Jaspar_motif_norm
Renee Matthews
2025-05-08
Last updated: 2025-05-15
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b3819a1. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/H3K27ac_integration_noM.Rmd
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/TFmapnames.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/datasave.RDS
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summarized_peaks_dataframe.txt
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Untracked files:
Untracked: RNA_seq_integration.Rmd
Untracked: analysis/.gitignore
Untracked: analysis/Diagnosis-tmm.Rmd
Untracked: analysis/Expressed_RNA_associations.Rmd
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/SVA.Rmd
Untracked: analysis/Tan2020.Rmd
Untracked: analysis/making_master_peaks_list.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/just_for_Fun.R
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: setup.RData
Unstaged changes:
Modified: ATAC_learning.Rproj
Modified: analysis/AF_HF_SNPs.Rmd
Modified: analysis/Cardiotox_SNPs.Rmd
Modified: analysis/H3K27ac_cormotif.Rmd
Modified: analysis/Jaspar_motif.Rmd
Modified: analysis/Jaspar_motif_ff.Rmd
Modified: analysis/RNA_seq_integration.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Jaspar_motif_norm.Rmd) and
HTML (docs/Jaspar_motif_norm.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 6638b27 | reneeisnowhere | 2025-05-12 | Build site. |
| Rmd | a88fc92 | reneeisnowhere | 2025-05-12 | wflow_publish("analysis/Jaspar_motif_norm.Rmd") |
| html | d377357 | reneeisnowhere | 2025-05-12 | Build site. |
| Rmd | fd6cfe1 | reneeisnowhere | 2025-05-12 | added supplemental data table code |
| html | a3f6c4c | reneeisnowhere | 2025-05-12 | Build site. |
| html | 7b1c18d | reneeisnowhere | 2025-05-08 | Build site. |
| Rmd | 0fb8e8b | reneeisnowhere | 2025-05-08 | updates to analysis |
library(tidyverse)
library(cowplot)
library(kableExtra)
library(broom)
library(RColorBrewer)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
library(Cormotif)
library(BiocParallel)
library(ggpubr)
library(devtools)
library(JASPAR2022)
library(TFBSTools)
library(MotifDb)
library(BSgenome.Hsapiens.UCSC.hg38)
library(data.table)
library(universalmotif)
library(ggseqlogo)
library(motifmatchr)
library(gridExtra)mrc_palette <- c(
"EAR_open" = "#F8766D",
"EAR_close" = "#f6483c",
"ESR_open" = "#7CAE00",
"ESR_close" = "#587b00",
"ESR_C"="grey40",
"ESR_opcl"="grey40",
"ESR_D"="tan",
"ESR_clop"="tan",
"LR_open" = "#00BFC4",
"LR_close" = "#008d91",
"NR" = "#C77CFF"
)EAR_open data
EAR_open_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_open_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
EAR_open_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_open_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
EAR_open_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_open_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
EAR_open_sea_all <- rbind(EAR_open_sea_disc, EAR_open_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)EAR_open_combo <- EAR_open_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., EAR_open_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ NA_character_
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="EAR_open")
EAR_open_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1721.1 (ZNF93) | MEME-3 | CGSSSSSSGCSSSGS | CGSSSSSSGCSSSGS | ZNF93 | 1.2e-28 | 9.74e-29 | 1270 | 28.8 | 16200 | 21.2 | 1.36 | EAR_open |
| 2 | 2 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 4.64e-27 | 1.89e-27 | 1480 | 33.4 | 19600 | 25.6 | 1.31 | EAR_open |
| 3 | 3 | 2-GCGSCGC | STREME-2 | 2-GCGSCGC | GCGSCGC | GCGSCGC | 2.15e-26 | 5.82e-27 | 1760 | 39.8 | 24200 | 31.6 | 1.26 | EAR_open |
| 4 | 4 | 4-GCGGCC | STREME-4 | 4-GCGGCC | GCGGCC | GCGGCC | 1.7e-24 | 3.46e-25 | 1560 | 35.4 | 21200 | 27.7 | 1.28 | EAR_open |
| 5 | 7 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 5.74e-24 | 9.32e-25 | 1220 | 27.7 | 15900 | 20.8 | 1.34 | EAR_open |
| 6 | 5 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 1.77e-23 | 2.4e-24 | 880 | 20 | 10700 | 14 | 1.43 | EAR_open |
| 7 | 6 | 5-CCCGGAG | STREME-5 | 5-CCCGGAG | CCCGGAG | CCCGGAG | 5.54e-23 | 5.85e-24 | 1060 | 24 | 13400 | 17.5 | 1.37 | EAR_open |
| 8 | 7 | MA1713.1 (ZNF610) | STREME-6 | 6-CGCTCC | CGCTCC | ZNF610 | 5.76e-23 | 5.85e-24 | 1360 | 30.9 | 18200 | 23.7 | 1.3 | EAR_open |
| 9 | 8 | 1-SCGGNCCGS | STREME-1 | 1-SCGGNCCGS | SCGGNCCGS | SCGGNCCGS | 2.47e-20 | 2.23e-21 | 837 | 19 | 10300 | 13.5 | 1.41 | EAR_open |
| 10 | 9 | MA0506.2 (Nrf1) | STREME-8 | 8-GCAGGCGC | GCAGGCGC | Nrf1 | 4.95e-20 | 4.02e-21 | 1750 | 39.8 | 24900 | 32.5 | 1.22 | EAR_open |
| 11 | 10 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 3.7e-19 | 2.73e-20 | 1290 | 29.2 | 17400 | 22.8 | 1.28 | EAR_open |
| 12 | 11 | 3-CCGGAGC | STREME-3 | 3-CCGGAGC | CCGGAGC | CCGGAGC | 6.82e-19 | 4.62e-20 | 1580 | 35.9 | 22300 | 29.1 | 1.24 | EAR_open |
| 13 | 12 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 9.22e-18 | 5.76e-19 | 1270 | 28.7 | 17300 | 22.5 | 1.28 | EAR_open |
| 14 | 13 | MA1990.1 (Gli1) | STREME-9 | 9-ACACACCACACACAC | ACACACCACACACAC | Gli1 | 3.81e-17 | 2.14e-18 | 65 | 1.47 | 257 | 0.34 | 4.44 | EAR_open |
| 15 | 1 | MA1721.1 (ZNF93) | MEME-5 | GGMGGSGGSGGCSGS | GGMGGSGGSGGCSGS | ZNF93 | 3.96e-17 | 2.14e-18 | 702 | 15.9 | 8590 | 11.2 | 1.42 | EAR_open |
| 16 | 14 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 6.01e-16 | 3.05e-17 | 1360 | 30.7 | 18900 | 24.7 | 1.25 | EAR_open |
| 17 | 10 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 8.37e-15 | 4e-16 | 1700 | 38.5 | 24700 | 32.2 | 1.19 | EAR_open |
| 18 | 15 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 9.74e-15 | 4.4e-16 | 2180 | 49.3 | 32800 | 42.8 | 1.15 | EAR_open |
| 19 | 15 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 1.59e-14 | 6.81e-16 | 1320 | 29.8 | 18500 | 24.1 | 1.24 | EAR_open |
| 20 | 12 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 1.49e-13 | 6.05e-15 | 774 | 17.6 | 10000 | 13.1 | 1.34 | EAR_open |
| 21 | 16 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 2.07e-13 | 8.02e-15 | 853 | 19.3 | 11300 | 14.7 | 1.32 | EAR_open |
| 22 | 17 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 3.2e-13 | 1.18e-14 | 1780 | 40.4 | 26300 | 34.4 | 1.18 | EAR_open |
| 23 | 18 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 7.97e-13 | 2.81e-14 | 808 | 18.3 | 10600 | 13.9 | 1.32 | EAR_open |
| 24 | 19 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 2.36e-12 | 8.01e-14 | 707 | 16 | 9130 | 11.9 | 1.35 | EAR_open |
| 25 | 20 | MA1971.1 | ZBED2 | MA1971.1 | NNMMCGAAACCNNV | ZBED2 | 8.97e-10 | 2.92e-11 | 771 | 17.5 | 10400 | 13.6 | 1.29 | EAR_open |
| 26 | 15 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 2.17e-09 | 6.78e-11 | 1550 | 35.1 | 23100 | 30.1 | 1.17 | EAR_open |
| 27 | 21 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 5.12e-09 | 1.5e-10 | 1250 | 28.4 | 18300 | 23.8 | 1.19 | EAR_open |
| 28 | 22 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 5.16e-09 | 1.5e-10 | 715 | 16.2 | 9640 | 12.6 | 1.29 | EAR_open |
| 29 | 23 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 1.36e-08 | 3.81e-10 | 1170 | 26.5 | 16900 | 22.1 | 1.2 | EAR_open |
| 30 | 24 | MA1684.1 | Foxn1 | MA1684.1 | RGAMGC | Foxn1 | 9.21e-08 | 2.49e-09 | 429 | 9.73 | 5410 | 7.06 | 1.38 | EAR_open |
| 31 | 15 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 1.95e-07 | 5.1e-09 | 1970 | 44.8 | 30600 | 40 | 1.12 | EAR_open |
| 32 | 25 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 2.76e-07 | 6.79e-09 | 1760 | 39.9 | 27100 | 35.3 | 1.13 | EAR_open |
| 33 | 13 | MA1107.2 (KLF9) | STREME-7 | 7-CATGCACACACACA | CATGCACACACACA | KLF9 | 2.76e-07 | 6.79e-09 | 62 | 1.41 | 417 | 0.54 | 2.62 | EAR_open |
| 34 | 15 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 3.55e-07 | 8.49e-09 | 977 | 22.2 | 14100 | 18.4 | 1.21 | EAR_open |
| 35 | 15 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 1.16e-06 | 2.69e-08 | 1520 | 34.6 | 23200 | 30.3 | 1.14 | EAR_open |
| 36 | 26 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 1.39e-06 | 3.13e-08 | 1540 | 34.8 | 23400 | 30.6 | 1.14 | EAR_open |
| 37 | 27 | MA0695.1 | ZBTB7C | MA0695.1 | RCGACCACCGAN | ZBTB7C | 1.99e-06 | 4.36e-08 | 722 | 16.4 | 10100 | 13.2 | 1.24 | EAR_open |
| 38 | 28 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 2.13e-06 | 4.55e-08 | 1370 | 31 | 20600 | 26.9 | 1.15 | EAR_open |
| 39 | 10 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 2.97e-06 | 6.18e-08 | 675 | 15.3 | 9390 | 12.2 | 1.25 | EAR_open |
| 40 | 13 | MA1107.2 (KLF9) | MEME-4 | ACAYRCACACACACA | ACAYRCACACACACA | KLF9 | 3.22e-06 | 6.55e-08 | 291 | 6.6 | 3520 | 4.59 | 1.44 | EAR_open |
| 41 | 15 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 4.43e-06 | 8.79e-08 | 1340 | 30.4 | 20200 | 26.4 | 1.15 | EAR_open |
| 42 | 29 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 9.57e-06 | 1.85e-07 | 1300 | 29.4 | 19600 | 25.5 | 1.15 | EAR_open |
| 43 | 13 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 1.85e-05 | 3.49e-07 | 1800 | 40.7 | 28000 | 36.6 | 1.11 | EAR_open |
| 44 | 30 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 1.98e-05 | 3.66e-07 | 1060 | 24 | 15700 | 20.5 | 1.17 | EAR_open |
| 45 | 12 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 2.51e-05 | 4.53e-07 | 1720 | 39.1 | 26900 | 35 | 1.12 | EAR_open |
| 46 | 17 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 4.07e-05 | 7.19e-07 | 776 | 17.6 | 11200 | 14.6 | 1.21 | EAR_open |
| 47 | 15 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 5.06e-05 | 8.74e-07 | 1800 | 40.8 | 28200 | 36.8 | 1.11 | EAR_open |
| 48 | 23 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 6.65e-05 | 1.13e-06 | 1090 | 24.6 | 16300 | 21.2 | 1.16 | EAR_open |
| 49 | 31 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 7.58e-05 | 1.26e-06 | 278 | 6.3 | 3460 | 4.51 | 1.4 | EAR_open |
| 50 | 15 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 0.000128 | 2.08e-06 | 1800 | 40.8 | 28300 | 37 | 1.1 | EAR_open |
| 51 | 32 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 0.000186 | 2.96e-06 | 955 | 21.7 | 14200 | 18.5 | 1.17 | EAR_open |
| 52 | 26 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 0.00026 | 4.07e-06 | 1060 | 24.1 | 16000 | 20.9 | 1.15 | EAR_open |
| 53 | 12 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 0.000268 | 4.11e-06 | 528 | 12 | 7360 | 9.61 | 1.25 | EAR_open |
| 54 | 33 | MA1941.1 | ETV2::FIGLA | MA1941.1 | RSCGGAARCAGSTGBN | ETV2::FIGLA | 0.000278 | 4.18e-06 | 798 | 18.1 | 11700 | 15.2 | 1.19 | EAR_open |
| 55 | 13 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 0.000338 | 5e-06 | 1140 | 25.9 | 17400 | 22.7 | 1.14 | EAR_open |
| 56 | 34 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 0.000405 | 5.88e-06 | 1460 | 33 | 22600 | 29.5 | 1.12 | EAR_open |
| 57 | 15 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 0.000543 | 7.73e-06 | 3020 | 68.4 | 49700 | 64.8 | 1.06 | EAR_open |
| 58 | 10 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 0.000603 | 8.45e-06 | 1910 | 43.3 | 30400 | 39.6 | 1.09 | EAR_open |
| 59 | 35 | MA1533.1 | NR1I2 | MA1533.1 | DYGAACTSVDTGAACTC | NR1I2 | 0.000832 | 1.15e-05 | 1300 | 29.5 | 20100 | 26.2 | 1.13 | EAR_open |
| 60 | 22 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 0.000909 | 1.23e-05 | 543 | 12.3 | 7680 | 10 | 1.23 | EAR_open |
| 61 | 36 | MA1602.1 | ZSCAN29 | MA1602.1 | YGTCTACRCNGN | ZSCAN29 | 0.000933 | 1.24e-05 | 1080 | 24.6 | 16500 | 21.5 | 1.14 | EAR_open |
| 62 | 22 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 0.000957 | 1.25e-05 | 1470 | 33.4 | 23000 | 30 | 1.11 | EAR_open |
| 63 | 15 | MA0006.1 | Ahr::Arnt | MA0006.1 | YGCGTG | Ahr::Arnt | 0.000995 | 1.27e-05 | 725 | 16.4 | 10600 | 13.8 | 1.19 | EAR_open |
| 64 | 10 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 0.001 | 1.27e-05 | 628 | 14.2 | 9040 | 11.8 | 1.21 | EAR_open |
| 65 | 15 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 0.00104 | 1.3e-05 | 372 | 8.44 | 5010 | 6.54 | 1.29 | EAR_open |
| 66 | 10 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 0.00111 | 1.36e-05 | 1850 | 41.9 | 29400 | 38.4 | 1.09 | EAR_open |
| 67 | 17 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 0.0013 | 1.57e-05 | 832 | 18.9 | 12400 | 16.1 | 1.17 | EAR_open |
| 68 | 17 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 0.00149 | 1.78e-05 | 1690 | 38.4 | 26800 | 34.9 | 1.1 | EAR_open |
| 69 | 26 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 0.0017 | 2e-05 | 1780 | 40.3 | 28200 | 36.9 | 1.09 | EAR_open |
| 70 | 37 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 0.00214 | 2.48e-05 | 1730 | 39.2 | 27400 | 35.8 | 1.1 | EAR_open |
| 13 | 13 | MA1107.2 (KLF9) | MEME-1 | ACACACABRCACACA | ACACACABRCACACA | KLF9 | 0.00226 | 0.000174 | 68 | 1.54 | 723 | 0.94 | 1.66 | EAR_open |
| 71 | 38 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 0.00288 | 3.3e-05 | 1180 | 26.7 | 18200 | 23.7 | 1.13 | EAR_open |
| 72 | 26 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 0.00339 | 3.83e-05 | 1750 | 39.8 | 27900 | 36.4 | 1.09 | EAR_open |
| 73 | 22 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 0.00453 | 5.05e-05 | 450 | 10.2 | 6320 | 8.25 | 1.24 | EAR_open |
| 74 | 39 | MA0736.1 | GLIS2 | MA0736.1 | GACCCCCCGCRAMG | GLIS2 | 0.00499 | 5.48e-05 | 933 | 21.2 | 14200 | 18.5 | 1.15 | EAR_open |
| 75 | 15 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 0.00567 | 6.15e-05 | 664 | 15.1 | 9760 | 12.7 | 1.18 | EAR_open |
| 76 | 15 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 0.00604 | 6.45e-05 | 1770 | 40 | 28200 | 36.8 | 1.09 | EAR_open |
| 77 | 29 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 0.00789 | 8.33e-05 | 1620 | 36.7 | 25700 | 33.5 | 1.09 | EAR_open |
| 78 | 32 | MA0645.1 | ETV6 | MA0645.1 | MSCGGAAGTR | ETV6 | 0.00855 | 8.82e-05 | 59 | 1.34 | 540 | 0.7 | 1.93 | EAR_open |
| 79 | 26 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 0.00867 | 8.82e-05 | 1040 | 23.5 | 15900 | 20.7 | 1.13 | EAR_open |
| 80 | 40 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 0.00868 | 8.82e-05 | 1950 | 44.2 | 31300 | 40.9 | 1.08 | EAR_open |
| 81 | 15 | MA0104.4 | MYCN | MA0104.4 | VVCCACGTGGBB | MYCN | 0.0143 | 0.000144 | 2380 | 53.8 | 38800 | 50.6 | 1.06 | EAR_open |
| 82 | 13 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 0.0149 | 0.000148 | 2250 | 51.1 | 36700 | 47.8 | 1.07 | EAR_open |
| 83 | 27 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 0.0188 | 0.000184 | 335 | 7.6 | 4620 | 6.02 | 1.26 | EAR_open |
| 84 | 26 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 0.02 | 0.000193 | 1010 | 22.8 | 15500 | 20.2 | 1.13 | EAR_open |
| 85 | 41 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 0.0214 | 0.000204 | 1690 | 38.2 | 27000 | 35.2 | 1.09 | EAR_open |
| 14 | 42 | MA1125.1 (ZNF384) | MEME-2 | TTTTTWTTTTTTTTT | TTTTTWTTTTTTTTT | ZNF384 | 14 | 1 | 127 | 2.88 | 2960 | 3.87 | 0.75 | EAR_open |
ER_rat <- 1.25
mrc_type <- "EAR_open"
EAR_open_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))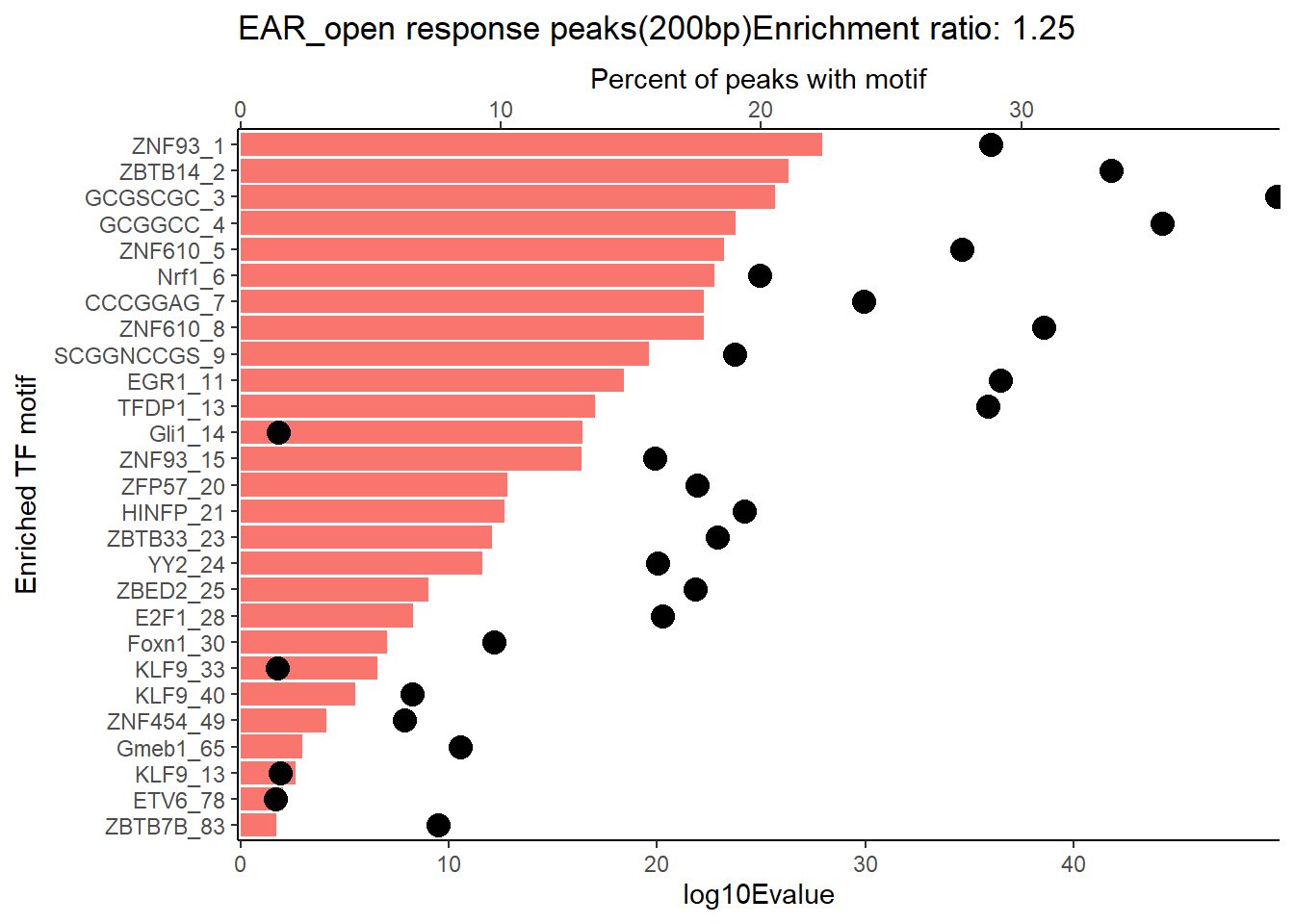
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
EAR_open_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*.5), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.5,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))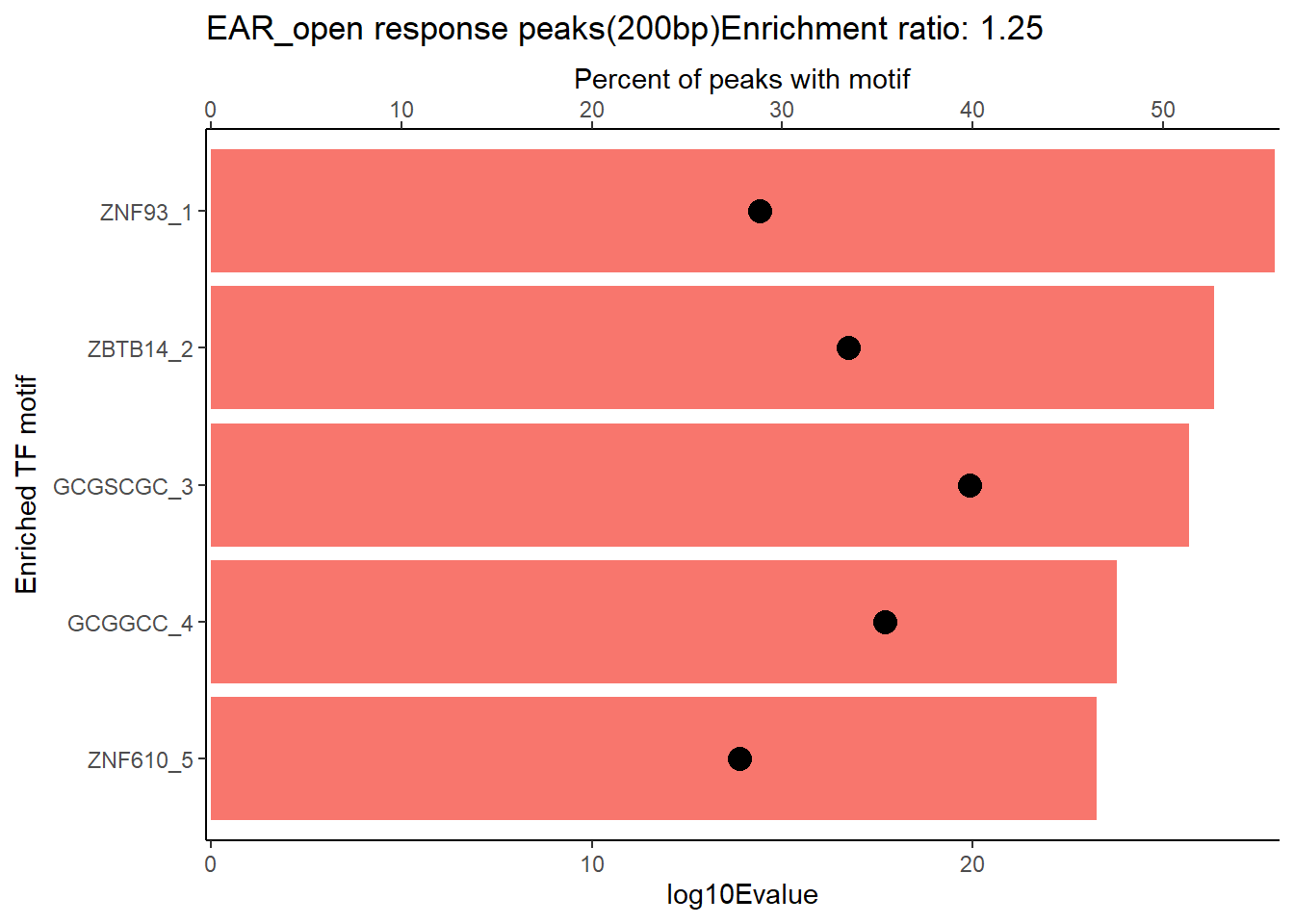
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
EAR_close data
EAR_close_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_close_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
EAR_close_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_close_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
EAR_close_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_close_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
EAR_close_sea_all <- rbind(EAR_close_sea_disc, EAR_close_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)EAR_close_combo <- EAR_close_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., EAR_close_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="EAR_close")
EAR_close_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0476.1 (FOS) | STREME-1 | 1-VTGASTCAB | VTGASTCAB | FOS | 4.82e-72 | 2.63e-72 | 1200 | 43.5 | 20700 | 27 | 1.61 | EAR_close |
| 2 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 8.33e-71 | 2.28e-71 | 1120 | 40.3 | 18700 | 24.3 | 1.66 | EAR_close |
| 3 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 3.77e-65 | 6.86e-66 | 1100 | 39.6 | 18600 | 24.3 | 1.63 | EAR_close |
| 4 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 5.26e-65 | 7.19e-66 | 1120 | 40.6 | 19300 | 25.2 | 1.61 | EAR_close |
| 5 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 8.59e-64 | 9.39e-65 | 1090 | 39.3 | 18600 | 24.2 | 1.62 | EAR_close |
| 6 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 1.15e-63 | 1.05e-64 | 1370 | 49.4 | 25400 | 33.2 | 1.49 | EAR_close |
| 7 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 2.53e-63 | 1.98e-64 | 1190 | 43 | 21000 | 27.5 | 1.57 | EAR_close |
| 8 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 6.88e-63 | 4.7e-64 | 1220 | 44 | 21800 | 28.4 | 1.55 | EAR_close |
| 9 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 9.64e-63 | 5.85e-64 | 918 | 33.2 | 14700 | 19.1 | 1.73 | EAR_close |
| 10 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 1.96e-62 | 1.07e-63 | 1040 | 37.4 | 17400 | 22.7 | 1.65 | EAR_close |
| 11 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 3.21e-62 | 1.6e-63 | 1380 | 49.7 | 25800 | 33.7 | 1.48 | EAR_close |
| 12 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 5.57e-62 | 2.54e-63 | 893 | 32.3 | 14200 | 18.5 | 1.75 | EAR_close |
| 13 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 1.38e-61 | 5.82e-63 | 941 | 34 | 15300 | 19.9 | 1.71 | EAR_close |
| 14 | 2 | MA0036.3 (GATA2) | STREME-2 | 2-GWGATAA | GWGATAA | GATA2 | 6.53e-61 | 2.55e-62 | 1970 | 71.1 | 42300 | 55.2 | 1.29 | EAR_close |
| 15 | 3 | MA0498.2 (MEIS1) | STREME-3 | 3-CTGTCAG | CTGTCAG | MEIS1 | 8.83e-60 | 3.22e-61 | 2680 | 96.8 | 67300 | 87.8 | 1.1 | EAR_close |
| 16 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 4.03e-59 | 1.38e-60 | 848 | 30.6 | 13400 | 17.4 | 1.76 | EAR_close |
| 17 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 9.69e-59 | 3.11e-60 | 879 | 31.8 | 14100 | 18.4 | 1.73 | EAR_close |
| 18 | 1 | MA1988.1 (Atf3) | MEME-1 | TGACTCAB | TGACTCAB | Atf3 | 1.19e-58 | 3.61e-60 | 753 | 27.2 | 11300 | 14.8 | 1.85 | EAR_close |
| 19 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 2.73e-58 | 7.85e-60 | 1220 | 43.9 | 22100 | 28.9 | 1.52 | EAR_close |
| 20 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 2.62e-55 | 7.15e-57 | 1460 | 52.6 | 28500 | 37.2 | 1.41 | EAR_close |
| 21 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 9.11e-55 | 2.37e-56 | 986 | 35.6 | 16900 | 22 | 1.62 | EAR_close |
| 22 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 1.14e-53 | 2.82e-55 | 1450 | 52.3 | 28500 | 37.2 | 1.41 | EAR_close |
| 23 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 1.25e-53 | 2.97e-55 | 1660 | 59.9 | 34200 | 44.7 | 1.34 | EAR_close |
| 24 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 1.7e-53 | 3.87e-55 | 756 | 27.3 | 11700 | 15.3 | 1.79 | EAR_close |
| 25 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 3.38e-53 | 7.38e-55 | 1410 | 50.9 | 27500 | 35.9 | 1.42 | EAR_close |
| 26 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 4.41e-53 | 9.27e-55 | 1300 | 47 | 24800 | 32.3 | 1.46 | EAR_close |
| 27 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 6.94e-52 | 1.4e-53 | 868 | 31.4 | 14400 | 18.7 | 1.67 | EAR_close |
| 28 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 2.95e-49 | 5.75e-51 | 714 | 25.8 | 11100 | 14.5 | 1.78 | EAR_close |
| 29 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 5.3e-44 | 9.99e-46 | 1700 | 61.3 | 36300 | 47.4 | 1.29 | EAR_close |
| 30 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 8.22e-44 | 1.5e-45 | 695 | 25.1 | 11100 | 14.5 | 1.73 | EAR_close |
| 31 | 4 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 2.89e-42 | 5.1e-44 | 2650 | 95.8 | 67500 | 88.1 | 1.09 | EAR_close |
| 32 | 2 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 5.92e-41 | 1.01e-42 | 1020 | 37 | 19100 | 24.9 | 1.49 | EAR_close |
| 33 | 5 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 8.35e-41 | 1.38e-42 | 2400 | 86.9 | 58400 | 76.2 | 1.14 | EAR_close |
| 34 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 1.65e-40 | 2.65e-42 | 674 | 24.4 | 10900 | 14.2 | 1.71 | EAR_close |
| 35 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 4.46e-37 | 6.96e-39 | 894 | 32.3 | 16300 | 21.3 | 1.52 | EAR_close |
| 36 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 4.96e-37 | 7.53e-39 | 940 | 34 | 17400 | 22.7 | 1.5 | EAR_close |
| 37 | 6 | 5-AASATA | STREME-5 | 5-AASATA | AASATA | AASATA | 8.84e-32 | 1.31e-33 | 2470 | 89.3 | 61800 | 80.6 | 1.11 | EAR_close |
| 38 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 2.38e-31 | 3.42e-33 | 700 | 25.3 | 12300 | 16 | 1.58 | EAR_close |
| 39 | 2 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 1.82e-28 | 2.55e-30 | 639 | 23.1 | 11200 | 14.6 | 1.58 | EAR_close |
| 40 | 7 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 7.65e-28 | 1.04e-29 | 2470 | 89.3 | 62300 | 81.2 | 1.1 | EAR_close |
| 41 | 8 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 9.08e-28 | 1.21e-29 | 2640 | 95.3 | 68300 | 89.1 | 1.07 | EAR_close |
| 42 | 2 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 1.03e-26 | 1.34e-28 | 555 | 20 | 9440 | 12.3 | 1.63 | EAR_close |
| 43 | 3 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 1.58e-26 | 2.01e-28 | 698 | 25.2 | 12700 | 16.6 | 1.52 | EAR_close |
| 44 | 8 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 1.9e-26 | 2.36e-28 | 1770 | 63.9 | 40700 | 53.1 | 1.2 | EAR_close |
| 45 | 2 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 6.52e-26 | 7.92e-28 | 445 | 16.1 | 7090 | 9.25 | 1.74 | EAR_close |
| 46 | 8 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 1.36e-25 | 1.61e-27 | 2490 | 89.9 | 63100 | 82.3 | 1.09 | EAR_close |
| 47 | 7 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 4.05e-25 | 4.7e-27 | 2150 | 77.6 | 52100 | 68 | 1.14 | EAR_close |
| 48 | 8 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 5.47e-25 | 6.22e-27 | 2500 | 90.5 | 63700 | 83.1 | 1.09 | EAR_close |
| 49 | 3 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 4.23e-24 | 4.72e-26 | 1240 | 44.9 | 26700 | 34.8 | 1.29 | EAR_close |
| 50 | 8 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 2.75e-23 | 3e-25 | 2480 | 89.4 | 62900 | 82.1 | 1.09 | EAR_close |
| 51 | 4 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 3.04e-23 | 3.26e-25 | 2680 | 96.6 | 70200 | 91.6 | 1.05 | EAR_close |
| 52 | 9 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 3.25e-23 | 3.41e-25 | 2420 | 87.2 | 60900 | 79.5 | 1.1 | EAR_close |
| 53 | 8 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 6.27e-23 | 6.46e-25 | 2320 | 83.7 | 57700 | 75.3 | 1.11 | EAR_close |
| 54 | 8 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 9.77e-23 | 9.89e-25 | 2420 | 87.4 | 61100 | 79.7 | 1.1 | EAR_close |
| 55 | 10 | MA1645.1 (NKX2-2) | STREME-7 | 7-ACTCAA | ACTCAA | NKX2-2 | 2.27e-22 | 2.25e-24 | 919 | 33.2 | 18600 | 24.3 | 1.37 | EAR_close |
| 56 | 8 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 2.55e-22 | 2.48e-24 | 2340 | 84.4 | 58500 | 76.3 | 1.11 | EAR_close |
| 57 | 8 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 2.9e-22 | 2.78e-24 | 2140 | 77.3 | 52300 | 68.3 | 1.13 | EAR_close |
| 58 | 11 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 4.28e-22 | 4.03e-24 | 2590 | 93.4 | 66900 | 87.4 | 1.07 | EAR_close |
| 59 | 2 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 4.47e-21 | 4.14e-23 | 333 | 12 | 5100 | 6.65 | 1.81 | EAR_close |
| 60 | 12 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 6.03e-21 | 5.49e-23 | 2690 | 97.3 | 71200 | 93 | 1.05 | EAR_close |
| 61 | 2 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 1.21e-20 | 1.09e-22 | 523 | 18.9 | 9310 | 12.2 | 1.56 | EAR_close |
| 62 | 8 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 1.32e-20 | 1.17e-22 | 2490 | 90 | 63800 | 83.3 | 1.08 | EAR_close |
| 63 | 7 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 1.8e-20 | 1.56e-22 | 2140 | 77.2 | 52600 | 68.6 | 1.13 | EAR_close |
| 64 | 8 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 2.98e-20 | 2.55e-22 | 2560 | 92.5 | 66300 | 86.5 | 1.07 | EAR_close |
| 65 | 13 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 2.86e-19 | 2.4e-21 | 2120 | 76.5 | 52100 | 68 | 1.12 | EAR_close |
| 66 | 8 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 3.66e-19 | 3.03e-21 | 2600 | 94 | 67900 | 88.6 | 1.06 | EAR_close |
| 67 | 7 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 6.64e-19 | 5.41e-21 | 2370 | 85.7 | 60200 | 78.5 | 1.09 | EAR_close |
| 68 | 14 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 1.43e-18 | 1.15e-20 | 2350 | 85 | 59600 | 77.8 | 1.09 | EAR_close |
| 69 | 15 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 2.65e-18 | 2.1e-20 | 2150 | 77.5 | 53200 | 69.4 | 1.12 | EAR_close |
| 70 | 8 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 5.14e-18 | 4.01e-20 | 1740 | 63 | 41400 | 54 | 1.17 | EAR_close |
| 71 | 16 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 7.35e-18 | 5.65e-20 | 615 | 22.2 | 11800 | 15.4 | 1.45 | EAR_close |
| 72 | 8 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 7.89e-18 | 5.98e-20 | 1490 | 53.9 | 34500 | 45 | 1.2 | EAR_close |
| 73 | 2 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 8.7e-18 | 6.51e-20 | 884 | 31.9 | 18400 | 24 | 1.33 | EAR_close |
| 74 | 7 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 1.24e-17 | 9.13e-20 | 2240 | 80.8 | 56100 | 73.2 | 1.1 | EAR_close |
| 75 | 8 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 1.76e-17 | 1.28e-19 | 2160 | 78 | 53700 | 70.1 | 1.11 | EAR_close |
| 76 | 8 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 1.81e-17 | 1.3e-19 | 2600 | 93.9 | 67900 | 88.6 | 1.06 | EAR_close |
| 77 | 9 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 2.01e-17 | 1.43e-19 | 2440 | 88.1 | 62500 | 81.5 | 1.08 | EAR_close |
| 78 | 11 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 2.24e-17 | 1.57e-19 | 2670 | 96.4 | 70500 | 92 | 1.05 | EAR_close |
| 79 | 13 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 3.31e-17 | 2.29e-19 | 2500 | 90.2 | 64500 | 84.1 | 1.07 | EAR_close |
| 80 | 9 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 3.95e-17 | 2.7e-19 | 2500 | 90.2 | 64500 | 84.2 | 1.07 | EAR_close |
| 81 | 16 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 4.96e-17 | 3.34e-19 | 2500 | 90.1 | 64400 | 84.1 | 1.07 | EAR_close |
| 82 | 9 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 6.15e-17 | 4.1e-19 | 2620 | 94.6 | 68800 | 89.8 | 1.05 | EAR_close |
| 83 | 15 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 8.49e-17 | 5.59e-19 | 2020 | 72.9 | 49600 | 64.7 | 1.13 | EAR_close |
| 84 | 9 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 9.38e-17 | 6.1e-19 | 2710 | 97.8 | 72200 | 94.3 | 1.04 | EAR_close |
| 85 | 9 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 1.08e-16 | 6.96e-19 | 2460 | 88.8 | 63300 | 82.6 | 1.08 | EAR_close |
| 86 | 8 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 1.96e-16 | 1.25e-18 | 2570 | 92.7 | 67000 | 87.4 | 1.06 | EAR_close |
| 87 | 9 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 2.5e-16 | 1.57e-18 | 2400 | 86.8 | 61600 | 80.3 | 1.08 | EAR_close |
| 88 | 8 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 3.15e-16 | 1.95e-18 | 1480 | 53.5 | 34400 | 44.9 | 1.19 | EAR_close |
| 89 | 8 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 6.4e-16 | 3.93e-18 | 1960 | 70.7 | 47900 | 62.6 | 1.13 | EAR_close |
| 90 | 17 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 7.84e-16 | 4.76e-18 | 2300 | 83.2 | 58400 | 76.2 | 1.09 | EAR_close |
| 91 | 7 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 9.28e-16 | 5.57e-18 | 2100 | 75.9 | 52300 | 68.2 | 1.11 | EAR_close |
| 92 | 8 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 9.59e-16 | 5.69e-18 | 2610 | 94.4 | 68700 | 89.6 | 1.05 | EAR_close |
| 93 | 8 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 1.92e-15 | 1.13e-17 | 2310 | 83.6 | 58800 | 76.7 | 1.09 | EAR_close |
| 94 | 15 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 1.97e-15 | 1.14e-17 | 2410 | 87.2 | 62000 | 80.9 | 1.08 | EAR_close |
| 95 | 15 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 2.6e-15 | 1.5e-17 | 1460 | 52.6 | 33900 | 44.2 | 1.19 | EAR_close |
| 96 | 8 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 3.72e-15 | 2.12e-17 | 1860 | 67.1 | 45200 | 59 | 1.14 | EAR_close |
| 97 | 8 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 3.81e-15 | 2.15e-17 | 2750 | 99.5 | 74500 | 97.2 | 1.02 | EAR_close |
| 98 | 8 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 5.81e-15 | 3.24e-17 | 2480 | 89.6 | 64300 | 83.9 | 1.07 | EAR_close |
| 99 | 9 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 8.92e-15 | 4.92e-17 | 2450 | 88.6 | 63400 | 82.7 | 1.07 | EAR_close |
| 100 | 8 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 9.57e-15 | 5.23e-17 | 1950 | 70.4 | 48000 | 62.6 | 1.13 | EAR_close |
| 101 | 15 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 1.08e-14 | 5.83e-17 | 2130 | 76.8 | 53200 | 69.5 | 1.11 | EAR_close |
| 102 | 9 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.3e-14 | 6.94e-17 | 2720 | 98.4 | 73100 | 95.4 | 1.03 | EAR_close |
| 103 | 18 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 1.73e-14 | 9.18e-17 | 915 | 33.1 | 19700 | 25.7 | 1.29 | EAR_close |
| 104 | 8 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 3.9e-14 | 2.05e-16 | 1500 | 54.2 | 35400 | 46.2 | 1.17 | EAR_close |
| 105 | 9 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 3.99e-14 | 2.08e-16 | 2260 | 81.8 | 57600 | 75.1 | 1.09 | EAR_close |
| 106 | 8 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 4.31e-14 | 2.22e-16 | 2240 | 80.7 | 56600 | 73.9 | 1.09 | EAR_close |
| 107 | 15 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 7.37e-14 | 3.77e-16 | 2210 | 79.7 | 55800 | 72.8 | 1.09 | EAR_close |
| 108 | 8 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 8.41e-14 | 4.25e-16 | 1910 | 68.9 | 46900 | 61.2 | 1.13 | EAR_close |
| 109 | 8 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 8.8e-14 | 4.41e-16 | 1450 | 52.4 | 34000 | 44.4 | 1.18 | EAR_close |
| 110 | 8 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 1.84e-13 | 9.16e-16 | 1950 | 70.5 | 48300 | 63 | 1.12 | EAR_close |
| 111 | 4 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 2.08e-13 | 1.02e-15 | 2590 | 93.6 | 68300 | 89.1 | 1.05 | EAR_close |
| 112 | 9 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 2.24e-13 | 1.09e-15 | 2360 | 85.4 | 60700 | 79.2 | 1.08 | EAR_close |
| 113 | 9 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 2.62e-13 | 1.26e-15 | 2690 | 97.2 | 71800 | 93.7 | 1.04 | EAR_close |
| 114 | 8 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 3.27e-13 | 1.57e-15 | 1570 | 56.6 | 37400 | 48.8 | 1.16 | EAR_close |
| 115 | 31 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 4.08e-13 | 1.94e-15 | 704 | 25.4 | 14600 | 19.1 | 1.33 | EAR_close |
| 116 | 19 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 4.3e-13 | 2.03e-15 | 2550 | 92 | 66700 | 87.1 | 1.06 | EAR_close |
| 117 | 20 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 6.49e-13 | 3.03e-15 | 2220 | 80.2 | 56400 | 73.6 | 1.09 | EAR_close |
| 118 | 8 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 7.71e-13 | 3.57e-15 | 2250 | 81.2 | 57200 | 74.7 | 1.09 | EAR_close |
| 119 | 8 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 1.34e-12 | 6.14e-15 | 1820 | 65.8 | 44700 | 58.3 | 1.13 | EAR_close |
| 120 | 3 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 1.67e-12 | 7.62e-15 | 992 | 35.8 | 22100 | 28.8 | 1.25 | EAR_close |
| 121 | 16 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 2.06e-12 | 9.32e-15 | 2590 | 93.7 | 68500 | 89.4 | 1.05 | EAR_close |
| 122 | 15 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 3.57e-12 | 1.6e-14 | 790 | 28.5 | 16900 | 22.1 | 1.29 | EAR_close |
| 123 | 8 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 4.99e-12 | 2.22e-14 | 1980 | 71.6 | 49500 | 64.6 | 1.11 | EAR_close |
| 124 | 7 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 5.26e-12 | 2.32e-14 | 2180 | 78.7 | 55300 | 72.2 | 1.09 | EAR_close |
| 125 | 8 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 6.11e-12 | 2.67e-14 | 2000 | 72.1 | 49900 | 65.1 | 1.11 | EAR_close |
| 126 | 21 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 9.86e-12 | 4.27e-14 | 2700 | 97.5 | 72400 | 94.4 | 1.03 | EAR_close |
| 127 | 8 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 1.43e-11 | 6.14e-14 | 1310 | 47.4 | 30700 | 40.1 | 1.18 | EAR_close |
| 128 | 8 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 1.62e-11 | 6.9e-14 | 2010 | 72.5 | 50400 | 65.7 | 1.1 | EAR_close |
| 129 | 22 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 1.7e-11 | 7.21e-14 | 2290 | 82.7 | 58800 | 76.7 | 1.08 | EAR_close |
| 130 | 8 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 1.83e-11 | 7.68e-14 | 1720 | 62 | 42000 | 54.8 | 1.13 | EAR_close |
| 131 | 9 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 1.96e-11 | 8.16e-14 | 2380 | 86 | 61600 | 80.4 | 1.07 | EAR_close |
| 132 | 17 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 2.17e-11 | 8.98e-14 | 2390 | 86.4 | 62000 | 80.9 | 1.07 | EAR_close |
| 133 | 7 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 3.19e-11 | 1.31e-13 | 2270 | 82.2 | 58400 | 76.2 | 1.08 | EAR_close |
| 134 | 15 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 3.28e-11 | 1.34e-13 | 531 | 19.2 | 10700 | 13.9 | 1.38 | EAR_close |
| 135 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 5.65e-11 | 2.29e-13 | 2000 | 72.2 | 50200 | 65.5 | 1.1 | EAR_close |
| 136 | 23 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 6.03e-11 | 2.42e-13 | 1850 | 67 | 46000 | 60 | 1.12 | EAR_close |
| 137 | 20 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 6.91e-11 | 2.76e-13 | 2150 | 77.6 | 54600 | 71.2 | 1.09 | EAR_close |
| 138 | 7 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 7.31e-11 | 2.89e-13 | 1960 | 71 | 49200 | 64.2 | 1.11 | EAR_close |
| 139 | 15 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 8.75e-11 | 3.44e-13 | 619 | 22.4 | 12900 | 16.8 | 1.33 | EAR_close |
| 140 | 15 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 9.36e-11 | 3.65e-13 | 1360 | 49 | 32200 | 42 | 1.17 | EAR_close |
| 141 | 18 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 2.19e-10 | 8.47e-13 | 916 | 33.1 | 20500 | 26.8 | 1.24 | EAR_close |
| 142 | 1 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 3.21e-10 | 1.24e-12 | 2540 | 91.9 | 67100 | 87.5 | 1.05 | EAR_close |
| 143 | 8 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 1.19e-09 | 4.54e-12 | 1900 | 68.6 | 47600 | 62.1 | 1.1 | EAR_close |
| 144 | 8 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 1.3e-09 | 4.92e-12 | 1950 | 70.5 | 49100 | 64.1 | 1.1 | EAR_close |
| 145 | 8 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 1.38e-09 | 5.21e-12 | 2560 | 92.5 | 67800 | 88.4 | 1.05 | EAR_close |
| 146 | 8 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 1.49e-09 | 5.56e-12 | 1980 | 71.6 | 50000 | 65.2 | 1.1 | EAR_close |
| 147 | 8 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 1.75e-09 | 6.5e-12 | 2020 | 72.9 | 51100 | 66.6 | 1.09 | EAR_close |
| 148 | 8 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 1.96e-09 | 7.25e-12 | 1780 | 64.4 | 44400 | 57.9 | 1.11 | EAR_close |
| 149 | 11 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 2.1e-09 | 7.71e-12 | 2070 | 74.9 | 52800 | 68.8 | 1.09 | EAR_close |
| 150 | 19 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 2.66e-09 | 9.7e-12 | 2560 | 92.6 | 67900 | 88.6 | 1.05 | EAR_close |
| 151 | 8 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 2.7e-09 | 9.75e-12 | 2080 | 75.1 | 52900 | 69.1 | 1.09 | EAR_close |
| 152 | 7 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 3.11e-09 | 1.12e-11 | 1670 | 60.3 | 41100 | 53.7 | 1.12 | EAR_close |
| 153 | 9 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 6.44e-09 | 2.3e-11 | 2710 | 97.9 | 73200 | 95.5 | 1.03 | EAR_close |
| 154 | 4 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 8.03e-09 | 2.85e-11 | 2350 | 85 | 61300 | 80 | 1.06 | EAR_close |
| 155 | 15 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 9.86e-09 | 3.47e-11 | 1840 | 66.3 | 46000 | 60 | 1.1 | EAR_close |
| 156 | 12 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 1.22e-08 | 4.26e-11 | 2670 | 96.5 | 71800 | 93.6 | 1.03 | EAR_close |
| 157 | 24 | MA0664.1 | MLXIPL | MA0664.1 | RTCACGTGAT | MLXIPL | 1.24e-08 | 4.31e-11 | 1560 | 56.4 | 38300 | 49.9 | 1.13 | EAR_close |
| 158 | 15 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 1.64e-08 | 5.68e-11 | 1370 | 49.5 | 33000 | 43.1 | 1.15 | EAR_close |
| 159 | 4 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 2.66e-08 | 9.13e-11 | 2530 | 91.3 | 66900 | 87.3 | 1.05 | EAR_close |
| 160 | 4 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 2.72e-08 | 9.29e-11 | 2020 | 72.8 | 51300 | 67 | 1.09 | EAR_close |
| 161 | 3 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 3.12e-08 | 1.06e-10 | 2500 | 90.3 | 66000 | 86.1 | 1.05 | EAR_close |
| 162 | 10 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 3.45e-08 | 1.16e-10 | 2270 | 82 | 58900 | 76.8 | 1.07 | EAR_close |
| 163 | 8 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 4.79e-08 | 1.61e-10 | 1560 | 56.4 | 38400 | 50.1 | 1.13 | EAR_close |
| 164 | 20 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 5.2e-08 | 1.73e-10 | 2120 | 76.5 | 54400 | 70.9 | 1.08 | EAR_close |
| 165 | 8 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 6.12e-08 | 2.03e-10 | 1340 | 48.3 | 32300 | 42.2 | 1.15 | EAR_close |
| 166 | 8 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 1.23e-07 | 4.06e-10 | 2100 | 75.7 | 53800 | 70.2 | 1.08 | EAR_close |
| 167 | 8 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 1.36e-07 | 4.45e-10 | 1320 | 47.5 | 31800 | 41.5 | 1.15 | EAR_close |
| 168 | 15 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 1.39e-07 | 4.53e-10 | 1300 | 46.9 | 31300 | 40.9 | 1.15 | EAR_close |
| 169 | 8 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 1.4e-07 | 4.54e-10 | 2170 | 78.5 | 56100 | 73.2 | 1.07 | EAR_close |
| 170 | 20 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 2.23e-07 | 7.18e-10 | 1740 | 63 | 43700 | 57 | 1.1 | EAR_close |
| 171 | 7 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 2.45e-07 | 7.83e-10 | 1760 | 63.6 | 44200 | 57.6 | 1.1 | EAR_close |
| 172 | 25 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 2.98e-07 | 9.45e-10 | 2400 | 86.8 | 63100 | 82.4 | 1.05 | EAR_close |
| 173 | 9 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 3.07e-07 | 9.69e-10 | 2730 | 98.8 | 74300 | 96.9 | 1.02 | EAR_close |
| 174 | 13 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 3.52e-07 | 1.1e-09 | 2370 | 85.7 | 62200 | 81.2 | 1.06 | EAR_close |
| 175 | 11 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 4.18e-07 | 1.31e-09 | 1910 | 69.2 | 48700 | 63.5 | 1.09 | EAR_close |
| 176 | 26 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 4.36e-07 | 1.35e-09 | 2690 | 97.1 | 72500 | 94.6 | 1.03 | EAR_close |
| 177 | 27 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 4.89e-07 | 1.51e-09 | 2320 | 83.6 | 60500 | 79 | 1.06 | EAR_close |
| 178 | 8 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 5.45e-07 | 1.67e-09 | 1440 | 52.2 | 35500 | 46.3 | 1.13 | EAR_close |
| 179 | 12 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 6.98e-07 | 2.13e-09 | 1440 | 52 | 35300 | 46.1 | 1.13 | EAR_close |
| 180 | 17 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 8.34e-07 | 2.53e-09 | 2400 | 86.9 | 63300 | 82.6 | 1.05 | EAR_close |
| 181 | 17 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 8.46e-07 | 2.55e-09 | 2080 | 75.1 | 53500 | 69.9 | 1.08 | EAR_close |
| 182 | 8 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 9.94e-07 | 2.98e-09 | 1630 | 59 | 40800 | 53.3 | 1.11 | EAR_close |
| 183 | 28 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 1.05e-06 | 3.15e-09 | 766 | 27.7 | 17400 | 22.7 | 1.22 | EAR_close |
| 184 | 8 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 1.07e-06 | 3.17e-09 | 1790 | 64.7 | 45300 | 59.1 | 1.1 | EAR_close |
| 185 | 1 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 1.3e-06 | 3.85e-09 | 2400 | 86.7 | 63200 | 82.5 | 1.05 | EAR_close |
| 186 | 29 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 1.43e-06 | 4.21e-09 | 1960 | 71 | 50300 | 65.6 | 1.08 | EAR_close |
| 187 | 15 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 1.44e-06 | 4.22e-09 | 550 | 19.9 | 11900 | 15.6 | 1.28 | EAR_close |
| 188 | 30 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 1.46e-06 | 4.23e-09 | 1150 | 41.6 | 27700 | 36.1 | 1.15 | EAR_close |
| 189 | 8 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 1.97e-06 | 5.69e-09 | 1580 | 56.9 | 39300 | 51.3 | 1.11 | EAR_close |
| 190 | 8 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 2.19e-06 | 6.29e-09 | 1390 | 50.1 | 34100 | 44.4 | 1.13 | EAR_close |
| 191 | 31 | MA0592.3 (ESRRA) | STREME-4 | 4-AGGTCA | AGGTCA | ESRRA | 4.66e-06 | 1.33e-08 | 2460 | 88.9 | 65200 | 85.1 | 1.04 | EAR_close |
| 192 | 32 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 5.66e-06 | 1.61e-08 | 708 | 25.6 | 16100 | 21 | 1.22 | EAR_close |
| 193 | 46 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 6.01e-06 | 1.7e-08 | 2460 | 88.9 | 65200 | 85.1 | 1.04 | EAR_close |
| 194 | 4 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 6.18e-06 | 1.74e-08 | 2420 | 87.6 | 64100 | 83.6 | 1.05 | EAR_close |
| 195 | 33 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 6.57e-06 | 1.84e-08 | 212 | 7.66 | 3880 | 5.07 | 1.52 | EAR_close |
| 196 | 10 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 6.68e-06 | 1.86e-08 | 1490 | 53.8 | 37000 | 48.3 | 1.11 | EAR_close |
| 197 | 8 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 7.82e-06 | 2.17e-08 | 1100 | 39.8 | 26500 | 34.6 | 1.15 | EAR_close |
| 198 | 32 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 8.35e-06 | 2.3e-08 | 345 | 12.5 | 7020 | 9.16 | 1.36 | EAR_close |
| 199 | 34 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 1.08e-05 | 2.97e-08 | 2110 | 76.2 | 54700 | 71.4 | 1.07 | EAR_close |
| 200 | 35 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 1.22e-05 | 3.33e-08 | 1330 | 47.9 | 32600 | 42.6 | 1.13 | EAR_close |
| 201 | 8 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 1.27e-05 | 3.45e-08 | 1470 | 53 | 36500 | 47.6 | 1.11 | EAR_close |
| 202 | 15 | MA0656.1 | JDP2 | MA0656.1 | GATGACGTCAYC | JDP2 | 1.28e-05 | 3.46e-08 | 235 | 8.49 | 4450 | 5.8 | 1.47 | EAR_close |
| 203 | 30 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 1.44e-05 | 3.88e-08 | 842 | 30.4 | 19700 | 25.6 | 1.19 | EAR_close |
| 204 | 27 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 1.77e-05 | 4.75e-08 | 1660 | 60 | 41900 | 54.7 | 1.1 | EAR_close |
| 205 | 15 | MA0834.1 | ATF7 | MA0834.1 | NDATGACGTCATMV | ATF7 | 2.38e-05 | 6.35e-08 | 318 | 11.5 | 6440 | 8.41 | 1.37 | EAR_close |
| 206 | 36 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 2.41e-05 | 6.4e-08 | 1720 | 62.3 | 43800 | 57.1 | 1.09 | EAR_close |
| 207 | 15 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 2.82e-05 | 7.44e-08 | 1280 | 46.1 | 31400 | 41 | 1.13 | EAR_close |
| 208 | 37 | MA1627.1 (Wt1) | STREME-6 | 6-CCTCCCCCA | CCTCCCCCA | Wt1 | 3.35e-05 | 8.8e-08 | 316 | 11.4 | 6420 | 8.37 | 1.37 | EAR_close |
| 209 | 8 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 3.94e-05 | 1.03e-07 | 1220 | 43.9 | 29800 | 38.8 | 1.13 | EAR_close |
| 210 | 4 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 4.49e-05 | 1.17e-07 | 2390 | 86.3 | 63200 | 82.5 | 1.05 | EAR_close |
| 211 | 32 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 4.79e-05 | 1.24e-07 | 621 | 22.4 | 14100 | 18.3 | 1.22 | EAR_close |
| 212 | 21 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 5.52e-05 | 1.42e-07 | 2560 | 92.5 | 68600 | 89.5 | 1.03 | EAR_close |
| 213 | 15 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 5.9e-05 | 1.51e-07 | 966 | 34.9 | 23100 | 30.1 | 1.16 | EAR_close |
| 214 | 15 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 5.98e-05 | 1.53e-07 | 1700 | 61.3 | 43100 | 56.2 | 1.09 | EAR_close |
| 215 | 15 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 6.79e-05 | 1.72e-07 | 1750 | 63.3 | 44700 | 58.3 | 1.09 | EAR_close |
| 216 | 33 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 8.17e-05 | 2.07e-07 | 225 | 8.13 | 4320 | 5.64 | 1.45 | EAR_close |
| 217 | 4 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 9.3e-05 | 2.34e-07 | 2230 | 80.6 | 58500 | 76.4 | 1.05 | EAR_close |
| 218 | 20 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 0.000125 | 3.14e-07 | 451 | 16.3 | 9840 | 12.8 | 1.27 | EAR_close |
| 219 | 45 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 0.00013 | 3.25e-07 | 2630 | 95 | 71000 | 92.6 | 1.03 | EAR_close |
| 220 | 15 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 0.000149 | 3.71e-07 | 1750 | 63.3 | 44800 | 58.4 | 1.08 | EAR_close |
| 221 | 38 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 0.000157 | 3.88e-07 | 1330 | 48.2 | 33200 | 43.3 | 1.11 | EAR_close |
| 222 | 33 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 0.000164 | 4.04e-07 | 143 | 5.17 | 2500 | 3.26 | 1.59 | EAR_close |
| 223 | 8 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 0.000169 | 4.13e-07 | 1730 | 62.5 | 44200 | 57.7 | 1.08 | EAR_close |
| 224 | 8 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 0.00017 | 4.15e-07 | 1810 | 65.3 | 46400 | 60.5 | 1.08 | EAR_close |
| 225 | 8 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 0.000184 | 4.46e-07 | 820 | 29.6 | 19400 | 25.3 | 1.17 | EAR_close |
| 226 | 18 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 0.000214 | 5.18e-07 | 577 | 20.8 | 13100 | 17.1 | 1.22 | EAR_close |
| 227 | 35 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 0.000223 | 5.36e-07 | 828 | 29.9 | 19600 | 25.6 | 1.17 | EAR_close |
| 228 | 15 | MA0840.1 | Creb5 | MA0840.1 | NATGACGTCAYH | Creb5 | 0.000285 | 6.83e-07 | 366 | 13.2 | 7800 | 10.2 | 1.3 | EAR_close |
| 229 | 8 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 0.00031 | 7.4e-07 | 1210 | 43.9 | 30000 | 39.1 | 1.12 | EAR_close |
| 230 | 15 | MA1951.1 | FOS | MA1951.1 | BGATGACGTCATCR | FOS | 0.000318 | 7.55e-07 | 610 | 22 | 14000 | 18.2 | 1.21 | EAR_close |
| 231 | 39 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 0.000324 | 7.67e-07 | 1900 | 68.6 | 49100 | 64.1 | 1.07 | EAR_close |
| 232 | 33 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 0.000363 | 8.55e-07 | 77 | 2.78 | 1130 | 1.48 | 1.91 | EAR_close |
| 233 | 8 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 0.000506 | 1.19e-06 | 1020 | 36.8 | 24800 | 32.4 | 1.14 | EAR_close |
| 234 | 21 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 0.000641 | 1.5e-06 | 2610 | 94.2 | 70300 | 91.7 | 1.03 | EAR_close |
| 235 | 4 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 0.000681 | 1.58e-06 | 2560 | 92.4 | 68800 | 89.7 | 1.03 | EAR_close |
| 236 | 40 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 0.000697 | 1.61e-06 | 1770 | 64 | 45600 | 59.5 | 1.08 | EAR_close |
| 237 | 24 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 0.000819 | 1.89e-06 | 906 | 32.7 | 21800 | 28.5 | 1.15 | EAR_close |
| 238 | 4 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 0.000919 | 2.11e-06 | 2360 | 85.1 | 62600 | 81.6 | 1.04 | EAR_close |
| 239 | 32 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 0.000952 | 2.18e-06 | 507 | 18.3 | 11400 | 14.9 | 1.23 | EAR_close |
| 240 | 8 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 0.00102 | 2.31e-06 | 608 | 22 | 14000 | 18.3 | 1.2 | EAR_close |
| 241 | 8 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 0.00115 | 2.61e-06 | 2000 | 72.4 | 52300 | 68.2 | 1.06 | EAR_close |
| 242 | 13 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 0.0013 | 2.92e-06 | 2310 | 83.6 | 61300 | 80 | 1.04 | EAR_close |
| 243 | 32 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 0.0018 | 4.06e-06 | 1640 | 59.2 | 42000 | 54.8 | 1.08 | EAR_close |
| 244 | 8 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 0.00182 | 4.07e-06 | 753 | 27.2 | 17900 | 23.4 | 1.17 | EAR_close |
| 245 | 15 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 0.00274 | 6.11e-06 | 2000 | 72.3 | 52300 | 68.3 | 1.06 | EAR_close |
| 246 | 7 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 0.00314 | 6.97e-06 | 1790 | 64.7 | 46300 | 60.4 | 1.07 | EAR_close |
| 247 | 32 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 0.00353 | 7.81e-06 | 473 | 17.1 | 10700 | 14 | 1.22 | EAR_close |
| 248 | 8 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 0.00357 | 7.86e-06 | 1210 | 43.7 | 30200 | 39.5 | 1.11 | EAR_close |
| 249 | 44 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 0.00362 | 7.94e-06 | 1010 | 36.4 | 24700 | 32.3 | 1.13 | EAR_close |
| 250 | 8 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 0.00442 | 9.65e-06 | 862 | 31.1 | 20900 | 27.3 | 1.14 | EAR_close |
| 9 | 44 | MA1596.1 (ZNF460) | MEME-3 | CCHCHSCCWCC | CCHCHSCCWCC | ZNF460 | 0.0045 | 5e-04 | 1000 | 36.3 | 25400 | 33.1 | 1.1 | EAR_close |
| 251 | 10 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 0.00452 | 9.84e-06 | 2040 | 73.7 | 53500 | 69.8 | 1.06 | EAR_close |
| 252 | 32 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 0.00459 | 9.94e-06 | 649 | 23.4 | 15300 | 20 | 1.18 | EAR_close |
| 253 | 8 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 0.00462 | 9.97e-06 | 992 | 35.8 | 24400 | 31.8 | 1.13 | EAR_close |
| 254 | 30 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 0.00478 | 1.03e-05 | 1160 | 41.9 | 28900 | 37.7 | 1.11 | EAR_close |
| 255 | 41 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 0.0054 | 1.16e-05 | 1990 | 72 | 52200 | 68.1 | 1.06 | EAR_close |
| 256 | 42 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 0.00659 | 1.41e-05 | 62 | 2.24 | 922 | 1.2 | 1.89 | EAR_close |
| 257 | 30 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.00719 | 1.53e-05 | 422 | 15.2 | 9500 | 12.4 | 1.23 | EAR_close |
| 258 | 32 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 0.00815 | 1.72e-05 | 377 | 13.6 | 8380 | 10.9 | 1.25 | EAR_close |
| 259 | 3 | MA0783.1 | PKNOX2 | MA0783.1 | TGACAGGTGTCA | PKNOX2 | 0.00852 | 1.8e-05 | 201 | 7.26 | 4060 | 5.3 | 1.38 | EAR_close |
| 260 | 20 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 0.00861 | 1.81e-05 | 2680 | 96.7 | 72800 | 95 | 1.02 | EAR_close |
| 10 | 45 | MA1731.1 (ZNF768) | MEME-6 | TCTCYYTMTCTCTYT | TCTCYYTMTCTCTYT | ZNF768 | 0.0091 | 0.00091 | 203 | 7.33 | 4440 | 5.79 | 1.27 | EAR_close |
| 261 | 31 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 0.011 | 2.31e-05 | 1000 | 36.2 | 24800 | 32.3 | 1.12 | EAR_close |
| 262 | 30 | MA0816.1 | Ascl2 | MA0816.1 | ARCAGCTGCY | Ascl2 | 0.0118 | 2.47e-05 | 2140 | 77.2 | 56500 | 73.7 | 1.05 | EAR_close |
| 263 | 15 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 0.0157 | 3.26e-05 | 766 | 27.7 | 18500 | 24.2 | 1.15 | EAR_close |
| 264 | 31 | MA0071.1 | RORA | MA0071.1 | AWMWAGGTCA | RORA | 0.0166 | 3.45e-05 | 497 | 18 | 11500 | 15 | 1.2 | EAR_close |
| 265 | 9 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 0.0169 | 3.49e-05 | 1550 | 55.8 | 39800 | 51.9 | 1.08 | EAR_close |
| 266 | 20 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 0.0182 | 3.75e-05 | 1780 | 64.3 | 46300 | 60.5 | 1.06 | EAR_close |
| 267 | 30 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 0.0187 | 3.82e-05 | 1460 | 52.7 | 37300 | 48.7 | 1.08 | EAR_close |
| 268 | 27 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 0.019 | 3.88e-05 | 2660 | 95.9 | 72100 | 94.1 | 1.02 | EAR_close |
| 269 | 31 | MA0017.2 | NR2F1 | MA0017.2 | CARAGGTCAMVNG | NR2F1 | 0.0199 | 4.05e-05 | 455 | 16.4 | 10500 | 13.6 | 1.21 | EAR_close |
| 270 | 31 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 0.0213 | 4.31e-05 | 1360 | 49.1 | 34600 | 45.2 | 1.09 | EAR_close |
| 271 | 30 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 0.024 | 4.83e-05 | 746 | 27 | 18100 | 23.6 | 1.14 | EAR_close |
| 272 | 32 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 0.028 | 5.62e-05 | 371 | 13.4 | 8360 | 10.9 | 1.23 | EAR_close |
| 273 | 43 | MA0164.1 | Nr2e3 | MA0164.1 | CAAGCTT | Nr2e3 | 0.0344 | 6.89e-05 | 595 | 21.5 | 14100 | 18.5 | 1.17 | EAR_close |
| 274 | 15 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 0.0346 | 6.91e-05 | 2380 | 86.2 | 63900 | 83.4 | 1.03 | EAR_close |
| 275 | 8 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 0.0421 | 8.36e-05 | 545 | 19.7 | 12900 | 16.8 | 1.17 | EAR_close |
| 276 | 15 | MA0828.2 | SREBF2 | MA0828.2 | WWATCACGTGATWW | SREBF2 | 0.048 | 9.5e-05 | 206 | 7.44 | 4310 | 5.62 | 1.33 | EAR_close |
| 11 | 46 | MA0090.3 (TEAD1) | MEME-4 | CAYTCCAGCCT | CAYTCCAGCCT | TEAD1 | 0.0508 | 0.00462 | 13 | 0.47 | 142 | 0.19 | 2.71 | EAR_close |
| 12 | 47 | GAGACRGRGTCTCRC | MEME-7 | GAGACRGRGTCTCRC | GAGACRGRGTCTCRC | GAGACRGRGTCTCRC | 0.0912 | 0.0076 | 331 | 12 | 7980 | 10.4 | 1.15 | EAR_close |
| 13 | 48 | 8-AACTGTGGAGA | STREME-8 | 8-AACTGTGGAGA | AACTGTGGAGA | AACTGTGGAGA | 0.192 | 0.0148 | 397 | 14.3 | 9840 | 12.8 | 1.12 | EAR_close |
| 14 | 49 | MA1125.1 (ZNF384) | MEME-2 | AAAAAAAAAAARAAA | AAAAAAAAAAARAAA | ZNF384 | 0.619 | 0.0442 | 28 | 1.01 | 533 | 0.7 | 1.5 | EAR_close |
| 15 | 50 | MA1581.1 (ZBTB6) | MEME-8 | GGCTCAAGYGATYCT | GGCTCAAGYGATYCT | ZBTB6 | 1.34 | 0.0896 | 124 | 4.48 | 3020 | 3.94 | 1.15 | EAR_close |
| 16 | 51 | GCCTGTARTCCCAGC | MEME-5 | GCCTGTARTCCCAGC | GCCTGTARTCCCAGC | GCCTGTARTCCCAGC | 1.9 | 0.119 | 7 | 0.25 | 111 | 0.14 | 1.98 | EAR_close |
ER_rat <- 1.25
mrc_type <- "EAR_close"
EAR_close_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))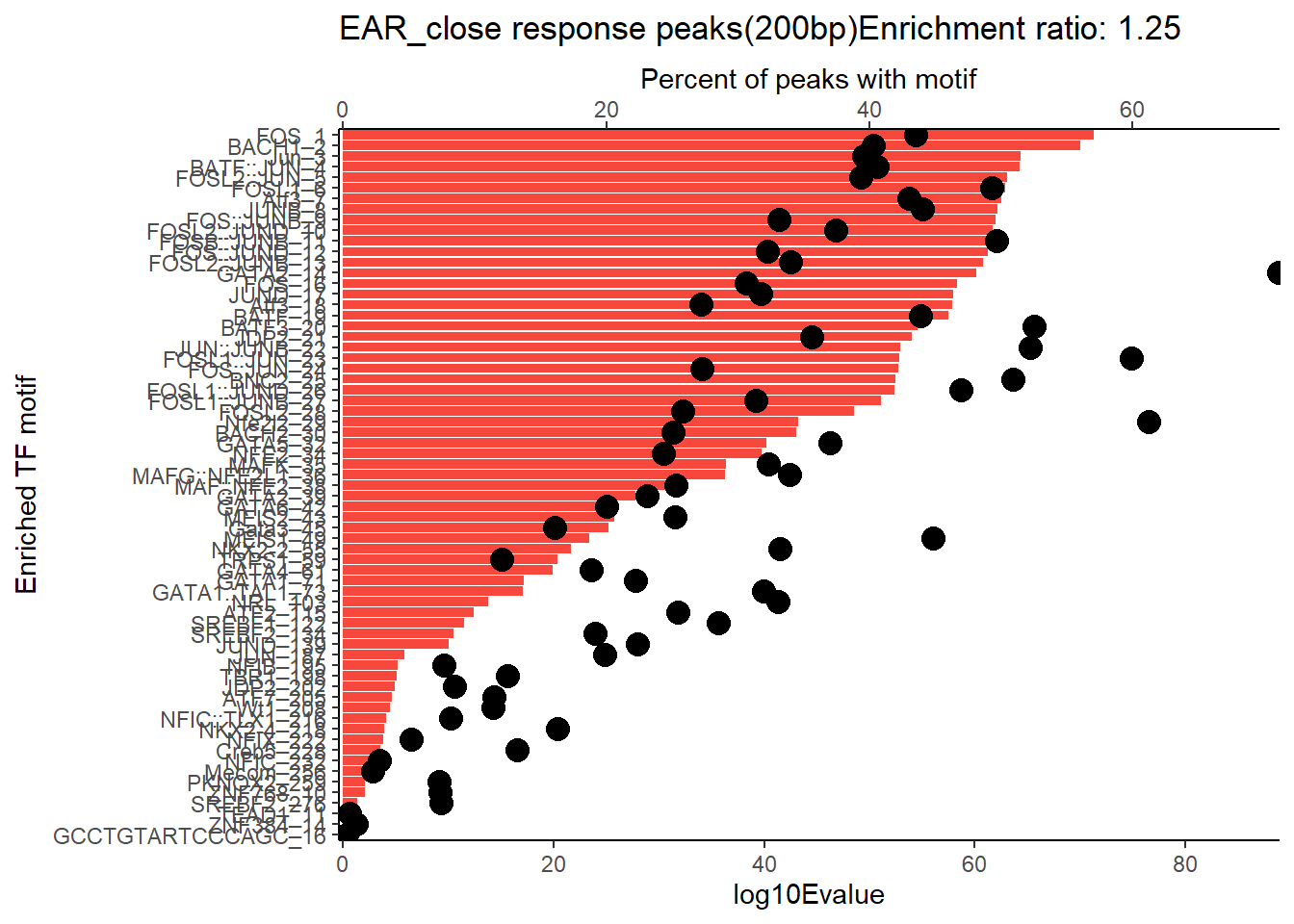
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
EAR_close_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*.8), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.8,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))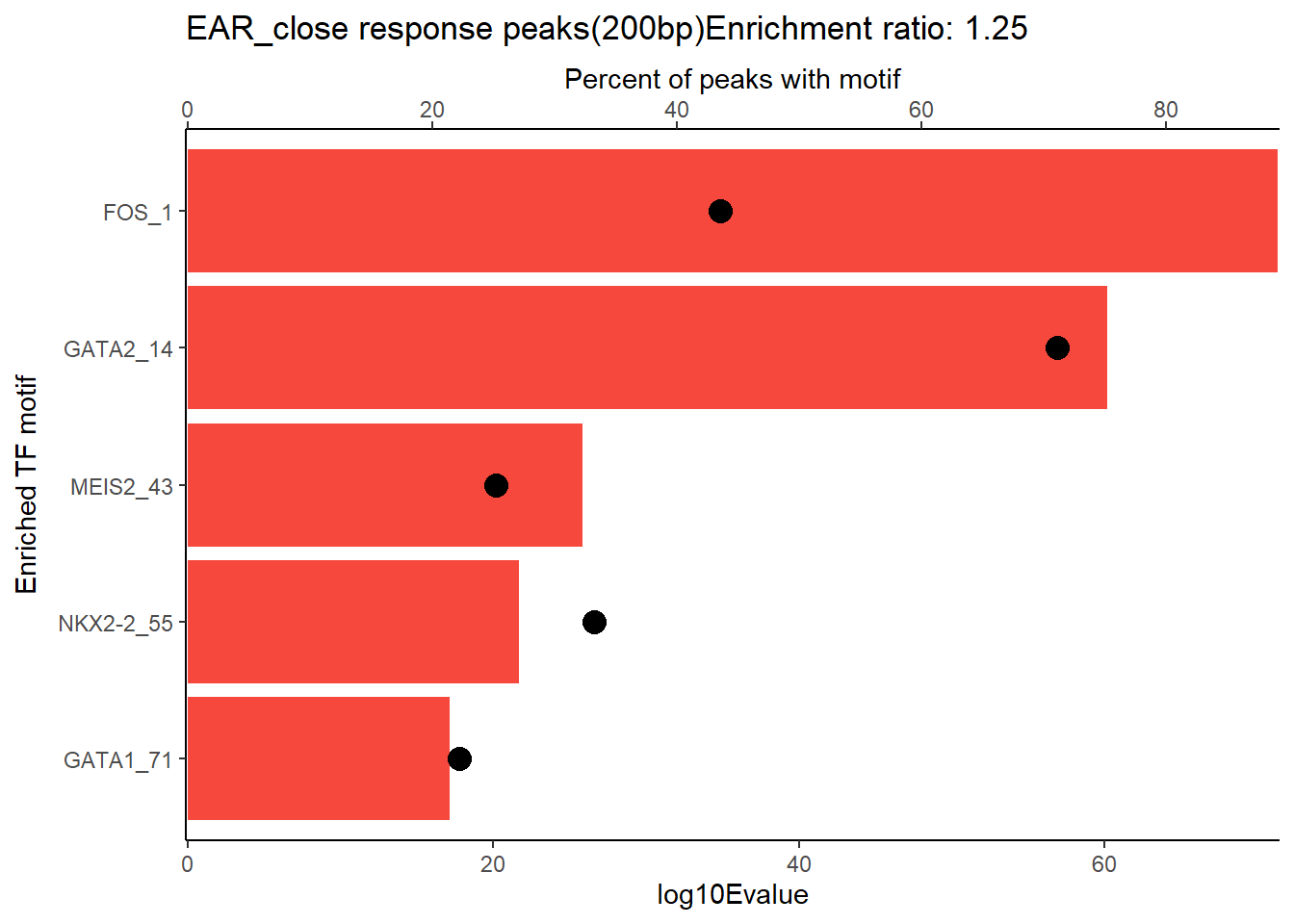
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_open data
ESR_open_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_open_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
ESR_open_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_open_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
ESR_open_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_open_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
ESR_open_sea_all <- rbind(ESR_open_sea_disc, ESR_open_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)ESR_open_combo <- ESR_open_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., ESR_open_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="ESR_open")
ESR_open_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 10-CACACATGCACACAC | STREME-10 | 10-CACACATGCACACAC | 10-CACACATGCACACAC | CACACATGCACACAC | 1.8e-189 | 1.3e-189 | 549 | 9.72 | 1370 | 1.78 | 5.46 | ESR_open |
| 2 | 1 | GTGYRTGTGTGTRTG | MEME-7 | GTGYRTGTGTGTRTG | GTGYRTGTGTGTRTG | GTGYRTGTGTGTRTG | 9.06e-176 | 3.27e-176 | 537 | 9.5 | 1420 | 1.85 | 5.14 | ESR_open |
| 3 | 1 | MA1107.2 (KLF9) | MEME-1 | TGTGTGTGTGTGTGT | TGTGTGTGTGTGTGT | KLF9 | 5.74e-169 | 1.38e-169 | 476 | 8.42 | 1140 | 1.48 | 5.68 | ESR_open |
| 4 | 1 | MA1107.2 (KLF9) | STREME-3 | 3-ATGTGTGTGTG | ATGTGTGTGTG | KLF9 | 8.48e-155 | 1.53e-155 | 499 | 8.83 | 1390 | 1.82 | 4.87 | ESR_open |
| 5 | 1 | 2-CACACACAYACACA | STREME-2 | 2-CACACACAYACACA | CACACACAYACACA | CACACACAYACACA | 1.42e-145 | 2.05e-146 | 409 | 7.24 | 967 | 1.26 | 5.74 | ESR_open |
| 6 | 1 | ACACAYRYACACACA | MEME-8 | ACACAYRYACACACA | ACACAYRYACACACA | ACACAYRYACACACA | 1.82e-144 | 2.19e-145 | 423 | 7.49 | 1050 | 1.37 | 5.46 | ESR_open |
| 7 | 2 | MA1990.1 (Gli1) | STREME-4 | 4-ACACACCACACACAC | ACACACCACACACAC | Gli1 | 3.29e-134 | 3.39e-135 | 267 | 4.73 | 375 | 0.49 | 9.67 | ESR_open |
| 8 | 2 | ACACACCACACACA | MEME-6 | ACACACCACACACA | ACACACCACACACA | ACACACCACACACA | 5.12e-133 | 4.62e-134 | 204 | 3.61 | 164 | 0.21 | 16.9 | ESR_open |
| 9 | 1 | 6-GTGTGTRTG | STREME-6 | 6-GTGTGTRTG | GTGTGTRTG | GTGTGTRTG | 9.95e-132 | 7.98e-133 | 757 | 13.4 | 3510 | 4.58 | 2.93 | ESR_open |
| 10 | 1 | MA1107.2 (KLF9) | MEME-11 | CAYACACACACACAY | CAYACACACACACAY | KLF9 | 1.58e-120 | 1.14e-121 | 360 | 6.37 | 910 | 1.19 | 5.37 | ESR_open |
| 11 | 3 | MA0491.2 (JUND) | STREME-1 | 1-DATGASTCATH | DATGASTCATH | JUND | 3.23e-120 | 2.12e-121 | 698 | 12.4 | 3250 | 4.24 | 2.92 | ESR_open |
| 12 | 3 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 1.1e-103 | 6.59e-105 | 765 | 13.5 | 4130 | 5.39 | 2.51 | ESR_open |
| 13 | 3 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 4.66e-103 | 2.59e-104 | 820 | 14.5 | 4630 | 6.04 | 2.41 | ESR_open |
| 14 | 3 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 4.3e-102 | 2.22e-103 | 785 | 13.9 | 4340 | 5.66 | 2.46 | ESR_open |
| 15 | 3 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 7.98e-99 | 3.84e-100 | 687 | 12.2 | 3570 | 4.65 | 2.62 | ESR_open |
| 16 | 3 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 1.41e-98 | 6.37e-100 | 609 | 10.8 | 2930 | 3.82 | 2.82 | ESR_open |
| 17 | 3 | MA0476.1 (FOS) | MEME-4 | TGASTCAT | TGASTCAT | FOS | 1.78e-98 | 7.55e-100 | 1150 | 20.3 | 7860 | 10.2 | 1.98 | ESR_open |
| 18 | 2 | TGTGTGGTGTGTGTG | MEME-2 | TGTGTGGTGTGTGTG | TGTGTGGTGTGTGTG | TGTGTGGTGTGTGTG | 2.06e-98 | 8.28e-100 | 331 | 5.86 | 949 | 1.24 | 4.74 | ESR_open |
| 19 | 3 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 1.36e-97 | 5.16e-99 | 572 | 10.1 | 2650 | 3.46 | 2.93 | ESR_open |
| 20 | 3 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 2.01e-97 | 7.25e-99 | 719 | 12.7 | 3870 | 5.05 | 2.52 | ESR_open |
| 21 | 3 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 1.95e-95 | 6.72e-97 | 864 | 15.3 | 5210 | 6.8 | 2.25 | ESR_open |
| 22 | 3 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 5.64e-95 | 1.85e-96 | 935 | 16.6 | 5890 | 7.69 | 2.15 | ESR_open |
| 23 | 3 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 6.01e-95 | 1.88e-96 | 699 | 12.4 | 3750 | 4.9 | 2.53 | ESR_open |
| 24 | 3 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 5.57e-94 | 1.67e-95 | 900 | 15.9 | 5590 | 7.29 | 2.19 | ESR_open |
| 25 | 3 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 1.94e-92 | 5.6e-94 | 828 | 14.6 | 4960 | 6.47 | 2.27 | ESR_open |
| 26 | 3 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 5.46e-92 | 1.52e-93 | 819 | 14.5 | 4890 | 6.38 | 2.27 | ESR_open |
| 27 | 3 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 7.93e-92 | 2.12e-93 | 643 | 11.4 | 3340 | 4.36 | 2.61 | ESR_open |
| 28 | 3 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 7.75e-89 | 2e-90 | 1060 | 18.8 | 7320 | 9.55 | 1.97 | ESR_open |
| 29 | 3 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 2.65e-87 | 6.59e-89 | 551 | 9.75 | 2670 | 3.49 | 2.8 | ESR_open |
| 30 | 3 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 2.12e-86 | 5.11e-88 | 1100 | 19.4 | 7720 | 10.1 | 1.92 | ESR_open |
| 31 | 3 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 4.78e-86 | 1.11e-87 | 530 | 9.38 | 2530 | 3.3 | 2.85 | ESR_open |
| 32 | 3 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 4.12e-85 | 9.3e-87 | 493 | 8.73 | 2250 | 2.94 | 2.97 | ESR_open |
| 33 | 3 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 4.93e-82 | 1.08e-83 | 890 | 15.8 | 5830 | 7.61 | 2.07 | ESR_open |
| 34 | 4 | MA1155.1 | ZSCAN4 | MA1155.1 | TGCACACACTGAAAA | ZSCAN4 | 1.85e-81 | 3.92e-83 | 481 | 8.51 | 2220 | 2.9 | 2.94 | ESR_open |
| 35 | 3 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 4.22e-79 | 8.7e-81 | 478 | 8.46 | 2240 | 2.93 | 2.89 | ESR_open |
| 36 | 3 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 5.19e-77 | 1.04e-78 | 707 | 12.5 | 4250 | 5.55 | 2.26 | ESR_open |
| 37 | 3 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 6.28e-76 | 1.23e-77 | 405 | 7.17 | 1740 | 2.27 | 3.17 | ESR_open |
| 38 | 3 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 5.1e-75 | 9.68e-77 | 1210 | 21.5 | 9380 | 12.2 | 1.76 | ESR_open |
| 39 | 3 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 8.47e-73 | 1.57e-74 | 1120 | 19.9 | 8520 | 11.1 | 1.79 | ESR_open |
| 40 | 5 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 6.85e-72 | 1.24e-73 | 519 | 9.19 | 2720 | 3.55 | 2.59 | ESR_open |
| 41 | 1 | 5-CATACA | STREME-5 | 5-CATACA | CATACA | CATACA | 1.45e-63 | 2.56e-65 | 2510 | 44.4 | 25300 | 33 | 1.35 | ESR_open |
| 42 | 3 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 1.08e-61 | 1.85e-63 | 625 | 11.1 | 3910 | 5.1 | 2.17 | ESR_open |
| 43 | 6 | MA0525.2 (TP63) | STREME-11 | 11-ACATGT | 11-ACATGT | TP63 | 1.71e-54 | 2.86e-56 | 3070 | 54.4 | 33300 | 43.4 | 1.25 | ESR_open |
| 44 | 7 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 1.41e-51 | 2.32e-53 | 2910 | 51.5 | 31300 | 40.9 | 1.26 | ESR_open |
| 45 | 8 | 7-ATGTAC | STREME-7 | 7-ATGTAC | ATGTAC | ATGTAC | 9.75e-43 | 1.56e-44 | 2920 | 51.7 | 32200 | 42 | 1.23 | ESR_open |
| 46 | 5 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 8.89e-41 | 1.4e-42 | 742 | 13.1 | 5800 | 7.57 | 1.74 | ESR_open |
| 47 | 9 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 2.58e-38 | 3.96e-40 | 2430 | 43 | 26100 | 34.1 | 1.26 | ESR_open |
| 48 | 6 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 6.52e-30 | 9.8e-32 | 138 | 2.44 | 511 | 0.67 | 3.68 | ESR_open |
| 49 | 10 | MA0006.1 | Ahr::Arnt | MA0006.1 | YGCGTG | Ahr::Arnt | 4.89e-29 | 7.21e-31 | 523 | 9.26 | 4030 | 5.25 | 1.76 | ESR_open |
| 50 | 11 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 2.99e-28 | 4.31e-30 | 3320 | 58.7 | 38900 | 50.8 | 1.16 | ESR_open |
| 51 | 11 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 2.03e-27 | 2.88e-29 | 2270 | 40.2 | 25100 | 32.7 | 1.23 | ESR_open |
| 52 | 12 | MA0073.1 | RREB1 | MA0073.1 | CCCCMAAMCAMCCMCMMMCV | RREB1 | 4.18e-27 | 5.8e-29 | 530 | 9.38 | 4180 | 5.46 | 1.72 | ESR_open |
| 53 | 3 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 4.74e-25 | 6.45e-27 | 549 | 9.72 | 4480 | 5.84 | 1.67 | ESR_open |
| 54 | 11 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 5.51e-25 | 7.36e-27 | 3870 | 68.4 | 46900 | 61.2 | 1.12 | ESR_open |
| 55 | 7 | MA0623.2 | NEUROG1 | MA0623.2 | RACATATGTY | NEUROG1 | 7e-24 | 9.19e-26 | 1550 | 27.4 | 16200 | 21.2 | 1.29 | ESR_open |
| 56 | 13 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 3.79e-23 | 4.89e-25 | 2380 | 42 | 26800 | 35 | 1.2 | ESR_open |
| 57 | 13 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 1.36e-22 | 1.73e-24 | 3480 | 61.6 | 41800 | 54.6 | 1.13 | ESR_open |
| 58 | 14 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 2.54e-22 | 3.16e-24 | 745 | 13.2 | 6790 | 8.85 | 1.49 | ESR_open |
| 59 | 7 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 2.6e-22 | 3.18e-24 | 2210 | 39.2 | 24800 | 32.4 | 1.21 | ESR_open |
| 60 | 11 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 4.79e-22 | 5.76e-24 | 2750 | 48.7 | 31900 | 41.7 | 1.17 | ESR_open |
| 61 | 7 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 1.2e-21 | 1.42e-23 | 3000 | 53.1 | 35300 | 46.1 | 1.15 | ESR_open |
| 62 | 3 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 1e-20 | 1.16e-22 | 520 | 9.2 | 4390 | 5.72 | 1.61 | ESR_open |
| 63 | 7 | MA0819.2 | CLOCK | MA0819.2 | NRACACGTGB | CLOCK | 1.27e-20 | 1.46e-22 | 3070 | 54.3 | 36400 | 47.4 | 1.14 | ESR_open |
| 64 | 9 | MA1718.1 | ZNF8 | MA1718.1 | GTATGGATATACCACAKTTT | ZNF8 | 1.3e-20 | 1.47e-22 | 520 | 9.2 | 4390 | 5.73 | 1.61 | ESR_open |
| 65 | 13 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 1.54e-20 | 1.71e-22 | 2100 | 37.3 | 23600 | 30.8 | 1.21 | ESR_open |
| 66 | 11 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 2.41e-20 | 2.64e-22 | 3080 | 54.5 | 36600 | 47.7 | 1.14 | ESR_open |
| 67 | 11 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 1.03e-18 | 1.1e-20 | 5120 | 90.6 | 66200 | 86.4 | 1.05 | ESR_open |
| 68 | 7 | MA0622.1 | Mlxip | MA0622.1 | BCACGTGK | Mlxip | 1.04e-18 | 1.1e-20 | 93 | 1.65 | 358 | 0.47 | 3.55 | ESR_open |
| 69 | 15 | MA1125.1 (ZNF384) | MEME-3 | WWTTTTTWTWWTTTT | WWTTTTTWTWWTTTT | ZNF384 | 1.75e-18 | 1.83e-20 | 1630 | 28.9 | 17800 | 23.2 | 1.24 | ESR_open |
| 70 | 3 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 1.82e-18 | 1.88e-20 | 2140 | 38 | 24400 | 31.8 | 1.19 | ESR_open |
| 71 | 11 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 1.95e-18 | 1.99e-20 | 2440 | 43.2 | 28300 | 36.9 | 1.17 | ESR_open |
| 72 | 7 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 2.65e-18 | 2.66e-20 | 507 | 8.97 | 4380 | 5.71 | 1.57 | ESR_open |
| 73 | 3 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 4.01e-18 | 3.97e-20 | 1560 | 27.7 | 17000 | 22.2 | 1.25 | ESR_open |
| 74 | 13 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 2.12e-17 | 2.07e-19 | 4400 | 77.9 | 55500 | 72.4 | 1.08 | ESR_open |
| 75 | 13 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 2.73e-17 | 2.62e-19 | 4160 | 73.6 | 51900 | 67.8 | 1.09 | ESR_open |
| 76 | 11 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 8.18e-17 | 7.77e-19 | 4870 | 86.2 | 62500 | 81.5 | 1.06 | ESR_open |
| 77 | 16 | 9-ATKHYWRDMAT | STREME-9 | 9-ATKHYWRDMAT | ATKHYWRDMAT | ATKHYWRDMAT | 1.58e-16 | 1.48e-18 | 1090 | 19.3 | 11300 | 14.7 | 1.31 | ESR_open |
| 78 | 11 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 2.3e-16 | 2.12e-18 | 4720 | 83.6 | 60300 | 78.7 | 1.06 | ESR_open |
| 79 | 11 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 5.79e-16 | 5.29e-18 | 2400 | 42.4 | 28000 | 36.5 | 1.16 | ESR_open |
| 80 | 17 | MA0052.4 (MEF2A) | STREME-8 | 8-AHAAATATTTT | AHAAATATTTT | MEF2A | 6.45e-16 | 5.81e-18 | 728 | 12.9 | 7030 | 9.17 | 1.41 | ESR_open |
| 81 | 3 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 1.31e-15 | 1.16e-17 | 4120 | 72.8 | 51600 | 67.3 | 1.08 | ESR_open |
| 82 | 11 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 1.81e-15 | 1.59e-17 | 1340 | 23.7 | 14400 | 18.8 | 1.26 | ESR_open |
| 83 | 7 | MA0059.1 | MAX::MYC | MA0059.1 | RASCACGTGGT | MAX::MYC | 2.99e-15 | 2.6e-17 | 2040 | 36.1 | 23400 | 30.5 | 1.18 | ESR_open |
| 84 | 11 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 3.82e-15 | 3.27e-17 | 4200 | 74.4 | 52900 | 69 | 1.08 | ESR_open |
| 85 | 11 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 3.85e-15 | 3.27e-17 | 1640 | 29 | 18300 | 23.8 | 1.22 | ESR_open |
| 86 | 11 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 6.74e-15 | 5.65e-17 | 2550 | 45.1 | 30100 | 39.3 | 1.15 | ESR_open |
| 87 | 11 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 9.6e-15 | 7.96e-17 | 4230 | 74.9 | 53400 | 69.6 | 1.08 | ESR_open |
| 88 | 18 | MA0804.1 | TBX19 | MA0804.1 | DTTMRCACMTAGGTGTGAAW | TBX19 | 7.04e-14 | 5.77e-16 | 1610 | 28.4 | 18000 | 23.5 | 1.21 | ESR_open |
| 89 | 11 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 3.59e-13 | 2.91e-15 | 728 | 12.9 | 7250 | 9.46 | 1.36 | ESR_open |
| 90 | 7 | MA0818.2 | BHLHE22 | MA0818.2 | AMCATATGKY | BHLHE22 | 6.01e-13 | 4.82e-15 | 1720 | 30.4 | 19500 | 25.5 | 1.19 | ESR_open |
| 91 | 7 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 1.36e-12 | 1.08e-14 | 1960 | 34.8 | 22800 | 29.7 | 1.17 | ESR_open |
| 92 | 11 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 2.83e-12 | 2.22e-14 | 2420 | 42.9 | 28800 | 37.6 | 1.14 | ESR_open |
| 93 | 11 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 7.65e-12 | 5.94e-14 | 2680 | 47.5 | 32400 | 42.2 | 1.12 | ESR_open |
| 94 | 7 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 1.34e-11 | 1.03e-13 | 172 | 3.04 | 1200 | 1.56 | 1.96 | ESR_open |
| 95 | 7 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 1.97e-11 | 1.5e-13 | 123 | 2.18 | 744 | 0.97 | 2.26 | ESR_open |
| 96 | 19 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 4.16e-11 | 3.12e-13 | 2020 | 35.7 | 23700 | 30.9 | 1.16 | ESR_open |
| 97 | 11 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 5.6e-11 | 4.17e-13 | 2090 | 37 | 24700 | 32.2 | 1.15 | ESR_open |
| 98 | 13 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 5.83e-11 | 4.29e-13 | 3940 | 69.7 | 49700 | 64.9 | 1.07 | ESR_open |
| 99 | 11 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 6.5e-11 | 4.74e-13 | 4120 | 72.9 | 52300 | 68.2 | 1.07 | ESR_open |
| 100 | 20 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 6.95e-11 | 5.01e-13 | 3810 | 67.4 | 47900 | 62.5 | 1.08 | ESR_open |
| 101 | 11 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 7.26e-11 | 5.19e-13 | 4970 | 88 | 64700 | 84.4 | 1.04 | ESR_open |
| 102 | 11 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 9.65e-11 | 6.83e-13 | 5370 | 95.1 | 71000 | 92.6 | 1.03 | ESR_open |
| 103 | 21 | CTCCCRARKWGCTGG | MEME-5 | CTCCCRARKWGCTGG | CTCCCRARKWGCTGG | CTCCCRARKWGCTGG | 1.42e-10 | 9.93e-13 | 181 | 3.2 | 1320 | 1.72 | 1.87 | ESR_open |
| 104 | 7 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 2.16e-10 | 1.5e-12 | 274 | 4.85 | 2290 | 2.99 | 1.63 | ESR_open |
| 105 | 22 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 2.25e-10 | 1.55e-12 | 3960 | 70.1 | 50200 | 65.5 | 1.07 | ESR_open |
| 106 | 18 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 2.91e-10 | 1.98e-12 | 3840 | 68 | 48500 | 63.3 | 1.07 | ESR_open |
| 107 | 23 | MA1602.1 | ZSCAN29 | MA1602.1 | YGTCTACRCNGN | ZSCAN29 | 3.97e-10 | 2.68e-12 | 1830 | 32.4 | 21400 | 27.9 | 1.16 | ESR_open |
| 108 | 11 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 4.18e-10 | 2.8e-12 | 1750 | 30.9 | 20300 | 26.5 | 1.17 | ESR_open |
| 109 | 3 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 5.13e-10 | 3.4e-12 | 2880 | 51 | 35300 | 46 | 1.11 | ESR_open |
| 110 | 3 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 7.31e-10 | 4.8e-12 | 2210 | 39.2 | 26400 | 34.5 | 1.14 | ESR_open |
| 111 | 11 | MA0030.1 | FOXF2 | MA0030.1 | BNAASGTAAACAAD | FOXF2 | 7.73e-10 | 5.03e-12 | 1230 | 21.7 | 13700 | 17.9 | 1.22 | ESR_open |
| 112 | 19 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 1.86e-09 | 1.2e-11 | 1430 | 25.4 | 16400 | 21.4 | 1.19 | ESR_open |
| 113 | 7 | MA0817.1 | BHLHE23 | MA0817.1 | AAACATATGBTN | BHLHE23 | 5.14e-09 | 3.28e-11 | 1150 | 20.3 | 12800 | 16.7 | 1.22 | ESR_open |
| 114 | 2 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 8.81e-09 | 5.58e-11 | 4540 | 80.4 | 58700 | 76.6 | 1.05 | ESR_open |
| 115 | 7 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 2.16e-08 | 1.35e-10 | 205 | 3.63 | 1660 | 2.17 | 1.68 | ESR_open |
| 116 | 18 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 3.28e-08 | 2.04e-10 | 524 | 9.27 | 5270 | 6.88 | 1.35 | ESR_open |
| 117 | 24 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 3.5e-08 | 2.16e-10 | 1660 | 29.4 | 19500 | 25.4 | 1.16 | ESR_open |
| 118 | 25 | TGGCTCAYGCCTGTA | MEME-9 | TGGCTCAYGCCTGTA | TGGCTCAYGCCTGTA | TGGCTCAYGCCTGTA | 7.52e-08 | 4.6e-10 | 178 | 3.15 | 1410 | 1.83 | 1.73 | ESR_open |
| 119 | 11 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 8.28e-08 | 5.02e-10 | 3080 | 54.5 | 38400 | 50.1 | 1.09 | ESR_open |
| 120 | 7 | MA1493.1 | HES6 | MA1493.1 | GGCACGTGTY | HES6 | 1.79e-07 | 1.08e-09 | 489 | 8.65 | 4920 | 6.42 | 1.35 | ESR_open |
| 121 | 11 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 5.6e-07 | 3.34e-09 | 2200 | 38.8 | 26700 | 34.8 | 1.12 | ESR_open |
| 122 | 14 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 8.1e-07 | 4.79e-09 | 1000 | 17.7 | 11300 | 14.7 | 1.21 | ESR_open |
| 123 | 26 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 1.35e-06 | 7.89e-09 | 777 | 13.8 | 8500 | 11.1 | 1.24 | ESR_open |
| 124 | 7 | MA0825.1 | MNT | MA0825.1 | RVCACGTGMH | MNT | 2.3e-06 | 1.34e-08 | 3630 | 64.3 | 46200 | 60.4 | 1.07 | ESR_open |
| 125 | 11 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 4.25e-06 | 2.45e-08 | 4320 | 76.5 | 56000 | 73.1 | 1.05 | ESR_open |
| 126 | 7 | MA0464.2 | BHLHE40 | MA0464.2 | VKCACGTGMC | BHLHE40 | 5.09e-06 | 2.92e-08 | 172 | 3.04 | 1430 | 1.87 | 1.64 | ESR_open |
| 127 | 21 | 12-GCACTTTGGGAGGC | STREME-12 | 12-GCACTTTGGGAGGC | 12-GCACTTTGGGAGGC | GCACTTTGGGAGGC | 5.58e-06 | 3.17e-08 | 65 | 1.15 | 377 | 0.49 | 2.37 | ESR_open |
| 128 | 11 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 5.91e-06 | 3.33e-08 | 1940 | 34.4 | 23600 | 30.7 | 1.12 | ESR_open |
| 129 | 20 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 6.92e-06 | 3.87e-08 | 3290 | 58.3 | 41700 | 54.4 | 1.07 | ESR_open |
| 130 | 7 | MA0871.2 | TFEC | MA0871.2 | NVCACGTGACN | TFEC | 1.09e-05 | 6.05e-08 | 1320 | 23.4 | 15500 | 20.3 | 1.16 | ESR_open |
| 131 | 3 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 1.18e-05 | 6.5e-08 | 4300 | 76 | 55700 | 72.7 | 1.05 | ESR_open |
| 132 | 7 | MA0608.1 | Creb3l2 | MA0608.1 | GCCACGTGD | Creb3l2 | 1.35e-05 | 7.38e-08 | 511 | 9.04 | 5370 | 7 | 1.29 | ESR_open |
| 133 | 20 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 1.73e-05 | 9.4e-08 | 3530 | 62.5 | 45000 | 58.8 | 1.06 | ESR_open |
| 134 | 7 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 2.65e-05 | 1.43e-07 | 1540 | 27.3 | 18500 | 24.1 | 1.14 | ESR_open |
| 135 | 2 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 2.75e-05 | 1.47e-07 | 358 | 6.34 | 3580 | 4.67 | 1.36 | ESR_open |
| 136 | 7 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 2.97e-05 | 1.58e-07 | 276 | 4.88 | 2630 | 3.43 | 1.43 | ESR_open |
| 137 | 7 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 5.08e-05 | 2.68e-07 | 238 | 4.21 | 2220 | 2.89 | 1.46 | ESR_open |
| 138 | 7 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 6.86e-05 | 3.59e-07 | 299 | 5.29 | 2920 | 3.82 | 1.39 | ESR_open |
| 139 | 7 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 7.65e-05 | 3.97e-07 | 2170 | 38.4 | 26800 | 35 | 1.1 | ESR_open |
| 140 | 11 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 7.72e-05 | 3.98e-07 | 1830 | 32.5 | 22300 | 29.1 | 1.11 | ESR_open |
| 141 | 18 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 9.54e-05 | 4.88e-07 | 1220 | 21.6 | 14400 | 18.7 | 1.15 | ESR_open |
| 142 | 11 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 0.000128 | 6.5e-07 | 3190 | 56.4 | 40500 | 52.9 | 1.07 | ESR_open |
| 143 | 7 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 0.000204 | 1.03e-06 | 3000 | 53.1 | 38000 | 49.6 | 1.07 | ESR_open |
| 144 | 14 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 0.000226 | 1.13e-06 | 528 | 9.35 | 5710 | 7.45 | 1.26 | ESR_open |
| 145 | 7 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 0.000259 | 1.28e-06 | 1170 | 20.7 | 13800 | 18 | 1.15 | ESR_open |
| 146 | 7 | MA0826.1 | OLIG1 | MA0826.1 | AMCATATGKT | OLIG1 | 0.00026 | 1.28e-06 | 536 | 9.49 | 5820 | 7.59 | 1.25 | ESR_open |
| 147 | 7 | MA0058.3 | MAX | MA0058.3 | AVCACGTGNY | MAX | 0.000303 | 1.49e-06 | 2760 | 48.8 | 34800 | 45.4 | 1.08 | ESR_open |
| 148 | 7 | MA1559.1 | SNAI3 | MA1559.1 | RRCAGGTGYA | SNAI3 | 0.000437 | 2.12e-06 | 1540 | 27.3 | 18600 | 24.3 | 1.12 | ESR_open |
| 149 | 13 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 0.000439 | 2.12e-06 | 2300 | 40.8 | 28700 | 37.5 | 1.09 | ESR_open |
| 150 | 13 | MA0619.1 | LIN54 | MA0619.1 | DTTTRAATH | LIN54 | 0.000714 | 3.44e-06 | 750 | 13.3 | 8530 | 11.1 | 1.19 | ESR_open |
| 151 | 7 | MA1621.1 | Rbpjl | MA1621.1 | NNVACACCTGTBNN | Rbpjl | 0.000781 | 3.73e-06 | 1120 | 19.8 | 13200 | 17.2 | 1.15 | ESR_open |
| 152 | 24 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 0.000829 | 3.94e-06 | 4610 | 81.6 | 60500 | 78.9 | 1.03 | ESR_open |
| 153 | 26 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 0.00105 | 4.96e-06 | 861 | 15.2 | 9960 | 13 | 1.17 | ESR_open |
| 154 | 3 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 0.00118 | 5.52e-06 | 2280 | 40.4 | 28600 | 37.2 | 1.08 | ESR_open |
| 155 | 7 | MA0603.1 | Arntl | MA0603.1 | NGTCACGTGH | Arntl | 0.00129 | 5.99e-06 | 398 | 7.04 | 4220 | 5.5 | 1.28 | ESR_open |
| 156 | 2 | MA0002.2 | Runx1 | MA0002.2 | BBYTGTGGTTT | Runx1 | 0.00151 | 6.96e-06 | 3710 | 65.6 | 47900 | 62.5 | 1.05 | ESR_open |
| 157 | 27 | 13-GTCTCTACTAAAA | STREME-13 | 13-GTCTCTACTAAAA | 13-GTCTCTACTAAAA | GTCTCTACTAAAA | 0.00163 | 7.49e-06 | 221 | 3.91 | 2140 | 2.79 | 1.41 | ESR_open |
| 158 | 3 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 0.00164 | 7.49e-06 | 2630 | 46.6 | 33300 | 43.4 | 1.07 | ESR_open |
| 159 | 14 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 0.00263 | 1.19e-05 | 501 | 8.87 | 5510 | 7.19 | 1.24 | ESR_open |
| 160 | 28 | MA1546.1 | PAX3 | MA1546.1 | YSGTCACGSYWATTAN | PAX3 | 0.00283 | 1.28e-05 | 3270 | 57.8 | 42000 | 54.7 | 1.06 | ESR_open |
| 161 | 29 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 0.00313 | 1.4e-05 | 3020 | 53.4 | 38600 | 50.3 | 1.06 | ESR_open |
| 162 | 14 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 0.00322 | 1.44e-05 | 2010 | 35.6 | 25100 | 32.7 | 1.09 | ESR_open |
| 163 | 30 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 0.00551 | 2.44e-05 | 1730 | 30.6 | 21400 | 27.9 | 1.1 | ESR_open |
| 164 | 7 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 0.00597 | 2.63e-05 | 2260 | 39.9 | 28400 | 37 | 1.08 | ESR_open |
| 165 | 31 | MA1580.1 | ZBTB32 | MA1580.1 | TGTACAGTAW | ZBTB32 | 0.0132 | 5.78e-05 | 1340 | 23.8 | 16400 | 21.4 | 1.11 | ESR_open |
| 24 | 34 | TTGAGSYCAGGAGTT | MEME-12 | TTGAGSYCAGGAGTT | TTGAGSYCAGGAGTT | TTGAGSYCAGGAGTT | 0.0135 | 0.000562 | 46 | 0.81 | 357 | 0.47 | 1.78 | ESR_open |
| 166 | 22 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 0.0151 | 6.56e-05 | 5140 | 91.1 | 68500 | 89.3 | 1.02 | ESR_open |
| 167 | 32 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 0.0174 | 7.53e-05 | 3570 | 63.2 | 46300 | 60.4 | 1.05 | ESR_open |
| 168 | 7 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 0.0192 | 8.23e-05 | 3140 | 55.6 | 40500 | 52.8 | 1.05 | ESR_open |
| 169 | 19 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 0.0283 | 0.000121 | 2080 | 36.8 | 26200 | 34.1 | 1.08 | ESR_open |
| 170 | 19 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 0.0285 | 0.000121 | 589 | 10.4 | 6760 | 8.81 | 1.18 | ESR_open |
| 171 | 32 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 0.0308 | 0.00013 | 4570 | 80.8 | 60200 | 78.6 | 1.03 | ESR_open |
| 172 | 33 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 0.0449 | 0.000188 | 1450 | 25.6 | 17900 | 23.3 | 1.1 | ESR_open |
| 173 | 18 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 0.0483 | 0.000201 | 368 | 6.51 | 4040 | 5.27 | 1.24 | ESR_open |
| 25 | 35 | MA0090.3 (TEAD1) | MEME-10 | AGGCTGGAGTGCAGT | AGGCTGGAGTGCAGT | TEAD1 | 2.57 | 0.103 | 177 | 3.13 | 2170 | 2.83 | 1.11 | ESR_open |
ER_rat <- 1.25
mrc_type <- "ESR_open"
ESR_open_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))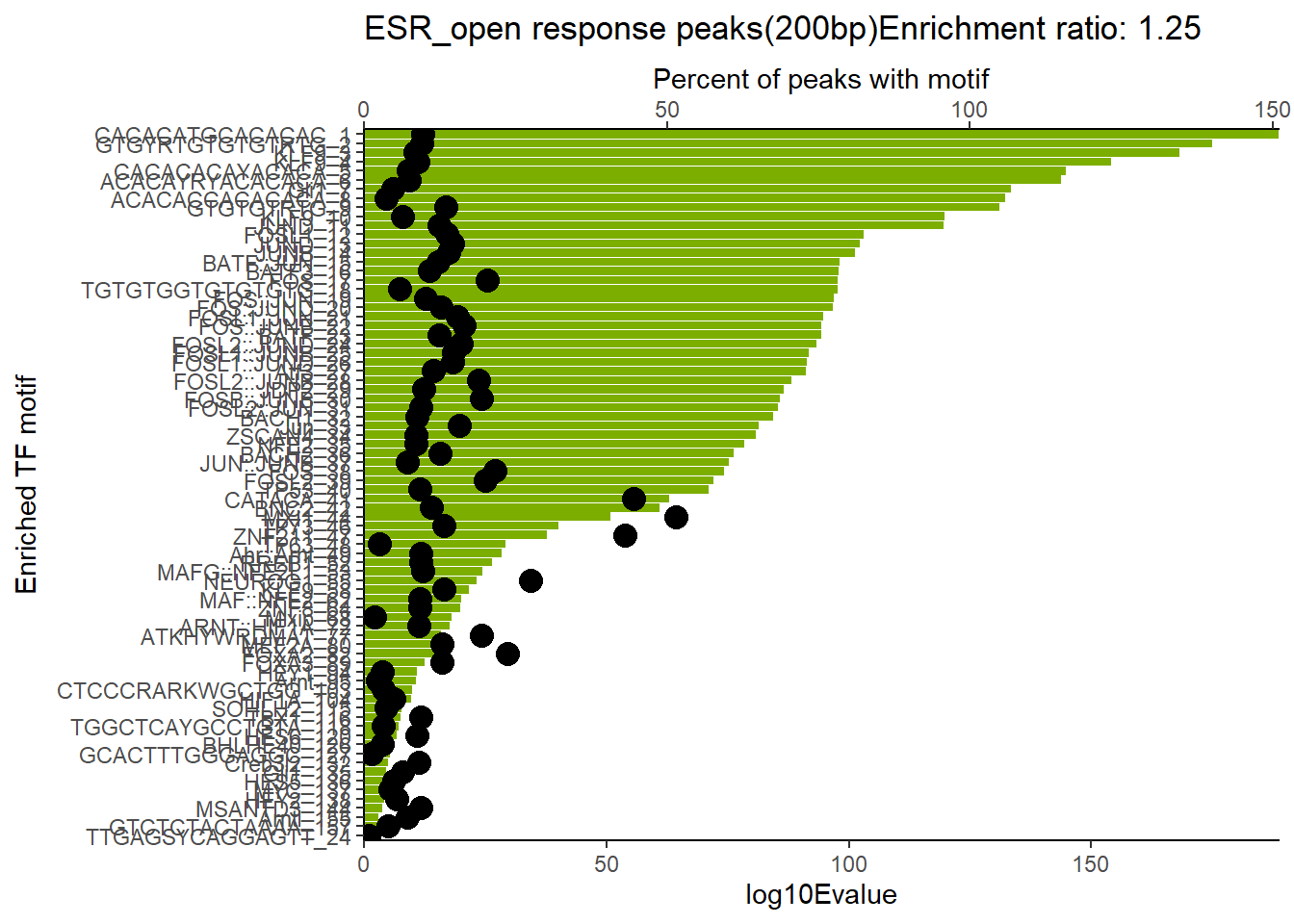
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_open_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*10), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./10,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))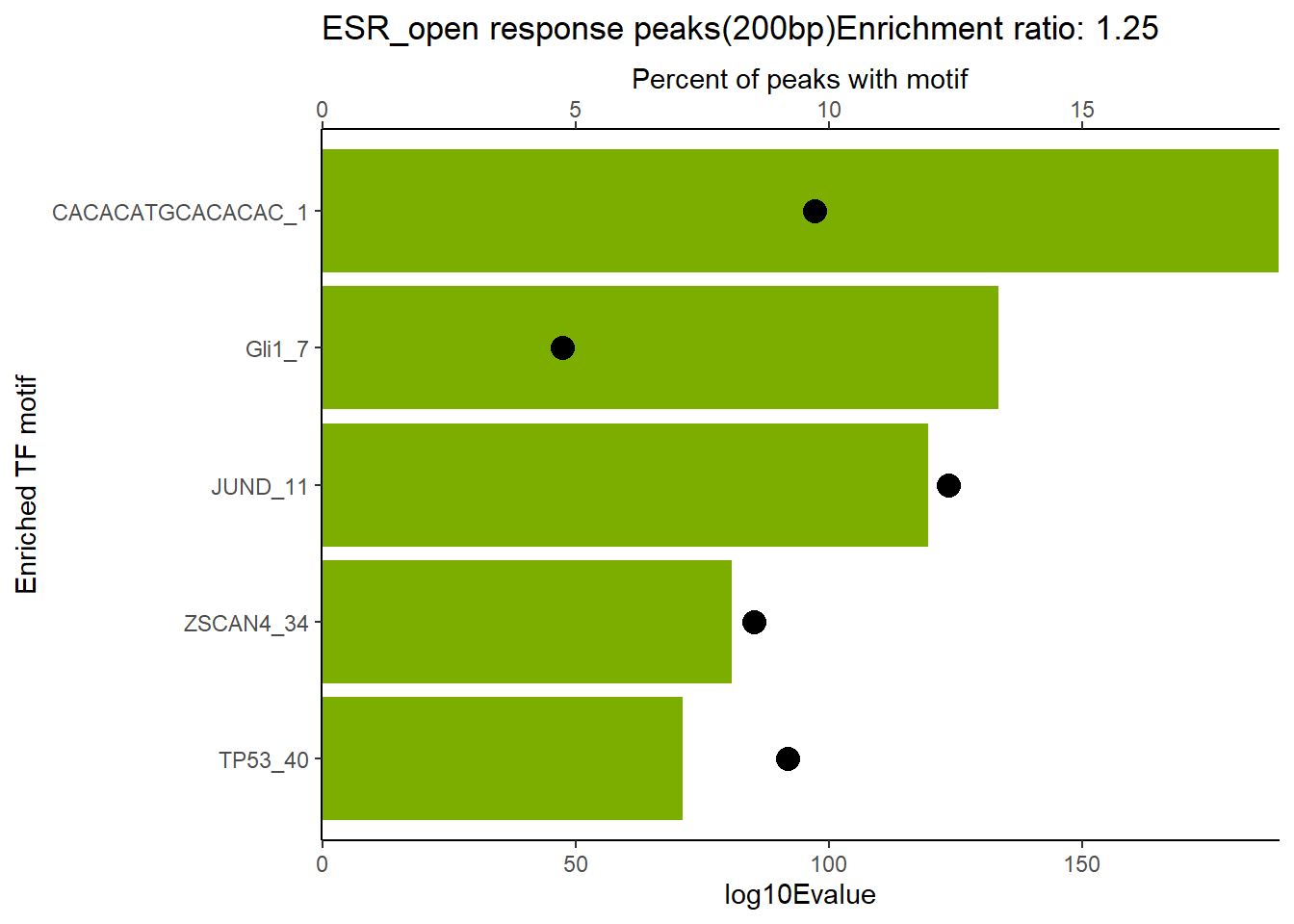
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_close data
ESR_close_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_close_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
ESR_close_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_close_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
ESR_close_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_close_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
ESR_close_sea_all <- rbind(ESR_close_sea_disc, ESR_close_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)ESR_close_combo <- ESR_close_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., ESR_close_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="ESR_close")
ESR_close_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0774.1 (MEIS2) | STREME-1 | 1-CTGWCAG | CTGWCAG | MEIS2 | 2.24e-135 | 1.58e-135 | 4660 | 65.3 | 38300 | 49.9 | 1.31 | ESR_close |
| 2 | 1 | MA0797.1 (TGIF2) | STREME-10 | 10-SCTGTCA | 10-SCTGTCA | TGIF2 | 4.08e-118 | 1.44e-118 | 2640 | 37 | 18400 | 24 | 1.54 | ESR_close |
| 3 | 2 | MA1653.1 (ZNF148) | STREME-2 | 2-CCCWSCCC | CCCWSCCC | ZNF148 | 1.01e-112 | 2.38e-113 | 5030 | 70.4 | 43500 | 56.8 | 1.24 | ESR_close |
| 4 | 1 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 5.18e-111 | 9.15e-112 | 3170 | 44.3 | 23700 | 30.9 | 1.43 | ESR_close |
| 5 | 3 | MA1565.1 (TBX18) | STREME-6 | 6-CACCTCY | CACCTCY | TBX18 | 7.08e-93 | 1e-93 | 2870 | 40.2 | 21600 | 28.2 | 1.43 | ESR_close |
| 6 | 2 | MA1522.1 (MAZ) | MEME-1 | GRGGWGGRGGSAGGG | GRGGWGGRGGSAGGG | MAZ | 3.53e-90 | 4.16e-91 | 2790 | 39.1 | 21000 | 27.4 | 1.43 | ESR_close |
| 7 | 4 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 8.13e-84 | 8.2e-85 | 2890 | 40.4 | 22200 | 29 | 1.4 | ESR_close |
| 8 | 4 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 4.77e-70 | 4.21e-71 | 2520 | 35.3 | 19300 | 25.2 | 1.4 | ESR_close |
| 9 | 5 | 3-CAGSCTG | STREME-3 | 3-CAGSCTG | CAGSCTG | CAGSCTG | 1.78e-67 | 1.32e-68 | 2560 | 35.9 | 19900 | 25.9 | 1.39 | ESR_close |
| 10 | 6 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 1.87e-67 | 1.32e-68 | 813 | 11.4 | 4300 | 5.61 | 2.03 | ESR_close |
| 11 | 7 | MA0808.1 (TEAD3) | MEME-3 | SWGGVAKG | SWGGVAKG | TEAD3 | 2.03e-65 | 1.31e-66 | 4150 | 58.1 | 36300 | 47.4 | 1.23 | ESR_close |
| 12 | 8 | MA1645.1 (NKX2-2) | STREME-11 | 11-CCTCTCA | 11-CCTCTCA | NKX2-2 | 1.84e-62 | 1.08e-63 | 5290 | 74 | 49200 | 64.2 | 1.15 | ESR_close |
| 13 | 4 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 2.3e-60 | 1.25e-61 | 1070 | 15 | 6600 | 8.61 | 1.75 | ESR_close |
| 14 | 4 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 5.84e-57 | 2.95e-58 | 3960 | 55.5 | 34800 | 45.4 | 1.22 | ESR_close |
| 15 | 1 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 6.44e-57 | 3.03e-58 | 2220 | 31 | 17100 | 22.3 | 1.39 | ESR_close |
| 16 | 1 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 8.27e-57 | 3.65e-58 | 4000 | 56 | 35200 | 45.9 | 1.22 | ESR_close |
| 17 | 9 | GCYCTGRG | MEME-9 | GCYCTGRG | GCYCTGRG | GCYCTGRG | 3.94e-56 | 1.64e-57 | 2870 | 40.2 | 23500 | 30.7 | 1.31 | ESR_close |
| 18 | 4 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 1.37e-55 | 5.38e-57 | 2600 | 36.5 | 20900 | 27.3 | 1.34 | ESR_close |
| 19 | 6 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 1.78e-55 | 6.6e-57 | 828 | 11.6 | 4750 | 6.2 | 1.87 | ESR_close |
| 20 | 4 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 7.04e-54 | 2.48e-55 | 6200 | 86.8 | 60700 | 79.2 | 1.09 | ESR_close |
| 21 | 4 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 1.4e-53 | 4.71e-55 | 2290 | 32.1 | 18000 | 23.5 | 1.37 | ESR_close |
| 22 | 10 | MA1721.1 (ZNF93) | MEME-2 | CWGCAGCH | CWGCAGCH | ZNF93 | 3.38e-53 | 1.08e-54 | 3480 | 48.8 | 30000 | 39.2 | 1.25 | ESR_close |
| 23 | 4 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 9.97e-53 | 3.06e-54 | 4470 | 62.6 | 40600 | 53 | 1.18 | ESR_close |
| 24 | 11 | 4-AGGCCAG | STREME-4 | 4-AGGCCAG | AGGCCAG | AGGCCAG | 2.38e-52 | 6.99e-54 | 6140 | 86 | 60200 | 78.5 | 1.1 | ESR_close |
| 25 | 4 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 2.12e-50 | 5.99e-52 | 4450 | 62.4 | 40600 | 53 | 1.18 | ESR_close |
| 26 | 12 | MA1726.1 (ZNF331) | STREME-8 | 8-AGKGCC | AGKGCC | ZNF331 | 2.46e-50 | 6.67e-52 | 2400 | 33.6 | 19200 | 25 | 1.34 | ESR_close |
| 27 | 4 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 2.88e-50 | 7.54e-52 | 4310 | 60.3 | 39000 | 50.9 | 1.19 | ESR_close |
| 28 | 4 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 8.38e-50 | 2.11e-51 | 4540 | 63.6 | 41600 | 54.3 | 1.17 | ESR_close |
| 29 | 4 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 1.54e-48 | 3.75e-50 | 4240 | 59.4 | 38400 | 50.2 | 1.19 | ESR_close |
| 30 | 4 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 4.47e-48 | 1.05e-49 | 2730 | 38.2 | 22600 | 29.5 | 1.29 | ESR_close |
| 31 | 13 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 3.63e-46 | 8.27e-48 | 6280 | 87.9 | 62200 | 81.2 | 1.08 | ESR_close |
| 32 | 4 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 1.78e-45 | 3.92e-47 | 4160 | 58.3 | 37800 | 49.3 | 1.18 | ESR_close |
| 33 | 14 | MA0672.1 (NKX2-3) | STREME-7 | 7-CCACTTGA | CCACTTGA | NKX2-3 | 5.33e-45 | 1.14e-46 | 941 | 13.2 | 6040 | 7.87 | 1.67 | ESR_close |
| 34 | 4 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 1.6e-43 | 3.31e-45 | 4400 | 61.7 | 40600 | 52.9 | 1.17 | ESR_close |
| 35 | 4 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 4.24e-43 | 8.55e-45 | 4650 | 65.1 | 43300 | 56.5 | 1.15 | ESR_close |
| 36 | 15 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 9.76e-43 | 1.88e-44 | 4090 | 57.3 | 37200 | 48.6 | 1.18 | ESR_close |
| 37 | 4 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 9.84e-43 | 1.88e-44 | 4340 | 60.8 | 39900 | 52 | 1.17 | ESR_close |
| 38 | 16 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 3.1e-42 | 5.76e-44 | 5440 | 76.2 | 52400 | 68.4 | 1.12 | ESR_close |
| 39 | 4 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 4.46e-42 | 8.08e-44 | 3120 | 43.7 | 27000 | 35.3 | 1.24 | ESR_close |
| 40 | 14 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 2.56e-40 | 4.51e-42 | 1040 | 14.6 | 7120 | 9.29 | 1.58 | ESR_close |
| 41 | 14 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 1.36e-37 | 2.34e-39 | 1700 | 23.8 | 13300 | 17.3 | 1.38 | ESR_close |
| 42 | 4 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 2.58e-37 | 4.34e-39 | 4900 | 68.6 | 46500 | 60.7 | 1.13 | ESR_close |
| 43 | 17 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 4.97e-37 | 8.17e-39 | 4310 | 60.4 | 40100 | 52.3 | 1.16 | ESR_close |
| 44 | 1 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 3.19e-36 | 5.13e-38 | 2200 | 30.9 | 18200 | 23.8 | 1.3 | ESR_close |
| 45 | 4 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 3.57e-35 | 5.6e-37 | 6220 | 87.1 | 62200 | 81.2 | 1.07 | ESR_close |
| 46 | 18 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 9.15e-34 | 1.4e-35 | 5980 | 83.7 | 59400 | 77.5 | 1.08 | ESR_close |
| 47 | 19 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 1.57e-33 | 2.35e-35 | 4850 | 67.9 | 46300 | 60.4 | 1.12 | ESR_close |
| 48 | 20 | MA1728.1 (ZNF549) | STREME-16 | 16-CTGCCCAC | 16-CTGCCCAC | ZNF549 | 1.04e-32 | 1.53e-34 | 3210 | 45 | 28700 | 37.4 | 1.2 | ESR_close |
| 49 | 4 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 6.01e-32 | 8.66e-34 | 5600 | 78.4 | 55000 | 71.7 | 1.09 | ESR_close |
| 50 | 37 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 1.05e-30 | 1.49e-32 | 3190 | 44.6 | 28600 | 37.3 | 1.2 | ESR_close |
| 51 | 21 | MA1981.1 (ZNF530) | MEME-5 | CAGRGSCWGGG | CAGRGSCWGGG | ZNF530 | 2.47e-30 | 3.43e-32 | 948 | 13.3 | 6720 | 8.77 | 1.51 | ESR_close |
| 52 | 22 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 1.13e-29 | 1.54e-31 | 5660 | 79.3 | 55900 | 72.9 | 1.09 | ESR_close |
| 53 | 23 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 2.83e-29 | 3.77e-31 | 4280 | 59.9 | 40400 | 52.7 | 1.14 | ESR_close |
| 54 | 24 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 1.12e-28 | 1.47e-30 | 6560 | 91.8 | 66900 | 87.3 | 1.05 | ESR_close |
| 55 | 4 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 1.24e-28 | 1.59e-30 | 4030 | 56.4 | 37700 | 49.2 | 1.15 | ESR_close |
| 56 | 6 | MA0739.1 | Hic1 | MA0739.1 | RTGCCAACY | Hic1 | 3.74e-28 | 4.72e-30 | 2440 | 34.1 | 21100 | 27.6 | 1.24 | ESR_close |
| 57 | 25 | MA0154.4 (EBF1) | STREME-9 | 9-CCCWGGG | CCCWGGG | EBF1 | 7.82e-28 | 9.69e-30 | 901 | 12.6 | 6430 | 8.39 | 1.51 | ESR_close |
| 58 | 26 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 9.18e-28 | 1.12e-29 | 6560 | 91.8 | 67000 | 87.4 | 1.05 | ESR_close |
| 59 | 27 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 1.32e-26 | 1.58e-28 | 5630 | 78.8 | 55800 | 72.8 | 1.08 | ESR_close |
| 60 | 28 | MA1535.1 | NR2C1 | MA1535.1 | NRRGGTCAN | NR2C1 | 7.04e-26 | 8.15e-28 | 6250 | 87.6 | 63300 | 82.6 | 1.06 | ESR_close |
| 61 | 29 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 7.04e-26 | 8.15e-28 | 4230 | 59.3 | 40200 | 52.4 | 1.13 | ESR_close |
| 62 | 24 | MA0522.3 | TCF3 | MA0522.3 | NVCACCTGCNN | TCF3 | 8.11e-26 | 9.24e-28 | 6780 | 94.9 | 70000 | 91.4 | 1.04 | ESR_close |
| 63 | 30 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 1.26e-25 | 1.41e-27 | 4840 | 67.8 | 46900 | 61.2 | 1.11 | ESR_close |
| 64 | 14 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 2.39e-24 | 2.64e-26 | 1110 | 15.5 | 8520 | 11.1 | 1.4 | ESR_close |
| 65 | 24 | MA0816.1 | Ascl2 | MA0816.1 | ARCAGCTGCY | Ascl2 | 5.45e-24 | 5.92e-26 | 4590 | 64.3 | 44300 | 57.8 | 1.11 | ESR_close |
| 66 | 18 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 1.85e-23 | 1.98e-25 | 6420 | 89.9 | 65500 | 85.5 | 1.05 | ESR_close |
| 67 | 24 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 2.7e-23 | 2.84e-25 | 1290 | 18 | 10200 | 13.4 | 1.35 | ESR_close |
| 68 | 50 | MA1628.1 | Zic1::Zic2 | MA1628.1 | CVCAGCAGGNV | Zic1::Zic2 | 5.31e-23 | 5.52e-25 | 6400 | 89.7 | 65400 | 85.3 | 1.05 | ESR_close |
| 69 | 24 | MA1648.1 | TCF12 | MA1648.1 | NNCACCTGCNN | TCF12 | 5.6e-23 | 5.73e-25 | 6100 | 85.5 | 61700 | 80.5 | 1.06 | ESR_close |
| 70 | 18 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 7.1e-23 | 7.16e-25 | 5870 | 82.2 | 59000 | 76.9 | 1.07 | ESR_close |
| 71 | 13 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 3.07e-22 | 3.05e-24 | 1220 | 17.1 | 9690 | 12.6 | 1.35 | ESR_close |
| 72 | 5 | CCAGSCWG | MEME-6 | CCAGSCWG | CCAGSCWG | CCAGSCWG | 4.12e-22 | 4.05e-24 | 766 | 10.7 | 5520 | 7.2 | 1.49 | ESR_close |
| 73 | 31 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 4.3e-22 | 4.15e-24 | 4770 | 66.7 | 46400 | 60.6 | 1.1 | ESR_close |
| 74 | 32 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 1.09e-21 | 1.04e-23 | 3470 | 48.6 | 32500 | 42.4 | 1.15 | ESR_close |
| 75 | 33 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 3.77e-21 | 3.55e-23 | 4230 | 59.2 | 40600 | 53 | 1.12 | ESR_close |
| 76 | 1 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 1.28e-20 | 1.19e-22 | 3000 | 42 | 27600 | 36 | 1.17 | ESR_close |
| 77 | 4 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 1.47e-20 | 1.35e-22 | 6580 | 92.2 | 67800 | 88.4 | 1.04 | ESR_close |
| 78 | 34 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 5.38e-20 | 4.87e-22 | 6470 | 90.6 | 66400 | 86.6 | 1.05 | ESR_close |
| 79 | 35 | MA0842.2 (NRL) | STREME-13 | 13-AAGTCTGCTGAAGC | 13-AAGTCTGCTGAAGC | NRL | 8.16e-20 | 7.29e-22 | 30 | 0.42 | 13 | 0.02 | 23.8 | ESR_close |
| 80 | 24 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 1.24e-19 | 1.1e-21 | 1110 | 15.5 | 8800 | 11.5 | 1.35 | ESR_close |
| 81 | 36 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 1.43e-19 | 1.25e-21 | 4620 | 64.8 | 45100 | 58.9 | 1.1 | ESR_close |
| 82 | 37 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 2.37e-19 | 2.04e-21 | 2480 | 34.7 | 22300 | 29.2 | 1.19 | ESR_close |
| 83 | 6 | MA0738.1 | HIC2 | MA0738.1 | RTGCCCRSB | HIC2 | 2.57e-19 | 2.18e-21 | 5610 | 78.6 | 56300 | 73.5 | 1.07 | ESR_close |
| 84 | 1 | MA0796.1 | TGIF1 | MA0796.1 | TGACAGCTGTCA | TGIF1 | 2.61e-19 | 2.19e-21 | 240 | 3.36 | 1240 | 1.62 | 2.09 | ESR_close |
| 85 | 24 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 5.47e-19 | 4.54e-21 | 1090 | 15.3 | 8710 | 11.4 | 1.35 | ESR_close |
| 86 | 4 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 9.01e-19 | 7.4e-21 | 3780 | 52.9 | 36000 | 47 | 1.13 | ESR_close |
| 87 | 24 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 1.04e-18 | 8.45e-21 | 3730 | 52.2 | 35500 | 46.4 | 1.13 | ESR_close |
| 88 | 24 | MA0521.2 | Tcf12 | MA0521.2 | NNACAGCTGTNN | Tcf12 | 2.06e-17 | 1.66e-19 | 1370 | 19.2 | 11500 | 15 | 1.28 | ESR_close |
| 89 | 37 | MA0482.2 (GATA4) | STREME-5 | 5-CAGATAAGG | CAGATAAGG | GATA4 | 3.9e-17 | 3.09e-19 | 469 | 6.57 | 3160 | 4.12 | 1.6 | ESR_close |
| 90 | 38 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 4.12e-17 | 3.24e-19 | 6120 | 85.7 | 62400 | 81.4 | 1.05 | ESR_close |
| 91 | 20 | MA1728.1 | ZNF549 | MA1728.1 | NNTGCTGCCCWR | ZNF549 | 5.44e-17 | 4.18e-19 | 6160 | 86.3 | 63000 | 82.2 | 1.05 | ESR_close |
| 92 | 6 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 5.45e-17 | 4.18e-19 | 227 | 3.18 | 1200 | 1.57 | 2.03 | ESR_close |
| 93 | 6 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 5.74e-17 | 4.36e-19 | 2550 | 35.7 | 23400 | 30.5 | 1.17 | ESR_close |
| 94 | 23 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 6.07e-17 | 4.56e-19 | 2850 | 39.9 | 26500 | 34.5 | 1.16 | ESR_close |
| 95 | 23 | MA0103.3 | ZEB1 | MA0103.3 | SNCACCTGSVN | ZEB1 | 9.03e-17 | 6.71e-19 | 6090 | 85.3 | 62200 | 81.1 | 1.05 | ESR_close |
| 96 | 34 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 9.49e-17 | 6.98e-19 | 4190 | 58.6 | 40700 | 53.1 | 1.1 | ESR_close |
| 97 | 24 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 3.02e-16 | 2.2e-18 | 839 | 11.8 | 6520 | 8.5 | 1.38 | ESR_close |
| 98 | 39 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 3.34e-16 | 2.4e-18 | 4780 | 66.9 | 47200 | 61.6 | 1.09 | ESR_close |
| 99 | 23 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 3.98e-16 | 2.84e-18 | 4770 | 66.8 | 47200 | 61.6 | 1.09 | ESR_close |
| 100 | 14 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 6.97e-16 | 4.92e-18 | 3290 | 46.1 | 31200 | 40.7 | 1.13 | ESR_close |
| 101 | 50 | MA0697.2 | Zic3 | MA0697.2 | CNCAGCAGGAGNN | Zic3 | 1.13e-15 | 7.88e-18 | 6460 | 90.5 | 66700 | 87 | 1.04 | ESR_close |
| 102 | 26 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 1.54e-15 | 1.07e-17 | 6570 | 92 | 68100 | 88.8 | 1.04 | ESR_close |
| 103 | 4 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 1.74e-15 | 1.19e-17 | 4480 | 62.7 | 44000 | 57.4 | 1.09 | ESR_close |
| 104 | 24 | MA1652.1 | ZKSCAN5 | MA1652.1 | NNRGGARGTGAGRR | ZKSCAN5 | 2.36e-15 | 1.6e-17 | 6150 | 86.2 | 63000 | 82.2 | 1.05 | ESR_close |
| 105 | 14 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 2.54e-15 | 1.71e-17 | 488 | 6.83 | 3410 | 4.44 | 1.54 | ESR_close |
| 106 | 1 | MA0783.1 | PKNOX2 | MA0783.1 | TGACAGGTGTCA | PKNOX2 | 2.93e-15 | 1.95e-17 | 191 | 2.67 | 980 | 1.28 | 2.1 | ESR_close |
| 107 | 40 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 1.7e-14 | 1.12e-16 | 6750 | 94.6 | 70400 | 91.9 | 1.03 | ESR_close |
| 108 | 41 | MA0145.2 | Tfcp2l1 | MA0145.2 | CCAGYYYVADCCRG | Tfcp2l1 | 3.93e-14 | 2.57e-16 | 6740 | 94.4 | 70300 | 91.7 | 1.03 | ESR_close |
| 109 | 23 | MA0745.2 | SNAI2 | MA0745.2 | NBGCACCTGTMNY | SNAI2 | 5.8e-14 | 3.76e-16 | 6000 | 84.1 | 61400 | 80.1 | 1.05 | ESR_close |
| 110 | 42 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 7.36e-14 | 4.73e-16 | 4550 | 63.7 | 45000 | 58.7 | 1.09 | ESR_close |
| 111 | 34 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 2.51e-13 | 1.6e-15 | 6450 | 90.3 | 66800 | 87.1 | 1.04 | ESR_close |
| 112 | 4 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 3.99e-13 | 2.52e-15 | 5900 | 82.6 | 60200 | 78.6 | 1.05 | ESR_close |
| 113 | 24 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 4.07e-13 | 2.54e-15 | 4400 | 61.6 | 43400 | 56.6 | 1.09 | ESR_close |
| 114 | 31 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 6.97e-13 | 4.31e-15 | 961 | 13.5 | 7910 | 10.3 | 1.31 | ESR_close |
| 115 | 1 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 8.75e-13 | 5.38e-15 | 2850 | 39.9 | 27000 | 35.2 | 1.14 | ESR_close |
| 116 | 43 | MA0751.1 | ZIC4 | MA0751.1 | GRCCCCCCGCKGYGH | ZIC4 | 1.38e-12 | 8.43e-15 | 5260 | 73.6 | 53000 | 69.2 | 1.06 | ESR_close |
| 117 | 28 | MA0504.1 | NR2C2 | MA0504.1 | VGRGGTCARAGGTCA | NR2C2 | 2.07e-12 | 1.25e-14 | 3150 | 44.1 | 30100 | 39.3 | 1.12 | ESR_close |
| 118 | 18 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 2.15e-12 | 1.29e-14 | 4650 | 65.1 | 46300 | 60.4 | 1.08 | ESR_close |
| 119 | 24 | MA1641.1 | MYF5 | MA1641.1 | NVACAGCTGTBN | MYF5 | 9.78e-12 | 5.8e-14 | 2410 | 33.8 | 22500 | 29.4 | 1.15 | ESR_close |
| 120 | 24 | MA0048.2 | NHLH1 | MA0048.2 | CGCAGCTGCK | NHLH1 | 1.46e-11 | 8.6e-14 | 1820 | 25.5 | 16500 | 21.5 | 1.18 | ESR_close |
| 121 | 23 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 1.53e-11 | 8.92e-14 | 5590 | 78.3 | 56900 | 74.2 | 1.05 | ESR_close |
| 122 | 44 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 2.2e-11 | 1.27e-13 | 3620 | 50.6 | 35200 | 46 | 1.1 | ESR_close |
| 123 | 34 | MA1987.1 | ZNF701 | MA1987.1 | GAGCACYAARGGGGGRADDDD | ZNF701 | 3.71e-11 | 2.13e-13 | 5160 | 72.3 | 52100 | 68 | 1.06 | ESR_close |
| 124 | 24 | MA1472.2 | Bhlha15 | MA1472.2 | NVACAGCTGTBN | Bhlha15 | 4.55e-11 | 2.58e-13 | 1850 | 25.9 | 16900 | 22 | 1.18 | ESR_close |
| 125 | 28 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 4.56e-11 | 2.58e-13 | 4490 | 62.9 | 44700 | 58.4 | 1.08 | ESR_close |
| 126 | 45 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 5.73e-11 | 3.21e-13 | 5970 | 83.6 | 61400 | 80.1 | 1.04 | ESR_close |
| 127 | 24 | MA2002.1 | Zfp335 | MA2002.1 | VNTCAGGCANH | Zfp335 | 1.05e-10 | 5.86e-13 | 1910 | 26.8 | 17500 | 22.9 | 1.17 | ESR_close |
| 128 | 23 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 1.64e-10 | 9.04e-13 | 4360 | 61.1 | 43400 | 56.7 | 1.08 | ESR_close |
| 129 | 23 | MA1558.1 | SNAI1 | MA1558.1 | DRCAGGTGYD | SNAI1 | 1.66e-10 | 9.04e-13 | 4750 | 66.5 | 47700 | 62.2 | 1.07 | ESR_close |
| 130 | 24 | MA0500.2 | MYOG | MA0500.2 | NDRCAGCTGYHN | MYOG | 1.66e-10 | 9.04e-13 | 981 | 13.7 | 8300 | 10.8 | 1.27 | ESR_close |
| 131 | 23 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 2.11e-10 | 1.14e-12 | 1760 | 24.7 | 16100 | 21 | 1.18 | ESR_close |
| 132 | 46 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 2.35e-10 | 1.25e-12 | 1380 | 19.3 | 12200 | 15.9 | 1.21 | ESR_close |
| 133 | 47 | 14-CTTATAGATAAAA | STREME-14 | 14-CTTATAGATAAAA | 14-CTTATAGATAAAA | CTTATAGATAAAA | 3.18e-10 | 1.69e-12 | 20 | 0.28 | 15 | 0.02 | 14.1 | ESR_close |
| 134 | 48 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 3.86e-10 | 2.04e-12 | 675 | 9.45 | 5410 | 7.06 | 1.34 | ESR_close |
| 135 | 50 | MA1629.1 | Zic2 | MA1629.1 | NDCACAGCAGGDRG | Zic2 | 4.27e-10 | 2.23e-12 | 6680 | 93.6 | 69900 | 91.2 | 1.03 | ESR_close |
| 136 | 30 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 6.61e-10 | 3.43e-12 | 4830 | 67.6 | 48600 | 63.4 | 1.07 | ESR_close |
| 137 | 30 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 1.04e-09 | 5.36e-12 | 5850 | 81.9 | 60100 | 78.4 | 1.04 | ESR_close |
| 138 | 49 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 1.47e-09 | 7.53e-12 | 6930 | 97.1 | 73100 | 95.4 | 1.02 | ESR_close |
| 139 | 24 | MA1109.1 | NEUROD1 | MA1109.1 | NRACAGATGGYNN | NEUROD1 | 1.51e-09 | 7.69e-12 | 6060 | 84.9 | 62600 | 81.7 | 1.04 | ESR_close |
| 140 | 50 | MA0697.2 (Zic3) | STREME-15 | 15-CAGCAGG | 15-CAGCAGG | Zic3 | 1.62e-09 | 8.15e-12 | 7050 | 98.7 | 74700 | 97.4 | 1.01 | ESR_close |
| 141 | 51 | MA1972.1 | ZFP14 | MA1972.1 | GGAGGCHCHGGARBG | ZFP14 | 2.15e-09 | 1.08e-11 | 5100 | 71.4 | 51700 | 67.5 | 1.06 | ESR_close |
| 142 | 52 | MA1153.1 | Smad4 | MA1153.1 | TGTCTRGM | Smad4 | 5.78e-09 | 2.87e-11 | 1430 | 20 | 12800 | 16.8 | 1.19 | ESR_close |
| 143 | 1 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 7.92e-09 | 3.91e-11 | 537 | 7.52 | 4220 | 5.51 | 1.37 | ESR_close |
| 144 | 24 | MA1559.1 | SNAI3 | MA1559.1 | RRCAGGTGYA | SNAI3 | 1.58e-08 | 7.73e-11 | 4200 | 58.9 | 42000 | 54.8 | 1.07 | ESR_close |
| 145 | 23 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 2.71e-08 | 1.32e-10 | 2140 | 29.9 | 20200 | 26.3 | 1.14 | ESR_close |
| 146 | 37 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 2.79e-08 | 1.35e-10 | 1090 | 15.3 | 9620 | 12.6 | 1.22 | ESR_close |
| 147 | 37 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 3e-08 | 1.44e-10 | 2140 | 30 | 20200 | 26.3 | 1.14 | ESR_close |
| 148 | 37 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 3.79e-08 | 1.81e-10 | 880 | 12.3 | 7540 | 9.84 | 1.25 | ESR_close |
| 149 | 53 | MA1600.1 | ZNF684 | MA1600.1 | RYACAGTCCACCCCTTKR | ZNF684 | 5.73e-08 | 2.72e-10 | 1930 | 27.1 | 18100 | 23.6 | 1.15 | ESR_close |
| 150 | 54 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 5.87e-08 | 2.77e-10 | 6490 | 90.9 | 67800 | 88.5 | 1.03 | ESR_close |
| 151 | 23 | MA1566.2 | TBX3 | MA1566.2 | RAGGTGTGAAV | TBX3 | 6.16e-08 | 2.88e-10 | 4740 | 66.3 | 47900 | 62.5 | 1.06 | ESR_close |
| 152 | 26 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 9.32e-08 | 4.33e-10 | 6700 | 93.8 | 70300 | 91.8 | 1.02 | ESR_close |
| 153 | 38 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 1.28e-07 | 5.89e-10 | 6350 | 89 | 66200 | 86.4 | 1.03 | ESR_close |
| 154 | 26 | MA1581.1 | ZBTB6 | MA1581.1 | DNSCTTGAGCCNV | ZBTB6 | 1.49e-07 | 6.84e-10 | 6750 | 94.5 | 70900 | 92.5 | 1.02 | ESR_close |
| 155 | 37 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 1.67e-07 | 7.59e-10 | 2080 | 29.1 | 19600 | 25.6 | 1.13 | ESR_close |
| 156 | 6 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 2.52e-07 | 1.14e-09 | 146 | 2.04 | 865 | 1.13 | 1.82 | ESR_close |
| 157 | 24 | MA1100.2 | ASCL1 | MA1100.2 | VGCAGCTGCN | ASCL1 | 2.9e-07 | 1.31e-09 | 6980 | 97.8 | 73900 | 96.4 | 1.01 | ESR_close |
| 158 | 55 | MA0831.3 | TFE3 | MA0831.3 | GTCACGTGAC | TFE3 | 3.5e-07 | 1.57e-09 | 1430 | 20 | 13100 | 17 | 1.17 | ESR_close |
| 159 | 56 | MA1726.1 | ZNF331 | MA1726.1 | NMYTGCAGAGCCCH | ZNF331 | 3.8e-07 | 1.69e-09 | 6100 | 85.4 | 63300 | 82.6 | 1.03 | ESR_close |
| 160 | 6 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 7.05e-07 | 3.11e-09 | 1920 | 26.8 | 18100 | 23.6 | 1.14 | ESR_close |
| 161 | 23 | MA0690.2 | TBX21 | MA0690.2 | TTTCACACCTT | TBX21 | 9.44e-07 | 4.14e-09 | 3990 | 55.9 | 40000 | 52.2 | 1.07 | ESR_close |
| 162 | 57 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 1.04e-06 | 4.53e-09 | 2960 | 41.4 | 29000 | 37.8 | 1.1 | ESR_close |
| 163 | 24 | MA0820.1 | FIGLA | MA0820.1 | WMCACCTGKW | FIGLA | 2.73e-06 | 1.18e-08 | 4690 | 65.7 | 47700 | 62.2 | 1.06 | ESR_close |
| 164 | 24 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 4.49e-06 | 1.93e-08 | 1570 | 21.9 | 14600 | 19.1 | 1.15 | ESR_close |
| 165 | 58 | MA0059.1 (MAX::MYC) | STREME-12 | 12-ACAGAGCACCTGG | 12-ACAGAGCACCTGG | MAX::MYC | 5.41e-06 | 2.32e-08 | 13 | 0.18 | 10 | 0.01 | 13.7 | ESR_close |
| 166 | 48 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 5.81e-06 | 2.47e-08 | 4240 | 59.4 | 42900 | 55.9 | 1.06 | ESR_close |
| 167 | 24 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 7.4e-06 | 3.13e-08 | 5960 | 83.4 | 61900 | 80.7 | 1.03 | ESR_close |
| 168 | 37 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 1.66e-05 | 6.96e-08 | 1830 | 25.6 | 17400 | 22.7 | 1.13 | ESR_close |
| 169 | 49 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 2.09e-05 | 8.73e-08 | 1170 | 16.4 | 10700 | 14 | 1.17 | ESR_close |
| 170 | 33 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 2.44e-05 | 1.01e-07 | 2390 | 33.5 | 23300 | 30.3 | 1.1 | ESR_close |
| 171 | 59 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 4.28e-05 | 1.77e-07 | 2310 | 32.4 | 22500 | 29.4 | 1.1 | ESR_close |
| 172 | 60 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 0.000114 | 4.67e-07 | 1370 | 19.1 | 12800 | 16.7 | 1.15 | ESR_close |
| 173 | 24 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 0.000162 | 6.59e-07 | 1080 | 15.1 | 9920 | 12.9 | 1.17 | ESR_close |
| 174 | 46 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 0.000184 | 7.45e-07 | 373 | 5.22 | 3020 | 3.94 | 1.33 | ESR_close |
| 175 | 28 | MA0159.1 | RARA::RXRA | MA0159.1 | RGKTCANVGRSAGGTCA | RARA::RXRA | 0.00019 | 7.67e-07 | 3220 | 45.1 | 32200 | 42 | 1.07 | ESR_close |
| 176 | 61 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 0.000193 | 7.73e-07 | 4580 | 64.2 | 46900 | 61.1 | 1.05 | ESR_close |
| 177 | 62 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 0.000253 | 1.01e-06 | 5930 | 83 | 61800 | 80.6 | 1.03 | ESR_close |
| 178 | 40 | MA0646.1 | GCM1 | MA0646.1 | BATGCGGGTAC | GCM1 | 0.000367 | 1.46e-06 | 5460 | 76.5 | 56600 | 73.8 | 1.04 | ESR_close |
| 179 | 15 | MA0736.1 | GLIS2 | MA0736.1 | GACCCCCCGCRAMG | GLIS2 | 0.000424 | 1.67e-06 | 2580 | 36.1 | 25400 | 33.2 | 1.09 | ESR_close |
| 180 | 63 | MA0065.2 | Pparg::Rxra | MA0065.2 | STRGGGCARAGGKCA | Pparg::Rxra | 0.000435 | 1.71e-06 | 4380 | 61.4 | 44700 | 58.4 | 1.05 | ESR_close |
| 181 | 24 | MA1619.1 | Ptf1A | MA1619.1 | NNACAGCTGTNN | Ptf1A | 0.00049 | 1.91e-06 | 2630 | 36.8 | 26000 | 34 | 1.09 | ESR_close |
| 182 | 59 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 0.000635 | 2.47e-06 | 4920 | 68.9 | 50700 | 66.1 | 1.04 | ESR_close |
| 183 | 24 | MA0104.4 | MYCN | MA0104.4 | VVCCACGTGGBB | MYCN | 0.00108 | 4.15e-06 | 5320 | 74.5 | 55100 | 71.9 | 1.04 | ESR_close |
| 184 | 24 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.00118 | 4.53e-06 | 3210 | 44.9 | 32200 | 42 | 1.07 | ESR_close |
| 185 | 24 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 0.00125 | 4.76e-06 | 1310 | 18.3 | 12400 | 16.2 | 1.14 | ESR_close |
| 186 | 18 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 0.00161 | 6.1e-06 | 1670 | 23.4 | 16100 | 21.1 | 1.11 | ESR_close |
| 187 | 64 | 17-CTCTGAAGAGAGCAG | STREME-17 | 17-CTCTGAAGAGAGCAG | 17-CTCTGAAGAGAGCAG | CTCTGAAGAGAGCAG | 0.00192 | 7.26e-06 | 84 | 1.18 | 503 | 0.66 | 1.81 | ESR_close |
| 188 | 23 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 0.00199 | 7.47e-06 | 1260 | 17.6 | 11900 | 15.5 | 1.14 | ESR_close |
| 189 | 28 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 0.00245 | 9.16e-06 | 6400 | 89.6 | 67300 | 87.8 | 1.02 | ESR_close |
| 190 | 65 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 0.00361 | 1.34e-05 | 6790 | 95 | 71800 | 93.8 | 1.01 | ESR_close |
| 191 | 66 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 0.00398 | 1.47e-05 | 4510 | 63.2 | 46400 | 60.5 | 1.04 | ESR_close |
| 192 | 67 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 0.00406 | 1.49e-05 | 3710 | 52 | 37700 | 49.2 | 1.06 | ESR_close |
| 193 | 66 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 0.00415 | 1.52e-05 | 6280 | 87.9 | 65900 | 86 | 1.02 | ESR_close |
| 194 | 23 | MA1565.1 | TBX18 | MA1565.1 | DRAGGTGTGAAR | TBX18 | 0.00784 | 2.85e-05 | 3810 | 53.4 | 38900 | 50.7 | 1.05 | ESR_close |
| 195 | 1 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 0.0082 | 2.97e-05 | 129 | 1.81 | 905 | 1.18 | 1.54 | ESR_close |
| 196 | 48 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 0.00898 | 3.24e-05 | 317 | 4.44 | 2630 | 3.43 | 1.3 | ESR_close |
| 197 | 65 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 0.0107 | 3.85e-05 | 619 | 8.67 | 5570 | 7.27 | 1.19 | ESR_close |
| 198 | 18 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 0.0111 | 3.96e-05 | 1750 | 24.4 | 17100 | 22.2 | 1.1 | ESR_close |
| 199 | 23 | MA1621.1 | Rbpjl | MA1621.1 | NNVACACCTGTBNN | Rbpjl | 0.0115 | 4.09e-05 | 4500 | 63 | 46300 | 60.5 | 1.04 | ESR_close |
| 200 | 18 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 0.0131 | 4.64e-05 | 1630 | 22.9 | 15900 | 20.7 | 1.1 | ESR_close |
| 201 | 28 | MA0730.1 | RARA | MA0730.1 | AGGTCAHSYAAAGGTCA | RARA | 0.0137 | 4.82e-05 | 106 | 1.48 | 718 | 0.94 | 1.6 | ESR_close |
| 202 | 39 | MA0056.2 | MZF1 | MA0056.2 | NNAATCCCCANNN | MZF1 | 0.0153 | 5.36e-05 | 5800 | 81.2 | 60700 | 79.2 | 1.03 | ESR_close |
| 203 | 66 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 0.0163 | 5.66e-05 | 982 | 13.8 | 9240 | 12 | 1.14 | ESR_close |
| 204 | 68 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 0.0195 | 6.74e-05 | 3670 | 51.3 | 37400 | 48.8 | 1.05 | ESR_close |
| 205 | 54 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 0.0229 | 7.89e-05 | 6860 | 96 | 72800 | 94.9 | 1.01 | ESR_close |
| 206 | 65 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 0.0294 | 0.000101 | 6900 | 96.6 | 73300 | 95.6 | 1.01 | ESR_close |
| 207 | 59 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 0.0301 | 0.000103 | 5010 | 70.1 | 52000 | 67.8 | 1.03 | ESR_close |
| 208 | 65 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 0.0377 | 0.000128 | 6950 | 97.4 | 74000 | 96.5 | 1.01 | ESR_close |
| 209 | 48 | MA1468.1 | ATOH7 | MA1468.1 | AVCATATGBY | ATOH7 | 0.0407 | 0.000137 | 6420 | 89.9 | 67700 | 88.3 | 1.02 | ESR_close |
| 210 | 48 | MA1730.1 | ZNF708 | MA1730.1 | NNRGCTGTRCCTCCY | ZNF708 | 0.043 | 0.000144 | 5340 | 74.8 | 55700 | 72.6 | 1.03 | ESR_close |
| 211 | 69 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 0.0488 | 0.000163 | 3990 | 55.9 | 41000 | 53.5 | 1.04 | ESR_close |
| 24 | 70 | MA1731.1 (ZNF768) | MEME-8 | WCTYTCTCTCTCTCT | WCTYTCTCTCTCTCT | ZNF768 | 0.456 | 0.019 | 312 | 4.37 | 2950 | 3.85 | 1.14 | ESR_close |
| 25 | 71 | MA1107.2 (KLF9) | MEME-7 | TGTGTGTGTGTGTGT | TGTGTGTGTGTGTGT | KLF9 | 1.16 | 0.0462 | 846 | 11.8 | 8560 | 11.2 | 1.06 | ESR_close |
| 26 | 72 | MA1125.1 (ZNF384) | MEME-4 | AAAAAAAAAAAAAAA | AAAAAAAAAAAAAAA | ZNF384 | 18.7 | 0.72 | 47 | 0.66 | 546 | 0.71 | 0.942 | ESR_close |
ER_rat <- 1.25
mrc_type <- "ESR_close"
ESR_close_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))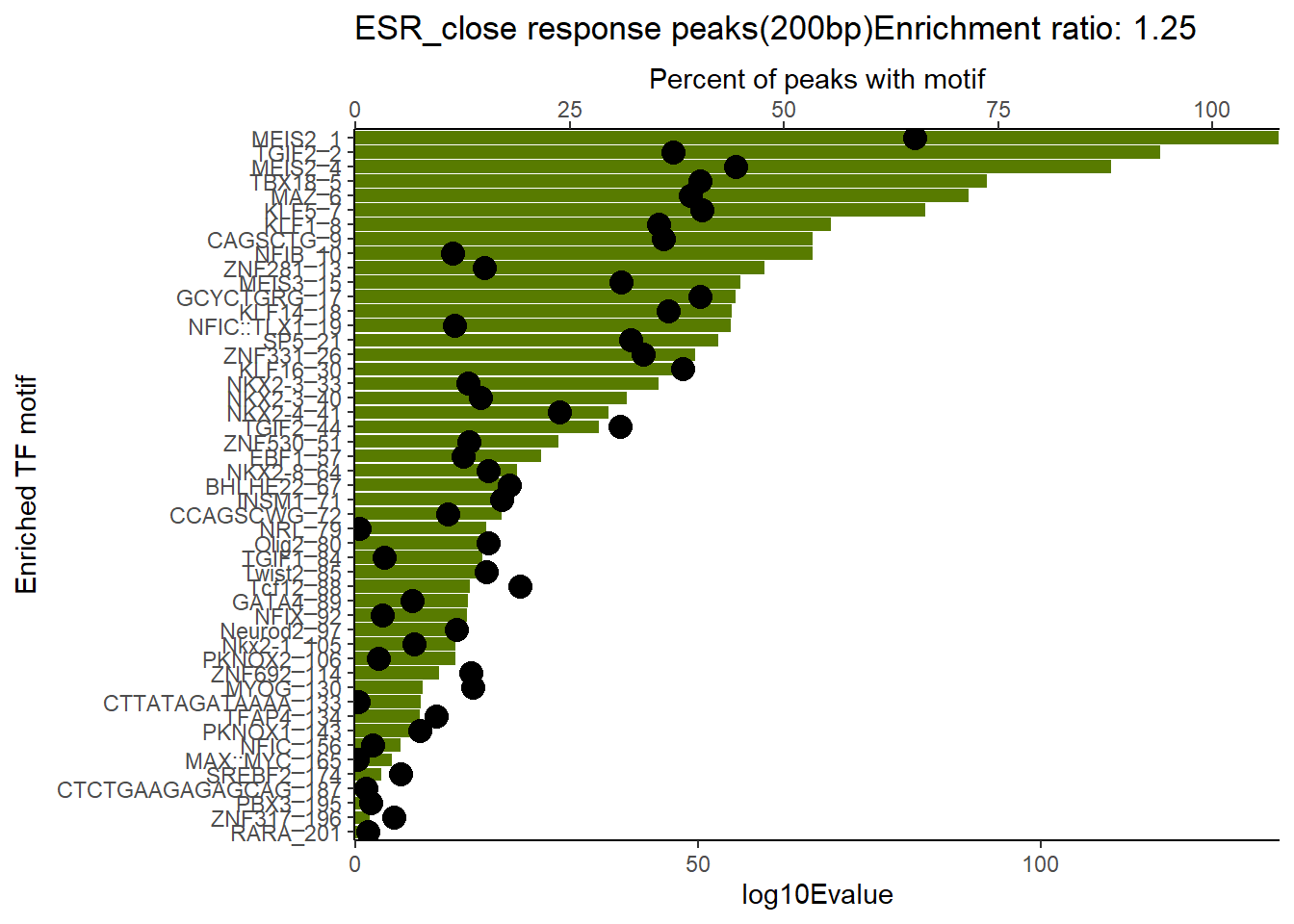
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_close_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.75), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.75,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))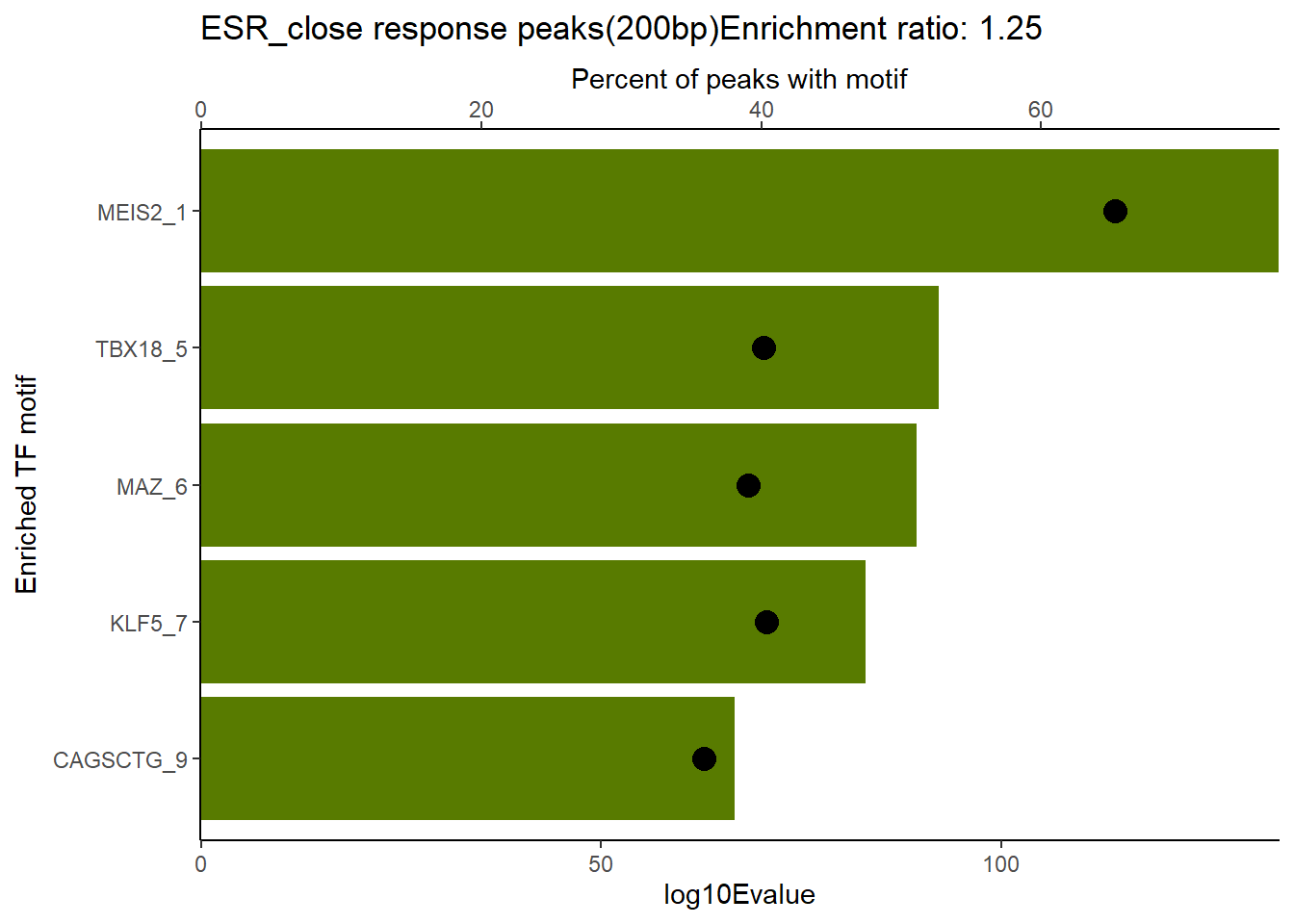
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_opcl data
ESR_opcl_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_opcl_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
ESR_opcl_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_opcl_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
ESR_opcl_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_opcl_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
ESR_opcl_sea_all <- rbind(ESR_opcl_sea_disc, ESR_opcl_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)ESR_opcl_combo <- ESR_opcl_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., ESR_opcl_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="ESR_opcl")
ESR_opcl_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0808.1 (TEAD3) | MEME-1 | TGGAATKY | TGGAATKY | TEAD3 | 7.39e-12 | 5.4e-12 | 76 | 41.5 | 13100 | 17.1 | 2.44 | ESR_opcl |
| 2 | 2 | MA1580.1 (ZBTB32) | STREME-2 | 2-TACAGT | TACAGT | ZBTB32 | 4.25e-08 | 1.12e-08 | 165 | 90.2 | 53600 | 70 | 1.29 | ESR_opcl |
| 3 | 3 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 4.61e-08 | 1.12e-08 | 154 | 84.2 | 47600 | 62.1 | 1.36 | ESR_opcl |
| 4 | 4 | 5-ATTAGAA | STREME-5 | 5-ATTAGAA | ATTAGAA | ATTAGAA | 5.33e-07 | 8.69e-08 | 165 | 90.2 | 54800 | 71.4 | 1.26 | ESR_opcl |
| 5 | 1 | MA0090.3 (TEAD1) | STREME-3 | 3-ATTCCATAM | ATTCCATAM | TEAD1 | 5.94e-07 | 8.69e-08 | 149 | 81.4 | 46200 | 60.3 | 1.35 | ESR_opcl |
| 6 | 5 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 8.16e-07 | 9.94e-08 | 152 | 83.1 | 47900 | 62.5 | 1.33 | ESR_opcl |
| 7 | 3 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 2.72e-06 | 2.85e-07 | 121 | 66.1 | 34100 | 44.5 | 1.49 | ESR_opcl |
| 8 | 1 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 1e-05 | 9.18e-07 | 89 | 48.6 | 22000 | 28.8 | 1.7 | ESR_opcl |
| 9 | 6 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 2.21e-05 | 1.79e-06 | 162 | 88.5 | 54800 | 71.5 | 1.24 | ESR_opcl |
| 10 | 7 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 3.81e-05 | 2.6e-06 | 135 | 73.8 | 41600 | 54.3 | 1.36 | ESR_opcl |
| 11 | 8 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 3.91e-05 | 2.6e-06 | 165 | 90.2 | 56800 | 74.1 | 1.22 | ESR_opcl |
| 12 | 3 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 8.09e-05 | 4.93e-06 | 165 | 90.2 | 57100 | 74.6 | 1.21 | ESR_opcl |
| 13 | 1 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 0.000122 | 6.88e-06 | 66 | 36.1 | 15000 | 19.6 | 1.86 | ESR_opcl |
| 14 | 9 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 0.000146 | 7.61e-06 | 123 | 67.2 | 37000 | 48.2 | 1.4 | ESR_opcl |
| 15 | 3 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 0.000244 | 1.12e-05 | 106 | 57.9 | 30100 | 39.3 | 1.48 | ESR_opcl |
| 16 | 8 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 0.000244 | 1.12e-05 | 140 | 76.5 | 44900 | 58.6 | 1.31 | ESR_opcl |
| 17 | 10 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 0.000322 | 1.38e-05 | 154 | 84.2 | 51900 | 67.8 | 1.24 | ESR_opcl |
| 18 | 11 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 0.000402 | 1.63e-05 | 113 | 61.8 | 33300 | 43.4 | 1.43 | ESR_opcl |
| 6 | 23 | 4-VWWDTAHWWB | STREME-4 | 4-VWWDTAHWWB | VWWDTAHWWB | VWWDTAHWWB | 0.000454 | 7.56e-05 | 73 | 39.9 | 20500 | 26.7 | 1.5 | ESR_opcl |
| 19 | 12 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 0.000513 | 1.97e-05 | 136 | 74.3 | 43500 | 56.7 | 1.31 | ESR_opcl |
| 20 | 5 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 0.000628 | 2.3e-05 | 135 | 73.8 | 43100 | 56.3 | 1.31 | ESR_opcl |
| 21 | 13 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 0.000806 | 2.81e-05 | 153 | 83.6 | 51900 | 67.8 | 1.24 | ESR_opcl |
| 22 | 13 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 0.000905 | 3.01e-05 | 140 | 76.5 | 45700 | 59.6 | 1.29 | ESR_opcl |
| 23 | 10 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 0.000978 | 3.11e-05 | 154 | 84.2 | 52500 | 68.6 | 1.23 | ESR_opcl |
| 24 | 10 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 0.00112 | 3.33e-05 | 158 | 86.3 | 54700 | 71.4 | 1.21 | ESR_opcl |
| 25 | 14 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 0.00114 | 3.33e-05 | 133 | 72.7 | 42600 | 55.5 | 1.31 | ESR_opcl |
| 26 | 13 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 0.00177 | 4.99e-05 | 164 | 89.6 | 58200 | 75.9 | 1.18 | ESR_opcl |
| 27 | 15 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 0.00189 | 5.13e-05 | 107 | 58.5 | 31700 | 41.3 | 1.42 | ESR_opcl |
| 28 | 13 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 0.00198 | 5.18e-05 | 121 | 66.1 | 37600 | 49 | 1.35 | ESR_opcl |
| 29 | 8 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 0.00238 | 6.01e-05 | 151 | 82.5 | 51500 | 67.3 | 1.23 | ESR_opcl |
| 30 | 13 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 0.00266 | 6.48e-05 | 135 | 73.8 | 44000 | 57.4 | 1.29 | ESR_opcl |
| 31 | 16 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 0.00282 | 6.66e-05 | 117 | 63.9 | 36100 | 47.1 | 1.36 | ESR_opcl |
| 32 | 10 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 0.00371 | 8.47e-05 | 128 | 70 | 41000 | 53.5 | 1.31 | ESR_opcl |
| 33 | 8 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 0.0039 | 8.64e-05 | 133 | 72.7 | 43300 | 56.5 | 1.29 | ESR_opcl |
| 34 | 8 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 0.00461 | 9.91e-05 | 150 | 82 | 51400 | 67.1 | 1.22 | ESR_opcl |
| 35 | 8 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 0.00505 | 0.000105 | 91 | 49.7 | 25800 | 33.7 | 1.48 | ESR_opcl |
| 36 | 17 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 0.00567 | 0.000115 | 163 | 89.1 | 58300 | 76.1 | 1.17 | ESR_opcl |
| 37 | 5 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 0.00596 | 0.000118 | 156 | 85.2 | 54600 | 71.3 | 1.2 | ESR_opcl |
| 38 | 8 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 0.00619 | 0.000119 | 166 | 90.7 | 60000 | 78.3 | 1.16 | ESR_opcl |
| 39 | 13 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 0.00686 | 0.000129 | 165 | 90.2 | 59500 | 77.6 | 1.16 | ESR_opcl |
| 40 | 13 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 0.00713 | 0.00013 | 133 | 72.7 | 43700 | 57 | 1.28 | ESR_opcl |
| 41 | 8 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 0.00808 | 0.000144 | 135 | 73.8 | 44700 | 58.3 | 1.27 | ESR_opcl |
| 42 | 8 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 0.00835 | 0.000145 | 148 | 80.9 | 50800 | 66.3 | 1.22 | ESR_opcl |
| 43 | 13 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 0.0118 | 2e-04 | 117 | 63.9 | 37000 | 48.2 | 1.33 | ESR_opcl |
| 44 | 13 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 0.0133 | 0.000221 | 114 | 62.3 | 35800 | 46.7 | 1.34 | ESR_opcl |
| 45 | 5 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 0.0143 | 0.000231 | 171 | 93.4 | 63400 | 82.7 | 1.13 | ESR_opcl |
| 46 | 13 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 0.0145 | 0.000231 | 101 | 55.2 | 30400 | 39.7 | 1.4 | ESR_opcl |
| 47 | 18 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 0.0184 | 0.000287 | 131 | 71.6 | 43400 | 56.6 | 1.27 | ESR_opcl |
| 48 | 19 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 0.0223 | 0.000339 | 120 | 65.6 | 38700 | 50.5 | 1.3 | ESR_opcl |
| 49 | 20 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 0.0234 | 0.000349 | 160 | 87.4 | 57500 | 75.1 | 1.17 | ESR_opcl |
| 50 | 21 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 0.0247 | 0.000357 | 54 | 29.5 | 13200 | 17.2 | 1.74 | ESR_opcl |
| 51 | 8 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 0.0253 | 0.000357 | 79 | 43.2 | 22200 | 28.9 | 1.5 | ESR_opcl |
| 52 | 17 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 0.0257 | 0.000357 | 127 | 69.4 | 41800 | 54.6 | 1.27 | ESR_opcl |
| 53 | 22 | 1-MAWWTK | STREME-1 | 1-MAWWTK | MAWWTK | MAWWTK | 0.0259 | 0.000357 | 102 | 55.7 | 31200 | 40.7 | 1.37 | ESR_opcl |
| 54 | 17 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 0.029 | 0.000392 | 110 | 60.1 | 34600 | 45.1 | 1.34 | ESR_opcl |
| 55 | 7 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 0.0307 | 0.000408 | 88 | 48.1 | 25700 | 33.6 | 1.44 | ESR_opcl |
| 56 | 12 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 0.0319 | 0.000417 | 114 | 62.3 | 36300 | 47.4 | 1.32 | ESR_opcl |
| 57 | 13 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 0.0331 | 0.000424 | 124 | 67.8 | 40700 | 53.1 | 1.28 | ESR_opcl |
| 58 | 9 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 0.0369 | 0.000465 | 98 | 53.6 | 29800 | 38.9 | 1.38 | ESR_opcl |
| 59 | 8 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 0.0394 | 0.000489 | 161 | 88 | 58400 | 76.2 | 1.16 | ESR_opcl |
| 60 | 8 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 0.043 | 0.000523 | 170 | 92.9 | 63400 | 82.7 | 1.12 | ESR_opcl |
| 61 | 8 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 0.0454 | 0.000544 | 146 | 79.8 | 51000 | 66.5 | 1.2 | ESR_opcl |
| 62 | 8 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 0.0463 | 0.000545 | 156 | 85.2 | 55900 | 73 | 1.17 | ESR_opcl |
| 63 | 5 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 0.047 | 0.000545 | 142 | 77.6 | 49100 | 64 | 1.21 | ESR_opcl |
ER_rat <- 1.25
mrc_type <- "ESR_opcl"
ESR_opcl_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))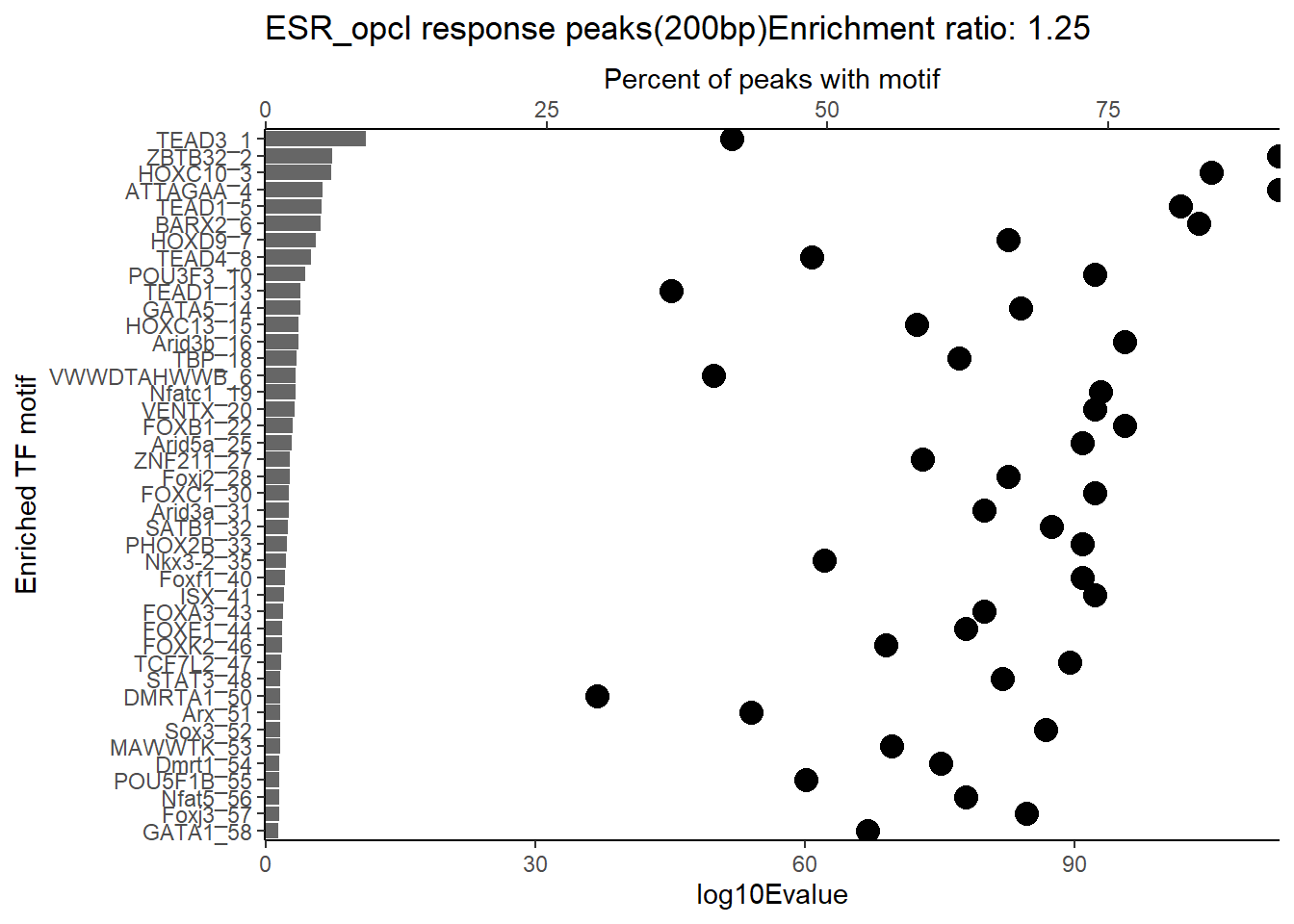
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_opcl_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*.1), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.1,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))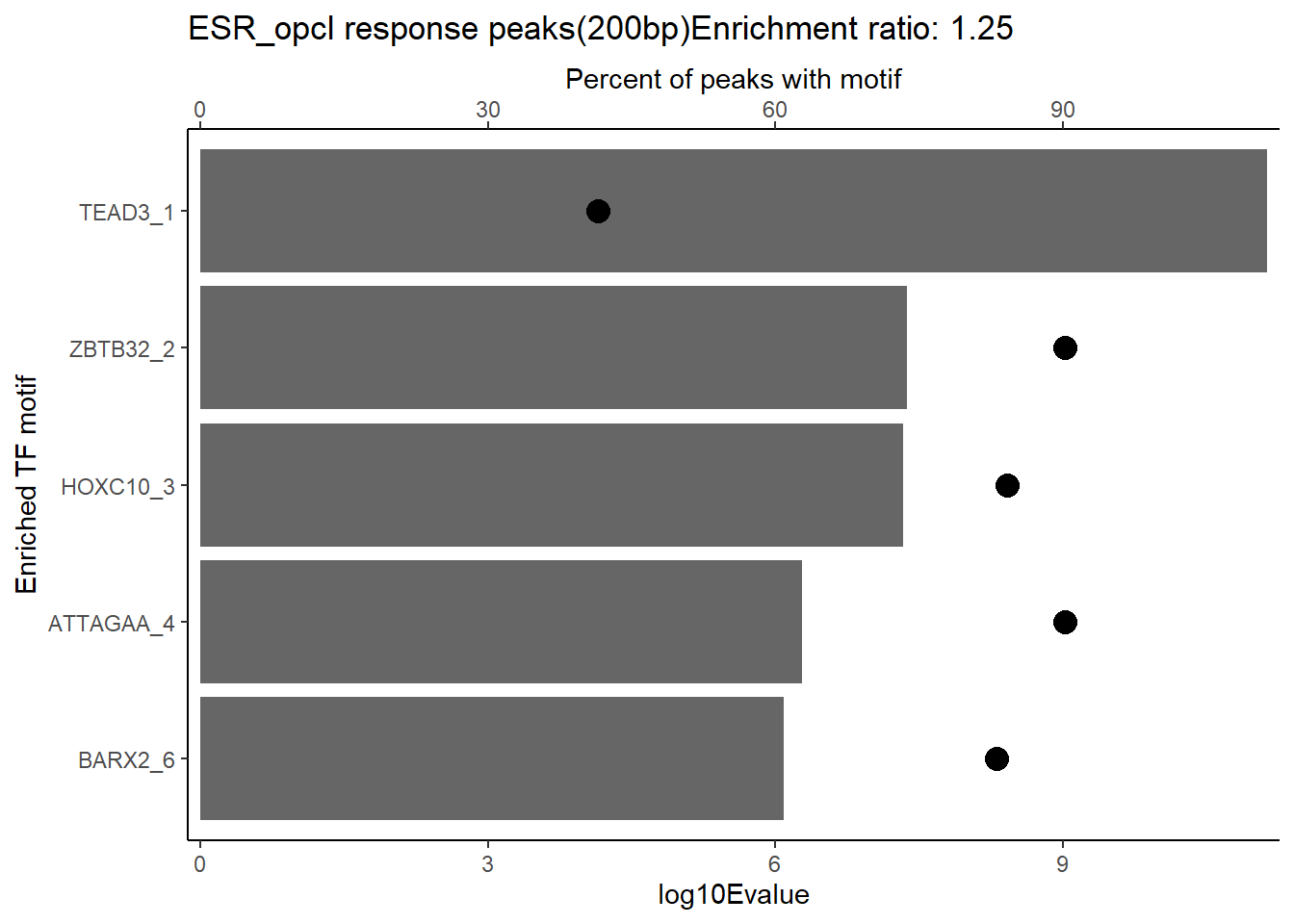
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_clop data
ESR_clop_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_clop_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
ESR_clop_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_clop_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
ESR_clop_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_clop_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
ESR_clop_sea_all <- rbind(ESR_clop_sea_disc, ESR_clop_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)ESR_clop_combo <- ESR_clop_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., ESR_clop_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="ESR_clop")
ESR_clop_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1138.1 (FOSL2::JUNB) | MEME-1 | TGASTCAT | TGASTCAT | FOSL2::JUNB | 6.64e-111 | 3.58e-111 | 342 | 53.2 | 11200 | 14.6 | 3.64 | ESR_clop |
| 2 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-DVTGASTCABH | DVTGASTCABH | FOS::JUNB | 1.11e-108 | 3e-109 | 321 | 49.9 | 9920 | 12.9 | 3.86 | ESR_clop |
| 3 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 4.53e-106 | 8.14e-107 | 308 | 47.9 | 9250 | 12.1 | 3.98 | ESR_clop |
| 4 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 7.21e-100 | 9.73e-101 | 325 | 50.5 | 11000 | 14.4 | 3.53 | ESR_clop |
| 5 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 6.71e-97 | 7.23e-98 | 327 | 50.9 | 11400 | 14.9 | 3.41 | ESR_clop |
| 6 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 6.26e-96 | 5.63e-97 | 312 | 48.5 | 10400 | 13.6 | 3.57 | ESR_clop |
| 7 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 9.26e-96 | 7.13e-97 | 283 | 44 | 8440 | 11 | 4 | ESR_clop |
| 8 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 1.09e-95 | 7.34e-97 | 296 | 46 | 9320 | 12.2 | 3.79 | ESR_clop |
| 9 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 1.5e-95 | 8.96e-97 | 288 | 44.8 | 8790 | 11.5 | 3.91 | ESR_clop |
| 10 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 2.34e-95 | 1.26e-96 | 319 | 49.6 | 11000 | 14.3 | 3.47 | ESR_clop |
| 11 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 9.45e-95 | 4.64e-96 | 309 | 48.1 | 10300 | 13.5 | 3.58 | ESR_clop |
| 12 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 2.03e-93 | 9.11e-95 | 309 | 48.1 | 10400 | 13.6 | 3.53 | ESR_clop |
| 13 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 5.96e-93 | 2.47e-94 | 287 | 44.6 | 8950 | 11.7 | 3.83 | ESR_clop |
| 14 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 2.88e-92 | 1.11e-93 | 319 | 49.6 | 11300 | 14.7 | 3.37 | ESR_clop |
| 15 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 1.4e-91 | 5.03e-93 | 275 | 42.8 | 8260 | 10.8 | 3.97 | ESR_clop |
| 16 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 1.01e-90 | 3.42e-92 | 272 | 42.3 | 8140 | 10.6 | 3.99 | ESR_clop |
| 17 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 1.57e-90 | 4.98e-92 | 358 | 55.7 | 14600 | 19 | 2.93 | ESR_clop |
| 18 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 3.54e-90 | 1.06e-91 | 267 | 41.5 | 7860 | 10.3 | 4.06 | ESR_clop |
| 19 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 1.72e-88 | 4.87e-90 | 390 | 60.6 | 17700 | 23 | 2.63 | ESR_clop |
| 20 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 8.49e-88 | 2.29e-89 | 269 | 41.8 | 8190 | 10.7 | 3.92 | ESR_clop |
| 21 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 1.19e-85 | 3.06e-87 | 385 | 59.9 | 17600 | 22.9 | 2.61 | ESR_clop |
| 22 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 4.19e-85 | 1.03e-86 | 255 | 39.7 | 7510 | 9.8 | 4.06 | ESR_clop |
| 23 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 2.89e-83 | 6.77e-85 | 264 | 41.1 | 8260 | 10.8 | 3.82 | ESR_clop |
| 24 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 1.11e-81 | 2.5e-83 | 294 | 45.7 | 10500 | 13.7 | 3.34 | ESR_clop |
| 25 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 6.96e-81 | 1.5e-82 | 240 | 37.3 | 6910 | 9.02 | 4.15 | ESR_clop |
| 26 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 7.34e-80 | 1.52e-81 | 296 | 46 | 10800 | 14.2 | 3.26 | ESR_clop |
| 27 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 1.49e-77 | 2.97e-79 | 293 | 45.6 | 10900 | 14.2 | 3.22 | ESR_clop |
| 28 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 4.35e-76 | 8.38e-78 | 215 | 33.4 | 5780 | 7.54 | 4.45 | ESR_clop |
| 29 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 1.58e-52 | 2.93e-54 | 360 | 56 | 20300 | 26.5 | 2.11 | ESR_clop |
| 30 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 1.21e-45 | 2.17e-47 | 304 | 47.3 | 16200 | 21.1 | 2.24 | ESR_clop |
| 31 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 8.08e-40 | 1.41e-41 | 252 | 39.2 | 12600 | 16.4 | 2.39 | ESR_clop |
| 32 | 2 | MA1970.1 (TRPS1) | STREME-2 | 2-WAGATRABNWA | WAGATRABNWA | TRPS1 | 1.26e-38 | 2.12e-40 | 344 | 53.5 | 21500 | 28 | 1.91 | ESR_clop |
| 33 | 1 | MA0676.1 (Nr2e1) | STREME-3 | 3-AGTCAW | AGTCAW | Nr2e1 | 2.34e-37 | 3.82e-39 | 515 | 80.1 | 42200 | 55 | 1.46 | ESR_clop |
| 34 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 1.52e-34 | 2.41e-36 | 161 | 25 | 6250 | 8.16 | 3.08 | ESR_clop |
| 35 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 6.19e-33 | 9.53e-35 | 477 | 74.2 | 38300 | 50 | 1.49 | ESR_clop |
| 36 | 2 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 5.56e-25 | 8.33e-27 | 372 | 57.8 | 28000 | 36.5 | 1.59 | ESR_clop |
| 37 | 2 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 8.37e-24 | 1.22e-25 | 334 | 51.9 | 24100 | 31.5 | 1.65 | ESR_clop |
| 38 | 2 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 2.28e-22 | 3.24e-24 | 588 | 91.4 | 57900 | 75.6 | 1.21 | ESR_clop |
| 39 | 3 | MA0018.4 (CREB1) | STREME-4 | 4-ACATCA | ACATCA | CREB1 | 5.02e-22 | 6.95e-24 | 447 | 69.5 | 37900 | 49.4 | 1.41 | ESR_clop |
| 40 | 4 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 5.22e-22 | 7.04e-24 | 513 | 79.8 | 46600 | 60.8 | 1.31 | ESR_clop |
| 41 | 2 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 5.51e-22 | 7.25e-24 | 445 | 69.2 | 37600 | 49.1 | 1.41 | ESR_clop |
| 42 | 5 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 6.83e-22 | 8.77e-24 | 524 | 81.5 | 48200 | 62.9 | 1.3 | ESR_clop |
| 43 | 6 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 3.83e-21 | 4.81e-23 | 539 | 83.8 | 50600 | 66 | 1.27 | ESR_clop |
| 44 | 5 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 5.5e-21 | 6.74e-23 | 411 | 63.9 | 33800 | 44.1 | 1.45 | ESR_clop |
| 45 | 7 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 7.56e-21 | 9.06e-23 | 588 | 91.4 | 58400 | 76.2 | 1.2 | ESR_clop |
| 46 | 8 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 9.14e-21 | 1.07e-22 | 565 | 87.9 | 54700 | 71.4 | 1.23 | ESR_clop |
| 47 | 7 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 9.41e-21 | 1.08e-22 | 483 | 75.1 | 42900 | 56 | 1.34 | ESR_clop |
| 48 | 2 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 1.23e-20 | 1.39e-22 | 460 | 71.5 | 40000 | 52.2 | 1.37 | ESR_clop |
| 49 | 5 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 1.26e-20 | 1.39e-22 | 535 | 83.2 | 50200 | 65.5 | 1.27 | ESR_clop |
| 50 | 5 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 1.44e-20 | 1.55e-22 | 480 | 74.6 | 42600 | 55.6 | 1.34 | ESR_clop |
| 51 | 9 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 8.37e-20 | 8.85e-22 | 485 | 75.4 | 43500 | 56.8 | 1.33 | ESR_clop |
| 52 | 5 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 1.42e-19 | 1.47e-21 | 472 | 73.4 | 41900 | 54.7 | 1.34 | ESR_clop |
| 53 | 10 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 1.67e-19 | 1.7e-21 | 534 | 83 | 50500 | 65.8 | 1.26 | ESR_clop |
| 54 | 5 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 1.76e-19 | 1.76e-21 | 550 | 85.5 | 52800 | 68.9 | 1.24 | ESR_clop |
| 55 | 11 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 2.22e-19 | 2.17e-21 | 567 | 88.2 | 55500 | 72.4 | 1.22 | ESR_clop |
| 56 | 5 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 3.73e-19 | 3.55e-21 | 592 | 92.1 | 59700 | 77.8 | 1.18 | ESR_clop |
| 57 | 5 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 3.75e-19 | 3.55e-21 | 608 | 94.6 | 62500 | 81.6 | 1.16 | ESR_clop |
| 58 | 5 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 5.66e-19 | 5.27e-21 | 591 | 91.9 | 59500 | 77.7 | 1.18 | ESR_clop |
| 59 | 12 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 8.36e-19 | 7.63e-21 | 559 | 86.9 | 54400 | 71 | 1.22 | ESR_clop |
| 60 | 5 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 8.49e-19 | 7.63e-21 | 431 | 67 | 37000 | 48.3 | 1.39 | ESR_clop |
| 61 | 13 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 1.06e-18 | 9.37e-21 | 612 | 95.2 | 63400 | 82.8 | 1.15 | ESR_clop |
| 62 | 4 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 2.15e-18 | 1.84e-20 | 482 | 75 | 43700 | 57 | 1.32 | ESR_clop |
| 63 | 12 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 2.15e-18 | 1.84e-20 | 629 | 97.8 | 67100 | 87.5 | 1.12 | ESR_clop |
| 64 | 12 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 2.65e-18 | 2.24e-20 | 630 | 98 | 67300 | 87.8 | 1.12 | ESR_clop |
| 65 | 12 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 4.17e-18 | 3.46e-20 | 610 | 94.9 | 63200 | 82.5 | 1.15 | ESR_clop |
| 66 | 5 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 1.11e-17 | 9.06e-20 | 467 | 72.6 | 42000 | 54.8 | 1.33 | ESR_clop |
| 67 | 5 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 1.42e-17 | 1.14e-19 | 574 | 89.3 | 57200 | 74.7 | 1.2 | ESR_clop |
| 68 | 5 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 1.68e-17 | 1.33e-19 | 599 | 93.2 | 61400 | 80.2 | 1.16 | ESR_clop |
| 69 | 10 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 2.38e-17 | 1.86e-19 | 594 | 92.4 | 60600 | 79.1 | 1.17 | ESR_clop |
| 70 | 5 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 3.87e-17 | 2.98e-19 | 496 | 77.1 | 46000 | 60 | 1.29 | ESR_clop |
| 71 | 5 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 6.05e-17 | 4.59e-19 | 491 | 76.4 | 45400 | 59.3 | 1.29 | ESR_clop |
| 72 | 6 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 6.88e-17 | 5.15e-19 | 499 | 77.6 | 46500 | 60.7 | 1.28 | ESR_clop |
| 73 | 5 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 8.18e-17 | 6.04e-19 | 596 | 92.7 | 61100 | 79.8 | 1.16 | ESR_clop |
| 74 | 2 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 1.02e-16 | 7.44e-19 | 596 | 92.7 | 61200 | 79.8 | 1.16 | ESR_clop |
| 75 | 2 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 1.07e-16 | 7.71e-19 | 452 | 70.3 | 40400 | 52.8 | 1.33 | ESR_clop |
| 76 | 5 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 1.28e-16 | 9.05e-19 | 576 | 89.6 | 57900 | 75.5 | 1.19 | ESR_clop |
| 77 | 5 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 1.38e-16 | 9.64e-19 | 621 | 96.6 | 65900 | 86 | 1.12 | ESR_clop |
| 78 | 5 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 2.24e-16 | 1.55e-18 | 551 | 85.7 | 54100 | 70.6 | 1.21 | ESR_clop |
| 79 | 14 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 2.99e-16 | 2.04e-18 | 451 | 70.1 | 40500 | 52.8 | 1.33 | ESR_clop |
| 80 | 5 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 3.2e-16 | 2.16e-18 | 576 | 89.6 | 58000 | 75.7 | 1.18 | ESR_clop |
| 81 | 5 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 1.63e-15 | 1.09e-17 | 603 | 93.8 | 62800 | 82 | 1.14 | ESR_clop |
| 82 | 5 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 2.85e-15 | 1.87e-17 | 549 | 85.4 | 54200 | 70.8 | 1.21 | ESR_clop |
| 83 | 5 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 3.03e-15 | 1.97e-17 | 510 | 79.3 | 48700 | 63.5 | 1.25 | ESR_clop |
| 84 | 5 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 5.2e-15 | 3.34e-17 | 436 | 67.8 | 39100 | 51 | 1.33 | ESR_clop |
| 85 | 5 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 5.56e-15 | 3.53e-17 | 554 | 86.2 | 55100 | 71.9 | 1.2 | ESR_clop |
| 86 | 5 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 8.74e-15 | 5.48e-17 | 623 | 96.9 | 66900 | 87.2 | 1.11 | ESR_clop |
| 87 | 5 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 9.45e-15 | 5.86e-17 | 547 | 85.1 | 54100 | 70.6 | 1.2 | ESR_clop |
| 88 | 5 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 1.23e-14 | 7.53e-17 | 593 | 92.2 | 61400 | 80.1 | 1.15 | ESR_clop |
| 89 | 10 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 1.87e-14 | 1.13e-16 | 537 | 83.5 | 52800 | 68.9 | 1.21 | ESR_clop |
| 90 | 5 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 2.25e-14 | 1.35e-16 | 616 | 95.8 | 65600 | 85.6 | 1.12 | ESR_clop |
| 91 | 5 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 2.61e-14 | 1.52e-16 | 393 | 61.1 | 34100 | 44.5 | 1.37 | ESR_clop |
| 92 | 5 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 2.61e-14 | 1.52e-16 | 604 | 93.9 | 63400 | 82.7 | 1.14 | ESR_clop |
| 93 | 5 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 2.62e-14 | 1.52e-16 | 370 | 57.5 | 31400 | 41 | 1.41 | ESR_clop |
| 94 | 15 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 2.92e-14 | 1.68e-16 | 569 | 88.5 | 57700 | 75.2 | 1.18 | ESR_clop |
| 95 | 5 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 3.72e-14 | 2.11e-16 | 607 | 94.4 | 64000 | 83.5 | 1.13 | ESR_clop |
| 96 | 10 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 4.71e-14 | 2.65e-16 | 515 | 80.1 | 49900 | 65 | 1.23 | ESR_clop |
| 97 | 5 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 1.15e-13 | 6.42e-16 | 601 | 93.5 | 63100 | 82.3 | 1.14 | ESR_clop |
| 98 | 12 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 1.55e-13 | 8.53e-16 | 588 | 91.4 | 61000 | 79.5 | 1.15 | ESR_clop |
| 99 | 5 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 1.69e-13 | 9.19e-16 | 607 | 94.4 | 64200 | 83.8 | 1.13 | ESR_clop |
| 100 | 5 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 1.94e-13 | 1.05e-15 | 621 | 96.6 | 66900 | 87.3 | 1.11 | ESR_clop |
| 101 | 10 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 2.33e-13 | 1.25e-15 | 552 | 85.8 | 55400 | 72.3 | 1.19 | ESR_clop |
| 102 | 13 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 3.35e-13 | 1.77e-15 | 588 | 91.4 | 61100 | 79.7 | 1.15 | ESR_clop |
| 103 | 12 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 3.6e-13 | 1.89e-15 | 484 | 75.3 | 46000 | 60.1 | 1.25 | ESR_clop |
| 104 | 5 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 3.75e-13 | 1.94e-15 | 522 | 81.2 | 51200 | 66.8 | 1.22 | ESR_clop |
| 105 | 5 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 3.97e-13 | 2.04e-15 | 565 | 87.9 | 57500 | 75 | 1.17 | ESR_clop |
| 106 | 1 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 4.74e-13 | 2.41e-15 | 292 | 45.4 | 23200 | 30.2 | 1.51 | ESR_clop |
| 107 | 16 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 5.49e-13 | 2.77e-15 | 600 | 93.3 | 63200 | 82.4 | 1.13 | ESR_clop |
| 108 | 5 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 5.97e-13 | 2.98e-15 | 603 | 93.8 | 63700 | 83.1 | 1.13 | ESR_clop |
| 109 | 17 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 6.69e-13 | 3.31e-15 | 602 | 93.6 | 63600 | 82.9 | 1.13 | ESR_clop |
| 110 | 5 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 7.8e-13 | 3.82e-15 | 560 | 87.1 | 56800 | 74.2 | 1.17 | ESR_clop |
| 111 | 12 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 7.93e-13 | 3.85e-15 | 590 | 91.8 | 61600 | 80.3 | 1.14 | ESR_clop |
| 112 | 5 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 8.9e-13 | 4.29e-15 | 556 | 86.5 | 56300 | 73.4 | 1.18 | ESR_clop |
| 113 | 5 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 1.08e-12 | 5.13e-15 | 611 | 95 | 65200 | 85.1 | 1.12 | ESR_clop |
| 114 | 5 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 1.32e-12 | 6.26e-15 | 494 | 76.8 | 47600 | 62.1 | 1.24 | ESR_clop |
| 115 | 5 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 1.39e-12 | 6.54e-15 | 321 | 49.9 | 26500 | 34.6 | 1.44 | ESR_clop |
| 116 | 5 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 1.56e-12 | 7.28e-15 | 501 | 77.9 | 48600 | 63.4 | 1.23 | ESR_clop |
| 117 | 5 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 1.79e-12 | 8.26e-15 | 508 | 79 | 49600 | 64.7 | 1.22 | ESR_clop |
| 118 | 5 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 2.18e-12 | 9.95e-15 | 616 | 95.8 | 66300 | 86.5 | 1.11 | ESR_clop |
| 119 | 5 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 2.22e-12 | 1.01e-14 | 516 | 80.2 | 50700 | 66.2 | 1.21 | ESR_clop |
| 120 | 5 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 2.33e-12 | 1.05e-14 | 521 | 81 | 51400 | 67.1 | 1.21 | ESR_clop |
| 121 | 18 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 2.44e-12 | 1.09e-14 | 545 | 84.8 | 54800 | 71.5 | 1.19 | ESR_clop |
| 122 | 2 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 3.16e-12 | 1.4e-14 | 295 | 45.9 | 23800 | 31.1 | 1.48 | ESR_clop |
| 123 | 5 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 3.69e-12 | 1.61e-14 | 357 | 55.5 | 30800 | 40.2 | 1.38 | ESR_clop |
| 124 | 5 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 3.71e-12 | 1.61e-14 | 604 | 93.9 | 64200 | 83.7 | 1.12 | ESR_clop |
| 125 | 12 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 3.93e-12 | 1.69e-14 | 564 | 87.7 | 57700 | 75.3 | 1.16 | ESR_clop |
| 126 | 10 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 4.06e-12 | 1.74e-14 | 479 | 74.5 | 45800 | 59.8 | 1.25 | ESR_clop |
| 127 | 5 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 4.13e-12 | 1.75e-14 | 567 | 88.2 | 58200 | 75.9 | 1.16 | ESR_clop |
| 128 | 5 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 4.47e-12 | 1.88e-14 | 417 | 64.8 | 38000 | 49.6 | 1.31 | ESR_clop |
| 129 | 12 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 5.64e-12 | 2.36e-14 | 625 | 97.2 | 68200 | 89 | 1.09 | ESR_clop |
| 130 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 6.87e-12 | 2.85e-14 | 296 | 46 | 24100 | 31.4 | 1.47 | ESR_clop |
| 131 | 12 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 7.55e-12 | 3.11e-14 | 618 | 96.1 | 66800 | 87.2 | 1.1 | ESR_clop |
| 132 | 5 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 8.21e-12 | 3.36e-14 | 533 | 82.9 | 53300 | 69.6 | 1.19 | ESR_clop |
| 133 | 12 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 8.94e-12 | 3.62e-14 | 597 | 92.8 | 63100 | 82.4 | 1.13 | ESR_clop |
| 134 | 5 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 1.55e-11 | 6.22e-14 | 620 | 96.4 | 67300 | 87.8 | 1.1 | ESR_clop |
| 135 | 6 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 1.67e-11 | 6.66e-14 | 343 | 53.3 | 29500 | 38.5 | 1.39 | ESR_clop |
| 136 | 5 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 2e-11 | 7.93e-14 | 618 | 96.1 | 67000 | 87.4 | 1.1 | ESR_clop |
| 137 | 5 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 3.48e-11 | 1.37e-13 | 564 | 87.7 | 58100 | 75.8 | 1.16 | ESR_clop |
| 138 | 5 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 3.85e-11 | 1.51e-13 | 589 | 91.6 | 62000 | 81 | 1.13 | ESR_clop |
| 139 | 5 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 3.96e-11 | 1.54e-13 | 404 | 62.8 | 36800 | 48.1 | 1.31 | ESR_clop |
| 140 | 9 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 9.48e-11 | 3.65e-13 | 382 | 59.4 | 34400 | 44.8 | 1.33 | ESR_clop |
| 141 | 5 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 1.04e-10 | 4e-13 | 501 | 77.9 | 49400 | 64.5 | 1.21 | ESR_clop |
| 142 | 6 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 1.24e-10 | 4.7e-13 | 338 | 52.6 | 29300 | 38.2 | 1.38 | ESR_clop |
| 143 | 5 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 1.39e-10 | 5.25e-13 | 381 | 59.2 | 34300 | 44.8 | 1.32 | ESR_clop |
| 144 | 19 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 1.5e-10 | 5.6e-13 | 573 | 89.1 | 59800 | 78 | 1.14 | ESR_clop |
| 145 | 5 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 1.65e-10 | 6.13e-13 | 540 | 84 | 54900 | 71.6 | 1.17 | ESR_clop |
| 146 | 20 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 1.89e-10 | 6.97e-13 | 533 | 82.9 | 53900 | 70.4 | 1.18 | ESR_clop |
| 147 | 12 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 2.61e-10 | 9.58e-13 | 494 | 76.8 | 48700 | 63.5 | 1.21 | ESR_clop |
| 148 | 10 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 2.84e-10 | 1.03e-12 | 512 | 79.6 | 51100 | 66.7 | 1.19 | ESR_clop |
| 149 | 5 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 4.61e-10 | 1.67e-12 | 423 | 65.8 | 39700 | 51.8 | 1.27 | ESR_clop |
| 150 | 17 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 6.44e-10 | 2.31e-12 | 512 | 79.6 | 51300 | 66.9 | 1.19 | ESR_clop |
| 151 | 21 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 8.88e-10 | 3.17e-12 | 237 | 36.9 | 18600 | 24.3 | 1.52 | ESR_clop |
| 152 | 12 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 1.16e-09 | 4.12e-12 | 495 | 77 | 49100 | 64.1 | 1.2 | ESR_clop |
| 153 | 5 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 1.38e-09 | 4.85e-12 | 594 | 92.4 | 63500 | 82.8 | 1.12 | ESR_clop |
| 154 | 5 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 1.78e-09 | 6.22e-12 | 437 | 68 | 41700 | 54.4 | 1.25 | ESR_clop |
| 155 | 11 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 2.65e-09 | 9.22e-12 | 586 | 91.1 | 62300 | 81.3 | 1.12 | ESR_clop |
| 156 | 5 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 3.66e-09 | 1.26e-11 | 579 | 90 | 61300 | 80 | 1.13 | ESR_clop |
| 157 | 5 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 3.94e-09 | 1.35e-11 | 547 | 85.1 | 56500 | 73.8 | 1.15 | ESR_clop |
| 158 | 12 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 7.81e-09 | 2.67e-11 | 465 | 72.3 | 45600 | 59.5 | 1.22 | ESR_clop |
| 159 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 1.05e-08 | 3.57e-11 | 184 | 28.6 | 13600 | 17.8 | 1.62 | ESR_clop |
| 160 | 10 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 1.13e-08 | 3.8e-11 | 496 | 77.1 | 49700 | 64.9 | 1.19 | ESR_clop |
| 161 | 22 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 1.23e-08 | 4.11e-11 | 563 | 87.6 | 59100 | 77.1 | 1.14 | ESR_clop |
| 162 | 5 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 1.48e-08 | 4.93e-11 | 286 | 44.5 | 24400 | 31.8 | 1.4 | ESR_clop |
| 163 | 19 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 1.6e-08 | 5.29e-11 | 618 | 96.1 | 68000 | 88.7 | 1.08 | ESR_clop |
| 164 | 21 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 1.87e-08 | 6.15e-11 | 452 | 70.3 | 44100 | 57.6 | 1.22 | ESR_clop |
| 165 | 5 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 2.22e-08 | 7.27e-11 | 428 | 66.6 | 41100 | 53.7 | 1.24 | ESR_clop |
| 166 | 10 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 2.84e-08 | 9.24e-11 | 541 | 84.1 | 56100 | 73.2 | 1.15 | ESR_clop |
| 167 | 5 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 2.88e-08 | 9.31e-11 | 532 | 82.7 | 54800 | 71.6 | 1.16 | ESR_clop |
| 168 | 17 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 3.67e-08 | 1.18e-10 | 468 | 72.8 | 46300 | 60.4 | 1.21 | ESR_clop |
| 169 | 17 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 4.22e-08 | 1.35e-10 | 499 | 77.6 | 50400 | 65.8 | 1.18 | ESR_clop |
| 170 | 21 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 4.59e-08 | 1.46e-10 | 547 | 85.1 | 57100 | 74.4 | 1.14 | ESR_clop |
| 171 | 21 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 5.15e-08 | 1.62e-10 | 523 | 81.3 | 53700 | 70.1 | 1.16 | ESR_clop |
| 172 | 23 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 5.6e-08 | 1.76e-10 | 286 | 44.5 | 24700 | 32.2 | 1.38 | ESR_clop |
| 173 | 24 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 5.79e-08 | 1.81e-10 | 598 | 93 | 64800 | 84.6 | 1.1 | ESR_clop |
| 174 | 5 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 6.21e-08 | 1.92e-10 | 583 | 90.7 | 62400 | 81.5 | 1.11 | ESR_clop |
| 175 | 20 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 8.84e-08 | 2.72e-10 | 571 | 88.8 | 60700 | 79.2 | 1.12 | ESR_clop |
| 176 | 13 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 1.32e-07 | 4.06e-10 | 585 | 91 | 62900 | 82.1 | 1.11 | ESR_clop |
| 177 | 11 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 4.76e-07 | 1.45e-09 | 392 | 61 | 37400 | 48.9 | 1.25 | ESR_clop |
| 178 | 5 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 7.06e-07 | 2.14e-09 | 397 | 61.7 | 38100 | 49.8 | 1.24 | ESR_clop |
| 179 | 9 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 7.54e-07 | 2.27e-09 | 411 | 63.9 | 39900 | 52 | 1.23 | ESR_clop |
| 180 | 13 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 7.67e-07 | 2.3e-09 | 537 | 83.5 | 56300 | 73.4 | 1.14 | ESR_clop |
| 181 | 21 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 8.59e-07 | 2.56e-09 | 433 | 67.3 | 42600 | 55.6 | 1.21 | ESR_clop |
| 182 | 21 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 1.09e-06 | 3.24e-09 | 400 | 62.2 | 38600 | 50.4 | 1.24 | ESR_clop |
| 183 | 21 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 1.17e-06 | 3.45e-09 | 528 | 82.1 | 55100 | 71.9 | 1.14 | ESR_clop |
| 184 | 5 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 1.22e-06 | 3.56e-09 | 499 | 77.6 | 51200 | 66.8 | 1.16 | ESR_clop |
| 185 | 20 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 2.12e-06 | 6.17e-09 | 79 | 12.3 | 4600 | 6 | 2.07 | ESR_clop |
| 186 | 13 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 2.71e-06 | 7.85e-09 | 468 | 72.8 | 47400 | 61.8 | 1.18 | ESR_clop |
| 187 | 1 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 3.23e-06 | 9.32e-09 | 366 | 56.9 | 34800 | 45.4 | 1.25 | ESR_clop |
| 188 | 25 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 3.27e-06 | 9.38e-09 | 553 | 86 | 58800 | 76.8 | 1.12 | ESR_clop |
| 189 | 5 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 4.09e-06 | 1.17e-08 | 329 | 51.2 | 30500 | 39.8 | 1.29 | ESR_clop |
| 190 | 24 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 4.27e-06 | 1.21e-08 | 575 | 89.4 | 62100 | 81 | 1.1 | ESR_clop |
| 191 | 5 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 5.23e-06 | 1.47e-08 | 500 | 77.8 | 51700 | 67.4 | 1.15 | ESR_clop |
| 192 | 5 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 5.25e-06 | 1.47e-08 | 408 | 63.4 | 40000 | 52.2 | 1.22 | ESR_clop |
| 193 | 13 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 5.27e-06 | 1.47e-08 | 406 | 63.1 | 39800 | 51.9 | 1.22 | ESR_clop |
| 194 | 17 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 5.53e-06 | 1.54e-08 | 259 | 40.3 | 22700 | 29.6 | 1.36 | ESR_clop |
| 195 | 17 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 8.66e-06 | 2.39e-08 | 530 | 82.4 | 55900 | 72.9 | 1.13 | ESR_clop |
| 196 | 5 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 1.14e-05 | 3.13e-08 | 603 | 93.8 | 66600 | 86.9 | 1.08 | ESR_clop |
| 197 | 26 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 1.38e-05 | 3.77e-08 | 440 | 68.4 | 44200 | 57.7 | 1.19 | ESR_clop |
| 198 | 27 | MA1581.1 (ZBTB6) | MEME-11 | CCTGGGCTCAAGYRA | CCTGGGCTCAAGYRA | ZBTB6 | 1.56e-05 | 4.24e-08 | 23 | 3.58 | 648 | 0.85 | 4.4 | ESR_clop |
| 199 | 5 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 1.76e-05 | 4.78e-08 | 521 | 81 | 54800 | 71.5 | 1.13 | ESR_clop |
| 200 | 9 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 1.85e-05 | 4.99e-08 | 244 | 38 | 21300 | 27.8 | 1.37 | ESR_clop |
| 201 | 23 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 2.12e-05 | 5.69e-08 | 550 | 85.5 | 58900 | 76.8 | 1.11 | ESR_clop |
| 202 | 28 | MA1125.1 (ZNF384) | MEME-2 | TTTTTTTTTTTTTTT | TTTTTTTTTTTTTTT | ZNF384 | 2.52e-05 | 6.73e-08 | 247 | 38.4 | 21700 | 28.4 | 1.36 | ESR_clop |
| 203 | 4 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 2.63e-05 | 6.99e-08 | 255 | 39.7 | 22600 | 29.5 | 1.35 | ESR_clop |
| 204 | 17 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 3.11e-05 | 8.23e-08 | 83 | 12.9 | 5250 | 6.86 | 1.9 | ESR_clop |
| 205 | 12 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 3.17e-05 | 8.35e-08 | 362 | 56.3 | 34900 | 45.6 | 1.24 | ESR_clop |
| 206 | 19 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 3.7e-05 | 9.68e-08 | 399 | 62 | 39400 | 51.4 | 1.21 | ESR_clop |
| 207 | 5 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 4.14e-05 | 1.08e-07 | 342 | 53.2 | 32600 | 42.6 | 1.25 | ESR_clop |
| 208 | 29 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 4.25e-05 | 1.1e-07 | 169 | 26.3 | 13600 | 17.7 | 1.49 | ESR_clop |
| 209 | 5 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 5.03e-05 | 1.3e-07 | 464 | 72.2 | 47600 | 62.1 | 1.16 | ESR_clop |
| 210 | 12 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 5.72e-05 | 1.47e-07 | 356 | 55.4 | 34400 | 44.9 | 1.24 | ESR_clop |
| 211 | 19 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 6.82e-05 | 1.74e-07 | 413 | 64.2 | 41300 | 53.9 | 1.19 | ESR_clop |
| 212 | 30 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 7.6e-05 | 1.93e-07 | 599 | 93.2 | 66400 | 86.6 | 1.08 | ESR_clop |
| 213 | 19 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 7.69e-05 | 1.94e-07 | 426 | 66.2 | 42900 | 56 | 1.18 | ESR_clop |
| 214 | 28 | MA1125.1 (ZNF384) | STREME-7 | 7-AGAAAAAAAAAW | AGAAAAAAAAAW | ZNF384 | 7.71e-05 | 1.94e-07 | 68 | 10.6 | 4060 | 5.3 | 2.02 | ESR_clop |
| 215 | 19 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 8.82e-05 | 2.21e-07 | 594 | 92.4 | 65600 | 85.6 | 1.08 | ESR_clop |
| 216 | 13 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 0.000102 | 2.55e-07 | 535 | 83.2 | 57200 | 74.6 | 1.12 | ESR_clop |
| 217 | 26 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 0.000125 | 3.12e-07 | 500 | 77.8 | 52500 | 68.5 | 1.14 | ESR_clop |
| 218 | 18 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 0.000149 | 3.69e-07 | 283 | 44 | 26200 | 34.2 | 1.29 | ESR_clop |
| 219 | 12 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 0.00015 | 3.7e-07 | 346 | 53.8 | 33500 | 43.7 | 1.23 | ESR_clop |
| 220 | 31 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 0.000197 | 4.82e-07 | 575 | 89.4 | 63000 | 82.2 | 1.09 | ESR_clop |
| 221 | 5 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 0.000238 | 5.81e-07 | 396 | 61.6 | 39600 | 51.6 | 1.19 | ESR_clop |
| 222 | 5 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 0.000298 | 7.24e-07 | 333 | 51.8 | 32100 | 41.9 | 1.24 | ESR_clop |
| 223 | 5 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 0.000365 | 8.82e-07 | 313 | 48.7 | 29900 | 39 | 1.25 | ESR_clop |
| 224 | 32 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 0.000382 | 9.21e-07 | 431 | 67 | 44000 | 57.4 | 1.17 | ESR_clop |
| 225 | 5 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 0.000608 | 1.46e-06 | 540 | 84 | 58300 | 76.1 | 1.1 | ESR_clop |
| 226 | 33 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 0.000612 | 1.46e-06 | 365 | 56.8 | 36100 | 47.2 | 1.21 | ESR_clop |
| 227 | 26 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 0.000618 | 1.47e-06 | 268 | 41.7 | 24900 | 32.5 | 1.29 | ESR_clop |
| 228 | 12 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 0.000629 | 1.49e-06 | 32 | 4.98 | 1400 | 1.83 | 2.79 | ESR_clop |
| 229 | 34 | MA1973.1 (ZKSCAN3) | MEME-3 | TGTYRCCCAGGCTGG | TGTYRCCCAGGCTGG | ZKSCAN3 | 0.000755 | 1.78e-06 | 91 | 14.2 | 6420 | 8.38 | 1.7 | ESR_clop |
| 230 | 23 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 0.000776 | 1.82e-06 | 495 | 77 | 52400 | 68.4 | 1.13 | ESR_clop |
| 231 | 23 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 0.000932 | 2.18e-06 | 540 | 84 | 58400 | 76.2 | 1.1 | ESR_clop |
| 232 | 19 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 0.000961 | 2.23e-06 | 387 | 60.2 | 38900 | 50.8 | 1.19 | ESR_clop |
| 233 | 35 | 6-AGGTTGCAAT | STREME-6 | 6-AGGTTGCAAT | AGGTTGCAAT | AGGTTGCAAT | 0.00104 | 2.41e-06 | 48 | 7.47 | 2670 | 3.49 | 2.18 | ESR_clop |
| 234 | 36 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 0.00111 | 2.55e-06 | 544 | 84.6 | 59000 | 77 | 1.1 | ESR_clop |
| 235 | 37 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 0.00115 | 2.65e-06 | 436 | 67.8 | 45000 | 58.7 | 1.16 | ESR_clop |
| 236 | 38 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 0.00117 | 2.68e-06 | 539 | 83.8 | 58400 | 76.2 | 1.1 | ESR_clop |
| 237 | 39 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 0.00164 | 3.74e-06 | 325 | 50.5 | 31700 | 41.4 | 1.22 | ESR_clop |
| 238 | 38 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 0.00173 | 3.93e-06 | 577 | 89.7 | 63800 | 83.2 | 1.08 | ESR_clop |
| 239 | 16 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 0.00192 | 4.34e-06 | 334 | 51.9 | 32800 | 42.8 | 1.21 | ESR_clop |
| 240 | 23 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 0.00198 | 4.45e-06 | 482 | 75 | 51000 | 66.5 | 1.13 | ESR_clop |
| 241 | 40 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 0.00209 | 4.67e-06 | 407 | 63.3 | 41600 | 54.3 | 1.17 | ESR_clop |
| 242 | 23 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 0.00222 | 4.96e-06 | 173 | 26.9 | 14900 | 19.4 | 1.39 | ESR_clop |
| 243 | 1 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 0.00224 | 4.96e-06 | 148 | 23 | 12300 | 16 | 1.45 | ESR_clop |
| 244 | 7 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 0.00231 | 5.1e-06 | 279 | 43.4 | 26500 | 34.6 | 1.26 | ESR_clop |
| 245 | 41 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 0.00238 | 5.24e-06 | 625 | 97.2 | 71300 | 93 | 1.04 | ESR_clop |
| 246 | 23 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 0.00248 | 5.44e-06 | 463 | 72 | 48600 | 63.4 | 1.14 | ESR_clop |
| 247 | 9 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 0.00282 | 6.16e-06 | 314 | 48.8 | 30600 | 39.9 | 1.22 | ESR_clop |
| 248 | 42 | 8-STAYCTCAT | STREME-8 | 8-STAYCTCAT | STAYCTCAT | STAYCTCAT | 0.00329 | 7.15e-06 | 519 | 80.7 | 56000 | 73 | 1.11 | ESR_clop |
| 249 | 21 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 0.00341 | 7.38e-06 | 551 | 85.7 | 60300 | 78.7 | 1.09 | ESR_clop |
| 12 | 46 | GYGAGACYCTGTCTC | MEME-7 | GYGAGACYCTGTCTC | GYGAGACYCTGTCTC | GYGAGACYCTGTCTC | 0.00344 | 0.000286 | 11 | 1.71 | 335 | 0.44 | 4.25 | ESR_clop |
| 250 | 21 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 0.00349 | 7.53e-06 | 240 | 37.3 | 22300 | 29 | 1.29 | ESR_clop |
| 251 | 23 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 0.00356 | 7.64e-06 | 381 | 59.2 | 38600 | 50.4 | 1.18 | ESR_clop |
| 252 | 17 | MA0837.2 | CEBPE | MA0837.2 | KATTGCGCAATM | CEBPE | 0.00366 | 7.83e-06 | 95 | 14.8 | 7060 | 9.22 | 1.62 | ESR_clop |
| 253 | 5 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 0.00369 | 7.87e-06 | 266 | 41.4 | 25200 | 32.9 | 1.26 | ESR_clop |
| 254 | 23 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 0.00381 | 8.09e-06 | 120 | 18.7 | 9530 | 12.4 | 1.51 | ESR_clop |
| 255 | 19 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 0.00387 | 8.18e-06 | 518 | 80.6 | 55900 | 72.9 | 1.11 | ESR_clop |
| 256 | 6 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 0.00396 | 8.33e-06 | 136 | 21.2 | 11200 | 14.6 | 1.46 | ESR_clop |
| 257 | 21 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 0.00418 | 8.77e-06 | 123 | 19.1 | 9850 | 12.8 | 1.5 | ESR_clop |
| 258 | 17 | MA0466.3 | CEBPB | MA0466.3 | KATTGCGCAATM | CEBPB | 0.0046 | 9.61e-06 | 64 | 9.95 | 4220 | 5.5 | 1.83 | ESR_clop |
| 259 | 12 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 0.00492 | 1.03e-05 | 232 | 36.1 | 21500 | 28 | 1.29 | ESR_clop |
| 260 | 43 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 0.00505 | 1.05e-05 | 112 | 17.4 | 8790 | 11.5 | 1.53 | ESR_clop |
| 13 | 47 | MA0682.2 (PITX1) | MEME-4 | GCTGGGATTACAGGC | GCTGGGATTACAGGC | PITX1 | 0.00531 | 0.000408 | 66 | 10.3 | 5030 | 6.57 | 1.58 | ESR_clop |
| 261 | 13 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 0.00582 | 1.2e-05 | 481 | 74.8 | 51200 | 66.8 | 1.12 | ESR_clop |
| 262 | 5 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 0.00587 | 1.21e-05 | 333 | 51.8 | 33100 | 43.1 | 1.2 | ESR_clop |
| 263 | 5 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 0.00588 | 1.21e-05 | 354 | 55 | 35500 | 46.4 | 1.19 | ESR_clop |
| 264 | 16 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 0.00855 | 1.75e-05 | 98 | 15.2 | 7510 | 9.8 | 1.57 | ESR_clop |
| 265 | 12 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 0.00985 | 2.01e-05 | 38 | 5.91 | 2080 | 2.72 | 2.23 | ESR_clop |
| 266 | 44 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 0.0112 | 2.26e-05 | 482 | 75 | 51500 | 67.2 | 1.12 | ESR_clop |
| 267 | 5 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 0.0114 | 2.3e-05 | 195 | 30.3 | 17600 | 23 | 1.32 | ESR_clop |
| 268 | 12 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 0.0116 | 2.33e-05 | 420 | 65.3 | 43800 | 57.1 | 1.14 | ESR_clop |
| 269 | 23 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 0.0121 | 2.42e-05 | 153 | 23.8 | 13200 | 17.2 | 1.39 | ESR_clop |
| 270 | 5 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 0.0154 | 3.08e-05 | 247 | 38.4 | 23500 | 30.6 | 1.26 | ESR_clop |
| 271 | 21 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 0.0167 | 3.32e-05 | 224 | 34.8 | 20900 | 27.3 | 1.28 | ESR_clop |
| 14 | 48 | MA1515.1 (KLF2) | MEME-10 | GCYRGGCRTGGTGGC | GCYRGGCRTGGTGGC | KLF2 | 0.0173 | 0.00124 | 28 | 4.35 | 1720 | 2.24 | 2.01 | ESR_clop |
| 272 | 41 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 0.0189 | 3.75e-05 | 583 | 90.7 | 65300 | 85.2 | 1.06 | ESR_clop |
| 273 | 16 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 0.0242 | 4.77e-05 | 406 | 63.1 | 42300 | 55.2 | 1.15 | ESR_clop |
| 274 | 17 | MA0843.1 | TEF | MA0843.1 | BRTTACRTAAYM | TEF | 0.0242 | 4.77e-05 | 220 | 34.2 | 20600 | 26.9 | 1.28 | ESR_clop |
| 275 | 21 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 0.0249 | 4.89e-05 | 126 | 19.6 | 10500 | 13.8 | 1.43 | ESR_clop |
| 276 | 5 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 0.0258 | 5.04e-05 | 286 | 44.5 | 28100 | 36.6 | 1.22 | ESR_clop |
| 277 | 45 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 0.0291 | 5.66e-05 | 417 | 64.8 | 43700 | 57 | 1.14 | ESR_clop |
| 278 | 29 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 0.0304 | 5.89e-05 | 611 | 95 | 69500 | 90.7 | 1.05 | ESR_clop |
| 279 | 23 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 0.0362 | 6.99e-05 | 347 | 54 | 35300 | 46.1 | 1.17 | ESR_clop |
| 280 | 21 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 0.0446 | 8.59e-05 | 227 | 35.3 | 21600 | 28.2 | 1.26 | ESR_clop |
| 15 | 49 | MA1107.2 (KLF9) | MEME-8 | GTGWGTGTRDRTGTG | GTGWGTGTRDRTGTG | KLF9 | 0.0519 | 0.00346 | 17 | 2.64 | 925 | 1.21 | 2.31 | ESR_clop |
| 16 | 50 | 5-ACTCCA | STREME-5 | 5-ACTCCA | ACTCCA | ACTCCA | 0.156 | 0.00975 | 568 | 88.3 | 65100 | 85 | 1.04 | ESR_clop |
| 17 | 35 | GCTCACTGCAACCTC | MEME-6 | GCTCACTGCAACCTC | GCTCACTGCAACCTC | GCTCACTGCAACCTC | 0.377 | 0.0222 | 4 | 0.62 | 119 | 0.16 | 4.96 | ESR_clop |
| 18 | 51 | MA1596.1 (ZNF460) | MEME-5 | CCTRYCTCWBCCTCY | CCTRYCTCWBCCTCY | ZNF460 | 0.545 | 0.0303 | 402 | 62.5 | 45000 | 58.8 | 1.07 | ESR_clop |
| 19 | 52 | GATYRYRCCACTGCA | MEME-9 | GATYRYRCCACTGCA | GATYRYRCCACTGCA | GATYRYRCCACTGCA | 19 | 1 | 0 | 0 | 2 | 0 | 39.7 | ESR_clop |
ER_rat <- 1.25
mrc_type <- "ESR_clop"
ESR_clop_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))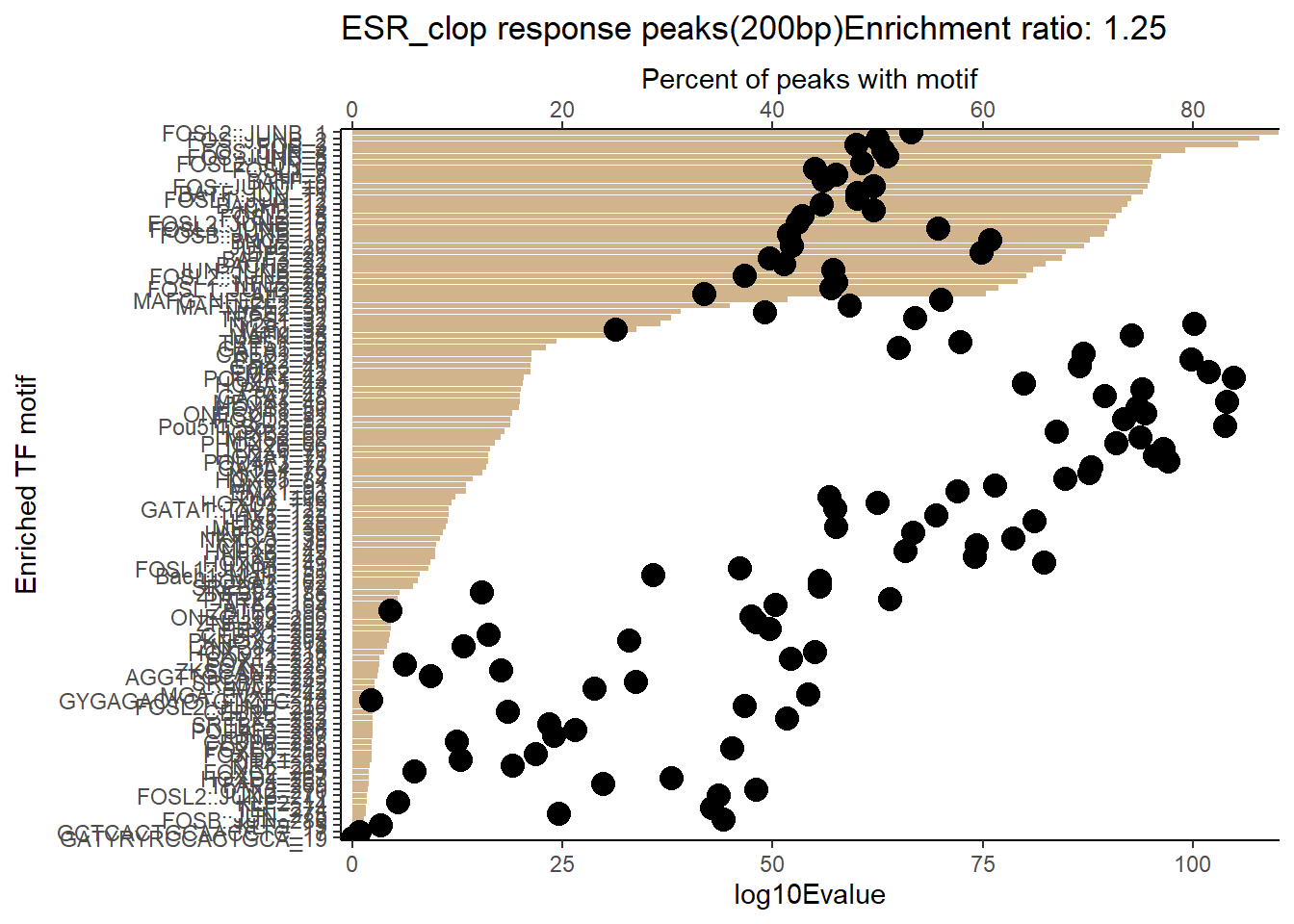
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
ESR_clop_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.1), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.1,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))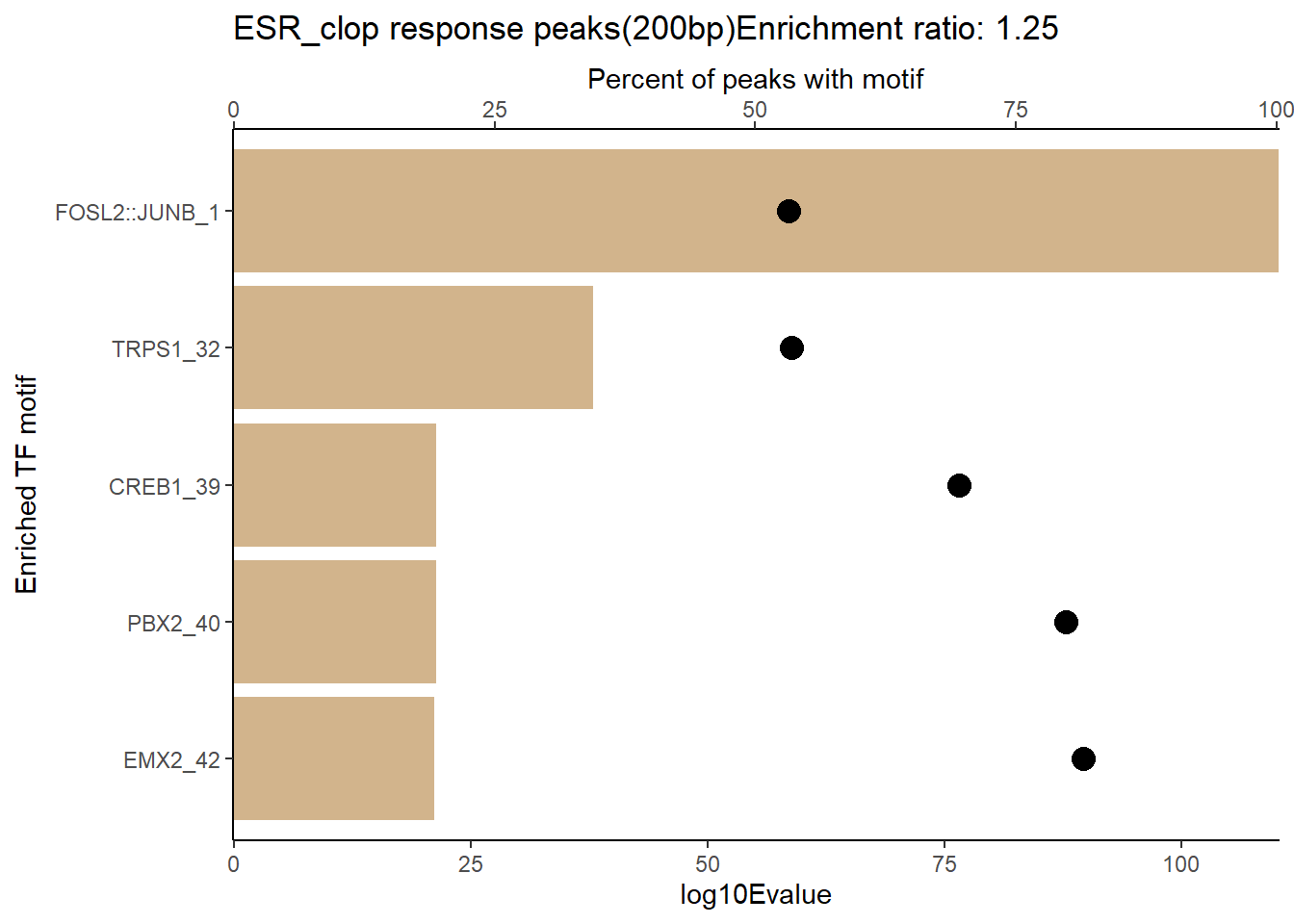
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
LR_open data
LR_open_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_open_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
LR_open_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_open_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
LR_open_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_open_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
LR_open_sea_all <- rbind(LR_open_sea_disc, LR_open_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)LR_open_combo <- LR_open_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., LR_open_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="LR_open")
LR_open_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-DATGASTCATH | DATGASTCATH | FOS::JUNB | 0 | 0 | 11700 | 50.9 | 12000 | 15.6 | 3.26 | LR_open |
| 2 | 1 | MA1988.1 (Atf3) | MEME-1 | DRTGACTCAYH | DRTGACTCAYH | Atf3 | 0 | 0 | 11500 | 49.9 | 11500 | 15.1 | 3.32 | LR_open |
| 3 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 11600 | 50.2 | 12000 | 15.6 | 3.22 | LR_open |
| 4 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 0 | 0 | 12100 | 52.4 | 13200 | 17.3 | 3.03 | LR_open |
| 5 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 0 | 0 | 11900 | 51.6 | 12800 | 16.7 | 3.09 | LR_open |
| 6 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0 | 0 | 11100 | 48 | 11000 | 14.3 | 3.36 | LR_open |
| 7 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 12400 | 53.7 | 14100 | 18.4 | 2.91 | LR_open |
| 8 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 0 | 0 | 12400 | 53.6 | 14200 | 18.5 | 2.9 | LR_open |
| 9 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 0 | 0 | 10300 | 44.5 | 9450 | 12.3 | 3.61 | LR_open |
| 10 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 0 | 0 | 11500 | 50 | 12300 | 16 | 3.13 | LR_open |
| 11 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 0 | 0 | 11900 | 51.7 | 13200 | 17.2 | 3.01 | LR_open |
| 12 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 0 | 0 | 12100 | 52.4 | 13600 | 17.7 | 2.95 | LR_open |
| 13 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 0 | 0 | 10900 | 47.4 | 10900 | 14.3 | 3.32 | LR_open |
| 14 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 0 | 0 | 10700 | 46.4 | 10400 | 13.6 | 3.4 | LR_open |
| 15 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 0 | 0 | 12000 | 52 | 13400 | 17.5 | 2.96 | LR_open |
| 16 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 0 | 0 | 12300 | 53.4 | 14300 | 18.6 | 2.87 | LR_open |
| 17 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 0 | 0 | 11700 | 50.8 | 12800 | 16.7 | 3.04 | LR_open |
| 18 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 0 | 0 | 11400 | 49.5 | 12200 | 15.9 | 3.11 | LR_open |
| 19 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 0 | 0 | 12100 | 52.6 | 14000 | 18.2 | 2.89 | LR_open |
| 20 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 0 | 0 | 12700 | 55 | 15400 | 20.1 | 2.73 | LR_open |
| 21 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 0 | 0 | 11300 | 49 | 12300 | 16 | 3.06 | LR_open |
| 22 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 0 | 0 | 11500 | 49.7 | 12700 | 16.6 | 2.99 | LR_open |
| 23 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 0 | 0 | 11600 | 50.4 | 13100 | 17.1 | 2.95 | LR_open |
| 24 | 1 | MA0476.1 (FOS) | STREME-10 | 10-TGANTCA | 10-TGANTCA | FOS | 0 | 0 | 10600 | 46.1 | 10900 | 14.2 | 3.24 | LR_open |
| 25 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 0 | 0 | 10000 | 43.6 | 9660 | 12.6 | 3.46 | LR_open |
| 26 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 0 | 0 | 9840 | 42.7 | 9300 | 12.1 | 3.52 | LR_open |
| 27 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 0 | 0 | 10800 | 46.9 | 12100 | 15.7 | 2.98 | LR_open |
| 28 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 0 | 0 | 9410 | 40.8 | 9010 | 11.8 | 3.47 | LR_open |
| 29 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 0 | 0 | 8980 | 38.9 | 8420 | 11 | 3.54 | LR_open |
| 30 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 0 | 0 | 12000 | 52.2 | 19300 | 25.1 | 2.08 | LR_open |
| 31 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 0 | 0 | 10200 | 44.2 | 15000 | 19.5 | 2.27 | LR_open |
| 32 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 0 | 0 | 11600 | 50.5 | 19700 | 25.7 | 1.97 | LR_open |
| 33 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 0 | 0 | 10900 | 47.1 | 19700 | 25.7 | 1.83 | LR_open |
| 34 | 2 | 2-ATKMAT | STREME-2 | 2-ATKMAT | ATKMAT | ATKMAT | 0 | 0 | 17900 | 77.8 | 45100 | 58.9 | 1.32 | LR_open |
| 35 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 0 | 0 | 9690 | 42 | 18100 | 23.7 | 1.78 | LR_open |
| 36 | 3 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 0 | 0 | 20300 | 87.9 | 55500 | 72.5 | 1.21 | LR_open |
| 37 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 0 | 0 | 17600 | 76.2 | 44800 | 58.4 | 1.3 | LR_open |
| 38 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 0 | 0 | 16300 | 70.7 | 40200 | 52.5 | 1.35 | LR_open |
| 39 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 0 | 0 | 20000 | 86.6 | 54700 | 71.3 | 1.21 | LR_open |
| 40 | 4 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 0 | 0 | 17700 | 76.9 | 45700 | 59.7 | 1.29 | LR_open |
| 41 | 5 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 0 | 0 | 19800 | 85.7 | 54200 | 70.7 | 1.21 | LR_open |
| 42 | 6 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 0 | 0 | 20300 | 87.9 | 56400 | 73.6 | 1.19 | LR_open |
| 43 | 7 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 0 | 0 | 18700 | 81.3 | 50100 | 65.4 | 1.24 | LR_open |
| 44 | 8 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 0 | 0 | 20000 | 86.7 | 55600 | 72.5 | 1.2 | LR_open |
| 45 | 8 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 0 | 0 | 20000 | 86.6 | 55600 | 72.6 | 1.19 | LR_open |
| 46 | 9 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 0 | 0 | 18300 | 79.6 | 49000 | 63.9 | 1.24 | LR_open |
| 47 | 9 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 0 | 0 | 17200 | 74.6 | 44700 | 58.3 | 1.28 | LR_open |
| 48 | 7 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 0 | 0 | 17600 | 76.5 | 46500 | 60.6 | 1.26 | LR_open |
| 49 | 6 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 0 | 0 | 21100 | 91.7 | 61100 | 79.7 | 1.15 | LR_open |
| 50 | 10 | 3-WTVWBAW | STREME-3 | 3-WTVWBAW | WTVWBAW | WTVWBAW | 0 | 0 | 21300 | 92.4 | 61900 | 80.8 | 1.14 | LR_open |
| 51 | 8 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 0 | 0 | 21000 | 91.1 | 60500 | 79 | 1.15 | LR_open |
| 52 | 6 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 0 | 0 | 21300 | 92.2 | 61800 | 80.6 | 1.14 | LR_open |
| 53 | 4 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 0 | 0 | 14900 | 64.8 | 37000 | 48.3 | 1.34 | LR_open |
| 54 | 6 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 0 | 0 | 20600 | 89.4 | 58900 | 76.8 | 1.16 | LR_open |
| 55 | 8 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 0 | 0 | 21500 | 93.1 | 63000 | 82.3 | 1.13 | LR_open |
| 56 | 7 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 0 | 0 | 21000 | 91.1 | 60900 | 79.4 | 1.15 | LR_open |
| 57 | 7 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 0 | 0 | 19200 | 83.4 | 53200 | 69.4 | 1.2 | LR_open |
| 58 | 11 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 0 | 0 | 20800 | 90.4 | 60100 | 78.4 | 1.15 | LR_open |
| 59 | 12 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 0 | 0 | 16200 | 70.5 | 42000 | 54.8 | 1.29 | LR_open |
| 60 | 8 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 0 | 0 | 18800 | 81.7 | 51800 | 67.6 | 1.21 | LR_open |
| 61 | 13 | 15-HHATDD | STREME-15 | 15-HHATDD | 15-HHATDD | HHATDD | 0 | 0 | 22000 | 95.2 | 65600 | 85.6 | 1.11 | LR_open |
| 62 | 7 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 0 | 0 | 18800 | 81.6 | 51800 | 67.6 | 1.21 | LR_open |
| 63 | 4 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 0 | 0 | 13300 | 57.8 | 32200 | 42 | 1.38 | LR_open |
| 64 | 7 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 0 | 0 | 19300 | 83.6 | 53700 | 70 | 1.19 | LR_open |
| 65 | 14 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 0 | 0 | 19500 | 84.5 | 54600 | 71.3 | 1.19 | LR_open |
| 66 | 7 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 0 | 0 | 19700 | 85.6 | 55700 | 72.7 | 1.18 | LR_open |
| 67 | 9 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 0 | 0 | 17000 | 73.9 | 45200 | 59 | 1.25 | LR_open |
| 68 | 8 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 0 | 0 | 19800 | 86 | 56100 | 73.2 | 1.17 | LR_open |
| 69 | 4 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 0 | 0 | 12900 | 56 | 31000 | 40.5 | 1.38 | LR_open |
| 70 | 8 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 0 | 0 | 21800 | 94.4 | 64900 | 84.7 | 1.11 | LR_open |
| 71 | 8 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 0 | 0 | 18300 | 79.2 | 50000 | 65.3 | 1.21 | LR_open |
| 72 | 15 | 4-TRTAYA | STREME-4 | 4-TRTAYA | TRTAYA | TRTAYA | 0 | 0 | 21600 | 93.7 | 64300 | 83.9 | 1.12 | LR_open |
| 73 | 9 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 0 | 0 | 18500 | 80.1 | 51000 | 66.5 | 1.2 | LR_open |
| 74 | 6 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 0 | 0 | 21000 | 91 | 61400 | 80.1 | 1.14 | LR_open |
| 75 | 7 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 0 | 0 | 20400 | 88.5 | 58900 | 76.8 | 1.15 | LR_open |
| 76 | 7 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 0 | 0 | 17900 | 77.7 | 48900 | 63.8 | 1.22 | LR_open |
| 77 | 8 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 0 | 0 | 18700 | 81 | 51800 | 67.6 | 1.2 | LR_open |
| 78 | 7 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 0 | 0 | 18100 | 78.5 | 49600 | 64.7 | 1.21 | LR_open |
| 79 | 6 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 0 | 0 | 20800 | 90.2 | 60700 | 79.2 | 1.14 | LR_open |
| 80 | 16 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 0 | 0 | 21500 | 93.4 | 64000 | 83.6 | 1.12 | LR_open |
| 81 | 17 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 0 | 0 | 19600 | 84.9 | 55600 | 72.6 | 1.17 | LR_open |
| 82 | 7 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 0 | 0 | 18100 | 78.4 | 49700 | 64.9 | 1.21 | LR_open |
| 83 | 7 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 0 | 0 | 20600 | 89.4 | 60000 | 78.3 | 1.14 | LR_open |
| 84 | 14 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 0 | 0 | 20600 | 89.2 | 59900 | 78.1 | 1.14 | LR_open |
| 85 | 7 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 0 | 0 | 16200 | 70.5 | 43100 | 56.2 | 1.25 | LR_open |
| 86 | 6 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 0 | 0 | 21800 | 94.4 | 65400 | 85.4 | 1.11 | LR_open |
| 87 | 6 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 0 | 0 | 21400 | 92.7 | 63600 | 82.9 | 1.12 | LR_open |
| 88 | 7 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 0 | 0 | 18900 | 81.9 | 53100 | 69.3 | 1.18 | LR_open |
| 89 | 18 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 0 | 0 | 20000 | 86.9 | 57800 | 75.4 | 1.15 | LR_open |
| 90 | 19 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 0 | 0 | 21300 | 92.4 | 63300 | 82.6 | 1.12 | LR_open |
| 91 | 7 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 2.01084717857387e-321 | 2.01084717857387e-321 | 15900 | 68.9 | 42000 | 54.8 | 1.26 | LR_open |
| 92 | 7 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 5.41001882196165e-319 | 2.46044691628941e-321 | 20700 | 90 | 60900 | 79.5 | 1.13 | LR_open |
| 93 | 20 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 2.05000188100679e-318 | 9.19950232556401e-321 | 19300 | 83.8 | 55000 | 71.7 | 1.17 | LR_open |
| 94 | 7 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 3.58000001561162e-316 | 1.5900020614463e-318 | 19300 | 83.7 | 54900 | 71.6 | 1.17 | LR_open |
| 95 | 21 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 9.99999998481684e-316 | 4.39000053349656e-318 | 17100 | 74.1 | 46400 | 60.5 | 1.22 | LR_open |
| 96 | 7 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 1.69000000005259e-314 | 7.32999991728063e-317 | 20700 | 89.8 | 60800 | 79.3 | 1.13 | LR_open |
| 97 | 7 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 1.94000000001886e-313 | 8.35000002412995e-316 | 21100 | 91.6 | 62600 | 81.7 | 1.12 | LR_open |
| 98 | 7 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 1.40999999999996e-311 | 6.01000000011395e-314 | 14000 | 60.5 | 35500 | 46.3 | 1.31 | LR_open |
| 99 | 6 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 1.38e-309 | 5.80999999999765e-312 | 21300 | 92.5 | 63600 | 83 | 1.11 | LR_open |
| 100 | 22 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 3.88e-308 | 1.61999999999998e-310 | 17000 | 73.8 | 46300 | 60.5 | 1.22 | LR_open |
| 101 | 7 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 2.54e-304 | 1.05e-306 | 19400 | 84 | 55400 | 72.2 | 1.16 | LR_open |
| 102 | 6 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 2.59e-300 | 1.06e-302 | 18800 | 81.3 | 53000 | 69.2 | 1.18 | LR_open |
| 103 | 7 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 8.19999999999999e-297 | 3.32e-299 | 20200 | 87.6 | 58800 | 76.8 | 1.14 | LR_open |
| 104 | 6 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 1.66e-296 | 6.67e-299 | 20700 | 89.9 | 61100 | 79.8 | 1.13 | LR_open |
| 105 | 23 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 1.78e-296 | 7.06e-299 | 21100 | 91.5 | 62800 | 81.9 | 1.12 | LR_open |
| 106 | 7 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 1.98e-296 | 7.78e-299 | 18400 | 79.7 | 51600 | 67.3 | 1.18 | LR_open |
| 107 | 16 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 2.79e-296 | 1.09e-298 | 21600 | 93.8 | 65200 | 85.1 | 1.1 | LR_open |
| 108 | 7 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 1.82e-293 | 7.03e-296 | 21200 | 92.2 | 63500 | 82.9 | 1.11 | LR_open |
| 109 | 7 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 5.02e-289 | 1.92e-291 | 16000 | 69.5 | 43100 | 56.2 | 1.24 | LR_open |
| 110 | 7 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 8.62e-289 | 3.27e-291 | 18500 | 80.1 | 52100 | 67.9 | 1.18 | LR_open |
| 111 | 24 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 9.99999999999999e-287 | 3.77e-289 | 14600 | 63.3 | 38100 | 49.7 | 1.27 | LR_open |
| 112 | 14 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 6.55e-286 | 2.44e-288 | 19100 | 82.7 | 54500 | 71.1 | 1.16 | LR_open |
| 113 | 21 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 9.44999999999999e-286 | 3.49e-288 | 17500 | 75.8 | 48400 | 63.1 | 1.2 | LR_open |
| 114 | 7 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.93e-285 | 7.07e-288 | 18100 | 78.6 | 50800 | 66.3 | 1.19 | LR_open |
| 115 | 6 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 2.76e-283 | 1e-285 | 17300 | 75.2 | 47900 | 62.5 | 1.2 | LR_open |
| 116 | 7 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 1.07e-282 | 3.84e-285 | 16700 | 72.4 | 45600 | 59.5 | 1.22 | LR_open |
| 117 | 7 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 2.84e-282 | 1.01e-284 | 13300 | 57.8 | 33900 | 44.2 | 1.31 | LR_open |
| 118 | 7 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 2.18e-280 | 7.7e-283 | 18500 | 80 | 52200 | 68.1 | 1.18 | LR_open |
| 119 | 7 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 1.49e-277 | 5.22e-280 | 19600 | 84.9 | 56700 | 74 | 1.15 | LR_open |
| 120 | 25 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 1.54e-275 | 5.34e-278 | 21200 | 92.1 | 63700 | 83.1 | 1.11 | LR_open |
| 121 | 7 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 2.32e-275 | 8e-278 | 20100 | 87.1 | 58700 | 76.6 | 1.14 | LR_open |
| 122 | 7 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 2.37e-275 | 8.11e-278 | 20900 | 90.6 | 62200 | 81.2 | 1.12 | LR_open |
| 123 | 6 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 4.31e-275 | 1.46e-277 | 20800 | 90.4 | 62000 | 80.9 | 1.12 | LR_open |
| 124 | 7 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 8.21e-275 | 2.76e-277 | 13200 | 57.2 | 33600 | 43.8 | 1.3 | LR_open |
| 125 | 25 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 1.45e-273 | 4.84e-276 | 21600 | 93.8 | 65600 | 85.5 | 1.1 | LR_open |
| 126 | 6 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 5.53e-273 | 1.83e-275 | 19900 | 86.4 | 58100 | 75.8 | 1.14 | LR_open |
| 127 | 7 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 4.31e-272 | 1.41e-274 | 15700 | 68 | 42100 | 55 | 1.24 | LR_open |
| 128 | 26 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 1.57e-271 | 5.1e-274 | 19300 | 83.5 | 55500 | 72.4 | 1.15 | LR_open |
| 129 | 7 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 6.84e-269 | 2.21e-271 | 15400 | 67 | 41400 | 54 | 1.24 | LR_open |
| 130 | 6 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 1.23e-268 | 3.94e-271 | 20200 | 87.6 | 59300 | 77.4 | 1.13 | LR_open |
| 131 | 11 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 1.23e-267 | 3.9e-270 | 17400 | 75.6 | 48600 | 63.4 | 1.19 | LR_open |
| 132 | 7 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 1.94e-267 | 6.12e-270 | 20600 | 89.4 | 61100 | 79.7 | 1.12 | LR_open |
| 133 | 27 | 6-TTAAAA | STREME-6 | 6-TTAAAA | TTAAAA | TTAAAA | 1.28e-265 | 4.01e-268 | 22000 | 95.3 | 67300 | 87.8 | 1.08 | LR_open |
| 134 | 9 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 1.96e-265 | 6.11e-268 | 9990 | 43.3 | 23600 | 30.8 | 1.41 | LR_open |
| 135 | 7 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 2.13e-263 | 6.57e-266 | 21900 | 95 | 67100 | 87.5 | 1.09 | LR_open |
| 136 | 6 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 5.67e-263 | 1.74e-265 | 22000 | 95.5 | 67600 | 88.2 | 1.08 | LR_open |
| 137 | 6 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 1.17e-262 | 3.57e-265 | 15300 | 66.5 | 41100 | 53.7 | 1.24 | LR_open |
| 138 | 7 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 2.74e-262 | 8.28e-265 | 14700 | 63.8 | 39000 | 50.9 | 1.25 | LR_open |
| 139 | 21 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 7.89e-260 | 2.37e-262 | 20600 | 89.3 | 61200 | 79.8 | 1.12 | LR_open |
| 140 | 7 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 1.31e-259 | 3.89e-262 | 19400 | 84 | 56200 | 73.3 | 1.15 | LR_open |
| 141 | 7 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 7.27e-259 | 2.15e-261 | 19400 | 84.3 | 56400 | 73.5 | 1.15 | LR_open |
| 142 | 16 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 1.43e-255 | 4.21e-258 | 22200 | 96.2 | 68600 | 89.5 | 1.08 | LR_open |
| 143 | 7 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 1.66e-254 | 4.84e-257 | 22000 | 95.3 | 67500 | 88.1 | 1.08 | LR_open |
| 144 | 7 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 1.86e-254 | 5.38e-257 | 14500 | 62.9 | 38400 | 50.2 | 1.25 | LR_open |
| 145 | 7 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 4.56e-254 | 1.31e-256 | 21300 | 92.2 | 64100 | 83.7 | 1.1 | LR_open |
| 146 | 25 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 1.93e-253 | 5.51e-256 | 20400 | 88.4 | 60300 | 78.7 | 1.12 | LR_open |
| 147 | 7 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 1.13e-251 | 3.2e-254 | 18600 | 80.8 | 53400 | 69.7 | 1.16 | LR_open |
| 148 | 14 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 1.41e-250 | 3.96e-253 | 14500 | 62.9 | 38500 | 50.3 | 1.25 | LR_open |
| 149 | 7 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 6.38e-248 | 1.79e-250 | 18600 | 80.8 | 53400 | 69.7 | 1.16 | LR_open |
| 150 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 1.88e-247 | 5.22e-250 | 4730 | 20.5 | 8800 | 11.5 | 1.79 | LR_open |
| 151 | 22 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 2.99e-247 | 8.26e-250 | 18700 | 81.2 | 53800 | 70.2 | 1.16 | LR_open |
| 152 | 7 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 8.81999999999999e-247 | 2.42e-249 | 20300 | 87.9 | 60000 | 78.2 | 1.12 | LR_open |
| 153 | 7 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 4.26e-246 | 1.16e-248 | 17800 | 77 | 50200 | 65.5 | 1.18 | LR_open |
| 154 | 22 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 2.43e-245 | 6.57e-248 | 16200 | 70.2 | 44500 | 58.1 | 1.21 | LR_open |
| 155 | 7 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 5.37e-245 | 1.45e-247 | 21100 | 91.5 | 63500 | 82.9 | 1.1 | LR_open |
| 156 | 7 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 6.97e-244 | 1.86e-246 | 20800 | 90.2 | 62300 | 81.3 | 1.11 | LR_open |
| 157 | 7 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 7.5e-244 | 1.99e-246 | 19800 | 85.8 | 58100 | 75.8 | 1.13 | LR_open |
| 158 | 7 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 1.9e-243 | 5e-246 | 15800 | 68.5 | 43100 | 56.2 | 1.22 | LR_open |
| 159 | 28 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 4.08e-243 | 1.07e-245 | 17900 | 77.6 | 50800 | 66.2 | 1.17 | LR_open |
| 160 | 7 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 1.08e-242 | 2.82e-245 | 20600 | 89.2 | 61300 | 80 | 1.12 | LR_open |
| 161 | 20 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 3.5e-242 | 9.07e-245 | 13100 | 56.8 | 34000 | 44.3 | 1.28 | LR_open |
| 162 | 7 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 1.07e-241 | 2.74e-244 | 17100 | 74.1 | 47700 | 62.3 | 1.19 | LR_open |
| 163 | 9 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 3.37e-241 | 8.62e-244 | 17100 | 74 | 47700 | 62.3 | 1.19 | LR_open |
| 164 | 7 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 8.72e-241 | 2.22e-243 | 16900 | 73.1 | 47000 | 61.3 | 1.19 | LR_open |
| 165 | 9 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 3.22e-239 | 8.15e-242 | 19600 | 85 | 57400 | 74.9 | 1.14 | LR_open |
| 166 | 20 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 1.04e-235 | 2.62e-238 | 19200 | 83.2 | 55800 | 72.8 | 1.14 | LR_open |
| 167 | 7 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 2.81e-235 | 7.02e-238 | 19100 | 82.9 | 55600 | 72.5 | 1.14 | LR_open |
| 168 | 7 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 6e-235 | 1.49e-237 | 15800 | 68.6 | 43400 | 56.6 | 1.21 | LR_open |
| 169 | 21 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 2.43e-234 | 5.98e-237 | 16200 | 70.1 | 44600 | 58.2 | 1.2 | LR_open |
| 170 | 16 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 1.4e-231 | 3.43e-234 | 22300 | 96.6 | 69400 | 90.5 | 1.07 | LR_open |
| 171 | 7 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 2.32e-231 | 5.66e-234 | 21500 | 93.3 | 65600 | 85.6 | 1.09 | LR_open |
| 172 | 7 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 7.65e-231 | 1.85e-233 | 21600 | 93.5 | 65800 | 85.9 | 1.09 | LR_open |
| 173 | 7 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 1.37e-230 | 3.29e-233 | 18200 | 79.1 | 52300 | 68.2 | 1.16 | LR_open |
| 174 | 25 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 5.23e-229 | 1.25e-231 | 21800 | 94.7 | 67100 | 87.6 | 1.08 | LR_open |
| 175 | 7 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 6.14e-228 | 1.46e-230 | 16800 | 73 | 47200 | 61.5 | 1.19 | LR_open |
| 176 | 6 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 3.35e-227 | 7.94e-230 | 21700 | 93.9 | 66300 | 86.6 | 1.08 | LR_open |
| 177 | 7 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 4.49e-227 | 1.06e-229 | 17800 | 77.1 | 50600 | 66 | 1.17 | LR_open |
| 178 | 7 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 3.68e-226 | 8.63e-229 | 15800 | 68.7 | 43600 | 56.9 | 1.21 | LR_open |
| 179 | 7 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 6.48e-225 | 1.51e-227 | 12900 | 55.8 | 33500 | 43.7 | 1.28 | LR_open |
| 180 | 7 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 5.78e-224 | 1.34e-226 | 18000 | 78.2 | 51600 | 67.4 | 1.16 | LR_open |
| 181 | 16 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 4.74e-223 | 1.09e-225 | 22000 | 95.5 | 68100 | 88.9 | 1.07 | LR_open |
| 182 | 29 | 5-AACMAA | STREME-5 | 5-AACMAA | AACMAA | AACMAA | 2.3e-219 | 5.26e-222 | 18500 | 80.1 | 53300 | 69.6 | 1.15 | LR_open |
| 183 | 6 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 1.12e-217 | 2.55e-220 | 12800 | 55.6 | 33500 | 43.7 | 1.27 | LR_open |
| 184 | 7 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 8.34e-216 | 1.89e-218 | 14100 | 61.1 | 37800 | 49.3 | 1.24 | LR_open |
| 185 | 7 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 6.5e-215 | 1.46e-217 | 12800 | 55.4 | 33400 | 43.6 | 1.27 | LR_open |
| 186 | 7 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 9.70999999999999e-215 | 2.18e-217 | 15800 | 68.7 | 43900 | 57.2 | 1.2 | LR_open |
| 187 | 30 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 1.63e-213 | 3.63e-216 | 19400 | 84.4 | 57300 | 74.7 | 1.13 | LR_open |
| 188 | 7 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 4.82e-212 | 1.07e-214 | 17100 | 74 | 48300 | 63.1 | 1.17 | LR_open |
| 189 | 6 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 8.24e-211 | 1.82e-213 | 19500 | 84.6 | 57600 | 75.1 | 1.13 | LR_open |
| 190 | 7 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 7.63e-210 | 1.67e-212 | 12500 | 54.3 | 32700 | 42.7 | 1.27 | LR_open |
| 191 | 7 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 1.13e-209 | 2.47e-212 | 18300 | 79.2 | 52800 | 68.8 | 1.15 | LR_open |
| 192 | 9 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 1.8e-209 | 3.9e-212 | 16400 | 71.2 | 46100 | 60.1 | 1.19 | LR_open |
| 193 | 6 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 2.38e-209 | 5.14e-212 | 15000 | 65.2 | 41100 | 53.7 | 1.21 | LR_open |
| 194 | 7 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 2.47e-209 | 5.31e-212 | 20500 | 88.8 | 61500 | 80.2 | 1.11 | LR_open |
| 195 | 25 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 1.47e-208 | 3.14e-211 | 21300 | 92.4 | 65100 | 84.9 | 1.09 | LR_open |
| 196 | 7 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 1.12e-206 | 2.38e-209 | 21900 | 95 | 67800 | 88.5 | 1.07 | LR_open |
| 197 | 7 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 1.75e-205 | 3.71e-208 | 10900 | 47.3 | 27600 | 36 | 1.32 | LR_open |
| 198 | 7 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 2.09e-205 | 4.41e-208 | 16800 | 72.9 | 47500 | 62 | 1.18 | LR_open |
| 199 | 14 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 1.02e-202 | 2.14e-205 | 15100 | 65.7 | 41700 | 54.4 | 1.21 | LR_open |
| 200 | 7 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 2.03e-202 | 4.22e-205 | 20200 | 87.7 | 60600 | 79.1 | 1.11 | LR_open |
| 201 | 31 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 5.56e-201 | 1.15e-203 | 21300 | 92.5 | 65200 | 85.1 | 1.09 | LR_open |
| 202 | 7 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.02e-198 | 2.11e-201 | 14800 | 64.3 | 40700 | 53.1 | 1.21 | LR_open |
| 203 | 7 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 6.93e-198 | 1.42e-200 | 17700 | 76.9 | 51000 | 66.6 | 1.15 | LR_open |
| 204 | 7 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 2.33e-196 | 4.77e-199 | 21500 | 93.2 | 66000 | 86.2 | 1.08 | LR_open |
| 205 | 7 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 7.31e-195 | 1.49e-197 | 11900 | 51.6 | 30900 | 40.4 | 1.28 | LR_open |
| 206 | 7 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 1.39e-193 | 2.81e-196 | 22300 | 96.6 | 69800 | 91.1 | 1.06 | LR_open |
| 207 | 21 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 1.39e-190 | 2.79e-193 | 18000 | 78.2 | 52300 | 68.3 | 1.15 | LR_open |
| 208 | 7 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 5.67e-188 | 1.14e-190 | 20800 | 90.3 | 63300 | 82.6 | 1.09 | LR_open |
| 209 | 73 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 4.77e-187 | 9.52e-190 | 14700 | 63.6 | 40400 | 52.7 | 1.21 | LR_open |
| 210 | 6 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 3.36e-186 | 6.67e-189 | 16800 | 72.7 | 47800 | 62.4 | 1.17 | LR_open |
| 211 | 32 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 6.74e-186 | 1.33e-188 | 13300 | 57.7 | 35800 | 46.8 | 1.24 | LR_open |
| 212 | 9 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 3.09e-184 | 6.08e-187 | 14500 | 63 | 40000 | 52.2 | 1.21 | LR_open |
| 213 | 21 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 3.66e-184 | 7.17e-187 | 14000 | 60.8 | 38300 | 50 | 1.22 | LR_open |
| 214 | 31 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 1.88e-183 | 3.66e-186 | 21800 | 94.5 | 67700 | 88.3 | 1.07 | LR_open |
| 215 | 7 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 1.05e-181 | 2.04e-184 | 15000 | 65 | 41600 | 54.3 | 1.2 | LR_open |
| 216 | 33 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 1.27e-181 | 2.45e-184 | 19500 | 84.6 | 58200 | 75.9 | 1.12 | LR_open |
| 217 | 7 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 3.79e-181 | 7.28e-184 | 17900 | 77.8 | 52200 | 68.1 | 1.14 | LR_open |
| 218 | 34 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 1.08e-179 | 2.06e-182 | 18400 | 79.9 | 54000 | 70.5 | 1.13 | LR_open |
| 219 | 7 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 1.17e-178 | 2.23e-181 | 15600 | 67.5 | 43700 | 57 | 1.18 | LR_open |
| 220 | 35 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 2.29e-177 | 4.34e-180 | 12700 | 55.2 | 34100 | 44.5 | 1.24 | LR_open |
| 221 | 36 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 2.33e-177 | 4.4e-180 | 21800 | 94.5 | 67700 | 88.3 | 1.07 | LR_open |
| 222 | 7 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 4.89e-176 | 9.19e-179 | 12700 | 55.2 | 34100 | 44.5 | 1.24 | LR_open |
| 223 | 33 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 8.19e-176 | 1.53e-178 | 19100 | 82.8 | 56700 | 73.9 | 1.12 | LR_open |
| 224 | 7 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 2.07e-175 | 3.85e-178 | 10000 | 43.4 | 25400 | 33.1 | 1.31 | LR_open |
| 225 | 16 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 3.28e-173 | 6.08e-176 | 19600 | 85 | 58700 | 76.6 | 1.11 | LR_open |
| 226 | 7 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 1.15e-171 | 2.12e-174 | 14100 | 61.1 | 38700 | 50.6 | 1.21 | LR_open |
| 227 | 7 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 2.33e-171 | 4.28e-174 | 11700 | 50.8 | 30900 | 40.3 | 1.26 | LR_open |
| 228 | 37 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 9.85e-171 | 1.8e-173 | 21900 | 94.9 | 68300 | 89.1 | 1.07 | LR_open |
| 229 | 8 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 1.19e-169 | 2.16e-172 | 8690 | 37.7 | 21400 | 27.9 | 1.35 | LR_open |
| 230 | 7 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 1.67e-169 | 3.03e-172 | 10400 | 44.9 | 26600 | 34.7 | 1.3 | LR_open |
| 231 | 33 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 1.25e-167 | 2.26e-170 | 19400 | 84 | 57900 | 75.5 | 1.11 | LR_open |
| 232 | 6 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 3.6e-167 | 6.46e-170 | 22200 | 96.4 | 70000 | 91.3 | 1.06 | LR_open |
| 233 | 7 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 1.09e-166 | 1.94e-169 | 13300 | 57.6 | 36200 | 47.2 | 1.22 | LR_open |
| 234 | 18 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 1.89e-166 | 3.36e-169 | 19700 | 85.5 | 59200 | 77.3 | 1.11 | LR_open |
| 235 | 38 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 7.17e-165 | 1.27e-167 | 21000 | 91.1 | 64500 | 84.2 | 1.08 | LR_open |
| 236 | 16 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 6.45e-164 | 1.14e-166 | 19000 | 82.3 | 56400 | 73.6 | 1.12 | LR_open |
| 237 | 25 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 2.31e-163 | 4.06e-166 | 20000 | 86.6 | 60300 | 78.7 | 1.1 | LR_open |
| 238 | 9 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 7.77e-163 | 1.36e-165 | 15200 | 65.9 | 42800 | 55.8 | 1.18 | LR_open |
| 239 | 22 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 1.36e-161 | 2.38e-164 | 9510 | 41.3 | 24100 | 31.4 | 1.31 | LR_open |
| 240 | 7 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 3.85e-158 | 6.68e-161 | 20100 | 87.2 | 61000 | 79.5 | 1.1 | LR_open |
| 241 | 14 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 2.03e-157 | 3.52e-160 | 11300 | 48.8 | 29700 | 38.8 | 1.26 | LR_open |
| 242 | 36 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 7.5e-156 | 1.29e-158 | 17000 | 73.6 | 49200 | 64.2 | 1.15 | LR_open |
| 243 | 39 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 1.82e-152 | 3.12e-155 | 17200 | 74.5 | 50100 | 65.4 | 1.14 | LR_open |
| 244 | 4 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 2.35e-152 | 4.02e-155 | 5570 | 24.2 | 12500 | 16.3 | 1.48 | LR_open |
| 245 | 40 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 5.34e-145 | 9.09e-148 | 18700 | 81 | 55700 | 72.7 | 1.11 | LR_open |
| 246 | 9 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 9.15e-145 | 1.55e-147 | 15600 | 67.6 | 44600 | 58.1 | 1.16 | LR_open |
| 247 | 41 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 1.46e-143 | 2.47e-146 | 19900 | 86.2 | 60300 | 78.7 | 1.09 | LR_open |
| 248 | 20 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 3.43e-142 | 5.77e-145 | 10200 | 44 | 26600 | 34.7 | 1.27 | LR_open |
| 249 | 19 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 8.45e-141 | 1.41e-143 | 20000 | 86.6 | 60800 | 79.3 | 1.09 | LR_open |
| 250 | 20 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 8.53e-141 | 1.42e-143 | 12500 | 54.2 | 34200 | 44.7 | 1.21 | LR_open |
| 251 | 23 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 6.36e-140 | 1.06e-142 | 18500 | 80.2 | 55100 | 72 | 1.11 | LR_open |
| 252 | 42 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 4.8e-137 | 7.93e-140 | 16900 | 73.2 | 49300 | 64.4 | 1.14 | LR_open |
| 253 | 43 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 1.46e-135 | 2.41e-138 | 19900 | 86.2 | 60600 | 79 | 1.09 | LR_open |
| 254 | 7 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 2.85e-135 | 4.68e-138 | 9650 | 41.8 | 25100 | 32.8 | 1.28 | LR_open |
| 255 | 9 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 8.61e-133 | 1.41e-135 | 7610 | 33 | 18900 | 24.6 | 1.34 | LR_open |
| 256 | 7 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 1.82e-132 | 2.97e-135 | 9710 | 42.1 | 25400 | 33.2 | 1.27 | LR_open |
| 257 | 23 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 6.55e-130 | 1.06e-132 | 22200 | 96.4 | 70500 | 92 | 1.05 | LR_open |
| 258 | 7 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 6.64e-130 | 1.07e-132 | 18300 | 79.2 | 54600 | 71.2 | 1.11 | LR_open |
| 259 | 12 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 2.81e-129 | 4.52e-132 | 18900 | 82.2 | 57100 | 74.5 | 1.1 | LR_open |
| 260 | 9 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 6.45e-129 | 1.03e-131 | 7540 | 32.7 | 18800 | 24.5 | 1.34 | LR_open |
| 261 | 7 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 7.96e-126 | 1.27e-128 | 8560 | 37.1 | 22000 | 28.7 | 1.29 | LR_open |
| 262 | 44 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 1.71e-125 | 2.72e-128 | 21000 | 90.9 | 65100 | 84.9 | 1.07 | LR_open |
| 263 | 9 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 1.48e-124 | 2.34e-127 | 11300 | 48.9 | 30600 | 39.9 | 1.22 | LR_open |
| 264 | 12 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.05e-123 | 1.66e-126 | 22100 | 95.7 | 69900 | 91.2 | 1.05 | LR_open |
| 265 | 45 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 1.72e-122 | 2.71e-125 | 5330 | 23.1 | 12400 | 16.1 | 1.43 | LR_open |
| 266 | 44 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 2.79e-120 | 4.37e-123 | 19600 | 85.2 | 60000 | 78.3 | 1.09 | LR_open |
| 267 | 46 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 1.48e-118 | 2.31e-121 | 16800 | 72.6 | 49400 | 64.4 | 1.13 | LR_open |
| 268 | 36 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 2.94e-116 | 4.57e-119 | 22100 | 95.7 | 70000 | 91.3 | 1.05 | LR_open |
| 269 | 30 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 5.54e-115 | 8.58e-118 | 18300 | 79.3 | 55000 | 71.8 | 1.1 | LR_open |
| 270 | 33 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 9.47e-114 | 1.46e-116 | 18300 | 79.5 | 55200 | 72 | 1.1 | LR_open |
| 271 | 23 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 3.76e-112 | 5.78e-115 | 13400 | 58.2 | 38000 | 49.6 | 1.17 | LR_open |
| 272 | 47 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 1.04e-109 | 1.59e-112 | 21000 | 91.2 | 65700 | 85.8 | 1.06 | LR_open |
| 273 | 12 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 2.06e-109 | 3.15e-112 | 18500 | 80.4 | 56100 | 73.3 | 1.1 | LR_open |
| 274 | 12 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 1.49e-105 | 2.26e-108 | 16400 | 70.9 | 48300 | 63.1 | 1.12 | LR_open |
| 275 | 12 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 8.8e-105 | 1.33e-107 | 17800 | 77.3 | 53700 | 70 | 1.1 | LR_open |
| 276 | 48 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 7.13e-103 | 1.08e-105 | 13500 | 58.7 | 38800 | 50.6 | 1.16 | LR_open |
| 277 | 19 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 9.71e-101 | 1.46e-103 | 9040 | 39.2 | 24100 | 31.5 | 1.24 | LR_open |
| 278 | 20 | MA0873.1 | HOXD12 | MA0873.1 | RGTCGTAAAAH | HOXD12 | 1.09e-99 | 1.63e-102 | 8810 | 38.2 | 23400 | 30.6 | 1.25 | LR_open |
| 279 | 6 | MA0593.1 | FOXP2 | MA0593.1 | RWGTAAACAVR | FOXP2 | 3.58e-99 | 5.36e-102 | 7350 | 31.9 | 18900 | 24.7 | 1.29 | LR_open |
| 280 | 12 | MA0843.1 | TEF | MA0843.1 | BRTTACRTAAYM | TEF | 7.54e-99 | 1.12e-101 | 8020 | 34.8 | 21000 | 27.4 | 1.27 | LR_open |
| 281 | 49 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 3.5e-97 | 5.19e-100 | 15300 | 66.3 | 44800 | 58.5 | 1.13 | LR_open |
| 282 | 50 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 6.52e-97 | 9.64e-100 | 18000 | 78.2 | 54600 | 71.2 | 1.1 | LR_open |
| 283 | 34 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 7.77e-97 | 1.14e-99 | 17100 | 74 | 51200 | 66.8 | 1.11 | LR_open |
| 284 | 7 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 9.15e-96 | 1.34e-98 | 5800 | 25.2 | 14300 | 18.7 | 1.35 | LR_open |
| 285 | 12 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 4.11e-95 | 6.01e-98 | 19300 | 83.9 | 59500 | 77.6 | 1.08 | LR_open |
| 286 | 12 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 3.3e-94 | 4.81e-97 | 14400 | 62.4 | 41900 | 54.6 | 1.14 | LR_open |
| 287 | 25 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 2.74e-93 | 3.98e-96 | 12700 | 55.2 | 36300 | 47.4 | 1.16 | LR_open |
| 288 | 30 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 9.88e-93 | 1.43e-95 | 20500 | 88.9 | 64000 | 83.4 | 1.07 | LR_open |
| 289 | 22 | MA0030.1 | FOXF2 | MA0030.1 | BNAASGTAAACAAD | FOXF2 | 1.87e-92 | 2.7e-95 | 5410 | 23.5 | 13200 | 17.3 | 1.36 | LR_open |
| 290 | 20 | MA0594.2 | HOXA9 | MA0594.2 | RTCGTWAHNN | HOXA9 | 9.94e-92 | 1.43e-94 | 22000 | 95.6 | 70300 | 91.8 | 1.04 | LR_open |
| 291 | 51 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 1.62e-91 | 2.32e-94 | 17000 | 73.7 | 51000 | 66.6 | 1.11 | LR_open |
| 292 | 52 | MA0520.1 | Stat6 | MA0520.1 | BDBTTCCWSAGAAVY | Stat6 | 2.59e-88 | 3.69e-91 | 9180 | 39.8 | 25000 | 32.6 | 1.22 | LR_open |
| 293 | 21 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 9.71e-88 | 1.38e-90 | 6870 | 29.8 | 17800 | 23.2 | 1.29 | LR_open |
| 294 | 53 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 5.09e-87 | 7.22e-90 | 15900 | 68.9 | 47300 | 61.7 | 1.12 | LR_open |
| 295 | 47 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 1.67e-85 | 2.36e-88 | 19500 | 84.5 | 60300 | 78.6 | 1.07 | LR_open |
| 296 | 38 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 7.89e-85 | 1.11e-87 | 22000 | 95.4 | 70300 | 91.7 | 1.04 | LR_open |
| 297 | 54 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 3.84e-82 | 5.39e-85 | 18700 | 80.9 | 57300 | 74.8 | 1.08 | LR_open |
| 298 | 25 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 9.04e-82 | 1.26e-84 | 8630 | 37.4 | 23400 | 30.5 | 1.23 | LR_open |
| 299 | 12 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 4.36e-81 | 6.08e-84 | 7230 | 31.4 | 19000 | 24.8 | 1.26 | LR_open |
| 300 | 55 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 4.44e-79 | 6.17e-82 | 15500 | 67 | 46100 | 60.1 | 1.12 | LR_open |
| 301 | 56 | MA0687.1 | SPIC | MA0687.1 | HAAAAGVGGAAGTA | SPIC | 2.15e-78 | 2.98e-81 | 19400 | 84.3 | 60300 | 78.7 | 1.07 | LR_open |
| 302 | 16 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 1.09e-77 | 1.5e-80 | 21100 | 91.5 | 66700 | 87 | 1.05 | LR_open |
| 303 | 50 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 2.08e-76 | 2.87e-79 | 20800 | 90.3 | 65600 | 85.6 | 1.05 | LR_open |
| 304 | 7 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 3.17e-76 | 4.34e-79 | 6120 | 26.6 | 15800 | 20.6 | 1.29 | LR_open |
| 305 | 12 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 4.02e-75 | 5.5e-78 | 17100 | 74 | 51800 | 67.6 | 1.09 | LR_open |
| 306 | 56 | MA1952.1 | FOXJ2::ELF1 | MA1952.1 | RTAAACMGGAAGTR | FOXJ2::ELF1 | 6.78e-75 | 9.24e-78 | 20300 | 87.9 | 63600 | 82.9 | 1.06 | LR_open |
| 307 | 57 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 2.31e-74 | 3.14e-77 | 14200 | 61.6 | 41900 | 54.7 | 1.13 | LR_open |
| 308 | 7 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 2.38e-74 | 3.23e-77 | 8060 | 35 | 21800 | 28.5 | 1.23 | LR_open |
| 309 | 50 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 4.38e-74 | 5.91e-77 | 22500 | 97.5 | 72600 | 94.8 | 1.03 | LR_open |
| 310 | 36 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 1.1e-73 | 1.49e-76 | 12700 | 54.9 | 36700 | 48 | 1.14 | LR_open |
| 311 | 73 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 4.53e-72 | 6.08e-75 | 10800 | 46.9 | 30700 | 40.1 | 1.17 | LR_open |
| 312 | 16 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 2.02e-70 | 2.7e-73 | 5940 | 25.8 | 15400 | 20.1 | 1.28 | LR_open |
| 313 | 49 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 4.09e-70 | 5.44e-73 | 15700 | 68 | 47200 | 61.5 | 1.11 | LR_open |
| 314 | 45 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 2.33e-69 | 3.09e-72 | 5280 | 22.9 | 13400 | 17.5 | 1.31 | LR_open |
| 315 | 12 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 9.06e-69 | 1.2e-71 | 20000 | 86.6 | 62600 | 81.7 | 1.06 | LR_open |
| 316 | 12 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 1.45e-68 | 1.91e-71 | 20100 | 87.2 | 63100 | 82.3 | 1.06 | LR_open |
| 317 | 55 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 4.56e-67 | 5.99e-70 | 12300 | 53.2 | 35700 | 46.6 | 1.14 | LR_open |
| 318 | 19 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 1.85e-65 | 2.43e-68 | 10300 | 44.8 | 29400 | 38.4 | 1.17 | LR_open |
| 319 | 38 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 2.5e-64 | 3.27e-67 | 19000 | 82.4 | 59100 | 77.2 | 1.07 | LR_open |
| 320 | 53 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 1.46e-63 | 1.9e-66 | 21100 | 91.3 | 66900 | 87.3 | 1.05 | LR_open |
| 321 | 42 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 4.9e-63 | 6.37e-66 | 20800 | 90.4 | 66000 | 86.2 | 1.05 | LR_open |
| 322 | 33 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 5.02e-63 | 6.5e-66 | 17400 | 75.4 | 53300 | 69.6 | 1.08 | LR_open |
| 323 | 12 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 8.9e-62 | 1.15e-64 | 19300 | 83.7 | 60300 | 78.7 | 1.06 | LR_open |
| 324 | 58 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 5.29e-61 | 6.8e-64 | 18100 | 78.4 | 55900 | 72.9 | 1.07 | LR_open |
| 325 | 20 | MA1506.1 | HOXD10 | MA1506.1 | RGTCGTAAAAH | HOXD10 | 7.92e-61 | 1.02e-63 | 5600 | 24.3 | 14700 | 19.1 | 1.27 | LR_open |
| 326 | 59 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 1.02e-60 | 1.31e-63 | 11500 | 49.8 | 33400 | 43.5 | 1.14 | LR_open |
| 327 | 60 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 7.85e-60 | 1e-62 | 18600 | 80.6 | 57700 | 75.3 | 1.07 | LR_open |
| 328 | 7 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 5.52e-59 | 7.02e-62 | 5960 | 25.8 | 15800 | 20.6 | 1.25 | LR_open |
| 329 | 55 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 1.25e-58 | 1.58e-61 | 16700 | 72.4 | 51100 | 66.7 | 1.09 | LR_open |
| 330 | 31 | MA0517.1 | STAT1::STAT2 | MA0517.1 | THAGTTTCRKTTTCY | STAT1::STAT2 | 3.23e-57 | 4.08e-60 | 11900 | 51.4 | 34700 | 45.3 | 1.14 | LR_open |
| 331 | 61 | MA1125.1 (ZNF384) | MEME-2 | AAAAAAAAAAAAAAA | AAAAAAAAAAAAAAA | ZNF384 | 5.1e-57 | 6.43e-60 | 3490 | 15.2 | 8490 | 11.1 | 1.37 | LR_open |
| 332 | 49 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 5.34e-57 | 6.7e-60 | 20800 | 90.1 | 66000 | 86.1 | 1.05 | LR_open |
| 333 | 62 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 4.49e-56 | 5.62e-59 | 5480 | 23.8 | 14500 | 18.9 | 1.26 | LR_open |
| 334 | 21 | MA0837.2 | CEBPE | MA0837.2 | KATTGCGCAATM | CEBPE | 5.07e-56 | 6.33e-59 | 3020 | 13.1 | 7140 | 9.32 | 1.4 | LR_open |
| 335 | 31 | MA0772.1 | IRF7 | MA0772.1 | HCGAAARYGAAAVT | IRF7 | 1.93e-55 | 2.4e-58 | 8140 | 35.3 | 22800 | 29.7 | 1.19 | LR_open |
| 336 | 63 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 1.71e-53 | 2.12e-56 | 6690 | 29 | 18200 | 23.8 | 1.22 | LR_open |
| 337 | 64 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 1.79e-53 | 2.22e-56 | 16300 | 70.5 | 49800 | 64.9 | 1.09 | LR_open |
| 338 | 65 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 1.64e-52 | 2.02e-55 | 19600 | 85.2 | 61800 | 80.7 | 1.06 | LR_open |
| 339 | 37 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 5.38e-52 | 6.61e-55 | 20900 | 90.4 | 66400 | 86.7 | 1.04 | LR_open |
| 340 | 73 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 8.57e-52 | 1.05e-54 | 6220 | 27 | 16900 | 22 | 1.23 | LR_open |
| 341 | 66 | MA1592.1 | ZNF274 | MA1592.1 | NRTRTGAGTTCTCGYN | ZNF274 | 2.59e-51 | 3.16e-54 | 6160 | 26.7 | 16700 | 21.8 | 1.23 | LR_open |
| 342 | 67 | MA0619.1 | LIN54 | MA0619.1 | DTTTRAATH | LIN54 | 5.32e-49 | 6.49e-52 | 5510 | 23.9 | 14800 | 19.3 | 1.24 | LR_open |
| 343 | 49 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 1.01e-48 | 1.23e-51 | 15500 | 67.4 | 47500 | 62 | 1.09 | LR_open |
| 344 | 68 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 1.61e-48 | 1.95e-51 | 21900 | 95.1 | 70700 | 92.3 | 1.03 | LR_open |
| 345 | 46 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 2.08e-45 | 2.51e-48 | 18200 | 79 | 57000 | 74.4 | 1.06 | LR_open |
| 346 | 69 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 2.91e-44 | 3.51e-47 | 21900 | 95.1 | 70800 | 92.4 | 1.03 | LR_open |
| 347 | 65 | MA0743.2 | SCRT1 | MA0743.2 | NDWKCAACAGGTGKNN | SCRT1 | 1.33e-42 | 1.59e-45 | 17600 | 76.3 | 54900 | 71.6 | 1.07 | LR_open |
| 348 | 7 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 2.96e-42 | 3.55e-45 | 21600 | 93.8 | 69700 | 91 | 1.03 | LR_open |
| 349 | 51 | MA1534.1 | NR1I3 | MA1534.1 | RTGAACTTT | NR1I3 | 5.09e-42 | 6.09e-45 | 15200 | 65.9 | 46600 | 60.8 | 1.08 | LR_open |
| 350 | 49 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 1.32e-41 | 1.57e-44 | 11900 | 51.7 | 35600 | 46.4 | 1.11 | LR_open |
| 351 | 19 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 5.47e-40 | 6.49e-43 | 4220 | 18.3 | 11100 | 14.5 | 1.26 | LR_open |
| 352 | 20 | MA1503.1 | HOXB9 | MA1503.1 | GTCGTAAAAH | HOXB9 | 1.47e-39 | 1.75e-42 | 2360 | 10.2 | 5670 | 7.4 | 1.39 | LR_open |
| 353 | 23 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 2.29e-39 | 2.71e-42 | 15100 | 65.5 | 46400 | 60.5 | 1.08 | LR_open |
| 354 | 55 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 2.78e-38 | 3.27e-41 | 13800 | 59.9 | 42100 | 54.9 | 1.09 | LR_open |
| 355 | 47 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 2.74e-37 | 3.22e-40 | 19500 | 84.5 | 61900 | 80.7 | 1.05 | LR_open |
| 356 | 7 | MA0784.2 | POU1F1 | MA0784.2 | ANCTMATTWGCATAWT | POU1F1 | 1.35e-35 | 1.58e-38 | 1960 | 8.51 | 4620 | 6.03 | 1.41 | LR_open |
| 357 | 70 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 1.96e-35 | 2.28e-38 | 1500 | 6.51 | 3340 | 4.35 | 1.5 | LR_open |
| 358 | 71 | MA0684.2 | RUNX3 | MA0684.2 | NHAACCTCAANN | RUNX3 | 2.01e-35 | 2.34e-38 | 18000 | 78.2 | 56700 | 74 | 1.06 | LR_open |
| 359 | 46 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 6.53e-35 | 7.59e-38 | 20000 | 86.5 | 63600 | 83 | 1.04 | LR_open |
| 360 | 68 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 1.1e-34 | 1.28e-37 | 18200 | 79 | 57400 | 75 | 1.05 | LR_open |
| 361 | 70 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 1.34e-34 | 1.55e-37 | 259 | 1.12 | 276 | 0.36 | 3.12 | LR_open |
| 362 | 12 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 2.82e-34 | 3.25e-37 | 4720 | 20.5 | 12900 | 16.8 | 1.22 | LR_open |
| 363 | 51 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 4.35e-34 | 4.99e-37 | 21700 | 94.2 | 70300 | 91.7 | 1.03 | LR_open |
| 364 | 49 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 9.29e-34 | 1.06e-36 | 14800 | 64 | 45500 | 59.4 | 1.08 | LR_open |
| 365 | 72 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 4.58e-33 | 5.23e-36 | 22700 | 98.6 | 74500 | 97.2 | 1.01 | LR_open |
| 366 | 44 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 1.07e-32 | 1.21e-35 | 11500 | 49.9 | 34700 | 45.2 | 1.1 | LR_open |
| 367 | 73 | MA0682.2 (PITX1) | MEME-3 | AAGTGCTGGGATTAC | AAGTGCTGGGATTAC | PITX1 | 2.12e-32 | 2.41e-35 | 1930 | 8.36 | 4600 | 6 | 1.39 | LR_open |
| 368 | 14 | MA1473.1 | CDX4 | MA1473.1 | GGYMATAAAAC | CDX4 | 5.78e-32 | 6.55e-35 | 2590 | 11.2 | 6540 | 8.53 | 1.32 | LR_open |
| 369 | 70 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 9e-32 | 1.02e-34 | 411 | 1.78 | 609 | 0.79 | 2.25 | LR_open |
| 370 | 73 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 1.58e-29 | 1.78e-32 | 1850 | 8.04 | 4460 | 5.82 | 1.38 | LR_open |
| 371 | 71 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 2.14e-29 | 2.4e-32 | 22900 | 99.5 | 75600 | 98.6 | 1.01 | LR_open |
| 372 | 21 | MA0466.3 | CEBPB | MA0466.3 | KATTGCGCAATM | CEBPB | 3.39e-29 | 3.8e-32 | 473 | 2.05 | 775 | 1.01 | 2.03 | LR_open |
| 373 | 11 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 3.99e-29 | 4.46e-32 | 3500 | 15.2 | 9330 | 12.2 | 1.25 | LR_open |
| 374 | 49 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 8.52e-29 | 9.5e-32 | 6940 | 30.1 | 20100 | 26.2 | 1.15 | LR_open |
| 375 | 49 | MA0826.1 | OLIG1 | MA0826.1 | AMCATATGKT | OLIG1 | 1.35e-28 | 1.5e-31 | 6340 | 27.5 | 18200 | 23.7 | 1.16 | LR_open |
| 376 | 12 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 4.02e-28 | 4.46e-31 | 3360 | 14.6 | 8960 | 11.7 | 1.25 | LR_open |
| 377 | 20 | MA0485.2 | HOXC9 | MA0485.2 | GTCGTAAAAH | HOXC9 | 1.4e-27 | 1.55e-30 | 1160 | 5.01 | 2550 | 3.33 | 1.51 | LR_open |
| 378 | 54 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 1.66e-27 | 1.83e-30 | 19500 | 84.6 | 62400 | 81.4 | 1.04 | LR_open |
| 379 | 73 | MA1547.2 (PITX2) | STREME-11 | 11-CTGGGAYTAC | 11-CTGGGAYTAC | PITX2 | 1.25e-26 | 1.37e-29 | 2780 | 12 | 7250 | 9.46 | 1.27 | LR_open |
| 380 | 47 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 1.34e-26 | 1.47e-29 | 11000 | 47.8 | 33400 | 43.6 | 1.1 | LR_open |
| 381 | 74 | MA1580.1 | ZBTB32 | MA1580.1 | TGTACAGTAW | ZBTB32 | 9.31e-26 | 1.02e-28 | 4260 | 18.5 | 11800 | 15.4 | 1.2 | LR_open |
| 382 | 65 | MA0647.1 | GRHL1 | MA0647.1 | AAAACCGGTTTD | GRHL1 | 2.36e-25 | 2.57e-28 | 6950 | 30.2 | 20300 | 26.5 | 1.14 | LR_open |
| 383 | 63 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 2.79e-25 | 3.04e-28 | 4430 | 19.2 | 12300 | 16.1 | 1.19 | LR_open |
| 384 | 20 | MA0651.2 | HOXC11 | MA0651.2 | AGGTCGTAAAAH | HOXC11 | 3.99e-25 | 4.33e-28 | 2340 | 10.1 | 5990 | 7.82 | 1.3 | LR_open |
| 385 | 75 | 9-CACTTTGGGAG | STREME-9 | 9-CACTTTGGGAG | CACTTTGGGAG | CACTTTGGGAG | 6.84e-25 | 7.4e-28 | 325 | 1.41 | 481 | 0.63 | 2.25 | LR_open |
| 386 | 46 | MA0137.3 | STAT1 | MA0137.3 | TTTCYRGGAAA | STAT1 | 7.71e-25 | 8.32e-28 | 11300 | 49 | 34400 | 44.9 | 1.09 | LR_open |
| 387 | 46 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 9.74e-25 | 1.05e-27 | 9190 | 39.9 | 27500 | 35.9 | 1.11 | LR_open |
| 388 | 76 | TAGAGAYRGGGTTTC | MEME-5 | TAGAGAYRGGGTTTC | TAGAGAYRGGGTTTC | TAGAGAYRGGGTTTC | 5.75e-24 | 6.18e-27 | 460 | 2 | 803 | 1.05 | 1.91 | LR_open |
| 389 | 42 | MA1956.1 | FOXO1::FLI1 | MA1956.1 | GTAAACAGGAAGTK | FOXO1::FLI1 | 7.75e-24 | 8.3e-27 | 3970 | 17.2 | 11000 | 14.3 | 1.2 | LR_open |
| 390 | 46 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 1.55e-23 | 1.66e-26 | 10800 | 46.7 | 32700 | 42.7 | 1.09 | LR_open |
| 391 | 12 | MA0840.1 | Creb5 | MA0840.1 | NATGACGTCAYH | Creb5 | 1.74e-22 | 1.86e-25 | 2660 | 11.6 | 7050 | 9.2 | 1.26 | LR_open |
| 392 | 36 | MA1421.1 | TCF7L1 | MA1421.1 | AAAGATCAAAGG | TCF7L1 | 3.78e-22 | 4.02e-25 | 3600 | 15.6 | 9910 | 12.9 | 1.21 | LR_open |
| 393 | 49 | MA0817.1 | BHLHE23 | MA0817.1 | AAACATATGBTN | BHLHE23 | 4.49e-22 | 4.76e-25 | 2340 | 10.1 | 6090 | 7.95 | 1.28 | LR_open |
| 394 | 25 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 6.14e-21 | 6.5e-24 | 2150 | 9.32 | 5570 | 7.27 | 1.28 | LR_open |
| 395 | 30 | MA0610.1 | DMRT3 | MA0610.1 | MATGTATCAAN | DMRT3 | 1.93e-20 | 2.03e-23 | 6940 | 30.1 | 20500 | 26.8 | 1.12 | LR_open |
| 396 | 77 | MA1585.1 | ZKSCAN1 | MA1585.1 | AYAGTAGGTS | ZKSCAN1 | 3.2e-20 | 3.37e-23 | 11100 | 48.1 | 34000 | 44.4 | 1.08 | LR_open |
| 397 | 46 | MA0463.2 | BCL6 | MA0463.2 | NYGCTTTCKAGGAAYN | BCL6 | 1.63e-19 | 1.72e-22 | 13500 | 58.4 | 42000 | 54.8 | 1.07 | LR_open |
| 398 | 58 | MA1148.1 | PPARA::RXRA | MA1148.1 | AWNTRGGTYAAAGGTCAN | PPARA::RXRA | 3.42e-19 | 3.58e-22 | 10600 | 46.1 | 32600 | 42.5 | 1.08 | LR_open |
| 399 | 78 | MA0164.1 | Nr2e3 | MA0164.1 | CAAGCTT | Nr2e3 | 8.6e-19 | 8.98e-22 | 5150 | 22.4 | 14900 | 19.5 | 1.15 | LR_open |
| 400 | 68 | MA0072.1 | RORA | MA0072.1 | NWWAWBTAGGTCAR | RORA | 2.02e-18 | 2.1e-21 | 1970 | 8.56 | 5130 | 6.7 | 1.28 | LR_open |
| 401 | 79 | MA1973.1 (ZKSCAN3) | MEME-6 | TGTYRCCCAGGCTGG | TGTYRCCCAGGCTGG | ZKSCAN3 | 2.27e-18 | 2.35e-21 | 383 | 1.66 | 685 | 0.89 | 1.86 | LR_open |
| 402 | 7 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 1.02e-17 | 1.06e-20 | 14100 | 61.1 | 44200 | 57.7 | 1.06 | LR_open |
| 403 | 50 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 1.57e-17 | 1.63e-20 | 17900 | 77.6 | 57200 | 74.6 | 1.04 | LR_open |
| 404 | 80 | MA0019.1 | Ddit3::Cebpa | MA0019.1 | VNRTGCAATMCC | Ddit3::Cebpa | 5.11e-17 | 5.28e-20 | 9860 | 42.8 | 30200 | 39.4 | 1.09 | LR_open |
| 405 | 54 | MA0804.1 | TBX19 | MA0804.1 | DTTMRCACMTAGGTGTGAAW | TBX19 | 4.27e-16 | 4.39e-19 | 6420 | 27.9 | 19100 | 24.9 | 1.12 | LR_open |
| 406 | 7 | MA1546.1 | PAX3 | MA1546.1 | YSGTCACGSYWATTAN | PAX3 | 4.4e-16 | 4.52e-19 | 12800 | 55.6 | 40100 | 52.3 | 1.06 | LR_open |
| 407 | 7 | MA0703.2 | LMX1B | MA0703.2 | AWTTTAATTAA | LMX1B | 1.47e-15 | 1.5e-18 | 1690 | 7.34 | 4400 | 5.74 | 1.28 | LR_open |
| 408 | 42 | MA1955.1 | FOXO1::ELK3 | MA1955.1 | RWMAACAGGAAGTN | FOXO1::ELK3 | 1.81e-15 | 1.85e-18 | 12400 | 54 | 38800 | 50.7 | 1.06 | LR_open |
| 409 | 47 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 2.07e-15 | 2.11e-18 | 17800 | 77.2 | 57000 | 74.4 | 1.04 | LR_open |
| 410 | 11 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 2.13e-15 | 2.17e-18 | 3010 | 13 | 8390 | 10.9 | 1.19 | LR_open |
| 411 | 51 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 2.69e-15 | 2.73e-18 | 15600 | 67.5 | 49400 | 64.4 | 1.05 | LR_open |
| 412 | 81 | MA1596.1 (ZNF460) | STREME-14 | 14-ACCTCAGCCTCC | 14-ACCTCAGCCTCC | ZNF460 | 2.93e-15 | 2.96e-18 | 309 | 1.34 | 544 | 0.71 | 1.89 | LR_open |
| 413 | 54 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 3.38e-15 | 3.41e-18 | 20000 | 86.9 | 64800 | 84.6 | 1.03 | LR_open |
| 414 | 82 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 1.23e-14 | 1.24e-17 | 3940 | 17.1 | 11300 | 14.8 | 1.16 | LR_open |
| 415 | 83 | MA1714.1 | ZNF675 | MA1714.1 | RGGVHYMGGAGGMCAAAATGD | ZNF675 | 4.09e-13 | 4.11e-16 | 11000 | 47.9 | 34400 | 44.9 | 1.07 | LR_open |
| 416 | 42 | MA1950.1 | FLI1::FOXI1 | MA1950.1 | RTAAACAGGAARYN | FLI1::FOXI1 | 8.14e-13 | 8.16e-16 | 16700 | 72.6 | 53600 | 69.9 | 1.04 | LR_open |
| 417 | 79 | MA1973.1 (ZKSCAN3) | STREME-8 | 8-CCAGCCTGGGYRAC | CCAGCCTGGGYRAC | ZKSCAN3 | 2.85e-12 | 2.85e-15 | 329 | 1.43 | 633 | 0.83 | 1.73 | LR_open |
| 418 | 50 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.99e-12 | 2.98e-15 | 16300 | 70.6 | 52000 | 67.8 | 1.04 | LR_open |
| 419 | 65 | MA0744.2 | SCRT2 | MA0744.2 | NNWGCAACAGGTGDNN | SCRT2 | 3.4e-12 | 3.38e-15 | 16700 | 72.3 | 53300 | 69.6 | 1.04 | LR_open |
| 420 | 84 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 7.78e-12 | 7.72e-15 | 20800 | 90.3 | 67800 | 88.5 | 1.02 | LR_open |
| 421 | 85 | MA1107.2 (KLF9) | STREME-13 | 13-ACCACACMCA | 13-ACCACACMCA | KLF9 | 1.29e-11 | 1.28e-14 | 478 | 2.07 | 1030 | 1.35 | 1.54 | LR_open |
| 422 | 86 | MA1949.1 | FLI1::DRGX | MA1949.1 | ACCGGAAGTAATTAT | FLI1::DRGX | 1.88e-11 | 1.86e-14 | 13600 | 59.1 | 43100 | 56.2 | 1.05 | LR_open |
| 423 | 87 | 12-CTCCTGGGCTCAA | STREME-12 | 12-CTCCTGGGCTCAA | 12-CTCCTGGGCTCAA | CTCCTGGGCTCAA | 2.42e-11 | 2.39e-14 | 197 | 0.85 | 323 | 0.42 | 2.03 | LR_open |
| 424 | 58 | MA1541.1 | NR6A1 | MA1541.1 | NYCAAGKTCAAGKTCAN | NR6A1 | 4.41e-11 | 4.34e-14 | 15700 | 68.2 | 50300 | 65.6 | 1.04 | LR_open |
| 425 | 47 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 1.26e-09 | 1.24e-12 | 8880 | 38.5 | 27600 | 36 | 1.07 | LR_open |
| 426 | 49 | MA1467.2 | Atoh1 | MA1467.2 | RVCAGATGGYN | Atoh1 | 2.92e-09 | 2.85e-12 | 21000 | 91 | 68600 | 89.5 | 1.02 | LR_open |
| 427 | 46 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 8.46e-09 | 8.26e-12 | 19400 | 84.1 | 63000 | 82.2 | 1.02 | LR_open |
| 428 | 55 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 1.54e-08 | 1.5e-11 | 4230 | 18.3 | 12600 | 16.4 | 1.11 | LR_open |
| 429 | 58 | MA0160.2 | NR4A2 | MA0160.2 | BWAAAGGTCA | NR4A2 | 1.69e-08 | 1.64e-11 | 13100 | 56.9 | 41700 | 54.5 | 1.05 | LR_open |
| 430 | 88 | MA1980.1 | ZNF418 | MA1980.1 | MDGAGGCTAAAAGCA | ZNF418 | 1.8e-08 | 1.74e-11 | 3520 | 15.2 | 10400 | 13.5 | 1.13 | LR_open |
| 431 | 89 | MA1594.1 | ZNF382 | MA1594.1 | AGGRGAYATCACYACMGAYCCCAC | ZNF382 | 4.24e-08 | 4.1e-11 | 2160 | 9.35 | 6120 | 7.99 | 1.17 | LR_open |
| 432 | 81 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 4.47e-08 | 4.31e-11 | 234 | 1.01 | 454 | 0.59 | 1.72 | LR_open |
| 433 | 49 | MA0607.2 | BHLHA15 | MA0607.2 | ACCATATGGT | BHLHA15 | 4.65e-08 | 4.48e-11 | 4680 | 20.3 | 14100 | 18.4 | 1.1 | LR_open |
| 434 | 71 | MA0002.2 | Runx1 | MA0002.2 | BBYTGTGGTTT | Runx1 | 8.42e-08 | 8.09e-11 | 2880 | 12.5 | 8400 | 11 | 1.14 | LR_open |
| 435 | 49 | MA1123.2 | TWIST1 | MA1123.2 | NNDCCAGATGTBN | TWIST1 | 9.32e-08 | 8.93e-11 | 22100 | 96 | 72900 | 95.1 | 1.01 | LR_open |
| 436 | 90 | MA0770.1 | HSF2 | MA0770.1 | TTCTAGAAYRTTC | HSF2 | 1.13e-07 | 1.08e-10 | 6730 | 29.2 | 20700 | 27 | 1.08 | LR_open |
| 437 | 12 | MA0656.1 | JDP2 | MA0656.1 | GATGACGTCAYC | JDP2 | 4.43e-07 | 4.22e-10 | 2100 | 9.09 | 5990 | 7.82 | 1.16 | LR_open |
| 438 | 58 | MA0071.1 | RORA | MA0071.1 | AWMWAGGTCA | RORA | 5.08e-07 | 4.83e-10 | 5910 | 25.6 | 18100 | 23.6 | 1.08 | LR_open |
| 439 | 42 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 7.16e-07 | 6.8e-10 | 22000 | 95.4 | 72400 | 94.4 | 1.01 | LR_open |
| 440 | 85 | MA0732.1 (EGR3) | STREME-7 | 7-CACCCACACAC | CACCCACACAC | EGR3 | 8.82e-07 | 8.36e-10 | 268 | 1.16 | 563 | 0.73 | 1.59 | LR_open |
| 441 | 65 | MA1968.1 | TFCP2 | MA1968.1 | AAACCGGTTY | TFCP2 | 1.3e-06 | 1.23e-09 | 16000 | 69.4 | 51600 | 67.3 | 1.03 | LR_open |
| 442 | 91 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 1.44e-06 | 1.35e-09 | 18400 | 79.7 | 59700 | 77.9 | 1.02 | LR_open |
| 443 | 38 | MA1525.2 | NFATC4 | MA1525.2 | AAYGGAAAMW | NFATC4 | 2.19e-06 | 2.07e-09 | 1880 | 8.15 | 5360 | 6.99 | 1.17 | LR_open |
| 444 | 20 | MA1958.1 | HOXD12::ELK1 | MA1958.1 | DRVCGGAAGTCGTAAAD | HOXD12::ELK1 | 8.81e-06 | 8.27e-09 | 988 | 4.29 | 2660 | 3.48 | 1.23 | LR_open |
| 445 | 12 | MA1951.1 | FOS | MA1951.1 | BGATGACGTCATCR | FOS | 8.97e-06 | 8.4e-09 | 4980 | 21.6 | 15300 | 19.9 | 1.09 | LR_open |
| 446 | 58 | MA0141.3 | ESRRB | MA0141.3 | TCAAGGTCAWW | ESRRB | 1.16e-05 | 1.08e-08 | 6540 | 28.4 | 20300 | 26.5 | 1.07 | LR_open |
| 447 | 55 | MA0831.3 | TFE3 | MA0831.3 | GTCACGTGAC | TFE3 | 1.94e-05 | 1.81e-08 | 4220 | 18.3 | 12800 | 16.8 | 1.09 | LR_open |
| 448 | 8 | MA0507.2 | POU2F2 | MA0507.2 | AWTATGCAAATKAG | POU2F2 | 4.59e-05 | 4.27e-08 | 274 | 1.19 | 612 | 0.8 | 1.49 | LR_open |
| 449 | 90 | MA0486.2 | HSF1 | MA0486.2 | TTCTAGAAYRTTC | HSF1 | 6.3e-05 | 5.85e-08 | 3310 | 14.4 | 9970 | 13 | 1.1 | LR_open |
| 450 | 55 | MA0828.2 | SREBF2 | MA0828.2 | WWATCACGTGATWW | SREBF2 | 8.43e-05 | 7.81e-08 | 1560 | 6.76 | 4460 | 5.81 | 1.16 | LR_open |
| 451 | 54 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 9.18e-05 | 8.49e-08 | 22300 | 96.6 | 73500 | 95.9 | 1.01 | LR_open |
| 452 | 54 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 0.00011 | 1.01e-07 | 2540 | 11 | 7520 | 9.82 | 1.12 | LR_open |
| 453 | 49 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.000121 | 1.11e-07 | 2520 | 10.9 | 7490 | 9.77 | 1.12 | LR_open |
| 454 | 58 | MA0643.1 | Esrrg | MA0643.1 | TCAAGGTCAH | Esrrg | 0.000125 | 1.15e-07 | 8370 | 36.3 | 26400 | 34.5 | 1.05 | LR_open |
| 455 | 71 | MA0511.2 | RUNX2 | MA0511.2 | WAACCGCAA | RUNX2 | 0.000168 | 1.54e-07 | 244 | 1.06 | 542 | 0.71 | 1.5 | LR_open |
| 456 | 49 | MA0818.2 | BHLHE22 | MA0818.2 | AMCATATGKY | BHLHE22 | 0.000277 | 2.53e-07 | 654 | 2.84 | 1730 | 2.25 | 1.26 | LR_open |
| 457 | 81 | MA1596.1 (ZNF460) | MEME-4 | RGGAGGCBGAGGHKG | RGGAGGCBGAGGHKG | ZNF460 | 0.00031 | 2.83e-07 | 196 | 0.85 | 418 | 0.55 | 1.56 | LR_open |
| 458 | 42 | MA1954.1 | FOXO1::ELK1 | MA1954.1 | RWMAACAGGAAGTD | FOXO1::ELK1 | 0.000317 | 2.88e-07 | 13100 | 57 | 42200 | 55.1 | 1.03 | LR_open |
| 459 | 54 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 0.000495 | 4.5e-07 | 1350 | 5.86 | 3860 | 5.04 | 1.16 | LR_open |
| 460 | 54 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 0.00132 | 1.19e-06 | 3760 | 16.3 | 11500 | 15 | 1.08 | LR_open |
| 461 | 86 | MA1937.1 | ERF::HOXB13 | MA1937.1 | VSCGGAARYNRTWAAN | ERF::HOXB13 | 0.00148 | 1.34e-06 | 8280 | 35.9 | 26200 | 34.2 | 1.05 | LR_open |
| 462 | 91 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 0.0017 | 1.54e-06 | 6690 | 29 | 21000 | 27.5 | 1.06 | LR_open |
| 463 | 12 | MA0605.2 | ATF3 | MA0605.2 | RRTGACGTCAYY | ATF3 | 0.0023 | 2.07e-06 | 670 | 2.91 | 1810 | 2.36 | 1.23 | LR_open |
| 464 | 81 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 0.00315 | 2.83e-06 | 928 | 4.02 | 2600 | 3.39 | 1.19 | LR_open |
| 465 | 54 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 0.0036 | 3.23e-06 | 1890 | 8.19 | 5600 | 7.3 | 1.12 | LR_open |
| 466 | 82 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 0.00585 | 5.23e-06 | 2230 | 9.67 | 6690 | 8.73 | 1.11 | LR_open |
| 467 | 58 | MA0592.3 | ESRRA | MA0592.3 | NBTCAAGGTCAHN | ESRRA | 0.00677 | 6.04e-06 | 8130 | 35.3 | 25900 | 33.7 | 1.05 | LR_open |
| 468 | 55 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 0.00807 | 7.19e-06 | 4500 | 19.5 | 14000 | 18.3 | 1.07 | LR_open |
| 469 | 92 | MA1983.1 | ZNF582 | MA1983.1 | TCTGTTACTTGCAGCCAAAWG | ZNF582 | 0.00898 | 7.98e-06 | 4190 | 18.2 | 13000 | 16.9 | 1.07 | LR_open |
| 470 | 33 | MA0870.1 | Sox1 | MA0870.1 | AACAATANCATTGTT | Sox1 | 0.0109 | 9.66e-06 | 662 | 2.87 | 1820 | 2.37 | 1.21 | LR_open |
| 471 | 53 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 0.012 | 1.06e-05 | 22500 | 97.6 | 74400 | 97.1 | 1.01 | LR_open |
| 472 | 91 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 0.0136 | 1.2e-05 | 7640 | 33.1 | 24300 | 31.7 | 1.05 | LR_open |
| 473 | 42 | MA1579.1 | ZBTB26 | MA1579.1 | NNCTCCAGAAANNNN | ZBTB26 | 0.0163 | 1.44e-05 | 1000 | 4.35 | 2860 | 3.74 | 1.16 | LR_open |
| 474 | 49 | MA0623.2 | NEUROG1 | MA0623.2 | RACATATGTY | NEUROG1 | 0.023 | 2.02e-05 | 601 | 2.61 | 1640 | 2.15 | 1.22 | LR_open |
| 475 | 31 | MA0653.1 | IRF9 | MA0653.1 | AACGAAACCGAAACT | IRF9 | 0.0293 | 2.57e-05 | 86 | 0.37 | 164 | 0.21 | 1.75 | LR_open |
| 476 | 54 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 0.0307 | 2.68e-05 | 2520 | 10.9 | 7680 | 10 | 1.09 | LR_open |
| 477 | 93 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 0.0356 | 3.11e-05 | 1010 | 4.39 | 2920 | 3.81 | 1.15 | LR_open |
ER_rat <- 1.25
mrc_type <- "LR_open"
LR_open_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
slice_head(n=80)%>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))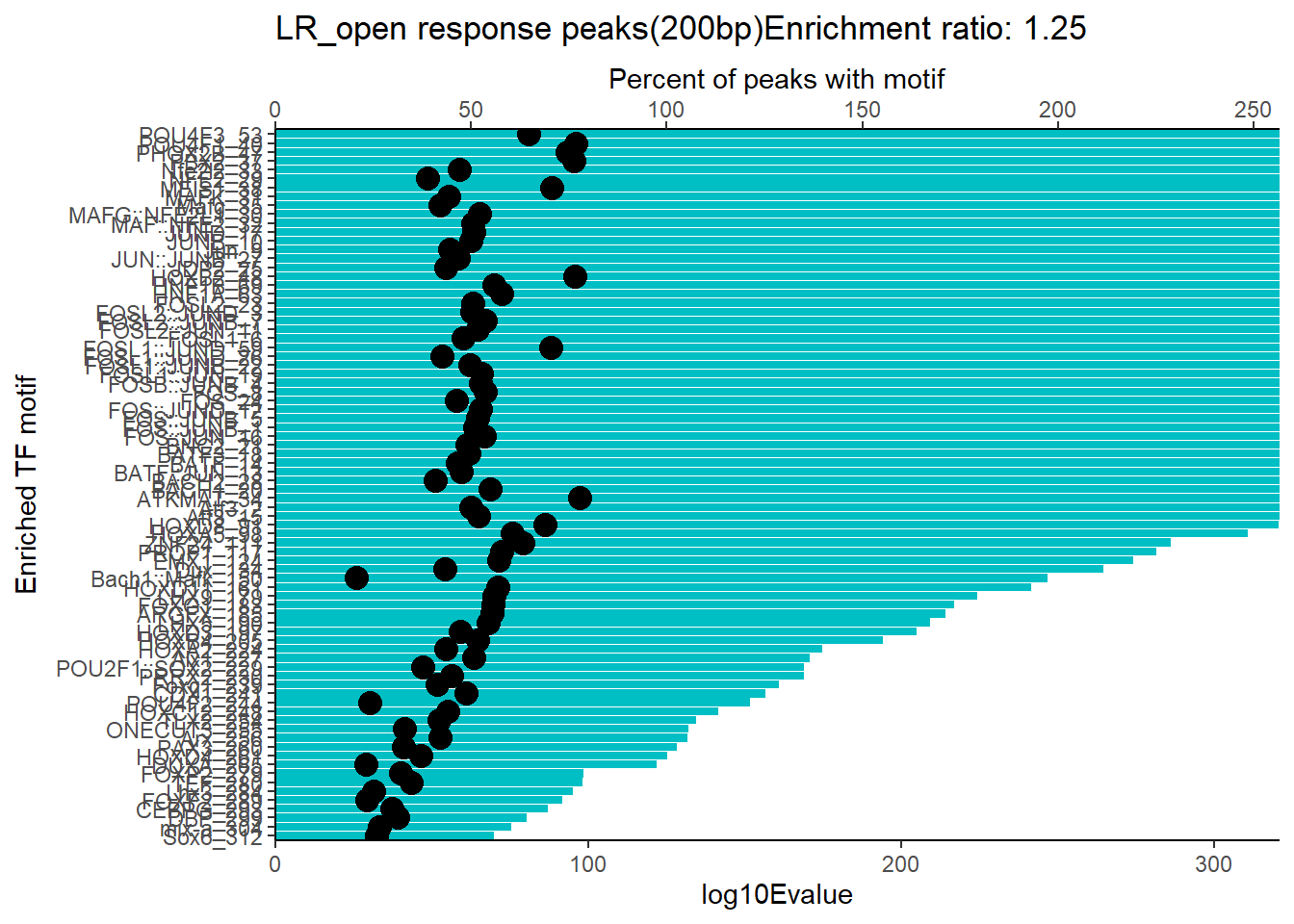
LR_open_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
# mutate(
# RANK = as.numeric(RANK),
# log10Evalue = -log10(if_else(EVALUE.y == 0, 1e-300, EVALUE.y)) + 1e-5 * (max(RANK, na.rm = TRUE) - RANK)
# ) %>%
mutate(log10Evalue = -log10(if_else(EVALUE.y == 0, 1e-300, EVALUE.y))) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*3), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./3,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))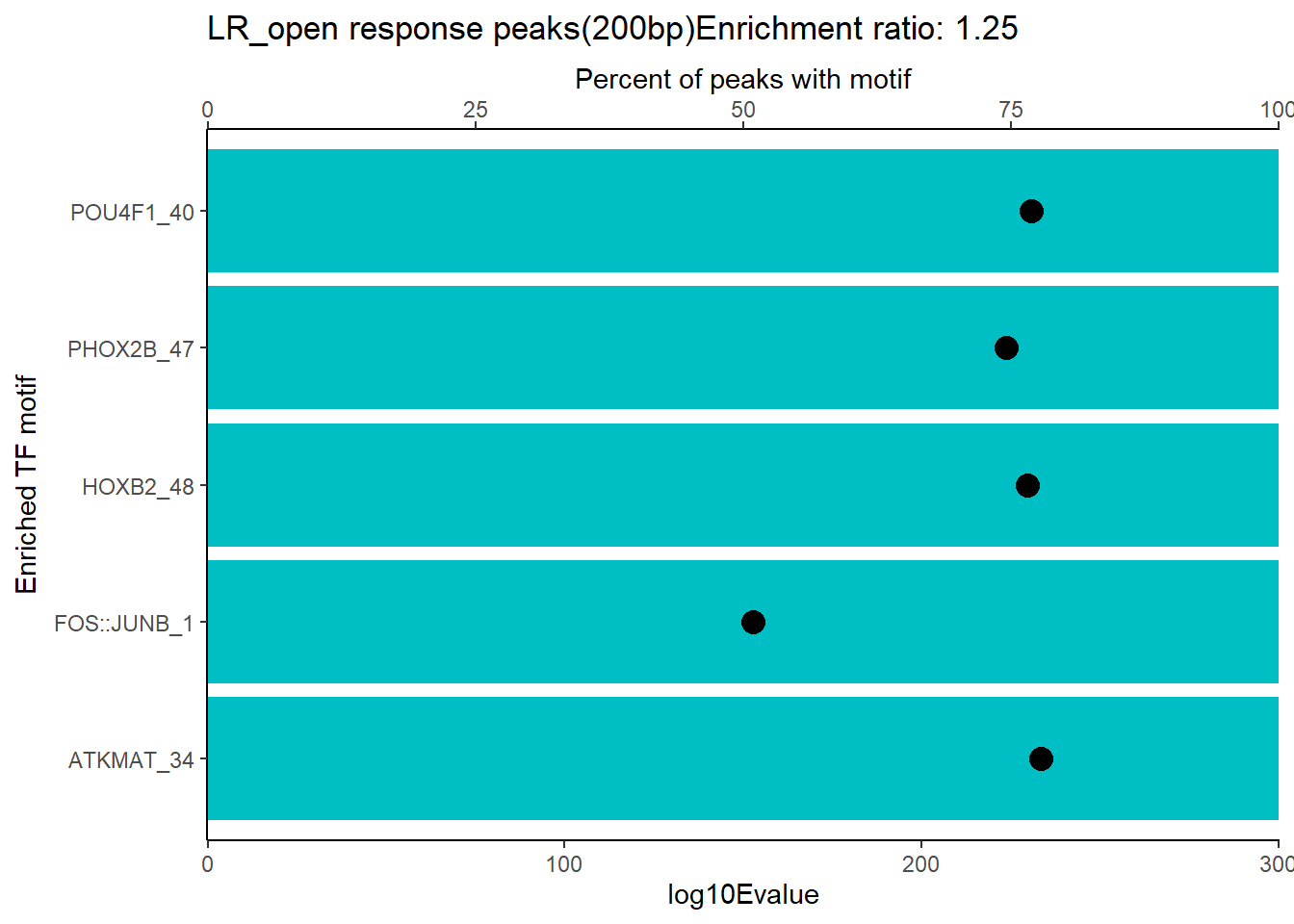
LR_close data
LR_close_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_close_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
LR_close_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_close_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
LR_close_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_close_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
LR_close_sea_all <- rbind(LR_close_sea_disc, LR_close_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)LR_close_combo <- LR_close_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., LR_close_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="LR_close")
LR_close_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EAR open v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0774.1 (MEIS2) | STREME-1 | 1-DGCTGTCA | DGCTGTCA | MEIS2 | 2.35e-143 | 1.55e-143 | 8140 | 48.6 | 29000 | 37.8 | 1.29 | LR_close |
| 2 | 1 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 3.53e-106 | 1.16e-106 | 6100 | 36.4 | 21200 | 27.7 | 1.32 | LR_close |
| 3 | 1 | MA0774.1 (MEIS2) | STREME-3 | 3-CTGTCWG | CTGTCWG | MEIS2 | 2.75e-79 | 6.03e-80 | 10800 | 64.4 | 43200 | 56.4 | 1.14 | LR_close |
| 4 | 2 | MA0672.1 (NKX2-3) | STREME-2 | 2-CCACTTRA | CCACTTRA | NKX2-3 | 7.25e-77 | 1.19e-77 | 5860 | 35 | 21200 | 27.6 | 1.27 | LR_close |
| 5 | 2 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 3.82e-63 | 5.03e-64 | 6830 | 40.8 | 25900 | 33.8 | 1.21 | LR_close |
| 6 | 1 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 1.42e-62 | 1.56e-63 | 2080 | 12.4 | 6200 | 8.08 | 1.53 | LR_close |
| 7 | 1 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 1.37e-57 | 1.29e-58 | 7070 | 42.2 | 27200 | 35.4 | 1.19 | LR_close |
| 8 | 3 | MA0739.1 (Hic1) | STREME-5 | 5-ATGCCA | ATGCCA | Hic1 | 6.58e-54 | 5.41e-55 | 12500 | 74.8 | 52600 | 68.6 | 1.09 | LR_close |
| 9 | 2 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 1.07e-47 | 7.84e-49 | 2700 | 16.1 | 9020 | 11.8 | 1.37 | LR_close |
| 10 | 4 | MA0140.2 (GATA1::TAL1) | STREME-6 | 6-WGATAA | WGATAA | GATA1::TAL1 | 1.41e-46 | 9.31e-48 | 10700 | 64.1 | 44400 | 57.9 | 1.11 | LR_close |
| 11 | 2 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 1.65e-45 | 9.9e-47 | 6820 | 40.7 | 26600 | 34.7 | 1.17 | LR_close |
| 12 | 5 | MA1655.1 (ZNF341) | STREME-11 | 11-ACAGCCA | 11-ACAGCCA | ZNF341 | 2.22e-38 | 1.22e-39 | 12200 | 73 | 51900 | 67.7 | 1.08 | LR_close |
| 13 | 2 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 2.88e-34 | 1.46e-35 | 4780 | 28.5 | 18200 | 23.8 | 1.2 | LR_close |
| 14 | 6 | MA1972.1 (ZFP14) | STREME-12 | 12-CAGGGCCT | 12-CAGGGCCT | ZFP14 | 1.36e-33 | 6.39e-35 | 8330 | 49.7 | 34000 | 44.4 | 1.12 | LR_close |
| 15 | 7 | 8-CTGCACA | STREME-8 | 8-CTGCACA | CTGCACA | CTGCACA | 1.95e-33 | 8.55e-35 | 14700 | 87.5 | 64200 | 83.7 | 1.05 | LR_close |
| 16 | 8 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 1.1e-31 | 4.51e-33 | 16000 | 95.4 | 71200 | 92.9 | 1.03 | LR_close |
| 17 | 9 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 2.26e-31 | 8.75e-33 | 9160 | 54.7 | 37900 | 49.5 | 1.1 | LR_close |
| 18 | 10 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 8.13e-31 | 2.97e-32 | 15500 | 92.4 | 68500 | 89.4 | 1.03 | LR_close |
| 19 | 11 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 2.48e-30 | 8.58e-32 | 15400 | 91.7 | 67900 | 88.6 | 1.03 | LR_close |
| 20 | 1 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 8.5e-30 | 2.8e-31 | 3760 | 22.5 | 14100 | 18.4 | 1.22 | LR_close |
| 21 | 1 | MA0739.1 | Hic1 | MA0739.1 | RTGCCAACY | Hic1 | 1.73e-29 | 5.39e-31 | 7070 | 42.2 | 28600 | 37.3 | 1.13 | LR_close |
| 22 | 8 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 1.8e-29 | 5.39e-31 | 15800 | 94.2 | 70200 | 91.6 | 1.03 | LR_close |
| 23 | 8 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 2.11e-29 | 6.05e-31 | 15500 | 92.5 | 68700 | 89.6 | 1.03 | LR_close |
| 24 | 12 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 1.03e-27 | 2.84e-29 | 10200 | 60.8 | 42900 | 56 | 1.09 | LR_close |
| 25 | 13 | 7-AGCACT | STREME-7 | 7-AGCACT | AGCACT | AGCACT | 1.59e-26 | 4.17e-28 | 15400 | 91.9 | 68300 | 89.1 | 1.03 | LR_close |
| 26 | 10 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 1.41e-25 | 3.57e-27 | 15800 | 94.6 | 70700 | 92.3 | 1.03 | LR_close |
| 27 | 8 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 2.16e-25 | 5.27e-27 | 15800 | 94.1 | 70200 | 91.6 | 1.03 | LR_close |
| 28 | 8 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 2.64e-25 | 6.2e-27 | 15800 | 94 | 70200 | 91.6 | 1.03 | LR_close |
| 29 | 8 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 3.64e-25 | 8.25e-27 | 15700 | 93.6 | 69800 | 91 | 1.03 | LR_close |
| 30 | 9 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 1.39e-24 | 3.06e-26 | 11200 | 66.8 | 47800 | 62.3 | 1.07 | LR_close |
| 31 | 8 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 1.46e-24 | 3.09e-26 | 15200 | 90.8 | 67400 | 88 | 1.03 | LR_close |
| 32 | 8 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 1.63e-24 | 3.27e-26 | 14500 | 86.6 | 63800 | 83.3 | 1.04 | LR_close |
| 33 | 8 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 1.64e-24 | 3.27e-26 | 15800 | 94.4 | 70600 | 92.1 | 1.03 | LR_close |
| 34 | 14 | 10-WGKGCMCW | STREME-10 | 10-WGKGCMCW | 10-WGKGCMCW | WGKGCMCW | 2.27e-24 | 4.39e-26 | 8290 | 49.5 | 34400 | 44.9 | 1.1 | LR_close |
| 35 | 15 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 3.53e-24 | 6.63e-26 | 15000 | 89.3 | 66200 | 86.3 | 1.04 | LR_close |
| 36 | 8 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 6.76e-24 | 1.24e-25 | 14500 | 86.3 | 63600 | 83 | 1.04 | LR_close |
| 37 | 16 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 1.65e-23 | 2.86e-25 | 8520 | 50.9 | 35500 | 46.4 | 1.1 | LR_close |
| 38 | 8 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 1.65e-23 | 2.86e-25 | 15000 | 89.3 | 66200 | 86.4 | 1.03 | LR_close |
| 39 | 8 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 3.18e-23 | 5.37e-25 | 15100 | 90.1 | 66800 | 87.2 | 1.03 | LR_close |
| 40 | 17 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 6.01e-23 | 9.88e-25 | 15300 | 91.2 | 67800 | 88.4 | 1.03 | LR_close |
| 41 | 18 | CTGCCTGCCTG | MEME-5 | CTGCCTGCCTG | CTGCCTGCCTG | CTGCCTGCCTG | 1.66e-22 | 2.67e-24 | 10700 | 63.9 | 45600 | 59.6 | 1.07 | LR_close |
| 42 | 12 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 3.36e-22 | 5.26e-24 | 6680 | 39.9 | 27300 | 35.6 | 1.12 | LR_close |
| 43 | 8 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 1.1e-21 | 1.68e-23 | 15200 | 90.5 | 67300 | 87.8 | 1.03 | LR_close |
| 44 | 8 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 1.66e-21 | 2.49e-23 | 15300 | 91.6 | 68200 | 89 | 1.03 | LR_close |
| 45 | 8 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 2.11e-21 | 3.09e-23 | 15900 | 95 | 71300 | 93 | 1.02 | LR_close |
| 46 | 12 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 1.52e-20 | 2.17e-22 | 9180 | 54.8 | 38800 | 50.6 | 1.08 | LR_close |
| 47 | 8 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 2.64e-20 | 3.7e-22 | 14700 | 87.8 | 65100 | 84.9 | 1.03 | LR_close |
| 48 | 19 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 3.21e-20 | 4.32e-22 | 15300 | 91.2 | 67900 | 88.6 | 1.03 | LR_close |
| 49 | 8 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 3.21e-20 | 4.32e-22 | 14500 | 86.8 | 64300 | 83.9 | 1.04 | LR_close |
| 50 | 10 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 7.33e-20 | 9.65e-22 | 16300 | 97.3 | 73400 | 95.8 | 1.02 | LR_close |
| 51 | 4 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 2.71e-19 | 3.5e-21 | 15300 | 91.3 | 68100 | 88.8 | 1.03 | LR_close |
| 52 | 19 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 7.26e-19 | 9.19e-21 | 15400 | 92 | 68700 | 89.6 | 1.03 | LR_close |
| 53 | 16 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 9.42e-19 | 1.17e-20 | 2800 | 16.7 | 10600 | 13.8 | 1.21 | LR_close |
| 54 | 8 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 1.19e-18 | 1.45e-20 | 14800 | 88.5 | 65800 | 85.8 | 1.03 | LR_close |
| 55 | 8 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 1.32e-18 | 1.58e-20 | 14900 | 88.9 | 66100 | 86.2 | 1.03 | LR_close |
| 56 | 8 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 2.21e-18 | 2.6e-20 | 14000 | 83.6 | 61700 | 80.6 | 1.04 | LR_close |
| 57 | 15 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 2.62e-18 | 3.03e-20 | 15200 | 91 | 67900 | 88.6 | 1.03 | LR_close |
| 58 | 16 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 3.63e-18 | 4.12e-20 | 2830 | 16.9 | 10700 | 14 | 1.2 | LR_close |
| 59 | 9 | MA0670.1 | NFIA | MA0670.1 | NDTGCCAAND | NFIA | 3.98e-18 | 4.42e-20 | 1670 | 9.98 | 5930 | 7.74 | 1.29 | LR_close |
| 60 | 8 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 4.03e-18 | 4.42e-20 | 14800 | 88.3 | 65600 | 85.6 | 1.03 | LR_close |
| 61 | 4 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 7.07e-18 | 7.62e-20 | 4260 | 25.4 | 16900 | 22.1 | 1.15 | LR_close |
| 62 | 16 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 7.21e-18 | 7.65e-20 | 2810 | 16.8 | 10700 | 13.9 | 1.2 | LR_close |
| 63 | 16 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 1.22e-17 | 1.28e-19 | 2770 | 16.6 | 10500 | 13.8 | 1.2 | LR_close |
| 64 | 19 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 3.08e-17 | 3.17e-19 | 13900 | 82.7 | 61100 | 79.7 | 1.04 | LR_close |
| 65 | 8 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 4.67e-17 | 4.73e-19 | 15500 | 92.8 | 69500 | 90.6 | 1.02 | LR_close |
| 66 | 20 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 6.2e-17 | 6.18e-19 | 14800 | 88.5 | 65800 | 85.9 | 1.03 | LR_close |
| 67 | 2 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 7.81e-17 | 7.67e-19 | 1360 | 8.14 | 4740 | 6.18 | 1.32 | LR_close |
| 68 | 8 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 8.53e-17 | 8.26e-19 | 15700 | 94 | 70500 | 92 | 1.02 | LR_close |
| 69 | 19 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 1.11e-16 | 1.06e-18 | 15100 | 89.9 | 67000 | 87.4 | 1.03 | LR_close |
| 70 | 19 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 1.4e-16 | 1.32e-18 | 15100 | 90 | 67100 | 87.5 | 1.03 | LR_close |
| 71 | 21 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 1.93e-16 | 1.79e-18 | 8310 | 49.6 | 35100 | 45.8 | 1.08 | LR_close |
| 72 | 8 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 5.56e-16 | 5.08e-18 | 16200 | 96.8 | 73000 | 95.3 | 1.02 | LR_close |
| 73 | 20 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 5.77e-16 | 5.2e-18 | 15000 | 89.4 | 66600 | 87 | 1.03 | LR_close |
| 74 | 4 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 6.79e-16 | 6.04e-18 | 5350 | 32 | 21900 | 28.5 | 1.12 | LR_close |
| 75 | 8 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1e-15 | 8.8e-18 | 13700 | 82 | 60600 | 79.1 | 1.04 | LR_close |
| 76 | 8 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 1.13e-15 | 9.8e-18 | 15800 | 94.1 | 70700 | 92.2 | 1.02 | LR_close |
| 77 | 9 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 4.8e-15 | 4.1e-17 | 3010 | 18 | 11700 | 15.3 | 1.18 | LR_close |
| 78 | 8 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 8.03e-15 | 6.78e-17 | 13700 | 81.9 | 60500 | 79 | 1.04 | LR_close |
| 79 | 4 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 1.09e-14 | 9.05e-17 | 14400 | 85.9 | 63800 | 83.3 | 1.03 | LR_close |
| 80 | 19 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 1.17e-14 | 9.64e-17 | 15600 | 93 | 69700 | 91 | 1.02 | LR_close |
| 81 | 10 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 1.87e-14 | 1.52e-16 | 16100 | 95.9 | 72300 | 94.3 | 1.02 | LR_close |
| 82 | 1 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 2e-14 | 1.61e-16 | 1400 | 8.33 | 4960 | 6.48 | 1.29 | LR_close |
| 83 | 22 | MA0018.4 (CREB1) | STREME-4 | 4-GTGATGACA | GTGATGACA | CREB1 | 2.17e-14 | 1.72e-16 | 380 | 2.27 | 1030 | 1.35 | 1.69 | LR_close |
| 84 | 1 | MA0783.1 | PKNOX2 | MA0783.1 | TGACAGGTGTCA | PKNOX2 | 4.23e-14 | 3.32e-16 | 587 | 3.5 | 1790 | 2.34 | 1.5 | LR_close |
| 85 | 23 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 5.05e-14 | 3.91e-16 | 15800 | 94.3 | 70900 | 92.5 | 1.02 | LR_close |
| 86 | 12 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 5.4e-14 | 4.13e-16 | 10100 | 60.2 | 43500 | 56.7 | 1.06 | LR_close |
| 87 | 8 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 6.31e-14 | 4.77e-16 | 13700 | 81.9 | 60600 | 79.1 | 1.04 | LR_close |
| 88 | 8 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 6.57e-14 | 4.87e-16 | 14600 | 87.1 | 64900 | 84.6 | 1.03 | LR_close |
| 89 | 19 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 6.59e-14 | 4.87e-16 | 15900 | 94.7 | 71300 | 93 | 1.02 | LR_close |
| 90 | 21 | MA0816.1 | Ascl2 | MA0816.1 | ARCAGCTGCY | Ascl2 | 8.08e-14 | 5.91e-16 | 6000 | 35.8 | 24900 | 32.5 | 1.1 | LR_close |
| 91 | 9 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 1.04e-13 | 7.43e-16 | 2500 | 14.9 | 9590 | 12.5 | 1.19 | LR_close |
| 92 | 8 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 1.05e-13 | 7.43e-16 | 16000 | 95.7 | 72200 | 94.2 | 1.02 | LR_close |
| 93 | 16 | MA1472.2 | Bhlha15 | MA1472.2 | NVACAGCTGTBN | Bhlha15 | 1.05e-13 | 7.43e-16 | 7560 | 45.2 | 32000 | 41.7 | 1.08 | LR_close |
| 94 | 8 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 1.28e-13 | 8.99e-16 | 15800 | 94.6 | 71200 | 92.9 | 1.02 | LR_close |
| 95 | 24 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 1.82e-13 | 1.26e-15 | 13600 | 81.1 | 60000 | 78.3 | 1.04 | LR_close |
| 96 | 19 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 2.96e-13 | 2.03e-15 | 14400 | 85.9 | 63900 | 83.4 | 1.03 | LR_close |
| 97 | 1 | MA0796.1 | TGIF1 | MA0796.1 | TGACAGCTGTCA | TGIF1 | 3.08e-13 | 2.09e-15 | 643 | 3.84 | 2030 | 2.64 | 1.45 | LR_close |
| 98 | 8 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 3.22e-13 | 2.16e-15 | 14900 | 89 | 66500 | 86.8 | 1.03 | LR_close |
| 99 | 19 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 3.39e-13 | 2.25e-15 | 14400 | 86.1 | 64100 | 83.6 | 1.03 | LR_close |
| 100 | 8 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 4.86e-13 | 3.2e-15 | 13400 | 79.8 | 59000 | 77 | 1.04 | LR_close |
| 101 | 25 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 1.02e-12 | 6.62e-15 | 14100 | 84 | 62400 | 81.4 | 1.03 | LR_close |
| 102 | 8 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 1.03e-12 | 6.62e-15 | 14100 | 84.2 | 62600 | 81.6 | 1.03 | LR_close |
| 103 | 26 | MA0738.1 | HIC2 | MA0738.1 | RTGCCCRSB | HIC2 | 2.89e-12 | 1.85e-14 | 7540 | 45 | 32000 | 41.7 | 1.08 | LR_close |
| 104 | 16 | MA1641.1 | MYF5 | MA1641.1 | NVACAGCTGTBN | MYF5 | 3.34e-12 | 2.12e-14 | 5420 | 32.4 | 22500 | 29.3 | 1.1 | LR_close |
| 105 | 8 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 5.67e-12 | 3.55e-14 | 13300 | 79.3 | 58700 | 76.6 | 1.04 | LR_close |
| 106 | 8 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 5.74e-12 | 3.56e-14 | 14900 | 88.9 | 66500 | 86.8 | 1.02 | LR_close |
| 107 | 1 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 8.56e-12 | 5.27e-14 | 2200 | 13.1 | 8440 | 11 | 1.19 | LR_close |
| 108 | 2 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 9.62e-12 | 5.86e-14 | 1260 | 7.5 | 4510 | 5.89 | 1.27 | LR_close |
| 109 | 8 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 1.22e-11 | 7.34e-14 | 13600 | 81.3 | 60300 | 78.7 | 1.03 | LR_close |
| 110 | 19 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.54e-11 | 9.2e-14 | 16200 | 96.9 | 73300 | 95.7 | 1.01 | LR_close |
| 111 | 8 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 2.27e-11 | 1.34e-13 | 15400 | 92 | 69100 | 90.1 | 1.02 | LR_close |
| 112 | 22 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 2.51e-11 | 1.47e-13 | 14000 | 83.8 | 62300 | 81.3 | 1.03 | LR_close |
| 113 | 27 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 4.59e-11 | 2.67e-13 | 16400 | 97.9 | 74200 | 96.8 | 1.01 | LR_close |
| 114 | 28 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 4.95e-11 | 2.85e-13 | 14900 | 88.8 | 66500 | 86.8 | 1.02 | LR_close |
| 115 | 21 | MA0665.1 | MSC | MA0665.1 | AACAGCTGTT | MSC | 5.14e-11 | 2.94e-13 | 7620 | 45.5 | 32500 | 42.4 | 1.07 | LR_close |
| 116 | 17 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 5.25e-11 | 2.98e-13 | 15500 | 92.3 | 69400 | 90.5 | 1.02 | LR_close |
| 117 | 19 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 5.57e-11 | 3.13e-13 | 14900 | 89 | 66600 | 87 | 1.02 | LR_close |
| 118 | 8 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 5.66e-11 | 3.16e-13 | 16300 | 97.4 | 73700 | 96.2 | 1.01 | LR_close |
| 119 | 2 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 5.72e-11 | 3.16e-13 | 9320 | 55.6 | 40200 | 52.5 | 1.06 | LR_close |
| 120 | 8 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 5.97e-11 | 3.28e-13 | 11800 | 70.6 | 51900 | 67.7 | 1.04 | LR_close |
| 121 | 8 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 6.86e-11 | 3.73e-13 | 15300 | 91.1 | 68400 | 89.2 | 1.02 | LR_close |
| 122 | 19 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 2.43e-10 | 1.31e-12 | 14400 | 86.2 | 64400 | 84 | 1.03 | LR_close |
| 123 | 15 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 2.71e-10 | 1.45e-12 | 11900 | 71 | 52300 | 68.2 | 1.04 | LR_close |
| 124 | 29 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 3.79e-10 | 2.01e-12 | 14500 | 86.6 | 64700 | 84.5 | 1.03 | LR_close |
| 125 | 30 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 4.71e-10 | 2.48e-12 | 15100 | 89.9 | 67400 | 88 | 1.02 | LR_close |
| 126 | 4 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 1.37e-09 | 7.13e-12 | 5300 | 31.7 | 22200 | 28.9 | 1.09 | LR_close |
| 127 | 25 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 1.5e-09 | 7.75e-12 | 14100 | 84.3 | 62900 | 82 | 1.03 | LR_close |
| 128 | 8 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 1.71e-09 | 8.81e-12 | 15800 | 94.1 | 71000 | 92.6 | 1.02 | LR_close |
| 129 | 31 | MA1566.2 | TBX3 | MA1566.2 | RAGGTGTGAAV | TBX3 | 1.73e-09 | 8.82e-12 | 6030 | 36 | 25400 | 33.2 | 1.08 | LR_close |
| 130 | 21 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 1.97e-09 | 9.95e-12 | 3360 | 20.1 | 13600 | 17.8 | 1.13 | LR_close |
| 131 | 9 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 2.21e-09 | 1.11e-11 | 1210 | 7.21 | 4420 | 5.77 | 1.25 | LR_close |
| 132 | 19 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 2.64e-09 | 1.32e-11 | 13800 | 82.4 | 61400 | 80.1 | 1.03 | LR_close |
| 133 | 8 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 2.86e-09 | 1.41e-11 | 16500 | 98.5 | 74800 | 97.7 | 1.01 | LR_close |
| 134 | 8 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 4.36e-09 | 2.14e-11 | 14500 | 86.6 | 64800 | 84.6 | 1.02 | LR_close |
| 135 | 8 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 5.67e-09 | 2.76e-11 | 13300 | 79.5 | 59100 | 77.1 | 1.03 | LR_close |
| 136 | 10 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 6.01e-09 | 2.91e-11 | 15500 | 92.8 | 69900 | 91.2 | 1.02 | LR_close |
| 137 | 8 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 6.28e-09 | 3.02e-11 | 12400 | 74.3 | 55000 | 71.7 | 1.04 | LR_close |
| 138 | 8 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 8.3e-09 | 3.96e-11 | 12800 | 76.6 | 56800 | 74.1 | 1.03 | LR_close |
| 139 | 29 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 1.96e-08 | 9.28e-11 | 14400 | 86.2 | 64500 | 84.2 | 1.02 | LR_close |
| 140 | 20 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 2.01e-08 | 9.43e-11 | 11500 | 68.7 | 50600 | 66.1 | 1.04 | LR_close |
| 141 | 47 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 3.39e-08 | 1.58e-10 | 14300 | 85.4 | 63900 | 83.4 | 1.02 | LR_close |
| 142 | 4 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 3.96e-08 | 1.84e-10 | 12300 | 73.6 | 54500 | 71.1 | 1.03 | LR_close |
| 143 | 26 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 5.27e-08 | 2.43e-10 | 15800 | 94.3 | 71300 | 93 | 1.01 | LR_close |
| 144 | 8 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 7.16e-08 | 3.27e-10 | 12700 | 76.1 | 56500 | 73.7 | 1.03 | LR_close |
| 145 | 21 | MA0669.1 | NEUROG2 | MA0669.1 | RACATATGTC | NEUROG2 | 7.49e-08 | 3.4e-10 | 6030 | 36 | 25600 | 33.4 | 1.08 | LR_close |
| 146 | 8 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 9.89e-08 | 4.46e-10 | 16300 | 97.6 | 74100 | 96.6 | 1.01 | LR_close |
| 147 | 32 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 1.33e-07 | 5.94e-10 | 14500 | 86.6 | 64900 | 84.7 | 1.02 | LR_close |
| 148 | 10 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 1.34e-07 | 5.94e-10 | 15600 | 93.2 | 70400 | 91.8 | 1.02 | LR_close |
| 149 | 23 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 1.49e-07 | 6.58e-10 | 16400 | 98.1 | 74600 | 97.3 | 1.01 | LR_close |
| 150 | 15 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 1.63e-07 | 7.15e-10 | 12700 | 75.6 | 56200 | 73.3 | 1.03 | LR_close |
| 151 | 16 | MA0521.2 | Tcf12 | MA0521.2 | NNACAGCTGTNN | Tcf12 | 1.66e-07 | 7.24e-10 | 4200 | 25.1 | 17500 | 22.8 | 1.1 | LR_close |
| 152 | 8 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 2.78e-07 | 1.2e-09 | 13400 | 80.2 | 59800 | 78 | 1.03 | LR_close |
| 153 | 19 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 3.84e-07 | 1.65e-09 | 14400 | 85.8 | 64300 | 83.9 | 1.02 | LR_close |
| 154 | 31 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 5.03e-07 | 2.15e-09 | 9790 | 58.5 | 42800 | 55.9 | 1.05 | LR_close |
| 155 | 2 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 1.26e-06 | 5.33e-09 | 15900 | 94.7 | 71600 | 93.4 | 1.01 | LR_close |
| 156 | 31 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 2.45e-06 | 1.03e-08 | 6060 | 36.2 | 25900 | 33.8 | 1.07 | LR_close |
| 157 | 26 | MA1728.1 | ZNF549 | MA1728.1 | NNTGCTGCCCWR | ZNF549 | 2.98e-06 | 1.25e-08 | 15400 | 91.9 | 69400 | 90.5 | 1.02 | LR_close |
| 158 | 8 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 5.33e-06 | 2.22e-08 | 11600 | 69 | 51100 | 66.7 | 1.03 | LR_close |
| 159 | 20 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 7.94e-06 | 3.29e-08 | 11800 | 70.2 | 52100 | 68 | 1.03 | LR_close |
| 160 | 8 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 9.6e-06 | 3.95e-08 | 14600 | 87.1 | 65500 | 85.4 | 1.02 | LR_close |
| 161 | 1 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 1.02e-05 | 4.17e-08 | 986 | 5.89 | 3700 | 4.82 | 1.22 | LR_close |
| 162 | 31 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 1.16e-05 | 4.7e-08 | 3400 | 20.3 | 14100 | 18.4 | 1.1 | LR_close |
| 163 | 8 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 1.5e-05 | 6.04e-08 | 13200 | 79 | 59000 | 77 | 1.03 | LR_close |
| 164 | 8 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 1.69e-05 | 6.79e-08 | 10800 | 64.7 | 47800 | 62.4 | 1.04 | LR_close |
| 165 | 31 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 1.74e-05 | 6.88e-08 | 3940 | 23.5 | 16500 | 21.6 | 1.09 | LR_close |
| 166 | 33 | MA1573.2 (Thap11) | STREME-9 | 9-CTGGGAAATGTAGTY | CTGGGAAATGTAGTY | Thap11 | 1.74e-05 | 6.88e-08 | 33 | 0.2 | 37 | 0.05 | 4.09 | LR_close |
| 167 | 8 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 1.92e-05 | 7.57e-08 | 4760 | 28.4 | 20200 | 26.4 | 1.08 | LR_close |
| 168 | 34 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 2.38e-05 | 9.33e-08 | 15700 | 93.9 | 71100 | 92.7 | 1.01 | LR_close |
| 169 | 8 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 2.61e-05 | 1.02e-07 | 13600 | 81.1 | 60800 | 79.3 | 1.02 | LR_close |
| 170 | 27 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 2.69e-05 | 1.04e-07 | 14800 | 88.4 | 66500 | 86.8 | 1.02 | LR_close |
| 171 | 16 | MA0500.2 | MYOG | MA0500.2 | NDRCAGCTGYHN | MYOG | 2.99e-05 | 1.15e-07 | 12500 | 74.4 | 55500 | 72.4 | 1.03 | LR_close |
| 172 | 23 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 4.51e-05 | 1.73e-07 | 15900 | 94.8 | 71900 | 93.8 | 1.01 | LR_close |
| 173 | 11 | MA1153.1 | Smad4 | MA1153.1 | TGTCTRGM | Smad4 | 6.81e-05 | 2.59e-07 | 2560 | 15.3 | 10500 | 13.7 | 1.11 | LR_close |
| 174 | 34 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 9.27e-05 | 3.51e-07 | 15200 | 91 | 68700 | 89.7 | 1.01 | LR_close |
| 175 | 32 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 0.000124 | 4.66e-07 | 13600 | 81.4 | 61100 | 79.7 | 1.02 | LR_close |
| 176 | 35 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 0.000149 | 5.56e-07 | 14200 | 84.9 | 63800 | 83.3 | 1.02 | LR_close |
| 177 | 33 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 0.000161 | 5.98e-07 | 49 | 0.29 | 84 | 0.11 | 2.69 | LR_close |
| 178 | 21 | MA0832.1 | Tcf21 | MA0832.1 | RYAACAGCTGTTRN | Tcf21 | 0.000165 | 6.06e-07 | 1660 | 9.89 | 6620 | 8.64 | 1.15 | LR_close |
| 179 | 23 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 0.000165 | 6.06e-07 | 10800 | 64.6 | 47900 | 62.5 | 1.03 | LR_close |
| 180 | 15 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 0.000166 | 6.06e-07 | 12900 | 77.3 | 57800 | 75.4 | 1.02 | LR_close |
| 181 | 8 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 0.000188 | 6.82e-07 | 10100 | 60.4 | 44700 | 58.3 | 1.04 | LR_close |
| 182 | 19 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 0.000202 | 7.32e-07 | 16500 | 98.4 | 75000 | 97.8 | 1.01 | LR_close |
| 183 | 31 | MA0690.2 | TBX21 | MA0690.2 | TTTCACACCTT | TBX21 | 0.000274 | 9.84e-07 | 5910 | 35.3 | 25500 | 33.3 | 1.06 | LR_close |
| 184 | 36 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 0.000367 | 1.31e-06 | 14500 | 86.3 | 65000 | 84.8 | 1.02 | LR_close |
| 185 | 8 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 0.000371 | 1.32e-06 | 10500 | 62.6 | 46400 | 60.5 | 1.03 | LR_close |
| 186 | 9 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 0.000375 | 1.33e-06 | 1070 | 6.41 | 4160 | 5.42 | 1.18 | LR_close |
| 187 | 31 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.00049 | 1.73e-06 | 4600 | 27.4 | 19600 | 25.6 | 1.07 | LR_close |
| 188 | 21 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.000493 | 1.73e-06 | 4260 | 25.4 | 18100 | 23.7 | 1.08 | LR_close |
| 189 | 8 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 0.000516 | 1.79e-06 | 9990 | 59.6 | 44100 | 57.6 | 1.04 | LR_close |
| 190 | 16 | MA0820.1 | FIGLA | MA0820.1 | WMCACCTGKW | FIGLA | 0.000606 | 2.1e-06 | 16000 | 95.4 | 72400 | 94.4 | 1.01 | LR_close |
| 191 | 16 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 0.000664 | 2.29e-06 | 15600 | 93.2 | 70600 | 92.1 | 1.01 | LR_close |
| 192 | 16 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 0.000721 | 2.47e-06 | 13200 | 78.5 | 58900 | 76.8 | 1.02 | LR_close |
| 193 | 9 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 0.000745 | 2.54e-06 | 12900 | 77.2 | 57800 | 75.5 | 1.02 | LR_close |
| 194 | 37 | MA0071.1 | RORA | MA0071.1 | AWMWAGGTCA | RORA | 0.000895 | 3.04e-06 | 2490 | 14.9 | 10300 | 13.5 | 1.1 | LR_close |
| 195 | 16 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 0.00114 | 3.84e-06 | 1910 | 11.4 | 7810 | 10.2 | 1.12 | LR_close |
| 196 | 8 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 0.00139 | 4.66e-06 | 13400 | 80.2 | 60200 | 78.6 | 1.02 | LR_close |
| 197 | 38 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 0.00141 | 4.71e-06 | 14000 | 83.8 | 63100 | 82.3 | 1.02 | LR_close |
| 198 | 16 | MA1619.1 | Ptf1A | MA1619.1 | NNACAGCTGTNN | Ptf1A | 0.00148 | 4.93e-06 | 8400 | 50.1 | 36900 | 48.2 | 1.04 | LR_close |
| 199 | 8 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 0.00218 | 7.21e-06 | 12300 | 73.5 | 55000 | 71.8 | 1.02 | LR_close |
| 200 | 39 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 0.00323 | 1.06e-05 | 15700 | 93.5 | 70900 | 92.5 | 1.01 | LR_close |
| 201 | 16 | MA1558.1 | SNAI1 | MA1558.1 | DRCAGGTGYD | SNAI1 | 0.00337 | 1.1e-05 | 13900 | 83.2 | 62600 | 81.7 | 1.02 | LR_close |
| 202 | 31 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 0.00363 | 1.18e-05 | 5920 | 35.4 | 25700 | 33.6 | 1.05 | LR_close |
| 203 | 8 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 0.00432 | 1.4e-05 | 10500 | 62.8 | 46800 | 61 | 1.03 | LR_close |
| 204 | 8 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 0.00449 | 1.45e-05 | 16500 | 98.8 | 75400 | 98.3 | 1 | LR_close |
| 205 | 16 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 0.00598 | 1.92e-05 | 14300 | 85.3 | 64300 | 83.9 | 1.02 | LR_close |
| 206 | 40 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 0.0062 | 1.98e-05 | 13100 | 78.4 | 58900 | 76.9 | 1.02 | LR_close |
| 207 | 20 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 0.00642 | 2.04e-05 | 10300 | 61.5 | 45800 | 59.7 | 1.03 | LR_close |
| 208 | 39 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 0.0088 | 2.78e-05 | 14900 | 89.2 | 67500 | 88.1 | 1.01 | LR_close |
| 209 | 16 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 0.00903 | 2.84e-05 | 4420 | 26.4 | 19000 | 24.8 | 1.06 | LR_close |
| 210 | 16 | MA1559.1 | SNAI3 | MA1559.1 | RRCAGGTGYA | SNAI3 | 0.00948 | 2.97e-05 | 10000 | 59.7 | 44400 | 57.9 | 1.03 | LR_close |
| 211 | 16 | MA0623.2 | NEUROG1 | MA0623.2 | RACATATGTY | NEUROG1 | 0.0106 | 3.31e-05 | 1140 | 6.83 | 4560 | 5.95 | 1.15 | LR_close |
| 212 | 16 | MA1100.2 | ASCL1 | MA1100.2 | VGCAGCTGCN | ASCL1 | 0.0135 | 4.19e-05 | 14000 | 83.9 | 63300 | 82.5 | 1.02 | LR_close |
| 213 | 41 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 0.0136 | 4.19e-05 | 15600 | 93.1 | 70600 | 92.2 | 1.01 | LR_close |
| 214 | 8 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 0.015 | 4.62e-05 | 11000 | 65.4 | 48900 | 63.7 | 1.03 | LR_close |
| 215 | 21 | MA0818.2 | BHLHE22 | MA0818.2 | AMCATATGKY | BHLHE22 | 0.018 | 5.5e-05 | 5050 | 30.1 | 21900 | 28.5 | 1.06 | LR_close |
| 216 | 27 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 0.0203 | 6.17e-05 | 5610 | 33.5 | 24400 | 31.9 | 1.05 | LR_close |
| 217 | 10 | MA0870.1 | Sox1 | MA0870.1 | AACAATANCATTGTT | Sox1 | 0.0204 | 6.19e-05 | 552 | 3.3 | 2080 | 2.71 | 1.22 | LR_close |
| 218 | 8 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 0.0211 | 6.37e-05 | 10800 | 64.6 | 48300 | 63 | 1.03 | LR_close |
| 219 | 37 | MA1535.1 | NR2C1 | MA1535.1 | NRRGGTCAN | NR2C1 | 0.0224 | 6.72e-05 | 14900 | 89 | 67300 | 87.8 | 1.01 | LR_close |
| 220 | 42 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 0.0251 | 7.52e-05 | 12800 | 76.4 | 57400 | 74.9 | 1.02 | LR_close |
| 221 | 31 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 0.0275 | 8.15e-05 | 1760 | 10.5 | 7260 | 9.47 | 1.11 | LR_close |
| 222 | 31 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 0.0275 | 8.15e-05 | 9800 | 58.5 | 43500 | 56.8 | 1.03 | LR_close |
| 223 | 21 | MA0607.2 | BHLHA15 | MA0607.2 | ACCATATGGT | BHLHA15 | 0.0283 | 8.34e-05 | 4590 | 27.4 | 19800 | 25.9 | 1.06 | LR_close |
| 224 | 16 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.03 | 8.8e-05 | 1590 | 9.51 | 6550 | 8.54 | 1.11 | LR_close |
| 225 | 19 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 0.0405 | 0.000119 | 10800 | 64.4 | 48200 | 62.8 | 1.03 | LR_close |
| 226 | 40 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 0.0419 | 0.000122 | 13200 | 78.9 | 59400 | 77.5 | 1.02 | LR_close |
| 227 | 8 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 0.0439 | 0.000127 | 13100 | 78.1 | 58800 | 76.7 | 1.02 | LR_close |
| 228 | 43 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 0.0468 | 0.000135 | 15200 | 90.6 | 68700 | 89.6 | 1.01 | LR_close |
| 14 | 44 | MA1107.2 (KLF9) | MEME-3 | ACACACACACACACA | ACACACACACACACA | KLF9 | 1.26 | 0.0902 | 22 | 0.13 | 68 | 0.09 | 1.53 | LR_close |
| 15 | 45 | MA1723.1 (PRDM9) | MEME-2 | DGGRGVWGGGRRGAG | DGGRGVWGGGRRGAG | PRDM9 | 11.3 | 0.75 | 501 | 2.99 | 2340 | 3.05 | 0.982 | LR_close |
| 16 | 46 | MA1125.1 (ZNF384) | MEME-1 | AAAAWAAAAAAARAA | AAAAWAAAAAAARAA | ZNF384 | 17 | 1 | 190 | 1.13 | 1100 | 1.44 | 0.792 | LR_close |
| 17 | 47 | MA0139.1 (CTCF) | MEME-4 | SYGCCMYCTGSTGGY | SYGCCMYCTGSTGGY | CTCF | 17 | 1 | 1890 | 11.3 | 10400 | 13.6 | 0.831 | LR_close |
ER_rat <- 1.25
mrc_type <- "LR_close"
LR_close_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))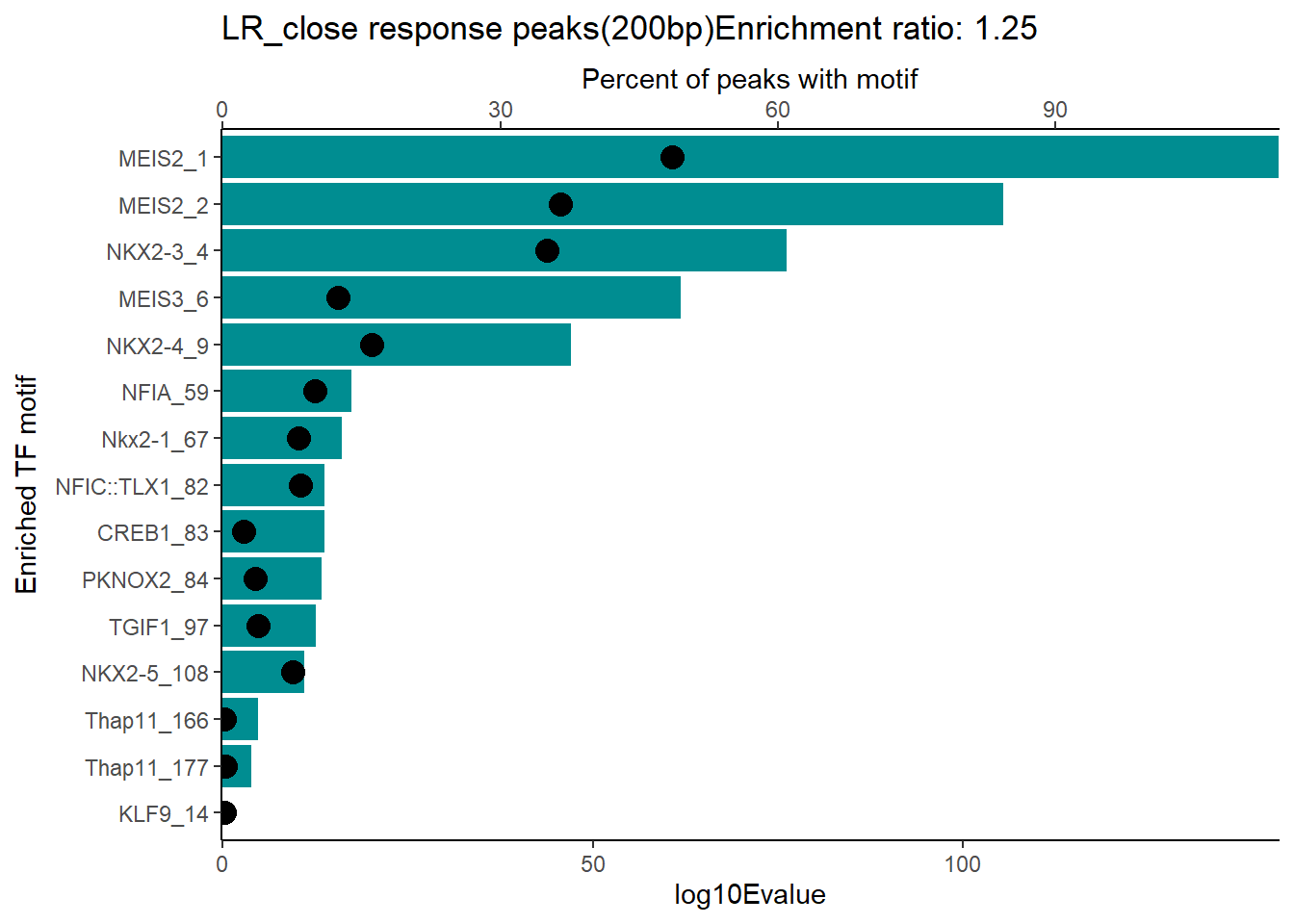
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
LR_close_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP.`*2.5), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./2.5,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))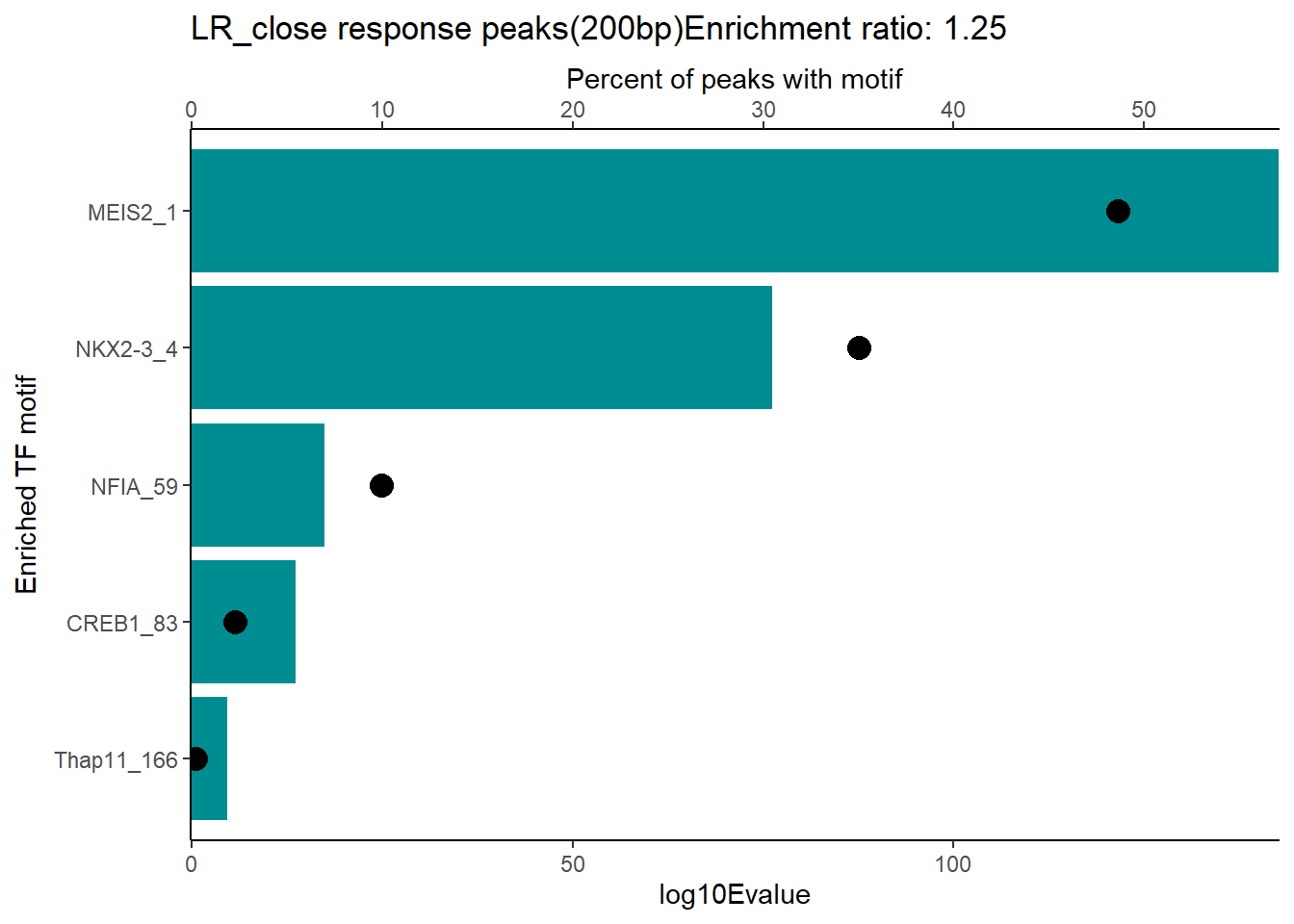
| Version | Author | Date |
|---|---|---|
| 7b1c18d | reneeisnowhere | 2025-05-08 |
meme_motifs_EAR_open <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_open_xstreme/xstreme.txt")
meme_motifs_ESR_open <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_open_xstreme/xstreme.txt")
meme_motifs_LR_open <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_open_xstreme/xstreme.txt")
meme_motifs_EAR_close <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_close_xstreme/xstreme.txt")
meme_motifs_ESR_close <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_close_xstreme/xstreme.txt")
meme_motifs_LR_close <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_close_xstreme/xstreme.txt")
meme_motifs_ESR_opcl <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_opcl_xstreme/xstreme.txt")
meme_motifs_ESR_clop <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_clop_xstreme/xstreme.txt")making table of significant
full_motifs_dataframe <- bind_rows(EAR_close_combo,
ESR_close_combo,
LR_close_combo,
EAR_open_combo,
ESR_open_combo,
LR_open_combo,
ESR_clop_combo,
ESR_opcl_combo)
for_supplemental <- full_motifs_dataframe %>%
dplyr::select(CLUSTER:EVALUE.y, ENR_RATIO,mrc_type) %>%
dplyr::filter(ENR_RATIO>1.25) %>%
dplyr::filter(EVALUE.y<0.05) %>%
group_by(mrc_type,CLUSTER) %>%
summarize(ID=paste(unique(ID), collapse = ";"),
ALT_ID=paste(unique(ALT_ID), collapse = ";"),
motif_name=paste(unique(motif_name),collapse="; "),
mrc=unique(mrc_type),
sig_val=paste0(min(EVALUE.y),"-",max(EVALUE.y)),
order_val=min(EVALUE.y)) %>%
arrange(mrc,order_val)
# write_delim(for_supplemental,"data/Final_four_data/re_analysis/motif_cluster_dataframe.txt",delim="\t")
for_supplemental# A tibble: 160 × 8
# Groups: mrc_type [8]
mrc_type CLUSTER ID ALT_ID motif_name mrc sig_val order_val
<chr> <int> <chr> <chr> <chr> <chr> <chr> <dbl>
1 EAR_close 1 1-VTGASTCAB;MA16… STREM… FOS; BACH… EAR_… 4.82e-… 4.82e-72
2 EAR_close 2 2-GWGATAA;MA0766… STREM… GATA2; GA… EAR_… 6.53e-… 6.53e-61
3 EAR_close 3 MA0774.1;MA0498.… MEIS2… MEIS2; ME… EAR_… 1.58e-… 1.58e-26
4 EAR_close 10 7-ACTCAA STREM… NKX2-2 EAR_… 2.27e-… 2.27e-22
5 EAR_close 16 MA0035.4 GATA1 GATA1 EAR_… 7.35e-… 7.35e-18
6 EAR_close 18 MA0842.2 NRL NRL EAR_… 1.73e-… 1.73e-14
7 EAR_close 31 MA1632.1 ATF2 ATF2 EAR_… 4.08e-… 4.08e-13
8 EAR_close 15 MA0595.1;MA0596.… SREBF… SREBF1; S… EAR_… 3.57e-… 3.57e-12
9 EAR_close 33 MA1643.1;MA0119.… NFIB;… NFIB; NFI… EAR_… 6.57e-… 6.57e- 6
10 EAR_close 32 MA0802.1 TBR1 TBR1 EAR_… 8.35e-… 8.35e- 6
# ℹ 150 more rows full_motifs_dataframe %>%
dplyr::filter(mrc_type=="LR_open") RANK CLUSTER SIM_MOTIF ALT_ID ID
1 1 1 MA1134.1 (FOS::JUNB) STREME-1 1-DATGASTCATH
2 2 1 MA1988.1 (Atf3) MEME-1 DRTGACTCAYH
3 3 1 MA1144.1 FOSL2::JUND MA1144.1
4 4 1 MA1135.1 FOSB::JUNB MA1135.1
5 5 1 MA1134.1 FOS::JUNB MA1134.1
6 6 1 MA0477.2 FOSL1 MA0477.2
7 7 1 MA1138.1 FOSL2::JUNB MA1138.1
8 8 1 MA0476.1 FOS MA0476.1
9 9 1 MA0489.2 Jun MA0489.2
10 10 1 MA0490.2 JUNB MA0490.2
11 11 1 MA1130.1 FOSL2::JUN MA1130.1
12 12 1 MA1141.1 FOS::JUND MA1141.1
13 13 1 MA0462.2 BATF::JUN MA0462.2
14 14 1 MA1634.1 BATF MA1634.1
15 15 1 MA1988.1 Atf3 MA1988.1
16 16 1 MA0099.3 FOS::JUN MA0099.3
17 17 1 MA0491.2 JUND MA0491.2
18 18 1 MA0835.2 BATF3 MA0835.2
19 19 1 MA1128.1 FOSL1::JUN MA1128.1
20 20 1 MA1633.2 BACH1 MA1633.2
21 21 1 MA1928.1 BNC2 MA1928.1
22 22 1 MA1137.1 FOSL1::JUNB MA1137.1
23 23 1 MA0478.1 FOSL2 MA0478.1
24 24 1 MA0476.1 (FOS) STREME-10 10-TGANTCA
25 25 1 MA0655.1 JDP2 MA0655.1
26 26 1 MA1142.1 FOSL1::JUND MA1142.1
27 27 1 MA1132.1 JUN::JUNB MA1132.1
28 28 1 MA1101.2 BACH2 MA1101.2
29 29 1 MA0841.1 NFE2 MA0841.1
30 30 1 MA0089.2 MAFG::NFE2L1 MA0089.2
31 31 1 MA0496.3 MAFK MA0496.3
32 32 1 MA0501.1 MAF::NFE2 MA0501.1
33 33 1 MA0150.2 Nfe2l2 MA0150.2
34 34 2 2-ATKMAT STREME-2 2-ATKMAT
35 35 1 MA0659.3 Mafg MA0659.3
36 36 3 MA0676.1 Nr2e1 MA0676.1
37 37 1 MA1113.2 PBX2 MA1113.2
38 38 1 MA1639.1 MEIS1 MA1639.1
39 39 1 MA1640.1 MEIS2 MA1640.1
40 40 4 MA0790.1 POU4F1 MA0790.1
41 41 5 MA1978.1 ZNF354A MA1978.1
42 42 6 MA0845.1 FOXB1 MA0845.1
43 43 7 MA0886.1 EMX2 MA0886.1
44 44 8 MA0788.1 POU3F3 MA0788.1
45 45 8 MA0786.1 POU3F1 MA0786.1
46 46 9 MA0680.2 Pax7 MA0680.2
47 47 9 MA0681.2 PHOX2B MA0681.2
48 48 7 MA0902.2 HOXB2 MA0902.2
49 49 6 MA0032.2 FOXC1 MA0032.2
50 50 10 3-WTVWBAW STREME-3 3-WTVWBAW
51 51 8 MA0785.1 POU2F1 MA0785.1
52 52 6 MA0041.2 FOXD3 MA0041.2
53 53 4 MA0791.1 POU4F3 MA0791.1
54 54 6 MA0846.1 FOXC2 MA0846.1
55 55 8 MA0792.1 POU5F1B MA0792.1
56 56 7 MA0710.1 NOTO MA0710.1
57 57 7 MA0628.1 POU6F1 MA0628.1
58 58 11 MA0052.4 MEF2A MA0052.4
59 59 12 MA1143.1 FOSL1::JUND MA1143.1
60 60 8 MA0787.1 POU3F2 MA0787.1
61 61 13 15-HHATDD STREME-15 15-HHATDD
62 62 7 MA1963.1 SATB1 MA1963.1
63 63 4 MA0046.2 HNF1A MA0046.2
64 64 7 MA0713.1 PHOX2A MA0713.1
65 65 14 MA0901.2 HOXB13 MA0901.2
66 66 7 MA0793.1 POU6F2 MA0793.1
67 67 9 MA0679.2 ONECUT1 MA0679.2
68 68 8 MA0627.2 POU2F3 MA0627.2
69 69 4 MA0153.2 HNF1B MA0153.2
70 70 8 MA1115.1 POU5F1 MA1115.1
71 71 8 MA0142.1 Pou5f1::Sox2 MA0142.1
72 72 15 4-TRTAYA STREME-4 4-TRTAYA
73 73 9 MA1471.1 BARX2 MA1471.1
74 74 6 MA0148.4 FOXA1 MA0148.4
75 75 7 MA0723.2 VAX2 MA0723.2
76 76 7 MA0722.1 VAX1 MA0722.1
77 77 8 MA0789.1 POU3F4 MA0789.1
78 78 7 MA1549.1 POU6F1 MA1549.1
79 79 6 MA1607.1 Foxl2 MA1607.1
80 80 16 MA0084.1 SRY MA0084.1
81 81 17 MA0602.1 Arid5a MA0602.1
82 82 7 MA0706.1 MEOX2 MA0706.1
83 83 7 MA0675.1 NKX6-2 MA0675.1
84 84 14 MA0465.2 CDX2 MA0465.2
85 85 7 MA1502.1 HOXB8 MA1502.1
86 86 6 MA0047.3 FOXA2 MA0047.3
87 87 6 MA0481.3 FOXP1 MA0481.3
88 88 7 MA1481.1 DRGX MA1481.1
89 89 18 MA1603.1 Dmrt1 MA1603.1
90 90 19 MA0495.3 MAFF MA0495.3
91 91 7 MA0910.2 HOXD8 MA0910.2
92 92 7 MA0027.2 EN1 MA0027.2
93 93 20 MA0905.1 HOXC10 MA0905.1
94 94 7 MA0718.1 RAX MA0718.1
95 95 21 MA0025.2 NFIL3 MA0025.2
96 96 7 MA0716.1 PRRX1 MA0716.1
97 97 7 MA0720.1 Shox2 MA0720.1
98 98 7 MA0158.2 HOXA5 MA0158.2
99 99 6 MA1683.1 FOXA3 MA1683.1
100 100 22 MA1487.2 FOXE1 MA1487.2
101 101 7 MA0704.1 Lhx4 MA0704.1
102 102 6 MA1606.1 Foxf1 MA1606.1
103 103 7 MA0644.2 ESX1 MA0644.2
104 104 6 MA1103.2 FOXK2 MA1103.2
105 105 23 MA1480.1 DPRX MA1480.1
106 106 7 MA0899.1 HOXA10 MA0899.1
107 107 16 MA1152.1 SOX15 MA1152.1
108 108 7 MA0882.1 DLX6 MA0882.1
109 109 7 MA0601.1 Arid3b MA0601.1
110 110 7 MA0658.1 LHX6 MA0658.1
111 111 24 MA1124.1 ZNF24 MA1124.1
112 112 14 MA0650.3 Hoxa13 MA0650.3
113 113 21 MA1636.1 CEBPG MA1636.1
114 114 7 MA0700.2 LHX2 MA0700.2
115 115 6 MA0031.1 FOXD1 MA0031.1
116 116 7 MA0654.1 ISX MA0654.1
117 117 7 MA0715.1 PROP1 MA0715.1
118 118 7 MA0674.1 NKX6-1 MA0674.1
119 119 7 MA0880.1 Dlx3 MA0880.1
120 120 25 MA1104.2 GATA6 MA1104.2
121 121 7 MA0661.1 MEOX1 MA0661.1
122 122 7 MA0881.1 Dlx4 MA0881.1
123 123 6 MA0033.2 FOXL1 MA0033.2
124 124 7 MA0612.2 EMX1 MA0612.2
125 125 25 MA0035.4 GATA1 MA0035.4
126 126 6 MA0042.2 FOXI1 MA0042.2
127 127 7 MA0707.2 MNX1 MA0707.2
128 128 26 MA0108.2 TBP MA0108.2
129 129 7 MA0903.1 HOXB3 MA0903.1
130 130 6 MA0848.1 FOXO4 MA0848.1
131 131 11 MA0497.1 MEF2C MA0497.1
132 132 7 MA0721.1 UNCX MA0721.1
133 133 27 6-TTAAAA STREME-6 6-TTAAAA
134 134 9 MA0611.2 Dux MA0611.2
135 135 7 MA0725.1 VSX1 MA0725.1
136 136 6 MA0157.3 Foxo3 MA0157.3
137 137 6 MA0851.1 Foxj3 MA0851.1
138 138 7 MA1495.1 HOXA1 MA1495.1
139 139 21 MA0102.4 CEBPA MA0102.4
140 140 7 MA0888.1 EVX2 MA0888.1
141 141 7 MA0879.2 DLX1 MA0879.2
142 142 16 MA0868.2 SOX8 MA0868.2
143 143 7 MA0726.1 VSX2 MA0726.1
144 144 7 MA1505.1 HOXC8 MA1505.1
145 145 7 MA0068.2 PAX4 MA0068.2
146 146 25 MA0036.3 GATA2 MA0036.3
147 147 7 MA0630.1 SHOX MA0630.1
148 148 14 MA1125.1 ZNF384 MA1125.1
149 149 7 MA0890.1 GBX2 MA0890.1
150 150 1 MA0591.1 Bach1::Mafk MA0591.1
151 151 22 MA1489.1 FOXN3 MA1489.1
152 152 7 MA0634.1 ALX3 MA0634.1
153 153 7 MA0125.1 Nobox MA0125.1
154 154 22 MA0847.3 FOXD2 MA0847.3
155 155 7 MA0876.1 BSX MA0876.1
156 156 7 MA1476.2 Dlx5 MA1476.2
157 157 7 MA0635.1 BARHL2 MA0635.1
158 158 7 MA0904.2 HOXB5 MA0904.2
159 159 28 MA1657.1 ZNF652 MA1657.1
160 160 7 MA0662.1 MIXL1 MA0662.1
161 161 20 MA0908.1 HOXD11 MA0908.1
162 162 7 MA0887.1 EVX1 MA0887.1
163 163 9 MA0755.1 CUX2 MA0755.1
164 164 7 MA0709.1 Msx3 MA0709.1
165 165 9 MA0122.3 Nkx3-2 MA0122.3
166 166 20 MA0911.1 Hoxa11 MA0911.1
167 167 7 MA1518.2 Lhx1 MA1518.2
168 168 7 MA0892.1 GSX1 MA0892.1
169 169 21 MA0043.3 HLF MA0043.3
170 170 16 MA0077.1 SOX9 MA0077.1
171 171 7 MA0648.1 GSC MA0648.1
172 172 7 MA0885.2 Dlx2 MA0885.2
173 173 7 MA0717.1 RAX2 MA0717.1
174 174 25 MA1970.1 TRPS1 MA1970.1
175 175 7 MA0909.3 Hoxd13 MA0909.3
176 176 6 MA0480.2 Foxo1 MA0480.2
177 177 7 MA0889.1 GBX1 MA0889.1
178 178 7 MA0724.1 VENTX MA0724.1
179 179 7 MA0701.2 LHX9 MA0701.2
180 180 7 MA0666.2 MSX1 MA0666.2
181 181 16 MA0143.4 SOX2 MA0143.4
182 182 29 5-AACMAA STREME-5 5-AACMAA
183 183 6 MA0613.1 FOXG1 MA0613.1
184 184 7 MA1530.1 NKX6-3 MA1530.1
185 185 7 MA1463.1 ARGFX MA1463.1
186 186 7 MA0132.2 PDX1 MA0132.2
187 187 30 MA1479.1 DMRTC2 MA1479.1
188 188 7 MA0875.1 BARX1 MA0875.1
189 189 6 MA0852.2 FOXK1 MA0852.2
190 190 7 MA1519.1 LHX5 MA1519.1
191 191 7 MA0708.2 MSX2 MA0708.2
192 192 9 MA0898.1 Hmx3 MA0898.1
193 193 6 MA0614.1 Foxj2 MA0614.1
194 194 7 MA0712.2 OTX2 MA0712.2
195 195 25 MA0037.4 Gata3 MA0037.4
196 196 7 MA0891.1 GSC2 MA0891.1
197 197 7 MA0912.2 HOXD3 MA0912.2
198 198 7 MA0894.1 HESX1 MA0894.1
199 199 14 MA0913.2 HOXD9 MA0913.2
200 200 7 MA0699.1 LBX2 MA0699.1
201 201 31 MA0050.3 Irf1 MA0050.3
202 202 7 MA1498.2 HOXA7 MA1498.2
203 203 7 MA1500.1 HOXB6 MA1500.1
204 204 7 MA1497.1 HOXA6 MA1497.1
205 205 7 MA1499.1 HOXB4 MA1499.1
206 206 7 MA0895.1 HMBOX1 MA0895.1
207 207 21 MA0836.2 CEBPD MA0836.2
208 208 7 MA1608.1 Isl1 MA1608.1
209 209 73 MA0682.2 PITX1 MA0682.2
210 210 6 MA0849.1 FOXO6 MA0849.1
211 211 32 MA1974.1 ZNF211 MA1974.1
212 212 9 MA0754.2 CUX1 MA0754.2
213 213 21 MA0833.2 ATF4 MA0833.2
214 214 31 MA1623.1 Stat2 MA1623.1
215 215 7 MA0705.1 Lhx8 MA0705.1
216 216 33 MA0514.2 Sox3 MA0514.2
217 217 7 MA1496.1 HOXA4 MA1496.1
218 218 34 MA1562.1 SOX14 MA1562.1
219 219 7 MA0642.2 EN2 MA0642.2
220 220 35 MA0151.1 Arid3a MA0151.1
221 221 36 MA0769.2 TCF7 MA0769.2
222 222 7 MA0853.1 Alx4 MA0853.1
223 223 33 MA0867.2 SOX4 MA0867.2
224 224 7 MA0900.2 HOXA2 MA0900.2
225 225 16 MA1120.1 SOX13 MA1120.1
226 226 7 MA1504.1 HOXC4 MA1504.1
227 227 7 MA0854.1 Alx1 MA0854.1
228 228 37 MA0914.1 ISL2 MA0914.1
229 229 8 MA1962.1 POU2F1::SOX2 MA1962.1
230 230 7 MA0075.3 PRRX2 MA0075.3
231 231 33 MA1563.2 SOX18 MA1563.2
232 232 6 MA0850.1 FOXP3 MA0850.1
233 233 7 MA0618.1 LBX1 MA0618.1
234 234 18 MA0442.2 SOX10 MA0442.2
235 235 38 MA0606.2 Nfat5 MA0606.2
236 236 16 MA0078.2 Sox17 MA0078.2
237 237 25 MA0482.2 GATA4 MA0482.2
238 238 9 MA0897.1 Hmx2 MA0897.1
239 239 22 MA0040.1 Foxq1 MA0040.1
240 240 7 MA0893.2 GSX2 MA0893.2
241 241 14 MA0878.3 CDX1 MA0878.3
242 242 36 MA0523.1 TCF7L2 MA0523.1
243 243 39 MA1729.1 ZNF680 MA1729.1
244 244 4 MA0683.1 POU4F2 MA0683.1
245 245 40 MA1960.1 MGA::EVX1 MA1960.1
246 246 9 MA0896.1 Hmx1 MA0896.1
247 247 41 MA0877.3 BARHL1 MA0877.3
248 248 20 MA0906.1 HOXC12 MA0906.1
249 249 19 MA0117.2 Mafb MA0117.2
250 250 20 MA0907.1 HOXC13 MA0907.1
251 251 23 MA0711.1 OTX1 MA0711.1
252 252 42 MA1709.1 ZIM3 MA1709.1
253 253 43 MA0629.1 Rhox11 MA0629.1
254 254 7 MA1577.1 TLX2 MA1577.1
255 255 9 MA0757.1 ONECUT3 MA0757.1
256 256 7 MA0874.1 Arx MA0874.1
257 257 23 MA0714.1 PITX3 MA0714.1
258 258 7 MA0467.2 Crx MA0467.2
259 259 12 MA1126.1 FOS::JUN MA1126.1
260 260 9 MA0780.1 PAX3 MA0780.1
261 261 7 MA1507.1 HOXD4 MA1507.1
262 262 44 MA1119.1 SIX2 MA1119.1
263 263 9 MA0070.1 PBX1 MA0070.1
264 264 12 MA0018.4 CREB1 MA0018.4
265 265 45 MA0884.2 DUXA MA0884.2
266 266 44 MA1118.1 SIX1 MA1118.1
267 267 46 MA0144.2 STAT3 MA0144.2
268 268 36 MA0768.2 Lef1 MA0768.2
269 269 30 MA1707.1 DMRTA1 MA1707.1
270 270 33 MA0095.3 Yy1 MA0095.3
271 271 23 MA0883.1 Dmbx1 MA0883.1
272 272 47 MA0809.2 TEAD4 MA0809.2
273 273 12 MA1150.1 RORB MA1150.1
274 274 12 MA1632.1 ATF2 MA1632.1
275 275 12 MA1131.1 FOSL2::JUN MA1131.1
276 276 48 MA1588.1 ZNF136 MA1588.1
277 277 19 MA0842.2 NRL MA0842.2
278 278 20 MA0873.1 HOXD12 MA0873.1
279 279 6 MA0593.1 FOXP2 MA0593.1
280 280 12 MA0843.1 TEF MA0843.1
281 281 49 MA0678.1 OLIG2 MA0678.1
282 282 50 MA0063.2 NKX2-5 MA0063.2
283 283 34 MA1561.1 SOX12 MA1561.1
284 284 7 MA0135.1 Lhx3 MA0135.1
285 285 12 MA1127.1 FOSB::JUN MA1127.1
286 286 12 MA1139.1 FOSL2::JUNB MA1139.1
287 287 25 MA0766.2 GATA5 MA0766.2
288 288 30 MA1478.1 DMRTA2 MA1478.1
289 289 22 MA0030.1 FOXF2 MA0030.1
290 290 20 MA0594.2 HOXA9 MA0594.2
291 291 51 MA0693.3 Vdr MA0693.3
292 292 52 MA0520.1 Stat6 MA0520.1
293 293 21 MA0838.1 CEBPG MA0838.1
294 294 53 MA0782.2 PKNOX1 MA0782.2
295 295 47 MA0090.3 TEAD1 MA0090.3
296 296 38 MA0152.2 Nfatc2 MA0152.2
297 297 54 MA0689.1 TBX20 MA0689.1
298 298 25 MA0140.2 GATA1::TAL1 MA0140.2
299 299 12 MA0639.1 DBP MA0639.1
300 300 55 MA0620.3 MITF MA0620.3
301 301 56 MA0687.1 SPIC MA0687.1
302 302 16 MA0087.2 Sox5 MA0087.2
303 303 50 MA1523.1 MSANTD3 MA1523.1
304 304 7 MA0621.1 mix-a MA0621.1
305 305 12 MA1145.1 FOSL2::JUND MA1145.1
306 306 56 MA1952.1 FOXJ2::ELF1 MA1952.1
307 307 57 MA0069.1 PAX6 MA0069.1
308 308 7 MA1501.1 HOXB7 MA1501.1
309 309 50 MA1994.1 Nkx2-1 MA1994.1
310 310 36 MA1991.1 Hnf1A MA1991.1
311 311 73 MA0483.1 Gfi1B MA0483.1
312 312 16 MA0515.1 Sox6 MA0515.1
313 313 49 MA0827.1 OLIG3 MA0827.1
314 314 45 MA0468.1 DUX4 MA0468.1
315 315 12 MA1129.1 FOSL1::JUN MA1129.1
316 316 12 MA1133.1 JUN::JUNB MA1133.1
317 317 55 MA0626.1 Npas2 MA0626.1
318 318 19 MA1520.1 MAF MA1520.1
319 319 38 MA0624.2 Nfatc1 MA0624.2
320 320 53 MA0498.2 MEIS1 MA0498.2
321 321 42 MA1953.1 FOXO1::ELF1 MA1953.1
322 322 33 MA0869.2 Sox11 MA0869.2
323 323 12 MA1136.1 FOSB::JUNB MA1136.1
324 324 58 MA1112.2 NR4A1 MA1112.2
325 325 20 MA1506.1 HOXD10 MA1506.1
326 326 59 MA0631.1 Six3 MA0631.1
327 327 60 MA1975.1 ZNF214 MA1975.1
328 328 7 MA0702.2 LMX1A MA0702.2
329 329 55 MA0093.3 USF1 MA0093.3
330 330 31 MA0517.1 STAT1::STAT2 MA0517.1
331 331 61 MA1125.1 (ZNF384) MEME-2 AAAAAAAAAAAAAAA
332 332 49 MA0461.2 Atoh1 MA0461.2
333 333 62 MA0479.1 FOXH1 MA0479.1
334 334 21 MA0837.2 CEBPE MA0837.2
335 335 31 MA0772.1 IRF7 MA0772.1
336 336 63 MA0596.1 SREBF2 MA0596.1
337 337 64 MA1720.1 ZNF85 MA1720.1
338 338 65 MA1105.2 GRHL2 MA1105.2
339 339 37 MA0124.2 Nkx3-1 MA0124.2
340 340 73 MA0038.2 GFI1 MA0038.2
341 341 66 MA1592.1 ZNF274 MA1592.1
342 342 67 MA0619.1 LIN54 MA0619.1
343 343 49 MA1108.2 MXI1 MA1108.2
344 344 68 MA1151.1 RORC MA1151.1
345 345 46 MA1625.1 Stat5b MA1625.1
346 346 69 MA0508.3 PRDM1 MA0508.3
347 347 65 MA0743.2 SCRT1 MA0743.2
348 348 7 MA1644.1 NFYC MA1644.1
349 349 51 MA1534.1 NR1I3 MA1534.1
350 350 49 MA1618.1 Ptf1a MA1618.1
351 351 19 MA1521.1 MAFA MA1521.1
352 352 20 MA1503.1 HOXB9 MA1503.1
353 353 23 MA0719.1 RHOXF1 MA0719.1
354 354 55 MA0829.2 SREBF1 MA0829.2
355 355 47 MA1121.1 TEAD2 MA1121.1
356 356 7 MA0784.2 POU1F1 MA0784.2
357 357 70 MA0861.1 TP73 MA0861.1
358 358 71 MA0684.2 RUNX3 MA0684.2
359 359 46 MA1624.1 Stat5a MA1624.1
360 360 68 MA0494.1 Nr1h3::Rxra MA0494.1
361 361 70 MA0106.3 TP53 MA0106.3
362 362 12 MA0492.1 JUND MA0492.1
363 363 51 MA1111.1 NR2F2 MA1111.1
364 364 49 MA1524.2 Msgn1 MA1524.2
365 365 72 MA1593.1 ZNF317 MA1593.1
366 366 44 MA2001.1 Six4 MA2001.1
367 367 73 MA0682.2 (PITX1) MEME-3 AAGTGCTGGGATTAC
368 368 14 MA1473.1 CDX4 MA1473.1
369 369 70 MA0525.2 TP63 MA0525.2
370 370 73 MA1547.2 PITX2 MA1547.2
371 371 71 MA1989.1 Bcl11B MA1989.1
372 372 21 MA0466.3 CEBPB MA0466.3
373 373 11 MA0773.1 MEF2D MA0773.1
374 374 49 MA1570.1 TFAP4 MA1570.1
375 375 49 MA0826.1 OLIG1 MA0826.1
376 376 12 MA0488.1 JUN MA0488.1
377 377 20 MA0485.2 HOXC9 MA0485.2
378 378 54 MA0800.1 EOMES MA0800.1
379 379 73 MA1547.2 (PITX2) STREME-11 11-CTGGGAYTAC
380 380 47 MA0808.1 TEAD3 MA0808.1
381 381 74 MA1580.1 ZBTB32 MA1580.1
382 382 65 MA0647.1 GRHL1 MA0647.1
383 383 63 MA0595.1 SREBF1 MA0595.1
384 384 20 MA0651.2 HOXC11 MA0651.2
385 385 75 9-CACTTTGGGAG STREME-9 9-CACTTTGGGAG
386 386 46 MA0137.3 STAT1 MA0137.3
387 387 46 MA0518.1 Stat4 MA0518.1
388 388 76 TAGAGAYRGGGTTTC MEME-5 TAGAGAYRGGGTTTC
389 389 42 MA1956.1 FOXO1::FLI1 MA1956.1
390 390 46 MA0519.1 Stat5a::Stat5b MA0519.1
391 391 12 MA0840.1 Creb5 MA0840.1
392 392 36 MA1421.1 TCF7L1 MA1421.1
393 393 49 MA0817.1 BHLHE23 MA0817.1
394 394 25 MA0029.1 Mecom MA0029.1
395 395 30 MA0610.1 DMRT3 MA0610.1
396 396 77 MA1585.1 ZKSCAN1 MA1585.1
397 397 46 MA0463.2 BCL6 MA0463.2
398 398 58 MA1148.1 PPARA::RXRA MA1148.1
399 399 78 MA0164.1 Nr2e3 MA0164.1
400 400 68 MA0072.1 RORA MA0072.1
401 401 79 MA1973.1 (ZKSCAN3) MEME-6 TGTYRCCCAGGCTGG
402 402 7 MA0060.3 NFYA MA0060.3
403 403 50 MA1645.1 NKX2-2 MA1645.1
404 404 80 MA0019.1 Ddit3::Cebpa MA0019.1
405 405 54 MA0804.1 TBX19 MA0804.1
406 406 7 MA1546.1 PAX3 MA1546.1
407 407 7 MA0703.2 LMX1B MA0703.2
408 408 42 MA1955.1 FOXO1::ELK3 MA1955.1
409 409 47 MA0114.4 HNF4A MA0114.4
410 410 11 MA0660.1 MEF2B MA0660.1
411 411 51 MA1996.1 Nr1H2 MA1996.1
412 412 81 MA1596.1 (ZNF460) STREME-14 14-ACCTCAGCCTCC
413 413 54 MA0802.1 TBR1 MA0802.1
414 414 82 MA0698.1 ZBTB18 MA0698.1
415 415 83 MA1714.1 ZNF675 MA1714.1
416 416 42 MA1950.1 FLI1::FOXI1 MA1950.1
417 417 79 MA1973.1 (ZKSCAN3) STREME-8 8-CCAGCCTGGGYRAC
418 418 50 MA1114.1 PBX3 MA1114.1
419 419 65 MA0744.2 SCRT2 MA0744.2
420 420 84 MA0101.1 REL MA0101.1
421 421 85 MA1107.2 (KLF9) STREME-13 13-ACCACACMCA
422 422 86 MA1949.1 FLI1::DRGX MA1949.1
423 423 87 12-CTCCTGGGCTCAA STREME-12 12-CTCCTGGGCTCAA
424 424 58 MA1541.1 NR6A1 MA1541.1
425 425 47 MA0484.2 HNF4G MA0484.2
426 426 49 MA1467.2 Atoh1 MA1467.2
427 427 46 MA1116.1 RBPJ MA1116.1
428 428 55 MA1995.1 Npas4 MA1995.1
429 429 58 MA0160.2 NR4A2 MA0160.2
430 430 88 MA1980.1 ZNF418 MA1980.1
431 431 89 MA1594.1 ZNF382 MA1594.1
432 432 81 MA1596.1 ZNF460 MA1596.1
433 433 49 MA0607.2 BHLHA15 MA0607.2
434 434 71 MA0002.2 Runx1 MA0002.2
435 435 49 MA1123.2 TWIST1 MA1123.2
436 436 90 MA0770.1 HSF2 MA0770.1
437 437 12 MA0656.1 JDP2 MA0656.1
438 438 58 MA0071.1 RORA MA0071.1
439 439 42 MA1508.1 IKZF1 MA1508.1
440 440 85 MA0732.1 (EGR3) STREME-7 7-CACCCACACAC
441 441 65 MA1968.1 TFCP2 MA1968.1
442 442 91 MA1572.1 TGIF2LY MA1572.1
443 443 38 MA1525.2 NFATC4 MA1525.2
444 444 20 MA1958.1 HOXD12::ELK1 MA1958.1
445 445 12 MA1951.1 FOS MA1951.1
446 446 58 MA0141.3 ESRRB MA0141.3
447 447 55 MA0831.3 TFE3 MA0831.3
448 448 8 MA0507.2 POU2F2 MA0507.2
449 449 90 MA0486.2 HSF1 MA0486.2
450 450 55 MA0828.2 SREBF2 MA0828.2
451 451 54 MA0688.1 TBX2 MA0688.1
452 452 54 MA0803.1 TBX15 MA0803.1
453 453 49 MA1638.1 HAND2 MA1638.1
454 454 58 MA0643.1 Esrrg MA0643.1
455 455 71 MA0511.2 RUNX2 MA0511.2
456 456 49 MA0818.2 BHLHE22 MA0818.2
457 457 81 MA1596.1 (ZNF460) MEME-4 RGGAGGCBGAGGHKG
458 458 42 MA1954.1 FOXO1::ELK1 MA1954.1
459 459 54 MA0805.1 TBX1 MA0805.1
460 460 54 MA0807.1 TBX5 MA0807.1
461 461 86 MA1937.1 ERF::HOXB13 MA1937.1
462 462 91 MA0797.1 TGIF2 MA0797.1
463 463 12 MA0605.2 ATF3 MA0605.2
464 464 81 MA1587.1 ZNF135 MA1587.1
465 465 54 MA0801.1 MGA MA0801.1
466 466 82 MA0691.1 TFAP4 MA0691.1
467 467 58 MA0592.3 ESRRA MA0592.3
468 468 55 MA0692.1 TFEB MA0692.1
469 469 92 MA1983.1 ZNF582 MA1983.1
470 470 33 MA0870.1 Sox1 MA0870.1
471 471 53 MA0775.1 MEIS3 MA0775.1
472 472 91 MA0667.1 MYF6 MA0667.1
473 473 42 MA1579.1 ZBTB26 MA1579.1
474 474 49 MA0623.2 NEUROG1 MA0623.2
475 475 31 MA0653.1 IRF9 MA0653.1
476 476 54 MA0806.1 TBX4 MA0806.1
477 477 93 MA0130.1 ZNF354C MA0130.1
CONSENSUS.x motif_name EVALUE.y QVALUE TP
1 DATGASTCATH FOS::JUNB 0.000000e+00 0.000000e+00 11729
2 DRTGACTCAYH Atf3 0.000000e+00 0.000000e+00 11509
3 KATGACTCAT FOSL2::JUND 0.000000e+00 0.000000e+00 11579
4 KRTGASTCAT FOSB::JUNB 0.000000e+00 0.000000e+00 12079
5 KATGASTCATHN FOS::JUNB 0.000000e+00 0.000000e+00 11894
6 NNATGACTCATNN FOSL1 0.000000e+00 0.000000e+00 11077
7 KRTGASTCAT FOSL2::JUNB 0.000000e+00 0.000000e+00 12375
8 DVTGASTCATB FOS 0.000000e+00 0.000000e+00 12368
9 NVTGACTCATNN Jun 0.000000e+00 0.000000e+00 10270
10 NNATGACTCATNN JUNB 0.000000e+00 0.000000e+00 11535
11 NNRTGAGTCAYN FOSL2::JUN 0.000000e+00 0.000000e+00 11914
12 NKATGAGTCATNN FOS::JUND 0.000000e+00 0.000000e+00 12070
13 DATGACTCATH BATF::JUN 0.000000e+00 0.000000e+00 10924
14 DATGACTCATH BATF 0.000000e+00 0.000000e+00 10696
15 NRTGACTCABN Atf3 0.000000e+00 0.000000e+00 11989
16 ATGAGTCAYM FOS::JUN 0.000000e+00 0.000000e+00 12324
17 NNATGACTCATNN JUND 0.000000e+00 0.000000e+00 11713
18 DATGACTCATH BATF3 0.000000e+00 0.000000e+00 11419
19 NKATGACTCATNN FOSL1::JUN 0.000000e+00 0.000000e+00 12116
20 ATGACTCAT BACH1 0.000000e+00 0.000000e+00 12677
21 NDRTGAGTCABN BNC2 0.000000e+00 0.000000e+00 11306
22 NDRTGACTCATNN FOSL1::JUNB 0.000000e+00 0.000000e+00 11464
23 KRRTGASTCAB FOSL2 0.000000e+00 0.000000e+00 11626
24 10-TGANTCA FOS 0.000000e+00 0.000000e+00 10636
25 ATGACTCAT JDP2 0.000000e+00 0.000000e+00 10045
26 NRTGACTCAT FOSL1::JUND 0.000000e+00 0.000000e+00 9837
27 KATGACKCAT JUN::JUNB 0.000000e+00 0.000000e+00 10803
28 DWANCATGASTCATSNTWH BACH2 0.000000e+00 0.000000e+00 9412
29 VATGACTCATS NFE2 0.000000e+00 0.000000e+00 8975
30 NVNATGACTCAGCADW MAFG::NFE2L1 0.000000e+00 0.000000e+00 12027
31 NGMTGACTCAGCMNH MAFK 0.000000e+00 0.000000e+00 10201
32 ATGACTCAGCANWTY MAF::NFE2 0.000000e+00 0.000000e+00 11642
33 CASNATGACTCAGCA Nfe2l2 0.000000e+00 0.000000e+00 10857
34 ATKMAT ATKMAT 0.000000e+00 0.000000e+00 17941
35 NWGMTGACTCAGCAN Mafg 0.000000e+00 0.000000e+00 9692
36 AAAAGTCAA Nr2e1 0.000000e+00 0.000000e+00 20274
37 NHCATAAATCAHN PBX2 0.000000e+00 0.000000e+00 17570
38 NHCATMAATCAHN MEIS1 0.000000e+00 0.000000e+00 16297
39 NNATGATTTATGDNH MEIS2 0.000000e+00 0.000000e+00 19957
40 ATGMATAATTAATG POU4F1 0.000000e+00 0.000000e+00 17723
41 HWAADWATAAATGRAYWAWTT ZNF354A 0.000000e+00 0.000000e+00 19759
42 WATGTAAATAT FOXB1 0.000000e+00 0.000000e+00 20261
43 VBTAATTARB EMX2 0.000000e+00 0.000000e+00 18738
44 WWTATGCWAATTW POU3F3 0.000000e+00 0.000000e+00 19999
45 WTATGCWAATKW POU3F1 0.000000e+00 0.000000e+00 19966
46 DWTAATCAATTAWNH Pax7 0.000000e+00 0.000000e+00 18342
47 WWTAATCAAATTAWWW PHOX2B 0.000000e+00 0.000000e+00 17189
48 SYAATTAS HOXB2 0.000000e+00 0.000000e+00 17648
49 WAWGTAAAYAW FOXC1 0.000000e+00 0.000000e+00 21146
50 WTVWBAW WTVWBAW 0.000000e+00 0.000000e+00 21308
51 AWTATGCWAATK POU2F1 0.000000e+00 0.000000e+00 21014
52 WAWGHAAATAAACARY FOXD3 0.000000e+00 0.000000e+00 21264
53 RTGMATWATTAATGAR POU4F3 0.000000e+00 0.000000e+00 14937
54 WAWGTAAACAWW FOXC2 0.000000e+00 0.000000e+00 20601
55 TATGCWAAT POU5F1B 0.000000e+00 0.000000e+00 21474
56 NYTAATTAVN NOTO 0.000000e+00 0.000000e+00 21007
57 AYTAATTART POU6F1 0.000000e+00 0.000000e+00 19220
58 DDCTAAAAATAGMHH MEF2A 0.000000e+00 0.000000e+00 20833
59 RTGACGTMAY FOSL1::JUND 0.000000e+00 0.000000e+00 16250
60 WTATGCWAATKA POU3F2 0.000000e+00 0.000000e+00 18845
61 15-HHATDD HHATDD 0.000000e+00 0.000000e+00 21956
62 WWDCTAATAAHWW SATB1 0.000000e+00 0.000000e+00 18821
63 DRTTAATNATTAACN HNF1A 0.000000e+00 0.000000e+00 13337
64 TAATYYAATTA PHOX2A 0.000000e+00 0.000000e+00 19263
65 WDCCAATAAAAWWW HOXB13 0.000000e+00 0.000000e+00 19479
66 ASCTMATTAW POU6F2 0.000000e+00 0.000000e+00 19745
67 WAAAAATCAATAAWWN ONECUT1 0.000000e+00 0.000000e+00 17033
68 NWTATGCAAATNH POU2F3 0.000000e+00 0.000000e+00 19816
69 GTTAATNATTAAY HNF1B 0.000000e+00 0.000000e+00 12899
70 WHATGCAAATN POU5F1 0.000000e+00 0.000000e+00 21753
71 CWTTGTYATGCAAAT Pou5f1::Sox2 0.000000e+00 0.000000e+00 18270
72 TRTAYA TRTAYA 0.000000e+00 0.000000e+00 21613
73 NWWAAYMATTAN BARX2 0.000000e+00 0.000000e+00 18474
74 WWGTAAACATNN FOXA1 0.000000e+00 0.000000e+00 20978
75 BYAATTAV VAX2 0.000000e+00 0.000000e+00 20411
76 YTAATTAH VAX1 0.000000e+00 0.000000e+00 17918
77 TATGCWAAT POU3F4 0.000000e+00 0.000000e+00 18676
78 RTAATGAGVN POU6F1 0.000000e+00 0.000000e+00 18089
79 DDWATGTAAACANN Foxl2 0.000000e+00 0.000000e+00 20808
80 NWWAACAAT SRY 0.000000e+00 0.000000e+00 21532
81 SYAATATTGVDANH Arid5a 0.000000e+00 0.000000e+00 19575
82 DSTAATTAWN MEOX2 0.000000e+00 0.000000e+00 18080
83 NYMATTAA NKX6-2 0.000000e+00 0.000000e+00 20603
84 NDGCAATAAAWN CDX2 0.000000e+00 0.000000e+00 20558
85 GYMATTAM HOXB8 0.000000e+00 0.000000e+00 16248
86 RWGTAAACANN FOXA2 0.000000e+00 0.000000e+00 21770
87 NDGTAAACANH FOXP1 0.000000e+00 0.000000e+00 21368
88 YYAATTAN DRGX 0.000000e+00 0.000000e+00 18888
89 GHTACAAAGTADC Dmrt1 0.000000e+00 0.000000e+00 20045
90 DWGTCAGCATTTTWHN MAFF 0.000000e+00 0.000000e+00 21298
91 GYMATTAM HOXD8 2.010847e-321 2.010847e-321 15882
92 SYAATTAV EN1 5.410019e-319 2.460447e-321 20748
93 GTCRTAAAAH HOXC10 2.050002e-318 9.199502e-321 19325
94 RYYAATTARH RAX 3.580000e-316 1.590002e-318 19294
95 NRTTATGYAATNN NFIL3 1.000000e-315 4.390001e-318 17076
96 YYAATTAR PRRX1 1.690000e-314 7.330000e-317 20702
97 YYAATTAR Shox2 1.940000e-313 8.350000e-316 21115
98 GYMATTAS HOXA5 1.410000e-311 6.010000e-314 13954
99 DWGTAAACANN FOXA3 1.380000e-309 5.810000e-312 21317
100 VYTAAAYAAACAAH FOXE1 3.880000e-308 1.620000e-310 17028
101 YTAATTAR Lhx4 2.540000e-304 1.050000e-306 19366
102 DWRTAAACANN Foxf1 2.590000e-300 1.060000e-302 18754
103 YYAATTAN ESX1 8.200000e-297 3.320000e-299 20186
104 DDGTAAACANN FOXK2 1.660000e-296 6.670000e-299 20724
105 AGATAATCCN DPRX 1.780000e-296 7.060000e-299 21091
106 DGYMATAAAAH HOXA10 1.980000e-296 7.780000e-299 18369
107 CYWTTGTTHW SOX15 2.790000e-296 1.090000e-298 21623
108 VYAATTAN DLX6 1.820000e-293 7.030000e-296 21247
109 ATATTAATWAN Arid3b 5.020000e-289 1.920000e-291 16029
110 RCTAATTARY LHX6 8.620000e-289 3.270000e-291 18460
111 CATTCATTCATTC ZNF24 1.000000e-286 3.770000e-289 14594
112 NNCCAATAAAAN Hoxa13 6.550000e-286 2.440000e-288 19065
113 NDATGATGCAATMHH CEBPG 9.450000e-286 3.490000e-288 17468
114 DDCCAATTADN LHX2 1.930000e-285 7.070000e-288 18117
115 GTAAACAW FOXD1 2.760000e-283 1.000000e-285 17329
116 YYAATTAR ISX 1.070000e-282 3.840000e-285 16688
117 TAATYWAATTA PROP1 2.840000e-282 1.010000e-284 13316
118 NTMATTAA NKX6-1 2.180000e-280 7.700000e-283 18456
119 NYAATTAN Dlx3 1.490000e-277 5.220000e-280 19584
120 HWWTCTTATCTNH GATA6 1.540000e-275 5.340000e-278 21231
121 VSTAATTAHC MEOX1 2.320000e-275 8.000000e-278 20073
122 VYAATTAN Dlx4 2.370000e-275 8.110000e-278 20899
123 RTAAACA FOXL1 4.310000e-275 1.460000e-277 20843
124 BTAATTAS EMX1 8.210000e-275 2.760000e-277 13183
125 NHCTAATCTDH GATA1 1.450000e-273 4.840000e-276 21627
126 GTAAACA FOXI1 5.530000e-273 1.830000e-275 19918
127 VYRATTAK MNX1 4.310000e-272 1.410000e-274 15668
128 STATAAAWRSVVVBN TBP 1.570000e-271 5.100000e-274 19259
129 NNYMATTARN HOXB3 6.840000e-269 2.210000e-271 15441
130 GTAAACA FOXO4 1.230000e-268 3.940000e-271 20185
131 DDDCYAAAAATAGMW MEF2C 1.230000e-267 3.900000e-270 17430
132 YYAATTAV UNCX 1.940000e-267 6.120000e-270 20615
133 TTAAAA TTAAAA 1.280000e-265 4.010000e-268 21969
134 NTGATTBAATCAVWNN Dux 1.960000e-265 6.110000e-268 9993
135 YTAATTAN VSX1 2.130000e-263 6.570000e-266 21910
136 NDGTAAACARNN Foxo3 5.670000e-263 1.740000e-265 22011
137 NDAADGTAAACAAANNM Foxj3 1.170000e-262 3.570000e-265 15334
138 STAATTAS HOXA1 2.740000e-262 8.280000e-265 14717
139 NDATTGCACAATHN CEBPA 7.890000e-260 2.370000e-262 20597
140 VVTAATTAVB EVX2 1.310000e-259 3.890000e-262 19377
141 VYAATTAN DLX1 7.270000e-259 2.150000e-261 19426
142 MGAACAATRG SOX8 1.430000e-255 4.210000e-258 22192
143 CTAATTAN VSX2 1.660000e-254 4.840000e-257 21982
144 KTAATTAM HOXC8 1.860000e-254 5.380000e-257 14506
145 CTAATTAG PAX4 4.560000e-254 1.310000e-256 21256
146 NBCTTATCTNH GATA2 1.930000e-253 5.510000e-256 20371
147 HTAATTRR SHOX 1.130000e-251 3.200000e-254 18641
148 DNWMAAAAAAAA ZNF384 1.410000e-250 3.960000e-253 14509
149 RSYAATTARN GBX2 6.380000e-248 1.790000e-250 18623
150 RSSATGACTCAGCAB Bach1::Mafk 1.880000e-247 5.220000e-250 4733
151 GTAAACAA FOXN3 2.990000e-247 8.260000e-250 18725
152 YYYAATTANV ALX3 8.820000e-247 2.420000e-249 20261
153 TAATTRSY Nobox 4.260000e-246 1.160000e-248 17763
154 SYTAARYAAACA FOXD2 2.430000e-245 6.570000e-248 16196
155 SYVATTAV BSX 5.370000e-245 1.450000e-247 21090
156 NDGCAATTAGNN Dlx5 6.970000e-244 1.860000e-246 20805
157 VHTAAAYGRT BARHL2 7.500000e-244 1.990000e-246 19789
158 STAATTAS HOXB5 1.900000e-243 5.000000e-246 15786
159 BAAAGRGTTAAN ZNF652 4.080000e-243 1.070000e-245 17899
160 NBYAATTAVN MIXL1 1.080000e-242 2.820000e-245 20568
161 RTCGTAAAAH HOXD11 3.500000e-242 9.070000e-245 13106
162 VVTAATTABS EVX1 1.070000e-241 2.740000e-244 17075
163 TRATCGATAW CUX2 3.370000e-241 8.620000e-244 17063
164 SCAATTAN Msx3 8.720000e-241 2.220000e-243 16854
165 WWAAMCACTTAAN Nkx3-2 3.220000e-239 8.150000e-242 19596
166 RGTCGTAAAANT Hoxa11 1.040000e-235 2.620000e-238 19184
167 NNNTGCTAATTAGCANNN Lhx1 2.810000e-235 7.020000e-238 19115
168 NCYMATTAVN GSX1 6.000000e-235 1.490000e-237 15820
169 NNRTTATGCAACHN HLF 2.430000e-234 5.980000e-237 16171
170 CYATTGTTY SOX9 1.400000e-231 3.430000e-234 22283
171 VYTAATCCSY GSC 2.320000e-231 5.660000e-234 21506
172 NDGCAATTAGNN Dlx2 7.650000e-231 1.850000e-233 21556
173 YYAATTAV RAX2 1.370000e-230 3.290000e-233 18237
174 NHTCTTATCTNH TRPS1 5.230000e-229 1.250000e-231 21829
175 NNCAATAAAW Hoxd13 6.140000e-228 1.460000e-230 16836
176 NDGTAAACANN Foxo1 3.350000e-227 7.940000e-230 21652
177 VCYAATTASY GBX1 4.490000e-227 1.060000e-229 17770
178 AMCGATTAR VENTX 3.680000e-226 8.630000e-229 15841
179 CYAATTAR LHX9 6.480000e-225 1.510000e-227 12863
180 SCAATTAR MSX1 5.780000e-224 1.340000e-226 18026
181 DNACAATGGNN SOX2 4.740000e-223 1.090000e-225 22015
182 AACMAA AACMAA 2.300000e-219 5.260000e-222 18458
183 RTAAACAW FOXG1 1.120000e-217 2.550000e-220 12824
184 VYCATTAWW NKX6-3 8.340000e-216 1.890000e-218 14086
185 DCTAATTARM ARGFX 6.500000e-215 1.460000e-217 12776
186 VYAATTAR PDX1 9.710000e-215 2.180000e-217 15840
187 AATTGMTACATT DMRTC2 1.630000e-213 3.630000e-216 19449
188 VCMATTAV BARX1 4.820000e-212 1.070000e-214 17072
189 NRDGTAAACAAGNN FOXK1 8.240000e-211 1.820000e-213 19507
190 STAATTAV LHX5 7.630000e-210 1.670000e-212 12529
191 SCAATTAV MSX2 1.130000e-209 2.470000e-212 18253
192 ASAAGCAATTAAWRRAT Hmx3 1.800000e-209 3.900000e-212 16428
193 RTAAACAA Foxj2 2.380000e-209 5.140000e-212 15021
194 NDRGGATTARNN OTX2 2.470000e-209 5.310000e-212 20471
195 NHTCTTATCTNH Gata3 1.470000e-208 3.140000e-211 21310
196 NYTAATCCBH GSC2 1.120000e-206 2.380000e-209 21902
197 CTAATTAC HOXD3 1.750000e-205 3.710000e-208 10910
198 GHTAATTRRH HESX1 2.090000e-205 4.410000e-208 16798
199 GYMATAAAAH HOXD9 1.020000e-202 2.140000e-205 15139
200 NCYAATTARN LBX2 2.030000e-202 4.220000e-205 20227
201 ANTGAAACTGAAASH Irf1 5.560000e-201 1.150000e-203 21317
202 SYMATTAV HOXA7 1.020000e-198 2.110000e-201 14825
203 BTAATKRC HOXB6 6.930000e-198 1.420000e-200 17723
204 BGYMATTANN HOXA6 2.330000e-196 4.770000e-199 21482
205 RTMATTAR HOXB4 7.310000e-195 1.490000e-197 11886
206 MYTAGTTAMS HMBOX1 1.390000e-193 2.810000e-196 22269
207 NRTTGCACAAYHN CEBPD 1.390000e-190 2.790000e-193 18035
208 NNCCATTAGNN Isl1 5.670000e-188 1.140000e-190 20813
209 YTAATCCH PITX1 4.770000e-187 9.520000e-190 14674
210 GTAAACA FOXO6 3.360000e-186 6.670000e-189 16765
211 HWYATATACCAYRYW ZNF211 6.740000e-186 1.330000e-188 13312
212 TAATCGATAM CUX1 3.090000e-184 6.080000e-187 14532
213 DDATGATGCAATMN ATF4 3.660000e-184 7.170000e-187 14029
214 RGAAACAGAAASH Stat2 1.880000e-183 3.660000e-186 21797
215 CTAATTAV Lhx8 1.050000e-181 2.040000e-184 14978
216 DNACAATGGNN Sox3 1.270000e-181 2.450000e-184 19517
217 RTMATTAV HOXA4 3.790000e-181 7.280000e-184 17940
218 CCGAACAATG SOX14 1.080000e-179 2.060000e-182 18429
219 VSYAATTA EN2 1.170000e-178 2.230000e-181 15562
220 ATYAAA Arid3a 2.290000e-177 4.340000e-180 12739
221 NNCTTTGAWVT TCF7 2.330000e-177 4.400000e-180 21781
222 CGCRYTAATTARYHNNY Alx4 4.890000e-176 9.190000e-179 12724
223 RAACAAAGRV SOX4 8.190000e-176 1.530000e-178 19098
224 STAATTAS HOXA2 2.070000e-175 3.850000e-178 10015
225 DVACAATGGNN SOX13 3.280000e-173 6.080000e-176 19610
226 VTMATTAN HOXC4 1.150000e-171 2.120000e-174 14080
227 CGMRYTAATTARTNMNY Alx1 2.330000e-171 4.280000e-174 11706
228 GCAMTTAR ISL2 9.850000e-171 1.800000e-173 21881
229 MATTTRCATMACAATRG POU2F1::SOX2 1.190000e-169 2.160000e-172 8694
230 CTAATTAN PRRX2 1.670000e-169 3.030000e-172 10355
231 AACAATDV SOX18 1.250000e-167 2.260000e-170 19364
232 RTAAACA FOXP3 3.600000e-167 6.460000e-170 22232
233 TTAATTAG LBX1 1.090000e-166 1.940000e-169 13289
234 NDAACAAAGVN SOX10 1.890000e-166 3.360000e-169 19710
235 NNATGGAAAAWN Nfat5 7.170000e-165 1.270000e-167 21006
236 NNAGAACAATGGNN Sox17 6.450000e-164 1.140000e-166 18966
237 NNCCTTATCTNH GATA4 2.310000e-163 4.060000e-166 19967
238 ASAAGCAATTAAHVVRT Hmx2 7.770000e-163 1.360000e-165 15196
239 HATTGTTTATW Foxq1 1.360000e-161 2.380000e-164 9512
240 SYMATTAR GSX2 3.850000e-158 6.680000e-161 20102
241 GGYMATAAAA CDX1 2.030000e-157 3.520000e-160 11251
242 DRASATCAAAGRVA TCF7L2 7.500000e-156 1.290000e-158 16960
243 GNCCAAGAAGAATDA ZNF680 1.820000e-152 3.120000e-155 17187
244 RTGCATAATTAATGAG POU4F2 2.350000e-152 4.020000e-155 5570
245 RGGTGWTAATKD MGA::EVX1 5.340000e-145 9.090000e-148 18677
246 VSVAGCAATTAAWGVDB Hmx1 9.150000e-145 1.550000e-147 15575
247 AMCGTTTA BARHL1 1.460000e-143 2.470000e-146 19862
248 RGTCGTAAAAH HOXC12 3.430000e-142 5.770000e-145 10150
249 AWWNTGCTGACD Mafb 8.450000e-141 1.410000e-143 19960
250 BCTCGTAAAAH HOXC13 8.530000e-141 1.420000e-143 12504
251 HTAATCCS OTX1 6.360000e-140 1.060000e-142 18484
252 RVMAACAGAAACCYM ZIM3 4.800000e-137 7.930000e-140 16874
253 ANKNCGCTGTWAWDVRW Rhox11 1.460000e-135 2.410000e-138 19879
254 BYAATTAR TLX2 2.850000e-135 4.680000e-138 9648
255 AAAAAATCRATAAH ONECUT3 8.610000e-133 1.410000e-135 7609
256 VTBCRYTAATTARTKSW Arx 1.820000e-132 2.970000e-135 9708
257 NYTAATCCC PITX3 6.550000e-130 1.060000e-132 22227
258 NDGGATTANN Crx 6.640000e-130 1.070000e-132 18269
259 DRTGACGTCATHNDTN FOS::JUN 2.810000e-129 4.520000e-132 18942
260 TAATYRATTA PAX3 6.450000e-129 1.030000e-131 7542
261 VTMATTAD HOXD4 7.960000e-126 1.270000e-128 8564
262 NNCTGAAACCTGATMY SIX2 1.710000e-125 2.720000e-128 20956
263 HCATCAATCAAW PBX1 1.480000e-124 2.340000e-127 11269
264 NNRTGATGTCAYN CREB1 1.050000e-123 1.660000e-126 22070
265 YTRATTWAATYAR DUXA 1.720000e-122 2.710000e-125 5331
266 GWAACCTGAKM SIX1 2.790000e-120 4.370000e-123 19641
267 YTTCYKGGAAD STAT3 1.480000e-118 2.310000e-121 16751
268 NNBCCTTTGATSTN Lef1 2.940000e-116 4.570000e-119 22060
269 AWTTGWTACAWT DMRTA1 5.540000e-115 8.580000e-118 18279
270 NNCAAAATGGCN Yy1 9.470000e-114 1.460000e-116 18324
271 WDRWNMGGATTADKNNW Dmbx1 3.760000e-112 5.780000e-115 13410
272 NNACATTCCARN TEAD4 1.040000e-109 1.590000e-112 21039
273 AWWTRGGTCAH RORB 2.060000e-109 3.150000e-112 18547
274 NNATGABGTCATN ATF2 1.490000e-105 2.260000e-108 16351
275 GRTGACGTMAT FOSL2::JUN 8.800000e-105 1.330000e-107 17832
276 RWATTCTTGGTTGRC ZNF136 7.130000e-103 1.080000e-105 13542
277 AAWWNTGCTGACG NRL 9.710000e-101 1.460000e-103 9042
278 RGTCGTAAAAH HOXD12 1.090000e-99 1.630000e-102 8811
279 RWGTAAACAVR FOXP2 3.580000e-99 5.360000e-102 7349
280 BRTTACRTAAYM TEF 7.540000e-99 1.120000e-101 8024
281 AMCATATGKT OLIG2 3.500000e-97 5.190000e-100 15276
282 NNCACTCAANN NKX2-5 6.520000e-97 9.640000e-100 18032
283 ACCGAACAATV SOX12 7.770000e-97 1.140000e-99 17072
284 WAATTAATTAWYY Lhx3 9.150000e-96 1.340000e-98 5802
285 GATGACGTCAT FOSB::JUN 4.110000e-95 6.010000e-98 19344
286 DATGACGTCATH FOSL2::JUNB 3.300000e-94 4.810000e-97 14380
287 SWGATAASRN GATA5 2.740000e-93 3.980000e-96 12727
288 AATTGHTACAWT DMRTA2 9.880000e-93 1.430000e-95 20493
289 BNAASGTAAACAAD FOXF2 1.870000e-92 2.700000e-95 5409
290 RTCGTWAHNN HOXA9 9.940000e-92 1.430000e-94 22042
291 NNGAGTTCANN Vdr 1.620000e-91 2.320000e-94 16995
292 BDBTTCCWSAGAAVY Stat6 2.590000e-88 3.690000e-91 9183
293 ATTRCGCAAY CEBPG 9.710000e-88 1.380000e-90 6874
294 NNTGAGTGACAGVNN PKNOX1 5.090000e-87 7.220000e-90 15879
295 NNACATTCCAGSN TEAD1 1.670000e-85 2.360000e-88 19480
296 NNRNAATGGAAANWNH Nfatc2 7.890000e-85 1.110000e-87 21996
297 WAGGTGTGAAR TBX20 3.840000e-82 5.390000e-85 18660
298 MTTATCWSNNNNNHVCAG GATA1::TAL1 9.040000e-82 1.260000e-84 8629
299 NRTTACGTAAYV DBP 4.360000e-81 6.080000e-84 7229
300 NNNDRTCACGTGAYHNNN MITF 4.440000e-79 6.170000e-82 15458
301 HAAAAGVGGAAGTA SPIC 2.150000e-78 2.980000e-81 19438
302 VRVAACAATGGNN Sox5 1.090000e-77 1.500000e-80 21089
303 STVCACTCAC MSANTD3 2.080000e-76 2.870000e-79 20816
304 NDTTAATTAGN mix-a 3.170000e-76 4.340000e-79 6122
305 NNRTGACGTCAYHSN FOSL2::JUND 4.020000e-75 5.500000e-78 17055
306 RTAAACMGGAAGTR FOXJ2::ELF1 6.780000e-75 9.240000e-78 20271
307 TTCACGCWTSANTK PAX6 2.310000e-74 3.140000e-77 14193
308 GGTAATTAVS HOXB7 2.380000e-74 3.230000e-77 8057
309 NVCACTTGARHNN Nkx2-1 4.380000e-74 5.910000e-77 22483
310 NBCCTTTGATSTBN Hnf1A 1.100000e-73 1.490000e-76 12655
311 AAATCWCWGCH Gfi1B 4.530000e-72 6.080000e-75 10809
312 CCWTTGTYYY Sox6 2.020000e-70 2.700000e-73 5942
313 AMCATATGBY OLIG3 4.090000e-70 5.440000e-73 15684
314 TAAYYYAATCA DUX4 2.330000e-69 3.090000e-72 5278
315 ATGACGTCAT FOSL1::JUN 9.060000e-69 1.200000e-71 19978
316 KRTGACGTCATN JUN::JUNB 1.450000e-68 1.910000e-71 20101
317 NSCACGTGTN Npas2 4.560000e-67 5.990000e-70 12271
318 NTGCTGASTCAGCAH MAF 1.850000e-65 2.430000e-68 10329
319 NVATGGAAAAWN Nfatc1 2.500000e-64 3.270000e-67 19005
320 HTGACAD MEIS1 1.460000e-63 1.900000e-66 21053
321 RTMAACAGGAAGTN FOXO1::ELF1 4.900000e-63 6.370000e-66 20831
322 NRGAACAAAGVV Sox11 5.020000e-63 6.500000e-66 17376
323 RTGACGTCAT FOSB::JUNB 8.900000e-62 1.150000e-64 19308
324 NNAAAGGTCANN NR4A1 5.290000e-61 6.800000e-64 18066
325 RGTCGTAAAAH HOXD10 7.920000e-61 1.020000e-63 5598
326 DATRGGGTATCAYNWNT Six3 1.020000e-60 1.310000e-63 11489
327 VWTCATCAABGTCCT ZNF214 7.850000e-60 1.000000e-62 18574
328 TTAATTAM LMX1A 5.520000e-59 7.020000e-62 5956
329 NDGTCATGTGACHN USF1 1.250000e-58 1.580000e-61 16704
330 THAGTTTCRKTTTCY STAT1::STAT2 3.230000e-57 4.080000e-60 11852
331 AAAAAAAAAAAAAAA ZNF384 5.100000e-57 6.430000e-60 3493
332 RMCATATGBY Atoh1 5.340000e-57 6.700000e-60 20774
333 BNSAATCCACA FOXH1 4.490000e-56 5.620000e-59 5485
334 KATTGCGCAATM CEBPE 5.070000e-56 6.330000e-59 3017
335 HCGAAARYGAAAVT IRF7 1.930000e-55 2.400000e-58 8144
336 RTGGGGTGAY SREBF2 1.710000e-53 2.120000e-56 6688
337 DHDGAGATTACWKCAK ZNF85 1.790000e-53 2.220000e-56 16256
338 NRAACAGGTTYN GRHL2 1.640000e-52 2.020000e-55 19634
339 RCCACTTAA Nkx3-1 5.380000e-52 6.610000e-55 20852
340 BMAATCACDGCA GFI1 8.570000e-52 1.050000e-54 6222
341 NRTRTGAGTTCTCGYN ZNF274 2.590000e-51 3.160000e-54 6163
342 DTTTRAATH LIN54 5.320000e-49 6.490000e-52 5514
343 NNCACATGBN MXI1 1.010000e-48 1.230000e-51 15546
344 NWAWNTRGGTCA RORC 1.610000e-48 1.950000e-51 21924
345 NNNTTCCCAGAANNN Stat5b 2.080000e-45 2.510000e-48 18218
346 YNCTTTCTCTH PRDM1 2.910000e-44 3.510000e-47 21918
347 NDWKCAACAGGTGKNN SCRT1 1.330000e-42 1.590000e-45 17594
348 RDCCAATCASN NFYC 2.960000e-42 3.550000e-45 21631
349 RTGAACTTT NR1I3 5.090000e-42 6.090000e-45 15197
350 NNACAGATGTTNN Ptf1a 1.320000e-41 1.570000e-44 11910
351 NTGCTGASTCAGCAN MAFA 5.470000e-40 6.490000e-43 4219
352 GTCGTAAAAH HOXB9 1.470000e-39 1.750000e-42 2364
353 NTRATCCN RHOXF1 2.290000e-39 2.710000e-42 15095
354 DNATCACSTGATNH SREBF1 2.780000e-38 3.270000e-41 13811
355 NYACATTCCWNNS TEAD2 2.740000e-37 3.220000e-40 19482
356 ANCTMATTWGCATAWT POU1F1 1.350000e-35 1.580000e-38 1963
357 DRCATGTCNNRACAYGYM TP73 1.960000e-35 2.280000e-38 1500
358 NHAACCTCAANN RUNX3 2.010000e-35 2.340000e-38 18023
359 NTTCCAAGAANH Stat5a 6.530000e-35 7.590000e-38 19950
360 TGACCTNNAGTRACCYYDN Nr1h3::Rxra 1.100000e-34 1.280000e-37 18217
361 RACATGYCCGGRCATGTY TP53 1.340000e-34 1.550000e-37 259
362 DDDRATGABGTCATN JUND 2.820000e-34 3.250000e-37 4725
363 NAAAGGTCANR NR2F2 4.350000e-34 4.990000e-37 21712
364 VRRRACAAATGGTNNN Msgn1 9.290000e-34 1.060000e-36 14760
365 WVACAGCAGAYW ZNF317 4.580000e-33 5.230000e-36 22721
366 GDAACCTGANH Six4 1.070000e-32 1.210000e-35 11505
367 AAGTGCTGGGATTAC PITX1 2.120000e-32 2.410000e-35 1927
368 GGYMATAAAAC CDX4 5.780000e-32 6.550000e-35 2593
369 RACATGTYGKGACATGTC TP63 9.000000e-32 1.020000e-34 411
370 YTAATCCY PITX2 1.580000e-29 1.780000e-32 1853
371 NNAAACCACAARNN Bcl11B 2.140000e-29 2.400000e-32 22938
372 KATTGCGCAATM CEBPB 3.390000e-29 3.800000e-32 473
373 DCTAWAAATAGM MEF2D 3.990000e-29 4.460000e-32 3499
374 AHCATRTGDT TFAP4 8.520000e-29 9.500000e-32 6943
375 AMCATATGKT OLIG1 1.350000e-28 1.500000e-31 6339
376 DDRATGATGTCAT JUN 4.020000e-28 4.460000e-31 3363
377 GTCGTAAAAH HOXC9 1.400000e-27 1.550000e-30 1155
378 RAGGTGTGAAAWN EOMES 1.660000e-27 1.830000e-30 19512
379 11-CTGGGAYTAC PITX2 1.250000e-26 1.370000e-29 2777
380 RCATTCCW TEAD3 1.340000e-26 1.470000e-29 11027
381 TGTACAGTAW ZBTB32 9.310000e-26 1.020000e-28 4261
382 AAAACCGGTTTD GRHL1 2.360000e-25 2.570000e-28 6954
383 VTCACCCCAY SREBF1 2.790000e-25 3.040000e-28 4429
384 AGGTCGTAAAAH HOXC11 3.990000e-25 4.330000e-28 2337
385 CACTTTGGGAG CACTTTGGGAG 6.840000e-25 7.400000e-28 325
386 TTTCYRGGAAA STAT1 7.710000e-25 8.320000e-28 11299
387 YTTCYRGGAARNVR Stat4 9.740000e-25 1.050000e-27 9193
388 TAGAGAYRGGGTTTC TAGAGAYRGGGTTTC 5.750000e-24 6.180000e-27 460
389 GTAAACAGGAAGTK FOXO1::FLI1 7.750000e-24 8.300000e-27 3968
390 DTTTCCMAGAA Stat5a::Stat5b 1.550000e-23 1.660000e-26 10758
391 NATGACGTCAYH Creb5 1.740000e-22 1.860000e-25 2662
392 AAAGATCAAAGG TCF7L1 3.780000e-22 4.020000e-25 3601
393 AAACATATGBTN BHLHE23 4.490000e-22 4.760000e-25 2337
394 AAGAYAAGATAANA Mecom 6.140000e-21 6.500000e-24 2148
395 MATGTATCAAN DMRT3 1.930000e-20 2.030000e-23 6941
396 AYAGTAGGTS ZKSCAN1 3.200000e-20 3.370000e-23 11092
397 NYGCTTTCKAGGAAYN BCL6 1.630000e-19 1.720000e-22 13462
398 AWNTRGGTYAAAGGTCAN PPARA::RXRA 3.420000e-19 3.580000e-22 10622
399 CAAGCTT Nr2e3 8.600000e-19 8.980000e-22 5154
400 NWWAWBTAGGTCAR RORA 2.020000e-18 2.100000e-21 1973
401 TGTYRCCCAGGCTGG ZKSCAN3 2.270000e-18 2.350000e-21 383
402 RRCCAATCAGM NFYA 1.020000e-17 1.060000e-20 14081
403 NNNCCACTCAANNN NKX2-2 1.570000e-17 1.630000e-20 17892
404 VNRTGCAATMCC Ddit3::Cebpa 5.110000e-17 5.280000e-20 9863
405 DTTMRCACMTAGGTGTGAAW TBX19 4.270000e-16 4.390000e-19 6424
406 YSGTCACGSYWATTAN PAX3 4.400000e-16 4.520000e-19 12818
407 AWTTTAATTAA LMX1B 1.470000e-15 1.500000e-18 1693
408 RWMAACAGGAAGTN FOXO1::ELK3 1.810000e-15 1.850000e-18 12438
409 DNCAAAGTCCANN HNF4A 2.070000e-15 2.110000e-18 17800
410 RCTAWAAATAGC MEF2B 2.130000e-15 2.170000e-18 3007
411 NNRAGGTCANN Nr1H2 2.690000e-15 2.730000e-18 15563
412 14-ACCTCAGCCTCC ZNF460 2.930000e-15 2.960000e-18 309
413 AGGTGTGAAA TBR1 3.380000e-15 3.410000e-18 20030
414 NATCCAGATGTKB ZBTB18 1.230000e-14 1.240000e-17 3938
415 RGGVHYMGGAGGMCAAAATGD ZNF675 4.090000e-13 4.110000e-16 11036
416 RTAAACAGGAARYN FLI1::FOXI1 8.140000e-13 8.160000e-16 16737
417 CCAGCCTGGGYRAC ZKSCAN3 2.850000e-12 2.850000e-15 329
418 NNNTGAGTGACAGGBNN PBX3 2.990000e-12 2.980000e-15 16266
419 NNWGCAACAGGTGDNN SCRT2 3.400000e-12 3.380000e-15 16661
420 BGGRNWTTCC REL 7.780000e-12 7.720000e-15 20818
421 13-ACCACACMCA KLF9 1.290000e-11 1.280000e-14 478
422 ACCGGAAGTAATTAT FLI1::DRGX 1.880000e-11 1.860000e-14 13616
423 12-CTCCTGGGCTCAA CTCCTGGGCTCAA 2.420000e-11 2.390000e-14 197
424 NYCAAGKTCAAGKTCAN NR6A1 4.410000e-11 4.340000e-14 15736
425 DNCAAAGTCCANN HNF4G 1.260000e-09 1.240000e-12 8880
426 RVCAGATGGYN Atoh1 2.920000e-09 2.850000e-12 20991
427 BVTGGGAANN RBPJ 8.460000e-09 8.260000e-12 19397
428 NNTCGTGACHN Npas4 1.540000e-08 1.500000e-11 4227
429 BWAAAGGTCA NR4A2 1.690000e-08 1.640000e-11 13126
430 MDGAGGCTAAAAGCA ZNF418 1.800000e-08 1.740000e-11 3516
431 AGGRGAYATCACYACMGAYCCCAC ZNF382 4.240000e-08 4.100000e-11 2156
432 GCCTCMGCCTCCCRAG ZNF460 4.470000e-08 4.310000e-11 234
433 ACCATATGGT BHLHA15 4.650000e-08 4.480000e-11 4685
434 BBYTGTGGTTT Runx1 8.420000e-08 8.090000e-11 2880
435 NNDCCAGATGTBN TWIST1 9.320000e-08 8.930000e-11 22146
436 TTCTAGAAYRTTC HSF2 1.130000e-07 1.080000e-10 6727
437 GATGACGTCAYC JDP2 4.430000e-07 4.220000e-10 2096
438 AWMWAGGTCA RORA 5.080000e-07 4.830000e-10 5907
439 VVAACAGGAARN IKZF1 7.160000e-07 6.800000e-10 22001
440 CACCCACACAC EGR3 8.820000e-07 8.360000e-10 268
441 AAACCGGTTY TFCP2 1.300000e-06 1.230000e-09 15992
442 TGACAGCTGTCA TGIF2LY 1.440000e-06 1.350000e-09 18384
443 AAYGGAAAMW NFATC4 2.190000e-06 2.070000e-09 1878
444 DRVCGGAAGTCGTAAAD HOXD12::ELK1 8.810000e-06 8.270000e-09 988
445 BGATGACGTCATCR FOS 8.970000e-06 8.400000e-09 4984
446 TCAAGGTCAWW ESRRB 1.160000e-05 1.080000e-08 6537
447 GTCACGTGAC TFE3 1.940000e-05 1.810000e-08 4223
448 AWTATGCAAATKAG POU2F2 4.590000e-05 4.270000e-08 274
449 TTCTAGAAYRTTC HSF1 6.300000e-05 5.850000e-08 3311
450 WWATCACGTGATWW SREBF2 8.430000e-05 7.810000e-08 1558
451 DAGGTGTGAAA TBX2 9.180000e-05 8.490000e-08 22280
452 AGGTGTGA TBX15 1.100000e-04 1.010000e-07 2535
453 NVCAGATGNN HAND2 1.210000e-04 1.110000e-07 2523
454 TCAAGGTCAH Esrrg 1.250000e-04 1.150000e-07 8374
455 WAACCGCAA RUNX2 1.680000e-04 1.540000e-07 244
456 AMCATATGKY BHLHE22 2.770000e-04 2.530000e-07 654
457 RGGAGGCBGAGGHKG ZNF460 3.100000e-04 2.830000e-07 196
458 RWMAACAGGAAGTD FOXO1::ELK1 3.170000e-04 2.880000e-07 13136
459 AGGTGTGA TBX1 4.950000e-04 4.500000e-07 1352
460 AGGTGTKA TBX5 1.320000e-03 1.190000e-06 3756
461 VSCGGAARYNRTWAAN ERF::HOXB13 1.480000e-03 1.340000e-06 8277
462 TGACAGSTGTCA TGIF2 1.700000e-03 1.540000e-06 6690
463 RRTGACGTCAYY ATF3 2.300000e-03 2.070000e-06 670
464 CCTCGACCTCCYRR ZNF135 3.150000e-03 2.830000e-06 928
465 AGGTGTGA MGA 3.600000e-03 3.230000e-06 1889
466 AWCAGCTGWT TFAP4 5.850000e-03 5.230000e-06 2230
467 NBTCAAGGTCAHN ESRRA 6.770000e-03 6.040000e-06 8134
468 RYCACGTGAC TFEB 8.070000e-03 7.190000e-06 4499
469 TCTGTTACTTGCAGCCAAAWG ZNF582 8.980000e-03 7.980000e-06 4186
470 AACAATANCATTGTT Sox1 1.090000e-02 9.660000e-06 662
471 DTGACAGS MEIS3 1.200000e-02 1.060000e-05 22507
472 AACARCTGTT MYF6 1.360000e-02 1.200000e-05 7641
473 NNCTCCAGAAANNNN ZBTB26 1.630000e-02 1.440000e-05 1002
474 RACATATGTY NEUROG1 2.300000e-02 2.020000e-05 601
475 AACGAAACCGAAACT IRF9 2.930000e-02 2.570000e-05 86
476 AGGTGTGA TBX4 3.070000e-02 2.680000e-05 2520
477 MTCCAC ZNF354C 3.560000e-02 3.110000e-05 1013
TP. FP FP. ENR_RATIO mrc_type
1 50.87 11954 15.60 3.26 LR_open
2 49.92 11540 15.06 3.32 LR_open
3 50.22 11965 15.61 3.22 LR_open
4 52.39 13240 17.28 3.03 LR_open
5 51.59 12807 16.71 3.09 LR_open
6 48.04 10955 14.29 3.36 LR_open
7 53.67 14117 18.42 2.91 LR_open
8 53.64 14175 18.50 2.90 LR_open
9 44.54 9446 12.33 3.61 LR_open
10 50.03 12256 15.99 3.13 LR_open
11 51.67 13176 17.19 3.01 LR_open
12 52.35 13585 17.73 2.95 LR_open
13 47.38 10927 14.26 3.32 LR_open
14 46.39 10446 13.63 3.40 LR_open
15 52.00 13444 17.54 2.96 LR_open
16 53.45 14287 18.64 2.87 LR_open
17 50.80 12819 16.73 3.04 LR_open
18 49.53 12208 15.93 3.11 LR_open
19 52.55 13955 18.21 2.89 LR_open
20 54.98 15420 20.12 2.73 LR_open
21 49.04 12273 16.01 3.06 LR_open
22 49.72 12727 16.61 2.99 LR_open
23 50.43 13114 17.11 2.95 LR_open
24 46.13 10907 14.23 3.24 LR_open
25 43.57 9655 12.60 3.46 LR_open
26 42.67 9298 12.13 3.52 LR_open
27 46.86 12055 15.73 2.98 LR_open
28 40.82 9007 11.75 3.47 LR_open
29 38.93 8416 10.98 3.54 LR_open
30 52.16 19256 25.13 2.08 LR_open
31 44.24 14969 19.53 2.27 LR_open
32 50.49 19693 25.70 1.97 LR_open
33 47.09 19691 25.69 1.83 LR_open
34 77.81 45112 58.86 1.32 LR_open
35 42.04 18134 23.66 1.78 LR_open
36 87.93 55534 72.46 1.21 LR_open
37 76.21 44757 58.40 1.30 LR_open
38 70.68 40224 52.49 1.35 LR_open
39 86.56 54657 71.32 1.21 LR_open
40 76.87 45739 59.68 1.29 LR_open
41 85.70 54198 70.72 1.21 LR_open
42 87.88 56409 73.60 1.19 LR_open
43 81.27 50092 65.36 1.24 LR_open
44 86.74 55579 72.52 1.20 LR_open
45 86.60 55598 72.55 1.19 LR_open
46 79.55 48999 63.93 1.24 LR_open
47 74.55 44690 58.31 1.28 LR_open
48 76.54 46467 60.63 1.26 LR_open
49 91.72 61083 79.70 1.15 LR_open
50 92.42 61894 80.76 1.14 LR_open
51 91.14 60526 78.98 1.15 LR_open
52 92.23 61809 80.65 1.14 LR_open
53 64.79 37027 48.31 1.34 LR_open
54 89.35 58861 76.80 1.16 LR_open
55 93.14 63044 82.26 1.13 LR_open
56 91.11 60864 79.42 1.15 LR_open
57 83.36 53212 69.43 1.20 LR_open
58 90.36 60085 78.40 1.15 LR_open
59 70.48 42005 54.81 1.29 LR_open
60 81.74 51818 67.61 1.21 LR_open
61 95.23 65642 85.65 1.11 LR_open
62 81.63 51812 67.61 1.21 LR_open
63 57.85 32225 42.05 1.38 LR_open
64 83.55 53680 70.04 1.19 LR_open
65 84.49 54615 71.26 1.19 LR_open
66 85.64 55749 72.74 1.18 LR_open
67 73.88 45202 58.98 1.25 LR_open
68 85.95 56139 73.25 1.17 LR_open
69 55.95 31017 40.47 1.38 LR_open
70 94.35 64882 84.66 1.11 LR_open
71 79.24 50016 65.26 1.21 LR_open
72 93.74 64330 83.94 1.12 LR_open
73 80.13 50982 66.52 1.20 LR_open
74 90.99 61390 80.10 1.14 LR_open
75 88.53 58887 76.84 1.15 LR_open
76 77.72 48872 63.77 1.22 LR_open
77 81.00 51820 67.62 1.20 LR_open
78 78.46 49584 64.70 1.21 LR_open
79 90.25 60700 79.20 1.14 LR_open
80 93.39 64048 83.57 1.12 LR_open
81 84.90 55599 72.55 1.17 LR_open
82 78.42 49747 64.91 1.21 LR_open
83 89.36 60012 78.30 1.14 LR_open
84 89.17 59859 78.11 1.14 LR_open
85 70.47 43091 56.23 1.25 LR_open
86 94.42 65448 85.40 1.11 LR_open
87 92.68 63567 82.94 1.12 LR_open
88 81.92 53088 69.27 1.18 LR_open
89 86.94 57779 75.39 1.15 LR_open
90 92.38 63291 82.58 1.12 LR_open
91 68.88 41988 54.79 1.26 LR_open
92 89.99 60902 79.47 1.13 LR_open
93 83.82 54958 71.71 1.17 LR_open
94 83.68 54870 71.60 1.17 LR_open
95 74.06 46393 60.53 1.22 LR_open
96 89.79 60767 79.29 1.13 LR_open
97 91.58 62619 81.71 1.12 LR_open
98 60.52 35505 46.33 1.31 LR_open
99 92.46 63598 82.98 1.11 LR_open
100 73.85 46349 60.48 1.22 LR_open
101 84.00 55352 72.22 1.16 LR_open
102 81.34 53000 69.16 1.18 LR_open
103 87.55 58832 76.77 1.14 LR_open
104 89.89 61133 79.77 1.13 LR_open
105 91.48 62756 81.89 1.12 LR_open
106 79.67 51578 67.30 1.18 LR_open
107 93.78 65220 85.10 1.10 LR_open
108 92.15 63506 82.86 1.11 LR_open
109 69.52 43086 56.22 1.24 LR_open
110 80.07 52060 67.93 1.18 LR_open
111 63.30 38123 49.74 1.27 LR_open
112 82.69 54464 71.07 1.16 LR_open
113 75.76 48369 63.11 1.20 LR_open
114 78.58 50809 66.30 1.19 LR_open
115 75.16 47899 62.50 1.20 LR_open
116 72.38 45566 59.46 1.22 LR_open
117 57.76 33896 44.23 1.31 LR_open
118 80.05 52192 68.10 1.18 LR_open
119 84.94 56674 73.95 1.15 LR_open
120 92.08 63695 83.11 1.11 LR_open
121 87.06 58707 76.60 1.14 LR_open
122 90.64 62220 81.19 1.12 LR_open
123 90.40 61979 80.87 1.12 LR_open
124 57.18 33592 43.83 1.30 LR_open
125 93.80 65554 85.54 1.10 LR_open
126 86.39 58108 75.82 1.14 LR_open
127 67.96 42123 54.96 1.24 LR_open
128 83.53 55478 72.39 1.15 LR_open
129 66.97 41387 54.00 1.24 LR_open
130 87.55 59282 77.35 1.13 LR_open
131 75.60 48559 63.36 1.19 LR_open
132 89.41 61114 79.74 1.12 LR_open
133 95.29 67321 87.84 1.08 LR_open
134 43.34 23574 30.76 1.41 LR_open
135 95.03 67059 87.50 1.09 LR_open
136 95.47 67566 88.16 1.08 LR_open
137 66.51 41132 53.67 1.24 LR_open
138 63.83 39003 50.89 1.25 LR_open
139 89.33 61159 79.80 1.12 LR_open
140 84.04 56154 73.27 1.15 LR_open
141 84.26 56362 73.54 1.15 LR_open
142 96.25 68585 89.49 1.08 LR_open
143 95.34 67535 88.12 1.08 LR_open
144 62.92 38431 50.15 1.25 LR_open
145 92.19 64129 83.68 1.10 LR_open
146 88.35 60309 78.69 1.12 LR_open
147 80.85 53420 69.70 1.16 LR_open
148 62.93 38517 50.26 1.25 LR_open
149 80.77 53420 69.70 1.16 LR_open
150 20.53 8798 11.48 1.79 LR_open
151 81.22 53825 70.23 1.16 LR_open
152 87.88 59962 78.24 1.12 LR_open
153 77.04 50201 65.50 1.18 LR_open
154 70.25 44508 58.07 1.21 LR_open
155 91.47 63530 82.90 1.10 LR_open
156 90.24 62307 81.30 1.11 LR_open
157 85.83 58081 75.79 1.13 LR_open
158 68.47 43092 56.23 1.22 LR_open
159 77.63 50766 66.24 1.17 LR_open
160 89.21 61316 80.01 1.12 LR_open
161 56.84 33959 44.31 1.28 LR_open
162 74.06 47749 62.30 1.19 LR_open
163 74.01 47715 62.26 1.19 LR_open
164 73.10 46963 61.28 1.19 LR_open
165 84.99 57388 74.88 1.14 LR_open
166 83.21 55822 72.84 1.14 LR_open
167 82.91 55560 72.50 1.14 LR_open
168 68.62 43382 56.61 1.21 LR_open
169 70.14 44638 58.24 1.20 LR_open
170 96.65 69369 90.51 1.07 LR_open
171 93.28 65606 85.60 1.09 LR_open
172 93.49 65843 85.91 1.09 LR_open
173 79.10 52278 68.21 1.16 LR_open
174 94.68 67147 87.61 1.08 LR_open
175 73.02 47154 61.53 1.19 LR_open
176 93.91 66341 86.56 1.08 LR_open
177 77.07 50597 66.02 1.17 LR_open
178 68.71 43635 56.94 1.21 LR_open
179 55.79 33501 43.71 1.28 LR_open
180 78.18 51614 67.35 1.16 LR_open
181 95.48 68130 88.90 1.07 LR_open
182 80.06 53334 69.59 1.15 LR_open
183 55.62 33521 43.74 1.27 LR_open
184 61.09 37783 49.30 1.24 LR_open
185 55.41 33420 43.61 1.27 LR_open
186 68.70 43869 57.24 1.20 LR_open
187 84.36 57278 74.74 1.13 LR_open
188 74.05 48336 63.07 1.17 LR_open
189 84.61 57558 75.10 1.13 LR_open
190 54.34 32713 42.68 1.27 LR_open
191 79.17 52752 68.83 1.15 LR_open
192 71.25 46068 60.11 1.19 LR_open
193 65.15 41123 53.66 1.21 LR_open
194 88.79 61481 80.22 1.11 LR_open
195 92.43 65081 84.92 1.09 LR_open
196 94.99 67819 88.49 1.07 LR_open
197 47.32 27571 35.98 1.32 LR_open
198 72.86 47483 61.96 1.18 LR_open
199 65.66 41675 54.38 1.21 LR_open
200 87.73 60602 79.07 1.11 LR_open
201 92.46 65235 85.12 1.09 LR_open
202 64.30 40677 53.08 1.21 LR_open
203 76.87 51019 66.57 1.15 LR_open
204 93.17 66043 86.17 1.08 LR_open
205 51.55 30932 40.36 1.28 LR_open
206 96.59 69803 91.08 1.06 LR_open
207 78.22 52331 68.28 1.15 LR_open
208 90.27 63289 82.58 1.09 LR_open
209 63.65 40418 52.74 1.21 LR_open
210 72.71 47781 62.35 1.17 LR_open
211 57.74 35830 46.75 1.24 LR_open
212 63.03 39995 52.19 1.21 LR_open
213 60.85 38283 49.95 1.22 LR_open
214 94.54 67678 88.31 1.07 LR_open
215 64.96 41587 54.26 1.20 LR_open
216 84.65 58168 75.90 1.12 LR_open
217 77.81 52181 68.09 1.14 LR_open
218 79.93 54038 70.51 1.13 LR_open
219 67.50 43687 57.00 1.18 LR_open
220 55.25 34119 44.52 1.24 LR_open
221 94.47 67700 88.34 1.07 LR_open
222 55.19 34100 44.49 1.24 LR_open
223 82.83 56663 73.93 1.12 LR_open
224 43.44 25376 33.11 1.31 LR_open
225 85.05 58703 76.60 1.11 LR_open
226 61.07 38746 50.56 1.21 LR_open
227 50.77 30871 40.28 1.26 LR_open
228 94.90 68265 89.07 1.07 LR_open
229 37.71 21399 27.92 1.35 LR_open
230 44.91 26579 34.68 1.30 LR_open
231 83.99 57861 75.50 1.11 LR_open
232 96.43 69990 91.32 1.06 LR_open
233 57.64 36199 47.23 1.22 LR_open
234 85.49 59234 77.29 1.11 LR_open
235 91.11 64519 84.19 1.08 LR_open
236 82.26 56412 73.61 1.12 LR_open
237 86.60 60311 78.69 1.10 LR_open
238 65.91 42786 55.83 1.18 LR_open
239 41.26 24104 31.45 1.31 LR_open
240 87.19 60954 79.53 1.10 LR_open
241 48.80 29724 38.78 1.26 LR_open
242 73.56 49183 64.17 1.15 LR_open
243 74.54 50083 65.35 1.14 LR_open
244 24.16 12480 16.28 1.48 LR_open
245 81.01 55745 72.74 1.11 LR_open
246 67.55 44553 58.13 1.16 LR_open
247 86.15 60314 78.70 1.09 LR_open
248 44.02 26566 34.66 1.27 LR_open
249 86.57 60758 79.28 1.09 LR_open
250 54.23 34231 44.67 1.21 LR_open
251 80.17 55143 71.95 1.11 LR_open
252 73.19 49340 64.38 1.14 LR_open
253 86.22 60556 79.01 1.09 LR_open
254 41.85 25147 32.81 1.28 LR_open
255 33.00 18893 24.65 1.34 LR_open
256 42.11 25406 33.15 1.27 LR_open
257 96.40 70526 92.02 1.05 LR_open
258 79.24 54590 71.23 1.11 LR_open
259 82.16 57114 74.52 1.10 LR_open
260 32.71 18778 24.50 1.34 LR_open
261 37.14 21989 28.69 1.29 LR_open
262 90.89 65072 84.91 1.07 LR_open
263 48.88 30613 39.94 1.22 LR_open
264 95.72 69895 91.20 1.05 LR_open
265 23.12 12359 16.13 1.43 LR_open
266 85.19 59986 78.27 1.09 LR_open
267 72.65 49390 64.44 1.13 LR_open
268 95.68 69975 91.30 1.05 LR_open
269 79.28 55009 71.78 1.10 LR_open
270 79.48 55207 72.04 1.10 LR_open
271 58.16 38036 49.63 1.17 LR_open
272 91.25 65741 85.78 1.06 LR_open
273 80.44 56144 73.26 1.10 LR_open
274 70.92 48342 63.08 1.12 LR_open
275 77.34 53654 70.01 1.10 LR_open
276 58.74 38756 50.57 1.16 LR_open
277 39.22 24144 31.50 1.24 LR_open
278 38.22 23446 30.59 1.25 LR_open
279 31.87 18918 24.68 1.29 LR_open
280 34.80 21010 27.41 1.27 LR_open
281 66.26 44845 58.51 1.13 LR_open
282 78.21 54603 71.25 1.10 LR_open
283 74.05 51153 66.75 1.11 LR_open
284 25.16 14326 18.69 1.35 LR_open
285 83.90 59494 77.63 1.08 LR_open
286 62.37 41864 54.62 1.14 LR_open
287 55.20 36325 47.40 1.16 LR_open
288 88.88 63951 83.44 1.07 LR_open
289 23.46 13239 17.27 1.36 LR_open
290 95.60 70331 91.77 1.04 LR_open
291 73.71 51040 66.60 1.11 LR_open
292 39.83 24954 32.56 1.22 LR_open
293 29.81 17769 23.19 1.29 LR_open
294 68.87 47258 61.66 1.12 LR_open
295 84.49 60267 78.64 1.07 LR_open
296 95.40 70261 91.68 1.04 LR_open
297 80.93 57327 74.80 1.08 LR_open
298 37.43 23401 30.53 1.23 LR_open
299 31.35 19048 24.85 1.26 LR_open
300 67.05 46061 60.10 1.12 LR_open
301 84.31 60312 78.70 1.07 LR_open
302 91.47 66668 86.99 1.05 LR_open
303 90.28 65622 85.62 1.05 LR_open
304 26.55 15795 20.61 1.29 LR_open
305 73.97 51778 67.56 1.09 LR_open
306 87.92 63554 82.93 1.06 LR_open
307 61.56 41893 54.66 1.13 LR_open
308 34.95 21833 28.49 1.23 LR_open
309 97.51 72639 94.78 1.03 LR_open
310 54.89 36745 47.95 1.14 LR_open
311 46.88 30720 40.08 1.17 LR_open
312 25.77 15413 20.11 1.28 LR_open
313 68.03 47156 61.53 1.11 LR_open
314 22.89 13437 17.53 1.31 LR_open
315 86.65 62616 81.70 1.06 LR_open
316 87.18 63086 82.32 1.06 LR_open
317 53.22 35710 46.60 1.14 LR_open
318 44.80 29397 38.36 1.17 LR_open
319 82.43 59146 77.17 1.07 LR_open
320 91.31 66883 87.27 1.05 LR_open
321 90.35 66033 86.16 1.05 LR_open
322 75.36 53334 69.59 1.08 LR_open
323 83.74 60340 78.73 1.06 LR_open
324 78.36 55863 72.89 1.07 LR_open
325 24.28 14651 19.12 1.27 LR_open
326 49.83 33368 43.54 1.14 LR_open
327 80.56 57731 75.33 1.07 LR_open
328 25.83 15802 20.62 1.25 LR_open
329 72.45 51131 66.72 1.09 LR_open
330 51.41 34704 45.28 1.14 LR_open
331 15.15 8488 11.08 1.37 LR_open
332 90.10 65983 86.10 1.05 LR_open
333 23.79 14454 18.86 1.26 LR_open
334 13.09 7141 9.32 1.40 LR_open
335 35.32 22757 29.69 1.19 LR_open
336 29.01 18249 23.81 1.22 LR_open
337 70.51 49771 64.94 1.09 LR_open
338 85.16 61847 80.70 1.06 LR_open
339 90.44 66428 86.68 1.04 LR_open
340 26.99 16861 22.00 1.23 LR_open
341 26.73 16695 21.78 1.23 LR_open
342 23.92 14774 19.28 1.24 LR_open
343 67.43 47515 62.00 1.09 LR_open
344 95.09 70739 92.30 1.03 LR_open
345 79.02 56991 74.36 1.06 LR_open
346 95.06 70820 92.41 1.03 LR_open
347 76.31 54899 71.63 1.07 LR_open
348 93.82 69729 90.98 1.03 LR_open
349 65.91 46616 60.83 1.08 LR_open
350 51.66 35574 46.42 1.11 LR_open
351 18.30 11132 14.53 1.26 LR_open
352 10.25 5668 7.40 1.39 LR_open
353 65.47 46392 60.53 1.08 LR_open
354 59.90 42089 54.92 1.09 LR_open
355 84.50 61851 80.70 1.05 LR_open
356 8.51 4623 6.03 1.41 LR_open
357 6.51 3336 4.35 1.50 LR_open
358 78.17 56725 74.02 1.06 LR_open
359 86.53 63645 83.05 1.04 LR_open
360 79.01 57443 74.95 1.05 LR_open
361 1.12 276 0.36 3.12 LR_open
362 20.49 12880 16.81 1.22 LR_open
363 94.17 70280 91.70 1.03 LR_open
364 64.02 45529 59.41 1.08 LR_open
365 98.55 74463 97.16 1.01 LR_open
366 49.90 34677 45.25 1.10 LR_open
367 8.36 4598 6.00 1.39 LR_open
368 11.25 6536 8.53 1.32 LR_open
369 1.78 609 0.79 2.25 LR_open
370 8.04 4459 5.82 1.38 LR_open
371 99.49 75572 98.61 1.01 LR_open
372 2.05 775 1.01 2.03 LR_open
373 15.18 9331 12.18 1.25 LR_open
374 30.11 20070 26.19 1.15 LR_open
375 27.49 18163 23.70 1.16 LR_open
376 14.59 8956 11.69 1.25 LR_open
377 5.01 2551 3.33 1.51 LR_open
378 84.63 62375 81.39 1.04 LR_open
379 12.04 7248 9.46 1.27 LR_open
380 47.83 33436 43.63 1.10 LR_open
381 18.48 11793 15.39 1.20 LR_open
382 30.16 20280 26.46 1.14 LR_open
383 19.21 12333 16.09 1.19 LR_open
384 10.14 5991 7.82 1.30 LR_open
385 1.41 481 0.63 2.25 LR_open
386 49.01 34438 44.94 1.09 LR_open
387 39.87 27535 35.93 1.11 LR_open
388 2.00 803 1.05 1.91 LR_open
389 17.21 10971 14.32 1.20 LR_open
390 46.66 32732 42.71 1.09 LR_open
391 11.55 7048 9.20 1.26 LR_open
392 15.62 9914 12.94 1.21 LR_open
393 10.14 6091 7.95 1.28 LR_open
394 9.32 5568 7.27 1.28 LR_open
395 30.10 20510 26.76 1.12 LR_open
396 48.11 34045 44.42 1.08 LR_open
397 58.39 41983 54.78 1.07 LR_open
398 46.07 32563 42.49 1.08 LR_open
399 22.35 14916 19.46 1.15 LR_open
400 8.56 5134 6.70 1.28 LR_open
401 1.66 685 0.89 1.86 LR_open
402 61.07 44187 57.66 1.06 LR_open
403 77.60 57206 74.64 1.04 LR_open
404 42.78 30217 39.43 1.09 LR_open
405 27.86 19117 24.94 1.12 LR_open
406 55.60 40070 52.28 1.06 LR_open
407 7.34 4402 5.74 1.28 LR_open
408 53.95 38848 50.69 1.06 LR_open
409 77.20 57024 74.41 1.04 LR_open
410 13.04 8387 10.94 1.19 LR_open
411 67.50 49372 64.42 1.05 LR_open
412 1.34 544 0.71 1.89 LR_open
413 86.88 64837 84.60 1.03 LR_open
414 17.08 11318 14.77 1.16 LR_open
415 47.87 34377 44.86 1.07 LR_open
416 72.59 53556 69.88 1.04 LR_open
417 1.43 633 0.83 1.73 LR_open
418 70.55 51993 67.84 1.04 LR_open
419 72.26 53344 69.60 1.04 LR_open
420 90.29 67831 88.51 1.02 LR_open
421 2.07 1033 1.35 1.54 LR_open
422 59.06 43109 56.25 1.05 LR_open
423 0.85 323 0.42 2.03 LR_open
424 68.25 50287 65.62 1.04 LR_open
425 38.51 27575 35.98 1.07 LR_open
426 91.04 68595 89.50 1.02 LR_open
427 84.13 63024 82.23 1.02 LR_open
428 18.33 12610 16.45 1.11 LR_open
429 56.93 41739 54.46 1.05 LR_open
430 15.25 10359 13.52 1.13 LR_open
431 9.35 6124 7.99 1.17 LR_open
432 1.01 454 0.59 1.72 LR_open
433 20.32 14108 18.41 1.10 LR_open
434 12.49 8399 10.96 1.14 LR_open
435 96.05 72852 95.06 1.01 LR_open
436 29.18 20727 27.04 1.08 LR_open
437 9.09 5994 7.82 1.16 LR_open
438 25.62 18128 23.65 1.08 LR_open
439 95.42 72362 94.42 1.01 LR_open
440 1.16 563 0.73 1.59 LR_open
441 69.36 51565 67.28 1.03 LR_open
442 79.74 59712 77.91 1.02 LR_open
443 8.15 5357 6.99 1.17 LR_open
444 4.29 2665 3.48 1.23 LR_open
445 21.62 15260 19.91 1.09 LR_open
446 28.35 20302 26.49 1.07 LR_open
447 18.32 12842 16.76 1.09 LR_open
448 1.19 612 0.80 1.49 LR_open
449 14.36 9968 13.01 1.10 LR_open
450 6.76 4455 5.81 1.16 LR_open
451 96.63 73486 95.89 1.01 LR_open
452 10.99 7523 9.82 1.12 LR_open
453 10.94 7488 9.77 1.12 LR_open
454 36.32 26423 34.48 1.05 LR_open
455 1.06 542 0.71 1.50 LR_open
456 2.84 1726 2.25 1.26 LR_open
457 0.85 418 0.55 1.56 LR_open
458 56.97 42247 55.12 1.03 LR_open
459 5.86 3861 5.04 1.16 LR_open
460 16.29 11510 15.02 1.08 LR_open
461 35.90 26237 34.23 1.05 LR_open
462 29.02 21042 27.46 1.06 LR_open
463 2.91 1810 2.36 1.23 LR_open
464 4.02 2599 3.39 1.19 LR_open
465 8.19 5595 7.30 1.12 LR_open
466 9.67 6690 8.73 1.11 LR_open
467 35.28 25855 33.74 1.05 LR_open
468 19.51 13991 18.26 1.07 LR_open
469 18.16 12983 16.94 1.07 LR_open
470 2.87 1815 2.37 1.21 LR_open
471 97.62 74422 97.11 1.01 LR_open
472 33.14 24277 31.68 1.05 LR_open
473 4.35 2865 3.74 1.16 LR_open
474 2.61 1645 2.15 1.22 LR_open
475 0.37 164 0.21 1.75 LR_open
476 10.93 7677 10.02 1.09 LR_open
477 4.39 2919 3.81 1.15 LR_open
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] motifmatchr_1.28.0
[2] ggseqlogo_0.2
[3] universalmotif_1.24.2
[4] data.table_1.17.0
[5] BSgenome.Hsapiens.UCSC.hg38_1.4.5
[6] BSgenome_1.74.0
[7] BiocIO_1.16.0
[8] MotifDb_1.48.0
[9] Biostrings_2.74.1
[10] XVector_0.46.0
[11] TFBSTools_1.44.0
[12] JASPAR2022_0.99.8
[13] BiocFileCache_2.14.0
[14] dbplyr_2.5.0
[15] devtools_2.4.5
[16] usethis_3.1.0
[17] ggpubr_0.6.0
[18] BiocParallel_1.40.0
[19] Cormotif_1.52.0
[20] affy_1.84.0
[21] scales_1.3.0
[22] VennDiagram_1.7.3
[23] futile.logger_1.4.3
[24] gridExtra_2.3
[25] ggfortify_0.4.17
[26] edgeR_4.4.2
[27] limma_3.62.2
[28] rtracklayer_1.66.0
[29] org.Hs.eg.db_3.20.0
[30] TxDb.Hsapiens.UCSC.hg38.knownGene_3.20.0
[31] GenomicFeatures_1.58.0
[32] AnnotationDbi_1.68.0
[33] Biobase_2.66.0
[34] GenomicRanges_1.58.0
[35] GenomeInfoDb_1.42.3
[36] IRanges_2.40.1
[37] S4Vectors_0.44.0
[38] BiocGenerics_0.52.0
[39] ChIPseeker_1.42.1
[40] RColorBrewer_1.1-3
[41] broom_1.0.7
[42] kableExtra_1.4.0
[43] cowplot_1.1.3
[44] lubridate_1.9.4
[45] forcats_1.0.0
[46] stringr_1.5.1
[47] dplyr_1.1.4
[48] purrr_1.0.4
[49] readr_2.1.5
[50] tidyr_1.3.1
[51] tibble_3.2.1
[52] ggplot2_3.5.1
[53] tidyverse_2.0.0
[54] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.5
[2] matrixStats_1.5.0
[3] bitops_1.0-9
[4] DirichletMultinomial_1.48.0
[5] enrichplot_1.26.6
[6] httr_1.4.7
[7] profvis_0.4.0
[8] tools_4.4.2
[9] backports_1.5.0
[10] utf8_1.2.4
[11] R6_2.6.1
[12] lazyeval_0.2.2
[13] urlchecker_1.0.1
[14] withr_3.0.2
[15] preprocessCore_1.68.0
[16] cli_3.6.4
[17] formatR_1.14
[18] labeling_0.4.3
[19] sass_0.4.9
[20] Rsamtools_2.22.0
[21] systemfonts_1.2.1
[22] yulab.utils_0.2.0
[23] DOSE_4.0.0
[24] svglite_2.1.3
[25] R.utils_2.13.0
[26] sessioninfo_1.2.3
[27] plotrix_3.8-4
[28] rstudioapi_0.17.1
[29] RSQLite_2.3.9
[30] generics_0.1.3
[31] gridGraphics_0.5-1
[32] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[33] gtools_3.9.5
[34] car_3.1-3
[35] GO.db_3.20.0
[36] Matrix_1.7-3
[37] abind_1.4-8
[38] R.methodsS3_1.8.2
[39] lifecycle_1.0.4
[40] whisker_0.4.1
[41] yaml_2.3.10
[42] carData_3.0-5
[43] SummarizedExperiment_1.36.0
[44] gplots_3.2.0
[45] qvalue_2.38.0
[46] SparseArray_1.6.2
[47] blob_1.2.4
[48] promises_1.3.2
[49] pwalign_1.2.0
[50] crayon_1.5.3
[51] miniUI_0.1.1.1
[52] ggtangle_0.0.6
[53] lattice_0.22-6
[54] annotate_1.84.0
[55] KEGGREST_1.46.0
[56] pillar_1.10.1
[57] knitr_1.49
[58] fgsea_1.32.2
[59] rjson_0.2.23
[60] boot_1.3-31
[61] codetools_0.2-20
[62] fastmatch_1.1-6
[63] glue_1.8.0
[64] getPass_0.2-4
[65] ggfun_0.1.8
[66] remotes_2.5.0
[67] vctrs_0.6.5
[68] png_0.1-8
[69] treeio_1.30.0
[70] poweRlaw_1.0.0
[71] gtable_0.3.6
[72] cachem_1.1.0
[73] xfun_0.51
[74] S4Arrays_1.6.0
[75] mime_0.12
[76] statmod_1.5.0
[77] ellipsis_0.3.2
[78] nlme_3.1-167
[79] ggtree_3.14.0
[80] bit64_4.6.0-1
[81] filelock_1.0.3
[82] rprojroot_2.0.4
[83] bslib_0.9.0
[84] affyio_1.76.0
[85] KernSmooth_2.23-26
[86] splitstackshape_1.4.8
[87] seqLogo_1.72.0
[88] colorspace_2.1-1
[89] DBI_1.2.3
[90] tidyselect_1.2.1
[91] processx_3.8.6
[92] bit_4.6.0
[93] compiler_4.4.2
[94] curl_6.2.1
[95] git2r_0.35.0
[96] xml2_1.3.7
[97] DelayedArray_0.32.0
[98] caTools_1.18.3
[99] callr_3.7.6
[100] digest_0.6.37
[101] rmarkdown_2.29
[102] htmltools_0.5.8.1
[103] pkgconfig_2.0.3
[104] MatrixGenerics_1.18.1
[105] fastmap_1.2.0
[106] rlang_1.1.5
[107] htmlwidgets_1.6.4
[108] UCSC.utils_1.2.0
[109] shiny_1.10.0
[110] farver_2.1.2
[111] jquerylib_0.1.4
[112] jsonlite_1.9.1
[113] GOSemSim_2.32.0
[114] R.oo_1.27.0
[115] RCurl_1.98-1.16
[116] magrittr_2.0.3
[117] Formula_1.2-5
[118] GenomeInfoDbData_1.2.13
[119] ggplotify_0.1.2
[120] patchwork_1.3.0
[121] munsell_0.5.1
[122] Rcpp_1.0.14
[123] ape_5.8-1
[124] stringi_1.8.4
[125] MASS_7.3-65
[126] zlibbioc_1.52.0
[127] plyr_1.8.9
[128] pkgbuild_1.4.6
[129] parallel_4.4.2
[130] ggrepel_0.9.6
[131] CNEr_1.42.0
[132] splines_4.4.2
[133] hms_1.1.3
[134] locfit_1.5-9.12
[135] ps_1.9.0
[136] igraph_2.1.4
[137] ggsignif_0.6.4
[138] reshape2_1.4.4
[139] pkgload_1.4.0
[140] TFMPvalue_0.0.9
[141] futile.options_1.0.1
[142] XML_3.99-0.18
[143] evaluate_1.0.3
[144] lambda.r_1.2.4
[145] BiocManager_1.30.25
[146] tzdb_0.4.0
[147] httpuv_1.6.15
[148] xtable_1.8-4
[149] restfulr_0.0.15
[150] tidytree_0.4.6
[151] rstatix_0.7.2
[152] later_1.4.1
[153] viridisLite_0.4.2
[154] aplot_0.2.5
[155] memoise_2.0.1
[156] GenomicAlignments_1.42.0
[157] timechange_0.3.0