Figure_S17
Last updated: 2025-08-13
Checks: 6 1
Knit directory: Paul_CX_2025/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250129) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e0ace73. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: 0.1 box.svg
Ignored: Rplot04.svg
Untracked files:
Untracked: analysis/Figure_S17.Rmd
Unstaged changes:
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
📌 Load Required Libraries
library(tidyverse)
library(ggfortify)
library(ggplot2)
library(cluster)
library(edgeR)
library(limma)
library(Homo.sapiens)
library(BiocParallel)
library(qvalue)
library(pheatmap)
library(clusterProfiler)
library(AnnotationDbi)
library(org.Hs.eg.db)
library(RColorBrewer)
library(readr)
library(TxDb.Hsapiens.UCSC.hg38.knownGene)
library(DOSE)
# Load UCSC transcript database
txdb <- TxDb.Hsapiens.UCSC.hg38.knownGene📌 0.1 Micromolar
📌 Read and Process Data
# Load the saved datasets
prob_1_0.1 <- read.csv("data/prob_1_0.1.csv")$Entrez_ID
prob_2_0.1 <- read.csv("data/prob_2_0.1.csv")$Entrez_ID
prob_3_0.1 <- read.csv("data/prob_3_0.1.csv")$Entrez_ID
prob_1_0.5 <- read.csv("data/prob_1_0.5.csv")$Entrez_ID
prob_2_0.5 <- read.csv("data/prob_2_0.5.csv")$Entrez_ID
prob_3_0.5 <- read.csv("data/prob_3_0.5.csv")$Entrez_ID
prob_4_0.5 <- read.csv("data/prob_4_0.5.csv")$Entrez_ID
prob_5_0.5 <- read.csv("data/prob_5_0.5.csv")$Entrez_ID
CX_0.1_3 <- read.csv("data/DEGs/Toptable_CX_0.1_3.csv")
background<-as.character(CX_0.1_3$Entrez_ID)📌 Non response (0.1 µM)
📌 GO Enrichment Clusterprofiler
# 📦 Load Libraries
library(tidyverse)
library(patchwork)Warning: package 'patchwork' was built under R version 4.3.3library(data.table)Warning: package 'data.table' was built under R version 4.3.3# 📁 Load File
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_Non_response_(0.1).csv")
# 🧠 Convert and filter (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# Create tibbles for each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plot function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🔧 Plot for each GO category
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")
#plot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
#plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine plots
combined_plot <- plot_BP
combined_plot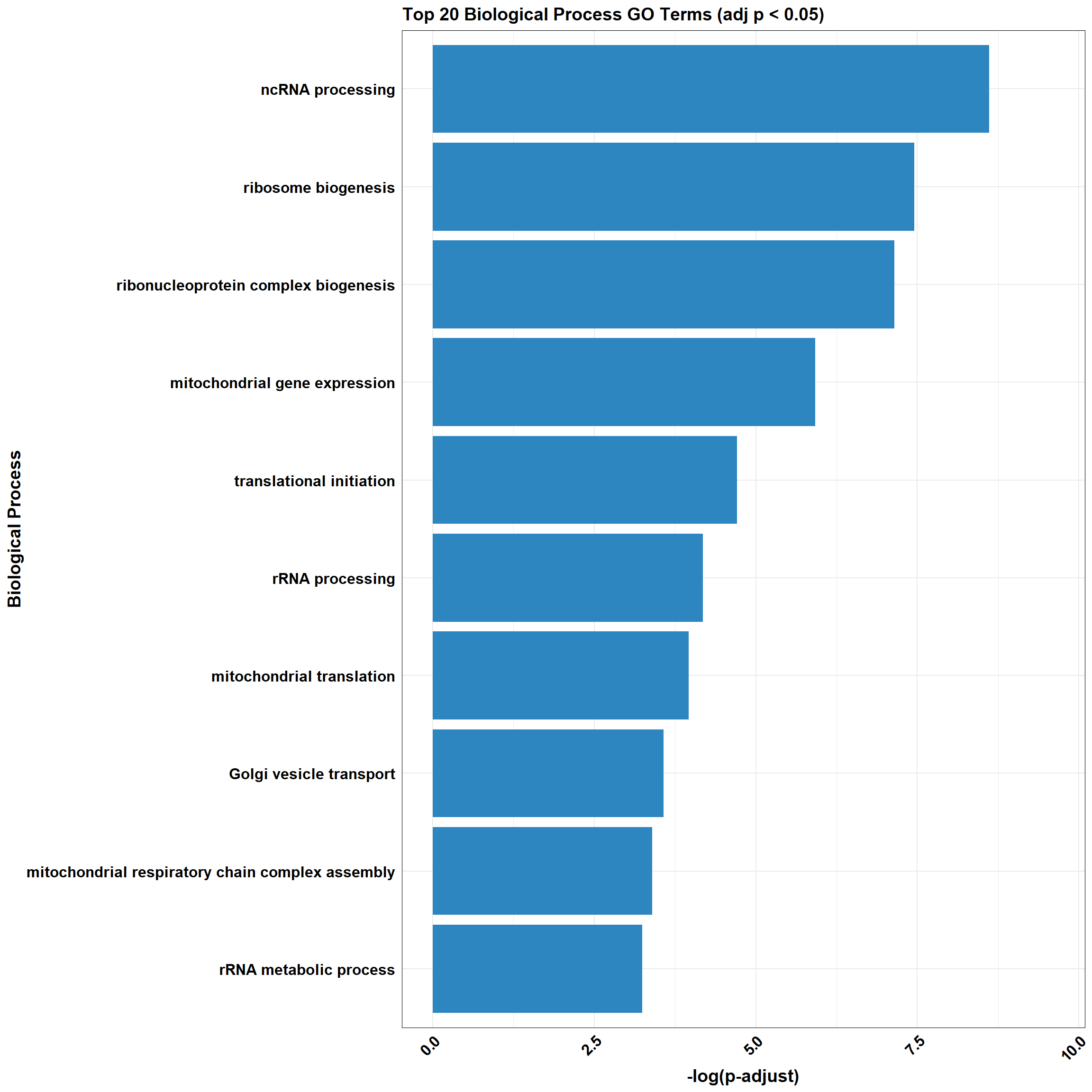
📌 CX_DOX_1
📌 GO Enrichment Clusterprofiler
# 📦 Load Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load GO CSV for CX-DOX mid-late response (0.1)
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_CX-DOX_mid-late_response_(0.1).csv")
# 🧠 Convert and filter (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# Create tibbles for each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plot function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🔧 Plot for each GO category
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")
plot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine plots
combined_plot <- plot_BP
combined_plot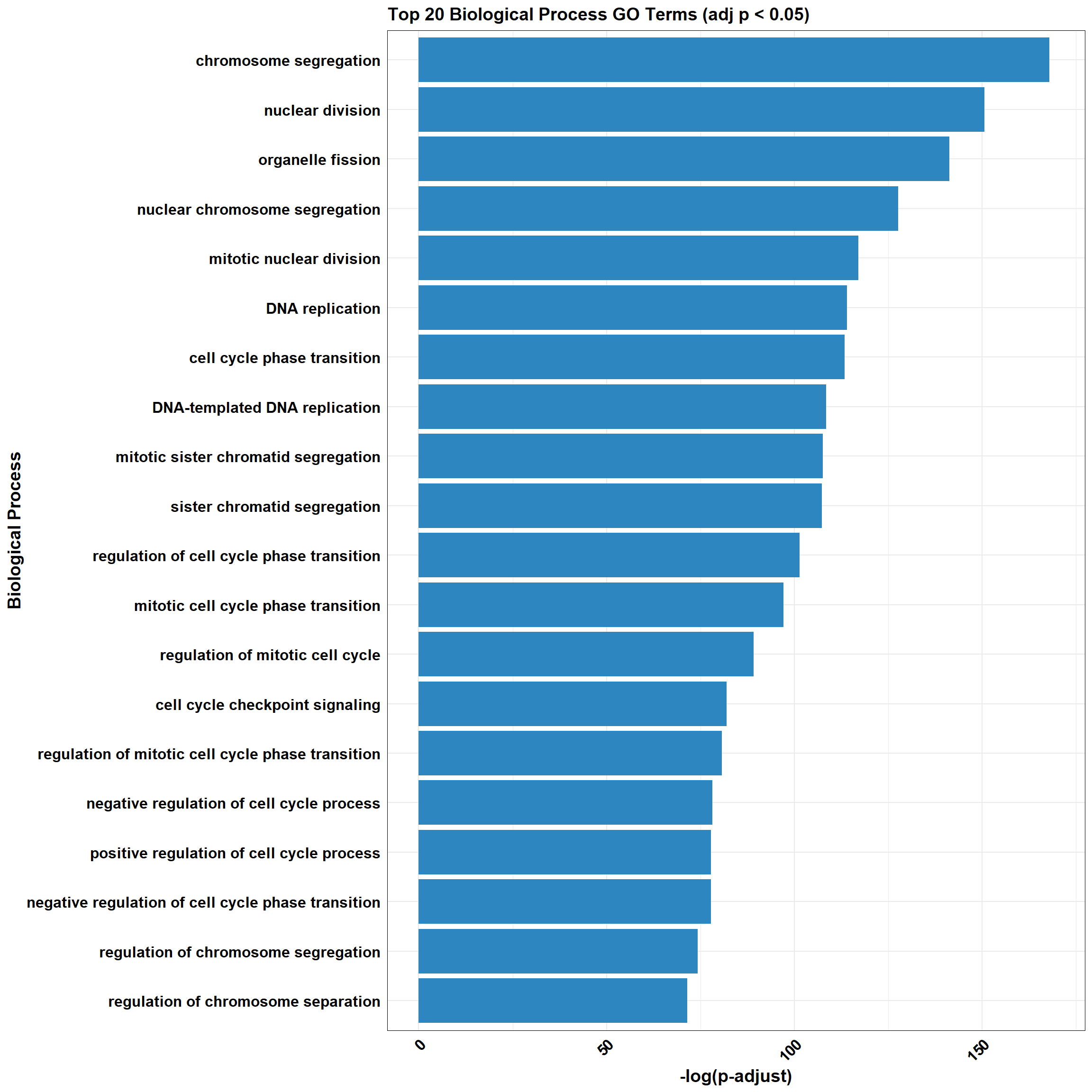
📌 DOX_sp_1
📌 GO Enrichment Clusterprofiler
# 📦 Load Required Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load GO Enrichment CSV
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_DOX_only_mid-late_(0.1).csv")
# 🧠 Convert and filter (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# Create tibbles for each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plot function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🔧 Plot for each GO category
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")Warning in max(tibble$neglog, na.rm = TRUE): no non-missing arguments to max;
returning -Infplot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine plots
combined_plot <- plot_BP
combined_plot
📌 Non response (0.5 µM)
📌 GO Enrichment Clusterprofiler
# 📦 Load Required Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load Enrichment File
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_Non_response_(0.5).csv")
# 🧠 Process Enrichment (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# ⛏️ Extract each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plotting Function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🎨 Generate GO Plots
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")
plot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine & Show
combined_plot <- plot_BP
combined_plot
📌 DOX_sp_2
📌 GO Enrichment Clusterprofiler
# 📦 Load Required Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load Enrichment File
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_DOX_specific_response_(0.5).csv")
# 🧠 Process Enrichment (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# ⛏️ Extract each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plotting Function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🎨 Generate GO Plots
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")Warning in max(tibble$neglog, na.rm = TRUE): no non-missing arguments to max;
returning -Infplot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine & Show
combined_plot <- plot_BP
combined_plot
📌 DOX_sp_3
📌 GO Enrichment Clusterprofiler
# 📦 Load Required Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load Enrichment File
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_DOX_only_mid-late_response_(0.5).csv")
# 🧠 Process Enrichment (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# ⛏️ Extract each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plotting Function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🎨 Generate GO Plots
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")
plot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")Warning in max(tibble$neglog, na.rm = TRUE): no non-missing arguments to max;
returning -Infplot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")Warning in max(tibble$neglog, na.rm = TRUE): no non-missing arguments to max;
returning -Inf# 🧩 Combine & Show
combined_plot <- plot_BP
combined_plot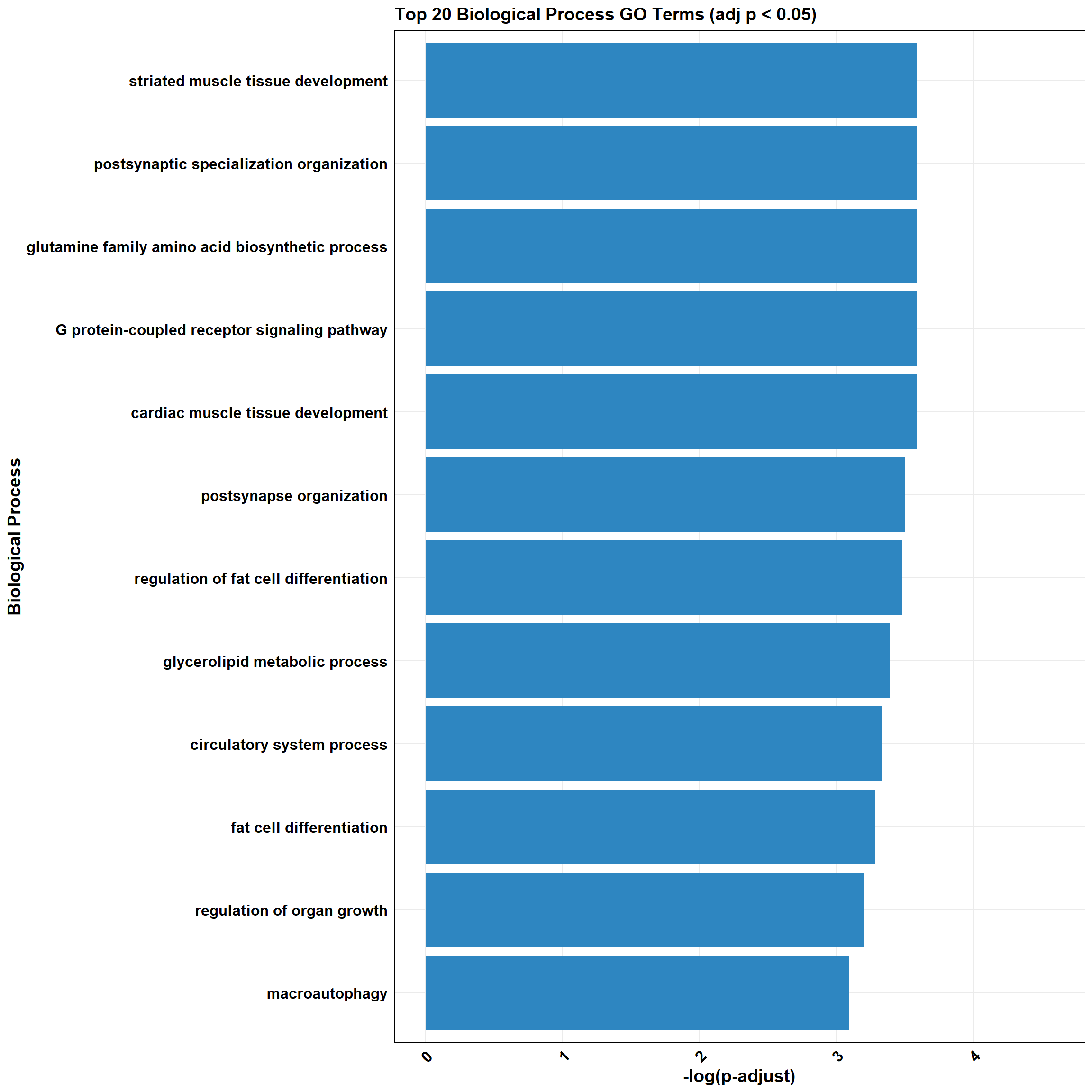
📌 CX_DOX_2
📌 GO Enrichment Clusterprofiler
# 📦 Load Required Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load Enrichment File
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_CX_total_+_DOX_early_response_(0.5).csv")
# 🧠 Process Enrichment (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# ⛏️ Extract each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plotting Function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🎨 Generate GO Plots
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")
plot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine & Show
combined_plot <- plot_BP
combined_plot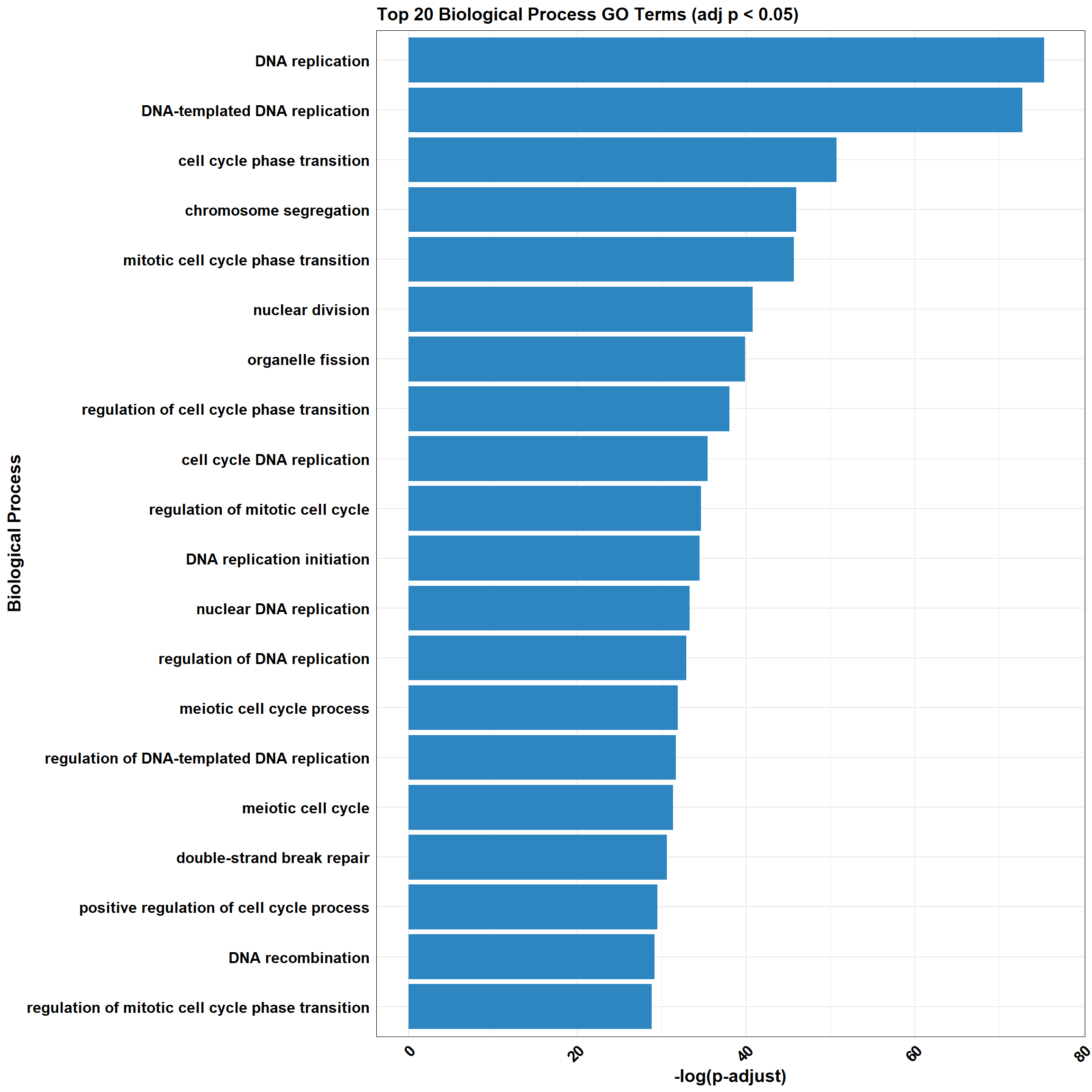
📌 CX_DOX_3
📌 GO Enrichment Clusterprofiler
# 📦 Load Required Libraries
library(tidyverse)
library(data.table)
library(patchwork)
# 📁 Load Enrichment File
go_df <- fread("data/BP/CorMotif_Terms_Combined/GO_All_DOX_early_+_CX-DOX_mid-late_response_(0.5).csv")
# 🧠 Process Enrichment (only adj p < 0.05)
process_enrichment_tibble <- function(df, ontology, category_label) {
df %>%
filter(Ontology == ontology & p.adjust < 0.05) %>%
mutate(neglog = -log(p.adjust)) %>%
arrange(desc(neglog)) %>%
slice(1:20) %>%
mutate(Category = category_label)
}
# ⛏️ Extract each GO class
BP_Tibble <- process_enrichment_tibble(go_df, "BP", "Biological Process")
MF_Tibble <- process_enrichment_tibble(go_df, "MF", "Molecular Function")
CC_Tibble <- process_enrichment_tibble(go_df, "CC", "Cellular Component")
# 📊 Plotting Function
process_enrichment_plot <- function(tibble, title, color) {
ggplot(tibble, aes(x = neglog, y = reorder(Description, neglog))) +
geom_bar(stat = "identity", fill = color) +
labs(
x = "-log(p-adjust)", y = title,
title = paste("Top 20", title, "GO Terms (adj p < 0.05)")
) +
theme_minimal() +
theme(
axis.text.x = element_text(size = 12, face = "bold", colour = "black", angle = 45, hjust = 1),
axis.text.y = element_text(size = 12, face = "bold", colour = "black"),
axis.title.x = element_text(size = 14, face = "bold", colour = "black"),
axis.title.y = element_text(size = 14, face = "bold", colour = "black"),
plot.title = element_text(size = 14, face = "bold", colour = "black"),
panel.border = element_rect(colour = "black", fill = NA, linewidth = 0.3)
) +
xlim(0, max(tibble$neglog, na.rm = TRUE) + 1)
}
# 🎨 Generate GO Plots
plot_BP <- process_enrichment_plot(BP_Tibble, "Biological Process", "#2E86C1")
plot_MF <- process_enrichment_plot(MF_Tibble, "Molecular Function", "#28B463")
plot_CC <- process_enrichment_plot(CC_Tibble, "Cellular Component", "#D35400")
# 🧩 Combine & Show
combined_plot <- plot_BP
combined_plot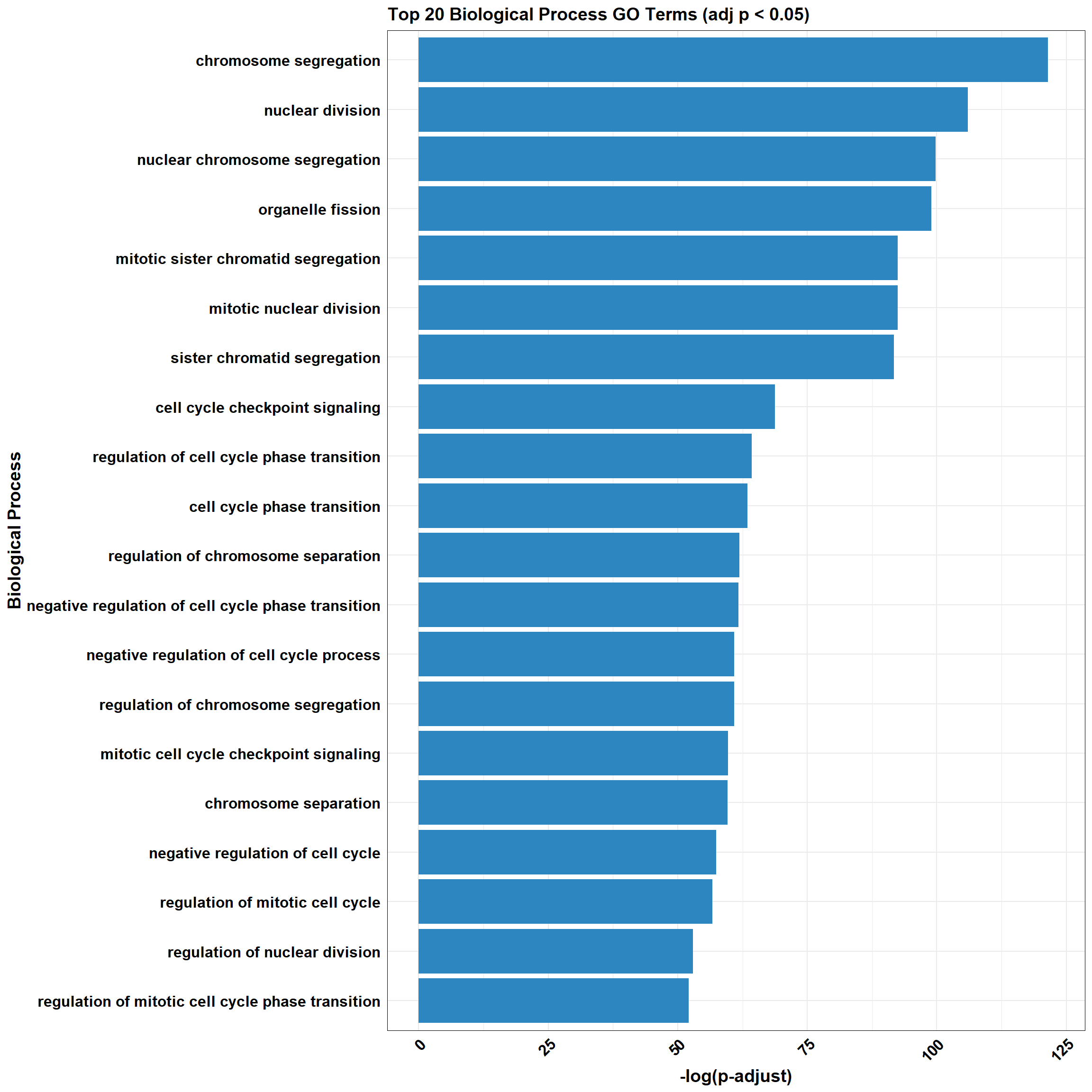
sessionInfo()R version 4.3.0 (2023-04-21 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] data.table_1.17.0
[2] patchwork_1.3.0
[3] DOSE_3.28.2
[4] TxDb.Hsapiens.UCSC.hg38.knownGene_3.18.0
[5] RColorBrewer_1.1-3
[6] clusterProfiler_4.10.1
[7] pheatmap_1.0.12
[8] qvalue_2.34.0
[9] BiocParallel_1.36.0
[10] Homo.sapiens_1.3.1
[11] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[12] org.Hs.eg.db_3.18.0
[13] GO.db_3.18.0
[14] OrganismDbi_1.44.0
[15] GenomicFeatures_1.54.4
[16] GenomicRanges_1.54.1
[17] GenomeInfoDb_1.38.8
[18] AnnotationDbi_1.64.1
[19] IRanges_2.36.0
[20] S4Vectors_0.40.2
[21] Biobase_2.62.0
[22] BiocGenerics_0.48.1
[23] edgeR_4.0.16
[24] limma_3.58.1
[25] cluster_2.1.8.1
[26] ggfortify_0.4.17
[27] lubridate_1.9.4
[28] forcats_1.0.0
[29] stringr_1.5.1
[30] dplyr_1.1.4
[31] purrr_1.0.4
[32] readr_2.1.5
[33] tidyr_1.3.1
[34] tibble_3.2.1
[35] ggplot2_3.5.2
[36] tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] splines_4.3.0 later_1.3.2
[3] BiocIO_1.12.0 bitops_1.0-9
[5] ggplotify_0.1.2 filelock_1.0.3
[7] polyclip_1.10-7 graph_1.80.0
[9] XML_3.99-0.18 lifecycle_1.0.4
[11] rprojroot_2.0.4 lattice_0.22-7
[13] MASS_7.3-60 magrittr_2.0.3
[15] sass_0.4.10 rmarkdown_2.29
[17] jquerylib_0.1.4 yaml_2.3.10
[19] httpuv_1.6.15 cowplot_1.1.3
[21] DBI_1.2.3 abind_1.4-8
[23] zlibbioc_1.48.2 ggraph_2.2.1
[25] RCurl_1.98-1.17 yulab.utils_0.2.0
[27] tweenr_2.0.3 rappdirs_0.3.3
[29] git2r_0.36.2 GenomeInfoDbData_1.2.11
[31] enrichplot_1.22.0 ggrepel_0.9.6
[33] tidytree_0.4.6 codetools_0.2-20
[35] DelayedArray_0.28.0 xml2_1.3.8
[37] ggforce_0.4.2 tidyselect_1.2.1
[39] aplot_0.2.5 farver_2.1.2
[41] viridis_0.6.5 matrixStats_1.5.0
[43] BiocFileCache_2.10.2 GenomicAlignments_1.38.2
[45] jsonlite_2.0.0 tidygraph_1.3.1
[47] tools_4.3.0 progress_1.2.3
[49] treeio_1.26.0 Rcpp_1.0.12
[51] glue_1.7.0 gridExtra_2.3
[53] SparseArray_1.2.4 xfun_0.52
[55] MatrixGenerics_1.14.0 withr_3.0.2
[57] BiocManager_1.30.25 fastmap_1.2.0
[59] digest_0.6.34 timechange_0.3.0
[61] R6_2.6.1 gridGraphics_0.5-1
[63] colorspace_2.1-0 biomaRt_2.58.2
[65] RSQLite_2.3.9 generics_0.1.3
[67] rtracklayer_1.62.0 prettyunits_1.2.0
[69] graphlayouts_1.2.2 httr_1.4.7
[71] S4Arrays_1.2.1 scatterpie_0.2.4
[73] pkgconfig_2.0.3 gtable_0.3.6
[75] blob_1.2.4 workflowr_1.7.1
[77] XVector_0.42.0 shadowtext_0.1.4
[79] htmltools_0.5.8.1 fgsea_1.28.0
[81] RBGL_1.78.0 scales_1.3.0
[83] png_0.1-8 ggfun_0.1.8
[85] knitr_1.50 rstudioapi_0.17.1
[87] tzdb_0.5.0 reshape2_1.4.4
[89] rjson_0.2.23 nlme_3.1-168
[91] curl_6.2.2 cachem_1.1.0
[93] parallel_4.3.0 HDO.db_0.99.1
[95] restfulr_0.0.15 pillar_1.10.2
[97] grid_4.3.0 vctrs_0.6.5
[99] promises_1.3.2 dbplyr_2.5.0
[101] evaluate_1.0.3 cli_3.6.1
[103] locfit_1.5-9.12 compiler_4.3.0
[105] Rsamtools_2.18.0 rlang_1.1.3
[107] crayon_1.5.3 labeling_0.4.3
[109] plyr_1.8.9 fs_1.6.3
[111] stringi_1.8.3 viridisLite_0.4.2
[113] munsell_0.5.1 Biostrings_2.70.3
[115] lazyeval_0.2.2 GOSemSim_2.28.1
[117] Matrix_1.6-1.1 hms_1.1.3
[119] bit64_4.6.0-1 KEGGREST_1.42.0
[121] statmod_1.5.0 SummarizedExperiment_1.32.0
[123] igraph_2.1.4 memoise_2.0.1
[125] bslib_0.9.0 ggtree_3.10.1
[127] fastmatch_1.1-6 bit_4.6.0
[129] gson_0.1.0 ape_5.8-1