CTCF_peak
EAR_close 53 3022
EAR_open 185 4714
ESR_close 289 7645
ESR_opcl 4 199
ESR_open 65 6212
LR_close 896 17714
LR_open 131 25486
NR 5280 79874dELS_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(dELS_status, mrc) %>%
tally %>%
mutate(mrc=factor(mrc, levels = c("EAR_open","ESR_open","LR_open","ESR_opcl", "EAR_close","ESR_close","LR_close","ESR_clop", "NR"))) %>%
pivot_wider(id_cols = mrc, names_from = dELS_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
dELS_mat dELS_peak not_dELS_peak
EAR_close 205 2870
EAR_open 208 4691
ESR_clop 14 700
ESR_close 1113 6821
ESR_opcl 14 189
ESR_open 186 6091
LR_close 1964 16646
LR_open 774 24843
NR 5686 79468pELS_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(pELS_status, mrc) %>%
tally %>%
mutate(mrc=factor(mrc, levels = c("EAR_open","ESR_open","LR_open","ESR_opcl", "EAR_close","ESR_close","LR_close","ESR_clop", "NR"))) %>%
pivot_wider(id_cols = mrc, names_from = pELS_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
pELS_mat not_pELS_peak pELS_peak
EAR_close 3020 55
EAR_open 4108 791
ESR_clop 710 4
ESR_close 7584 350
ESR_opcl 196 7
ESR_open 6090 187
LR_close 17499 1111
LR_open 25391 226
NR 77156 7998PLS_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(PLS_status, mrc) %>%
tally %>%
mutate(mrc=factor(mrc, levels = c("EAR_open","ESR_open","LR_open","ESR_opcl", "EAR_close","ESR_close","LR_close","ESR_clop", "NR"))) %>%
pivot_wider(id_cols = mrc, names_from = PLS_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
PLS_mat PLS_peak not_PLS_peak
EAR_close 15 3060
EAR_open 946 3953
ESR_close 194 7740
ESR_opcl 1 202
ESR_open 115 6162
LR_close 1174 17436
LR_open 124 25493
NR 9443 75711odds ratio results
matrix_list <- list("All cREs"= cRE_mat,"PLS"=PLS_mat, "dELS"=dELS_mat, "pELS"=pELS_mat,"CTCF"= CTCF_mat)
results_or <- data.frame(Matrix_Name = character(),
Row_Compared = character(),
Odds_Ratio = numeric(),
Lower_CI = numeric(),
Upper_CI = numeric(),
P_Value = numeric(),
stringsAsFactors = FALSE)
# Loop through each matrix in the list
for (matrix_name in names(matrix_list)) {
current_matrix <- matrix_list[[matrix_name]]
n_rows <- nrow(current_matrix)
# Loop through each row of the current matrix (except the last row)
for (i in 1:(n_rows - 1)) {
# Perform odds ratio test between row i and the last row using epitools
test_result <- tryCatch({
contingency_table <- rbind(current_matrix[i, ], current_matrix[n_rows, ])
# Check if any row in the contingency table contains only zeros
if (any(rowSums(contingency_table) == 0)) {
stop("Contingency table contains empty rows.")
}
oddsratio_result <- oddsratio(contingency_table)
# Ensure the oddsratio result has at least 2 rows
if (nrow(oddsratio_result$measure) < 2) {
stop("oddsratio result does not have enough data.")
}
list(oddsratio = oddsratio_result, p.value = oddsratio_result$p.value[2,"chi.square"])
}, error = function(e) {
cat("Error in odds ratio test for row", i, "in matrix", matrix_name, ":", e$message, "\n")
return(NULL)
})
# Only store the result if test_result is valid (i.e., not NULL)
if (!is.null(test_result)) {
or_value <- test_result$oddsratio$measure[2, "estimate"]
lower_ci <- test_result$oddsratio$measure[2, "lower"]
upper_ci <- test_result$oddsratio$measure[2, "upper"]
p_value <- test_result$oddsratio$p.value[2,"chi.square"]
# Check if the values are numeric and valid (not NA)
if (!is.na(or_value) && !is.na(lower_ci) && !is.na(upper_ci) && !is.na(p_value)) {
# Store the results in the dataframe
results_or <- rbind(results_or, data.frame(Matrix_Name = matrix_name,
Row_Compared = rownames(current_matrix)[i],
Odds_Ratio = or_value,
Lower_CI = lower_ci,
Upper_CI = upper_ci,
P_Value = p_value))
}
}
}
}
# Print the resulting dataframe
print(results_or) %>%
kable(., caption = "Odd ratio results and significance values of TSS and CpG enrichment compared to No response group") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 14) %>%
scroll_box(width = "100%", height = "400px") Matrix_Name Row_Compared Odds_Ratio Lower_CI Upper_CI
estimate All cREs EAR_close 0.33413089 0.296852431 0.37476753
estimate1 All cREs EAR_open 1.28208081 1.204176799 1.36444549
estimate2 All cREs ESR_clop 0.07449141 0.044881181 0.11526243
estimate3 All cREs ESR_close 0.86583822 0.819919213 0.91394794
estimate4 All cREs ESR_opcl 0.38513592 0.244965612 0.57776218
estimate5 All cREs ESR_open 0.24129445 0.219499033 0.26468097
estimate6 All cREs LR_close 0.88706240 0.854672238 0.92051932
estimate7 All cREs LR_open 0.14109037 0.132835268 0.14971335
estimate8 PLS EAR_close 0.03975658 0.022797655 0.06349348
estimate9 PLS EAR_open 1.91893859 1.781028384 2.06546507
estimate10 PLS ESR_close 0.20112102 0.173599679 0.23158497
estimate11 PLS ESR_opcl 0.04547902 0.001960244 0.19763155
estimate12 PLS ESR_open 0.14985132 0.123743139 0.17948544
estimate13 PLS LR_close 0.53990850 0.506787691 0.57468362
estimate14 PLS LR_open 0.03905158 0.032501508 0.04641806
estimate15 dELS EAR_close 0.99900769 0.862333112 1.15089561
estimate16 dELS EAR_open 0.62013489 0.536764284 0.71242916
estimate17 dELS ESR_clop 0.28276093 0.158262160 0.46103441
estimate18 dELS ESR_close 2.28074889 2.128141997 2.44231013
estimate19 dELS ESR_opcl 1.04649971 0.578818829 1.73536017
estimate20 dELS ESR_open 0.42714993 0.367018246 0.49390476
estimate21 dELS LR_close 1.64905922 1.562134953 1.74023431
estimate22 dELS LR_open 0.43551277 0.403167198 0.46979014
estimate23 pELS EAR_close 5.67510713 4.389460507 7.50639156
estimate24 pELS EAR_open 0.53827209 0.497411548 0.58317755
estimate25 pELS ESR_clop 17.69111986 7.596472261 58.02448266
estimate26 pELS ESR_close 2.24519918 2.015399481 2.50961461
estimate27 pELS ESR_opcl 2.83866273 1.442613956 6.71725577
estimate28 pELS ESR_open 3.37301551 2.920119420 3.92119045
estimate29 pELS LR_close 1.63251469 1.530689086 1.74282701
estimate30 pELS LR_open 11.63779496 10.215900039 13.32964519
estimate31 CTCF EAR_close 0.26612181 0.200005260 0.34577686
estimate32 CTCF EAR_open 0.59417924 0.509991703 0.68785377
estimate33 CTCF ESR_close 0.57214560 0.506046669 0.64430623
estimate34 CTCF ESR_opcl 0.31605227 0.095743813 0.74376750
estimate35 CTCF ESR_open 0.15868945 0.122843865 0.20107860
estimate36 CTCF LR_close 0.76528751 0.711262119 0.82250029
estimate37 CTCF LR_open 0.07785612 0.065111148 0.09222061
P_Value
estimate 1.802343e-83
estimate1 5.599162e-15
estimate2 4.381627e-46
estimate3 1.927568e-07
estimate4 4.541572e-06
estimate5 3.683956e-227
estimate6 2.371758e-10
estimate7 0.000000e+00
estimate8 8.886292e-78
estimate9 1.110003e-68
estimate10 4.276917e-129
estimate11 1.529496e-06
estimate12 2.101411e-118
estimate13 1.211921e-84
estimate14 0.000000e+00
estimate15 9.814613e-01
estimate16 2.206703e-11
estimate17 4.621167e-07
estimate18 5.432381e-128
estimate19 9.005404e-01
estimate20 4.973085e-31
estimate21 4.719946e-75
estimate22 3.078418e-106
estimate23 6.585935e-47
estimate24 4.096888e-54
estimate25 6.233974e-16
estimate26 6.780993e-50
estimate27 3.712335e-03
estimate28 4.040218e-66
estimate29 1.646500e-50
estimate30 0.000000e+00
estimate31 1.391090e-24
estimate32 4.816608e-12
estimate33 3.977173e-20
estimate34 1.248910e-02
estimate35 1.403504e-63
estimate36 4.517144e-13
estimate37 2.775286e-300| Matrix_Name | Row_Compared | Odds_Ratio | Lower_CI | Upper_CI | P_Value | |
|---|---|---|---|---|---|---|
| estimate | All cREs | EAR_close | 0.3341309 | 0.2968524 | 0.3747675 | 0.0000000 |
| estimate1 | All cREs | EAR_open | 1.2820808 | 1.2041768 | 1.3644455 | 0.0000000 |
| estimate2 | All cREs | ESR_clop | 0.0744914 | 0.0448812 | 0.1152624 | 0.0000000 |
| estimate3 | All cREs | ESR_close | 0.8658382 | 0.8199192 | 0.9139479 | 0.0000002 |
| estimate4 | All cREs | ESR_opcl | 0.3851359 | 0.2449656 | 0.5777622 | 0.0000045 |
| estimate5 | All cREs | ESR_open | 0.2412944 | 0.2194990 | 0.2646810 | 0.0000000 |
| estimate6 | All cREs | LR_close | 0.8870624 | 0.8546722 | 0.9205193 | 0.0000000 |
| estimate7 | All cREs | LR_open | 0.1410904 | 0.1328353 | 0.1497134 | 0.0000000 |
| estimate8 | PLS | EAR_close | 0.0397566 | 0.0227977 | 0.0634935 | 0.0000000 |
| estimate9 | PLS | EAR_open | 1.9189386 | 1.7810284 | 2.0654651 | 0.0000000 |
| estimate10 | PLS | ESR_close | 0.2011210 | 0.1735997 | 0.2315850 | 0.0000000 |
| estimate11 | PLS | ESR_opcl | 0.0454790 | 0.0019602 | 0.1976316 | 0.0000015 |
| estimate12 | PLS | ESR_open | 0.1498513 | 0.1237431 | 0.1794854 | 0.0000000 |
| estimate13 | PLS | LR_close | 0.5399085 | 0.5067877 | 0.5746836 | 0.0000000 |
| estimate14 | PLS | LR_open | 0.0390516 | 0.0325015 | 0.0464181 | 0.0000000 |
| estimate15 | dELS | EAR_close | 0.9990077 | 0.8623331 | 1.1508956 | 0.9814613 |
| estimate16 | dELS | EAR_open | 0.6201349 | 0.5367643 | 0.7124292 | 0.0000000 |
| estimate17 | dELS | ESR_clop | 0.2827609 | 0.1582622 | 0.4610344 | 0.0000005 |
| estimate18 | dELS | ESR_close | 2.2807489 | 2.1281420 | 2.4423101 | 0.0000000 |
| estimate19 | dELS | ESR_opcl | 1.0464997 | 0.5788188 | 1.7353602 | 0.9005404 |
| estimate20 | dELS | ESR_open | 0.4271499 | 0.3670182 | 0.4939048 | 0.0000000 |
| estimate21 | dELS | LR_close | 1.6490592 | 1.5621350 | 1.7402343 | 0.0000000 |
| estimate22 | dELS | LR_open | 0.4355128 | 0.4031672 | 0.4697901 | 0.0000000 |
| estimate23 | pELS | EAR_close | 5.6751071 | 4.3894605 | 7.5063916 | 0.0000000 |
| estimate24 | pELS | EAR_open | 0.5382721 | 0.4974115 | 0.5831775 | 0.0000000 |
| estimate25 | pELS | ESR_clop | 17.6911199 | 7.5964723 | 58.0244827 | 0.0000000 |
| estimate26 | pELS | ESR_close | 2.2451992 | 2.0153995 | 2.5096146 | 0.0000000 |
| estimate27 | pELS | ESR_opcl | 2.8386627 | 1.4426140 | 6.7172558 | 0.0037123 |
| estimate28 | pELS | ESR_open | 3.3730155 | 2.9201194 | 3.9211904 | 0.0000000 |
| estimate29 | pELS | LR_close | 1.6325147 | 1.5306891 | 1.7428270 | 0.0000000 |
| estimate30 | pELS | LR_open | 11.6377950 | 10.2159000 | 13.3296452 | 0.0000000 |
| estimate31 | CTCF | EAR_close | 0.2661218 | 0.2000053 | 0.3457769 | 0.0000000 |
| estimate32 | CTCF | EAR_open | 0.5941792 | 0.5099917 | 0.6878538 | 0.0000000 |
| estimate33 | CTCF | ESR_close | 0.5721456 | 0.5060467 | 0.6443062 | 0.0000000 |
| estimate34 | CTCF | ESR_opcl | 0.3160523 | 0.0957438 | 0.7437675 | 0.0124891 |
| estimate35 | CTCF | ESR_open | 0.1586895 | 0.1228439 | 0.2010786 | 0.0000000 |
| estimate36 | CTCF | LR_close | 0.7652875 | 0.7112621 | 0.8225003 | 0.0000000 |
| estimate37 | CTCF | LR_open | 0.0778561 | 0.0651111 | 0.0922206 | 0.0000000 |
col_fun_OR = colorRamp2(c(0,1,1.5,5), c("blueviolet","white","lightgreen","green3" ))
sig_mat_OR <- results_or %>%
as.data.frame() %>%
dplyr::select( Matrix_Name,Row_Compared,P_Value) %>%
group_by(Row_Compared) %>%
mutate(rank_val=rank(P_Value, ties.method = "first")) %>%
mutate(BH_correction= p.adjust(P_Value,method= "BH")) %>%
pivot_wider(., id_cols = Matrix_Name, names_from = Row_Compared, values_from = BH_correction) %>%
dplyr::select(Matrix_Name,EAR_open,ESR_open,LR_open,ESR_opcl,EAR_close,ESR_close,LR_close,ESR_clop) %>%
column_to_rownames("Matrix_Name") %>%
as.matrix()
# saveRDS(results_or,"data/Final_four_data/re_analysis/OR_results_cREs_df_1bp.RDS")results_or %>%
as.data.frame() %>%
dplyr::select( Matrix_Name,Row_Compared,Odds_Ratio) %>%
pivot_wider(., id_cols = Matrix_Name, names_from = Row_Compared, values_from = Odds_Ratio) %>%
dplyr::select(Matrix_Name,EAR_open,ESR_open,LR_open,ESR_opcl,EAR_close,ESR_close,LR_close,ESR_clop) %>%
column_to_rownames("Matrix_Name") %>%
as.matrix() %>%
ComplexHeatmap::Heatmap(. ,col = col_fun_OR,
cluster_rows=FALSE,
cluster_columns=FALSE,
column_names_side = "top",
column_names_rot = 45,
# na_col = "black",
cell_fun = function(j, i, x, y, width, height, fill) {if (!is.na(sig_mat_OR[i, j]) && sig_mat_OR[i, j] < 0.05 && .[i, j] > 1) {
grid.text("*", x, y, gp = gpar(fontsize = 20))}})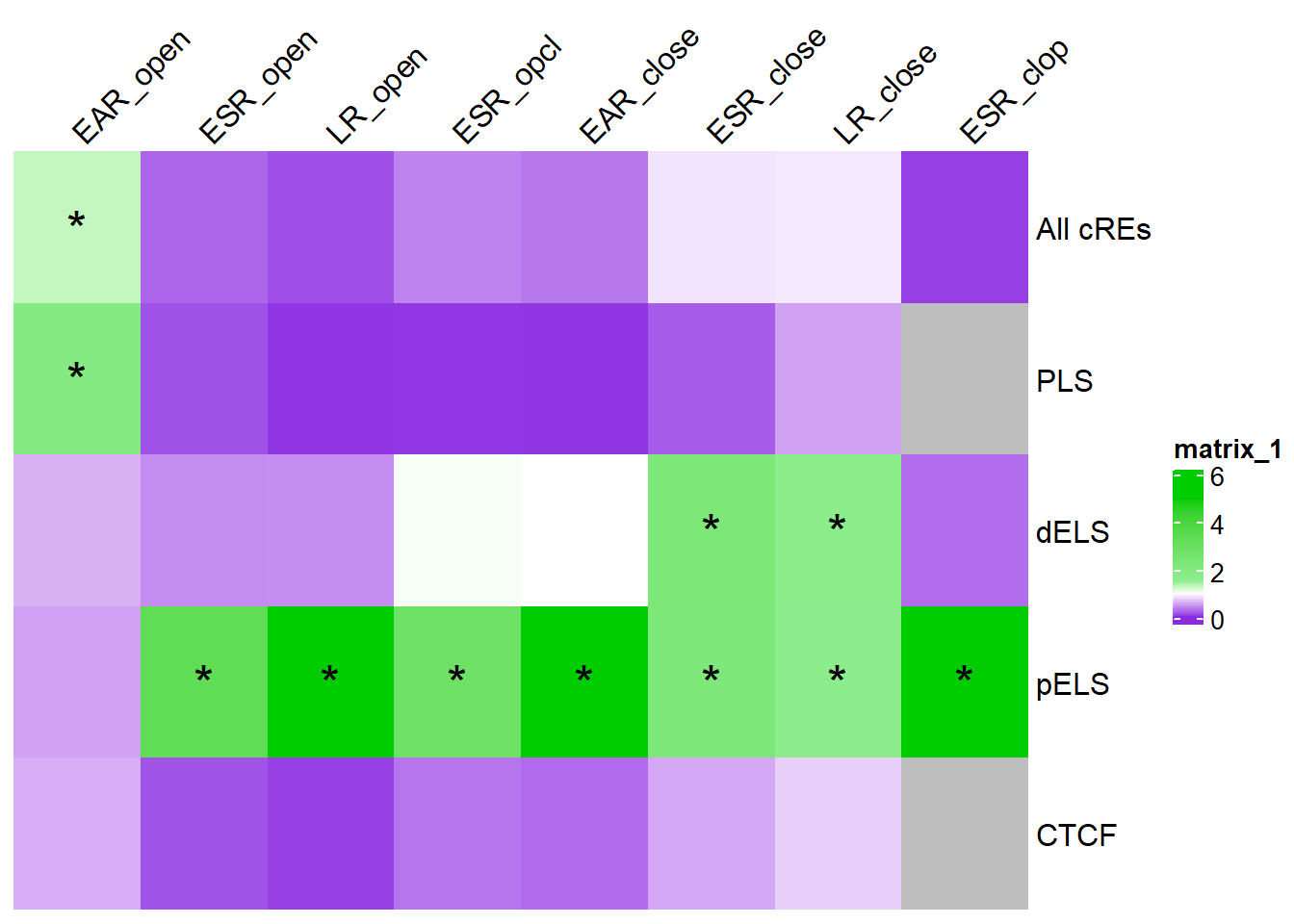
| Version | Author | Date |
|---|---|---|
| 5e6e462 | reneeisnowhere | 2025-05-07 |
note, this is corrected for multiple testing across categories within each motif cluster.
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] circlize_0.4.16
[2] epitools_0.5-10.1
[3] ggrepel_0.9.6
[4] plyranges_1.26.0
[5] ggsignif_0.6.4
[6] genomation_1.38.0
[7] smplot2_0.2.5
[8] eulerr_7.0.2
[9] biomaRt_2.62.1
[10] devtools_2.4.5
[11] usethis_3.1.0
[12] ggpubr_0.6.0
[13] BiocParallel_1.40.0
[14] scales_1.3.0
[15] VennDiagram_1.7.3
[16] futile.logger_1.4.3
[17] gridExtra_2.3
[18] ggfortify_0.4.17
[19] edgeR_4.4.2
[20] limma_3.62.2
[21] rtracklayer_1.66.0
[22] org.Hs.eg.db_3.20.0
[23] TxDb.Hsapiens.UCSC.hg38.knownGene_3.20.0
[24] GenomicFeatures_1.58.0
[25] AnnotationDbi_1.68.0
[26] Biobase_2.66.0
[27] GenomicRanges_1.58.0
[28] GenomeInfoDb_1.42.3
[29] IRanges_2.40.1
[30] S4Vectors_0.44.0
[31] BiocGenerics_0.52.0
[32] ChIPseeker_1.42.1
[33] RColorBrewer_1.1-3
[34] broom_1.0.7
[35] kableExtra_1.4.0
[36] lubridate_1.9.4
[37] forcats_1.0.0
[38] stringr_1.5.1
[39] dplyr_1.1.4
[40] purrr_1.0.4
[41] readr_2.1.5
[42] tidyr_1.3.1
[43] tibble_3.2.1
[44] ggplot2_3.5.1
[45] tidyverse_2.0.0
[46] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.5
[2] matrixStats_1.5.0
[3] bitops_1.0-9
[4] enrichplot_1.26.6
[5] doParallel_1.0.17
[6] httr_1.4.7
[7] profvis_0.4.0
[8] tools_4.4.2
[9] backports_1.5.0
[10] R6_2.6.1
[11] lazyeval_0.2.2
[12] GetoptLong_1.0.5
[13] urlchecker_1.0.1
[14] withr_3.0.2
[15] prettyunits_1.2.0
[16] cli_3.6.4
[17] formatR_1.14
[18] Cairo_1.6-2
[19] sass_0.4.9
[20] Rsamtools_2.22.0
[21] systemfonts_1.2.1
[22] yulab.utils_0.2.0
[23] foreign_0.8-88
[24] DOSE_4.0.0
[25] svglite_2.1.3
[26] R.utils_2.13.0
[27] sessioninfo_1.2.3
[28] plotrix_3.8-4
[29] BSgenome_1.74.0
[30] pwr_1.3-0
[31] impute_1.80.0
[32] rstudioapi_0.17.1
[33] RSQLite_2.3.9
[34] shape_1.4.6.1
[35] generics_0.1.3
[36] gridGraphics_0.5-1
[37] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[38] BiocIO_1.16.0
[39] vroom_1.6.5
[40] gtools_3.9.5
[41] car_3.1-3
[42] GO.db_3.20.0
[43] Matrix_1.7-3
[44] abind_1.4-8
[45] R.methodsS3_1.8.2
[46] lifecycle_1.0.4
[47] whisker_0.4.1
[48] yaml_2.3.10
[49] carData_3.0-5
[50] SummarizedExperiment_1.36.0
[51] gplots_3.2.0
[52] qvalue_2.38.0
[53] SparseArray_1.6.2
[54] BiocFileCache_2.14.0
[55] blob_1.2.4
[56] promises_1.3.2
[57] crayon_1.5.3
[58] miniUI_0.1.1.1
[59] ggtangle_0.0.6
[60] lattice_0.22-6
[61] cowplot_1.1.3
[62] KEGGREST_1.46.0
[63] magick_2.8.5
[64] ComplexHeatmap_2.22.0
[65] pillar_1.10.1
[66] knitr_1.49
[67] fgsea_1.32.2
[68] rjson_0.2.23
[69] boot_1.3-31
[70] codetools_0.2-20
[71] fastmatch_1.1-6
[72] glue_1.8.0
[73] getPass_0.2-4
[74] ggfun_0.1.8
[75] data.table_1.17.0
[76] remotes_2.5.0
[77] vctrs_0.6.5
[78] png_0.1-8
[79] treeio_1.30.0
[80] gtable_0.3.6
[81] cachem_1.1.0
[82] xfun_0.51
[83] S4Arrays_1.6.0
[84] mime_0.12
[85] iterators_1.0.14
[86] statmod_1.5.0
[87] ellipsis_0.3.2
[88] nlme_3.1-167
[89] ggtree_3.14.0
[90] bit64_4.6.0-1
[91] filelock_1.0.3
[92] progress_1.2.3
[93] rprojroot_2.0.4
[94] bslib_0.9.0
[95] rpart_4.1.24
[96] KernSmooth_2.23-26
[97] Hmisc_5.2-2
[98] colorspace_2.1-1
[99] DBI_1.2.3
[100] seqPattern_1.38.0
[101] nnet_7.3-20
[102] tidyselect_1.2.1
[103] processx_3.8.6
[104] bit_4.6.0
[105] compiler_4.4.2
[106] curl_6.2.1
[107] git2r_0.35.0
[108] httr2_1.1.1
[109] htmlTable_2.4.3
[110] xml2_1.3.7
[111] DelayedArray_0.32.0
[112] checkmate_2.3.2
[113] caTools_1.18.3
[114] callr_3.7.6
[115] rappdirs_0.3.3
[116] digest_0.6.37
[117] rmarkdown_2.29
[118] XVector_0.46.0
[119] base64enc_0.1-3
[120] htmltools_0.5.8.1
[121] pkgconfig_2.0.3
[122] MatrixGenerics_1.18.1
[123] dbplyr_2.5.0
[124] fastmap_1.2.0
[125] GlobalOptions_0.1.2
[126] rlang_1.1.5
[127] htmlwidgets_1.6.4
[128] UCSC.utils_1.2.0
[129] shiny_1.10.0
[130] farver_2.1.2
[131] jquerylib_0.1.4
[132] zoo_1.8-13
[133] jsonlite_1.9.1
[134] GOSemSim_2.32.0
[135] R.oo_1.27.0
[136] RCurl_1.98-1.16
[137] magrittr_2.0.3
[138] Formula_1.2-5
[139] GenomeInfoDbData_1.2.13
[140] ggplotify_0.1.2
[141] patchwork_1.3.0
[142] munsell_0.5.1
[143] Rcpp_1.0.14
[144] ape_5.8-1
[145] stringi_1.8.4
[146] zlibbioc_1.52.0
[147] plyr_1.8.9
[148] pkgbuild_1.4.6
[149] parallel_4.4.2
[150] Biostrings_2.74.1
[151] splines_4.4.2
[152] hms_1.1.3
[153] locfit_1.5-9.12
[154] ps_1.9.0
[155] igraph_2.1.4
[156] reshape2_1.4.4
[157] pkgload_1.4.0
[158] futile.options_1.0.1
[159] XML_3.99-0.18
[160] evaluate_1.0.3
[161] lambda.r_1.2.4
[162] foreach_1.5.2
[163] tzdb_0.4.0
[164] httpuv_1.6.15
[165] clue_0.3-66
[166] gridBase_0.4-7
[167] xtable_1.8-4
[168] restfulr_0.0.15
[169] tidytree_0.4.6
[170] rstatix_0.7.2
[171] later_1.4.1
[172] viridisLite_0.4.2
[173] aplot_0.2.5
[174] memoise_2.0.1
[175] GenomicAlignments_1.42.0
[176] cluster_2.1.8.1
[177] timechange_0.3.0